Upload maisi_ct_generative version 1.0.0
Browse files- LICENSE +247 -0
- configs/all_anatomy_size_condtions.json +0 -0
- configs/candidate_masks_flexible_size_and_spacing_3000.json +0 -0
- configs/image_median_statistics.json +72 -0
- configs/inference.json +312 -0
- configs/inference_trt.json +19 -0
- configs/integration_test_masks.json +98 -0
- configs/label_dict.json +134 -0
- configs/label_dict_124_to_132.json +502 -0
- configs/logging.conf +21 -0
- configs/metadata.json +269 -0
- configs/multi_gpu_train.json +34 -0
- configs/train.json +271 -0
- datasets/C4KC-KiTS_subset.json +814 -0
- datasets/C4KC-KiTS_subset.zip +3 -0
- datasets/IntegrationTest-AbdomenCT.nii.gz +3 -0
- datasets/all_masks_flexible_size_and_spacing_3000.zip +3 -0
- docs/README.md +129 -0
- docs/data_license.txt +49 -0
- models/autoencoder.pt +3 -0
- models/controlnet-20datasets-e20wl100fold0bc_noi_dia_fsize_current.pt +3 -0
- models/controlnet.pt +3 -0
- models/diffusion_unet.pt +3 -0
- models/input_unet3d_data-all_steps1000size512ddpm_random_current_inputx_v1.pt +3 -0
- models/mask_generation_autoencoder.pt +3 -0
- models/mask_generation_diffusion_unet.pt +3 -0
- scripts/__init__.py +12 -0
- scripts/augmentation.py +373 -0
- scripts/find_masks.py +137 -0
- scripts/quality_check.py +149 -0
- scripts/rectified_flow.py +163 -0
- scripts/sample.py +1036 -0
- scripts/trainer.py +246 -0
- scripts/utils.py +696 -0
LICENSE
ADDED
|
@@ -0,0 +1,247 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Code License
|
| 2 |
+
|
| 3 |
+
This license applies to all files except the model weights in the directory.
|
| 4 |
+
|
| 5 |
+
Apache License
|
| 6 |
+
Version 2.0, January 2004
|
| 7 |
+
http://www.apache.org/licenses/
|
| 8 |
+
|
| 9 |
+
TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
|
| 10 |
+
|
| 11 |
+
1. Definitions.
|
| 12 |
+
|
| 13 |
+
"License" shall mean the terms and conditions for use, reproduction,
|
| 14 |
+
and distribution as defined by Sections 1 through 9 of this document.
|
| 15 |
+
|
| 16 |
+
"Licensor" shall mean the copyright owner or entity authorized by
|
| 17 |
+
the copyright owner that is granting the License.
|
| 18 |
+
|
| 19 |
+
"Legal Entity" shall mean the union of the acting entity and all
|
| 20 |
+
other entities that control, are controlled by, or are under common
|
| 21 |
+
control with that entity. For the purposes of this definition,
|
| 22 |
+
"control" means (i) the power, direct or indirect, to cause the
|
| 23 |
+
direction or management of such entity, whether by contract or
|
| 24 |
+
otherwise, or (ii) ownership of fifty percent (50%) or more of the
|
| 25 |
+
outstanding shares, or (iii) beneficial ownership of such entity.
|
| 26 |
+
|
| 27 |
+
"You" (or "Your") shall mean an individual or Legal Entity
|
| 28 |
+
exercising permissions granted by this License.
|
| 29 |
+
|
| 30 |
+
"Source" form shall mean the preferred form for making modifications,
|
| 31 |
+
including but not limited to software source code, documentation
|
| 32 |
+
source, and configuration files.
|
| 33 |
+
|
| 34 |
+
"Object" form shall mean any form resulting from mechanical
|
| 35 |
+
transformation or translation of a Source form, including but
|
| 36 |
+
not limited to compiled object code, generated documentation,
|
| 37 |
+
and conversions to other media types.
|
| 38 |
+
|
| 39 |
+
"Work" shall mean the work of authorship, whether in Source or
|
| 40 |
+
Object form, made available under the License, as indicated by a
|
| 41 |
+
copyright notice that is included in or attached to the work
|
| 42 |
+
(an example is provided in the Appendix below).
|
| 43 |
+
|
| 44 |
+
"Derivative Works" shall mean any work, whether in Source or Object
|
| 45 |
+
form, that is based on (or derived from) the Work and for which the
|
| 46 |
+
editorial revisions, annotations, elaborations, or other modifications
|
| 47 |
+
represent, as a whole, an original work of authorship. For the purposes
|
| 48 |
+
of this License, Derivative Works shall not include works that remain
|
| 49 |
+
separable from, or merely link (or bind by name) to the interfaces of,
|
| 50 |
+
the Work and Derivative Works thereof.
|
| 51 |
+
|
| 52 |
+
"Contribution" shall mean any work of authorship, including
|
| 53 |
+
the original version of the Work and any modifications or additions
|
| 54 |
+
to that Work or Derivative Works thereof, that is intentionally
|
| 55 |
+
submitted to Licensor for inclusion in the Work by the copyright owner
|
| 56 |
+
or by an individual or Legal Entity authorized to submit on behalf of
|
| 57 |
+
the copyright owner. For the purposes of this definition, "submitted"
|
| 58 |
+
means any form of electronic, verbal, or written communication sent
|
| 59 |
+
to the Licensor or its representatives, including but not limited to
|
| 60 |
+
communication on electronic mailing lists, source code control systems,
|
| 61 |
+
and issue tracking systems that are managed by, or on behalf of, the
|
| 62 |
+
Licensor for the purpose of discussing and improving the Work, but
|
| 63 |
+
excluding communication that is conspicuously marked or otherwise
|
| 64 |
+
designated in writing by the copyright owner as "Not a Contribution."
|
| 65 |
+
|
| 66 |
+
"Contributor" shall mean Licensor and any individual or Legal Entity
|
| 67 |
+
on behalf of whom a Contribution has been received by Licensor and
|
| 68 |
+
subsequently incorporated within the Work.
|
| 69 |
+
|
| 70 |
+
2. Grant of Copyright License. Subject to the terms and conditions of
|
| 71 |
+
this License, each Contributor hereby grants to You a perpetual,
|
| 72 |
+
worldwide, non-exclusive, no-charge, royalty-free, irrevocable
|
| 73 |
+
copyright license to reproduce, prepare Derivative Works of,
|
| 74 |
+
publicly display, publicly perform, sublicense, and distribute the
|
| 75 |
+
Work and such Derivative Works in Source or Object form.
|
| 76 |
+
|
| 77 |
+
3. Grant of Patent License. Subject to the terms and conditions of
|
| 78 |
+
this License, each Contributor hereby grants to You a perpetual,
|
| 79 |
+
worldwide, non-exclusive, no-charge, royalty-free, irrevocable
|
| 80 |
+
(except as stated in this section) patent license to make, have made,
|
| 81 |
+
use, offer to sell, sell, import, and otherwise transfer the Work,
|
| 82 |
+
where such license applies only to those patent claims licensable
|
| 83 |
+
by such Contributor that are necessarily infringed by their
|
| 84 |
+
Contribution(s) alone or by combination of their Contribution(s)
|
| 85 |
+
with the Work to which such Contribution(s) was submitted. If You
|
| 86 |
+
institute patent litigation against any entity (including a
|
| 87 |
+
cross-claim or counterclaim in a lawsuit) alleging that the Work
|
| 88 |
+
or a Contribution incorporated within the Work constitutes direct
|
| 89 |
+
or contributory patent infringement, then any patent licenses
|
| 90 |
+
granted to You under this License for that Work shall terminate
|
| 91 |
+
as of the date such litigation is filed.
|
| 92 |
+
|
| 93 |
+
4. Redistribution. You may reproduce and distribute copies of the
|
| 94 |
+
Work or Derivative Works thereof in any medium, with or without
|
| 95 |
+
modifications, and in Source or Object form, provided that You
|
| 96 |
+
meet the following conditions:
|
| 97 |
+
|
| 98 |
+
(a) You must give any other recipients of the Work or
|
| 99 |
+
Derivative Works a copy of this License; and
|
| 100 |
+
|
| 101 |
+
(b) You must cause any modified files to carry prominent notices
|
| 102 |
+
stating that You changed the files; and
|
| 103 |
+
|
| 104 |
+
(c) You must retain, in the Source form of any Derivative Works
|
| 105 |
+
that You distribute, all copyright, patent, trademark, and
|
| 106 |
+
attribution notices from the Source form of the Work,
|
| 107 |
+
excluding those notices that do not pertain to any part of
|
| 108 |
+
the Derivative Works; and
|
| 109 |
+
|
| 110 |
+
(d) If the Work includes a "NOTICE" text file as part of its
|
| 111 |
+
distribution, then any Derivative Works that You distribute must
|
| 112 |
+
include a readable copy of the attribution notices contained
|
| 113 |
+
within such NOTICE file, excluding those notices that do not
|
| 114 |
+
pertain to any part of the Derivative Works, in at least one
|
| 115 |
+
of the following places: within a NOTICE text file distributed
|
| 116 |
+
as part of the Derivative Works; within the Source form or
|
| 117 |
+
documentation, if provided along with the Derivative Works; or,
|
| 118 |
+
within a display generated by the Derivative Works, if and
|
| 119 |
+
wherever such third-party notices normally appear. The contents
|
| 120 |
+
of the NOTICE file are for informational purposes only and
|
| 121 |
+
do not modify the License. You may add Your own attribution
|
| 122 |
+
notices within Derivative Works that You distribute, alongside
|
| 123 |
+
or as an addendum to the NOTICE text from the Work, provided
|
| 124 |
+
that such additional attribution notices cannot be construed
|
| 125 |
+
as modifying the License.
|
| 126 |
+
|
| 127 |
+
You may add Your own copyright statement to Your modifications and
|
| 128 |
+
may provide additional or different license terms and conditions
|
| 129 |
+
for use, reproduction, or distribution of Your modifications, or
|
| 130 |
+
for any such Derivative Works as a whole, provided Your use,
|
| 131 |
+
reproduction, and distribution of the Work otherwise complies with
|
| 132 |
+
the conditions stated in this License.
|
| 133 |
+
|
| 134 |
+
5. Submission of Contributions. Unless You explicitly state otherwise,
|
| 135 |
+
any Contribution intentionally submitted for inclusion in the Work
|
| 136 |
+
by You to the Licensor shall be under the terms and conditions of
|
| 137 |
+
this License, without any additional terms or conditions.
|
| 138 |
+
Notwithstanding the above, nothing herein shall supersede or modify
|
| 139 |
+
the terms of any separate license agreement you may have executed
|
| 140 |
+
with Licensor regarding such Contributions.
|
| 141 |
+
|
| 142 |
+
6. Trademarks. This License does not grant permission to use the trade
|
| 143 |
+
names, trademarks, service marks, or product names of the Licensor,
|
| 144 |
+
except as required for reasonable and customary use in describing the
|
| 145 |
+
origin of the Work and reproducing the content of the NOTICE file.
|
| 146 |
+
|
| 147 |
+
7. Disclaimer of Warranty. Unless required by applicable law or
|
| 148 |
+
agreed to in writing, Licensor provides the Work (and each
|
| 149 |
+
Contributor provides its Contributions) on an "AS IS" BASIS,
|
| 150 |
+
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
|
| 151 |
+
implied, including, without limitation, any warranties or conditions
|
| 152 |
+
of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
|
| 153 |
+
PARTICULAR PURPOSE. You are solely responsible for determining the
|
| 154 |
+
appropriateness of using or redistributing the Work and assume any
|
| 155 |
+
risks associated with Your exercise of permissions under this License.
|
| 156 |
+
|
| 157 |
+
8. Limitation of Liability. In no event and under no legal theory,
|
| 158 |
+
whether in tort (including negligence), contract, or otherwise,
|
| 159 |
+
unless required by applicable law (such as deliberate and grossly
|
| 160 |
+
negligent acts) or agreed to in writing, shall any Contributor be
|
| 161 |
+
liable to You for damages, including any direct, indirect, special,
|
| 162 |
+
incidental, or consequential damages of any character arising as a
|
| 163 |
+
result of this License or out of the use or inability to use the
|
| 164 |
+
Work (including but not limited to damages for loss of goodwill,
|
| 165 |
+
work stoppage, computer failure or malfunction, or any and all
|
| 166 |
+
other commercial damages or losses), even if such Contributor
|
| 167 |
+
has been advised of the possibility of such damages.
|
| 168 |
+
|
| 169 |
+
9. Accepting Warranty or Additional Liability. While redistributing
|
| 170 |
+
the Work or Derivative Works thereof, You may choose to offer,
|
| 171 |
+
and charge a fee for, acceptance of support, warranty, indemnity,
|
| 172 |
+
or other liability obligations and/or rights consistent with this
|
| 173 |
+
License. However, in accepting such obligations, You may act only
|
| 174 |
+
on Your own behalf and on Your sole responsibility, not on behalf
|
| 175 |
+
of any other Contributor, and only if You agree to indemnify,
|
| 176 |
+
defend, and hold each Contributor harmless for any liability
|
| 177 |
+
incurred by, or claims asserted against, such Contributor by reason
|
| 178 |
+
of your accepting any such warranty or additional liability.
|
| 179 |
+
|
| 180 |
+
END OF TERMS AND CONDITIONS
|
| 181 |
+
|
| 182 |
+
APPENDIX: How to apply the Apache License to your work.
|
| 183 |
+
|
| 184 |
+
To apply the Apache License to your work, attach the following
|
| 185 |
+
boilerplate notice, with the fields enclosed by brackets "[]"
|
| 186 |
+
replaced with your own identifying information. (Don't include
|
| 187 |
+
the brackets!) The text should be enclosed in the appropriate
|
| 188 |
+
comment syntax for the file format. We also recommend that a
|
| 189 |
+
file or class name and description of purpose be included on the
|
| 190 |
+
same "printed page" as the copyright notice for easier
|
| 191 |
+
identification within third-party archives.
|
| 192 |
+
|
| 193 |
+
Copyright [yyyy] [name of copyright owner]
|
| 194 |
+
|
| 195 |
+
Licensed under the Apache License, Version 2.0 (the "License");
|
| 196 |
+
you may not use this file except in compliance with the License.
|
| 197 |
+
You may obtain a copy of the License at
|
| 198 |
+
|
| 199 |
+
http://www.apache.org/licenses/LICENSE-2.0
|
| 200 |
+
|
| 201 |
+
Unless required by applicable law or agreed to in writing, software
|
| 202 |
+
distributed under the License is distributed on an "AS IS" BASIS,
|
| 203 |
+
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 204 |
+
See the License for the specific language governing permissions and
|
| 205 |
+
limitations under the License.
|
| 206 |
+
|
| 207 |
+
------------------------------------------------------------------------------
|
| 208 |
+
|
| 209 |
+
Model Weights License
|
| 210 |
+
|
| 211 |
+
This license applies to model weights in the directory.
|
| 212 |
+
|
| 213 |
+
NVIDIA License
|
| 214 |
+
|
| 215 |
+
1. Definitions
|
| 216 |
+
|
| 217 |
+
“Licensor” means any person or entity that distributes its Work.
|
| 218 |
+
“Work” means (a) the original work of authorship made available under this license, which may include software, documentation, or other files, and (b) any additions to or derivative works thereof that are made available under this license.
|
| 219 |
+
The terms “reproduce,” “reproduction,” “derivative works,” and “distribution” have the meaning as provided under U.S. copyright law; provided, however, that for the purposes of this license, derivative works shall not include works that remain separable from, or merely link (or bind by name) to the interfaces of, the Work.
|
| 220 |
+
Works are “made available” under this license by including in or with the Work either (a) a copyright notice referencing the applicability of this license to the Work, or (b) a copy of this license.
|
| 221 |
+
|
| 222 |
+
2. License Grant
|
| 223 |
+
|
| 224 |
+
2.1 Copyright Grant. Subject to the terms and conditions of this license, each Licensor grants to you a perpetual, worldwide, non-exclusive, royalty-free, copyright license to use, reproduce, prepare derivative works of, publicly display, publicly perform, sublicense and distribute its Work and any resulting derivative works in any form.
|
| 225 |
+
|
| 226 |
+
3. Limitations
|
| 227 |
+
|
| 228 |
+
3.1 Redistribution. You may reproduce or distribute the Work only if (a) you do so under this license, (b) you include a complete copy of this license with your distribution, and (c) you retain without modification any copyright, patent, trademark, or attribution notices that are present in the Work.
|
| 229 |
+
|
| 230 |
+
3.2 Derivative Works. You may specify that additional or different terms apply to the use, reproduction, and distribution of your derivative works of the Work (“Your Terms”) only if (a) Your Terms provide that the use limitation in Section 3.3 applies to your derivative works, and (b) you identify the specific derivative works that are subject to Your Terms. Notwithstanding Your Terms, this license (including the redistribution requirements in Section 3.1) will continue to apply to the Work itself.
|
| 231 |
+
|
| 232 |
+
3.3 Use Limitation. The Work and any derivative works thereof only may be used or intended for use non-commercially. Notwithstanding the foregoing, NVIDIA Corporation and its affiliates may use the Work and any derivative works commercially. As used herein, “non-commercially” means for research or evaluation purposes only.
|
| 233 |
+
|
| 234 |
+
3.4 Patent Claims. If you bring or threaten to bring a patent claim against any Licensor (including any claim, cross-claim or counterclaim in a lawsuit) to enforce any patents that you allege are infringed by any Work, then your rights under this license from such Licensor (including the grant in Section 2.1) will terminate immediately.
|
| 235 |
+
|
| 236 |
+
3.5 Trademarks. This license does not grant any rights to use any Licensor’s or its affiliates’ names, logos, or trademarks, except as necessary to reproduce the notices described in this license.
|
| 237 |
+
|
| 238 |
+
3.6 Termination. If you violate any term of this license, then your rights under this license (including the grant in Section 2.1) will terminate immediately.
|
| 239 |
+
|
| 240 |
+
4. Disclaimer of Warranty.
|
| 241 |
+
|
| 242 |
+
THE WORK IS PROVIDED “AS IS” WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, EITHER EXPRESS OR IMPLIED, INCLUDING WARRANTIES OR CONDITIONS OF
|
| 243 |
+
MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE, TITLE OR NON-INFRINGEMENT. YOU BEAR THE RISK OF UNDERTAKING ANY ACTIVITIES UNDER THIS LICENSE.
|
| 244 |
+
|
| 245 |
+
5. Limitation of Liability.
|
| 246 |
+
|
| 247 |
+
EXCEPT AS PROHIBITED BY APPLICABLE LAW, IN NO EVENT AND UNDER NO LEGAL THEORY, WHETHER IN TORT (INCLUDING NEGLIGENCE), CONTRACT, OR OTHERWISE SHALL ANY LICENSOR BE LIABLE TO YOU FOR DAMAGES, INCLUDING ANY DIRECT, INDIRECT, SPECIAL, INCIDENTAL, OR CONSEQUENTIAL DAMAGES ARISING OUT OF OR RELATED TO THIS LICENSE, THE USE OR INABILITY TO USE THE WORK (INCLUDING BUT NOT LIMITED TO LOSS OF GOODWILL, BUSINESS INTERRUPTION, LOST PROFITS OR DATA, COMPUTER FAILURE OR MALFUNCTION, OR ANY OTHER DAMAGES OR LOSSES), EVEN IF THE LICENSOR HAS BEEN ADVISED OF THE POSSIBILITY OF SUCH DAMAGES.
|
configs/all_anatomy_size_condtions.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
configs/candidate_masks_flexible_size_and_spacing_3000.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
configs/image_median_statistics.json
ADDED
|
@@ -0,0 +1,72 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"liver": {
|
| 3 |
+
"min_median": -14.0,
|
| 4 |
+
"max_median": 1000.0,
|
| 5 |
+
"percentile_0_5": 9.530000000000001,
|
| 6 |
+
"percentile_99_5": 162.0,
|
| 7 |
+
"sigma_6_low": -21.596463547885904,
|
| 8 |
+
"sigma_6_high": 156.27881534763367,
|
| 9 |
+
"sigma_12_low": -110.53410299564568,
|
| 10 |
+
"sigma_12_high": 245.21645479539342
|
| 11 |
+
},
|
| 12 |
+
"spleen": {
|
| 13 |
+
"min_median": -69.0,
|
| 14 |
+
"max_median": 1000.0,
|
| 15 |
+
"percentile_0_5": 16.925000000000004,
|
| 16 |
+
"percentile_99_5": 184.07500000000073,
|
| 17 |
+
"sigma_6_low": -43.133891656525165,
|
| 18 |
+
"sigma_6_high": 177.40494997185993,
|
| 19 |
+
"sigma_12_low": -153.4033124707177,
|
| 20 |
+
"sigma_12_high": 287.6743707860525
|
| 21 |
+
},
|
| 22 |
+
"pancreas": {
|
| 23 |
+
"min_median": -124.0,
|
| 24 |
+
"max_median": 1000.0,
|
| 25 |
+
"percentile_0_5": -29.0,
|
| 26 |
+
"percentile_99_5": 145.92000000000007,
|
| 27 |
+
"sigma_6_low": -56.59382515620725,
|
| 28 |
+
"sigma_6_high": 149.50627399318438,
|
| 29 |
+
"sigma_12_low": -159.64387473090306,
|
| 30 |
+
"sigma_12_high": 252.5563235678802
|
| 31 |
+
},
|
| 32 |
+
"kidney": {
|
| 33 |
+
"min_median": -165.5,
|
| 34 |
+
"max_median": 819.0,
|
| 35 |
+
"percentile_0_5": -40.0,
|
| 36 |
+
"percentile_99_5": 254.61999999999898,
|
| 37 |
+
"sigma_6_low": -130.56375604853028,
|
| 38 |
+
"sigma_6_high": 267.28163511081016,
|
| 39 |
+
"sigma_12_low": -329.4864516282005,
|
| 40 |
+
"sigma_12_high": 466.20433069048045
|
| 41 |
+
},
|
| 42 |
+
"lung": {
|
| 43 |
+
"min_median": -1000.0,
|
| 44 |
+
"max_median": 65.0,
|
| 45 |
+
"percentile_0_5": -937.0,
|
| 46 |
+
"percentile_99_5": -366.9500000000007,
|
| 47 |
+
"sigma_6_low": -1088.5583843889117,
|
| 48 |
+
"sigma_6_high": -551.8503346949108,
|
| 49 |
+
"sigma_12_low": -1356.912409235912,
|
| 50 |
+
"sigma_12_high": -283.4963098479103
|
| 51 |
+
},
|
| 52 |
+
"bone": {
|
| 53 |
+
"min_median": 77.5,
|
| 54 |
+
"max_median": 1000.0,
|
| 55 |
+
"percentile_0_5": 136.45499999999998,
|
| 56 |
+
"percentile_99_5": 551.6350000000002,
|
| 57 |
+
"sigma_6_low": 71.39901958080469,
|
| 58 |
+
"sigma_6_high": 471.9957615639765,
|
| 59 |
+
"sigma_12_low": -128.8993514107812,
|
| 60 |
+
"sigma_12_high": 672.2941325555623
|
| 61 |
+
},
|
| 62 |
+
"brain": {
|
| 63 |
+
"min_median": -1000.0,
|
| 64 |
+
"max_median": 238.0,
|
| 65 |
+
"percentile_0_5": -951.0,
|
| 66 |
+
"percentile_99_5": 126.25,
|
| 67 |
+
"sigma_6_low": -304.8208236135867,
|
| 68 |
+
"sigma_6_high": 369.5118535139189,
|
| 69 |
+
"sigma_12_low": -641.9871621773394,
|
| 70 |
+
"sigma_12_high": 706.6781920776717
|
| 71 |
+
}
|
| 72 |
+
}
|
configs/inference.json
ADDED
|
@@ -0,0 +1,312 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import torch",
|
| 4 |
+
"$from pathlib import Path",
|
| 5 |
+
"$import scripts"
|
| 6 |
+
],
|
| 7 |
+
"bundle_root": ".",
|
| 8 |
+
"model_dir": "$@bundle_root + '/models'",
|
| 9 |
+
"output_dir": "$@bundle_root + '/output'",
|
| 10 |
+
"create_output_dir": "$Path(@output_dir).mkdir(exist_ok=True)",
|
| 11 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 12 |
+
"trained_autoencoder_path": "$@model_dir + '/autoencoder.pt'",
|
| 13 |
+
"trained_diffusion_path": "$@model_dir + '/diffusion_unet.pt'",
|
| 14 |
+
"trained_controlnet_path": "$@model_dir + '/controlnet.pt'",
|
| 15 |
+
"trained_mask_generation_autoencoder_path": "$@model_dir + '/mask_generation_autoencoder.pt'",
|
| 16 |
+
"trained_mask_generation_diffusion_path": "$@model_dir + '/mask_generation_diffusion_unet.pt'",
|
| 17 |
+
"all_mask_files_base_dir": "$@bundle_root + '/datasets/all_masks_flexible_size_and_spacing_3000'",
|
| 18 |
+
"all_mask_files_json": "$@bundle_root + '/configs/candidate_masks_flexible_size_and_spacing_3000.json'",
|
| 19 |
+
"all_anatomy_size_condtions_json": "$@bundle_root + '/configs/all_anatomy_size_condtions.json'",
|
| 20 |
+
"label_dict_json": "$@bundle_root + '/configs/label_dict.json'",
|
| 21 |
+
"label_dict_remap_json": "$@bundle_root + '/configs/label_dict_124_to_132.json'",
|
| 22 |
+
"real_img_median_statistics_file": "$@bundle_root + '/configs/image_median_statistics.json'",
|
| 23 |
+
"num_output_samples": 1,
|
| 24 |
+
"body_region": [],
|
| 25 |
+
"anatomy_list": [
|
| 26 |
+
"liver"
|
| 27 |
+
],
|
| 28 |
+
"modality": "ct",
|
| 29 |
+
"controllable_anatomy_size": [],
|
| 30 |
+
"num_inference_steps": 30,
|
| 31 |
+
"mask_generation_num_inference_steps": 1000,
|
| 32 |
+
"random_seed": null,
|
| 33 |
+
"spatial_dims": 3,
|
| 34 |
+
"image_channels": 1,
|
| 35 |
+
"latent_channels": 4,
|
| 36 |
+
"output_size_xy": 512,
|
| 37 |
+
"output_size_z": 512,
|
| 38 |
+
"output_size": [
|
| 39 |
+
"@output_size_xy",
|
| 40 |
+
"@output_size_xy",
|
| 41 |
+
"@output_size_z"
|
| 42 |
+
],
|
| 43 |
+
"image_output_ext": ".nii.gz",
|
| 44 |
+
"label_output_ext": ".nii.gz",
|
| 45 |
+
"spacing_xy": 1.0,
|
| 46 |
+
"spacing_z": 1.0,
|
| 47 |
+
"spacing": [
|
| 48 |
+
"@spacing_xy",
|
| 49 |
+
"@spacing_xy",
|
| 50 |
+
"@spacing_z"
|
| 51 |
+
],
|
| 52 |
+
"latent_shape": [
|
| 53 |
+
"@latent_channels",
|
| 54 |
+
"$@output_size[0]//4",
|
| 55 |
+
"$@output_size[1]//4",
|
| 56 |
+
"$@output_size[2]//4"
|
| 57 |
+
],
|
| 58 |
+
"mask_generation_latent_shape": [
|
| 59 |
+
4,
|
| 60 |
+
64,
|
| 61 |
+
64,
|
| 62 |
+
64
|
| 63 |
+
],
|
| 64 |
+
"autoencoder_sliding_window_infer_size": [
|
| 65 |
+
80,
|
| 66 |
+
80,
|
| 67 |
+
80
|
| 68 |
+
],
|
| 69 |
+
"autoencoder_sliding_window_infer_overlap": 0.4,
|
| 70 |
+
"autoencoder_def": {
|
| 71 |
+
"_target_": "monai.apps.generation.maisi.networks.autoencoderkl_maisi.AutoencoderKlMaisi",
|
| 72 |
+
"spatial_dims": "@spatial_dims",
|
| 73 |
+
"in_channels": "@image_channels",
|
| 74 |
+
"out_channels": "@image_channels",
|
| 75 |
+
"latent_channels": "@latent_channels",
|
| 76 |
+
"num_channels": [
|
| 77 |
+
64,
|
| 78 |
+
128,
|
| 79 |
+
256
|
| 80 |
+
],
|
| 81 |
+
"num_res_blocks": [
|
| 82 |
+
2,
|
| 83 |
+
2,
|
| 84 |
+
2
|
| 85 |
+
],
|
| 86 |
+
"norm_num_groups": 32,
|
| 87 |
+
"norm_eps": 1e-06,
|
| 88 |
+
"attention_levels": [
|
| 89 |
+
false,
|
| 90 |
+
false,
|
| 91 |
+
false
|
| 92 |
+
],
|
| 93 |
+
"with_encoder_nonlocal_attn": false,
|
| 94 |
+
"with_decoder_nonlocal_attn": false,
|
| 95 |
+
"use_checkpointing": false,
|
| 96 |
+
"use_convtranspose": false,
|
| 97 |
+
"norm_float16": true,
|
| 98 |
+
"num_splits": 2,
|
| 99 |
+
"dim_split": 1
|
| 100 |
+
},
|
| 101 |
+
"diffusion_unet_def": {
|
| 102 |
+
"_target_": "monai.apps.generation.maisi.networks.diffusion_model_unet_maisi.DiffusionModelUNetMaisi",
|
| 103 |
+
"spatial_dims": "@spatial_dims",
|
| 104 |
+
"in_channels": "@latent_channels",
|
| 105 |
+
"out_channels": "@latent_channels",
|
| 106 |
+
"num_channels": [
|
| 107 |
+
64,
|
| 108 |
+
128,
|
| 109 |
+
256,
|
| 110 |
+
512
|
| 111 |
+
],
|
| 112 |
+
"attention_levels": [
|
| 113 |
+
false,
|
| 114 |
+
false,
|
| 115 |
+
true,
|
| 116 |
+
true
|
| 117 |
+
],
|
| 118 |
+
"num_head_channels": [
|
| 119 |
+
0,
|
| 120 |
+
0,
|
| 121 |
+
32,
|
| 122 |
+
32
|
| 123 |
+
],
|
| 124 |
+
"num_res_blocks": 2,
|
| 125 |
+
"use_flash_attention": true,
|
| 126 |
+
"include_top_region_index_input": false,
|
| 127 |
+
"include_bottom_region_index_input": false,
|
| 128 |
+
"include_spacing_input": true,
|
| 129 |
+
"num_class_embeds": 128,
|
| 130 |
+
"resblock_updown": true,
|
| 131 |
+
"include_fc": true
|
| 132 |
+
},
|
| 133 |
+
"controlnet_def": {
|
| 134 |
+
"_target_": "monai.apps.generation.maisi.networks.controlnet_maisi.ControlNetMaisi",
|
| 135 |
+
"spatial_dims": "@spatial_dims",
|
| 136 |
+
"in_channels": "@latent_channels",
|
| 137 |
+
"num_channels": [
|
| 138 |
+
64,
|
| 139 |
+
128,
|
| 140 |
+
256,
|
| 141 |
+
512
|
| 142 |
+
],
|
| 143 |
+
"attention_levels": [
|
| 144 |
+
false,
|
| 145 |
+
false,
|
| 146 |
+
true,
|
| 147 |
+
true
|
| 148 |
+
],
|
| 149 |
+
"num_head_channels": [
|
| 150 |
+
0,
|
| 151 |
+
0,
|
| 152 |
+
32,
|
| 153 |
+
32
|
| 154 |
+
],
|
| 155 |
+
"num_res_blocks": 2,
|
| 156 |
+
"use_flash_attention": true,
|
| 157 |
+
"conditioning_embedding_in_channels": 8,
|
| 158 |
+
"conditioning_embedding_num_channels": [
|
| 159 |
+
8,
|
| 160 |
+
32,
|
| 161 |
+
64
|
| 162 |
+
],
|
| 163 |
+
"num_class_embeds": 128,
|
| 164 |
+
"resblock_updown": true,
|
| 165 |
+
"include_fc": true
|
| 166 |
+
},
|
| 167 |
+
"mask_generation_autoencoder_def": {
|
| 168 |
+
"_target_": "monai.apps.generation.maisi.networks.autoencoderkl_maisi.AutoencoderKlMaisi",
|
| 169 |
+
"spatial_dims": "@spatial_dims",
|
| 170 |
+
"in_channels": 8,
|
| 171 |
+
"out_channels": 125,
|
| 172 |
+
"latent_channels": "@latent_channels",
|
| 173 |
+
"num_channels": [
|
| 174 |
+
32,
|
| 175 |
+
64,
|
| 176 |
+
128
|
| 177 |
+
],
|
| 178 |
+
"num_res_blocks": [
|
| 179 |
+
1,
|
| 180 |
+
2,
|
| 181 |
+
2
|
| 182 |
+
],
|
| 183 |
+
"norm_num_groups": 32,
|
| 184 |
+
"norm_eps": 1e-06,
|
| 185 |
+
"attention_levels": [
|
| 186 |
+
false,
|
| 187 |
+
false,
|
| 188 |
+
false
|
| 189 |
+
],
|
| 190 |
+
"with_encoder_nonlocal_attn": false,
|
| 191 |
+
"with_decoder_nonlocal_attn": false,
|
| 192 |
+
"use_flash_attention": false,
|
| 193 |
+
"use_checkpointing": true,
|
| 194 |
+
"use_convtranspose": true,
|
| 195 |
+
"norm_float16": true,
|
| 196 |
+
"num_splits": 8,
|
| 197 |
+
"dim_split": 1
|
| 198 |
+
},
|
| 199 |
+
"mask_generation_diffusion_def": {
|
| 200 |
+
"_target_": "monai.networks.nets.diffusion_model_unet.DiffusionModelUNet",
|
| 201 |
+
"spatial_dims": "@spatial_dims",
|
| 202 |
+
"in_channels": "@latent_channels",
|
| 203 |
+
"out_channels": "@latent_channels",
|
| 204 |
+
"channels": [
|
| 205 |
+
64,
|
| 206 |
+
128,
|
| 207 |
+
256,
|
| 208 |
+
512
|
| 209 |
+
],
|
| 210 |
+
"attention_levels": [
|
| 211 |
+
false,
|
| 212 |
+
false,
|
| 213 |
+
true,
|
| 214 |
+
true
|
| 215 |
+
],
|
| 216 |
+
"num_head_channels": [
|
| 217 |
+
0,
|
| 218 |
+
0,
|
| 219 |
+
32,
|
| 220 |
+
32
|
| 221 |
+
],
|
| 222 |
+
"num_res_blocks": 2,
|
| 223 |
+
"use_flash_attention": true,
|
| 224 |
+
"with_conditioning": true,
|
| 225 |
+
"upcast_attention": true,
|
| 226 |
+
"cross_attention_dim": 10
|
| 227 |
+
},
|
| 228 |
+
"autoencoder": "$@autoencoder_def.to(@device)",
|
| 229 |
+
"checkpoint_autoencoder": "$torch.load(@trained_autoencoder_path, weights_only=True)",
|
| 230 |
+
"load_autoencoder": "[email protected]_state_dict(@checkpoint_autoencoder)",
|
| 231 |
+
"diffusion_unet": "$@diffusion_unet_def.to(@device)",
|
| 232 |
+
"checkpoint_diffusion_unet": "$torch.load(@trained_diffusion_path, weights_only=False)",
|
| 233 |
+
"load_diffusion": "$@diffusion_unet.load_state_dict(@checkpoint_diffusion_unet['unet_state_dict'])",
|
| 234 |
+
"controlnet": "$@controlnet_def.to(@device)",
|
| 235 |
+
"copy_controlnet_state": "$monai.networks.utils.copy_model_state(@controlnet, @diffusion_unet.state_dict())",
|
| 236 |
+
"checkpoint_controlnet": "$torch.load(@trained_controlnet_path, weights_only=False)",
|
| 237 |
+
"load_controlnet": "[email protected]_state_dict(@checkpoint_controlnet['controlnet_state_dict'], strict=True)",
|
| 238 |
+
"scale_factor": "$@checkpoint_diffusion_unet['scale_factor'].to(@device)",
|
| 239 |
+
"mask_generation_autoencoder": "$@mask_generation_autoencoder_def.to(@device)",
|
| 240 |
+
"checkpoint_mask_generation_autoencoder": "$torch.load(@trained_mask_generation_autoencoder_path, weights_only=True)",
|
| 241 |
+
"load_mask_generation_autoencoder": "$@mask_generation_autoencoder.load_state_dict(@checkpoint_mask_generation_autoencoder, strict=True)",
|
| 242 |
+
"mask_generation_diffusion_unet": "$@mask_generation_diffusion_def.to(@device)",
|
| 243 |
+
"checkpoint_mask_generation_diffusion_unet": "$torch.load(@trained_mask_generation_diffusion_path, weights_only=True)",
|
| 244 |
+
"load_mask_generation_diffusion": "$@mask_generation_diffusion_unet.load_state_dict(@checkpoint_mask_generation_diffusion_unet['unet_state_dict'], strict=True)",
|
| 245 |
+
"mask_generation_scale_factor": "$@checkpoint_mask_generation_diffusion_unet['scale_factor']",
|
| 246 |
+
"noise_scheduler": {
|
| 247 |
+
"_target_": "scripts.rectified_flow.RFlowScheduler",
|
| 248 |
+
"num_train_timesteps": 1000,
|
| 249 |
+
"use_discrete_timesteps": false,
|
| 250 |
+
"use_timestep_transform": true,
|
| 251 |
+
"sample_method": "uniform"
|
| 252 |
+
},
|
| 253 |
+
"mask_generation_noise_scheduler": {
|
| 254 |
+
"_target_": "monai.networks.schedulers.ddpm.DDPMScheduler",
|
| 255 |
+
"num_train_timesteps": 1000,
|
| 256 |
+
"beta_start": 0.0015,
|
| 257 |
+
"beta_end": 0.0195,
|
| 258 |
+
"schedule": "scaled_linear_beta",
|
| 259 |
+
"clip_sample": false
|
| 260 |
+
},
|
| 261 |
+
"check_input": "$scripts.sample.check_input(@body_region,@anatomy_list,@label_dict_json,@output_size,@spacing,@controllable_anatomy_size)",
|
| 262 |
+
"ldm_sampler": {
|
| 263 |
+
"_target_": "scripts.sample.LDMSampler",
|
| 264 |
+
"_requires_": [
|
| 265 |
+
"@create_output_dir",
|
| 266 |
+
"@load_diffusion",
|
| 267 |
+
"@load_autoencoder",
|
| 268 |
+
"@copy_controlnet_state",
|
| 269 |
+
"@load_controlnet",
|
| 270 |
+
"@load_mask_generation_autoencoder",
|
| 271 |
+
"@load_mask_generation_diffusion",
|
| 272 |
+
"@check_input"
|
| 273 |
+
],
|
| 274 |
+
"body_region": "@body_region",
|
| 275 |
+
"anatomy_list": "@anatomy_list",
|
| 276 |
+
"modality": "@modality",
|
| 277 |
+
"all_mask_files_json": "@all_mask_files_json",
|
| 278 |
+
"all_anatomy_size_condtions_json": "@all_anatomy_size_condtions_json",
|
| 279 |
+
"all_mask_files_base_dir": "@all_mask_files_base_dir",
|
| 280 |
+
"label_dict_json": "@label_dict_json",
|
| 281 |
+
"label_dict_remap_json": "@label_dict_remap_json",
|
| 282 |
+
"autoencoder": "@autoencoder",
|
| 283 |
+
"diffusion_unet": "@diffusion_unet",
|
| 284 |
+
"controlnet": "@controlnet",
|
| 285 |
+
"scale_factor": "@scale_factor",
|
| 286 |
+
"noise_scheduler": "@noise_scheduler",
|
| 287 |
+
"mask_generation_autoencoder": "@mask_generation_autoencoder",
|
| 288 |
+
"mask_generation_diffusion_unet": "@mask_generation_diffusion_unet",
|
| 289 |
+
"mask_generation_scale_factor": "@mask_generation_scale_factor",
|
| 290 |
+
"mask_generation_noise_scheduler": "@mask_generation_noise_scheduler",
|
| 291 |
+
"controllable_anatomy_size": "@controllable_anatomy_size",
|
| 292 |
+
"image_output_ext": "@image_output_ext",
|
| 293 |
+
"label_output_ext": "@label_output_ext",
|
| 294 |
+
"real_img_median_statistics": "@real_img_median_statistics_file",
|
| 295 |
+
"device": "@device",
|
| 296 |
+
"latent_shape": "@latent_shape",
|
| 297 |
+
"mask_generation_latent_shape": "@mask_generation_latent_shape",
|
| 298 |
+
"output_size": "@output_size",
|
| 299 |
+
"spacing": "@spacing",
|
| 300 |
+
"output_dir": "@output_dir",
|
| 301 |
+
"num_inference_steps": "@num_inference_steps",
|
| 302 |
+
"mask_generation_num_inference_steps": "@mask_generation_num_inference_steps",
|
| 303 |
+
"random_seed": "@random_seed",
|
| 304 |
+
"autoencoder_sliding_window_infer_size": "@autoencoder_sliding_window_infer_size",
|
| 305 |
+
"autoencoder_sliding_window_infer_overlap": "@autoencoder_sliding_window_infer_overlap"
|
| 306 |
+
},
|
| 307 |
+
"run": [
|
| 308 |
+
"$monai.utils.set_determinism(seed=@random_seed)",
|
| 309 |
+
"$@ldm_sampler.sample_multiple_images(@num_output_samples)"
|
| 310 |
+
],
|
| 311 |
+
"evaluator": null
|
| 312 |
+
}
|
configs/inference_trt.json
ADDED
|
@@ -0,0 +1,19 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"+imports": [
|
| 3 |
+
"$from monai.networks import trt_compile"
|
| 4 |
+
],
|
| 5 |
+
"c_trt_args": {
|
| 6 |
+
"export_args": {
|
| 7 |
+
"dynamo": "$False",
|
| 8 |
+
"report": "$True"
|
| 9 |
+
},
|
| 10 |
+
"output_lists": [
|
| 11 |
+
[
|
| 12 |
+
-1
|
| 13 |
+
],
|
| 14 |
+
[]
|
| 15 |
+
]
|
| 16 |
+
},
|
| 17 |
+
"controlnet": "$trt_compile(@controlnet_def.to(@device), @trained_controlnet_path, @c_trt_args)",
|
| 18 |
+
"diffusion_unet": "$trt_compile(@diffusion_unet_def.to(@device), @trained_diffusion_path)"
|
| 19 |
+
}
|
configs/integration_test_masks.json
ADDED
|
@@ -0,0 +1,98 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[
|
| 2 |
+
{
|
| 3 |
+
"bottom_region_index": [
|
| 4 |
+
0,
|
| 5 |
+
0,
|
| 6 |
+
0,
|
| 7 |
+
1
|
| 8 |
+
],
|
| 9 |
+
"dim": [
|
| 10 |
+
512,
|
| 11 |
+
512,
|
| 12 |
+
512
|
| 13 |
+
],
|
| 14 |
+
"label_list": [
|
| 15 |
+
1,
|
| 16 |
+
3,
|
| 17 |
+
4,
|
| 18 |
+
5,
|
| 19 |
+
6,
|
| 20 |
+
7,
|
| 21 |
+
8,
|
| 22 |
+
9,
|
| 23 |
+
10,
|
| 24 |
+
11,
|
| 25 |
+
12,
|
| 26 |
+
13,
|
| 27 |
+
14,
|
| 28 |
+
15,
|
| 29 |
+
17,
|
| 30 |
+
19,
|
| 31 |
+
25,
|
| 32 |
+
28,
|
| 33 |
+
29,
|
| 34 |
+
31,
|
| 35 |
+
32,
|
| 36 |
+
33,
|
| 37 |
+
34,
|
| 38 |
+
35,
|
| 39 |
+
36,
|
| 40 |
+
37,
|
| 41 |
+
38,
|
| 42 |
+
39,
|
| 43 |
+
40,
|
| 44 |
+
41,
|
| 45 |
+
42,
|
| 46 |
+
58,
|
| 47 |
+
59,
|
| 48 |
+
60,
|
| 49 |
+
61,
|
| 50 |
+
62,
|
| 51 |
+
69,
|
| 52 |
+
70,
|
| 53 |
+
71,
|
| 54 |
+
72,
|
| 55 |
+
73,
|
| 56 |
+
74,
|
| 57 |
+
81,
|
| 58 |
+
82,
|
| 59 |
+
83,
|
| 60 |
+
84,
|
| 61 |
+
85,
|
| 62 |
+
86,
|
| 63 |
+
93,
|
| 64 |
+
94,
|
| 65 |
+
95,
|
| 66 |
+
96,
|
| 67 |
+
97,
|
| 68 |
+
98,
|
| 69 |
+
99,
|
| 70 |
+
100,
|
| 71 |
+
101,
|
| 72 |
+
102,
|
| 73 |
+
103,
|
| 74 |
+
104,
|
| 75 |
+
105,
|
| 76 |
+
106,
|
| 77 |
+
107,
|
| 78 |
+
114,
|
| 79 |
+
115,
|
| 80 |
+
118,
|
| 81 |
+
121,
|
| 82 |
+
122,
|
| 83 |
+
127
|
| 84 |
+
],
|
| 85 |
+
"pseudo_label_filename": "./IntegrationTest-AbdomenCT.nii.gz",
|
| 86 |
+
"spacing": [
|
| 87 |
+
1.0,
|
| 88 |
+
1.0,
|
| 89 |
+
1.0
|
| 90 |
+
],
|
| 91 |
+
"top_region_index": [
|
| 92 |
+
0,
|
| 93 |
+
1,
|
| 94 |
+
0,
|
| 95 |
+
0
|
| 96 |
+
]
|
| 97 |
+
}
|
| 98 |
+
]
|
configs/label_dict.json
ADDED
|
@@ -0,0 +1,134 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"liver": 1,
|
| 3 |
+
"dummy1": 2,
|
| 4 |
+
"spleen": 3,
|
| 5 |
+
"pancreas": 4,
|
| 6 |
+
"right kidney": 5,
|
| 7 |
+
"aorta": 6,
|
| 8 |
+
"inferior vena cava": 7,
|
| 9 |
+
"right adrenal gland": 8,
|
| 10 |
+
"left adrenal gland": 9,
|
| 11 |
+
"gallbladder": 10,
|
| 12 |
+
"esophagus": 11,
|
| 13 |
+
"stomach": 12,
|
| 14 |
+
"duodenum": 13,
|
| 15 |
+
"left kidney": 14,
|
| 16 |
+
"bladder": 15,
|
| 17 |
+
"dummy2": 16,
|
| 18 |
+
"portal vein and splenic vein": 17,
|
| 19 |
+
"dummy3": 18,
|
| 20 |
+
"small bowel": 19,
|
| 21 |
+
"dummy4": 20,
|
| 22 |
+
"dummy5": 21,
|
| 23 |
+
"brain": 22,
|
| 24 |
+
"lung tumor": 23,
|
| 25 |
+
"pancreatic tumor": 24,
|
| 26 |
+
"hepatic vessel": 25,
|
| 27 |
+
"hepatic tumor": 26,
|
| 28 |
+
"colon cancer primaries": 27,
|
| 29 |
+
"left lung upper lobe": 28,
|
| 30 |
+
"left lung lower lobe": 29,
|
| 31 |
+
"right lung upper lobe": 30,
|
| 32 |
+
"right lung middle lobe": 31,
|
| 33 |
+
"right lung lower lobe": 32,
|
| 34 |
+
"vertebrae L5": 33,
|
| 35 |
+
"vertebrae L4": 34,
|
| 36 |
+
"vertebrae L3": 35,
|
| 37 |
+
"vertebrae L2": 36,
|
| 38 |
+
"vertebrae L1": 37,
|
| 39 |
+
"vertebrae T12": 38,
|
| 40 |
+
"vertebrae T11": 39,
|
| 41 |
+
"vertebrae T10": 40,
|
| 42 |
+
"vertebrae T9": 41,
|
| 43 |
+
"vertebrae T8": 42,
|
| 44 |
+
"vertebrae T7": 43,
|
| 45 |
+
"vertebrae T6": 44,
|
| 46 |
+
"vertebrae T5": 45,
|
| 47 |
+
"vertebrae T4": 46,
|
| 48 |
+
"vertebrae T3": 47,
|
| 49 |
+
"vertebrae T2": 48,
|
| 50 |
+
"vertebrae T1": 49,
|
| 51 |
+
"vertebrae C7": 50,
|
| 52 |
+
"vertebrae C6": 51,
|
| 53 |
+
"vertebrae C5": 52,
|
| 54 |
+
"vertebrae C4": 53,
|
| 55 |
+
"vertebrae C3": 54,
|
| 56 |
+
"vertebrae C2": 55,
|
| 57 |
+
"vertebrae C1": 56,
|
| 58 |
+
"trachea": 57,
|
| 59 |
+
"left iliac artery": 58,
|
| 60 |
+
"right iliac artery": 59,
|
| 61 |
+
"left iliac vena": 60,
|
| 62 |
+
"right iliac vena": 61,
|
| 63 |
+
"colon": 62,
|
| 64 |
+
"left rib 1": 63,
|
| 65 |
+
"left rib 2": 64,
|
| 66 |
+
"left rib 3": 65,
|
| 67 |
+
"left rib 4": 66,
|
| 68 |
+
"left rib 5": 67,
|
| 69 |
+
"left rib 6": 68,
|
| 70 |
+
"left rib 7": 69,
|
| 71 |
+
"left rib 8": 70,
|
| 72 |
+
"left rib 9": 71,
|
| 73 |
+
"left rib 10": 72,
|
| 74 |
+
"left rib 11": 73,
|
| 75 |
+
"left rib 12": 74,
|
| 76 |
+
"right rib 1": 75,
|
| 77 |
+
"right rib 2": 76,
|
| 78 |
+
"right rib 3": 77,
|
| 79 |
+
"right rib 4": 78,
|
| 80 |
+
"right rib 5": 79,
|
| 81 |
+
"right rib 6": 80,
|
| 82 |
+
"right rib 7": 81,
|
| 83 |
+
"right rib 8": 82,
|
| 84 |
+
"right rib 9": 83,
|
| 85 |
+
"right rib 10": 84,
|
| 86 |
+
"right rib 11": 85,
|
| 87 |
+
"right rib 12": 86,
|
| 88 |
+
"left humerus": 87,
|
| 89 |
+
"right humerus": 88,
|
| 90 |
+
"left scapula": 89,
|
| 91 |
+
"right scapula": 90,
|
| 92 |
+
"left clavicula": 91,
|
| 93 |
+
"right clavicula": 92,
|
| 94 |
+
"left femur": 93,
|
| 95 |
+
"right femur": 94,
|
| 96 |
+
"left hip": 95,
|
| 97 |
+
"right hip": 96,
|
| 98 |
+
"sacrum": 97,
|
| 99 |
+
"left gluteus maximus": 98,
|
| 100 |
+
"right gluteus maximus": 99,
|
| 101 |
+
"left gluteus medius": 100,
|
| 102 |
+
"right gluteus medius": 101,
|
| 103 |
+
"left gluteus minimus": 102,
|
| 104 |
+
"right gluteus minimus": 103,
|
| 105 |
+
"left autochthon": 104,
|
| 106 |
+
"right autochthon": 105,
|
| 107 |
+
"left iliopsoas": 106,
|
| 108 |
+
"right iliopsoas": 107,
|
| 109 |
+
"left atrial appendage": 108,
|
| 110 |
+
"brachiocephalic trunk": 109,
|
| 111 |
+
"left brachiocephalic vein": 110,
|
| 112 |
+
"right brachiocephalic vein": 111,
|
| 113 |
+
"left common carotid artery": 112,
|
| 114 |
+
"right common carotid artery": 113,
|
| 115 |
+
"costal cartilages": 114,
|
| 116 |
+
"heart": 115,
|
| 117 |
+
"left kidney cyst": 116,
|
| 118 |
+
"right kidney cyst": 117,
|
| 119 |
+
"prostate": 118,
|
| 120 |
+
"pulmonary vein": 119,
|
| 121 |
+
"skull": 120,
|
| 122 |
+
"spinal cord": 121,
|
| 123 |
+
"sternum": 122,
|
| 124 |
+
"left subclavian artery": 123,
|
| 125 |
+
"right subclavian artery": 124,
|
| 126 |
+
"superior vena cava": 125,
|
| 127 |
+
"thyroid gland": 126,
|
| 128 |
+
"vertebrae S1": 127,
|
| 129 |
+
"bone lesion": 128,
|
| 130 |
+
"dummy6": 129,
|
| 131 |
+
"dummy7": 130,
|
| 132 |
+
"dummy8": 131,
|
| 133 |
+
"airway": 132
|
| 134 |
+
}
|
configs/label_dict_124_to_132.json
ADDED
|
@@ -0,0 +1,502 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"background": [
|
| 3 |
+
0,
|
| 4 |
+
0
|
| 5 |
+
],
|
| 6 |
+
"liver": [
|
| 7 |
+
1,
|
| 8 |
+
1
|
| 9 |
+
],
|
| 10 |
+
"spleen": [
|
| 11 |
+
2,
|
| 12 |
+
3
|
| 13 |
+
],
|
| 14 |
+
"pancreas": [
|
| 15 |
+
3,
|
| 16 |
+
4
|
| 17 |
+
],
|
| 18 |
+
"right kidney": [
|
| 19 |
+
4,
|
| 20 |
+
5
|
| 21 |
+
],
|
| 22 |
+
"aorta": [
|
| 23 |
+
5,
|
| 24 |
+
6
|
| 25 |
+
],
|
| 26 |
+
"inferior vena cava": [
|
| 27 |
+
6,
|
| 28 |
+
7
|
| 29 |
+
],
|
| 30 |
+
"right adrenal gland": [
|
| 31 |
+
7,
|
| 32 |
+
8
|
| 33 |
+
],
|
| 34 |
+
"left adrenal gland": [
|
| 35 |
+
8,
|
| 36 |
+
9
|
| 37 |
+
],
|
| 38 |
+
"gallbladder": [
|
| 39 |
+
9,
|
| 40 |
+
10
|
| 41 |
+
],
|
| 42 |
+
"esophagus": [
|
| 43 |
+
10,
|
| 44 |
+
11
|
| 45 |
+
],
|
| 46 |
+
"stomach": [
|
| 47 |
+
11,
|
| 48 |
+
12
|
| 49 |
+
],
|
| 50 |
+
"duodenum": [
|
| 51 |
+
12,
|
| 52 |
+
13
|
| 53 |
+
],
|
| 54 |
+
"left kidney": [
|
| 55 |
+
13,
|
| 56 |
+
14
|
| 57 |
+
],
|
| 58 |
+
"bladder": [
|
| 59 |
+
14,
|
| 60 |
+
15
|
| 61 |
+
],
|
| 62 |
+
"portal vein and splenic vein": [
|
| 63 |
+
15,
|
| 64 |
+
17
|
| 65 |
+
],
|
| 66 |
+
"small bowel": [
|
| 67 |
+
16,
|
| 68 |
+
19
|
| 69 |
+
],
|
| 70 |
+
"brain": [
|
| 71 |
+
17,
|
| 72 |
+
22
|
| 73 |
+
],
|
| 74 |
+
"lung tumor": [
|
| 75 |
+
18,
|
| 76 |
+
23
|
| 77 |
+
],
|
| 78 |
+
"pancreatic tumor": [
|
| 79 |
+
19,
|
| 80 |
+
24
|
| 81 |
+
],
|
| 82 |
+
"hepatic vessel": [
|
| 83 |
+
20,
|
| 84 |
+
25
|
| 85 |
+
],
|
| 86 |
+
"hepatic tumor": [
|
| 87 |
+
21,
|
| 88 |
+
26
|
| 89 |
+
],
|
| 90 |
+
"colon cancer primaries": [
|
| 91 |
+
22,
|
| 92 |
+
27
|
| 93 |
+
],
|
| 94 |
+
"left lung upper lobe": [
|
| 95 |
+
23,
|
| 96 |
+
28
|
| 97 |
+
],
|
| 98 |
+
"left lung lower lobe": [
|
| 99 |
+
24,
|
| 100 |
+
29
|
| 101 |
+
],
|
| 102 |
+
"right lung upper lobe": [
|
| 103 |
+
25,
|
| 104 |
+
30
|
| 105 |
+
],
|
| 106 |
+
"right lung middle lobe": [
|
| 107 |
+
26,
|
| 108 |
+
31
|
| 109 |
+
],
|
| 110 |
+
"right lung lower lobe": [
|
| 111 |
+
27,
|
| 112 |
+
32
|
| 113 |
+
],
|
| 114 |
+
"vertebrae L5": [
|
| 115 |
+
28,
|
| 116 |
+
33
|
| 117 |
+
],
|
| 118 |
+
"vertebrae L4": [
|
| 119 |
+
29,
|
| 120 |
+
34
|
| 121 |
+
],
|
| 122 |
+
"vertebrae L3": [
|
| 123 |
+
30,
|
| 124 |
+
35
|
| 125 |
+
],
|
| 126 |
+
"vertebrae L2": [
|
| 127 |
+
31,
|
| 128 |
+
36
|
| 129 |
+
],
|
| 130 |
+
"vertebrae L1": [
|
| 131 |
+
32,
|
| 132 |
+
37
|
| 133 |
+
],
|
| 134 |
+
"vertebrae T12": [
|
| 135 |
+
33,
|
| 136 |
+
38
|
| 137 |
+
],
|
| 138 |
+
"vertebrae T11": [
|
| 139 |
+
34,
|
| 140 |
+
39
|
| 141 |
+
],
|
| 142 |
+
"vertebrae T10": [
|
| 143 |
+
35,
|
| 144 |
+
40
|
| 145 |
+
],
|
| 146 |
+
"vertebrae T9": [
|
| 147 |
+
36,
|
| 148 |
+
41
|
| 149 |
+
],
|
| 150 |
+
"vertebrae T8": [
|
| 151 |
+
37,
|
| 152 |
+
42
|
| 153 |
+
],
|
| 154 |
+
"vertebrae T7": [
|
| 155 |
+
38,
|
| 156 |
+
43
|
| 157 |
+
],
|
| 158 |
+
"vertebrae T6": [
|
| 159 |
+
39,
|
| 160 |
+
44
|
| 161 |
+
],
|
| 162 |
+
"vertebrae T5": [
|
| 163 |
+
40,
|
| 164 |
+
45
|
| 165 |
+
],
|
| 166 |
+
"vertebrae T4": [
|
| 167 |
+
41,
|
| 168 |
+
46
|
| 169 |
+
],
|
| 170 |
+
"vertebrae T3": [
|
| 171 |
+
42,
|
| 172 |
+
47
|
| 173 |
+
],
|
| 174 |
+
"vertebrae T2": [
|
| 175 |
+
43,
|
| 176 |
+
48
|
| 177 |
+
],
|
| 178 |
+
"vertebrae T1": [
|
| 179 |
+
44,
|
| 180 |
+
49
|
| 181 |
+
],
|
| 182 |
+
"vertebrae C7": [
|
| 183 |
+
45,
|
| 184 |
+
50
|
| 185 |
+
],
|
| 186 |
+
"vertebrae C6": [
|
| 187 |
+
46,
|
| 188 |
+
51
|
| 189 |
+
],
|
| 190 |
+
"vertebrae C5": [
|
| 191 |
+
47,
|
| 192 |
+
52
|
| 193 |
+
],
|
| 194 |
+
"vertebrae C4": [
|
| 195 |
+
48,
|
| 196 |
+
53
|
| 197 |
+
],
|
| 198 |
+
"vertebrae C3": [
|
| 199 |
+
49,
|
| 200 |
+
54
|
| 201 |
+
],
|
| 202 |
+
"vertebrae C2": [
|
| 203 |
+
50,
|
| 204 |
+
55
|
| 205 |
+
],
|
| 206 |
+
"vertebrae C1": [
|
| 207 |
+
51,
|
| 208 |
+
56
|
| 209 |
+
],
|
| 210 |
+
"trachea": [
|
| 211 |
+
52,
|
| 212 |
+
57
|
| 213 |
+
],
|
| 214 |
+
"left iliac artery": [
|
| 215 |
+
53,
|
| 216 |
+
58
|
| 217 |
+
],
|
| 218 |
+
"right iliac artery": [
|
| 219 |
+
54,
|
| 220 |
+
59
|
| 221 |
+
],
|
| 222 |
+
"left iliac vena": [
|
| 223 |
+
55,
|
| 224 |
+
60
|
| 225 |
+
],
|
| 226 |
+
"right iliac vena": [
|
| 227 |
+
56,
|
| 228 |
+
61
|
| 229 |
+
],
|
| 230 |
+
"colon": [
|
| 231 |
+
57,
|
| 232 |
+
62
|
| 233 |
+
],
|
| 234 |
+
"left rib 1": [
|
| 235 |
+
58,
|
| 236 |
+
63
|
| 237 |
+
],
|
| 238 |
+
"left rib 2": [
|
| 239 |
+
59,
|
| 240 |
+
64
|
| 241 |
+
],
|
| 242 |
+
"left rib 3": [
|
| 243 |
+
60,
|
| 244 |
+
65
|
| 245 |
+
],
|
| 246 |
+
"left rib 4": [
|
| 247 |
+
61,
|
| 248 |
+
66
|
| 249 |
+
],
|
| 250 |
+
"left rib 5": [
|
| 251 |
+
62,
|
| 252 |
+
67
|
| 253 |
+
],
|
| 254 |
+
"left rib 6": [
|
| 255 |
+
63,
|
| 256 |
+
68
|
| 257 |
+
],
|
| 258 |
+
"left rib 7": [
|
| 259 |
+
64,
|
| 260 |
+
69
|
| 261 |
+
],
|
| 262 |
+
"left rib 8": [
|
| 263 |
+
65,
|
| 264 |
+
70
|
| 265 |
+
],
|
| 266 |
+
"left rib 9": [
|
| 267 |
+
66,
|
| 268 |
+
71
|
| 269 |
+
],
|
| 270 |
+
"left rib 10": [
|
| 271 |
+
67,
|
| 272 |
+
72
|
| 273 |
+
],
|
| 274 |
+
"left rib 11": [
|
| 275 |
+
68,
|
| 276 |
+
73
|
| 277 |
+
],
|
| 278 |
+
"left rib 12": [
|
| 279 |
+
69,
|
| 280 |
+
74
|
| 281 |
+
],
|
| 282 |
+
"right rib 1": [
|
| 283 |
+
70,
|
| 284 |
+
75
|
| 285 |
+
],
|
| 286 |
+
"right rib 2": [
|
| 287 |
+
71,
|
| 288 |
+
76
|
| 289 |
+
],
|
| 290 |
+
"right rib 3": [
|
| 291 |
+
72,
|
| 292 |
+
77
|
| 293 |
+
],
|
| 294 |
+
"right rib 4": [
|
| 295 |
+
73,
|
| 296 |
+
78
|
| 297 |
+
],
|
| 298 |
+
"right rib 5": [
|
| 299 |
+
74,
|
| 300 |
+
79
|
| 301 |
+
],
|
| 302 |
+
"right rib 6": [
|
| 303 |
+
75,
|
| 304 |
+
80
|
| 305 |
+
],
|
| 306 |
+
"right rib 7": [
|
| 307 |
+
76,
|
| 308 |
+
81
|
| 309 |
+
],
|
| 310 |
+
"right rib 8": [
|
| 311 |
+
77,
|
| 312 |
+
82
|
| 313 |
+
],
|
| 314 |
+
"right rib 9": [
|
| 315 |
+
78,
|
| 316 |
+
83
|
| 317 |
+
],
|
| 318 |
+
"right rib 10": [
|
| 319 |
+
79,
|
| 320 |
+
84
|
| 321 |
+
],
|
| 322 |
+
"right rib 11": [
|
| 323 |
+
80,
|
| 324 |
+
85
|
| 325 |
+
],
|
| 326 |
+
"right rib 12": [
|
| 327 |
+
81,
|
| 328 |
+
86
|
| 329 |
+
],
|
| 330 |
+
"left humerus": [
|
| 331 |
+
82,
|
| 332 |
+
87
|
| 333 |
+
],
|
| 334 |
+
"right humerus": [
|
| 335 |
+
83,
|
| 336 |
+
88
|
| 337 |
+
],
|
| 338 |
+
"left scapula": [
|
| 339 |
+
84,
|
| 340 |
+
89
|
| 341 |
+
],
|
| 342 |
+
"right scapula": [
|
| 343 |
+
85,
|
| 344 |
+
90
|
| 345 |
+
],
|
| 346 |
+
"left clavicula": [
|
| 347 |
+
86,
|
| 348 |
+
91
|
| 349 |
+
],
|
| 350 |
+
"right clavicula": [
|
| 351 |
+
87,
|
| 352 |
+
92
|
| 353 |
+
],
|
| 354 |
+
"left femur": [
|
| 355 |
+
88,
|
| 356 |
+
93
|
| 357 |
+
],
|
| 358 |
+
"right femur": [
|
| 359 |
+
89,
|
| 360 |
+
94
|
| 361 |
+
],
|
| 362 |
+
"left hip": [
|
| 363 |
+
90,
|
| 364 |
+
95
|
| 365 |
+
],
|
| 366 |
+
"right hip": [
|
| 367 |
+
91,
|
| 368 |
+
96
|
| 369 |
+
],
|
| 370 |
+
"sacrum": [
|
| 371 |
+
92,
|
| 372 |
+
97
|
| 373 |
+
],
|
| 374 |
+
"left gluteus maximus": [
|
| 375 |
+
93,
|
| 376 |
+
98
|
| 377 |
+
],
|
| 378 |
+
"right gluteus maximus": [
|
| 379 |
+
94,
|
| 380 |
+
99
|
| 381 |
+
],
|
| 382 |
+
"left gluteus medius": [
|
| 383 |
+
95,
|
| 384 |
+
100
|
| 385 |
+
],
|
| 386 |
+
"right gluteus medius": [
|
| 387 |
+
96,
|
| 388 |
+
101
|
| 389 |
+
],
|
| 390 |
+
"left gluteus minimus": [
|
| 391 |
+
97,
|
| 392 |
+
102
|
| 393 |
+
],
|
| 394 |
+
"right gluteus minimus": [
|
| 395 |
+
98,
|
| 396 |
+
103
|
| 397 |
+
],
|
| 398 |
+
"left autochthon": [
|
| 399 |
+
99,
|
| 400 |
+
104
|
| 401 |
+
],
|
| 402 |
+
"right autochthon": [
|
| 403 |
+
100,
|
| 404 |
+
105
|
| 405 |
+
],
|
| 406 |
+
"left iliopsoas": [
|
| 407 |
+
101,
|
| 408 |
+
106
|
| 409 |
+
],
|
| 410 |
+
"right iliopsoas": [
|
| 411 |
+
102,
|
| 412 |
+
107
|
| 413 |
+
],
|
| 414 |
+
"left atrial appendage": [
|
| 415 |
+
103,
|
| 416 |
+
108
|
| 417 |
+
],
|
| 418 |
+
"brachiocephalic trunk": [
|
| 419 |
+
104,
|
| 420 |
+
109
|
| 421 |
+
],
|
| 422 |
+
"left brachiocephalic vein": [
|
| 423 |
+
105,
|
| 424 |
+
110
|
| 425 |
+
],
|
| 426 |
+
"right brachiocephalic vein": [
|
| 427 |
+
106,
|
| 428 |
+
111
|
| 429 |
+
],
|
| 430 |
+
"left common carotid artery": [
|
| 431 |
+
107,
|
| 432 |
+
112
|
| 433 |
+
],
|
| 434 |
+
"right common carotid artery": [
|
| 435 |
+
108,
|
| 436 |
+
113
|
| 437 |
+
],
|
| 438 |
+
"costal cartilages": [
|
| 439 |
+
109,
|
| 440 |
+
114
|
| 441 |
+
],
|
| 442 |
+
"heart": [
|
| 443 |
+
110,
|
| 444 |
+
115
|
| 445 |
+
],
|
| 446 |
+
"prostate": [
|
| 447 |
+
111,
|
| 448 |
+
118
|
| 449 |
+
],
|
| 450 |
+
"pulmonary vein": [
|
| 451 |
+
112,
|
| 452 |
+
119
|
| 453 |
+
],
|
| 454 |
+
"skull": [
|
| 455 |
+
113,
|
| 456 |
+
120
|
| 457 |
+
],
|
| 458 |
+
"spinal cord": [
|
| 459 |
+
114,
|
| 460 |
+
121
|
| 461 |
+
],
|
| 462 |
+
"sternum": [
|
| 463 |
+
115,
|
| 464 |
+
122
|
| 465 |
+
],
|
| 466 |
+
"left subclavian artery": [
|
| 467 |
+
116,
|
| 468 |
+
123
|
| 469 |
+
],
|
| 470 |
+
"right subclavian artery": [
|
| 471 |
+
117,
|
| 472 |
+
124
|
| 473 |
+
],
|
| 474 |
+
"superior vena cava": [
|
| 475 |
+
118,
|
| 476 |
+
125
|
| 477 |
+
],
|
| 478 |
+
"thyroid gland": [
|
| 479 |
+
119,
|
| 480 |
+
126
|
| 481 |
+
],
|
| 482 |
+
"vertebrae S1": [
|
| 483 |
+
120,
|
| 484 |
+
127
|
| 485 |
+
],
|
| 486 |
+
"bone lesion": [
|
| 487 |
+
121,
|
| 488 |
+
128
|
| 489 |
+
],
|
| 490 |
+
"kidney mass": [
|
| 491 |
+
122,
|
| 492 |
+
129
|
| 493 |
+
],
|
| 494 |
+
"airway": [
|
| 495 |
+
123,
|
| 496 |
+
132
|
| 497 |
+
],
|
| 498 |
+
"body": [
|
| 499 |
+
124,
|
| 500 |
+
200
|
| 501 |
+
]
|
| 502 |
+
}
|
configs/logging.conf
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[loggers]
|
| 2 |
+
keys=root
|
| 3 |
+
|
| 4 |
+
[handlers]
|
| 5 |
+
keys=consoleHandler
|
| 6 |
+
|
| 7 |
+
[formatters]
|
| 8 |
+
keys=fullFormatter
|
| 9 |
+
|
| 10 |
+
[logger_root]
|
| 11 |
+
level=INFO
|
| 12 |
+
handlers=consoleHandler
|
| 13 |
+
|
| 14 |
+
[handler_consoleHandler]
|
| 15 |
+
class=StreamHandler
|
| 16 |
+
level=INFO
|
| 17 |
+
formatter=fullFormatter
|
| 18 |
+
args=(sys.stdout,)
|
| 19 |
+
|
| 20 |
+
[formatter_fullFormatter]
|
| 21 |
+
format=%(asctime)s - %(name)s - %(levelname)s - %(message)s
|
configs/metadata.json
ADDED
|
@@ -0,0 +1,269 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_generator_ldm_20240318.json",
|
| 3 |
+
"version": "1.0.0",
|
| 4 |
+
"changelog": {
|
| 5 |
+
"1.0.0": "accelerated maisi, inference only, is not compartible with previous maisi diffusion model weights",
|
| 6 |
+
"0.4.6": "add TensorRT support",
|
| 7 |
+
"0.4.5": "update README",
|
| 8 |
+
"0.4.4": "update issue for IgniteInfo",
|
| 9 |
+
"0.4.3": "remove download large files, add weights_only when loading weights and add label_dict to large files",
|
| 10 |
+
"0.4.2": "update train.json to fix finetune ckpt bug",
|
| 11 |
+
"0.4.1": "update large files",
|
| 12 |
+
"0.4.0": "update to use monai 1.4, model ckpt updated, rm GenerativeAI repo, add quality check",
|
| 13 |
+
"0.3.6": "first oss version"
|
| 14 |
+
},
|
| 15 |
+
"monai_version": "1.4.0",
|
| 16 |
+
"pytorch_version": "2.4.0",
|
| 17 |
+
"numpy_version": "1.24.4",
|
| 18 |
+
"optional_packages_version": {
|
| 19 |
+
"fire": "0.6.0",
|
| 20 |
+
"nibabel": "5.2.1",
|
| 21 |
+
"tqdm": "4.66.4"
|
| 22 |
+
},
|
| 23 |
+
"supported_apps": {
|
| 24 |
+
"maisi-nim": ""
|
| 25 |
+
},
|
| 26 |
+
"name": "CT image latent diffusion generation",
|
| 27 |
+
"task": "CT image synthesis",
|
| 28 |
+
"description": "A generative model for creating 3D CT from Gaussian noise",
|
| 29 |
+
"authors": "MONAI team",
|
| 30 |
+
"copyright": "Copyright (c) MONAI Consortium",
|
| 31 |
+
"data_source": "http://medicaldecathlon.com/",
|
| 32 |
+
"data_type": "nibabel",
|
| 33 |
+
"image_classes": "Flair brain MRI with 1.1x1.1x1.1 mm voxel size",
|
| 34 |
+
"eval_metrics": {},
|
| 35 |
+
"intended_use": "This is a research tool/prototype and not to be used clinically",
|
| 36 |
+
"references": [],
|
| 37 |
+
"autoencoder_data_format": {
|
| 38 |
+
"inputs": {
|
| 39 |
+
"image": {
|
| 40 |
+
"type": "feature",
|
| 41 |
+
"format": "image",
|
| 42 |
+
"num_channels": 4,
|
| 43 |
+
"spatial_shape": [
|
| 44 |
+
128,
|
| 45 |
+
128,
|
| 46 |
+
128
|
| 47 |
+
],
|
| 48 |
+
"dtype": "float16",
|
| 49 |
+
"value_range": [
|
| 50 |
+
0,
|
| 51 |
+
1
|
| 52 |
+
],
|
| 53 |
+
"is_patch_data": true
|
| 54 |
+
},
|
| 55 |
+
"body_region": {
|
| 56 |
+
"type": "array",
|
| 57 |
+
"value_range": [
|
| 58 |
+
"head",
|
| 59 |
+
"abdomen",
|
| 60 |
+
"chest/thorax",
|
| 61 |
+
"pelvis/lower"
|
| 62 |
+
]
|
| 63 |
+
},
|
| 64 |
+
"anatomy_list": {
|
| 65 |
+
"type": "array",
|
| 66 |
+
"value_range": [
|
| 67 |
+
"liver",
|
| 68 |
+
"spleen",
|
| 69 |
+
"pancreas",
|
| 70 |
+
"right kidney",
|
| 71 |
+
"aorta",
|
| 72 |
+
"inferior vena cava",
|
| 73 |
+
"right adrenal gland",
|
| 74 |
+
"left adrenal gland",
|
| 75 |
+
"gallbladder",
|
| 76 |
+
"esophagus",
|
| 77 |
+
"stomach",
|
| 78 |
+
"duodenum",
|
| 79 |
+
"left kidney",
|
| 80 |
+
"bladder",
|
| 81 |
+
"portal vein and splenic vein",
|
| 82 |
+
"small bowel",
|
| 83 |
+
"brain",
|
| 84 |
+
"lung tumor",
|
| 85 |
+
"pancreatic tumor",
|
| 86 |
+
"hepatic vessel",
|
| 87 |
+
"hepatic tumor",
|
| 88 |
+
"colon cancer primaries",
|
| 89 |
+
"left lung upper lobe",
|
| 90 |
+
"left lung lower lobe",
|
| 91 |
+
"right lung upper lobe",
|
| 92 |
+
"right lung middle lobe",
|
| 93 |
+
"right lung lower lobe",
|
| 94 |
+
"vertebrae L5",
|
| 95 |
+
"vertebrae L4",
|
| 96 |
+
"vertebrae L3",
|
| 97 |
+
"vertebrae L2",
|
| 98 |
+
"vertebrae L1",
|
| 99 |
+
"vertebrae T12",
|
| 100 |
+
"vertebrae T11",
|
| 101 |
+
"vertebrae T10",
|
| 102 |
+
"vertebrae T9",
|
| 103 |
+
"vertebrae T8",
|
| 104 |
+
"vertebrae T7",
|
| 105 |
+
"vertebrae T6",
|
| 106 |
+
"vertebrae T5",
|
| 107 |
+
"vertebrae T4",
|
| 108 |
+
"vertebrae T3",
|
| 109 |
+
"vertebrae T2",
|
| 110 |
+
"vertebrae T1",
|
| 111 |
+
"vertebrae C7",
|
| 112 |
+
"vertebrae C6",
|
| 113 |
+
"vertebrae C5",
|
| 114 |
+
"vertebrae C4",
|
| 115 |
+
"vertebrae C3",
|
| 116 |
+
"vertebrae C2",
|
| 117 |
+
"vertebrae C1",
|
| 118 |
+
"trachea",
|
| 119 |
+
"left iliac artery",
|
| 120 |
+
"right iliac artery",
|
| 121 |
+
"left iliac vena",
|
| 122 |
+
"right iliac vena",
|
| 123 |
+
"colon",
|
| 124 |
+
"left rib 1",
|
| 125 |
+
"left rib 2",
|
| 126 |
+
"left rib 3",
|
| 127 |
+
"left rib 4",
|
| 128 |
+
"left rib 5",
|
| 129 |
+
"left rib 6",
|
| 130 |
+
"left rib 7",
|
| 131 |
+
"left rib 8",
|
| 132 |
+
"left rib 9",
|
| 133 |
+
"left rib 10",
|
| 134 |
+
"left rib 11",
|
| 135 |
+
"left rib 12",
|
| 136 |
+
"right rib 1",
|
| 137 |
+
"right rib 2",
|
| 138 |
+
"right rib 3",
|
| 139 |
+
"right rib 4",
|
| 140 |
+
"right rib 5",
|
| 141 |
+
"right rib 6",
|
| 142 |
+
"right rib 7",
|
| 143 |
+
"right rib 8",
|
| 144 |
+
"right rib 9",
|
| 145 |
+
"right rib 10",
|
| 146 |
+
"right rib 11",
|
| 147 |
+
"right rib 12",
|
| 148 |
+
"left humerus",
|
| 149 |
+
"right humerus",
|
| 150 |
+
"left scapula",
|
| 151 |
+
"right scapula",
|
| 152 |
+
"left clavicula",
|
| 153 |
+
"right clavicula",
|
| 154 |
+
"left femur",
|
| 155 |
+
"right femur",
|
| 156 |
+
"left hip",
|
| 157 |
+
"right hip",
|
| 158 |
+
"sacrum",
|
| 159 |
+
"left gluteus maximus",
|
| 160 |
+
"right gluteus maximus",
|
| 161 |
+
"left gluteus medius",
|
| 162 |
+
"right gluteus medius",
|
| 163 |
+
"left gluteus minimus",
|
| 164 |
+
"right gluteus minimus",
|
| 165 |
+
"left autochthon",
|
| 166 |
+
"right autochthon",
|
| 167 |
+
"left iliopsoas",
|
| 168 |
+
"right iliopsoas",
|
| 169 |
+
"left atrial appendage",
|
| 170 |
+
"brachiocephalic trunk",
|
| 171 |
+
"left brachiocephalic vein",
|
| 172 |
+
"right brachiocephalic vein",
|
| 173 |
+
"left common carotid artery",
|
| 174 |
+
"right common carotid artery",
|
| 175 |
+
"costal cartilages",
|
| 176 |
+
"heart",
|
| 177 |
+
"left kidney cyst",
|
| 178 |
+
"right kidney cyst",
|
| 179 |
+
"prostate",
|
| 180 |
+
"pulmonary vein",
|
| 181 |
+
"skull",
|
| 182 |
+
"spinal cord",
|
| 183 |
+
"sternum",
|
| 184 |
+
"left subclavian artery",
|
| 185 |
+
"right subclavian artery",
|
| 186 |
+
"superior vena cava",
|
| 187 |
+
"thyroid gland",
|
| 188 |
+
"vertebrae S1",
|
| 189 |
+
"bone lesion",
|
| 190 |
+
"airway"
|
| 191 |
+
]
|
| 192 |
+
}
|
| 193 |
+
},
|
| 194 |
+
"outputs": {
|
| 195 |
+
"pred": {
|
| 196 |
+
"type": "image",
|
| 197 |
+
"format": "image",
|
| 198 |
+
"num_channels": 1,
|
| 199 |
+
"spatial_shape": [
|
| 200 |
+
512,
|
| 201 |
+
512,
|
| 202 |
+
512
|
| 203 |
+
],
|
| 204 |
+
"dtype": "float16",
|
| 205 |
+
"value_range": [
|
| 206 |
+
0,
|
| 207 |
+
1
|
| 208 |
+
],
|
| 209 |
+
"is_patch_data": true,
|
| 210 |
+
"channel_def": {
|
| 211 |
+
"0": "image"
|
| 212 |
+
}
|
| 213 |
+
}
|
| 214 |
+
}
|
| 215 |
+
},
|
| 216 |
+
"generator_data_format": {
|
| 217 |
+
"inputs": {
|
| 218 |
+
"latent": {
|
| 219 |
+
"type": "noise",
|
| 220 |
+
"format": "image",
|
| 221 |
+
"num_channels": 4,
|
| 222 |
+
"spatial_shape": [
|
| 223 |
+
128,
|
| 224 |
+
128,
|
| 225 |
+
128
|
| 226 |
+
],
|
| 227 |
+
"dtype": "float16",
|
| 228 |
+
"value_range": [
|
| 229 |
+
0,
|
| 230 |
+
1
|
| 231 |
+
],
|
| 232 |
+
"is_patch_data": true
|
| 233 |
+
},
|
| 234 |
+
"condition": {
|
| 235 |
+
"type": "timesteps",
|
| 236 |
+
"format": "timesteps",
|
| 237 |
+
"num_channels": 1,
|
| 238 |
+
"spatial_shape": [],
|
| 239 |
+
"dtype": "long",
|
| 240 |
+
"value_range": [
|
| 241 |
+
0,
|
| 242 |
+
1000
|
| 243 |
+
],
|
| 244 |
+
"is_patch_data": false
|
| 245 |
+
}
|
| 246 |
+
},
|
| 247 |
+
"outputs": {
|
| 248 |
+
"pred": {
|
| 249 |
+
"type": "feature",
|
| 250 |
+
"format": "image",
|
| 251 |
+
"num_channels": 4,
|
| 252 |
+
"spatial_shape": [
|
| 253 |
+
128,
|
| 254 |
+
128,
|
| 255 |
+
128
|
| 256 |
+
],
|
| 257 |
+
"dtype": "float16",
|
| 258 |
+
"value_range": [
|
| 259 |
+
0,
|
| 260 |
+
1
|
| 261 |
+
],
|
| 262 |
+
"is_patch_data": true,
|
| 263 |
+
"channel_def": {
|
| 264 |
+
"0": "image"
|
| 265 |
+
}
|
| 266 |
+
}
|
| 267 |
+
}
|
| 268 |
+
}
|
| 269 |
+
}
|
configs/multi_gpu_train.json
ADDED
|
@@ -0,0 +1,34 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"device": "$torch.device('cuda:' + os.environ['LOCAL_RANK'])",
|
| 3 |
+
"use_tensorboard": "$dist.get_rank() == 0",
|
| 4 |
+
"controlnet": {
|
| 5 |
+
"_target_": "torch.nn.parallel.DistributedDataParallel",
|
| 6 |
+
"module": "$@controlnet_def.to(@device)",
|
| 7 |
+
"find_unused_parameters": true,
|
| 8 |
+
"device_ids": [
|
| 9 |
+
"@device"
|
| 10 |
+
]
|
| 11 |
+
},
|
| 12 |
+
"load_controlnet": "[email protected]_state_dict(@checkpoint_controlnet['controlnet_state_dict'], strict=True)",
|
| 13 |
+
"train#sampler": {
|
| 14 |
+
"_target_": "DistributedSampler",
|
| 15 |
+
"dataset": "@train#dataset",
|
| 16 |
+
"even_divisible": true,
|
| 17 |
+
"shuffle": true
|
| 18 |
+
},
|
| 19 |
+
"train#dataloader#sampler": "@train#sampler",
|
| 20 |
+
"train#dataloader#shuffle": false,
|
| 21 |
+
"train#trainer#train_handlers": "$@train#handlers[: -1 if dist.get_rank() > 0 else None]",
|
| 22 |
+
"initialize": [
|
| 23 |
+
"$import torch.distributed as dist",
|
| 24 |
+
"$dist.is_initialized() or dist.init_process_group(backend='nccl')",
|
| 25 |
+
"$torch.cuda.set_device(@device)",
|
| 26 |
+
"$monai.utils.set_determinism(seed=123)"
|
| 27 |
+
],
|
| 28 |
+
"run": [
|
| 29 |
+
"$@train#trainer.run()"
|
| 30 |
+
],
|
| 31 |
+
"finalize": [
|
| 32 |
+
"$dist.is_initialized() and dist.destroy_process_group()"
|
| 33 |
+
]
|
| 34 |
+
}
|
configs/train.json
ADDED
|
@@ -0,0 +1,271 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import glob",
|
| 4 |
+
"$import os",
|
| 5 |
+
"$import scripts",
|
| 6 |
+
"$import ignite"
|
| 7 |
+
],
|
| 8 |
+
"bundle_root": ".",
|
| 9 |
+
"ckpt_dir": "$@bundle_root + '/models'",
|
| 10 |
+
"output_dir": "$@bundle_root + '/output'",
|
| 11 |
+
"data_list_file_path": "$@bundle_root + '/datasets/C4KC-KiTS_subset.json'",
|
| 12 |
+
"dataset_dir": "$@bundle_root + '/datasets/C4KC-KiTS_subset'",
|
| 13 |
+
"trained_diffusion_path": "$@ckpt_dir + '/input_unet3d_data-all_steps1000size512ddpm_random_current_inputx_v1.pt'",
|
| 14 |
+
"trained_controlnet_path": "$@ckpt_dir + '/controlnet-20datasets-e20wl100fold0bc_noi_dia_fsize_current.pt'",
|
| 15 |
+
"use_tensorboard": true,
|
| 16 |
+
"fold": 0,
|
| 17 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 18 |
+
"epochs": 100,
|
| 19 |
+
"batch_size": 1,
|
| 20 |
+
"val_at_start": false,
|
| 21 |
+
"learning_rate": 0.0001,
|
| 22 |
+
"weighted_loss_label": [
|
| 23 |
+
129
|
| 24 |
+
],
|
| 25 |
+
"weighted_loss": 100,
|
| 26 |
+
"amp": true,
|
| 27 |
+
"train_datalist": "$scripts.utils.maisi_datafold_read(json_list=@data_list_file_path, data_base_dir=@dataset_dir, fold=@fold)[0]",
|
| 28 |
+
"spatial_dims": 3,
|
| 29 |
+
"image_channels": 1,
|
| 30 |
+
"latent_channels": 4,
|
| 31 |
+
"diffusion_unet_def": {
|
| 32 |
+
"_target_": "monai.apps.generation.maisi.networks.diffusion_model_unet_maisi.DiffusionModelUNetMaisi",
|
| 33 |
+
"spatial_dims": "@spatial_dims",
|
| 34 |
+
"in_channels": "@latent_channels",
|
| 35 |
+
"out_channels": "@latent_channels",
|
| 36 |
+
"num_channels": [
|
| 37 |
+
64,
|
| 38 |
+
128,
|
| 39 |
+
256,
|
| 40 |
+
512
|
| 41 |
+
],
|
| 42 |
+
"attention_levels": [
|
| 43 |
+
false,
|
| 44 |
+
false,
|
| 45 |
+
true,
|
| 46 |
+
true
|
| 47 |
+
],
|
| 48 |
+
"num_head_channels": [
|
| 49 |
+
0,
|
| 50 |
+
0,
|
| 51 |
+
32,
|
| 52 |
+
32
|
| 53 |
+
],
|
| 54 |
+
"num_res_blocks": 2,
|
| 55 |
+
"use_flash_attention": true,
|
| 56 |
+
"include_top_region_index_input": true,
|
| 57 |
+
"include_bottom_region_index_input": true,
|
| 58 |
+
"include_spacing_input": true
|
| 59 |
+
},
|
| 60 |
+
"controlnet_def": {
|
| 61 |
+
"_target_": "monai.apps.generation.maisi.networks.controlnet_maisi.ControlNetMaisi",
|
| 62 |
+
"spatial_dims": "@spatial_dims",
|
| 63 |
+
"in_channels": "@latent_channels",
|
| 64 |
+
"num_channels": [
|
| 65 |
+
64,
|
| 66 |
+
128,
|
| 67 |
+
256,
|
| 68 |
+
512
|
| 69 |
+
],
|
| 70 |
+
"attention_levels": [
|
| 71 |
+
false,
|
| 72 |
+
false,
|
| 73 |
+
true,
|
| 74 |
+
true
|
| 75 |
+
],
|
| 76 |
+
"num_head_channels": [
|
| 77 |
+
0,
|
| 78 |
+
0,
|
| 79 |
+
32,
|
| 80 |
+
32
|
| 81 |
+
],
|
| 82 |
+
"num_res_blocks": 2,
|
| 83 |
+
"use_flash_attention": true,
|
| 84 |
+
"conditioning_embedding_in_channels": 8,
|
| 85 |
+
"conditioning_embedding_num_channels": [
|
| 86 |
+
8,
|
| 87 |
+
32,
|
| 88 |
+
64
|
| 89 |
+
]
|
| 90 |
+
},
|
| 91 |
+
"noise_scheduler": {
|
| 92 |
+
"_target_": "monai.networks.schedulers.ddpm.DDPMScheduler",
|
| 93 |
+
"num_train_timesteps": 1000,
|
| 94 |
+
"beta_start": 0.0015,
|
| 95 |
+
"beta_end": 0.0195,
|
| 96 |
+
"schedule": "scaled_linear_beta",
|
| 97 |
+
"clip_sample": false
|
| 98 |
+
},
|
| 99 |
+
"unzip_dataset": "$scripts.utils.unzip_dataset(@dataset_dir)",
|
| 100 |
+
"diffusion_unet": "$@diffusion_unet_def.to(@device)",
|
| 101 |
+
"checkpoint_diffusion_unet": "$torch.load(@trained_diffusion_path, weights_only=False)",
|
| 102 |
+
"load_diffusion": "$@diffusion_unet.load_state_dict(@checkpoint_diffusion_unet['unet_state_dict'])",
|
| 103 |
+
"controlnet": "$@controlnet_def.to(@device)",
|
| 104 |
+
"copy_controlnet_state": "$monai.networks.utils.copy_model_state(@controlnet, @diffusion_unet.state_dict())",
|
| 105 |
+
"checkpoint_controlnet": "$torch.load(@trained_controlnet_path, weights_only=False)",
|
| 106 |
+
"load_controlnet": "[email protected]_state_dict(@checkpoint_controlnet['controlnet_state_dict'], strict=True)",
|
| 107 |
+
"scale_factor": "$@checkpoint_diffusion_unet['scale_factor'].to(@device)",
|
| 108 |
+
"loss": {
|
| 109 |
+
"_target_": "torch.nn.L1Loss",
|
| 110 |
+
"reduction": "none"
|
| 111 |
+
},
|
| 112 |
+
"optimizer": {
|
| 113 |
+
"_target_": "torch.optim.AdamW",
|
| 114 |
+
"params": "[email protected]()",
|
| 115 |
+
"lr": "@learning_rate",
|
| 116 |
+
"weight_decay": 1e-05
|
| 117 |
+
},
|
| 118 |
+
"lr_schedule": {
|
| 119 |
+
"activate": true,
|
| 120 |
+
"lr_scheduler": {
|
| 121 |
+
"_target_": "torch.optim.lr_scheduler.PolynomialLR",
|
| 122 |
+
"optimizer": "@optimizer",
|
| 123 |
+
"total_iters": "$(@epochs * len(@train#dataloader.dataset)) / @batch_size",
|
| 124 |
+
"power": 2.0
|
| 125 |
+
}
|
| 126 |
+
},
|
| 127 |
+
"train": {
|
| 128 |
+
"deterministic_transforms": [
|
| 129 |
+
{
|
| 130 |
+
"_target_": "LoadImaged",
|
| 131 |
+
"keys": [
|
| 132 |
+
"image",
|
| 133 |
+
"label"
|
| 134 |
+
],
|
| 135 |
+
"image_only": true,
|
| 136 |
+
"ensure_channel_first": true
|
| 137 |
+
},
|
| 138 |
+
{
|
| 139 |
+
"_target_": "Orientationd",
|
| 140 |
+
"keys": [
|
| 141 |
+
"label"
|
| 142 |
+
],
|
| 143 |
+
"axcodes": "RAS"
|
| 144 |
+
},
|
| 145 |
+
{
|
| 146 |
+
"_target_": "EnsureTyped",
|
| 147 |
+
"keys": [
|
| 148 |
+
"label"
|
| 149 |
+
],
|
| 150 |
+
"dtype": "$torch.uint8",
|
| 151 |
+
"track_meta": true
|
| 152 |
+
},
|
| 153 |
+
{
|
| 154 |
+
"_target_": "Lambdad",
|
| 155 |
+
"keys": "top_region_index",
|
| 156 |
+
"func": "$lambda x: torch.FloatTensor(x)"
|
| 157 |
+
},
|
| 158 |
+
{
|
| 159 |
+
"_target_": "Lambdad",
|
| 160 |
+
"keys": "bottom_region_index",
|
| 161 |
+
"func": "$lambda x: torch.FloatTensor(x)"
|
| 162 |
+
},
|
| 163 |
+
{
|
| 164 |
+
"_target_": "Lambdad",
|
| 165 |
+
"keys": "spacing",
|
| 166 |
+
"func": "$lambda x: torch.FloatTensor(x)"
|
| 167 |
+
},
|
| 168 |
+
{
|
| 169 |
+
"_target_": "Lambdad",
|
| 170 |
+
"keys": "top_region_index",
|
| 171 |
+
"func": "$lambda x: x * 1e2"
|
| 172 |
+
},
|
| 173 |
+
{
|
| 174 |
+
"_target_": "Lambdad",
|
| 175 |
+
"keys": "bottom_region_index",
|
| 176 |
+
"func": "$lambda x: x * 1e2"
|
| 177 |
+
},
|
| 178 |
+
{
|
| 179 |
+
"_target_": "Lambdad",
|
| 180 |
+
"keys": "spacing",
|
| 181 |
+
"func": "$lambda x: x * 1e2"
|
| 182 |
+
}
|
| 183 |
+
],
|
| 184 |
+
"inferer": {
|
| 185 |
+
"_target_": "SimpleInferer"
|
| 186 |
+
},
|
| 187 |
+
"preprocessing": {
|
| 188 |
+
"_target_": "Compose",
|
| 189 |
+
"transforms": "$@train#deterministic_transforms"
|
| 190 |
+
},
|
| 191 |
+
"dataset": {
|
| 192 |
+
"_target_": "Dataset",
|
| 193 |
+
"data": "@train_datalist",
|
| 194 |
+
"transform": "@train#preprocessing"
|
| 195 |
+
},
|
| 196 |
+
"dataloader": {
|
| 197 |
+
"_target_": "DataLoader",
|
| 198 |
+
"dataset": "@train#dataset",
|
| 199 |
+
"batch_size": "@batch_size",
|
| 200 |
+
"shuffle": true,
|
| 201 |
+
"num_workers": 4,
|
| 202 |
+
"pin_memory": true,
|
| 203 |
+
"persistent_workers": true
|
| 204 |
+
},
|
| 205 |
+
"handlers": [
|
| 206 |
+
{
|
| 207 |
+
"_target_": "LrScheduleHandler",
|
| 208 |
+
"_disabled_": "$not @lr_schedule#activate",
|
| 209 |
+
"lr_scheduler": "@lr_schedule#lr_scheduler",
|
| 210 |
+
"epoch_level": false,
|
| 211 |
+
"print_lr": true
|
| 212 |
+
},
|
| 213 |
+
{
|
| 214 |
+
"_target_": "CheckpointSaver",
|
| 215 |
+
"save_dir": "@ckpt_dir",
|
| 216 |
+
"save_dict": {
|
| 217 |
+
"controlnet_state_dict": "@controlnet",
|
| 218 |
+
"optimizer": "@optimizer"
|
| 219 |
+
},
|
| 220 |
+
"save_interval": 1,
|
| 221 |
+
"n_saved": 5
|
| 222 |
+
},
|
| 223 |
+
{
|
| 224 |
+
"_target_": "TensorBoardStatsHandler",
|
| 225 |
+
"_disabled_": "$not @use_tensorboard",
|
| 226 |
+
"log_dir": "@output_dir",
|
| 227 |
+
"tag_name": "train_loss",
|
| 228 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 229 |
+
},
|
| 230 |
+
{
|
| 231 |
+
"_target_": "StatsHandler",
|
| 232 |
+
"tag_name": "train_loss",
|
| 233 |
+
"name": "StatsHandler",
|
| 234 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 235 |
+
}
|
| 236 |
+
],
|
| 237 |
+
"trainer": {
|
| 238 |
+
"_target_": "scripts.trainer.MAISIControlNetTrainer",
|
| 239 |
+
"_requires_": [
|
| 240 |
+
"@load_diffusion",
|
| 241 |
+
"@copy_controlnet_state",
|
| 242 |
+
"@load_controlnet",
|
| 243 |
+
"@unzip_dataset"
|
| 244 |
+
],
|
| 245 |
+
"max_epochs": "@epochs",
|
| 246 |
+
"device": "@device",
|
| 247 |
+
"train_data_loader": "@train#dataloader",
|
| 248 |
+
"diffusion_unet": "@diffusion_unet",
|
| 249 |
+
"controlnet": "@controlnet",
|
| 250 |
+
"noise_scheduler": "@noise_scheduler",
|
| 251 |
+
"loss_function": "@loss",
|
| 252 |
+
"optimizer": "@optimizer",
|
| 253 |
+
"inferer": "@train#inferer",
|
| 254 |
+
"key_train_metric": null,
|
| 255 |
+
"train_handlers": "@train#handlers",
|
| 256 |
+
"amp": "@amp",
|
| 257 |
+
"hyper_kwargs": {
|
| 258 |
+
"weighted_loss": "@weighted_loss",
|
| 259 |
+
"weighted_loss_label": "@weighted_loss_label",
|
| 260 |
+
"scale_factor": "@scale_factor"
|
| 261 |
+
}
|
| 262 |
+
}
|
| 263 |
+
},
|
| 264 |
+
"initialize": [
|
| 265 |
+
"$monai.utils.set_determinism(seed=0)"
|
| 266 |
+
],
|
| 267 |
+
"run": [
|
| 268 |
+
"$@train#trainer.add_event_handler(ignite.engine.Events.ITERATION_COMPLETED, ignite.handlers.TerminateOnNan())",
|
| 269 |
+
"$@train#trainer.run()"
|
| 270 |
+
]
|
| 271 |
+
}
|
datasets/C4KC-KiTS_subset.json
ADDED
|
@@ -0,0 +1,814 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"training": [
|
| 3 |
+
{
|
| 4 |
+
"image": "KiTS-00186/2_arterial_emb_zs99.nii.gz",
|
| 5 |
+
"label": "KiTS-00186/mask_combined_label_zs99_wbdm.nii.gz",
|
| 6 |
+
"fold": 0,
|
| 7 |
+
"dim": [
|
| 8 |
+
512,
|
| 9 |
+
512,
|
| 10 |
+
512
|
| 11 |
+
],
|
| 12 |
+
"spacing": [
|
| 13 |
+
1.0,
|
| 14 |
+
1.0,
|
| 15 |
+
1.0
|
| 16 |
+
],
|
| 17 |
+
"top_region_index": [
|
| 18 |
+
0,
|
| 19 |
+
1,
|
| 20 |
+
0,
|
| 21 |
+
0
|
| 22 |
+
],
|
| 23 |
+
"bottom_region_index": [
|
| 24 |
+
0,
|
| 25 |
+
0,
|
| 26 |
+
0,
|
| 27 |
+
1
|
| 28 |
+
]
|
| 29 |
+
},
|
| 30 |
+
{
|
| 31 |
+
"image": "KiTS-00066/3_arterial_emb_zs11.nii.gz",
|
| 32 |
+
"label": "KiTS-00066/mask_combined_label_zs11_wbdm.nii.gz",
|
| 33 |
+
"fold": 0,
|
| 34 |
+
"dim": [
|
| 35 |
+
512,
|
| 36 |
+
512,
|
| 37 |
+
512
|
| 38 |
+
],
|
| 39 |
+
"spacing": [
|
| 40 |
+
1.0,
|
| 41 |
+
1.0,
|
| 42 |
+
1.0
|
| 43 |
+
],
|
| 44 |
+
"top_region_index": [
|
| 45 |
+
0,
|
| 46 |
+
1,
|
| 47 |
+
0,
|
| 48 |
+
0
|
| 49 |
+
],
|
| 50 |
+
"bottom_region_index": [
|
| 51 |
+
0,
|
| 52 |
+
0,
|
| 53 |
+
0,
|
| 54 |
+
1
|
| 55 |
+
]
|
| 56 |
+
},
|
| 57 |
+
{
|
| 58 |
+
"image": "KiTS-00012/2_arterial_emb_zs0.nii.gz",
|
| 59 |
+
"label": "KiTS-00012/mask_combined_label_zs0_wbdm.nii.gz",
|
| 60 |
+
"fold": 0,
|
| 61 |
+
"dim": [
|
| 62 |
+
512,
|
| 63 |
+
512,
|
| 64 |
+
512
|
| 65 |
+
],
|
| 66 |
+
"spacing": [
|
| 67 |
+
1.0,
|
| 68 |
+
1.0,
|
| 69 |
+
1.0
|
| 70 |
+
],
|
| 71 |
+
"top_region_index": [
|
| 72 |
+
0,
|
| 73 |
+
1,
|
| 74 |
+
0,
|
| 75 |
+
0
|
| 76 |
+
],
|
| 77 |
+
"bottom_region_index": [
|
| 78 |
+
0,
|
| 79 |
+
0,
|
| 80 |
+
0,
|
| 81 |
+
1
|
| 82 |
+
]
|
| 83 |
+
},
|
| 84 |
+
{
|
| 85 |
+
"image": "KiTS-00055/2_arterial_emb_zs0.nii.gz",
|
| 86 |
+
"label": "KiTS-00055/mask_combined_label_zs0_wbdm.nii.gz",
|
| 87 |
+
"fold": 0,
|
| 88 |
+
"dim": [
|
| 89 |
+
512,
|
| 90 |
+
512,
|
| 91 |
+
512
|
| 92 |
+
],
|
| 93 |
+
"spacing": [
|
| 94 |
+
1.0,
|
| 95 |
+
1.0,
|
| 96 |
+
1.0
|
| 97 |
+
],
|
| 98 |
+
"top_region_index": [
|
| 99 |
+
0,
|
| 100 |
+
1,
|
| 101 |
+
0,
|
| 102 |
+
0
|
| 103 |
+
],
|
| 104 |
+
"bottom_region_index": [
|
| 105 |
+
0,
|
| 106 |
+
0,
|
| 107 |
+
0,
|
| 108 |
+
1
|
| 109 |
+
]
|
| 110 |
+
},
|
| 111 |
+
{
|
| 112 |
+
"image": "KiTS-00193/100_arterial_emb_zs0.nii.gz",
|
| 113 |
+
"label": "KiTS-00193/mask_combined_label_zs0_wbdm.nii.gz",
|
| 114 |
+
"fold": 0,
|
| 115 |
+
"dim": [
|
| 116 |
+
512,
|
| 117 |
+
512,
|
| 118 |
+
512
|
| 119 |
+
],
|
| 120 |
+
"spacing": [
|
| 121 |
+
1.0,
|
| 122 |
+
1.0,
|
| 123 |
+
1.0
|
| 124 |
+
],
|
| 125 |
+
"top_region_index": [
|
| 126 |
+
0,
|
| 127 |
+
1,
|
| 128 |
+
0,
|
| 129 |
+
0
|
| 130 |
+
],
|
| 131 |
+
"bottom_region_index": [
|
| 132 |
+
0,
|
| 133 |
+
0,
|
| 134 |
+
1,
|
| 135 |
+
0
|
| 136 |
+
]
|
| 137 |
+
},
|
| 138 |
+
{
|
| 139 |
+
"image": "KiTS-00142/7_arterial_emb_zs0.nii.gz",
|
| 140 |
+
"label": "KiTS-00142/mask_combined_label_zs0_wbdm.nii.gz",
|
| 141 |
+
"fold": 0,
|
| 142 |
+
"dim": [
|
| 143 |
+
512,
|
| 144 |
+
512,
|
| 145 |
+
512
|
| 146 |
+
],
|
| 147 |
+
"spacing": [
|
| 148 |
+
1.0,
|
| 149 |
+
1.0,
|
| 150 |
+
1.0
|
| 151 |
+
],
|
| 152 |
+
"top_region_index": [
|
| 153 |
+
0,
|
| 154 |
+
1,
|
| 155 |
+
0,
|
| 156 |
+
0
|
| 157 |
+
],
|
| 158 |
+
"bottom_region_index": [
|
| 159 |
+
0,
|
| 160 |
+
0,
|
| 161 |
+
1,
|
| 162 |
+
0
|
| 163 |
+
]
|
| 164 |
+
},
|
| 165 |
+
{
|
| 166 |
+
"image": "KiTS-00069/3_arterial_emb_zs0.nii.gz",
|
| 167 |
+
"label": "KiTS-00069/mask_combined_label_zs0_wbdm.nii.gz",
|
| 168 |
+
"fold": 0,
|
| 169 |
+
"dim": [
|
| 170 |
+
512,
|
| 171 |
+
512,
|
| 172 |
+
512
|
| 173 |
+
],
|
| 174 |
+
"spacing": [
|
| 175 |
+
1.0,
|
| 176 |
+
1.0,
|
| 177 |
+
1.0
|
| 178 |
+
],
|
| 179 |
+
"top_region_index": [
|
| 180 |
+
0,
|
| 181 |
+
1,
|
| 182 |
+
0,
|
| 183 |
+
0
|
| 184 |
+
],
|
| 185 |
+
"bottom_region_index": [
|
| 186 |
+
0,
|
| 187 |
+
0,
|
| 188 |
+
0,
|
| 189 |
+
1
|
| 190 |
+
]
|
| 191 |
+
},
|
| 192 |
+
{
|
| 193 |
+
"image": "KiTS-00124/2_arterial_emb_zs0.nii.gz",
|
| 194 |
+
"label": "KiTS-00124/mask_combined_label_zs0_wbdm.nii.gz",
|
| 195 |
+
"fold": 0,
|
| 196 |
+
"dim": [
|
| 197 |
+
512,
|
| 198 |
+
512,
|
| 199 |
+
512
|
| 200 |
+
],
|
| 201 |
+
"spacing": [
|
| 202 |
+
1.0,
|
| 203 |
+
1.0,
|
| 204 |
+
1.0
|
| 205 |
+
],
|
| 206 |
+
"top_region_index": [
|
| 207 |
+
0,
|
| 208 |
+
1,
|
| 209 |
+
0,
|
| 210 |
+
0
|
| 211 |
+
],
|
| 212 |
+
"bottom_region_index": [
|
| 213 |
+
0,
|
| 214 |
+
0,
|
| 215 |
+
0,
|
| 216 |
+
1
|
| 217 |
+
]
|
| 218 |
+
},
|
| 219 |
+
{
|
| 220 |
+
"image": "KiTS-00208/2_arterial_emb_zs0.nii.gz",
|
| 221 |
+
"label": "KiTS-00208/mask_combined_label_zs0_wbdm.nii.gz",
|
| 222 |
+
"fold": 0,
|
| 223 |
+
"dim": [
|
| 224 |
+
512,
|
| 225 |
+
512,
|
| 226 |
+
512
|
| 227 |
+
],
|
| 228 |
+
"spacing": [
|
| 229 |
+
1.0,
|
| 230 |
+
1.0,
|
| 231 |
+
1.0
|
| 232 |
+
],
|
| 233 |
+
"top_region_index": [
|
| 234 |
+
0,
|
| 235 |
+
1,
|
| 236 |
+
0,
|
| 237 |
+
0
|
| 238 |
+
],
|
| 239 |
+
"bottom_region_index": [
|
| 240 |
+
0,
|
| 241 |
+
0,
|
| 242 |
+
0,
|
| 243 |
+
1
|
| 244 |
+
]
|
| 245 |
+
},
|
| 246 |
+
{
|
| 247 |
+
"image": "KiTS-00116/9_arterial_emb_zs0.nii.gz",
|
| 248 |
+
"label": "KiTS-00116/mask_combined_label_zs0_wbdm.nii.gz",
|
| 249 |
+
"fold": 0,
|
| 250 |
+
"dim": [
|
| 251 |
+
512,
|
| 252 |
+
512,
|
| 253 |
+
512
|
| 254 |
+
],
|
| 255 |
+
"spacing": [
|
| 256 |
+
1.0,
|
| 257 |
+
1.0,
|
| 258 |
+
1.0
|
| 259 |
+
],
|
| 260 |
+
"top_region_index": [
|
| 261 |
+
0,
|
| 262 |
+
1,
|
| 263 |
+
0,
|
| 264 |
+
0
|
| 265 |
+
],
|
| 266 |
+
"bottom_region_index": [
|
| 267 |
+
0,
|
| 268 |
+
0,
|
| 269 |
+
1,
|
| 270 |
+
0
|
| 271 |
+
]
|
| 272 |
+
},
|
| 273 |
+
{
|
| 274 |
+
"image": "KiTS-00061/4_arterial_emb_zs0.nii.gz",
|
| 275 |
+
"label": "KiTS-00061/mask_combined_label_zs0_wbdm.nii.gz",
|
| 276 |
+
"fold": 1,
|
| 277 |
+
"dim": [
|
| 278 |
+
512,
|
| 279 |
+
512,
|
| 280 |
+
512
|
| 281 |
+
],
|
| 282 |
+
"spacing": [
|
| 283 |
+
1.0,
|
| 284 |
+
1.0,
|
| 285 |
+
1.0
|
| 286 |
+
],
|
| 287 |
+
"top_region_index": [
|
| 288 |
+
0,
|
| 289 |
+
0,
|
| 290 |
+
1,
|
| 291 |
+
0
|
| 292 |
+
],
|
| 293 |
+
"bottom_region_index": [
|
| 294 |
+
0,
|
| 295 |
+
0,
|
| 296 |
+
1,
|
| 297 |
+
0
|
| 298 |
+
]
|
| 299 |
+
},
|
| 300 |
+
{
|
| 301 |
+
"image": "KiTS-00040/3_arterial_emb_zs0.nii.gz",
|
| 302 |
+
"label": "KiTS-00040/mask_combined_label_zs0_wbdm.nii.gz",
|
| 303 |
+
"fold": 1,
|
| 304 |
+
"dim": [
|
| 305 |
+
512,
|
| 306 |
+
512,
|
| 307 |
+
512
|
| 308 |
+
],
|
| 309 |
+
"spacing": [
|
| 310 |
+
1.0,
|
| 311 |
+
1.0,
|
| 312 |
+
1.0
|
| 313 |
+
],
|
| 314 |
+
"top_region_index": [
|
| 315 |
+
0,
|
| 316 |
+
1,
|
| 317 |
+
0,
|
| 318 |
+
0
|
| 319 |
+
],
|
| 320 |
+
"bottom_region_index": [
|
| 321 |
+
0,
|
| 322 |
+
0,
|
| 323 |
+
0,
|
| 324 |
+
1
|
| 325 |
+
]
|
| 326 |
+
},
|
| 327 |
+
{
|
| 328 |
+
"image": "KiTS-00068/7_arterial_emb_zs0.nii.gz",
|
| 329 |
+
"label": "KiTS-00068/mask_combined_label_zs0_wbdm.nii.gz",
|
| 330 |
+
"fold": 1,
|
| 331 |
+
"dim": [
|
| 332 |
+
512,
|
| 333 |
+
512,
|
| 334 |
+
512
|
| 335 |
+
],
|
| 336 |
+
"spacing": [
|
| 337 |
+
1.0,
|
| 338 |
+
1.0,
|
| 339 |
+
1.0
|
| 340 |
+
],
|
| 341 |
+
"top_region_index": [
|
| 342 |
+
0,
|
| 343 |
+
1,
|
| 344 |
+
0,
|
| 345 |
+
0
|
| 346 |
+
],
|
| 347 |
+
"bottom_region_index": [
|
| 348 |
+
0,
|
| 349 |
+
0,
|
| 350 |
+
1,
|
| 351 |
+
0
|
| 352 |
+
]
|
| 353 |
+
},
|
| 354 |
+
{
|
| 355 |
+
"image": "KiTS-00036/2_arterial_emb_zs0.nii.gz",
|
| 356 |
+
"label": "KiTS-00036/mask_combined_label_zs0_wbdm.nii.gz",
|
| 357 |
+
"fold": 1,
|
| 358 |
+
"dim": [
|
| 359 |
+
512,
|
| 360 |
+
512,
|
| 361 |
+
512
|
| 362 |
+
],
|
| 363 |
+
"spacing": [
|
| 364 |
+
1.0,
|
| 365 |
+
1.0,
|
| 366 |
+
1.0
|
| 367 |
+
],
|
| 368 |
+
"top_region_index": [
|
| 369 |
+
0,
|
| 370 |
+
1,
|
| 371 |
+
0,
|
| 372 |
+
0
|
| 373 |
+
],
|
| 374 |
+
"bottom_region_index": [
|
| 375 |
+
0,
|
| 376 |
+
0,
|
| 377 |
+
0,
|
| 378 |
+
1
|
| 379 |
+
]
|
| 380 |
+
},
|
| 381 |
+
{
|
| 382 |
+
"image": "KiTS-00153/8_arterial_emb_zs0.nii.gz",
|
| 383 |
+
"label": "KiTS-00153/mask_combined_label_zs0_wbdm.nii.gz",
|
| 384 |
+
"fold": 1,
|
| 385 |
+
"dim": [
|
| 386 |
+
512,
|
| 387 |
+
512,
|
| 388 |
+
512
|
| 389 |
+
],
|
| 390 |
+
"spacing": [
|
| 391 |
+
1.0,
|
| 392 |
+
1.0,
|
| 393 |
+
1.0
|
| 394 |
+
],
|
| 395 |
+
"top_region_index": [
|
| 396 |
+
0,
|
| 397 |
+
1,
|
| 398 |
+
0,
|
| 399 |
+
0
|
| 400 |
+
],
|
| 401 |
+
"bottom_region_index": [
|
| 402 |
+
0,
|
| 403 |
+
0,
|
| 404 |
+
1,
|
| 405 |
+
0
|
| 406 |
+
]
|
| 407 |
+
},
|
| 408 |
+
{
|
| 409 |
+
"image": "KiTS-00189/2_arterial_emb_zs107.nii.gz",
|
| 410 |
+
"label": "KiTS-00189/mask_combined_label_zs107_wbdm.nii.gz",
|
| 411 |
+
"fold": 1,
|
| 412 |
+
"dim": [
|
| 413 |
+
512,
|
| 414 |
+
512,
|
| 415 |
+
512
|
| 416 |
+
],
|
| 417 |
+
"spacing": [
|
| 418 |
+
1.0,
|
| 419 |
+
1.0,
|
| 420 |
+
1.0
|
| 421 |
+
],
|
| 422 |
+
"top_region_index": [
|
| 423 |
+
0,
|
| 424 |
+
1,
|
| 425 |
+
0,
|
| 426 |
+
0
|
| 427 |
+
],
|
| 428 |
+
"bottom_region_index": [
|
| 429 |
+
0,
|
| 430 |
+
0,
|
| 431 |
+
0,
|
| 432 |
+
1
|
| 433 |
+
]
|
| 434 |
+
},
|
| 435 |
+
{
|
| 436 |
+
"image": "KiTS-00091/7_arterial_emb_zs0.nii.gz",
|
| 437 |
+
"label": "KiTS-00091/mask_combined_label_zs0_wbdm.nii.gz",
|
| 438 |
+
"fold": 1,
|
| 439 |
+
"dim": [
|
| 440 |
+
512,
|
| 441 |
+
512,
|
| 442 |
+
512
|
| 443 |
+
],
|
| 444 |
+
"spacing": [
|
| 445 |
+
1.0,
|
| 446 |
+
1.0,
|
| 447 |
+
1.0
|
| 448 |
+
],
|
| 449 |
+
"top_region_index": [
|
| 450 |
+
0,
|
| 451 |
+
1,
|
| 452 |
+
0,
|
| 453 |
+
0
|
| 454 |
+
],
|
| 455 |
+
"bottom_region_index": [
|
| 456 |
+
0,
|
| 457 |
+
0,
|
| 458 |
+
1,
|
| 459 |
+
0
|
| 460 |
+
]
|
| 461 |
+
},
|
| 462 |
+
{
|
| 463 |
+
"image": "KiTS-00110/3_arterial_emb_zs0.nii.gz",
|
| 464 |
+
"label": "KiTS-00110/mask_combined_label_zs0_wbdm.nii.gz",
|
| 465 |
+
"fold": 1,
|
| 466 |
+
"dim": [
|
| 467 |
+
512,
|
| 468 |
+
512,
|
| 469 |
+
512
|
| 470 |
+
],
|
| 471 |
+
"spacing": [
|
| 472 |
+
1.0,
|
| 473 |
+
1.0,
|
| 474 |
+
1.0
|
| 475 |
+
],
|
| 476 |
+
"top_region_index": [
|
| 477 |
+
0,
|
| 478 |
+
1,
|
| 479 |
+
0,
|
| 480 |
+
0
|
| 481 |
+
],
|
| 482 |
+
"bottom_region_index": [
|
| 483 |
+
0,
|
| 484 |
+
0,
|
| 485 |
+
1,
|
| 486 |
+
0
|
| 487 |
+
]
|
| 488 |
+
},
|
| 489 |
+
{
|
| 490 |
+
"image": "KiTS-00046/2_arterial_emb_zs0.nii.gz",
|
| 491 |
+
"label": "KiTS-00046/mask_combined_label_zs0_wbdm.nii.gz",
|
| 492 |
+
"fold": 1,
|
| 493 |
+
"dim": [
|
| 494 |
+
512,
|
| 495 |
+
512,
|
| 496 |
+
512
|
| 497 |
+
],
|
| 498 |
+
"spacing": [
|
| 499 |
+
1.0,
|
| 500 |
+
1.0,
|
| 501 |
+
1.0
|
| 502 |
+
],
|
| 503 |
+
"top_region_index": [
|
| 504 |
+
0,
|
| 505 |
+
1,
|
| 506 |
+
0,
|
| 507 |
+
0
|
| 508 |
+
],
|
| 509 |
+
"bottom_region_index": [
|
| 510 |
+
0,
|
| 511 |
+
0,
|
| 512 |
+
0,
|
| 513 |
+
1
|
| 514 |
+
]
|
| 515 |
+
},
|
| 516 |
+
{
|
| 517 |
+
"image": "KiTS-00178/3_arterial_emb_zs0.nii.gz",
|
| 518 |
+
"label": "KiTS-00178/mask_combined_label_zs0_wbdm.nii.gz",
|
| 519 |
+
"fold": 1,
|
| 520 |
+
"dim": [
|
| 521 |
+
512,
|
| 522 |
+
512,
|
| 523 |
+
512
|
| 524 |
+
],
|
| 525 |
+
"spacing": [
|
| 526 |
+
1.0,
|
| 527 |
+
1.0,
|
| 528 |
+
1.0
|
| 529 |
+
],
|
| 530 |
+
"top_region_index": [
|
| 531 |
+
0,
|
| 532 |
+
1,
|
| 533 |
+
0,
|
| 534 |
+
0
|
| 535 |
+
],
|
| 536 |
+
"bottom_region_index": [
|
| 537 |
+
0,
|
| 538 |
+
0,
|
| 539 |
+
0,
|
| 540 |
+
1
|
| 541 |
+
]
|
| 542 |
+
},
|
| 543 |
+
{
|
| 544 |
+
"image": "KiTS-00075/2_arterial_emb_zs0.nii.gz",
|
| 545 |
+
"label": "KiTS-00075/mask_combined_label_zs0_wbdm.nii.gz",
|
| 546 |
+
"fold": 1,
|
| 547 |
+
"dim": [
|
| 548 |
+
512,
|
| 549 |
+
512,
|
| 550 |
+
512
|
| 551 |
+
],
|
| 552 |
+
"spacing": [
|
| 553 |
+
1.0,
|
| 554 |
+
1.0,
|
| 555 |
+
1.0
|
| 556 |
+
],
|
| 557 |
+
"top_region_index": [
|
| 558 |
+
0,
|
| 559 |
+
1,
|
| 560 |
+
0,
|
| 561 |
+
0
|
| 562 |
+
],
|
| 563 |
+
"bottom_region_index": [
|
| 564 |
+
0,
|
| 565 |
+
0,
|
| 566 |
+
0,
|
| 567 |
+
1
|
| 568 |
+
]
|
| 569 |
+
},
|
| 570 |
+
{
|
| 571 |
+
"image": "KiTS-00037/6_arterial_emb_zs0.nii.gz",
|
| 572 |
+
"label": "KiTS-00037/mask_combined_label_zs0_wbdm.nii.gz",
|
| 573 |
+
"fold": 1,
|
| 574 |
+
"dim": [
|
| 575 |
+
512,
|
| 576 |
+
512,
|
| 577 |
+
512
|
| 578 |
+
],
|
| 579 |
+
"spacing": [
|
| 580 |
+
1.0,
|
| 581 |
+
1.0,
|
| 582 |
+
1.0
|
| 583 |
+
],
|
| 584 |
+
"top_region_index": [
|
| 585 |
+
0,
|
| 586 |
+
1,
|
| 587 |
+
0,
|
| 588 |
+
0
|
| 589 |
+
],
|
| 590 |
+
"bottom_region_index": [
|
| 591 |
+
0,
|
| 592 |
+
0,
|
| 593 |
+
0,
|
| 594 |
+
1
|
| 595 |
+
]
|
| 596 |
+
},
|
| 597 |
+
{
|
| 598 |
+
"image": "KiTS-00130/9_arterial_emb_zs0.nii.gz",
|
| 599 |
+
"label": "KiTS-00130/mask_combined_label_zs0_wbdm.nii.gz",
|
| 600 |
+
"fold": 1,
|
| 601 |
+
"dim": [
|
| 602 |
+
512,
|
| 603 |
+
512,
|
| 604 |
+
512
|
| 605 |
+
],
|
| 606 |
+
"spacing": [
|
| 607 |
+
1.0,
|
| 608 |
+
1.0,
|
| 609 |
+
1.0
|
| 610 |
+
],
|
| 611 |
+
"top_region_index": [
|
| 612 |
+
0,
|
| 613 |
+
1,
|
| 614 |
+
0,
|
| 615 |
+
0
|
| 616 |
+
],
|
| 617 |
+
"bottom_region_index": [
|
| 618 |
+
0,
|
| 619 |
+
0,
|
| 620 |
+
1,
|
| 621 |
+
0
|
| 622 |
+
]
|
| 623 |
+
},
|
| 624 |
+
{
|
| 625 |
+
"image": "KiTS-00063/6_arterial_emb_zs0.nii.gz",
|
| 626 |
+
"label": "KiTS-00063/mask_combined_label_zs0_wbdm.nii.gz",
|
| 627 |
+
"fold": 1,
|
| 628 |
+
"dim": [
|
| 629 |
+
512,
|
| 630 |
+
512,
|
| 631 |
+
512
|
| 632 |
+
],
|
| 633 |
+
"spacing": [
|
| 634 |
+
1.0,
|
| 635 |
+
1.0,
|
| 636 |
+
1.0
|
| 637 |
+
],
|
| 638 |
+
"top_region_index": [
|
| 639 |
+
0,
|
| 640 |
+
1,
|
| 641 |
+
0,
|
| 642 |
+
0
|
| 643 |
+
],
|
| 644 |
+
"bottom_region_index": [
|
| 645 |
+
0,
|
| 646 |
+
0,
|
| 647 |
+
1,
|
| 648 |
+
0
|
| 649 |
+
]
|
| 650 |
+
},
|
| 651 |
+
{
|
| 652 |
+
"image": "KiTS-00205/4_arterial_emb_zs0.nii.gz",
|
| 653 |
+
"label": "KiTS-00205/mask_combined_label_zs0_wbdm.nii.gz",
|
| 654 |
+
"fold": 1,
|
| 655 |
+
"dim": [
|
| 656 |
+
512,
|
| 657 |
+
512,
|
| 658 |
+
512
|
| 659 |
+
],
|
| 660 |
+
"spacing": [
|
| 661 |
+
1.0,
|
| 662 |
+
1.0,
|
| 663 |
+
1.0
|
| 664 |
+
],
|
| 665 |
+
"top_region_index": [
|
| 666 |
+
0,
|
| 667 |
+
1,
|
| 668 |
+
0,
|
| 669 |
+
0
|
| 670 |
+
],
|
| 671 |
+
"bottom_region_index": [
|
| 672 |
+
0,
|
| 673 |
+
0,
|
| 674 |
+
0,
|
| 675 |
+
1
|
| 676 |
+
]
|
| 677 |
+
},
|
| 678 |
+
{
|
| 679 |
+
"image": "KiTS-00167/2_arterial_emb_zs0.nii.gz",
|
| 680 |
+
"label": "KiTS-00167/mask_combined_label_zs0_wbdm.nii.gz",
|
| 681 |
+
"fold": 1,
|
| 682 |
+
"dim": [
|
| 683 |
+
512,
|
| 684 |
+
512,
|
| 685 |
+
512
|
| 686 |
+
],
|
| 687 |
+
"spacing": [
|
| 688 |
+
1.0,
|
| 689 |
+
1.0,
|
| 690 |
+
1.0
|
| 691 |
+
],
|
| 692 |
+
"top_region_index": [
|
| 693 |
+
0,
|
| 694 |
+
1,
|
| 695 |
+
0,
|
| 696 |
+
0
|
| 697 |
+
],
|
| 698 |
+
"bottom_region_index": [
|
| 699 |
+
0,
|
| 700 |
+
0,
|
| 701 |
+
0,
|
| 702 |
+
1
|
| 703 |
+
]
|
| 704 |
+
},
|
| 705 |
+
{
|
| 706 |
+
"image": "KiTS-00059/8_arterial_emb_zs0.nii.gz",
|
| 707 |
+
"label": "KiTS-00059/mask_combined_label_zs0_wbdm.nii.gz",
|
| 708 |
+
"fold": 1,
|
| 709 |
+
"dim": [
|
| 710 |
+
512,
|
| 711 |
+
512,
|
| 712 |
+
512
|
| 713 |
+
],
|
| 714 |
+
"spacing": [
|
| 715 |
+
1.0,
|
| 716 |
+
1.0,
|
| 717 |
+
1.0
|
| 718 |
+
],
|
| 719 |
+
"top_region_index": [
|
| 720 |
+
0,
|
| 721 |
+
1,
|
| 722 |
+
0,
|
| 723 |
+
0
|
| 724 |
+
],
|
| 725 |
+
"bottom_region_index": [
|
| 726 |
+
0,
|
| 727 |
+
0,
|
| 728 |
+
1,
|
| 729 |
+
0
|
| 730 |
+
]
|
| 731 |
+
},
|
| 732 |
+
{
|
| 733 |
+
"image": "KiTS-00172/3_arterial_emb_zs0.nii.gz",
|
| 734 |
+
"label": "KiTS-00172/mask_combined_label_zs0_wbdm.nii.gz",
|
| 735 |
+
"fold": 1,
|
| 736 |
+
"dim": [
|
| 737 |
+
512,
|
| 738 |
+
512,
|
| 739 |
+
512
|
| 740 |
+
],
|
| 741 |
+
"spacing": [
|
| 742 |
+
1.0,
|
| 743 |
+
1.0,
|
| 744 |
+
1.0
|
| 745 |
+
],
|
| 746 |
+
"top_region_index": [
|
| 747 |
+
0,
|
| 748 |
+
1,
|
| 749 |
+
0,
|
| 750 |
+
0
|
| 751 |
+
],
|
| 752 |
+
"bottom_region_index": [
|
| 753 |
+
0,
|
| 754 |
+
0,
|
| 755 |
+
0,
|
| 756 |
+
1
|
| 757 |
+
]
|
| 758 |
+
},
|
| 759 |
+
{
|
| 760 |
+
"image": "KiTS-00093/7_arterial_emb_zs0.nii.gz",
|
| 761 |
+
"label": "KiTS-00093/mask_combined_label_zs0_wbdm.nii.gz",
|
| 762 |
+
"fold": 1,
|
| 763 |
+
"dim": [
|
| 764 |
+
512,
|
| 765 |
+
512,
|
| 766 |
+
512
|
| 767 |
+
],
|
| 768 |
+
"spacing": [
|
| 769 |
+
1.0,
|
| 770 |
+
1.0,
|
| 771 |
+
1.0
|
| 772 |
+
],
|
| 773 |
+
"top_region_index": [
|
| 774 |
+
0,
|
| 775 |
+
1,
|
| 776 |
+
0,
|
| 777 |
+
0
|
| 778 |
+
],
|
| 779 |
+
"bottom_region_index": [
|
| 780 |
+
0,
|
| 781 |
+
0,
|
| 782 |
+
0,
|
| 783 |
+
1
|
| 784 |
+
]
|
| 785 |
+
},
|
| 786 |
+
{
|
| 787 |
+
"image": "KiTS-00197/2_arterial_emb_zs0.nii.gz",
|
| 788 |
+
"label": "KiTS-00197/mask_combined_label_zs0_wbdm.nii.gz",
|
| 789 |
+
"fold": 1,
|
| 790 |
+
"dim": [
|
| 791 |
+
512,
|
| 792 |
+
512,
|
| 793 |
+
512
|
| 794 |
+
],
|
| 795 |
+
"spacing": [
|
| 796 |
+
1.0,
|
| 797 |
+
1.0,
|
| 798 |
+
1.0
|
| 799 |
+
],
|
| 800 |
+
"top_region_index": [
|
| 801 |
+
0,
|
| 802 |
+
1,
|
| 803 |
+
0,
|
| 804 |
+
0
|
| 805 |
+
],
|
| 806 |
+
"bottom_region_index": [
|
| 807 |
+
0,
|
| 808 |
+
0,
|
| 809 |
+
0,
|
| 810 |
+
1
|
| 811 |
+
]
|
| 812 |
+
}
|
| 813 |
+
]
|
| 814 |
+
}
|
datasets/C4KC-KiTS_subset.zip
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:8bb65d194571c8db8e26ac911b04898cd54376f3c76a0303be70c4f883102088
|
| 3 |
+
size 3155140827
|
datasets/IntegrationTest-AbdomenCT.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:50b4a84769a31aeadd5f6d1a2bece82ba138bfb0eabe94ab13894fc8eb5dac90
|
| 3 |
+
size 7493659
|
datasets/all_masks_flexible_size_and_spacing_3000.zip
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6d89ebad0762448eca5b01e7b2e3199439111af50519fd4b8f124bc9e62968eb
|
| 3 |
+
size 9028952285
|
docs/README.md
ADDED
|
@@ -0,0 +1,129 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Model Overview
|
| 2 |
+
This bundle is for Nvidia MAISI (Medical AI for Synthetic Imaging), a 3D Latent Diffusion Model that can generate large CT images with paired segmentation masks, variable volume size and voxel size, as well as controllable organ/tumor size.
|
| 3 |
+
|
| 4 |
+
The inference workflow of MAISI is depicted in the figure below. It first generates latent features from random noise by applying multiple denoising steps using the trained diffusion model. Then it decodes the denoised latent features into images using the trained autoencoder.
|
| 5 |
+
|
| 6 |
+
<p align="center">
|
| 7 |
+
<img src="https://developer.download.nvidia.com/assets/Clara/Images/maisi_workflow_1.0.1.png" alt="MAISI inference scheme">
|
| 8 |
+
</p>
|
| 9 |
+
|
| 10 |
+
MAISI is based on the following papers:
|
| 11 |
+
|
| 12 |
+
[**Latent Diffusion:** Rombach, Robin, et al. "High-resolution image synthesis with latent diffusion models." CVPR 2022.](https://openaccess.thecvf.com/content/CVPR2022/papers/Rombach_High-Resolution_Image_Synthesis_With_Latent_Diffusion_Models_CVPR_2022_paper.pdf)
|
| 13 |
+
|
| 14 |
+
[**ControlNet:** Lvmin Zhang, Anyi Rao, Maneesh Agrawala; “Adding Conditional Control to Text-to-Image Diffusion Models.” ICCV 2023.](https://openaccess.thecvf.com/content/ICCV2023/papers/Zhang_Adding_Conditional_Control_to_Text-to-Image_Diffusion_Models_ICCV_2023_paper.pdf)
|
| 15 |
+
|
| 16 |
+
[**Rectified Flow:** Liu, Xingchao, and Chengyue Gong. "Flow Straight and Fast: Learning to Generate and Transfer Data with Rectified Flow." ICLR 2023.](https://arxiv.org/pdf/2209.03003)
|
| 17 |
+
|
| 18 |
+
#### Example synthetic image
|
| 19 |
+
An example result from inference is shown below:
|
| 20 |
+
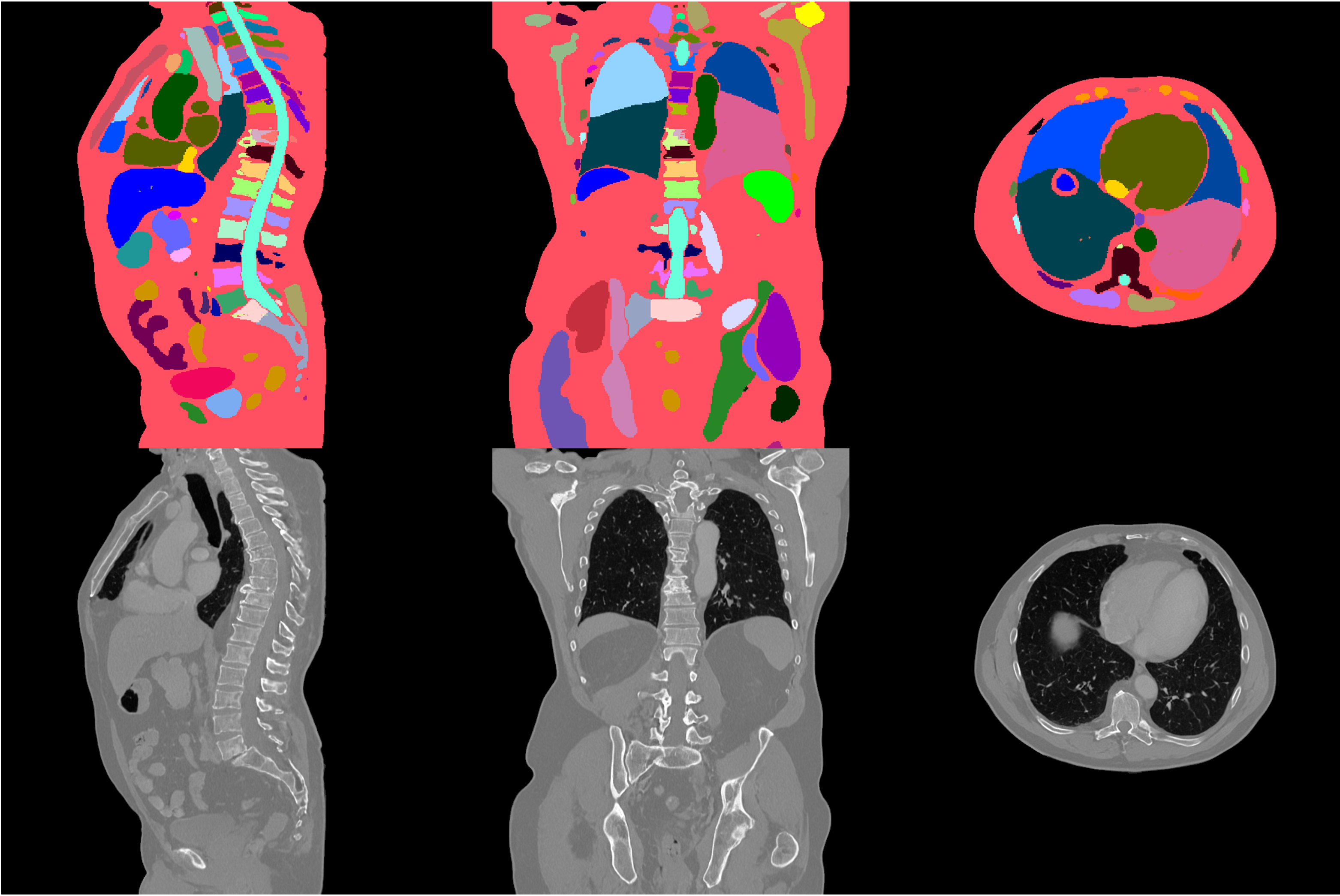
|
| 21 |
+
|
| 22 |
+
### Inference configuration
|
| 23 |
+
The inference requires:
|
| 24 |
+
- GPU: at least 58GB GPU memory for 512 x 512 x 512
|
| 25 |
+
- Disk Memory: at least 21GB disk memory
|
| 26 |
+
|
| 27 |
+
#### Inference parameters:
|
| 28 |
+
The information for the inference input, like body region and anatomy to generate, is stored in [./configs/inference.json](../configs/inference.json). Please feel free to play with it. Here are the details of the parameters.
|
| 29 |
+
|
| 30 |
+
- `"num_output_samples"`: int, the number of output image/mask pairs it will generate.
|
| 31 |
+
- `"spacing"`: voxel size of generated images. E.g., if set to `[1.5, 1.5, 2.0]`, it will generate images with a resolution of 1.5×1.5×2.0 mm. The spacing for x and y axes has to be between 0.5 and 3.0 mm and the spacing for the z axis has to be between 0.5 and 5.0 mm.
|
| 32 |
+
- `"output_size"`: volume size of generated images. E.g., if set to `[512, 512, 256]`, it will generate images with size of 512×512×256. They need to be divisible by 16. If you have a small GPU memory size, you should adjust it to small numbers. Note that `"spacing"` and `"output_size"` together decide the output field of view (FOV). For eample, if set them to `[1.5, 1.5, 2.0]`mm and `[512, 512, 256]`, the FOV is 768×768×512 mm. We recommend output_size is the FOV in x and y axis are same and to be at least 256mm for head, at least 384mm for other body regions like abdomen, and no larger than 640mm. The output size for the x and y axes can be selected from [256, 384, 512], while for the z axis, it can be chosen from [128, 256, 384, 512, 640, 768].
|
| 33 |
+
- `"controllable_anatomy_size"`: a list of controllable anatomy and its size scale (0--1). E.g., if set to `[["liver", 0.5],["hepatic tumor", 0.3]]`, the generated image will contain liver that have a median size, with size around 50% percentile, and hepatic tumor that is relatively small, with around 30% percentile. In addition, if the size scale is set to -1, it indicates that the organ does not exist or should be removed. The output will contain paired image and segmentation mask for the controllable anatomy.
|
| 34 |
+
The following organs support generation with a controllable size: ``["liver", "gallbladder", "stomach", "pancreas", "colon", "lung tumor", "bone lesion", "hepatic tumor", "colon cancer primaries", "pancreatic tumor"]``.
|
| 35 |
+
The raw output of the current mask generation model has a fixed size of $256^3$ voxels with a spacing of $1.5^3$ mm. If the "output_size" differs from this default, the generated masks will be resampled to the desired `"output_size"` and `"spacing"`. Note that resampling may degrade the quality of the generated masks and could trigger multiple inference attempts if the images fail to pass the [image quality check](../scripts/quality_check.py).
|
| 36 |
+
- `"body_region"`: Deprecated, please leave it as empty `"[]"`.
|
| 37 |
+
- `"anatomy_list"`: If "controllable_anatomy_size" is not specified, the output will contain paired image and segmentation mask for the anatomy in "./configs/label_dict.json".
|
| 38 |
+
- `"autoencoder_sliding_window_infer_size"`: in order to save GPU memory, we use sliding window inference when decoding latents to image when `"output_size"` is large. This is the patch size of the sliding window. Small value will reduce GPU memory but increase time cost. They need to be divisible by 16.
|
| 39 |
+
- `"autoencoder_sliding_window_infer_overlap"`: float between 0 and 1. Large value will reduce the stitching artifacts when stitching patches during sliding window inference, but increase time cost. If you do not observe seam lines in the generated image result, you can use a smaller value to save inference time.
|
| 40 |
+
|
| 41 |
+
To generate images with substantial dimensions, such as 512 × 512 × 512 or larger, using GPUs with 80GB of memory, it is advisable to configure the `"num_splits"` parameter in [the auto-encoder configuration](./configs/config_maisi.json#L11-L37) to 16. This adjustment is crucial to avoid out-of-memory issues during inference.
|
| 42 |
+
|
| 43 |
+
#### Recommended spacing for different output sizes:
|
| 44 |
+
|
| 45 |
+
|`"output_size"`| Recommended `"spacing"`|
|
| 46 |
+
|:-----:|:-----:|
|
| 47 |
+
[256, 256, 256] | [1.5, 1.5, 1.5] |
|
| 48 |
+
[512, 512, 128] | [0.8, 0.8, 2.5] |
|
| 49 |
+
[512, 512, 512] | [1.0, 1.0, 1.0] |
|
| 50 |
+
|
| 51 |
+
### Execute inference
|
| 52 |
+
The following code generates a synthetic image from a random sampled noise.
|
| 53 |
+
```
|
| 54 |
+
python -m monai.bundle run --config_file configs/inference.json
|
| 55 |
+
```
|
| 56 |
+
|
| 57 |
+
## Execute Finetuning
|
| 58 |
+
|
| 59 |
+
### Training configuration
|
| 60 |
+
The training was performed with the following:
|
| 61 |
+
- GPU: at least 60GB GPU memory for 512 x 512 x 512 volume
|
| 62 |
+
- Actual Model Input (the size of image embedding in latent space): 128 x 128 x 128
|
| 63 |
+
- AMP: True
|
| 64 |
+
|
| 65 |
+
### Run finetuning:
|
| 66 |
+
This config executes finetuning for pretrained ControlNet with with a new class (i.e., Kidney Tumor). When finetuning with new class names, please update `configs/train.json`'s `weighted_loss_label` and `configs/label_dict.json` accordingly. There are 8 dummy labels as placeholders in default `configs/label_dict.json` that can be used for finetuning.
|
| 67 |
+
```
|
| 68 |
+
python -m monai.bundle run --config_file configs/train.json
|
| 69 |
+
```
|
| 70 |
+
|
| 71 |
+
### Override the `train` config to execute multi-GPU training:
|
| 72 |
+
|
| 73 |
+
```
|
| 74 |
+
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
|
| 75 |
+
```
|
| 76 |
+
|
| 77 |
+
### Data:
|
| 78 |
+
The preprocessed subset of [C4KC-KiTS](https://www.cancerimagingarchive.net/collection/c4kc-kits/) dataset used in this finetuning config is provided in `./dataset/C4KC-KiTS_subset`.
|
| 79 |
+
```
|
| 80 |
+
|-*arterial*.nii.gz # original image
|
| 81 |
+
|-*arterial_emb*.nii.gz # encoded image embedding
|
| 82 |
+
KiTS-000* --|-mask*.nii.gz # original labels
|
| 83 |
+
|-mask_pseudo_label*.nii.gz # pseudo labels
|
| 84 |
+
|-mask_combined_label*.nii.gz # combined mask of original and pseudo labels
|
| 85 |
+
|
| 86 |
+
```
|
| 87 |
+
An example combined mask of original and pseudo labels is shown below:
|
| 88 |
+
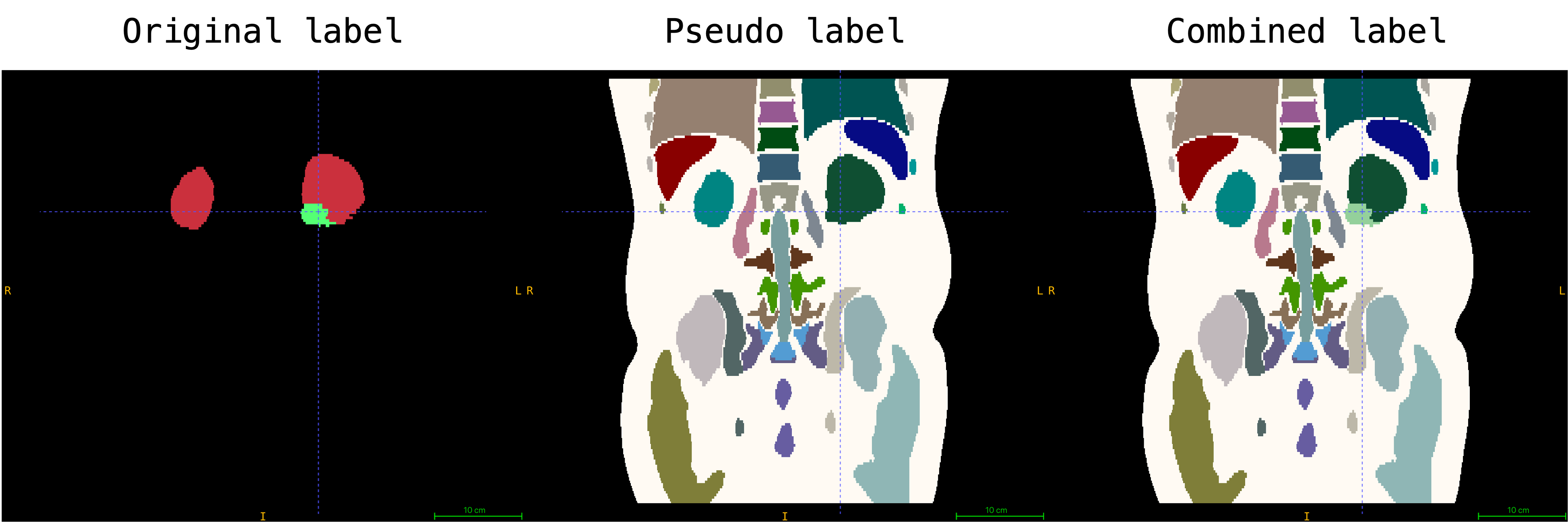
|
| 89 |
+
|
| 90 |
+
Please note that the label of Kidney Tumor is mapped to index `129` in this preprocessed dataset. The encoded image embedding is generated by provided `Autoencoder` in `./models/autoencoder_epoch273.pt` during preprocessing to save memeory usage for training. The pseudo labels are generated by [VISTA 3D](https://github.com/Project-MONAI/VISTA). In addition, the dimension of each volume and corresponding pseudo label is resampled to the closest multiple of 128 (e.g., 128, 256, 384, 512, ...).
|
| 91 |
+
|
| 92 |
+
The training workflow requires one JSON file to specify the image embedding and segmentation pairs. The example file is located in the `./dataset/C4KC-KiTS_subset.json`.
|
| 93 |
+
|
| 94 |
+
The JSON file has the following structure:
|
| 95 |
+
```python
|
| 96 |
+
{
|
| 97 |
+
"training": [
|
| 98 |
+
{
|
| 99 |
+
"image": "*/*arterial_emb*.nii.gz", # relative path to the image embedding file
|
| 100 |
+
"label": "*/mask_combined_label*.nii.gz", # relative path to the combined label file
|
| 101 |
+
"dim": [512, 512, 512], # the dimension of image
|
| 102 |
+
"spacing": [1.0, 1.0, 1.0], # the spacing of image
|
| 103 |
+
"top_region_index": [0, 1, 0, 0], # the top region index of the image
|
| 104 |
+
"bottom_region_index": [0, 0, 0, 1], # the bottom region index of the image
|
| 105 |
+
"fold": 0 # fold index for cross validation, fold 0 is used for training
|
| 106 |
+
},
|
| 107 |
+
|
| 108 |
+
...
|
| 109 |
+
]
|
| 110 |
+
}
|
| 111 |
+
```
|
| 112 |
+
|
| 113 |
+
# References
|
| 114 |
+
[1] Rombach, Robin, et al. "High-resolution image synthesis with latent diffusion models." Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022. https://openaccess.thecvf.com/content/CVPR2022/papers/Rombach_High-Resolution_Image_Synthesis_With_Latent_Diffusion_Models_CVPR_2022_paper.pdf
|
| 115 |
+
|
| 116 |
+
# License
|
| 117 |
+
|
| 118 |
+
## Code License
|
| 119 |
+
|
| 120 |
+
This project includes code licensed under the Apache License 2.0.
|
| 121 |
+
You may obtain a copy of the License at
|
| 122 |
+
|
| 123 |
+
http://www.apache.org/licenses/LICENSE-2.0
|
| 124 |
+
|
| 125 |
+
## Model Weights License
|
| 126 |
+
|
| 127 |
+
The model weights included in this project are licensed under the NCLS v1 License.
|
| 128 |
+
|
| 129 |
+
Both licenses' full texts have been combined into a single `LICENSE` file. Please refer to this `LICENSE` file for more details about the terms and conditions of both licenses.
|
docs/data_license.txt
ADDED
|
@@ -0,0 +1,49 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Third Party Licenses
|
| 2 |
+
-----------------------------------------------------------------------
|
| 3 |
+
|
| 4 |
+
/*********************************************************************/
|
| 5 |
+
i. Multimodal Brain Tumor Segmentation Challenge 2018
|
| 6 |
+
https://www.med.upenn.edu/sbia/brats2018/data.html
|
| 7 |
+
/*********************************************************************/
|
| 8 |
+
|
| 9 |
+
Data Usage Agreement / Citations
|
| 10 |
+
|
| 11 |
+
You are free to use and/or refer to the BraTS datasets in your own
|
| 12 |
+
research, provided that you always cite the following two manuscripts:
|
| 13 |
+
|
| 14 |
+
[1] Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby
|
| 15 |
+
[J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber
|
| 16 |
+
[MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N,
|
| 17 |
+
[Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Γ, Durst CR,
|
| 18 |
+
[Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P,
|
| 19 |
+
[Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E,
|
| 20 |
+
[Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv
|
| 21 |
+
[TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J,
|
| 22 |
+
[Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM,
|
| 23 |
+
[Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B,
|
| 24 |
+
[Zikic D, Prastawa M, Reyes M, Van Leemput K. "The Multimodal Brain
|
| 25 |
+
[Tumor Image Segmentation Benchmark (BRATS)", IEEE Transactions on
|
| 26 |
+
[Medical Imaging 34(10), 1993-2024 (2015) DOI:
|
| 27 |
+
[10.1109/TMI.2014.2377694
|
| 28 |
+
|
| 29 |
+
[2] Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS,
|
| 30 |
+
[Freymann JB, Farahani K, Davatzikos C. "Advancing The Cancer Genome
|
| 31 |
+
[Atlas glioma MRI collections with expert segmentation labels and
|
| 32 |
+
[radiomic features", Nature Scientific Data, 4:170117 (2017) DOI:
|
| 33 |
+
[10.1038/sdata.2017.117
|
| 34 |
+
|
| 35 |
+
In addition, if there are no restrictions imposed from the
|
| 36 |
+
journal/conference you submit your paper about citing "Data
|
| 37 |
+
Citations", please be specific and also cite the following:
|
| 38 |
+
|
| 39 |
+
[3] Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J,
|
| 40 |
+
[Freymann J, Farahani K, Davatzikos C. "Segmentation Labels and
|
| 41 |
+
[Radiomic Features for the Pre-operative Scans of the TCGA-GBM
|
| 42 |
+
[collection", The Cancer Imaging Archive, 2017. DOI:
|
| 43 |
+
[10.7937/K9/TCIA.2017.KLXWJJ1Q
|
| 44 |
+
|
| 45 |
+
[4] Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J,
|
| 46 |
+
[Freymann J, Farahani K, Davatzikos C. "Segmentation Labels and
|
| 47 |
+
[Radiomic Features for the Pre-operative Scans of the TCGA-LGG
|
| 48 |
+
[collection", The Cancer Imaging Archive, 2017. DOI:
|
| 49 |
+
[10.7937/K9/TCIA.2017.GJQ7R0EF
|
models/autoencoder.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:1f8a7a056d0ebc00486edc43c26768bf1c12eaa6df9dd172e34598003be95eb3
|
| 3 |
+
size 83831868
|
models/controlnet-20datasets-e20wl100fold0bc_noi_dia_fsize_current.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:05fda7bfffde524d56cc2dc5b990f901216bc46c4b5e261404aebc409d27b78b
|
| 3 |
+
size 278366962
|
models/controlnet.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:468c7c1d358530b9ebbdd643e4c1b1c1e4037df154e0bc15d21fc49e56a57f75
|
| 3 |
+
size 288255799
|
models/diffusion_unet.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:cfc1ec59782f5ac7c0d22dd501654732109a971b93cbaa6607c4106a7f29066f
|
| 3 |
+
size 2166600232
|
models/input_unet3d_data-all_steps1000size512ddpm_random_current_inputx_v1.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:b32be13118f9d6a077d42dd250c09c5e41673b48dbf2f35f2c587a7a9ebe5686
|
| 3 |
+
size 685298858
|
models/mask_generation_autoencoder.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:539175f6ede3cb1e6f01bfd6347cd446d601cf4a508fc632a1e36362b1428a5d
|
| 3 |
+
size 21072774
|
models/mask_generation_diffusion_unet.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:b7d668b1356e9b94b8269decadf8f8116dc4ee2d365580d05349b4ddf6739155
|
| 3 |
+
size 788941780
|
scripts/__init__.py
ADDED
|
@@ -0,0 +1,12 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
from . import sample, utils
|
scripts/augmentation.py
ADDED
|
@@ -0,0 +1,373 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
import numpy as np
|
| 13 |
+
import torch
|
| 14 |
+
import torch.nn.functional as F
|
| 15 |
+
from monai.transforms import Rand3DElastic, RandAffine, RandZoom
|
| 16 |
+
from monai.utils import ensure_tuple_rep
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
def erode3d(input_tensor, erosion=3):
|
| 20 |
+
# Define the structuring element
|
| 21 |
+
erosion = ensure_tuple_rep(erosion, 3)
|
| 22 |
+
structuring_element = torch.ones(1, 1, erosion[0], erosion[1], erosion[2]).to(input_tensor.device)
|
| 23 |
+
|
| 24 |
+
# Pad the input tensor to handle border pixels
|
| 25 |
+
input_padded = F.pad(
|
| 26 |
+
input_tensor.float().unsqueeze(0).unsqueeze(0),
|
| 27 |
+
(erosion[0] // 2, erosion[0] // 2, erosion[1] // 2, erosion[1] // 2, erosion[2] // 2, erosion[2] // 2),
|
| 28 |
+
mode="constant",
|
| 29 |
+
value=1.0,
|
| 30 |
+
)
|
| 31 |
+
|
| 32 |
+
# Apply erosion operation
|
| 33 |
+
output = F.conv3d(input_padded, structuring_element, padding=0)
|
| 34 |
+
|
| 35 |
+
# Set output values based on the minimum value within the structuring element
|
| 36 |
+
output = torch.where(output == torch.sum(structuring_element), 1.0, 0.0)
|
| 37 |
+
|
| 38 |
+
return output.squeeze(0).squeeze(0)
|
| 39 |
+
|
| 40 |
+
|
| 41 |
+
def dilate3d(input_tensor, erosion=3):
|
| 42 |
+
# Define the structuring element
|
| 43 |
+
erosion = ensure_tuple_rep(erosion, 3)
|
| 44 |
+
structuring_element = torch.ones(1, 1, erosion[0], erosion[1], erosion[2]).to(input_tensor.device)
|
| 45 |
+
|
| 46 |
+
# Pad the input tensor to handle border pixels
|
| 47 |
+
input_padded = F.pad(
|
| 48 |
+
input_tensor.float().unsqueeze(0).unsqueeze(0),
|
| 49 |
+
(erosion[0] // 2, erosion[0] // 2, erosion[1] // 2, erosion[1] // 2, erosion[2] // 2, erosion[2] // 2),
|
| 50 |
+
mode="constant",
|
| 51 |
+
value=1.0,
|
| 52 |
+
)
|
| 53 |
+
|
| 54 |
+
# Apply erosion operation
|
| 55 |
+
output = F.conv3d(input_padded, structuring_element, padding=0)
|
| 56 |
+
|
| 57 |
+
# Set output values based on the minimum value within the structuring element
|
| 58 |
+
output = torch.where(output > 0, 1.0, 0.0)
|
| 59 |
+
|
| 60 |
+
return output.squeeze(0).squeeze(0)
|
| 61 |
+
|
| 62 |
+
|
| 63 |
+
def augmentation_tumor_bone(pt_nda, output_size, random_seed):
|
| 64 |
+
volume = pt_nda.squeeze(0)
|
| 65 |
+
real_l_volume_ = torch.zeros_like(volume)
|
| 66 |
+
real_l_volume_[volume == 128] = 1
|
| 67 |
+
real_l_volume_ = real_l_volume_.to(torch.uint8)
|
| 68 |
+
|
| 69 |
+
elastic = RandAffine(
|
| 70 |
+
mode="nearest",
|
| 71 |
+
prob=1.0,
|
| 72 |
+
translate_range=(5, 5, 0),
|
| 73 |
+
rotate_range=(0, 0, 0.1),
|
| 74 |
+
scale_range=(0.15, 0.15, 0),
|
| 75 |
+
padding_mode="zeros",
|
| 76 |
+
)
|
| 77 |
+
elastic.set_random_state(seed=random_seed)
|
| 78 |
+
|
| 79 |
+
tumor_szie = torch.sum((real_l_volume_ > 0).float())
|
| 80 |
+
###########################
|
| 81 |
+
# remove pred in pseudo_label in real lesion region
|
| 82 |
+
volume[real_l_volume_ > 0] = 200
|
| 83 |
+
###########################
|
| 84 |
+
if tumor_szie > 0:
|
| 85 |
+
# get organ mask
|
| 86 |
+
organ_mask = (
|
| 87 |
+
torch.logical_and(33 <= volume, volume <= 56).float()
|
| 88 |
+
+ torch.logical_and(63 <= volume, volume <= 97).float()
|
| 89 |
+
+ (volume == 127).float()
|
| 90 |
+
+ (volume == 114).float()
|
| 91 |
+
+ real_l_volume_
|
| 92 |
+
)
|
| 93 |
+
organ_mask = (organ_mask > 0).float()
|
| 94 |
+
cnt = 0
|
| 95 |
+
while True:
|
| 96 |
+
threshold = 0.8 if cnt < 40 else 0.75
|
| 97 |
+
real_l_volume = real_l_volume_
|
| 98 |
+
# random distor mask
|
| 99 |
+
distored_mask = elastic((real_l_volume > 0).cuda(), spatial_size=tuple(output_size)).as_tensor()
|
| 100 |
+
real_l_volume = distored_mask * organ_mask
|
| 101 |
+
cnt += 1
|
| 102 |
+
print(torch.sum(real_l_volume), "|", tumor_szie * threshold)
|
| 103 |
+
if torch.sum(real_l_volume) >= tumor_szie * threshold:
|
| 104 |
+
real_l_volume = dilate3d(real_l_volume.squeeze(0), erosion=5)
|
| 105 |
+
real_l_volume = erode3d(real_l_volume, erosion=5).unsqueeze(0).to(torch.uint8)
|
| 106 |
+
break
|
| 107 |
+
else:
|
| 108 |
+
real_l_volume = real_l_volume_
|
| 109 |
+
|
| 110 |
+
volume[real_l_volume == 1] = 128
|
| 111 |
+
|
| 112 |
+
pt_nda = volume.unsqueeze(0)
|
| 113 |
+
return pt_nda
|
| 114 |
+
|
| 115 |
+
|
| 116 |
+
def augmentation_tumor_liver(pt_nda, output_size, random_seed):
|
| 117 |
+
volume = pt_nda.squeeze(0)
|
| 118 |
+
real_l_volume_ = torch.zeros_like(volume)
|
| 119 |
+
real_l_volume_[volume == 1] = 1
|
| 120 |
+
real_l_volume_[volume == 26] = 2
|
| 121 |
+
real_l_volume_ = real_l_volume_.to(torch.uint8)
|
| 122 |
+
|
| 123 |
+
elastic = Rand3DElastic(
|
| 124 |
+
mode="nearest",
|
| 125 |
+
prob=1.0,
|
| 126 |
+
sigma_range=(5, 8),
|
| 127 |
+
magnitude_range=(100, 200),
|
| 128 |
+
translate_range=(10, 10, 10),
|
| 129 |
+
rotate_range=(np.pi / 36, np.pi / 36, np.pi / 36),
|
| 130 |
+
scale_range=(0.2, 0.2, 0.2),
|
| 131 |
+
padding_mode="zeros",
|
| 132 |
+
)
|
| 133 |
+
elastic.set_random_state(seed=random_seed)
|
| 134 |
+
|
| 135 |
+
tumor_szie = torch.sum(real_l_volume_ == 2)
|
| 136 |
+
###########################
|
| 137 |
+
# remove pred organ labels
|
| 138 |
+
volume[volume == 1] = 0
|
| 139 |
+
volume[volume == 26] = 0
|
| 140 |
+
# before move tumor maks, full the original location by organ labels
|
| 141 |
+
volume[real_l_volume_ == 1] = 1
|
| 142 |
+
volume[real_l_volume_ == 2] = 1
|
| 143 |
+
###########################
|
| 144 |
+
while True:
|
| 145 |
+
real_l_volume = real_l_volume_
|
| 146 |
+
# random distor mask
|
| 147 |
+
real_l_volume = elastic((real_l_volume == 2).cuda(), spatial_size=tuple(output_size)).as_tensor()
|
| 148 |
+
# get organ mask
|
| 149 |
+
organ_mask = (real_l_volume_ == 1).float() + (real_l_volume_ == 2).float()
|
| 150 |
+
|
| 151 |
+
organ_mask = dilate3d(organ_mask.squeeze(0), erosion=5)
|
| 152 |
+
organ_mask = erode3d(organ_mask, erosion=5).unsqueeze(0)
|
| 153 |
+
real_l_volume = real_l_volume * organ_mask
|
| 154 |
+
print(torch.sum(real_l_volume), "|", tumor_szie * 0.80)
|
| 155 |
+
if torch.sum(real_l_volume) >= tumor_szie * 0.80:
|
| 156 |
+
real_l_volume = dilate3d(real_l_volume.squeeze(0), erosion=5)
|
| 157 |
+
real_l_volume = erode3d(real_l_volume, erosion=5).unsqueeze(0)
|
| 158 |
+
break
|
| 159 |
+
|
| 160 |
+
volume[real_l_volume == 1] = 26
|
| 161 |
+
|
| 162 |
+
pt_nda = volume.unsqueeze(0)
|
| 163 |
+
return pt_nda
|
| 164 |
+
|
| 165 |
+
|
| 166 |
+
def augmentation_tumor_lung(pt_nda, output_size, random_seed):
|
| 167 |
+
volume = pt_nda.squeeze(0)
|
| 168 |
+
real_l_volume_ = torch.zeros_like(volume)
|
| 169 |
+
real_l_volume_[volume == 23] = 1
|
| 170 |
+
real_l_volume_ = real_l_volume_.to(torch.uint8)
|
| 171 |
+
|
| 172 |
+
elastic = Rand3DElastic(
|
| 173 |
+
mode="nearest",
|
| 174 |
+
prob=1.0,
|
| 175 |
+
sigma_range=(5, 8),
|
| 176 |
+
magnitude_range=(100, 200),
|
| 177 |
+
translate_range=(20, 20, 20),
|
| 178 |
+
rotate_range=(np.pi / 36, np.pi / 36, np.pi),
|
| 179 |
+
scale_range=(0.15, 0.15, 0.15),
|
| 180 |
+
padding_mode="zeros",
|
| 181 |
+
)
|
| 182 |
+
elastic.set_random_state(seed=random_seed)
|
| 183 |
+
|
| 184 |
+
tumor_szie = torch.sum(real_l_volume_)
|
| 185 |
+
# before move lung tumor maks, full the original location by lung labels
|
| 186 |
+
new_real_l_volume_ = dilate3d(real_l_volume_.squeeze(0), erosion=3)
|
| 187 |
+
new_real_l_volume_ = new_real_l_volume_.unsqueeze(0)
|
| 188 |
+
new_real_l_volume_[real_l_volume_ > 0] = 0
|
| 189 |
+
new_real_l_volume_[volume < 28] = 0
|
| 190 |
+
new_real_l_volume_[volume > 32] = 0
|
| 191 |
+
tmp = volume[(volume * new_real_l_volume_).nonzero(as_tuple=True)].view(-1)
|
| 192 |
+
|
| 193 |
+
mode = torch.mode(tmp, 0)[0].item()
|
| 194 |
+
print(mode)
|
| 195 |
+
assert 28 <= mode <= 32
|
| 196 |
+
volume[real_l_volume_.bool()] = mode
|
| 197 |
+
###########################
|
| 198 |
+
if tumor_szie > 0:
|
| 199 |
+
# aug
|
| 200 |
+
while True:
|
| 201 |
+
real_l_volume = real_l_volume_
|
| 202 |
+
# random distor mask
|
| 203 |
+
real_l_volume = elastic(real_l_volume, spatial_size=tuple(output_size)).as_tensor()
|
| 204 |
+
# get lung mask v2 (133 order)
|
| 205 |
+
lung_mask = (
|
| 206 |
+
(volume == 28).float()
|
| 207 |
+
+ (volume == 29).float()
|
| 208 |
+
+ (volume == 30).float()
|
| 209 |
+
+ (volume == 31).float()
|
| 210 |
+
+ (volume == 32).float()
|
| 211 |
+
)
|
| 212 |
+
|
| 213 |
+
lung_mask = dilate3d(lung_mask.squeeze(0), erosion=5)
|
| 214 |
+
lung_mask = erode3d(lung_mask, erosion=5).unsqueeze(0)
|
| 215 |
+
real_l_volume = real_l_volume * lung_mask
|
| 216 |
+
print(torch.sum(real_l_volume), "|", tumor_szie * 0.85)
|
| 217 |
+
if torch.sum(real_l_volume) >= tumor_szie * 0.85:
|
| 218 |
+
real_l_volume = dilate3d(real_l_volume.squeeze(0), erosion=5)
|
| 219 |
+
real_l_volume = erode3d(real_l_volume, erosion=5).unsqueeze(0).to(torch.uint8)
|
| 220 |
+
break
|
| 221 |
+
else:
|
| 222 |
+
real_l_volume = real_l_volume_
|
| 223 |
+
|
| 224 |
+
volume[real_l_volume == 1] = 23
|
| 225 |
+
|
| 226 |
+
pt_nda = volume.unsqueeze(0)
|
| 227 |
+
return pt_nda
|
| 228 |
+
|
| 229 |
+
|
| 230 |
+
def augmentation_tumor_pancreas(pt_nda, output_size, random_seed):
|
| 231 |
+
volume = pt_nda.squeeze(0)
|
| 232 |
+
real_l_volume_ = torch.zeros_like(volume)
|
| 233 |
+
real_l_volume_[volume == 4] = 1
|
| 234 |
+
real_l_volume_[volume == 24] = 2
|
| 235 |
+
real_l_volume_ = real_l_volume_.to(torch.uint8)
|
| 236 |
+
|
| 237 |
+
elastic = Rand3DElastic(
|
| 238 |
+
mode="nearest",
|
| 239 |
+
prob=1.0,
|
| 240 |
+
sigma_range=(5, 8),
|
| 241 |
+
magnitude_range=(100, 200),
|
| 242 |
+
translate_range=(15, 15, 15),
|
| 243 |
+
rotate_range=(np.pi / 36, np.pi / 36, np.pi / 36),
|
| 244 |
+
scale_range=(0.1, 0.1, 0.1),
|
| 245 |
+
padding_mode="zeros",
|
| 246 |
+
)
|
| 247 |
+
elastic.set_random_state(seed=random_seed)
|
| 248 |
+
|
| 249 |
+
tumor_szie = torch.sum(real_l_volume_ == 2)
|
| 250 |
+
###########################
|
| 251 |
+
# remove pred organ labels
|
| 252 |
+
volume[volume == 24] = 0
|
| 253 |
+
volume[volume == 4] = 0
|
| 254 |
+
# before move tumor maks, full the original location by organ labels
|
| 255 |
+
volume[real_l_volume_ == 1] = 4
|
| 256 |
+
volume[real_l_volume_ == 2] = 4
|
| 257 |
+
###########################
|
| 258 |
+
while True:
|
| 259 |
+
real_l_volume = real_l_volume_
|
| 260 |
+
# random distor mask
|
| 261 |
+
real_l_volume = elastic((real_l_volume == 2).cuda(), spatial_size=tuple(output_size)).as_tensor()
|
| 262 |
+
# get organ mask
|
| 263 |
+
organ_mask = (real_l_volume_ == 1).float() + (real_l_volume_ == 2).float()
|
| 264 |
+
|
| 265 |
+
organ_mask = dilate3d(organ_mask.squeeze(0), erosion=5)
|
| 266 |
+
organ_mask = erode3d(organ_mask, erosion=5).unsqueeze(0)
|
| 267 |
+
real_l_volume = real_l_volume * organ_mask
|
| 268 |
+
print(torch.sum(real_l_volume), "|", tumor_szie * 0.80)
|
| 269 |
+
if torch.sum(real_l_volume) >= tumor_szie * 0.80:
|
| 270 |
+
real_l_volume = dilate3d(real_l_volume.squeeze(0), erosion=5)
|
| 271 |
+
real_l_volume = erode3d(real_l_volume, erosion=5).unsqueeze(0)
|
| 272 |
+
break
|
| 273 |
+
|
| 274 |
+
volume[real_l_volume == 1] = 24
|
| 275 |
+
|
| 276 |
+
pt_nda = volume.unsqueeze(0)
|
| 277 |
+
return pt_nda
|
| 278 |
+
|
| 279 |
+
|
| 280 |
+
def augmentation_tumor_colon(pt_nda, output_size, random_seed):
|
| 281 |
+
volume = pt_nda.squeeze(0)
|
| 282 |
+
real_l_volume_ = torch.zeros_like(volume)
|
| 283 |
+
real_l_volume_[volume == 27] = 1
|
| 284 |
+
real_l_volume_ = real_l_volume_.to(torch.uint8)
|
| 285 |
+
|
| 286 |
+
elastic = Rand3DElastic(
|
| 287 |
+
mode="nearest",
|
| 288 |
+
prob=1.0,
|
| 289 |
+
sigma_range=(5, 8),
|
| 290 |
+
magnitude_range=(100, 200),
|
| 291 |
+
translate_range=(5, 5, 5),
|
| 292 |
+
rotate_range=(np.pi / 36, np.pi / 36, np.pi / 36),
|
| 293 |
+
scale_range=(0.1, 0.1, 0.1),
|
| 294 |
+
padding_mode="zeros",
|
| 295 |
+
)
|
| 296 |
+
elastic.set_random_state(seed=random_seed)
|
| 297 |
+
|
| 298 |
+
tumor_szie = torch.sum(real_l_volume_)
|
| 299 |
+
###########################
|
| 300 |
+
# before move tumor maks, full the original location by organ labels
|
| 301 |
+
volume[real_l_volume_.bool()] = 62
|
| 302 |
+
###########################
|
| 303 |
+
if tumor_szie > 0:
|
| 304 |
+
# get organ mask
|
| 305 |
+
organ_mask = (volume == 62).float()
|
| 306 |
+
organ_mask = dilate3d(organ_mask.squeeze(0), erosion=5)
|
| 307 |
+
organ_mask = erode3d(organ_mask, erosion=5).unsqueeze(0)
|
| 308 |
+
# cnt = 0
|
| 309 |
+
cnt = 0
|
| 310 |
+
while True:
|
| 311 |
+
threshold = 0.8
|
| 312 |
+
real_l_volume = real_l_volume_
|
| 313 |
+
if cnt < 20:
|
| 314 |
+
# random distor mask
|
| 315 |
+
distored_mask = elastic((real_l_volume == 1).cuda(), spatial_size=tuple(output_size)).as_tensor()
|
| 316 |
+
real_l_volume = distored_mask * organ_mask
|
| 317 |
+
elif 20 <= cnt < 40:
|
| 318 |
+
threshold = 0.75
|
| 319 |
+
else:
|
| 320 |
+
break
|
| 321 |
+
|
| 322 |
+
real_l_volume = real_l_volume * organ_mask
|
| 323 |
+
print(torch.sum(real_l_volume), "|", tumor_szie * threshold)
|
| 324 |
+
cnt += 1
|
| 325 |
+
if torch.sum(real_l_volume) >= tumor_szie * threshold:
|
| 326 |
+
real_l_volume = dilate3d(real_l_volume.squeeze(0), erosion=5)
|
| 327 |
+
real_l_volume = erode3d(real_l_volume, erosion=5).unsqueeze(0).to(torch.uint8)
|
| 328 |
+
break
|
| 329 |
+
else:
|
| 330 |
+
real_l_volume = real_l_volume_
|
| 331 |
+
# break
|
| 332 |
+
volume[real_l_volume == 1] = 27
|
| 333 |
+
|
| 334 |
+
pt_nda = volume.unsqueeze(0)
|
| 335 |
+
return pt_nda
|
| 336 |
+
|
| 337 |
+
|
| 338 |
+
def augmentation_body(pt_nda, random_seed):
|
| 339 |
+
volume = pt_nda.squeeze(0)
|
| 340 |
+
|
| 341 |
+
zoom = RandZoom(min_zoom=0.99, max_zoom=1.01, mode="nearest", align_corners=None, prob=1.0)
|
| 342 |
+
zoom.set_random_state(seed=random_seed)
|
| 343 |
+
|
| 344 |
+
volume = zoom(volume)
|
| 345 |
+
|
| 346 |
+
pt_nda = volume.unsqueeze(0)
|
| 347 |
+
return pt_nda
|
| 348 |
+
|
| 349 |
+
|
| 350 |
+
def augmentation(pt_nda, output_size, random_seed):
|
| 351 |
+
label_list = torch.unique(pt_nda)
|
| 352 |
+
label_list = list(label_list.cpu().numpy())
|
| 353 |
+
|
| 354 |
+
if 128 in label_list:
|
| 355 |
+
print("augmenting bone lesion/tumor")
|
| 356 |
+
pt_nda = augmentation_tumor_bone(pt_nda, output_size, random_seed)
|
| 357 |
+
elif 26 in label_list:
|
| 358 |
+
print("augmenting liver tumor")
|
| 359 |
+
pt_nda = augmentation_tumor_liver(pt_nda, output_size, random_seed)
|
| 360 |
+
elif 23 in label_list:
|
| 361 |
+
print("augmenting lung tumor")
|
| 362 |
+
pt_nda = augmentation_tumor_lung(pt_nda, output_size, random_seed)
|
| 363 |
+
elif 24 in label_list:
|
| 364 |
+
print("augmenting pancreas tumor")
|
| 365 |
+
pt_nda = augmentation_tumor_pancreas(pt_nda, output_size, random_seed)
|
| 366 |
+
elif 27 in label_list:
|
| 367 |
+
print("augmenting colon tumor")
|
| 368 |
+
pt_nda = augmentation_tumor_colon(pt_nda, output_size, random_seed)
|
| 369 |
+
else:
|
| 370 |
+
print("augmenting body")
|
| 371 |
+
pt_nda = augmentation_body(pt_nda, random_seed)
|
| 372 |
+
|
| 373 |
+
return pt_nda
|
scripts/find_masks.py
ADDED
|
@@ -0,0 +1,137 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
import json
|
| 14 |
+
import os
|
| 15 |
+
from typing import Sequence
|
| 16 |
+
|
| 17 |
+
from monai.apps.utils import extractall
|
| 18 |
+
from monai.utils import ensure_tuple_rep
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
def convert_body_region(body_region: str | Sequence[str]) -> Sequence[int]:
|
| 22 |
+
"""
|
| 23 |
+
Convert body region string to body region index.
|
| 24 |
+
Args:
|
| 25 |
+
body_region: list of input body region string. If single str, will be converted to list of str.
|
| 26 |
+
Return:
|
| 27 |
+
body_region_indices, list of input body region index.
|
| 28 |
+
"""
|
| 29 |
+
if type(body_region) is str:
|
| 30 |
+
body_region = [body_region]
|
| 31 |
+
|
| 32 |
+
# body region mapping for maisi
|
| 33 |
+
region_mapping_maisi = {
|
| 34 |
+
"head": 0,
|
| 35 |
+
"chest": 1,
|
| 36 |
+
"thorax": 1,
|
| 37 |
+
"chest/thorax": 1,
|
| 38 |
+
"abdomen": 2,
|
| 39 |
+
"pelvis": 3,
|
| 40 |
+
"lower": 3,
|
| 41 |
+
"pelvis/lower": 3,
|
| 42 |
+
}
|
| 43 |
+
|
| 44 |
+
# perform mapping
|
| 45 |
+
body_region_indices = []
|
| 46 |
+
for region in body_region:
|
| 47 |
+
normalized_region = region.lower() # norm str to lower case
|
| 48 |
+
if normalized_region not in region_mapping_maisi:
|
| 49 |
+
raise ValueError(f"Invalid region: {normalized_region}")
|
| 50 |
+
body_region_indices.append(region_mapping_maisi[normalized_region])
|
| 51 |
+
|
| 52 |
+
return body_region_indices
|
| 53 |
+
|
| 54 |
+
|
| 55 |
+
def find_masks(
|
| 56 |
+
anatomy_list: int | Sequence[int],
|
| 57 |
+
spacing: Sequence[float] | float = 1.0,
|
| 58 |
+
output_size: Sequence[int] = (512, 512, 512),
|
| 59 |
+
check_spacing_and_output_size: bool = False,
|
| 60 |
+
database_filepath: str = "./configs/database.json",
|
| 61 |
+
mask_foldername: str = "./datasets/masks/",
|
| 62 |
+
):
|
| 63 |
+
"""
|
| 64 |
+
Find candidate masks that fullfills all the requirements.
|
| 65 |
+
They shoud contain all the anatomies in `anatomy_list`.
|
| 66 |
+
If there is no tumor specified in `anatomy_list`, we also expect the candidate masks to be tumor free.
|
| 67 |
+
If check_spacing_and_output_size is True, the candidate masks need to have the expected `spacing` and `output_size`.
|
| 68 |
+
Args:
|
| 69 |
+
anatomy_list: list of input anatomy. The found candidate mask will include these anatomies.
|
| 70 |
+
spacing: list of three floats, voxel spacing. If providing a single number, will use it for all the three dimensions.
|
| 71 |
+
output_size: list of three int, expected candidate mask spatial size.
|
| 72 |
+
check_spacing_and_output_size: whether we expect candidate mask to have spatial size of `output_size`
|
| 73 |
+
and voxel size of `spacing`.
|
| 74 |
+
database_filepath: path for the json file that stores the information of all the candidate masks.
|
| 75 |
+
mask_foldername: directory that saves all the candidate masks.
|
| 76 |
+
Return:
|
| 77 |
+
candidate_masks, list of dict, each dict contains information of one candidate mask that fullfills all the requirements.
|
| 78 |
+
"""
|
| 79 |
+
# check and preprocess input
|
| 80 |
+
if isinstance(anatomy_list, int):
|
| 81 |
+
anatomy_list = [anatomy_list]
|
| 82 |
+
|
| 83 |
+
spacing = ensure_tuple_rep(spacing, 3)
|
| 84 |
+
|
| 85 |
+
if not os.path.exists(mask_foldername):
|
| 86 |
+
zip_file_path = mask_foldername + ".zip"
|
| 87 |
+
|
| 88 |
+
if not os.path.isfile(zip_file_path):
|
| 89 |
+
raise ValueError(f"Please download {zip_file_path} following the instruction in ./datasets/README.md.")
|
| 90 |
+
|
| 91 |
+
print(f"Extracting {zip_file_path} to {os.path.dirname(zip_file_path)}")
|
| 92 |
+
extractall(filepath=zip_file_path, output_dir=os.path.dirname(zip_file_path), file_type="zip")
|
| 93 |
+
print(f"Unzipped {zip_file_path} to {mask_foldername}.")
|
| 94 |
+
|
| 95 |
+
if not os.path.isfile(database_filepath):
|
| 96 |
+
raise ValueError(f"Please download {database_filepath} following the instruction in ./datasets/README.md.")
|
| 97 |
+
with open(database_filepath, "r") as f:
|
| 98 |
+
db = json.load(f)
|
| 99 |
+
|
| 100 |
+
# select candidate_masks
|
| 101 |
+
candidate_masks = []
|
| 102 |
+
for _item in db:
|
| 103 |
+
if not set(anatomy_list).issubset(_item["label_list"]):
|
| 104 |
+
continue
|
| 105 |
+
|
| 106 |
+
# whether to keep this mask, default to be True.
|
| 107 |
+
keep_mask = True
|
| 108 |
+
|
| 109 |
+
for tumor_label in [23, 24, 26, 27, 128]:
|
| 110 |
+
# we skip those mask with tumors if users do not provide tumor label in anatomy_list
|
| 111 |
+
if tumor_label not in anatomy_list and tumor_label in _item["label_list"]:
|
| 112 |
+
keep_mask = False
|
| 113 |
+
|
| 114 |
+
if check_spacing_and_output_size:
|
| 115 |
+
# if the output_size and spacing are different with user's input, skip it
|
| 116 |
+
for axis in range(3):
|
| 117 |
+
if _item["dim"][axis] != output_size[axis] or _item["spacing"][axis] != spacing[axis]:
|
| 118 |
+
keep_mask = False
|
| 119 |
+
|
| 120 |
+
if keep_mask:
|
| 121 |
+
# if decide to keep this mask, we pack the information of this mask and add to final output.
|
| 122 |
+
candidate = {
|
| 123 |
+
"pseudo_label": os.path.join(mask_foldername, _item["pseudo_label_filename"]),
|
| 124 |
+
"spacing": _item["spacing"],
|
| 125 |
+
"dim": _item["dim"],
|
| 126 |
+
}
|
| 127 |
+
|
| 128 |
+
# Conditionally add the label to the candidate dictionary
|
| 129 |
+
if "label_filename" in _item:
|
| 130 |
+
candidate["label"] = os.path.join(mask_foldername, _item["label_filename"])
|
| 131 |
+
|
| 132 |
+
candidate_masks.append(candidate)
|
| 133 |
+
|
| 134 |
+
if len(candidate_masks) == 0 and not check_spacing_and_output_size:
|
| 135 |
+
raise ValueError("Cannot find body region with given anatomy list.")
|
| 136 |
+
|
| 137 |
+
return candidate_masks
|
scripts/quality_check.py
ADDED
|
@@ -0,0 +1,149 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
import numpy as np
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
def get_masked_data(label_data, image_data, labels):
|
| 16 |
+
"""
|
| 17 |
+
Extracts and returns the image data corresponding to specified labels within a 3D volume.
|
| 18 |
+
|
| 19 |
+
This function efficiently masks the `image_data` array based on the provided `labels` in the `label_data` array.
|
| 20 |
+
The function handles cases with both a large and small number of labels, optimizing performance accordingly.
|
| 21 |
+
|
| 22 |
+
Args:
|
| 23 |
+
label_data (np.ndarray): A NumPy array containing label data, representing different anatomical
|
| 24 |
+
regions or classes in a 3D medical image.
|
| 25 |
+
image_data (np.ndarray): A NumPy array containing the image data from which the relevant regions
|
| 26 |
+
will be extracted.
|
| 27 |
+
labels (list of int): A list of integers representing the label values to be used for masking.
|
| 28 |
+
|
| 29 |
+
Returns:
|
| 30 |
+
np.ndarray: A NumPy array containing the elements of `image_data` that correspond to the specified
|
| 31 |
+
labels in `label_data`. If no labels are provided, an empty array is returned.
|
| 32 |
+
|
| 33 |
+
Raises:
|
| 34 |
+
ValueError: If `image_data` and `label_data` do not have the same shape.
|
| 35 |
+
|
| 36 |
+
Example:
|
| 37 |
+
label_int_dict = {"liver": [1], "kidney": [5, 14]}
|
| 38 |
+
masked_data = get_masked_data(label_data, image_data, label_int_dict["kidney"])
|
| 39 |
+
"""
|
| 40 |
+
|
| 41 |
+
# Check if the shapes of image_data and label_data match
|
| 42 |
+
if image_data.shape != label_data.shape:
|
| 43 |
+
raise ValueError(
|
| 44 |
+
f"Shape mismatch: image_data has shape {image_data.shape}, "
|
| 45 |
+
f"but label_data has shape {label_data.shape}. They must be the same."
|
| 46 |
+
)
|
| 47 |
+
|
| 48 |
+
if not labels:
|
| 49 |
+
return np.array([]) # Return an empty array if no labels are provided
|
| 50 |
+
|
| 51 |
+
labels = list(set(labels)) # remove duplicate items
|
| 52 |
+
|
| 53 |
+
# Optimize performance based on the number of labels
|
| 54 |
+
num_label_acceleration_thresh = 3
|
| 55 |
+
if len(labels) >= num_label_acceleration_thresh:
|
| 56 |
+
# if many labels, np.isin is faster
|
| 57 |
+
mask = np.isin(label_data, labels)
|
| 58 |
+
else:
|
| 59 |
+
# Use logical OR to combine masks if the number of labels is small
|
| 60 |
+
mask = np.zeros_like(label_data, dtype=bool)
|
| 61 |
+
for label in labels:
|
| 62 |
+
mask = np.logical_or(mask, label_data == label)
|
| 63 |
+
|
| 64 |
+
# Retrieve the masked data
|
| 65 |
+
masked_data = image_data[mask.astype(bool)]
|
| 66 |
+
|
| 67 |
+
return masked_data
|
| 68 |
+
|
| 69 |
+
|
| 70 |
+
def is_outlier(statistics, image_data, label_data, label_int_dict):
|
| 71 |
+
"""
|
| 72 |
+
Perform a quality check on the generated image by comparing its statistics with precomputed thresholds.
|
| 73 |
+
|
| 74 |
+
Args:
|
| 75 |
+
statistics (dict): Dictionary containing precomputed statistics including mean +/- 3sigma ranges.
|
| 76 |
+
image_data (np.ndarray): The image data to be checked, typically a 3D NumPy array.
|
| 77 |
+
label_data (np.ndarray): The label data corresponding to the image, used for masking regions of interest.
|
| 78 |
+
label_int_dict (dict): Dictionary mapping label names to their corresponding integer lists.
|
| 79 |
+
e.g., label_int_dict = {"liver": [1], "kidney": [5, 14]}
|
| 80 |
+
|
| 81 |
+
Returns:
|
| 82 |
+
dict: A dictionary with labels as keys, each containing the quality check result,
|
| 83 |
+
including whether it's an outlier, the median value, and the thresholds used.
|
| 84 |
+
If no data is found for a label, the median value will be `None` and `is_outlier` will be `False`.
|
| 85 |
+
|
| 86 |
+
Example:
|
| 87 |
+
# Example input data
|
| 88 |
+
statistics = {
|
| 89 |
+
"liver": {
|
| 90 |
+
"sigma_6_low": -21.596463547885904,
|
| 91 |
+
"sigma_6_high": 156.27881534763367
|
| 92 |
+
},
|
| 93 |
+
"kidney": {
|
| 94 |
+
"sigma_6_low": -15.0,
|
| 95 |
+
"sigma_6_high": 120.0
|
| 96 |
+
}
|
| 97 |
+
}
|
| 98 |
+
label_int_dict = {
|
| 99 |
+
"liver": [1],
|
| 100 |
+
"kidney": [5, 14]
|
| 101 |
+
}
|
| 102 |
+
image_data = np.random.rand(100, 100, 100) # Replace with actual image data
|
| 103 |
+
label_data = np.zeros((100, 100, 100)) # Replace with actual label data
|
| 104 |
+
label_data[40:60, 40:60, 40:60] = 1 # Example region for liver
|
| 105 |
+
label_data[70:90, 70:90, 70:90] = 5 # Example region for kidney
|
| 106 |
+
result = is_outlier(statistics, image_data, label_data, label_int_dict)
|
| 107 |
+
"""
|
| 108 |
+
outlier_results = {}
|
| 109 |
+
|
| 110 |
+
for label_name, stats in statistics.items():
|
| 111 |
+
# Get the thresholds from the statistics
|
| 112 |
+
low_thresh = min(stats["sigma_6_low"], stats["percentile_0_5"]) # or "sigma_12_low" depending on your needs
|
| 113 |
+
high_thresh = max(stats["sigma_6_high"], stats["percentile_99_5"]) # or "sigma_12_high" depending on your needs
|
| 114 |
+
|
| 115 |
+
if label_name == "bone":
|
| 116 |
+
high_thresh = 1000.0
|
| 117 |
+
|
| 118 |
+
# Retrieve the corresponding label integers
|
| 119 |
+
labels = label_int_dict.get(label_name, [])
|
| 120 |
+
masked_data = get_masked_data(label_data, image_data, labels)
|
| 121 |
+
masked_data = masked_data[~np.isnan(masked_data)]
|
| 122 |
+
|
| 123 |
+
if len(masked_data) == 0 or masked_data.size == 0:
|
| 124 |
+
outlier_results[label_name] = {
|
| 125 |
+
"is_outlier": False,
|
| 126 |
+
"median_value": None,
|
| 127 |
+
"low_thresh": low_thresh,
|
| 128 |
+
"high_thresh": high_thresh,
|
| 129 |
+
}
|
| 130 |
+
continue
|
| 131 |
+
|
| 132 |
+
# Compute the median of the masked region
|
| 133 |
+
median_value = np.nanmedian(masked_data)
|
| 134 |
+
|
| 135 |
+
if np.isnan(median_value):
|
| 136 |
+
median_value = None
|
| 137 |
+
is_outlier = False
|
| 138 |
+
else:
|
| 139 |
+
# Determine if the median value is an outlier
|
| 140 |
+
is_outlier = median_value < low_thresh or median_value > high_thresh
|
| 141 |
+
|
| 142 |
+
outlier_results[label_name] = {
|
| 143 |
+
"is_outlier": is_outlier,
|
| 144 |
+
"median_value": median_value,
|
| 145 |
+
"low_thresh": low_thresh,
|
| 146 |
+
"high_thresh": high_thresh,
|
| 147 |
+
}
|
| 148 |
+
|
| 149 |
+
return outlier_results
|
scripts/rectified_flow.py
ADDED
|
@@ -0,0 +1,163 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from typing import Any
|
| 2 |
+
|
| 3 |
+
import numpy as np
|
| 4 |
+
import torch
|
| 5 |
+
from monai.networks.schedulers import Scheduler
|
| 6 |
+
from torch.distributions import LogisticNormal
|
| 7 |
+
|
| 8 |
+
# code modified from https://github.com/hpcaitech/Open-Sora/blob/main/opensora/schedulers/rf/rectified_flow.py
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
def timestep_transform(
|
| 12 |
+
t, input_img_size, base_img_size=32 * 32 * 32, scale=1.0, num_train_timesteps=1000, spatial_dim=3
|
| 13 |
+
):
|
| 14 |
+
t = t / num_train_timesteps
|
| 15 |
+
ratio_space = (input_img_size / base_img_size).pow(1.0 / spatial_dim)
|
| 16 |
+
|
| 17 |
+
ratio = ratio_space * scale
|
| 18 |
+
new_t = ratio * t / (1 + (ratio - 1) * t)
|
| 19 |
+
|
| 20 |
+
new_t = new_t * num_train_timesteps
|
| 21 |
+
return new_t
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
class RFlowScheduler(Scheduler):
|
| 25 |
+
def __init__(
|
| 26 |
+
self,
|
| 27 |
+
num_train_timesteps=1000,
|
| 28 |
+
num_inference_steps=10,
|
| 29 |
+
use_discrete_timesteps=False,
|
| 30 |
+
sample_method="uniform",
|
| 31 |
+
loc=0.0,
|
| 32 |
+
scale=1.0,
|
| 33 |
+
use_timestep_transform=False,
|
| 34 |
+
transform_scale=1.0,
|
| 35 |
+
steps_offset: int = 0,
|
| 36 |
+
):
|
| 37 |
+
self.num_train_timesteps = num_train_timesteps
|
| 38 |
+
self.num_inference_steps = num_inference_steps
|
| 39 |
+
self.use_discrete_timesteps = use_discrete_timesteps
|
| 40 |
+
|
| 41 |
+
# sample method
|
| 42 |
+
assert sample_method in ["uniform", "logit-normal"]
|
| 43 |
+
# assert (
|
| 44 |
+
# sample_method == "uniform" or not use_discrete_timesteps
|
| 45 |
+
# ), "Only uniform sampling is supported for discrete timesteps"
|
| 46 |
+
self.sample_method = sample_method
|
| 47 |
+
if sample_method == "logit-normal":
|
| 48 |
+
self.distribution = LogisticNormal(torch.tensor([loc]), torch.tensor([scale]))
|
| 49 |
+
self.sample_t = lambda x: self.distribution.sample((x.shape[0],))[:, 0].to(x.device)
|
| 50 |
+
|
| 51 |
+
# timestep transform
|
| 52 |
+
self.use_timestep_transform = use_timestep_transform
|
| 53 |
+
self.transform_scale = transform_scale
|
| 54 |
+
self.steps_offset = steps_offset
|
| 55 |
+
|
| 56 |
+
def add_noise(
|
| 57 |
+
self, original_samples: torch.FloatTensor, noise: torch.FloatTensor, timesteps: torch.IntTensor
|
| 58 |
+
) -> torch.FloatTensor:
|
| 59 |
+
"""
|
| 60 |
+
compatible with diffusers add_noise()
|
| 61 |
+
"""
|
| 62 |
+
timepoints = timesteps.float() / self.num_train_timesteps
|
| 63 |
+
timepoints = 1 - timepoints # [1,1/1000]
|
| 64 |
+
|
| 65 |
+
# timepoint (bsz) noise: (bsz, 4, frame, w ,h)
|
| 66 |
+
# expand timepoint to noise shape
|
| 67 |
+
timepoints = timepoints.unsqueeze(1).unsqueeze(1).unsqueeze(1).unsqueeze(1)
|
| 68 |
+
timepoints = timepoints.repeat(1, noise.shape[1], noise.shape[2], noise.shape[3], noise.shape[4])
|
| 69 |
+
|
| 70 |
+
return timepoints * original_samples + (1 - timepoints) * noise
|
| 71 |
+
|
| 72 |
+
def set_timesteps(
|
| 73 |
+
self,
|
| 74 |
+
num_inference_steps: int,
|
| 75 |
+
device: str | torch.device | None = None,
|
| 76 |
+
input_img_size: int | None = None,
|
| 77 |
+
base_img_size: int = 32 * 32 * 32,
|
| 78 |
+
) -> None:
|
| 79 |
+
"""
|
| 80 |
+
Sets the discrete timesteps used for the diffusion chain. Supporting function to be run before inference.
|
| 81 |
+
|
| 82 |
+
Args:
|
| 83 |
+
num_inference_steps: number of diffusion steps used when generating samples with a pre-trained model.
|
| 84 |
+
device: target device to put the data.
|
| 85 |
+
input_img_size: int, H*W*D of the image, used with self.use_timestep_transform is True.
|
| 86 |
+
base_img_size: int, reference H*W*D size, used with self.use_timestep_transform is True.
|
| 87 |
+
"""
|
| 88 |
+
if num_inference_steps > self.num_train_timesteps:
|
| 89 |
+
raise ValueError(
|
| 90 |
+
f"`num_inference_steps`: {num_inference_steps} cannot be larger than `self.num_train_timesteps`:"
|
| 91 |
+
f" {self.num_train_timesteps} as the unet model trained with this scheduler can only handle"
|
| 92 |
+
f" maximal {self.num_train_timesteps} timesteps."
|
| 93 |
+
)
|
| 94 |
+
|
| 95 |
+
self.num_inference_steps = num_inference_steps
|
| 96 |
+
# prepare timesteps
|
| 97 |
+
timesteps = [
|
| 98 |
+
(1.0 - i / self.num_inference_steps) * self.num_train_timesteps for i in range(self.num_inference_steps)
|
| 99 |
+
]
|
| 100 |
+
if self.use_discrete_timesteps:
|
| 101 |
+
timesteps = [int(round(t)) for t in timesteps]
|
| 102 |
+
if self.use_timestep_transform:
|
| 103 |
+
timesteps = [
|
| 104 |
+
timestep_transform(
|
| 105 |
+
t,
|
| 106 |
+
input_img_size=input_img_size,
|
| 107 |
+
base_img_size=base_img_size,
|
| 108 |
+
num_train_timesteps=self.num_train_timesteps,
|
| 109 |
+
)
|
| 110 |
+
for t in timesteps
|
| 111 |
+
]
|
| 112 |
+
timesteps = np.array(timesteps).astype(np.float16)
|
| 113 |
+
if self.use_discrete_timesteps:
|
| 114 |
+
timesteps = timesteps.astype(np.int64)
|
| 115 |
+
self.timesteps = torch.from_numpy(timesteps).to(device)
|
| 116 |
+
self.timesteps += self.steps_offset
|
| 117 |
+
print(self.timesteps)
|
| 118 |
+
|
| 119 |
+
def sample_timesteps(self, x_start):
|
| 120 |
+
if self.sample_method == "uniform":
|
| 121 |
+
t = torch.rand((x_start.shape[0],), device=x_start.device) * self.num_train_timesteps
|
| 122 |
+
elif self.sample_method == "logit-normal":
|
| 123 |
+
t = self.sample_t(x_start) * self.num_train_timesteps
|
| 124 |
+
|
| 125 |
+
if self.use_discrete_timesteps:
|
| 126 |
+
t = t.long()
|
| 127 |
+
|
| 128 |
+
if self.use_timestep_transform:
|
| 129 |
+
input_img_size = torch.prod(torch.tensor(x_start.shape[-3:]))
|
| 130 |
+
base_img_size = 32 * 32 * 32
|
| 131 |
+
t = timestep_transform(
|
| 132 |
+
t,
|
| 133 |
+
input_img_size=input_img_size,
|
| 134 |
+
base_img_size=base_img_size,
|
| 135 |
+
num_train_timesteps=self.num_train_timesteps,
|
| 136 |
+
)
|
| 137 |
+
|
| 138 |
+
return t
|
| 139 |
+
|
| 140 |
+
def step(
|
| 141 |
+
self, model_output: torch.Tensor, timestep: int, sample: torch.Tensor, next_timestep=None
|
| 142 |
+
) -> tuple[torch.Tensor, Any]:
|
| 143 |
+
"""
|
| 144 |
+
Predict the sample at the previous timestep. Core function to propagate the diffusion
|
| 145 |
+
process from the learned model outputs.
|
| 146 |
+
|
| 147 |
+
Args:
|
| 148 |
+
model_output: direct output from learned diffusion model.
|
| 149 |
+
timestep: current discrete timestep in the diffusion chain.
|
| 150 |
+
sample: current instance of sample being created by diffusion process.
|
| 151 |
+
Returns:
|
| 152 |
+
pred_prev_sample: Predicted previous sample
|
| 153 |
+
None
|
| 154 |
+
"""
|
| 155 |
+
v_pred = model_output
|
| 156 |
+
if next_timestep is None:
|
| 157 |
+
dt = 1.0 / self.num_inference_steps
|
| 158 |
+
else:
|
| 159 |
+
dt = timestep - next_timestep
|
| 160 |
+
dt = dt / self.num_train_timesteps
|
| 161 |
+
z = sample + v_pred * dt
|
| 162 |
+
|
| 163 |
+
return z, None
|
scripts/sample.py
ADDED
|
@@ -0,0 +1,1036 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
import json
|
| 13 |
+
import logging
|
| 14 |
+
import os
|
| 15 |
+
import random
|
| 16 |
+
import time
|
| 17 |
+
from datetime import datetime
|
| 18 |
+
|
| 19 |
+
import monai
|
| 20 |
+
import torch
|
| 21 |
+
from monai.data import MetaTensor
|
| 22 |
+
from monai.inferers.inferer import DiffusionInferer, SlidingWindowInferer
|
| 23 |
+
from monai.transforms import Compose, SaveImage
|
| 24 |
+
from monai.utils import set_determinism
|
| 25 |
+
from tqdm import tqdm
|
| 26 |
+
|
| 27 |
+
from .augmentation import augmentation
|
| 28 |
+
from .find_masks import find_masks
|
| 29 |
+
from .quality_check import is_outlier
|
| 30 |
+
from .utils import binarize_labels, dynamic_infer, general_mask_generation_post_process, remap_labels
|
| 31 |
+
|
| 32 |
+
modality_mapping = {
|
| 33 |
+
"unknown": 0,
|
| 34 |
+
"ct": 1,
|
| 35 |
+
"ct_wo_contrast": 2,
|
| 36 |
+
"ct_contrast": 3,
|
| 37 |
+
"mri": 8,
|
| 38 |
+
"mri_t1": 9,
|
| 39 |
+
"mri_t2": 10,
|
| 40 |
+
"mri_flair": 11,
|
| 41 |
+
"mri_pd": 12,
|
| 42 |
+
"mri_dwi": 13,
|
| 43 |
+
"mri_adc": 14,
|
| 44 |
+
"mri_ssfp": 15,
|
| 45 |
+
"mri_mra": 16,
|
| 46 |
+
} # current version only support "ct"
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
class ReconModel(torch.nn.Module):
|
| 50 |
+
"""
|
| 51 |
+
A PyTorch module for reconstructing images from latent representations.
|
| 52 |
+
|
| 53 |
+
Attributes:
|
| 54 |
+
autoencoder: The autoencoder model used for decoding.
|
| 55 |
+
scale_factor: Scaling factor applied to the input before decoding.
|
| 56 |
+
"""
|
| 57 |
+
|
| 58 |
+
def __init__(self, autoencoder, scale_factor):
|
| 59 |
+
super().__init__()
|
| 60 |
+
self.autoencoder = autoencoder
|
| 61 |
+
self.scale_factor = scale_factor
|
| 62 |
+
|
| 63 |
+
def forward(self, z):
|
| 64 |
+
"""
|
| 65 |
+
Decode the input latent representation to an image.
|
| 66 |
+
|
| 67 |
+
Args:
|
| 68 |
+
z (torch.Tensor): The input latent representation.
|
| 69 |
+
|
| 70 |
+
Returns:
|
| 71 |
+
torch.Tensor: The reconstructed image.
|
| 72 |
+
"""
|
| 73 |
+
recon_pt_nda = self.autoencoder.decode_stage_2_outputs(z / self.scale_factor)
|
| 74 |
+
return recon_pt_nda
|
| 75 |
+
|
| 76 |
+
|
| 77 |
+
def initialize_noise_latents(latent_shape, device):
|
| 78 |
+
"""
|
| 79 |
+
Initialize random noise latents for image generation with float16.
|
| 80 |
+
|
| 81 |
+
Args:
|
| 82 |
+
latent_shape (tuple): The shape of the latent space.
|
| 83 |
+
device (torch.device): The device to create the tensor on.
|
| 84 |
+
|
| 85 |
+
Returns:
|
| 86 |
+
torch.Tensor: Initialized noise latents.
|
| 87 |
+
"""
|
| 88 |
+
return torch.randn([1] + list(latent_shape)).half().to(device)
|
| 89 |
+
|
| 90 |
+
|
| 91 |
+
def ldm_conditional_sample_one_mask(
|
| 92 |
+
autoencoder,
|
| 93 |
+
diffusion_unet,
|
| 94 |
+
noise_scheduler,
|
| 95 |
+
scale_factor,
|
| 96 |
+
anatomy_size,
|
| 97 |
+
device,
|
| 98 |
+
latent_shape,
|
| 99 |
+
label_dict_remap_json,
|
| 100 |
+
num_inference_steps=1000,
|
| 101 |
+
autoencoder_sliding_window_infer_size=(96, 96, 96),
|
| 102 |
+
autoencoder_sliding_window_infer_overlap=0.6667,
|
| 103 |
+
):
|
| 104 |
+
"""
|
| 105 |
+
Generate a single synthetic mask using a latent diffusion model.
|
| 106 |
+
|
| 107 |
+
Args:
|
| 108 |
+
autoencoder (nn.Module): The autoencoder model.
|
| 109 |
+
diffusion_unet (nn.Module): The diffusion U-Net model.
|
| 110 |
+
noise_scheduler: The noise scheduler for the diffusion process.
|
| 111 |
+
scale_factor (float): Scaling factor for the latent space.
|
| 112 |
+
anatomy_size (torch.Tensor): Tensor specifying the desired anatomy sizes.
|
| 113 |
+
device (torch.device): The device to run the computation on.
|
| 114 |
+
latent_shape (tuple): The shape of the latent space.
|
| 115 |
+
label_dict_remap_json (str): Path to the JSON file for label remapping.
|
| 116 |
+
num_inference_steps (int): Number of inference steps for the diffusion process.
|
| 117 |
+
autoencoder_sliding_window_infer_size (list, optional): Size of the sliding window for inference. Defaults to [96, 96, 96].
|
| 118 |
+
autoencoder_sliding_window_infer_overlap (float, optional): Overlap ratio for sliding window inference. Defaults to 0.6667.
|
| 119 |
+
|
| 120 |
+
Returns:
|
| 121 |
+
torch.Tensor: The generated synthetic mask.
|
| 122 |
+
"""
|
| 123 |
+
recon_model = ReconModel(autoencoder=autoencoder, scale_factor=scale_factor).to(device)
|
| 124 |
+
|
| 125 |
+
with torch.no_grad(), torch.amp.autocast("cuda"):
|
| 126 |
+
# Generate random noise
|
| 127 |
+
latents = initialize_noise_latents(latent_shape, device)
|
| 128 |
+
anatomy_size = torch.FloatTensor(anatomy_size).unsqueeze(0).unsqueeze(0).half().to(device)
|
| 129 |
+
# synthesize latents
|
| 130 |
+
noise_scheduler.set_timesteps(num_inference_steps=num_inference_steps)
|
| 131 |
+
inferer_ddpm = DiffusionInferer(noise_scheduler)
|
| 132 |
+
latents = inferer_ddpm.sample(
|
| 133 |
+
input_noise=latents,
|
| 134 |
+
diffusion_model=diffusion_unet,
|
| 135 |
+
scheduler=noise_scheduler,
|
| 136 |
+
verbose=True,
|
| 137 |
+
conditioning=anatomy_size.to(device),
|
| 138 |
+
)
|
| 139 |
+
# decode latents to synthesized masks
|
| 140 |
+
inferer = SlidingWindowInferer(
|
| 141 |
+
roi_size=autoencoder_sliding_window_infer_size,
|
| 142 |
+
sw_batch_size=1,
|
| 143 |
+
progress=True,
|
| 144 |
+
mode="gaussian",
|
| 145 |
+
overlap=autoencoder_sliding_window_infer_overlap,
|
| 146 |
+
device=torch.device("cpu"),
|
| 147 |
+
sw_device=device,
|
| 148 |
+
)
|
| 149 |
+
synthetic_mask = dynamic_infer(inferer, recon_model, latents)
|
| 150 |
+
synthetic_mask = torch.softmax(synthetic_mask, dim=1)
|
| 151 |
+
synthetic_mask = torch.argmax(synthetic_mask, dim=1, keepdim=True)
|
| 152 |
+
# mapping raw index to 132 labels
|
| 153 |
+
synthetic_mask = remap_labels(synthetic_mask, label_dict_remap_json)
|
| 154 |
+
|
| 155 |
+
# post process
|
| 156 |
+
data = synthetic_mask.squeeze().cpu().detach().numpy()
|
| 157 |
+
|
| 158 |
+
labels = [23, 24, 26, 27, 128]
|
| 159 |
+
target_tumor_label = None
|
| 160 |
+
for index, size in enumerate(anatomy_size[0, 0, 5:10]):
|
| 161 |
+
if size.item() != -1.0:
|
| 162 |
+
target_tumor_label = labels[index]
|
| 163 |
+
|
| 164 |
+
logging.info(f"target_tumor_label for postprocess:{target_tumor_label}")
|
| 165 |
+
data = general_mask_generation_post_process(data, target_tumor_label=target_tumor_label, device=device)
|
| 166 |
+
synthetic_mask = torch.from_numpy(data).unsqueeze(0).unsqueeze(0).to(device)
|
| 167 |
+
|
| 168 |
+
return synthetic_mask
|
| 169 |
+
|
| 170 |
+
|
| 171 |
+
def ldm_conditional_sample_one_image(
|
| 172 |
+
autoencoder,
|
| 173 |
+
diffusion_unet,
|
| 174 |
+
controlnet,
|
| 175 |
+
noise_scheduler,
|
| 176 |
+
scale_factor,
|
| 177 |
+
device,
|
| 178 |
+
combine_label_or,
|
| 179 |
+
modality_tensor,
|
| 180 |
+
spacing_tensor,
|
| 181 |
+
latent_shape,
|
| 182 |
+
output_size,
|
| 183 |
+
noise_factor,
|
| 184 |
+
num_inference_steps=1000,
|
| 185 |
+
autoencoder_sliding_window_infer_size=(96, 96, 96),
|
| 186 |
+
autoencoder_sliding_window_infer_overlap=0.6667,
|
| 187 |
+
):
|
| 188 |
+
"""
|
| 189 |
+
Generate a single synthetic image using a latent diffusion model with controlnet.
|
| 190 |
+
|
| 191 |
+
Args:
|
| 192 |
+
autoencoder (nn.Module): The autoencoder model.
|
| 193 |
+
diffusion_unet (nn.Module): The diffusion U-Net model.
|
| 194 |
+
controlnet (nn.Module): The controlnet model.
|
| 195 |
+
noise_scheduler: The noise scheduler for the diffusion process.
|
| 196 |
+
scale_factor (float): Scaling factor for the latent space.
|
| 197 |
+
device (torch.device): The device to run the computation on.
|
| 198 |
+
combine_label_or (torch.Tensor): The combined label tensor.
|
| 199 |
+
spacing_tensor (torch.Tensor): Tensor specifying the spacing.
|
| 200 |
+
latent_shape (tuple): The shape of the latent space.
|
| 201 |
+
output_size (tuple): The desired output size of the image.
|
| 202 |
+
noise_factor (float): Factor to scale the initial noise.
|
| 203 |
+
num_inference_steps (int): Number of inference steps for the diffusion process.
|
| 204 |
+
autoencoder_sliding_window_infer_size (list, optional): Size of the sliding window for inference. Defaults to [96, 96, 96].
|
| 205 |
+
autoencoder_sliding_window_infer_overlap (float, optional): Overlap ratio for sliding window inference. Defaults to 0.6667.
|
| 206 |
+
|
| 207 |
+
Returns:
|
| 208 |
+
tuple: A tuple containing the synthetic image and its corresponding label.
|
| 209 |
+
"""
|
| 210 |
+
# CT image intensity range
|
| 211 |
+
a_min = -1000
|
| 212 |
+
a_max = 1000
|
| 213 |
+
# autoencoder output intensity range
|
| 214 |
+
b_min = 0.0
|
| 215 |
+
b_max = 1
|
| 216 |
+
|
| 217 |
+
recon_model = ReconModel(autoencoder=autoencoder, scale_factor=scale_factor).to(device)
|
| 218 |
+
|
| 219 |
+
with torch.no_grad(), torch.amp.autocast("cuda", enabled=True):
|
| 220 |
+
logging.info("---- Start generating latent features... ----")
|
| 221 |
+
start_time = time.time()
|
| 222 |
+
# generate segmentation mask
|
| 223 |
+
combine_label = combine_label_or.to(device)
|
| 224 |
+
if (
|
| 225 |
+
output_size[0] != combine_label.shape[2]
|
| 226 |
+
or output_size[1] != combine_label.shape[3]
|
| 227 |
+
or output_size[2] != combine_label.shape[4]
|
| 228 |
+
):
|
| 229 |
+
logging.info(
|
| 230 |
+
"output_size is not a desired value. Need to interpolate the mask to match "
|
| 231 |
+
"with output_size. The result image will be very low quality."
|
| 232 |
+
)
|
| 233 |
+
combine_label = torch.nn.functional.interpolate(combine_label, size=output_size, mode="nearest")
|
| 234 |
+
|
| 235 |
+
controlnet_cond_vis = binarize_labels(combine_label.as_tensor().long()).half()
|
| 236 |
+
|
| 237 |
+
# Generate random noise
|
| 238 |
+
latents = initialize_noise_latents(latent_shape, device) * noise_factor
|
| 239 |
+
|
| 240 |
+
# synthesize latents
|
| 241 |
+
noise_scheduler.set_timesteps(
|
| 242 |
+
num_inference_steps=num_inference_steps, input_img_size=torch.prod(torch.tensor(latent_shape[-3:]))
|
| 243 |
+
)
|
| 244 |
+
# synthesize latents
|
| 245 |
+
guidance_scale = 0 # API for classifier-free guidence, not used in this version
|
| 246 |
+
all_next_timesteps = torch.cat(
|
| 247 |
+
(noise_scheduler.timesteps[1:], torch.tensor([0], dtype=noise_scheduler.timesteps.dtype))
|
| 248 |
+
)
|
| 249 |
+
for t, next_t in tqdm(
|
| 250 |
+
zip(noise_scheduler.timesteps, all_next_timesteps),
|
| 251 |
+
total=min(len(noise_scheduler.timesteps), len(all_next_timesteps)),
|
| 252 |
+
):
|
| 253 |
+
timesteps = torch.Tensor((t,)).to(device)
|
| 254 |
+
if guidance_scale == 0:
|
| 255 |
+
down_block_res_samples, mid_block_res_sample = controlnet(
|
| 256 |
+
x=latents, timesteps=timesteps, controlnet_cond=controlnet_cond_vis, class_labels=modality_tensor
|
| 257 |
+
)
|
| 258 |
+
predicted_velocity = diffusion_unet(
|
| 259 |
+
x=latents,
|
| 260 |
+
timesteps=timesteps,
|
| 261 |
+
spacing_tensor=spacing_tensor,
|
| 262 |
+
class_labels=modality_tensor,
|
| 263 |
+
down_block_additional_residuals=down_block_res_samples,
|
| 264 |
+
mid_block_additional_residual=mid_block_res_sample,
|
| 265 |
+
)
|
| 266 |
+
else:
|
| 267 |
+
down_block_res_samples, mid_block_res_sample = controlnet(
|
| 268 |
+
x=torch.cat([latents] * 2),
|
| 269 |
+
timesteps=torch.cat([timesteps] * 2),
|
| 270 |
+
controlnet_cond=torch.cat([controlnet_cond_vis] * 2),
|
| 271 |
+
class_labels=torch.cat([modality_tensor, torch.zeros_like(modality_tensor)]),
|
| 272 |
+
)
|
| 273 |
+
model_t, model_uncond = diffusion_unet(
|
| 274 |
+
x=torch.cat([latents] * 2),
|
| 275 |
+
timesteps=timesteps,
|
| 276 |
+
spacing_tensor=torch.cat([timesteps] * 2),
|
| 277 |
+
class_labels=torch.cat([modality_tensor, torch.zeros_like(modality_tensor)]),
|
| 278 |
+
down_block_additional_residuals=down_block_res_samples,
|
| 279 |
+
mid_block_additional_residual=mid_block_res_sample,
|
| 280 |
+
).chunk(2)
|
| 281 |
+
predicted_velocity = model_uncond + guidance_scale * (model_t - model_uncond)
|
| 282 |
+
latents, _ = noise_scheduler.step(predicted_velocity, t, latents, next_timestep=next_t)
|
| 283 |
+
end_time = time.time()
|
| 284 |
+
logging.info(f"---- Latent features generation time: {end_time - start_time} seconds ----")
|
| 285 |
+
del predicted_velocity
|
| 286 |
+
torch.cuda.empty_cache()
|
| 287 |
+
|
| 288 |
+
# decode latents to synthesized images
|
| 289 |
+
logging.info("---- Start decoding latent features into images... ----")
|
| 290 |
+
inferer = SlidingWindowInferer(
|
| 291 |
+
roi_size=autoencoder_sliding_window_infer_size,
|
| 292 |
+
sw_batch_size=1,
|
| 293 |
+
progress=True,
|
| 294 |
+
mode="gaussian",
|
| 295 |
+
overlap=autoencoder_sliding_window_infer_overlap,
|
| 296 |
+
device=torch.device("cpu"),
|
| 297 |
+
sw_device=device,
|
| 298 |
+
)
|
| 299 |
+
start_time = time.time()
|
| 300 |
+
synthetic_images = dynamic_infer(inferer, recon_model, latents)
|
| 301 |
+
synthetic_images = torch.clip(synthetic_images, b_min, b_max).cpu()
|
| 302 |
+
end_time = time.time()
|
| 303 |
+
logging.info(f"---- Image decoding time: {end_time - start_time} seconds ----")
|
| 304 |
+
|
| 305 |
+
# post processing:
|
| 306 |
+
# project output to [0, 1]
|
| 307 |
+
synthetic_images = (synthetic_images - b_min) / (b_max - b_min)
|
| 308 |
+
# project output to [-1000, 1000]
|
| 309 |
+
synthetic_images = synthetic_images * (a_max - a_min) + a_min
|
| 310 |
+
# regularize background intensities
|
| 311 |
+
synthetic_images = crop_img_body_mask(synthetic_images, combine_label)
|
| 312 |
+
torch.cuda.empty_cache()
|
| 313 |
+
|
| 314 |
+
return synthetic_images, combine_label
|
| 315 |
+
|
| 316 |
+
|
| 317 |
+
def filter_mask_with_organs(combine_label, anatomy_list):
|
| 318 |
+
"""
|
| 319 |
+
Filter a mask to only include specified organs.
|
| 320 |
+
|
| 321 |
+
Args:
|
| 322 |
+
combine_label (torch.Tensor): The input mask.
|
| 323 |
+
anatomy_list (list): List of organ labels to keep.
|
| 324 |
+
|
| 325 |
+
Returns:
|
| 326 |
+
torch.Tensor: The filtered mask.
|
| 327 |
+
"""
|
| 328 |
+
# final output mask file has shape of output_size, contains labels in anatomy_list
|
| 329 |
+
# it is already interpolated to target size
|
| 330 |
+
combine_label = combine_label.long()
|
| 331 |
+
# filter out the organs that are not in anatomy_list
|
| 332 |
+
for i in range(len(anatomy_list)):
|
| 333 |
+
organ = anatomy_list[i]
|
| 334 |
+
# replace it with a negative value so it will get mixed
|
| 335 |
+
combine_label[combine_label == organ] = -(i + 1)
|
| 336 |
+
# zero-out voxels with value not in anatomy_list
|
| 337 |
+
combine_label[combine_label > 0] = 0
|
| 338 |
+
# output positive values
|
| 339 |
+
combine_label = -combine_label
|
| 340 |
+
return combine_label
|
| 341 |
+
|
| 342 |
+
|
| 343 |
+
def crop_img_body_mask(synthetic_images, combine_label):
|
| 344 |
+
"""
|
| 345 |
+
Crop the synthetic image using a body mask.
|
| 346 |
+
|
| 347 |
+
Args:
|
| 348 |
+
synthetic_images (torch.Tensor): The synthetic images.
|
| 349 |
+
combine_label (torch.Tensor): The body mask.
|
| 350 |
+
|
| 351 |
+
Returns:
|
| 352 |
+
torch.Tensor: The cropped synthetic images.
|
| 353 |
+
"""
|
| 354 |
+
synthetic_images[combine_label == 0] = -1000
|
| 355 |
+
return synthetic_images
|
| 356 |
+
|
| 357 |
+
|
| 358 |
+
def check_input(body_region, anatomy_list, label_dict_json, output_size, spacing, controllable_anatomy_size):
|
| 359 |
+
"""
|
| 360 |
+
Validate input parameters for image generation.
|
| 361 |
+
|
| 362 |
+
Args:
|
| 363 |
+
body_region (list): List of body regions.
|
| 364 |
+
anatomy_list (list): List of anatomical structures.
|
| 365 |
+
label_dict_json (str): Path to the label dictionary JSON file.
|
| 366 |
+
output_size (tuple): Desired output size of the image.
|
| 367 |
+
spacing (tuple): Desired voxel spacing.
|
| 368 |
+
controllable_anatomy_size (list): List of tuples specifying controllable anatomy sizes.
|
| 369 |
+
|
| 370 |
+
Raises:
|
| 371 |
+
ValueError: If any input parameter is invalid.
|
| 372 |
+
"""
|
| 373 |
+
# check output_size and spacing format
|
| 374 |
+
if output_size[0] != output_size[1]:
|
| 375 |
+
raise ValueError(f"The first two components of output_size need to be equal, yet got {output_size}.")
|
| 376 |
+
if (output_size[0] not in [256, 384, 512]) or (output_size[2] not in [128, 256, 384, 512, 640, 768]):
|
| 377 |
+
raise ValueError(
|
| 378 |
+
(
|
| 379 |
+
"The output_size[0] have to be chosen from [256, 384, 512], and output_size[2] "
|
| 380 |
+
f"have to be chosen from [128, 256, 384, 512, 640, 768], yet got {output_size}."
|
| 381 |
+
)
|
| 382 |
+
)
|
| 383 |
+
|
| 384 |
+
if spacing[0] != spacing[1]:
|
| 385 |
+
raise ValueError(f"The first two components of spacing need to be equal, yet got {spacing}.")
|
| 386 |
+
if spacing[0] < 0.5 or spacing[0] > 3.0 or spacing[2] < 0.5 or spacing[2] > 5.0:
|
| 387 |
+
raise ValueError(
|
| 388 |
+
f"spacing[0] have to be between 0.5 and 3.0 mm, spacing[2] have to be between 0.5 and 5.0 mm, yet got {spacing}."
|
| 389 |
+
)
|
| 390 |
+
|
| 391 |
+
if (
|
| 392 |
+
output_size[0] * spacing[0] < 256
|
| 393 |
+
or output_size[2] * spacing[2] < 128
|
| 394 |
+
or output_size[0] * spacing[0] > 640
|
| 395 |
+
or output_size[2] * spacing[2] > 2000
|
| 396 |
+
):
|
| 397 |
+
fov = [output_size[axis] * spacing[axis] for axis in range(3)]
|
| 398 |
+
raise ValueError(
|
| 399 |
+
(
|
| 400 |
+
f"`'spacing'({spacing}mm) and 'output_size'({output_size}) together decide the output field of view (FOV). "
|
| 401 |
+
f"The FOV will be {fov}mm. We recommend the FOV in x and y axis to be at least 256mm for head, and at least "
|
| 402 |
+
"384mm for other body regions like abdomen, and less than 640mm. "
|
| 403 |
+
"For z-axis, we require it to be at least 128mm and less than 2000mm."
|
| 404 |
+
)
|
| 405 |
+
)
|
| 406 |
+
|
| 407 |
+
# check controllable_anatomy_size format
|
| 408 |
+
if len(controllable_anatomy_size) > 10:
|
| 409 |
+
raise ValueError(
|
| 410 |
+
(
|
| 411 |
+
"The output_size[0] have to be chosen from [256, 384, 512], and output_size[2] "
|
| 412 |
+
f"have to be chosen from [128, 256, 384, 512, 640, 768], yet got {output_size}."
|
| 413 |
+
)
|
| 414 |
+
)
|
| 415 |
+
available_controllable_organ = ["liver", "gallbladder", "stomach", "pancreas", "colon"]
|
| 416 |
+
available_controllable_tumor = [
|
| 417 |
+
"hepatic tumor",
|
| 418 |
+
"bone lesion",
|
| 419 |
+
"lung tumor",
|
| 420 |
+
"colon cancer primaries",
|
| 421 |
+
"pancreatic tumor",
|
| 422 |
+
]
|
| 423 |
+
available_controllable_anatomy = available_controllable_organ + available_controllable_tumor
|
| 424 |
+
controllable_tumor = []
|
| 425 |
+
controllable_organ = []
|
| 426 |
+
for controllable_anatomy_size_pair in controllable_anatomy_size:
|
| 427 |
+
if controllable_anatomy_size_pair[0] not in available_controllable_anatomy:
|
| 428 |
+
raise ValueError(
|
| 429 |
+
(
|
| 430 |
+
f"The controllable_anatomy have to be chosen from {available_controllable_anatomy}, "
|
| 431 |
+
f"yet got {controllable_anatomy_size_pair[0]}."
|
| 432 |
+
)
|
| 433 |
+
)
|
| 434 |
+
if controllable_anatomy_size_pair[0] in available_controllable_tumor:
|
| 435 |
+
controllable_tumor += [controllable_anatomy_size_pair[0]]
|
| 436 |
+
if controllable_anatomy_size_pair[0] in available_controllable_organ:
|
| 437 |
+
controllable_organ += [controllable_anatomy_size_pair[0]]
|
| 438 |
+
if controllable_anatomy_size_pair[1] == -1:
|
| 439 |
+
continue
|
| 440 |
+
if controllable_anatomy_size_pair[1] < 0 or controllable_anatomy_size_pair[1] > 1.0:
|
| 441 |
+
raise ValueError(
|
| 442 |
+
(
|
| 443 |
+
"The controllable size scale have to be between 0 and 1,0, or equal to -1, "
|
| 444 |
+
f"yet got {controllable_anatomy_size_pair[1]}."
|
| 445 |
+
)
|
| 446 |
+
)
|
| 447 |
+
if len(controllable_tumor + controllable_organ) != len(list(set(controllable_tumor + controllable_organ))):
|
| 448 |
+
raise ValueError(f"Please do not repeat controllable_anatomy. Got {controllable_tumor + controllable_organ}.")
|
| 449 |
+
if len(controllable_tumor) > 1:
|
| 450 |
+
raise ValueError(f"Only one controllable tumor is supported. Yet got {controllable_tumor}.")
|
| 451 |
+
|
| 452 |
+
if len(controllable_anatomy_size) > 0:
|
| 453 |
+
logging.info(
|
| 454 |
+
(
|
| 455 |
+
"`controllable_anatomy_size` is not empty.\nWe will ignore `body_region` and `anatomy_list` "
|
| 456 |
+
f"and synthesize based on `controllable_anatomy_size`: ({controllable_anatomy_size})."
|
| 457 |
+
)
|
| 458 |
+
)
|
| 459 |
+
else:
|
| 460 |
+
logging.info(
|
| 461 |
+
(f"`controllable_anatomy_size` is empty.\nWe will synthesize based on `anatomy_list`: ({anatomy_list}).")
|
| 462 |
+
)
|
| 463 |
+
# check body_region format
|
| 464 |
+
available_body_region = ["head", "chest", "thorax", "abdomen", "pelvis", "lower"]
|
| 465 |
+
for region in body_region:
|
| 466 |
+
if region not in available_body_region:
|
| 467 |
+
raise ValueError(
|
| 468 |
+
f"The components in body_region have to be chosen from {available_body_region}, yet got {region}."
|
| 469 |
+
)
|
| 470 |
+
|
| 471 |
+
# check anatomy_list format
|
| 472 |
+
with open(label_dict_json) as f:
|
| 473 |
+
label_dict = json.load(f)
|
| 474 |
+
for anatomy in anatomy_list:
|
| 475 |
+
if anatomy not in label_dict.keys():
|
| 476 |
+
raise ValueError(
|
| 477 |
+
f"The components in anatomy_list have to be chosen from {label_dict.keys()}, yet got {anatomy}."
|
| 478 |
+
)
|
| 479 |
+
logging.info(f"The generate results will have voxel size to be {spacing} mm, volume size to be {output_size}.")
|
| 480 |
+
|
| 481 |
+
return
|
| 482 |
+
|
| 483 |
+
|
| 484 |
+
class LDMSampler:
|
| 485 |
+
"""
|
| 486 |
+
A sampler class for generating synthetic medical images and masks using latent diffusion models.
|
| 487 |
+
|
| 488 |
+
Attributes:
|
| 489 |
+
Various attributes related to model configuration, input parameters, and generation settings.
|
| 490 |
+
"""
|
| 491 |
+
|
| 492 |
+
def __init__(
|
| 493 |
+
self,
|
| 494 |
+
body_region,
|
| 495 |
+
anatomy_list,
|
| 496 |
+
modality,
|
| 497 |
+
all_mask_files_json,
|
| 498 |
+
all_anatomy_size_condtions_json,
|
| 499 |
+
all_mask_files_base_dir,
|
| 500 |
+
label_dict_json,
|
| 501 |
+
label_dict_remap_json,
|
| 502 |
+
autoencoder,
|
| 503 |
+
diffusion_unet,
|
| 504 |
+
controlnet,
|
| 505 |
+
noise_scheduler,
|
| 506 |
+
scale_factor,
|
| 507 |
+
mask_generation_autoencoder,
|
| 508 |
+
mask_generation_diffusion_unet,
|
| 509 |
+
mask_generation_scale_factor,
|
| 510 |
+
mask_generation_noise_scheduler,
|
| 511 |
+
device,
|
| 512 |
+
latent_shape,
|
| 513 |
+
mask_generation_latent_shape,
|
| 514 |
+
output_size,
|
| 515 |
+
output_dir,
|
| 516 |
+
controllable_anatomy_size,
|
| 517 |
+
image_output_ext=".nii.gz",
|
| 518 |
+
label_output_ext=".nii.gz",
|
| 519 |
+
real_img_median_statistics="./configs/image_median_statistics.json",
|
| 520 |
+
spacing=(1, 1, 1),
|
| 521 |
+
num_inference_steps=None,
|
| 522 |
+
mask_generation_num_inference_steps=None,
|
| 523 |
+
random_seed=None,
|
| 524 |
+
autoencoder_sliding_window_infer_size=(96, 96, 96),
|
| 525 |
+
autoencoder_sliding_window_infer_overlap=0.6667,
|
| 526 |
+
) -> None:
|
| 527 |
+
"""
|
| 528 |
+
Initialize the LDMSampler with various parameters and models.
|
| 529 |
+
|
| 530 |
+
Args:
|
| 531 |
+
Various parameters related to model configuration, input settings, and output specifications.
|
| 532 |
+
"""
|
| 533 |
+
self.random_seed = random_seed
|
| 534 |
+
if random_seed is not None:
|
| 535 |
+
set_determinism(seed=random_seed)
|
| 536 |
+
|
| 537 |
+
with open(label_dict_json, "r") as f:
|
| 538 |
+
label_dict = json.load(f)
|
| 539 |
+
self.all_anatomy_size_condtions_json = all_anatomy_size_condtions_json
|
| 540 |
+
|
| 541 |
+
# intialize variables
|
| 542 |
+
self.body_region = body_region
|
| 543 |
+
self.anatomy_list = [label_dict[organ] for organ in anatomy_list]
|
| 544 |
+
self.modality_int = modality_mapping[modality]
|
| 545 |
+
self.all_mask_files_json = all_mask_files_json
|
| 546 |
+
self.data_root = all_mask_files_base_dir
|
| 547 |
+
self.label_dict_remap_json = label_dict_remap_json
|
| 548 |
+
self.autoencoder = autoencoder
|
| 549 |
+
self.diffusion_unet = diffusion_unet
|
| 550 |
+
self.controlnet = controlnet
|
| 551 |
+
self.noise_scheduler = noise_scheduler
|
| 552 |
+
self.scale_factor = scale_factor
|
| 553 |
+
self.mask_generation_autoencoder = mask_generation_autoencoder
|
| 554 |
+
self.mask_generation_diffusion_unet = mask_generation_diffusion_unet
|
| 555 |
+
self.mask_generation_scale_factor = mask_generation_scale_factor
|
| 556 |
+
self.mask_generation_noise_scheduler = mask_generation_noise_scheduler
|
| 557 |
+
self.device = device
|
| 558 |
+
self.latent_shape = latent_shape
|
| 559 |
+
self.mask_generation_latent_shape = mask_generation_latent_shape
|
| 560 |
+
self.output_size = output_size
|
| 561 |
+
self.output_dir = output_dir
|
| 562 |
+
self.noise_factor = 1.0
|
| 563 |
+
self.controllable_anatomy_size = controllable_anatomy_size
|
| 564 |
+
if len(self.controllable_anatomy_size):
|
| 565 |
+
logging.info("controllable_anatomy_size is given, mask generation is triggered!")
|
| 566 |
+
# overwrite the anatomy_list by given organs in self.controllable_anatomy_size
|
| 567 |
+
self.anatomy_list = [label_dict[organ_and_size[0]] for organ_and_size in self.controllable_anatomy_size]
|
| 568 |
+
self.image_output_ext = image_output_ext
|
| 569 |
+
self.label_output_ext = label_output_ext
|
| 570 |
+
# Set the default value for number of inference steps to 1000
|
| 571 |
+
self.num_inference_steps = num_inference_steps if num_inference_steps is not None else 1000
|
| 572 |
+
self.mask_generation_num_inference_steps = (
|
| 573 |
+
mask_generation_num_inference_steps if mask_generation_num_inference_steps is not None else 1000
|
| 574 |
+
)
|
| 575 |
+
|
| 576 |
+
if any(size % 16 != 0 for size in autoencoder_sliding_window_infer_size):
|
| 577 |
+
raise ValueError(
|
| 578 |
+
f"autoencoder_sliding_window_infer_size must be divisible by 16.\n Got {autoencoder_sliding_window_infer_size}"
|
| 579 |
+
)
|
| 580 |
+
if not (0 <= autoencoder_sliding_window_infer_overlap <= 1):
|
| 581 |
+
raise ValueError(
|
| 582 |
+
(
|
| 583 |
+
"Value of autoencoder_sliding_window_infer_overlap must be between 0 "
|
| 584 |
+
f"and 1.\n Got {autoencoder_sliding_window_infer_overlap}"
|
| 585 |
+
)
|
| 586 |
+
)
|
| 587 |
+
self.autoencoder_sliding_window_infer_size = autoencoder_sliding_window_infer_size
|
| 588 |
+
self.autoencoder_sliding_window_infer_overlap = autoencoder_sliding_window_infer_overlap
|
| 589 |
+
|
| 590 |
+
# quality check args
|
| 591 |
+
self.max_try_time = 3 # if not pass quality check, will try self.max_try_time times
|
| 592 |
+
with open(real_img_median_statistics, "r") as json_file:
|
| 593 |
+
self.median_statistics = json.load(json_file)
|
| 594 |
+
self.label_int_dict = {
|
| 595 |
+
"liver": [1],
|
| 596 |
+
"spleen": [3],
|
| 597 |
+
"pancreas": [4],
|
| 598 |
+
"kidney": [5, 14],
|
| 599 |
+
"lung": [28, 29, 30, 31, 31],
|
| 600 |
+
"brain": [22],
|
| 601 |
+
"hepatic tumor": [26],
|
| 602 |
+
"bone lesion": [128],
|
| 603 |
+
"lung tumor": [23],
|
| 604 |
+
"colon cancer primaries": [27],
|
| 605 |
+
"pancreatic tumor": [24],
|
| 606 |
+
"bone": list(range(33, 57)) + list(range(63, 98)) + [120, 122, 127],
|
| 607 |
+
}
|
| 608 |
+
|
| 609 |
+
# networks
|
| 610 |
+
self.autoencoder.eval()
|
| 611 |
+
self.diffusion_unet.eval()
|
| 612 |
+
self.controlnet.eval()
|
| 613 |
+
self.mask_generation_autoencoder.eval()
|
| 614 |
+
self.mask_generation_diffusion_unet.eval()
|
| 615 |
+
|
| 616 |
+
self.spacing = spacing
|
| 617 |
+
|
| 618 |
+
self.val_transforms = Compose(
|
| 619 |
+
[
|
| 620 |
+
monai.transforms.LoadImaged(keys=["pseudo_label"]),
|
| 621 |
+
monai.transforms.EnsureChannelFirstd(keys=["pseudo_label"]),
|
| 622 |
+
monai.transforms.Orientationd(keys=["pseudo_label"], axcodes="RAS"),
|
| 623 |
+
monai.transforms.EnsureTyped(keys=["pseudo_label"], dtype=torch.uint8),
|
| 624 |
+
monai.transforms.Lambdad(keys="spacing", func=lambda x: torch.FloatTensor(x)),
|
| 625 |
+
monai.transforms.Lambdad(keys="spacing", func=lambda x: x * 1e2),
|
| 626 |
+
]
|
| 627 |
+
)
|
| 628 |
+
logging.info("LDM sampler initialized.")
|
| 629 |
+
|
| 630 |
+
def sample_multiple_images(self, num_img):
|
| 631 |
+
"""
|
| 632 |
+
Generate multiple synthetic images and masks.
|
| 633 |
+
|
| 634 |
+
Args:
|
| 635 |
+
num_img (int): Number of images to generate.
|
| 636 |
+
"""
|
| 637 |
+
output_filenames = []
|
| 638 |
+
if len(self.controllable_anatomy_size) > 0:
|
| 639 |
+
# we will use mask generation instead of finding candidate masks
|
| 640 |
+
# create a dummy selected_mask_files for placeholder
|
| 641 |
+
selected_mask_files = list(range(num_img))
|
| 642 |
+
# prerpare organ size conditions
|
| 643 |
+
anatomy_size_condtion = self.prepare_anatomy_size_condtion(self.controllable_anatomy_size)
|
| 644 |
+
else:
|
| 645 |
+
need_resample = False
|
| 646 |
+
# find candidate mask and save to candidate_mask_files
|
| 647 |
+
candidate_mask_files = find_masks(
|
| 648 |
+
self.anatomy_list, self.spacing, self.output_size, True, self.all_mask_files_json, self.data_root
|
| 649 |
+
)
|
| 650 |
+
if len(candidate_mask_files) < num_img:
|
| 651 |
+
# if we cannot find enough masks based on the exact match of anatomy list, spacing, and output size,
|
| 652 |
+
# then we will try to find the closest mask in terms of spacing, and output size.
|
| 653 |
+
logging.info("Resample mask file to get desired output size and spacing")
|
| 654 |
+
candidate_mask_files = self.find_closest_masks(num_img)
|
| 655 |
+
need_resample = True
|
| 656 |
+
|
| 657 |
+
selected_mask_files = self.select_mask(candidate_mask_files, num_img)
|
| 658 |
+
if len(selected_mask_files) < num_img:
|
| 659 |
+
raise ValueError(
|
| 660 |
+
(
|
| 661 |
+
f"len(selected_mask_files) ({len(selected_mask_files)}) < num_img ({num_img}). "
|
| 662 |
+
"This should not happen. Please revisit function select_mask(self, candidate_mask_files, num_img)."
|
| 663 |
+
)
|
| 664 |
+
)
|
| 665 |
+
num_generated_img = 0
|
| 666 |
+
for index_s in range(len(selected_mask_files)):
|
| 667 |
+
item = selected_mask_files[index_s]
|
| 668 |
+
if num_generated_img >= num_img:
|
| 669 |
+
break
|
| 670 |
+
logging.info("---- Start preparing masks... ----")
|
| 671 |
+
start_time = time.time()
|
| 672 |
+
logging.info(f"Image will be generated based on {item}.")
|
| 673 |
+
if len(self.controllable_anatomy_size) > 0:
|
| 674 |
+
# generate a synthetic mask
|
| 675 |
+
(combine_label_or, spacing_tensor) = self.prepare_one_mask_and_meta_info(anatomy_size_condtion)
|
| 676 |
+
else:
|
| 677 |
+
# read in mask file
|
| 678 |
+
mask_file = item["mask_file"]
|
| 679 |
+
if_aug = item["if_aug"]
|
| 680 |
+
(combine_label_or, spacing_tensor) = self.read_mask_information(mask_file)
|
| 681 |
+
if need_resample:
|
| 682 |
+
combine_label_or = self.ensure_output_size_and_spacing(combine_label_or)
|
| 683 |
+
# mask augmentation
|
| 684 |
+
if if_aug:
|
| 685 |
+
combine_label_or = augmentation(combine_label_or, self.output_size, random_seed=self.random_seed)
|
| 686 |
+
end_time = time.time()
|
| 687 |
+
logging.info(f"---- Mask preparation time: {end_time - start_time} seconds ----")
|
| 688 |
+
torch.cuda.empty_cache()
|
| 689 |
+
# generate image/label pairs
|
| 690 |
+
modality_tensor = torch.ones_like(spacing_tensor[:, 0]).long() * self.modality_int
|
| 691 |
+
# start generation
|
| 692 |
+
synthetic_images, synthetic_labels = self.sample_one_pair(combine_label_or, modality_tensor, spacing_tensor)
|
| 693 |
+
# synthetic image quality check
|
| 694 |
+
pass_quality_check = self.quality_check(
|
| 695 |
+
synthetic_images.cpu().detach().numpy(), combine_label_or.cpu().detach().numpy()
|
| 696 |
+
)
|
| 697 |
+
if pass_quality_check or (num_img - num_generated_img) >= (len(selected_mask_files) - index_s):
|
| 698 |
+
if not pass_quality_check:
|
| 699 |
+
logging.info(
|
| 700 |
+
"Generated image/label pair did not pass quality check, but will still save them. "
|
| 701 |
+
"Please consider changing spacing and output_size to facilitate a more realistic setting."
|
| 702 |
+
)
|
| 703 |
+
num_generated_img = num_generated_img + 1
|
| 704 |
+
# save image/label pairs
|
| 705 |
+
output_postfix = datetime.now().strftime("%Y%m%d_%H%M%S_%f")
|
| 706 |
+
synthetic_labels.meta["filename_or_obj"] = "sample.nii.gz"
|
| 707 |
+
synthetic_images = MetaTensor(synthetic_images, meta=synthetic_labels.meta)
|
| 708 |
+
img_saver = SaveImage(
|
| 709 |
+
output_dir=self.output_dir,
|
| 710 |
+
output_postfix=output_postfix + "_image",
|
| 711 |
+
output_ext=self.image_output_ext,
|
| 712 |
+
separate_folder=False,
|
| 713 |
+
)
|
| 714 |
+
img_saver(synthetic_images[0])
|
| 715 |
+
synthetic_images_filename = os.path.join(
|
| 716 |
+
self.output_dir, "sample_" + output_postfix + "_image" + self.image_output_ext
|
| 717 |
+
)
|
| 718 |
+
# filter out the organs that are not in anatomy_list
|
| 719 |
+
synthetic_labels = filter_mask_with_organs(synthetic_labels, self.anatomy_list)
|
| 720 |
+
label_saver = SaveImage(
|
| 721 |
+
output_dir=self.output_dir,
|
| 722 |
+
output_postfix=output_postfix + "_label",
|
| 723 |
+
output_ext=self.label_output_ext,
|
| 724 |
+
separate_folder=False,
|
| 725 |
+
)
|
| 726 |
+
label_saver(synthetic_labels[0])
|
| 727 |
+
synthetic_labels_filename = os.path.join(
|
| 728 |
+
self.output_dir, "sample_" + output_postfix + "_label" + self.label_output_ext
|
| 729 |
+
)
|
| 730 |
+
output_filenames.append([synthetic_images_filename, synthetic_labels_filename])
|
| 731 |
+
else:
|
| 732 |
+
logging.info("Generated image/label pair did not pass quality check, will re-generate another pair.")
|
| 733 |
+
return output_filenames
|
| 734 |
+
|
| 735 |
+
def select_mask(self, candidate_mask_files, num_img):
|
| 736 |
+
"""
|
| 737 |
+
Select mask files for image generation.
|
| 738 |
+
|
| 739 |
+
Args:
|
| 740 |
+
candidate_mask_files (list): List of candidate mask files.
|
| 741 |
+
num_img (int): Number of images to generate.
|
| 742 |
+
|
| 743 |
+
Returns:
|
| 744 |
+
list: Selected mask files with augmentation flags.
|
| 745 |
+
"""
|
| 746 |
+
selected_mask_files = []
|
| 747 |
+
random.shuffle(candidate_mask_files)
|
| 748 |
+
|
| 749 |
+
for n in range(num_img * self.max_try_time):
|
| 750 |
+
mask_file = candidate_mask_files[n % len(candidate_mask_files)]
|
| 751 |
+
selected_mask_files.append({"mask_file": mask_file, "if_aug": True})
|
| 752 |
+
return selected_mask_files
|
| 753 |
+
|
| 754 |
+
def sample_one_pair(self, combine_label_or_aug, modality_tensor, spacing_tensor):
|
| 755 |
+
"""
|
| 756 |
+
Generate a single pair of synthetic image and mask.
|
| 757 |
+
|
| 758 |
+
Args:
|
| 759 |
+
combine_label_or_aug (torch.Tensor): Combined label tensor or augmented label.
|
| 760 |
+
modality_tensor (torch.Tensor): Tensor specifying the image modality.
|
| 761 |
+
spacing_tensor (torch.Tensor): Tensor specifying the spacing.
|
| 762 |
+
|
| 763 |
+
Returns:
|
| 764 |
+
tuple: A tuple containing the synthetic image and its corresponding label.
|
| 765 |
+
"""
|
| 766 |
+
# generate image/label pairs
|
| 767 |
+
synthetic_images, synthetic_labels = ldm_conditional_sample_one_image(
|
| 768 |
+
autoencoder=self.autoencoder,
|
| 769 |
+
diffusion_unet=self.diffusion_unet,
|
| 770 |
+
controlnet=self.controlnet,
|
| 771 |
+
noise_scheduler=self.noise_scheduler,
|
| 772 |
+
scale_factor=self.scale_factor,
|
| 773 |
+
device=self.device,
|
| 774 |
+
combine_label_or=combine_label_or_aug,
|
| 775 |
+
modality_tensor=modality_tensor,
|
| 776 |
+
spacing_tensor=spacing_tensor,
|
| 777 |
+
latent_shape=self.latent_shape,
|
| 778 |
+
output_size=self.output_size,
|
| 779 |
+
noise_factor=self.noise_factor,
|
| 780 |
+
num_inference_steps=self.num_inference_steps,
|
| 781 |
+
autoencoder_sliding_window_infer_size=self.autoencoder_sliding_window_infer_size,
|
| 782 |
+
autoencoder_sliding_window_infer_overlap=self.autoencoder_sliding_window_infer_overlap,
|
| 783 |
+
)
|
| 784 |
+
return synthetic_images, synthetic_labels
|
| 785 |
+
|
| 786 |
+
def prepare_anatomy_size_condtion(self, controllable_anatomy_size):
|
| 787 |
+
"""
|
| 788 |
+
Prepare anatomy size conditions for mask generation.
|
| 789 |
+
|
| 790 |
+
Args:
|
| 791 |
+
controllable_anatomy_size (list): List of tuples specifying controllable anatomy sizes.
|
| 792 |
+
|
| 793 |
+
Returns:
|
| 794 |
+
list: Prepared anatomy size conditions.
|
| 795 |
+
"""
|
| 796 |
+
anatomy_size_idx = {
|
| 797 |
+
"gallbladder": 0,
|
| 798 |
+
"liver": 1,
|
| 799 |
+
"stomach": 2,
|
| 800 |
+
"pancreas": 3,
|
| 801 |
+
"colon": 4,
|
| 802 |
+
"lung tumor": 5,
|
| 803 |
+
"pancreatic tumor": 6,
|
| 804 |
+
"hepatic tumor": 7,
|
| 805 |
+
"colon cancer primaries": 8,
|
| 806 |
+
"bone lesion": 9,
|
| 807 |
+
}
|
| 808 |
+
provide_anatomy_size = [None for _ in range(10)]
|
| 809 |
+
logging.info(f"controllable_anatomy_size: {controllable_anatomy_size}")
|
| 810 |
+
for element in controllable_anatomy_size:
|
| 811 |
+
anatomy_name, anatomy_size = element
|
| 812 |
+
provide_anatomy_size[anatomy_size_idx[anatomy_name]] = anatomy_size
|
| 813 |
+
|
| 814 |
+
with open(self.all_anatomy_size_condtions_json, "r") as f:
|
| 815 |
+
all_anatomy_size_condtions = json.load(f)
|
| 816 |
+
|
| 817 |
+
# loop through the database and find closest combinations
|
| 818 |
+
candidate_list = []
|
| 819 |
+
for anatomy_size in all_anatomy_size_condtions:
|
| 820 |
+
size = anatomy_size["organ_size"]
|
| 821 |
+
diff = 0
|
| 822 |
+
for db_size, provide_size in zip(size, provide_anatomy_size):
|
| 823 |
+
if provide_size is None:
|
| 824 |
+
continue
|
| 825 |
+
diff += abs(provide_size - db_size)
|
| 826 |
+
candidate_list.append((size, diff))
|
| 827 |
+
candidate_condition = sorted(candidate_list, key=lambda x: x[1])[0][0]
|
| 828 |
+
|
| 829 |
+
# overwrite the anatomy size provided by users
|
| 830 |
+
for element in controllable_anatomy_size:
|
| 831 |
+
anatomy_name, anatomy_size = element
|
| 832 |
+
candidate_condition[anatomy_size_idx[anatomy_name]] = anatomy_size
|
| 833 |
+
|
| 834 |
+
return candidate_condition
|
| 835 |
+
|
| 836 |
+
def prepare_one_mask_and_meta_info(self, anatomy_size_condtion):
|
| 837 |
+
"""
|
| 838 |
+
Prepare a single mask and its associated meta information.
|
| 839 |
+
|
| 840 |
+
Args:
|
| 841 |
+
anatomy_size_condtion (list): Anatomy size conditions.
|
| 842 |
+
|
| 843 |
+
Returns:
|
| 844 |
+
tuple: A tuple containing the prepared mask and associated tensors.
|
| 845 |
+
"""
|
| 846 |
+
combine_label_or = self.sample_one_mask(anatomy_size=anatomy_size_condtion)
|
| 847 |
+
# TODO: current mask generation model only can generate 256^3 volumes with 1.5 mm spacing.
|
| 848 |
+
affine = torch.zeros((4, 4))
|
| 849 |
+
affine[0, 0] = 1.5
|
| 850 |
+
affine[1, 1] = 1.5
|
| 851 |
+
affine[2, 2] = 1.5
|
| 852 |
+
affine[3, 3] = 1.0 # dummy
|
| 853 |
+
combine_label_or = MetaTensor(combine_label_or, affine=affine)
|
| 854 |
+
combine_label_or = self.ensure_output_size_and_spacing(combine_label_or)
|
| 855 |
+
|
| 856 |
+
spacing_tensor = torch.FloatTensor(self.spacing).unsqueeze(0).half().to(self.device) * 1e2
|
| 857 |
+
|
| 858 |
+
return combine_label_or, spacing_tensor
|
| 859 |
+
|
| 860 |
+
def sample_one_mask(self, anatomy_size):
|
| 861 |
+
"""
|
| 862 |
+
Generate a single synthetic mask.
|
| 863 |
+
|
| 864 |
+
Args:
|
| 865 |
+
anatomy_size (list): Anatomy size specifications.
|
| 866 |
+
|
| 867 |
+
Returns:
|
| 868 |
+
torch.Tensor: The generated synthetic mask.
|
| 869 |
+
"""
|
| 870 |
+
# generate one synthetic mask
|
| 871 |
+
synthetic_mask = ldm_conditional_sample_one_mask(
|
| 872 |
+
self.mask_generation_autoencoder,
|
| 873 |
+
self.mask_generation_diffusion_unet,
|
| 874 |
+
self.mask_generation_noise_scheduler,
|
| 875 |
+
self.mask_generation_scale_factor,
|
| 876 |
+
anatomy_size,
|
| 877 |
+
self.device,
|
| 878 |
+
self.mask_generation_latent_shape,
|
| 879 |
+
label_dict_remap_json=self.label_dict_remap_json,
|
| 880 |
+
num_inference_steps=self.mask_generation_num_inference_steps,
|
| 881 |
+
autoencoder_sliding_window_infer_size=self.autoencoder_sliding_window_infer_size,
|
| 882 |
+
autoencoder_sliding_window_infer_overlap=self.autoencoder_sliding_window_infer_overlap,
|
| 883 |
+
)
|
| 884 |
+
return synthetic_mask
|
| 885 |
+
|
| 886 |
+
def ensure_output_size_and_spacing(self, labels, check_contains_target_labels=True):
|
| 887 |
+
"""
|
| 888 |
+
Ensure the output mask has the correct size and spacing.
|
| 889 |
+
|
| 890 |
+
Args:
|
| 891 |
+
labels (torch.Tensor): Input label tensor.
|
| 892 |
+
check_contains_target_labels (bool): Whether to check if the resampled mask contains target labels.
|
| 893 |
+
|
| 894 |
+
Returns:
|
| 895 |
+
torch.Tensor: Resampled label tensor.
|
| 896 |
+
|
| 897 |
+
Raises:
|
| 898 |
+
ValueError: If the resampled mask doesn't contain required class labels.
|
| 899 |
+
"""
|
| 900 |
+
current_spacing = [labels.affine[0, 0], labels.affine[1, 1], labels.affine[2, 2]]
|
| 901 |
+
current_shape = list(labels.squeeze().shape)
|
| 902 |
+
|
| 903 |
+
need_resample = False
|
| 904 |
+
# check spacing
|
| 905 |
+
for i, j in zip(current_spacing, self.spacing):
|
| 906 |
+
if i != j:
|
| 907 |
+
need_resample = True
|
| 908 |
+
# check output size
|
| 909 |
+
for i, j in zip(current_shape, self.output_size):
|
| 910 |
+
if i != j:
|
| 911 |
+
need_resample = True
|
| 912 |
+
# resample to target size and spacing
|
| 913 |
+
if need_resample:
|
| 914 |
+
logging.info("Resampling mask to target shape and spacing")
|
| 915 |
+
logging.info(f"Resize Spacing: {current_spacing} -> {self.spacing}")
|
| 916 |
+
logging.info(f"Output size: {current_shape} -> {self.output_size}")
|
| 917 |
+
spacing = monai.transforms.Spacing(pixdim=tuple(self.spacing), mode="nearest")
|
| 918 |
+
pad_crop = monai.transforms.ResizeWithPadOrCrop(spatial_size=tuple(self.output_size))
|
| 919 |
+
labels = pad_crop(spacing(labels.squeeze(0))).unsqueeze(0).to(labels.dtype)
|
| 920 |
+
|
| 921 |
+
contained_labels = torch.unique(labels)
|
| 922 |
+
if check_contains_target_labels:
|
| 923 |
+
# check if the resampled mask still contains those target labels
|
| 924 |
+
for anatomy_label in self.anatomy_list:
|
| 925 |
+
if anatomy_label not in contained_labels:
|
| 926 |
+
raise ValueError(
|
| 927 |
+
(
|
| 928 |
+
f"Resampled mask does not contain required class labels {anatomy_label}. "
|
| 929 |
+
"Please consider increasing the output spacing or specifying a larger output size."
|
| 930 |
+
)
|
| 931 |
+
)
|
| 932 |
+
return labels
|
| 933 |
+
|
| 934 |
+
def read_mask_information(self, mask_file):
|
| 935 |
+
"""
|
| 936 |
+
Read mask information from a file.
|
| 937 |
+
|
| 938 |
+
Args:
|
| 939 |
+
mask_file (str): Path to the mask file.
|
| 940 |
+
|
| 941 |
+
Returns:
|
| 942 |
+
tuple: A tuple containing the mask tensor and associated information.
|
| 943 |
+
"""
|
| 944 |
+
val_data = self.val_transforms(mask_file)
|
| 945 |
+
|
| 946 |
+
for key in ["pseudo_label", "spacing"]:
|
| 947 |
+
val_data[key] = val_data[key].unsqueeze(0).to(self.device)
|
| 948 |
+
|
| 949 |
+
return (val_data["pseudo_label"], val_data["spacing"])
|
| 950 |
+
|
| 951 |
+
def find_closest_masks(self, num_img):
|
| 952 |
+
"""
|
| 953 |
+
Find the closest matching masks from the database.
|
| 954 |
+
|
| 955 |
+
Args:
|
| 956 |
+
num_img (int): Number of images to generate.
|
| 957 |
+
|
| 958 |
+
Returns:
|
| 959 |
+
list: List of closest matching mask candidates.
|
| 960 |
+
|
| 961 |
+
Raises:
|
| 962 |
+
ValueError: If suitable candidates cannot be found.
|
| 963 |
+
"""
|
| 964 |
+
# first check the database based on anatomy list
|
| 965 |
+
candidates = find_masks(
|
| 966 |
+
self.anatomy_list, self.spacing, self.output_size, False, self.all_mask_files_json, self.data_root
|
| 967 |
+
)
|
| 968 |
+
|
| 969 |
+
if len(candidates) < num_img:
|
| 970 |
+
raise ValueError(f"candidate masks are less than {num_img}).")
|
| 971 |
+
|
| 972 |
+
# loop through the database and find closest combinations
|
| 973 |
+
new_candidates = []
|
| 974 |
+
for c in candidates:
|
| 975 |
+
diff = 0
|
| 976 |
+
include_c = True
|
| 977 |
+
for axis in range(3):
|
| 978 |
+
if abs(c["dim"][axis]) < self.output_size[axis] - 64:
|
| 979 |
+
# we cannot upsample the mask too much
|
| 980 |
+
include_c = False
|
| 981 |
+
break
|
| 982 |
+
# check diff in FOV, major metric
|
| 983 |
+
diff += abs(
|
| 984 |
+
(abs(c["dim"][axis] * c["spacing"][axis]) - self.output_size[axis] * self.spacing[axis]) / 10
|
| 985 |
+
)
|
| 986 |
+
# check diff in dim
|
| 987 |
+
diff += abs((abs(c["dim"][axis]) - self.output_size[axis]) / 100)
|
| 988 |
+
# check diff in spacing
|
| 989 |
+
diff += abs(abs(c["spacing"][axis]) - self.spacing[axis])
|
| 990 |
+
if include_c:
|
| 991 |
+
new_candidates.append((c, diff))
|
| 992 |
+
|
| 993 |
+
# choose top-2*num_img candidates (at least 5)
|
| 994 |
+
new_candidates = sorted(new_candidates, key=lambda x: x[1])[: max(2 * num_img, 5)]
|
| 995 |
+
final_candidates = []
|
| 996 |
+
|
| 997 |
+
# check top-2*num_img candidates and update spacing after resampling
|
| 998 |
+
image_loader = monai.transforms.LoadImage(image_only=True, ensure_channel_first=True)
|
| 999 |
+
for c, _ in new_candidates:
|
| 1000 |
+
label = image_loader(c["pseudo_label"])
|
| 1001 |
+
try:
|
| 1002 |
+
label = self.ensure_output_size_and_spacing(label.unsqueeze(0))
|
| 1003 |
+
except ValueError as e:
|
| 1004 |
+
if "Resampled mask does not contain required class labels" in str(e):
|
| 1005 |
+
continue
|
| 1006 |
+
else:
|
| 1007 |
+
raise e
|
| 1008 |
+
# get region_index after resample
|
| 1009 |
+
c["spacing"] = self.spacing
|
| 1010 |
+
c["dim"] = self.output_size
|
| 1011 |
+
|
| 1012 |
+
final_candidates.append(c)
|
| 1013 |
+
if len(final_candidates) == 0:
|
| 1014 |
+
raise ValueError("Cannot find body region with given anatomy list.")
|
| 1015 |
+
return final_candidates
|
| 1016 |
+
|
| 1017 |
+
def quality_check(self, image_data, label_data):
|
| 1018 |
+
"""
|
| 1019 |
+
Perform a quality check on the generated image.
|
| 1020 |
+
Args:
|
| 1021 |
+
image_data (np.ndarray): The generated image.
|
| 1022 |
+
label_data (np.ndarray): The corresponding whole body mask.
|
| 1023 |
+
Returns:
|
| 1024 |
+
bool: True if the image passes the quality check, False otherwise.
|
| 1025 |
+
"""
|
| 1026 |
+
outlier_results = is_outlier(self.median_statistics, image_data, label_data, self.label_int_dict)
|
| 1027 |
+
for label, result in outlier_results.items():
|
| 1028 |
+
if result.get("is_outlier", False):
|
| 1029 |
+
logging.info(
|
| 1030 |
+
(
|
| 1031 |
+
f"Generated image quality check for label '{label}' failed: median value {result['median_value']} "
|
| 1032 |
+
f"is outside the acceptable range ({result['low_thresh']} - {result['high_thresh']})."
|
| 1033 |
+
)
|
| 1034 |
+
)
|
| 1035 |
+
return False
|
| 1036 |
+
return True
|
scripts/trainer.py
ADDED
|
@@ -0,0 +1,246 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
from __future__ import annotations
|
| 13 |
+
|
| 14 |
+
from typing import TYPE_CHECKING, Any, Callable, Iterable, Sequence
|
| 15 |
+
|
| 16 |
+
import torch
|
| 17 |
+
import torch.nn.functional as F
|
| 18 |
+
from monai.engines.trainer import Trainer
|
| 19 |
+
from monai.engines.utils import IterationEvents, PrepareBatchExtraInput, default_metric_cmp_fn
|
| 20 |
+
from monai.inferers import Inferer
|
| 21 |
+
from monai.networks.schedulers import Scheduler
|
| 22 |
+
from monai.transforms import Transform
|
| 23 |
+
from monai.utils import IgniteInfo, RankFilter, min_version, optional_import
|
| 24 |
+
from monai.utils.enums import CommonKeys as Keys
|
| 25 |
+
from torch.optim.optimizer import Optimizer
|
| 26 |
+
from torch.utils.data import DataLoader
|
| 27 |
+
|
| 28 |
+
from .utils import binarize_labels
|
| 29 |
+
|
| 30 |
+
if TYPE_CHECKING:
|
| 31 |
+
from ignite.engine import Engine, EventEnum
|
| 32 |
+
from ignite.metrics import Metric
|
| 33 |
+
else:
|
| 34 |
+
Engine, _ = optional_import("ignite.engine", IgniteInfo.OPT_IMPORT_VERSION, min_version, "Engine")
|
| 35 |
+
Metric, _ = optional_import("ignite.metrics", IgniteInfo.OPT_IMPORT_VERSION, min_version, "Metric")
|
| 36 |
+
EventEnum, _ = optional_import("ignite.engine", IgniteInfo.OPT_IMPORT_VERSION, min_version, "EventEnum")
|
| 37 |
+
|
| 38 |
+
__all__ = ["MAISIControlNetTrainer"]
|
| 39 |
+
|
| 40 |
+
# Module-level variable for prepare_batch default value
|
| 41 |
+
DEFAULT_PREPARE_BATCH = PrepareBatchExtraInput(extra_keys=("dim", "spacing", "top_region_index", "bottom_region_index"))
|
| 42 |
+
|
| 43 |
+
|
| 44 |
+
class MAISIControlNetTrainer(Trainer):
|
| 45 |
+
"""
|
| 46 |
+
Supervised training method with image and label, inherits from ``Trainer`` and ``Workflow``.
|
| 47 |
+
Args:
|
| 48 |
+
device: an object representing the device on which to run.
|
| 49 |
+
max_epochs: the total epoch number for trainer to run.
|
| 50 |
+
train_data_loader: Ignite engine use data_loader to run, must be Iterable or torch.DataLoader.
|
| 51 |
+
controlnet: controlnet to train in the trainer, should be regular PyTorch `torch.nn.Module`.
|
| 52 |
+
diffusion_unet: diffusion_unet used in the trainer, should be regular PyTorch `torch.nn.Module`.
|
| 53 |
+
optimizer: the optimizer associated to the detector, should be regular PyTorch optimizer from `torch.optim`
|
| 54 |
+
or its subclass.
|
| 55 |
+
epoch_length: number of iterations for one epoch, default to `len(train_data_loader)`.
|
| 56 |
+
non_blocking: if True and this copy is between CPU and GPU, the copy may occur asynchronously
|
| 57 |
+
with respect to the host. For other cases, this argument has no effect.
|
| 58 |
+
prepare_batch: function to parse expected data (usually `image`,`box`, `label` and other detector args)
|
| 59 |
+
from `engine.state.batch` for every iteration, for more details please refer to:
|
| 60 |
+
https://pytorch.org/ignite/generated/ignite.engine.create_supervised_trainer.html.
|
| 61 |
+
iteration_update: the callable function for every iteration, expect to accept `engine`
|
| 62 |
+
and `engine.state.batch` as inputs, return data will be stored in `engine.state.output`.
|
| 63 |
+
if not provided, use `self._iteration()` instead. for more details please refer to:
|
| 64 |
+
https://pytorch.org/ignite/generated/ignite.engine.engine.Engine.html.
|
| 65 |
+
inferer: inference method that execute model forward on input data, like: SlidingWindow, etc.
|
| 66 |
+
postprocessing: execute additional transformation for the model output data.
|
| 67 |
+
Typically, several Tensor based transforms composed by `Compose`.
|
| 68 |
+
key_train_metric: compute metric when every iteration completed, and save average value to
|
| 69 |
+
engine.state.metrics when epoch completlabel_set = np.arange(output_classes).tolist()d.
|
| 70 |
+
key_train_metric is the main metric to compare and save the checkpoint into files.
|
| 71 |
+
additional_metrics: more Ignite metrics that also attach to Ignite Engine.
|
| 72 |
+
metric_cmp_fn: function to compare current key metric with previous best key metric value,
|
| 73 |
+
it must accept 2 args (current_metric, previous_best) and return a bool result: if `True`, will update
|
| 74 |
+
`best_metric` and `best_metric_epoch` with current metric and epoch, default to `greater than`.
|
| 75 |
+
train_handlers: every handler is a set of Ignite Event-Handlers, must have `attach` function, like:
|
| 76 |
+
CheckpointHandler, StatsHandler, etc.
|
| 77 |
+
amp: whether to enable auto-mixed-precision training, default is False.
|
| 78 |
+
event_names: additional custom ignite events that will register to the engine.
|
| 79 |
+
new events can be a list of str or `ignite.engine.events.EventEnum`.
|
| 80 |
+
event_to_attr: a dictionary to map an event to a state attribute, then add to `engine.state`.
|
| 81 |
+
for more details, check: https://pytorch.org/ignite/generated/ignite.engine.engine.Engine.html
|
| 82 |
+
#ignite.engine.engine.Engine.register_events.
|
| 83 |
+
decollate: whether to decollate the batch-first data to a list of data after model computation,
|
| 84 |
+
recommend `decollate=True` when `postprocessing` uses components from `monai.transforms`.
|
| 85 |
+
default to `True`.
|
| 86 |
+
optim_set_to_none: when calling `optimizer.zero_grad()`, instead of setting to zero, set the grads to None.
|
| 87 |
+
more details: https://pytorch.org/docs/stable/generated/torch.optim.Optimizer.zero_grad.html.
|
| 88 |
+
to_kwargs: dict of other args for `prepare_batch` API when converting the input data, except for
|
| 89 |
+
`device`, `non_blocking`.
|
| 90 |
+
amp_kwargs: dict of the args for `torch.cuda.amp.autocast()` API, for more details:
|
| 91 |
+
https://pytorch.org/docs/stable/amp.html#torch.cuda.amp.autocast.
|
| 92 |
+
"""
|
| 93 |
+
|
| 94 |
+
def __init__(
|
| 95 |
+
self,
|
| 96 |
+
device: torch.device,
|
| 97 |
+
max_epochs: int,
|
| 98 |
+
train_data_loader: Iterable | DataLoader,
|
| 99 |
+
controlnet: torch.nn.Module,
|
| 100 |
+
diffusion_unet: torch.nn.Module,
|
| 101 |
+
optimizer: Optimizer,
|
| 102 |
+
loss_function: Callable,
|
| 103 |
+
inferer: Inferer,
|
| 104 |
+
noise_scheduler: Scheduler,
|
| 105 |
+
epoch_length: int | None = None,
|
| 106 |
+
non_blocking: bool = False,
|
| 107 |
+
prepare_batch: Callable = DEFAULT_PREPARE_BATCH,
|
| 108 |
+
iteration_update: Callable[[Engine, Any], Any] | None = None,
|
| 109 |
+
postprocessing: Transform | None = None,
|
| 110 |
+
key_train_metric: dict[str, Metric] | None = None,
|
| 111 |
+
additional_metrics: dict[str, Metric] | None = None,
|
| 112 |
+
metric_cmp_fn: Callable = default_metric_cmp_fn,
|
| 113 |
+
train_handlers: Sequence | None = None,
|
| 114 |
+
amp: bool = False,
|
| 115 |
+
event_names: list[str | EventEnum] | None = None,
|
| 116 |
+
event_to_attr: dict | None = None,
|
| 117 |
+
decollate: bool = True,
|
| 118 |
+
optim_set_to_none: bool = False,
|
| 119 |
+
to_kwargs: dict | None = None,
|
| 120 |
+
amp_kwargs: dict | None = None,
|
| 121 |
+
hyper_kwargs: dict | None = None,
|
| 122 |
+
) -> None:
|
| 123 |
+
super().__init__(
|
| 124 |
+
device=device,
|
| 125 |
+
max_epochs=max_epochs,
|
| 126 |
+
data_loader=train_data_loader,
|
| 127 |
+
epoch_length=epoch_length,
|
| 128 |
+
non_blocking=non_blocking,
|
| 129 |
+
prepare_batch=prepare_batch,
|
| 130 |
+
iteration_update=iteration_update,
|
| 131 |
+
postprocessing=postprocessing,
|
| 132 |
+
key_metric=key_train_metric,
|
| 133 |
+
additional_metrics=additional_metrics,
|
| 134 |
+
metric_cmp_fn=metric_cmp_fn,
|
| 135 |
+
handlers=train_handlers,
|
| 136 |
+
amp=amp,
|
| 137 |
+
event_names=event_names,
|
| 138 |
+
event_to_attr=event_to_attr,
|
| 139 |
+
decollate=decollate,
|
| 140 |
+
to_kwargs=to_kwargs,
|
| 141 |
+
amp_kwargs=amp_kwargs,
|
| 142 |
+
)
|
| 143 |
+
|
| 144 |
+
self.controlnet = controlnet
|
| 145 |
+
self.diffusion_unet = diffusion_unet
|
| 146 |
+
self.optimizer = optimizer
|
| 147 |
+
self.loss_function = loss_function
|
| 148 |
+
self.inferer = inferer
|
| 149 |
+
self.optim_set_to_none = optim_set_to_none
|
| 150 |
+
self.hyper_kwargs = hyper_kwargs
|
| 151 |
+
self.noise_scheduler = noise_scheduler
|
| 152 |
+
self.logger.addFilter(RankFilter())
|
| 153 |
+
for p in self.diffusion_unet.parameters():
|
| 154 |
+
p.requires_grad = False
|
| 155 |
+
print("freeze the parameters of the diffusion unet model.")
|
| 156 |
+
|
| 157 |
+
def _iteration(self, engine, batchdata: dict[str, torch.Tensor]):
|
| 158 |
+
"""
|
| 159 |
+
Callback function for the Supervised Training processing logic of 1 iteration in Ignite Engine.
|
| 160 |
+
Return below items in a dictionary:
|
| 161 |
+
- IMAGE: image Tensor data for model input, already moved to device.
|
| 162 |
+
Args:
|
| 163 |
+
engine: `Vista3DTrainer` to execute operation for an iteration.
|
| 164 |
+
batchdata: input data for this iteration, usually can be dictionary or tuple of Tensor data.
|
| 165 |
+
Raises:
|
| 166 |
+
ValueError: When ``batchdata`` is None.
|
| 167 |
+
"""
|
| 168 |
+
|
| 169 |
+
if batchdata is None:
|
| 170 |
+
raise ValueError("Must provide batch data for current iteration.")
|
| 171 |
+
|
| 172 |
+
inputs, labels, (dim, spacing, top_region_index, bottom_region_index), _ = engine.prepare_batch(
|
| 173 |
+
batchdata, engine.state.device, engine.non_blocking, **engine.to_kwargs
|
| 174 |
+
)
|
| 175 |
+
engine.state.output = {Keys.IMAGE: inputs, Keys.LABEL: labels}
|
| 176 |
+
weighted_loss_label = engine.hyper_kwargs["weighted_loss_label"]
|
| 177 |
+
weighted_loss = engine.hyper_kwargs["weighted_loss"]
|
| 178 |
+
scale_factor = engine.hyper_kwargs["scale_factor"]
|
| 179 |
+
# scale image embedding by the provided scale_factor
|
| 180 |
+
inputs = inputs * scale_factor
|
| 181 |
+
|
| 182 |
+
def _compute_pred_loss():
|
| 183 |
+
# generate random noise
|
| 184 |
+
noise_shape = list(inputs.shape)
|
| 185 |
+
noise = torch.randn(noise_shape, dtype=inputs.dtype).to(inputs.device)
|
| 186 |
+
|
| 187 |
+
# use binary encoding to encode segmentation mask
|
| 188 |
+
controlnet_cond = binarize_labels(labels.as_tensor().to(torch.uint8)).float()
|
| 189 |
+
|
| 190 |
+
# create timesteps
|
| 191 |
+
timesteps = torch.randint(
|
| 192 |
+
0, engine.noise_scheduler.num_train_timesteps, (inputs.shape[0],), device=inputs.device
|
| 193 |
+
).long()
|
| 194 |
+
|
| 195 |
+
# Create noisy latent
|
| 196 |
+
noisy_latent = engine.noise_scheduler.add_noise(original_samples=inputs, noise=noise, timesteps=timesteps)
|
| 197 |
+
|
| 198 |
+
# Get controlnet output
|
| 199 |
+
down_block_res_samples, mid_block_res_sample = engine.controlnet(
|
| 200 |
+
x=noisy_latent, timesteps=timesteps, controlnet_cond=controlnet_cond
|
| 201 |
+
)
|
| 202 |
+
noise_pred = engine.diffusion_unet(
|
| 203 |
+
x=noisy_latent,
|
| 204 |
+
timesteps=timesteps,
|
| 205 |
+
top_region_index_tensor=top_region_index,
|
| 206 |
+
bottom_region_index_tensor=bottom_region_index,
|
| 207 |
+
spacing_tensor=spacing,
|
| 208 |
+
down_block_additional_residuals=down_block_res_samples,
|
| 209 |
+
mid_block_additional_residual=mid_block_res_sample,
|
| 210 |
+
)
|
| 211 |
+
|
| 212 |
+
engine.state.output[Keys.PRED] = noise_pred
|
| 213 |
+
engine.fire_event(IterationEvents.FORWARD_COMPLETED)
|
| 214 |
+
|
| 215 |
+
if weighted_loss > 1.0:
|
| 216 |
+
weights = torch.ones_like(inputs).to(inputs.device)
|
| 217 |
+
roi = torch.zeros([noise_shape[0]] + [1] + noise_shape[2:]).to(inputs.device)
|
| 218 |
+
interpolate_label = F.interpolate(labels, size=inputs.shape[2:], mode="nearest")
|
| 219 |
+
# assign larger weights for ROI (tumor)
|
| 220 |
+
for label in weighted_loss_label:
|
| 221 |
+
roi[interpolate_label == label] = 1
|
| 222 |
+
weights[roi.repeat(1, inputs.shape[1], 1, 1, 1) == 1] = weighted_loss
|
| 223 |
+
loss = (F.l1_loss(noise_pred.float(), noise.float(), reduction="none") * weights).mean()
|
| 224 |
+
else:
|
| 225 |
+
loss = F.l1_loss(noise_pred.float(), noise.float())
|
| 226 |
+
|
| 227 |
+
engine.state.output[Keys.LOSS] = loss
|
| 228 |
+
engine.fire_event(IterationEvents.LOSS_COMPLETED)
|
| 229 |
+
|
| 230 |
+
engine.controlnet.train()
|
| 231 |
+
engine.optimizer.zero_grad(set_to_none=engine.optim_set_to_none)
|
| 232 |
+
|
| 233 |
+
if engine.amp and engine.scaler is not None:
|
| 234 |
+
with torch.amp.autocast("cuda", **engine.amp_kwargs):
|
| 235 |
+
_compute_pred_loss()
|
| 236 |
+
engine.scaler.scale(engine.state.output[Keys.LOSS]).backward()
|
| 237 |
+
engine.fire_event(IterationEvents.BACKWARD_COMPLETED)
|
| 238 |
+
engine.scaler.step(engine.optimizer)
|
| 239 |
+
engine.scaler.update()
|
| 240 |
+
else:
|
| 241 |
+
_compute_pred_loss()
|
| 242 |
+
engine.state.output[Keys.LOSS].backward()
|
| 243 |
+
engine.fire_event(IterationEvents.BACKWARD_COMPLETED)
|
| 244 |
+
engine.optimizer.step()
|
| 245 |
+
engine.fire_event(IterationEvents.MODEL_COMPLETED)
|
| 246 |
+
return engine.state.output
|
scripts/utils.py
ADDED
|
@@ -0,0 +1,696 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
import copy
|
| 11 |
+
import json
|
| 12 |
+
import math
|
| 13 |
+
import os
|
| 14 |
+
import zipfile
|
| 15 |
+
from argparse import Namespace
|
| 16 |
+
from datetime import timedelta
|
| 17 |
+
from typing import Any, Sequence
|
| 18 |
+
|
| 19 |
+
import numpy as np
|
| 20 |
+
import skimage
|
| 21 |
+
import torch
|
| 22 |
+
import torch.distributed as dist
|
| 23 |
+
from monai.bundle import ConfigParser
|
| 24 |
+
from monai.config import DtypeLike, NdarrayOrTensor
|
| 25 |
+
from monai.data import CacheDataset, DataLoader, partition_dataset
|
| 26 |
+
from monai.transforms import Compose, EnsureTyped, Lambdad, LoadImaged, Orientationd
|
| 27 |
+
from monai.transforms.utils_morphological_ops import dilate, erode
|
| 28 |
+
from monai.utils import TransformBackends, convert_data_type, convert_to_dst_type, get_equivalent_dtype
|
| 29 |
+
from scipy import stats
|
| 30 |
+
from torch import Tensor
|
| 31 |
+
|
| 32 |
+
|
| 33 |
+
def unzip_dataset(dataset_dir):
|
| 34 |
+
if dist.is_available() and dist.is_initialized():
|
| 35 |
+
rank = dist.get_rank()
|
| 36 |
+
else:
|
| 37 |
+
rank = 0
|
| 38 |
+
|
| 39 |
+
if rank == 0:
|
| 40 |
+
if not os.path.exists(dataset_dir):
|
| 41 |
+
zip_file_path = dataset_dir + ".zip"
|
| 42 |
+
if not os.path.isfile(zip_file_path):
|
| 43 |
+
raise ValueError(f"Please download {zip_file_path}.")
|
| 44 |
+
with zipfile.ZipFile(zip_file_path, "r") as zip_ref:
|
| 45 |
+
zip_ref.extractall(path=os.path.dirname(dataset_dir))
|
| 46 |
+
print(f"Unzipped {zip_file_path} to {dataset_dir}.")
|
| 47 |
+
|
| 48 |
+
if dist.is_available() and dist.is_initialized():
|
| 49 |
+
dist.barrier() # Synchronize all processes
|
| 50 |
+
|
| 51 |
+
return
|
| 52 |
+
|
| 53 |
+
|
| 54 |
+
def add_data_dir2path(list_files: list, data_dir: str, fold: int = None) -> tuple[list, list]:
|
| 55 |
+
"""
|
| 56 |
+
Read a list of data dictionary.
|
| 57 |
+
|
| 58 |
+
Args:
|
| 59 |
+
list_files (list): input data to load and transform to generate dataset for model.
|
| 60 |
+
data_dir (str): directory of files.
|
| 61 |
+
fold (int, optional): fold index for cross validation. Defaults to None.
|
| 62 |
+
|
| 63 |
+
Returns:
|
| 64 |
+
tuple[list, list]: A tuple of two arrays (training, validation).
|
| 65 |
+
"""
|
| 66 |
+
new_list_files = copy.deepcopy(list_files)
|
| 67 |
+
if fold is not None:
|
| 68 |
+
new_list_files_train = []
|
| 69 |
+
new_list_files_val = []
|
| 70 |
+
for d in new_list_files:
|
| 71 |
+
d["image"] = os.path.join(data_dir, d["image"])
|
| 72 |
+
|
| 73 |
+
if "label" in d:
|
| 74 |
+
d["label"] = os.path.join(data_dir, d["label"])
|
| 75 |
+
|
| 76 |
+
if fold is not None:
|
| 77 |
+
if d["fold"] == fold:
|
| 78 |
+
new_list_files_val.append(copy.deepcopy(d))
|
| 79 |
+
else:
|
| 80 |
+
new_list_files_train.append(copy.deepcopy(d))
|
| 81 |
+
|
| 82 |
+
if fold is not None:
|
| 83 |
+
return new_list_files_train, new_list_files_val
|
| 84 |
+
else:
|
| 85 |
+
return new_list_files, []
|
| 86 |
+
|
| 87 |
+
|
| 88 |
+
def maisi_datafold_read(json_list, data_base_dir, fold=None):
|
| 89 |
+
with open(json_list, "r") as f:
|
| 90 |
+
filenames_train = json.load(f)["training"]
|
| 91 |
+
# training data
|
| 92 |
+
train_files, val_files = add_data_dir2path(filenames_train, data_base_dir, fold=fold)
|
| 93 |
+
print(f"dataset: {data_base_dir}, num_training_files: {len(train_files)}, num_val_files: {len(val_files)}")
|
| 94 |
+
return train_files, val_files
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
def remap_labels(mask, label_dict_remap_json):
|
| 98 |
+
"""
|
| 99 |
+
Remap labels in the mask according to the provided label dictionary.
|
| 100 |
+
|
| 101 |
+
This function reads a JSON file containing label mapping information and applies
|
| 102 |
+
the mapping to the input mask.
|
| 103 |
+
|
| 104 |
+
Args:
|
| 105 |
+
mask (Tensor): The input mask tensor to be remapped.
|
| 106 |
+
label_dict_remap_json (str): Path to the JSON file containing the label mapping dictionary.
|
| 107 |
+
|
| 108 |
+
Returns:
|
| 109 |
+
Tensor: The remapped mask tensor.
|
| 110 |
+
"""
|
| 111 |
+
with open(label_dict_remap_json, "r") as f:
|
| 112 |
+
mapping_dict = json.load(f)
|
| 113 |
+
mapper = MapLabelValue(
|
| 114 |
+
orig_labels=[pair[0] for pair in mapping_dict.values()],
|
| 115 |
+
target_labels=[pair[1] for pair in mapping_dict.values()],
|
| 116 |
+
dtype=torch.uint8,
|
| 117 |
+
)
|
| 118 |
+
return mapper(mask[0, ...])[None, ...].to(mask.device)
|
| 119 |
+
|
| 120 |
+
|
| 121 |
+
def get_index_arr(img):
|
| 122 |
+
"""
|
| 123 |
+
Generate an index array for the given image.
|
| 124 |
+
|
| 125 |
+
This function creates a 3D array of indices corresponding to the dimensions of the input image.
|
| 126 |
+
|
| 127 |
+
Args:
|
| 128 |
+
img (ndarray): The input image array.
|
| 129 |
+
|
| 130 |
+
Returns:
|
| 131 |
+
ndarray: A 3D array containing the indices for each dimension of the input image.
|
| 132 |
+
"""
|
| 133 |
+
return np.moveaxis(
|
| 134 |
+
np.moveaxis(
|
| 135 |
+
np.stack(np.meshgrid(np.arange(img.shape[0]), np.arange(img.shape[1]), np.arange(img.shape[2]))), 0, 3
|
| 136 |
+
),
|
| 137 |
+
0,
|
| 138 |
+
1,
|
| 139 |
+
)
|
| 140 |
+
|
| 141 |
+
|
| 142 |
+
def supress_non_largest_components(img, target_label, default_val=0):
|
| 143 |
+
"""
|
| 144 |
+
Suppress all components except the largest one(s) for specified target labels.
|
| 145 |
+
|
| 146 |
+
This function identifies the largest component(s) for each target label and
|
| 147 |
+
suppresses all other smaller components.
|
| 148 |
+
|
| 149 |
+
Args:
|
| 150 |
+
img (ndarray): The input image array.
|
| 151 |
+
target_label (list): List of label values to process.
|
| 152 |
+
default_val (int, optional): Value to assign to suppressed voxels. Defaults to 0.
|
| 153 |
+
|
| 154 |
+
Returns:
|
| 155 |
+
tuple: A tuple containing:
|
| 156 |
+
- ndarray: Modified image with non-largest components suppressed.
|
| 157 |
+
- int: Number of voxels that were changed.
|
| 158 |
+
"""
|
| 159 |
+
index_arr = get_index_arr(img)
|
| 160 |
+
img_mod = copy.deepcopy(img)
|
| 161 |
+
new_background = np.zeros(img.shape, dtype=np.bool_)
|
| 162 |
+
for label in target_label:
|
| 163 |
+
label_cc = skimage.measure.label(img == label, connectivity=3)
|
| 164 |
+
uv, uc = np.unique(label_cc, return_counts=True)
|
| 165 |
+
dominant_vals = uv[np.argsort(uc)[::-1][:2]]
|
| 166 |
+
if len(dominant_vals) >= 2: # Case: no predictions
|
| 167 |
+
new_background = np.logical_or(
|
| 168 |
+
new_background,
|
| 169 |
+
np.logical_not(np.logical_or(label_cc == dominant_vals[0], label_cc == dominant_vals[1])),
|
| 170 |
+
)
|
| 171 |
+
|
| 172 |
+
for voxel in index_arr[new_background]:
|
| 173 |
+
img_mod[tuple(voxel)] = default_val
|
| 174 |
+
diff = np.sum((img - img_mod) > 0)
|
| 175 |
+
|
| 176 |
+
return img_mod, diff
|
| 177 |
+
|
| 178 |
+
|
| 179 |
+
def erode_one_img(mask_t: Tensor, filter_size: int | Sequence[int] = 3, pad_value: float = 1.0) -> Tensor:
|
| 180 |
+
"""
|
| 181 |
+
Erode 2D/3D binary mask with data type as torch tensor.
|
| 182 |
+
|
| 183 |
+
Args:
|
| 184 |
+
mask_t: input 2D/3D binary mask, [M,N] or [M,N,P] torch tensor.
|
| 185 |
+
filter_size: erosion filter size, has to be odd numbers, default to be 3.
|
| 186 |
+
pad_value: the filled value for padding. We need to pad the input before filtering
|
| 187 |
+
to keep the output with the same size as input. Usually use default value
|
| 188 |
+
and not changed.
|
| 189 |
+
|
| 190 |
+
Return:
|
| 191 |
+
Tensor: eroded mask, same shape as input.
|
| 192 |
+
"""
|
| 193 |
+
return erode(mask_t.float().unsqueeze(0).unsqueeze(0), filter_size, pad_value=pad_value).squeeze(0).squeeze(0)
|
| 194 |
+
|
| 195 |
+
|
| 196 |
+
def dilate_one_img(mask_t: Tensor, filter_size: int | Sequence[int] = 3, pad_value: float = 0.0) -> Tensor:
|
| 197 |
+
"""
|
| 198 |
+
Dilate 2D/3D binary mask with data type as torch tensor.
|
| 199 |
+
|
| 200 |
+
Args:
|
| 201 |
+
mask_t: input 2D/3D binary mask, [M,N] or [M,N,P] torch tensor.
|
| 202 |
+
filter_size: dilation filter size, has to be odd numbers, default to be 3.
|
| 203 |
+
pad_value: the filled value for padding. We need to pad the input before filtering
|
| 204 |
+
to keep the output with the same size as input. Usually use default value
|
| 205 |
+
and not changed.
|
| 206 |
+
|
| 207 |
+
Return:
|
| 208 |
+
Tensor: dilated mask, same shape as input.
|
| 209 |
+
"""
|
| 210 |
+
return dilate(mask_t.float().unsqueeze(0).unsqueeze(0), filter_size, pad_value=pad_value).squeeze(0).squeeze(0)
|
| 211 |
+
|
| 212 |
+
|
| 213 |
+
def binarize_labels(x: Tensor, bits: int = 8) -> Tensor:
|
| 214 |
+
"""
|
| 215 |
+
Convert input tensor to binary representation.
|
| 216 |
+
|
| 217 |
+
This function takes an input tensor and converts it to a binary representation
|
| 218 |
+
using the specified number of bits.
|
| 219 |
+
|
| 220 |
+
Args:
|
| 221 |
+
x (Tensor): Input tensor with shape (B, 1, H, W, D).
|
| 222 |
+
bits (int, optional): Number of bits to use for binary representation. Defaults to 8.
|
| 223 |
+
|
| 224 |
+
Returns:
|
| 225 |
+
Tensor: Binary representation of the input tensor with shape (B, bits, H, W, D).
|
| 226 |
+
"""
|
| 227 |
+
mask = 2 ** torch.arange(bits).to(x.device, x.dtype)
|
| 228 |
+
return x.unsqueeze(-1).bitwise_and(mask).ne(0).byte().squeeze(1).permute(0, 4, 1, 2, 3)
|
| 229 |
+
|
| 230 |
+
|
| 231 |
+
def setup_ddp(rank: int, world_size: int) -> torch.device:
|
| 232 |
+
"""
|
| 233 |
+
Initialize the distributed process group.
|
| 234 |
+
|
| 235 |
+
Args:
|
| 236 |
+
rank (int): rank of the current process.
|
| 237 |
+
world_size (int): number of processes participating in the job.
|
| 238 |
+
|
| 239 |
+
Returns:
|
| 240 |
+
torch.device: device of the current process.
|
| 241 |
+
"""
|
| 242 |
+
dist.init_process_group(
|
| 243 |
+
backend="nccl", init_method="env://", timeout=timedelta(seconds=36000), rank=rank, world_size=world_size
|
| 244 |
+
)
|
| 245 |
+
dist.barrier()
|
| 246 |
+
device = torch.device(f"cuda:{rank}")
|
| 247 |
+
return device
|
| 248 |
+
|
| 249 |
+
|
| 250 |
+
def define_instance(args: Namespace, instance_def_key: str) -> Any:
|
| 251 |
+
"""
|
| 252 |
+
Define and instantiate an object based on the provided arguments and instance definition key.
|
| 253 |
+
|
| 254 |
+
This function uses a ConfigParser to parse the arguments and instantiate an object
|
| 255 |
+
defined by the instance_def_key.
|
| 256 |
+
|
| 257 |
+
Args:
|
| 258 |
+
args: An object containing the arguments to be parsed.
|
| 259 |
+
instance_def_key (str): The key used to retrieve the instance definition from the parsed content.
|
| 260 |
+
|
| 261 |
+
Returns:
|
| 262 |
+
The instantiated object as defined by the instance_def_key in the parsed configuration.
|
| 263 |
+
"""
|
| 264 |
+
parser = ConfigParser(vars(args))
|
| 265 |
+
parser.parse(True)
|
| 266 |
+
return parser.get_parsed_content(instance_def_key, instantiate=True)
|
| 267 |
+
|
| 268 |
+
|
| 269 |
+
def prepare_maisi_controlnet_json_dataloader(
|
| 270 |
+
json_data_list: list | str,
|
| 271 |
+
data_base_dir: list | str,
|
| 272 |
+
batch_size: int = 1,
|
| 273 |
+
fold: int = 0,
|
| 274 |
+
cache_rate: float = 0.0,
|
| 275 |
+
rank: int = 0,
|
| 276 |
+
world_size: int = 1,
|
| 277 |
+
) -> tuple[DataLoader, DataLoader]:
|
| 278 |
+
"""
|
| 279 |
+
Prepare dataloaders for training and validation.
|
| 280 |
+
|
| 281 |
+
Args:
|
| 282 |
+
json_data_list (list | str): the name of JSON files listing the data.
|
| 283 |
+
data_base_dir (list | str): directory of files.
|
| 284 |
+
batch_size (int, optional): how many samples per batch to load . Defaults to 1.
|
| 285 |
+
fold (int, optional): fold index for cross validation. Defaults to 0.
|
| 286 |
+
cache_rate (float, optional): percentage of cached data in total. Defaults to 0.0.
|
| 287 |
+
rank (int, optional): rank of the current process. Defaults to 0.
|
| 288 |
+
world_size (int, optional): number of processes participating in the job. Defaults to 1.
|
| 289 |
+
|
| 290 |
+
Returns:
|
| 291 |
+
tuple[DataLoader, DataLoader]: A tuple of two dataloaders (training, validation).
|
| 292 |
+
"""
|
| 293 |
+
use_ddp = world_size > 1
|
| 294 |
+
if isinstance(json_data_list, list):
|
| 295 |
+
assert isinstance(data_base_dir, list)
|
| 296 |
+
list_train = []
|
| 297 |
+
list_valid = []
|
| 298 |
+
for data_list, data_root in zip(json_data_list, data_base_dir):
|
| 299 |
+
with open(data_list, "r") as f:
|
| 300 |
+
json_data = json.load(f)["training"]
|
| 301 |
+
train, val = add_data_dir2path(json_data, data_root, fold)
|
| 302 |
+
list_train += train
|
| 303 |
+
list_valid += val
|
| 304 |
+
else:
|
| 305 |
+
with open(json_data_list, "r") as f:
|
| 306 |
+
json_data = json.load(f)["training"]
|
| 307 |
+
list_train, list_valid = add_data_dir2path(json_data, data_base_dir, fold)
|
| 308 |
+
|
| 309 |
+
common_transform = [
|
| 310 |
+
LoadImaged(keys=["image", "label"], image_only=True, ensure_channel_first=True),
|
| 311 |
+
Orientationd(keys=["label"], axcodes="RAS"),
|
| 312 |
+
EnsureTyped(keys=["label"], dtype=torch.uint8, track_meta=True),
|
| 313 |
+
Lambdad(keys="top_region_index", func=lambda x: torch.FloatTensor(x)),
|
| 314 |
+
Lambdad(keys="bottom_region_index", func=lambda x: torch.FloatTensor(x)),
|
| 315 |
+
Lambdad(keys="spacing", func=lambda x: torch.FloatTensor(x)),
|
| 316 |
+
Lambdad(keys=["top_region_index", "bottom_region_index", "spacing"], func=lambda x: x * 1e2),
|
| 317 |
+
]
|
| 318 |
+
train_transforms, val_transforms = Compose(common_transform), Compose(common_transform)
|
| 319 |
+
|
| 320 |
+
train_loader = None
|
| 321 |
+
|
| 322 |
+
if use_ddp:
|
| 323 |
+
list_train = partition_dataset(data=list_train, shuffle=True, num_partitions=world_size, even_divisible=True)[
|
| 324 |
+
rank
|
| 325 |
+
]
|
| 326 |
+
train_ds = CacheDataset(data=list_train, transform=train_transforms, cache_rate=cache_rate, num_workers=8)
|
| 327 |
+
train_loader = DataLoader(train_ds, batch_size=batch_size, shuffle=True, num_workers=8, pin_memory=True)
|
| 328 |
+
if use_ddp:
|
| 329 |
+
list_valid = partition_dataset(data=list_valid, shuffle=True, num_partitions=world_size, even_divisible=False)[
|
| 330 |
+
rank
|
| 331 |
+
]
|
| 332 |
+
val_ds = CacheDataset(data=list_valid, transform=val_transforms, cache_rate=cache_rate, num_workers=8)
|
| 333 |
+
val_loader = DataLoader(val_ds, batch_size=batch_size, shuffle=False, num_workers=2, pin_memory=False)
|
| 334 |
+
return train_loader, val_loader
|
| 335 |
+
|
| 336 |
+
|
| 337 |
+
def organ_fill_by_closing(data, target_label, device, close_times=2, filter_size=3, pad_value=0.0):
|
| 338 |
+
"""
|
| 339 |
+
Fill holes in an organ mask using morphological closing operations.
|
| 340 |
+
|
| 341 |
+
This function performs a series of dilation and erosion operations to fill holes
|
| 342 |
+
in the organ mask identified by the target label.
|
| 343 |
+
|
| 344 |
+
Args:
|
| 345 |
+
data (ndarray): The input data containing organ labels.
|
| 346 |
+
target_label (int): The label of the organ to be processed.
|
| 347 |
+
device (str): The device to perform the operations on (e.g., 'cuda:0').
|
| 348 |
+
close_times (int, optional): Number of times to perform the closing operation. Defaults to 2.
|
| 349 |
+
filter_size (int, optional): Size of the filter for dilation and erosion. Defaults to 3.
|
| 350 |
+
pad_value (float, optional): Value used for padding in dilation and erosion. Defaults to 0.0.
|
| 351 |
+
|
| 352 |
+
Returns:
|
| 353 |
+
ndarray: Boolean mask of the filled organ.
|
| 354 |
+
"""
|
| 355 |
+
mask = (data == target_label).astype(np.uint8)
|
| 356 |
+
mask = torch.from_numpy(mask).to(device)
|
| 357 |
+
for _ in range(close_times):
|
| 358 |
+
mask = dilate_one_img(mask, filter_size=filter_size, pad_value=pad_value)
|
| 359 |
+
mask = erode_one_img(mask, filter_size=filter_size, pad_value=pad_value)
|
| 360 |
+
return mask.cpu().numpy().astype(np.bool_)
|
| 361 |
+
|
| 362 |
+
|
| 363 |
+
def organ_fill_by_removed_mask(data, target_label, remove_mask, device):
|
| 364 |
+
"""
|
| 365 |
+
Fill an organ mask in regions where it was previously removed.
|
| 366 |
+
|
| 367 |
+
Args:
|
| 368 |
+
data (ndarray): The input data containing organ labels.
|
| 369 |
+
target_label (int): The label of the organ to be processed.
|
| 370 |
+
remove_mask (ndarray): Boolean mask indicating regions where the organ was removed.
|
| 371 |
+
device (str): The device to perform the operations on (e.g., 'cuda:0').
|
| 372 |
+
|
| 373 |
+
Returns:
|
| 374 |
+
ndarray: Boolean mask of the filled organ in previously removed regions.
|
| 375 |
+
"""
|
| 376 |
+
mask = (data == target_label).astype(np.uint8)
|
| 377 |
+
mask = dilate_one_img(torch.from_numpy(mask).to(device), filter_size=3, pad_value=0.0)
|
| 378 |
+
mask = dilate_one_img(mask, filter_size=3, pad_value=0.0)
|
| 379 |
+
roi_oragn_mask = dilate_one_img(mask, filter_size=3, pad_value=0.0).cpu().numpy()
|
| 380 |
+
return (roi_oragn_mask * remove_mask).astype(np.bool_)
|
| 381 |
+
|
| 382 |
+
|
| 383 |
+
def get_body_region_index_from_mask(input_mask):
|
| 384 |
+
"""
|
| 385 |
+
Determine the top and bottom body region indices from an input mask.
|
| 386 |
+
|
| 387 |
+
Args:
|
| 388 |
+
input_mask (Tensor): Input mask tensor containing body region labels.
|
| 389 |
+
|
| 390 |
+
Returns:
|
| 391 |
+
tuple: Two lists representing the top and bottom region indices.
|
| 392 |
+
"""
|
| 393 |
+
region_indices = {}
|
| 394 |
+
# head and neck
|
| 395 |
+
region_indices["region_0"] = [22, 120]
|
| 396 |
+
# thorax
|
| 397 |
+
region_indices["region_1"] = [28, 29, 30, 31, 32]
|
| 398 |
+
# abdomen
|
| 399 |
+
region_indices["region_2"] = [1, 2, 3, 4, 5, 14]
|
| 400 |
+
# pelvis and lower
|
| 401 |
+
region_indices["region_3"] = [93, 94]
|
| 402 |
+
|
| 403 |
+
nda = input_mask.cpu().numpy().squeeze()
|
| 404 |
+
unique_elements = np.lib.arraysetops.unique(nda)
|
| 405 |
+
unique_elements = list(unique_elements)
|
| 406 |
+
# print(f"nda: {nda.shape} {unique_elements}.")
|
| 407 |
+
overlap_array = np.zeros(len(region_indices), dtype=np.uint8)
|
| 408 |
+
for _j in range(len(region_indices)):
|
| 409 |
+
overlap = any(element in region_indices[f"region_{_j}"] for element in unique_elements)
|
| 410 |
+
overlap_array[_j] = np.uint8(overlap)
|
| 411 |
+
overlap_array_indices = np.nonzero(overlap_array)[0]
|
| 412 |
+
top_region_index = np.eye(len(region_indices), dtype=np.uint8)[np.amin(overlap_array_indices), ...]
|
| 413 |
+
top_region_index = list(top_region_index)
|
| 414 |
+
top_region_index = [int(_k) for _k in top_region_index]
|
| 415 |
+
bottom_region_index = np.eye(len(region_indices), dtype=np.uint8)[np.amax(overlap_array_indices), ...]
|
| 416 |
+
bottom_region_index = list(bottom_region_index)
|
| 417 |
+
bottom_region_index = [int(_k) for _k in bottom_region_index]
|
| 418 |
+
# print(f"{top_region_index} {bottom_region_index}")
|
| 419 |
+
return top_region_index, bottom_region_index
|
| 420 |
+
|
| 421 |
+
|
| 422 |
+
def general_mask_generation_post_process(volume_t, target_tumor_label=None, device="cuda:0"):
|
| 423 |
+
"""
|
| 424 |
+
Perform post-processing on a generated mask volume.
|
| 425 |
+
|
| 426 |
+
This function applies various refinement steps to improve the quality of the generated mask,
|
| 427 |
+
including body mask refinement, tumor prediction refinement, and organ-specific processing.
|
| 428 |
+
|
| 429 |
+
Args:
|
| 430 |
+
volume_t (ndarray): Input volume containing organ and tumor labels.
|
| 431 |
+
target_tumor_label (int, optional): Label of the target tumor. Defaults to None.
|
| 432 |
+
device (str, optional): Device to perform operations on. Defaults to "cuda:0".
|
| 433 |
+
|
| 434 |
+
Returns:
|
| 435 |
+
ndarray: Post-processed volume with refined organ and tumor labels.
|
| 436 |
+
"""
|
| 437 |
+
# assume volume_t is np array with shape (H,W,D)
|
| 438 |
+
hepatic_vessel = volume_t == 25
|
| 439 |
+
airway = volume_t == 132
|
| 440 |
+
|
| 441 |
+
# ------------ refine body mask pred
|
| 442 |
+
body_region_mask = (
|
| 443 |
+
erode_one_img(torch.from_numpy((volume_t > 0)).to(device), filter_size=3, pad_value=0.0).cpu().numpy()
|
| 444 |
+
)
|
| 445 |
+
body_region_mask, _ = supress_non_largest_components(body_region_mask, [1])
|
| 446 |
+
body_region_mask = (
|
| 447 |
+
dilate_one_img(torch.from_numpy(body_region_mask).to(device), filter_size=3, pad_value=0.0)
|
| 448 |
+
.cpu()
|
| 449 |
+
.numpy()
|
| 450 |
+
.astype(np.uint8)
|
| 451 |
+
)
|
| 452 |
+
volume_t = volume_t * body_region_mask
|
| 453 |
+
|
| 454 |
+
# ------------ refine tumor pred
|
| 455 |
+
tumor_organ_dict = {23: 28, 24: 4, 26: 1, 27: 62, 128: 200}
|
| 456 |
+
for t in [23, 24, 26, 27, 128]:
|
| 457 |
+
if t != target_tumor_label:
|
| 458 |
+
volume_t[volume_t == t] = tumor_organ_dict[t]
|
| 459 |
+
else:
|
| 460 |
+
volume_t[organ_fill_by_closing(volume_t, target_label=t, device=device)] = t
|
| 461 |
+
volume_t[organ_fill_by_closing(volume_t, target_label=t, device=device)] = t
|
| 462 |
+
# we only keep the largest connected componet for tumors except hepatic tumor and bone lesion
|
| 463 |
+
if target_tumor_label != 26 and target_tumor_label != 128:
|
| 464 |
+
volume_t, _ = supress_non_largest_components(volume_t, [target_tumor_label], default_val=200)
|
| 465 |
+
target_tumor = volume_t == target_tumor_label
|
| 466 |
+
|
| 467 |
+
# ------------ remove undesired organ pred
|
| 468 |
+
# general post-process non-largest components suppression
|
| 469 |
+
# process 4 ROI organs + spleen + 2 kidney + 5 lung lobes + duodenum + inferior vena cava
|
| 470 |
+
oran_list = [1, 4, 10, 12, 3, 28, 29, 30, 31, 32, 5, 14, 13, 6, 7, 8, 9, 10]
|
| 471 |
+
if target_tumor_label != 128:
|
| 472 |
+
oran_list += list(range(33, 60)) # + list(range(63,87))
|
| 473 |
+
data, _ = supress_non_largest_components(volume_t, oran_list, default_val=200) # 200 is body region
|
| 474 |
+
organ_remove_mask = (volume_t - data).astype(np.bool_)
|
| 475 |
+
# process intestinal system (stomach 12, duodenum 13, small bowel 19, colon 62)
|
| 476 |
+
intestinal_mask_ = (
|
| 477 |
+
(data == 12).astype(np.uint8)
|
| 478 |
+
+ (data == 13).astype(np.uint8)
|
| 479 |
+
+ (data == 19).astype(np.uint8)
|
| 480 |
+
+ (data == 62).astype(np.uint8)
|
| 481 |
+
)
|
| 482 |
+
intestinal_mask, _ = supress_non_largest_components(intestinal_mask_, [1], default_val=0)
|
| 483 |
+
# process small bowel 19
|
| 484 |
+
small_bowel_remove_mask = (data == 19).astype(np.uint8) - (data == 19).astype(np.uint8) * intestinal_mask
|
| 485 |
+
# process colon 62
|
| 486 |
+
colon_remove_mask = (data == 62).astype(np.uint8) - (data == 62).astype(np.uint8) * intestinal_mask
|
| 487 |
+
intestinal_remove_mask = (small_bowel_remove_mask + colon_remove_mask).astype(np.bool_)
|
| 488 |
+
data[intestinal_remove_mask] = 200
|
| 489 |
+
|
| 490 |
+
# ------------ full correponding organ in removed regions
|
| 491 |
+
for organ_label in oran_list:
|
| 492 |
+
data[organ_fill_by_closing(data, target_label=organ_label, device=device)] = organ_label
|
| 493 |
+
|
| 494 |
+
if target_tumor_label == 23 and np.sum(target_tumor) > 0:
|
| 495 |
+
# speical process for cases with lung tumor
|
| 496 |
+
dia_lung_tumor_mask = (
|
| 497 |
+
dilate_one_img(torch.from_numpy((data == 23)).to(device), filter_size=3, pad_value=0.0).cpu().numpy()
|
| 498 |
+
)
|
| 499 |
+
tmp = (
|
| 500 |
+
(data * (dia_lung_tumor_mask.astype(np.uint8) - (data == 23).astype(np.uint8))).astype(np.float32).flatten()
|
| 501 |
+
)
|
| 502 |
+
tmp[tmp == 0] = float("nan")
|
| 503 |
+
mode = int(stats.mode(tmp.flatten(), nan_policy="omit")[0])
|
| 504 |
+
if mode in [28, 29, 30, 31, 32]:
|
| 505 |
+
dia_lung_tumor_mask = (
|
| 506 |
+
dilate_one_img(torch.from_numpy(dia_lung_tumor_mask).to(device), filter_size=3, pad_value=0.0)
|
| 507 |
+
.cpu()
|
| 508 |
+
.numpy()
|
| 509 |
+
)
|
| 510 |
+
lung_remove_mask = dia_lung_tumor_mask.astype(np.uint8) - (data == 23).astype(np.uint8).astype(np.uint8)
|
| 511 |
+
data[organ_fill_by_removed_mask(data, target_label=mode, remove_mask=lung_remove_mask, device=device)] = (
|
| 512 |
+
mode
|
| 513 |
+
)
|
| 514 |
+
dia_lung_tumor_mask = (
|
| 515 |
+
dilate_one_img(torch.from_numpy(dia_lung_tumor_mask).to(device), filter_size=3, pad_value=0.0).cpu().numpy()
|
| 516 |
+
)
|
| 517 |
+
data[
|
| 518 |
+
organ_fill_by_removed_mask(
|
| 519 |
+
data, target_label=23, remove_mask=dia_lung_tumor_mask * organ_remove_mask, device=device
|
| 520 |
+
)
|
| 521 |
+
] = 23
|
| 522 |
+
for organ_label in [28, 29, 30, 31, 32]:
|
| 523 |
+
data[organ_fill_by_closing(data, target_label=organ_label, device=device)] = organ_label
|
| 524 |
+
data[organ_fill_by_closing(data, target_label=organ_label, device=device)] = organ_label
|
| 525 |
+
data[organ_fill_by_closing(data, target_label=organ_label, device=device)] = organ_label
|
| 526 |
+
|
| 527 |
+
if target_tumor_label == 26 and np.sum(target_tumor) > 0:
|
| 528 |
+
# speical process for cases with hepatic tumor
|
| 529 |
+
# process liver 1
|
| 530 |
+
data[organ_fill_by_removed_mask(data, target_label=1, remove_mask=intestinal_remove_mask, device=device)] = 1
|
| 531 |
+
data[organ_fill_by_removed_mask(data, target_label=1, remove_mask=intestinal_remove_mask, device=device)] = 1
|
| 532 |
+
# process spleen 2
|
| 533 |
+
data[organ_fill_by_removed_mask(data, target_label=3, remove_mask=organ_remove_mask, device=device)] = 3
|
| 534 |
+
data[organ_fill_by_removed_mask(data, target_label=3, remove_mask=organ_remove_mask, device=device)] = 3
|
| 535 |
+
dia_tumor_mask = (
|
| 536 |
+
dilate_one_img(torch.from_numpy((data == target_tumor_label)).to(device), filter_size=3, pad_value=0.0)
|
| 537 |
+
.cpu()
|
| 538 |
+
.numpy()
|
| 539 |
+
)
|
| 540 |
+
dia_tumor_mask = (
|
| 541 |
+
dilate_one_img(torch.from_numpy(dia_tumor_mask).to(device), filter_size=3, pad_value=0.0).cpu().numpy()
|
| 542 |
+
)
|
| 543 |
+
data[
|
| 544 |
+
organ_fill_by_removed_mask(
|
| 545 |
+
data, target_label=target_tumor_label, remove_mask=dia_tumor_mask * organ_remove_mask, device=device
|
| 546 |
+
)
|
| 547 |
+
] = target_tumor_label
|
| 548 |
+
# refine hepatic tumor
|
| 549 |
+
hepatic_tumor_vessel_liver_mask_ = (
|
| 550 |
+
(data == 26).astype(np.uint8) + (data == 25).astype(np.uint8) + (data == 1).astype(np.uint8)
|
| 551 |
+
)
|
| 552 |
+
hepatic_tumor_vessel_liver_mask_ = (hepatic_tumor_vessel_liver_mask_ > 1).astype(np.uint8)
|
| 553 |
+
hepatic_tumor_vessel_liver_mask, _ = supress_non_largest_components(
|
| 554 |
+
hepatic_tumor_vessel_liver_mask_, [1], default_val=0
|
| 555 |
+
)
|
| 556 |
+
removed_region = (hepatic_tumor_vessel_liver_mask_ - hepatic_tumor_vessel_liver_mask).astype(np.bool_)
|
| 557 |
+
data[removed_region] = 200
|
| 558 |
+
target_tumor = (target_tumor * hepatic_tumor_vessel_liver_mask).astype(np.bool_)
|
| 559 |
+
# refine liver
|
| 560 |
+
data[organ_fill_by_closing(data, target_label=1, device=device)] = 1
|
| 561 |
+
data[organ_fill_by_closing(data, target_label=1, device=device)] = 1
|
| 562 |
+
data[organ_fill_by_closing(data, target_label=1, device=device)] = 1
|
| 563 |
+
|
| 564 |
+
if target_tumor_label == 27 and np.sum(target_tumor) > 0:
|
| 565 |
+
# speical process for cases with colon tumor
|
| 566 |
+
dia_tumor_mask = (
|
| 567 |
+
dilate_one_img(torch.from_numpy((data == target_tumor_label)).to(device), filter_size=3, pad_value=0.0)
|
| 568 |
+
.cpu()
|
| 569 |
+
.numpy()
|
| 570 |
+
)
|
| 571 |
+
dia_tumor_mask = (
|
| 572 |
+
dilate_one_img(torch.from_numpy(dia_tumor_mask).to(device), filter_size=3, pad_value=0.0).cpu().numpy()
|
| 573 |
+
)
|
| 574 |
+
data[
|
| 575 |
+
organ_fill_by_removed_mask(
|
| 576 |
+
data, target_label=target_tumor_label, remove_mask=dia_tumor_mask * organ_remove_mask, device=device
|
| 577 |
+
)
|
| 578 |
+
] = target_tumor_label
|
| 579 |
+
|
| 580 |
+
if target_tumor_label == 129 and np.sum(target_tumor) > 0:
|
| 581 |
+
# speical process for cases with kidney tumor
|
| 582 |
+
for organ_label in [5, 14]:
|
| 583 |
+
data[organ_fill_by_closing(data, target_label=organ_label, device=device)] = organ_label
|
| 584 |
+
data[organ_fill_by_closing(data, target_label=organ_label, device=device)] = organ_label
|
| 585 |
+
data[organ_fill_by_closing(data, target_label=organ_label, device=device)] = organ_label
|
| 586 |
+
# TODO: current model does not support hepatic vessel by size control.
|
| 587 |
+
# we treat it as liver for better visiaulization
|
| 588 |
+
print(
|
| 589 |
+
"Current model does not support hepatic vessel by size control, "
|
| 590 |
+
"so we treat generated hepatic vessel as part of liver for better visiaulization."
|
| 591 |
+
)
|
| 592 |
+
data[hepatic_vessel] = 1
|
| 593 |
+
data[airway] = 132
|
| 594 |
+
if target_tumor_label is not None:
|
| 595 |
+
data[target_tumor] = target_tumor_label
|
| 596 |
+
|
| 597 |
+
return data
|
| 598 |
+
|
| 599 |
+
|
| 600 |
+
class MapLabelValue:
|
| 601 |
+
"""
|
| 602 |
+
Utility to map label values to another set of values.
|
| 603 |
+
For example, map [3, 2, 1] to [0, 1, 2], [1, 2, 3] -> [0.5, 1.5, 2.5], ["label3", "label2", "label1"] -> [0, 1, 2],
|
| 604 |
+
[3.5, 2.5, 1.5] -> ["label0", "label1", "label2"], etc.
|
| 605 |
+
The label data must be numpy array or array-like data and the output data will be numpy array.
|
| 606 |
+
|
| 607 |
+
"""
|
| 608 |
+
|
| 609 |
+
backend = [TransformBackends.NUMPY, TransformBackends.TORCH]
|
| 610 |
+
|
| 611 |
+
def __init__(self, orig_labels: Sequence, target_labels: Sequence, dtype: DtypeLike = np.float32) -> None:
|
| 612 |
+
"""
|
| 613 |
+
Args:
|
| 614 |
+
orig_labels: original labels that map to others.
|
| 615 |
+
target_labels: expected label values, 1: 1 map to the `orig_labels`.
|
| 616 |
+
dtype: convert the output data to dtype, default to float32.
|
| 617 |
+
if dtype is from PyTorch, the transform will use the pytorch backend, else with numpy backend.
|
| 618 |
+
|
| 619 |
+
"""
|
| 620 |
+
if len(orig_labels) != len(target_labels):
|
| 621 |
+
raise ValueError("orig_labels and target_labels must have the same length.")
|
| 622 |
+
|
| 623 |
+
self.orig_labels = orig_labels
|
| 624 |
+
self.target_labels = target_labels
|
| 625 |
+
self.pair = tuple((o, t) for o, t in zip(self.orig_labels, self.target_labels) if o != t)
|
| 626 |
+
type_dtype = type(dtype)
|
| 627 |
+
if getattr(type_dtype, "__module__", "") == "torch":
|
| 628 |
+
self.use_numpy = False
|
| 629 |
+
self.dtype = get_equivalent_dtype(dtype, data_type=torch.Tensor)
|
| 630 |
+
else:
|
| 631 |
+
self.use_numpy = True
|
| 632 |
+
self.dtype = get_equivalent_dtype(dtype, data_type=np.ndarray)
|
| 633 |
+
|
| 634 |
+
def __call__(self, img: NdarrayOrTensor):
|
| 635 |
+
"""
|
| 636 |
+
Apply the label mapping to the input image.
|
| 637 |
+
|
| 638 |
+
Args:
|
| 639 |
+
img (NdarrayOrTensor): Input image to be remapped.
|
| 640 |
+
|
| 641 |
+
Returns:
|
| 642 |
+
NdarrayOrTensor: Remapped image.
|
| 643 |
+
"""
|
| 644 |
+
if self.use_numpy:
|
| 645 |
+
img_np, *_ = convert_data_type(img, np.ndarray)
|
| 646 |
+
_out_shape = img_np.shape
|
| 647 |
+
img_flat = img_np.flatten()
|
| 648 |
+
try:
|
| 649 |
+
out_flat = img_flat.astype(self.dtype)
|
| 650 |
+
except ValueError:
|
| 651 |
+
# can't copy unchanged labels as the expected dtype is not supported, must map all the label values
|
| 652 |
+
out_flat = np.zeros(shape=img_flat.shape, dtype=self.dtype)
|
| 653 |
+
for o, t in self.pair:
|
| 654 |
+
out_flat[img_flat == o] = t
|
| 655 |
+
out_t = out_flat.reshape(_out_shape)
|
| 656 |
+
else:
|
| 657 |
+
img_t, *_ = convert_data_type(img, torch.Tensor)
|
| 658 |
+
out_t = img_t.detach().clone().to(self.dtype) # type: ignore
|
| 659 |
+
for o, t in self.pair:
|
| 660 |
+
out_t[img_t == o] = t
|
| 661 |
+
out, *_ = convert_to_dst_type(src=out_t, dst=img, dtype=self.dtype)
|
| 662 |
+
return out
|
| 663 |
+
|
| 664 |
+
|
| 665 |
+
def dynamic_infer(inferer, model, images):
|
| 666 |
+
"""
|
| 667 |
+
Perform dynamic inference using a model and an inferer, typically a monai SlidingWindowInferer.
|
| 668 |
+
|
| 669 |
+
This function determines whether to use the model directly or to use the provided inferer
|
| 670 |
+
(such as a sliding window inferer) based on the size of the input images.
|
| 671 |
+
|
| 672 |
+
Args:
|
| 673 |
+
inferer: An inference object, typically a monai SlidingWindowInferer, which handles patch-based inference.
|
| 674 |
+
model (torch.nn.Module): The model used for inference.
|
| 675 |
+
images (torch.Tensor): The input images for inference, shape [N,C,H,W,D] or [N,C,H,W].
|
| 676 |
+
|
| 677 |
+
Returns:
|
| 678 |
+
torch.Tensor: The output from the model or the inferer, depending on the input size.
|
| 679 |
+
"""
|
| 680 |
+
if torch.numel(images[0:1, 0:1, ...]) < math.prod(inferer.roi_size):
|
| 681 |
+
return model(images)
|
| 682 |
+
else:
|
| 683 |
+
# Extract the spatial dimensions from the images tensor (H, W, D)
|
| 684 |
+
spatial_dims = images.shape[2:]
|
| 685 |
+
orig_roi = inferer.roi_size
|
| 686 |
+
|
| 687 |
+
# Check that roi has the same number of dimensions as spatial_dims
|
| 688 |
+
if len(orig_roi) != len(spatial_dims):
|
| 689 |
+
raise ValueError(f"ROI length ({len(orig_roi)}) does not match spatial dimensions ({len(spatial_dims)}).")
|
| 690 |
+
|
| 691 |
+
# Iterate and adjust each ROI dimension
|
| 692 |
+
adjusted_roi = [min(roi_dim, img_dim) for roi_dim, img_dim in zip(orig_roi, spatial_dims)]
|
| 693 |
+
inferer.roi_size = adjusted_roi
|
| 694 |
+
output = inferer(network=model, inputs=images)
|
| 695 |
+
inferer.roi_size = orig_roi
|
| 696 |
+
return output
|