Upload folder using huggingface_hub
Browse files- .gitattributes +1 -1
- README.md +203 -3
- autogluon/metrics/feature_importance.csv +98 -0
- autogluon/metrics/metrics.json +8 -0
- autogluon/model/feature_importance.csv +98 -0
- autogluon/model/leaderboard.csv +3 -0
- autogluon/model/learner.pkl +3 -0
- autogluon/model/metadata.json +352 -0
- autogluon/model/model_info.txt +11 -0
- autogluon/model/models/LightGBMBasic/model.pkl +3 -0
- autogluon/model/models/WeightedEnsemble_L2/model.pkl +3 -0
- autogluon/model/models/trainer.pkl +3 -0
- autogluon/model/predictor.pkl +3 -0
- autogluon/model/version.txt +1 -0
- autogluon/plots/confusion_matrix.png +0 -0
- autogluon/plots/feature_importance.png +0 -0
- autogluon/plots/roc_curve.png +0 -0
- config.json +23 -0
- logistic_regression/metrics/feature_importance.csv +96 -0
- logistic_regression/metrics/metrics.json +14 -0
- logistic_regression/model/model.joblib +3 -0
- logistic_regression/model/model_summary.txt +2 -0
- logistic_regression/model/params.json +5 -0
- logistic_regression/model/scaler.joblib +3 -0
- logistic_regression/plots/confusion_matrix.png +0 -0
- logistic_regression/plots/roc_curve.png +0 -0
- neural_network/metrics/metrics.json +25 -0
- neural_network/model/model.keras +0 -0
- neural_network/model/model_summary.txt +24 -0
- neural_network/model/params.json +10 -0
- neural_network/model/scaler.joblib +3 -0
- neural_network/plots/confusion_matrix.png +0 -0
- neural_network/plots/model_architecture.png +0 -0
- neural_network/plots/roc_curve.png +0 -0
- preprocessing_config.json +189 -0
- tokenizer_config.json +7 -0
.gitattributes
CHANGED
|
@@ -32,4 +32,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 32 |
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
-
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
| 32 |
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
+
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
README.md
CHANGED
|
@@ -1,3 +1,203 @@
|
|
| 1 |
-
---
|
| 2 |
-
|
| 3 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
language: en
|
| 3 |
+
license: mit
|
| 4 |
+
model-index:
|
| 5 |
+
- name: aai540-group3/diabetes-readmission
|
| 6 |
+
results:
|
| 7 |
+
- task:
|
| 8 |
+
type: binary-classification
|
| 9 |
+
dataset:
|
| 10 |
+
name: Diabetes 130-US Hospitals
|
| 11 |
+
type: hospital-readmission
|
| 12 |
+
metrics:
|
| 13 |
+
- type: accuracy
|
| 14 |
+
value: 0.8865474882652552
|
| 15 |
+
name: accuracy
|
| 16 |
+
- type: auc
|
| 17 |
+
value: 0.6467403398083669
|
| 18 |
+
name: auc
|
| 19 |
+
---
|
| 20 |
+
|
| 21 |
+
# aai540-group3/diabetes-readmission
|
| 22 |
+
|
| 23 |
+
## Model Description
|
| 24 |
+
|
| 25 |
+
This model predicts 30-day hospital readmissions for diabetic patients using historical patient data
|
| 26 |
+
and machine learning techniques. The model aims to identify high-risk individuals enabling targeted
|
| 27 |
+
interventions and improved healthcare resource allocation.
|
| 28 |
+
|
| 29 |
+
## Overview
|
| 30 |
+
|
| 31 |
+
- **Task:** Binary Classification (Hospital Readmission Prediction)
|
| 32 |
+
- **Model Type:** autogluon
|
| 33 |
+
- **Framework:** Python Autogluon
|
| 34 |
+
- **License:** MIT
|
| 35 |
+
- **Last Updated:** 2024-10-29
|
| 36 |
+
|
| 37 |
+
## Performance Metrics
|
| 38 |
+
|
| 39 |
+
- **Test Accuracy:** 0.8865
|
| 40 |
+
- **Test ROC-AUC:** 0.6467
|
| 41 |
+
|
| 42 |
+
## Feature Importance
|
| 43 |
+
|
| 44 |
+
Significant features and their importance scores:
|
| 45 |
+
|
| 46 |
+
| Feature | Importance | p-value | 99% CI |
|
| 47 |
+
|---------|------------|----------|----------|
|
| 48 |
+
| 0 | 0.0563 | 3.24e-04 | [0.0294, 0.0832] |
|
| 49 |
+
| 1 | 0.0358 | 8.45e-06 | [0.0290, 0.0426] |
|
| 50 |
+
| 2 | 0.0080 | 0.0083 | [-0.0013, 0.0173] |
|
| 51 |
+
| 3 | 0.0046 | 1.96e-04 | [0.0027, 0.0065] |
|
| 52 |
+
| 4 | 0.0023 | 0.0055 | [-0.0001, 0.0046] |
|
| 53 |
+
| 5 | 0.0008 | 0.1840 | [-0.0027, 0.0043] |
|
| 54 |
+
|
| 55 |
+
*Note: Only features with non-zero importance are shown. The confidence intervals (CI) are calculated at the 99% level. Features with p-value < 0.05 are considered statistically significant.*
|
| 56 |
+
|
| 57 |
+
## Features
|
| 58 |
+
|
| 59 |
+
### Numeric Features
|
| 60 |
+
- Patient demographics (age)
|
| 61 |
+
- Hospital stay metrics (time_in_hospital, num_procedures, num_lab_procedures)
|
| 62 |
+
- Medication metrics (num_medications, total_medications)
|
| 63 |
+
- Service utilization (number_outpatient, number_emergency, number_inpatient)
|
| 64 |
+
- Diagnostic information (number_diagnoses)
|
| 65 |
+
|
| 66 |
+
### Binary Features
|
| 67 |
+
- Patient characteristics (gender)
|
| 68 |
+
- Medication flags (diabetesmed, change, insulin_with_oral)
|
| 69 |
+
|
| 70 |
+
### Interaction Features
|
| 71 |
+
- Time-based interactions (medications × time, procedures × time)
|
| 72 |
+
- Complexity indicators (age × diagnoses, medications × procedures)
|
| 73 |
+
- Resource utilization (lab procedures × time, medications × changes)
|
| 74 |
+
|
| 75 |
+
### Ratio Features
|
| 76 |
+
- Resource efficiency (procedure/medication ratio, lab/procedure ratio)
|
| 77 |
+
- Diagnostic density (diagnosis/procedure ratio)
|
| 78 |
+
|
| 79 |
+
## Intended Use
|
| 80 |
+
|
| 81 |
+
This model is designed for healthcare professionals to assess the risk of 30-day readmission
|
| 82 |
+
for diabetic patients. It should be used as a supportive tool in conjunction with clinical judgment.
|
| 83 |
+
|
| 84 |
+
### Primary Intended Uses
|
| 85 |
+
- Predict likelihood of 30-day hospital readmission
|
| 86 |
+
- Support resource allocation and intervention planning
|
| 87 |
+
- Aid in identifying high-risk patients
|
| 88 |
+
- Assist in care management decision-making
|
| 89 |
+
|
| 90 |
+
### Out-of-Scope Uses
|
| 91 |
+
- Non-diabetic patient populations
|
| 92 |
+
- Predicting readmissions beyond 30 days
|
| 93 |
+
- Making final decisions without clinical oversight
|
| 94 |
+
- Use as sole determinant for patient care decisions
|
| 95 |
+
- Emergency or critical care decision-making
|
| 96 |
+
|
| 97 |
+
## Training Data
|
| 98 |
+
|
| 99 |
+
The model was trained on the [Diabetes 130-US Hospitals Dataset](https://doi.org/10.24432/C5230J)
|
| 100 |
+
(1999-2008) from UCI ML Repository. This dataset includes:
|
| 101 |
+
|
| 102 |
+
- Over 100,000 hospital admissions
|
| 103 |
+
- 50+ features including patient demographics, diagnoses, procedures
|
| 104 |
+
- Binary outcome: readmission within 30 days
|
| 105 |
+
- Comprehensive medication tracking
|
| 106 |
+
- Detailed hospital utilization metrics
|
| 107 |
+
|
| 108 |
+
## Training Procedure
|
| 109 |
+
|
| 110 |
+
### Data Preprocessing
|
| 111 |
+
- Missing value imputation using mean/mode
|
| 112 |
+
- Outlier handling using 5-sigma clipping
|
| 113 |
+
- Feature scaling using StandardScaler
|
| 114 |
+
- Categorical encoding using one-hot encoding
|
| 115 |
+
- Log transformation for skewed features
|
| 116 |
+
|
| 117 |
+
### Feature Engineering
|
| 118 |
+
- Created interaction terms between key variables
|
| 119 |
+
- Generated resource utilization ratios
|
| 120 |
+
- Aggregated medication usage metrics
|
| 121 |
+
- Developed time-based interaction features
|
| 122 |
+
- Constructed diagnostic density metrics
|
| 123 |
+
|
| 124 |
+
### Model Training
|
| 125 |
+
- Data split: 70% training, 15% validation, 15% test
|
| 126 |
+
- Cross-validation for model selection
|
| 127 |
+
- Hyperparameter optimization via grid search
|
| 128 |
+
- Early stopping to prevent overfitting
|
| 129 |
+
- Model selection based on ROC-AUC performance
|
| 130 |
+
|
| 131 |
+
## Limitations & Biases
|
| 132 |
+
|
| 133 |
+
### Known Limitations
|
| 134 |
+
- Model performance depends on data quality and completeness
|
| 135 |
+
- Limited to the scope of training data timeframe (1999-2008)
|
| 136 |
+
- May not generalize to significantly different healthcare systems
|
| 137 |
+
- Requires standardized input data format
|
| 138 |
+
|
| 139 |
+
### Potential Biases
|
| 140 |
+
- May exhibit demographic biases present in training data
|
| 141 |
+
- Performance may vary across different hospital systems
|
| 142 |
+
- Could be influenced by regional healthcare practices
|
| 143 |
+
- Might show temporal biases due to historical data
|
| 144 |
+
|
| 145 |
+
### Recommendations
|
| 146 |
+
- Regular model monitoring and retraining
|
| 147 |
+
- Careful validation in new deployment contexts
|
| 148 |
+
- Assessment of performance across demographic groups
|
| 149 |
+
- Integration with existing clinical workflows
|
| 150 |
+
|
| 151 |
+
## Monitoring & Maintenance
|
| 152 |
+
|
| 153 |
+
### Monitoring Requirements
|
| 154 |
+
- Track prediction accuracy across different patient groups
|
| 155 |
+
- Monitor input data distribution shifts
|
| 156 |
+
- Assess feature importance stability
|
| 157 |
+
- Evaluate performance metrics over time
|
| 158 |
+
|
| 159 |
+
### Maintenance Schedule
|
| 160 |
+
- Quarterly performance reviews recommended
|
| 161 |
+
- Annual retraining with updated data
|
| 162 |
+
- Regular bias assessments
|
| 163 |
+
- Ongoing validation against current practices
|
| 164 |
+
|
| 165 |
+
## Citation
|
| 166 |
+
|
| 167 |
+
```bibtex
|
| 168 |
+
@misc{diabetes-readmission-model,
|
| 169 |
+
title = {Hospital Readmission Prediction Model for Diabetic Patients},
|
| 170 |
+
author = {Agustin, Jonathan and Robertson, Zack and Vo, Lisa},
|
| 171 |
+
year = {2024},
|
| 172 |
+
publisher = {Hugging Face},
|
| 173 |
+
howpublished = {\url{https://huggingface.co/{REPO_ID}}}
|
| 174 |
+
}
|
| 175 |
+
|
| 176 |
+
@misc{diabetes-dataset,
|
| 177 |
+
title = {Diabetes 130-US Hospitals for Years 1999-2008 Data Set},
|
| 178 |
+
author = {Strack, B. and DeShazo, J. and Gennings, C. and Olmo, J. and
|
| 179 |
+
Ventura, S. and Cios, K. and Clore, J.},
|
| 180 |
+
year = {2014},
|
| 181 |
+
publisher = {UCI Machine Learning Repository},
|
| 182 |
+
doi = {10.24432/C5230J}
|
| 183 |
+
}
|
| 184 |
+
```
|
| 185 |
+
|
| 186 |
+
## Model Card Authors
|
| 187 |
+
|
| 188 |
+
Jonathan Agustin, Zack Robertson, Lisa Vo
|
| 189 |
+
|
| 190 |
+
## For Questions, Issues, or Feedback
|
| 191 |
+
|
| 192 |
+
- GitHub Issues: [Repository Issues](https://github.com/aai540-group3/diabetes-readmission/issues)
|
| 193 |
+
- Email: [team contact information]
|
| 194 |
+
|
| 195 |
+
## Updates and Versions
|
| 196 |
+
|
| 197 |
+
- {pd.Timestamp.now().strftime('%Y-%m-%d')}: Initial model release
|
| 198 |
+
- Feature engineering pipeline implemented
|
| 199 |
+
- Comprehensive preprocessing system added
|
| 200 |
+
- Model evaluation and selection completed
|
| 201 |
+
|
| 202 |
+
---
|
| 203 |
+
Last updated: {pd.Timestamp.now().strftime('%Y-%m-%d')}
|
autogluon/metrics/feature_importance.csv
ADDED
|
@@ -0,0 +1,98 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
,importance,stddev,p_value,n,p99_high,p99_low
|
| 2 |
+
total_encounters,0.05633188246384744,0.01306857727918546,0.0003239944497075829,5,0.08324026370862368,0.029423501219071196
|
| 3 |
+
discharge_disposition_id_2.0,0.03579614353303893,0.0032878240066241274,8.445030543368168e-06,5,0.042565818605235,0.029026468460842857
|
| 4 |
+
encounter_per_time,0.008011873284703586,0.004518928602209438,0.008307777731853096,5,0.017316409734883593,-0.0012926631654764202
|
| 5 |
+
age_x_number_diagnoses,0.004567182369341038,0.0009303464717592386,0.00019566488400355028,5,0.006482778613448061,0.002651586125234015
|
| 6 |
+
number_diagnoses_x_time_in_hospital,0.0022883946798970545,0.00114142152620697,0.005482230034090644,5,0.0046385973784792405,-6.180801868513114e-05
|
| 7 |
+
age_x_num_medications,0.0007673725098147121,0.0016926127397477618,0.1840177606988291,5,0.004252485535349559,-0.002717740515720135
|
| 8 |
+
gender,0.0,0.0,0.5,5,0.0,0.0
|
| 9 |
+
num_medications_x_numchange,0.0,0.0,0.5,5,0.0,0.0
|
| 10 |
+
admission_type_id_5,0.0,0.0,0.5,5,0.0,0.0
|
| 11 |
+
admission_type_id_4,0.0,0.0,0.5,5,0.0,0.0
|
| 12 |
+
admission_type_id_3,0.0,0.0,0.5,5,0.0,0.0
|
| 13 |
+
diagnosis_procedure_ratio,0.0,0.0,0.5,5,0.0,0.0
|
| 14 |
+
lab_procedure_ratio,0.0,0.0,0.5,5,0.0,0.0
|
| 15 |
+
procedure_medication_ratio,0.0,0.0,0.5,5,0.0,0.0
|
| 16 |
+
time_in_hospital_x_num_lab_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 17 |
+
change_x_num_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 18 |
+
num_medications_x_num_lab_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 19 |
+
admission_source_id_4,0.0,0.0,0.5,5,0.0,0.0
|
| 20 |
+
num_medications_x_num_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 21 |
+
total_medications_x_number_diagnoses,0.0,0.0,0.5,5,0.0,0.0
|
| 22 |
+
num_lab_procedures_x_time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 23 |
+
num_procedures_x_time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 24 |
+
num_medications_x_time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 25 |
+
discharge_disposition_id_3.5,0.0,0.0,0.5,5,0.0,0.0
|
| 26 |
+
admission_source_id_7,0.0,0.0,0.5,5,0.0,0.0
|
| 27 |
+
number_emergency_log1p,0.0,0.0,0.5,5,0.0,0.0
|
| 28 |
+
admission_source_id_8,0.0,0.0,0.5,5,0.0,0.0
|
| 29 |
+
event_timestamp,0.0,0.0,0.5,5,0.0,0.0
|
| 30 |
+
level1_diag1_8.0,0.0,0.0,0.5,5,0.0,0.0
|
| 31 |
+
A1Cresult_1,0.0,0.0,0.5,5,0.0,0.0
|
| 32 |
+
A1Cresult_0,0.0,0.0,0.5,5,0.0,0.0
|
| 33 |
+
max_glu_serum_1,0.0,0.0,0.5,5,0.0,0.0
|
| 34 |
+
max_glu_serum_0,0.0,0.0,0.5,5,0.0,0.0
|
| 35 |
+
discharge_disposition_id_18,0.0,0.0,0.5,5,0.0,0.0
|
| 36 |
+
discharge_disposition_id_10,0.0,0.0,0.5,5,0.0,0.0
|
| 37 |
+
discharge_disposition_id_7,0.0,0.0,0.5,5,0.0,0.0
|
| 38 |
+
discharge_disposition_id_2,0.0,0.0,0.5,5,0.0,0.0
|
| 39 |
+
gender_1,0.0,0.0,0.5,5,0.0,0.0
|
| 40 |
+
level1_diag1_7.5,0.0,0.0,0.5,5,0.0,0.0
|
| 41 |
+
level1_diag1_7.0,0.0,0.0,0.5,5,0.0,0.0
|
| 42 |
+
level1_diag1_6.0,0.0,0.0,0.5,5,0.0,0.0
|
| 43 |
+
level1_diag1_5.0,0.0,0.0,0.5,5,0.0,0.0
|
| 44 |
+
level1_diag1_4.0,0.0,0.0,0.5,5,0.0,0.0
|
| 45 |
+
level1_diag1_3.0,0.0,0.0,0.5,5,0.0,0.0
|
| 46 |
+
level1_diag1_2.0,0.0,0.0,0.5,5,0.0,0.0
|
| 47 |
+
level1_diag1_1.0,0.0,0.0,0.5,5,0.0,0.0
|
| 48 |
+
admission_source_id_11,0.0,0.0,0.5,5,0.0,0.0
|
| 49 |
+
admission_source_id_9,0.0,0.0,0.5,5,0.0,0.0
|
| 50 |
+
number_inpatient_log1p,0.0,0.0,0.5,5,0.0,0.0
|
| 51 |
+
diagnoses_per_encounter,0.0,0.0,0.5,5,0.0,0.0
|
| 52 |
+
number_outpatient_log1p,0.0,0.0,0.5,5,0.0,0.0
|
| 53 |
+
metformin,0.0,0.0,0.5,5,0.0,0.0
|
| 54 |
+
pioglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 55 |
+
tolbutamide,0.0,0.0,0.5,5,0.0,0.0
|
| 56 |
+
glyburide,0.0,0.0,0.5,5,0.0,0.0
|
| 57 |
+
glipizide,0.0,0.0,0.5,5,0.0,0.0
|
| 58 |
+
acetohexamide,0.0,0.0,0.5,5,0.0,0.0
|
| 59 |
+
glimepiride,0.0,0.0,0.5,5,0.0,0.0
|
| 60 |
+
chlorpropamide,0.0,0.0,0.5,5,0.0,0.0
|
| 61 |
+
nateglinide,0.0,0.0,0.5,5,0.0,0.0
|
| 62 |
+
repaglinide,0.0,0.0,0.5,5,0.0,0.0
|
| 63 |
+
a1cresult,0.0,0.0,0.5,5,0.0,0.0
|
| 64 |
+
acarbose,0.0,0.0,0.5,5,0.0,0.0
|
| 65 |
+
max_glu_serum,0.0,0.0,0.5,5,0.0,0.0
|
| 66 |
+
number_diagnoses,0.0,0.0,0.5,5,0.0,0.0
|
| 67 |
+
diag_3,0.0,0.0,0.5,5,0.0,0.0
|
| 68 |
+
diag_2,0.0,0.0,0.5,5,0.0,0.0
|
| 69 |
+
diag_1,0.0,0.0,0.5,5,0.0,0.0
|
| 70 |
+
num_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 71 |
+
num_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 72 |
+
num_lab_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 73 |
+
time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 74 |
+
rosiglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 75 |
+
miglitol,0.0,0.0,0.5,5,0.0,0.0
|
| 76 |
+
age,0.0,0.0,0.5,5,0.0,0.0
|
| 77 |
+
level1_diag2,0.0,0.0,0.5,5,0.0,0.0
|
| 78 |
+
procedures_to_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 79 |
+
lab_procedures_per_day,0.0,0.0,0.5,5,0.0,0.0
|
| 80 |
+
procedures_per_day,0.0,0.0,0.5,5,0.0,0.0
|
| 81 |
+
nummed,0.0,0.0,0.5,5,0.0,0.0
|
| 82 |
+
numchange,0.0,0.0,0.5,5,0.0,0.0
|
| 83 |
+
insulin_with_oral,0.0,0.0,0.5,5,0.0,0.0
|
| 84 |
+
medication_density,0.0,0.0,0.5,5,0.0,0.0
|
| 85 |
+
total_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 86 |
+
level1_diag3,0.0,0.0,0.5,5,0.0,0.0
|
| 87 |
+
id,0.0,0.0,0.5,5,0.0,0.0
|
| 88 |
+
troglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 89 |
+
diabetesmed,0.0,0.0,0.5,5,0.0,0.0
|
| 90 |
+
change,0.0,0.0,0.5,5,0.0,0.0
|
| 91 |
+
metformin-pioglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 92 |
+
metformin-rosiglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 93 |
+
glimepiride-pioglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 94 |
+
glipizide-metformin,0.0,0.0,0.5,5,0.0,0.0
|
| 95 |
+
glyburide-metformin,0.0,0.0,0.5,5,0.0,0.0
|
| 96 |
+
insulin,0.0,0.0,0.5,5,0.0,0.0
|
| 97 |
+
tolazamide,0.0,0.0,0.5,5,0.0,0.0
|
| 98 |
+
created_timestamp,0.0,0.0,0.5,5,0.0,0.0
|
autogluon/metrics/metrics.json
ADDED
|
@@ -0,0 +1,8 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"test_accuracy": 0.8865474882652552,
|
| 3 |
+
"test_precision": 0.0,
|
| 4 |
+
"test_recall": 0.0,
|
| 5 |
+
"test_f1": 0.0,
|
| 6 |
+
"test_auc": 0.6467403398083669,
|
| 7 |
+
"test_pr_auc": 0.18117429407499608
|
| 8 |
+
}
|
autogluon/model/feature_importance.csv
ADDED
|
@@ -0,0 +1,98 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
,importance,stddev,p_value,n,p99_high,p99_low
|
| 2 |
+
total_encounters,0.05633188246384744,0.01306857727918546,0.0003239944497075829,5,0.08324026370862368,0.029423501219071196
|
| 3 |
+
discharge_disposition_id_2.0,0.03579614353303893,0.0032878240066241274,8.445030543368168e-06,5,0.042565818605235,0.029026468460842857
|
| 4 |
+
encounter_per_time,0.008011873284703586,0.004518928602209438,0.008307777731853096,5,0.017316409734883593,-0.0012926631654764202
|
| 5 |
+
age_x_number_diagnoses,0.004567182369341038,0.0009303464717592386,0.00019566488400355028,5,0.006482778613448061,0.002651586125234015
|
| 6 |
+
number_diagnoses_x_time_in_hospital,0.0022883946798970545,0.00114142152620697,0.005482230034090644,5,0.0046385973784792405,-6.180801868513114e-05
|
| 7 |
+
age_x_num_medications,0.0007673725098147121,0.0016926127397477618,0.1840177606988291,5,0.004252485535349559,-0.002717740515720135
|
| 8 |
+
gender,0.0,0.0,0.5,5,0.0,0.0
|
| 9 |
+
num_medications_x_numchange,0.0,0.0,0.5,5,0.0,0.0
|
| 10 |
+
admission_type_id_5,0.0,0.0,0.5,5,0.0,0.0
|
| 11 |
+
admission_type_id_4,0.0,0.0,0.5,5,0.0,0.0
|
| 12 |
+
admission_type_id_3,0.0,0.0,0.5,5,0.0,0.0
|
| 13 |
+
diagnosis_procedure_ratio,0.0,0.0,0.5,5,0.0,0.0
|
| 14 |
+
lab_procedure_ratio,0.0,0.0,0.5,5,0.0,0.0
|
| 15 |
+
procedure_medication_ratio,0.0,0.0,0.5,5,0.0,0.0
|
| 16 |
+
time_in_hospital_x_num_lab_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 17 |
+
change_x_num_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 18 |
+
num_medications_x_num_lab_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 19 |
+
admission_source_id_4,0.0,0.0,0.5,5,0.0,0.0
|
| 20 |
+
num_medications_x_num_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 21 |
+
total_medications_x_number_diagnoses,0.0,0.0,0.5,5,0.0,0.0
|
| 22 |
+
num_lab_procedures_x_time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 23 |
+
num_procedures_x_time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 24 |
+
num_medications_x_time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 25 |
+
discharge_disposition_id_3.5,0.0,0.0,0.5,5,0.0,0.0
|
| 26 |
+
admission_source_id_7,0.0,0.0,0.5,5,0.0,0.0
|
| 27 |
+
number_emergency_log1p,0.0,0.0,0.5,5,0.0,0.0
|
| 28 |
+
admission_source_id_8,0.0,0.0,0.5,5,0.0,0.0
|
| 29 |
+
event_timestamp,0.0,0.0,0.5,5,0.0,0.0
|
| 30 |
+
level1_diag1_8.0,0.0,0.0,0.5,5,0.0,0.0
|
| 31 |
+
A1Cresult_1,0.0,0.0,0.5,5,0.0,0.0
|
| 32 |
+
A1Cresult_0,0.0,0.0,0.5,5,0.0,0.0
|
| 33 |
+
max_glu_serum_1,0.0,0.0,0.5,5,0.0,0.0
|
| 34 |
+
max_glu_serum_0,0.0,0.0,0.5,5,0.0,0.0
|
| 35 |
+
discharge_disposition_id_18,0.0,0.0,0.5,5,0.0,0.0
|
| 36 |
+
discharge_disposition_id_10,0.0,0.0,0.5,5,0.0,0.0
|
| 37 |
+
discharge_disposition_id_7,0.0,0.0,0.5,5,0.0,0.0
|
| 38 |
+
discharge_disposition_id_2,0.0,0.0,0.5,5,0.0,0.0
|
| 39 |
+
gender_1,0.0,0.0,0.5,5,0.0,0.0
|
| 40 |
+
level1_diag1_7.5,0.0,0.0,0.5,5,0.0,0.0
|
| 41 |
+
level1_diag1_7.0,0.0,0.0,0.5,5,0.0,0.0
|
| 42 |
+
level1_diag1_6.0,0.0,0.0,0.5,5,0.0,0.0
|
| 43 |
+
level1_diag1_5.0,0.0,0.0,0.5,5,0.0,0.0
|
| 44 |
+
level1_diag1_4.0,0.0,0.0,0.5,5,0.0,0.0
|
| 45 |
+
level1_diag1_3.0,0.0,0.0,0.5,5,0.0,0.0
|
| 46 |
+
level1_diag1_2.0,0.0,0.0,0.5,5,0.0,0.0
|
| 47 |
+
level1_diag1_1.0,0.0,0.0,0.5,5,0.0,0.0
|
| 48 |
+
admission_source_id_11,0.0,0.0,0.5,5,0.0,0.0
|
| 49 |
+
admission_source_id_9,0.0,0.0,0.5,5,0.0,0.0
|
| 50 |
+
number_inpatient_log1p,0.0,0.0,0.5,5,0.0,0.0
|
| 51 |
+
diagnoses_per_encounter,0.0,0.0,0.5,5,0.0,0.0
|
| 52 |
+
number_outpatient_log1p,0.0,0.0,0.5,5,0.0,0.0
|
| 53 |
+
metformin,0.0,0.0,0.5,5,0.0,0.0
|
| 54 |
+
pioglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 55 |
+
tolbutamide,0.0,0.0,0.5,5,0.0,0.0
|
| 56 |
+
glyburide,0.0,0.0,0.5,5,0.0,0.0
|
| 57 |
+
glipizide,0.0,0.0,0.5,5,0.0,0.0
|
| 58 |
+
acetohexamide,0.0,0.0,0.5,5,0.0,0.0
|
| 59 |
+
glimepiride,0.0,0.0,0.5,5,0.0,0.0
|
| 60 |
+
chlorpropamide,0.0,0.0,0.5,5,0.0,0.0
|
| 61 |
+
nateglinide,0.0,0.0,0.5,5,0.0,0.0
|
| 62 |
+
repaglinide,0.0,0.0,0.5,5,0.0,0.0
|
| 63 |
+
a1cresult,0.0,0.0,0.5,5,0.0,0.0
|
| 64 |
+
acarbose,0.0,0.0,0.5,5,0.0,0.0
|
| 65 |
+
max_glu_serum,0.0,0.0,0.5,5,0.0,0.0
|
| 66 |
+
number_diagnoses,0.0,0.0,0.5,5,0.0,0.0
|
| 67 |
+
diag_3,0.0,0.0,0.5,5,0.0,0.0
|
| 68 |
+
diag_2,0.0,0.0,0.5,5,0.0,0.0
|
| 69 |
+
diag_1,0.0,0.0,0.5,5,0.0,0.0
|
| 70 |
+
num_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 71 |
+
num_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 72 |
+
num_lab_procedures,0.0,0.0,0.5,5,0.0,0.0
|
| 73 |
+
time_in_hospital,0.0,0.0,0.5,5,0.0,0.0
|
| 74 |
+
rosiglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 75 |
+
miglitol,0.0,0.0,0.5,5,0.0,0.0
|
| 76 |
+
age,0.0,0.0,0.5,5,0.0,0.0
|
| 77 |
+
level1_diag2,0.0,0.0,0.5,5,0.0,0.0
|
| 78 |
+
procedures_to_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 79 |
+
lab_procedures_per_day,0.0,0.0,0.5,5,0.0,0.0
|
| 80 |
+
procedures_per_day,0.0,0.0,0.5,5,0.0,0.0
|
| 81 |
+
nummed,0.0,0.0,0.5,5,0.0,0.0
|
| 82 |
+
numchange,0.0,0.0,0.5,5,0.0,0.0
|
| 83 |
+
insulin_with_oral,0.0,0.0,0.5,5,0.0,0.0
|
| 84 |
+
medication_density,0.0,0.0,0.5,5,0.0,0.0
|
| 85 |
+
total_medications,0.0,0.0,0.5,5,0.0,0.0
|
| 86 |
+
level1_diag3,0.0,0.0,0.5,5,0.0,0.0
|
| 87 |
+
id,0.0,0.0,0.5,5,0.0,0.0
|
| 88 |
+
troglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 89 |
+
diabetesmed,0.0,0.0,0.5,5,0.0,0.0
|
| 90 |
+
change,0.0,0.0,0.5,5,0.0,0.0
|
| 91 |
+
metformin-pioglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 92 |
+
metformin-rosiglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 93 |
+
glimepiride-pioglitazone,0.0,0.0,0.5,5,0.0,0.0
|
| 94 |
+
glipizide-metformin,0.0,0.0,0.5,5,0.0,0.0
|
| 95 |
+
glyburide-metformin,0.0,0.0,0.5,5,0.0,0.0
|
| 96 |
+
insulin,0.0,0.0,0.5,5,0.0,0.0
|
| 97 |
+
tolazamide,0.0,0.0,0.5,5,0.0,0.0
|
| 98 |
+
created_timestamp,0.0,0.0,0.5,5,0.0,0.0
|
autogluon/model/leaderboard.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
model,score_test,score_val,eval_metric,pred_time_test,pred_time_val,fit_time,pred_time_test_marginal,pred_time_val_marginal,fit_time_marginal,stack_level,can_infer,fit_order
|
| 2 |
+
LightGBMBasic,0.6467403398083669,0.6342024393654344,roc_auc,0.03393197059631348,0.03538703918457031,1.146812915802002,0.03393197059631348,0.03538703918457031,1.146812915802002,1,True,1
|
| 3 |
+
WeightedEnsemble_L2,0.6467403398083669,0.6342024393654344,roc_auc,0.0435791015625,0.035985708236694336,1.149925947189331,0.009647130966186523,0.0005986690521240234,0.0031130313873291016,2,True,2
|
autogluon/model/learner.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:15ff306c94de1c67d7c49887f2ddda86584e9575df3afdac54b8c4008c615cfa
|
| 3 |
+
size 6740
|
autogluon/model/metadata.json
ADDED
|
@@ -0,0 +1,352 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"system": "Linux",
|
| 3 |
+
"version": "1.1.1",
|
| 4 |
+
"lite": false,
|
| 5 |
+
"py_version": "3.10",
|
| 6 |
+
"py_version_micro": "3.10.15",
|
| 7 |
+
"packages": {
|
| 8 |
+
"gitpython": "3.1.43",
|
| 9 |
+
"mako": "1.3.6",
|
| 10 |
+
"markdown": "3.7",
|
| 11 |
+
"markupsafe": "3.0.2",
|
| 12 |
+
"pydrive2": "1.20.0",
|
| 13 |
+
"pyjwt": "2.9.0",
|
| 14 |
+
"pysocks": "1.7.1",
|
| 15 |
+
"pyyaml": "6.0.2",
|
| 16 |
+
"sqlalchemy": "2.0.36",
|
| 17 |
+
"absl-py": "2.1.0",
|
| 18 |
+
"accelerate": "0.21.0",
|
| 19 |
+
"adlfs": "2024.7.0",
|
| 20 |
+
"aiobotocore": "2.15.2",
|
| 21 |
+
"aiohappyeyeballs": "2.4.3",
|
| 22 |
+
"aiohttp": "3.10.10",
|
| 23 |
+
"aiohttp-cors": "0.7.0",
|
| 24 |
+
"aiohttp-retry": "2.9.0",
|
| 25 |
+
"aioitertools": "0.12.0",
|
| 26 |
+
"aiooss2": "0.2.10",
|
| 27 |
+
"aiosignal": "1.3.1",
|
| 28 |
+
"alembic": "1.13.3",
|
| 29 |
+
"aliyun-python-sdk-core": "2.16.0",
|
| 30 |
+
"aliyun-python-sdk-kms": "2.16.5",
|
| 31 |
+
"amqp": "5.2.0",
|
| 32 |
+
"annotated-types": "0.7.0",
|
| 33 |
+
"antlr4-python3-runtime": "4.9.3",
|
| 34 |
+
"anyio": "4.6.2.post1",
|
| 35 |
+
"appdirs": "1.4.4",
|
| 36 |
+
"argcomplete": "3.5.1",
|
| 37 |
+
"async-timeout": "4.0.3",
|
| 38 |
+
"asyncssh": "2.18.0",
|
| 39 |
+
"atpublic": "5.0",
|
| 40 |
+
"attrs": "24.2.0",
|
| 41 |
+
"autogluon": "1.1.1",
|
| 42 |
+
"autogluon.common": "1.1.1",
|
| 43 |
+
"autogluon.core": "1.1.1",
|
| 44 |
+
"autogluon.features": "1.1.1",
|
| 45 |
+
"autogluon.multimodal": "1.1.1",
|
| 46 |
+
"autogluon.tabular": "1.1.1",
|
| 47 |
+
"autogluon.timeseries": "1.1.1",
|
| 48 |
+
"azure-core": "1.31.0",
|
| 49 |
+
"azure-datalake-store": "0.0.53",
|
| 50 |
+
"azure-identity": "1.19.0",
|
| 51 |
+
"azure-storage-blob": "12.23.1",
|
| 52 |
+
"bcrypt": "4.2.0",
|
| 53 |
+
"beautifulsoup4": "4.12.3",
|
| 54 |
+
"billiard": "4.2.1",
|
| 55 |
+
"blis": "0.7.11",
|
| 56 |
+
"boto3": "1.35.36",
|
| 57 |
+
"botocore": "1.35.36",
|
| 58 |
+
"cachetools": "5.5.0",
|
| 59 |
+
"catalogue": "2.0.10",
|
| 60 |
+
"catboost": "1.2.7",
|
| 61 |
+
"celery": "5.4.0",
|
| 62 |
+
"certifi": "2024.8.30",
|
| 63 |
+
"cffi": "1.17.1",
|
| 64 |
+
"charset-normalizer": "3.4.0",
|
| 65 |
+
"click": "8.1.7",
|
| 66 |
+
"click-didyoumean": "0.3.1",
|
| 67 |
+
"click-plugins": "1.1.1",
|
| 68 |
+
"click-repl": "0.3.0",
|
| 69 |
+
"cloudpathlib": "0.20.0",
|
| 70 |
+
"cloudpickle": "3.1.0",
|
| 71 |
+
"colorama": "0.4.6",
|
| 72 |
+
"coloredlogs": "15.0.1",
|
| 73 |
+
"colorful": "0.5.6",
|
| 74 |
+
"colorlog": "6.8.2",
|
| 75 |
+
"confection": "0.1.5",
|
| 76 |
+
"configobj": "5.0.9",
|
| 77 |
+
"contourpy": "1.3.0",
|
| 78 |
+
"crcmod": "1.7",
|
| 79 |
+
"cryptography": "43.0.3",
|
| 80 |
+
"cycler": "0.12.1",
|
| 81 |
+
"cymem": "2.0.8",
|
| 82 |
+
"datasets": "2.14.4",
|
| 83 |
+
"decorator": "5.1.1",
|
| 84 |
+
"defusedxml": "0.7.1",
|
| 85 |
+
"dictdiffer": "0.9.0",
|
| 86 |
+
"dill": "0.3.7",
|
| 87 |
+
"diskcache": "5.6.3",
|
| 88 |
+
"distlib": "0.3.9",
|
| 89 |
+
"distro": "1.9.0",
|
| 90 |
+
"dpath": "2.2.0",
|
| 91 |
+
"dulwich": "0.22.3",
|
| 92 |
+
"dvc": "3.56.0",
|
| 93 |
+
"dvc-azure": "3.1.0",
|
| 94 |
+
"dvc-data": "3.16.6",
|
| 95 |
+
"dvc-gdrive": "3.0.1",
|
| 96 |
+
"dvc-gs": "3.0.1",
|
| 97 |
+
"dvc-hdfs": "3.0.0",
|
| 98 |
+
"dvc-http": "2.32.0",
|
| 99 |
+
"dvc-objects": "5.1.0",
|
| 100 |
+
"dvc-oss": "3.0.0",
|
| 101 |
+
"dvc-render": "1.0.2",
|
| 102 |
+
"dvc-s3": "3.2.0",
|
| 103 |
+
"dvc-ssh": "4.1.1",
|
| 104 |
+
"dvc-studio-client": "0.21.0",
|
| 105 |
+
"dvc-task": "0.40.2",
|
| 106 |
+
"dvc-webdav": "3.0.0",
|
| 107 |
+
"dvc-webhdfs": "3.1.0",
|
| 108 |
+
"dvclive": "3.48.0",
|
| 109 |
+
"entrypoints": "0.4",
|
| 110 |
+
"evaluate": "0.4.3",
|
| 111 |
+
"exceptiongroup": "1.2.2",
|
| 112 |
+
"fastai": "2.7.18",
|
| 113 |
+
"fastcore": "1.7.19",
|
| 114 |
+
"fastdownload": "0.0.7",
|
| 115 |
+
"fastprogress": "1.0.3",
|
| 116 |
+
"filelock": "3.16.1",
|
| 117 |
+
"flatbuffers": "24.3.25",
|
| 118 |
+
"flatten-dict": "0.4.2",
|
| 119 |
+
"flufl-lock": "8.1.0",
|
| 120 |
+
"fonttools": "4.54.1",
|
| 121 |
+
"frozenlist": "1.5.0",
|
| 122 |
+
"fsspec": "2024.10.0",
|
| 123 |
+
"funcy": "2.0",
|
| 124 |
+
"future": "1.0.0",
|
| 125 |
+
"gcsfs": "2024.10.0",
|
| 126 |
+
"gdown": "5.2.0",
|
| 127 |
+
"gitdb": "4.0.11",
|
| 128 |
+
"gluonts": "0.15.1",
|
| 129 |
+
"google-api-core": "2.22.0",
|
| 130 |
+
"google-api-python-client": "2.149.0",
|
| 131 |
+
"google-auth": "2.35.0",
|
| 132 |
+
"google-auth-httplib2": "0.2.0",
|
| 133 |
+
"google-auth-oauthlib": "1.2.1",
|
| 134 |
+
"google-cloud-core": "2.4.1",
|
| 135 |
+
"google-cloud-storage": "2.18.2",
|
| 136 |
+
"google-crc32c": "1.6.0",
|
| 137 |
+
"google-resumable-media": "2.7.2",
|
| 138 |
+
"googleapis-common-protos": "1.65.0",
|
| 139 |
+
"grandalf": "0.8",
|
| 140 |
+
"graphviz": "0.20.3",
|
| 141 |
+
"greenlet": "3.1.1",
|
| 142 |
+
"grpcio": "1.67.1",
|
| 143 |
+
"gto": "1.7.1",
|
| 144 |
+
"h11": "0.14.0",
|
| 145 |
+
"httpcore": "1.0.6",
|
| 146 |
+
"httplib2": "0.22.0",
|
| 147 |
+
"httpx": "0.27.2",
|
| 148 |
+
"huggingface-hub": "0.26.2",
|
| 149 |
+
"humanfriendly": "10.0",
|
| 150 |
+
"hydra-core": "1.3.2",
|
| 151 |
+
"hyperopt": "0.2.7",
|
| 152 |
+
"idna": "3.10",
|
| 153 |
+
"imageio": "2.36.0",
|
| 154 |
+
"isodate": "0.7.2",
|
| 155 |
+
"iterative-telemetry": "0.0.9",
|
| 156 |
+
"jinja2": "3.1.4",
|
| 157 |
+
"jmespath": "0.10.0",
|
| 158 |
+
"joblib": "1.4.2",
|
| 159 |
+
"jsonschema": "4.21.1",
|
| 160 |
+
"jsonschema-specifications": "2024.10.1",
|
| 161 |
+
"kiwisolver": "1.4.7",
|
| 162 |
+
"knack": "0.12.0",
|
| 163 |
+
"kombu": "5.4.2",
|
| 164 |
+
"langcodes": "3.4.1",
|
| 165 |
+
"language-data": "1.2.0",
|
| 166 |
+
"lazy-loader": "0.4",
|
| 167 |
+
"lightgbm": "4.3.0",
|
| 168 |
+
"lightning": "2.3.3",
|
| 169 |
+
"lightning-utilities": "0.11.8",
|
| 170 |
+
"llvmlite": "0.43.0",
|
| 171 |
+
"marisa-trie": "1.2.1",
|
| 172 |
+
"markdown-it-py": "3.0.0",
|
| 173 |
+
"matplotlib": "3.9.2",
|
| 174 |
+
"mdurl": "0.1.2",
|
| 175 |
+
"mlforecast": "0.10.0",
|
| 176 |
+
"model-index": "0.1.11",
|
| 177 |
+
"mpmath": "1.3.0",
|
| 178 |
+
"msal": "1.31.0",
|
| 179 |
+
"msal-extensions": "1.2.0",
|
| 180 |
+
"msgpack": "1.1.0",
|
| 181 |
+
"multidict": "6.1.0",
|
| 182 |
+
"multiprocess": "0.70.15",
|
| 183 |
+
"murmurhash": "1.0.10",
|
| 184 |
+
"networkx": "3.4.2",
|
| 185 |
+
"nlpaug": "1.1.11",
|
| 186 |
+
"nltk": "3.9.1",
|
| 187 |
+
"nptyping": "2.4.1",
|
| 188 |
+
"numba": "0.60.0",
|
| 189 |
+
"numpy": "1.26.4",
|
| 190 |
+
"nvidia-cublas-cu12": "12.1.3.1",
|
| 191 |
+
"nvidia-cuda-cupti-cu12": "12.1.105",
|
| 192 |
+
"nvidia-cuda-nvrtc-cu12": "12.1.105",
|
| 193 |
+
"nvidia-cuda-runtime-cu12": "12.1.105",
|
| 194 |
+
"nvidia-cudnn-cu12": "8.9.2.26",
|
| 195 |
+
"nvidia-cufft-cu12": "11.0.2.54",
|
| 196 |
+
"nvidia-curand-cu12": "10.3.2.106",
|
| 197 |
+
"nvidia-cusolver-cu12": "11.4.5.107",
|
| 198 |
+
"nvidia-cusparse-cu12": "12.1.0.106",
|
| 199 |
+
"nvidia-ml-py3": "7.352.0",
|
| 200 |
+
"nvidia-nccl-cu12": "2.20.5",
|
| 201 |
+
"nvidia-nvjitlink-cu12": "12.6.77",
|
| 202 |
+
"nvidia-nvtx-cu12": "12.1.105",
|
| 203 |
+
"oauth2client": "4.1.3",
|
| 204 |
+
"oauthlib": "3.2.2",
|
| 205 |
+
"omegaconf": "2.2.3",
|
| 206 |
+
"onnx": "1.17.0",
|
| 207 |
+
"onnxruntime": "1.19.2",
|
| 208 |
+
"opencensus": "0.11.4",
|
| 209 |
+
"opencensus-context": "0.1.3",
|
| 210 |
+
"opendatalab": "0.0.10",
|
| 211 |
+
"openmim": "0.3.9",
|
| 212 |
+
"openxlab": "0.0.11",
|
| 213 |
+
"optimum": "1.17.1",
|
| 214 |
+
"optuna": "4.0.0",
|
| 215 |
+
"ordered-set": "4.1.0",
|
| 216 |
+
"orjson": "3.10.10",
|
| 217 |
+
"oss2": "2.18.1",
|
| 218 |
+
"ossfs": "2023.12.0",
|
| 219 |
+
"packaging": "24.1",
|
| 220 |
+
"pandas": "2.2.3",
|
| 221 |
+
"pathspec": "0.12.1",
|
| 222 |
+
"patsy": "0.5.6",
|
| 223 |
+
"pdf2image": "1.17.0",
|
| 224 |
+
"pillow": "10.4.0",
|
| 225 |
+
"pip": "24.3.1",
|
| 226 |
+
"platformdirs": "3.11.0",
|
| 227 |
+
"plotly": "5.24.1",
|
| 228 |
+
"portalocker": "2.10.1",
|
| 229 |
+
"preshed": "3.0.9",
|
| 230 |
+
"prometheus-client": "0.21.0",
|
| 231 |
+
"prompt-toolkit": "3.0.48",
|
| 232 |
+
"propcache": "0.2.0",
|
| 233 |
+
"proto-plus": "1.25.0",
|
| 234 |
+
"protobuf": "5.28.3",
|
| 235 |
+
"psutil": "5.9.8",
|
| 236 |
+
"py4j": "0.10.9.7",
|
| 237 |
+
"pyopenssl": "24.2.1",
|
| 238 |
+
"py-spy": "0.3.14",
|
| 239 |
+
"pyarrow": "18.0.0",
|
| 240 |
+
"pyasn1": "0.6.1",
|
| 241 |
+
"pyasn1-modules": "0.4.1",
|
| 242 |
+
"pycparser": "2.22",
|
| 243 |
+
"pycryptodome": "3.21.0",
|
| 244 |
+
"pydantic": "2.9.2",
|
| 245 |
+
"pydantic-core": "2.23.4",
|
| 246 |
+
"pydot": "3.0.2",
|
| 247 |
+
"pygit2": "1.16.0",
|
| 248 |
+
"pygments": "2.18.0",
|
| 249 |
+
"pygraphviz": "1.14",
|
| 250 |
+
"pygtrie": "2.5.0",
|
| 251 |
+
"pynvml": "11.5.3",
|
| 252 |
+
"pyparsing": "3.2.0",
|
| 253 |
+
"pytesseract": "0.3.10",
|
| 254 |
+
"python-dateutil": "2.9.0.post0",
|
| 255 |
+
"pytorch-lightning": "2.3.3",
|
| 256 |
+
"pytorch-metric-learning": "2.3.0",
|
| 257 |
+
"pytz": "2024.2",
|
| 258 |
+
"pywavelets": "1.7.0",
|
| 259 |
+
"ray": "2.10.0",
|
| 260 |
+
"referencing": "0.35.1",
|
| 261 |
+
"regex": "2024.9.11",
|
| 262 |
+
"requests": "2.32.3",
|
| 263 |
+
"requests-oauthlib": "2.0.0",
|
| 264 |
+
"rich": "13.9.3",
|
| 265 |
+
"rpds-py": "0.20.0",
|
| 266 |
+
"rsa": "4.9",
|
| 267 |
+
"ruamel.yaml": "0.18.6",
|
| 268 |
+
"ruamel.yaml.clib": "0.2.12",
|
| 269 |
+
"s3fs": "2024.10.0",
|
| 270 |
+
"s3transfer": "0.10.3",
|
| 271 |
+
"safetensors": "0.4.5",
|
| 272 |
+
"scikit-image": "0.20.0",
|
| 273 |
+
"scikit-learn": "1.4.0",
|
| 274 |
+
"scipy": "1.12.0",
|
| 275 |
+
"scmrepo": "3.3.8",
|
| 276 |
+
"seaborn": "0.13.2",
|
| 277 |
+
"semver": "3.0.2",
|
| 278 |
+
"sentencepiece": "0.2.0",
|
| 279 |
+
"seqeval": "1.2.2",
|
| 280 |
+
"setuptools": "75.2.0",
|
| 281 |
+
"shellingham": "1.5.4",
|
| 282 |
+
"shortuuid": "1.0.13",
|
| 283 |
+
"shtab": "1.7.1",
|
| 284 |
+
"six": "1.16.0",
|
| 285 |
+
"smart-open": "7.0.5",
|
| 286 |
+
"smmap": "5.0.1",
|
| 287 |
+
"sniffio": "1.3.1",
|
| 288 |
+
"soupsieve": "2.6",
|
| 289 |
+
"spacy": "3.7.5",
|
| 290 |
+
"spacy-legacy": "3.0.12",
|
| 291 |
+
"spacy-loggers": "1.0.5",
|
| 292 |
+
"sqltrie": "0.11.1",
|
| 293 |
+
"srsly": "2.4.8",
|
| 294 |
+
"sshfs": "2024.9.0",
|
| 295 |
+
"statsforecast": "1.4.0",
|
| 296 |
+
"statsmodels": "0.14.4",
|
| 297 |
+
"sympy": "1.13.3",
|
| 298 |
+
"tabulate": "0.9.0",
|
| 299 |
+
"tenacity": "9.0.0",
|
| 300 |
+
"tensorboard": "2.18.0",
|
| 301 |
+
"tensorboardx": "2.6.2.2",
|
| 302 |
+
"tensorboard-data-server": "0.7.2",
|
| 303 |
+
"text-unidecode": "1.3",
|
| 304 |
+
"thinc": "8.2.5",
|
| 305 |
+
"threadpoolctl": "3.5.0",
|
| 306 |
+
"tifffile": "2024.9.20",
|
| 307 |
+
"timm": "0.9.16",
|
| 308 |
+
"tokenizers": "0.19.1",
|
| 309 |
+
"tomlkit": "0.13.2",
|
| 310 |
+
"toolz": "0.12.1",
|
| 311 |
+
"torch": "2.3.1",
|
| 312 |
+
"torchmetrics": "1.2.1",
|
| 313 |
+
"torchvision": "0.18.1",
|
| 314 |
+
"tqdm": "4.66.6",
|
| 315 |
+
"transformers": "4.40.2",
|
| 316 |
+
"triton": "2.3.1",
|
| 317 |
+
"typer": "0.12.5",
|
| 318 |
+
"typing-extensions": "4.12.2",
|
| 319 |
+
"tzdata": "2024.2",
|
| 320 |
+
"uritemplate": "4.1.1",
|
| 321 |
+
"urllib3": "2.2.3",
|
| 322 |
+
"utilsforecast": "0.0.10",
|
| 323 |
+
"vine": "5.1.0",
|
| 324 |
+
"virtualenv": "20.27.1",
|
| 325 |
+
"voluptuous": "0.15.2",
|
| 326 |
+
"wasabi": "1.1.3",
|
| 327 |
+
"wcwidth": "0.2.13",
|
| 328 |
+
"weasel": "0.4.1",
|
| 329 |
+
"webdav4": "0.10.0",
|
| 330 |
+
"werkzeug": "3.0.6",
|
| 331 |
+
"window-ops": "0.0.15",
|
| 332 |
+
"wrapt": "1.16.0",
|
| 333 |
+
"xgboost": "2.0.3",
|
| 334 |
+
"xxhash": "3.5.0",
|
| 335 |
+
"yarl": "1.17.0",
|
| 336 |
+
"zc.lockfile": "3.0.post1",
|
| 337 |
+
"autocommand": "2.2.2",
|
| 338 |
+
"backports.tarfile": "1.2.0",
|
| 339 |
+
"importlib-metadata": "8.0.0",
|
| 340 |
+
"importlib-resources": "6.4.0",
|
| 341 |
+
"inflect": "7.3.1",
|
| 342 |
+
"jaraco.collections": "5.1.0",
|
| 343 |
+
"jaraco.context": "5.3.0",
|
| 344 |
+
"jaraco.functools": "4.0.1",
|
| 345 |
+
"jaraco.text": "3.12.1",
|
| 346 |
+
"more-itertools": "10.3.0",
|
| 347 |
+
"tomli": "2.0.1",
|
| 348 |
+
"typeguard": "4.3.0",
|
| 349 |
+
"wheel": "0.43.0",
|
| 350 |
+
"zipp": "3.19.2"
|
| 351 |
+
}
|
| 352 |
+
}
|
autogluon/model/model_info.txt
ADDED
|
@@ -0,0 +1,11 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"type_of_learner": "",
|
| 3 |
+
"model_types": "[]",
|
| 4 |
+
"model_performance": "{}",
|
| 5 |
+
"model_best": "",
|
| 6 |
+
"model_paths": "{}",
|
| 7 |
+
"model_fit_times": "{}",
|
| 8 |
+
"model_pred_times": "{}",
|
| 9 |
+
"num_bag_folds": "0",
|
| 10 |
+
"max_stack_level": "2"
|
| 11 |
+
}
|
autogluon/model/models/LightGBMBasic/model.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:490527fd79d7ecb074d5629d05a7ed4684748df2a64284d49a6359833b0e2599
|
| 3 |
+
size 18255
|
autogluon/model/models/WeightedEnsemble_L2/model.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:8b1c58a255a92233bd6a40a3deedad052d8a33ab8f1a24ac99b6b2309f573b48
|
| 3 |
+
size 6266
|
autogluon/model/models/trainer.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:b0ae12b06443f3b81b7bfb4c054957b48a823e49e21cafc7cf45f74fbe585e31
|
| 3 |
+
size 4548
|
autogluon/model/predictor.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:11b0e97edfad17ac8de2c788d60d0970dabc449b5d13d5bb94a21f92936ff57e
|
| 3 |
+
size 498
|
autogluon/model/version.txt
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
1.1.1
|
autogluon/plots/confusion_matrix.png
ADDED
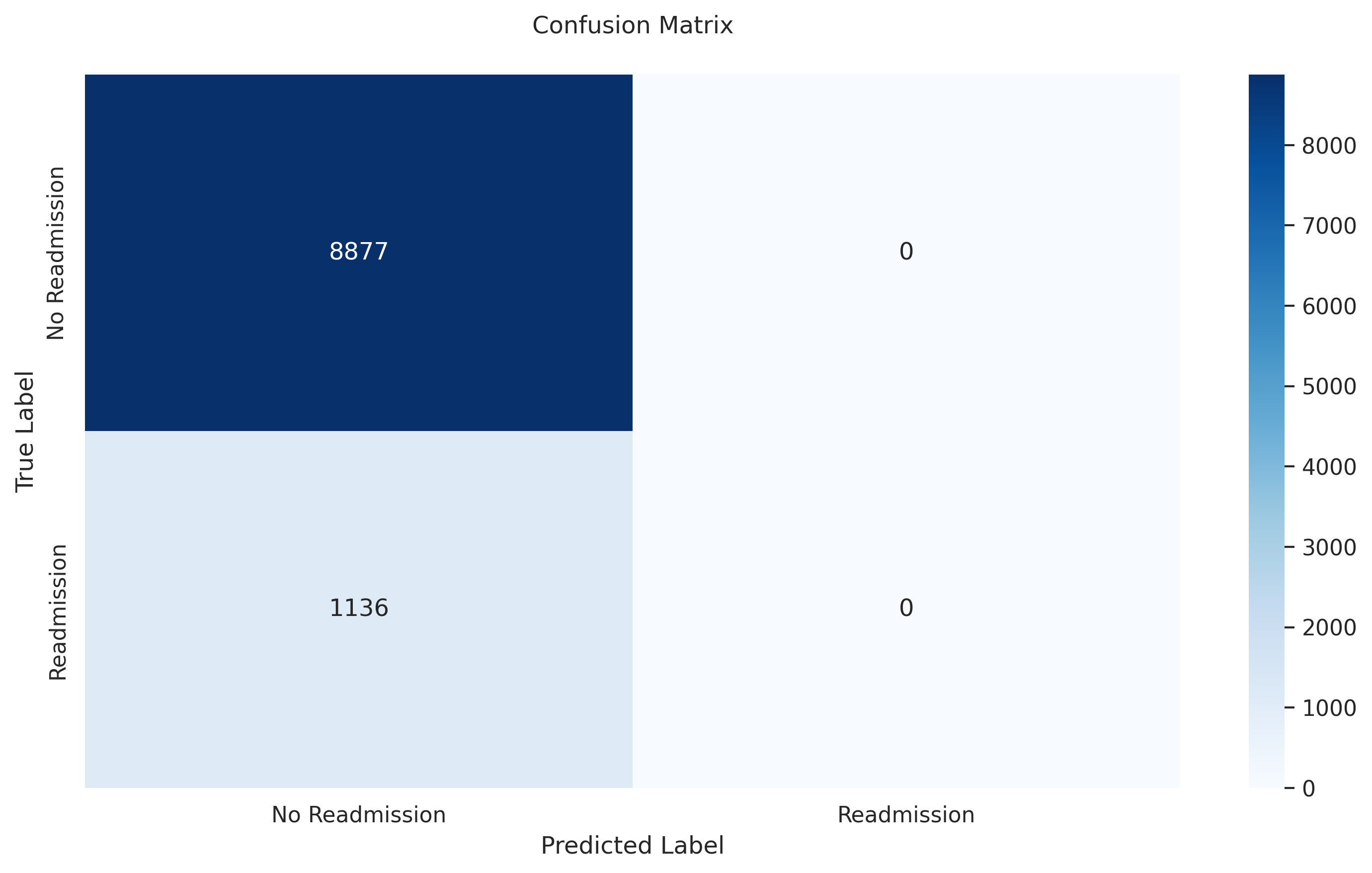
|
autogluon/plots/feature_importance.png
ADDED
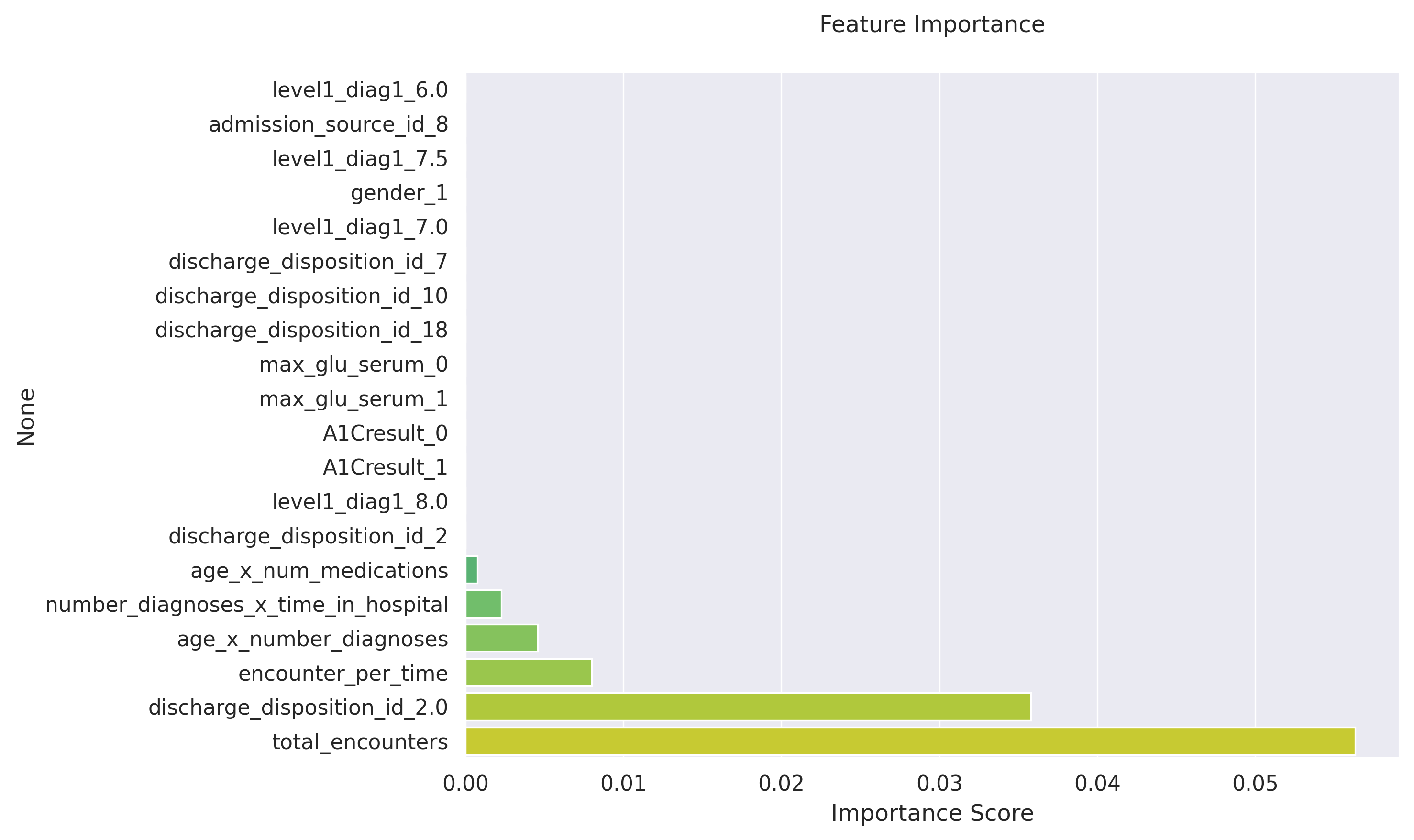
|
autogluon/plots/roc_curve.png
ADDED
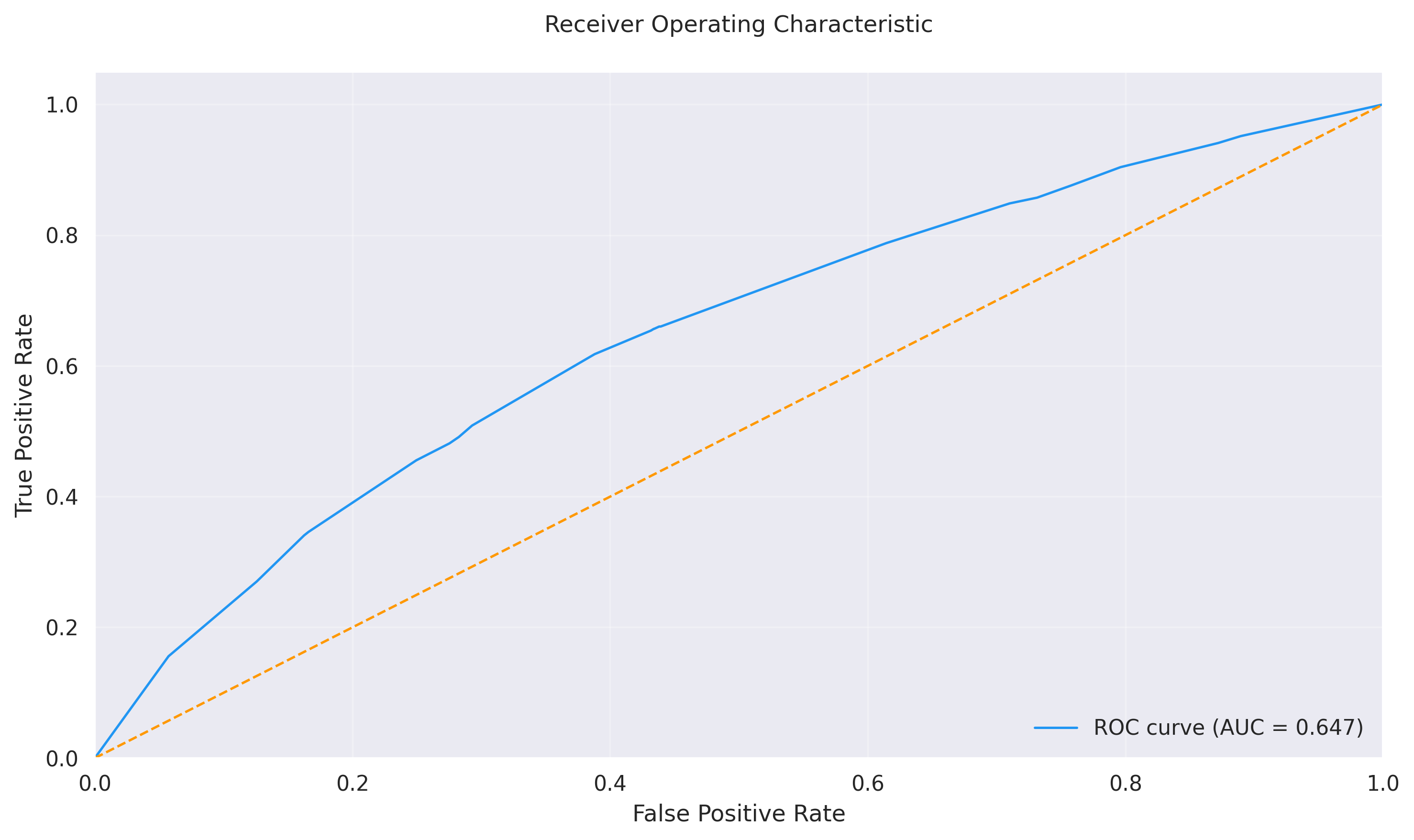
|
config.json
ADDED
|
@@ -0,0 +1,23 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"architectures": [
|
| 3 |
+
"TabularBinaryClassification"
|
| 4 |
+
],
|
| 5 |
+
"model_type": "autogluon",
|
| 6 |
+
"num_classes": 2,
|
| 7 |
+
"id2label": {
|
| 8 |
+
"0": "NO_READMISSION",
|
| 9 |
+
"1": "READMISSION"
|
| 10 |
+
},
|
| 11 |
+
"label2id": {
|
| 12 |
+
"NO_READMISSION": 0,
|
| 13 |
+
"READMISSION": 1
|
| 14 |
+
},
|
| 15 |
+
"task_specific_params": {
|
| 16 |
+
"classification": {
|
| 17 |
+
"problem_type": "binary_classification"
|
| 18 |
+
}
|
| 19 |
+
},
|
| 20 |
+
"preprocessing": {
|
| 21 |
+
"featurization_config": "preprocessing_config.json"
|
| 22 |
+
}
|
| 23 |
+
}
|
logistic_regression/metrics/feature_importance.csv
ADDED
|
@@ -0,0 +1,96 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
0
|
| 2 |
+
0.006981576326524651
|
| 3 |
+
-0.05324149411400925
|
| 4 |
+
0.14520246099157075
|
| 5 |
+
0.16641223168656358
|
| 6 |
+
-0.1849432098152513
|
| 7 |
+
-0.11971438258491879
|
| 8 |
+
-0.6673136921817721
|
| 9 |
+
0.0074340512164186865
|
| 10 |
+
0.00710484910980806
|
| 11 |
+
0.0208036887896313
|
| 12 |
+
-0.06866704970640153
|
| 13 |
+
0.02239663469454034
|
| 14 |
+
-0.04780275329131699
|
| 15 |
+
-0.001653654097302643
|
| 16 |
+
-0.005213524709277947
|
| 17 |
+
-0.038513975558627074
|
| 18 |
+
-0.04872273341141308
|
| 19 |
+
-0.021360986979047168
|
| 20 |
+
-0.0034907486285829445
|
| 21 |
+
-0.0357575743039899
|
| 22 |
+
-0.01159127633728281
|
| 23 |
+
-0.02893996449233495
|
| 24 |
+
-0.025228955046709828
|
| 25 |
+
-0.04067449776032111
|
| 26 |
+
-0.030644765633352167
|
| 27 |
+
-0.03515763759629512
|
| 28 |
+
-0.016977946801395048
|
| 29 |
+
0.09623559368069025
|
| 30 |
+
-0.012484082699882501
|
| 31 |
+
-0.0009575349297166111
|
| 32 |
+
6.65410045567994e-20
|
| 33 |
+
-0.028620120717707788
|
| 34 |
+
-0.019723707791475878
|
| 35 |
+
0.11631781014500936
|
| 36 |
+
0.0
|
| 37 |
+
-0.07245758942528399
|
| 38 |
+
-0.0656703695598517
|
| 39 |
+
-0.033527097500422244
|
| 40 |
+
-0.013496060253957467
|
| 41 |
+
-0.03848636989252564
|
| 42 |
+
-0.06888678626860888
|
| 43 |
+
-0.013496060253957467
|
| 44 |
+
-0.013496060253957467
|
| 45 |
+
0.4826929696848854
|
| 46 |
+
-0.033688000688812766
|
| 47 |
+
0.012021279187115997
|
| 48 |
+
-0.10097152621946577
|
| 49 |
+
-0.05616962767498837
|
| 50 |
+
0.37548621562715806
|
| 51 |
+
0.0
|
| 52 |
+
0.0
|
| 53 |
+
0.3189040988295721
|
| 54 |
+
-0.033023665719576566
|
| 55 |
+
0.16943919206646788
|
| 56 |
+
-0.135015724007822
|
| 57 |
+
-0.10070402041672873
|
| 58 |
+
-0.1747234071221618
|
| 59 |
+
-0.053920085325419025
|
| 60 |
+
0.044547618656999764
|
| 61 |
+
-0.0623980238417873
|
| 62 |
+
-0.135015724007822
|
| 63 |
+
0.109933062277
|
| 64 |
+
-0.07844982806285462
|
| 65 |
+
0.05400868882081664
|
| 66 |
+
-0.05616962767498837
|
| 67 |
+
-0.1295288236523361
|
| 68 |
+
-0.01614939687456253
|
| 69 |
+
0.7051654943458252
|
| 70 |
+
0.021261903585148515
|
| 71 |
+
0.2950273268755952
|
| 72 |
+
0.49860678225720706
|
| 73 |
+
0.20411937062125285
|
| 74 |
+
0.3154294773153667
|
| 75 |
+
0.8437534157758343
|
| 76 |
+
0.017755007787785892
|
| 77 |
+
0.2207871318828695
|
| 78 |
+
-0.013672481533416214
|
| 79 |
+
0.944583524149922
|
| 80 |
+
0.73223783295433
|
| 81 |
+
0.6556707014460541
|
| 82 |
+
0.04423504672748837
|
| 83 |
+
0.7594858589066283
|
| 84 |
+
0.5685208571153414
|
| 85 |
+
0.4973672369575105
|
| 86 |
+
0.2100659929803077
|
| 87 |
+
0.0
|
| 88 |
+
0.0
|
| 89 |
+
0.0
|
| 90 |
+
0.0
|
| 91 |
+
0.0
|
| 92 |
+
0.0
|
| 93 |
+
0.0
|
| 94 |
+
0.0
|
| 95 |
+
0.0
|
| 96 |
+
0.0
|
logistic_regression/metrics/metrics.json
ADDED
|
@@ -0,0 +1,14 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"val_accuracy": 0.7306731921693967,
|
| 3 |
+
"val_precision": 0.17053609117771212,
|
| 4 |
+
"val_recall": 0.35579040070453544,
|
| 5 |
+
"val_f1": 0.23056070766157796,
|
| 6 |
+
"val_auc": 0.5940456525813896,
|
| 7 |
+
"val_avg_precision": 0.159584262357455,
|
| 8 |
+
"test_accuracy": 0.7294881398252184,
|
| 9 |
+
"test_precision": 0.1630248500428449,
|
| 10 |
+
"test_recall": 0.33509467195068254,
|
| 11 |
+
"test_f1": 0.21933996253062402,
|
| 12 |
+
"test_auc": 0.5861944545016542,
|
| 13 |
+
"test_avg_precision": 0.15440710835201596
|
| 14 |
+
}
|
logistic_regression/model/model.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:4f399e7348b5eb61e6baade37068ffa8d7a5e9445227e15a9294eb9f54b65e56
|
| 3 |
+
size 1631
|
logistic_regression/model/model_summary.txt
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Model Parameters:
|
| 2 |
+
{'C': 1.0, 'class_weight': None, 'dual': False, 'fit_intercept': True, 'intercept_scaling': 1, 'l1_ratio': None, 'max_iter': 1000, 'multi_class': 'deprecated', 'n_jobs': None, 'penalty': 'l2', 'random_state': 42, 'solver': 'saga', 'tol': 0.0001, 'verbose': 0, 'warm_start': False}
|
logistic_regression/model/params.json
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"C": 1.0,
|
| 3 |
+
"penalty": "l2",
|
| 4 |
+
"solver": "saga"
|
| 5 |
+
}
|
logistic_regression/model/scaler.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:cc54552b9983586c38e93f9623af8ecf307363a1083289b8b52260d5ac009c14
|
| 3 |
+
size 5519
|
logistic_regression/plots/confusion_matrix.png
ADDED
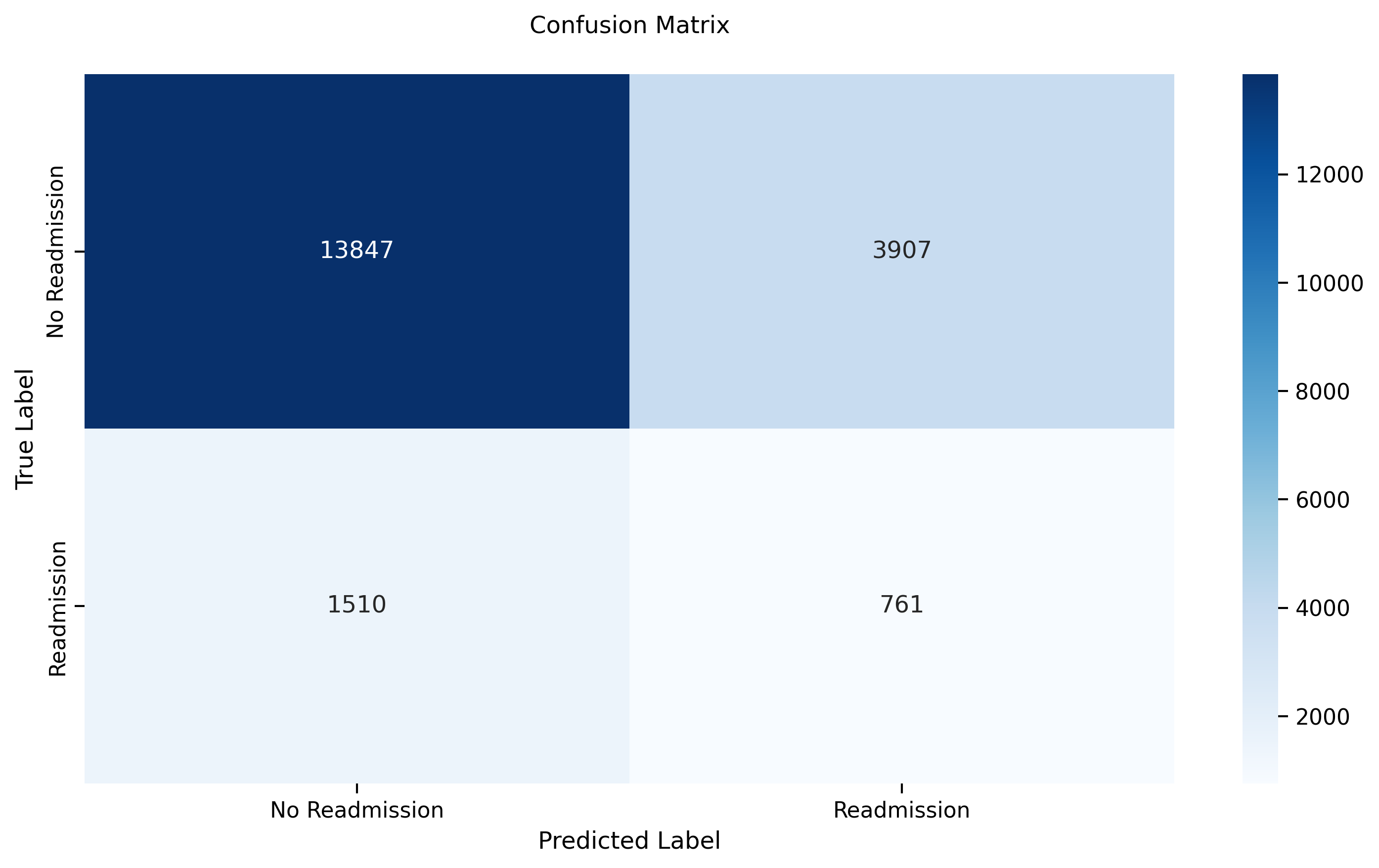
|
logistic_regression/plots/roc_curve.png
ADDED
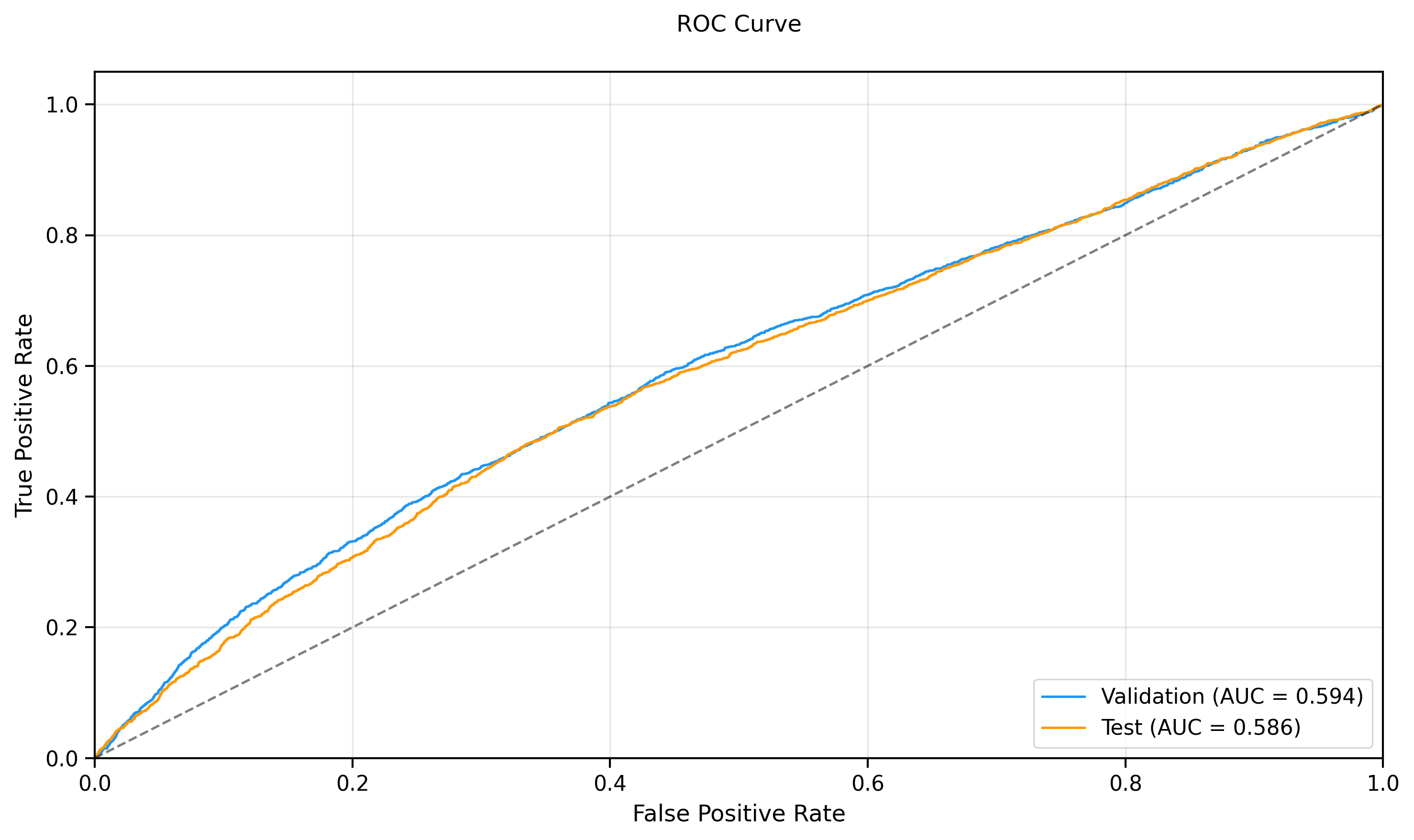
|
neural_network/metrics/metrics.json
ADDED
|
@@ -0,0 +1,25 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"train": {
|
| 3 |
+
"accuracy": 0.7377527952194214,
|
| 4 |
+
"auc": 0.8147459626197815,
|
| 5 |
+
"loss": 0.5141830444335938
|
| 6 |
+
},
|
| 7 |
+
"eval": {
|
| 8 |
+
"accuracy": 0.7731721997261047,
|
| 9 |
+
"auc": 0.6272097826004028,
|
| 10 |
+
"loss": 0.5016754269599915
|
| 11 |
+
},
|
| 12 |
+
"step": 0,
|
| 13 |
+
"val_accuracy": 0.7731721933679585,
|
| 14 |
+
"val_precision": 0.19533136570968607,
|
| 15 |
+
"val_recall": 0.32056362835755176,
|
| 16 |
+
"val_f1": 0.24274758252750916,
|
| 17 |
+
"val_auc": 0.6272456279863441,
|
| 18 |
+
"val_avg_precision": 0.17174880265376707,
|
| 19 |
+
"test_accuracy": 0.7703870162297128,
|
| 20 |
+
"test_precision": 0.18009348364036293,
|
| 21 |
+
"test_recall": 0.2884191985909291,
|
| 22 |
+
"test_f1": 0.22173324306025727,
|
| 23 |
+
"test_auc": 0.6234149477766672,
|
| 24 |
+
"test_avg_precision": 0.1627787212516569
|
| 25 |
+
}
|
neural_network/model/model.keras
ADDED
|
Binary file (123 kB). View file
|
|
|
neural_network/model/model_summary.txt
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Model: "sequential_1"
|
| 2 |
+
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━┓
|
| 3 |
+
┃ Layer (type) ┃ Output Shape ┃ Param # ┃
|
| 4 |
+
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━┩
|
| 5 |
+
│ dense_3 (Dense) │ (None, 48) │ 4,608 │
|
| 6 |
+
├──────────────────────────────────────┼─────────────────────────────┼─────────────────┤
|
| 7 |
+
│ batch_normalization_2 │ (None, 48) │ 192 │
|
| 8 |
+
│ (BatchNormalization) │ │ │
|
| 9 |
+
├──────────────────────────────────────┼─────────────────────────────┼─────────────────┤
|
| 10 |
+
│ dropout_2 (Dropout) │ (None, 48) │ 0 │
|
| 11 |
+
├──────────────────────────────────────┼─────────────────────────────┼─────────────────┤
|
| 12 |
+
│ dense_4 (Dense) │ (None, 38) │ 1,862 │
|
| 13 |
+
├──────────────────────────────────────┼─────────────────────────────┼─────────────────┤
|
| 14 |
+
│ batch_normalization_3 │ (None, 38) │ 152 │
|
| 15 |
+
│ (BatchNormalization) │ │ │
|
| 16 |
+
├──────────────────────────────────────┼─────────────────────────────┼─────────────────┤
|
| 17 |
+
│ dropout_3 (Dropout) │ (None, 38) │ 0 │
|
| 18 |
+
├──────────────────────────────────────┼─────────────────────────────┼─────────────────┤
|
| 19 |
+
│ dense_5 (Dense) │ (None, 1) │ 39 │
|
| 20 |
+
└──────────────────────────────────────┴─────────────────────────────┴─────────────────┘
|
| 21 |
+
Total params: 6,853 (26.77 KB)
|
| 22 |
+
Trainable params: 6,681 (26.10 KB)
|
| 23 |
+
Non-trainable params: 172 (688.00 B)
|
| 24 |
+
|
neural_network/model/params.json
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"n_layers": 2,
|
| 3 |
+
"units_first": 48,
|
| 4 |
+
"units_factor": 0.7950714306409916,
|
| 5 |
+
"dropout": 0.2731993941811405,
|
| 6 |
+
"learning_rate": 0.003968793330444372,
|
| 7 |
+
"batch_size": 32,
|
| 8 |
+
"activation": "relu",
|
| 9 |
+
"optimizer": "adam"
|
| 10 |
+
}
|
neural_network/model/scaler.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0ee48355f8a8cc48ceade28dc3ecd4944699c612fe75252bb66ddce52091ebb5
|
| 3 |
+
size 5519
|
neural_network/plots/confusion_matrix.png
ADDED
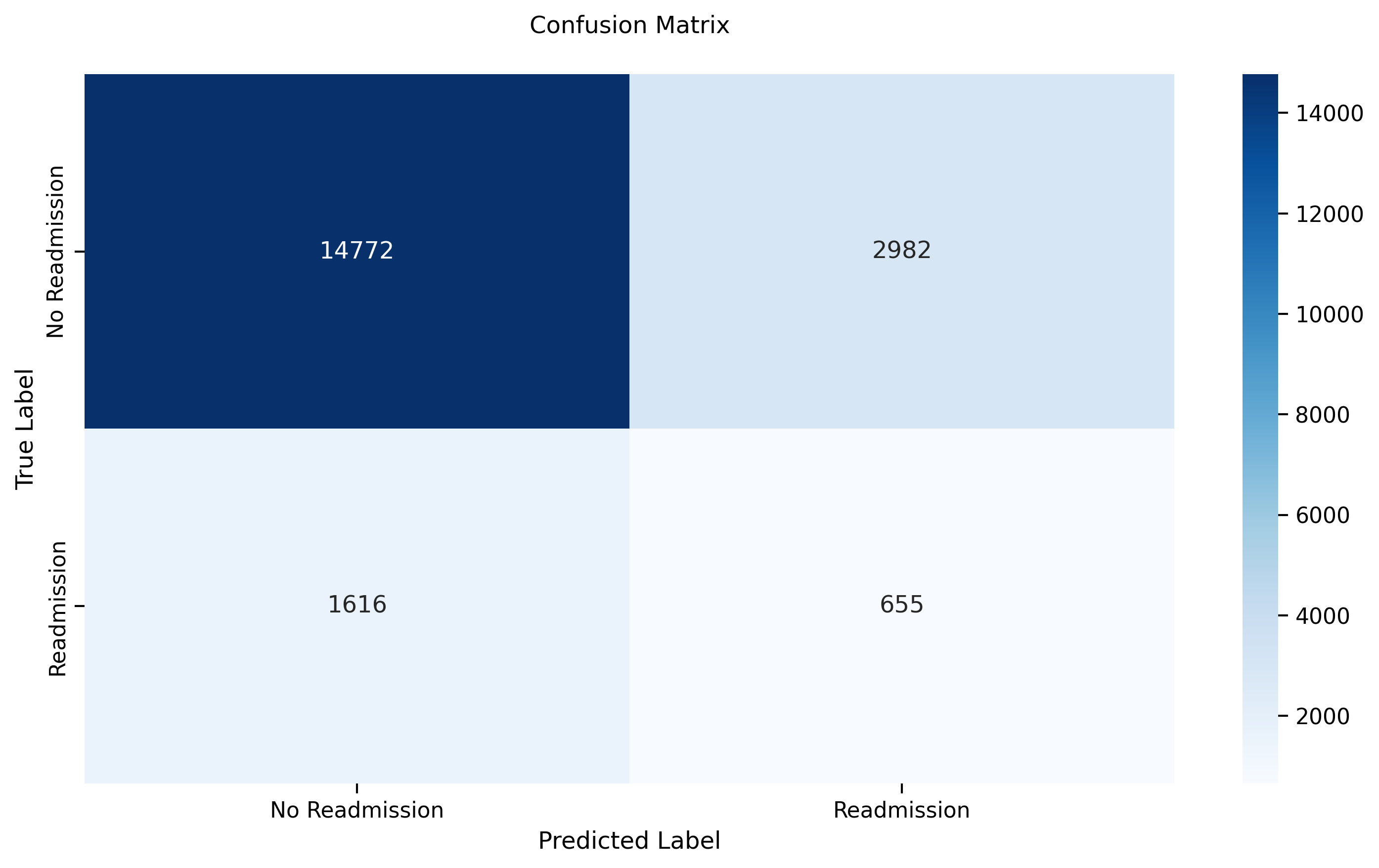
|
neural_network/plots/model_architecture.png
ADDED
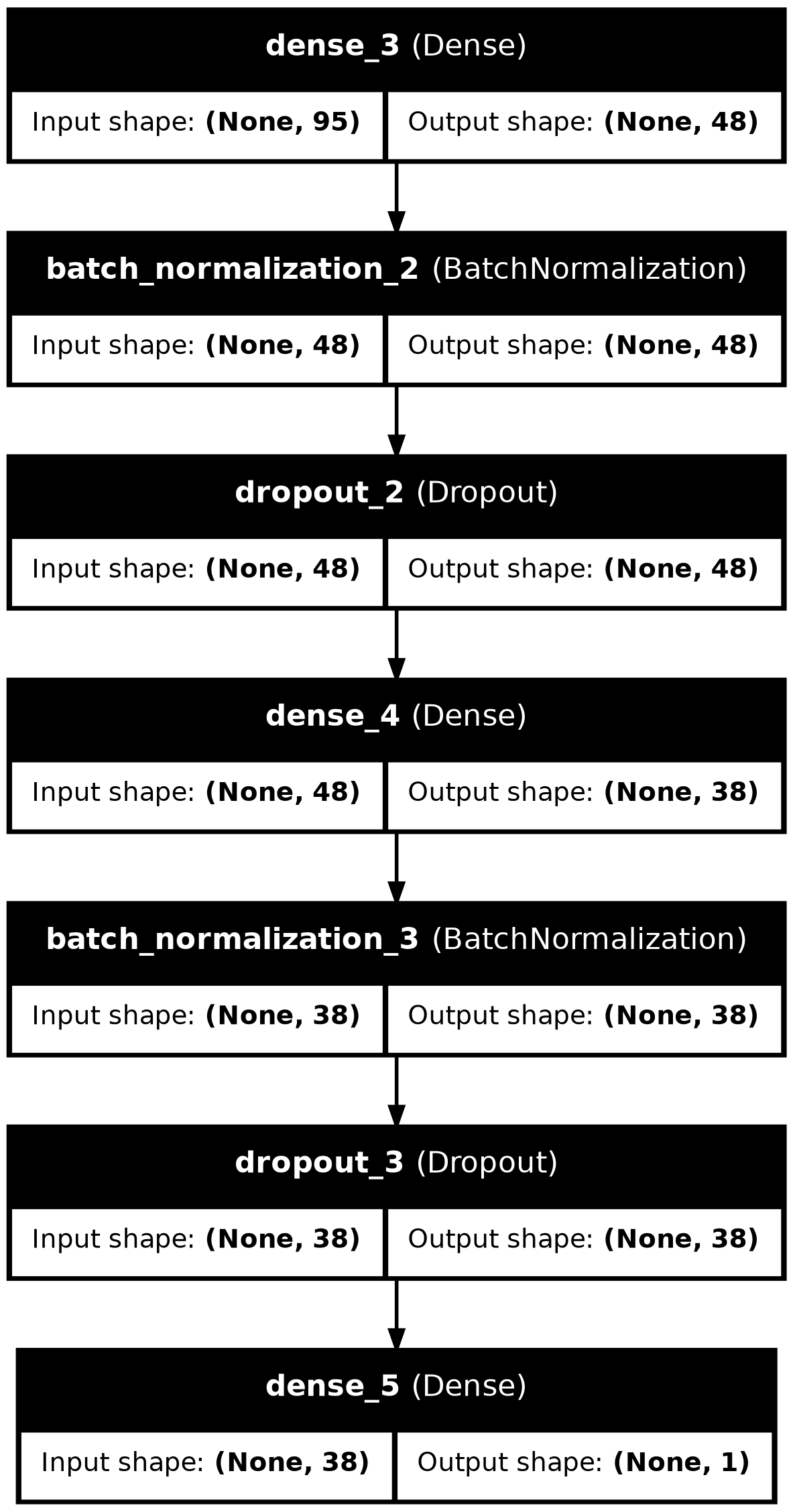
|
neural_network/plots/roc_curve.png
ADDED
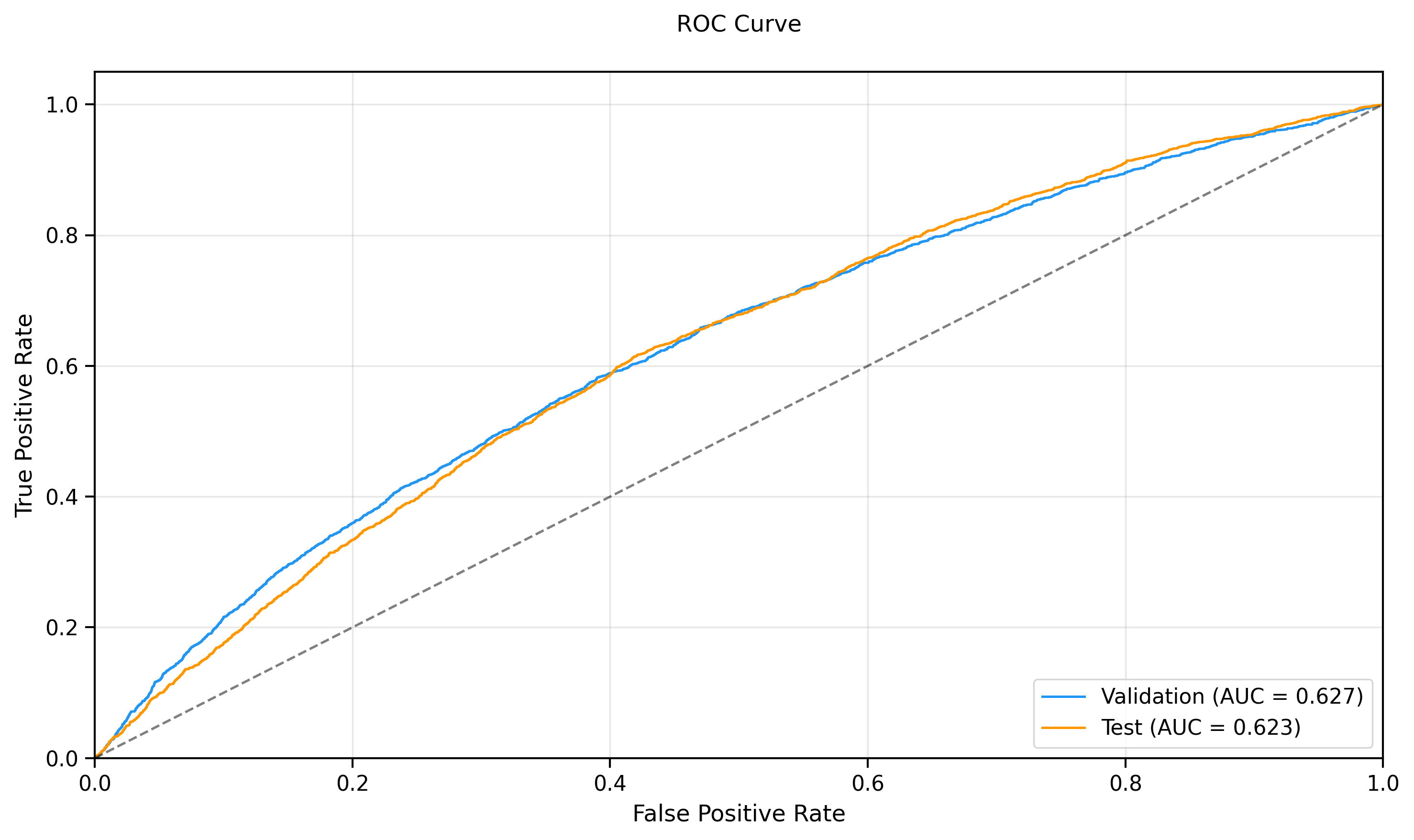
|
preprocessing_config.json
ADDED
|
@@ -0,0 +1,189 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"preprocessor": {
|
| 3 |
+
"numeric_features": [
|
| 4 |
+
"age",
|
| 5 |
+
"time_in_hospital",
|
| 6 |
+
"num_lab_procedures",
|
| 7 |
+
"num_procedures",
|
| 8 |
+
"num_medications",
|
| 9 |
+
"number_diagnoses",
|
| 10 |
+
"total_medications",
|
| 11 |
+
"medication_density",
|
| 12 |
+
"numchange",
|
| 13 |
+
"nummed",
|
| 14 |
+
"total_encounters",
|
| 15 |
+
"encounter_per_time",
|
| 16 |
+
"procedures_per_day",
|
| 17 |
+
"lab_procedures_per_day",
|
| 18 |
+
"procedures_to_medications",
|
| 19 |
+
"diagnoses_per_encounter",
|
| 20 |
+
"number_outpatient_log1p",
|
| 21 |
+
"number_emergency_log1p",
|
| 22 |
+
"number_inpatient_log1p"
|
| 23 |
+
],
|
| 24 |
+
"binary_features": [
|
| 25 |
+
"gender",
|
| 26 |
+
"diabetesmed",
|
| 27 |
+
"change",
|
| 28 |
+
"insulin_with_oral"
|
| 29 |
+
],
|
| 30 |
+
"medication_features": [
|
| 31 |
+
"metformin",
|
| 32 |
+
"repaglinide",
|
| 33 |
+
"nateglinide",
|
| 34 |
+
"chlorpropamide",
|
| 35 |
+
"glimepiride",
|
| 36 |
+
"glipizide",
|
| 37 |
+
"glyburide",
|
| 38 |
+
"pioglitazone",
|
| 39 |
+
"rosiglitazone",
|
| 40 |
+
"acarbose",
|
| 41 |
+
"miglitol",
|
| 42 |
+
"insulin",
|
| 43 |
+
"glyburide-metformin",
|
| 44 |
+
"tolazamide",
|
| 45 |
+
"metformin-pioglitazone",
|
| 46 |
+
"metformin-rosiglitazone",
|
| 47 |
+
"glimepiride-pioglitazone",
|
| 48 |
+
"glipizide-metformin",
|
| 49 |
+
"troglitazone",
|
| 50 |
+
"tolbutamide",
|
| 51 |
+
"acetohexamide"
|
| 52 |
+
],
|
| 53 |
+
"interaction_features": [
|
| 54 |
+
"num_medications_x_time_in_hospital",
|
| 55 |
+
"num_procedures_x_time_in_hospital",
|
| 56 |
+
"num_lab_procedures_x_time_in_hospital",
|
| 57 |
+
"number_diagnoses_x_time_in_hospital",
|
| 58 |
+
"age_x_number_diagnoses",
|
| 59 |
+
"age_x_num_medications",
|
| 60 |
+
"total_medications_x_number_diagnoses",
|
| 61 |
+
"num_medications_x_num_procedures",
|
| 62 |
+
"time_in_hospital_x_num_lab_procedures",
|
| 63 |
+
"num_medications_x_num_lab_procedures",
|
| 64 |
+
"change_x_num_medications",
|
| 65 |
+
"num_medications_x_numchange"
|
| 66 |
+
],
|
| 67 |
+
"ratio_features": [
|
| 68 |
+
"procedure_medication_ratio",
|
| 69 |
+
"lab_procedure_ratio",
|
| 70 |
+
"diagnosis_procedure_ratio"
|
| 71 |
+
],
|
| 72 |
+
"categorical_features": {
|
| 73 |
+
"admission_type_id": [
|
| 74 |
+
1,
|
| 75 |
+
2,
|
| 76 |
+
3,
|
| 77 |
+
4,
|
| 78 |
+
5,
|
| 79 |
+
6,
|
| 80 |
+
7,
|
| 81 |
+
8
|
| 82 |
+
],
|
| 83 |
+
"discharge_disposition_id": [
|
| 84 |
+
1,
|
| 85 |
+
2,
|
| 86 |
+
3,
|
| 87 |
+
4,
|
| 88 |
+
5,
|
| 89 |
+
6,
|
| 90 |
+
7,
|
| 91 |
+
8,
|
| 92 |
+
9,
|
| 93 |
+
10,
|
| 94 |
+
11,
|
| 95 |
+
12,
|
| 96 |
+
13,
|
| 97 |
+
14,
|
| 98 |
+
15,
|
| 99 |
+
16,
|
| 100 |
+
17,
|
| 101 |
+
18,
|
| 102 |
+
19,
|
| 103 |
+
20,
|
| 104 |
+
21,
|
| 105 |
+
22,
|
| 106 |
+
23,
|
| 107 |
+
24,
|
| 108 |
+
25,
|
| 109 |
+
26
|
| 110 |
+
],
|
| 111 |
+
"admission_source_id": [
|
| 112 |
+
1,
|
| 113 |
+
2,
|
| 114 |
+
3,
|
| 115 |
+
4,
|
| 116 |
+
5,
|
| 117 |
+
6,
|
| 118 |
+
7,
|
| 119 |
+
8,
|
| 120 |
+
9,
|
| 121 |
+
10,
|
| 122 |
+
11,
|
| 123 |
+
12,
|
| 124 |
+
13,
|
| 125 |
+
14,
|
| 126 |
+
15,
|
| 127 |
+
16,
|
| 128 |
+
17,
|
| 129 |
+
18,
|
| 130 |
+
19,
|
| 131 |
+
20,
|
| 132 |
+
21,
|
| 133 |
+
22,
|
| 134 |
+
23,
|
| 135 |
+
24,
|
| 136 |
+
25
|
| 137 |
+
],
|
| 138 |
+
"level1_diag1": [
|
| 139 |
+
0,
|
| 140 |
+
1,
|
| 141 |
+
2,
|
| 142 |
+
3,
|
| 143 |
+
4,
|
| 144 |
+
5,
|
| 145 |
+
6,
|
| 146 |
+
7,
|
| 147 |
+
8
|
| 148 |
+
]
|
| 149 |
+
},
|
| 150 |
+
"lab_features": {
|
| 151 |
+
"a1cresult": {
|
| 152 |
+
"mapping": {
|
| 153 |
+
">7": 1,
|
| 154 |
+
">8": 1,
|
| 155 |
+
"Norm": 0,
|
| 156 |
+
"None": -99
|
| 157 |
+
}
|
| 158 |
+
},
|
| 159 |
+
"max_glu_serum": {
|
| 160 |
+
"mapping": {
|
| 161 |
+
">200": 1,
|
| 162 |
+
">300": 1,
|
| 163 |
+
"Norm": 0,
|
| 164 |
+
"None": -99
|
| 165 |
+
}
|
| 166 |
+
}
|
| 167 |
+
}
|
| 168 |
+
},
|
| 169 |
+
"transformations": {
|
| 170 |
+
"numeric_scaling": "standard",
|
| 171 |
+
"outlier_handling": {
|
| 172 |
+
"method": "clip",
|
| 173 |
+
"std_multiplier": 5
|
| 174 |
+
},
|
| 175 |
+
"missing_values": {
|
| 176 |
+
"numeric": "mean",
|
| 177 |
+
"categorical": "mode"
|
| 178 |
+
}
|
| 179 |
+
},
|
| 180 |
+
"target": {
|
| 181 |
+
"name": "readmitted",
|
| 182 |
+
"type": "binary",
|
| 183 |
+
"mapping": {
|
| 184 |
+
">30": 0,
|
| 185 |
+
"<30": 1,
|
| 186 |
+
"NO": 0
|
| 187 |
+
}
|
| 188 |
+
}
|
| 189 |
+
}
|
tokenizer_config.json
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"feature_extractor_type": "tabular",
|
| 3 |
+
"framework": "pt",
|
| 4 |
+
"num_features": null,
|
| 5 |
+
"requires_preprocessing": true,
|
| 6 |
+
"preprocessing_config": "preprocessing_config.json"
|
| 7 |
+
}
|