Upload folder using huggingface_hub
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- .gitattributes +1 -0
- .ipynb_checkpoints/lecture_intro_cn-checkpoint.ipynb +14 -1
- 01-data_env/.ipynb_checkpoints/3-dataset-use-checkpoint.ipynb +4 -1
- 01-data_env/1-env-intro.ipynb +42 -1
- 01-data_env/2-data-intro.ipynb +3 -3
- 01-data_env/3-dataset-use.ipynb +7 -2
- 01-data_env/img/.ipynb_checkpoints/zhushi-checkpoint.png +0 -0
- 01-data_env/img/datasets_dnagpt.png +0 -0
- 02-gpt2_bert/.ipynb_checkpoints/1-dna-bpe-checkpoint.ipynb +528 -0
- 02-gpt2_bert/.ipynb_checkpoints/2-dna-gpt-checkpoint.ipynb +0 -0
- 02-gpt2_bert/.ipynb_checkpoints/3-dna-bert-checkpoint.ipynb +253 -0
- 02-gpt2_bert/.ipynb_checkpoints/4-gene-feature-checkpoint.ipynb +489 -0
- 02-gpt2_bert/.ipynb_checkpoints/5-multi-seq-gpt-checkpoint.ipynb +261 -0
- 02-gpt2_bert/.ipynb_checkpoints/dna_wordpiece_dict-checkpoint.json +0 -0
- 02-gpt2_bert/1-dna-bpe.ipynb +528 -0
- 02-gpt2_bert/2-dna-gpt.ipynb +0 -0
- 02-gpt2_bert/3-dna-bert.ipynb +0 -0
- 02-gpt2_bert/4-gene-feature.ipynb +489 -0
- 02-gpt2_bert/5-multi-seq-gpt.ipynb +261 -0
- 02-gpt2_bert/dna_bert_v0/config.json +28 -0
- 02-gpt2_bert/dna_bert_v0/generation_config.json +7 -0
- 02-gpt2_bert/dna_bert_v0/model.safetensors +3 -0
- 02-gpt2_bert/dna_bert_v0/training_args.bin +3 -0
- 02-gpt2_bert/dna_bpe_dict.json +0 -0
- 02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/merges-checkpoint.txt +3 -0
- 02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/special_tokens_map-checkpoint.json +5 -0
- 02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/tokenizer-checkpoint.json +0 -0
- 02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/tokenizer_config-checkpoint.json +20 -0
- 02-gpt2_bert/dna_bpe_dict/merges.txt +3 -0
- 02-gpt2_bert/dna_bpe_dict/special_tokens_map.json +5 -0
- 02-gpt2_bert/dna_bpe_dict/tokenizer.json +0 -0
- 02-gpt2_bert/dna_bpe_dict/tokenizer_config.json +20 -0
- 02-gpt2_bert/dna_bpe_dict/vocab.json +0 -0
- 02-gpt2_bert/dna_gpt2_v0/config.json +39 -0
- 02-gpt2_bert/dna_gpt2_v0/generation_config.json +6 -0
- 02-gpt2_bert/dna_gpt2_v0/merges.txt +3 -0
- 02-gpt2_bert/dna_gpt2_v0/model.safetensors +3 -0
- 02-gpt2_bert/dna_gpt2_v0/special_tokens_map.json +23 -0
- 02-gpt2_bert/dna_gpt2_v0/tokenizer.json +0 -0
- 02-gpt2_bert/dna_gpt2_v0/tokenizer_config.json +20 -0
- 02-gpt2_bert/dna_gpt2_v0/training_args.bin +3 -0
- 02-gpt2_bert/dna_gpt2_v0/vocab.json +0 -0
- 02-gpt2_bert/dna_wordpiece_dict.json +0 -0
- 02-gpt2_bert/dna_wordpiece_dict/special_tokens_map.json +7 -0
- 02-gpt2_bert/dna_wordpiece_dict/tokenizer.json +0 -0
- 02-gpt2_bert/dna_wordpiece_dict/tokenizer_config.json +53 -0
- 02-gpt2_bert/img/.ipynb_checkpoints/gpt2-stru-checkpoint.png +0 -0
- 02-gpt2_bert/img/gpt2-netron.png +0 -0
- 02-gpt2_bert/img/gpt2-stru.png +0 -0
- 02-gpt2_bert/img/llm-visual.png +0 -0
.gitattributes
CHANGED
|
@@ -35,3 +35,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
*.psd filter=lfs diff=lfs merge=lfs -text
|
| 37 |
*.txt filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
*.psd filter=lfs diff=lfs merge=lfs -text
|
| 37 |
*.txt filter=lfs diff=lfs merge=lfs -text
|
| 38 |
+
img/gpt2_bridge.png filter=lfs diff=lfs merge=lfs -text
|
.ipynb_checkpoints/lecture_intro_cn-checkpoint.ipynb
CHANGED
|
@@ -5,7 +5,7 @@
|
|
| 5 |
"id": "2365faf7-39fb-4e53-a810-2e28c4f6b4c1",
|
| 6 |
"metadata": {},
|
| 7 |
"source": [
|
| 8 |
-
"# DNAGTP2
|
| 9 |
"\n",
|
| 10 |
"## 1 概要\n",
|
| 11 |
"自然语言大模型早已超出NLP研究领域,正在成为AI for science的基石。生物信息学中的基因序列,则是和自然语言最类似的,把大模型应用于生物序列研究,就成了最近一两年的热门研究方向,特别是2024年预测蛋白质结构的alphaFold获得诺贝尔化学奖,更是为生物学的研究指明了未来的方向。\n",
|
|
@@ -18,6 +18,9 @@
|
|
| 18 |
"\n",
|
| 19 |
"DNAGTP2就是这样的梯子,仅望能抛砖引玉,让更多的生物学工作者能够越过大模型的门槛,戴上大模型的翅膀,卷过同行。\n",
|
| 20 |
"\n",
|
|
|
|
|
|
|
|
|
|
| 21 |
"## 2 教程特色\n",
|
| 22 |
"本教程主要有以下特色:\n",
|
| 23 |
"\n",
|
|
@@ -39,7 +42,17 @@
|
|
| 39 |
"\n",
|
| 40 |
"2 大模型学习入门。不仅是生物学领域的,都可以看看,和一般大模型入门没啥差别,只是数据不同。\n",
|
| 41 |
"\n",
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 42 |
"## 3 教程大纲\n",
|
|
|
|
|
|
|
|
|
|
|
|
|
| 43 |
"1 数据和环境\n",
|
| 44 |
"\n",
|
| 45 |
"1.1 大模型运行环境简介\n",
|
|
|
|
| 5 |
"id": "2365faf7-39fb-4e53-a810-2e28c4f6b4c1",
|
| 6 |
"metadata": {},
|
| 7 |
"source": [
|
| 8 |
+
"# DNAGTP2-基因序列大模型最佳入门\n",
|
| 9 |
"\n",
|
| 10 |
"## 1 概要\n",
|
| 11 |
"自然语言大模型早已超出NLP研究领域,正在成为AI for science的基石。生物信息学中的基因序列,则是和自然语言最类似的,把大模型应用于生物序列研究,就成了最近一两年的热门研究方向,特别是2024年预测蛋白质结构的alphaFold获得诺贝尔化学奖,更是为生物学的研究指明了未来的方向。\n",
|
|
|
|
| 18 |
"\n",
|
| 19 |
"DNAGTP2就是这样的梯子,仅望能抛砖引玉,让更多的生物学工作者能够越过大模型的门槛,戴上大模型的翅膀,卷过同行。\n",
|
| 20 |
"\n",
|
| 21 |
+
"\n",
|
| 22 |
+
"<<img src='img/gpt2_bridge.png' width=\"600px\" />\n",
|
| 23 |
+
"\n",
|
| 24 |
"## 2 教程特色\n",
|
| 25 |
"本教程主要有以下特色:\n",
|
| 26 |
"\n",
|
|
|
|
| 42 |
"\n",
|
| 43 |
"2 大模型学习入门。不仅是生物学领域的,都可以看看,和一般大模型入门没啥差别,只是数据不同。\n",
|
| 44 |
"\n",
|
| 45 |
+
"\n",
|
| 46 |
+
"huggingface: https://huggingface.co/dnagpt/dnagpt2\n",
|
| 47 |
+
"\n",
|
| 48 |
+
"github: https://github.com/maris205/dnagpt2\n",
|
| 49 |
+
"\n",
|
| 50 |
+
"\n",
|
| 51 |
"## 3 教程大纲\n",
|
| 52 |
+
"\n",
|
| 53 |
+
"<img src='img/DNAGPT2.png' width=\"600px\" />\n",
|
| 54 |
+
"\n",
|
| 55 |
+
"\n",
|
| 56 |
"1 数据和环境\n",
|
| 57 |
"\n",
|
| 58 |
"1.1 大模型运行环境简介\n",
|
01-data_env/.ipynb_checkpoints/3-dataset-use-checkpoint.ipynb
CHANGED
|
@@ -134,6 +134,7 @@
|
|
| 134 |
]
|
| 135 |
},
|
| 136 |
{
|
|
|
|
| 137 |
"cell_type": "markdown",
|
| 138 |
"id": "17a1fa7c-ff4b-419f-8a82-e58cc5777cd4",
|
| 139 |
"metadata": {},
|
|
@@ -142,7 +143,9 @@
|
|
| 142 |
"\n",
|
| 143 |
"当然,数据也可以直接从huggingface的线上仓库读取,这时候需要注意科学上网问题。\n",
|
| 144 |
"\n",
|
| 145 |
-
"具体使用函数也是load_dataset"
|
|
|
|
|
|
|
| 146 |
]
|
| 147 |
},
|
| 148 |
{
|
|
|
|
| 134 |
]
|
| 135 |
},
|
| 136 |
{
|
| 137 |
+
"attachments": {},
|
| 138 |
"cell_type": "markdown",
|
| 139 |
"id": "17a1fa7c-ff4b-419f-8a82-e58cc5777cd4",
|
| 140 |
"metadata": {},
|
|
|
|
| 143 |
"\n",
|
| 144 |
"当然,数据也可以直接从huggingface的线上仓库读取,这时候需要注意科学上网问题。\n",
|
| 145 |
"\n",
|
| 146 |
+
"具体使用函数也是load_dataset\n",
|
| 147 |
+
"\n",
|
| 148 |
+
"<img src='img/datasets_dnagpt.png' width='800px' />"
|
| 149 |
]
|
| 150 |
},
|
| 151 |
{
|
01-data_env/1-env-intro.ipynb
CHANGED
|
@@ -62,7 +62,48 @@
|
|
| 62 |
"id": "444adc87-78c8-4209-8260-0c5c4a668ea0",
|
| 63 |
"metadata": {},
|
| 64 |
"outputs": [],
|
| 65 |
-
"source": [
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 66 |
}
|
| 67 |
],
|
| 68 |
"metadata": {
|
|
|
|
| 62 |
"id": "444adc87-78c8-4209-8260-0c5c4a668ea0",
|
| 63 |
"metadata": {},
|
| 64 |
"outputs": [],
|
| 65 |
+
"source": [
|
| 66 |
+
"import os\n",
|
| 67 |
+
"\n",
|
| 68 |
+
"# 设置环境变量, autodl专区 其他idc\n",
|
| 69 |
+
"os.environ['HF_ENDPOINT'] = 'https://hf-mirror.com'\n",
|
| 70 |
+
"\n",
|
| 71 |
+
"# 打印环境变量以确认设置成功\n",
|
| 72 |
+
"print(os.environ.get('HF_ENDPOINT'))"
|
| 73 |
+
]
|
| 74 |
+
},
|
| 75 |
+
{
|
| 76 |
+
"cell_type": "code",
|
| 77 |
+
"execution_count": null,
|
| 78 |
+
"id": "06d9dc67-dbd4-4d37-bbdb-ccf59c8fdbf9",
|
| 79 |
+
"metadata": {},
|
| 80 |
+
"outputs": [],
|
| 81 |
+
"source": [
|
| 82 |
+
"import subprocess\n",
|
| 83 |
+
"import os\n",
|
| 84 |
+
"# 设置环境变量, autodl一般区域\n",
|
| 85 |
+
"result = subprocess.run('bash -c \"source /etc/network_turbo && env | grep proxy\"', shell=True, capture_output=True, text=True)\n",
|
| 86 |
+
"output = result.stdout\n",
|
| 87 |
+
"for line in output.splitlines():\n",
|
| 88 |
+
" if '=' in line:\n",
|
| 89 |
+
" var, value = line.split('=', 1)\n",
|
| 90 |
+
" os.environ[var] = value"
|
| 91 |
+
]
|
| 92 |
+
},
|
| 93 |
+
{
|
| 94 |
+
"cell_type": "code",
|
| 95 |
+
"execution_count": null,
|
| 96 |
+
"id": "2168e365-8254-4063-98bd-27afdbdb2f32",
|
| 97 |
+
"metadata": {},
|
| 98 |
+
"outputs": [],
|
| 99 |
+
"source": [
|
| 100 |
+
"#lfs 支持\n",
|
| 101 |
+
"!apt-get update\n",
|
| 102 |
+
"\n",
|
| 103 |
+
"!apt-get install git-lfs\n",
|
| 104 |
+
"\n",
|
| 105 |
+
"!git lfs install"
|
| 106 |
+
]
|
| 107 |
}
|
| 108 |
],
|
| 109 |
"metadata": {
|
01-data_env/2-data-intro.ipynb
CHANGED
|
@@ -15,9 +15,9 @@
|
|
| 15 |
"source": [
|
| 16 |
"本教程主要关注基因相关的生物序列数据,包括主要的DNA和蛋白质序列,data目录下数据如下:\n",
|
| 17 |
"\n",
|
| 18 |
-
"* dna_1g.txt DNA序列数据,大小1G,从
|
| 19 |
-
"* potein_1g.txt 蛋白质序列数据,大小1G,从pdb
|
| 20 |
-
"* english_500m.txt 英文数据,大小500M,就是英文百科"
|
| 21 |
]
|
| 22 |
},
|
| 23 |
{
|
|
|
|
| 15 |
"source": [
|
| 16 |
"本教程主要关注基因相关的生物序列数据,包括主要的DNA和蛋白质序列,data目录下数据如下:\n",
|
| 17 |
"\n",
|
| 18 |
+
"* dna_1g.txt DNA序列数据,大小1G,从GUE数据集中抽取,具体可参考dnabert2的论文,包括多个模式生物的数据(https://github.com/MAGICS-LAB/DNABERT_2)\n",
|
| 19 |
+
"* potein_1g.txt 蛋白质序列数据,大小1G,从pdb/uniprot数据库中抽取(https://www.uniprot.org/help/downloads)\n",
|
| 20 |
+
"* english_500m.txt 英文数据,大小500M,就是英文百科(https://huggingface.co/datasets/Salesforce/wikitext, https://huggingface.co/datasets/iohadrubin/wikitext-103-raw-v1)"
|
| 21 |
]
|
| 22 |
},
|
| 23 |
{
|
01-data_env/3-dataset-use.ipynb
CHANGED
|
@@ -117,7 +117,9 @@
|
|
| 117 |
"dna_dataset_sample = DatasetDict(\n",
|
| 118 |
" {\n",
|
| 119 |
" \"train\": dna_dataset[\"train\"].shuffle().select(range(50000)), \n",
|
| 120 |
-
" \"valid\": dna_dataset[\"test\"].shuffle().select(range(500))
|
|
|
|
|
|
|
| 121 |
" }\n",
|
| 122 |
")\n",
|
| 123 |
"dna_dataset_sample"
|
|
@@ -134,6 +136,7 @@
|
|
| 134 |
]
|
| 135 |
},
|
| 136 |
{
|
|
|
|
| 137 |
"cell_type": "markdown",
|
| 138 |
"id": "17a1fa7c-ff4b-419f-8a82-e58cc5777cd4",
|
| 139 |
"metadata": {},
|
|
@@ -142,7 +145,9 @@
|
|
| 142 |
"\n",
|
| 143 |
"当然,数据也可以直接从huggingface的线上仓库读取,这时候需要注意科学上网问题。\n",
|
| 144 |
"\n",
|
| 145 |
-
"具体使用函数也是load_dataset"
|
|
|
|
|
|
|
| 146 |
]
|
| 147 |
},
|
| 148 |
{
|
|
|
|
| 117 |
"dna_dataset_sample = DatasetDict(\n",
|
| 118 |
" {\n",
|
| 119 |
" \"train\": dna_dataset[\"train\"].shuffle().select(range(50000)), \n",
|
| 120 |
+
" \"valid\": dna_dataset[\"test\"].shuffle().select(range(500)),\n",
|
| 121 |
+
" \"evla\": dna_dataset[\"test\"].shuffle().select(range(500))\n",
|
| 122 |
+
"\n",
|
| 123 |
" }\n",
|
| 124 |
")\n",
|
| 125 |
"dna_dataset_sample"
|
|
|
|
| 136 |
]
|
| 137 |
},
|
| 138 |
{
|
| 139 |
+
"attachments": {},
|
| 140 |
"cell_type": "markdown",
|
| 141 |
"id": "17a1fa7c-ff4b-419f-8a82-e58cc5777cd4",
|
| 142 |
"metadata": {},
|
|
|
|
| 145 |
"\n",
|
| 146 |
"当然,数据也可以直接从huggingface的线上仓库读取,这时候需要注意科学上网问题。\n",
|
| 147 |
"\n",
|
| 148 |
+
"具体使用函数也是load_dataset\n",
|
| 149 |
+
"\n",
|
| 150 |
+
"<img src='img/datasets_dnagpt.png' width='800px' />"
|
| 151 |
]
|
| 152 |
},
|
| 153 |
{
|
01-data_env/img/.ipynb_checkpoints/zhushi-checkpoint.png
ADDED
|
01-data_env/img/datasets_dnagpt.png
ADDED
|
02-gpt2_bert/.ipynb_checkpoints/1-dna-bpe-checkpoint.ipynb
ADDED
|
@@ -0,0 +1,528 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"id": "a9fffce5-83e3-4838-8335-acb2e3b50c35",
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"source": [
|
| 8 |
+
"# 2.1 DNA分词器构建"
|
| 9 |
+
]
|
| 10 |
+
},
|
| 11 |
+
{
|
| 12 |
+
"cell_type": "markdown",
|
| 13 |
+
"id": "f28b0950-37dc-4f78-ae6c-9fca33d513fc",
|
| 14 |
+
"metadata": {},
|
| 15 |
+
"source": [
|
| 16 |
+
"## **分词算法**\n",
|
| 17 |
+
"\n",
|
| 18 |
+
"### **什么是分词**\n",
|
| 19 |
+
"分词就是把一个文本序列,分成一个一个的token/词,对于英文这种天生带空格的语言,一般使用空格和标点分词就行了,而对于中文等语言,并没有特殊的符号来分词,因此,一般需要设计专门的分词算法,对于大模型而言,一般需要处理多种语言,因此,也需要专门的分词算法。\n",
|
| 20 |
+
"\n",
|
| 21 |
+
"在大模型(如 BERT、GPT 系列、T5 等)中,分词器(tokenizer)扮演着至关重要的角色。它负责将原始文本转换为模型可以处理的格式,即将文本分解成 token 序列,并将这些 token 映射到模型词汇表中的唯一 ID。分词器的选择和配置直接影响模型的性能和效果。以下是几种常见的分词器及其特点,特别关注它们在大型语言模型中的应用。\n",
|
| 22 |
+
"\n",
|
| 23 |
+
"### 1. **WordPiece 分词器**\n",
|
| 24 |
+
"\n",
|
| 25 |
+
"- **使用场景**:广泛应用于 BERT 及其变体。\n",
|
| 26 |
+
"- **工作原理**:基于频率统计,从语料库中学习最有效的词汇表。它根据子词(subword)在文本中的出现频率来决定如何分割单词。例如,“playing” 可能被分为 “play” 和 “##ing”,其中“##”表示该部分是前一个 token 的延续。\n",
|
| 27 |
+
"- **优点**:\n",
|
| 28 |
+
" - 处理未知词汇能力强,能够将未见过的词汇分解为已知的子词。\n",
|
| 29 |
+
" - 兼容性好,适合多种语言任务。\n",
|
| 30 |
+
"- **缺点**:\n",
|
| 31 |
+
" - 需要额外的标记(如 `##`)来指示子词,可能影响某些应用场景下的可读性。\n",
|
| 32 |
+
"\n",
|
| 33 |
+
"### 2. **Byte Pair Encoding (BPE)**\n",
|
| 34 |
+
"\n",
|
| 35 |
+
"- **使用场景**:广泛应用于 GPT 系列、RoBERTa、XLM-R 等模型。\n",
|
| 36 |
+
"- **工作原理**:通过迭代地合并最常见的字符对来构建词汇表。BPE 是一种无监督的学习方法,能够在不依赖于预先定义的词汇表的情况下进行分词。\n",
|
| 37 |
+
"- **优点**:\n",
|
| 38 |
+
" - 灵活性高,适应性强,尤其适用于多语言模型。\n",
|
| 39 |
+
" - 不需要特殊标记,生成的词汇表更简洁。\n",
|
| 40 |
+
"- **缺点**:\n",
|
| 41 |
+
" - 对于某些语言或领域特定的词汇,可能会产生较短的子词,导致信息丢失。\n",
|
| 42 |
+
"\n",
|
| 43 |
+
"### 3. **SentencePiece**\n",
|
| 44 |
+
"\n",
|
| 45 |
+
"- **使用场景**:常见于 T5、mBART 等多语言模型。\n",
|
| 46 |
+
"- **工作原理**:结合了 BPE 和 WordPiece 的优点,同时支持字符级和词汇级分词。它可以在没有空格的语言(如中文、日文)中表现良好。\n",
|
| 47 |
+
"- **优点**:\n",
|
| 48 |
+
" - 支持无空格语言,适合多语言处理。\n",
|
| 49 |
+
" - 学习速度快,适应性强。\n",
|
| 50 |
+
"- **缺点**:\n",
|
| 51 |
+
" - 对于某些特定领域的专业术语,可能需要额外的预处理步骤。\n",
|
| 52 |
+
"\n",
|
| 53 |
+
"### 4. **Character-Level Tokenizer**\n",
|
| 54 |
+
"\n",
|
| 55 |
+
"- **使用场景**:较少用于大型语言模型,但在某些特定任务(如拼写检查、手写识别)中有应用。\n",
|
| 56 |
+
"- **工作原理**:直接将每个字符视为一个 token。这种方式简单直接,但通常会导致较大的词汇表。\n",
|
| 57 |
+
"- **优点**:\n",
|
| 58 |
+
" - 简单易实现,不需要复杂的训练过程。\n",
|
| 59 |
+
" - 对于字符级别的任务非常有效。\n",
|
| 60 |
+
"- **缺点**:\n",
|
| 61 |
+
" - 词汇表较大,计算资源消耗较多。\n",
|
| 62 |
+
" - 捕捉上下文信息的能力较弱。\n",
|
| 63 |
+
"\n",
|
| 64 |
+
"### 5. **Unigram Language Model**\n",
|
| 65 |
+
"\n",
|
| 66 |
+
"- **使用场景**:主要用于 SentencePiece 中。\n",
|
| 67 |
+
"- **工作原理**:基于概率分布,选择最优的分词方案以最大化似然函数。这种方法类似于 BPE,但在构建词汇表时考虑了更多的统计信息。\n",
|
| 68 |
+
"- **优点**:\n",
|
| 69 |
+
" - 统计基础强,优化效果好。\n",
|
| 70 |
+
" - 适应性强,适用于多种语言和任务。\n",
|
| 71 |
+
"- **缺点**:\n",
|
| 72 |
+
" - 计算复杂度较高,训练时间较长。\n",
|
| 73 |
+
"\n",
|
| 74 |
+
"### 分词器的关键特性\n",
|
| 75 |
+
"\n",
|
| 76 |
+
"无论选择哪种分词器,以下几个关键特性都是设计和应用中需要考虑的:\n",
|
| 77 |
+
"\n",
|
| 78 |
+
"- **词汇表大小**:决定了模型所能识别的词汇量。较大的词汇表可以捕捉更多细节,但也增加了内存和计算需求。\n",
|
| 79 |
+
"- **处理未知词汇的能力**:好的分词器应该能够有效地处理未登录词(OOV, Out-Of-Vocabulary),将其分解为已知的子词。\n",
|
| 80 |
+
"- **多语言支持**:对于多语言模型,分词器应能处理不同语言的文本,尤其是那些没有明显分隔符的语言。\n",
|
| 81 |
+
"- **效率和速度**:分词器的执行速度直接影响整个数据处理管道的效率,尤其是在大规模数据集上。\n",
|
| 82 |
+
"- **兼容性和灵活性**:分词器应与目标模型架构兼容,并且能够灵活适应不同的任务需求。"
|
| 83 |
+
]
|
| 84 |
+
},
|
| 85 |
+
{
|
| 86 |
+
"cell_type": "markdown",
|
| 87 |
+
"id": "165e2594-277d-44d0-b582-77859a0bc0b2",
|
| 88 |
+
"metadata": {},
|
| 89 |
+
"source": [
|
| 90 |
+
"## DNA等生物序列分词\n",
|
| 91 |
+
"在生物信息学中,DNA 和蛋白质序列的处理与自然语言处理(NLP)有相似之处,但也有其独特性。为了提取这些生物序列的特征并用于机器学习或深度学习模型,通常需要将长序列分解成更小的片段(类似于 NLP 中的“分词”),以便更好地捕捉局部和全局特征。以下是几种常见的方法,用于对 DNA 和蛋白质序列进行“分词”,以提取有用的特征。\n",
|
| 92 |
+
"\n",
|
| 93 |
+
"### 1. **K-mer 分解**\n",
|
| 94 |
+
"\n",
|
| 95 |
+
"**定义**:K-mer 是指长度为 k 的连续子序列。例如,在 DNA 序列中,一个 3-mer 可能是 \"ATG\" 或 \"CGA\"。\n",
|
| 96 |
+
"\n",
|
| 97 |
+
"**应用**:\n",
|
| 98 |
+
"- **DNA 序列**:常用的 k 值范围从 3 到 6。较小的 k 值可以捕捉到更细粒度的信息,而较大的 k 值则有助于识别更长的模式。\n",
|
| 99 |
+
"- **蛋白质序列**:k 值通常较大,因为氨基酸的数量较多(20 种),较长的 k-mer 可以捕捉到重要的结构域或功能区域。\n",
|
| 100 |
+
"\n",
|
| 101 |
+
"**优点**:\n",
|
| 102 |
+
"- 简单且直观,易于实现。\n",
|
| 103 |
+
"- 可以捕捉到短序列中的局部特征。\n",
|
| 104 |
+
"\n",
|
| 105 |
+
"**缺点**:\n",
|
| 106 |
+
"- 对于非常长的序列,生成的 k-mer 数量会非常大,导致维度爆炸问题。\n",
|
| 107 |
+
"- 不同位置的 k-mer 之间缺乏上下文关系。"
|
| 108 |
+
]
|
| 109 |
+
},
|
| 110 |
+
{
|
| 111 |
+
"cell_type": "code",
|
| 112 |
+
"execution_count": 2,
|
| 113 |
+
"id": "29c390ef-2e9d-493e-9991-69ecb835b52b",
|
| 114 |
+
"metadata": {},
|
| 115 |
+
"outputs": [
|
| 116 |
+
{
|
| 117 |
+
"name": "stdout",
|
| 118 |
+
"output_type": "stream",
|
| 119 |
+
"text": [
|
| 120 |
+
"DNA 3-mers: ['ATG', 'TGC', 'GCG', 'CGT', 'GTA', 'TAC', 'ACG', 'CGT', 'GTA']\n",
|
| 121 |
+
"Protein 4-mers: ['MKQH', 'KQHK', 'QHKA', 'HKAM', 'KAMI', 'AMIV', 'MIVA', 'IVAL', 'VALI', 'ALIV', 'LIVL', 'IVLI', 'VLIT', 'LITA', 'ITAY']\n"
|
| 122 |
+
]
|
| 123 |
+
}
|
| 124 |
+
],
|
| 125 |
+
"source": [
|
| 126 |
+
"#示例代码(Python)\n",
|
| 127 |
+
"\n",
|
| 128 |
+
"def k_mer(seq, k):\n",
|
| 129 |
+
" return [seq[i:i+k] for i in range(len(seq) - k + 1)]\n",
|
| 130 |
+
"\n",
|
| 131 |
+
"dna_sequence = \"ATGCGTACGTA\"\n",
|
| 132 |
+
"protein_sequence = \"MKQHKAMIVALIVLITAY\"\n",
|
| 133 |
+
"\n",
|
| 134 |
+
"print(\"DNA 3-mers:\", k_mer(dna_sequence, 3))\n",
|
| 135 |
+
"print(\"Protein 4-mers:\", k_mer(protein_sequence, 4))"
|
| 136 |
+
]
|
| 137 |
+
},
|
| 138 |
+
{
|
| 139 |
+
"cell_type": "markdown",
|
| 140 |
+
"id": "7ced2bfb-bd42-425a-a3ad-54c9573609c5",
|
| 141 |
+
"metadata": {},
|
| 142 |
+
"source": [
|
| 143 |
+
"### 2. **滑动窗口**\n",
|
| 144 |
+
"\n",
|
| 145 |
+
"**定义**:滑动窗口方法通过设定一个固定大小的窗口沿着序列移动,并在每个位置提取窗口内的子序列。这与 K-mer 类似,但允许重叠。\n",
|
| 146 |
+
"\n",
|
| 147 |
+
"**应用**:\n",
|
| 148 |
+
"- **DNA 和蛋白质序列**:窗口大小可以根据具体任务调整,如基因预测、蛋白质结构预测等。\n",
|
| 149 |
+
"\n",
|
| 150 |
+
"**优点**:\n",
|
| 151 |
+
"- 提供了更多的灵活性,可以控制窗口的步长和大小。\n",
|
| 152 |
+
"- 有助于捕捉局部和全局特征。\n",
|
| 153 |
+
"\n",
|
| 154 |
+
"**缺点**:\n",
|
| 155 |
+
"- 计算复杂度较高,尤其是当窗口大小较大时。"
|
| 156 |
+
]
|
| 157 |
+
},
|
| 158 |
+
{
|
| 159 |
+
"cell_type": "code",
|
| 160 |
+
"execution_count": 4,
|
| 161 |
+
"id": "82cecf91-0076-4c12-b11c-b35120581ef9",
|
| 162 |
+
"metadata": {},
|
| 163 |
+
"outputs": [
|
| 164 |
+
{
|
| 165 |
+
"name": "stdout",
|
| 166 |
+
"output_type": "stream",
|
| 167 |
+
"text": [
|
| 168 |
+
"Sliding window (DNA, size=3, step=1): ['ATG', 'TGC', 'GCG', 'CGT', 'GTA', 'TAC', 'ACG', 'CGT', 'GTA']\n",
|
| 169 |
+
"Sliding window (Protein, size=4, step=2): ['MKQH', 'QHKA', 'KAMI', 'MIVA', 'VALI', 'LIVL', 'VLIT', 'ITAY']\n"
|
| 170 |
+
]
|
| 171 |
+
}
|
| 172 |
+
],
|
| 173 |
+
"source": [
|
| 174 |
+
"def sliding_window(seq, window_size, step=1):\n",
|
| 175 |
+
" return [seq[i:i+window_size] for i in range(0, len(seq) - window_size + 1, step)]\n",
|
| 176 |
+
"\n",
|
| 177 |
+
"dna_sequence = \"ATGCGTACGTA\"\n",
|
| 178 |
+
"protein_sequence = \"MKQHKAMIVALIVLITAY\"\n",
|
| 179 |
+
"\n",
|
| 180 |
+
"print(\"Sliding window (DNA, size=3, step=1):\", sliding_window(dna_sequence, 3))\n",
|
| 181 |
+
"print(\"Sliding window (Protein, size=4, step=2):\", sliding_window(protein_sequence, 4, step=2))"
|
| 182 |
+
]
|
| 183 |
+
},
|
| 184 |
+
{
|
| 185 |
+
"cell_type": "markdown",
|
| 186 |
+
"id": "c33ab920-b451-4846-93d4-20da5a4e1001",
|
| 187 |
+
"metadata": {},
|
| 188 |
+
"source": [
|
| 189 |
+
"### 3. **词表分词和嵌入式表示**\n",
|
| 190 |
+
"\n",
|
| 191 |
+
"**定义**:使用预训练的嵌入模型(如 Word2Vec、BERT 等)来将每个 token 映射到高维向量空间中。对于生物序列,可以使用专门设计的嵌入模型,如 ProtTrans、ESM 等。\n",
|
| 192 |
+
"\n",
|
| 193 |
+
"**应用**:\n",
|
| 194 |
+
"- **DNA 和蛋白质序列**:嵌入模型可以捕捉到序列中的语义信息和上下文依赖关系。\n",
|
| 195 |
+
"\n",
|
| 196 |
+
"**优点**:\n",
|
| 197 |
+
"- 捕捉到丰富的语义信息,适合复杂的下游任务。\n",
|
| 198 |
+
"- 可以利用大规模预训练模型的优势。\n",
|
| 199 |
+
"\n",
|
| 200 |
+
"**缺点**:\n",
|
| 201 |
+
"- 需要大量的计算资源来进行预训练。\n",
|
| 202 |
+
"- 模型复杂度较高,解释性较差。"
|
| 203 |
+
]
|
| 204 |
+
},
|
| 205 |
+
{
|
| 206 |
+
"cell_type": "code",
|
| 207 |
+
"execution_count": 5,
|
| 208 |
+
"id": "02bf2af0-6077-4b27-8822-f1c3f22914fa",
|
| 209 |
+
"metadata": {},
|
| 210 |
+
"outputs": [],
|
| 211 |
+
"source": [
|
| 212 |
+
"import subprocess\n",
|
| 213 |
+
"import os\n",
|
| 214 |
+
"# 设置环境变量, autodl一般区域\n",
|
| 215 |
+
"result = subprocess.run('bash -c \"source /etc/network_turbo && env | grep proxy\"', shell=True, capture_output=True, text=True)\n",
|
| 216 |
+
"output = result.stdout\n",
|
| 217 |
+
"for line in output.splitlines():\n",
|
| 218 |
+
" if '=' in line:\n",
|
| 219 |
+
" var, value = line.split('=', 1)\n",
|
| 220 |
+
" os.environ[var] = value\n",
|
| 221 |
+
"\n",
|
| 222 |
+
"\"\"\"\n",
|
| 223 |
+
"import os\n",
|
| 224 |
+
"\n",
|
| 225 |
+
"# 设置环境变量, autodl专区 其他idc\n",
|
| 226 |
+
"os.environ['HF_ENDPOINT'] = 'https://hf-mirror.com'\n",
|
| 227 |
+
"\n",
|
| 228 |
+
"# 打印环境变量以确认设置成功\n",
|
| 229 |
+
"print(os.environ.get('HF_ENDPOINT'))\n",
|
| 230 |
+
"\"\"\""
|
| 231 |
+
]
|
| 232 |
+
},
|
| 233 |
+
{
|
| 234 |
+
"cell_type": "code",
|
| 235 |
+
"execution_count": 15,
|
| 236 |
+
"id": "d43b60ee-67f2-4d06-95ea-966c01084fc4",
|
| 237 |
+
"metadata": {
|
| 238 |
+
"scrolled": true
|
| 239 |
+
},
|
| 240 |
+
"outputs": [
|
| 241 |
+
{
|
| 242 |
+
"name": "stderr",
|
| 243 |
+
"output_type": "stream",
|
| 244 |
+
"text": [
|
| 245 |
+
"huggingface/tokenizers: The current process just got forked, after parallelism has already been used. Disabling parallelism to avoid deadlocks...\n",
|
| 246 |
+
"To disable this warning, you can either:\n",
|
| 247 |
+
"\t- Avoid using `tokenizers` before the fork if possible\n",
|
| 248 |
+
"\t- Explicitly set the environment variable TOKENIZERS_PARALLELISM=(true | false)\n"
|
| 249 |
+
]
|
| 250 |
+
},
|
| 251 |
+
{
|
| 252 |
+
"name": "stdout",
|
| 253 |
+
"output_type": "stream",
|
| 254 |
+
"text": [
|
| 255 |
+
"['ATGCG', 'TACG', 'T', 'A']\n",
|
| 256 |
+
"Embeddings shape: torch.Size([1, 4, 768])\n"
|
| 257 |
+
]
|
| 258 |
+
}
|
| 259 |
+
],
|
| 260 |
+
"source": [
|
| 261 |
+
"from transformers import AutoTokenizer, AutoModel\n",
|
| 262 |
+
"import torch\n",
|
| 263 |
+
"\n",
|
| 264 |
+
"# 加载预训练的蛋白质嵌入模型\n",
|
| 265 |
+
"tokenizer = AutoTokenizer.from_pretrained(\"dnagpt/gpt_dna_v0\")\n",
|
| 266 |
+
"model = AutoModel.from_pretrained(\"dnagpt/gpt_dna_v0\")\n",
|
| 267 |
+
"\n",
|
| 268 |
+
"dna_sequence = \"ATGCGTACGTA\"\n",
|
| 269 |
+
"print(tokenizer.tokenize(dna_sequence))\n",
|
| 270 |
+
"\n",
|
| 271 |
+
"# 编码序列\n",
|
| 272 |
+
"inputs = tokenizer(dna_sequence, return_tensors=\"pt\")\n",
|
| 273 |
+
"\n",
|
| 274 |
+
"# 获取嵌入\n",
|
| 275 |
+
"with torch.no_grad():\n",
|
| 276 |
+
" outputs = model(**inputs)\n",
|
| 277 |
+
" embeddings = outputs.last_hidden_state\n",
|
| 278 |
+
"\n",
|
| 279 |
+
"print(\"Embeddings shape:\", embeddings.shape)"
|
| 280 |
+
]
|
| 281 |
+
},
|
| 282 |
+
{
|
| 283 |
+
"cell_type": "markdown",
|
| 284 |
+
"id": "c24f10dc-1117-4493-9333-5ed6d898f44a",
|
| 285 |
+
"metadata": {},
|
| 286 |
+
"source": [
|
| 287 |
+
"### 训练DNA BPE分词器\n",
|
| 288 |
+
"\n",
|
| 289 |
+
"以上方法展示了如何对 DNA 和蛋白质序列进行“分词”,以提取有用的特征。选择哪种方法取决于具体的任务需求和数据特性。对于简单的分类或回归任务,K-mer 分解或滑动窗口可能是足够的;而对于更复杂的任务,如序列标注或结构预测,基于词汇表的方法或嵌入表示可能会提供更好的性能。\n",
|
| 290 |
+
"\n",
|
| 291 |
+
"目前大部分生物序列大模型的论文中,使用最多的依然是传统的K-mer,但一些SOTA的论文则以BEP为主。而BEP分词也是目前GPT、llama等主流自然语言大模型使用的基础分词器。\n",
|
| 292 |
+
"\n",
|
| 293 |
+
"因此,我们也演示下从头训练一个DNA BPE分词器的方法。\n",
|
| 294 |
+
"\n",
|
| 295 |
+
"我们首先看下GPT2模型,默认的分词器,对DNA序列分词的结果:"
|
| 296 |
+
]
|
| 297 |
+
},
|
| 298 |
+
{
|
| 299 |
+
"cell_type": "code",
|
| 300 |
+
"execution_count": 10,
|
| 301 |
+
"id": "43f1eb8b-1cc2-4ab5-aa8e-2a63132be98c",
|
| 302 |
+
"metadata": {},
|
| 303 |
+
"outputs": [],
|
| 304 |
+
"source": [
|
| 305 |
+
"from tokenizers import (\n",
|
| 306 |
+
" decoders,\n",
|
| 307 |
+
" models,\n",
|
| 308 |
+
" normalizers,\n",
|
| 309 |
+
" pre_tokenizers,\n",
|
| 310 |
+
" processors,\n",
|
| 311 |
+
" trainers,\n",
|
| 312 |
+
" Tokenizer,\n",
|
| 313 |
+
")\n",
|
| 314 |
+
"from transformers import AutoTokenizer"
|
| 315 |
+
]
|
| 316 |
+
},
|
| 317 |
+
{
|
| 318 |
+
"cell_type": "code",
|
| 319 |
+
"execution_count": 15,
|
| 320 |
+
"id": "27e88f7b-1399-418b-9b91-f970762fac0c",
|
| 321 |
+
"metadata": {},
|
| 322 |
+
"outputs": [],
|
| 323 |
+
"source": [
|
| 324 |
+
"gpt2_tokenizer = AutoTokenizer.from_pretrained('gpt2')\n",
|
| 325 |
+
"gpt2_tokenizer.pad_token = gpt2_tokenizer.eos_token"
|
| 326 |
+
]
|
| 327 |
+
},
|
| 328 |
+
{
|
| 329 |
+
"cell_type": "code",
|
| 330 |
+
"execution_count": 16,
|
| 331 |
+
"id": "4b015db7-63ba-4909-b02f-07634b3d5584",
|
| 332 |
+
"metadata": {},
|
| 333 |
+
"outputs": [
|
| 334 |
+
{
|
| 335 |
+
"data": {
|
| 336 |
+
"text/plain": [
|
| 337 |
+
"['T', 'GG', 'C', 'GT', 'GA', 'AC', 'CC', 'GG', 'G', 'AT', 'C', 'GG', 'G']"
|
| 338 |
+
]
|
| 339 |
+
},
|
| 340 |
+
"execution_count": 16,
|
| 341 |
+
"metadata": {},
|
| 342 |
+
"output_type": "execute_result"
|
| 343 |
+
}
|
| 344 |
+
],
|
| 345 |
+
"source": [
|
| 346 |
+
"gpt2_tokenizer.tokenize(\"TGGCGTGAACCCGGGATCGGG\")"
|
| 347 |
+
]
|
| 348 |
+
},
|
| 349 |
+
{
|
| 350 |
+
"cell_type": "markdown",
|
| 351 |
+
"id": "a246fbc9-9e29-4b63-bdf7-f80635d06d1e",
|
| 352 |
+
"metadata": {},
|
| 353 |
+
"source": [
|
| 354 |
+
"可以看到,gpt2模型因为是以英文为主的BPE分词模型,分解的都是1到2个字母的结果,这样显然很难充分表达生物语义,因此,我们使用DNA序列来训练1个BPE分词器,代码也非常简单:"
|
| 355 |
+
]
|
| 356 |
+
},
|
| 357 |
+
{
|
| 358 |
+
"cell_type": "code",
|
| 359 |
+
"execution_count": 2,
|
| 360 |
+
"id": "8357a695-1c29-4b5c-8099-d2e337189410",
|
| 361 |
+
"metadata": {},
|
| 362 |
+
"outputs": [],
|
| 363 |
+
"source": [
|
| 364 |
+
"tokenizer = Tokenizer(models.BPE())\n",
|
| 365 |
+
"tokenizer.pre_tokenizer = pre_tokenizers.ByteLevel(add_prefix_space=False, use_regex=False) #use_regex=False,空格当成一般字符串\n",
|
| 366 |
+
"trainer = trainers.BpeTrainer(vocab_size=30000, special_tokens=[\"<|endoftext|>\"]) #3w words"
|
| 367 |
+
]
|
| 368 |
+
},
|
| 369 |
+
{
|
| 370 |
+
"cell_type": "code",
|
| 371 |
+
"execution_count": 3,
|
| 372 |
+
"id": "32c95888-1498-45cf-8453-421219cc7d45",
|
| 373 |
+
"metadata": {},
|
| 374 |
+
"outputs": [
|
| 375 |
+
{
|
| 376 |
+
"name": "stdout",
|
| 377 |
+
"output_type": "stream",
|
| 378 |
+
"text": [
|
| 379 |
+
"\n",
|
| 380 |
+
"\n",
|
| 381 |
+
"\n"
|
| 382 |
+
]
|
| 383 |
+
}
|
| 384 |
+
],
|
| 385 |
+
"source": [
|
| 386 |
+
"tokenizer.train([\"../01-data_env/data/dna_1g.txt\"], trainer=trainer) #all file list, take 10-20 min"
|
| 387 |
+
]
|
| 388 |
+
},
|
| 389 |
+
{
|
| 390 |
+
"cell_type": "code",
|
| 391 |
+
"execution_count": 4,
|
| 392 |
+
"id": "5ffdd717-72ed-4a37-bafc-b4a0f61f8ff1",
|
| 393 |
+
"metadata": {},
|
| 394 |
+
"outputs": [
|
| 395 |
+
{
|
| 396 |
+
"name": "stdout",
|
| 397 |
+
"output_type": "stream",
|
| 398 |
+
"text": [
|
| 399 |
+
"['TG', 'GCGTGAA', 'CCCGG', 'GATCGG', 'G']\n"
|
| 400 |
+
]
|
| 401 |
+
}
|
| 402 |
+
],
|
| 403 |
+
"source": [
|
| 404 |
+
"encoding = tokenizer.encode(\"TGGCGTGAACCCGGGATCGGG\")\n",
|
| 405 |
+
"print(encoding.tokens)"
|
| 406 |
+
]
|
| 407 |
+
},
|
| 408 |
+
{
|
| 409 |
+
"cell_type": "markdown",
|
| 410 |
+
"id": "a96e7838-6c23-4446-bf86-b098cd93214a",
|
| 411 |
+
"metadata": {},
|
| 412 |
+
"source": [
|
| 413 |
+
"可以看到,以DNA数据训练的分词器,分词效果明显要好的多,各种长度的词都有。"
|
| 414 |
+
]
|
| 415 |
+
},
|
| 416 |
+
{
|
| 417 |
+
"cell_type": "code",
|
| 418 |
+
"execution_count": 5,
|
| 419 |
+
"id": "f1d757c1-702b-4147-9207-471f422f67b2",
|
| 420 |
+
"metadata": {},
|
| 421 |
+
"outputs": [],
|
| 422 |
+
"source": [
|
| 423 |
+
"tokenizer.save(\"dna_bpe_dict.json\")"
|
| 424 |
+
]
|
| 425 |
+
},
|
| 426 |
+
{
|
| 427 |
+
"cell_type": "code",
|
| 428 |
+
"execution_count": 6,
|
| 429 |
+
"id": "caf8ecea-359e-487b-b456-fab546b9da0d",
|
| 430 |
+
"metadata": {},
|
| 431 |
+
"outputs": [],
|
| 432 |
+
"source": [
|
| 433 |
+
"#然后我们可以使用from_file() 方法从该文件里重新加载 Tokenizer 对象:\n",
|
| 434 |
+
"new_tokenizer = Tokenizer.from_file(\"dna_bpe_dict.json\")"
|
| 435 |
+
]
|
| 436 |
+
},
|
| 437 |
+
{
|
| 438 |
+
"cell_type": "code",
|
| 439 |
+
"execution_count": 7,
|
| 440 |
+
"id": "8ec6f045-bc30-4012-8027-a879df8def3a",
|
| 441 |
+
"metadata": {},
|
| 442 |
+
"outputs": [
|
| 443 |
+
{
|
| 444 |
+
"data": {
|
| 445 |
+
"text/plain": [
|
| 446 |
+
"('dna_bpe_dict/tokenizer_config.json',\n",
|
| 447 |
+
" 'dna_bpe_dict/special_tokens_map.json',\n",
|
| 448 |
+
" 'dna_bpe_dict/vocab.json',\n",
|
| 449 |
+
" 'dna_bpe_dict/merges.txt',\n",
|
| 450 |
+
" 'dna_bpe_dict/added_tokens.json',\n",
|
| 451 |
+
" 'dna_bpe_dict/tokenizer.json')"
|
| 452 |
+
]
|
| 453 |
+
},
|
| 454 |
+
"execution_count": 7,
|
| 455 |
+
"metadata": {},
|
| 456 |
+
"output_type": "execute_result"
|
| 457 |
+
}
|
| 458 |
+
],
|
| 459 |
+
"source": [
|
| 460 |
+
"#要在 🤗 Transformers 中使用这个标记器,我们必须将它包裹在一个 PreTrainedTokenizerFast 类中\n",
|
| 461 |
+
"from transformers import GPT2TokenizerFast\n",
|
| 462 |
+
"dna_tokenizer = GPT2TokenizerFast(tokenizer_object=new_tokenizer)\n",
|
| 463 |
+
"dna_tokenizer.save_pretrained(\"dna_bpe_dict\")\n",
|
| 464 |
+
"#dna_tokenizer.push_to_hub(\"dna_bpe_dict_1g\", organization=\"dnagpt\", use_auth_token=\"hf_*****\") # push to huggingface"
|
| 465 |
+
]
|
| 466 |
+
},
|
| 467 |
+
{
|
| 468 |
+
"cell_type": "code",
|
| 469 |
+
"execution_count": 11,
|
| 470 |
+
"id": "f84506d8-6208-4027-aad7-2b68a1bc16d6",
|
| 471 |
+
"metadata": {},
|
| 472 |
+
"outputs": [],
|
| 473 |
+
"source": [
|
| 474 |
+
"tokenizer_new = AutoTokenizer.from_pretrained('dna_bpe_dict')"
|
| 475 |
+
]
|
| 476 |
+
},
|
| 477 |
+
{
|
| 478 |
+
"cell_type": "code",
|
| 479 |
+
"execution_count": 12,
|
| 480 |
+
"id": "d40d4d53-6fed-445c-afb5-c0346ab854c8",
|
| 481 |
+
"metadata": {},
|
| 482 |
+
"outputs": [
|
| 483 |
+
{
|
| 484 |
+
"data": {
|
| 485 |
+
"text/plain": [
|
| 486 |
+
"['TG', 'GCGTGAA', 'CCCGG', 'GATCGG', 'G']"
|
| 487 |
+
]
|
| 488 |
+
},
|
| 489 |
+
"execution_count": 12,
|
| 490 |
+
"metadata": {},
|
| 491 |
+
"output_type": "execute_result"
|
| 492 |
+
}
|
| 493 |
+
],
|
| 494 |
+
"source": [
|
| 495 |
+
"tokenizer_new.tokenize(\"TGGCGTGAACCCGGGATCGGG\")"
|
| 496 |
+
]
|
| 497 |
+
},
|
| 498 |
+
{
|
| 499 |
+
"cell_type": "code",
|
| 500 |
+
"execution_count": null,
|
| 501 |
+
"id": "640302f6-f740-41a4-ae92-ca4c43d97493",
|
| 502 |
+
"metadata": {},
|
| 503 |
+
"outputs": [],
|
| 504 |
+
"source": []
|
| 505 |
+
}
|
| 506 |
+
],
|
| 507 |
+
"metadata": {
|
| 508 |
+
"kernelspec": {
|
| 509 |
+
"display_name": "Python 3 (ipykernel)",
|
| 510 |
+
"language": "python",
|
| 511 |
+
"name": "python3"
|
| 512 |
+
},
|
| 513 |
+
"language_info": {
|
| 514 |
+
"codemirror_mode": {
|
| 515 |
+
"name": "ipython",
|
| 516 |
+
"version": 3
|
| 517 |
+
},
|
| 518 |
+
"file_extension": ".py",
|
| 519 |
+
"mimetype": "text/x-python",
|
| 520 |
+
"name": "python",
|
| 521 |
+
"nbconvert_exporter": "python",
|
| 522 |
+
"pygments_lexer": "ipython3",
|
| 523 |
+
"version": "3.12.3"
|
| 524 |
+
}
|
| 525 |
+
},
|
| 526 |
+
"nbformat": 4,
|
| 527 |
+
"nbformat_minor": 5
|
| 528 |
+
}
|
02-gpt2_bert/.ipynb_checkpoints/2-dna-gpt-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/.ipynb_checkpoints/3-dna-bert-checkpoint.ipynb
ADDED
|
@@ -0,0 +1,253 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": null,
|
| 6 |
+
"id": "a3ec4b86-2029-4d50-9bbf-64b208249165",
|
| 7 |
+
"metadata": {},
|
| 8 |
+
"outputs": [],
|
| 9 |
+
"source": [
|
| 10 |
+
"from tokenizers import Tokenizer\n",
|
| 11 |
+
"from tokenizers.models import WordPiece\n",
|
| 12 |
+
"from tokenizers.trainers import WordPieceTrainer\n",
|
| 13 |
+
"from tokenizers.pre_tokenizers import Whitespace"
|
| 14 |
+
]
|
| 15 |
+
},
|
| 16 |
+
{
|
| 17 |
+
"cell_type": "code",
|
| 18 |
+
"execution_count": null,
|
| 19 |
+
"id": "47b3fc92-df22-4e4b-bdf9-671bda924c49",
|
| 20 |
+
"metadata": {},
|
| 21 |
+
"outputs": [],
|
| 22 |
+
"source": [
|
| 23 |
+
"# 初始化一个空的 WordPiece 模型\n",
|
| 24 |
+
"tokenizer = Tokenizer(WordPiece(unk_token=\"[UNK]\"))"
|
| 25 |
+
]
|
| 26 |
+
},
|
| 27 |
+
{
|
| 28 |
+
"cell_type": "code",
|
| 29 |
+
"execution_count": null,
|
| 30 |
+
"id": "73f59aa6-8cce-4124-a3ee-7a5617b91ea7",
|
| 31 |
+
"metadata": {},
|
| 32 |
+
"outputs": [],
|
| 33 |
+
"source": [
|
| 34 |
+
"# 设置训练参数\n",
|
| 35 |
+
"trainer = WordPieceTrainer(\n",
|
| 36 |
+
" vocab_size=90000, # 词汇表大小\n",
|
| 37 |
+
" min_frequency=2, # 最小词频\n",
|
| 38 |
+
" special_tokens=[\n",
|
| 39 |
+
" \"[PAD]\", \"[UNK]\", \"[CLS]\", \"[SEP]\", \"[MASK]\"\n",
|
| 40 |
+
" ]\n",
|
| 41 |
+
")\n",
|
| 42 |
+
"\n",
|
| 43 |
+
"tokenizer.train(files=[\"../01-data_env/data/dna_1g.txt\"], trainer=trainer)"
|
| 44 |
+
]
|
| 45 |
+
},
|
| 46 |
+
{
|
| 47 |
+
"cell_type": "code",
|
| 48 |
+
"execution_count": null,
|
| 49 |
+
"id": "7a0ccd64-5172-4f40-9868-cdf02687ae10",
|
| 50 |
+
"metadata": {},
|
| 51 |
+
"outputs": [],
|
| 52 |
+
"source": [
|
| 53 |
+
"tokenizer.save(\"dna_wordpiece_dict.json\")"
|
| 54 |
+
]
|
| 55 |
+
},
|
| 56 |
+
{
|
| 57 |
+
"cell_type": "markdown",
|
| 58 |
+
"id": "eea3c48a-2245-478e-a2ce-f5d1af399d83",
|
| 59 |
+
"metadata": {},
|
| 60 |
+
"source": [
|
| 61 |
+
"## GPT2和bert配置的关键区别\n",
|
| 62 |
+
"* 最大长度:\n",
|
| 63 |
+
"在 GPT-2 中,n_ctx 参数指定了模型的最大上下文窗口大小。\n",
|
| 64 |
+
"在 BERT 中,你应该设置 max_position_embeddings 来指定最大位置嵌入数,这限制了输入序列的最大长度。\n",
|
| 65 |
+
"* 特殊 token ID:\n",
|
| 66 |
+
"GPT-2 使用 bos_token_id 和 eos_token_id 分别表示句子的开始和结束。\n",
|
| 67 |
+
"BERT 使用 [CLS] (cls_token_id) 表示句子的开始,用 [SEP] (sep_token_id) 表示句子的结束。BERT 还有专门的填充 token [PAD] (pad_token_id)。\n",
|
| 68 |
+
"* 模型类选择:\n",
|
| 69 |
+
"对于 GPT-2,你使用了 GPT2LMHeadModel,它适合生成任务或语言建模。\n",
|
| 70 |
+
"对于 BERT,如果你打算进行预训练(例如 Masked Language Modeling),应该使用 BertForMaskedLM。\n",
|
| 71 |
+
"* 预训练权重:\n",
|
| 72 |
+
"如果你想从头开始训练,像上面的例子中那样直接从配置创建模型即可。\n",
|
| 73 |
+
"如果你希望基于现有预训练模型微调,则可以使用 from_pretrained 方法加载预训练权重。"
|
| 74 |
+
]
|
| 75 |
+
},
|
| 76 |
+
{
|
| 77 |
+
"cell_type": "code",
|
| 78 |
+
"execution_count": null,
|
| 79 |
+
"id": "48e1f20b-cd1a-49fa-be2b-aba30a24e706",
|
| 80 |
+
"metadata": {},
|
| 81 |
+
"outputs": [],
|
| 82 |
+
"source": [
|
| 83 |
+
"new_tokenizer = Tokenizer.from_file(\"dna_wordpiece_dict.json\")\n",
|
| 84 |
+
"\n",
|
| 85 |
+
"wrapped_tokenizer = PreTrainedTokenizerFast(\n",
|
| 86 |
+
" tokenizer_object=new_tokenizer,\n",
|
| 87 |
+
" unk_token=\"[UNK]\",\n",
|
| 88 |
+
" pad_token=\"[PAD]\",\n",
|
| 89 |
+
" cls_token=\"[CLS]\",\n",
|
| 90 |
+
" sep_token=\"[SEP]\",\n",
|
| 91 |
+
" mask_token=\"[MASK]\",\n",
|
| 92 |
+
")\n",
|
| 93 |
+
"wrapped_tokenizer.save_pretrained(\"dna_wordpiece_dict\")"
|
| 94 |
+
]
|
| 95 |
+
},
|
| 96 |
+
{
|
| 97 |
+
"cell_type": "code",
|
| 98 |
+
"execution_count": null,
|
| 99 |
+
"id": "c94dc601-86ec-421c-8638-c8d8b5078682",
|
| 100 |
+
"metadata": {},
|
| 101 |
+
"outputs": [],
|
| 102 |
+
"source": [
|
| 103 |
+
"from transformers import AutoTokenizer, GPT2LMHeadModel, AutoConfig,GPT2Tokenizer\n",
|
| 104 |
+
"from transformers import GPT2Tokenizer,GPT2Model,AutoModel\n",
|
| 105 |
+
"from transformers import DataCollatorForLanguageModeling\n",
|
| 106 |
+
"from transformers import Trainer, TrainingArguments\n",
|
| 107 |
+
"from transformers import LineByLineTextDataset\n",
|
| 108 |
+
"from tokenizers import Tokenizer\n",
|
| 109 |
+
"from datasets import load_dataset\n",
|
| 110 |
+
"from transformers import BertConfig, BertModel"
|
| 111 |
+
]
|
| 112 |
+
},
|
| 113 |
+
{
|
| 114 |
+
"cell_type": "code",
|
| 115 |
+
"execution_count": null,
|
| 116 |
+
"id": "b2658cd2-0ac5-483e-b04d-2716993770e3",
|
| 117 |
+
"metadata": {},
|
| 118 |
+
"outputs": [],
|
| 119 |
+
"source": [
|
| 120 |
+
"tokenizer = AutoTokenizer.from_pretrained(\"dna_wordpiece_dict\")\n",
|
| 121 |
+
"#tokenizer.pad_token = tokenizer.eos_token"
|
| 122 |
+
]
|
| 123 |
+
},
|
| 124 |
+
{
|
| 125 |
+
"cell_type": "code",
|
| 126 |
+
"execution_count": null,
|
| 127 |
+
"id": "a7d0b7b8-b6dc-422a-9133-1d51ec40adbe",
|
| 128 |
+
"metadata": {},
|
| 129 |
+
"outputs": [],
|
| 130 |
+
"source": [
|
| 131 |
+
"max_length = 256 #最大输入长度\n",
|
| 132 |
+
"\n",
|
| 133 |
+
"# Building the config\n",
|
| 134 |
+
"#config = BertConfig()\n",
|
| 135 |
+
"\n",
|
| 136 |
+
"\n",
|
| 137 |
+
"# 构建配置\n",
|
| 138 |
+
"config = AutoConfig.from_pretrained(\n",
|
| 139 |
+
" \"bert-base-uncased\", # 或者其他预训练 BERT 模型名称,这里只是为了获取默认配置\n",
|
| 140 |
+
" vocab_size=len(tokenizer),\n",
|
| 141 |
+
" max_position_embeddings=max_length, # 对应于最大位置嵌入数\n",
|
| 142 |
+
" pad_token_id=tokenizer.pad_token_id,\n",
|
| 143 |
+
" bos_token_id=tokenizer.cls_token_id, # BERT 使用 [CLS] 作为句子开始标记\n",
|
| 144 |
+
" eos_token_id=tokenizer.sep_token_id # BERT 使用 [SEP] 作为句子结束标记\n",
|
| 145 |
+
")\n",
|
| 146 |
+
"\n",
|
| 147 |
+
"\n",
|
| 148 |
+
"# Building the model from the config\n",
|
| 149 |
+
"model = AutoModelForMaskedLM.from_config(config)"
|
| 150 |
+
]
|
| 151 |
+
},
|
| 152 |
+
{
|
| 153 |
+
"cell_type": "code",
|
| 154 |
+
"execution_count": null,
|
| 155 |
+
"id": "afc2cdd1-228e-4ee7-95f5-07718f00723d",
|
| 156 |
+
"metadata": {},
|
| 157 |
+
"outputs": [],
|
| 158 |
+
"source": [
|
| 159 |
+
"# 1. load dna dataset\n",
|
| 160 |
+
"raw_dataset = load_dataset('text', data_files=\"../01-data_env/data/dna_1g.txt\")\n",
|
| 161 |
+
"dataset = raw_dataset[\"train\"].train_test_split(test_size=0.1, shuffle=True)\n",
|
| 162 |
+
"\n",
|
| 163 |
+
"# 2. tokenize\n",
|
| 164 |
+
"def tokenize_function(examples):\n",
|
| 165 |
+
" return tokenizer(examples['text'], truncation=True, padding='max_length', max_length=max_length)\n",
|
| 166 |
+
"\n",
|
| 167 |
+
"# 3. 对数据集应用分词函数\n",
|
| 168 |
+
"tokenized_datasets = dataset.map(tokenize_function, batched=True, remove_columns=['text'], num_proc=15) # 设置为你的 CPU 核心数或根据需要调整\n",
|
| 169 |
+
"\n",
|
| 170 |
+
"# 4. 创建一个数据收集器,用于动态填充和遮蔽,注意mlm=true\n",
|
| 171 |
+
"data_collator = DataCollatorForLanguageModeling(\n",
|
| 172 |
+
" tokenizer=tokenizer, mlm=True,mlm_probability=0.15\n",
|
| 173 |
+
")"
|
| 174 |
+
]
|
| 175 |
+
},
|
| 176 |
+
{
|
| 177 |
+
"cell_type": "code",
|
| 178 |
+
"execution_count": null,
|
| 179 |
+
"id": "604491f9-2ee7-4722-aad6-02e98457b5ee",
|
| 180 |
+
"metadata": {},
|
| 181 |
+
"outputs": [],
|
| 182 |
+
"source": [
|
| 183 |
+
"run_path = \"bert_run\"\n",
|
| 184 |
+
"train_epoches = 5\n",
|
| 185 |
+
"batch_size = 10\n",
|
| 186 |
+
"\n",
|
| 187 |
+
"\n",
|
| 188 |
+
"training_args = TrainingArguments(\n",
|
| 189 |
+
" output_dir=run_path,\n",
|
| 190 |
+
" overwrite_output_dir=True,\n",
|
| 191 |
+
" num_train_epochs=train_epoches,\n",
|
| 192 |
+
" per_device_train_batch_size=batch_size,\n",
|
| 193 |
+
" save_steps=2000,\n",
|
| 194 |
+
" save_total_limit=2,\n",
|
| 195 |
+
" prediction_loss_only=True,\n",
|
| 196 |
+
" fp16=True, #v100没法用\n",
|
| 197 |
+
" )\n",
|
| 198 |
+
"\n",
|
| 199 |
+
"\n",
|
| 200 |
+
"trainer = Trainer(\n",
|
| 201 |
+
" model=model,\n",
|
| 202 |
+
" args=training_args,\n",
|
| 203 |
+
" train_dataset=tokenized_datasets[\"train\"],\n",
|
| 204 |
+
" eval_dataset=tokenized_datasets[\"test\"],\n",
|
| 205 |
+
" data_collator=data_collator,\n",
|
| 206 |
+
")"
|
| 207 |
+
]
|
| 208 |
+
},
|
| 209 |
+
{
|
| 210 |
+
"cell_type": "code",
|
| 211 |
+
"execution_count": null,
|
| 212 |
+
"id": "d91a8bfb-f3ff-4031-a0d7-ebedc200d65a",
|
| 213 |
+
"metadata": {},
|
| 214 |
+
"outputs": [],
|
| 215 |
+
"source": [
|
| 216 |
+
"trainer.train()\n",
|
| 217 |
+
"trainer.save_model(\"dna_bert_v0\")"
|
| 218 |
+
]
|
| 219 |
+
},
|
| 220 |
+
{
|
| 221 |
+
"cell_type": "code",
|
| 222 |
+
"execution_count": null,
|
| 223 |
+
"id": "fc4ad6ad-6433-471f-8510-1ae46558d4ce",
|
| 224 |
+
"metadata": {},
|
| 225 |
+
"outputs": [],
|
| 226 |
+
"source": [
|
| 227 |
+
"#upload model\n",
|
| 228 |
+
"#model.push_to_hub(\"dna_bert_v0\", organization=\"dnagpt\", use_auth_token=\"hf_*******\")"
|
| 229 |
+
]
|
| 230 |
+
}
|
| 231 |
+
],
|
| 232 |
+
"metadata": {
|
| 233 |
+
"kernelspec": {
|
| 234 |
+
"display_name": "Python 3 (ipykernel)",
|
| 235 |
+
"language": "python",
|
| 236 |
+
"name": "python3"
|
| 237 |
+
},
|
| 238 |
+
"language_info": {
|
| 239 |
+
"codemirror_mode": {
|
| 240 |
+
"name": "ipython",
|
| 241 |
+
"version": 3
|
| 242 |
+
},
|
| 243 |
+
"file_extension": ".py",
|
| 244 |
+
"mimetype": "text/x-python",
|
| 245 |
+
"name": "python",
|
| 246 |
+
"nbconvert_exporter": "python",
|
| 247 |
+
"pygments_lexer": "ipython3",
|
| 248 |
+
"version": "3.12.3"
|
| 249 |
+
}
|
| 250 |
+
},
|
| 251 |
+
"nbformat": 4,
|
| 252 |
+
"nbformat_minor": 5
|
| 253 |
+
}
|
02-gpt2_bert/.ipynb_checkpoints/4-gene-feature-checkpoint.ipynb
ADDED
|
@@ -0,0 +1,489 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"id": "b1b37ca8-25a3-440c-9b68-7f72ce670ade",
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"source": [
|
| 8 |
+
"# 2.4 基因大模型的生物序列特征提取"
|
| 9 |
+
]
|
| 10 |
+
},
|
| 11 |
+
{
|
| 12 |
+
"cell_type": "markdown",
|
| 13 |
+
"id": "d3d04215-2b6c-41fb-92a4-90c82d322ba4",
|
| 14 |
+
"metadata": {},
|
| 15 |
+
"source": [
|
| 16 |
+
"使用 GPT-2 模型获取文本的特征向量是一个常见的需求,尤其是在进行文本分类、相似度计算或其他下游任务时。Hugging Face 的 transformers 库提供了简单易用的接口来实现这一点。以下是详细的步骤和代码示例,帮助你从 GPT-2 模型中提取文本的特征向量。"
|
| 17 |
+
]
|
| 18 |
+
},
|
| 19 |
+
{
|
| 20 |
+
"cell_type": "markdown",
|
| 21 |
+
"id": "3ff5b7c6-e57c-4839-8510-f764154faa65",
|
| 22 |
+
"metadata": {},
|
| 23 |
+
"source": [
|
| 24 |
+
"使用 GPT-2 模型获取文本的特征向量是一个常见的需求,尤其是在进行文本分类、相似度计算或其他下游任务时。Hugging Face 的 `transformers` 库提供了简单易用的接口来实现这一点。以下是详细的步骤和代码示例,帮助你从 GPT-2 模型中提取文本的特征向量。\n",
|
| 25 |
+
"\n",
|
| 26 |
+
"### 方法 1: 使用隐藏状态(Hidden States)\n",
|
| 27 |
+
"\n",
|
| 28 |
+
"GPT-2 是一个基于 Transformer 的语言模型,它在每一层都有隐藏状态(hidden states),这些隐藏状态可以作为文本的特征表示。你可以选择最后一层的隐藏状态作为最终的特征向量,或者对多层的隐藏状态进行平均或拼接。\n",
|
| 29 |
+
"\n",
|
| 30 |
+
"\n",
|
| 31 |
+
"### 方法 2: 使用池化策略\n",
|
| 32 |
+
"\n",
|
| 33 |
+
"另一种方法是通过对所有 token 的隐藏状态进行池化操作来获得句子级别的特征向量。常见的池化方法包括:\n",
|
| 34 |
+
"\n",
|
| 35 |
+
"- **均值池化**(Mean Pooling):对所有 token 的隐藏状态求平均。\n",
|
| 36 |
+
"- **最大池化**(Max Pooling):对每个维度取最大值。"
|
| 37 |
+
]
|
| 38 |
+
},
|
| 39 |
+
{
|
| 40 |
+
"cell_type": "code",
|
| 41 |
+
"execution_count": 43,
|
| 42 |
+
"id": "e7fe053b-d6da-488a-9c62-24e4b40a992d",
|
| 43 |
+
"metadata": {},
|
| 44 |
+
"outputs": [
|
| 45 |
+
{
|
| 46 |
+
"name": "stdout",
|
| 47 |
+
"output_type": "stream",
|
| 48 |
+
"text": [
|
| 49 |
+
"{'input_ids': tensor([[ 1, 191, 29, 753, 1241, 2104, 12297, 357, 85, 4395,\n",
|
| 50 |
+
" 26392, 16]]), 'attention_mask': tensor([[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]])}\n",
|
| 51 |
+
"torch.Size([768])\n",
|
| 52 |
+
"torch.Size([768])\n",
|
| 53 |
+
"torch.Size([768])\n"
|
| 54 |
+
]
|
| 55 |
+
}
|
| 56 |
+
],
|
| 57 |
+
"source": [
|
| 58 |
+
"from transformers import AutoTokenizer, AutoModel\n",
|
| 59 |
+
"tokenizer = AutoTokenizer.from_pretrained('dna_bpe_dict')\n",
|
| 60 |
+
"tokenizer.tokenize(\"GAGCACATTCGCCTGCGTGCGCACTCACACACACGTTCAAAAAGAGTCCATTCGATTCTGGCAGTAG\")\n",
|
| 61 |
+
"#result: [G','AGCAC','ATTCGCC',....]\n",
|
| 62 |
+
"\n",
|
| 63 |
+
"model = AutoModel.from_pretrained('dna_gpt2_v0')\n",
|
| 64 |
+
"import torch\n",
|
| 65 |
+
"dna = \"ACGTAGCATCGGATCTATCTATCGACACTTGGTTATCGATCTACGAGCATCTCGTTAGC\"\n",
|
| 66 |
+
"inputs = tokenizer(dna, return_tensors = 'pt')\n",
|
| 67 |
+
"print(inputs)\n",
|
| 68 |
+
"\n",
|
| 69 |
+
"outputs = model(inputs[\"input_ids\"])\n",
|
| 70 |
+
"#outputs = model(**inputs)\n",
|
| 71 |
+
"\n",
|
| 72 |
+
"hidden_states = outputs.last_hidden_state # [1, sequence_length, 768] outputs.last_hidden_state or outputs[0]\n",
|
| 73 |
+
"\n",
|
| 74 |
+
"# embedding with mean pooling\n",
|
| 75 |
+
"embedding_mean = torch.mean(hidden_states[0], dim=0)\n",
|
| 76 |
+
"print(embedding_mean.shape) # expect to be 768\n",
|
| 77 |
+
"\n",
|
| 78 |
+
"# embedding with max pooling\n",
|
| 79 |
+
"embedding_max = torch.max(hidden_states[0], dim=0)[0]\n",
|
| 80 |
+
"print(embedding_max.shape) # expect to be 768\n",
|
| 81 |
+
"\n",
|
| 82 |
+
"# embedding with first token\n",
|
| 83 |
+
"embedding_first_token = hidden_states[0][0]\n",
|
| 84 |
+
"print(embedding_first_token.shape) # expect to be 768"
|
| 85 |
+
]
|
| 86 |
+
},
|
| 87 |
+
{
|
| 88 |
+
"cell_type": "code",
|
| 89 |
+
"execution_count": 44,
|
| 90 |
+
"id": "a1f2b545-283a-4613-a953-beb82f427826",
|
| 91 |
+
"metadata": {},
|
| 92 |
+
"outputs": [
|
| 93 |
+
{
|
| 94 |
+
"name": "stderr",
|
| 95 |
+
"output_type": "stream",
|
| 96 |
+
"text": [
|
| 97 |
+
"Some weights of BertModel were not initialized from the model checkpoint at dna_bert_v0 and are newly initialized: ['bert.pooler.dense.bias', 'bert.pooler.dense.weight']\n",
|
| 98 |
+
"You should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.\n"
|
| 99 |
+
]
|
| 100 |
+
},
|
| 101 |
+
{
|
| 102 |
+
"name": "stdout",
|
| 103 |
+
"output_type": "stream",
|
| 104 |
+
"text": [
|
| 105 |
+
"{'input_ids': tensor([[ 6, 200, 16057, 10, 1256, 2123, 12294, 366, 13138, 7826,\n",
|
| 106 |
+
" 82, 25]]), 'token_type_ids': tensor([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]]), 'attention_mask': tensor([[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]])}\n",
|
| 107 |
+
"torch.Size([768])\n",
|
| 108 |
+
"torch.Size([768])\n",
|
| 109 |
+
"torch.Size([768])\n"
|
| 110 |
+
]
|
| 111 |
+
}
|
| 112 |
+
],
|
| 113 |
+
"source": [
|
| 114 |
+
"from transformers import AutoTokenizer, AutoModel\n",
|
| 115 |
+
"import torch\n",
|
| 116 |
+
"\n",
|
| 117 |
+
"tokenizer = AutoTokenizer.from_pretrained('dna_wordpiece_dict')\n",
|
| 118 |
+
"tokenizer.tokenize(\"GAGCACATTCGCCTGCGTGCGCACTCACACACACGTTCAAAAAGAGTCCATTCGATTCTGGCAGTAG\")\n",
|
| 119 |
+
"#result: [G','AGCAC','ATTCGCC',....]\n",
|
| 120 |
+
"\n",
|
| 121 |
+
"model = AutoModel.from_pretrained('dna_bert_v0')\n",
|
| 122 |
+
"dna = \"ACGTAGCATCGGATCTATCTATCGACACTTGGTTATCGATCTACGAGCATCTCGTTAGC\"\n",
|
| 123 |
+
"inputs = tokenizer(dna, return_tensors = 'pt')\n",
|
| 124 |
+
"print(inputs)\n",
|
| 125 |
+
"\n",
|
| 126 |
+
"outputs = model(inputs[\"input_ids\"])\n",
|
| 127 |
+
"#outputs = model(**inputs)\n",
|
| 128 |
+
"\n",
|
| 129 |
+
"hidden_states = outputs.last_hidden_state # [1, sequence_length, 768] outputs.last_hidden_state or outputs[0]\n",
|
| 130 |
+
"\n",
|
| 131 |
+
"# embedding with mean pooling\n",
|
| 132 |
+
"embedding_mean = torch.mean(hidden_states[0], dim=0)\n",
|
| 133 |
+
"print(embedding_mean.shape) # expect to be 768\n",
|
| 134 |
+
"\n",
|
| 135 |
+
"# embedding with max pooling\n",
|
| 136 |
+
"embedding_max = torch.max(hidden_states[0], dim=0)[0]\n",
|
| 137 |
+
"print(embedding_max.shape) # expect to be 768\n",
|
| 138 |
+
"\n",
|
| 139 |
+
"# embedding with first token\n",
|
| 140 |
+
"embedding_first_token = hidden_states[0][0]\n",
|
| 141 |
+
"print(embedding_first_token.shape) # expect to be 768"
|
| 142 |
+
]
|
| 143 |
+
},
|
| 144 |
+
{
|
| 145 |
+
"cell_type": "markdown",
|
| 146 |
+
"id": "56761874-9af7-4b90-aa8b-131e5b8c69b6",
|
| 147 |
+
"metadata": {},
|
| 148 |
+
"source": [
|
| 149 |
+
"## 特征提取并分类\n",
|
| 150 |
+
"\n",
|
| 151 |
+
"我们使用第一章中的\"dnagpt/dna_core_promoter\"数据集,演示下使用我们训练的DNA GPT2或者DNA bert模型,提取序列特征,然使用最基础的逻辑回归分类方法,对序列进行分类。"
|
| 152 |
+
]
|
| 153 |
+
},
|
| 154 |
+
{
|
| 155 |
+
"cell_type": "code",
|
| 156 |
+
"execution_count": 45,
|
| 157 |
+
"id": "f1ca177c-a80f-48a1-b2f9-16c13b3350db",
|
| 158 |
+
"metadata": {},
|
| 159 |
+
"outputs": [
|
| 160 |
+
{
|
| 161 |
+
"data": {
|
| 162 |
+
"text/plain": [
|
| 163 |
+
"\"\\nimport os\\n\\n# 设置环境变量, autodl专区 其他idc\\nos.environ['HF_ENDPOINT'] = 'https://hf-mirror.com'\\n\\n# 打印环境变量以确认设置成功\\nprint(os.environ.get('HF_ENDPOINT'))\\n\""
|
| 164 |
+
]
|
| 165 |
+
},
|
| 166 |
+
"execution_count": 45,
|
| 167 |
+
"metadata": {},
|
| 168 |
+
"output_type": "execute_result"
|
| 169 |
+
}
|
| 170 |
+
],
|
| 171 |
+
"source": [
|
| 172 |
+
"import subprocess\n",
|
| 173 |
+
"import os\n",
|
| 174 |
+
"# 设置环境变量, autodl一般区域\n",
|
| 175 |
+
"result = subprocess.run('bash -c \"source /etc/network_turbo && env | grep proxy\"', shell=True, capture_output=True, text=True)\n",
|
| 176 |
+
"output = result.stdout\n",
|
| 177 |
+
"for line in output.splitlines():\n",
|
| 178 |
+
" if '=' in line:\n",
|
| 179 |
+
" var, value = line.split('=', 1)\n",
|
| 180 |
+
" os.environ[var] = value\n",
|
| 181 |
+
"\n",
|
| 182 |
+
"#或者\n",
|
| 183 |
+
"\"\"\"\n",
|
| 184 |
+
"import os\n",
|
| 185 |
+
"\n",
|
| 186 |
+
"# 设置环境变量, autodl专区 其他idc\n",
|
| 187 |
+
"os.environ['HF_ENDPOINT'] = 'https://hf-mirror.com'\n",
|
| 188 |
+
"\n",
|
| 189 |
+
"# 打印环境变量以确认设置成功\n",
|
| 190 |
+
"print(os.environ.get('HF_ENDPOINT'))\n",
|
| 191 |
+
"\"\"\""
|
| 192 |
+
]
|
| 193 |
+
},
|
| 194 |
+
{
|
| 195 |
+
"cell_type": "code",
|
| 196 |
+
"execution_count": 46,
|
| 197 |
+
"id": "2295739c-e80a-47be-9400-88bfab4b0bb6",
|
| 198 |
+
"metadata": {},
|
| 199 |
+
"outputs": [
|
| 200 |
+
{
|
| 201 |
+
"data": {
|
| 202 |
+
"text/plain": [
|
| 203 |
+
"DatasetDict({\n",
|
| 204 |
+
" train: Dataset({\n",
|
| 205 |
+
" features: ['sequence', 'label'],\n",
|
| 206 |
+
" num_rows: 59196\n",
|
| 207 |
+
" })\n",
|
| 208 |
+
"})"
|
| 209 |
+
]
|
| 210 |
+
},
|
| 211 |
+
"execution_count": 46,
|
| 212 |
+
"metadata": {},
|
| 213 |
+
"output_type": "execute_result"
|
| 214 |
+
}
|
| 215 |
+
],
|
| 216 |
+
"source": [
|
| 217 |
+
"from datasets import load_dataset\n",
|
| 218 |
+
"dna_data = load_dataset(\"dnagpt/dna_core_promoter\")\n",
|
| 219 |
+
"dna_data"
|
| 220 |
+
]
|
| 221 |
+
},
|
| 222 |
+
{
|
| 223 |
+
"cell_type": "markdown",
|
| 224 |
+
"id": "c804bced-f151-43a7-8a95-156db358da3e",
|
| 225 |
+
"metadata": {},
|
| 226 |
+
"source": [
|
| 227 |
+
"这里,我们不需要关注这个数据的具体生物学含义,只需知道sequence是具体的DNA序列,label是分类标签,有两个类别0和1即可"
|
| 228 |
+
]
|
| 229 |
+
},
|
| 230 |
+
{
|
| 231 |
+
"cell_type": "code",
|
| 232 |
+
"execution_count": 47,
|
| 233 |
+
"id": "9a47a1b1-21f2-4d71-801c-50f88e326ed3",
|
| 234 |
+
"metadata": {},
|
| 235 |
+
"outputs": [
|
| 236 |
+
{
|
| 237 |
+
"data": {
|
| 238 |
+
"text/plain": [
|
| 239 |
+
"{'sequence': 'CATGCGGGTCGATATCCTATCTGAATCTCTCAGCCCAAGAGGGAGTCCGCTCATCTATTCGGCAGTACTG',\n",
|
| 240 |
+
" 'label': 0}"
|
| 241 |
+
]
|
| 242 |
+
},
|
| 243 |
+
"execution_count": 47,
|
| 244 |
+
"metadata": {},
|
| 245 |
+
"output_type": "execute_result"
|
| 246 |
+
}
|
| 247 |
+
],
|
| 248 |
+
"source": [
|
| 249 |
+
"dna_data[\"train\"][0]"
|
| 250 |
+
]
|
| 251 |
+
},
|
| 252 |
+
{
|
| 253 |
+
"cell_type": "markdown",
|
| 254 |
+
"id": "cde7986d-a225-41ca-8f11-614d079fd2bf",
|
| 255 |
+
"metadata": {},
|
| 256 |
+
"source": [
|
| 257 |
+
"这里使用scikit-learn库来构建逻辑回归分类器。首先是特征提取:"
|
| 258 |
+
]
|
| 259 |
+
},
|
| 260 |
+
{
|
| 261 |
+
"cell_type": "code",
|
| 262 |
+
"execution_count": 52,
|
| 263 |
+
"id": "4010d991-056a-43ce-8cca-30eeec8678f5",
|
| 264 |
+
"metadata": {},
|
| 265 |
+
"outputs": [],
|
| 266 |
+
"source": [
|
| 267 |
+
"import numpy as np\n",
|
| 268 |
+
"from sklearn.model_selection import train_test_split\n",
|
| 269 |
+
"from sklearn.linear_model import LogisticRegression\n",
|
| 270 |
+
"from sklearn.datasets import load_iris\n",
|
| 271 |
+
"from sklearn.metrics import accuracy_score\n",
|
| 272 |
+
"\n",
|
| 273 |
+
"\n",
|
| 274 |
+
"def get_gpt2_feature(sequence):\n",
|
| 275 |
+
" return \n",
|
| 276 |
+
"\n",
|
| 277 |
+
"# 加载数据集\n",
|
| 278 |
+
"data = load_iris()\n",
|
| 279 |
+
"X = data.data[data.target < 2] # 只选择前两个类别\n",
|
| 280 |
+
"y = data.target[data.target < 2]\n",
|
| 281 |
+
"\n",
|
| 282 |
+
"X = []\n",
|
| 283 |
+
"Y = []\n",
|
| 284 |
+
"\n",
|
| 285 |
+
"for item in dna_data[\"train\"]:\n",
|
| 286 |
+
" sequence = item[\"sequence\"]\n",
|
| 287 |
+
" label = item[\"label\"]\n",
|
| 288 |
+
" x_v = get_gpt2_feature(sequence)\n",
|
| 289 |
+
" y_v = label\n",
|
| 290 |
+
" X.append(x_v)\n",
|
| 291 |
+
" Y.append(y_v)"
|
| 292 |
+
]
|
| 293 |
+
},
|
| 294 |
+
{
|
| 295 |
+
"cell_type": "code",
|
| 296 |
+
"execution_count": 49,
|
| 297 |
+
"id": "8af0effa-b2b6-4e49-9256-cead146d848c",
|
| 298 |
+
"metadata": {},
|
| 299 |
+
"outputs": [
|
| 300 |
+
{
|
| 301 |
+
"data": {
|
| 302 |
+
"text/plain": [
|
| 303 |
+
"array([[5.1, 3.5, 1.4, 0.2],\n",
|
| 304 |
+
" [4.9, 3. , 1.4, 0.2],\n",
|
| 305 |
+
" [4.7, 3.2, 1.3, 0.2],\n",
|
| 306 |
+
" [4.6, 3.1, 1.5, 0.2],\n",
|
| 307 |
+
" [5. , 3.6, 1.4, 0.2],\n",
|
| 308 |
+
" [5.4, 3.9, 1.7, 0.4],\n",
|
| 309 |
+
" [4.6, 3.4, 1.4, 0.3],\n",
|
| 310 |
+
" [5. , 3.4, 1.5, 0.2],\n",
|
| 311 |
+
" [4.4, 2.9, 1.4, 0.2],\n",
|
| 312 |
+
" [4.9, 3.1, 1.5, 0.1],\n",
|
| 313 |
+
" [5.4, 3.7, 1.5, 0.2],\n",
|
| 314 |
+
" [4.8, 3.4, 1.6, 0.2],\n",
|
| 315 |
+
" [4.8, 3. , 1.4, 0.1],\n",
|
| 316 |
+
" [4.3, 3. , 1.1, 0.1],\n",
|
| 317 |
+
" [5.8, 4. , 1.2, 0.2],\n",
|
| 318 |
+
" [5.7, 4.4, 1.5, 0.4],\n",
|
| 319 |
+
" [5.4, 3.9, 1.3, 0.4],\n",
|
| 320 |
+
" [5.1, 3.5, 1.4, 0.3],\n",
|
| 321 |
+
" [5.7, 3.8, 1.7, 0.3],\n",
|
| 322 |
+
" [5.1, 3.8, 1.5, 0.3],\n",
|
| 323 |
+
" [5.4, 3.4, 1.7, 0.2],\n",
|
| 324 |
+
" [5.1, 3.7, 1.5, 0.4],\n",
|
| 325 |
+
" [4.6, 3.6, 1. , 0.2],\n",
|
| 326 |
+
" [5.1, 3.3, 1.7, 0.5],\n",
|
| 327 |
+
" [4.8, 3.4, 1.9, 0.2],\n",
|
| 328 |
+
" [5. , 3. , 1.6, 0.2],\n",
|
| 329 |
+
" [5. , 3.4, 1.6, 0.4],\n",
|
| 330 |
+
" [5.2, 3.5, 1.5, 0.2],\n",
|
| 331 |
+
" [5.2, 3.4, 1.4, 0.2],\n",
|
| 332 |
+
" [4.7, 3.2, 1.6, 0.2],\n",
|
| 333 |
+
" [4.8, 3.1, 1.6, 0.2],\n",
|
| 334 |
+
" [5.4, 3.4, 1.5, 0.4],\n",
|
| 335 |
+
" [5.2, 4.1, 1.5, 0.1],\n",
|
| 336 |
+
" [5.5, 4.2, 1.4, 0.2],\n",
|
| 337 |
+
" [4.9, 3.1, 1.5, 0.2],\n",
|
| 338 |
+
" [5. , 3.2, 1.2, 0.2],\n",
|
| 339 |
+
" [5.5, 3.5, 1.3, 0.2],\n",
|
| 340 |
+
" [4.9, 3.6, 1.4, 0.1],\n",
|
| 341 |
+
" [4.4, 3. , 1.3, 0.2],\n",
|
| 342 |
+
" [5.1, 3.4, 1.5, 0.2],\n",
|
| 343 |
+
" [5. , 3.5, 1.3, 0.3],\n",
|
| 344 |
+
" [4.5, 2.3, 1.3, 0.3],\n",
|
| 345 |
+
" [4.4, 3.2, 1.3, 0.2],\n",
|
| 346 |
+
" [5. , 3.5, 1.6, 0.6],\n",
|
| 347 |
+
" [5.1, 3.8, 1.9, 0.4],\n",
|
| 348 |
+
" [4.8, 3. , 1.4, 0.3],\n",
|
| 349 |
+
" [5.1, 3.8, 1.6, 0.2],\n",
|
| 350 |
+
" [4.6, 3.2, 1.4, 0.2],\n",
|
| 351 |
+
" [5.3, 3.7, 1.5, 0.2],\n",
|
| 352 |
+
" [5. , 3.3, 1.4, 0.2],\n",
|
| 353 |
+
" [7. , 3.2, 4.7, 1.4],\n",
|
| 354 |
+
" [6.4, 3.2, 4.5, 1.5],\n",
|
| 355 |
+
" [6.9, 3.1, 4.9, 1.5],\n",
|
| 356 |
+
" [5.5, 2.3, 4. , 1.3],\n",
|
| 357 |
+
" [6.5, 2.8, 4.6, 1.5],\n",
|
| 358 |
+
" [5.7, 2.8, 4.5, 1.3],\n",
|
| 359 |
+
" [6.3, 3.3, 4.7, 1.6],\n",
|
| 360 |
+
" [4.9, 2.4, 3.3, 1. ],\n",
|
| 361 |
+
" [6.6, 2.9, 4.6, 1.3],\n",
|
| 362 |
+
" [5.2, 2.7, 3.9, 1.4],\n",
|
| 363 |
+
" [5. , 2. , 3.5, 1. ],\n",
|
| 364 |
+
" [5.9, 3. , 4.2, 1.5],\n",
|
| 365 |
+
" [6. , 2.2, 4. , 1. ],\n",
|
| 366 |
+
" [6.1, 2.9, 4.7, 1.4],\n",
|
| 367 |
+
" [5.6, 2.9, 3.6, 1.3],\n",
|
| 368 |
+
" [6.7, 3.1, 4.4, 1.4],\n",
|
| 369 |
+
" [5.6, 3. , 4.5, 1.5],\n",
|
| 370 |
+
" [5.8, 2.7, 4.1, 1. ],\n",
|
| 371 |
+
" [6.2, 2.2, 4.5, 1.5],\n",
|
| 372 |
+
" [5.6, 2.5, 3.9, 1.1],\n",
|
| 373 |
+
" [5.9, 3.2, 4.8, 1.8],\n",
|
| 374 |
+
" [6.1, 2.8, 4. , 1.3],\n",
|
| 375 |
+
" [6.3, 2.5, 4.9, 1.5],\n",
|
| 376 |
+
" [6.1, 2.8, 4.7, 1.2],\n",
|
| 377 |
+
" [6.4, 2.9, 4.3, 1.3],\n",
|
| 378 |
+
" [6.6, 3. , 4.4, 1.4],\n",
|
| 379 |
+
" [6.8, 2.8, 4.8, 1.4],\n",
|
| 380 |
+
" [6.7, 3. , 5. , 1.7],\n",
|
| 381 |
+
" [6. , 2.9, 4.5, 1.5],\n",
|
| 382 |
+
" [5.7, 2.6, 3.5, 1. ],\n",
|
| 383 |
+
" [5.5, 2.4, 3.8, 1.1],\n",
|
| 384 |
+
" [5.5, 2.4, 3.7, 1. ],\n",
|
| 385 |
+
" [5.8, 2.7, 3.9, 1.2],\n",
|
| 386 |
+
" [6. , 2.7, 5.1, 1.6],\n",
|
| 387 |
+
" [5.4, 3. , 4.5, 1.5],\n",
|
| 388 |
+
" [6. , 3.4, 4.5, 1.6],\n",
|
| 389 |
+
" [6.7, 3.1, 4.7, 1.5],\n",
|
| 390 |
+
" [6.3, 2.3, 4.4, 1.3],\n",
|
| 391 |
+
" [5.6, 3. , 4.1, 1.3],\n",
|
| 392 |
+
" [5.5, 2.5, 4. , 1.3],\n",
|
| 393 |
+
" [5.5, 2.6, 4.4, 1.2],\n",
|
| 394 |
+
" [6.1, 3. , 4.6, 1.4],\n",
|
| 395 |
+
" [5.8, 2.6, 4. , 1.2],\n",
|
| 396 |
+
" [5. , 2.3, 3.3, 1. ],\n",
|
| 397 |
+
" [5.6, 2.7, 4.2, 1.3],\n",
|
| 398 |
+
" [5.7, 3. , 4.2, 1.2],\n",
|
| 399 |
+
" [5.7, 2.9, 4.2, 1.3],\n",
|
| 400 |
+
" [6.2, 2.9, 4.3, 1.3],\n",
|
| 401 |
+
" [5.1, 2.5, 3. , 1.1],\n",
|
| 402 |
+
" [5.7, 2.8, 4.1, 1.3]])"
|
| 403 |
+
]
|
| 404 |
+
},
|
| 405 |
+
"execution_count": 49,
|
| 406 |
+
"metadata": {},
|
| 407 |
+
"output_type": "execute_result"
|
| 408 |
+
}
|
| 409 |
+
],
|
| 410 |
+
"source": [
|
| 411 |
+
"X"
|
| 412 |
+
]
|
| 413 |
+
},
|
| 414 |
+
{
|
| 415 |
+
"cell_type": "code",
|
| 416 |
+
"execution_count": 51,
|
| 417 |
+
"id": "868a3cab-e991-4990-9ec5-3e632a41a599",
|
| 418 |
+
"metadata": {},
|
| 419 |
+
"outputs": [
|
| 420 |
+
{
|
| 421 |
+
"data": {
|
| 422 |
+
"text/plain": [
|
| 423 |
+
"array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n",
|
| 424 |
+
" 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n",
|
| 425 |
+
" 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,\n",
|
| 426 |
+
" 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,\n",
|
| 427 |
+
" 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1])"
|
| 428 |
+
]
|
| 429 |
+
},
|
| 430 |
+
"execution_count": 51,
|
| 431 |
+
"metadata": {},
|
| 432 |
+
"output_type": "execute_result"
|
| 433 |
+
}
|
| 434 |
+
],
|
| 435 |
+
"source": [
|
| 436 |
+
"y"
|
| 437 |
+
]
|
| 438 |
+
},
|
| 439 |
+
{
|
| 440 |
+
"cell_type": "code",
|
| 441 |
+
"execution_count": null,
|
| 442 |
+
"id": "5ab0c188-6476-43c4-b361-a2bfe0ec7a8a",
|
| 443 |
+
"metadata": {},
|
| 444 |
+
"outputs": [],
|
| 445 |
+
"source": [
|
| 446 |
+
"# 将数据分为训练集和测试集\n",
|
| 447 |
+
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)\n",
|
| 448 |
+
"\n",
|
| 449 |
+
"# 创建逻辑回归模型\n",
|
| 450 |
+
"model = LogisticRegression()\n",
|
| 451 |
+
"\n",
|
| 452 |
+
"# 训练模型\n",
|
| 453 |
+
"model.fit(X_train, y_train)\n",
|
| 454 |
+
"\n",
|
| 455 |
+
"# 在测试集上进行预测\n",
|
| 456 |
+
"y_pred = model.predict(X_test)\n",
|
| 457 |
+
"\n",
|
| 458 |
+
"# 计算准确率\n",
|
| 459 |
+
"accuracy = accuracy_score(y_test, y_pred)\n",
|
| 460 |
+
"print(f\"Accuracy: {accuracy * 100:.2f}%\")\n",
|
| 461 |
+
"\n",
|
| 462 |
+
"# 输出部分预测结果与真实标签对比\n",
|
| 463 |
+
"for i in range(5):\n",
|
| 464 |
+
" print(f\"True: {y_test[i]}, Predicted: {y_pred[i]}\")"
|
| 465 |
+
]
|
| 466 |
+
}
|
| 467 |
+
],
|
| 468 |
+
"metadata": {
|
| 469 |
+
"kernelspec": {
|
| 470 |
+
"display_name": "Python 3 (ipykernel)",
|
| 471 |
+
"language": "python",
|
| 472 |
+
"name": "python3"
|
| 473 |
+
},
|
| 474 |
+
"language_info": {
|
| 475 |
+
"codemirror_mode": {
|
| 476 |
+
"name": "ipython",
|
| 477 |
+
"version": 3
|
| 478 |
+
},
|
| 479 |
+
"file_extension": ".py",
|
| 480 |
+
"mimetype": "text/x-python",
|
| 481 |
+
"name": "python",
|
| 482 |
+
"nbconvert_exporter": "python",
|
| 483 |
+
"pygments_lexer": "ipython3",
|
| 484 |
+
"version": "3.12.3"
|
| 485 |
+
}
|
| 486 |
+
},
|
| 487 |
+
"nbformat": 4,
|
| 488 |
+
"nbformat_minor": 5
|
| 489 |
+
}
|
02-gpt2_bert/.ipynb_checkpoints/5-multi-seq-gpt-checkpoint.ipynb
ADDED
|
@@ -0,0 +1,261 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"id": "9131f25f-227b-4dbe-b28d-c5006df092c6",
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"source": [
|
| 8 |
+
"# 2.5 基于多模态数据构建大模型"
|
| 9 |
+
]
|
| 10 |
+
},
|
| 11 |
+
{
|
| 12 |
+
"cell_type": "code",
|
| 13 |
+
"execution_count": null,
|
| 14 |
+
"id": "1a30b35c-1f5f-41e6-8fe1-5f522c700e9e",
|
| 15 |
+
"metadata": {},
|
| 16 |
+
"outputs": [],
|
| 17 |
+
"source": [
|
| 18 |
+
"from tokenizers import (\n",
|
| 19 |
+
" decoders,\n",
|
| 20 |
+
" models,\n",
|
| 21 |
+
" normalizers,\n",
|
| 22 |
+
" pre_tokenizers,\n",
|
| 23 |
+
" processors,\n",
|
| 24 |
+
" trainers,\n",
|
| 25 |
+
" Tokenizer,\n",
|
| 26 |
+
")\n",
|
| 27 |
+
"from transformers import AutoTokenizer"
|
| 28 |
+
]
|
| 29 |
+
},
|
| 30 |
+
{
|
| 31 |
+
"cell_type": "code",
|
| 32 |
+
"execution_count": null,
|
| 33 |
+
"id": "688fa3b1-f2ca-457a-abde-117c79b54fa9",
|
| 34 |
+
"metadata": {},
|
| 35 |
+
"outputs": [],
|
| 36 |
+
"source": [
|
| 37 |
+
"tokenizer = Tokenizer(models.BPE())\n",
|
| 38 |
+
"tokenizer.pre_tokenizer = pre_tokenizers.ByteLevel(add_prefix_space=False, use_regex=False) #use_regex=False,空格当成一般字符串\n",
|
| 39 |
+
"trainer = trainers.BpeTrainer(vocab_size=90000, special_tokens=[\"<|endoftext|>\"]) #9w words"
|
| 40 |
+
]
|
| 41 |
+
},
|
| 42 |
+
{
|
| 43 |
+
"cell_type": "code",
|
| 44 |
+
"execution_count": null,
|
| 45 |
+
"id": "7d680700-1051-4af4-94d6-2ce3071a5979",
|
| 46 |
+
"metadata": {},
|
| 47 |
+
"outputs": [],
|
| 48 |
+
"source": [
|
| 49 |
+
"tokenizer.train([\"../01-data_env/data/dna_1g.txt\",\"../01-data_env/data/protein_1g.txt\",\"../01-data_env/data/english_500m.txt\"]\n",
|
| 50 |
+
" , trainer=trainer) #all file list, take 10-20 min"
|
| 51 |
+
]
|
| 52 |
+
},
|
| 53 |
+
{
|
| 54 |
+
"cell_type": "code",
|
| 55 |
+
"execution_count": null,
|
| 56 |
+
"id": "74434ece-2f6e-46fa-9a9e-ff88e9364de8",
|
| 57 |
+
"metadata": {},
|
| 58 |
+
"outputs": [],
|
| 59 |
+
"source": [
|
| 60 |
+
"tokenizer.save(\"gene_eng_dict.json\")"
|
| 61 |
+
]
|
| 62 |
+
},
|
| 63 |
+
{
|
| 64 |
+
"cell_type": "code",
|
| 65 |
+
"execution_count": null,
|
| 66 |
+
"id": "8ea34e18-6cee-40b9-ba96-d8734153eb9f",
|
| 67 |
+
"metadata": {},
|
| 68 |
+
"outputs": [],
|
| 69 |
+
"source": [
|
| 70 |
+
"#然后我们可以使用from_file() 方法从该文件里重新加载 Tokenizer 对象:\n",
|
| 71 |
+
"new_tokenizer = Tokenizer.from_file(\"gene_eng_dict.json\")\n",
|
| 72 |
+
"\n",
|
| 73 |
+
"#要在 🤗 Transformers 中使用这个标记器,我们必须将它包裹在一个 PreTrainedTokenizerFast 类中\n",
|
| 74 |
+
"from transformers import GPT2TokenizerFast\n",
|
| 75 |
+
"gene_eng_tokenizer = GPT2TokenizerFast(tokenizer_object=new_tokenizer)\n",
|
| 76 |
+
"gene_eng_tokenizer.save_pretrained(\"gene_eng_dict\")\n",
|
| 77 |
+
"#dna_tokenizer.push_to_hub(\"dna_bpe_dict_1g\", organization=\"dnagpt\", use_auth_token=\"hf_*****\") # push to huggingface"
|
| 78 |
+
]
|
| 79 |
+
},
|
| 80 |
+
{
|
| 81 |
+
"cell_type": "code",
|
| 82 |
+
"execution_count": null,
|
| 83 |
+
"id": "16c7a3ef-c924-4fbb-b8ab-c12fab43f019",
|
| 84 |
+
"metadata": {},
|
| 85 |
+
"outputs": [],
|
| 86 |
+
"source": [
|
| 87 |
+
"tokenizer_new = AutoTokenizer.from_pretrained('gene_eng_dict')\n",
|
| 88 |
+
"tokenizer_new.tokenize(\"TGGCGTGAACCCGGGATCGGG,hello world hello gene, MANITWMANHTGWSDFILLGLFRQSKHPALLCVVIFVVFLMAL\")"
|
| 89 |
+
]
|
| 90 |
+
},
|
| 91 |
+
{
|
| 92 |
+
"cell_type": "markdown",
|
| 93 |
+
"id": "0ca0b2e3-f270-4645-abbb-cb8535e97a0a",
|
| 94 |
+
"metadata": {},
|
| 95 |
+
"source": [
|
| 96 |
+
"## 训练混合模型"
|
| 97 |
+
]
|
| 98 |
+
},
|
| 99 |
+
{
|
| 100 |
+
"cell_type": "code",
|
| 101 |
+
"execution_count": null,
|
| 102 |
+
"id": "c9b1c9b4-57a8-4711-912d-307e55481f8a",
|
| 103 |
+
"metadata": {},
|
| 104 |
+
"outputs": [],
|
| 105 |
+
"source": [
|
| 106 |
+
"from transformers import AutoTokenizer, GPT2LMHeadModel, AutoConfig,GPT2Tokenizer\n",
|
| 107 |
+
"from transformers import GPT2Tokenizer,GPT2Model,AutoModel\n",
|
| 108 |
+
"from transformers import DataCollatorForLanguageModeling\n",
|
| 109 |
+
"from transformers import Trainer, TrainingArguments\n",
|
| 110 |
+
"from transformers import LineByLineTextDataset\n",
|
| 111 |
+
"from tokenizers import Tokenizer\n",
|
| 112 |
+
"from datasets import load_dataset"
|
| 113 |
+
]
|
| 114 |
+
},
|
| 115 |
+
{
|
| 116 |
+
"cell_type": "code",
|
| 117 |
+
"execution_count": null,
|
| 118 |
+
"id": "3926a959-4224-4d78-9413-dc47a58087e0",
|
| 119 |
+
"metadata": {},
|
| 120 |
+
"outputs": [],
|
| 121 |
+
"source": [
|
| 122 |
+
"tokenizer = GPT2Tokenizer.from_pretrained(\"gene_eng_dict\")\n",
|
| 123 |
+
"tokenizer.pad_token = tokenizer.eos_token"
|
| 124 |
+
]
|
| 125 |
+
},
|
| 126 |
+
{
|
| 127 |
+
"cell_type": "code",
|
| 128 |
+
"execution_count": null,
|
| 129 |
+
"id": "1c2f5a6d-d405-40dc-a802-f0c1dff50a1e",
|
| 130 |
+
"metadata": {},
|
| 131 |
+
"outputs": [],
|
| 132 |
+
"source": [
|
| 133 |
+
"max_length = 256 #最大输入长度\n",
|
| 134 |
+
"\n",
|
| 135 |
+
"config = AutoConfig.from_pretrained(\n",
|
| 136 |
+
" \"gpt2\",\n",
|
| 137 |
+
" vocab_size=len(tokenizer),\n",
|
| 138 |
+
" n_ctx=max_length, #最大长度\n",
|
| 139 |
+
" bos_token_id=tokenizer.bos_token_id,\n",
|
| 140 |
+
" eos_token_id=tokenizer.eos_token_id,\n",
|
| 141 |
+
")\n",
|
| 142 |
+
"\n",
|
| 143 |
+
"model = GPT2LMHeadModel(config) #for pretrain,从头预训练"
|
| 144 |
+
]
|
| 145 |
+
},
|
| 146 |
+
{
|
| 147 |
+
"cell_type": "code",
|
| 148 |
+
"execution_count": null,
|
| 149 |
+
"id": "c8a47141-56a7-4e41-8cfd-1b381a64e2c0",
|
| 150 |
+
"metadata": {},
|
| 151 |
+
"outputs": [],
|
| 152 |
+
"source": [
|
| 153 |
+
"# 1. load dna dataset\n",
|
| 154 |
+
"raw_dataset = load_dataset('text', \n",
|
| 155 |
+
" data_files=[\"../01-data_env/data/dna_1g.txt\",\"../01-data_env/data/protein_1g.txt\",\"../01-data_env/data/english_500m.txt\"])\n",
|
| 156 |
+
"\n",
|
| 157 |
+
"dataset = raw_dataset[\"train\"].train_test_split(test_size=0.05, shuffle=True)\n",
|
| 158 |
+
"\n",
|
| 159 |
+
"# 2. tokenize\n",
|
| 160 |
+
"def tokenize_function(examples):\n",
|
| 161 |
+
" return tokenizer(examples['text'], truncation=True, padding='max_length', max_length=max_length)\n",
|
| 162 |
+
"\n",
|
| 163 |
+
"# 3. 对数据集应用分词函数\n",
|
| 164 |
+
"tokenized_datasets = dataset.map(tokenize_function, batched=True, remove_columns=['text'], num_proc=15) # 设置为你的 CPU 核心数或根据需要调整\n",
|
| 165 |
+
"\n",
|
| 166 |
+
"# 4. 创建一个数据收集器,用于动态填充和遮蔽\n",
|
| 167 |
+
"data_collator = DataCollatorForLanguageModeling(\n",
|
| 168 |
+
" tokenizer=tokenizer, mlm=False\n",
|
| 169 |
+
")"
|
| 170 |
+
]
|
| 171 |
+
},
|
| 172 |
+
{
|
| 173 |
+
"cell_type": "code",
|
| 174 |
+
"execution_count": null,
|
| 175 |
+
"id": "f4f802a2-88e2-49c2-a654-9d6e0996433a",
|
| 176 |
+
"metadata": {},
|
| 177 |
+
"outputs": [],
|
| 178 |
+
"source": [
|
| 179 |
+
"run_path = \"gpt2_run\"\n",
|
| 180 |
+
"train_epoches = 5\n",
|
| 181 |
+
"batch_size = 10\n",
|
| 182 |
+
"\n",
|
| 183 |
+
"\n",
|
| 184 |
+
"training_args = TrainingArguments(\n",
|
| 185 |
+
" output_dir=run_path,\n",
|
| 186 |
+
" overwrite_output_dir=True,\n",
|
| 187 |
+
" num_train_epochs=train_epoches,\n",
|
| 188 |
+
" per_device_train_batch_size=batch_size,\n",
|
| 189 |
+
" save_steps=2000,\n",
|
| 190 |
+
" save_total_limit=2,\n",
|
| 191 |
+
" prediction_loss_only=True,\n",
|
| 192 |
+
" fp16=True, #v100没法用\n",
|
| 193 |
+
" )\n",
|
| 194 |
+
"\n",
|
| 195 |
+
"\n",
|
| 196 |
+
"trainer = Trainer(\n",
|
| 197 |
+
" model=model,\n",
|
| 198 |
+
" args=training_args,\n",
|
| 199 |
+
" train_dataset=tokenized_datasets[\"train\"],\n",
|
| 200 |
+
" eval_dataset=tokenized_datasets[\"test\"],\n",
|
| 201 |
+
" data_collator=data_collator,\n",
|
| 202 |
+
")"
|
| 203 |
+
]
|
| 204 |
+
},
|
| 205 |
+
{
|
| 206 |
+
"cell_type": "code",
|
| 207 |
+
"execution_count": null,
|
| 208 |
+
"id": "13fa4a99-ee7c-4d6a-853f-4be04a4ee43c",
|
| 209 |
+
"metadata": {},
|
| 210 |
+
"outputs": [],
|
| 211 |
+
"source": [
|
| 212 |
+
"trainer.train()\n",
|
| 213 |
+
"trainer.save_model(\"gene_eng_gpt2_v0\")"
|
| 214 |
+
]
|
| 215 |
+
},
|
| 216 |
+
{
|
| 217 |
+
"cell_type": "code",
|
| 218 |
+
"execution_count": null,
|
| 219 |
+
"id": "ca452721-3914-49be-a577-d4c257946578",
|
| 220 |
+
"metadata": {},
|
| 221 |
+
"outputs": [],
|
| 222 |
+
"source": [
|
| 223 |
+
"import math\n",
|
| 224 |
+
"eval_results = trainer.evaluate()\n",
|
| 225 |
+
"print(f\"Perplexity: {math.exp(eval_results['eval_loss']):.2f}\")"
|
| 226 |
+
]
|
| 227 |
+
},
|
| 228 |
+
{
|
| 229 |
+
"cell_type": "code",
|
| 230 |
+
"execution_count": null,
|
| 231 |
+
"id": "b7e7a455-0e08-4a75-87c1-0f909829b1c1",
|
| 232 |
+
"metadata": {},
|
| 233 |
+
"outputs": [],
|
| 234 |
+
"source": [
|
| 235 |
+
"#upload model\n",
|
| 236 |
+
"#model.push_to_hub(\"gene_eng_gpt2_v0\", organization=\"dnagpt\", use_auth_token=\"hf_*******\")"
|
| 237 |
+
]
|
| 238 |
+
}
|
| 239 |
+
],
|
| 240 |
+
"metadata": {
|
| 241 |
+
"kernelspec": {
|
| 242 |
+
"display_name": "Python 3 (ipykernel)",
|
| 243 |
+
"language": "python",
|
| 244 |
+
"name": "python3"
|
| 245 |
+
},
|
| 246 |
+
"language_info": {
|
| 247 |
+
"codemirror_mode": {
|
| 248 |
+
"name": "ipython",
|
| 249 |
+
"version": 3
|
| 250 |
+
},
|
| 251 |
+
"file_extension": ".py",
|
| 252 |
+
"mimetype": "text/x-python",
|
| 253 |
+
"name": "python",
|
| 254 |
+
"nbconvert_exporter": "python",
|
| 255 |
+
"pygments_lexer": "ipython3",
|
| 256 |
+
"version": "3.12.3"
|
| 257 |
+
}
|
| 258 |
+
},
|
| 259 |
+
"nbformat": 4,
|
| 260 |
+
"nbformat_minor": 5
|
| 261 |
+
}
|
02-gpt2_bert/.ipynb_checkpoints/dna_wordpiece_dict-checkpoint.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/1-dna-bpe.ipynb
ADDED
|
@@ -0,0 +1,528 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"id": "a9fffce5-83e3-4838-8335-acb2e3b50c35",
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"source": [
|
| 8 |
+
"# 2.1 DNA分词器构建"
|
| 9 |
+
]
|
| 10 |
+
},
|
| 11 |
+
{
|
| 12 |
+
"cell_type": "markdown",
|
| 13 |
+
"id": "f28b0950-37dc-4f78-ae6c-9fca33d513fc",
|
| 14 |
+
"metadata": {},
|
| 15 |
+
"source": [
|
| 16 |
+
"## **分词算法**\n",
|
| 17 |
+
"\n",
|
| 18 |
+
"### **什么是分词**\n",
|
| 19 |
+
"分词就是把一个文本序列,分成一个一个的token/词,对于英文这种天生带空格的语言,一般使用空格和标点分词就行了,而对于中文等语言,并没有特殊的符号来分词,因此,一般需要设计专门的分词算法,对于大模型而言,一般需要处理多种语言,因此,也需要专门的分词算法。\n",
|
| 20 |
+
"\n",
|
| 21 |
+
"在大模型(如 BERT、GPT 系列、T5 等)中,分词器(tokenizer)扮演着至关重要的角色。它负责将原始文本转换为模型可以处理的格式,即将文本分解成 token 序列,并将这些 token 映射到模型词汇表中的唯一 ID。分词器的选择和配置直接影响模型的性能和效果。以下是几种常见的分词器及其特点,特别关注它们在大型语言模型中的应用。\n",
|
| 22 |
+
"\n",
|
| 23 |
+
"### 1. **WordPiece 分词器**\n",
|
| 24 |
+
"\n",
|
| 25 |
+
"- **使用场景**:广泛应用于 BERT 及其变体。\n",
|
| 26 |
+
"- **工作原理**:基于频率统计,从语料库中学习最有效的词汇表。它根据子词(subword)在文本中的出现频率来决定如何分割单词。例如,“playing” 可能被分为 “play” 和 “##ing”,其中“##”表示该部分是前一个 token 的延续。\n",
|
| 27 |
+
"- **优点**:\n",
|
| 28 |
+
" - 处理未知词汇能力强,能够将未见过的词汇分解为已知的子词。\n",
|
| 29 |
+
" - 兼容性好,适合多种语言任务。\n",
|
| 30 |
+
"- **缺点**:\n",
|
| 31 |
+
" - 需要额外的标记(如 `##`)来指示子词,可能影响某些应用场景下的可读性。\n",
|
| 32 |
+
"\n",
|
| 33 |
+
"### 2. **Byte Pair Encoding (BPE)**\n",
|
| 34 |
+
"\n",
|
| 35 |
+
"- **使用场景**:广泛应用于 GPT 系列、RoBERTa、XLM-R 等模型。\n",
|
| 36 |
+
"- **工作原理**:通过迭代地合并最常见的字符对来构建词汇表。BPE 是一种无监督的学习方法,能够在不依赖于预先定义的词汇表的情况下进行分词。\n",
|
| 37 |
+
"- **优点**:\n",
|
| 38 |
+
" - 灵活性高,适应性强,尤其适用于多语言模型。\n",
|
| 39 |
+
" - 不需要特殊标记,生成的词汇表更简洁。\n",
|
| 40 |
+
"- **缺点**:\n",
|
| 41 |
+
" - 对于某些语言或领域特定的词汇,可能会产生较短的子词,导致信息丢失。\n",
|
| 42 |
+
"\n",
|
| 43 |
+
"### 3. **SentencePiece**\n",
|
| 44 |
+
"\n",
|
| 45 |
+
"- **使用场景**:常见于 T5、mBART 等多语言模型。\n",
|
| 46 |
+
"- **工作原理**:结合了 BPE 和 WordPiece 的优点,同时支持字符级和词汇级分词。它可以在没有空格的语言(如中文、日文)中表现良好。\n",
|
| 47 |
+
"- **优点**:\n",
|
| 48 |
+
" - 支持无空格语言,适合多语言处理。\n",
|
| 49 |
+
" - 学习速度快,适应性强。\n",
|
| 50 |
+
"- **缺点**:\n",
|
| 51 |
+
" - 对于某些特定领域的专业术语,可能需要额外的预处理步骤。\n",
|
| 52 |
+
"\n",
|
| 53 |
+
"### 4. **Character-Level Tokenizer**\n",
|
| 54 |
+
"\n",
|
| 55 |
+
"- **使用场景**:较少用于大型语言模型,但在某些特定任务(如拼写检查、手写识别)中有应用。\n",
|
| 56 |
+
"- **工作原理**:直接将每个字符视为一个 token。这种方式简单直接,但通常会导致较大的词汇表。\n",
|
| 57 |
+
"- **优点**:\n",
|
| 58 |
+
" - 简单易实现,不需要复杂的训练过程。\n",
|
| 59 |
+
" - 对于字符级别的任务非常有效。\n",
|
| 60 |
+
"- **缺点**:\n",
|
| 61 |
+
" - 词汇表较大,计算资源消耗较多。\n",
|
| 62 |
+
" - 捕捉上下文信息的能力较弱。\n",
|
| 63 |
+
"\n",
|
| 64 |
+
"### 5. **Unigram Language Model**\n",
|
| 65 |
+
"\n",
|
| 66 |
+
"- **使用场景**:主要用于 SentencePiece 中。\n",
|
| 67 |
+
"- **工作原理**:基于概率分布,选择最优的分词方案以最大化似然函数。这种方法类似于 BPE,但在构建词汇表时考虑了更多的统计信息。\n",
|
| 68 |
+
"- **优点**:\n",
|
| 69 |
+
" - 统计基础强,优化效果好。\n",
|
| 70 |
+
" - 适应性强,适用于多种语言和任务。\n",
|
| 71 |
+
"- **缺点**:\n",
|
| 72 |
+
" - 计算复杂度较高,训练时间较长。\n",
|
| 73 |
+
"\n",
|
| 74 |
+
"### 分词器的关键特性\n",
|
| 75 |
+
"\n",
|
| 76 |
+
"无论选择哪种分词器,以下几个关键特性都是设计和应用中需要考虑的:\n",
|
| 77 |
+
"\n",
|
| 78 |
+
"- **词汇表大小**:决定了模型所能识别的词汇量。较大的词汇表可以捕捉更多细节,但也增加了内存和计算需求。\n",
|
| 79 |
+
"- **处理未知词汇的能力**:好的分词器应该能够有效地处理未登录词(OOV, Out-Of-Vocabulary),将其分解为已知的子词。\n",
|
| 80 |
+
"- **多语言支持**:对于多语言模型,分词器应能处理不同语言的文本,尤其是那些没有明显分隔符的语言。\n",
|
| 81 |
+
"- **效率和速度**:分词器的执行速度直接影响整个数据处理管道的效率,尤其是在大规模数据集上。\n",
|
| 82 |
+
"- **兼容性和灵活性**:分词器应与目标模型架构兼容,并且能够灵活适应不同的任务需求。"
|
| 83 |
+
]
|
| 84 |
+
},
|
| 85 |
+
{
|
| 86 |
+
"cell_type": "markdown",
|
| 87 |
+
"id": "165e2594-277d-44d0-b582-77859a0bc0b2",
|
| 88 |
+
"metadata": {},
|
| 89 |
+
"source": [
|
| 90 |
+
"## DNA等生物序列分词\n",
|
| 91 |
+
"在生物信息学中,DNA 和蛋白质序列的处理与自然语言处理(NLP)有相似之处,但也有其独特性。为了提取这些生物序列的特征并用于机器学习或深度学习模型,通常需要将长序列分解成更小的片段(类似于 NLP 中的“分词”),以便更好地捕捉局部和全局特征。以下是几种常见的方法,用于对 DNA 和蛋白质序列进行“分词”,以提取有用的特征。\n",
|
| 92 |
+
"\n",
|
| 93 |
+
"### 1. **K-mer 分解**\n",
|
| 94 |
+
"\n",
|
| 95 |
+
"**定义**:K-mer 是指长度为 k 的连续子序列。例如,在 DNA 序列中,一个 3-mer 可能是 \"ATG\" 或 \"CGA\"。\n",
|
| 96 |
+
"\n",
|
| 97 |
+
"**应用**:\n",
|
| 98 |
+
"- **DNA 序列**:常用的 k 值范围从 3 到 6。较小的 k 值可以捕捉到更细粒度的信息,而较大的 k 值则有助于识别更长的模式。\n",
|
| 99 |
+
"- **蛋白质序列**:k 值通常较大,因为氨基酸的数量较多(20 种),较长的 k-mer 可以捕捉到重要的结构域或功能区域。\n",
|
| 100 |
+
"\n",
|
| 101 |
+
"**优点**:\n",
|
| 102 |
+
"- 简单且直观,易于实现。\n",
|
| 103 |
+
"- 可以捕捉到短序列中的局部特征。\n",
|
| 104 |
+
"\n",
|
| 105 |
+
"**缺点**:\n",
|
| 106 |
+
"- 对于非常长的序列,生成的 k-mer 数量会非常大,导致维度爆炸问题。\n",
|
| 107 |
+
"- 不同位置的 k-mer 之间缺乏上下文关系。"
|
| 108 |
+
]
|
| 109 |
+
},
|
| 110 |
+
{
|
| 111 |
+
"cell_type": "code",
|
| 112 |
+
"execution_count": 2,
|
| 113 |
+
"id": "29c390ef-2e9d-493e-9991-69ecb835b52b",
|
| 114 |
+
"metadata": {},
|
| 115 |
+
"outputs": [
|
| 116 |
+
{
|
| 117 |
+
"name": "stdout",
|
| 118 |
+
"output_type": "stream",
|
| 119 |
+
"text": [
|
| 120 |
+
"DNA 3-mers: ['ATG', 'TGC', 'GCG', 'CGT', 'GTA', 'TAC', 'ACG', 'CGT', 'GTA']\n",
|
| 121 |
+
"Protein 4-mers: ['MKQH', 'KQHK', 'QHKA', 'HKAM', 'KAMI', 'AMIV', 'MIVA', 'IVAL', 'VALI', 'ALIV', 'LIVL', 'IVLI', 'VLIT', 'LITA', 'ITAY']\n"
|
| 122 |
+
]
|
| 123 |
+
}
|
| 124 |
+
],
|
| 125 |
+
"source": [
|
| 126 |
+
"#示例代码(Python)\n",
|
| 127 |
+
"\n",
|
| 128 |
+
"def k_mer(seq, k):\n",
|
| 129 |
+
" return [seq[i:i+k] for i in range(len(seq) - k + 1)]\n",
|
| 130 |
+
"\n",
|
| 131 |
+
"dna_sequence = \"ATGCGTACGTA\"\n",
|
| 132 |
+
"protein_sequence = \"MKQHKAMIVALIVLITAY\"\n",
|
| 133 |
+
"\n",
|
| 134 |
+
"print(\"DNA 3-mers:\", k_mer(dna_sequence, 3))\n",
|
| 135 |
+
"print(\"Protein 4-mers:\", k_mer(protein_sequence, 4))"
|
| 136 |
+
]
|
| 137 |
+
},
|
| 138 |
+
{
|
| 139 |
+
"cell_type": "markdown",
|
| 140 |
+
"id": "7ced2bfb-bd42-425a-a3ad-54c9573609c5",
|
| 141 |
+
"metadata": {},
|
| 142 |
+
"source": [
|
| 143 |
+
"### 2. **滑动窗口**\n",
|
| 144 |
+
"\n",
|
| 145 |
+
"**定义**:滑动窗口方法通过设定一个固定大小的窗口沿着序列移动,并在每个位置提取窗口内的子序列。这与 K-mer 类似,但允许重叠。\n",
|
| 146 |
+
"\n",
|
| 147 |
+
"**应用**:\n",
|
| 148 |
+
"- **DNA 和蛋白质序列**:窗口大小可以根据具体任务调整,如基因预测、蛋白质结构预测等。\n",
|
| 149 |
+
"\n",
|
| 150 |
+
"**优点**:\n",
|
| 151 |
+
"- 提供了更多的灵活性,可以控制窗口的步长和大小。\n",
|
| 152 |
+
"- 有助于捕捉局部和全局特征。\n",
|
| 153 |
+
"\n",
|
| 154 |
+
"**缺点**:\n",
|
| 155 |
+
"- 计算复杂度较高,尤其是当窗口大小较大时。"
|
| 156 |
+
]
|
| 157 |
+
},
|
| 158 |
+
{
|
| 159 |
+
"cell_type": "code",
|
| 160 |
+
"execution_count": 4,
|
| 161 |
+
"id": "82cecf91-0076-4c12-b11c-b35120581ef9",
|
| 162 |
+
"metadata": {},
|
| 163 |
+
"outputs": [
|
| 164 |
+
{
|
| 165 |
+
"name": "stdout",
|
| 166 |
+
"output_type": "stream",
|
| 167 |
+
"text": [
|
| 168 |
+
"Sliding window (DNA, size=3, step=1): ['ATG', 'TGC', 'GCG', 'CGT', 'GTA', 'TAC', 'ACG', 'CGT', 'GTA']\n",
|
| 169 |
+
"Sliding window (Protein, size=4, step=2): ['MKQH', 'QHKA', 'KAMI', 'MIVA', 'VALI', 'LIVL', 'VLIT', 'ITAY']\n"
|
| 170 |
+
]
|
| 171 |
+
}
|
| 172 |
+
],
|
| 173 |
+
"source": [
|
| 174 |
+
"def sliding_window(seq, window_size, step=1):\n",
|
| 175 |
+
" return [seq[i:i+window_size] for i in range(0, len(seq) - window_size + 1, step)]\n",
|
| 176 |
+
"\n",
|
| 177 |
+
"dna_sequence = \"ATGCGTACGTA\"\n",
|
| 178 |
+
"protein_sequence = \"MKQHKAMIVALIVLITAY\"\n",
|
| 179 |
+
"\n",
|
| 180 |
+
"print(\"Sliding window (DNA, size=3, step=1):\", sliding_window(dna_sequence, 3))\n",
|
| 181 |
+
"print(\"Sliding window (Protein, size=4, step=2):\", sliding_window(protein_sequence, 4, step=2))"
|
| 182 |
+
]
|
| 183 |
+
},
|
| 184 |
+
{
|
| 185 |
+
"cell_type": "markdown",
|
| 186 |
+
"id": "c33ab920-b451-4846-93d4-20da5a4e1001",
|
| 187 |
+
"metadata": {},
|
| 188 |
+
"source": [
|
| 189 |
+
"### 3. **词表分词和嵌入式表示**\n",
|
| 190 |
+
"\n",
|
| 191 |
+
"**定义**:使用预训练的嵌入模型(如 Word2Vec、BERT 等)来将每个 token 映射到高维向量空间中。对于生物序列,可以使用专门设计的嵌入模型,如 ProtTrans、ESM 等。\n",
|
| 192 |
+
"\n",
|
| 193 |
+
"**应用**:\n",
|
| 194 |
+
"- **DNA 和蛋白质序列**:嵌入模型可以捕捉到序列中的语义信息和上下文依赖关系。\n",
|
| 195 |
+
"\n",
|
| 196 |
+
"**优点**:\n",
|
| 197 |
+
"- 捕捉到丰富的语义信息,适合复杂的下游任务。\n",
|
| 198 |
+
"- 可以利用大规模预训练模型的优势。\n",
|
| 199 |
+
"\n",
|
| 200 |
+
"**缺点**:\n",
|
| 201 |
+
"- 需要大量的计算资源来进行预训练。\n",
|
| 202 |
+
"- 模型复杂度较高,解释性较差。"
|
| 203 |
+
]
|
| 204 |
+
},
|
| 205 |
+
{
|
| 206 |
+
"cell_type": "code",
|
| 207 |
+
"execution_count": 5,
|
| 208 |
+
"id": "02bf2af0-6077-4b27-8822-f1c3f22914fa",
|
| 209 |
+
"metadata": {},
|
| 210 |
+
"outputs": [],
|
| 211 |
+
"source": [
|
| 212 |
+
"import subprocess\n",
|
| 213 |
+
"import os\n",
|
| 214 |
+
"# 设置环境变量, autodl一般区域\n",
|
| 215 |
+
"result = subprocess.run('bash -c \"source /etc/network_turbo && env | grep proxy\"', shell=True, capture_output=True, text=True)\n",
|
| 216 |
+
"output = result.stdout\n",
|
| 217 |
+
"for line in output.splitlines():\n",
|
| 218 |
+
" if '=' in line:\n",
|
| 219 |
+
" var, value = line.split('=', 1)\n",
|
| 220 |
+
" os.environ[var] = value\n",
|
| 221 |
+
"\n",
|
| 222 |
+
"\"\"\"\n",
|
| 223 |
+
"import os\n",
|
| 224 |
+
"\n",
|
| 225 |
+
"# 设置环境变量, autodl专区 其他idc\n",
|
| 226 |
+
"os.environ['HF_ENDPOINT'] = 'https://hf-mirror.com'\n",
|
| 227 |
+
"\n",
|
| 228 |
+
"# 打印环境变量以确认设置成功\n",
|
| 229 |
+
"print(os.environ.get('HF_ENDPOINT'))\n",
|
| 230 |
+
"\"\"\""
|
| 231 |
+
]
|
| 232 |
+
},
|
| 233 |
+
{
|
| 234 |
+
"cell_type": "code",
|
| 235 |
+
"execution_count": 15,
|
| 236 |
+
"id": "d43b60ee-67f2-4d06-95ea-966c01084fc4",
|
| 237 |
+
"metadata": {
|
| 238 |
+
"scrolled": true
|
| 239 |
+
},
|
| 240 |
+
"outputs": [
|
| 241 |
+
{
|
| 242 |
+
"name": "stderr",
|
| 243 |
+
"output_type": "stream",
|
| 244 |
+
"text": [
|
| 245 |
+
"huggingface/tokenizers: The current process just got forked, after parallelism has already been used. Disabling parallelism to avoid deadlocks...\n",
|
| 246 |
+
"To disable this warning, you can either:\n",
|
| 247 |
+
"\t- Avoid using `tokenizers` before the fork if possible\n",
|
| 248 |
+
"\t- Explicitly set the environment variable TOKENIZERS_PARALLELISM=(true | false)\n"
|
| 249 |
+
]
|
| 250 |
+
},
|
| 251 |
+
{
|
| 252 |
+
"name": "stdout",
|
| 253 |
+
"output_type": "stream",
|
| 254 |
+
"text": [
|
| 255 |
+
"['ATGCG', 'TACG', 'T', 'A']\n",
|
| 256 |
+
"Embeddings shape: torch.Size([1, 4, 768])\n"
|
| 257 |
+
]
|
| 258 |
+
}
|
| 259 |
+
],
|
| 260 |
+
"source": [
|
| 261 |
+
"from transformers import AutoTokenizer, AutoModel\n",
|
| 262 |
+
"import torch\n",
|
| 263 |
+
"\n",
|
| 264 |
+
"# 加载预训练的蛋白质嵌入模型\n",
|
| 265 |
+
"tokenizer = AutoTokenizer.from_pretrained(\"dnagpt/gpt_dna_v0\")\n",
|
| 266 |
+
"model = AutoModel.from_pretrained(\"dnagpt/gpt_dna_v0\")\n",
|
| 267 |
+
"\n",
|
| 268 |
+
"dna_sequence = \"ATGCGTACGTA\"\n",
|
| 269 |
+
"print(tokenizer.tokenize(dna_sequence))\n",
|
| 270 |
+
"\n",
|
| 271 |
+
"# 编码序列\n",
|
| 272 |
+
"inputs = tokenizer(dna_sequence, return_tensors=\"pt\")\n",
|
| 273 |
+
"\n",
|
| 274 |
+
"# 获取嵌入\n",
|
| 275 |
+
"with torch.no_grad():\n",
|
| 276 |
+
" outputs = model(**inputs)\n",
|
| 277 |
+
" embeddings = outputs.last_hidden_state\n",
|
| 278 |
+
"\n",
|
| 279 |
+
"print(\"Embeddings shape:\", embeddings.shape)"
|
| 280 |
+
]
|
| 281 |
+
},
|
| 282 |
+
{
|
| 283 |
+
"cell_type": "markdown",
|
| 284 |
+
"id": "c24f10dc-1117-4493-9333-5ed6d898f44a",
|
| 285 |
+
"metadata": {},
|
| 286 |
+
"source": [
|
| 287 |
+
"### 训练DNA BPE分词器\n",
|
| 288 |
+
"\n",
|
| 289 |
+
"以上方法展示了如何对 DNA 和蛋白质序列进行“分词”,以提取有用的特征。选择哪种方法取决于具体的任务需求和数据特性。对于简单的分类或回归任务,K-mer 分解或滑动窗口可能是足够的;而对于更复杂的任务,如序列标注或结构预测,基于词汇表的方法或嵌入表示可能会提供更好的性能。\n",
|
| 290 |
+
"\n",
|
| 291 |
+
"目前大部分生物序列大模型的论文中,使用最多的依然是传统的K-mer,但一些SOTA的论文则以BEP为主。而BEP分词也是目前GPT、llama等主流自然语言大模型使用的基础分词器。\n",
|
| 292 |
+
"\n",
|
| 293 |
+
"因此,我们也演示下从头训练一个DNA BPE分词器的方法。\n",
|
| 294 |
+
"\n",
|
| 295 |
+
"我们首先看下GPT2模型,默认的分词器,对DNA序列分词的结果:"
|
| 296 |
+
]
|
| 297 |
+
},
|
| 298 |
+
{
|
| 299 |
+
"cell_type": "code",
|
| 300 |
+
"execution_count": 10,
|
| 301 |
+
"id": "43f1eb8b-1cc2-4ab5-aa8e-2a63132be98c",
|
| 302 |
+
"metadata": {},
|
| 303 |
+
"outputs": [],
|
| 304 |
+
"source": [
|
| 305 |
+
"from tokenizers import (\n",
|
| 306 |
+
" decoders,\n",
|
| 307 |
+
" models,\n",
|
| 308 |
+
" normalizers,\n",
|
| 309 |
+
" pre_tokenizers,\n",
|
| 310 |
+
" processors,\n",
|
| 311 |
+
" trainers,\n",
|
| 312 |
+
" Tokenizer,\n",
|
| 313 |
+
")\n",
|
| 314 |
+
"from transformers import AutoTokenizer"
|
| 315 |
+
]
|
| 316 |
+
},
|
| 317 |
+
{
|
| 318 |
+
"cell_type": "code",
|
| 319 |
+
"execution_count": 15,
|
| 320 |
+
"id": "27e88f7b-1399-418b-9b91-f970762fac0c",
|
| 321 |
+
"metadata": {},
|
| 322 |
+
"outputs": [],
|
| 323 |
+
"source": [
|
| 324 |
+
"gpt2_tokenizer = AutoTokenizer.from_pretrained('gpt2')\n",
|
| 325 |
+
"gpt2_tokenizer.pad_token = gpt2_tokenizer.eos_token"
|
| 326 |
+
]
|
| 327 |
+
},
|
| 328 |
+
{
|
| 329 |
+
"cell_type": "code",
|
| 330 |
+
"execution_count": 16,
|
| 331 |
+
"id": "4b015db7-63ba-4909-b02f-07634b3d5584",
|
| 332 |
+
"metadata": {},
|
| 333 |
+
"outputs": [
|
| 334 |
+
{
|
| 335 |
+
"data": {
|
| 336 |
+
"text/plain": [
|
| 337 |
+
"['T', 'GG', 'C', 'GT', 'GA', 'AC', 'CC', 'GG', 'G', 'AT', 'C', 'GG', 'G']"
|
| 338 |
+
]
|
| 339 |
+
},
|
| 340 |
+
"execution_count": 16,
|
| 341 |
+
"metadata": {},
|
| 342 |
+
"output_type": "execute_result"
|
| 343 |
+
}
|
| 344 |
+
],
|
| 345 |
+
"source": [
|
| 346 |
+
"gpt2_tokenizer.tokenize(\"TGGCGTGAACCCGGGATCGGG\")"
|
| 347 |
+
]
|
| 348 |
+
},
|
| 349 |
+
{
|
| 350 |
+
"cell_type": "markdown",
|
| 351 |
+
"id": "a246fbc9-9e29-4b63-bdf7-f80635d06d1e",
|
| 352 |
+
"metadata": {},
|
| 353 |
+
"source": [
|
| 354 |
+
"可以看到,gpt2模型因为是以英文为主的BPE分词模型,分解的都是1到2个字母的结果,这样显然很难充分表达生物语义,因此,我们使用DNA序列来训练1个BPE分词器,代码也非常简单:"
|
| 355 |
+
]
|
| 356 |
+
},
|
| 357 |
+
{
|
| 358 |
+
"cell_type": "code",
|
| 359 |
+
"execution_count": 2,
|
| 360 |
+
"id": "8357a695-1c29-4b5c-8099-d2e337189410",
|
| 361 |
+
"metadata": {},
|
| 362 |
+
"outputs": [],
|
| 363 |
+
"source": [
|
| 364 |
+
"tokenizer = Tokenizer(models.BPE())\n",
|
| 365 |
+
"tokenizer.pre_tokenizer = pre_tokenizers.ByteLevel(add_prefix_space=False, use_regex=False) #use_regex=False,空格当成一般字符串\n",
|
| 366 |
+
"trainer = trainers.BpeTrainer(vocab_size=30000, special_tokens=[\"<|endoftext|>\"]) #3w words"
|
| 367 |
+
]
|
| 368 |
+
},
|
| 369 |
+
{
|
| 370 |
+
"cell_type": "code",
|
| 371 |
+
"execution_count": 3,
|
| 372 |
+
"id": "32c95888-1498-45cf-8453-421219cc7d45",
|
| 373 |
+
"metadata": {},
|
| 374 |
+
"outputs": [
|
| 375 |
+
{
|
| 376 |
+
"name": "stdout",
|
| 377 |
+
"output_type": "stream",
|
| 378 |
+
"text": [
|
| 379 |
+
"\n",
|
| 380 |
+
"\n",
|
| 381 |
+
"\n"
|
| 382 |
+
]
|
| 383 |
+
}
|
| 384 |
+
],
|
| 385 |
+
"source": [
|
| 386 |
+
"tokenizer.train([\"../01-data_env/data/dna_1g.txt\"], trainer=trainer) #all file list, take 10-20 min"
|
| 387 |
+
]
|
| 388 |
+
},
|
| 389 |
+
{
|
| 390 |
+
"cell_type": "code",
|
| 391 |
+
"execution_count": 4,
|
| 392 |
+
"id": "5ffdd717-72ed-4a37-bafc-b4a0f61f8ff1",
|
| 393 |
+
"metadata": {},
|
| 394 |
+
"outputs": [
|
| 395 |
+
{
|
| 396 |
+
"name": "stdout",
|
| 397 |
+
"output_type": "stream",
|
| 398 |
+
"text": [
|
| 399 |
+
"['TG', 'GCGTGAA', 'CCCGG', 'GATCGG', 'G']\n"
|
| 400 |
+
]
|
| 401 |
+
}
|
| 402 |
+
],
|
| 403 |
+
"source": [
|
| 404 |
+
"encoding = tokenizer.encode(\"TGGCGTGAACCCGGGATCGGG\")\n",
|
| 405 |
+
"print(encoding.tokens)"
|
| 406 |
+
]
|
| 407 |
+
},
|
| 408 |
+
{
|
| 409 |
+
"cell_type": "markdown",
|
| 410 |
+
"id": "a96e7838-6c23-4446-bf86-b098cd93214a",
|
| 411 |
+
"metadata": {},
|
| 412 |
+
"source": [
|
| 413 |
+
"可以看到,以DNA数据训练的分词器,分词效果明显要好的多,各种长度的词都有。"
|
| 414 |
+
]
|
| 415 |
+
},
|
| 416 |
+
{
|
| 417 |
+
"cell_type": "code",
|
| 418 |
+
"execution_count": 5,
|
| 419 |
+
"id": "f1d757c1-702b-4147-9207-471f422f67b2",
|
| 420 |
+
"metadata": {},
|
| 421 |
+
"outputs": [],
|
| 422 |
+
"source": [
|
| 423 |
+
"tokenizer.save(\"dna_bpe_dict.json\")"
|
| 424 |
+
]
|
| 425 |
+
},
|
| 426 |
+
{
|
| 427 |
+
"cell_type": "code",
|
| 428 |
+
"execution_count": 6,
|
| 429 |
+
"id": "caf8ecea-359e-487b-b456-fab546b9da0d",
|
| 430 |
+
"metadata": {},
|
| 431 |
+
"outputs": [],
|
| 432 |
+
"source": [
|
| 433 |
+
"#然后我们可以使用from_file() 方法从该文件里重新加载 Tokenizer 对象:\n",
|
| 434 |
+
"new_tokenizer = Tokenizer.from_file(\"dna_bpe_dict.json\")"
|
| 435 |
+
]
|
| 436 |
+
},
|
| 437 |
+
{
|
| 438 |
+
"cell_type": "code",
|
| 439 |
+
"execution_count": 7,
|
| 440 |
+
"id": "8ec6f045-bc30-4012-8027-a879df8def3a",
|
| 441 |
+
"metadata": {},
|
| 442 |
+
"outputs": [
|
| 443 |
+
{
|
| 444 |
+
"data": {
|
| 445 |
+
"text/plain": [
|
| 446 |
+
"('dna_bpe_dict/tokenizer_config.json',\n",
|
| 447 |
+
" 'dna_bpe_dict/special_tokens_map.json',\n",
|
| 448 |
+
" 'dna_bpe_dict/vocab.json',\n",
|
| 449 |
+
" 'dna_bpe_dict/merges.txt',\n",
|
| 450 |
+
" 'dna_bpe_dict/added_tokens.json',\n",
|
| 451 |
+
" 'dna_bpe_dict/tokenizer.json')"
|
| 452 |
+
]
|
| 453 |
+
},
|
| 454 |
+
"execution_count": 7,
|
| 455 |
+
"metadata": {},
|
| 456 |
+
"output_type": "execute_result"
|
| 457 |
+
}
|
| 458 |
+
],
|
| 459 |
+
"source": [
|
| 460 |
+
"#要在 🤗 Transformers 中使用这个标记器,我们必须将它包裹在一个 PreTrainedTokenizerFast 类中\n",
|
| 461 |
+
"from transformers import GPT2TokenizerFast\n",
|
| 462 |
+
"dna_tokenizer = GPT2TokenizerFast(tokenizer_object=new_tokenizer)\n",
|
| 463 |
+
"dna_tokenizer.save_pretrained(\"dna_bpe_dict\")\n",
|
| 464 |
+
"#dna_tokenizer.push_to_hub(\"dna_bpe_dict_1g\", organization=\"dnagpt\", use_auth_token=\"hf_*****\") # push to huggingface"
|
| 465 |
+
]
|
| 466 |
+
},
|
| 467 |
+
{
|
| 468 |
+
"cell_type": "code",
|
| 469 |
+
"execution_count": 11,
|
| 470 |
+
"id": "f84506d8-6208-4027-aad7-2b68a1bc16d6",
|
| 471 |
+
"metadata": {},
|
| 472 |
+
"outputs": [],
|
| 473 |
+
"source": [
|
| 474 |
+
"tokenizer_new = AutoTokenizer.from_pretrained('dna_bpe_dict')"
|
| 475 |
+
]
|
| 476 |
+
},
|
| 477 |
+
{
|
| 478 |
+
"cell_type": "code",
|
| 479 |
+
"execution_count": 12,
|
| 480 |
+
"id": "d40d4d53-6fed-445c-afb5-c0346ab854c8",
|
| 481 |
+
"metadata": {},
|
| 482 |
+
"outputs": [
|
| 483 |
+
{
|
| 484 |
+
"data": {
|
| 485 |
+
"text/plain": [
|
| 486 |
+
"['TG', 'GCGTGAA', 'CCCGG', 'GATCGG', 'G']"
|
| 487 |
+
]
|
| 488 |
+
},
|
| 489 |
+
"execution_count": 12,
|
| 490 |
+
"metadata": {},
|
| 491 |
+
"output_type": "execute_result"
|
| 492 |
+
}
|
| 493 |
+
],
|
| 494 |
+
"source": [
|
| 495 |
+
"tokenizer_new.tokenize(\"TGGCGTGAACCCGGGATCGGG\")"
|
| 496 |
+
]
|
| 497 |
+
},
|
| 498 |
+
{
|
| 499 |
+
"cell_type": "code",
|
| 500 |
+
"execution_count": null,
|
| 501 |
+
"id": "640302f6-f740-41a4-ae92-ca4c43d97493",
|
| 502 |
+
"metadata": {},
|
| 503 |
+
"outputs": [],
|
| 504 |
+
"source": []
|
| 505 |
+
}
|
| 506 |
+
],
|
| 507 |
+
"metadata": {
|
| 508 |
+
"kernelspec": {
|
| 509 |
+
"display_name": "Python 3 (ipykernel)",
|
| 510 |
+
"language": "python",
|
| 511 |
+
"name": "python3"
|
| 512 |
+
},
|
| 513 |
+
"language_info": {
|
| 514 |
+
"codemirror_mode": {
|
| 515 |
+
"name": "ipython",
|
| 516 |
+
"version": 3
|
| 517 |
+
},
|
| 518 |
+
"file_extension": ".py",
|
| 519 |
+
"mimetype": "text/x-python",
|
| 520 |
+
"name": "python",
|
| 521 |
+
"nbconvert_exporter": "python",
|
| 522 |
+
"pygments_lexer": "ipython3",
|
| 523 |
+
"version": "3.12.3"
|
| 524 |
+
}
|
| 525 |
+
},
|
| 526 |
+
"nbformat": 4,
|
| 527 |
+
"nbformat_minor": 5
|
| 528 |
+
}
|
02-gpt2_bert/2-dna-gpt.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/3-dna-bert.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/4-gene-feature.ipynb
ADDED
|
@@ -0,0 +1,489 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"id": "b1b37ca8-25a3-440c-9b68-7f72ce670ade",
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"source": [
|
| 8 |
+
"# 2.4 基因大模型的生物序列特征提取"
|
| 9 |
+
]
|
| 10 |
+
},
|
| 11 |
+
{
|
| 12 |
+
"cell_type": "markdown",
|
| 13 |
+
"id": "d3d04215-2b6c-41fb-92a4-90c82d322ba4",
|
| 14 |
+
"metadata": {},
|
| 15 |
+
"source": [
|
| 16 |
+
"使用 GPT-2 模型获取文本的特征向量是一个常见的需求,尤其是在进行文本分类、相似度计算或其他下游任务时。Hugging Face 的 transformers 库提供了简单易用的接口来实现这一点。以下是详细的步骤和代码示例,帮助你从 GPT-2 模型中提取文本的特征向量。"
|
| 17 |
+
]
|
| 18 |
+
},
|
| 19 |
+
{
|
| 20 |
+
"cell_type": "markdown",
|
| 21 |
+
"id": "3ff5b7c6-e57c-4839-8510-f764154faa65",
|
| 22 |
+
"metadata": {},
|
| 23 |
+
"source": [
|
| 24 |
+
"使用 GPT-2 模型获取文本的特征向量是一个常见的需求,尤其是在进行文本分类、相似度计算或其他下游任务时。Hugging Face 的 `transformers` 库提供了简单易用的接口来实现这一点。以下是详细的步骤和代码示例,帮助你从 GPT-2 模型中提取文本的特征向量。\n",
|
| 25 |
+
"\n",
|
| 26 |
+
"### 方法 1: 使用隐藏状态(Hidden States)\n",
|
| 27 |
+
"\n",
|
| 28 |
+
"GPT-2 是一个基于 Transformer 的语言模型,它在每一层都有隐藏状态(hidden states),这些隐藏状态可以作为文本的特征表示。你可以选择最后一层的隐藏状态作为最终的特征向量,或者对多层的隐藏状态进行平均或拼接。\n",
|
| 29 |
+
"\n",
|
| 30 |
+
"\n",
|
| 31 |
+
"### 方法 2: 使用池化策略\n",
|
| 32 |
+
"\n",
|
| 33 |
+
"另一种方法是通过对所有 token 的隐藏状态进行池化操作来获得句子级别的特征向量。常见的池化方法包括:\n",
|
| 34 |
+
"\n",
|
| 35 |
+
"- **均值池化**(Mean Pooling):对所有 token 的隐藏状态求平均。\n",
|
| 36 |
+
"- **最大池化**(Max Pooling):对每个维度取最大值。"
|
| 37 |
+
]
|
| 38 |
+
},
|
| 39 |
+
{
|
| 40 |
+
"cell_type": "code",
|
| 41 |
+
"execution_count": 43,
|
| 42 |
+
"id": "e7fe053b-d6da-488a-9c62-24e4b40a992d",
|
| 43 |
+
"metadata": {},
|
| 44 |
+
"outputs": [
|
| 45 |
+
{
|
| 46 |
+
"name": "stdout",
|
| 47 |
+
"output_type": "stream",
|
| 48 |
+
"text": [
|
| 49 |
+
"{'input_ids': tensor([[ 1, 191, 29, 753, 1241, 2104, 12297, 357, 85, 4395,\n",
|
| 50 |
+
" 26392, 16]]), 'attention_mask': tensor([[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]])}\n",
|
| 51 |
+
"torch.Size([768])\n",
|
| 52 |
+
"torch.Size([768])\n",
|
| 53 |
+
"torch.Size([768])\n"
|
| 54 |
+
]
|
| 55 |
+
}
|
| 56 |
+
],
|
| 57 |
+
"source": [
|
| 58 |
+
"from transformers import AutoTokenizer, AutoModel\n",
|
| 59 |
+
"tokenizer = AutoTokenizer.from_pretrained('dna_bpe_dict')\n",
|
| 60 |
+
"tokenizer.tokenize(\"GAGCACATTCGCCTGCGTGCGCACTCACACACACGTTCAAAAAGAGTCCATTCGATTCTGGCAGTAG\")\n",
|
| 61 |
+
"#result: [G','AGCAC','ATTCGCC',....]\n",
|
| 62 |
+
"\n",
|
| 63 |
+
"model = AutoModel.from_pretrained('dna_gpt2_v0')\n",
|
| 64 |
+
"import torch\n",
|
| 65 |
+
"dna = \"ACGTAGCATCGGATCTATCTATCGACACTTGGTTATCGATCTACGAGCATCTCGTTAGC\"\n",
|
| 66 |
+
"inputs = tokenizer(dna, return_tensors = 'pt')\n",
|
| 67 |
+
"print(inputs)\n",
|
| 68 |
+
"\n",
|
| 69 |
+
"outputs = model(inputs[\"input_ids\"])\n",
|
| 70 |
+
"#outputs = model(**inputs)\n",
|
| 71 |
+
"\n",
|
| 72 |
+
"hidden_states = outputs.last_hidden_state # [1, sequence_length, 768] outputs.last_hidden_state or outputs[0]\n",
|
| 73 |
+
"\n",
|
| 74 |
+
"# embedding with mean pooling\n",
|
| 75 |
+
"embedding_mean = torch.mean(hidden_states[0], dim=0)\n",
|
| 76 |
+
"print(embedding_mean.shape) # expect to be 768\n",
|
| 77 |
+
"\n",
|
| 78 |
+
"# embedding with max pooling\n",
|
| 79 |
+
"embedding_max = torch.max(hidden_states[0], dim=0)[0]\n",
|
| 80 |
+
"print(embedding_max.shape) # expect to be 768\n",
|
| 81 |
+
"\n",
|
| 82 |
+
"# embedding with first token\n",
|
| 83 |
+
"embedding_first_token = hidden_states[0][0]\n",
|
| 84 |
+
"print(embedding_first_token.shape) # expect to be 768"
|
| 85 |
+
]
|
| 86 |
+
},
|
| 87 |
+
{
|
| 88 |
+
"cell_type": "code",
|
| 89 |
+
"execution_count": 44,
|
| 90 |
+
"id": "a1f2b545-283a-4613-a953-beb82f427826",
|
| 91 |
+
"metadata": {},
|
| 92 |
+
"outputs": [
|
| 93 |
+
{
|
| 94 |
+
"name": "stderr",
|
| 95 |
+
"output_type": "stream",
|
| 96 |
+
"text": [
|
| 97 |
+
"Some weights of BertModel were not initialized from the model checkpoint at dna_bert_v0 and are newly initialized: ['bert.pooler.dense.bias', 'bert.pooler.dense.weight']\n",
|
| 98 |
+
"You should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.\n"
|
| 99 |
+
]
|
| 100 |
+
},
|
| 101 |
+
{
|
| 102 |
+
"name": "stdout",
|
| 103 |
+
"output_type": "stream",
|
| 104 |
+
"text": [
|
| 105 |
+
"{'input_ids': tensor([[ 6, 200, 16057, 10, 1256, 2123, 12294, 366, 13138, 7826,\n",
|
| 106 |
+
" 82, 25]]), 'token_type_ids': tensor([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]]), 'attention_mask': tensor([[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]])}\n",
|
| 107 |
+
"torch.Size([768])\n",
|
| 108 |
+
"torch.Size([768])\n",
|
| 109 |
+
"torch.Size([768])\n"
|
| 110 |
+
]
|
| 111 |
+
}
|
| 112 |
+
],
|
| 113 |
+
"source": [
|
| 114 |
+
"from transformers import AutoTokenizer, AutoModel\n",
|
| 115 |
+
"import torch\n",
|
| 116 |
+
"\n",
|
| 117 |
+
"tokenizer = AutoTokenizer.from_pretrained('dna_wordpiece_dict')\n",
|
| 118 |
+
"tokenizer.tokenize(\"GAGCACATTCGCCTGCGTGCGCACTCACACACACGTTCAAAAAGAGTCCATTCGATTCTGGCAGTAG\")\n",
|
| 119 |
+
"#result: [G','AGCAC','ATTCGCC',....]\n",
|
| 120 |
+
"\n",
|
| 121 |
+
"model = AutoModel.from_pretrained('dna_bert_v0')\n",
|
| 122 |
+
"dna = \"ACGTAGCATCGGATCTATCTATCGACACTTGGTTATCGATCTACGAGCATCTCGTTAGC\"\n",
|
| 123 |
+
"inputs = tokenizer(dna, return_tensors = 'pt')\n",
|
| 124 |
+
"print(inputs)\n",
|
| 125 |
+
"\n",
|
| 126 |
+
"outputs = model(inputs[\"input_ids\"])\n",
|
| 127 |
+
"#outputs = model(**inputs)\n",
|
| 128 |
+
"\n",
|
| 129 |
+
"hidden_states = outputs.last_hidden_state # [1, sequence_length, 768] outputs.last_hidden_state or outputs[0]\n",
|
| 130 |
+
"\n",
|
| 131 |
+
"# embedding with mean pooling\n",
|
| 132 |
+
"embedding_mean = torch.mean(hidden_states[0], dim=0)\n",
|
| 133 |
+
"print(embedding_mean.shape) # expect to be 768\n",
|
| 134 |
+
"\n",
|
| 135 |
+
"# embedding with max pooling\n",
|
| 136 |
+
"embedding_max = torch.max(hidden_states[0], dim=0)[0]\n",
|
| 137 |
+
"print(embedding_max.shape) # expect to be 768\n",
|
| 138 |
+
"\n",
|
| 139 |
+
"# embedding with first token\n",
|
| 140 |
+
"embedding_first_token = hidden_states[0][0]\n",
|
| 141 |
+
"print(embedding_first_token.shape) # expect to be 768"
|
| 142 |
+
]
|
| 143 |
+
},
|
| 144 |
+
{
|
| 145 |
+
"cell_type": "markdown",
|
| 146 |
+
"id": "56761874-9af7-4b90-aa8b-131e5b8c69b6",
|
| 147 |
+
"metadata": {},
|
| 148 |
+
"source": [
|
| 149 |
+
"## 特征提取并分类\n",
|
| 150 |
+
"\n",
|
| 151 |
+
"我们使用第一章中的\"dnagpt/dna_core_promoter\"数据集,演示下使用我们训练的DNA GPT2或者DNA bert模型,提取序列特征,然使用最基础的逻辑回归分类方法,对序列进行分类。"
|
| 152 |
+
]
|
| 153 |
+
},
|
| 154 |
+
{
|
| 155 |
+
"cell_type": "code",
|
| 156 |
+
"execution_count": 45,
|
| 157 |
+
"id": "f1ca177c-a80f-48a1-b2f9-16c13b3350db",
|
| 158 |
+
"metadata": {},
|
| 159 |
+
"outputs": [
|
| 160 |
+
{
|
| 161 |
+
"data": {
|
| 162 |
+
"text/plain": [
|
| 163 |
+
"\"\\nimport os\\n\\n# 设置环境变量, autodl专区 其他idc\\nos.environ['HF_ENDPOINT'] = 'https://hf-mirror.com'\\n\\n# 打印环境变量以确认设置成功\\nprint(os.environ.get('HF_ENDPOINT'))\\n\""
|
| 164 |
+
]
|
| 165 |
+
},
|
| 166 |
+
"execution_count": 45,
|
| 167 |
+
"metadata": {},
|
| 168 |
+
"output_type": "execute_result"
|
| 169 |
+
}
|
| 170 |
+
],
|
| 171 |
+
"source": [
|
| 172 |
+
"import subprocess\n",
|
| 173 |
+
"import os\n",
|
| 174 |
+
"# 设置环境变量, autodl一般区域\n",
|
| 175 |
+
"result = subprocess.run('bash -c \"source /etc/network_turbo && env | grep proxy\"', shell=True, capture_output=True, text=True)\n",
|
| 176 |
+
"output = result.stdout\n",
|
| 177 |
+
"for line in output.splitlines():\n",
|
| 178 |
+
" if '=' in line:\n",
|
| 179 |
+
" var, value = line.split('=', 1)\n",
|
| 180 |
+
" os.environ[var] = value\n",
|
| 181 |
+
"\n",
|
| 182 |
+
"#或者\n",
|
| 183 |
+
"\"\"\"\n",
|
| 184 |
+
"import os\n",
|
| 185 |
+
"\n",
|
| 186 |
+
"# 设置环境变量, autodl专区 其他idc\n",
|
| 187 |
+
"os.environ['HF_ENDPOINT'] = 'https://hf-mirror.com'\n",
|
| 188 |
+
"\n",
|
| 189 |
+
"# 打印环境变量以确认设置成功\n",
|
| 190 |
+
"print(os.environ.get('HF_ENDPOINT'))\n",
|
| 191 |
+
"\"\"\""
|
| 192 |
+
]
|
| 193 |
+
},
|
| 194 |
+
{
|
| 195 |
+
"cell_type": "code",
|
| 196 |
+
"execution_count": 46,
|
| 197 |
+
"id": "2295739c-e80a-47be-9400-88bfab4b0bb6",
|
| 198 |
+
"metadata": {},
|
| 199 |
+
"outputs": [
|
| 200 |
+
{
|
| 201 |
+
"data": {
|
| 202 |
+
"text/plain": [
|
| 203 |
+
"DatasetDict({\n",
|
| 204 |
+
" train: Dataset({\n",
|
| 205 |
+
" features: ['sequence', 'label'],\n",
|
| 206 |
+
" num_rows: 59196\n",
|
| 207 |
+
" })\n",
|
| 208 |
+
"})"
|
| 209 |
+
]
|
| 210 |
+
},
|
| 211 |
+
"execution_count": 46,
|
| 212 |
+
"metadata": {},
|
| 213 |
+
"output_type": "execute_result"
|
| 214 |
+
}
|
| 215 |
+
],
|
| 216 |
+
"source": [
|
| 217 |
+
"from datasets import load_dataset\n",
|
| 218 |
+
"dna_data = load_dataset(\"dnagpt/dna_core_promoter\")\n",
|
| 219 |
+
"dna_data"
|
| 220 |
+
]
|
| 221 |
+
},
|
| 222 |
+
{
|
| 223 |
+
"cell_type": "markdown",
|
| 224 |
+
"id": "c804bced-f151-43a7-8a95-156db358da3e",
|
| 225 |
+
"metadata": {},
|
| 226 |
+
"source": [
|
| 227 |
+
"这里,我们不需要关注这个数据的具体生物学含义,只需知道sequence是具体的DNA序列,label是分类标签,有两个类别0和1即可"
|
| 228 |
+
]
|
| 229 |
+
},
|
| 230 |
+
{
|
| 231 |
+
"cell_type": "code",
|
| 232 |
+
"execution_count": 47,
|
| 233 |
+
"id": "9a47a1b1-21f2-4d71-801c-50f88e326ed3",
|
| 234 |
+
"metadata": {},
|
| 235 |
+
"outputs": [
|
| 236 |
+
{
|
| 237 |
+
"data": {
|
| 238 |
+
"text/plain": [
|
| 239 |
+
"{'sequence': 'CATGCGGGTCGATATCCTATCTGAATCTCTCAGCCCAAGAGGGAGTCCGCTCATCTATTCGGCAGTACTG',\n",
|
| 240 |
+
" 'label': 0}"
|
| 241 |
+
]
|
| 242 |
+
},
|
| 243 |
+
"execution_count": 47,
|
| 244 |
+
"metadata": {},
|
| 245 |
+
"output_type": "execute_result"
|
| 246 |
+
}
|
| 247 |
+
],
|
| 248 |
+
"source": [
|
| 249 |
+
"dna_data[\"train\"][0]"
|
| 250 |
+
]
|
| 251 |
+
},
|
| 252 |
+
{
|
| 253 |
+
"cell_type": "markdown",
|
| 254 |
+
"id": "cde7986d-a225-41ca-8f11-614d079fd2bf",
|
| 255 |
+
"metadata": {},
|
| 256 |
+
"source": [
|
| 257 |
+
"这里使用scikit-learn库来构建逻辑回归分类器。首先是特征提取:"
|
| 258 |
+
]
|
| 259 |
+
},
|
| 260 |
+
{
|
| 261 |
+
"cell_type": "code",
|
| 262 |
+
"execution_count": 52,
|
| 263 |
+
"id": "4010d991-056a-43ce-8cca-30eeec8678f5",
|
| 264 |
+
"metadata": {},
|
| 265 |
+
"outputs": [],
|
| 266 |
+
"source": [
|
| 267 |
+
"import numpy as np\n",
|
| 268 |
+
"from sklearn.model_selection import train_test_split\n",
|
| 269 |
+
"from sklearn.linear_model import LogisticRegression\n",
|
| 270 |
+
"from sklearn.datasets import load_iris\n",
|
| 271 |
+
"from sklearn.metrics import accuracy_score\n",
|
| 272 |
+
"\n",
|
| 273 |
+
"\n",
|
| 274 |
+
"def get_gpt2_feature(sequence):\n",
|
| 275 |
+
" return \n",
|
| 276 |
+
"\n",
|
| 277 |
+
"# 加载数据集\n",
|
| 278 |
+
"data = load_iris()\n",
|
| 279 |
+
"X = data.data[data.target < 2] # 只选择前两个类别\n",
|
| 280 |
+
"y = data.target[data.target < 2]\n",
|
| 281 |
+
"\n",
|
| 282 |
+
"X = []\n",
|
| 283 |
+
"Y = []\n",
|
| 284 |
+
"\n",
|
| 285 |
+
"for item in dna_data[\"train\"]:\n",
|
| 286 |
+
" sequence = item[\"sequence\"]\n",
|
| 287 |
+
" label = item[\"label\"]\n",
|
| 288 |
+
" x_v = get_gpt2_feature(sequence)\n",
|
| 289 |
+
" y_v = label\n",
|
| 290 |
+
" X.append(x_v)\n",
|
| 291 |
+
" Y.append(y_v)"
|
| 292 |
+
]
|
| 293 |
+
},
|
| 294 |
+
{
|
| 295 |
+
"cell_type": "code",
|
| 296 |
+
"execution_count": 49,
|
| 297 |
+
"id": "8af0effa-b2b6-4e49-9256-cead146d848c",
|
| 298 |
+
"metadata": {},
|
| 299 |
+
"outputs": [
|
| 300 |
+
{
|
| 301 |
+
"data": {
|
| 302 |
+
"text/plain": [
|
| 303 |
+
"array([[5.1, 3.5, 1.4, 0.2],\n",
|
| 304 |
+
" [4.9, 3. , 1.4, 0.2],\n",
|
| 305 |
+
" [4.7, 3.2, 1.3, 0.2],\n",
|
| 306 |
+
" [4.6, 3.1, 1.5, 0.2],\n",
|
| 307 |
+
" [5. , 3.6, 1.4, 0.2],\n",
|
| 308 |
+
" [5.4, 3.9, 1.7, 0.4],\n",
|
| 309 |
+
" [4.6, 3.4, 1.4, 0.3],\n",
|
| 310 |
+
" [5. , 3.4, 1.5, 0.2],\n",
|
| 311 |
+
" [4.4, 2.9, 1.4, 0.2],\n",
|
| 312 |
+
" [4.9, 3.1, 1.5, 0.1],\n",
|
| 313 |
+
" [5.4, 3.7, 1.5, 0.2],\n",
|
| 314 |
+
" [4.8, 3.4, 1.6, 0.2],\n",
|
| 315 |
+
" [4.8, 3. , 1.4, 0.1],\n",
|
| 316 |
+
" [4.3, 3. , 1.1, 0.1],\n",
|
| 317 |
+
" [5.8, 4. , 1.2, 0.2],\n",
|
| 318 |
+
" [5.7, 4.4, 1.5, 0.4],\n",
|
| 319 |
+
" [5.4, 3.9, 1.3, 0.4],\n",
|
| 320 |
+
" [5.1, 3.5, 1.4, 0.3],\n",
|
| 321 |
+
" [5.7, 3.8, 1.7, 0.3],\n",
|
| 322 |
+
" [5.1, 3.8, 1.5, 0.3],\n",
|
| 323 |
+
" [5.4, 3.4, 1.7, 0.2],\n",
|
| 324 |
+
" [5.1, 3.7, 1.5, 0.4],\n",
|
| 325 |
+
" [4.6, 3.6, 1. , 0.2],\n",
|
| 326 |
+
" [5.1, 3.3, 1.7, 0.5],\n",
|
| 327 |
+
" [4.8, 3.4, 1.9, 0.2],\n",
|
| 328 |
+
" [5. , 3. , 1.6, 0.2],\n",
|
| 329 |
+
" [5. , 3.4, 1.6, 0.4],\n",
|
| 330 |
+
" [5.2, 3.5, 1.5, 0.2],\n",
|
| 331 |
+
" [5.2, 3.4, 1.4, 0.2],\n",
|
| 332 |
+
" [4.7, 3.2, 1.6, 0.2],\n",
|
| 333 |
+
" [4.8, 3.1, 1.6, 0.2],\n",
|
| 334 |
+
" [5.4, 3.4, 1.5, 0.4],\n",
|
| 335 |
+
" [5.2, 4.1, 1.5, 0.1],\n",
|
| 336 |
+
" [5.5, 4.2, 1.4, 0.2],\n",
|
| 337 |
+
" [4.9, 3.1, 1.5, 0.2],\n",
|
| 338 |
+
" [5. , 3.2, 1.2, 0.2],\n",
|
| 339 |
+
" [5.5, 3.5, 1.3, 0.2],\n",
|
| 340 |
+
" [4.9, 3.6, 1.4, 0.1],\n",
|
| 341 |
+
" [4.4, 3. , 1.3, 0.2],\n",
|
| 342 |
+
" [5.1, 3.4, 1.5, 0.2],\n",
|
| 343 |
+
" [5. , 3.5, 1.3, 0.3],\n",
|
| 344 |
+
" [4.5, 2.3, 1.3, 0.3],\n",
|
| 345 |
+
" [4.4, 3.2, 1.3, 0.2],\n",
|
| 346 |
+
" [5. , 3.5, 1.6, 0.6],\n",
|
| 347 |
+
" [5.1, 3.8, 1.9, 0.4],\n",
|
| 348 |
+
" [4.8, 3. , 1.4, 0.3],\n",
|
| 349 |
+
" [5.1, 3.8, 1.6, 0.2],\n",
|
| 350 |
+
" [4.6, 3.2, 1.4, 0.2],\n",
|
| 351 |
+
" [5.3, 3.7, 1.5, 0.2],\n",
|
| 352 |
+
" [5. , 3.3, 1.4, 0.2],\n",
|
| 353 |
+
" [7. , 3.2, 4.7, 1.4],\n",
|
| 354 |
+
" [6.4, 3.2, 4.5, 1.5],\n",
|
| 355 |
+
" [6.9, 3.1, 4.9, 1.5],\n",
|
| 356 |
+
" [5.5, 2.3, 4. , 1.3],\n",
|
| 357 |
+
" [6.5, 2.8, 4.6, 1.5],\n",
|
| 358 |
+
" [5.7, 2.8, 4.5, 1.3],\n",
|
| 359 |
+
" [6.3, 3.3, 4.7, 1.6],\n",
|
| 360 |
+
" [4.9, 2.4, 3.3, 1. ],\n",
|
| 361 |
+
" [6.6, 2.9, 4.6, 1.3],\n",
|
| 362 |
+
" [5.2, 2.7, 3.9, 1.4],\n",
|
| 363 |
+
" [5. , 2. , 3.5, 1. ],\n",
|
| 364 |
+
" [5.9, 3. , 4.2, 1.5],\n",
|
| 365 |
+
" [6. , 2.2, 4. , 1. ],\n",
|
| 366 |
+
" [6.1, 2.9, 4.7, 1.4],\n",
|
| 367 |
+
" [5.6, 2.9, 3.6, 1.3],\n",
|
| 368 |
+
" [6.7, 3.1, 4.4, 1.4],\n",
|
| 369 |
+
" [5.6, 3. , 4.5, 1.5],\n",
|
| 370 |
+
" [5.8, 2.7, 4.1, 1. ],\n",
|
| 371 |
+
" [6.2, 2.2, 4.5, 1.5],\n",
|
| 372 |
+
" [5.6, 2.5, 3.9, 1.1],\n",
|
| 373 |
+
" [5.9, 3.2, 4.8, 1.8],\n",
|
| 374 |
+
" [6.1, 2.8, 4. , 1.3],\n",
|
| 375 |
+
" [6.3, 2.5, 4.9, 1.5],\n",
|
| 376 |
+
" [6.1, 2.8, 4.7, 1.2],\n",
|
| 377 |
+
" [6.4, 2.9, 4.3, 1.3],\n",
|
| 378 |
+
" [6.6, 3. , 4.4, 1.4],\n",
|
| 379 |
+
" [6.8, 2.8, 4.8, 1.4],\n",
|
| 380 |
+
" [6.7, 3. , 5. , 1.7],\n",
|
| 381 |
+
" [6. , 2.9, 4.5, 1.5],\n",
|
| 382 |
+
" [5.7, 2.6, 3.5, 1. ],\n",
|
| 383 |
+
" [5.5, 2.4, 3.8, 1.1],\n",
|
| 384 |
+
" [5.5, 2.4, 3.7, 1. ],\n",
|
| 385 |
+
" [5.8, 2.7, 3.9, 1.2],\n",
|
| 386 |
+
" [6. , 2.7, 5.1, 1.6],\n",
|
| 387 |
+
" [5.4, 3. , 4.5, 1.5],\n",
|
| 388 |
+
" [6. , 3.4, 4.5, 1.6],\n",
|
| 389 |
+
" [6.7, 3.1, 4.7, 1.5],\n",
|
| 390 |
+
" [6.3, 2.3, 4.4, 1.3],\n",
|
| 391 |
+
" [5.6, 3. , 4.1, 1.3],\n",
|
| 392 |
+
" [5.5, 2.5, 4. , 1.3],\n",
|
| 393 |
+
" [5.5, 2.6, 4.4, 1.2],\n",
|
| 394 |
+
" [6.1, 3. , 4.6, 1.4],\n",
|
| 395 |
+
" [5.8, 2.6, 4. , 1.2],\n",
|
| 396 |
+
" [5. , 2.3, 3.3, 1. ],\n",
|
| 397 |
+
" [5.6, 2.7, 4.2, 1.3],\n",
|
| 398 |
+
" [5.7, 3. , 4.2, 1.2],\n",
|
| 399 |
+
" [5.7, 2.9, 4.2, 1.3],\n",
|
| 400 |
+
" [6.2, 2.9, 4.3, 1.3],\n",
|
| 401 |
+
" [5.1, 2.5, 3. , 1.1],\n",
|
| 402 |
+
" [5.7, 2.8, 4.1, 1.3]])"
|
| 403 |
+
]
|
| 404 |
+
},
|
| 405 |
+
"execution_count": 49,
|
| 406 |
+
"metadata": {},
|
| 407 |
+
"output_type": "execute_result"
|
| 408 |
+
}
|
| 409 |
+
],
|
| 410 |
+
"source": [
|
| 411 |
+
"X"
|
| 412 |
+
]
|
| 413 |
+
},
|
| 414 |
+
{
|
| 415 |
+
"cell_type": "code",
|
| 416 |
+
"execution_count": 51,
|
| 417 |
+
"id": "868a3cab-e991-4990-9ec5-3e632a41a599",
|
| 418 |
+
"metadata": {},
|
| 419 |
+
"outputs": [
|
| 420 |
+
{
|
| 421 |
+
"data": {
|
| 422 |
+
"text/plain": [
|
| 423 |
+
"array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n",
|
| 424 |
+
" 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n",
|
| 425 |
+
" 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,\n",
|
| 426 |
+
" 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,\n",
|
| 427 |
+
" 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1])"
|
| 428 |
+
]
|
| 429 |
+
},
|
| 430 |
+
"execution_count": 51,
|
| 431 |
+
"metadata": {},
|
| 432 |
+
"output_type": "execute_result"
|
| 433 |
+
}
|
| 434 |
+
],
|
| 435 |
+
"source": [
|
| 436 |
+
"y"
|
| 437 |
+
]
|
| 438 |
+
},
|
| 439 |
+
{
|
| 440 |
+
"cell_type": "code",
|
| 441 |
+
"execution_count": null,
|
| 442 |
+
"id": "5ab0c188-6476-43c4-b361-a2bfe0ec7a8a",
|
| 443 |
+
"metadata": {},
|
| 444 |
+
"outputs": [],
|
| 445 |
+
"source": [
|
| 446 |
+
"# 将数据分为训练集和测试集\n",
|
| 447 |
+
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)\n",
|
| 448 |
+
"\n",
|
| 449 |
+
"# 创建逻辑回归模型\n",
|
| 450 |
+
"model = LogisticRegression()\n",
|
| 451 |
+
"\n",
|
| 452 |
+
"# 训练模型\n",
|
| 453 |
+
"model.fit(X_train, y_train)\n",
|
| 454 |
+
"\n",
|
| 455 |
+
"# 在测试集上进行预测\n",
|
| 456 |
+
"y_pred = model.predict(X_test)\n",
|
| 457 |
+
"\n",
|
| 458 |
+
"# 计算准确率\n",
|
| 459 |
+
"accuracy = accuracy_score(y_test, y_pred)\n",
|
| 460 |
+
"print(f\"Accuracy: {accuracy * 100:.2f}%\")\n",
|
| 461 |
+
"\n",
|
| 462 |
+
"# 输出部分预测结果与真实标签对比\n",
|
| 463 |
+
"for i in range(5):\n",
|
| 464 |
+
" print(f\"True: {y_test[i]}, Predicted: {y_pred[i]}\")"
|
| 465 |
+
]
|
| 466 |
+
}
|
| 467 |
+
],
|
| 468 |
+
"metadata": {
|
| 469 |
+
"kernelspec": {
|
| 470 |
+
"display_name": "Python 3 (ipykernel)",
|
| 471 |
+
"language": "python",
|
| 472 |
+
"name": "python3"
|
| 473 |
+
},
|
| 474 |
+
"language_info": {
|
| 475 |
+
"codemirror_mode": {
|
| 476 |
+
"name": "ipython",
|
| 477 |
+
"version": 3
|
| 478 |
+
},
|
| 479 |
+
"file_extension": ".py",
|
| 480 |
+
"mimetype": "text/x-python",
|
| 481 |
+
"name": "python",
|
| 482 |
+
"nbconvert_exporter": "python",
|
| 483 |
+
"pygments_lexer": "ipython3",
|
| 484 |
+
"version": "3.12.3"
|
| 485 |
+
}
|
| 486 |
+
},
|
| 487 |
+
"nbformat": 4,
|
| 488 |
+
"nbformat_minor": 5
|
| 489 |
+
}
|
02-gpt2_bert/5-multi-seq-gpt.ipynb
ADDED
|
@@ -0,0 +1,261 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"id": "9131f25f-227b-4dbe-b28d-c5006df092c6",
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"source": [
|
| 8 |
+
"# 2.5 基于多模态数据构建大模型"
|
| 9 |
+
]
|
| 10 |
+
},
|
| 11 |
+
{
|
| 12 |
+
"cell_type": "code",
|
| 13 |
+
"execution_count": null,
|
| 14 |
+
"id": "1a30b35c-1f5f-41e6-8fe1-5f522c700e9e",
|
| 15 |
+
"metadata": {},
|
| 16 |
+
"outputs": [],
|
| 17 |
+
"source": [
|
| 18 |
+
"from tokenizers import (\n",
|
| 19 |
+
" decoders,\n",
|
| 20 |
+
" models,\n",
|
| 21 |
+
" normalizers,\n",
|
| 22 |
+
" pre_tokenizers,\n",
|
| 23 |
+
" processors,\n",
|
| 24 |
+
" trainers,\n",
|
| 25 |
+
" Tokenizer,\n",
|
| 26 |
+
")\n",
|
| 27 |
+
"from transformers import AutoTokenizer"
|
| 28 |
+
]
|
| 29 |
+
},
|
| 30 |
+
{
|
| 31 |
+
"cell_type": "code",
|
| 32 |
+
"execution_count": null,
|
| 33 |
+
"id": "688fa3b1-f2ca-457a-abde-117c79b54fa9",
|
| 34 |
+
"metadata": {},
|
| 35 |
+
"outputs": [],
|
| 36 |
+
"source": [
|
| 37 |
+
"tokenizer = Tokenizer(models.BPE())\n",
|
| 38 |
+
"tokenizer.pre_tokenizer = pre_tokenizers.ByteLevel(add_prefix_space=False, use_regex=False) #use_regex=False,空格当成一般字符串\n",
|
| 39 |
+
"trainer = trainers.BpeTrainer(vocab_size=90000, special_tokens=[\"<|endoftext|>\"]) #9w words"
|
| 40 |
+
]
|
| 41 |
+
},
|
| 42 |
+
{
|
| 43 |
+
"cell_type": "code",
|
| 44 |
+
"execution_count": null,
|
| 45 |
+
"id": "7d680700-1051-4af4-94d6-2ce3071a5979",
|
| 46 |
+
"metadata": {},
|
| 47 |
+
"outputs": [],
|
| 48 |
+
"source": [
|
| 49 |
+
"tokenizer.train([\"../01-data_env/data/dna_1g.txt\",\"../01-data_env/data/protein_1g.txt\",\"../01-data_env/data/english_500m.txt\"]\n",
|
| 50 |
+
" , trainer=trainer) #all file list, take 10-20 min"
|
| 51 |
+
]
|
| 52 |
+
},
|
| 53 |
+
{
|
| 54 |
+
"cell_type": "code",
|
| 55 |
+
"execution_count": null,
|
| 56 |
+
"id": "74434ece-2f6e-46fa-9a9e-ff88e9364de8",
|
| 57 |
+
"metadata": {},
|
| 58 |
+
"outputs": [],
|
| 59 |
+
"source": [
|
| 60 |
+
"tokenizer.save(\"gene_eng_dict.json\")"
|
| 61 |
+
]
|
| 62 |
+
},
|
| 63 |
+
{
|
| 64 |
+
"cell_type": "code",
|
| 65 |
+
"execution_count": null,
|
| 66 |
+
"id": "8ea34e18-6cee-40b9-ba96-d8734153eb9f",
|
| 67 |
+
"metadata": {},
|
| 68 |
+
"outputs": [],
|
| 69 |
+
"source": [
|
| 70 |
+
"#然后我们可以使用from_file() 方法从该文件里重新加载 Tokenizer 对象:\n",
|
| 71 |
+
"new_tokenizer = Tokenizer.from_file(\"gene_eng_dict.json\")\n",
|
| 72 |
+
"\n",
|
| 73 |
+
"#要在 🤗 Transformers 中使用这个标记器,我们必须将它包裹在一个 PreTrainedTokenizerFast 类中\n",
|
| 74 |
+
"from transformers import GPT2TokenizerFast\n",
|
| 75 |
+
"gene_eng_tokenizer = GPT2TokenizerFast(tokenizer_object=new_tokenizer)\n",
|
| 76 |
+
"gene_eng_tokenizer.save_pretrained(\"gene_eng_dict\")\n",
|
| 77 |
+
"#dna_tokenizer.push_to_hub(\"dna_bpe_dict_1g\", organization=\"dnagpt\", use_auth_token=\"hf_*****\") # push to huggingface"
|
| 78 |
+
]
|
| 79 |
+
},
|
| 80 |
+
{
|
| 81 |
+
"cell_type": "code",
|
| 82 |
+
"execution_count": null,
|
| 83 |
+
"id": "16c7a3ef-c924-4fbb-b8ab-c12fab43f019",
|
| 84 |
+
"metadata": {},
|
| 85 |
+
"outputs": [],
|
| 86 |
+
"source": [
|
| 87 |
+
"tokenizer_new = AutoTokenizer.from_pretrained('gene_eng_dict')\n",
|
| 88 |
+
"tokenizer_new.tokenize(\"TGGCGTGAACCCGGGATCGGG,hello world hello gene, MANITWMANHTGWSDFILLGLFRQSKHPALLCVVIFVVFLMAL\")"
|
| 89 |
+
]
|
| 90 |
+
},
|
| 91 |
+
{
|
| 92 |
+
"cell_type": "markdown",
|
| 93 |
+
"id": "0ca0b2e3-f270-4645-abbb-cb8535e97a0a",
|
| 94 |
+
"metadata": {},
|
| 95 |
+
"source": [
|
| 96 |
+
"## 训练混合模型"
|
| 97 |
+
]
|
| 98 |
+
},
|
| 99 |
+
{
|
| 100 |
+
"cell_type": "code",
|
| 101 |
+
"execution_count": null,
|
| 102 |
+
"id": "c9b1c9b4-57a8-4711-912d-307e55481f8a",
|
| 103 |
+
"metadata": {},
|
| 104 |
+
"outputs": [],
|
| 105 |
+
"source": [
|
| 106 |
+
"from transformers import AutoTokenizer, GPT2LMHeadModel, AutoConfig,GPT2Tokenizer\n",
|
| 107 |
+
"from transformers import GPT2Tokenizer,GPT2Model,AutoModel\n",
|
| 108 |
+
"from transformers import DataCollatorForLanguageModeling\n",
|
| 109 |
+
"from transformers import Trainer, TrainingArguments\n",
|
| 110 |
+
"from transformers import LineByLineTextDataset\n",
|
| 111 |
+
"from tokenizers import Tokenizer\n",
|
| 112 |
+
"from datasets import load_dataset"
|
| 113 |
+
]
|
| 114 |
+
},
|
| 115 |
+
{
|
| 116 |
+
"cell_type": "code",
|
| 117 |
+
"execution_count": null,
|
| 118 |
+
"id": "3926a959-4224-4d78-9413-dc47a58087e0",
|
| 119 |
+
"metadata": {},
|
| 120 |
+
"outputs": [],
|
| 121 |
+
"source": [
|
| 122 |
+
"tokenizer = GPT2Tokenizer.from_pretrained(\"gene_eng_dict\")\n",
|
| 123 |
+
"tokenizer.pad_token = tokenizer.eos_token"
|
| 124 |
+
]
|
| 125 |
+
},
|
| 126 |
+
{
|
| 127 |
+
"cell_type": "code",
|
| 128 |
+
"execution_count": null,
|
| 129 |
+
"id": "1c2f5a6d-d405-40dc-a802-f0c1dff50a1e",
|
| 130 |
+
"metadata": {},
|
| 131 |
+
"outputs": [],
|
| 132 |
+
"source": [
|
| 133 |
+
"max_length = 256 #最大输入长度\n",
|
| 134 |
+
"\n",
|
| 135 |
+
"config = AutoConfig.from_pretrained(\n",
|
| 136 |
+
" \"gpt2\",\n",
|
| 137 |
+
" vocab_size=len(tokenizer),\n",
|
| 138 |
+
" n_ctx=max_length, #最大长度\n",
|
| 139 |
+
" bos_token_id=tokenizer.bos_token_id,\n",
|
| 140 |
+
" eos_token_id=tokenizer.eos_token_id,\n",
|
| 141 |
+
")\n",
|
| 142 |
+
"\n",
|
| 143 |
+
"model = GPT2LMHeadModel(config) #for pretrain,从头预训练"
|
| 144 |
+
]
|
| 145 |
+
},
|
| 146 |
+
{
|
| 147 |
+
"cell_type": "code",
|
| 148 |
+
"execution_count": null,
|
| 149 |
+
"id": "c8a47141-56a7-4e41-8cfd-1b381a64e2c0",
|
| 150 |
+
"metadata": {},
|
| 151 |
+
"outputs": [],
|
| 152 |
+
"source": [
|
| 153 |
+
"# 1. load dna dataset\n",
|
| 154 |
+
"raw_dataset = load_dataset('text', \n",
|
| 155 |
+
" data_files=[\"../01-data_env/data/dna_1g.txt\",\"../01-data_env/data/protein_1g.txt\",\"../01-data_env/data/english_500m.txt\"])\n",
|
| 156 |
+
"\n",
|
| 157 |
+
"dataset = raw_dataset[\"train\"].train_test_split(test_size=0.05, shuffle=True)\n",
|
| 158 |
+
"\n",
|
| 159 |
+
"# 2. tokenize\n",
|
| 160 |
+
"def tokenize_function(examples):\n",
|
| 161 |
+
" return tokenizer(examples['text'], truncation=True, padding='max_length', max_length=max_length)\n",
|
| 162 |
+
"\n",
|
| 163 |
+
"# 3. 对数据集应用分词函数\n",
|
| 164 |
+
"tokenized_datasets = dataset.map(tokenize_function, batched=True, remove_columns=['text'], num_proc=15) # 设置为你的 CPU 核心数或根据需要调整\n",
|
| 165 |
+
"\n",
|
| 166 |
+
"# 4. 创建一个数据收集器,用于动态填充和遮蔽\n",
|
| 167 |
+
"data_collator = DataCollatorForLanguageModeling(\n",
|
| 168 |
+
" tokenizer=tokenizer, mlm=False\n",
|
| 169 |
+
")"
|
| 170 |
+
]
|
| 171 |
+
},
|
| 172 |
+
{
|
| 173 |
+
"cell_type": "code",
|
| 174 |
+
"execution_count": null,
|
| 175 |
+
"id": "f4f802a2-88e2-49c2-a654-9d6e0996433a",
|
| 176 |
+
"metadata": {},
|
| 177 |
+
"outputs": [],
|
| 178 |
+
"source": [
|
| 179 |
+
"run_path = \"gpt2_run\"\n",
|
| 180 |
+
"train_epoches = 5\n",
|
| 181 |
+
"batch_size = 10\n",
|
| 182 |
+
"\n",
|
| 183 |
+
"\n",
|
| 184 |
+
"training_args = TrainingArguments(\n",
|
| 185 |
+
" output_dir=run_path,\n",
|
| 186 |
+
" overwrite_output_dir=True,\n",
|
| 187 |
+
" num_train_epochs=train_epoches,\n",
|
| 188 |
+
" per_device_train_batch_size=batch_size,\n",
|
| 189 |
+
" save_steps=2000,\n",
|
| 190 |
+
" save_total_limit=2,\n",
|
| 191 |
+
" prediction_loss_only=True,\n",
|
| 192 |
+
" fp16=True, #v100没法用\n",
|
| 193 |
+
" )\n",
|
| 194 |
+
"\n",
|
| 195 |
+
"\n",
|
| 196 |
+
"trainer = Trainer(\n",
|
| 197 |
+
" model=model,\n",
|
| 198 |
+
" args=training_args,\n",
|
| 199 |
+
" train_dataset=tokenized_datasets[\"train\"],\n",
|
| 200 |
+
" eval_dataset=tokenized_datasets[\"test\"],\n",
|
| 201 |
+
" data_collator=data_collator,\n",
|
| 202 |
+
")"
|
| 203 |
+
]
|
| 204 |
+
},
|
| 205 |
+
{
|
| 206 |
+
"cell_type": "code",
|
| 207 |
+
"execution_count": null,
|
| 208 |
+
"id": "13fa4a99-ee7c-4d6a-853f-4be04a4ee43c",
|
| 209 |
+
"metadata": {},
|
| 210 |
+
"outputs": [],
|
| 211 |
+
"source": [
|
| 212 |
+
"trainer.train()\n",
|
| 213 |
+
"trainer.save_model(\"gene_eng_gpt2_v0\")"
|
| 214 |
+
]
|
| 215 |
+
},
|
| 216 |
+
{
|
| 217 |
+
"cell_type": "code",
|
| 218 |
+
"execution_count": null,
|
| 219 |
+
"id": "ca452721-3914-49be-a577-d4c257946578",
|
| 220 |
+
"metadata": {},
|
| 221 |
+
"outputs": [],
|
| 222 |
+
"source": [
|
| 223 |
+
"import math\n",
|
| 224 |
+
"eval_results = trainer.evaluate()\n",
|
| 225 |
+
"print(f\"Perplexity: {math.exp(eval_results['eval_loss']):.2f}\")"
|
| 226 |
+
]
|
| 227 |
+
},
|
| 228 |
+
{
|
| 229 |
+
"cell_type": "code",
|
| 230 |
+
"execution_count": null,
|
| 231 |
+
"id": "b7e7a455-0e08-4a75-87c1-0f909829b1c1",
|
| 232 |
+
"metadata": {},
|
| 233 |
+
"outputs": [],
|
| 234 |
+
"source": [
|
| 235 |
+
"#upload model\n",
|
| 236 |
+
"#model.push_to_hub(\"gene_eng_gpt2_v0\", organization=\"dnagpt\", use_auth_token=\"hf_*******\")"
|
| 237 |
+
]
|
| 238 |
+
}
|
| 239 |
+
],
|
| 240 |
+
"metadata": {
|
| 241 |
+
"kernelspec": {
|
| 242 |
+
"display_name": "Python 3 (ipykernel)",
|
| 243 |
+
"language": "python",
|
| 244 |
+
"name": "python3"
|
| 245 |
+
},
|
| 246 |
+
"language_info": {
|
| 247 |
+
"codemirror_mode": {
|
| 248 |
+
"name": "ipython",
|
| 249 |
+
"version": 3
|
| 250 |
+
},
|
| 251 |
+
"file_extension": ".py",
|
| 252 |
+
"mimetype": "text/x-python",
|
| 253 |
+
"name": "python",
|
| 254 |
+
"nbconvert_exporter": "python",
|
| 255 |
+
"pygments_lexer": "ipython3",
|
| 256 |
+
"version": "3.12.3"
|
| 257 |
+
}
|
| 258 |
+
},
|
| 259 |
+
"nbformat": 4,
|
| 260 |
+
"nbformat_minor": 5
|
| 261 |
+
}
|
02-gpt2_bert/dna_bert_v0/config.json
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"_name_or_path": "bert-base-uncased",
|
| 3 |
+
"architectures": [
|
| 4 |
+
"BertForMaskedLM"
|
| 5 |
+
],
|
| 6 |
+
"attention_probs_dropout_prob": 0.1,
|
| 7 |
+
"bos_token_id": 2,
|
| 8 |
+
"classifier_dropout": null,
|
| 9 |
+
"eos_token_id": 3,
|
| 10 |
+
"gradient_checkpointing": false,
|
| 11 |
+
"hidden_act": "gelu",
|
| 12 |
+
"hidden_dropout_prob": 0.1,
|
| 13 |
+
"hidden_size": 768,
|
| 14 |
+
"initializer_range": 0.02,
|
| 15 |
+
"intermediate_size": 3072,
|
| 16 |
+
"layer_norm_eps": 1e-12,
|
| 17 |
+
"max_position_embeddings": 256,
|
| 18 |
+
"model_type": "bert",
|
| 19 |
+
"num_attention_heads": 12,
|
| 20 |
+
"num_hidden_layers": 12,
|
| 21 |
+
"pad_token_id": 0,
|
| 22 |
+
"position_embedding_type": "absolute",
|
| 23 |
+
"torch_dtype": "float32",
|
| 24 |
+
"transformers_version": "4.47.1",
|
| 25 |
+
"type_vocab_size": 2,
|
| 26 |
+
"use_cache": true,
|
| 27 |
+
"vocab_size": 30000
|
| 28 |
+
}
|
02-gpt2_bert/dna_bert_v0/generation_config.json
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"_from_model_config": true,
|
| 3 |
+
"bos_token_id": 2,
|
| 4 |
+
"eos_token_id": 3,
|
| 5 |
+
"pad_token_id": 0,
|
| 6 |
+
"transformers_version": "4.47.1"
|
| 7 |
+
}
|
02-gpt2_bert/dna_bert_v0/model.safetensors
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e4ddcda268020f781f23a80f4c0e6ab047189dd5f1cf61418429b08ad7ede68c
|
| 3 |
+
size 435688784
|
02-gpt2_bert/dna_bert_v0/training_args.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:4c59e1911412cf624ecf026e2f285ee02c53693e2822a16004df0254bd63561d
|
| 3 |
+
size 5304
|
02-gpt2_bert/dna_bpe_dict.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/merges-checkpoint.txt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6c79d89982c6ffe11f99a9830590377eba204aa277e9e00da4b44db9a758babd
|
| 3 |
+
size 323115
|
02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/special_tokens_map-checkpoint.json
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"bos_token": "<|endoftext|>",
|
| 3 |
+
"eos_token": "<|endoftext|>",
|
| 4 |
+
"unk_token": "<|endoftext|>"
|
| 5 |
+
}
|
02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/tokenizer-checkpoint.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_bpe_dict/.ipynb_checkpoints/tokenizer_config-checkpoint.json
ADDED
|
@@ -0,0 +1,20 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"add_prefix_space": false,
|
| 3 |
+
"added_tokens_decoder": {
|
| 4 |
+
"0": {
|
| 5 |
+
"content": "<|endoftext|>",
|
| 6 |
+
"lstrip": false,
|
| 7 |
+
"normalized": false,
|
| 8 |
+
"rstrip": false,
|
| 9 |
+
"single_word": false,
|
| 10 |
+
"special": true
|
| 11 |
+
}
|
| 12 |
+
},
|
| 13 |
+
"bos_token": "<|endoftext|>",
|
| 14 |
+
"clean_up_tokenization_spaces": false,
|
| 15 |
+
"eos_token": "<|endoftext|>",
|
| 16 |
+
"extra_special_tokens": {},
|
| 17 |
+
"model_max_length": 1000000000000000019884624838656,
|
| 18 |
+
"tokenizer_class": "GPT2Tokenizer",
|
| 19 |
+
"unk_token": "<|endoftext|>"
|
| 20 |
+
}
|
02-gpt2_bert/dna_bpe_dict/merges.txt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6c79d89982c6ffe11f99a9830590377eba204aa277e9e00da4b44db9a758babd
|
| 3 |
+
size 323115
|
02-gpt2_bert/dna_bpe_dict/special_tokens_map.json
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"bos_token": "<|endoftext|>",
|
| 3 |
+
"eos_token": "<|endoftext|>",
|
| 4 |
+
"unk_token": "<|endoftext|>"
|
| 5 |
+
}
|
02-gpt2_bert/dna_bpe_dict/tokenizer.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_bpe_dict/tokenizer_config.json
ADDED
|
@@ -0,0 +1,20 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"add_prefix_space": false,
|
| 3 |
+
"added_tokens_decoder": {
|
| 4 |
+
"0": {
|
| 5 |
+
"content": "<|endoftext|>",
|
| 6 |
+
"lstrip": false,
|
| 7 |
+
"normalized": false,
|
| 8 |
+
"rstrip": false,
|
| 9 |
+
"single_word": false,
|
| 10 |
+
"special": true
|
| 11 |
+
}
|
| 12 |
+
},
|
| 13 |
+
"bos_token": "<|endoftext|>",
|
| 14 |
+
"clean_up_tokenization_spaces": false,
|
| 15 |
+
"eos_token": "<|endoftext|>",
|
| 16 |
+
"extra_special_tokens": {},
|
| 17 |
+
"model_max_length": 1000000000000000019884624838656,
|
| 18 |
+
"tokenizer_class": "GPT2Tokenizer",
|
| 19 |
+
"unk_token": "<|endoftext|>"
|
| 20 |
+
}
|
02-gpt2_bert/dna_bpe_dict/vocab.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_gpt2_v0/config.json
ADDED
|
@@ -0,0 +1,39 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"_name_or_path": "gpt2",
|
| 3 |
+
"activation_function": "gelu_new",
|
| 4 |
+
"architectures": [
|
| 5 |
+
"GPT2LMHeadModel"
|
| 6 |
+
],
|
| 7 |
+
"attn_pdrop": 0.1,
|
| 8 |
+
"bos_token_id": 0,
|
| 9 |
+
"embd_pdrop": 0.1,
|
| 10 |
+
"eos_token_id": 0,
|
| 11 |
+
"initializer_range": 0.02,
|
| 12 |
+
"layer_norm_epsilon": 1e-05,
|
| 13 |
+
"model_type": "gpt2",
|
| 14 |
+
"n_ctx": 256,
|
| 15 |
+
"n_embd": 768,
|
| 16 |
+
"n_head": 12,
|
| 17 |
+
"n_inner": null,
|
| 18 |
+
"n_layer": 12,
|
| 19 |
+
"n_positions": 1024,
|
| 20 |
+
"reorder_and_upcast_attn": false,
|
| 21 |
+
"resid_pdrop": 0.1,
|
| 22 |
+
"scale_attn_by_inverse_layer_idx": false,
|
| 23 |
+
"scale_attn_weights": true,
|
| 24 |
+
"summary_activation": null,
|
| 25 |
+
"summary_first_dropout": 0.1,
|
| 26 |
+
"summary_proj_to_labels": true,
|
| 27 |
+
"summary_type": "cls_index",
|
| 28 |
+
"summary_use_proj": true,
|
| 29 |
+
"task_specific_params": {
|
| 30 |
+
"text-generation": {
|
| 31 |
+
"do_sample": true,
|
| 32 |
+
"max_length": 50
|
| 33 |
+
}
|
| 34 |
+
},
|
| 35 |
+
"torch_dtype": "float32",
|
| 36 |
+
"transformers_version": "4.47.1",
|
| 37 |
+
"use_cache": true,
|
| 38 |
+
"vocab_size": 30000
|
| 39 |
+
}
|
02-gpt2_bert/dna_gpt2_v0/generation_config.json
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"_from_model_config": true,
|
| 3 |
+
"bos_token_id": 0,
|
| 4 |
+
"eos_token_id": 0,
|
| 5 |
+
"transformers_version": "4.47.1"
|
| 6 |
+
}
|
02-gpt2_bert/dna_gpt2_v0/merges.txt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6c79d89982c6ffe11f99a9830590377eba204aa277e9e00da4b44db9a758babd
|
| 3 |
+
size 323115
|
02-gpt2_bert/dna_gpt2_v0/model.safetensors
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:4df0047ff1933c1cae760e550e58e593b9da1b9ad3cd9461bc665e9c17bc0416
|
| 3 |
+
size 435544704
|
02-gpt2_bert/dna_gpt2_v0/special_tokens_map.json
ADDED
|
@@ -0,0 +1,23 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"bos_token": {
|
| 3 |
+
"content": "<|endoftext|>",
|
| 4 |
+
"lstrip": false,
|
| 5 |
+
"normalized": false,
|
| 6 |
+
"rstrip": false,
|
| 7 |
+
"single_word": false
|
| 8 |
+
},
|
| 9 |
+
"eos_token": {
|
| 10 |
+
"content": "<|endoftext|>",
|
| 11 |
+
"lstrip": false,
|
| 12 |
+
"normalized": false,
|
| 13 |
+
"rstrip": false,
|
| 14 |
+
"single_word": false
|
| 15 |
+
},
|
| 16 |
+
"unk_token": {
|
| 17 |
+
"content": "<|endoftext|>",
|
| 18 |
+
"lstrip": false,
|
| 19 |
+
"normalized": false,
|
| 20 |
+
"rstrip": false,
|
| 21 |
+
"single_word": false
|
| 22 |
+
}
|
| 23 |
+
}
|
02-gpt2_bert/dna_gpt2_v0/tokenizer.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_gpt2_v0/tokenizer_config.json
ADDED
|
@@ -0,0 +1,20 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"add_prefix_space": false,
|
| 3 |
+
"added_tokens_decoder": {
|
| 4 |
+
"0": {
|
| 5 |
+
"content": "<|endoftext|>",
|
| 6 |
+
"lstrip": false,
|
| 7 |
+
"normalized": false,
|
| 8 |
+
"rstrip": false,
|
| 9 |
+
"single_word": false,
|
| 10 |
+
"special": true
|
| 11 |
+
}
|
| 12 |
+
},
|
| 13 |
+
"bos_token": "<|endoftext|>",
|
| 14 |
+
"clean_up_tokenization_spaces": false,
|
| 15 |
+
"eos_token": "<|endoftext|>",
|
| 16 |
+
"extra_special_tokens": {},
|
| 17 |
+
"model_max_length": 1000000000000000019884624838656,
|
| 18 |
+
"tokenizer_class": "GPT2Tokenizer",
|
| 19 |
+
"unk_token": "<|endoftext|>"
|
| 20 |
+
}
|
02-gpt2_bert/dna_gpt2_v0/training_args.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:bcbeec6a01e5f602600f5a54d50cadadebc1e9685dec8b7b5998b4798129b24b
|
| 3 |
+
size 5304
|
02-gpt2_bert/dna_gpt2_v0/vocab.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_wordpiece_dict.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_wordpiece_dict/special_tokens_map.json
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cls_token": "[CLS]",
|
| 3 |
+
"mask_token": "[MASK]",
|
| 4 |
+
"pad_token": "[PAD]",
|
| 5 |
+
"sep_token": "[SEP]",
|
| 6 |
+
"unk_token": "[UNK]"
|
| 7 |
+
}
|
02-gpt2_bert/dna_wordpiece_dict/tokenizer.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
02-gpt2_bert/dna_wordpiece_dict/tokenizer_config.json
ADDED
|
@@ -0,0 +1,53 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"added_tokens_decoder": {
|
| 3 |
+
"0": {
|
| 4 |
+
"content": "[PAD]",
|
| 5 |
+
"lstrip": false,
|
| 6 |
+
"normalized": false,
|
| 7 |
+
"rstrip": false,
|
| 8 |
+
"single_word": false,
|
| 9 |
+
"special": true
|
| 10 |
+
},
|
| 11 |
+
"1": {
|
| 12 |
+
"content": "[UNK]",
|
| 13 |
+
"lstrip": false,
|
| 14 |
+
"normalized": false,
|
| 15 |
+
"rstrip": false,
|
| 16 |
+
"single_word": false,
|
| 17 |
+
"special": true
|
| 18 |
+
},
|
| 19 |
+
"2": {
|
| 20 |
+
"content": "[CLS]",
|
| 21 |
+
"lstrip": false,
|
| 22 |
+
"normalized": false,
|
| 23 |
+
"rstrip": false,
|
| 24 |
+
"single_word": false,
|
| 25 |
+
"special": true
|
| 26 |
+
},
|
| 27 |
+
"3": {
|
| 28 |
+
"content": "[SEP]",
|
| 29 |
+
"lstrip": false,
|
| 30 |
+
"normalized": false,
|
| 31 |
+
"rstrip": false,
|
| 32 |
+
"single_word": false,
|
| 33 |
+
"special": true
|
| 34 |
+
},
|
| 35 |
+
"4": {
|
| 36 |
+
"content": "[MASK]",
|
| 37 |
+
"lstrip": false,
|
| 38 |
+
"normalized": false,
|
| 39 |
+
"rstrip": false,
|
| 40 |
+
"single_word": false,
|
| 41 |
+
"special": true
|
| 42 |
+
}
|
| 43 |
+
},
|
| 44 |
+
"clean_up_tokenization_spaces": false,
|
| 45 |
+
"cls_token": "[CLS]",
|
| 46 |
+
"extra_special_tokens": {},
|
| 47 |
+
"mask_token": "[MASK]",
|
| 48 |
+
"model_max_length": 1000000000000000019884624838656,
|
| 49 |
+
"pad_token": "[PAD]",
|
| 50 |
+
"sep_token": "[SEP]",
|
| 51 |
+
"tokenizer_class": "PreTrainedTokenizerFast",
|
| 52 |
+
"unk_token": "[UNK]"
|
| 53 |
+
}
|
02-gpt2_bert/img/.ipynb_checkpoints/gpt2-stru-checkpoint.png
ADDED

|
02-gpt2_bert/img/gpt2-netron.png
ADDED

|
02-gpt2_bert/img/gpt2-stru.png
ADDED
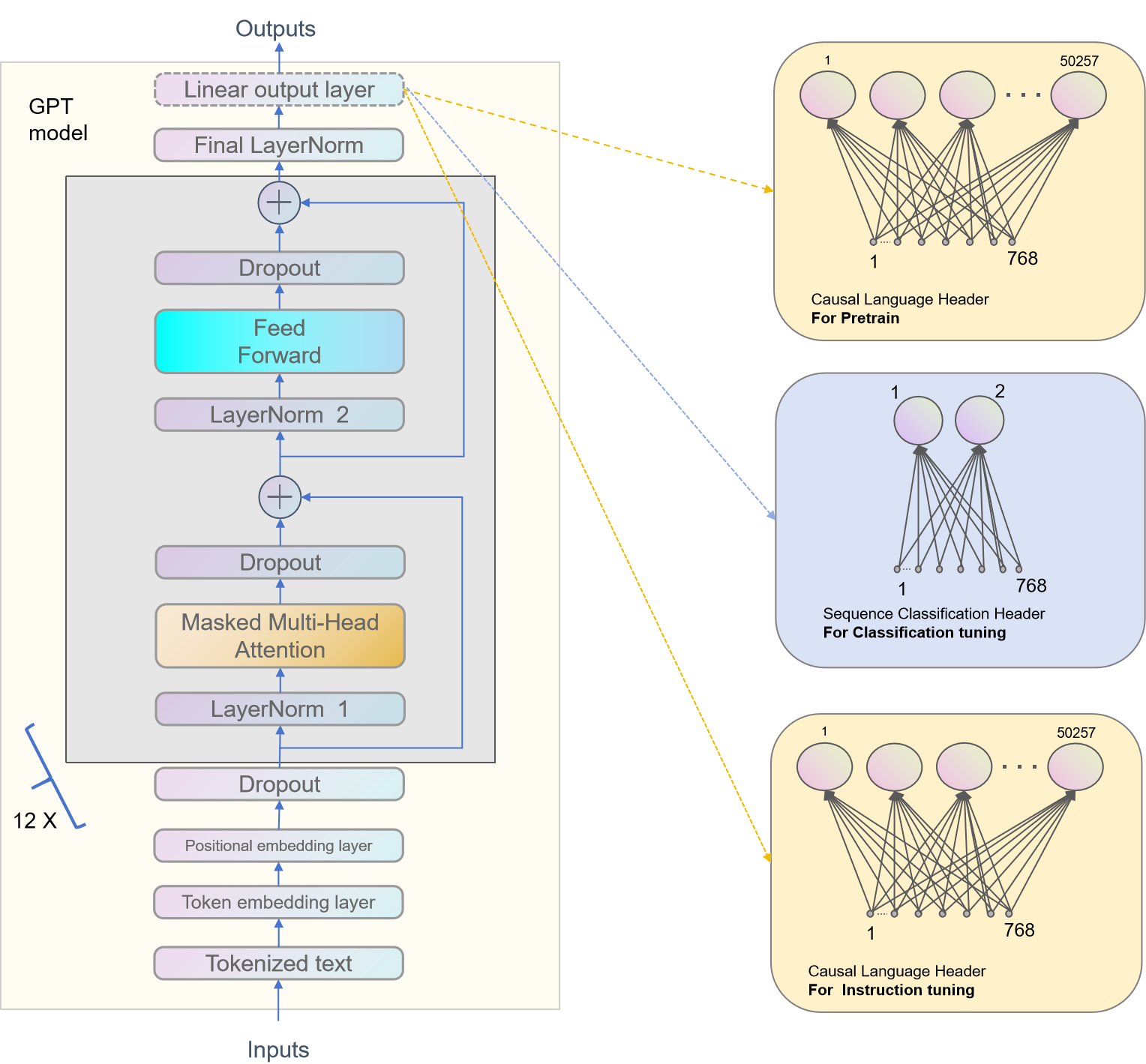
|
02-gpt2_bert/img/llm-visual.png
ADDED

|