Push model using huggingface_hub.
Browse files
README.md
CHANGED
|
@@ -26,7 +26,7 @@ This model contains the implementation of the Multi-view Molecular Embedding wit
|
|
| 26 |
|
| 27 |
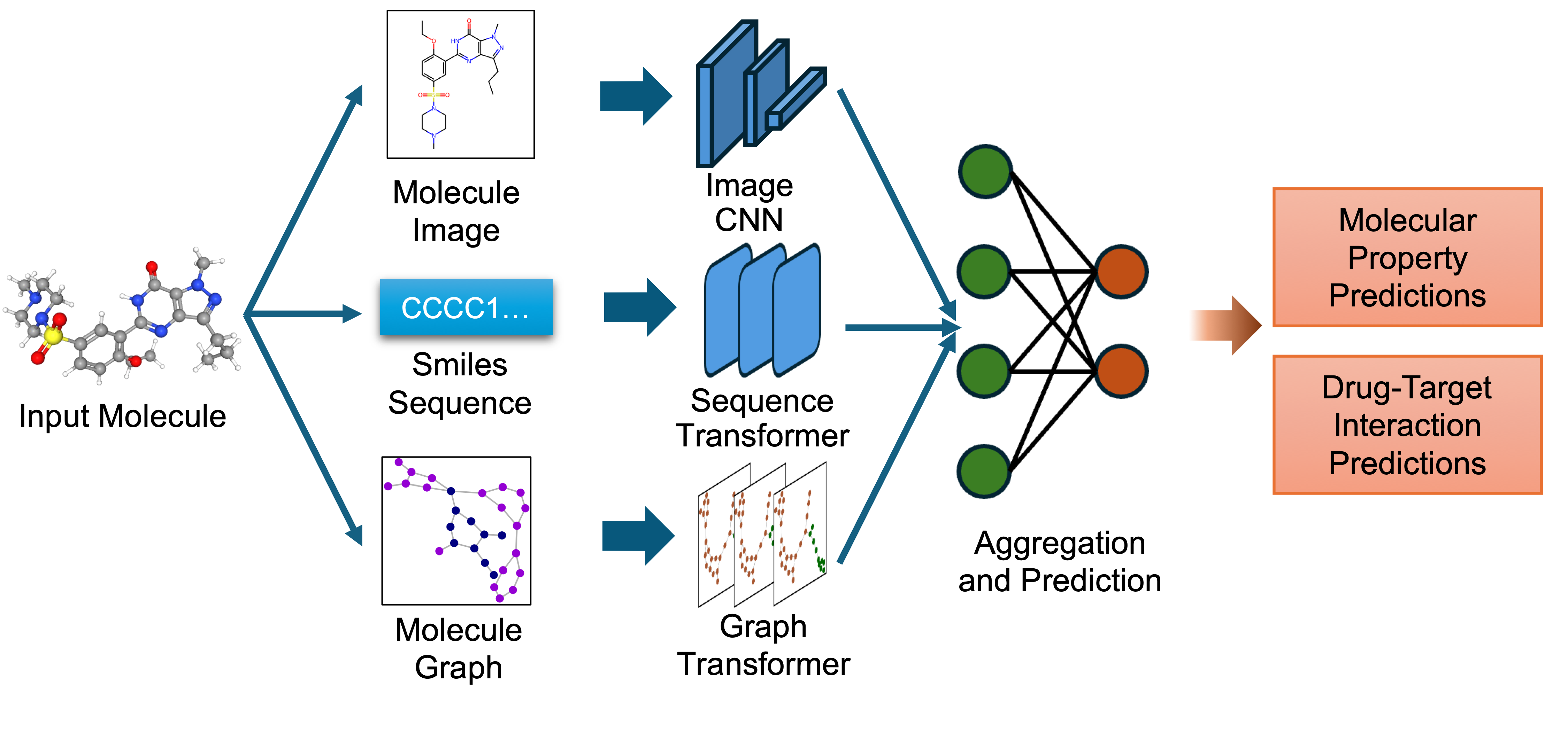
It was introduced in the paper [Multi-view biomedical foundation models for molecule-target and property prediction](https://arxiv.org/) by authors and first released in [this repository](https://github.com/BiomedSciAI/biomed-multi-view).
|
| 28 |
|
| 29 |
-

|
| 30 |
|
| 31 |
* Image Representation: Captures the 2D visual depiction of molecular structures, highlighting features like symmetry, bond angles, and functional groups. Molecular images are generated using RDKit and undergo data augmentation during training to enhance robustness.
|
| 32 |
* Graph Representation: Encodes molecules as undirected graphs where nodes represent atoms and edges represent bonds. Atom-specific properties (e.g., atomic number, chirality) and bond-specific properties (e.g., bond type, stereochemistry) are embedded using categorical embedding techniques.
|
|
|
|
| 26 |
|
| 27 |
It was introduced in the paper [Multi-view biomedical foundation models for molecule-target and property prediction](https://arxiv.org/) by authors and first released in [this repository](https://github.com/BiomedSciAI/biomed-multi-view).
|
| 28 |
|
| 29 |
+
|
| 30 |
|
| 31 |
* Image Representation: Captures the 2D visual depiction of molecular structures, highlighting features like symmetry, bond angles, and functional groups. Molecular images are generated using RDKit and undergo data augmentation during training to enhance robustness.
|
| 32 |
* Graph Representation: Encodes molecules as undirected graphs where nodes represent atoms and edges represent bonds. Atom-specific properties (e.g., atomic number, chirality) and bond-specific properties (e.g., bond type, stereochemistry) are embedded using categorical embedding techniques.
|