-------------------------------------------------------------------------------------
Fig.1 - The demonstration of T1w MRI images registered in MNI space and the whole brain segmentation labels with 133 classes
# Model Overview A pre-trained larger UNEST base model [1] for volumetric (3D) whole brain segmentation with T1w MR images. To leverage information across embedded sequences, ”shifted window” transformers are proposed for dense predictions and modeling multi-scale features. However, these attempts that aim to complicate the self-attention range often yield high computation complexity and data inefficiency. Inspired by the aggregation function in the nested ViT, we propose a new design of a 3D U-shape medical segmentation model with Nested Transformers (UNesT) hierarchically with the 3D block aggregation function, that learn locality behaviors for small structures or small dataset. This design retains the original global self-attention mechanism and achieves information communication across patches by stacking transformer encoders hierarchically. 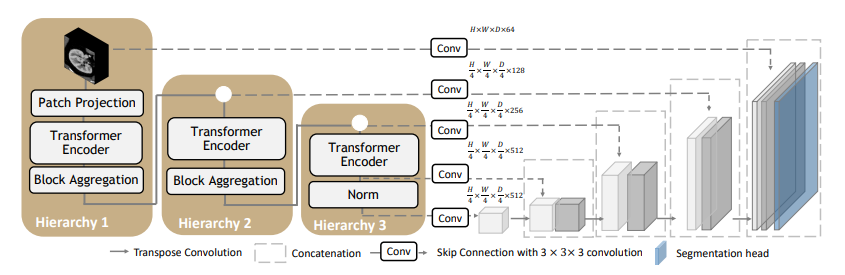Fig.2 - The network architecture of UNEST Base model
## Data The training data is from the Vanderbilt University and Vanderbilt University Medical Center with public released OASIS and CANDI datsets. Training and testing data are MRI T1-weighted (T1w) 3D volumes coming from 3 different sites. There are a total of 133 classes in the whole brain segmentation task. Among 50 T1w MRI scans from Open Access Series on Imaging Studies (OASIS) (Marcus et al., 2007) dataset, 45 scans are used for training and the other 5 for validation. The testing cohort contains Colin27 T1w scan (Aubert-Broche et al., 2006) and 13 T1w MRI scans from the Child and Adolescent Neuro Development Initiative (CANDI) (Kennedy et al., 2012). All data are registered to the MNI space using the MNI305 (Evans et al., 1993) template and preprocessed follow the method in (Huo et al., 2019). Input images are randomly cropped to the size of 96 × 96 × 96. ### Important ```diff + All the brain MRI images for training are registered to Affine registration from the target image to the MNI305 template using NiftyReg. + The data should be in the MNI305 space before inference. Registration to MNI Space: Sample suggestion. E.g., use ANTS or other tools for registering T1 MRI image to MNI305 Space. ``` pip install antspyx ``` Sample ANTS registration ``` import ants import sys import os fixed_image = ants.image_read('Fig.3 - The output prediction comparison with variant and ground truth
## Complete ROI of the whole brain segmentation 133 brain structures are segmented. | #1 | #2 | #3 | #4 | | :------------ | :---------- | :-------- | :-------- | | 0: background | 1 : 3rd-Ventricle | 2 : 4th-Ventricle | 3 : Right-Accumbens-Area | | 4 : Left-Accumbens-Area | 5 : Right-Amygdala | 6 : Left-Amygdala | 7 : Brain-Stem | | 8 : Right-Caudate | 9 : Left-Caudate | 10 : Right-Cerebellum-Exterior | 11 : Left-Cerebellum-Exterior | | 12 : Right-Cerebellum-White-Matter | 13 : Left-Cerebellum-White-Matter | 14 : Right-Cerebral-White-Matter | 15 : Left-Cerebral-White-Matter | | 16 : Right-Hippocampus | 17 : Left-Hippocampus | 18 : Right-Inf-Lat-Vent | 19 : Left-Inf-Lat-Vent | | 20 : Right-Lateral-Ventricle | 21 : Left-Lateral-Ventricle | 22 : Right-Pallidum | 23 : Left-Pallidum | | 24 : Right-Putamen | 25 : Left-Putamen | 26 : Right-Thalamus-Proper | 27 : Left-Thalamus-Proper | | 28 : Right-Ventral-DC | 29 : Left-Ventral-DC | 30 : Cerebellar-Vermal-Lobules-I-V | 31 : Cerebellar-Vermal-Lobules-VI-VII | | 32 : Cerebellar-Vermal-Lobules-VIII-X | 33 : Left-Basal-Forebrain | 34 : Right-Basal-Forebrain | 35 : Right-ACgG--anterior-cingulate-gyrus | | 36 : Left-ACgG--anterior-cingulate-gyrus | 37 : Right-AIns--anterior-insula | 38 : Left-AIns--anterior-insula | 39 : Right-AOrG--anterior-orbital-gyrus | | 40 : Left-AOrG--anterior-orbital-gyrus | 41 : Right-AnG---angular-gyrus | 42 : Left-AnG---angular-gyrus | 43 : Right-Calc--calcarine-cortex | | 44 : Left-Calc--calcarine-cortex | 45 : Right-CO----central-operculum | 46 : Left-CO----central-operculum | 47 : Right-Cun---cuneus | | 48 : Left-Cun---cuneus | 49 : Right-Ent---entorhinal-area | 50 : Left-Ent---entorhinal-area | 51 : Right-FO----frontal-operculum | | 52 : Left-FO----frontal-operculum | 53 : Right-FRP---frontal-pole | 54 : Left-FRP---frontal-pole | 55 : Right-FuG---fusiform-gyrus | | 56 : Left-FuG---fusiform-gyrus | 57 : Right-GRe---gyrus-rectus | 58 : Left-GRe---gyrus-rectus | 59 : Right-IOG---inferior-occipital-gyrus , | 60 : Left-IOG---inferior-occipital-gyrus | 61 : Right-ITG---inferior-temporal-gyrus | 62 : Left-ITG---inferior-temporal-gyrus | 63 : Right-LiG---lingual-gyrus | | 64 : Left-LiG---lingual-gyrus | 65 : Right-LOrG--lateral-orbital-gyrus | 66 : Left-LOrG--lateral-orbital-gyrus | 67 : Right-MCgG--middle-cingulate-gyrus | | 68 : Left-MCgG--middle-cingulate-gyrus | 69 : Right-MFC---medial-frontal-cortex | 70 : Left-MFC---medial-frontal-cortex | 71 : Right-MFG---middle-frontal-gyrus | | 72 : Left-MFG---middle-frontal-gyrus | 73 : Right-MOG---middle-occipital-gyrus | 74 : Left-MOG---middle-occipital-gyrus | 75 : Right-MOrG--medial-orbital-gyrus | | 76 : Left-MOrG--medial-orbital-gyrus | 77 : Right-MPoG--postcentral-gyrus | 78 : Left-MPoG--postcentral-gyrus | 79 : Right-MPrG--precentral-gyrus | | 80 : Left-MPrG--precentral-gyrus | 81 : Right-MSFG--superior-frontal-gyrus | 82 : Left-MSFG--superior-frontal-gyrus | 83 : Right-MTG---middle-temporal-gyrus | | 84 : Left-MTG---middle-temporal-gyrus | 85 : Right-OCP---occipital-pole | 86 : Left-OCP---occipital-pole | 87 : Right-OFuG--occipital-fusiform-gyrus | | 88 : Left-OFuG--occipital-fusiform-gyrus | 89 : Right-OpIFG-opercular-part-of-the-IFG | 90 : Left-OpIFG-opercular-part-of-the-IFG | 91 : Right-OrIFG-orbital-part-of-the-IFG | | 92 : Left-OrIFG-orbital-part-of-the-IFG | 93 : Right-PCgG--posterior-cingulate-gyrus | 94 : Left-PCgG--posterior-cingulate-gyrus | 95 : Right-PCu---precuneus | | 96 : Left-PCu---precuneus | 97 : Right-PHG---parahippocampal-gyrus | 98 : Left-PHG---parahippocampal-gyrus | 99 : Right-PIns--posterior-insula | | 100 : Left-PIns--posterior-insula | 101 : Right-PO----parietal-operculum | 102 : Left-PO----parietal-operculum | 103 : Right-PoG---postcentral-gyrus | | 104 : Left-PoG---postcentral-gyrus | 105 : Right-POrG--posterior-orbital-gyrus | 106 : Left-POrG--posterior-orbital-gyrus | 107 : Right-PP----planum-polare | | 108 : Left-PP----planum-polare | 109 : Right-PrG---precentral-gyrus | 110 : Left-PrG---precentral-gyrus | 111 : Right-PT----planum-temporale | | 112 : Left-PT----planum-temporale | 113 : Right-SCA---subcallosal-area | 114 : Left-SCA---subcallosal-area | 115 : Right-SFG---superior-frontal-gyrus | | 116 : Left-SFG---superior-frontal-gyrus | 117 : Right-SMC---supplementary-motor-cortex | 118 : Left-SMC---supplementary-motor-cortex | 119 : Right-SMG---supramarginal-gyrus | | 120 : Left-SMG---supramarginal-gyrus | 121 : Right-SOG---superior-occipital-gyrus | 122 : Left-SOG---superior-occipital-gyrus | 123 : Right-SPL---superior-parietal-lobule | | 124 : Left-SPL---superior-parietal-lobule | 125 : Right-STG---superior-temporal-gyrus | 126 : Left-STG---superior-temporal-gyrus | 127 : Right-TMP---temporal-pole | | 128 : Left-TMP---temporal-pole | 129 : Right-TrIFG-triangular-part-of-the-IFG | 130 : Left-TrIFG-triangular-part-of-the-IFG | 131 : Right-TTG---transverse-temporal-gyrus | | 132 : Left-TTG---transverse-temporal-gyrus | ## Bundle Integration in MONAI Lable The inference pipleine can be easily used by the MONAI Label server and 3D Slicer for fast labeling T1w MRI images in MNI space. 