---
license: apache-2.0
---
# Positive Transfer Of The Whisper Speech Transformer To Human And Animal Voice Activity Detection
We proposed WhisperSeg, utilizing the Whisper Transformer pre-trained for Automatic Speech Recognition (ASR) for both human and animal Voice Activity Detection (VAD). For more details, please refer to our paper
> [Positive Transfer of the Whisper Speech Transformer to Human and Animal Voice Activity Detection](https://doi.org/10.1101/2023.09.30.560270)
>
> Nianlong Gu, Kanghwi Lee, Maris Basha, Sumit Kumar Ram, Guanghao You, Richard H. R. Hahnloser
> University of Zurich and ETH Zurich
The model "nccratliri/whisperseg-large-ms" is the checkpoint of the multi-species WhisperSeg-large that was finetuned on the vocal segmentation datasets of five species.
## Usage
### Clone the GitHub repo and install dependencies
```bash
git clone https://github.com/nianlonggu/WhisperSeg.git
cd WhisperSeg; pip install -r requirements.txt
```
Then in the folder "WhisperSeg", run the following python script:
```python
from model import WhisperSegmenter
import librosa
import json
segmenter = WhisperSegmenter( "nccratliri/whisperseg-large-ms", device="cuda" )
sr = 32000
min_frequency = 0
spec_time_step = 0.0025
min_segment_length = 0.01
eps = 0.02
num_trials = 3
audio, _ = librosa.load( "data/example_subset/Zebra_finch/test_adults/zebra_finch_g17y2U-f00007.wav",
sr = sr )
prediction = segmenter.segment( audio, sr = sr, min_frequency = min_frequency, spec_time_step = spec_time_step,
min_segment_length = min_segment_length, eps = eps,num_trials = num_trials )
print(prediction)
```
{'onset': [0.01, 0.38, 0.603, 0.758, 0.912, 1.813, 1.967, 2.073, 2.838, 2.982, 3.112, 3.668, 3.828, 3.953, 5.158, 5.323, 5.467], 'offset': [0.073, 0.447, 0.673, 0.83, 1.483, 1.882, 2.037, 2.643, 2.893, 3.063, 3.283, 3.742, 3.898, 4.523, 5.223, 5.393, 6.043], 'cluster': ['zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0']}
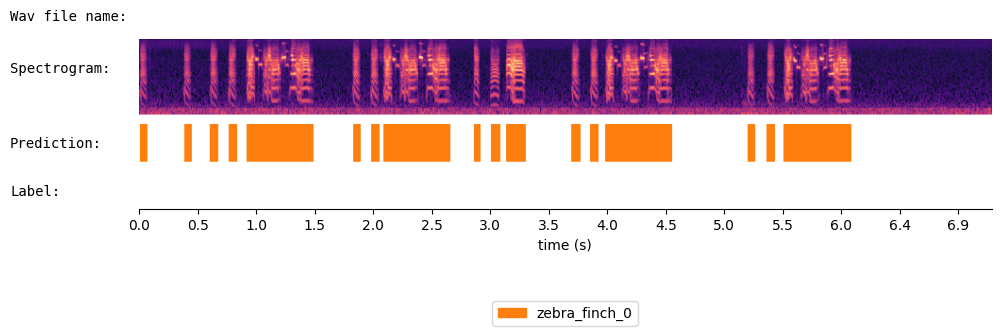
Visualize the results of WhisperSeg:
```python
from audio_utils import SpecViewer
spec_viewer = SpecViewer()
spec_viewer.visualize( audio = audio, sr = sr, min_frequency= min_frequency, prediction = prediction,
window_size=8, precision_bits=1
)
```
Run it in Google Colab:  For more details, please refer to the GitHub repository: https://github.com/nianlonggu/WhisperSeg
## Citation
When using our code or models for your work, please cite the following paper:
```
@article {Gu2023.09.30.560270,
author = {Nianlong Gu and Kanghwi Lee and Maris Basha and Sumit Kumar Ram and Guanghao You and Richard Hahnloser},
title = {Positive Transfer of the Whisper Speech Transformer to Human and Animal Voice Activity Detection},
elocation-id = {2023.09.30.560270},
year = {2023},
doi = {10.1101/2023.09.30.560270},
publisher = {Cold Spring Harbor Laboratory},
abstract = {This paper introduces WhisperSeg, utilizing the Whisper Transformer pre-trained for Automatic Speech Recognition (ASR) for human and animal Voice Activity Detection (VAD). Contrary to traditional methods that detect human voice or animal vocalizations from a short audio frame and rely on careful threshold selection, WhisperSeg processes entire spectrograms of long audio and generates plain text representations of onset, offset, and type of voice activity. Processing a longer audio context with a larger network greatly improves detection accuracy from few labeled examples. We further demonstrate a positive transfer of detection performance to new animal species, making our approach viable in the data-scarce multi-species setting.Competing Interest StatementThe authors have declared no competing interest.},
URL = {https://www.biorxiv.org/content/early/2023/10/02/2023.09.30.560270},
eprint = {https://www.biorxiv.org/content/early/2023/10/02/2023.09.30.560270.full.pdf},
journal = {bioRxiv}
}
```
## Contact
nianlong.gu@uzh.ch
For more details, please refer to the GitHub repository: https://github.com/nianlonggu/WhisperSeg
## Citation
When using our code or models for your work, please cite the following paper:
```
@article {Gu2023.09.30.560270,
author = {Nianlong Gu and Kanghwi Lee and Maris Basha and Sumit Kumar Ram and Guanghao You and Richard Hahnloser},
title = {Positive Transfer of the Whisper Speech Transformer to Human and Animal Voice Activity Detection},
elocation-id = {2023.09.30.560270},
year = {2023},
doi = {10.1101/2023.09.30.560270},
publisher = {Cold Spring Harbor Laboratory},
abstract = {This paper introduces WhisperSeg, utilizing the Whisper Transformer pre-trained for Automatic Speech Recognition (ASR) for human and animal Voice Activity Detection (VAD). Contrary to traditional methods that detect human voice or animal vocalizations from a short audio frame and rely on careful threshold selection, WhisperSeg processes entire spectrograms of long audio and generates plain text representations of onset, offset, and type of voice activity. Processing a longer audio context with a larger network greatly improves detection accuracy from few labeled examples. We further demonstrate a positive transfer of detection performance to new animal species, making our approach viable in the data-scarce multi-species setting.Competing Interest StatementThe authors have declared no competing interest.},
URL = {https://www.biorxiv.org/content/early/2023/10/02/2023.09.30.560270},
eprint = {https://www.biorxiv.org/content/early/2023/10/02/2023.09.30.560270.full.pdf},
journal = {bioRxiv}
}
```
## Contact
nianlong.gu@uzh.ch