Spaces:
Running
Running
first commit
Browse files- ai_models.html +71 -0
- contact.html +82 -0
- documentation.html +110 -0
- img/contour_matching.png +0 -0
- img/index/code.png +0 -0
- img/index/comparison.png +0 -0
- img/index/ct-scan.png +0 -0
- img/index/dicom.PNG +0 -0
- img/index/edit.png +0 -0
- img/index/evaluation.png +0 -0
- img/index/innovation.png +0 -0
- img/index/parrot-logo.png +0 -0
- img/index/personal-data.png +0 -0
- img/index/research-and-development.png +0 -0
- img/python_code_editor.png +0 -0
- img/screen_ai_models_management.png +0 -0
- img/screen_patient_modeling.png +0 -0
- img/screen_plan_evaluation.png +0 -0
- img/screen_study_management.png +0 -0
- index.html +158 -17
ai_models.html
ADDED
|
@@ -0,0 +1,71 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
<!DOCTYPE html>
|
| 2 |
+
<html lang="en">
|
| 3 |
+
<head>
|
| 4 |
+
<meta charset="UTF-8">
|
| 5 |
+
<meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no">
|
| 6 |
+
<title>PARROT - Documentation</title>
|
| 7 |
+
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/css/bootstrap.min.css">
|
| 8 |
+
</head>
|
| 9 |
+
<body>
|
| 10 |
+
|
| 11 |
+
<nav class="navbar navbar-expand-lg navbar-dark bg-dark fixed-top">
|
| 12 |
+
<a class="navbar-brand" href="#">PARROT</a>
|
| 13 |
+
<button class="navbar-toggler" type="button" data-toggle="collapse" data-target="#navbarNav"
|
| 14 |
+
aria-controls="navbarNav" aria-expanded="false" aria-label="Toggle navigation">
|
| 15 |
+
<span class="navbar-toggler-icon"></span>
|
| 16 |
+
</button>
|
| 17 |
+
<div class="collapse navbar-collapse" id="navbarNav">
|
| 18 |
+
<ul class="navbar-nav">
|
| 19 |
+
<li class="nav-item">
|
| 20 |
+
<a class="nav-link" href="index.html">Home</a>
|
| 21 |
+
</li>
|
| 22 |
+
<li class="nav-item active">
|
| 23 |
+
<a class="nav-link" href="#">Documentation</a>
|
| 24 |
+
</li>
|
| 25 |
+
<li class="nav-item">
|
| 26 |
+
<a class="nav-link" href="ai_models.html">AI models</a>
|
| 27 |
+
</li>
|
| 28 |
+
<li class="nav-item">
|
| 29 |
+
<a class="nav-link" href="contact.html">Contact</a>
|
| 30 |
+
</li>
|
| 31 |
+
</ul>
|
| 32 |
+
</div>
|
| 33 |
+
</nav>
|
| 34 |
+
|
| 35 |
+
<div class="container mt-5">
|
| 36 |
+
<div class="row">
|
| 37 |
+
<div class="col-lg-8">
|
| 38 |
+
<h1 class="mb-4 mt-4">AI models</h1>
|
| 39 |
+
|
| 40 |
+
<section id="segmentation-models">
|
| 41 |
+
<h2>Segmentation models</h2>
|
| 42 |
+
</section>
|
| 43 |
+
|
| 44 |
+
<section id="dose-prediction">
|
| 45 |
+
<h2>Dose prediction</h2>
|
| 46 |
+
</section>
|
| 47 |
+
|
| 48 |
+
<section id="add-your-models">
|
| 49 |
+
<h2>Add your models</h2>
|
| 50 |
+
</section>
|
| 51 |
+
</div>
|
| 52 |
+
|
| 53 |
+
|
| 54 |
+
<div class="col-lg-4">
|
| 55 |
+
<nav id="right-menu" class="navbar navbar-light bg-light">
|
| 56 |
+
<nav class="nav flex-column">
|
| 57 |
+
<a class="nav-link" href="#segmentation-models">Segmentation models</a>
|
| 58 |
+
<a class="nav-link" href="#dose-prediction">Dose prediction</a>
|
| 59 |
+
<a class="nav-link" href="#add-your-models">Add your models</a>
|
| 60 |
+
</nav>
|
| 61 |
+
</nav>
|
| 62 |
+
</div>
|
| 63 |
+
</div>
|
| 64 |
+
</div>
|
| 65 |
+
|
| 66 |
+
<script src="https://code.jquery.com/jquery-3.5.1.slim.min.js"></script>
|
| 67 |
+
<script src="https://cdn.jsdelivr.net/npm/@popperjs/[email protected]/dist/umd/popper.min.js"></script>
|
| 68 |
+
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/js/bootstrap.min.js"></script>
|
| 69 |
+
|
| 70 |
+
</body>
|
| 71 |
+
</html>
|
contact.html
ADDED
|
@@ -0,0 +1,82 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
<!DOCTYPE html>
|
| 2 |
+
<html lang="en">
|
| 3 |
+
<head>
|
| 4 |
+
<meta charset="UTF-8">
|
| 5 |
+
<meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no">
|
| 6 |
+
<title>PARROT - Contact</title>
|
| 7 |
+
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/css/bootstrap.min.css">
|
| 8 |
+
</head>
|
| 9 |
+
<body>
|
| 10 |
+
|
| 11 |
+
<nav class="navbar navbar-expand-lg navbar-dark bg-dark fixed-top">
|
| 12 |
+
<a class="navbar-brand" href="#">PARROT</a>
|
| 13 |
+
<button class="navbar-toggler" type="button" data-toggle="collapse" data-target="#navbarNav"
|
| 14 |
+
aria-controls="navbarNav" aria-expanded="false" aria-label="Toggle navigation">
|
| 15 |
+
<span class="navbar-toggler-icon"></span>
|
| 16 |
+
</button>
|
| 17 |
+
<div class="collapse navbar-collapse" id="navbarNav">
|
| 18 |
+
<ul class="navbar-nav">
|
| 19 |
+
<li class="nav-item">
|
| 20 |
+
<a class="nav-link" href="index.html">Home</a>
|
| 21 |
+
</li>
|
| 22 |
+
<li class="nav-item">
|
| 23 |
+
<a class="nav-link" href="documentation.html">Documentation</a>
|
| 24 |
+
</li>
|
| 25 |
+
<li class="nav-item">
|
| 26 |
+
<a class="nav-link" href="ai_models.html">AI models</a>
|
| 27 |
+
</li>
|
| 28 |
+
<li class="nav-item active">
|
| 29 |
+
<a class="nav-link" href="#">Contact</a>
|
| 30 |
+
</li>
|
| 31 |
+
</ul>
|
| 32 |
+
</div>
|
| 33 |
+
</nav>
|
| 34 |
+
|
| 35 |
+
<div class="container mt-5">
|
| 36 |
+
<div class="row">
|
| 37 |
+
<div class="col-lg-8">
|
| 38 |
+
<h1>Contact Us</h1>
|
| 39 |
+
<p>If you have any questions or want to share information about your AI model, please use the form below.</p>
|
| 40 |
+
|
| 41 |
+
<form action="#" method="post">
|
| 42 |
+
<div class="form-group">
|
| 43 |
+
<label for="name">Your Name</label>
|
| 44 |
+
<input type="text" class="form-control" id="name" name="name" required>
|
| 45 |
+
</div>
|
| 46 |
+
<div class="form-group">
|
| 47 |
+
<label for="email">Your Email</label>
|
| 48 |
+
<input type="email" class="form-control" id="email" name="email" required>
|
| 49 |
+
</div>
|
| 50 |
+
<div class="form-group">
|
| 51 |
+
<label for="inputDescription">Describe Your AI Model</label>
|
| 52 |
+
<textarea class="form-control" id="inputDescription" name="description" rows="5" required></textarea>
|
| 53 |
+
</div>
|
| 54 |
+
<div class="form-group">
|
| 55 |
+
<label for="inputInput">Input Details</label>
|
| 56 |
+
<input type="text" class="form-control" id="inputInput" name="input" required>
|
| 57 |
+
</div>
|
| 58 |
+
<div class="form-group">
|
| 59 |
+
<label for="outputOutput">Output Details</label>
|
| 60 |
+
<input type="text" class="form-control" id="outputOutput" name="output" required>
|
| 61 |
+
</div>
|
| 62 |
+
<div class="form-group">
|
| 63 |
+
<label for="trainingSetSize">Training Set Size</label>
|
| 64 |
+
<input type="text" class="form-control" id="trainingSetSize" name="trainingSetSize" required>
|
| 65 |
+
</div>
|
| 66 |
+
<div class="form-group">
|
| 67 |
+
<label for="numberOfEpochs">Number of Epochs</label>
|
| 68 |
+
<input type="text" class="form-control" id="numberOfEpochs" name="numberOfEpochs" required>
|
| 69 |
+
</div>
|
| 70 |
+
|
| 71 |
+
<button type="submit" class="btn btn-primary">Submit</button>
|
| 72 |
+
</form>
|
| 73 |
+
</div>
|
| 74 |
+
</div>
|
| 75 |
+
</div>
|
| 76 |
+
|
| 77 |
+
<script src="https://code.jquery.com/jquery-3.5.1.slim.min.js"></script>
|
| 78 |
+
<script src="https://cdn.jsdelivr.net/npm/@popperjs/[email protected]/dist/umd/popper.min.js"></script>
|
| 79 |
+
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/js/bootstrap.min.js"></script>
|
| 80 |
+
|
| 81 |
+
</body>
|
| 82 |
+
</html>
|
documentation.html
ADDED
|
@@ -0,0 +1,110 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
<!DOCTYPE html>
|
| 2 |
+
<html lang="en">
|
| 3 |
+
<head>
|
| 4 |
+
<meta charset="UTF-8">
|
| 5 |
+
<meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no">
|
| 6 |
+
<title>PARROT - Documentation</title>
|
| 7 |
+
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/css/bootstrap.min.css">
|
| 8 |
+
</head>
|
| 9 |
+
<body>
|
| 10 |
+
|
| 11 |
+
<nav class="navbar navbar-expand-lg navbar-dark bg-dark fixed-top">
|
| 12 |
+
<a class="navbar-brand" href="#">PARROT</a>
|
| 13 |
+
<button class="navbar-toggler" type="button" data-toggle="collapse" data-target="#navbarNav"
|
| 14 |
+
aria-controls="navbarNav" aria-expanded="false" aria-label="Toggle navigation">
|
| 15 |
+
<span class="navbar-toggler-icon"></span>
|
| 16 |
+
</button>
|
| 17 |
+
<div class="collapse navbar-collapse" id="navbarNav">
|
| 18 |
+
<ul class="navbar-nav">
|
| 19 |
+
<li class="nav-item">
|
| 20 |
+
<a class="nav-link" href="index.html">Home</a>
|
| 21 |
+
</li>
|
| 22 |
+
<li class="nav-item active">
|
| 23 |
+
<a class="nav-link" href="#">Documentation</a>
|
| 24 |
+
</li>
|
| 25 |
+
<li class="nav-item">
|
| 26 |
+
<a class="nav-link" href="ai_models.html">AI models</a>
|
| 27 |
+
</li>
|
| 28 |
+
<li class="nav-item">
|
| 29 |
+
<a class="nav-link" href="contact.html">Contact</a>
|
| 30 |
+
</li>
|
| 31 |
+
</ul>
|
| 32 |
+
</div>
|
| 33 |
+
</nav>
|
| 34 |
+
|
| 35 |
+
<div class="container mt-5">
|
| 36 |
+
<div class="row">
|
| 37 |
+
<div class="col-lg-8">
|
| 38 |
+
<h1 class="mb-4 mt-4">Documentation</h1>
|
| 39 |
+
|
| 40 |
+
<section id="installation">
|
| 41 |
+
<h2>Installation</h2>
|
| 42 |
+
<p>Instructions for installing PARROT on your system... Warning: The Orthanc server will run on port 8042 of your local computer and the nginx server on port 2000.</p>
|
| 43 |
+
<p>The installation package includes all servers. However, the Orthanc server is a manual installation. The installation instructions can be found below.</p>
|
| 44 |
+
</section>
|
| 45 |
+
|
| 46 |
+
<section id="study-management">
|
| 47 |
+
<h2>Study Management</h2>
|
| 48 |
+
<p>Users can import patient data from this screen to a local Orthanc daemon—a lightweight, standalone DICOM server, which ensures the confidentiality of patient files. A list of patient data present in the Orthanc server is displayed with summarized information. Users can load the data into the app by clicking on the Load button. The left icon displays more complete information from DICOM tags and users can delete a patient study from the server with the red arrow on the right.</p>
|
| 49 |
+
<figure>
|
| 50 |
+
<img src="img/screen_study_management.png" alt="Study Management Image" class="img-fluid mb-3">
|
| 51 |
+
<figcaption>Figure ?: Study Management screen.</figcaption>
|
| 52 |
+
</figure>
|
| 53 |
+
</section>
|
| 54 |
+
|
| 55 |
+
<section id="ai-models-management">
|
| 56 |
+
<h2>AI Models Management</h2>
|
| 57 |
+
<figure>
|
| 58 |
+
<img src="img/screen_ai_models_management.png" alt="AI Models Management Image" class="img-fluid mb-3">
|
| 59 |
+
<figcaption>Figure ?: AI Models Management screen.</figcaption>
|
| 60 |
+
</figure>
|
| 61 |
+
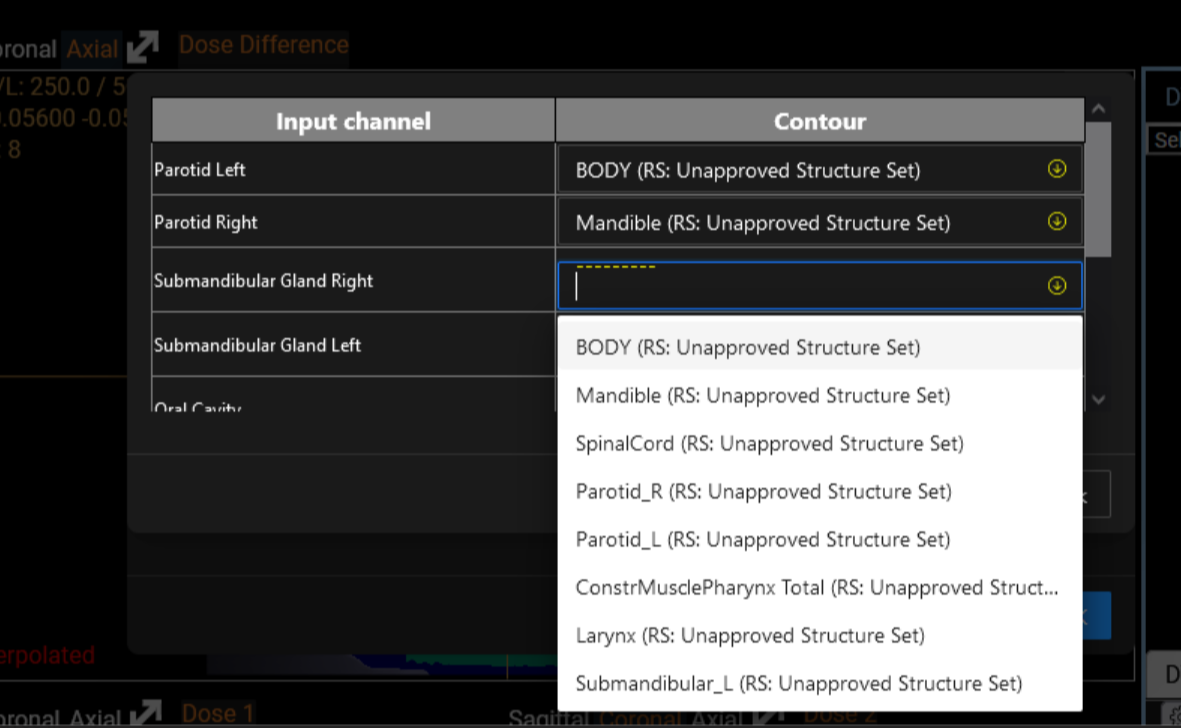
<p>Eight segmentation models and six dose prediction models are made available and listed on the screen (Figure 2). These models are trained from state-of-the-art architectures such as nnUNet, SwinUNETR and HDUNet (Table 1). On the left hand side of each line, a standardized description of the model is available. The PDF description file contains details about the architecture with a link of the original research article, details on the training process including dataset, and performance on our test set (Figure 3). Users also have the flexibility to run their own models using a Python scripting editor and display the output in the viewer (Figure 4). We distinguish two categories of models: the segmentation models and the dose prediction models. A key difference between the two categories is that segmentation models require only the anatomy of the patient (CT scan or MRI image) whereas dose prediction models also require binary masks of tumor volumes (TV) and organs at risk (OARs). For that latter, we implemented a dialog that allows users to map structures present in the current patient data and the input required for the AI dose prediction model (Figure 5).</p>
|
| 62 |
+
<iframe width="560" height="315" src="https://www.youtube.com/embed/example_video_code" frameborder="0" allowfullscreen class="mb-4"></iframe>
|
| 63 |
+
|
| 64 |
+
</section>
|
| 65 |
+
|
| 66 |
+
<section id="patient-modeling">
|
| 67 |
+
<h2>Patient Modeling</h2>
|
| 68 |
+
<figure>
|
| 69 |
+
<img src="img/screen_patient_modeling.png" alt="Patient Modeling Image" class="img-fluid mb-3">
|
| 70 |
+
<figcaption>Figure ?: Patient Modeling screen.</figcaption>
|
| 71 |
+
</figure>
|
| 72 |
+
<p>Eight segmentation models and six dose prediction models are made available and listed on the screen (Figure 3). These models are trained from state-of-the-art architectures such as nnUNet [2], SwinUNETR [3] and HDUNet [4] (Table 1). On the left hand side of each line, a standardized description of the model is available. The PDF description file contains details about the architecture with a link of the original research article, details on the training process including dataset, and performance on our test set (Figure 4).
|
| 73 |
+
|
| 74 |
+
To run a model prediction, the user simply clicks on the yellow Prediction button on the right hand side of the model line. This will open a Python scripting editor to run the code (Figure 5). Two choices are available to the user: either run the code locally or in one of the Python environments provided. The latter option provides a Python version with a set of libraries. Users can select a Python environment with the drop-down menu in the upper right corner and by hovering on it, a description of the libraries they contain is displayed. Once the prediction is done, users can upload the result to the study to visualize the output in the Patient Modeling screen (Figure 7). Users also have the flexibility to run their own models by loading their own code in the Python scripting editor. On our website, we provide a description and examples using the API to retrieve the patient information loaded on PARROT and upload the result.
|
| 75 |
+
|
| 76 |
+
We distinguish two categories of models: the segmentation models and the dose prediction models. A key difference between the two categories is that segmentation models require only the anatomy of the patient (CT scan or MRI image) whereas dose prediction models also require binary masks of tumor volumes (TV) and organs at risk (OARs). For that latter, we implemented a dialog that allows users to map structures present in the current patient data and the input required for the AI dose prediction model (Figure 6).
|
| 77 |
+
</p>
|
| 78 |
+
</section>
|
| 79 |
+
|
| 80 |
+
<section id="plan-evaluation">
|
| 81 |
+
<h2>Plan Evaluation</h2>
|
| 82 |
+
<figure>
|
| 83 |
+
<img src="img/screen_plan_evaluation.png" alt="Plan Evaluation Image" class="img-fluid mb-3">
|
| 84 |
+
<figcaption>Figure ?: Plan Evaluation screen.</figcaption>
|
| 85 |
+
</figure>
|
| 86 |
+
<p>A comprehensive display of two dose distributions comparison is presented on the last screen. The two dose distributions can originate from the importation step or result from the inference of ai models. We provide a set of tools for comparison, such as DVH curves, dose statistics, and the fulfillment of clinical goals. Clinical goals can be uploaded following an Excel template so that it fits any clinical requirements of the users institutions. The platform incorporates treatment indications based on NTCP protocols. It has been shown in multiple studies that predicted dose distributions yield meaningful output when used with NTCP models for treatment selection decision support [5] [6]. This is a useful decision support tool for the clinical community.
|
| 87 |
+
</p>
|
| 88 |
+
</section>
|
| 89 |
+
</div>
|
| 90 |
+
|
| 91 |
+
<div class="col-lg-4">
|
| 92 |
+
<nav id="right-menu" class="navbar navbar-light bg-light">
|
| 93 |
+
<nav class="nav flex-column">
|
| 94 |
+
<a class="nav-link" href="#installation">Installation</a>
|
| 95 |
+
<a class="nav-link" href="#study-management">Study Management</a>
|
| 96 |
+
<a class="nav-link" href="#ai-models-management">AI Models Management</a>
|
| 97 |
+
<a class="nav-link" href="#patient-modeling">Patient Modeling</a>
|
| 98 |
+
<a class="nav-link" href="#plan-evaluation">Plan Evaluation</a>
|
| 99 |
+
</nav>
|
| 100 |
+
</nav>
|
| 101 |
+
</div>
|
| 102 |
+
</div>
|
| 103 |
+
</div>
|
| 104 |
+
|
| 105 |
+
<script src="https://code.jquery.com/jquery-3.5.1.slim.min.js"></script>
|
| 106 |
+
<script src="https://cdn.jsdelivr.net/npm/@popperjs/[email protected]/dist/umd/popper.min.js"></script>
|
| 107 |
+
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/js/bootstrap.min.js"></script>
|
| 108 |
+
|
| 109 |
+
</body>
|
| 110 |
+
</html>
|
img/contour_matching.png
ADDED
|
img/index/code.png
ADDED

|
img/index/comparison.png
ADDED

|
img/index/ct-scan.png
ADDED

|
img/index/dicom.PNG
ADDED
|
img/index/edit.png
ADDED

|
img/index/evaluation.png
ADDED

|
img/index/innovation.png
ADDED
|
img/index/parrot-logo.png
ADDED

|
img/index/personal-data.png
ADDED

|
img/index/research-and-development.png
ADDED
|
img/python_code_editor.png
ADDED

|
img/screen_ai_models_management.png
ADDED

|
img/screen_patient_modeling.png
ADDED

|
img/screen_plan_evaluation.png
ADDED

|
img/screen_study_management.png
ADDED

|
index.html
CHANGED
|
@@ -1,19 +1,160 @@
|
|
| 1 |
<!DOCTYPE html>
|
| 2 |
-
<html>
|
| 3 |
-
|
| 4 |
-
|
| 5 |
-
|
| 6 |
-
|
| 7 |
-
|
| 8 |
-
|
| 9 |
-
|
| 10 |
-
|
| 11 |
-
|
| 12 |
-
|
| 13 |
-
|
| 14 |
-
|
| 15 |
-
|
| 16 |
-
|
| 17 |
-
|
| 18 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 19 |
</html>
|
|
|
|
| 1 |
<!DOCTYPE html>
|
| 2 |
+
<html lang="en">
|
| 3 |
+
<head>
|
| 4 |
+
<meta charset="UTF-8">
|
| 5 |
+
<meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no">
|
| 6 |
+
<title>PARROT - Platform for AI-guided Radiation Oncology Treatment</title>
|
| 7 |
+
<script src="https://code.jquery.com/jquery-3.6.0.min.js"></script>
|
| 8 |
+
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/css/bootstrap.min.css">
|
| 9 |
+
<style>
|
| 10 |
+
body {
|
| 11 |
+
padding-top: 56px;
|
| 12 |
+
}
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
</style>
|
| 16 |
+
</head>
|
| 17 |
+
<body>
|
| 18 |
+
|
| 19 |
+
<nav class="navbar navbar-expand-lg navbar-dark bg-dark fixed-top">
|
| 20 |
+
<a class="navbar-brand" href="#">PARROT</a>
|
| 21 |
+
<button class="navbar-toggler" type="button" data-toggle="collapse" data-target="#navbarNav"
|
| 22 |
+
aria-controls="navbarNav" aria-expanded="false" aria-label="Toggle navigation">
|
| 23 |
+
<span class="navbar-toggler-icon"></span>
|
| 24 |
+
</button>
|
| 25 |
+
<div class="collapse navbar-collapse" id="navbarNav">
|
| 26 |
+
<ul class="navbar-nav">
|
| 27 |
+
<li class="nav-item active">
|
| 28 |
+
<a class="nav-link" href="#">Home</a>
|
| 29 |
+
</li>
|
| 30 |
+
<li class="nav-item">
|
| 31 |
+
<a class="nav-link" href="documentation.html">Documentation</a>
|
| 32 |
+
</li>
|
| 33 |
+
<li class="nav-item">
|
| 34 |
+
<a class="nav-link" href="ai_models.html">AI models</a>
|
| 35 |
+
</li>
|
| 36 |
+
<li class="nav-item">
|
| 37 |
+
<a class="nav-link" href="contact.html">Contact</a>
|
| 38 |
+
</li>
|
| 39 |
+
</ul>
|
| 40 |
+
</div>
|
| 41 |
+
</nav>
|
| 42 |
+
|
| 43 |
+
<div class="container mt-5">
|
| 44 |
+
<div class="row">
|
| 45 |
+
<div class="col-lg-8">
|
| 46 |
+
<h1>Welcome to PARROT</h1>
|
| 47 |
+
<p>
|
| 48 |
+
PARROT, which stands for Platform for ARtificial intelligence guided Radiation Oncology Treatment,
|
| 49 |
+
is a user-friendly, free, and open-source web platform. It allows users to visualize DICOM files, run AI models,
|
| 50 |
+
display and evaluate predictions easily. The platform includes several trained state-of-the-art dose prediction
|
| 51 |
+
and contour segmentation models. Users can also add their own models using the embedded code editor.
|
| 52 |
+
</p>
|
| 53 |
+
<p>
|
| 54 |
+
The app consists of a frontend built in React JavaScript, including the graphical interface with a DICOM viewer,
|
| 55 |
+
a selection panel of AI models, an editor of contours predictions, and evaluation tools. The backend is built with Flask
|
| 56 |
+
to handle the predictions of AI models.
|
| 57 |
+
</p>
|
| 58 |
+
</div>
|
| 59 |
+
<div class="col">
|
| 60 |
+
<img src="img/index/parrot-logo.png" alt="PARROT Logo" class="index/parrot-logo img-fluid">
|
| 61 |
+
</div>
|
| 62 |
+
</div>
|
| 63 |
+
<div class="col-lg text-center">
|
| 64 |
+
<button id="downloadButton" class="btn btn-primary">Download Latest Release</button>
|
| 65 |
+
</div>
|
| 66 |
+
</div>
|
| 67 |
+
<div class="container mt-5">
|
| 68 |
+
<div class="row">
|
| 69 |
+
<div class="col text-center mb-5">
|
| 70 |
+
<h2>
|
| 71 |
+
PARROT allows you to:
|
| 72 |
+
</h2>
|
| 73 |
+
</div>
|
| 74 |
+
</div>
|
| 75 |
+
<div class="row">
|
| 76 |
+
<div class="col-lg-3 text-center">
|
| 77 |
+
<img src="img/index/research-and-development.png" alt="State-of-the-art models" class="img-fluid w-50 mb-3">
|
| 78 |
+
<p>Run state-of-the-art AI models</p>
|
| 79 |
+
</div>
|
| 80 |
+
<div class="col-lg-3 text-center">
|
| 81 |
+
<img src="img/index/ct-scan.png" alt="ct scan" class="img-fluid w-50 mb-3">
|
| 82 |
+
<p>Visualize contours segmented with AI on CT and MR images</p>
|
| 83 |
+
</div>
|
| 84 |
+
<div class="col-lg-3 text-center">
|
| 85 |
+
<img src="img/index/comparison.png" alt="ct scan" class="img-fluid w-50 mb-3">
|
| 86 |
+
<p>Compare uploaded and/or predicted dose distributions</p>
|
| 87 |
+
</div>
|
| 88 |
+
<div class="col-lg-3 text-center">
|
| 89 |
+
<img src="img/index/edit.png" alt="ct scan" class="img-fluid w-50 mb-3">
|
| 90 |
+
<p>Correct contours predicted by AI model</p>
|
| 91 |
+
</div>
|
| 92 |
+
</div>
|
| 93 |
+
<div class="row">
|
| 94 |
+
<div class="col-lg-3 text-center">
|
| 95 |
+
<img src="img/index/dicom.PNG" alt="ct scan" class="img-fluid w-50 mb-3">
|
| 96 |
+
<p>Full DICOM to DICOM workflow</p>
|
| 97 |
+
</div>
|
| 98 |
+
<div class="col-lg-3 text-center">
|
| 99 |
+
<img src="img/index/personal-data.png" alt="ct scan" class="img-fluid w-50 mb-3">
|
| 100 |
+
<p>Local web application to ensure patient data privacy</p>
|
| 101 |
+
</div>
|
| 102 |
+
<div class="col-lg-3 text-center">
|
| 103 |
+
<img src="img/index/innovation.png" alt="ct scan" class="img-fluid w-50 mb-3">
|
| 104 |
+
<p>Run your own AI models</p>
|
| 105 |
+
</div>
|
| 106 |
+
<div class="col-lg-3 text-center">
|
| 107 |
+
<img src="img/index/evaluation.png" alt="ct scan" class="img-fluid w-50 mb-3">
|
| 108 |
+
<p>Clinical evaluation tools: NTCP, DVH curves, clinical objectives, dose statistics</p>
|
| 109 |
+
</div>
|
| 110 |
+
</div>
|
| 111 |
+
|
| 112 |
+
|
| 113 |
+
|
| 114 |
+
</div>
|
| 115 |
+
|
| 116 |
+
|
| 117 |
+
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/js/bootstrap.min.js"></script>
|
| 118 |
+
|
| 119 |
+
<script>
|
| 120 |
+
$(document).ready(function () {
|
| 121 |
+
// Replace 'YOUR_PROJECT_ID' and 'YOUR_REPO_NAME' with your GitLab project ID and repository name
|
| 122 |
+
var projectId = '25472370';
|
| 123 |
+
var repoName = 'charp-viewer';
|
| 124 |
+
// https://gitlab.com/zhaosadre/charp-viewer/-/releases -->
|
| 125 |
+
// GitLab API endpoint for releases
|
| 126 |
+
var apiUrl = 'https://gitlab.com/api/v4/projects/'+projectId+'/releases/'//+tag_name+'/assets/links'
|
| 127 |
+
//var apiUrl = 'https://gitlab.com/api/v4/projects/' + projectId + '/repository/tags'; -->
|
| 128 |
+
|
| 129 |
+
// Fetch the latest release information
|
| 130 |
+
|
| 131 |
+
$.ajax({
|
| 132 |
+
url: apiUrl,
|
| 133 |
+
method: 'GET',
|
| 134 |
+
success: function (data) {
|
| 135 |
+
console.log("data",data);
|
| 136 |
+
if (data.length > 0) {
|
| 137 |
+
var latestRelease = data[0];
|
| 138 |
+
console.log("latestRelease",latestRelease);
|
| 139 |
+
var releaseAssets = latestRelease.assets;
|
| 140 |
+
console.log("releaseAssets",releaseAssets);
|
| 141 |
+
// Assuming the first asset is the desired download (e.g., a zip file)
|
| 142 |
+
var downloadUrl = releaseAssets.sources.length > 0 ? releaseAssets.sources[0].url : '';
|
| 143 |
+
|
| 144 |
+
// Update the download button href attribute
|
| 145 |
+
$('#downloadButton').attr('onclick', 'window.location.href="' + downloadUrl + '"');
|
| 146 |
+
}
|
| 147 |
+
},
|
| 148 |
+
error: function (error) {
|
| 149 |
+
console.error('Error fetching release information:', error);
|
| 150 |
+
}
|
| 151 |
+
});
|
| 152 |
+
});
|
| 153 |
+
</script>
|
| 154 |
+
|
| 155 |
+
<script src="https://code.jquery.com/jquery-3.6.0.min.js"></script>
|
| 156 |
+
<script src="https://cdn.jsdelivr.net/npm/@popperjs/[email protected]/dist/umd/popper.min.js"></script>
|
| 157 |
+
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.5.2/js/bootstrap.min.js"></script>
|
| 158 |
+
|
| 159 |
+
</body>
|
| 160 |
</html>
|