Upload 7 files
Browse files
documentation/architecture.png
ADDED
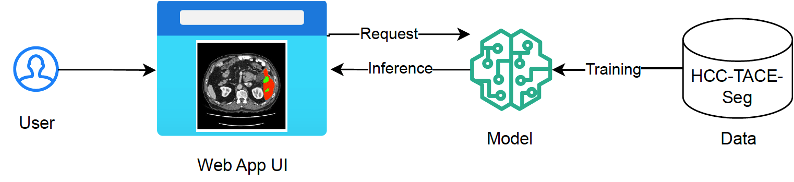
|
documentation/deep_learning_model.md
ADDED
|
@@ -0,0 +1,38 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Deep Learning Model for Liver Tumor Segmentation
|
| 2 |
+
|
| 3 |
+
Deep learning algorithms has showed promise for producing automat-ed liver and tumor segmentation. While most algorithms achieved excellent per-formance for liver segmentation (0.95 dice score), how to improve the perfor-mance on liver tumor segmentation is still an active area of research (0.65 dice score).
|
| 4 |
+
|
| 5 |
+
This work tried several innovative techniques attempting to improve liver tumor segmentation, including: a
|
| 6 |
+
|
| 7 |
+
* Global and Local Segmentation Pipeline: We design a sequential pipeline that starts with a global-view liver segmentation model followed by a localview tumor segmentation model. We show that this two-step approach is more flexible than a multi-class approach, allowing preprocessing, training, and post-processing to be optimized for each segmentation task.
|
| 8 |
+
|
| 9 |
+
* Weak Supervision Incorporation from Clinical Data: Leveraging clinical data, we derive an indicator of tumor severity at the population level. This indicator serves to weakly supervise our tumor segmentation model, which is trained on patient-specific CT scans. By integrating this population-level insight, we regularize the model's learning process, effectively minimizing the risk of overfitting.
|
| 10 |
+
|
| 11 |
+
* Active Tumor ContoAur Adjustment: We employ a morphological algorithm to refine the segmented tumor boundaries in the post-processing stage, improving the segmentation results for tumors with imprecise margins.
|
| 12 |
+
|
| 13 |
+
## Data
|
| 14 |
+
|
| 15 |
+
We utilized the HCC-TACE-Seg dataset that is publicly available from the Cancer
|
| 16 |
+
Imaging Archive (TCIA). This dataset contains pre-procedural CT scans of liver tumors from 98 patients along with corresponding segmentation labels for five classes of objects (normal region, liver, tumor mass, portal vein, and abdominal aorta). Each CT scan has spatial dimensions 512 x 512 x N, where the number of slices N varies by patient. Additionally, a clinical data file containing demographics, related medical history and diagnosis is provided in HCC-TACE-Seg.
|
| 17 |
+
|
| 18 |
+
## Data Preprocessing
|
| 19 |
+

|
| 20 |
+
|
| 21 |
+
## Model
|
| 22 |
+
|
| 23 |
+
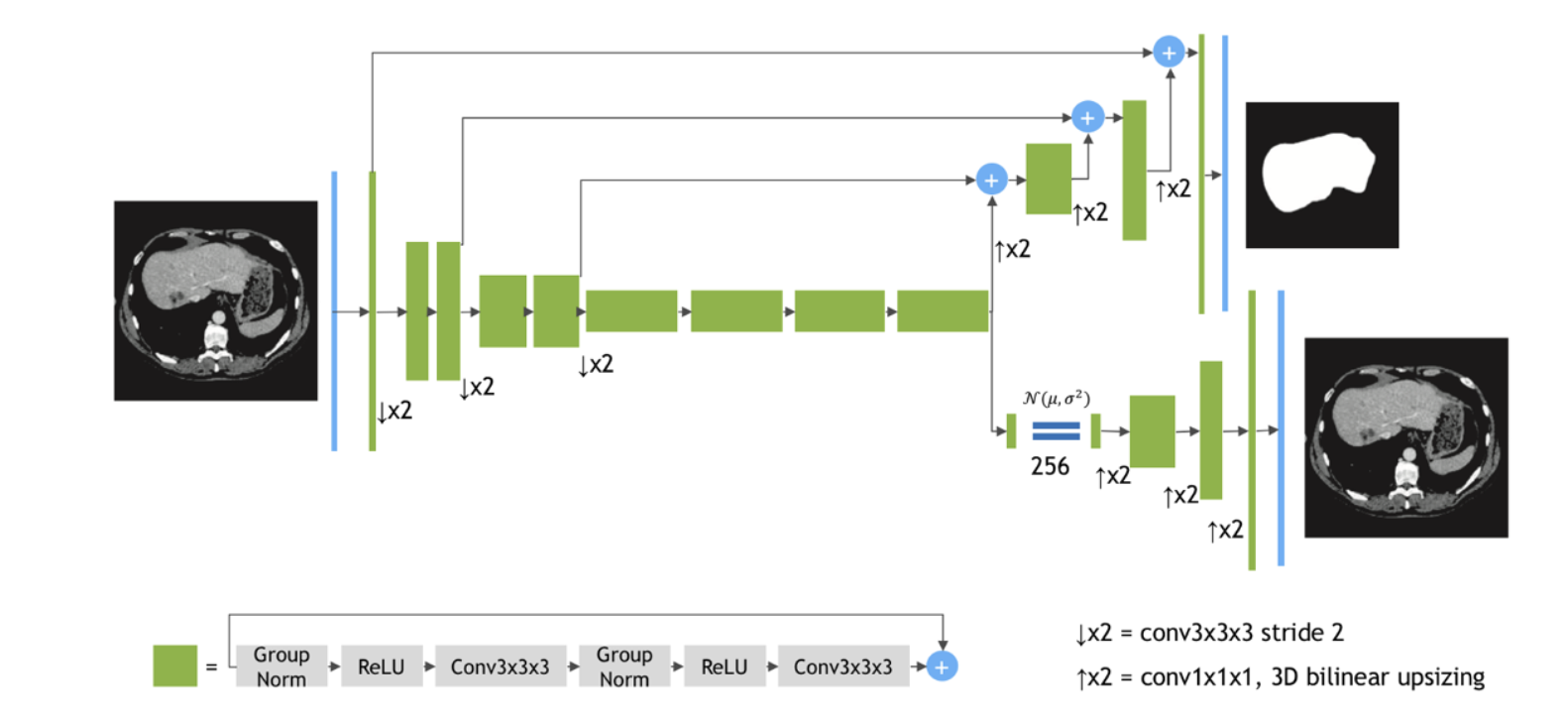
We used the SegResNetVAE as the backbone model. This model is built upon an asymmetrical encoderdecoder
|
| 24 |
+
based Convolutional Neural Networks (CNNs) architecture. It features an encoder designed for comprehensive image feature extraction and a compact decoder to reconstruct segmentation masks. To augment this structure, an additional variational auto-encoder (VAE) branch is integrated at the encoder's endpoint. The architecture is shown in figure below.
|
| 25 |
+
|
| 26 |
+

|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
## Results
|
| 30 |
+
|
| 31 |
+
Using randomly sampled 80% of patients as training and 20% as test, our model achieved the following test performance on the dataset:
|
| 32 |
+
|
| 33 |
+
| | Accuracy |Dice| |Sensitivity |IoU|
|
| 34 |
+
| -------- | ------- |------- |------- |------- |
|
| 35 |
+
|Liver |0.994 |0.942| 0.955 |0.895|
|
| 36 |
+
|Tumor |0.995 |0.570| 0.801 |0.460|
|
| 37 |
+
|
| 38 |
+
|
documentation/preprocessing.png
ADDED

|
documentation/requirements.txt
ADDED
|
@@ -0,0 +1,8 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
gradio==4.27.0
|
| 2 |
+
torchvision==0.17.2
|
| 3 |
+
monai==1.3.0
|
| 4 |
+
pynrrd==1.0.0
|
| 5 |
+
nibabel==5.2.1
|
| 6 |
+
scikit-image==0.23.2
|
| 7 |
+
morphsnakes==2.0.1
|
| 8 |
+
opencv-python==4.9.0.80
|
documentation/research.md
ADDED
|
@@ -0,0 +1,36 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Liver Cancer
|
| 2 |
+
|
| 3 |
+
Liver cancer is a significant global health concern, with more than 800,000 people diagnosed with this cancer each year worldwide and with primary liver cancer being a leading cause of cancer deaths worldwide (American Cancer Society, 2024). Liver is also a common destination for metastatic cancer cells originating from various abdominal organs, including the colon, rectum, pancreas, as well as distant organs such as the breast and lung. Consequently, meticulous examination of the liver and its lesions is integral to comprehensive tumor staging and management strategies. Standard protocols for tumor assessment, such as the Response Evaluation Criteria in Solid Tumor (RECIST), necessitate precise measurement of the diameter of the largest target lesion (Eisenhauer et al., 2009). Therefore, achieving accurate localization and precise segmentation of liver tumors within CT scans is imperative for the diagnosis, treatment planning, and monitoring of treatment response in patients with liver cancer (Shiina et al., 2018; Virdis et al., 2019).
|
| 4 |
+
|
| 5 |
+
## Deep Learning for Liver Tumor Segmentation
|
| 6 |
+
|
| 7 |
+
Manual delineation of target lesions in CT scans is fraught with challenges, being both time-consuming and prone to poor reproducibility and operator-dependent variability. Automated liver tumor segmentation can provide clinicians with rapid and consistent tumor delineation. Recently, deep learning algorithms have showed promise for producing automated liver and tumor segmentation (Gul et al., 2002). While most algorithms
|
| 8 |
+
achieved exceptional performance in liver segmentation, with dice scores ranging from 0.90 to 0.96, enhancing liver tumor segmentation remains a challenge, currently standing at dice scores from 0.41 to 0.67 according to the recent Liver Tumor Segmentation Benchmark (LiTS) (Bilic et al.,2023).
|
| 9 |
+
|
| 10 |
+
## Challenges in Liver Tumor Segmentation
|
| 11 |
+
|
| 12 |
+
Liver tumor segmentation is an inherently challenging task. Tumors vary significantly in size, shape, and location across different patients, leading to a wide range of tumor characteristics and making it challenging for models to generalize (Sabir et al., 2022). Moreover, margins of some tumors are imprecise as CT scans exhibit low gentile brightness and roughness making it difficult to distinguish between tumor and healthy tissue (Sabir et al., 2022).
|
| 13 |
+
|
| 14 |
+
# Similar Tools on AI-assisted Segmentation
|
| 15 |
+
|
| 16 |
+
In recent years, AI-assisted segmentation has been trendy in research and industry.
|
| 17 |
+
|
| 18 |
+
A notable publicaly available online tool is [MedSeg](https://htmlsegmentation.s3.eu-north-1.amazonaws.com/index.html). This tool provides a number of pre-trained models to perform segmentation as well as annotate, edit, and save their segmentations. The tool has capability for organ segmentation of liver, lung, spleen, pancreas, and ventricles.
|
| 19 |
+
|
| 20 |
+
Another great work is [MD.ai Annotator](https://md.ai/) that provides web-based annotation tools optimized for medical deep learning. It allows real-time collaboration within a team. The users can easily export the annotated images or prototype deep learning models using annotated data.
|
| 21 |
+
|
| 22 |
+
None of the existing tools has a focus on liver tumor segmentation, which is a very challenging task still demanding research for more accurate models.
|
| 23 |
+
|
| 24 |
+
|
| 25 |
+
# References
|
| 26 |
+
|
| 27 |
+
1. American Cancer Society. (2024). Key Statistics About Liver Cancer. All about Cancer. https://www.cancer.org/cancer/types/liver-cancer/about/what-is-key-statistics.html
|
| 28 |
+
2. Bilic, P., Christ, P., Li, H. B., Vorontsov, E., Ben-Cohen, A., Kaissis, G., Szeskin, A., Jacobs, C., Mamani, G. E. H., Chartrand, G., Lohöfer, F., Holch, J. W., Sommer, W., Hofmann, F., Hostettler, A., Lev-Cohain, N., Drozdzal, M., Amitai, M. M., Vivanti, R., … Menze, B. (2023). The Liver Tumor Seg-mentation Benchmark (LiTS). Medical Image Analysis, 84, 102680. https://doi.org/10.1016/j.media.2022.102680
|
| 29 |
+
3. Eisenhauer, E. A., Therasse, P., Bogaerts, J., Schwartz, L. H., Sargent, D., Ford, R., Dancey, J., Arbuck, S., Gwyther, S., Mooney, M., Rubinstein, L., Shankar, L., Dodd, L., Kaplan, R., Lacombe, D., & Verweij, J. (2009). New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). European Journal of Cancer, 45(2), 228–247. https://doi.org/10.1016/j.ejca.2008.10.026
|
| 30 |
+
4. Moawad, A. W., Fuentes, D., Morshid, A., Khalaf, A. M., Elmohr, M. M., Abusaif, A., Hazle, J. D., Kaseb, A. O., Hassan, M., Mahvash, A., Szklaruk, J., Qayyom, A., & Elsayes, K. (2021). Multimodality annotated HCC cases with and without advanced imaging segmentation (Version 1) [dataset]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.5FNA-0924
|
| 31 |
+
5. Moghbel, M., Mashohor, S., Mahmud, R., & Saripan, M. I. B. (2018). Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artificial Intelligence Review, 50(4), 497–537. https://doi.org/10.1007/s10462-017-9550-x
|
| 32 |
+
6. Shiina, S., Sato, K., Tateishi, R., Shimizu, M., Ohama, H., Hatanaka, T., Takawa, M., Nagamatsu, H., & Imai, Y. (2018). Percutaneous Ablation for Hepatocellular Carcinoma: Comparison of Various Ablation Techniques and Surgery. Canadian Journal of Gastroenterology and Hepatology, 2018, e4756147. https://doi.org/10.1155/2018/4756147
|
| 33 |
+
7. Virdis, F., Reccia, I., Di Saverio, S., Tugnoli, G., Kwan, S. H., Kumar, J., Atzeni, J., & Podda, M. (2019). Clinical outcomes of primary arterial embo-lization in severe hepatic trauma: A systematic review. Diagnostic and Inter-ventional Imaging, 100(2), 65–75. https://doi.org/10.1016/j.diii.2018.10.004
|
| 34 |
+
|
| 35 |
+
|
| 36 |
+
|
documentation/segresnetvae.png
ADDED
|
documentation/user_manual.md
ADDED
|
@@ -0,0 +1,71 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# User Instructions Manual
|
| 2 |
+
|
| 3 |
+
This tool is designed to assist in the identification, segmentation, and analysis of lung and tumor from medical images. By uploading a CT scan image, a pre-trained machine learning model will automatically segment the lung and tumor regions. Segmented tumor's characteristics such as shape, size, and location are then analyzed to produce an AI-generated diagnosis report of the lung cancer.
|
| 4 |
+
|
| 5 |
+
|
| 6 |
+
## Deployment
|
| 7 |
+
|
| 8 |
+
### Run locally
|
| 9 |
+
|
| 10 |
+
To locally deploy this tool, you need to follow these steps:
|
| 11 |
+
1. (optional) Setup a local environment. This can be done in multiple ways. For example, via Python's [venv](https://realpython.com/python-virtual-environments-a-primer/) module:
|
| 12 |
+
```python -m venv .venv```
|
| 13 |
+
```source .venv/bin/activate```
|
| 14 |
+
|
| 15 |
+
2. Install required dependencies. Note that Python>=3.11 is required.
|
| 16 |
+
```pip install -r requirements.txt ```
|
| 17 |
+
|
| 18 |
+
3. Launch the app by running. Your app will be running on local URL http://127.0.0.1:7860.
|
| 19 |
+
```python app.py```
|
| 20 |
+
|
| 21 |
+
If you want to create a shareable link, you can edit the following line in app.py:
|
| 22 |
+
```app.launch(share=True)```
|
| 23 |
+
|
| 24 |
+
|
| 25 |
+
### Deploy through HF Hosting
|
| 26 |
+
|
| 27 |
+
Hugging Faces (HF) has a nice integration with Gradio. This tool has been deployed through HF Spaces by running the following steps:
|
| 28 |
+
|
| 29 |
+
1. Create a free HF account
|
| 30 |
+
2. Create a repo in HF Spaces for this project
|
| 31 |
+
3. Git clone this repo in a local directory
|
| 32 |
+
4. (Option 1) From the terminal and at the project directory run
|
| 33 |
+
```gradio deploy```
|
| 34 |
+
5. (Option 2) From the browser, simply drag and drop all directory files including the requirements.txt. This will automatically deploy the model.
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
### Deploy through Github Pages
|
| 38 |
+
|
| 39 |
+
1. Create a Github Pages
|
| 40 |
+
2. Add the following embed to the HTML file:
|
| 41 |
+
```
|
| 42 |
+
<script
|
| 43 |
+
type="module"
|
| 44 |
+
src="https://gradio.s3-us-west-2.amazonaws.com/4.27.0/gradio.js"
|
| 45 |
+
></script>
|
| 46 |
+
```
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
## Quick Start
|
| 50 |
+
|
| 51 |
+
Using this tool consists of four simple steps:
|
| 52 |
+
|
| 53 |
+
1. **Upload a CT scan image**. Currently we only accept files in .nrrd format. Alternatively, the user can select one of the example images from the bottom of the page. Once selected, click 'Upload' button. The image will be displayed in the image viewer.
|
| 54 |
+
|
| 55 |
+
2. **Generate segmentation**. In the multi-choice menu, select either 'tumor' and/or 'liver' to generate tumor and/or liver segmentations, Click 'Generate Segmentation' to run inference using the pretrained deep learning model. This may take a few minutes.
|
| 56 |
+
|
| 57 |
+
3. **Visualize and download segmentations**. Once completed, the segmented masks can be visualized in the image viewer overlaid on the images. The user can use the slicer to view different z-axis slices. The buttons 'Download liver mask' and 'Download tumor mask' can be used to export the segmentations as .npy files.
|
| 58 |
+
|
| 59 |
+
4. **Generate AI diagnostic report**. Lastly, clicking the 'Generate summary' button can generate an GPT-generated summary report for the image.
|
| 60 |
+
|
| 61 |
+
|
| 62 |
+
## Main Features
|
| 63 |
+
|
| 64 |
+
This tool consists of two main features:
|
| 65 |
+
|
| 66 |
+
* **Liver and tumor segmentation**. Given a 3D CT scan, two pre-trained deep learning model are run sequentially, one generating segmentation of liver and the other generaitng segmentation of tumor within the liver. These models were developed in-house using ~100 patients from the [HCC-TASE-Seg dataset](https://www.cancerimagingarchive.net/collection/hcc-tace-seg/). The models achieved 0.954 dice score for lung segmentation and 0.570 dice score for tumor segmentation. The source code of model development can be found under ```model_development/run_best_model_notebook.ipynb```.
|
| 67 |
+
|
| 68 |
+
* **AI-generated summary report**. Given the segmented liver and tumor masks, the model extracts tumor descriptors including leiosn size (cm), lesion, shape, lesion density, and involvement of adjacent organs. These information are given to a GPT model to generate a summary of lung cancer diagnosis.
|
| 69 |
+
|
| 70 |
+
|
| 71 |
+
|