metadata
license: apache-2.0
tags:
- 3D medical CLIP
- Image-text retrieval
metrics:
- accuracy
pipeline_tag: image-feature-extraction
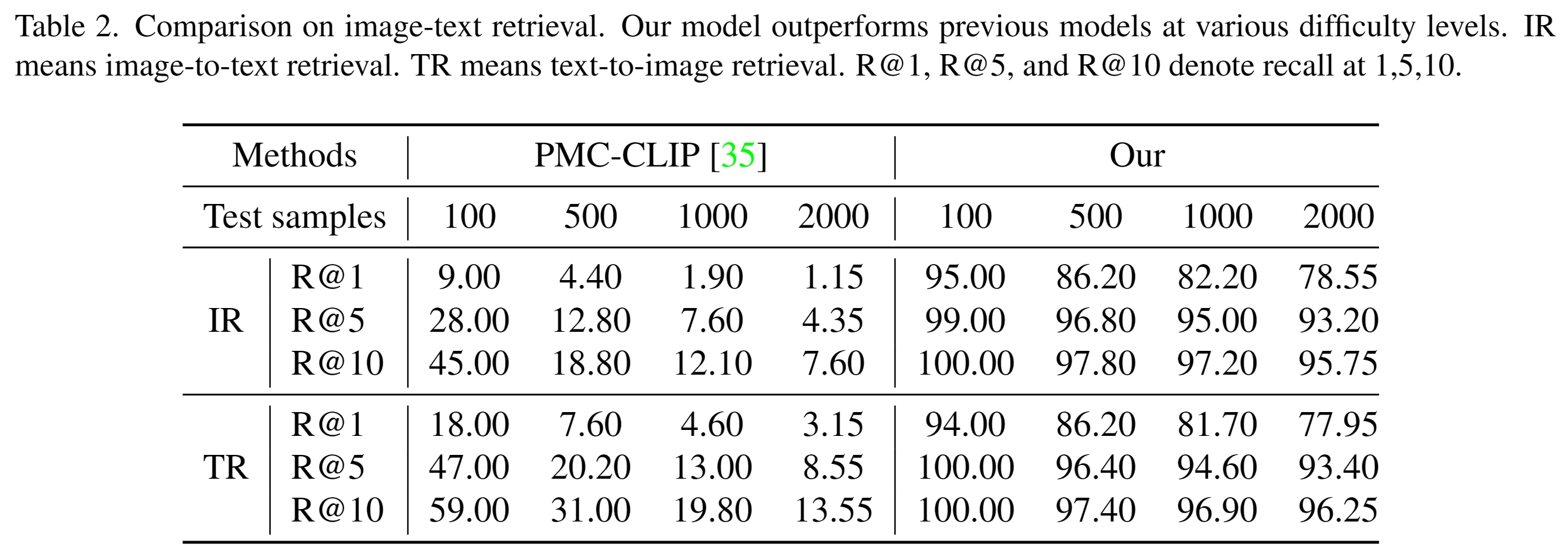
M3D-CLIP is one of the works in the M3D series. It is a 3D medical CLIP model that aligns vision and language through contrastive loss on the M3D-Cap dataset. The vision encoder uses 3D ViT with 32*256*256 image size and 4*16*16 patch size. The language encoder utilizes a pre-trained BERT as initialization.
The uses of M3D-CLIP:
- 3D medical image and text retrieval task.
- Aligned and powerful image and text features for downstream tasks.
- Text-aligned visual encoders are excellent pre-trained models for visual and multi-modal tasks.
Quickstart
device = torch.device("cuda") # or cpu
tokenizer = AutoTokenizer.from_pretrained(
"GoodBaiBai88/M3D-CLIP",
model_max_length=512,
padding_side="right",
use_fast=False
)
model = AutoModel.from_pretrained(
"GoodBaiBai88/M3D-CLIP",
trust_remote_code=True
)
model = model.to(device=device)
# Prepare your 3D medical image:
# 1. The image shape needs to be processed as 1*32*256*256, considering resize and other methods.
# 2. The image needs to be normalized to 0-1, considering Min-Max Normalization.
# 3. The image format needs to be converted to .npy
# 4. Although we did not train on 2D images, in theory, the 2D image can be interpolated to the shape of 1*32*256*256 for input.
image_path = ""
input_txt = ""
text_tensor = tokenizer(input_txt, max_length=512, truncation=True, padding="max_length", return_tensors="pt")
input_id = text_tensor["input_ids"].to(device=device)
attention_mask = text_tensor["attention_mask"].to(device=device)
image = np.load(image_path).to(device=device)
with torch.inference_mode():
image_features = model.encode_image(image)[:, 0]
text_features = model.encode_text(input_id, attention_mask)[:, 0]
Citation
If you feel helpful from our work, please consider citing the following work:
@misc{bai2024m3d,
title={M3D: Advancing 3D Medical Image Analysis with Multi-Modal Large Language Models},
author={Fan Bai and Yuxin Du and Tiejun Huang and Max Q. -H. Meng and Bo Zhao},
year={2024},
eprint={2404.00578},
archivePrefix={arXiv},
primaryClass={cs.CV}
}