repo
stringclasses 358
values | pull_number
int64 6
67.9k
| instance_id
stringlengths 12
49
| issue_numbers
sequencelengths 1
7
| base_commit
stringlengths 40
40
| patch
stringlengths 87
101M
| test_patch
stringlengths 72
22.3M
| problem_statement
stringlengths 3
256k
| hints_text
stringlengths 0
545k
| created_at
stringlengths 20
20
| PASS_TO_PASS
sequencelengths 0
0
| FAIL_TO_PASS
sequencelengths 0
0
|
|---|---|---|---|---|---|---|---|---|---|---|---|
huggingface/datasets | 3,019 | huggingface__datasets-3019 | [
"3010"
] | 592f1fd2141b88906cc7c75e0b742f6f5c48e605 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -2347,7 +2347,7 @@ def init_buffer_and_writer():
return self
@transmit_format
- @fingerprint_transform(inplace=False, ignore_kwargs=["load_from_cache_file", "cache_file_name"], version="2.0.0")
+ @fingerprint_transform(inplace=False, ignore_kwargs=["load_from_cache_file", "cache_file_name"], version="2.0.1")
def filter(
self,
function: Optional[Callable] = None,
@@ -2413,7 +2413,9 @@ def filter(
raise ValueError("Parameter `remove_columns` passed to .filter() is no longer supported.")
indices = self.map(
- function=partial(get_indices_from_mask_function, function, batched, with_indices, input_columns),
+ function=partial(
+ get_indices_from_mask_function, function, batched, with_indices, input_columns, self._indices
+ ),
with_indices=True,
features=Features({"indices": Value("uint64")}),
batched=True,
@@ -3607,6 +3609,7 @@ def get_indices_from_mask_function(
batched: bool,
with_indices: bool,
input_columns: Optional[Union[str, List[str]]],
+ indices_mapping: Optional[Table] = None,
*args,
**fn_kwargs,
):
@@ -3635,4 +3638,9 @@ def get_indices_from_mask_function(
mask.append(
function(*input, indices[i], **fn_kwargs) if with_indices else function(*input, **fn_kwargs)
)
- return {"indices": [i for i, to_keep in zip(indices, mask) if to_keep]}
+ indices_array = [i for i, to_keep in zip(indices, mask) if to_keep]
+ if indices_mapping is not None:
+ indices_array = pa.array(indices_array, type=pa.uint64())
+ indices_array = indices_mapping.column(0).take(indices_array)
+ indices_array = indices_array.to_pylist()
+ return {"indices": indices_array}
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -1175,6 +1175,15 @@ def test_filter(self, in_memory):
self.assertNotEqual(dset_filter_even_num._fingerprint, fingerprint)
self.assertEqual(dset_filter_even_num.format["type"], "numpy")
+ def test_filter_with_indices_mapping(self, in_memory):
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ dset = Dataset.from_dict({"col": [0, 1, 2]})
+ with self._to(in_memory, tmp_dir, dset) as dset:
+ with dset.filter(lambda x: x["col"] > 0) as dset:
+ self.assertListEqual(dset["col"], [1, 2])
+ with dset.filter(lambda x: x["col"] < 2) as dset:
+ self.assertListEqual(dset["col"], [1])
+
def test_filter_fn_kwargs(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict({"id": range(10)}) as dset:
| Chain filtering is leaking
## Describe the bug
As there's no support for lists within dataset fields, I convert my lists to json-string format. However, the bug described is occurring even when the data format is 'string'.
These samples show that filtering behavior diverges from what's expected when chaining filterings.
On sample 2 the second filtering leads to "leaking" of data that should've been filtered on the first filtering into the results.
## Steps to reproduce the bug
Sample 1:
```python
import datasets
import json
items = [[1, 2], [3], [4]]
jsoned_items = map(json.dumps, [[1, 2], [3], [4]])
ds = datasets.Dataset.from_dict({'a': jsoned_items})
print(list(ds))
# > Prints: [{'a': '[1, 2]'}, {'a': '[3]'}, {'a': '[4]'}] as expected
filtered = ds
# get all lists that are shorter than 2
filtered = filtered.filter(lambda x: len(json.loads(x['a'])) < 2, load_from_cache_file=False)
print(list(filtered))
# > Prints: [{'a': '[3]'}, {'a': '[4]'}] as expected
# get all lists, which have a value bigger than 3 on its zero index
filtered = filtered.filter(lambda x: json.loads(x['a'])[0] > 3, load_from_cache_file=False)
print(list(filtered))
# > Should be: [{'a': [4]}]
# > Prints: [{'a': [3]}]
```
Sample 2:
```python
import datasets
import json
items = [[1, 2], [3], [4]]
jsoned_items = map(json.dumps, [[1, 2], [3], [4]])
ds = datasets.Dataset.from_dict({'a': jsoned_items})
print(list(ds))
# > Prints: [{'a': '[1, 2]'}, {'a': '[3]'}, {'a': '[4]'}]
filtered = ds
# get all lists, which have a value bigger than 3 on its zero index
filtered = filtered.filter(lambda x: json.loads(x['a'])[0] > 3, load_from_cache_file=False)
print(list(filtered))
# > Prints: [{'a': '[4]'}] as expected
# get all lists that are shorter than 2
filtered = filtered.filter(lambda x: len(json.loads(x['a'])) < 2, load_from_cache_file=False)
print(list(filtered))
# > Prints: [{'a': '[1, 2]'}]
# > Should be: [{'a': '[4]'}] (remain intact)
```
## Expected results
Expected and actual results are attached to the code snippets.
## Actual results
Expected and actual results are attached to the code snippets.
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 1.12.1
- Platform: Windows-10-10.0.19042-SP0
- Python version: 3.9.7
- PyArrow version: 5.0.0
| ### Update:
I wrote a bit cleaner code snippet (without transforming to json) that can expose leaking.
```python
import datasets
import json
items = ['ab', 'c', 'df']
ds = datasets.Dataset.from_dict({'col': items})
print(list(ds))
# > Prints: [{'col': 'ab'}, {'col': 'c'}, {'col': 'df'}]
filtered = ds
# get all items that are starting with a character with ascii code bigger than 'a'
filtered = filtered.filter(lambda x: x['col'][0] > 'a', load_from_cache_file=False)
print(list(filtered))
# > Prints: [{'col': 'c'}, {'col': 'df'}] as expected
# get all items that are shorter than 2
filtered = filtered.filter(lambda x: len(x['col']) < 2, load_from_cache_file=False)
print(list(filtered))
# > Prints: [{'col': 'ab'}] -> this is a leaked item from the first filter
# > Should be: [{'col': 'c'}]
```
Thanks for reporting. I'm looking into it | 2021-10-04T15:42:58Z | [] | [] |
huggingface/datasets | 3,077 | huggingface__datasets-3077 | [
"3076"
] | f839338d619ad7adbd2729e94d7cbaeebe322ff6 | diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -581,7 +581,7 @@ def get_module(self) -> MetricModule:
imports = get_imports(local_path)
local_imports = _download_additional_modules(
name=self.name,
- base_path=hf_github_url(path=self.name, name="", revision=revision),
+ base_path=hf_github_url(path=self.name, name="", revision=revision, dataset=False),
imports=imports,
download_config=self.download_config,
)
| diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -181,7 +181,15 @@ def test_CanonicalDatasetModuleFactory(self):
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
- def test_CanonicalMetricModuleFactory(self):
+ def test_CanonicalMetricModuleFactory_with_internal_import(self):
+ # "squad_v2" requires additional imports (internal)
+ factory = CanonicalMetricModuleFactory(
+ "squad_v2", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
+ )
+ module_factory_result = factory.get_module()
+ assert importlib.import_module(module_factory_result.module_path) is not None
+
+ def test_CanonicalMetricModuleFactory_with_external_import(self):
# "bleu" requires additional imports (external from github)
factory = CanonicalMetricModuleFactory(
"bleu", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
| Error when loading a metric
## Describe the bug
As reported by @sgugger, after last release, exception is thrown when loading a metric.
## Steps to reproduce the bug
```python
from datasets import load_metric
metric = load_metric("squad_v2")
```
## Actual results
```
FileNotFoundError Traceback (most recent call last)
<ipython-input-1-e612a8cab787> in <module>
1 from datasets import load_metric
----> 2 metric = load_metric("squad_v2")
d:\projects\huggingface\datasets\src\datasets\load.py in load_metric(path, config_name, process_id, num_process, cache_dir, experiment_id, keep_in_memory, download_config, download_mode, revision, script_version, **metric_init_kwargs)
1336 )
1337 revision = script_version
-> 1338 metric_module = metric_module_factory(
1339 path, revision=revision, download_config=download_config, download_mode=download_mode
1340 ).module_path
d:\projects\huggingface\datasets\src\datasets\load.py in metric_module_factory(path, revision, download_config, download_mode, force_local_path, dynamic_modules_path, **download_kwargs)
1237 if not isinstance(e1, FileNotFoundError):
1238 raise e1 from None
-> 1239 raise FileNotFoundError(
1240 f"Couldn't find a metric script at {relative_to_absolute_path(combined_path)}. "
1241 f"Metric '{path}' doesn't exist on the Hugging Face Hub either."
FileNotFoundError: Couldn't find a metric script at D:\projects\huggingface\datasets\squad_v2\squad_v2.py. Metric 'squad_v2' doesn't exist on the Hugging Face Hub either.
```
| 2021-10-14T09:06:58Z | [] | [] |
|
huggingface/datasets | 3,086 | huggingface__datasets-3086 | [
"3083"
] | 2da1203a3e1a7307a49cd67c69e16bd0d2787656 | diff --git a/src/datasets/features/audio.py b/src/datasets/features/audio.py
--- a/src/datasets/features/audio.py
+++ b/src/datasets/features/audio.py
@@ -21,7 +21,6 @@ class Audio:
dtype: ClassVar[str] = "dict"
pa_type: ClassVar[Any] = None
_type: str = field(default="Audio", init=False, repr=False)
- _resampler: Any = field(default=None, init=False, repr=False, compare=False)
def __call__(self):
return pa.string()
@@ -69,7 +68,7 @@ def _decode_example_with_torchaudio(self, value):
if self.mono:
array = array.mean(axis=0)
if self.sampling_rate and self.sampling_rate != sampling_rate:
- if not self._resampler:
+ if not hasattr(self, "_resampler"):
self._resampler = T.Resample(sampling_rate, self.sampling_rate)
array = self._resampler(array, sampling_rate, self.sampling_rate)
sampling_rate = self.sampling_rate
| diff --git a/tests/features/test_audio.py b/tests/features/test_audio.py
--- a/tests/features/test_audio.py
+++ b/tests/features/test_audio.py
@@ -4,7 +4,7 @@
import pytest
-from datasets import Dataset
+from datasets import Dataset, load_dataset
from datasets.features import Audio, Features, Value
@@ -250,3 +250,12 @@ def test_formatted_dataset_with_audio_feature(shared_datadir):
assert column[0]["path"] == audio_path
assert column[0]["array"].shape == (202311,)
assert column[0]["sampling_rate"] == 44100
+
+
+@require_sndfile
+def test_dataset_with_audio_feature_loaded_from_cache():
+ # load first time
+ ds = load_dataset("patrickvonplaten/librispeech_asr_dummy", "clean")
+ # load from cache
+ ds = load_dataset("patrickvonplaten/librispeech_asr_dummy", "clean", split="validation")
+ assert isinstance(ds, Dataset)
| Datasets with Audio feature raise error when loaded from cache due to _resampler parameter
## Describe the bug
As reported by @patrickvonplaten, when loaded from the cache, datasets containing the Audio feature raise TypeError.
## Steps to reproduce the bug
```python
from datasets import load_dataset
# load first time works
ds = load_dataset("patrickvonplaten/librispeech_asr_dummy", "clean")
# load from cache breaks
ds = load_dataset("patrickvonplaten/librispeech_asr_dummy", "clean")
```
## Actual results
```
TypeError: __init__() got an unexpected keyword argument '_resampler'
```
| 2021-10-14T14:38:50Z | [] | [] |
|
huggingface/datasets | 3,088 | huggingface__datasets-3088 | [
"3087"
] | 568db594d51110da9e23d224abded2a976b3c8c7 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -503,12 +503,12 @@ def wrapper(*args, **kwargs):
out: Union["Dataset", "DatasetDict"] = func(self, *args, **kwargs)
datasets: List["Dataset"] = list(out.values()) if isinstance(out, dict) else [out]
for dataset in datasets:
- # Remove task templates if a feature of the template has changed
+ # Remove task templates if a column mapping of the template is no longer valid
if self.info.task_templates is not None:
dataset.info.task_templates = [
template
for template in self.info.task_templates
- if all(dataset.features.get(k) == self.features.get(k) for k in template.features.keys())
+ if all(dataset.features.get(k) == self.features.get(k) for k in template.column_mapping.keys())
]
return out
| diff --git a/tests/test_tasks.py b/tests/test_tasks.py
--- a/tests/test_tasks.py
+++ b/tests/test_tasks.py
@@ -158,3 +158,16 @@ def dont_keep_task(x):
# reload from cache
mapped_dataset = dataset.map(dont_keep_task)
assert mapped_dataset.info.task_templates == []
+
+ def test_remove_and_map_on_task_template(self):
+ features = Features({"text": Value("string"), "label": ClassLabel(names=("pos", "neg"))})
+ task_templates = TextClassification(text_column="text", label_column="label")
+ info = DatasetInfo(features=features, task_templates=task_templates)
+ dataset = Dataset.from_dict({"text": ["A sentence."], "label": ["pos"]}, info=info)
+
+ def process(example):
+ return example
+
+ modified_dataset = dataset.remove_columns("label")
+ mapped_dataset = modified_dataset.map(process)
+ assert mapped_dataset.info.task_templates == []
| Removing label column in a text classification dataset yields to errors
## Describe the bug
This looks like #3059 but it's not linked to the cache this time. Removing the `label` column from a text classification dataset and then performing any processing will result in an error.
To reproduce:
```py
from datasets import load_dataset
from transformers import AutoTokenizer
raw_datasets = load_dataset("imdb")
raw_datasets = raw_datasets.remove_columns("label")
model_checkpoint = "distilbert-base-cased"
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
context_length = 128
def tokenize_pad_and_truncate(texts):
return tokenizer(texts["text"], truncation=True, padding="max_length", max_length=context_length)
tokenized_datasets = raw_datasets.map(tokenize_pad_and_truncate, batched=True)
```
Traceback:
```
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
<ipython-input-1-ba61bb32f786> in <module>
12 return tokenizer(texts["text"], truncation=True, padding="max_length", max_length=context_length)
13
---> 14 tokenized_datasets = raw_datasets.map(tokenize_pad_and_truncate, batched=True)
~/git/datasets/src/datasets/dataset_dict.py in map(self, function, with_indices, input_columns, batched, batch_size, remove_columns, keep_in_memory, load_from_cache_file, cache_file_names, writer_batch_size, features, disable_nullable, fn_kwargs, num_proc, desc)
500 desc=desc,
501 )
--> 502 for k, dataset in self.items()
503 }
504 )
~/git/datasets/src/datasets/dataset_dict.py in <dictcomp>(.0)
500 desc=desc,
501 )
--> 502 for k, dataset in self.items()
503 }
504 )
~/git/datasets/src/datasets/arrow_dataset.py in map(self, function, with_indices, input_columns, batched, batch_size, drop_last_batch, remove_columns, keep_in_memory, load_from_cache_file, cache_file_name, writer_batch_size, features, disable_nullable, fn_kwargs, num_proc, suffix_template, new_fingerprint, desc)
2051 new_fingerprint=new_fingerprint,
2052 disable_tqdm=disable_tqdm,
-> 2053 desc=desc,
2054 )
2055 else:
~/git/datasets/src/datasets/arrow_dataset.py in wrapper(*args, **kwargs)
501 self: "Dataset" = kwargs.pop("self")
502 # apply actual function
--> 503 out: Union["Dataset", "DatasetDict"] = func(self, *args, **kwargs)
504 datasets: List["Dataset"] = list(out.values()) if isinstance(out, dict) else [out]
505 for dataset in datasets:
~/git/datasets/src/datasets/arrow_dataset.py in wrapper(*args, **kwargs)
468 }
469 # apply actual function
--> 470 out: Union["Dataset", "DatasetDict"] = func(self, *args, **kwargs)
471 datasets: List["Dataset"] = list(out.values()) if isinstance(out, dict) else [out]
472 # re-apply format to the output
~/git/datasets/src/datasets/fingerprint.py in wrapper(*args, **kwargs)
404 # Call actual function
405
--> 406 out = func(self, *args, **kwargs)
407
408 # Update fingerprint of in-place transforms + update in-place history of transforms
~/git/datasets/src/datasets/arrow_dataset.py in _map_single(self, function, with_indices, input_columns, batched, batch_size, drop_last_batch, remove_columns, keep_in_memory, load_from_cache_file, cache_file_name, writer_batch_size, features, disable_nullable, fn_kwargs, new_fingerprint, rank, offset, disable_tqdm, desc, cache_only)
2243 if os.path.exists(cache_file_name) and load_from_cache_file:
2244 logger.warning("Loading cached processed dataset at %s", cache_file_name)
-> 2245 info = self.info.copy()
2246 info.features = features
2247 info.task_templates = None
~/git/datasets/src/datasets/info.py in copy(self)
278
279 def copy(self) -> "DatasetInfo":
--> 280 return self.__class__(**{k: copy.deepcopy(v) for k, v in self.__dict__.items()})
281
282
~/git/datasets/src/datasets/info.py in __init__(self, description, citation, homepage, license, features, post_processed, supervised_keys, task_templates, builder_name, config_name, version, splits, download_checksums, download_size, post_processing_size, dataset_size, size_in_bytes)
~/git/datasets/src/datasets/info.py in __post_init__(self)
177 for idx, template in enumerate(self.task_templates):
178 if isinstance(template, TextClassification):
--> 179 labels = self.features[template.label_column].names
180 self.task_templates[idx] = TextClassification(
181 text_column=template.text_column, label_column=template.label_column, labels=labels
KeyError: 'label'
```
| 2021-10-14T23:49:40Z | [] | [] |
|
huggingface/datasets | 3,096 | huggingface__datasets-3096 | [
"3095"
] | 324c84e8d9038dc9c707112dda7b29f5e5e36dd6 | diff --git a/src/datasets/features/audio.py b/src/datasets/features/audio.py
--- a/src/datasets/features/audio.py
+++ b/src/datasets/features/audio.py
@@ -64,14 +64,14 @@ def _decode_example_with_torchaudio(self, value):
raise ImportError("To support decoding 'mp3' audio files, please install 'sox'.") from err
array, sampling_rate = torchaudio.load(value)
- array = array.numpy()
- if self.mono:
- array = array.mean(axis=0)
if self.sampling_rate and self.sampling_rate != sampling_rate:
if not hasattr(self, "_resampler"):
self._resampler = T.Resample(sampling_rate, self.sampling_rate)
- array = self._resampler(array, sampling_rate, self.sampling_rate)
+ array = self._resampler(array)
sampling_rate = self.sampling_rate
+ array = array.numpy()
+ if self.mono:
+ array = array.mean(axis=0)
return array, sampling_rate
def decode_batch(self, values):
| diff --git a/tests/features/test_audio.py b/tests/features/test_audio.py
--- a/tests/features/test_audio.py
+++ b/tests/features/test_audio.py
@@ -122,6 +122,34 @@ def test_resampling_at_loading_dataset_with_audio_feature(shared_datadir):
assert column[0]["sampling_rate"] == 16000
+@require_sox
+@require_sndfile
+def test_resampling_at_loading_dataset_with_audio_feature_mp3(shared_datadir):
+ audio_path = str(shared_datadir / "test_audio_44100.mp3")
+ data = {"audio": [audio_path]}
+ features = Features({"audio": Audio(sampling_rate=16000)})
+ dset = Dataset.from_dict(data, features=features)
+ item = dset[0]
+ assert item.keys() == {"audio"}
+ assert item["audio"].keys() == {"path", "array", "sampling_rate"}
+ assert item["audio"]["path"] == audio_path
+ assert item["audio"]["array"].shape == (39707,)
+ assert item["audio"]["sampling_rate"] == 16000
+ batch = dset[:1]
+ assert batch.keys() == {"audio"}
+ assert len(batch["audio"]) == 1
+ assert batch["audio"][0].keys() == {"path", "array", "sampling_rate"}
+ assert batch["audio"][0]["path"] == audio_path
+ assert batch["audio"][0]["array"].shape == (39707,)
+ assert batch["audio"][0]["sampling_rate"] == 16000
+ column = dset["audio"]
+ assert len(column) == 1
+ assert column[0].keys() == {"path", "array", "sampling_rate"}
+ assert column[0]["path"] == audio_path
+ assert column[0]["array"].shape == (39707,)
+ assert column[0]["sampling_rate"] == 16000
+
+
@require_sndfile
def test_resampling_after_loading_dataset_with_audio_feature(shared_datadir):
audio_path = str(shared_datadir / "test_audio_44100.wav")
@@ -152,6 +180,37 @@ def test_resampling_after_loading_dataset_with_audio_feature(shared_datadir):
assert column[0]["sampling_rate"] == 16000
+@require_sox
+@require_sndfile
+def test_resampling_after_loading_dataset_with_audio_feature_mp3(shared_datadir):
+ audio_path = str(shared_datadir / "test_audio_44100.mp3")
+ data = {"audio": [audio_path]}
+ features = Features({"audio": Audio()})
+ dset = Dataset.from_dict(data, features=features)
+ item = dset[0]
+ assert item["audio"]["sampling_rate"] == 44100
+ dset = dset.cast_column("audio", Audio(sampling_rate=16000))
+ item = dset[0]
+ assert item.keys() == {"audio"}
+ assert item["audio"].keys() == {"path", "array", "sampling_rate"}
+ assert item["audio"]["path"] == audio_path
+ assert item["audio"]["array"].shape == (39707,)
+ assert item["audio"]["sampling_rate"] == 16000
+ batch = dset[:1]
+ assert batch.keys() == {"audio"}
+ assert len(batch["audio"]) == 1
+ assert batch["audio"][0].keys() == {"path", "array", "sampling_rate"}
+ assert batch["audio"][0]["path"] == audio_path
+ assert batch["audio"][0]["array"].shape == (39707,)
+ assert batch["audio"][0]["sampling_rate"] == 16000
+ column = dset["audio"]
+ assert len(column) == 1
+ assert column[0].keys() == {"path", "array", "sampling_rate"}
+ assert column[0]["path"] == audio_path
+ assert column[0]["array"].shape == (39707,)
+ assert column[0]["sampling_rate"] == 16000
+
+
@require_sndfile
def test_dataset_with_audio_feature_map_is_not_decoded(shared_datadir):
audio_path = str(shared_datadir / "test_audio_44100.wav")
| `cast_column` makes audio decoding fail
## Describe the bug
After changing the sampling rate automatic decoding fails.
## Steps to reproduce the bug
```python
from datasets import load_dataset
import datasets
ds = load_dataset("common_voice", "ab", split="train")
ds = ds.cast_column("audio", datasets.features.Audio(sampling_rate=16_000))
print(ds[0]["audio"]) # <- this fails currently
```
yields:
```
TypeError: forward() takes 2 positional arguments but 4 were given
```
## Expected results
no failure
## Actual results
Specify the actual results or traceback.
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
Copy-and-paste the text below in your GitHub issue.
- `datasets` version: 1.13.2 (master)
- Platform: Linux-5.11.0-1019-aws-x86_64-with-glibc2.29
- Python version: 3.8.10
- PyArrow version: 5.0.0
| cc @anton-l @albertvillanova | 2021-10-15T15:05:19Z | [] | [] |
huggingface/datasets | 3,120 | huggingface__datasets-3120 | [
"3111"
] | c98c23c4260edadab00f997d1a5d66b7f2e93ce9 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -3650,7 +3650,10 @@ def concatenate_datasets(
return dsets[0]
table = concat_tables(tables_to_concat, axis=axis)
if axis == 1:
- table = update_metadata_with_features(table, None)
+ # Merge features (ignore duplicated columns for now and let Dataset.__init__ check for those)
+ table = update_metadata_with_features(
+ table, Features({k: v for dset in dsets for k, v in dset.features.items()})
+ )
def apply_offset_to_indices_table(table, offset):
if offset == 0:
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -2190,6 +2190,22 @@ def test_concatenate_datasets(dataset_type, axis, expected_shape, dataset_dict,
assert_arrow_metadata_are_synced_with_dataset_features(dataset)
[email protected]("axis", [0, 1])
+def test_concatenate_datasets_complex_features(axis):
+ n = 5
+ dataset1 = Dataset.from_dict(
+ {"col_1": [0] * n, "col_2": list(range(n))},
+ features=Features({"col_1": Value("int32"), "col_2": ClassLabel(num_classes=n)}),
+ )
+ if axis == 1:
+ dataset2 = dataset1.rename_columns({col: col + "_" for col in dataset1.column_names})
+ expected_features = Features({**dataset1.features, **dataset2.features})
+ else:
+ dataset2 = dataset1
+ expected_features = dataset1.features
+ assert concatenate_datasets([dataset1, dataset2], axis=axis).features == expected_features
+
+
@pytest.mark.parametrize("other_dataset_type", ["in_memory", "memory_mapped", "concatenation"])
@pytest.mark.parametrize("axis, expected_shape", [(0, (8, 3)), (1, (4, 6))])
def test_concatenate_datasets_with_concatenation_tables(
| concatenate_datasets removes ClassLabel typing.
## Describe the bug
When concatenating two datasets, we lose typing of ClassLabel columns.
I can work on this if this is a legitimate bug,
## Steps to reproduce the bug
```python
import datasets
from datasets import Dataset, ClassLabel, Value, concatenate_datasets
DS_LEN = 100
my_dataset = Dataset.from_dict(
{
"sentence": [f"{chr(i % 10)}" for i in range(DS_LEN)],
"label": [i % 2 for i in range(DS_LEN)]
}
)
my_predictions = Dataset.from_dict(
{
"pred": [(i + 1) % 2 for i in range(DS_LEN)]
}
)
my_dataset = my_dataset.cast(datasets.Features({"sentence": Value("string"), "label": ClassLabel(2, names=["POS", "NEG"])}))
print("Original")
print(my_dataset)
print(my_dataset.features)
concat_ds = concatenate_datasets([my_dataset, my_predictions], axis=1)
print("Concatenated")
print(concat_ds)
print(concat_ds.features)
```
## Expected results
The features of `concat_ds` should contain ClassLabel.
## Actual results
On master, I get:
```
{'sentence': Value(dtype='string', id=None), 'label': Value(dtype='int64', id=None), 'pred': Value(dtype='int64', id=None)}
```
## Environment info
- `datasets` version: 1.14.1.dev0
- Platform: macOS-10.15.7-x86_64-i386-64bit
- Python version: 3.8.11
- PyArrow version: 4.0.1
| Something like this would fix it I think: https://github.com/huggingface/datasets/compare/master...Dref360:HF-3111/concatenate_types?expand=1 | 2021-10-20T15:54:58Z | [] | [] |
huggingface/datasets | 3,129 | huggingface__datasets-3129 | [
"3128"
] | ed1b4921a126ce007e6d2bb859730eb6a0fedbb8 | diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -211,7 +211,7 @@ def __init__(
raise ValueError("At least one of path and stream must be provided.")
if features is not None:
self._features = features
- self._schema = pa.schema(features.type)
+ self._schema = None
elif schema is not None:
self._schema: pa.Schema = schema
self._features = Features.from_arrow_schema(self._schema)
@@ -226,9 +226,7 @@ def __init__(
self._hasher = KeyHasher("")
self._check_duplicates = check_duplicates
-
- if disable_nullable and self._schema is not None:
- self._schema = pa.schema(pa.field(field.name, field.type, nullable=False) for field in self._schema)
+ self._disable_nullable = disable_nullable
self._path = path
if stream is None:
@@ -273,6 +271,7 @@ def close(self):
self.stream.close() # This also closes self.pa_writer if it is opened
def _build_writer(self, inferred_schema: pa.Schema):
+ schema = self.schema
inferred_features = Features.from_arrow_schema(inferred_schema)
if self._features is not None:
if self.update_features: # keep original features it they match, or update them
@@ -283,21 +282,27 @@ def _build_writer(self, inferred_schema: pa.Schema):
if inferred_field == fields[name]:
inferred_features[name] = self._features[name]
self._features = inferred_features
- self._schema: pa.Schema = inferred_schema
+ schema: pa.Schema = inferred_schema
else:
self._features = inferred_features
- self._schema: pa.Schema = inferred_schema
+ schema: pa.Schema = inferred_schema
if self.disable_nullable:
- self._schema = pa.schema(pa.field(field.name, field.type, nullable=False) for field in self._schema)
+ schema = pa.schema(pa.field(field.name, field.type, nullable=False) for field in schema)
if self.with_metadata:
- self._schema = self._schema.with_metadata(
- self._build_metadata(DatasetInfo(features=self._features), self.fingerprint)
- )
- self.pa_writer = pa.RecordBatchStreamWriter(self.stream, self._schema)
+ schema = schema.with_metadata(self._build_metadata(DatasetInfo(features=self._features), self.fingerprint))
+ self._schema = schema
+ self.pa_writer = pa.RecordBatchStreamWriter(self.stream, schema)
@property
def schema(self):
- return self._schema if self._schema is not None else []
+ _schema = (
+ self._schema
+ if self._schema is not None
+ else (pa.schema(self._features.type) if self._features is not None else None)
+ )
+ if self._disable_nullable and _schema is not None:
+ _schema = pa.schema(pa.field(field.name, field.type, nullable=False) for field in _schema)
+ return _schema if _schema is not None else []
@staticmethod
def _build_metadata(info: DatasetInfo, fingerprint: Optional[str] = None) -> Dict[str, str]:
@@ -316,18 +321,18 @@ def write_examples_on_file(self):
# Since current_examples contains (example, key) tuples
cols = (
- [col for col in self._schema.names if col in self.current_examples[0][0]]
- + [col for col in self.current_examples[0][0].keys() if col not in self._schema.names]
- if self._schema
+ [col for col in self.schema.names if col in self.current_examples[0][0]]
+ + [col for col in self.current_examples[0][0].keys() if col not in self.schema.names]
+ if self.schema
else self.current_examples[0][0].keys()
)
- schema = None if self.pa_writer is None and self.update_features else self._schema
- try_schema = self._schema if self.pa_writer is None and self.update_features else None
+ schema = None if self.pa_writer is None and self.update_features else self.schema
+ try_schema = self.schema if self.pa_writer is None and self.update_features else None
arrays = []
inferred_types = []
for col in cols:
- col_type = schema.field(col).type if schema is not None else None
+ col_type = schema.field(col).type if schema else None
col_try_type = try_schema.field(col).type if try_schema is not None and col in try_schema.names else None
typed_sequence = OptimizedTypedSequence(
[row[0][col] for row in self.current_examples], type=col_type, try_type=col_try_type, col=col
@@ -345,7 +350,7 @@ def write_examples_on_file(self):
)
arrays.append(pa_array)
inferred_types.append(inferred_type)
- schema = pa.schema(zip(cols, inferred_types)) if self.pa_writer is None else self._schema
+ schema = pa.schema(zip(cols, inferred_types)) if self.pa_writer is None else self.schema
table = pa.Table.from_arrays(arrays, schema=schema)
self.write_table(table)
self.current_examples = []
@@ -426,11 +431,11 @@ def write_batch(
"""
if batch_examples and len(next(iter(batch_examples.values()))) == 0:
return
- schema = None if self.pa_writer is None and self.update_features else self._schema
- try_schema = self._schema if self.pa_writer is None and self.update_features else None
+ schema = None if self.pa_writer is None and self.update_features else self.schema
+ try_schema = self.schema if self.pa_writer is None and self.update_features else None
typed_sequence_examples = {}
for col in sorted(batch_examples.keys()):
- col_type = schema.field(col).type if schema is not None else None
+ col_type = schema.field(col).type if schema else None
col_try_type = try_schema.field(col).type if try_schema is not None and col in try_schema.names else None
typed_sequence = OptimizedTypedSequence(batch_examples[col], type=col_type, try_type=col_try_type, col=col)
typed_sequence_examples[col] = typed_sequence
@@ -465,8 +470,8 @@ def finalize(self, close_stream=True):
self.hkey_record = []
self.write_examples_on_file()
if self.pa_writer is None:
- if self._schema is not None:
- self._build_writer(self._schema)
+ if self.schema:
+ self._build_writer(self.schema)
else:
raise ValueError("Please pass `features` or at least one example when writing data")
self.pa_writer.close()
diff --git a/src/datasets/features/audio.py b/src/datasets/features/audio.py
--- a/src/datasets/features/audio.py
+++ b/src/datasets/features/audio.py
@@ -1,5 +1,6 @@
from collections import defaultdict
from dataclasses import dataclass, field
+from io import BytesIO
from typing import Any, ClassVar, Optional
import pyarrow as pa
@@ -11,13 +12,24 @@
class Audio:
"""Audio Feature to extract audio data from an audio file.
+ Input: The Audio feature accepts as input:
+ - A :obj:`str`: Absolute path to the audio file (i.e. random access is allowed).
+ - A :obj:`dict` with the keys:
+
+ - path: String with relative path of the audio file to the archive file.
+ - bytes: Bytes content of the audio file.
+
+ This is useful for archived files with sequential access.
+
Args:
sampling_rate (:obj:`int`, optional): Target sampling rate. If `None`, the native sampling rate is used.
- mono (:obj:`bool`, default ```True``): Whether to convert the audio signal to mono by averaging samples across channels.
+ mono (:obj:`bool`, default ``True``): Whether to convert the audio signal to mono by averaging samples across
+ channels.
"""
sampling_rate: Optional[int] = None
mono: bool = True
+ _storage_dtype: str = "string"
id: Optional[str] = None
# Automatically constructed
dtype: ClassVar[str] = "dict"
@@ -25,36 +37,73 @@ class Audio:
_type: str = field(default="Audio", init=False, repr=False)
def __call__(self):
- return pa.string()
+ return (
+ pa.struct({"path": pa.string(), "bytes": pa.binary()}) if self._storage_dtype == "struct" else pa.string()
+ )
+
+ def encode_example(self, value):
+ """Encode example into a format for Arrow.
+
+ Args:
+ value (:obj:`str` or :obj:`dict`): Data passed as input to Audio feature.
+
+ Returns:
+ :obj:`str` or :obj:`dict`
+ """
+ if isinstance(value, dict):
+ self._storage_dtype = "struct"
+ return value
def decode_example(self, value):
"""Decode example audio file into audio data.
Args:
- value: Audio file path.
+ value (obj:`str` or :obj:`dict`): Either a string with the absolute audio file path or a dictionary with
+ keys:
+
+ - path: String with relative audio file path.
+ - bytes: Bytes of the audio file.
Returns:
dict
"""
- # TODO: backard compatibility for users without audio dependencies
- array, sampling_rate = (
- self._decode_example_with_torchaudio(value)
- if value.endswith(".mp3")
- else self._decode_example_with_librosa(value)
- )
- return {"path": value, "array": array, "sampling_rate": sampling_rate}
+ path, file = (value["path"], BytesIO(value["bytes"])) if isinstance(value, dict) else (value, None)
+ if path.endswith("mp3"):
+ array, sampling_rate = self._decode_mp3(file if file else path)
+ else:
+ if file:
+ array, sampling_rate = self._decode_non_mp3_file_like(file)
+ else:
+ array, sampling_rate = self._decode_non_mp3_path_like(path)
+ return {"path": path, "array": array, "sampling_rate": sampling_rate}
- def _decode_example_with_librosa(self, value):
+ def _decode_non_mp3_path_like(self, path):
try:
import librosa
except ImportError as err:
raise ImportError("To support decoding audio files, please install 'librosa'.") from err
- with xopen(value, "rb") as f:
+ with xopen(path, "rb") as f:
array, sampling_rate = librosa.load(f, sr=self.sampling_rate, mono=self.mono)
return array, sampling_rate
- def _decode_example_with_torchaudio(self, value):
+ def _decode_non_mp3_file_like(self, file):
+ try:
+ import librosa
+ import soundfile as sf
+ except ImportError as err:
+ raise ImportError("To support decoding audio files, please install 'librosa'.") from err
+
+ array, sampling_rate = sf.read(file)
+ array = array.T
+ if self.mono:
+ array = librosa.to_mono(array)
+ if self.sampling_rate and self.sampling_rate != sampling_rate:
+ array = librosa.resample(array, sampling_rate, self.sampling_rate, res_type="kaiser_best")
+ sampling_rate = self.sampling_rate
+ return array, sampling_rate
+
+ def _decode_mp3(self, path_or_file):
try:
import torchaudio
import torchaudio.transforms as T
@@ -65,7 +114,7 @@ def _decode_example_with_torchaudio(self, value):
except RuntimeError as err:
raise ImportError("To support decoding 'mp3' audio files, please install 'sox'.") from err
- array, sampling_rate = torchaudio.load(value)
+ array, sampling_rate = torchaudio.load(path_or_file, format="mp3")
if self.sampling_rate and self.sampling_rate != sampling_rate:
if not hasattr(self, "_resampler"):
self._resampler = T.Resample(sampling_rate, self.sampling_rate)
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -851,7 +851,7 @@ def encode_nested_example(schema, obj):
return list(obj)
# Object with special encoding:
# ClassLabel will convert from string to int, TranslationVariableLanguages does some checks
- elif isinstance(schema, (ClassLabel, TranslationVariableLanguages, Value, _ArrayXD)):
+ elif isinstance(schema, (Audio, ClassLabel, TranslationVariableLanguages, Value, _ArrayXD)):
return schema.encode_example(obj)
# Other object should be directly convertible to a native Arrow type (like Translation and Translation)
return obj
@@ -963,7 +963,8 @@ class Features(dict):
:class:`datasets.Sequence`.
- a :class:`Array2D`, :class:`Array3D`, :class:`Array4D` or :class:`Array5D` feature for multidimensional arrays
- - a :class:`datasets.Audio` stores the path to an audio file and can extract audio data from it
+ - an :class:`Audio` feature to store the absolute path to an audio file or a dictionary with the relative path
+ to an audio file ("path" key) and its bytes content ("bytes" key). This feature extracts the audio data.
- :class:`datasets.Translation` and :class:`datasets.TranslationVariableLanguages`, the two features specific to Machine Translation
"""
| diff --git a/tests/features/test_audio.py b/tests/features/test_audio.py
--- a/tests/features/test_audio.py
+++ b/tests/features/test_audio.py
@@ -1,4 +1,6 @@
+import os
import sys
+import tarfile
from ctypes.util import find_library
from importlib.util import find_spec
@@ -26,12 +28,41 @@
require_torchaudio = pytest.mark.skipif(find_spec("torchaudio") is None, reason="Test requires 'torchaudio'")
[email protected]()
+def tar_wav_path(shared_datadir, tmp_path_factory):
+ audio_path = str(shared_datadir / "test_audio_44100.wav")
+ path = tmp_path_factory.mktemp("data") / "audio_data.wav.tar"
+ with tarfile.TarFile(path, "w") as f:
+ f.add(audio_path, arcname=os.path.basename(audio_path))
+ return path
+
+
[email protected]()
+def tar_mp3_path(shared_datadir, tmp_path_factory):
+ audio_path = str(shared_datadir / "test_audio_44100.mp3")
+ path = tmp_path_factory.mktemp("data") / "audio_data.mp3.tar"
+ with tarfile.TarFile(path, "w") as f:
+ f.add(audio_path, arcname=os.path.basename(audio_path))
+ return path
+
+
+def iter_archive(archive_path):
+ with tarfile.open(archive_path) as tar:
+ for tarinfo in tar:
+ file_path = tarinfo.name
+ file_obj = tar.extractfile(tarinfo)
+ yield file_path, file_obj
+
+
def test_audio_instantiation():
audio = Audio()
+ assert audio.sampling_rate is None
+ assert audio.mono is True
assert audio.id is None
assert audio.dtype == "dict"
assert audio.pa_type is None
assert audio._type == "Audio"
+ assert audio._storage_dtype == "string"
@require_sndfile
@@ -95,6 +126,67 @@ def test_dataset_with_audio_feature(shared_datadir):
assert column[0]["sampling_rate"] == 44100
+@require_sndfile
+def test_dataset_with_audio_feature_tar_wav(tar_wav_path):
+ audio_filename = "test_audio_44100.wav"
+ data = {"audio": []}
+ for file_path, file_obj in iter_archive(tar_wav_path):
+ data["audio"].append({"path": file_path, "bytes": file_obj.read()})
+ break
+ features = Features({"audio": Audio()})
+ dset = Dataset.from_dict(data, features=features)
+ item = dset[0]
+ assert item.keys() == {"audio"}
+ assert item["audio"].keys() == {"path", "array", "sampling_rate"}
+ assert item["audio"]["path"] == audio_filename
+ assert item["audio"]["array"].shape == (202311,)
+ assert item["audio"]["sampling_rate"] == 44100
+ batch = dset[:1]
+ assert batch.keys() == {"audio"}
+ assert len(batch["audio"]) == 1
+ assert batch["audio"][0].keys() == {"path", "array", "sampling_rate"}
+ assert batch["audio"][0]["path"] == audio_filename
+ assert batch["audio"][0]["array"].shape == (202311,)
+ assert batch["audio"][0]["sampling_rate"] == 44100
+ column = dset["audio"]
+ assert len(column) == 1
+ assert column[0].keys() == {"path", "array", "sampling_rate"}
+ assert column[0]["path"] == audio_filename
+ assert column[0]["array"].shape == (202311,)
+ assert column[0]["sampling_rate"] == 44100
+
+
+@require_sox
+@require_torchaudio
+def test_dataset_with_audio_feature_tar_mp3(tar_mp3_path):
+ audio_filename = "test_audio_44100.mp3"
+ data = {"audio": []}
+ for file_path, file_obj in iter_archive(tar_mp3_path):
+ data["audio"].append({"path": file_path, "bytes": file_obj.read()})
+ break
+ features = Features({"audio": Audio()})
+ dset = Dataset.from_dict(data, features=features)
+ item = dset[0]
+ assert item.keys() == {"audio"}
+ assert item["audio"].keys() == {"path", "array", "sampling_rate"}
+ assert item["audio"]["path"] == audio_filename
+ assert item["audio"]["array"].shape == (109440,)
+ assert item["audio"]["sampling_rate"] == 44100
+ batch = dset[:1]
+ assert batch.keys() == {"audio"}
+ assert len(batch["audio"]) == 1
+ assert batch["audio"][0].keys() == {"path", "array", "sampling_rate"}
+ assert batch["audio"][0]["path"] == audio_filename
+ assert batch["audio"][0]["array"].shape == (109440,)
+ assert batch["audio"][0]["sampling_rate"] == 44100
+ column = dset["audio"]
+ assert len(column) == 1
+ assert column[0].keys() == {"path", "array", "sampling_rate"}
+ assert column[0]["path"] == audio_filename
+ assert column[0]["array"].shape == (109440,)
+ assert column[0]["sampling_rate"] == 44100
+
+
@require_sndfile
def test_resampling_at_loading_dataset_with_audio_feature(shared_datadir):
audio_path = str(shared_datadir / "test_audio_44100.wav")
| Support Audio feature for TAR archives in sequential access
Currently, Audio feature accesses each audio file by their file path.
However, streamed TAR archive files do not allow random access to their archived files.
Therefore, we should enhance the Audio feature to support TAR archived files in sequential access.
| 2021-10-21T08:56:51Z | [] | [] |
|
huggingface/datasets | 3,133 | huggingface__datasets-3133 | [
"3132"
] | c98c23c4260edadab00f997d1a5d66b7f2e93ce9 | diff --git a/src/datasets/iterable_dataset.py b/src/datasets/iterable_dataset.py
--- a/src/datasets/iterable_dataset.py
+++ b/src/datasets/iterable_dataset.py
@@ -341,7 +341,10 @@ def __iter__(self):
for key, example in self._iter():
if self.features:
# we encode the example for ClassLabel feature types for example
- yield self.features.encode_example(example)
+ encoded_example = self.features.encode_example(example)
+ # Decode example for Audio feature, e.g.
+ decoded_example = self.features.decode_example(encoded_example)
+ yield decoded_example
else:
yield example
| diff --git a/tests/features/test_audio.py b/tests/features/test_audio.py
--- a/tests/features/test_audio.py
+++ b/tests/features/test_audio.py
@@ -311,6 +311,34 @@ def test_formatted_dataset_with_audio_feature(shared_datadir):
assert column[0]["sampling_rate"] == 44100
[email protected]
+def jsonl_audio_dataset_path(shared_datadir, tmp_path_factory):
+ import json
+
+ audio_path = str(shared_datadir / "test_audio_44100.wav")
+ data = [{"audio": audio_path, "text": "Hello world!"}]
+ path = str(tmp_path_factory.mktemp("data") / "audio_dataset.jsonl")
+ with open(path, "w") as f:
+ for item in data:
+ f.write(json.dumps(item) + "\n")
+ return path
+
+
+@require_sndfile
[email protected]("streaming", [False, True])
+def test_load_dataset_with_audio_feature(streaming, jsonl_audio_dataset_path, shared_datadir):
+ audio_path = str(shared_datadir / "test_audio_44100.wav")
+ data_files = jsonl_audio_dataset_path
+ features = Features({"audio": Audio(), "text": Value("string")})
+ dset = load_dataset("json", split="train", data_files=data_files, features=features, streaming=streaming)
+ item = dset[0] if not streaming else next(iter(dset))
+ assert item.keys() == {"audio", "text"}
+ assert item["audio"].keys() == {"path", "array", "sampling_rate"}
+ assert item["audio"]["path"] == audio_path
+ assert item["audio"]["array"].shape == (202311,)
+ assert item["audio"]["sampling_rate"] == 44100
+
+
@require_sndfile
def test_dataset_with_audio_feature_loaded_from_cache():
# load first time
| Support Audio feature in streaming mode
Currently, Audio feature is only supported for non-streaming datasets.
Due to the large size of many speech datasets, we should also support Audio feature in streaming mode.
| 2021-10-21T13:37:57Z | [] | [] |
|
huggingface/datasets | 3,151 | huggingface__datasets-3151 | [
"3150"
] | 87c71b9c29a40958973004910f97e4892559dfed | diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -127,7 +127,7 @@
"aiobotocore",
"boto3",
"botocore",
- "faiss-cpu",
+ "faiss-cpu>=1.6.4",
"fsspec[s3]",
"moto[s3,server]==2.0.4",
"rarfile>=4.0",
@@ -167,9 +167,7 @@
"importlib_resources;python_version<'3.7'",
]
-if os.name == "nt": # windows
- TESTS_REQUIRE.remove("faiss-cpu") # faiss doesn't exist on windows
-else:
+if os.name != "nt":
# dependencies of unbabel-comet
# only test if not on windows since there're issues installing fairseq on windows
TESTS_REQUIRE.extend(
| diff --git a/tests/test_search.py b/tests/test_search.py
--- a/tests/test_search.py
+++ b/tests/test_search.py
@@ -1,3 +1,4 @@
+import os
import tempfile
from functools import partial
from unittest import TestCase
@@ -50,9 +51,16 @@ def test_serialization(self):
index_name="vecs",
metric_type=faiss.METRIC_INNER_PRODUCT,
)
- with tempfile.NamedTemporaryFile() as tmp_file:
+
+ # Setting delete=False and unlinking manually is not pretty... but it is required on Windows to
+ # ensure somewhat stable behaviour. If we don't, we get PermissionErrors. This is an age-old issue.
+ # see https://bugs.python.org/issue14243 and
+ # https://stackoverflow.com/questions/23212435/permission-denied-to-write-to-my-temporary-file/23212515
+ with tempfile.NamedTemporaryFile(delete=False) as tmp_file:
dset.save_faiss_index("vecs", tmp_file.name)
dset.load_faiss_index("vecs2", tmp_file.name)
+ os.unlink(tmp_file.name)
+
scores, examples = dset.get_nearest_examples("vecs2", np.ones(5, dtype=np.float32))
self.assertEqual(examples["filename"][0], "my_name-train_29")
@@ -133,9 +141,16 @@ def test_serialization(self):
index = FaissIndex(metric_type=faiss.METRIC_INNER_PRODUCT)
index.add_vectors(np.eye(5, dtype=np.float32))
- with tempfile.NamedTemporaryFile() as tmp_file:
+
+ # Setting delete=False and unlinking manually is not pretty... but it is required on Windows to
+ # ensure somewhat stable behaviour. If we don't, we get PermissionErrors. This is an age-old issue.
+ # see https://bugs.python.org/issue14243 and
+ # https://stackoverflow.com/questions/23212435/permission-denied-to-write-to-my-temporary-file/23212515
+ with tempfile.NamedTemporaryFile(delete=False) as tmp_file:
index.save(tmp_file.name)
index = FaissIndex.load(tmp_file.name)
+ os.unlink(tmp_file.name)
+
query = np.zeros(5, dtype=np.float32)
query[1] = 1
scores, indices = index.search(query)
| Faiss _is_ available on Windows
In the setup file, I find the following:
https://github.com/huggingface/datasets/blob/87c71b9c29a40958973004910f97e4892559dfed/setup.py#L171
However, FAISS does install perfectly fine on Windows on my system. You can also confirm this on the [PyPi page](https://pypi.org/project/faiss-cpu/#files), where Windows wheels are available. Maybe this was true for older versions? For current versions, this can be removed I think.
(This isn't really a bug but didn't know how else to tag.)
If you agree I can do a quick PR and remove that line.
| Sure, feel free to open a PR. | 2021-10-22T19:34:29Z | [] | [] |
huggingface/datasets | 3,159 | huggingface__datasets-3159 | [
"3135"
] | 87c71b9c29a40958973004910f97e4892559dfed | diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -398,11 +398,11 @@ def _create_builder_config(self, name=None, custom_features=None, **config_kwarg
@utils.memoize()
def builder_configs(cls):
"""Pre-defined list of configurations for this builder class."""
- config_dict = {config.name: config for config in cls.BUILDER_CONFIGS}
- if len(config_dict) != len(cls.BUILDER_CONFIGS):
+ configs = {config.name: config for config in cls.BUILDER_CONFIGS}
+ if len(configs) != len(cls.BUILDER_CONFIGS):
names = [config.name for config in cls.BUILDER_CONFIGS]
raise ValueError("Names in BUILDER_CONFIGS must not be duplicated. Got %s" % names)
- return config_dict
+ return configs
@property
def cache_dir(self):
diff --git a/src/datasets/inspect.py b/src/datasets/inspect.py
--- a/src/datasets/inspect.py
+++ b/src/datasets/inspect.py
@@ -20,7 +20,7 @@
from .features import Features
from .hf_api import HfApi
-from .load import import_main_class, load_dataset_builder, prepare_module
+from .load import dataset_module_factory, import_main_class, load_dataset_builder, prepare_module
from .utils import DownloadConfig
from .utils.download_manager import GenerateMode
from .utils.logging import get_logger
@@ -194,9 +194,8 @@ def get_dataset_config_names(
download_kwargs: optional attributes for DownloadConfig() which will override the attributes in download_config if supplied,
for example ``use_auth_token``
"""
- module_path, _ = prepare_module(
+ dataset_module = dataset_module_factory(
path,
- dataset=True,
revision=revision,
download_config=download_config,
download_mode=download_mode,
@@ -205,8 +204,8 @@ def get_dataset_config_names(
data_files=data_files,
**download_kwargs,
)
- builder_cls = import_main_class(module_path, dataset=True)
- return list(builder_cls.builder_configs.keys())
+ builder_cls = import_main_class(dataset_module.module_path)
+ return list(builder_cls.builder_configs.keys()) or [dataset_module.builder_kwargs.get("name", "default")]
def get_dataset_split_names(
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -1438,14 +1438,13 @@ def load_dataset_builder(
if use_auth_token is not None:
download_config = download_config.copy() if download_config else DownloadConfig()
download_config.use_auth_token = use_auth_token
- dataset_module_factory_result = dataset_module_factory(
+ dataset_module = dataset_module_factory(
path, revision=revision, download_config=download_config, download_mode=download_mode, data_files=data_files
)
# Get dataset builder class from the processing script
- dataset_module = dataset_module_factory_result.module_path
- builder_cls = import_main_class(dataset_module)
- builder_kwargs = dataset_module_factory_result.builder_kwargs
+ builder_cls = import_main_class(dataset_module.module_path)
+ builder_kwargs = dataset_module.builder_kwargs
data_files = builder_kwargs.pop("data_files", data_files)
name = builder_kwargs.pop("name", name)
hash = builder_kwargs.pop("hash")
| diff --git a/tests/test_inspect.py b/tests/test_inspect.py
new file mode 100644
--- /dev/null
+++ b/tests/test_inspect.py
@@ -0,0 +1,12 @@
+import pytest
+
+from datasets import get_dataset_config_names
+
+
[email protected](
+ "path, expected",
+ [("squad", "plain_text"), ("acronym_identification", "default"), ("Check/region_1", "Check___region_1")],
+)
+def test_get_dataset_config_names(path, expected):
+ config_names = get_dataset_config_names(path)
+ assert expected in config_names
| Make inspect.get_dataset_config_names always return a non-empty list of configs
**Is your feature request related to a problem? Please describe.**
Currently, some datasets have a configuration, while others don't. It would be simpler for the user to always have configuration names to refer to
**Describe the solution you'd like**
In that sense inspect.get_dataset_config_names should always return at least one configuration name, be it `default` or `Check___region_1` (for community datasets like `Check/region_1`).
https://github.com/huggingface/datasets/blob/c5747a5e1dde2670b7f2ca6e79e2ffd99dff85af/src/datasets/inspect.py#L161
| Hi @severo, I guess this issue requests not only to be able to access the configuration name (by using `inspect.get_dataset_config_names`), but the configuration itself as well (I mean you use the name to get the configuration afterwards, maybe using `builder_cls.builder_configs`), is this right?
Yes, maybe the issue could be reformulated. As a user, I want to avoid having to manage special cases:
- I want to be able to get the names of a dataset's configs, and use them in the rest of the API (get the data, get the split names, etc).
- I don't want to have to manage datasets with named configs (`glue`) differently from datasets without named configs (`acronym_identification`, `Check/region_1`) | 2021-10-25T13:59:43Z | [] | [] |
huggingface/datasets | 3,166 | huggingface__datasets-3166 | [
"3165"
] | dfdd2f949c1840926c02ae47f0f0c43083ef0b1f | diff --git a/src/datasets/commands/dummy_data.py b/src/datasets/commands/dummy_data.py
--- a/src/datasets/commands/dummy_data.py
+++ b/src/datasets/commands/dummy_data.py
@@ -10,7 +10,7 @@
from datasets import config
from datasets.commands import BaseDatasetsCLICommand
-from datasets.load import import_main_class, prepare_module
+from datasets.load import dataset_module_factory, import_main_class
from datasets.utils import MockDownloadManager
from datasets.utils.download_manager import DownloadManager
from datasets.utils.file_utils import DownloadConfig
@@ -287,8 +287,8 @@ def __init__(
def run(self):
set_verbosity_warning()
- module_path, hash = prepare_module(self._path_to_dataset)
- builder_cls = import_main_class(module_path)
+ dataset_module = dataset_module_factory(self._path_to_dataset)
+ builder_cls = import_main_class(dataset_module.module_path)
# use `None` as config if no configs
builder_configs = builder_cls.BUILDER_CONFIGS or [None]
@@ -302,7 +302,7 @@ def run(self):
version = builder_config.version
name = builder_config.name
- dataset_builder = builder_cls(name=name, hash=hash, cache_dir=tmp_dir)
+ dataset_builder = builder_cls(name=name, hash=dataset_module.hash, cache_dir=tmp_dir)
mock_dl_manager = MockDownloadManager(
dataset_name=self._dataset_name,
config=builder_config,
diff --git a/src/datasets/commands/run_beam.py b/src/datasets/commands/run_beam.py
--- a/src/datasets/commands/run_beam.py
+++ b/src/datasets/commands/run_beam.py
@@ -7,7 +7,7 @@
from datasets import config
from datasets.builder import DatasetBuilder
from datasets.commands import BaseDatasetsCLICommand
-from datasets.load import import_main_class, prepare_module
+from datasets.load import dataset_module_factory, import_main_class
from datasets.utils.download_manager import DownloadConfig, GenerateMode
@@ -86,12 +86,8 @@ def run(self):
print("Both parameters `name` and `all_configs` can't be used at once.")
exit(1)
path, name = self._dataset, self._name
- module_path, hash, base_path, namespace = prepare_module(
- path,
- return_associated_base_path=True,
- return_namespace=True,
- )
- builder_cls = import_main_class(module_path)
+ dataset_module = dataset_module_factory(path)
+ builder_cls = import_main_class(dataset_module.module_path)
builders: List[DatasetBuilder] = []
if self._beam_pipeline_options:
beam_options = beam.options.pipeline_options.PipelineOptions(
@@ -105,11 +101,11 @@ def run(self):
builder_cls(
name=builder_config.name,
data_dir=self._data_dir,
- hash=hash,
+ hash=dataset_module.hash,
beam_options=beam_options,
cache_dir=self._cache_dir,
- base_path=base_path,
- namespace=namespace,
+ base_path=dataset_module.builder_kwargs.get("base_path"),
+ namespace=dataset_module.builder_kwargs.get("namespace"),
)
)
else:
@@ -119,8 +115,8 @@ def run(self):
data_dir=self._data_dir,
beam_options=beam_options,
cache_dir=self._cache_dir,
- base_path=base_path,
- namespace=namespace,
+ base_path=dataset_module.builder_kwargs.get("base_path"),
+ namespace=dataset_module.builder_kwargs.get("namespace"),
)
)
diff --git a/src/datasets/inspect.py b/src/datasets/inspect.py
--- a/src/datasets/inspect.py
+++ b/src/datasets/inspect.py
@@ -20,7 +20,7 @@
from .features import Features
from .hf_api import HfApi
-from .load import dataset_module_factory, import_main_class, load_dataset_builder, prepare_module
+from .load import dataset_module_factory, import_main_class, load_dataset_builder, metric_module_factory
from .utils import DownloadConfig
from .utils.download_manager import GenerateMode
from .utils.logging import get_logger
@@ -72,12 +72,12 @@ def inspect_dataset(path: str, local_path: str, download_config: Optional[Downlo
download_config (Optional ``datasets.DownloadConfig``: specific download configuration parameters.
**download_kwargs: optional attributes for DownloadConfig() which will override the attributes in download_config if supplied.
"""
- module_path, _ = prepare_module(
- path, download_config=download_config, dataset=True, force_local_path=local_path, **download_kwargs
+ dataset_module = dataset_module_factory(
+ path, download_config=download_config, force_local_path=local_path, **download_kwargs
)
print(
f"The processing script for dataset {path} can be inspected at {local_path}. "
- f"The main class is in {module_path}. "
+ f"The main class is in {dataset_module.module_path}. "
f"You can modify this processing script and use it with `datasets.load_dataset({local_path})`."
)
@@ -97,12 +97,12 @@ def inspect_metric(path: str, local_path: str, download_config: Optional[Downloa
download_config (Optional ``datasets.DownloadConfig``: specific download configuration parameters.
**download_kwargs: optional attributes for DownloadConfig() which will override the attributes in download_config if supplied.
"""
- module_path, _ = prepare_module(
- path, download_config=download_config, dataset=False, force_local_path=local_path, **download_kwargs
+ metric_module = metric_module_factory(
+ path, download_config=download_config, force_local_path=local_path, **download_kwargs
)
print(
f"The processing scripts for metric {path} can be inspected at {local_path}. "
- f"The main class is in {module_path}. "
+ f"The main class is in {metric_module.module_path}. "
f"You can modify this processing scripts and use it with `datasets.load_metric({local_path})`."
)
@@ -143,9 +143,8 @@ def get_dataset_infos(
download_kwargs: optional attributes for DownloadConfig() which will override the attributes in download_config if supplied,
for example ``use_auth_token``
"""
- module_path, _ = prepare_module(
+ dataset_module = dataset_module_factory(
path,
- dataset=True,
revision=revision,
download_config=download_config,
download_mode=download_mode,
@@ -154,7 +153,7 @@ def get_dataset_infos(
data_files=data_files,
**download_kwargs,
)
- builder_cls = import_main_class(module_path, dataset=True)
+ builder_cls = import_main_class(dataset_module.module_path, dataset=True)
return builder_cls.get_all_exported_dataset_infos()
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -47,6 +47,7 @@
from .splits import Split
from .streaming import extend_module_for_streaming
from .tasks import TaskTemplate
+from .utils.deprecation_utils import deprecated
from .utils.download_manager import GenerateMode
from .utils.file_utils import (
DownloadConfig,
@@ -1250,6 +1251,7 @@ def metric_module_factory(
raise FileNotFoundError(f"Couldn't find a metric script at {relative_to_absolute_path(combined_path)}.")
+@deprecated("Use dataset_module_factory or metric_module_factory instead.")
def prepare_module(
path: str,
revision: Optional[Union[str, Version]] = None,
@@ -1262,14 +1264,18 @@ def prepare_module(
script_version="deprecated",
**download_kwargs,
) -> Union[Tuple[str, str], Tuple[str, str, Optional[str]]]:
- """For backward compatibility. Please use dataset_module_factory or metric_module_factory instead."""
+ """
+ .. deprecated:: 1.13
+ `prepare_module` was deprecated in version 1.13 and will be removed in the next major version.
+ For backward compatibility, please use :func:`dataset_module_factory` or :func:`metric_module_factory` instead.
+ """
if script_version != "deprecated":
warnings.warn(
"'script_version' was renamed to 'revision' in version 1.13 and will be removed in 1.15.", FutureWarning
)
revision = script_version
- if dataset:
- results = dataset_module_factory(
+ module = (
+ dataset_module_factory(
path,
revision=revision,
download_config=download_config,
@@ -1279,9 +1285,8 @@ def prepare_module(
data_files=data_files,
**download_kwargs,
)
- return results.module_path, results.hash
- else:
- results = metric_module_factory(
+ if dataset
+ else metric_module_factory(
path,
revision=revision,
download_config=download_config,
@@ -1290,7 +1295,8 @@ def prepare_module(
dynamic_modules_path=dynamic_modules_path,
**download_kwargs,
)
- return results.module_path, results.hash
+ )
+ return module.module_path, module.hash
def load_metric(
diff --git a/src/datasets/utils/patching.py b/src/datasets/utils/patching.py
--- a/src/datasets/utils/patching.py
+++ b/src/datasets/utils/patching.py
@@ -22,11 +22,11 @@ class patch_submodule:
Examples:
>>> import importlib
- >>> from datasets.load import prepare_module
+ >>> from datasets.load import dataset_module_factory
>>> from datasets.streaming import patch_submodule, xjoin
>>>
- >>> snli_module_path, _ = prepare_module("snli")
- >>> snli_module = importlib.import_module(snli_module_path)
+ >>> dataset_module = dataset_module_factory("snli")
+ >>> snli_module = importlib.import_module(dataset_module.module_path)
>>> patcher = patch_submodule(snli_module, "os.path.join", xjoin)
>>> patcher.start()
>>> assert snli_module.os.path.join is xjoin
| diff --git a/tests/test_dataset_common.py b/tests/test_dataset_common.py
--- a/tests/test_dataset_common.py
+++ b/tests/test_dataset_common.py
@@ -25,13 +25,13 @@
from absl.testing import parameterized
import datasets
-from datasets import cached_path, import_main_class, load_dataset, prepare_module
from datasets.builder import BuilderConfig, DatasetBuilder
from datasets.features import ClassLabel, Features, Value
+from datasets.load import dataset_module_factory, import_main_class, load_dataset
from datasets.packaged_modules import _PACKAGED_DATASETS_MODULES
from datasets.search import _has_faiss
from datasets.utils.download_manager import GenerateMode
-from datasets.utils.file_utils import DownloadConfig, is_remote_url
+from datasets.utils.file_utils import DownloadConfig, cached_path, is_remote_url
from datasets.utils.logging import get_logger
from datasets.utils.mock_download_manager import MockDownloadManager
@@ -100,11 +100,11 @@ def __init__(self, parent):
def load_builder_class(self, dataset_name, is_local=False):
# Download/copy dataset script
if is_local is True:
- module_path, _ = prepare_module(os.path.join("datasets", dataset_name))
+ dataset_module = dataset_module_factory(os.path.join("datasets", dataset_name))
else:
- module_path, _ = prepare_module(dataset_name, download_config=DownloadConfig(force_download=True))
+ dataset_module = dataset_module_factory(dataset_name, download_config=DownloadConfig(force_download=True))
# Get dataset builder class
- builder_cls = import_main_class(module_path)
+ builder_cls = import_main_class(dataset_module.module_path)
return builder_cls
def load_all_configs(self, dataset_name, is_local=False) -> List[Optional[BuilderConfig]]:
@@ -254,8 +254,8 @@ def test_load_dataset_all_configs(self, dataset_name):
@slow
def test_load_real_dataset(self, dataset_name):
path = "./datasets/" + dataset_name
- module_path, hash = prepare_module(path, download_config=DownloadConfig(local_files_only=True), dataset=True)
- builder_cls = import_main_class(module_path, dataset=True)
+ dataset_module = dataset_module_factory(path, download_config=DownloadConfig(local_files_only=True))
+ builder_cls = import_main_class(dataset_module.module_path)
name = builder_cls.BUILDER_CONFIGS[0].name if builder_cls.BUILDER_CONFIGS else None
with tempfile.TemporaryDirectory() as temp_cache_dir:
dataset = load_dataset(
@@ -268,8 +268,8 @@ def test_load_real_dataset(self, dataset_name):
@slow
def test_load_real_dataset_all_configs(self, dataset_name):
path = "./datasets/" + dataset_name
- module_path, hash = prepare_module(path, download_config=DownloadConfig(local_files_only=True), dataset=True)
- builder_cls = import_main_class(module_path, dataset=True)
+ dataset_module = dataset_module_factory(path, download_config=DownloadConfig(local_files_only=True))
+ builder_cls = import_main_class(dataset_module.module_path)
config_names = (
[config.name for config in builder_cls.BUILDER_CONFIGS] if len(builder_cls.BUILDER_CONFIGS) > 0 else [None]
)
diff --git a/tests/test_hf_gcp.py b/tests/test_hf_gcp.py
--- a/tests/test_hf_gcp.py
+++ b/tests/test_hf_gcp.py
@@ -7,7 +7,7 @@
from datasets import config
from datasets.arrow_reader import HF_GCP_BASE_URL
from datasets.builder import DatasetBuilder
-from datasets.load import import_main_class, prepare_module
+from datasets.load import dataset_module_factory, import_main_class
from datasets.utils import cached_path
@@ -52,16 +52,16 @@ class TestDatasetOnHfGcp(TestCase):
def test_dataset_info_available(self, dataset, config_name):
with TemporaryDirectory() as tmp_dir:
- local_module_path, local_hash = prepare_module(
- os.path.join("datasets", dataset), dataset=True, cache_dir=tmp_dir, local_files_only=True
+ dataset_module = dataset_module_factory(
+ os.path.join("datasets", dataset), cache_dir=tmp_dir, local_files_only=True
)
- builder_cls = import_main_class(local_module_path, dataset=True)
+ builder_cls = import_main_class(dataset_module.module_path, dataset=True)
builder_instance: DatasetBuilder = builder_cls(
cache_dir=tmp_dir,
name=config_name,
- hash=local_hash,
+ hash=dataset_module.hash,
)
dataset_info_url = os.path.join(
diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -295,58 +295,58 @@ def _dummy_module_dir(self, modules_dir, dummy_module_name, dummy_code):
f.write(dummy_code)
return module_dir
- def test_prepare_module(self):
+ def test_dataset_module_factory(self):
with tempfile.TemporaryDirectory() as tmp_dir:
# prepare module from directory path
dummy_code = "MY_DUMMY_VARIABLE = 'hello there'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name1__", dummy_code)
- importable_module_path, module_hash = datasets.load.prepare_module(
+ dataset_module = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
- dummy_module = importlib.import_module(importable_module_path)
+ dummy_module = importlib.import_module(dataset_module.module_path)
self.assertEqual(dummy_module.MY_DUMMY_VARIABLE, "hello there")
- self.assertEqual(module_hash, sha256(dummy_code.encode("utf-8")).hexdigest())
+ self.assertEqual(dataset_module.hash, sha256(dummy_code.encode("utf-8")).hexdigest())
# prepare module from file path + check resolved_file_path
dummy_code = "MY_DUMMY_VARIABLE = 'general kenobi'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name1__", dummy_code)
module_path = os.path.join(module_dir, "__dummy_module_name1__.py")
- importable_module_path, module_hash = datasets.load.prepare_module(
+ dataset_module = datasets.load.dataset_module_factory(
module_path, dynamic_modules_path=self.dynamic_modules_path
)
- dummy_module = importlib.import_module(importable_module_path)
+ dummy_module = importlib.import_module(dataset_module.module_path)
self.assertEqual(dummy_module.MY_DUMMY_VARIABLE, "general kenobi")
- self.assertEqual(module_hash, sha256(dummy_code.encode("utf-8")).hexdigest())
+ self.assertEqual(dataset_module.hash, sha256(dummy_code.encode("utf-8")).hexdigest())
# missing module
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
with self.assertRaises((FileNotFoundError, ConnectionError, requests.exceptions.ConnectionError)):
- datasets.load.prepare_module(
+ datasets.load.dataset_module_factory(
"__missing_dummy_module_name__", dynamic_modules_path=self.dynamic_modules_path
)
- def test_offline_prepare_module(self):
+ def test_offline_dataset_module_factory(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_code = "MY_DUMMY_VARIABLE = 'hello there'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name2__", dummy_code)
- importable_module_path1, _ = datasets.load.prepare_module(
+ dataset_module_1 = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
time.sleep(0.1) # make sure there's a difference in the OS update time of the python file
dummy_code = "MY_DUMMY_VARIABLE = 'general kenobi'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name2__", dummy_code)
- importable_module_path2, _ = datasets.load.prepare_module(
+ dataset_module_2 = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
self._caplog.clear()
# allow provide the module name without an explicit path to remote or local actual file
- importable_module_path3, _ = datasets.load.prepare_module(
+ dataset_module_3 = datasets.load.dataset_module_factory(
"__dummy_module_name2__", dynamic_modules_path=self.dynamic_modules_path
)
# it loads the most recent version of the module
- self.assertEqual(importable_module_path2, importable_module_path3)
- self.assertNotEqual(importable_module_path1, importable_module_path3)
+ self.assertEqual(dataset_module_2.module_path, dataset_module_3.module_path)
+ self.assertNotEqual(dataset_module_1.module_path, dataset_module_3.module_path)
self.assertIn("Using the latest cached version of the module", self._caplog.text)
def test_load_dataset_canonical(self):
diff --git a/tests/test_metric_common.py b/tests/test_metric_common.py
--- a/tests/test_metric_common.py
+++ b/tests/test_metric_common.py
@@ -63,7 +63,7 @@ class LocalMetricTest(parameterized.TestCase):
def test_load_metric(self, metric_name):
doctest.ELLIPSIS_MARKER = "[...]"
metric_module = importlib.import_module(
- datasets.load.prepare_module(os.path.join("metrics", metric_name), dataset=False)[0]
+ datasets.load.metric_module_factory(os.path.join("metrics", metric_name)).module_path
)
metric = datasets.load.import_main_class(metric_module.__name__, dataset=False)
# check parameters
@@ -81,7 +81,9 @@ def test_load_metric(self, metric_name):
@slow
def test_load_real_metric(self, metric_name):
doctest.ELLIPSIS_MARKER = "[...]"
- metric_module = importlib.import_module(datasets.load.prepare_module(os.path.join("metrics", metric_name))[0])
+ metric_module = importlib.import_module(
+ datasets.load.metric_module_factory(os.path.join("metrics", metric_name)).module_path
+ )
# run doctest
with self.use_local_metrics():
results = doctest.testmod(metric_module, verbose=True, raise_on_error=True)
| Deprecate prepare_module
In version 1.13, `prepare_module` was deprecated.
Add deprecation warning and remove its usage from all the library.
| 2021-10-26T15:28:24Z | [] | [] |
|
huggingface/datasets | 3,195 | huggingface__datasets-3195 | [
"3181"
] | 50658bb4eeee4e0a78e2a74310cbbfadee66c2b3 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -62,7 +62,6 @@
from .search import IndexableMixin
from .splits import NamedSplit, Split
from .table import (
- ConcatenationTable,
InMemoryTable,
MemoryMappedTable,
Table,
@@ -106,7 +105,7 @@ class Example(LazyDict):
def __getitem__(self, key):
value = super().__getitem__(key)
if self.decoding and self.features and key in self.features:
- value = self.features[key].decode_example(value)
+ value = self.features[key].decode_example(value) if value is not None else None
self[key] = value
del self.features[key]
return value
@@ -116,7 +115,7 @@ class Batch(LazyDict):
def __getitem__(self, key):
values = super().__getitem__(key)
if self.decoding and self.features and key in self.features:
- values = [self.features[key].decode_example(value) for value in values]
+ values = [self.features[key].decode_example(value) if value is not None else None for value in values]
self[key] = values
del self.features[key]
return values
@@ -533,6 +532,7 @@ def wrapper(*args, **kwargs):
def update_metadata_with_features(table: Table, features: Features):
"""To be used in dataset transforms that modify the features of the dataset, in order to update the features stored in the metadata of its schema."""
+ features = Features({col_name: features[col_name] for col_name in table.column_names})
if table.schema.metadata is None or "huggingface".encode("utf-8") not in table.schema.metadata:
pa_metadata = ArrowWriter._build_metadata(DatasetInfo(features=features))
else:
@@ -558,6 +558,44 @@ def _check_table(table) -> Table:
raise TypeError(f"Expected a pyarrow.Table or a datasets.table.Table object, but got {table}.")
+def _check_column_names(column_names: List[str]):
+ """Check the column names to make sure they don't contain duplicates."""
+ counter = Counter(column_names)
+ if not all(count == 1 for count in counter.values()):
+ duplicated_columns = [col for col in counter if counter[col] > 1]
+ raise ValueError(f"The table can't have duplicated columns but columns {duplicated_columns} are duplicated.")
+
+
+def _check_if_features_can_be_aligned(features_list: List[Features]):
+ """Check if the dictionaries of features can be aligned.
+
+ Two dictonaries of features can be aligned if the keys they share have the same type or some of them is of type `Value("null")`.
+ """
+ name2feature = {}
+ for features in features_list:
+ for k, v in features.items():
+ if k not in name2feature or (isinstance(name2feature[k], Value) and name2feature[k].dtype == "null"):
+ name2feature[k] = v
+
+ for features in features_list:
+ for k, v in features.items():
+ if not (isinstance(v, Value) and v.dtype == "null") and name2feature[k] != v:
+ raise ValueError(
+ f'The features can\'t be aligned because the key {k} of features {features} has unexpected type - {v} (expected either {name2feature[k]} or Value("null").'
+ )
+
+
+def _align_features(features_list: List[Features]) -> List[Features]:
+ """Align dictionaries of features so that the keys that are found in multiple dictionaries share the same feature."""
+ name2feature = {}
+ for features in features_list:
+ for k, v in features.items():
+ if k not in name2feature or (isinstance(name2feature[k], Value) and name2feature[k].dtype == "null"):
+ name2feature[k] = v
+
+ return [Features({k: name2feature[k] for k in features.keys()}) for features in features_list]
+
+
class NonExistentDatasetError(Exception):
"""Used when we expect the existence of a dataset"""
@@ -625,14 +663,7 @@ def __init__(
assert pa.types.is_unsigned_integer(
self._indices.column(0)[0].type
), f"indices must be an Arrow table of unsigned integers, current type is {self._indices.column(0)[0].type}"
- counter = Counter(self._data.column_names)
- if not all(count == 1 for count in counter.values()):
- duplicated_columns = [col for col in counter if counter[col] > 1]
- raise ValueError(
- f"The table can't have duplicated columns but columns {duplicated_columns} are duplicated."
- )
-
- # Update metadata
+ _check_column_names(self._data.column_names)
self._data = update_metadata_with_features(self._data, self.features)
@@ -1141,11 +1172,15 @@ def unique(self, column: str) -> List[Any]:
return dataset._data.column(column).unique().to_pylist()
- def class_encode_column(self, column: str) -> "Dataset":
+ def class_encode_column(self, column: str, include_nulls: bool = False) -> "Dataset":
"""Casts the given column as :obj:``datasets.features.ClassLabel`` and updates the table.
Args:
column (`str`): The name of the column to cast (list all the column names with :func:`datasets.Dataset.column_names`)
+ include_nulls (`bool`, default `False`):
+ Whether to include null values in the class labels. If True, the null values will be encoded as the `"None"` class label.
+
+ .. versionadded:: 1.14.2
"""
# Sanity checks
if column not in self._data.column_names:
@@ -1153,13 +1188,19 @@ def class_encode_column(self, column: str) -> "Dataset":
src_feat = self.features[column]
if not isinstance(src_feat, Value):
raise ValueError(
- f"Class encoding is only supported for {type(Value)} column, and column {column} is {type(src_feat)}."
+ f"Class encoding is only supported for {Value.__name__} column, and column {column} is {type(src_feat).__name__}."
)
- if src_feat.dtype != "string":
+ if src_feat.dtype != "string" or (include_nulls and None in self.unique(column)):
+
+ def stringify_column(batch):
+ batch[column] = [
+ str(sample) if include_nulls or sample is not None else None for sample in batch[column]
+ ]
+ return batch
+
dset = self.map(
- lambda batch: {column: [str(sample) for sample in batch]},
- input_columns=column,
+ stringify_column,
batched=True,
desc="Stringifying the column",
)
@@ -1167,15 +1208,20 @@ def class_encode_column(self, column: str) -> "Dataset":
dset = self
# Create the new feature
- class_names = sorted(dset.unique(column))
+ class_names = sorted(sample for sample in dset.unique(column) if include_nulls or sample is not None)
dst_feat = ClassLabel(names=class_names)
+
+ def cast_to_class_labels(batch):
+ batch[column] = [
+ dst_feat.str2int(sample) if include_nulls or sample is not None else None for sample in batch[column]
+ ]
+ return batch
+
dset = dset.map(
- lambda batch: {column: dst_feat.str2int(batch)},
- input_columns=column,
+ cast_to_class_labels,
batched=True,
desc="Casting to class labels",
)
- dset = concatenate_datasets([self.remove_columns([column]), dset], axis=1)
new_features = dset.features.copy()
new_features[column] = dst_feat
@@ -1591,7 +1637,7 @@ def rename(columns):
)
dataset._data = dataset._data.rename_columns(new_column_names)
- dataset._data = update_metadata_with_features(dataset._data, self.features)
+ dataset._data = update_metadata_with_features(dataset._data, dataset.features)
dataset._fingerprint = new_fingerprint
return dataset
@@ -2701,6 +2747,7 @@ def sort(
column: str,
reverse: bool = False,
kind: str = None,
+ null_placement: str = "last",
keep_in_memory: bool = False,
load_from_cache_file: bool = True,
indices_cache_file_name: Optional[str] = None,
@@ -2709,16 +2756,20 @@ def sort(
) -> "Dataset":
"""Create a new dataset sorted according to a column.
- Currently sorting according to a column name uses numpy sorting algorithm under the hood.
- The column should thus be a numpy compatible type (in particular not a nested type).
+ Currently sorting according to a column name uses pandas sorting algorithm under the hood.
+ The column should thus be a pandas compatible type (in particular not a nested type).
This also means that the column used for sorting is fully loaded in memory (which should be fine in most cases).
Args:
column (:obj:`str`): column name to sort by.
reverse (:obj:`bool`, default `False`): If True, sort by descending order rather then ascending.
- kind (:obj:`str`, optional): Numpy algorithm for sorting selected in {‘quicksort’, ‘mergesort’, ‘heapsort’, ‘stable’},
+ kind (:obj:`str`, optional): Pandas algorithm for sorting selected in {‘quicksort’, ‘mergesort’, ‘heapsort’, ‘stable’},
The default is ‘quicksort’. Note that both ‘stable’ and ‘mergesort’ use timsort under the covers and, in general,
the actual implementation will vary with data type. The ‘mergesort’ option is retained for backwards compatibility.
+ null_placement (:obj:`str`, default `last`):
+ Put `None` values at the beginning if ‘first‘; ‘last‘ puts `None` values at the end.
+
+ .. versionadded:: 1.14.2
keep_in_memory (:obj:`bool`, default `False`): Keep the sorted indices in memory instead of writing it to a cache file.
load_from_cache_file (:obj:`bool`, default `True`): If a cache file storing the sorted indices
can be identified, use it instead of recomputing.
@@ -2755,11 +2806,13 @@ def sort(
)
column_data = self._getitem(
- column, format_type="numpy", format_columns=None, output_all_columns=False, format_kwargs=None
+ column, format_type="pandas", format_columns=None, output_all_columns=False, format_kwargs=None
+ )
+
+ df_sorted = column_data.to_frame().sort_values(
+ column, ascending=not reverse, kind=kind, na_position=null_placement
)
- indices = np.argsort(column_data, kind=kind)
- if reverse:
- indices = indices[::-1]
+ indices = df_sorted.index.to_numpy()
return self.select(
indices=indices,
@@ -3477,8 +3530,9 @@ def add_column(self, name: str, column: Union[list, np.array], new_fingerprint:
:class:`Dataset`
"""
column_table = InMemoryTable.from_pydict({name: column})
+ _check_column_names(self._data.column_names + column_table.column_names)
# Concatenate tables horizontally
- table = ConcatenationTable.from_tables([self._data, column_table], axis=1)
+ table = concat_tables([self._data, column_table], axis=1)
# Update features
info = self.info.copy()
info.features.update(Features.from_arrow_schema(column_table.schema))
@@ -3692,21 +3746,28 @@ def add_item(self, item: dict, new_fingerprint: str):
Returns:
:class:`Dataset`
"""
- item_table = InMemoryTable.from_pydict({k: [item[k]] for k in self.features.keys() if k in item})
- # Cast item
- schema = pa.schema(self.features.type)
- item_table = item_table.cast(schema)
- # Concatenate tables
- table = concat_tables([self._data, item_table])
+ item_table = InMemoryTable.from_pydict({k: [v] for k, v in item.items()})
+ # We don't call _check_if_features_can_be_aligned here so this cast is "unsafe"
+ dset_features, item_features = _align_features([self.features, Features.from_arrow_schema(item_table.schema)])
+ # Cast to align the schemas of the tables and concatenate the tables
+ table = concat_tables(
+ [
+ self._data.cast(pa.schema(dset_features.type)) if self.features != dset_features else self._data,
+ item_table.cast(pa.schema(item_features.type)),
+ ]
+ )
if self._indices is None:
indices_table = None
else:
item_indices_array = pa.array([len(self._data)], type=pa.uint64())
item_indices_table = InMemoryTable.from_arrays([item_indices_array], names=["indices"])
indices_table = concat_tables([self._indices, item_indices_table])
+ info = self.info.copy()
+ info.features.update(item_features)
+ table = update_metadata_with_features(table, info.features)
return Dataset(
table,
- info=self.info.copy(),
+ info=info,
split=self.split,
indices_table=indices_table,
fingerprint=new_fingerprint,
@@ -3750,8 +3811,13 @@ def align_labels_with_mapping(self, label2id: Dict, label_column: str) -> "Datas
int2str_function = label_feature.int2str
def process_label_ids(batch):
- dset_label_names = [int2str_function(label_id).lower() for label_id in batch[label_column]]
- batch[label_column] = [label2id[label_name] for label_name in dset_label_names]
+ dset_label_names = [
+ int2str_function(label_id).lower() if label_id is not None else None
+ for label_id in batch[label_column]
+ ]
+ batch[label_column] = [
+ label2id[label_name] if label_name is not None else None for label_name in dset_label_names
+ ]
return batch
features = self.features.copy()
@@ -3778,10 +3844,20 @@ def concatenate_datasets(
.. versionadded:: 1.6.0
"""
- if axis == 0 and not all([dset.features.type == dsets[0].features.type for dset in dsets]):
- raise ValueError("Features must match for all datasets")
- elif axis == 1 and not all([dset.num_rows == dsets[0].num_rows for dset in dsets]):
- raise ValueError("Number of rows must match for all datasets")
+ # Ignore datasets with no rows
+ if any(dset.num_rows > 0 for dset in dsets):
+ dsets = [dset for dset in dsets if dset.num_rows > 0]
+ else:
+ # Return first dataset if all datasets are empty
+ return dsets[0]
+
+ # Perform checks (and a potentional cast if axis=0)
+ if axis == 0:
+ _check_if_features_can_be_aligned([dset.features for dset in dsets])
+ else:
+ if not all([dset.num_rows == dsets[0].num_rows for dset in dsets]):
+ raise ValueError("Number of rows must match for all datasets")
+ _check_column_names([col_name for dset in dsets for col_name in dset._data.column_names])
# Find common format or reset format
format = dsets[0].format
@@ -3820,26 +3896,22 @@ def apply_offset_to_indices_table(table, offset):
indices_table = concat_tables(indices_tables)
else:
indices_table = InMemoryTable.from_batches([], schema=pa.schema({"indices": pa.int64()}))
- elif axis == 1 and len(dsets) == 1:
- indices_table = dsets[0]._indices
- elif axis == 1 and len(dsets) > 1:
- for i in range(len(dsets)):
- dsets[i] = dsets[i].flatten_indices()
- indices_table = None
+ else:
+ if len(dsets) == 1:
+ indices_table = dsets[0]._indices
+ else:
+ for i in range(len(dsets)):
+ dsets[i] = dsets[i].flatten_indices()
+ indices_table = None
else:
indices_table = None
- # Concatenate tables
- tables_to_concat = [dset._data for dset in dsets if len(dset._data) > 0]
- # There might be no table with data left hence return first empty table
- if not tables_to_concat:
- return dsets[0]
- table = concat_tables(tables_to_concat, axis=axis)
- if axis == 1:
- # Merge features (ignore duplicated columns for now and let Dataset.__init__ check for those)
- table = update_metadata_with_features(
- table, Features({k: v for dset in dsets for k, v in dset.features.items()})
- )
+ table = concat_tables([dset._data for dset in dsets], axis=axis)
+ if axis == 0:
+ features_list = _align_features([dset.features for dset in dsets])
+ else:
+ features_list = [dset.features for dset in dsets]
+ table = update_metadata_with_features(table, {k: v for features in features_list for k, v in features.items()})
# Concatenate infos
if info is None:
diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -38,6 +38,7 @@
from .keyhash import DuplicatedKeysError, KeyHasher
from .utils import logging
from .utils.file_utils import hash_url_to_filename
+from .utils.py_utils import first_non_null_value
logger = logging.get_logger(__name__)
@@ -111,14 +112,15 @@ def __arrow_array__(self, type=None):
else:
type = self.type
trying_int_optimization = False
+ non_null_idx, non_null_value = first_non_null_value(self.data)
if type is None: # automatic type inference for custom objects
- if config.PIL_AVAILABLE and "PIL" in sys.modules and isinstance(self.data[0], PIL.Image.Image):
+ if config.PIL_AVAILABLE and "PIL" in sys.modules and isinstance(non_null_value, PIL.Image.Image):
type = ImageExtensionType()
try:
if isinstance(type, _ArrayXDExtensionType):
if isinstance(self.data, np.ndarray):
storage = numpy_to_pyarrow_listarray(self.data, type=type.value_type)
- elif isinstance(self.data, list) and self.data and isinstance(self.data[0], np.ndarray):
+ elif isinstance(self.data, list) and self.data and isinstance(non_null_value, np.ndarray):
storage = list_of_np_array_to_pyarrow_listarray(self.data, type=type.value_type)
else:
storage = pa.array(self.data, type.storage_dtype)
@@ -130,17 +132,17 @@ def __arrow_array__(self, type=None):
out = numpy_to_pyarrow_listarray(self.data)
if type is not None:
out = out.cast(type)
- elif isinstance(self.data, list) and self.data and isinstance(self.data[0], np.ndarray):
+ elif isinstance(self.data, list) and self.data and isinstance(non_null_value, np.ndarray):
out = list_of_np_array_to_pyarrow_listarray(self.data)
if type is not None:
out = out.cast(type)
else:
out = pa.array(cast_to_python_objects(self.data, only_1d_for_numpy=True), type=type)
- if trying_type and not isinstance(type, ImageExtensionType):
+ if trying_type and not isinstance(type, ImageExtensionType) and non_null_idx != -1:
is_equal = (
- np.array_equal(np.array(out[0].as_py()), self.data[0])
- if isinstance(self.data[0], np.ndarray)
- else out[0].as_py() == self.data[0]
+ np.array_equal(np.array(out[non_null_idx].as_py()), self.data[non_null_idx])
+ if isinstance(self.data[non_null_idx], np.ndarray)
+ else out[non_null_idx].as_py() == self.data[non_null_idx]
)
if not is_equal:
raise TypeError(
@@ -161,7 +163,11 @@ def __arrow_array__(self, type=None):
try: # second chance
if isinstance(self.data, np.ndarray):
return numpy_to_pyarrow_listarray(self.data)
- elif isinstance(self.data, list) and self.data and isinstance(self.data[0], np.ndarray):
+ elif (
+ isinstance(self.data, list)
+ and self.data
+ and any(isinstance(value, np.ndarray) for value in self.data)
+ ):
return list_of_np_array_to_pyarrow_listarray(self.data)
else:
return pa.array(cast_to_python_objects(self.data, only_1d_for_numpy=True))
diff --git a/src/datasets/combine.py b/src/datasets/combine.py
--- a/src/datasets/combine.py
+++ b/src/datasets/combine.py
@@ -119,15 +119,6 @@ def _interleave_map_style_datasets(
"""
from .arrow_dataset import concatenate_datasets
- if not all([dset.features.type == datasets[0].features.type for dset in datasets]):
- raise ValueError("Features must match for all datasets")
-
- # Find common format or reset format
- format = datasets[0].format
- if any(dset.format != format for dset in datasets):
- format = {}
- logger.info("Some of the datasets have disparate format. Resetting the format of the interleaved dataset.")
-
# To interleave the datasets, we concatenate them and then we re-order the indices
concatenated_datasets = concatenate_datasets(datasets, info=info, split=split)
diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -267,14 +267,20 @@ def rename_column(self, original_column_name: str, new_column_name: str) -> "Dat
}
)
- def class_encode_column(self, column: str) -> "DatasetDict":
+ def class_encode_column(self, column: str, include_nulls: bool = False) -> "DatasetDict":
"""Casts the given column as :obj:``datasets.features.ClassLabel`` and updates the tables.
Args:
column (`str`): The name of the column to cast
+ include_nulls (`bool`, default `False`):
+ Whether to include null values in the class labels. If True, the null values will be encoded as the `"None"` class label.
+
+ .. versionadded:: 1.14.2
"""
self._check_values_type()
- return DatasetDict({k: dataset.class_encode_column(column=column) for k, dataset in self.items()})
+ return DatasetDict(
+ {k: dataset.class_encode_column(column=column, include_nulls=include_nulls) for k, dataset in self.items()}
+ )
@contextlib.contextmanager
def formatted_as(
@@ -577,6 +583,7 @@ def sort(
column: str,
reverse: bool = False,
kind: str = None,
+ null_placement: str = "last",
keep_in_memory: bool = False,
load_from_cache_file: bool = True,
indices_cache_file_names: Optional[Dict[str, Optional[str]]] = None,
@@ -585,25 +592,28 @@ def sort(
"""Create a new dataset sorted according to a column.
The transformation is applied to all the datasets of the dataset dictionary.
- Currently sorting according to a column name uses numpy sorting algorithm under the hood.
- The column should thus be a numpy compatible type (in particular not a nested type).
+ Currently sorting according to a column name uses pandas sorting algorithm under the hood.
+ The column should thus be a pandas compatible type (in particular not a nested type).
This also means that the column used for sorting is fully loaded in memory (which should be fine in most cases).
Args:
- column (`str`): column name to sort by.
- reverse: (`bool`, defaults to `False`): If True, sort by descending order rather then ascending.
- kind (Optional `str`): Numpy algorithm for sorting selected in {‘quicksort’, ‘mergesort’, ‘heapsort’, ‘stable’},
+ column (:obj:`str`): column name to sort by.
+ reverse (:obj:`bool`, default `False`): If True, sort by descending order rather then ascending.
+ kind (:obj:`str`, optional): Pandas algorithm for sorting selected in {‘quicksort’, ‘mergesort’, ‘heapsort’, ‘stable’},
The default is ‘quicksort’. Note that both ‘stable’ and ‘mergesort’ use timsort under the covers and, in general,
the actual implementation will vary with data type. The ‘mergesort’ option is retained for backwards compatibility.
- keep_in_memory (`bool`, defaults to `False`): Keep the dataset in memory instead of writing it to a cache file.
- load_from_cache_file (`bool`, defaults to `True`): If a cache file storing the current computation from `function`
+ null_placement (:obj:`str`, default `last`):
+ Put `None` values at the beginning if ‘first‘; ‘last‘ puts `None` values at the end.
+
+ .. versionadded:: 1.14.2
+ keep_in_memory (:obj:`bool`, default `False`): Keep the sorted indices in memory instead of writing it to a cache file.
+ load_from_cache_file (:obj:`bool`, default `True`): If a cache file storing the sorted indices
can be identified, use it instead of recomputing.
indices_cache_file_names (`Optional[Dict[str, str]]`, defaults to `None`): Provide the name of a path for the cache file. It is used to store the
indices mapping instead of the automatically generated cache file name.
You have to provide one :obj:`cache_file_name` per dataset in the dataset dictionary.
writer_batch_size (:obj:`int`, default `1000`): Number of rows per write operation for the cache file writer.
- This value is a good trade-off between memory usage during the processing, and processing speed.
- Higher value makes the processing do fewer lookups, lower value consume less temporary memory while running `.map()`.
+ Higher value gives smaller cache files, lower value consume less temporary memory.
"""
self._check_values_type()
if indices_cache_file_names is None:
@@ -614,6 +624,7 @@ def sort(
column=column,
reverse=reverse,
kind=kind,
+ null_placement=null_placement,
keep_in_memory=keep_in_memory,
load_from_cache_file=load_from_cache_file,
indices_cache_file_name=indices_cache_file_names[k],
diff --git a/src/datasets/features/audio.py b/src/datasets/features/audio.py
--- a/src/datasets/features/audio.py
+++ b/src/datasets/features/audio.py
@@ -1,4 +1,3 @@
-from collections import defaultdict
from dataclasses import dataclass, field
from io import BytesIO
from typing import Any, ClassVar, Optional
@@ -124,11 +123,3 @@ def _decode_mp3(self, path_or_file):
if self.mono:
array = array.mean(axis=0)
return array, sampling_rate
-
- def decode_batch(self, values):
- decoded_batch = defaultdict(list)
- for value in values:
- decoded_example = self.decode_example(value)
- for k, v in decoded_example.items():
- decoded_batch[k].append(v)
- return dict(decoded_batch)
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -33,7 +33,6 @@
from pandas.api.extensions import ExtensionArray as PandasExtensionArray
from pandas.api.extensions import ExtensionDtype as PandasExtensionDtype
from pyarrow.lib import TimestampType
-from pyarrow.types import is_boolean, is_primitive
from datasets import config, utils
from datasets.features.audio import Audio
@@ -369,7 +368,7 @@ def __init__(self, shape: tuple, dtype: str):
assert (
self.ndims is not None and self.ndims > 1
), "You must instantiate an array type with a value for dim that is > 1"
- assert len(shape) == self.ndims, f"shape={shape} and ndims={self.ndims} dom't match"
+ assert len(shape) == self.ndims, f"shape={shape} and ndims={self.ndims} don't match"
self.shape = tuple(shape)
self.value_type = dtype
self.storage_dtype = self._generate_dtype(self.value_type)
@@ -412,7 +411,7 @@ class Array5DExtensionType(_ArrayXDExtensionType):
ndims = 5
-def _is_zero_copy_only(pa_type: pa.DataType) -> bool:
+def _is_zero_copy_only(pa_type: pa.DataType, unnest: bool = False) -> bool:
"""
When converting a pyarrow array to a numpy array, we must know whether this could be done in zero-copy or not.
This function returns the value of the ``zero_copy_only`` parameter to pass to ``.to_numpy()``, given the type of the pyarrow array.
@@ -423,12 +422,20 @@ def _is_zero_copy_only(pa_type: pa.DataType) -> bool:
# see https://arrow.apache.org/docs/python/generated/pyarrow.Array.html#pyarrow.Array.to_numpy
# and https://issues.apache.org/jira/browse/ARROW-2871?jql=text%20~%20%22boolean%20to_numpy%22
"""
- return is_primitive(pa_type) and not is_boolean(pa_type)
+
+ def _unnest_pa_type(pa_type: pa.DataType) -> pa.DataType:
+ if pa.types.is_list(pa_type):
+ return _unnest_pa_type(pa_type.value_type)
+ return pa_type
+
+ if unnest:
+ pa_type = _unnest_pa_type(pa_type)
+ return pa.types.is_primitive(pa_type) and not pa.types.is_boolean(pa_type)
class ArrayExtensionArray(pa.ExtensionArray):
def __array__(self):
- zero_copy_only = _is_zero_copy_only(self.storage.type)
+ zero_copy_only = _is_zero_copy_only(self.storage.type, unnest=True)
return self.to_numpy(zero_copy_only=zero_copy_only)
def __getitem__(self, i):
@@ -437,11 +444,18 @@ def __getitem__(self, i):
def to_numpy(self, zero_copy_only=True):
storage: pa.ListArray = self.storage
size = 1
+
+ null_indices = np.arange(len(storage))[storage.is_null().to_numpy(zero_copy_only=False)]
+
for i in range(self.type.ndims):
size *= self.type.shape[i]
storage = storage.flatten()
numpy_arr = storage.to_numpy(zero_copy_only=zero_copy_only)
- numpy_arr = numpy_arr.reshape(len(self), *self.type.shape)
+ numpy_arr = numpy_arr.reshape(len(self) - len(null_indices), *self.type.shape)
+
+ if len(null_indices):
+ numpy_arr = np.insert(numpy_arr.astype(np.float64), null_indices, np.nan, axis=0)
+
return numpy_arr
def to_list_of_numpy(self, zero_copy_only=True):
@@ -454,20 +468,23 @@ def to_list_of_numpy(self, zero_copy_only=True):
arrays = []
first_dim_offsets = np.array([off.as_py() for off in storage.offsets])
- for i in range(len(storage)):
- storage_el = storage[i : i + 1]
- first_dim = first_dim_offsets[i + 1] - first_dim_offsets[i]
- # flatten storage
- for _ in range(ndims):
- storage_el = storage_el.flatten()
+ for i, is_null in enumerate(storage.is_null().to_numpy(zero_copy_only=False)):
+ if is_null:
+ arrays.append(np.nan)
+ else:
+ storage_el = storage[i : i + 1]
+ first_dim = first_dim_offsets[i + 1] - first_dim_offsets[i]
+ # flatten storage
+ for _ in range(ndims):
+ storage_el = storage_el.flatten()
- numpy_arr = storage_el.to_numpy(zero_copy_only=zero_copy_only)
- arrays.append(numpy_arr.reshape(first_dim, *shape[1:]))
+ numpy_arr = storage_el.to_numpy(zero_copy_only=zero_copy_only)
+ arrays.append(numpy_arr.reshape(first_dim, *shape[1:]))
return arrays
def to_pylist(self):
- zero_copy_only = _is_zero_copy_only(self.storage.type)
+ zero_copy_only = _is_zero_copy_only(self.storage.type, unnest=True)
if self.type.shape[0] is None:
return self.to_list_of_numpy(zero_copy_only=zero_copy_only)
else:
@@ -486,7 +503,7 @@ def __from_arrow__(self, array: Union[pa.Array, pa.ChunkedArray]):
"Dynamic first dimension is not supported for "
f"PandasArrayExtensionDtype, dimension: {array.type.shape}"
)
- zero_copy_only = _is_zero_copy_only(array.type)
+ zero_copy_only = _is_zero_copy_only(array.type, unnest=True)
if isinstance(array, pa.ChunkedArray):
numpy_arr = np.vstack([chunk.to_numpy(zero_copy_only=zero_copy_only) for chunk in array.chunks])
else:
@@ -959,8 +976,14 @@ def numpy_to_pyarrow_listarray(arr: np.ndarray, type: pa.DataType = None) -> pa.
return values
-def list_of_pa_arrays_to_pyarrow_listarray(l_arr: List[pa.Array]) -> pa.ListArray:
- offsets = pa.array(np.cumsum([0] + [len(arr) for arr in l_arr]), type=pa.int32())
+def list_of_pa_arrays_to_pyarrow_listarray(l_arr: List[Optional[pa.Array]]) -> pa.ListArray:
+ null_indices = [i for i, arr in enumerate(l_arr) if arr is None]
+ l_arr = [arr for arr in l_arr if arr is not None]
+ offsets = np.cumsum(
+ [0] + [len(arr) for arr in l_arr], dtype=np.object
+ ) # convert to dtype object to allow None insertion
+ offsets = np.insert(offsets, null_indices, None)
+ offsets = pa.array(offsets, type=pa.int32())
values = pa.concat_arrays(l_arr)
return pa.ListArray.from_arrays(offsets, values)
@@ -968,7 +991,9 @@ def list_of_pa_arrays_to_pyarrow_listarray(l_arr: List[pa.Array]) -> pa.ListArra
def list_of_np_array_to_pyarrow_listarray(l_arr: List[np.ndarray], type: pa.DataType = None) -> pa.ListArray:
"""Build a PyArrow ListArray from a possibly nested list of NumPy arrays"""
if len(l_arr) > 0:
- return list_of_pa_arrays_to_pyarrow_listarray([numpy_to_pyarrow_listarray(arr, type=type) for arr in l_arr])
+ return list_of_pa_arrays_to_pyarrow_listarray(
+ [numpy_to_pyarrow_listarray(arr, type=type) if arr is not None else None for arr in l_arr]
+ )
else:
return pa.array([], type=type)
@@ -1097,7 +1122,9 @@ def decode_example(self, example: dict):
:obj:`dict[str, Any]`
"""
return {
- column: feature.decode_example(value) if hasattr(feature, "decode_example") else value
+ column: feature.decode_example(value)
+ if hasattr(feature, "decode_example") and value is not None
+ else value
for column, (feature, value) in utils.zip_dict(
{key: value for key, value in self.items() if key in example}, example
)
@@ -1114,7 +1141,7 @@ def decode_column(self, column: list, column_name: str):
:obj:`list[Any]`
"""
return (
- [self[column_name].decode_example(value) for value in column]
+ [self[column_name].decode_example(value) if value is not None else None for value in column]
if hasattr(self[column_name], "decode_example")
else column
)
@@ -1131,7 +1158,7 @@ def decode_batch(self, batch: dict):
decoded_batch = {}
for column_name, column in batch.items():
decoded_batch[column_name] = (
- [self[column_name].decode_example(value) for value in column]
+ [self[column_name].decode_example(value) if value is not None else None for value in column]
if hasattr(self[column_name], "decode_example")
else column
)
diff --git a/src/datasets/features/image.py b/src/datasets/features/image.py
--- a/src/datasets/features/image.py
+++ b/src/datasets/features/image.py
@@ -10,6 +10,7 @@
from .. import config
from ..utils.file_utils import is_local_path
+from ..utils.py_utils import first_non_null_value, no_op_if_value_is_null
from ..utils.streaming_download_manager import xopen
@@ -273,18 +274,23 @@ def objects_to_list_of_image_dicts(
raise ImportError("To support encoding images, please install 'Pillow'.")
if objs:
- obj = objs[0]
+ _, obj = first_non_null_value(objs)
if isinstance(obj, str):
- return [{"path": obj, "bytes": None} for obj in objs]
+ return [{"path": obj, "bytes": None} if obj is not None else None for obj in objs]
if isinstance(obj, np.ndarray):
- return [{"path": None, "bytes": image_to_bytes(PIL.Image.fromarray(obj.astype(np.uint8)))} for obj in objs]
- elif isinstance(obj, PIL.Image.Image):
return [
- {"path": obj.filename, "bytes": None}
- if hasattr(obj, "filename") and obj.filename != ""
- else {"path": None, "bytes": image_to_bytes(obj)}
+ {"path": None, "bytes": image_to_bytes(PIL.Image.fromarray(obj.astype(np.uint8)))}
+ if obj is not None
+ else None
for obj in objs
]
+ elif isinstance(obj, PIL.Image.Image):
+ obj_to_image_dict_func = no_op_if_value_is_null(
+ lambda obj: {"path": obj.filename, "bytes": None}
+ if hasattr(obj, "filename") and obj.filename != ""
+ else {"path": None, "bytes": image_to_bytes(obj)}
+ )
+ return [obj_to_image_dict_func(obj) for obj in objs]
else:
return objs
else:
diff --git a/src/datasets/formatting/formatting.py b/src/datasets/formatting/formatting.py
--- a/src/datasets/formatting/formatting.py
+++ b/src/datasets/formatting/formatting.py
@@ -22,6 +22,7 @@
from ..features import _ArrayXDExtensionType, _is_zero_copy_only, pandas_types_mapper
from ..table import Table
+from ..utils import no_op_if_value_is_null
T = TypeVar("T")
@@ -96,6 +97,10 @@ def _query_table(table: Table, key: Union[int, slice, range, str, Iterable]) ->
_raise_bad_key_type(key)
+def _is_array_with_nulls(pa_array: pa.Array) -> bool:
+ return pa_array.null_count > 0
+
+
class BaseArrowExtractor(Generic[RowFormat, ColumnFormat, BatchFormat]):
"""
Arrow extractor are used to extract data from pyarrow tables.
@@ -154,26 +159,44 @@ def extract_batch(self, pa_table: pa.Table) -> dict:
return {col: self._arrow_array_to_numpy(pa_table[col]) for col in pa_table.column_names}
def _arrow_array_to_numpy(self, pa_array: pa.Array) -> np.ndarray:
- zero_copy_only = _is_zero_copy_only(pa_array.type)
if isinstance(pa_array, pa.ChunkedArray):
- # don't call to_numpy() directly or we end up with a np.array with dtype object
- # call to_numpy on the chunks instead
- # for ArrayExtensionArray call py_list directly to support dynamic dimensions
if isinstance(pa_array.type, _ArrayXDExtensionType):
- array: List = [row for chunk in pa_array.chunks for row in chunk.to_pylist()]
+ # don't call to_pylist() to preserve dtype of the fixed-size array
+ zero_copy_only = _is_zero_copy_only(pa_array.type.storage_dtype, unnest=True)
+ if pa_array.type.shape[0] is None:
+ array: List = [
+ row
+ for chunk in pa_array.chunks
+ for row in chunk.to_list_of_numpy(zero_copy_only=zero_copy_only)
+ ]
+ else:
+ array: List = [
+ row for chunk in pa_array.chunks for row in chunk.to_numpy(zero_copy_only=zero_copy_only)
+ ]
else:
+ zero_copy_only = _is_zero_copy_only(pa_array.type) and all(
+ not _is_array_with_nulls(chunk) for chunk in pa_array.chunks
+ )
array: List = [
row for chunk in pa_array.chunks for row in chunk.to_numpy(zero_copy_only=zero_copy_only)
]
else:
- # cast to list of arrays or we end up with a np.array with dtype object
- # for ArrayExtensionArray call py_list directly to support dynamic dimensions
if isinstance(pa_array.type, _ArrayXDExtensionType):
- array: List = pa_array.to_pylist()
+ # don't call to_pylist() to preserve dtype of the fixed-size array
+ zero_copy_only = _is_zero_copy_only(pa_array.type.storage_dtype, unnest=True)
+ if pa_array.type.shape[0] is None:
+ array: List = pa_array.to_list_of_numpy(zero_copy_only=zero_copy_only)
+ else:
+ array: List = pa_array.to_numpy(zero_copy_only=zero_copy_only)
else:
+ zero_copy_only = _is_zero_copy_only(pa_array.type) and not _is_array_with_nulls(pa_array)
array: List = pa_array.to_numpy(zero_copy_only=zero_copy_only).tolist()
if len(array) > 0:
- if any(isinstance(x, np.ndarray) and (x.dtype == np.object or x.shape != array[0].shape) for x in array):
+ if any(
+ (isinstance(x, np.ndarray) and (x.dtype == np.object or x.shape != array[0].shape))
+ or (isinstance(x, float) and np.isnan(x))
+ for x in array
+ ):
return np.array(array, copy=False, **{**self.np_array_kwargs, "dtype": np.object})
return np.array(array, copy=False, **self.np_array_kwargs)
@@ -210,7 +233,7 @@ def __init__(self, features):
def decode_row(self, row: pd.DataFrame) -> pd.DataFrame:
decode = (
{
- column_name: feature.decode_example
+ column_name: no_op_if_value_is_null(feature.decode_example)
for column_name, feature in self.features.items()
if column_name in row.columns and hasattr(feature, "decode_example")
}
@@ -223,7 +246,7 @@ def decode_row(self, row: pd.DataFrame) -> pd.DataFrame:
def decode_column(self, column: pd.Series, column_name: str) -> pd.Series:
decode = (
- self.features[column_name].decode_example
+ no_op_if_value_is_null(self.features[column_name].decode_example)
if self.features and column_name in self.features and hasattr(self.features[column_name], "decode_example")
else None
)
diff --git a/src/datasets/table.py b/src/datasets/table.py
--- a/src/datasets/table.py
+++ b/src/datasets/table.py
@@ -610,15 +610,7 @@ def __setstate__(self, state):
def _concat_blocks(blocks: List[Union[TableBlock, pa.Table]], axis: int = 0) -> pa.Table:
pa_tables = [table.table if hasattr(table, "table") else table for table in blocks]
if axis == 0:
- # Align schemas: re-order the columns to make the schemas match before concatenating over rows
- schema = pa_tables[0].schema
- pa_tables = [
- table
- if table.schema == schema
- else pa.Table.from_arrays([table[name] for name in schema.names], names=schema.names)
- for table in pa_tables
- ]
- return pa.concat_tables(pa_tables)
+ return pa.concat_tables(pa_tables, promote=True)
elif axis == 1:
for i, table in enumerate(pa_tables):
if i == 0:
diff --git a/src/datasets/utils/__init__.py b/src/datasets/utils/__init__.py
--- a/src/datasets/utils/__init__.py
+++ b/src/datasets/utils/__init__.py
@@ -26,10 +26,12 @@
classproperty,
copyfunc,
dumps,
+ first_non_null_value,
flatten_nest_dict,
has_sufficient_disk_space,
map_nested,
memoize,
+ no_op_if_value_is_null,
size_str,
temporary_assignment,
unique_values,
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -129,6 +129,23 @@ def unique_values(values):
yield value
+def no_op_if_value_is_null(func):
+ """If the value is None, return None, else call `func`."""
+
+ def wrapper(value):
+ return func(value) if value is not None else None
+
+ return wrapper
+
+
+def first_non_null_value(iterable):
+ """Return the index and the value of the first non-null value in the iterable. If all values are None, return -1 as index."""
+ for i, value in enumerate(iterable):
+ if value is not None:
+ return i, value
+ return -1, None
+
+
def zip_dict(*dicts):
"""Iterate over items of dictionaries grouped by their keys."""
for key in unique_values(itertools.chain(*dicts)): # set merge all keys
| diff --git a/tests/features/test_array_xd.py b/tests/features/test_array_xd.py
--- a/tests/features/test_array_xd.py
+++ b/tests/features/test_array_xd.py
@@ -319,11 +319,33 @@ def test_array_xd_numpy_arrow_extractor(dtype, dummy_value):
np.testing.assert_equal(arr, np.array([[[dummy_value] * 2] * 2], dtype=np.dtype(dtype)))
-def test_dataset_map():
+def test_array_xd_with_none():
+ # Fixed shape
+ features = datasets.Features({"foo": datasets.Array2D(dtype="int32", shape=(2, 2))})
+ dummy_array = np.array([[1, 2], [3, 4]], dtype="int32")
+ dataset = datasets.Dataset.from_dict({"foo": [dummy_array, None, dummy_array]}, features=features)
+ arr = NumpyArrowExtractor().extract_column(dataset._data)
+ assert isinstance(arr, np.ndarray) and arr.dtype == np.float64 and arr.shape == (3, 2, 2)
+ assert np.allclose(arr[0], dummy_array) and np.allclose(arr[2], dummy_array)
+ assert np.all(np.isnan(arr[1])) # broadcasted np.nan - use np.all
+
+ # Dynamic shape
+ features = datasets.Features({"foo": datasets.Array2D(dtype="int32", shape=(None, 2))})
+ dummy_array = np.array([[1, 2], [3, 4]], dtype="int32")
+ dataset = datasets.Dataset.from_dict({"foo": [dummy_array, None, dummy_array]}, features=features)
+ arr = NumpyArrowExtractor().extract_column(dataset._data)
+ assert isinstance(arr, np.ndarray) and arr.dtype == np.object and arr.shape == (3,)
+ np.testing.assert_equal(arr[0], dummy_array)
+ np.testing.assert_equal(arr[2], dummy_array)
+ assert np.isnan(arr[1]) # a single np.nan value - np.all not needed
+
+
[email protected]("with_none", [False, True])
+def test_dataset_map(with_none):
ds = datasets.Dataset.from_dict({"path": ["path1", "path2"]})
def process_data(batch):
- return {
+ batch = {
"image": [
np.array(
[
@@ -335,11 +357,17 @@ def process_data(batch):
for _ in batch["path"]
]
}
+ if with_none:
+ batch["image"][0] = None
+ return batch
features = datasets.Features({"image": Array3D(dtype="int32", shape=(3, 3, 3))})
processed_ds = ds.map(process_data, batched=True, remove_columns=ds.column_names, features=features)
assert processed_ds.shape == (2, 1)
with processed_ds.with_format("numpy") as pds:
- for example in pds:
+ for i, example in enumerate(pds):
assert "image" in example
assert isinstance(example["image"], np.ndarray)
+ assert example["image"].shape == (3, 3, 3)
+ if with_none and i == 0:
+ assert np.all(np.isnan(example["image"]))
diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -2238,6 +2238,28 @@ def test_cast_with_sliced_list():
assert casted_dataset.features == new_features
[email protected]("include_nulls", [False, True])
+def test_class_encode_column_with_none(include_nulls):
+ dataset = Dataset.from_dict({"col_1": ["a", "b", "c", None, "d", None]})
+ dataset = dataset.class_encode_column("col_1", include_nulls=include_nulls)
+ class_names = ["a", "b", "c", "d"]
+ if include_nulls:
+ class_names += ["None"]
+ assert isinstance(dataset.features["col_1"], ClassLabel)
+ assert set(dataset.features["col_1"].names) == set(class_names)
+ assert (None in dataset.unique("col_1")) == (not include_nulls)
+
+
[email protected]("null_placement", ["first", "last"])
+def test_sort_with_none(null_placement):
+ dataset = Dataset.from_dict({"col_1": ["item_2", "item_3", "item_1", None, "item_4", None]})
+ dataset = dataset.sort("col_1", null_placement=null_placement)
+ if null_placement == "first":
+ assert dataset["col_1"] == [None, None, "item_1", "item_2", "item_3", "item_4"]
+ else:
+ assert dataset["col_1"] == ["item_1", "item_2", "item_3", "item_4", None, None]
+
+
def test_update_metadata_with_features(dataset_dict):
table1 = pa.Table.from_pydict(dataset_dict)
features1 = Features.from_arrow_schema(table1.schema)
@@ -2273,6 +2295,24 @@ def test_concatenate_datasets(dataset_type, axis, expected_shape, dataset_dict,
assert_arrow_metadata_are_synced_with_dataset_features(dataset)
+def test_concatenate_datasets_new_columns():
+ dataset1 = Dataset.from_dict({"col_1": ["a", "b", "c"]})
+ dataset2 = Dataset.from_dict({"col_1": ["d", "e", "f"], "col_2": [True, False, True]})
+ dataset = concatenate_datasets([dataset1, dataset2])
+ assert dataset.data.shape == (6, 2)
+ assert dataset.features == Features({"col_1": Value("string"), "col_2": Value("bool")})
+ assert dataset[:] == {"col_1": ["a", "b", "c", "d", "e", "f"], "col_2": [None, None, None, True, False, True]}
+ dataset3 = Dataset.from_dict({"col_3": ["a_1"]})
+ dataset = concatenate_datasets([dataset, dataset3])
+ assert dataset.data.shape == (7, 3)
+ assert dataset.features == Features({"col_1": Value("string"), "col_2": Value("bool"), "col_3": Value("string")})
+ assert dataset[:] == {
+ "col_1": ["a", "b", "c", "d", "e", "f", None],
+ "col_2": [None, None, None, True, False, True, None],
+ "col_3": [None, None, None, None, None, None, "a_1"],
+ }
+
+
@pytest.mark.parametrize("axis", [0, 1])
def test_concatenate_datasets_complex_features(axis):
n = 5
@@ -2456,6 +2496,30 @@ def test_dataset_add_item(item, in_memory, dataset_dict, arrow_path, transform):
assert dataset_indices == dataset_to_test_indices + [len(dataset_to_test._data)]
+def test_dataset_add_item_new_columns():
+ dataset = Dataset.from_dict({"col_1": [0, 1, 2]}, features=Features({"col_1": Value("uint8")}))
+ dataset = dataset.add_item({"col_1": 3, "col_2": "a"})
+ assert dataset.data.shape == (4, 2)
+ assert dataset.features == Features({"col_1": Value("uint8"), "col_2": Value("string")})
+ assert dataset[:] == {"col_1": [0, 1, 2, 3], "col_2": [None, None, None, "a"]}
+ dataset = dataset.add_item({"col_3": True})
+ assert dataset.data.shape == (5, 3)
+ assert dataset.features == Features({"col_1": Value("uint8"), "col_2": Value("string"), "col_3": Value("bool")})
+ assert dataset[:] == {
+ "col_1": [0, 1, 2, 3, None],
+ "col_2": [None, None, None, "a", None],
+ "col_3": [None, None, None, None, True],
+ }
+
+
+def test_dataset_add_item_introduce_feature_type():
+ dataset = Dataset.from_dict({"col_1": [None, None, None]})
+ dataset = dataset.add_item({"col_1": "a"})
+ assert dataset.data.shape == (4, 1)
+ assert dataset.features == Features({"col_1": Value("string")})
+ assert dataset[:] == {"col_1": [None, None, None, "a"]}
+
+
@pytest.mark.parametrize("in_memory", [False, True])
def test_dataset_from_file(in_memory, dataset, arrow_file):
filename = arrow_file
diff --git a/tests/test_table.py b/tests/test_table.py
--- a/tests/test_table.py
+++ b/tests/test_table.py
@@ -91,6 +91,10 @@ def assert_index_attributes_equal(table: Table, other: Table):
assert table._schema == other._schema
+def add_suffix_to_column_names(table, suffix):
+ return table.rename_columns([f"{name}{suffix}" for name in table.column_names])
+
+
def test_inject_arrow_table_documentation(in_memory_pa_table):
method = pa.Table.slice
@@ -636,9 +640,11 @@ def test_concatenation_table_from_tables(axis, in_memory_pa_table, arrow_file):
if axis == 0:
expected_table = pa.concat_tables([in_memory_pa_table] * len(tables))
else:
+ # avoids error due to duplicate column names
+ tables[1:] = [add_suffix_to_column_names(table, i) for i, table in enumerate(tables[1:], 1)]
expected_table = in_memory_pa_table
- for _ in range(1, len(tables)):
- for name, col in zip(in_memory_pa_table.column_names, in_memory_pa_table.columns):
+ for table in tables[1:]:
+ for name, col in zip(table.column_names, table.columns):
expected_table = expected_table.append_column(name, col)
with assert_arrow_memory_doesnt_increase():
@@ -911,7 +917,10 @@ def test_concat_tables(arrow_file, in_memory_pa_table):
assert isinstance(concatenated_table.blocks[0][0], InMemoryTable)
assert isinstance(concatenated_table.blocks[1][0], MemoryMappedTable)
assert isinstance(concatenated_table.blocks[2][0], InMemoryTable)
- concatenated_table = concat_tables(tables, axis=1)
+ # add suffix to avoid error due to duplicate column names
+ concatenated_table = concat_tables(
+ [add_suffix_to_column_names(table, i) for i, table in enumerate(tables)], axis=1
+ )
assert concatenated_table.table.shape == (10, 16)
assert len(concatenated_table.blocks[0]) == 3 # t0 and t1 are consolidated as a single InMemoryTable
assert isinstance(concatenated_table.blocks[0][0], InMemoryTable)
| `None` converted to `"None"` when loading a dataset
## Describe the bug
When loading a dataset `None` values of the type `NoneType` are converted to `'None'` of the type `str`.
## Steps to reproduce the bug
```python
from datasets import load_dataset
qasper = load_dataset("qasper", split="train", download_mode="reuse_cache_if_exists")
print(qasper[60]["full_text"]["section_name"])
```
When installing version 1.1.40, the output is
`[None, 'Introduction', 'Benchmark Datasets', ...]`
When installing from the master branch, the output is
`['None', 'Introduction', 'Benchmark Datasets', ...]`
Notice how the first element was changed from `NoneType` to `str`.
## Expected results
`None` should stay as is.
## Actual results
`None` is converted to a string.
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: master
- Platform: Linux-4.4.0-19041-Microsoft-x86_64-with-glibc2.17
- Python version: 3.8.10
- PyArrow version: 4.0.1
| Hi @eladsegal, thanks for reporting.
@mariosasko I saw you are already working on this, but maybe my comment will be useful to you.
All values are casted to their corresponding feature type (including `None` values). For example if the feature type is `Value("bool")`, `None` is casted to `False`.
It is true that strings were an exception, but this was recently fixed by @lhoestq (see #3158).
Thanks for reporting.
This is actually a breaking change that I think can cause issues when users preprocess their data. String columns used to be nullable. Maybe we can correct https://github.com/huggingface/datasets/pull/3158 to keep the None values and avoid this breaking change ?
EDIT: the other types (bool, int, etc) can also become nullable IMO
So what would be the best way to handle a feature that can have a null value in some of the instances? So far I used `None`.
Using the empty string won't be a good option, as it can be an actual value in the data and is not the same as not having a value at all.
Hi @eladsegal,
Use `None`. As @albertvillanova correctly pointed out, this change in conversion was introduced (by mistake) in #3158. To avoid it, install the earlier revision with:
```
pip install git+https://github.com/huggingface/datasets.git@8107844ec0e7add005db0585c772ee20adc01a5e
```
I'm making all the feature types nullable as we speak, and the fix will be merged probably early next week. | 2021-11-02T11:15:10Z | [] | [] |
huggingface/datasets | 3,196 | huggingface__datasets-3196 | [
"3040"
] | 03afcab3210e3e31491244847a7d0d02c32508a2 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -937,18 +937,6 @@ def save_to_disk(self, dataset_path: str, fs=None):
Saves a dataset to a dataset directory, or in a filesystem using either :class:`~filesystems.S3FileSystem` or
any implementation of ``fsspec.spec.AbstractFileSystem``.
-
- Note regarding sliced datasets:
-
- If you sliced the dataset in some way (using shard, train_test_split or select for example), then an indices mapping
- is added to avoid having to rewrite a new arrow Table (save time + disk/memory usage).
- It maps the indices used by __getitem__ to the right rows if the arrow Table.
- By default save_to_disk does save the full dataset table + the mapping.
-
- If you want to only save the shard of the dataset instead of the original arrow file and the indices,
- then you have to call :func:`datasets.Dataset.flatten_indices` before saving.
- This will create a new arrow table by using the right rows of the original table.
-
Args:
dataset_path (:obj:`str`): Path (e.g. `dataset/train`) or remote URI (e.g. `s3://my-bucket/dataset/train`)
of the dataset directory where the dataset will be saved to.
@@ -959,6 +947,8 @@ def save_to_disk(self, dataset_path: str, fs=None):
not self.list_indexes()
), "please remove all the indexes using `dataset.drop_index` before saving a dataset"
+ dataset = self.flatten_indices() if self._indices is not None else self
+
if is_remote_filesystem(fs):
dataset_path = extract_path_from_uri(dataset_path)
else:
@@ -976,7 +966,7 @@ def save_to_disk(self, dataset_path: str, fs=None):
# Get json serializable state
state = {
- key: self.__dict__[key]
+ key: dataset.__dict__[key]
for key in [
"_fingerprint",
"_format_columns",
@@ -987,13 +977,10 @@ def save_to_disk(self, dataset_path: str, fs=None):
]
}
- split = self.__dict__["_split"]
+ split = dataset.__dict__["_split"]
state["_split"] = str(split) if split is not None else split
state["_data_files"] = [{"filename": config.DATASET_ARROW_FILENAME}]
- state["_indices_data_files"] = (
- [{"filename": config.DATASET_INDICES_FILENAME}] if self._indices is not None else None
- )
for k in state["_format_kwargs"].keys():
try:
json.dumps(state["_format_kwargs"][k])
@@ -1003,19 +990,14 @@ def save_to_disk(self, dataset_path: str, fs=None):
) from None
# Get json serializable dataset info
- dataset_info = asdict(self._info)
+ dataset_info = asdict(dataset._info)
# Save dataset + indices + state + info
fs.makedirs(dataset_path, exist_ok=True)
with fs.open(Path(dataset_path, config.DATASET_ARROW_FILENAME).as_posix(), "wb") as dataset_file:
with ArrowWriter(stream=dataset_file) as writer:
- writer.write_table(self._data)
+ writer.write_table(dataset._data)
writer.finalize()
- if self._indices is not None:
- with fs.open(Path(dataset_path, config.DATASET_INDICES_FILENAME).as_posix(), "wb") as indices_file:
- with ArrowWriter(stream=indices_file) as writer:
- writer.write_table(self._indices)
- writer.finalize()
with fs.open(
Path(dataset_path, config.DATASET_STATE_JSON_FILENAME).as_posix(), "w", encoding="utf-8"
) as state_file:
@@ -1083,20 +1065,12 @@ def load_from_disk(dataset_path: str, fs=None, keep_in_memory: Optional[bool] =
table_cls.from_file(Path(dataset_path, data_file["filename"]).as_posix())
for data_file in state["_data_files"]
)
- if state.get("_indices_data_files"):
- indices_table = concat_tables(
- table_cls.from_file(Path(dataset_path, indices_file["filename"]).as_posix())
- for indices_file in state["_indices_data_files"]
- )
- else:
- indices_table = None
split = state["_split"]
split = Split(split) if split is not None else split
return Dataset(
arrow_table=arrow_table,
- indices_table=indices_table,
info=dataset_info,
split=split,
fingerprint=state["_fingerprint"],
@@ -1154,13 +1128,11 @@ def unique(self, column: str) -> List[Any]:
raise ValueError(f"Column ({column}) not in table columns ({self._data.column_names}).")
if self._indices is not None and self._indices.num_rows != self._data.num_rows:
- raise ValueError(
- f"This dataset is a shallow copy using an indices mapping of another Datset {self._data.num_rows}."
- f"The `Dataset.unique()` method is currently not handled on shallow copy. Please use `Dataset.flatten_indices()` "
- f"to create a deep copy of the dataset and be able to use `Dataset.unique()`."
- )
+ dataset = self.flatten_indices()
+ else:
+ dataset = self
- return self._data.column(column).unique().to_pylist()
+ return dataset._data.column(column).unique().to_pylist()
def class_encode_column(self, column: str) -> "Dataset":
"""Casts the given column as :obj:``datasets.features.ClassLabel`` and updates the table.
@@ -1177,10 +1149,12 @@ def class_encode_column(self, column: str) -> "Dataset":
f"Class encoding is only supported for {type(Value)} column, and column {column} is {type(src_feat)}."
)
- # Stringify the column
if src_feat.dtype != "string":
dset = self.map(
- lambda batch: {column: [str(sample) for sample in batch]}, input_columns=column, batched=True
+ lambda batch: {column: [str(sample) for sample in batch]},
+ input_columns=column,
+ batched=True,
+ desc="Stringifying the column",
)
else:
dset = self
@@ -1188,7 +1162,12 @@ def class_encode_column(self, column: str) -> "Dataset":
# Create the new feature
class_names = sorted(dset.unique(column))
dst_feat = ClassLabel(names=class_names)
- dset = dset.map(lambda batch: {column: dst_feat.str2int(batch)}, input_columns=column, batched=True)
+ dset = dset.map(
+ lambda batch: {column: dst_feat.str2int(batch)},
+ input_columns=column,
+ batched=True,
+ desc="Casting to class labels",
+ )
dset = concatenate_datasets([self.remove_columns([column]), dset], axis=1)
new_features = dset.features.copy()
@@ -1319,6 +1298,7 @@ def cast_(
writer_batch_size=writer_batch_size,
num_proc=num_proc,
features=features,
+ desc="Casting the dataset",
)
self._data = dataset._data
self._info = dataset._info
@@ -1380,6 +1360,7 @@ def cast(
writer_batch_size=writer_batch_size,
num_proc=num_proc,
features=features,
+ desc="Casting the dataset",
)
dataset = dataset.with_format(**format)
return dataset
@@ -2593,6 +2574,7 @@ def flatten_indices(
features=features,
disable_nullable=disable_nullable,
new_fingerprint=new_fingerprint,
+ desc="Flattening the indices",
)
def _new_dataset_with_indices(
@@ -3619,21 +3601,31 @@ def align_labels_with_mapping(self, label2id: Dict, label_column: str) -> "Datas
label2id = {'CONTRADICTION': 0, 'NEUTRAL': 1, 'ENTAILMENT': 2}
ds_aligned = ds.align_labels_with_mapping(label2id, "label")
"""
- features = self.features.copy()
- int2str_function = features[label_column].int2str
+ # Sanity checks
+ if label_column not in self._data.column_names:
+ raise ValueError(f"Column ({label_column}) not in table columns ({self._data.column_names}).")
+
+ label_feature = self.features[label_column]
+ if not isinstance(label_feature, ClassLabel):
+ raise ValueError(
+ f"Aligning labels with a mapping is only supported for {ClassLabel.__name__} column, and column {label_feature} is {type(label_feature).__name__}."
+ )
+
# Sort input mapping by ID value to ensure the label names are aligned
label2id = dict(sorted(label2id.items(), key=lambda item: item[1]))
label_names = list(label2id.keys())
- features[label_column] = ClassLabel(num_classes=len(label_names), names=label_names)
# Some label mappings use uppercase label names so we lowercase them during alignment
label2id = {k.lower(): v for k, v in label2id.items()}
+ int2str_function = label_feature.int2str
def process_label_ids(batch):
dset_label_names = [int2str_function(label_id).lower() for label_id in batch[label_column]]
batch[label_column] = [label2id[label_name] for label_name in dset_label_names]
return batch
- return self.map(process_label_ids, features=features, batched=True)
+ features = self.features.copy()
+ features[label_column] = ClassLabel(num_classes=len(label_names), names=label_names)
+ return self.map(process_label_ids, features=features, batched=True, desc="Aligning the labels")
def concatenate_datasets(
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -612,8 +612,6 @@ class ClassLabel:
* `names`: a list of label strings
* `names_file`: a file containing the list of labels.
- Note: On python2, the strings are encoded as utf-8.
-
Args:
num_classes: `int`, number of classes. All labels must be < num_classes.
names: `list<str>`, string names for the integer classes. The
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -1823,10 +1823,6 @@ def test_flatten_indices(self, in_memory):
self.assertNotEqual(dset._indices, None)
- # Test unique fail
- with self.assertRaises(ValueError):
- dset.unique(dset.column_names[0])
-
tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
fingerprint = dset._fingerprint
dset.set_format("numpy")
| [save_to_disk] Using `select()` followed by `save_to_disk` saves complete dataset making it hard to create dummy dataset
## Describe the bug
When only keeping a dummy size of a dataset (say the first 100 samples), and then saving it to disk to upload it in the following to the hub for easy demo/use - not just the small dataset is saved but the whole dataset with an indices file. The problem with this is that the dataset is still very big.
## Steps to reproduce the bug
E.g. run the following:
```python
from datasets import load_dataset, save_to_disk
nlp = load_dataset("glue", "mnli", split="train")
nlp.save_to_disk("full")
nlp = nlp.select(range(100))
nlp.save_to_disk("dummy")
```
Now one can see that both `"dummy"` and `"full"` have the same size. This shouldn't be the case IMO.
## Expected results
IMO `"dummy"` should be much smaller so that one can easily play around with the dataset on the hub.
## Actual results
Specify the actual results or traceback.
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 1.12.2.dev0
- Platform: Linux-5.11.0-34-generic-x86_64-with-glibc2.10
- Python version: 3.8.5
- PyArrow version: 5.0.0
| Hi,
the `save_to_disk` docstring explains that `flatten_indices` has to be called on a dataset before saving it to save only the shard/slice of the dataset.
That works! Thansk!
Might be worth doing that automatically actually in case the `save_to_disk` is called on a dataset that has an indices mapping :-)
I agree with @patrickvonplaten: this issue is reported recurrently, so better if we implement the `.flatten_indices()` automatically?
That would be great indeed - I don't really see a use case where one would not like to call `.flatten_indices()` before calling `save_to_disk`
+1 on this ! | 2021-11-02T11:28:50Z | [] | [] |
huggingface/datasets | 3,234 | huggingface__datasets-3234 | [
"2206"
] | f6dcafce996f39b6a4bbe3a9833287346f4a4b68 | diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -100,6 +100,7 @@ def __arrow_array__(self, type=None):
trying_type = True
else:
type = self.type
+ trying_int_optimization = False
try:
if isinstance(type, _ArrayXDExtensionType):
if isinstance(self.data, np.ndarray):
@@ -130,6 +131,7 @@ def __arrow_array__(self, type=None):
"Specified try_type alters data. Please check that the type/feature that you provided match the type/features of the data."
)
if self.optimized_int_type and self.type is None and self.try_type is None:
+ trying_int_optimization = True
if pa.types.is_int64(out.type):
out = out.cast(self.optimized_int_type)
elif pa.types.is_list(out.type):
@@ -154,6 +156,10 @@ def __arrow_array__(self, type=None):
type_(self.data), e
)
) from None
+ elif trying_int_optimization and "not in range" in str(e):
+ optimized_int_type_str = np.dtype(self.optimized_int_type.to_pandas_dtype()).name
+ logger.info(f"Failed to cast a sequence to {optimized_int_type_str}. Falling back to int64.")
+ return out
else:
raise
elif "overflow" in str(e):
@@ -162,6 +168,10 @@ def __arrow_array__(self, type=None):
type_(self.data), e
)
) from None
+ elif trying_int_optimization and "not in range" in str(e):
+ optimized_int_type_str = np.dtype(self.optimized_int_type.to_pandas_dtype()).name
+ logger.info(f"Failed to cast a sequence to {optimized_int_type_str}. Falling back to int64.")
+ return out
else:
raise
| diff --git a/tests/test_arrow_writer.py b/tests/test_arrow_writer.py
--- a/tests/test_arrow_writer.py
+++ b/tests/test_arrow_writer.py
@@ -1,7 +1,9 @@
+import copy
import os
import tempfile
from unittest import TestCase
+import numpy as np
import pyarrow as pa
import pytest
@@ -211,6 +213,13 @@ def get_base_dtype(arr_type):
return arr_type
+def change_first_primitive_element_in_list(lst, value):
+ if isinstance(lst[0], list):
+ change_first_primitive_element_in_list(lst[0], value)
+ else:
+ lst[0] = value
+
+
@pytest.mark.parametrize("optimized_int_type, expected_dtype", [(None, pa.int64()), (pa.int32(), pa.int32())])
@pytest.mark.parametrize("sequence", [[1, 2, 3], [[1, 2, 3]], [[[1, 2, 3]]]])
def test_optimized_int_type_for_typed_sequence(sequence, optimized_int_type, expected_dtype):
@@ -230,9 +239,19 @@ def test_optimized_int_type_for_typed_sequence(sequence, optimized_int_type, exp
)
@pytest.mark.parametrize("sequence", [[1, 2, 3], [[1, 2, 3]], [[[1, 2, 3]]]])
def test_optimized_typed_sequence(sequence, col, expected_dtype):
+ # in range
arr = pa.array(OptimizedTypedSequence(sequence, col=col))
assert get_base_dtype(arr.type) == expected_dtype
+ # not in range
+ if col != "other":
+ # avoids errors due to in-place modifications
+ sequence = copy.deepcopy(sequence)
+ value = np.iinfo(expected_dtype.to_pandas_dtype()).max + 1
+ change_first_primitive_element_in_list(sequence, value)
+ arr = pa.array(OptimizedTypedSequence(sequence, col=col))
+ assert get_base_dtype(arr.type) == pa.int64()
+
@pytest.mark.parametrize("raise_exception", [False, True])
def test_arrow_writer_closes_stream(raise_exception, tmp_path):
| Got pyarrow error when loading a dataset while adding special tokens into the tokenizer
I added five more special tokens into the GPT2 tokenizer. But after that, when I try to pre-process the data using my previous code, I got an error shown below:
Traceback (most recent call last):
File "/home/xuyan/anaconda3/envs/convqa/lib/python3.7/site-packages/datasets/arrow_dataset.py", line 1687, in _map_single
writer.write(example)
File "/home/xuyan/anaconda3/envs/convqa/lib/python3.7/site-packages/datasets/arrow_writer.py", line 296, in write
self.write_on_file()
File "/home/xuyan/anaconda3/envs/convqa/lib/python3.7/site-packages/datasets/arrow_writer.py", line 270, in write_on_file
pa_array = pa.array(typed_sequence)
File "pyarrow/array.pxi", line 222, in pyarrow.lib.array
File "pyarrow/array.pxi", line 110, in pyarrow.lib._handle_arrow_array_protocol
File "/home/xuyan/anaconda3/envs/convqa/lib/python3.7/site-packages/datasets/arrow_writer.py", line 108, in __arrow_array__
out = out.cast(pa.list_(self.optimized_int_type))
File "pyarrow/array.pxi", line 810, in pyarrow.lib.Array.cast
File "/home/xuyan/anaconda3/envs/convqa/lib/python3.7/site-packages/pyarrow/compute.py", line 281, in cast
return call_function("cast", [arr], options)
File "pyarrow/_compute.pyx", line 465, in pyarrow._compute.call_function
File "pyarrow/_compute.pyx", line 294, in pyarrow._compute.Function.call
File "pyarrow/error.pxi", line 122, in pyarrow.lib.pyarrow_internal_check_status
File "pyarrow/error.pxi", line 84, in pyarrow.lib.check_status
pyarrow.lib.ArrowInvalid: Integer value 50259 not in range: -128 to 127
Do you have any idea about it?
| Hi,
the output of the tokenizers is treated specially in the lib to optimize the dataset size (see the code [here](https://github.com/huggingface/datasets/blob/master/src/datasets/arrow_writer.py#L138-L141)). It looks like that one of the values in a dictionary returned by the tokenizer is out of the assumed range.
Can you please provide a minimal reproducible example for more help?
Hi @yana-xuyan, thanks for reporting.
As clearly @mariosasko explained, `datasets` performs some optimizations in order to reduce the size of the dataset cache files. And one of them is storing the field `special_tokens_mask` as `int8`, which means that this field can only contain integers between `-128` to `127`. As your message error states, one of the values of this field is `50259`, and therefore it cannot be stored as an `int8`.
Maybe we could implement a way to disable this optimization and allow using any integer value; although the size of the cache files would be much larger.
I'm facing same issue @mariosasko @albertvillanova
```
ArrowInvalid: Integer value 50260 not in range: -128 to 127
```
To reproduce:
```python
SPECIAL_TOKENS = ['<bos>','<eos>','<speaker1>','<speaker2>','<pad>']
ATTR_TO_SPECIAL_TOKEN = {
'bos_token': '<bos>',
'eos_token': '<eos>',
'pad_token': '<pad>',
'additional_special_tokens': ['<speaker1>', '<speaker2>']
}
tokenizer = AutoTokenizer.from_pretrained("gpt2", use_fast=False)
num_added_tokens =tokenizer.add_special_tokens(ATTR_TO_SPECIAL_TOKEN)
vocab_size = len(self.tokenizer.encoder) + num_added_tokens
vocab =tokenizer.get_vocab()
pad_index = tokenizer.pad_token_id
eos_index = tokenizer.eos_token_id
bos_index = tokenizer.bos_token_id
speaker1_index = vocab["<speaker1>"]
speaker2_index = vocab["<speaker2>"]
```
```python
tokenizer.decode(['50260'])
'<speaker1>'
```
@mariosasko
I am hitting this bug in the Bert tokenizer too. I see that @albertvillanova labeled this as a bug back in April. Has there been a fix released yet?
What I did for now is to just disable the optimization in the HF library. @yana-xuyan and @thomas-happify, is that what you did and did that work for you?
Hi @gregg-ADP,
This is still a bug.
As @albertvillanova has suggested, maybe it's indeed worth adding a variable to `config.py` to have a way to disable this behavior.
In the meantime, this forced optimization can be disabled by specifying `features` (of the returned examples) in the `map` call:
```python
from datasets import *
... # dataset init
ds.map(process_example, features=Features({"special_tokens_mask": Sequence(Value("int32")), ... rest of the features})
```
cc @lhoestq so he is also aware of this issue
Thanks for the quick reply @mariosasko. What I did was to changed the optimizer to use int32 instead of int8.
What you're suggesting specifies the type for each feature explicitly without changing the HF code. This is definitely a better option. However, we are hitting a new error later:
```
File "/Users/ccccc/PycharmProjects/aaaa-ml/venv-source/lib/python3.8/site-packages/torch/nn/modules/module.py", line 1051, in _call_impl
return forward_call(*input, **kwargs)
TypeError: forward() got an unexpected keyword argument 'pos'
```
Where 'pos' is the name of a new feature we added. Do you agree that your way of fixing the optimizer issue will not fix our new issue? If not, I will continue with this optimizer fix until we resolve our other issue.
| 2021-11-08T16:10:27Z | [] | [] |
huggingface/datasets | 3,277 | huggingface__datasets-3277 | [
"3257"
] | d8a998cf2deb18fe0b55361ec4218f91ffdb8be9 | diff --git a/benchmarks/format.py b/benchmarks/format.py
--- a/benchmarks/format.py
+++ b/benchmarks/format.py
@@ -24,12 +24,12 @@ def format_json_to_md(input_json_file, output_md_file):
old_val = metric_vals.get("old", None)
dif_val = metric_vals.get("diff", None)
- val_str = " {:f}".format(new_val) if isinstance(new_val, (int, float)) else "None"
+ val_str = f" {new_val:f}" if isinstance(new_val, (int, float)) else "None"
if old_val is not None:
- val_str += " / {:f}".format(old_val) if isinstance(old_val, (int, float)) else "None"
+ val_str += f" / {new_val:f}" if isinstance(old_val, (int, float)) else "None"
if dif_val is not None:
- val_str += " ({:f})".format(dif_val) if isinstance(dif_val, (int, float)) else "None"
+ val_str += f" ({dif_val:f})" if isinstance(dif_val, (int, float)) else "None"
title += " " + metric_name + " |"
lines += "---|"
diff --git a/metrics/coval/coval.py b/metrics/coval/coval.py
--- a/metrics/coval/coval.py
+++ b/metrics/coval/coval.py
@@ -210,17 +210,17 @@ def get_coref_infos(
if remove_nested:
logger.info(
"Number of removed nested coreferring mentions in the key "
- "annotation: %s; and system annotation: %s" % (key_nested_coref_num, sys_nested_coref_num)
+ f"annotation: {key_nested_coref_num}; and system annotation: {sys_nested_coref_num}"
)
logger.info(
"Number of resulting singleton clusters in the key "
- "annotation: %s; and system annotation: %s" % (key_removed_nested_clusters, sys_removed_nested_clusters)
+ f"annotation: {key_removed_nested_clusters}; and system annotation: {sys_removed_nested_clusters}"
)
if not keep_singletons:
logger.info(
- "%d and %d singletons are removed from the key and system "
- "files, respectively" % (key_singletons_num, sys_singletons_num)
+ f"{key_singletons_num:d} and {sys_singletons_num:d} singletons are removed from the key and system "
+ "files, respectively"
)
return doc_coref_infos
@@ -242,14 +242,14 @@ def evaluate(key_lines, sys_lines, metrics, NP_only, remove_nested, keep_singlet
logger.info(
name.ljust(10),
- "Recall: %.2f" % (recall * 100),
- " Precision: %.2f" % (precision * 100),
- " F1: %.2f" % (f1 * 100),
+ f"Recall: {recall * 100:.2f}",
+ f" Precision: {precision * 100:.2f}",
+ f" F1: {f1 * 100:.2f}",
)
if conll_subparts_num == 3:
conll = (conll / 3) * 100
- logger.info("CoNLL score: %.2f" % conll)
+ logger.info(f"CoNLL score: {conll:.2f}")
output_scores.update({"conll_score": conll})
return output_scores
diff --git a/metrics/squad_v2/evaluate.py b/metrics/squad_v2/evaluate.py
--- a/metrics/squad_v2/evaluate.py
+++ b/metrics/squad_v2/evaluate.py
@@ -113,7 +113,7 @@ def get_raw_scores(dataset, preds):
# For unanswerable questions, only correct answer is empty string
gold_answers = [""]
if qid not in preds:
- print("Missing prediction for %s" % qid)
+ print(f"Missing prediction for {qid}")
continue
a_pred = preds[qid]
# Take max over all gold answers
@@ -156,7 +156,7 @@ def make_eval_dict(exact_scores, f1_scores, qid_list=None):
def merge_eval(main_eval, new_eval, prefix):
for k in new_eval:
- main_eval["%s_%s" % (prefix, k)] = new_eval[k]
+ main_eval[f"{prefix}_{k}"] = new_eval[k]
def plot_pr_curve(precisions, recalls, out_image, title):
@@ -238,8 +238,8 @@ def histogram_na_prob(na_probs, qid_list, image_dir, name):
plt.hist(x, weights=weights, bins=20, range=(0.0, 1.0))
plt.xlabel("Model probability of no-answer")
plt.ylabel("Proportion of dataset")
- plt.title("Histogram of no-answer probability: %s" % name)
- plt.savefig(os.path.join(image_dir, "na_prob_hist_%s.png" % name))
+ plt.title(f"Histogram of no-answer probability: {name}")
+ plt.savefig(os.path.join(image_dir, f"na_prob_hist_{name}.png"))
plt.clf()
diff --git a/metrics/super_glue/record_evaluation.py b/metrics/super_glue/record_evaluation.py
--- a/metrics/super_glue/record_evaluation.py
+++ b/metrics/super_glue/record_evaluation.py
@@ -63,7 +63,7 @@ def evaluate(dataset, predictions):
for qa in passage["qas"]:
total += 1
if qa["id"] not in predictions:
- message = "Unanswered question {} will receive score 0.".format(qa["id"])
+ message = f'Unanswered question {qa["id"]} will receive score 0.'
print(message, file=sys.stderr)
continue
@@ -95,7 +95,7 @@ def evaluate(dataset, predictions):
dataset_json = json.load(data_file)
if dataset_json["version"] != expected_version:
print(
- "Evaluation expects v-{}, but got dataset with v-{}".format(expected_version, dataset_json["version"]),
+ f'Evaluation expects v-{expected_version}, but got dataset with v-{dataset_json["version"]}',
file=sys.stderr,
)
dataset = dataset_json["data"]
@@ -106,6 +106,6 @@ def evaluate(dataset, predictions):
metrics, correct_ids = evaluate(dataset, predictions)
if args.output_correct_ids:
- print("Output {} correctly answered question IDs.".format(len(correct_ids)))
+ print(f"Output {len(correct_ids)} correctly answered question IDs.")
with open("correct_ids.json", "w") as f:
json.dump(correct_ids, f)
diff --git a/metrics/super_glue/super_glue.py b/metrics/super_glue/super_glue.py
--- a/metrics/super_glue/super_glue.py
+++ b/metrics/super_glue/super_glue.py
@@ -125,7 +125,7 @@ def evaluate_multirc(ids_preds, labels):
"""
question_map = {}
for id_pred, label in zip(ids_preds, labels):
- question_id = "{}-{}".format(id_pred["idx"]["paragraph"], id_pred["idx"]["question"])
+ question_id = f'{id_pred["idx"]["paragraph"]}-{id_pred["idx"]["question"]}'
pred = id_pred["prediction"]
if question_id in question_map:
question_map[question_id].append((pred, label))
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -621,9 +621,7 @@ def __init__(
assert self._fingerprint is not None, "Fingerprint can't be None in a Dataset object"
if self.info.features.type != inferred_features.type:
raise ValueError(
- "External features info don't match the dataset:\nGot\n{}\nwith type\n{}\n\nbut expected something like\n{}\nwith type\n{}".format(
- self.info.features, self.info.features.type, inferred_features, inferred_features.type
- )
+ f"External features info don't match the dataset:\nGot\n{self.info.features}\nwith type\n{self.info.features.type}\n\nbut expected something like\n{inferred_features}\nwith type\n{inferred_features.type}"
)
if self._indices is not None:
@@ -735,9 +733,7 @@ def from_pandas(
"""
if info is not None and features is not None and info.features != features:
raise ValueError(
- "Features specified in `features` and `info.features` can't be different:\n{}\n{}".format(
- features, info.features
- )
+ f"Features specified in `features` and `info.features` can't be different:\n{features}\n{info.features}"
)
features = features if features is not None else info.features if info is not None else None
if info is None:
@@ -768,9 +764,7 @@ def from_dict(
"""
if info is not None and features is not None and info.features != features:
raise ValueError(
- "Features specified in `features` and `info.features` can't be different:\n{}\n{}".format(
- features, info.features
- )
+ f"Features specified in `features` and `info.features` can't be different:\n{features}\n{info.features}"
)
features = features if features is not None else info.features if info is not None else None
if info is None:
@@ -1008,7 +1002,7 @@ def save_to_disk(self, dataset_path: str, fs=None):
# Sort only the first level of keys, or we might shuffle fields of nested features if we use sort_keys=True
sorted_keys_dataset_info = {key: dataset_info[key] for key in sorted(dataset_info)}
json.dump(sorted_keys_dataset_info, dataset_info_file, indent=2)
- logger.info("Dataset saved in {}".format(dataset_path))
+ logger.info(f"Dataset saved in {dataset_path}")
@staticmethod
def load_from_disk(dataset_path: str, fs=None, keep_in_memory: Optional[bool] = None) -> "Dataset":
@@ -1216,9 +1210,7 @@ def flatten_(self, max_depth=16):
break
self.info.features = Features.from_arrow_schema(self._data.schema)
self._data = update_metadata_with_features(self._data, self.features)
- logger.info(
- "Flattened dataset from depth {} to depth {}.".format(depth, 1 if depth + 1 < max_depth else "unknown")
- )
+ logger.info(f'Flattened dataset from depth {depth} to depth { 1 if depth + 1 < max_depth else "unknown"}.')
@fingerprint_transform(inplace=False)
def flatten(self, new_fingerprint, max_depth=16) -> "Dataset":
@@ -1237,9 +1229,7 @@ def flatten(self, new_fingerprint, max_depth=16) -> "Dataset":
break
dataset.info.features = Features.from_arrow_schema(dataset._data.schema)
dataset._data = update_metadata_with_features(dataset._data, dataset.features)
- logger.info(
- "Flattened dataset from depth {} to depth {}.".format(depth, 1 if depth + 1 < max_depth else "unknown")
- )
+ logger.info(f'Flattened dataset from depth {depth} to depth {1 if depth + 1 < max_depth else "unknown"}.')
dataset._fingerprint = new_fingerprint
return dataset
@@ -1688,9 +1678,7 @@ def set_format(
columns = list(columns)
if columns is not None and any(col not in self._data.column_names for col in columns):
raise ValueError(
- "Columns {} not in the dataset. Current columns in the dataset: {}".format(
- list(filter(lambda col: col not in self._data.column_names, columns)), self._data.column_names
- )
+ f"Columns {list(filter(lambda col: col not in self._data.column_names, columns))} not in the dataset. Current columns in the dataset: {self._data.column_names}"
)
if columns is not None:
columns = columns.copy() # Ensures modifications made to the list after this call don't cause bugs
@@ -2002,9 +1990,7 @@ def decorated(item, *args, **kwargs):
for input_column in input_columns:
if input_column not in self._data.column_names:
raise ValueError(
- "Input column {} not in the dataset. Current columns in the dataset: {}".format(
- input_column, self._data.column_names
- )
+ f"Input column {input_column} not in the dataset. Current columns in the dataset: {self._data.column_names}"
)
if isinstance(remove_columns, str):
@@ -2012,10 +1998,7 @@ def decorated(item, *args, **kwargs):
if remove_columns is not None and any(col not in self._data.column_names for col in remove_columns):
raise ValueError(
- "Column to remove {} not in the dataset. Current columns in the dataset: {}".format(
- list(filter(lambda col: col not in self._data.column_names, remove_columns)),
- self._data.column_names,
- )
+ f"Column to remove {list(filter(lambda col: col not in self._data.column_names, remove_columns))} not in the dataset. Current columns in the dataset: {self._data.column_names}"
)
load_from_cache_file = load_from_cache_file if load_from_cache_file is not None else is_caching_enabled()
@@ -2057,7 +2040,7 @@ def format_cache_file_name(cache_file_name, rank):
sep = cache_file_name.rindex(".")
base_name, extension = cache_file_name[:sep], cache_file_name[sep:]
cache_file_name = base_name + suffix_template.format(rank=rank, num_proc=num_proc) + extension
- logger.info("Process #{} will write at {}".format(rank, cache_file_name))
+ logger.info(f"Process #{rank} will write at {cache_file_name}")
return cache_file_name
prev_env = deepcopy(os.environ)
@@ -2126,7 +2109,7 @@ def catch_non_existent_error(func, kwargs):
if nb_of_missing_shards > 0:
with Pool(nb_of_missing_shards, initargs=initargs, initializer=initializer) as pool:
os.environ = prev_env
- logger.info("Spawning {} processes".format(num_proc))
+ logger.info(f"Spawning {num_proc} processes")
results = {
i: pool.apply_async(self.__class__._map_single, kwds=kwds)
for i, (kwds, cached_shard) in enumerate(zip(kwds_per_shard, transformed_shards))
@@ -2143,7 +2126,7 @@ def catch_non_existent_error(func, kwargs):
transformed_shards.count(None) == 0
), "All shards have to be defined Datasets, none should still be missing."
- logger.info("Concatenating {} shards".format(num_proc))
+ logger.info(f"Concatenating {num_proc} shards")
result = concatenate_datasets(transformed_shards)
if new_fingerprint is not None:
result._fingerprint = new_fingerprint
@@ -2240,7 +2223,7 @@ def _map_single(
# current dataset file and the mapping args
cache_file_name = self._get_cache_file_path(new_fingerprint)
if os.path.exists(cache_file_name) and load_from_cache_file:
- logger.warning("Loading cached processed dataset at %s", cache_file_name)
+ logger.warning(f"Loading cached processed dataset at {cache_file_name}")
info = self.info.copy()
info.features = features
info.task_templates = None
@@ -2262,9 +2245,7 @@ def validate_function_output(processed_inputs, indices):
"""Validate output of the map function."""
if processed_inputs is not None and not isinstance(processed_inputs, (Mapping, pa.Table)):
raise TypeError(
- "Provided `function` which is applied to all elements of table returns a variable of type {}. Make sure provided `function` returns a variable of type `dict` (or a pyarrow table) to update the dataset or `None` if you are only interested in side effects.".format(
- type(processed_inputs)
- )
+ f"Provided `function` which is applied to all elements of table returns a variable of type {type(processed_inputs)}. Make sure provided `function` returns a variable of type `dict` (or a pyarrow table) to update the dataset or `None` if you are only interested in side effects."
)
elif isinstance(indices, list) and isinstance(processed_inputs, Mapping):
allowed_batch_return_types = (list, np.ndarray)
@@ -2273,9 +2254,7 @@ def validate_function_output(processed_inputs, indices):
)
if all_dict_values_are_lists is False:
raise TypeError(
- "Provided `function` which is applied to all elements of table returns a `dict` of types {}. When using `batched=True`, make sure provided `function` returns a `dict` of types like `{}`.".format(
- [type(x) for x in processed_inputs.values()], allowed_batch_return_types
- )
+ "Provided `function` which is applied to all elements of table returns a `dict` of types {[type(x) for x in processed_inputs.values()]}. When using `batched=True`, make sure provided `function` returns a `dict` of types like `{allowed_batch_return_types}`."
)
def apply_function_on_filtered_inputs(inputs, indices, check_same_num_examples=False, offset=0):
@@ -2338,7 +2317,7 @@ def init_buffer_and_writer():
)
else:
buf_writer = None
- logger.info("Caching processed dataset at %s", cache_file_name)
+ logger.info(f"Caching processed dataset at {cache_file_name}")
tmp_file = tempfile.NamedTemporaryFile("wb", dir=os.path.dirname(cache_file_name), delete=False)
writer = ArrowWriter(
features=writer_features,
@@ -2653,7 +2632,7 @@ def select(
)
else:
buf_writer = None
- logger.info("Caching indices mapping at %s", indices_cache_file_name)
+ logger.info(f"Caching indices mapping at {indices_cache_file_name}")
tmp_file = tempfile.NamedTemporaryFile("wb", dir=os.path.dirname(indices_cache_file_name), delete=False)
writer = ArrowWriter(
path=tmp_file.name, writer_batch_size=writer_batch_size, fingerprint=new_fingerprint, unit="indices"
@@ -2738,10 +2717,7 @@ def sort(
# Check the column name
if not isinstance(column, str) or column not in self._data.column_names:
raise ValueError(
- "Column '{}' not found in the dataset. Please provide a column selected in: {}".format(
- column,
- self._data.column_names,
- )
+ f"Column '{column}' not found in the dataset. Please provide a column selected in: {self._data.column_names}"
)
# Check if we've already cached this computation (indexed by a hash)
@@ -2750,7 +2726,7 @@ def sort(
# we create a unique hash from the function, current dataset file and the mapping args
indices_cache_file_name = self._get_cache_file_path(new_fingerprint)
if os.path.exists(indices_cache_file_name) and load_from_cache_file:
- logger.warning("Loading cached sorted indices for dataset at %s", indices_cache_file_name)
+ logger.warning(f"Loading cached sorted indices for dataset at {indices_cache_file_name}")
return self._new_dataset_with_indices(
fingerprint=new_fingerprint, indices_cache_file_name=indices_cache_file_name
)
@@ -2833,7 +2809,7 @@ def shuffle(
# we create a unique hash from the function, current dataset file and the mapping args
indices_cache_file_name = self._get_cache_file_path(new_fingerprint)
if os.path.exists(indices_cache_file_name) and load_from_cache_file:
- logger.warning("Loading cached shuffled indices for dataset at %s", indices_cache_file_name)
+ logger.warning(f"Loading cached shuffled indices for dataset at {indices_cache_file_name}")
return self._new_dataset_with_indices(
fingerprint=new_fingerprint, indices_cache_file_name=indices_cache_file_name
)
@@ -3008,9 +2984,7 @@ def train_test_split(
and load_from_cache_file
):
logger.warning(
- "Loading cached split indices for dataset at %s and %s",
- train_indices_cache_file_name,
- test_indices_cache_file_name,
+ f"Loading cached split indices for dataset at {train_indices_cache_file_name} and {test_indices_cache_file_name}"
)
return DatasetDict(
{
diff --git a/src/datasets/arrow_reader.py b/src/datasets/arrow_reader.py
--- a/src/datasets/arrow_reader.py
+++ b/src/datasets/arrow_reader.py
@@ -44,9 +44,9 @@
HF_GCP_BASE_URL = "https://storage.googleapis.com/huggingface-nlp/cache/datasets"
_SUB_SPEC_RE = re.compile(
- r"""
+ fr"""
^
- (?P<split>{split_re})
+ (?P<split>{_split_re[1:-1]})
(\[
((?P<from>-?\d+)
(?P<from_pct>%)?)?
@@ -55,9 +55,7 @@
(?P<to_pct>%)?)?
\])?(\((?P<rounding>[^\)]*)\))?
$
-""".format(
- split_re=_split_re[1:-1]
- ), # remove ^ and $
+""", # remove ^ and $
re.X,
)
@@ -212,7 +210,7 @@ def read(
files = self.get_file_instructions(name, instructions, split_infos)
if not files:
- msg = 'Instruction "%s" corresponds to no data!' % instructions
+ msg = f'Instruction "{instructions}" corresponds to no data!'
raise AssertionError(msg)
return self.read_files(files=files, original_instructions=instructions, in_memory=in_memory)
@@ -394,7 +392,7 @@ def _str_to_read_instruction(spec):
"""Returns ReadInstruction for given string."""
res = _SUB_SPEC_RE.match(spec)
if not res:
- raise AssertionError("Unrecognized instruction format: %s" % spec)
+ raise AssertionError(f"Unrecognized instruction format: {spec}")
unit = "%" if res.group("from_pct") or res.group("to_pct") else "abs"
return ReadInstruction(
split_name=res.group("split"),
@@ -430,7 +428,7 @@ def _rel_to_abs_instr(rel_instr, name2len):
pct_to_abs = _pct_to_abs_closest if rel_instr.rounding == "closest" else _pct_to_abs_pct1
split = rel_instr.splitname
if split not in name2len:
- raise ValueError('Unknown split "{}". Should be one of {}.'.format(split, list(name2len)))
+ raise ValueError(f'Unknown split "{split}". Should be one of {list(name2len)}.')
num_examples = name2len[split]
from_ = rel_instr.from_
to = rel_instr.to
@@ -441,7 +439,7 @@ def _rel_to_abs_instr(rel_instr, name2len):
from_ = 0 if from_ is None else from_
to = num_examples if to is None else to
if abs(from_) > num_examples or abs(to) > num_examples:
- msg = "Requested slice [%s:%s] incompatible with %s examples." % (from_ or "", to or "", num_examples)
+ msg = f'Requested slice [{from_ or ""}:{to or ""}] incompatible with {num_examples} examples.'
raise AssertionError(msg)
if from_ < 0:
from_ = num_examples + from_
@@ -559,7 +557,7 @@ def from_spec(cls, spec):
spec = str(spec) # Need to convert to str in case of NamedSplit instance.
subs = _ADDITION_SEP_RE.split(spec)
if not subs:
- raise AssertionError("No instructions could be built out of %s" % spec)
+ raise AssertionError(f"No instructions could be built out of {spec}")
instruction = _str_to_read_instruction(subs[0])
return sum([_str_to_read_instruction(sub) for sub in subs[1:]], instruction)
@@ -602,7 +600,7 @@ def __str__(self):
return self.to_spec()
def __repr__(self):
- return "ReadInstruction(%s)" % self._relative_instructions
+ return f"ReadInstruction({self._relative_instructions})"
def to_absolute(self, name2len):
"""Translate instruction into a list of absolute instructions.
diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -152,9 +152,7 @@ def __arrow_array__(self, type=None):
except pa.lib.ArrowInvalid as e:
if "overflow" in str(e):
raise OverflowError(
- "There was an overflow with type {}. Try to reduce writer_batch_size to have batches smaller than 2GB.\n({})".format(
- type_(self.data), e
- )
+ f"There was an overflow with type {type_(self.data)}. Try to reduce writer_batch_size to have batches smaller than 2GB.\n({e})"
) from None
elif trying_int_optimization and "not in range" in str(e):
optimized_int_type_str = np.dtype(self.optimized_int_type.to_pandas_dtype()).name
@@ -164,9 +162,7 @@ def __arrow_array__(self, type=None):
raise
elif "overflow" in str(e):
raise OverflowError(
- "There was an overflow with type {}. Try to reduce writer_batch_size to have batches smaller than 2GB.\n({})".format(
- type_(self.data), e
- )
+ f"There was an overflow with type {type_(self.data)}. Try to reduce writer_batch_size to have batches smaller than 2GB.\n({e})"
) from None
elif trying_int_optimization and "not in range" in str(e):
optimized_int_type_str = np.dtype(self.optimized_int_type.to_pandas_dtype()).name
@@ -339,9 +335,7 @@ def write_examples_on_file(self):
# This check fails with FloatArrays with nans, which is not what we want, so account for that:
if not isinstance(pa_array[0], pa.lib.FloatScalar):
raise OverflowError(
- "There was an overflow in the {}. Try to reduce writer_batch_size to have batches smaller than 2GB".format(
- type(pa_array)
- )
+ f"There was an overflow in the {type(pa_array)}. Try to reduce writer_batch_size to have batches smaller than 2GB"
)
arrays.append(pa_array)
inferred_types.append(inferred_type)
@@ -473,11 +467,7 @@ def finalize(self, close_stream=True):
if close_stream:
self.stream.close()
logger.debug(
- "Done writing %s %s in %s bytes %s.",
- self._num_examples,
- self.unit,
- self._num_bytes,
- self._path if self._path else "",
+ f"Done writing {self._num_examples} {self.unit} in {self._num_bytes} bytes {self._path if self._path else ''}."
)
return self._num_examples, self._num_bytes
@@ -550,7 +540,7 @@ def finalize(self, metrics_query_result: dict):
from .utils import beam_utils
# Convert to arrow
- logger.info("Converting parquet file {} to arrow {}".format(self._parquet_path, self._path))
+ logger.info(f"Converting parquet file {self._parquet_path} to arrow {self._path}")
shards = [
metadata.path
for metadata in beam.io.filesystems.FileSystems.match([self._parquet_path + "*.parquet"])[0].metadata_list
diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -289,9 +289,7 @@ def __init__(
self.info = DatasetInfo.from_directory(self._cache_dir)
else: # dir exists but no data, remove the empty dir as data aren't available anymore
logger.warning(
- "Old caching folder {} for dataset {} exists but not data were found. Removing it. ".format(
- self._cache_dir, self.name
- )
+ f"Old caching folder {self._cache_dir} for dataset {self.name} exists but not data were found. Removing it. "
)
os.rmdir(self._cache_dir)
@@ -331,25 +329,23 @@ def _create_builder_config(self, name=None, custom_features=None, **config_kwarg
if name is None and self.BUILDER_CONFIGS and not config_kwargs:
if self.DEFAULT_CONFIG_NAME is not None:
builder_config = self.builder_configs.get(self.DEFAULT_CONFIG_NAME)
- logger.warning("No config specified, defaulting to: %s/%s", self.name, builder_config.name)
+ logger.warning(f"No config specified, defaulting to: {self.name}/{builder_config.name}")
else:
if len(self.BUILDER_CONFIGS) > 1:
- example_of_usage = "load_dataset('{}', '{}')".format(self.name, self.BUILDER_CONFIGS[0].name)
+ example_of_usage = f"load_dataset('{self.name}', '{self.BUILDER_CONFIGS[0].name}')"
raise ValueError(
"Config name is missing."
- "\nPlease pick one among the available configs: %s" % list(self.builder_configs.keys())
- + "\nExample of usage:\n\t`{}`".format(example_of_usage)
+ f"\nPlease pick one among the available configs: {list(self.builder_configs.keys())}"
+ + f"\nExample of usage:\n\t`{example_of_usage}`"
)
builder_config = self.BUILDER_CONFIGS[0]
- logger.info("No config specified, defaulting to first: %s/%s", self.name, builder_config.name)
+ logger.info(f"No config specified, defaulting to first: {self.name}/{builder_config.name}")
# try get config by name
if isinstance(name, str):
builder_config = self.builder_configs.get(name)
if builder_config is None and self.BUILDER_CONFIGS:
- raise ValueError(
- "BuilderConfig %s not found. Available: %s" % (name, list(self.builder_configs.keys()))
- )
+ raise ValueError(f"BuilderConfig {name} not found. Available: {list(self.builder_configs.keys())}")
# if not using an existing config, then create a new config on the fly with config_kwargs
if not builder_config:
@@ -369,7 +365,7 @@ def _create_builder_config(self, name=None, custom_features=None, **config_kwarg
setattr(builder_config, key, value)
if not builder_config.name:
- raise ValueError("BuilderConfig must have a name, got %s" % builder_config.name)
+ raise ValueError(f"BuilderConfig must have a name, got {builder_config.name}")
# compute the config id that is going to be used for caching
config_id = builder_config.create_config_id(
@@ -378,18 +374,17 @@ def _create_builder_config(self, name=None, custom_features=None, **config_kwarg
)
is_custom = config_id not in self.builder_configs
if is_custom:
- logger.warning("Using custom data configuration %s", config_id)
+ logger.warning(f"Using custom data configuration {config_id}")
else:
if builder_config != self.builder_configs[builder_config.name]:
raise ValueError(
"Cannot name a custom BuilderConfig the same as an available "
- "BuilderConfig. Change the name. Available BuilderConfigs: %s"
- % (list(self.builder_configs.keys()))
+ f"BuilderConfig. Change the name. Available BuilderConfigs: {list(self.builder_configs.keys())}"
)
if not builder_config.version:
- raise ValueError("BuilderConfig %s must have a version" % builder_config.name)
+ raise ValueError(f"BuilderConfig {builder_config.name} must have a version")
# if not builder_config.description:
- # raise ValueError("BuilderConfig %s must have a description" % builder_config.name)
+ # raise ValueError(f"BuilderConfig {builder_config.name} must have a description" )
return builder_config, config_id
@@ -401,7 +396,7 @@ def builder_configs(cls):
configs = {config.name: config for config in cls.BUILDER_CONFIGS}
if len(configs) != len(cls.BUILDER_CONFIGS):
names = [config.name for config in cls.BUILDER_CONFIGS]
- raise ValueError("Names in BUILDER_CONFIGS must not be duplicated. Got %s" % names)
+ raise ValueError(f"Names in BUILDER_CONFIGS must not be duplicated. Got {names}")
return configs
@property
@@ -453,14 +448,9 @@ def _other_versions_on_disk():
other_version = version_dirs[0][0]
if other_version != self.config.version:
warn_msg = (
- "Found a different version {other_version} of dataset {name} in "
- "cache_dir {cache_dir}. Using currently defined version "
- "{cur_version}.".format(
- other_version=str(other_version),
- name=self.name,
- cache_dir=self._cache_dir_root,
- cur_version=str(self.config.version),
- )
+ f"Found a different version {str(other_version)} of dataset {self.name} in "
+ f"cache_dir {self._cache_dir_root}. Using currently defined version "
+ f"{str(self.config.version)}."
)
logger.warning(warn_msg)
@@ -537,22 +527,17 @@ def download_and_prepare(
with FileLock(lock_path):
data_exists = os.path.exists(self._cache_dir)
if data_exists and download_mode == GenerateMode.REUSE_DATASET_IF_EXISTS:
- logger.warning("Reusing dataset %s (%s)", self.name, self._cache_dir)
+ logger.warning(f"Reusing dataset {self.name} ({self._cache_dir})")
# We need to update the info in case some splits were added in the meantime
# for example when calling load_dataset from multiple workers.
self.info = self._load_info()
self.download_post_processing_resources(dl_manager)
return
- logger.info("Generating dataset %s (%s)", self.name, self._cache_dir)
+ logger.info(f"Generating dataset {self.name} ({self._cache_dir})")
if not is_remote_url(self._cache_dir_root): # if cache dir is local, check for available space
if not utils.has_sufficient_disk_space(self.info.size_in_bytes or 0, directory=self._cache_dir_root):
raise IOError(
- "Not enough disk space. Needed: {} (download: {}, generated: {}, post-processed: {})".format(
- utils.size_str(self.info.size_in_bytes or 0),
- utils.size_str(self.info.download_size or 0),
- utils.size_str(self.info.dataset_size or 0),
- utils.size_str(self.info.post_processing_size or 0),
- )
+ f"Not enough disk space. Needed: {utils.size_str(self.info.size_in_bytes or 0)} (download: {utils.size_str(self.info.download_size or 0)}, generated: {utils.size_str(self.info.dataset_size or 0)}, post-processed: {utils.size_str(self.info.post_processing_size or 0)})"
)
@contextlib.contextmanager
@@ -647,14 +632,12 @@ def _download_prepared_from_hf_gcs(self, download_config: DownloadConfig):
for split in self.info.splits:
for resource_file_name in self._post_processing_resources(split).values():
if os.sep in resource_file_name:
- raise ValueError("Resources shouldn't be in a sub-directory: {}".format(resource_file_name))
+ raise ValueError(f"Resources shouldn't be in a sub-directory: {resource_file_name}")
try:
resource_path = utils.cached_path(remote_cache_dir + "/" + resource_file_name)
shutil.move(resource_path, os.path.join(self._cache_dir, resource_file_name))
except ConnectionError:
- logger.info(
- "Couldn't download resourse file {} from Hf google storage.".format(resource_file_name)
- )
+ logger.info(f"Couldn't download resourse file {resource_file_name} from Hf google storage.")
logger.info("Dataset downloaded from Hf google storage.")
def _download_and_prepare(self, dl_manager, verify_infos, **prepare_split_kwargs):
@@ -690,7 +673,7 @@ def _download_and_prepare(self, dl_manager, verify_infos, **prepare_split_kwargs
"._split_generator()."
)
- logger.info("Generating split %s", split_generator.split_info.name)
+ logger.info(f"Generating split {split_generator.split_info.name}")
split_dict.add(split_generator.split_info)
try:
@@ -717,16 +700,14 @@ def download_post_processing_resources(self, dl_manager):
for split in self.info.splits:
for resource_name, resource_file_name in self._post_processing_resources(split).items():
if os.sep in resource_file_name:
- raise ValueError("Resources shouldn't be in a sub-directory: {}".format(resource_file_name))
+ raise ValueError(f"Resources shouldn't be in a sub-directory: {resource_file_name}")
resource_path = os.path.join(self._cache_dir, resource_file_name)
if not os.path.exists(resource_path):
downloaded_resource_path = self._download_post_processing_resources(
split, resource_name, dl_manager
)
if downloaded_resource_path:
- logger.info(
- "Downloaded post-processing resource {} as {}".format(resource_name, resource_file_name)
- )
+ logger.info(f"Downloaded post-processing resource {resource_name} as {resource_file_name}")
shutil.move(downloaded_resource_path, resource_path)
def _load_info(self) -> DatasetInfo:
@@ -766,16 +747,13 @@ def as_dataset(
if not os.path.exists(self._cache_dir):
raise AssertionError(
(
- "Dataset %s: could not find data in %s. Please make sure to call "
+ f"Dataset {self.name}: could not find data in {self._cache_dir_root}. Please make sure to call "
"builder.download_and_prepare(), or pass download=True to "
"datasets.load_dataset() before trying to access the Dataset object."
)
- % (self.name, self._cache_dir_root)
)
- logger.debug(
- "Constructing Dataset for split %s, from %s", split or ", ".join(self.info.splits), self._cache_dir
- )
+ logger.debug(f'Constructing Dataset for split {split or ", ".join(self.info.splits)}, from {self._cache_dir}')
# By default, return all splits
if split is None:
@@ -820,7 +798,7 @@ def _build_single_dataset(
if run_post_process:
for resource_file_name in self._post_processing_resources(split).values():
if os.sep in resource_file_name:
- raise ValueError("Resources shouldn't be in a sub-directory: {}".format(resource_file_name))
+ raise ValueError(f"Resources shouldn't be in a sub-directory: {resource_file_name}")
resources_paths = {
resource_name: os.path.join(self._cache_dir, resource_file_name)
for resource_name, resource_file_name in self._post_processing_resources(split).items()
@@ -859,9 +837,7 @@ def _build_single_dataset(
if self.info.post_processed.features is not None:
if self.info.post_processed.features.type != ds.features.type:
raise ValueError(
- "Post-processed features info don't match the dataset:\nGot\n{}\nbut expected something like\n{}".format(
- self.info.post_processed.features, ds.features
- )
+ f"Post-processed features info don't match the dataset:\nGot\n{self.info.post_processed.features}\nbut expected something like\n{ds.features}"
)
else:
ds.info.features = self.info.post_processed.features
@@ -1081,7 +1057,7 @@ def _generate_examples(self, **kwargs):
def _prepare_split(self, split_generator):
split_info = split_generator.split_info
- fname = "{}-{}.arrow".format(self.name, split_generator.name)
+ fname = f"{self.name}-{split_generator.name}.arrow"
fpath = os.path.join(self._cache_dir, fname)
generator = self._generate_examples(**split_generator.gen_kwargs)
@@ -1149,7 +1125,7 @@ def _generate_tables(self, **kwargs):
raise NotImplementedError()
def _prepare_split(self, split_generator):
- fname = "{}-{}.arrow".format(self.name, split_generator.name)
+ fname = f"{self.name}-{split_generator.name}.arrow"
fpath = os.path.join(self._cache_dir, fname)
generator = self._generate_tables(**split_generator.gen_kwargs)
@@ -1253,7 +1229,7 @@ def _download_and_prepare(self, dl_manager, verify_infos):
"\nIf you really want to run it locally because you feel like the "
"Dataset is small enough, you can use the local beam runner called "
"`DirectRunner` (you may run out of memory). \nExample of usage: "
- "\n\t`{}`".format(usage_example)
+ f"\n\t`{usage_example}`"
)
beam_options = beam_options or beam.options.pipeline_options.PipelineOptions()
@@ -1304,7 +1280,7 @@ def _prepare_split(self, split_generator, pipeline):
output_prefix = os.path.join(self._cache_dir, output_prefix)
# To write examples to disk:
- fname = "{}-{}.arrow".format(self.name, split_name)
+ fname = f"{self.name}-{split_name}.arrow"
fpath = os.path.join(self._cache_dir, fname)
beam_writer = BeamWriter(
features=self.info.features, path=fpath, namespace=split_name, cache_dir=self._cache_dir
diff --git a/src/datasets/commands/convert.py b/src/datasets/commands/convert.py
--- a/src/datasets/commands/convert.py
+++ b/src/datasets/commands/convert.py
@@ -88,7 +88,7 @@ def run(self):
abs_datasets_path = os.path.abspath(self._datasets_directory)
- self._logger.info("Converting datasets from %s to %s", abs_tfds_path, abs_datasets_path)
+ self._logger.info(f"Converting datasets from {abs_tfds_path} to {abs_datasets_path}")
utils_files = []
with_manual_update = []
@@ -100,7 +100,7 @@ def run(self):
file_names = [os.path.basename(self._tfds_path)]
for f_name in file_names:
- self._logger.info("Looking at file %s", f_name)
+ self._logger.info(f"Looking at file {f_name}")
input_file = os.path.join(abs_tfds_path, f_name)
output_file = os.path.join(abs_datasets_path, f_name)
@@ -167,7 +167,7 @@ def run(self):
output_dir = os.path.join(abs_datasets_path, dir_name)
output_file = os.path.join(output_dir, f_name)
os.makedirs(output_dir, exist_ok=True)
- self._logger.info("Adding directory %s", output_dir)
+ self._logger.info(f"Adding directory {output_dir}")
imports_to_builder_map.update({imp: output_dir for imp in tfds_imports})
else:
# Utilities will be moved at the end
@@ -178,13 +178,13 @@ def run(self):
with open(output_file, "w", encoding="utf-8") as f:
f.writelines(out_lines)
- self._logger.info("Converted in %s", output_file)
+ self._logger.info(f"Converted in {output_file}")
for utils_file in utils_files:
try:
f_name = os.path.basename(utils_file)
dest_folder = imports_to_builder_map[f_name.replace(".py", "")]
- self._logger.info("Moving %s to %s", utils_file, dest_folder)
+ self._logger.info(f"Moving {dest_folder} to {utils_file}")
shutil.copy(utils_file, dest_folder)
except KeyError:
self._logger.error(f"Cannot find destination folder for {utils_file}. Please copy manually.")
diff --git a/src/datasets/commands/env.py b/src/datasets/commands/env.py
--- a/src/datasets/commands/env.py
+++ b/src/datasets/commands/env.py
@@ -32,4 +32,4 @@ def run(self):
@staticmethod
def format_dict(d):
- return "\n".join(["- {}: {}".format(prop, val) for prop, val in d.items()]) + "\n"
+ return "\n".join([f"- {prop}: {val}" for prop, val in d.items()]) + "\n"
diff --git a/src/datasets/commands/run_beam.py b/src/datasets/commands/run_beam.py
--- a/src/datasets/commands/run_beam.py
+++ b/src/datasets/commands/run_beam.py
@@ -91,7 +91,7 @@ def run(self):
builders: List[DatasetBuilder] = []
if self._beam_pipeline_options:
beam_options = beam.options.pipeline_options.PipelineOptions(
- flags=["--%s" % opt.strip() for opt in self._beam_pipeline_options.split(",") if opt]
+ flags=[f"--{opt.strip()}" for opt in self._beam_pipeline_options.split(",") if opt]
)
else:
beam_options = None
@@ -147,10 +147,10 @@ def run(self):
elif os.path.isfile(combined_path):
dataset_dir = path
else: # in case of a remote dataset
- print("Dataset Infos file saved at {}".format(dataset_infos_path))
+ print(f"Dataset Infos file saved at {dataset_infos_path}")
exit(1)
# Move datasetinfo back to the user
user_dataset_infos_path = os.path.join(dataset_dir, config.DATASETDICT_INFOS_FILENAME)
copyfile(dataset_infos_path, user_dataset_infos_path)
- print("Dataset Infos file saved at {}".format(user_dataset_infos_path))
+ print(f"Dataset Infos file saved at {user_dataset_infos_path}")
diff --git a/src/datasets/config.py b/src/datasets/config.py
--- a/src/datasets/config.py
+++ b/src/datasets/config.py
@@ -116,7 +116,7 @@
try:
BEAM_VERSION = version.parse(importlib_metadata.version("apache_beam"))
BEAM_AVAILABLE = True
- logger.info("Apache Beam version {} available.".format(BEAM_VERSION))
+ logger.info(f"Apache Beam version {BEAM_VERSION} available.")
except importlib_metadata.PackageNotFoundError:
pass
else:
diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -28,9 +28,7 @@ class DatasetDict(dict):
def _check_values_type(self):
for dataset in self.values():
if not isinstance(dataset, Dataset):
- raise TypeError(
- "Values in `DatasetDict` should of type `Dataset` but got type '{}'".format(type(dataset))
- )
+ raise TypeError(f"Values in `DatasetDict` should of type `Dataset` but got type '{type(dataset)}'")
def __getitem__(self, k) -> Dataset:
if isinstance(k, (str, NamedSplit)) or len(self) == 0:
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -360,7 +360,7 @@ def __init__(self, shape: tuple, dtype: str):
assert (
self.ndims is not None and self.ndims > 1
), "You must instantiate an array type with a value for dim that is > 1"
- assert len(shape) == self.ndims, "shape={} and ndims={} dom't match".format(shape, self.ndims)
+ assert len(shape) == self.ndims, f"shape={shape} and ndims={self.ndims} dom't match"
self.shape = tuple(shape)
self.value_type = dtype
self.storage_dtype = self._generate_dtype(self.value_type)
@@ -475,7 +475,7 @@ def __from_arrow__(self, array):
if array.type.shape[0] is None:
raise NotImplementedError(
"Dynamic first dimension is not supported for "
- "PandasArrayExtensionDtype, dimension: {}".format(array.type.shape)
+ f"PandasArrayExtensionDtype, dimension: {array.type.shape}"
)
zero_copy_only = _is_zero_copy_only(array.type)
if isinstance(array, pa.ChunkedArray):
@@ -592,7 +592,7 @@ def __len__(self) -> int:
def __eq__(self, other) -> np.ndarray:
if not isinstance(other, PandasArrayExtensionArray):
- raise NotImplementedError("Invalid type to compare to: {}".format(type(other)))
+ raise NotImplementedError(f"Invalid type to compare to: {type(other)}")
return (self._data == other._data).all()
@@ -648,7 +648,7 @@ def __post_init__(self):
elif self.num_classes != len(self.names):
raise ValueError(
"ClassLabel number of names do not match the defined num_classes. "
- "Got {} names VS {} num_classes".format(len(self.names), self.num_classes)
+ f"Got {len(self.names)} names VS {self.num_classes} num_classes"
)
# Prepare mappings
self._int2str = [str(name) for name in self.names]
@@ -684,7 +684,7 @@ def str2int(self, values: Union[str, Iterable]):
except ValueError:
failed_parse = True
if failed_parse or not 0 <= value < self.num_classes:
- raise ValueError("Invalid string class label %s" % value)
+ raise ValueError(f"Invalid string class label {value}")
return output if return_list else output[0]
def int2str(self, values: Union[int, Iterable]):
@@ -699,7 +699,7 @@ def int2str(self, values: Union[int, Iterable]):
for v in values:
if not 0 <= v < self.num_classes:
- raise ValueError("Invalid integer class label %d" % v)
+ raise ValueError(f"Invalid integer class label {v:d}")
if self._int2str:
output = [self._int2str[int(v)] for v in values]
@@ -721,9 +721,7 @@ def encode_example(self, example_data):
# Allowing -1 to mean no label.
if not -1 <= example_data < self.num_classes:
- raise ValueError(
- "Class label %d greater than configured num_classes %d" % (example_data, self.num_classes)
- )
+ raise ValueError(f"Class label {example_data:d} greater than configured num_classes {self.num_classes}")
return example_data
@staticmethod
@@ -837,7 +835,7 @@ def encode_nested_example(schema, obj):
return list_dict
# schema.feature is not a dict
if isinstance(obj, str): # don't interpret a string as a list
- raise ValueError("Got a string but expected a list instead: '{}'".format(obj))
+ raise ValueError(f"Got a string but expected a list instead: '{obj}'")
if obj is None:
return None
else:
@@ -1044,7 +1042,7 @@ def encode_batch(self, batch):
"""
encoded_batch = {}
if set(batch) != set(self):
- raise ValueError("Column mismatch between batch {} and features {}".format(set(batch), set(self)))
+ raise ValueError(f"Column mismatch between batch {set(batch)} and features {set(self)}")
for key, column in batch.items():
column = cast_to_python_objects(column)
encoded_batch[key] = [encode_nested_example(self[key], obj) for obj in column]
diff --git a/src/datasets/features/translation.py b/src/datasets/features/translation.py
--- a/src/datasets/features/translation.py
+++ b/src/datasets/features/translation.py
@@ -97,9 +97,7 @@ def encode_example(self, translation_dict):
lang_set = set(self.languages)
if self.languages and set(translation_dict) - lang_set:
raise ValueError(
- "Some languages in example ({0}) are not in valid set ({1}).".format(
- ", ".join(sorted(set(translation_dict) - lang_set)), ", ".join(lang_set)
- )
+ f'Some languages in example ({", ".join(sorted(set(translation_dict) - lang_set))}) are not in valid set ({", ".join(lang_set)}).'
)
# Convert dictionary into tuples, splitting out cases where there are
diff --git a/src/datasets/fingerprint.py b/src/datasets/fingerprint.py
--- a/src/datasets/fingerprint.py
+++ b/src/datasets/fingerprint.py
@@ -335,12 +335,12 @@ def fingerprint_transform(
It should be in the format "MAJOR.MINOR.PATCH".
"""
- assert use_kwargs is None or isinstance(use_kwargs, list), "use_kwargs is supposed to be a list, not {}".format(
- type(use_kwargs)
- )
+ assert use_kwargs is None or isinstance(
+ use_kwargs, list
+ ), f"use_kwargs is supposed to be a list, not {type(use_kwargs)}"
assert ignore_kwargs is None or isinstance(
ignore_kwargs, list
- ), "ignore_kwargs is supposed to be a list, not {}".format(type(use_kwargs))
+ ), f"ignore_kwargs is supposed to be a list, not {type(use_kwargs)}"
assert not inplace or not fingerprint_names, "fingerprint_names are only used when inplace is False"
fingerprint_names = fingerprint_names if fingerprint_names is not None else ["new_fingerprint"]
@@ -348,10 +348,10 @@ def _fingerprint(func):
assert inplace or all( # check that not in-place functions require fingerprint parameters
name in func.__code__.co_varnames for name in fingerprint_names
- ), "function {} is missing parameters {} in signature".format(func, fingerprint_names)
+ ), f"function {func} is missing parameters {fingerprint_names} in signature"
if randomized_function: # randomized function have seed and generator parameters
- assert "seed" in func.__code__.co_varnames, "'seed' must be in {}'s signature".format(func)
- assert "generator" in func.__code__.co_varnames, "'generator' must be in {}'s signature".format(func)
+ assert "seed" in func.__code__.co_varnames, f"'seed' must be in {func}'s signature"
+ assert "generator" in func.__code__.co_varnames, f"'generator' must be in {func}'s signature"
# this has to be outside the wrapper or since __qualname__ changes in multiprocessing
transform = f"{func.__module__}.{func.__qualname__}"
if version is not None:
diff --git a/src/datasets/formatting/__init__.py b/src/datasets/formatting/__init__.py
--- a/src/datasets/formatting/__init__.py
+++ b/src/datasets/formatting/__init__.py
@@ -124,7 +124,5 @@ def get_formatter(format_type: Optional[str], **format_kwargs) -> Formatter:
raise _FORMAT_TYPES_ALIASES_UNAVAILABLE[format_type]
else:
raise ValueError(
- "Return type should be None or selected in {}, but got '{}'".format(
- list(type for type in _FORMAT_TYPES.keys() if type != None), format_type
- )
+ f"Return type should be None or selected in {list(type for type in _FORMAT_TYPES.keys() if type != None)}, but got '{format_type}'"
)
diff --git a/src/datasets/formatting/formatting.py b/src/datasets/formatting/formatting.py
--- a/src/datasets/formatting/formatting.py
+++ b/src/datasets/formatting/formatting.py
@@ -396,7 +396,7 @@ def format_batch(self, pa_table: pa.Table) -> dict:
def _check_valid_column_key(key: str, columns: List[str]) -> None:
if key not in columns:
- raise KeyError("Column {} not in the dataset. Current columns in the dataset: {}".format(key, columns))
+ raise KeyError(f"Column {key} not in the dataset. Current columns in the dataset: {columns}")
def _check_valid_index_key(key: Union[int, slice, range, Iterable], size: int) -> None:
diff --git a/src/datasets/info.py b/src/datasets/info.py
--- a/src/datasets/info.py
+++ b/src/datasets/info.py
@@ -247,7 +247,7 @@ def from_directory(cls, dataset_info_dir: str) -> "DatasetInfo":
dataset_info_dir (`str`): The directory containing the metadata file. This
should be the root directory of a specific dataset version.
"""
- logger.info("Loading Dataset info from %s", dataset_info_dir)
+ logger.info(f"Loading Dataset info from {dataset_info_dir}")
if not dataset_info_dir:
raise ValueError("Calling DatasetInfo.from_directory() with undefined dataset_info_dir.")
@@ -279,17 +279,17 @@ def write_to_directory(self, dataset_infos_dir, overwrite=False):
total_dataset_infos = {}
dataset_infos_path = os.path.join(dataset_infos_dir, config.DATASETDICT_INFOS_FILENAME)
if os.path.exists(dataset_infos_path) and not overwrite:
- logger.info("Dataset Infos already exists in {}. Completing it with new infos.".format(dataset_infos_dir))
+ logger.info(f"Dataset Infos already exists in {dataset_infos_dir}. Completing it with new infos.")
total_dataset_infos = self.from_directory(dataset_infos_dir)
else:
- logger.info("Writing new Dataset Infos in {}".format(dataset_infos_dir))
+ logger.info(f"Writing new Dataset Infos in {dataset_infos_dir}")
total_dataset_infos.update(self)
with open(dataset_infos_path, "w", encoding="utf-8") as f:
json.dump({config_name: asdict(dset_info) for config_name, dset_info in total_dataset_infos.items()}, f)
@classmethod
def from_directory(cls, dataset_infos_dir):
- logger.info("Loading Dataset Infos from {}".format(dataset_infos_dir))
+ logger.info(f"Loading Dataset Infos from {dataset_infos_dir}")
with open(os.path.join(dataset_infos_dir, config.DATASETDICT_INFOS_FILENAME), "r", encoding="utf-8") as f:
dataset_infos_dict = {
config_name: DatasetInfo.from_dict(dataset_info_dict)
@@ -354,7 +354,7 @@ def from_directory(cls, metric_info_dir) -> "MetricInfo":
metric_info_dir: `str` The directory containing the metadata file. This
should be the root directory of a specific dataset version.
"""
- logger.info("Loading Metric info from %s", metric_info_dir)
+ logger.info(f"Loading Metric info from {metric_info_dir}")
if not metric_info_dir:
raise ValueError("Calling MetricInfo.from_directory() with undefined metric_info_dir.")
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -159,7 +159,7 @@ def convert_github_url(url_path: str) -> Tuple[str, Optional[str]]:
github_path = parsed.path[1:]
repo_info, branch = github_path.split("/tree/") if "/tree/" in github_path else (github_path, "master")
repo_owner, repo_name = repo_info.split("/")
- url_path = "https://github.com/{}/{}/archive/{}.zip".format(repo_owner, repo_name, branch)
+ url_path = f"https://github.com/{repo_owner}/{repo_name}/archive/{branch}.zip"
sub_directory = f"{repo_name}-{branch}"
return url_path, sub_directory
@@ -197,7 +197,7 @@ def get_imports(file_path: str) -> Tuple[str, str, str, str]:
with open(file_path, mode="r", encoding="utf-8") as f:
lines.extend(f.readlines())
- logger.debug("Checking %s for additional imports.", file_path)
+ logger.debug(f"Checking {file_path} for additional imports.")
imports: List[Tuple[str, str, str, Optional[str]]] = []
is_in_docstring = False
for line in lines:
@@ -1712,12 +1712,12 @@ def load_from_disk(dataset_path: str, fs=None, keep_in_memory: Optional[bool] =
dest_dataset_path = dataset_path
if not fs.exists(dest_dataset_path):
- raise FileNotFoundError("Directory {} not found".format(dataset_path))
+ raise FileNotFoundError(f"Directory {dataset_path} not found")
if fs.isfile(Path(dest_dataset_path, config.DATASET_INFO_FILENAME).as_posix()):
return Dataset.load_from_disk(dataset_path, fs, keep_in_memory=keep_in_memory)
elif fs.isfile(Path(dest_dataset_path, config.DATASETDICT_JSON_FILENAME).as_posix()):
return DatasetDict.load_from_disk(dataset_path, fs, keep_in_memory=keep_in_memory)
else:
raise FileNotFoundError(
- "Directory {} is neither a dataset directory nor a dataset dict directory.".format(dataset_path)
+ f"Directory {dataset_path} is neither a dataset directory nor a dataset dict directory."
)
diff --git a/src/datasets/naming.py b/src/datasets/naming.py
--- a/src/datasets/naming.py
+++ b/src/datasets/naming.py
@@ -46,30 +46,30 @@ def snakecase_to_camelcase(name):
def filename_prefix_for_name(name):
if os.path.basename(name) != name:
- raise ValueError("Should be a dataset name, not a path: %s" % name)
+ raise ValueError(f"Should be a dataset name, not a path: {name}")
return camelcase_to_snakecase(name)
def filename_prefix_for_split(name, split):
if os.path.basename(name) != name:
- raise ValueError("Should be a dataset name, not a path: %s" % name)
+ raise ValueError(f"Should be a dataset name, not a path: {name}")
if not re.match(_split_re, split):
raise ValueError(f"Split name should match '{_split_re}'' but got '{split}'.")
- return "%s-%s" % (filename_prefix_for_name(name), split)
+ return f"{filename_prefix_for_name(name)}-{split}"
def filepattern_for_dataset_split(dataset_name, split, data_dir, filetype_suffix=None):
prefix = filename_prefix_for_split(dataset_name, split)
if filetype_suffix:
- prefix += ".%s" % filetype_suffix
+ prefix += f".{filetype_suffix}"
filepath = os.path.join(data_dir, prefix)
- return "%s*" % filepath
+ return f"{filepath}*"
def filename_for_dataset_split(dataset_name, split, filetype_suffix=None):
prefix = filename_prefix_for_split(dataset_name, split)
if filetype_suffix:
- prefix += ".%s" % filetype_suffix
+ prefix += f".{filetype_suffix}"
return prefix
diff --git a/src/datasets/search.py b/src/datasets/search.py
--- a/src/datasets/search.py
+++ b/src/datasets/search.py
@@ -168,7 +168,7 @@ def passage_generator():
logger.warning(
f"Some documents failed to be added to ElasticSearch. Failures: {len(documents)-successes}/{len(documents)}"
)
- logger.info("Indexed %d documents" % (successes,))
+ logger.info(f"Indexed {successes:d} documents")
def search(self, query: str, k=10) -> SearchResults:
"""Find the nearest examples indices to the query.
@@ -267,7 +267,7 @@ def add_vectors(
self.faiss_res = faiss.StandardGpuResources()
index = faiss.index_cpu_to_gpu(self.faiss_res, self.device, index)
self.faiss_index = index
- logger.info("Created faiss index of type {}".format(type(self.faiss_index)))
+ logger.info(f"Created faiss index of type {type(self.faiss_index)}")
# Set verbosity level
if faiss_verbose is not None:
@@ -282,13 +282,13 @@ def add_vectors(
# Train
if train_size is not None:
train_vecs = vectors[:train_size] if column is None else vectors[:train_size][column]
- logger.info("Training the index with the first {} vectors".format(len(train_vecs)))
+ logger.info(f"Training the index with the first {len(train_vecs)} vectors")
self.faiss_index.train(train_vecs)
else:
logger.info("Ignored the training step of the faiss index as `train_size` is None.")
# Add vectors
- logger.info("Adding {} vectors to the faiss index".format(len(vectors)))
+ logger.info(f"Adding {len(vectors)} vectors to the faiss index")
for i in utils.tqdm(
range(0, len(vectors), batch_size), disable=bool(logging.get_verbosity() == logging.NOTSET)
):
@@ -474,9 +474,9 @@ def save_faiss_index(self, index_name: str, file: Union[str, PurePath]):
"""
index = self.get_index(index_name)
if not isinstance(index, FaissIndex):
- raise ValueError("Index '{}' is not a FaissIndex but a '{}'".format(index_name, type(index)))
+ raise ValueError(f"Index '{index_name}' is not a FaissIndex but a '{type(index)}'")
index.save(file)
- logger.info("Saved FaissIndex {} at {}".format(index_name, file))
+ logger.info(f"Saved FaissIndex {index_name} at {file}")
def load_faiss_index(
self,
@@ -498,11 +498,9 @@ def load_faiss_index(
index = FaissIndex.load(file, device=device)
assert index.faiss_index.ntotal == len(
self
- ), "Index size should match Dataset size, but Index '{}' at {} has {} elements while the dataset has {} examples.".format(
- index_name, file, index.faiss_index.ntotal, len(self)
- )
+ ), f"Index size should match Dataset size, but Index '{index_name}' at {file} has {index.faiss_index.ntotal} elements while the dataset has {len(self)} examples."
self._indexes[index_name] = index
- logger.info("Loaded FaissIndex {} from {}".format(index_name, file))
+ logger.info(f"Loaded FaissIndex {index_name} from {file}")
def add_elasticsearch_index(
self,
diff --git a/src/datasets/splits.py b/src/datasets/splits.py
--- a/src/datasets/splits.py
+++ b/src/datasets/splits.py
@@ -193,8 +193,8 @@ def subsplit(self, arg=None, k=None, percent=None, weighted=None): # pylint: di
if not (k or percent or weighted):
raise ValueError(
- "Invalid split argument {}. Only list, slice and int supported. "
- "One of k, weighted or percent should be set to a non empty value.".format(arg)
+ f"Invalid split argument {arg}. Only list, slice and int supported. "
+ "One of k, weighted or percent should be set to a non empty value."
)
def assert_slices_coverage(slices):
@@ -203,7 +203,7 @@ def assert_slices_coverage(slices):
if k:
if not 0 < k <= 100:
- raise ValueError("Subsplit k should be between 0 and 100, got {}".format(k))
+ raise ValueError(f"Subsplit k should be between 0 and 100, got {k}")
shift = 100 // k
slices = [slice(i * shift, (i + 1) * shift) for i in range(k)]
# Round up last element to ensure all elements are taken
@@ -244,7 +244,7 @@ def assert_slices_coverage(slices):
class PercentSliceMeta(type):
def __getitem__(cls, slice_value):
if not isinstance(slice_value, slice):
- raise ValueError("datasets.percent should only be called with slice, not {}".format(slice_value))
+ raise ValueError(f"datasets.percent should only be called with slice, not {slice_value}")
return slice_value
@@ -276,7 +276,7 @@ def get_read_instruction(self, split_dict):
return read_instruction1 + read_instruction2
def __repr__(self):
- return "({!r} + {!r})".format(self._split1, self._split2)
+ return f"({repr(self._split1)} + {repr(self._split2)})"
class _SubSplit(SplitBase):
@@ -298,7 +298,7 @@ def __repr__(self):
stop="" if self._slice_value.stop is None else self._slice_value.stop,
step=self._slice_value.step,
)
- return "{!r}(datasets.percent[{}])".format(self._split, slice_str)
+ return f"{repr(self._split)}(datasets.percent[{slice_str}])"
class NamedSplit(SplitBase):
@@ -361,7 +361,7 @@ def __eq__(self, other):
elif isinstance(other, str): # Other should be string
return self._name == other
else:
- raise ValueError("Equality not supported between split {} and {}".format(self, other))
+ raise ValueError(f"Equality not supported between split {self} and {other}")
def __hash__(self):
return hash(self._name)
@@ -470,7 +470,7 @@ def __getitem__(self, slice_value):
split_instruction = SplitReadInstruction()
for v in self._splits.values():
if v.slice_value is not None:
- raise ValueError("Trying to slice Split {} which has already been sliced".format(v.split_info.name))
+ raise ValueError(f"Trying to slice Split {v.split_info.name} which has already been sliced")
v = v._asdict()
v["slice_value"] = slice_value
split_instruction.add(SlicedSplitInfo(**v))
@@ -502,15 +502,15 @@ def __getitem__(self, key: Union[SplitBase, str]):
def __setitem__(self, key: Union[SplitBase, str], value: SplitInfo):
if key != value.name:
- raise ValueError("Cannot add elem. (key mismatch: '{}' != '{}')".format(key, value.name))
+ raise ValueError(f"Cannot add elem. (key mismatch: '{key}' != '{value.name}')")
if key in self:
- raise ValueError("Split {} already present".format(key))
+ raise ValueError(f"Split {key} already present")
super(SplitDict, self).__setitem__(key, value)
def add(self, split_info: SplitInfo):
"""Add the split info."""
if split_info.name in self:
- raise ValueError("Split {} already present".format(split_info.name))
+ raise ValueError(f"Split {split_info.name} already present")
split_info.dataset_name = self.dataset_name
super(SplitDict, self).__setitem__(split_info.name, split_info)
diff --git a/src/datasets/utils/beam_utils.py b/src/datasets/utils/beam_utils.py
--- a/src/datasets/utils/beam_utils.py
+++ b/src/datasets/utils/beam_utils.py
@@ -29,9 +29,9 @@ def upload_local_to_remote(local_file_path, remote_file_path, force_upload=False
fs = FileSystems
if fs.exists(remote_file_path):
if force_upload:
- logger.info("Remote path already exist: {}. Overwriting it as force_upload=True.".format(remote_file_path))
+ logger.info(f"Remote path already exist: {remote_file_path}. Overwriting it as force_upload=True.")
else:
- logger.info("Remote path already exist: {}. Skipping it as force_upload=False.".format(remote_file_path))
+ logger.info(f"Remote path already exist: {remote_file_path}. Skipping it as force_upload=False.")
return
with fs.create(remote_file_path) as remote_file:
with open(local_file_path, "rb") as local_file:
@@ -46,9 +46,9 @@ def download_remote_to_local(remote_file_path, local_file_path, force_download=F
fs = FileSystems
if os.path.exists(local_file_path):
if force_download:
- logger.info("Local path already exist: {}. Overwriting it as force_upload=True.".format(remote_file_path))
+ logger.info(f"Local path already exist: {remote_file_path}. Overwriting it as force_upload=True.")
else:
- logger.info("Local path already exist: {}. Skipping it as force_upload=False.".format(remote_file_path))
+ logger.info(f"Local path already exist: {remote_file_path}. Skipping it as force_upload=False.")
return
with fs.open(remote_file_path) as remote_file:
with open(local_file_path, "wb") as local_file:
@@ -112,7 +112,7 @@ def __init__(
.. testcleanup::
- for output in glob.glob('{}*'.format(filename)):
+ for output in glob.glob(f'{filename}*'):
os.remove(output)
For more information on supported types and schema, please see the pyarrow
diff --git a/src/datasets/utils/download_manager.py b/src/datasets/utils/download_manager.py
--- a/src/datasets/utils/download_manager.py
+++ b/src/datasets/utils/download_manager.py
@@ -116,9 +116,7 @@ def upload(local_file_path):
remote_dir, config.DOWNLOADED_DATASETS_DIR, os.path.basename(local_file_path)
)
logger.info(
- "Uploading {} ({}) to {}.".format(
- local_file_path, size_str(os.path.getsize(local_file_path)), remote_file_path
- )
+ f"Uploading {local_file_path} ({size_str(os.path.getsize(local_file_path))}) to {remote_file_path}."
)
upload_local_to_remote(local_file_path, remote_file_path)
return remote_file_path
@@ -198,7 +196,7 @@ def download(self, url_or_urls):
download_func, url_or_urls, map_tuple=True, num_proc=download_config.num_proc, disable_tqdm=False
)
duration = datetime.now() - start_time
- logger.info("Downloading took {} min".format(duration.total_seconds() // 60))
+ logger.info(f"Downloading took {duration.total_seconds() // 60} min")
url_or_urls = NestedDataStructure(url_or_urls)
downloaded_path_or_paths = NestedDataStructure(downloaded_path_or_paths)
self.downloaded_paths.update(dict(zip(url_or_urls.flatten(), downloaded_path_or_paths.flatten())))
@@ -206,7 +204,7 @@ def download(self, url_or_urls):
start_time = datetime.now()
self._record_sizes_checksums(url_or_urls, downloaded_path_or_paths)
duration = datetime.now() - start_time
- logger.info("Checksum Computation took {} min".format(duration.total_seconds() // 60))
+ logger.info(f"Checksum Computation took {duration.total_seconds() // 60} min")
return downloaded_path_or_paths.data
diff --git a/src/datasets/utils/file_utils.py b/src/datasets/utils/file_utils.py
--- a/src/datasets/utils/file_utils.py
+++ b/src/datasets/utils/file_utils.py
@@ -309,10 +309,10 @@ def cached_path(
output_path = url_or_filename
elif is_local_path(url_or_filename):
# File, but it doesn't exist.
- raise FileNotFoundError("Local file {} doesn't exist".format(url_or_filename))
+ raise FileNotFoundError(f"Local file {url_or_filename} doesn't exist")
else:
# Something unknown
- raise ValueError("unable to parse {} as a URL or as a local path".format(url_or_filename))
+ raise ValueError(f"unable to parse {url_or_filename} as a URL or as a local path")
if output_path is None:
return output_path
@@ -326,18 +326,18 @@ def cached_path(
def get_datasets_user_agent(user_agent: Optional[Union[str, dict]] = None) -> str:
- ua = "datasets/{}; python/{}".format(__version__, config.PY_VERSION)
- ua += "; pyarrow/{}".format(config.PYARROW_VERSION)
+ ua = f"datasets/{__version__}; python/{config.PY_VERSION}"
+ ua += f"; pyarrow/{config.PYARROW_VERSION}"
if config.TORCH_AVAILABLE:
- ua += "; torch/{}".format(config.TORCH_VERSION)
+ ua += f"; torch/{config.TORCH_VERSION}"
if config.TF_AVAILABLE:
- ua += "; tensorflow/{}".format(config.TF_VERSION)
+ ua += f"; tensorflow/{config.TF_VERSION}"
if config.JAX_AVAILABLE:
- ua += "; jax/{}".format(config.JAX_VERSION)
+ ua += f"; jax/{config.JAX_VERSION}"
if config.BEAM_AVAILABLE:
- ua += "; apache_beam/{}".format(config.BEAM_VERSION)
+ ua += f"; apache_beam/{config.BEAM_VERSION}"
if isinstance(user_agent, dict):
- ua += "; " + "; ".join("{}/{}".format(k, v) for k, v in user_agent.items())
+ ua += f"; {'; '.join(f'{k}/{v}' for k, v in user_agent.items())}"
elif isinstance(user_agent, str):
ua += "; " + user_agent
return ua
@@ -355,7 +355,7 @@ def get_authentication_headers_for_url(url: str, use_auth_token: Optional[Union[
token = hf_api.HfFolder.get_token()
if token:
- headers["authorization"] = "Bearer {}".format(token)
+ headers["authorization"] = f"Bearer {token}"
return headers
@@ -434,7 +434,7 @@ def http_get(url, temp_file, proxies=None, resume_size=0, headers=None, cookies=
headers = copy.deepcopy(headers) or {}
headers["user-agent"] = get_datasets_user_agent(user_agent=headers.get("user-agent"))
if resume_size > 0:
- headers["Range"] = "bytes=%d-" % (resume_size,)
+ headers["Range"] = f"bytes={resume_size:d}-"
response = _request_with_retry(
method="GET",
url=url,
@@ -577,7 +577,7 @@ def get_from_cache(
or (response.status_code == 403 and "ndownloader.figstatic.com" in url)
):
connected = True
- logger.info("Couldn't get ETag version for url {}".format(url))
+ logger.info(f"Couldn't get ETag version for url {url}")
except (EnvironmentError, requests.exceptions.Timeout):
# not connected
pass
@@ -593,9 +593,9 @@ def get_from_cache(
" disabled. To enable file online look-ups, set 'local_files_only' to False."
)
elif response is not None and response.status_code == 404:
- raise FileNotFoundError("Couldn't find file at {}".format(url))
+ raise FileNotFoundError(f"Couldn't find file at {url}")
_raise_if_offline_mode_is_enabled(f"Tried to reach {url}")
- raise ConnectionError("Couldn't reach {}".format(url))
+ raise ConnectionError(f"Couldn't reach {url}")
# Try a second time
filename = hash_url_to_filename(original_url, etag)
@@ -629,7 +629,7 @@ def _resumable_file_manager():
# Download to temporary file, then copy to cache dir once finished.
# Otherwise you get corrupt cache entries if the download gets interrupted.
with temp_file_manager() as temp_file:
- logger.info("%s not found in cache or force_download set to True, downloading to %s", url, temp_file.name)
+ logger.info(f"{url} not found in cache or force_download set to True, downloading to {temp_file.name}")
# GET file object
if url.startswith("ftp://"):
@@ -645,10 +645,10 @@ def _resumable_file_manager():
max_retries=max_retries,
)
- logger.info("storing %s in cache at %s", url, cache_path)
+ logger.info(f"storing {url} in cache at {cache_path}")
shutil.move(temp_file.name, cache_path)
- logger.info("creating metadata file for %s", cache_path)
+ logger.info(f"creating metadata file for {cache_path}")
meta = {"url": url, "etag": etag}
meta_path = cache_path + ".json"
with open(meta_path, "w", encoding="utf-8") as meta_file:
diff --git a/src/datasets/utils/filelock.py b/src/datasets/utils/filelock.py
--- a/src/datasets/utils/filelock.py
+++ b/src/datasets/utils/filelock.py
@@ -99,7 +99,7 @@ def __init__(self, lock_file):
return None
def __str__(self):
- temp = "The file lock '{}' could not be acquired.".format(self.lock_file)
+ temp = f"The file lock '{self.lock_file}' could not be acquired."
return temp
@@ -269,18 +269,18 @@ def acquire(self, timeout=None, poll_intervall=0.05):
while True:
with self._thread_lock:
if not self.is_locked:
- logger().debug("Attempting to acquire lock %s on %s", lock_id, lock_filename)
+ logger().debug(f"Attempting to acquire lock {lock_id} on {lock_filename}")
self._acquire()
if self.is_locked:
- logger().debug("Lock %s acquired on %s", lock_id, lock_filename)
+ logger().debug(f"Lock {lock_id} acquired on {lock_filename}")
break
elif timeout >= 0 and time.time() - start_time > timeout:
- logger().debug("Timeout on acquiring lock %s on %s", lock_id, lock_filename)
+ logger().debug(f"Timeout on acquiring lock {lock_id} on {lock_filename}")
raise Timeout(self._lock_file)
else:
logger().debug(
- "Lock %s not acquired on %s, waiting %s seconds ...", lock_id, lock_filename, poll_intervall
+ f"Lock {lock_id} not acquired on {lock_filename}, waiting {poll_intervall} seconds ..."
)
time.sleep(poll_intervall)
except: # noqa
@@ -313,10 +313,10 @@ def release(self, force=False):
lock_id = id(self)
lock_filename = self._lock_file
- logger().debug("Attempting to release lock %s on %s", lock_id, lock_filename)
+ logger().debug(f"Attempting to release lock {lock_id} on {lock_filename}")
self._release()
self._lock_counter = 0
- logger().debug("Lock %s released on %s", lock_id, lock_filename)
+ logger().debug(f"Lock {lock_id} released on {lock_filename}")
return None
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -78,8 +78,8 @@ def size_str(size_in_bytes):
for (name, size_bytes) in _NAME_LIST:
value = size_in_bytes / size_bytes
if value >= 1.0:
- return "{:.2f} {}".format(value, name)
- return "{} {}".format(int(size_in_bytes), "bytes")
+ return f"{value:.2f} {name}"
+ return f"{int(size_in_bytes)} bytes"
def string_to_dict(string: str, pattern: str) -> Dict[str, str]:
@@ -259,18 +259,16 @@ def map_nested(
f"length: {sum(len(i[1]) for i in split_kwds)}"
)
logger.info(
- "Spawning {} processes for {} objects in slices of {}".format(
- num_proc, len(iterable), [len(i[1]) for i in split_kwds]
- )
+ f"Spawning {num_proc} processes for {len(iterable)} objects in slices of {[len(i[1]) for i in split_kwds]}"
)
initargs, initializer = None, None
if not disable_tqdm:
initargs, initializer = (RLock(),), tqdm.set_lock
with Pool(num_proc, initargs=initargs, initializer=initializer) as pool:
mapped = pool.map(_single_map_nested, split_kwds)
- logger.info("Finished {} processes".format(num_proc))
+ logger.info(f"Finished {num_proc} processes")
mapped = [obj for proc_res in mapped for obj in proc_res]
- logger.info("Unpacked {} objects".format(len(mapped)))
+ logger.info(f"Unpacked {len(mapped)} objects")
if isinstance(data_struct, dict):
return dict(zip(data_struct.keys(), mapped))
@@ -305,7 +303,7 @@ def flatten_nest_dict(d):
flat_dict = NonMutableDict()
for k, v in d.items():
if isinstance(v, dict):
- flat_dict.update({"{}/{}".format(k, k2): v2 for k2, v2 in flatten_nest_dict(v).items()})
+ flat_dict.update({f"{k}/{k2}": v2 for k2, v2 in flatten_nest_dict(v).items()})
else:
flat_dict[k] = v
return flat_dict
@@ -433,7 +431,7 @@ def _save_parametrized_type_hint(pickler, obj):
else:
initargs = (obj.__origin__, (list(args[:-1]), args[-1]))
else: # pragma: no cover
- raise pickle.PicklingError("Datasets pickle Error: Unknown type {}".format(type(obj)))
+ raise pickle.PicklingError(f"Datasets pickle Error: Unknown type {type(obj)}")
pickler.save_reduce(_CloudPickleTypeHintFix._create_parametrized_type_hint, initargs, obj=obj)
@@ -444,7 +442,7 @@ def _save_code(pickler, obj):
This is a modified version that removes the origin (filename + line no.)
of functions created in notebooks or shells for example.
"""
- dill._dill.log.info("Co: %s" % obj)
+ dill._dill.log.info(f"Co: {obj}")
# The filename of a function is the .py file where it is defined.
# Filenames of functions created in notebooks or shells start with '<'
# ex: <ipython-input-13-9ed2afe61d25> for ipython, and <stdin> for shell
@@ -532,7 +530,7 @@ def save_function(pickler, obj):
the keys in the output dictionary of globalvars can change.
"""
if not dill._dill._locate_function(obj):
- dill._dill.log.info("F1: %s" % obj)
+ dill._dill.log.info(f"F1: {obj}")
if getattr(pickler, "_recurse", False):
# recurse to get all globals referred to by obj
globalvars = dill.detect.globalvars
@@ -591,7 +589,7 @@ def save_function(pickler, obj):
pickler.clear_memo()
dill._dill.log.info("# F1")
else:
- dill._dill.log.info("F2: %s" % obj)
+ dill._dill.log.info(f"F2: {obj}")
name = getattr(obj, "__qualname__", getattr(obj, "__name__", None))
dill._dill.StockPickler.save_global(pickler, obj, name=name)
dill._dill.log.info("# F2")
@@ -609,7 +607,7 @@ def copyfunc(func):
@pklregister(type(regex.Regex("", 0)))
def _save_regex(pickler, obj):
- dill._dill.log.info("Re: %s" % obj)
+ dill._dill.log.info(f"Re: {obj}")
args = (
obj.pattern,
obj.flags,
diff --git a/src/datasets/utils/version.py b/src/datasets/utils/version.py
--- a/src/datasets/utils/version.py
+++ b/src/datasets/utils/version.py
@@ -53,7 +53,7 @@ def __post_init__(self):
self.major, self.minor, self.patch = _str_to_version(self.version_str)
def __repr__(self):
- return "{}.{}.{}".format(*self.tuple)
+ return f"{self.tuple[0]}.{self.tuple[1]}.{self.tuple[2]}"
@property
def tuple(self):
@@ -64,7 +64,7 @@ def _validate_operand(self, other):
return Version(other)
elif isinstance(other, Version):
return other
- raise AssertionError("{} (type {}) cannot be compared to version.".format(other, type(other)))
+ raise AssertionError(f"{other} (type {type(other)}) cannot be compared to version.")
def __eq__(self, other):
other = self._validate_operand(other)
@@ -111,7 +111,7 @@ def _str_to_version(version_str, allow_wildcard=False):
reg = _VERSION_WILDCARD_REG if allow_wildcard else _VERSION_RESOLVED_REG
res = reg.match(version_str)
if not res:
- msg = "Invalid version '{}'. Format should be x.y.z".format(version_str)
+ msg = f"Invalid version '{version_str}'. Format should be x.y.z"
if allow_wildcard:
msg += " with {x,y,z} being digits or wildcard."
else:
diff --git a/templates/new_metric_script.py b/templates/new_metric_script.py
--- a/templates/new_metric_script.py
+++ b/templates/new_metric_script.py
@@ -97,7 +97,7 @@ def _compute(self, predictions, references):
second_score = sum(abs(len(i) - len(j)) for i, j in zip(predictions, references) if i not in self.bad_words)
second_score /= sum(i not in self.bad_words for i in predictions)
else:
- raise ValueError("Invalid config name for NewMetric: {}. Please use 'max' or 'mean'.".format(self.config_name))
+ raise ValueError(f"Invalid config name for NewMetric: {self.config_name}. Please use 'max' or 'mean'.")
return {
"accuracy": accuracy,
| diff --git a/src/datasets/commands/test.py b/src/datasets/commands/test.py
--- a/src/datasets/commands/test.py
+++ b/src/datasets/commands/test.py
@@ -169,13 +169,13 @@ def get_builders() -> Generator[DatasetBuilder, None, None]:
dataset_dir = path
else: # in case of a remote dataset
dataset_dir = None
- print("Dataset Infos file saved at {}".format(dataset_infos_path))
+ print(f"Dataset Infos file saved at {dataset_infos_path}")
# Move dataset_info back to the user
if dataset_dir is not None:
user_dataset_infos_path = os.path.join(dataset_dir, datasets.config.DATASETDICT_INFOS_FILENAME)
copyfile(dataset_infos_path, user_dataset_infos_path)
- print("Dataset Infos file saved at {}".format(user_dataset_infos_path))
+ print(f"Dataset Infos file saved at {user_dataset_infos_path}")
# If clear_cache=True, the download folder and the dataset builder cache directory are deleted
if self._clear_cache:
diff --git a/tests/hub_fixtures.py b/tests/hub_fixtures.py
--- a/tests/hub_fixtures.py
+++ b/tests/hub_fixtures.py
@@ -31,7 +31,7 @@ def hf_token(hf_api: HfApi):
@pytest.fixture(scope="session")
def hf_private_dataset_repo_txt_data_(hf_api: HfApi, hf_token, text_file):
- repo_name = "repo_txt_data-{}".format(int(time.time() * 10e3))
+ repo_name = f"repo_txt_data-{int(time.time() * 10e3)}"
hf_api.create_repo(token=hf_token, name=repo_name, repo_type="dataset", private=True)
repo_id = f"{USER}/{repo_name}"
hf_api.upload_file(
@@ -57,7 +57,7 @@ def hf_private_dataset_repo_txt_data(hf_private_dataset_repo_txt_data_):
@pytest.fixture(scope="session")
def hf_private_dataset_repo_zipped_txt_data_(hf_api: HfApi, hf_token, zip_csv_path):
- repo_name = "repo_zipped_txt_data-{}".format(int(time.time() * 10e3))
+ repo_name = f"repo_zipped_txt_data-{int(time.time() * 10e3)}"
hf_api.create_repo(token=hf_token, name=repo_name, repo_type="dataset", private=True)
repo_id = f"{USER}/{repo_name}"
hf_api.upload_file(
diff --git a/tests/s3_fixtures.py b/tests/s3_fixtures.py
--- a/tests/s3_fixtures.py
+++ b/tests/s3_fixtures.py
@@ -9,7 +9,7 @@
s3_test_bucket_name = "test"
s3_port = 5555
-s3_endpoint_uri = "http://127.0.0.1:%s/" % s3_port
+s3_endpoint_uri = f"http://127.0.0.1:{s3_port}/"
S3_FAKE_ENV_VARS = {
"AWS_ACCESS_KEY_ID": "fake_access_key",
@@ -29,7 +29,7 @@ def s3_base():
old_environ = os.environ.copy()
os.environ.update(S3_FAKE_ENV_VARS)
- proc = subprocess.Popen(shlex.split("moto_server s3 -p %s" % s3_port))
+ proc = subprocess.Popen(shlex.split(f"moto_server s3 -p {s3_port}"))
timeout = 5
while timeout > 0:
diff --git a/tests/test_array_xd.py b/tests/test_array_xd.py
--- a/tests/test_array_xd.py
+++ b/tests/test_array_xd.py
@@ -129,7 +129,7 @@ def get_array_feature_types():
shape_2 = [3, 4, 5, 6, 7]
return [
{
- "testcase_name": "{}d".format(d),
+ "testcase_name": f"{d}d",
"array_feature": array_feature,
"shape_1": tuple(shape_1[:d]),
"shape_2": tuple(shape_2[:d]),
diff --git a/tests/test_builder.py b/tests/test_builder.py
--- a/tests/test_builder.py
+++ b/tests/test_builder.py
@@ -28,7 +28,7 @@ def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN)]
def _prepare_split(self, split_generator, **kwargs):
- fname = "{}-{}.arrow".format(self.name, split_generator.name)
+ fname = f"{self.name}-{split_generator.name}.arrow"
with ArrowWriter(features=self.info.features, path=os.path.join(self._cache_dir, fname)) as writer:
writer.write_batch({"text": ["foo"] * 100})
num_examples, num_bytes = writer.finalize()
@@ -203,7 +203,7 @@ def char_tokenize(example):
return dataset.map(char_tokenize, cache_file_name=resources_paths["tokenized_dataset"])
def _post_processing_resources(self, split):
- return {"tokenized_dataset": "tokenized_dataset-{split}.arrow".format(split=split)}
+ return {"tokenized_dataset": f"tokenized_dataset-{split}.arrow"}
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
@@ -344,7 +344,7 @@ def _post_process(self, dataset, resources_paths):
return dataset
def _post_processing_resources(self, split):
- return {"index": "Flat-{split}.faiss".format(split=split)}
+ return {"index": f"Flat-{split}.faiss"}
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
@@ -412,7 +412,7 @@ def char_tokenize(example):
return dataset.map(char_tokenize, cache_file_name=resources_paths["tokenized_dataset"])
def _post_processing_resources(self, split):
- return {"tokenized_dataset": "tokenized_dataset-{split}.arrow".format(split=split)}
+ return {"tokenized_dataset": f"tokenized_dataset-{split}.arrow"}
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
@@ -464,7 +464,7 @@ def _post_process(self, dataset, resources_paths):
return dataset
def _post_processing_resources(self, split):
- return {"index": "Flat-{split}.faiss".format(split=split)}
+ return {"index": f"Flat-{split}.faiss"}
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
diff --git a/tests/test_dataset_dict.py b/tests/test_dataset_dict.py
--- a/tests/test_dataset_dict.py
+++ b/tests/test_dataset_dict.py
@@ -30,7 +30,7 @@ def _create_dummy_dataset(self, multiple_columns=False):
dset = Dataset.from_dict(data)
else:
dset = Dataset.from_dict(
- {"filename": ["my_name-train" + "_" + "{:03d}".format(x) for x in np.arange(30).tolist()]}
+ {"filename": ["my_name-train" + "_" + f"{x:03d}" for x in np.arange(30).tolist()]}
)
return dset
@@ -320,7 +320,7 @@ def test_sort(self):
self.assertListEqual(list(dsets.keys()), list(sorted_dsets_1.keys()))
self.assertListEqual(
[f.split("_")[-1] for f in sorted_dsets_1["train"]["filename"]],
- sorted("{:03d}".format(x) for x in range(30)),
+ sorted(f"{x:03d}" for x in range(30)),
)
indices_cache_file_names = {
@@ -333,7 +333,7 @@ def test_sort(self):
self.assertListEqual(list(dsets.keys()), list(sorted_dsets_2.keys()))
self.assertListEqual(
[f.split("_")[-1] for f in sorted_dsets_2["train"]["filename"]],
- sorted(("{:03d}".format(x) for x in range(30)), reverse=True),
+ sorted((f"{x:03d}" for x in range(30)), reverse=True),
)
del dsets, sorted_dsets_1, sorted_dsets_2
diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -87,7 +87,7 @@ def __getitem__(self, name):
for item in self._fs_contents:
if item["name"] == name:
return item
- raise IndexError("{name} not found!".format(name=name))
+ raise IndexError(f"{name} not found!")
def ls(self, path, detail=True, refresh=True, **kwargs):
if kwargs.pop("strip_proto", True):
diff --git a/tests/utils.py b/tests/utils.py
--- a/tests/utils.py
+++ b/tests/utils.py
@@ -24,7 +24,7 @@ def parse_flag_from_env(key, default=False):
_value = strtobool(value)
except ValueError:
# More values are supported, but let's keep the message simple.
- raise ValueError("If set, {} must be yes or no.".format(key))
+ raise ValueError(f"If set, {key} must be yes or no.")
return _value
| Use f-strings for string formatting
f-strings offer better readability/performance than `str.format` and `%`, so we should use them in all places in our codebase unless there is good reason to keep the older syntax.
> **NOTE FOR CONTRIBUTORS**: To avoid large PRs and possible merge conflicts, do 1-3 modules per PR. Also, feel free to ignore the files located under `datasets/*`.
| Hi, I would be glad to help with this. Is there anyone else working on it?
Hi, I would be glad to work on this too.
#self-assign | 2021-11-15T21:37:05Z | [] | [] |
huggingface/datasets | 3,288 | huggingface__datasets-3288 | [
"3273"
] | b29fb550c31de337b952035a7584147e0f18c0cf | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -3660,18 +3660,6 @@ def concatenate_datasets(
format = {}
logger.info("Some of the datasets have disparate format. Resetting the format of the concatenated dataset.")
- # Concatenate tables
- tables_to_concat = [dset._data for dset in dsets if len(dset._data) > 0]
- # There might be no table with data left hence return first empty table
- if not tables_to_concat:
- return dsets[0]
- table = concat_tables(tables_to_concat, axis=axis)
- if axis == 1:
- # Merge features (ignore duplicated columns for now and let Dataset.__init__ check for those)
- table = update_metadata_with_features(
- table, Features({k: v for dset in dsets for k, v in dset.features.items()})
- )
-
def apply_offset_to_indices_table(table, offset):
if offset == 0:
return table
@@ -3682,30 +3670,48 @@ def apply_offset_to_indices_table(table, offset):
# Concatenate indices if they exist
if any(dset._indices is not None for dset in dsets):
-
- # Datasets with no indices tables are replaced with a dataset with an indices table in memory.
- # Applying an offset to an indices table also brings the table in memory.
- for i in range(len(dsets)):
- if dsets[i]._indices is None:
- dsets[i] = dsets[i].select(range(len(dsets[i])))
- assert all(dset._indices is not None for dset in dsets), "each dataset should have an indices table"
-
- # An offset needs to be applied to the indices before concatenating
- indices_tables = []
- offset = 0
- for dset in dsets:
- indices_tables.append(apply_offset_to_indices_table(dset._indices, offset))
- offset += len(dset._data)
-
- # Concatenate indices
- indices_tables = [t for t in indices_tables if len(t) > 0]
- if indices_tables:
- indices_table = concat_tables(indices_tables)
- else:
- indices_table = InMemoryTable.from_batches([], schema=pa.schema({"indices": pa.int64()}))
+ if axis == 0:
+ # Datasets with no indices tables are replaced with a dataset with an indices table in memory.
+ # Applying an offset to an indices table also brings the table in memory.
+ indices_tables = []
+ for i in range(len(dsets)):
+ if dsets[i]._indices is None:
+ dsets[i] = dsets[i].select(range(len(dsets[i])))
+ indices_tables.append(dsets[i]._indices)
+
+ # An offset needs to be applied to the indices before concatenating
+ offset = 0
+ for i in range(len(dsets)):
+ indices_tables[i] = apply_offset_to_indices_table(indices_tables[i], offset)
+ offset += len(dsets[i]._data)
+
+ # Concatenate indices
+ indices_tables = [t for t in indices_tables if len(t) > 0]
+ if indices_tables:
+ indices_table = concat_tables(indices_tables)
+ else:
+ indices_table = InMemoryTable.from_batches([], schema=pa.schema({"indices": pa.int64()}))
+ elif axis == 1 and len(dsets) == 1:
+ indices_table = dsets[0]._indices
+ elif axis == 1 and len(dsets) > 1:
+ for i in range(len(dsets)):
+ dsets[i] = dsets[i].flatten_indices()
+ indices_table = None
else:
indices_table = None
+ # Concatenate tables
+ tables_to_concat = [dset._data for dset in dsets if len(dset._data) > 0]
+ # There might be no table with data left hence return first empty table
+ if not tables_to_concat:
+ return dsets[0]
+ table = concat_tables(tables_to_concat, axis=axis)
+ if axis == 1:
+ # Merge features (ignore duplicated columns for now and let Dataset.__init__ check for those)
+ table = update_metadata_with_features(
+ table, Features({k: v for dset in dsets for k, v in dset.features.items()})
+ )
+
# Concatenate infos
if info is None:
info = DatasetInfo.from_merge([dset.info for dset in dsets])
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -603,7 +603,7 @@ def test_concatenate_formatted(self, in_memory):
del dset1, dset2, dset3
def test_concatenate_with_indices(self, in_memory):
- data1, data2, data3 = {"id": [0, 1, 2] * 2}, {"id": [3, 4, 5] * 2}, {"id": [6, 7]}
+ data1, data2, data3 = {"id": [0, 1, 2] * 2}, {"id": [3, 4, 5] * 2}, {"id": [6, 7, 8]}
info1 = DatasetInfo(description="Dataset1")
info2 = DatasetInfo(description="Dataset2")
with tempfile.TemporaryDirectory() as tmp_dir:
@@ -616,16 +616,46 @@ def test_concatenate_with_indices(self, in_memory):
dset1, dset2, dset3 = dset1.select([0, 1, 2]), dset2.select([0, 1, 2]), dset3
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
- self.assertEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 2))
+ self.assertEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 3))
self.assertEqual(len(dset_concat), len(dset1) + len(dset2) + len(dset3))
- self.assertListEqual(dset_concat["id"], [0, 1, 2, 3, 4, 5, 6, 7])
+ self.assertListEqual(dset_concat["id"], [0, 1, 2, 3, 4, 5, 6, 7, 8])
# in_memory = False:
- # 3 cache files for the dset_concat._data table, and 1 for the dset_concat._indices_table
- # no cache file for the indices
+ # 3 cache files for the dset_concat._data table
+ # no cache file for the indices because it's in memory
# in_memory = True:
# no cache files since both dset_concat._data and dset_concat._indices are in memory
self.assertEqual(len(dset_concat.cache_files), 0 if in_memory else 3)
self.assertEqual(dset_concat.info.description, "Dataset1\n\nDataset2\n\n")
+
+ dset1 = dset1.rename_columns({"id": "id1"})
+ dset2 = dset2.rename_columns({"id": "id2"})
+ dset3 = dset3.rename_columns({"id": "id3"})
+ with concatenate_datasets([dset1, dset2, dset3], axis=1) as dset_concat:
+ self.assertEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 3))
+ self.assertEqual(len(dset_concat), len(dset1))
+ self.assertListEqual(dset_concat["id1"], [0, 1, 2])
+ self.assertListEqual(dset_concat["id2"], [3, 4, 5])
+ self.assertListEqual(dset_concat["id3"], [6, 7, 8])
+ # in_memory = False:
+ # 3 cache files for the dset_concat._data table
+ # no cache file for the indices because it's None
+ # in_memory = True:
+ # no cache files since dset_concat._data is in memory and dset_concat._indices is None
+ self.assertEqual(len(dset_concat.cache_files), 0 if in_memory else 3)
+ self.assertIsNone(dset_concat._indices)
+ self.assertEqual(dset_concat.info.description, "Dataset1\n\nDataset2\n\n")
+
+ with concatenate_datasets([dset1], axis=1) as dset_concat:
+ self.assertEqual(len(dset_concat), len(dset1))
+ self.assertListEqual(dset_concat["id1"], [0, 1, 2])
+ # in_memory = False:
+ # 1 cache file for the dset_concat._data table
+ # no cache file for the indices because it's in memory
+ # in_memory = True:
+ # no cache files since both dset_concat._data and dset_concat._indices are in memory
+ self.assertEqual(len(dset_concat.cache_files), 0 if in_memory else 1)
+ self.assertTrue(dset_concat._indices == dset1._indices)
+ self.assertEqual(dset_concat.info.description, "Dataset1")
del dset1, dset2, dset3
def test_concatenate_with_indices_from_disk(self, in_memory):
| Respect row ordering when concatenating datasets along axis=1
Currently, there is a bug when concatenating datasets along `axis=1` if more than one dataset has the `_indices` attribute defined. In that scenario, all indices mappings except the first one get ignored.
A minimal reproducible example:
```python
>>> from datasets import Dataset, concatenate_datasets
>>> a = Dataset.from_dict({"a": [30, 20, 10]})
>>> b = Dataset.from_dict({"b": [2, 1, 3]})
>>> d = concatenate_datasets([a.sort("a"), b.sort("b")], axis=1)
>>> print(d[:3]) # expected: {'a': [10, 20, 30], 'b': [1, 2, 3]}
{'a': [10, 20, 30], 'b': [3, 1, 2]}
```
I've noticed the bug while working on #3195.
| 2021-11-17T13:41:28Z | [] | [] |
|
huggingface/datasets | 3,296 | huggingface__datasets-3296 | [
"3295"
] | d3c7b9481d427ce41256edaf6773c47570f06f3b | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -1004,6 +1004,23 @@ def save_to_disk(self, dataset_path: str, fs=None):
json.dump(sorted_keys_dataset_info, dataset_info_file, indent=2)
logger.info(f"Dataset saved in {dataset_path}")
+ @staticmethod
+ def _build_local_temp_path(uri_or_path: str) -> Path:
+ """
+ Builds and returns a Path concatenating a local temporary dir with the dir path (or absolute/relative
+ path extracted from the uri) passed.
+
+ Args:
+ uri_or_path (:obj:`str`): Path (e.g. `"dataset/train"`) or remote URI (e.g.
+ `"s3://my-bucket/dataset/train"`) to concatenate.
+
+ Returns:
+ :class:`Path`: the concatenated path (temp dir + path)
+ """
+ src_dataset_path = Path(uri_or_path)
+ tmp_dir = get_temporary_cache_files_directory()
+ return Path(tmp_dir, src_dataset_path.relative_to(src_dataset_path.anchor))
+
@staticmethod
def load_from_disk(dataset_path: str, fs=None, keep_in_memory: Optional[bool] = None) -> "Dataset":
"""
@@ -1037,8 +1054,7 @@ def load_from_disk(dataset_path: str, fs=None, keep_in_memory: Optional[bool] =
if is_remote_filesystem(fs):
src_dataset_path = extract_path_from_uri(dataset_path)
- tmp_dir = get_temporary_cache_files_directory()
- dataset_path = Path(tmp_dir, src_dataset_path)
+ dataset_path = Dataset._build_local_temp_path(src_dataset_path)
fs.download(src_dataset_path, dataset_path.as_posix(), recursive=True)
with open(
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -20,6 +20,7 @@
from datasets.arrow_dataset import Dataset, transmit_format, update_metadata_with_features
from datasets.dataset_dict import DatasetDict
from datasets.features import Array2D, Array3D, ClassLabel, Features, Sequence, Value
+from datasets.filesystems import extract_path_from_uri
from datasets.info import DatasetInfo
from datasets.splits import NamedSplit
from datasets.table import ConcatenationTable, InMemoryTable, MemoryMappedTable
@@ -2769,6 +2770,29 @@ def test_dummy_dataset_serialize_s3(s3, dataset):
assert dataset["id"][0] == 0
[email protected](
+ "uri_or_path",
+ [
+ "relative/path",
+ "/absolute/path",
+ "s3://bucket/relative/path",
+ "hdfs://relative/path",
+ "hdfs:///absolute/path",
+ ],
+)
+def test_build_local_temp_path(uri_or_path):
+ extracted_path = extract_path_from_uri(uri_or_path)
+ local_temp_path = Dataset._build_local_temp_path(extracted_path)
+
+ assert (
+ "tmp" in local_temp_path.as_posix()
+ and "hdfs" not in local_temp_path.as_posix()
+ and "s3" not in local_temp_path.as_posix()
+ and not local_temp_path.as_posix().startswith(extracted_path)
+ and local_temp_path.as_posix().endswith(extracted_path)
+ ), f"Local temp path: {local_temp_path.as_posix()}"
+
+
class TaskTemplatesTest(TestCase):
def test_task_text_classification(self):
labels = sorted(["pos", "neg"])
| Temporary dataset_path for remote fs URIs not built properly in arrow_dataset.py::load_from_disk
## Describe the bug
When trying to build a temporary dataset path from a remote URI in this block of code:
https://github.com/huggingface/datasets/blob/42f6b1d18a4a1b6009b6e62d115491be16dfca22/src/datasets/arrow_dataset.py#L1038-L1042
the result is not the expected when passing an absolute path in an URI like `hdfs:///absolute/path`.
## Steps to reproduce the bug
```python
dataset_path = "hdfs:///absolute/path"
src_dataset_path = extract_path_from_uri(dataset_path)
tmp_dir = get_temporary_cache_files_directory()
dataset_path = Path(tmp_dir, src_dataset_path)
print(dataset_path)
```
## Expected results
With the code above, we would expect a value in `dataset_path` similar to:
`/tmp/tmpnwxyvao5/absolute/path`
## Actual results
However, we get a `dataset_path` value like:
`/absolute/path`
This is because this line here: https://github.com/huggingface/datasets/blob/42f6b1d18a4a1b6009b6e62d115491be16dfca22/src/datasets/arrow_dataset.py#L1041
returns the last absolute path when two absolute paths (the one in `tmp_dir` and the one extracted from the URI in `src_dataset_path`) are passed as arguments.
## Environment info
- `datasets` version: 1.13.3
- Platform: Linux-3.10.0-1160.15.2.el7.x86_64-x86_64-with-glibc2.33
- Python version: 3.9.7
- PyArrow version: 5.0.0
| 2021-11-18T23:32:45Z | [] | [] |
|
huggingface/datasets | 3,375 | huggingface__datasets-3375 | [
"3373"
] | aa1ba6b68f8c12801bdd156707ea12061545361a | diff --git a/src/datasets/packaged_modules/csv/csv.py b/src/datasets/packaged_modules/csv/csv.py
--- a/src/datasets/packaged_modules/csv/csv.py
+++ b/src/datasets/packaged_modules/csv/csv.py
@@ -1,6 +1,4 @@
# coding=utf-8
-import glob
-import os
from dataclasses import dataclass
from typing import List, Optional, Union
@@ -19,16 +17,6 @@
_PANDAS_READ_CSV_NEW_1_3_0_PARAMETERS = ["encoding_errors", "on_bad_lines"]
-def _iter_files(files):
- for file in files:
- if os.path.isfile(file):
- yield file
- else:
- for subfile in glob.glob(os.path.join(file, "**", "*"), recursive=True):
- if os.path.isfile(subfile):
- yield subfile
-
-
@dataclass
class CsvConfig(datasets.BuilderConfig):
"""BuilderConfig for CSV."""
@@ -150,16 +138,14 @@ def _split_generators(self, dl_manager):
files = data_files
if isinstance(files, str):
files = [files]
- if any(os.path.isdir(file) for file in files):
- files = [file for file in _iter_files(files)]
- return [datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": files})]
+ return [
+ datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": dl_manager.iter_files(files)})
+ ]
splits = []
for split_name, files in data_files.items():
if isinstance(files, str):
files = [files]
- if any(os.path.isdir(file) for file in files):
- files = [file for file in _iter_files(files)]
- splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": files}))
+ splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": dl_manager.iter_files(files)}))
return splits
def _generate_tables(self, files):
diff --git a/src/datasets/packaged_modules/json/json.py b/src/datasets/packaged_modules/json/json.py
--- a/src/datasets/packaged_modules/json/json.py
+++ b/src/datasets/packaged_modules/json/json.py
@@ -54,12 +54,14 @@ def _split_generators(self, dl_manager):
files = data_files
if isinstance(files, str):
files = [files]
- return [datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": files})]
+ return [
+ datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": dl_manager.iter_files(files)})
+ ]
splits = []
for split_name, files in data_files.items():
if isinstance(files, str):
files = [files]
- splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": files}))
+ splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": dl_manager.iter_files(files)}))
return splits
def _cast_classlabels(self, pa_table: pa.Table) -> pa.Table:
diff --git a/src/datasets/packaged_modules/text/text.py b/src/datasets/packaged_modules/text/text.py
--- a/src/datasets/packaged_modules/text/text.py
+++ b/src/datasets/packaged_modules/text/text.py
@@ -38,12 +38,14 @@ def _split_generators(self, dl_manager):
files = data_files
if isinstance(files, str):
files = [files]
- return [datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": files})]
+ return [
+ datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": dl_manager.iter_files(files)})
+ ]
splits = []
for split_name, files in data_files.items():
if isinstance(files, str):
files = [files]
- splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": files}))
+ splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": dl_manager.iter_files(files)}))
return splits
def _generate_tables(self, files):
diff --git a/src/datasets/utils/download_manager.py b/src/datasets/utils/download_manager.py
--- a/src/datasets/utils/download_manager.py
+++ b/src/datasets/utils/download_manager.py
@@ -241,6 +241,23 @@ def iter_archive(self, path):
stream.members = []
del stream
+ def iter_files(self, paths):
+ """Iterate over file paths.
+
+ Args:
+ paths (list): Root paths.
+
+ Yields:
+ str: File path.
+ """
+ for path in paths:
+ if os.path.isfile(path):
+ yield path
+ else:
+ for dirpath, _, filenames in os.walk(path):
+ for filename in filenames:
+ yield os.path.join(dirpath, filename)
+
def extract(self, path_or_paths, num_proc=None):
"""Extract given path(s).
diff --git a/src/datasets/utils/mock_download_manager.py b/src/datasets/utils/mock_download_manager.py
--- a/src/datasets/utils/mock_download_manager.py
+++ b/src/datasets/utils/mock_download_manager.py
@@ -213,3 +213,7 @@ def iter_archive(self, path):
for file_path in path.rglob("*"):
if file_path.is_file() and not file_path.name.startswith(".") and not file_path.name.startswith("__"):
yield file_path.relative_to(path).as_posix(), file_path.open("rb")
+
+ def iter_files(self, paths):
+ for path in paths:
+ yield path
diff --git a/src/datasets/utils/streaming_download_manager.py b/src/datasets/utils/streaming_download_manager.py
--- a/src/datasets/utils/streaming_download_manager.py
+++ b/src/datasets/utils/streaming_download_manager.py
@@ -433,6 +433,32 @@ def xpathsuffix(path: Path):
return PurePosixPath(_as_posix(path).split("::")[0]).suffix
+def xwalk(urlpath, use_auth_token: Optional[Union[str, bool]] = None):
+ """Extend `os.walk` function to support remote files.
+
+ Args:
+ urlpath: URL root path.
+
+ Yields:
+ tuple: 3-tuple (dirpath, dirnames, filenames).
+ """
+ main_hop, *rest_hops = urlpath.split("::")
+ if is_local_path(main_hop):
+ return os.walk(main_hop)
+ else:
+ # walking inside a zip in a private repo requires authentication
+ if rest_hops and (rest_hops[0].startswith("http://") or rest_hops[0].startswith("https://")):
+ url = rest_hops[0]
+ url, kwargs = _prepare_http_url_kwargs(url, use_auth_token=use_auth_token)
+ storage_options = {"https": kwargs}
+ urlpath = "::".join([main_hop, url, *rest_hops[1:]])
+ else:
+ storage_options = None
+ fs, *_ = fsspec.get_fs_token_paths(urlpath, storage_options=storage_options)
+ for dirpath, dirnames, filenames in fs.walk(main_hop):
+ yield "::".join([f"{fs.protocol}://{dirpath}"] + rest_hops), dirnames, filenames
+
+
def xpandas_read_csv(filepath_or_buffer, use_auth_token: Optional[Union[str, bool]] = None, **kwargs):
import pandas as pd
@@ -527,3 +553,20 @@ def iter_archive(self, urlpath: str):
yield (file_path, file_obj)
stream.members = []
del stream
+
+ def iter_files(self, urlpaths):
+ """Iterate over files.
+
+ Args:
+ urlpaths (list): Root URL paths.
+
+ Yields:
+ str: File URL path.
+ """
+ for urlpath in urlpaths:
+ if "://::" not in urlpath: # workaround for os.path.isfile(urlpath):
+ yield urlpath
+ else:
+ for dirpath, _, filenames in xwalk(urlpath, use_auth_token=self.download_config.use_auth_token):
+ for filename in filenames:
+ yield xjoin(dirpath, filename)
| diff --git a/tests/conftest.py b/tests/conftest.py
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -273,6 +273,17 @@ def zip_csv_path(csv_path, csv2_path, tmp_path_factory):
return path
[email protected](scope="session")
+def zip_csv_with_dir_path(csv_path, csv2_path, tmp_path_factory):
+ import zipfile
+
+ path = tmp_path_factory.mktemp("data") / "dataset_with_dir.csv.zip"
+ with zipfile.ZipFile(path, "w") as f:
+ f.write(csv_path, arcname=os.path.join("main_dir", os.path.basename(csv_path)))
+ f.write(csv2_path, arcname=os.path.join("main_dir", os.path.basename(csv2_path)))
+ return path
+
+
@pytest.fixture(scope="session")
def parquet_path(tmp_path_factory):
path = str(tmp_path_factory.mktemp("data") / "dataset.parquet")
@@ -318,6 +329,15 @@ def jsonl_path(tmp_path_factory):
return path
[email protected](scope="session")
+def jsonl2_path(tmp_path_factory):
+ path = str(tmp_path_factory.mktemp("data") / "dataset2.jsonl")
+ with open(path, "w") as f:
+ for item in DATA:
+ f.write(json.dumps(item) + "\n")
+ return path
+
+
@pytest.fixture(scope="session")
def jsonl_312_path(tmp_path_factory):
path = str(tmp_path_factory.mktemp("data") / "dataset_312.jsonl")
@@ -336,16 +356,6 @@ def jsonl_str_path(tmp_path_factory):
return path
[email protected](scope="session")
-def text_path(tmp_path_factory):
- data = ["0", "1", "2", "3"]
- path = str(tmp_path_factory.mktemp("data") / "dataset.txt")
- with open(path, "w") as f:
- for item in data:
- f.write(item + "\n")
- return path
-
-
@pytest.fixture(scope="session")
def text_gz_path(tmp_path_factory, text_path):
import gzip
@@ -366,3 +376,67 @@ def jsonl_gz_path(tmp_path_factory, jsonl_path):
with gzip.open(path, "wb") as zipped_file:
zipped_file.writelines(orig_file)
return path
+
+
[email protected](scope="session")
+def zip_jsonl_path(jsonl_path, jsonl2_path, tmp_path_factory):
+ import zipfile
+
+ path = tmp_path_factory.mktemp("data") / "dataset.jsonl.zip"
+ with zipfile.ZipFile(path, "w") as f:
+ f.write(jsonl_path, arcname=os.path.basename(jsonl_path))
+ f.write(jsonl2_path, arcname=os.path.basename(jsonl2_path))
+ return path
+
+
[email protected](scope="session")
+def zip_jsonl_with_dir_path(jsonl_path, jsonl2_path, tmp_path_factory):
+ import zipfile
+
+ path = tmp_path_factory.mktemp("data") / "dataset_with_dir.jsonl.zip"
+ with zipfile.ZipFile(path, "w") as f:
+ f.write(jsonl_path, arcname=os.path.join("main_dir", os.path.basename(jsonl_path)))
+ f.write(jsonl2_path, arcname=os.path.join("main_dir", os.path.basename(jsonl2_path)))
+ return path
+
+
[email protected](scope="session")
+def text_path(tmp_path_factory):
+ data = ["0", "1", "2", "3"]
+ path = str(tmp_path_factory.mktemp("data") / "dataset.txt")
+ with open(path, "w") as f:
+ for item in data:
+ f.write(item + "\n")
+ return path
+
+
[email protected](scope="session")
+def text2_path(tmp_path_factory):
+ data = ["0", "1", "2", "3"]
+ path = str(tmp_path_factory.mktemp("data") / "dataset2.txt")
+ with open(path, "w") as f:
+ for item in data:
+ f.write(item + "\n")
+ return path
+
+
[email protected](scope="session")
+def zip_text_path(text_path, text2_path, tmp_path_factory):
+ import zipfile
+
+ path = tmp_path_factory.mktemp("data") / "dataset.text.zip"
+ with zipfile.ZipFile(path, "w") as f:
+ f.write(text_path, arcname=os.path.basename(text_path))
+ f.write(text2_path, arcname=os.path.basename(text2_path))
+ return path
+
+
[email protected](scope="session")
+def zip_text_with_dir_path(text_path, text2_path, tmp_path_factory):
+ import zipfile
+
+ path = tmp_path_factory.mktemp("data") / "dataset_with_dir.text.zip"
+ with zipfile.ZipFile(path, "w") as f:
+ f.write(text_path, arcname=os.path.join("main_dir", os.path.basename(text_path)))
+ f.write(text2_path, arcname=os.path.join("main_dir", os.path.basename(text2_path)))
+ return path
diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -157,17 +157,6 @@ def metric_loading_script_dir(tmp_path):
return str(script_dir)
[email protected](scope="session")
-def zip_csv_with_dir_path(csv_path, csv2_path, tmp_path_factory):
- import zipfile
-
- path = tmp_path_factory.mktemp("data") / "dataset_with_dir.csv.zip"
- with zipfile.ZipFile(path, "w") as f:
- f.write(csv_path, arcname=os.path.join("main_dir", os.path.basename(csv_path)))
- f.write(csv2_path, arcname=os.path.join("main_dir", os.path.basename(csv2_path)))
- return path
-
-
@pytest.mark.parametrize("data_file, expected_module", [("zip_csv_path", "csv"), ("zip_csv_with_dir_path", "csv")])
def test_infer_module_for_data_files_in_archives(data_file, expected_module, zip_csv_path, zip_csv_with_dir_path):
data_file_paths = {"zip_csv_path": zip_csv_path, "zip_csv_with_dir_path": zip_csv_with_dir_path}
@@ -555,12 +544,78 @@ def test_load_dataset_streaming_csv(path_extension, streaming, csv_path, bz2_csv
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
-def test_load_dataset_zip_csv(zip_csv_path):
- data_files = str(zip_csv_path)
[email protected]("streaming", [False, True])
[email protected]("data_file", ["zip_csv_path", "zip_csv_with_dir_path", "csv_path"])
+def test_load_dataset_zip_csv(data_file, streaming, zip_csv_path, zip_csv_with_dir_path, csv_path):
+ data_file_paths = {
+ "zip_csv_path": zip_csv_path,
+ "zip_csv_with_dir_path": zip_csv_with_dir_path,
+ "csv_path": csv_path,
+ }
+ data_files = str(data_file_paths[data_file])
+ expected_size = 8 if data_file.startswith("zip") else 4
features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
- ds = load_dataset("csv", split="train", data_files=data_files, features=features)
- ds_item = next(iter(ds))
- assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
+ ds = load_dataset("csv", split="train", data_files=data_files, features=features, streaming=streaming)
+ if streaming:
+ ds_item_counter = 0
+ for ds_item in ds:
+ if ds_item_counter == 0:
+ assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
+ ds_item_counter += 1
+ assert ds_item_counter == expected_size
+ else:
+ assert ds.shape[0] == expected_size
+ ds_item = next(iter(ds))
+ assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
+
+
[email protected]("streaming", [False, True])
[email protected]("data_file", ["zip_jsonl_path", "zip_jsonl_with_dir_path", "jsonl_path"])
+def test_load_dataset_zip_jsonl(data_file, streaming, zip_jsonl_path, zip_jsonl_with_dir_path, jsonl_path):
+ data_file_paths = {
+ "zip_jsonl_path": zip_jsonl_path,
+ "zip_jsonl_with_dir_path": zip_jsonl_with_dir_path,
+ "jsonl_path": jsonl_path,
+ }
+ data_files = str(data_file_paths[data_file])
+ expected_size = 8 if data_file.startswith("zip") else 4
+ features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
+ ds = load_dataset("json", split="train", data_files=data_files, features=features, streaming=streaming)
+ if streaming:
+ ds_item_counter = 0
+ for ds_item in ds:
+ if ds_item_counter == 0:
+ assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
+ ds_item_counter += 1
+ assert ds_item_counter == expected_size
+ else:
+ assert ds.shape[0] == expected_size
+ ds_item = next(iter(ds))
+ assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
+
+
[email protected]("streaming", [False, True])
[email protected]("data_file", ["zip_text_path", "zip_text_with_dir_path", "text_path"])
+def test_load_dataset_zip_text(data_file, streaming, zip_text_path, zip_text_with_dir_path, text_path):
+ data_file_paths = {
+ "zip_text_path": zip_text_path,
+ "zip_text_with_dir_path": zip_text_with_dir_path,
+ "text_path": text_path,
+ }
+ data_files = str(data_file_paths[data_file])
+ expected_size = 8 if data_file.startswith("zip") else 4
+ ds = load_dataset("text", split="train", data_files=data_files, streaming=streaming)
+ if streaming:
+ ds_item_counter = 0
+ for ds_item in ds:
+ if ds_item_counter == 0:
+ assert ds_item == {"text": "0"}
+ ds_item_counter += 1
+ assert ds_item_counter == expected_size
+ else:
+ assert ds.shape[0] == expected_size
+ ds_item = next(iter(ds))
+ assert ds_item == {"text": "0"}
def test_loading_from_the_datasets_hub():
| Support streaming zipped CSV dataset repo by passing only repo name
Given a community 🤗 dataset repository containing only a zipped CSV file (only raw data, no loading script), I would like to load it in streaming mode without passing `data_files`:
```
ds_name = "bigscience-catalogue-data/vietnamese_poetry_from_fsoft_ai_lab"
ds = load_dataset(ds_name, split="train", streaming=True, use_auth_token=True)
item = next(iter(ds))
```
Currently, it gives a `FileNotFoundError` because there is no glob (no "\*" after "zip://": "zip://*") in the passed URL:
```
'zip://::https://huggingface.co/datasets/bigscience-catalogue-data/vietnamese_poetry_from_fsoft_ai_lab/resolve/e5d45f1bd9a8a798cc14f0a45ebc1ce91907c792/poems_dataset.zip'
```
| 2021-12-03T10:43:05Z | [] | [] |
|
huggingface/datasets | 3,382 | huggingface__datasets-3382 | [
"3337"
] | 6090f3cfb5c819f441dd4a4bb635e037c875b044 | diff --git a/datasets/big_patent/big_patent.py b/datasets/big_patent/big_patent.py
--- a/datasets/big_patent/big_patent.py
+++ b/datasets/big_patent/big_patent.py
@@ -101,7 +101,7 @@ class BigPatent(datasets.GeneratorBasedBuilder):
BigPatentConfig( # pylint:disable=g-complex-comprehension
cpc_codes=[k],
name=k,
- description=(f"Patents under Cooperative Patent Classification (CPC)" "{k}: {v}"),
+ description=("Patents under Cooperative Patent Classification (CPC)" f"{k}: {v}"),
)
for k, v in sorted(_CPC_DESCRIPTION.items())
]
diff --git a/datasets/gem/gem.py b/datasets/gem/gem.py
--- a/datasets/gem/gem.py
+++ b/datasets/gem/gem.py
@@ -883,7 +883,7 @@ def _split_generators(self, dl_manager):
("challenge_test_bfp_02", "test_xsum_ButterFingersPerturbation_p=0.02_500.json"),
("challenge_test_bfp_05", "test_xsum_ButterFingersPerturbation_p=0.05_500.json"),
("challenge_test_nopunc", "test_xsum_WithoutPunctuation500.json"),
- ("challenge_test_covid", f"en_test_covid19.jsonl"),
+ ("challenge_test_covid", "en_test_covid19.jsonl"),
]
return [
datasets.SplitGenerator(
diff --git a/datasets/multilingual_librispeech/multilingual_librispeech.py b/datasets/multilingual_librispeech/multilingual_librispeech.py
--- a/datasets/multilingual_librispeech/multilingual_librispeech.py
+++ b/datasets/multilingual_librispeech/multilingual_librispeech.py
@@ -127,7 +127,7 @@ def _generate_examples(self, data_dir, sub_folder=""):
all_ids = []
for path in all_ids_paths:
with open(path, "r", encoding="utf-8") as f:
- all_ids += [l.strip() for l in f.readlines()]
+ all_ids += [line.strip() for line in f.readlines()]
all_ids = set(all_ids)
diff --git a/datasets/sem_eval_2020_task_11/sem_eval_2020_task_11.py b/datasets/sem_eval_2020_task_11/sem_eval_2020_task_11.py
--- a/datasets/sem_eval_2020_task_11/sem_eval_2020_task_11.py
+++ b/datasets/sem_eval_2020_task_11/sem_eval_2020_task_11.py
@@ -236,7 +236,7 @@ def _process_tc_labels_template(self, tc_labels_template):
with open(tc_labels_template, encoding="utf-8") as f:
tc_labels_test = f.readlines()
- tc_labels_test = [l.rstrip("\n").split("\t") for l in tc_labels_test]
+ tc_labels_test = [line.rstrip("\n").split("\t") for line in tc_labels_test]
tc_test_template = defaultdict(lambda: [])
diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -183,7 +183,7 @@
]
)
-QUALITY_REQUIRE = ["black==21.4b0", "flake8==3.7.9", "isort>=5.0.0", "pyyaml>=5.3.1"]
+QUALITY_REQUIRE = ["black==21.4b0", "flake8>=3.8.3", "isort>=5.0.0", "pyyaml>=5.3.1"]
EXTRAS_REQUIRE = {
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -24,14 +24,27 @@
import tempfile
import weakref
from collections import Counter, UserDict
-from collections.abc import Iterable, Mapping
+from collections.abc import Mapping
from copy import deepcopy
from dataclasses import asdict
from functools import partial, wraps
from io import BytesIO
from math import ceil, floor
from pathlib import Path
-from typing import TYPE_CHECKING, Any, BinaryIO, Callable, Dict, Iterator, List, Optional, Tuple, Union
+from typing import (
+ TYPE_CHECKING,
+ Any,
+ BinaryIO,
+ Callable,
+ Dict,
+ Iterable,
+ Iterator,
+ List,
+ Optional,
+ Tuple,
+ Union,
+ overload,
+)
import fsspec
import numpy as np
@@ -1887,7 +1900,15 @@ def _getitem(self, key: Union[int, slice, str], decoded: bool = True, **kwargs)
)
return formatted_output
- def __getitem__(self, key: Union[int, slice, str]) -> Union[Dict, List]:
+ @overload
+ def __getitem__(self, key: Union[int, slice, Iterable[int]]) -> Dict: # noqa: F811
+ ...
+
+ @overload
+ def __getitem__(self, key: str) -> List: # noqa: F811
+ ...
+
+ def __getitem__(self, key): # noqa: F811
"""Can be used to index columns (by string names) or rows (by integer index or iterable of indices or bools)."""
return self._getitem(
key,
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -1228,7 +1228,7 @@ def recursive_reorder(source, target, stack=""):
raise ValueError(f"Type mismatch: between {source} and {target}" + stack_position)
if len(source) != len(target):
raise ValueError(f"Length mismatch: between {source} and {target}" + stack_position)
- return [recursive_reorder(source[i], target[i], stack + f".<list>") for i in range(len(target))]
+ return [recursive_reorder(source[i], target[i], stack + ".<list>") for i in range(len(target))]
else:
return source
diff --git a/src/datasets/fingerprint.py b/src/datasets/fingerprint.py
--- a/src/datasets/fingerprint.py
+++ b/src/datasets/fingerprint.py
@@ -342,7 +342,7 @@ def fingerprint_transform(
raise ValueError(f"ignore_kwargs is supposed to be a list, not {type(use_kwargs)}")
if inplace and fingerprint_names:
- raise ValueError(f"fingerprint_names are only used when inplace is False")
+ raise ValueError("fingerprint_names are only used when inplace is False")
fingerprint_names = fingerprint_names if fingerprint_names is not None else ["new_fingerprint"]
diff --git a/src/datasets/utils/metadata.py b/src/datasets/utils/metadata.py
--- a/src/datasets/utils/metadata.py
+++ b/src/datasets/utils/metadata.py
@@ -363,7 +363,7 @@ def validate_paperswithcode_id_errors(paperswithcode_id: Optional[str]) -> Valid
def validate_pretty_name(pretty_name: Union[str, Dict[str, str]]):
if isinstance(pretty_name, str):
if len(pretty_name) == 0:
- return None, f"The pretty name must have a length greater than 0 but got an empty string."
+ return None, "The pretty name must have a length greater than 0 but got an empty string."
else:
error_string = ""
for key, value in pretty_name.items():
diff --git a/src/datasets/utils/readme.py b/src/datasets/utils/readme.py
--- a/src/datasets/utils/readme.py
+++ b/src/datasets/utils/readme.py
@@ -246,7 +246,7 @@ def _validate(self, readme_structure):
elif num_first_level_keys < 1:
# If less than one, append error.
error_list.append(
- f"The README has no first-level headings. One heading is expected. Skipping further validation for this README."
+ "The README has no first-level headings. One heading is expected. Skipping further validation for this README."
)
else:
@@ -263,7 +263,7 @@ def _validate(self, readme_structure):
else:
# If not found, append error
error_list.append(
- f"No first-level heading starting with `Dataset Card for` found in README. Skipping further validation for this README."
+ "No first-level heading starting with `Dataset Card for` found in README. Skipping further validation for this README."
)
if error_list:
# If there are errors, do not return the dictionary as it is invalid
| diff --git a/tests/test_packaged_modules.py b/tests/test_packaged_modules.py
--- a/tests/test_packaged_modules.py
+++ b/tests/test_packaged_modules.py
@@ -62,7 +62,7 @@ def test_csv_generate_tables_raises_error_with_malformed_csv(csv_file, malformed
pass
assert any(
record.levelname == "ERROR"
- and f"Failed to read file" in record.message
+ and "Failed to read file" in record.message
and os.path.basename(malformed_csv_file) in record.message
for record in caplog.records
)
| Typing of Dataset.__getitem__ could be improved.
## Describe the bug
The newly added typing for Dataset.__getitem__ is Union[Dict, List]. This makes tools like mypy a bit awkward to use as we need to check the type manually. We could use type overloading to make this easier. [Documentation](https://docs.python.org/3/library/typing.html#typing.overload)
## Steps to reproduce the bug
Let's have a file `test.py`
```python
from typing import List, Dict, Any
from datasets import Dataset
ds = Dataset.from_dict({
'a': [1,2,3],
'b': ["1", "2", "3"]
})
one_colum: List[str] = ds['a']
some_index: Dict[Any, Any] = ds[1]
```
## Expected results
Running `mypy test.py` should not give any error.
## Actual results
```
test.py:10: error: Incompatible types in assignment (expression has type "Union[Dict[Any, Any], List[Any]]", variable has type "List[str]")
test.py:11: error: Incompatible types in assignment (expression has type "Union[Dict[Any, Any], List[Any]]", variable has type "Dict[Any, Any]")
Found 2 errors in 1 file (checked 1 source file)
```
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 1.13.3
- Platform: macOS-10.16-x86_64-i386-64bit
- Python version: 3.8.8
- PyArrow version: 6.0.1
| Hi ! Thanks for the suggestion, I didn't know about this decorator.
If you are interesting in contributing, feel free to open a pull request to add the overload methods for each typing combination :) To assign you to this issue, you can comment `#self-assign` in this thread.
`Dataset.__getitem__` is defined right here: https://github.com/huggingface/datasets/blob/e6f1352fe19679de897f3d962e616936a17094f5/src/datasets/arrow_dataset.py#L1840
#self-assign | 2021-12-04T20:54:49Z | [] | [] |
huggingface/datasets | 3,402 | huggingface__datasets-3402 | [
"3306"
] | 0009a8ed208ff8f7b91869769ec795f831471b81 | diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -152,7 +152,7 @@ def _cast_to_python_objects(obj: Any, only_1d_for_numpy: bool) -> Tuple[Any, boo
Cast pytorch/tensorflow/pandas objects to python numpy array/lists.
It works recursively.
- To avoid iterating over possibly long lists, it first checks if the first element that is not None has to be casted.
+ To avoid iterating over possibly long lists, it first checks (recursively) if the first element that is not None or empty (if it is a sequence) has to be casted.
If the first element needs to be casted, then all the elements of the list will be casted, otherwise they'll stay the same.
This trick allows to cast objects that contain tokenizers outputs without iterating over every single token for example.
@@ -221,7 +221,7 @@ def _cast_to_python_objects(obj: Any, only_1d_for_numpy: bool) -> Tuple[Any, boo
elif isinstance(obj, (list, tuple)):
if len(obj) > 0:
for first_elmt in obj:
- if first_elmt is not None:
+ if _check_non_null_non_empty_recursive(first_elmt):
break
casted_first_elmt, has_changed_first_elmt = _cast_to_python_objects(
first_elmt, only_1d_for_numpy=only_1d_for_numpy
@@ -244,7 +244,7 @@ def cast_to_python_objects(obj: Any, only_1d_for_numpy=False) -> Any:
Cast numpy/pytorch/tensorflow/pandas objects to python lists.
It works recursively.
- To avoid iterating over possibly long lists, it first checks if the first element that is not None has to be casted.
+ To avoid iterating over possibly long lists, it first checks (recursively) if the first element that is not None or empty (if it is a sequence) has to be casted.
If the first element needs to be casted, then all the elements of the list will be casted, otherwise they'll stay the same.
This trick allows to cast objects that contain tokenizers outputs without iterating over every single token for example.
@@ -774,6 +774,28 @@ class Sequence:
]
+def _check_non_null_non_empty_recursive(obj, schema: Optional[FeatureType] = None) -> bool:
+ """
+ Check if the object is not None.
+ If the object is a list or a tuple, recursively check the first element of the sequence and stop if at any point the first element is not a sequence or is an empty sequence.
+ """
+ if obj is None:
+ return False
+ elif isinstance(obj, (list, tuple)) and (schema is None or isinstance(schema, (list, tuple, Sequence))):
+ if len(obj) > 0:
+ if schema is None:
+ pass
+ elif isinstance(schema, (list, tuple)):
+ schema = schema[0]
+ else:
+ schema = schema.feature
+ return _check_non_null_non_empty_recursive(obj[0], schema)
+ else:
+ return False
+ else:
+ return True
+
+
def get_nested_type(schema: FeatureType) -> pa.DataType:
"""
get_nested_type() converts a datasets.FeatureType into a pyarrow.DataType, and acts as the inverse of
@@ -810,7 +832,7 @@ def encode_nested_example(schema, obj):
"""Encode a nested example.
This is used since some features (in particular ClassLabel) have some logic during encoding.
- To avoid iterating over possibly long lists, it first checks if the first element that is not None has to be encoded.
+ To avoid iterating over possibly long lists, it first checks (recursively) if the first element that is not None or empty (if it is a sequence) has to be encoded.
If the first element needs to be encoded, then all the elements of the list will be encoded, otherwise they'll stay the same.
"""
# Nested structures: we allow dict, list/tuples, sequences
@@ -825,7 +847,7 @@ def encode_nested_example(schema, obj):
else:
if len(obj) > 0:
for first_elmt in obj:
- if first_elmt is not None:
+ if _check_non_null_non_empty_recursive(first_elmt, sub_schema):
break
if encode_nested_example(sub_schema, first_elmt) != first_elmt:
return [encode_nested_example(sub_schema, o) for o in obj]
@@ -853,7 +875,7 @@ def encode_nested_example(schema, obj):
else:
if len(obj) > 0:
for first_elmt in obj:
- if first_elmt is not None:
+ if _check_non_null_non_empty_recursive(first_elmt, schema.feature):
break
# be careful when comparing tensors here
if not isinstance(first_elmt, list) or encode_nested_example(schema.feature, first_elmt) != first_elmt:
| diff --git a/tests/features/test_features.py b/tests/features/test_features.py
--- a/tests/features/test_features.py
+++ b/tests/features/test_features.py
@@ -244,6 +244,23 @@ def test_encode_nested_example_sequence_with_none():
assert result is None
+def test_encode_batch_with_example_with_empty_first_elem():
+ features = Features(
+ {
+ "x": Sequence(Sequence(ClassLabel(names=["a", "b"]))),
+ }
+ )
+ encoded_batch = features.encode_batch(
+ {
+ "x": [
+ [["a"], ["b"]],
+ [[], ["b"]],
+ ]
+ }
+ )
+ assert encoded_batch == {"x": [[[0], [1]], [[], [1]]]}
+
+
def iternumpy(key1, value1, value2):
if value1.dtype != value2.dtype: # check only for dtype
raise AssertionError(
| nested sequence feature won't encode example if the first item of the outside sequence is an empty list
## Describe the bug
As the title, nested sequence feature won't encode example if the first item of the outside sequence is an empty list.
## Steps to reproduce the bug
```python
from datasets import Features, Sequence, ClassLabel
features = Features({
'x': Sequence(Sequence(ClassLabel(names=['a', 'b']))),
})
print(features.encode_batch({
'x': [
[['a'], ['b']],
[[], ['b']],
]
}))
```
## Expected results
print `{'x': [[[0], [1]], [[], ['1']]]}`
## Actual results
print `{'x': [[[0], [1]], [[], ['b']]]}`
## Environment info
- `datasets` version: 1.15.1
- Platform: Linux-5.13.0-21-generic-x86_64-with-glibc2.34
- Python version: 3.9.7
- PyArrow version: 6.0.0
## Additional information
I think the issue stems from [here](https://github.com/huggingface/datasets/blob/8555197a3fe826e98bd0206c2d031c4488c53c5c/src/datasets/features/features.py#L847-L848).
| knock knock | 2021-12-07T17:48:16Z | [] | [] |
huggingface/datasets | 3,406 | huggingface__datasets-3406 | [
"3405"
] | 49bb250458117feab0fd7e85326d0de6398c3ba4 | diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -437,7 +437,7 @@ def infer_module_for_data_files_in_archives(
archived_files = []
for filepath in data_files_list:
if str(filepath).endswith(".zip"):
- extracted = xjoin(StreamingDownloadManager().extract(filepath), "*")
+ extracted = xjoin(StreamingDownloadManager().extract(filepath), "**")
archived_files += [
f.split("::")[0] for f in xglob(extracted, recursive=True, use_auth_token=use_auth_token)
]
| diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -31,6 +31,7 @@
LocalDatasetModuleFactoryWithScript,
LocalMetricModuleFactory,
PackagedDatasetModuleFactory,
+ infer_module_for_data_files_in_archives,
)
from datasets.utils.file_utils import DownloadConfig, is_remote_url
@@ -156,6 +157,25 @@ def metric_loading_script_dir(tmp_path):
return str(script_dir)
[email protected](scope="session")
+def zip_csv_with_dir_path(csv_path, csv2_path, tmp_path_factory):
+ import zipfile
+
+ path = tmp_path_factory.mktemp("data") / "dataset_with_dir.csv.zip"
+ with zipfile.ZipFile(path, "w") as f:
+ f.write(csv_path, arcname=os.path.join("main_dir", os.path.basename(csv_path)))
+ f.write(csv2_path, arcname=os.path.join("main_dir", os.path.basename(csv2_path)))
+ return path
+
+
[email protected]("data_file, expected_module", [("zip_csv_path", "csv"), ("zip_csv_with_dir_path", "csv")])
+def test_infer_module_for_data_files_in_archives(data_file, expected_module, zip_csv_path, zip_csv_with_dir_path):
+ data_file_paths = {"zip_csv_path": zip_csv_path, "zip_csv_with_dir_path": zip_csv_with_dir_path}
+ data_files = [str(data_file_paths[data_file])]
+ inferred_module = infer_module_for_data_files_in_archives(data_files, False)
+ assert inferred_module == expected_module
+
+
class ModuleFactoryTest(TestCase):
@pytest.fixture(autouse=True)
def inject_fixtures(self, jsonl_path, data_dir, dataset_loading_script_dir, metric_loading_script_dir):
| ZIP format inference does not work when files located in a dir inside the archive
## Describe the bug
When a zipped file contains archived files within a directory, the function `infer_module_for_data_files_in_archives` does not work.
It only works for files located in the root directory of the ZIP file.
## Steps to reproduce the bug
```python
infer_module_for_data_files_in_archives(["path/to/zip/file.zip"], False)
```
| 2021-12-08T12:39:12Z | [] | [] |
|
huggingface/datasets | 3,439 | huggingface__datasets-3439 | [
"3369"
] | 624b013776e7769c29437ce2635da61599262f5d | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -59,7 +59,7 @@
from . import config, utils
from .arrow_reader import ArrowReader
from .arrow_writer import ArrowWriter, OptimizedTypedSequence
-from .features import ClassLabel, Features, Sequence, Value, _ArrayXD, pandas_types_mapper
+from .features import ClassLabel, Features, FeatureType, Sequence, Value, _ArrayXD, pandas_types_mapper
from .filesystems import extract_path_from_uri, is_remote_filesystem
from .fingerprint import (
fingerprint_transform,
@@ -1416,7 +1416,7 @@ def cast(
return dataset
@fingerprint_transform(inplace=False)
- def cast_column(self, column: str, feature, new_fingerprint: str) -> "Dataset":
+ def cast_column(self, column: str, feature: FeatureType, new_fingerprint: str) -> "Dataset":
"""Cast column to feature for decoding.
Args:
diff --git a/src/datasets/iterable_dataset.py b/src/datasets/iterable_dataset.py
--- a/src/datasets/iterable_dataset.py
+++ b/src/datasets/iterable_dataset.py
@@ -7,7 +7,7 @@
import pyarrow as pa
from .arrow_dataset import DatasetInfoMixin
-from .features import Features
+from .features import Features, FeatureType
from .formatting import PythonFormatter
from .info import DatasetInfo
from .splits import NamedSplit
@@ -494,6 +494,26 @@ def remove_fn(example):
return self.map(remove_fn)
+ def cast_column(self, column: str, feature: FeatureType) -> "IterableDataset":
+ """Cast column to feature for decoding.
+
+ Args:
+ column (:obj:`str`): Column name.
+ feature (:class:`Feature`): Target feature.
+
+ Returns:
+ :class:`IterableDataset`
+ """
+ info = copy.deepcopy(self._info)
+ info.features[column] = feature
+ return iterable_dataset(
+ ex_iterable=self._ex_iterable,
+ info=info,
+ split=self._split,
+ format_type=self._format_type,
+ shuffling=copy.deepcopy(self._shuffling),
+ )
+
def iterable_dataset(
ex_iterable: Iterable,
| diff --git a/tests/test_iterable_dataset.py b/tests/test_iterable_dataset.py
--- a/tests/test_iterable_dataset.py
+++ b/tests/test_iterable_dataset.py
@@ -335,7 +335,7 @@ def test_iterable_dataset_shuffle(dataset: IterableDataset, generate_examples_fn
),
],
)
-def test_terable_dataset_features(generate_examples_fn, features):
+def test_iterable_dataset_features(generate_examples_fn, features):
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": 0})
dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
if features:
@@ -386,6 +386,16 @@ def test_iterable_dataset_shuffle_after_skip_or_take(generate_examples_fn, metho
assert sorted(dataset, key=key) == sorted(shuffled_dataset, key=key)
+def test_iterable_dataset_cast_column(generate_examples_fn):
+ ex_iterable = ExamplesIterable(generate_examples_fn, {"label": 10})
+ features = Features({"id": Value("int64"), "label": Value("int64")})
+ dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
+ casted_dataset = dataset.cast_column("label", Value("bool"))
+ casted_features = features.copy()
+ casted_features["label"] = Value("bool")
+ assert list(casted_dataset) == [casted_features.encode_example(ex) for _, ex in ex_iterable]
+
+
@pytest.mark.parametrize(
"probas, seed, expected_length",
[
| [Audio] Allow resampling for audio datasets in streaming mode
Many audio datasets like Common Voice always need to be resampled. This can very easily be done in non-streaming mode as follows:
```python
from datasets import load_dataset
ds = load_dataset("common_voice", "ab", split="test")
ds = ds.cast_column("audio", Audio(sampling_rate=16_000))
```
However in streaming mode it fails currently:
```python
from datasets import load_dataset
ds = load_dataset("common_voice", "ab", split="test", streaming=True)
ds = ds.cast_column("audio", Audio(sampling_rate=16_000))
```
with the following error:
```
AttributeError: 'IterableDataset' object has no attribute 'cast_column'
```
It would be great if we could add such a feature (I'm not 100% sure though how complex this would be)
| This requires implementing `cast_column` for iterable datasets, it could be a very nice addition !
<s>It can also be useful to be able to disable the audio/image decoding for the dataset viewer (see PR https://github.com/huggingface/datasets/pull/3430) cc @severo </s>
EDIT: actually following https://github.com/huggingface/datasets/issues/3145 the dataset viewer might not need it anymore
Just to clarify a bit. This feature is **always** needed when using the common voice dataset in streaming mode. So I think it's quite important | 2021-12-15T19:00:45Z | [] | [] |
huggingface/datasets | 3,454 | huggingface__datasets-3454 | [
"3453"
] | 6a7467be15428c3de46702e1bc2d86cc1a7c8e37 | diff --git a/src/datasets/utils/download_manager.py b/src/datasets/utils/download_manager.py
--- a/src/datasets/utils/download_manager.py
+++ b/src/datasets/utils/download_manager.py
@@ -239,15 +239,15 @@ def _iter_archive(f):
# skipping hidden files
continue
file_obj = stream.extractfile(tarinfo)
- yield (file_path, file_obj)
+ yield file_path, file_obj
stream.members = []
del stream
if hasattr(path_or_buf, "read"):
- return _iter_archive(path_or_buf)
+ yield from _iter_archive(path_or_buf)
else:
with open(path_or_buf, "rb") as f:
- return _iter_archive(f)
+ yield from _iter_archive(f)
def iter_files(self, paths):
"""Iterate over file paths.
diff --git a/src/datasets/utils/streaming_download_manager.py b/src/datasets/utils/streaming_download_manager.py
--- a/src/datasets/utils/streaming_download_manager.py
+++ b/src/datasets/utils/streaming_download_manager.py
@@ -586,15 +586,15 @@ def _iter_archive(f):
# skipping hidden files
continue
file_obj = stream.extractfile(tarinfo)
- yield (file_path, file_obj)
+ yield file_path, file_obj
stream.members = []
del stream
if hasattr(urlpath_or_buf, "read"):
- return _iter_archive(urlpath_or_buf)
+ yield from _iter_archive(urlpath_or_buf)
else:
with xopen(urlpath_or_buf, "rb", use_auth_token=self.download_config.use_auth_token) as f:
- return _iter_archive(f)
+ yield from _iter_archive(f)
def iter_files(self, urlpaths):
"""Iterate over files.
| diff --git a/tests/conftest.py b/tests/conftest.py
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -2,6 +2,7 @@
import json
import lzma
import os
+import tarfile
import textwrap
import pyarrow as pa
@@ -400,6 +401,23 @@ def zip_jsonl_with_dir_path(jsonl_path, jsonl2_path, tmp_path_factory):
return path
[email protected](scope="session")
+def tar_jsonl_path(jsonl_path, jsonl2_path, tmp_path_factory):
+ path = tmp_path_factory.mktemp("data") / "dataset.jsonl.tar"
+ with tarfile.TarFile(path, "w") as f:
+ f.add(jsonl_path, arcname=os.path.basename(jsonl_path))
+ f.add(jsonl2_path, arcname=os.path.basename(jsonl2_path))
+ return path
+
+
[email protected](scope="session")
+def tar_nested_jsonl_path(tar_jsonl_path, jsonl_path, jsonl2_path, tmp_path_factory):
+ path = tmp_path_factory.mktemp("data") / "dataset_nested.jsonl.tar"
+ with tarfile.TarFile(path, "w") as f:
+ f.add(tar_jsonl_path, arcname=os.path.join("nested", os.path.basename(tar_jsonl_path)))
+ return path
+
+
@pytest.fixture(scope="session")
def text_path(tmp_path_factory):
data = ["0", "1", "2", "3"]
diff --git a/tests/test_download_manager.py b/tests/test_download_manager.py
--- a/tests/test_download_manager.py
+++ b/tests/test_download_manager.py
@@ -111,3 +111,27 @@ def test_download_manager_extract(paths_type, xz_file, text_file):
extracted_file_content = extracted_path.read_text()
expected_file_content = text_file.read_text()
assert extracted_file_content == expected_file_content
+
+
+def _test_jsonl(path, file):
+ assert path.endswith(".jsonl")
+ for num_items, line in enumerate(file, start=1):
+ item = json.loads(line.decode("utf-8"))
+ assert item.keys() == {"col_1", "col_2", "col_3"}
+ assert num_items == 4
+
+
+def test_iter_archive_path(tar_jsonl_path):
+ dl_manager = DownloadManager()
+ for num_jsonl, (path, file) in enumerate(dl_manager.iter_archive(tar_jsonl_path), start=1):
+ _test_jsonl(path, file)
+ assert num_jsonl == 2
+
+
+def test_iter_archive_file(tar_nested_jsonl_path):
+ dl_manager = DownloadManager()
+ for num_tar, (path, file) in enumerate(dl_manager.iter_archive(tar_nested_jsonl_path), start=1):
+ for num_jsonl, (subpath, subfile) in enumerate(dl_manager.iter_archive(file), start=1):
+ _test_jsonl(subpath, subfile)
+ assert num_tar == 1
+ assert num_jsonl == 2
diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -1,3 +1,4 @@
+import json
import os
import re
from pathlib import Path
@@ -573,3 +574,27 @@ def test_streaming_gg_drive_zipped():
assert xbasename(all_files[0]) == TEST_GG_DRIVE_FILENAME
with xopen(all_files[0]) as f:
assert f.read() == TEST_GG_DRIVE_CONTENT
+
+
+def _test_jsonl(path, file):
+ assert path.endswith(".jsonl")
+ for num_items, line in enumerate(file, start=1):
+ item = json.loads(line.decode("utf-8"))
+ assert item.keys() == {"col_1", "col_2", "col_3"}
+ assert num_items == 4
+
+
+def test_iter_archive_path(tar_jsonl_path):
+ dl_manager = StreamingDownloadManager()
+ for num_jsonl, (path, file) in enumerate(dl_manager.iter_archive(str(tar_jsonl_path)), start=1):
+ _test_jsonl(path, file)
+ assert num_jsonl == 2
+
+
+def test_iter_archive_file(tar_nested_jsonl_path):
+ dl_manager = StreamingDownloadManager()
+ for num_tar, (path, file) in enumerate(dl_manager.iter_archive(str(tar_nested_jsonl_path)), start=1):
+ for num_jsonl, (subpath, subfile) in enumerate(dl_manager.iter_archive(file), start=1):
+ _test_jsonl(subpath, subfile)
+ assert num_tar == 1
+ assert num_jsonl == 2
| ValueError while iter_archive
## Describe the bug
After the merge of:
- #3443
the method `iter_archive` throws a ValueError:
```
ValueError: read of closed file
```
## Steps to reproduce the bug
```python
for path, file in dl_manager.iter_archive(archive_path):
pass
```
| 2021-12-20T08:50:15Z | [] | [] |
|
huggingface/datasets | 3,556 | huggingface__datasets-3556 | [
"3505"
] | bb3e6012609df6d45231729c78d7b0806ed41c6c | diff --git a/src/datasets/combine.py b/src/datasets/combine.py
--- a/src/datasets/combine.py
+++ b/src/datasets/combine.py
@@ -174,14 +174,15 @@ def _interleave_iterable_datasets(
"""
from .iterable_dataset import (
CyclingMultiSourcesExamplesIterable,
- MappedExamplesIterable,
RandomlyCyclingMultiSourcesExamplesIterable,
+ TypedExamplesIterable,
iterable_dataset,
)
- # Keep individual features formatting
ex_iterables = [
- MappedExamplesIterable(d._ex_iterable, d.features.encode_example) if d.features is not None else d._ex_iterable
+ TypedExamplesIterable(d._ex_iterable, d.features)
+ if not isinstance(d._ex_iterable, TypedExamplesIterable) and d.features is not None
+ else d._ex_iterable
for d in datasets
]
# Use cycling or random cycling or sources
diff --git a/src/datasets/iterable_dataset.py b/src/datasets/iterable_dataset.py
--- a/src/datasets/iterable_dataset.py
+++ b/src/datasets/iterable_dataset.py
@@ -281,6 +281,31 @@ def n_shards(self) -> int:
return self.ex_iterable.n_shards
+class TypedExamplesIterable(_BaseExamplesIterable):
+ def __init__(self, ex_iterable: _BaseExamplesIterable, features: Features):
+ self.ex_iterable = ex_iterable
+ self.features = features
+
+ def __iter__(self):
+ for key, example in self.ex_iterable:
+ # we encode the example for ClassLabel feature types for example
+ encoded_example = self.features.encode_example(example)
+ # Decode example for Audio feature, e.g.
+ decoded_example = self.features.decode_example(encoded_example)
+ yield key, decoded_example
+
+ def shuffle_data_sources(self, seed: Optional[int]) -> "TypedExamplesIterable":
+ """Shuffle the wrapped examples iterable."""
+ return TypedExamplesIterable(
+ self.ex_iterable.shuffle_data_sources(seed),
+ features=self.features,
+ )
+
+ @property
+ def n_shards(self) -> int:
+ return self.ex_iterable.n_shards
+
+
def _generate_examples_from_tables_wrapper(generate_tables_fn):
def wrapper(**kwargs):
python_formatter = PythonFormatter()
@@ -397,7 +422,12 @@ def map(self, function: Callable, batched: bool = False, batch_size: int = 1000)
info = copy.deepcopy(self._info)
info.features = None
ex_iterable = MappedExamplesIterable(
- self._ex_iterable, function=function, batched=batched, batch_size=batch_size
+ TypedExamplesIterable(self._ex_iterable, self._info.features)
+ if self._info.features is not None
+ else self._ex_iterable,
+ function=function,
+ batched=batched,
+ batch_size=batch_size,
)
return iterable_dataset(
ex_iterable=ex_iterable,
| diff --git a/tests/test_iterable_dataset.py b/tests/test_iterable_dataset.py
--- a/tests/test_iterable_dataset.py
+++ b/tests/test_iterable_dataset.py
@@ -16,6 +16,7 @@
ShufflingConfig,
SkipExamplesIterable,
TakeExamplesIterable,
+ TypedExamplesIterable,
_batch_to_examples,
_examples_to_batch,
iterable_dataset,
@@ -290,6 +291,25 @@ def test_iterable_dataset_map_batched(dataset: IterableDataset, generate_example
assert next(iter(dataset)) == _func_unbatched(next(iter(generate_examples_fn()))[1])
+def test_iterable_dataset_map_complex_features(dataset: IterableDataset, generate_examples_fn):
+ # https://github.com/huggingface/datasets/issues/3505
+ ex_iterable = ExamplesIterable(generate_examples_fn, {"label": "positive"})
+ features = Features(
+ {
+ "id": Value("int64"),
+ "label": Value("string"),
+ }
+ )
+ dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
+ dataset = dataset.cast_column("label", ClassLabel(names=["negative", "positive"]))
+ dataset = dataset.map(lambda x: {"id+1": x["id"] + 1, **x})
+ assert isinstance(dataset._ex_iterable, MappedExamplesIterable)
+ features["label"] = ClassLabel(names=["negative", "positive"])
+ assert [{k: v for k, v in ex.items() if k != "id+1"} for ex in dataset] == [
+ features.encode_example(ex) for _, ex in ex_iterable
+ ]
+
+
@pytest.mark.parametrize("seed", [42, 1337, 101010, 123456])
@pytest.mark.parametrize("epoch", [None, 0, 1])
def test_iterable_dataset_shuffle(dataset: IterableDataset, generate_examples_fn, seed, epoch):
@@ -454,6 +474,6 @@ def test_interleave_datasets_with_features(dataset: IterableDataset, generate_ex
merged_dataset = interleave_datasets([dataset, dataset_with_features], probabilities=[0, 1])
assert isinstance(merged_dataset._ex_iterable, CyclingMultiSourcesExamplesIterable)
- assert isinstance(merged_dataset._ex_iterable.ex_iterables[1], MappedExamplesIterable)
- assert merged_dataset._ex_iterable.ex_iterables[1].function == features.encode_example
+ assert isinstance(merged_dataset._ex_iterable.ex_iterables[1], TypedExamplesIterable)
+ assert merged_dataset._ex_iterable.ex_iterables[1].features == features
assert next(iter(merged_dataset)) == next(iter(dataset_with_features))
| cast_column function not working with map function in streaming mode for Audio features
## Describe the bug
I am trying to use Audio class for loading audio features using custom dataset. I am able to cast 'audio' feature into 'Audio' format with cast_column function. On using map function, I am not getting 'Audio' casted feature but getting path of audio file only.
I am getting features of 'audio' of string type with load_dataset call. After using cast_column 'audio' feature is converted into 'Audio' type. But in map function I am not able to get Audio type for audio feature & getting string type data containing path of file only. So I am not able to use processor in encode function.
## Steps to reproduce the bug
```python
# Sample code to reproduce the bug
from datasets import load_dataset, Audio
from transformers import Wav2Vec2Processor
def encode(batch, processor):
print("Audio: ",batch['audio'])
batch["input_values"] = processor(batch["audio"]['array'], sampling_rate=16000).input_values
return batch
def print_ds(ds):
iterator = iter(ds)
for d in iterator:
print("Data: ",d)
break
processor = Wav2Vec2Processor.from_pretrained(pretrained_model_path)
dataset = load_dataset("custom_dataset.py","train",data_files={'train':'train_path.txt'},
data_dir="data", streaming=True, split="train")
print("Features: ",dataset.features)
print_ds(dataset)
dataset = dataset.cast_column("audio", Audio(sampling_rate=16_000))
print("Features: ",dataset.features)
print_ds(dataset)
dataset = dataset.map(lambda x: encode(x,processor))
print("Features: ",dataset.features)
print_ds(dataset)
```
## Expected results
map function not printing Audio type features be used with processor function and getting error in processor call due to this.
## Actual results
# after load_dataset call
Features: {'sentence': Value(dtype='string', id=None), 'audio': Value(dtype='string', id=None)}
Data: {'sentence': 'और अपने पेट को माँ की स्वादिष्ट गरमगरम जलेबियाँ हड़पते\n', 'audio': 'data/0116_003.wav'}
# after cast_column call
Features: {'sentence': Value(dtype='string', id=None), 'audio': Audio(sampling_rate=16000, mono=True, _storage_dtype='string', id=None)}
Data: {'sentence': 'और अपने पेट को माँ की स्वादिष्ट गरमगरम जलेबियाँ हड़पते\n', 'audio': {'path': 'data/0116_003.wav', 'array': array([ 1.2662281e-06, 1.0264218e-06, -1.3615092e-06, ...,
1.3017889e-02, 1.0085563e-02, 4.8155054e-03], dtype=float32), 'sampling_rate': 16000}}
# after map call
Features: None
Audio: data/0116_003.wav
Traceback (most recent call last):
File "demo2.py", line 36, in <module>
print_ds(dataset)
File "demo2.py", line 11, in print_ds
for d in iterator:
File "/opt/conda/lib/python3.7/site-packages/datasets/iterable_dataset.py", line 341, in __iter__
for key, example in self._iter():
File "/opt/conda/lib/python3.7/site-packages/datasets/iterable_dataset.py", line 338, in _iter
yield from ex_iterable
File "/opt/conda/lib/python3.7/site-packages/datasets/iterable_dataset.py", line 192, in __iter__
yield key, self.function(example)
File "demo2.py", line 32, in <lambda>
dataset = dataset.map(lambda x: batch_encode(x,processor))
File "demo2.py", line 6, in batch_encode
batch["input_values"] = processor(batch["audio"]['array'], sampling_rate=16000).input_values
TypeError: string indices must be integers
## Environment info
- `datasets` version: 1.17.0
- Platform: Linux-4.14.243 with-debian-bullseye-sid
- Python version: 3.7.9
- PyArrow version: 6.0.1
| Hi! This is probably due to the fact that `IterableDataset.map` sets `features` to `None` before mapping examples. We can fix the issue by passing the old `features` dict to the map generator and performing encoding/decoding there (before calling the map transform function). | 2022-01-10T13:32:20Z | [] | [] |
huggingface/datasets | 3,623 | huggingface__datasets-3623 | [
"3622"
] | 300ddd779dc4078f82274959baf18209a5e99dea | diff --git a/src/datasets/streaming.py b/src/datasets/streaming.py
--- a/src/datasets/streaming.py
+++ b/src/datasets/streaming.py
@@ -25,6 +25,7 @@
xpathrglob,
xpathstem,
xpathsuffix,
+ xrelpath,
xsio_loadmat,
xsplitext,
xwalk,
@@ -75,6 +76,7 @@ def wrapper(*args, **kwargs):
patch_submodule(module, "os.path.join", xjoin).start()
patch_submodule(module, "os.path.dirname", xdirname).start()
patch_submodule(module, "os.path.basename", xbasename).start()
+ patch_submodule(module, "os.path.relpath", xrelpath).start()
patch_submodule(module, "os.path.splitext", xsplitext).start()
# allow checks on paths
patch_submodule(module, "os.path.isdir", wrap_auth(xisdir)).start()
diff --git a/src/datasets/utils/streaming_download_manager.py b/src/datasets/utils/streaming_download_manager.py
--- a/src/datasets/utils/streaming_download_manager.py
+++ b/src/datasets/utils/streaming_download_manager.py
@@ -234,6 +234,23 @@ def xisdir(path, use_auth_token: Optional[Union[str, bool]] = None) -> bool:
return fs.isdir(main_hop)
+def xrelpath(path, start=None):
+ """Extend `os.path.relpath` function to support remote files.
+
+ Args:
+ path (:obj:`str`): URL path.
+ start (:obj:`str`): Start URL directory path.
+
+ Returns:
+ :obj:`str`
+ """
+ main_hop, *rest_hops = path.split("::")
+ if is_local_path(main_hop):
+ return os.path.relpath(main_hop, start=start) if start else os.path.relpath(main_hop)
+ else:
+ return posixpath.relpath(main_hop, start=start.split("::")[0]) if start else os.path.relpath(main_hop)
+
+
def _as_posix(path: Path):
"""Extend :meth:`pathlib.PurePath.as_posix` to fix missing slashes after protocol.
| diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -27,6 +27,7 @@
xpathrglob,
xpathstem,
xpathsuffix,
+ xrelpath,
xsplitext,
)
@@ -352,6 +353,29 @@ def test_xglob(input_path, expected_paths, tmp_path, mock_fsspec):
assert output_paths == expected_paths
[email protected](
+ "input_path, start_path, expected_path",
+ [
+ ("dir1/dir2/file.txt".replace("/", os.path.sep), "dir1", "dir2/file.txt".replace("/", os.path.sep)),
+ ("dir1/dir2/file.txt".replace("/", os.path.sep), "dir1/dir2".replace("/", os.path.sep), "file.txt"),
+ ("zip://file.txt::https://host.com/archive.zip", "zip://::https://host.com/archive.zip", "file.txt"),
+ (
+ "zip://folder/file.txt::https://host.com/archive.zip",
+ "zip://::https://host.com/archive.zip",
+ "folder/file.txt",
+ ),
+ (
+ "zip://folder/file.txt::https://host.com/archive.zip",
+ "zip://folder::https://host.com/archive.zip",
+ "file.txt",
+ ),
+ ],
+)
+def test_xrelpath(input_path, start_path, expected_path):
+ outut_path = xrelpath(input_path, start=start_path)
+ assert outut_path == expected_path
+
+
@pytest.mark.parametrize(
"input_path, pattern, expected_paths",
[
| Extend support for streaming datasets that use os.path.relpath
Extend support for streaming datasets that use `os.path.relpath`.
This feature will also be useful to yield the relative path of audio or image files.
| 2022-01-24T16:00:52Z | [] | [] |
|
huggingface/datasets | 3,642 | huggingface__datasets-3642 | [
"3611"
] | e2e96ff94c9ac1ddff53bcc5241d60515c7dfaa7 | diff --git a/src/datasets/formatting/formatting.py b/src/datasets/formatting/formatting.py
--- a/src/datasets/formatting/formatting.py
+++ b/src/datasets/formatting/formatting.py
@@ -57,7 +57,7 @@ def _query_table_with_indices_mapping(
key = indices.fast_slice(key % indices.num_rows, 1).column(0)[0].as_py()
return _query_table(table, key)
if isinstance(key, slice):
- key = range(*key.indices(table.num_rows))
+ key = range(*key.indices(indices.num_rows))
if isinstance(key, range):
if _is_range_contiguous(key) and key.start >= 0:
return _query_table(
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -195,6 +195,10 @@ def test_dataset_getitem(self, in_memory):
self.assertListEqual(dset[[0, -1]]["filename"], ["my_name-train_0", "my_name-train_29"])
self.assertListEqual(dset[np.array([0, -1])]["filename"], ["my_name-train_0", "my_name-train_29"])
+ with dset.select(range(2)) as dset_subset:
+ self.assertListEqual(dset_subset[-1:]["filename"], ["my_name-train_1"])
+ self.assertListEqual(dset_subset["filename"][-1:], ["my_name-train_1"])
+
def test_dummy_dataset_deepcopy(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
| Indexing bug after dataset.select()
## Describe the bug
A clear and concise description of what the bug is.
Dataset indexing is not working as expected after `dataset.select(range(100))`
## Steps to reproduce the bug
```python
# Sample code to reproduce the bug
import datasets
task_to_keys = {
"cola": ("sentence", None),
"mnli": ("premise", "hypothesis"),
"mrpc": ("sentence1", "sentence2"),
"qnli": ("question", "sentence"),
"qqp": ("question1", "question2"),
"rte": ("sentence1", "sentence2"),
"sst2": ("sentence", None),
"stsb": ("sentence1", "sentence2"),
"wnli": ("sentence1", "sentence2"),
}
task_name = "sst2"
raw_datasets = datasets.load_dataset("glue", task_name)
train_dataset = raw_datasets["train"]
print("before select: ",train_dataset[-2:])
# before select: {'sentence': ['a patient viewer ', 'this new jangle of noise , mayhem and stupidity must be a serious contender for the title . '], 'label': [1, 0], 'idx': [67347, 67348]}
train_dataset = train_dataset.select(range(100))
print("after select: ",train_dataset[-2:])
# after select: {'sentence': [], 'label': [], 'idx': []}
```
link to colab: https://colab.research.google.com/drive/1LngeRC9f0jE7eSQ4Kh1cIeb411lRXQD-?usp=sharing
## Expected results
A clear and concise description of the expected results.
showing 98, 99 index data
## Actual results
Specify the actual results or traceback.
empty
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 1.17.0
- Platform: Linux-5.4.144+-x86_64-with-Ubuntu-18.04-bionic
- Python version: 3.7.12
- PyArrow version: 3.0.0
| 2022-01-27T14:45:53Z | [] | [] |
|
huggingface/datasets | 3,647 | huggingface__datasets-3647 | [
"3599"
] | 3adc314fbf25bddc5ef3d5e3979f669a173f3374 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -3649,13 +3649,14 @@ def add_column(self, name: str, column: Union[list, np.array], new_fingerprint:
"""
column_table = InMemoryTable.from_pydict({name: column})
_check_column_names(self._data.column_names + column_table.column_names)
+ dataset = self.flatten_indices() if self._indices is not None else self
# Concatenate tables horizontally
- table = concat_tables([self._data, column_table], axis=1)
+ table = concat_tables([dataset._data, column_table], axis=1)
# Update features
- info = self.info.copy()
+ info = dataset.info.copy()
info.features.update(Features.from_arrow_schema(column_table.schema))
table = update_metadata_with_features(table, info.features)
- return Dataset(table, info=info, split=self.split, indices_table=self._indices, fingerprint=new_fingerprint)
+ return Dataset(table, info=info, split=self.split, indices_table=None, fingerprint=new_fingerprint)
def add_faiss_index(
self,
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -2491,7 +2491,13 @@ def test_interleave_datasets_probabilities():
@pytest.mark.parametrize("in_memory", [False, True])
@pytest.mark.parametrize(
"transform",
- [None, ("shuffle", (42,), {}), ("with_format", ("pandas",), {}), ("class_encode_column", ("col_2",), {})],
+ [
+ None,
+ ("shuffle", (42,), {}),
+ ("with_format", ("pandas",), {}),
+ ("class_encode_column", ("col_2",), {}),
+ ("select", (range(3),), {}),
+ ],
)
def test_dataset_add_column(column, expected_dtype, in_memory, transform, dataset_dict, arrow_path):
column_name = "col_4"
@@ -2503,8 +2509,9 @@ def test_dataset_add_column(column, expected_dtype, in_memory, transform, datase
if transform is not None:
transform_name, args, kwargs = transform
original_dataset: Dataset = getattr(original_dataset, transform_name)(*args, **kwargs)
+ column = column[:3] if transform is not None and transform_name == "select" else column
dataset = original_dataset.add_column(column_name, column)
- assert dataset.data.shape == (4, 4)
+ assert dataset.data.shape == (3, 4) if transform is not None and transform_name == "select" else (4, 4)
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
# Sort expected features as in the original dataset
expected_features = {feature: expected_features[feature] for feature in original_dataset.features}
| The `add_column()` method does not work if used on dataset sliced with `select()`
Hello, I posted this as a question on the forums ([here](https://discuss.huggingface.co/t/add-column-does-not-work-if-used-on-dataset-sliced-with-select/13893)):
I have a dataset with 2000 entries
> dataset = Dataset.from_dict({'colA': list(range(2000))})
and from which I want to extract the first one thousand rows, create a new dataset with these and also add a new column to it:
> dataset2 = dataset.select(list(range(1000)))
> final_dataset = dataset2.add_column('colB', list(range(1000)))
This gives an error
>ArrowInvalid: Added column's length must match table's length. Expected length 2000 but got length 1000
So it looks like even though it is a dataset with 1000 rows, it "remembers" the shape of the one it was sliced from.
## Actual results
```
ArrowInvalid Traceback (most recent call last)
<ipython-input-138-e806860f3ce3> in <module>
----> 1 final_dataset = dataset2.add_column('colB', list(range(1000)))
~/.local/lib/python3.8/site-packages/datasets/arrow_dataset.py in wrapper(*args, **kwargs)
468 }
469 # apply actual function
--> 470 out: Union["Dataset", "DatasetDict"] = func(self, *args, **kwargs)
471 datasets: List["Dataset"] = list(out.values()) if isinstance(out, dict) else [out]
472 # re-apply format to the output
~/.local/lib/python3.8/site-packages/datasets/fingerprint.py in wrapper(*args, **kwargs)
404 # Call actual function
405
--> 406 out = func(self, *args, **kwargs)
407
408 # Update fingerprint of in-place transforms + update in-place history of transforms
~/.local/lib/python3.8/site-packages/datasets/arrow_dataset.py in add_column(self, name, column, new_fingerprint)
3343 column_table = InMemoryTable.from_pydict({name: column})
3344 # Concatenate tables horizontally
-> 3345 table = ConcatenationTable.from_tables([self._data, column_table], axis=1)
3346 # Update features
3347 info = self.info.copy()
~/.local/lib/python3.8/site-packages/datasets/table.py in from_tables(cls, tables, axis)
729 table_blocks = to_blocks(table)
730 blocks = _extend_blocks(blocks, table_blocks, axis=axis)
--> 731 return cls.from_blocks(blocks)
732
733 @property
~/.local/lib/python3.8/site-packages/datasets/table.py in from_blocks(cls, blocks)
668 @classmethod
669 def from_blocks(cls, blocks: TableBlockContainer) -> "ConcatenationTable":
--> 670 blocks = cls._consolidate_blocks(blocks)
671 if isinstance(blocks, TableBlock):
672 table = blocks
~/.local/lib/python3.8/site-packages/datasets/table.py in _consolidate_blocks(cls, blocks)
664 return cls._merge_blocks(blocks, axis=0)
665 else:
--> 666 return cls._merge_blocks(blocks)
667
668 @classmethod
~/.local/lib/python3.8/site-packages/datasets/table.py in _merge_blocks(cls, blocks, axis)
650 merged_blocks += list(block_group)
651 else: # both
--> 652 merged_blocks = [cls._merge_blocks(row_block, axis=1) for row_block in blocks]
653 if all(len(row_block) == 1 for row_block in merged_blocks):
654 merged_blocks = cls._merge_blocks(
~/.local/lib/python3.8/site-packages/datasets/table.py in <listcomp>(.0)
650 merged_blocks += list(block_group)
651 else: # both
--> 652 merged_blocks = [cls._merge_blocks(row_block, axis=1) for row_block in blocks]
653 if all(len(row_block) == 1 for row_block in merged_blocks):
654 merged_blocks = cls._merge_blocks(
~/.local/lib/python3.8/site-packages/datasets/table.py in _merge_blocks(cls, blocks, axis)
647 for is_in_memory, block_group in groupby(blocks, key=lambda x: isinstance(x, InMemoryTable)):
648 if is_in_memory:
--> 649 block_group = [InMemoryTable(cls._concat_blocks(list(block_group), axis=axis))]
650 merged_blocks += list(block_group)
651 else: # both
~/.local/lib/python3.8/site-packages/datasets/table.py in _concat_blocks(blocks, axis)
626 else:
627 for name, col in zip(table.column_names, table.columns):
--> 628 pa_table = pa_table.append_column(name, col)
629 return pa_table
630 else:
~/.local/lib/python3.8/site-packages/pyarrow/table.pxi in pyarrow.lib.Table.append_column()
~/.local/lib/python3.8/site-packages/pyarrow/table.pxi in pyarrow.lib.Table.add_column()
~/.local/lib/python3.8/site-packages/pyarrow/error.pxi in pyarrow.lib.pyarrow_internal_check_status()
~/.local/lib/python3.8/site-packages/pyarrow/error.pxi in pyarrow.lib.check_status()
ArrowInvalid: Added column's length must match table's length. Expected length 2000 but got length 1000
```
A solution provided by @mariosasko is to use `dataset2.flatten_indices()` after the `select()` and before attempting to add the new column:
> dataset = Dataset.from_dict({'colA': list(range(2000))})
> dataset2 = dataset.select(list(range(1000)))
> dataset2 = dataset2.flatten_indices()
> final_dataset = dataset2.add_column('colB', list(range(1000)))
which works.
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 1.13.2 (note: also checked with version 1.17.0, still the same error)
- Platform: Ubuntu 20.04.3
- Python version: 3.8.10
- PyArrow version: 6.0.0
| similar #3611 | 2022-01-28T13:06:29Z | [] | [] |
huggingface/datasets | 3,654 | huggingface__datasets-3654 | [
"2630"
] | 4c417d52def6e20359ca16c6723e0a2855e5c3fd | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -2096,7 +2096,7 @@ def decorated(item, *args, **kwargs):
f"num_proc must be <= {len(self)}. Reducing num_proc to {num_proc} for dataset of size {len(self)}."
)
- disable_tqdm = bool(logging.get_verbosity() == logging.NOTSET) or not utils.is_progress_bar_enabled()
+ disable_tqdm = not utils.is_progress_bar_enabled()
if num_proc is None or num_proc == 1:
return self._map_single(
@@ -2300,7 +2300,7 @@ def _map_single(
logging.set_verbosity_warning()
# Print at least one thing to fix tqdm in notebooks in multiprocessing
# see https://github.com/tqdm/tqdm/issues/485#issuecomment-473338308
- if rank is not None and not disable_tqdm and "notebook" in tqdm.__name__:
+ if rank is not None and not disable_tqdm and any("notebook" in tqdm_cls.__name__ for tqdm_cls in tqdm.__mro__):
print(" ", end="", flush=True)
if fn_kwargs is None:
@@ -2443,13 +2443,20 @@ def init_buffer_and_writer():
input_dataset = self
# Loop over single examples or batches and write to buffer/file if examples are to be updated
- pbar_iterable = (
- input_dataset._iter(decoded=False) if not batched else range(0, len(input_dataset), batch_size)
- )
+ if not batched:
+ pbar_iterable = input_dataset._iter(decoded=False)
+ pbar_total = len(input_dataset)
+ else:
+ num_rows = (
+ len(input_dataset) if not drop_last_batch else len(input_dataset) // batch_size * batch_size
+ )
+ pbar_iterable = range(0, num_rows, batch_size)
+ pbar_total = (num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size
pbar_unit = "ex" if not batched else "ba"
- pbar_desc = (desc or "") + " #" + str(rank) if rank is not None else desc
+ pbar_desc = (desc + " " if desc is not None else "") + "#" + str(rank) if rank is not None else desc
pbar = utils.tqdm(
pbar_iterable,
+ total=pbar_total,
disable=disable_tqdm,
position=rank,
unit=pbar_unit,
@@ -2468,8 +2475,6 @@ def init_buffer_and_writer():
writer.write(example)
else:
for i in pbar:
- if drop_last_batch and i + batch_size > input_dataset.num_rows:
- continue
batch = input_dataset._getitem(
slice(i, i + batch_size),
decoded=False,
@@ -3539,7 +3544,7 @@ def delete_file(file):
file_shards_to_delete,
desc="Deleting unused files from dataset repository",
total=len(file_shards_to_delete),
- disable=bool(logging.get_verbosity() == logging.NOTSET) or not utils.is_progress_bar_enabled(),
+ disable=not utils.is_progress_bar_enabled(),
):
delete_file(file)
@@ -3548,7 +3553,7 @@ def delete_file(file):
enumerate(shards),
desc="Pushing dataset shards to the dataset hub",
total=num_shards,
- disable=bool(logging.get_verbosity() == logging.NOTSET),
+ disable=not utils.is_progress_bar_enabled(),
):
buffer = BytesIO()
shard.to_parquet(buffer)
diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -640,7 +640,7 @@ def finalize(self, metrics_query_result: dict):
def parquet_to_arrow(sources, destination):
"""Convert parquet files to arrow file. Inputs can be str paths or file-like objects"""
stream = None if isinstance(destination, str) else destination
- disable = bool(logging.get_verbosity() == logging.NOTSET)
+ disable = not utils.is_progress_bar_enabled()
with ArrowWriter(path=destination, stream=stream) as writer:
for source in utils.tqdm(sources, unit="sources", disable=disable):
pf = pa.parquet.ParquetFile(source)
diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -675,7 +675,7 @@ def _download_and_prepare(self, dl_manager, verify_infos, **prepare_split_kwargs
"._split_generator()."
)
- logger.info(f"Generating split {split_generator.split_info.name}")
+ logger.info(f"Generating {split_generator.split_info.name} split")
split_dict.add(split_generator.split_info)
try:
@@ -769,7 +769,7 @@ def as_dataset(
),
split,
map_tuple=True,
- disable_tqdm=False,
+ disable_tqdm=not utils.is_progress_bar_enabled(),
)
if isinstance(datasets, dict):
datasets = DatasetDict(datasets)
@@ -1055,7 +1055,10 @@ def _generate_examples(self, **kwargs):
raise NotImplementedError()
def _prepare_split(self, split_generator):
- split_info = split_generator.split_info
+ if self.info.splits is not None:
+ split_info = self.info.splits[split_generator.name]
+ else:
+ split_info = split_generator.split_info
fname = f"{self.name}-{split_generator.name}.arrow"
fpath = os.path.join(self._cache_dir, fname)
@@ -1075,7 +1078,8 @@ def _prepare_split(self, split_generator):
unit=" examples",
total=split_info.num_examples,
leave=False,
- disable=bool(logging.get_verbosity() == logging.NOTSET),
+ disable=not utils.is_progress_bar_enabled(),
+ desc=f"Generating {split_info.name} split",
):
example = self.info.features.encode_example(record)
writer.write(example, key)
@@ -1131,7 +1135,7 @@ def _prepare_split(self, split_generator):
generator = self._generate_tables(**split_generator.gen_kwargs)
with ArrowWriter(features=self.info.features, path=fpath) as writer:
for key, table in utils.tqdm(
- generator, unit=" tables", leave=False, disable=True # bool(logging.get_verbosity() == logging.NOTSET)
+ generator, unit=" tables", leave=False, disable=True # not utils.is_progress_bar_enabled()
):
writer.write_table(table)
num_examples, num_bytes = writer.finalize()
diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -13,7 +13,7 @@
from .utils import logging
from .utils.file_utils import hf_hub_url, is_remote_url, request_etag
from .utils.py_utils import string_to_dict
-from .utils.tqdm_utils import tqdm
+from .utils.tqdm_utils import is_progress_bar_enabled, tqdm
DEFAULT_SPLIT = str(Split.TRAIN)
@@ -497,7 +497,7 @@ def _get_origin_metadata_locally_or_by_urls(
max_workers=max_workers,
tqdm_class=tqdm,
desc="Resolving data files",
- disable=len(data_files) <= 16 or logging.get_verbosity() == logging.NOTSET,
+ disable=len(data_files) <= 16 or not is_progress_bar_enabled(),
)
diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -435,11 +435,13 @@ def with_transform(
def map(
self,
- function,
+ function: Optional[Callable] = None,
with_indices: bool = False,
+ with_rank: bool = False,
input_columns: Optional[Union[str, List[str]]] = None,
batched: bool = False,
batch_size: Optional[int] = 1000,
+ drop_last_batch: bool = False,
remove_columns: Optional[Union[str, List[str]]] = None,
keep_in_memory: bool = False,
load_from_cache_file: bool = True,
@@ -462,11 +464,15 @@ def map(
- `function(batch: Dict[List]) -> Union[Dict, Any]` if `batched=True` and `with_indices=False`
- `function(batch: Dict[List], indices: List[int]) -> Union[Dict, Any]` if `batched=True` and `with_indices=True`
with_indices (`bool`, defaults to `False`): Provide example indices to `function`. Note that in this case the signature of `function` should be `def function(example, idx): ...`.
+ with_rank (:obj:`bool`, default `False`): Provide process rank to `function`. Note that in this case the
+ signature of `function` should be `def function(example[, idx], rank): ...`.
input_columns (`Optional[Union[str, List[str]]]`, defaults to `None`): The columns to be passed into `function` as
positional arguments. If `None`, a dict mapping to all formatted columns is passed as one argument.
batched (`bool`, defaults to `False`): Provide batch of examples to `function`
batch_size (`Optional[int]`, defaults to `1000`): Number of examples per batch provided to `function` if `batched=True`
`batch_size <= 0` or `batch_size == None`: Provide the full dataset as a single batch to `function`
+ drop_last_batch (:obj:`bool`, default `False`): Whether a last batch smaller than the batch_size should be
+ dropped instead of being processed by the function.
remove_columns (`Optional[Union[str, List[str]]]`, defaults to `None`): Remove a selection of columns while doing the mapping.
Columns will be removed before updating the examples with the output of `function`, i.e. if `function` is adding
columns with names in `remove_columns`, these columns will be kept.
@@ -495,9 +501,11 @@ def map(
k: dataset.map(
function=function,
with_indices=with_indices,
+ with_rank=with_rank,
input_columns=input_columns,
batched=batched,
batch_size=batch_size,
+ drop_last_batch=drop_last_batch,
remove_columns=remove_columns,
keep_in_memory=keep_in_memory,
load_from_cache_file=load_from_cache_file,
diff --git a/src/datasets/io/csv.py b/src/datasets/io/csv.py
--- a/src/datasets/io/csv.py
+++ b/src/datasets/io/csv.py
@@ -5,7 +5,6 @@
from .. import Dataset, Features, NamedSplit, config, utils
from ..formatting import query_table
from ..packaged_modules.csv.csv import Csv
-from ..utils import logging
from ..utils.typing import NestedDataStructureLike, PathLike
from .abc import AbstractDatasetReader
@@ -108,22 +107,23 @@ def _write(self, file_obj: BinaryIO, header: bool = True, **to_csv_kwargs) -> in
for offset in utils.tqdm(
range(0, len(self.dataset), self.batch_size),
unit="ba",
- disable=bool(logging.get_verbosity() == logging.NOTSET),
+ disable=not utils.is_progress_bar_enabled(),
desc="Creating CSV from Arrow format",
):
csv_str = self._batch_csv((offset, header, to_csv_kwargs))
written += file_obj.write(csv_str)
else:
+ num_rows, batch_size = len(self.dataset), self.batch_size
with multiprocessing.Pool(self.num_proc) as pool:
for csv_str in utils.tqdm(
pool.imap(
self._batch_csv,
- [(offset, header, to_csv_kwargs) for offset in range(0, len(self.dataset), self.batch_size)],
+ [(offset, header, to_csv_kwargs) for offset in range(0, num_rows, batch_size)],
),
- total=(len(self.dataset) // self.batch_size) + 1,
+ total=(num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size,
unit="ba",
- disable=bool(logging.get_verbosity() == logging.NOTSET),
+ disable=not utils.is_progress_bar_enabled(),
desc="Creating CSV from Arrow format",
):
written += file_obj.write(csv_str)
diff --git a/src/datasets/io/json.py b/src/datasets/io/json.py
--- a/src/datasets/io/json.py
+++ b/src/datasets/io/json.py
@@ -5,7 +5,6 @@
from .. import Dataset, Features, NamedSplit, config, utils
from ..formatting import query_table
from ..packaged_modules.json.json import Json
-from ..utils import logging
from ..utils.typing import NestedDataStructureLike, PathLike
from .abc import AbstractDatasetReader
from .handler import IOHandler
@@ -124,24 +123,22 @@ def _write(
for offset in utils.tqdm(
range(0, len(self.dataset), self.batch_size),
unit="ba",
- disable=bool(logging.get_verbosity() == logging.NOTSET),
+ disable=not utils.is_progress_bar_enabled(),
desc="Creating json from Arrow format",
):
json_str = self._batch_json((offset, orient, lines, to_json_kwargs))
written += file_obj.write(json_str)
else:
+ num_rows, batch_size = len(self.dataset), self.batch_size
with multiprocessing.Pool(self.num_proc) as pool:
for json_str in utils.tqdm(
pool.imap(
self._batch_json,
- [
- (offset, orient, lines, to_json_kwargs)
- for offset in range(0, len(self.dataset), self.batch_size)
- ],
+ [(offset, orient, lines, to_json_kwargs) for offset in range(0, num_rows, batch_size)],
),
- total=(len(self.dataset) // self.batch_size) + 1,
+ total=(num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size,
unit="ba",
- disable=bool(logging.get_verbosity() == logging.NOTSET),
+ disable=not utils.is_progress_bar_enabled(),
desc="Creating json from Arrow format",
):
written += file_obj.write(json_str)
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -259,6 +259,9 @@ def _download_additional_modules(
"""
local_imports = []
library_imports = []
+ download_config = download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading extra modules"
for import_type, import_name, import_path, sub_directory in imports:
if import_type == "library":
library_imports.append((import_name, import_path)) # Import from a library
@@ -494,7 +497,7 @@ def __init__(
):
self.name = name
self.revision = revision
- self.download_config = download_config
+ self.download_config = download_config or DownloadConfig()
self.download_mode = download_mode
self.dynamic_modules_path = dynamic_modules_path
assert self.name.count("/") == 0
@@ -502,15 +505,21 @@ def __init__(
def download_loading_script(self, revision: Optional[str]) -> str:
file_path = hf_github_url(path=self.name, name=self.name + ".py", revision=revision)
- return cached_path(file_path, download_config=self.download_config)
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading builder script"
+ return cached_path(file_path, download_config=download_config)
def download_dataset_infos_file(self, revision: Optional[str]) -> str:
dataset_infos = hf_github_url(path=self.name, name=config.DATASETDICT_INFOS_FILENAME, revision=revision)
# Download the dataset infos file if available
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading metadata"
try:
return cached_path(
dataset_infos,
- download_config=self.download_config,
+ download_config=download_config,
)
except (FileNotFoundError, ConnectionError):
return None
@@ -569,7 +578,7 @@ def __init__(
):
self.name = name
self.revision = revision
- self.download_config = download_config
+ self.download_config = download_config or DownloadConfig()
self.download_mode = download_mode
self.dynamic_modules_path = dynamic_modules_path
assert self.name.count("/") == 0
@@ -577,7 +586,10 @@ def __init__(
def download_loading_script(self, revision: Optional[str]) -> str:
file_path = hf_github_url(path=self.name, name=self.name + ".py", revision=revision, dataset=False)
- return cached_path(file_path, download_config=self.download_config)
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading builder script"
+ return cached_path(file_path, download_config=download_config)
def get_module(self) -> MetricModule:
# get script and other files
@@ -630,7 +642,7 @@ def __init__(
):
self.path = path
self.name = Path(path).stem
- self.download_config = download_config
+ self.download_config = download_config or DownloadConfig()
self.download_mode = download_mode
self.dynamic_modules_path = dynamic_modules_path
@@ -671,7 +683,7 @@ def __init__(
):
self.path = path
self.name = Path(path).stem
- self.download_config = download_config
+ self.download_config = download_config or DownloadConfig()
self.download_mode = download_mode
self.dynamic_modules_path = dynamic_modules_path
@@ -836,6 +848,9 @@ def get_module(self) -> DatasetModule:
"name": self.name.replace("/", "--"),
"base_path": hf_hub_url(self.name, "", revision=self.revision),
}
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading metadata"
try:
dataset_infos_path = cached_path(
hf_hub_url(self.name, config.DATASETDICT_INFOS_FILENAME, revision=self.revision),
@@ -863,7 +878,7 @@ def __init__(
):
self.name = name
self.revision = revision
- self.download_config = download_config
+ self.download_config = download_config or DownloadConfig()
self.download_mode = download_mode
self.dynamic_modules_path = dynamic_modules_path
assert self.name.count("/") == 1
@@ -871,15 +886,21 @@ def __init__(
def download_loading_script(self) -> str:
file_path = hf_hub_url(path=self.name, name=self.name.split("/")[1] + ".py", revision=self.revision)
- return cached_path(file_path, download_config=self.download_config)
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading builder script"
+ return cached_path(file_path, download_config=download_config)
def download_dataset_infos_file(self) -> str:
dataset_infos = hf_hub_url(path=self.name, name=config.DATASETDICT_INFOS_FILENAME, revision=self.revision)
# Download the dataset infos file if available
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading metadata"
try:
return cached_path(
dataset_infos,
- download_config=self.download_config,
+ download_config=download_config,
)
except (FileNotFoundError, ConnectionError):
return None
diff --git a/src/datasets/search.py b/src/datasets/search.py
--- a/src/datasets/search.py
+++ b/src/datasets/search.py
@@ -150,9 +150,7 @@ def add_documents(self, documents: Union[List[str], "Dataset"], column: Optional
index_config = self.es_index_config
self.es_client.indices.create(index=index_name, body=index_config)
number_of_docs = len(documents)
- progress = utils.tqdm(
- unit="docs", total=number_of_docs, disable=bool(logging.get_verbosity() == logging.NOTSET)
- )
+ progress = utils.tqdm(unit="docs", total=number_of_docs, disable=not utils.is_progress_bar_enabled())
successes = 0
def passage_generator():
@@ -297,9 +295,7 @@ def add_vectors(
# Add vectors
logger.info(f"Adding {len(vectors)} vectors to the faiss index")
- for i in utils.tqdm(
- range(0, len(vectors), batch_size), disable=bool(logging.get_verbosity() == logging.NOTSET)
- ):
+ for i in utils.tqdm(range(0, len(vectors), batch_size), disable=not utils.is_progress_bar_enabled()):
vecs = vectors[i : i + batch_size] if column is None else vectors[i : i + batch_size][column]
self.faiss_index.add(vecs)
diff --git a/src/datasets/utils/download_manager.py b/src/datasets/utils/download_manager.py
--- a/src/datasets/utils/download_manager.py
+++ b/src/datasets/utils/download_manager.py
@@ -23,7 +23,7 @@
from functools import partial
from typing import Dict, Optional, Union
-from .. import config
+from .. import config, utils
from .file_utils import (
DownloadConfig,
cached_path,
@@ -122,7 +122,9 @@ def upload(local_file_path):
return remote_file_path
uploaded_path_or_paths = map_nested(
- lambda local_file_path: upload(local_file_path), downloaded_path_or_paths, disable_tqdm=False
+ lambda local_file_path: upload(local_file_path),
+ downloaded_path_or_paths,
+ disable_tqdm=not utils.is_progress_bar_enabled(),
)
return uploaded_path_or_paths
@@ -152,7 +154,9 @@ def download_custom(self, url_or_urls, custom_download):
def url_to_downloaded_path(url):
return os.path.join(cache_dir, hash_url_to_filename(url))
- downloaded_path_or_paths = map_nested(url_to_downloaded_path, url_or_urls, disable_tqdm=False)
+ downloaded_path_or_paths = map_nested(
+ url_to_downloaded_path, url_or_urls, disable_tqdm=not utils.is_progress_bar_enabled()
+ )
url_or_urls = NestedDataStructure(url_or_urls)
downloaded_path_or_paths = NestedDataStructure(downloaded_path_or_paths)
for url, path in zip(url_or_urls.flatten(), downloaded_path_or_paths.flatten()):
@@ -188,12 +192,19 @@ def download(self, url_or_urls):
# Note that if we have less than 16 files, multi-processing is not activated
if download_config.num_proc is None:
download_config.num_proc = 16
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading data"
download_func = partial(self._download, download_config=download_config)
start_time = datetime.now()
downloaded_path_or_paths = map_nested(
- download_func, url_or_urls, map_tuple=True, num_proc=download_config.num_proc, disable_tqdm=False
+ download_func,
+ url_or_urls,
+ map_tuple=True,
+ num_proc=download_config.num_proc,
+ disable_tqdm=not utils.is_progress_bar_enabled(),
+ desc="Downloading data files",
)
duration = datetime.now() - start_time
logger.info(f"Downloading took {duration.total_seconds() // 60} min")
@@ -280,8 +291,15 @@ def extract(self, path_or_paths, num_proc=None):
"""
download_config = self.download_config.copy()
download_config.extract_compressed_file = True
+ # Extract downloads the file first if it is not already downloaded
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading data"
extracted_paths = map_nested(
- partial(cached_path, download_config=download_config), path_or_paths, num_proc=num_proc, disable_tqdm=False
+ partial(cached_path, download_config=download_config),
+ path_or_paths,
+ num_proc=num_proc,
+ disable_tqdm=not utils.is_progress_bar_enabled(),
+ desc="Extracting data files",
)
path_or_paths = NestedDataStructure(path_or_paths)
extracted_paths = NestedDataStructure(extracted_paths)
diff --git a/src/datasets/utils/file_utils.py b/src/datasets/utils/file_utils.py
--- a/src/datasets/utils/file_utils.py
+++ b/src/datasets/utils/file_utils.py
@@ -241,6 +241,7 @@ class DownloadConfig:
for remote files on the Datasets Hub. If True, will get token from ~/.huggingface.
ignore_url_params (:obj:`bool`, default ``False``): Whether to strip all query parameters and #fragments from
the download URL before using it for caching the file.
+ download_desc (:obj:`str`, optional): A description to be displayed alongside with the progress bar while downloading the files.
"""
cache_dir: Optional[Union[str, Path]] = None
@@ -257,6 +258,7 @@ class DownloadConfig:
max_retries: int = 1
use_auth_token: Optional[Union[str, bool]] = None
ignore_url_params: bool = False
+ download_desc: Optional[str] = None
def copy(self) -> "DownloadConfig":
return self.__class__(**{k: copy.deepcopy(v) for k, v in self.__dict__.items()})
@@ -307,6 +309,7 @@ def cached_path(
max_retries=download_config.max_retries,
use_auth_token=download_config.use_auth_token,
ignore_url_params=download_config.ignore_url_params,
+ download_desc=download_config.download_desc,
)
elif os.path.exists(url_or_filename):
# File, and it exists.
@@ -434,7 +437,9 @@ def ftp_get(url, temp_file, timeout=10.0):
raise ConnectionError(e) from None
-def http_get(url, temp_file, proxies=None, resume_size=0, headers=None, cookies=None, timeout=100.0, max_retries=0):
+def http_get(
+ url, temp_file, proxies=None, resume_size=0, headers=None, cookies=None, timeout=100.0, max_retries=0, desc=None
+):
headers = copy.deepcopy(headers) or {}
headers["user-agent"] = get_datasets_user_agent(user_agent=headers.get("user-agent"))
if resume_size > 0:
@@ -453,19 +458,17 @@ def http_get(url, temp_file, proxies=None, resume_size=0, headers=None, cookies=
return
content_length = response.headers.get("Content-Length")
total = resume_size + int(content_length) if content_length is not None else None
- progress = utils.tqdm(
+ with utils.tqdm(
unit="B",
unit_scale=True,
total=total,
initial=resume_size,
- desc="Downloading",
- disable=bool(logging.get_verbosity() == logging.NOTSET),
- )
- for chunk in response.iter_content(chunk_size=1024):
- if chunk: # filter out keep-alive new chunks
+ desc=desc or "Downloading",
+ disable=not utils.is_progress_bar_enabled(),
+ ) as progress:
+ for chunk in response.iter_content(chunk_size=1024):
progress.update(len(chunk))
temp_file.write(chunk)
- progress.close()
def http_head(
@@ -507,6 +510,7 @@ def get_from_cache(
max_retries=0,
use_auth_token=None,
ignore_url_params=False,
+ download_desc=None,
) -> str:
"""
Given a URL, look for the corresponding file in the local cache.
@@ -664,6 +668,7 @@ def _resumable_file_manager():
headers=headers,
cookies=cookies,
max_retries=max_retries,
+ desc=download_desc,
)
logger.info(f"storing {url} in cache at {cache_path}")
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -189,7 +189,7 @@ def __get__(self, obj, objtype=None):
def _single_map_nested(args):
"""Apply a function recursively to each element of a nested data struct."""
- function, data_struct, types, rank, disable_tqdm = args
+ function, data_struct, types, rank, disable_tqdm, desc = args
# Singleton first to spare some computation
if not isinstance(data_struct, dict) and not isinstance(data_struct, types):
@@ -200,18 +200,18 @@ def _single_map_nested(args):
logging.set_verbosity_warning()
# Print at least one thing to fix tqdm in notebooks in multiprocessing
# see https://github.com/tqdm/tqdm/issues/485#issuecomment-473338308
- if rank is not None and not disable_tqdm and "notebook" in tqdm.__name__:
+ if rank is not None and not disable_tqdm and any("notebook" in tqdm_cls.__name__ for tqdm_cls in tqdm.__mro__):
print(" ", end="", flush=True)
# Loop over single examples or batches and write to buffer/file if examples are to be updated
pbar_iterable = data_struct.items() if isinstance(data_struct, dict) else data_struct
- pbar_desc = "#" + str(rank) if rank is not None else None
+ pbar_desc = (desc + " " if desc is not None else "") + "#" + str(rank) if rank is not None else desc
pbar = utils.tqdm(pbar_iterable, disable=disable_tqdm, position=rank, unit="obj", desc=pbar_desc)
if isinstance(data_struct, dict):
- return {k: _single_map_nested((function, v, types, None, True)) for k, v in pbar}
+ return {k: _single_map_nested((function, v, types, None, True, None)) for k, v in pbar}
else:
- mapped = [_single_map_nested((function, v, types, None, True)) for v in pbar]
+ mapped = [_single_map_nested((function, v, types, None, True, None)) for v in pbar]
if isinstance(data_struct, list):
return mapped
elif isinstance(data_struct, tuple):
@@ -230,6 +230,7 @@ def map_nested(
num_proc: Optional[int] = None,
types=None,
disable_tqdm: bool = True,
+ desc: Optional[str] = None,
):
"""Apply a function recursively to each element of a nested data struct.
If num_proc > 1 and the length of data_struct is longer than num_proc: use multi-processing
@@ -249,17 +250,15 @@ def map_nested(
if not isinstance(data_struct, dict) and not isinstance(data_struct, types):
return function(data_struct)
- disable_tqdm = (
- disable_tqdm or bool(logging.get_verbosity() == logging.NOTSET) or not utils.is_progress_bar_enabled()
- )
+ disable_tqdm = disable_tqdm or not utils.is_progress_bar_enabled()
iterable = list(data_struct.values()) if isinstance(data_struct, dict) else data_struct
if num_proc is None:
num_proc = 1
if num_proc <= 1 or len(iterable) <= num_proc:
mapped = [
- _single_map_nested((function, obj, types, None, True))
- for obj in utils.tqdm(iterable, disable=disable_tqdm)
+ _single_map_nested((function, obj, types, None, True, None))
+ for obj in utils.tqdm(iterable, disable=disable_tqdm, desc=desc)
]
else:
split_kwds = [] # We organize the splits ourselve (contiguous splits)
@@ -268,7 +267,7 @@ def map_nested(
mod = len(iterable) % num_proc
start = div * index + min(index, mod)
end = start + div + (1 if index < mod else 0)
- split_kwds.append((function, iterable[start:end], types, index, disable_tqdm))
+ split_kwds.append((function, iterable[start:end], types, index, disable_tqdm, desc))
if len(iterable) != sum(len(i[1]) for i in split_kwds):
raise ValueError(
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -1011,6 +1011,21 @@ def map_batched(example):
)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_batched)
+ # change batch size and drop the last batch
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
+ batch_size = 4
+ with dset.map(
+ map_batched, batched=True, batch_size=batch_size, drop_last_batch=True
+ ) as dset_test_batched:
+ self.assertEqual(len(dset_test_batched), 30 // batch_size * batch_size)
+ self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
+ self.assertDictEqual(
+ dset_test_batched.features,
+ Features({"filename": Value("string"), "filename_new": Value("string")}),
+ )
+ assert_arrow_metadata_are_synced_with_dataset_features(dset_test_batched)
+
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.formatted_as("numpy", columns=["filename"]):
| Progress bars are not properly rendered in Jupyter notebook
## Describe the bug
The progress bars are not Jupyter widgets; regular progress bars appear (like in a terminal).
## Steps to reproduce the bug
```python
ds.map(tokenize, num_proc=10)
```
## Expected results
Jupyter widgets displaying the progress bars.
## Actual results
Simple plane progress bars.
cc: Reported by @thomwolf
| To add my experience when trying to debug this issue:
Seems like previously the workaround given [here](https://github.com/tqdm/tqdm/issues/485#issuecomment-473338308) worked around this issue. But with the latest version of jupyter/tqdm I still get terminal warnings that IPython tried to send a message from a forked process.
Hi @mludv, thanks for the hint!!! :)
We will definitely take it into account to try to fix this issue... It seems somehow related to `multiprocessing` and `tqdm`... | 2022-01-31T17:22:43Z | [] | [] |
huggingface/datasets | 3,695 | huggingface__datasets-3695 | [
"3631"
] | 36db39c75179a0a491c69a4491f7ae7e4615e66f | diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -19,7 +19,7 @@
import re
import sys
from collections.abc import Iterable
-from dataclasses import _asdict_inner, dataclass, field, fields
+from dataclasses import InitVar, _asdict_inner, dataclass, field, fields
from functools import reduce
from operator import mul
from typing import Any, ClassVar, Dict, List, Optional
@@ -761,7 +761,7 @@ class ClassLabel:
num_classes: int = None
names: List[str] = None
- names_file: Optional[str] = None
+ names_file: InitVar[Optional[str]] = None # Pseudo-field: ignored by asdict and fields when converting to/from dict
id: Optional[str] = None
# Automatically constructed
dtype: ClassVar[str] = "int64"
@@ -770,7 +770,8 @@ class ClassLabel:
_int2str: ClassVar[Dict[int, int]] = None
_type: str = field(default="ClassLabel", init=False, repr=False)
- def __post_init__(self):
+ def __post_init__(self, names_file):
+ self.names_file = names_file
if self.names_file is not None and self.names is not None:
raise ValueError("Please provide either names or names_file but not both.")
# Set self.names
| diff --git a/tests/features/test_features.py b/tests/features/test_features.py
--- a/tests/features/test_features.py
+++ b/tests/features/test_features.py
@@ -17,6 +17,7 @@
_cast_to_python_objects,
cast_to_python_objects,
encode_nested_example,
+ generate_from_dict,
string_to_arrow,
)
from datasets.info import DatasetInfo
@@ -269,6 +270,20 @@ def test_classlabel_int2str():
classlabel.int2str(len(names))
[email protected]("class_label_arg", ["names", "names_file"])
+def test_class_label_to_and_from_dict(class_label_arg, tmp_path_factory):
+ names = ["negative", "positive"]
+ names_file = str(tmp_path_factory.mktemp("features") / "labels.txt")
+ with open(names_file, "w", encoding="utf-8") as f:
+ f.write("\n".join(names))
+ if class_label_arg == "names":
+ class_label = ClassLabel(names=names)
+ elif class_label_arg == "names_file":
+ class_label = ClassLabel(names_file=names_file)
+ generated_class_label = generate_from_dict(asdict(class_label))
+ assert generated_class_label == class_label
+
+
def test_encode_nested_example_sequence_with_none():
schema = Sequence(Value("int32"))
obj = None
| Labels conflict when loading a local CSV file.
## Describe the bug
I am trying to load a local CSV file with a separate file containing label names. It is successfully loaded for the first time, but when I try to load it again, there is a conflict between provided labels and the cached dataset info. Disabling caching globally and/or using `download_mode="force_redownload"` did not help.
## Steps to reproduce the bug
```python
load_dataset('csv', data_files='data/my_data.csv',
features=Features(text=Value(dtype='string'),
label=ClassLabel(names_file='data/my_data_labels.txt')))
```
`my_data.csv` file has the following structure:
```
text,label
"example1",0
"example2",1
...
```
and the `my_data_labels.txt` looks like this:
```
label1
label2
...
```
## Expected results
Successfully loaded dataset.
## Actual results
```python
File "/usr/local/lib/python3.8/site-packages/datasets/load.py", line 1706, in load_dataset
ds = builder_instance.as_dataset(split=split, ignore_verifications=ignore_verifications, in_memory=keep_in_memory)
File "/usr/local/lib/python3.8/site-packages/datasets/builder.py", line 766, in as_dataset
datasets = utils.map_nested(
File "/usr/local/lib/python3.8/site-packages/datasets/utils/py_utils.py", line 261, in map_nested
mapped = [
File "/usr/local/lib/python3.8/site-packages/datasets/utils/py_utils.py", line 262, in <listcomp>
_single_map_nested((function, obj, types, None, True))
File "/usr/local/lib/python3.8/site-packages/datasets/utils/py_utils.py", line 197, in _single_map_nested
return function(data_struct)
File "/usr/local/lib/python3.8/site-packages/datasets/builder.py", line 797, in _build_single_dataset
ds = self._as_dataset(
File "/usr/local/lib/python3.8/site-packages/datasets/builder.py", line 872, in _as_dataset
return Dataset(fingerprint=fingerprint, **dataset_kwargs)
File "/usr/local/lib/python3.8/site-packages/datasets/arrow_dataset.py", line 638, in __init__
inferred_features = Features.from_arrow_schema(arrow_table.schema)
File "/usr/local/lib/python3.8/site-packages/datasets/features/features.py", line 1242, in from_arrow_schema
return Features.from_dict(metadata["info"]["features"])
File "/usr/local/lib/python3.8/site-packages/datasets/features/features.py", line 1271, in from_dict
obj = generate_from_dict(dic)
File "/usr/local/lib/python3.8/site-packages/datasets/features/features.py", line 1076, in generate_from_dict
return {key: generate_from_dict(value) for key, value in obj.items()}
File "/usr/local/lib/python3.8/site-packages/datasets/features/features.py", line 1076, in <dictcomp>
return {key: generate_from_dict(value) for key, value in obj.items()}
File "/usr/local/lib/python3.8/site-packages/datasets/features/features.py", line 1083, in generate_from_dict
return class_type(**{k: v for k, v in obj.items() if k in field_names})
File "<string>", line 7, in __init__
File "/usr/local/lib/python3.8/site-packages/datasets/features/features.py", line 776, in __post_init__
raise ValueError("Please provide either names or names_file but not both.")
ValueError: Please provide either names or names_file but not both.
```
## Environment info
- `datasets` version: 1.18.0
- Python version: 3.8.2
| 2022-02-10T09:47:10Z | [] | [] |
|
huggingface/datasets | 3,719 | huggingface__datasets-3719 | [
"3707"
] | 52fe45b7f8d69422f2e77d68d71ce836dc678a72 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -567,6 +567,13 @@ def _check_column_names(column_names: List[str]):
raise ValueError(f"The table can't have duplicated columns but columns {duplicated_columns} are duplicated.")
+def _check_valid_indices_value(value, size):
+ if (value < 0 and value + size < 0) or (value >= size):
+ raise IndexError(
+ f"Invalid value {value} in indices iterable. All values must be within range [-{size}, {size - 1}]."
+ )
+
+
def _check_if_features_can_be_aligned(features_list: List[Features]):
"""Check if the dictionaries of features can be aligned.
@@ -2748,6 +2755,12 @@ def select(
path=tmp_file.name, writer_batch_size=writer_batch_size, fingerprint=new_fingerprint, unit="indices"
)
+ indices = list(indices)
+
+ size = len(self)
+ _check_valid_indices_value(int(max(indices)), size=size)
+ _check_valid_indices_value(int(min(indices)), size=size)
+
indices_array = pa.array(indices, type=pa.uint64())
# Check if we need to convert indices
if self._indices is not None:
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -1492,7 +1492,21 @@ def test_select(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
bad_indices = list(range(5))
- bad_indices[3] = "foo"
+ bad_indices[-1] = len(dset) + 10 # out of bounds
+ tmp_file = os.path.join(tmp_dir, "test.arrow")
+ self.assertRaises(
+ Exception,
+ dset.select,
+ indices=bad_indices,
+ indices_cache_file_name=tmp_file,
+ writer_batch_size=2,
+ )
+ self.assertFalse(os.path.exists(tmp_file))
+
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
+ bad_indices = list(range(5))
+ bad_indices[3] = "foo" # wrong type
tmp_file = os.path.join(tmp_dir, "test.arrow")
self.assertRaises(
Exception,
| `.select`: unexpected behavior with `indices`
## Describe the bug
The `.select` method will not throw when sending `indices` bigger than the dataset length; `indices` will be wrapped instead. This behavior is not documented anywhere, and is not intuitive.
## Steps to reproduce the bug
```python
from datasets import Dataset
ds = Dataset.from_dict({"text": ["d", "e", "f"], "label": [4, 5, 6]})
res1 = ds.select([1, 2, 3])['text']
res2 = ds.select([1000])['text']
```
## Expected results
Both results should throw an `Error`.
## Actual results
`res1` will give `['e', 'f', 'd']`
`res2` will give `['e']`
## Environment info
Bug found from this environment:
- `datasets` version: 1.16.1
- Platform: macOS-10.16-x86_64-i386-64bit
- Python version: 3.8.7
- PyArrow version: 6.0.1
It was also replicated on `master`.
| Hi! Currently, we compute the final index as `index % len(dset)`. I agree this behavior is somewhat unexpected and that it would be more appropriate to raise an error instead (this is what `df.iloc` in Pandas does, for instance).
@albertvillanova @lhoestq wdyt?
I agree. I think `index % len(dset)` was used to support negative indices.
I think this needs to be fixed in `datasets.formatting.formatting._check_valid_index_key` if I'm not mistaken | 2022-02-14T12:31:41Z | [] | [] |
huggingface/datasets | 3,723 | huggingface__datasets-3723 | [
"3686"
] | 87cb7ee8cddff35539e24f4ea8434da4fad5c17d | diff --git a/src/datasets/features/audio.py b/src/datasets/features/audio.py
--- a/src/datasets/features/audio.py
+++ b/src/datasets/features/audio.py
@@ -1,6 +1,6 @@
from dataclasses import dataclass, field
from io import BytesIO
-from typing import Any, ClassVar, Optional, Union
+from typing import TYPE_CHECKING, Any, ClassVar, Dict, Optional, Union
import pyarrow as pa
from packaging import version
@@ -10,6 +10,10 @@
from ..utils.streaming_download_manager import xopen
+if TYPE_CHECKING:
+ from .features import FeatureType
+
+
@dataclass
class Audio:
"""Audio Feature to extract audio data from an audio file.
@@ -109,6 +113,17 @@ def decode_example(self, value: dict) -> dict:
array, sampling_rate = self._decode_non_mp3_path_like(path)
return {"path": path, "array": array, "sampling_rate": sampling_rate}
+ def flatten(self) -> Union["FeatureType", Dict[str, "FeatureType"]]:
+ """If in the decodable state, raise an error, otherwise flatten the feature into a dictionary."""
+ from .features import Value
+
+ if self.decode:
+ raise ValueError("Cannot flatten a decoded Audio feature.")
+ return {
+ "bytes": Value("binary"),
+ "path": Value("string"),
+ }
+
def cast_storage(self, storage: Union[pa.StringArray, pa.StructArray]) -> pa.StructArray:
"""Cast an Arrow array to the Audio arrow storage type.
The Arrow types that can be converted to the Audio pyarrow storage type are:
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -1207,6 +1207,20 @@ def __setitem__(self, column_name: str, feature: FeatureType):
super().__setitem__(column_name, feature)
self._column_requires_decoding[column_name] = require_decoding(feature)
+ def __delitem__(self, column_name: str):
+ super().__delitem__(column_name)
+ del self._column_requires_decoding[column_name]
+
+ def update(self, iterable, **kwds):
+ if hasattr(iterable, "keys"):
+ for key in iterable.keys():
+ self[key] = iterable[key]
+ else:
+ for key, value in iterable:
+ self[key] = value
+ for key in kwds:
+ self[key] = kwds[key]
+
def __reduce__(self):
return Features, (dict(self),)
@@ -1459,6 +1473,10 @@ def flatten(self, max_depth=16) -> "Features":
no_change = False
flattened.update({f"{column_name}.{k}": Sequence(v) for k, v in subfeature.feature.items()})
del flattened[column_name]
+ elif hasattr(subfeature, "flatten") and subfeature.flatten() != subfeature:
+ no_change = False
+ flattened.update({f"{column_name}.{k}": v for k, v in subfeature.flatten().items()})
+ del flattened[column_name]
self = flattened
if no_change:
break
diff --git a/src/datasets/features/image.py b/src/datasets/features/image.py
--- a/src/datasets/features/image.py
+++ b/src/datasets/features/image.py
@@ -1,6 +1,6 @@
from dataclasses import dataclass, field
from io import BytesIO
-from typing import TYPE_CHECKING, Any, ClassVar, List, Optional, Union
+from typing import TYPE_CHECKING, Any, ClassVar, Dict, List, Optional, Union
import numpy as np
import pyarrow as pa
@@ -15,6 +15,8 @@
if TYPE_CHECKING:
import PIL.Image
+ from .features import FeatureType
+
_IMAGE_COMPRESSION_FORMATS: Optional[List[str]] = None
@@ -114,6 +116,19 @@ def decode_example(self, value: dict) -> "PIL.Image.Image":
image = PIL.Image.open(BytesIO(bytes_))
return image
+ def flatten(self) -> Union["FeatureType", Dict[str, "FeatureType"]]:
+ """If in the decodable state, return the feature itself, otherwise flatten the feature into a dictionary."""
+ from .features import Value
+
+ return (
+ self
+ if self.decode
+ else {
+ "bytes": Value("binary"),
+ "path": Value("string"),
+ }
+ )
+
def cast_storage(self, storage: Union[pa.StringArray, pa.StructArray, pa.ListArray]) -> pa.StructArray:
"""Cast an Arrow array to the Image arrow storage type.
The Arrow types that can be converted to the Image pyarrow storage type are:
diff --git a/src/datasets/features/translation.py b/src/datasets/features/translation.py
--- a/src/datasets/features/translation.py
+++ b/src/datasets/features/translation.py
@@ -1,9 +1,13 @@
from dataclasses import dataclass, field
-from typing import Any, ClassVar, List, Optional
+from typing import TYPE_CHECKING, Any, ClassVar, Dict, List, Optional, Union
import pyarrow as pa
+if TYPE_CHECKING:
+ from .features import FeatureType
+
+
@dataclass
class Translation:
"""`FeatureConnector` for translations with fixed languages per example.
@@ -40,6 +44,12 @@ class Translation:
def __call__(self):
return pa.struct({lang: pa.string() for lang in sorted(self.languages)})
+ def flatten(self) -> Union["FeatureType", Dict[str, "FeatureType"]]:
+ """Flatten the Translation feature into a dictionary."""
+ from .features import Value
+
+ return {k: Value("string") for k in sorted(self.languages)}
+
@dataclass
class TranslationVariableLanguages:
@@ -113,3 +123,12 @@ def encode_example(self, translation_dict):
languages, translations = zip(*sorted(translation_tuples))
return {"language": languages, "translation": translations}
+
+ def flatten(self) -> Union["FeatureType", Dict[str, "FeatureType"]]:
+ """Flatten the TranslationVariableLanguages feature into a dictionary."""
+ from .features import Sequence, Value
+
+ return {
+ "language": Sequence(Value("string")),
+ "translation": Sequence(Value("string")),
+ }
diff --git a/src/datasets/table.py b/src/datasets/table.py
--- a/src/datasets/table.py
+++ b/src/datasets/table.py
@@ -799,7 +799,7 @@ def flatten(self, *args, **kwargs):
Returns:
:class:`datasets.table.Table`:
"""
- return InMemoryTable(self.table.flatten(*args, **kwargs))
+ return InMemoryTable(table_flatten(self.table, *args, **kwargs))
def combine_chunks(self, *args, **kwargs):
"""
@@ -993,6 +993,8 @@ def _apply_replays(table: pa.Table, replays: Optional[List[Replay]] = None) -> p
for name, args, kwargs in replays:
if name == "cast":
table = table_cast(table, *args, **kwargs)
+ elif name == "flatten":
+ table = table_flatten(table, *args, **kwargs)
else:
table = getattr(table, name)(*args, **kwargs)
return table
@@ -1043,7 +1045,7 @@ def flatten(self, *args, **kwargs):
"""
replay = ("flatten", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
- return MemoryMappedTable(self.table.flatten(*args, **kwargs), self.path, replays)
+ return MemoryMappedTable(table_flatten(self.table, *args, **kwargs), self.path, replays)
def combine_chunks(self, *args, **kwargs):
"""
@@ -1432,7 +1434,7 @@ def flatten(self, *args, **kwargs):
Returns:
:class:`datasets.table.Table`:
"""
- table = self.table.flatten(*args, **kwargs)
+ table = table_flatten(self.table, *args, **kwargs)
blocks = []
for tables in self.blocks:
blocks.append([t.flatten(*args, **kwargs) for t in tables])
@@ -1843,7 +1845,7 @@ def cast_table_to_schema(table: pa.Table, schema: pa.Schema):
def table_cast(table: pa.Table, schema: pa.Schema):
- """Improved version of pa.Table.cast
+ """Improved version of pa.Table.cast.
It supports casting to feature types stored in the schema metadata.
@@ -1862,6 +1864,47 @@ def table_cast(table: pa.Table, schema: pa.Schema):
return table
+def table_flatten(table: pa.Table):
+ """Improved version of pa.Table.flatten.
+
+ It behaves as pa.Table.flatten in a sense it does 1-step flatten of the columns with a struct type into one column per struct field,
+ but updates the metadata and skips decodable features unless the `decode` attribute of these features is set to False.
+
+ Args:
+ table (Table): PyArrow table to flatten
+
+ Returns:
+ Table: the flattened table
+ """
+ from .features import Features
+
+ features = Features.from_arrow_schema(table.schema)
+ if any(hasattr(subfeature, "flatten") and subfeature.flatten() == subfeature for subfeature in features.values()):
+ flat_arrays = []
+ flat_column_names = []
+ for field in table.schema:
+ array = table.column(field.name)
+ subfeature = features[field.name]
+ if pa.types.is_struct(field.type) and (
+ not hasattr(subfeature, "flatten") or subfeature.flatten() != subfeature
+ ):
+ flat_arrays.extend(array.flatten())
+ flat_column_names.extend([f"{field.name}.{subfield.name}" for subfield in field.type])
+ else:
+ flat_arrays.append(array)
+ flat_column_names.append(field.name)
+ flat_table = pa.Table.from_arrays(
+ flat_arrays,
+ names=flat_column_names,
+ )
+ else:
+ flat_table = table.flatten()
+ # Preserve complex types in the metadata
+ flat_features = features.flatten(max_depth=2)
+ flat_features = Features({column_name: flat_features[column_name] for column_name in flat_table.column_names})
+ return flat_table.replace_schema_metadata(flat_features.arrow_schema.metadata)
+
+
def table_visitor(table: pa.Table, function: Callable[[pa.Array], None]):
"""Visit all arrays in a table and apply a function to them.
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -20,7 +20,17 @@
from datasets import concatenate_datasets, interleave_datasets, load_from_disk
from datasets.arrow_dataset import Dataset, transmit_format, update_metadata_with_features
from datasets.dataset_dict import DatasetDict
-from datasets.features import Array2D, Array3D, ClassLabel, Features, Sequence, Value
+from datasets.features import (
+ Array2D,
+ Array3D,
+ ClassLabel,
+ Features,
+ Image,
+ Sequence,
+ Translation,
+ TranslationVariableLanguages,
+ Value,
+)
from datasets.filesystems import extract_path_from_uri
from datasets.info import DatasetInfo
from datasets.splits import NamedSplit
@@ -40,6 +50,7 @@
assert_arrow_memory_doesnt_increase,
assert_arrow_memory_increases,
require_jax,
+ require_pil,
require_s3,
require_tf,
require_torch,
@@ -737,6 +748,118 @@ def test_flatten(self, in_memory):
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with Dataset.from_dict(
+ {"a": [{"en": "Thank you", "fr": "Merci"}] * 10, "foo": [1] * 10},
+ features=Features({"a": Translation(languages=["en", "fr"]), "foo": Value("int64")}),
+ ) as dset:
+ with self._to(in_memory, tmp_dir, dset) as dset:
+ fingerprint = dset._fingerprint
+ with dset.flatten() as dset:
+ self.assertListEqual(sorted(dset.column_names), ["a.en", "a.fr", "foo"])
+ self.assertListEqual(sorted(dset.features.keys()), ["a.en", "a.fr", "foo"])
+ self.assertDictEqual(
+ dset.features,
+ Features({"a.en": Value("string"), "a.fr": Value("string"), "foo": Value("int64")}),
+ )
+ self.assertNotEqual(dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(dset)
+
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with Dataset.from_dict(
+ {"a": [{"en": "the cat", "fr": ["le chat", "la chatte"], "de": "die katze"}] * 10, "foo": [1] * 10},
+ features=Features(
+ {"a": TranslationVariableLanguages(languages=["en", "fr", "de"]), "foo": Value("int64")}
+ ),
+ ) as dset:
+ with self._to(in_memory, tmp_dir, dset) as dset:
+ fingerprint = dset._fingerprint
+ with dset.flatten() as dset:
+ self.assertListEqual(sorted(dset.column_names), ["a.language", "a.translation", "foo"])
+ self.assertListEqual(sorted(dset.features.keys()), ["a.language", "a.translation", "foo"])
+ self.assertDictEqual(
+ dset.features,
+ Features(
+ {
+ "a.language": Sequence(Value("string")),
+ "a.translation": Sequence(Value("string")),
+ "foo": Value("int64"),
+ }
+ ),
+ )
+ self.assertNotEqual(dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(dset)
+
+ @require_pil
+ def test_flatten_complex_image(self, in_memory):
+ # decoding turned on
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with Dataset.from_dict(
+ {"a": [np.arange(4 * 4 * 3).reshape(4, 4, 3)] * 10, "foo": [1] * 10},
+ features=Features({"a": Image(), "foo": Value("int64")}),
+ ) as dset:
+ with self._to(in_memory, tmp_dir, dset) as dset:
+ fingerprint = dset._fingerprint
+ with dset.flatten() as dset:
+ self.assertListEqual(sorted(dset.column_names), ["a", "foo"])
+ self.assertListEqual(sorted(dset.features.keys()), ["a", "foo"])
+ self.assertDictEqual(dset.features, Features({"a": Image(), "foo": Value("int64")}))
+ self.assertNotEqual(dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(dset)
+
+ # decoding turned on + nesting
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with Dataset.from_dict(
+ {"a": [{"b": np.arange(4 * 4 * 3).reshape(4, 4, 3)}] * 10, "foo": [1] * 10},
+ features=Features({"a": {"b": Image()}, "foo": Value("int64")}),
+ ) as dset:
+ with self._to(in_memory, tmp_dir, dset) as dset:
+ fingerprint = dset._fingerprint
+ with dset.flatten() as dset:
+ self.assertListEqual(sorted(dset.column_names), ["a.b", "foo"])
+ self.assertListEqual(sorted(dset.features.keys()), ["a.b", "foo"])
+ self.assertDictEqual(dset.features, Features({"a.b": Image(), "foo": Value("int64")}))
+ self.assertNotEqual(dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(dset)
+
+ # decoding turned off
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with Dataset.from_dict(
+ {"a": [np.arange(4 * 4 * 3).reshape(4, 4, 3)] * 10, "foo": [1] * 10},
+ features=Features({"a": Image(decode=False), "foo": Value("int64")}),
+ ) as dset:
+ with self._to(in_memory, tmp_dir, dset) as dset:
+ fingerprint = dset._fingerprint
+ with dset.flatten() as dset:
+ self.assertListEqual(sorted(dset.column_names), ["a.bytes", "a.path", "foo"])
+ self.assertListEqual(sorted(dset.features.keys()), ["a.bytes", "a.path", "foo"])
+ self.assertDictEqual(
+ dset.features,
+ Features({"a.bytes": Value("binary"), "a.path": Value("string"), "foo": Value("int64")}),
+ )
+ self.assertNotEqual(dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(dset)
+
+ # decoding turned off + nesting
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with Dataset.from_dict(
+ {"a": [{"b": np.arange(4 * 4 * 3).reshape(4, 4, 3)}] * 10, "foo": [1] * 10},
+ features=Features({"a": {"b": Image(decode=False)}, "foo": Value("int64")}),
+ ) as dset:
+ with self._to(in_memory, tmp_dir, dset) as dset:
+ fingerprint = dset._fingerprint
+ with dset.flatten() as dset:
+ self.assertListEqual(sorted(dset.column_names), ["a.b.bytes", "a.b.path", "foo"])
+ self.assertListEqual(sorted(dset.features.keys()), ["a.b.bytes", "a.b.path", "foo"])
+ self.assertDictEqual(
+ dset.features,
+ Features(
+ {"a.b.bytes": Value("binary"), "a.b.path": Value("string"), "foo": Value("int64")}
+ ),
+ )
+ self.assertNotEqual(dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(dset)
+
def test_map(self, in_memory):
# standard
with tempfile.TemporaryDirectory() as tmp_dir:
| `Translation` features cannot be `flatten`ed
## Describe the bug
(`Dataset.flatten`)[https://github.com/huggingface/datasets/blob/master/src/datasets/arrow_dataset.py#L1265] fails for columns with feature (`Translation`)[https://github.com/huggingface/datasets/blob/3edbeb0ec6519b79f1119adc251a1a6b379a2c12/src/datasets/features/translation.py#L8]
## Steps to reproduce the bug
```python
from datasets import load_dataset
dataset = load_dataset("europa_ecdc_tm", "en2fr", split="train[:10]")
print(dataset.features)
# {'translation': Translation(languages=['en', 'fr'], id=None)}
print(dataset[0])
# {'translation': {'en': 'Vaccination against hepatitis C is not yet available.', 'fr': 'Aucune vaccination contre l’hépatite C n’est encore disponible.'}}
dataset.flatten()
```
## Expected results
`dataset.flatten` should flatten the `Translation` column as if it were a dict of `Value("string")`
```python
dataset[0]
# {'translation.en': 'Vaccination against hepatitis C is not yet available.', 'translation.fr': 'Aucune vaccination contre l’hépatite C n’est encore disponible.' }
dataset.features
# {'translation.en': Value("string"), 'translation.fr': Value("string")}
```
## Actual results
```python
In [31]: dset.flatten()
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
<ipython-input-31-bb88eb5276ee> in <module>
----> 1 dset.flatten()
[...]\site-packages\datasets\fingerprint.py in wrapper(*args, **kwargs)
411 # Call actual function
412
--> 413 out = func(self, *args, **kwargs)
414
415 # Update fingerprint of in-place transforms + update in-place history of transforms
[...]\site-packages\datasets\arrow_dataset.py in flatten(self, new_fingerprint, max_depth)
1294 break
1295 dataset.info.features = self.features.flatten(max_depth=max_depth)
-> 1296 dataset._data = update_metadata_with_features(dataset._data, dataset.features)
1297 logger.info(f'Flattened dataset from depth {depth} to depth {1 if depth + 1 < max_depth else "unknown"}.')
1298 dataset._fingerprint = new_fingerprint
[...]\site-packages\datasets\arrow_dataset.py in update_metadata_with_features(table, features)
534 def update_metadata_with_features(table: Table, features: Features):
535 """To be used in dataset transforms that modify the features of the dataset, in order to update the features stored in the metadata of its schema."""
--> 536 features = Features({col_name: features[col_name] for col_name in table.column_names})
537 if table.schema.metadata is None or b"huggingface" not in table.schema.metadata:
538 pa_metadata = ArrowWriter._build_metadata(DatasetInfo(features=features))
[...]\site-packages\datasets\arrow_dataset.py in <dictcomp>(.0)
534 def update_metadata_with_features(table: Table, features: Features):
535 """To be used in dataset transforms that modify the features of the dataset, in order to update the features stored in the metadata of its schema."""
--> 536 features = Features({col_name: features[col_name] for col_name in table.column_names})
537 if table.schema.metadata is None or b"huggingface" not in table.schema.metadata:
538 pa_metadata = ArrowWriter._build_metadata(DatasetInfo(features=features))
KeyError: 'translation.en'
```
## Environment info
- `datasets` version: 1.18.3
- Platform: Windows-10-10.0.19041-SP0
- Python version: 3.7.10
- PyArrow version: 3.0.0
| 2022-02-15T14:45:33Z | [] | [] |
|
huggingface/datasets | 3,759 | huggingface__datasets-3759 | [
"769"
] | b55c590809381d7822ad76c2bebbb06e895393fa | diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -48,7 +48,7 @@
from .naming import camelcase_to_snakecase, filename_prefix_for_split
from .splits import Split, SplitDict, SplitGenerator
from .utils import logging
-from .utils.download_manager import DownloadManager, GenerateMode
+from .utils.download_manager import DownloadManager, DownloadMode
from .utils.file_utils import DownloadConfig, is_remote_url
from .utils.filelock import FileLock
from .utils.info_utils import get_size_checksum_dict, verify_checksums, verify_splits
@@ -477,7 +477,7 @@ def get_imported_module_dir(cls):
def download_and_prepare(
self,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
ignore_verifications: bool = False,
try_from_hf_gcs: bool = True,
dl_manager: Optional[DownloadManager] = None,
@@ -489,7 +489,7 @@ def download_and_prepare(
Args:
download_config (Optional ``datasets.DownloadConfig``: specific download configuration parameters.
- download_mode (Optional `datasets.GenerateMode`): select the download/generate mode - Default to REUSE_DATASET_IF_EXISTS
+ download_mode (Optional `datasets.DownloadMode`): select the download/generate mode - Default to REUSE_DATASET_IF_EXISTS
ignore_verifications (bool): Ignore the verifications of the downloaded/processed dataset information (checksums/size/splits/...)
save_infos (bool): Save the dataset information (checksums/size/splits/...)
try_from_hf_gcs (bool): If True, it will try to download the already prepared dataset from the Hf google cloud storage
@@ -500,15 +500,15 @@ def download_and_prepare(
If True, will get token from ~/.huggingface.
"""
- download_mode = GenerateMode(download_mode or GenerateMode.REUSE_DATASET_IF_EXISTS)
+ download_mode = DownloadMode(download_mode or DownloadMode.REUSE_DATASET_IF_EXISTS)
verify_infos = not ignore_verifications
base_path = base_path if base_path is not None else self.base_path
if dl_manager is None:
if download_config is None:
download_config = DownloadConfig(
cache_dir=self._cache_downloaded_dir,
- force_download=bool(download_mode == GenerateMode.FORCE_REDOWNLOAD),
- force_extract=bool(download_mode == GenerateMode.FORCE_REDOWNLOAD),
+ force_download=bool(download_mode == DownloadMode.FORCE_REDOWNLOAD),
+ force_extract=bool(download_mode == DownloadMode.FORCE_REDOWNLOAD),
use_etag=False,
use_auth_token=use_auth_token,
) # We don't use etag for data files to speed up the process
@@ -527,7 +527,7 @@ def download_and_prepare(
lock_path = os.path.join(self._cache_dir_root, self._cache_dir.replace(os.sep, "_") + ".lock")
with FileLock(lock_path):
data_exists = os.path.exists(self._cache_dir)
- if data_exists and download_mode == GenerateMode.REUSE_DATASET_IF_EXISTS:
+ if data_exists and download_mode == DownloadMode.REUSE_DATASET_IF_EXISTS:
logger.warning(f"Reusing dataset {self.name} ({self._cache_dir})")
# We need to update the info in case some splits were added in the meantime
# for example when calling load_dataset from multiple workers.
diff --git a/src/datasets/commands/run_beam.py b/src/datasets/commands/run_beam.py
--- a/src/datasets/commands/run_beam.py
+++ b/src/datasets/commands/run_beam.py
@@ -8,7 +8,7 @@
from datasets.builder import DatasetBuilder
from datasets.commands import BaseDatasetsCLICommand
from datasets.load import dataset_module_factory, import_main_class
-from datasets.utils.download_manager import DownloadConfig, GenerateMode
+from datasets.utils.download_manager import DownloadConfig, DownloadMode
def run_beam_command_factory(args):
@@ -122,9 +122,9 @@ def run(self):
for builder in builders:
builder.download_and_prepare(
- download_mode=GenerateMode.REUSE_CACHE_IF_EXISTS
+ download_mode=DownloadMode.REUSE_CACHE_IF_EXISTS
if not self._force_redownload
- else GenerateMode.FORCE_REDOWNLOAD,
+ else DownloadMode.FORCE_REDOWNLOAD,
download_config=DownloadConfig(cache_dir=config.DOWNLOADED_DATASETS_PATH),
save_infos=self._save_infos,
ignore_verifications=self._ignore_verifications,
diff --git a/src/datasets/inspect.py b/src/datasets/inspect.py
--- a/src/datasets/inspect.py
+++ b/src/datasets/inspect.py
@@ -28,7 +28,7 @@
metric_module_factory,
)
from .utils import DownloadConfig
-from .utils.download_manager import GenerateMode
+from .utils.download_manager import DownloadMode
from .utils.logging import get_logger
from .utils.streaming_download_manager import StreamingDownloadManager
from .utils.version import Version
@@ -125,7 +125,7 @@ def get_dataset_infos(
path: str,
data_files: Optional[Union[Dict, List, str]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
revision: Optional[Union[str, Version]] = None,
use_auth_token: Optional[Union[bool, str]] = None,
**config_kwargs,
@@ -146,7 +146,7 @@ def get_dataset_infos(
- it will also try to load it from the master branch if it's not available at the local version of the lib.
Specifying a version that is different from your local version of the lib might cause compatibility issues.
download_config (:class:`DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
data_files (:obj:`Union[Dict, List, str]`, optional): Defining the data_files of the dataset configuration.
use_auth_token (``str`` or ``bool``, optional): Optional string or boolean to use as Bearer token for remote files on the Datasets Hub.
If True, will get token from `"~/.huggingface"`.
@@ -178,7 +178,7 @@ def get_dataset_config_names(
path: str,
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
force_local_path: Optional[str] = None,
dynamic_modules_path: Optional[str] = None,
data_files: Optional[Union[Dict, List, str]] = None,
@@ -200,7 +200,7 @@ def get_dataset_config_names(
- it will also try to load it from the master branch if it's not available at the local version of the lib.
Specifying a version that is different from your local version of the lib might cause compatibility issues.
download_config (:class:`DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
force_local_path (Optional str): Optional path to a local path to download and prepare the script to.
Used to inspect or modify the script folder.
dynamic_modules_path (Optional str, defaults to HF_MODULES_CACHE / "datasets_modules", i.e. ~/.cache/huggingface/modules/datasets_modules):
@@ -229,7 +229,7 @@ def get_dataset_config_info(
config_name: Optional[str] = None,
data_files: Optional[Union[str, Sequence[str], Mapping[str, Union[str, Sequence[str]]]]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
revision: Optional[Union[str, Version]] = None,
use_auth_token: Optional[Union[bool, str]] = None,
**config_kwargs,
@@ -246,7 +246,7 @@ def get_dataset_config_info(
config_name (:obj:`str`, optional): Defining the name of the dataset configuration.
data_files (:obj:`str` or :obj:`Sequence` or :obj:`Mapping`, optional): Path(s) to source data file(s).
download_config (:class:`~utils.DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
revision (:class:`~utils.Version` or :obj:`str`, optional): Version of the dataset script to load:
- For canonical datasets in the `huggingface/datasets` library like "squad", the default version of the module is the local version of the lib.
@@ -291,7 +291,7 @@ def get_dataset_split_names(
config_name: Optional[str] = None,
data_files: Optional[Union[str, Sequence[str], Mapping[str, Union[str, Sequence[str]]]]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
revision: Optional[Union[str, Version]] = None,
use_auth_token: Optional[Union[bool, str]] = None,
**config_kwargs,
@@ -308,7 +308,7 @@ def get_dataset_split_names(
config_name (:obj:`str`, optional): Defining the name of the dataset configuration.
data_files (:obj:`str` or :obj:`Sequence` or :obj:`Mapping`, optional): Path(s) to source data file(s).
download_config (:class:`~utils.DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
revision (:class:`~utils.Version` or :obj:`str`, optional): Version of the dataset script to load:
- For canonical datasets in the `huggingface/datasets` library like "squad", the default version of the module is the local version of the lib.
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -54,7 +54,7 @@
from .streaming import extend_module_for_streaming
from .tasks import TaskTemplate
from .utils.deprecation_utils import deprecated
-from .utils.download_manager import GenerateMode
+from .utils.download_manager import DownloadMode
from .utils.file_utils import (
DownloadConfig,
OfflineModeIsEnabled,
@@ -312,7 +312,7 @@ def _copy_script_and_other_resources_in_importable_dir(
original_local_path: str,
local_imports: List[Tuple[str, str]],
additional_files: List[Tuple[str, str]],
- download_mode: Optional[GenerateMode],
+ download_mode: Optional[DownloadMode],
) -> str:
"""Copy a script and its required imports to an importable directory
@@ -323,7 +323,7 @@ def _copy_script_and_other_resources_in_importable_dir(
original_local_path (str): local path to the resource script
local_imports (List[Tuple[str, str]]): list of (destination_filename, import_file_to_copy)
additional_files (List[Tuple[str, str]]): list of (destination_filename, additional_file_to_copy)
- download_mode (Optional[GenerateMode]): download mode
+ download_mode (Optional[DownloadMode]): download mode
Return:
importable_local_file: path to an importable module with importlib.import_module
@@ -339,7 +339,7 @@ def _copy_script_and_other_resources_in_importable_dir(
lock_path = importable_directory_path + ".lock"
with FileLock(lock_path):
# Create main dataset/metrics folder if needed
- if download_mode == GenerateMode.FORCE_REDOWNLOAD and os.path.exists(importable_directory_path):
+ if download_mode == DownloadMode.FORCE_REDOWNLOAD and os.path.exists(importable_directory_path):
shutil.rmtree(importable_directory_path)
os.makedirs(importable_directory_path, exist_ok=True)
@@ -399,7 +399,7 @@ def _create_importable_file(
dynamic_modules_path: str,
module_namespace: str,
name: str,
- download_mode: GenerateMode,
+ download_mode: DownloadMode,
) -> Tuple[str, str]:
importable_directory_path = os.path.join(dynamic_modules_path, module_namespace, name.replace("/", "--"))
Path(importable_directory_path).mkdir(parents=True, exist_ok=True)
@@ -492,7 +492,7 @@ def __init__(
name: str,
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
dynamic_modules_path: Optional[str] = None,
):
self.name = name
@@ -573,7 +573,7 @@ def __init__(
name: str,
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
dynamic_modules_path: Optional[str] = None,
):
self.name = name
@@ -637,7 +637,7 @@ def __init__(
self,
path: str,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
dynamic_modules_path: Optional[str] = None,
):
self.path = path
@@ -678,7 +678,7 @@ def __init__(
self,
path: str,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
dynamic_modules_path: Optional[str] = None,
):
self.path = path
@@ -724,7 +724,7 @@ def __init__(
self,
path: str,
data_files: Optional[Union[str, List, Dict]] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
):
self.path = path
self.name = Path(path).stem
@@ -769,7 +769,7 @@ def __init__(
name: str,
data_files: Optional[Union[str, List, Dict]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
):
self.name = name
self.data_files = data_files
@@ -801,7 +801,7 @@ def __init__(
revision: Optional[Union[str, Version]] = None,
data_files: Optional[Union[str, List, Dict]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
):
self.name = name
self.revision = revision
@@ -873,7 +873,7 @@ def __init__(
name: str,
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
dynamic_modules_path: Optional[str] = None,
):
self.name = name
@@ -1041,7 +1041,7 @@ def dataset_module_factory(
path: str,
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
force_local_path: Optional[str] = None,
dynamic_modules_path: Optional[str] = None,
data_files: Optional[Union[Dict, List, str, DataFilesDict]] = None,
@@ -1088,7 +1088,7 @@ def dataset_module_factory(
- it will also try to load it from the master branch if it's not available at the local version of the lib.
Specifying a version that is different from your local version of the lib might cause compatibility issues.
download_config (:class:`DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
force_local_path (Optional str): Optional path to a local path to download and prepare the script to.
Used to inspect or modify the script folder.
dynamic_modules_path (Optional str, defaults to HF_MODULES_CACHE / "datasets_modules", i.e. ~/.cache/huggingface/modules/datasets_modules):
@@ -1108,7 +1108,7 @@ def dataset_module_factory(
download_config.extract_compressed_file = True
download_config.force_extract = True
download_config.force_download = download_mode = (
- GenerateMode(download_mode or GenerateMode.REUSE_DATASET_IF_EXISTS) == GenerateMode.FORCE_REDOWNLOAD
+ DownloadMode(download_mode or DownloadMode.REUSE_DATASET_IF_EXISTS) == DownloadMode.FORCE_REDOWNLOAD
)
filename = list(filter(lambda x: x, path.replace(os.sep, "/").split("/")))[-1]
@@ -1232,7 +1232,7 @@ def metric_module_factory(
path: str,
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
force_local_path: Optional[str] = None,
dynamic_modules_path: Optional[str] = None,
**download_kwargs,
@@ -1260,7 +1260,7 @@ def metric_module_factory(
- it will also try to load it from the master branch if it's not available at the local version of the lib.
Specifying a version that is different from your local version of the lib might cause compatibility issues.
download_config (:class:`DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
force_local_path (Optional str): Optional path to a local path to download and prepare the script to.
Used to inspect or modify the script folder.
dynamic_modules_path (Optional str, defaults to HF_MODULES_CACHE / "datasets_modules", i.e. ~/.cache/huggingface/modules/datasets_modules):
@@ -1342,7 +1342,7 @@ def prepare_module(
path: str,
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
dataset: bool = True,
force_local_path: Optional[str] = None,
dynamic_modules_path: Optional[str] = None,
@@ -1394,7 +1394,7 @@ def load_metric(
experiment_id: Optional[str] = None,
keep_in_memory: bool = False,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
revision: Optional[Union[str, Version]] = None,
script_version="deprecated",
**metric_init_kwargs,
@@ -1417,7 +1417,7 @@ def load_metric(
This is useful to compute metrics in distributed setups (in particular non-additive metrics like F1).
keep_in_memory (bool): Whether to store the temporary results in memory (defaults to False)
download_config (Optional ``datasets.DownloadConfig``: specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
revision (Optional ``Union[str, datasets.Version]``): if specified, the module will be loaded from the datasets repository
at this version. By default it is set to the local version of the lib. Specifying a version that is different from
your local version of the lib might cause compatibility issues.
@@ -1461,7 +1461,7 @@ def load_dataset_builder(
cache_dir: Optional[str] = None,
features: Optional[Features] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
revision: Optional[Union[str, Version]] = None,
use_auth_token: Optional[Union[bool, str]] = None,
script_version="deprecated",
@@ -1505,7 +1505,7 @@ def load_dataset_builder(
cache_dir (:obj:`str`, optional): Directory to read/write data. Defaults to "~/.cache/huggingface/datasets".
features (:class:`Features`, optional): Set the features type to use for this dataset.
download_config (:class:`~utils.DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
revision (:class:`~utils.Version` or :obj:`str`, optional): Version of the dataset script to load:
- For canonical datasets in the `huggingface/datasets` library like "squad", the default version of the module is the local version of the lib.
@@ -1575,7 +1575,7 @@ def load_dataset(
cache_dir: Optional[str] = None,
features: Optional[Features] = None,
download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[GenerateMode] = None,
+ download_mode: Optional[DownloadMode] = None,
ignore_verifications: bool = False,
keep_in_memory: Optional[bool] = None,
save_infos: bool = False,
@@ -1648,7 +1648,7 @@ def load_dataset(
cache_dir (:obj:`str`, optional): Directory to read/write data. Defaults to "~/.cache/huggingface/datasets".
features (:class:`Features`, optional): Set the features type to use for this dataset.
download_config (:class:`~utils.DownloadConfig`, optional): Specific download configuration parameters.
- download_mode (:class:`GenerateMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
+ download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
ignore_verifications (:obj:`bool`, default ``False``): Ignore the verifications of the downloaded/processed dataset information (checksums/size/splits/...).
keep_in_memory (:obj:`bool`, default ``None``): Whether to copy the dataset in-memory. If `None`, the dataset
will not be copied in-memory unless explicitly enabled by setting `datasets.config.IN_MEMORY_MAX_SIZE` to
diff --git a/src/datasets/utils/__init__.py b/src/datasets/utils/__init__.py
--- a/src/datasets/utils/__init__.py
+++ b/src/datasets/utils/__init__.py
@@ -17,7 +17,7 @@
"""Util import."""
from . import logging
-from .download_manager import DownloadManager, GenerateMode
+from .download_manager import DownloadManager, DownloadMode, GenerateMode
from .file_utils import DownloadConfig, cached_path, hf_bucket_url, is_remote_url, relative_to_absolute_path, temp_seed
from .mock_download_manager import MockDownloadManager
from .py_utils import (
diff --git a/src/datasets/utils/deprecation_utils.py b/src/datasets/utils/deprecation_utils.py
--- a/src/datasets/utils/deprecation_utils.py
+++ b/src/datasets/utils/deprecation_utils.py
@@ -1,3 +1,4 @@
+import enum
import warnings
from functools import wraps
from typing import Callable, Optional
@@ -41,3 +42,52 @@ def wrapper(*args, **kwargs):
return wrapper
return decorator
+
+
+class OnAccess(enum.EnumMeta):
+ """
+ Enum metaclass that calls a user-specified function whenever a member is accessed.
+ """
+
+ def __getattribute__(cls, name):
+ obj = super().__getattribute__(name)
+ if isinstance(obj, enum.Enum) and obj._on_access:
+ obj._on_access()
+ return obj
+
+ def __getitem__(cls, name):
+ member = super().__getitem__(name)
+ if member._on_access:
+ member._on_access()
+ return member
+
+ def __call__(cls, value, names=None, *, module=None, qualname=None, type=None, start=1):
+ obj = super().__call__(value, names, module=module, qualname=qualname, type=type, start=start)
+ if isinstance(obj, enum.Enum) and obj._on_access:
+ obj._on_access()
+ return obj
+
+
+class DeprecatedEnum(enum.Enum, metaclass=OnAccess):
+ """
+ Enum class that calls `deprecate` method whenever a member is accessed.
+ """
+
+ def __new__(cls, value):
+ member = object.__new__(cls)
+ member._value_ = value
+ member._on_access = member.deprecate
+ return member
+
+ @property
+ def help_message(self):
+ return ""
+
+ def deprecate(self):
+ help_message = f" {self.help_message}" if self.help_message else ""
+ warnings.warn(
+ f"'{self.__objclass__.__name__}' is deprecated and will be removed in the next major version of datasets."
+ + help_message,
+ FutureWarning,
+ stacklevel=3,
+ )
diff --git a/src/datasets/utils/download_manager.py b/src/datasets/utils/download_manager.py
--- a/src/datasets/utils/download_manager.py
+++ b/src/datasets/utils/download_manager.py
@@ -24,6 +24,7 @@
from typing import Dict, Optional, Union
from .. import config, utils
+from .deprecation_utils import DeprecatedEnum
from .file_utils import (
DownloadConfig,
cached_path,
@@ -40,7 +41,7 @@
logger = get_logger(__name__)
-class GenerateMode(enum.Enum):
+class DownloadMode(enum.Enum):
"""`Enum` for how to treat pre-existing downloads and data.
The default mode is `REUSE_DATASET_IF_EXISTS`, which will reuse both
@@ -64,6 +65,16 @@ class GenerateMode(enum.Enum):
FORCE_REDOWNLOAD = "force_redownload"
+class GenerateMode(DeprecatedEnum):
+ REUSE_DATASET_IF_EXISTS = "reuse_dataset_if_exists"
+ REUSE_CACHE_IF_EXISTS = "reuse_cache_if_exists"
+ FORCE_REDOWNLOAD = "force_redownload"
+
+ @property
+ def help_message(self):
+ return "Use 'DownloadMode' instead."
+
+
class DownloadManager:
is_streaming = False
| diff --git a/src/datasets/commands/test.py b/src/datasets/commands/test.py
--- a/src/datasets/commands/test.py
+++ b/src/datasets/commands/test.py
@@ -8,7 +8,7 @@
from datasets.builder import DatasetBuilder
from datasets.commands import BaseDatasetsCLICommand
from datasets.load import dataset_module_factory, import_main_class
-from datasets.utils.download_manager import GenerateMode
+from datasets.utils.download_manager import DownloadMode
from datasets.utils.filelock import logger as fl_logger
from datasets.utils.logging import ERROR, get_logger
@@ -154,9 +154,9 @@ def get_builders() -> Generator[DatasetBuilder, None, None]:
for j, builder in enumerate(get_builders()):
print(f"Testing builder '{builder.config.name}' ({j + 1}/{n_builders})")
builder.download_and_prepare(
- download_mode=GenerateMode.REUSE_CACHE_IF_EXISTS
+ download_mode=DownloadMode.REUSE_CACHE_IF_EXISTS
if not self._force_redownload
- else GenerateMode.FORCE_REDOWNLOAD,
+ else DownloadMode.FORCE_REDOWNLOAD,
ignore_verifications=self._ignore_verifications,
try_from_hf_gcs=False,
)
diff --git a/tests/test_builder.py b/tests/test_builder.py
--- a/tests/test_builder.py
+++ b/tests/test_builder.py
@@ -15,7 +15,7 @@
from datasets.features import Features, Value
from datasets.info import DatasetInfo, PostProcessedInfo
from datasets.splits import Split, SplitDict, SplitGenerator, SplitInfo
-from datasets.utils.download_manager import GenerateMode
+from datasets.utils.download_manager import DownloadMode
from .utils import assert_arrow_memory_doesnt_increase, assert_arrow_memory_increases, require_faiss
@@ -119,7 +119,7 @@ def _split_generators(self, dl_manager):
def _run_concurrent_download_and_prepare(tmp_dir):
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.REUSE_DATASET_IF_EXISTS)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.REUSE_DATASET_IF_EXISTS)
return dummy_builder
@@ -127,7 +127,7 @@ class BuilderTest(TestCase):
def test_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
)
@@ -166,13 +166,13 @@ def test_download_and_prepare_with_base_path(self):
dummy_builder = DummyBuilderWithDownload(cache_dir=tmp_dir, name="dummy", rel_path=rel_path)
with self.assertRaises(FileNotFoundError):
dummy_builder.download_and_prepare(
- try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD, base_path=tmp_dir
+ try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
# test absolute path is missing
dummy_builder = DummyBuilderWithDownload(cache_dir=tmp_dir, name="dummy", abs_path=abs_path)
with self.assertRaises(FileNotFoundError):
dummy_builder.download_and_prepare(
- try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD, base_path=tmp_dir
+ try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
# test that they are both properly loaded when they exist
open(os.path.join(tmp_dir, rel_path), "w")
@@ -181,7 +181,7 @@ def test_download_and_prepare_with_base_path(self):
cache_dir=tmp_dir, name="dummy", rel_path=rel_path, abs_path=abs_path
)
dummy_builder.download_and_prepare(
- try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD, base_path=tmp_dir
+ try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
self.assertTrue(
os.path.exists(
@@ -421,7 +421,7 @@ def _post_processing_resources(self, split):
)
dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
dummy_builder._post_processing_resources = types.MethodType(_post_processing_resources, dummy_builder)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
)
@@ -441,7 +441,7 @@ def _post_process(self, dataset, resources_paths):
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
)
@@ -470,7 +470,7 @@ def _post_processing_resources(self, split):
dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
dummy_builder._post_processing_resources = types.MethodType(_post_processing_resources, dummy_builder)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
)
@@ -492,14 +492,14 @@ def _prepare_split(self, split_generator, **kwargs):
ValueError,
dummy_builder.download_and_prepare,
try_from_hf_gcs=False,
- download_mode=GenerateMode.FORCE_REDOWNLOAD,
+ download_mode=DownloadMode.FORCE_REDOWNLOAD,
)
self.assertRaises(AssertionError, dummy_builder.as_dataset)
def test_generator_based_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(
os.path.join(
@@ -718,7 +718,7 @@ def test_generator_based_builder_as_dataset(in_memory, tmp_path):
cache_dir.mkdir()
cache_dir = str(cache_dir)
dummy_builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
with assert_arrow_memory_increases() if in_memory else assert_arrow_memory_doesnt_increase():
dataset = dummy_builder.as_dataset("train", in_memory=in_memory)
assert dataset.data.to_pydict() == {"text": ["foo"] * 100}
@@ -733,6 +733,6 @@ def test_custom_writer_batch_size(tmp_path, writer_batch_size, default_writer_ba
DummyGeneratorBasedBuilder.DEFAULT_WRITER_BATCH_SIZE = default_writer_batch_size
dummy_builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir, name="dummy", writer_batch_size=writer_batch_size)
assert dummy_builder._writer_batch_size == (writer_batch_size or default_writer_batch_size)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=GenerateMode.FORCE_REDOWNLOAD)
+ dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
dataset = dummy_builder.as_dataset("train")
assert len(dataset.data[0].chunks) == expected_chunks
diff --git a/tests/test_dataset_common.py b/tests/test_dataset_common.py
--- a/tests/test_dataset_common.py
+++ b/tests/test_dataset_common.py
@@ -28,7 +28,7 @@
from datasets.load import dataset_module_factory, import_main_class, load_dataset
from datasets.packaged_modules import _PACKAGED_DATASETS_MODULES
from datasets.search import _has_faiss
-from datasets.utils.download_manager import GenerateMode
+from datasets.utils.download_manager import DownloadMode
from datasets.utils.file_utils import DownloadConfig, cached_path, is_remote_url
from datasets.utils.logging import get_logger
from datasets.utils.mock_download_manager import MockDownloadManager
@@ -169,7 +169,7 @@ def check_if_url_is_valid(url):
# generate examples from dummy data
dataset_builder.download_and_prepare(
dl_manager=mock_dl_manager,
- download_mode=GenerateMode.FORCE_REDOWNLOAD,
+ download_mode=DownloadMode.FORCE_REDOWNLOAD,
ignore_verifications=True,
try_from_hf_gcs=False,
)
@@ -257,7 +257,7 @@ def test_load_real_dataset(self, dataset_name):
name = builder_cls.BUILDER_CONFIGS[0].name if builder_cls.BUILDER_CONFIGS else None
with tempfile.TemporaryDirectory() as temp_cache_dir:
dataset = load_dataset(
- path, name=name, cache_dir=temp_cache_dir, download_mode=GenerateMode.FORCE_REDOWNLOAD
+ path, name=name, cache_dir=temp_cache_dir, download_mode=DownloadMode.FORCE_REDOWNLOAD
)
for split in dataset.keys():
self.assertTrue(len(dataset[split]) > 0)
@@ -274,7 +274,7 @@ def test_load_real_dataset_all_configs(self, dataset_name):
for name in config_names:
with tempfile.TemporaryDirectory() as temp_cache_dir:
dataset = load_dataset(
- path, name=name, cache_dir=temp_cache_dir, download_mode=GenerateMode.FORCE_REDOWNLOAD
+ path, name=name, cache_dir=temp_cache_dir, download_mode=DownloadMode.FORCE_REDOWNLOAD
)
for split in dataset.keys():
self.assertTrue(len(dataset[split]) > 0)
| How to choose proper download_mode in function load_dataset?
Hi, I am a beginner to datasets and I try to use datasets to load my csv file.
my csv file looks like this
```
text,label
"Effective but too-tepid biopic",3
"If you sometimes like to go to the movies to have fun , Wasabi is a good place to start .",4
"Emerges as something rare , an issue movie that 's so honest and keenly observed that it does n't feel like one .",5
```
First I try to use this command to load my csv file .
``` python
dataset=load_dataset('csv', data_files=['sst_test.csv'])
```
It seems good, but when i try to overwrite the convert_options to convert 'label' columns from int64 to float32 like this.
``` python
import pyarrow as pa
from pyarrow import csv
read_options = csv.ReadOptions(block_size=1024*1024)
parse_options = csv.ParseOptions()
convert_options = csv.ConvertOptions(column_types={'text': pa.string(), 'label': pa.float32()})
dataset = load_dataset('csv', data_files=['sst_test.csv'], read_options=read_options,
parse_options=parse_options, convert_options=convert_options)
```
It keeps the same:
```shell
Dataset(features: {'text': Value(dtype='string', id=None), 'label': Value(dtype='int64', id=None)}, num_rows: 2210)
```
I think this issue is caused by the parameter "download_mode" Default to REUSE_DATASET_IF_EXISTS because after I delete the cache_dir, it seems right.
Is it a bug? How to choose proper download_mode to avoid this issue?
| `download_mode=datasets.GenerateMode.FORCE_REDOWNLOAD` should work.
This makes me think we we should rename this to DownloadMode.FORCE_REDOWNLOAD. Currently that's confusing
Can we just use `features=...` in `load_dataset` for this @lhoestq?
Indeed you should use `features` in this case.
```python
features = Features({'text': Value('string'), 'label': Value('float32')})
dataset = load_dataset('csv', data_files=['sst_test.csv'], features=features)
```
Note that because of an issue with the caching when you change the features (see #750 ) you still need to specify the `FORCE_REDOWNLOAD ` flag. I'm working on a fix for this one
https://github.com/huggingface/datasets/issues/769#issuecomment-717837832
> This makes me think we we should rename this to DownloadMode.FORCE_REDOWNLOAD. Currently that's confusing
@lhoestq do you still think we should rename it?
It's no big deal, but since it can be confusing to users I think it's worth renaming it, and deprecate `GenerateMode` until `datasets` 2.0 at least. IMO it's confusing to have `download_mode=GenerateMode.something` | 2022-02-18T16:53:53Z | [] | [] |
huggingface/datasets | 3,824 | huggingface__datasets-3824 | [
"3818"
] | d58415637bf5456bbb1beecebe73d9ce4068f106 | diff --git a/metrics/sari/sari.py b/metrics/sari/sari.py
--- a/metrics/sari/sari.py
+++ b/metrics/sari/sari.py
@@ -265,6 +265,7 @@ def _info(self):
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
+ "sources": datasets.Value("string", id="sequence"),
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="sequence"), id="references"),
}
diff --git a/src/datasets/metric.py b/src/datasets/metric.py
--- a/src/datasets/metric.py
+++ b/src/datasets/metric.py
@@ -61,6 +61,18 @@ def _release(self):
self._lock_file_fd = None
+# lists - summarize long lists similarly to NumPy
+# arrays/tensors - let the frameworks control formatting
+def summarize_if_long_list(obj):
+ if not type(obj) == list or len(obj) <= 6:
+ return f"{obj}"
+
+ def format_chunk(chunk):
+ return ", ".join(repr(x) for x in chunk)
+
+ return f"[{format_chunk(obj[:3])}, ..., {format_chunk(obj[-3:])}]"
+
+
class MetricInfoMixin:
"""This base class exposes some attributes of MetricInfo
at the base level of the Metric for easy access.
@@ -391,11 +403,15 @@ def compute(self, *, predictions=None, references=None, **kwargs) -> Optional[di
- None if the metric is not run on the main process (``process_id != 0``).
"""
all_kwargs = {"predictions": predictions, "references": references, **kwargs}
- missing_inputs = [k for k in self.features if k not in all_kwargs]
- if missing_inputs:
- raise ValueError(
- f"Metric inputs are missing: {missing_inputs}. All required inputs are {list(self.features)}"
- )
+ if predictions is None and references is None:
+ missing_kwargs = {k: None for k in self.features if k not in all_kwargs}
+ all_kwargs.update(missing_kwargs)
+ else:
+ missing_inputs = [k for k in self.features if k not in all_kwargs]
+ if missing_inputs:
+ raise ValueError(
+ f"Metric inputs are missing: {missing_inputs}. All required inputs are {list(self.features)}"
+ )
inputs = {input_name: all_kwargs[input_name] for input_name in self.features}
compute_kwargs = {k: kwargs[k] for k in kwargs if k not in self.features}
@@ -450,18 +466,6 @@ def add_batch(self, *, predictions=None, references=None, **kwargs):
try:
self.writer.write_batch(batch)
except pa.ArrowInvalid:
-
- # lists - summarize long lists similarly to NumPy
- # arrays/tensors - let the frameworks control formatting
- def summarize_if_long_list(obj):
- if not type(obj) == list or len(obj) <= 6:
- return f"{obj}"
-
- def format_chunk(chunk):
- return ", ".join(repr(x) for x in chunk)
-
- return f"[{format_chunk(obj[:3])}, ..., {format_chunk(obj[-3:])}]"
-
if any(len(batch[c]) != len(next(iter(batch.values()))) for c in batch):
col0 = next(iter(batch))
bad_col = [c for c in batch if len(batch[c]) != len(batch[col0])][0]
@@ -470,8 +474,10 @@ def format_chunk(chunk):
)
elif sorted(self.features) != ["references", "predictions"]:
error_msg = f"Metric inputs don't match the expected format.\n" f"Expected format: {self.features},\n"
- for input_name in self.features:
- error_msg += f"Input {input_name}: {summarize_if_long_list(batch[input_name])},\n"
+ error_msg_inputs = ",\n".join(
+ f"Input {input_name}: {summarize_if_long_list(batch[input_name])}" for input_name in self.features
+ )
+ error_msg += error_msg_inputs
else:
error_msg = (
f"Predictions and/or references don't match the expected format.\n"
@@ -500,8 +506,10 @@ def add(self, *, prediction=None, reference=None, **kwargs):
self.writer.write(example)
except pa.ArrowInvalid:
error_msg = f"Metric inputs don't match the expected format.\n" f"Expected format: {self.features},\n"
- for input_name in self.features:
- error_msg += f"Input {input_name}: {example[input_name]},\n"
+ error_msg_inputs = ",\n".join(
+ f"Input {input_name}: {summarize_if_long_list(example[input_name])}" for input_name in self.features
+ )
+ error_msg += error_msg_inputs
raise ValueError(error_msg) from None
def _init_writer(self, timeout=1):
| diff --git a/tests/test_metric.py b/tests/test_metric.py
--- a/tests/test_metric.py
+++ b/tests/test_metric.py
@@ -526,3 +526,56 @@ def test_safety_checks_process_vars():
with pytest.raises(ValueError):
_ = DummyMetric(num_process=2, process_id=3)
+
+
+class AccuracyWithNonStandardFeatureNames(Metric):
+ def _info(self):
+ return MetricInfo(
+ description="dummy metric for tests",
+ citation="insert citation here",
+ features=Features({"inputs": Value("int64"), "targets": Value("int64")}),
+ )
+
+ def _compute(self, inputs, targets):
+ return (
+ {
+ "accuracy": sum(i == j for i, j in zip(inputs, targets)) / len(targets),
+ }
+ if targets
+ else {}
+ )
+
+ @classmethod
+ def inputs_and_targets(cls):
+ return ([1, 2, 3, 4], [1, 2, 4, 3])
+
+ @classmethod
+ def expected_results(cls):
+ return {"accuracy": 0.5}
+
+
+def test_metric_with_non_standard_feature_names_add(tmp_path):
+ cache_dir = tmp_path / "cache"
+ inputs, targets = AccuracyWithNonStandardFeatureNames.inputs_and_targets()
+ metric = AccuracyWithNonStandardFeatureNames(cache_dir=cache_dir)
+ for input, target in zip(inputs, targets):
+ metric.add(inputs=input, targets=target)
+ results = metric.compute()
+ assert results == AccuracyWithNonStandardFeatureNames.expected_results()
+
+
+def test_metric_with_non_standard_feature_names_add_batch(tmp_path):
+ cache_dir = tmp_path / "cache"
+ inputs, targets = AccuracyWithNonStandardFeatureNames.inputs_and_targets()
+ metric = AccuracyWithNonStandardFeatureNames(cache_dir=cache_dir)
+ metric.add_batch(inputs=inputs, targets=targets)
+ results = metric.compute()
+ assert results == AccuracyWithNonStandardFeatureNames.expected_results()
+
+
+def test_metric_with_non_standard_feature_names_compute(tmp_path):
+ cache_dir = tmp_path / "cache"
+ inputs, targets = AccuracyWithNonStandardFeatureNames.inputs_and_targets()
+ metric = AccuracyWithNonStandardFeatureNames(cache_dir=cache_dir)
+ results = metric.compute(inputs=inputs, targets=targets)
+ assert results == AccuracyWithNonStandardFeatureNames.expected_results()
| Support for "sources" parameter in the add() and add_batch() methods in datasets.metric - SARI
**Is your feature request related to a problem? Please describe.**
The methods `add_batch` and `add` from the `Metric` [class](https://github.com/huggingface/datasets/blob/1675ad6a958435b675a849eafa8a7f10fe0f43bc/src/datasets/metric.py) does not work with [SARI](https://github.com/huggingface/datasets/blob/master/metrics/sari/sari.py) metric. This metric not only relies on the predictions and references, but also in the input.
For example, when the `add_batch` method is used, then the `compute()` method fails:
```
metric = load_metric("sari")
metric.add_batch(
predictions=["About 95 you now get in ."],
references=[["About 95 species are currently known .","About 95 species are now accepted .","95 species are now accepted ."]])
metric.compute()
> TypeError: _compute() missing 1 required positional argument: 'sources'
```
Therefore, the `compute() `method can only be used standalone:
```
metric = load_metric("sari")
result = metric.compute(
sources=["About 95 species are currently accepted ."],
predictions=["About 95 you now get in ."],
references=[["About 95 species are currently known .","About 95 species are now accepted .","95 species are now accepted ."]])
> {'sari': 26.953601953601954}
```
**Describe the solution you'd like**
Support for an additional parameter `sources` in the `add_batch` and `add` of the `Metric` class.
```
add_batch(*, sources=None, predictions=None, references=None, **kwargs)
add(*, sources=None, predictions=None, references=None, **kwargs)
compute()
```
**Describe alternatives you've considered**
I've tried to override the `add_batch` and `add`, however, these are highly dependent to the `Metric` class. We could also write a simple function that compute the scores of a sentences list, but then we lose the functionality from the original [add](https://huggingface.co/docs/datasets/_modules/datasets/metric.html#Metric.add) and [add_batch method](https://huggingface.co/docs/datasets/_modules/datasets/metric.html#Metric.add_batch).
**Additional context**
These methods are used in the transformers [pytorch examples](https://github.com/huggingface/transformers/blob/master/examples/pytorch/summarization/run_summarization_no_trainer.py).
| Hi, thanks for reporting! We can add a `sources: datasets.Value("string")` feature to the `Features` dict in the `SARI` script to fix this. Would you be interested in submitting a PR?
Hi Mario,
Thanks for your message. I did try to add `sources` into the `Features` dict using a script for the metric:
```
features=datasets.Features(
{
"sources": datasets.Value("string", id="sequence"),
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="sequence"), id="references"),
}
),
```
But that only avoids a failure in `encode_batch` in the `add_batch` method:
```
batch = {"predictions": predictions, "references": references}
batch = self.info.features.encode_batch(batch)
```
The real problem is that `add_batch()`, `add()` and `compute()` does not receive a `sources` param:
```
def add_batch(self, *, predictions=None, references=None):
def add(self, *, prediction=None, reference=None):
def compute(self, *, predictions=None, references=None, **kwargs)
```
And then, it fails:
`TypeError: add_batch() got an unexpected keyword argument sources`
I need this for adding any metric based on SARI or alike, not only for sari.py :)
Let me know if I understood correctly the proposed solution.
The `Metric` class has been modified recently to support this use-case, but the `add_batch` + `compute` pattern still doesn't work correctly. I'll open a PR. | 2022-03-04T12:04:40Z | [] | [] |
huggingface/datasets | 3,892 | huggingface__datasets-3892 | [
"3848"
] | 82e8e9136dd632a2e28d2487c16736af207da751 | diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -297,6 +297,9 @@ def __init__(
# Set download manager
self.dl_manager = None
+ # Record infos even if verify_infos=False; used by "datasets-cli test" to generate dataset_infos.json
+ self._record_infos = False
+
# Must be set for datasets that use 'data_dir' functionality - the ones
# that require users to do additional steps to download the data
# (this is usually due to some external regulations / rules).
@@ -518,7 +521,7 @@ def download_and_prepare(
download_config=download_config,
data_dir=self.config.data_dir,
base_path=base_path,
- record_checksums=verify_infos,
+ record_checksums=self._record_infos or verify_infos,
)
elif isinstance(dl_manager, MockDownloadManager):
try_from_hf_gcs = False
| diff --git a/src/datasets/commands/test.py b/src/datasets/commands/test.py
--- a/src/datasets/commands/test.py
+++ b/src/datasets/commands/test.py
@@ -153,6 +153,7 @@ def get_builders() -> Generator[DatasetBuilder, None, None]:
for j, builder in enumerate(get_builders()):
print(f"Testing builder '{builder.config.name}' ({j + 1}/{n_builders})")
+ builder._record_infos = True
builder.download_and_prepare(
download_mode=DownloadMode.REUSE_CACHE_IF_EXISTS
if not self._force_redownload
diff --git a/tests/commands/__init__.py b/tests/commands/__init__.py
new file mode 100644
diff --git a/tests/commands/conftest.py b/tests/commands/conftest.py
new file mode 100644
--- /dev/null
+++ b/tests/commands/conftest.py
@@ -0,0 +1,74 @@
+import pytest
+
+
+DATASET_LOADING_SCRIPT_NAME = "__dummy_dataset1__"
+
+DATASET_LOADING_SCRIPT_CODE = """
+import json
+import os
+
+import datasets
+
+
+REPO_URL = "https://huggingface.co/datasets/albertvillanova/tests-raw-jsonl/resolve/main/"
+URLS = {"train": REPO_URL + "wikiann-bn-train.jsonl", "validation": REPO_URL + "wikiann-bn-validation.jsonl"}
+
+
+class __DummyDataset1__(datasets.GeneratorBasedBuilder):
+
+ def _info(self):
+ features = datasets.Features(
+ {
+ "tokens": datasets.Sequence(datasets.Value("string")),
+ "ner_tags": datasets.Sequence(
+ datasets.features.ClassLabel(
+ names=[
+ "O",
+ "B-PER",
+ "I-PER",
+ "B-ORG",
+ "I-ORG",
+ "B-LOC",
+ "I-LOC",
+ ]
+ )
+ ),
+ "langs": datasets.Sequence(datasets.Value("string")),
+ "spans": datasets.Sequence(datasets.Value("string")),
+ }
+ )
+ return datasets.DatasetInfo(features=features)
+
+ def _split_generators(self, dl_manager):
+ dl_path = dl_manager.download(URLS)
+ return [
+ datasets.SplitGenerator(datasets.Split.TRAIN, gen_kwargs={"filepath": dl_path["train"]}),
+ datasets.SplitGenerator(datasets.Split.VALIDATION, gen_kwargs={"filepath": dl_path["validation"]}),
+ ]
+
+ def _generate_examples(self, filepath):
+ with open(filepath, "r", encoding="utf-8") as f:
+ for i, line in enumerate(f):
+ yield i, json.loads(line)
+"""
+
+
[email protected]
+def dataset_loading_script_name():
+ return DATASET_LOADING_SCRIPT_NAME
+
+
[email protected]
+def dataset_loading_script_code():
+ return DATASET_LOADING_SCRIPT_CODE
+
+
[email protected]
+def dataset_loading_script_dir(dataset_loading_script_name, dataset_loading_script_code, tmp_path):
+ script_name = dataset_loading_script_name
+ script_dir = tmp_path / script_name
+ script_dir.mkdir()
+ script_path = script_dir / f"{script_name}.py"
+ with open(script_path, "w") as f:
+ f.write(dataset_loading_script_code)
+ return str(script_dir)
diff --git a/tests/commands/test_test.py b/tests/commands/test_test.py
new file mode 100644
--- /dev/null
+++ b/tests/commands/test_test.py
@@ -0,0 +1,171 @@
+import json
+import os
+from collections import namedtuple
+from dataclasses import dataclass
+
+from packaging import version
+
+from datasets import config
+from datasets.commands.test import TestCommand
+
+
+if config.PY_VERSION >= version.parse("3.7"):
+ TestCommandArgs = namedtuple(
+ "TestCommandArgs",
+ [
+ "dataset",
+ "name",
+ "cache_dir",
+ "data_dir",
+ "all_configs",
+ "save_infos",
+ "ignore_verifications",
+ "force_redownload",
+ "clear_cache",
+ "proc_rank",
+ "num_proc",
+ ],
+ defaults=[None, None, None, False, False, False, False, False, 0, 1],
+ )
+else:
+
+ @dataclass
+ class TestCommandArgs:
+ dataset: str
+ name: str = None
+ cache_dir: str = None
+ data_dir: str = None
+ all_configs: bool = False
+ save_infos: bool = False
+ ignore_verifications: bool = False
+ force_redownload: bool = False
+ clear_cache: bool = False
+ proc_rank: int = 0
+ num_proc: int = 1
+
+ def __iter__(self):
+ return iter(self.__dict__.values())
+
+
+def test_test_command(dataset_loading_script_dir):
+ args = TestCommandArgs(dataset=dataset_loading_script_dir, all_configs=True, save_infos=True)
+ test_command = TestCommand(*args)
+ test_command.run()
+ dataset_infos_path = os.path.join(dataset_loading_script_dir, config.DATASETDICT_INFOS_FILENAME)
+ assert os.path.exists(dataset_infos_path)
+ with open(dataset_infos_path, encoding="utf-8") as f:
+ dataset_infos = json.load(f)
+ expected_dataset_infos = {
+ "default": {
+ "description": "",
+ "citation": "",
+ "homepage": "",
+ "license": "",
+ "features": {
+ "tokens": {
+ "feature": {"dtype": "string", "id": None, "_type": "Value"},
+ "length": -1,
+ "id": None,
+ "_type": "Sequence",
+ },
+ "ner_tags": {
+ "feature": {
+ "num_classes": 7,
+ "names": ["O", "B-PER", "I-PER", "B-ORG", "I-ORG", "B-LOC", "I-LOC"],
+ "id": None,
+ "_type": "ClassLabel",
+ },
+ "length": -1,
+ "id": None,
+ "_type": "Sequence",
+ },
+ "langs": {
+ "feature": {"dtype": "string", "id": None, "_type": "Value"},
+ "length": -1,
+ "id": None,
+ "_type": "Sequence",
+ },
+ "spans": {
+ "feature": {"dtype": "string", "id": None, "_type": "Value"},
+ "length": -1,
+ "id": None,
+ "_type": "Sequence",
+ },
+ },
+ "post_processed": None,
+ "supervised_keys": None,
+ "task_templates": None,
+ "builder_name": "__dummy_dataset1__",
+ "config_name": "default",
+ "version": {"version_str": "0.0.0", "description": None, "major": 0, "minor": 0, "patch": 0},
+ "splits": {
+ "train": {
+ "name": "train",
+ "num_bytes": 2351591,
+ "num_examples": 10000,
+ "dataset_name": "__dummy_dataset1__",
+ },
+ "validation": {
+ "name": "validation",
+ "num_bytes": 238446,
+ "num_examples": 1000,
+ "dataset_name": "__dummy_dataset1__",
+ },
+ },
+ "download_checksums": {
+ "https://huggingface.co/datasets/albertvillanova/tests-raw-jsonl/resolve/main/wikiann-bn-train.jsonl": {
+ "num_bytes": 3578339,
+ "checksum": "6fbe6dbdcb3c9c3a98b0ab4d56b1c8b73baab9293d603064a5ab5230ab4f366b",
+ },
+ "https://huggingface.co/datasets/albertvillanova/tests-raw-jsonl/resolve/main/wikiann-bn-validation.jsonl": {
+ "num_bytes": 362341,
+ "checksum": "2ddd0c090a8ccb721d7aa8477ed7323750683822c247015d5cfab1af1c8c8b3f",
+ },
+ },
+ "download_size": 3940680,
+ "post_processing_size": None,
+ "dataset_size": 2590037,
+ "size_in_bytes": 6530717,
+ }
+ }
+ assert dataset_infos.keys() == expected_dataset_infos.keys()
+ assert dataset_infos["default"].keys() == expected_dataset_infos["default"].keys()
+ for key in dataset_infos["default"].keys():
+ if key in [
+ "description",
+ "citation",
+ "homepage",
+ "license",
+ "features",
+ "post_processed",
+ "supervised_keys",
+ "task_templates",
+ "builder_name",
+ "config_name",
+ "version",
+ "download_checksums",
+ "download_size",
+ "post_processing_size",
+ ]:
+ assert dataset_infos["default"][key] == expected_dataset_infos["default"][key]
+ elif key in ["dataset_size", "size_in_bytes"]:
+ assert round(dataset_infos["default"][key] / 10**5) == round(
+ expected_dataset_infos["default"][key] / 10**5
+ )
+ elif key == "splits":
+ assert dataset_infos["default"]["splits"].keys() == expected_dataset_infos["default"]["splits"].keys()
+ for split in dataset_infos["default"]["splits"].keys():
+ assert (
+ dataset_infos["default"]["splits"][split].keys()
+ == expected_dataset_infos["default"]["splits"][split].keys()
+ )
+ for subkey in dataset_infos["default"]["splits"][split].keys():
+ if subkey == "num_bytes":
+ assert round(dataset_infos["default"]["splits"][split][subkey] / 10**2) == round(
+ expected_dataset_infos["default"]["splits"][split][subkey] / 10**2
+ )
+ else:
+ assert (
+ dataset_infos["default"]["splits"][split][subkey]
+ == expected_dataset_infos["default"]["splits"][split][subkey]
+ )
| NonMatchingChecksumError when checksum is None
I ran into the following error when adding a new dataset:
```bash
expected_checksums = {'https://adversarialglue.github.io/dataset/dev.zip': {'checksum': None, 'num_bytes': 40662}}
recorded_checksums = {'https://adversarialglue.github.io/dataset/dev.zip': {'checksum': 'efb4cbd3aa4a87bfaffc310ae951981cc0a36c6c71c6425dd74e5b55f2f325c9', 'num_bytes': 40662}}
verification_name = 'dataset source files'
def verify_checksums(expected_checksums: Optional[dict], recorded_checksums: dict, verification_name=None):
if expected_checksums is None:
logger.info("Unable to verify checksums.")
return
if len(set(expected_checksums) - set(recorded_checksums)) > 0:
raise ExpectedMoreDownloadedFiles(str(set(expected_checksums) - set(recorded_checksums)))
if len(set(recorded_checksums) - set(expected_checksums)) > 0:
raise UnexpectedDownloadedFile(str(set(recorded_checksums) - set(expected_checksums)))
bad_urls = [url for url in expected_checksums if expected_checksums[url] != recorded_checksums[url]]
for_verification_name = " for " + verification_name if verification_name is not None else ""
if len(bad_urls) > 0:
error_msg = "Checksums didn't match" + for_verification_name + ":\n"
> raise NonMatchingChecksumError(error_msg + str(bad_urls))
E datasets.utils.info_utils.NonMatchingChecksumError: Checksums didn't match for dataset source files:
E ['https://adversarialglue.github.io/dataset/dev.zip']
src/datasets/utils/info_utils.py:40: NonMatchingChecksumError
```
## Expected results
The dataset downloads correctly, and there is no error.
## Actual results
Datasets library is looking for a checksum of None, and it gets a non-None checksum, and throws an error. This is clearly a bug.
| Hi @jxmorris12, thanks for reporting.
The objective of `verify_checksums` is to check that both checksums are equal. Therefore if one is None and the other is non-None, they are not equal, and the function accordingly raises a NonMatchingChecksumError. That behavior is expected.
The question is: how did you generate the expected checksum? Normally, it should not be None. To properly generate it (it is contained in the `dataset_infos.json` file), you should have runned: https://github.com/huggingface/datasets/blob/master/ADD_NEW_DATASET.md
```shell
datasets-cli test <your-dataset-folder> --save_infos --all_configs
```
On the other hand, you should take into account that the generation of this file is NOT mandatory for personal/community datasets (we only require it for "canonical" datasets, i.e., datasets added to our library GitHub repository: https://github.com/huggingface/datasets/tree/master/datasets). Therefore, other option would be just to delete the `dataset_infos.json` file. If that file is not present, the function `verify_checksums` is not executed.
Finally, you can circumvent the `verify_checksums` function by passing `ignore_verifications=True` to `load_dataset`:
```python
load_dataset(..., ignore_verifications=True)
```
Thanks @albertvillanova!
That's fine. I did run that command when I was adding a new dataset. Maybe because the command crashed in the middle, the checksum wasn't stored properly. I don't know where the bug is happening. But either (i) `verify_checksums` should properly handle this edge case, where the passed checksum is None or (ii) the `datasets-cli test` shouldn't generate a corrupted dataset_infos.json file.
Just a more high-level thing, I was trying to follow the instructions for adding a dataset in the CONTRIBUTING.md, so if running that command isn't even necessary, that should probably be mentioned in the document, right? But that's somewhat of a moot point, since something isn't working quite right internally if I was able to get into this corrupted state in the first place, just by following those instructions.
Hi @jxmorris12,
Definitely, your `dataset_infos.json` was corrupted (and wrongly contains expected None checksum).
While we further investigate how this can happen and fix it, feel free to delete your `dataset_infos.json` file and recreate it with:
```shell
datasets-cli test <your-dataset-folder> --save_infos --all_configs
```
Also note that `verify_checksum` is working as expected: if it receives a None and and a non-None checksums as input pair, it must raise an exception: they are not equal. That is not a bug.
At a higher level, also note that we are preparing the release of `datasets` version 2.0, and some docs are being updated...
In order to add a dataset, I think the most updated instructions are in our official documentation pages: https://huggingface.co/docs/datasets/share
Thanks for the info. Maybe you can update the contributing.md if it's not up-to-date. | 2022-03-11T10:04:04Z | [] | [] |
huggingface/datasets | 3,897 | huggingface__datasets-3897 | [
"3586"
] | 3dfdb054e3636a5784d365bcd65759e5bd697b03 | diff --git a/metrics/perplexity/perplexity.py b/metrics/perplexity/perplexity.py
--- a/metrics/perplexity/perplexity.py
+++ b/metrics/perplexity/perplexity.py
@@ -17,7 +17,7 @@
from transformers import AutoModelForCausalLM, AutoTokenizer
import datasets
-from datasets import utils
+from datasets import logging
_CITATION = """\
@@ -113,7 +113,7 @@ def _compute(self, input_texts, model_id, stride=512, device=None):
ppls = []
- for text_index in utils.tqdm_utils.tqdm(range(0, len(encoded_texts))):
+ for text_index in logging.tqdm(range(0, len(encoded_texts))):
encoded_text = encoded_texts[text_index]
special_tokens_mask = special_tokens_masks[text_index]
diff --git a/src/datasets/__init__.py b/src/datasets/__init__.py
--- a/src/datasets/__init__.py
+++ b/src/datasets/__init__.py
@@ -40,7 +40,7 @@
from .combine import interleave_datasets
from .dataset_dict import DatasetDict, IterableDatasetDict
from .features import *
-from .fingerprint import is_caching_enabled, set_caching_enabled
+from .fingerprint import disable_caching, enable_caching, is_caching_enabled, set_caching_enabled
from .info import DatasetInfo, MetricInfo
from .inspect import (
get_dataset_config_info,
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -55,7 +55,7 @@
from requests import HTTPError
from tqdm.auto import tqdm
-from . import config, utils
+from . import config
from .arrow_reader import ArrowReader
from .arrow_writer import ArrowWriter, OptimizedTypedSequence
from .features import Audio, ClassLabel, Features, Image, Sequence, Value
@@ -1947,7 +1947,7 @@ def decorated(item, *args, **kwargs):
f"num_proc must be <= {len(self)}. Reducing num_proc to {num_proc} for dataset of size {len(self)}."
)
- disable_tqdm = not utils.is_progress_bar_enabled()
+ disable_tqdm = not logging.is_progress_bar_enabled()
if num_proc is None or num_proc == 1:
return self._map_single(
@@ -2305,7 +2305,7 @@ def init_buffer_and_writer():
pbar_total = (num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size
pbar_unit = "ex" if not batched else "ba"
pbar_desc = (desc + " " if desc is not None else "") + "#" + str(rank) if rank is not None else desc
- pbar = utils.tqdm_utils.tqdm(
+ pbar = logging.tqdm(
pbar_iterable,
total=pbar_total,
disable=disable_tqdm,
@@ -3455,20 +3455,20 @@ def delete_file(file):
api.delete_file(file, repo_id=repo_id, token=token, repo_type="dataset", revision=branch)
if len(file_shards_to_delete):
- for file in utils.tqdm_utils.tqdm(
+ for file in logging.tqdm(
file_shards_to_delete,
desc="Deleting unused files from dataset repository",
total=len(file_shards_to_delete),
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
):
delete_file(file)
uploaded_size = 0
- for index, shard in utils.tqdm_utils.tqdm(
+ for index, shard in logging.tqdm(
enumerate(shards),
desc="Pushing dataset shards to the dataset hub",
total=num_shards,
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
):
buffer = BytesIO()
shard.to_parquet(buffer)
diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -22,7 +22,7 @@
import numpy as np
import pyarrow as pa
-from . import config, utils
+from . import config
from .features import Features, Image, Value
from .features.features import (
FeatureType,
@@ -637,11 +637,11 @@ def finalize(self, metrics_query_result: dict):
def parquet_to_arrow(sources, destination):
"""Convert parquet files to arrow file. Inputs can be str paths or file-like objects"""
stream = None if isinstance(destination, str) else destination
- disable = not utils.is_progress_bar_enabled()
+ disable = not logging.is_progress_bar_enabled()
with ArrowWriter(path=destination, stream=stream) as writer:
- for source in utils.tqdm_utils.tqdm(sources, unit="sources", disable=disable):
+ for source in logging.tqdm(sources, unit="sources", disable=disable):
pf = pa.parquet.ParquetFile(source)
- for i in utils.tqdm_utils.tqdm(range(pf.num_row_groups), unit="row_groups", leave=False, disable=disable):
+ for i in logging.tqdm(range(pf.num_row_groups), unit="row_groups", leave=False, disable=disable):
df = pf.read_row_group(i).to_pandas()
for col in df.columns:
df[col] = df[col].apply(json.loads)
diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -777,7 +777,7 @@ def as_dataset(
),
split,
map_tuple=True,
- disable_tqdm=not utils.is_progress_bar_enabled(),
+ disable_tqdm=not logging.is_progress_bar_enabled(),
)
if isinstance(datasets, dict):
datasets = DatasetDict(datasets)
@@ -1081,12 +1081,12 @@ def _prepare_split(self, split_generator, check_duplicate_keys):
check_duplicates=check_duplicate_keys,
) as writer:
try:
- for key, record in utils.tqdm_utils.tqdm(
+ for key, record in logging.tqdm(
generator,
unit=" examples",
total=split_info.num_examples,
leave=False,
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
desc=f"Generating {split_info.name} split",
):
example = self.info.features.encode_example(record)
@@ -1145,8 +1145,8 @@ def _prepare_split(self, split_generator):
generator = self._generate_tables(**split_generator.gen_kwargs)
with ArrowWriter(features=self.info.features, path=fpath) as writer:
- for key, table in utils.tqdm_utils.tqdm(
- generator, unit=" tables", leave=False, disable=True # not utils.is_progress_bar_enabled()
+ for key, table in logging.tqdm(
+ generator, unit=" tables", leave=False, disable=True # not logging.is_progress_bar_enabled()
):
writer.write_table(table)
num_examples, num_bytes = writer.finalize()
diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -12,7 +12,6 @@
from .utils import logging
from .utils.file_utils import hf_hub_url, is_remote_url, request_etag
from .utils.py_utils import string_to_dict
-from .utils.tqdm_utils import is_progress_bar_enabled, tqdm
DEFAULT_SPLIT = str(Split.TRAIN)
@@ -494,9 +493,9 @@ def _get_origin_metadata_locally_or_by_urls(
partial(_get_single_origin_metadata_locally_or_by_urls, use_auth_token=use_auth_token),
data_files,
max_workers=max_workers,
- tqdm_class=tqdm,
+ tqdm_class=logging.tqdm,
desc="Resolving data files",
- disable=len(data_files) <= 16 or not is_progress_bar_enabled(),
+ disable=len(data_files) <= 16 or not logging.is_progress_bar_enabled(),
)
diff --git a/src/datasets/fingerprint.py b/src/datasets/fingerprint.py
--- a/src/datasets/fingerprint.py
+++ b/src/datasets/fingerprint.py
@@ -16,6 +16,7 @@
from .info import DatasetInfo
from .table import ConcatenationTable, InMemoryTable, MemoryMappedTable, Table
+from .utils.deprecation_utils import deprecated
from .utils.logging import get_logger
from .utils.py_utils import dumps
@@ -92,6 +93,51 @@ def get_datasets_with_cache_file_in_temp_dir():
return list(_DATASETS_WITH_TABLE_IN_TEMP_DIR) if _DATASETS_WITH_TABLE_IN_TEMP_DIR is not None else []
+def enable_caching():
+ """
+ When applying transforms on a dataset, the data are stored in cache files.
+ The caching mechanism allows to reload an existing cache file if it's already been computed.
+
+ Reloading a dataset is possible since the cache files are named using the dataset fingerprint, which is updated
+ after each transform.
+
+ If disabled, the library will no longer reload cached datasets files when applying transforms to the datasets.
+ More precisely, if the caching is disabled:
+ - cache files are always recreated
+ - cache files are written to a temporary directory that is deleted when session closes
+ - cache files are named using a random hash instead of the dataset fingerprint
+ - use :func:`datasets.Dataset.save_to_disk` to save a transformed dataset or it will be deleted when session closes
+ - caching doesn't affect :func:`datasets.load_dataset`. If you want to regenerate a dataset from scratch you should use
+ the ``download_mode`` parameter in :func:`datasets.load_dataset`.
+ """
+ global _CACHING_ENABLED
+ _CACHING_ENABLED = True
+
+
+def disable_caching():
+ """
+ When applying transforms on a dataset, the data are stored in cache files.
+ The caching mechanism allows to reload an existing cache file if it's already been computed.
+
+ Reloading a dataset is possible since the cache files are named using the dataset fingerprint, which is updated
+ after each transform.
+
+ If disabled, the library will no longer reload cached datasets files when applying transforms to the datasets.
+ More precisely, if the caching is disabled:
+ - cache files are always recreated
+ - cache files are written to a temporary directory that is deleted when session closes
+ - cache files are named using a random hash instead of the dataset fingerprint
+ - use :func:`datasets.Dataset.save_to_disk` to save a transformed dataset or it will be deleted when session closes
+ - caching doesn't affect :func:`datasets.load_dataset`. If you want to regenerate a dataset from scratch you should use
+ the ``download_mode`` parameter in :func:`datasets.load_dataset`.
+ """
+ global _CACHING_ENABLED
+ _CACHING_ENABLED = False
+
+
+@deprecated(
+ "Use datasets.enable_caching() or datasets.disable_caching() instead. This function will be removed in a future version of datasets."
+)
def set_caching_enabled(boolean: bool):
"""
When applying transforms on a dataset, the data are stored in cache files.
diff --git a/src/datasets/io/csv.py b/src/datasets/io/csv.py
--- a/src/datasets/io/csv.py
+++ b/src/datasets/io/csv.py
@@ -2,9 +2,10 @@
import os
from typing import BinaryIO, Optional, Union
-from .. import Dataset, Features, NamedSplit, config, utils
+from .. import Dataset, Features, NamedSplit, config
from ..formatting import query_table
from ..packaged_modules.csv.csv import Csv
+from ..utils import logging
from ..utils.typing import NestedDataStructureLike, PathLike
from .abc import AbstractDatasetReader
@@ -104,10 +105,10 @@ def _write(self, file_obj: BinaryIO, header: bool = True, **to_csv_kwargs) -> in
written = 0
if self.num_proc is None or self.num_proc == 1:
- for offset in utils.tqdm_utils.tqdm(
+ for offset in logging.tqdm(
range(0, len(self.dataset), self.batch_size),
unit="ba",
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
desc="Creating CSV from Arrow format",
):
csv_str = self._batch_csv((offset, header, to_csv_kwargs))
@@ -116,14 +117,14 @@ def _write(self, file_obj: BinaryIO, header: bool = True, **to_csv_kwargs) -> in
else:
num_rows, batch_size = len(self.dataset), self.batch_size
with multiprocessing.Pool(self.num_proc) as pool:
- for csv_str in utils.tqdm_utils.tqdm(
+ for csv_str in logging.tqdm(
pool.imap(
self._batch_csv,
[(offset, header, to_csv_kwargs) for offset in range(0, num_rows, batch_size)],
),
total=(num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size,
unit="ba",
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
desc="Creating CSV from Arrow format",
):
written += file_obj.write(csv_str)
diff --git a/src/datasets/io/json.py b/src/datasets/io/json.py
--- a/src/datasets/io/json.py
+++ b/src/datasets/io/json.py
@@ -4,9 +4,10 @@
import fsspec
-from .. import Dataset, Features, NamedSplit, config, utils
+from .. import Dataset, Features, NamedSplit, config
from ..formatting import query_table
from ..packaged_modules.json.json import Json
+from ..utils import logging
from ..utils.typing import NestedDataStructureLike, PathLike
from .abc import AbstractDatasetReader
@@ -126,10 +127,10 @@ def _write(
written = 0
if self.num_proc is None or self.num_proc == 1:
- for offset in utils.tqdm_utils.tqdm(
+ for offset in logging.tqdm(
range(0, len(self.dataset), self.batch_size),
unit="ba",
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
desc="Creating json from Arrow format",
):
json_str = self._batch_json((offset, orient, lines, to_json_kwargs))
@@ -137,14 +138,14 @@ def _write(
else:
num_rows, batch_size = len(self.dataset), self.batch_size
with multiprocessing.Pool(self.num_proc) as pool:
- for json_str in utils.tqdm_utils.tqdm(
+ for json_str in logging.tqdm(
pool.imap(
self._batch_json,
[(offset, orient, lines, to_json_kwargs) for offset in range(0, num_rows, batch_size)],
),
total=(num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size,
unit="ba",
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
desc="Creating json from Arrow format",
):
written += file_obj.write(json_str)
diff --git a/src/datasets/search.py b/src/datasets/search.py
--- a/src/datasets/search.py
+++ b/src/datasets/search.py
@@ -6,7 +6,6 @@
import numpy as np
-from . import utils
from .utils import logging
@@ -150,9 +149,7 @@ def add_documents(self, documents: Union[List[str], "Dataset"], column: Optional
index_config = self.es_index_config
self.es_client.indices.create(index=index_name, body=index_config)
number_of_docs = len(documents)
- progress = utils.tqdm_utils.tqdm(
- unit="docs", total=number_of_docs, disable=not utils.is_progress_bar_enabled()
- )
+ progress = logging.tqdm(unit="docs", total=number_of_docs, disable=not logging.is_progress_bar_enabled())
successes = 0
def passage_generator():
@@ -295,9 +292,7 @@ def add_vectors(
# Add vectors
logger.info(f"Adding {len(vectors)} vectors to the faiss index")
- for i in utils.tqdm_utils.tqdm(
- range(0, len(vectors), batch_size), disable=not utils.is_progress_bar_enabled()
- ):
+ for i in logging.tqdm(range(0, len(vectors), batch_size), disable=not logging.is_progress_bar_enabled()):
vecs = vectors[i : i + batch_size] if column is None else vectors[i : i + batch_size][column]
self.faiss_index.add(vecs)
diff --git a/src/datasets/utils/__init__.py b/src/datasets/utils/__init__.py
--- a/src/datasets/utils/__init__.py
+++ b/src/datasets/utils/__init__.py
@@ -21,11 +21,11 @@
"DownloadManager",
"DownloadMode",
"disable_progress_bar",
+ "enable_progress_bar",
"is_progress_bar_enabled",
- "set_progress_bar_enabled",
"Version",
]
from .download_manager import DownloadConfig, DownloadManager, DownloadMode
-from .tqdm_utils import disable_progress_bar, is_progress_bar_enabled, set_progress_bar_enabled
+from .logging import disable_progress_bar, enable_progress_bar, is_progress_bar_enabled
from .version import Version
diff --git a/src/datasets/utils/download_manager.py b/src/datasets/utils/download_manager.py
--- a/src/datasets/utils/download_manager.py
+++ b/src/datasets/utils/download_manager.py
@@ -23,7 +23,7 @@
from functools import partial
from typing import Callable, Dict, Generator, Iterable, List, Optional, Tuple, Union
-from .. import config, utils
+from .. import config
from .deprecation_utils import DeprecatedEnum
from .file_utils import (
DownloadConfig,
@@ -34,7 +34,7 @@
url_or_path_join,
)
from .info_utils import get_size_checksum_dict
-from .logging import get_logger
+from .logging import get_logger, is_progress_bar_enabled
from .py_utils import NestedDataStructure, map_nested, size_str
@@ -203,7 +203,7 @@ def upload(local_file_path):
uploaded_path_or_paths = map_nested(
lambda local_file_path: upload(local_file_path),
downloaded_path_or_paths,
- disable_tqdm=not utils.is_progress_bar_enabled(),
+ disable_tqdm=not is_progress_bar_enabled(),
)
return uploaded_path_or_paths
@@ -236,7 +236,7 @@ def url_to_downloaded_path(url):
return os.path.join(cache_dir, hash_url_to_filename(url))
downloaded_path_or_paths = map_nested(
- url_to_downloaded_path, url_or_urls, disable_tqdm=not utils.is_progress_bar_enabled()
+ url_to_downloaded_path, url_or_urls, disable_tqdm=not is_progress_bar_enabled()
)
url_or_urls = NestedDataStructure(url_or_urls)
downloaded_path_or_paths = NestedDataStructure(downloaded_path_or_paths)
@@ -284,7 +284,7 @@ def download(self, url_or_urls):
url_or_urls,
map_tuple=True,
num_proc=download_config.num_proc,
- disable_tqdm=not utils.is_progress_bar_enabled(),
+ disable_tqdm=not is_progress_bar_enabled(),
desc="Downloading data files",
)
duration = datetime.now() - start_time
@@ -356,7 +356,7 @@ def extract(self, path_or_paths, num_proc=None):
partial(cached_path, download_config=download_config),
path_or_paths,
num_proc=num_proc,
- disable_tqdm=not utils.is_progress_bar_enabled(),
+ disable_tqdm=not is_progress_bar_enabled(),
desc="Extracting data files",
)
path_or_paths = NestedDataStructure(path_or_paths)
diff --git a/src/datasets/utils/file_utils.py b/src/datasets/utils/file_utils.py
--- a/src/datasets/utils/file_utils.py
+++ b/src/datasets/utils/file_utils.py
@@ -25,7 +25,7 @@
import requests
-from .. import __version__, config, utils
+from .. import __version__, config
from . import logging
from .extract import ExtractManager
from .filelock import FileLock
@@ -392,13 +392,13 @@ def http_get(
return
content_length = response.headers.get("Content-Length")
total = resume_size + int(content_length) if content_length is not None else None
- with utils.tqdm_utils.tqdm(
+ with logging.tqdm(
unit="B",
unit_scale=True,
total=total,
initial=resume_size,
desc=desc or "Downloading",
- disable=not utils.is_progress_bar_enabled(),
+ disable=not logging.is_progress_bar_enabled(),
) as progress:
for chunk in response.iter_content(chunk_size=1024):
progress.update(len(chunk))
diff --git a/src/datasets/utils/logging.py b/src/datasets/utils/logging.py
--- a/src/datasets/utils/logging.py
+++ b/src/datasets/utils/logging.py
@@ -25,6 +25,8 @@
from logging import WARNING # NOQA
from typing import Optional
+from tqdm import auto as tqdm_lib
+
log_levels = {
"debug": logging.DEBUG,
@@ -167,3 +169,68 @@ def enable_propagation() -> None:
# Configure the library root logger at the module level (singleton-like)
_configure_library_root_logger()
+
+
+class EmptyTqdm:
+ """Dummy tqdm which doesn't do anything."""
+
+ def __init__(self, *args, **kwargs): # pylint: disable=unused-argument
+ self._iterator = args[0] if args else None
+
+ def __iter__(self):
+ return iter(self._iterator)
+
+ def __getattr__(self, _):
+ """Return empty function."""
+
+ def empty_fn(*args, **kwargs): # pylint: disable=unused-argument
+ return
+
+ return empty_fn
+
+ def __enter__(self):
+ return self
+
+ def __exit__(self, type_, value, traceback):
+ return
+
+
+_tqdm_active = True
+
+
+class _tqdm_cls:
+ def __call__(self, *args, **kwargs):
+ if _tqdm_active:
+ return tqdm_lib.tqdm(*args, **kwargs)
+ else:
+ return EmptyTqdm(*args, **kwargs)
+
+ def set_lock(self, *args, **kwargs):
+ self._lock = None
+ if _tqdm_active:
+ return tqdm_lib.tqdm.set_lock(*args, **kwargs)
+
+ def get_lock(self):
+ if _tqdm_active:
+ return tqdm_lib.tqdm.get_lock()
+
+
+tqdm = _tqdm_cls()
+
+
+def is_progress_bar_enabled() -> bool:
+ """Return a boolean indicating whether tqdm progress bars are enabled."""
+ global _tqdm_active
+ return bool(_tqdm_active)
+
+
+def enable_progress_bar():
+ """Enable tqdm progress bar."""
+ global _tqdm_active
+ _tqdm_active = True
+
+
+def disable_progress_bar():
+ """Enable tqdm progress bar."""
+ global _tqdm_active
+ _tqdm_active = False
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -36,7 +36,7 @@
import numpy as np
from tqdm.auto import tqdm
-from .. import config, utils
+from .. import config
from . import logging
@@ -261,7 +261,7 @@ def _single_map_nested(args):
# Loop over single examples or batches and write to buffer/file if examples are to be updated
pbar_iterable = data_struct.items() if isinstance(data_struct, dict) else data_struct
pbar_desc = (desc + " " if desc is not None else "") + "#" + str(rank) if rank is not None else desc
- pbar = utils.tqdm_utils.tqdm(pbar_iterable, disable=disable_tqdm, position=rank, unit="obj", desc=pbar_desc)
+ pbar = logging.tqdm(pbar_iterable, disable=disable_tqdm, position=rank, unit="obj", desc=pbar_desc)
if isinstance(data_struct, dict):
return {k: _single_map_nested((function, v, types, None, True, None)) for k, v in pbar}
@@ -305,7 +305,7 @@ def map_nested(
if not isinstance(data_struct, dict) and not isinstance(data_struct, types):
return function(data_struct)
- disable_tqdm = disable_tqdm or not utils.is_progress_bar_enabled()
+ disable_tqdm = disable_tqdm or not logging.is_progress_bar_enabled()
iterable = list(data_struct.values()) if isinstance(data_struct, dict) else data_struct
if num_proc is None:
@@ -313,7 +313,7 @@ def map_nested(
if num_proc <= 1 or len(iterable) <= num_proc:
mapped = [
_single_map_nested((function, obj, types, None, True, None))
- for obj in utils.tqdm_utils.tqdm(iterable, disable=disable_tqdm, desc=desc)
+ for obj in logging.tqdm(iterable, disable=disable_tqdm, desc=desc)
]
else:
split_kwds = [] # We organize the splits ourselve (contiguous splits)
diff --git a/src/datasets/utils/tqdm_utils.py b/src/datasets/utils/tqdm_utils.py
deleted file mode 100644
--- a/src/datasets/utils/tqdm_utils.py
+++ /dev/null
@@ -1,94 +0,0 @@
-# Copyright 2020 The HuggingFace Datasets Authors and the TensorFlow Datasets Authors.
-#
-# Licensed under the Apache License, Version 2.0 (the "License");
-# you may not use this file except in compliance with the License.
-# You may obtain a copy of the License at
-#
-# http://www.apache.org/licenses/LICENSE-2.0
-#
-# Unless required by applicable law or agreed to in writing, software
-# distributed under the License is distributed on an "AS IS" BASIS,
-# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
-# See the License for the specific language governing permissions and
-# limitations under the License.
-
-# Lint as: python3
-"""Wrapper around tqdm.
-"""
-from tqdm import auto as tqdm_lib
-
-from .deprecation_utils import deprecated
-
-
-class EmptyTqdm:
- """Dummy tqdm which doesn't do anything."""
-
- def __init__(self, *args, **kwargs): # pylint: disable=unused-argument
- self._iterator = args[0] if args else None
-
- def __iter__(self):
- return iter(self._iterator)
-
- def __getattr__(self, _):
- """Return empty function."""
-
- def empty_fn(*args, **kwargs): # pylint: disable=unused-argument
- return
-
- return empty_fn
-
- def __enter__(self):
- return self
-
- def __exit__(self, type_, value, traceback):
- return
-
-
-_active = True
-
-
-class _tqdm_cls:
- def __call__(self, *args, **kwargs):
- if _active:
- return tqdm_lib.tqdm(*args, **kwargs)
- else:
- return EmptyTqdm(*args, **kwargs)
-
- def set_lock(self, *args, **kwargs):
- self._lock = None
- if _active:
- return tqdm_lib.tqdm.set_lock(*args, **kwargs)
-
- def get_lock(self):
- if _active:
- return tqdm_lib.tqdm.get_lock()
-
-
-tqdm = _tqdm_cls()
-
-
-def set_progress_bar_enabled(boolean: bool):
- """Enable/disable tqdm progress bars."""
- global _active
- _active = bool(boolean)
-
-
-def is_progress_bar_enabled() -> bool:
- """Return a boolean indicating whether tqdm progress bars are enabled."""
- global _active
- return bool(_active)
-
-
-@deprecated("Use set_progress_bar_enabled(False) instead.")
-def disable_progress_bar():
- """Disable tqdm progress bar.
-
- .. deprecated:: 1.12.2
- Use set_progress_bar_enabled(False) instead.
-
- Usage:
-
- datasets.disable_progress_bar()
- """
- global _active
- _active = False
| diff --git a/tests/conftest.py b/tests/conftest.py
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -36,7 +36,7 @@ def set_test_cache_config(tmp_path_factory, monkeypatch):
@pytest.fixture(autouse=True, scope="session")
def disable_tqdm_output():
- datasets.set_progress_bar_enabled(False)
+ datasets.disable_progress_bar()
@pytest.fixture(autouse=True)
diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -1100,7 +1100,7 @@ def test_map_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._caplog.at_level(WARNING):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
- datasets.set_caching_enabled(False)
+ datasets.disable_caching()
with dset.map(lambda x: {"foo": "bar"}) as dset_test1:
with dset.map(lambda x: {"foo": "bar"}) as dset_test2:
self.assertNotEqual(dset_test1.cache_files, dset_test2.cache_files)
@@ -1111,7 +1111,7 @@ def test_map_caching(self, in_memory):
self.assertIn("tmp", dset_test1.cache_files[0]["filename"])
self.assertIn("tmp", dset_test2.cache_files[0]["filename"])
finally:
- datasets.set_caching_enabled(True)
+ datasets.enable_caching()
@require_torch
def test_map_torch(self, in_memory):
diff --git a/tests/test_tqdm_utils.py b/tests/test_logging.py
similarity index 74%
rename from tests/test_tqdm_utils.py
rename to tests/test_logging.py
--- a/tests/test_tqdm_utils.py
+++ b/tests/test_logging.py
@@ -4,16 +4,16 @@
from datasets import Dataset
-def test_set_progress_bar_enabled():
+def test_enable_disable_progress_bar():
dset = Dataset.from_dict({"col_1": [3, 2, 0, 1]})
with patch("tqdm.auto.tqdm") as mock_tqdm:
- datasets.set_progress_bar_enabled(True)
+ datasets.disable_progress_bar()
dset.map(lambda x: {"col_2": x["col_1"] + 1})
- mock_tqdm.assert_called()
+ mock_tqdm.assert_not_called()
mock_tqdm.reset_mock()
- datasets.set_progress_bar_enabled(False)
+ datasets.enable_progress_bar()
dset.map(lambda x: {"col_2": x["col_1"] + 1})
- mock_tqdm.assert_not_called()
+ mock_tqdm.assert_called()
| Revisit `enable/disable_` toggle function prefix
As discussed in https://github.com/huggingface/transformers/pull/15167, we should revisit the `enable/disable_` toggle function prefix, potentially in favor of `set_enabled_`. Concretely, this translates to
- De-deprecating `disable_progress_bar()`
- Adding `enable_progress_bar()`
- On the caching side, adding `enable_caching` and `disable_caching`
Additional decisions have to be made with regards to the existing `set_enabled_X` functions; that is, whether to keep them as is or deprecate them in favor of the aforementioned functions.
cc @mariosasko @lhoestq
| 2022-03-11T18:12:22Z | [] | [] |
|
huggingface/datasets | 3,910 | huggingface__datasets-3910 | [
"3729"
] | 6cf0abaed1c2911e5bf23e48e4908929b43d85ae | diff --git a/src/datasets/packaged_modules/text/text.py b/src/datasets/packaged_modules/text/text.py
--- a/src/datasets/packaged_modules/text/text.py
+++ b/src/datasets/packaged_modules/text/text.py
@@ -1,4 +1,5 @@
from dataclasses import dataclass
+from io import StringIO
from typing import Optional
import pyarrow as pa
@@ -52,7 +53,7 @@ def _split_generators(self, dl_manager):
def _generate_tables(self, files):
schema = pa.schema(self.config.features.type if self.config.features is not None else {"text": pa.string()})
for file_idx, file in enumerate(files):
- batch_idx = 0
+ # open in text mode, by default translates universal newlines ("\n", "\r\n" and "\r") into "\n"
with open(file, encoding=self.config.encoding) as f:
if self.config.sample_by == "line":
batch_idx = 0
@@ -61,7 +62,10 @@ def _generate_tables(self, files):
if not batch:
break
batch += f.readline() # finish current line
- batch = batch.splitlines(keepends=self.config.keep_linebreaks)
+ # StringIO.readlines, by default splits only on "\n" (and keeps line breaks)
+ batch = StringIO(batch).readlines()
+ if not self.config.keep_linebreaks:
+ batch = [line.rstrip("\n") for line in batch]
pa_table = pa.Table.from_arrays([pa.array(batch)], schema=schema)
# Uncomment for debugging (will print the Arrow table size and elements)
# logger.warning(f"pa_table: {pa_table} num rows: {pa_table.num_rows}")
| diff --git a/tests/conftest.py b/tests/conftest.py
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -458,3 +458,12 @@ def zip_text_with_dir_path(text_path, text2_path, tmp_path_factory):
f.write(text_path, arcname=os.path.join("main_dir", os.path.basename(text_path)))
f.write(text2_path, arcname=os.path.join("main_dir", os.path.basename(text2_path)))
return path
+
+
[email protected](scope="session")
+def text_path_with_unicode_new_lines(tmp_path_factory):
+ text = "\n".join(["First", "Second\u2029with Unicode new line", "Third"])
+ path = str(tmp_path_factory.mktemp("data") / "dataset_with_unicode_new_lines.txt")
+ with open(path, "w", encoding="utf-8") as f:
+ f.write(text)
+ return path
diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -651,6 +651,12 @@ def test_load_dataset_zip_text(data_file, streaming, zip_text_path, zip_text_wit
assert ds_item == {"text": "0"}
+def test_load_dataset_text_with_unicode_new_lines(text_path_with_unicode_new_lines):
+ data_files = str(text_path_with_unicode_new_lines)
+ ds = load_dataset("text", split="train", data_files=data_files)
+ assert ds.num_rows == 3
+
+
def test_loading_from_the_datasets_hub():
with tempfile.TemporaryDirectory() as tmp_dir:
dataset = load_dataset(SAMPLE_DATASET_IDENTIFIER, cache_dir=tmp_dir)
diff --git a/tests/test_packaged_modules.py b/tests/test_packaged_modules.py
--- a/tests/test_packaged_modules.py
+++ b/tests/test_packaged_modules.py
@@ -50,7 +50,7 @@ def text_file(tmp_path):
Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
"""
)
- with open(filename, "w") as f:
+ with open(filename, "w", encoding="utf-8") as f:
f.write(data)
return str(filename)
@@ -76,9 +76,9 @@ def test_csv_generate_tables_raises_error_with_malformed_csv(csv_file, malformed
@pytest.mark.parametrize("keep_linebreaks", [True, False])
def test_text_linebreaks(text_file, keep_linebreaks):
- with open(text_file) as f:
+ with open(text_file, encoding="utf-8") as f:
expected_content = f.read().splitlines(keepends=keep_linebreaks)
- text = Text(keep_linebreaks=keep_linebreaks)
+ text = Text(keep_linebreaks=keep_linebreaks, encoding="utf-8")
generator = text._generate_tables([text_file])
generated_content = pa.concat_tables([table for _, table in generator]).to_pydict()["text"]
assert generated_content == expected_content
| Wrong number of examples when loading a text dataset
## Describe the bug
when I use load_dataset to read a txt file I find that the number of the samples is incorrect
## Steps to reproduce the bug
```
fr = open('train.txt','r',encoding='utf-8').readlines()
print(len(fr)) # 1199637
datasets = load_dataset('text', data_files={'train': ['train.txt']}, streaming=False)
print(len(datasets['train'])) # 1199649
```
I also use command line operation to verify it
```
$ wc -l train.txt
1199637 train.txt
```
## Expected results
please fix that issue
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 1.8.3
- Platform:windows&linux
- Python version:3.7
- PyArrow version:6.0.1
| Hi @kg-nlp, thanks for reporting.
That is weird... I guess we would need some sample data file where this behavior appears to reproduce the bug for further investigation...
ok, I found the reason why that two results are not same.
there is /u2029 in the text, the datasets will split sentence according to the /u2029,but when I use open function will not do that .
so I want to know which function shell do that
thanks | 2022-03-14T15:54:58Z | [] | [] |
huggingface/datasets | 3,998 | huggingface__datasets-3998 | [
"3996"
] | b73f7157935a2a93dbc664a2625c701791d83d58 | diff --git a/src/datasets/features/audio.py b/src/datasets/features/audio.py
--- a/src/datasets/features/audio.py
+++ b/src/datasets/features/audio.py
@@ -72,7 +72,7 @@ def encode_example(self, value: Union[str, dict]) -> dict:
return {"bytes": None, "path": value}
elif isinstance(value, dict) and "array" in value:
buffer = BytesIO()
- sf.write(buffer, value["array"], value["sampling_rate"])
+ sf.write(buffer, value["array"], value["sampling_rate"], format="wav")
return {"bytes": buffer.getvalue(), "path": value.get("path")}
elif value.get("bytes") is not None or value.get("path") is not None:
return {"bytes": value.get("bytes"), "path": value.get("path")}
| diff --git a/tests/features/test_audio.py b/tests/features/test_audio.py
--- a/tests/features/test_audio.py
+++ b/tests/features/test_audio.py
@@ -64,6 +64,7 @@ def test_audio_feature_type_to_arrow():
lambda audio_path: {"path": audio_path, "bytes": open(audio_path, "rb").read()},
lambda audio_path: {"path": None, "bytes": open(audio_path, "rb").read()},
lambda audio_path: {"bytes": open(audio_path, "rb").read()},
+ lambda audio_path: {"array": [0.1, 0.2, 0.3], "sampling_rate": 16_000},
],
)
def test_audio_feature_encode_example(shared_datadir, build_example):
| Audio.encode_example() throws an error when writing example from array
## Describe the bug
When trying to do `Audio().encode_example()` with preexisting array (see [this line](https://github.com/huggingface/datasets/blob/master/src/datasets/features/audio.py#L73)), `sf.write()` throws you an error:
`TypeError: No format specified and unable to get format from file extension: <_io.BytesIO object at 0x7f4218c0db30>`
## Steps to reproduce the bug
### Sample code to reproduce the bug
```python
# download sample file
!wget https://huggingface.co/datasets/polinaeterna/test_encode_example/resolve/main/common_voice_vi_21824030.mp3
arr, sr = librosa.load("common_voice_vi_21824030.mp3")
Audio().encode_example({
"path": "common_voice_vi_21824030.mp3",
"array": arr,
"sampling_rate":sr
})
```
## Expected results
An encoded example (`{"bytes": b'....', "path": 'path'}`)
## Actual results
```python
TypeError Traceback (most recent call last)
Input In [3], in <module>
1 arr, sr = librosa.load("common_voice_vi_21824030.mp3")
----> 3 Audio().encode_example({
4 "path": "common_voice_vi_21824030.mp3",
5 "array": arr,
6 "sampling_rate":sr
7 })
File ~/workspace/datasets/src/datasets/features/audio.py:75, in Audio.encode_example(self, value)
73 elif isinstance(value, dict) and "array" in value:
74 buffer = BytesIO()
---> 75 sf.write(buffer, value["array"], value["sampling_rate"])
76 return {"bytes": buffer.getvalue(), "path": value.get("path")}
77 elif value.get("bytes") is not None or value.get("path") is not None:
File ~/miniconda3/envs/datasets/lib/python3.8/site-packages/soundfile.py:314, in write(file, data, samplerate, subtype, endian, format, closefd)
312 else:
313 channels = data.shape[1]
--> 314 with SoundFile(file, 'w', samplerate, channels,
315 subtype, endian, format, closefd) as f:
316 f.write(data)
File ~/miniconda3/envs/datasets/lib/python3.8/site-packages/soundfile.py:627, in SoundFile.__init__(self, file, mode, samplerate, channels, subtype, endian, format, closefd)
625 mode_int = _check_mode(mode)
626 self._mode = mode
--> 627 self._info = _create_info_struct(file, mode, samplerate, channels,
628 format, subtype, endian)
629 self._file = self._open(file, mode_int, closefd)
630 if set(mode).issuperset('r+') and self.seekable():
631 # Move write position to 0 (like in Python file objects)
File ~/miniconda3/envs/datasets/lib/python3.8/site-packages/soundfile.py:1416, in _create_info_struct(file, mode, samplerate, channels, format, subtype, endian)
1414 original_format = format
1415 if format is None:
-> 1416 format = _get_format_from_filename(file, mode)
1417 assert isinstance(format, (_unicode, str))
1418 else:
File ~/miniconda3/envs/datasets/lib/python3.8/site-packages/soundfile.py:1457, in _get_format_from_filename(file, mode)
1455 pass
1456 if format.upper() not in _formats and 'r' not in mode:
-> 1457 raise TypeError("No format specified and unable to get format from "
1458 "file extension: {0!r}".format(file))
1459 return format
TypeError: No format specified and unable to get format from file extension: <_io.BytesIO object at 0x7fd8daf88180>
```
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: datasets master
- Platform: Ubuntu 20.04
- Python version: python 3.8.12
- PyArrow version: 6.0.1
## Solution
I guess we just need to add `format` arg in [this line](https://github.com/huggingface/datasets/blob/master/src/datasets/features/audio.py#L75) like this:
```python
sf.write(buffer, value["array"], value["sampling_rate"], format="wav")
```
BTW discovered this when trying to decode audio in mp3 format without torchaudio (would be useful for TensorFlow users), like this:
```python
from datasets import load_dataset, Features, Audio
ds = load_dataset("common_voice", "vi", split="test")
ds = ds.remove_columns("audio")
ds.select(range(3)) # 3 samples just for testing
def load_mp3_with_librosa(example):
arr, sr = librosa.load(example["path"])
example["audio"] = {
"path": example["path"],
"array": arr,
"sampling_rate": sr
}
return example
updated_dataset = ds.map(lambda example: load_mp3_with_librosa(example),
features=Features(
{"audio": Audio(decode=False)}
))
```
@lhoestq @mariosasko @albertvillanova am I right in my logic? do we agree that we can set wav as the format? 🤗
| Good catch ! Yes I think passing `format="wav"` is the right thing to do | 2022-03-23T20:32:13Z | [] | [] |
huggingface/datasets | 4,045 | huggingface__datasets-4045 | [
"4044"
] | 8171c740351a645c5b93ebd340e4169aa814c4c0 | diff --git a/src/datasets/commands/dummy_data.py b/src/datasets/commands/dummy_data.py
--- a/src/datasets/commands/dummy_data.py
+++ b/src/datasets/commands/dummy_data.py
@@ -358,7 +358,7 @@ def _autogenerate_dummy_data(self, dataset_builder, mock_dl_manager, keep_uncomp
try:
split_generators = dataset_builder._split_generators(mock_dl_manager)
for split_generator in split_generators:
- dataset_builder._prepare_split(split_generator)
+ dataset_builder._prepare_split(split_generator, check_duplicate_keys=False)
n_examples_per_split[split_generator.name] = split_generator.split_info.num_examples
except OSError as e:
logger.error(
| diff --git a/tests/commands/conftest.py b/tests/commands/conftest.py
--- a/tests/commands/conftest.py
+++ b/tests/commands/conftest.py
@@ -66,8 +66,8 @@ def dataset_loading_script_code():
@pytest.fixture
def dataset_loading_script_dir(dataset_loading_script_name, dataset_loading_script_code, tmp_path):
script_name = dataset_loading_script_name
- script_dir = tmp_path / script_name
- script_dir.mkdir()
+ script_dir = tmp_path / "datasets" / script_name
+ script_dir.mkdir(parents=True)
script_path = script_dir / f"{script_name}.py"
with open(script_path, "w") as f:
f.write(dataset_loading_script_code)
diff --git a/tests/commands/test_dummy_data.py b/tests/commands/test_dummy_data.py
new file mode 100644
--- /dev/null
+++ b/tests/commands/test_dummy_data.py
@@ -0,0 +1,58 @@
+import os
+from collections import namedtuple
+from dataclasses import dataclass
+
+from packaging import version
+
+from datasets import config
+from datasets.commands.dummy_data import DummyDataCommand
+
+
+if config.PY_VERSION >= version.parse("3.7"):
+ DummyDataCommandArgs = namedtuple(
+ "DummyDataCommandArgs",
+ [
+ "path_to_dataset",
+ "auto_generate",
+ "n_lines",
+ "json_field",
+ "xml_tag",
+ "match_text_files",
+ "keep_uncompressed",
+ "cache_dir",
+ "encoding",
+ ],
+ defaults=[False, 5, None, None, None, False, None, None],
+ )
+else:
+
+ @dataclass
+ class DummyDataCommandArgs:
+ path_to_dataset: str
+ auto_generate: bool = False
+ n_lines: int = 5
+ json_field: str = None
+ xml_tag: str = None
+ match_text_files: str = None
+ keep_uncompressed: bool = False
+ cache_dir: str = None
+ encoding: str = None
+
+ def __iter__(self):
+ return iter(self.__dict__.values())
+
+
+class MockDummyDataCommand(DummyDataCommand):
+ def _autogenerate_dummy_data(self, dataset_builder, mock_dl_manager, keep_uncompressed):
+ mock_dl_manager.datasets_scripts_dir = os.path.abspath(os.path.join(self._path_to_dataset, os.pardir))
+ return super()._autogenerate_dummy_data(dataset_builder, mock_dl_manager, keep_uncompressed)
+
+
+def test_dummy_data_command(dataset_loading_script_dir, capfd):
+ args = DummyDataCommandArgs(path_to_dataset=dataset_loading_script_dir, auto_generate=True)
+ dummy_data_command = MockDummyDataCommand(*args)
+ _ = capfd.readouterr()
+ dummy_data_command.run()
+ assert os.path.exists(os.path.join(dataset_loading_script_dir, "dummy", "0.0.0", "dummy_data.zip"))
+ captured = capfd.readouterr()
+ assert captured.out.startswith("Automatic dummy data generation succeeded for all configs of")
| CLI dummy data generation is broken
## Describe the bug
We get a TypeError when running CLI dummy data generation:
```shell
datasets-cli dummy_data datasets/<your-dataset-folder> --auto_generate
```
gives:
```
File ".../huggingface/datasets/src/datasets/commands/dummy_data.py", line 361, in _autogenerate_dummy_data
dataset_builder._prepare_split(split_generator)
TypeError: _prepare_split() missing 1 required positional argument: 'check_duplicate_keys'
```
| 2022-03-28T16:09:15Z | [] | [] |
|
huggingface/datasets | 4,059 | huggingface__datasets-4059 | [
"2048"
] | 51aef08ad7053c0bfe8f9a961207b26df15850d3 | diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -424,93 +424,6 @@ def get_module(self) -> MetricModule:
raise NotImplementedError
-class GithubDatasetModuleFactory(_DatasetModuleFactory):
- """
- Get the module of a dataset from GitHub (legacy).
- The dataset script is downloaded from GitHub.
- This class will eventually be removed and a HubDatasetModuleFactory will be used instead.
- """
-
- def __init__(
- self,
- name: str,
- revision: Optional[Union[str, Version]] = None,
- download_config: Optional[DownloadConfig] = None,
- download_mode: Optional[DownloadMode] = None,
- dynamic_modules_path: Optional[str] = None,
- ):
- self.name = name
- self.revision = revision
- self.download_config = download_config.copy() if download_config else DownloadConfig()
- if self.download_config.max_retries < 3:
- self.download_config.max_retries = 3
- self.download_mode = download_mode
- self.dynamic_modules_path = dynamic_modules_path
- assert self.name.count("/") == 0
- increase_load_count(name, resource_type="dataset")
-
- def download_loading_script(self, revision: Optional[str]) -> str:
- file_path = hf_github_url(path=self.name, name=self.name + ".py", revision=revision)
- download_config = self.download_config.copy()
- if download_config.download_desc is None:
- download_config.download_desc = "Downloading builder script"
- return cached_path(file_path, download_config=download_config)
-
- def download_dataset_infos_file(self, revision: Optional[str]) -> str:
- dataset_infos = hf_github_url(path=self.name, name=config.DATASETDICT_INFOS_FILENAME, revision=revision)
- # Download the dataset infos file if available
- download_config = self.download_config.copy()
- if download_config.download_desc is None:
- download_config.download_desc = "Downloading metadata"
- try:
- return cached_path(
- dataset_infos,
- download_config=download_config,
- )
- except (FileNotFoundError, ConnectionError):
- return None
-
- def get_module(self) -> DatasetModule:
- # get script and other files
- revision = self.revision
- try:
- local_path = self.download_loading_script(revision)
- except FileNotFoundError:
- if revision is not None or os.getenv("HF_SCRIPTS_VERSION", None) is not None:
- raise
- else:
- revision = "main"
- local_path = self.download_loading_script(revision)
- logger.warning(
- f"Couldn't find a directory or a dataset named '{self.name}' in this version. "
- f"It was picked from the main branch on github instead."
- )
- dataset_infos_path = self.download_dataset_infos_file(revision)
- imports = get_imports(local_path)
- local_imports = _download_additional_modules(
- name=self.name,
- base_path=hf_github_url(path=self.name, name="", revision=revision),
- imports=imports,
- download_config=self.download_config,
- )
- additional_files = [(config.DATASETDICT_INFOS_FILENAME, dataset_infos_path)] if dataset_infos_path else []
- # copy the script and the files in an importable directory
- dynamic_modules_path = self.dynamic_modules_path if self.dynamic_modules_path else init_dynamic_modules()
- module_path, hash = _create_importable_file(
- local_path=local_path,
- local_imports=local_imports,
- additional_files=additional_files,
- dynamic_modules_path=dynamic_modules_path,
- module_namespace="datasets",
- name=self.name,
- download_mode=self.download_mode,
- )
- # make the new module to be noticed by the import system
- importlib.invalidate_caches()
- builder_kwargs = {"hash": hash, "base_path": hf_hub_url(self.name, "", revision=self.revision)}
- return DatasetModule(module_path, hash, builder_kwargs)
-
-
class GithubMetricModuleFactory(_MetricModuleFactory):
"""Get the module of a metric. The metric script is downloaded from GitHub.
@@ -917,11 +830,10 @@ def __init__(
self.download_config = download_config or DownloadConfig()
self.download_mode = download_mode
self.dynamic_modules_path = dynamic_modules_path
- assert self.name.count("/") == 1
increase_load_count(name, resource_type="dataset")
def download_loading_script(self) -> str:
- file_path = hf_hub_url(repo_id=self.name, path=self.name.split("/")[1] + ".py", revision=self.revision)
+ file_path = hf_hub_url(repo_id=self.name, path=self.name.split("/")[-1] + ".py", revision=self.revision)
download_config = self.download_config.copy()
if download_config.download_desc is None:
download_config.download_desc = "Downloading builder script"
@@ -1197,67 +1109,57 @@ def dataset_module_factory(
elif is_relative_path(path) and path.count("/") <= 1:
try:
_raise_if_offline_mode_is_enabled()
- if path.count("/") == 0: # even though the dataset is on the Hub, we get it from GitHub for now
- # TODO(QL): use a Hub dataset module factory instead of GitHub
- return GithubDatasetModuleFactory(
+ hf_api = HfApi(config.HF_ENDPOINT)
+ try:
+ if isinstance(download_config.use_auth_token, bool):
+ token = HfFolder.get_token() if download_config.use_auth_token else None
+ else:
+ token = download_config.use_auth_token
+ dataset_info = hf_api.dataset_info(
+ repo_id=path,
+ revision=revision,
+ token=token if token else "no-token",
+ timeout=100.0,
+ )
+ except Exception as e: # noqa: catch any exception of hf_hub and consider that the dataset doesn't exist
+ if isinstance(
+ e,
+ (
+ OfflineModeIsEnabled,
+ requests.exceptions.ConnectTimeout,
+ requests.exceptions.ConnectionError,
+ ),
+ ):
+ raise ConnectionError(f"Couldn't reach '{path}' on the Hub ({type(e).__name__})")
+ elif "404" in str(e):
+ msg = f"Dataset '{path}' doesn't exist on the Hub"
+ raise FileNotFoundError(msg + f" at revision '{revision}'" if revision else msg)
+ elif "401" in str(e):
+ msg = f"Dataset '{path}' doesn't exist on the Hub"
+ msg = msg + f" at revision '{revision}'" if revision else msg
+ raise FileNotFoundError(
+ msg
+ + ". If the repo is private, make sure you are authenticated with `use_auth_token=True` after logging in with `huggingface-cli login`."
+ )
+ else:
+ raise e
+ if filename in [sibling.rfilename for sibling in dataset_info.siblings]:
+ return HubDatasetModuleFactoryWithScript(
path,
revision=revision,
download_config=download_config,
download_mode=download_mode,
dynamic_modules_path=dynamic_modules_path,
).get_module()
- elif path.count("/") == 1: # community dataset on the Hub
- hf_api = HfApi(config.HF_ENDPOINT)
- try:
- if isinstance(download_config.use_auth_token, bool):
- token = HfFolder.get_token() if download_config.use_auth_token else None
- else:
- token = download_config.use_auth_token
- dataset_info = hf_api.dataset_info(
- repo_id=path,
- revision=revision,
- token=token if token else "no-token",
- timeout=100.0,
- )
- except Exception as e: # noqa: catch any exception of hf_hub and consider that the dataset doesn't exist
- if isinstance(
- e,
- (
- OfflineModeIsEnabled,
- requests.exceptions.ConnectTimeout,
- requests.exceptions.ConnectionError,
- ),
- ):
- raise ConnectionError(f"Couldn't reach '{path}' on the Hub ({type(e).__name__})")
- elif "404" in str(e):
- msg = f"Dataset '{path}' doesn't exist on the Hub"
- raise FileNotFoundError(msg + f" at revision '{revision}'" if revision else msg)
- elif "401" in str(e):
- msg = f"Dataset '{path}' doesn't exist on the Hub"
- msg = msg + f" at revision '{revision}'" if revision else msg
- raise FileNotFoundError(
- msg
- + ". If the repo is private, make sure you are authenticated with `use_auth_token=True` after logging in with `huggingface-cli login`."
- )
- else:
- raise e
- if filename in [sibling.rfilename for sibling in dataset_info.siblings]:
- return HubDatasetModuleFactoryWithScript(
- path,
- revision=revision,
- download_config=download_config,
- download_mode=download_mode,
- dynamic_modules_path=dynamic_modules_path,
- ).get_module()
- else:
- return HubDatasetModuleFactoryWithoutScript(
- path,
- revision=revision,
- data_dir=data_dir,
- data_files=data_files,
- download_config=download_config,
- download_mode=download_mode,
- ).get_module()
+ else:
+ return HubDatasetModuleFactoryWithoutScript(
+ path,
+ revision=revision,
+ data_dir=data_dir,
+ data_files=data_files,
+ download_config=download_config,
+ download_mode=download_mode,
+ ).get_module()
except Exception as e1: # noqa: all the attempts failed, before raising the error we should check if the module is already cached.
try:
return CachedDatasetModuleFactory(path, dynamic_modules_path=dynamic_modules_path).get_module()
| diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -1,6 +1,5 @@
import importlib
import os
-import re
import shutil
import tempfile
import time
@@ -13,7 +12,7 @@
import requests
import datasets
-from datasets import SCRIPTS_VERSION, config, load_dataset, load_from_disk
+from datasets import config, load_dataset, load_from_disk
from datasets.arrow_dataset import Dataset
from datasets.builder import DatasetBuilder
from datasets.data_files import DataFilesDict
@@ -24,7 +23,6 @@
from datasets.load import (
CachedDatasetModuleFactory,
CachedMetricModuleFactory,
- GithubDatasetModuleFactory,
GithubMetricModuleFactory,
HubDatasetModuleFactoryWithoutScript,
HubDatasetModuleFactoryWithScript,
@@ -35,7 +33,6 @@
infer_module_for_data_files,
infer_module_for_data_files_in_archives,
)
-from datasets.utils.file_utils import is_remote_url
from .utils import (
OfflineSimulationMode,
@@ -255,9 +252,9 @@ def setUp(self):
hf_modules_cache=self.hf_modules_cache,
)
- def test_GithubDatasetModuleFactory(self):
+ def test_HubDatasetModuleFactoryWithScript_with_github_dataset(self):
# "wmt_t2t" has additional imports (internal)
- factory = GithubDatasetModuleFactory(
+ factory = HubDatasetModuleFactoryWithScript(
"wmt_t2t", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
@@ -479,7 +476,6 @@ def test_CachedMetricModuleFactory(self):
[
CachedDatasetModuleFactory,
CachedMetricModuleFactory,
- GithubDatasetModuleFactory,
GithubMetricModuleFactory,
HubDatasetModuleFactoryWithoutScript,
HubDatasetModuleFactoryWithScript,
@@ -577,17 +573,16 @@ def test_offline_dataset_module_factory(self):
self.assertIn("Using the latest cached version of the module", self._caplog.text)
def test_load_dataset_from_github(self):
- scripts_version = os.getenv("HF_SCRIPTS_VERSION", SCRIPTS_VERSION)
with self.assertRaises(FileNotFoundError) as context:
datasets.load_dataset("_dummy")
self.assertIn(
- "https://raw.githubusercontent.com/huggingface/datasets/main/datasets/_dummy/_dummy.py",
+ "Dataset '_dummy' doesn't exist on the Hub",
str(context.exception),
)
with self.assertRaises(FileNotFoundError) as context:
datasets.load_dataset("_dummy", revision="0.0.0")
self.assertIn(
- "https://raw.githubusercontent.com/huggingface/datasets/0.0.0/datasets/_dummy/_dummy.py",
+ "Dataset '_dummy' doesn't exist on the Hub at revision '0.0.0'",
str(context.exception),
)
for offline_simulation_mode in list(OfflineSimulationMode):
@@ -596,7 +591,7 @@ def test_load_dataset_from_github(self):
datasets.load_dataset("_dummy")
if offline_simulation_mode != OfflineSimulationMode.HF_DATASETS_OFFLINE_SET_TO_1:
self.assertIn(
- f"https://raw.githubusercontent.com/huggingface/datasets/{scripts_version}/datasets/_dummy/_dummy.py",
+ "Couldn't reach '_dummy' on the Hub",
str(context.exception),
)
@@ -708,11 +703,7 @@ def test_load_dataset_local(dataset_loading_script_dir, data_dir, keep_in_memory
assert "Using the latest cached version of the module" in caplog.text
with pytest.raises(FileNotFoundError) as exc_info:
datasets.load_dataset(SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST)
- m_combined_path = re.search(
- rf"http\S*{re.escape(SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST + '/' + SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST + '.py')}\b",
- str(exc_info.value),
- )
- assert m_combined_path is not None and is_remote_url(m_combined_path.group())
+ assert f"Dataset '{SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST}' doesn't exist on the Hub" in str(exc_info.value)
assert os.path.abspath(SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST) in str(exc_info.value)
| github is not always available - probably need a back up
Yesterday morning github wasn't working:
```
:/tmp$ wget https://raw.githubusercontent.com/huggingface/datasets/1.4.1/metrics/sacrebleu/sacrebleu.py--2021-03-12 18:35:59-- https://raw.githubusercontent.com/huggingface/datasets/1.4.1/metrics/sacrebleu/sacrebleu.py
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.111.133, 185.199.109.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.
HTTP request sent, awaiting response... 500 Internal Server Error
2021-03-12 18:36:11 ERROR 500: Internal Server Error.
```
Suggestion: have a failover system and replicate the data on another system and reach there if gh isn't reachable? perhaps gh can be a master and the replicate a slave - so there is only one true source.
| 2022-03-30T09:21:56Z | [] | [] |
|
huggingface/datasets | 4,144 | huggingface__datasets-4144 | [
"4150"
] | e74d69c1d41dd320e77ca7244c624592f1a9fa3d | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -296,10 +296,13 @@ def get_patterns_locally(base_path: str) -> Dict[str, List[str]]:
def _resolve_single_pattern_in_dataset_repository(
dataset_info: huggingface_hub.hf_api.DatasetInfo,
pattern: str,
+ base_path: Optional[str] = None,
allowed_extensions: Optional[list] = None,
) -> List[PurePath]:
data_files_ignore = FILES_TO_IGNORE
fs = HfFileSystem(repo_info=dataset_info)
+ if base_path:
+ pattern = f"{base_path}/{pattern}"
glob_iter = [PurePath(filepath) for filepath in fs.glob(PurePath(pattern).as_posix()) if fs.isfile(filepath)]
matched_paths = [
filepath
@@ -330,6 +333,7 @@ def _resolve_single_pattern_in_dataset_repository(
def resolve_patterns_in_dataset_repository(
dataset_info: huggingface_hub.hf_api.DatasetInfo,
patterns: List[str],
+ base_path: Optional[str] = None,
allowed_extensions: Optional[list] = None,
) -> List[Url]:
"""
@@ -364,6 +368,9 @@ def resolve_patterns_in_dataset_repository(
dataset_info (huggingface_hub.hf_api.DatasetInfo): dataset info obtained using the hugginggace_hub.HfApi
patterns (List[str]): Unix patterns or paths of the files in the dataset repository.
The paths should be relative to the root of the repository.
+ base_path (Optional[str], optional): Path inside a repo to use when resolving relative paths.
+ Defaults to None (search from a repository's root). Used if files only from a specific
+ directory should be resolved.
allowed_extensions (Optional[list], optional): White-list of file extensions to use. Defaults to None (all extensions).
For example: allowed_extensions=["csv", "json", "txt", "parquet"]
@@ -372,7 +379,9 @@ def resolve_patterns_in_dataset_repository(
"""
data_files_urls: List[Url] = []
for pattern in patterns:
- for rel_path in _resolve_single_pattern_in_dataset_repository(dataset_info, pattern, allowed_extensions):
+ for rel_path in _resolve_single_pattern_in_dataset_repository(
+ dataset_info, pattern, base_path, allowed_extensions
+ ):
data_files_urls.append(Url(hf_hub_url(dataset_info.id, rel_path.as_posix(), revision=dataset_info.sha)))
if not data_files_urls:
error_msg = f"Unable to resolve any data file that matches {patterns} in dataset repository {dataset_info.id}"
@@ -382,7 +391,9 @@ def resolve_patterns_in_dataset_repository(
return data_files_urls
-def get_patterns_in_dataset_repository(dataset_info: huggingface_hub.hf_api.DatasetInfo) -> Dict[str, List[str]]:
+def get_patterns_in_dataset_repository(
+ dataset_info: huggingface_hub.hf_api.DatasetInfo, base_path: str
+) -> Dict[str, List[str]]:
"""
Get the default pattern from a repository by testing all the supported patterns.
The first patterns to return a non-empty list of data files is returned.
@@ -466,7 +477,7 @@ def get_patterns_in_dataset_repository(dataset_info: huggingface_hub.hf_api.Data
In order, it first tests if SPLIT_PATTERN_SHARDED works, otherwise it tests the patterns in ALL_DEFAULT_PATTERNS.
"""
- resolver = partial(_resolve_single_pattern_in_dataset_repository, dataset_info)
+ resolver = partial(_resolve_single_pattern_in_dataset_repository, dataset_info, base_path=base_path)
try:
return _get_data_files_patterns(resolver)
except FileNotFoundError:
@@ -526,9 +537,10 @@ def from_hf_repo(
cls,
patterns: List[str],
dataset_info: huggingface_hub.hf_api.DatasetInfo,
+ base_path: Optional[str] = None,
allowed_extensions: Optional[List[str]] = None,
) -> "DataFilesList":
- data_files = resolve_patterns_in_dataset_repository(dataset_info, patterns, allowed_extensions)
+ data_files = resolve_patterns_in_dataset_repository(dataset_info, patterns, base_path, allowed_extensions)
origin_metadata = [(dataset_info.id, dataset_info.sha) for _ in patterns]
return cls(data_files, origin_metadata)
@@ -589,13 +601,17 @@ def from_hf_repo(
cls,
patterns: Dict[str, Union[List[str], DataFilesList]],
dataset_info: huggingface_hub.hf_api.DatasetInfo,
+ base_path: Optional[str] = None,
allowed_extensions: Optional[List[str]] = None,
) -> "DataFilesDict":
out = cls()
for key, patterns_for_key in patterns.items():
out[key] = (
DataFilesList.from_hf_repo(
- patterns_for_key, dataset_info=dataset_info, allowed_extensions=allowed_extensions
+ patterns_for_key,
+ dataset_info=dataset_info,
+ base_path=base_path,
+ allowed_extensions=allowed_extensions,
)
if not isinstance(patterns_for_key, DataFilesList)
else patterns_for_key
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -733,20 +733,27 @@ def __init__(
data_files: Optional[Union[str, List, Dict]] = None,
download_mode: Optional[DownloadMode] = None,
):
- if data_files is None and data_dir is not None:
- data_files = os.path.join(data_dir, "**")
+ if data_dir and os.path.isabs(data_dir):
+ raise ValueError(f"`data_dir` must be relative to a dataset directory's root: {path}")
self.path = path
self.name = Path(path).stem
self.data_files = data_files
+ self.data_dir = data_dir
self.download_mode = download_mode
def get_module(self) -> DatasetModule:
patterns = (
- sanitize_patterns(self.data_files) if self.data_files is not None else get_patterns_locally(self.path)
+ sanitize_patterns(self.data_files)
+ if self.data_files is not None
+ else get_patterns_locally(os.path.join(self.path, self.data_dir))
+ if self.data_dir is not None
+ else get_patterns_locally(self.path)
)
data_files = DataFilesDict.from_local_or_remote(
- patterns, base_path=self.path, allowed_extensions=ALL_ALLOWED_EXTENSIONS
+ patterns,
+ base_path=os.path.join(self.path, self.data_dir) if self.data_dir else self.path,
+ allowed_extensions=ALL_ALLOWED_EXTENSIONS,
)
infered_module_names = {
key: infer_module_for_data_files(data_files_list) for key, data_files_list in data_files.items()
@@ -782,12 +789,11 @@ def __init__(
download_config: Optional[DownloadConfig] = None,
download_mode: Optional[DownloadMode] = None,
):
- if data_files is None and data_dir is not None:
- data_files = os.path.join(data_dir, "**")
self.name = name
self.data_files = data_files
- self.downnload_config = download_config
+ self.data_dir = data_dir
+ self.download_config = download_config
self.download_mode = download_mode
increase_load_count(name, resource_type="dataset")
@@ -795,9 +801,15 @@ def get_module(self) -> DatasetModule:
patterns = (
sanitize_patterns(self.data_files)
if self.data_files is not None
+ else get_patterns_locally(str(Path(self.data_dir).resolve()))
+ if self.data_dir is not None
else get_patterns_locally(str(Path().resolve()))
)
- data_files = DataFilesDict.from_local_or_remote(patterns, use_auth_token=self.downnload_config.use_auth_token)
+ data_files = DataFilesDict.from_local_or_remote(
+ patterns,
+ use_auth_token=self.download_config.use_auth_token,
+ base_path=str(Path(self.data_dir).resolve()) if self.data_dir else None,
+ )
module_path, hash = _PACKAGED_DATASETS_MODULES[self.name]
builder_kwargs = {"hash": hash, "data_files": data_files}
return DatasetModule(module_path, hash, builder_kwargs)
@@ -818,12 +830,11 @@ def __init__(
download_config: Optional[DownloadConfig] = None,
download_mode: Optional[DownloadMode] = None,
):
- if data_files is None and data_dir is not None:
- data_files = os.path.join(data_dir, "**")
self.name = name
self.revision = revision
self.data_files = data_files
+ self.data_dir = data_dir
self.download_config = download_config or DownloadConfig()
self.download_mode = download_mode
assert self.name.count("/") == 1
@@ -843,11 +854,12 @@ def get_module(self) -> DatasetModule:
patterns = (
sanitize_patterns(self.data_files)
if self.data_files is not None
- else get_patterns_in_dataset_repository(hfh_dataset_info)
+ else get_patterns_in_dataset_repository(hfh_dataset_info, self.data_dir)
)
data_files = DataFilesDict.from_hf_repo(
patterns,
dataset_info=hfh_dataset_info,
+ base_path=self.data_dir,
allowed_extensions=ALL_ALLOWED_EXTENSIONS,
)
infered_module_names = {
| diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -22,7 +22,16 @@
_TEST_PATTERNS = ["*", "**", "**/*", "*.txt", "data/*", "**/*.txt", "**/train.txt"]
_FILES_TO_IGNORE = {".dummy", "README.md", "dummy_data.zip", "dataset_infos.json"}
_TEST_PATTERNS_SIZES = dict(
- [("*", 0), ("**", 2), ("**/*", 2), ("*.txt", 0), ("data/*", 2), ("**/*.txt", 2), ("**/train.txt", 1)]
+ [
+ ("*", 0),
+ ("**", 4),
+ ("**/*", 4),
+ ("*.txt", 0),
+ ("data/*", 2),
+ ("data/**", 4),
+ ("**/*.txt", 4),
+ ("**/train.txt", 2),
+ ]
)
_TEST_URL = "https://raw.githubusercontent.com/huggingface/datasets/9675a5a1e7b99a86f9c250f6ea5fa5d1e6d5cc7d/setup.py"
@@ -41,6 +50,13 @@ def complex_data_dir(tmp_path):
f.write("This is a readme")
with open(data_dir / ".dummy", "w") as f:
f.write("this is a dummy file that is not a data file")
+
+ (data_dir / "data" / "subdir").mkdir()
+ with open(data_dir / "data" / "subdir" / "train.txt", "w") as f:
+ f.write("foo\n" * 10)
+ with open(data_dir / "data" / "subdir" / "test.txt", "w") as f:
+ f.write("bar\n" * 10)
+
return str(data_dir)
@@ -116,7 +132,7 @@ def test_resolve_patterns_locally_or_by_urls_with_absolute_path(tmp_path, comple
assert len(resolved_data_files) == 1
[email protected]("pattern,size,extensions", [("**", 2, ["txt"]), ("**", 2, None), ("**", 0, ["blablabla"])])
[email protected]("pattern,size,extensions", [("**", 4, ["txt"]), ("**", 4, None), ("**", 0, ["blablabla"])])
def test_resolve_patterns_locally_or_by_urls_with_extensions(complex_data_dir, pattern, size, extensions):
if size > 0:
resolved_data_files = resolve_patterns_locally_or_by_urls(
@@ -125,7 +141,7 @@ def test_resolve_patterns_locally_or_by_urls_with_extensions(complex_data_dir, p
assert len(resolved_data_files) == size
else:
with pytest.raises(FileNotFoundError):
- resolve_patterns_locally_or_by_urls(complex_data_dir, pattern, allowed_extensions=extensions)
+ resolve_patterns_locally_or_by_urls(complex_data_dir, [pattern], allowed_extensions=extensions)
def test_fail_resolve_patterns_locally_or_by_urls(complex_data_dir):
@@ -154,7 +170,21 @@ def test_resolve_patterns_in_dataset_repository(hub_dataset_info, pattern, hub_d
assert len(hub_dataset_info_patterns_results[pattern]) == 0
[email protected]("pattern,size,extensions", [("**", 2, ["txt"]), ("**", 2, None), ("**", 0, ["blablabla"])])
[email protected](
+ "pattern,size,base_path", [("**", 4, None), ("**", 4, "data"), ("**", 2, "data/subdir"), ("**", 0, "data/subdir2")]
+)
+def test_resolve_patterns_in_dataset_repository_with_base_path(hub_dataset_info, pattern, size, base_path):
+ if size > 0:
+ resolved_data_files = resolve_patterns_in_dataset_repository(hub_dataset_info, [pattern], base_path=base_path)
+ assert len(resolved_data_files) == size
+ else:
+ with pytest.raises(FileNotFoundError):
+ resolved_data_files = resolve_patterns_in_dataset_repository(
+ hub_dataset_info, [pattern], base_path=base_path
+ )
+
+
[email protected]("pattern,size,extensions", [("**", 4, ["txt"]), ("**", 4, None), ("**", 0, ["blablabla"])])
def test_resolve_patterns_in_dataset_repository_with_extensions(hub_dataset_info, pattern, size, extensions):
if size > 0:
resolved_data_files = resolve_patterns_in_dataset_repository(
@@ -222,6 +252,26 @@ def test_DataFilesDict_from_hf_repo(hub_dataset_info, hub_dataset_info_patterns_
assert len(hub_dataset_info_patterns_results[pattern]) == 0
[email protected](
+ "pattern,size,base_path,split_name",
+ [
+ ("**", 4, None, "train"),
+ ("**", 4, "data", "train"),
+ ("**", 2, "data/subdir", "train"),
+ ("**train*", 1, "data/subdir", "train"),
+ ("**test*", 1, "data/subdir", "test"),
+ ("**", 0, "data/subdir2", "train"),
+ ],
+)
+def test_DataFilesDict_from_hf_repo_with_base_path(hub_dataset_info, pattern, size, base_path, split_name):
+ if size > 0:
+ data_files = DataFilesDict.from_hf_repo({split_name: [pattern]}, hub_dataset_info, base_path=base_path)
+ assert len(data_files[split_name]) == size
+ else:
+ with pytest.raises(FileNotFoundError):
+ data_files = DataFilesDict.from_hf_repo({split_name: [pattern]}, hub_dataset_info, base_path=base_path)
+
+
@pytest.mark.parametrize("pattern", _TEST_PATTERNS)
def test_DataFilesDict_from_local_or_remote(complex_data_dir, pattern_results, pattern):
split_name = "train"
@@ -243,11 +293,11 @@ def test_DataFilesDict_from_hf_repo_hashing(hub_dataset_info):
data_files2 = DataFilesDict(sorted(data_files1.items(), reverse=True))
assert Hasher.hash(data_files1) == Hasher.hash(data_files2)
- patterns2 = {"train": ["data/train.txt"], "test": ["data/test.txt"]}
+ patterns2 = {"train": ["data/**train.txt"], "test": ["data/**test.txt"]}
data_files2 = DataFilesDict.from_hf_repo(patterns2, hub_dataset_info)
assert Hasher.hash(data_files1) == Hasher.hash(data_files2)
- patterns2 = {"train": ["data/train.txt"], "test": ["data/train.txt"]}
+ patterns2 = {"train": ["data/**train.txt"], "test": ["data/**train.txt"]}
data_files2 = DataFilesDict.from_hf_repo(patterns2, hub_dataset_info)
assert Hasher.hash(data_files1) != Hasher.hash(data_files2)
diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -105,6 +105,28 @@ def data_dir(tmp_path):
return str(data_dir)
[email protected]
+def sub_data_dirs(tmp_path):
+ data_dir2 = tmp_path / "data_dir2"
+ relative_subdir1 = "subdir1"
+ sub_data_dir1 = data_dir2 / relative_subdir1
+ sub_data_dir1.mkdir(parents=True)
+ with open(sub_data_dir1 / "train.txt", "w") as f:
+ f.write("foo\n" * 10)
+ with open(sub_data_dir1 / "test.txt", "w") as f:
+ f.write("bar\n" * 10)
+
+ relative_subdir2 = "subdir2"
+ sub_data_dir2 = tmp_path / data_dir2 / relative_subdir2
+ sub_data_dir2.mkdir(parents=True)
+ with open(sub_data_dir2 / "train.txt", "w") as f:
+ f.write("foo\n" * 10)
+ with open(sub_data_dir2 / "test.txt", "w") as f:
+ f.write("bar\n" * 10)
+
+ return str(data_dir2), relative_subdir1
+
+
@pytest.fixture
def complex_data_dir(tmp_path):
data_dir = tmp_path / "complex_data_dir"
@@ -168,9 +190,13 @@ def test_infer_module_for_data_files_in_archives(data_file, expected_module, zip
class ModuleFactoryTest(TestCase):
@pytest.fixture(autouse=True)
- def inject_fixtures(self, jsonl_path, data_dir, dataset_loading_script_dir, metric_loading_script_dir):
+ def inject_fixtures(
+ self, jsonl_path, data_dir, sub_data_dirs, dataset_loading_script_dir, metric_loading_script_dir
+ ):
self._jsonl_path = jsonl_path
self._data_dir = data_dir
+ self._data_dir2 = sub_data_dirs[0]
+ self._sub_data_dir = sub_data_dirs[1]
self._dataset_loading_script_dir = dataset_loading_script_dir
self._metric_loading_script_dir = metric_loading_script_dir
@@ -231,6 +257,21 @@ def test_LocalDatasetModuleFactoryWithoutScript(self):
assert importlib.import_module(module_factory_result.module_path) is not None
assert os.path.isdir(module_factory_result.builder_kwargs["base_path"])
+ def test_LocalDatasetModuleFactoryWithoutScript_with_data_dir(self):
+ factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir2, data_dir=self._sub_data_dir)
+ module_factory_result = factory.get_module()
+ assert importlib.import_module(module_factory_result.module_path) is not None
+ assert (
+ module_factory_result.builder_kwargs["data_files"] is not None
+ and len(module_factory_result.builder_kwargs["data_files"]["train"]) == 1
+ and len(module_factory_result.builder_kwargs["data_files"]["test"]) == 1
+ )
+ assert all(
+ self._sub_data_dir in Path(data_file).parts
+ for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
+ + module_factory_result.builder_kwargs["data_files"]["test"]
+ )
+
def test_PackagedDatasetModuleFactory(self):
factory = PackagedDatasetModuleFactory(
"json", data_files=self._jsonl_path, download_config=self.download_config
@@ -245,8 +286,10 @@ def test_PackagedDatasetModuleFactory_with_data_dir(self):
assert (
module_factory_result.builder_kwargs["data_files"] is not None
and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
+ and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
)
assert Path(module_factory_result.builder_kwargs["data_files"]["train"][0]).parent.samefile(self._data_dir)
+ assert Path(module_factory_result.builder_kwargs["data_files"]["test"][0]).parent.samefile(self._data_dir)
def test_HubDatasetModuleFactoryWithoutScript(self):
factory = HubDatasetModuleFactoryWithoutScript(
@@ -266,11 +309,13 @@ def test_HubDatasetModuleFactoryWithoutScript_with_data_dir(self):
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
assert (
module_factory_result.builder_kwargs["data_files"] is not None
- and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
+ and len(module_factory_result.builder_kwargs["data_files"]["train"]) == 1
+ and len(module_factory_result.builder_kwargs["data_files"]["test"]) == 1
)
assert all(
data_dir in Path(data_file).parts
for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
+ + module_factory_result.builder_kwargs["data_files"]["test"]
)
def test_HubDatasetModuleFactoryWithScript(self):
| Inconsistent splits generation for datasets without loading script (packaged dataset puts everything into a single split)
## Describe the bug
Splits for dataset loaders without scripts are prepared inconsistently. I think it might be confusing for users.
## Steps to reproduce the bug
* If you load a packaged datasets from Hub, it infers splits from directory structure / filenames (check out the data [here](https://huggingface.co/datasets/nateraw/test-imagefolder-dataset)):
```python
ds = load_dataset("nateraw/test-imagefolder-dataset")
print(ds)
### Output:
DatasetDict({
train: Dataset({
features: ['image', 'label'],
num_rows: 6
})
test: Dataset({
features: ['image', 'label'],
num_rows: 4
})
})
```
* If you do the same from locally stored data specifying only directory path you'll get the same:
```python
ds = load_dataset("/path/to/local/data/test-imagefolder-dataset")
print(ds)
### Output:
DatasetDict({
train: Dataset({
features: ['image', 'label'],
num_rows: 6
})
test: Dataset({
features: ['image', 'label'],
num_rows: 4
})
})
```
* However, if you explicitely specify package name (like `imagefolder`, `csv`, `json`), all the data is put into a single split:
```python
ds = load_dataset("imagefolder", data_dir="/path/to/local/data/test-imagefolder-dataset")
print(ds)
### Output:
DatasetDict({
train: Dataset({
features: ['image', 'label'],
num_rows: 10
})
})
```
## Expected results
For `load_dataset("imagefolder", data_dir="/path/to/local/data/test-imagefolder-dataset")` I expect the same output as of the two first options.
| 2022-04-11T13:57:33Z | [] | [] |
|
huggingface/datasets | 4,194 | huggingface__datasets-4194 | [
"4191"
] | b564af7c40d8df6da482af27fe1766e873aa2dd2 | diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -32,6 +32,7 @@
get_nested_type,
list_of_np_array_to_pyarrow_listarray,
numpy_to_pyarrow_listarray,
+ to_pyarrow_listarray,
)
from .info import DatasetInfo
from .keyhash import DuplicatedKeysError, KeyHasher
@@ -169,12 +170,7 @@ def __arrow_array__(self, type: Optional[pa.DataType] = None):
try:
# custom pyarrow types
if isinstance(pa_type, _ArrayXDExtensionType):
- if isinstance(data, np.ndarray):
- storage = numpy_to_pyarrow_listarray(data, type=pa_type.value_type)
- elif isinstance(data, list) and data and isinstance(first_non_null_value(data)[1], np.ndarray):
- storage = list_of_np_array_to_pyarrow_listarray(data, type=pa_type.value_type)
- else:
- storage = pa.array(data, pa_type.storage_dtype)
+ storage = to_pyarrow_listarray(data, pa_type)
return pa.ExtensionArray.from_storage(pa_type, storage)
# efficient np array to pyarrow array
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -35,7 +35,7 @@
from .. import config
from ..utils import logging
-from ..utils.py_utils import zip_dict
+from ..utils.py_utils import first_non_null_value, zip_dict
from .audio import Audio
from .image import Image, encode_pil_image
from .translation import Translation, TranslationVariableLanguages
@@ -1160,7 +1160,7 @@ def list_of_pa_arrays_to_pyarrow_listarray(l_arr: List[Optional[pa.Array]]) -> p
null_indices = [i for i, arr in enumerate(l_arr) if arr is None]
l_arr = [arr for arr in l_arr if arr is not None]
offsets = np.cumsum(
- [0] + [len(arr) for arr in l_arr], dtype=np.object
+ [0] + [len(arr) for arr in l_arr], dtype=object
) # convert to dtype object to allow None insertion
offsets = np.insert(offsets, null_indices, None)
offsets = pa.array(offsets, type=pa.int32())
@@ -1178,6 +1178,55 @@ def list_of_np_array_to_pyarrow_listarray(l_arr: List[np.ndarray], type: pa.Data
return pa.array([], type=type)
+def contains_any_np_array(data: Any):
+ """Return `True` if data is a NumPy ndarray or (recursively) if first non-null value in list is a NumPy ndarray.
+
+ Args:
+ data (Any): Data.
+
+ Returns:
+ bool
+ """
+ if isinstance(data, np.ndarray):
+ return True
+ elif isinstance(data, list):
+ return contains_any_np_array(first_non_null_value(data)[1])
+ else:
+ return False
+
+
+def any_np_array_to_pyarrow_listarray(data: Union[np.ndarray, List], type: pa.DataType = None) -> pa.ListArray:
+ """Convert to PyArrow ListArray either a NumPy ndarray or (recursively) a list that may contain any NumPy ndarray.
+
+ Args:
+ data (Union[np.ndarray, List]): Data.
+ type (pa.DataType): Explicit PyArrow DataType passed to coerce the ListArray data type.
+
+ Returns:
+ pa.ListArray
+ """
+ if isinstance(data, np.ndarray):
+ return numpy_to_pyarrow_listarray(data, type=type)
+ elif isinstance(data, list):
+ return list_of_pa_arrays_to_pyarrow_listarray([any_np_array_to_pyarrow_listarray(i, type=type) for i in data])
+
+
+def to_pyarrow_listarray(data: Any, pa_type: _ArrayXDExtensionType) -> pa.Array:
+ """Convert to PyArrow ListArray.
+
+ Args:
+ data (Any): Sequence, iterable, np.ndarray or pd.Series.
+ pa_type (_ArrayXDExtensionType): Any of the ArrayNDExtensionType.
+
+ Returns:
+ pyarrow.Array
+ """
+ if contains_any_np_array(data):
+ return any_np_array_to_pyarrow_listarray(data, type=pa_type.value_type)
+ else:
+ return pa.array(data, pa_type.storage_dtype)
+
+
def require_decoding(feature: FeatureType, ignore_decode_attribute: bool = False) -> bool:
"""Check if a (possibly nested) feature requires decoding.
| diff --git a/tests/features/test_array_xd.py b/tests/features/test_array_xd.py
--- a/tests/features/test_array_xd.py
+++ b/tests/features/test_array_xd.py
@@ -341,6 +341,24 @@ def test_array_xd_with_none():
assert np.isnan(arr[1]) # a single np.nan value - np.all not needed
[email protected](
+ "data, feature, expected",
+ [
+ (np.zeros((2, 2)), None, [[0.0, 0.0], [0.0, 0.0]]),
+ (np.zeros((2, 3)), datasets.Array2D(shape=(2, 3), dtype="float32"), [[0.0, 0.0, 0.0], [0.0, 0.0, 0.0]]),
+ ([np.zeros(2)], datasets.Array2D(shape=(1, 2), dtype="float32"), [[0.0, 0.0]]),
+ (
+ [np.zeros((2, 3))],
+ datasets.Array3D(shape=(1, 2, 3), dtype="float32"),
+ [[[0.0, 0.0, 0.0], [0.0, 0.0, 0.0]]],
+ ),
+ ],
+)
+def test_array_xd_with_np(data, feature, expected):
+ ds = datasets.Dataset.from_dict({"col": [data]}, features=datasets.Features({"col": feature}) if feature else None)
+ assert ds[0]["col"] == expected
+
+
@pytest.mark.parametrize("with_none", [False, True])
def test_dataset_map(with_none):
ds = datasets.Dataset.from_dict({"path": ["path1", "path2"]})
| feat: create an `Array3D` column from a list of arrays of dimension 2
**Is your feature request related to a problem? Please describe.**
It is possible to create an `Array2D` column from a list of arrays of dimension 1. Similarly, I think it might be nice to be able to create a `Array3D` column from a list of lists of arrays of dimension 1.
To illustrate my proposal, let's take the following toy dataset t:
```python
import numpy as np
from datasets import Dataset, features
data_map = {
1: np.array([[0.2, 0,4],[0.19, 0,3]]),
2: np.array([[0.1, 0,4],[0.19, 0,3]]),
}
def create_toy_ds():
my_dict = {"id":[1, 2]}
return Dataset.from_dict(my_dict)
ds = create_toy_ds()
```
The following 2D processing works without any errors raised:
```python
def prepare_dataset_2D(batch):
batch["pixel_values"] = [data_map[index] for index in batch["id"]]
return batch
ds_2D = ds.map(
prepare_dataset_2D,
batched=True,
remove_columns=ds.column_names,
features=features.Features({"pixel_values": features.Array2D(shape=(2, 3), dtype="float32")})
)
```
The following 3D processing doesn't work:
```python
def prepare_dataset_3D(batch):
batch["pixel_values"] = [[data_map[index]] for index in batch["id"]]
return batch
ds_3D = ds.map(
prepare_dataset_3D,
batched=True,
remove_columns=ds.column_names,
features=features.Features({"pixel_values": features.Array3D(shape=(1, 2, 3, dtype="float32")})
)
```
The error raised is:
```
---------------------------------------------------------------------------
ArrowInvalid Traceback (most recent call last)
[<ipython-input-6-676547e4cd41>](https://localhost:8080/#) in <module>()
3 batched=True,
4 remove_columns=ds.column_names,
----> 5 features=features.Features({"pixel_values": features.Array3D(shape=(1, 2, 3), dtype="float32")})
6 )
12 frames
[/usr/local/lib/python3.7/dist-packages/datasets/arrow_dataset.py](https://localhost:8080/#) in map(self, function, with_indices, with_rank, input_columns, batched, batch_size, drop_last_batch, remove_columns, keep_in_memory, load_from_cache_file, cache_file_name, writer_batch_size, features, disable_nullable, fn_kwargs, num_proc, suffix_template, new_fingerprint, desc)
1971 new_fingerprint=new_fingerprint,
1972 disable_tqdm=disable_tqdm,
-> 1973 desc=desc,
1974 )
1975 else:
[/usr/local/lib/python3.7/dist-packages/datasets/arrow_dataset.py](https://localhost:8080/#) in wrapper(*args, **kwargs)
518 self: "Dataset" = kwargs.pop("self")
519 # apply actual function
--> 520 out: Union["Dataset", "DatasetDict"] = func(self, *args, **kwargs)
521 datasets: List["Dataset"] = list(out.values()) if isinstance(out, dict) else [out]
522 for dataset in datasets:
[/usr/local/lib/python3.7/dist-packages/datasets/arrow_dataset.py](https://localhost:8080/#) in wrapper(*args, **kwargs)
485 }
486 # apply actual function
--> 487 out: Union["Dataset", "DatasetDict"] = func(self, *args, **kwargs)
488 datasets: List["Dataset"] = list(out.values()) if isinstance(out, dict) else [out]
489 # re-apply format to the output
[/usr/local/lib/python3.7/dist-packages/datasets/fingerprint.py](https://localhost:8080/#) in wrapper(*args, **kwargs)
456 # Call actual function
457
--> 458 out = func(self, *args, **kwargs)
459
460 # Update fingerprint of in-place transforms + update in-place history of transforms
[/usr/local/lib/python3.7/dist-packages/datasets/arrow_dataset.py](https://localhost:8080/#) in _map_single(self, function, with_indices, with_rank, input_columns, batched, batch_size, drop_last_batch, remove_columns, keep_in_memory, load_from_cache_file, cache_file_name, writer_batch_size, features, disable_nullable, fn_kwargs, new_fingerprint, rank, offset, disable_tqdm, desc, cache_only)
2354 writer.write_table(batch)
2355 else:
-> 2356 writer.write_batch(batch)
2357 if update_data and writer is not None:
2358 writer.finalize() # close_stream=bool(buf_writer is None)) # We only close if we are writing in a file
[/usr/local/lib/python3.7/dist-packages/datasets/arrow_writer.py](https://localhost:8080/#) in write_batch(self, batch_examples, writer_batch_size)
505 col_try_type = try_features[col] if try_features is not None and col in try_features else None
506 typed_sequence = OptimizedTypedSequence(batch_examples[col], type=col_type, try_type=col_try_type, col=col)
--> 507 arrays.append(pa.array(typed_sequence))
508 inferred_features[col] = typed_sequence.get_inferred_type()
509 schema = inferred_features.arrow_schema if self.pa_writer is None else self.schema
/usr/local/lib/python3.7/dist-packages/pyarrow/array.pxi in pyarrow.lib.array()
/usr/local/lib/python3.7/dist-packages/pyarrow/array.pxi in pyarrow.lib._handle_arrow_array_protocol()
[/usr/local/lib/python3.7/dist-packages/datasets/arrow_writer.py](https://localhost:8080/#) in __arrow_array__(self, type)
175 storage = list_of_np_array_to_pyarrow_listarray(data, type=pa_type.value_type)
176 else:
--> 177 storage = pa.array(data, pa_type.storage_dtype)
178 return pa.ExtensionArray.from_storage(pa_type, storage)
179
/usr/local/lib/python3.7/dist-packages/pyarrow/array.pxi in pyarrow.lib.array()
/usr/local/lib/python3.7/dist-packages/pyarrow/array.pxi in pyarrow.lib._sequence_to_array()
/usr/local/lib/python3.7/dist-packages/pyarrow/error.pxi in pyarrow.lib.pyarrow_internal_check_status()
/usr/local/lib/python3.7/dist-packages/pyarrow/error.pxi in pyarrow.lib.check_status()
ArrowInvalid: Can only convert 1-dimensional array values
```
**Describe the solution you'd like**
No error in the second scenario and an identical result to the following snippets.
**Describe alternatives you've considered**
There are other alternatives that work such as:
```python
def prepare_dataset_3D_bis(batch):
batch["pixel_values"] = [[data_map[index].tolist()] for index in batch["id"]]
return batch
ds_3D_bis = ds.map(
prepare_dataset_3D_bis,
batched=True,
remove_columns=ds.column_names,
features=features.Features({"pixel_values": features.Array3D(shape=(1, 2, 3), dtype="float32")})
)
```
or
```python
def prepare_dataset_3D_ter(batch):
batch["pixel_values"] = [data_map[index][np.newaxis, :, :] for index in batch["id"]]
return batch
ds_3D_ter = ds.map(
prepare_dataset_3D_ter,
batched=True,
remove_columns=ds.column_names,
features=features.Features({"pixel_values": features.Array3D(shape=(1, 2, 3), dtype="float32")})
)
```
But both solutions require the user to be aware that `data_map[index]` is an `np.array` type.
cc @lhoestq as we discuss this offline :smile:
| Hi @SaulLu, thanks for your proposal.
Just I got a bit confused about the dimensions...
- For the 2D case, you mention it is possible to create an `Array2D` from a list of arrays of dimension 1
- However, you give an example of creating an `Array2D` from arrays of dimension 2:
- the values of `data_map` are arrays of dimension 2
- the outer list in `prepare_dataset_2D` should not be taken into account in the dimension counting, as it is used because in `map` you pass `batched=True`
Note that for the 3D alternatives you mention:
- In `prepare_dataset_3D_ter`, you create an `Array3D` from arrays of dimension 3:
- the array `data_map[index][np.newaxis, :, :]` has dimension 3
- the outer list in `prepare_dataset_3D_ter` is the one used by `batched=True`
- In `prepare_dataset_3D_bis`, you create an `Array3D` from a list of list of lists:
- the value of `data_map[index].tolist()` is a list of lists
- it is enclosed by another list `[data_map[index].tolist()]`, thus giving a list of list of lists
- the outer list is the one used by `batched=True`
Therefore, if I understand correctly, your request would be to be able to create an `Array3D` from a list of an array of dimension 2:
- In `prepare_dataset_3D`, `data_map[index]` is an array of dimension 2
- it is enclosed by a list `[data_map[index]]`, thus giving a list of an array of dimension 2
- the outer list is the one used by `batched=True`
Please, feel free to tell me if I did not understand you correctly.
Hi @albertvillanova ,
Indeed my message was confusing and you guessed right :smile: : I think would be interesting to be able to create an Array3D from a list of an array of dimension 2.
For the 2D case I should have given as a "similar" example:
```python
data_map_1D = {
1: np.array([0.2, 0.4]),
2: np.array([0.1, 0.4]),
}
def prepare_dataset_2D(batch):
batch["pixel_values"] = [[data_map_1D[index]] for index in batch["id"]]
return batch
ds_2D = ds.map(
prepare_dataset_2D,
batched=True,
remove_columns=ds.column_names,
features=features.Features({"pixel_values": features.Array2D(shape=(1, 2), dtype="float32")})
)
``` | 2022-04-21T12:22:26Z | [] | [] |
huggingface/datasets | 4,322 | huggingface__datasets-4322 | [
"3452"
] | c1dce72ff259d3e7abee34900e48578ec685fb00 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -93,6 +93,7 @@
from .utils.file_utils import _retry, estimate_dataset_size
from .utils.info_utils import is_small_dataset
from .utils.py_utils import convert_file_size_to_int, temporary_assignment, unique_values
+from .utils.stratify import stratified_shuffle_split_generate_indices
from .utils.typing import PathLike
@@ -3255,6 +3256,7 @@ def train_test_split(
test_size: Union[float, int, None] = None,
train_size: Union[float, int, None] = None,
shuffle: bool = True,
+ stratify_by_column: Optional[str] = None,
seed: Optional[int] = None,
generator: Optional[np.random.Generator] = None,
keep_in_memory: bool = False,
@@ -3281,6 +3283,7 @@ def train_test_split(
If int, represents the absolute number of train samples.
If None, the value is automatically set to the complement of the test size.
shuffle (:obj:`bool`, optional, default `True`): Whether or not to shuffle the data before splitting.
+ stratify_by_column (:obj:`str`, optional, default `None`): The column name of labels to be used to perform stratified split of data.
seed (:obj:`int`, optional): A seed to initialize the default BitGenerator if ``generator=None``.
If None, then fresh, unpredictable entropy will be pulled from the OS.
If an int or array_like[ints] is passed, then it will be passed to SeedSequence to derive the initial BitGenerator state.
@@ -3320,6 +3323,24 @@ def train_test_split(
# set a seed
>>> ds = ds.train_test_split(test_size=0.2, seed=42)
+
+ # stratified split
+ >>> ds = load_dataset("imdb",split="train")
+ Dataset({
+ features: ['text', 'label'],
+ num_rows: 25000
+ })
+ >>> ds = ds.train_test_split(test_size=0.2, stratify_by_column="label")
+ DatasetDict({
+ train: Dataset({
+ features: ['text', 'label'],
+ num_rows: 20000
+ })
+ test: Dataset({
+ features: ['text', 'label'],
+ num_rows: 5000
+ })
+ })
```
"""
from .dataset_dict import DatasetDict # import here because of circular dependency
@@ -3437,15 +3458,42 @@ def train_test_split(
),
}
)
-
if not shuffle:
+ if stratify_by_column is not None:
+ raise ValueError("Stratified train/test split is not implemented for `shuffle=False`")
train_indices = np.arange(n_train)
test_indices = np.arange(n_train, n_train + n_test)
else:
+ # stratified partition
+ if stratify_by_column is not None:
+ if stratify_by_column not in self.features.keys():
+ raise ValueError(f"Key {stratify_by_column} not found in {self.features.keys()}")
+ if not isinstance(self.features[stratify_by_column], ClassLabel):
+ raise ValueError(
+ f"Stratifying by column is only supported for {ClassLabel.__name__} column, and column {stratify_by_column} is {type(self.features[stratify_by_column]).__name__}."
+ )
+ try:
+ train_indices, test_indices = next(
+ stratified_shuffle_split_generate_indices(
+ self.with_format("numpy")[stratify_by_column], n_train, n_test, rng=generator
+ )
+ )
+ except Exception as error:
+ if str(error) == "Minimum class count error":
+ raise ValueError(
+ f"The least populated class in {stratify_by_column} column has only 1"
+ " member, which is too few. The minimum"
+ " number of groups for any class cannot"
+ " be less than 2."
+ )
+ else:
+ raise error
+
# random partition
- permutation = generator.permutation(len(self))
- test_indices = permutation[:n_test]
- train_indices = permutation[n_test : (n_test + n_train)]
+ else:
+ permutation = generator.permutation(len(self))
+ test_indices = permutation[:n_test]
+ train_indices = permutation[n_test : (n_test + n_train)]
train_split = self.select(
indices=train_indices,
diff --git a/src/datasets/utils/stratify.py b/src/datasets/utils/stratify.py
new file mode 100644
--- /dev/null
+++ b/src/datasets/utils/stratify.py
@@ -0,0 +1,107 @@
+import numpy as np
+
+
+def approximate_mode(class_counts, n_draws, rng):
+ """Computes approximate mode of multivariate hypergeometric.
+ This is an approximation to the mode of the multivariate
+ hypergeometric given by class_counts and n_draws.
+ It shouldn't be off by more than one.
+ It is the mostly likely outcome of drawing n_draws many
+ samples from the population given by class_counts.
+ Args
+ ----------
+ class_counts : ndarray of int
+ Population per class.
+ n_draws : int
+ Number of draws (samples to draw) from the overall population.
+ rng : random state
+ Used to break ties.
+ Returns
+ -------
+ sampled_classes : ndarray of int
+ Number of samples drawn from each class.
+ np.sum(sampled_classes) == n_draws
+
+ """
+ # this computes a bad approximation to the mode of the
+ # multivariate hypergeometric given by class_counts and n_draws
+ continuous = n_draws * class_counts / class_counts.sum()
+ # floored means we don't overshoot n_samples, but probably undershoot
+ floored = np.floor(continuous)
+ # we add samples according to how much "left over" probability
+ # they had, until we arrive at n_samples
+ need_to_add = int(n_draws - floored.sum())
+ if need_to_add > 0:
+ remainder = continuous - floored
+ values = np.sort(np.unique(remainder))[::-1]
+ # add according to remainder, but break ties
+ # randomly to avoid biases
+ for value in values:
+ (inds,) = np.where(remainder == value)
+ # if we need_to_add less than what's in inds
+ # we draw randomly from them.
+ # if we need to add more, we add them all and
+ # go to the next value
+ add_now = min(len(inds), need_to_add)
+ inds = rng.choice(inds, size=add_now, replace=False)
+ floored[inds] += 1
+ need_to_add -= add_now
+ if need_to_add == 0:
+ break
+ return floored.astype(np.int)
+
+
+def stratified_shuffle_split_generate_indices(y, n_train, n_test, rng, n_splits=10):
+ """
+
+ Provides train/test indices to split data in train/test sets.
+ It's reference is taken from StratifiedShuffleSplit implementation
+ of scikit-learn library.
+
+ Args
+ ----------
+
+ n_train : int,
+ represents the absolute number of train samples.
+
+ n_test : int,
+ represents the absolute number of test samples.
+
+ random_state : int or RandomState instance, default=None
+ Controls the randomness of the training and testing indices produced.
+ Pass an int for reproducible output across multiple function calls.
+
+ n_splits : int, default=10
+ Number of re-shuffling & splitting iterations.
+ """
+ classes, y_indices = np.unique(y, return_inverse=True)
+ n_classes = classes.shape[0]
+ class_counts = np.bincount(y_indices)
+ if np.min(class_counts) < 2:
+ raise ValueError("Minimum class count error")
+ if n_train < n_classes:
+ raise ValueError(
+ "The train_size = %d should be greater or " "equal to the number of classes = %d" % (n_train, n_classes)
+ )
+ if n_test < n_classes:
+ raise ValueError(
+ "The test_size = %d should be greater or " "equal to the number of classes = %d" % (n_test, n_classes)
+ )
+ class_indices = np.split(np.argsort(y_indices, kind="mergesort"), np.cumsum(class_counts)[:-1])
+ for _ in range(n_splits):
+ n_i = approximate_mode(class_counts, n_train, rng)
+ class_counts_remaining = class_counts - n_i
+ t_i = approximate_mode(class_counts_remaining, n_test, rng)
+
+ train = []
+ test = []
+
+ for i in range(n_classes):
+ permutation = rng.permutation(class_counts[i])
+ perm_indices_class_i = class_indices[i].take(permutation, mode="clip")
+ train.extend(perm_indices_class_i[: n_i[i]])
+ test.extend(perm_indices_class_i[n_i[i] : n_i[i] + t_i[i]])
+ train = rng.permutation(train)
+ test = rng.permutation(test)
+
+ yield train, test
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -11,6 +11,7 @@
from unittest.mock import patch
import numpy as np
+import numpy.testing as npt
import pandas as pd
import pyarrow as pa
import pytest
@@ -3553,3 +3554,69 @@ def test_task_text_classification_when_columns_removed(self):
with Dataset.from_dict(data, info=info) as dset:
with dset.map(lambda x: {"new_column": 0}, remove_columns=dset.column_names) as dset:
self.assertDictEqual(dset.features, features_after_map)
+
+
+class StratifiedTest(TestCase):
+ def test_errors_train_test_split_stratify(self):
+ ys = [
+ np.array([0, 0, 0, 0, 1, 1, 1, 2, 2, 2, 2, 2]),
+ np.array([0, 1, 1, 1, 2, 2, 2, 3, 3, 3]),
+ np.array([0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2] * 2),
+ np.array([0, 0, 1, 1, 2, 2, 3, 3, 4, 4, 5, 5]),
+ np.array([0, 0, 1, 1, 2, 2, 3, 3, 4, 4, 5, 5]),
+ ]
+ for i in range(len(ys)):
+ features = Features({"text": Value("int64"), "label": ClassLabel(len(np.unique(ys[i])))})
+ data = {"text": np.ones(len(ys[i])), "label": ys[i]}
+ d1 = Dataset.from_dict(data, features=features)
+
+ # For checking stratify_by_column exist as key in self.features.keys()
+ if i == 0:
+ self.assertRaises(ValueError, d1.train_test_split, 0.33, stratify_by_column="labl")
+
+ # For checking minimum class count error
+ elif i == 1:
+ self.assertRaises(ValueError, d1.train_test_split, 0.33, stratify_by_column="label")
+
+ # For check typeof label as ClassLabel type
+ elif i == 2:
+ d1 = Dataset.from_dict(data)
+ self.assertRaises(ValueError, d1.train_test_split, 0.33, stratify_by_column="label")
+
+ # For checking test_size should be greater than or equal to number of classes
+ elif i == 3:
+ self.assertRaises(ValueError, d1.train_test_split, 0.30, stratify_by_column="label")
+
+ # For checking train_size should be greater than or equal to number of classes
+ elif i == 4:
+ self.assertRaises(ValueError, d1.train_test_split, 0.60, stratify_by_column="label")
+
+ def test_train_test_split_startify(self):
+ ys = [
+ np.array([0, 0, 0, 0, 1, 1, 1, 2, 2, 2, 2, 2]),
+ np.array([0, 0, 0, 1, 1, 1, 2, 2, 2, 3, 3, 3]),
+ np.array([0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2] * 2),
+ np.array([0, 0, 1, 1, 1, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3]),
+ np.array([0] * 800 + [1] * 50),
+ ]
+ for y in ys:
+ features = Features({"text": Value("int64"), "label": ClassLabel(len(np.unique(y)))})
+ data = {"text": np.ones(len(y)), "label": y}
+ d1 = Dataset.from_dict(data, features=features)
+ d1 = d1.train_test_split(test_size=0.33, stratify_by_column="label")
+ y = np.asanyarray(y) # To make it indexable for y[train]
+ test_size = np.ceil(0.33 * len(y))
+ train_size = len(y) - test_size
+ npt.assert_array_equal(np.unique(d1["train"]["label"]), np.unique(d1["test"]["label"]))
+
+ # checking classes proportion
+ p_train = np.bincount(np.unique(d1["train"]["label"], return_inverse=True)[1]) / float(
+ len(d1["train"]["label"])
+ )
+ p_test = np.bincount(np.unique(d1["test"]["label"], return_inverse=True)[1]) / float(
+ len(d1["test"]["label"])
+ )
+ npt.assert_array_almost_equal(p_train, p_test, 1)
+ assert len(d1["train"]["text"]) + len(d1["test"]["text"]) == y.size
+ assert len(d1["train"]["text"]) == train_size
+ assert len(d1["test"]["text"]) == test_size
| why the stratify option is omitted from test_train_split function?
why the stratify option is omitted from test_train_split function?
is there any other way implement the stratify option while splitting the dataset? as it is important point to be considered while splitting the dataset.
| Hi ! It's simply not added yet :)
If someone wants to contribute to add the `stratify` parameter I'd be happy to give some pointers.
In the meantime, I guess you can use `sklearn` or other tools to do a stratified train/test split over the **indices** of your dataset and then do
```
train_dataset = dataset.select(train_indices)
test_dataset = dataset.select(test_indices)
```
Hi @lhoestq I would like to add `stratify` parameter, can you give me some pointers for adding the same ?
Hi ! Sure :)
The `train_test_split` method is defined here:
https://github.com/huggingface/datasets/blob/dc62232fa1b3bcfe2fbddcb721f2d141f8908943/src/datasets/arrow_dataset.py#L3253-L3253
and inside `train_test_split ` we need to create the right `train_indices` and `test_indices` that are passed here to `.select()`:
https://github.com/huggingface/datasets/blob/dc62232fa1b3bcfe2fbddcb721f2d141f8908943/src/datasets/arrow_dataset.py#L3450-L3464
For example if your dataset is like
| | label |
|---:|--------:|
| 0 | 1 |
| 1 | 1 |
| 2 | 0 |
| 3 | 0 |
and the user passes `stratify=dataset["label"]`, then you should get indices that look like this
```
train_indices = [0, 2]
test_indices = [1, 3]
```
these indices will be passed to `.select` to return the stratified train and test splits :)
Feel free to îng me if you have any question ! | 2022-05-12T08:00:31Z | [] | [] |
huggingface/datasets | 4,372 | huggingface__datasets-4372 | [
"4211"
] | b053f8bfb2865af17198cb34c3b63d98525d72d0 | diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -35,7 +35,15 @@ class DatasetDict(dict):
def _check_values_type(self):
for dataset in self.values():
if not isinstance(dataset, Dataset):
- raise TypeError(f"Values in `DatasetDict` should of type `Dataset` but got type '{type(dataset)}'")
+ raise TypeError(f"Values in `DatasetDict` should be of type `Dataset` but got type '{type(dataset)}'")
+
+ def _check_values_features(self):
+ items = list(self.items())
+ for item_a, item_b in zip(items[:-1], items[1:]):
+ if item_a[1].features != item_b[1].features:
+ raise ValueError(
+ f"All datasets in `DatasetDict` should have the same features but features for '{item_a[0]}' and '{item_b[0]}' don't match: {item_a[1].features} != {item_b[1].features}"
+ )
def __getitem__(self, k) -> Dataset:
if isinstance(k, (str, NamedSplit)) or len(self) == 0:
@@ -1330,6 +1338,7 @@ def push_to_hub(
max_shard_size = shard_size
self._check_values_type()
+ self._check_values_features()
total_uploaded_size = 0
total_dataset_nbytes = 0
info_to_dump: DatasetInfo = next(iter(self.values())).info.copy()
| diff --git a/tests/test_upstream_hub.py b/tests/test_upstream_hub.py
--- a/tests/test_upstream_hub.py
+++ b/tests/test_upstream_hub.py
@@ -101,6 +101,20 @@ def test_push_dataset_dict_to_hub_name_without_namespace(self):
finally:
self.cleanup_repo(ds_name)
+ def test_push_dataset_dict_to_hub_datasets_with_different_features(self):
+ ds_train = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
+ ds_test = Dataset.from_dict({"x": [True, False, True], "y": ["a", "b", "c"]})
+
+ local_ds = DatasetDict({"train": ds_train, "test": ds_test})
+
+ ds_name = f"{USER}/test-{int(time.time() * 10e3)}"
+ try:
+ with self.assertRaises(ValueError):
+ local_ds.push_to_hub(ds_name.split("/")[-1], token=self._token)
+ except AssertionError:
+ self.cleanup_repo(ds_name)
+ raise
+
def test_push_dataset_dict_to_hub_private(self):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
| DatasetDict containing Datasets with different features when pushed to hub gets remapped features
Hi there,
I am trying to load a dataset to the Hub. This dataset is a `DatasetDict` composed of various splits. Some splits have a different `Feature` mapping. Locally, the DatasetDict preserves the individual features but if I `push_to_hub` and then `load_dataset`, the features are all the same.
Dataset and code to reproduce available [here](https://huggingface.co/datasets/pietrolesci/robust_nli).
In short:
I have 3 feature mapping
```python
Tri_features = Features(
{
"idx": Value(dtype="int64"),
"premise": Value(dtype="string"),
"hypothesis": Value(dtype="string"),
"label": ClassLabel(num_classes=3, names=["entailment", "neutral", "contradiction"]),
}
)
Ent_features = Features(
{
"idx": Value(dtype="int64"),
"premise": Value(dtype="string"),
"hypothesis": Value(dtype="string"),
"label": ClassLabel(num_classes=2, names=["non-entailment", "entailment"]),
}
)
Con_features = Features(
{
"idx": Value(dtype="int64"),
"premise": Value(dtype="string"),
"hypothesis": Value(dtype="string"),
"label": ClassLabel(num_classes=2, names=["non-contradiction", "contradiction"]),
}
)
```
Then I create different datasets
```python
dataset_splits = {}
for split in df["split"].unique():
print(split)
df_split = df.loc[df["split"] == split].copy()
if split in Tri_dataset:
df_split["label"] = df_split["label"].map({"entailment": 0, "neutral": 1, "contradiction": 2})
ds = Dataset.from_pandas(df_split, features=Tri_features)
elif split in Ent_bin_dataset:
df_split["label"] = df_split["label"].map({"non-entailment": 0, "entailment": 1})
ds = Dataset.from_pandas(df_split, features=Ent_features)
elif split in Con_bin_dataset:
df_split["label"] = df_split["label"].map({"non-contradiction": 0, "contradiction": 1})
ds = Dataset.from_pandas(df_split, features=Con_features)
else:
print("ERROR:", split)
dataset_splits[split] = ds
datasets = DatasetDict(dataset_splits)
```
I then push to hub
```python
datasets.push_to_hub("pietrolesci/robust_nli", token="<token>")
```
Finally, I load it from the hub
```python
datasets_loaded_from_hub = load_dataset("pietrolesci/robust_nli")
```
And I get that
```python
datasets["LI_TS"].features != datasets_loaded_from_hub["LI_TS"].features
```
since
```python
"label": ClassLabel(num_classes=2, names=["non-contradiction", "contradiction"])
```
gets remapped to
```python
"label": ClassLabel(num_classes=3, names=["entailment", "neutral", "contradiction"])
```
| Hi @pietrolesci, thanks for reporting.
Please note that this is a design purpose: a `DatasetDict` has the same features for all its datasets. Normally, a `DatasetDict` is composed of several sub-datasets each corresponding to a different **split**.
To handle sub-datasets with different features, we use another approach: use different **configurations** instead of **splits**.
However, for the moment `push_to_hub` does not support specifying different configurations. IMHO, we should implement this.
Hi @albertvillanova,
Thanks a lot for your reply! I got it now. The strange thing for me was to have it correctly working (i.e., DatasetDict with different features in some datasets) locally and not on the Hub. It would be great to have configuration supported by `push_to_hub`. Personally, this latter functionality allowed me to iterate rather quickly on dataset curation.
Again, thanks for your time @albertvillanova!
Best,
Pietro
Hi! Yes, we should override `DatasetDict.__setitem__` and throw an error if features dictionaries are different. `DatasetDict` is a subclass of `dict`, so `DatasetDict.{update/setdefault}` need to be overridden as well. We could avoid this by subclassing `UserDict`, but then we would get the name collision - `DatasetDict.data` vs. `UserDict.data`. This makes me think we should rename the `data` attribute of `DatasetDict`/`Dataset` for easier dict subclassing (would also simplify https://github.com/huggingface/datasets/pull/3997) and to follow good Python practices. Another option is to have a custom `UserDict` class in `py_utils`, but it can be hard to keep this class consistent with the built-in `UserDict`.
@albertvillanova @lhoestq wdyt?
I would keep things simple and keep subclassing dict. Regarding the features check, I guess this can be done only for `push_to_hub` right ? It is the only function right now that requires the underlying datasets to be splits (e.g. train/test) and have the same features.
Note that later you will be able to push datasets with different features as different dataset **configurations** (similarly to the [GLUE subsets](https://huggingface.co/datasets/glue) for example). We will work on this soon | 2022-05-19T12:32:30Z | [] | [] |
huggingface/datasets | 4,388 | huggingface__datasets-4388 | [
"4381"
] | 95193ae61e92aa537d0c65d37a1fd9d2393aae89 | diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -248,7 +248,7 @@ def __init__(
"""
# DatasetBuilder name
- self.name: str = camelcase_to_snakecase(self.__class__.__name__)
+ self.name: str = camelcase_to_snakecase(self.__module__.split(".")[-1])
self.hash: Optional[str] = hash
self.base_path = base_path
self.use_auth_token = use_auth_token
| diff --git a/tests/test_beam.py b/tests/test_beam.py
--- a/tests/test_beam.py
+++ b/tests/test_beam.py
@@ -65,9 +65,7 @@ def test_download_and_prepare(self):
builder.download_and_prepare()
self.assertTrue(
os.path.exists(
- os.path.join(
- tmp_cache_dir, "dummy_beam_dataset", "default", "0.0.0", "dummy_beam_dataset-train.arrow"
- )
+ os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", f"{builder.name}-train.arrow")
)
)
self.assertDictEqual(builder.info.features, datasets.Features({"content": datasets.Value("string")}))
@@ -79,9 +77,7 @@ def test_download_and_prepare(self):
dset["train"][expected_num_examples - 1], get_test_dummy_examples()[expected_num_examples - 1][1]
)
self.assertTrue(
- os.path.exists(
- os.path.join(tmp_cache_dir, "dummy_beam_dataset", "default", "0.0.0", "dataset_info.json")
- )
+ os.path.exists(os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", "dataset_info.json"))
)
del dset
@@ -99,9 +95,7 @@ def test_nested_features(self):
builder.download_and_prepare()
self.assertTrue(
os.path.exists(
- os.path.join(
- tmp_cache_dir, "nested_beam_dataset", "default", "0.0.0", "nested_beam_dataset-train.arrow"
- )
+ os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", f"{builder.name}-train.arrow")
)
)
self.assertDictEqual(
@@ -115,8 +109,6 @@ def test_nested_features(self):
dset["train"][expected_num_examples - 1], get_test_nested_examples()[expected_num_examples - 1][1]
)
self.assertTrue(
- os.path.exists(
- os.path.join(tmp_cache_dir, "nested_beam_dataset", "default", "0.0.0", "dataset_info.json")
- )
+ os.path.exists(os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", "dataset_info.json"))
)
del dset
diff --git a/tests/test_builder.py b/tests/test_builder.py
--- a/tests/test_builder.py
+++ b/tests/test_builder.py
@@ -123,40 +123,35 @@ def _split_generators(self, dl_manager):
def _run_concurrent_download_and_prepare(tmp_dir):
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.REUSE_DATASET_IF_EXISTS)
- return dummy_builder
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.REUSE_DATASET_IF_EXISTS)
+ return builder
class BuilderTest(TestCase):
def test_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
- )
- self.assertDictEqual(dummy_builder.info.features, Features({"text": Value("string")}))
- self.assertEqual(dummy_builder.info.splits["train"].num_examples, 100)
- self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dataset_info.json"))
+ os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", f"{builder.name}-train.arrow"))
)
+ self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
+ self.assertEqual(builder.info.splits["train"].num_examples, 100)
+ self.assertTrue(os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", "dataset_info.json")))
def test_download_and_prepare_checksum_computation(self):
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
- self.assertTrue(all(v["checksum"] is not None for _, v in dummy_builder.info.download_checksums.items()))
- dummy_builder_skip_checksum_computation = DummyBuilderSkipChecksumComputation(
- cache_dir=tmp_dir, name="dummy"
- )
- dummy_builder_skip_checksum_computation.download_and_prepare(
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ self.assertTrue(all(v["checksum"] is not None for _, v in builder.info.download_checksums.items()))
+ builder_skip_checksum_computation = DummyBuilderSkipChecksumComputation(cache_dir=tmp_dir, name="dummy")
+ builder_skip_checksum_computation.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD
)
self.assertTrue(
all(
- v["checksum"] is None
- for _, v in dummy_builder_skip_checksum_computation.info.download_checksums.items()
+ v["checksum"] is None for _, v in builder_skip_checksum_computation.info.download_checksums.items()
)
)
@@ -168,17 +163,17 @@ def test_concurrent_download_and_prepare(self):
pool.apply_async(_run_concurrent_download_and_prepare, kwds={"tmp_dir": tmp_dir})
for _ in range(processes)
]
- dummy_builders = [job.get() for job in jobs]
- for dummy_builder in dummy_builders:
+ builders = [job.get() for job in jobs]
+ for builder in builders:
self.assertTrue(
os.path.exists(
- os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow")
+ os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", f"{builder.name}-train.arrow")
)
)
- self.assertDictEqual(dummy_builder.info.features, Features({"text": Value("string")}))
- self.assertEqual(dummy_builder.info.splits["train"].num_examples, 100)
+ self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
+ self.assertEqual(builder.info.splits["train"].num_examples, 100)
self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dataset_info.json"))
+ os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", "dataset_info.json"))
)
def test_download_and_prepare_with_base_path(self):
@@ -186,34 +181,32 @@ def test_download_and_prepare_with_base_path(self):
rel_path = "dummy1.data"
abs_path = os.path.join(tmp_dir, "dummy2.data")
# test relative path is missing
- dummy_builder = DummyBuilderWithDownload(cache_dir=tmp_dir, name="dummy", rel_path=rel_path)
+ builder = DummyBuilderWithDownload(cache_dir=tmp_dir, name="dummy", rel_path=rel_path)
with self.assertRaises(FileNotFoundError):
- dummy_builder.download_and_prepare(
+ builder.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
# test absolute path is missing
- dummy_builder = DummyBuilderWithDownload(cache_dir=tmp_dir, name="dummy", abs_path=abs_path)
+ builder = DummyBuilderWithDownload(cache_dir=tmp_dir, name="dummy", abs_path=abs_path)
with self.assertRaises(FileNotFoundError):
- dummy_builder.download_and_prepare(
+ builder.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
# test that they are both properly loaded when they exist
open(os.path.join(tmp_dir, rel_path), "w")
open(abs_path, "w")
- dummy_builder = DummyBuilderWithDownload(
- cache_dir=tmp_dir, name="dummy", rel_path=rel_path, abs_path=abs_path
- )
- dummy_builder.download_and_prepare(
+ builder = DummyBuilderWithDownload(cache_dir=tmp_dir, name="dummy", rel_path=rel_path, abs_path=abs_path)
+ builder.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir,
- "dummy_builder_with_download",
+ builder.name,
"dummy",
"0.0.0",
- "dummy_builder_with_download-train.arrow",
+ f"{builder.name}-train.arrow",
)
)
)
@@ -229,34 +222,34 @@ def _post_processing_resources(self, split):
return {"tokenized_dataset": f"tokenized_dataset-{split}.arrow"}
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.info.post_processed = PostProcessedInfo(
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder.info.post_processed = PostProcessedInfo(
features=Features({"text": Value("string"), "tokens": [Value("string")]})
)
- dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
- dummy_builder._post_processing_resources = types.MethodType(_post_processing_resources, dummy_builder)
- os.makedirs(dummy_builder.cache_dir)
+ builder._post_process = types.MethodType(_post_process, builder)
+ builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
+ os.makedirs(builder.cache_dir)
- dummy_builder.info.splits = SplitDict()
- dummy_builder.info.splits.add(SplitInfo("train", num_examples=10))
- dummy_builder.info.splits.add(SplitInfo("test", num_examples=10))
+ builder.info.splits = SplitDict()
+ builder.info.splits.add(SplitInfo("train", num_examples=10))
+ builder.info.splits.add(SplitInfo("test", num_examples=10))
- for split in dummy_builder.info.splits:
+ for split in builder.info.splits:
with ArrowWriter(
- path=os.path.join(dummy_builder.cache_dir, f"dummy_builder-{split}.arrow"),
+ path=os.path.join(builder.cache_dir, f"{builder.name}-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with ArrowWriter(
- path=os.path.join(dummy_builder.cache_dir, f"tokenized_dataset-{split}.arrow"),
+ path=os.path.join(builder.cache_dir, f"tokenized_dataset-{split}.arrow"),
features=Features({"text": Value("string"), "tokens": [Value("string")]}),
) as writer:
writer.write_batch({"text": ["foo"] * 10, "tokens": [list("foo")] * 10})
writer.finalize()
- dsets = dummy_builder.as_dataset()
+ dsets = builder.as_dataset()
self.assertIsInstance(dsets, DatasetDict)
self.assertListEqual(list(dsets.keys()), ["train", "test"])
self.assertEqual(len(dsets["train"]), 10)
@@ -271,19 +264,19 @@ def _post_processing_resources(self, split):
self.assertListEqual(dsets["test"].column_names, ["text", "tokens"])
del dsets
- dset = dummy_builder.as_dataset("train")
+ dset = builder.as_dataset("train")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train")
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"text": Value("string"), "tokens": [Value("string")]}))
self.assertListEqual(dset.column_names, ["text", "tokens"])
- self.assertGreater(dummy_builder.info.post_processing_size, 0)
+ self.assertGreater(builder.info.post_processing_size, 0)
self.assertGreater(
- dummy_builder.info.post_processed.resources_checksums["train"]["tokenized_dataset"]["num_bytes"], 0
+ builder.info.post_processed.resources_checksums["train"]["tokenized_dataset"]["num_bytes"], 0
)
del dset
- dset = dummy_builder.as_dataset("train+test[:30%]")
+ dset = builder.as_dataset("train+test[:30%]")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test[:30%]")
self.assertEqual(len(dset), 13)
@@ -291,7 +284,7 @@ def _post_processing_resources(self, split):
self.assertListEqual(dset.column_names, ["text", "tokens"])
del dset
- dset = dummy_builder.as_dataset("all")
+ dset = builder.as_dataset("all")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test")
self.assertEqual(len(dset), 20)
@@ -303,30 +296,30 @@ def _post_process(self, dataset, resources_paths):
return dataset.select([0, 1], keep_in_memory=True)
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
- os.makedirs(dummy_builder.cache_dir)
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder._post_process = types.MethodType(_post_process, builder)
+ os.makedirs(builder.cache_dir)
- dummy_builder.info.splits = SplitDict()
- dummy_builder.info.splits.add(SplitInfo("train", num_examples=10))
- dummy_builder.info.splits.add(SplitInfo("test", num_examples=10))
+ builder.info.splits = SplitDict()
+ builder.info.splits.add(SplitInfo("train", num_examples=10))
+ builder.info.splits.add(SplitInfo("test", num_examples=10))
- for split in dummy_builder.info.splits:
+ for split in builder.info.splits:
with ArrowWriter(
- path=os.path.join(dummy_builder.cache_dir, f"dummy_builder-{split}.arrow"),
+ path=os.path.join(builder.cache_dir, f"{builder.name}-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with ArrowWriter(
- path=os.path.join(dummy_builder.cache_dir, f"small_dataset-{split}.arrow"),
+ path=os.path.join(builder.cache_dir, f"small_dataset-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 2})
writer.finalize()
- dsets = dummy_builder.as_dataset()
+ dsets = builder.as_dataset()
self.assertIsInstance(dsets, DatasetDict)
self.assertListEqual(list(dsets.keys()), ["train", "test"])
self.assertEqual(len(dsets["train"]), 2)
@@ -337,7 +330,7 @@ def _post_process(self, dataset, resources_paths):
self.assertListEqual(dsets["test"].column_names, ["text"])
del dsets
- dset = dummy_builder.as_dataset("train")
+ dset = builder.as_dataset("train")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train")
self.assertEqual(len(dset), 2)
@@ -345,7 +338,7 @@ def _post_process(self, dataset, resources_paths):
self.assertListEqual(dset.column_names, ["text"])
del dset
- dset = dummy_builder.as_dataset("train+test[:30%]")
+ dset = builder.as_dataset("train+test[:30%]")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test[:30%]")
self.assertEqual(len(dset), 2)
@@ -370,31 +363,31 @@ def _post_processing_resources(self, split):
return {"index": f"Flat-{split}.faiss"}
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
- dummy_builder._post_processing_resources = types.MethodType(_post_processing_resources, dummy_builder)
- os.makedirs(dummy_builder.cache_dir)
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder._post_process = types.MethodType(_post_process, builder)
+ builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
+ os.makedirs(builder.cache_dir)
- dummy_builder.info.splits = SplitDict()
- dummy_builder.info.splits.add(SplitInfo("train", num_examples=10))
- dummy_builder.info.splits.add(SplitInfo("test", num_examples=10))
+ builder.info.splits = SplitDict()
+ builder.info.splits.add(SplitInfo("train", num_examples=10))
+ builder.info.splits.add(SplitInfo("test", num_examples=10))
- for split in dummy_builder.info.splits:
+ for split in builder.info.splits:
with ArrowWriter(
- path=os.path.join(dummy_builder.cache_dir, f"dummy_builder-{split}.arrow"),
+ path=os.path.join(builder.cache_dir, f"{builder.name}-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with ArrowWriter(
- path=os.path.join(dummy_builder.cache_dir, f"small_dataset-{split}.arrow"),
+ path=os.path.join(builder.cache_dir, f"small_dataset-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 2})
writer.finalize()
- dsets = dummy_builder.as_dataset()
+ dsets = builder.as_dataset()
self.assertIsInstance(dsets, DatasetDict)
self.assertListEqual(list(dsets.keys()), ["train", "test"])
self.assertEqual(len(dsets["train"]), 10)
@@ -405,11 +398,11 @@ def _post_processing_resources(self, split):
self.assertListEqual(dsets["test"].column_names, ["text"])
self.assertListEqual(dsets["train"].list_indexes(), ["my_index"])
self.assertListEqual(dsets["test"].list_indexes(), ["my_index"])
- self.assertGreater(dummy_builder.info.post_processing_size, 0)
- self.assertGreater(dummy_builder.info.post_processed.resources_checksums["train"]["index"]["num_bytes"], 0)
+ self.assertGreater(builder.info.post_processing_size, 0)
+ self.assertGreater(builder.info.post_processed.resources_checksums["train"]["index"]["num_bytes"], 0)
del dsets
- dset = dummy_builder.as_dataset("train")
+ dset = builder.as_dataset("train")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train")
self.assertEqual(len(dset), 10)
@@ -418,7 +411,7 @@ def _post_processing_resources(self, split):
self.assertListEqual(dset.list_indexes(), ["my_index"])
del dset
- dset = dummy_builder.as_dataset("train+test[:30%]")
+ dset = builder.as_dataset("train+test[:30%]")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test[:30%]")
self.assertEqual(len(dset), 13)
@@ -438,42 +431,38 @@ def _post_processing_resources(self, split):
return {"tokenized_dataset": f"tokenized_dataset-{split}.arrow"}
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.info.post_processed = PostProcessedInfo(
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder.info.post_processed = PostProcessedInfo(
features=Features({"text": Value("string"), "tokens": [Value("string")]})
)
- dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
- dummy_builder._post_processing_resources = types.MethodType(_post_processing_resources, dummy_builder)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ builder._post_process = types.MethodType(_post_process, builder)
+ builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
+ os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", f"{builder.name}-train.arrow"))
)
- self.assertDictEqual(dummy_builder.info.features, Features({"text": Value("string")}))
+ self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
self.assertDictEqual(
- dummy_builder.info.post_processed.features,
+ builder.info.post_processed.features,
Features({"text": Value("string"), "tokens": [Value("string")]}),
)
- self.assertEqual(dummy_builder.info.splits["train"].num_examples, 100)
- self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dataset_info.json"))
- )
+ self.assertEqual(builder.info.splits["train"].num_examples, 100)
+ self.assertTrue(os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", "dataset_info.json")))
def _post_process(self, dataset, resources_paths):
return dataset.select([0, 1], keep_in_memory=True)
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
- self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
- )
- self.assertDictEqual(dummy_builder.info.features, Features({"text": Value("string")}))
- self.assertIsNone(dummy_builder.info.post_processed)
- self.assertEqual(dummy_builder.info.splits["train"].num_examples, 100)
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder._post_process = types.MethodType(_post_process, builder)
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dataset_info.json"))
+ os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", f"{builder.name}-train.arrow"))
)
+ self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
+ self.assertIsNone(builder.info.post_processed)
+ self.assertEqual(builder.info.splits["train"].num_examples, 100)
+ self.assertTrue(os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", "dataset_info.json")))
def _post_process(self, dataset, resources_paths):
if os.path.exists(resources_paths["index"]):
@@ -490,81 +479,73 @@ def _post_processing_resources(self, split):
return {"index": f"Flat-{split}.faiss"}
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder._post_process = types.MethodType(_post_process, dummy_builder)
- dummy_builder._post_processing_resources = types.MethodType(_post_processing_resources, dummy_builder)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder._post_process = types.MethodType(_post_process, builder)
+ builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dummy_builder-train.arrow"))
- )
- self.assertDictEqual(dummy_builder.info.features, Features({"text": Value("string")}))
- self.assertIsNone(dummy_builder.info.post_processed)
- self.assertEqual(dummy_builder.info.splits["train"].num_examples, 100)
- self.assertTrue(
- os.path.exists(os.path.join(tmp_dir, "dummy_builder", "dummy", "0.0.0", "dataset_info.json"))
+ os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", f"{builder.name}-train.arrow"))
)
+ self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
+ self.assertIsNone(builder.info.post_processed)
+ self.assertEqual(builder.info.splits["train"].num_examples, 100)
+ self.assertTrue(os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", "dataset_info.json")))
def test_error_download_and_prepare(self):
def _prepare_split(self, split_generator, **kwargs):
raise ValueError()
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder._prepare_split = types.MethodType(_prepare_split, dummy_builder)
+ builder = DummyBuilder(cache_dir=tmp_dir, name="dummy")
+ builder._prepare_split = types.MethodType(_prepare_split, builder)
self.assertRaises(
ValueError,
- dummy_builder.download_and_prepare,
+ builder.download_and_prepare,
try_from_hf_gcs=False,
download_mode=DownloadMode.FORCE_REDOWNLOAD,
)
- self.assertRaises(AssertionError, dummy_builder.as_dataset)
+ self.assertRaises(AssertionError, builder.as_dataset)
def test_generator_based_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy")
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir,
- "dummy_generator_based_builder",
+ builder.name,
"dummy",
"0.0.0",
- "dummy_generator_based_builder-train.arrow",
+ f"{builder.name}-train.arrow",
)
)
)
- self.assertDictEqual(dummy_builder.info.features, Features({"text": Value("string")}))
- self.assertEqual(dummy_builder.info.splits["train"].num_examples, 100)
- self.assertTrue(
- os.path.exists(
- os.path.join(tmp_dir, "dummy_generator_based_builder", "dummy", "0.0.0", "dataset_info.json")
- )
- )
+ self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
+ self.assertEqual(builder.info.splits["train"].num_examples, 100)
+ self.assertTrue(os.path.exists(os.path.join(tmp_dir, builder.name, "dummy", "0.0.0", "dataset_info.json")))
# Test that duplicated keys are ignored if ignore_verifications is True
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy")
+ builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy")
with patch("datasets.builder.ArrowWriter", side_effect=ArrowWriter) as mock_arrow_writer:
- dummy_builder.download_and_prepare(download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ builder.download_and_prepare(download_mode=DownloadMode.FORCE_REDOWNLOAD)
mock_arrow_writer.assert_called_once()
args, kwargs = mock_arrow_writer.call_args_list[0]
self.assertTrue(kwargs["check_duplicates"])
mock_arrow_writer.reset_mock()
- dummy_builder.download_and_prepare(
- download_mode=DownloadMode.FORCE_REDOWNLOAD, ignore_verifications=True
- )
+ builder.download_and_prepare(download_mode=DownloadMode.FORCE_REDOWNLOAD, ignore_verifications=True)
mock_arrow_writer.assert_called_once()
args, kwargs = mock_arrow_writer.call_args_list[0]
self.assertFalse(kwargs["check_duplicates"])
def test_cache_dir_no_args(self):
with tempfile.TemporaryDirectory() as tmp_dir:
- dummy_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_dir=None, data_files=None)
- relative_cache_dir_parts = Path(dummy_builder._relative_data_dir()).parts
- self.assertEqual(relative_cache_dir_parts, ("dummy_generator_based_builder", "dummy", "0.0.0"))
+ builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_dir=None, data_files=None)
+ relative_cache_dir_parts = Path(builder._relative_data_dir()).parts
+ self.assertEqual(relative_cache_dir_parts, (builder.name, "dummy", "0.0.0"))
def test_cache_dir_for_data_files(self):
with tempfile.TemporaryDirectory() as tmp_dir:
@@ -575,112 +556,110 @@ def test_cache_dir_for_data_files(self):
with open(dummy_data2, "w", encoding="utf-8") as f:
f.writelines("foo bar\n")
- dummy_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_files=dummy_data1)
+ builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_files=dummy_data1)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_files=dummy_data1)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_files=[dummy_data1])
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"train": dummy_data1}
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={Split.TRAIN: dummy_data1}
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"train": [dummy_data1]}
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"test": dummy_data1}
)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_files=dummy_data2)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, name="dummy", data_files=[dummy_data2])
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files=[dummy_data1, dummy_data2]
)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
- dummy_builder = DummyGeneratorBasedBuilder(
+ builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files=[dummy_data1, dummy_data2]
)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files=[dummy_data1, dummy_data2]
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files=[dummy_data2, dummy_data1]
)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
- dummy_builder = DummyGeneratorBasedBuilder(
+ builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"train": dummy_data1, "test": dummy_data2}
)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"train": dummy_data1, "test": dummy_data2}
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"train": [dummy_data1], "test": dummy_data2}
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"test": dummy_data2, "train": dummy_data1}
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"train": dummy_data1, "validation": dummy_data2}
)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, name="dummy", data_files={"train": [dummy_data1, dummy_data2], "test": dummy_data2}
)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
def test_cache_dir_for_features(self):
with tempfile.TemporaryDirectory() as tmp_dir:
f1 = Features({"id": Value("int8")})
f2 = Features({"id": Value("int32")})
- dummy_builder = DummyGeneratorBasedBuilderWithIntegers(cache_dir=tmp_dir, name="dummy", features=f1)
+ builder = DummyGeneratorBasedBuilderWithIntegers(cache_dir=tmp_dir, name="dummy", features=f1)
other_builder = DummyGeneratorBasedBuilderWithIntegers(cache_dir=tmp_dir, name="dummy", features=f1)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilderWithIntegers(cache_dir=tmp_dir, name="dummy", features=f2)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
def test_cache_dir_for_config_kwargs(self):
with tempfile.TemporaryDirectory() as tmp_dir:
# create config on the fly
- dummy_builder = DummyGeneratorBasedBuilderWithConfig(
- cache_dir=tmp_dir, name="dummy", content="foo", times=2
- )
+ builder = DummyGeneratorBasedBuilderWithConfig(cache_dir=tmp_dir, name="dummy", content="foo", times=2)
other_builder = DummyGeneratorBasedBuilderWithConfig(
cache_dir=tmp_dir, name="dummy", times=2, content="foo"
)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
- self.assertIn("content=foo", dummy_builder.cache_dir)
- self.assertIn("times=2", dummy_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
+ self.assertIn("content=foo", builder.cache_dir)
+ self.assertIn("times=2", builder.cache_dir)
other_builder = DummyGeneratorBasedBuilderWithConfig(
cache_dir=tmp_dir, name="dummy", content="bar", times=2
)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilderWithConfig(cache_dir=tmp_dir, name="dummy", content="foo")
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
with tempfile.TemporaryDirectory() as tmp_dir:
# overwrite an existing config
- dummy_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="a", content="foo", times=2)
+ builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="a", content="foo", times=2)
other_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="a", times=2, content="foo")
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
- self.assertIn("content=foo", dummy_builder.cache_dir)
- self.assertIn("times=2", dummy_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
+ self.assertIn("content=foo", builder.cache_dir)
+ self.assertIn("times=2", builder.cache_dir)
other_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="a", content="bar", times=2)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="a", content="foo")
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
def test_config_names(self):
with tempfile.TemporaryDirectory() as tmp_dir:
@@ -689,25 +668,25 @@ def test_config_names(self):
DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, data_files=None, data_dir=None)
self.assertIn("Please pick one among the available configs", str(error_context.exception))
- dummy_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="a")
- self.assertEqual(dummy_builder.config.name, "a")
+ builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="a")
+ self.assertEqual(builder.config.name, "a")
- dummy_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="b")
- self.assertEqual(dummy_builder.config.name, "b")
+ builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, name="b")
+ self.assertEqual(builder.config.name, "b")
with self.assertRaises(ValueError):
DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir)
- dummy_builder = DummyBuilderWithDefaultConfig(cache_dir=tmp_dir)
- self.assertEqual(dummy_builder.config.name, "a")
+ builder = DummyBuilderWithDefaultConfig(cache_dir=tmp_dir)
+ self.assertEqual(builder.config.name, "a")
def test_cache_dir_for_data_dir(self):
with tempfile.TemporaryDirectory() as tmp_dir, tempfile.TemporaryDirectory() as data_dir:
- dummy_builder = DummyBuilderWithManualDownload(cache_dir=tmp_dir, name="a", data_dir=data_dir)
+ builder = DummyBuilderWithManualDownload(cache_dir=tmp_dir, name="a", data_dir=data_dir)
other_builder = DummyBuilderWithManualDownload(cache_dir=tmp_dir, name="a", data_dir=data_dir)
- self.assertEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyBuilderWithManualDownload(cache_dir=tmp_dir, name="a", data_dir=tmp_dir)
- self.assertNotEqual(dummy_builder.cache_dir, other_builder.cache_dir)
+ self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
@pytest.mark.parametrize(
@@ -721,23 +700,23 @@ def test_cache_dir_for_data_dir(self):
@pytest.mark.parametrize("in_memory", [False, True])
def test_builder_as_dataset(split, expected_dataset_class, expected_dataset_length, in_memory, tmp_path):
cache_dir = str(tmp_path)
- dummy_builder = DummyBuilder(cache_dir=cache_dir, name="dummy")
- os.makedirs(dummy_builder.cache_dir)
+ builder = DummyBuilder(cache_dir=cache_dir, name="dummy")
+ os.makedirs(builder.cache_dir)
- dummy_builder.info.splits = SplitDict()
- dummy_builder.info.splits.add(SplitInfo("train", num_examples=10))
- dummy_builder.info.splits.add(SplitInfo("test", num_examples=10))
+ builder.info.splits = SplitDict()
+ builder.info.splits.add(SplitInfo("train", num_examples=10))
+ builder.info.splits.add(SplitInfo("test", num_examples=10))
- for info_split in dummy_builder.info.splits:
+ for info_split in builder.info.splits:
with ArrowWriter(
- path=os.path.join(dummy_builder.cache_dir, f"dummy_builder-{info_split}.arrow"),
+ path=os.path.join(builder.cache_dir, f"{builder.name}-{info_split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with assert_arrow_memory_increases() if in_memory else assert_arrow_memory_doesnt_increase():
- dataset = dummy_builder.as_dataset(split=split, in_memory=in_memory)
+ dataset = builder.as_dataset(split=split, in_memory=in_memory)
assert isinstance(dataset, expected_dataset_class)
if isinstance(dataset, DatasetDict):
assert list(dataset.keys()) == ["train", "test"]
@@ -758,10 +737,10 @@ def test_generator_based_builder_as_dataset(in_memory, tmp_path):
cache_dir = tmp_path / "data"
cache_dir.mkdir()
cache_dir = str(cache_dir)
- dummy_builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir, name="dummy")
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir, name="dummy")
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
with assert_arrow_memory_increases() if in_memory else assert_arrow_memory_doesnt_increase():
- dataset = dummy_builder.as_dataset("train", in_memory=in_memory)
+ dataset = builder.as_dataset("train", in_memory=in_memory)
assert dataset.data.to_pydict() == {"text": ["foo"] * 100}
@@ -772,8 +751,8 @@ def test_custom_writer_batch_size(tmp_path, writer_batch_size, default_writer_ba
cache_dir = str(tmp_path)
if default_writer_batch_size:
DummyGeneratorBasedBuilder.DEFAULT_WRITER_BATCH_SIZE = default_writer_batch_size
- dummy_builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir, name="dummy", writer_batch_size=writer_batch_size)
- assert dummy_builder._writer_batch_size == (writer_batch_size or default_writer_batch_size)
- dummy_builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
- dataset = dummy_builder.as_dataset("train")
+ builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir, name="dummy", writer_batch_size=writer_batch_size)
+ assert builder._writer_batch_size == (writer_batch_size or default_writer_batch_size)
+ builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
+ dataset = builder.as_dataset("train")
assert len(dataset.data[0].chunks) == expected_chunks
| Bug in caching 2 datasets both with the same builder class name
## Describe the bug
The two datasets `mteb/mtop_intent` and `mteb/mtop_domain `use both the same cache folder `.cache/huggingface/datasets/mteb___mtop`. So if you first load `mteb/mtop_intent` then datasets will not load `mteb/mtop_domain`.
If you delete this cache folder and flip the order how you load the two datasets , you will get the opposite datasets loaded (difference is here in terms of the label and label_text).
## Steps to reproduce the bug
```python
import datasets
dataset = datasets.load_dataset("mteb/mtop_intent", "en")
print(dataset['train'][0])
dataset = datasets.load_dataset("mteb/mtop_domain", "en")
print(dataset['train'][0])
```
## Expected results
```
Reusing dataset mtop (/home/nouamane/.cache/huggingface/datasets/mteb___mtop_intent/en/0.0.0/f930e32a294fed424f70263d8802390e350fff17862266e5fc156175c07d9c35)
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 920.14it/s]
{'id': 3232343436343136, 'text': 'Has Angelika Kratzer video messaged me?', 'label': 1, 'label_text': 'GET_MESSAGE'}
Reusing dataset mtop (/home/nouamane/.cache/huggingface/datasets/mteb___mtop_domain/en/0.0.0/f930e32a294fed424f70263d8802390e350fff17862266e5fc156175c07d9c35)
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 1307.59it/s]
{'id': 3232343436343136, 'text': 'Has Angelika Kratzer video messaged me?', 'label': 0, 'label_text': 'messaging'}
```
## Actual results
```
Reusing dataset mtop (/home/nouamane/.cache/huggingface/datasets/mteb___mtop/en/0.0.0/f930e32a294fed424f70263d8802390e350fff17862266e5fc156175c07d9c35)
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 920.14it/s]
{'id': 3232343436343136, 'text': 'Has Angelika Kratzer video messaged me?', 'label': 1, 'label_text': 'GET_MESSAGE'}
Reusing dataset mtop (/home/nouamane/.cache/huggingface/datasets/mteb___mtop/en/0.0.0/f930e32a294fed424f70263d8802390e350fff17862266e5fc156175c07d9c35)
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 1307.59it/s]
{'id': 3232343436343136, 'text': 'Has Angelika Kratzer video messaged me?', 'label': 1, 'label_text': 'GET_MESSAGE'}
```
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 2.2.1
- Platform: macOS-12.1-arm64-arm-64bit
- Python version: 3.9.12
- PyArrow version: 8.0.0
- Pandas version: 1.4.2
| Hi @NouamaneTazi, thanks for reporting.
Please note that both datasets are cached in the same directory because their loading builder classes have the same name: `class MTOP(datasets.GeneratorBasedBuilder)`.
You should name their builder classes differently, e.g.:
- `MtopDomain`
- `MtopIntent` | 2022-05-23T06:26:35Z | [] | [] |
huggingface/datasets | 4,412 | huggingface__datasets-4412 | [
"4115"
] | 0bb47271910c8a0b628dba157988372307fca1d2 | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -122,7 +122,7 @@ def _resolve_single_pattern_locally(
matched_paths = [
Path(filepath).resolve()
for filepath in glob_iter
- if filepath.name not in data_files_ignore and not filepath.name.startswith(".")
+ if filepath.name not in data_files_ignore and not any(part.startswith((".", "__")) for part in filepath.parts)
]
if allowed_extensions is not None:
out = [
@@ -307,7 +307,7 @@ def _resolve_single_pattern_in_dataset_repository(
matched_paths = [
filepath
for filepath in glob_iter
- if filepath.name not in data_files_ignore and not filepath.name.startswith(".")
+ if filepath.name not in data_files_ignore and not any(part.startswith((".", "__")) for part in filepath.parts)
]
if allowed_extensions is not None:
out = [
diff --git a/src/datasets/download/download_manager.py b/src/datasets/download/download_manager.py
--- a/src/datasets/download/download_manager.py
+++ b/src/datasets/download/download_manager.py
@@ -122,10 +122,23 @@ def _iter_from_paths(cls, urlpaths: Union[str, List[str]]) -> Generator[str, Non
urlpaths = [urlpaths]
for urlpath in urlpaths:
if os.path.isfile(urlpath):
+ if os.path.basename(urlpath).startswith((".", "__")):
+ # skipping hidden files
+ return
yield urlpath
else:
- for dirpath, _, filenames in os.walk(urlpath):
+ for dirpath, dirnames, filenames in os.walk(urlpath):
+ for i, dirname in enumerate(dirnames):
+ if dirname.startswith((".", "__")):
+ # skipping hidden directories; prune the search
+ del dirnames[i]
+ if os.path.basename(dirpath).startswith((".", "__")):
+ # skipping hidden directories
+ continue
for filename in filenames:
+ if filename.startswith((".", "__")):
+ # skipping hidden files
+ continue
yield os.path.join(dirpath, filename)
@classmethod
diff --git a/src/datasets/download/mock_download_manager.py b/src/datasets/download/mock_download_manager.py
--- a/src/datasets/download/mock_download_manager.py
+++ b/src/datasets/download/mock_download_manager.py
@@ -211,7 +211,7 @@ def manage_extracted_files(self):
def iter_archive(self, path):
path = Path(path)
for file_path in path.rglob("*"):
- if file_path.is_file() and not file_path.name.startswith(".") and not file_path.name.startswith("__"):
+ if file_path.is_file() and not file_path.name.startswith((".", "__")):
yield file_path.relative_to(path).as_posix(), file_path.open("rb")
def iter_files(self, paths):
@@ -219,8 +219,14 @@ def iter_files(self, paths):
paths = [paths]
for path in paths:
if os.path.isfile(path):
+ if os.path.basename(path).startswith((".", "__")):
+ return
yield path
else:
for dirpath, _, filenames in os.walk(path):
+ if os.path.basename(dirpath).startswith((".", "__")):
+ continue
for filename in filenames:
+ if filename.startswith((".", "__")):
+ continue
yield os.path.join(dirpath, filename)
diff --git a/src/datasets/download/streaming_download_manager.py b/src/datasets/download/streaming_download_manager.py
--- a/src/datasets/download/streaming_download_manager.py
+++ b/src/datasets/download/streaming_download_manager.py
@@ -636,7 +636,7 @@ def xwalk(urlpath, use_auth_token: Optional[Union[str, bool]] = None):
"""
main_hop, *rest_hops = str(urlpath).split("::")
if is_local_path(main_hop):
- return os.walk(main_hop)
+ yield from os.walk(main_hop)
else:
# walking inside a zip in a private repo requires authentication
if rest_hops and (rest_hops[0].startswith("http://") or rest_hops[0].startswith("https://")):
@@ -718,7 +718,7 @@ def _iter_from_fileobj(cls, f) -> Generator[Tuple, None, None]:
continue
if file_path is None:
continue
- if os.path.basename(file_path).startswith(".") or os.path.basename(file_path).startswith("__"):
+ if os.path.basename(file_path).startswith((".", "__")):
# skipping hidden files
continue
file_obj = stream.extractfile(tarinfo)
@@ -753,10 +753,24 @@ def _iter_from_urlpaths(
urlpaths = [urlpaths]
for urlpath in urlpaths:
if xisfile(urlpath, use_auth_token=use_auth_token):
+ if xbasename(urlpath).startswith((".", "__")):
+ # skipping hidden files
+ return
yield urlpath
else:
- for dirpath, _, filenames in xwalk(urlpath, use_auth_token=use_auth_token):
+ for dirpath, dirnames, filenames in xwalk(urlpath, use_auth_token=use_auth_token):
+ for i, dirname in enumerate(dirnames):
+ if dirname.startswith((".", "__")):
+ # skipping hidden directories; prune the search
+ # (only works for local paths as fsspec's walk doesn't support the in-place modification)
+ del dirnames[i]
+ if xbasename(dirpath).startswith((".", "__")):
+ # skipping hidden directories
+ continue
for filename in filenames:
+ if filename.startswith((".", "__")):
+ # skipping hidden files
+ continue
yield xjoin(dirpath, filename)
@classmethod
diff --git a/src/datasets/packaged_modules/pandas/pandas.py b/src/datasets/packaged_modules/pandas/pandas.py
--- a/src/datasets/packaged_modules/pandas/pandas.py
+++ b/src/datasets/packaged_modules/pandas/pandas.py
@@ -17,12 +17,16 @@ def _split_generators(self, dl_manager):
files = data_files
if isinstance(files, str):
files = [files]
- return [datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": files})]
+ # Use `dl_manager.iter_files` to skip hidden files in an extracted archive
+ return [
+ datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": dl_manager.iter_files(files)})
+ ]
splits = []
for split_name, files in data_files.items():
if isinstance(files, str):
files = [files]
- splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": files}))
+ # Use `dl_manager.iter_files` to skip hidden files in an extracted archive
+ splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": dl_manager.iter_files(files)}))
return splits
def _generate_tables(self, files):
diff --git a/src/datasets/packaged_modules/parquet/parquet.py b/src/datasets/packaged_modules/parquet/parquet.py
--- a/src/datasets/packaged_modules/parquet/parquet.py
+++ b/src/datasets/packaged_modules/parquet/parquet.py
@@ -34,12 +34,16 @@ def _split_generators(self, dl_manager):
files = data_files
if isinstance(files, str):
files = [files]
- return [datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": files})]
+ # Use `dl_manager.iter_files` to skip hidden files in an extracted archive
+ return [
+ datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"files": dl_manager.iter_files(files)})
+ ]
splits = []
for split_name, files in data_files.items():
if isinstance(files, str):
files = [files]
- splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": files}))
+ # Use `dl_manager.iter_files` to skip hidden files in an extracted archive
+ splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": dl_manager.iter_files(files)}))
return splits
def _generate_tables(self, files):
| diff --git a/tests/conftest.py b/tests/conftest.py
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -483,3 +483,26 @@ def zip_image_path(image_path, tmp_path_factory):
f.write(image_path, arcname=os.path.basename(image_path))
f.write(image_path, arcname=os.path.basename(image_path).replace(".jpg", "2.jpg"))
return path
+
+
[email protected](scope="session")
+def data_dir_with_hidden_files(tmp_path_factory):
+ data_dir = tmp_path_factory.mktemp("data_dir")
+
+ (data_dir / "subdir").mkdir()
+ with open(data_dir / "subdir" / "train.txt", "w") as f:
+ f.write("foo\n" * 10)
+ with open(data_dir / "subdir" / "test.txt", "w") as f:
+ f.write("bar\n" * 10)
+ # hidden file
+ with open(data_dir / "subdir" / ".test.txt", "w") as f:
+ f.write("bar\n" * 10)
+
+ # hidden directory
+ (data_dir / ".subdir").mkdir()
+ with open(data_dir / ".subdir" / "train.txt", "w") as f:
+ f.write("foo\n" * 10)
+ with open(data_dir / ".subdir" / "test.txt", "w") as f:
+ f.write("bar\n" * 10)
+
+ return data_dir
diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -21,6 +21,7 @@
_TEST_PATTERNS = ["*", "**", "**/*", "*.txt", "data/*", "**/*.txt", "**/train.txt"]
_FILES_TO_IGNORE = {".dummy", "README.md", "dummy_data.zip", "dataset_infos.json"}
+_DIRS_TO_IGNORE = {"data/.dummy_subdir"}
_TEST_PATTERNS_SIZES = dict(
[
("*", 0),
@@ -57,9 +58,24 @@ def complex_data_dir(tmp_path):
with open(data_dir / "data" / "subdir" / "test.txt", "w") as f:
f.write("bar\n" * 10)
+ (data_dir / "data" / ".dummy_subdir").mkdir()
+ with open(data_dir / "data" / ".dummy_subdir" / "train.txt", "w") as f:
+ f.write("foo\n" * 10)
+ with open(data_dir / "data" / ".dummy_subdir" / "test.txt", "w") as f:
+ f.write("bar\n" * 10)
+
return str(data_dir)
+def is_relative_to(path, *other):
+ # A built-in method in Python 3.9+
+ try:
+ path.relative_to(*other)
+ return True
+ except ValueError:
+ return False
+
+
@pytest.fixture
def pattern_results(complex_data_dir):
# We use fsspec glob as a reference for data files resolution from patterns.
@@ -78,7 +94,11 @@ def pattern_results(complex_data_dir):
pattern: sorted(
str(Path(path).resolve())
for path in fsspec.filesystem("file").glob(os.path.join(complex_data_dir, pattern))
- if Path(path).name not in _FILES_TO_IGNORE and Path(path).is_file()
+ if Path(path).name not in _FILES_TO_IGNORE
+ and not any(
+ is_relative_to(Path(path), os.path.join(complex_data_dir, dir_path)) for dir_path in _DIRS_TO_IGNORE
+ )
+ and Path(path).is_file()
)
for pattern in _TEST_PATTERNS
}
diff --git a/tests/test_download_manager.py b/tests/test_download_manager.py
--- a/tests/test_download_manager.py
+++ b/tests/test_download_manager.py
@@ -136,3 +136,10 @@ def test_iter_archive_file(tar_nested_jsonl_path):
_test_jsonl(subpath, subfile)
assert num_tar == 1
assert num_jsonl == 2
+
+
+def test_iter_files(data_dir_with_hidden_files):
+ dl_manager = DownloadManager()
+ for num_file, file in enumerate(dl_manager.iter_files(data_dir_with_hidden_files), start=1):
+ pass
+ assert num_file == 2
diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -750,3 +750,10 @@ def test_iter_archive_file(tar_nested_jsonl_path):
_test_jsonl(subpath, subfile)
assert num_tar == 1
assert num_jsonl == 2
+
+
+def test_iter_files(data_dir_with_hidden_files):
+ dl_manager = StreamingDownloadManager()
+ for num_file, file in enumerate(dl_manager.iter_files(data_dir_with_hidden_files), start=1):
+ pass
+ assert num_file == 2
| ImageFolder add option to ignore some folders like '.ipynb_checkpoints'
**Is your feature request related to a problem? Please describe.**
I sometimes like to peek at the dataset images from jupyterlab. thus '.ipynb_checkpoints' folder appears where my dataset is and (just realized) leads to accidental duplicate image additions. I think this is an easy enough thing to miss especially if the dataset is very large.
**Describe the solution you'd like**
maybe have an option `ignore` or something .gitignore style
`dataset = load_dataset("imagefolder", data_dir="./data/original", ignore="regex?")`
**Describe alternatives you've considered**
Could filter out manually
| Maybe it would be nice to ignore private dirs like this one (ones starting with `.`) by default.
CC @mariosasko
Maybe we can add a `ignore_hidden_files` flag to the builder configs of our packaged loaders (to be consistent across all of them), wdyt @lhoestq @albertvillanova?
I think they should always ignore them actually ! Not sure if adding a flag would be helpful
@lhoestq But what if the user explicitly requests those files via regex?
`glob.glob` ignores hidden files (files starting with ".") by default unless they are explicitly requested, but fsspec's `glob` doesn't follow this behavior, which is probably a bug, so maybe we can raise an issue or open a PR in their repo?
> @lhoestq But what if the user explicitly requests those files via regex?
Usually hidden files are meant to be ignored. If they are data files, they must be placed outside a hidden directory in the first place right ? I think it's more sensible to explain this than adding a flag.
> glob.glob ignores hidden files (files starting with ".") by default unless they are explicitly requested, but fsspec's glob doesn't follow this behavior, which is probably a bug, so maybe we can raise an issue or open a PR in their repo?
After globbing using `fsspec`, we already ignore files that start with a `.` in `_resolve_single_pattern_locally` and `_resolve_single_pattern_in_dataset_repository`, I guess we can just account for parent directories as well ?
We could open an issue on `fsspec` but I think they won't change this since it's an important breaking change for them. | 2022-05-26T12:10:28Z | [] | [] |
huggingface/datasets | 4,433 | huggingface__datasets-4433 | [
"4348"
] | a442dfc883c3952fe0cc28bc8ac05011f83cd346 | diff --git a/src/datasets/download/download_manager.py b/src/datasets/download/download_manager.py
--- a/src/datasets/download/download_manager.py
+++ b/src/datasets/download/download_manager.py
@@ -128,10 +128,9 @@ def _iter_from_paths(cls, urlpaths: Union[str, List[str]]) -> Generator[str, Non
yield urlpath
else:
for dirpath, dirnames, filenames in os.walk(urlpath):
- for i, dirname in enumerate(dirnames):
- if dirname.startswith((".", "__")):
- # skipping hidden directories; prune the search
- del dirnames[i]
+ # skipping hidden directories; prune the search
+ # [:] for the in-place list modification required by os.walk
+ dirnames[:] = [dirname for dirname in dirnames if not dirname.startswith((".", "__"))]
if os.path.basename(dirpath).startswith((".", "__")):
# skipping hidden directories
continue
diff --git a/src/datasets/download/streaming_download_manager.py b/src/datasets/download/streaming_download_manager.py
--- a/src/datasets/download/streaming_download_manager.py
+++ b/src/datasets/download/streaming_download_manager.py
@@ -759,11 +759,10 @@ def _iter_from_urlpaths(
yield urlpath
else:
for dirpath, dirnames, filenames in xwalk(urlpath, use_auth_token=use_auth_token):
- for i, dirname in enumerate(dirnames):
- if dirname.startswith((".", "__")):
- # skipping hidden directories; prune the search
- # (only works for local paths as fsspec's walk doesn't support the in-place modification)
- del dirnames[i]
+ # skipping hidden directories; prune the search
+ # [:] for the in-place list modification required by os.walk
+ # (only works for local paths as fsspec's walk doesn't support the in-place modification)
+ dirnames[:] = [dirname for dirname in dirnames if not dirname.startswith((".", "__"))]
if xbasename(dirpath).startswith((".", "__")):
# skipping hidden directories
continue
diff --git a/src/datasets/inspect.py b/src/datasets/inspect.py
--- a/src/datasets/inspect.py
+++ b/src/datasets/inspect.py
@@ -15,6 +15,10 @@
# Lint as: python3
""" List and inspect datasets and metrics."""
+import inspect
+import os
+import shutil
+from pathlib import PurePath
from typing import Dict, List, Mapping, Optional, Sequence, Union
import huggingface_hub
@@ -30,6 +34,7 @@
load_dataset_builder,
metric_module_factory,
)
+from .utils.file_utils import relative_to_absolute_path
from .utils.logging import get_logger
from .utils.version import Version
@@ -118,13 +123,24 @@ def inspect_dataset(path: str, local_path: str, download_config: Optional[Downlo
**download_kwargs (additional keyword arguments): Optional arguments for [`DownloadConfig`] which will override
the attributes of `download_config` if supplied.
"""
- dataset_module = dataset_module_factory(
- path, download_config=download_config, force_local_path=local_path, **download_kwargs
- )
+ dataset_module = dataset_module_factory(path, download_config=download_config, **download_kwargs)
+ builder_cls = import_main_class(dataset_module.module_path, dataset=True)
+ module_source_path = inspect.getsourcefile(builder_cls)
+ module_source_dirpath = os.path.dirname(module_source_path)
+ for dirpath, dirnames, filenames in os.walk(module_source_dirpath):
+ dst_dirpath = os.path.join(local_path, os.path.relpath(dirpath, module_source_dirpath))
+ os.makedirs(dst_dirpath, exist_ok=True)
+ # skipping hidden directories; prune the search
+ # [:] for the in-place list modification required by os.walk
+ dirnames[:] = [dirname for dirname in dirnames if not dirname.startswith((".", "__"))]
+ for filename in filenames:
+ shutil.copy2(os.path.join(dirpath, filename), os.path.join(dst_dirpath, filename))
+ shutil.copystat(dirpath, dst_dirpath)
+ local_path = relative_to_absolute_path(local_path)
print(
f"The processing script for dataset {path} can be inspected at {local_path}. "
- f"The main class is in {dataset_module.module_path}. "
- f"You can modify this processing script and use it with `datasets.load_dataset({local_path})`."
+ f"The main class is in {module_source_dirpath}. "
+ f'You can modify this processing script and use it with `datasets.load_dataset("{PurePath(local_path).as_posix()}")`.'
)
@@ -143,13 +159,23 @@ def inspect_metric(path: str, local_path: str, download_config: Optional[Downloa
download_config (Optional ``datasets.DownloadConfig``): specific download configuration parameters.
**download_kwargs (additional keyword arguments): optional attributes for DownloadConfig() which will override the attributes in download_config if supplied.
"""
- metric_module = metric_module_factory(
- path, download_config=download_config, force_local_path=local_path, **download_kwargs
- )
+ metric_module = metric_module_factory(path, download_config=download_config, **download_kwargs)
+ builder_cls = import_main_class(metric_module.module_path, dataset=False)
+ module_source_path = inspect.getsourcefile(builder_cls)
+ module_source_dirpath = os.path.dirname(module_source_path)
+ for dirpath, dirnames, filenames in os.walk(module_source_dirpath):
+ dst_dirpath = os.path.join(local_path, os.path.relpath(dirpath, module_source_dirpath))
+ os.makedirs(dst_dirpath, exist_ok=True)
+ # skipping hidden directories; prune the search
+ dirnames[:] = [dirname for dirname in dirnames if not dirname.startswith((".", "__"))]
+ for filename in filenames:
+ shutil.copy2(os.path.join(dirpath, filename), os.path.join(dst_dirpath, filename))
+ shutil.copystat(dirpath, dst_dirpath)
+ local_path = relative_to_absolute_path(local_path)
print(
f"The processing scripts for metric {path} can be inspected at {local_path}. "
- f"The main class is in {metric_module.module_path}. "
- f"You can modify this processing scripts and use it with `datasets.load_metric({local_path})`."
+ f"The main class is in {module_source_dirpath}. "
+ f'You can modify this processing scripts and use it with `datasets.load_metric("{PurePath(local_path).as_posix()}")`.'
)
@@ -219,7 +245,6 @@ def get_dataset_config_names(
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
download_mode: Optional[DownloadMode] = None,
- force_local_path: Optional[str] = None,
dynamic_modules_path: Optional[str] = None,
data_files: Optional[Union[Dict, List, str]] = None,
**download_kwargs,
@@ -241,8 +266,6 @@ def get_dataset_config_names(
Specifying a version that is different from your local version of the lib might cause compatibility issues.
download_config (:class:`DownloadConfig`, optional): Specific download configuration parameters.
download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
- force_local_path (Optional str): Optional path to a local path to download and prepare the script to.
- Used to inspect or modify the script folder.
dynamic_modules_path (Optional str, defaults to HF_MODULES_CACHE / "datasets_modules", i.e. ~/.cache/huggingface/modules/datasets_modules):
Optional path to the directory in which the dynamic modules are saved. It must have been initialized with :obj:`init_dynamic_modules`.
By default the datasets and metrics are stored inside the `datasets_modules` module.
@@ -274,7 +297,6 @@ def get_dataset_config_names(
revision=revision,
download_config=download_config,
download_mode=download_mode,
- force_local_path=force_local_path,
dynamic_modules_path=dynamic_modules_path,
data_files=data_files,
**download_kwargs,
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -1100,7 +1100,6 @@ def dataset_module_factory(
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
download_mode: Optional[DownloadMode] = None,
- force_local_path: Optional[str] = None,
dynamic_modules_path: Optional[str] = None,
data_dir: Optional[str] = None,
data_files: Optional[Union[Dict, List, str, DataFilesDict]] = None,
@@ -1142,8 +1141,6 @@ def dataset_module_factory(
You can specify a different version that the default "main" by using a commit sha or a git tag of the dataset repository.
download_config (:class:`DownloadConfig`, optional): Specific download configuration parameters.
download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
- force_local_path (Optional str): Optional path to a local path to download and prepare the script to.
- Used to inspect or modify the script folder.
dynamic_modules_path (Optional str, defaults to HF_MODULES_CACHE / "datasets_modules", i.e. ~/.cache/huggingface/modules/datasets_modules):
Optional path to the directory in which the dynamic modules are saved. It must have been initialized with :obj:`init_dynamic_modules`.
By default the datasets and metrics are stored inside the `datasets_modules` module.
@@ -1212,7 +1209,7 @@ def dataset_module_factory(
path, data_dir=data_dir, data_files=data_files, download_mode=download_mode
).get_module()
# Try remotely
- elif is_relative_path(path) and path.count("/") <= 1 and not force_local_path:
+ elif is_relative_path(path) and path.count("/") <= 1:
try:
_raise_if_offline_mode_is_enabled()
if path.count("/") == 0: # even though the dataset is on the Hub, we get it from GitHub for now
@@ -1292,7 +1289,6 @@ def metric_module_factory(
revision: Optional[Union[str, Version]] = None,
download_config: Optional[DownloadConfig] = None,
download_mode: Optional[DownloadMode] = None,
- force_local_path: Optional[str] = None,
dynamic_modules_path: Optional[str] = None,
**download_kwargs,
) -> MetricModule:
@@ -1320,8 +1316,6 @@ def metric_module_factory(
Specifying a version that is different from your local version of the lib might cause compatibility issues.
download_config (:class:`DownloadConfig`, optional): Specific download configuration parameters.
download_mode (:class:`DownloadMode`, default ``REUSE_DATASET_IF_EXISTS``): Download/generate mode.
- force_local_path (Optional str): Optional path to a local path to download and prepare the script to.
- Used to inspect or modify the script folder.
dynamic_modules_path (Optional str, defaults to HF_MODULES_CACHE / "datasets_modules", i.e. ~/.cache/huggingface/modules/datasets_modules):
Optional path to the directory in which the dynamic modules are saved. It must have been initialized with :obj:`init_dynamic_modules`.
By default, the datasets and metrics are stored inside the `datasets_modules` module.
@@ -1353,7 +1347,7 @@ def metric_module_factory(
return LocalMetricModuleFactory(
combined_path, download_mode=download_mode, dynamic_modules_path=dynamic_modules_path
).get_module()
- elif is_relative_path(path) and path.count("/") == 0 and not force_local_path:
+ elif is_relative_path(path) and path.count("/") == 0:
try:
return GithubMetricModuleFactory(
path,
| diff --git a/tests/test_inspect.py b/tests/test_inspect.py
--- a/tests/test_inspect.py
+++ b/tests/test_inspect.py
@@ -1,6 +1,31 @@
+import os
+
import pytest
-from datasets import get_dataset_config_info, get_dataset_config_names, get_dataset_infos, get_dataset_split_names
+from datasets import (
+ get_dataset_config_info,
+ get_dataset_config_names,
+ get_dataset_infos,
+ get_dataset_split_names,
+ inspect_dataset,
+ inspect_metric,
+)
+
+
[email protected]("path", ["paws", "csv"])
+def test_inspect_dataset(path, tmp_path):
+ inspect_dataset(path, tmp_path)
+ script_name = path + ".py"
+ assert script_name in os.listdir(tmp_path)
+ assert "__pycache__" not in os.listdir(tmp_path)
+
+
[email protected]("path", ["accuracy"])
+def test_inspect_metric(path, tmp_path):
+ inspect_metric(path, tmp_path)
+ script_name = path + ".py"
+ assert script_name in os.listdir(tmp_path)
+ assert "__pycache__" not in os.listdir(tmp_path)
@pytest.mark.parametrize(
| `inspect` functions can't fetch dataset script from the Hub
The `inspect_dataset` and `inspect_metric` functions are unable to retrieve a dataset or metric script from the Hub and store it locally at the specified `local_path`:
```py
>>> from datasets import inspect_dataset
>>> inspect_dataset('rotten_tomatoes', local_path='path/to/my/local/folder')
FileNotFoundError: Couldn't find a dataset script at /content/rotten_tomatoes/rotten_tomatoes.py or any data file in the same directory.
```
| Hi, thanks for reporting! `git bisect` points to #2986 as the PR that introduced the bug. Since then, there have been some additional changes to the loading logic, and in the current state, `force_local_path` (set via `local_path`) forbids pulling a script from the internet instead of downloading it: https://github.com/huggingface/datasets/blob/cfae0545b2ba05452e16136cacc7d370b4b186a1/src/datasets/inspect.py#L89-L91
cc @lhoestq: `force_local_path` is only used in `inspect_dataset` and `inspect_metric`. Is it OK if we revert the behavior to match the old one?
Good catch ! Yea I think it's fine :) | 2022-06-01T12:09:56Z | [] | [] |
huggingface/datasets | 4,530 | huggingface__datasets-4530 | [
"3964"
] | fdcb8b144ce3ef241410281e125bd03e87b8caa1 | diff --git a/src/datasets/packaged_modules/__init__.py b/src/datasets/packaged_modules/__init__.py
--- a/src/datasets/packaged_modules/__init__.py
+++ b/src/datasets/packaged_modules/__init__.py
@@ -3,6 +3,7 @@
from hashlib import sha256
from typing import List
+from .audiofolder import audiofolder
from .csv import csv
from .imagefolder import imagefolder
from .json import json
@@ -32,6 +33,7 @@ def _hash_python_lines(lines: List[str]) -> str:
"parquet": (parquet.__name__, _hash_python_lines(inspect.getsource(parquet).splitlines())),
"text": (text.__name__, _hash_python_lines(inspect.getsource(text).splitlines())),
"imagefolder": (imagefolder.__name__, _hash_python_lines(inspect.getsource(imagefolder).splitlines())),
+ "audiofolder": (audiofolder.__name__, _hash_python_lines(inspect.getsource(audiofolder).splitlines())),
}
_EXTENSION_TO_MODULE = {
@@ -42,7 +44,8 @@ def _hash_python_lines(lines: List[str]) -> str:
"parquet": ("parquet", {}),
"txt": ("text", {}),
}
-_EXTENSION_TO_MODULE.update({ext[1:]: ("imagefolder", {}) for ext in imagefolder.ImageFolder.IMAGE_EXTENSIONS})
-_EXTENSION_TO_MODULE.update({ext[1:].upper(): ("imagefolder", {}) for ext in imagefolder.ImageFolder.IMAGE_EXTENSIONS})
-
-_MODULE_SUPPORTS_METADATA = {"imagefolder"}
+_EXTENSION_TO_MODULE.update({ext[1:]: ("imagefolder", {}) for ext in imagefolder.ImageFolder.EXTENSIONS})
+_EXTENSION_TO_MODULE.update({ext[1:].upper(): ("imagefolder", {}) for ext in imagefolder.ImageFolder.EXTENSIONS})
+_EXTENSION_TO_MODULE.update({ext[1:]: ("audiofolder", {}) for ext in audiofolder.AudioFolder.EXTENSIONS})
+_EXTENSION_TO_MODULE.update({ext[1:].upper(): ("audiofolder", {}) for ext in audiofolder.AudioFolder.EXTENSIONS})
+_MODULE_SUPPORTS_METADATA = {"imagefolder", "audiofolder"}
diff --git a/src/datasets/packaged_modules/audiofolder/__init__.py b/src/datasets/packaged_modules/audiofolder/__init__.py
new file mode 100644
diff --git a/src/datasets/packaged_modules/audiofolder/audiofolder.py b/src/datasets/packaged_modules/audiofolder/audiofolder.py
new file mode 100644
--- /dev/null
+++ b/src/datasets/packaged_modules/audiofolder/audiofolder.py
@@ -0,0 +1,66 @@
+from typing import List
+
+import datasets
+
+from ..folder_based_builder import folder_based_builder
+
+
+logger = datasets.utils.logging.get_logger(__name__)
+
+
+class AudioFolderConfig(folder_based_builder.FolderBasedBuilderConfig):
+ """Builder Config for AudioFolder."""
+
+ drop_labels: bool = None
+ drop_metadata: bool = None
+
+
+class AudioFolder(folder_based_builder.FolderBasedBuilder):
+ BASE_FEATURE = datasets.Audio()
+ BASE_COLUMN_NAME = "audio"
+ BUILDER_CONFIG_CLASS = AudioFolderConfig
+ EXTENSIONS: List[str] # definition at the bottom of the script
+
+
+# Obtained with:
+# ```
+# import soundfile as sf
+#
+# AUDIO_EXTENSIONS = [f".{format.lower()}" for format in sf.available_formats().keys()]
+#
+# # .mp3 is currently decoded via `torchaudio`, .opus decoding is supported if version of `libsndfile` >= 1.0.30:
+# AUDIO_EXTENSIONS.extend([".mp3", ".opus"])
+# ```
+# We intentionally do not run this code on launch because:
+# (1) Soundfile is an optional dependency, so importing it in global namespace is not allowed
+# (2) To ensure the list of supported extensions is deterministic
+AUDIO_EXTENSIONS = [
+ ".aiff",
+ ".au",
+ ".avr",
+ ".caf",
+ ".flac",
+ ".htk",
+ ".svx",
+ ".mat4",
+ ".mat5",
+ ".mpc2k",
+ ".ogg",
+ ".paf",
+ ".pvf",
+ ".raw",
+ ".rf64",
+ ".sd2",
+ ".sds",
+ ".ircam",
+ ".voc",
+ ".w64",
+ ".wav",
+ ".nist",
+ ".wavex",
+ ".wve",
+ ".xi",
+ ".mp3",
+ ".opus",
+]
+AudioFolder.EXTENSIONS = AUDIO_EXTENSIONS
diff --git a/src/datasets/packaged_modules/folder_based_builder/__init__.py b/src/datasets/packaged_modules/folder_based_builder/__init__.py
new file mode 100644
diff --git a/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
new file mode 100644
--- /dev/null
+++ b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
@@ -0,0 +1,391 @@
+import collections
+import itertools
+import os
+from dataclasses import dataclass
+from typing import Any, List, Optional, Tuple
+
+import pyarrow.compute as pc
+import pyarrow.json as paj
+
+import datasets
+
+
+logger = datasets.utils.logging.get_logger(__name__)
+
+
+if datasets.config.PYARROW_VERSION.major >= 7:
+
+ def pa_table_to_pylist(table):
+ return table.to_pylist()
+
+else:
+
+ def pa_table_to_pylist(table):
+ keys = table.column_names
+ values = table.to_pydict().values()
+ return [{k: v for k, v in zip(keys, row_values)} for row_values in zip(*values)]
+
+
+def count_path_segments(path):
+ cnt = 0
+ while True:
+ parts = os.path.split(path)
+ if parts[0] == path:
+ break
+ elif parts[1] == path:
+ break
+ else:
+ path = parts[0]
+ cnt += 1
+ return cnt
+
+
+@dataclass
+class FolderBasedBuilderConfig(datasets.BuilderConfig):
+ """BuilderConfig for AutoFolder."""
+
+ features: Optional[datasets.Features] = None
+ drop_labels: bool = None
+ drop_metadata: bool = None
+
+
+class FolderBasedBuilder(datasets.GeneratorBasedBuilder):
+ """
+ Base class for generic data loaders for vision and image data.
+
+
+ Abstract class attributes to be overridden by a child class:
+ BASE_FEATURE: feature object to decode data (i.e. datasets.Image(), datasets.Audio(), ...)
+ BASE_COLUMN_NAME: string key name of a base feature (i.e. "image", "audio", ...)
+ BUILDER_CONFIG_CLASS: builder config inherited from `folder_based_builder.FolderBasedBuilderConfig`
+ EXTENSIONS: list of allowed extensions (only files with these extensions and METADATA_FILENAME files
+ will be included in a dataset)
+ """
+
+ BASE_FEATURE: Any
+ BASE_COLUMN_NAME: str
+ BUILDER_CONFIG_CLASS: FolderBasedBuilderConfig
+ EXTENSIONS: List[str]
+
+ SKIP_CHECKSUM_COMPUTATION_BY_DEFAULT: bool = True
+ METADATA_FILENAME: str = "metadata.jsonl"
+
+ def _info(self):
+ return datasets.DatasetInfo(features=self.config.features)
+
+ def _split_generators(self, dl_manager):
+ if not self.config.data_files:
+ raise ValueError(f"At least one data file must be specified, but got data_files={self.config.data_files}")
+
+ # Do an early pass if:
+ # * `drop_labels` is None (default) or False, to infer the class labels
+ # * `drop_metadata` is None (default) or False, to find the metadata files
+ do_analyze = not self.config.drop_labels or not self.config.drop_metadata
+ labels = set()
+ metadata_files = collections.defaultdict(set)
+
+ def analyze(files_or_archives, downloaded_files_or_dirs, split):
+ if len(downloaded_files_or_dirs) == 0:
+ return
+ # The files are separated from the archives at this point, so check the first sample
+ # to see if it's a file or a directory and iterate accordingly
+ if os.path.isfile(downloaded_files_or_dirs[0]):
+ original_files, downloaded_files = files_or_archives, downloaded_files_or_dirs
+ for original_file, downloaded_file in zip(original_files, downloaded_files):
+ original_file, downloaded_file = str(original_file), str(downloaded_file)
+ _, original_file_ext = os.path.splitext(original_file)
+ if original_file_ext.lower() in self.EXTENSIONS:
+ if not self.config.drop_labels:
+ labels.add(os.path.basename(os.path.dirname(original_file)))
+ elif os.path.basename(original_file) == self.METADATA_FILENAME:
+ metadata_files[split].add((original_file, downloaded_file))
+ else:
+ original_file_name = os.path.basename(original_file)
+ logger.debug(
+ f"The file '{original_file_name}' was ignored: it is not an {self.BASE_COLUMN_NAME}, and is not {self.METADATA_FILENAME} either."
+ )
+ else:
+ archives, downloaded_dirs = files_or_archives, downloaded_files_or_dirs
+ for archive, downloaded_dir in zip(archives, downloaded_dirs):
+ archive, downloaded_dir = str(archive), str(downloaded_dir)
+ for downloaded_dir_file in dl_manager.iter_files(downloaded_dir):
+ _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
+ if downloaded_dir_file_ext in self.EXTENSIONS:
+ if not self.config.drop_labels:
+ labels.add(os.path.basename(os.path.dirname(downloaded_dir_file)))
+ elif os.path.basename(downloaded_dir_file) == self.METADATA_FILENAME:
+ metadata_files[split].add((None, downloaded_dir_file))
+ else:
+ archive_file_name = os.path.basename(archive)
+ original_file_name = os.path.basename(downloaded_dir_file)
+ logger.debug(
+ f"The file '{original_file_name}' from the archive '{archive_file_name}' was ignored: it is not an {self.BASE_COLUMN_NAME}, and is not {self.METADATA_FILENAME} either."
+ )
+
+ data_files = self.config.data_files
+ splits = []
+ for split_name, files in data_files.items():
+ if isinstance(files, str):
+ files = [files]
+ files, archives = self._split_files_and_archives(files)
+ downloaded_files = dl_manager.download(files)
+ downloaded_dirs = dl_manager.download_and_extract(archives)
+ if do_analyze: # drop_metadata is None or False, drop_labels is None or False
+ logger.info(f"Searching for labels and/or metadata files in {split_name} data files...")
+ analyze(files, downloaded_files, split_name)
+ analyze(archives, downloaded_dirs, split_name)
+
+ if metadata_files:
+ # add metadata if `metadata_files` are found and `drop_metadata` is None (default) or False
+ add_metadata = not (self.config.drop_metadata is True)
+ # if `metadata_files` are found, add labels only if
+ # `drop_labels` is set up to False explicitly (not-default behavior)
+ add_labels = self.config.drop_labels is False
+ else:
+ # if `metadata_files` are not found, don't add metadata
+ add_metadata = False
+ # if `metadata_files` are not found but `drop_labels` is None (default) or False, add them
+ add_labels = not (self.config.drop_labels is True)
+
+ if add_labels:
+ logger.info("Adding the labels inferred from data directories to the dataset's features...")
+ if add_metadata:
+ logger.info("Adding metadata to the dataset...")
+ else:
+ add_labels, add_metadata, metadata_files = False, False, {}
+
+ splits.append(
+ datasets.SplitGenerator(
+ name=split_name,
+ gen_kwargs={
+ "files": [(file, downloaded_file) for file, downloaded_file in zip(files, downloaded_files)]
+ + [(None, dl_manager.iter_files(downloaded_dir)) for downloaded_dir in downloaded_dirs],
+ "metadata_files": metadata_files,
+ "split_name": split_name,
+ "add_labels": add_labels,
+ "add_metadata": add_metadata,
+ },
+ )
+ )
+
+ if add_metadata:
+ # Verify that:
+ # * all metadata files have the same set of features
+ # * the `file_name` key is one of the metadata keys and is of type string
+ features_per_metadata_file: List[Tuple[str, datasets.Features]] = []
+ for _, downloaded_metadata_file in itertools.chain.from_iterable(metadata_files.values()):
+ with open(downloaded_metadata_file, "rb") as f:
+ pa_metadata_table = paj.read_json(f)
+ features_per_metadata_file.append(
+ (downloaded_metadata_file, datasets.Features.from_arrow_schema(pa_metadata_table.schema))
+ )
+ for downloaded_metadata_file, metadata_features in features_per_metadata_file:
+ if metadata_features != features_per_metadata_file[0][1]:
+ raise ValueError(
+ f"Metadata files {downloaded_metadata_file} and {features_per_metadata_file[0][0]} have different features: {features_per_metadata_file[0]} != {metadata_features}"
+ )
+ metadata_features = features_per_metadata_file[0][1]
+ if "file_name" not in metadata_features:
+ raise ValueError("`file_name` must be present as dictionary key in metadata files")
+ if metadata_features["file_name"] != datasets.Value("string"):
+ raise ValueError("`file_name` key must be a string")
+ del metadata_features["file_name"]
+ else:
+ metadata_features = None
+
+ # Normally, we would do this in _info, but we need to know the labels and/or metadata
+ # before building the features
+ if self.config.features is None:
+ if add_labels:
+ self.info.features = datasets.Features(
+ {
+ self.BASE_COLUMN_NAME: self.BASE_FEATURE,
+ "label": datasets.ClassLabel(names=sorted(labels)),
+ }
+ )
+ else:
+ self.info.features = datasets.Features({self.BASE_COLUMN_NAME: self.BASE_FEATURE})
+
+ if add_metadata:
+ # Warn if there are duplicated keys in metadata compared to the existing features
+ # (`BASE_COLUMN_NAME`, optionally "label")
+ duplicated_keys = set(self.info.features) & set(metadata_features)
+ if duplicated_keys:
+ logger.warning(
+ f"Ignoring metadata columns {list(duplicated_keys)} as they are already present in "
+ f"the features dictionary."
+ )
+ # skip metadata duplicated keys
+ self.info.features.update(
+ {
+ feature: metadata_features[feature]
+ for feature in metadata_features
+ if feature not in duplicated_keys
+ }
+ )
+
+ return splits
+
+ def _split_files_and_archives(self, data_files):
+ files, archives = [], []
+ for data_file in data_files:
+ _, data_file_ext = os.path.splitext(data_file)
+ if data_file_ext.lower() in self.EXTENSIONS:
+ files.append(data_file)
+ elif os.path.basename(data_file) == self.METADATA_FILENAME:
+ files.append(data_file)
+ else:
+ archives.append(data_file)
+ return files, archives
+
+ def _generate_examples(self, files, metadata_files, split_name, add_metadata, add_labels):
+ split_metadata_files = metadata_files.get(split_name, [])
+ sample_empty_metadata = (
+ {k: None for k in self.info.features if k != self.BASE_COLUMN_NAME} if self.info.features else {}
+ )
+ last_checked_dir = None
+ metadata_dir = None
+ metadata_dict = None
+ downloaded_metadata_file = None
+
+ file_idx = 0
+ for original_file, downloaded_file_or_dir in files:
+ if original_file is not None:
+ _, original_file_ext = os.path.splitext(original_file)
+ if original_file_ext.lower() in self.EXTENSIONS:
+ if add_metadata:
+ # If the file is a file of a needed type, and we've just entered a new directory,
+ # find the nereast metadata file (by counting path segments) for the directory
+ current_dir = os.path.dirname(original_file)
+ if last_checked_dir is None or last_checked_dir != current_dir:
+ last_checked_dir = current_dir
+ metadata_file_candidates = [
+ (
+ os.path.relpath(original_file, os.path.dirname(metadata_file_candidate)),
+ metadata_file_candidate,
+ downloaded_metadata_file,
+ )
+ for metadata_file_candidate, downloaded_metadata_file in split_metadata_files
+ if metadata_file_candidate
+ is not None # ignore metadata_files that are inside archives
+ and not os.path.relpath(
+ original_file, os.path.dirname(metadata_file_candidate)
+ ).startswith("..")
+ ]
+ if metadata_file_candidates:
+ _, metadata_file, downloaded_metadata_file = min(
+ metadata_file_candidates, key=lambda x: count_path_segments(x[0])
+ )
+ with open(downloaded_metadata_file, "rb") as f:
+ pa_metadata_table = paj.read_json(f)
+ pa_file_name_array = pa_metadata_table["file_name"]
+ pa_file_name_array = pc.replace_substring(
+ pa_file_name_array, pattern="\\", replacement="/"
+ )
+ pa_metadata_table = pa_metadata_table.drop(["file_name"])
+ metadata_dir = os.path.dirname(metadata_file)
+ metadata_dict = {
+ file_name: sample_metadata
+ for file_name, sample_metadata in zip(
+ pa_file_name_array.to_pylist(), pa_table_to_pylist(pa_metadata_table)
+ )
+ }
+ else:
+ raise ValueError(
+ f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
+ )
+ if metadata_dir is not None and downloaded_metadata_file is not None:
+ file_relpath = os.path.relpath(original_file, metadata_dir)
+ file_relpath = file_relpath.replace("\\", "/")
+ if file_relpath not in metadata_dict:
+ raise ValueError(
+ f"{self.BASE_COLUMN_NAME} at {file_relpath} doesn't have metadata in {downloaded_metadata_file}."
+ )
+ sample_metadata = metadata_dict[file_relpath]
+ else:
+ raise ValueError(
+ f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
+ )
+ else:
+ sample_metadata = {}
+ if add_labels:
+ sample_label = {"label": os.path.basename(os.path.dirname(original_file))}
+ else:
+ sample_label = {}
+ yield file_idx, {
+ **sample_empty_metadata,
+ self.BASE_COLUMN_NAME: downloaded_file_or_dir,
+ **sample_metadata,
+ **sample_label,
+ }
+ file_idx += 1
+ else:
+ for downloaded_dir_file in downloaded_file_or_dir:
+ _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
+ if downloaded_dir_file_ext.lower() in self.EXTENSIONS:
+ if add_metadata:
+ current_dir = os.path.dirname(downloaded_dir_file)
+ if last_checked_dir is None or last_checked_dir != current_dir:
+ last_checked_dir = current_dir
+ metadata_file_candidates = [
+ (
+ os.path.relpath(
+ downloaded_dir_file, os.path.dirname(downloaded_metadata_file)
+ ),
+ metadata_file_candidate,
+ downloaded_metadata_file,
+ )
+ for metadata_file_candidate, downloaded_metadata_file in split_metadata_files
+ if metadata_file_candidate
+ is None # ignore metadata_files that are not inside archives
+ and not os.path.relpath(
+ downloaded_dir_file, os.path.dirname(downloaded_metadata_file)
+ ).startswith("..")
+ ]
+ if metadata_file_candidates:
+ _, metadata_file, downloaded_metadata_file = min(
+ metadata_file_candidates, key=lambda x: count_path_segments(x[0])
+ )
+ with open(downloaded_metadata_file, "rb") as f:
+ pa_metadata_table = paj.read_json(f)
+ pa_file_name_array = pa_metadata_table["file_name"]
+ pa_file_name_array = pc.replace_substring(
+ pa_file_name_array, pattern="\\", replacement="/"
+ )
+ pa_metadata_table = pa_metadata_table.drop(["file_name"])
+ metadata_dir = os.path.dirname(downloaded_metadata_file)
+ metadata_dict = {
+ file_name: sample_metadata
+ for file_name, sample_metadata in zip(
+ pa_file_name_array.to_pylist(), pa_table_to_pylist(pa_metadata_table)
+ )
+ }
+ else:
+ raise ValueError(
+ f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
+ )
+ if metadata_dir is not None and downloaded_metadata_file is not None:
+ downloaded_dir_file_relpath = os.path.relpath(downloaded_dir_file, metadata_dir)
+ downloaded_dir_file_relpath = downloaded_dir_file_relpath.replace("\\", "/")
+ if downloaded_dir_file_relpath not in metadata_dict:
+ raise ValueError(
+ f"{self.BASE_COLUMN_NAME} at {downloaded_dir_file_relpath} doesn't have metadata in {downloaded_metadata_file}."
+ )
+ sample_metadata = metadata_dict[downloaded_dir_file_relpath]
+ else:
+ raise ValueError(
+ f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
+ )
+ else:
+ sample_metadata = {}
+ if add_labels:
+ sample_label = {"label": os.path.basename(os.path.dirname(downloaded_dir_file))}
+ else:
+ sample_label = {}
+ yield file_idx, {
+ **sample_empty_metadata,
+ self.BASE_COLUMN_NAME: downloaded_dir_file,
+ **sample_metadata,
+ **sample_label,
+ }
+ file_idx += 1
diff --git a/src/datasets/packaged_modules/imagefolder/imagefolder.py b/src/datasets/packaged_modules/imagefolder/imagefolder.py
--- a/src/datasets/packaged_modules/imagefolder/imagefolder.py
+++ b/src/datasets/packaged_modules/imagefolder/imagefolder.py
@@ -1,373 +1,25 @@
-import collections
-import itertools
-import os
-from dataclasses import dataclass
-from typing import List, Optional, Tuple
-
-import pyarrow.compute as pc
-import pyarrow.json as paj
+from typing import List
import datasets
-from datasets.tasks import ImageClassification
-
-
-logger = datasets.utils.logging.get_logger(__name__)
-
-
-if datasets.config.PYARROW_VERSION.major >= 7:
-
- def pa_table_to_pylist(table):
- return table.to_pylist()
-else:
+from ..folder_based_builder import folder_based_builder
- def pa_table_to_pylist(table):
- keys = table.column_names
- values = table.to_pydict().values()
- return [{k: v for k, v in zip(keys, row_values)} for row_values in zip(*values)]
-
-def count_path_segments(path):
- cnt = 0
- while True:
- parts = os.path.split(path)
- if parts[0] == path:
- break
- elif parts[1] == path:
- break
- else:
- path = parts[0]
- cnt += 1
- return cnt
+logger = datasets.utils.logging.get_logger(__name__)
-@dataclass
-class ImageFolderConfig(datasets.BuilderConfig):
+class ImageFolderConfig(folder_based_builder.FolderBasedBuilderConfig):
"""BuilderConfig for ImageFolder."""
- features: Optional[datasets.Features] = None
drop_labels: bool = None
drop_metadata: bool = None
-class ImageFolder(datasets.GeneratorBasedBuilder):
+class ImageFolder(folder_based_builder.FolderBasedBuilder):
+ BASE_FEATURE = datasets.Image()
+ BASE_COLUMN_NAME = "image"
BUILDER_CONFIG_CLASS = ImageFolderConfig
-
- IMAGE_EXTENSIONS: List[str] = [] # definition at the bottom of the script
- SKIP_CHECKSUM_COMPUTATION_BY_DEFAULT = True
- METADATA_FILENAME: str = "metadata.jsonl"
-
- def _info(self):
- return datasets.DatasetInfo(features=self.config.features)
-
- def _split_generators(self, dl_manager):
- if not self.config.data_files:
- raise ValueError(f"At least one data file must be specified, but got data_files={self.config.data_files}")
-
- # Do an early pass if:
- # * `drop_labels` is None (default) or False, to infer the class labels
- # * `drop_metadata` is None (default) or False, to find the metadata files
- do_analyze = not self.config.drop_labels or not self.config.drop_metadata
- labels = set()
- metadata_files = collections.defaultdict(set)
-
- def analyze(files_or_archives, downloaded_files_or_dirs, split):
- if len(downloaded_files_or_dirs) == 0:
- return
- # The files are separated from the archives at this point, so check the first sample
- # to see if it's a file or a directory and iterate accordingly
- if os.path.isfile(downloaded_files_or_dirs[0]):
- original_files, downloaded_files = files_or_archives, downloaded_files_or_dirs
- for original_file, downloaded_file in zip(original_files, downloaded_files):
- original_file, downloaded_file = str(original_file), str(downloaded_file)
- _, original_file_ext = os.path.splitext(original_file)
- if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
- if not self.config.drop_labels:
- labels.add(os.path.basename(os.path.dirname(original_file)))
- elif os.path.basename(original_file) == self.METADATA_FILENAME:
- metadata_files[split].add((original_file, downloaded_file))
- else:
- original_file_name = os.path.basename(original_file)
- logger.debug(
- f"The file '{original_file_name}' was ignored: it is not an image, and is not {self.METADATA_FILENAME} either."
- )
- else:
- archives, downloaded_dirs = files_or_archives, downloaded_files_or_dirs
- for archive, downloaded_dir in zip(archives, downloaded_dirs):
- archive, downloaded_dir = str(archive), str(downloaded_dir)
- for downloaded_dir_file in dl_manager.iter_files(downloaded_dir):
- _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
- if downloaded_dir_file_ext in self.IMAGE_EXTENSIONS:
- if not self.config.drop_labels:
- labels.add(os.path.basename(os.path.dirname(downloaded_dir_file)))
- elif os.path.basename(downloaded_dir_file) == self.METADATA_FILENAME:
- metadata_files[split].add((None, downloaded_dir_file))
- else:
- archive_file_name = os.path.basename(archive)
- original_file_name = os.path.basename(downloaded_dir_file)
- logger.debug(
- f"The file '{original_file_name}' from the archive '{archive_file_name}' was ignored: it is not an image, and is not {self.METADATA_FILENAME} either."
- )
-
- data_files = self.config.data_files
- splits = []
- for split_name, files in data_files.items():
- if isinstance(files, str):
- files = [files]
- files, archives = self._split_files_and_archives(files)
- downloaded_files = dl_manager.download(files)
- downloaded_dirs = dl_manager.download_and_extract(archives)
- if do_analyze: # drop_metadata is None or False, drop_labels is None or False
- logger.info(f"Searching for labels and/or metadata files in {split_name} data files...")
- analyze(files, downloaded_files, split_name)
- analyze(archives, downloaded_dirs, split_name)
-
- if metadata_files:
- # add metadata if `metadata_files` are found and `drop_metadata` is None (default) or False
- add_metadata = not (self.config.drop_metadata is True)
- # if `metadata_files` are found, add labels only if
- # `drop_labels` is set up to False explicitly (not-default behavior)
- add_labels = self.config.drop_labels is False
- else:
- # if `metadata_files` are not found, don't add metadata
- add_metadata = False
- # if `metadata_files` are not found but `drop_labels` is None (default) or False, add them
- add_labels = not (self.config.drop_labels is True)
-
- if add_labels:
- logger.info("Adding the labels inferred from data directories to the dataset's features...")
- if add_metadata:
- logger.info("Adding metadata to the dataset...")
- else:
- add_labels, add_metadata, metadata_files = False, False, {}
-
- splits.append(
- datasets.SplitGenerator(
- name=split_name,
- gen_kwargs={
- "files": [(file, downloaded_file) for file, downloaded_file in zip(files, downloaded_files)]
- + [(None, dl_manager.iter_files(downloaded_dir)) for downloaded_dir in downloaded_dirs],
- "metadata_files": metadata_files,
- "split_name": split_name,
- "add_labels": add_labels,
- "add_metadata": add_metadata,
- },
- )
- )
-
- if add_metadata:
- # Verify that:
- # * all metadata files have the same set of features
- # * the `file_name` key is one of the metadata keys and is of type string
- features_per_metadata_file: List[Tuple[str, datasets.Features]] = []
- for _, downloaded_metadata_file in itertools.chain.from_iterable(metadata_files.values()):
- with open(downloaded_metadata_file, "rb") as f:
- pa_metadata_table = paj.read_json(f)
- features_per_metadata_file.append(
- (downloaded_metadata_file, datasets.Features.from_arrow_schema(pa_metadata_table.schema))
- )
- for downloaded_metadata_file, metadata_features in features_per_metadata_file:
- if metadata_features != features_per_metadata_file[0][1]:
- raise ValueError(
- f"Metadata files {downloaded_metadata_file} and {features_per_metadata_file[0][0]} have different features: {features_per_metadata_file[0]} != {metadata_features}"
- )
- metadata_features = features_per_metadata_file[0][1]
- if "file_name" not in metadata_features:
- raise ValueError("`file_name` must be present as dictionary key in metadata files")
- if metadata_features["file_name"] != datasets.Value("string"):
- raise ValueError("`file_name` key must be a string")
- del metadata_features["file_name"]
- else:
- metadata_features = None
-
- # Normally, we would do this in _info, but we need to know the labels and/or metadata
- # before building the features
- if self.config.features is None:
- if add_labels:
- self.info.features = datasets.Features(
- {"image": datasets.Image(), "label": datasets.ClassLabel(names=sorted(labels))}
- )
- task_template = ImageClassification(image_column="image", label_column="label")
- task_template = task_template.align_with_features(self.info.features)
- self.info.task_templates = [task_template]
- else:
- self.info.features = datasets.Features({"image": datasets.Image()})
-
- if add_metadata:
- # Warn if there are duplicated keys in metadata compared to the existing features ("image", optionally "label")
- duplicated_keys = set(self.info.features) & set(metadata_features)
- if duplicated_keys:
- logger.warning(
- f"Ignoring metadata columns {list(duplicated_keys)} as they are already present in "
- f"the features dictionary."
- )
- # skip metadata duplicated keys
- self.info.features.update(
- {
- feature: metadata_features[feature]
- for feature in metadata_features
- if feature not in duplicated_keys
- }
- )
-
- return splits
-
- def _split_files_and_archives(self, data_files):
- files, archives = [], []
- for data_file in data_files:
- _, data_file_ext = os.path.splitext(data_file)
- if data_file_ext.lower() in self.IMAGE_EXTENSIONS:
- files.append(data_file)
- elif os.path.basename(data_file) == self.METADATA_FILENAME:
- files.append(data_file)
- else:
- archives.append(data_file)
- return files, archives
-
- def _generate_examples(self, files, metadata_files, split_name, add_metadata, add_labels):
- split_metadata_files = metadata_files.get(split_name, [])
- image_empty = {k: None for k in self.info.features if k != "image"} if self.info.features else {}
- last_checked_dir = None
- metadata_dir = None
- metadata_dict = None
- downloaded_metadata_file = None
-
- file_idx = 0
- for original_file, downloaded_file_or_dir in files:
- if original_file is not None:
- _, original_file_ext = os.path.splitext(original_file)
- if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
- if add_metadata:
- # If the file is an image, and we've just entered a new directory,
- # find the nereast metadata file (by counting path segments) for the directory
- current_dir = os.path.dirname(original_file)
- if last_checked_dir is None or last_checked_dir != current_dir:
- last_checked_dir = current_dir
- metadata_file_candidates = [
- (
- os.path.relpath(original_file, os.path.dirname(metadata_file_candidate)),
- metadata_file_candidate,
- downloaded_metadata_file,
- )
- for metadata_file_candidate, downloaded_metadata_file in split_metadata_files
- if metadata_file_candidate
- is not None # ignore metadata_files that are inside archives
- and not os.path.relpath(
- original_file, os.path.dirname(metadata_file_candidate)
- ).startswith("..")
- ]
- if metadata_file_candidates:
- _, metadata_file, downloaded_metadata_file = min(
- metadata_file_candidates, key=lambda x: count_path_segments(x[0])
- )
- with open(downloaded_metadata_file, "rb") as f:
- pa_metadata_table = paj.read_json(f)
- pa_file_name_array = pa_metadata_table["file_name"]
- pa_file_name_array = pc.replace_substring(
- pa_file_name_array, pattern="\\", replacement="/"
- )
- pa_metadata_table = pa_metadata_table.drop(["file_name"])
- metadata_dir = os.path.dirname(metadata_file)
- metadata_dict = {
- file_name: image_metadata
- for file_name, image_metadata in zip(
- pa_file_name_array.to_pylist(), pa_table_to_pylist(pa_metadata_table)
- )
- }
- else:
- raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
- )
- if metadata_dir is not None and downloaded_metadata_file is not None:
- file_relpath = os.path.relpath(original_file, metadata_dir)
- file_relpath = file_relpath.replace("\\", "/")
- if file_relpath not in metadata_dict:
- raise ValueError(
- f"Image at {file_relpath} doesn't have metadata in {downloaded_metadata_file}."
- )
- image_metadata = metadata_dict[file_relpath]
- else:
- raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
- )
- else:
- image_metadata = {}
- if add_labels:
- image_label = {"label": os.path.basename(os.path.dirname(original_file))}
- else:
- image_label = {}
- yield file_idx, {**image_empty, "image": downloaded_file_or_dir, **image_metadata, **image_label}
- file_idx += 1
- else:
- for downloaded_dir_file in downloaded_file_or_dir:
- _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
- if downloaded_dir_file_ext.lower() in self.IMAGE_EXTENSIONS:
- if add_metadata:
- current_dir = os.path.dirname(downloaded_dir_file)
- if last_checked_dir is None or last_checked_dir != current_dir:
- last_checked_dir = current_dir
- metadata_file_candidates = [
- (
- os.path.relpath(
- downloaded_dir_file, os.path.dirname(downloaded_metadata_file)
- ),
- metadata_file_candidate,
- downloaded_metadata_file,
- )
- for metadata_file_candidate, downloaded_metadata_file in split_metadata_files
- if metadata_file_candidate
- is None # ignore metadata_files that are not inside archives
- and not os.path.relpath(
- downloaded_dir_file, os.path.dirname(downloaded_metadata_file)
- ).startswith("..")
- ]
- if metadata_file_candidates:
- _, metadata_file, downloaded_metadata_file = min(
- metadata_file_candidates, key=lambda x: count_path_segments(x[0])
- )
- with open(downloaded_metadata_file, "rb") as f:
- pa_metadata_table = paj.read_json(f)
- pa_file_name_array = pa_metadata_table["file_name"]
- pa_file_name_array = pc.replace_substring(
- pa_file_name_array, pattern="\\", replacement="/"
- )
- pa_metadata_table = pa_metadata_table.drop(["file_name"])
- metadata_dir = os.path.dirname(downloaded_metadata_file)
- metadata_dict = {
- file_name: image_metadata
- for file_name, image_metadata in zip(
- pa_file_name_array.to_pylist(), pa_table_to_pylist(pa_metadata_table)
- )
- }
- else:
- raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
- )
- if metadata_dir is not None and downloaded_metadata_file is not None:
- downloaded_dir_file_relpath = os.path.relpath(downloaded_dir_file, metadata_dir)
- downloaded_dir_file_relpath = downloaded_dir_file_relpath.replace("\\", "/")
- if downloaded_dir_file_relpath not in metadata_dict:
- raise ValueError(
- f"Image at {downloaded_dir_file_relpath} doesn't have metadata in {downloaded_metadata_file}."
- )
- image_metadata = metadata_dict[downloaded_dir_file_relpath]
- else:
- raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
- )
- else:
- image_metadata = {}
- if add_labels:
- image_label = {"label": os.path.basename(os.path.dirname(downloaded_dir_file))}
- else:
- image_label = {}
- yield file_idx, {
- **image_empty,
- "image": downloaded_dir_file,
- **image_metadata,
- **image_label,
- }
- file_idx += 1
+ EXTENSIONS: List[str] # definition at the bottom of the script
# Obtained with:
@@ -382,7 +34,7 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
# We intentionally do not run this code on launch because:
# (1) Pillow is an optional dependency, so importing Pillow in global namespace is not allowed
# (2) To ensure the list of supported extensions is deterministic
-ImageFolder.IMAGE_EXTENSIONS = [
+IMAGE_EXTENSIONS = [
".blp",
".bmp",
".dib",
@@ -447,3 +99,4 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
".xbm",
".xpm",
]
+ImageFolder.EXTENSIONS = IMAGE_EXTENSIONS
diff --git a/src/datasets/streaming.py b/src/datasets/streaming.py
--- a/src/datasets/streaming.py
+++ b/src/datasets/streaming.py
@@ -52,7 +52,8 @@ def extend_module_for_streaming(module_path, use_auth_token: Optional[Union[str,
Args:
module_path: Path to the module to be extended.
- use_auth_token: Whether to use authentication token.
+ use_auth_token (``str`` or :obj:`bool`, optional): Optional string or boolean to use as Bearer token for remote files on the Datasets Hub.
+ If True, will get token from `"~/.huggingface"`.
"""
module = importlib.import_module(module_path)
@@ -100,8 +101,6 @@ def extend_dataset_builder_for_streaming(builder: "DatasetBuilder"):
Args:
builder (:class:`DatasetBuilder`): Dataset builder instance.
- use_auth_token (``str`` or :obj:`bool`, optional): Optional string or boolean to use as Bearer token for remote files on the Datasets Hub.
- If True, will get token from `"~/.huggingface"`.
"""
# this extends the open and os.path.join functions for data streaming
extend_module_for_streaming(builder.__module__, use_auth_token=builder.use_auth_token)
@@ -112,3 +111,16 @@ def extend_dataset_builder_for_streaming(builder: "DatasetBuilder"):
internal_import_name = imports[1]
internal_module_name = ".".join(builder.__module__.split(".")[:-1] + [internal_import_name])
extend_module_for_streaming(internal_module_name, use_auth_token=builder.use_auth_token)
+
+ # builders can inherit from other builders that might use streaming functionality
+ # (for example, ImageFolder and AudioFolder inherit from FolderBuilder which implements examples generation)
+ # but these parents builders are not patched automatically as they are not instantiated, so we patch them here
+ from .builder import DatasetBuilder
+
+ parent_builder_modules = [
+ cls.__module__
+ for cls in type(builder).__mro__[1:] # make sure it's not the same module we've already patched
+ if issubclass(cls, DatasetBuilder) and cls.__module__ != DatasetBuilder.__module__
+ ] # check it's not a standard builder from datasets.builder
+ for module in parent_builder_modules:
+ extend_module_for_streaming(module, use_auth_token=builder.use_auth_token)
| diff --git a/tests/fixtures/files.py b/tests/fixtures/files.py
--- a/tests/fixtures/files.py
+++ b/tests/fixtures/files.py
@@ -472,6 +472,11 @@ def image_file():
return os.path.join("tests", "features", "data", "test_image_rgb.jpg")
[email protected](scope="session")
+def audio_file():
+ return os.path.join("tests", "features", "data", "test_audio_44100.wav")
+
+
@pytest.fixture(scope="session")
def zip_image_path(image_file, tmp_path_factory):
path = tmp_path_factory.mktemp("data") / "dataset.img.zip"
diff --git a/tests/packaged_modules/test_audiofolder.py b/tests/packaged_modules/test_audiofolder.py
new file mode 100644
--- /dev/null
+++ b/tests/packaged_modules/test_audiofolder.py
@@ -0,0 +1,444 @@
+import shutil
+import textwrap
+
+import librosa
+import numpy as np
+import pytest
+import soundfile as sf
+
+from datasets import Audio, ClassLabel, Features, Value
+from datasets.data_files import DataFilesDict, get_data_patterns_locally
+from datasets.download.streaming_download_manager import StreamingDownloadManager
+from datasets.packaged_modules.audiofolder.audiofolder import AudioFolder
+
+from ..utils import require_sndfile
+
+
[email protected]
+def cache_dir(tmp_path):
+ return str(tmp_path / "audiofolder_cache_dir")
+
+
[email protected]
+def data_files_with_labels_no_metadata(tmp_path, audio_file):
+ data_dir = tmp_path / "data_files_with_labels_no_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir_class_0 = data_dir / "fr"
+ subdir_class_0.mkdir(parents=True, exist_ok=True)
+ # data dirs can be nested but audiofolder should care only about the last part of the path:
+ subdir_class_1 = data_dir / "subdir" / "uk"
+ subdir_class_1.mkdir(parents=True, exist_ok=True)
+
+ audio_filename = subdir_class_0 / "audio_fr.wav"
+ shutil.copyfile(audio_file, audio_filename)
+ audio_filename2 = subdir_class_1 / "audio_uk.wav"
+ shutil.copyfile(audio_file, audio_filename2)
+
+ data_files_with_labels_no_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+
+ return data_files_with_labels_no_metadata
+
+
[email protected]
+def audio_files_with_labels_and_duplicated_label_key_in_metadata(tmp_path, audio_file):
+ data_dir = tmp_path / "audio_files_with_labels_and_label_key_in_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir_class_0 = data_dir / "fr"
+ subdir_class_0.mkdir(parents=True, exist_ok=True)
+ subdir_class_1 = data_dir / "uk"
+ subdir_class_1.mkdir(parents=True, exist_ok=True)
+
+ audio_filename = subdir_class_0 / "audio_fr.wav"
+ shutil.copyfile(audio_file, audio_filename)
+ audio_filename2 = subdir_class_1 / "audio_uk.wav"
+ shutil.copyfile(audio_file, audio_filename2)
+
+ audio_metadata_filename = tmp_path / data_dir / "metadata.jsonl"
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "fr/audio_fr.wav", "text": "Audio in French", "label": "Fr"}
+ {"file_name": "uk/audio_uk.wav", "text": "Audio in Ukrainian", "label": "Uk"}
+ """
+ )
+ with open(audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+
+ return str(audio_filename), str(audio_filename2), str(audio_metadata_filename)
+
+
[email protected]
+def audio_file_with_metadata(tmp_path, audio_file):
+ audio_filename = tmp_path / "audio_file.wav"
+ shutil.copyfile(audio_file, audio_filename)
+ audio_metadata_filename = tmp_path / "metadata.jsonl"
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "audio_file.wav", "text": "Audio transcription"}
+ """
+ )
+ with open(audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+ return str(audio_filename), str(audio_metadata_filename)
+
+
[email protected]
+def audio_files_with_metadata_that_misses_one_audio(tmp_path, audio_file):
+ audio_filename = tmp_path / "audio_file.wav"
+ shutil.copyfile(audio_file, audio_filename)
+ audio_filename2 = tmp_path / "audio_file2.wav"
+ shutil.copyfile(audio_file, audio_filename2)
+ audio_metadata_filename = tmp_path / "metadata.jsonl"
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "audio_file.wav", "text": "Audio transcription"}
+ """
+ )
+ with open(audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+ return str(audio_filename), str(audio_filename2), str(audio_metadata_filename)
+
+
[email protected]
+def data_files_with_one_split_and_metadata(tmp_path, audio_file):
+ data_dir = tmp_path / "audiofolder_data_dir_with_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir = data_dir / "subdir"
+ subdir.mkdir(parents=True, exist_ok=True)
+
+ audio_filename = data_dir / "audio_file.wav"
+ shutil.copyfile(audio_file, audio_filename)
+ audio_filename2 = data_dir / "audio_file2.wav"
+ shutil.copyfile(audio_file, audio_filename2)
+ audio_filename3 = subdir / "audio_file3.wav" # in subdir
+ shutil.copyfile(audio_file, audio_filename3)
+
+ audio_metadata_filename = data_dir / "metadata.jsonl"
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "audio_file.wav", "text": "First audio transcription"}
+ {"file_name": "audio_file2.wav", "text": "Second audio transcription"}
+ {"file_name": "subdir/audio_file3.wav", "text": "Third audio transcription (in subdir)"}
+ """
+ )
+ with open(audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+ data_files_with_one_split_and_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+ assert len(data_files_with_one_split_and_metadata) == 1
+ assert len(data_files_with_one_split_and_metadata["train"]) == 4
+ return data_files_with_one_split_and_metadata
+
+
[email protected]
+def data_files_with_two_splits_and_metadata(tmp_path, audio_file):
+ data_dir = tmp_path / "audiofolder_data_dir_with_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ train_dir = data_dir / "train"
+ train_dir.mkdir(parents=True, exist_ok=True)
+ test_dir = data_dir / "test"
+ test_dir.mkdir(parents=True, exist_ok=True)
+
+ audio_filename = train_dir / "audio_file.wav" # train audio
+ shutil.copyfile(audio_file, audio_filename)
+ audio_filename2 = train_dir / "audio_file2.wav" # train audio
+ shutil.copyfile(audio_file, audio_filename2)
+ audio_filename3 = test_dir / "audio_file3.wav" # test audio
+ shutil.copyfile(audio_file, audio_filename3)
+
+ train_audio_metadata_filename = train_dir / "metadata.jsonl"
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "audio_file.wav", "text": "First train audio transcription"}
+ {"file_name": "audio_file2.wav", "text": "Second train audio transcription"}
+ """
+ )
+ with open(train_audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+ test_audio_metadata_filename = test_dir / "metadata.jsonl"
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "audio_file3.wav", "text": "Test audio transcription"}
+ """
+ )
+ with open(test_audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+ data_files_with_two_splits_and_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+ assert len(data_files_with_two_splits_and_metadata) == 2
+ assert len(data_files_with_two_splits_and_metadata["train"]) == 3
+ assert len(data_files_with_two_splits_and_metadata["test"]) == 2
+ return data_files_with_two_splits_and_metadata
+
+
[email protected]
+def data_files_with_zip_archives(tmp_path, audio_file):
+ data_dir = tmp_path / "audiofolder_data_dir_with_zip_archives"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ archive_dir = data_dir / "archive"
+ archive_dir.mkdir(parents=True, exist_ok=True)
+ subdir = archive_dir / "subdir"
+ subdir.mkdir(parents=True, exist_ok=True)
+
+ audio_filename = archive_dir / "audio_file.wav"
+ shutil.copyfile(audio_file, audio_filename)
+ audio_filename2 = subdir / "audio_file2.wav" # in subdir
+ # make sure they're two different audios
+ # Indeed we won't be able to compare the audio filenames, since the archive is not extracted in streaming mode
+ array, sampling_rate = librosa.load(str(audio_filename), sr=16000) # original sampling rate is 44100
+ sf.write(str(audio_filename2), array, samplerate=16000)
+
+ audio_metadata_filename = archive_dir / "metadata.jsonl"
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "audio_file.wav", "text": "First audio transcription"}
+ {"file_name": "subdir/audio_file2.wav", "text": "Second audio transcription (in subdir)"}
+ """
+ )
+
+ with open(audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+
+ shutil.make_archive(str(archive_dir), "zip", archive_dir)
+ shutil.rmtree(str(archive_dir))
+
+ data_files_with_zip_archives = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+
+ assert len(data_files_with_zip_archives) == 1
+ assert len(data_files_with_zip_archives["train"]) == 1
+ return data_files_with_zip_archives
+
+
+@require_sndfile
+# check that labels are inferred correctly from dir names
+def test_generate_examples_with_labels(data_files_with_labels_no_metadata, cache_dir):
+ # there are no metadata.jsonl files in this test case
+ audiofolder = AudioFolder(data_files=data_files_with_labels_no_metadata, cache_dir=cache_dir, drop_labels=False)
+ audiofolder.download_and_prepare()
+ assert audiofolder.info.features == Features({"audio": Audio(), "label": ClassLabel(names=["fr", "uk"])})
+ dataset = list(audiofolder.as_dataset()["train"])
+ label_feature = audiofolder.info.features["label"]
+
+ assert dataset[0]["label"] == label_feature._str2int["fr"]
+ assert dataset[1]["label"] == label_feature._str2int["uk"]
+
+
+@require_sndfile
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_generate_examples_duplicated_label_key(
+ audio_files_with_labels_and_duplicated_label_key_in_metadata, drop_metadata, drop_labels, cache_dir, caplog
+):
+ fr_audio_file, uk_audio_file, audio_metadata_file = audio_files_with_labels_and_duplicated_label_key_in_metadata
+ audiofolder = AudioFolder(
+ drop_metadata=drop_metadata,
+ drop_labels=drop_labels,
+ data_files=[fr_audio_file, uk_audio_file, audio_metadata_file],
+ cache_dir=cache_dir,
+ )
+ if drop_labels is False:
+ # infer labels from directories even if metadata files are found
+ audiofolder.download_and_prepare()
+ warning_in_logs = any("ignoring metadata columns" in record.msg.lower() for record in caplog.records)
+ assert warning_in_logs if drop_metadata is not True else not warning_in_logs
+ dataset = audiofolder.as_dataset()["train"]
+ assert audiofolder.info.features["label"] == ClassLabel(names=["fr", "uk"])
+ assert all(example["label"] in audiofolder.info.features["label"]._str2int.values() for example in dataset)
+ else:
+ audiofolder.download_and_prepare()
+ dataset = audiofolder.as_dataset()["train"]
+ if drop_metadata is not True:
+ # labels are from metadata
+ assert audiofolder.info.features["label"] == Value("string")
+ assert all(example["label"] in ["Fr", "Uk"] for example in dataset)
+ else:
+ # drop both labels and metadata
+ assert audiofolder.info.features == Features({"audio": Audio()})
+ assert all(example.keys() == {"audio"} for example in dataset)
+
+
+@require_sndfile
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_generate_examples_drop_labels(audio_file, drop_metadata, drop_labels):
+ audiofolder = AudioFolder(drop_metadata=drop_metadata, drop_labels=drop_labels, data_files={"train": [audio_file]})
+ gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ # removing the labels explicitly requires drop_labels=True
+ assert gen_kwargs["add_labels"] is not bool(drop_labels)
+ assert gen_kwargs["add_metadata"] is False # metadata files is not present in this case
+ generator = audiofolder._generate_examples(**gen_kwargs)
+ if not drop_labels:
+ assert all(
+ example.keys() == {"audio", "label"} and all(val is not None for val in example.values())
+ for _, example in generator
+ )
+ else:
+ assert all(
+ example.keys() == {"audio"} and all(val is not None for val in example.values())
+ for _, example in generator
+ )
+
+
+@require_sndfile
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_generate_examples_drop_metadata(audio_file_with_metadata, drop_metadata, drop_labels):
+ audio_file, audio_metadata_file = audio_file_with_metadata
+ audiofolder = AudioFolder(
+ drop_metadata=drop_metadata, drop_labels=drop_labels, data_files={"train": [audio_file, audio_metadata_file]}
+ )
+ gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ # since the dataset has metadata, removing the metadata explicitly requires drop_metadata=True
+ assert gen_kwargs["add_metadata"] is not bool(drop_metadata)
+ # since the dataset has metadata, adding the labels explicitly requires drop_labels=False
+ assert gen_kwargs["add_labels"] is (drop_labels is False)
+ generator = audiofolder._generate_examples(**gen_kwargs)
+ expected_columns = {"audio"}
+ if gen_kwargs["add_metadata"]:
+ expected_columns.add("text")
+ if gen_kwargs["add_labels"]:
+ expected_columns.add("label")
+ result = [example for _, example in generator]
+ assert len(result) == 1
+ example = result[0]
+ assert example.keys() == expected_columns
+ for column in expected_columns:
+ assert example[column] is not None
+
+
+@require_sndfile
[email protected]("drop_metadata", [None, True, False])
+def test_generate_examples_with_metadata_in_wrong_location(audio_file, audio_file_with_metadata, drop_metadata):
+ _, audio_metadata_file = audio_file_with_metadata
+ audiofolder = AudioFolder(drop_metadata=drop_metadata, data_files={"train": [audio_file, audio_metadata_file]})
+ gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ generator = audiofolder._generate_examples(**gen_kwargs)
+ if not drop_metadata:
+ with pytest.raises(ValueError):
+ list(generator)
+ else:
+ assert all(
+ example.keys() == {"audio"} and all(val is not None for val in example.values())
+ for _, example in generator
+ )
+
+
+@require_sndfile
[email protected]("drop_metadata", [None, True, False])
+def test_generate_examples_with_metadata_that_misses_one_audio(
+ audio_files_with_metadata_that_misses_one_audio, drop_metadata
+):
+ audio_file, audio_file2, audio_metadata_file = audio_files_with_metadata_that_misses_one_audio
+ if not drop_metadata:
+ features = Features({"audio": Audio(), "text": Value("string")})
+ else:
+ features = Features({"audio": Audio()})
+ audiofolder = AudioFolder(
+ drop_metadata=drop_metadata,
+ features=features,
+ data_files={"train": [audio_file, audio_file2, audio_metadata_file]},
+ )
+ gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ generator = audiofolder._generate_examples(**gen_kwargs)
+ if not drop_metadata:
+ with pytest.raises(ValueError):
+ _ = list(generator)
+ else:
+ assert all(
+ example.keys() == {"audio"} and all(val is not None for val in example.values())
+ for _, example in generator
+ )
+
+
+@require_sndfile
[email protected]("streaming", [False, True])
[email protected]("n_splits", [1, 2])
+def test_data_files_with_metadata_and_splits(
+ streaming, cache_dir, n_splits, data_files_with_one_split_and_metadata, data_files_with_two_splits_and_metadata
+):
+ data_files = data_files_with_one_split_and_metadata if n_splits == 1 else data_files_with_two_splits_and_metadata
+ audiofolder = AudioFolder(data_files=data_files, cache_dir=cache_dir)
+ audiofolder.download_and_prepare()
+ datasets = audiofolder.as_streaming_dataset() if streaming else audiofolder.as_dataset()
+ for split, data_files in data_files.items():
+ expected_num_of_audios = len(data_files) - 1 # don't count the metadata file
+ assert split in datasets
+ dataset = list(datasets[split])
+ assert len(dataset) == expected_num_of_audios
+ # make sure each sample has its own audio and metadata
+ assert len(set(example["audio"]["path"] for example in dataset)) == expected_num_of_audios
+ assert len(set(example["text"] for example in dataset)) == expected_num_of_audios
+ assert all(example["text"] is not None for example in dataset)
+
+
+@require_sndfile
[email protected]("streaming", [False, True])
+def test_data_files_with_metadata_and_archives(streaming, cache_dir, data_files_with_zip_archives):
+ audiofolder = AudioFolder(data_files=data_files_with_zip_archives, cache_dir=cache_dir)
+ audiofolder.download_and_prepare()
+ datasets = audiofolder.as_streaming_dataset() if streaming else audiofolder.as_dataset()
+ for split, data_files in data_files_with_zip_archives.items():
+ num_of_archives = len(data_files) # the metadata file is inside the archive
+ expected_num_of_audios = 2 * num_of_archives
+ assert split in datasets
+ dataset = list(datasets[split])
+ assert len(dataset) == expected_num_of_audios
+ # make sure each sample has its own audio (all arrays are different) and metadata
+ assert (
+ sum(np.array_equal(dataset[0]["audio"]["array"], example["audio"]["array"]) for example in dataset[1:])
+ == 0
+ )
+ assert len(set(example["text"] for example in dataset)) == expected_num_of_audios
+ assert all(example["text"] is not None for example in dataset)
+
+
+@require_sndfile
+def test_data_files_with_wrong_metadata_file_name(cache_dir, tmp_path, audio_file):
+ data_dir = tmp_path / "data_dir_with_bad_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ shutil.copyfile(audio_file, data_dir / "audio_file.wav")
+ audio_metadata_filename = data_dir / "bad_metadata.jsonl" # bad file
+ audio_metadata = textwrap.dedent(
+ """\
+ {"file_name": "audio_file.wav", "text": "Audio transcription"}
+ """
+ )
+ with open(audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+ audiofolder = AudioFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
+ audiofolder.download_and_prepare()
+ dataset = audiofolder.as_dataset(split="train")
+ # check that there are no metadata, since the metadata file name doesn't have the right name
+ assert "text" not in dataset.column_names
+
+
+@require_sndfile
+def test_data_files_with_wrong_audio_file_name_column_in_metadata_file(cache_dir, tmp_path, audio_file):
+ data_dir = tmp_path / "data_dir_with_bad_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ shutil.copyfile(audio_file, data_dir / "audio_file.wav")
+ audio_metadata_filename = data_dir / "metadata.jsonl"
+ audio_metadata = textwrap.dedent( # with bad column "bad_file_name" instead of "file_name"
+ """\
+ {"bad_file_name_column": "audio_file.wav", "text": "Audio transcription"}
+ """
+ )
+ with open(audio_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(audio_metadata)
+
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+ audiofolder = AudioFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
+ with pytest.raises(ValueError) as exc_info:
+ audiofolder.download_and_prepare()
+ assert "`file_name` must be present" in str(exc_info.value)
diff --git a/tests/packaged_modules/test_folder_based_builder.py b/tests/packaged_modules/test_folder_based_builder.py
new file mode 100644
--- /dev/null
+++ b/tests/packaged_modules/test_folder_based_builder.py
@@ -0,0 +1,435 @@
+import importlib
+import shutil
+import textwrap
+
+import pytest
+
+from datasets import ClassLabel, DownloadManager, Features, Value
+from datasets.data_files import DataFilesDict, get_data_patterns_locally
+from datasets.download.streaming_download_manager import StreamingDownloadManager
+from datasets.packaged_modules.folder_based_builder.folder_based_builder import (
+ FolderBasedBuilder,
+ FolderBasedBuilderConfig,
+)
+
+
+class DummyFolderBasedBuilder(FolderBasedBuilder):
+ BASE_FEATURE = None
+ BASE_COLUMN_NAME = "base"
+ BUILDER_CONFIG_CLASS = FolderBasedBuilderConfig
+ EXTENSIONS = [".txt"]
+
+
[email protected]
+def cache_dir(tmp_path):
+ return str(tmp_path / "autofolder_cache_dir")
+
+
[email protected]
+def auto_text_file(text_file):
+ return str(text_file)
+
+
[email protected]
+def data_files_with_labels_no_metadata(tmp_path, auto_text_file):
+ data_dir = tmp_path / "data_files_with_labels_no_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir_class_0 = data_dir / "class0"
+ subdir_class_0.mkdir(parents=True, exist_ok=True)
+ # data dirs can be nested but FolderBasedBuilder should care only about the last part of the path:
+ subdir_class_1 = data_dir / "subdir" / "class1"
+ subdir_class_1.mkdir(parents=True, exist_ok=True)
+
+ filename = subdir_class_0 / "file0.txt"
+ shutil.copyfile(auto_text_file, filename)
+ filename2 = subdir_class_1 / "file1.txt"
+ shutil.copyfile(auto_text_file, filename2)
+
+ data_files_with_labels_no_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+
+ return data_files_with_labels_no_metadata
+
+
[email protected]
+def files_with_labels_and_duplicated_label_key_in_metadata(tmp_path, auto_text_file):
+ data_dir = tmp_path / "files_with_labels_and_label_key_in_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir_class_0 = data_dir / "class0"
+ subdir_class_0.mkdir(parents=True, exist_ok=True)
+ subdir_class_1 = data_dir / "class1"
+ subdir_class_1.mkdir(parents=True, exist_ok=True)
+
+ filename = subdir_class_0 / "file_class0.txt"
+ shutil.copyfile(auto_text_file, filename)
+ filename2 = subdir_class_1 / "file_class1.txt"
+ shutil.copyfile(auto_text_file, filename2)
+
+ metadata_filename = tmp_path / data_dir / "metadata.jsonl"
+ metadata = textwrap.dedent(
+ """\
+ {"file_name": "class0/file_class0.txt", "additional_feature": "First dummy file", "label": "CLASS_0"}
+ {"file_name": "class1/file_class1.txt", "additional_feature": "Second dummy file", "label": "CLASS_1"}
+ """
+ )
+ with open(metadata_filename, "w", encoding="utf-8") as f:
+ f.write(metadata)
+
+ return str(filename), str(filename2), str(metadata_filename)
+
+
[email protected]
+def file_with_metadata(tmp_path, text_file):
+ filename = tmp_path / "file.txt"
+ shutil.copyfile(text_file, filename)
+ metadata_filename = tmp_path / "metadata.jsonl"
+ metadata = textwrap.dedent(
+ """\
+ {"file_name": "file.txt", "additional_feature": "Dummy file"}
+ """
+ )
+ with open(metadata_filename, "w", encoding="utf-8") as f:
+ f.write(metadata)
+ return str(filename), str(metadata_filename)
+
+
[email protected]()
+def files_with_metadata_that_misses_one_sample(tmp_path, auto_text_file):
+ filename = tmp_path / "file.txt"
+ shutil.copyfile(auto_text_file, filename)
+ filename2 = tmp_path / "file2.txt"
+ shutil.copyfile(auto_text_file, filename2)
+ metadata_filename = tmp_path / "metadata.jsonl"
+ metadata = textwrap.dedent(
+ """\
+ {"file_name": "file.txt", "additional_feature": "Dummy file"}
+ """
+ )
+ with open(metadata_filename, "w", encoding="utf-8") as f:
+ f.write(metadata)
+ return str(filename), str(filename2), str(metadata_filename)
+
+
[email protected]
+def data_files_with_one_split_and_metadata(tmp_path, auto_text_file):
+ data_dir = tmp_path / "autofolder_data_dir_with_metadata_one_split"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir = data_dir / "subdir"
+ subdir.mkdir(parents=True, exist_ok=True)
+
+ filename = data_dir / "file.txt"
+ shutil.copyfile(auto_text_file, filename)
+ filename2 = data_dir / "file2.txt"
+ shutil.copyfile(auto_text_file, filename2)
+ filename3 = subdir / "file3.txt" # in subdir
+ shutil.copyfile(auto_text_file, filename3)
+
+ metadata_filename = data_dir / "metadata.jsonl"
+ metadata = textwrap.dedent(
+ """\
+ {"file_name": "file.txt", "additional_feature": "Dummy file"}
+ {"file_name": "file2.txt", "additional_feature": "Second dummy file"}
+ {"file_name": "subdir/file3.txt", "additional_feature": "Third dummy file"}
+ """
+ )
+ with open(metadata_filename, "w", encoding="utf-8") as f:
+ f.write(metadata)
+ data_files_with_one_split_and_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(data_dir), data_dir
+ )
+ assert len(data_files_with_one_split_and_metadata) == 1
+ assert len(data_files_with_one_split_and_metadata["train"]) == 4
+ return data_files_with_one_split_and_metadata
+
+
[email protected]
+def data_files_with_two_splits_and_metadata(tmp_path, auto_text_file):
+ data_dir = tmp_path / "autofolder_data_dir_with_metadata_two_splits"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ train_dir = data_dir / "train"
+ train_dir.mkdir(parents=True, exist_ok=True)
+ test_dir = data_dir / "test"
+ test_dir.mkdir(parents=True, exist_ok=True)
+
+ filename = train_dir / "file.txt" # train
+ shutil.copyfile(auto_text_file, filename)
+ filename2 = train_dir / "file2.txt" # train
+ shutil.copyfile(auto_text_file, filename2)
+ filename3 = test_dir / "file3.txt" # test
+ shutil.copyfile(auto_text_file, filename3)
+
+ train_metadata_filename = train_dir / "metadata.jsonl"
+ train_metadata = textwrap.dedent(
+ """\
+ {"file_name": "file.txt", "additional_feature": "Train dummy file"}
+ {"file_name": "file2.txt", "additional_feature": "Second train dummy file"}
+ """
+ )
+ with open(train_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(train_metadata)
+ test_metadata_filename = test_dir / "metadata.jsonl"
+ test_metadata = textwrap.dedent(
+ """\
+ {"file_name": "file3.txt", "additional_feature": "Test dummy file"}
+ """
+ )
+ with open(test_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(test_metadata)
+ data_files_with_two_splits_and_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(data_dir), data_dir
+ )
+ assert len(data_files_with_two_splits_and_metadata) == 2
+ assert len(data_files_with_two_splits_and_metadata["train"]) == 3
+ assert len(data_files_with_two_splits_and_metadata["test"]) == 2
+ return data_files_with_two_splits_and_metadata
+
+
[email protected]
+def data_files_with_zip_archives(tmp_path, auto_text_file):
+ data_dir = tmp_path / "autofolder_data_dir_with_zip_archives"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ archive_dir = data_dir / "archive"
+ archive_dir.mkdir(parents=True, exist_ok=True)
+ subdir = archive_dir / "subdir"
+ subdir.mkdir(parents=True, exist_ok=True)
+
+ filename = archive_dir / "file.txt"
+ shutil.copyfile(auto_text_file, filename)
+ filename2 = subdir / "file2.txt" # in subdir
+ shutil.copyfile(auto_text_file, filename2)
+
+ metadata_filename = archive_dir / "metadata.jsonl"
+ metadata = textwrap.dedent(
+ """\
+ {"file_name": "file.txt", "additional_feature": "Dummy file"}
+ {"file_name": "subdir/file2.txt", "additional_feature": "Second dummy file"}
+ """
+ )
+ with open(metadata_filename, "w", encoding="utf-8") as f:
+ f.write(metadata)
+
+ shutil.make_archive(archive_dir, "zip", archive_dir)
+ shutil.rmtree(str(archive_dir))
+
+ data_files_with_zip_archives = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
+
+ assert len(data_files_with_zip_archives) == 1
+ assert len(data_files_with_zip_archives["train"]) == 1
+ return data_files_with_zip_archives
+
+
+def test_inferring_labels_from_data_dirs(data_files_with_labels_no_metadata, cache_dir):
+ autofolder = DummyFolderBasedBuilder(
+ data_files=data_files_with_labels_no_metadata, cache_dir=cache_dir, drop_labels=False
+ )
+ gen_kwargs = autofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ assert autofolder.info.features == Features({"base": None, "label": ClassLabel(names=["class0", "class1"])})
+ generator = autofolder._generate_examples(**gen_kwargs)
+ assert all(example["label"] in {"class0", "class1"} for _, example in generator)
+
+
+def test_default_autofolder_not_usable(data_files_with_labels_no_metadata, cache_dir):
+ # builder would try to access non-existing attributes of a default `BuilderConfig` class
+ # as a custom one is not provided
+ with pytest.raises(AttributeError):
+ _ = FolderBasedBuilder(
+ data_files=data_files_with_labels_no_metadata,
+ cache_dir=cache_dir,
+ )
+
+
+# test that AutoFolder is extended for streaming when it's child class is instantiated:
+# see line 115 in src/datasets/streaming.py
+def test_streaming_patched():
+ _ = DummyFolderBasedBuilder()
+ module = importlib.import_module(FolderBasedBuilder.__module__)
+ assert hasattr(module, "_patched_for_streaming")
+ assert module._patched_for_streaming
+
+
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_prepare_generate_examples_duplicated_label_key(
+ files_with_labels_and_duplicated_label_key_in_metadata, drop_metadata, drop_labels, cache_dir, caplog
+):
+ class0_file, class1_file, metadata_file = files_with_labels_and_duplicated_label_key_in_metadata
+ autofolder = DummyFolderBasedBuilder(
+ data_files=[class0_file, class1_file, metadata_file],
+ cache_dir=cache_dir,
+ drop_metadata=drop_metadata,
+ drop_labels=drop_labels,
+ )
+ gen_kwargs = autofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ generator = autofolder._generate_examples(**gen_kwargs)
+ if drop_labels is False:
+ # infer labels from directories even if metadata files are found
+ warning_in_logs = any("ignoring metadata columns" in record.msg.lower() for record in caplog.records)
+ assert warning_in_logs if drop_metadata is not True else not warning_in_logs
+ assert autofolder.info.features["label"] == ClassLabel(names=["class0", "class1"])
+ assert all(example["label"] in ["class0", "class1"] for _, example in generator)
+
+ else:
+ if drop_metadata is not True:
+ # labels are from metadata
+ assert autofolder.info.features["label"] == Value("string")
+ assert all(example["label"] in ["CLASS_0", "CLASS_1"] for _, example in generator)
+ else:
+ # drop both labels and metadata
+ assert autofolder.info.features == Features({"base": None})
+ assert all(example.keys() == {"base"} for _, example in generator)
+
+
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_prepare_generate_examples_drop_labels(auto_text_file, drop_metadata, drop_labels):
+ autofolder = DummyFolderBasedBuilder(
+ data_files={"train": [auto_text_file]},
+ drop_metadata=drop_metadata,
+ drop_labels=drop_labels,
+ )
+ gen_kwargs = autofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ # removing the labels explicitly requires drop_labels=True
+ assert gen_kwargs["add_labels"] is not bool(drop_labels)
+ assert gen_kwargs["add_metadata"] is False
+ generator = autofolder._generate_examples(**gen_kwargs)
+ if not drop_labels:
+ assert all(
+ example.keys() == {"base", "label"} and all(val is not None for val in example.values())
+ for _, example in generator
+ )
+ else:
+ assert all(
+ example.keys() == {"base"} and all(val is not None for val in example.values()) for _, example in generator
+ )
+
+
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_prepare_generate_examples_drop_metadata(file_with_metadata, drop_metadata, drop_labels):
+ file, metadata_file = file_with_metadata
+ autofolder = DummyFolderBasedBuilder(
+ data_files=[file, metadata_file],
+ drop_metadata=drop_metadata,
+ drop_labels=drop_labels,
+ )
+ gen_kwargs = autofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ # since the dataset has metadata, removing the metadata explicitly requires drop_metadata=True
+ assert gen_kwargs["add_metadata"] is not bool(drop_metadata)
+ # since the dataset has metadata, adding the labels explicitly requires drop_labels=False
+ assert gen_kwargs["add_labels"] is (drop_labels is False)
+ generator = autofolder._generate_examples(**gen_kwargs)
+ expected_columns = {"base"}
+ if gen_kwargs["add_metadata"]:
+ expected_columns.add("additional_feature")
+ if gen_kwargs["add_labels"]:
+ expected_columns.add("label")
+ result = [example for _, example in generator]
+ assert len(result) == 1
+ example = result[0]
+ assert example.keys() == expected_columns
+ for column in expected_columns:
+ assert example[column] is not None
+
+
[email protected]("drop_metadata", [None, True, False])
+def test_prepare_generate_examples_with_metadata_that_misses_one_sample(
+ files_with_metadata_that_misses_one_sample, drop_metadata
+):
+ file, file2, metadata_file = files_with_metadata_that_misses_one_sample
+ if not drop_metadata:
+ features = Features({"base": None, "additional_feature": Value("string")})
+ else:
+ features = Features({"base": None})
+ autofolder = DummyFolderBasedBuilder(
+ data_files=[file, file2, metadata_file],
+ drop_metadata=drop_metadata,
+ features=features,
+ )
+ gen_kwargs = autofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ generator = autofolder._generate_examples(**gen_kwargs)
+ if not drop_metadata:
+ with pytest.raises(ValueError):
+ list(generator)
+ else:
+ assert all(
+ example.keys() == {"base"} and all(val is not None for val in example.values()) for _, example in generator
+ )
+
+
[email protected]("streaming", [False, True])
[email protected]("n_splits", [1, 2])
+def test_data_files_with_metadata_and_splits(
+ streaming, cache_dir, n_splits, data_files_with_one_split_and_metadata, data_files_with_two_splits_and_metadata
+):
+ data_files = data_files_with_one_split_and_metadata if n_splits == 1 else data_files_with_two_splits_and_metadata
+ autofolder = DummyFolderBasedBuilder(
+ data_files=data_files,
+ cache_dir=cache_dir,
+ )
+ download_manager = StreamingDownloadManager() if streaming else DownloadManager()
+ generated_splits = autofolder._split_generators(download_manager)
+ for (split, files), generated_split in zip(data_files.items(), generated_splits):
+ assert split == generated_split.name
+ expected_num_of_examples = len(files) - 1
+ generated_examples = list(autofolder._generate_examples(**generated_split.gen_kwargs))
+ assert len(generated_examples) == expected_num_of_examples
+ assert len(set(example["base"] for _, example in generated_examples)) == expected_num_of_examples
+ assert len(set(example["additional_feature"] for _, example in generated_examples)) == expected_num_of_examples
+ assert all(example["additional_feature"] is not None for _, example in generated_examples)
+
+
[email protected]("streaming", [False, True])
+def test_data_files_with_metadata_and_archives(streaming, cache_dir, data_files_with_zip_archives):
+ autofolder = DummyFolderBasedBuilder(data_files=data_files_with_zip_archives, cache_dir=cache_dir)
+ download_manager = StreamingDownloadManager() if streaming else DownloadManager()
+ generated_splits = autofolder._split_generators(download_manager)
+ for (split, files), generated_split in zip(data_files_with_zip_archives.items(), generated_splits):
+ assert split == generated_split.name
+ num_of_archives = len(files)
+ expected_num_of_examples = 2 * num_of_archives
+ generated_examples = list(autofolder._generate_examples(**generated_split.gen_kwargs))
+ assert len(generated_examples) == expected_num_of_examples
+ assert len(set(example["base"] for _, example in generated_examples)) == expected_num_of_examples
+ assert len(set(example["additional_feature"] for _, example in generated_examples)) == expected_num_of_examples
+ assert all(example["additional_feature"] is not None for _, example in generated_examples)
+
+
+def test_data_files_with_wrong_metadata_file_name(cache_dir, tmp_path, auto_text_file):
+ data_dir = tmp_path / "data_dir_with_bad_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ shutil.copyfile(auto_text_file, data_dir / "file.txt")
+ metadata_filename = data_dir / "bad_metadata.jsonl" # bad file
+ metadata = textwrap.dedent(
+ """\
+ {"file_name": "file.txt", "additional_feature": "Dummy file"}
+ """
+ )
+ with open(metadata_filename, "w", encoding="utf-8") as f:
+ f.write(metadata)
+
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
+ autofolder = DummyFolderBasedBuilder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
+ gen_kwargs = autofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ generator = autofolder._generate_examples(**gen_kwargs)
+ assert all("additional_feature" not in example for _, example in generator)
+
+
+def test_data_files_with_wrong_file_name_column_in_metadata_file(cache_dir, tmp_path, auto_text_file):
+ data_dir = tmp_path / "data_dir_with_bad_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ shutil.copyfile(auto_text_file, data_dir / "file.txt")
+ metadata_filename = data_dir / "metadata.jsonl"
+ metadata = textwrap.dedent( # with bad column "bad_file_name" instead of "file_name"
+ """\
+ {"bad_file_name": "file.txt", "additional_feature": "Dummy file"}
+ """
+ )
+ with open(metadata_filename, "w", encoding="utf-8") as f:
+ f.write(metadata)
+
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
+ autofolder = DummyFolderBasedBuilder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
+ with pytest.raises(ValueError) as exc_info:
+ _ = autofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ assert "`file_name` must be present" in str(exc_info.value)
diff --git a/tests/packaged_modules/test_imagefolder.py b/tests/packaged_modules/test_imagefolder.py
--- a/tests/packaged_modules/test_imagefolder.py
+++ b/tests/packaged_modules/test_imagefolder.py
@@ -8,7 +8,6 @@
from datasets.data_files import DataFilesDict, get_data_patterns_locally
from datasets.download.streaming_download_manager import StreamingDownloadManager
from datasets.packaged_modules.imagefolder.imagefolder import ImageFolder
-from datasets.streaming import extend_module_for_streaming
from ..utils import require_pil
@@ -376,8 +375,6 @@ def test_data_files_with_metadata_and_splits(
@require_pil
@pytest.mark.parametrize("streaming", [False, True])
def test_data_files_with_metadata_and_archives(streaming, cache_dir, data_files_with_zip_archives):
- if streaming:
- extend_module_for_streaming(ImageFolder.__module__)
imagefolder = ImageFolder(data_files=data_files_with_zip_archives, cache_dir=cache_dir)
imagefolder.download_and_prepare()
datasets = imagefolder.as_streaming_dataset() if streaming else imagefolder.as_dataset()
diff --git a/tests/test_dataset_common.py b/tests/test_dataset_common.py
--- a/tests/test_dataset_common.py
+++ b/tests/test_dataset_common.py
@@ -82,6 +82,7 @@ def get_packaged_dataset_dummy_data_files(dataset_name, path_to_dummy_data):
"csv": ".csv",
"parquet": ".parquet",
"imagefolder": "/",
+ "audiofolder": "/",
}
return {
"train": os.path.join(path_to_dummy_data, "train" + extensions[dataset_name]),
| Add default Audio Loader
**Is your feature request related to a problem? Please describe.**
Writing a custom loading dataset script might be a bit challenging for users.
**Describe the solution you'd like**
Add default Audio loader (analogous to ImageFolder) for small datasets with standard directory structure.
**Describe alternatives you've considered**
Create a custom loading script? that's what users doing now.
| 2022-06-20T12:54:02Z | [] | [] |
|
huggingface/datasets | 4,551 | huggingface__datasets-4551 | [
"4549"
] | bbe338d1f766a3069f00dc391f97b61777a94d96 | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -10,7 +10,7 @@
from .filesystems.hffilesystem import HfFileSystem
from .splits import Split
from .utils import logging
-from .utils.file_utils import hf_hub_url, is_remote_url, request_etag
+from .utils.file_utils import hf_hub_url, is_relative_path, is_remote_url, request_etag
from .utils.py_utils import string_to_dict
@@ -76,6 +76,101 @@ def sanitize_patterns(patterns: Union[Dict, List, str]) -> Dict[str, Union[List[
return {DEFAULT_SPLIT: list(patterns)}
+def _is_inside_unrequested_special_dir(matched_rel_path: str, pattern: str) -> bool:
+ """
+ When a path matches a pattern, we additionnally check if it's inside a special directory
+ we ignore by default (if it starts with a double underscore).
+
+ Users can still explicitly request a filepath inside such a directory if "__pycache__" is
+ mentioned explicitly in the requested pattern.
+
+ Some examples:
+
+ base directory:
+
+ ./
+ └── __pycache__
+ └── b.txt
+
+ >>> _is_inside_unrequested_special_dir("__pycache__/b.txt", "**")
+ True
+ >>> _is_inside_unrequested_special_dir("__pycache__/b.txt", "*/b.txt")
+ True
+ >>> _is_inside_unrequested_special_dir("__pycache__/b.txt", "__pycache__/*")
+ False
+ >>> _is_inside_unrequested_special_dir("__pycache__/b.txt", "__*/*")
+ False
+ """
+ # We just need to check if every special directories from the path is present explicly in the pattern.
+ # Since we assume that the path matches the pattern, it's equivalent to counting that both
+ # the parent path and the parent pattern have the same number of special directories.
+ data_dirs_to_ignore_in_path = [part for part in PurePath(matched_rel_path).parent.parts if part.startswith("__")]
+ data_dirs_to_ignore_in_pattern = [part for part in PurePath(pattern).parent.parts if part.startswith("__")]
+ return len(data_dirs_to_ignore_in_path) != len(data_dirs_to_ignore_in_pattern)
+
+
+def _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(matched_rel_path: str, pattern: str) -> bool:
+ """
+ When a path matches a pattern, we additionnally check if it's a hidden file or if it's inside
+ a hidden directory we ignore by default, i.e. if the file name or a parent directory name starts with a dot.
+
+ Users can still explicitly request a filepath that is hidden or is inside a hidden directory
+ if the hidden part is mentioned explicitly in the requested pattern.
+
+ Some examples:
+
+ base directory:
+
+ ./
+ └── .hidden_file.txt
+
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_file.txt", "**")
+ True
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_file.txt", ".*")
+ False
+
+ base directory:
+
+ ./
+ └── .hidden_dir
+ └── a.txt
+
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/a.txt", "**")
+ True
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/a.txt", ".*/*")
+ False
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/a.txt", ".hidden_dir/*")
+ False
+
+ base directory:
+
+ ./
+ └── .hidden_dir
+ └── .hidden_file.txt
+
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/.hidden_file.txt", "**")
+ True
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/.hidden_file.txt", ".*/*")
+ True
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/.hidden_file.txt", ".*/.*")
+ False
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/.hidden_file.txt", ".hidden_dir/*")
+ True
+ >>> _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(".hidden_dir/.hidden_file.txt", ".hidden_dir/.*")
+ False
+ """
+ # We just need to check if every hidden part from the path is present explicly in the pattern.
+ # Since we assume that the path matches the pattern, it's equivalent to counting that both
+ # the path and the pattern have the same number of hidden parts.
+ hidden_directories_in_path = [
+ part for part in PurePath(matched_rel_path).parts if part.startswith(".") and not set(part) == {"."}
+ ]
+ hidden_directories_in_pattern = [
+ part for part in PurePath(pattern).parts if part.startswith(".") and not set(part) == {"."}
+ ]
+ return len(hidden_directories_in_path) != len(hidden_directories_in_pattern)
+
+
def _get_data_files_patterns(pattern_resolver: Callable[[str], List[PurePath]]) -> Dict[str, List[str]]:
"""
Get the default pattern from a directory or repository by testing all the supported patterns.
@@ -133,15 +228,22 @@ def _resolve_single_pattern_locally(
It also supports absolute paths in patterns.
If an URL is passed, it is returned as is.
"""
- pattern = os.path.join(base_path, pattern)
- data_files_ignore = FILES_TO_IGNORE
+ if is_relative_path(pattern):
+ pattern = os.path.join(base_path, pattern)
+ else:
+ base_path = "/"
fs = LocalFileSystem()
glob_iter = [PurePath(filepath) for filepath in fs.glob(pattern) if fs.isfile(filepath)]
matched_paths = [
Path(filepath).resolve()
for filepath in glob_iter
- if filepath.name not in data_files_ignore
- and not any(part.startswith((".", "__")) and set(part) != {"."} for part in filepath.parts)
+ if (filepath.name not in FILES_TO_IGNORE or PurePath(pattern).name == filepath.name)
+ and not _is_inside_unrequested_special_dir(
+ os.path.relpath(filepath, base_path), os.path.relpath(pattern, base_path)
+ )
+ and not _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(
+ os.path.relpath(filepath, base_path), os.path.relpath(pattern, base_path)
+ )
] # ignore .ipynb and __pycache__, but keep /../
if allowed_extensions is not None:
out = [
@@ -187,9 +289,15 @@ def resolve_patterns_locally_or_by_urls(
- '*' matches any character except a forward-slash (to match just the file or directory name)
- '**' matches any character including a forward-slash /
+ Hidden files and directories (i.e. whose names start with a dot) are ignored, unless they are explicitly requested.
+ The same applies to special directories that start with a double underscore like "__pycache__".
+ You can still include one if the pattern explicilty mentions it:
+ - to include a hidden file: "*/.hidden.txt" or "*/.*"
+ - to include a hidden directory: ".hidden/*" or ".*/*"
+ - to include a special directory: "__special__/*" or "__*/*"
+
Example::
- >>> import huggingface_hub
>>> from datasets.data_files import resolve_patterns_locally_or_by_urls
>>> base_path = "."
>>> resolve_patterns_locally_or_by_urls(base_path, ["src/**/*.yaml"])
@@ -329,16 +437,22 @@ def _resolve_single_pattern_in_dataset_repository(
base_path: Optional[str] = None,
allowed_extensions: Optional[list] = None,
) -> List[PurePath]:
- data_files_ignore = FILES_TO_IGNORE
fs = HfFileSystem(repo_info=dataset_info)
if base_path:
pattern = f"{base_path}/{pattern}"
+ else:
+ base_path = "/"
glob_iter = [PurePath(filepath) for filepath in fs.glob(PurePath(pattern).as_posix()) if fs.isfile(filepath)]
matched_paths = [
filepath
for filepath in glob_iter
- if filepath.name not in data_files_ignore
- and not any(part.startswith((".", "__")) and set(part) != {"."} for part in filepath.parts)
+ if (filepath.name not in FILES_TO_IGNORE or PurePath(pattern).name == filepath.name)
+ and not _is_inside_unrequested_special_dir(
+ os.path.relpath(filepath, base_path), os.path.relpath(pattern, base_path)
+ )
+ and not _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(
+ os.path.relpath(filepath, base_path), os.path.relpath(pattern, base_path)
+ )
] # ignore .ipynb and __pycache__, but keep /../
if allowed_extensions is not None:
out = [
@@ -386,6 +500,13 @@ def resolve_patterns_in_dataset_repository(
- '*' matches any character except a forward-slash (to match just the file or directory name)
- '**' matches any character including a forward-slash /
+ Hidden files and directories (i.e. whose names start with a dot) are ignored, unless they are explicitly requested.
+ The same applies to special directories that start with a double underscore like "__pycache__".
+ You can still include one if the pattern explicilty mentions it:
+ - to include a hidden file: "*/.hidden.txt" or "*/.*"
+ - to include a hidden directory: ".hidden/*" or ".*/*"
+ - to include a special directory: "__special__/*" or "__*/*"
+
Example::
>>> import huggingface_hub
| diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -13,6 +13,8 @@
Url,
_get_data_files_patterns,
_get_metadata_files_patterns,
+ _is_inside_unrequested_special_dir,
+ _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir,
resolve_patterns_in_dataset_repository,
resolve_patterns_locally_or_by_urls,
)
@@ -22,7 +24,7 @@
_TEST_PATTERNS = ["*", "**", "**/*", "*.txt", "data/*", "**/*.txt", "**/train.txt"]
_FILES_TO_IGNORE = {".dummy", "README.md", "dummy_data.zip", "dataset_infos.json"}
-_DIRS_TO_IGNORE = {"data/.dummy_subdir"}
+_DIRS_TO_IGNORE = {"data/.dummy_subdir", "__pycache__"}
_TEST_PATTERNS_SIZES = dict(
[
("*", 0),
@@ -65,6 +67,10 @@ def complex_data_dir(tmp_path):
with open(data_dir / "data" / ".dummy_subdir" / "test.txt", "w") as f:
f.write("bar\n" * 10)
+ (data_dir / "__pycache__").mkdir()
+ with open(data_dir / "__pycache__" / "script.py", "w") as f:
+ f.write("foo\n" * 10)
+
return str(data_dir)
@@ -131,6 +137,42 @@ def hub_dataset_info_patterns_results(hub_dataset_info, complex_data_dir, patter
}
+def test_is_inside_unrequested_special_dir(complex_data_dir, pattern_results):
+ # usual patterns outside special dir work fine
+ for pattern, result in pattern_results.items():
+ if result:
+ matched_rel_path = str(Path(result[0]).relative_to(complex_data_dir))
+ assert _is_inside_unrequested_special_dir(matched_rel_path, pattern) is False
+ # check behavior for special dir
+ f = _is_inside_unrequested_special_dir
+ assert f("__pycache__/b.txt", "**") is True
+ assert f("__pycache__/b.txt", "*/b.txt") is True
+ assert f("__pycache__/b.txt", "__pycache__/*") is False
+ assert f("__pycache__/__b.txt", "__pycache__/*") is False
+ assert f("__pycache__/__b.txt", "__*/*") is False
+ assert f("__b.txt", "*") is False
+
+
+def test_is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir(complex_data_dir, pattern_results):
+ # usual patterns outside hidden dir work fine
+ for pattern, result in pattern_results.items():
+ if result:
+ matched_rel_path = str(Path(result[0]).relative_to(complex_data_dir))
+ assert _is_inside_unrequested_special_dir(matched_rel_path, pattern) is False
+ # check behavior for hidden dir and file
+ f = _is_unrequested_hidden_file_or_is_inside_unrequested_hidden_dir
+ assert f(".hidden_file.txt", "**") is True
+ assert f(".hidden_file.txt", ".*") is False
+ assert f(".hidden_dir/a.txt", "**") is True
+ assert f(".hidden_dir/a.txt", ".*/*") is False
+ assert f(".hidden_dir/a.txt", ".hidden_dir/*") is False
+ assert f(".hidden_dir/.hidden_file.txt", "**") is True
+ assert f(".hidden_dir/.hidden_file.txt", ".*/*") is True
+ assert f(".hidden_dir/.hidden_file.txt", ".*/.*") is False
+ assert f(".hidden_dir/.hidden_file.txt", ".hidden_dir/*") is True
+ assert f(".hidden_dir/.hidden_file.txt", ".hidden_dir/.*") is False
+
+
@pytest.mark.parametrize("pattern", _TEST_PATTERNS)
def test_pattern_results_fixture(pattern_results, pattern):
assert len(pattern_results[pattern]) == _TEST_PATTERNS_SIZES[pattern]
@@ -147,6 +189,14 @@ def test_resolve_patterns_locally_or_by_urls(complex_data_dir, pattern, pattern_
assert len(pattern_results[pattern]) == 0
+def test_resolve_patterns_locally_or_by_urls_with_dot_in_base_path(complex_data_dir):
+ base_path_with_dot = os.path.join(complex_data_dir, "data", ".dummy_subdir")
+ resolved_data_files = resolve_patterns_locally_or_by_urls(
+ base_path_with_dot, [os.path.join(base_path_with_dot, "train.txt")]
+ )
+ assert len(resolved_data_files) == 1
+
+
def test_resolve_patterns_locally_or_by_urls_with_absolute_path(tmp_path, complex_data_dir):
abs_path = os.path.join(complex_data_dir, "data", "train.txt")
resolved_data_files = resolve_patterns_locally_or_by_urls(str(tmp_path / "blabla"), [abs_path])
@@ -159,6 +209,47 @@ def test_resolve_patterns_locally_or_by_urls_with_double_dots(tmp_path, complex_
assert len(resolved_data_files) == 1
+def test_resolve_patterns_locally_or_by_urls_returns_hidden_file_only_if_requested(complex_data_dir):
+ with pytest.raises(FileNotFoundError):
+ resolve_patterns_locally_or_by_urls(complex_data_dir, ["*dummy"])
+ resolved_data_files = resolve_patterns_locally_or_by_urls(complex_data_dir, [".dummy"])
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_locally_or_by_urls_hidden_base_path(tmp_path):
+ hidden = tmp_path / ".test_hidden_base_path"
+ hidden.mkdir()
+ (tmp_path / ".test_hidden_base_path" / "a.txt").touch()
+ resolved_data_files = resolve_patterns_locally_or_by_urls(str(hidden), ["*"])
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_locally_or_by_urls_returns_hidden_dir_only_if_requested(complex_data_dir):
+ with pytest.raises(FileNotFoundError):
+ resolve_patterns_locally_or_by_urls(complex_data_dir, ["data/*dummy_subdir/train.txt"])
+ resolved_data_files = resolve_patterns_locally_or_by_urls(complex_data_dir, ["data/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+ resolved_data_files = resolve_patterns_locally_or_by_urls(complex_data_dir, ["*/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_locally_or_by_urls_returns_special_dir_only_if_requested(complex_data_dir):
+ with pytest.raises(FileNotFoundError):
+ resolve_patterns_locally_or_by_urls(complex_data_dir, ["data/*dummy_subdir/train.txt"])
+ resolved_data_files = resolve_patterns_locally_or_by_urls(complex_data_dir, ["data/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+ resolved_data_files = resolve_patterns_locally_or_by_urls(complex_data_dir, ["*/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_locally_or_by_urls_special_base_path(tmp_path):
+ special = tmp_path / "__test_special_base_path__"
+ special.mkdir()
+ (tmp_path / "__test_special_base_path__" / "a.txt").touch()
+ resolved_data_files = resolve_patterns_locally_or_by_urls(str(special), ["*"])
+ assert len(resolved_data_files) == 1
+
+
@pytest.mark.parametrize("pattern,size,extensions", [("**", 4, ["txt"]), ("**", 4, None), ("**", 0, ["blablabla"])])
def test_resolve_patterns_locally_or_by_urls_with_extensions(complex_data_dir, pattern, size, extensions):
if size > 0:
@@ -239,6 +330,45 @@ def test_resolve_patterns_in_dataset_repository_sorted_files():
assert resolved_names == sorted(unsorted_names)
+def test_resolve_patterns_in_dataset_repository_returns_hidden_file_only_if_requested(hub_dataset_info):
+ with pytest.raises(FileNotFoundError):
+ resolve_patterns_in_dataset_repository(hub_dataset_info, ["*dummy"])
+ resolved_data_files = resolve_patterns_in_dataset_repository(hub_dataset_info, [".dummy"])
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_in_dataset_repository_hidden_base_path():
+ siblings = [{"rfilename": ".hidden/a.txt"}]
+ datasets_infos = DatasetInfo(id="test_hidden_base_path", siblings=siblings, sha="foobar")
+ resolved_data_files = resolve_patterns_in_dataset_repository(datasets_infos, ["*"], base_path=".hidden")
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_in_dataset_repository_returns_hidden_dir_only_if_requested(hub_dataset_info):
+ with pytest.raises(FileNotFoundError):
+ resolve_patterns_in_dataset_repository(hub_dataset_info, ["data/*dummy_subdir/train.txt"])
+ resolved_data_files = resolve_patterns_in_dataset_repository(hub_dataset_info, ["data/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+ resolved_data_files = resolve_patterns_in_dataset_repository(hub_dataset_info, ["*/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_in_dataset_repository_returns_special_dir_only_if_requested(hub_dataset_info):
+ with pytest.raises(FileNotFoundError):
+ resolve_patterns_in_dataset_repository(hub_dataset_info, ["data/*dummy_subdir/train.txt"])
+ resolved_data_files = resolve_patterns_in_dataset_repository(hub_dataset_info, ["data/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+ resolved_data_files = resolve_patterns_in_dataset_repository(hub_dataset_info, ["*/.dummy_subdir/train.txt"])
+ assert len(resolved_data_files) == 1
+
+
+def test_resolve_patterns_in_dataset_repository_special_base_path():
+ siblings = [{"rfilename": "__special__/a.txt"}]
+ datasets_infos = DatasetInfo(id="test_hidden_base_path", siblings=siblings, sha="foobar")
+ resolved_data_files = resolve_patterns_in_dataset_repository(datasets_infos, ["*"], base_path="__special__")
+ assert len(resolved_data_files) == 1
+
+
@pytest.mark.parametrize("pattern", _TEST_PATTERNS)
def test_DataFilesList_from_hf_repo(hub_dataset_info, hub_dataset_info_patterns_results, pattern):
try:
| FileNotFoundError when passing a data_file inside a directory starting with double underscores
Bug experienced in the `accelerate` CI: https://github.com/huggingface/accelerate/runs/7016055148?check_suite_focus=true
This is related to https://github.com/huggingface/datasets/pull/4505 and the changes from https://github.com/huggingface/datasets/pull/4412
| 2022-06-23T14:49:11Z | [] | [] |
|
huggingface/datasets | 4,576 | huggingface__datasets-4576 | [
"4548"
] | f6de6dcac81764c365423f5822f1989897146e64 | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -48,6 +48,7 @@ class Url(str):
DEFAULT_PATTERNS_SPLIT_IN_DIR_NAME,
DEFAULT_PATTERNS_ALL,
]
+METADATA_PATTERNS = ["metadata.jsonl", "**/metadata.jsonl"] # metadata file for ImageFolder and AudioFolder
WILDCARD_CHARACTERS = "*[]"
FILES_TO_IGNORE = ["README.md", "config.json", "dataset_infos.json", "dummy_data.zip", "dataset_dict.json"]
@@ -107,6 +108,23 @@ def _get_data_files_patterns(pattern_resolver: Callable[[str], List[PurePath]])
raise FileNotFoundError(f"Couldn't resolve pattern {pattern} with resolver {pattern_resolver}")
+def _get_metadata_files_patterns(pattern_resolver: Callable[[str], List[PurePath]]) -> Dict[str, List[str]]:
+ """
+ Get the supported metadata patterns from a directory or repository.
+ """
+ non_empty_patterns = []
+ for pattern in METADATA_PATTERNS:
+ try:
+ metadata_files = pattern_resolver(pattern)
+ if len(metadata_files) > 0:
+ non_empty_patterns.append(pattern)
+ except FileNotFoundError:
+ pass
+ if non_empty_patterns:
+ return non_empty_patterns
+ raise FileNotFoundError(f"Couldn't resolve pattern {pattern} with resolver {pattern_resolver}")
+
+
def _resolve_single_pattern_locally(
base_path: str, pattern: str, allowed_extensions: Optional[List[str]] = None
) -> List[Path]:
@@ -203,7 +221,7 @@ def resolve_patterns_locally_or_by_urls(
return data_files
-def get_patterns_locally(base_path: str) -> Dict[str, List[str]]:
+def get_data_patterns_locally(base_path: str) -> Dict[str, List[str]]:
"""
Get the default pattern from a directory testing all the supported patterns.
The first patterns to return a non-empty list of data files is returned.
@@ -294,6 +312,17 @@ def get_patterns_locally(base_path: str) -> Dict[str, List[str]]:
raise FileNotFoundError(f"The directory at {base_path} doesn't contain any data file") from None
+def get_metadata_patterns_locally(base_path: str) -> List[str]:
+ """
+ Get the supported metadata patterns from a local directory.
+ """
+ resolver = partial(_resolve_single_pattern_locally, base_path)
+ try:
+ return _get_metadata_files_patterns(resolver)
+ except FileNotFoundError:
+ raise FileNotFoundError(f"The directory at {base_path} doesn't contain any metadata file") from None
+
+
def _resolve_single_pattern_in_dataset_repository(
dataset_info: huggingface_hub.hf_api.DatasetInfo,
pattern: str,
@@ -393,7 +422,7 @@ def resolve_patterns_in_dataset_repository(
return data_files_urls
-def get_patterns_in_dataset_repository(
+def get_data_patterns_in_dataset_repository(
dataset_info: huggingface_hub.hf_api.DatasetInfo, base_path: str
) -> Dict[str, List[str]]:
"""
@@ -488,6 +517,21 @@ def get_patterns_in_dataset_repository(
) from None
+def get_metadata_patterns_in_dataset_repository(
+ dataset_info: huggingface_hub.hf_api.DatasetInfo, base_path: str
+) -> List[str]:
+ """
+ Get the supported metadata patterns from a remote repository.
+ """
+ resolver = partial(_resolve_single_pattern_in_dataset_repository, dataset_info, base_path=base_path)
+ try:
+ return _get_metadata_files_patterns(resolver)
+ except FileNotFoundError:
+ raise FileNotFoundError(
+ f"The dataset repository at '{dataset_info.id}' doesn't contain any metadata file."
+ ) from None
+
+
def _get_single_origin_metadata_locally_or_by_urls(
data_file: Union[Path, Url], use_auth_token: Optional[Union[bool, str]] = None
) -> Tuple[str]:
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -34,10 +34,13 @@
from .arrow_dataset import Dataset
from .builder import DatasetBuilder
from .data_files import (
+ DEFAULT_PATTERNS_ALL,
DataFilesDict,
DataFilesList,
- get_patterns_in_dataset_repository,
- get_patterns_locally,
+ get_data_patterns_in_dataset_repository,
+ get_data_patterns_locally,
+ get_metadata_patterns_in_dataset_repository,
+ get_metadata_patterns_locally,
sanitize_patterns,
)
from .dataset_dict import DatasetDict, IterableDatasetDict
@@ -49,7 +52,12 @@
from .info import DatasetInfo, DatasetInfosDict
from .iterable_dataset import IterableDataset
from .metric import Metric
-from .packaged_modules import _EXTENSION_TO_MODULE, _PACKAGED_DATASETS_MODULES, _hash_python_lines
+from .packaged_modules import (
+ _EXTENSION_TO_MODULE,
+ _MODULE_SUPPORTS_METADATA,
+ _PACKAGED_DATASETS_MODULES,
+ _hash_python_lines,
+)
from .splits import Split
from .tasks import TaskTemplate
from .utils.file_utils import (
@@ -73,8 +81,6 @@
logger = get_logger(__name__)
-DEFAULT_SPLIT = str(Split.TRAIN)
-
ALL_ALLOWED_EXTENSIONS = list(_EXTENSION_TO_MODULE.keys()) + ["zip"]
@@ -677,16 +683,17 @@ def __init__(
self.download_mode = download_mode
def get_module(self) -> DatasetModule:
+ base_path = os.path.join(self.path, self.data_dir) if self.data_dir else self.path
patterns = (
sanitize_patterns(self.data_files)
if self.data_files is not None
- else get_patterns_locally(os.path.join(self.path, self.data_dir))
+ else get_data_patterns_locally(base_path)
if self.data_dir is not None
- else get_patterns_locally(self.path)
+ else get_data_patterns_locally(base_path)
)
data_files = DataFilesDict.from_local_or_remote(
patterns,
- base_path=os.path.join(self.path, self.data_dir) if self.data_dir else self.path,
+ base_path=base_path,
allowed_extensions=ALL_ALLOWED_EXTENSIONS,
)
module_names = {
@@ -697,6 +704,19 @@ def get_module(self) -> DatasetModule:
module_name, builder_kwargs = next(iter(module_names.values()))
if not module_name:
raise FileNotFoundError(f"No data files or dataset script found in {self.path}")
+ # Collect metadata files if the module supports them
+ if self.data_files is None and module_name in _MODULE_SUPPORTS_METADATA and patterns != DEFAULT_PATTERNS_ALL:
+ try:
+ metadata_patterns = get_metadata_patterns_locally(base_path)
+ except FileNotFoundError:
+ metadata_patterns = None
+ if metadata_patterns is not None:
+ metadata_files = DataFilesList.from_local_or_remote(metadata_patterns, base_path=base_path)
+ for key in data_files:
+ data_files[key] = DataFilesList(
+ data_files[key] + metadata_files,
+ data_files[key].origin_metadata + metadata_files.origin_metadata,
+ )
module_path, hash = _PACKAGED_DATASETS_MODULES[module_name]
builder_kwargs = {
"hash": hash,
@@ -733,18 +753,33 @@ def __init__(
increase_load_count(name, resource_type="dataset")
def get_module(self) -> DatasetModule:
+ base_path = str(Path(self.data_dir).resolve()) if self.data_dir is not None else str(Path().resolve())
patterns = (
sanitize_patterns(self.data_files)
if self.data_files is not None
- else get_patterns_locally(str(Path(self.data_dir).resolve()))
+ else get_data_patterns_locally(base_path)
if self.data_dir is not None
- else get_patterns_locally(str(Path().resolve()))
+ else get_data_patterns_locally(base_path)
)
data_files = DataFilesDict.from_local_or_remote(
patterns,
use_auth_token=self.download_config.use_auth_token,
- base_path=str(Path(self.data_dir).resolve()) if self.data_dir else None,
+ base_path=base_path,
)
+ if self.data_files is None and self.name in _MODULE_SUPPORTS_METADATA and patterns != DEFAULT_PATTERNS_ALL:
+ try:
+ metadata_patterns = get_metadata_patterns_locally(base_path)
+ except FileNotFoundError:
+ metadata_patterns = None
+ if metadata_patterns is not None:
+ metadata_files = DataFilesList.from_local_or_remote(
+ metadata_patterns, use_auth_token=self.download_config.use_auth_token, base_path=base_path
+ )
+ for key in data_files:
+ data_files[key] = DataFilesList(
+ data_files[key] + metadata_files,
+ data_files[key].origin_metadata + metadata_files.origin_metadata,
+ )
module_path, hash = _PACKAGED_DATASETS_MODULES[self.name]
builder_kwargs = {"hash": hash, "data_files": data_files}
return DatasetModule(module_path, hash, builder_kwargs)
@@ -789,7 +824,7 @@ def get_module(self) -> DatasetModule:
patterns = (
sanitize_patterns(self.data_files)
if self.data_files is not None
- else get_patterns_in_dataset_repository(hfh_dataset_info, self.data_dir)
+ else get_data_patterns_in_dataset_repository(hfh_dataset_info, self.data_dir)
)
data_files = DataFilesDict.from_hf_repo(
patterns,
@@ -806,6 +841,21 @@ def get_module(self) -> DatasetModule:
module_name, builder_kwargs = next(iter(module_names.values()))
if not module_name:
raise FileNotFoundError(f"No data files or dataset script found in {self.name}")
+ # Collect metadata files if the module supports them
+ if self.data_files is None and module_name in _MODULE_SUPPORTS_METADATA and patterns != DEFAULT_PATTERNS_ALL:
+ try:
+ metadata_patterns = get_metadata_patterns_in_dataset_repository(hfh_dataset_info, self.data_dir)
+ except FileNotFoundError:
+ metadata_patterns = None
+ if metadata_patterns is not None:
+ metadata_files = DataFilesList.from_hf_repo(
+ metadata_patterns, dataset_info=hfh_dataset_info, base_path=self.data_dir
+ )
+ for key in data_files:
+ data_files[key] = DataFilesList(
+ data_files[key] + metadata_files,
+ data_files[key].origin_metadata + metadata_files.origin_metadata,
+ )
module_path, hash = _PACKAGED_DATASETS_MODULES[module_name]
builder_kwargs = {
"hash": hash,
diff --git a/src/datasets/packaged_modules/__init__.py b/src/datasets/packaged_modules/__init__.py
--- a/src/datasets/packaged_modules/__init__.py
+++ b/src/datasets/packaged_modules/__init__.py
@@ -44,3 +44,5 @@ def _hash_python_lines(lines: List[str]) -> str:
}
_EXTENSION_TO_MODULE.update({ext[1:]: ("imagefolder", {}) for ext in imagefolder.ImageFolder.IMAGE_EXTENSIONS})
_EXTENSION_TO_MODULE.update({ext[1:].upper(): ("imagefolder", {}) for ext in imagefolder.ImageFolder.IMAGE_EXTENSIONS})
+
+_MODULE_SUPPORTS_METADATA = {"imagefolder"}
diff --git a/src/datasets/packaged_modules/imagefolder/imagefolder.py b/src/datasets/packaged_modules/imagefolder/imagefolder.py
--- a/src/datasets/packaged_modules/imagefolder/imagefolder.py
+++ b/src/datasets/packaged_modules/imagefolder/imagefolder.py
@@ -70,7 +70,7 @@ def _split_generators(self, dl_manager):
do_analyze = (self.config.features is None and not self.config.drop_labels) or not self.config.drop_metadata
if do_analyze:
labels = set()
- metadata_files = collections.defaultdict(list)
+ metadata_files = collections.defaultdict(set)
def analyze(files_or_archives, downloaded_files_or_dirs, split):
if len(downloaded_files_or_dirs) == 0:
@@ -85,7 +85,7 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
labels.add(os.path.basename(os.path.dirname(original_file)))
elif os.path.basename(original_file) == self.METADATA_FILENAME:
- metadata_files[split].append((original_file, downloaded_file))
+ metadata_files[split].add((original_file, downloaded_file))
else:
original_file_name = os.path.basename(original_file)
logger.debug(
@@ -100,7 +100,7 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
if downloaded_dir_file_ext in self.IMAGE_EXTENSIONS:
labels.add(os.path.basename(os.path.dirname(downloaded_dir_file)))
elif os.path.basename(downloaded_dir_file) == self.METADATA_FILENAME:
- metadata_files[split].append((None, downloaded_dir_file))
+ metadata_files[split].add((None, downloaded_dir_file))
else:
archive_file_name = os.path.basename(archive)
original_file_name = os.path.basename(downloaded_dir_file)
| diff --git a/tests/packaged_modules/test_imagefolder.py b/tests/packaged_modules/test_imagefolder.py
--- a/tests/packaged_modules/test_imagefolder.py
+++ b/tests/packaged_modules/test_imagefolder.py
@@ -5,7 +5,7 @@
import pytest
from datasets import Features, Image, Value
-from datasets.data_files import DataFilesDict, get_patterns_locally
+from datasets.data_files import DataFilesDict, get_data_patterns_locally
from datasets.packaged_modules.imagefolder.imagefolder import ImageFolder
from datasets.streaming import extend_module_for_streaming
@@ -51,7 +51,7 @@ def image_files_with_metadata_that_misses_one_image(tmp_path, image_file):
@pytest.fixture
def data_files_with_one_split_and_metadata(tmp_path, image_file):
- data_dir = tmp_path / "imagefolder_data_dir_with_metadata"
+ data_dir = tmp_path / "imagefolder_data_dir_with_metadata_one_split"
data_dir.mkdir(parents=True, exist_ok=True)
subdir = data_dir / "subdir"
subdir.mkdir(parents=True, exist_ok=True)
@@ -74,7 +74,7 @@ def data_files_with_one_split_and_metadata(tmp_path, image_file):
with open(image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
data_files_with_one_split_and_metadata = DataFilesDict.from_local_or_remote(
- get_patterns_locally(data_dir), data_dir
+ get_data_patterns_locally(data_dir), data_dir
)
assert len(data_files_with_one_split_and_metadata) == 1
assert len(data_files_with_one_split_and_metadata["train"]) == 4
@@ -83,7 +83,7 @@ def data_files_with_one_split_and_metadata(tmp_path, image_file):
@pytest.fixture
def data_files_with_two_splits_and_metadata(tmp_path, image_file):
- data_dir = tmp_path / "imagefolder_data_dir_with_metadata"
+ data_dir = tmp_path / "imagefolder_data_dir_with_metadata_two_splits"
data_dir.mkdir(parents=True, exist_ok=True)
train_dir = data_dir / "train"
train_dir.mkdir(parents=True, exist_ok=True)
@@ -106,16 +106,16 @@ def data_files_with_two_splits_and_metadata(tmp_path, image_file):
)
with open(train_image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
- train_image_metadata_filename = test_dir / "metadata.jsonl"
+ test_image_metadata_filename = test_dir / "metadata.jsonl"
image_metadata = textwrap.dedent(
"""\
{"file_name": "image_rgb3.jpg", "caption": "Nice test image"}
"""
)
- with open(train_image_metadata_filename, "w", encoding="utf-8") as f:
+ with open(test_image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
data_files_with_two_splits_and_metadata = DataFilesDict.from_local_or_remote(
- get_patterns_locally(data_dir), data_dir
+ get_data_patterns_locally(data_dir), data_dir
)
assert len(data_files_with_two_splits_and_metadata) == 2
assert len(data_files_with_two_splits_and_metadata["train"]) == 3
@@ -154,7 +154,7 @@ def data_files_with_zip_archives(tmp_path, image_file):
shutil.make_archive(archive_dir, "zip", archive_dir)
shutil.rmtree(str(archive_dir))
- data_files_with_zip_archives = DataFilesDict.from_local_or_remote(get_patterns_locally(data_dir), data_dir)
+ data_files_with_zip_archives = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
assert len(data_files_with_zip_archives) == 1
assert len(data_files_with_zip_archives["train"]) == 1
@@ -305,7 +305,7 @@ def test_data_files_with_wrong_metadata_file_name(cache_dir, tmp_path, image_fil
with open(image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
- data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_patterns_locally(data_dir), data_dir)
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
imagefolder = ImageFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
imagefolder.download_and_prepare()
dataset = imagefolder.as_dataset(split="train")
@@ -327,7 +327,7 @@ def test_data_files_with_wrong_image_file_name_column_in_metadata_file(cache_dir
with open(image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
- data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_patterns_locally(data_dir), data_dir)
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
imagefolder = ImageFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
with pytest.raises(ValueError) as exc_info:
imagefolder.download_and_prepare()
diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -12,6 +12,7 @@
DataFilesList,
Url,
_get_data_files_patterns,
+ _get_metadata_files_patterns,
resolve_patterns_in_dataset_repository,
resolve_patterns_locally_or_by_urls,
)
@@ -364,7 +365,7 @@ def test_DataFilesDict_from_hf_local_or_remote_hashing(text_file):
"data_file_per_split",
[
# === Main cases ===
- # file named afetr split at the root
+ # file named after split at the root
{"train": "train.txt", "test": "test.txt", "validation": "valid.txt"},
# file named after split in a directory
{
@@ -392,6 +393,7 @@ def test_DataFilesDict_from_hf_local_or_remote_hashing(text_file):
# Default to train split
{"train": "dataset.txt"},
{"train": "data/dataset.txt"},
+ {"train": ["data/image.jpg", "metadata.jsonl"]},
# With prefix or suffix in directory or file names
{"train": "my_train_dir/dataset.txt"},
{"train": "data/my_train_file.txt"},
@@ -428,3 +430,23 @@ def resolver(pattern):
]
assert len(matched) == len(data_file_per_split[split])
assert matched == data_file_per_split[split]
+
+
[email protected](
+ "metadata_files",
+ [
+ # metadata files at the root
+ ["metadata.jsonl"],
+ # nested metadata files
+ ["data/metadata.jsonl", "data/train/metadata.jsonl"],
+ ],
+)
+def test_get_metadata_files_patterns(metadata_files):
+ def resolver(pattern):
+ return [PurePath(path) for path in set(metadata_files) if PurePath(path).match(pattern)]
+
+ patterns = _get_metadata_files_patterns(resolver)
+ matched = [path for path in metadata_files for pattern in patterns if PurePath(path).match(pattern)]
+ # Use set to remove the difference between in behavior between PurePath.match and mathcing via fsspec.glob
+ assert len(set(matched)) == len(metadata_files)
+ assert sorted(set(matched)) == sorted(metadata_files)
diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -76,6 +76,7 @@ def _generate_examples(self, filepath, **kwargs):
SAMPLE_DATASET_IDENTIFIER = "lhoestq/test" # has dataset script
SAMPLE_DATASET_IDENTIFIER2 = "lhoestq/test2" # only has data files
SAMPLE_DATASET_IDENTIFIER3 = "mariosasko/test_multi_dir_dataset" # has multiple data directories
+SAMPLE_DATASET_IDENTIFIER4 = "mariosasko/test_imagefolder_with_metadata" # imagefolder with a metadata file outside of the train/test directories
SAMPLE_NOT_EXISTING_DATASET_IDENTIFIER = "lhoestq/_dummy"
SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST = "_dummy"
@@ -108,6 +109,24 @@ def data_dir(tmp_path):
return str(data_dir)
[email protected]
+def data_dir_with_metadata(tmp_path):
+ data_dir = tmp_path / "data_dir_with_metadata"
+ data_dir.mkdir()
+ with open(data_dir / "train.jpg", "wb") as f:
+ f.write(b"train_image_bytes")
+ with open(data_dir / "test.jpg", "wb") as f:
+ f.write(b"test_image_bytes")
+ with open(data_dir / "metadata.jsonl", "w") as f:
+ f.write(
+ """\
+ {"file_name": "train.jpg", "caption": "Cool tran image"}
+ {"file_name": "test.jpg", "caption": "Cool test image"}
+ """
+ )
+ return str(data_dir)
+
+
@pytest.fixture
def sub_data_dirs(tmp_path):
data_dir2 = tmp_path / "data_dir2"
@@ -211,10 +230,17 @@ def test_infer_module_for_data_files_in_archives(data_file, expected_module, zip
class ModuleFactoryTest(TestCase):
@pytest.fixture(autouse=True)
def inject_fixtures(
- self, jsonl_path, data_dir, sub_data_dirs, dataset_loading_script_dir, metric_loading_script_dir
+ self,
+ jsonl_path,
+ data_dir,
+ data_dir_with_metadata,
+ sub_data_dirs,
+ dataset_loading_script_dir,
+ metric_loading_script_dir,
):
self._jsonl_path = jsonl_path
self._data_dir = data_dir
+ self._data_dir_with_metadata = data_dir_with_metadata
self._data_dir2 = sub_data_dirs[0]
self._sub_data_dir = sub_data_dirs[1]
self._dataset_loading_script_dir = dataset_loading_script_dir
@@ -292,6 +318,24 @@ def test_LocalDatasetModuleFactoryWithoutScript_with_data_dir(self):
+ module_factory_result.builder_kwargs["data_files"]["test"]
)
+ def test_LocalDatasetModuleFactoryWithoutScript_with_metadata(self):
+ factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir_with_metadata)
+ module_factory_result = factory.get_module()
+ assert importlib.import_module(module_factory_result.module_path) is not None
+ assert (
+ module_factory_result.builder_kwargs["data_files"] is not None
+ and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
+ and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
+ )
+ assert any(
+ data_file.name == "metadata.jsonl"
+ for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
+ )
+ assert any(
+ data_file.name == "metadata.jsonl"
+ for data_file in module_factory_result.builder_kwargs["data_files"]["test"]
+ )
+
def test_PackagedDatasetModuleFactory(self):
factory = PackagedDatasetModuleFactory(
"json", data_files=self._jsonl_path, download_config=self.download_config
@@ -311,6 +355,32 @@ def test_PackagedDatasetModuleFactory_with_data_dir(self):
assert Path(module_factory_result.builder_kwargs["data_files"]["train"][0]).parent.samefile(self._data_dir)
assert Path(module_factory_result.builder_kwargs["data_files"]["test"][0]).parent.samefile(self._data_dir)
+ def test_PackagedDatasetModuleFactory_with_data_dir_and_metadata(self):
+ factory = PackagedDatasetModuleFactory(
+ "imagefolder", data_dir=self._data_dir_with_metadata, download_config=self.download_config
+ )
+ module_factory_result = factory.get_module()
+ assert importlib.import_module(module_factory_result.module_path) is not None
+ assert (
+ module_factory_result.builder_kwargs["data_files"] is not None
+ and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
+ and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
+ )
+ assert Path(module_factory_result.builder_kwargs["data_files"]["train"][0]).parent.samefile(
+ self._data_dir_with_metadata
+ )
+ assert Path(module_factory_result.builder_kwargs["data_files"]["test"][0]).parent.samefile(
+ self._data_dir_with_metadata
+ )
+ assert any(
+ data_file.name == "metadata.jsonl"
+ for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
+ )
+ assert any(
+ data_file.name == "metadata.jsonl"
+ for data_file in module_factory_result.builder_kwargs["data_files"]["test"]
+ )
+
def test_HubDatasetModuleFactoryWithoutScript(self):
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER2, download_config=self.download_config
@@ -338,6 +408,27 @@ def test_HubDatasetModuleFactoryWithoutScript_with_data_dir(self):
+ module_factory_result.builder_kwargs["data_files"]["test"]
)
+ def test_HubDatasetModuleFactoryWithoutScript_with_metadata(self):
+ factory = HubDatasetModuleFactoryWithoutScript(
+ SAMPLE_DATASET_IDENTIFIER4, download_config=self.download_config
+ )
+ module_factory_result = factory.get_module()
+ assert importlib.import_module(module_factory_result.module_path) is not None
+ assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
+ assert (
+ module_factory_result.builder_kwargs["data_files"] is not None
+ and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
+ and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
+ )
+ assert any(
+ Path(data_file).name == "metadata.jsonl"
+ for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
+ )
+ assert any(
+ Path(data_file).name == "metadata.jsonl"
+ for data_file in module_factory_result.builder_kwargs["data_files"]["test"]
+ )
+
def test_HubDatasetModuleFactoryWithScript(self):
factory = HubDatasetModuleFactoryWithScript(
SAMPLE_DATASET_IDENTIFIER,
| Metadata.jsonl for Imagefolder is ignored if it's in a parent directory to the splits directories/do not have "{split}_" prefix
If data contains a single `metadata.jsonl` file for several splits, it won't be included in a dataset's `data_files` and therefore ignored.
This happens when a directory is structured like as follows:
```
train/
file_1.jpg
file_2.jpg
test/
file_3.jpg
file_4.jpg
metadata.jsonl
```
or like as follows:
```
train_file_1.jpg
train_file_2.jpg
test_file_3.jpg
test_file_4.jpg
metadata.jsonl
```
The same for HF repos.
because it's ignored by the patterns [here](https://github.com/huggingface/datasets/blob/master/src/datasets/data_files.py#L29)
@lhoestq @mariosasko Do you think it's better to add this functionality in `data_files.py` or just specifically in imagefolder/audiofolder code? In `data_files.py` would me more general but I don't know if there are any other cases when that might be needed.
| I agree it would be nice to support this. It doesn't fit really well in the current data_files.py, where files of each splits are separated in different folder though, maybe we have to modify a bit the logic here.
One idea would be to extend `get_patterns_in_dataset_repository` and `get_patterns_locally` to additionally check for `metadata.json`, but feel free to comment if you have better ideas (I feel like we're reaching the limits of what the current implementation IMO, so we could think of a different way of resolving the data files if necessary) | 2022-06-27T12:01:29Z | [] | [] |
huggingface/datasets | 4,615 | huggingface__datasets-4615 | [
"4591"
] | e662d75291c86cfcb2e0f8de93305815166bc98d | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -84,8 +84,8 @@
InMemoryTable,
MemoryMappedTable,
Table,
- cast_table_to_features,
concat_tables,
+ embed_table_storage,
list_table_cache_files,
table_cast,
table_visitor,
@@ -95,7 +95,7 @@
from .utils._hf_hub_fixes import create_repo
from .utils.file_utils import _retry, cached_path, estimate_dataset_size, hf_hub_url
from .utils.info_utils import is_small_dataset
-from .utils.py_utils import convert_file_size_to_int, temporary_assignment, unique_values
+from .utils.py_utils import convert_file_size_to_int, unique_values
from .utils.stratify import stratified_shuffle_split_generate_indices
from .utils.tf_utils import minimal_tf_collate_fn
from .utils.typing import PathLike
@@ -4150,26 +4150,17 @@ def extra_nbytes_visitor(array, feature):
if decodable_columns:
def shards_with_embedded_external_files(shards):
- # Temporarily assign the modified version of `cast_storage` before the cast to the decodable
- # feature types to delete path information and embed file content in the arrow file.
- with contextlib.ExitStack() as stack:
- for decodable_feature_type in [Audio, Image]:
- stack.enter_context(
- temporary_assignment(
- decodable_feature_type, "cast_storage", decodable_feature_type.embed_storage
- )
- )
- for shard in shards:
- format = shard.format
- shard = shard.with_format("arrow")
- shard = shard.map(
- partial(cast_table_to_features, features=shard.features),
- batched=True,
- batch_size=1000,
- keep_in_memory=True,
- )
- shard = shard.with_format(**format)
- yield shard
+ for shard in shards:
+ format = shard.format
+ shard = shard.with_format("arrow")
+ shard = shard.map(
+ embed_table_storage,
+ batched=True,
+ batch_size=1000,
+ keep_in_memory=True,
+ )
+ shard = shard.with_format(**format)
+ yield shard
shards = shards_with_embedded_external_files(shards)
@@ -4224,7 +4215,9 @@ def path_in_repo(_index, shard):
for data_file in data_files
if data_file.startswith(f"data/{split}-") and data_file not in shards_path_in_repo
]
- deleted_size = sum(xgetsize(hf_hub_url(repo_id, data_file)) for data_file in data_files_to_delete)
+ deleted_size = sum(
+ xgetsize(hf_hub_url(repo_id, data_file), use_auth_token=token) for data_file in data_files_to_delete
+ )
def delete_file(file):
api.delete_file(file, repo_id=repo_id, token=token, repo_type="dataset", revision=branch)
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -1438,6 +1438,24 @@ def require_storage_cast(feature: FeatureType) -> bool:
return hasattr(feature, "cast_storage")
+def require_storage_embed(feature: FeatureType) -> bool:
+ """Check if a (possibly nested) feature requires embedding data into storage.
+
+ Args:
+ feature (FeatureType): the feature type to be checked
+ Returns:
+ :obj:`bool`
+ """
+ if isinstance(feature, dict):
+ return any(require_storage_cast(f) for f in feature.values())
+ elif isinstance(feature, (list, tuple)):
+ return require_storage_cast(feature[0])
+ elif isinstance(feature, Sequence):
+ return require_storage_cast(feature.feature)
+ else:
+ return hasattr(feature, "embed_storage")
+
+
def keep_features_dicts_synced(func):
"""
Wrapper to keep the secondary dictionary, which tracks whether keys are decodable, of the :class:`datasets.Features` object
diff --git a/src/datasets/table.py b/src/datasets/table.py
--- a/src/datasets/table.py
+++ b/src/datasets/table.py
@@ -1779,7 +1779,7 @@ def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_
= if casting from numbers to strings and allow_number_to_str is False
Returns:
- array (:obj:`pyarrow.Array`): the casted array
+ array (:obj:`pyarrow.Array`): the casted array
"""
from .features.features import Sequence, get_nested_type
@@ -1850,8 +1850,89 @@ def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_
raise TypeError(f"Couldn't cast array of type\n{array.type}\nto\n{feature}")
+@_wrap_for_chunked_arrays
+def embed_array_storage(array: pa.Array, feature: "FeatureType"):
+ """Embed data into an arrays's storage.
+ For custom features like Audio or Image, it takes into account the "embed_storage" methods
+ they defined to enable embedding external data (e.g. an image file) into an other arrow types.
+
+ Args:
+ array (pa.Array): the PyArrow array in which to embed data
+ feature (FeatureType): array features
+
+ Raises:
+ TypeError: if the target type is not supported according, e.g.
+
+ - if a field is missing
+
+ Returns:
+ array (:obj:`pyarrow.Array`): the casted array
+ """
+ from .features import Sequence
+
+ _e = embed_array_storage
+
+ if isinstance(array, pa.ExtensionArray):
+ array = array.storage
+ if hasattr(feature, "embed_storage"):
+ return feature.embed_storage(array)
+ elif pa.types.is_struct(array.type):
+ # feature must be a dict or Sequence(subfeatures_dict)
+ if isinstance(feature, Sequence) and isinstance(feature.feature, dict):
+ feature = {
+ name: Sequence(subfeature, length=feature.length) for name, subfeature in feature.feature.items()
+ }
+ if isinstance(feature, dict):
+ arrays = [_e(array.field(name), subfeature) for name, subfeature in feature.items()]
+ return pa.StructArray.from_arrays(arrays, names=list(feature), mask=array.is_null())
+ elif pa.types.is_list(array.type):
+ # feature must be either [subfeature] or Sequence(subfeature)
+ if isinstance(feature, list):
+ if array.null_count > 0:
+ warnings.warn(
+ f"None values are converted to empty lists when embedding array storage with {feature}. More info: https://github.com/huggingface/datasets/issues/3676. This will raise an error in a future major version of `datasets`"
+ )
+ return pa.ListArray.from_arrays(array.offsets, _e(array.values, feature[0]))
+ elif isinstance(feature, Sequence):
+ if feature.length > -1:
+ if feature.length * len(array) == len(array.values):
+ return pa.FixedSizeListArray.from_arrays(_e(array.values, feature.feature), feature.length)
+ else:
+ casted_values = _e(array.values, feature.feature)
+ if casted_values.type == array.values.type:
+ return array
+ else:
+ if array.null_count > 0:
+ warnings.warn(
+ f"None values are converted to empty lists when embedding array storage with {feature}. More info: https://github.com/huggingface/datasets/issues/3676. This will raise an error in a future major version of `datasets`"
+ )
+ return pa.ListArray.from_arrays(array.offsets, _e(array.values, feature.feature))
+ elif pa.types.is_fixed_size_list(array.type):
+ # feature must be either [subfeature] or Sequence(subfeature)
+ if isinstance(feature, list):
+ if array.null_count > 0:
+ warnings.warn(
+ f"None values are converted to empty lists when embedding array storage with {feature}. More info: https://github.com/huggingface/datasets/issues/3676. This will raise an error in a future major version of `datasets`"
+ )
+ return pa.ListArray.from_arrays(array.offsets, _e(array.values, feature[0]))
+ elif isinstance(feature, Sequence):
+ if feature.length > -1:
+ if feature.length * len(array) == len(array.values):
+ return pa.FixedSizeListArray.from_arrays(_e(array.values, feature.feature), feature.length)
+ else:
+ offsets_arr = pa.array(range(len(array) + 1), pa.int32())
+ if array.null_count > 0:
+ warnings.warn(
+ f"None values are converted to empty lists when embedding array storage with {feature}. More info: https://github.com/huggingface/datasets/issues/3676. This will raise an error in a future major version of `datasets`"
+ )
+ return pa.ListArray.from_arrays(offsets_arr, _e(array.values, feature.feature))
+ if not isinstance(feature, (Sequence, dict, list, tuple)):
+ return array
+ raise TypeError(f"Couldn't embed array of type\n{array.type}\nwith\n{feature}")
+
+
def cast_table_to_features(table: pa.Table, features: "Features"):
- """Cast an table to the arrow schema that corresponds to the requested features.
+ """Cast a table to the arrow schema that corresponds to the requested features.
Args:
table (:obj:`pyarrow.Table`): PyArrow table to cast
@@ -1885,6 +1966,25 @@ def cast_table_to_schema(table: pa.Table, schema: pa.Schema):
return pa.Table.from_arrays(arrays, schema=schema)
+def embed_table_storage(table: pa.Table):
+ """Embed external data into a table's storage.
+
+ Args:
+ table (:obj:`pyarrow.Table`): PyArrow table in which to embed data
+
+ Returns:
+ table (:obj:`pyarrow.Table`): the table with embedded data
+ """
+ from .features.features import Features, require_storage_embed
+
+ features = Features.from_arrow_schema(table.schema)
+ arrays = [
+ embed_array_storage(table[name], feature) if require_storage_embed(feature) else table[name]
+ for name, feature in features.items()
+ ]
+ return pa.Table.from_arrays(arrays, schema=features.arrow_schema)
+
+
def table_cast(table: pa.Table, schema: pa.Schema):
"""Improved version of pa.Table.cast.
| diff --git a/tests/test_table.py b/tests/test_table.py
--- a/tests/test_table.py
+++ b/tests/test_table.py
@@ -7,7 +7,7 @@
import pytest
from datasets import Sequence, Value
-from datasets.features.features import ClassLabel, Features
+from datasets.features.features import ClassLabel, Features, Image
from datasets.table import (
ConcatenationTable,
InMemoryTable,
@@ -20,7 +20,10 @@
_memory_mapped_arrow_table_from_file,
cast_array_to_feature,
concat_tables,
+ embed_array_storage,
+ embed_table_storage,
inject_arrow_table_documentation,
+ table_cast,
)
from .utils import assert_arrow_memory_doesnt_increase, assert_arrow_memory_increases, slow
@@ -1045,3 +1048,29 @@ def test_cast_array_to_features_to_null_type():
arr = pa.array([[None, 1]])
with pytest.raises(TypeError):
cast_array_to_feature(arr, Sequence(Value("null")))
+
+
+def test_embed_array_storage(image_file):
+ array = pa.array([{"bytes": None, "path": image_file}], type=Image.pa_type)
+ embedded_images_array = embed_array_storage(array, Image())
+ assert embedded_images_array.to_pylist()[0]["path"] is None
+ assert isinstance(embedded_images_array.to_pylist()[0]["bytes"], bytes)
+
+
+def test_embed_array_storage_nested(image_file):
+ array = pa.array([[{"bytes": None, "path": image_file}]], type=pa.list_(Image.pa_type))
+ embedded_images_array = embed_array_storage(array, [Image()])
+ assert embedded_images_array.to_pylist()[0][0]["path"] is None
+ assert isinstance(embedded_images_array.to_pylist()[0][0]["bytes"], bytes)
+ array = pa.array([{"foo": {"bytes": None, "path": image_file}}], type=pa.struct({"foo": Image.pa_type}))
+ embedded_images_array = embed_array_storage(array, {"foo": Image()})
+ assert embedded_images_array.to_pylist()[0]["foo"]["path"] is None
+ assert isinstance(embedded_images_array.to_pylist()[0]["foo"]["bytes"], bytes)
+
+
+def test_embed_table_storage(image_file):
+ features = Features({"image": Image()})
+ table = table_cast(pa.table({"image": [image_file]}), features.arrow_schema)
+ embedded_images_table = embed_table_storage(table)
+ assert embedded_images_table.to_pydict()["image"][0]["path"] is None
+ assert isinstance(embedded_images_table.to_pydict()["image"][0]["bytes"], bytes)
diff --git a/tests/test_upstream_hub.py b/tests/test_upstream_hub.py
--- a/tests/test_upstream_hub.py
+++ b/tests/test_upstream_hub.py
@@ -417,6 +417,31 @@ def test_push_dataset_to_hub_custom_features_image(self):
finally:
self.cleanup_repo(ds_name)
+ @require_pil
+ def test_push_dataset_to_hub_custom_features_image_list(self):
+ image_path = os.path.join(os.path.dirname(__file__), "features", "data", "test_image_rgb.jpg")
+ data = {"x": [[image_path], [image_path, image_path]], "y": [0, -1]}
+ features = Features({"x": [Image()], "y": Value("int32")})
+ ds = Dataset.from_dict(data, features=features)
+
+ for embed_external_files in [True, False]:
+ ds_name = f"{USER}/test-{int(time.time() * 10e3)}"
+ try:
+ ds.push_to_hub(ds_name, embed_external_files=embed_external_files, token=self._token)
+ hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
+
+ self.assertListEqual(ds.column_names, hub_ds.column_names)
+ self.assertListEqual(list(ds.features.keys()), list(hub_ds.features.keys()))
+ self.assertDictEqual(ds.features, hub_ds.features)
+ self.assertEqual(ds[:], hub_ds[:])
+ hub_ds = hub_ds.cast_column("x", [Image(decode=False)])
+ elem = hub_ds[0]["x"][0]
+ path, bytes_ = elem["path"], elem["bytes"]
+ self.assertTrue(bool(path) == (not embed_external_files))
+ self.assertTrue(bool(bytes_) == embed_external_files)
+ finally:
+ self.cleanup_repo(ds_name)
+
def test_push_dataset_dict_to_hub_custom_features(self):
features = Features({"x": Value("int64"), "y": ClassLabel(names=["neg", "pos"])})
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [0, 0, 1]}, features=features)
| Can't push Images to hub with manual Dataset
## Describe the bug
If I create a dataset including an 'Image' feature manually, when pushing to hub decoded images are not pushed,
instead it looks for image where image local path is/used to be.
This doesn't (at least didn't used to) happen with imagefolder. I want to build dataset manually because it is complicated.
This happens even though the dataset is looking like decoded images:
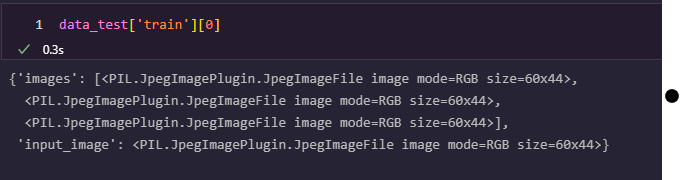
and I use `embed_external_files=True` while `push_to_hub` (same with false)
## Steps to reproduce the bug
```python
from PIL import Image
from datasets import Image as ImageFeature
from datasets import Features,Dataset
#manually create dataset
feats=Features(
{
"images": [ImageFeature()], #same even if explicitly ImageFeature(decode=True)
"input_image": ImageFeature(),
}
)
test_data={"images":[[Image.open("test.jpg"),Image.open("test.jpg"),Image.open("test.jpg")]], "input_image":[Image.open("test.jpg")]}
test_dataset=Dataset.from_dict(test_data,features=feats)
print(test_dataset)
test_dataset.push_to_hub("ceyda/image_test_public",private=False,token="",embed_external_files=True)
# clear cache rm -r ~/.cache/huggingface
# remove "test.jpg" # remove to see that it is looking for image on the local path
test_dataset=load_dataset("ceyda/image_test_public",use_auth_token="")
print(test_dataset)
print(test_dataset['train'][0])
```
## Expected results
should be able to push image bytes if dataset has `Image(decode=True)`
## Actual results
errors because it is trying to decode file from the non existing local path.
```
----> print(test_dataset['train'][0])
File ~/.local/lib/python3.8/site-packages/datasets/arrow_dataset.py:2154, in Dataset.__getitem__(self, key)
2152 def __getitem__(self, key): # noqa: F811
2153 """Can be used to index columns (by string names) or rows (by integer index or iterable of indices or bools)."""
-> 2154 return self._getitem(
2155 key,
2156 )
File ~/.local/lib/python3.8/site-packages/datasets/arrow_dataset.py:2139, in Dataset._getitem(self, key, decoded, **kwargs)
2137 formatter = get_formatter(format_type, features=self.features, decoded=decoded, **format_kwargs)
2138 pa_subtable = query_table(self._data, key, indices=self._indices if self._indices is not None else None)
-> 2139 formatted_output = format_table(
2140 pa_subtable, key, formatter=formatter, format_columns=format_columns, output_all_columns=output_all_columns
2141 )
2142 return formatted_output
File ~/.local/lib/python3.8/site-packages/datasets/formatting/formatting.py:532, in format_table(table, key, formatter, format_columns, output_all_columns)
530 python_formatter = PythonFormatter(features=None)
531 if format_columns is None:
...
-> 3068 fp = builtins.open(filename, "rb")
3069 exclusive_fp = True
3071 try:
FileNotFoundError: [Errno 2] No such file or directory: 'test.jpg'
```
## Environment info
- `datasets` version: 2.3.2
- Platform: Linux-5.4.0-1074-azure-x86_64-with-glibc2.29
- Python version: 3.8.10
- PyArrow version: 8.0.0
- Pandas version: 1.4.2
| Hi, thanks for reporting! This issue stems from the changes introduced in https://github.com/huggingface/datasets/pull/4282 (cc @lhoestq), in which list casts are ignored if they don't change the list type (required to preserve `null` values). And `push_to_hub` does a special cast to embed external image files but doesn't change the types, hence the failure. | 2022-07-01T11:52:08Z | [] | [] |
huggingface/datasets | 4,622 | huggingface__datasets-4622 | [
"4621"
] | e0fa1bfea54e425107b6c90fd1cd961233c07e8f | diff --git a/src/datasets/packaged_modules/imagefolder/imagefolder.py b/src/datasets/packaged_modules/imagefolder/imagefolder.py
--- a/src/datasets/packaged_modules/imagefolder/imagefolder.py
+++ b/src/datasets/packaged_modules/imagefolder/imagefolder.py
@@ -46,8 +46,8 @@ class ImageFolderConfig(datasets.BuilderConfig):
"""BuilderConfig for ImageFolder."""
features: Optional[datasets.Features] = None
- drop_labels: bool = False
- drop_metadata: bool = False
+ drop_labels: bool = None
+ drop_metadata: bool = None
class ImageFolder(datasets.GeneratorBasedBuilder):
@@ -65,53 +65,49 @@ def _split_generators(self, dl_manager):
raise ValueError(f"At least one data file must be specified, but got data_files={self.config.data_files}")
# Do an early pass if:
- # * `features` are not specified, to infer the class labels
- # * `drop_metadata` is False, to find the metadata files
- do_analyze = (self.config.features is None and not self.config.drop_labels) or not self.config.drop_metadata
- if do_analyze:
- labels = set()
- metadata_files = collections.defaultdict(set)
-
- def analyze(files_or_archives, downloaded_files_or_dirs, split):
- if len(downloaded_files_or_dirs) == 0:
- return
- # The files are separated from the archives at this point, so check the first sample
- # to see if it's a file or a directory and iterate accordingly
- if os.path.isfile(downloaded_files_or_dirs[0]):
- original_files, downloaded_files = files_or_archives, downloaded_files_or_dirs
- for original_file, downloaded_file in zip(original_files, downloaded_files):
- original_file, downloaded_file = str(original_file), str(downloaded_file)
- _, original_file_ext = os.path.splitext(original_file)
- if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
+ # * `drop_labels` is None (default) or False, to infer the class labels
+ # * `drop_metadata` is None (default) or False, to find the metadata files
+ do_analyze = not self.config.drop_labels or not self.config.drop_metadata
+ labels = set()
+ metadata_files = collections.defaultdict(set)
+
+ def analyze(files_or_archives, downloaded_files_or_dirs, split):
+ if len(downloaded_files_or_dirs) == 0:
+ return
+ # The files are separated from the archives at this point, so check the first sample
+ # to see if it's a file or a directory and iterate accordingly
+ if os.path.isfile(downloaded_files_or_dirs[0]):
+ original_files, downloaded_files = files_or_archives, downloaded_files_or_dirs
+ for original_file, downloaded_file in zip(original_files, downloaded_files):
+ original_file, downloaded_file = str(original_file), str(downloaded_file)
+ _, original_file_ext = os.path.splitext(original_file)
+ if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
+ if not self.config.drop_labels:
labels.add(os.path.basename(os.path.dirname(original_file)))
- elif os.path.basename(original_file) == self.METADATA_FILENAME:
- metadata_files[split].add((original_file, downloaded_file))
+ elif os.path.basename(original_file) == self.METADATA_FILENAME:
+ metadata_files[split].add((original_file, downloaded_file))
+ else:
+ original_file_name = os.path.basename(original_file)
+ logger.debug(
+ f"The file '{original_file_name}' was ignored: it is not an image, and is not {self.METADATA_FILENAME} either."
+ )
+ else:
+ archives, downloaded_dirs = files_or_archives, downloaded_files_or_dirs
+ for archive, downloaded_dir in zip(archives, downloaded_dirs):
+ archive, downloaded_dir = str(archive), str(downloaded_dir)
+ for downloaded_dir_file in dl_manager.iter_files(downloaded_dir):
+ _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
+ if downloaded_dir_file_ext in self.IMAGE_EXTENSIONS:
+ if not self.config.drop_labels:
+ labels.add(os.path.basename(os.path.dirname(downloaded_dir_file)))
+ elif os.path.basename(downloaded_dir_file) == self.METADATA_FILENAME:
+ metadata_files[split].add((None, downloaded_dir_file))
else:
- original_file_name = os.path.basename(original_file)
+ archive_file_name = os.path.basename(archive)
+ original_file_name = os.path.basename(downloaded_dir_file)
logger.debug(
- f"The file '{original_file_name}' was ignored: it is not an image, and is not {self.METADATA_FILENAME} either."
+ f"The file '{original_file_name}' from the archive '{archive_file_name}' was ignored: it is not an image, and is not {self.METADATA_FILENAME} either."
)
- else:
- archives, downloaded_dirs = files_or_archives, downloaded_files_or_dirs
- for archive, downloaded_dir in zip(archives, downloaded_dirs):
- archive, downloaded_dir = str(archive), str(downloaded_dir)
- for downloaded_dir_file in dl_manager.iter_files(downloaded_dir):
- _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
- if downloaded_dir_file_ext in self.IMAGE_EXTENSIONS:
- labels.add(os.path.basename(os.path.dirname(downloaded_dir_file)))
- elif os.path.basename(downloaded_dir_file) == self.METADATA_FILENAME:
- metadata_files[split].add((None, downloaded_dir_file))
- else:
- archive_file_name = os.path.basename(archive)
- original_file_name = os.path.basename(downloaded_dir_file)
- logger.debug(
- f"The file '{original_file_name}' from the archive '{archive_file_name}' was ignored: it is not an image, and is not {self.METADATA_FILENAME} either."
- )
-
- if not self.config.drop_labels:
- logger.info("Inferring labels from data files...")
- if not self.config.drop_metadata:
- logger.info("Analyzing metadata files...")
data_files = self.config.data_files
splits = []
@@ -121,22 +117,45 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
files, archives = self._split_files_and_archives(files)
downloaded_files = dl_manager.download(files)
downloaded_dirs = dl_manager.download_and_extract(archives)
- if do_analyze:
+ if do_analyze: # drop_metadata is None or False, drop_labels is None or False
+ logger.info(f"Searching for labels and/or metadata files in {split_name} data files...")
analyze(files, downloaded_files, split_name)
analyze(archives, downloaded_dirs, split_name)
+
+ if metadata_files:
+ # add metadata if `metadata_files` are found and `drop_metadata` is None (default) or False
+ add_metadata = not (self.config.drop_metadata is True)
+ # if `metadata_files` are found, add labels only if
+ # `drop_labels` is set up to False explicitly (not-default behavior)
+ add_labels = self.config.drop_labels is False
+ else:
+ # if `metadata_files` are not found, don't add metadata
+ add_metadata = False
+ # if `metadata_files` are not found but `drop_labels` is None (default) or False, add them
+ add_labels = not (self.config.drop_labels is True)
+
+ if add_labels:
+ logger.info("Adding the labels inferred from data directories to the dataset's features...")
+ if add_metadata:
+ logger.info("Adding metadata to the dataset...")
+ else:
+ add_labels, add_metadata, metadata_files = False, False, {}
+
splits.append(
datasets.SplitGenerator(
name=split_name,
gen_kwargs={
"files": [(file, downloaded_file) for file, downloaded_file in zip(files, downloaded_files)]
+ [(None, dl_manager.iter_files(downloaded_dir)) for downloaded_dir in downloaded_dirs],
- "metadata_files": metadata_files if not self.config.drop_metadata else None,
+ "metadata_files": metadata_files,
"split_name": split_name,
+ "add_labels": add_labels,
+ "add_metadata": add_metadata,
},
)
)
- if not self.config.drop_metadata and metadata_files:
+ if add_metadata:
# Verify that:
# * all metadata files have the same set of features
# * the `file_name` key is one of the metadata keys and is of type string
@@ -164,7 +183,7 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
# Normally, we would do this in _info, but we need to know the labels and/or metadata
# before building the features
if self.config.features is None:
- if not self.config.drop_labels and not metadata_files:
+ if add_labels:
self.info.features = datasets.Features(
{"image": datasets.Image(), "label": datasets.ClassLabel(names=sorted(labels))}
)
@@ -174,14 +193,22 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
else:
self.info.features = datasets.Features({"image": datasets.Image()})
- if not self.config.drop_metadata and metadata_files:
- # Verify that there are no duplicated keys when compared to the existing features ("image", optionally "label")
+ if add_metadata:
+ # Warn if there are duplicated keys in metadata compared to the existing features ("image", optionally "label")
duplicated_keys = set(self.info.features) & set(metadata_features)
if duplicated_keys:
- raise ValueError(
- f"Metadata feature keys {list(duplicated_keys)} are already present as the image features"
+ logger.warning(
+ f"Ignoring metadata columns {list(duplicated_keys)} as they are already present in "
+ f"the features dictionary."
)
- self.info.features.update(metadata_features)
+ # skip metadata duplicated keys
+ self.info.features.update(
+ {
+ feature: metadata_features[feature]
+ for feature in metadata_features
+ if feature not in duplicated_keys
+ }
+ )
return splits
@@ -197,21 +224,20 @@ def _split_files_and_archives(self, data_files):
archives.append(data_file)
return files, archives
- def _generate_examples(self, files, metadata_files, split_name):
- if not self.config.drop_metadata and metadata_files:
- split_metadata_files = metadata_files.get(split_name, [])
- image_empty_metadata = {k: None for k in self.info.features if k != "image"}
-
- last_checked_dir = None
- metadata_dir = None
- metadata_dict = None
- downloaded_metadata_file = None
-
- file_idx = 0
- for original_file, downloaded_file_or_dir in files:
- if original_file is not None:
- _, original_file_ext = os.path.splitext(original_file)
- if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
+ def _generate_examples(self, files, metadata_files, split_name, add_metadata, add_labels):
+ split_metadata_files = metadata_files.get(split_name, [])
+ image_empty = {k: None for k in self.info.features if k != "image"} if self.info.features else {}
+ last_checked_dir = None
+ metadata_dir = None
+ metadata_dict = None
+ downloaded_metadata_file = None
+
+ file_idx = 0
+ for original_file, downloaded_file_or_dir in files:
+ if original_file is not None:
+ _, original_file_ext = os.path.splitext(original_file)
+ if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
+ if add_metadata:
# If the file is an image, and we've just entered a new directory,
# find the nereast metadata file (by counting path segments) for the directory
current_dir = os.path.dirname(original_file)
@@ -264,16 +290,19 @@ def _generate_examples(self, files, metadata_files, split_name):
raise ValueError(
f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
)
- yield file_idx, {
- **image_empty_metadata,
- "image": downloaded_file_or_dir,
- **image_metadata,
- }
- file_idx += 1
- else:
- for downloaded_dir_file in downloaded_file_or_dir:
- _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
- if downloaded_dir_file_ext.lower() in self.IMAGE_EXTENSIONS:
+ else:
+ image_metadata = {}
+ if add_labels:
+ image_label = {"label": os.path.basename(os.path.dirname(original_file))}
+ else:
+ image_label = {}
+ yield file_idx, {**image_empty, "image": downloaded_file_or_dir, **image_metadata, **image_label}
+ file_idx += 1
+ else:
+ for downloaded_dir_file in downloaded_file_or_dir:
+ _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
+ if downloaded_dir_file_ext.lower() in self.IMAGE_EXTENSIONS:
+ if add_metadata:
current_dir = os.path.dirname(downloaded_dir_file)
if last_checked_dir is None or last_checked_dir != current_dir:
last_checked_dir = current_dir
@@ -326,42 +355,19 @@ def _generate_examples(self, files, metadata_files, split_name):
raise ValueError(
f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
)
- yield file_idx, {
- **image_empty_metadata,
- "image": downloaded_dir_file,
- **image_metadata,
- }
- file_idx += 1
- else:
- file_idx = 0
- for original_file, downloaded_file_or_dir in files:
- if original_file is not None:
- _, original_file_ext = os.path.splitext(original_file)
- if original_file_ext.lower() in self.IMAGE_EXTENSIONS:
- if self.config.drop_labels or metadata_files:
- yield file_idx, {
- "image": downloaded_file_or_dir,
- }
else:
- yield file_idx, {
- "image": downloaded_file_or_dir,
- "label": os.path.basename(os.path.dirname(original_file)),
- }
+ image_metadata = {}
+ if add_labels:
+ image_label = {"label": os.path.basename(os.path.dirname(downloaded_dir_file))}
+ else:
+ image_label = {}
+ yield file_idx, {
+ **image_empty,
+ "image": downloaded_dir_file,
+ **image_metadata,
+ **image_label,
+ }
file_idx += 1
- else:
- for downloaded_dir_file in downloaded_file_or_dir:
- _, downloaded_dir_file_ext = os.path.splitext(downloaded_dir_file)
- if downloaded_dir_file_ext.lower() in self.IMAGE_EXTENSIONS:
- if self.config.drop_labels or metadata_files:
- yield file_idx, {
- "image": downloaded_dir_file,
- }
- else:
- yield file_idx, {
- "image": downloaded_dir_file,
- "label": os.path.basename(os.path.dirname(downloaded_dir_file)),
- }
- file_idx += 1
# Obtained with:
| diff --git a/tests/packaged_modules/test_imagefolder.py b/tests/packaged_modules/test_imagefolder.py
--- a/tests/packaged_modules/test_imagefolder.py
+++ b/tests/packaged_modules/test_imagefolder.py
@@ -4,8 +4,9 @@
import numpy as np
import pytest
-from datasets import Features, Image, Value
+from datasets import ClassLabel, Features, Image, Value
from datasets.data_files import DataFilesDict, get_data_patterns_locally
+from datasets.download.streaming_download_manager import StreamingDownloadManager
from datasets.packaged_modules.imagefolder.imagefolder import ImageFolder
from datasets.streaming import extend_module_for_streaming
@@ -17,6 +18,55 @@ def cache_dir(tmp_path):
return str(tmp_path / "imagefolder_cache_dir")
[email protected]
+def data_files_with_labels_no_metadata(tmp_path, image_file):
+ data_dir = tmp_path / "data_files_with_labels_no_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir_class_0 = data_dir / "cat"
+ subdir_class_0.mkdir(parents=True, exist_ok=True)
+ # data dirs can be nested but imagefolder should care only about the last part of the path:
+ subdir_class_1 = data_dir / "subdir" / "dog"
+ subdir_class_1.mkdir(parents=True, exist_ok=True)
+
+ image_filename = subdir_class_0 / "image_cat.jpg"
+ shutil.copyfile(image_file, image_filename)
+ image_filename2 = subdir_class_1 / "image_dog.jpg"
+ shutil.copyfile(image_file, image_filename2)
+
+ data_files_with_labels_no_metadata = DataFilesDict.from_local_or_remote(
+ get_data_patterns_locally(str(data_dir)), str(data_dir)
+ )
+
+ return data_files_with_labels_no_metadata
+
+
[email protected]
+def image_files_with_labels_and_duplicated_label_key_in_metadata(tmp_path, image_file):
+ data_dir = tmp_path / "image_files_with_labels_and_label_key_in_metadata"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ subdir_class_0 = data_dir / "cat"
+ subdir_class_0.mkdir(parents=True, exist_ok=True)
+ subdir_class_1 = data_dir / "dog"
+ subdir_class_1.mkdir(parents=True, exist_ok=True)
+
+ image_filename = subdir_class_0 / "image_cat.jpg"
+ shutil.copyfile(image_file, image_filename)
+ image_filename2 = subdir_class_1 / "image_dog.jpg"
+ shutil.copyfile(image_file, image_filename2)
+
+ image_metadata_filename = tmp_path / data_dir / "metadata.jsonl"
+ image_metadata = textwrap.dedent(
+ """\
+ {"file_name": "cat/image_cat.jpg", "caption": "Nice image of a cat", "label": "Cat"}
+ {"file_name": "dog/image_dog.jpg", "caption": "Nice image of a dog", "label": "Dog"}
+ """
+ )
+ with open(image_metadata_filename, "w", encoding="utf-8") as f:
+ f.write(image_metadata)
+
+ return str(image_filename), str(image_filename2), str(image_metadata_filename)
+
+
@pytest.fixture
def image_file_with_metadata(tmp_path, image_file):
image_filename = tmp_path / "image_rgb.jpg"
@@ -162,10 +212,63 @@ def data_files_with_zip_archives(tmp_path, image_file):
@require_pil
[email protected]("drop_labels", [True, False])
-def test_generate_examples_drop_labels(image_file, drop_labels):
- imagefolder = ImageFolder(drop_labels=drop_labels)
- generator = imagefolder._generate_examples([(image_file, image_file)], None, "train")
+# check that labels are inferred correctly from dir names
+def test_generate_examples_with_labels(data_files_with_labels_no_metadata, cache_dir):
+ # there are no metadata.jsonl files in this test case
+ imagefolder = ImageFolder(data_files=data_files_with_labels_no_metadata, cache_dir=cache_dir, drop_labels=False)
+ imagefolder.download_and_prepare()
+ assert imagefolder.info.features == Features({"image": Image(), "label": ClassLabel(names=["cat", "dog"])})
+ dataset = list(imagefolder.as_dataset()["train"])
+ label_feature = imagefolder.info.features["label"]
+
+ assert dataset[0]["label"] == label_feature._str2int["cat"]
+ assert dataset[1]["label"] == label_feature._str2int["dog"]
+
+
+@require_pil
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_generate_examples_duplicated_label_key(
+ image_files_with_labels_and_duplicated_label_key_in_metadata, drop_metadata, drop_labels, cache_dir, caplog
+):
+ cat_image_file, dog_image_file, image_metadata_file = image_files_with_labels_and_duplicated_label_key_in_metadata
+ imagefolder = ImageFolder(
+ drop_metadata=drop_metadata,
+ drop_labels=drop_labels,
+ data_files=[cat_image_file, dog_image_file, image_metadata_file],
+ cache_dir=cache_dir,
+ )
+ if drop_labels is False:
+ # infer labels from directories even if metadata files are found
+ imagefolder.download_and_prepare()
+ warning_in_logs = any("ignoring metadata columns" in record.msg.lower() for record in caplog.records)
+ assert warning_in_logs if drop_metadata is not True else not warning_in_logs
+ dataset = imagefolder.as_dataset()["train"]
+ assert imagefolder.info.features["label"] == ClassLabel(names=["cat", "dog"])
+ assert all(example["label"] in imagefolder.info.features["label"]._str2int.values() for example in dataset)
+ else:
+ imagefolder.download_and_prepare()
+ dataset = imagefolder.as_dataset()["train"]
+ if drop_metadata is not True:
+ # labels are from metadata
+ assert imagefolder.info.features["label"] == Value("string")
+ assert all(example["label"] in ["Cat", "Dog"] for example in dataset)
+ else:
+ # drop both labels and metadata
+ assert imagefolder.info.features == Features({"image": Image()})
+ assert all(example.keys() == {"image"} for example in dataset)
+
+
+@require_pil
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_generate_examples_drop_labels(image_file, drop_metadata, drop_labels):
+ imagefolder = ImageFolder(drop_metadata=drop_metadata, drop_labels=drop_labels, data_files={"train": [image_file]})
+ gen_kwargs = imagefolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ # removing the labels explicitly requires drop_labels=True
+ assert gen_kwargs["add_labels"] is not bool(drop_labels)
+ assert gen_kwargs["add_metadata"] is False
+ generator = imagefolder._generate_examples(**gen_kwargs)
if not drop_labels:
assert all(
example.keys() == {"image", "label"} and all(val is not None for val in example.values())
@@ -179,41 +282,39 @@ def test_generate_examples_drop_labels(image_file, drop_labels):
@require_pil
[email protected]("drop_metadata", [True, False])
-def test_generate_examples_drop_metadata(image_file_with_metadata, drop_metadata):
[email protected]("drop_metadata", [None, True, False])
[email protected]("drop_labels", [None, True, False])
+def test_generate_examples_drop_metadata(image_file_with_metadata, drop_metadata, drop_labels):
image_file, image_metadata_file = image_file_with_metadata
- if not drop_metadata:
- features = Features({"image": Image(), "caption": Value("string")})
- else:
- features = Features({"image": Image()})
- imagefolder = ImageFolder(drop_metadata=drop_metadata, features=features)
- generator = imagefolder._generate_examples(
- [(image_file, image_file)], {"train": [(image_metadata_file, image_metadata_file)]}, "train"
+ imagefolder = ImageFolder(
+ drop_metadata=drop_metadata, drop_labels=drop_labels, data_files={"train": [image_file, image_metadata_file]}
)
- if not drop_metadata:
- assert all(
- example.keys() == {"image", "caption"} and all(val is not None for val in example.values())
- for _, example in generator
- )
- else:
- assert all(
- example.keys() == {"image"} and all(val is not None for val in example.values())
- for _, example in generator
- )
+ gen_kwargs = imagefolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ # since the dataset has metadata, removing the metadata explicitly requires drop_metadata=True
+ assert gen_kwargs["add_metadata"] is not bool(drop_metadata)
+ # since the dataset has metadata, adding the labels explicitly requires drop_labels=False
+ assert gen_kwargs["add_labels"] is (drop_labels is False)
+ generator = imagefolder._generate_examples(**gen_kwargs)
+ expected_columns = {"image"}
+ if gen_kwargs["add_metadata"]:
+ expected_columns.add("caption")
+ if gen_kwargs["add_labels"]:
+ expected_columns.add("label")
+ result = [example for _, example in generator]
+ assert len(result) == 1
+ example = result[0]
+ assert example.keys() == expected_columns
+ for column in expected_columns:
+ assert example[column] is not None
@require_pil
[email protected]("drop_metadata", [True, False])
[email protected]("drop_metadata", [None, True, False])
def test_generate_examples_with_metadata_in_wrong_location(image_file, image_file_with_metadata, drop_metadata):
_, image_metadata_file = image_file_with_metadata
- if not drop_metadata:
- features = Features({"image": Image(), "caption": Value("string")})
- else:
- features = Features({"image": Image()})
- imagefolder = ImageFolder(drop_metadata=drop_metadata, features=features)
- generator = imagefolder._generate_examples(
- [(image_file, image_file)], {"train": [(image_metadata_file, image_metadata_file)]}, "train"
- )
+ imagefolder = ImageFolder(drop_metadata=drop_metadata, data_files={"train": [image_file, image_metadata_file]})
+ gen_kwargs = imagefolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ generator = imagefolder._generate_examples(**gen_kwargs)
if not drop_metadata:
with pytest.raises(ValueError):
list(generator)
@@ -225,7 +326,7 @@ def test_generate_examples_with_metadata_in_wrong_location(image_file, image_fil
@require_pil
[email protected]("drop_metadata", [True, False])
[email protected]("drop_metadata", [None, True, False])
def test_generate_examples_with_metadata_that_misses_one_image(
image_files_with_metadata_that_misses_one_image, drop_metadata
):
@@ -234,12 +335,13 @@ def test_generate_examples_with_metadata_that_misses_one_image(
features = Features({"image": Image(), "caption": Value("string")})
else:
features = Features({"image": Image()})
- imagefolder = ImageFolder(drop_metadata=drop_metadata, features=features)
- generator = imagefolder._generate_examples(
- [(image_file, image_file), (image_file2, image_file2)],
- {"train": [(image_metadata_file, image_metadata_file)]},
- "train",
+ imagefolder = ImageFolder(
+ drop_metadata=drop_metadata,
+ features=features,
+ data_files={"train": [image_file, image_file2, image_metadata_file]},
)
+ gen_kwargs = imagefolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
+ generator = imagefolder._generate_examples(**gen_kwargs)
if not drop_metadata:
with pytest.raises(ValueError):
list(generator)
| ImageFolder raises an error with parameters drop_metadata=True and drop_labels=False when metadata.jsonl is present
## Describe the bug
If you pass `drop_metadata=True` and `drop_labels=False` when a `data_dir` contains at least one `matadata.jsonl` file, you will get a KeyError. This is probably not a very useful case but we shouldn't get an error anyway. Asking users to move metadata files manually outside `data_dir` or pass features manually (when there is a tool that can infer them automatically) don't look like a good idea to me either.
## Steps to reproduce the bug
### Clone an example dataset from the Hub
```bash
git clone https://huggingface.co/datasets/nateraw/test-imagefolder-metadata
```
### Try to load it
```python
from datasets import load_dataset
ds = load_dataset("test-imagefolder-metadata", drop_metadata=True, drop_labels=False)
```
or even just
```python
ds = load_dataset("test-imagefolder-metadata", drop_metadata=True)
```
as `drop_labels=False` is a default value.
## Expected results
A DatasetDict object with two features: `"image"` and `"label"`.
## Actual results
```
Traceback (most recent call last):
File "/home/polina/workspace/datasets/debug.py", line 18, in <module>
ds = load_dataset(
File "/home/polina/workspace/datasets/src/datasets/load.py", line 1732, in load_dataset
builder_instance.download_and_prepare(
File "/home/polina/workspace/datasets/src/datasets/builder.py", line 704, in download_and_prepare
self._download_and_prepare(
File "/home/polina/workspace/datasets/src/datasets/builder.py", line 1227, in _download_and_prepare
super()._download_and_prepare(dl_manager, verify_infos, check_duplicate_keys=verify_infos)
File "/home/polina/workspace/datasets/src/datasets/builder.py", line 793, in _download_and_prepare
self._prepare_split(split_generator, **prepare_split_kwargs)
File "/home/polina/workspace/datasets/src/datasets/builder.py", line 1218, in _prepare_split
example = self.info.features.encode_example(record)
File "/home/polina/workspace/datasets/src/datasets/features/features.py", line 1596, in encode_example
return encode_nested_example(self, example)
File "/home/polina/workspace/datasets/src/datasets/features/features.py", line 1165, in encode_nested_example
{
File "/home/polina/workspace/datasets/src/datasets/features/features.py", line 1165, in <dictcomp>
{
File "/home/polina/workspace/datasets/src/datasets/utils/py_utils.py", line 249, in zip_dict
yield key, tuple(d[key] for d in dicts)
File "/home/polina/workspace/datasets/src/datasets/utils/py_utils.py", line 249, in <genexpr>
yield key, tuple(d[key] for d in dicts)
KeyError: 'label'
```
## Environment info
`datasets` master branch
- `datasets` version: 2.3.3.dev0
- Platform: Linux-5.14.0-1042-oem-x86_64-with-glibc2.17
- Python version: 3.8.12
- PyArrow version: 6.0.1
- Pandas version: 1.4.1
| 2022-07-04T11:23:20Z | [] | [] |
|
huggingface/datasets | 4,628 | huggingface__datasets-4628 | [
"4620"
] | 84fc3ad73c85de4eda5d152dfede7671491449cb | diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -78,9 +78,9 @@ def _arrow_to_datasets_dtype(arrow_type: pa.DataType) -> str:
elif pyarrow.types.is_float64(arrow_type):
return "float64" # pyarrow dtype is "double"
elif pyarrow.types.is_time32(arrow_type):
- return f"time32[{arrow_type.unit}]"
+ return f"time32[{pa.type_for_alias(str(arrow_type)).unit}]"
elif pyarrow.types.is_time64(arrow_type):
- return f"time64[{arrow_type.unit}]"
+ return f"time64[{pa.type_for_alias(str(arrow_type)).unit}]"
elif pyarrow.types.is_timestamp(arrow_type):
if arrow_type.tz is None:
return f"timestamp[{arrow_type.unit}]"
| diff --git a/tests/features/test_features.py b/tests/features/test_features.py
--- a/tests/features/test_features.py
+++ b/tests/features/test_features.py
@@ -1,3 +1,4 @@
+import datetime
from dataclasses import asdict
from unittest import TestCase
from unittest.mock import patch
@@ -58,6 +59,7 @@ def test_string_to_arrow_bijection_for_primitive_types(self):
pa.string(),
pa.int32(),
pa.float64(),
+ pa.array([datetime.time(1, 1, 1)]).type, # arrow type: DataType(time64[us])
]
for dt in supported_pyarrow_datatypes:
self.assertEqual(dt, string_to_arrow(_arrow_to_datasets_dtype(dt)))
| Data type is not recognized when using datetime.time
## Describe the bug
Creating a dataset from a pandas dataframe with `datetime.time` format generates an error.
## Steps to reproduce the bug
```python
import pandas as pd
from datetime import time
from datasets import Dataset
df = pd.DataFrame({"feature_name": [time(1, 1, 1)]})
dataset = Dataset.from_pandas(df)
```
## Expected results
The dataset should be created.
## Actual results
```
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/home/slesage/hf/datasets-server/services/worker/.venv/lib/python3.9/site-packages/datasets/arrow_dataset.py", line 823, in from_pandas
return cls(table, info=info, split=split)
File "/home/slesage/hf/datasets-server/services/worker/.venv/lib/python3.9/site-packages/datasets/arrow_dataset.py", line 679, in __init__
inferred_features = Features.from_arrow_schema(arrow_table.schema)
File "/home/slesage/hf/datasets-server/services/worker/.venv/lib/python3.9/site-packages/datasets/features/features.py", line 1551, in from_arrow_schema
obj = {field.name: generate_from_arrow_type(field.type) for field in pa_schema}
File "/home/slesage/hf/datasets-server/services/worker/.venv/lib/python3.9/site-packages/datasets/features/features.py", line 1551, in <dictcomp>
obj = {field.name: generate_from_arrow_type(field.type) for field in pa_schema}
File "/home/slesage/hf/datasets-server/services/worker/.venv/lib/python3.9/site-packages/datasets/features/features.py", line 1315, in generate_from_arrow_type
return Value(dtype=_arrow_to_datasets_dtype(pa_type))
File "/home/slesage/hf/datasets-server/services/worker/.venv/lib/python3.9/site-packages/datasets/features/features.py", line 83, in _arrow_to_datasets_dtype
return f"time64[{arrow_type.unit}]"
AttributeError: 'pyarrow.lib.DataType' object has no attribute 'unit'
```
## Environment info
- `datasets` version: 2.3.3.dev0
- Platform: Linux-5.13.0-1031-aws-x86_64-with-glibc2.31
- Python version: 3.9.6
- PyArrow version: 7.0.0
- Pandas version: 1.4.2
| cc @mariosasko
Hi, thanks for reporting! I'm investigating the issue. | 2022-07-04T16:20:15Z | [] | [] |
huggingface/datasets | 4,672 | huggingface__datasets-4672 | [
"4670"
] | f3b6697011cb6fc568b8f8b32f53501a8f2e8967 | diff --git a/src/datasets/config.py b/src/datasets/config.py
--- a/src/datasets/config.py
+++ b/src/datasets/config.py
@@ -133,6 +133,7 @@
RARFILE_AVAILABLE = importlib.util.find_spec("rarfile") is not None
ZSTANDARD_AVAILABLE = importlib.util.find_spec("zstandard") is not None
LZ4_AVAILABLE = importlib.util.find_spec("lz4") is not None
+PY7ZR_AVAILABLE = importlib.util.find_spec("py7zr") is not None
# Cache location
diff --git a/src/datasets/utils/extract.py b/src/datasets/utils/extract.py
--- a/src/datasets/utils/extract.py
+++ b/src/datasets/utils/extract.py
@@ -178,9 +178,41 @@ def extract(input_path, output_path):
shutil.copyfileobj(compressed_file, extracted_file)
+class SevenZipExtractor:
+ magic_number = b"\x37\x7A\xBC\xAF\x27\x1C"
+
+ @classmethod
+ def is_extractable(cls, path):
+ with open(path, "rb") as f:
+ try:
+ magic_number = f.read(len(cls.magic_number))
+ except OSError:
+ return False
+ return True if magic_number == cls.magic_number else False
+
+ @staticmethod
+ def extract(input_path: str, output_path: str):
+ if not config.PY7ZR_AVAILABLE:
+ raise OSError("Please pip install py7zr")
+ import py7zr
+
+ os.makedirs(output_path, exist_ok=True)
+ with py7zr.SevenZipFile(input_path, "r") as archive:
+ archive.extractall(output_path)
+
+
class Extractor:
# Put zip file to the last, b/c it is possible wrongly detected as zip (I guess it means: as tar or gzip)
- extractors = [TarExtractor, GzipExtractor, ZipExtractor, XzExtractor, RarExtractor, ZstdExtractor, Bzip2Extractor]
+ extractors = [
+ TarExtractor,
+ GzipExtractor,
+ ZipExtractor,
+ XzExtractor,
+ RarExtractor,
+ ZstdExtractor,
+ Bzip2Extractor,
+ SevenZipExtractor,
+ ]
@classmethod
def is_extractable(cls, path, return_extractor=False):
| diff --git a/tests/conftest.py b/tests/conftest.py
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -149,6 +149,17 @@ def lz4_file(tmp_path_factory):
return path
[email protected](scope="session")
+def seven_zip_file(tmp_path_factory, text_file):
+ if config.PY7ZR_AVAILABLE:
+ import py7zr
+
+ path = tmp_path_factory.mktemp("data") / "file.txt.7z"
+ with py7zr.SevenZipFile(path, "w") as archive:
+ archive.write(text_file, arcname=os.path.basename(text_file))
+ return path
+
+
@pytest.fixture(scope="session")
def xml_file(tmp_path_factory):
filename = tmp_path_factory.mktemp("data") / "file.xml"
diff --git a/tests/test_extract.py b/tests/test_extract.py
--- a/tests/test_extract.py
+++ b/tests/test_extract.py
@@ -1,8 +1,22 @@
import pytest
-from datasets.utils.extract import Extractor, ZstdExtractor
+from datasets.utils.extract import Extractor, SevenZipExtractor, ZstdExtractor
-from .utils import require_zstandard
+from .utils import require_py7zr, require_zstandard
+
+
+@require_py7zr
+def test_seven_zip_extractor(seven_zip_file, tmp_path, text_file):
+ input_path = seven_zip_file
+ assert SevenZipExtractor.is_extractable(input_path)
+ output_path = tmp_path / "extracted"
+ SevenZipExtractor.extract(input_path, output_path)
+ assert output_path.is_dir()
+ for file_path in output_path.iterdir():
+ assert file_path.name == text_file.name
+ extracted_file_content = file_path.read_text(encoding="utf-8")
+ expected_file_content = text_file.read_text(encoding="utf-8")
+ assert extracted_file_content == expected_file_content
@require_zstandard
@@ -19,15 +33,29 @@ def test_zstd_extractor(zstd_file, tmp_path, text_file):
@require_zstandard
[email protected]("compression_format", ["gzip", "xz", "zstd", "bz2"])
-def test_extractor(compression_format, gz_file, xz_file, zstd_file, bz2_file, tmp_path, text_file):
- input_paths = {"gzip": gz_file, "xz": xz_file, "zstd": zstd_file, "bz2": bz2_file}
- input_path = str(input_paths[compression_format])
- output_path = str(tmp_path / "extracted.txt")
[email protected](
+ "compression_format, is_archive", [("gzip", False), ("xz", False), ("zstd", False), ("bz2", False), ("7z", True)]
+)
+def test_extractor(
+ compression_format, is_archive, gz_file, xz_file, zstd_file, bz2_file, seven_zip_file, tmp_path, text_file
+):
+ input_paths = {"gzip": gz_file, "xz": xz_file, "zstd": zstd_file, "bz2": bz2_file, "7z": seven_zip_file}
+ input_path = input_paths[compression_format]
+ if input_path is None:
+ reason = f"for '{compression_format}' compression_format, "
+ if compression_format == "7z":
+ reason += require_py7zr.kwargs["reason"]
+ pytest.skip(reason)
+ input_path = str(input_path)
assert Extractor.is_extractable(input_path)
+ output_path = tmp_path / ("extracted" if is_archive else "extracted.txt")
Extractor.extract(input_path, output_path)
- with open(output_path) as f:
- extracted_file_content = f.read()
- with open(text_file) as f:
- expected_file_content = f.read()
+ if is_archive:
+ assert output_path.is_dir()
+ for file_path in output_path.iterdir():
+ assert file_path.name == text_file.name
+ extracted_file_content = file_path.read_text(encoding="utf-8")
+ else:
+ extracted_file_content = output_path.read_text(encoding="utf-8")
+ expected_file_content = text_file.read_text(encoding="utf-8")
assert extracted_file_content == expected_file_content
diff --git a/tests/utils.py b/tests/utils.py
--- a/tests/utils.py
+++ b/tests/utils.py
@@ -12,6 +12,7 @@
from unittest.mock import patch
import pyarrow as pa
+import pytest
from packaging import version
from datasets import config
@@ -39,6 +40,9 @@ def parse_flag_from_env(key, default=False):
_run_packaged_tests = parse_flag_from_env("RUN_PACKAGED", default=True)
+require_py7zr = pytest.mark.skipif(not config.PY7ZR_AVAILABLE, reason="test requires py7zr")
+
+
def require_beam(test_case):
"""
Decorator marking a test that requires Apache Beam.
| Can't extract files from `.7z` zipfile using `download_and_extract`
## Describe the bug
I'm adding a new dataset which is a `.7z` zip file in Google drive and contains 3 json files inside. I'm able to download the data files using `download_and_extract` but after downloading it throws this error:
```
>>> dataset = load_dataset("./datasets/mantis/")
Using custom data configuration default
Downloading and preparing dataset mantis/default to /Users/bhavitvyamalik/.cache/huggingface/datasets/mantis/default/1.1.0/611affa804ec53e2055a335cc1b8b213bb5a0b5142d919967729d5ee23c6bab4...
Downloading data: 100%|█████████████████████████████████████████████████████████| 77.2M/77.2M [00:23<00:00, 3.28MB/s]
/Users/bhavitvyamalik/.cache/huggingface/datasets/downloads/fc3d70123c9de8407587a59aa426c37819cf2bf016795d33270e8a1d558a34e6
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/Users/bhavitvyamalik/Desktop/work/hf/datasets/src/datasets/load.py", line 1745, in load_dataset
use_auth_token=use_auth_token,
File "/Users/bhavitvyamalik/Desktop/work/hf/datasets/src/datasets/builder.py", line 595, in download_and_prepare
dl_manager=dl_manager, verify_infos=verify_infos, **download_and_prepare_kwargs
File "/Users/bhavitvyamalik/Desktop/work/hf/datasets/src/datasets/builder.py", line 690, in _download_and_prepare
) from None
OSError: Cannot find data file.
Original error:
[Errno 20] Not a directory: '/Users/bhavitvyamalik/.cache/huggingface/datasets/downloads/fc3d70123c9de8407587a59aa426c37819cf2bf016795d33270e8a1d558a34e6/merged_train.json'
```
just before generating the splits. I checked `fc3d70123c9de8407587a59aa426c37819cf2bf016795d33270e8a1d558a34e6` file and it's `7z` zip file (similar to downloaded Google drive file) which means it didn't get unzip. Do I need to unzip it separately and then pass the paths for train,dev,test files in `SplitGenerator`?
## Environment info
- `datasets` version: 1.18.4.dev0
- Platform: Darwin-19.6.0-x86_64-i386-64bit
- Python version: 3.7.8
- PyArrow version: 5.0.0
| Hi @bhavitvyamalik, thanks for reporting.
Yes, currently we do not support 7zip archive compression: I think we should.
As a workaround, you could uncompress it explicitly, like done in e.g. `samsum` dataset:
https://github.com/huggingface/datasets/blob/fedf891a08bfc77041d575fad6c26091bc0fce52/datasets/samsum/samsum.py#L106-L110
Related to this issue: https://github.com/huggingface/datasets/issues/3541 | 2022-07-11T15:56:51Z | [] | [] |
huggingface/datasets | 4,740 | huggingface__datasets-4740 | [
"4636"
] | 2f71b9c9fedfa3a5bb31070199b0a8d45badd82d | diff --git a/src/datasets/download/download_manager.py b/src/datasets/download/download_manager.py
--- a/src/datasets/download/download_manager.py
+++ b/src/datasets/download/download_manager.py
@@ -278,15 +278,16 @@ def url_to_downloaded_path(url):
return downloaded_path_or_paths.data
def download(self, url_or_urls):
- """Download given url(s).
+ """Download given URL(s).
+
+ By default, if there is more than one URL to download, multiprocessing is used with maximum `num_proc = 16`.
+ Pass customized `download_config.num_proc` to change this behavior.
Args:
- url_or_urls: url or `list`/`dict` of urls to download and extract. Each
- url is a `str`.
+ url_or_urls (`str` or `list` or `dict`): URL or list/dict of URLs to download. Each URL is a `str`.
Returns:
- downloaded_path(s): `str`, The downloaded paths matching the given input
- url_or_urls.
+ `str` or `list` or `dict`: The downloaded paths matching the given input `url_or_urls`.
Example:
@@ -297,7 +298,7 @@ def download(self, url_or_urls):
download_config = self.download_config.copy()
download_config.extract_compressed_file = False
# Default to using 16 parallel thread for downloading
- # Note that if we have less than 16 files, multi-processing is not activated
+ # Note that if we have less than or equal to 16 files, multi-processing is not activated
if download_config.num_proc is None:
download_config.num_proc = 16
if download_config.download_desc is None:
@@ -311,6 +312,7 @@ def download(self, url_or_urls):
url_or_urls,
map_tuple=True,
num_proc=download_config.num_proc,
+ parallel_min_length=16,
disable_tqdm=not is_progress_bar_enabled(),
desc="Downloading data files",
)
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -30,7 +30,7 @@
from multiprocessing import Pool, RLock
from shutil import disk_usage
from types import CodeType, FunctionType
-from typing import Dict, List, Optional, Tuple, Union
+from typing import Any, Callable, Dict, List, Optional, Tuple, Union
from urllib.parse import urlparse
import dill
@@ -353,19 +353,53 @@ def _single_map_nested(args):
def map_nested(
- function,
- data_struct,
+ function: Callable[[Any], Any],
+ data_struct: Any,
dict_only: bool = False,
map_list: bool = True,
map_tuple: bool = False,
map_numpy: bool = False,
num_proc: Optional[int] = None,
- types=None,
+ parallel_min_length: int = 2,
+ types: Optional[tuple] = None,
disable_tqdm: bool = True,
desc: Optional[str] = None,
-):
+) -> Any:
"""Apply a function recursively to each element of a nested data struct.
- If num_proc > 1 and the length of data_struct is longer than num_proc: use multi-processing
+
+ Use multiprocessing if num_proc > 1 and the length of data_struct is greater than or equal to
+ `parallel_min_length`.
+
+ <Changed version="2.4.0">
+
+ Before version 2.4.0, multiprocessing was not used if `num_proc` was greater than or equal to ``len(iterable)``.
+
+ Now, if `num_proc` is greater than or equal to ``len(iterable)``, `num_proc` is set to ``len(iterable)`` and
+ multiprocessing is used.
+
+ </Changed>
+
+ Args:
+ function (`Callable`): Function to be applied to `data_struct`.
+ data_struct (`Any`): Data structure to apply `function` to.
+ dict_only (`bool`, default `False`): Whether only apply `function` recursively to `dict` values in
+ `data_struct`.
+ map_list (`bool`, default `True`): Whether also apply `function` recursively to `list` elements (besides `dict`
+ values).
+ map_tuple (`bool`, default `False`): Whether also apply `function` recursively to `tuple` elements (besides
+ `dict` values).
+ map_numpy (`bool, default `False`): Whether also apply `function` recursively to `numpy.array` elements (besides
+ `dict` values).
+ num_proc (`int`, *optional*): Number of processes.
+ parallel_min_length (`int`, default `2`): Minimum length of `data_struct` required for parallel
+ processing.
+ types (`tuple`, *optional*): Additional types (besides `dict` values) to apply `function` recursively to their
+ elements.
+ disable_tqdm (`bool`, default `True`): Whether to disable the tqdm progressbar.
+ desc (`str`, *optional*): Prefix for the tqdm progressbar.
+
+ Returns:
+ `Any`
"""
if types is None:
types = []
@@ -387,12 +421,13 @@ def map_nested(
if num_proc is None:
num_proc = 1
- if num_proc <= 1 or len(iterable) <= num_proc:
+ if num_proc <= 1 or len(iterable) < parallel_min_length:
mapped = [
_single_map_nested((function, obj, types, None, True, None))
for obj in logging.tqdm(iterable, disable=disable_tqdm, desc=desc)
]
else:
+ num_proc = num_proc if num_proc <= len(iterable) else len(iterable)
split_kwds = [] # We organize the splits ourselve (contiguous splits)
for index in range(num_proc):
div = len(iterable) // num_proc
| diff --git a/tests/test_py_utils.py b/tests/test_py_utils.py
--- a/tests/test_py_utils.py
+++ b/tests/test_py_utils.py
@@ -1,5 +1,6 @@
from dataclasses import dataclass
from unittest import TestCase
+from unittest.mock import patch
import numpy as np
import pytest
@@ -98,6 +99,36 @@ class Foo:
self.assertEqual(foo.my_attr, "bar")
[email protected](
+ "iterable_length, num_proc, expected_num_proc",
+ [
+ (1, None, 1),
+ (1, 1, 1),
+ (2, None, 1),
+ (2, 1, 1),
+ (2, 2, 1),
+ (2, 3, 1),
+ (3, 2, 1),
+ (16, 16, 16),
+ (16, 17, 16),
+ (17, 16, 16),
+ ],
+)
+def test_map_nested_num_proc(iterable_length, num_proc, expected_num_proc):
+ with patch("datasets.utils.py_utils._single_map_nested") as mock_single_map_nested, patch(
+ "datasets.utils.py_utils.Pool"
+ ) as mock_multiprocessing_pool:
+ data_struct = {f"{i}": i for i in range(iterable_length)}
+ _ = map_nested(lambda x: x + 10, data_struct, num_proc=num_proc, parallel_min_length=16)
+ if expected_num_proc == 1:
+ assert mock_single_map_nested.called
+ assert not mock_multiprocessing_pool.called
+ else:
+ assert not mock_single_map_nested.called
+ assert mock_multiprocessing_pool.called
+ assert mock_multiprocessing_pool.call_args[0][0] == expected_num_proc
+
+
class TempSeedTest(TestCase):
@require_tf
def test_tensorflow(self):
| Add info in docs about behavior of download_config.num_proc
**Is your feature request related to a problem? Please describe.**
I went to override `download_config.num_proc` and was confused about what was happening under the hood. It would be nice to have the behavior documented a bit better so folks know what's happening when they use it.
**Describe the solution you'd like**
- Add note about how the default number of workers is 16. Related code:
https://github.com/huggingface/datasets/blob/7bcac0a6a0fc367cc068f184fa132b8de8dfa11d/src/datasets/download/download_manager.py#L299-L302
- Add note that if the number of workers is higher than the number of files to download, it won't use multiprocessing.
**Describe alternatives you've considered**
maybe it would also be nice to set `num_proc` = `num_files` when `num_proc` > `num_files`.
**Additional context**
...
| 2022-07-25T08:44:19Z | [] | [] |
|
huggingface/datasets | 4,741 | huggingface__datasets-4741 | [
"4681"
] | fcfcc951a73efbc677f9def9a8707d0af93d5890 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -27,7 +27,6 @@
from collections import Counter, UserDict
from collections.abc import Mapping
from copy import deepcopy
-from dataclasses import asdict
from functools import partial, wraps
from io import BytesIO
from math import ceil, floor
@@ -95,7 +94,7 @@
from .utils._hf_hub_fixes import create_repo
from .utils.file_utils import _retry, cached_path, estimate_dataset_size, hf_hub_url
from .utils.info_utils import is_small_dataset
-from .utils.py_utils import convert_file_size_to_int, unique_values
+from .utils.py_utils import asdict, convert_file_size_to_int, unique_values
from .utils.stratify import stratified_shuffle_split_generate_indices
from .utils.tf_utils import minimal_tf_collate_fn
from .utils.typing import PathLike
diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -16,7 +16,6 @@
import json
import os
import sys
-from dataclasses import asdict
from typing import Any, Dict, Iterable, List, Optional, Tuple, Union
import numpy as np
@@ -39,7 +38,7 @@
from .table import array_cast, cast_array_to_feature, table_cast
from .utils import logging
from .utils.file_utils import hash_url_to_filename
-from .utils.py_utils import first_non_null_value
+from .utils.py_utils import asdict, first_non_null_value
logger = logging.get_logger(__name__)
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -19,7 +19,7 @@
import re
import sys
from collections.abc import Iterable, Mapping
-from dataclasses import InitVar, _asdict_inner, dataclass, field, fields
+from dataclasses import InitVar, dataclass, field, fields
from functools import reduce, wraps
from operator import mul
from typing import Any, ClassVar, Dict, List, Optional
@@ -37,7 +37,7 @@
from .. import config
from ..table import array_cast
from ..utils import logging
-from ..utils.py_utils import first_non_null_value, zip_dict
+from ..utils.py_utils import asdict, first_non_null_value, zip_dict
from .audio import Audio
from .image import Image, encode_pil_image
from .translation import Translation, TranslationVariableLanguages
@@ -1598,7 +1598,7 @@ def from_dict(cls, dic) -> "Features":
return cls(**obj)
def to_dict(self):
- return _asdict_inner(self, dict)
+ return asdict(self)
def encode_example(self, example):
"""
diff --git a/src/datasets/fingerprint.py b/src/datasets/fingerprint.py
--- a/src/datasets/fingerprint.py
+++ b/src/datasets/fingerprint.py
@@ -5,7 +5,6 @@
import shutil
import tempfile
import weakref
-from dataclasses import asdict
from functools import wraps
from pathlib import Path
from typing import TYPE_CHECKING, Any, Dict, List, Optional, Union
@@ -19,7 +18,7 @@
from .table import ConcatenationTable, InMemoryTable, MemoryMappedTable, Table
from .utils.deprecation_utils import deprecated
from .utils.logging import get_logger
-from .utils.py_utils import dumps
+from .utils.py_utils import asdict, dumps
if TYPE_CHECKING:
diff --git a/src/datasets/info.py b/src/datasets/info.py
--- a/src/datasets/info.py
+++ b/src/datasets/info.py
@@ -32,7 +32,7 @@
import dataclasses
import json
import os
-from dataclasses import asdict, dataclass, field
+from dataclasses import dataclass, field
from typing import Dict, List, Optional, Union
from . import config
@@ -41,7 +41,7 @@
from .tasks import TaskTemplate, task_template_from_dict
from .utils import Version
from .utils.logging import get_logger
-from .utils.py_utils import unique_values
+from .utils.py_utils import asdict, unique_values
logger = get_logger(__name__)
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -18,6 +18,7 @@
"""
import contextlib
+import copy
import functools
import itertools
import os
@@ -26,6 +27,7 @@
import sys
import types
from contextlib import contextmanager
+from dataclasses import fields, is_dataclass
from io import BytesIO as StringIO
from multiprocessing import Pool, RLock
from shutil import disk_usage
@@ -151,6 +153,41 @@ def string_to_dict(string: str, pattern: str) -> Dict[str, str]:
return _dict
+def asdict(obj):
+ """Convert an object to its dictionary representation recursively."""
+
+ # Implementation based on https://docs.python.org/3/library/dataclasses.html#dataclasses.asdict
+
+ def _is_dataclass_instance(obj):
+ # https://docs.python.org/3/library/dataclasses.html#dataclasses.is_dataclass
+ return is_dataclass(obj) and not isinstance(obj, type)
+
+ def _asdict_inner(obj):
+ if _is_dataclass_instance(obj):
+ result = {}
+ for f in fields(obj):
+ value = _asdict_inner(getattr(obj, f.name))
+ result[f.name] = value
+ return result
+ elif isinstance(obj, tuple) and hasattr(obj, "_fields"):
+ # obj is a namedtuple
+ return type(obj)(*[_asdict_inner(v) for v in obj])
+ elif isinstance(obj, (list, tuple)):
+ # Assume we can create an object of this type by passing in a
+ # generator (which is not true for namedtuples, handled
+ # above).
+ return type(obj)(_asdict_inner(v) for v in obj)
+ elif isinstance(obj, dict):
+ return {_asdict_inner(k): _asdict_inner(v) for k, v in obj.items()}
+ else:
+ return copy.deepcopy(obj)
+
+ if not isinstance(obj, dict) and not _is_dataclass_instance(obj):
+ raise TypeError(f"{obj} is not a dict or a dataclass")
+
+ return _asdict_inner(obj)
+
+
@contextlib.contextmanager
def temporary_assignment(obj, attr, value):
"""Temporarily assign obj.attr to value."""
| diff --git a/tests/features/test_features.py b/tests/features/test_features.py
--- a/tests/features/test_features.py
+++ b/tests/features/test_features.py
@@ -1,5 +1,4 @@
import datetime
-from dataclasses import asdict
from unittest import TestCase
from unittest.mock import patch
@@ -20,6 +19,7 @@
)
from datasets.features.translation import Translation, TranslationVariableLanguages
from datasets.info import DatasetInfo
+from datasets.utils.py_utils import asdict
from ..utils import require_jax, require_tf, require_torch
@@ -101,6 +101,13 @@ def test_feature_named_type(self):
reloaded_features = Features.from_dict(asdict(ds_info)["features"])
assert features == reloaded_features
+ def test_class_label_feature_with_no_labels(self):
+ """reference: issue #4681"""
+ features = Features({"label": ClassLabel(names=[])})
+ ds_info = DatasetInfo(features=features)
+ reloaded_features = Features.from_dict(asdict(ds_info)["features"])
+ assert features == reloaded_features
+
def test_reorder_fields_as(self):
features = Features(
{
diff --git a/tests/test_py_utils.py b/tests/test_py_utils.py
--- a/tests/test_py_utils.py
+++ b/tests/test_py_utils.py
@@ -1,9 +1,10 @@
+from dataclasses import dataclass
from unittest import TestCase
import numpy as np
import pytest
-from datasets.utils.py_utils import NestedDataStructure, map_nested, temp_seed, temporary_assignment, zip_dict
+from datasets.utils.py_utils import NestedDataStructure, asdict, map_nested, temp_seed, temporary_assignment, zip_dict
from .utils import require_tf, require_torch
@@ -16,6 +17,12 @@ def add_one(i): # picklable for multiprocessing
return i + 1
+@dataclass
+class A:
+ x: int
+ y: str
+
+
class PyUtilsTest(TestCase):
def test_map_nested(self):
s1 = {}
@@ -175,3 +182,16 @@ def test_nested_data_structure_data(input_data):
def test_flatten(data, expected_output):
output = NestedDataStructure(data).flatten()
assert output == expected_output
+
+
+def test_asdict():
+ input = A(x=1, y="foobar")
+ expected_output = {"x": 1, "y": "foobar"}
+ assert asdict(input) == expected_output
+
+ input = {"a": {"b": A(x=10, y="foo")}, "c": [A(x=20, y="bar")]}
+ expected_output = {"a": {"b": {"x": 10, "y": "foo"}}, "c": [{"x": 20, "y": "bar"}]}
+ assert asdict(input) == expected_output
+
+ with pytest.raises(TypeError):
+ asdict([1, A(x=10, y="foo")])
| IndexError when loading ImageFolder
## Describe the bug
Loading an image dataset with `imagefolder` throws `IndexError: list index out of range` when the given folder contains a non-image file (like a csv).
## Steps to reproduce the bug
Put a csv file in a folder with images and load it:
```python
import datasets
datasets.load_dataset("imagefolder", data_dir=path/to/folder)
```
## Expected results
I would expect a better error message, like `Unsupported file` or even the dataset loader just ignoring every file that is not an image in that case.
## Actual results
Here is the whole traceback:
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 2.3.2
- Platform: Linux-5.11.0-051100-generic-x86_64-with-glibc2.27
- Python version: 3.9.9
- PyArrow version: 8.0.0
- Pandas version: 1.4.3
| Hi, thanks for reporting! If there are no examples in ImageFolder, the `label` column is of type `ClassLabel(names=[])`, which leads to an error in [this line](https://github.com/huggingface/datasets/blob/c15b391942764152f6060b59921b09cacc5f22a6/src/datasets/arrow_writer.py#L387) as `asdict(info)` calls `Features({..., "label": {'num_classes': 0, 'names': [], 'id': None, '_type': 'ClassLabel'}})`, which then calls `require_decoding` [here](https://github.com/huggingface/datasets/blob/c15b391942764152f6060b59921b09cacc5f22a6/src/datasets/features/features.py#L1516) on the dict value it does not expect.
I see two ways to fix this:
* custom `asdict` where `dict_factory` is also applied on the `dict` object itself besides dataclasses (the built-in implementation calls `type(dict_obj)` - this means we also need to fix `Features.to_dict` btw)
* implement `DatasetInfo.to_dict` (though adding `to_dict` to a data class is a bit weird IMO)
@lhoestq Which one of these approaches do you like more?
Small pref for the first option, it feels weird to know that `Features()` can be called with a dictionary of types defined as dictionaries instead of type instances. | 2022-07-25T10:41:27Z | [] | [] |
huggingface/datasets | 4,781 | huggingface__datasets-4781 | [
"4772"
] | 08a7b389cdd6fb49264a72aa8ccfc49a233494b6 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -241,6 +241,7 @@ def _get_output_signature(
`collate_fn`.
batch_size (:obj:`int`, optional): The size of batches loaded from the dataset. Used for shape inference.
Can be None, which indicates that batch sizes can be variable.
+ num_test_batches (:obj:`int`): The number of batches to load from the dataset for shape inference.
Returns:
:obj:`dict`: Dict mapping column names to tf.Tensorspec objects
@@ -257,16 +258,15 @@ def _get_output_signature(
batch_size = min(len(dataset), batch_size)
test_batch_size = min(len(dataset), 2)
+ if cols_to_retain is not None:
+ cols_to_retain = list(set(cols_to_retain + ["label_ids", "label", "labels"]))
+
test_batches = []
for _ in range(num_test_batches):
indices = sample(range(len(dataset)), test_batch_size)
test_batch = dataset[indices]
if cols_to_retain is not None:
- test_batch = {
- key: value
- for key, value in test_batch.items()
- if key in cols_to_retain or key in ("label_ids", "label")
- }
+ test_batch = {key: value for key, value in test_batch.items() if key in cols_to_retain}
test_batch = [{key: value[i] for key, value in test_batch.items()} for i in range(test_batch_size)]
test_batch = collate_fn(test_batch, **collate_fn_args)
test_batches.append(test_batch)
@@ -397,19 +397,16 @@ def to_tf_dataset(
raise ValueError("List of columns contains duplicates.")
cols_to_retain = list(set(columns + label_cols))
else:
- cols_to_retain = None # Indicates keeping all non-numerical columns
+ cols_to_retain = None # Indicates keeping all valid columns
columns = []
if self.format["type"] != "custom":
dataset = self.with_format("numpy")
else:
dataset = self
- # If the user hasn't specified columns, give them all columns. This may break some data collators if columns
- # are non-numeric!
- # If drop_remainder is True then all batches will have the same size, so this can be included in the
- # output shape. If drop_remainder is False then batch size can be variable, so that dimension should
- # be listed as None
+ # TODO(Matt, QL): deprecate the retention of label_ids and label
+
output_signature, columns_to_np_types = dataset._get_output_signature(
dataset,
collate_fn=collate_fn,
@@ -418,19 +415,36 @@ def to_tf_dataset(
batch_size=batch_size if drop_remainder else None,
)
+ if "labels" in output_signature:
+ if ("label_ids" in columns or "label" in columns) and "labels" not in columns:
+ columns = [col for col in columns if col not in ["label_ids", "label"]] + ["labels"]
+ if ("label_ids" in label_cols or "label" in label_cols) and "labels" not in label_cols:
+ label_cols = [col for col in label_cols if col not in ["label_ids", "label"]] + ["labels"]
+
+ for col in columns:
+ if col not in output_signature:
+ raise ValueError(f"Column {col} not found in dataset!")
+
+ for col in label_cols:
+ if col not in output_signature:
+ raise ValueError(f"Label column {col} not found in dataset!")
+
def np_get_batch(indices):
- # Following the logic in `transformers.Trainer`, we do not drop `label_ids` or `label` even if they
- # are not in the list of requested columns, because the collator may rename them
- # This might work better if moved to a method attached to our transformers Model objects, but doing so
- # could break backward compatibility
- # TODO(Matt, QL): deprecate the retention of label_ids and label
- batch = dataset[indices]
+ # Optimization - if we're loading a sequential batch, do it with slicing instead of a list of indices
+ if np.all(np.diff(indices) == 1):
+ batch = dataset[indices[0] : indices[-1] + 1]
+ else:
+ batch = dataset[indices]
+
if cols_to_retain is not None:
batch = {
key: value
for key, value in batch.items()
- if key in cols_to_retain or key in ("label_ids", "label")
+ if key in cols_to_retain or key in ("label", "label_ids", "labels")
}
+ elif cols_to_retain is not None:
+ batch = {key: value for key, value in batch.items() if key in cols_to_retain}
+
actual_size = len(list(batch.values())[0]) # Get the length of one of the arrays, assume all same
# Our collators expect a list of dicts, not a dict of lists/arrays, so we invert
batch = [{key: value[i] for key, value in batch.items()} for i in range(actual_size)]
@@ -465,24 +479,22 @@ def ensure_shapes(input_dict):
tf_dataset = tf_dataset.map(ensure_shapes)
- if label_cols:
-
- def split_features_and_labels(input_batch):
- features = {key: tensor for key, tensor in input_batch.items() if key in columns}
- labels = {key: tensor for key, tensor in input_batch.items() if key in label_cols}
- assert set(features.keys()).union(labels.keys()) == set(input_batch.keys())
- if len(features) == 1:
- features = list(features.values())[0]
- if len(labels) == 1:
- labels = list(labels.values())[0]
+ def split_features_and_labels(input_batch):
+ # TODO(Matt, QL): deprecate returning the dict content when there's only one key
+ features = {key: tensor for key, tensor in input_batch.items() if key in columns}
+ labels = {key: tensor for key, tensor in input_batch.items() if key in label_cols}
+ if len(features) == 1:
+ features = list(features.values())[0]
+ if len(labels) == 1:
+ labels = list(labels.values())[0]
+ if isinstance(labels, dict) and len(labels) == 0:
+ return features
+ else:
return features, labels
+ if cols_to_retain is not None:
tf_dataset = tf_dataset.map(split_features_and_labels)
- # TODO(Matt, QL): deprecate returning the dict content when there's only one key
- elif isinstance(tf_dataset.element_spec, dict) and len(tf_dataset.element_spec) == 1:
- tf_dataset = tf_dataset.map(lambda x: list(x.values())[0])
-
if prefetch:
tf_dataset = tf_dataset.prefetch(tf.data.experimental.AUTOTUNE)
diff --git a/src/datasets/utils/tf_utils.py b/src/datasets/utils/tf_utils.py
--- a/src/datasets/utils/tf_utils.py
+++ b/src/datasets/utils/tf_utils.py
@@ -38,6 +38,14 @@ def minimal_tf_collate_fn(features):
return batch
+def minimal_tf_collate_fn_with_renaming(features):
+ batch = minimal_tf_collate_fn(features)
+ if "label" in batch:
+ batch["labels"] = batch["label"]
+ del batch["label"]
+ return batch
+
+
def is_numeric_pa_type(pa_type):
if pa.types.is_list(pa_type):
return is_numeric_pa_type(pa_type.value_type)
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -2388,6 +2388,66 @@ def test_tf_dataset_conversion(self, in_memory):
del transform_dset
del tf_dataset # For correct cleanup
+ @require_tf
+ def test_tf_label_renaming(self, in_memory):
+ # Protect TF-specific imports in here
+ import tensorflow as tf
+
+ from datasets.utils.tf_utils import minimal_tf_collate_fn_with_renaming
+
+ tmp_dir = tempfile.TemporaryDirectory()
+ with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
+ with dset.rename_columns({"col_1": "features", "col_2": "label"}) as new_dset:
+ tf_dataset = new_dset.to_tf_dataset(collate_fn=minimal_tf_collate_fn_with_renaming, batch_size=4)
+ batch = next(iter(tf_dataset))
+ self.assertTrue("labels" in batch and "features" in batch)
+
+ tf_dataset = new_dset.to_tf_dataset(
+ columns=["features", "labels"], collate_fn=minimal_tf_collate_fn_with_renaming, batch_size=4
+ )
+ batch = next(iter(tf_dataset))
+ self.assertTrue("labels" in batch and "features" in batch)
+
+ tf_dataset = new_dset.to_tf_dataset(
+ columns=["features", "label"], collate_fn=minimal_tf_collate_fn_with_renaming, batch_size=4
+ )
+ batch = next(iter(tf_dataset))
+ self.assertTrue("labels" in batch and "features" in batch) # Assert renaming was handled correctly
+
+ tf_dataset = new_dset.to_tf_dataset(
+ columns=["features"],
+ label_cols=["labels"],
+ collate_fn=minimal_tf_collate_fn_with_renaming,
+ batch_size=4,
+ )
+ batch = next(iter(tf_dataset))
+ self.assertEqual(len(batch), 2)
+ # Assert that we don't have any empty entries here
+ self.assertTrue(isinstance(batch[0], tf.Tensor) and isinstance(batch[1], tf.Tensor))
+
+ tf_dataset = new_dset.to_tf_dataset(
+ columns=["features"],
+ label_cols=["label"],
+ collate_fn=minimal_tf_collate_fn_with_renaming,
+ batch_size=4,
+ )
+ batch = next(iter(tf_dataset))
+ self.assertEqual(len(batch), 2)
+ # Assert that we don't have any empty entries here
+ self.assertTrue(isinstance(batch[0], tf.Tensor) and isinstance(batch[1], tf.Tensor))
+
+ tf_dataset = new_dset.to_tf_dataset(
+ columns=["features"],
+ collate_fn=minimal_tf_collate_fn_with_renaming,
+ batch_size=4,
+ )
+ batch = next(iter(tf_dataset))
+ # Assert that labels didn't creep in when we don't ask for them
+ # just because the collate_fn added them
+ self.assertTrue(isinstance(batch, tf.Tensor))
+
+ del tf_dataset # For correct cleanup
+
@require_tf
def test_tf_dataset_options(self, in_memory):
tmp_dir = tempfile.TemporaryDirectory()
| AssertionError when using label_cols in to_tf_dataset
## Describe the bug
An incorrect `AssertionError` is raised when using `label_cols` in `to_tf_dataset` and the label's key name is `label`.
The assertion is in this line:
https://github.com/huggingface/datasets/blob/2.4.0/src/datasets/arrow_dataset.py#L475
## Steps to reproduce the bug
```python
from datasets import load_dataset
from transformers import DefaultDataCollator
dataset = load_dataset('glue', 'mrpc', split='train')
tf_dataset = dataset.to_tf_dataset(
columns=["sentence1", "sentence2", "idx"],
label_cols=["label"],
batch_size=16,
collate_fn=DefaultDataCollator(return_tensors="tf"),
)
```
## Expected results
No assertion error.
## Actual results
```
AssertionError: in user code:
File "/opt/conda/lib/python3.8/site-packages/datasets/arrow_dataset.py", line 475, in split_features_and_labels *
assert set(features.keys()).union(labels.keys()) == set(input_batch.keys())
```
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 2.4.0
- Platform: Linux-4.18.0-305.45.1.el8_4.ppc64le-ppc64le-with-glibc2.17
- Python version: 3.8.13
- PyArrow version: 7.0.0
- Pandas version: 1.4.3
| cc @Rocketknight1
Hi @lehrig, this is caused by the data collator renaming "label" to "labels". If you set `label_cols=["labels"]` in the call it will work correctly. However, I agree that the cause of the bug is not obvious, so I'll see if I can make a PR to clarify things when the collator renames columns.
Thanks - and wow, that appears like a strange side-effect of the data collator. Is that really needed?
Why not make it more explicit? For example, extend `DefaultDataCollator` with an optional property `label_col_name` to be used as label column; only when it is not provided default to `labels` (and document that this happens) for backwards-compatibility?
Haha, I honestly have no idea why our data collators rename `"label"` (the standard label column name in our datasets) to `"labels"` (the standard label column name input to our models). It's been a pain point when I design TF data pipelines, though, because I don't want to hardcode things like that - especially in `datasets`, because the renaming is something that happens purely at the `transformers` end. I don't think I could make the change in the data collators themselves at this point, because it would break backward compatibility for everything in PyTorch as well as TF.
In the most recent version of `transformers` we added a [prepare_tf_dataset](https://huggingface.co/docs/transformers/main_classes/model#transformers.TFPreTrainedModel.prepare_tf_dataset) method to our models which takes care of these details for you, and even chooses appropriate columns and labels for the model you're using. In future we might make that the officially recommended way to convert HF datasets to `tf.data.Dataset`.
Interesting, that'd be great especially for clarity. https://huggingface.co/docs/datasets/use_with_tensorflow#data-loading already improved clarity, yet, all those options will still confuse people. Looking forward to those advances in the hope there'll be only 1 way in the future ;)
Anyways, I am happy for the time being with the work-around you provided. Thank you! | 2022-08-02T16:42:07Z | [] | [] |
huggingface/datasets | 4,837 | huggingface__datasets-4837 | [
"4814"
] | 2e7142a3c6500b560da45e8d5128e320a09fcbd4 | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -79,7 +79,12 @@ class Url(str):
DEFAULT_PATTERNS_SPLIT_IN_DIR_NAME,
DEFAULT_PATTERNS_ALL,
]
-METADATA_PATTERNS = ["metadata.jsonl", "**/metadata.jsonl"] # metadata file for ImageFolder and AudioFolder
+METADATA_PATTERNS = [
+ "metadata.csv",
+ "**/metadata.csv",
+ "metadata.jsonl",
+ "**/metadata.jsonl",
+] # metadata file for ImageFolder and AudioFolder
WILDCARD_CHARACTERS = "*[]"
FILES_TO_IGNORE = ["README.md", "config.json", "dataset_infos.json", "dummy_data.zip", "dataset_dict.json"]
diff --git a/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
--- a/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
+++ b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
@@ -4,6 +4,8 @@
from dataclasses import dataclass
from typing import Any, List, Optional, Tuple
+import pandas as pd
+import pyarrow as pa
import pyarrow.compute as pc
import pyarrow.json as paj
@@ -68,7 +70,7 @@ class FolderBasedBuilder(datasets.GeneratorBasedBuilder):
EXTENSIONS: List[str]
SKIP_CHECKSUM_COMPUTATION_BY_DEFAULT: bool = True
- METADATA_FILENAME: str = "metadata.jsonl"
+ METADATA_FILENAMES: List[str] = ["metadata.csv", "metadata.jsonl"]
def _info(self):
return datasets.DatasetInfo(features=self.config.features)
@@ -97,12 +99,12 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
if original_file_ext.lower() in self.EXTENSIONS:
if not self.config.drop_labels:
labels.add(os.path.basename(os.path.dirname(original_file)))
- elif os.path.basename(original_file) == self.METADATA_FILENAME:
+ elif os.path.basename(original_file) in self.METADATA_FILENAMES:
metadata_files[split].add((original_file, downloaded_file))
else:
original_file_name = os.path.basename(original_file)
logger.debug(
- f"The file '{original_file_name}' was ignored: it is not an {self.BASE_COLUMN_NAME}, and is not {self.METADATA_FILENAME} either."
+ f"The file '{original_file_name}' was ignored: it is not an image, and is not {self.METADATA_FILENAMES} either."
)
else:
archives, downloaded_dirs = files_or_archives, downloaded_files_or_dirs
@@ -113,13 +115,13 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
if downloaded_dir_file_ext in self.EXTENSIONS:
if not self.config.drop_labels:
labels.add(os.path.basename(os.path.dirname(downloaded_dir_file)))
- elif os.path.basename(downloaded_dir_file) == self.METADATA_FILENAME:
+ elif os.path.basename(downloaded_dir_file) in self.METADATA_FILENAMES:
metadata_files[split].add((None, downloaded_dir_file))
else:
archive_file_name = os.path.basename(archive)
original_file_name = os.path.basename(downloaded_dir_file)
logger.debug(
- f"The file '{original_file_name}' from the archive '{archive_file_name}' was ignored: it is not an {self.BASE_COLUMN_NAME}, and is not {self.METADATA_FILENAME} either."
+ f"The file '{original_file_name}' from the archive '{archive_file_name}' was ignored: it is not an {self.BASE_COLUMN_NAME}, and is not {self.METADATA_FILENAMES} either."
)
data_files = self.config.data_files
@@ -173,9 +175,18 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
# * all metadata files have the same set of features
# * the `file_name` key is one of the metadata keys and is of type string
features_per_metadata_file: List[Tuple[str, datasets.Features]] = []
+
+ # Check that all metadata files share the same format
+ metadata_ext = set(
+ os.path.splitext(downloaded_metadata_file)[1][1:]
+ for _, downloaded_metadata_file in itertools.chain.from_iterable(metadata_files.values())
+ )
+ if len(metadata_ext) > 1:
+ raise ValueError(f"Found metadata files with different extensions: {list(metadata_ext)}")
+ metadata_ext = metadata_ext.pop()
+
for _, downloaded_metadata_file in itertools.chain.from_iterable(metadata_files.values()):
- with open(downloaded_metadata_file, "rb") as f:
- pa_metadata_table = paj.read_json(f)
+ pa_metadata_table = self._read_metadata(downloaded_metadata_file)
features_per_metadata_file.append(
(downloaded_metadata_file, datasets.Features.from_arrow_schema(pa_metadata_table.schema))
)
@@ -232,12 +243,21 @@ def _split_files_and_archives(self, data_files):
_, data_file_ext = os.path.splitext(data_file)
if data_file_ext.lower() in self.EXTENSIONS:
files.append(data_file)
- elif os.path.basename(data_file) == self.METADATA_FILENAME:
+ elif os.path.basename(data_file) in self.METADATA_FILENAMES:
files.append(data_file)
else:
archives.append(data_file)
return files, archives
+ def _read_metadata(self, metadata_file):
+ metadata_file_ext = os.path.splitext(metadata_file)[1][1:]
+ if metadata_file_ext == "csv":
+ # Use `pd.read_csv` (although slower) instead of `pyarrow.csv.read_csv` for reading CSV files for consistency with the CSV packaged module
+ return pa.Table.from_pandas(pd.read_csv(metadata_file))
+ else:
+ with open(metadata_file, "rb") as f:
+ return paj.read_json(f)
+
def _generate_examples(self, files, metadata_files, split_name, add_metadata, add_labels):
split_metadata_files = metadata_files.get(split_name, [])
sample_empty_metadata = (
@@ -248,6 +268,13 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
metadata_dict = None
downloaded_metadata_file = None
+ if split_metadata_files:
+ metadata_ext = set(
+ os.path.splitext(downloaded_metadata_file)[1][1:]
+ for _, downloaded_metadata_file in split_metadata_files
+ )
+ metadata_ext = metadata_ext.pop()
+
file_idx = 0
for original_file, downloaded_file_or_dir in files:
if original_file is not None:
@@ -276,8 +303,7 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
_, metadata_file, downloaded_metadata_file = min(
metadata_file_candidates, key=lambda x: count_path_segments(x[0])
)
- with open(downloaded_metadata_file, "rb") as f:
- pa_metadata_table = paj.read_json(f)
+ pa_metadata_table = self._read_metadata(downloaded_metadata_file)
pa_file_name_array = pa_metadata_table["file_name"]
pa_file_name_array = pc.replace_substring(
pa_file_name_array, pattern="\\", replacement="/"
@@ -292,7 +318,7 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
}
else:
raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
+ f"One or several metadata.{metadata_ext} were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
)
if metadata_dir is not None and downloaded_metadata_file is not None:
file_relpath = os.path.relpath(original_file, metadata_dir)
@@ -304,7 +330,7 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
sample_metadata = metadata_dict[file_relpath]
else:
raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
+ f"One or several metadata.{metadata_ext} were found, but not in the same directory or in a parent directory of {downloaded_file_or_dir}."
)
else:
sample_metadata = {}
@@ -346,8 +372,7 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
_, metadata_file, downloaded_metadata_file = min(
metadata_file_candidates, key=lambda x: count_path_segments(x[0])
)
- with open(downloaded_metadata_file, "rb") as f:
- pa_metadata_table = paj.read_json(f)
+ pa_metadata_table = self._read_metadata(downloaded_metadata_file)
pa_file_name_array = pa_metadata_table["file_name"]
pa_file_name_array = pc.replace_substring(
pa_file_name_array, pattern="\\", replacement="/"
@@ -362,7 +387,7 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
}
else:
raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
+ f"One or several metadata.{metadata_ext} were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
)
if metadata_dir is not None and downloaded_metadata_file is not None:
downloaded_dir_file_relpath = os.path.relpath(downloaded_dir_file, metadata_dir)
@@ -374,7 +399,7 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
sample_metadata = metadata_dict[downloaded_dir_file_relpath]
else:
raise ValueError(
- f"One or several metadata.jsonl were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
+ f"One or several metadata.{metadata_ext} were found, but not in the same directory or in a parent directory of {downloaded_dir_file}."
)
else:
sample_metadata = {}
| diff --git a/tests/packaged_modules/test_audiofolder.py b/tests/packaged_modules/test_audiofolder.py
--- a/tests/packaged_modules/test_audiofolder.py
+++ b/tests/packaged_modules/test_audiofolder.py
@@ -132,8 +132,8 @@ def data_files_with_one_split_and_metadata(tmp_path, audio_file):
return data_files_with_one_split_and_metadata
[email protected]
-def data_files_with_two_splits_and_metadata(tmp_path, audio_file):
[email protected](params=["jsonl", "csv"])
+def data_files_with_two_splits_and_metadata(request, tmp_path, audio_file):
data_dir = tmp_path / "audiofolder_data_dir_with_metadata"
data_dir.mkdir(parents=True, exist_ok=True)
train_dir = data_dir / "train"
@@ -148,20 +148,39 @@ def data_files_with_two_splits_and_metadata(tmp_path, audio_file):
audio_filename3 = test_dir / "audio_file3.wav" # test audio
shutil.copyfile(audio_file, audio_filename3)
- train_audio_metadata_filename = train_dir / "metadata.jsonl"
- audio_metadata = textwrap.dedent(
- """\
+ train_audio_metadata_filename = train_dir / f"metadata.{request.param}"
+ audio_metadata = (
+ textwrap.dedent(
+ """\
{"file_name": "audio_file.wav", "text": "First train audio transcription"}
{"file_name": "audio_file2.wav", "text": "Second train audio transcription"}
"""
+ )
+ if request.param == "jsonl"
+ else textwrap.dedent(
+ """\
+ file_name,text
+ audio_file.wav,First train audio transcription
+ audio_file2.wav,Second train audio transcription
+ """
+ )
)
with open(train_audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
- test_audio_metadata_filename = test_dir / "metadata.jsonl"
- audio_metadata = textwrap.dedent(
- """\
+ test_audio_metadata_filename = test_dir / f"metadata.{request.param}"
+ audio_metadata = (
+ textwrap.dedent(
+ """\
{"file_name": "audio_file3.wav", "text": "Test audio transcription"}
"""
+ )
+ if request.param == "jsonl"
+ else textwrap.dedent(
+ """\
+ file_name,text
+ audio_file3.wav,Test audio transcription
+ """
+ )
)
with open(test_audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
@@ -357,11 +376,26 @@ def test_generate_examples_with_metadata_that_misses_one_audio(
@require_sndfile
@pytest.mark.parametrize("streaming", [False, True])
[email protected]("n_splits", [1, 2])
-def test_data_files_with_metadata_and_splits(
- streaming, cache_dir, n_splits, data_files_with_one_split_and_metadata, data_files_with_two_splits_and_metadata
-):
- data_files = data_files_with_one_split_and_metadata if n_splits == 1 else data_files_with_two_splits_and_metadata
+def test_data_files_with_metadata_and_single_split(streaming, cache_dir, data_files_with_one_split_and_metadata):
+ data_files = data_files_with_one_split_and_metadata
+ audiofolder = AudioFolder(data_files=data_files, cache_dir=cache_dir)
+ audiofolder.download_and_prepare()
+ datasets = audiofolder.as_streaming_dataset() if streaming else audiofolder.as_dataset()
+ for split, data_files in data_files.items():
+ expected_num_of_audios = len(data_files) - 1 # don't count the metadata file
+ assert split in datasets
+ dataset = list(datasets[split])
+ assert len(dataset) == expected_num_of_audios
+ # make sure each sample has its own audio and metadata
+ assert len(set(example["audio"]["path"] for example in dataset)) == expected_num_of_audios
+ assert len(set(example["text"] for example in dataset)) == expected_num_of_audios
+ assert all(example["text"] is not None for example in dataset)
+
+
+@require_sndfile
[email protected]("streaming", [False, True])
+def test_data_files_with_metadata_and_multiple_splits(streaming, cache_dir, data_files_with_two_splits_and_metadata):
+ data_files = data_files_with_two_splits_and_metadata
audiofolder = AudioFolder(data_files=data_files, cache_dir=cache_dir)
audiofolder.download_and_prepare()
datasets = audiofolder.as_streaming_dataset() if streaming else audiofolder.as_dataset()
@@ -442,3 +476,33 @@ def test_data_files_with_wrong_audio_file_name_column_in_metadata_file(cache_dir
with pytest.raises(ValueError) as exc_info:
audiofolder.download_and_prepare()
assert "`file_name` must be present" in str(exc_info.value)
+
+
+@require_sndfile
+def test_data_files_with_with_metadata_in_different_formats(cache_dir, tmp_path, audio_file):
+ data_dir = tmp_path / "data_dir_with_metadata_in_different_format"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ shutil.copyfile(audio_file, data_dir / "audio_file.wav")
+ audio_metadata_filename_jsonl = data_dir / "metadata.jsonl"
+ audio_metadata_jsonl = textwrap.dedent(
+ """\
+ {"file_name": "audio_file.wav", "text": "Audio transcription"}
+ """
+ )
+ with open(audio_metadata_filename_jsonl, "w", encoding="utf-8") as f:
+ f.write(audio_metadata_jsonl)
+ audio_metadata_filename_csv = data_dir / "metadata.csv"
+ audio_metadata_csv = textwrap.dedent(
+ """\
+ file_name,text
+ audio_file.wav,Audio transcription
+ """
+ )
+ with open(audio_metadata_filename_csv, "w", encoding="utf-8") as f:
+ f.write(audio_metadata_csv)
+
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
+ audiofolder = AudioFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
+ with pytest.raises(ValueError) as exc_info:
+ audiofolder.download_and_prepare()
+ assert "metadata files with different extensions" in str(exc_info.value)
diff --git a/tests/packaged_modules/test_imagefolder.py b/tests/packaged_modules/test_imagefolder.py
--- a/tests/packaged_modules/test_imagefolder.py
+++ b/tests/packaged_modules/test_imagefolder.py
@@ -98,8 +98,8 @@ def image_files_with_metadata_that_misses_one_image(tmp_path, image_file):
return str(image_filename), str(image_filename2), str(image_metadata_filename)
[email protected]
-def data_files_with_one_split_and_metadata(tmp_path, image_file):
[email protected](params=["jsonl", "csv"])
+def data_files_with_one_split_and_metadata(request, tmp_path, image_file):
data_dir = tmp_path / "imagefolder_data_dir_with_metadata_one_split"
data_dir.mkdir(parents=True, exist_ok=True)
subdir = data_dir / "subdir"
@@ -112,13 +112,24 @@ def data_files_with_one_split_and_metadata(tmp_path, image_file):
image_filename3 = subdir / "image_rgb3.jpg" # in subdir
shutil.copyfile(image_file, image_filename3)
- image_metadata_filename = data_dir / "metadata.jsonl"
- image_metadata = textwrap.dedent(
- """\
+ image_metadata_filename = data_dir / f"metadata.{request.param}"
+ image_metadata = (
+ textwrap.dedent(
+ """\
{"file_name": "image_rgb.jpg", "caption": "Nice image"}
{"file_name": "image_rgb2.jpg", "caption": "Nice second image"}
{"file_name": "subdir/image_rgb3.jpg", "caption": "Nice third image"}
"""
+ )
+ if request.param == "jsonl"
+ else textwrap.dedent(
+ """\
+ file_name,caption
+ image_rgb.jpg,Nice image
+ image_rgb2.jpg,Nice second image
+ subdir/image_rgb3.jpg,Nice third image
+ """
+ )
)
with open(image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
@@ -130,8 +141,8 @@ def data_files_with_one_split_and_metadata(tmp_path, image_file):
return data_files_with_one_split_and_metadata
[email protected]
-def data_files_with_two_splits_and_metadata(tmp_path, image_file):
[email protected](params=["jsonl", "csv"])
+def data_files_with_two_splits_and_metadata(request, tmp_path, image_file):
data_dir = tmp_path / "imagefolder_data_dir_with_metadata_two_splits"
data_dir.mkdir(parents=True, exist_ok=True)
train_dir = data_dir / "train"
@@ -146,20 +157,39 @@ def data_files_with_two_splits_and_metadata(tmp_path, image_file):
image_filename3 = test_dir / "image_rgb3.jpg" # test image
shutil.copyfile(image_file, image_filename3)
- train_image_metadata_filename = train_dir / "metadata.jsonl"
- image_metadata = textwrap.dedent(
- """\
+ train_image_metadata_filename = train_dir / f"metadata.{request.param}"
+ image_metadata = (
+ textwrap.dedent(
+ """\
{"file_name": "image_rgb.jpg", "caption": "Nice train image"}
{"file_name": "image_rgb2.jpg", "caption": "Nice second train image"}
"""
+ )
+ if request.param == "jsonl"
+ else textwrap.dedent(
+ """\
+ file_name,caption
+ image_rgb.jpg,Nice train image
+ image_rgb2.jpg,Nice second train image
+ """
+ )
)
with open(train_image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
- test_image_metadata_filename = test_dir / "metadata.jsonl"
- image_metadata = textwrap.dedent(
- """\
+ test_image_metadata_filename = test_dir / f"metadata.{request.param}"
+ image_metadata = (
+ textwrap.dedent(
+ """\
{"file_name": "image_rgb3.jpg", "caption": "Nice test image"}
"""
+ )
+ if request.param == "jsonl"
+ else textwrap.dedent(
+ """\
+ file_name,caption
+ image_rgb3.jpg,Nice test image
+ """
+ )
)
with open(test_image_metadata_filename, "w", encoding="utf-8") as f:
f.write(image_metadata)
@@ -353,11 +383,26 @@ def test_generate_examples_with_metadata_that_misses_one_image(
@require_pil
@pytest.mark.parametrize("streaming", [False, True])
[email protected]("n_splits", [1, 2])
-def test_data_files_with_metadata_and_splits(
- streaming, cache_dir, n_splits, data_files_with_one_split_and_metadata, data_files_with_two_splits_and_metadata
-):
- data_files = data_files_with_one_split_and_metadata if n_splits == 1 else data_files_with_two_splits_and_metadata
+def test_data_files_with_metadata_and_single_split(streaming, cache_dir, data_files_with_one_split_and_metadata):
+ data_files = data_files_with_one_split_and_metadata
+ imagefolder = ImageFolder(data_files=data_files, cache_dir=cache_dir)
+ imagefolder.download_and_prepare()
+ datasets = imagefolder.as_streaming_dataset() if streaming else imagefolder.as_dataset()
+ for split, data_files in data_files.items():
+ expected_num_of_images = len(data_files) - 1 # don't count the metadata file
+ assert split in datasets
+ dataset = list(datasets[split])
+ assert len(dataset) == expected_num_of_images
+ # make sure each sample has its own image and metadata
+ assert len(set(example["image"].filename for example in dataset)) == expected_num_of_images
+ assert len(set(example["caption"] for example in dataset)) == expected_num_of_images
+ assert all(example["caption"] is not None for example in dataset)
+
+
+@require_pil
[email protected]("streaming", [False, True])
+def test_data_files_with_metadata_and_multiple_splits(streaming, cache_dir, data_files_with_two_splits_and_metadata):
+ data_files = data_files_with_two_splits_and_metadata
imagefolder = ImageFolder(data_files=data_files, cache_dir=cache_dir)
imagefolder.download_and_prepare()
datasets = imagefolder.as_streaming_dataset() if streaming else imagefolder.as_dataset()
@@ -431,3 +476,33 @@ def test_data_files_with_wrong_image_file_name_column_in_metadata_file(cache_dir
with pytest.raises(ValueError) as exc_info:
imagefolder.download_and_prepare()
assert "`file_name` must be present" in str(exc_info.value)
+
+
+@require_pil
+def test_data_files_with_with_metadata_in_different_formats(cache_dir, tmp_path, image_file):
+ data_dir = tmp_path / "data_dir_with_metadata_in_different_format"
+ data_dir.mkdir(parents=True, exist_ok=True)
+ shutil.copyfile(image_file, data_dir / "image_rgb.jpg")
+ image_metadata_filename_jsonl = data_dir / "metadata.jsonl"
+ image_metadata_jsonl = textwrap.dedent(
+ """\
+ {"file_name": "image_rgb.jpg", "caption": "Nice image"}
+ """
+ )
+ with open(image_metadata_filename_jsonl, "w", encoding="utf-8") as f:
+ f.write(image_metadata_jsonl)
+ image_metadata_filename_csv = data_dir / "metadata.csv"
+ image_metadata_csv = textwrap.dedent(
+ """\
+ file_name,caption
+ image_rgb.jpg,Nice image
+ """
+ )
+ with open(image_metadata_filename_csv, "w", encoding="utf-8") as f:
+ f.write(image_metadata_csv)
+
+ data_files_with_bad_metadata = DataFilesDict.from_local_or_remote(get_data_patterns_locally(data_dir), data_dir)
+ imagefolder = ImageFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
+ with pytest.raises(ValueError) as exc_info:
+ imagefolder.download_and_prepare()
+ assert "metadata files with different extensions" in str(exc_info.value)
diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -566,6 +566,7 @@ def ls(self, path, detail=True, refresh=True, **kwargs):
{"train": "dataset.txt"},
{"train": "data/dataset.txt"},
{"train": ["data/image.jpg", "metadata.jsonl"]},
+ {"train": ["data/image.jpg", "metadata.csv"]},
# With prefix or suffix in directory or file names
{"train": "my_train_dir/dataset.txt"},
{"train": "data/my_train_file.txt"},
@@ -615,8 +616,10 @@ def resolver(pattern):
[
# metadata files at the root
["metadata.jsonl"],
+ ["metadata.csv"],
# nested metadata files
["data/metadata.jsonl", "data/train/metadata.jsonl"],
+ ["data/metadata.csv", "data/train/metadata.csv"],
],
)
def test_get_metadata_files_patterns(metadata_files):
| Support CSV as metadata file format in AudioFolder/ImageFolder
Requested here: https://discuss.huggingface.co/t/how-to-structure-an-image-dataset-repo-using-the-image-folder-approach/21004. CSV is also used in AutoTrain for specifying metadata in image datasets.
| 2022-08-12T11:19:18Z | [] | [] |
|
huggingface/datasets | 4,844 | huggingface__datasets-4844 | [
"4839"
] | 965a537a151dc1d78719791cc40eaaa599563c08 | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -28,7 +28,7 @@ class Url(str):
TRAIN_KEYWORDS = ["train", "training"]
TEST_KEYWORDS = ["test", "testing", "eval", "evaluation"]
-VALIDATION_KEYWORDS = ["validation", "valid", "dev"]
+VALIDATION_KEYWORDS = ["validation", "valid", "dev", "val"]
NON_WORDS_CHARS = "-._ 0-9"
KEYWORDS_IN_FILENAME_BASE_PATTERNS = ["**[{sep}/]{keyword}[{sep}]*", "{keyword}[{sep}]*"]
KEYWORDS_IN_DIR_NAME_BASE_PATTERNS = ["{keyword}[{sep}/]**", "**[{sep}/]{keyword}[{sep}/]**"]
| diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -581,6 +581,9 @@ def ls(self, path, detail=True, refresh=True, **kwargs):
{"validation": "dev.txt"},
{"validation": "data/dev.txt"},
{"validation": "dev/dataset.txt"},
+ # With valid<>val aliases
+ {"validation": "val.txt"},
+ {"validation": "data/val.txt"},
# With other extensions
{"train": "train.parquet", "test": "test.parquet", "validation": "valid.parquet"},
# With "dev" or "eval" without separators
| ImageFolder dataset builder does not read the validation data set if it is named as "val"
**Is your feature request related to a problem? Please describe.**
Currently, the `'imagefolder'` data set builder in [`load_dataset()`](https://github.com/huggingface/datasets/blob/2.4.0/src/datasets/load.py#L1541] ) only [supports](https://github.com/huggingface/datasets/blob/6c609a322da994de149b2c938f19439bca99408e/src/datasets/data_files.py#L31) the following names as the validation data set directory name: `["validation", "valid", "dev"]`. When the validation directory is named as `'val'`, the Data set will not have a validation split. I expected this to be a trivial task but ended up spending a lot of time before knowing that only the above names are supported.
Here's a minimal example of `val` not being recognized:
```python
import os
import numpy as np
import cv2
from datasets import load_dataset
# creating a dummy data set with the following structure:
# ROOT
# | -- train
# | ---- class_1
# | ---- class_2
# | -- val
# | ---- class_1
# | ---- class_2
ROOT = "data"
for which in ["train", "val"]:
for class_name in ["class_1", "class_2"]:
dir_name = os.path.join(ROOT, which, class_name)
if not os.path.exists(dir_name):
os.makedirs(dir_name)
for i in range(10):
cv2.imwrite(
os.path.join(dir_name, f"{i}.png"),
np.random.random((224, 224))
)
# trying to create a data set
dataset = load_dataset(
"imagefolder",
data_dir=ROOT
)
>> dataset
DatasetDict({
train: Dataset({
features: ['image', 'label'],
num_rows: 20
})
})
# ^ note how the dataset only has a 'train' subset
```
**Describe the solution you'd like**
The suggestion is to include `"val"` to [that list ](https://github.com/huggingface/datasets/blob/6c609a322da994de149b2c938f19439bca99408e/src/datasets/data_files.py#L31) as that's a commonly used phrase to name the validation directory.
Also, In the documentation, explicitly mention that only such directory names are supported as train/val/test directories to avoid confusion.
**Describe alternatives you've considered**
In the documentation, explicitly mention that only such directory names are supported as train/val/test directories without adding `val` to the above list.
**Additional context**
A question asked in the forum: [
Loading an imagenet-style image dataset with train/val directories](https://discuss.huggingface.co/t/loading-an-imagenet-style-image-dataset-with-train-val-directories/21554)
| 2022-08-13T06:49:41Z | [] | [] |
|
huggingface/datasets | 4,926 | huggingface__datasets-4926 | [
"2773"
] | 7a8a79ab1af0b68aff1c7b5c43f273b3f583b5e9 | diff --git a/datasets/conll2000/conll2000.py b/datasets/conll2000/conll2000.py
--- a/datasets/conll2000/conll2000.py
+++ b/datasets/conll2000/conll2000.py
@@ -53,25 +53,9 @@
_TEST_FILE = "test.txt"
-class Conll2000Config(datasets.BuilderConfig):
- """BuilderConfig for Conll2000"""
-
- def __init__(self, **kwargs):
- """BuilderConfig forConll2000.
-
- Args:
- **kwargs: keyword arguments forwarded to super.
- """
- super(Conll2000Config, self).__init__(**kwargs)
-
-
class Conll2000(datasets.GeneratorBasedBuilder):
"""Conll2000 dataset."""
- BUILDER_CONFIGS = [
- Conll2000Config(name="conll2000", version=datasets.Version("1.0.0"), description="Conll2000 dataset"),
- ]
-
def _info(self):
return datasets.DatasetInfo(
description=_DESCRIPTION,
diff --git a/datasets/crime_and_punish/crime_and_punish.py b/datasets/crime_and_punish/crime_and_punish.py
--- a/datasets/crime_and_punish/crime_and_punish.py
+++ b/datasets/crime_and_punish/crime_and_punish.py
@@ -8,36 +8,7 @@
_DATA_URL = "https://raw.githubusercontent.com/patrickvonplaten/datasets/master/crime_and_punishment.txt"
-class CrimeAndPunishConfig(datasets.BuilderConfig):
- """BuilderConfig for Crime and Punish."""
-
- def __init__(self, data_url, **kwargs):
- """BuilderConfig for BlogAuthorship
-
- Args:
- data_url: `string`, url to the dataset (word or raw level)
- **kwargs: keyword arguments forwarded to super.
- """
- super(CrimeAndPunishConfig, self).__init__(
- version=datasets.Version(
- "1.0.0",
- ),
- **kwargs,
- )
- self.data_url = data_url
-
-
class CrimeAndPunish(datasets.GeneratorBasedBuilder):
-
- VERSION = datasets.Version("0.1.0")
- BUILDER_CONFIGS = [
- CrimeAndPunishConfig(
- name="crime-and-punish",
- data_url=_DATA_URL,
- description="word level dataset. No processing is needed other than replacing newlines with <eos> tokens.",
- ),
- ]
-
def _info(self):
return datasets.DatasetInfo(
# This is the description that will appear on the datasets page.
@@ -58,17 +29,14 @@ def _info(self):
def _split_generators(self, dl_manager):
"""Returns SplitGenerators."""
- if self.config.name == "crime-and-punish":
- data = dl_manager.download_and_extract(self.config.data_url)
+ data = dl_manager.download_and_extract(_DATA_URL)
- return [
- datasets.SplitGenerator(
- name=datasets.Split.TRAIN,
- gen_kwargs={"data_file": data, "split": "train"},
- ),
- ]
- else:
- raise ValueError(f"{self.config.name} does not exist")
+ return [
+ datasets.SplitGenerator(
+ name=datasets.Split.TRAIN,
+ gen_kwargs={"data_file": data, "split": "train"},
+ ),
+ ]
def _generate_examples(self, data_file, split):
diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -94,6 +94,8 @@
# Utilities from PyPA to e.g., compare versions
"packaging",
"responses<0.19",
+ # To parse YAML metadata from dataset cards
+ "pyyaml>=5.1",
]
AUDIO_REQUIRE = [
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -88,7 +88,7 @@
from .info import DatasetInfo, DatasetInfosDict
from .naming import _split_re
from .search import IndexableMixin
-from .splits import NamedSplit, Split, SplitInfo
+from .splits import NamedSplit, Split, SplitDict, SplitInfo
from .table import (
InMemoryTable,
MemoryMappedTable,
@@ -105,6 +105,7 @@
from .utils._hf_hub_fixes import list_repo_files as hf_api_list_repo_files
from .utils.file_utils import _retry, cached_path, estimate_dataset_size, hf_hub_url
from .utils.info_utils import is_small_dataset
+from .utils.metadata import DatasetMetadata
from .utils.py_utils import asdict, convert_file_size_to_int, unique_values
from .utils.stratify import stratified_shuffle_split_generate_indices
from .utils.tf_utils import minimal_tf_collate_fn
@@ -4433,13 +4434,25 @@ def push_to_hub(
info_to_dump.download_size = uploaded_size
info_to_dump.dataset_size = dataset_nbytes
info_to_dump.size_in_bytes = uploaded_size + dataset_nbytes
- info_to_dump.splits = {
- split: SplitInfo(split, num_bytes=dataset_nbytes, num_examples=len(self), dataset_name=dataset_name)
- }
- if config.DATASETDICT_INFOS_FILENAME in repo_files:
+ info_to_dump.splits = SplitDict(
+ {split: SplitInfo(split, num_bytes=dataset_nbytes, num_examples=len(self), dataset_name=dataset_name)}
+ )
+ # get the info from the README to update them
+ if "README.md" in repo_files:
+ download_config = DownloadConfig()
+ download_config.download_desc = "Downloading metadata"
+ dataset_readme_path = cached_path(
+ hf_hub_url(repo_id, "README.md"),
+ download_config=download_config,
+ )
+ dataset_metadata = DatasetMetadata.from_readme(Path(dataset_readme_path))
+ dataset_infos: DatasetInfosDict = DatasetInfosDict.from_metadata(dataset_metadata)
+ repo_info = dataset_infos[next(iter(dataset_infos))]
+ # get the deprecated dataset_infos.json to uodate them
+ elif config.DATASETDICT_INFOS_FILENAME in repo_files:
+ dataset_metadata = DatasetMetadata()
download_config = DownloadConfig()
download_config.download_desc = "Downloading metadata"
- download_config.use_auth_token = token if token is not None else HfFolder.get_token()
dataset_infos_path = cached_path(
hf_hub_url(repo_id, config.DATASETDICT_INFOS_FILENAME),
download_config=download_config,
@@ -4447,8 +4460,12 @@ def push_to_hub(
with open(dataset_infos_path, encoding="utf-8") as f:
dataset_infos: DatasetInfosDict = json.load(f)
repo_info = DatasetInfo.from_dict(dataset_infos[next(iter(dataset_infos))])
+ else:
+ dataset_metadata = DatasetMetadata()
+ repo_info = None
+ # update the total info to dump from existing info
+ if repo_info is not None:
logger.warning("Updating downloaded metadata with the new split.")
-
if repo_info.splits and list(repo_info.splits) != [split]:
if self.features != repo_info.features:
raise ValueError(
@@ -4457,23 +4474,40 @@ def push_to_hub(
if split in repo_info.splits:
repo_info.download_size -= deleted_size
- repo_info.dataset_size -= repo_info.splits[split].num_bytes
+ repo_info.dataset_size -= repo_info.splits.get(split, SplitInfo()).num_bytes or 0
repo_info.download_checksums = None
- repo_info.download_size += uploaded_size
- repo_info.dataset_size += dataset_nbytes
+ repo_info.download_size = (repo_info.download_size or 0) + uploaded_size
+ repo_info.dataset_size = (repo_info.dataset_size or 0) + dataset_nbytes
repo_info.size_in_bytes = repo_info.download_size + repo_info.dataset_size
repo_info.splits[split] = SplitInfo(
split, num_bytes=dataset_nbytes, num_examples=len(self), dataset_name=dataset_name
)
info_to_dump = repo_info
- buffer = BytesIO()
- buffer.write(f'{{"{organization}--{dataset_name}": '.encode())
- info_to_dump._dump_info(buffer, pretty_print=True)
- buffer.write(b"}")
+ # push to the deprecated dataset_infos.json
+ if config.DATASETDICT_INFOS_FILENAME in repo_files:
+ buffer = BytesIO()
+ buffer.write(b'{"default": ')
+ info_to_dump._dump_info(buffer, pretty_print=True)
+ buffer.write(b"}")
+ HfApi(endpoint=config.HF_ENDPOINT).upload_file(
+ path_or_fileobj=buffer.getvalue(),
+ path_in_repo=config.DATASETDICT_INFOS_FILENAME,
+ repo_id=repo_id,
+ token=token,
+ repo_type="dataset",
+ revision=branch,
+ )
+ # push to README
+ DatasetInfosDict({"default": info_to_dump}).to_metadata(dataset_metadata)
+ if "README.md" in repo_files:
+ with open(dataset_readme_path, encoding="utf-8") as readme_file:
+ readme_content = readme_file.read()
+ else:
+ readme_content = f'# Dataset Card for "{repo_id.split("/")[-1]}"\n\n[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)'
HfApi(endpoint=config.HF_ENDPOINT).upload_file(
- path_or_fileobj=buffer.getvalue(),
- path_in_repo=config.DATASETDICT_INFOS_FILENAME,
+ path_or_fileobj=dataset_metadata._to_readme(readme_content).encode(),
+ path_in_repo="README.md",
repo_id=repo_id,
token=token,
repo_type="dataset",
diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -358,7 +358,7 @@ def __init__(
# Set download manager
self.dl_manager = None
- # Record infos even if verify_infos=False; used by "datasets-cli test" to generate dataset_infos.json
+ # Record infos even if verify_infos=False; used by "datasets-cli test" to generate file checksums for (deprecated) dataset_infos.json
self._record_infos = False
# Enable streaming (e.g. it patches "open" to work with remote files)
@@ -383,7 +383,7 @@ def manual_download_instructions(self) -> Optional[str]:
return None
@classmethod
- def get_all_exported_dataset_infos(cls) -> dict:
+ def get_all_exported_dataset_infos(cls) -> DatasetInfosDict:
"""Empty dict if doesn't exist
Example:
@@ -395,10 +395,7 @@ def get_all_exported_dataset_infos(cls) -> dict:
{'default': DatasetInfo(description="Movie Review Dataset.\nThis is a dataset of containing 5,331 positive and 5,331 negative processed\nsentences from Rotten Tomatoes movie reviews. This data was first used in Bo\nPang and Lillian Lee, ``Seeing stars: Exploiting class relationships for\nsentiment categorization with respect to rating scales.'', Proceedings of the\nACL, 2005.\n", citation='@InProceedings{Pang+Lee:05a,\n author = {Bo Pang and Lillian Lee},\n title = {Seeing stars: Exploiting class relationships for sentiment\n categorization with respect to rating scales},\n booktitle = {Proceedings of the ACL},\n year = 2005\n}\n', homepage='http://www.cs.cornell.edu/people/pabo/movie-review-data/', license='', features={'text': Value(dtype='string', id=None), 'label': ClassLabel(num_classes=2, names=['neg', 'pos'], id=None)}, post_processed=None, supervised_keys=SupervisedKeysData(input='', output=''), task_templates=[TextClassification(task='text-classification', text_column='text', label_column='label')], builder_name='rotten_tomatoes_movie_review', config_name='default', version=1.0.0, splits={'train': SplitInfo(name='train', num_bytes=1074810, num_examples=8530, dataset_name='rotten_tomatoes_movie_review'), 'validation': SplitInfo(name='validation', num_bytes=134679, num_examples=1066, dataset_name='rotten_tomatoes_movie_review'), 'test': SplitInfo(name='test', num_bytes=135972, num_examples=1066, dataset_name='rotten_tomatoes_movie_review')}, download_checksums={'https://storage.googleapis.com/seldon-datasets/sentence_polarity_v1/rt-polaritydata.tar.gz': {'num_bytes': 487770, 'checksum': 'a05befe52aafda71d458d188a1c54506a998b1308613ba76bbda2e5029409ce9'}}, download_size=487770, post_processing_size=None, dataset_size=1345461, size_in_bytes=1833231)}
```
"""
- dset_infos_file_path = os.path.join(cls.get_imported_module_dir(), config.DATASETDICT_INFOS_FILENAME)
- if os.path.exists(dset_infos_file_path):
- return DatasetInfosDict.from_directory(cls.get_imported_module_dir())
- return {}
+ return DatasetInfosDict.from_directory(cls.get_imported_module_dir())
def get_exported_dataset_info(self) -> DatasetInfo:
"""Empty DatasetInfo if doesn't exist
@@ -468,11 +465,14 @@ def _create_builder_config(self, name=None, custom_features=None, **config_kwarg
config_kwargs,
custom_features=custom_features,
)
- is_custom = config_id not in self.builder_configs
+ is_custom = (config_id not in self.builder_configs) and config_id != "default"
if is_custom:
logger.warning(f"Using custom data configuration {config_id}")
else:
- if builder_config != self.builder_configs[builder_config.name]:
+ if (
+ builder_config.name in self.builder_configs
+ and builder_config != self.builder_configs[builder_config.name]
+ ):
raise ValueError(
"Cannot name a custom BuilderConfig the same as an available "
f"BuilderConfig. Change the name. Available BuilderConfigs: {list(self.builder_configs.keys())}"
diff --git a/src/datasets/commands/run_beam.py b/src/datasets/commands/run_beam.py
--- a/src/datasets/commands/run_beam.py
+++ b/src/datasets/commands/run_beam.py
@@ -20,7 +20,7 @@ def run_beam_command_factory(args, **kwargs):
args.beam_pipeline_options,
args.data_dir,
args.all_configs,
- args.save_infos,
+ args.save_info or args.save_infos,
args.ignore_verifications,
args.force_redownload,
**kwargs,
@@ -52,11 +52,13 @@ def register_subcommand(parser: ArgumentParser):
help="Can be used to specify a manual directory to get the files from",
)
run_beam_parser.add_argument("--all_configs", action="store_true", help="Test all dataset configurations")
- run_beam_parser.add_argument("--save_infos", action="store_true", help="Save the dataset infos file")
+ run_beam_parser.add_argument("--save_info", action="store_true", help="Save the dataset infos file")
run_beam_parser.add_argument(
"--ignore_verifications", action="store_true", help="Run the test without checksums and splits checks"
)
run_beam_parser.add_argument("--force_redownload", action="store_true", help="Force dataset redownload")
+ # aliases
+ run_beam_parser.add_argument("--save_infos", action="store_true", help="alias for save_info")
run_beam_parser.set_defaults(func=run_beam_command_factory)
def __init__(
diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -12,18 +12,23 @@
import numpy as np
from huggingface_hub import HfApi
+from datasets.utils.metadata import DatasetMetadata
+
from . import config
from .arrow_dataset import Dataset
+from .download import DownloadConfig
from .features import Features
from .features.features import FeatureType
from .filesystems import extract_path_from_uri, is_remote_filesystem
-from .info import DatasetInfo
+from .info import DatasetInfo, DatasetInfosDict
from .naming import _split_re
from .splits import NamedSplit, Split, SplitDict, SplitInfo
from .table import Table
from .tasks import TaskTemplate
from .utils import logging
+from .utils._hf_hub_fixes import list_repo_files as hf_api_list_repo_files
from .utils.doc_utils import is_documented_by
+from .utils.file_utils import cached_path, hf_hub_url
from .utils.typing import PathLike
@@ -1343,8 +1348,7 @@ def push_to_hub(
total_uploaded_size = 0
total_dataset_nbytes = 0
info_to_dump: DatasetInfo = next(iter(self.values())).info.copy()
- dataset_name = repo_id.split("/")[-1]
- info_to_dump.splits = SplitDict(dataset_name=dataset_name)
+ info_to_dump.splits = SplitDict()
for split in self.keys():
if not re.match(_split_re, split):
@@ -1364,21 +1368,47 @@ def push_to_hub(
)
total_uploaded_size += uploaded_size
total_dataset_nbytes += dataset_nbytes
- info_to_dump.splits[split] = SplitInfo(
- str(split), num_bytes=dataset_nbytes, num_examples=len(self[split]), dataset_name=dataset_name
- )
- organization, dataset_name = repo_id.split("/")
+ info_to_dump.splits[split] = SplitInfo(str(split), num_bytes=dataset_nbytes, num_examples=len(self[split]))
info_to_dump.download_checksums = None
info_to_dump.download_size = total_uploaded_size
info_to_dump.dataset_size = total_dataset_nbytes
info_to_dump.size_in_bytes = total_uploaded_size + total_dataset_nbytes
- buffer = BytesIO()
- buffer.write(f'{{"{organization}--{dataset_name}": '.encode())
- info_to_dump._dump_info(buffer)
- buffer.write(b"}")
+
+ api = HfApi(endpoint=config.HF_ENDPOINT)
+ repo_files = hf_api_list_repo_files(api, repo_id, repo_type="dataset", revision=branch, token=token)
+
+ # push to the deprecated dataset_infos.json
+ if config.DATASETDICT_INFOS_FILENAME in repo_files:
+ buffer = BytesIO()
+ buffer.write(b'{"default": ')
+ info_to_dump._dump_info(buffer, pretty_print=True)
+ buffer.write(b"}")
+ HfApi(endpoint=config.HF_ENDPOINT).upload_file(
+ path_or_fileobj=buffer.getvalue(),
+ path_in_repo=config.DATASETDICT_INFOS_FILENAME,
+ repo_id=repo_id,
+ token=token,
+ repo_type="dataset",
+ revision=branch,
+ )
+ # push to README
+ if "README.md" in repo_files:
+ download_config = DownloadConfig()
+ download_config.download_desc = "Downloading metadata"
+ dataset_readme_path = cached_path(
+ hf_hub_url(repo_id, "README.md"),
+ download_config=download_config,
+ )
+ dataset_metadata = DatasetMetadata.from_readme(Path(dataset_readme_path))
+ with open(dataset_readme_path, encoding="utf-8") as readme_file:
+ readme_content = readme_file.read()
+ else:
+ dataset_metadata = DatasetMetadata()
+ readme_content = f'# Dataset Card for "{repo_id.split("/")[-1]}"\n\n[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)'
+ DatasetInfosDict({"default": info_to_dump}).to_metadata(dataset_metadata)
HfApi(endpoint=config.HF_ENDPOINT).upload_file(
- path_or_fileobj=buffer.getvalue(),
- path_in_repo=config.DATASETDICT_INFOS_FILENAME,
+ path_or_fileobj=dataset_metadata._to_readme(readme_content).encode(),
+ path_in_repo="README.md",
repo_id=repo_id,
token=token,
repo_type="dataset",
diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -35,6 +35,7 @@
from pandas.api.extensions import ExtensionDtype as PandasExtensionDtype
from .. import config
+from ..naming import camelcase_to_snakecase, snakecase_to_camelcase
from ..table import array_cast
from ..utils import logging
from ..utils.py_utils import asdict, first_non_null_value, zip_dict
@@ -887,7 +888,7 @@ class ClassLabel:
```
"""
- num_classes: int = None
+ num_classes: InitVar[Optional[int]] = None # Pseudo-field: ignored by asdict/fields when converting to/from dict
names: List[str] = None
names_file: InitVar[Optional[str]] = None # Pseudo-field: ignored by asdict/fields when converting to/from dict
id: Optional[str] = None
@@ -898,7 +899,8 @@ class ClassLabel:
_int2str: ClassVar[Dict[int, int]] = None
_type: str = field(default="ClassLabel", init=False, repr=False)
- def __post_init__(self, names_file):
+ def __post_init__(self, num_classes, names_file):
+ self.num_classes = num_classes
self.names_file = names_file
if self.names_file is not None and self.names is not None:
raise ValueError("Please provide either names or names_file but not both.")
@@ -1281,7 +1283,7 @@ def generate_from_dict(obj: Any):
class_type = globals()[obj.pop("_type")]
if class_type == Sequence:
- return Sequence(feature=generate_from_dict(obj["feature"]), length=obj["length"])
+ return Sequence(feature=generate_from_dict(obj["feature"]), length=obj.get("length", -1))
field_names = {f.name for f in fields(class_type)}
return class_type(**{k: v for k, v in obj.items() if k in field_names})
@@ -1600,6 +1602,148 @@ def from_dict(cls, dic) -> "Features":
def to_dict(self):
return asdict(self)
+ def _to_yaml_list(self) -> list:
+ # we compute the YAML list from the dict representation that is used for JSON dump
+ yaml_data = self.to_dict()
+
+ def simplify(feature: dict) -> dict:
+ if not isinstance(feature, dict):
+ raise TypeError(f"Expected a dict but got a {type(feature)}: {feature}")
+
+ #
+ # sequence: -> sequence: int32
+ # dtype: int32 ->
+ #
+ if isinstance(feature.get("sequence"), dict) and list(feature["sequence"]) == ["dtype"]:
+ feature["sequence"] = feature["sequence"]["dtype"]
+
+ #
+ # sequence: -> sequence:
+ # struct: -> - name: foo
+ # - name: foo -> dtype: int32
+ # dtype: int32 ->
+ #
+ if isinstance(feature.get("sequence"), dict) and list(feature["sequence"]) == ["struct"]:
+ feature["sequence"] = feature["sequence"]["struct"]
+
+ #
+ # list: -> list: int32
+ # dtype: int32 ->
+ #
+ if isinstance(feature.get("list"), dict) and list(feature["list"]) == ["dtype"]:
+ feature["list"] = feature["list"]["dtype"]
+
+ #
+ # list: -> list:
+ # struct: -> - name: foo
+ # - name: foo -> dtype: int32
+ # dtype: int32 ->
+ #
+ if isinstance(feature.get("list"), dict) and list(feature["list"]) == ["struct"]:
+ feature["list"] = feature["list"]["struct"]
+
+ #
+ # class_label: -> class_label:
+ # names: -> names:
+ # - negative -> 0: negative
+ # - positive -> 1: positive
+ #
+ if isinstance(feature.get("class_label"), dict) and isinstance(feature["class_label"].get("names"), list):
+ feature["class_label"]["names"] = dict(enumerate(feature["class_label"]["names"]))
+ return feature
+
+ def to_yaml_inner(obj: Union[dict, list]) -> dict:
+ if isinstance(obj, dict):
+ _type = obj.pop("_type", None)
+ if _type == "Sequence":
+ _feature = obj.pop("feature")
+ return simplify({"sequence": to_yaml_inner(_feature), **obj})
+ elif _type == "Value":
+ return obj
+ elif _type and not obj:
+ return {"dtype": camelcase_to_snakecase(_type)}
+ elif _type:
+ return {"dtype": simplify({camelcase_to_snakecase(_type): obj})}
+ else:
+ return {"struct": [{"name": name, **to_yaml_inner(_feature)} for name, _feature in obj.items()]}
+ elif isinstance(obj, list):
+ return simplify({"list": simplify(to_yaml_inner(obj[0]))})
+ else:
+ raise TypeError(f"Expected a dict or a list but got {type(obj)}: {obj}")
+
+ return to_yaml_inner(yaml_data)["struct"]
+
+ @classmethod
+ def _from_yaml_list(cls, yaml_data: list) -> "Features":
+ yaml_data = copy.deepcopy(yaml_data)
+
+ # we convert the list obtained from YAML data into the dict representation that is used for JSON dump
+
+ def unsimplify(feature: dict) -> dict:
+ if not isinstance(feature, dict):
+ raise TypeError(f"Expected a dict but got a {type(feature)}: {feature}")
+ #
+ # sequence: int32 -> sequence:
+ # -> dtype: int32
+ #
+ if isinstance(feature.get("sequence"), str):
+ feature["sequence"] = {"dtype": feature["sequence"]}
+ #
+ # list: int32 -> list:
+ # -> dtype: int32
+ #
+ if isinstance(feature.get("list"), str):
+ feature["list"] = {"dtype": feature["list"]}
+
+ #
+ # class_label: -> class_label:
+ # names: -> names:
+ # 0: negative -> - negative
+ # 1: positive -> - positive
+ #
+ if isinstance(feature.get("class_label"), dict) and isinstance(feature["class_label"].get("names"), dict):
+ label_ids = sorted(feature["class_label"]["names"])
+ if label_ids and label_ids != list(range(label_ids[-1] + 1)):
+ raise ValueError(
+ f"ClassLabel expected a value for all label ids [0:{label_ids[-1] + 1}] but some ids are missing."
+ )
+ feature["class_label"]["names"] = [feature["class_label"]["names"][label_id] for label_id in label_ids]
+ return feature
+
+ def from_yaml_inner(obj: Union[dict, list]) -> Union[dict, list]:
+
+ if isinstance(obj, dict):
+ if not obj:
+ return {}
+ _type = next(iter(obj))
+ if _type == "sequence":
+ _feature = unsimplify(obj).pop(_type)
+ return {"feature": from_yaml_inner(_feature), **obj, "_type": "Sequence"}
+ if _type == "list":
+ return [from_yaml_inner(unsimplify(obj)[_type])]
+ if _type == "struct":
+ return from_yaml_inner(obj["struct"])
+ elif _type == "dtype":
+ if isinstance(obj["dtype"], str):
+ # e.g. int32, float64, string, audio, image
+ try:
+ Value(obj["dtype"])
+ return {**obj, "_type": "Value"}
+ except ValueError:
+ # for audio and image that are Audio and Image types, not Value
+ return {"_type": snakecase_to_camelcase(obj["dtype"])}
+ else:
+ return from_yaml_inner(obj["dtype"])
+ else:
+ return {"_type": snakecase_to_camelcase(_type), **unsimplify(obj)[_type]}
+ elif isinstance(obj, list):
+ names = [_feature.pop("name") for _feature in obj]
+ return {name: from_yaml_inner(_feature) for name, _feature in zip(names, obj)}
+ else:
+ raise TypeError(f"Expected a dict or a list but got {type(obj)}: {obj}")
+
+ return cls.from_dict(from_yaml_inner(yaml_data))
+
def encode_example(self, example):
"""
Encode example into a format for Arrow.
diff --git a/src/datasets/info.py b/src/datasets/info.py
--- a/src/datasets/info.py
+++ b/src/datasets/info.py
@@ -33,8 +33,9 @@
import json
import os
import posixpath
-from dataclasses import dataclass, field
-from typing import Dict, List, Optional, Union
+from dataclasses import dataclass
+from pathlib import Path
+from typing import ClassVar, Dict, List, Optional, Union
from fsspec.implementations.local import LocalFileSystem
@@ -45,6 +46,7 @@
from .tasks import TaskTemplate, task_template_from_dict
from .utils import Version
from .utils.logging import get_logger
+from .utils.metadata import DatasetMetadata
from .utils.py_utils import asdict, unique_values
@@ -118,10 +120,10 @@ class DatasetInfo:
"""
# Set in the dataset scripts
- description: str = field(default_factory=str)
- citation: str = field(default_factory=str)
- homepage: str = field(default_factory=str)
- license: str = field(default_factory=str)
+ description: str = dataclasses.field(default_factory=str)
+ citation: str = dataclasses.field(default_factory=str)
+ homepage: str = dataclasses.field(default_factory=str)
+ license: str = dataclasses.field(default_factory=str)
features: Optional[Features] = None
post_processed: Optional[PostProcessedInfo] = None
supervised_keys: Optional[SupervisedKeysData] = None
@@ -139,6 +141,14 @@ class DatasetInfo:
dataset_size: Optional[int] = None
size_in_bytes: Optional[int] = None
+ _INCLUDED_INFO_IN_YAML: ClassVar[List[str]] = [
+ "config_name",
+ "download_size",
+ "dataset_size",
+ "features",
+ "splits",
+ ]
+
def __post_init__(self):
# Convert back to the correct classes when we reload from dict
if self.features is not None and not isinstance(self.features, Features):
@@ -301,33 +311,112 @@ def update(self, other_dataset_info: "DatasetInfo", ignore_none=True):
def copy(self) -> "DatasetInfo":
return self.__class__(**{k: copy.deepcopy(v) for k, v in self.__dict__.items()})
+ def _to_yaml_dict(self) -> dict:
+ yaml_dict = {}
+ dataset_info_dict = asdict(self)
+ for key in dataset_info_dict:
+ if key in self._INCLUDED_INFO_IN_YAML:
+ value = getattr(self, key)
+ if hasattr(value, "_to_yaml_list"): # Features, SplitDict
+ yaml_dict[key] = value._to_yaml_list()
+ elif hasattr(value, "_to_yaml_string"): # Version
+ yaml_dict[key] = value._to_yaml_string()
+ else:
+ yaml_dict[key] = value
+ return yaml_dict
+
+ @classmethod
+ def _from_yaml_dict(cls, yaml_data: dict) -> "DatasetInfo":
+ yaml_data = copy.deepcopy(yaml_data)
+ if yaml_data.get("features") is not None:
+ yaml_data["features"] = Features._from_yaml_list(yaml_data["features"])
+ if yaml_data.get("splits") is not None:
+ yaml_data["splits"] = SplitDict._from_yaml_list(yaml_data["splits"])
+ field_names = {f.name for f in dataclasses.fields(cls)}
+ return cls(**{k: v for k, v in yaml_data.items() if k in field_names})
+
class DatasetInfosDict(Dict[str, DatasetInfo]):
def write_to_directory(self, dataset_infos_dir, overwrite=False, pretty_print=False):
total_dataset_infos = {}
dataset_infos_path = os.path.join(dataset_infos_dir, config.DATASETDICT_INFOS_FILENAME)
- if os.path.exists(dataset_infos_path) and not overwrite:
- logger.info(f"Dataset Infos already exists in {dataset_infos_dir}. Completing it with new infos.")
+ dataset_readme_path = os.path.join(dataset_infos_dir, "README.md")
+ if not overwrite:
total_dataset_infos = self.from_directory(dataset_infos_dir)
- else:
- logger.info(f"Writing new Dataset Infos in {dataset_infos_dir}")
total_dataset_infos.update(self)
- with open(dataset_infos_path, "w", encoding="utf-8") as f:
- json.dump(
- {config_name: asdict(dset_info) for config_name, dset_info in total_dataset_infos.items()},
- f,
- indent=4 if pretty_print else None,
- )
+ if os.path.exists(dataset_infos_path):
+ # for backward compatibility, let's update the JSON file if it exists
+ with open(dataset_infos_path, "w", encoding="utf-8") as f:
+ dataset_infos_dict = {
+ config_name: asdict(dset_info) for config_name, dset_info in total_dataset_infos.items()
+ }
+ json.dump(dataset_infos_dict, f, indent=4 if pretty_print else None)
+ # Dump the infos in the YAML part of the README.md file
+ if os.path.exists(dataset_readme_path):
+ dataset_metadata = DatasetMetadata.from_readme(Path(dataset_readme_path))
+ else:
+ dataset_metadata = DatasetMetadata()
+ if total_dataset_infos:
+ total_dataset_infos.to_metadata(dataset_metadata)
+ dataset_metadata.to_readme(Path(dataset_readme_path))
@classmethod
def from_directory(cls, dataset_infos_dir):
logger.info(f"Loading Dataset Infos from {dataset_infos_dir}")
- with open(os.path.join(dataset_infos_dir, config.DATASETDICT_INFOS_FILENAME), encoding="utf-8") as f:
- dataset_infos_dict = {
- config_name: DatasetInfo.from_dict(dataset_info_dict)
- for config_name, dataset_info_dict in json.load(f).items()
- }
- return cls(**dataset_infos_dict)
+ # Load the info from the YAML part of README.md
+ if os.path.exists(os.path.join(dataset_infos_dir, "README.md")):
+ dataset_metadata = DatasetMetadata.from_readme(Path(dataset_infos_dir) / "README.md")
+ return cls.from_metadata(dataset_metadata)
+ elif os.path.exists(os.path.join(dataset_infos_dir, config.DATASETDICT_INFOS_FILENAME)):
+ # this is just to have backward compatibility with dataset_infos.json files
+ with open(os.path.join(dataset_infos_dir, config.DATASETDICT_INFOS_FILENAME), encoding="utf-8") as f:
+ return cls(
+ {
+ config_name: DatasetInfo.from_dict(dataset_info_dict)
+ for config_name, dataset_info_dict in json.load(f).items()
+ }
+ )
+ else:
+ return cls()
+
+ @classmethod
+ def from_metadata(cls, dataset_metadata: DatasetMetadata):
+ if isinstance(dataset_metadata.get("dataset_info"), (list, dict)):
+ if isinstance(dataset_metadata["dataset_info"], list):
+ return cls(
+ {
+ dataset_info_yaml_dict.get("config_name", "default"): DatasetInfo._from_yaml_dict(
+ dataset_info_yaml_dict
+ )
+ for dataset_info_yaml_dict in dataset_metadata["dataset_info"]
+ }
+ )
+ else:
+ dataset_info = DatasetInfo._from_yaml_dict(dataset_metadata["dataset_info"])
+ dataset_info.config_name = dataset_metadata["dataset_info"].get("config_name", "default")
+ return cls({dataset_info.config_name: dataset_info})
+ else:
+ return cls()
+
+ def to_metadata(self, dataset_metadata: DatasetMetadata) -> None:
+ if self:
+ total_dataset_infos = {config_name: dset_info._to_yaml_dict() for config_name, dset_info in self.items()}
+ # the config_name from the dataset_infos_dict takes over the config_name of the DatasetInfo
+ for config_name, dset_info_yaml_dict in total_dataset_infos.items():
+ dset_info_yaml_dict["config_name"] = config_name
+ if len(total_dataset_infos) == 1:
+ # use a struct instead of a list of configurations, since there's only one
+ dataset_metadata["dataset_info"] = next(iter(total_dataset_infos.values()))
+ # no need to include the configuration name when there's only one configuration and it's called "default"
+ if dataset_metadata["dataset_info"].get("config_name") == "default":
+ dataset_metadata["dataset_info"].pop("config_name", None)
+ else:
+ dataset_metadata["dataset_info"] = []
+ for config_name, dataset_info_yaml_dict in total_dataset_infos.items():
+ # add the config_name field in first position
+ dataset_info_yaml_dict.pop("config_name", None)
+ dataset_info_yaml_dict = {"config_name": config_name, **dataset_info_yaml_dict}
+ dataset_metadata["dataset_info"].append(dataset_info_yaml_dict)
@dataclass
@@ -344,11 +433,11 @@ class MetricInfo:
description: str
citation: str
features: Features
- inputs_description: str = field(default_factory=str)
- homepage: str = field(default_factory=str)
- license: str = field(default_factory=str)
- codebase_urls: List[str] = field(default_factory=list)
- reference_urls: List[str] = field(default_factory=list)
+ inputs_description: str = dataclasses.field(default_factory=str)
+ homepage: str = dataclasses.field(default_factory=str)
+ license: str = dataclasses.field(default_factory=str)
+ codebase_urls: List[str] = dataclasses.field(default_factory=list)
+ reference_urls: List[str] = dataclasses.field(default_factory=list)
streamable: bool = False
format: Optional[str] = None
diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -79,6 +79,7 @@
from .utils.filelock import FileLock
from .utils.info_utils import is_small_dataset
from .utils.logging import get_logger
+from .utils.metadata import DatasetMetadata
from .utils.py_utils import get_imports
from .utils.version import Version
@@ -569,6 +570,7 @@ def __init__(
def get_module(self) -> DatasetModule:
# get script and other files
dataset_infos_path = Path(self.path).parent / config.DATASETDICT_INFOS_FILENAME
+ dataset_readme_path = Path(self.path).parent / "README.md"
imports = get_imports(self.path)
local_imports = _download_additional_modules(
name=self.name,
@@ -576,9 +578,11 @@ def get_module(self) -> DatasetModule:
imports=imports,
download_config=self.download_config,
)
- additional_files = (
- [(config.DATASETDICT_INFOS_FILENAME, str(dataset_infos_path))] if dataset_infos_path.is_file() else []
- )
+ additional_files = []
+ if dataset_infos_path.is_file():
+ additional_files.append((config.DATASETDICT_INFOS_FILENAME, str(dataset_infos_path)))
+ if dataset_readme_path.is_file():
+ additional_files.append(("README.md", dataset_readme_path))
# copy the script and the files in an importable directory
dynamic_modules_path = self.dynamic_modules_path if self.dynamic_modules_path else init_dynamic_modules()
module_path, hash = _create_importable_file(
@@ -658,8 +662,21 @@ def get_module(self) -> DatasetModule:
if os.path.isfile(os.path.join(self.path, config.DATASETDICT_INFOS_FILENAME)):
with open(os.path.join(self.path, config.DATASETDICT_INFOS_FILENAME), encoding="utf-8") as f:
dataset_infos: DatasetInfosDict = json.load(f)
- builder_kwargs["config_name"] = next(iter(dataset_infos))
- builder_kwargs["info"] = DatasetInfo.from_dict(dataset_infos[builder_kwargs["config_name"]])
+ if dataset_infos:
+ builder_kwargs["config_name"] = next(iter(dataset_infos))
+ builder_kwargs["info"] = DatasetInfo.from_dict(next(iter(dataset_infos.values())))
+ if os.path.isfile(os.path.join(self.path, "README.md")):
+ dataset_metadata = DatasetMetadata.from_readme(Path(self.path) / "README.md")
+ if isinstance(dataset_metadata.get("dataset_info"), list) and dataset_metadata["dataset_info"]:
+ dataset_info_dict = dataset_metadata["dataset_info"][0]
+ builder_kwargs["info"] = DatasetInfo._from_yaml_dict(dataset_info_dict)
+ if "config_name" in dataset_info_dict:
+ builder_kwargs["config_name"] = dataset_info_dict["config_name"]
+ elif isinstance(dataset_metadata.get("dataset_info"), dict) and dataset_metadata["dataset_info"]:
+ dataset_info_dict = dataset_metadata["dataset_info"]
+ builder_kwargs["info"] = DatasetInfo._from_yaml_dict(dataset_info_dict)
+ if "config_name" in dataset_info_dict:
+ builder_kwargs["config_name"] = dataset_info_dict["config_name"]
return DatasetModule(module_path, hash, builder_kwargs)
@@ -798,8 +815,31 @@ def get_module(self) -> DatasetModule:
)
with open(dataset_infos_path, encoding="utf-8") as f:
dataset_infos: DatasetInfosDict = json.load(f)
- builder_kwargs["config_name"] = next(iter(dataset_infos))
- builder_kwargs["info"] = DatasetInfo.from_dict(dataset_infos[builder_kwargs["config_name"]])
+ if dataset_infos:
+ builder_kwargs["config_name"] = next(iter(dataset_infos))
+ builder_kwargs["info"] = DatasetInfo.from_dict(next(iter(dataset_infos.values())))
+ except FileNotFoundError:
+ pass
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading readme"
+ try:
+ dataset_readme_path = cached_path(
+ hf_hub_url(self.name, "README.md", revision=self.revision),
+ download_config=self.download_config,
+ )
+ dataset_metadata = DatasetMetadata.from_readme(Path(dataset_readme_path))
+ if isinstance(dataset_metadata.get("dataset_info"), list) and dataset_metadata["dataset_info"]:
+ dataset_info_dict = dataset_metadata["dataset_info"][0]
+ builder_kwargs["info"] = DatasetInfo._from_yaml_dict(dataset_info_dict)
+ if "config_name" in dataset_info_dict:
+ builder_kwargs["config_name"] = dataset_info_dict["config_name"]
+ elif isinstance(dataset_metadata.get("dataset_info"), dict) and dataset_metadata["dataset_info"]:
+ dataset_info_dict = dataset_metadata["dataset_info"]
+ builder_kwargs["info"] = DatasetInfo._from_yaml_dict(dataset_info_dict)
+ if "config_name" in dataset_info_dict:
+ builder_kwargs["config_name"] = dataset_info_dict["config_name"]
+
except FileNotFoundError:
pass
return DatasetModule(module_path, hash, builder_kwargs)
@@ -844,10 +884,25 @@ def download_dataset_infos_file(self) -> str:
except (FileNotFoundError, ConnectionError):
return None
+ def download_dataset_readme_file(self) -> str:
+ readme_url = hf_hub_url(repo_id=self.name, path="README.md", revision=self.revision)
+ # Download the dataset infos file if available
+ download_config = self.download_config.copy()
+ if download_config.download_desc is None:
+ download_config.download_desc = "Downloading readme"
+ try:
+ return cached_path(
+ readme_url,
+ download_config=download_config,
+ )
+ except (FileNotFoundError, ConnectionError):
+ return None
+
def get_module(self) -> DatasetModule:
# get script and other files
local_path = self.download_loading_script()
dataset_infos_path = self.download_dataset_infos_file()
+ dataset_readme_path = self.download_dataset_readme_file()
imports = get_imports(local_path)
local_imports = _download_additional_modules(
name=self.name,
@@ -855,7 +910,11 @@ def get_module(self) -> DatasetModule:
imports=imports,
download_config=self.download_config,
)
- additional_files = [(config.DATASETDICT_INFOS_FILENAME, dataset_infos_path)] if dataset_infos_path else []
+ additional_files = []
+ if dataset_infos_path:
+ additional_files.append((config.DATASETDICT_INFOS_FILENAME, dataset_infos_path))
+ if dataset_readme_path:
+ additional_files.append(("README.md", dataset_readme_path))
# copy the script and the files in an importable directory
dynamic_modules_path = self.dynamic_modules_path if self.dynamic_modules_path else init_dynamic_modules()
module_path, hash = _create_importable_file(
diff --git a/src/datasets/splits.py b/src/datasets/splits.py
--- a/src/datasets/splits.py
+++ b/src/datasets/splits.py
@@ -18,13 +18,15 @@
import abc
import collections
+import copy
+import dataclasses
import re
-from dataclasses import dataclass, field
+from dataclasses import dataclass
from typing import Dict, List, Optional, Union
from .arrow_reader import FileInstructions, make_file_instructions
from .naming import _split_re
-from .utils.py_utils import NonMutableDict
+from .utils.py_utils import NonMutableDict, asdict
@dataclass
@@ -32,7 +34,14 @@ class SplitInfo:
name: str = ""
num_bytes: int = 0
num_examples: int = 0
- dataset_name: Optional[str] = None
+
+ # Deprecated
+ # For backward compatibility, this field needs to always be included in files like
+ # dataset_infos.json and dataset_info.json files
+ # To do so, we always include it in the output of datasets.utils.py_utils.asdict(split_info)
+ dataset_name: Optional[str] = dataclasses.field(
+ default=None, metadata={"include_in_asdict_even_if_is_default": True}
+ )
@property
def file_instructions(self):
@@ -545,7 +554,7 @@ def from_split_dict(cls, split_infos: Union[List, Dict], dataset_name: Optional[
split_infos = list(split_infos.values())
if dataset_name is None:
- dataset_name = split_infos[0]["dataset_name"] if split_infos else None
+ dataset_name = split_infos[0].get("dataset_name") if split_infos else None
split_dict = cls(dataset_name=dataset_name)
@@ -559,11 +568,27 @@ def from_split_dict(cls, split_infos: Union[List, Dict], dataset_name: Optional[
def to_split_dict(self):
"""Returns a list of SplitInfo protos that we have."""
# Return the SplitInfo, sorted by name
- return sorted((s for s in self.values()), key=lambda s: s.name)
+ out = []
+ for split_name, split_info in sorted(self.items()):
+ split_info = copy.deepcopy(split_info)
+ split_info.name = split_name
+ out.append(split_info)
+ return out
def copy(self):
return SplitDict.from_split_dict(self.to_split_dict(), self.dataset_name)
+ def _to_yaml_list(self) -> list:
+ out = [asdict(s) for s in self.to_split_dict()]
+ # we don't need the dataset_name attribute that is deprecated
+ for split_info_dict in out:
+ split_info_dict.pop("dataset_name", None)
+ return out
+
+ @classmethod
+ def _from_yaml_list(cls, yaml_data: list) -> "SplitDict":
+ return cls.from_split_dict(yaml_data)
+
@dataclass
class SplitGenerator:
@@ -591,8 +616,8 @@ class SplitGenerator:
"""
name: str
- gen_kwargs: Dict = field(default_factory=dict)
- split_info: SplitInfo = field(init=False)
+ gen_kwargs: Dict = dataclasses.field(default_factory=dict)
+ split_info: SplitInfo = dataclasses.field(init=False)
def __post_init__(self):
self.name = str(self.name) # Make sure we convert NamedSplits in strings
diff --git a/src/datasets/utils/metadata.py b/src/datasets/utils/metadata.py
--- a/src/datasets/utils/metadata.py
+++ b/src/datasets/utils/metadata.py
@@ -1,58 +1,11 @@
-import json
-import logging
-import re
-import warnings
from collections import Counter
-from dataclasses import dataclass, fields
from pathlib import Path
-from typing import Any, Callable, ClassVar, Dict, List, Optional, Tuple, Type, Union
-
-
-try: # Python >= 3.8
- from typing import get_args, get_origin
-except ImportError:
-
- def get_args(tp):
- return getattr(tp, "__args__", ())
-
- def get_origin(tp):
- return getattr(tp, "__origin__", None)
-
-
-# loading package files: https://stackoverflow.com/a/20885799
-try:
- import importlib.resources as pkg_resources
-except ImportError:
- # Try backported to PY<37 `importlib_resources`.
- import importlib_resources as pkg_resources
+from typing import Optional, Tuple
import yaml
-from . import resources
-
-
-BASE_REF_URL = "https://github.com/huggingface/datasets/tree/main/src/datasets/utils"
-this_url = f"{BASE_REF_URL}/{__file__}"
-logger = logging.getLogger(__name__)
-
-
-def load_json_resource(resource: str) -> Tuple[Any, str]:
- content = pkg_resources.read_text(resources, resource)
- return json.loads(content), f"{BASE_REF_URL}/resources/{resource}"
-
-# Source of languages.json:
-# https://datahub.io/core/language-codes/r/ietf-language-tags.csv
-# Language names were obtained with langcodes: https://github.com/LuminosoInsight/langcodes
-known_language_codes, known_language_codes_url = load_json_resource("languages.json")
-known_task_ids, known_task_ids_url = load_json_resource("tasks.json")
-known_creators, known_creators_url = load_json_resource("creators.json")
-known_size_categories, known_size_categories_url = load_json_resource("size_categories.json")
-known_multilingualities, known_multilingualities_url = load_json_resource("multilingualities.json")
-known_source_datasets, known_source_datasets_url = ["original", "extended", r"extended\|.*"], this_url
-
-
-class NoDuplicateSafeLoader(yaml.SafeLoader):
+class _NoDuplicateSafeLoader(yaml.SafeLoader):
def _check_no_duplicates_on_constructed_node(self, node):
keys = [self.constructed_objects[key_node] for key_node, _ in node.value]
keys = [tuple(key) if isinstance(key, list) else key for key in keys]
@@ -67,231 +20,20 @@ def construct_mapping(self, node, deep=False):
return mapping
-def yaml_block_from_readme(path: Path) -> Optional[str]:
- with open(path, encoding="utf-8") as readme_file:
- content = [line.rstrip("\n") for line in readme_file]
-
- if content[0] == "---" and "---" in content[1:]:
- yamlblock = "\n".join(content[1 : content[1:].index("---") + 1])
- return yamlblock
-
- return None
-
-
-def metadata_dict_from_readme(path: Path) -> Optional[Dict[str, List[str]]]:
- """Loads a dataset's metadata from the dataset card (REAMDE.md), as a Python dict"""
- yaml_block = yaml_block_from_readme(path=path)
- if yaml_block is None:
- return None
- metada_dict = yaml.load(yaml_block, Loader=NoDuplicateSafeLoader) or dict()
- return metada_dict
+def _split_yaml_from_readme(readme_content: str) -> Tuple[Optional[str], str]:
+ full_content = [line for line in readme_content.splitlines()]
+ if full_content[0] == "---" and "---" in full_content[1:]:
+ sep_idx = full_content[1:].index("---") + 1
+ yamlblock = "\n".join(full_content[1:sep_idx])
+ return yamlblock, "\n".join(full_content[sep_idx + 1 :])
+ return None, "\n".join(full_content)
-ValidatorOutput = Tuple[List[str], Optional[str]]
-
-
-def tagset_validator(
- items: Union[List[str], Dict[str, List[str]]],
- reference_values: List[str],
- name: str,
- url: str,
- escape_validation_predicate_fn: Optional[Callable[[Any], bool]] = None,
-) -> ValidatorOutput:
- reference_values = re.compile("^(?:" + "|".join(reference_values) + ")$")
- if isinstance(items, list):
- if escape_validation_predicate_fn is not None:
- invalid_values = [
- v for v in items if not reference_values.match(v) and escape_validation_predicate_fn(v) is False
- ]
- else:
- invalid_values = [v for v in items if not reference_values.match(v)]
-
- else:
- invalid_values = []
- if escape_validation_predicate_fn is not None:
- for config_name, values in items.items():
- invalid_values += [
- v for v in values if not reference_values.match(v) and escape_validation_predicate_fn(v) is False
- ]
- else:
- for config_name, values in items.items():
- invalid_values += [v for v in values if not reference_values.match(v)]
- if len(invalid_values) > 0:
- return [], f"{invalid_values} are not registered tags for '{name}', reference at {url}"
- return items, None
-
-
-def validate_type(value: Any, expected_type: Type):
- error_string = ""
- NoneType = type(None)
- if expected_type is NoneType:
- if not isinstance(value, NoneType):
- return f"Expected `{NoneType}`. Found value: `{value}` of type `{type(value)}`.\n"
- else:
- return error_string
- if expected_type is str:
- if not isinstance(value, str):
- return f"Expected `{str}`. Found value: `{value}` of type: `{type(value)}`.\n"
-
- elif isinstance(value, str) and len(value) == 0:
- return (
- f"Expected `{str}` with length > 0. Found value: `{value}` of type: `{type(value)}` with length: 0.\n"
- )
- else:
- return error_string
- # Add more `elif` statements if primitive type checking is needed
- else:
- expected_type_origin = get_origin(expected_type) # typing.List[str] -> list
- expected_type_args = get_args(expected_type) # typing.List[str] -> (str, )
-
- if expected_type_origin is Union:
- for type_arg in expected_type_args:
- temp_error_string = validate_type(value, type_arg)
- if temp_error_string == "": # at least one type is successfully validated
- return temp_error_string
- else:
- if error_string == "":
- error_string = "(" + temp_error_string + ")"
- else:
- error_string += "\nOR\n" + "(" + temp_error_string + ")"
- elif value is None:
- return f"Expected `{expected_type}`. Found value: `{value}` of type: `{type(value)}`.\n"
- else:
- # Assuming non empty `List`/`Dict`/`Tuple`
- if expected_type is EmptyList:
- if len(value) == 0:
- return ""
- else:
- return f"Expected `{expected_type}` of length 0. Found value of type: `{type(value)}`, with length: {len(value)}.\n"
-
- # Assuming non empty
- if not isinstance(value, (dict, tuple, list)) or len(value) == 0:
- return f"Expected `{expected_type}` with length > 0. Found value of type: `{type(value)}`, with length: {len(value)}.\n"
-
- if expected_type_origin is dict:
- if not isinstance(value, dict):
- return f"Expected `{expected_type}` with length > 0. Found value of type: `{type(value)}`, with length: {len(value)}.\n"
- if expected_type_args != get_args(Dict): # if we specified types for keys and values
- key_type, value_type = expected_type_args
- key_error_string = ""
- value_error_string = ""
- for k, v in value.items():
- key_error_string += validate_type(k, key_type)
- value_error_string += validate_type(v, value_type)
- if key_error_string != "" or value_error_string != "":
- return f"Typing errors with keys:\n {key_error_string} and values:\n {value_error_string}"
-
- else: # `List`/`Tuple`
- if not isinstance(value, (list, tuple)):
- return f"Expected `{expected_type}` with length > 0. Found value of type: `{type(value)}`, with length: {len(value)}.\n"
- if expected_type_args != get_args(List): # if we specified types for the items in the list
- value_type = expected_type_args[0]
- value_error_string = ""
- for v in value:
- value_error_string += validate_type(v, value_type)
- if value_error_string != "":
- return f"Typing errors with values:\n {value_error_string}"
-
- return error_string
-
-
-def validate_metadata_type(metadata_dict: dict):
- field_types = {field.name: field.type for field in fields(DatasetMetadata)}
-
- typing_errors = {}
- for field_name, field_value in metadata_dict.items():
- field_type_error = validate_type(
- metadata_dict[field_name], field_types.get(field_name, Union[List[str], Dict[str, List[str]]])
- )
- if field_type_error != "":
- typing_errors[field_name] = field_type_error
- if len(typing_errors) > 0:
- raise TypeError(f"The following typing errors are found: {typing_errors}")
-
-
-class _nothing:
- pass
-
-
-EmptyList = List[_nothing]
-
-
-@dataclass
-class DatasetMetadata:
- annotations_creators: List[str]
- language_creators: Union[EmptyList, List[str]]
- language: Union[EmptyList, List[str]]
- license: List[str]
- multilinguality: List[str]
- pretty_name: str
- size_categories: List[str]
- source_datasets: List[str]
- task_categories: List[str]
- task_ids: Union[EmptyList, List[str]]
- language_details: Optional[str] = None
- language_bcp47: Optional[List[str]] = None
- paperswithcode_id: Optional[str] = None
- train_eval_index: Optional[List[Dict]] = None
- configs: Optional[List[str]] = None
- extra_gated_fields: Optional[Dict] = None
- extra_gated_prompt: Optional[str] = None
- license_details: Optional[str] = None
- tags: Optional[List[str]] = None
- licenses: Optional[Union[EmptyList, List[str]]] = None # deprecated
- languages: Optional[Union[EmptyList, List[str]]] = None # deprecated
+class DatasetMetadata(dict):
# class attributes
- _FIELDS_WITH_DASHES: ClassVar[set] = {"train_eval_index"} # train-eval-index in the YAML metadata
- _ALLOWED_YAML_KEYS: ClassVar[set] = set() # populated later
- _DEPRECATED_YAML_KEYS = ["licenses", "languages"]
-
- def __post_init__(self):
- if self.licenses is not None:
- warnings.warning("The 'licenses' YAML field is deprecated, please use 'license' instead.")
- if self.languages is not None:
- warnings.warning("The 'languages' YAML field is deprecated, please use 'language' instead.")
-
- def validate(self):
- validate_metadata_type(metadata_dict=vars(self))
-
- self.annotations_creators, annotations_creators_errors = self.validate_annotations_creators(
- self.annotations_creators
- )
- self.language_creators, language_creators_errors = self.validate_language_creators(self.language_creators)
- self.language, language_errors = self.validate_language_codes(self.language or self.languages)
- self.multilinguality, multilinguality_errors = self.validate_mulitlinguality(self.multilinguality)
- self.size_categories, size_categories_errors = self.validate_size_catgeories(self.size_categories)
- self.source_datasets, source_datasets_errors = self.validate_source_datasets(self.source_datasets)
- self.task_categories, task_categories_errors = self.validate_task_categories(self.task_categories)
- self.task_ids, task_ids_errors = self.validate_task_ids(self.task_ids)
- self.paperswithcode_id, paperswithcode_id_errors = self.validate_paperswithcode_id_errors(
- self.paperswithcode_id
- )
- self.train_eval_index, train_eval_index_errors = self.validate_train_eval_index(self.train_eval_index)
-
- errors = {
- "annotations_creators": annotations_creators_errors,
- "language_creators": language_creators_errors,
- "multilinguality": multilinguality_errors,
- "size_categories": size_categories_errors,
- "source_datasets": source_datasets_errors,
- "task_categories": task_categories_errors,
- "task_ids": task_ids_errors,
- "language": language_errors,
- "paperswithcode_id": paperswithcode_id_errors,
- "train_eval_index": train_eval_index_errors,
- }
-
- exception_msg_dict = dict()
- for yaml_field, errs in errors.items():
- if errs is not None:
- exception_msg_dict[yaml_field] = errs
- if len(exception_msg_dict) > 0:
- raise TypeError(
- "Could not validate the metadata, found the following errors:\n"
- + "\n".join(f"* field '{fieldname}':\n\t{err}" for fieldname, err in exception_msg_dict.items())
- )
+ _FIELDS_WITH_DASHES = {"train_eval_index"} # train-eval-index in the YAML metadata
@classmethod
def from_readme(cls, path: Path) -> "DatasetMetadata":
@@ -304,172 +46,65 @@ def from_readme(cls, path: Path) -> "DatasetMetadata":
:class:`DatasetMetadata`: The dataset's metadata
Raises:
- :obj:`TypeError`: If the dataset card has no metadata (no YAML header)
:obj:`TypeError`: If the dataset's metadata is invalid
"""
- yaml_string = yaml_block_from_readme(path)
+ with open(path, encoding="utf-8") as readme_file:
+ yaml_string, _ = _split_yaml_from_readme(readme_file.read())
if yaml_string is not None:
return cls.from_yaml_string(yaml_string)
else:
- raise TypeError(f"Unable to find a yaml block in '{path}'")
+ return cls()
+
+ def to_readme(self, path: Path):
+ if path.exists():
+ with open(path, encoding="utf-8") as readme_file:
+ readme_content = readme_file.read()
+ else:
+ readme_content = None
+ updated_readme_content = self._to_readme(readme_content)
+ with open(path, "w", encoding="utf-8") as readme_file:
+ readme_file.write(updated_readme_content)
+
+ def _to_readme(self, readme_content: Optional[str] = None) -> str:
+ if readme_content is not None:
+ _, content = _split_yaml_from_readme(readme_content)
+ full_content = "---\n" + self.to_yaml_string() + "---\n" + content
+ else:
+ full_content = "---\n" + self.to_yaml_string() + "---\n"
+ return full_content
@classmethod
- def _metadata_dict_from_yaml_string(cls, string: str) -> dict:
- """Loads and validates the dataset metadat from a YAML string
+ def from_yaml_string(cls, string: str) -> "DatasetMetadata":
+ """Loads and validates the dataset metadata from a YAML string
Args:
string (:obj:`str`): The YAML string
Returns:
- :class:`dict`: The dataset's metadata as a dictionary
+ :class:`DatasetMetadata`: The dataset's metadata
Raises:
:obj:`TypeError`: If the dataset's metadata is invalid
"""
- metadata_dict = yaml.load(string, Loader=NoDuplicateSafeLoader) or dict()
-
- # Check if the YAML keys are all correct
- bad_keys = [k for k in metadata_dict if k not in cls._ALLOWED_YAML_KEYS]
- if bad_keys:
- raise ValueError(f"Bad YAML keys: {bad_keys}. Allowed fields: {cls._ALLOWED_YAML_KEYS}")
-
- # Check if config names are valid
- bad_keys = [k for k in metadata_dict if k not in cls._ALLOWED_YAML_KEYS]
- if bad_keys:
- raise ValueError(f"Bad YAML keys: {bad_keys}. Allowed fields: {cls._ALLOWED_YAML_KEYS}")
+ metadata_dict = yaml.load(string, Loader=_NoDuplicateSafeLoader) or dict()
# Convert the YAML keys to DatasetMetadata fields
metadata_dict = {
(key.replace("-", "_") if key.replace("-", "_") in cls._FIELDS_WITH_DASHES else key): value
for key, value in metadata_dict.items()
}
- return metadata_dict
-
- @classmethod
- def from_yaml_string(cls, string: str) -> "DatasetMetadata":
- """Loads and validates the dataset metadata from a YAML string
-
- Args:
- string (:obj:`str`): The YAML string
-
- Returns:
- :class:`DatasetMetadata`: The dataset's metadata
-
- Raises:
- :obj:`TypeError`: If the dataset's metadata is invalid
- :obj:`ValueError`: If the dataset's metadata is invalid
- """
- metadata_dict = cls._metadata_dict_from_yaml_string(string)
return cls(**metadata_dict)
- @staticmethod
- def validate_annotations_creators(annotations_creators: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- return tagset_validator(
- annotations_creators, known_creators["annotations"], "annotations_creators", known_creators_url
- )
-
- @staticmethod
- def validate_language_creators(language_creators: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- return tagset_validator(language_creators, known_creators["language"], "language_creators", known_creators_url)
-
- @staticmethod
- def validate_language_codes(languages: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- return tagset_validator(
- languages,
- known_language_codes.keys(),
- "language",
- known_language_codes_url,
- lambda lang: lang == "unknown",
- )
-
- @staticmethod
- def validate_task_categories(task_categories: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- # TODO: we're currently ignoring all values starting with 'other' as our task taxonomy is bound to change
- # in the near future and we don't want to waste energy in tagging against a moving taxonomy.
- known_set = list(known_task_ids.keys())
- validated, error = tagset_validator(
- task_categories, known_set, "task_categories", known_task_ids_url, lambda e: e.startswith("other-")
- )
- return validated, error
-
- @staticmethod
- def validate_task_ids(task_ids: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- # TODO: we're currently ignoring all values starting with 'other' as our task taxonomy is bound to change
- # in the near future and we don't want to waste energy in tagging against a moving taxonomy.
- known_set = [tid for _cat, d in known_task_ids.items() for tid in d.get("subtasks", [])]
- validated, error = tagset_validator(
- task_ids,
- known_set,
- "task_ids",
- known_task_ids_url,
- lambda e: not e or "-other-" in e or e.startswith("other-"),
- )
- return validated, error
-
- @staticmethod
- def validate_mulitlinguality(multilinguality: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- validated, error = tagset_validator(
- multilinguality,
- list(known_multilingualities.keys()),
- "multilinguality",
- known_size_categories_url,
- lambda e: e.startswith("other-"),
- )
- return validated, error
-
- @staticmethod
- def validate_size_catgeories(size_cats: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- return tagset_validator(size_cats, known_size_categories, "size_categories", known_size_categories_url)
-
- @staticmethod
- def validate_source_datasets(sources: Union[List[str], Dict[str, List[str]]]) -> ValidatorOutput:
- return tagset_validator(sources, known_source_datasets, "source_datasets", known_source_datasets_url)
-
- @staticmethod
- def validate_paperswithcode_id_errors(paperswithcode_id: Optional[str]) -> ValidatorOutput:
- if paperswithcode_id is None:
- return paperswithcode_id, None
- else:
- if " " in paperswithcode_id or paperswithcode_id.lower() != paperswithcode_id:
- return (
- None,
- f"The paperswithcode_id must be lower case and not contain spaces but got {paperswithcode_id}. You can find the paperswithcode_id in the URL of the dataset page on paperswithcode.com.",
- )
- else:
- return paperswithcode_id, None
-
- @staticmethod
- def validate_pretty_name(pretty_name: Union[str, Dict[str, str]]):
- if isinstance(pretty_name, str):
- if len(pretty_name) == 0:
- return None, "The pretty name must have a length greater than 0 but got an empty string."
- else:
- error_string = ""
- for key, value in pretty_name.items():
- if len(value) == 0:
- error_string += f"The pretty name must have a length greater than 0 but got an empty string for config: {key}.\n"
-
- if error_string == "":
- return None, error_string
- else:
- return pretty_name, None
-
- @staticmethod
- def validate_train_eval_index(train_eval_index: Optional[Dict]):
- if train_eval_index is not None and not isinstance(train_eval_index, list):
- return None, f"train-eval-index must be a list, but got {type(train_eval_index)} instead."
- else:
- return train_eval_index, None
-
-
-# In general the allowed YAML keys are the fields of the DatasetMetadata dataclass.
-# However it is not the case certain fields like train_eval_index,
-# for which the YAML key must use dashes and not underscores.
-# Fields that corresponds to YAML keys with dashes are defined in DatasetMetadata._FIELDS_WITH_DASHES
-DatasetMetadata._ALLOWED_YAML_KEYS = {
- field.name.replace("_", "-") if field.name in DatasetMetadata._FIELDS_WITH_DASHES else field.name
- for field in fields(DatasetMetadata)
-}
+ def to_yaml_string(self) -> str:
+ return yaml.safe_dump(
+ {
+ (key.replace("_", "-") if key in self._FIELDS_WITH_DASHES else key): value
+ for key, value in self.items()
+ },
+ sort_keys=False,
+ allow_unicode=True,
+ encoding="utf-8",
+ ).decode("utf-8")
if __name__ == "__main__":
@@ -481,4 +116,5 @@ def validate_train_eval_index(train_eval_index: Optional[Dict]):
readme_filepath = Path(args.readme_filepath)
dataset_metadata = DatasetMetadata.from_readme(readme_filepath)
- dataset_metadata.validate()
+ print(dataset_metadata)
+ dataset_metadata.to_readme(readme_filepath)
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -167,7 +167,8 @@ def _asdict_inner(obj):
result = {}
for f in fields(obj):
value = _asdict_inner(getattr(obj, f.name))
- result[f.name] = value
+ if not f.init or value != f.default or f.metadata.get("include_in_asdict_even_if_is_default", False):
+ result[f.name] = value
return result
elif isinstance(obj, tuple) and hasattr(obj, "_fields"):
# obj is a namedtuple
diff --git a/src/datasets/utils/version.py b/src/datasets/utils/version.py
--- a/src/datasets/utils/version.py
+++ b/src/datasets/utils/version.py
@@ -113,6 +113,9 @@ def from_dict(cls, dic):
field_names = {f.name for f in dataclasses.fields(cls)}
return cls(**{k: v for k, v in dic.items() if k in field_names})
+ def _to_yaml_string(self) -> str:
+ return self.version_str
+
def _str_to_version(version_str, allow_wildcard=False):
"""Return the tuple (major, minor, patch) version extracted from the str."""
| diff --git a/src/datasets/commands/test.py b/src/datasets/commands/test.py
--- a/src/datasets/commands/test.py
+++ b/src/datasets/commands/test.py
@@ -23,7 +23,7 @@ def _test_command_factory(args):
args.cache_dir,
args.data_dir,
args.all_configs,
- args.save_infos,
+ args.save_info or args.save_infos,
args.ignore_verifications,
args.force_redownload,
args.clear_cache,
@@ -50,7 +50,9 @@ def register_subcommand(parser: ArgumentParser):
help="Can be used to specify a manual directory to get the files from.",
)
test_parser.add_argument("--all_configs", action="store_true", help="Test all dataset configurations")
- test_parser.add_argument("--save_infos", action="store_true", help="Save the dataset infos file")
+ test_parser.add_argument(
+ "--save_info", action="store_true", help="Save the dataset infos in the dataset card (README.md)"
+ )
test_parser.add_argument(
"--ignore_verifications", action="store_true", help="Run the test without checksums and splits checks"
)
@@ -60,6 +62,8 @@ def register_subcommand(parser: ArgumentParser):
action="store_true",
help="Remove downloaded files and cached datasets after each config test",
)
+ # aliases
+ test_parser.add_argument("--save_infos", action="store_true", help="alias to save_info")
test_parser.add_argument("dataset", type=str, help="Name of the dataset to download")
test_parser.set_defaults(func=_test_command_factory)
@@ -145,13 +149,13 @@ def get_builders() -> Generator[DatasetBuilder, None, None]:
if self._save_infos:
builder._save_infos()
- # If save_infos=True, the dataset infos file is created next to the loaded module file.
+ # If save_infos=True, the dataset card (README.md) is created next to the loaded module file.
+ # The dataset_infos are saved in the YAML part of the README.md
+
# Let's move it to the original directory of the dataset script, to allow the user to
# upload them on S3 at the same time afterwards.
if self._save_infos:
- dataset_infos_path = os.path.join(
- builder_cls.get_imported_module_dir(), datasets.config.DATASETDICT_INFOS_FILENAME
- )
+ dataset_readme_path = os.path.join(builder_cls.get_imported_module_dir(), "README.md")
name = Path(path).name + ".py"
combined_path = os.path.join(path, name)
if os.path.isfile(path):
@@ -162,13 +166,13 @@ def get_builders() -> Generator[DatasetBuilder, None, None]:
dataset_dir = path
else: # in case of a remote dataset
dataset_dir = None
- print(f"Dataset Infos file saved at {dataset_infos_path}")
+ print(f"Dataset card saved at {dataset_readme_path}")
# Move dataset_info back to the user
if dataset_dir is not None:
- user_dataset_infos_path = os.path.join(dataset_dir, datasets.config.DATASETDICT_INFOS_FILENAME)
- copyfile(dataset_infos_path, user_dataset_infos_path)
- print(f"Dataset Infos file saved at {user_dataset_infos_path}")
+ user_dataset_readme_path = os.path.join(dataset_dir, "README.md")
+ copyfile(dataset_readme_path, user_dataset_readme_path)
+ print(f"Dataset card saved at {user_dataset_readme_path}")
# If clear_cache=True, the download folder and the dataset builder cache directory are deleted
if self._clear_cache:
diff --git a/tests/commands/test_test.py b/tests/commands/test_test.py
--- a/tests/commands/test_test.py
+++ b/tests/commands/test_test.py
@@ -1,11 +1,11 @@
-import json
import os
from collections import namedtuple
import pytest
-from datasets import config
+from datasets import ClassLabel, Features, Sequence, Value
from datasets.commands.test import TestCommand
+from datasets.info import DatasetInfo, DatasetInfosDict
_TestCommandArgs = namedtuple(
@@ -25,126 +25,58 @@
)
+def is_1percent_close(source, target):
+ return (abs(source - target) / target) < 0.01
+
+
@pytest.mark.integration
def test_test_command(dataset_loading_script_dir):
args = _TestCommandArgs(dataset=dataset_loading_script_dir, all_configs=True, save_infos=True)
test_command = TestCommand(*args)
test_command.run()
- dataset_infos_path = os.path.join(dataset_loading_script_dir, config.DATASETDICT_INFOS_FILENAME)
- assert os.path.exists(dataset_infos_path)
- with open(dataset_infos_path, encoding="utf-8") as f:
- dataset_infos = json.load(f)
- expected_dataset_infos = {
- "default": {
- "description": "",
- "citation": "",
- "homepage": "",
- "license": "",
- "features": {
- "tokens": {
- "feature": {"dtype": "string", "id": None, "_type": "Value"},
- "length": -1,
- "id": None,
- "_type": "Sequence",
- },
- "ner_tags": {
- "feature": {
- "num_classes": 7,
- "names": ["O", "B-PER", "I-PER", "B-ORG", "I-ORG", "B-LOC", "I-LOC"],
- "id": None,
- "_type": "ClassLabel",
+ dataset_readme_path = os.path.join(dataset_loading_script_dir, "README.md")
+ assert os.path.exists(dataset_readme_path)
+ dataset_infos = DatasetInfosDict.from_directory(dataset_loading_script_dir)
+ expected_dataset_infos = DatasetInfosDict(
+ {
+ "default": DatasetInfo(
+ features=Features(
+ {
+ "tokens": Sequence(Value("string")),
+ "ner_tags": Sequence(
+ ClassLabel(names=["O", "B-PER", "I-PER", "B-ORG", "I-ORG", "B-LOC", "I-LOC"])
+ ),
+ "langs": Sequence(Value("string")),
+ "spans": Sequence(Value("string")),
+ }
+ ),
+ splits=[
+ {
+ "name": "train",
+ "num_bytes": 2351563,
+ "num_examples": 10000,
+ },
+ {
+ "name": "validation",
+ "num_bytes": 238418,
+ "num_examples": 1000,
},
- "length": -1,
- "id": None,
- "_type": "Sequence",
- },
- "langs": {
- "feature": {"dtype": "string", "id": None, "_type": "Value"},
- "length": -1,
- "id": None,
- "_type": "Sequence",
- },
- "spans": {
- "feature": {"dtype": "string", "id": None, "_type": "Value"},
- "length": -1,
- "id": None,
- "_type": "Sequence",
- },
- },
- "post_processed": None,
- "supervised_keys": None,
- "task_templates": None,
- "builder_name": "__dummy_dataset1__",
- "config_name": "default",
- "version": {"version_str": "0.0.0", "description": None, "major": 0, "minor": 0, "patch": 0},
- "splits": {
- "train": {
- "name": "train",
- "num_bytes": 2351591,
- "num_examples": 10000,
- "dataset_name": "__dummy_dataset1__",
- },
- "validation": {
- "name": "validation",
- "num_bytes": 238446,
- "num_examples": 1000,
- "dataset_name": "__dummy_dataset1__",
- },
- },
- "download_checksums": {
- "https://huggingface.co/datasets/albertvillanova/tests-raw-jsonl/resolve/main/wikiann-bn-train.jsonl": {
- "num_bytes": 3578339,
- "checksum": "6fbe6dbdcb3c9c3a98b0ab4d56b1c8b73baab9293d603064a5ab5230ab4f366b",
- },
- "https://huggingface.co/datasets/albertvillanova/tests-raw-jsonl/resolve/main/wikiann-bn-validation.jsonl": {
- "num_bytes": 362341,
- "checksum": "2ddd0c090a8ccb721d7aa8477ed7323750683822c247015d5cfab1af1c8c8b3f",
- },
- },
- "download_size": 3940680,
- "post_processing_size": None,
- "dataset_size": 2590037,
- "size_in_bytes": 6530717,
+ ],
+ download_size=3940680,
+ dataset_size=2589981,
+ )
}
- }
+ )
assert dataset_infos.keys() == expected_dataset_infos.keys()
- assert dataset_infos["default"].keys() == expected_dataset_infos["default"].keys()
- for key in dataset_infos["default"].keys():
- if key in [
- "description",
- "citation",
- "homepage",
- "license",
- "features",
- "post_processed",
- "supervised_keys",
- "task_templates",
- "builder_name",
- "config_name",
- "version",
- "download_checksums",
- "download_size",
- "post_processing_size",
- ]:
- assert dataset_infos["default"][key] == expected_dataset_infos["default"][key]
- elif key in ["dataset_size", "size_in_bytes"]:
- assert round(dataset_infos["default"][key] / 10**5) == round(
- expected_dataset_infos["default"][key] / 10**5
- )
+ for key in DatasetInfo._INCLUDED_INFO_IN_YAML:
+ result, expected = getattr(dataset_infos["default"], key), getattr(expected_dataset_infos["default"], key)
+ if key == "num_bytes":
+ assert is_1percent_close(result, expected)
elif key == "splits":
- assert dataset_infos["default"]["splits"].keys() == expected_dataset_infos["default"]["splits"].keys()
- for split in dataset_infos["default"]["splits"].keys():
- assert (
- dataset_infos["default"]["splits"][split].keys()
- == expected_dataset_infos["default"]["splits"][split].keys()
- )
- for subkey in dataset_infos["default"]["splits"][split].keys():
- if subkey == "num_bytes":
- assert round(dataset_infos["default"]["splits"][split][subkey] / 10**2) == round(
- expected_dataset_infos["default"]["splits"][split][subkey] / 10**2
- )
- else:
- assert (
- dataset_infos["default"]["splits"][split][subkey]
- == expected_dataset_infos["default"]["splits"][split][subkey]
- )
+ assert list(result) == list(expected)
+ for split in result:
+ assert result[split].name == expected[split].name
+ assert result[split].num_examples == expected[split].num_examples
+ assert is_1percent_close(result[split].num_bytes, expected[split].num_bytes)
+ else:
+ result == expected
diff --git a/tests/features/test_features.py b/tests/features/test_features.py
--- a/tests/features/test_features.py
+++ b/tests/features/test_features.py
@@ -7,8 +7,9 @@
import pyarrow as pa
import pytest
+from datasets import Array2D
from datasets.arrow_dataset import Dataset
-from datasets.features import ClassLabel, Features, Image, Sequence, Value
+from datasets.features import Audio, ClassLabel, Features, Image, Sequence, Value
from datasets.features.features import (
_arrow_to_datasets_dtype,
_cast_to_python_objects,
@@ -562,3 +563,65 @@ def test_dont_iterate_over_each_element_in_a_list(self, mocked_cast):
obj = {"col_1": [[1, 2], [3, 4], [5, 6]]}
cast_to_python_objects(obj)
self.assertEqual(mocked_cast.call_count, 4) # 4 = depth of obj
+
+
+SIMPLE_FEATURES = [
+ Features(),
+ Features({"a": Value("int32")}),
+ Features({"a": Value("int32", id="my feature")}),
+ Features({"a": Value("int32"), "b": Value("float64"), "c": Value("string")}),
+]
+
+CUSTOM_FEATURES = [
+ Features({"label": ClassLabel(names=["negative", "positive"])}),
+ Features({"array": Array2D(dtype="float32", shape=(4, 4))}),
+ Features({"image": Image()}),
+ Features({"audio": Audio()}),
+ Features({"image": Image(decode=False)}),
+ Features({"audio": Audio(decode=False)}),
+ Features({"translation": Translation(["en", "fr"])}),
+ Features({"translation": TranslationVariableLanguages(["en", "fr"])}),
+]
+
+NESTED_FEATURES = [
+ Features({"foo": {}}),
+ Features({"foo": {"bar": Value("int32")}}),
+ Features({"foo": {"bar1": Value("int32"), "bar2": Value("float64")}}),
+ Features({"foo": Sequence(Value("int32"))}),
+ Features({"foo": Sequence({})}),
+ Features({"foo": Sequence({"bar": Value("int32")})}),
+ Features({"foo": [Value("int32")]}),
+ Features({"foo": [{"bar": Value("int32")}]}),
+]
+
+NESTED_CUSTOM_FEATURES = [
+ Features({"foo": {"bar": ClassLabel(names=["negative", "positive"])}}),
+ Features({"foo": Sequence(ClassLabel(names=["negative", "positive"]))}),
+ Features({"foo": Sequence({"bar": ClassLabel(names=["negative", "positive"])})}),
+ Features({"foo": [ClassLabel(names=["negative", "positive"])]}),
+ Features({"foo": [{"bar": ClassLabel(names=["negative", "positive"])}]}),
+]
+
+
[email protected]("features", SIMPLE_FEATURES + CUSTOM_FEATURES + NESTED_FEATURES + NESTED_CUSTOM_FEATURES)
+def test_features_to_dict(features: Features):
+ features_dict = features.to_dict()
+ assert isinstance(features_dict, dict)
+ reloaded = Features.from_dict(features_dict)
+ assert features == reloaded
+
+
[email protected]("features", SIMPLE_FEATURES + CUSTOM_FEATURES + NESTED_FEATURES + NESTED_CUSTOM_FEATURES)
+def test_features_to_yaml_list(features: Features):
+ features_yaml_list = features._to_yaml_list()
+ assert isinstance(features_yaml_list, list)
+ reloaded = Features._from_yaml_list(features_yaml_list)
+ assert features == reloaded
+
+
[email protected]("features", SIMPLE_FEATURES + CUSTOM_FEATURES + NESTED_FEATURES + NESTED_CUSTOM_FEATURES)
+def test_features_to_arrow_schema(features: Features):
+ arrow_schema = features.arrow_schema
+ assert isinstance(arrow_schema, pa.Schema)
+ reloaded = Features.from_arrow_schema(arrow_schema)
+ assert features == reloaded
diff --git a/tests/test_dataset_cards.py b/tests/test_dataset_cards.py
--- a/tests/test_dataset_cards.py
+++ b/tests/test_dataset_cards.py
@@ -20,7 +20,7 @@
from datasets.packaged_modules import _PACKAGED_DATASETS_MODULES
from datasets.utils.logging import get_logger
-from datasets.utils.metadata import DatasetMetadata, validate_metadata_type, yaml_block_from_readme
+from datasets.utils.metadata import DatasetMetadata
from datasets.utils.readme import ReadMe
from .utils import slow
@@ -46,7 +46,11 @@ def get_changed_datasets(repo_path: Path) -> List[Path]:
def get_all_datasets(repo_path: Path) -> List[Path]:
- dataset_names = [path.parts[-1] for path in (repo_path / "datasets").iterdir() if path.is_dir()]
+ dataset_names = [
+ path.parts[-1]
+ for path in (repo_path / "datasets").iterdir()
+ if path.is_dir() and (path / path.name).with_suffix(".py").is_file()
+ ]
return [dataset_name for dataset_name in dataset_names if dataset_name not in _PACKAGED_DATASETS_MODULES]
@@ -57,21 +61,20 @@ def test_changed_dataset_card(dataset_name):
assert card_path.exists()
error_messages = []
try:
- readme = ReadMe.from_readme(card_path)
+ ReadMe.from_readme(card_path)
except Exception as readme_parsing_error:
error_messages.append(
f"The following issues have been found in the dataset cards:\nREADME Parsing:\n{readme_parsing_error}"
)
try:
- readme = ReadMe.from_readme(card_path, suppress_parsing_errors=True)
- readme.validate()
+ ReadMe.from_readme(card_path, suppress_parsing_errors=True)
except Exception as readme_validation_error:
error_messages.append(
f"The following issues have been found in the dataset cards:\nREADME Validation:\n{readme_validation_error}"
)
try:
metadata = DatasetMetadata.from_readme(card_path)
- metadata.validate()
+ assert metadata, "empty metadata"
except Exception as metadata_error:
error_messages.append(
f"The following issues have been found in the dataset cards:\nYAML tags:\n{metadata_error}"
@@ -89,10 +92,8 @@ def test_dataset_card_yaml_structure(dataset_name):
"""
card_path = repo_path / "datasets" / dataset_name / "README.md"
assert card_path.exists()
- yaml_string = yaml_block_from_readme(card_path)
- metadata_dict = DatasetMetadata._metadata_dict_from_yaml_string(yaml_string)
+ metadata_dict = DatasetMetadata.from_readme(card_path)
assert len(metadata_dict) > 0
- validate_metadata_type(metadata_dict)
@slow
@@ -117,7 +118,7 @@ def test_dataset_card(dataset_name):
)
try:
metadata = DatasetMetadata.from_readme(card_path)
- metadata.validate()
+ assert metadata
except Exception as metadata_error:
error_messages.append(
f"The following issues have been found in the dataset cards:\nYAML tags:\n{metadata_error}"
diff --git a/tests/test_info.py b/tests/test_info.py
new file mode 100644
--- /dev/null
+++ b/tests/test_info.py
@@ -0,0 +1,110 @@
+import os
+
+import pytest
+import yaml
+
+from datasets.features.features import Features, Value
+from datasets.info import DatasetInfo, DatasetInfosDict
+
+
[email protected](
+ "dataset_info",
+ [
+ DatasetInfo(),
+ DatasetInfo(
+ description="foo",
+ features=Features({"a": Value("int32")}),
+ builder_name="builder",
+ config_name="config",
+ version="1.0.0",
+ splits=[{"name": "train"}],
+ download_size=42,
+ ),
+ ],
+)
+def test_dataset_info_dump_and_reload(tmp_path, dataset_info: DatasetInfo):
+ tmp_path = str(tmp_path)
+ dataset_info.write_to_directory(tmp_path)
+ reloaded = DatasetInfo.from_directory(tmp_path)
+ assert dataset_info == reloaded
+ assert os.path.exists(os.path.join(tmp_path, "dataset_info.json"))
+
+
+def test_dataset_info_to_yaml_dict():
+ dataset_info = DatasetInfo(
+ description="foo",
+ citation="bar",
+ homepage="https://foo.bar",
+ license="CC0",
+ features=Features({"a": Value("int32")}),
+ post_processed={},
+ supervised_keys=tuple(),
+ task_templates=[],
+ builder_name="builder",
+ config_name="config",
+ version="1.0.0",
+ splits=[{"name": "train", "num_examples": 42}],
+ download_checksums={},
+ download_size=1337,
+ post_processing_size=442,
+ dataset_size=1234,
+ size_in_bytes=1337 + 442 + 1234,
+ )
+ dataset_info_yaml_dict = dataset_info._to_yaml_dict()
+ assert sorted(dataset_info_yaml_dict) == sorted(DatasetInfo._INCLUDED_INFO_IN_YAML)
+ for key in DatasetInfo._INCLUDED_INFO_IN_YAML:
+ assert key in dataset_info_yaml_dict
+ assert isinstance(dataset_info_yaml_dict[key], (list, dict, int, str))
+ dataset_info_yaml = yaml.safe_dump(dataset_info_yaml_dict)
+ reloaded = yaml.safe_load(dataset_info_yaml)
+ assert dataset_info_yaml_dict == reloaded
+
+
+def test_dataset_info_to_yaml_dict_empty():
+ dataset_info = DatasetInfo()
+ dataset_info_yaml_dict = dataset_info._to_yaml_dict()
+ assert dataset_info_yaml_dict == {}
+
+
[email protected](
+ "dataset_infos_dict",
+ [
+ DatasetInfosDict(),
+ DatasetInfosDict({"default": DatasetInfo()}),
+ DatasetInfosDict({"my_config_name": DatasetInfo()}),
+ DatasetInfosDict(
+ {
+ "default": DatasetInfo(
+ description="foo",
+ features=Features({"a": Value("int32")}),
+ builder_name="builder",
+ config_name="config",
+ version="1.0.0",
+ splits=[{"name": "train"}],
+ download_size=42,
+ )
+ }
+ ),
+ DatasetInfosDict(
+ {
+ "v1": DatasetInfo(dataset_size=42),
+ "v2": DatasetInfo(dataset_size=1337),
+ }
+ ),
+ ],
+)
+def test_dataset_infos_dict_dump_and_reload(tmp_path, dataset_infos_dict: DatasetInfosDict):
+ tmp_path = str(tmp_path)
+ dataset_infos_dict.write_to_directory(tmp_path)
+ reloaded = DatasetInfosDict.from_directory(tmp_path)
+
+ # the config_name of the dataset_infos_dict take over the attribute
+ for config_name, dataset_info in dataset_infos_dict.items():
+ dataset_info.config_name = config_name
+ # the yaml representation doesn't include fields like description or citation
+ # so we just test that we can recover what we can from the yaml
+ dataset_infos_dict[config_name] = DatasetInfo._from_yaml_dict(dataset_info._to_yaml_dict())
+ assert dataset_infos_dict == reloaded
+
+ if dataset_infos_dict:
+ assert os.path.exists(os.path.join(tmp_path, "README.md"))
diff --git a/tests/test_metadata_util.py b/tests/test_metadata_util.py
--- a/tests/test_metadata_util.py
+++ b/tests/test_metadata_util.py
@@ -1,16 +1,9 @@
import re
import tempfile
import unittest
-from dataclasses import _MISSING_TYPE, asdict, fields
from pathlib import Path
-from datasets.utils.metadata import (
- DatasetMetadata,
- metadata_dict_from_readme,
- tagset_validator,
- validate_metadata_type,
- yaml_block_from_readme,
-)
+from datasets.utils.metadata import DatasetMetadata
def _dedent(string: str) -> str:
@@ -48,157 +41,26 @@ def _dedent(string: str) -> str:
class TestMetadataUtils(unittest.TestCase):
- def test_validate_metadata_type(self):
- metadata_dict = {
- "tag": ["list", "of", "values"],
- "another tag": ["Another", {"list"}, ["of"], 0x646D46736457567A],
- }
- with self.assertRaises(TypeError):
- validate_metadata_type(metadata_dict)
-
- metadata_dict = {"tag1": []}
- with self.assertRaises(TypeError):
- validate_metadata_type(metadata_dict)
-
- metadata_dict = {"tag1": None}
- with self.assertRaises(TypeError):
- validate_metadata_type(metadata_dict)
-
- def test_tagset_validator(self):
- name = "test_tag"
- url = "https://dummy.hf.co"
-
- items = ["tag1", "tag2", "tag2", "tag3"]
- reference_values = ["tag1", "tag2", "tag3"]
- returned_values, error = tagset_validator(items=items, reference_values=reference_values, name=name, url=url)
- self.assertListEqual(returned_values, items)
- self.assertIsNone(error)
-
- items = []
- reference_values = ["tag1", "tag2", "tag3"]
- items, error = tagset_validator(items=items, reference_values=reference_values, name=name, url=url)
- self.assertListEqual(items, [])
- self.assertIsNone(error)
-
- items = []
- reference_values = []
- returned_values, error = tagset_validator(items=items, reference_values=reference_values, name=name, url=url)
- self.assertListEqual(returned_values, [])
- self.assertIsNone(error)
-
- items = ["tag1", "tag2", "tag2", "tag3", "unknown tag"]
- reference_values = ["tag1", "tag2", "tag3"]
- returned_values, error = tagset_validator(items=items, reference_values=reference_values, name=name, url=url)
- self.assertListEqual(returned_values, [])
- self.assertEqual(error, f"{['unknown tag']} are not registered tags for '{name}', reference at {url}")
-
- def predicate_fn(string):
- return "ignore" in string
-
- items = ["process me", "process me too", "ignore me"]
- reference_values = ["process me too"]
- returned_values, error = tagset_validator(
- items=items,
- reference_values=reference_values,
- name=name,
- url=url,
- escape_validation_predicate_fn=predicate_fn,
- )
- self.assertListEqual(returned_values, [])
- self.assertEqual(error, f"{['process me']} are not registered tags for '{name}', reference at {url}")
-
- items = ["process me", "process me too", "ignore me"]
- reference_values = ["process me too", "process me"]
- returned_values, error = tagset_validator(
- items=items,
- reference_values=reference_values,
- name=name,
- url=url,
- escape_validation_predicate_fn=predicate_fn,
- )
- self.assertListEqual(returned_values, items)
- self.assertIsNone(error)
-
- items = ["ignore me too", "ignore me"]
- reference_values = ["process me too"]
- returned_values, error = tagset_validator(
- items=items,
- reference_values=reference_values,
- name=name,
- url=url,
- escape_validation_predicate_fn=predicate_fn,
- )
- self.assertListEqual(returned_values, items)
- self.assertIsNone(error)
-
- def test_yaml_block_from_readme(self):
- with tempfile.TemporaryDirectory() as tmp_dir:
- path = Path(tmp_dir) / "README.md"
-
- with open(path, "w+") as readme_file:
- readme_file.write(README_YAML)
- yaml_block = yaml_block_from_readme(path=path)
- self.assertEqual(
- yaml_block,
- _dedent(
- """\
- language:
- - zh
- - en
- task_ids:
- - sentiment-classification
- """
- ),
- )
-
- with open(path, "w+") as readme_file:
- readme_file.write(README_EMPTY_YAML)
- yaml_block = yaml_block_from_readme(path=path)
- self.assertEqual(
- yaml_block,
- _dedent(
- """\
- """
- ),
- )
-
- with open(path, "w+") as readme_file:
- readme_file.write(README_NO_YAML)
- yaml_block = yaml_block_from_readme(path=path)
- self.assertIsNone(yaml_block)
-
def test_metadata_dict_from_readme(self):
with tempfile.TemporaryDirectory() as tmp_dir:
path = Path(tmp_dir) / "README.md"
with open(path, "w+") as readme_file:
readme_file.write(README_YAML)
- metadata_dict = metadata_dict_from_readme(path)
+ metadata_dict = DatasetMetadata.from_readme(path)
self.assertDictEqual(metadata_dict, {"language": ["zh", "en"], "task_ids": ["sentiment-classification"]})
with open(path, "w+") as readme_file:
readme_file.write(README_EMPTY_YAML)
- metadata_dict = metadata_dict_from_readme(path)
+ metadata_dict = DatasetMetadata.from_readme(path)
self.assertDictEqual(metadata_dict, {})
with open(path, "w+") as readme_file:
readme_file.write(README_NO_YAML)
- metadata_dict = metadata_dict_from_readme(path)
- self.assertIsNone(metadata_dict)
+ metadata_dict = DatasetMetadata.from_readme(path)
+ self.assertEqual(metadata_dict, {})
def test_from_yaml_string(self):
- default_optional_keys = {
- field.name: field.default
- for field in fields(DatasetMetadata)
- if type(field.default) is not _MISSING_TYPE and field.name not in DatasetMetadata._DEPRECATED_YAML_KEYS
- }
-
- default_deprecated_keys = {
- field.name: field.default
- for field in fields(DatasetMetadata)
- if field.name in DatasetMetadata._DEPRECATED_YAML_KEYS
- }
-
valid_yaml_string = _dedent(
"""\
annotations_creators:
@@ -222,87 +84,7 @@ def test_from_yaml_string(self):
- open-domain-qa
"""
)
- DatasetMetadata.from_yaml_string(valid_yaml_string)
-
- valid_yaml_string_with_configs = _dedent(
- """\
- annotations_creators:
- - found
- language_creators:
- - found
- language:
- en:
- - en
- fr:
- - fr
- license:
- - unknown
- multilinguality:
- - monolingual
- pretty_name: Test Dataset
- size_categories:
- - 10K<n<100K
- source_datasets:
- - extended|other-yahoo-webscope-l6
- task_categories:
- - question-answering
- task_ids:
- - open-domain-qa
- """
- )
- DatasetMetadata.from_yaml_string(valid_yaml_string_with_configs)
-
- invalid_tag_yaml = _dedent(
- """\
- annotations_creators:
- - found
- language_creators:
- - some guys in Panama
- language:
- - en
- license:
- - unknown
- multilinguality:
- - monolingual
- pretty_name: Test Dataset
- size_categories:
- - 10K<n<100K
- source_datasets:
- - extended|other-yahoo-webscope-l6
- task_categories:
- - question-answering
- task_ids:
- - open-domain-qa
- """
- )
- with self.assertRaises(TypeError):
- metadata = DatasetMetadata.from_yaml_string(invalid_tag_yaml)
- metadata.validate()
-
- missing_tag_yaml = _dedent(
- """\
- annotations_creators:
- - found
- language:
- - en
- license:
- - unknown
- multilinguality:
- - monolingual
- pretty_name: Test Dataset
- size_categories:
- - 10K<n<100K
- source_datasets:
- - extended|other-yahoo-webscope-l6
- task_categories:
- - question-answering
- task_ids:
- - open-domain-qa
- """
- )
- with self.assertRaises(TypeError):
- metadata = DatasetMetadata.from_yaml_string(missing_tag_yaml)
- metadata.validate()
+ assert DatasetMetadata.from_yaml_string(valid_yaml_string)
duplicate_yaml_keys = _dedent(
"""\
@@ -328,119 +110,7 @@ def test_from_yaml_string(self):
"""
)
with self.assertRaises(TypeError):
- metadata = DatasetMetadata.from_yaml_string(duplicate_yaml_keys)
- metadata.validate()
-
- valid_yaml_string_with_duplicate_configs = _dedent(
- """\
- annotations_creators:
- - found
- language_creators:
- - found
- language:
- en:
- - en
- en:
- - en
- license:
- - unknown
- multilinguality:
- - monolingual
- pretty_name: Test Dataset
- size_categories:
- - 10K<n<100K
- source_datasets:
- - extended|other-yahoo-webscope-l6
- task_categories:
- - question-answering
- task_ids:
- - open-domain-qa
- """
- )
- with self.assertRaises(TypeError):
- metadata = DatasetMetadata.from_yaml_string(valid_yaml_string_with_duplicate_configs)
- metadata.validate()
-
- valid_yaml_string_with_paperswithcode_id = _dedent(
- """\
- annotations_creators:
- - found
- language_creators:
- - found
- language:
- - en
- license:
- - unknown
- multilinguality:
- - monolingual
- pretty_name: Test Dataset
- size_categories:
- - 10K<n<100K
- source_datasets:
- - extended|other-yahoo-webscope-l6
- task_categories:
- - question-answering
- task_ids:
- - open-domain-qa
- paperswithcode_id: squad
- """
- )
- DatasetMetadata.from_yaml_string(valid_yaml_string_with_paperswithcode_id)
-
- valid_yaml_string_with_null_paperswithcode_id = _dedent(
- """\
- annotations_creators:
- - found
- language_creators:
- - found
- language:
- - en
- license:
- - unknown
- multilinguality:
- - monolingual
- pretty_name: Test Dataset
- size_categories:
- - 10K<n<100K
- source_datasets:
- - extended|other-yahoo-webscope-l6
- task_categories:
- - question-answering
- task_ids:
- - open-domain-qa
- paperswithcode_id: null
- """
- )
- DatasetMetadata.from_yaml_string(valid_yaml_string_with_null_paperswithcode_id)
-
- valid_yaml_string_with_list_paperswithcode_id = _dedent(
- """\
- annotations_creators:
- - found
- language_creators:
- - found
- language:
- - en
- license:
- - unknown
- multilinguality:
- - monolingual
- pretty_name: Test Dataset
- size_categories:
- - 10K<n<100K
- source_datasets:
- - extended|other-yahoo-webscope-l6
- task_categories:
- - question-answering
- task_ids:
- - open-domain-qa
- paperswithcode_id:
- - squad
- """
- )
- with self.assertRaises(TypeError):
- metadata = DatasetMetadata.from_yaml_string(valid_yaml_string_with_list_paperswithcode_id)
- metadata.validate()
+ DatasetMetadata.from_yaml_string(duplicate_yaml_keys)
valid_yaml_with_optional_keys = _dedent(
"""\
@@ -490,53 +160,4 @@ def test_from_yaml_string(self):
I agree to use this model for non-commerical use ONLY: checkbox
"""
)
-
- metadata = DatasetMetadata.from_yaml_string(valid_yaml_with_optional_keys)
- metadata_dict = asdict(metadata)
- expected = {
- **default_optional_keys,
- **default_deprecated_keys,
- "annotations_creators": ["found"],
- "language_creators": ["found"],
- "language": ["en"],
- "license": ["unknown"],
- "multilinguality": ["monolingual"],
- "pretty_name": "Test Dataset",
- "size_categories": ["10K<n<100K"],
- "source_datasets": ["extended|other-yahoo-webscope-l6"],
- "task_categories": ["text-classification"],
- "task_ids": ["multi-class-classification"],
- "paperswithcode_id": ["squad"],
- "configs": ["en"],
- "train_eval_index": [
- {
- "config": "en",
- "task": "text-classification",
- "task_id": "multi_class_classification",
- "splits": {"train_split": "train", "eval_split": "test"},
- "col_mapping": {"text": "text", "label": "target"},
- "metrics": [{"type": "accuracy", "name": "Accuracy"}],
- },
- ],
- "extra_gated_prompt": (
- "By clicking on “Access repository” below, you also agree to ImageNet Terms of Access:\n"
- '[RESEARCHER_FULLNAME] (the "Researcher") has requested permission to use the ImageNet database '
- '(the "Database") at Princeton University and Stanford University. In exchange for such permission, '
- "Researcher hereby agrees to the following terms and conditions:\n"
- "1. Researcher shall use the Database only for non-commercial research and educational purposes.\n"
- ),
- "extra_gated_fields": {
- "Company": "text",
- "Country": "text",
- "I agree to use this model for non-commerical use ONLY": "checkbox",
- },
- }
- self.assertEqual(sorted(metadata_dict), sorted(expected))
- for key in expected:
- if key == "train_eval_index":
- self.assertEqual(len(metadata_dict[key]), len(expected[key]))
- for tei, expected_tei in zip(metadata_dict[key], expected[key]):
- for subkey in expected_tei:
- self.assertEqual(tei[subkey], expected_tei[subkey], msg=f"failed at {subkey}")
- else:
- self.assertEqual(metadata_dict[key], expected[key], msg=f"failed at {key}")
+ assert DatasetMetadata.from_yaml_string(valid_yaml_with_optional_keys)
diff --git a/tests/test_splits.py b/tests/test_splits.py
new file mode 100644
--- /dev/null
+++ b/tests/test_splits.py
@@ -0,0 +1,36 @@
+import pytest
+
+from datasets.splits import SplitDict, SplitInfo
+from datasets.utils.py_utils import asdict
+
+
[email protected](
+ "split_dict",
+ [
+ SplitDict(),
+ SplitDict({"train": SplitInfo(name="train", num_bytes=1337, num_examples=42, dataset_name="my_dataset")}),
+ SplitDict({"train": SplitInfo(name="train", num_bytes=1337, num_examples=42)}),
+ SplitDict({"train": SplitInfo()}),
+ ],
+)
+def test_split_dict_to_yaml_list(split_dict: SplitDict):
+ split_dict_yaml_list = split_dict._to_yaml_list()
+ assert len(split_dict_yaml_list) == len(split_dict)
+ reloaded = SplitDict._from_yaml_list(split_dict_yaml_list)
+ for split_name, split_info in split_dict.items():
+ # dataset_name field is deprecated, and is therefore not part of the YAML dump
+ split_info.dataset_name = None
+ # the split name of split_dict takes over the name of the split info object
+ split_info.name = split_name
+ assert split_dict == reloaded
+
+
[email protected](
+ "split_info", [SplitInfo(), SplitInfo(dataset_name=None), SplitInfo(dataset_name="my_dataset")]
+)
+def test_split_dict_asdict_has_dataset_name(split_info):
+ # For backward compatibility, we need asdict(split_dict) to return split info dictrionaries with the "dataset_name"
+ # field even if it's deprecated. This way old versionso of `datasets` can still reload dataset_infos.json files
+ split_dict_asdict = asdict(SplitDict({"train": split_info}))
+ assert "dataset_name" in split_dict_asdict["train"]
+ assert split_dict_asdict["train"]["dataset_name"] == split_info.dataset_name
diff --git a/tests/test_upstream_hub.py b/tests/test_upstream_hub.py
--- a/tests/test_upstream_hub.py
+++ b/tests/test_upstream_hub.py
@@ -42,7 +42,7 @@ def test_push_dataset_dict_to_hub_no_token(self, temporary_repo):
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
- files, [".gitattributes", "data/train-00000-of-00001-*.parquet", "dataset_infos.json"]
+ files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
@@ -64,7 +64,7 @@ def test_push_dataset_dict_to_hub_name_without_namespace(self, temporary_repo):
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
- files, [".gitattributes", "data/train-00000-of-00001-*.parquet", "dataset_infos.json"]
+ files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
@@ -100,7 +100,7 @@ def test_push_dataset_dict_to_hub_private(self, temporary_repo):
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
- files, [".gitattributes", "data/train-00000-of-00001-*.parquet", "dataset_infos.json"]
+ files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
@@ -122,7 +122,7 @@ def test_push_dataset_dict_to_hub(self, temporary_repo):
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
- files, [".gitattributes", "data/train-00000-of-00001-*.parquet", "dataset_infos.json"]
+ files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
@@ -148,9 +148,9 @@ def test_push_dataset_dict_to_hub_multiple_files(self, temporary_repo):
files,
[
".gitattributes",
+ "README.md",
"data/train-00000-of-00002-*.parquet",
"data/train-00001-of-00002-*.parquet",
- "dataset_infos.json",
],
)
)
@@ -176,9 +176,9 @@ def test_push_dataset_dict_to_hub_multiple_files_with_max_shard_size(self, tempo
files,
[
".gitattributes",
+ "README.md",
"data/train-00000-of-00002-*.parquet",
"data/train-00001-of-00002-*.parquet",
- "dataset_infos.json",
],
)
)
@@ -221,11 +221,11 @@ def test_push_dataset_dict_to_hub_overwrite_files(self, temporary_repo):
files,
[
".gitattributes",
+ "README.md",
"data/random-00000-of-00001-*.parquet",
"data/train-00000-of-00002-*.parquet",
"data/train-00001-of-00002-*.parquet",
"datafile.txt",
- "dataset_infos.json",
],
)
)
@@ -268,10 +268,10 @@ def test_push_dataset_dict_to_hub_overwrite_files(self, temporary_repo):
files,
[
".gitattributes",
+ "README.md",
"data/random-00000-of-00001-*.parquet",
"data/train-00000-of-00001-*.parquet",
"datafile.txt",
- "dataset_infos.json",
],
)
)
| Remove dataset_infos.json
**Is your feature request related to a problem? Please describe.**
As discussed, there are infos in the `dataset_infos.json` which are redundant and we could have them only in the README file.
Others could be migrated to the README, like: "dataset_size", "size_in_bytes", "download_size", "splits.split_name.[num_bytes, num_examples]",...
However, there are others that do not seem too meaningful in the README, like the checksums.
**Describe the solution you'd like**
Open a discussion to decide what to do with the `dataset_infos.json` files: which information to be migrated and/or which information to be kept.
cc: @julien-c @lhoestq
| 2022-09-02T16:10:05Z | [] | [] |
|
huggingface/datasets | 4,928 | huggingface__datasets-4928 | [
"3094"
] | 98dec7013288df9230b69cd851dfa7952ed361c9 | diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -155,6 +155,7 @@
"scipy",
"sentencepiece", # for bleurt
"seqeval",
+ "sqlalchemy",
"tldextract",
# to speed up pip backtracking
"toml>=0.10.1",
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -111,6 +111,10 @@
if TYPE_CHECKING:
+ import sqlite3
+
+ import sqlalchemy
+
from .dataset_dict import DatasetDict
logger = logging.get_logger(__name__)
@@ -1092,6 +1096,56 @@ def from_text(
path_or_paths, split=split, features=features, cache_dir=cache_dir, keep_in_memory=keep_in_memory, **kwargs
).read()
+ @staticmethod
+ def from_sql(
+ sql: Union[str, "sqlalchemy.sql.Selectable"],
+ con: str,
+ features: Optional[Features] = None,
+ cache_dir: str = None,
+ keep_in_memory: bool = False,
+ **kwargs,
+ ):
+ """Create Dataset from SQL query or database table.
+
+ Args:
+ sql (`str` or :obj:`sqlalchemy.sql.Selectable`): SQL query to be executed or a table name.
+ con (`str`): A connection URI string used to instantiate a database connection.
+ features (:class:`Features`, optional): Dataset features.
+ cache_dir (:obj:`str`, optional, default ``"~/.cache/huggingface/datasets"``): Directory to cache data.
+ keep_in_memory (:obj:`bool`, default ``False``): Whether to copy the data in-memory.
+ **kwargs (additional keyword arguments): Keyword arguments to be passed to :class:`SqlConfig`.
+
+ Returns:
+ :class:`Dataset`
+
+ Example:
+
+ ```py
+ >>> # Fetch a database table
+ >>> ds = Dataset.from_sql("test_data", "postgres:///db_name")
+ >>> # Execute a SQL query on the table
+ >>> ds = Dataset.from_sql("SELECT sentence FROM test_data", "postgres:///db_name")
+ >>> # Use a Selectable object to specify the query
+ >>> from sqlalchemy import select, text
+ >>> stmt = select([text("sentence")]).select_from(text("test_data"))
+ >>> ds = Dataset.from_sql(stmt, "postgres:///db_name")
+ ```
+
+ <Tip {warning=true}>
+ `sqlalchemy` needs to be installed to use this function.
+ </Tip>
+ """
+ from .io.sql import SqlDatasetReader
+
+ return SqlDatasetReader(
+ sql,
+ con,
+ features=features,
+ cache_dir=cache_dir,
+ keep_in_memory=keep_in_memory,
+ **kwargs,
+ ).read()
+
def __del__(self):
if hasattr(self, "_data"):
del self._data
@@ -4098,6 +4152,43 @@ def to_parquet(
return ParquetDatasetWriter(self, path_or_buf, batch_size=batch_size, **parquet_writer_kwargs).write()
+ def to_sql(
+ self,
+ name: str,
+ con: Union[str, "sqlalchemy.engine.Connection", "sqlalchemy.engine.Engine", "sqlite3.Connection"],
+ batch_size: Optional[int] = None,
+ **sql_writer_kwargs,
+ ) -> int:
+ """Exports the dataset to a SQL database.
+
+ Args:
+ name (`str`): Name of SQL table.
+ con (`str` or `sqlite3.Connection` or `sqlalchemy.engine.Connection` or `sqlalchemy.engine.Connection`):
+ A database connection URI string or an existing SQLite3/SQLAlchemy connection used to write to a database.
+ batch_size (:obj:`int`, optional): Size of the batch to load in memory and write at once.
+ Defaults to :obj:`datasets.config.DEFAULT_MAX_BATCH_SIZE`.
+ **sql_writer_kwargs (additional keyword arguments): Parameters to pass to pandas's :function:`Dataframe.to_sql`
+
+ Returns:
+ int: The number of records written.
+
+ Example:
+
+ ```py
+ >>> # con provided as a connection URI string
+ >>> ds.to_sql("data", "sqlite:///my_own_db.sql")
+ >>> # con provided as a sqlite3 connection object
+ >>> import sqlite3
+ >>> con = sqlite3.connect("my_own_db.sql")
+ >>> with con:
+ ... ds.to_sql("data", con)
+ ```
+ """
+ # Dynamic import to avoid circular dependency
+ from .io.sql import SqlDatasetWriter
+
+ return SqlDatasetWriter(self, name, con, batch_size=batch_size, **sql_writer_kwargs).write()
+
def _push_parquet_shards_to_hub(
self,
repo_id: str,
diff --git a/src/datasets/config.py b/src/datasets/config.py
--- a/src/datasets/config.py
+++ b/src/datasets/config.py
@@ -125,6 +125,9 @@
logger.info("Disabling Apache Beam because USE_BEAM is set to False")
+# Optional tools for data loading
+SQLALCHEMY_AVAILABLE = importlib.util.find_spec("sqlalchemy") is not None
+
# Optional tools for feature decoding
PIL_AVAILABLE = importlib.util.find_spec("PIL") is not None
diff --git a/src/datasets/io/sql.py b/src/datasets/io/sql.py
new file mode 100644
--- /dev/null
+++ b/src/datasets/io/sql.py
@@ -0,0 +1,129 @@
+import multiprocessing
+from typing import TYPE_CHECKING, Optional, Union
+
+from .. import Dataset, Features, config
+from ..formatting import query_table
+from ..packaged_modules.sql.sql import Sql
+from ..utils import logging
+from .abc import AbstractDatasetInputStream
+
+
+if TYPE_CHECKING:
+ import sqlite3
+
+ import sqlalchemy
+
+
+class SqlDatasetReader(AbstractDatasetInputStream):
+ def __init__(
+ self,
+ sql: Union[str, "sqlalchemy.sql.Selectable"],
+ con: str,
+ features: Optional[Features] = None,
+ cache_dir: str = None,
+ keep_in_memory: bool = False,
+ **kwargs,
+ ):
+ super().__init__(features=features, cache_dir=cache_dir, keep_in_memory=keep_in_memory, **kwargs)
+ self.builder = Sql(
+ cache_dir=cache_dir,
+ features=features,
+ sql=sql,
+ con=con,
+ **kwargs,
+ )
+
+ def read(self):
+ download_config = None
+ download_mode = None
+ ignore_verifications = False
+ use_auth_token = None
+ base_path = None
+
+ self.builder.download_and_prepare(
+ download_config=download_config,
+ download_mode=download_mode,
+ ignore_verifications=ignore_verifications,
+ # try_from_hf_gcs=try_from_hf_gcs,
+ base_path=base_path,
+ use_auth_token=use_auth_token,
+ )
+
+ # Build dataset for splits
+ dataset = self.builder.as_dataset(
+ split="train", ignore_verifications=ignore_verifications, in_memory=self.keep_in_memory
+ )
+ return dataset
+
+
+class SqlDatasetWriter:
+ def __init__(
+ self,
+ dataset: Dataset,
+ name: str,
+ con: Union[str, "sqlalchemy.engine.Connection", "sqlalchemy.engine.Engine", "sqlite3.Connection"],
+ batch_size: Optional[int] = None,
+ num_proc: Optional[int] = None,
+ **to_sql_kwargs,
+ ):
+
+ if num_proc is not None and num_proc <= 0:
+ raise ValueError(f"num_proc {num_proc} must be an integer > 0.")
+
+ self.dataset = dataset
+ self.name = name
+ self.con = con
+ self.batch_size = batch_size if batch_size else config.DEFAULT_MAX_BATCH_SIZE
+ self.num_proc = num_proc
+ self.to_sql_kwargs = to_sql_kwargs
+
+ def write(self) -> int:
+ _ = self.to_sql_kwargs.pop("sql", None)
+ _ = self.to_sql_kwargs.pop("con", None)
+
+ written = self._write(**self.to_sql_kwargs)
+ return written
+
+ def _batch_sql(self, args):
+ offset, to_sql_kwargs = args
+ to_sql_kwargs = {**to_sql_kwargs, "if_exists": "append"} if offset > 0 else to_sql_kwargs
+ batch = query_table(
+ table=self.dataset.data,
+ key=slice(offset, offset + self.batch_size),
+ indices=self.dataset._indices,
+ )
+ df = batch.to_pandas()
+ num_rows = df.to_sql(self.name, self.con, **to_sql_kwargs)
+ return num_rows or len(df)
+
+ def _write(self, **to_sql_kwargs) -> int:
+ """Writes the pyarrow table as SQL to a database.
+
+ Caller is responsible for opening and closing the SQL connection.
+ """
+ written = 0
+
+ if self.num_proc is None or self.num_proc == 1:
+ for offset in logging.tqdm(
+ range(0, len(self.dataset), self.batch_size),
+ unit="ba",
+ disable=not logging.is_progress_bar_enabled(),
+ desc="Creating SQL from Arrow format",
+ ):
+ written += self._batch_sql((offset, to_sql_kwargs))
+ else:
+ num_rows, batch_size = len(self.dataset), self.batch_size
+ with multiprocessing.Pool(self.num_proc) as pool:
+ for num_rows in logging.tqdm(
+ pool.imap(
+ self._batch_sql,
+ [(offset, to_sql_kwargs) for offset in range(0, num_rows, batch_size)],
+ ),
+ total=(num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size,
+ unit="ba",
+ disable=not logging.is_progress_bar_enabled(),
+ desc="Creating SQL from Arrow format",
+ ):
+ written += num_rows
+
+ return written
diff --git a/src/datasets/packaged_modules/__init__.py b/src/datasets/packaged_modules/__init__.py
--- a/src/datasets/packaged_modules/__init__.py
+++ b/src/datasets/packaged_modules/__init__.py
@@ -9,6 +9,7 @@
from .json import json
from .pandas import pandas
from .parquet import parquet
+from .sql import sql # noqa F401
from .text import text
diff --git a/src/datasets/packaged_modules/csv/csv.py b/src/datasets/packaged_modules/csv/csv.py
--- a/src/datasets/packaged_modules/csv/csv.py
+++ b/src/datasets/packaged_modules/csv/csv.py
@@ -70,8 +70,8 @@ def __post_init__(self):
self.names = self.column_names
@property
- def read_csv_kwargs(self):
- read_csv_kwargs = dict(
+ def pd_read_csv_kwargs(self):
+ pd_read_csv_kwargs = dict(
sep=self.sep,
header=self.header,
names=self.names,
@@ -112,16 +112,16 @@ def read_csv_kwargs(self):
# some kwargs must not be passed if they don't have a default value
# some others are deprecated and we can also not pass them if they are the default value
- for read_csv_parameter in _PANDAS_READ_CSV_NO_DEFAULT_PARAMETERS + _PANDAS_READ_CSV_DEPRECATED_PARAMETERS:
- if read_csv_kwargs[read_csv_parameter] == getattr(CsvConfig(), read_csv_parameter):
- del read_csv_kwargs[read_csv_parameter]
+ for pd_read_csv_parameter in _PANDAS_READ_CSV_NO_DEFAULT_PARAMETERS + _PANDAS_READ_CSV_DEPRECATED_PARAMETERS:
+ if pd_read_csv_kwargs[pd_read_csv_parameter] == getattr(CsvConfig(), pd_read_csv_parameter):
+ del pd_read_csv_kwargs[pd_read_csv_parameter]
# Remove 1.3 new arguments
if not (datasets.config.PANDAS_VERSION.major >= 1 and datasets.config.PANDAS_VERSION.minor >= 3):
- for read_csv_parameter in _PANDAS_READ_CSV_NEW_1_3_0_PARAMETERS:
- del read_csv_kwargs[read_csv_parameter]
+ for pd_read_csv_parameter in _PANDAS_READ_CSV_NEW_1_3_0_PARAMETERS:
+ del pd_read_csv_kwargs[pd_read_csv_parameter]
- return read_csv_kwargs
+ return pd_read_csv_kwargs
class Csv(datasets.ArrowBasedBuilder):
@@ -172,7 +172,7 @@ def _generate_tables(self, files):
else None
)
for file_idx, file in enumerate(itertools.chain.from_iterable(files)):
- csv_file_reader = pd.read_csv(file, iterator=True, dtype=dtype, **self.config.read_csv_kwargs)
+ csv_file_reader = pd.read_csv(file, iterator=True, dtype=dtype, **self.config.pd_read_csv_kwargs)
try:
for batch_idx, df in enumerate(csv_file_reader):
pa_table = pa.Table.from_pandas(df)
diff --git a/src/datasets/packaged_modules/sql/__init__.py b/src/datasets/packaged_modules/sql/__init__.py
new file mode 100644
diff --git a/src/datasets/packaged_modules/sql/sql.py b/src/datasets/packaged_modules/sql/sql.py
new file mode 100644
--- /dev/null
+++ b/src/datasets/packaged_modules/sql/sql.py
@@ -0,0 +1,108 @@
+import sys
+from dataclasses import dataclass
+from typing import TYPE_CHECKING, Dict, List, Optional, Tuple, Union
+
+import pandas as pd
+import pyarrow as pa
+
+import datasets
+import datasets.config
+from datasets.features.features import require_storage_cast
+from datasets.table import table_cast
+
+
+if TYPE_CHECKING:
+ import sqlalchemy
+
+
+@dataclass
+class SqlConfig(datasets.BuilderConfig):
+ """BuilderConfig for SQL."""
+
+ sql: Union[str, "sqlalchemy.sql.Selectable"] = None
+ con: str = None
+ index_col: Optional[Union[str, List[str]]] = None
+ coerce_float: bool = True
+ params: Optional[Union[List, Tuple, Dict]] = None
+ parse_dates: Optional[Union[List, Dict]] = None
+ columns: Optional[List[str]] = None
+ chunksize: Optional[int] = 10_000
+ features: Optional[datasets.Features] = None
+
+ def __post_init__(self):
+ if self.sql is None:
+ raise ValueError("sql must be specified")
+ if self.con is None:
+ raise ValueError("con must be specified")
+ if not isinstance(self.con, str):
+ raise ValueError(f"con must be a database URI string, but got {self.con} with type {type(self.con)}.")
+
+ def create_config_id(
+ self,
+ config_kwargs: dict,
+ custom_features: Optional[datasets.Features] = None,
+ ) -> str:
+ # We need to stringify the Selectable object to make its hash deterministic
+
+ # The process of stringifying is explained here: http://docs.sqlalchemy.org/en/latest/faq/sqlexpressions.html
+ sql = config_kwargs["sql"]
+ if not isinstance(sql, str):
+ if datasets.config.SQLALCHEMY_AVAILABLE and "sqlalchemy" in sys.modules:
+ import sqlalchemy
+
+ if isinstance(sql, sqlalchemy.sql.Selectable):
+ config_kwargs = config_kwargs.copy()
+ engine = sqlalchemy.create_engine(config_kwargs["con"].split("://")[0] + "://")
+ sql_str = str(sql.compile(dialect=engine.dialect))
+ config_kwargs["sql"] = sql_str
+ else:
+ raise TypeError(
+ f"Supported types for 'sql' are string and sqlalchemy.sql.Selectable but got {type(sql)}: {sql}"
+ )
+ else:
+ raise TypeError(
+ f"Supported types for 'sql' are string and sqlalchemy.sql.Selectable but got {type(sql)}: {sql}"
+ )
+ return super().create_config_id(config_kwargs, custom_features=custom_features)
+
+ @property
+ def pd_read_sql_kwargs(self):
+ pd_read_sql_kwargs = dict(
+ index_col=self.index_col,
+ columns=self.columns,
+ params=self.params,
+ coerce_float=self.coerce_float,
+ parse_dates=self.parse_dates,
+ )
+ return pd_read_sql_kwargs
+
+
+class Sql(datasets.ArrowBasedBuilder):
+ BUILDER_CONFIG_CLASS = SqlConfig
+
+ def _info(self):
+ return datasets.DatasetInfo(features=self.config.features)
+
+ def _split_generators(self, dl_manager):
+ return [datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={})]
+
+ def _cast_table(self, pa_table: pa.Table) -> pa.Table:
+ if self.config.features is not None:
+ schema = self.config.features.arrow_schema
+ if all(not require_storage_cast(feature) for feature in self.config.features.values()):
+ # cheaper cast
+ pa_table = pa.Table.from_arrays([pa_table[field.name] for field in schema], schema=schema)
+ else:
+ # more expensive cast; allows str <-> int/float or str to Audio for example
+ pa_table = table_cast(pa_table, schema)
+ return pa_table
+
+ def _generate_tables(self):
+ chunksize = self.config.chunksize
+ sql_reader = pd.read_sql(
+ self.config.sql, self.config.con, chunksize=chunksize, **self.config.pd_read_sql_kwargs
+ )
+ sql_reader = [sql_reader] if chunksize is None else sql_reader
+ for chunk_idx, df in enumerate(sql_reader):
+ pa_table = pa.Table.from_pandas(df)
+ yield chunk_idx, self._cast_table(pa_table)
| diff --git a/tests/fixtures/files.py b/tests/fixtures/files.py
--- a/tests/fixtures/files.py
+++ b/tests/fixtures/files.py
@@ -1,6 +1,8 @@
+import contextlib
import csv
import json
import os
+import sqlite3
import tarfile
import textwrap
import zipfile
@@ -238,6 +240,18 @@ def arrow_path(tmp_path_factory):
return path
[email protected](scope="session")
+def sqlite_path(tmp_path_factory):
+ path = str(tmp_path_factory.mktemp("data") / "dataset.sqlite")
+ with contextlib.closing(sqlite3.connect(path)) as con:
+ cur = con.cursor()
+ cur.execute("CREATE TABLE dataset(col_1 text, col_2 int, col_3 real)")
+ for item in DATA:
+ cur.execute("INSERT INTO dataset(col_1, col_2, col_3) VALUES (?, ?, ?)", tuple(item.values()))
+ con.commit()
+ return path
+
+
@pytest.fixture(scope="session")
def csv_path(tmp_path_factory):
path = str(tmp_path_factory.mktemp("data") / "dataset.csv")
diff --git a/tests/io/test_sql.py b/tests/io/test_sql.py
new file mode 100644
--- /dev/null
+++ b/tests/io/test_sql.py
@@ -0,0 +1,98 @@
+import contextlib
+import os
+import sqlite3
+
+import pytest
+
+from datasets import Dataset, Features, Value
+from datasets.io.sql import SqlDatasetReader, SqlDatasetWriter
+
+from ..utils import assert_arrow_memory_doesnt_increase, assert_arrow_memory_increases, require_sqlalchemy
+
+
+def _check_sql_dataset(dataset, expected_features):
+ assert isinstance(dataset, Dataset)
+ assert dataset.num_rows == 4
+ assert dataset.num_columns == 3
+ assert dataset.column_names == ["col_1", "col_2", "col_3"]
+ for feature, expected_dtype in expected_features.items():
+ assert dataset.features[feature].dtype == expected_dtype
+
+
+@require_sqlalchemy
[email protected]("keep_in_memory", [False, True])
+def test_dataset_from_sql_keep_in_memory(keep_in_memory, sqlite_path, tmp_path):
+ cache_dir = tmp_path / "cache"
+ expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
+ with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
+ dataset = SqlDatasetReader(
+ "dataset", "sqlite:///" + sqlite_path, cache_dir=cache_dir, keep_in_memory=keep_in_memory
+ ).read()
+ _check_sql_dataset(dataset, expected_features)
+
+
+@require_sqlalchemy
[email protected](
+ "features",
+ [
+ None,
+ {"col_1": "string", "col_2": "int64", "col_3": "float64"},
+ {"col_1": "string", "col_2": "string", "col_3": "string"},
+ {"col_1": "int32", "col_2": "int32", "col_3": "int32"},
+ {"col_1": "float32", "col_2": "float32", "col_3": "float32"},
+ ],
+)
+def test_dataset_from_sql_features(features, sqlite_path, tmp_path):
+ cache_dir = tmp_path / "cache"
+ default_expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
+ expected_features = features.copy() if features else default_expected_features
+ features = (
+ Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
+ )
+ dataset = SqlDatasetReader("dataset", "sqlite:///" + sqlite_path, features=features, cache_dir=cache_dir).read()
+ _check_sql_dataset(dataset, expected_features)
+
+
+def iter_sql_file(sqlite_path):
+ with contextlib.closing(sqlite3.connect(sqlite_path)) as con:
+ cur = con.cursor()
+ cur.execute("SELECT * FROM dataset")
+ for row in cur:
+ yield row
+
+
+@require_sqlalchemy
+def test_dataset_to_sql(sqlite_path, tmp_path):
+ cache_dir = tmp_path / "cache"
+ output_sqlite_path = os.path.join(cache_dir, "tmp.sql")
+ dataset = SqlDatasetReader("dataset", "sqlite:///" + sqlite_path, cache_dir=cache_dir).read()
+ SqlDatasetWriter(dataset, "dataset", "sqlite:///" + output_sqlite_path, index=False, num_proc=1).write()
+
+ original_sql = iter_sql_file(sqlite_path)
+ expected_sql = iter_sql_file(output_sqlite_path)
+
+ for row1, row2 in zip(original_sql, expected_sql):
+ assert row1 == row2
+
+
+@require_sqlalchemy
+def test_dataset_to_sql_multiproc(sqlite_path, tmp_path):
+ cache_dir = tmp_path / "cache"
+ output_sqlite_path = os.path.join(cache_dir, "tmp.sql")
+ dataset = SqlDatasetReader("dataset", "sqlite:///" + sqlite_path, cache_dir=cache_dir).read()
+ SqlDatasetWriter(dataset, "dataset", "sqlite:///" + output_sqlite_path, index=False, num_proc=2).write()
+
+ original_sql = iter_sql_file(sqlite_path)
+ expected_sql = iter_sql_file(output_sqlite_path)
+
+ for row1, row2 in zip(original_sql, expected_sql):
+ assert row1 == row2
+
+
+@require_sqlalchemy
+def test_dataset_to_sql_invalidproc(sqlite_path, tmp_path):
+ cache_dir = tmp_path / "cache"
+ output_sqlite_path = os.path.join(cache_dir, "tmp.sql")
+ dataset = SqlDatasetReader("dataset", "sqlite:///" + sqlite_path, cache_dir=cache_dir).read()
+ with pytest.raises(ValueError):
+ SqlDatasetWriter(dataset, "dataset", "sqlite:///" + output_sqlite_path, index=False, num_proc=0).write()
diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -1,3 +1,4 @@
+import contextlib
import copy
import itertools
import json
@@ -53,6 +54,7 @@
require_jax,
require_pil,
require_s3,
+ require_sqlalchemy,
require_tf,
require_torch,
require_transformers,
@@ -2049,6 +2051,68 @@ def test_to_parquet(self, in_memory):
self.assertEqual(parquet_dset.shape, dset.shape)
self.assertListEqual(list(parquet_dset.columns), list(dset.column_names))
+ @require_sqlalchemy
+ def test_to_sql(self, in_memory):
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ # Destionation specified as database URI string
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ file_path = os.path.join(tmp_dir, "test_path.sqlite")
+ _ = dset.to_sql("data", "sqlite:///" + file_path, index=False)
+
+ self.assertTrue(os.path.isfile(file_path))
+ sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
+
+ self.assertEqual(sql_dset.shape, dset.shape)
+ self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
+
+ # Destionation specified as sqlite3 connection
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ import sqlite3
+
+ file_path = os.path.join(tmp_dir, "test_path.sqlite")
+ with contextlib.closing(sqlite3.connect(file_path)) as con:
+ _ = dset.to_sql("data", con, index=False, if_exists="replace")
+
+ self.assertTrue(os.path.isfile(file_path))
+ sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
+
+ self.assertEqual(sql_dset.shape, dset.shape)
+ self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
+
+ # Test writing to a database in chunks
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ file_path = os.path.join(tmp_dir, "test_path.sqlite")
+ _ = dset.to_sql("data", "sqlite:///" + file_path, batch_size=1, index=False, if_exists="replace")
+
+ self.assertTrue(os.path.isfile(file_path))
+ sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
+
+ self.assertEqual(sql_dset.shape, dset.shape)
+ self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
+
+ # After a select/shuffle transform
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ dset = dset.select(range(0, len(dset), 2)).shuffle()
+ file_path = os.path.join(tmp_dir, "test_path.sqlite")
+ _ = dset.to_sql("data", "sqlite:///" + file_path, index=False, if_exists="replace")
+
+ self.assertTrue(os.path.isfile(file_path))
+ sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
+
+ self.assertEqual(sql_dset.shape, dset.shape)
+ self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
+
+ # With array features
+ with self._create_dummy_dataset(in_memory, tmp_dir, array_features=True) as dset:
+ file_path = os.path.join(tmp_dir, "test_path.sqlite")
+ _ = dset.to_sql("data", "sqlite:///" + file_path, index=False, if_exists="replace")
+
+ self.assertTrue(os.path.isfile(file_path))
+ sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
+
+ self.assertEqual(sql_dset.shape, dset.shape)
+ self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
+
def test_train_test_split(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
@@ -3249,6 +3313,49 @@ def test_dataset_from_generator_features(features, data_generator, tmp_path):
_check_generator_dataset(dataset, expected_features)
+def _check_sql_dataset(dataset, expected_features):
+ assert isinstance(dataset, Dataset)
+ assert dataset.num_rows == 4
+ assert dataset.num_columns == 3
+ assert dataset.column_names == ["col_1", "col_2", "col_3"]
+ for feature, expected_dtype in expected_features.items():
+ assert dataset.features[feature].dtype == expected_dtype
+
+
+@require_sqlalchemy
[email protected](
+ "features",
+ [
+ None,
+ {"col_1": "string", "col_2": "int64", "col_3": "float64"},
+ {"col_1": "string", "col_2": "string", "col_3": "string"},
+ {"col_1": "int32", "col_2": "int32", "col_3": "int32"},
+ {"col_1": "float32", "col_2": "float32", "col_3": "float32"},
+ ],
+)
+def test_dataset_from_sql_features(features, sqlite_path, tmp_path):
+ cache_dir = tmp_path / "cache"
+ default_expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
+ expected_features = features.copy() if features else default_expected_features
+ features = (
+ Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
+ )
+ dataset = Dataset.from_sql("dataset", "sqlite:///" + sqlite_path, features=features, cache_dir=cache_dir)
+ _check_sql_dataset(dataset, expected_features)
+
+
+@require_sqlalchemy
[email protected]("keep_in_memory", [False, True])
+def test_dataset_from_sql_keep_in_memory(keep_in_memory, sqlite_path, tmp_path):
+ cache_dir = tmp_path / "cache"
+ expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
+ with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
+ dataset = Dataset.from_sql(
+ "dataset", "sqlite:///" + sqlite_path, cache_dir=cache_dir, keep_in_memory=keep_in_memory
+ )
+ _check_sql_dataset(dataset, expected_features)
+
+
def test_dataset_to_json(dataset, tmp_path):
file_path = tmp_path / "test_path.jsonl"
bytes_written = dataset.to_json(path_or_buf=file_path)
diff --git a/tests/utils.py b/tests/utils.py
--- a/tests/utils.py
+++ b/tests/utils.py
@@ -130,6 +130,20 @@ def require_elasticsearch(test_case):
return test_case
+def require_sqlalchemy(test_case):
+ """
+ Decorator marking a test that requires SQLAlchemy.
+
+ These tests are skipped when SQLAlchemy isn't installed.
+
+ """
+ try:
+ import sqlalchemy # noqa
+ except ImportError:
+ test_case = unittest.skip("test requires sqlalchemy")(test_case)
+ return test_case
+
+
def require_torch(test_case):
"""
Decorator marking a test that requires PyTorch.
| Support loading a dataset from SQLite files
As requested by @julien-c, we could eventually support loading a dataset from SQLite files, like it is the case for JSON/CSV files.
| for reference Kaggle has a good number of open source datasets stored in sqlite
Alternatively a tutorial or tool on how to convert from sqlite to parquet would be cool too
Hello, could we leverage [`pandas.read_sql`](https://pandas.pydata.org/docs/reference/api/pandas.read_sql.html) for this?
This would be basically the same as [`CSVBuilder`](https://github.com/huggingface/datasets/blob/7380140accf522a4363bb56c0b77a4190f49bed6/src/datasets/packaged_modules/csv/csv.py#L127)
, but uses `pandas.read_sql(..., chunksize=1)` instead of `pandas.read_csv(..., iterator=True)`
I'm happy to work on this :)
self-assign | 2022-09-03T19:09:08Z | [] | [] |
huggingface/datasets | 4,956 | huggingface__datasets-4956 | [
"4953"
] | 62f9243324296d2d5d1350a9c2fe89ace4dde998 | diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -124,7 +124,7 @@
"moto[s3,server]==2.0.4",
"rarfile>=4.0",
"s3fs>=2021.11.1", # aligned with fsspec[http]>=2021.11.1
- "tensorflow>=2.3,!=2.6.0,!=2.6.1,<2.10", # temporarily pin <2.10
+ "tensorflow>=2.3,!=2.6.0,!=2.6.1",
"torch",
"torchaudio<0.12.0",
"soundfile",
| diff --git a/tests/test_py_utils.py b/tests/test_py_utils.py
--- a/tests/test_py_utils.py
+++ b/tests/test_py_utils.py
@@ -135,8 +135,9 @@ def test_tensorflow(self):
import tensorflow as tf
from tensorflow.keras import layers
+ model = layers.Dense(2)
+
def gen_random_output():
- model = layers.Dense(2)
x = tf.random.uniform((1, 3))
return model(x).numpy()
| CI test of TensorFlow is failing
## Describe the bug
The following CI test fails: https://github.com/huggingface/datasets/runs/8246722693?check_suite_focus=true
```
FAILED tests/test_py_utils.py::TempSeedTest::test_tensorflow - AssertionError:
```
Details:
```
_________________________ TempSeedTest.test_tensorflow _________________________
[gw0] linux -- Python 3.7.13 /opt/hostedtoolcache/Python/3.7.13/x64/bin/python
self = <tests.test_py_utils.TempSeedTest testMethod=test_tensorflow>
@require_tf
def test_tensorflow(self):
import tensorflow as tf
from tensorflow.keras import layers
def gen_random_output():
model = layers.Dense(2)
x = tf.random.uniform((1, 3))
return model(x).numpy()
with temp_seed(42, set_tensorflow=True):
out1 = gen_random_output()
with temp_seed(42, set_tensorflow=True):
out2 = gen_random_output()
out3 = gen_random_output()
> np.testing.assert_equal(out1, out2)
E AssertionError:
E Arrays are not equal
E
E Mismatched elements: 2 / 2 (100%)
E Max absolute difference: 0.84619296
E Max relative difference: 16.083529
E x: array([[-0.793581, 0.333286]], dtype=float32)
E y: array([[0.052612, 0.539708]], dtype=float32)
tests/test_py_utils.py:149: AssertionError
```
| 2022-09-08T14:39:10Z | [] | [] |
|
huggingface/datasets | 5,043 | huggingface__datasets-5043 | [
"5038"
] | 1ea4d091b7a4b83a85b2eeb8df65115d39af3766 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -2383,8 +2383,15 @@ def map(
if num_proc is not None and num_proc <= 0:
raise ValueError("num_proc must be an integer > 0.")
- # If the array is empty we do nothing (but we make sure to remove the requested columns anyway)
+ # If the array is empty we do nothing (but we make sure to handle an empty indices mapping and remove the requested columns anyway)
if len(self) == 0:
+ if self._indices is not None: # empty incides mapping
+ self = Dataset(
+ self.data.slice(0, 0),
+ info=self.info.copy(),
+ split=self.split,
+ fingerprint=new_fingerprint,
+ )
if remove_columns:
return self.remove_columns(remove_columns)
else:
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -2167,20 +2167,45 @@ def test_shard(self, in_memory):
def test_flatten_indices(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
- self.assertEqual(dset._indices, None)
+ self.assertIsNone(dset._indices)
tmp_file = os.path.join(tmp_dir, "test.arrow")
with dset.select(range(0, 10, 2), indices_cache_file_name=tmp_file) as dset:
self.assertEqual(len(dset), 5)
- self.assertNotEqual(dset._indices, None)
+ self.assertIsNotNone(dset._indices)
tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
fingerprint = dset._fingerprint
dset.set_format("numpy")
with dset.flatten_indices(cache_file_name=tmp_file_2) as dset:
self.assertEqual(len(dset), 5)
- self.assertEqual(dset._indices, None)
+ self.assertEqual(len(dset.data), len(dset))
+ self.assertIsNone(dset._indices)
+ self.assertNotEqual(dset._fingerprint, fingerprint)
+ self.assertEqual(dset.format["type"], "numpy")
+ # Test unique works
+ dset.unique(dset.column_names[0])
+ assert_arrow_metadata_are_synced_with_dataset_features(dset)
+
+ # Empty indices mapping
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
+ self.assertIsNone(dset._indices, None)
+
+ tmp_file = os.path.join(tmp_dir, "test.arrow")
+ with dset.filter(lambda _: False, cache_file_name=tmp_file) as dset:
+ self.assertEqual(len(dset), 0)
+
+ self.assertIsNotNone(dset._indices, None)
+
+ tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
+ fingerprint = dset._fingerprint
+ dset.set_format("numpy")
+ with dset.flatten_indices(cache_file_name=tmp_file_2) as dset:
+ self.assertEqual(len(dset), 0)
+ self.assertEqual(len(dset.data), len(dset))
+ self.assertIsNone(dset._indices, None)
self.assertNotEqual(dset._fingerprint, fingerprint)
self.assertEqual(dset.format["type"], "numpy")
# Test unique works
| `Dataset.unique` showing wrong output after filtering
## Describe the bug
After filtering a dataset, and if no samples remain, `Dataset.unique` will return the unique values of the unfiltered dataset.
## Steps to reproduce the bug
```python
from datasets import Dataset
dataset = Dataset.from_dict({'id': [0]})
dataset = dataset.filter(lambda _: False)
print(dataset.unique('id'))
```
## Expected results
The above code should return an empty list since the dataset is empty.
## Actual results
```bash
[0]
```
## Environment info
- `datasets` version: 2.5.1
- Platform: Linux-5.18.19-100.fc35.x86_64-x86_64-with-glibc2.34
- Python version: 3.9.14
- PyArrow version: 7.0.0
- Pandas version: 1.3.5
| Hi! It seems like `flatten_indices` (called in `unique`) doesn't know how to handle empty indices mappings. I'm working on the fix. | 2022-09-29T16:17:28Z | [] | [] |
huggingface/datasets | 5,079 | huggingface__datasets-5079 | [
"5074"
] | 6ad430ba0cdeeb601170f732d4bd977f5c04594d | diff --git a/src/datasets/arrow_reader.py b/src/datasets/arrow_reader.py
--- a/src/datasets/arrow_reader.py
+++ b/src/datasets/arrow_reader.py
@@ -211,7 +211,7 @@ def read(
files = self.get_file_instructions(name, instructions, split_infos)
if not files:
msg = f'Instruction "{instructions}" corresponds to no data!'
- raise AssertionError(msg)
+ raise ValueError(msg)
return self.read_files(files=files, original_instructions=instructions, in_memory=in_memory)
def read_files(
@@ -381,11 +381,11 @@ def __post_init__(self):
if self.rounding is not None and self.rounding not in ["closest", "pct1_dropremainder"]:
raise ValueError("rounding must be either closest or pct1_dropremainder")
if self.unit != "%" and self.rounding is not None:
- raise AssertionError("It is forbidden to specify rounding if not using percent slicing.")
+ raise ValueError("It is forbidden to specify rounding if not using percent slicing.")
if self.unit == "%" and self.from_ is not None and abs(self.from_) > 100:
- raise AssertionError("Percent slice boundaries must be > -100 and < 100.")
+ raise ValueError("Percent slice boundaries must be > -100 and < 100.")
if self.unit == "%" and self.to is not None and abs(self.to) > 100:
- raise AssertionError("Percent slice boundaries must be > -100 and < 100.")
+ raise ValueError("Percent slice boundaries must be > -100 and < 100.")
# Update via __dict__ due to instance being "frozen"
self.__dict__["rounding"] = "closest" if self.rounding is None and self.unit == "%" else self.rounding
@@ -394,7 +394,7 @@ def _str_to_read_instruction(spec):
"""Returns ReadInstruction for given string."""
res = _SUB_SPEC_RE.match(spec)
if not res:
- raise AssertionError(f"Unrecognized instruction format: {spec}")
+ raise ValueError(f"Unrecognized instruction format: {spec}")
unit = "%" if res.group("from_pct") or res.group("to_pct") else "abs"
return ReadInstruction(
split_name=res.group("split"),
@@ -412,7 +412,7 @@ def _pct_to_abs_pct1(boundary, num_examples):
'Using "pct1_dropremainder" rounding on a split with less than 100 '
"elements is forbidden: it always results in an empty dataset."
)
- raise AssertionError(msg)
+ raise ValueError(msg)
return boundary * math.trunc(num_examples / 100.0)
@@ -442,7 +442,7 @@ def _rel_to_abs_instr(rel_instr, name2len):
to = num_examples if to is None else to
if abs(from_) > num_examples or abs(to) > num_examples:
msg = f'Requested slice [{from_ or ""}:{to or ""}] incompatible with {num_examples} examples.'
- raise AssertionError(msg)
+ raise ValueError(msg)
if from_ < 0:
from_ = num_examples + from_
elif from_ == 0:
@@ -557,7 +557,7 @@ def from_spec(cls, spec):
spec = str(spec) # Need to convert to str in case of NamedSplit instance.
subs = _ADDITION_SEP_RE.split(spec)
if not subs:
- raise AssertionError(f"No instructions could be built out of {spec}")
+ raise ValueError(f"No instructions could be built out of {spec}")
instruction = _str_to_read_instruction(subs[0])
return sum((_str_to_read_instruction(sub) for sub in subs[1:]), instruction)
@@ -585,7 +585,7 @@ def __add__(self, other):
"""Returns a new ReadInstruction obj, result of appending other to self."""
if not isinstance(other, ReadInstruction):
msg = "ReadInstruction can only be added to another ReadInstruction obj."
- raise AssertionError(msg)
+ raise TypeError(msg)
self_ris = self._relative_instructions
other_ris = other._relative_instructions # pylint: disable=protected-access
if (
@@ -593,7 +593,7 @@ def __add__(self, other):
and other_ris[0].unit != "abs"
and self._relative_instructions[0].rounding != other_ris[0].rounding
):
- raise AssertionError("It is forbidden to sum ReadInstruction instances with different rounding values.")
+ raise ValueError("It is forbidden to sum ReadInstruction instances with different rounding values.")
return self._read_instruction_from_relative_instructions(self_ris + other_ris)
def __str__(self):
diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -991,7 +991,7 @@ def as_dataset(
if not is_local:
raise NotImplementedError(f"Loading a dataset cached in a {type(self._fs).__name__} is not supported.")
if not os.path.exists(self._output_dir):
- raise AssertionError(
+ raise FileNotFoundError(
f"Dataset {self.name}: could not find data in {self._output_dir}. Please make sure to call "
"builder.download_and_prepare(), or use "
"datasets.load_dataset() before trying to access the Dataset object."
diff --git a/src/datasets/utils/version.py b/src/datasets/utils/version.py
--- a/src/datasets/utils/version.py
+++ b/src/datasets/utils/version.py
@@ -69,12 +69,12 @@ def _validate_operand(self, other):
return Version(other)
elif isinstance(other, Version):
return other
- raise AssertionError(f"{other} (type {type(other)}) cannot be compared to version.")
+ raise TypeError(f"{other} (type {type(other)}) cannot be compared to version.")
def __eq__(self, other):
try:
other = self._validate_operand(other)
- except (AssertionError, ValueError):
+ except (TypeError, ValueError):
return False
else:
return self.tuple == other.tuple
| diff --git a/tests/test_builder.py b/tests/test_builder.py
--- a/tests/test_builder.py
+++ b/tests/test_builder.py
@@ -560,7 +560,7 @@ def _prepare_split(self, split_generator, **kwargs):
try_from_hf_gcs=False,
download_mode=DownloadMode.FORCE_REDOWNLOAD,
)
- self.assertRaises(AssertionError, builder.as_dataset)
+ self.assertRaises(FileNotFoundError, builder.as_dataset)
def test_generator_based_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
| Replace AssertionErrors with more meaningful errors
Replace the AssertionErrors with more meaningful errors such as ValueError, TypeError, etc.
The files with AssertionErrors that need to be replaced:
```
src/datasets/arrow_reader.py
src/datasets/builder.py
src/datasets/utils/version.py
```
| Hi, can I pick up this issue?
#self-assign | 2022-10-06T01:39:35Z | [] | [] |
huggingface/datasets | 5,087 | huggingface__datasets-5087 | [
"5085"
] | efaeee6eb0e9e8c8d610ed4dd62d2fabf66e2149 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -2491,7 +2491,7 @@ def map(
# If the array is empty we do nothing (but we make sure to handle an empty indices mapping and remove the requested columns anyway)
if len(self) == 0:
- if self._indices is not None: # empty incides mapping
+ if self._indices is not None: # empty indices mapping
self = Dataset(
self.data.slice(0, 0),
info=self.info.copy(),
@@ -3084,6 +3084,9 @@ def filter(
if function is None:
function = lambda x: True # noqa: E731
+ if len(self) == 0:
+ return self
+
indices = self.map(
function=partial(
get_indices_from_mask_function, function, batched, with_indices, input_columns, self._indices
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -1469,6 +1469,21 @@ def test_filter_with_indices_mapping(self, in_memory):
with dset.filter(lambda x: x["col"] < 2) as dset:
self.assertListEqual(dset["col"], [1])
+ def test_filter_empty(self, in_memory):
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
+ self.assertIsNone(dset._indices, None)
+
+ tmp_file = os.path.join(tmp_dir, "test.arrow")
+ with dset.filter(lambda _: False, cache_file_name=tmp_file) as dset:
+ self.assertEqual(len(dset), 0)
+ self.assertIsNotNone(dset._indices, None)
+
+ tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
+ with dset.filter(lambda _: False, cache_file_name=tmp_file_2) as dset2:
+ self.assertEqual(len(dset2), 0)
+ self.assertEqual(dset._indices, dset2._indices)
+
def test_filter_batched(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
dset = Dataset.from_dict({"col": [0, 1, 2]})
| Filtering on an empty dataset returns a corrupted dataset.
## Describe the bug
When filtering a dataset twice, where the first result is an empty dataset, the second dataset seems corrupted.
## Steps to reproduce the bug
```python
datasets = load_dataset("glue", "sst2")
dataset_split = datasets['validation']
ds_filter_1 = dataset_split.filter(lambda x: False) # Some filtering condition that leads to an empty dataset
assert ds_filter_1.num_rows == 0
sentences = ds_filter_1['sentence']
assert len(sentences) == 0
ds_filter_2 = ds_filter_1.filter(lambda x: False) # Some other filtering condition
assert ds_filter_2.num_rows == 0
assert 'sentence' in ds_filter_2.column_names
sentences = ds_filter_2['sentence']
```
## Expected results
The last line should be returning an empty list, same as 4 lines above.
## Actual results
The last line currently raises `IndexError: index out of bounds`.
## Environment info
<!-- You can run the command `datasets-cli env` and copy-and-paste its output below. -->
- `datasets` version: 2.5.2
- Platform: macOS-11.6.6-x86_64-i386-64bit
- Python version: 3.9.11
- PyArrow version: 7.0.0
- Pandas version: 1.4.1
| ~~It seems like #5043 fix (merged recently) is the root cause of such behaviour. When we empty indices mapping (because the dataset length equals to zero), we can no longer get column item like: `ds_filter_2['sentence']` which uses
`ds_filter_1._indices.column(0)`~~
**UPDATE:**
Empty datasets are returned without going through partial function on `map` method, which will not work to get indices for `filter`: we need to run `get_indices_from_mask_function` partial function on the dataset to get output = `{"indices": []}`. But this is complicated since functions used in args, in particular `get_indices_from_mask_function`, do not support empty datasets.
We can just handle empty datasets aside on filter method.
#self-assign | 2022-10-07T01:07:00Z | [] | [] |
huggingface/datasets | 5,113 | huggingface__datasets-5113 | [
"5112"
] | 3ad9644b9a2e4558dd1d0f1e43c67658674e6228 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -2961,7 +2961,7 @@ def init_buffer_and_writer():
else:
writer.write(example)
else:
- for i, batch in enumerate(pbar):
+ for i, batch in zip(range(0, num_rows, batch_size), pbar):
indices = list(
range(*(slice(i, i + batch_size).indices(input_dataset.num_rows)))
) # Something simpler?
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -3081,6 +3081,12 @@ def test_dataset_add_item_introduce_feature_type():
assert dataset[:] == {"col_1": [None, None, None, "a"]}
+def test_dataset_filter_batched_indices():
+ ds = Dataset.from_dict({"num": [0, 1, 2, 3]})
+ ds = ds.filter(lambda num: num % 2 == 0, input_columns="num", batch_size=2)
+ assert all(item["num"] % 2 == 0 for item in ds)
+
+
@pytest.mark.parametrize("in_memory", [False, True])
def test_dataset_from_file(in_memory, dataset, arrow_file):
filename = arrow_file
| Bug with filtered indices
## Describe the bug
As reported by @PartiallyTyped (and by @Muennighoff):
- https://github.com/huggingface/datasets/issues/5111#issuecomment-1278652524
There is an issue with the indices of a filtered dataset.
## Steps to reproduce the bug
```python
ds = Dataset.from_dict({"num": [0, 1, 2, 3]})
ds = ds.filter(lambda num: num % 2 == 0, input_columns="num", batch_size=2)
assert all(item["num"] % 2 == 0 for item in ds)
```
## Expected results
The indices of the filtered dataset should correspond to the examples with "language" equals to "english".
## Actual results
Indices to items with other languages are included in the filtered dataset indices
## Preliminar investigation
It seems a bug introduced by:
- #5030
| The issue is here:
https://github.com/huggingface/datasets/blob/3ad9644b9a2e4558dd1d0f1e43c67658674e6228/src/datasets/arrow_dataset.py#L2964 | 2022-10-14T11:30:03Z | [] | [] |
huggingface/datasets | 5,126 | huggingface__datasets-5126 | [
"5098"
] | 85cd129bde605cd9acacdff0d065fc02e39e09b1 | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -275,7 +275,7 @@ def _resolve_single_pattern_locally(
fs = LocalFileSystem()
glob_iter = [PurePath(filepath) for filepath in fs.glob(pattern) if fs.isfile(filepath)]
matched_paths = [
- Path(filepath).resolve()
+ Path(os.path.abspath(filepath))
for filepath in glob_iter
if (filepath.name not in FILES_TO_IGNORE or PurePath(pattern).name == filepath.name)
and not _is_inside_unrequested_special_dir(
| diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -46,11 +46,13 @@
def complex_data_dir(tmp_path):
data_dir = tmp_path / "complex_data_dir"
data_dir.mkdir()
+
(data_dir / "data").mkdir()
with open(data_dir / "data" / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(data_dir / "data" / "test.txt", "w") as f:
f.write("bar\n" * 10)
+
with open(data_dir / "README.md", "w") as f:
f.write("This is a readme")
with open(data_dir / ".dummy", "w") as f:
@@ -100,7 +102,7 @@ def pattern_results(complex_data_dir):
return {
pattern: sorted(
- str(Path(path).resolve())
+ str(Path(os.path.abspath(path)))
for path in fsspec.filesystem("file").glob(os.path.join(complex_data_dir, pattern))
if Path(path).name not in _FILES_TO_IGNORE
and not any(
@@ -268,6 +270,14 @@ def test_fail_resolve_patterns_locally_or_by_urls(complex_data_dir):
resolve_patterns_locally_or_by_urls(complex_data_dir, ["blablabla"])
[email protected](os.name == "nt", reason="Windows does not support symlinks in the default mode")
+def test_resolve_patterns_locally_or_by_urls_does_not_resolve_symbolic_links(tmp_path, complex_data_dir):
+ (tmp_path / "train_data_symlink.txt").symlink_to(os.path.join(complex_data_dir, "data", "train.txt"))
+ resolved_data_files = resolve_patterns_locally_or_by_urls(str(tmp_path), ["train_data_symlink.txt"])
+ assert len(resolved_data_files) == 1
+ assert resolved_data_files[0] == tmp_path / "train_data_symlink.txt"
+
+
def test_resolve_patterns_locally_or_by_urls_sorted_files(tmp_path_factory):
path = str(tmp_path_factory.mktemp("unsorted_text_files"))
unsorted_names = ["0.txt", "2.txt", "3.txt"]
| Classes label error when loading symbolic links using imagefolder
**Is your feature request related to a problem? Please describe.**
Like this: #4015
When there are **symbolic links** to pictures in the data folder, the parent folder name of the **real file** will be used as the class name instead of the parent folder of the symbolic link itself. Can you give an option to decide whether to enable symbolic link tracking?
This is inconsistent with the `torchvision.datasets.ImageFolder` behavior.
For example:
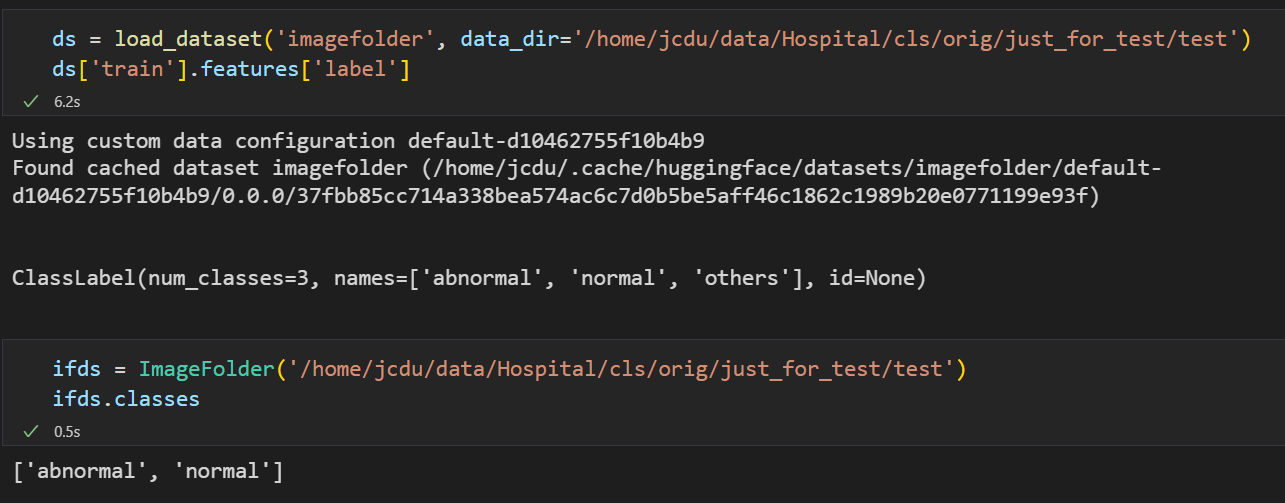
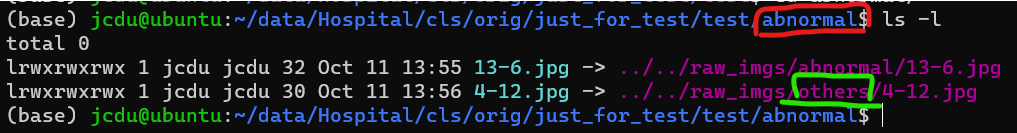
It use `others` in green circle as class label but not `abnormal`, I wish `load_dataset` not use the real file parent as label.
**Describe the solution you'd like**
A clear and concise description of what you want to happen.
**Describe alternatives you've considered**
A clear and concise description of any alternative solutions or features you've considered.
**Additional context**
Add any other context about the feature request here.
| It can be solved temporarily by remove `resolve` in
https://github.com/huggingface/datasets/blob/bef23be3d9543b1ca2da87ab2f05070201044ddc/src/datasets/data_files.py#L278
Hi, thanks for reporting and suggesting a fix! We still need to account for `.`/`..` in the file path, so a more robust fix would be `Path(os.path.abspath(filepath))`.
> Hi, thanks for reporting and suggesting a fix! We still need to account for `.`/`..` in the file path, so a more robust fix would be `Path(os.path.abspath(filepath))`.
Thanks for your reply! | 2022-10-17T15:11:02Z | [] | [] |
huggingface/datasets | 5,128 | huggingface__datasets-5128 | [
"5046"
] | 85cd129bde605cd9acacdff0d065fc02e39e09b1 | diff --git a/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
--- a/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
+++ b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
@@ -6,7 +6,6 @@
import pandas as pd
import pyarrow as pa
-import pyarrow.compute as pc
import pyarrow.json as paj
import datasets
@@ -310,13 +309,10 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
)
pa_metadata_table = self._read_metadata(downloaded_metadata_file)
pa_file_name_array = pa_metadata_table["file_name"]
- pa_file_name_array = pc.replace_substring(
- pa_file_name_array, pattern="\\", replacement="/"
- )
pa_metadata_table = pa_metadata_table.drop(["file_name"])
metadata_dir = os.path.dirname(metadata_file)
metadata_dict = {
- file_name: sample_metadata
+ os.path.normpath(file_name).replace("\\", "/"): sample_metadata
for file_name, sample_metadata in zip(
pa_file_name_array.to_pylist(), pa_table_to_pylist(pa_metadata_table)
)
@@ -379,13 +375,10 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
)
pa_metadata_table = self._read_metadata(downloaded_metadata_file)
pa_file_name_array = pa_metadata_table["file_name"]
- pa_file_name_array = pc.replace_substring(
- pa_file_name_array, pattern="\\", replacement="/"
- )
pa_metadata_table = pa_metadata_table.drop(["file_name"])
metadata_dir = os.path.dirname(downloaded_metadata_file)
metadata_dict = {
- file_name: sample_metadata
+ os.path.normpath(file_name).replace("\\", "/"): sample_metadata
for file_name, sample_metadata in zip(
pa_file_name_array.to_pylist(), pa_table_to_pylist(pa_metadata_table)
)
| diff --git a/tests/packaged_modules/test_folder_based_builder.py b/tests/packaged_modules/test_folder_based_builder.py
--- a/tests/packaged_modules/test_folder_based_builder.py
+++ b/tests/packaged_modules/test_folder_based_builder.py
@@ -132,7 +132,7 @@ def data_files_with_one_split_and_metadata(tmp_path, auto_text_file):
"""\
{"file_name": "file.txt", "additional_feature": "Dummy file"}
{"file_name": "file2.txt", "additional_feature": "Second dummy file"}
- {"file_name": "subdir/file3.txt", "additional_feature": "Third dummy file"}
+ {"file_name": "./subdir/file3.txt", "additional_feature": "Third dummy file"}
"""
)
with open(metadata_filename, "w", encoding="utf-8") as f:
| Audiofolder creates empty Dataset if files same level as metadata
## Describe the bug
When audio files are at the same level as the metadata (`metadata.csv` or `metadata.jsonl` ), the `load_dataset` returns a `DatasetDict` with no rows but the correct columns.
https://github.com/huggingface/datasets/blob/1ea4d091b7a4b83a85b2eeb8df65115d39af3766/docs/source/audio_dataset.mdx?plain=1#L88
## Steps to reproduce the bug
`metadata.csv`:
```csv
file_name,duration,transcription
./2063_fe9936e7-62b2-4e62-a276-acbd344480ce_1.wav,10.768,hello
```
```python
>>> audio_dataset = load_dataset("audiofolder", data_dir="/audio-data/")
>>> audio_dataset
DatasetDict({
train: Dataset({
features: ['audio', 'duration', 'transcription'],
num_rows: 0
})
validation: Dataset({
features: ['audio', 'duration', 'transcription'],
num_rows: 0
})
})
```
I've tried, with no success,:
- setting `split` to something else so I don't get a `DatasetDict`,
- removing the `./`,
- using `.jsonl`.
## Expected results
```
Dataset({
features: ['audio', 'duration', 'transcription'],
num_rows: 1
})
```
## Actual results
```
DatasetDict({
train: Dataset({
features: ['audio', 'duration', 'transcription'],
num_rows: 0
})
validation: Dataset({
features: ['audio', 'duration', 'transcription'],
num_rows: 0
})
})
```
## Environment info
- `datasets` version: 2.5.1
- Platform: Linux-5.13.0-1025-aws-x86_64-with-glibc2.29
- Python version: 3.8.10
- PyArrow version: 9.0.0
- Pandas version: 1.5.0
| Hi! Unfortunately, I can't reproduce this behavior. Instead, I get `ValueError: audio at 2063_fe9936e7-62b2-4e62-a276-acbd344480ce_1.wav doesn't have metadata in /audio-data/metadata.csv`, which can be fixed by removing the `./` from the file name.
(Link to a Colab that tries to reproduce this behavior: https://colab.research.google.com/drive/1IhQzULYi0Van1xLrN_SddBX1JF7mLZZK?usp=sharing)
I think we can make the file name matching part more robust by replacing `file_name` with `os.path.normpath(file_name)`, to ignore "./" among other things, in these two places:
* https://github.com/huggingface/datasets/blob/85cd129bde605cd9acacdff0d065fc02e39e09b1/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py#L319
* https://github.com/huggingface/datasets/blob/85cd129bde605cd9acacdff0d065fc02e39e09b1/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py#L388 | 2022-10-18T08:22:48Z | [] | [] |
huggingface/datasets | 5,149 | huggingface__datasets-5149 | [
"5145"
] | f09f781be3278156ce3aa6ec90c1926b1846a78f | diff --git a/src/datasets/download/download_manager.py b/src/datasets/download/download_manager.py
--- a/src/datasets/download/download_manager.py
+++ b/src/datasets/download/download_manager.py
@@ -130,11 +130,11 @@ def _iter_from_paths(cls, urlpaths: Union[str, List[str]]) -> Generator[str, Non
for dirpath, dirnames, filenames in os.walk(urlpath):
# skipping hidden directories; prune the search
# [:] for the in-place list modification required by os.walk
- dirnames[:] = [dirname for dirname in dirnames if not dirname.startswith((".", "__"))]
+ dirnames[:] = sorted([dirname for dirname in dirnames if not dirname.startswith((".", "__"))])
if os.path.basename(dirpath).startswith((".", "__")):
# skipping hidden directories
continue
- for filename in filenames:
+ for filename in sorted(filenames):
if filename.startswith((".", "__")):
# skipping hidden files
continue
diff --git a/src/datasets/download/mock_download_manager.py b/src/datasets/download/mock_download_manager.py
--- a/src/datasets/download/mock_download_manager.py
+++ b/src/datasets/download/mock_download_manager.py
@@ -236,10 +236,11 @@ def iter_files(self, paths):
return
yield path
else:
- for dirpath, _, filenames in os.walk(path):
+ for dirpath, dirnames, filenames in os.walk(path):
if os.path.basename(dirpath).startswith((".", "__")):
continue
- for filename in filenames:
+ dirnames.sort()
+ for filename in sorted(filenames):
if filename.startswith((".", "__")):
continue
yield os.path.join(dirpath, filename)
diff --git a/src/datasets/download/streaming_download_manager.py b/src/datasets/download/streaming_download_manager.py
--- a/src/datasets/download/streaming_download_manager.py
+++ b/src/datasets/download/streaming_download_manager.py
@@ -835,11 +835,11 @@ def _iter_from_urlpaths(
# skipping hidden directories; prune the search
# [:] for the in-place list modification required by os.walk
# (only works for local paths as fsspec's walk doesn't support the in-place modification)
- dirnames[:] = [dirname for dirname in dirnames if not dirname.startswith((".", "__"))]
+ dirnames[:] = sorted([dirname for dirname in dirnames if not dirname.startswith((".", "__"))])
if xbasename(dirpath).startswith((".", "__")):
# skipping hidden directories
continue
- for filename in filenames:
+ for filename in sorted(filenames):
if filename.startswith((".", "__")):
# skipping hidden files
continue
| diff --git a/tests/test_download_manager.py b/tests/test_download_manager.py
--- a/tests/test_download_manager.py
+++ b/tests/test_download_manager.py
@@ -141,5 +141,5 @@ def test_iter_archive_file(tar_nested_jsonl_path):
def test_iter_files(data_dir_with_hidden_files):
dl_manager = DownloadManager()
for num_file, file in enumerate(dl_manager.iter_files(data_dir_with_hidden_files), start=1):
- pass
+ assert os.path.basename(file) == ("test.txt" if num_file == 1 else "train.txt")
assert num_file == 2
diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -834,5 +834,5 @@ def test_iter_archive_file(tar_nested_jsonl_path):
def test_iter_files(data_dir_with_hidden_files):
dl_manager = StreamingDownloadManager()
for num_file, file in enumerate(dl_manager.iter_files(data_dir_with_hidden_files), start=1):
- pass
+ assert os.path.basename(file) == ("test.txt" if num_file == 1 else "train.txt")
assert num_file == 2
| Dataset order is not deterministic with ZIP archives and `iter_files`
### Describe the bug
For the `beans` dataset (did not try on other), the order of samples is not the same on different machines. Tested on my local laptop, github actions machine, and ec2 instance. The three yield a different order.
### Steps to reproduce the bug
In a clean docker container or conda environment with datasets==2.6.1, run
```python
from datasets import load_dataset
from pprint import pprint
data = load_dataset("beans", split="validation")
pprint(data["image_file_path"])
```
### Expected behavior
The order of the images is the same on all machines.
### Environment info
On the EC2 instance:
```
- `datasets` version: 2.6.1
- Platform: Linux-4.14.291-218.527.amzn2.x86_64-x86_64-with-glibc2.2.5
- Python version: 3.7.10
- PyArrow version: 9.0.0
- Pandas version: 1.3.5
- Numpy version: not checked
```
On my local laptop:
```
- `datasets` version: 2.6.1
- Platform: Linux-5.15.0-50-generic-x86_64-with-glibc2.35
- Python version: 3.9.12
- PyArrow version: 7.0.0
- Pandas version: 1.3.5
- Numpy version: 1.23.1
```
On github actions:
```
- `datasets` version: 2.6.1
- Platform: Linux-5.15.0-1022-azure-x86_64-with-glibc2.2.5
- Python version: 3.8.14
- PyArrow version: 9.0.0
- Pandas version: 1.5.1
- Numpy version: 1.23.4
```
| Thanks for reporting ! The issue doesn't come from shuffling, but from `beans` row order not being deterministic:
https://huggingface.co/datasets/beans/blob/main/beans.py uses `dl_manager.iter_files` on ZIP archives and the file order doesn't seen to be deterministic and changes across machines
Thank you for noticing indeed!
This is still a bug, so I'd keep this one open if you don't mind ;) | 2022-10-24T08:16:27Z | [] | [] |
huggingface/datasets | 5,166 | huggingface__datasets-5166 | [
"5162"
] | 11f1fdea2c0b4aa4010b15d5ee585507d9ad40b0 | diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -70,7 +70,7 @@
# Minimum 6.0.0 to support wrap_array which is needed for ArrayND features
"pyarrow>=6.0.0",
# For smart caching dataset processing
- "dill<0.3.6", # tmp pin until 0.3.6 release: see https://github.com/huggingface/datasets/pull/4397
+ "dill<0.3.7", # tmp pin until next 0.3.7 release: see https://github.com/huggingface/datasets/pull/5166
# For performance gains with apache arrow
"pandas",
# for downloading datasets over HTTPS
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -634,39 +634,76 @@ def proxy(func):
return proxy
-@pklregister(CodeType)
-def _save_code(pickler, obj):
- """
- From dill._dill.save_code
- This is a modified version that removes the origin (filename + line no.)
- of functions created in notebooks or shells for example.
- """
- dill._dill.log.info(f"Co: {obj}")
- # The filename of a function is the .py file where it is defined.
- # Filenames of functions created in notebooks or shells start with '<'
- # ex: <ipython-input-13-9ed2afe61d25> for ipython, and <stdin> for shell
- # Moreover lambda functions have a special name: '<lambda>'
- # ex: (lambda x: x).__code__.co_name == "<lambda>" # True
- #
- # For the hashing mechanism we ignore where the function has been defined
- # More specifically:
- # - we ignore the filename of special functions (filename starts with '<')
- # - we always ignore the line number
- # - we only use the base name of the file instead of the whole path,
- # to be robust in case a script is moved for example.
- #
- # Only those two lines are different from the original implementation:
- co_filename = (
- "" if obj.co_filename.startswith("<") or obj.co_name == "<lambda>" else os.path.basename(obj.co_filename)
- )
- co_firstlineno = 1
- # The rest is the same as in the original dill implementation
- if dill._dill.PY3:
- if hasattr(obj, "co_posonlyargcount"):
+if config.DILL_VERSION < version.parse("0.3.6"):
+
+ @pklregister(CodeType)
+ def _save_code(pickler, obj):
+ """
+ From dill._dill.save_code
+ This is a modified version that removes the origin (filename + line no.)
+ of functions created in notebooks or shells for example.
+ """
+ dill._dill.log.info(f"Co: {obj}")
+ # The filename of a function is the .py file where it is defined.
+ # Filenames of functions created in notebooks or shells start with '<'
+ # ex: <ipython-input-13-9ed2afe61d25> for ipython, and <stdin> for shell
+ # Moreover lambda functions have a special name: '<lambda>'
+ # ex: (lambda x: x).__code__.co_name == "<lambda>" # True
+ #
+ # For the hashing mechanism we ignore where the function has been defined
+ # More specifically:
+ # - we ignore the filename of special functions (filename starts with '<')
+ # - we always ignore the line number
+ # - we only use the base name of the file instead of the whole path,
+ # to be robust in case a script is moved for example.
+ #
+ # Only those two lines are different from the original implementation:
+ co_filename = (
+ "" if obj.co_filename.startswith("<") or obj.co_name == "<lambda>" else os.path.basename(obj.co_filename)
+ )
+ co_firstlineno = 1
+ # The rest is the same as in the original dill implementation
+ if dill._dill.PY3:
+ if hasattr(obj, "co_posonlyargcount"):
+ args = (
+ obj.co_argcount,
+ obj.co_posonlyargcount,
+ obj.co_kwonlyargcount,
+ obj.co_nlocals,
+ obj.co_stacksize,
+ obj.co_flags,
+ obj.co_code,
+ obj.co_consts,
+ obj.co_names,
+ obj.co_varnames,
+ co_filename,
+ obj.co_name,
+ co_firstlineno,
+ obj.co_lnotab,
+ obj.co_freevars,
+ obj.co_cellvars,
+ )
+ else:
+ args = (
+ obj.co_argcount,
+ obj.co_kwonlyargcount,
+ obj.co_nlocals,
+ obj.co_stacksize,
+ obj.co_flags,
+ obj.co_code,
+ obj.co_consts,
+ obj.co_names,
+ obj.co_varnames,
+ co_filename,
+ obj.co_name,
+ co_firstlineno,
+ obj.co_lnotab,
+ obj.co_freevars,
+ obj.co_cellvars,
+ )
+ else:
args = (
obj.co_argcount,
- obj.co_posonlyargcount,
- obj.co_kwonlyargcount,
obj.co_nlocals,
obj.co_stacksize,
obj.co_flags,
@@ -681,9 +718,47 @@ def _save_code(pickler, obj):
obj.co_freevars,
obj.co_cellvars,
)
- else:
+ pickler.save_reduce(CodeType, args, obj=obj)
+ dill._dill.log.info("# Co")
+ return
+
+elif config.DILL_VERSION.release[:3] == version.parse("0.3.6").release:
+
+ # From: https://github.com/uqfoundation/dill/blob/dill-0.3.6/dill/_dill.py#L1104
+ @pklregister(CodeType)
+ def save_code(pickler, obj):
+ dill._dill.logger.trace(pickler, "Co: %s", obj)
+
+ ############################################################################################################
+ # Modification here for huggingface/datasets
+ # The filename of a function is the .py file where it is defined.
+ # Filenames of functions created in notebooks or shells start with '<'
+ # ex: <ipython-input-13-9ed2afe61d25> for ipython, and <stdin> for shell
+ # Moreover lambda functions have a special name: '<lambda>'
+ # ex: (lambda x: x).__code__.co_name == "<lambda>" # True
+ #
+ # For the hashing mechanism we ignore where the function has been defined
+ # More specifically:
+ # - we ignore the filename of special functions (filename starts with '<')
+ # - we always ignore the line number
+ # - we only use the base name of the file instead of the whole path,
+ # to be robust in case a script is moved for example.
+ #
+ # Only those two lines are different from the original implementation:
+ co_filename = (
+ "" if obj.co_filename.startswith("<") or obj.co_name == "<lambda>" else os.path.basename(obj.co_filename)
+ )
+ co_firstlineno = 1
+ # The rest is the same as in the original dill implementation, except for the replacements:
+ # - obj.co_filename => co_filename
+ # - obj.co_firstlineno => co_firstlineno
+ ############################################################################################################
+
+ if hasattr(obj, "co_endlinetable"): # python 3.11a (20 args)
args = (
+ obj.co_lnotab, # for < python 3.10 [not counted in args]
obj.co_argcount,
+ obj.co_posonlyargcount,
obj.co_kwonlyargcount,
obj.co_nlocals,
obj.co_stacksize,
@@ -692,33 +767,100 @@ def _save_code(pickler, obj):
obj.co_consts,
obj.co_names,
obj.co_varnames,
- co_filename,
+ co_filename, # Modification for huggingface/datasets ############################################
obj.co_name,
- co_firstlineno,
+ obj.co_qualname,
+ co_firstlineno, # Modification for huggingface/datasets #########################################
+ obj.co_linetable,
+ obj.co_endlinetable,
+ obj.co_columntable,
+ obj.co_exceptiontable,
+ obj.co_freevars,
+ obj.co_cellvars,
+ )
+ elif hasattr(obj, "co_exceptiontable"): # python 3.11 (18 args)
+ args = (
+ obj.co_lnotab, # for < python 3.10 [not counted in args]
+ obj.co_argcount,
+ obj.co_posonlyargcount,
+ obj.co_kwonlyargcount,
+ obj.co_nlocals,
+ obj.co_stacksize,
+ obj.co_flags,
+ obj.co_code,
+ obj.co_consts,
+ obj.co_names,
+ obj.co_varnames,
+ co_filename, # Modification for huggingface/datasets ############################################
+ obj.co_name,
+ obj.co_qualname,
+ co_firstlineno, # Modification for huggingface/datasets #########################################
+ obj.co_linetable,
+ obj.co_exceptiontable,
+ obj.co_freevars,
+ obj.co_cellvars,
+ )
+ elif hasattr(obj, "co_linetable"): # python 3.10 (16 args)
+ args = (
+ obj.co_lnotab, # for < python 3.10 [not counted in args]
+ obj.co_argcount,
+ obj.co_posonlyargcount,
+ obj.co_kwonlyargcount,
+ obj.co_nlocals,
+ obj.co_stacksize,
+ obj.co_flags,
+ obj.co_code,
+ obj.co_consts,
+ obj.co_names,
+ obj.co_varnames,
+ co_filename, # Modification for huggingface/datasets ############################################
+ obj.co_name,
+ co_firstlineno, # Modification for huggingface/datasets #########################################
+ obj.co_linetable,
+ obj.co_freevars,
+ obj.co_cellvars,
+ )
+ elif hasattr(obj, "co_posonlyargcount"): # python 3.8 (16 args)
+ args = (
+ obj.co_argcount,
+ obj.co_posonlyargcount,
+ obj.co_kwonlyargcount,
+ obj.co_nlocals,
+ obj.co_stacksize,
+ obj.co_flags,
+ obj.co_code,
+ obj.co_consts,
+ obj.co_names,
+ obj.co_varnames,
+ co_filename, # Modification for huggingface/datasets ############################################
+ obj.co_name,
+ co_firstlineno, # Modification for huggingface/datasets #########################################
+ obj.co_lnotab,
+ obj.co_freevars,
+ obj.co_cellvars,
+ )
+ else: # python 3.7 (15 args)
+ args = (
+ obj.co_argcount,
+ obj.co_kwonlyargcount,
+ obj.co_nlocals,
+ obj.co_stacksize,
+ obj.co_flags,
+ obj.co_code,
+ obj.co_consts,
+ obj.co_names,
+ obj.co_varnames,
+ co_filename, # Modification for huggingface/datasets ############################################
+ obj.co_name,
+ co_firstlineno, # Modification for huggingface/datasets #########################################
obj.co_lnotab,
obj.co_freevars,
obj.co_cellvars,
)
- else:
- args = (
- obj.co_argcount,
- obj.co_nlocals,
- obj.co_stacksize,
- obj.co_flags,
- obj.co_code,
- obj.co_consts,
- obj.co_names,
- obj.co_varnames,
- co_filename,
- obj.co_name,
- co_firstlineno,
- obj.co_lnotab,
- obj.co_freevars,
- obj.co_cellvars,
- )
- pickler.save_reduce(CodeType, args, obj=obj)
- dill._dill.log.info("# Co")
- return
+
+ pickler.save_reduce(dill._dill._create_code, args, obj=obj)
+ dill._dill.logger.trace(pickler, "# Co")
+ return
if config.DILL_VERSION < version.parse("0.3.5"):
@@ -796,7 +938,7 @@ def save_function(pickler, obj):
dill._dill.log.info("# F2")
return
-else: # config.DILL_VERSION >= version.parse("0.3.5")
+elif config.DILL_VERSION.release[:3] == version.parse("0.3.5").release: # 0.3.5, 0.3.5.1
# https://github.com/uqfoundation/dill/blob/dill-0.3.5.1/dill/_dill.py
@pklregister(FunctionType)
@@ -804,7 +946,6 @@ def save_function(pickler, obj):
if not dill._dill._locate_function(obj, pickler):
dill._dill.log.info("F1: %s" % obj)
_recurse = getattr(pickler, "_recurse", None)
- # _byref = getattr(pickler, "_byref", None) # TODO: not used
_postproc = getattr(pickler, "_postproc", None)
_main_modified = getattr(pickler, "_main_modified", None)
_original_main = getattr(pickler, "_original_main", dill._dill.__builtin__) # 'None'
@@ -941,6 +1082,147 @@ def save_function(pickler, obj):
dill._dill.log.info("# F2")
return
+elif config.DILL_VERSION.release[:3] == version.parse("0.3.6").release:
+
+ # From: https://github.com/uqfoundation/dill/blob/dill-0.3.6/dill/_dill.py#L1739
+ @pklregister(FunctionType)
+ def save_function(pickler, obj):
+ if not dill._dill._locate_function(obj, pickler):
+ if type(obj.__code__) is not CodeType:
+ # Some PyPy builtin functions have no module name, and thus are not
+ # able to be located
+ module_name = getattr(obj, "__module__", None)
+ if module_name is None:
+ module_name = dill._dill.__builtin__.__name__
+ module = dill._dill._import_module(module_name, safe=True)
+ _pypy_builtin = False
+ try:
+ found, _ = dill._dill._getattribute(module, obj.__qualname__)
+ if getattr(found, "__func__", None) is obj:
+ _pypy_builtin = True
+ except AttributeError:
+ pass
+
+ if _pypy_builtin:
+ dill._dill.logger.trace(pickler, "F3: %s", obj)
+ pickler.save_reduce(getattr, (found, "__func__"), obj=obj)
+ dill._dill.logger.trace(pickler, "# F3")
+ return
+
+ dill._dill.logger.trace(pickler, "F1: %s", obj)
+ _recurse = getattr(pickler, "_recurse", None)
+ _postproc = getattr(pickler, "_postproc", None)
+ _main_modified = getattr(pickler, "_main_modified", None)
+ _original_main = getattr(pickler, "_original_main", dill._dill.__builtin__) # 'None'
+ postproc_list = []
+ if _recurse:
+ # recurse to get all globals referred to by obj
+ from dill.detect import globalvars
+
+ globs_copy = globalvars(obj, recurse=True, builtin=True)
+
+ # Add the name of the module to the globs dictionary to prevent
+ # the duplication of the dictionary. Pickle the unpopulated
+ # globals dictionary and set the remaining items after the function
+ # is created to correctly handle recursion.
+ globs = {"__name__": obj.__module__}
+ else:
+ globs_copy = obj.__globals__
+
+ # If the globals is the __dict__ from the module being saved as a
+ # session, substitute it by the dictionary being actually saved.
+ if _main_modified and globs_copy is _original_main.__dict__:
+ globs_copy = getattr(pickler, "_main", _original_main).__dict__
+ globs = globs_copy
+ # If the globals is a module __dict__, do not save it in the pickle.
+ elif (
+ globs_copy is not None
+ and obj.__module__ is not None
+ and getattr(dill._dill._import_module(obj.__module__, True), "__dict__", None) is globs_copy
+ ):
+ globs = globs_copy
+ else:
+ globs = {"__name__": obj.__module__}
+
+ ########################################################################################################
+ # Modification here for huggingface/datasets
+ # - globs is a dictionary with keys = var names (str) and values = python objects
+ # - globs_copy is a dictionary with keys = var names (str) and values = ids of the python objects
+ # However the dictionary is not always loaded in the same order,
+ # therefore we have to sort the keys to make deterministic.
+ # This is important to make `dump` deterministic.
+ # Only these line are different from the original implementation:
+ # START
+ globs_is_globs_copy = globs is globs_copy
+ globs = dict(sorted(globs.items()))
+ if globs_is_globs_copy:
+ globs_copy = globs
+ elif globs_copy is not None:
+ globs_copy = dict(sorted(globs_copy.items()))
+ # END
+ ########################################################################################################
+
+ if globs_copy is not None and globs is not globs_copy:
+ # In the case that the globals are copied, we need to ensure that
+ # the globals dictionary is updated when all objects in the
+ # dictionary are already created.
+ glob_ids = {id(g) for g in globs_copy.values()}
+ for stack_element in _postproc:
+ if stack_element in glob_ids:
+ _postproc[stack_element].append((dill._dill._setitems, (globs, globs_copy)))
+ break
+ else:
+ postproc_list.append((dill._dill._setitems, (globs, globs_copy)))
+
+ closure = obj.__closure__
+ state_dict = {}
+ for fattrname in ("__doc__", "__kwdefaults__", "__annotations__"):
+ fattr = getattr(obj, fattrname, None)
+ if fattr is not None:
+ state_dict[fattrname] = fattr
+ if obj.__qualname__ != obj.__name__:
+ state_dict["__qualname__"] = obj.__qualname__
+ if "__name__" not in globs or obj.__module__ != globs["__name__"]:
+ state_dict["__module__"] = obj.__module__
+
+ state = obj.__dict__
+ if type(state) is not dict:
+ state_dict["__dict__"] = state
+ state = None
+ if state_dict:
+ state = state, state_dict
+
+ dill._dill._save_with_postproc(
+ pickler,
+ (dill._dill._create_function, (obj.__code__, globs, obj.__name__, obj.__defaults__, closure), state),
+ obj=obj,
+ postproc_list=postproc_list,
+ )
+
+ # Lift closure cell update to earliest function (#458)
+ if _postproc:
+ topmost_postproc = next(iter(_postproc.values()), None)
+ if closure and topmost_postproc:
+ for cell in closure:
+ possible_postproc = (setattr, (cell, "cell_contents", obj))
+ try:
+ topmost_postproc.remove(possible_postproc)
+ except ValueError:
+ continue
+
+ # Change the value of the cell
+ pickler.save_reduce(*possible_postproc)
+ # pop None created by calling preprocessing step off stack
+ pickler.write(bytes("0", "UTF-8"))
+
+ dill._dill.logger.trace(pickler, "# F1")
+ else:
+ dill._dill.logger.trace(pickler, "F2: %s", obj)
+ name = getattr(obj, "__qualname__", getattr(obj, "__name__", None))
+ dill._dill.StockPickler.save_global(pickler, obj, name=name)
+ dill._dill.logger.trace(pickler, "# F2")
+ return
+
def copyfunc(func):
result = types.FunctionType(func.__code__, func.__globals__, func.__name__, func.__defaults__, func.__closure__)
@@ -951,16 +1233,31 @@ def copyfunc(func):
try:
import regex
- @pklregister(type(regex.Regex("", 0)))
- def _save_regex(pickler, obj):
- dill._dill.log.info(f"Re: {obj}")
- args = (
- obj.pattern,
- obj.flags,
- )
- pickler.save_reduce(regex.compile, args, obj=obj)
- dill._dill.log.info("# Re")
- return
+ if config.DILL_VERSION < version.parse("0.3.6"):
+
+ @pklregister(type(regex.Regex("", 0)))
+ def _save_regex(pickler, obj):
+ dill._dill.log.info(f"Re: {obj}")
+ args = (
+ obj.pattern,
+ obj.flags,
+ )
+ pickler.save_reduce(regex.compile, args, obj=obj)
+ dill._dill.log.info("# Re")
+ return
+
+ elif config.DILL_VERSION.release[:3] == version.parse("0.3.6").release:
+
+ @pklregister(type(regex.Regex("", 0)))
+ def _save_regex(pickler, obj):
+ dill._dill.logger.trace(pickler, "Re: %s", obj)
+ args = (
+ obj.pattern,
+ obj.flags,
+ )
+ pickler.save_reduce(regex.compile, args, obj=obj)
+ dill._dill.logger.trace(pickler, "# Re")
+ return
except ImportError:
pass
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -9,7 +9,7 @@
from functools import partial
from pathlib import Path
from unittest import TestCase
-from unittest.mock import patch
+from unittest.mock import MagicMock, patch
import numpy as np
import numpy.testing as npt
@@ -62,6 +62,11 @@
)
+class PickableMagicMock(MagicMock):
+ def __reduce__(self):
+ return MagicMock, ()
+
+
class Unpicklable:
def __getstate__(self):
raise pickle.PicklingError()
@@ -105,14 +110,6 @@ def assert_arrow_metadata_are_synced_with_dataset_features(dataset: Dataset):
@parameterized.named_parameters(IN_MEMORY_PARAMETERS)
class BaseDatasetTest(TestCase):
- def setUp(self):
- # google colab doesn't allow to pickle loggers
- # so we want to make sure each tests passes without pickling the logger
- def reduce_ex(self):
- raise pickle.PicklingError()
-
- datasets.arrow_dataset.logger.__reduce_ex__ = reduce_ex
-
@pytest.fixture(autouse=True)
def inject_fixtures(self, caplog):
self._caplog = caplog
@@ -1255,7 +1252,11 @@ def test_map_caching(self, in_memory):
self._caplog.clear()
with self._caplog.at_level(WARNING):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
- with patch("datasets.arrow_dataset.Pool", side_effect=datasets.arrow_dataset.Pool) as mock_pool:
+ with patch(
+ "datasets.arrow_dataset.Pool",
+ new_callable=PickableMagicMock,
+ side_effect=datasets.arrow_dataset.Pool,
+ ) as mock_pool:
with dset.map(lambda x: {"foo": "bar"}, num_proc=2) as dset_test1:
dset_test1_data_files = list(dset_test1.cache_files)
self.assertEqual(mock_pool.call_count, 1)
| Pip-compile: Could not find a version that matches dill<0.3.6,>=0.3.6
### Describe the bug
When using `pip-compile` (part of `pip-tools`) to generate a pinned requirements file that includes `datasets`, a version conflict of `dill` appears.
It is caused by a transitive dependency conflict between `datasets` and `multiprocess`.
### Steps to reproduce the bug
```bash
$ echo "datasets" > requirements.in
$ pip install pip-tools
$ pip-compile requirements.in
Could not find a version that matches dill<0.3.6,>=0.3.6 (from datasets==2.6.1->-r requirements.in (line 1))
Tried: 0.2, 0.2, 0.2.1, 0.2.1, 0.2.2, 0.2.2, 0.2.3, 0.2.3, 0.2.4, 0.2.4, 0.2.5, 0.2.5, 0.2.6, 0.2.7, 0.2.7.1, 0.2.8, 0.2.8.1, 0.2.8.2, 0.2.9, 0.3.0, 0.3.1, 0.3.1.1, 0.3.2, 0.3.3, 0.3.3, 0.3.4, 0.3.4, 0.3.5, 0.3.5, 0.3.5.1, 0.3.5.1, 0.3.6, 0.3.6
Skipped pre-versions: 0.1a1, 0.2a1, 0.2a1, 0.2b1, 0.2b1
There are incompatible versions in the resolved dependencies:
dill<0.3.6 (from datasets==2.6.1->-r requirements.in (line 1))
dill>=0.3.6 (from multiprocess==0.70.14->datasets==2.6.1->-r requirements.in (line 1))
```
### Expected behavior
A correctly generated file `requirements.txt` with pinned dependencies
### Environment info
Tested with versions `2.6.1, 2.6.0, 2.5.2` on Python 3.8 and 3.10 on Ubuntu 20.04LTS and Python 3.10 on MacOS 12.6 (M1).
| Thanks for reporting, @Rijgersberg.
We were waiting for the release of `dill` 0.3.6, that happened 2 days ago (24 Oct 2022): https://github.com/uqfoundation/dill/releases/tag/dill-0.3.6
- See comment: https://github.com/huggingface/datasets/pull/4397#discussion_r880629543
Also `multiprocess` 0.70.14 was released 2 days ago: https://github.com/uqfoundation/multiprocess/releases/tag/multiprocess-0.70.14
We are addressing this issue to align dependencies. | 2022-10-26T08:24:59Z | [] | [] |
huggingface/datasets | 5,169 | huggingface__datasets-5169 | [
"5157"
] | ff5bd84fac270095f908eae36faf5dcec496567a | diff --git a/src/datasets/features/audio.py b/src/datasets/features/audio.py
--- a/src/datasets/features/audio.py
+++ b/src/datasets/features/audio.py
@@ -18,6 +18,9 @@
from .features import FeatureType
+_ffmpeg_warned, _librosa_warned, _audioread_warned = False, False, False
+
+
@dataclass
class Audio:
"""Audio Feature to extract audio data from an audio file.
@@ -295,12 +298,15 @@ def _decode_mp3(self, path_or_file):
try: # try torchaudio anyway because sometimes it works (depending on the os and os packages installed)
array, sampling_rate = self._decode_mp3_torchaudio(path_or_file)
except RuntimeError:
- warnings.warn(
- "\nTo support 'mp3' decoding with `torchaudio>=0.12.0`, please install `ffmpeg4` system package. On Google Colab you can run:\n\n"
- "\t!add-apt-repository -y ppa:jonathonf/ffmpeg-4 && apt update && apt install -y ffmpeg\n\n"
- "and restart your runtime. Alternatively, you can downgrade `torchaudio`:\n\n"
- "\tpip install \"torchaudio<0.12\"`.\n\nOtherwise 'mp3' files will be decoded with `librosa`."
- )
+ global _ffmpeg_warned
+ if not _ffmpeg_warned:
+ warnings.warn(
+ "\nTo support 'mp3' decoding with `torchaudio>=0.12.0`, please install `ffmpeg4` system package. On Google Colab you can run:\n\n"
+ "\t!add-apt-repository -y ppa:jonathonf/ffmpeg-4 && apt update && apt install -y ffmpeg\n\n"
+ "and restart your runtime. Alternatively, you can downgrade `torchaudio`:\n\n"
+ "\tpip install \"torchaudio<0.12\"`.\n\nOtherwise 'mp3' files will be decoded with `librosa`."
+ )
+ _ffmpeg_warned = True
try:
# flake8: noqa
import librosa
@@ -309,11 +315,14 @@ def _decode_mp3(self, path_or_file):
"To support 'mp3' decoding with `torchaudio>=0.12.0`, please install `ffmpeg4` system package. On Google Colab you can run:\n\n"
"\t!add-apt-repository -y ppa:jonathonf/ffmpeg-4 && apt update && apt install -y ffmpeg\n\n"
"and restart your runtime. Alternatively, you can downgrade `torchaudio`:\n\n"
- "\tpip install \"torchaudio<0.12\"`.\n\nOtherwise 'mp3' files will be decoded with `librosa`:\n\n"
- "\tpip install librosa\n\nNote that decoding will be extremely slow in that case."
+ "\tpip install \"torchaudio<0.12\".\n\nTo decode 'mp3' files without `torchaudio`, please install `librosa`:\n\n"
+ "\tpip install librosa\n\nNote that decoding might be extremely slow in that case."
) from err
# try to decode with librosa for torchaudio>=0.12.0 as a workaround
- warnings.warn("Decoding mp3 with `librosa` instead of `torchaudio`, decoding is slow.")
+ global _librosa_warned
+ if not _librosa_warned:
+ warnings.warn("Decoding mp3 with `librosa` instead of `torchaudio`, decoding might be slow.")
+ _librosa_warned = True
try:
array, sampling_rate = self._decode_mp3_librosa(path_or_file)
except RuntimeError as err:
@@ -342,5 +351,13 @@ def _decode_mp3_torchaudio(self, path_or_file):
def _decode_mp3_librosa(self, path_or_file):
import librosa
- array, sampling_rate = librosa.load(path_or_file, mono=self.mono, sr=self.sampling_rate)
+ global _audioread_warned
+
+ with warnings.catch_warnings():
+ if _audioread_warned:
+ warnings.filterwarnings("ignore", "pysoundfile failed.+?", UserWarning, module=librosa.__name__)
+ else:
+ _audioread_warned = True
+ array, sampling_rate = librosa.load(path_or_file, mono=self.mono, sr=self.sampling_rate)
+
return array, sampling_rate
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -657,6 +657,8 @@ def _save_code(pickler, obj):
# The filename of a function is the .py file where it is defined.
# Filenames of functions created in notebooks or shells start with '<'
# ex: <ipython-input-13-9ed2afe61d25> for ipython, and <stdin> for shell
+ # Filenames of functions created in ipykernel the filename
+ # look like f"{tempdir}/ipykernel_{id1}/{id2}.py"
# Moreover lambda functions have a special name: '<lambda>'
# ex: (lambda x: x).__code__.co_name == "<lambda>" # True
#
@@ -669,7 +671,14 @@ def _save_code(pickler, obj):
#
# Only those two lines are different from the original implementation:
co_filename = (
- "" if obj.co_filename.startswith("<") or obj.co_name == "<lambda>" else os.path.basename(obj.co_filename)
+ ""
+ if obj.co_filename.startswith("<")
+ or (
+ len(obj.co_filename.split(os.path.sep)) > 1
+ and obj.co_filename.split(os.path.sep)[-2].startswith("ipykernel_")
+ )
+ or obj.co_name == "<lambda>"
+ else os.path.basename(obj.co_filename)
)
co_firstlineno = 1
# The rest is the same as in the original dill implementation
@@ -744,6 +753,8 @@ def save_code(pickler, obj):
# The filename of a function is the .py file where it is defined.
# Filenames of functions created in notebooks or shells start with '<'
# ex: <ipython-input-13-9ed2afe61d25> for ipython, and <stdin> for shell
+ # Filenames of functions created in ipykernel the filename
+ # look like f"{tempdir}/ipykernel_{id1}/{id2}.py"
# Moreover lambda functions have a special name: '<lambda>'
# ex: (lambda x: x).__code__.co_name == "<lambda>" # True
#
@@ -756,7 +767,14 @@ def save_code(pickler, obj):
#
# Only those two lines are different from the original implementation:
co_filename = (
- "" if obj.co_filename.startswith("<") or obj.co_name == "<lambda>" else os.path.basename(obj.co_filename)
+ ""
+ if obj.co_filename.startswith("<")
+ or (
+ len(obj.co_filename.split(os.path.sep)) > 1
+ and obj.co_filename.split(os.path.sep)[-2].startswith("ipykernel_")
+ )
+ or obj.co_name == "<lambda>"
+ else os.path.basename(obj.co_filename)
)
co_firstlineno = 1
# The rest is the same as in the original dill implementation, except for the replacements:
| diff --git a/tests/features/test_audio.py b/tests/features/test_audio.py
--- a/tests/features/test_audio.py
+++ b/tests/features/test_audio.py
@@ -150,9 +150,7 @@ def test_audio_decode_example_mp3_torchaudio_latest(shared_datadir, torchaudio_f
audio_path = str(shared_datadir / "test_audio_44100.mp3")
audio = Audio()
- with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock, pytest.warns(
- UserWarning, match=r"Decoding mp3 with `librosa` instead of `torchaudio`.+?"
- ) if torchaudio_failed else nullcontext():
+ with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock:
if torchaudio_failed:
load_mock.side_effect = RuntimeError()
@@ -215,9 +213,7 @@ def test_audio_resampling_mp3_different_sampling_rates_torchaudio_latest(shared_
audio = Audio(sampling_rate=48000)
# if torchaudio>=0.12 failed, mp3 must be decoded anyway (with librosa)
- with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock, pytest.warns(
- UserWarning, match=r"Decoding mp3 with `librosa` instead of `torchaudio`.+?"
- ) if torchaudio_failed else nullcontext():
+ with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock:
if torchaudio_failed:
load_mock.side_effect = RuntimeError()
@@ -453,9 +449,7 @@ def test_resampling_at_loading_dataset_with_audio_feature_mp3_torchaudio_latest(
dset = Dataset.from_dict(data, features=features)
# if torchaudio>=0.12 failed, mp3 must be decoded anyway (with librosa)
- with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock, pytest.warns(
- UserWarning, match=r"Decoding mp3 with `librosa` instead of `torchaudio`.+?"
- ) if torchaudio_failed else nullcontext():
+ with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock:
if torchaudio_failed:
load_mock.side_effect = RuntimeError()
@@ -551,9 +545,7 @@ def test_resampling_after_loading_dataset_with_audio_feature_mp3_torchaudio_late
dset = Dataset.from_dict(data, features=features)
# if torchaudio>=0.12 failed, mp3 must be decoded anyway (with librosa)
- with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock, pytest.warns(
- UserWarning, match=r"Decoding mp3 with `librosa` instead of `torchaudio`.+?"
- ) if torchaudio_failed else nullcontext():
+ with patch("torchaudio.load") if torchaudio_failed else nullcontext() as load_mock:
if torchaudio_failed:
load_mock.side_effect = RuntimeError()
diff --git a/tests/test_fingerprint.py b/tests/test_fingerprint.py
--- a/tests/test_fingerprint.py
+++ b/tests/test_fingerprint.py
@@ -1,8 +1,10 @@
import json
+import os
import pickle
import subprocess
from hashlib import md5
from pathlib import Path
+from tempfile import gettempdir
from textwrap import dedent
from types import CodeType, FunctionType
from unittest import TestCase
@@ -206,6 +208,15 @@ def func():
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
+ co_filename, returned_obj = os.path.join(gettempdir(), "ipykernel_12345", "321456789.py"), [0]
+ hash4 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
+ co_filename, returned_obj = os.path.join(gettempdir(), "ipykernel_12345", "321456789.py"), [1]
+ hash5 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
+ co_filename, returned_obj = os.path.join(gettempdir(), "ipykernel_12345", "654123987.py"), [0]
+ hash6 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
+ self.assertEqual(hash4, hash6)
+ self.assertNotEqual(hash4, hash5)
+
def test_recurse_dump_for_function_with_shuffled_globals(self):
foo, bar = [0], [1]
| Consistent caching between python and jupyter
### Feature request
I hope this is not my mistake, currently if I use `load_dataset` from a python session on a custom dataset to do the preprocessing, it will be saved in the cache and in other python sessions it will be loaded from the cache, however calling the same from a jupyter notebook does not work, meaning the preprocessing starts from scratch.
If adjusting the hashes is impossible, is there a way to manually set dataset fingerprint to "force" this behaviour?
### Motivation
If this is not already the case and I am doing something wrong, it would be useful to have the two fingerprints consistent so one can create the dataset once and then try small things on jupyter without preprocessing everything again.
### Your contribution
I am happy to try a PR if you give me some pointers where the changes should happen
| Hi ! Maybe it's possible to have a consistent hash for a function defined in `__main__` and a function define in a notebook.
However for functions imported from another location, pickle uses the location to identify the code, so in that case we can't do much I believe.
Would it be ok for you if we only try to do this for functions in `__main__` / jupyter ?
If you'd like to contribute, you can read this part of the code and let me know if you have questions:
https://github.com/huggingface/datasets/blob/7feeb5648a63b6135a8259dedc3b1e19185ee4c7/src/datasets/utils/py_utils.py#L617-L643
I think the key here would be to also ignore the "co_filename" of functions defined in `__main__`
Seems like a good solution, I will start a PR and see if I understood the changes needed. Thanks! | 2022-10-27T05:56:17Z | [] | [] |
huggingface/datasets | 5,268 | huggingface__datasets-5268 | [
"5263"
] | 4377c5b5735d62d009878d44d43402b1151f7953 | diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -128,7 +128,7 @@
"importlib_metadata;python_version<'3.8'",
# to save datasets locally or on any filesystem
# minimum 2021.11.1 so that BlockSizeError is fixed: see https://github.com/fsspec/filesystem_spec/pull/830
- "fsspec[http]>=2021.11.1", # aligned s3fs with this
+ "fsspec[http]>=2021.11.1",
# for data streaming via http
"aiohttp",
# To get datasets from the Datasets Hub on huggingface.co
@@ -165,13 +165,8 @@
# optional dependencies
"apache-beam>=2.26.0;python_version<'3.8'", # doesn't support recent dill versions for recent python versions
"elasticsearch<8.0.0", # 8.0 asks users to provide hosts or cloud_id when instantiating ElasticSearch()
- "aiobotocore>=2.0.1;python_version<'3.8'", # required by s3fs>=2021.11.1
- "boto3>=1.19.8;python_version<'3.8'", # to be compatible with aiobotocore>=2.0.1 - both have strong dependencies on botocore
- "botocore>=1.22.8;python_version<'3.8'", # to be compatible with aiobotocore and boto3
"faiss-cpu>=1.6.4",
- "fsspec",
"lz4",
- "moto[s3,server]==2.0.4;python_version<'3.8'",
"py7zr",
"rarfile>=4.0",
"s3fs>=2021.11.1;python_version<'3.8'", # aligned with fsspec[http]>=2021.11.1; test only on python 3.7 for now
@@ -232,12 +227,7 @@
],
"tensorflow_gpu": ["tensorflow-gpu>=2.2.0,!=2.6.0,!=2.6.1"],
"torch": ["torch"],
- "s3": [
- "fsspec",
- "boto3",
- "botocore",
- "s3fs",
- ],
+ "s3": ["s3fs"],
"streaming": [], # for backward compatibility
"dev": TESTS_REQUIRE + QUALITY_REQUIRE + DOCS_REQUIRE,
"tests": TESTS_REQUIRE,
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -20,10 +20,12 @@
import itertools
import json
import os
+import posixpath
import re
import shutil
import sys
import tempfile
+import time
import warnings
import weakref
from collections import Counter
@@ -107,7 +109,7 @@
from .utils.hub import hf_hub_url
from .utils.info_utils import is_small_dataset
from .utils.metadata import DatasetMetadata
-from .utils.py_utils import asdict, convert_file_size_to_int, unique_values
+from .utils.py_utils import asdict, convert_file_size_to_int, iflatmap_unordered, unique_values
from .utils.stratify import stratified_shuffle_split_generate_indices
from .utils.tf_utils import minimal_tf_collate_fn
from .utils.typing import PathLike
@@ -1256,64 +1258,103 @@ def __exit__(self, exc_type, exc_val, exc_tb):
# Here `del` is used to del the pyarrow tables. This properly closes the files used for memory mapped tables
self.__del__()
- def save_to_disk(self, dataset_path: str, fs=None):
+ def save_to_disk(
+ self,
+ dataset_path: PathLike,
+ fs="deprecated",
+ max_shard_size: Optional[Union[str, int]] = None,
+ num_shards: Optional[int] = None,
+ num_proc: Optional[int] = None,
+ storage_options: Optional[dict] = None,
+ ):
"""
- Saves a dataset to a dataset directory, or in a filesystem using either `filesystems.S3FileSystem` or
+ Saves a dataset to a dataset directory, or in a filesystem using either `s3fs.S3FileSystem` or
any implementation of `fsspec.spec.AbstractFileSystem`.
For [`Image`] and [`Audio`] data:
- If your images and audio files are local files, then the resulting arrow file will store paths to these files.
- If you want to include the bytes or your images or audio files instead, you must `read()` those files first.
- This can be done by storing the "bytes" instead of the "path" of the images or audio files:
-
- ```python
- >>> def read_image_file(example):
- ... with open(example["image"].filename, "rb") as f:
- ... return {"image": {"bytes": f.read()}}
- >>> ds = ds.map(read_image_file)
- >>> ds.save_to_disk("path/to/dataset/dir")
- ```
-
- ```python
- >>> def read_audio_file(example):
- ... with open(example["audio"]["path"], "rb") as f:
- ... return {"audio": {"bytes": f.read()}}
- >>> ds = ds.map(read_audio_file)
- >>> ds.save_to_disk("path/to/dataset/dir")
- ```
+ All the Image() and Audio() data are stored in the arrow files.
+ If you want to store paths or urls, please use the Value("string") type.
Args:
dataset_path (`str`):
Path (e.g. `dataset/train`) or remote URI (e.g. `s3://my-bucket/dataset/train`)
of the dataset directory where the dataset will be saved to.
- fs (`filesystems.S3FileSystem`, `fsspec.spec.AbstractFileSystem`, *optional*, defaults to `None`):
- Instance of the remote filesystem used to download the files from.
+ fs (`fsspec.spec.AbstractFileSystem`, *optional*):
+ Instance of the remote filesystem where the dataset will be saved to.
+
+ <Deprecated version="2.8.0">
+
+ `fs` was deprecated in version 2.8.0 and will be removed in 3.0.0.
+ Please use `storage_options` instead, e.g. `storage_options=fs.storage_options`
+
+ </Deprecated>
+
+ max_shard_size (`int` or `str`, *optional*, defaults to `"500MB"`):
+ The maximum size of the dataset shards to be uploaded to the hub. If expressed as a string, needs to be digits followed by a unit
+ (like `"50MB"`).
+ num_shards (`int`, *optional*):
+ Number of shards to write. By default the number of shards depends on `max_shard_size`.
+
+ <Added version="2.8.0"/>
+ num_proc (`int`, *optional*):
+ Number of processes when downloading and generating the dataset locally.
+ Multiprocessing is disabled by default.
+
+ <Added version="2.8.0"/>
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.8.0"/>
Example:
```py
- >>> saved_ds = ds.save_to_disk("path/to/dataset/directory")
+ >>> ds.save_to_disk("path/to/dataset/directory")
+ >>> ds.save_to_disk("path/to/dataset/directory", max_shard_size="1GB")
+ >>> ds.save_to_disk("path/to/dataset/directory", num_shards=1024)
```
"""
+ if max_shard_size is not None and num_shards is not None:
+ raise ValueError(
+ "Failed to push_to_hub: please specify either max_shard_size or num_shards, but not both."
+ )
+ if fs != "deprecated":
+ warnings.warn(
+ "'fs' was is deprecated in favor of 'storage_options' in version 2.8.0 and will be removed in 3.0.0.\n"
+ "You can remove this warning by passing 'storage_options=fs.storage_options' instead.",
+ FutureWarning,
+ )
+ storage_options = fs.storage_options
+
+ if num_shards is None:
+ dataset_nbytes = self._estimate_nbytes()
+ max_shard_size = convert_file_size_to_int(max_shard_size or config.MAX_SHARD_SIZE)
+ num_shards = int(dataset_nbytes / max_shard_size) + 1
+ num_shards = max(num_shards, num_proc or 1)
+
+ num_proc = num_proc if num_proc is not None else 1
+ num_shards = num_shards if num_shards is not None else num_proc
+
+ fs_token_paths = fsspec.get_fs_token_paths(dataset_path, storage_options=storage_options)
+ fs: fsspec.AbstractFileSystem = fs_token_paths[0]
+ is_local = not is_remote_filesystem(fs)
+ path_join = os.path.join if is_local else posixpath.join
+
if self.list_indexes():
raise ValueError("please remove all the indexes using `dataset.drop_index` before saving a dataset")
- dataset = self.flatten_indices() if self._indices is not None else self
+ dataset = self.flatten_indices(num_proc=num_proc) if self._indices is not None else self
- if is_remote_filesystem(fs):
- dataset_path = extract_path_from_uri(dataset_path)
- else:
- fs = fsspec.filesystem("file")
- cache_files_paths = [Path(cache_filename["filename"]) for cache_filename in self.cache_files]
+ if is_local:
+ Path(dataset_path).resolve().mkdir(parents=True, exist_ok=True)
+ parent_cache_files_paths = set(
+ Path(cache_filename["filename"]).resolve().parent for cache_filename in self.cache_files
+ )
# Check that the dataset doesn't overwrite iself. It can cause a permission error on Windows and a segfault on linux.
- if Path(dataset_path, config.DATASET_ARROW_FILENAME) in cache_files_paths:
+ if Path(dataset_path).resolve() in parent_cache_files_paths:
raise PermissionError(
- f"Tried to overwrite {Path(dataset_path, config.DATASET_ARROW_FILENAME)} but a dataset can't overwrite itself."
- )
- if Path(dataset_path, config.DATASET_INDICES_FILENAME) in cache_files_paths:
- raise PermissionError(
- f"Tried to overwrite {Path(dataset_path, config.DATASET_INDICES_FILENAME)} but a dataset can't overwrite itself."
+ f"Tried to overwrite {Path(dataset_path).resolve()} but a dataset can't overwrite itself."
)
# Get json serializable state
@@ -1324,15 +1365,13 @@ def save_to_disk(self, dataset_path: str, fs=None):
"_format_columns",
"_format_kwargs",
"_format_type",
- "_indexes",
"_output_all_columns",
]
}
-
- split = dataset.__dict__["_split"]
- state["_split"] = str(split) if split is not None else split
-
- state["_data_files"] = [{"filename": config.DATASET_ARROW_FILENAME}]
+ state["_split"] = str(dataset.split) if dataset.split is not None else dataset.split
+ state["_data_files"] = [
+ {"filename": f"data-{shard_idx:05d}-of-{num_shards:05d}.arrow"} for shard_idx in range(num_shards)
+ ]
for k in state["_format_kwargs"].keys():
try:
json.dumps(state["_format_kwargs"][k])
@@ -1340,27 +1379,91 @@ def save_to_disk(self, dataset_path: str, fs=None):
raise TypeError(
str(e) + f"\nThe format kwargs must be JSON serializable, but key '{k}' isn't."
) from None
-
# Get json serializable dataset info
dataset_info = asdict(dataset._info)
- # Save dataset + state + info
- fs.makedirs(dataset_path, exist_ok=True)
- with fs.open(Path(dataset_path, config.DATASET_ARROW_FILENAME).as_posix(), "wb") as dataset_file:
- with ArrowWriter(stream=dataset_file) as writer:
- writer.write_table(dataset._data.table)
- writer.finalize()
- with fs.open(
- Path(dataset_path, config.DATASET_STATE_JSON_FILENAME).as_posix(), "w", encoding="utf-8"
- ) as state_file:
+ shards_done = 0
+ pbar = logging.tqdm(
+ disable=not logging.is_progress_bar_enabled(),
+ unit=" examples",
+ total=len(dataset),
+ leave=False,
+ desc=f"Saving the dataset ({shards_done}/{num_shards} shards)",
+ )
+ kwargs_per_job = (
+ {
+ "job_id": shard_idx,
+ "shard": dataset.shard(num_shards=num_shards, index=shard_idx, contiguous=True),
+ "fpath": path_join(dataset_path, f"data-{shard_idx:05d}-of-{num_shards:05d}.arrow"),
+ "storage_options": storage_options,
+ }
+ for shard_idx in range(num_shards)
+ )
+ shard_lengths = [None] * num_shards
+ shard_sizes = [None] * num_shards
+ if num_proc > 1:
+ with Pool(num_proc) as pool:
+ for job_id, done, content in iflatmap_unordered(
+ pool, Dataset._save_to_disk_single, kwargs_iterable=kwargs_per_job
+ ):
+ if done:
+ shards_done += 1
+ pbar.set_description(f"Saving the dataset ({shards_done}/{num_shards} shards)")
+ logger.debug(f"Finished writing shard number {job_id} of {num_shards}.")
+ shard_lengths[job_id], shard_sizes[job_id] = content
+ else:
+ pbar.update(content)
+ else:
+ for kwargs in kwargs_per_job:
+ for job_id, done, content in Dataset._save_to_disk_single(**kwargs):
+ if done:
+ shards_done += 1
+ pbar.set_description(f"Saving the dataset ({shards_done}/{num_shards} shards)")
+ logger.debug(f"Finished writing shard number {job_id} of {num_shards}.")
+ shard_lengths[job_id], shard_sizes[job_id] = content
+ else:
+ pbar.update(content)
+ with fs.open(path_join(dataset_path, config.DATASET_STATE_JSON_FILENAME), "w", encoding="utf-8") as state_file:
json.dump(state, state_file, indent=2, sort_keys=True)
with fs.open(
- Path(dataset_path, config.DATASET_INFO_FILENAME).as_posix(), "w", encoding="utf-8"
+ path_join(dataset_path, config.DATASET_INFO_FILENAME), "w", encoding="utf-8"
) as dataset_info_file:
# Sort only the first level of keys, or we might shuffle fields of nested features if we use sort_keys=True
sorted_keys_dataset_info = {key: dataset_info[key] for key in sorted(dataset_info)}
json.dump(sorted_keys_dataset_info, dataset_info_file, indent=2)
- logger.info(f"Dataset saved in {dataset_path}")
+
+ @staticmethod
+ def _save_to_disk_single(job_id: int, shard: "Dataset", fpath: str, storage_options: Optional[dict]):
+ batch_size = config.DEFAULT_MAX_BATCH_SIZE
+
+ if shard._indices is not None:
+ raise ValueError(
+ "`_save_to_disk_single` only support shards with flattened indices. "
+ "Please call ds.flatten_indices() before saving to disk."
+ )
+
+ num_examples_progress_update = 0
+ writer = ArrowWriter(
+ features=shard.features,
+ path=fpath,
+ storage_options=storage_options,
+ embed_local_files=True,
+ )
+ try:
+ _time = time.time()
+ for pa_table in table_iter(shard.data, batch_size=batch_size):
+ writer.write_table(pa_table)
+ num_examples_progress_update += len(pa_table)
+ if time.time() > _time + config.PBAR_REFRESH_TIME_INTERVAL:
+ _time = time.time()
+ yield job_id, False, num_examples_progress_update
+ num_examples_progress_update = 0
+ finally:
+ yield job_id, False, num_examples_progress_update
+ num_examples, num_bytes = writer.finalize()
+ writer.close()
+
+ yield job_id, True, (num_examples, num_bytes)
@staticmethod
def _build_local_temp_path(uri_or_path: str) -> Path:
@@ -1380,23 +1483,40 @@ def _build_local_temp_path(uri_or_path: str) -> Path:
return Path(tmp_dir, src_dataset_path.relative_to(src_dataset_path.anchor))
@staticmethod
- def load_from_disk(dataset_path: str, fs=None, keep_in_memory: Optional[bool] = None) -> "Dataset":
+ def load_from_disk(
+ dataset_path: str,
+ fs="deprecated",
+ keep_in_memory: Optional[bool] = None,
+ storage_options: Optional[dict] = None,
+ ) -> "Dataset":
"""
Loads a dataset that was previously saved using [`save_to_disk`] from a dataset directory, or from a
- filesystem using either [`filesystems.S3FileSystem`] or any implementation of
+ filesystem using either `s3fs.S3FileSystem` or any implementation of
`fsspec.spec.AbstractFileSystem`.
Args:
dataset_path (`str`):
- Path (e.g. `"dataset/train"`) or remote URI (e.g.
- `"s3//my-bucket/dataset/train"`) of the dataset directory where the dataset will be loaded from.
- fs ([`filesystems.S3FileSystem`], `fsspec.spec.AbstractFileSystem`, *optional*, defaults to `None`):
- Instance of the remote filesystem used to download the files from.
+ Path (e.g. `"dataset/train"`) or remote URI (e.g. `"s3//my-bucket/dataset/train"`)
+ of the dataset directory where the dataset will be loaded from.
+ fs (`fsspec.spec.AbstractFileSystem`, *optional*):
+ Instance of the remote filesystem where the dataset will be saved to.
+
+ <Deprecated version="2.8.0">
+
+ `fs` was deprecated in version 2.8.0 and will be removed in 3.0.0.
+ Please use `storage_options` instead, e.g. `storage_options=fs.storage_options`
+
+ </Deprecated>
+
keep_in_memory (`bool`, defaults to `None`):
Whether to copy the dataset in-memory. If `None`, the
dataset will not be copied in-memory unless explicitly enabled by setting
`datasets.config.IN_MEMORY_MAX_SIZE` to nonzero. See more details in the
[improve performance](./cache#improve-performance) section.
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.8.0"/>
Returns:
[`Dataset`] or [`DatasetDict`]:
@@ -1409,8 +1529,17 @@ def load_from_disk(dataset_path: str, fs=None, keep_in_memory: Optional[bool] =
>>> ds = load_from_disk("path/to/dataset/directory")
```
"""
+ if fs != "deprecated":
+ warnings.warn(
+ "'fs' was is deprecated in favor of 'storage_options' in version 2.8.0 and will be removed in 3.0.0.\n"
+ "You can remove this warning by passing 'storage_options=fs.storage_options' instead.",
+ FutureWarning,
+ )
+ storage_options = fs.storage_options
+
+ fs_token_paths = fsspec.get_fs_token_paths(dataset_path, storage_options=storage_options)
+ fs: fsspec.AbstractFileSystem = fs_token_paths[0]
# copies file from filesystem if it is remote filesystem to local filesystem and modifies dataset_path to temp directory containing local copies
- fs = fsspec.filesystem("file") if fs is None else fs
dataset_dict_json_path = Path(dataset_path, config.DATASETDICT_JSON_FILENAME).as_posix()
dataset_info_path = Path(dataset_path, config.DATASET_INFO_FILENAME).as_posix()
if not fs.isfile(dataset_info_path) and fs.isfile(dataset_dict_json_path):
@@ -2044,7 +2173,7 @@ def __iter__(self):
If a formatting is set with :meth:`Dataset.set_format` rows will be returned with the
selected format.
"""
- if self._indices is None and config.PYARROW_VERSION.major >= 8:
+ if self._indices is None:
# Fast iteration
# Benchmark: https://gist.github.com/mariosasko/0248288a2e3a7556873969717c1fe52b (fast_iter_batch)
format_kwargs = self._format_kwargs if self._format_kwargs is not None else {}
@@ -3280,6 +3409,7 @@ def flatten_indices(
writer_batch_size: Optional[int] = 1000,
features: Optional[Features] = None,
disable_nullable: bool = False,
+ num_proc: Optional[int] = None,
new_fingerprint: Optional[str] = None,
) -> "Dataset":
"""Create and cache a new Dataset by flattening the indices mapping.
@@ -3299,6 +3429,8 @@ def flatten_indices(
instead of the automatically generated one.
disable_nullable (`bool`, defaults to `False`):
Allow null values in the table.
+ num_proc (`int`, optional, default `None`):
+ Max number of processes when generating cache. Already cached shards are loaded sequentially
new_fingerprint (`str`, *optional*, defaults to `None`):
The new fingerprint of the dataset after transform.
If `None`, the new fingerprint is computed using a hash of the previous fingerprint, and the transform arguments
@@ -3313,6 +3445,7 @@ def flatten_indices(
disable_nullable=disable_nullable,
new_fingerprint=new_fingerprint,
desc="Flattening the indices",
+ num_proc=num_proc,
)
def _new_dataset_with_indices(
@@ -4478,6 +4611,36 @@ def to_sql(
return SqlDatasetWriter(self, name, con, batch_size=batch_size, **sql_writer_kwargs).write()
+ def _estimate_nbytes(self) -> int:
+ dataset_nbytes = self.data.nbytes
+
+ # Find decodable columns, because if there are any, we need to
+ # adjust the dataset size computation (needed for sharding) to account for possible external files
+ decodable_columns = [k for k, v in self.features.items() if require_decoding(v, ignore_decode_attribute=True)]
+
+ if decodable_columns:
+ # Approximate the space needed to store the bytes from the external files by analyzing the first 1000 examples
+ extra_nbytes = 0
+
+ def extra_nbytes_visitor(array, feature):
+ nonlocal extra_nbytes
+ if isinstance(feature, (Audio, Image)):
+ for x in array.to_pylist():
+ if x is not None and x["bytes"] is None and x["path"] is not None:
+ size = xgetsize(x["path"])
+ extra_nbytes += size
+ extra_nbytes -= array.field("path").nbytes
+
+ table = self.with_format("arrow")[:1000]
+ table_visitor(table, extra_nbytes_visitor)
+
+ extra_nbytes = extra_nbytes * len(self.data) / len(table)
+ dataset_nbytes = dataset_nbytes + extra_nbytes
+
+ if self._indices is not None:
+ dataset_nbytes = dataset_nbytes * len(self._indices) / len(self.data)
+ return dataset_nbytes
+
def _push_parquet_shards_to_hub(
self,
repo_id: str,
@@ -4486,6 +4649,7 @@ def _push_parquet_shards_to_hub(
token: Optional[str] = None,
branch: Optional[str] = None,
max_shard_size: Optional[Union[int, str]] = None,
+ num_shards: Optional[int] = None,
embed_external_files: bool = True,
) -> Tuple[str, str, int, int]:
"""Pushes the dataset to the hub.
@@ -4509,8 +4673,12 @@ def _push_parquet_shards_to_hub(
The git branch on which to push the dataset. This defaults to the default branch as specified
in your repository, which defaults to `"main"`.
max_shard_size (`int` or `str`, *optional*, defaults to `"500MB"`):
- The maximum size of the dataset shards to be uploaded to the hub. If expressed as a string, needs to be digits followed by a unit
- (like `"5MB"`).
+ The maximum size of the dataset shards to be uploaded to the hub. If expressed as a string, needs to be digits followed by a
+ a unit (like `"5MB"`).
+ num_shards (`int`, *optional*):
+ Number of shards to write. By default the number of shards depends on `max_shard_size`.
+
+ <Added version="2.8.0"/>
embed_external_files (`bool`, default ``True``):
Whether to embed file bytes in the shards.
In particular, this will do the following before the push for the fields of type:
@@ -4531,7 +4699,10 @@ def _push_parquet_shards_to_hub(
>>> dataset.push_to_hub("<organization>/<dataset_id>", split="evaluation")
```
"""
- max_shard_size = convert_file_size_to_int(max_shard_size or config.MAX_SHARD_SIZE)
+ if max_shard_size is not None and num_shards is not None:
+ raise ValueError(
+ "Failed to push_to_hub: please specify either max_shard_size or num_shards, but not both."
+ )
api = HfApi(endpoint=config.HF_ENDPOINT)
token = token if token is not None else HfFolder.get_token()
@@ -4569,40 +4740,20 @@ def _push_parquet_shards_to_hub(
)
# Find decodable columns, because if there are any, we need to:
- # (1) adjust the dataset size computation (needed for sharding) to account for possible external files
- # (2) embed the bytes from the files in the shards
+ # embed the bytes from the files in the shards
decodable_columns = (
[k for k, v in self.features.items() if require_decoding(v, ignore_decode_attribute=True)]
if embed_external_files
else []
)
- dataset_nbytes = self.data.nbytes
-
- if decodable_columns:
- # Approximate the space needed to store the bytes from the external files by analyzing the first 1000 examples
- extra_nbytes = 0
-
- def extra_nbytes_visitor(array, feature):
- nonlocal extra_nbytes
- if isinstance(feature, (Audio, Image)):
- for x in array.to_pylist():
- if x is not None and x["bytes"] is None and x["path"] is not None:
- size = xgetsize(x["path"])
- extra_nbytes += size
- extra_nbytes -= array.field("path").nbytes
-
- table = self.with_format("arrow")[:1000]
- table_visitor(table, extra_nbytes_visitor)
+ dataset_nbytes = self._estimate_nbytes()
- extra_nbytes = extra_nbytes * len(self.data) / len(table)
- dataset_nbytes = dataset_nbytes + extra_nbytes
+ if num_shards is None:
+ max_shard_size = convert_file_size_to_int(max_shard_size or config.MAX_SHARD_SIZE)
+ num_shards = int(dataset_nbytes / max_shard_size) + 1
+ num_shards = max(num_shards, 1)
- if self._indices is not None:
- dataset_nbytes = dataset_nbytes * len(self._indices) / len(self.data)
-
- num_shards = int(dataset_nbytes / max_shard_size) + 1
- num_shards = max(num_shards, 1)
shards = (self.shard(num_shards=num_shards, index=i, contiguous=True) for i in range(num_shards))
if decodable_columns:
@@ -4700,6 +4851,7 @@ def push_to_hub(
token: Optional[str] = None,
branch: Optional[str] = None,
max_shard_size: Optional[Union[int, str]] = None,
+ num_shards: Optional[int] = None,
shard_size: Optional[int] = "deprecated",
embed_external_files: bool = True,
):
@@ -4728,8 +4880,11 @@ def push_to_hub(
The git branch on which to push the dataset. This defaults to the default branch as specified
in your repository, which defaults to `"main"`.
max_shard_size (`int` or `str`, *optional*, defaults to `"500MB"`):
- The maximum size of the dataset shards to be uploaded to the hub. If expressed as a string, needs to be digits followed by a unit
- (like `"5MB"`).
+ The maximum size of the dataset shards to be uploaded to the hub. If expressed as a string, needs to be digits followed by
+ a unit (like `"5MB"`).
+ num_shards (`int`, *optional*): Number of shards to write. By default the number of shards depends on `max_shard_size`.
+
+ <Added version="2.8.0"/>
shard_size (`int`, *optional*):
<Deprecated version="2.4.0">
@@ -4747,7 +4902,10 @@ def push_to_hub(
Example:
```python
- >>> dataset.push_to_hub("<organization>/<dataset_id>", split="evaluation")
+ >>> dataset.push_to_hub("<organization>/<dataset_id>")
+ >>> dataset.push_to_hub("<organization>/<dataset_id>", split="validation")
+ >>> dataset.push_to_hub("<organization>/<dataset_id>", max_shard_size="1GB")
+ >>> dataset.push_to_hub("<organization>/<dataset_id>", num_shards=1024)
```
"""
if shard_size != "deprecated":
@@ -4757,6 +4915,11 @@ def push_to_hub(
)
max_shard_size = shard_size
+ if max_shard_size is not None and num_shards is not None:
+ raise ValueError(
+ "Failed to push_to_hub: please specify either max_shard_size or num_shards, but not both."
+ )
+
repo_id, split, uploaded_size, dataset_nbytes, repo_files, deleted_size = self._push_parquet_shards_to_hub(
repo_id=repo_id,
split=split,
@@ -4764,6 +4927,7 @@ def push_to_hub(
token=token,
branch=branch,
max_shard_size=max_shard_size,
+ num_shards=num_shards,
embed_external_files=embed_external_files,
)
organization, dataset_name = repo_id.split("/")
diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -1447,7 +1447,7 @@ def _prepare_split(
gen_kwargs = split_generator.gen_kwargs
job_id = 0
for job_id, done, content in self._prepare_split_single(
- {"gen_kwargs": gen_kwargs, "job_id": job_id, **_prepare_split_args}
+ gen_kwargs=gen_kwargs, job_id=job_id, **_prepare_split_args
):
if done:
result = content
@@ -1459,13 +1459,13 @@ def _prepare_split(
[item] for item in result
]
else:
- args_per_job = [
+ kwargs_per_job = [
{"gen_kwargs": gen_kwargs, "job_id": job_id, **_prepare_split_args}
for job_id, gen_kwargs in enumerate(
_split_gen_kwargs(split_generator.gen_kwargs, max_num_jobs=num_proc)
)
]
- num_jobs = len(args_per_job)
+ num_jobs = len(kwargs_per_job)
examples_per_job = [None] * num_jobs
bytes_per_job = [None] * num_jobs
@@ -1474,7 +1474,9 @@ def _prepare_split(
shard_lengths_per_job = [None] * num_jobs
with Pool(num_proc) as pool:
- for job_id, done, content in iflatmap_unordered(pool, self._prepare_split_single, args_per_job):
+ for job_id, done, content in iflatmap_unordered(
+ pool, self._prepare_split_single, kwargs_iterable=kwargs_per_job
+ ):
if done:
# the content is the result of the job
(
@@ -1534,15 +1536,16 @@ def _rename_shard(shard_and_job: Tuple[int]):
if self.info.features is None:
self.info.features = features
- def _prepare_split_single(self, arg: dict) -> Iterable[Tuple[int, bool, Union[int, tuple]]]:
- gen_kwargs: dict = arg["gen_kwargs"]
- fpath: str = arg["fpath"]
- file_format: str = arg["file_format"]
- max_shard_size: int = arg["max_shard_size"]
- split_info: SplitInfo = arg["split_info"]
- check_duplicate_keys: bool = arg["check_duplicate_keys"]
- job_id: int = arg["job_id"]
- refresh_rate = 0.05 # 20 progress updates per sec
+ def _prepare_split_single(
+ self,
+ gen_kwargs: dict,
+ fpath: str,
+ file_format: str,
+ max_shard_size: int,
+ split_info: SplitInfo,
+ check_duplicate_keys: bool,
+ job_id: int,
+ ) -> Iterable[Tuple[int, bool, Union[int, tuple]]]:
generator = self._generate_examples(**gen_kwargs)
writer_class = ParquetWriter if file_format == "parquet" else ArrowWriter
@@ -1584,7 +1587,7 @@ def _prepare_split_single(self, arg: dict) -> Iterable[Tuple[int, bool, Union[in
example = self.info.features.encode_example(record) if self.info.features is not None else record
writer.write(example, key)
num_examples_progress_update += 1
- if time.time() > _time + refresh_rate:
+ if time.time() > _time + config.PBAR_REFRESH_TIME_INTERVAL:
_time = time.time()
yield job_id, False, num_examples_progress_update
num_examples_progress_update = 0
@@ -1694,7 +1697,6 @@ def _prepare_split(
"fpath": fpath,
"file_format": file_format,
"max_shard_size": max_shard_size,
- "split_info": split_info,
}
if num_proc is None or num_proc == 1:
@@ -1702,7 +1704,7 @@ def _prepare_split(
gen_kwargs = split_generator.gen_kwargs
job_id = 0
for job_id, done, content in self._prepare_split_single(
- {"gen_kwargs": gen_kwargs, "job_id": job_id, **_prepare_split_args}
+ gen_kwargs=gen_kwargs, job_id=job_id, **_prepare_split_args
):
if done:
result = content
@@ -1714,13 +1716,13 @@ def _prepare_split(
[item] for item in result
]
else:
- args_per_job = [
+ kwargs_per_job = [
{"gen_kwargs": gen_kwargs, "job_id": job_id, **_prepare_split_args}
for job_id, gen_kwargs in enumerate(
_split_gen_kwargs(split_generator.gen_kwargs, max_num_jobs=num_proc)
)
]
- num_jobs = len(args_per_job)
+ num_jobs = len(kwargs_per_job)
examples_per_job = [None] * num_jobs
bytes_per_job = [None] * num_jobs
@@ -1729,7 +1731,9 @@ def _prepare_split(
shard_lengths_per_job = [None] * num_jobs
with Pool(num_proc) as pool:
- for job_id, done, content in iflatmap_unordered(pool, self._prepare_split_single, args_per_job):
+ for job_id, done, content in iflatmap_unordered(
+ pool, self._prepare_split_single, kwargs_iterable=kwargs_per_job
+ ):
if done:
# the content is the result of the job
(
@@ -1789,13 +1793,9 @@ def _rename_shard(shard_id_and_job: Tuple[int]):
if self.info.features is None:
self.info.features = features
- def _prepare_split_single(self, arg: dict) -> Iterable[Tuple[int, bool, Union[int, tuple]]]:
- gen_kwargs: dict = arg["gen_kwargs"]
- fpath: str = arg["fpath"]
- file_format: str = arg["file_format"]
- max_shard_size: int = arg["max_shard_size"]
- job_id: int = arg["job_id"]
- refresh_rate = 0.05 # 20 progress updates per sec
+ def _prepare_split_single(
+ self, gen_kwargs: dict, fpath: str, file_format: str, max_shard_size: int, job_id: int
+ ) -> Iterable[Tuple[int, bool, Union[int, tuple]]]:
generator = self._generate_tables(**gen_kwargs)
writer_class = ParquetWriter if file_format == "parquet" else ArrowWriter
@@ -1830,7 +1830,7 @@ def _prepare_split_single(self, arg: dict) -> Iterable[Tuple[int, bool, Union[in
)
writer.write_table(table)
num_examples_progress_update += len(table)
- if time.time() > _time + refresh_rate:
+ if time.time() > _time + config.PBAR_REFRESH_TIME_INTERVAL:
_time = time.time()
yield job_id, False, num_examples_progress_update
num_examples_progress_update = 0
diff --git a/src/datasets/config.py b/src/datasets/config.py
--- a/src/datasets/config.py
+++ b/src/datasets/config.py
@@ -210,3 +210,6 @@
DATA_FILES_MAX_NUMBER_FOR_MODULE_INFERENCE = 200
GLOBBED_DATA_FILES_MAX_NUMBER_FOR_MODULE_INFERENCE = 10
ARCHIVED_DATA_FILES_MAX_NUMBER_FOR_MODULE_INFERENCE = 200
+
+# Progress bars
+PBAR_REFRESH_TIME_INTERVAL = 0.05 # 20 progress updates per sec
diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -2,6 +2,7 @@
import copy
import json
import os
+import posixpath
import re
import warnings
from io import BytesIO
@@ -1096,73 +1097,136 @@ def shuffle(
}
)
- def save_to_disk(self, dataset_dict_path: str, fs=None):
+ def save_to_disk(
+ self,
+ dataset_dict_path: PathLike,
+ fs="deprecated",
+ max_shard_size: Optional[Union[str, int]] = None,
+ num_shards: Optional[Dict[str, int]] = None,
+ num_proc: Optional[int] = None,
+ storage_options: Optional[dict] = None,
+ ):
"""
Saves a dataset dict to a filesystem using either [`~filesystems.S3FileSystem`] or
`fsspec.spec.AbstractFileSystem`.
For [`Image`] and [`Audio`] data:
- If your images and audio files are local files, then the resulting arrow file will store paths to these files.
- If you want to include the bytes or your images or audio files instead, you must `read()` those files first.
- This can be done by storing the "bytes" instead of the "path" of the images or audio files:
-
- ```python
- >>> def read_image_file(example):
- ... with open(example["image"].filename, "rb") as f:
- ... return {"image": {"bytes": f.read()}}
- >>> ds = ds.map(read_image_file)
- >>> ds.save_to_disk("path/to/dataset/dir")
- ```
-
- ```python
- >>> def read_audio_file(example):
- ... with open(example["audio"]["path"], "rb") as f:
- ... return {"audio": {"bytes": f.read()}}
- >>> ds = ds.map(read_audio_file)
- >>> ds.save_to_disk("path/to/dataset/dir")
- ```
+ All the Image() and Audio() data are stored in the arrow files.
+ If you want to store paths or urls, please use the Value("string") type.
Args:
dataset_dict_path (`str`):
Path (e.g. `dataset/train`) or remote URI
(e.g. `s3://my-bucket/dataset/train`) of the dataset dict directory where the dataset dict will be
saved to.
- fs ([`~filesystems.S3FileSystem`], `fsspec.spec.AbstractFileSystem`, *optional*, defaults to `None`):
- Instance of the remote filesystem used to download the files from.
+ fs (`fsspec.spec.AbstractFileSystem`, *optional*):
+ Instance of the remote filesystem where the dataset will be saved to.
+
+ <Deprecated version="2.8.0">
+
+ `fs` was deprecated in version 2.8.0 and will be removed in 3.0.0.
+ Please use `storage_options` instead, e.g. `storage_options=fs.storage_options`
+
+ </Deprecated>
+
+ max_shard_size (`int` or `str`, *optional*, defaults to `"500MB"`):
+ The maximum size of the dataset shards to be uploaded to the hub. If expressed as a string, needs to be digits followed by a unit
+ (like `"50MB"`).
+ num_shards (`Dict[str, int]`, *optional*):
+ Number of shards to write. By default the number of shards depends on `max_shard_size`.
+ You need to provide the number of shards for each dataset in the dataset dictionary.
+ Use a dictionary to define a different num_shards for each split.
+
+ <Added version="2.8.0"/>
+ num_proc (`int`, *optional*, default `None`):
+ Number of processes when downloading and generating the dataset locally.
+ Multiprocessing is disabled by default.
+
+ <Added version="2.8.0"/>
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.8.0"/>
+
+ Example:
+
+ ```python
+ >>> dataset_dict.save_to_disk("path/to/dataset/directory")
+ >>> dataset_dict.save_to_disk("path/to/dataset/directory", max_shard_size="1GB")
+ >>> dataset_dict.save_to_disk("path/to/dataset/directory", num_shards={"train": 1024, "test": 8})
+ ```
"""
- if is_remote_filesystem(fs):
- dest_dataset_dict_path = extract_path_from_uri(dataset_dict_path)
- else:
- fs = fsspec.filesystem("file")
- dest_dataset_dict_path = dataset_dict_path
- os.makedirs(dest_dataset_dict_path, exist_ok=True)
+ if fs != "deprecated":
+ warnings.warn(
+ "'fs' was is deprecated in favor of 'storage_options' in version 2.8.0 and will be removed in 3.0.0.\n"
+ "You can remove this warning by passing 'storage_options=fs.storage_options' instead.",
+ FutureWarning,
+ )
+ storage_options = fs.storage_options
+
+ fs_token_paths = fsspec.get_fs_token_paths(dataset_dict_path, storage_options=storage_options)
+ fs: fsspec.AbstractFileSystem = fs_token_paths[0]
+ is_local = not is_remote_filesystem(fs)
+ path_join = os.path.join if is_local else posixpath.join
+
+ if num_shards is None:
+ num_shards = {k: None for k in self}
+ elif not isinstance(num_shards, dict):
+ raise ValueError(
+ "Please provide one `num_shards` per dataset in the dataset dictionary, e.g. {{'train': 128, 'test': 4}}"
+ )
+
+ if is_local:
+ Path(dataset_dict_path).resolve().mkdir(parents=True, exist_ok=True)
json.dump(
{"splits": list(self)},
- fs.open(Path(dest_dataset_dict_path, config.DATASETDICT_JSON_FILENAME).as_posix(), "w", encoding="utf-8"),
+ fs.open(path_join(dataset_dict_path, config.DATASETDICT_JSON_FILENAME), "w", encoding="utf-8"),
)
for k, dataset in self.items():
- dataset.save_to_disk(Path(dest_dataset_dict_path, k).as_posix(), fs)
+ dataset.save_to_disk(
+ path_join(dataset_dict_path, k),
+ num_shards=num_shards.get(k),
+ max_shard_size=max_shard_size,
+ num_proc=num_proc,
+ storage_options=storage_options,
+ )
@staticmethod
- def load_from_disk(dataset_dict_path: str, fs=None, keep_in_memory: Optional[bool] = None) -> "DatasetDict":
+ def load_from_disk(
+ dataset_dict_path: PathLike,
+ fs="deprecated",
+ keep_in_memory: Optional[bool] = None,
+ storage_options: Optional[dict] = None,
+ ) -> "DatasetDict":
"""
Load a dataset that was previously saved using [`save_to_disk`] from a filesystem using either
[`~filesystems.S3FileSystem`] or `fsspec.spec.AbstractFileSystem`.
Args:
dataset_dict_path (`str`):
- Path (e.g. `"dataset/train"`) or remote URI (e.g.
- `"s3//my-bucket/dataset/train"`) of the dataset dict directory where the dataset dict will be loaded
- from.
- fs ([`~filesystems.S3FileSystem`] or `fsspec.spec.AbstractFileSystem`, *optional*, defaults to `None`):
- Instance of the remote filesystem used to download the files from.
+ Path (e.g. `"dataset/train"`) or remote URI (e.g. `"s3//my-bucket/dataset/train"`)
+ of the dataset dict directory where the dataset dict will be loaded from.
+ fs (`fsspec.spec.AbstractFileSystem`, *optional*):
+ Instance of the remote filesystem where the dataset will be saved to.
+
+ <Deprecated version="2.8.0">
+
+ `fs` was deprecated in version 2.8.0 and will be removed in 3.0.0.
+ Please use `storage_options` instead, e.g. `storage_options=fs.storage_options`
+
+ </Deprecated>
+
keep_in_memory (`bool`, defaults to `None`):
Whether to copy the dataset in-memory. If `None`, the
dataset will not be copied in-memory unless explicitly enabled by setting
`datasets.config.IN_MEMORY_MAX_SIZE` to nonzero. See more details in the
[improve performance](./cache#improve-performance) section.
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.8.0"/>
Returns:
[`DatasetDict`]
@@ -1173,6 +1237,17 @@ def load_from_disk(dataset_dict_path: str, fs=None, keep_in_memory: Optional[boo
>>> ds = load_from_disk('path/to/dataset/directory')
```
"""
+ if fs != "deprecated":
+ warnings.warn(
+ "'fs' was is deprecated in favor of 'storage_options' in version 2.8.0 and will be removed in 3.0.0.\n"
+ "You can remove this warning by passing 'storage_options=fs.storage_options' instead.",
+ FutureWarning,
+ )
+ storage_options = fs.storage_options
+
+ fs_token_paths = fsspec.get_fs_token_paths(dataset_dict_path, storage_options=storage_options)
+ fs: fsspec.AbstractFileSystem = fs_token_paths[0]
+
dataset_dict = DatasetDict()
if is_remote_filesystem(fs):
dest_dataset_dict_path = extract_path_from_uri(dataset_dict_path)
@@ -1191,7 +1266,9 @@ def load_from_disk(dataset_dict_path: str, fs=None, keep_in_memory: Optional[boo
if is_remote_filesystem(fs)
else Path(dest_dataset_dict_path, k).as_posix()
)
- dataset_dict[k] = Dataset.load_from_disk(dataset_dict_split_path, fs, keep_in_memory=keep_in_memory)
+ dataset_dict[k] = Dataset.load_from_disk(
+ dataset_dict_split_path, keep_in_memory=keep_in_memory, storage_options=storage_options
+ )
return dataset_dict
@staticmethod
@@ -1382,7 +1459,8 @@ def push_to_hub(
token: Optional[str] = None,
branch: Optional[None] = None,
max_shard_size: Optional[Union[int, str]] = None,
- shard_size: Optional[int] = "deprecated",
+ num_shards: Optional[Dict[str, int]] = None,
+ shard_size: Optional[Union[int, str]] = "deprecated",
embed_external_files: bool = True,
):
"""Pushes the [`DatasetDict`] to the hub as a Parquet dataset.
@@ -1411,7 +1489,12 @@ def push_to_hub(
max_shard_size (`int` or `str`, *optional*, defaults to `"500MB"`):
The maximum size of the dataset shards to be uploaded to the hub. If expressed as a string, needs to be digits followed by a unit
(like `"500MB"` or `"1GB"`).
- shard_size (`int`, *optional*):
+ num_shards (`Dict[str, int]`, *optional*):
+ Number of shards to write. By default the number of shards depends on `max_shard_size`.
+ Use a dictionary to define a different num_shards for each split.
+
+ <Added version="2.8.0"/>
+ shard_size (`int` or `str`, *optional*):
<Deprecated version="2.4.0">
@@ -1429,6 +1512,9 @@ def push_to_hub(
```python
>>> dataset_dict.push_to_hub("<organization>/<dataset_id>")
+ >>> dataset_dict.push_to_hub("<organization>/<dataset_id>", private=True)
+ >>> dataset_dict.push_to_hub("<organization>/<dataset_id>", max_shard_size="1GB")
+ >>> dataset_dict.push_to_hub("<organization>/<dataset_id>", num_shards={"train": 1024, "test": 8})
```
"""
if shard_size != "deprecated":
@@ -1438,6 +1524,13 @@ def push_to_hub(
)
max_shard_size = shard_size
+ if num_shards is None:
+ num_shards = {k: None for k in self}
+ elif not isinstance(num_shards, dict):
+ raise ValueError(
+ "Please provide one `num_shards` per dataset in the dataset dictionary, e.g. {{'train': 128, 'test': 4}}"
+ )
+
self._check_values_type()
self._check_values_features()
total_uploaded_size = 0
@@ -1459,6 +1552,7 @@ def push_to_hub(
token=token,
branch=branch,
max_shard_size=max_shard_size,
+ num_shards=num_shards.get(split),
embed_external_files=embed_external_files,
)
total_uploaded_size += uploaded_size
diff --git a/src/datasets/table.py b/src/datasets/table.py
--- a/src/datasets/table.py
+++ b/src/datasets/table.py
@@ -4,7 +4,7 @@
import warnings
from functools import partial
from itertools import groupby
-from typing import TYPE_CHECKING, Callable, List, Optional, Tuple, TypeVar, Union
+from typing import TYPE_CHECKING, Callable, Iterator, List, Optional, Tuple, TypeVar, Union
import numpy as np
import pyarrow as pa
@@ -101,8 +101,10 @@ def _interpolation_search(arr: List[int], x: int) -> int:
class IndexedTableMixin:
def __init__(self, table: pa.Table):
- self._schema = table.schema
- self._batches = [recordbatch for recordbatch in table.to_batches() if len(recordbatch) > 0]
+ self._schema: pa.Schema = table.schema
+ self._batches: List[pa.RecordBatch] = [
+ recordbatch for recordbatch in table.to_batches() if len(recordbatch) > 0
+ ]
self._offsets: np.ndarray = np.cumsum([0] + [len(b) for b in self._batches], dtype=np.int64)
def fast_gather(self, indices: Union[List[int], np.ndarray]) -> pa.Table:
@@ -146,6 +148,20 @@ def fast_slice(self, offset=0, length=None) -> pa.Table:
return pa.Table.from_batches(batches, schema=self._schema)
+class _RecordBatchReader:
+ def __init__(self, table: "Table", max_chunksize: Optional[int] = None):
+ self.table = table
+ self.max_chunksize = max_chunksize
+
+ def __iter__(self):
+ for batch in self.table._batches:
+ if self.max_chunksize is None or len(batch) <= self.max_chunksize:
+ yield batch
+ else:
+ for offset in range(0, len(batch), self.max_chunksize):
+ yield batch.slice(offset, self.max_chunksize)
+
+
class Table(IndexedTableMixin):
"""
Wraps a pyarrow Table by using composition.
@@ -331,7 +347,7 @@ def to_pandas(self, *args, **kwargs):
def to_string(self, *args, **kwargs):
return self.table.to_string(*args, **kwargs)
- def to_reader(self, *args, **kwargs):
+ def to_reader(self, max_chunksize: Optional[int] = None):
"""
Convert the Table to a RecordBatchReader.
@@ -343,17 +359,11 @@ def to_reader(self, *args, **kwargs):
on the chunk layout of individual columns.
Returns:
- `pyarrow.RecordBatchReader`
-
- <Tip warning={true}>
-
- pyarrow >= 8.0.0 needs to be installed to use this method.
-
- </Tip>
+ `pyarrow.RecordBatchReader` if pyarrow>=8.0.0, otherwise a `pyarrow.RecordBatch` iterable
"""
if config.PYARROW_VERSION.major < 8:
- raise NotImplementedError("`pyarrow>=8.0.0` is required to use this method")
- return self.table.to_reader(*args, **kwargs)
+ return _RecordBatchReader(self, max_chunksize=max_chunksize)
+ return self.table.to_reader(max_chunksize=max_chunksize)
def field(self, *args, **kwargs):
"""
@@ -2357,11 +2367,9 @@ def _visit(array, feature):
_visit(table[name], feature)
-def table_iter(pa_table: pa.Table, batch_size: int, drop_last_batch=False):
+def table_iter(table: Table, batch_size: int, drop_last_batch=False) -> Iterator[pa.Table]:
"""Iterate over sub-tables of size `batch_size`.
- Requires pyarrow>=8.0.0
-
Args:
table (`pyarrow.Table`):
PyArrow table to iterate over.
@@ -2370,11 +2378,9 @@ def table_iter(pa_table: pa.Table, batch_size: int, drop_last_batch=False):
drop_last_batch (`bool`, defaults to `False`):
Drop the last batch if it is smaller than `batch_size`.
"""
- if config.PYARROW_VERSION.major < 8:
- raise RuntimeError(f"pyarrow>=8.0.0 is needed to use table_iter but you have {config.PYARROW_VERSION}")
chunks_buffer = []
chunks_buffer_size = 0
- for chunk in pa_table.to_reader(max_chunksize=batch_size):
+ for chunk in table.to_reader(max_chunksize=batch_size):
if len(chunk) == 0:
continue
elif chunks_buffer_size + len(chunk) < batch_size:
diff --git a/src/datasets/utils/py_utils.py b/src/datasets/utils/py_utils.py
--- a/src/datasets/utils/py_utils.py
+++ b/src/datasets/utils/py_utils.py
@@ -90,7 +90,7 @@ def size_str(size_in_bytes):
def convert_file_size_to_int(size: Union[int, str]) -> int:
"""
- Converts a size expressed as a string with digits an unit (like `"5MB"`) to an integer (in bytes).
+ Converts a size expressed as a string with digits an unit (like `"50MB"`) to an integer (in bytes).
Args:
size (`int` or `str`): The size to convert. Will be directly returned if an `int`.
@@ -1335,25 +1335,27 @@ def copyfunc(func):
return result
-X = TypeVar("X")
Y = TypeVar("Y")
-def _write_generator_to_queue(queue: queue.Queue, func: Callable[[X], Iterable[Y]], arg: X) -> int:
- for i, result in enumerate(func(arg)):
+def _write_generator_to_queue(queue: queue.Queue, func: Callable[..., Iterable[Y]], kwargs: dict) -> int:
+ for i, result in enumerate(func(**kwargs)):
queue.put(result)
return i
def iflatmap_unordered(
pool: Union[multiprocessing.pool.Pool, multiprocess.pool.Pool],
- func: Callable[[X], Iterable[Y]],
- iterable: Iterable[X],
+ func: Callable[..., Iterable[Y]],
+ *,
+ kwargs_iterable: Iterable[dict],
) -> Iterable[Y]:
manager_cls = Manager if isinstance(pool, multiprocessing.pool.Pool) else multiprocess.Manager
with manager_cls() as manager:
queue = manager.Queue()
- async_results = [pool.apply_async(_write_generator_to_queue, (queue, func, arg)) for arg in iterable]
+ async_results = [
+ pool.apply_async(_write_generator_to_queue, (queue, func, kwargs)) for kwargs in kwargs_iterable
+ ]
while True:
try:
yield queue.get(timeout=0.05)
| diff --git a/tests/conftest.py b/tests/conftest.py
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -4,7 +4,7 @@
# Import fixture modules as plugins
-pytest_plugins = ["tests.fixtures.files", "tests.fixtures.hub", "tests.fixtures.s3", "tests.fixtures.fsspec"]
+pytest_plugins = ["tests.fixtures.files", "tests.fixtures.hub", "tests.fixtures.fsspec"]
def pytest_collection_modifyitems(config, items):
diff --git a/tests/fixtures/fsspec.py b/tests/fixtures/fsspec.py
--- a/tests/fixtures/fsspec.py
+++ b/tests/fixtures/fsspec.py
@@ -81,4 +81,4 @@ def mock_fsspec(monkeypatch):
@pytest.fixture
def mockfs(tmp_path_factory, mock_fsspec):
local_fs_dir = tmp_path_factory.mktemp("mockfs")
- return MockFileSystem(local_root_dir=local_fs_dir)
+ return MockFileSystem(local_root_dir=local_fs_dir, auto_mkdir=True)
diff --git a/tests/fixtures/s3.py b/tests/fixtures/s3.py
deleted file mode 100644
--- a/tests/fixtures/s3.py
+++ /dev/null
@@ -1,74 +0,0 @@
-import os
-import time
-
-import pytest
-import requests
-
-
-# From: https://github.com/dask/s3fs/blob/ffe3a5293524869df56e74973af0d2c204ae9cbf/s3fs/tests/test_s3fs.py#L25-L141
-
-S3_TEST_BUCKET_NAME = "test"
-s3_port = 5555
-s3_endpoint_uri = f"http://127.0.0.1:{s3_port}/"
-
-S3_FAKE_ENV_VARS = {
- "AWS_ACCESS_KEY_ID": "fake_access_key",
- "AWS_SECRET_ACCESS_KEY": "fake_secret_key",
- "AWS_SECURITY_TOKEN": "fake_secrurity_token",
- "AWS_SESSION_TOKEN": "fake_session_token",
-}
-
-
[email protected](scope="session")
-def s3_test_bucket_name():
- return S3_TEST_BUCKET_NAME
-
-
[email protected]()
-def s3_base():
- # writable local S3 system
- import shlex
- import subprocess
-
- # Mocked AWS Credentials for moto.
- old_environ = os.environ.copy()
- os.environ.update(S3_FAKE_ENV_VARS)
-
- proc = subprocess.Popen(shlex.split(f"moto_server s3 -p {s3_port}"))
-
- timeout = 5
- while timeout > 0:
- try:
- r = requests.get(s3_endpoint_uri)
- if r.ok:
- break
- except: # noqa
- pass
- timeout -= 0.1
- time.sleep(0.1)
- yield
- proc.terminate()
- proc.wait()
- os.environ.clear()
- os.environ.update(old_environ)
-
-
-def get_boto3_client():
- from botocore.session import Session
-
- # NB: we use the sync botocore client for setup
- session = Session()
- return session.create_client("s3", endpoint_url=s3_endpoint_uri)
-
-
[email protected]()
-def s3(s3_base, s3_test_bucket_name):
- client = get_boto3_client()
- client.create_bucket(Bucket=s3_test_bucket_name, ACL="public-read")
-
- from s3fs.core import S3FileSystem
-
- S3FileSystem.clear_instance_cache()
- s3 = S3FileSystem(anon=False, client_kwargs={"endpoint_url": s3_endpoint_uri})
- s3.invalidate_cache()
- yield s3
diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -53,7 +53,6 @@
assert_arrow_memory_increases,
require_jax,
require_pil,
- require_s3,
require_sqlalchemy,
require_tf,
require_torch,
@@ -266,6 +265,7 @@ def test_dummy_dataset_serialize(self, in_memory):
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
+ expected = dset.to_dict()
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
dataset_path = os.path.join(tmp_dir, "my_dataset") # abs path
@@ -302,6 +302,47 @@ def test_dummy_dataset_serialize(self, in_memory):
self.assertDictEqual(dset[0]["nested"], {"a": 1, "c": 100, "x": 10})
self.assertDictEqual(dset["nested"][0], {"a": 1, "c": 100, "x": 10})
+ with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
+ with assert_arrow_memory_doesnt_increase():
+ dset.save_to_disk(dataset_path, num_shards=4)
+
+ with Dataset.load_from_disk(dataset_path) as dset:
+ self.assertEqual(len(dset), 10)
+ self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
+ self.assertDictEqual(dset.to_dict(), expected)
+ self.assertEqual(len(dset.cache_files), 4)
+
+ with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
+ with assert_arrow_memory_doesnt_increase():
+ dset.save_to_disk(dataset_path, num_proc=2)
+
+ with Dataset.load_from_disk(dataset_path) as dset:
+ self.assertEqual(len(dset), 10)
+ self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
+ self.assertDictEqual(dset.to_dict(), expected)
+ self.assertEqual(len(dset.cache_files), 2)
+
+ with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
+ with assert_arrow_memory_doesnt_increase():
+ dset.save_to_disk(dataset_path, num_shards=7, num_proc=2)
+
+ with Dataset.load_from_disk(dataset_path) as dset:
+ self.assertEqual(len(dset), 10)
+ self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
+ self.assertDictEqual(dset.to_dict(), expected)
+ self.assertEqual(len(dset.cache_files), 7)
+
+ with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
+ with assert_arrow_memory_doesnt_increase():
+ max_shard_size = dset._estimate_nbytes() // 2 + 1
+ dset.save_to_disk(dataset_path, max_shard_size=max_shard_size)
+
+ with Dataset.load_from_disk(dataset_path) as dset:
+ self.assertEqual(len(dset), 10)
+ self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
+ self.assertDictEqual(dset.to_dict(), expected)
+ self.assertEqual(len(dset.cache_files), 2)
+
def test_dummy_dataset_load_from_disk(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
@@ -3543,25 +3584,15 @@ def test_pickle_dataset_after_transforming_the_table(in_memory, method_and_param
assert dataset._data.table == reloaded_dataset._data.table
[email protected](
- os.name in ["nt", "posix"] and (os.getenv("CIRCLECI") == "true" or os.getenv("GITHUB_ACTIONS") == "true"),
- reason='On Windows CircleCI or GitHub Actions, it raises botocore.exceptions.EndpointConnectionError: Could not connect to the endpoint URL: "http://127.0.0.1:5555/test"',
-) # TODO: find what's wrong with CircleCI / GitHub Actions
-@require_s3
[email protected]
-def test_dummy_dataset_serialize_s3(s3, dataset, s3_test_bucket_name):
- mock_bucket = s3_test_bucket_name
- dataset_path = f"s3://{mock_bucket}/my_dataset"
- features = dataset.features
- dataset.save_to_disk(dataset_path, s3)
- dataset = dataset.load_from_disk(dataset_path, s3)
- assert os.path.isfile(dataset.cache_files[0]["filename"])
-
- assert len(dataset) == 10
- assert len(dataset.shuffle()) == 10
- assert dataset.features == features
- assert dataset[0]["id"] == 0
- assert dataset["id"][0] == 0
+def test_dummy_dataset_serialize_fs(dataset, mockfs):
+ dataset_path = "mock://my_dataset"
+ dataset.save_to_disk(dataset_path, storage_options=mockfs.storage_options)
+ assert mockfs.isdir(dataset_path)
+ assert mockfs.glob(dataset_path + "/*")
+ reloaded = dataset.load_from_disk(dataset_path, storage_options=mockfs.storage_options)
+ assert len(reloaded) == len(dataset)
+ assert reloaded.features == dataset.features
+ assert reloaded.to_dict() == dataset.to_dict()
@pytest.mark.parametrize(
@@ -4109,6 +4140,23 @@ def test_train_test_split_startify(self):
assert len(d1["test"]["text"]) == test_size
+def test_dataset_estimate_nbytes():
+ ds = Dataset.from_dict({"a": ["0" * 100] * 100})
+ assert 0.9 * ds._estimate_nbytes() < 100 * 100, "must be smaller than full dataset size"
+
+ ds = Dataset.from_dict({"a": ["0" * 100] * 100}).select([0])
+ assert 0.9 * ds._estimate_nbytes() < 100 * 100, "must be smaller than one chunk"
+
+ ds = Dataset.from_dict({"a": ["0" * 100] * 100})
+ ds = concatenate_datasets([ds] * 100)
+ assert 0.9 * ds._estimate_nbytes() < 100 * 100 * 100, "must be smaller than full dataset size"
+ assert 1.1 * ds._estimate_nbytes() > 100 * 100 * 100, "must be bigger than full dataset size"
+
+ ds = Dataset.from_dict({"a": ["0" * 100] * 100})
+ ds = concatenate_datasets([ds] * 100).select([0])
+ assert 0.9 * ds._estimate_nbytes() < 100 * 100, "must be smaller than one chunk"
+
+
@pytest.mark.parametrize("return_lazy_dict", [True, False, "mix"])
def test_map_cases(return_lazy_dict):
def f(x):
diff --git a/tests/test_dataset_dict.py b/tests/test_dataset_dict.py
--- a/tests/test_dataset_dict.py
+++ b/tests/test_dataset_dict.py
@@ -12,13 +12,7 @@
from datasets.features import ClassLabel, Features, Sequence, Value
from datasets.splits import NamedSplit
-from .utils import (
- assert_arrow_memory_doesnt_increase,
- assert_arrow_memory_increases,
- require_s3,
- require_tf,
- require_torch,
-)
+from .utils import assert_arrow_memory_doesnt_increase, assert_arrow_memory_increases, require_tf, require_torch
class DatasetDictTest(TestCase):
@@ -394,6 +388,30 @@ def test_serialization(self):
self.assertListEqual(reloaded_dsets["train"].column_names, ["filename"])
del dsets, reloaded_dsets
+ dsets = self._create_dummy_dataset_dict()
+ dsets.save_to_disk(tmp_dir, num_shards={"train": 3, "test": 2})
+ reloaded_dsets = DatasetDict.load_from_disk(tmp_dir)
+ self.assertListEqual(sorted(reloaded_dsets), ["test", "train"])
+ self.assertEqual(len(reloaded_dsets["train"]), 30)
+ self.assertListEqual(reloaded_dsets["train"].column_names, ["filename"])
+ self.assertEqual(len(reloaded_dsets["train"].cache_files), 3)
+ self.assertEqual(len(reloaded_dsets["test"]), 30)
+ self.assertListEqual(reloaded_dsets["test"].column_names, ["filename"])
+ self.assertEqual(len(reloaded_dsets["test"].cache_files), 2)
+ del reloaded_dsets
+
+ dsets = self._create_dummy_dataset_dict()
+ dsets.save_to_disk(tmp_dir, num_proc=2)
+ reloaded_dsets = DatasetDict.load_from_disk(tmp_dir)
+ self.assertListEqual(sorted(reloaded_dsets), ["test", "train"])
+ self.assertEqual(len(reloaded_dsets["train"]), 30)
+ self.assertListEqual(reloaded_dsets["train"].column_names, ["filename"])
+ self.assertEqual(len(reloaded_dsets["train"].cache_files), 2)
+ self.assertEqual(len(reloaded_dsets["test"]), 30)
+ self.assertListEqual(reloaded_dsets["test"].column_names, ["filename"])
+ self.assertEqual(len(reloaded_dsets["test"].cache_files), 2)
+ del reloaded_dsets
+
def test_load_from_disk(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dsets = self._create_dummy_dataset_dict()
@@ -447,6 +465,24 @@ def test_align_labels_with_mapping(self):
self.assertListEqual(test_expected_label_names, test_aligned_label_names)
+def test_dummy_datasetdict_serialize_fs(mockfs):
+ dataset_dict = DatasetDict(
+ {
+ "train": Dataset.from_dict({"a": range(30)}),
+ "test": Dataset.from_dict({"a": range(10)}),
+ }
+ )
+ dataset_path = "mock://my_dataset"
+ dataset_dict.save_to_disk(dataset_path, storage_options=mockfs.storage_options)
+ assert mockfs.isdir(dataset_path)
+ assert mockfs.glob(dataset_path + "/*")
+ reloaded = dataset_dict.load_from_disk(dataset_path, storage_options=mockfs.storage_options)
+ assert list(reloaded) == list(dataset_dict)
+ for k in dataset_dict:
+ assert reloaded[k].features == dataset_dict[k].features
+ assert reloaded[k].to_dict() == dataset_dict[k].to_dict()
+
+
def _check_csv_datasetdict(dataset_dict, expected_features, splits=("train",)):
assert isinstance(dataset_dict, DatasetDict)
for split in splits:
@@ -665,24 +701,3 @@ def test_datasetdict_from_text_split(split, text_path, tmp_path):
dataset = DatasetDict.from_text(path, cache_dir=cache_dir)
_check_text_datasetdict(dataset, expected_features, splits=list(path.keys()))
assert all(dataset[split].split == split for split in path.keys())
-
-
[email protected](
- os.name in ["nt", "posix"] and (os.getenv("CIRCLECI") == "true" or os.getenv("GITHUB_ACTIONS") == "true"),
- reason='On Windows CircleCI or GitHub Actions, it raises botocore.exceptions.EndpointConnectionError: Could not connect to the endpoint URL: "http://127.0.0.1:5555/test"',
-) # TODO: find what's wrong with CircleCI / GitHub Actions
-@require_s3
[email protected]
-def test_dummy_dataset_serialize_s3(s3, dataset, s3_test_bucket_name):
- dsets = DatasetDict({"train": dataset, "test": dataset.select(range(2))})
- mock_bucket = s3_test_bucket_name
- dataset_path = f"s3://{mock_bucket}/datasets/dict"
- column_names = dsets["train"].column_names
- lengths = [len(dset) for dset in dsets.values()]
- dataset.save_to_disk(dataset_path, s3)
- dataset = dataset.load_from_disk(dataset_path, s3)
-
- assert sorted(dsets) == ["test", "train"]
- assert [len(dset) for dset in dsets.values()] == lengths
- assert dsets["train"].column_names == column_names
- assert dsets["test"].column_names == column_names
diff --git a/tests/test_filesystem.py b/tests/test_filesystem.py
--- a/tests/test_filesystem.py
+++ b/tests/test_filesystem.py
@@ -11,10 +11,8 @@
def test_extract_path_from_uri():
- mock_bucket = "moto-mock-s3-bucket"
-
+ mock_bucket = "mock-s3-bucket"
dataset_path = f"s3://{mock_bucket}"
-
dataset_path = extract_path_from_uri(dataset_path)
assert dataset_path.startswith("s3://") is False
diff --git a/tests/test_py_utils.py b/tests/test_py_utils.py
--- a/tests/test_py_utils.py
+++ b/tests/test_py_utils.py
@@ -240,6 +240,10 @@ def test_asdict():
asdict([1, A(x=10, y="foo")])
+def _split_text(text: str):
+ return text.split()
+
+
def _2seconds_generator_of_2items_with_timing(content):
yield (time.time(), content)
time.sleep(2)
@@ -249,14 +253,14 @@ def _2seconds_generator_of_2items_with_timing(content):
def test_iflatmap_unordered():
with Pool(2) as pool:
- out = list(iflatmap_unordered(pool, str.split, ["hello there"] * 10))
+ out = list(iflatmap_unordered(pool, _split_text, kwargs_iterable=[{"text": "hello there"}] * 10))
assert out.count("hello") == 10
assert out.count("there") == 10
assert len(out) == 20
# check multiprocess from pathos (uses dill for pickling)
with multiprocess.Pool(2) as pool:
- out = list(iflatmap_unordered(pool, str.split, ["hello there"] * 10))
+ out = list(iflatmap_unordered(pool, _split_text, kwargs_iterable=[{"text": "hello there"}] * 10))
assert out.count("hello") == 10
assert out.count("there") == 10
assert len(out) == 20
@@ -264,7 +268,9 @@ def test_iflatmap_unordered():
# check that we get items as fast as possible
with Pool(2) as pool:
out = []
- for yield_time, content in iflatmap_unordered(pool, _2seconds_generator_of_2items_with_timing, ["a", "b"]):
+ for yield_time, content in iflatmap_unordered(
+ pool, _2seconds_generator_of_2items_with_timing, kwargs_iterable=[{"content": "a"}, {"content": "b"}]
+ ):
assert yield_time < time.time() + 0.1, "we should each item directly after it was yielded"
out.append(content)
assert out.count("a") == 2
diff --git a/tests/test_table.py b/tests/test_table.py
--- a/tests/test_table.py
+++ b/tests/test_table.py
@@ -1162,21 +1162,20 @@ def test_embed_table_storage(image_file):
assert isinstance(embedded_images_table.to_pydict()["image"][0]["bytes"], bytes)
[email protected](datasets.config.PYARROW_VERSION.major < 8, reason="only available on pyarrow>=8")
@pytest.mark.parametrize(
- "pa_table",
+ "table",
[
- pa.table({"foo": range(10)}),
- pa.concat_tables([pa.table({"foo": range(0, 5)}), pa.table({"foo": range(5, 10)})]),
- pa.concat_tables([pa.table({"foo": [i]}) for i in range(10)]),
+ InMemoryTable(pa.table({"foo": range(10)})),
+ InMemoryTable(pa.concat_tables([pa.table({"foo": range(0, 5)}), pa.table({"foo": range(5, 10)})])),
+ InMemoryTable(pa.concat_tables([pa.table({"foo": [i]}) for i in range(10)])),
],
)
@pytest.mark.parametrize("batch_size", [1, 2, 3, 9, 10, 11, 20])
@pytest.mark.parametrize("drop_last_batch", [False, True])
-def test_table_iter(pa_table, batch_size, drop_last_batch):
- num_rows = len(pa_table) if not drop_last_batch else len(pa_table) // batch_size * batch_size
+def test_table_iter(table, batch_size, drop_last_batch):
+ num_rows = len(table) if not drop_last_batch else len(table) // batch_size * batch_size
num_batches = (num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size
- subtables = list(table_iter(pa_table, batch_size=batch_size, drop_last_batch=drop_last_batch))
+ subtables = list(table_iter(table, batch_size=batch_size, drop_last_batch=drop_last_batch))
assert len(subtables) == num_batches
if drop_last_batch:
assert all(len(subtable) == batch_size for subtable in subtables)
@@ -1185,7 +1184,7 @@ def test_table_iter(pa_table, batch_size, drop_last_batch):
assert len(subtables[-1]) <= batch_size
if num_rows > 0:
reloaded = pa.concat_tables(subtables)
- assert pa_table.slice(0, num_rows).to_pydict() == reloaded.to_pydict()
+ assert table.slice(0, num_rows).to_pydict() == reloaded.to_pydict()
@pytest.mark.parametrize(
diff --git a/tests/test_upstream_hub.py b/tests/test_upstream_hub.py
--- a/tests/test_upstream_hub.py
+++ b/tests/test_upstream_hub.py
@@ -184,6 +184,34 @@ def test_push_dataset_dict_to_hub_multiple_files_with_max_shard_size(self, tempo
)
)
+ def test_push_dataset_dict_to_hub_multiple_files_with_num_shards(self, temporary_repo):
+ ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
+
+ local_ds = DatasetDict({"train": ds})
+
+ with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
+ local_ds.push_to_hub(ds_name, token=self._token, num_shards={"train": 2})
+ hub_ds = load_dataset(ds_name, download_mode="force_redownload")
+
+ assert local_ds.column_names == hub_ds.column_names
+ assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
+ assert local_ds["train"].features == hub_ds["train"].features
+
+ # Ensure that there are two files on the repository that have the correct name
+ files = sorted(list_repo_files(self._api, ds_name, repo_type="dataset", use_auth_token=self._token))
+ assert all(
+ fnmatch.fnmatch(file, expected_file)
+ for file, expected_file in zip(
+ files,
+ [
+ ".gitattributes",
+ "README.md",
+ "data/train-00000-of-00002-*.parquet",
+ "data/train-00001-of-00002-*.parquet",
+ ],
+ )
+ )
+
def test_push_dataset_dict_to_hub_overwrite_files(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
ds2 = Dataset.from_dict({"x": list(range(100)), "y": list(range(100))})
diff --git a/tests/utils.py b/tests/utils.py
--- a/tests/utils.py
+++ b/tests/utils.py
@@ -201,22 +201,6 @@ def require_transformers(test_case):
return test_case
-def require_s3(test_case):
- """
- Decorator marking a test that requires s3fs and moto to mock s3.
-
- These tests are skipped when they aren't installed.
-
- """
- try:
- import moto # noqa F401
- import s3fs # noqa F401
- except ImportError:
- return unittest.skip("test requires s3fs and moto")(test_case)
- else:
- return test_case
-
-
def require_spacy(test_case):
"""
Decorator marking a test that requires spacy.
| Save a dataset in a determined number of shards
This is useful to distribute the shards to training nodes.
This can be implemented in `save_to_disk` and can also leverage multiprocessing to speed up the process
| 2022-11-18T18:50:01Z | [] | [] |
|
huggingface/datasets | 5,294 | huggingface__datasets-5294 | [
"5293"
] | 494a3d8356e09af6c69ded33dc7f2e1a7d239ab9 | diff --git a/src/datasets/download/streaming_download_manager.py b/src/datasets/download/streaming_download_manager.py
--- a/src/datasets/download/streaming_download_manager.py
+++ b/src/datasets/download/streaming_download_manager.py
@@ -691,6 +691,10 @@ def joinpath(self, *p: Tuple[str, ...]) -> "xPath":
def __truediv__(self, p: str) -> "xPath":
return self.joinpath(p)
+ def with_suffix(self, suffix):
+ main_hop, *rest_hops = _as_posix(self).split("::")
+ return type(self)("::".join([_as_posix(PurePosixPath(main_hop).with_suffix(suffix))] + rest_hops))
+
def xgzip_open(filepath_or_buffer, *args, use_auth_token: Optional[Union[str, bool]] = None, **kwargs):
import gzip
| diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -640,6 +640,18 @@ def test_xpathsuffix(input_path, expected):
assert xPath(input_path).suffix == expected
[email protected](
+ "input_path, suffix, expected",
+ [
+ ("zip://file.txt::https://host.com/archive.zip", ".ann", "zip://file.ann::https://host.com/archive.zip"),
+ ("file.txt", ".ann", "file.ann"),
+ ((Path().resolve() / "file.txt").as_posix(), ".ann", (Path().resolve() / "file.ann").as_posix()),
+ ],
+)
+def test_xpath_with_suffix(input_path, suffix, expected):
+ assert xPath(input_path).with_suffix(suffix) == xPath(expected)
+
+
@pytest.mark.parametrize("urlpath", [r"C:\\foo\bar.txt", "/foo/bar.txt", "https://f.oo/bar.txt"])
def test_streaming_dl_manager_download_dummy_path(urlpath):
dl_manager = StreamingDownloadManager()
| Support streaming datasets with pathlib.Path.with_suffix
Extend support for streaming datasets that use `pathlib.Path.with_suffix`.
This feature will be useful e.g. for datasets containing text files and annotated files with the same name but different extension.
| 2022-11-24T18:04:38Z | [] | [] |
|
huggingface/datasets | 5,297 | huggingface__datasets-5297 | [
"5296"
] | 494a3d8356e09af6c69ded33dc7f2e1a7d239ab9 | diff --git a/src/datasets/download/streaming_download_manager.py b/src/datasets/download/streaming_download_manager.py
--- a/src/datasets/download/streaming_download_manager.py
+++ b/src/datasets/download/streaming_download_manager.py
@@ -104,10 +104,10 @@ def xjoin(a, *p):
"""
a, *b = str(a).split("::")
if is_local_path(a):
- a = Path(a, *p).as_posix()
+ return os.path.join(a, *p)
else:
a = posixpath.join(a, *p)
- return "::".join([a] + b)
+ return "::".join([a] + b)
def xdirname(a):
| diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -165,8 +165,11 @@ def test_as_posix(input_path, expected_path):
@pytest.mark.parametrize(
"input_path, paths_to_join, expected_path",
[
- (str(Path(__file__).resolve().parent), (Path(__file__).name,), str(Path(__file__).resolve())),
- ("https://host.com/archive.zip", ("file.txt",), "https://host.com/archive.zip/file.txt"),
+ (
+ "https://host.com/archive.zip",
+ ("file.txt",),
+ "https://host.com/archive.zip/file.txt",
+ ),
(
"zip://::https://host.com/archive.zip",
("file.txt",),
@@ -180,19 +183,18 @@ def test_as_posix(input_path, expected_path):
(
".",
("file.txt",),
- "file.txt",
+ os.path.join(".", "file.txt"),
),
(
- Path().resolve().as_posix(),
+ str(Path().resolve()),
("file.txt",),
- (Path().resolve() / "file.txt").as_posix(),
+ str((Path().resolve() / "file.txt")),
),
],
)
def test_xjoin(input_path, paths_to_join, expected_path):
output_path = xjoin(input_path, *paths_to_join)
- output_path = _readd_double_slash_removed_by_path(Path(output_path).as_posix())
- assert output_path == _readd_double_slash_removed_by_path(Path(expected_path).as_posix())
+ assert output_path == expected_path
output_path = xPath(input_path).joinpath(*paths_to_join)
assert output_path == xPath(expected_path)
| Bug in xjoin with Windows pathnames
Currently, `xjoin` function has a bug with local Windows pathnames: instead of returning the OS-dependent join pathname, it always returns it in POSIX format.
```python
from datasets.download.streaming_download_manager import xjoin
path = xjoin("C:\\Users\\USERNAME", "filename.txt")
```
Join path should be:
```python
"C:\\Users\\USERNAME\\filename.txt"
```
However it is:
```python
"C:/Users/USERNAME/filename.txt"
```
| 2022-11-25T13:30:17Z | [] | [] |
|
huggingface/datasets | 5,319 | huggingface__datasets-5319 | [
"5316"
] | 617d15b2edb74485f8cd7aef249ab9dc49797bca | diff --git a/src/datasets/packaged_modules/text/text.py b/src/datasets/packaged_modules/text/text.py
--- a/src/datasets/packaged_modules/text/text.py
+++ b/src/datasets/packaged_modules/text/text.py
@@ -93,9 +93,10 @@ def _generate_tables(self, files):
batch_idx = 0
batch = ""
while True:
- batch += f.read(self.config.chunksize)
- if not batch:
+ new_batch = f.read(self.config.chunksize)
+ if not new_batch:
break
+ batch += new_batch
batch += f.readline() # finish current line
batch = batch.split("\n\n")
pa_table = pa.Table.from_arrays(
@@ -107,6 +108,9 @@ def _generate_tables(self, files):
yield (file_idx, batch_idx), self._cast_table(pa_table)
batch_idx += 1
batch = batch[-1]
+ if batch:
+ pa_table = pa.Table.from_arrays([pa.array([batch])], names=pa_table_names)
+ yield (file_idx, batch_idx), self._cast_table(pa_table)
elif self.config.sample_by == "document":
text = f.read()
pa_table = pa.Table.from_arrays([pa.array([text])], names=pa_table_names)
| diff --git a/tests/packaged_modules/test_text.py b/tests/packaged_modules/test_text.py
--- a/tests/packaged_modules/test_text.py
+++ b/tests/packaged_modules/test_text.py
@@ -18,6 +18,12 @@ def text_file(tmp_path):
Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat.
Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur.
Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
+
+ Second paragraph:
+ Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
+ Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat.
+ Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur.
+ Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
"""
)
with open(filename, "w", encoding="utf-8") as f:
@@ -53,3 +59,19 @@ def test_text_cast_image(text_file_with_image):
assert pa_table.schema.field("image").type == Image()()
generated_content = pa_table.to_pydict()["image"]
assert generated_content == [{"path": image_file, "bytes": None}]
+
+
[email protected]("sample_by", ["line", "paragraph", "document"])
+def test_text_sample_by(sample_by, text_file):
+ with open(text_file, encoding="utf-8") as f:
+ expected_content = f.read()
+ if sample_by == "line":
+ expected_content = expected_content.splitlines()
+ elif sample_by == "paragraph":
+ expected_content = expected_content.split("\n\n")
+ elif sample_by == "document":
+ expected_content = [expected_content]
+ text = Text(sample_by=sample_by, encoding="utf-8", chunksize=100)
+ generator = text._generate_tables([[text_file]])
+ generated_content = pa.concat_tables([table for _, table in generator]).to_pydict()["text"]
+ assert generated_content == expected_content
| Bug in sample_by="paragraph"
### Describe the bug
I think [this line](https://github.com/huggingface/datasets/blob/main/src/datasets/packaged_modules/text/text.py#L96) is wrong and should be `batch = f.read(self.config.chunksize)`. Otherwise it will never terminate because even when `f` is finished reading, `batch` will still be truthy from the last iteration.
### Steps to reproduce the bug
```
> cat test.txt
a b c
d e f
````
```python
>>> import datasets
>>> datasets.load_dataset("text", data_files={"train":"test.txt"}, sample_by="paragraph")
```
This will go on forever.
### Expected behavior
Terminates very quickly.
### Environment info
`version = "2.6.1"` but I think the bug is still there on main.
| Thanks for reporting, @adampauls.
We are having a look at it. | 2022-12-01T09:08:09Z | [] | [] |
huggingface/datasets | 5,359 | huggingface__datasets-5359 | [
"5332"
] | c902456677116a081f762fa2b4aad13a0aa04d6e | diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -19,6 +19,7 @@
import re
import sys
from collections.abc import Iterable, Mapping
+from collections.abc import Sequence as SequenceABC
from dataclasses import InitVar, dataclass, field, fields
from functools import reduce, wraps
from operator import mul
@@ -944,6 +945,8 @@ def __post_init__(self, num_classes, names_file):
self.names = [str(i) for i in range(self.num_classes)]
else:
raise ValueError("Please provide either num_classes, names or names_file.")
+ elif not isinstance(self.names, SequenceABC):
+ raise TypeError(f"Please provide names as a list, is {type(self.names)}")
# Set self.num_classes
if self.num_classes is None:
self.num_classes = len(self.names)
| diff --git a/tests/features/test_features.py b/tests/features/test_features.py
--- a/tests/features/test_features.py
+++ b/tests/features/test_features.py
@@ -287,6 +287,8 @@ def test_classlabel_init(tmp_path_factory):
classlabel = ClassLabel(names=names, names_file=names_file)
with pytest.raises(ValueError):
classlabel = ClassLabel()
+ with pytest.raises(TypeError):
+ classlabel = ClassLabel(names=np.array(names))
def test_classlabel_str2int():
| Passing numpy array to ClassLabel names causes ValueError
### Describe the bug
If a numpy array is passed to the names argument of ClassLabel, creating a dataset with those features causes an error.
### Steps to reproduce the bug
https://colab.research.google.com/drive/1cV_es1PWZiEuus17n-2C-w0KEoEZ68IX
TLDR:
If I define my classes as:
```
my_classes = np.array(['one', 'two', 'three'])
```
Then this errors:
```py
features = Features({'value': Value('string'), 'label': ClassLabel(names=my_classes)})
dataset = Dataset.from_list(my_data, features=features)
```
```
ValueError Traceback (most recent call last)
[<ipython-input-8-a8a9d53ec82f>](https://localhost:8080/#) in <module>
----> 1 dataset = Dataset.from_list(my_data, features=features)
11 frames
[/usr/local/lib/python3.8/dist-packages/datasets/utils/py_utils.py](https://localhost:8080/#) in _asdict_inner(obj)
183 for f in fields(obj):
184 value = _asdict_inner(getattr(obj, f.name))
--> 185 if not f.init or value != f.default or f.metadata.get("include_in_asdict_even_if_is_default", False):
186 result[f.name] = value
187 return result
ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
```
But this works:
```
features2 = Features({'value': Value('string'), 'label': ClassLabel(names=list(my_classes))})
dataset2 = Dataset.from_list(my_data, features=features2)
```
### Expected behavior
If I provide a numpy array of class names, I would expect either an error that the names list is the wrong type, or for it to be cast internally.
### Environment info
- `datasets` version: 2.7.1
- Platform: Linux-5.15.0-56-generic-x86_64-with-glibc2.10
- Python version: 3.8.15
- PyArrow version: 10.0.1
- Pandas version: 1.5.2
Additionally:
- Numpy version: 1.23.5
| Should `datasets` allow `ClassLabel` input parameter to be an `np.array` even though internally we need to cast it to a Python list? @lhoestq @mariosasko
Hi! No, I don't think so. The `names` parameter is [annotated](https://github.com/huggingface/datasets/blob/582236640b9109988e5f7a16a8353696ffa09a16/src/datasets/features/features.py#L892) as `List[str]` (**NumPy arrays are not lists**), and considering that type checking is not a common practice in Python, I think we can leave the code as-is.
I appreciate it is the wrong type, and that type checking is not common, but I think there's a few circumstances that make it a good idea from a usability perspective.
It's quite a difficult error to debug because it comes from a utility function (so it's not immediately obvious which parameter caused it). What makes it even more difficult is the exception happens when the features instance is used to instantiate the dataset, **not** when when the wrong type is actually passed when the features is instantiated. When I was debugging the error, I didn't really consider it could be an issue with the features instance because it had instantiated fine. It's also not one of the more common exceptions caused by trying to use a non-list as a list.
It's also relatively easy to accidentally get a numpy array of class types (e.g. calling `unique()` on a pandas dataframe column). Additionally, passing in a `set` instead of the list (again, relatively easy because people may run `set(classes)` to generate uniques) causes an error when the features instance is used, albeit a slightly more obvious one.
The names list is already being processed and validated in the `__post_init__` method anyway, so it would not really be adding any complexity to check it is actually a list here too. I'm happy to contribute this change if you change your mind about whether it's worthwhile.
I agree that it's not easy to debug this issue, so perhaps we could add some basic type checking (e.g. `not isinstance(names, list)` -> error) to make debugging easier. Feel free to submit a PR.
> Additionally, passing in a set instead of the list (again, relatively easy because people may run set(classes) to generate uniques) causes an error when the features instance is used, albeit a slightly more obvious one.
`set` is an unordered structure (it's ordered in Python 3.6+, but this is CPython's implementation detail), and the order of ClassLabel `names` matters, so this doesn't require a fix. | 2022-12-13T23:04:06Z | [] | [] |
huggingface/datasets | 5,416 | huggingface__datasets-5416 | [
"5415",
"5414"
] | 87c2d111503a22b5ac0395777bdb2f60359e7fb4 | diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -1414,17 +1414,18 @@ def _prepare_split(
fname = f"{self.name}-{split_generator.name}{SUFFIX}.{file_format}"
fpath = path_join(self._output_dir, fname)
- num_input_shards = _number_of_shards_in_gen_kwargs(split_generator.gen_kwargs)
- if num_input_shards <= 1 and num_proc is not None:
- logger.warning(
- f"Setting num_proc from {num_proc} back to 1 for the {split_info.name} split to disable multiprocessing as it only contains one shard."
- )
- num_proc = 1
- elif num_proc is not None and num_input_shards < num_proc:
- logger.info(
- f"Setting num_proc from {num_proc} to {num_input_shards} for the {split_info.name} split as it only contains {num_input_shards} shards."
- )
- num_proc = num_input_shards
+ if num_proc and num_proc > 1:
+ num_input_shards = _number_of_shards_in_gen_kwargs(split_generator.gen_kwargs)
+ if num_input_shards <= 1 and num_proc is not None:
+ logger.warning(
+ f"Setting num_proc from {num_proc} back to 1 for the {split_info.name} split to disable multiprocessing as it only contains one shard."
+ )
+ num_proc = 1
+ elif num_proc is not None and num_input_shards < num_proc:
+ logger.info(
+ f"Setting num_proc from {num_proc} to {num_input_shards} for the {split_info.name} split as it only contains {num_input_shards} shards."
+ )
+ num_proc = num_input_shards
pbar = logging.tqdm(
disable=not logging.is_progress_bar_enabled(),
@@ -1673,17 +1674,18 @@ def _prepare_split(
fname = f"{self.name}-{split_generator.name}{SUFFIX}.{file_format}"
fpath = path_join(self._output_dir, fname)
- num_input_shards = _number_of_shards_in_gen_kwargs(split_generator.gen_kwargs)
- if num_input_shards <= 1 and num_proc is not None:
- logger.warning(
- f"Setting num_proc from {num_proc} back to 1 for the {split_info.name} split to disable multiprocessing as it only contains one shard."
- )
- num_proc = 1
- elif num_proc is not None and num_input_shards < num_proc:
- logger.info(
- f"Setting num_proc from {num_proc} to {num_input_shards} for the {split_info.name} split as it only contains {num_input_shards} shards."
- )
- num_proc = num_input_shards
+ if num_proc and num_proc > 1:
+ num_input_shards = _number_of_shards_in_gen_kwargs(split_generator.gen_kwargs)
+ if num_input_shards <= 1 and num_proc is not None:
+ logger.warning(
+ f"Setting num_proc from {num_proc} back to 1 for the {split_info.name} split to disable multiprocessing as it only contains one shard."
+ )
+ num_proc = 1
+ elif num_proc is not None and num_input_shards < num_proc:
+ logger.info(
+ f"Setting num_proc from {num_proc} to {num_input_shards} for the {split_info.name} split as it only contains {num_input_shards} shards."
+ )
+ num_proc = num_input_shards
pbar = logging.tqdm(
disable=not logging.is_progress_bar_enabled(),
| diff --git a/tests/test_builder.py b/tests/test_builder.py
--- a/tests/test_builder.py
+++ b/tests/test_builder.py
@@ -2,6 +2,7 @@
import os
import tempfile
import types
+from contextlib import nullcontext as does_not_raise
from multiprocessing import Process
from pathlib import Path
from unittest import TestCase
@@ -194,6 +195,52 @@ def _generate_examples(self, filepaths):
idx += 1
+class DummyArrowBasedBuilderWithAmbiguousShards(ArrowBasedBuilder):
+ def _info(self):
+ return DatasetInfo(features=Features({"id": Value("int8"), "filepath": Value("string")}))
+
+ def _split_generators(self, dl_manager):
+ return [
+ SplitGenerator(
+ name=Split.TRAIN,
+ gen_kwargs={
+ "filepaths": [f"data{i}.txt" for i in range(4)],
+ "dummy_kwarg_with_different_length": [f"dummy_data{i}.txt" for i in range(3)],
+ },
+ )
+ ]
+
+ def _generate_tables(self, filepaths, dummy_kwarg_with_different_length):
+ idx = 0
+ for filepath in filepaths:
+ for i in range(10):
+ yield idx, pa.table({"id": range(10 * i, 10 * (i + 1)), "filepath": [filepath] * 10})
+ idx += 1
+
+
+class DummyGeneratorBasedBuilderWithAmbiguousShards(GeneratorBasedBuilder):
+ def _info(self):
+ return DatasetInfo(features=Features({"id": Value("int8"), "filepath": Value("string")}))
+
+ def _split_generators(self, dl_manager):
+ return [
+ SplitGenerator(
+ name=Split.TRAIN,
+ gen_kwargs={
+ "filepaths": [f"data{i}.txt" for i in range(4)],
+ "dummy_kwarg_with_different_length": [f"dummy_data{i}.txt" for i in range(3)],
+ },
+ )
+ ]
+
+ def _generate_examples(self, filepaths, dummy_kwarg_with_different_length):
+ idx = 0
+ for filepath in filepaths:
+ for i in range(100):
+ yield idx, {"id": i, "filepath": filepath}
+ idx += 1
+
+
def _run_concurrent_download_and_prepare(tmp_dir):
builder = DummyBuilder(cache_dir=tmp_dir)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.REUSE_DATASET_IF_EXISTS)
@@ -1068,6 +1115,15 @@ def test_generator_based_builder_download_and_prepare_with_num_proc(tmp_path):
}
[email protected](
+ "num_proc, expectation", [(None, does_not_raise()), (1, does_not_raise()), (2, pytest.raises(RuntimeError))]
+)
+def test_generator_based_builder_download_and_prepare_with_ambiguous_shards(num_proc, expectation, tmp_path):
+ builder = DummyGeneratorBasedBuilderWithAmbiguousShards(cache_dir=tmp_path)
+ with expectation:
+ builder.download_and_prepare(num_proc=num_proc)
+
+
def test_arrow_based_builder_download_and_prepare_as_parquet(tmp_path):
builder = DummyArrowBasedBuilder(cache_dir=tmp_path)
builder.download_and_prepare(file_format="parquet")
@@ -1130,6 +1186,15 @@ def test_arrow_based_builder_download_and_prepare_with_num_proc(tmp_path):
}
[email protected](
+ "num_proc, expectation", [(None, does_not_raise()), (1, does_not_raise()), (2, pytest.raises(RuntimeError))]
+)
+def test_arrow_based_builder_download_and_prepare_with_ambiguous_shards(num_proc, expectation, tmp_path):
+ builder = DummyArrowBasedBuilderWithAmbiguousShards(cache_dir=tmp_path)
+ with expectation:
+ builder.download_and_prepare(num_proc=num_proc)
+
+
@require_beam
def test_beam_based_builder_download_and_prepare_as_parquet(tmp_path):
builder = DummyBeamBasedBuilder(cache_dir=tmp_path, beam_runner="DirectRunner")
| RuntimeError: Sharding is ambiguous for this dataset
### Describe the bug
When loading some datasets, a RuntimeError is raised.
For example, for "ami" dataset: https://huggingface.co/datasets/ami/discussions/3
```
.../huggingface/datasets/src/datasets/builder.py in _prepare_split(self, split_generator, check_duplicate_keys, file_format, num_proc, max_shard_size)
1415 fpath = path_join(self._output_dir, fname)
1416
-> 1417 num_input_shards = _number_of_shards_in_gen_kwargs(split_generator.gen_kwargs)
1418 if num_input_shards <= 1 and num_proc is not None:
1419 logger.warning(
.../huggingface/datasets/src/datasets/utils/sharding.py in _number_of_shards_in_gen_kwargs(gen_kwargs)
10 lists_lengths = {key: len(value) for key, value in gen_kwargs.items() if isinstance(value, list)}
11 if len(set(lists_lengths.values())) > 1:
---> 12 raise RuntimeError(
13 (
14 "Sharding is ambiguous for this dataset: "
RuntimeError: Sharding is ambiguous for this dataset: we found several data sources lists of different lengths, and we don't know over which list we should parallelize:
- key samples_paths has length 6
- key ids has length 7
- key verification_ids has length 6
To fix this, check the 'gen_kwargs' and make sure to use lists only for data sources, and use tuples otherwise. In the end there should only be one single list, or several lists with the same length.
```
This behavior was introduced when implementing multiprocessing by PR:
- #5107
### Steps to reproduce the bug
```python
ds = load_dataset("ami", "microphone-single", split="train", revision="2d7620bb7c3f1aab9f329615c3bdb598069d907a")
```
### Expected behavior
No error raised.
### Environment info
Since datasets 2.7.0
Sharding error with Multilingual LibriSpeech
### Describe the bug
Loading the German Multilingual LibriSpeech dataset results in a RuntimeError regarding sharding with the following stacktrace:
```
Downloading and preparing dataset multilingual_librispeech/german to /home/nithin/datadrive/cache/huggingface/datasets/facebook___multilingual_librispeech/german/2.1.0/1904af50f57a5c370c9364cc337699cfe496d4e9edcae6648a96be23086362d0...
Downloading data files: 100%
3/3 [00:00<00:00, 107.23it/s]
Downloading data files: 100%
1/1 [00:00<00:00, 35.08it/s]
Downloading data files: 100%
6/6 [00:00<00:00, 303.36it/s]
Downloading data files: 100%
3/3 [00:00<00:00, 130.37it/s]
Downloading data files: 100%
1049/1049 [00:00<00:00, 4491.40it/s]
Downloading data files: 100%
37/37 [00:00<00:00, 1096.78it/s]
Downloading data files: 100%
40/40 [00:00<00:00, 1003.93it/s]
Extracting data files: 100%
3/3 [00:11<00:00, 2.62s/it]
Generating train split:
469942/0 [34:13<00:00, 273.21 examples/s]
Output exceeds the size limit. Open the full output data in a text editor
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
<ipython-input-14-74fa6d092bdc> in <module>
----> 1 mls = load_dataset(MLS_DATASET,
2 LANGUAGE,
3 cache_dir="~/datadrive/cache/huggingface/datasets",
4 ignore_verifications=True)
/anaconda/envs/py38_default/lib/python3.8/site-packages/datasets/load.py in load_dataset(path, name, data_dir, data_files, split, cache_dir, features, download_config, download_mode, ignore_verifications, keep_in_memory, save_infos, revision, use_auth_token, task, streaming, num_proc, **config_kwargs)
1755
1756 # Download and prepare data
-> 1757 builder_instance.download_and_prepare(
1758 download_config=download_config,
1759 download_mode=download_mode,
/anaconda/envs/py38_default/lib/python3.8/site-packages/datasets/builder.py in download_and_prepare(self, output_dir, download_config, download_mode, ignore_verifications, try_from_hf_gcs, dl_manager, base_path, use_auth_token, file_format, max_shard_size, num_proc, storage_options, **download_and_prepare_kwargs)
858 if num_proc is not None:
859 prepare_split_kwargs["num_proc"] = num_proc
--> 860 self._download_and_prepare(
861 dl_manager=dl_manager,
862 verify_infos=verify_infos,
/anaconda/envs/py38_default/lib/python3.8/site-packages/datasets/builder.py in _download_and_prepare(self, dl_manager, verify_infos, **prepare_splits_kwargs)
1609
1610 def _download_and_prepare(self, dl_manager, verify_infos, **prepare_splits_kwargs):
...
RuntimeError: Sharding is ambiguous for this dataset: we found several data sources lists of different lengths, and we don't know over which list we should parallelize:
- key audio_archives has length 1049
- key local_extracted_archive has length 1049
- key limited_ids_paths has length 1
To fix this, check the 'gen_kwargs' and make sure to use lists only for data sources, and use tuples otherwise. In the end there should only be one single list, or several lists with the same length.
```
### Steps to reproduce the bug
Here is the code to reproduce it:
```python
from datasets import load_dataset
MLS_DATASET = "facebook/multilingual_librispeech"
LANGUAGE = "german"
mls = load_dataset(MLS_DATASET,
LANGUAGE,
cache_dir="~/datadrive/cache/huggingface/datasets",
ignore_verifications=True)
```
### Expected behavior
The expected behaviour is that the dataset is successfully loaded.
### Environment info
- `datasets` version: 2.8.0
- Platform: Linux-5.4.0-1094-azure-x86_64-with-glibc2.10
- Python version: 3.8.8
- PyArrow version: 10.0.1
- Pandas version: 1.2.4
|
Thanks for reporting, @Nithin-Holla.
This is a known issue for multiple datasets and we are investigating it:
- See e.g.: https://huggingface.co/datasets/ami/discussions/3
Main issue:
- #5415
@albertvillanova Thanks! As a workaround for now, can I use the dataset in streaming mode? | 2023-01-10T08:43:19Z | [] | [] |
huggingface/datasets | 5,449 | huggingface__datasets-5449 | [
"5448"
] | 5f819ba3d0306748aaf9fd8ea040b981dd08e5e5 | diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -128,7 +128,7 @@
"importlib_metadata;python_version<'3.8'",
# to save datasets locally or on any filesystem
# minimum 2021.11.1 so that BlockSizeError is fixed: see https://github.com/fsspec/filesystem_spec/pull/830
- "fsspec[http]>=2021.11.1,<2023.1.0",
+ "fsspec[http]>=2021.11.1",
# for data streaming via http
"aiohttp",
# To get datasets from the Datasets Hub on huggingface.co
| diff --git a/tests/fixtures/fsspec.py b/tests/fixtures/fsspec.py
--- a/tests/fixtures/fsspec.py
+++ b/tests/fixtures/fsspec.py
@@ -74,8 +74,11 @@ def _strip_protocol(cls, path):
@pytest.fixture
-def mock_fsspec(monkeypatch):
- monkeypatch.setitem(fsspec.registry.target, "mock", MockFileSystem)
+def mock_fsspec():
+ original_registry = fsspec.registry.copy()
+ fsspec.register_implementation("mock", MockFileSystem)
+ yield
+ fsspec.registry = original_registry
@pytest.fixture
diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -128,8 +128,11 @@ def _open(
@pytest.fixture
-def mock_fsspec(monkeypatch):
- monkeypatch.setitem(fsspec.registry.target, "mock", DummyTestFS)
+def mock_fsspec():
+ original_registry = fsspec.registry.copy()
+ fsspec.register_implementation("mock", DummyTestFS)
+ yield
+ fsspec.registry = original_registry
def _readd_double_slash_removed_by_path(path_as_posix: str) -> str:
| Support fsspec 2023.1.0 in CI
Once we find out the root cause of:
- #5445
we should revert the temporary pin on fsspec introduced by:
- #5447
| 2023-01-20T12:53:17Z | [] | [] |
|
huggingface/datasets | 5,480 | huggingface__datasets-5480 | [
"5474"
] | c4a4f96ef0a4ec4b25f0872f160fa1eb9d2e711c | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -2145,6 +2145,55 @@ def rename(columns):
dataset._fingerprint = new_fingerprint
return dataset
+ @transmit_tasks
+ @transmit_format
+ @fingerprint_transform(inplace=False)
+ def select_columns(self, column_names: Union[str, List[str]], new_fingerprint: Optional[str] = None) -> "Dataset":
+ """Select one or several column(s) in the dataset and the features
+ associated to them.
+
+ Args:
+ column_names (`Union[str, List[str]]`):
+ Name of the column(s) to keep.
+ new_fingerprint (`str`, *optional*):
+ The new fingerprint of the dataset after transform. If `None`,
+ the new fingerprint is computed using a hash of the previous
+ fingerprint, and the transform arguments.
+
+ Returns:
+ [`Dataset`]: A copy of the dataset object which only consists of
+ selected columns.
+
+ Example:
+
+ ```py
+ >>> from datasets import load_dataset
+ >>> ds = load_dataset("rotten_tomatoes", split="validation")
+ >>> ds.select_columns(['text'])
+ Dataset({
+ features: ['text'],
+ num_rows: 1066
+ })
+ ```
+ """
+ if isinstance(column_names, str):
+ column_names = [column_names]
+
+ for column_name in column_names:
+ if column_name not in self._data.column_names:
+ raise ValueError(
+ f"Column name {column_name} not in the "
+ "dataset. Current columns in the dataset: "
+ f"{self._data.column_names}."
+ )
+
+ dataset = copy.deepcopy(self)
+ dataset._info.features = Features({k: v for k, v in dataset._info.features.items() if k in column_names})
+ dataset._data = dataset._data.select(column_names)
+ dataset._data = update_metadata_with_features(dataset._data, dataset.features)
+ dataset._fingerprint = new_fingerprint
+ return dataset
+
def __len__(self):
"""Number of rows in the dataset.
diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -432,6 +432,42 @@ def rename_columns(self, column_mapping: Dict[str, str]) -> "DatasetDict":
self._check_values_type()
return DatasetDict({k: dataset.rename_columns(column_mapping=column_mapping) for k, dataset in self.items()})
+ def select_columns(self, column_names: Union[str, List[str]]) -> "DatasetDict":
+ """Select one or several column(s) from each split in the dataset and
+ the features associated to the column(s).
+
+ The transformation is applied to all the splits of the dataset
+ dictionary.
+
+ Args:
+ column_names (`Union[str, List[str]]`):
+ Name of the column(s) to keep.
+
+ Example:
+
+ ```py
+ >>> from datasets import load_dataset
+ >>> ds = load_dataset("rotten_tomatoes")
+ >>> ds.select_columns("text")
+ DatasetDict({
+ train: Dataset({
+ features: ['text'],
+ num_rows: 8530
+ })
+ validation: Dataset({
+ features: ['text'],
+ num_rows: 1066
+ })
+ test: Dataset({
+ features: ['text'],
+ num_rows: 1066
+ })
+ })
+ ```
+ """
+ self._check_values_type()
+ return DatasetDict({k: dataset.select_columns(column_names=column_names) for k, dataset in self.items()})
+
def class_encode_column(self, column: str, include_nulls: bool = False) -> "DatasetDict":
"""Casts the given column as `datasets.features.ClassLabel` and updates the tables.
@@ -1911,6 +1947,32 @@ def remove_columns(self, column_names: Union[str, List[str]]) -> "IterableDatase
"""
return IterableDatasetDict({k: dataset.remove_columns(column_names) for k, dataset in self.items()})
+ def select_columns(self, column_names: Union[str, List[str]]) -> "IterableDatasetDict":
+ """Select one or several column(s) in the dataset and the features
+ associated to them. The selection is done on-the-fly on the examples
+ when iterating over the dataset. The selection is applied to all the
+ datasets of the dataset dictionary.
+
+
+ Args:
+ column_names (`Union[str, List[str]]`):
+ Name of the column(s) to keep.
+
+ Returns:
+ [`IterableDatasetDict`]: A copy of the dataset object with only selected columns.
+
+ Example:
+
+ ```py
+ >>> from datasets import load_dataset
+ >>> ds = load_dataset("rotten_tomatoes", streaming=True)
+ >>> ds = ds.select("text")
+ >>> next(iter(ds["train"]))
+ {'text': 'the rock is destined to be the 21st century\'s new " conan " and that he\'s going to make a splash even greater than arnold schwarzenegger , jean-claud van damme or steven segal .'}
+ ```
+ """
+ return IterableDatasetDict({k: dataset.select_columns(column_names) for k, dataset in self.items()})
+
def cast_column(self, column: str, feature: FeatureType) -> "IterableDatasetDict":
"""Cast column to feature for decoding.
The type casting is applied to all the datasets of the dataset dictionary.
diff --git a/src/datasets/iterable_dataset.py b/src/datasets/iterable_dataset.py
--- a/src/datasets/iterable_dataset.py
+++ b/src/datasets/iterable_dataset.py
@@ -146,6 +146,26 @@ def shard_data_sources(self, shard_indices: List[int]) -> "ExamplesIterable":
)
+class SelectColumnsIterable(_BaseExamplesIterable):
+ def __init__(self, ex_iterable: _BaseExamplesIterable, column_names: List[str]):
+ self.ex_iterable = ex_iterable
+ self.column_names = column_names
+
+ def __iter__(self):
+ for idx, row in self.ex_iterable:
+ yield idx, {c: row[c] for c in self.column_names}
+
+ def shuffle_data_sources(self, generator: np.random.Generator) -> "SelectColumnsIterable":
+ return SelectColumnsIterable(self.ex_iterable.shuffle_data_sources(generator), self.column_names)
+
+ def shard_data_sources(self, shard_indices: List[int]) -> "SelectColumnsIterable":
+ return SelectColumnsIterable(self.ex_iterable.shard_data_sources(shard_indices), self.column_names)
+
+ @property
+ def n_shards(self) -> int:
+ return self.ex_iterable.n_shards
+
+
class StepExamplesIterable(_BaseExamplesIterable):
def __init__(self, ex_iterable: _BaseExamplesIterable, step: int, offset: int):
self.ex_iterable = ex_iterable
@@ -1494,6 +1514,57 @@ def remove_columns(self, column_names: Union[str, List[str]]) -> "IterableDatase
del ds_iterable._info.features[col]
return ds_iterable
+ def select_columns(self, column_names: Union[str, List[str]]) -> "IterableDataset":
+ """Select one or several column(s) in the dataset and the features
+ associated to them. The selection is done on-the-fly on the examples
+ when iterating over the dataset.
+
+
+ Args:
+ column_names (`Union[str, List[str]]`):
+ Name of the column(s) to select.
+
+ Returns:
+ `IterableDataset`: A copy of the dataset object with selected columns.
+
+ Example:
+
+ ```py
+ >>> from datasets import load_dataset
+ >>> ds = load_dataset("rotten_tomatoes", split="train", streaming=True)
+ >>> next(iter(ds))
+ {'text': 'the rock is destined to be the 21st century\'s new " conan " and that he\'s going to make a splash even greater than arnold schwarzenegger , jean-claud van damme or steven segal .', 'label': 1}
+ >>> ds = ds.select_columns("text")
+ >>> next(iter(ds))
+ {'text': 'the rock is destined to be the 21st century\'s new " conan " and that he\'s going to make a splash even greater than arnold schwarzenegger , jean-claud van damme or steven segal .'}
+ ```
+ """
+ if isinstance(column_names, str):
+ column_names = [column_names]
+
+ if self._info:
+ info = copy.deepcopy(self._info)
+ if self._info.features is not None:
+ for column_name in column_names:
+ if column_name not in self._info.features:
+ raise ValueError(
+ f"Column name {column_name} not in the "
+ "dataset. Columns in the dataset: "
+ f"{list(self._info.features.keys())}."
+ )
+ info.features = Features({c: info.features[c] for c in column_names})
+
+ ex_iterable = SelectColumnsIterable(self._ex_iterable, column_names)
+ return IterableDataset(
+ ex_iterable=ex_iterable,
+ info=info,
+ split=self._split,
+ format_type=self._format_type,
+ shuffling=self._shuffling,
+ distributed=self._distributed,
+ token_per_repo_id=self._token_per_repo_id,
+ )
+
def cast_column(self, column: str, feature: FeatureType) -> "IterableDataset":
"""Cast column to feature for decoding.
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -622,6 +622,40 @@ def test_rename_columns(self, in_memory):
with self.assertRaises(ValueError):
dset.rename_columns({"col_1": "new_name", "col_2": "new_name"})
+ def test_select_columns(self, in_memory):
+ with tempfile.TemporaryDirectory() as tmp_dir:
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ fingerprint = dset._fingerprint
+ with dset.select_columns(column_names=[]) as new_dset:
+ self.assertEqual(new_dset.num_columns, 0)
+ self.assertListEqual(list(new_dset.column_names), [])
+ self.assertNotEqual(new_dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
+
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ fingerprint = dset._fingerprint
+ with dset.select_columns(column_names="col_1") as new_dset:
+ self.assertEqual(new_dset.num_columns, 1)
+ self.assertListEqual(list(new_dset.column_names), ["col_1"])
+ self.assertNotEqual(new_dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
+
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ with dset.select_columns(column_names=["col_1", "col_2", "col_3"]) as new_dset:
+ self.assertEqual(new_dset.num_columns, 3)
+ self.assertListEqual(list(new_dset.column_names), ["col_1", "col_2", "col_3"])
+ self.assertNotEqual(new_dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
+
+ with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
+ dset._format_columns = ["col_1", "col_2", "col_3"]
+ with dset.select_columns(column_names=["col_1"]) as new_dset:
+ self.assertListEqual(new_dset._format_columns, ["col_1"])
+ self.assertEqual(new_dset.num_columns, 1)
+ self.assertListEqual(list(new_dset.column_names), ["col_1"])
+ self.assertNotEqual(new_dset._fingerprint, fingerprint)
+ assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
+
def test_concatenate(self, in_memory):
data1, data2, data3 = {"id": [0, 1, 2]}, {"id": [3, 4, 5]}, {"id": [6, 7]}
info1 = DatasetInfo(description="Dataset1")
diff --git a/tests/test_dataset_dict.py b/tests/test_dataset_dict.py
--- a/tests/test_dataset_dict.py
+++ b/tests/test_dataset_dict.py
@@ -233,6 +233,32 @@ def test_rename_column(self):
self.assertListEqual(list(dset_split.column_names), ["new_name", "col_2"])
del dset
+ def test_select_columns(self):
+ dset = self._create_dummy_dataset_dict(multiple_columns=True)
+ dset = dset.select_columns(column_names=[])
+ for dset_split in dset.values():
+ self.assertEqual(dset_split.num_columns, 0)
+
+ dset = self._create_dummy_dataset_dict(multiple_columns=True)
+ dset = dset.select_columns(column_names="col_1")
+ for dset_split in dset.values():
+ self.assertEqual(dset_split.num_columns, 1)
+ self.assertListEqual(list(dset_split.column_names), ["col_1"])
+
+ dset = self._create_dummy_dataset_dict(multiple_columns=True)
+ dset = dset.select_columns(column_names=["col_1", "col_2"])
+ for dset_split in dset.values():
+ self.assertEqual(dset_split.num_columns, 2)
+
+ dset = self._create_dummy_dataset_dict(multiple_columns=True)
+ for dset_split in dset.values():
+ dset_split._format_columns = ["col_1", "col_2"]
+ dset = dset.select_columns(column_names=["col_1"])
+ for dset_split in dset.values():
+ self.assertEqual(dset_split.num_columns, 1)
+ self.assertListEqual(list(dset_split.column_names), ["col_1"])
+ self.assertListEqual(dset_split._format_columns, ["col_1"])
+
def test_map(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dsets = self._create_dummy_dataset_dict()
diff --git a/tests/test_iterable_dataset.py b/tests/test_iterable_dataset.py
--- a/tests/test_iterable_dataset.py
+++ b/tests/test_iterable_dataset.py
@@ -1009,6 +1009,23 @@ def test_iterable_dataset_remove_columns(dataset_with_several_columns):
assert all(c not in new_dataset.features for c in ["id", "filepath"])
+def test_iterable_dataset_select_columns(dataset_with_several_columns):
+ new_dataset = dataset_with_several_columns.select_columns("id")
+ assert list(new_dataset) == [
+ {k: v for k, v in example.items() if k == "id"} for example in dataset_with_several_columns
+ ]
+ assert new_dataset.features is None
+ new_dataset = dataset_with_several_columns.select_columns(["id", "filepath"])
+ assert list(new_dataset) == [
+ {k: v for k, v in example.items() if k in ("id", "filepath")} for example in dataset_with_several_columns
+ ]
+ assert new_dataset.features is None
+ # remove the columns if ds.features was not None
+ new_dataset = dataset_with_several_columns._resolve_features().select_columns(["id", "filepath"])
+ assert new_dataset.features is not None
+ assert all(c in new_dataset.features for c in ["id", "filepath"])
+
+
def test_iterable_dataset_cast_column():
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": 10})
features = Features({"id": Value("int64"), "label": Value("int64")})
| Column project operation on `datasets.Dataset`
### Feature request
There is no operation to select a subset of columns of original dataset. Expected API follows.
```python
a = Dataset.from_dict({
'int': [0, 1, 2]
'char': ['a', 'b', 'c'],
'none': [None] * 3,
})
b = a.project('int', 'char') # usually, .select()
print(a.column_names) # stdout: ['int', 'char', 'none']
print(b.column_names) # stdout: ['int', 'char']
```
Method project can easily accept not only column names (as a `str)` but univariant function applied to corresponding column as an example. Or keyword arguments can be used in order to rename columns in advance (see `pandas`, `pyspark`, `pyarrow`, and SQL)..
### Motivation
Projection is a typical operation in every data processing library. And it is a basic block of a well-known data manipulation language like SQL. Without this operation `datasets.Dataset` interface is not complete.
### Your contribution
Not sure. Some of my PRs are still open and some do not have any discussions.
| Hi ! This would be a nice addition indeed :) This sounds like a duplicate of https://github.com/huggingface/datasets/issues/5468
> Not sure. Some of my PRs are still open and some do not have any discussions.
Sorry to hear that, feel free to ping me on those PRs | 2023-01-27T20:06:16Z | [] | [] |
huggingface/datasets | 5,516 | huggingface__datasets-5516 | [
"5482"
] | b065547654efa0ec633cf373ac1512884c68b2e1 | diff --git a/src/datasets/io/parquet.py b/src/datasets/io/parquet.py
--- a/src/datasets/io/parquet.py
+++ b/src/datasets/io/parquet.py
@@ -1,7 +1,6 @@
import os
from typing import BinaryIO, Optional, Union
-import pyarrow as pa
import pyarrow.parquet as pq
from .. import Dataset, Features, NamedSplit, config
@@ -100,7 +99,8 @@ def _write(self, file_obj: BinaryIO, batch_size: int, **parquet_writer_kwargs) -
"""
written = 0
_ = parquet_writer_kwargs.pop("path_or_buf", None)
- schema = pa.schema(self.dataset.features.type)
+ schema = self.dataset.features.arrow_schema
+
writer = pq.ParquetWriter(file_obj, schema=schema, **parquet_writer_kwargs)
for offset in logging.tqdm(
| diff --git a/tests/io/test_parquet.py b/tests/io/test_parquet.py
--- a/tests/io/test_parquet.py
+++ b/tests/io/test_parquet.py
@@ -2,6 +2,7 @@
import pytest
from datasets import Dataset, DatasetDict, Features, NamedSplit, Value
+from datasets.features.image import Image
from datasets.io.parquet import ParquetDatasetReader, ParquetDatasetWriter
from ..utils import assert_arrow_memory_doesnt_increase, assert_arrow_memory_increases
@@ -130,3 +131,16 @@ def test_parquer_write(dataset, tmp_path):
pf = pq.ParquetFile(tmp_path / "foo.parquet")
output_table = pf.read()
assert dataset.data.table == output_table
+
+
+def test_dataset_to_parquet_keeps_features(shared_datadir, tmp_path):
+ image_path = str(shared_datadir / "test_image_rgb.jpg")
+ data = {"image": [image_path]}
+ features = Features({"image": Image()})
+ dataset = Dataset.from_dict(data, features=features)
+ writer = ParquetDatasetWriter(dataset, tmp_path / "foo.parquet")
+ assert writer.write() > 0
+
+ reloaded_dataset = Dataset.from_parquet(str(tmp_path / "foo.parquet"))
+
+ assert dataset.features == reloaded_dataset.features
| Reload features from Parquet metadata
The idea would be to allow this :
```python
ds.to_parquet("my_dataset/ds.parquet")
reloaded = load_dataset("my_dataset")
assert ds.features == reloaded.features
```
And it should also work with Image and Audio types (right now they're reloaded as a dict type)
This can be implemented by storing and reading the feature types in the parquet metadata, as we do for arrow files.
| I'd be happy to have a look, if nobody else has started working on this yet @lhoestq.
It seems to me that for the `arrow` format features are currently attached as metadata [in `datasets.arrow_writer`](https://github.com/huggingface/datasets/blob/5f810b7011a8a4ab077a1847c024d2d9e267b065/src/datasets/arrow_writer.py#L412) and retrieved from the metadata at `load_dataset` time using [`datasets.features.features.from_arrow_schema`](https://github.com/huggingface/datasets/blob/5f810b7011a8a4ab077a1847c024d2d9e267b065/src/datasets/features/features.py#L1602).
This will need to be replicated for `parquet` via calls to [this api](https://arrow.apache.org/docs/python/generated/pyarrow.parquet.write_metadata.html) from `io.parquet.ParquetWriter` and `io.parquet.ParquetReader` [respectively](https://github.com/huggingface/datasets/blob/5f810b7011a8a4ab077a1847c024d2d9e267b065/src/datasets/io/parquet.py#L104).
Any other important considerations?
Thanks @MFreidank ! That's correct :)
Reading the metadata to infer the features can be ideally done in the `parquet.py` file in `packaged_builder` when a parquet file is read. You can cast the arrow table to the schema you get from the features.arrow_schema
#self-assign | 2023-02-09T10:52:15Z | [] | [] |
huggingface/datasets | 5,526 | huggingface__datasets-5526 | [
"5428"
] | 939b2332115c7ec3dd56f58169800ed81cc4a982 | diff --git a/src/datasets/search.py b/src/datasets/search.py
--- a/src/datasets/search.py
+++ b/src/datasets/search.py
@@ -4,6 +4,7 @@
from pathlib import PurePath
from typing import TYPE_CHECKING, Dict, List, NamedTuple, Optional, Union
+import fsspec
import numpy as np
from .utils import logging
@@ -375,7 +376,7 @@ def search_batch(self, queries: np.array, k=10) -> BatchedSearchResults:
scores, indices = self.faiss_index.search(queries, k)
return BatchedSearchResults(scores, indices.astype(int))
- def save(self, file: Union[str, PurePath]):
+ def save(self, file: Union[str, PurePath], storage_options: Optional[Dict] = None):
"""Serialize the FaissIndex on disk"""
import faiss # noqa: F811
@@ -384,20 +385,23 @@ def save(self, file: Union[str, PurePath]):
else:
index = self.faiss_index
- faiss.write_index(index, str(file))
+ with fsspec.open(str(file), "wb", **(storage_options or {})) as f:
+ faiss.write_index(index, faiss.BufferedIOWriter(faiss.PyCallbackIOWriter(f.write)))
@classmethod
def load(
cls,
file: Union[str, PurePath],
device: Optional[Union[int, List[int]]] = None,
+ storage_options: Optional[Dict] = None,
) -> "FaissIndex":
"""Deserialize the FaissIndex from disk"""
import faiss # noqa: F811
# Instances of FaissIndex is essentially just a wrapper for faiss indices.
faiss_index = cls(device=device)
- index = faiss.read_index(str(file))
+ with fsspec.open(str(file), "rb", **(storage_options or {})) as f:
+ index = faiss.read_index(faiss.BufferedIOReader(faiss.PyCallbackIOReader(f.read)))
faiss_index.faiss_index = faiss_index._faiss_index_to_device(index, faiss_index.device)
return faiss_index
@@ -520,17 +524,22 @@ def add_faiss_index_from_external_arrays(
)
self._indexes[index_name] = faiss_index
- def save_faiss_index(self, index_name: str, file: Union[str, PurePath]):
+ def save_faiss_index(self, index_name: str, file: Union[str, PurePath], storage_options: Optional[Dict] = None):
"""Save a FaissIndex on disk.
Args:
index_name (`str`): The index_name/identifier of the index. This is the index_name that is used to call `.get_nearest` or `.search`.
- file (`str`): The path to the serialized faiss index on disk.
+ file (`str`): The path to the serialized faiss index on disk or remote URI (e.g. `"s3://my-bucket/index.faiss"`).
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.11.0"/>
+
"""
index = self.get_index(index_name)
if not isinstance(index, FaissIndex):
raise ValueError(f"Index '{index_name}' is not a FaissIndex but a '{type(index)}'")
- index.save(file)
+ index.save(file, storage_options=storage_options)
logger.info(f"Saved FaissIndex {index_name} at {file}")
def load_faiss_index(
@@ -538,6 +547,7 @@ def load_faiss_index(
index_name: str,
file: Union[str, PurePath],
device: Optional[Union[int, List[int]]] = None,
+ storage_options: Optional[Dict] = None,
):
"""Load a FaissIndex from disk.
@@ -547,11 +557,16 @@ def load_faiss_index(
Args:
index_name (`str`): The index_name/identifier of the index. This is the index_name that is used to
call `.get_nearest` or `.search`.
- file (`str`): The path to the serialized faiss index on disk.
+ file (`str`): The path to the serialized faiss index on disk or remote URI (e.g. `"s3://my-bucket/index.faiss"`).
device (Optional `Union[int, List[int]]`): If positive integer, this is the index of the GPU to use. If negative integer, use all GPUs.
If a list of positive integers is passed in, run only on those GPUs. By default it uses the CPU.
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.11.0"/>
+
"""
- index = FaissIndex.load(file, device=device)
+ index = FaissIndex.load(file, device=device, storage_options=storage_options)
if index.faiss_index.ntotal != len(self):
raise ValueError(
f"Index size should match Dataset size, but Index '{index_name}' at {file} has {index.faiss_index.ntotal} elements while the dataset has {len(self)} examples."
| diff --git a/tests/test_search.py b/tests/test_search.py
--- a/tests/test_search.py
+++ b/tests/test_search.py
@@ -167,6 +167,25 @@ def test_serialization(self):
self.assertEqual(indices[0], 1)
+@require_faiss
+def test_serialization_fs(mockfs):
+ import faiss
+
+ index = FaissIndex(metric_type=faiss.METRIC_INNER_PRODUCT)
+ index.add_vectors(np.eye(5, dtype=np.float32))
+
+ index_name = "index.faiss"
+ path = f"mock://{index_name}"
+ index.save(path, storage_options=mockfs.storage_options)
+ index = FaissIndex.load(path, storage_options=mockfs.storage_options)
+
+ query = np.zeros(5, dtype=np.float32)
+ query[1] = 1
+ scores, indices = index.search(query)
+ assert scores[0] > 0
+ assert indices[0] == 1
+
+
@require_elasticsearch
class ElasticSearchIndexTest(TestCase):
def test_elasticsearch(self):
| Load/Save FAISS index using fsspec
### Feature request
From what I understand `faiss` already support this [link](https://github.com/facebookresearch/faiss/wiki/Index-IO,-cloning-and-hyper-parameter-tuning#generic-io-support)
I would like to use a stream as input to `Dataset.load_faiss_index` and `Dataset.save_faiss_index`.
### Motivation
In my case, I'm saving faiss index in cloud storage and use `fsspec` to load them. It would be ideal if I could send the stream directly instead of copying the file locally (or mounting the bucket) and then load the index.
### Your contribution
I can submit the PR
| Hi! Sure, feel free to submit a PR. Maybe if we want to be consistent with the existing API, it would be cleaner to directly add support for `fsspec` paths in `Dataset.load_faiss_index`/`Dataset.save_faiss_index` in the same manner as it was done in `Dataset.load_from_disk`/`Dataset.save_to_disk`.
That's a great idea! I'll do that instead. | 2023-02-10T23:37:14Z | [] | [] |
huggingface/datasets | 5,549 | huggingface__datasets-5549 | [
"5548"
] | c480083958126c40bb7bdba8e1eeb3945a8fe6ea | diff --git a/metrics/code_eval/execute.py b/metrics/code_eval/execute.py
--- a/metrics/code_eval/execute.py
+++ b/metrics/code_eval/execute.py
@@ -45,12 +45,12 @@ def check_correctness(check_program, timeout, task_id, completion_id):
if not result:
result.append("timed out")
- return dict(
- task_id=task_id,
- passed=result[0] == "passed",
- result=result[0],
- completion_id=completion_id,
- )
+ return {
+ "task_id": task_id,
+ "passed": result[0] == "passed",
+ "result": result[0],
+ "completion_id": completion_id,
+ }
def unsafe_execute(check_program, result, timeout):
diff --git a/metrics/cuad/evaluate.py b/metrics/cuad/evaluate.py
--- a/metrics/cuad/evaluate.py
+++ b/metrics/cuad/evaluate.py
@@ -159,7 +159,7 @@ def evaluate(dataset, predictions):
message = "Unanswered question " + qa["id"] + " will receive score 0."
print(message, file=sys.stderr)
continue
- ground_truths = list(map(lambda x: x["text"], qa["answers"]))
+ ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
precision, recall = compute_precision_recall(prediction, ground_truths, qa["id"])
diff --git a/metrics/mean_iou/mean_iou.py b/metrics/mean_iou/mean_iou.py
--- a/metrics/mean_iou/mean_iou.py
+++ b/metrics/mean_iou/mean_iou.py
@@ -252,7 +252,7 @@ def mean_iou(
)
# compute metrics
- metrics = dict()
+ metrics = {}
all_acc = total_area_intersect.sum() / total_area_label.sum()
iou = total_area_intersect / total_area_union
diff --git a/metrics/rouge/rouge.py b/metrics/rouge/rouge.py
--- a/metrics/rouge/rouge.py
+++ b/metrics/rouge/rouge.py
@@ -125,6 +125,6 @@ def _compute(self, predictions, references, rouge_types=None, use_aggregator=Tru
else:
result = {}
for key in scores[0]:
- result[key] = list(score[key] for score in scores)
+ result[key] = [score[key] for score in scores]
return result
diff --git a/metrics/sacrebleu/sacrebleu.py b/metrics/sacrebleu/sacrebleu.py
--- a/metrics/sacrebleu/sacrebleu.py
+++ b/metrics/sacrebleu/sacrebleu.py
@@ -151,7 +151,7 @@ def _compute(
force=force,
lowercase=lowercase,
use_effective_order=use_effective_order,
- **(dict(tokenize=tokenize) if tokenize else {}),
+ **({"tokenize": tokenize} if tokenize else {}),
)
output_dict = {
"score": output.score,
diff --git a/metrics/squad/evaluate.py b/metrics/squad/evaluate.py
--- a/metrics/squad/evaluate.py
+++ b/metrics/squad/evaluate.py
@@ -62,7 +62,7 @@ def evaluate(dataset, predictions):
message = "Unanswered question " + qa["id"] + " will receive score 0."
print(message, file=sys.stderr)
continue
- ground_truths = list(map(lambda x: x["text"], qa["answers"]))
+ ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
exact_match += metric_max_over_ground_truths(exact_match_score, prediction, ground_truths)
f1 += metric_max_over_ground_truths(f1_score, prediction, ground_truths)
diff --git a/metrics/super_glue/record_evaluation.py b/metrics/super_glue/record_evaluation.py
--- a/metrics/super_glue/record_evaluation.py
+++ b/metrics/super_glue/record_evaluation.py
@@ -67,7 +67,7 @@ def evaluate(dataset, predictions):
print(message, file=sys.stderr)
continue
- ground_truths = list(map(lambda x: x["text"], qa["answers"]))
+ ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
_exact_match = metric_max_over_ground_truths(exact_match_score, prediction, ground_truths)
diff --git a/metrics/xtreme_s/xtreme_s.py b/metrics/xtreme_s/xtreme_s.py
--- a/metrics/xtreme_s/xtreme_s.py
+++ b/metrics/xtreme_s/xtreme_s.py
@@ -179,7 +179,7 @@ def bleu(
force=force,
lowercase=lowercase,
use_effective_order=use_effective_order,
- **(dict(tokenize=tokenize) if tokenize else {}),
+ **({"tokenize": tokenize} if tokenize else {}),
)
return {"bleu": output.score}
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -298,7 +298,7 @@ def _get_output_signature(
shapes = [array.shape for array in np_arrays]
static_shape = []
for dim in range(len(shapes[0])):
- sizes = set([shape[dim] for shape in shapes])
+ sizes = {shape[dim] for shape in shapes}
if dim == 0:
static_shape.append(batch_size)
continue
@@ -1349,9 +1349,9 @@ def save_to_disk(
if is_local:
Path(dataset_path).resolve().mkdir(parents=True, exist_ok=True)
- parent_cache_files_paths = set(
+ parent_cache_files_paths = {
Path(cache_filename["filename"]).resolve().parent for cache_filename in self.cache_files
- )
+ }
# Check that the dataset doesn't overwrite iself. It can cause a permission error on Windows and a segfault on linux.
if Path(dataset_path).resolve() in parent_cache_files_paths:
raise PermissionError(
@@ -2883,22 +2883,22 @@ def map(
f"num_proc must be <= {len(self)}. Reducing num_proc to {num_proc} for dataset of size {len(self)}."
)
- dataset_kwargs = dict(
- shard=self,
- function=function,
- with_indices=with_indices,
- with_rank=with_rank,
- input_columns=input_columns,
- batched=batched,
- batch_size=batch_size,
- drop_last_batch=drop_last_batch,
- remove_columns=remove_columns,
- keep_in_memory=keep_in_memory,
- writer_batch_size=writer_batch_size,
- features=features,
- disable_nullable=disable_nullable,
- fn_kwargs=fn_kwargs,
- )
+ dataset_kwargs = {
+ "shard": self,
+ "function": function,
+ "with_indices": with_indices,
+ "with_rank": with_rank,
+ "input_columns": input_columns,
+ "batched": batched,
+ "batch_size": batch_size,
+ "drop_last_batch": drop_last_batch,
+ "remove_columns": remove_columns,
+ "keep_in_memory": keep_in_memory,
+ "writer_batch_size": writer_batch_size,
+ "features": features,
+ "disable_nullable": disable_nullable,
+ "fn_kwargs": fn_kwargs,
+ }
if new_fingerprint is None:
# we create a unique hash from the function,
@@ -5109,14 +5109,14 @@ def path_in_repo(_index, shard):
uploaded_size += buffer.tell()
_retry(
api.upload_file,
- func_kwargs=dict(
- path_or_fileobj=buffer.getvalue(),
- path_in_repo=shard_path_in_repo,
- repo_id=repo_id,
- token=token,
- repo_type="dataset",
- revision=branch,
- ),
+ func_kwargs={
+ "path_or_fileobj": buffer.getvalue(),
+ "path_in_repo": shard_path_in_repo,
+ "repo_id": repo_id,
+ "token": token,
+ "repo_type": "dataset",
+ "revision": branch,
+ },
exceptions=HTTPError,
status_codes=[504],
base_wait_time=2.0,
diff --git a/src/datasets/arrow_reader.py b/src/datasets/arrow_reader.py
--- a/src/datasets/arrow_reader.py
+++ b/src/datasets/arrow_reader.py
@@ -265,7 +265,7 @@ def read_files(
split = Split(str(original_instructions))
else:
split = None
- dataset_kwargs = dict(arrow_table=pa_table, info=self._info, split=split)
+ dataset_kwargs = {"arrow_table": pa_table, "info": self._info, "split": split}
return dataset_kwargs
def download_from_hf_gcs(self, download_config: DownloadConfig, relative_data_dir):
diff --git a/src/datasets/arrow_writer.py b/src/datasets/arrow_writer.py
--- a/src/datasets/arrow_writer.py
+++ b/src/datasets/arrow_writer.py
@@ -667,10 +667,9 @@ def finalize(self, metrics_query_result: dict):
from .utils import beam_utils
- shards_metadata = [
- metadata
- for metadata in beam.io.filesystems.FileSystems.match([self._parquet_path + "*.parquet"])[0].metadata_list
- ]
+ shards_metadata = list(
+ beam.io.filesystems.FileSystems.match([self._parquet_path + "*.parquet"])[0].metadata_list
+ )
shards = [metadata.path for metadata in shards_metadata]
num_bytes = sum([metadata.size_in_bytes for metadata in shards_metadata])
shard_lengths = get_parquet_lengths(shards)
diff --git a/src/datasets/features/translation.py b/src/datasets/features/translation.py
--- a/src/datasets/features/translation.py
+++ b/src/datasets/features/translation.py
@@ -90,7 +90,7 @@ class TranslationVariableLanguages:
_type: str = field(default="TranslationVariableLanguages", init=False, repr=False)
def __post_init__(self):
- self.languages = list(sorted(list(set(self.languages)))) if self.languages else None
+ self.languages = sorted(set(self.languages)) if self.languages else None
self.num_languages = len(self.languages) if self.languages else None
def __call__(self):
diff --git a/src/datasets/filesystems/hffilesystem.py b/src/datasets/filesystems/hffilesystem.py
--- a/src/datasets/filesystems/hffilesystem.py
+++ b/src/datasets/filesystems/hffilesystem.py
@@ -89,4 +89,4 @@ def ls(self, path, detail=False, **kwargs):
if detail:
return out
else:
- return list(sorted(f["name"] for f in out))
+ return sorted(f["name"] for f in out)
diff --git a/src/datasets/packaged_modules/csv/csv.py b/src/datasets/packaged_modules/csv/csv.py
--- a/src/datasets/packaged_modules/csv/csv.py
+++ b/src/datasets/packaged_modules/csv/csv.py
@@ -72,45 +72,45 @@ def __post_init__(self):
@property
def pd_read_csv_kwargs(self):
- pd_read_csv_kwargs = dict(
- sep=self.sep,
- header=self.header,
- names=self.names,
- index_col=self.index_col,
- usecols=self.usecols,
- prefix=self.prefix,
- mangle_dupe_cols=self.mangle_dupe_cols,
- engine=self.engine,
- converters=self.converters,
- true_values=self.true_values,
- false_values=self.false_values,
- skipinitialspace=self.skipinitialspace,
- skiprows=self.skiprows,
- nrows=self.nrows,
- na_values=self.na_values,
- keep_default_na=self.keep_default_na,
- na_filter=self.na_filter,
- verbose=self.verbose,
- skip_blank_lines=self.skip_blank_lines,
- thousands=self.thousands,
- decimal=self.decimal,
- lineterminator=self.lineterminator,
- quotechar=self.quotechar,
- quoting=self.quoting,
- escapechar=self.escapechar,
- comment=self.comment,
- encoding=self.encoding,
- dialect=self.dialect,
- error_bad_lines=self.error_bad_lines,
- warn_bad_lines=self.warn_bad_lines,
- skipfooter=self.skipfooter,
- doublequote=self.doublequote,
- memory_map=self.memory_map,
- float_precision=self.float_precision,
- chunksize=self.chunksize,
- encoding_errors=self.encoding_errors,
- on_bad_lines=self.on_bad_lines,
- )
+ pd_read_csv_kwargs = {
+ "sep": self.sep,
+ "header": self.header,
+ "names": self.names,
+ "index_col": self.index_col,
+ "usecols": self.usecols,
+ "prefix": self.prefix,
+ "mangle_dupe_cols": self.mangle_dupe_cols,
+ "engine": self.engine,
+ "converters": self.converters,
+ "true_values": self.true_values,
+ "false_values": self.false_values,
+ "skipinitialspace": self.skipinitialspace,
+ "skiprows": self.skiprows,
+ "nrows": self.nrows,
+ "na_values": self.na_values,
+ "keep_default_na": self.keep_default_na,
+ "na_filter": self.na_filter,
+ "verbose": self.verbose,
+ "skip_blank_lines": self.skip_blank_lines,
+ "thousands": self.thousands,
+ "decimal": self.decimal,
+ "lineterminator": self.lineterminator,
+ "quotechar": self.quotechar,
+ "quoting": self.quoting,
+ "escapechar": self.escapechar,
+ "comment": self.comment,
+ "encoding": self.encoding,
+ "dialect": self.dialect,
+ "error_bad_lines": self.error_bad_lines,
+ "warn_bad_lines": self.warn_bad_lines,
+ "skipfooter": self.skipfooter,
+ "doublequote": self.doublequote,
+ "memory_map": self.memory_map,
+ "float_precision": self.float_precision,
+ "chunksize": self.chunksize,
+ "encoding_errors": self.encoding_errors,
+ "on_bad_lines": self.on_bad_lines,
+ }
# some kwargs must not be passed if they don't have a default value
# some others are deprecated and we can also not pass them if they are the default value
diff --git a/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
--- a/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
+++ b/src/datasets/packaged_modules/folder_based_builder/folder_based_builder.py
@@ -176,10 +176,10 @@ def analyze(files_or_archives, downloaded_files_or_dirs, split):
features_per_metadata_file: List[Tuple[str, datasets.Features]] = []
# Check that all metadata files share the same format
- metadata_ext = set(
+ metadata_ext = {
os.path.splitext(downloaded_metadata_file)[1][1:]
for _, downloaded_metadata_file in itertools.chain.from_iterable(metadata_files.values())
- )
+ }
if len(metadata_ext) > 1:
raise ValueError(f"Found metadata files with different extensions: {list(metadata_ext)}")
metadata_ext = metadata_ext.pop()
@@ -269,10 +269,10 @@ def _generate_examples(self, files, metadata_files, split_name, add_metadata, ad
downloaded_metadata_file = None
if split_metadata_files:
- metadata_ext = set(
+ metadata_ext = {
os.path.splitext(downloaded_metadata_file)[1][1:]
for _, downloaded_metadata_file in split_metadata_files
- )
+ }
metadata_ext = metadata_ext.pop()
file_idx = 0
diff --git a/src/datasets/packaged_modules/sql/sql.py b/src/datasets/packaged_modules/sql/sql.py
--- a/src/datasets/packaged_modules/sql/sql.py
+++ b/src/datasets/packaged_modules/sql/sql.py
@@ -77,13 +77,13 @@ def create_config_id(
@property
def pd_read_sql_kwargs(self):
- pd_read_sql_kwargs = dict(
- index_col=self.index_col,
- columns=self.columns,
- params=self.params,
- coerce_float=self.coerce_float,
- parse_dates=self.parse_dates,
- )
+ pd_read_sql_kwargs = {
+ "index_col": self.index_col,
+ "columns": self.columns,
+ "params": self.params,
+ "coerce_float": self.coerce_float,
+ "parse_dates": self.parse_dates,
+ }
return pd_read_sql_kwargs
diff --git a/src/datasets/table.py b/src/datasets/table.py
--- a/src/datasets/table.py
+++ b/src/datasets/table.py
@@ -1872,7 +1872,7 @@ def array_concat(arrays: List[pa.Array]):
array (:obj:`pyarrow.Array`): the concatenated array
"""
arrays = list(arrays)
- array_types = set(array.type for array in arrays)
+ array_types = {array.type for array in arrays}
if not array_types:
raise ValueError("Couldn't concatenate empty list of arrays")
@@ -1966,9 +1966,7 @@ def array_cast(array: pa.Array, pa_type: pa.DataType, allow_number_to_str=True):
elif array.type == pa_type:
return array
elif pa.types.is_struct(array.type):
- if pa.types.is_struct(pa_type) and (
- set(field.name for field in pa_type) == set(field.name for field in array.type)
- ):
+ if pa.types.is_struct(pa_type) and ({field.name for field in pa_type} == {field.name for field in array.type}):
arrays = [_c(array.field(field.name), field.type) for field in pa_type]
return pa.StructArray.from_arrays(arrays, fields=list(pa_type), mask=array.is_null())
elif pa.types.is_list(array.type):
@@ -2061,7 +2059,7 @@ def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_
feature = {
name: Sequence(subfeature, length=feature.length) for name, subfeature in feature.feature.items()
}
- if isinstance(feature, dict) and set(field.name for field in array.type) == set(feature):
+ if isinstance(feature, dict) and {field.name for field in array.type} == set(feature):
arrays = [_c(array.field(name), subfeature) for name, subfeature in feature.items()]
return pa.StructArray.from_arrays(arrays, names=list(feature), mask=array.is_null())
elif pa.types.is_list(array.type):
diff --git a/src/datasets/utils/metadata.py b/src/datasets/utils/metadata.py
--- a/src/datasets/utils/metadata.py
+++ b/src/datasets/utils/metadata.py
@@ -21,7 +21,7 @@ def construct_mapping(self, node, deep=False):
def _split_yaml_from_readme(readme_content: str) -> Tuple[Optional[str], str]:
- full_content = [line for line in readme_content.splitlines()]
+ full_content = list(readme_content.splitlines())
if full_content and full_content[0] == "---" and "---" in full_content[1:]:
sep_idx = full_content[1:].index("---") + 1
yamlblock = "\n".join(full_content[1:sep_idx])
@@ -85,7 +85,7 @@ def from_yaml_string(cls, string: str) -> "DatasetMetadata":
Raises:
:obj:`TypeError`: If the dataset's metadata is invalid
"""
- metadata_dict = yaml.load(string, Loader=_NoDuplicateSafeLoader) or dict()
+ metadata_dict = yaml.load(string, Loader=_NoDuplicateSafeLoader) or {}
# Convert the YAML keys to DatasetMetadata fields
metadata_dict = {
diff --git a/src/datasets/utils/readme.py b/src/datasets/utils/readme.py
--- a/src/datasets/utils/readme.py
+++ b/src/datasets/utils/readme.py
@@ -189,7 +189,7 @@ def validate(self):
else:
content, error_list, warning_list = self._validate(self.structure)
if error_list != [] or warning_list != []:
- errors = "\n".join(list(map(lambda x: "-\t" + x, error_list + warning_list)))
+ errors = "\n".join(["-\t" + x for x in error_list + warning_list])
error_string = f"The following issues were found for the README at `{self.name}`:\n" + errors
raise ValueError(error_string)
diff --git a/src/datasets/utils/sharding.py b/src/datasets/utils/sharding.py
--- a/src/datasets/utils/sharding.py
+++ b/src/datasets/utils/sharding.py
@@ -83,7 +83,7 @@ def _shuffle_gen_kwargs(rng: np.random.Generator, gen_kwargs: dict) -> dict:
# This way entangled lists of (shard, shard_metadata) are still in the right order.
# First, let's generate the shuffled indices per list size
- list_sizes = set(len(value) for value in gen_kwargs.values() if isinstance(value, list))
+ list_sizes = {len(value) for value in gen_kwargs.values() if isinstance(value, list)}
indices_per_size = {}
for size in list_sizes:
indices_per_size[size] = list(range(size))
diff --git a/src/datasets/utils/tf_utils.py b/src/datasets/utils/tf_utils.py
--- a/src/datasets/utils/tf_utils.py
+++ b/src/datasets/utils/tf_utils.py
@@ -103,7 +103,7 @@ def np_get_batch(
batch = [{key: value[i] for key, value in batch.items()} for i in range(actual_size)]
batch = collate_fn(batch, **collate_fn_args)
if return_dict:
- out_batch = dict()
+ out_batch = {}
for col, cast_dtype in columns_to_np_types.items():
# In case the collate_fn returns something strange
array = np.array(batch[col])
@@ -395,7 +395,7 @@ def send_batch_to_parent(indices):
)
# Now begins the fun part where we start shovelling shared memory at the parent process
- out_arrays = dict()
+ out_arrays = {}
with SharedMemoryContext() as batch_shm_ctx:
# The batch shared memory context exists only as long as it takes for the parent process
# to read everything, after which it cleans everything up again
| diff --git a/tests/packaged_modules/test_audiofolder.py b/tests/packaged_modules/test_audiofolder.py
--- a/tests/packaged_modules/test_audiofolder.py
+++ b/tests/packaged_modules/test_audiofolder.py
@@ -388,8 +388,8 @@ def test_data_files_with_metadata_and_single_split(streaming, cache_dir, data_fi
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_audios
# make sure each sample has its own audio and metadata
- assert len(set(example["audio"]["path"] for example in dataset)) == expected_num_of_audios
- assert len(set(example["text"] for example in dataset)) == expected_num_of_audios
+ assert len({example["audio"]["path"] for example in dataset}) == expected_num_of_audios
+ assert len({example["text"] for example in dataset}) == expected_num_of_audios
assert all(example["text"] is not None for example in dataset)
@@ -406,8 +406,8 @@ def test_data_files_with_metadata_and_multiple_splits(streaming, cache_dir, data
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_audios
# make sure each sample has its own audio and metadata
- assert len(set(example["audio"]["path"] for example in dataset)) == expected_num_of_audios
- assert len(set(example["text"] for example in dataset)) == expected_num_of_audios
+ assert len({example["audio"]["path"] for example in dataset}) == expected_num_of_audios
+ assert len({example["text"] for example in dataset}) == expected_num_of_audios
assert all(example["text"] is not None for example in dataset)
@@ -428,7 +428,7 @@ def test_data_files_with_metadata_and_archives(streaming, cache_dir, data_files_
sum(np.array_equal(dataset[0]["audio"]["array"], example["audio"]["array"]) for example in dataset[1:])
== 0
)
- assert len(set(example["text"] for example in dataset)) == expected_num_of_audios
+ assert len({example["text"] for example in dataset}) == expected_num_of_audios
assert all(example["text"] is not None for example in dataset)
diff --git a/tests/packaged_modules/test_folder_based_builder.py b/tests/packaged_modules/test_folder_based_builder.py
--- a/tests/packaged_modules/test_folder_based_builder.py
+++ b/tests/packaged_modules/test_folder_based_builder.py
@@ -471,8 +471,8 @@ def test_data_files_with_metadata_and_splits(
expected_num_of_examples = len(files) - 1
generated_examples = list(autofolder._generate_examples(**generated_split.gen_kwargs))
assert len(generated_examples) == expected_num_of_examples
- assert len(set(example["base"] for _, example in generated_examples)) == expected_num_of_examples
- assert len(set(example["additional_feature"] for _, example in generated_examples)) == expected_num_of_examples
+ assert len({example["base"] for _, example in generated_examples}) == expected_num_of_examples
+ assert len({example["additional_feature"] for _, example in generated_examples}) == expected_num_of_examples
assert all(example["additional_feature"] is not None for _, example in generated_examples)
@@ -487,8 +487,8 @@ def test_data_files_with_metadata_and_archives(streaming, cache_dir, data_files_
expected_num_of_examples = 2 * num_of_archives
generated_examples = list(autofolder._generate_examples(**generated_split.gen_kwargs))
assert len(generated_examples) == expected_num_of_examples
- assert len(set(example["base"] for _, example in generated_examples)) == expected_num_of_examples
- assert len(set(example["additional_feature"] for _, example in generated_examples)) == expected_num_of_examples
+ assert len({example["base"] for _, example in generated_examples}) == expected_num_of_examples
+ assert len({example["additional_feature"] for _, example in generated_examples}) == expected_num_of_examples
assert all(example["additional_feature"] is not None for _, example in generated_examples)
diff --git a/tests/packaged_modules/test_imagefolder.py b/tests/packaged_modules/test_imagefolder.py
--- a/tests/packaged_modules/test_imagefolder.py
+++ b/tests/packaged_modules/test_imagefolder.py
@@ -395,8 +395,8 @@ def test_data_files_with_metadata_and_single_split(streaming, cache_dir, data_fi
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_images
# make sure each sample has its own image and metadata
- assert len(set(example["image"].filename for example in dataset)) == expected_num_of_images
- assert len(set(example["caption"] for example in dataset)) == expected_num_of_images
+ assert len({example["image"].filename for example in dataset}) == expected_num_of_images
+ assert len({example["caption"] for example in dataset}) == expected_num_of_images
assert all(example["caption"] is not None for example in dataset)
@@ -413,8 +413,8 @@ def test_data_files_with_metadata_and_multiple_splits(streaming, cache_dir, data
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_images
# make sure each sample has its own image and metadata
- assert len(set(example["image"].filename for example in dataset)) == expected_num_of_images
- assert len(set(example["caption"] for example in dataset)) == expected_num_of_images
+ assert len({example["image"].filename for example in dataset}) == expected_num_of_images
+ assert len({example["caption"] for example in dataset}) == expected_num_of_images
assert all(example["caption"] is not None for example in dataset)
@@ -431,8 +431,8 @@ def test_data_files_with_metadata_and_archives(streaming, cache_dir, data_files_
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_images
# make sure each sample has its own image and metadata
- assert len(set([np.array(example["image"])[0, 0, 0] for example in dataset])) == expected_num_of_images
- assert len(set(example["caption"] for example in dataset)) == expected_num_of_images
+ assert len({np.array(example["image"])[0, 0, 0] for example in dataset}) == expected_num_of_images
+ assert len({example["caption"] for example in dataset}) == expected_num_of_images
assert all(example["caption"] is not None for example in dataset)
diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -3686,11 +3686,11 @@ def test_dataset_to_json(dataset, tmp_path):
@pytest.mark.parametrize(
"method_and_params",
[
- ("rename_column", tuple(), {"original_column_name": "labels", "new_column_name": "label"}),
- ("remove_columns", tuple(), {"column_names": "labels"}),
+ ("rename_column", (), {"original_column_name": "labels", "new_column_name": "label"}),
+ ("remove_columns", (), {"column_names": "labels"}),
(
"cast",
- tuple(),
+ (),
{
"features": Features(
{
@@ -3707,7 +3707,7 @@ def test_dataset_to_json(dataset, tmp_path):
)
},
),
- ("flatten", tuple(), {}),
+ ("flatten", (), {}),
],
)
def test_pickle_dataset_after_transforming_the_table(in_memory, method_and_params, arrow_file):
diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -26,18 +26,16 @@
_TEST_PATTERNS = ["*", "**", "**/*", "*.txt", "data/*", "**/*.txt", "**/train.txt"]
_FILES_TO_IGNORE = {".dummy", "README.md", "dummy_data.zip", "dataset_infos.json"}
_DIRS_TO_IGNORE = {"data/.dummy_subdir", "__pycache__"}
-_TEST_PATTERNS_SIZES = dict(
- [
- ("*", 0),
- ("**", 4),
- ("**/*", 4),
- ("*.txt", 0),
- ("data/*", 2),
- ("data/**", 4),
- ("**/*.txt", 4),
- ("**/train.txt", 2),
- ]
-)
+_TEST_PATTERNS_SIZES = {
+ "*": 0,
+ "**": 4,
+ "**/*": 4,
+ "*.txt": 0,
+ "data/*": 2,
+ "data/**": 4,
+ "**/*.txt": 4,
+ "**/train.txt": 2,
+}
_TEST_URL = "https://raw.githubusercontent.com/huggingface/datasets/9675a5a1e7b99a86f9c250f6ea5fa5d1e6d5cc7d/setup.py"
diff --git a/tests/test_distributed.py b/tests/test_distributed.py
--- a/tests/test_distributed.py
+++ b/tests/test_distributed.py
@@ -18,7 +18,7 @@ def test_split_dataset_by_node_map_style():
split_dataset_by_node(full_ds, rank=rank, world_size=world_size) for rank in range(world_size)
]
assert sum(len(ds) for ds in datasets_per_rank) == full_size
- assert len(set(tuple(x.values()) for ds in datasets_per_rank for x in ds)) == full_size
+ assert len({tuple(x.values()) for ds in datasets_per_rank for x in ds}) == full_size
def test_split_dataset_by_node_iterable():
@@ -32,7 +32,7 @@ def gen():
split_dataset_by_node(full_ds, rank=rank, world_size=world_size) for rank in range(world_size)
]
assert sum(len(list(ds)) for ds in datasets_per_rank) == full_size
- assert len(set(tuple(x.values()) for ds in datasets_per_rank for x in ds)) == full_size
+ assert len({tuple(x.values()) for ds in datasets_per_rank for x in ds}) == full_size
@pytest.mark.parametrize("shards_per_node", [1, 2, 3])
@@ -52,7 +52,7 @@ def gen(shards):
]
assert [ds.n_shards for ds in datasets_per_rank] == [shards_per_node] * world_size
assert sum(len(list(ds)) for ds in datasets_per_rank) == full_size
- assert len(set(tuple(x.values()) for ds in datasets_per_rank for x in ds)) == full_size
+ assert len({tuple(x.values()) for ds in datasets_per_rank for x in ds}) == full_size
@pytest.mark.parametrize("streaming", [False, True])
diff --git a/tests/test_info.py b/tests/test_info.py
--- a/tests/test_info.py
+++ b/tests/test_info.py
@@ -64,7 +64,7 @@ def test_dataset_info_to_yaml_dict():
license="CC0",
features=Features({"a": Value("int32")}),
post_processed={},
- supervised_keys=tuple(),
+ supervised_keys=(),
task_templates=[],
builder_name="builder",
config_name="config",
diff --git a/tests/test_iterable_dataset.py b/tests/test_iterable_dataset.py
--- a/tests/test_iterable_dataset.py
+++ b/tests/test_iterable_dataset.py
@@ -141,7 +141,7 @@ def test_buffer_shuffled_examples_iterable(seed):
assert next(iter(ex_iterable)) == expected[0]
assert list(ex_iterable) == expected
- assert sorted(list(ex_iterable)) == sorted(all_examples)
+ assert sorted(ex_iterable) == sorted(all_examples)
def test_cycling_multi_sources_examples_iterable():
@@ -223,7 +223,7 @@ def test_mapped_examples_iterable(n, func, batched, batch_size):
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
@@ -271,7 +271,7 @@ def test_mapped_examples_iterable_drop_last_batch(n, func, batched, batch_size):
if not is_empty:
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
else:
with pytest.raises(StopIteration):
next(iter(ex_iterable))
@@ -315,7 +315,7 @@ def test_mapped_examples_iterable_with_indices(n, func, batched, batch_size):
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
@@ -358,7 +358,7 @@ def test_mapped_examples_iterable_remove_columns(n, func, batched, batch_size, r
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
@@ -396,7 +396,7 @@ def test_mapped_examples_iterable_fn_kwargs(n, func, batched, batch_size, fn_kwa
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
@@ -432,7 +432,7 @@ def test_mapped_examples_iterable_input_columns(n, func, batched, batch_size, in
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
@@ -466,7 +466,7 @@ def test_filtered_examples_iterable(n, func, batched, batch_size):
expected.extend([x for x, to_keep in zip(examples, mask) if to_keep])
if expected:
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
@@ -499,7 +499,7 @@ def test_filtered_examples_iterable_with_indices(n, func, batched, batch_size):
mask = func(batch, indices)
expected.extend([x for x, to_keep in zip(examples, mask) if to_keep])
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
@@ -532,7 +532,7 @@ def test_filtered_examples_iterable_input_columns(n, func, batched, batch_size,
mask = func(*[batch[col] for col in columns_to_input])
expected.extend([x for x, to_keep in zip(examples, mask) if to_keep])
assert next(iter(ex_iterable))[1] == expected[0]
- assert list(x for _, x in ex_iterable) == expected
+ assert [x for _, x in ex_iterable] == expected
def test_skip_examples_iterable():
@@ -561,8 +561,8 @@ def test_vertically_concatenated_examples_iterable():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label": 5})
concatenated_ex_iterable = VerticallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
- expected = list(x for _, x in ex_iterable1) + list(x for _, x in ex_iterable2)
- assert list(x for _, x in concatenated_ex_iterable) == expected
+ expected = [x for _, x in ex_iterable1] + [x for _, x in ex_iterable2]
+ assert [x for _, x in concatenated_ex_iterable] == expected
def test_vertically_concatenated_examples_iterable_with_different_columns():
@@ -571,8 +571,8 @@ def test_vertically_concatenated_examples_iterable_with_different_columns():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {})
concatenated_ex_iterable = VerticallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
- expected = list(x for _, x in ex_iterable1) + list(x for _, x in ex_iterable2)
- assert list(x for _, x in concatenated_ex_iterable) == expected
+ expected = [x for _, x in ex_iterable1] + [x for _, x in ex_iterable2]
+ assert [x for _, x in concatenated_ex_iterable] == expected
def test_vertically_concatenated_examples_iterable_shuffle_data_sources():
@@ -582,10 +582,10 @@ def test_vertically_concatenated_examples_iterable_shuffle_data_sources():
rng = np.random.default_rng(42)
shuffled_ex_iterable = concatenated_ex_iterable.shuffle_data_sources(rng)
# make sure the list of examples iterables is shuffled, and each examples iterable is shuffled
- expected = list(x for _, x in ex_iterable2.shuffle_data_sources(rng)) + list(
+ expected = [x for _, x in ex_iterable2.shuffle_data_sources(rng)] + [
x for _, x in ex_iterable1.shuffle_data_sources(rng)
- )
- assert list(x for _, x in shuffled_ex_iterable) == expected
+ ]
+ assert [x for _, x in shuffled_ex_iterable] == expected
def test_horizontally_concatenated_examples_iterable():
@@ -596,8 +596,8 @@ def test_horizontally_concatenated_examples_iterable():
list(concatenated_ex_iterable)
ex_iterable2 = MappedExamplesIterable(ex_iterable2, lambda x: x, remove_columns=["id"])
concatenated_ex_iterable = HorizontallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
- expected = list({**x, **y} for (_, x), (_, y) in zip(ex_iterable1, ex_iterable2))
- assert list(x for _, x in concatenated_ex_iterable) == expected
+ expected = [{**x, **y} for (_, x), (_, y) in zip(ex_iterable1, ex_iterable2)]
+ assert [x for _, x in concatenated_ex_iterable] == expected
assert (
concatenated_ex_iterable.shuffle_data_sources(np.random.default_rng(42)) is concatenated_ex_iterable
), "horizontally concatenated examples makes the shards order fixed"
@@ -692,7 +692,7 @@ def test_iterable_dataset_torch_dataloader_parallel():
result = list(dataloader)
expected = [example for _, example in ex_iterable]
assert len(result) == len(expected)
- assert set(str(x) for x in result) == set(str(x) for x in expected)
+ assert {str(x) for x in result} == {str(x) for x in expected}
@require_torch
@@ -706,7 +706,7 @@ def test_sharded_iterable_dataset_torch_dataloader_parallel(n_shards, num_worker
result = list(dataloader)
expected = [example for _, example in ex_iterable]
assert len(result) == len(expected)
- assert set(str(x) for x in result) == set(str(x) for x in expected)
+ assert {str(x) for x in result} == {str(x) for x in expected}
@require_torch
@@ -728,7 +728,7 @@ def test_iterable_dataset_from_hub_torch_dataloader_parallel(num_workers, tmp_pa
def test_iterable_dataset_iter_batch(batch_size, drop_last_batch):
n = 25
dataset = IterableDataset(ExamplesIterable(generate_examples_fn, {"n": n}))
- all_examples = list(ex for _, ex in generate_examples_fn(n=n))
+ all_examples = [ex for _, ex in generate_examples_fn(n=n)]
expected = []
for i in range(0, len(all_examples), batch_size):
if len(all_examples[i : i + batch_size]) < batch_size and drop_last_batch:
diff --git a/tests/test_py_utils.py b/tests/test_py_utils.py
--- a/tests/test_py_utils.py
+++ b/tests/test_py_utils.py
@@ -97,7 +97,7 @@ def test_zip_dict(self):
d2 = {"a": 3, "b": 4}
d3 = {"a": 5, "b": 6}
expected_zip_dict_result = sorted([("a", (1, 3, 5)), ("b", (2, 4, 6))])
- self.assertEqual(sorted(list(zip_dict(d1, d2, d3))), expected_zip_dict_result)
+ self.assertEqual(sorted(zip_dict(d1, d2, d3)), expected_zip_dict_result)
def test_temporary_assignment(self):
class Foo:
diff --git a/tests/test_table.py b/tests/test_table.py
--- a/tests/test_table.py
+++ b/tests/test_table.py
@@ -374,7 +374,7 @@ def test_in_memory_table_replace_schema_metadata(in_memory_pa_table):
def test_in_memory_table_add_column(in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
table = InMemoryTable(in_memory_pa_table).add_column(i, field_, column)
assert table.table == in_memory_pa_table.add_column(i, field_, column)
assert isinstance(table, InMemoryTable)
@@ -382,7 +382,7 @@ def test_in_memory_table_add_column(in_memory_pa_table):
def test_in_memory_table_append_column(in_memory_pa_table):
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
table = InMemoryTable(in_memory_pa_table).append_column(field_, column)
assert table.table == in_memory_pa_table.append_column(field_, column)
assert isinstance(table, InMemoryTable)
@@ -397,7 +397,7 @@ def test_in_memory_table_remove_column(in_memory_pa_table):
def test_in_memory_table_set_column(in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
table = InMemoryTable(in_memory_pa_table).set_column(i, field_, column)
assert table.table == in_memory_pa_table.set_column(i, field_, column)
assert isinstance(table, InMemoryTable)
@@ -436,7 +436,7 @@ def test_memory_mapped_table_from_file(arrow_file, in_memory_pa_table):
def test_memory_mapped_table_from_file_with_replay(arrow_file, in_memory_pa_table):
- replays = [("slice", (0, 1), {}), ("flatten", tuple(), {})]
+ replays = [("slice", (0, 1), {}), ("flatten", (), {})]
with assert_arrow_memory_doesnt_increase():
table = MemoryMappedTable.from_file(arrow_file, replays=replays)
assert len(table) == 1
@@ -475,7 +475,7 @@ def test_memory_mapped_table_pickle_doesnt_fill_memory(arrow_file):
def test_memory_mapped_table_pickle_applies_replay(arrow_file):
- replays = [("slice", (0, 1), {}), ("flatten", tuple(), {})]
+ replays = [("slice", (0, 1), {}), ("flatten", (), {})]
with assert_arrow_memory_doesnt_increase():
table = MemoryMappedTable.from_file(arrow_file, replays=replays)
assert isinstance(table, MemoryMappedTable)
@@ -509,7 +509,7 @@ def test_memory_mapped_table_flatten(arrow_file, in_memory_pa_table):
table = MemoryMappedTable.from_file(arrow_file).flatten()
assert table.table == in_memory_pa_table.flatten()
assert isinstance(table, MemoryMappedTable)
- assert table.replays == [("flatten", tuple(), {})]
+ assert table.replays == [("flatten", (), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
@@ -518,7 +518,7 @@ def test_memory_mapped_table_combine_chunks(arrow_file, in_memory_pa_table):
table = MemoryMappedTable.from_file(arrow_file).combine_chunks()
assert table.table == in_memory_pa_table.combine_chunks()
assert isinstance(table, MemoryMappedTable)
- assert table.replays == [("combine_chunks", tuple(), {})]
+ assert table.replays == [("combine_chunks", (), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
@@ -554,7 +554,7 @@ def test_memory_mapped_table_replace_schema_metadata(arrow_file, in_memory_pa_ta
def test_memory_mapped_table_add_column(arrow_file, in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
table = MemoryMappedTable.from_file(arrow_file).add_column(i, field_, column)
assert table.table == in_memory_pa_table.add_column(i, field_, column)
assert isinstance(table, MemoryMappedTable)
@@ -565,7 +565,7 @@ def test_memory_mapped_table_add_column(arrow_file, in_memory_pa_table):
def test_memory_mapped_table_append_column(arrow_file, in_memory_pa_table):
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
table = MemoryMappedTable.from_file(arrow_file).append_column(field_, column)
assert table.table == in_memory_pa_table.append_column(field_, column)
assert isinstance(table, MemoryMappedTable)
@@ -586,7 +586,7 @@ def test_memory_mapped_table_remove_column(arrow_file, in_memory_pa_table):
def test_memory_mapped_table_set_column(arrow_file, in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
table = MemoryMappedTable.from_file(arrow_file).set_column(i, field_, column)
assert table.table == in_memory_pa_table.set_column(i, field_, column)
assert isinstance(table, MemoryMappedTable)
@@ -912,7 +912,7 @@ def test_concatenation_table_add_column(
}[blocks_type]
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
with pytest.raises(NotImplementedError):
ConcatenationTable.from_blocks(blocks).add_column(i, field_, column)
# assert table.table == in_memory_pa_table.add_column(i, field_, column)
@@ -930,7 +930,7 @@ def test_concatenation_table_append_column(
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
with pytest.raises(NotImplementedError):
ConcatenationTable.from_blocks(blocks).append_column(field_, column)
# assert table.table == in_memory_pa_table.append_column(field_, column)
@@ -963,7 +963,7 @@ def test_concatenation_table_set_column(
}[blocks_type]
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
- column = pa.array([i for i in range(len(in_memory_pa_table))])
+ column = pa.array(list(range(len(in_memory_pa_table))))
with pytest.raises(NotImplementedError):
ConcatenationTable.from_blocks(blocks).set_column(i, field_, column)
# assert table.table == in_memory_pa_table.set_column(i, field_, column)
| Apply flake8-comprehensions to codebase
### Feature request
Apply ruff flake8 comprehension checks to codebase.
### Motivation
This should strictly improve the performance / readability of the codebase by removing unnecessary iteration, function calls, etc. This should generate better Python bytecode which should strictly improve performance.
I already applied this fixes to PyTorch and Sympy with little issue and have opened PRs to diffusers and transformers todo this as well.
### Your contribution
Making a PR.
| 2023-02-19T20:09:28Z | [] | [] |
|
huggingface/datasets | 5,582 | huggingface__datasets-5582 | [
"5383"
] | a940972a9a38543b2066129dc6e7987e08dca082 | diff --git a/src/datasets/iterable_dataset.py b/src/datasets/iterable_dataset.py
--- a/src/datasets/iterable_dataset.py
+++ b/src/datasets/iterable_dataset.py
@@ -1374,6 +1374,21 @@ def take(self, n) -> "IterableDataset":
token_per_repo_id=self._token_per_repo_id,
)
+ @property
+ def column_names(self) -> Optional[List[str]]:
+ """Names of the columns in the dataset.
+
+ Example:
+
+ ```py
+ >>> from datasets import load_dataset
+ >>> ds = load_dataset("rotten_tomatoes", split="validation", streaming=True)
+ >>> ds.column_names
+ ['text', 'label']
+ ```
+ """
+ return list(self._info.features.keys()) if self._info.features is not None else None
+
def add_column(self, name: str, column: Union[list, np.array]) -> "IterableDataset":
"""Add column to Dataset.
| diff --git a/tests/test_iterable_dataset.py b/tests/test_iterable_dataset.py
--- a/tests/test_iterable_dataset.py
+++ b/tests/test_iterable_dataset.py
@@ -966,6 +966,8 @@ def test_iterable_dataset_add_column(dataset_with_several_columns):
assert list(new_dataset) == [
{**example, "new_column": idx} for idx, example in enumerate(dataset_with_several_columns)
]
+ new_dataset = new_dataset._resolve_features()
+ assert "new_column" in new_dataset.column_names
def test_iterable_dataset_rename_column(dataset_with_several_columns):
@@ -974,9 +976,13 @@ def test_iterable_dataset_rename_column(dataset_with_several_columns):
{("new_id" if k == "id" else k): v for k, v in example.items()} for example in dataset_with_several_columns
]
assert new_dataset.features is None
+ assert new_dataset.column_names is None
# rename the column if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().rename_column("id", "new_id")
assert new_dataset.features is not None
+ assert new_dataset.column_names is not None
+ assert "id" not in new_dataset.column_names
+ assert "new_id" in new_dataset.column_names
def test_iterable_dataset_rename_columns(dataset_with_several_columns):
@@ -986,9 +992,13 @@ def test_iterable_dataset_rename_columns(dataset_with_several_columns):
{column_mapping.get(k, k): v for k, v in example.items()} for example in dataset_with_several_columns
]
assert new_dataset.features is None
+ assert new_dataset.column_names is None
# rename the columns if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().rename_columns(column_mapping)
assert new_dataset.features is not None
+ assert new_dataset.column_names is not None
+ assert all(c not in new_dataset.column_names for c in ["id", "filepath"])
+ assert all(c in new_dataset.column_names for c in ["new_id", "filename"])
def test_iterable_dataset_remove_columns(dataset_with_several_columns):
@@ -1002,10 +1012,13 @@ def test_iterable_dataset_remove_columns(dataset_with_several_columns):
{k: v for k, v in example.items() if k != "id" and k != "filepath"} for example in dataset_with_several_columns
]
assert new_dataset.features is None
+ assert new_dataset.column_names is None
# remove the columns if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().remove_columns(["id", "filepath"])
assert new_dataset.features is not None
+ assert new_dataset.column_names is not None
assert all(c not in new_dataset.features for c in ["id", "filepath"])
+ assert all(c not in new_dataset.column_names for c in ["id", "filepath"])
def test_iterable_dataset_select_columns(dataset_with_several_columns):
@@ -1019,10 +1032,12 @@ def test_iterable_dataset_select_columns(dataset_with_several_columns):
{k: v for k, v in example.items() if k in ("id", "filepath")} for example in dataset_with_several_columns
]
assert new_dataset.features is None
- # remove the columns if ds.features was not None
+ # select the columns if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().select_columns(["id", "filepath"])
assert new_dataset.features is not None
+ assert new_dataset.column_names is not None
assert all(c in new_dataset.features for c in ["id", "filepath"])
+ assert all(c in new_dataset.column_names for c in ["id", "filepath"])
def test_iterable_dataset_cast_column():
@@ -1046,12 +1061,16 @@ def test_iterable_dataset_cast():
def test_iterable_dataset_resolve_features():
ex_iterable = ExamplesIterable(generate_examples_fn, {})
- dataset = IterableDataset(ex_iterable)._resolve_features()
+ dataset = IterableDataset(ex_iterable)
+ assert dataset.features is None
+ assert dataset.column_names is None
+ dataset = dataset._resolve_features()
assert dataset.features == Features(
{
"id": Value("int64"),
}
)
+ assert dataset.column_names == ["id"]
def test_iterable_dataset_resolve_features_keep_order():
@@ -1062,6 +1081,7 @@ def gen():
dataset = IterableDataset(ex_iterable)._resolve_features()
# columns appear in order of appearance in the dataset
assert list(dataset.features) == ["a", "c", "b"]
+ assert dataset.column_names == ["a", "c", "b"]
def test_iterable_dataset_with_features_fill_with_none():
| IterableDataset missing column_names, differs from Dataset interface
### Describe the bug
The documentation on [Stream](https://huggingface.co/docs/datasets/v1.18.2/stream.html) seems to imply that IterableDataset behaves just like a Dataset. However, examples like
```
dataset.map(augment_data, batched=True, remove_columns=dataset.column_names, ...)
```
will not work because `.column_names` does not exist on IterableDataset. I cannot find any clear explanation on why this is not available, is it an oversight? We do have `iterable_ds.features` available.
### Steps to reproduce the bug
See above
### Expected behavior
Dataset and IterableDataset would be expected to have the same interface, with any differences noted in the documentation.
### Environment info
n/a
| Another example is that `IterableDataset.map` does not have `fn_kwargs`, among other arguments. It makes it harder to convert code from Dataset to IterableDataset.
Hi! `fn_kwargs` was added to `IterableDataset.map` in `datasets 2.5.0`, so please update your installation (`pip install -U datasets`) to use it.
Regarding `column_names`, I agree we should add this property to `IterableDataset`. In the meantime, you can use `list(dataset.features.keys())` instead.
Thanks! That's great news.
On Thu, Dec 22, 2022, 07:48 Mario Šaško ***@***.***> wrote:
> Hi! fn_kwargs was added to IterableDataset.map in datasets 2.5.0, so
> please update your installation (pip install -U datasets) to use it.
>
> Regarding column_names, I agree we should add this property to
> IterableDataset. In the meantime, you can use
> list(dataset.features.keys()) instead.
>
> —
> Reply to this email directly, view it on GitHub
> <https://github.com/huggingface/datasets/issues/5383#issuecomment-1362993633>,
> or unsubscribe
> <https://github.com/notifications/unsubscribe-auth/AAHD6N2EQUFEOUFDW3VHSILWORZ45ANCNFSM6AAAAAATGKWVGM>
> .
> You are receiving this because you authored the thread.Message ID:
> ***@***.***>
>
I'm marking this issue as a "good first issue", as it makes sense to have `IterableDataset.column_names` in the API. Besides the case when `features` are `None` (e.g., `features` are `None` after `map`), in which we can also return `column_names` as `None`, adding this property should be straightforward,
Hi @mariosasko, I can work on this if that's ok?
Yes! I've assigned you the issue. | 2023-02-27T10:50:07Z | [] | [] |
huggingface/datasets | 5,646 | huggingface__datasets-5646 | [
"5641"
] | f3d26e74898e0a9dc0d78490104e2e173269ef5b | diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -1559,8 +1559,12 @@ class Features(dict):
- [`~datasets.Translation`] and [`~datasets.TranslationVariableLanguages`], the two features specific to Machine Translation.
"""
- def __init__(self, *args, **kwargs):
- super().__init__(*args, **kwargs)
+ def __init__(*args, **kwargs):
+ # self not in the signature to allow passing self as a kwarg
+ if not args:
+ raise TypeError("descriptor '__init__' of 'Features' object needs an argument")
+ self, *args = args
+ super(Features, self).__init__(*args, **kwargs)
self._column_requires_decoding: Dict[str, bool] = {
col: require_decoding(feature) for col, feature in self.items()
}
| diff --git a/tests/features/test_features.py b/tests/features/test_features.py
--- a/tests/features/test_features.py
+++ b/tests/features/test_features.py
@@ -102,6 +102,13 @@ def test_feature_named_type(self):
reloaded_features = Features.from_dict(asdict(ds_info)["features"])
assert features == reloaded_features
+ def test_feature_named_self_as_kwarg(self):
+ """reference: issue #5641"""
+ features = Features(self=Value("string"))
+ ds_info = DatasetInfo(features=features)
+ reloaded_features = Features.from_dict(asdict(ds_info)["features"])
+ assert features == reloaded_features
+
def test_class_label_feature_with_no_labels(self):
"""reference: issue #4681"""
features = Features({"label": ClassLabel(names=[])})
| Features cannot be named "self"
### Describe the bug
Hi,
I noticed that we cannot create a HuggingFace dataset from Pandas DataFrame with a column named `self`.
The error seems to be coming from arguments validation in the `Features.from_dict` function.
### Steps to reproduce the bug
```python
import datasets
dummy_pandas = pd.DataFrame([0,1,2,3], columns = ["self"])
datasets.arrow_dataset.Dataset.from_pandas(dummy_pandas)
```
### Expected behavior
No error thrown
### Environment info
- `datasets` version: 2.8.0
- Python version: 3.9.5
- PyArrow version: 6.0.1
- Pandas version: 1.4.1
| 2023-03-16T16:17:03Z | [] | [] |
|
huggingface/datasets | 5,714 | huggingface__datasets-5714 | [
"5711"
] | 0803a006db1c395ac715662cc6079651f77c11ea | diff --git a/src/datasets/download/streaming_download_manager.py b/src/datasets/download/streaming_download_manager.py
--- a/src/datasets/download/streaming_download_manager.py
+++ b/src/datasets/download/streaming_download_manager.py
@@ -774,8 +774,7 @@ def xnumpy_load(filepath_or_buffer, *args, use_auth_token: Optional[Union[str, b
return np.load(filepath_or_buffer, *args, **kwargs)
else:
filepath_or_buffer = str(filepath_or_buffer)
- with xopen(filepath_or_buffer, "rb", use_auth_token=use_auth_token) as f:
- return np.load(f, *args, **kwargs)
+ return np.load(xopen(filepath_or_buffer, "rb", use_auth_token=use_auth_token), *args, **kwargs)
def xpandas_read_csv(filepath_or_buffer, use_auth_token: Optional[Union[str, bool]] = None, **kwargs):
| diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -18,6 +18,7 @@
xisfile,
xjoin,
xlistdir,
+ xnumpy_load,
xopen,
xPath,
xrelpath,
@@ -914,3 +915,19 @@ def test_iter_files(data_dir_with_hidden_files):
for num_file, file in enumerate(dl_manager.iter_files(data_dir_with_hidden_files), start=1):
assert os.path.basename(file) == ("test.txt" if num_file == 1 else "train.txt")
assert num_file == 2
+
+
+def test_xnumpy_load(tmp_path):
+ import numpy as np
+
+ expected_x = np.arange(10)
+ npy_path = tmp_path / "data-x.npy"
+ np.save(npy_path, expected_x)
+ x = xnumpy_load(npy_path)
+ assert np.array_equal(x, expected_x)
+
+ npz_path = tmp_path / "data.npz"
+ np.savez(npz_path, x=expected_x)
+ with xnumpy_load(npz_path) as f:
+ x = f["x"]
+ assert np.array_equal(x, expected_x)
| load_dataset in v2.11.0 raises "ValueError: seek of closed file" in np.load()
### Describe the bug
Hi,
I have some `dataset_load()` code of a custom offline dataset that works with datasets v2.10.1.
```python
ds = datasets.load_dataset(path=dataset_dir,
name=configuration,
data_dir=dataset_dir,
cache_dir=cache_dir,
aux_dir=aux_dir,
# download_mode=datasets.DownloadMode.FORCE_REDOWNLOAD,
num_proc=18)
```
When upgrading datasets to 2.11.0, it fails with error
```
Traceback (most recent call last):
File "<string>", line 2, in <module>
File "/home/ramon.casero/opt/miniconda3/envs/myenv/lib/python3.10/site-packages/datasets/load.py", line 1791, in load_dataset
builder_instance.download_and_prepare(
File "/home/ramon.casero/opt/miniconda3/envs/myenv/lib/python3.10/site-packages/datasets/builder.py", line 891, in download_and_prepare
self._download_and_prepare(
File "/home/ramon.casero/opt/miniconda3/envs/myenv/lib/python3.10/site-packages/datasets/builder.py", line 1651, in _download_and_prepare
super()._download_and_prepare(
File "/home/ramon.casero/opt/miniconda3/envs/myenv/lib/python3.10/site-packages/datasets/builder.py", line 964, in _download_and_prepare
split_generators = self._split_generators(dl_manager, **split_generators_kwargs)
File "/home/ramon.casero/.cache/huggingface/modules/datasets_modules/datasets/71f67f69e6e00e139903a121f96b71f39b65a6b6aaeb0862e6a5da3a3f565b4c/mydataset.py", line 682, in _split_generators
self.some_function()
File "/home/ramon.casero/.cache/huggingface/modules/datasets_modules/datasets/71f67f69e6e00e139903a121f96b71f39b65a6b6aaeb0862e6a5da3a3f565b4c/mydataset.py", line 1314, in some_function()
x_df = pd.DataFrame({'cell_type_descriptor': fp['x'].tolist()})
File "/home/ramon.casero/opt/miniconda3/envs/myenv/lib/python3.10/site-packages/numpy/lib/npyio.py", line 248, in __getitem__
bytes = self.zip.open(key)
File "/home/ramon.casero/opt/miniconda3/envs/myenv/lib/python3.10/zipfile.py", line 1530, in open
fheader = zef_file.read(sizeFileHeader)
File "/home/ramon.casero/opt/miniconda3/envs/myenv/lib/python3.10/zipfile.py", line 744, in read
self._file.seek(self._pos)
ValueError: seek of closed file
```
### Steps to reproduce the bug
Sorry, I cannot share the data or code because they are not mine to share, but the point of failure is a call in `some_function()`
```python
with np.load(embedding_filename) as fp:
x_df = pd.DataFrame({'feature': fp['x'].tolist()})
```
I'll try to generate a short snippet that reproduces the error.
### Expected behavior
I would expect that `load_dataset` works on the custom datasets generation script for v2.11.0 the same way it works for 2.10.1, without making `np.load()` give a `ValueError: seek of closed file` error.
### Environment info
- `datasets` version: 2.11.0
- Platform: Linux-4.18.0-483.el8.x86_64-x86_64-with-glibc2.28
- Python version: 3.10.8
- Huggingface_hub version: 0.12.0
- PyArrow version: 11.0.0
- Pandas version: 1.5.2
- numpy: 1.24.2
- This is an offline dataset that uses `datasets.config.HF_DATASETS_OFFLINE = True` in the generation script.
| It seems like https://github.com/huggingface/datasets/pull/5626 has introduced this error.
cc @albertvillanova
I think replacing:
https://github.com/huggingface/datasets/blob/0803a006db1c395ac715662cc6079651f77c11ea/src/datasets/download/streaming_download_manager.py#L777-L778
with:
```python
return np.load(xopen(filepath_or_buffer, "rb", use_auth_token=use_auth_token), *args, **kwargs)
```
should fix the issue.
(Maybe this is also worth doing a patch release afterward)
Thanks for reporting, @rcasero.
I can have a look... | 2023-04-06T13:01:45Z | [] | [] |
huggingface/datasets | 5,729 | huggingface__datasets-5729 | [
"5728"
] | 273392966e434286f4f5ba2ad596730bff11056d | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -38,6 +38,7 @@ class EmptyDatasetError(FileNotFoundError):
KEYWORDS_IN_FILENAME_BASE_PATTERNS = ["**[{sep}/]{keyword}[{sep}]*", "{keyword}[{sep}]*"]
KEYWORDS_IN_DIR_NAME_BASE_PATTERNS = ["{keyword}[{sep}/]**", "**[{sep}/]{keyword}[{sep}/]**"]
+DEFAULT_SPLITS = [Split.TRAIN, Split.TEST, Split.VALIDATION]
DEFAULT_PATTERNS_SPLIT_IN_FILENAME = {
Split.TRAIN: [
pattern.format(keyword=keyword, sep=NON_WORDS_CHARS)
@@ -226,7 +227,10 @@ def _get_data_files_patterns(pattern_resolver: Callable[[str], List[PurePath]])
if len(data_files) > 0:
data_files = [p.as_posix() for p in data_files]
splits: Set[str] = {string_to_dict(p, split_pattern)["split"] for p in data_files}
- return {split: [split_pattern.format(split=split)] for split in splits}
+ sorted_splits = [str(split) for split in DEFAULT_SPLITS if split in splits] + sorted(
+ splits - set(DEFAULT_SPLITS)
+ )
+ return {split: [split_pattern.format(split=split)] for split in sorted_splits}
# then check the default patterns based on train/valid/test splits
for patterns_dict in ALL_DEFAULT_PATTERNS:
non_empty_splits = []
| diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -616,7 +616,7 @@ def resolver(pattern):
return [PurePath(file_path) for file_path in fs.glob(pattern) if fs.isfile(file_path)]
patterns_per_split = _get_data_files_patterns(resolver)
- assert patterns_per_split.keys() == data_file_per_split.keys()
+ assert list(patterns_per_split.keys()) == list(data_file_per_split.keys()) # Test split order with list()
for split, patterns in patterns_per_split.items():
matched = [file_path.as_posix() for pattern in patterns for file_path in resolver(pattern)]
assert matched == data_file_per_split[split]
| The order of data split names is nondeterministic
After this CI error: https://github.com/huggingface/datasets/actions/runs/4639528358/jobs/8210492953?pr=5718
```
FAILED tests/test_data_files.py::test_get_data_files_patterns[data_file_per_split4] - AssertionError: assert ['random', 'train'] == ['train', 'random']
At index 0 diff: 'random' != 'train'
Full diff:
- ['train', 'random']
+ ['random', 'train']
```
I have checked locally and found out that the data split order is nondeterministic.
This is caused by the use of `set` for sharded splits.
| 2023-04-11T07:34:20Z | [] | [] |
|
huggingface/datasets | 5,733 | huggingface__datasets-5733 | [
"5734"
] | 273392966e434286f4f5ba2ad596730bff11056d | diff --git a/setup.py b/setup.py
--- a/setup.py
+++ b/setup.py
@@ -128,7 +128,7 @@
"importlib_metadata;python_version<'3.8'",
# to save datasets locally or on any filesystem
# minimum 2021.11.1 so that BlockSizeError is fixed: see https://github.com/fsspec/filesystem_spec/pull/830
- "fsspec[http]>=2021.11.1,<2023.4.0", # Temporary pin
+ "fsspec[http]>=2021.11.1",
# for data streaming via http
"aiohttp",
# To get datasets from the Datasets Hub on huggingface.co
| diff --git a/tests/fixtures/fsspec.py b/tests/fixtures/fsspec.py
--- a/tests/fixtures/fsspec.py
+++ b/tests/fixtures/fsspec.py
@@ -93,8 +93,8 @@ def _strip_protocol(cls, path):
@pytest.fixture
def mock_fsspec():
original_registry = fsspec.registry.copy()
- fsspec.register_implementation("mock", MockFileSystem)
- fsspec.register_implementation("tmp", TmpDirFileSystem)
+ fsspec.register_implementation("mock", MockFileSystem, clobber=True)
+ fsspec.register_implementation("tmp", TmpDirFileSystem, clobber=True)
yield
fsspec.registry = original_registry
diff --git a/tests/test_streaming_download_manager.py b/tests/test_streaming_download_manager.py
--- a/tests/test_streaming_download_manager.py
+++ b/tests/test_streaming_download_manager.py
@@ -131,7 +131,7 @@ def _open(
@pytest.fixture
def mock_fsspec():
original_registry = fsspec.registry.copy()
- fsspec.register_implementation("mock", DummyTestFS)
+ fsspec.register_implementation("mock", DummyTestFS, clobber=True)
yield
fsspec.registry = original_registry
| Remove temporary pin of fsspec
Once root cause is found and fixed, remove the temporary pin introduced by:
- #5731
| 2023-04-11T08:52:12Z | [] | [] |
|
huggingface/datasets | 5,772 | huggingface__datasets-5772 | [
"5726"
] | 61db0e9c936bc67c18b37b0960e2f0bb1f8ffdcd | diff --git a/src/datasets/packaged_modules/json/json.py b/src/datasets/packaged_modules/json/json.py
--- a/src/datasets/packaged_modules/json/json.py
+++ b/src/datasets/packaged_modules/json/json.py
@@ -84,10 +84,11 @@ def _generate_tables(self, files):
# We accept two format: a list of dicts or a dict of lists
if isinstance(dataset, (list, tuple)):
- mapping = {col: [dataset[i][col] for i in range(len(dataset))] for col in dataset[0].keys()}
+ keys = set().union(*[row.keys() for row in dataset])
+ mapping = {col: [row.get(col) for row in dataset] for col in keys}
else:
mapping = dataset
- pa_table = pa.Table.from_pydict(mapping=mapping)
+ pa_table = pa.Table.from_pydict(mapping)
yield file_idx, self._cast_table(pa_table)
# If the file has one json object per line
@@ -137,7 +138,9 @@ def _generate_tables(self, files):
# If possible, parse the file as a list of json objects and exit the loop
if isinstance(dataset, list): # list is the only sequence type supported in JSON
try:
- pa_table = pa.Table.from_pylist(dataset)
+ keys = set().union(*[row.keys() for row in dataset])
+ mapping = {col: [row.get(col) for row in dataset] for col in keys}
+ pa_table = pa.Table.from_pydict(mapping)
except (pa.ArrowInvalid, AttributeError) as e:
logger.error(f"Failed to read file '{file}' with error {type(e)}: {e}")
raise ValueError(f"Not able to read records in the JSON file at {file}.") from None
| diff --git a/tests/packaged_modules/test_json.py b/tests/packaged_modules/test_json.py
--- a/tests/packaged_modules/test_json.py
+++ b/tests/packaged_modules/test_json.py
@@ -12,6 +12,7 @@ def jsonl_file(tmp_path):
filename = tmp_path / "file.jsonl"
data = textwrap.dedent(
"""\
+ {"col_1": -1}
{"col_1": 1, "col_2": 2}
{"col_1": 10, "col_2": 20}
"""
@@ -27,6 +28,7 @@ def json_file_with_list_of_dicts(tmp_path):
data = textwrap.dedent(
"""\
[
+ {"col_1": -1},
{"col_1": 1, "col_2": 2},
{"col_1": 10, "col_2": 20}
]
@@ -46,6 +48,7 @@ def json_file_with_list_of_dicts_field(tmp_path):
"field1": 1,
"field2": "aabb",
"field3": [
+ {"col_1": -1},
{"col_1": 1, "col_2": 2},
{"col_1": 10, "col_2": 20}
]
@@ -69,7 +72,7 @@ def test_json_generate_tables(file_fixture, config_kwargs, request):
json = Json(**config_kwargs)
generator = json._generate_tables([[request.getfixturevalue(file_fixture)]])
pa_table = pa.concat_tables([table for _, table in generator])
- assert pa_table.to_pydict() == {"col_1": [1, 10], "col_2": [2, 20]}
+ assert pa_table.to_pydict() == {"col_1": [-1, 1, 10], "col_2": [None, 2, 20]}
@pytest.mark.parametrize(
@@ -98,4 +101,4 @@ def test_json_generate_tables_with_missing_features(file_fixture, config_kwargs,
json = Json(**config_kwargs)
generator = json._generate_tables([[request.getfixturevalue(file_fixture)]])
pa_table = pa.concat_tables([table for _, table in generator])
- assert pa_table.to_pydict() == {"col_1": [1, 10], "col_2": [2, 20], "missing_col": [None, None]}
+ assert pa_table.to_pydict() == {"col_1": [-1, 1, 10], "col_2": [None, 2, 20], "missing_col": [None, None, None]}
| Fallback JSON Dataset loading does not load all values when features specified manually
### Describe the bug
The fallback JSON dataset loader located here:
https://github.com/huggingface/datasets/blob/1c4ec00511868bd881e84a6f7e0333648d833b8e/src/datasets/packaged_modules/json/json.py#L130-L153
does not load the values of features correctly when features are specified manually and not all features have a value in the first entry of the dataset. I'm pretty sure this is not supposed to be expected bahavior?
To fix this you'd have to change this line:
https://github.com/huggingface/datasets/blob/1c4ec00511868bd881e84a6f7e0333648d833b8e/src/datasets/packaged_modules/json/json.py#L140
To pass a schema to pyarrow which has the same structure as the features argument passed to the load_dataset() method.
### Steps to reproduce the bug
Consider a dataset JSON like this:
```
[
{
"instruction": "Do stuff",
"output": "Answer stuff"
},
{
"instruction": "Do stuff2",
"input": "Additional Input2",
"output": "Answer stuff2"
}
]
```
Using this code to load the dataset:
```
from datasets import load_dataset, Features, Value
features = {
"instruction": Value("string"),
"input": Value("string"),
"output": Value("string")
}
features = Features(features)
ds = load_dataset("json", data_files="./ds.json", features=features)
for row in ds["train"]:
print(row)
```
we get a dataset that looks like this:
| **Instruction** | **Input** | **Output** |
|-----------------|--------------------|-----------------|
| "Do stuff" | None | "Answer Stuff" |
| "Do stuff2" | None | "Answer Stuff2" |
### Expected behavior
The input column should contain values other than None for dataset entries that have the "input" attribute set:
| **Instruction** | **Input** | **Output** |
|-----------------|--------------------|-----------------|
| "Do stuff" | None | "Answer Stuff" |
| "Do stuff2" | "Additional Input2" | "Answer Stuff2" |
### Environment info
Python 3.10.10
Datasets 2.11.0
Windows 10
| 2023-04-19T14:32:57Z | [] | [] |
|
huggingface/datasets | 5,787 | huggingface__datasets-5787 | [
"5785"
] | 649d5a3315f9e7666713b6affe318ee00c7163a0 | diff --git a/src/datasets/load.py b/src/datasets/load.py
--- a/src/datasets/load.py
+++ b/src/datasets/load.py
@@ -368,6 +368,7 @@ def infer_module_for_data_files(
return _EXTENSION_TO_MODULE[ext]
elif ext == "zip":
return infer_module_for_data_files_in_archives(data_files_list, use_auth_token=use_auth_token)
+ return None, {}
def infer_module_for_data_files_in_archives(
@@ -404,6 +405,7 @@ def infer_module_for_data_files_in_archives(
most_common = extensions_counter.most_common(1)[0][0]
if most_common in _EXTENSION_TO_MODULE:
return _EXTENSION_TO_MODULE[most_common]
+ return None, {}
@dataclass
@@ -632,14 +634,14 @@ def get_module(self) -> DatasetModule:
base_path=base_path,
allowed_extensions=ALL_ALLOWED_EXTENSIONS,
)
- module_names = {
- key: infer_module_for_data_files(data_files_list) for key, data_files_list in data_files.items()
+ split_modules = {
+ split: infer_module_for_data_files(data_files_list) for split, data_files_list in data_files.items()
}
- if len(set(list(zip(*module_names.values()))[0])) > 1:
- raise ValueError(f"Couldn't infer the same data file format for all splits. Got {module_names}")
- module_name, builder_kwargs = next(iter(module_names.values()))
+ module_name, builder_kwargs = next(iter(split_modules.values()))
+ if any((module_name, builder_kwargs) != split_module for split_module in split_modules.values()):
+ raise ValueError(f"Couldn't infer the same data file format for all splits. Got {split_modules}")
if not module_name:
- raise FileNotFoundError(f"No data files or dataset script found in {self.path}")
+ raise FileNotFoundError(f"No (supported) data files or dataset script found in {self.path}")
# Collect metadata files if the module supports them
if self.data_files is None and module_name in _MODULE_SUPPORTS_METADATA and patterns != DEFAULT_PATTERNS_ALL:
try:
@@ -772,15 +774,15 @@ def get_module(self) -> DatasetModule:
base_path=self.data_dir,
allowed_extensions=ALL_ALLOWED_EXTENSIONS,
)
- module_names = {
- key: infer_module_for_data_files(data_files_list, use_auth_token=self.download_config.use_auth_token)
- for key, data_files_list in data_files.items()
+ split_modules = {
+ split: infer_module_for_data_files(data_files_list, use_auth_token=self.download_config.use_auth_token)
+ for split, data_files_list in data_files.items()
}
- if len(set(list(zip(*module_names.values()))[0])) > 1:
- raise ValueError(f"Couldn't infer the same data file format for all splits. Got {module_names}")
- module_name, builder_kwargs = next(iter(module_names.values()))
+ module_name, builder_kwargs = next(iter(split_modules.values()))
+ if any((module_name, builder_kwargs) != split_module for split_module in split_modules.values()):
+ raise ValueError(f"Couldn't infer the same data file format for all splits. Got {split_modules}")
if not module_name:
- raise FileNotFoundError(f"No data files or dataset script found in {self.name}")
+ raise FileNotFoundError(f"No (supported) data files or dataset script found in {self.name}")
# Collect metadata files if the module supports them
if self.data_files is None and module_name in _MODULE_SUPPORTS_METADATA and patterns != DEFAULT_PATTERNS_ALL:
try:
| diff --git a/tests/fixtures/files.py b/tests/fixtures/files.py
--- a/tests/fixtures/files.py
+++ b/tests/fixtures/files.py
@@ -480,6 +480,15 @@ def zip_text_with_dir_path(text_path, text2_path, tmp_path_factory):
return path
[email protected](scope="session")
+def zip_unsupported_ext_path(text_path, text2_path, tmp_path_factory):
+ path = tmp_path_factory.mktemp("data") / "dataset.ext.zip"
+ with zipfile.ZipFile(path, "w") as f:
+ f.write(text_path, arcname=os.path.basename("unsupported.ext"))
+ f.write(text2_path, arcname=os.path.basename("unsupported_2.ext"))
+ return path
+
+
@pytest.fixture(scope="session")
def text_path_with_unicode_new_lines(tmp_path_factory):
text = "\n".join(["First", "Second\u2029with Unicode new line", "Third"])
diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -209,6 +209,8 @@ def metric_loading_script_dir(tmp_path):
(["train.jsonl"], "json", {}),
(["train.parquet"], "parquet", {}),
(["train.txt"], "text", {}),
+ (["unsupported.ext"], None, {}),
+ ([""], None, {}),
],
)
def test_infer_module_for_data_files(data_files, expected_module, expected_builder_kwargs):
@@ -217,9 +219,18 @@ def test_infer_module_for_data_files(data_files, expected_module, expected_build
assert builder_kwargs == expected_builder_kwargs
[email protected]("data_file, expected_module", [("zip_csv_path", "csv"), ("zip_csv_with_dir_path", "csv")])
-def test_infer_module_for_data_files_in_archives(data_file, expected_module, zip_csv_path, zip_csv_with_dir_path):
- data_file_paths = {"zip_csv_path": zip_csv_path, "zip_csv_with_dir_path": zip_csv_with_dir_path}
[email protected](
+ "data_file, expected_module",
+ [("zip_csv_path", "csv"), ("zip_csv_with_dir_path", "csv"), ("zip_unsupported_ext_path", None)],
+)
+def test_infer_module_for_data_files_in_archives(
+ data_file, expected_module, zip_csv_path, zip_csv_with_dir_path, zip_unsupported_ext_path
+):
+ data_file_paths = {
+ "zip_csv_path": zip_csv_path,
+ "zip_csv_with_dir_path": zip_csv_with_dir_path,
+ "zip_unsupported_ext_path": zip_unsupported_ext_path,
+ }
data_files = [str(data_file_paths[data_file])]
inferred_module, _ = infer_module_for_data_files_in_archives(data_files, False)
assert inferred_module == expected_module
| Unsupported data files raise TypeError: 'NoneType' object is not iterable
Currently, we raise a TypeError for unsupported data files:
```
TypeError: 'NoneType' object is not iterable
```
See:
- https://github.com/huggingface/datasets-server/issues/1073
We should give a more informative error message.
| 2023-04-24T10:44:50Z | [] | [] |
|
huggingface/datasets | 5,859 | huggingface__datasets-5859 | [
"5858"
] | db56f7f0d2f0b99af4da17d388c205152504c7d9 | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -115,7 +115,7 @@
from .utils.py_utils import Literal, asdict, convert_file_size_to_int, iflatmap_unordered, unique_values
from .utils.stratify import stratified_shuffle_split_generate_indices
from .utils.tf_utils import dataset_to_tf, minimal_tf_collate_fn, multiprocess_dataset_to_tf
-from .utils.typing import PathLike
+from .utils.typing import ListLike, PathLike
if TYPE_CHECKING:
@@ -2742,10 +2742,12 @@ def prepare_for_task(self, task: Union[str, TaskTemplate], id: int = 0) -> "Data
dataset = dataset.cast(features=template.features)
return dataset
- def _getitem(self, key: Union[int, slice, str], **kwargs) -> Union[Dict, List]:
+ def _getitem(self, key: Union[int, slice, str, ListLike[int]], **kwargs) -> Union[Dict, List]:
"""
- Can be used to index columns (by string names) or rows (by integer index, slices, or iter of indices or bools)
+ Can be used to index columns (by string names) or rows (by integer, slice, or list-like of integer indices)
"""
+ if isinstance(key, bool):
+ raise TypeError("dataset index must be int, str, slice or collection of int, not bool")
format_type = kwargs["format_type"] if "format_type" in kwargs else self._format_type
format_columns = kwargs["format_columns"] if "format_columns" in kwargs else self._format_columns
output_all_columns = (
diff --git a/src/datasets/utils/typing.py b/src/datasets/utils/typing.py
--- a/src/datasets/utils/typing.py
+++ b/src/datasets/utils/typing.py
@@ -1,8 +1,9 @@
import os
-from typing import Dict, List, TypeVar, Union
+from typing import Dict, List, Tuple, TypeVar, Union
T = TypeVar("T")
+ListLike = Union[List[T], Tuple[T, ...]]
NestedDataStructureLike = Union[T, List[T], Dict[str, T]]
PathLike = Union[str, bytes, os.PathLike]
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -209,7 +209,9 @@ def test_dataset_getitem(self, in_memory):
self.assertEqual(dset["filename"][:-1][-1], "my_name-train_28")
self.assertListEqual(dset[[0, -1]]["filename"], ["my_name-train_0", "my_name-train_29"])
+ self.assertListEqual(dset[range(0, -2, -1)]["filename"], ["my_name-train_0", "my_name-train_29"])
self.assertListEqual(dset[np.array([0, -1])]["filename"], ["my_name-train_0", "my_name-train_29"])
+ self.assertListEqual(dset[pd.Series([0, -1])]["filename"], ["my_name-train_0", "my_name-train_29"])
with dset.select(range(2)) as dset_subset:
self.assertListEqual(dset_subset[-1:]["filename"], ["my_name-train_1"])
@@ -4520,3 +4522,11 @@ def f(x):
ds = ds.map(f)
outputs = ds[:]
assert outputs == {"a": [{"nested": [[i]]} for i in [-1, -1, 2, 3]]}
+
+
+def test_dataset_getitem_raises():
+ ds = Dataset.from_dict({"a": [0, 1, 2, 3]})
+ with pytest.raises(TypeError):
+ ds[False]
+ with pytest.raises(TypeError):
+ ds._getitem(True)
| Throw an error when dataset improperly indexed
### Describe the bug
Pandas-style subset indexing on dataset does not throw an error, when maybe it should. Instead returns the first instance of the dataset regardless of index condition.
### Steps to reproduce the bug
Steps to reproduce the behavior:
1. `squad = datasets.load_dataset("squad_v2", split="validation")`
2. `item = squad[squad['question'] == "Who was the Norse leader?"]`
or `it = squad[squad['id'] == '56ddde6b9a695914005b962b']`
3. returns the first item in the dataset, which does not satisfy the above conditions:
`{'id': '56ddde6b9a695914005b9628', 'title': 'Normans', 'context': 'The Normans (Norman: Nourmands; French: Normands; Latin: Normanni) were the people who in the 10th and 11th centuries gave their name to Normandy, a region in France. They were descended from Norse ("Norman" comes from "Norseman") raiders and pirates from Denmark, Iceland and Norway who, under their leader Rollo, agreed to swear fealty to King Charles III of West Francia. Through generations of assimilation and mixing with the native Frankish and Roman-Gaulish populations, their descendants would gradually merge with the Carolingian-based cultures of West Francia. The distinct cultural and ethnic identity of the Normans emerged initially in the first half of the 10th century, and it continued to evolve over the succeeding centuries.', 'question': 'In what country is Normandy located?', 'answers': {'text': ['France', 'France', 'France', 'France'], 'answer_start': [159, 159, 159, 159]}}`
### Expected behavior
Should either throw an error message, or return the dataset item that satisfies the condition.
### Environment info
- `datasets` version: 2.9.0
- Platform: macOS-13.3.1-arm64-arm-64bit
- Python version: 3.10.8
- PyArrow version: 10.0.1
- Pandas version: 1.5.3
| Thanks for reporting, @sarahwie.
Please note that in `datasets` we do not have vectorized operation like `pandas`. Therefore, your equality comparisons above are `False`:
- For example: `squad['question']` returns a `list`, and this list is not equal to `"Who was the Norse leader?"`
The `False` value is equivalent to `0` when indexing a dataset, thus the reason why you get the first element (with index 0):
- For example: `squad[False]` is equivalent to `squad[0]`
Maybe we should an exception instead of assuming that `False` is equivalent to `0` (and `True` is equivalent to `1`) in the context of indexing. | 2023-05-15T08:08:42Z | [] | [] |
huggingface/datasets | 5,863 | huggingface__datasets-5863 | [
"5855"
] | a129219a48c1b07c06d4bc1db32c317bf513089d | diff --git a/src/datasets/utils/tf_utils.py b/src/datasets/utils/tf_utils.py
--- a/src/datasets/utils/tf_utils.py
+++ b/src/datasets/utils/tf_utils.py
@@ -15,6 +15,7 @@
"""TF-specific utils import."""
import os
+import warnings
from functools import partial
from math import ceil
from uuid import uuid4
@@ -173,6 +174,21 @@ def dataset_to_tf(
else:
raise ImportError("Called a Tensorflow-specific function but Tensorflow is not installed.")
+ # TODO Matt: When our minimum Python version is 3.8 or higher, we can delete all of this and move everything
+ # to the NumPy multiprocessing path.
+ if hasattr(tf, "random_index_shuffle"):
+ random_index_shuffle = tf.random_index_shuffle
+ elif hasattr(tf.random.experimental, "index_shuffle"):
+ random_index_shuffle = tf.random.experimental.index_shuffle
+ else:
+ if len(dataset) > 10_000_000:
+ warnings.warn(
+ "to_tf_dataset() can be memory-inefficient on versions of TensorFlow older than 2.9. "
+ "If you are iterating over a dataset with a very large number of samples, consider "
+ "upgrading to TF >= 2.9."
+ )
+ random_index_shuffle = None
+
getter_fn = partial(
np_get_batch,
dataset=dataset,
@@ -195,10 +211,22 @@ def fetch_function(indices):
)
return {key: output[i] for i, key in enumerate(columns_to_np_types.keys())}
- tf_dataset = tf.data.Dataset.from_tensor_slices(np.arange(len(dataset), dtype=np.int64))
+ tf_dataset = tf.data.Dataset.range(len(dataset))
+
+ if shuffle and random_index_shuffle is not None:
+ base_seed = tf.fill((3,), value=tf.cast(-1, dtype=tf.int64))
+
+ def scan_random_index(state, index):
+ if tf.reduce_all(state == -1):
+ # This generates a new random seed once per epoch only,
+ # to ensure that we iterate over each sample exactly once per epoch
+ state = tf.random.uniform(shape=(3,), maxval=2**62, dtype=tf.int64)
+ shuffled_index = random_index_shuffle(index=index, seed=state, max_index=len(dataset) - 1)
+ return state, shuffled_index
- if shuffle:
- tf_dataset = tf_dataset.shuffle(len(dataset))
+ tf_dataset = tf_dataset.scan(base_seed, scan_random_index)
+ elif shuffle:
+ tf_dataset = tf_dataset.shuffle(tf_dataset.cardinality())
if batch_size is not None:
tf_dataset = tf_dataset.batch(batch_size, drop_remainder=drop_remainder)
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -2755,6 +2755,8 @@ def test_tf_index_reshuffling(self, in_memory):
second_indices.append(batch["col_1"])
second_indices = np.concatenate([arr.numpy() for arr in second_indices])
self.assertFalse(np.array_equal(indices, second_indices))
+ self.assertEqual(len(indices), len(np.unique(indices)))
+ self.assertEqual(len(second_indices), len(np.unique(second_indices)))
tf_dataset = dset.to_tf_dataset(batch_size=1, shuffle=False, num_workers=num_workers)
for i, batch in enumerate(tf_dataset):
| `to_tf_dataset` consumes too much memory
### Describe the bug
Hi, I'm using `to_tf_dataset` to convert a _large_ dataset to `tf.data.Dataset`. I observed that the data loading *before* training took a lot of time and memory, even with `batch_size=1`.
After some digging, i believe the reason lies in the shuffle behavior. The [source code](https://github.com/huggingface/datasets/blob/main/src/datasets/utils/tf_utils.py#L185) uses `len(dataset)` as the `buffer_size`, which may load all the data into the memory, and the [tf.data doc](https://www.tensorflow.org/guide/data#randomly_shuffling_input_data) also states that "While large buffer_sizes shuffle more thoroughly, they can take a lot of memory, and significant time to fill".
### Steps to reproduce the bug
```python
from datasets import Dataset
def gen(): # some large data
for i in range(50000000):
yield {"data": i}
ds = Dataset.from_generator(gen, cache_dir="./huggingface")
tf_ds = ds.to_tf_dataset(
batch_size=64,
shuffle=False, # no shuffle
drop_remainder=False,
prefetch=True,
)
# fast and memory friendly 🤗
for batch in tf_ds:
...
tf_ds_shuffle = ds.to_tf_dataset(
batch_size=64,
shuffle=True,
drop_remainder=False,
prefetch=True,
)
# slow and memory hungry for simple iteration 😱
for batch in tf_ds_shuffle:
...
```
### Expected behavior
Shuffling should not load all the data into the memory. Would adding a `buffer_size` parameter in the `to_tf_dataset` API alleviate the problem?
### Environment info
- `datasets` version: 2.11.0
- Platform: Linux-5.17.1-051701-generic-x86_64-with-glibc2.17
- Python version: 3.8.13
- Huggingface_hub version: 0.13.4
- PyArrow version: 11.0.0
- Pandas version: 1.4.3
| Cc @amyeroberts @Rocketknight1
Indded I think it's because it does something like this under the hood when there's no multiprocessing:
```python
tf_dataset = tf_dataset.shuffle(len(dataset))
```
PS: with multiprocessing it appears to be different:
```python
indices = np.arange(len(dataset))
if shuffle:
np.random.shuffle(indices)
```
Hi @massquantity, the dataset being shuffled there is not the full dataset. If you look at [the line above](https://github.com/huggingface/datasets/blob/main/src/datasets/utils/tf_utils.py#L182), the dataset is actually just a single indices array at that point, and that array is the only thing that gets fully loaded into memory and shuffled. We then load samples from the dataset by applying a transform function to the shuffled dataset, which fetches samples based on the indices it receives.
If your dataset is **really** gigantic, then this index tensor might be a memory issue, but since it's just an int64 tensor it will only use 1GB of memory per 125 million samples.
Still, if you're encountering memory issues, there might be another cause here - can you share some code to reproduce the error, or does it depend on some internal/proprietary dataset?
Hi @Rocketknight1, you're right and I also noticed that only indices are used in shuffling. My data has shape (50000000, 10), but really the problem doesn't relate to a specific dataset. Simply running the following code costs me 10GB of memory.
```python
from datasets import Dataset
def gen():
for i in range(50000000):
yield {"data": i}
ds = Dataset.from_generator(gen, cache_dir="./huggingface")
tf_ds = ds.to_tf_dataset(
batch_size=1,
shuffle=True,
drop_remainder=False,
prefetch=True,
)
tf_ds = iter(tf_ds)
next(tf_ds)
# {'data': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([0])>}
```
I just realized maybe it was an issue from tensorflow (I'm using tf 2.12). So I tried the following code, and it used 10GB of memory too.
```python
import numpy as np
import tensorflow as tf
data_size = 50000000
tf_dataset = tf.data.Dataset.from_tensor_slices(np.arange(data_size))
tf_dataset = iter(tf_dataset.shuffle(data_size))
next(tf_dataset)
# <tf.Tensor: shape=(), dtype=int64, numpy=24774043>
```
By the way, as @lhoestq mentioned, multiprocessing uses numpy shuffling, and it uses less than 1 GB of memory:
```python
tf_ds_mp = ds.to_tf_dataset(
batch_size=1,
shuffle=True,
drop_remainder=False,
prefetch=True,
num_workers=2,
)
```
Thanks for that reproduction script - I've confirmed the same issue is occurring for me. Investigating it now!
Update: The memory usage is occurring in creation of the index and shuffle buffer. You can reproduce it very simply with:
```python
import tensorflow as tf
indices = tf.range(50_000_000, dtype=tf.int64)
dataset = tf.data.Dataset.from_tensor_slices(indices)
dataset = dataset.shuffle(len(dataset))
print(next(iter(dataset))
```
When I wrote this code I thought `tf.data` had an optimization for shuffling an entire tensor that wouldn't create the entire shuffle buffer, but evidently it's just creating the enormous buffer in memory. I'll see if I can find a more efficient way to do this - we might end up moving everything to the `numpy` multiprocessing path to avoid it. | 2023-05-15T15:28:34Z | [] | [] |
huggingface/datasets | 5,891 | huggingface__datasets-5891 | [
"1774",
"5875"
] | 5d9dfa9a8c077c783729a279623926faa9e2f3f1 | diff --git a/src/datasets/arrow_reader.py b/src/datasets/arrow_reader.py
--- a/src/datasets/arrow_reader.py
+++ b/src/datasets/arrow_reader.py
@@ -143,8 +143,11 @@ def make_file_instructions(
to = split_length if abs_instr.to is None else abs_instr.to
if shard_lengths is None: # not sharded
for filename in filenames:
- num_examples += to - from_
- file_instructions.append({"filename": filename, "skip": from_, "take": to - from_})
+ take = to - from_
+ if take == 0:
+ continue
+ num_examples += take
+ file_instructions.append({"filename": filename, "skip": from_, "take": take})
else: # sharded
index_start = 0 # Beginning (included) of moving window.
index_end = 0 # End (excluded) of moving window.
@@ -480,17 +483,12 @@ def _rel_to_abs_instr(rel_instr, name2len):
else:
from_ = 0 if from_ is None else from_
to = num_examples if to is None else to
- if abs(from_) > num_examples or abs(to) > num_examples:
- msg = f'Requested slice [{from_ or ""}:{to or ""}] incompatible with {num_examples} examples.'
- raise ValueError(msg)
if from_ < 0:
- from_ = num_examples + from_
- elif from_ == 0:
- from_ = None
+ from_ = max(num_examples + from_, 0)
if to < 0:
- to = num_examples + to
- elif to == num_examples:
- to = None
+ to = max(num_examples + to, 0)
+ from_ = min(from_, num_examples)
+ to = min(to, num_examples)
return _AbsoluteInstruction(split, from_, to)
| diff --git a/tests/test_arrow_reader.py b/tests/test_arrow_reader.py
--- a/tests/test_arrow_reader.py
+++ b/tests/test_arrow_reader.py
@@ -158,7 +158,7 @@ def test_read_instruction_spec():
assert ReadInstruction.from_spec(spec_train_test_pct_rounding).to_spec() == spec_train_test_pct_rounding
-def test_make_file_instructions():
+def test_make_file_instructions_basic():
name = "dummy"
split_infos = [SplitInfo(name="train", num_examples=100)]
instruction = "train[:33%]"
@@ -184,6 +184,81 @@ def test_make_file_instructions():
]
[email protected](
+ "split_name, instruction, shard_lengths, read_range",
+ [
+ ("train", "train[-20%:]", 100, (80, 100)),
+ ("train", "train[:200]", 100, (0, 100)),
+ ("train", "train[:-200]", 100, None),
+ ("train", "train[-200:]", 100, (0, 100)),
+ ("train", "train[-20%:]", [10] * 10, (80, 100)),
+ ("train", "train[:200]", [10] * 10, (0, 100)),
+ ("train", "train[:-200]", [10] * 10, None),
+ ("train", "train[-200:]", [10] * 10, (0, 100)),
+ ],
+)
+def test_make_file_instructions(split_name, instruction, shard_lengths, read_range):
+ name = "dummy"
+ split_infos = split_infos = [
+ SplitInfo(
+ name="train",
+ num_examples=shard_lengths if not isinstance(shard_lengths, list) else sum(shard_lengths),
+ shard_lengths=shard_lengths if isinstance(shard_lengths, list) else None,
+ )
+ ]
+ filetype_suffix = "arrow"
+ prefix_path = "prefix"
+ file_instructions = make_file_instructions(name, split_infos, instruction, filetype_suffix, prefix_path)
+ assert isinstance(file_instructions, FileInstructions)
+ assert file_instructions.num_examples == (read_range[1] - read_range[0] if read_range is not None else 0)
+ if read_range is None:
+ assert file_instructions.file_instructions == []
+ else:
+ if not isinstance(shard_lengths, list):
+ assert file_instructions.file_instructions == [
+ {
+ "filename": os.path.join(prefix_path, f"{name}-{split_name}.arrow"),
+ "skip": read_range[0],
+ "take": read_range[1] - read_range[0],
+ }
+ ]
+ else:
+ file_instructions_list = []
+ shard_offset = 0
+ for i, shard_length in enumerate(shard_lengths):
+ filename = os.path.join(prefix_path, f"{name}-{split_name}-{i:05d}-of-{len(shard_lengths):05d}.arrow")
+ if shard_offset <= read_range[0] < shard_offset + shard_length:
+ file_instructions_list.append(
+ {
+ "filename": filename,
+ "skip": read_range[0] - shard_offset,
+ "take": read_range[1] - read_range[0]
+ if read_range[1] < shard_offset + shard_length
+ else -1,
+ }
+ )
+ elif shard_offset < read_range[1] <= shard_offset + shard_length:
+ file_instructions_list.append(
+ {
+ "filename": filename,
+ "skip": 0,
+ "take": read_range[1] - shard_offset
+ if read_range[1] < shard_offset + shard_length
+ else -1,
+ }
+ )
+ elif read_range[0] < shard_offset and read_range[1] > shard_offset + shard_length:
+ file_instructions_list.append(
+ {
+ "filename": filename,
+ "skip": 0,
+ "take": -1,
+ }
+ )
+ shard_offset += shard_length
+ assert file_instructions.file_instructions == file_instructions_list
+
+
@pytest.mark.parametrize("name, expected_exception", [(None, TypeError), ("", ValueError)])
def test_make_file_instructions_raises(name, expected_exception):
split_infos = [SplitInfo(name="train", num_examples=100)]
| is it possible to make slice to be more compatible like python list and numpy?
Hi,
see below error:
```
AssertionError: Requested slice [:10000000000000000] incompatible with 20 examples.
```
Why split slicing doesn't behave like list slicing ?
### Describe the bug
If I want to get the first 10 samples of my dataset, I can do :
```
ds = datasets.load_dataset('mnist', split='train[:10]')
```
But if I exceed the number of samples in the dataset, an exception is raised :
```
ds = datasets.load_dataset('mnist', split='train[:999999999]')
```
> ValueError: Requested slice [:999999999] incompatible with 60000 examples.
### Steps to reproduce the bug
```
ds = datasets.load_dataset('mnist', split='train[:999999999]')
```
### Expected behavior
I would expect it to behave like python lists (no exception raised, the whole list is kept) :
```
d = list(range(1000))[:999999]
print(len(d)) # > 1000
```
### Environment info
- `datasets` version: 2.9.0
- Platform: macOS-12.6-arm64-arm-64bit
- Python version: 3.9.12
- PyArrow version: 11.0.0
- Pandas version: 1.5.3
| Hi ! Thanks for reporting.
I am working on changes in the way data are sliced from arrow. I can probably fix your issue with the changes I'm doing.
If you have some code to reproduce the issue it would be nice so I can make sure that this case will be supported :)
I'll make a PR in a few days
Good if you can take care at your side.
Here is the [colab notebook](https://colab.research.google.com/drive/19c-abm87RTRYgW9G1D8ktfwRW95zDYBZ?usp=sharing)
A duplicate of https://github.com/huggingface/datasets/issues/1774 | 2023-05-23T16:04:33Z | [] | [] |
huggingface/datasets | 5,894 | huggingface__datasets-5894 | [
"5876"
] | 22d1d533e8ab831b1aa1aab3e7d3c72ba42a83e8 | diff --git a/src/datasets/filesystems/__init__.py b/src/datasets/filesystems/__init__.py
--- a/src/datasets/filesystems/__init__.py
+++ b/src/datasets/filesystems/__init__.py
@@ -1,6 +1,7 @@
import importlib
import shutil
import threading
+import warnings
from typing import List
import fsspec
@@ -25,7 +26,9 @@
# Register custom filesystems
for fs_class in COMPRESSION_FILESYSTEMS + [HfFileSystem]:
- fsspec.register_implementation(fs_class.protocol, fs_class)
+ if fs_class.protocol in fsspec.registry and fsspec.registry[fs_class.protocol] is not fs_class:
+ warnings.warn(f"A filesystem protocol was already set for {fs_class.protocol} and will be overwritten.")
+ fsspec.register_implementation(fs_class.protocol, fs_class, clobber=True)
def extract_path_from_uri(dataset_path: str) -> str:
| diff --git a/tests/test_filesystem.py b/tests/test_filesystem.py
--- a/tests/test_filesystem.py
+++ b/tests/test_filesystem.py
@@ -1,7 +1,9 @@
+import importlib
import os
import fsspec
import pytest
+from fsspec import register_implementation
from fsspec.registry import _registry as _fsspec_registry
from datasets.filesystems import COMPRESSION_FILESYSTEMS, HfFileSystem, extract_path_from_uri, is_remote_filesystem
@@ -80,3 +82,21 @@ def test_hf_filesystem(hf_token, hf_api, hf_private_dataset_repo_txt_data, text_
assert hffs.isfile(".gitattributes") and hffs.isfile("data/text_data.txt")
with open(text_file) as f:
assert hffs.open("data/text_data.txt", "r").read() == f.read()
+
+
+def test_fs_overwrites():
+ protocol = "bz2"
+
+ # Import module
+ import datasets.filesystems
+
+ # Overwrite protocol and reload
+ register_implementation(protocol, None, clobber=True)
+ with pytest.warns(UserWarning) as warning_info:
+ importlib.reload(datasets.filesystems)
+
+ assert len(warning_info) == 1
+ assert (
+ str(warning_info[0].message)
+ == f"A filesystem protocol was already set for {protocol} and will be overwritten."
+ )
| Incompatibility with DataLab
### Describe the bug
Hello,
I am currently working on a project where both [DataLab](https://github.com/ExpressAI/DataLab) and [datasets](https://github.com/huggingface/datasets) are subdependencies.
I noticed that I cannot import both libraries, as they both register FileSystems in `fsspec`, expecting the FileSystems not being registered before.
When running the code below, I get the following error:
```
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\datalabs\__init__.py", line 28, in <module>
from datalabs.arrow_dataset import concatenate_datasets, Dataset
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\datalabs\arrow_dataset.py", line 60, in <module>
from datalabs.arrow_writer import ArrowWriter, OptimizedTypedSequence
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\datalabs\arrow_writer.py", line 28, in <module>
from datalabs.features import (
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\datalabs\features\__init__.py", line 2, in <module>
from datalabs.features.audio import Audio
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\datalabs\features\audio.py", line 21, in <module>
from datalabs.utils.streaming_download_manager import xopen
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\datalabs\utils\streaming_download_manager.py", line 16, in <module>
from datalabs.filesystems import COMPRESSION_FILESYSTEMS
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\datalabs\filesystems\__init__.py", line 37, in <module>
fsspec.register_implementation(fs_class.protocol, fs_class)
File "C:\Users\Bened\anaconda3\envs\ner-eval-dashboard2\lib\site-packages\fsspec\registry.py", line 51, in register_implementation
raise ValueError(
ValueError: Name (bz2) already in the registry and clobber is False
```
I think as simple solution would be to just set `clobber=True` in https://github.com/huggingface/datasets/blob/main/src/datasets/filesystems/__init__.py#L28. This allows the register to discard previous registrations. This should work, as the datalabs FileSystems are copies of the datasets FileSystems. However, I don't know if it is guaranteed to be compatible with other libraries that might use the same protocols.
I am linking the symmetric issue on [DataLab](https://github.com/ExpressAI/DataLab/issues/425) as ideally the issue is solved in both libraries the same way. Otherwise, it could lead to different behaviors depending on which library gets imported first.
### Steps to reproduce the bug
1. Run `pip install datalabs==0.4.15 datasets==2.12.0`
2. Run the following python code:
```
import datalabs
import datasets
```
### Expected behavior
It should be possible to import both libraries without getting a Value Error
### Environment info
datalabs==0.4.15
datasets==2.12.0
| Indeed, `clobber=True` (with a warning if the existing protocol will be overwritten) should fix the issue, but maybe a better solution is to register our compression filesystem before the script is executed and unregister them afterward. WDYT @lhoestq @albertvillanova?
I think we should use clobber and show a warning if it overwrote a registered filesystem indeed ! This way the user can re-register the filesystems if needed. Though they should probably be compatible (and maybe do the exact same thing) so I wouldn't de-register the `datasets` filesystems | 2023-05-24T21:41:52Z | [] | [] |
huggingface/datasets | 5,897 | huggingface__datasets-5897 | [
"5866"
] | 680162303f4c5dae6ad2edef6b3efadded7d37bd | diff --git a/src/datasets/table.py b/src/datasets/table.py
--- a/src/datasets/table.py
+++ b/src/datasets/table.py
@@ -1902,10 +1902,18 @@ def _concat_arrays(arrays):
_concat_arrays([array.values for array in arrays]),
)
elif pa.types.is_fixed_size_list(array_type):
- return pa.FixedSizeListArray.from_arrays(
- _concat_arrays([array.values for array in arrays]),
- array_type.list_size,
- )
+ if config.PYARROW_VERSION.major < 13:
+ # PyArrow bug: https://github.com/apache/arrow/issues/35360
+ return pa.FixedSizeListArray.from_arrays(
+ _concat_arrays([array.values[array.offset * array.type.list_size :] for array in arrays]),
+ array_type.list_size,
+ )
+ else:
+ return pa.FixedSizeListArray.from_arrays(
+ _concat_arrays([array.values for array in arrays]),
+ array_type.value_type,
+ array_type.list_size,
+ )
return pa.concat_arrays(arrays)
return _concat_arrays(arrays)
@@ -1968,9 +1976,13 @@ def array_cast(array: pa.Array, pa_type: pa.DataType, allow_number_to_str=True):
)
return pa.ListArray.from_arrays(array.offsets, _c(array.values, pa_type.value_type))
elif pa.types.is_fixed_size_list(array.type):
+ array_values = array.values
+ if config.PYARROW_VERSION.major < 13:
+ # PyArrow bug: https://github.com/apache/arrow/issues/35360
+ array_values = array.values[array.offset * array.type.list_size :]
if pa.types.is_fixed_size_list(pa_type):
return pa.FixedSizeListArray.from_arrays(
- _c(array.values, pa_type.value_type),
+ _c(array_values, pa_type.value_type),
pa_type.list_size,
)
elif pa.types.is_list(pa_type):
@@ -1982,9 +1994,9 @@ def array_cast(array: pa.Array, pa_type: pa.DataType, allow_number_to_str=True):
)
else:
return pa.ListArray.from_arrays(
- offsets_arr, _c(array.values, pa_type.value_type), mask=array.is_null()
+ offsets_arr, _c(array_values, pa_type.value_type), mask=array.is_null()
)
- return pa.ListArray.from_arrays(offsets_arr, _c(array.values, pa_type.value_type))
+ return pa.ListArray.from_arrays(offsets_arr, _c(array_values, pa_type.value_type))
else:
if (
not allow_number_to_str
@@ -2078,6 +2090,10 @@ def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_
return pa.ListArray.from_arrays(array.offsets, _c(array.values, feature.feature))
elif pa.types.is_fixed_size_list(array.type):
# feature must be either [subfeature] or Sequence(subfeature)
+ array_values = array.values
+ if config.PYARROW_VERSION.major < 13:
+ # PyArrow bug: https://github.com/apache/arrow/issues/35360
+ array_values = array.values[array.offset * array.type.list_size :]
if isinstance(feature, list):
if array.null_count > 0:
if config.PYARROW_VERSION.major < 10:
@@ -2085,12 +2101,12 @@ def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_
f"None values are converted to empty lists when converting array to {feature}. Install `pyarrow>=10.0.0` to avoid this behavior. More info: https://github.com/huggingface/datasets/issues/3676. This will raise an error in a future major version of `datasets`"
)
else:
- return pa.ListArray.from_arrays(array.offsets, _c(array.values, feature[0]), mask=array.is_null())
- return pa.ListArray.from_arrays(array.offsets, _c(array.values, feature[0]))
+ return pa.ListArray.from_arrays(array.offsets, _c(array_values, feature[0]), mask=array.is_null())
+ return pa.ListArray.from_arrays(array.offsets, _c(array_values, feature[0]))
elif isinstance(feature, Sequence):
if feature.length > -1:
- if feature.length * len(array) == len(array.values):
- return pa.FixedSizeListArray.from_arrays(_c(array.values, feature.feature), feature.length)
+ if feature.length * len(array) == len(array_values):
+ return pa.FixedSizeListArray.from_arrays(_c(array_values, feature.feature), feature.length)
else:
offsets_arr = pa.array(range(len(array) + 1), pa.int32())
if array.null_count > 0:
@@ -2100,9 +2116,9 @@ def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_
)
else:
return pa.ListArray.from_arrays(
- offsets_arr, _c(array.values, feature.feature), mask=array.is_null()
+ offsets_arr, _c(array_values, feature.feature), mask=array.is_null()
)
- return pa.ListArray.from_arrays(offsets_arr, _c(array.values, feature.feature))
+ return pa.ListArray.from_arrays(offsets_arr, _c(array_values, feature.feature))
if pa.types.is_null(array.type):
return array_cast(array, get_nested_type(feature), allow_number_to_str=allow_number_to_str)
elif not isinstance(feature, (Sequence, dict, list, tuple)):
@@ -2181,6 +2197,10 @@ def embed_array_storage(array: pa.Array, feature: "FeatureType"):
return pa.ListArray.from_arrays(array.offsets, _e(array.values, feature.feature))
elif pa.types.is_fixed_size_list(array.type):
# feature must be either [subfeature] or Sequence(subfeature)
+ array_values = array.values
+ if config.PYARROW_VERSION.major < 13:
+ # PyArrow bug: https://github.com/apache/arrow/issues/35360
+ array_values = array.values[array.offset * array.type.list_size :]
if isinstance(feature, list):
if array.null_count > 0:
if config.PYARROW_VERSION.major < 10:
@@ -2188,12 +2208,12 @@ def embed_array_storage(array: pa.Array, feature: "FeatureType"):
f"None values are converted to empty lists when embedding array storage with {feature}. Install `pyarrow>=10.0.0` to avoid this behavior. More info: https://github.com/huggingface/datasets/issues/3676. This will raise an error in a future major version of `datasets`"
)
else:
- return pa.ListArray.from_arrays(array.offsets, _e(array.values, feature[0]), mask=array.is_null())
- return pa.ListArray.from_arrays(array.offsets, _e(array.values, feature[0]))
+ return pa.ListArray.from_arrays(array.offsets, _e(array_values, feature[0]), mask=array.is_null())
+ return pa.ListArray.from_arrays(array.offsets, _e(array_values, feature[0]))
elif isinstance(feature, Sequence):
if feature.length > -1:
- if feature.length * len(array) == len(array.values):
- return pa.FixedSizeListArray.from_arrays(_e(array.values, feature.feature), feature.length)
+ if feature.length * len(array) == len(array_values):
+ return pa.FixedSizeListArray.from_arrays(_e(array_values, feature.feature), feature.length)
else:
offsets_arr = pa.array(range(len(array) + 1), pa.int32())
if array.null_count > 0:
@@ -2203,9 +2223,9 @@ def embed_array_storage(array: pa.Array, feature: "FeatureType"):
)
else:
return pa.ListArray.from_arrays(
- offsets_arr, _e(array.values, feature.feature), mask=array.is_null()
+ offsets_arr, _e(array_values, feature.feature), mask=array.is_null()
)
- return pa.ListArray.from_arrays(offsets_arr, _e(array.values, feature.feature))
+ return pa.ListArray.from_arrays(offsets_arr, _e(array_values, feature.feature))
if not isinstance(feature, (Sequence, dict, list, tuple)):
return array
raise TypeError(f"Couldn't embed array of type\n{array.type}\nwith\n{feature}")
| diff --git a/tests/test_table.py b/tests/test_table.py
--- a/tests/test_table.py
+++ b/tests/test_table.py
@@ -1183,6 +1183,13 @@ def test_cast_array_to_features_sequence_classlabel():
assert cast_array_to_feature(arr, Sequence(ClassLabel(names=["foo", "bar"])))
+def test_cast_sliced_fixed_size_array_to_features():
+ arr = pa.array([[0, 1, 2], [3, 4, 5], [6, 7, 8]], pa.list_(pa.int32(), 3))
+ casted_array = cast_array_to_feature(arr[1:], Sequence(Value("int64"), length=3))
+ assert casted_array.type == pa.list_(pa.int64(), 3)
+ assert casted_array.to_pylist() == arr[1:].to_pylist()
+
+
def test_embed_array_storage(image_file):
array = pa.array([{"bytes": None, "path": image_file}], type=Image.pa_type)
embedded_images_array = embed_array_storage(array, Image())
| Issue with Sequence features
### Describe the bug
Sequences features sometimes causes errors when the specified length is not -1
### Steps to reproduce the bug
```python
import numpy as np
from datasets import Features, ClassLabel, Sequence, Value, Dataset
feats = Features(**{'target': ClassLabel(names=[0, 1]),'x': Sequence(feature=Value(dtype='float64',id=None), length=2, id=None)})
Dataset.from_dict({"target": np.ones(2000).astype(int), "x": np.random.rand(2000,2)},features = feats).flatten_indices()
```
Throws:
```
TypeError: Couldn't cast array of type
fixed_size_list<item: double>[2]
to
Sequence(feature=Value(dtype='float64', id=None), length=2, id=None)
```
The same code works without any issues when `length = -1`
EDIT: The error seems to happen only when the length of the dataset is bigger than 1000 for some reason
### Expected behavior
No exception
### Environment info
- `datasets` version: 2.10.1
- Python version: 3.9.5
- PyArrow version: 11.0.0
- Pandas version: 1.4.1
| 2023-05-25T16:26:33Z | [] | [] |
|
huggingface/datasets | 5,933 | huggingface__datasets-5933 | [
"5927"
] | a129219a48c1b07c06d4bc1db32c317bf513089d | diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -714,10 +714,11 @@ def __getitem__(self, i):
def to_numpy(self, zero_copy_only=True):
storage: pa.ListArray = self.storage
+ null_mask = storage.is_null().to_numpy(zero_copy_only=False)
if self.type.shape[0] is not None:
size = 1
- null_indices = np.arange(len(storage))[storage.is_null().to_numpy(zero_copy_only=False)]
+ null_indices = np.arange(len(storage))[null_mask] - np.arange(np.sum(null_mask))
for i in range(self.type.ndims):
size *= self.type.shape[i]
@@ -733,7 +734,7 @@ def to_numpy(self, zero_copy_only=True):
ndims = self.type.ndims
arrays = []
first_dim_offsets = np.array([off.as_py() for off in storage.offsets])
- for i, is_null in enumerate(storage.is_null().to_numpy(zero_copy_only=False)):
+ for i, is_null in enumerate(null_mask):
if is_null:
arrays.append(np.nan)
else:
| diff --git a/tests/features/test_array_xd.py b/tests/features/test_array_xd.py
--- a/tests/features/test_array_xd.py
+++ b/tests/features/test_array_xd.py
@@ -380,21 +380,21 @@ def test_array_xd_with_none():
# Fixed shape
features = datasets.Features({"foo": datasets.Array2D(dtype="int32", shape=(2, 2))})
dummy_array = np.array([[1, 2], [3, 4]], dtype="int32")
- dataset = datasets.Dataset.from_dict({"foo": [dummy_array, None, dummy_array]}, features=features)
+ dataset = datasets.Dataset.from_dict({"foo": [dummy_array, None, dummy_array, None]}, features=features)
arr = NumpyArrowExtractor().extract_column(dataset._data)
- assert isinstance(arr, np.ndarray) and arr.dtype == np.float64 and arr.shape == (3, 2, 2)
+ assert isinstance(arr, np.ndarray) and arr.dtype == np.float64 and arr.shape == (4, 2, 2)
assert np.allclose(arr[0], dummy_array) and np.allclose(arr[2], dummy_array)
- assert np.all(np.isnan(arr[1])) # broadcasted np.nan - use np.all
+ assert np.all(np.isnan(arr[1])) and np.all(np.isnan(arr[3])) # broadcasted np.nan - use np.all
# Dynamic shape
features = datasets.Features({"foo": datasets.Array2D(dtype="int32", shape=(None, 2))})
dummy_array = np.array([[1, 2], [3, 4]], dtype="int32")
- dataset = datasets.Dataset.from_dict({"foo": [dummy_array, None, dummy_array]}, features=features)
+ dataset = datasets.Dataset.from_dict({"foo": [dummy_array, None, dummy_array, None]}, features=features)
arr = NumpyArrowExtractor().extract_column(dataset._data)
- assert isinstance(arr, np.ndarray) and arr.dtype == object and arr.shape == (3,)
+ assert isinstance(arr, np.ndarray) and arr.dtype == object and arr.shape == (4,)
np.testing.assert_equal(arr[0], dummy_array)
np.testing.assert_equal(arr[2], dummy_array)
- assert np.isnan(arr[1]) # a single np.nan value - np.all not needed
+ assert np.isnan(arr[1]) and np.isnan(arr[3]) # a single np.nan value - np.all not needed
@pytest.mark.parametrize(
| `IndexError` when indexing `Sequence` of `Array2D` with `None` values
### Describe the bug
Having `None` values in a `Sequence` of `ArrayND` fails.
### Steps to reproduce the bug
```python
from datasets import Array2D, Dataset, Features, Sequence
data = [
[
[[0]],
None,
None,
]
]
feature = Sequence(Array2D((1, 1), dtype="int64"))
dataset = Dataset.from_dict({"a": data}, features=Features({"a": feature}))
dataset[0] # error raised only when indexing
```
```
Traceback (most recent call last):
File "/Users/quentingallouedec/gia/c.py", line 13, in <module>
dataset[0] # error raised only when indexing
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/arrow_dataset.py", line 2658, in __getitem__
return self._getitem(key)
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/arrow_dataset.py", line 2643, in _getitem
formatted_output = format_table(
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/formatting/formatting.py", line 634, in format_table
return formatter(pa_table, query_type=query_type)
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/formatting/formatting.py", line 406, in __call__
return self.format_row(pa_table)
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/formatting/formatting.py", line 441, in format_row
row = self.python_arrow_extractor().extract_row(pa_table)
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/formatting/formatting.py", line 144, in extract_row
return _unnest(pa_table.to_pydict())
File "pyarrow/table.pxi", line 4146, in pyarrow.lib.Table.to_pydict
File "pyarrow/table.pxi", line 1312, in pyarrow.lib.ChunkedArray.to_pylist
File "pyarrow/array.pxi", line 1521, in pyarrow.lib.Array.to_pylist
File "pyarrow/scalar.pxi", line 675, in pyarrow.lib.ListScalar.as_py
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/features/features.py", line 760, in to_pylist
return self.to_numpy(zero_copy_only=zero_copy_only).tolist()
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/datasets/features/features.py", line 725, in to_numpy
numpy_arr = np.insert(numpy_arr.astype(np.float64), null_indices, np.nan, axis=0)
File "<__array_function__ internals>", line 200, in insert
File "/Users/quentingallouedec/gia/env/lib/python3.10/site-packages/numpy/lib/function_base.py", line 5426, in insert
old_mask[indices] = False
IndexError: index 3 is out of bounds for axis 0 with size 3
```
AFAIK, the problem only occurs when you use a `Sequence` of `ArrayND`.
I strongly suspect that the problem comes from this line, or `np.insert` is misused:
https://github.com/huggingface/datasets/blob/02ee418831aba68d0be93227bce8b3f42ef8980f/src/datasets/features/features.py#L729
To put t simply, you want something that do that:
```python
import numpy as np
numpy_arr = np.zeros((1, 1, 1))
null_indices = np.array([1, 2])
np.insert(numpy_arr, null_indices, np.nan, axis=0)
# raise an error, instead of outputting
# array([[[ 0.]],
# [[nan]],
# [[nan]]])
```
### Expected behavior
The previous code should not raise an error.
### Environment info
- Python 3.10.11
- datasets 2.10.0
- pyarrow 12.0.0
| Easy fix would be to add:
```python
null_indices -= np.arange(len(null_indices))
```
before L279, but I'm not sure it's the most intuitive way to fix it. | 2023-06-08T08:38:56Z | [] | [] |
huggingface/datasets | 5,948 | huggingface__datasets-5948 | [
"5936"
] | 7fcbe5b1575c8d162b65b9397b3dfda995a4e048 | diff --git a/src/datasets/features/features.py b/src/datasets/features/features.py
--- a/src/datasets/features/features.py
+++ b/src/datasets/features/features.py
@@ -521,8 +521,6 @@ def __call__(self):
return pa_type
def encode_example(self, value):
- if isinstance(value, np.ndarray):
- value = value.tolist()
return value
@@ -1390,7 +1388,8 @@ def numpy_to_pyarrow_listarray(arr: np.ndarray, type: pa.DataType = None) -> pa.
def list_of_pa_arrays_to_pyarrow_listarray(l_arr: List[Optional[pa.Array]]) -> pa.ListArray:
- null_indices = [i for i, arr in enumerate(l_arr) if arr is None]
+ null_mask = np.array([arr is None for arr in l_arr])
+ null_indices = np.arange(len(null_mask))[null_mask] - np.arange(np.sum(null_mask))
l_arr = [arr for arr in l_arr if arr is not None]
offsets = np.cumsum(
[0] + [len(arr) for arr in l_arr], dtype=object
| diff --git a/tests/features/test_array_xd.py b/tests/features/test_array_xd.py
--- a/tests/features/test_array_xd.py
+++ b/tests/features/test_array_xd.py
@@ -397,21 +397,38 @@ def test_array_xd_with_none():
assert np.isnan(arr[1]) and np.isnan(arr[3]) # a single np.nan value - np.all not needed
[email protected]("seq_type", ["no_sequence", "sequence", "sequence_of_sequence"])
@pytest.mark.parametrize(
- "data, feature, expected",
+ "dtype",
[
- (np.zeros((2, 2)), None, [[0.0, 0.0], [0.0, 0.0]]),
- (np.zeros((2, 3)), datasets.Array2D(shape=(2, 3), dtype="float32"), [[0.0, 0.0, 0.0], [0.0, 0.0, 0.0]]),
- ([np.zeros(2)], datasets.Array2D(shape=(1, 2), dtype="float32"), [[0.0, 0.0]]),
- (
- [np.zeros((2, 3))],
- datasets.Array3D(shape=(1, 2, 3), dtype="float32"),
- [[[0.0, 0.0, 0.0], [0.0, 0.0, 0.0]]],
- ),
+ "bool",
+ "int8",
+ "int16",
+ "int32",
+ "int64",
+ "uint8",
+ "uint16",
+ "uint32",
+ "uint64",
+ "float16",
+ "float32",
+ "float64",
],
)
-def test_array_xd_with_np(data, feature, expected):
- ds = datasets.Dataset.from_dict({"col": [data]}, features=datasets.Features({"col": feature}) if feature else None)
[email protected]("shape, feature_class", [((2, 3), datasets.Array2D), ((2, 3, 4), datasets.Array3D)])
+def test_array_xd_with_np(seq_type, dtype, shape, feature_class):
+ feature = feature_class(dtype=dtype, shape=shape)
+ data = np.zeros(shape, dtype=dtype)
+ expected = data.tolist()
+ if seq_type == "sequence":
+ feature = datasets.Sequence(feature)
+ data = [data]
+ expected = [expected]
+ elif seq_type == "sequence_of_sequence":
+ feature = datasets.Sequence(datasets.Sequence(feature))
+ data = [[data]]
+ expected = [[expected]]
+ ds = datasets.Dataset.from_dict({"col": [data]}, features=datasets.Features({"col": feature}))
assert ds[0]["col"] == expected
| Sequence of array not supported for most dtype
### Describe the bug
Create a dataset composed of sequence of array fails for most dtypes (see code below).
### Steps to reproduce the bug
```python
from datasets import Sequence, Array2D, Features, Dataset
import numpy as np
for dtype in [
"bool", # ok
"int8", # failed
"int16", # failed
"int32", # failed
"int64", # ok
"uint8", # failed
"uint16", # failed
"uint32", # failed
"uint64", # failed
"float16", # failed
"float32", # failed
"float64", # ok
]:
features = Features({"foo": Sequence(Array2D(dtype=dtype, shape=(2, 2)))})
sequence = [
[[1.0, 2.0], [3.0, 4.0]],
[[5.0, 6.0], [7.0, 8.0]],
]
array = np.array(sequence, dtype=dtype)
try:
dataset = Dataset.from_dict({"foo": [array]}, features=features)
except Exception as e:
print(f"Failed for dtype={dtype}")
```
Traceback for `dtype="int8"`:
```
Traceback (most recent call last):
File "/home/qgallouedec/datasets/a.py", line 29, in <module>
raise e
File "/home/qgallouedec/datasets/a.py", line 26, in <module>
dataset = Dataset.from_dict({"foo": [array]}, features=features)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/arrow_dataset.py", line 899, in from_dict
pa_table = InMemoryTable.from_pydict(mapping=mapping)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/table.py", line 799, in from_pydict
return cls(pa.Table.from_pydict(*args, **kwargs))
File "pyarrow/table.pxi", line 3725, in pyarrow.lib.Table.from_pydict
File "pyarrow/table.pxi", line 5254, in pyarrow.lib._from_pydict
File "pyarrow/array.pxi", line 350, in pyarrow.lib.asarray
File "pyarrow/array.pxi", line 236, in pyarrow.lib.array
File "pyarrow/array.pxi", line 110, in pyarrow.lib._handle_arrow_array_protocol
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/arrow_writer.py", line 204, in __arrow_array__
out = cast_array_to_feature(out, type, allow_number_to_str=not self.trying_type)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/table.py", line 1833, in wrapper
return func(array, *args, **kwargs)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/table.py", line 2091, in cast_array_to_feature
casted_values = _c(array.values, feature.feature)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/table.py", line 1833, in wrapper
return func(array, *args, **kwargs)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/table.py", line 2139, in cast_array_to_feature
return array_cast(array, feature(), allow_number_to_str=allow_number_to_str)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/table.py", line 1833, in wrapper
return func(array, *args, **kwargs)
File "/home/qgallouedec/env/lib/python3.10/site-packages/datasets/table.py", line 1967, in array_cast
return pa_type.wrap_array(array)
File "pyarrow/types.pxi", line 879, in pyarrow.lib.BaseExtensionType.wrap_array
TypeError: Incompatible storage type for extension<arrow.py_extension_type<Array2DExtensionType>>: expected list<item: list<item: int8>>, got list<item: list<item: int64>>
```
### Expected behavior
Not to fail.
### Environment info
- Python 3.10.6
- datasets: master branch
- Numpy: 1.23.4
| Related, `float16` is the only dtype not supported by `Array2D` (probably by every `ArrayND`):
```python
from datasets import Array2D, Features, Dataset
import numpy as np
for dtype in [
"bool", # ok
"int8", # ok
"int16", # ok
"int32", # ok
"int64", # ok
"uint8", # ok
"uint16", # ok
"uint32", # ok
"uint64", # ok
"float16", # failed
"float32", # ok
"float64", # ok
]:
features = Features({"foo": Array2D(dtype=dtype, shape=(3, 4))})
array = np.zeros((3, 4), dtype=dtype)
try:
dataset = Dataset.from_dict({"foo": [array]}, features=features)
except Exception as e:
print(f"Failed for dtype={dtype}")
```
Here's something I can't explain:
When an array is encoded in the `from_dict` method, the numpy array is converted to a list (thus losing the original dtype, which is transfromed to the nearest builtin Python type)
https://github.com/huggingface/datasets/blob/6ee61e6e695b1df9f232d47faf3a5e2b30b33737/src/datasets/features/features.py#L524-L525
However, later on, this same data is written to memory, and it seems authorized that the data is an array (or in this case, a list of arrays).
https://github.com/huggingface/datasets/blob/6ee61e6e695b1df9f232d47faf3a5e2b30b33737/src/datasets/arrow_writer.py#L185-L186
So the question is: why convert it to a Python list? This seems to be quite expensive both in terms of write time (all data is copied) and memory (e.g., an int8 is converted to an int64).
Finally, if I try to remove this step, it solves all the previous problems, and it seems to me that it doesn't break anything (the CI passes without problem).
Arrow only support 1d numpy arrays, so we convert multidim arrays to lists of 1s arrays (and keep the dtype).
Though you noticed that it's concerting to lists and lose the dtype. If it's the case then it's a bug.
Ok the conversion to list shouldn't be there indeed ! Could you open a PR to remove it ? | 2023-06-13T12:38:59Z | [] | [] |
huggingface/datasets | 6,009 | huggingface__datasets-6009 | [
"5677"
] | 3e34d06d746688dd5d26e4c85517b7e1a2f361ca | diff --git a/src/datasets/table.py b/src/datasets/table.py
--- a/src/datasets/table.py
+++ b/src/datasets/table.py
@@ -1969,6 +1969,8 @@ def array_cast(array: pa.Array, pa_type: pa.DataType, allow_number_to_str=True):
return array
elif pa.types.is_struct(array.type):
if pa.types.is_struct(pa_type) and ({field.name for field in pa_type} == {field.name for field in array.type}):
+ if array.type.num_fields == 0:
+ return array
arrays = [_c(array.field(field.name), field.type) for field in pa_type]
return pa.StructArray.from_arrays(arrays, fields=list(pa_type), mask=array.is_null())
elif pa.types.is_list(array.type):
@@ -2066,6 +2068,8 @@ def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_
name: Sequence(subfeature, length=feature.length) for name, subfeature in feature.feature.items()
}
if isinstance(feature, dict) and {field.name for field in array.type} == set(feature):
+ if array.type.num_fields == 0:
+ return array
arrays = [_c(array.field(name), subfeature) for name, subfeature in feature.items()]
return pa.StructArray.from_arrays(arrays, names=list(feature), mask=array.is_null())
elif pa.types.is_list(array.type):
| diff --git a/tests/test_table.py b/tests/test_table.py
--- a/tests/test_table.py
+++ b/tests/test_table.py
@@ -1124,6 +1124,12 @@ def test_cast_array_to_features_nested():
)
+def test_cast_array_to_features_to_nested_with_no_fields():
+ arr = pa.array([{}])
+ assert cast_array_to_feature(arr, {}).type == pa.struct({})
+ assert cast_array_to_feature(arr, {}).to_pylist() == arr.to_pylist()
+
+
def test_cast_array_to_features_nested_with_null_values():
# same type
arr = pa.array([{"foo": [None, [0]]}], pa.struct({"foo": pa.list_(pa.list_(pa.int64()))}))
| Dataset.map() crashes when any column contains more than 1000 empty dictionaries
### Describe the bug
`Dataset.map()` crashes any time any column contains more than `writer_batch_size` (default 1000) empty dictionaries, regardless of whether the column is being operated on. The error does not occur if the dictionaries are non-empty.
### Steps to reproduce the bug
Example:
```
import datasets
def add_one(example):
example["col2"] += 1
return example
n = 1001 # crashes
# n = 999 # works
ds = datasets.Dataset.from_dict({"col1": [{}] * n, "col2": [1] * n})
ds = ds.map(add_one, writer_batch_size=1000)
```
### Expected behavior
Above code should not crash
### Environment info
- `datasets` version: 2.10.1
- Platform: Linux-5.4.0-120-generic-x86_64-with-glibc2.10
- Python version: 3.8.15
- PyArrow version: 9.0.0
- Pandas version: 1.5.3
| 2023-07-06T18:48:14Z | [] | [] |
|
huggingface/datasets | 6,019 | huggingface__datasets-6019 | [
"2832"
] | b8067c0262073891180869f700ebef5ac3dc5cce | diff --git a/metrics/perplexity/perplexity.py b/metrics/perplexity/perplexity.py
--- a/metrics/perplexity/perplexity.py
+++ b/metrics/perplexity/perplexity.py
@@ -71,8 +71,7 @@
>>> perplexity = datasets.load_metric("perplexity")
>>> input_texts = datasets.load_dataset("wikitext",
... "wikitext-2-raw-v1",
- ... split="test")["text"][:50] # doctest:+ELLIPSIS
- [...]
+ ... split="test")["text"][:50]
>>> input_texts = [s for s in input_texts if s!='']
>>> results = perplexity.compute(model_id='gpt2',
... input_texts=input_texts) # doctest:+ELLIPSIS
diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -1486,7 +1486,6 @@ def save_to_disk(
disable=not logging.is_progress_bar_enabled(),
unit=" examples",
total=len(self),
- leave=False,
desc=f"Saving the dataset ({shards_done}/{num_shards} shards)",
)
kwargs_per_job = (
@@ -3070,7 +3069,7 @@ def load_processed_shard_from_cache(shard_kwargs):
transformed_dataset = None
try:
transformed_dataset = load_processed_shard_from_cache(dataset_kwargs)
- logger.warning(f"Loading cached processed dataset at {dataset_kwargs['cache_file_name']}")
+ logger.info(f"Loading cached processed dataset at {dataset_kwargs['cache_file_name']}")
except NonExistentDatasetError:
pass
if transformed_dataset is None:
@@ -3078,7 +3077,6 @@ def load_processed_shard_from_cache(shard_kwargs):
disable=not logging.is_progress_bar_enabled(),
unit=" examples",
total=pbar_total,
- leave=False,
desc=desc or "Map",
) as pbar:
for rank, done, content in Dataset._map_single(**dataset_kwargs):
@@ -3171,7 +3169,6 @@ def format_new_fingerprint(new_fingerprint: str, rank: int) -> str:
disable=not logging.is_progress_bar_enabled(),
unit=" examples",
total=pbar_total,
- leave=False,
desc=(desc or "Map") + f" (num_proc={num_proc})",
) as pbar:
for rank, done, content in iflatmap_unordered(
@@ -3187,7 +3184,7 @@ def format_new_fingerprint(new_fingerprint: str, rank: int) -> str:
for kwargs in kwargs_per_job:
del kwargs["shard"]
else:
- logger.warning(f"Loading cached processed dataset at {format_cache_file_name(cache_file_name, '*')}")
+ logger.info(f"Loading cached processed dataset at {format_cache_file_name(cache_file_name, '*')}")
assert (
None not in transformed_shards
), f"Failed to retrieve results from map: result list {transformed_shards} still contains None - at least one worker failed to return its results"
@@ -4086,7 +4083,7 @@ def sort(
# we create a unique hash from the function, current dataset file and the mapping args
indices_cache_file_name = self._get_cache_file_path(new_fingerprint)
if os.path.exists(indices_cache_file_name) and load_from_cache_file:
- logger.warning(f"Loading cached sorted indices for dataset at {indices_cache_file_name}")
+ logger.info(f"Loading cached sorted indices for dataset at {indices_cache_file_name}")
return self._new_dataset_with_indices(
fingerprint=new_fingerprint, indices_cache_file_name=indices_cache_file_name
)
@@ -4228,7 +4225,7 @@ def shuffle(
# we create a unique hash from the function, current dataset file and the mapping args
indices_cache_file_name = self._get_cache_file_path(new_fingerprint)
if os.path.exists(indices_cache_file_name) and load_from_cache_file:
- logger.warning(f"Loading cached shuffled indices for dataset at {indices_cache_file_name}")
+ logger.info(f"Loading cached shuffled indices for dataset at {indices_cache_file_name}")
return self._new_dataset_with_indices(
fingerprint=new_fingerprint, indices_cache_file_name=indices_cache_file_name
)
@@ -4459,7 +4456,7 @@ def train_test_split(
and os.path.exists(test_indices_cache_file_name)
and load_from_cache_file
):
- logger.warning(
+ logger.info(
f"Loading cached split indices for dataset at {train_indices_cache_file_name} and {test_indices_cache_file_name}"
)
return DatasetDict(
@@ -5273,7 +5270,7 @@ def path_in_repo(_index, shard):
first_shard = next(shards_iter)
first_shard_path_in_repo = path_in_repo(0, first_shard)
if first_shard_path_in_repo in data_files and num_shards < len(data_files):
- logger.warning("Resuming upload of the dataset shards.")
+ logger.info("Resuming upload of the dataset shards.")
uploaded_size = 0
shards_path_in_repo = []
@@ -5450,7 +5447,7 @@ def push_to_hub(
repo_info = None
# update the total info to dump from existing info
if repo_info is not None:
- logger.warning("Updating downloaded metadata with the new split.")
+ logger.info("Updating downloaded metadata with the new split.")
if repo_info.splits and list(repo_info.splits) != [split]:
if self._info.features != repo_info.features:
raise ValueError(
diff --git a/src/datasets/builder.py b/src/datasets/builder.py
--- a/src/datasets/builder.py
+++ b/src/datasets/builder.py
@@ -410,7 +410,7 @@ def __init__(
self.info = DatasetInfo.from_directory(self._cache_dir)
else: # dir exists but no data, remove the empty dir as data aren't available anymore
logger.warning(
- f"Old caching folder {self._cache_dir} for dataset {self.name} exists but no data were found. Removing it. "
+ f"Old caching folder {self._cache_dir} for dataset {self.name} exists but no data were found. Removing it."
)
os.rmdir(self._cache_dir)
@@ -490,7 +490,7 @@ def _create_builder_config(
if config_name is None and self.BUILDER_CONFIGS and not config_kwargs:
if self.DEFAULT_CONFIG_NAME is not None:
builder_config = self.builder_configs.get(self.DEFAULT_CONFIG_NAME)
- logger.warning(f"No config specified, defaulting to: {self.name}/{builder_config.name}")
+ logger.info(f"No config specified, defaulting to: {self.name}/{builder_config.name}")
else:
if len(self.BUILDER_CONFIGS) > 1:
example_of_usage = f"load_dataset('{self.name}', '{self.BUILDER_CONFIGS[0].name}')"
@@ -843,7 +843,7 @@ def download_and_prepare(
path_join = os.path.join if is_local else posixpath.join
data_exists = self._fs.exists(path_join(self._output_dir, config.DATASET_INFO_FILENAME))
if data_exists and download_mode == DownloadMode.REUSE_DATASET_IF_EXISTS:
- logger.warning(f"Found cached dataset {self.name} ({self._output_dir})")
+ logger.info(f"Found cached dataset {self.name} ({self._output_dir})")
# We need to update the info in case some splits were added in the meantime
# for example when calling load_dataset from multiple workers.
self.info = self._load_info()
@@ -882,7 +882,7 @@ def incomplete_dir(dirname):
# information needed to cancel download/preparation if needed.
# This comes right before the progress bar.
if self.info.size_in_bytes:
- print(
+ logger.info(
f"Downloading and preparing dataset {self.info.builder_name}/{self.info.config_name} "
f"(download: {size_str(self.info.download_size)}, generated: {size_str(self.info.dataset_size)}, "
f"post-processed: {size_str(self.info.post_processing_size)}, "
@@ -890,7 +890,7 @@ def incomplete_dir(dirname):
)
else:
_dest = self._fs._strip_protocol(self._output_dir) if is_local else self._output_dir
- print(
+ logger.info(
f"Downloading and preparing dataset {self.info.builder_name}/{self.info.config_name} to {_dest}..."
)
@@ -933,7 +933,7 @@ def incomplete_dir(dirname):
# Download post processing resources
self.download_post_processing_resources(dl_manager)
- print(
+ logger.info(
f"Dataset {self.name} downloaded and prepared to {self._output_dir}. "
f"Subsequent calls will reuse this data."
)
@@ -1162,7 +1162,7 @@ def as_dataset(
),
split,
map_tuple=True,
- disable_tqdm=not logging.is_progress_bar_enabled(),
+ disable_tqdm=True,
)
if isinstance(datasets, dict):
datasets = DatasetDict(datasets)
@@ -1490,7 +1490,7 @@ def _prepare_split(
)
num_proc = 1
elif num_proc is not None and num_input_shards < num_proc:
- logger.info(
+ logger.warning(
f"Setting num_proc from {num_proc} to {num_input_shards} for the {split_info.name} split as it only contains {num_input_shards} shards."
)
num_proc = num_input_shards
@@ -1499,7 +1499,6 @@ def _prepare_split(
disable=not logging.is_progress_bar_enabled(),
unit=" examples",
total=split_info.num_examples,
- leave=False,
desc=f"Generating {split_info.name} split",
)
@@ -1751,7 +1750,7 @@ def _prepare_split(
)
num_proc = 1
elif num_proc is not None and num_input_shards < num_proc:
- logger.info(
+ logger.warning(
f"Setting num_proc from {num_proc} to {num_input_shards} for the {split_info.name} split as it only contains {num_input_shards} shards."
)
num_proc = num_input_shards
@@ -1760,7 +1759,6 @@ def _prepare_split(
disable=not logging.is_progress_bar_enabled(),
unit=" examples",
total=split_info.num_examples,
- leave=False,
desc=f"Generating {split_info.name} split",
)
diff --git a/src/datasets/dataset_dict.py b/src/datasets/dataset_dict.py
--- a/src/datasets/dataset_dict.py
+++ b/src/datasets/dataset_dict.py
@@ -1628,7 +1628,7 @@ def push_to_hub(
raise ValueError(f"Split name should match '{_split_re}' but got '{split}'.")
for split in self.keys():
- logger.warning(f"Pushing split {split} to the Hub.")
+ logger.info(f"Pushing split {split} to the Hub.")
# The split=key needs to be removed before merging
repo_id, split, uploaded_size, dataset_nbytes, _, _ = self[split]._push_parquet_shards_to_hub(
repo_id,
diff --git a/src/datasets/iterable_dataset.py b/src/datasets/iterable_dataset.py
--- a/src/datasets/iterable_dataset.py
+++ b/src/datasets/iterable_dataset.py
@@ -1233,7 +1233,7 @@ def _iter_pytorch(self):
f"Too many dataloader workers: {worker_info.num_workers} (max is dataset.n_shards={ex_iterable.n_shards}). "
f"Stopping {worker_info.num_workers - ex_iterable.n_shards} dataloader workers."
)
- logger.warning(
+ logger.info(
f"To parallelize data loading, we give each process some shards (or data sources) to process. "
f"Therefore it's unnecessary to have a number of workers greater than dataset.n_shards={ex_iterable.n_shards}. "
f"To enable more parallelism, please split the dataset in more files than {ex_iterable.n_shards}."
@@ -1304,13 +1304,13 @@ def _prepare_ex_iterable_for_iteration(self) -> _BaseExamplesIterable:
if self._is_main_process():
n_shards_per_node = ex_iterable.n_shards // world_size
plural = "s" if n_shards_per_node > 1 else ""
- logger.warning(
+ logger.info(
f"Assigning {n_shards_per_node} shard{plural} (or data source{plural}) of the dataset to each node."
)
ex_iterable = ex_iterable.shard_data_sources(rank, world_size)
else:
if self._is_main_process():
- logger.warning(
+ logger.info(
f"Assigning 1 out of {world_size} examples of the dataset to each node. The others are skipped during the iteration."
)
logger.info(
diff --git a/src/datasets/utils/logging.py b/src/datasets/utils/logging.py
--- a/src/datasets/utils/logging.py
+++ b/src/datasets/utils/logging.py
@@ -69,6 +69,7 @@ def _get_library_root_logger() -> logging.Logger:
def _configure_library_root_logger() -> None:
# Apply our default configuration to the library root logger.
library_root_logger = _get_library_root_logger()
+ library_root_logger.addHandler(logging.StreamHandler())
library_root_logger.setLevel(_get_default_logging_level())
| diff --git a/tests/test_arrow_dataset.py b/tests/test_arrow_dataset.py
--- a/tests/test_arrow_dataset.py
+++ b/tests/test_arrow_dataset.py
@@ -48,7 +48,7 @@
Summarization,
TextClassification,
)
-from datasets.utils.logging import WARNING
+from datasets.utils.logging import INFO, get_logger
from datasets.utils.py_utils import temp_seed
from .utils import (
@@ -1320,7 +1320,7 @@ def test_map_fn_kwargs(self, in_memory):
def test_map_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
- with self._caplog.at_level(WARNING):
+ with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with patch(
"datasets.arrow_dataset.Dataset._map_single",
@@ -1338,7 +1338,7 @@ def test_map_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
- with self._caplog.at_level(WARNING):
+ with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(lambda x: {"foo": "bar"}) as dset_test1:
dset_test1_data_files = list(dset_test1.cache_files)
@@ -1349,7 +1349,7 @@ def test_map_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
- with self._caplog.at_level(WARNING):
+ with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with patch(
"datasets.arrow_dataset.Pool",
@@ -1369,7 +1369,7 @@ def test_map_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
- with self._caplog.at_level(WARNING):
+ with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(lambda x: {"foo": "bar"}, num_proc=2) as dset_test1:
dset_test1_data_files = list(dset_test1.cache_files)
@@ -1382,7 +1382,7 @@ def test_map_caching(self, in_memory):
try:
self._caplog.clear()
with tempfile.TemporaryDirectory() as tmp_dir:
- with self._caplog.at_level(WARNING):
+ with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
datasets.disable_caching()
with dset.map(lambda x: {"foo": "bar"}) as dset_test1:
@@ -1733,7 +1733,7 @@ def test_filter_multiprocessing(self, in_memory):
def test_filter_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
- with self._caplog.at_level(WARNING):
+ with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.filter(lambda x, i: i < 5, with_indices=True) as dset_filter_first_five1:
dset_test1_data_files = list(dset_filter_first_five1.cache_files)
diff --git a/tests/test_builder.py b/tests/test_builder.py
--- a/tests/test_builder.py
+++ b/tests/test_builder.py
@@ -27,6 +27,7 @@
from datasets.streaming import xjoin
from datasets.utils.file_utils import is_local_path
from datasets.utils.info_utils import VerificationMode
+from datasets.utils.logging import INFO, get_logger
from .utils import (
assert_arrow_memory_doesnt_increase,
@@ -1060,7 +1061,8 @@ def test_builder_with_filesystem_download_and_prepare_reload(tmp_path, mockfs, c
DatasetInfo().write_to_directory("mock://my_dataset", storage_options=mockfs.storage_options)
mockfs.touch(f"my_dataset/{builder.name}-train.arrow")
caplog.clear()
- builder.download_and_prepare("mock://my_dataset", storage_options=mockfs.storage_options)
+ with caplog.at_level(INFO, logger=get_logger().name):
+ builder.download_and_prepare("mock://my_dataset", storage_options=mockfs.storage_options)
assert "Found cached dataset" in caplog.text
diff --git a/tests/test_load.py b/tests/test_load.py
--- a/tests/test_load.py
+++ b/tests/test_load.py
@@ -36,6 +36,7 @@
infer_module_for_data_files,
infer_module_for_data_files_in_archives,
)
+from datasets.utils.logging import INFO, get_logger
from .utils import (
OfflineSimulationMode,
@@ -965,7 +966,8 @@ def test_load_dataset_then_move_then_reload(dataset_loading_script_dir, data_dir
del dataset
os.rename(cache_dir1, cache_dir2)
caplog.clear()
- dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir2)
+ with caplog.at_level(INFO, logger=get_logger().name):
+ dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir2)
assert "Found cached dataset" in caplog.text
assert dataset._fingerprint == fingerprint1, "for the caching mechanism to work, fingerprint should stay the same"
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="test", cache_dir=cache_dir2)
| Logging levels not taken into account
## Describe the bug
The `logging` module isn't working as intended relative to the levels to set.
## Steps to reproduce the bug
```python
from datasets import logging
logging.set_verbosity_debug()
logger = logging.get_logger()
logger.error("ERROR")
logger.warning("WARNING")
logger.info("INFO")
logger.debug("DEBUG"
```
## Expected results
I expect all logs to be output since I'm putting a `debug` level.
## Actual results
Only the two first logs are output.
## Environment info
- `datasets` version: 1.11.0
- Platform: Linux-5.13.9-arch1-1-x86_64-with-glibc2.33
- Python version: 3.9.6
- PyArrow version: 5.0.0
## To go further
This logging issue appears in `datasets` but not in `transformers`. It happens because there is no handler defined for the logger. When no handler is defined, the `logging` library will output a one-off error to stderr, using a `StderrHandler` with level `WARNING`.
`transformers` sets a default `StreamHandler` [here](https://github.com/huggingface/transformers/blob/5c6eca71a983bae2589eed01e5c04fcf88ba5690/src/transformers/utils/logging.py#L86)
| I just take a look at all the outputs produced by `datasets` using the different log-levels.
As far as i can tell using `datasets==1.17.0` they overall issue seems to be fixed.
However, I noticed that there is one tqdm based progress indicator appearing on STDERR that I can simply not suppress.
```
Resolving data files: 100%|██████████| 652/652 [00:00<00:00, 1604.52it/s]
```
According to _get_origin_metadata_locally_or_by_urls it shold be supressable by using the `NOTSET` log-level
https://github.com/huggingface/datasets/blob/1406a04c3e911cec2680d8bc513653e0cafcaaa4/src/datasets/data_files.py#L491-L501
Sadly when specifiing the log-level `NOTSET` it seems to has no effect.
But appart from it not having any effect I must admit that it seems unintuitive to me.
I would suggest changing this such that it is only shown when the log-level is greater or equal to INFO.
This would conform better to INFO according to the [documentation](https://huggingface.co/docs/datasets/v1.0.0/package_reference/logging_methods.html#datasets.logging.set_verbosity_info).
> This will display most of the logging information and tqdm bars.
Any inputs on this?
I will be happy to supply a PR if desired 👍
Hi! This should disable the tqdm output:
```python
import datasets
datasets.set_progress_bar_enabled(False)
```
On a side note: I believe the issue with logging (not tqdm) is still relevant on master. | 2023-07-11T18:30:23Z | [] | [] |
huggingface/datasets | 6,045 | huggingface__datasets-6045 | [
"6039"
] | ae126ac974cad3050f90106e5909232140786811 | diff --git a/src/datasets/packaged_modules/parquet/parquet.py b/src/datasets/packaged_modules/parquet/parquet.py
--- a/src/datasets/packaged_modules/parquet/parquet.py
+++ b/src/datasets/packaged_modules/parquet/parquet.py
@@ -45,11 +45,16 @@ def _split_generators(self, dl_manager):
files = [files]
# Use `dl_manager.iter_files` to skip hidden files in an extracted archive
files = [dl_manager.iter_files(file) for file in files]
- # Infer features is they are stoed in the arrow schema
+ # Infer features if they are stored in the arrow schema
if self.info.features is None:
for file in itertools.chain.from_iterable(files):
with open(file, "rb") as f:
- self.info.features = datasets.Features.from_arrow_schema(pq.read_schema(f))
+ features = datasets.Features.from_arrow_schema(pq.read_schema(f))
+ if self.config.columns is not None:
+ self.info.features = datasets.Features(
+ {col: feat for col, feat in features.items() if col in self.config.columns}
+ )
+ self.info.features = features
break
splits.append(datasets.SplitGenerator(name=split_name, gen_kwargs={"files": files}))
return splits
@@ -62,9 +67,8 @@ def _cast_table(self, pa_table: pa.Table) -> pa.Table:
return pa_table
def _generate_tables(self, files):
- schema = self.info.features.arrow_schema if self.info.features is not None else None
- if self.info.features is not None and self.config.columns is not None:
- if sorted(field.name for field in schema) != sorted(self.config.columns):
+ if self.config.features is not None and self.config.columns is not None:
+ if sorted(field.name for field in self.info.features.arrow_schema) != sorted(self.config.columns):
raise ValueError(
f"Tried to load parquet data with columns '{self.config.columns}' with mismatching features '{self.info.features}'"
)
| diff --git a/tests/io/test_parquet.py b/tests/io/test_parquet.py
--- a/tests/io/test_parquet.py
+++ b/tests/io/test_parquet.py
@@ -125,7 +125,7 @@ def test_parquet_datasetdict_reader_split(split, parquet_path, tmp_path):
assert all(dataset[split].split == split for split in path.keys())
-def test_parquer_write(dataset, tmp_path):
+def test_parquet_write(dataset, tmp_path):
writer = ParquetDatasetWriter(dataset, tmp_path / "foo.parquet")
assert writer.write() > 0
pf = pq.ParquetFile(tmp_path / "foo.parquet")
| Loading column subset from parquet file produces error since version 2.13
### Describe the bug
`load_dataset` allows loading a subset of columns from a parquet file with the `columns` argument. Since version 2.13, this produces the following error:
```
Traceback (most recent call last):
File "/usr/lib/python3.10/site-packages/datasets/builder.py", line 1879, in _prepare_split_single
for _, table in generator:
File "/usr/lib/python3.10/site-packages/datasets/packaged_modules/parquet/parquet.py", line 68, in _generate_tables
raise ValueError(
ValueError: Tried to load parquet data with columns '['sepal_length']' with mismatching features '{'sepal_length': Value(dtype='float64', id=None), 'sepal_width': Value(dtype='float64', id=None), 'petal_length': Value(dtype='float64', id=None), 'petal_width': Value(dtype='float64', id=None), 'species': Value(dtype='string', id=None)}'
```
This seems to occur because `datasets` is checking whether the columns in the schema exactly match the provided list of columns, instead of whether they are a subset.
### Steps to reproduce the bug
```python
# Prepare some sample data
import pandas as pd
iris = pd.read_csv('https://raw.githubusercontent.com/mwaskom/seaborn-data/master/iris.csv')
iris.to_parquet('iris.parquet')
# ['sepal_length', 'sepal_width', 'petal_length', 'petal_width', 'species']
print(iris.columns)
# Load data with datasets
from datasets import load_dataset
# Load full parquet file
dataset = load_dataset('parquet', data_files='iris.parquet')
# Load column subset; throws error for datasets>=2.13
dataset = load_dataset('parquet', data_files='iris.parquet', columns=['sepal_length'])
```
### Expected behavior
No error should be thrown and the given column subset should be loaded.
### Environment info
- `datasets` version: 2.13.0
- Platform: Linux-5.15.0-76-generic-x86_64-with-glibc2.35
- Python version: 3.10.9
- Huggingface_hub version: 0.16.4
- PyArrow version: 12.0.1
- Pandas version: 1.5.3
| 2023-07-17T15:50:15Z | [] | [] |
|
huggingface/datasets | 6,096 | huggingface__datasets-6096 | [
"6086"
] | 8247202a7ed1c3164c88f8f183513c5f003aa2af | diff --git a/src/datasets/arrow_dataset.py b/src/datasets/arrow_dataset.py
--- a/src/datasets/arrow_dataset.py
+++ b/src/datasets/arrow_dataset.py
@@ -4729,13 +4729,15 @@ def to_csv(
path_or_buf: Union[PathLike, BinaryIO],
batch_size: Optional[int] = None,
num_proc: Optional[int] = None,
+ storage_options: Optional[dict] = None,
**to_csv_kwargs,
) -> int:
"""Exports the dataset to csv
Args:
path_or_buf (`PathLike` or `FileOrBuffer`):
- Either a path to a file or a BinaryIO.
+ Either a path to a file (e.g. `file.csv`), a remote URI (e.g. `hf://datasets/username/my_dataset_name/data.csv`),
+ or a BinaryIO, where the dataset will be saved to in the specified format.
batch_size (`int`, *optional*):
Size of the batch to load in memory and write at once.
Defaults to `datasets.config.DEFAULT_MAX_BATCH_SIZE`.
@@ -4744,6 +4746,10 @@ def to_csv(
use multiprocessing. `batch_size` in this case defaults to
`datasets.config.DEFAULT_MAX_BATCH_SIZE` but feel free to make it 5x or 10x of the default
value if you have sufficient compute power.
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.19.0"/>
**to_csv_kwargs (additional keyword arguments):
Parameters to pass to pandas's [`pandas.DataFrame.to_csv`](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_json.html).
@@ -4768,7 +4774,14 @@ def to_csv(
# Dynamic import to avoid circular dependency
from .io.csv import CsvDatasetWriter
- return CsvDatasetWriter(self, path_or_buf, batch_size=batch_size, num_proc=num_proc, **to_csv_kwargs).write()
+ return CsvDatasetWriter(
+ self,
+ path_or_buf,
+ batch_size=batch_size,
+ num_proc=num_proc,
+ storage_options=storage_options,
+ **to_csv_kwargs,
+ ).write()
def to_dict(self, batch_size: Optional[int] = None, batched="deprecated") -> Union[dict, Iterator[dict]]:
"""Returns the dataset as a Python dict. Can also return a generator for large datasets.
@@ -4844,13 +4857,15 @@ def to_json(
path_or_buf: Union[PathLike, BinaryIO],
batch_size: Optional[int] = None,
num_proc: Optional[int] = None,
+ storage_options: Optional[dict] = None,
**to_json_kwargs,
) -> int:
"""Export the dataset to JSON Lines or JSON.
Args:
path_or_buf (`PathLike` or `FileOrBuffer`):
- Either a path to a file or a BinaryIO.
+ Either a path to a file (e.g. `file.json`), a remote URI (e.g. `hf://datasets/username/my_dataset_name/data.json`),
+ or a BinaryIO, where the dataset will be saved to in the specified format.
batch_size (`int`, *optional*):
Size of the batch to load in memory and write at once.
Defaults to `datasets.config.DEFAULT_MAX_BATCH_SIZE`.
@@ -4859,6 +4874,10 @@ def to_json(
use multiprocessing. `batch_size` in this case defaults to
`datasets.config.DEFAULT_MAX_BATCH_SIZE` but feel free to make it 5x or 10x of the default
value if you have sufficient compute power.
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.19.0"/>
**to_json_kwargs (additional keyword arguments):
Parameters to pass to pandas's [`pandas.DataFrame.to_json`](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_json.html).
@@ -4882,7 +4901,14 @@ def to_json(
# Dynamic import to avoid circular dependency
from .io.json import JsonDatasetWriter
- return JsonDatasetWriter(self, path_or_buf, batch_size=batch_size, num_proc=num_proc, **to_json_kwargs).write()
+ return JsonDatasetWriter(
+ self,
+ path_or_buf,
+ batch_size=batch_size,
+ num_proc=num_proc,
+ storage_options=storage_options,
+ **to_json_kwargs,
+ ).write()
def to_pandas(
self, batch_size: Optional[int] = None, batched: bool = False
@@ -4927,16 +4953,22 @@ def to_parquet(
self,
path_or_buf: Union[PathLike, BinaryIO],
batch_size: Optional[int] = None,
+ storage_options: Optional[dict] = None,
**parquet_writer_kwargs,
) -> int:
"""Exports the dataset to parquet
Args:
path_or_buf (`PathLike` or `FileOrBuffer`):
- Either a path to a file or a BinaryIO.
+ Either a path to a file (e.g. `file.parquet`), a remote URI (e.g. `hf://datasets/username/my_dataset_name/data.parquet`),
+ or a BinaryIO, where the dataset will be saved to in the specified format.
batch_size (`int`, *optional*):
Size of the batch to load in memory and write at once.
Defaults to `datasets.config.DEFAULT_MAX_BATCH_SIZE`.
+ storage_options (`dict`, *optional*):
+ Key/value pairs to be passed on to the file-system backend, if any.
+
+ <Added version="2.19.0"/>
**parquet_writer_kwargs (additional keyword arguments):
Parameters to pass to PyArrow's `pyarrow.parquet.ParquetWriter`.
@@ -4952,7 +4984,9 @@ def to_parquet(
# Dynamic import to avoid circular dependency
from .io.parquet import ParquetDatasetWriter
- return ParquetDatasetWriter(self, path_or_buf, batch_size=batch_size, **parquet_writer_kwargs).write()
+ return ParquetDatasetWriter(
+ self, path_or_buf, batch_size=batch_size, storage_options=storage_options, **parquet_writer_kwargs
+ ).write()
def to_sql(
self,
diff --git a/src/datasets/io/csv.py b/src/datasets/io/csv.py
--- a/src/datasets/io/csv.py
+++ b/src/datasets/io/csv.py
@@ -2,6 +2,8 @@
import os
from typing import BinaryIO, Optional, Union
+import fsspec
+
from .. import Dataset, Features, NamedSplit, config
from ..formatting import query_table
from ..packaged_modules.csv.csv import Csv
@@ -72,6 +74,7 @@ def __init__(
path_or_buf: Union[PathLike, BinaryIO],
batch_size: Optional[int] = None,
num_proc: Optional[int] = None,
+ storage_options: Optional[dict] = None,
**to_csv_kwargs,
):
if num_proc is not None and num_proc <= 0:
@@ -82,6 +85,7 @@ def __init__(
self.batch_size = batch_size if batch_size else config.DEFAULT_MAX_BATCH_SIZE
self.num_proc = num_proc
self.encoding = "utf-8"
+ self.storage_options = storage_options or {}
self.to_csv_kwargs = to_csv_kwargs
def write(self) -> int:
@@ -90,7 +94,7 @@ def write(self) -> int:
index = self.to_csv_kwargs.pop("index", False)
if isinstance(self.path_or_buf, (str, bytes, os.PathLike)):
- with open(self.path_or_buf, "wb+") as buffer:
+ with fsspec.open(self.path_or_buf, "wb", **(self.storage_options or {})) as buffer:
written = self._write(file_obj=buffer, header=header, index=index, **self.to_csv_kwargs)
else:
written = self._write(file_obj=self.path_or_buf, header=header, index=index, **self.to_csv_kwargs)
diff --git a/src/datasets/io/json.py b/src/datasets/io/json.py
--- a/src/datasets/io/json.py
+++ b/src/datasets/io/json.py
@@ -77,6 +77,7 @@ def __init__(
path_or_buf: Union[PathLike, BinaryIO],
batch_size: Optional[int] = None,
num_proc: Optional[int] = None,
+ storage_options: Optional[dict] = None,
**to_json_kwargs,
):
if num_proc is not None and num_proc <= 0:
@@ -87,6 +88,7 @@ def __init__(
self.batch_size = batch_size if batch_size else config.DEFAULT_MAX_BATCH_SIZE
self.num_proc = num_proc
self.encoding = "utf-8"
+ self.storage_options = storage_options or {}
self.to_json_kwargs = to_json_kwargs
def write(self) -> int:
@@ -104,7 +106,9 @@ def write(self) -> int:
raise NotImplementedError(f"`datasets` currently does not support {compression} compression")
if isinstance(self.path_or_buf, (str, bytes, os.PathLike)):
- with fsspec.open(self.path_or_buf, "wb", compression=compression) as buffer:
+ with fsspec.open(
+ self.path_or_buf, "wb", compression=compression, **(self.storage_options or {})
+ ) as buffer:
written = self._write(file_obj=buffer, orient=orient, lines=lines, **self.to_json_kwargs)
else:
if compression:
diff --git a/src/datasets/io/parquet.py b/src/datasets/io/parquet.py
--- a/src/datasets/io/parquet.py
+++ b/src/datasets/io/parquet.py
@@ -1,6 +1,7 @@
import os
from typing import BinaryIO, Optional, Union
+import fsspec
import numpy as np
import pyarrow.parquet as pq
@@ -112,18 +113,20 @@ def __init__(
dataset: Dataset,
path_or_buf: Union[PathLike, BinaryIO],
batch_size: Optional[int] = None,
+ storage_options: Optional[dict] = None,
**parquet_writer_kwargs,
):
self.dataset = dataset
self.path_or_buf = path_or_buf
self.batch_size = batch_size or get_writer_batch_size(dataset.features)
+ self.storage_options = storage_options or {}
self.parquet_writer_kwargs = parquet_writer_kwargs
def write(self) -> int:
batch_size = self.batch_size if self.batch_size else config.DEFAULT_MAX_BATCH_SIZE
if isinstance(self.path_or_buf, (str, bytes, os.PathLike)):
- with open(self.path_or_buf, "wb+") as buffer:
+ with fsspec.open(self.path_or_buf, "wb", **(self.storage_options or {})) as buffer:
written = self._write(file_obj=buffer, batch_size=batch_size, **self.parquet_writer_kwargs)
else:
written = self._write(file_obj=self.path_or_buf, batch_size=batch_size, **self.parquet_writer_kwargs)
| diff --git a/tests/io/test_csv.py b/tests/io/test_csv.py
--- a/tests/io/test_csv.py
+++ b/tests/io/test_csv.py
@@ -1,6 +1,7 @@
import csv
import os
+import fsspec
import pytest
from datasets import Dataset, DatasetDict, Features, NamedSplit, Value
@@ -162,3 +163,13 @@ def test_dataset_to_csv_invalidproc(csv_path, tmp_path):
dataset = CsvDatasetReader({"train": csv_path}, cache_dir=cache_dir).read()
with pytest.raises(ValueError):
CsvDatasetWriter(dataset["train"], output_csv, num_proc=0)
+
+
+def test_dataset_to_csv_fsspec(dataset, mockfs):
+ dataset_path = "mock://my_dataset.csv"
+ writer = CsvDatasetWriter(dataset, dataset_path, storage_options=mockfs.storage_options)
+ assert writer.write() > 0
+ assert mockfs.isfile(dataset_path)
+
+ with fsspec.open(dataset_path, "rb", **mockfs.storage_options) as f:
+ assert f.read()
diff --git a/tests/io/test_json.py b/tests/io/test_json.py
--- a/tests/io/test_json.py
+++ b/tests/io/test_json.py
@@ -268,3 +268,12 @@ def test_dataset_to_json_compression(self, shared_datadir, tmp_path_factory, ext
with fsspec.open(original_path, "rb", compression="infer") as f:
original_content = f.read()
assert exported_content == original_content
+
+ def test_dataset_to_json_fsspec(self, dataset, mockfs):
+ dataset_path = "mock://my_dataset.json"
+ writer = JsonDatasetWriter(dataset, dataset_path, storage_options=mockfs.storage_options)
+ assert writer.write() > 0
+ assert mockfs.isfile(dataset_path)
+
+ with fsspec.open(dataset_path, "rb", **mockfs.storage_options) as f:
+ assert f.read()
diff --git a/tests/io/test_parquet.py b/tests/io/test_parquet.py
--- a/tests/io/test_parquet.py
+++ b/tests/io/test_parquet.py
@@ -1,3 +1,4 @@
+import fsspec
import pyarrow.parquet as pq
import pytest
@@ -213,3 +214,13 @@ def test_dataset_to_parquet_keeps_features(shared_datadir, tmp_path):
)
def test_get_writer_batch_size(feature, expected):
assert get_writer_batch_size(feature) == expected
+
+
+def test_dataset_to_parquet_fsspec(dataset, mockfs):
+ dataset_path = "mock://my_dataset.csv"
+ writer = ParquetDatasetWriter(dataset, dataset_path, storage_options=mockfs.storage_options)
+ assert writer.write() > 0
+ assert mockfs.isfile(dataset_path)
+
+ with fsspec.open(dataset_path, "rb", **mockfs.storage_options) as f:
+ assert f.read()
| Support `fsspec` in `Dataset.to_<format>` methods
Supporting this should be fairly easy.
Requested on the forum [here](https://discuss.huggingface.co/t/how-can-i-convert-a-loaded-dataset-in-to-a-parquet-file-and-save-it-to-the-s3/48353).
| Hi @mariosasko unless someone's already working on it, I guess I can tackle it!
Hi! Sure, feel free to tackle this.
#self-assign
I'm assuming this should just cover `to_csv`, `to_parquet`, and `to_json`, right? As `to_list` and `to_dict` just return Python objects, `to_pandas` returns a `pandas.DataFrame` and `to_sql` just inserts into a SQL DB, is that right? | 2023-07-28T16:36:59Z | [] | [] |
huggingface/datasets | 6,105 | huggingface__datasets-6105 | [
"6100"
] | b20f6a82410dd47e89585bb932616a22e0eaf2e6 | diff --git a/src/datasets/data_files.py b/src/datasets/data_files.py
--- a/src/datasets/data_files.py
+++ b/src/datasets/data_files.py
@@ -332,8 +332,9 @@ def resolve_pattern(
fs, _, _ = get_fs_token_paths(pattern, storage_options=storage_options)
fs_base_path = base_path.split("::")[0].split("://")[-1] or fs.root_marker
fs_pattern = pattern.split("::")[0].split("://")[-1]
- protocol_prefix = fs.protocol + "://" if fs.protocol != "file" else ""
files_to_ignore = set(FILES_TO_IGNORE) - {xbasename(pattern)}
+ protocol = fs.protocol if isinstance(fs.protocol, str) else fs.protocol[0]
+ protocol_prefix = protocol + "://" if protocol != "file" else ""
matched_paths = [
filepath if filepath.startswith(protocol_prefix) else protocol_prefix + filepath
for filepath, info in fs.glob(pattern, detail=True).items()
| diff --git a/tests/test_data_files.py b/tests/test_data_files.py
--- a/tests/test_data_files.py
+++ b/tests/test_data_files.py
@@ -5,6 +5,7 @@
import fsspec
import pytest
+from fsspec.registry import _registry as _fsspec_registry
from fsspec.spec import AbstractFileSystem
from datasets.data_files import (
@@ -346,6 +347,21 @@ def test_resolve_pattern_in_dataset_repository_special_base_path(tmpfs):
assert len(resolved_data_files) == 1
[email protected]
+def dummy_fs():
+ DummyTestFS = mock_fs(["train.txt", "test.txt"])
+ _fsspec_registry["mock"] = DummyTestFS
+ _fsspec_registry["dummy"] = DummyTestFS
+ yield
+ del _fsspec_registry["mock"]
+ del _fsspec_registry["dummy"]
+
+
+def test_resolve_pattern_fs(dummy_fs):
+ resolved_data_files = resolve_pattern("mock://train.txt", base_path="")
+ assert resolved_data_files == ["mock://train.txt"]
+
+
@pytest.mark.parametrize("pattern", _TEST_PATTERNS)
def test_DataFilesList_from_patterns_in_dataset_repository_(
hub_dataset_repo_path, hub_dataset_repo_patterns_results, pattern
@@ -478,7 +494,7 @@ def mock_fs(file_paths: List[str]):
]
class DummyTestFS(AbstractFileSystem):
- protocol = "mock"
+ protocol = ("mock", "dummy")
_fs_contents = fs_contents
def ls(self, path, detail=True, refresh=True, **kwargs):
@@ -495,7 +511,7 @@ def ls(self, path, detail=True, refresh=True, **kwargs):
return files
return [file["name"] for file in files]
- return DummyTestFS()
+ return DummyTestFS
@pytest.mark.parametrize(
@@ -570,7 +586,8 @@ def ls(self, path, detail=True, refresh=True, **kwargs):
def test_get_data_files_patterns(data_file_per_split):
data_file_per_split = {k: v if isinstance(v, list) else [v] for k, v in data_file_per_split.items()}
file_paths = [file_path for split_file_paths in data_file_per_split.values() for file_path in split_file_paths]
- fs = mock_fs(file_paths)
+ DummyTestFS = mock_fs(file_paths)
+ fs = DummyTestFS()
def resolver(pattern):
return [file_path for file_path in fs.glob(pattern) if fs.isfile(file_path)]
| TypeError when loading from GCP bucket
### Describe the bug
Loading a dataset from a GCP bucket raises a type error. This bug was introduced recently (either in 2.14 or 2.14.1), and appeared during a migration from 2.13.1.
### Steps to reproduce the bug
Load any file from a GCP bucket:
```python
import datasets
datasets.load_dataset("json", data_files=["gs://..."])
```
The following exception is raised:
```python
Traceback (most recent call last):
...
packages/datasets/data_files.py", line 335, in resolve_pattern
protocol_prefix = fs.protocol + "://" if fs.protocol != "file" else ""
TypeError: can only concatenate tuple (not "str") to tuple
```
With a `GoogleFileSystem`, the attribute `fs.protocol` is a tuple `('gs', 'gcs')` and hence cannot be concatenated with a string.
### Expected behavior
The file should be loaded without exception.
### Environment info
- `datasets` version: 2.14.1
- Platform: macOS-13.2.1-x86_64-i386-64bit
- Python version: 3.10.12
- Huggingface_hub version: 0.16.4
- PyArrow version: 12.0.1
- Pandas version: 2.0.3
| Thanks for reporting, @bilelomrani1.
We are fixing it. | 2023-07-31T11:44:46Z | [] | [] |