markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
Run the evaluation stepUse the checkpointed model to run the evaluation step. | estimator_eval = RLEstimator(
role=role,
source_dir="src/",
dependencies=["common/sagemaker_rl"],
toolkit=RLToolkit.COACH,
toolkit_version="1.0.0",
framework=RLFramework.TENSORFLOW,
entry_point="evaluate-coach.py",
instance_count=1,
instance_type=instance_type,
base_job_name=job_name_prefix + "-evaluation",
hyperparameters={
"RLCOACH_PRESET": "preset-tsp-easy",
"evaluate_steps": 200, # max 4 episodes
},
)
estimator_eval.fit({"checkpoint": checkpoint_path}) | _____no_output_____ | Apache-2.0 | reinforcement_learning/rl_traveling_salesman_vehicle_routing_coach/rl_traveling_salesman_vehicle_routing_coach.ipynb | Amirosimani/amazon-sagemaker-examples |
Medium version of TSP We make the problem much harder in this version by randomizing the location of destiations each episode. Hence, RL agent has to come up with a general strategy to navigate the grid. Parameters, states, actions, and rewards are identical to the Easy version of TSP. StatesAt each time step, our agent is aware of the following information:1. For the Restuarant: 1. Location (x,y coordinates) 2. For the Driver 1. Location (x,y coordinates) 2. Is driver at restaurant (yes/no)3. For each Order: 1. Location (x,y coordinates) 2. Status (Delivered or Not Delivered) 3. Time (Time taken to deliver reach order -- incrementing until delivered)4. Miscellaneous 1. Time since start of episode 2. Time remaining until end of episode (i.e. until max time) ActionsAt each time step, our agent can take the following steps:- Up - Move one step up in the map- Down - Move one step down in the map- Right - Move one step right in the map- Left - Move one step left in the map RewardsAgent gets a reward of -1 for each time step. If an order is delivered in that timestep, it gets a positive reward inversely proportional to the time taken to deliver. If all the orders are delivered and the agent is back to the restaurant, it gets an additional reward inversely proportional to time since start of episode. Using AWS SageMaker for RL Train the model using Python SDK/Script modeSkipping through the basic setup, assuming you did that already for the easy version. For good results, we suggest you train for at least 1,000,000 steps. You can edit this either as a hyperparameter in the cell or directly change the preset file. | %%time
# run in local mode?
local_mode = False
# create unique job name
job_name_prefix = "rl-tsp-medium"
%%time
if local_mode:
instance_type = "local"
else:
instance_type = "ml.m4.4xlarge"
estimator = RLEstimator(
entry_point="train-coach.py",
source_dir="src",
dependencies=["common/sagemaker_rl"],
toolkit=RLToolkit.COACH,
toolkit_version="1.0.0",
framework=RLFramework.TENSORFLOW,
role=role,
instance_type=instance_type,
instance_count=1,
output_path=s3_output_path,
base_job_name=job_name_prefix,
hyperparameters={
"RLCOACH_PRESET": "preset-tsp-medium",
},
)
estimator.fit(wait=local_mode) | _____no_output_____ | Apache-2.0 | reinforcement_learning/rl_traveling_salesman_vehicle_routing_coach/rl_traveling_salesman_vehicle_routing_coach.ipynb | Amirosimani/amazon-sagemaker-examples |
Visualize, Compare with Baseline and EvaluateYou can follow the same set of code used for TSP easy version. Vehicle Routing Problem with Reinforcement Learning Vehicle Routing Problem (VRP) is a similar problem where the algorithm optimizes the movement of a fleet of vehicles. Our VRP formulation is a bit different, we have a delivery driver who accepts orders from customers, picks up food from a restaurant and delivers it to the customer. The driver optimizes to increase the number of successful deliveries within a time limit.Key differences from TSP: - Pathing is now automatic. Instead of choosing "left, right, up, down", now you just select your destination as your action. The environment will get you there in the fewest steps possible.- Since the routing/pathing is now taken care of, we add in complexity elsewhere...- There can be more than one restaurant, each with a different type of order (e.g. Pizzas vs. Burritos). Each order will have a different type, and you have to visit the correct restuarant to pick up an order before dropping it off.- Drivers have a limited capacity; they cannot pick up an infinite number of orders. Instead, they can only have (e.g. 5) orders in the car at any given time. This means they will have to return to the restaurant(s) in between deliveries to pick up more supply.- Orders now come in dynamically over time, rather than all being known at time zero. Each time step, there is some probability that an order will be generated. - As the driver/agent, we now have the choice to fulfill an order or not -- there's no penalty associated with not accepting an order, but a potential penalty if we accept an order and fail to deliver it before Timeout. StatesAt each time step, our agent is aware of the following information:1. For each Restuarant: 1. Location (x,y coordinates) 1. For the Driver 1. Location (x,y coordinates) 2. Capacity (maximum of orders you can carry, at one time, on the driver) 3. Used Capacity ( of orders you currently carry on the driver)1. For each Order: 1. Location (x,y coordinates) 2. Status (Accepted, Not Accepted, Delivered, Not Delivered) 3. Type (Which restuarant the order belongs to, like Pizza, or Burrito) 4. Time (Time since order was generated) 5. Promise (If you deliver the order by this time, you get a bonus reward) 6. Timeout (If you deliver the order after this time, you get a penalty) ActionsAt each time step, our agent can do ONE of the following:- Choose a restaurant to visit (incremental step L,R,U,D, will be auto-pathed)- Choose an order to visit (incremental step L,R,U,D, will be auto-pathed)- Accept an order (no movement will occur)- Do nothing Rewards- Driver gets a reward of -1 for each time step. - If driver delivers order, get a reward proportional to the time taken to deliver (extra bonus for beating Promise time)- If order expires (reaches Timeout), get a penalty Using AWS SageMaker RL Train the model using Python SDK/Script modeSkipping through the basic setup, assuming you did that already for the easy version. For good results, we suggest a minimum of 5,000,000 steps of training. | %%time
# run in local mode?
local_mode = False
# create unique job name
job_name_prefix = "rl-vrp-easy"
%%time
if local_mode:
instance_type = "local"
else:
instance_type = "ml.m4.4xlarge"
estimator = RLEstimator(
entry_point="train-coach.py",
source_dir="src",
dependencies=["common/sagemaker_rl"],
toolkit=RLToolkit.COACH,
toolkit_version="1.0.0",
framework=RLFramework.TENSORFLOW,
role=role,
instance_type=instance_type,
instance_count=1,
output_path=s3_output_path,
base_job_name=job_name_prefix,
hyperparameters={
"RLCOACH_PRESET": "preset-vrp-easy",
},
)
estimator.fit(wait=local_mode) | _____no_output_____ | Apache-2.0 | reinforcement_learning/rl_traveling_salesman_vehicle_routing_coach/rl_traveling_salesman_vehicle_routing_coach.ipynb | Amirosimani/amazon-sagemaker-examples |
!pip install tf-nightly-2.0-preview
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
print(tf.__version__)
dataset = tf.data.Dataset.range(10)
for val in dataset:
print(val.numpy())
dataset = tf.data.Dataset.range(10)
dataset = dataset.window(5, shift=1)
for window_dataset in dataset:
for val in window_dataset:
print(val.numpy(), end=" ")
print()
dataset = tf.data.Dataset.range(10)
dataset = dataset.window(5, shift=1, drop_remainder=True)
for window_dataset in dataset:
for val in window_dataset:
print(val.numpy(), end=" ")
print()
dataset = tf.data.Dataset.range(10)
dataset = dataset.window(5, shift=1, drop_remainder=True)
dataset = dataset.flat_map(lambda window: window.batch(5))
for window in dataset:
print(window.numpy())
dataset = tf.data.Dataset.range(10)
dataset = dataset.window(5, shift=1, drop_remainder=True)
dataset = dataset.flat_map(lambda window: window.batch(5))
dataset = dataset.map(lambda window: (window[:-1], window[-1:]))
for x,y in dataset:
print(x.numpy(), y.numpy())
dataset = tf.data.Dataset.range(10)
dataset = dataset.window(5, shift=1, drop_remainder=True)
dataset = dataset.flat_map(lambda window: window.batch(5))
dataset = dataset.map(lambda window: (window[:-1], window[-1:]))
dataset = dataset.shuffle(buffer_size=10)
for x,y in dataset:
print(x.numpy(), y.numpy())
dataset = tf.data.Dataset.range(10)
dataset = dataset.window(5, shift=1, drop_remainder=True)
dataset = dataset.flat_map(lambda window: window.batch(5))
dataset = dataset.map(lambda window: (window[:-1], window[-1:]))
dataset = dataset.shuffle(buffer_size=10)
dataset = dataset.batch(2).prefetch(1)
for x,y in dataset:
print("x = ", x.numpy())
print("y = ", y.numpy())
| x = [[0 1 2 3]
[4 5 6 7]]
y = [[4]
[8]]
x = [[2 3 4 5]
[3 4 5 6]]
y = [[6]
[7]]
x = [[5 6 7 8]
[1 2 3 4]]
y = [[9]
[5]]
| Apache-2.0 | S+P_Week_2_Lesson_1.ipynb | EgorBEremeev/SoloLearmML-coursera-deeplearning.ai |
|
MEDC0106: Bioinformatics in Applied Biomedical Science --------------------------------------------------------------- 06 - Introduction to Pandas*Written by:* Oliver Scott**This notebook provides a general introduction to Pandas.**Do not be afraid to make changes to the code cells to explore how things work! What is Pandas?**[Pandas](https://pandas.pydata.org/)** is a Python package for data analysis, providing functions for analysing, cleaning and manipulating data. Pandas is probably one of the most important tools for data scientists and is the backbone of most data science projects using Python.Pandas is built on top of NumPy, hence the Numpy structure is used a lot in the Pandas interface. Data manipulation often prefaces further analysis using other Python packages such as statistical analysis using [SciPy](https://www.scipy.org/), visualisation using tools such as [Matplotlib](https://matplotlib.org/) and machine learning using [scikit-learn](https://scikit-learn.org/stable/). These tools and others make up the Python scientific stack and are essential to learn for a career in informatics or data-science. To be effective in pandas it is essential to have a good grasp of the core concepts in Python (these concepts are outlined in the first session) along with some familiarity with NumPy. If you get lost with some concepts it might be a good idea to take a look through the previous material across the sessions.In this notebook we will learn the basics of Pandas. Pandas is a huge package and is deserving of an entire lecture series itself, so here we will learn tyhe fundamentals from which you will be able to build upon if you want to learn more.----- Contents1. [The Basics](The-Basics)2. [Creating DataFrames](Creating-DataFrames)3. [Reading Data](Reading-Data)4. [Essential Operations](Essential-Operations)5. [Slicing and Selecting](Slicing-and-Selecting)5. [Arithmetic](Arithmetic)6. [Applying Functions](Applying-Functions)7. [Time-Series](Time-Series)8. [Plotting](Plotting)----- Extra Resources:- [Pandas Getting Started Guide](https://pandas.pydata.org/pandas-docs/stable/getting_started/index.html)- [RealPython-01](https://realpython.com/pandas-python-explore-dataset/)- [RealPython-02](https://realpython.com/pandas-dataframe/)----- References:- [Pandas Documentation](https://pandas.pydata.org/docs/)- [Learn Data Science](https://www.learndatasci.com/tutorials/python-pandas-tutorial-complete-introduction-for-beginners/)----- The BasicsImporting Pandas is no different to any other package/module. Pandas users often use the `pd` alias to keep code clean: | import pandas as pd
s = pd.Series([1.0, 2.0, 3.0, 5.0])
s | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Core ComponentsPandas has two core components, the `Series` and the `DataFrame`.The `Series` can be imagined as a single column in a data table, whereas the `DataFrame` can be imagined as a full data table made up of multiple `Series`. Both types have a similar interface allowing a user to perform similar operations. DataFrames are similar to spreadsheets that you may have interacted with in software such as Excel. DataFrames are often faster, easier to use and more powerful than spreadsheets. [Image source](https://www.datasciencemadesimple.com/wp-content/uploads/2020/05/create-series-in-python-pandas-0.png?ezimgfmt=rs%3Adevice%2Frscb1-1)----- Creating DataFramesThere are numerous ways to create a DataFrame using the Pandas package. In most cases it is likely that you will want to read in data from a paticular file, however DataFrames can also be constructed from scratch from lists, tuples, NumPy arrays or Pandas series. Probably the most simple way however is from a simple Python dictionary `dict`. Suppose we wanted to construct a table like the one below:| PatientID | Gender | Age | Outcome ||-----------|--------|-----|----------|| 556785 | M | 19 | Negative || 998764 | F | 38 | Positive || 477822 | M | 54 | Positive || 678329 | M | 22 | Negative || 675859 | F | 41 | Negative |We can construct this using a Python dictionary where the key corresponds to the column name and the list the data present in the rows. For this we can use the default constructor `pd.DataFrame()`. Notice how there is also an unnamed column containing the numbers 0-4, this is the **index** of each row. In fact you may also specify a custom index when contructing a dataframe; (`pd.DataFrame(data, index=['Tom', 'Joanne', 'Joe', 'Xander', 'Selena'])`) In this case the index is the names of the patients. | # This is or dictionary containing the raw data
data = {
'PatientID': [556785, 998764, 477822, 678329, 675859],
'Gender': ['M', 'F', 'M', 'M', 'F'],
'Age': [19, 38, 54, 22, 41],
'Outcome': ['Negative', 'Poisitive', 'Positive', 'Negative', 'Negative']
}
# We can now construct a DataFrame like so:
df = pd.DataFrame(data)
df # show the data | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Often you will be working with very large tables of data making it impractical to view the whole table. Pandas provides a method `.head()` to display the first few n items or `.tail()` for the last few: | # Display the first three rows
df.head(n=3)
# Display the last two rows
df.tail(n=2) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Accessing an individual column is easy using the same syntax as a Python dictionary `dict`: | gender_column = df['Gender']
gender_column | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
If the column label is a string you may also use **dot-syntax** to access the column: | age_column = df.Age
age_column | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Reading DataReading and writing data from/to files in multiple formats is an essential part of the data analysis pipeline. Pandas can read data from file including; CSV, JSON, Excel, SQL and [many more](https://pandas.pydata.org/pandas-docs/stable/reference/io.html).In the folder `data` we have provided a dataset downloaded from the [UK government](https://coronavirus.data.gov.uk/details/cases?areaType=overview&areaName=United%20Kingdom) detailing the number of reported positive COVID-19 test results in the United Kingdom by date reported (up to Oct-31-21). The file is in the CSV format and can be read using Pandas with the function `.read_csv()`: | cv_data_path = './data/data_2021-Oct-31.csv' # This is the path to our data
cv_data = pd.read_csv(cv_data_path)
cv_data.head(n=10) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We could also easily write this DataFrame to a new CSV file using the method `df.to_csv()`:```pythoncv_data.to_csv('./data/coronavirus_testing_results.csv')```Give it a go. Maybe also saving to a different [format](https://pandas.pydata.org/pandas-docs/stable/reference/io.html)! Essential OperationsNow that we have loaded some data into a `DataFrame` we can perform operations for performing analysis. Typically once you have loaded some data you should view your data to make sure that it looks correct and to get an idea of what values you will be dealing with. Since we have already coovered visualising the data using `.head()`/`.tail()`, the next function you should probbaly run is `.info()` which provide essential details about your dataset including the number of rows/columns the number of none-null values (None), what type of data is in each column and how much memory the data is taking up: | cv_data.info() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Notice that we have 6 columns of which four are of type `object` (this could be something like a string) and two that are `int64` (integers) (these types correspond the the types used in NumPy). The info also tells us that we have 2466 non-null values and no null-values in this case. Knowing the datatype of ourt columns is very important as it will determine what operations we can perform on each column (we wouldnt want to calculate the mean of a column containing strings). Just like NumPy you can also use `.shape` to see the number of (rows/columns): | cv_data.shape | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Removing duplicate dataOften input data is noisy and needs cleaning up before we do any further analysis. It is often the case that data contains duplicated rows which is not great when we are trying to do statitical analysis. Luckily Pandas has utilities for dealing with this problem easily. The data we have read does not contain any duplicated rows so we will arbritrarily create some by duplicating the data and adding it to itself: | duplicated = cv_data.append(cv_data) # here we have copied the data and added it to itself
duplicated.shape | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Notice that we have to assign the result of the `append` to a new variable. Here we have copied the data so we wont do anything to the original DataFrame. We can now easily drop the duplicates using the `.drop_duplicates()` [method](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.drop_duplicates.html). It is always a good idea to look at the [documentation](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.drop_duplicates.html) to see what other arguments these functions accept. | duplicated = duplicated.drop_duplicates()
duplicated.shape | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Notice that the shape is now the same as the original data. Also notice that again we assigned the result to a new variable (with the same name). This technique can get quite annoying so Pandas often offers an argument `inplace` which if we set to `True` allows pandas to perform the operation modifying the original data rather than a copy.```pythonduplicated.drop_duplicates(inplace=True) no need to assign to a new variable``` Removing Null values (None)Data before cleaning commonly has missing values that you will need to deal with before further analysis. Missing values are represented by `None` or `np.nan`. There is usually two options to dealing with missing values:1. Remove all rows with missing data2. Imputing the missing valuesIn this tutorial we will stick to the first case.Again as our data is nice and clean it contains no null values so for this example we will inject a new column containing some null values, let's do this first: | import numpy as np
n_rows = cv_data.shape[0]
# p is for weighting the choice here it is more likely to choose 1 than None
null_containing_data = np.random.choice([None, 1], n_rows, p=[0.2, 0.8])
null_containing_data | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Now add a row to the data containing our constructed data: | cv_data['RandomData'] = null_containing_data # make a colum called 'RandomData'
cv_data.head(10) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We can also see now that we have null values: | cv_data.info() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We can also check the number of null values in each column using `.isnull()`. This returns a dataframe with boolean columns where `True` indicated a null value. We can then use `.sum()` to count the number of `True` values in each column: | cv_data.isnull().sum() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
When performing data analysis you often have to make the choice to remove missing values or impute them in some way. Removing data is only really recommended if the number of missing data points is relatively small. To remove null values you can simply use the [method](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.dropna.html) `.dropna()`. This operation will remove any row with at least 1 null value, returning a new DataFrame unless you specified inplace. Instead of dropping rows we could instead drop columns with null values by changing the axis of operation. Columns are represented by `axis=1` (The axes are defined in the same way as NumPy!): | # First lets remove rows with null values
remove_rows = cv_data.dropna()
remove_rows.head(10)
remove_rows.shape | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Now lets change the axis and remove the colum we injected: | cv_data.dropna(axis=1, inplace=True) # We can do it inplace since we do not care about this column
cv_data.head(10) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Understanding DataNow that your data is clean(er) than when we started, it is time to do some basic stats to understand the data that we have in each column. This may help inform us how to continue with our analysis and maybe how to plot the data. Pandas provides us with an easy way to get a quick summary of the distribution of our continuous variables `.describe()`: | cv_data.describe() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We can also do the same for categorical columns but we will have to do it seperately: | cv_data.areaName.describe() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
This shows us that in this dataset there are four unique area names with 'England' being the most frequent with a frequency of 640. We can also check the unique values using the `.unique()` method: | cv_data.areaName.unique() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We can see that the dataset contains data for:- England- Northern Ireland- Scotland- WalesBut how many times are these values recorded? We can use the method `.value_counts()` to find out: | cv_data.areaName.value_counts() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Slicing and SelectingIn the previous section we saw how to produce summaries of the entire data which is useful however, sometimes we will want to perform analysis on certain subsets of data. We have already seen how to extract a column of data using square brackets and dot-syntax `df['col'] / df.col` and now we will dive deeper into the Pandas selction language. When selecting parts of a DataFrame we may be returning either a `DataFrame` or a `Series`, it is important to know which so that yopu use the correct syntax. Selecting by Column(s)Using the square-bracket syntax we mentioned previously will return a Pandas `Series` | type(cv_data['areaName']) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
If you wish to access it as a dataframe you can supply the column name as a list: | type(cv_data[['areaName']]) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Adding another column to our selection is a simple as adding another column name to the list. Obviosuly inh this case our code will return a `DataFrame`: | selection = cv_data[['areaName', 'areaCode']]
selection.head(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Selecting by rowsSelecting rows is a little trickier with two methods:- `.loc`: locate by name- `.iloc`: locate by numerical indexConsidering that our data has a numerical index it makes sense for us to use `.iloc`. If our data has an index using strings `.loc` would be the correct solution if we want to select using the string. Of course `.iloc` will also work returning the data at the numerical position instead of the name.Both methods are similar to indexing lists or NumPy arrays: | cv_data.loc[222] # Return the row with index 222 | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Since Pandas is backed by NumPy we can also use slices to select a range of data: | cv_data.loc[222:226] | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Conditional SelectionsSelecting data by index can be useful, but if we do not know what dat the indexes correspond too this can be limiting. Perhaps we are only interested in the data from Wales, we can use conditional selections to make informed selections.Pandas like numpy can be indexed using a boolean array/Series/DataFrame generated using a conditional expression: | ind = cv_data['areaName'] == 'Wales' # A boolean Series
ind.tail(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Using this boolean Series we can index the DataFrame! | wales_data = cv_data[ind]
wales_data.head(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We can simplify this quite nicely into a one line expression: | wales_data = cv_data[cv_data['areaName'] == 'Wales']
wales_data.head(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Of course we can apply this to numerical columns also: | # Select rows where reported positives is less than 100
cv_data[cv_data['newCasesByPublishDate'] < 100].head(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Chaining conditional expressions allows us to create powerful selections. For this we can use the logical operators `|` and `&`. Remeber to put seperate conditions in brackets! | # Count dates in england with reported positive results > 10,000
cv_data[(cv_data['areaName'] == 'England') & (cv_data['newCasesByPublishDate'] > 10000)].shape | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
ArithmeticBasic arithmentic operations can be applied in the same way as NumPy arrays, so we will quickly brush over it: | cv_data.newCasesByPublishDate / 100 # divide a column by 100 return a Series | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
You may also perform arithmetic between columns: | cv_data.newCasesByPublishDate + cv_data.cumCasesByPublishDate | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
You can insert a new column with the result: | cv_data['Rubbish'] = cv_data.newCasesByPublishDate * 0.3 / cv_data.cumCasesByPublishDate
cv_data.head(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Pandas also provides some handy utility functions: | print(cv_data.newCasesByPublishDate.mean())
print(cv_data.newCasesByPublishDate.std()) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Applying FunctionsWhile it is possible to iterate over a DataFrame/Series like a NumPy array, it is slow in Python so instead we can use the `.apply()` function to apply a function to each element in a column or across columns. We can also save this result to a new column. Let's create an arbritary function that we can apply to the data we have: | def categorize_cases(x):
if x >= 10000:
return 'High'
elif x <= 200:
return 'Low'
else:
return 'Medium' | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
The above function categorises a case count into arbritarty categories: 'High', 'Medium' and 'Low'. Now we can apply this to the column 'newCasesByPublishDate': | cv_data['Category'] = cv_data['newCasesByPublishDate'].apply(categorize_cases)
cv_data.head(10) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Users often will use anonymous functions instead of defining an explicit function like above: | cv_data['newCategory'] = cv_data['newCasesByPublishDate'].apply(lambda x: 'Red' if x >= 20000 else 'Amber')
cv_data.head(10) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Time-SeriesSome of you may have notices that one of the columns contains dates as a string (object). This isnt paticularly useful to us in this form. Pandas however has a datetime type which we can use to make some more intelligent selections based on time spans. First we need to tell pandas that our column is a datetime column: | cv_data['date'] = pd.to_datetime(cv_data['date'])
cv_data.info() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Now we have the date in this form we can make selections within time ranges using the `.between()` method: | # Lets select data between the 20th and the 30th October 2021 and restrict it to England
selection = cv_data[(cv_data.date.between('2021-10-20','2021-10-30')) & (cv_data.areaName == 'England')]
selection.head(10) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Working with time-series data is even more powerful if we use the time as our index. Lets first only consider 'Scotland' in our analysis | scotland_data = pd.DataFrame(cv_data[cv_data.areaName == 'Scotland']) # also copy into a new DataFrame | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Now we can set the index of the 'scotland_data' DataFrame as the index: | scotland_data.set_index('date', inplace=True)
scotland_data.head(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
You may have noticed that the data is in time-decending order, often we we will want to reverse this ordering. Now that the index is the date we can sort it easilt using the `.sort_index()` method: | scotland_data.sort_index(inplace=True)
scotland_data.head(5) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Also we can simply use slicing to select a data range with `.loc`! | scotland_data.loc['2021-10-20':'2021-10-30'] | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We can resample time-series data into different intervals and get a mean value for that interval. Below we resmaple the data into 10-day intervals and calculate the mean of 'newCasesByPublishDate': | scotland_data.resample(rule='10d')['newCasesByPublishDate'].mean() | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Instead of mean you could use other functions such as `min()`, `max()`, `sum()` etc. Indeed you can also calculate a rolling statistic using `.rolling()` and a window size. Here we will calculate a rolling average using a ten day window: | scotland_data['rollingAvgTenDay'] = scotland_data.rolling(10)['newCasesByPublishDate'].mean()
scotland_data.head(20) | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
PlottingPandas allows the visualisation of data in DataFrames/Series interfacing with the plotting package [matplotlib](https://matplotlib.org/). Displaying the plots will first require that import matplotlib: | # We also add this 'Jupyter magic' to display plots in the notebook.
%matplotlib inline
import matplotlib.pyplot as plt | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Now creating a plot with pandas is as simple as calling `.plot()` on some selected data! | scotland_data.newCasesByPublishDate.plot(); # We also add the semicolon when plotting in Jupyter | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We could have also achieved the same result using the syntax:```pythonscotland_data.newCasesByPublishDate.plot.line()```These plotting functions also have many [arguments](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.plot.html) which you can specify to tune the look of your plots. Thes arguments are passed to the underlying matplotlib methods. We can also specify other types of plot. For example we could visualise the data as a box plot, do you notice anything unusual?: | # Select a time window (1-month)
window = scotland_data['2021-09-30':'2021-10-30']
window.newCasesByPublishDate.plot.box(); | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
How about we plot the raw data along with the 10 day rolling average: | scotland_data.newCasesByPublishDate.plot(figsize=(12, 8)) # also specify the size!
scotland_data.rollingAvgTenDay.plot()
plt.legend(); # We can also add a legend using matplotlib! | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
We can also save figures using `.savefig()`, check the data directory! | figure = scotland_data.newCasesByPublishDate.plot(figsize=(12, 8)).get_figure()
figure.savefig('./data/Scotland_2021-Oct-31.png'); | _____no_output_____ | CC-BY-4.0 | workshop/session_2/06_introduction_to_pandas.ipynb | MEDC0106/PythonWorkshop |
Introduction to GalSim HubAuthors: [@EiffL](https://github.com/EiffL)This notebook contains a short introduction to using GalSim-Hub for samplingrandomly generated galaxy light profiles, and drawing them using GalSim. Setting up the environmentBesides GalSim, GalSim-Hub requires TensorFlow (version 1.15, for stability reasons), and TensorFlow Hub. In a Colab environment, TensorFlow is already installed, so we only need to install GalSim, using some conda magic. | # Activating TensorFlow v1.15 environment on Colab
%tensorflow_version 1.x
# Installing and updating conda for Python 3.6
!wget https://repo.continuum.io/miniconda/Miniconda3-4.5.4-Linux-x86_64.sh && bash Miniconda3-4.5.4-Linux-x86_64.sh -bfp /usr/local
!conda install --channel defaults conda python=3.6 --yes
!conda update --channel defaults --all --yes
# Installing GalSim
!conda install -y -q -c conda-forge galsim=2.2.4=py36hbfbe4e9_0
# Adding conda packages to Python path
import sys
sys.path.append('/usr/local/lib/python3.6/site-packages') | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
And finally, installing GalSim Hub itself: | !pip install --no-deps git+https://github.com/McWilliamsCenter/galsim_hub.git | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
Loading and Using generative models from PythonThe first step is to load a generative model. This is done by creating an instance of the `galsim_hub.GenerativeGalaxyModel` class, either by providing a local path to a model directory, or much more conveniently, byusing the `hub:xxxxxx` syntax, where `xxxxxx` is the name of a published model, hosted on the GalSim Hub repository, see [here](https://github.com/McWilliamsCenter/galsim_hub/tree/master/hub).As an example, we will load the generative model presented in Lanusse et al. 2020: | import galsim
import galsim_hub
model = galsim_hub.GenerativeGalaxyModel('hub:Lanusse2020') | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
Behind the scene, the generative model has been downloaded from the repository,and is now ready to use.Models can be conditional, i.e. generating a light profile given some particular attributes as inputs. To introspect the model, and see what inputs are expected, you can use the `quantities` attribute: | model.quantities | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
We see that this model generates light profiles, given a particular magnitude, redshift, and size.Other interesting properties saved with the model (but knowledge of which is not necessary) are the native stamp size, and native pixel size at which the generative model is producing light profiles: | # Pixel size, in arcsec
model.pixel_size
# Stamp size
model.stamp_size | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
Now that the model is loaded, and that we know what inputs it expects, we can create a catalog listing some desired input quantities: | from astropy.table import Table
catalog = Table([[5., 10. ,20.], [24., 24., 24.], [0.5, 0.5, 0.5] ],
names=['flux_radius', 'mag_auto', 'zphot']) | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
In this example, we want 3 galaxies, all at the same i-band magnitude of 24, and redshift 0.5, but with different and increasing size.We can now sample actual GalSim light profiles with those properties from the model using the `sample()` method: | # Sample light profiles for these parameters
profiles = model.sample(catalog) | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
This returns a list of 3 profiles, represented by `galsim.InterpolatedImage` objects: | profiles[0] | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
**These objects can now be manipulated inside GalSim as any other light profile.**For instance, we can convolve these images with a PSF and add some observational noise: | %pylab inline
PSF = galsim.Gaussian(fwhm=0.06)
figure(figsize=(10,5))
for i in range(3):
# Convolving light profile with PSF
gal = galsim.Convolve(profiles[i], PSF)
# Drawing postage stamp of any desired size and pixel scale
im = gal.drawImage(nx=96, ny=96, scale=0.03)
# Adding some noise for realism
im.addNoise(galsim.getCOSMOSNoise())
# Drawing the image
subplot(1,3,i+1)
imshow(im.array) | Populating the interactive namespace from numpy and matplotlib
| MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
And voila! Using generative models directly from YamlGalSim Hub also provides a driver for using generative models direclty from a GalSim Yaml script. In this section, we will illustrate how to write such a script and execute it from the command line.Let's retrieve a script from the example folder of GalSim Hub: | !wget -q https://raw.githubusercontent.com/McWilliamsCenter/galsim_hub/master/examples/demo14.yaml
!cat demo14.yaml | modules:
- galsim_hub
psf :
type : Gaussian
sigma : 0.06 # arcsec
# Define the galaxy profile
gal :
type : GenerativeModelGalaxy
flux_radius : { type : Random , min : 5, max : 10 }
mag_auto : { type : Random , min : 24., max : 25. }
# The image field specifies some other information about the image to be drawn.
image :
type : Tiled
nx_tiles : 10
ny_tiles : 10
stamp_size : 64 # pixels
pixel_scale : 0.03 # arcsec / pixel
noise :
type : COSMOS
output :
dir : output_yaml
file_name : demo14.fits
# Define the input files
input :
generative_model :
file_name : 'hub:cosmos_size_mag'
| MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
Note the following points that are directly related to generative models: - In the preamble of the file, we load the `galsim_hub` module - In the galaxy section, we use the `GenerativeModelGalaxy` type, and provide some input distributions for the input quantities used by the model - In the `input` section, we provide the path to the generative model, or as in this case, only its hub tag, so that it can be automatically downloaded.We direct the interested reader to the GalSim documentation for further details on how to compose such a Yaml file.We can now execute that file from the command line: | !python /galsim demo14.yaml | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
This should generate a fits file corresponding to the Yaml description. Unfortunately, couldn't manage to get it to work yet on this hybrid conda/google environment... Suggestions welcome :-) | _____no_output_____ | MIT | notebooks/GalsimHubDemo.ipynb | Hbretonniere/galsim_hub |
|
CST_ALL_PTMs_NormalizationThis notebook will make the case for normalizing the distributions of PTM levels in all cell lines. I will combine the PTM data from all cell lines and look at the average properties of all PTM distributions in all cell lines. imports and function definitions | # imports and plotting defaults
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import matplotlib
matplotlib.style.use('ggplot')
from copy import deepcopy
# use clustergrammer module to load/process (source code in clustergrammer directory)
from clustergrammer import Network
# load data data and export as pandas dataframe: inst_df
def load_data(filename):
'''
load data using clustergrammer and export as pandas dataframe
'''
net = deepcopy(Network())
net.load_file(filename)
tmp_df = net.dat_to_df()
inst_df = tmp_df['mat']
# # simplify column names (remove categories)
# col_names = inst_df.columns.tolist()
# simple_col_names = []
# for inst_name in col_names:
# simple_col_names.append(inst_name[0])
# inst_df.columns = simple_col_names
print(inst_df.shape)
return inst_df
def plot_cl_boxplot_with_missing_data(inst_df):
'''
Make a box plot of the cell lines where the cell lines are ranked based
on their average PTM levels
'''
# get the order of the cell lines based on their mean
sorter = inst_df.mean().sort_values().index.tolist()
# reorder based on ascending mean values
sort_df = inst_df[sorter]
# box plot of PTM values ordered based on increasing mean
sort_df.plot(kind='box', figsize=(10,3), rot=90, ylim=(-4,4))
def plot_cl_boxplot_no_missing_data(inst_df):
# get the order of the cell lines based on their mean
sorter = inst_df.mean().sort_values().index.tolist()
# reorder based on ascending mean values
sort_df = inst_df[sorter]
# transpose to get PTMs as columns
tmp_df = sort_df.transpose()
# keep only PTMs that are measured in all cell lines
ptm_num_meas = tmp_df.count()
ptm_all_meas = ptm_num_meas[ptm_num_meas == 45]
ptm_all_meas = ptm_all_meas.index.tolist()
print('There are ' + str(len(ptm_all_meas)) + ' PTMs measured in all cell lines')
# only keep ptms that are measured in all cell lines
# I will call this full_df as in no missing measurements
full_df = tmp_df[ptm_all_meas]
# transpose back to PTMs as rows
full_df = full_df.transpose()
full_df.plot(kind='box', figsize=(10,3), rot=900, ylim=(-8,8))
num_ptm_all_meas = len(ptm_all_meas) | _____no_output_____ | MIT | notebooks/CST_ALL_PTMs_Normalization.ipynb | MaayanLab/cst_drug_treatment |
Load all PTM data and combine into single dataframe | filename = '../lung_cellline_3_1_16/lung_cl_all_ptm/all_ptm_ratios.tsv'
df_all = load_data(filename)
df_all.count().sort_values().plot(kind='bar', figsize=(10,2))
plot_cl_boxplot_with_missing_data(df_all) | _____no_output_____ | MIT | notebooks/CST_ALL_PTMs_Normalization.ipynb | MaayanLab/cst_drug_treatment |
Merge Plex-duplicate cell lines | filename = '../lung_cellline_3_1_16/lung_cl_all_ptm/all_ptm_ratios_uni_cl.tsv'
df_all = load_data(filename)
df_all.count().sort_values().plot(kind='bar', figsize=(10,2))
plot_cl_boxplot_with_missing_data(df_all) | _____no_output_____ | MIT | notebooks/CST_ALL_PTMs_Normalization.ipynb | MaayanLab/cst_drug_treatment |
Zscore rows | df_tmp = deepcopy(df_all)
df_tmp = df_tmp.transpose()
zdf_all = (df_tmp - df_tmp.mean())/df_tmp.std()
zdf_all = zdf_all.transpose()
print(zdf_all.shape)
zdf_all.count().sort_values().plot(kind='bar', figsize=(10,2))
plot_cl_boxplot_with_missing_data(zdf_all) | _____no_output_____ | MIT | notebooks/CST_ALL_PTMs_Normalization.ipynb | MaayanLab/cst_drug_treatment |
Run the following cell to generate an index sorted alphabetically by lowercase term local name. Omit this index if the terms have opaque local names. | # generate the index of terms grouped by category and sorted alphabetically by lowercase term local name
text = '### 3.1 Index By Term Name\n\n'
text += '(See also [3.2 Index By Label](#32-index-by-label))\n\n'
for category in range(0,len(display_order)):
text += '**' + display_label[category] + '**\n'
text += '\n'
if organized_in_categories:
filtered_table = terms_sorted_by_localname[terms_sorted_by_localname['skos_inScheme']==display_order[category]]
filtered_table.reset_index(drop=True, inplace=True)
else:
filtered_table = terms_sorted_by_localname
filtered_table.reset_index(drop=True, inplace=True)
for row_index,row in filtered_table.iterrows():
curie = row['pref_ns_prefix'] + ":" + row['term_localName']
curie_anchor = curie.replace(':','_')
text += '[' + row['label'] + '](#' + curie_anchor + ') |\n'
text = text[:len(text)-2] # remove final trailing vertical bar and newline
text += '\n\n' # put back removed newline
index_by_name = text
print(index_by_name) | _____no_output_____ | CC-BY-4.0 | code/build_format_cv/build-page-categories.ipynb | edwbaker/ac |
Run the following cell to generate an index by term label | text = '\n\n'
# Comment out the following two lines if there is no index by local names
#text = '### 3.2 Index By Label\n\n'
#text += '(See also [3.1 Index By Term Name](#31-index-by-term-name))\n\n'
for category in range(0,len(display_order)):
if organized_in_categories:
text += '**' + display_label[category] + '**\n'
text += '\n'
filtered_table = terms_sorted_by_label[terms_sorted_by_label['skos_inScheme']==display_order[category]]
filtered_table.reset_index(drop=True, inplace=True)
else:
filtered_table = terms_sorted_by_label
filtered_table.reset_index(drop=True, inplace=True)
for row_index,row in filtered_table.iterrows():
if row_index == 0 or (row_index != 0 and row['label'] != filtered_table.iloc[row_index - 1].loc['label']): # this is a hack to prevent duplicate labels
curie_anchor = row['pref_ns_prefix'] + "_" + row['term_localName']
text += '[' + row['label'] + '](#' + curie_anchor + ') |\n'
text = text[:len(text)-2] # remove final trailing vertical bar and newline
text += '\n\n' # put back removed newline
index_by_label = text
print(index_by_label)
decisions_df = pd.read_csv('https://raw.githubusercontent.com/tdwg/rs.tdwg.org/master/decisions/decisions-links.csv', na_filter=False)
# generate a table for each term, with terms grouped by category
# generate the Markdown for the terms table
text = '## 4 Vocabulary\n'
for category in range(0,len(display_order)):
if organized_in_categories:
text += '### 4.' + str(category + 1) + ' ' + display_label[category] + '\n'
text += '\n'
text += display_comments[category] # insert the comments for the category, if any.
filtered_table = terms_sorted_by_localname[terms_sorted_by_localname['skos_inScheme']==display_order[category]]
filtered_table.reset_index(drop=True, inplace=True)
else:
filtered_table = terms_sorted_by_localname
filtered_table.reset_index(drop=True, inplace=True)
for row_index,row in filtered_table.iterrows():
text += '<table>\n'
curie = row['pref_ns_prefix'] + ":" + row['term_localName']
curieAnchor = curie.replace(':','_')
text += '\t<thead>\n'
text += '\t\t<tr>\n'
text += '\t\t\t<th colspan="2"><a id="' + curieAnchor + '"></a>Term Name ' + curie + '</th>\n'
text += '\t\t</tr>\n'
text += '\t</thead>\n'
text += '\t<tbody>\n'
text += '\t\t<tr>\n'
text += '\t\t\t<td>Term IRI</td>\n'
uri = row['pref_ns_uri'] + row['term_localName']
text += '\t\t\t<td><a href="' + uri + '">' + uri + '</a></td>\n'
text += '\t\t</tr>\n'
text += '\t\t<tr>\n'
text += '\t\t\t<td>Modified</td>\n'
text += '\t\t\t<td>' + row['term_modified'] + '</td>\n'
text += '\t\t</tr>\n'
if row['version_iri'] != '':
text += '\t\t<tr>\n'
text += '\t\t\t<td>Term version IRI</td>\n'
text += '\t\t\t<td><a href="' + row['version_iri'] + '">' + row['version_iri'] + '</a></td>\n'
text += '\t\t</tr>\n'
text += '\t\t<tr>\n'
text += '\t\t\t<td>Label</td>\n'
text += '\t\t\t<td>' + row['label'] + '</td>\n'
text += '\t\t</tr>\n'
if row['term_deprecated'] != '':
text += '\t\t<tr>\n'
text += '\t\t\t<td></td>\n'
text += '\t\t\t<td><strong>This term is deprecated and should no longer be used.</strong></td>\n'
text += '\t\t</tr>\n'
text += '\t\t<tr>\n'
text += '\t\t\t<td>Definition</td>\n'
text += '\t\t\t<td>' + row['definition'] + '</td>\n'
text += '\t\t</tr>\n'
if row['usage'] != '':
text += '\t\t<tr>\n'
text += '\t\t\t<td>Usage</td>\n'
text += '\t\t\t<td>' + convert_link(convert_code(row['usage'])) + '</td>\n'
text += '\t\t</tr>\n'
if row['notes'] != '':
text += '\t\t<tr>\n'
text += '\t\t\t<td>Notes</td>\n'
text += '\t\t\t<td>' + convert_link(convert_code(row['notes'])) + '</td>\n'
text += '\t\t</tr>\n'
if row['examples'] != '':
text += '\t\t<tr>\n'
text += '\t\t\t<td>Examples</td>\n'
text += '\t\t\t<td>' + convert_link(convert_code(row['examples'])) + '</td>\n'
text += '\t\t</tr>\n'
if (vocab_type == 2 or vocab_type == 3) and row['controlled_value_string'] != '': # controlled vocabulary
text += '\t\t<tr>\n'
text += '\t\t\t<td>Controlled value</td>\n'
text += '\t\t\t<td>' + row['controlled_value_string'] + '</td>\n'
text += '\t\t</tr>\n'
if vocab_type == 3 and row['skos_broader'] != '': # controlled vocabulary with skos:broader relationships
text += '\t\t<tr>\n'
text += '\t\t\t<td>Has broader concept</td>\n'
curieAnchor = row['skos_broader'].replace(':','_')
text += '\t\t\t<td><a href="#' + curieAnchor + '">' + row['skos_broader'] + '</a></td>\n'
text += '\t\t</tr>\n'
if vocab_type == 3 and row['skos_exactMatch'] != '': # controlled vocabulary with skos:exactMatch relationships
text += '\t\t<tr>\n'
text += '\t\t\t<td>Has exact match</td>\n'
curieAnchor = row['skos_exactMatch'].replace(':','_')
text += '\t\t\t<td><a href="#' + curieAnchor + '">' + row['skos_exactMatch'] + '</a></td>\n'
text += '\t\t</tr>\n'
text += '\t\t<tr>\n'
text += '\t\t\t<td>Type</td>\n'
if row['type'] == 'http://www.w3.org/1999/02/22-rdf-syntax-ns#Property':
text += '\t\t\t<td>Property</td>\n'
elif row['type'] == 'http://www.w3.org/2000/01/rdf-schema#Class':
text += '\t\t\t<td>Class</td>\n'
elif row['type'] == 'http://www.w3.org/2004/02/skos/core#Concept':
text += '\t\t\t<td>Concept</td>\n'
else:
text += '\t\t\t<td>' + row['type'] + '</td>\n' # this should rarely happen
text += '\t\t</tr>\n'
# Look up decisions related to this term
for drow_index,drow in decisions_df.iterrows():
if drow['linked_affected_resource'] == uri:
text += '\t\t<tr>\n'
text += '\t\t\t<td>Executive Committee decision</td>\n'
text += '\t\t\t<td><a href="http://rs.tdwg.org/decisions/' + drow['decision_localName'] + '">http://rs.tdwg.org/decisions/' + drow['decision_localName'] + '</a></td>\n'
text += '\t\t</tr>\n'
text += '\t</tbody>\n'
text += '</table>\n'
text += '\n'
text += '\n'
term_table = text
print(term_table) | _____no_output_____ | CC-BY-4.0 | code/build_format_cv/build-page-categories.ipynb | edwbaker/ac |
Modify to display the indices that you want | text = index_by_label + term_table
#text = index_by_name + index_by_label + term_table
# read in header and footer, merge with terms table, and output
headerObject = open(headerFileName, 'rt', encoding='utf-8')
header = headerObject.read()
headerObject.close()
footerObject = open(footerFileName, 'rt', encoding='utf-8')
footer = footerObject.read()
footerObject.close()
output = header + text + footer
outputObject = open(outFileName, 'wt', encoding='utf-8')
outputObject.write(output)
outputObject.close()
print('done') | _____no_output_____ | CC-BY-4.0 | code/build_format_cv/build-page-categories.ipynb | edwbaker/ac |
**Download** (right-click, save target as ...) this page as a jupyterlab notebook from: [Lab14](http://54.243.252.9/engr-1330-webroot/8-Labs/Lab14/Lab14.ipynb)___ Laboratory 14: Causality, Simulation, and Probability LAST NAME, FIRST NAMER00000000ENGR 1330 Laboratory 14 - In-Lab | # Preamble script block to identify host, user, and kernel
import sys
! hostname
! whoami
print(sys.executable)
print(sys.version)
print(sys.version_info) | atomickitty
sensei
/opt/jupyterhub/bin/python3
3.8.10 (default, Sep 28 2021, 16:10:42)
[GCC 9.3.0]
sys.version_info(major=3, minor=8, micro=10, releaselevel='final', serial=0)
| CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
--- Python for Simulation What is Russian roulette?>Russian roulette (Russian: русская рулетка, russkaya ruletka) is a lethal game of chance in which a player places a single round in a revolver, spins the cylinder, places the muzzle against their head, and pulls the trigger in hopes that the loaded chamber does not align with the primer percussion mechanism and the barrel, causing the weapon to discharge. Russian refers to the supposed country of origin, and roulette to the element of risk-taking and the spinning of the revolver's cylinder, which is reminiscent of a spinning roulette wheel. - Wikipedia @ https://en.wikipedia.org/wiki/Russian_roulette  >A game of dafts, a game of chance One where revolver's the one to dance Rounds and rounds, it goes and spins Makes you regret all those sins \A game of fools, one of lethality With a one to six probability There were two guys and a gun With six chambers but only one... \CLICK, one pushed the gun CLICK, one missed the fun CLICK, "that awful sound" ... BANG!, one had his brains all around! ___ Example: Simulate a game of Russian Roulette:- For 2 rounds- For 5 rounds- For 10 rounds | import numpy as np #import numpy
revolver = np.array([1,0,0,0,0,0]) #create a numpy array with 1 bullet and 5 empty chambers
print(np.random.choice(revolver,2)) #randomly select a value from revolver - simulation
print(np.random.choice(revolver,5))
print(np.random.choice(revolver,10)) | [0 0 0 1 0 0 0 0 0 0]
| CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
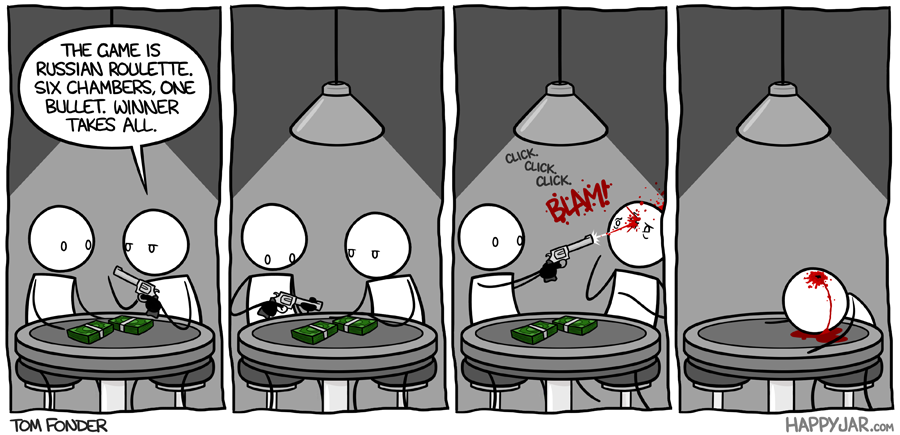 ___ Example: Simulate the results of throwing a D6 (regular dice) for 10 times. | import numpy as np #import numpy
dice = np.array([1,2,3,4,5,6]) #create a numpy array with values of a D6
np.random.choice(dice,10) #randomly selecting a value from dice for 10 times- simulation | _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: Assume the following rules:- If the dice shows 1 or 2 spots, my net gain is -1 dollar.- If the dice shows 3 or 4 spots, my net gain is 0 dollars.- If the dice shows 5 or 6 spots, my net gain is 1 dollar.__Define a function to simulate a game with the above rules, assuming a D6, and compute the net gain of the player over any given number of rolls. Compute the net gain for 5, 50, and 500 rolls__ | def D6game(nrolls):
import numpy as np #import numpy
dice = np.array([1,2,3,4,5,6]) #create a numpy array with values of a D6
rolls = np.random.choice(dice,nrolls) #randomly selecting a value from dice for nrolls times- simulation
gainlist =[] #create an empty list for gains|losses
for i in np.arange(len(rolls)): #Apply the rules
if rolls[i]<=2:
gainlist.append(-1)
elif rolls[i]<=4:
gainlist.append(0)
elif rolls[i]<=6:
gainlist.append(+1)
return (np.sum(gainlist)) #sum up all gains|losses
# return (gainlist,"The net gain is equal to:",np.sum(gainlist))
D6game(5)
D6game(50)
D6game(500) | _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
 Let's Make A Deal Game Show and Monty Hall Problem __The Monty Hall problem is a brain teaser, in the form of a probability puzzle, loosely based on the American television game show Let's Make a Deal and named after its original host, Monty Hall. The problem was originally posed (and solved) in a letter by Steve Selvin to the American Statistician in 1975 (Selvin 1975a), (Selvin 1975b).__>"Suppose you're on a game show, and you're given the choice of three doors: Behind one door is a car; behind the others, goats. You pick a door, say No. 1, and the host, who knows what's behind the doors, opens another door, say No. 3, which has a goat. He then says to you, "Do you want to pick door No. 2?" Is it to your advantage to switch your choice?"__*From Wikipedia: https://en.wikipedia.org/wiki/Monty_Hall_problem*__  /data/img1.png) 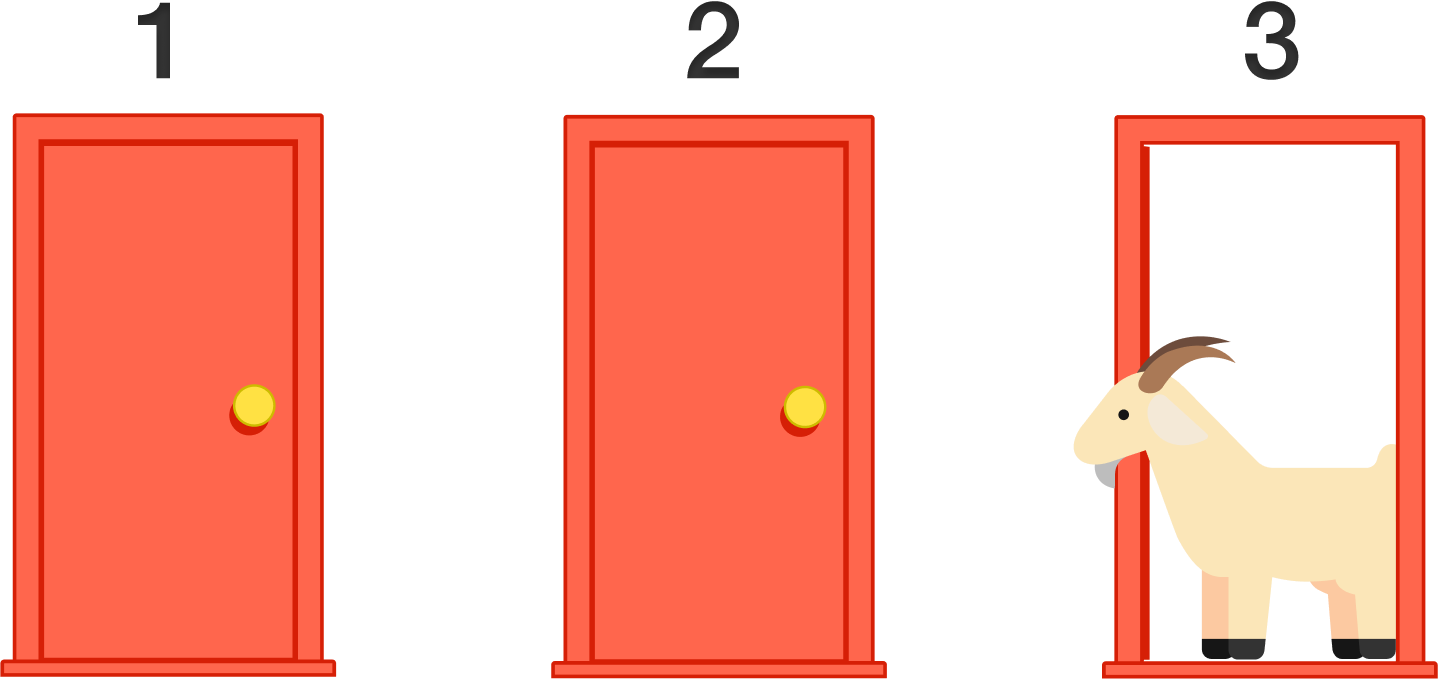 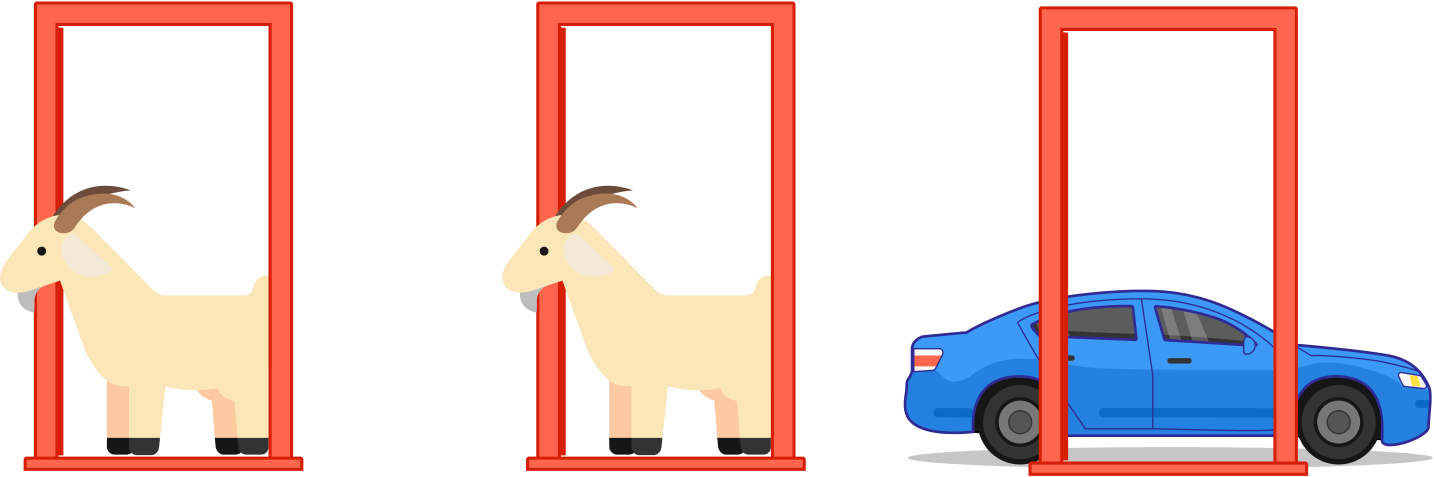 ___ Example: Simulate Monty Hall Game for 1000 times. Use a barplot and discuss whether players are better off sticking to their initial choice, or switching doors? | def othergoat(x): #Define a function to return "the other goat"!
if x == "Goat 1":
return "Goat 2"
elif x == "Goat 2":
return "Goat 1"
Doors = np.array(["Car","Goat 1","Goat 2"]) #Define a list for objects behind the doors
goats = np.array(["Goat 1" , "Goat 2"]) #Define a list for goats!
def MHgame():
#Function to simulate the Monty Hall Game
#For each guess, return ["the guess","the revealed", "the remaining"]
userguess=np.random.choice(Doors) #randomly selects a door as userguess
if userguess == "Goat 1":
return [userguess, "Goat 2","Car"]
if userguess == "Goat 2":
return [userguess, "Goat 1","Car"]
if userguess == "Car":
revealed = np.random.choice(goats)
return [userguess, revealed,othergoat(revealed)]
# Check and see if the MHgame function is doing what it is supposed to do:
for i in np.arange(1):
a =MHgame()
print(a)
print(a[0])
print(a[1])
print(a[2])
c1 = [] #Create an empty list for the userguess
c2 = [] #Create an empty list for the revealed
c3 = [] #Create an empty list for the remaining
for i in np.arange(1000): #Simulate the game for 1000 rounds - or any other number of rounds you desire
game = MHgame()
c1.append(game[0]) #In each round, add the first element to the userguess list
c2.append(game[1]) #In each round, add the second element to the revealed list
c3.append(game[2]) #In each round, add the third element to the remaining list
import pandas as pd
#Create a data frame (gamedf) with 3 columns ("Guess","Revealed", "Remaining") and 1000 (or how many number of rounds) rows
gamedf = pd.DataFrame({'Guess':c1,
'Revealed':c2,
'Remaining':c3})
gamedf
# Get the count of each item in the first and 3rd column
original_car =gamedf[gamedf.Guess == 'Car'].shape[0]
remaining_car =gamedf[gamedf.Remaining == 'Car'].shape[0]
original_g1 =gamedf[gamedf.Guess == 'Goat 1'].shape[0]
remaining_g1 =gamedf[gamedf.Remaining == 'Goat 1'].shape[0]
original_g2 =gamedf[gamedf.Guess == 'Goat 2'].shape[0]
remaining_g2 =gamedf[gamedf.Remaining == 'Goat 2'].shape[0]
# Let's plot a grouped barplot
import matplotlib.pyplot as plt
# set width of bar
barWidth = 0.25
# set height of bar
bars1 = [original_car,original_g1,original_g2]
bars2 = [remaining_car,remaining_g1,remaining_g2]
# Set position of bar on X axis
r1 = np.arange(len(bars1))
r2 = [x + barWidth for x in r1]
# Make the plot
plt.bar(r1, bars1, color='darkorange', width=barWidth, edgecolor='white', label='Original Guess')
plt.bar(r2, bars2, color='midnightblue', width=barWidth, edgecolor='white', label='Remaining Door')
# Add xticks on the middle of the group bars
plt.xlabel('Item', fontweight='bold')
plt.xticks([r + barWidth/2 for r in range(len(bars1))], ['Car', 'Goat 1', 'Goat 2'])
# Create legend & Show graphic
plt.legend()
plt.show()
| _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
__According to the plot, it is statitically beneficial for the players to switch doors because the initial chance for being correct is only 1/3__ 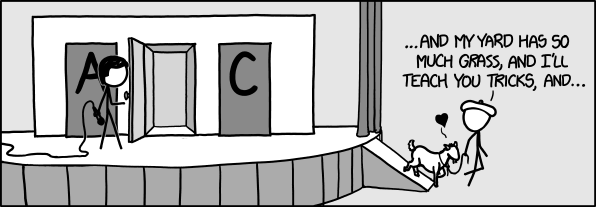 Python for Probability  Important Terminology: __Experiment:__ An occurrence with an uncertain outcome that we can observe. *For example, rolling a die.*__Outcome:__ The result of an experiment; one particular state of the world. What Laplace calls a "case."*For example: 4.*__Sample Space:__ The set of all possible outcomes for the experiment.*For example, {1, 2, 3, 4, 5, 6}.*__Event:__ A subset of possible outcomes that together have some property we are interested in.*For example, the event "even die roll" is the set of outcomes {2, 4, 6}.*__Probability:__ As Laplace said, the probability of an event with respect to a sample space is the number of favorable cases (outcomes from the sample space that are in the event) divided by the total number of cases in the sample space. (This assumes that all outcomes in the sample space are equally likely.) Since it is a ratio, probability will always be a number between 0 (representing an impossible event) and 1 (representing a certain event).*For example, the probability of an even die roll is 3/6 = 1/2.*__*From https://people.math.ethz.ch/~jteichma/probability.html*__ | import numpy as np
import pandas as pd
import matplotlib.pyplot as plt | _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: In a game of Russian Roulette, the chance of surviving each round is 5/6 which is almost 83%. Using a for loop, compute probability of surviving - For 2 rounds- For 5 rounds- For 10 rounds | nrounds =[]
probs =[]
for i in range(3):
nrounds.append(i)
probs.append((5/6)**i) #probability of surviving- not getting the bullet!
RRDF = pd.DataFrame({"# of Rounds": nrounds, "Probability of Surviving": probs})
RRDF
nrounds =[]
probs =[]
for i in range(6):
nrounds.append(i)
probs.append((5/6)**i) #probability of surviving- not getting the bullet!
RRDF = pd.DataFrame({"# of Rounds": nrounds, "Probability of Surviving": probs})
RRDF
nrounds =[]
probs =[]
for i in range(11):
nrounds.append(i)
probs.append((5/6)**i) #probability of surviving- not getting the bullet!
RRDF = pd.DataFrame({"# of Rounds": nrounds, "Probability of Surviving": probs})
RRDF
RRDF.plot.scatter(x="# of Rounds", y="Probability of Surviving",color="red") | _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: What will be the probability of constantly throwing an even number with a D20 in- For 2 rolls- For 5 rolls- For 10 rolls- For 15 rolls | nrolls =[]
probs =[]
for i in range(1,16,1):
nrolls.append(i)
probs.append((1/2)**i) #probability of throwing an even number-10/20 or 1/2
DRDF = pd.DataFrame({"# of Rolls": nrolls, "Probability of constantly throwing an even number": probs})
DRDF
DRDF.plot.scatter(x="# of Rolls", y="Probability of constantly throwing an even number",color="crimson") | _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: What will be the probability of throwing at least one 6 with a D6:- For 2 rolls- For 5 rolls- For 10 rolls- For 50 rolls - Make a scatter plot for this one! | nRolls =[]
probs =[]
for i in range(1,3,1):
nRolls.append(i)
probs.append(1-(5/6)**i) #probability of at least one 6: 1-(5/6)
rollsDF = pd.DataFrame({"# of Rolls": nRolls, "Probability of rolling at least one 6": probs})
rollsDF
nRolls =[]
probs =[]
for i in range(1,6,1):
nRolls.append(i)
probs.append(1-(5/6)**i) #probability of at least one 6: 1-(5/6)
rollsDF = pd.DataFrame({"# of Rolls": nRolls, "Probability of rolling at least one 6": probs})
rollsDF
nRolls =[]
probs =[]
for i in range(1,11,1):
nRolls.append(i)
probs.append(1-(5/6)**i) #probability of at least one 6: 1-(5/6)
rollsDF = pd.DataFrame({"# of Rolls": nRolls, "Probability of rolling at least one 6": probs})
rollsDF
nRolls =[]
probs =[]
for i in range(1,51,1):
nRolls.append(i)
probs.append(1-(5/6)**i) #probability of at least one 6: 1-(5/6)
rollsDF = pd.DataFrame({"# of Rolls": nRolls, "Probability of rolling at least one 6": probs})
rollsDF.plot.scatter(x="# of Rolls", y="Probability of rolling at least one 6") | _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: What is the probability of drawing an ace at least once (with replacement):- in 2 tries- in 5 tries- in 10 tries- in 20 tries - make a scatter plot. | nDraws =[]
probs =[]
for i in range(1,3,1):
nDraws.append(i)
probs.append(1-(48/52)**i) #probability of drawing an ace least once : 1-(48/52)
DrawsDF = pd.DataFrame({"# of Draws": nDraws, "Probability of drawing an ace at least once": probs})
DrawsDF
nDraws =[]
probs =[]
for i in range(1,6,1):
nDraws.append(i)
probs.append(1-(48/52)**i) #probability of drawing an ace least once : 1-(48/52)
DrawsDF = pd.DataFrame({"# of Draws": nDraws, "Probability of drawing an ace at least once": probs})
DrawsDF
nDraws =[]
probs =[]
for i in range(1,11,1):
nDraws.append(i)
probs.append(1-(48/52)**i) #probability of drawing an ace least once : 1-(48/52)
DrawsDF = pd.DataFrame({"# of Draws": nDraws, "Probability of drawing an ace at least once": probs})
DrawsDF
nDraws =[]
probs =[]
for i in range(1,21,1):
nDraws.append(i)
probs.append(1-(48/52)**i) #probability of drawing an ace at least once : 1-(48/52)
DrawsDF = pd.DataFrame({"# of Draws": nDraws, "Probability of drawing an ace at least once": probs})
DrawsDF
DrawsDF.plot.scatter(x="# of Draws", y="Probability of drawing an ace at least once") | _____no_output_____ | CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: - A) Write a function to find the probability of an event in percentage form based on given outcomes and sample space- B) Use the function and compute the probability of rolling a 4 with a D6- C) Use the function and compute the probability of drawing a King from a standard deck of cards- D) Use the function and compute the probability of drawing the King of Hearts from a standard deck of cards- E) Use the function and compute the probability of drawing an ace after drawing a king- F) Use the function and compute the probability of drawing an ace after drawing an ace- G) Use the function and compute the probability of drawing a heart OR a club- F) Use the function and compute the probability of drawing a Royal Flush *hint: (in poker) a straight flush including ace, king, queen, jack, and ten all in the same suit, which is the hand of the highest possible value__This problem is designed based on an example by *Daniel Poston* from DataCamp, accessible @ *https://www.datacamp.com/community/tutorials/statistics-python-tutorial-probability-1*__ | # A
# Create function that returns probability percent rounded to one decimal place
def Prob(outcome, sampspace):
probability = (outcome / sampspace) * 100
return round(probability, 1)
# B
outcome = 1 #Rolling a 4 is only one of the possible outcomes
space = 6 #Rolling a D6 can have 6 different outcomes
Prob(outcome, space)
# C
outcome = 4 #Drawing a king is four of the possible outcomes
space = 52 #Drawing from a standard deck of cards can have 52 different outcomes
Prob(outcome, space)
# D
outcome = 1 #Drawing the king of hearts is only 1 of the possible outcomes
space = 52 #Drawing from a standard deck of cards can have 52 different outcomes
Prob(outcome, space)
# E
outcome = 4 #Drawing an ace is 4 of the possible outcomes
space = 51 #One card has been drawn
Prob(outcome, space)
# F
outcome = 3 #Once Ace is already drawn
space = 51 #One card has been drawn
Prob(outcome, space)
# G
hearts = 13 #13 cards of hearts in a deck
space = 52 #total number of cards in a deck
clubs = 13 #13 cards of clubs in a deck
Prob_heartsORclubs= Prob(hearts, space) + Prob(clubs, space)
print("Probability of drawing a heart or a club is",Prob_heartsORclubs,"%")
# F
draw1 = 5 #5 cards are needed
space1 = 52 #out of the possible 52 cards
draw2 = 4 #4 cards are needed
space2 = 51 #out of the possible 51 cards
draw3 = 3 #3 cards are needed
space3 = 50 #out of the possible 50 cards
draw4 = 2 #2 cards are needed
space4 = 49 #out of the possible 49 cards
draw5 = 1 #1 cards is needed
space5 = 48 #out of the possible 48 cards
#Probability of a getting a Royal Flush
Prob_RF= 4*(Prob(draw1, space1)/100) * (Prob(draw2, space2)/100) * (Prob(draw3, space3)/100) * (Prob(draw4, space4)/100) * (Prob(draw5, space5)/100)
print("Probability of drawing a royal flush is",Prob_RF,"%") | Probability of drawing a royal flush is 1.5473203199999998e-06 %
| CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: Two unbiased dice are thrown once and the total score is observed. Define an appropriate function and use a simulation to find the estimated probability that :- the total score is greater than 10?- the total score is even and greater than 7?__This problem is designed based on an example by *Elliott Saslow*from Medium.com, accessible @ *https://medium.com/future-vision/simulating-probability-events-in-python-5dd29e34e381*__ | import numpy as np
def DiceRoll1(nSimulation):
count =0
dice = np.array([1,2,3,4,5,6]) #create a numpy array with values of a D6
for i in range(nSimulation):
die1 = np.random.choice(dice,1) #randomly selecting a value from dice - throw the D6 once
die2 = np.random.choice(dice,1) #randomly selecting a value from dice - throw the D6 once again!
score = die1 + die2 #summing them up
if score > 10: #if it meets our desired condition:
count +=1 #add one to the "count"
return count/nSimulation #compute the probability of the desired event by dividing count by the total number of trials
nSimulation = 10000
print("The probability of rolling a number greater than 10 after",nSimulation,"rolld is:",DiceRoll1(nSimulation)*100,"%")
import numpy as np
def DiceRoll2(nSimulation):
count =0
dice = np.array([1,2,3,4,5,6]) #create a numpy array with values of a D6
for i in range(nSimulation):
die1 = np.random.choice(dice,1) #randomly selecting a value from dice - throw the D6 once
die2 = np.random.choice(dice,1) #randomly selecting a value from dice - throw the D6 once again!
score = die1 + die2
if score %2 ==0 and score > 7: #the total score is even and greater than 7
count +=1
return count/nSimulation
nSimulation = 10000
print("The probability of rolling an even number and greater than 7 after",nSimulation," rolls is:",DiceRoll2(nSimulation)*100,"%") | The probability of rolling an even number and greater than 7 after 10000 rolls is: 24.77 %
| CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
___ Example: An urn contains 10 white balls, 20 reds and 30 greens. We want to draw 5 balls with replacement. Use a simulation (10000 trials) to find the estimated probability that:- we draw 3 white and 2 red balls- we draw 5 balls of the same color__This problem is designed based on an example by *Elliott Saslow*from Medium.com, accessible @ *https://medium.com/future-vision/simulating-probability-events-in-python-5dd29e34e381*__ | # A
import numpy as np
import random
d = {} #Create an empty dictionary to associate numbers and colors
for i in range(0,60,1): #total of 60 balls
if i <10: #10 white balls
d[i]="White"
elif i>9 and i<30: #20 red balls
d[i]="Red"
else: #60-30=30 green balls
d[i]="Green"
#
nSimulation= 10000 #How many trials?
outcome1= 0 #initial value on the desired outcome counter
for i in range(nSimulation):
draw=[] #an empty list for the draws
for i in range(5): #how many balls we want to draw?
draw.append(d[random.randint(0,59)]) #randomly choose a number from 0 to 59- simulation of drawing balls
drawarray = np.array(draw) #convert the list into a numpy array
white = sum(drawarray== "White") #count the white balls
red = sum(drawarray== "Red") #count the red balls
green = sum(drawarray== "Green") #count the green balls
if white ==3 and red==2: #If the desired condition is met, add one to the counter
outcome1 +=1
print("The probability of drawing 3 white and 2 red balls is",(outcome1/nSimulation)*100,"%")
# B
import numpy as np
import random
d = {}
for i in range(0,60,1):
if i <10:
d[i]="White"
elif i>9 and i<30:
d[i]="Red"
else:
d[i]="Green"
#
nSimulation= 10000
outcome1= 0
outcome2= 0 #we can consider multiple desired outcomes
for i in range(nSimulation):
draw=[]
for i in range(5):
draw.append(d[random.randint(0,59)])
drawarray = np.array(draw)
white = sum(drawarray== "White")
red = sum(drawarray== "Red")
green = sum(drawarray== "Green")
if white ==3 and red==2:
outcome1 +=1
if white ==5 or red==5 or green==5:
outcome2 +=1
print("The probability of drawing 3 white and 2 red balls is",(outcome1/nSimulation)*100,"%")
print("The probability of drawing 5 balls of the same color is",(outcome2/nSimulation)*100,"%")
| The probability of drawing 3 white and 2 red balls is 0.53 %
The probability of drawing 5 balls of the same color is 3.8 %
| CC0-1.0 | 8-Labs/Lab14/old_src/.ipynb_checkpoints/Lab14-checkpoint.ipynb | dustykat/engr-1330-psuedo-course |
NOMIS API DevelopmentShortly after this paper was accepted, I received a very helpful reply from NOMIS relating to the fact that I couldn't seem to get their API helper to give me the query needed to retrieve each of the data sets used in this research directly (which would make replication much, much faster and easier). I've not had time to refactor the code to incorporate their guidance but here is an overview of the process that will need to be followed:* Select your dataset* Select only the region in your query and not the LSOAs - so in the case of your query, choose the "London" region as an individual selection (not using "areas within").* Continue to select other options to build up your query as normal* Select the "Nomis API" download format and you should get the page with the API links (ignore the first tab and go to the "Complete list of API links" page to get the URL)* When you get the URL, modify it as in the example belowExample of modifying the URL is below, the change involves simply telling the API you want all LSOAs in the area you have selected, you do this by adding the text "TYPE304" (which is the code for 2001 LSOAs) to the geography code. The code for 2011 LSOAs is TYPE298.Original URL for the query London region:http://www.nomisweb.co.uk/api/v01/dataset/NM_1673_1.bulk.csv?date=latest&geography=2013265927&dwelling_type=0,3...5,7...9&measures=20100Modified URL for the query selecting all LSOAs in London region:http://www.nomisweb.co.uk/api/v01/dataset/NM_1673_1.bulk.csv?date=latest&geography=2013265927TYPE304&dwelling_type=0,3...5,7...9&measures=20100 | # 2011 TTW Data (need a different geography TYPE304 for 2001)
url = ('http://www.nomisweb.co.uk/api/v01/dataset/',
'NM_568_1.bulk.csv?',
'date=latest&',
'geography=2013265927TYPE298&',
'rural_urban=0&',
'cell=0...12&',
'measures=20100&',
'select=date_name,geography_name,geography_code,rural_urban_name,cell_name,measures_name,obs_value,obs_status_name')
print("".join(url)) | http://www.nomisweb.co.uk/api/v01/dataset/NM_568_1.bulk.csv?date=latest&geography=2013265927TYPE304&rural_urban=0&cell=0...12&measures=20100&select=date_name,geography_name,geography_code,rural_urban_name,cell_name,measures_name,obs_value,obs_status_name
| MIT | NOMIS API Test.ipynb | FoxyAI/urb-studies-predicting-gentrification |
Exercise 1 - Toffoli gate | from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit
from qiskit import IBMQ, Aer, execute
from ibm_quantum_widgets import CircuitComposer
editorEx = CircuitComposer()
editorEx
import math
pi=math.pi
# You can also build your circuit programmatically using Qiskit code
circuit = QuantumCircuit(3)
theta = pi/4 # Theta can be anything (pi chosen arbitrarily)
# WRITE YOUR CODE BETWEEN THESE LINES - START
### Hadamard ###
circuit.rz(pi/2, 2)
circuit.sx(2)
circuit.rz(pi/2, 2)
circuit.cx(1,2)
circuit.rz(-theta, 2)
circuit.cx(0,2)
circuit.rz(theta, 2)
circuit.cx(1,2)
circuit.rz(-theta, 2)
circuit.cx(0,2)
circuit.rz(theta, 1)
circuit.rz(theta, 2)
### Hadamard ###
circuit.rz(pi/2, 2)
circuit.sx(2)
circuit.rz(pi/2, 2)
circuit.cnot(0,1)
circuit.rz(theta, 0)
circuit.rz(-theta, 1)
circuit.cnot(0,1)
# WRITE YOUR CODE BETWEEN THESE LINES - END
# Checking the resulting circuit
qc = editorEx.circuit
qc = circuit # Uncomment this line if you want to submit the circuit built using Qiskit code
qc.draw(output='mpl') | _____no_output_____ | Apache-2.0 | solutions by participants/ex1/ex1-InnanNouhaila-79cost.ipynb | fazliberkordek/ibm-quantum-challenge-2021 |
1. Load data- I'm using the files that were updated at **April 21st**- ref : https://github.com/jihoo-kim/Data-Science-for-COVID-19- datasets - Region.csv → Region_sido_addPop_addHospital.csv - PatientInfo.csv - 흡연율-성-연령별.csv | # Prepare dataset
Region_df = pd.read_csv('../dataset/Region_sido_addPop_addHospital.csv')[['province', 'safe_hospitals_count', 'infection_hospitals_count', 'infection_hospitals_bed_num']]
Smoke_df = pd.read_csv('../dataset/흡연율_성_연령별.csv')
Smoke_df = Smoke_df.rename(columns={'성별':'sex','연령별':'age','분율':'smoking_rate'})
Smoke_df.sex.replace('여성','female',inplace=True)
Smoke_df.sex.replace('남성','male',inplace=True)
Smoke_df = Smoke_df[['sex','age','smoking_rate']]
PatientInfo_df = pd.read_csv('../dataset/Patient/PatientInfo.csv')
PatientInfo_df = pd.merge(PatientInfo_df, Smoke_df, how='left', on=['sex','age'])
print(f'PatientInfo.csv shape : {PatientInfo_df.shape}')
PatientInfo_df = PatientInfo_df[PatientInfo_df.state.isin(['released', 'deceased'])]
print(f' → datset shape : {PatientInfo_df.shape}')
PatientInfo_df['symptom_onset_date'] = pd.to_datetime(PatientInfo_df['symptom_onset_date']) # convert data type
PatientInfo_df['confirmed_date'] = pd.to_datetime(PatientInfo_df['confirmed_date']) # convert data type
PatientInfo_df['disease'] = PatientInfo_df['disease'].astype(float) # convert data type
display(PatientInfo_df.head(3))
# Save patient_id list
present_patients = PatientInfo_df.patient_id.astype(str).tolist()
with open('patients_id_0421.txt', 'w') as fp:
fp.write('\n'.join(present_patients))
# Check Null values
PatientInfo_df.contact_number.value_counts(dropna=False, normalize=True) * 100
PatientInfo_df.symptom_onset_date.value_counts(dropna=False, normalize=True) * 100
PatientInfo_df.infection_order.value_counts(dropna=False, normalize=True) * 100
# PatientInfo_df.disease.value_counts(dropna=False, normalize=True) * 100
# PatientInfo_df.country.value_counts(dropna=False, normalize=True) * 100 | _____no_output_____ | MIT | dacon_covid-19/ML/COVID_ML_05_XGBoost-GridSearchCV-final-model.ipynb | sam351/competitions |
2. Preprocess data- selected features : **'sex', 'birth_year', 'age', 'province', 'disease (98.9% NaN)', 'infection_case (34.1% NaN)', 'symptom_onset_date (86.2% NaN)', 'confirmed_date', 'contact_number (75.4% NaN)''**- dropped features after validation : 'country', 'city', 'infection_order (98.3% NaN)- handling nan - drop nan from 'sex' & 'age' - replace with mean of same age group in birth_year - replace with 'non-reported' in infection_case - fill -1 in disease - fill -1 in contact_number - fill -1 in days_btw_symptom_confirm- new features : 'years_after_birth', 'days_after_first_date', 'days_btw_symptom_confirm', 'days_after_first_date_province', 'province_safe_hospitals_count', 'province_infection_hospitals_count', 'province_infection_bed_count' - (ohter new features were created but deleted after evaluation - 'smoking rate')- feature encoding (categorical) - age : convert to integer (0~10) - label encoding - other categorical columns : one-hot encoding | # Select features
X_features = PatientInfo_df[['sex', 'birth_year', 'age', 'province', 'disease', 'infection_case',
'symptom_onset_date', 'confirmed_date', 'contact_number']].copy()
y_target = PatientInfo_df[['state']].copy()
print(f'X_features.shape : {X_features.shape}')
print(f'y_target.shape : {y_target.shape}')
X_features.head(3)
print('\n<< no of nan table (before handling) >>')
print(X_features.isna().sum())
# Handle nan - sex & age
y_target = y_target[~X_features.sex.isna() & ~X_features.age.isna()]
X_features = X_features[~X_features.sex.isna() & ~X_features.age.isna()]
# Handle nan - birth_year
mean_year_list = dict(X_features.groupby('age')['birth_year'].mean().round().reset_index().values)
X_features.loc[X_features.birth_year.isna(), 'birth_year'] = X_features.loc[X_features.birth_year.isna(), 'age'].map( lambda x : mean_year_list[ x ] )
# Handle nan - infection_case
X_features.loc[X_features.infection_case.isna(), 'infection_case'] = 'not-reported'
# Handle nan - disease, contact_number
X_features.disease = X_features.disease.fillna(-1)
X_features.contact_number = X_features.contact_number.fillna(-1)
print('\n<< no of nan table (after handling) >>')
print(X_features.isna().sum())
print(f'\n\nX_features.shape : {X_features.shape}')
print(f'y_target.shape : {y_target.shape}')
# Create new features
X_features['years_after_birth'] = datetime.date.today().year - X_features['birth_year']
X_features['days_after_first_date'] = X_features['confirmed_date'] - X_features['confirmed_date'].min()
X_features['days_after_first_date'] = X_features['days_after_first_date'].dt.days
X_features['days_btw_symptom_confirm'] = (X_features['confirmed_date'] - X_features['symptom_onset_date']).dt.days
X_features['days_btw_symptom_confirm'] = X_features['days_btw_symptom_confirm'].fillna(-1) # Handle nan
# deleted after evaluation - days_after_first_date_province
first_date_province_dict = dict(PatientInfo_df.groupby('province')['confirmed_date'].min().reset_index().values)
X_features['first_date_province'] = X_features['province'].map( lambda x : first_date_province_dict[x] )
X_features['days_after_first_date_province'] = X_features['confirmed_date'] - X_features['first_date_province']
X_features['days_after_first_date_province'] = X_features['days_after_first_date_province'].dt.days
# deleted after evaluation - province infomations
province_info_dict = { item_list[0]:item_list[1:] for item_list in Region_df.values }
X_features['province_safe_hospitals_count'] = X_features.province.map(lambda x : province_info_dict[x][0])
X_features['province_infection_hospitals_count'] = X_features.province.map(lambda x : province_info_dict[x][1])
X_features['province_infection_bed_count'] = X_features.province.map(lambda x : province_info_dict[x][2])
X_features = X_features.drop(columns=['birth_year', 'symptom_onset_date', 'confirmed_date', 'first_date_province'])
X_features.head(3)
# feature encoding - X_features
X_features_processed = X_features.copy()
X_features_processed = pd.concat([X_features_processed, pd.get_dummies(X_features_processed[['sex', 'province', 'infection_case']])],
axis=1) # one-hot encoding
X_features_processed = X_features_processed.drop(columns=['sex', 'province', 'infection_case'])
X_features_processed['age'] = X_features.age.str.replace('s','').astype(int)//10 # label encoding
display(X_features_processed.head(3))
print()
# feature encoding - y_target
y_target_processed = pd.get_dummies(y_target)['state_deceased']
display(y_target_processed.head(3)) | _____no_output_____ | MIT | dacon_covid-19/ML/COVID_ML_05_XGBoost-GridSearchCV-final-model.ipynb | sam351/competitions |
3. Split data into train, val, test- It's important that **labels are highly unbalanced** (only about 3% is deceased)- Since the dataset is quite small(1704 records), I will split the date into **6:2:2** for now (test data could be added from the next file update)- Since the labels are highly imbalanced, it's better to use **stratified random sampling**. | # Get train dataset
X_train_val, X_test, y_train_val, y_test = train_test_split(X_features_processed, y_target_processed, test_size=0.2,
random_state=0, stratify=y_target_processed)
# Get val & test dataset
X_train, X_val, y_train, y_val = train_test_split(X_train_val, y_train_val, test_size=0.33, random_state=0, stratify=y_train_val)
# Check the labels of each dataset
print('< Percentage of each label (whole dataset) >')
print(y_target_processed.value_counts(normalize=True) * 100)
print('\n< Percentage of each label (Train dataset) > - size of dataset :', y_train.shape[0])
print(y_train.value_counts(normalize=True) * 100)
print('\n< Percentage of each label (Validation dataset) > - size of dataset :', y_val.shape[0])
print(y_val.value_counts(normalize=True) * 100)
print('\n< Percentage of each label (Test dataset) > - size of dataset :', y_test.shape[0])
print(y_test.value_counts(normalize=True) * 100) | < Percentage of each label (whole dataset) >
0 96.044864
1 3.955136
Name: state_deceased, dtype: float64
< Percentage of each label (Train dataset) > - size of dataset : 907
0 96.030871
1 3.969129
Name: state_deceased, dtype: float64
< Percentage of each label (Validation dataset) > - size of dataset : 448
0 95.982143
1 4.017857
Name: state_deceased, dtype: float64
| MIT | dacon_covid-19/ML/COVID_ML_05_XGBoost-GridSearchCV-final-model.ipynb | sam351/competitions |
4. Train model - XGBoost with GridSearchCV | # Look for best hyper-parameters
start = time.time()
# Train model
xgb = XGBClassifier(random_state=0, n_jobs=-1)
xgb_param = {
'n_estimators': [100, 200, 300, 400],
'min_child_weight': [1, 2, 3],
'gamma': [1.5, 2, 2.5, 3],
'colsample_bytree': [0.7, 0.8, 0.9],
'max_depth': [5, 6, 7, 8, 9, 10]
}
grid_xgb = GridSearchCV(xgb, param_grid=xgb_param, scoring='f1', cv=5)
grid_xgb.fit(X_train_val, y_train_val)
print('time elapsed :', time.time()-start)
print('GridSearchCV 최적 파라미터:', grid_xgb.best_params_)
print('GridSearchCV 최고 정확도: {0:.4f}'.format(grid_xgb.best_score_))
scores_df = pd.DataFrame(grid_xgb.cv_results_).sort_values('rank_test_score')
scores_df[['params', 'mean_test_score', 'rank_test_score',
'split0_test_score', 'split2_test_score', 'split4_test_score']]
# Train final model
xgb_final = XGBClassifier(random_state=0, n_jobs=-1, n_estimators=100, colsample_bytree=0.8, gamma=2, max_depth=6, min_child_weight=1, subsample=0.6)
xgb_final.fit(X_train, y_train)
# Evaluate final model
def get_clf_eval(y_test , pred):
from sklearn.metrics import confusion_matrix, accuracy_score
from sklearn.metrics import precision_score, recall_score
from sklearn.metrics import f1_score, roc_auc_score
confusion = confusion_matrix( y_test, pred)
accuracy = accuracy_score(y_test , pred)
precision = precision_score(y_test , pred)
recall = recall_score(y_test , pred)
f1 = f1_score(y_test,pred)
roc_auc = roc_auc_score(y_test, pred)
print('오차 행렬')
print(confusion)
print('정확도: {0:.4f}, 정밀도: {1:.4f}, 재현율: {2:.4f},\
F1: {3:.4f}, AUC:{4:.4f}'.format(accuracy, precision, recall, f1, roc_auc))
pass
xgb_preds = xgb_final.predict(X_val)
get_clf_eval(y_val , xgb_preds)
# Visualize the feature importance
fig, ax = plt.subplots(figsize=(10, 7))
plot_importance(xgb_final, ax=ax)
ax.set_title('Feature importance', size=13)
ax.tick_params(axis='y', labelsize=15)
plt.show() | _____no_output_____ | MIT | dacon_covid-19/ML/COVID_ML_05_XGBoost-GridSearchCV-final-model.ipynb | sam351/competitions |
Style TransferWe are optimising the input image to reduce loss between output of convolutional layers(compared with output from layers when we pass the style/content image) of VGG16 model(we don't optimise weights since they are pretrained). Also we use the optimized input image as final prediction instead of output from convolutional layers. | import tensorflow as tf
if tf.__version__.startswith('2'):
tf.compat.v1.disable_eager_execution()
from tensorflow.keras.layers import Input, Lambda, Dense, Flatten
from tensorflow.keras.layers import AveragePooling2D, MaxPooling2D
from tensorflow.keras.layers import Conv2D
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras.applications.vgg16 import VGG16
from tensorflow.keras.applications.vgg16 import preprocess_input
from tensorflow.keras.preprocessing import image
import tensorflow.keras.backend as K
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import fmin_l_bfgs_b
from datetime import datetime | _____no_output_____ | MIT | NeuralStyleTransfer/StyleTransfer.ipynb | Frostday/Small-Tensorflow-Projects |
Transfer Learning Part | def VGG16_AvgPool(shape):
# we want to account for features across the entire image so get rid of the maxpool which throws away information and use average # pooling instead.
vgg = VGG16(input_shape=shape, weights='imagenet', include_top=False)
i = vgg.input
x = i
for layer in vgg.layers:
if layer.__class__ == MaxPooling2D:
# replace it with average pooling
x = AveragePooling2D()(x)
else:
x = layer(x)
return Model(i, x)
def VGG16_AvgPool_CutOff(shape, num_convs):
# this function creates a partial model because we don't need the full VGG network instead we need to stop at an intermediate
# convolution. Therefore this function allows us to specify how many convolutions we need
# there are 13 convolutions in total we can pick any of them as the "output" of our content model
if num_convs < 1 or num_convs > 13:
print("num_convs must be in the range [1, 13]")
return None
model = VGG16_AvgPool(shape)
n = 0
output = None
for layer in model.layers:
if layer.__class__ == Conv2D:
n += 1
if n >= num_convs:
output = layer.output
break
return Model(model.input, output) | _____no_output_____ | MIT | NeuralStyleTransfer/StyleTransfer.ipynb | Frostday/Small-Tensorflow-Projects |
Processing | # load the content image
def load_img_and_preprocess(path, shape=None):
img = image.load_img(path, target_size=shape)
# convert image to array and preprocess for vgg
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
return x
# since VGG accepts BGR this function allows us to convert our values back to RGB so we can plot it using matplotlib
# so this basically reverses the keras function - preprocess input
def unpreprocess(img):
img[..., 0] += 103.939
img[..., 1] += 116.779
img[..., 2] += 126.68
img = img[..., ::-1]
return img
def scale_img(x):
x = x - x.min()
x = x / x.max()
return x | _____no_output_____ | MIT | NeuralStyleTransfer/StyleTransfer.ipynb | Frostday/Small-Tensorflow-Projects |
Style lossConverting output to gram matrix and calculating style loss | def gram_matrix(img):
# input is (H, W, C) (C = # feature maps)
# we first need to convert it to (C, H*W)
X = K.batch_flatten(K.permute_dimensions(img, (2, 0, 1)))
# now, calculate the gram matrix
# gram = XX^T / N
# the constant is not important since we'll be weighting these
G = K.dot(X, K.transpose(X)) / img.get_shape().num_elements()
return G
def style_loss(y, t):
return K.mean(K.square(gram_matrix(y) - gram_matrix(t))) | _____no_output_____ | MIT | NeuralStyleTransfer/StyleTransfer.ipynb | Frostday/Small-Tensorflow-Projects |
Minimizing loss and optimising image(training) | # function to minimise loss by optimising input image
def minimize(fn, epochs, batch_shape, content_image):
t0 = datetime.now()
losses = []
# x = np.random.randn(np.prod(batch_shape))
x = content_image
for i in range(epochs):
x, l, _ = fmin_l_bfgs_b(
func=fn,
x0=x,
maxfun=20
)
x = np.clip(x, -127, 127)
print("iter=%s, loss=%s" % (i+1, l))
losses.append(l)
print("duration:", datetime.now() - t0)
plt.plot(losses)
plt.show()
newimg = x.reshape(*batch_shape)
final_img = unpreprocess(newimg)
return final_img[0] | _____no_output_____ | MIT | NeuralStyleTransfer/StyleTransfer.ipynb | Frostday/Small-Tensorflow-Projects |
Modelling | content_path = 'images/content/elephant.jpg'
style_path = 'images/style/lesdemoisellesdavignon.jpg'
x = load_img_and_preprocess(content_path)
h, w = x.shape[1:3]
# reduce image size while keeping ratio of dimensions same
i = 2
h_new = h
w_new = w
while h_new > 400 or w_new > 400:
h_new = h/i
w_new = w/i
i += 1
h = int(h_new)
w = int(w_new)
print(h, w)
fig = plt.figure()
img_content = image.load_img(content_path, target_size=(h, w))
ax1 = fig.add_subplot(1, 2, 1)
ax1.imshow(img_content)
img_style = image.load_img(style_path, target_size=(h, w))
ax2 = fig.add_subplot(1, 2, 2)
ax2.imshow(img_style)
plt.show()
# loading and preprocessing input images
content_img = load_img_and_preprocess(content_path, (h, w))
style_img = load_img_and_preprocess(style_path, (h, w))
print(content_img.shape)
print(style_img.shape)
batch_shape = content_img.shape
shape = content_img.shape[1:]
print(batch_shape)
print(shape)
# load the complete model
vgg = VGG16_AvgPool(shape)
# load the content model and we only want one output from this which is from 13th layer
content_model = Model(vgg.input, vgg.layers[13].get_output_at(0))
# target outputs from content image
content_target = K.variable(content_model.predict(content_img))
content_model.summary()
# index 0 correspond to the original vgg with maxpool so we do get_output_at(1) which corresponds to vgg with avg pool
symbolic_conv_outputs = [
layer.get_output_at(1) for layer in vgg.layers \
if layer.name.endswith('conv1')
]
# we collect all the convolutional layers in this list because we will need to take output from all of them
symbolic_conv_outputs
# make a big model that outputs multiple layers' outputs(outputs from all layers stored in list symbolic_conv_outputs)
style_model = Model(vgg.input, symbolic_conv_outputs)
# calculate the targets from convolutional outputs at each layer in symbolic_conv_outputs
style_layers_outputs = [K.variable(y) for y in style_model.predict(style_img)]
style_model.summary()
# we will assume the weight of the content loss is 1 and only weight the style losses
# style_weights = [0.2, 0.4, 0.3, 0.5, 0.2]
style_weights = [5, 4, 8, 7, 9]
# create the total loss which is the sum of content + style loss
loss = K.mean(K.square(content_model.output - content_target))
for w, symbolic, actual in zip(style_weights, symbolic_conv_outputs, style_layers_outputs):
# gram_matrix() expects a (H, W, C) as input
loss += w * style_loss(symbolic[0], actual[0])
content_model.input
style_model.input
content_model.output
style_model.output
# NOTE: it doesn't matter which model's input you use they are both pointing to the same keras Input layer in memory
grads = K.gradients(loss, vgg.input)
get_loss_and_grads = K.function(
inputs=[vgg.input],
outputs=[loss] + grads
)
def get_loss_and_grads_wrapper(x_vec):
l, g = get_loss_and_grads([x_vec.reshape(*batch_shape)])
return l.astype(np.float64), g.flatten().astype(np.float64)
# converting image shape to 1d array
img = np.reshape(content_img, (-1))
img.shape
final_img = minimize(get_loss_and_grads_wrapper, 10, batch_shape, img)
plt.imshow(scale_img(final_img))
plt.show() | _____no_output_____ | MIT | NeuralStyleTransfer/StyleTransfer.ipynb | Frostday/Small-Tensorflow-Projects |
ST0256 - Análisis NuméricoCapítulo 5: Diferenciación e integración numérica2021/01MEDELLÍN - COLOMBIA Text provided under a Creative Commons Attribution license, CC-BY. All code is made available under the FSF-approved MIT license.(c) Carlos Alberto Alvarez Henao *** ***Docente:*** Carlos Alberto Álvarez Henao, I.C. D.Sc.***e-mail:*** [email protected]***skype:*** carlos.alberto.alvarez.henao***Herramienta:*** [Jupyter notebook](http://jupyter.org/)***Kernel:*** Python 3.8*** Tabla de Contenidos1 Diferenciación Numérica1.1 Introducción1.2 Series de Taylor1.3 Esquemas de diferencias finitas para la primera derivada1.3.1 Esquema de primer orden hacia adelante (forward)1.3.2 Esquema de primer orden hacia atrás (backward)1.3.3 Esquema de segundo orden (central)1.3.4 Resumen esquemas diferencias finitas para la primera derivada1.4 Esquemas de diferencias finitas para la segunda derivada1.5 Implementación computacional de algunos esquemas de diferencias finitas2 Integración Numérica2.1 Introducción2.2 Fórmulas de integración de Newton - Cotes2.3 Regla trapezoidal2.3.1 Regla trapezoidal de aplicación simple2.3.2 Regla trapezoidal de aplicación múltiple2.3.3 Implementación computacional2.3.4 Error en la aplicación de la regla trapezoidal2.4 Reglas de Simpson2.4.1 Regla de Simpson1/3 de aplicación simple2.4.2 Error en la regla de Simpson 1/3 de aplicación simple2.4.3 Regla de simpson1/3 de aplicación múltiple2.4.4 Implementación computacional regla de Simpson1/3 de aplicación múltiple2.4.5 Regla de Simpson 3/8 de aplicación simple2.4.6 Regla de Simpson3/8 de aplicación múltiple2.4.7 Implementación computacional de la regla de Simpson3/8 de aplicación múltiple2.5 Cuadratura de Gauss2.5.1 Introducción2.5.2 Determinación de los coeficientes2.5.3 Cambios de los límites de integración2.5.4 Fórmulas de punto superior2.5.5 Ejemplo Cuadratura de Gauss Diferenciación Numérica Introducción La [diferenciación numérica](https://en.wikipedia.org/wiki/Numerical_differentiation) se emplea para determinar (estimar) el valor de la derivada de una función en un punto específico. No confundir con la derivada de una función, pues lo que se obtendrá es un valor puntual y no una función. En este capítulo nos centraremos únicamente en ecuiaciones unidimensionales. [Volver a la Tabla de Contenido](TOC) Series de Taylor De la [serie de Taylor](https://en.wikipedia.org/wiki/Taylor_series) \begin{equation*}f(x_{i \pm 1}) = f(x_i) \pm f'(x_i)h + \frac{f''(x_i)h^2}{2!} \pm \frac{f'''(x_i)h^3}{3!} + \ldots\label{eq:Ec5_1} \tag{5.1}\end{equation*}con $h=\Delta x = x_{i+1}-x_i$ siendo el tamaño de paso.Dada que la serie contiene infinitos términos, partir de la ecuación ($5.1$) se pueden obtener infinitos esquemas numéricos para determinar cada una de las infinitas derivadas de dicho polinomio. En este curso usaremos la técnica de [Diferencias Finitas](https://en.wikipedia.org/wiki/Finite_difference) para desarrollarlas. [Volver a la Tabla de Contenido](TOC) Esquemas de diferencias finitas para la primera derivada Esquema de primer orden hacia adelante (forward) De la ecuación [(5.1)](Ec5_1) tomando los valores positivos, que involucran únicamente términos hacia adelante, se trunca la serie hasta la primera derivada y se realiza un despeje algebraico para llegar a:\begin{equation*}f'(x_i) = \frac{f(x_{i+1})-f(x_i)}{h} + \mathcal{O}(h)\label{eq:Ec5_2} \tag{5.2}\end{equation*}se puede observar que el término $\mathcal{O}(h)$ indica que el error es de orden lineal, es decir, si se reduce el tamaño de paso, $h$, a la mitad, el error se reducirá a la mitad. Si se reduc el tamaño de paso a una cuarta parte, el error se reducirá, linealmente, una cuarta parte. [Volver a la Tabla de Contenido](TOC) Esquema de primer orden hacia atrás (backward) De la ecuación [(5.1)](Ec5_1) tomando los valores negativos, que involucran únicamente términos hacia atrás (backward), se trunca la serie hasta la primera derivada y se realiza un despeje algebraico para llegar a:\begin{equation*}f'(x_i) = \frac{f(x_{i})-f(x_{i-1})}{h} + \mathcal{O}(h)\label{eq:Ec5_3} \tag{5.3}\end{equation*}se observa que se llega a una expresión similar a la de la ecuación [(5.2)](Ec5_2), pero de esta vez, se tiene en cuenta es el valor anterior al punto $x_i$. También se observa que el error es de orden lineal, por lo que se mantiene un esquema de primer orden. [Volver a la Tabla de Contenido](TOC) Esquema de segundo orden (central) Una forma de aumentar el orden de estos esquemas, es realizar el truncamiento de la *serie de Taylor* hasta la segunda derivada, hacia adelante y hacia atras, y realizar su resta aritmética.\begin{equation*}\begin{split}f(x_{i+1}) & = f(x_i) + f'(x_i)h + \frac{f''(x_i)h^2}{2!} \\- \\f(x_{i-1}) & = f(x_i) - f'(x_i)h + \frac{f''(x_i)h^2}{2!} \\\hline \\f(x_{i+1}) - f(x_{i-1}) & = 2 f'(x_i)h\end{split}\label{eq:Ec5_4} \tag{5.4}\end{equation*} de la anterior ecuación, despejando el término que corresponde a la primera derivada queda:\begin{equation*}\begin{split}f'(x_i) = \frac{f(x_{i+1}) - f(x_{i-1})}{2h} + \mathcal{O}(h^2)\end{split}\label{eq:Ec5_5} \tag{5.5}\end{equation*}se llega al esquema de diferencias finitas central para la primera derivada, que es de orden dos, es decir, si se disminuye el tamaño de paso, $h$, a la mitad, el error se disminuye una cuarta partes. En principio, esta es una mejor aproximación que los dos esquemas anteriores. La selección del esquema dependerá de la disponibilidad de puntos y del fenómeno físico a tratar. [Volver a la Tabla de Contenido](TOC) Resumen esquemas diferencias finitas para la primera derivada Como la serie de Taylor es infinita, se podrían determinar infinitos esquemas de diferentes ordenes para la primera derivada. En la siguiente tabla se presentan algunos esquemas de diferencias finitas para la primera derivada de diferentes órdenes. Se deja al estudiante la consulta de otros esquemas.|***Esquema***|***Función***|***Error***||:-----:|:-----:|:---:||***Forward***|$$f´(x_0)=\frac{f(x_0+h)-f(x_0)}{h}$$|$$\mathcal{O}(h)$$|| |$$f´(x_0)=\frac{-3f(x_0)+4f(x_0+h)-f(x_0+2h)}{2h}$$|$$\mathcal{O}(h^2)$$||***Central***|$$f´(x_0)=\frac{f(x_0+h)-f(x_0-h)}{2h}$$|$$\mathcal{O}(h^2)$$|| |$$f´(x_0)=\frac{f(x_0-2h)-8f(x_0-h)+8f(x_0+h)-f(x_0+2h)}{12h}$$|$$\mathcal{O}(h^4)$$||***Backward***|$$f´(x_0)=\frac{f(x_0)-f(x_0-h)}{h}$$|$$\mathcal{O}(h)$$|| |$$f´(x_0)=\frac{f(x_0-2h)-4f(x_0-h)+3f(x_0)}{2h}$$|$$\mathcal{O}(h^2)$$| [Volver a la Tabla de Contenido](TOC) Esquemas de diferencias finitas para la segunda derivada Siguiendo con la misma forma de abordar el problema para la primera derivada, si se amplian los términos en la serie de Taylor hasta la tercera derivada tanto hacia adelante como hacia atrás, y se suman, se llega a:\begin{equation*}\begin{split}f(x_{i+1}) & = f(x_i) + f'(x_i)h + \frac{f''(x_i)h^2}{2!} + \frac{f'''(x_i)h^3}{3!}\\+ \\f(x_{i-1}) & = f(x_i) - f'(x_i)h + \frac{f''(x_i)h^2}{2!} - \frac{f'''(x_i)h^3}{3!}\\\hline \\f(x_{i+1}) + f(x_{i-1}) & = 2 f(x_i) + 2f''(x_i)\frac{h^2}{2!} + \mathcal{O}(h^3)\end{split}\label{eq:Ec5_6} \tag{5.6}\end{equation*} Despejando para el término de la segunda derivada, se llega a:\begin{equation*}\begin{split}f''(x_i) = \frac{f(x_{i+1}) - 2f(x_i) + f(x_{i-1})}{h^2} + \mathcal{O}(h^3)\end{split}\label{eq:Ec5_7} \tag{5.7}\end{equation*}Que corresponde a un esquema de diferencias finitas de segundo orden para la segunda derivada. A este esquema también se le llama "*molécula de tres puntos*"Igual que para la primera derivada, se pueden determinar infinitos esquemas de diferentes órdenes para la segunda derivada, y derivadas superiores. A continuación se muestra un cuadro resumen de algunos esquemas de diferencias finitas para la segunda derivada. Se deja al estudiante la revisión de esquemas de mayor orden para la segunda derivada y derivadas superiores.|***Esquema***|***Función***|***Error***||:-----:|:-----:|:---:||***Forward***|$$f''(x_0)=\frac{f(x_0)-2f(x_0+h)+f(x_0+2h)}{h^2}$$|$$\mathcal{O}(h)$$|| |$$f''(x_0)=\frac{2f(x_0)-5f(x_0+h)+4f(x_0+2h)-f(x_0+3h)}{h^2}$$|$$\mathcal{O}(h^2)$$||***Central***|$$f''(x_0)=\frac{f(x_0-h)-2f(x_0)+f(x_0+h)}{h^2}$$|$$\mathcal{O}(h^2)$$|| |$$f''(x_0)=\frac{-f(x_0-2h)+16f(x_0-h)-30f(x_0)+16f(x_0+h)-f(x_0+2h)}{12h^2}$$|$$\mathcal{O}(h^4)$$||***Backward***|$$f''(x_0)=\frac{f(x_0-2h)-2f(x_0-h)+f(x_0)}{h}$$|$$\mathcal{O}(h^2)$$|| |$$f''(x_0)=\frac{-f(x_0-3h)+4f(x_0-2h)-5f(x_0-h)+2f(x_0)}{h^2}$$|$$\mathcal{O}(h^2)$$| [Volver a la Tabla de Contenido](TOC) Implementación computacional de algunos esquemas de diferencias finitas A manera de ejemplo, se implementarán algunos esquemas simples de diferencias finitas para la primera derivada. Se deja como actividad a los estudiantes la implementación de otros esquemas para las diferentes derivadas. | import numpy as np
import matplotlib.pyplot as plt
import sympy as sym
sym.init_printing()
#Esquemas de diferencias finitas para la primera derivada
def df1df(x0, h):
# Esquema de diferencias finitas para la primera derivada hacia adelante (forward)
return (f(x0 + h) - f(x0)) / h
def df1db(x0, h):
# Esquema de diferencias finitas para la primera derivada hacia atrás (backward)
return (f(x0) - f(x0 - h) ) / h
def df1dc(x0,h):
# Esquema de diferencias finitas para la primera derivada central (central)
return (f(x0 + h) - f(x0 - h) ) / (2 * h)
#funcion a determinar el valor de la derivada
def f(x):
return 2*x**3 - 3*x**2 + 5*x+0.8
#cálculo y evaluación de la primera derivada empleando cálculo simbólico
def df1de(x0):
x = sym.Symbol('x')
df = sym.diff(f(x), x)
#print(df)
df1 = df.evalf(subs={x:x0})
return df1
h = 0.1
x0 = 0.8
print("1st derivative \t Value \t\t Error(%)")
print('---------------------------------------')
pde = df1de(x0)
pdf = df1df(x0, h)
epdf = abs((pde - pdf) / pde * 100)
print("forward \t {0:6.4f} \t {1:6.2f}".format(pdf,epdf))
pdb = df1db(x0, h)
epdb = abs((pde - pdb) / pde * 100)
print("backward \t {0:6.4f} \t {1:6.2f}".format(pdb,epdb))
pdc = df1dc(x0,h)
epdc = abs((pde - pdc) / pde * 100)
print("central \t {0:6.4f} \t {1:6.2f}".format(pdc, epdc))
print("exacta \t\t {0:6.4f} \t {1}".format(pde, ' -')) | _____no_output_____ | MIT | Cap05_IntegracionNumerica.ipynb | Youngermaster/Numerical-Analysis |
[Volver a la Tabla de Contenido](TOC) Integración Numérica Introducción La [integración numérica](https://en.wikipedia.org/wiki/Numerical_integration) aborda una amplia gama de algoritmos para determinar el valor numérico (aproximado) de una integral definida. En este curso nos centraremos principalmente en los métodos de cuadratura, tanto de interpolación como [gaussiana](https://en.wikipedia.org/wiki/Gaussian_quadrature), como dos ejemplos de dichos algoritmos. El problema a tratar en este capítulo es la solución aproximada de la función\begin{equation*}\begin{split}I = \int_a^b f(x) dx\end{split}\label{eq:Ec5_8} \tag{5.8}\end{equation*} [Volver a la Tabla de Contenido](TOC) Fórmulas de integración de *Newton - Cotes* La idea básica en la integración numérica es cambiar una función difícil de integrar, $f(x)$, dada por la ecuación [(5.8)](Ec5_8), por una función más simple, $p_n(x)$,\begin{equation*}\begin{split}\widetilde{I} \approx \int_{a=x_0}^{b=x_n} p_{n}(x) dx\end{split}\label{eq:Ec5_9} \tag{5.9}\end{equation*}Cabe resaltar que en integración numérica no se conocerá la función a integrar, solo se dispondrá de una serie de $n+1$ puntos $(x_i, y_i), i = 0, 1, 2, \ldots, n$, y a partir de ellos se construye un polinomio interpolante de grado $n$, $p_n$, entre los valores de los límites de integración $a = x_0$ y $b=x_n$. $p_n(x)$ es un polinomio de interpolación de la forma\begin{equation*}\begin{split}p_n(x)=a_0+a_1x+a_2x^2+\ldots+a_{n-1}x^{n-1}+a_nx^n\end{split}\label{eq:Ec5_10} \tag{5.10}\end{equation*}Las fórmulas de integración de [*Newton - Cotes*](https://en.wikipedia.org/wiki/Newton%E2%80%93Cotes_formulas), también llamadas de [cuadratura](https://en.wikipedia.org/wiki/Quadrature_(mathematics)), son un grupo de fórmulas de integración numérica de tipo interpolación, evaluando la función en puntos equidistantes, para determinar un valor aproximado de la integral. Si no se tienen puntos espaciados, otros métodos deben ser usados, como por ejemplo cuadratura gaussiana, que se verá al final del capítulo.La forma general de las fórmulas de Newton - Cotes está dada por la función:\begin{equation*}\begin{split}p_n(x)=\sum \limits_{i=0}^n f(x_i)L_{in}(x)\end{split}\label{eq:Ec5_11} \tag{5.11}\end{equation*}donde\begin{equation*}\begin{split}L_{in}(x)=\frac{(x-x_0)\ldots(x-x_{i-1})(x-x_{i+1})\ldots(x-x_n)}{(x_i-x_0)\ldots(x_i-x_{i-1})(x_i-x_{i+1})\ldots(x_i-x_n)}\end{split}\label{eq:Ec5_12} \tag{5.12}\end{equation*}es el polinomio de Lagrange, de donde se deduce que:\begin{equation*}\begin{split}\int_a^b p(x)dx=(b-a)\sum \limits_{i=0}^n f(x_i) \frac{1}{(b-a)} \int_a^b L_{in}(x)dx\end{split}\label{eq:Ec5_13} \tag{5.13}\end{equation*}entonces,\begin{equation*}\begin{split}\int_a^b f(x)dx \approx \int_a^b p(x)dx=(b-a)\sum \limits_{i=0}^n w_if(x_i) \end{split}\label{eq:Ec5_14} \tag{5.14}\end{equation*}donde los pesos, $w_i$ de la función son representados por\begin{equation*}\begin{split}w_i=\frac{1}{(b-a)} \int_a^b L_{in}(x)dx\end{split}\label{eq:Ec5_15} \tag{5.15}\end{equation*}A partir de esta idea se obtienen los diferentes esquemas de integración numérica de *Newton - Cotes* [Volver a la Tabla de Contenido](TOC) Regla trapezoidal Regla trapezoidal de aplicación simple La [regla trapezoidal](https://en.wikipedia.org/wiki/Trapezoidal_rule) emplea una aproximación de la función mediante una línea recta Fuente: wikipedia.com y corresponde al caso en el que el polinomio en la ecuación [(5.11)](Ec5_11) es de primer orden\begin{equation*}\begin{split}I=\int_{a}^{b}f(x)dx \approx \int_a^b \left[ f(a) + \frac{f(b)-f(a)}{b-a}(x-a)\right]dx= (b-a)\frac{f(a)+f(b)}{2}\end{split}\label{eq:Ec5_16} \tag{5.16}\end{equation*}Geométricamente, es equivalente a aproximar el área del trapezoide bajo la línea recta que conecta $f(a)$ y $f(b)$. La integral se representa como:$$I ≈ \text{ancho} \times \text{altura promedio}$$El error en la regla trapezoidal simple se puede determinar como:\begin{equation*}\begin{split}E_t=-\frac{1}{12}f''(\xi)(b-a)^3\end{split}\label{eq:Ec5_17} \tag{5.17}\end{equation*} [Volver a la Tabla de Contenido](TOC) Regla trapezoidal de aplicación múltiple Una manera de mejorar la exactitud de la regla trapezoidal es dividir el intervalo de integración de $a$ a $b$ en un número $n$ de segmentos y aplicar el método a cada uno de ellos. Las ecuaciones resultantes son llamadas fórmulas de integración de múltiple aplicación o compuestas. Fuente: wikipedia.com Hay $n+1$ puntos base igualmente espaciados $(x_0, x_1, x_2, \ldots, x_n)$. En consecuencia hay $n$ segmentos de igual anchura: $h = (b–a) / n$. Si $a$ y $b$ son designados como $x_0$ y $x_n$ respectivamente, la integral total se representará como:\begin{equation*}\begin{split}I=\int_{x_0}^{x_1}f(x)dx+\int_{x_1}^{x_2}f(x)dx+\int_{x_2}^{x_3}f(x)dx+\ldots+\int_{x_{n-2}}^{x_{n-1}}f(x)dx+\int_{x_{n-1}}^{x_n}f(x)dx\end{split}\label{eq:Ec5_18} \tag{5.18}\end{equation*}Al sustituir la regla trapezoidal simple en cada integrando, se tiene\begin{equation*}\begin{split}I\approx \left(f(x_0)+f(x_1)\right)\frac{h}{2}+\left(f(x_1)+f(x_2)\right)\frac{h}{2}+\left(f(x_2)+f(x_3)\right)\frac{h}{2}+\ldots\left(f(x_{n-2})+f(x_{n-1})\right)\frac{h}{2}+\left(f(x_{n-1})+f(x_n)\right)\frac{h}{2}\end{split}\label{eq:Ec5_19} \tag{5.19}\end{equation*}ahora, agrupando términos\begin{equation*}\begin{split}I\approx \frac{h}{2}\left[ f(x_0) + 2\sum_{i=1}^{n-1}f(x_i)+f(x_n) \right]\end{split}\label{eq:Ec5_20} \tag{5.20}\end{equation*}donde $h=(b-a)/n$ [Volver a la Tabla de Contenido](TOC) Implementación computacional | import numpy as np
import matplotlib.pyplot as plt
def trapezoidal(x):
n = len(x)
h = (x[-1] - x[0]) / n
suma = 0
for i in range(1, n-1):
suma += funcion(x[i])
return h * (funcion(x[0]) + 2 * suma + funcion(x[-1])) / 2
def funcion(x):
return 4 / (1 + x**2)
a = 0
b = 1
n = 1000
x = np.linspace(a, b, n+1)
I = trapezoidal(x)
I | _____no_output_____ | MIT | Cap05_IntegracionNumerica.ipynb | Youngermaster/Numerical-Analysis |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.