markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
Let's see how many hotspot mutations there are in the Cholangiocarcinoma (TCGA, PanCancer Atlas) study with study id `chol_tcga_pan_can_atlas_2018` from the cBioPortal: | %%time
cbioportal = SwaggerClient.from_url('https://www.cbioportal.org/api/api-docs',
config={"validate_requests":False,"validate_responses":False})
mutations = cbioportal.K_Mutations.getMutationsInMolecularProfileBySampleListIdUsingGET(
molecularProfileId='chol_tcga_pan_can_atlas_2018_mutations',
sampleListId='chol_tcga_pan_can_atlas_2018_all',
projection='DETAILED'
).result() | CPU times: user 766 ms, sys: 20.1 ms, total: 786 ms
Wall time: 1.03 s
| MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Convert the results to a dataframe again: | import pandas as pd
mdf = pd.DataFrame.from_dict([
# python magic that combines two dictionaries:
dict(
m.__dict__['_Model__dict'],
**m.__dict__['_Model__dict']['gene'].__dict__['_Model__dict'])
# create one item in the list for each mutation
for m in mutations
]) | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Then get only the unique mutations, to avoid calling the web service with the same variants: | variants = mdf['chromosome startPosition endPosition referenceAllele variantAllele'.split()]\
.drop_duplicates()\
.dropna(how='any',axis=0)\
.reset_index() | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Convert them to input that genome nexus will understand: | variants = variants.rename(columns={'startPosition':'start','endPosition':'end'})\
.to_dict(orient='records')
# remove the index field
for v in variants:
del v['index']
print("There are {} mutations left to annotate".format(len(variants))) | There are 1991 mutations left to annotate
| MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Annotate them with genome nexus: | %%time
variants_annotated = gn.annotation_controller.fetchVariantAnnotationByGenomicLocationPOST(
genomicLocations=variants,
fields='hotspots annotation_summary'.split()
).result() | CPU times: user 3.22 s, sys: 522 ms, total: 3.75 s
Wall time: 6.61 s
| MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Index the variants to make it easier to query them: | gn_dict = {
"{},{},{},{},{}".format(
v.annotation_summary.genomicLocation.chromosome,
v.annotation_summary.genomicLocation.start,
v.annotation_summary.genomicLocation.end,
v.annotation_summary.genomicLocation.referenceAllele,
v.annotation_summary.genomicLocation.variantAllele)
:
v for v in variants_annotated
} | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Add a new column to indicate whether something is a hotspot | def is_hotspot(x):
"""TODO: Current structure for hotspots in Genome Nexus is a little funky.
Need to check whether all lists in the annotation field are empty."""
if x:
return sum([len(a) for a in x.hotspots.annotation]) > 0
else:
return False
def create_dict_query_key(x):
return "{},{},{},{},{}".format(
x.chromosome, x.startPosition, x.endPosition, x.referenceAllele, x.variantAllele
)
mdf['is_hotspot'] = mdf.apply(lambda x: is_hotspot(gn_dict.get(create_dict_query_key(x), None)), axis=1) | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Then plot the results: | %matplotlib inline
import seaborn as sns
sns.set_style("white")
sns.set_context('notebook')
import matplotlib.pyplot as plt
mdf.groupby('hugoGeneSymbol').is_hotspot.sum().sort_values(ascending=False).head(10).plot(kind='bar')
sns.despine(trim=False)
plt.xlabel('')
plt.xticks(rotation=300)
plt.ylabel('Number of non-unique hotspots',labelpad=20)
plt.title('Hotspots in Cholangiocarcinoma (TCGA, PanCancer Atlas)',pad=25) | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
OncoKB API [OncoKB](https://oncokb.org) is is a precision oncology knowledge base and contains information about the effects and treatment implications of specific cancer gene alterations. Similarly to cBioPortal and Genome Nexus it provides a REST API following the [Swagger / OpenAPI specification](https://swagger.io/specification/). | oncokb = SwaggerClient.from_url('https://www.oncokb.org/api/v1/v2/api-docs',
config={"validate_requests":False,
"validate_responses":False,
"validate_swagger_spec":False})
print(oncokb) | SwaggerClient(https://www.oncokb.org:443/api/v1)
| MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
To look up annotations for a variant, one can use the following endpoint: | variant = oncokb.Annotations.annotateMutationsByGenomicChangeGetUsingGET(
genomicLocation='7,140453136,140453136,A,T',
).result()
drugs = oncokb.Drugs.drugsGetUsingGET().result() | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
You can see a lot of information is provided for that particular variant if you type tab after `variant.`: | drugs.count
variant.hotspot | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
For instance we can see the summary information about it: | variant.variantSummary | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
If you look up this variant on the OncoKB website: https://www.oncokb.org/gene/BRAF/V600E. You can see that there are various combinations of drugs and their level of evidence listed. This is a classification system for indicating how much we know about whether or not a patient might respond to a particular treatment. Please see https://www.oncokb.org/levels for more information about the levels of evidence for therapeutic biomarkers.We can use the same `variants` we pulled from cBioPortal in the previous section to figure out the highest level of each variant. | %%time
variants_annotated = oncokb.Annotations.annotateMutationsByGenomicChangePostUsingPOST(
body=[
{"genomicLocation":"{chromosome},{start},{end},{referenceAllele},{variantAllele}".format(**v)}
for v in variants
],
).result() | CPU times: user 363 ms, sys: 16.4 ms, total: 379 ms
Wall time: 9.89 s
| MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Count the highes level for each variant | from collections import Counter
counts_per_level = Counter([va.highestSensitiveLevel for va in variants_annotated if va.highestSensitiveLevel]) | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Then plot them | pd.DataFrame(counts_per_level,index=[0]).plot(kind='bar', colors=['#4D8834','#2E2E2C','#753579'])
plt.xticks([])
plt.ylabel('Number of variants')
plt.title('Actionable variants in chol_tcga_pan_can_atlas_2018')
sns.despine() | _____no_output_____ | MIT | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon |
Fetching the raw data from google places API by passing the coordinates of the cluster centroid and delivery radius as 5km | import pandas as pd
import googlemaps
import pprint
import json
import time
import xlsxwriter
import functools
import operator
from collections import Counter
from itertools import chain
#IMPORTING DATA
df = pd.read_excel('lat_long_google_api.xlsx')
rslt_df= df.copy()
# Define our API Key
API_KEY = 'Enter your API key'
# Define our Client
gmaps = googlemaps.Client(key = API_KEY) | _____no_output_____ | MIT | Fetching_data_google_places_API.ipynb | vivek1240/Fetching-the-raw-data-from-google-places-API |
Below code is the Call for the Places API and the result will be stored in a dictionary, we will take the key = store_id, value = result fetched for the corresponding store from the places API Dict "d" will contain the raw data corresponding to the coordinate(lat,long) | d= dict() #ALL THE RAW DATA WOULD BE STORED IN DICTIONARY CORRESPONDING TO THE LATITUDE OF THE STORE
d[rslt_df['store_id'][0]]=dict() #Taking store latitude as the
for i in range(len(rslt_df)):
lat= rslt_df['store_latitude'][i]
lon= rslt_df['store_longitude'][i]
d[rslt_df['store_id'][i]] = gmaps.places_nearby(location='{},{}'.format(lat,lon), radius = 5000, open_now =False ) #not giving type parameter so it will give all types in the result
time.sleep(5)
print(d[rslt_df['store_id'][i]]) | _____no_output_____ | MIT | Fetching_data_google_places_API.ipynb | vivek1240/Fetching-the-raw-data-from-google-places-API |
Fetched data with the index as cluster number | def no_of_lodges_or_eqv(d,key):
rawdata=[]
for i in range(len(d[key]['results'])):
rawdata.append(d[key]['results'][i]['types'])
rawdata = functools.reduce(operator.iconcat, rawdata, [])
rawdata = CountFrequency(rawdata)
return rawdata
def CountFrequency(my_list):
# Creating an empty dictionary
freq = {}
for item in my_list:
if (item in freq):
freq[item] += 1
else:
freq[item] = 1
return freq
#############
df_podcast=pd.DataFrame(columns=['atm', 'bakery', 'bank', 'establishment', 'finance', 'food', 'health', 'hospital', 'locality', 'point_of_interest', 'political', 'real_estate_agency', 'spa', 'store', 'sublocality', 'sublocality_level_1'])
###########
df_podcast.head() | _____no_output_____ | MIT | Fetching_data_google_places_API.ipynb | vivek1240/Fetching-the-raw-data-from-google-places-API |
MobileNetSSD with OpenCV- you can get trained model and prototxt : https://www.pyimagesearch.com/2017/09/11/object-detection-with-deep-learning-and-opencv/ | %matplotlib inline
# import the necessary packages
import numpy as np
import sys
from logging import getLogger, DEBUG, StreamHandler
import matplotlib.pyplot as plt
import cv2
def deep_learning_object_detection(image, prototxt, model):
logger = getLogger(__name__)
logger.setLevel(DEBUG)
handler = StreamHandler(sys.stderr)
handler.setLevel(DEBUG)
logger.addHandler(handler)
# construct the argument parse and parse the arguments
CONFIDENCE = 0.2
# initialize the list of class labels MobileNet SSD was trained to
# detect, then generate a set of bounding box colors for each class
CLASSES = ["background", "aeroplane", "bicycle", "bird", "boat","bottle", "bus", "car", "cat", "chair", "cow", "diningtable",
"dog", "horse", "motorbike", "person", "pottedplant", "sheep", "sofa", "train", "tvmonitor"]
COLORS = np.random.uniform(0, 255, size=(len(CLASSES), 3))
# load our serialized model from disk
logger.info("Loading model...")
net = cv2.dnn.readNetFromCaffe(prototxt, model)
# load the input image and construct an input blob for the image
# by resizing to a fixed 300x300 pixels and then normalizing it
# (note: normalization is done via the authors of the MobileNet SSD
# implementation)
image = cv2.imread(image)
(h, w) = image.shape[:2]
blob = cv2.dnn.blobFromImage(cv2.resize(image, (300, 300)), 0.007843, (300, 300), 127.5)
# pass the blob through the network and obtain the detections and
# predictions
logger.info("computing object detections...")
net.setInput(blob)
detections = net.forward()
# loop over the detections
for i in np.arange(0, detections.shape[2]):
# extract the confidence (i.e., probability) associated with the
# prediction
confidence = detections[0, 0, i, 2]
# filter out weak detections by ensuring the `confidence` is
# greater than the minimum confidence
if confidence > CONFIDENCE:
# extract the index of the class label from the `detections`,
# then compute the (x, y)-coordinates of the bounding box for
# the object
idx = int(detections[0, 0, i, 1])
box = detections[0, 0, i, 3:7] * np.array([w, h, w, h])
(startX, startY, endX, endY) = box.astype("int")
# display the prediction
label = "{}: {:.2f}%".format(CLASSES[idx], confidence * 100)
logger.info(label)
cv2.rectangle(image, (startX, startY), (endX, endY), COLORS[idx], 2)
y = startY - 15 if startY - 15 > 15 else startY + 15
cv2.putText(image, label, (startX, y), cv2.FONT_HERSHEY_SIMPLEX, 0.5, COLORS[idx], 2)
# show the output image
plt.imshow(image)
image = "images/example_01.jpg"
prototxt="MobileNetSSD_deploy.prototxt.txt"
model="MobileNetSSD_deploy.caffemodel"
deep_learning_object_detection(image, prototxt, model) | Loading model...
computing object detections...
car: 99.96%
car: 95.68%
| MIT | MobileNetSSD_OpenCV.ipynb | hurutoriya/yolov2_api |
try 1 | page_x_inches: float = 11. # inches
page_y_inches: float = 8.5 # inches
border:float = 0.
perlin_grid_params = {
'xstep':3,
'ystep':3,
'lod':10,
'falloff':None,
'noise_scale':0.0073,
'noiseSeed':6
}
particle_init_grid_params = {
'xstep':16,
'ystep':16,
}
buffer_style = 2
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
pg = gp.PerlinGrid(drawbox, **perlin_grid_params)
start_area = sa.scale(drawbox.centroid.buffer(brad*0.45), xfact=1.3)
start_area = drawbox.buffer(-20)
xcs, ycs = gp.overlay_grid(start_area, xstep=19, ystep=15)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=20, xscale=4, yscale=4)
pc.init_particles()
n_steps = np.random.randint(low=10, high=50, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=1.3) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.45, high=-0.25),
angles=-90,
)
stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)
P.fill_scale_trans(**stp.prms)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except :
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills = []
for p in gpolys:
try:
ifills.append(p.intersection_fill)
except:
pass
splits = utils.random_split(ifills, n_layers=5)
layers = [utils.merge_LineStrings(split) for split in splits]
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0083_perlin_flow_erode_frays_occlude.svg'
sk.save(savepath)
vpype_commands = 'linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 2 | page_x_inches: float = 11. # inches
page_y_inches: float = 8.5 # inches
border:float = 0.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0073,
'noiseSeed':8
}
buffer_style = 2
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
pg = gp.PerlinGrid(drawbox, **perlin_grid_params)
start_area = drawbox
xcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=30, xscale=7, yscale=7)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=10, high=90, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=1.3) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.4, high=-0.2),
d_translate_factor=0.7,
angles=-240,
)
stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)
P.fill_scale_trans(**stp.prms)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills = []
for p in gpolys:
ifills.append(p.intersection_fill)
splits = utils.random_split(ifills, n_layers=4)
layers = [utils.merge_LineStrings(split) for split in splits]
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0084_perlin_flow_erode_frays_occlude.svg'
sk.save(savepath)
vpype_commands = 'linesimplify --tolerance 0.05mm linemerge --tolerance 0.1mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 3 | page_x_inches: float = 6 # inches
page_y_inches: float = 6 # inches
border:float = 20.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0063,
'noiseSeed':8
}
buffer_style = 2
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
pg = gp.PerlinGrid(drawbox, **perlin_grid_params)
start_area = drawbox
xcs, ycs = gp.overlay_grid(start_area, xstep=45, ystep=45)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=30, xscale=6, yscale=6)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=80, high=190, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=1.3) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.4, high=-0.2),
d_translate_factor=0.7,
angles=-240,
)
stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)
P.fill0 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.4, high=-0.2),
d_translate_factor=0.7,
angles=120,
)
stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)
P.fill1 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills0 = []
ifills1 = []
for p in gpolys:
ifills0.append(p.fill0.intersection(p.p.buffer(1e-6)))
ifills1.append(p.fill1.intersection(p.p.buffer(1e-6)))
layers = []
layers.append(gp.merge_LineStrings(ifills0))
layers.append(gp.merge_LineStrings(ifills1))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0085_perlin_flow_erode_frays_color_mix.svg'
sk.save(savepath)
vpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 4 for fabiano black black |
border:float = 25.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0063,
'noiseSeed':8
}
buffer_style = 2
px = 200
py = 200
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
start_area = drawbox.centroid.buffer(brad/2)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=11, ystep=11)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=35, xscale=4, yscale=4)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=10, high=60, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=1.7) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.45, high=-0.25),
d_translate_factor=0.7,
angles=-240,
)
stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)
P.fill0 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills0 = []
for p in gpolys:
ifills0.append(p.fill0.intersection(p.p.buffer(1e-6)))
ifills0 = [l for l in ifills0 if l.length > 1e-1]
layers = []
layers.append(gp.merge_LineStrings(ifills0))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0086_perlin_flow_erode_frays_color_mix.svg'
sk.save(savepath)
vpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 5 two color | page_x_inches: float = 8.5 # inches
page_y_inches: float = 11 # inches
border:float = 0.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0063,
'noiseSeed':8
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
start_area = drawbox.centroid.buffer(brad*0.47)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=30, xscale=7, yscale=7)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=10, high=100, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=2.7) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.5, high=-0.25),
d_translate_factor=0.7,
angles=np.radians(-60),
)
stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)
P.fill0 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.5, high=-0.25),
d_translate_factor=0.7,
angles=np.radians(-120),
)
stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)
P.fill1 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills0 = []
ifills1 = []
for p in gpolys:
ifills0.append(p.fill0.intersection(p.p.buffer(1e-6)))
ifills1.append(p.fill1.intersection(p.p.buffer(1e-6)))
ifills0 = [l for l in ifills0 if l.length > 1e-1]
ifills1 = [l for l in ifills1 if l.length > 1e-1]
layers = []
layers.append(gp.merge_LineStrings(ifills0))
layers.append(gp.merge_LineStrings(ifills1))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0087_perlin_flow_erode_frays_color_mix.svg'
sk.save(savepath)
vpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 6 three color | page_x_inches: float = 8.5 # inches
page_y_inches: float = 11 # inches
border:float = 0.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0063,
'noiseSeed':8
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
start_area = drawbox.centroid.buffer(brad*0.47)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
n_fills = 3
fill_angles = [-30, -90, -150]
n_iter_choices = np.array(list(itertools.product(*[[2, 100]] * n_fills)))
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=30, xscale=7, yscale=7)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=10, high=100, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=2.7) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
P.fills = []
n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]
for i in range(n_fills):
stp = ScaleTransPrms(
n_iters=n_iter_choice[i],
d_buffer=np.random.uniform(low=-0.6, high=-0.25),
d_translate_factor=0.7,
angles=np.radians(fill_angles[i]),)
stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)
fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
P.fills.append(fill)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
fill_sets = []
for i in range(n_fills):
fill_sets.append([])
for p in gpolys:
for i in range(n_fills):
fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))
layers = []
for fill_set in fill_sets:
filter_fills = [l for l in fill_set if l.length > 0.2]
layers.append(gp.merge_LineStrings(filter_fills))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0088_perlin_flow_erode_frays_color_mix.svg'
sk.save(savepath)
vpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 7 | page_x_inches: float = 8.5 # inches
page_y_inches: float = 11 # inches
border:float = 0.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0063,
'noiseSeed':8
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
# start_area = drawbox.centroid.buffer(brad*0.47)
start_area = drawbox
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=35, ystep=35)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
n_fills = 3
fill_angles = [-30, -90, -150]
n_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))
n_iter_choices = n_iter_choices[1:-1,:]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=60, xscale=6, yscale=6)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=4, high=7, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=0.7) + np.random.uniform(low=0., high=3.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
P.fills = []
n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]
for i in range(n_fills):
stp = ScaleTransPrms(
n_iters=n_iter_choice[i],
d_buffer=np.random.uniform(low=-0.6, high=-0.25),
d_translate_factor=0.7,
angles=np.radians(fill_angles[i]),)
stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)
fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
P.fills.append(fill)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
fill_sets = []
for i in range(n_fills):
fill_sets.append([])
for p in gpolys:
for i in range(n_fills):
fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))
layers = []
for fill_set in fill_sets:
filter_fills = [l for l in fill_set if l.length > 0.2]
layers.append(gp.merge_LineStrings(filter_fills))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0090_perlin_flow_erode_frays_color_mix.svg'
sk.save(savepath)
vpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 8 | page_x_inches: float = 8.5 # inches
page_y_inches: float = 11 # inches
border:float = 0.
perlin_grid_params = {
'xstep':3,
'ystep':3,
'lod':10,
'falloff':None,
'noise_scale':0.0013,
'noiseSeed':8
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
# start_area = drawbox.centroid.buffer(brad*0.47)
start_area = drawbox.buffer(-5, cap_style=3, join_style=3)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
f,ax = plt.subplots(figsize=(6,6))
ax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)
n_fills = 3
fill_angles = [-30, -40, -50]
n_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))
n_iter_choices = n_iter_choices[1:-1,:]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=15, xscale=3, yscale=3)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.1, high=1.7) + np.random.uniform(low=0., high=0.3, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
P.fills = []
n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]
for i in range(n_fills):
stp = ScaleTransPrms(
n_iters=n_iter_choice[i],
d_buffer=np.random.uniform(low=-0.6, high=-0.2),
d_translate_factor=0.8,
angles=np.radians(fill_angles[i]),)
stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)
fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
P.fills.append(fill)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
fill_sets = []
for i in range(n_fills):
fill_sets.append([])
for p in gpolys:
for i in range(n_fills):
fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))
layers = []
for fill_set in fill_sets:
filter_fills = [l for l in fill_set if l.length > 0.2]
layers.append(gp.merge_LineStrings(filter_fills))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0091_perlin_flow_erode_frays_color_mix.svg'
sk.save(savepath)
vpype_commands = 'reloop linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 9 | page_x_inches: float = 6 # inches
page_y_inches: float = 6 # inches
border:float = 0.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0063,
'noiseSeed':8
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
# start_area = drawbox.centroid.buffer(brad*0.47)
start_area = drawbox.buffer(-10)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=25, ystep=25)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
n_fills = 3
fill_angles = [-30, -90, -150]
n_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))
n_iter_choices = n_iter_choices[1:-1,:]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=30, xscale=5, yscale=5)
pc.init_particles(start_bounds=start_area)
n_steps = np.random.randint(low=4, high=17, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=0.7) + np.random.uniform(low=0., high=3.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
P.fills = []
n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]
for i in range(n_fills):
stp = ScaleTransPrms(
n_iters=n_iter_choice[i],
d_buffer=np.random.uniform(low=-0.6, high=-0.25),
d_translate_factor=0.7,
angles=np.radians(fill_angles[i]),)
stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)
fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
P.fills.append(fill)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
fill_sets = []
for i in range(n_fills):
fill_sets.append([])
for p in gpolys:
for i in range(n_fills):
fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))
layers = []
for fill_set in fill_sets:
filter_fills = [l for l in fill_set if l.length > 0.2]
layers.append(gp.merge_LineStrings(filter_fills))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0092_perlin_flow_erode_frays_color_mix.svg'
sk.save(savepath)
vpype_commands = 'reloop linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 10 | page_x_inches: float = 8.5 # inches
page_y_inches: float = 11 # inches
border:float = 0.
perlin_grid_params = {
'xstep':3,
'ystep':3,
'lod':10,
'falloff':None,
'noise_scale':0.0013,
'noiseSeed':8
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
# start_area = drawbox.centroid.buffer(brad*0.47)
start_area = drawbox.buffer(-5, cap_style=3, join_style=3)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=25, ystep=25)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
f,ax = plt.subplots(figsize=(6,6))
ax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)
n_fills = 2
fill_angles = [-30, -40, ]
n_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))
n_iter_choices = n_iter_choices[[-1], :]
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=35, xscale=5, yscale=5)
pc.init_particles(start_bounds=drawbox)
n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.1, high=1.7) + np.random.uniform(low=0., high=0.3, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
P.fills = []
n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]
for i in range(n_fills):
stp = ScaleTransPrms(
n_iters=n_iter_choice[i],
d_buffer=np.random.uniform(low=-0.6, high=-0.2),
d_translate_factor=0.8,
angles=np.radians(fill_angles[i]),)
stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)
fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])
P.fills.append(fill)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except:
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
fill_sets = []
for i in range(n_fills):
fill_sets.append([])
for p in gpolys:
for i in range(n_fills):
fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))
layers = []
for fill_set in fill_sets:
filter_fills = [l for l in fill_set if l.length > 0.2]
layers.append(gp.merge_LineStrings(filter_fills))
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0093_perlin_flow_erode_frays.svg'
sk.save(savepath)
vpype_commands = 'reloop linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 11 | page_x_inches: float = 11 # inches
page_y_inches: float = 8.5 # inches
border:float = 0.
perlin_grid_params = {
'xstep':3,
'ystep':3,
'lod':10,
'falloff':None,
'noise_scale':0.0053,
'noiseSeed':3
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
start_area = drawbox.buffer(-10, cap_style=3, join_style=3)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=25, ystep=35)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
f,ax = plt.subplots(figsize=(6,6))
ax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=40, xscale=5, yscale=5)
pc.init_particles(start_bounds=start_area)
n_steps = np.random.randint(low=3, high=20, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=3.3) + np.random.uniform(low=0., high=1.7, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.75, high=-0.3),
angles=-90,
)
stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)
P.fill_scale_trans(**stp.prms)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except :
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills = []
for p in gpolys:
try:
ifills.append(p.intersection_fill)
except:
pass
splits = utils.random_split(ifills, n_layers=5)
layers = [gp.merge_LineStrings(split) for split in splits]
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='layer')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0100_perlin_flow_erode_frays_occlude.svg'
sk.save(savepath)
vpype_commands = 'reloop linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 12 | page_x_inches: float = 11 # inches
page_y_inches: float = 8.5 # inches
border:float = 0.
perlin_grid_params = {
'xstep':3,
'ystep':3,
'lod':10,
'falloff':None,
'noise_scale':0.0083,
'noiseSeed':3
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
start_area = drawbox.buffer(-10, cap_style=3, join_style=3)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=55)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
f,ax = plt.subplots(figsize=(6,6))
ax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=60, xscale=7, yscale=12)
pc.init_particles(start_bounds=start_area)
n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=6.3) + np.random.uniform(low=0., high=0.5, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
bd = np.interp(line.centroid.x, [10, 270], [0.1, 3])
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.65, high=-0.3),
angles=-90,
)
stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)
P.fill_scale_trans(**stp.prms)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except :
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills = []
for p in gpolys:
try:
ifills.append(p.intersection_fill)
except:
pass
splits = utils.random_split(ifills, n_layers=5)
layers = [gp.merge_LineStrings(split) for split in splits]
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='none')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0101_perlin_flow_erode_frays_occlude.svg'
sk.save(savepath)
vpype_commands = 'reloop linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
try 13 | page_x_inches: float = 11 # inches
page_y_inches: float = 8.5 # inches
border:float = 0.
perlin_grid_params = {
'xstep':1,
'ystep':1,
'lod':10,
'falloff':None,
'noise_scale':0.0193,
'noiseSeed':3
}
buffer_style = 3
px = utils.DistanceConverter(page_x_inches, 'inches').mm
py = utils.DistanceConverter(page_y_inches, 'inches').mm
page_format = f'{px}mmx{py}mm'
drawbox = sg.box(border, border, px-border, py-border)
xmin, ymin, xmax, ymax = drawbox.bounds
brad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])
start_area = drawbox.buffer(-10, cap_style=3, join_style=3)
pg = gp.PerlinGrid(start_area, **perlin_grid_params)
xcs, ycs = gp.overlay_grid(start_area, xstep=20, ystep=75)
start_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]
start_pts = [p for p in start_pts if start_area.contains(p)]
# f,ax = plt.subplots(figsize=(6,6))
# ax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)
gpolys = []
for p in start_pts:
pc = ParticleCluster(pos=p, perlin_grid=pg)
pc.gen_start_pts_gaussian(n_particles=60, xscale=8, yscale=13)
pc.init_particles(start_bounds=start_area)
n_steps = int(np.interp(pc.pos.centroid.x, [1, 270], [1, 30]))
# n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))
pc.step(n_steps)
buffer_distances = np.random.uniform(low=0.3, high=6.3) + np.random.uniform(low=0., high=0.5, size=len(pc.lines))
polys = []
for line, bd in zip(pc.lines, buffer_distances):
bd = np.interp(line.centroid.x, [10, 270], [0.1, 3.5])
poly = line.buffer(bd,
cap_style=buffer_style,
join_style=buffer_style)
polys.append(poly)
P = gp.Poly(so.unary_union(polys))
stp = ScaleTransPrms(
d_buffer=np.random.uniform(low=-0.4, high=-0.2),
angles=-60,
)
stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)
P.fill_scale_trans(**stp.prms)
gpolys.append(P)
zorder = np.random.permutation(len(gpolys))
for z, gpoly in zip(zorder, gpolys):
gpoly.z = z
for gp0, gp1 in itertools.combinations(gpolys, r=2):
try:
overlaps = gp0.p.overlaps(gp1.p)
except :
gp0.p = gp0.p.buffer(1e-6)
gp1.p = gp1.p.buffer(1e-6)
overlaps = gp0.p.overlaps(gp1.p)
if overlaps:
if gp0.z > gp1.z:
gp1.p = occlude(top=gp0.p, bottom=gp1.p)
elif gp0.z < gp1.z:
gp0.p = occlude(top=gp1.p, bottom=gp0.p)
ifills = []
for p in gpolys:
try:
ifills.append(p.intersection_fill)
except:
pass
splits = utils.random_split(ifills, n_layers=5)
layers = [gp.merge_LineStrings(split) for split in splits]
sk = vsketch.Vsketch()
sk.size(page_format)
sk.scale('1mm')
sk.penWidth('0.25mm')
for i, layer in enumerate(layers):
sk.stroke(i+1)
sk.geometry(layer)
sk.display(color_mode='none')
savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0102_perlin_flow_erode_frays_occlude.svg'
sk.save(savepath)
vpype_commands = 'reloop linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'
vpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write {savepath}'
os.system(vpype_str) | _____no_output_____ | MIT | scratch/002_test_perlin.ipynb | ANaka/genpen |
Assignment 7: Groundwater and the Woburn Toxics Trial*Due 4/25/17 5 pts *Please **submit your assignment as an html export**, and for written responses, please type them in a cell that is of type `Markdown.` The final part of the exercise involves drawing a flow net by hand (actually, you could tackle this part of the assignment first). For that, you may turn in a hard copy of your answer. | # Import numerical tools
import numpy as np
#Import pandas for reading in and managing data
import pandas as pd
# Import pyplot for plotting
import matplotlib.pyplot as plt
#Import seaborn (useful for plotting)
import seaborn as sns
# Magic function to make matplotlib inline; other style specs must come AFTER
%matplotlib inline
%config InlineBackend.figure_formats = {'svg',} | _____no_output_____ | BSD-3-Clause | Assignment7Groundwater.ipynb | LaurelOak/hydro-teaching-resources |
BackgroundThis investigation of groundwater flow and drawdown in wells is based on the lawsuit described in the book and movie A Civil Action (a true story). For the background behind this story, read [the Wikipedia page](http://en.wikipedia.org/wiki/A_Civil_Action). Then look at the map and animation of the study site [here](http://researchnews.osu.edu/archive/woburnpics.htm). Wells G and H shown on the map are the municipal wells from which Woburn’s water supply was withdrawn. In this assignment you will be investigating some of the very problems that the hydrologists hired to the case worked on. As usual, you may work with another person, but each person must turn in separate assignments and specify whom they worked with. Below I provide additional information specific to this assignment. IntroductionOne of the important questions raised during the trial centered on the drawdown of the water table when wells G and H (see map linked above) were operating and the effects of the Riley tannery well (also included in the model). If, when wells G and/or H operated, the water table was drawn down on the east side of the Aberjona River, then contaminants from west side of the river valley (Beatrice 15 acres and Olympia Trucking) could flow under the river to wells G and H. If the water table was not drawn down on the east side of the river when wells G, H, and the Riley Tannery well operated, then it is unlikely that contaminants on the west side of the river valley could flow to wells G and H. Thus, the interaction of these three wells with the Aberjona River and the groundwater flow system was critical to determining which of the contaminated properties contributed TCE and PCE to wells G and H. Evaluating sources of impact to wells G and HProving the connection between the contaminants migrating from the defendants’ properties to wells G and H during the first phase of the trial was a requirement to proceed to the next phase. To make arguments regarding the potential sources of TCE to wells G and H, the plaintiffs' expert had to define the subsurface geology, the surface and subsurface, and show that TCE was used at both the Riley/Beatrice and W.R. Grace properties. Counsel for the plaintiffs was able to identify Grace employees involved with on-site TCE disposal activities. However, the plaintiffs' counsel was not able to directly link disposal of wastes on the Riley 15-acre property with activities at the tannery itself. The [ATSDR report](http://www.atsdr.cdc.gov/HAC/PHA/wellsgh/wgh_p1.html), which was prepared three years after the trial, is a good illustration of how difficult it is to conclusively assess risk from a specific contaminant source at the Woburn site. Understanding groundwater flow to wellsThe U.S. Geological Survey first performed a seven-hour pumping test to characterize aquifer properties, and then, in December 1985-January 1986, a 30-day pumping test to evaluate the effects of pumping on the distribution of hydraulic head. This was the first time wells G and H operated since their closure in May 1979. It was also the first time synoptic sets of water levels had ever been measured in the network of monitoring wells surrounding wells G and H. During the 30-day pumping test, wells G and H operated at their average rates of 700 gpm (well G) and 400 gpm (well H). However, pumping records kept by the City of Woburn showed that between 1964 and 1979 the wells rarely operated together and that the two municipal wells were frequently not in use for months at a time. As a result of changes in the pumping rates of wells G and H and their periodic (discontinuous) use, the water table in the buried valley aquifer was dynamic, rising and falling as wells G and H were turned on and off and pumped heavily or lightly. These dynamic changes in water levels result in the groundwater flow system being transient in character – as opposed to being steady state in character.Under steady-state conditions, water levels at any location in the flow system do not change with time, which results in hydraulic gradients and flow velocities at any location not changing with time, and therein no net change in the amount of groundwater in storage. Under transient conditions, water levels at any location in the flow system change with time, which causes hydraulic gradients, flow velocities, and the amount of groundwater in storage (water levels) to change with time. In this exercise we assume that the wells G and H have been pumping together for a long enough time that the water table is stationary and flow is steady state. This is an assumption that we recognize is not true over periods of time greater than a few months. This simplistic assumption lets us learn about drawdown created by a single pumping well, the formation of a cone of depression, superposition of drawdowns from one or more pumping wells, and the creation of groundwater divides between pumping wells, all of which are important to understanding the responses of the groundwater flow system to the pumping history of wells G and H. The operation of the Riley well was estimated by de Lima and Olimpio ([USGS, 1989](https://pubs.usgs.gov/wri/1989/4059/report.pdf)) to operate at an average pumping rate of 200 gallons per minute, continuously. This was based on the wastewater discharge emanating from the tannery; actual pumping rates were not available. It is believed that pumping volumes fluctuated depending on the number of shifts working, the volume of leather orders, actual type of leather being processed, etc. Part I of assignment: Solving for aquifer properties1. The hydrologists who worked on the Woburn case conducted a pumping test in well H to determine the aquifer’s properties (storativity and hydraulic conductivity). Although I couldn’t access their data, I generated synthetic data that will give you the same results, in the cell below. During the pumping test, well H was pumped at a rate of 475 gallons per minute. Drawdown was measured in an observation well 75 feet away from the pumping well. The thickness of the aquifer was measured to be 140 ft. Using the Cooper-Jacob equation and the regression method we discussed in class, determine the aquifer’s hydraulic conductivity (in feet per day) and storativity. In your answer, show the plot of your regression (don't forget to label the axes, or to take the log of time), and mathematically show how you arrived at your final answer. Make sure to keep track of your units! *[1 pt.]*Helpful commands for linear regression: `np.polyfit()`, `np.polyval()`.See an example of their usage in the Feb 2 tutorial, found under *Files/Materials* on bCouses. | # The data
time_minutes = np.arange(170,570,10) #Times (minutes) at which drawdown was observed after the start of pumping
s = np.array([0.110256896, 0.122567503, 0.180480293, 0.214489433, 0.356304352, 0.554603882, 0.49240574, 0.524209467, 0.562727644, 0.754849464, 0.718713002, 0.752910019, 0.73903658, 0.89009263, 0.967712464, 0.910807162, 0.986238396, 1.042178187, 1.081114186, 1.080825045, 1.196565491, 1.264971986, 1.430805272, 1.377858223, 1.376787182, 1.340970634, 1.466832052, 1.528405086, 1.44136224, 1.610936499, 1.503519725, 1.55398487, 1.628028833, 1.675649643, 1.672772239, 1.730410501, 1.730935188, 1.756850444, 1.731143013, 1.818924173])#Drawdown in observation well, ft | _____no_output_____ | BSD-3-Clause | Assignment7Groundwater.ipynb | LaurelOak/hydro-teaching-resources |
Part II: Estimating hydraulic heads with the Thiem EquationWith knowledge of the hydraulic conductivity, you can now solve the Thiem equation for the equilibrium potentiometric surface under different combinations of well pumping rates. For this part of the assignment, you will be working with a module that solves the Thiem equation over a two-dimensional spatial domain.The module requires the user to input several parameters, as described in the red text. When you first run it, use the hydraulic conductivity that you just solved for. Assume that the Riley well (QR) pumps at an average rate of 200 gallons per minute (gpm), Well G (QG) pumps at an average rate of 700 gpm, and Well H (QH) pumps at an average rate of 400 gpm. When you run the module, you will see two plots. The first plot is a filled contour plot that depicts the drawdown relative to non-pumping conditions. The value shown is the composite amount of drawdown related to pumping from each well (drawdowns are additive). Note how the values increase in the cells immediately around the wells and decrease toward the margins of the graph. The second plot is a cross section that shows the drawdown associated with the north-south transect that passes through wells G and H under steady state conditions.**2.** Take a look at the model below. What is a key assumption that is being made in the implementation of the Theim equation, beyond the assumptions we talked about in class? Hint: compare the formulas under the `Calculate Drawdown for Each Well` section to Eq. 5.49 in the Fetter handout. *[1/3 pt.]* | def PlotWoburnDD(K,b,QG,QH,QR, returnval=0):
"""
This routine uses the Thiem equation for unCONFINED aquifers to generate a plot of drawdown
contours around the Aberjona River in Woburn, Massachusetts. The Riley well is the source of
the contamination. Wells G and H are wells for the town's municipal water supply. Users can
evaluate how the potentiometric surface changes with changes in hydraulic conductivity (K),
saturated thickness (b), and well pumping rates (QG for well G, QH for well H, and QR for the
Riley well). Note that K is in units of ft/day, b is in units of ft, and QG, QH, and QR are
in units of gallons per minute. returnval is an optional argument. If you set it equal to 1,
the function will return the value of drawdown at well G. If unspecified, the function returns
just the plots.
"""
#CONVERT GALLONS PER MINUTE TO CUBIC FEET PER DAY
QG = QG*192.5
QH = QH*192.5
QR = QR*192.5
#OTHER CONSTANTS
delx = 100 #Cell size in feet
r0 = 2700 #You figure this out! (It is in feet)
#SET UP THE EXPERIMENTAL GRID
nrows = 25 #number of rows
ncols = 27 #number of columns
rowvect = np.arange(nrows) #A vector of row coordinates
colvect = np.arange(ncols) #A vector of column coordinates
colcoords, rowcoords = np.meshgrid(colvect,rowvect) #Creates two matrices. In colcoords, each
#entry is the column index of the cell (i.e., point in space). In rowcoords, each entry is the
#row index of the cell (i.e., point in space).
#SPECIFY WELL LOCATIONS AND RIVER COORDINATES WITHIN THE EXPERIMENTAL GRID
loc_R = np.array([20,8]) #Index locations for the Riley well (row, column)
loc_H = np.array([10,13]) #Index locations for H well
loc_G = np.array([17,13]) #Index locations for G well
river_row = np.array([0, 1, 2, 3, 4, 5, 6, 6.4, 7, 8, 9, 9.9, 10, 11, 11.5, 12, 13, 14, 15, 16, 16.1, 17, 17.5, 18, 18.4, 19, 19.6, 20, 20.3, 21, 21.7, 22, 22.4, 23, 23.1, 23.9, 24, 24.8, 25])
river_col = np.array([8.6, 9, 9.1, 9.3, 9.5, 9.6, 9.9, 10, 10.3, 11, 11.7, 12, 12.1, 12.7, 12.9, 12.8, 12.6, 12, 11.6, 11.1, 11, 10.8, 10.7, 10.8, 11, 11.4, 12, 12.6, 13, 14, 15, 15.5, 16, 16.9, 17, 18, 18.1, 19, 19.2])
#CALCULATE DISTANCES BETWEEN EACH WELL AND EVERY OTHER CELL
d_R = np.sqrt((rowcoords-loc_R[0])**2+(colcoords-loc_R[1])**2)*delx #Solve for distance (feet) using the Pythagorean theorem
d_H = np.sqrt((rowcoords-loc_H[0])**2+(colcoords-loc_H[1])**2)*delx #Solve for distance (feet) using the Pythagorean theorem
d_G = np.sqrt((rowcoords-loc_G[0])**2+(colcoords-loc_G[1])**2)*delx #Solve for distance (feet) using the Pythagorean theorem
#SET DISTANCES TO WELL IN CELLS WITH WELL TO 25 FT (TO AVOID SINGULARITY AT 0)
d_R[20,8] = 25 #feet
d_H[10,13] = 25 #feet
d_G[17,13] = 25 #feet
#CALCULATE DRAWDOWN FROM EACH WELL
s_R = b-np.sqrt(b**2-QR*np.log(r0/d_R)/(np.pi*K)) #Drawdown from the Riley well
s_H = b-np.sqrt(b**2-QH*np.log(r0/d_H)/(np.pi*K)) #Drawdown from well H
s_G = b-np.sqrt(b**2-QG*np.log(r0/d_G)/(np.pi*K)) #Drawadown from well G
s_total = s_R+s_H+s_G #Combined drawdown from all of the wells, feet
#NOW GENERATE PLOTS
plt.figure(figsize=(10,10))
s_plot = plt.contourf(np.transpose((colcoords+0.5)*delx), np.transpose((rowcoords+0.5)*delx), np.transpose(s_total), cmap=plt.cm.plasma)
plt.gca().invert_yaxis() #This puts the origin of the plot on the upper left
cb = plt.colorbar(s_plot, orientation='horizontal')
cb.set_label('Drawdown, ft')
riv_plot = plt.plot(river_col*delx, river_row*delx, 'b-',linewidth=3.0)
wH = plt.plot((loc_H[1]+0.5)*delx, (loc_H[0]+0.5)*delx, 'ko')
ax = plt.gca()
ax.annotate('H', xy = ((loc_H[1]+0.5)*delx, (loc_H[0]+0.5)*delx))
wG = plt.plot((loc_G[1]+0.5)*delx, (loc_G[0]+0.5)*delx, 'ko')
wR = plt.plot((loc_R[1]+0.5)*delx, (loc_R[0]+0.5)*delx, 'ko')
ax.annotate('G', xy = ((loc_G[1]+0.5)*delx, (loc_G[0]+0.5)*delx))
ax.annotate('Riley', xy = ((loc_R[1]+0.5)*delx, (loc_R[0]+0.5)*delx))
#plt.axis('equal') #Uncomment this to make the x- and y- axis display on the same scale.
plt.title('Map of Aberjona River and Drawdown Contours')
plt.show()
plt.figure()
plt.fill_between(rowvect*delx, max(np.ceil(s_total[:,13])), s_total[:,13])
plt.plot([loc_H[0]*delx, loc_H[0]*delx], [min(s_total[:,13]), max(s_total[:,13])], 'r')
plt.plot([loc_G[0]*delx, loc_G[0]*delx], [min(s_total[:,13]), max(s_total[:,13])], 'r')
plt.gca().invert_yaxis()
plt.xlabel('y distance, feet')
plt.ylabel('Drawdown, feet')
plt.title('Cross-section across column with wells G and H')
if returnval==1:
return s_total[17,13] | _____no_output_____ | BSD-3-Clause | Assignment7Groundwater.ipynb | LaurelOak/hydro-teaching-resources |
Applied Probability Theory From Scratch Simpson's Paradox Bruno Gonçalves www.data4sci.com @bgoncalves, @data4sci | import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import sklearn
from sklearn.linear_model import LinearRegression
import watermark
%load_ext watermark
%matplotlib inline
%watermark -i -n -v -m -g -iv
plt.style.use('./d4sci.mplstyle') | _____no_output_____ | MIT | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads |
Load the iris dataset | iris = pd.read_csv('data/iris.csv')
iris | _____no_output_____ | MIT | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads |
Split the dataset across species for convenience | setosa = iris[['sepal_width', 'petal_width']][iris['species'] == 'setosa']
versicolor = iris[['sepal_width', 'petal_width']][iris['species'] == 'versicolor']
virginica = iris[['sepal_width', 'petal_width']][iris['species'] == 'virginica'] | _____no_output_____ | MIT | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads |
Perform the fits | lm_setosa = LinearRegression()
lm_setosa.fit(setosa['sepal_width'].values.reshape(-1,1), setosa['petal_width'])
y_setosa = lm_setosa.predict(setosa['sepal_width'].values.reshape(-1,1))
lm_versicolor = LinearRegression()
lm_versicolor.fit(versicolor['sepal_width'].values.reshape(-1,1), versicolor['petal_width'])
y_versicolor = lm_versicolor.predict(versicolor['sepal_width'].values.reshape(-1,1))
lm_virginica = LinearRegression()
lm_virginica.fit(virginica['sepal_width'].values.reshape(-1,1), virginica['petal_width'])
y_virginica = lm_virginica.predict(virginica['sepal_width'].values.reshape(-1,1))
lm_full = LinearRegression()
lm_full.fit(iris['sepal_width'].values.reshape(-1,1), iris['petal_width'])
y_full = lm_full.predict(iris['sepal_width'].values.reshape(-1,1)) | _____no_output_____ | MIT | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads |
Generate the plot | fig, axs = plt.subplots(ncols=2, sharey=True)
colors = plt.rcParams['axes.prop_cycle'].by_key()['color']
setosa.plot.scatter(x='sepal_width', y='petal_width', label='setosa', ax=axs[0], c=colors[0])
versicolor.plot.scatter(x='sepal_width', y='petal_width', label='versicolor', ax=axs[0], c=colors[1])
virginica.plot.scatter(x='sepal_width', y='petal_width', label='virginica', ax=axs[0], c=colors[2])
l4, = axs[0].plot(iris['sepal_width'].values.reshape(-1,1), y_full, '-', c=colors[3])
setosa.plot.scatter(x='sepal_width', y='petal_width', ax=axs[1], c=colors[0])
versicolor.plot.scatter(x='sepal_width', y='petal_width', ax=axs[1], c=colors[1])
virginica.plot.scatter(x='sepal_width', y='petal_width', ax=axs[1], c=colors[2])
l1, = axs[1].plot(setosa['sepal_width'].values.reshape(-1,1), y_setosa, '-', c=colors[0])
l2, = axs[1].plot(versicolor['sepal_width'].values.reshape(-1,1), y_versicolor, '-', c=colors[1])
l3, = axs[1].plot(virginica['sepal_width'].values.reshape(-1,1), y_virginica, '-', c=colors[2])
axs[0].set_xlabel('Sepal Width')
axs[1].set_xlabel('Sepal Width')
axs[0].set_ylabel('Petal Width')
fig.subplots_adjust(bottom=0.3, wspace=0.33)
axs[0].legend(handles = [l1, l2, l3, l4] , labels=['Setosa', 'Versicolor', 'Virginica', 'Total'],
loc='lower left', bbox_to_anchor=(0, -0.4), ncol=2, fancybox=True, shadow=False) | _____no_output_____ | MIT | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads |
Removing setosa | reduced = iris[iris['species'] != 'setosa'].copy()
lm_reduced = LinearRegression()
lm_reduced.fit(reduced['sepal_width'].values.reshape(-1,1), reduced['petal_width'])
y_reduced = lm_reduced.predict(reduced['sepal_width'].values.reshape(-1,1))
fig, axs = plt.subplots(ncols=1, sharey=True)
colors = plt.rcParams['axes.prop_cycle'].by_key()['color']
versicolor.plot.scatter(x='sepal_width', y='petal_width', ax=axs, c=colors[1])
virginica.plot.scatter(x='sepal_width', y='petal_width', ax=axs, c=colors[2])
axs.plot(reduced['sepal_width'].values.reshape(-1,1), y_reduced, '-', c=colors[3], label='reduced')
axs.plot(versicolor['sepal_width'].values.reshape(-1,1), y_versicolor, '-', c=colors[1], label='versicolor')
axs.plot(virginica['sepal_width'].values.reshape(-1,1), y_virginica, '-', c=colors[2], label='virginica')
axs.set_xlabel('Sepal Width')
axs.set_ylabel('Petal Width')
plt.legend() | _____no_output_____ | MIT | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads |
With rgb images Load data | import numpy as np
import pandas as pd
from glob import glob
from tqdm import tqdm
from sklearn.utils import shuffle
df = pd.read_csv('sample/Data_Entry_2017.csv')
diseases = ['Cardiomegaly','Emphysema','Effusion','Hernia','Nodule','Pneumothorax','Atelectasis','Pleural_Thickening','Mass','Edema','Consolidation','Infiltration','Fibrosis','Pneumonia']
#Number diseases
for disease in diseases :
df[disease] = df['Finding Labels'].apply(lambda x: 1 if disease in x else 0)
# #test to perfect
# df = df.drop(df[df['Emphysema']==0][:-127].index.values)
#remove Y after age
df['Age']=df['Patient Age'].apply(lambda x: x[:-1]).astype(int)
df['Age Type']=df['Patient Age'].apply(lambda x: x[-1:])
df.loc[df['Age Type']=='M',['Age']] = df[df['Age Type']=='M']['Age'].apply(lambda x: round(x/12.)).astype(int)
df.loc[df['Age Type']=='D',['Age']] = df[df['Age Type']=='D']['Age'].apply(lambda x: round(x/365.)).astype(int)
# remove outliers
df = df.drop(df['Age'].sort_values(ascending=False).head(16).index)
df['Age'] = df['Age']/df['Age'].max()
#one hot data
# df = df.drop(df.index[4242])
df = df.join(pd.get_dummies(df['Patient Gender']))
df = df.join(pd.get_dummies(df['View Position']))
#random samples
df = shuffle(df)
#get other data
data = df[['Age', 'F', 'M', 'AP', 'PA']]
data = np.array(data)
labels = df[diseases].as_matrix()
files_list = ('sample/images/' + df['Image Index']).tolist()
# #test to perfect
# labelB = df['Emphysema'].tolist()
labelB = (df[diseases].sum(axis=1)>0).tolist()
labelB = np.array(labelB, dtype=int)
from keras.preprocessing import image
from tqdm import tqdm
def path_to_tensor(img_path, shape):
# loads RGB image as PIL.Image.Image type
img = image.load_img(img_path, target_size=shape)
# convert PIL.Image.Image type to 3D tensor with shape (224, 224, 3)
x = image.img_to_array(img)/255
# convert 3D tensor to 4D tensor with shape (1, 224, 224, 3) and return 4D tensor
return np.expand_dims(x, axis=0)
def paths_to_tensor(img_paths, shape):
list_of_tensors = [path_to_tensor(img_path, shape) for img_path in tqdm(img_paths)]
return np.vstack(list_of_tensors)
train_labels = labelB[:89600][:, np.newaxis]
valid_labels = labelB[89600:100800][:, np.newaxis]
test_labels = labelB[100800:][:, np.newaxis]
train_data = data[:89600]
valid_data = data[89600:100800]
test_data = data[100800:]
img_shape = (64, 64)
train_tensors = paths_to_tensor(files_list[:89600], shape = img_shape)
valid_tensors = paths_to_tensor(files_list[89600:100800], shape = img_shape)
test_tensors = paths_to_tensor(files_list[100800:], shape = img_shape) | Using TensorFlow backend.
/home/aind2/anaconda3/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: compiletime version 3.5 of module 'tensorflow.python.framework.fast_tensor_util' does not match runtime version 3.6
return f(*args, **kwds)
100%|██████████| 89600/89600 [20:26<00:00, 73.03it/s]
100%|██████████| 11200/11200 [02:33<00:00, 73.10it/s]
100%|██████████| 11319/11319 [02:38<00:00, 71.35it/s]
| Apache-2.0 | vanilla CNN - FullDataset.ipynb | subha231/cancer |
CNN model | import time
from keras.layers import Conv2D, MaxPooling2D, GlobalAveragePooling2D, Dropout, Flatten, Dense
from keras.models import Sequential
from keras.layers.normalization import BatchNormalization
from keras import regularizers, initializers, optimizers
model = Sequential()
model.add(Conv2D(filters=16,
kernel_size=7,
padding='same',
activation='relu',
input_shape=train_tensors.shape[1:]))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=32,
kernel_size=5,
padding='same',
activation='relu'))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=64,
kernel_size=5,
padding='same',
activation='relu'))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=128,
kernel_size=5,
strides=2,
padding='same',
activation='relu'))
model.add(MaxPooling2D(pool_size=2))
model.add(Flatten())
model.add(Dense(100, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.summary()
from keras import backend as K
def binary_accuracy(y_true, y_pred):
return K.mean(K.equal(y_true, K.round(y_pred)))
def precision_threshold(threshold = 0.5):
def precision(y_true, y_pred):
threshold_value = threshold
y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())
true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))
predicted_positives = K.sum(y_pred)
precision_ratio = true_positives / (predicted_positives + K.epsilon())
return precision_ratio
return precision
def recall_threshold(threshold = 0.5):
def recall(y_true, y_pred):
threshold_value = threshold
y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())
true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.clip(y_true, 0, 1))
recall_ratio = true_positives / (possible_positives + K.epsilon())
return recall_ratio
return recall
def fbeta_score_threshold(beta = 1, threshold = 0.5):
def fbeta_score(y_true, y_pred):
threshold_value = threshold
beta_value = beta
p = precision_threshold(threshold_value)(y_true, y_pred)
r = recall_threshold(threshold_value)(y_true, y_pred)
bb = beta_value ** 2
fbeta_score = (1 + bb) * (p * r) / (bb * p + r + K.epsilon())
return fbeta_score
return fbeta_score
model.compile(optimizer='sgd', loss='binary_crossentropy',
metrics=[precision_threshold(threshold = 0.5),
recall_threshold(threshold = 0.5),
fbeta_score_threshold(beta=0.5, threshold = 0.5),
'accuracy'])
from keras.callbacks import ModelCheckpoint, CSVLogger, EarlyStopping
import numpy as np
epochs = 20
batch_size = 32
earlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=3, verbose=1, mode='auto')
log = CSVLogger('saved_models/log_bCNN_rgb.csv')
checkpointer = ModelCheckpoint(filepath='saved_models/bCNN.best.from_scratch.hdf5',
verbose=1, save_best_only=True)
start = time.time()
model.fit(train_tensors, train_labels,
validation_data=(valid_tensors, valid_labels),
epochs=epochs, batch_size=batch_size, callbacks=[checkpointer, log, earlystop], verbose=1)
# Show total training time
print("training time: %.2f minutes"%((time.time()-start)/60)) | Train on 89600 samples, validate on 11200 samples
Epoch 1/20
89536/89600 [============================>.] - ETA: 0s - loss: 0.6561 - precision: 0.5974 - recall: 0.4632 - fbeta_score: 0.5386 - acc: 0.6178Epoch 00000: val_loss improved from inf to 0.65672, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 52s - loss: 0.6561 - precision: 0.5974 - recall: 0.4632 - fbeta_score: 0.5387 - acc: 0.6178 - val_loss: 0.6567 - val_precision: 0.5574 - val_recall: 0.7224 - val_fbeta_score: 0.5805 - val_acc: 0.6148
Epoch 2/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6416 - precision: 0.6351 - recall: 0.5353 - fbeta_score: 0.6006 - acc: 0.6412Epoch 00001: val_loss improved from 0.65672 to 0.63831, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 51s - loss: 0.6416 - precision: 0.6353 - recall: 0.5353 - fbeta_score: 0.6007 - acc: 0.6412 - val_loss: 0.6383 - val_precision: 0.6061 - val_recall: 0.6122 - val_fbeta_score: 0.6025 - val_acc: 0.6460
Epoch 3/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6354 - precision: 0.6417 - recall: 0.5549 - fbeta_score: 0.6115 - acc: 0.6487Epoch 00002: val_loss improved from 0.63831 to 0.63043, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 51s - loss: 0.6354 - precision: 0.6417 - recall: 0.5549 - fbeta_score: 0.6115 - acc: 0.6487 - val_loss: 0.6304 - val_precision: 0.6347 - val_recall: 0.5533 - val_fbeta_score: 0.6106 - val_acc: 0.6556
Epoch 4/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6303 - precision: 0.6453 - recall: 0.5640 - fbeta_score: 0.6173 - acc: 0.6537Epoch 00003: val_loss improved from 0.63043 to 0.63033, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 51s - loss: 0.6303 - precision: 0.6455 - recall: 0.5640 - fbeta_score: 0.6174 - acc: 0.6538 - val_loss: 0.6303 - val_precision: 0.6627 - val_recall: 0.4711 - val_fbeta_score: 0.6032 - val_acc: 0.6525
Epoch 5/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6267 - precision: 0.6506 - recall: 0.5799 - fbeta_score: 0.6259 - acc: 0.6603Epoch 00004: val_loss improved from 0.63033 to 0.62570, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 52s - loss: 0.6267 - precision: 0.6506 - recall: 0.5800 - fbeta_score: 0.6260 - acc: 0.6603 - val_loss: 0.6257 - val_precision: 0.6350 - val_recall: 0.5960 - val_fbeta_score: 0.6213 - val_acc: 0.6642
Epoch 6/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6232 - precision: 0.6519 - recall: 0.5841 - fbeta_score: 0.6287 - acc: 0.6627Epoch 00005: val_loss improved from 0.62570 to 0.62252, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 52s - loss: 0.6232 - precision: 0.6519 - recall: 0.5839 - fbeta_score: 0.6287 - acc: 0.6627 - val_loss: 0.6225 - val_precision: 0.6655 - val_recall: 0.5105 - val_fbeta_score: 0.6197 - val_acc: 0.6628
Epoch 7/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6204 - precision: 0.6558 - recall: 0.5892 - fbeta_score: 0.6332 - acc: 0.6668Epoch 00006: val_loss did not improve
89600/89600 [==============================] - 51s - loss: 0.6204 - precision: 0.6558 - recall: 0.5893 - fbeta_score: 0.6333 - acc: 0.6669 - val_loss: 0.6237 - val_precision: 0.6176 - val_recall: 0.6639 - val_fbeta_score: 0.6217 - val_acc: 0.6624
Epoch 8/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6179 - precision: 0.6574 - recall: 0.5978 - fbeta_score: 0.6364 - acc: 0.6693Epoch 00007: val_loss did not improve
89600/89600 [==============================] - 51s - loss: 0.6180 - precision: 0.6574 - recall: 0.5979 - fbeta_score: 0.6364 - acc: 0.6692 - val_loss: 0.6243 - val_precision: 0.6157 - val_recall: 0.6677 - val_fbeta_score: 0.6206 - val_acc: 0.6618
Epoch 9/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6151 - precision: 0.6565 - recall: 0.5990 - fbeta_score: 0.6364 - acc: 0.6702Epoch 00008: val_loss improved from 0.62252 to 0.61709, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 51s - loss: 0.6152 - precision: 0.6564 - recall: 0.5988 - fbeta_score: 0.6362 - acc: 0.6701 - val_loss: 0.6171 - val_precision: 0.6748 - val_recall: 0.5154 - val_fbeta_score: 0.6274 - val_acc: 0.6702
Epoch 10/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6125 - precision: 0.6618 - recall: 0.6044 - fbeta_score: 0.6417 - acc: 0.6739Epoch 00009: val_loss improved from 0.61709 to 0.61645, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 51s - loss: 0.6125 - precision: 0.6618 - recall: 0.6043 - fbeta_score: 0.6416 - acc: 0.6739 - val_loss: 0.6164 - val_precision: 0.6585 - val_recall: 0.5817 - val_fbeta_score: 0.6353 - val_acc: 0.6746
Epoch 11/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6102 - precision: 0.6636 - recall: 0.6092 - fbeta_score: 0.6443 - acc: 0.6762Epoch 00010: val_loss improved from 0.61645 to 0.61598, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 52s - loss: 0.6102 - precision: 0.6636 - recall: 0.6092 - fbeta_score: 0.6443 - acc: 0.6762 - val_loss: 0.6160 - val_precision: 0.6263 - val_recall: 0.6632 - val_fbeta_score: 0.6286 - val_acc: 0.6686
Epoch 12/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6075 - precision: 0.6644 - recall: 0.6140 - fbeta_score: 0.6461 - acc: 0.6782Epoch 00011: val_loss improved from 0.61598 to 0.61276, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 53s - loss: 0.6074 - precision: 0.6644 - recall: 0.6140 - fbeta_score: 0.6461 - acc: 0.6782 - val_loss: 0.6128 - val_precision: 0.6400 - val_recall: 0.6476 - val_fbeta_score: 0.6363 - val_acc: 0.6753
Epoch 13/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6052 - precision: 0.6678 - recall: 0.6175 - fbeta_score: 0.6498 - acc: 0.6812Epoch 00012: val_loss improved from 0.61276 to 0.60942, saving model to saved_models/bCNN.best.from_scratch.hdf5
89600/89600 [==============================] - 53s - loss: 0.6052 - precision: 0.6678 - recall: 0.6174 - fbeta_score: 0.6498 - acc: 0.6812 - val_loss: 0.6094 - val_precision: 0.6545 - val_recall: 0.5914 - val_fbeta_score: 0.6348 - val_acc: 0.6748
Epoch 14/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.6021 - precision: 0.6698 - recall: 0.6210 - fbeta_score: 0.6520 - acc: 0.6836Epoch 00013: val_loss did not improve
89600/89600 [==============================] - 53s - loss: 0.6022 - precision: 0.6699 - recall: 0.6210 - fbeta_score: 0.6520 - acc: 0.6836 - val_loss: 0.6187 - val_precision: 0.6155 - val_recall: 0.6913 - val_fbeta_score: 0.6249 - val_acc: 0.6645
Epoch 15/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.5997 - precision: 0.6727 - recall: 0.6242 - fbeta_score: 0.6547 - acc: 0.6855Epoch 00014: val_loss did not improve
89600/89600 [==============================] - 53s - loss: 0.5999 - precision: 0.6724 - recall: 0.6239 - fbeta_score: 0.6544 - acc: 0.6852 - val_loss: 0.6147 - val_precision: 0.6373 - val_recall: 0.6321 - val_fbeta_score: 0.6307 - val_acc: 0.6702
Epoch 16/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.5969 - precision: 0.6754 - recall: 0.6313 - fbeta_score: 0.6586 - acc: 0.6883Epoch 00015: val_loss did not improve
89600/89600 [==============================] - 53s - loss: 0.5970 - precision: 0.6754 - recall: 0.6312 - fbeta_score: 0.6586 - acc: 0.6883 - val_loss: 0.6095 - val_precision: 0.6442 - val_recall: 0.6437 - val_fbeta_score: 0.6389 - val_acc: 0.6779
Epoch 17/20
89504/89600 [============================>.] - ETA: 0s - loss: 0.5937 - precision: 0.6794 - recall: 0.6334 - fbeta_score: 0.6622 - acc: 0.6916Epoch 00016: val_loss did not improve
89600/89600 [==============================] - 53s - loss: 0.5937 - precision: 0.6793 - recall: 0.6334 - fbeta_score: 0.6622 - acc: 0.6916 - val_loss: 0.6133 - val_precision: 0.6844 - val_recall: 0.5338 - val_fbeta_score: 0.6403 - val_acc: 0.6790
Epoch 00016: early stopping
training time: 14.89 minutes
| Apache-2.0 | vanilla CNN - FullDataset.ipynb | subha231/cancer |
Metric | model.load_weights('saved_models/bCNN.best.from_scratch.hdf5')
prediction = model.predict(test_tensors)
threshold = 0.5
beta = 0.5
pre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
rec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
fsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
print ("Precision: %f %%\nRecall: %f %%\nFscore: %f %%"% (pre, rec, fsc))
K.eval(binary_accuracy(K.variable(value=test_labels),
K.variable(value=prediction)))
prediction[:30]
threshold = 0.4
beta = 0.5
pre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
rec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
fsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
print ("Precision: %f %%\nRecall: %f %%\nFscore: %f %%"% (pre, rec, fsc))
threshold = 0.6
beta = 0.5
pre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
rec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
fsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
print ("Precision: %f %%\nRecall: %f %%\nFscore: %f %%"% (pre, rec, fsc)) | Precision: 0.712731 %
Recall: 0.404833 %
Fscore: 0.618630 %
| Apache-2.0 | vanilla CNN - FullDataset.ipynb | subha231/cancer |
With gray images | import numpy as np
import pandas as pd
from glob import glob
from tqdm import tqdm
from sklearn.utils import shuffle
df = pd.read_csv('sample/Data_Entry_2017.csv')
diseases = ['Cardiomegaly','Emphysema','Effusion','Hernia','Nodule','Pneumothorax','Atelectasis','Pleural_Thickening','Mass','Edema','Consolidation','Infiltration','Fibrosis','Pneumonia']
#Number diseases
for disease in diseases :
df[disease] = df['Finding Labels'].apply(lambda x: 1 if disease in x else 0)
# #test to perfect
# df = df.drop(df[df['Emphysema']==0][:-127].index.values)
#remove Y after age
df['Age']=df['Patient Age'].apply(lambda x: x[:-1]).astype(int)
df['Age Type']=df['Patient Age'].apply(lambda x: x[-1:])
df.loc[df['Age Type']=='M',['Age']] = df[df['Age Type']=='M']['Age'].apply(lambda x: round(x/12.)).astype(int)
df.loc[df['Age Type']=='D',['Age']] = df[df['Age Type']=='D']['Age'].apply(lambda x: round(x/365.)).astype(int)
# remove outliers
df = df.drop(df['Age'].sort_values(ascending=False).head(16).index)
df['Age'] = df['Age']/df['Age'].max()
#one hot data
# df = df.drop(df.index[4242])
df = df.join(pd.get_dummies(df['Patient Gender']))
df = df.join(pd.get_dummies(df['View Position']))
#random samples
df = shuffle(df)
#get other data
data = df[['Age', 'F', 'M', 'AP', 'PA']]
data = np.array(data)
labels = df[diseases].as_matrix()
files_list = ('sample/images/' + df['Image Index']).tolist()
# #test to perfect
# labelB = df['Emphysema'].tolist()
labelB = (df[diseases].sum(axis=1)>0).tolist()
labelB = np.array(labelB, dtype=int)
from keras.preprocessing import image
from tqdm import tqdm
def path_to_tensor(img_path, shape):
# loads RGB image as PIL.Image.Image type
img = image.load_img(img_path, grayscale=True, target_size=shape)
# convert PIL.Image.Image type to 3D tensor with shape (224, 224, 1)
x = image.img_to_array(img)/255
# convert 3D tensor to 4D tensor with shape (1, 224, 224, 1) and return 4D tensor
return np.expand_dims(x, axis=0)
def paths_to_tensor(img_paths, shape):
list_of_tensors = [path_to_tensor(img_path, shape) for img_path in tqdm(img_paths)]
return np.vstack(list_of_tensors)
train_labels = labelB[:89600][:, np.newaxis]
valid_labels = labelB[89600:100800][:, np.newaxis]
test_labels = labelB[100800:][:, np.newaxis]
train_data = data[:89600]
valid_data = data[89600:100800]
test_data = data[100800:]
img_shape = (64, 64)
train_tensors = paths_to_tensor(files_list[:89600], shape = img_shape)
valid_tensors = paths_to_tensor(files_list[89600:100800], shape = img_shape)
test_tensors = paths_to_tensor(files_list[100800:], shape = img_shape)
import time
from keras.layers import Conv2D, MaxPooling2D, GlobalAveragePooling2D, Dropout, Flatten, Dense
from keras.models import Sequential
from keras.layers.normalization import BatchNormalization
from keras import regularizers, initializers, optimizers
model = Sequential()
model.add(Conv2D(filters=16,
kernel_size=7,
padding='same',
activation='relu',
input_shape=train_tensors.shape[1:]))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=32,
kernel_size=5,
padding='same',
activation='relu'))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=64,
kernel_size=5,
padding='same',
activation='relu'))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=128,
kernel_size=5,
strides=2,
padding='same',
activation='relu'))
model.add(MaxPooling2D(pool_size=2))
model.add(Flatten())
model.add(Dense(100, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.summary()
from keras import backend as K
def binary_accuracy(y_true, y_pred):
return K.mean(K.equal(y_true, K.round(y_pred)))
def precision_threshold(threshold = 0.5):
def precision(y_true, y_pred):
threshold_value = threshold
y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())
true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))
predicted_positives = K.sum(y_pred)
precision_ratio = true_positives / (predicted_positives + K.epsilon())
return precision_ratio
return precision
def recall_threshold(threshold = 0.5):
def recall(y_true, y_pred):
threshold_value = threshold
y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())
true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.clip(y_true, 0, 1))
recall_ratio = true_positives / (possible_positives + K.epsilon())
return recall_ratio
return recall
def fbeta_score_threshold(beta = 1, threshold = 0.5):
def fbeta_score(y_true, y_pred):
threshold_value = threshold
beta_value = beta
p = precision_threshold(threshold_value)(y_true, y_pred)
r = recall_threshold(threshold_value)(y_true, y_pred)
bb = beta_value ** 2
fbeta_score = (1 + bb) * (p * r) / (bb * p + r + K.epsilon())
return fbeta_score
return fbeta_score
model.compile(optimizer='sgd', loss='binary_crossentropy',
metrics=[precision_threshold(threshold = 0.5),
recall_threshold(threshold = 0.5),
fbeta_score_threshold(beta=0.5, threshold = 0.5),
'accuracy'])
from keras.callbacks import ModelCheckpoint, CSVLogger, EarlyStopping
import numpy as np
epochs = 20
batch_size = 32
earlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=3, verbose=1, mode='auto')
log = CSVLogger('saved_models/log_bCNN_gray.csv')
checkpointer = ModelCheckpoint(filepath='saved_models/bCNN_gray.best.from_scratch.hdf5',
verbose=1, save_best_only=True)
start = time.time()
model.fit(train_tensors, train_labels,
validation_data=(valid_tensors, valid_labels),
epochs=epochs, batch_size=batch_size, callbacks=[checkpointer, log, earlystop], verbose=1)
# Show total training time
print("training time: %.2f minutes"%((time.time()-start)/60))
model.load_weights('saved_models/bCNN_gray.best.from_scratch.hdf5')
prediction = model.predict(test_tensors)
threshold = 0.5
beta = 0.5
pre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
rec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
fsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
print ("Precision: %f %%\nRecall: %f %%\nFscore: %f %%"% (pre, rec, fsc))
K.eval(binary_accuracy(K.variable(value=test_labels),
K.variable(value=prediction)))
threshold = 0.4
beta = 0.5
pre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
rec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
fsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),
K.variable(value=prediction)))
print ("Precision: %f %%\nRecall: %f %%\nFscore: %f %%"% (pre, rec, fsc)) | Precision: 0.627903 %
Recall: 0.710935 %
Fscore: 0.642921 %
| Apache-2.0 | vanilla CNN - FullDataset.ipynb | subha231/cancer |
WeatherPy---- Note* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps. | # Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import time
from scipy.stats import linregress
from pprint import pprint
# Import API key
from api_keys import weather_api_key
# Incorporated citipy to determine city based on latitude and longitude
from citipy import citipy
# Output File (CSV)
output_data_file = "../output_data/cities.csv"
# Range of latitudes and longitudes
lat_range = (-90, 90)
lng_range = (-180, 180) | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Generate Cities List | # List for holding lat_lngs and cities
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(lat_range[0], lat_range[1], size=1750) # increased the amount due to my logic below
lngs = np.random.uniform(lng_range[0], lng_range[1], size=1750) # increased the amount due to my logic below
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city)
# Print the city count to confirm sufficient count
len(cities) | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Perform API Calls* Perform a weather check on each city using a series of successive API calls.* Include a print log of each city as it'sbeing processed (with the city number and city name). | # Save config information.
url = "http://api.openweathermap.org/data/2.5/weather?"
units = "imperial"
# Build partial query URL
query_url = f"{url}appid={weather_api_key}&units={units}&q="
# Set up lists to hold reponse information
city_list = []
city_lat = []
city_lng = []
city_temp = []
city_humidity = []
city_clouds = []
city_wind = []
city_country = []
city_date = []
i = 1
setof = 1
# Loop through the list of cities and perform a request for data on each
print("""Beginning Data Retrieval\n-----------------------------""")
for city in cities:
if i < 51:
response = requests.get(query_url + city).json()
try:
if response['name'].casefold() == city: # casefold helps us ignore case
city_list.append(response['name'])
city_lat.append(response['coord']['lat'])
city_lng.append(response['coord']['lon'])
city_temp.append(response['main']['temp_max'])
city_humidity.append(response['main']['humidity'])
city_clouds.append(response['clouds']['all'])
city_wind.append(response['wind']['speed'])
city_country.append(response['sys']['country'])
city_date.append(response['dt'])
print(f"Processing Record {i} of Set {setof} | {city}")
else:
print("City not found. Skipping...") # filtering these out to be extra sure everything matches
except KeyError:
print("City not found. Skipping...")
i += 1
else:
time.sleep(60) #https://docs.python.org/3/library/time.html#time.sleep
setof += 1 # add one to the set
i = 1 # reset the ticker
response = requests.get(query_url + city).json()
try:
if response['name'].casefold() == city: # casefold helps us ignore case
city_list.append(response['name'])
city_lat.append(response['coord']['lat'])
city_lng.append(response['coord']['lon'])
city_temp.append(response['main']['temp_max'])
city_humidity.append(response['main']['humidity'])
city_clouds.append(response['clouds']['all'])
city_wind.append(response['wind']['speed'])
city_country.append(response['sys']['country'])
city_date.append(response['dt'])
print(f"Processing Record {i} of Set {setof} | {city}")
else:
print("City not found. Skipping...") # filtering these out to be extra sure everything matches
except KeyError:
print("City not found. Skipping...")
i += 1
print("""-----------------------------\nData Retrieval Complete\n-----------------------------""") | Beginning Data Retrieval
-----------------------------
Processing Record 1 of Set 1 | norman wells
Processing Record 2 of Set 1 | touros
City not found. Skipping...
Processing Record 4 of Set 1 | qaanaaq
City not found. Skipping...
Processing Record 6 of Set 1 | punta arenas
Processing Record 7 of Set 1 | ushuaia
Processing Record 8 of Set 1 | vaini
Processing Record 9 of Set 1 | busselton
Processing Record 10 of Set 1 | poum
Processing Record 11 of Set 1 | sheregesh
Processing Record 12 of Set 1 | mataura
Processing Record 13 of Set 1 | khatanga
Processing Record 14 of Set 1 | saint-augustin
Processing Record 15 of Set 1 | kavieng
Processing Record 16 of Set 1 | richards bay
Processing Record 17 of Set 1 | kahului
City not found. Skipping...
Processing Record 19 of Set 1 | pisco
City not found. Skipping...
Processing Record 21 of Set 1 | dalvik
Processing Record 22 of Set 1 | port hardy
Processing Record 23 of Set 1 | vostok
Processing Record 24 of Set 1 | airai
City not found. Skipping...
City not found. Skipping...
Processing Record 27 of Set 1 | rikitea
City not found. Skipping...
Processing Record 29 of Set 1 | kerman
Processing Record 30 of Set 1 | puerto ayora
Processing Record 31 of Set 1 | hasaki
Processing Record 32 of Set 1 | oussouye
Processing Record 33 of Set 1 | kapaa
City not found. Skipping...
Processing Record 35 of Set 1 | nome
Processing Record 36 of Set 1 | hofn
City not found. Skipping...
City not found. Skipping...
Processing Record 39 of Set 1 | bull savanna
City not found. Skipping...
Processing Record 41 of Set 1 | jamestown
Processing Record 42 of Set 1 | bang saphan
City not found. Skipping...
City not found. Skipping...
Processing Record 45 of Set 1 | san cristobal
Processing Record 46 of Set 1 | bluff
City not found. Skipping...
Processing Record 48 of Set 1 | albany
Processing Record 49 of Set 1 | bandarbeyla
Processing Record 50 of Set 1 | punta gorda
City not found. Skipping...
Processing Record 2 of Set 2 | lagoa
Processing Record 3 of Set 2 | port elizabeth
Processing Record 4 of Set 2 | hilo
Processing Record 5 of Set 2 | araouane
Processing Record 6 of Set 2 | alice springs
Processing Record 7 of Set 2 | vite
Processing Record 8 of Set 2 | nishihara
Processing Record 9 of Set 2 | tasiilaq
Processing Record 10 of Set 2 | inta
City not found. Skipping...
City not found. Skipping...
Processing Record 13 of Set 2 | grindavik
Processing Record 14 of Set 2 | hermanus
Processing Record 15 of Set 2 | garden city
Processing Record 16 of Set 2 | anju
Processing Record 17 of Set 2 | nadadores
Processing Record 18 of Set 2 | tuktoyaktuk
Processing Record 19 of Set 2 | motril
City not found. Skipping...
Processing Record 21 of Set 2 | new norfolk
Processing Record 22 of Set 2 | verkhoyansk
Processing Record 23 of Set 2 | bethel
Processing Record 24 of Set 2 | benguela
Processing Record 25 of Set 2 | ponta do sol
Processing Record 26 of Set 2 | hirara
City not found. Skipping...
City not found. Skipping...
City not found. Skipping...
Processing Record 30 of Set 2 | hvolsvollur
Processing Record 31 of Set 2 | akureyri
Processing Record 32 of Set 2 | verkhnyaya inta
Processing Record 33 of Set 2 | tabriz
Processing Record 34 of Set 2 | ribeira grande
City not found. Skipping...
Processing Record 36 of Set 2 | hobart
Processing Record 37 of Set 2 | castro
Processing Record 38 of Set 2 | rehoboth
Processing Record 39 of Set 2 | mount isa
City not found. Skipping...
Processing Record 41 of Set 2 | sisimiut
Processing Record 42 of Set 2 | matara
Processing Record 43 of Set 2 | ko samui
Processing Record 44 of Set 2 | dikson
City not found. Skipping...
City not found. Skipping...
Processing Record 47 of Set 2 | naze
Processing Record 48 of Set 2 | isperih
Processing Record 49 of Set 2 | nanortalik
Processing Record 50 of Set 2 | palmer
Processing Record 1 of Set 3 | bubaque
Processing Record 2 of Set 3 | cherskiy
Processing Record 3 of Set 3 | nouadhibou
Processing Record 4 of Set 3 | hithadhoo
Processing Record 5 of Set 3 | rhyl
Processing Record 6 of Set 3 | stornoway
City not found. Skipping...
City not found. Skipping...
Processing Record 9 of Set 3 | atuona
Processing Record 10 of Set 3 | samarai
Processing Record 11 of Set 3 | marchena
Processing Record 12 of Set 3 | mattru
City not found. Skipping...
Processing Record 14 of Set 3 | carnarvon
City not found. Skipping...
Processing Record 16 of Set 3 | namie
Processing Record 17 of Set 3 | mercedes
City not found. Skipping...
Processing Record 19 of Set 3 | saskylakh
Processing Record 20 of Set 3 | clyde river
City not found. Skipping...
Processing Record 22 of Set 3 | fortuna
Processing Record 23 of Set 3 | yumen
Processing Record 24 of Set 3 | mar del plata
Processing Record 25 of Set 3 | vao
Processing Record 26 of Set 3 | harnai
Processing Record 27 of Set 3 | pangnirtung
Processing Record 28 of Set 3 | avarua
Processing Record 29 of Set 3 | bereda
Processing Record 30 of Set 3 | longview
Processing Record 31 of Set 3 | genhe
Processing Record 32 of Set 3 | saint-philippe
Processing Record 33 of Set 3 | faanui
Processing Record 34 of Set 3 | meulaboh
Processing Record 35 of Set 3 | barra do corda
Processing Record 36 of Set 3 | sungai padi
Processing Record 37 of Set 3 | raglan
Processing Record 38 of Set 3 | yellowknife
City not found. Skipping...
City not found. Skipping...
City not found. Skipping...
Processing Record 42 of Set 3 | juneau
Processing Record 43 of Set 3 | carbonia
Processing Record 44 of Set 3 | saryg-sep
Processing Record 45 of Set 3 | sioux lookout
Processing Record 46 of Set 3 | gainesville
Processing Record 47 of Set 3 | cape town
Processing Record 48 of Set 3 | kaoma
City not found. Skipping...
Processing Record 50 of Set 3 | east london
City not found. Skipping...
City not found. Skipping...
Processing Record 3 of Set 4 | loa janan
Processing Record 4 of Set 4 | okha
Processing Record 5 of Set 4 | canchungo
Processing Record 6 of Set 4 | commerce
Processing Record 7 of Set 4 | bredasdorp
City not found. Skipping...
City not found. Skipping...
City not found. Skipping...
City not found. Skipping...
City not found. Skipping...
Processing Record 13 of Set 4 | arraial do cabo
Processing Record 14 of Set 4 | brae
City not found. Skipping...
Processing Record 16 of Set 4 | kruisfontein
Processing Record 17 of Set 4 | butaritari
City not found. Skipping...
Processing Record 19 of Set 4 | leningradskiy
Processing Record 20 of Set 4 | kismayo
Processing Record 21 of Set 4 | lebu
Processing Record 22 of Set 4 | saint-pierre
City not found. Skipping...
Processing Record 24 of Set 4 | lorengau
Processing Record 25 of Set 4 | katav-ivanovsk
Processing Record 26 of Set 4 | jalalabad
Processing Record 27 of Set 4 | pyu
City not found. Skipping...
Processing Record 29 of Set 4 | yaan
Processing Record 30 of Set 4 | tolaga bay
Processing Record 31 of Set 4 | beidao
Processing Record 32 of Set 4 | west wendover
Processing Record 33 of Set 4 | kavaratti
Processing Record 34 of Set 4 | geraldton
Processing Record 35 of Set 4 | guerrero negro
Processing Record 36 of Set 4 | port alfred
Processing Record 37 of Set 4 | tiksi
Processing Record 38 of Set 4 | eureka
City not found. Skipping...
Processing Record 40 of Set 4 | husavik
Processing Record 41 of Set 4 | bathsheba
Processing Record 42 of Set 4 | oskemen
City not found. Skipping...
Processing Record 44 of Set 4 | fernley
Processing Record 45 of Set 4 | longyearbyen
Processing Record 46 of Set 4 | senneterre
City not found. Skipping...
City not found. Skipping...
Processing Record 49 of Set 4 | torbay
City not found. Skipping...
Processing Record 1 of Set 5 | rawson
City not found. Skipping...
City not found. Skipping...
Processing Record 4 of Set 5 | aklavik
Processing Record 5 of Set 5 | chokurdakh
Processing Record 6 of Set 5 | buritizeiro
Processing Record 7 of Set 5 | normal
Processing Record 8 of Set 5 | manzhouli
Processing Record 9 of Set 5 | esperance
Processing Record 10 of Set 5 | balikpapan
Processing Record 11 of Set 5 | inuvik
Processing Record 12 of Set 5 | coquimbo
City not found. Skipping...
Processing Record 14 of Set 5 | gushikawa
Processing Record 15 of Set 5 | shimoda
Processing Record 16 of Set 5 | zhangjiakou
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Convert Raw Data to DataFrame* Export the city data into a .csv.* Display the DataFrame | # Create a data frame from cities info
weather_dict = {
"City": city_list,
"Lat": city_lat,
"Lng": city_lng,
"Max Temp": city_temp,
"Humidity": city_humidity,
"Cloudiness": city_clouds,
"Wind Speed": city_wind,
"Country": city_country,
"Date": city_date
}
weather_data = pd.DataFrame(weather_dict)
weather_data
# Exporting to csv file
weather_data.to_csv(output_data_file, index = False)
# Importing file, if needed
weather_data = pd.read_csv("../output_data/cities.csv") | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Inspect the data and remove the cities where the humidity > 100%.----Skip this step if there are no cities that have humidity > 100%. | # No cities have humidity > 100%, although a few are exactly 100%
humid_weather_data = weather_data.loc[weather_data["Humidity"] >= 100]
humid_weather_data | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Plotting the Data* Use proper labeling of the plots using plot titles (including date of analysis) and axes labels.* Save the plotted figures as .pngs. Latitude vs. Temperature Plot | # Setting x and y values
x_values = weather_data["Lat"]
y_values = weather_data["Max Temp"]
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="teal", edgecolors="black")
plt.title("City Latitude vs. Max Temperature")
plt.ylabel("Max Temperature (F)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show()
# Save the image
#fig_path = os.path.abspath() # Figures out the absolute path for you in case your working directory moves around.
#fig.savefig(my_path + '/Sub Directory/graph.png') | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
ONE-SENTENCE DESCRIPTION: The graph above is displaying the max temperature (y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). This graph suggests that cities north of the equator (>0 Lat) might have a lower max temp. Latitude vs. Humidity Plot | # Setting x and y values
x_values = weather_data["Lat"]
y_values = weather_data["Humidity"]
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="teal", edgecolors="black")
plt.title("City Latitude vs. Humidity")
plt.ylabel("Humidity (%)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
ONE-SENTENCE DESCRIPTION: The graph above is displaying the recent humidity (%)(y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). This graph does not have an immediately discernible trend, other than cities tending to cluster at or greater than 60% humidity. Latitude vs. Cloudiness Plot | # Setting x and y values
x_values = weather_data["Lat"]
y_values = weather_data["Cloudiness"]
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="teal", edgecolors="black")
plt.title("City Latitude vs. Cloudiness")
plt.ylabel("Cloudiness (%)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
ONE-SENTENCE DESCRIPTION:The graph above is displaying the recent cloudiness (%)(y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). This graph also does not have an immediately discernible trend. Latitude vs. Wind Speed Plot | # Setting x and y values
x_values = weather_data["Lat"]
y_values = weather_data["Wind Speed"]
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="teal", edgecolors="black")
plt.title("City Latitude vs. Wind Speed")
plt.ylabel("Wind Speed (mph)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
ONE-SENTENCE DESCRIPTION:The graph above is displaying the recent wind speed (mph)(y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). Perhaps one observable trend from this graph: it appears that there is greater range in the wind speeds of cities north of equator (>Lat 0). Linear Regression After each pair of plots, take the time to explain what the linear regression is modeling. For example, describe any relationships you notice and any other analysis you may have.Your final notebook must:* Randomly select **at least** 500 unique (non-repeat) cities based on latitude and longitude.* Perform a weather check on each of the cities using a series of successive API calls.* Include a print log of each city as it's being processed with the city number and city name.* Save a CSV of all retrieved data and a PNG image for each scatter plot. | # Making two separate dfs for north and south hemisphere
north_hemi = weather_data.loc[weather_data["Lat"] >= 0]
south_hemi = weather_data.loc[weather_data["Lat"] < 0] | _____no_output_____ | ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Northern Hemisphere - Max Temp vs. Latitude Linear Regression | # Setting x and y values
x_values = north_hemi["Lat"]
y_values = north_hemi["Max Temp"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (0,-10), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Northern Hemisphere: Max Temp vs. Latitude")
plt.ylabel("Max Temp (F)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: -0.8765697068804407
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Southern Hemisphere - Max Temp vs. Latitude Linear Regression | # Setting x and y values
x_values = south_hemi["Lat"]
y_values = south_hemi["Max Temp"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (-30,50), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Southern Hemisphere: Max Temp vs. Latitude")
plt.ylabel("Max Temp (F)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: 0.44590906959636567
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
ANALYSIS OF NORTH/SOUTH-HEMI MAX TEMP:The graphs above are displaying the max temperature (y-axis) of cities in the dataset arranged by latitude (x-axis) and divided into two groups--the southern hemisphere and the northern hemisphere. What we see from both of these graphs is that max temperatures are higher for cities closer to the equator. This relationship is weakly positively correlated for the southern hemisphere (moving towards the equator), and strongly negatively correlated for the northern hemisphere (moving away from the equator). It is plausible that the seasons may be playing a role, as the northern hemisphere is currently experiencing winter. Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression | # Setting x and y values
x_values = north_hemi["Lat"]
y_values = north_hemi["Humidity"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (40,20), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Northern Hemisphere: Humidity vs. Latitude")
plt.ylabel("Humidity (%)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: 0.30945504958053144
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression | # Setting x and y values
x_values = south_hemi["Lat"]
y_values = south_hemi["Humidity"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (-30,20), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Southern Hemisphere: Humidity vs. Latitude")
plt.ylabel("Humidity (%)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: 0.22520675986668628
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
ANALYSIS OF NORTH/SOUTH-HEMI HUMIDITY:The graphs above are displaying the humidity (y-axis) of cities in the dataset arranged by latitude (x-axis) and divided into two groups--the southern hemisphere and the northern hemisphere. These graphs reflect a weak positive correlation between humidity and latitude. Rather than being centered around the equator as shown in the previous section, the humidity seems to increase the more north a city is located. As speculated on previously, the seasons may be playing a role, however it is difficult to produce a logical explanation as colder air tends to hold less humidity. Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression | # Setting x and y values
x_values = north_hemi["Lat"]
y_values = north_hemi["Cloudiness"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (45,15), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Northern Hemisphere: Cloudiness vs. Latitude")
plt.ylabel("Cloudiness (%)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: 0.2989895417151042
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression | # Setting x and y values
x_values = south_hemi["Lat"]
y_values = south_hemi["Cloudiness"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (-50,30), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Southern Hemisphere: Cloudiness vs. Latitude")
plt.ylabel("Cloudiness (%)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: 0.23659107153505674
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
ANALYSIS OF NORTH/SOUTH-HEMI CLOUDINESS:The graphs above are displaying the cloudiness (y-axis) of cities in the dataset arranged by latitude (x-axis) and divided into two groups--the southern hemisphere and the northern hemisphere. These graphs also reflect a very weak positive correlation, if any, between cloudiness and latitude. Cloudiness seems to increase the more north a city is located. Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression | # Setting x and y values
x_values = north_hemi["Lat"]
y_values = north_hemi["Wind Speed"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (10,40), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Northern Hemisphere: Wind Speed vs. Latitude")
plt.ylabel("Wind Speed (mph)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: 0.04083196500915729
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Southern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression | # Setting x and y values
x_values = south_hemi["Lat"]
y_values = south_hemi["Wind Speed"]
# Linear Regression
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.plot(x_values, regress_values, "r-")
plt.annotate(line_eq, (-55,20), fontsize=15, color="red")
print(f"The r-value is: {rvalue}")
# Plot the scatter plot
plt.scatter(x_values, y_values, marker="o", facecolors="aqua", edgecolors="black")
plt.title("Southern Hemisphere: Wind Speed vs. Latitude")
plt.ylabel("Wind Speed (mph)", fontsize=12)
plt.xlabel("Latitude", fontsize=12)
plt.grid(alpha=0.2)
plt.show() | The r-value is: -0.1644284507948641
| ADSL | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge |
Copyright 2020 The TensorFlow Authors. | #@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License. | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Introduction to graphs and tf.function View on TensorFlow.org Run in Google Colab View source on GitHub Download notebook OverviewThis guide goes beneath the surface of TensorFlow and Keras to demonstrate how TensorFlow works. If you instead want to immediately get started with Keras, check out the [collection of Keras guides](keras/).In this guide, you'll learn how TensorFlow allows you to make simple changes to your code to get graphs, how graphs are stored and represented, and how you can use them to accelerate your models.Note: For those of you who are only familiar with TensorFlow 1.x, this guide demonstrates a very different view of graphs.**This is a big-picture overview that covers how `tf.function` allows you to switch from eager execution to graph execution.** For a more complete specification of `tf.function`, go to the [`tf.function` guide](function). What are graphs?In the previous three guides, you ran TensorFlow **eagerly**. This means TensorFlow operations are executed by Python, operation by operation, and returning results back to Python.While eager execution has several unique advantages, graph execution enables portability outside Python and tends to offer better performance. **Graph execution** means that tensor computations are executed as a *TensorFlow graph*, sometimes referred to as a `tf.Graph` or simply a "graph."**Graphs are data structures that contain a set of `tf.Operation` objects, which represent units of computation; and `tf.Tensor` objects, which represent the units of data that flow between operations.** They are defined in a `tf.Graph` context. Since these graphs are data structures, they can be saved, run, and restored all without the original Python code.This is what a TensorFlow graph representing a two-layer neural network looks like when visualized in TensorBoard. The benefits of graphsWith a graph, you have a great deal of flexibility. You can use your TensorFlow graph in environments that don't have a Python interpreter, like mobile applications, embedded devices, and backend servers. TensorFlow uses graphs as the format for [saved models](saved_model) when it exports them from Python.Graphs are also easily optimized, allowing the compiler to do transformations like:* Statically infer the value of tensors by folding constant nodes in your computation *("constant folding")*.* Separate sub-parts of a computation that are independent and split them between threads or devices.* Simplify arithmetic operations by eliminating common subexpressions. There is an entire optimization system, [Grappler](./graph_optimization.ipynb), to perform this and other speedups.In short, graphs are extremely useful and let your TensorFlow run **fast**, run **in parallel**, and run efficiently **on multiple devices**.However, you still want to define your machine learning models (or other computations) in Python for convenience, and then automatically construct graphs when you need them. Non-strict executionGraph execution only executes the operations necessary to produce the observable effects, which includes:- the return value of the function,- documented well-known side-effects: * input/output operations, `tf.print` * debugging operations, such as the assert functions in `tf.debugging`, * mutations of `tf.Variable`.This behavior is usually known as [Non-strict execution](https://en.wikipedia.org/wiki/Evaluation_strategyNon-strict_evaluation), and differs from eager execution, which steps through all of the program operations, needed or not. In particular, runtime error checking does not count as an observable effect. If an operation is skipped because it is unnecessary it cannot raise any runtime errors. In the following example, the "unnecessary" operation `tf.math.bincount` is skipped during graph execution, so the runtime error `InvalidArgumentError` is not raised as it would be in eager execution. Do not rely on an error being raised while executing a graph. | import tensorflow as tf
import timeit
from datetime import datetime
@tf.function
def unused_return_graph(x):
_ = tf.math.bincount(x)
return x
def unused_return_eager(x):
_ = tf.math.bincount(x)
return x
# `tf.math.bincount` in eager execution raises an error.
try:
_ = unused_return_eager([-1])
raise None
except tf.errors.InvalidArgumentError as e:
assert "Input arr must be non-negative" in str(e)
# Only needed operations are run during graph exection. The error is not raised.
_ = unused_return_graph([-1]) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Taking advantage of graphsYou create and run a graph in TensorFlow by using `tf.function`, either as a direct call or as a decorator. `tf.function` takes a regular function as input and returns a `Function`. **A `Function` is a Python callable that builds TensorFlow graphs from the Python function. You use a `Function` in the same way as its Python equivalent.** | # Define a Python function.
def a_regular_function(x, y, b):
x = tf.matmul(x, y)
x = x + b
return x
# `a_function_that_uses_a_graph` is a TensorFlow `Function`.
a_function_that_uses_a_graph = tf.function(a_regular_function)
# Make some tensors.
x1 = tf.constant([[1.0, 2.0]])
y1 = tf.constant([[2.0], [3.0]])
b1 = tf.constant(4.0)
orig_value = a_regular_function(x1, y1, b1).numpy()
# Call a `Function` like a Python function.
tf_function_value = a_function_that_uses_a_graph(x1, y1, b1).numpy()
assert(orig_value == tf_function_value) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
On the outside, a `Function` looks like a regular function you write using TensorFlow operations. [Underneath](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/python/eager/def_function.py), however, it is *very different*. A `Function` **encapsulates [several `tf.Graph`s behind one API](polymorphism_one_function_many_graphs).** That is how `Function` is able to give you the [benefits of graph execution](the_benefits_of_graphs), like speed and deployability. `tf.function` applies to a function *and all other functions it calls*: | def inner_function(x, y, b):
x = tf.matmul(x, y)
x = x + b
return x
# Use the decorator to make `outer_function` a `Function`.
@tf.function
def outer_function(x):
y = tf.constant([[2.0], [3.0]])
b = tf.constant(4.0)
return inner_function(x, y, b)
# Note that the callable will create a graph that
# includes `inner_function` as well as `outer_function`.
outer_function(tf.constant([[1.0, 2.0]])).numpy() | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
If you have used TensorFlow 1.x, you will notice that at no time did you need to define a `Placeholder` or `tf.Session`. Converting Python functions to graphsAny function you write with TensorFlow will contain a mixture of built-in TF operations and Python logic, such as `if-then` clauses, loops, `break`, `return`, `continue`, and more. While TensorFlow operations are easily captured by a `tf.Graph`, Python-specific logic needs to undergo an extra step in order to become part of the graph. `tf.function` uses a library called AutoGraph (`tf.autograph`) to convert Python code into graph-generating code. | def simple_relu(x):
if tf.greater(x, 0):
return x
else:
return 0
# `tf_simple_relu` is a TensorFlow `Function` that wraps `simple_relu`.
tf_simple_relu = tf.function(simple_relu)
print("First branch, with graph:", tf_simple_relu(tf.constant(1)).numpy())
print("Second branch, with graph:", tf_simple_relu(tf.constant(-1)).numpy()) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Though it is unlikely that you will need to view graphs directly, you can inspect the outputs to check the exact results. These are not easy to read, so no need to look too carefully! | # This is the graph-generating output of AutoGraph.
print(tf.autograph.to_code(simple_relu))
# This is the graph itself.
print(tf_simple_relu.get_concrete_function(tf.constant(1)).graph.as_graph_def()) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Most of the time, `tf.function` will work without special considerations. However, there are some caveats, and the [tf.function guide](./function.ipynb) can help here, as well as the [complete AutoGraph reference](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/python/autograph/g3doc/reference/index.md) Polymorphism: one `Function`, many graphsA `tf.Graph` is specialized to a specific type of inputs (for example, tensors with a specific [`dtype`](https://www.tensorflow.org/api_docs/python/tf/dtypes/DType) or objects with the same [`id()`](https://docs.python.org/3/library/functions.htmlid])).Each time you invoke a `Function` with new `dtypes` and shapes in its arguments, `Function` creates a new `tf.Graph` for the new arguments. The `dtypes` and shapes of a `tf.Graph`'s inputs are known as an **input signature** or just a **signature**.The `Function` stores the `tf.Graph` corresponding to that signature in a `ConcreteFunction`. **A `ConcreteFunction` is a wrapper around a `tf.Graph`.** | @tf.function
def my_relu(x):
return tf.maximum(0., x)
# `my_relu` creates new graphs as it observes more signatures.
print(my_relu(tf.constant(5.5)))
print(my_relu([1, -1]))
print(my_relu(tf.constant([3., -3.]))) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
If the `Function` has already been called with that signature, `Function` does not create a new `tf.Graph`. | # These two calls do *not* create new graphs.
print(my_relu(tf.constant(-2.5))) # Signature matches `tf.constant(5.5)`.
print(my_relu(tf.constant([-1., 1.]))) # Signature matches `tf.constant([3., -3.])`. | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Because it's backed by multiple graphs, a `Function` is **polymorphic**. That enables it to support more input types than a single `tf.Graph` could represent, as well as to optimize each `tf.Graph` for better performance. | # There are three `ConcreteFunction`s (one for each graph) in `my_relu`.
# The `ConcreteFunction` also knows the return type and shape!
print(my_relu.pretty_printed_concrete_signatures()) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Using `tf.function`So far, you've learned how to convert a Python function into a graph simply by using `tf.function` as a decorator or wrapper. But in practice, getting `tf.function` to work correctly can be tricky! In the following sections, you'll learn how you can make your code work as expected with `tf.function`. Graph execution vs. eager executionThe code in a `Function` can be executed both eagerly and as a graph. By default, `Function` executes its code as a graph: | @tf.function
def get_MSE(y_true, y_pred):
sq_diff = tf.pow(y_true - y_pred, 2)
return tf.reduce_mean(sq_diff)
y_true = tf.random.uniform([5], maxval=10, dtype=tf.int32)
y_pred = tf.random.uniform([5], maxval=10, dtype=tf.int32)
print(y_true)
print(y_pred)
get_MSE(y_true, y_pred) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
To verify that your `Function`'s graph is doing the same computation as its equivalent Python function, you can make it execute eagerly with `tf.config.run_functions_eagerly(True)`. This is a switch that **turns off `Function`'s ability to create and run graphs**, instead executing the code normally. | tf.config.run_functions_eagerly(True)
get_MSE(y_true, y_pred)
# Don't forget to set it back when you are done.
tf.config.run_functions_eagerly(False) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
However, `Function` can behave differently under graph and eager execution. The Python [`print`](https://docs.python.org/3/library/functions.htmlprint) function is one example of how these two modes differ. Let's check out what happens when you insert a `print` statement to your function and call it repeatedly. | @tf.function
def get_MSE(y_true, y_pred):
print("Calculating MSE!")
sq_diff = tf.pow(y_true - y_pred, 2)
return tf.reduce_mean(sq_diff) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Observe what is printed: | error = get_MSE(y_true, y_pred)
error = get_MSE(y_true, y_pred)
error = get_MSE(y_true, y_pred) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Is the output surprising? **`get_MSE` only printed once even though it was called *three* times.**To explain, the `print` statement is executed when `Function` runs the original code in order to create the graph in a process known as ["tracing"](function.ipynbtracing). **Tracing captures the TensorFlow operations into a graph, and `print` is not captured in the graph.** That graph is then executed for all three calls **without ever running the Python code again**.As a sanity check, let's turn off graph execution to compare: | # Now, globally set everything to run eagerly to force eager execution.
tf.config.run_functions_eagerly(True)
# Observe what is printed below.
error = get_MSE(y_true, y_pred)
error = get_MSE(y_true, y_pred)
error = get_MSE(y_true, y_pred)
tf.config.run_functions_eagerly(False) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
`print` is a *Python side effect*, and there are [other differences](functionlimitations) that you should be aware of when converting a function into a `Function`. Note: If you would like to print values in both eager and graph execution, use `tf.print` instead. `tf.function` best practicesIt may take some time to get used to the behavior of `Function`. To get started quickly, first-time users should play around with decorating toy functions with `@tf.function` to get experience with going from eager to graph execution.*Designing for `tf.function`* may be your best bet for writing graph-compatible TensorFlow programs. Here are some tips:- Toggle between eager and graph execution early and often with `tf.config.run_functions_eagerly` to pinpoint if/ when the two modes diverge.- Create `tf.Variable`soutside the Python function and modify them on the inside. The same goes for objects that use `tf.Variable`, like `keras.layers`, `keras.Model`s and `tf.optimizers`.- Avoid writing functions that [depend on outer Python variables](functiondepending_on_python_global_and_free_variables), excluding `tf.Variable`s and Keras objects.- Prefer to write functions which take tensors and other TensorFlow types as input. You can pass in other object types but [be careful](functiondepending_on_python_objects)!- Include as much computation as possible under a `tf.function` to maximize the performance gain. For example, decorate a whole training step or the entire training loop. Seeing the speed-up `tf.function` usually improves the performance of your code, but the amount of speed-up depends on the kind of computation you run. Small computations can be dominated by the overhead of calling a graph. You can measure the difference in performance like so: | x = tf.random.uniform(shape=[10, 10], minval=-1, maxval=2, dtype=tf.dtypes.int32)
def power(x, y):
result = tf.eye(10, dtype=tf.dtypes.int32)
for _ in range(y):
result = tf.matmul(x, result)
return result
print("Eager execution:", timeit.timeit(lambda: power(x, 100), number=1000))
power_as_graph = tf.function(power)
print("Graph execution:", timeit.timeit(lambda: power_as_graph(x, 100), number=1000)) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
`tf.function` is commonly used to speed up training loops, and you can learn more about it in [Writing a training loop from scratch](keras/writing_a_training_loop_from_scratchspeeding-up_your_training_step_with_tffunction) with Keras.Note: You can also try [`tf.function(jit_compile=True)`](https://www.tensorflow.org/xlaexplicit_compilation_with_tffunctionjit_compiletrue) for a more significant performance boost, especially if your code is heavy on TF control flow and uses many small tensors. Performance and trade-offsGraphs can speed up your code, but the process of creating them has some overhead. For some functions, the creation of the graph takes more time than the execution of the graph. **This investment is usually quickly paid back with the performance boost of subsequent executions, but it's important to be aware that the first few steps of any large model training can be slower due to tracing.**No matter how large your model, you want to avoid tracing frequently. The `tf.function` guide discusses [how to set input specifications and use tensor arguments](functioncontrolling_retracing) to avoid retracing. If you find you are getting unusually poor performance, it's a good idea to check if you are retracing accidentally. When is a `Function` tracing?To figure out when your `Function` is tracing, add a `print` statement to its code. As a rule of thumb, `Function` will execute the `print` statement every time it traces. | @tf.function
def a_function_with_python_side_effect(x):
print("Tracing!") # An eager-only side effect.
return x * x + tf.constant(2)
# This is traced the first time.
print(a_function_with_python_side_effect(tf.constant(2)))
# The second time through, you won't see the side effect.
print(a_function_with_python_side_effect(tf.constant(3)))
# This retraces each time the Python argument changes,
# as a Python argument could be an epoch count or other
# hyperparameter.
print(a_function_with_python_side_effect(2))
print(a_function_with_python_side_effect(3)) | _____no_output_____ | Apache-2.0 | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n |
Day 3 Part 1 | """
Right 3, down 1.
"""
i=1
trees=0
with open('input_day3.txt','r') as file:
for line in file:
if line.strip()[i%31-1] == '#':
trees+=1
i+=3
trees | _____no_output_____ | MIT | 2020/paula/day3/advent_code_day3.ipynb | bbglab/adventofcode |
Part 2 | """
Right 1, down 1.
"""
i=1
trees1=0
with open('input_day3.txt','r') as file:
for line in file:
if line.strip()[i%31-1] == '#':
trees1+=1
i+=1
"""
Right 5, down 1.
"""
i=1
trees2=0
with open('input_day3.txt','r') as file:
for line in file:
if line.strip()[i%31-1] == '#':
trees2+=1
i+=5
"""
Right 7, down 1.
"""
i=1
trees3=0
with open('input_day3.txt','r') as file:
for line in file:
if line.strip()[i%31-1] == '#':
trees3+=1
i+=7
"""
Right 1, down 2.
"""
i=1
trees4=0
j=0
with open('input_day3.txt','r') as file:
for line in file:
if j%2 == 0:
if line.strip()[i%31-1] == '#':
trees4+=1
i+=1
j+=1
print(trees,trees1,trees2,trees3,trees4)
trees*trees1*trees2*trees3*trees4 | _____no_output_____ | MIT | 2020/paula/day3/advent_code_day3.ipynb | bbglab/adventofcode |
ICPE 639 Introduction to Machine Learning ------ With Energy ApplicationsSome of the examples and exercises of this course are based on several books as well as open-access materials on machine learning, including [Hands-on Machine Learning with Scikit-Learn, Keras and TensorFlow](https://www.oreilly.com/library/view/hands-on-machine-learning/9781492032632/) &169; 2021: Xiaoning Qian [Homepage](http://xqian37.github.io/)**[Note]** This is currently a work in progress, will be updated as the material is tested in the class room.All material open source under a Creative Commons license and free for use in non-commercial applications.Source material used under the Creative Commons Attribution-NonCommercial 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc/3.0/ or send a letter to Creative Commons, PO Box 1866, Mountain View, CA 94042, USA. Support Vector MachineThis section will cover the content listed below: - [1 Support Vector Machine](1-Support-Vector-Machine)- [2 Support Vector Regression](2-Support-Vector-Regression)- [3 One-class SVM](3-One-class-SVM)- [4 Hands-on Exercise](4-Exercise)- [Reference](Reference) | import warnings
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from IPython.display import Image
from IPython.core.display import HTML
%matplotlib inline
warnings.filterwarnings('ignore') | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
1 Support Vector Machine 1.1 IntroductionWe will start the introduction of Support Vector Machine (SVM) for classification problems---Support Vector Classifier (SVC). Consider a simple binary classification problem. Assume we have a linearly separable data in 2-d feature space. We try to find a boundary that divides the data into two regions such that the misclassification can be minimized. Notice that different lines can be used as separators between samples. Depending on the line we choose, a new point marked by 'x' in the plot will be assigned to a different class label. The problem is how *well* the derived boundaries *generalize* to the new testing points. | from scipy import stats
import seaborn as sns
from sklearn.datasets.samples_generator import make_blobs
X, y = make_blobs(n_samples = 30, centers = 2, random_state = 0, cluster_std = 0.6)
plt.figure(figsize=(9, 7))
plt.scatter(X[:, 0], X[:, 1], c = y, s = 50, cmap = "icefire")
xfit = np.arange(-0.5, 3.0, 0.1)
for m, b in [(0.0, 2.5), (0.5, 1.8), (-0.2, 3.0)]:
plt.plot(xfit, m * xfit + b, '--k')
plt.plot([0.0], [2.0], 'x', color = 'red', markersize = 10)
plt.show()
make_blobs?? | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
SVMs provide a way to achieve good generalizability with the intuition: rather than simply drawing a zero-width line between the classes, consider each line with a margin of certain width, meaning that we do not worry about the errors as long as the errors fall within the margin. In SVMs, the line that maximizes this margin is the one to be chosen as the optimal model. | plt.figure(figsize=(9, 7))
plt.scatter(X[:, 0], X[:, 1], c = y, s = 50, cmap = "icefire")
xfit = np.arange(-0.5, 3.0, 0.1)
for m, b, d in [(0.0, 2.5, 0.4), (0.5, 1.8, 0.2), (-0.2, 3.0, 0.1)]:
yfit = m * xfit + b
plt.plot(xfit, yfit, '-k')
plt.fill_between(xfit, yfit - d, yfit + d, edgecolor='none',
color='#AAAAAA', alpha=0.4)
plt.plot([0.0], [2.0], 'x', color = 'red', markersize = 10)
plt.show() | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
To fit a SVM model on this generated dataset: | # for visualization
def plot_svc_decision_function(model, ax=None, plot_support=True):
"""Plot the decision function for a 2D SVC"""
if ax is None:
ax = plt.gca()
xlim = ax.get_xlim()
ylim = ax.get_ylim()
# create grid to evaluate model
x = np.linspace(xlim[0], xlim[1], 30)
y = np.linspace(ylim[0], ylim[1], 30)
Y, X = np.meshgrid(y, x)
xy = np.vstack([X.ravel(), Y.ravel()]).T
P = model.decision_function(xy).reshape(X.shape)
# plot decision boundary and margins
ax.contour(X, Y, P, colors='k',
levels=[-1, 0, 1], alpha=0.5,
linestyles=['--', '-', '--'])
# plot support vectors
if plot_support:
ax.scatter(model.support_vectors_[:, 0],
model.support_vectors_[:, 1],
s=300, linewidth=1, facecolors='none');
ax.set_xlim(xlim)
ax.set_ylim(ylim)
from sklearn.svm import SVC # "Support vector classifier"
model = SVC(kernel='linear', C=1E10)
model.fit(X, y)
plt.figure(figsize = (9, 7))
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='icefire')
plot_svc_decision_function(model) | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
This is the dividing line that maximizes the margin between two sets of points. There are some points touching the margin which are the pivotal elements of this fit known as the support vectors and can be returned by `support_vectors_`. A key to this classifier is that only the position of the support vectors matter. The points that are further away from the margin on the correct side do not change the fit. | model.support_vectors_ | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
This method can be extended to nonlinear boundaries with kernels which gives the Kernel SVM where we can map the data into higher-dimensional space defined by basis function and find a linear classifier for the nonlinear relationship. 1.2 Math Formulation of SVCLet $w$ denote the model coefficient vector and $b$ intercept, which define the linear boundary. The original SVC formulation can be written as: $$\max_{w,b} \frac{1}{\|w\|},$$$$\mbox{subject to: }\quad y_i (w^T x_i - b) \geq 1, \quad \forall i \mbox{ in training data set.}$$It can be rewritten as follows (allowing linearly nonseparable data):$$\min_{w,b} \|w\|^2 + C \sum_i \epsilon_i, $$$$\mbox{subject to: }\quad y_i (w^T x_i - b) \geq 1 - \epsilon_i, \quad \forall i \mbox{ in training data set.}$$ Primal-Dual (Optimization)The above is a **convex programming** formulation, which can be equivalently solved in the dual form: $$\max_{\alpha} \sum_i \alpha_i - \frac{1}{2}\sum_{i,j} \alpha_iy_i y_j \alpha_j, $$$$\mbox{subject to: }\quad \sum_i \alpha_i y_i = 0; \quad \alpha_i \geq 0, \quad \forall i \mbox{ in training data set.}$$Other **Karush–Kuhn–Tucker (KKT) conditions**: $w=\sum_i \alpha_i y_i x_i$ and $b$ derived by **support vectors**.**[Note]** Solving the dual form only requires the **inner product** term of the input features $$, which can be replaced by any kernel to extend it nonlinear SVC. 1.3 ExampleUse the labeled faces of various public figures in the Wild dataset as an example. Eight public figures are included. Each image is of size $62 \times 47$. We can use the pixels directly as a feature, but it's more efficient to do some preprocessing before hand, e.g. extract some fundamental components. | from sklearn.datasets import fetch_lfw_people
from sklearn.svm import SVC
from sklearn.decomposition import PCA
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.metrics import classification_report, confusion_matrix
faces = fetch_lfw_people(min_faces_per_person=60)
print(faces.target_names)
print(faces.images.shape)
fig, ax = plt.subplots(3, 5)
for i, axi in enumerate(ax.flat):
axi.imshow(faces.images[i], cmap='bone')
axi.set(xticks=[], yticks=[],
xlabel=faces.target_names[faces.target[i]])
pca = PCA(n_components=150, whiten=True, random_state=42)
svc = SVC(kernel='rbf', class_weight='balanced')
model = make_pipeline(pca, svc)
Xtrain, Xtest, ytrain, ytest = train_test_split(faces.data, faces.target,
random_state=42)
param_grid = {'svc__C': [1, 5, 10],
'svc__gamma': [0.0001, 0.001, 0.005]}
grid = GridSearchCV(model, param_grid)
%time grid.fit(Xtrain, ytrain)
print(grid.best_params_)
model = grid.best_estimator_
yfit = model.predict(Xtest)
fig, ax = plt.subplots(4, 6)
for i, axi in enumerate(ax.flat):
axi.imshow(Xtest[i].reshape(62, 47), cmap='bone')
axi.set(xticks=[], yticks=[])
axi.set_ylabel(faces.target_names[yfit[i]].split()[-1],
color='black' if yfit[i] == ytest[i] else 'red')
fig.suptitle('Predicted Names; Incorrect Labels in Red', size=14);
print(classification_report(ytest, yfit,
target_names=faces.target_names))
mat = confusion_matrix(ytest, yfit)
sns.heatmap(mat.T, square=True, annot=True, fmt='d', cbar=False,
xticklabels=faces.target_names,
yticklabels=faces.target_names)
plt.xlabel('true label')
plt.ylabel('predicted label'); | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
2 Support Vector RegressionThe Support Vector Regression uses the same principles as the SVM for classification. The differences are as follows:* The output is a real number, inifinite possibilities * A margin of tolerance is set in approximation to the SVM However, if we only need to reduce the errors to a certain degree, meaning as long as the errors fall within an acceptable range.For illustrution, consider the relationship between TV and Sales. The plot below shows the results of a trained SVR model on the Advertising dataset. The red line represents the line of simple linear regression fit. The gray dashed lines represent the margin of error $\epsilon = 5$. | from sklearn import linear_model
advertising = pd.read_csv('https://raw.githubusercontent.com/XiaomengYan/MachineLearning_dataset/main/Advertising.csv', usecols=[1,2,3,4])
# Visualization
X = advertising.TV
X = X.values.reshape(-1, 1)
y = advertising.Sales
# simple linear regression
regr = linear_model.LinearRegression()
regr.fit(X,y)
xfit = np.linspace(X.min(), X.max(), 1000).reshape(-1, 1)
yfit = regr.predict(xfit)
yfit_ub = yfit + 5
yfit_lb = yfit - 5
plt.figure(figsize = (10, 6))
plt.suptitle('TV vs Sales')
plt.scatter(X,y)
plt.xlim(-10, 330)
plt.xlabel("TV")
plt.ylabel("sales")
plt.plot(xfit, yfit, 'r',linewidth = 3)
plt.plot(xfit, yfit_lb, 'g--', linewidth = 2)
plt.plot(xfit, yfit_ub, 'g--', linewidth = 2)
plt.vlines(x= xfit[50], ymin=yfit[50], ymax=yfit_ub[50], colors='gray', ls=':', lw=2)
s = 'epsilon = 5'
plt.text(xfit[50]-20, 0.5 * (yfit[50] + yfit_ub[50]), s)
plt.vlines(xfit[50], ymin=yfit_lb[50], ymax=yfit[50], colors='gray', ls=':', lw=2)
plt.text(xfit[50]-20, 0.5 * (yfit[50] + yfit_lb[50]), s)
s = 'beta_i x_i'
plt.text(300, yfit[-1], s, fontsize = 15)
s = 'beta_i x_i + epsilon'
plt.text(300, yfit_ub[-1], s, fontsize = 15)
s = 'beta_i x_i - epsilon'
plt.text(300, yfit_lb[-1], s, fontsize = 15)
ax = plt.axes()
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
plt.show() | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
SVR gives us the flexibility to define how much error is acceptable in our model and will find an appropriate line (or hyperplane in higher dimensions) to fit the data. The objective function of SVR is to minimize the $l_2$-norm of the coefficients,$$\min \frac{1}{2}||w||^2$$and use the error term as the constraints as following,$$|y_i - w^Tx_i|\leq \epsilon$$**[Note]** The math formulation of SVR is similar as SVC. Based on the same *primal-dual* trick, kernel-based SVR can be derived for nonlinear problems. 3 One-class SVMIn addition to **anomaly detection** based on the probabilistic methods, for exam these steming from hypothesis testing, SVM can also be extended for that. One of such formulations is **one-class SVM**: $$\min_{R, c} R^2 + \frac{1}{C}\sum_i \xi_i $$subject to the following constraints: $$\|x_i - c \|^2 \leq R^2 + \xi_i, \quad \forall i; $$and $$\xi_i \geq 0, \quad \forall i.$$Or another formulation **$\nu$-SVM**: $$\min_{w,\xi,\rho} \frac{1}{2}||w||^2 + \frac{1}{\nu n}\sum_i \xi_i -\rho$$subject to the following constraints: $$w^Tx_i \geq \rho - \xi_i, \quad \forall i; $$and $$\xi_i \geq 0, \quad \forall i.$$http://rvlasveld.github.io/blog/2013/07/12/introduction-to-one-class-support-vector-machines/ | from sklearn.svm import OneClassSVM
#from sklearn.datasets import make_blobs
from numpy import quantile, where, random
#import matplotlib.pyplot as plt
random.seed(13)
x, _ = make_blobs(n_samples=200, centers=1, cluster_std=.3, center_box=(8, 8))
plt.scatter(x[:,0], x[:,1])
plt.show()
svm = OneClassSVM(kernel='rbf', gamma='auto', nu=0.05)
print(svm)
svm.fit(x)
pred = svm.predict(x)
anom_index = where(pred==-1)
values = x[anom_index]
plt.scatter(x[:,0], x[:,1])
plt.scatter(values[:,0], values[:,1], color='r')
plt.show()
svm = OneClassSVM(kernel='rbf', gamma='auto', nu=0.03)
print(svm)
pred = svm.fit_predict(x)
scores = svm.score_samples(x)
thresh = quantile(scores, 0.03)
print(thresh)
index = where(scores<=thresh)
values = x[index]
plt.scatter(x[:,0], x[:,1])
plt.scatter(values[:,0], values[:,1], color='r')
plt.show() | OneClassSVM(cache_size=200, coef0=0.0, degree=3, gamma='auto', kernel='rbf',
max_iter=-1, nu=0.03, shrinking=True, tol=0.001, verbose=False)
3.577526406228678
| MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
Kitchen Power Usage Example[REDD](http://redd.csail.mit.edu/) dataset contains several weeks of power data for 6 different homes. Here we'll extract one house's kitchen power useage as a simple example for one-class SVM. For more implementations, please refer to [minhup's repo](https://github.com/minhup/Energy-Disaggregation) . | # Download the dataset
!wget http://redd:[email protected]/data/low_freq.tar.bz2
!tar -xf low_freq.tar.bz2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
from IPython.display import display
import datetime
import time
import math
import warnings
warnings.filterwarnings("ignore")
import glob
def read_label():
label = {}
for i in range(1, 7):
hi = 'low_freq/house_{}/labels.dat'.format(i)
label[i] = {}
with open(hi) as f:
for line in f:
splitted_line = line.split(' ')
label[i][int(splitted_line[0])] = splitted_line[1].strip() + '_' + splitted_line[0]
return label
def read_merge_data(house):
path = 'low_freq/house_{}/'.format(house)
file = path + 'channel_1.dat'
df = pd.read_table(file, sep = ' ', names = ['unix_time', labels[house][1]],
dtype = {'unix_time': 'int64', labels[house][1]:'float64'})
num_apps = len(glob.glob(path + 'channel*'))
for i in range(2, num_apps + 1):
file = path + 'channel_{}.dat'.format(i)
data = pd.read_table(file, sep = ' ', names = ['unix_time', labels[house][i]],
dtype = {'unix_time': 'int64', labels[house][i]:'float64'})
df = pd.merge(df, data, how = 'inner', on = 'unix_time')
df['timestamp'] = df['unix_time'].astype("datetime64[s]")
df = df.set_index(df['timestamp'].values)
df.drop(['unix_time','timestamp'], axis=1, inplace=True)
return df
# Extract labels and data from the dataset
labels = read_label()
df = {}
for i in range(1,2):
print('House {}: '.format(i), labels[i], '\n')
df[i] = read_merge_data(i)
# Extract the time index
dates = {}
for i in range(1, 2):
dates[i] = [str(time)[:10] for time in df[i].index.values]
dates[i] = sorted(list(set(dates[i])))
print('House {0} data contain {1} days from {2} to {3}.'.format(i,len(dates[i]),dates[i][0], dates[i][-1]))
print(dates[i], '\n')
# Plot the first 3 days power usage of house 1's kitchen
house = 1
n_days = 3
df1 = df[house].loc[:dates[house][n_days - 1]]
plt.figure(figsize=(18,8))
plt.title('kitchen_outlets_7', fontsize='15')
plt.ylabel('Power Usage', fontsize='15')
plt.xlabel('Time', fontsize='15')
plt.plot(df1['kitchen_outlets_7'])
from sklearn.svm import OneClassSVM
from numpy import quantile, where, random
x = np.array(df1['kitchen_outlets_7']).reshape(-1, 1)
time = np.array(df1['kitchen_outlets_7'].index)
svm = OneClassSVM(kernel='rbf', gamma=0.001, nu=0.01)
print(svm)
svm.fit(x)
pred = svm.predict(x)
anom_index = where(pred==-1)
values = x[anom_index]
# Plot the prediction
plt.figure(figsize=(18,8))
plt.title('kitchen_outlets_7', fontsize='15')
plt.ylabel('Power Usage', fontsize='15')
plt.xlabel('Time', fontsize='15')
plt.plot(time, x)
plt.scatter(np.array(time)[anom_index], values, color='r')
plt.show() | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
Hands-on Exercise Please try to implement the SVM for classification of MNIST hand written digits dataset. Remember that different hyperparameters can have affect the results. 1. Prepare data: Load the MNIST dataset using `load_digits` from `sklearn.datasets`2. Prepare the tool: load `svm` from `sklearn`3. Split the data into training set and test set: use 70% for training and the remaining for testing, get help from `train_test_split` from `sklearn.model_selection`4. Select the evaluation metric to evaluate the classification result5. Try SVM with different settings and save the accuracy score in a dictionary with key being `kernel name_C` * $C = [0.001, 0.1, 0.5, 1, 10, 100]$ (Regularization parameter. The strength of the regularization is inversely proportional to C. Must be strictly positive. The penalty is a squared l2 penalty.) * kernel = \{‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’\} (Specifies the kernel type to be used in the algorithm.) 6. Visualize the first 4 results in the test set using polynomial kernel with $C = 0.1$. | from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
# loading data
from sklearn.datasets import load_digits
data = load_digits()
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=17 #test_size controls the proportion of test data in the whole data
)
n_samples = len(data.images)
n_train = X_train.shape[0]
print(n_samples, n_train)
# Use loop to do the cross-validation
C_list = [0.001, 0.1, 0.5, 1, 10, 100]
kernel_list = ['linear', 'poly', 'rbf', 'sigmoid']
accuracy_score_dict = dict()
for cc in C_list:
for kern in kernel_list:
acc_i = []
for i in np.arange(5):
if i == 0:
train_idx = np.arange(0, n_train) >= (i+1) * 251
else:
train_idx = (np.arange(0, n_train) < i * 251) + (np.arange(0, n_train) >= (i+1) * 251)
train_idx = train_idx > 0
val_idx = (np.arange(0, n_train) >= i * 251) * (np.arange(0, n_train) < (i+1) * 251)
clf_svm = SVC(C = cc, kernel = kern)
clf_svm.fit(X_train[train_idx, :], y_train[train_idx])
svm_pred = clf_svm.predict(X_train[val_idx, :])
acc_i.append(accuracy_score(y_train[val_idx], svm_pred))
accuracy_score_dict[kern + '_' + str(cc)] = np.mean(acc_i)
accuracy_score_dict
clf_svm = SVC(C = 1, kernel = 'poly')
clf_svm.fit(X_train, y_train)
svm_pred = clf_svm.predict(X_test)
_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))
for ax, image, prediction in zip(axes, X_test, svm_pred):
ax.set_axis_off()
image = image.reshape(8, 8)
ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
ax.set_title(f'Prediction: {prediction}')
# Use GridSearchCV
params_grid = {
'C': [0.001, 0.1, 0.5, 1, 10, 100],
'kernel': ['linear', 'poly', 'rbf', 'sigmoid']
}
grid = GridSearchCV(SVC(), params_grid, cv = 5, scoring = 'accuracy')
grid.fit(X_train, y_train)
selsvc = grid.best_estimator_
svm_pred = selsvc.predict(X_test)
print(grid.best_estimator_)
_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))
for ax, image, prediction in zip(axes, X_test, svm_pred):
ax.set_axis_off()
image = image.reshape(8, 8)
ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
ax.set_title(f'Prediction: {prediction}') | SVC(C=1, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma='scale', kernel='poly',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False)
| MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
Reference* [An Idiot's guide to Support vector machines - MIT](https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwiRtuO39ervAhXVK80KHTjLDGIQFjAMegQIERAD&url=http%3A%2F%2Fweb.mit.edu%2F6.034%2Fwwwbob%2Fsvm-notes-long-08.pdf&usg=AOvVaw3_uFIYSaBhhk_23fPFso52)* [Support Vector Machine — Simply Explained](https://towardsdatascience.com/support-vector-machine-simply-explained-fee28eba5496)* [Understaing support vector machine algorithm with example](https://www.analyticsvidhya.com/blog/2017/09/understaing-support-vector-machine-example-code/) Questions? | Image(url= "https://mirrors.creativecommons.org/presskit/buttons/88x31/png/by-nc-sa.png", width=100) | _____no_output_____ | MIT | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 |
Identify face mesh landmarks> Identify a subset of face mesh landmarks. How can we identify all the landmarks around the mouth?We could use [mesh_map.jpg](https://github.com/tensorflow/tfjs-models/blob/master/facemesh/mesh_map.jpg) and type out IDs of all the landmarks we're interested in but ... that'll take a while and will be hard to do without making any mistakes.How about we,- specify just 4 landmarks - to specify the left, top, right and bottom of a bounding box- then find all other landmarks that are in this bounding box? | from expoco.core import *
import numpy as np
import cv2, time, math
import win32api, win32con
import mediapipe as mp
mp_face_mesh = mp.solutions.face_mesh
from collections import namedtuple
BoundingLandmarks = namedtuple('BoundingLandmarks', 'left, top, right, bottom')
mouth_bounding_landmarks = BoundingLandmarks(57, 164, 287, 18)
class FacePointHelper:
def __init__(self, image_height, image_width, bounding_landmarks):
self.image_height, self.image_width = image_height, image_width
self.bounding_landmarks = bounding_landmarks
self.face_mesh = mp_face_mesh.FaceMesh(max_num_faces=1)
def process(self, image):
self.results = self.face_mesh.process(image) # cv2.cvtColor(image, cv2.COLOR_BGR2RGB) already done
return self.results
def get_bounding_box(self, pixel_coordinates=True):
fn = self._landmark_to_pixel_coordinates if pixel_coordinates else self._landmark_to_x_y
bls = BoundingLandmarks(*[fn(i) for i in self.bounding_landmarks])
return [bls.left[0], bls.top[1]], [bls.right[0], bls.bottom[1]]
def get_bound_landmarks(self):
result = []
[left, top], [right, bottom] = self.get_bounding_box(False)
for i in range(468): # len(self.results.multi_face_landmarks[0])
landmark = self._landmark_to_x_y(i)
if left <= landmark[0] <= right and top <= landmark[1] <= bottom:
result.append(i)
return result
def _landmark(self, i):
return self.results.multi_face_landmarks[0].landmark[i] # [0] is OK as we're running with max_num_faces=1
def _is_valid_normalized_value(self, value):
return (value > 0 or math.isclose(0, value)) and (value < 1 or math.isclose(1, value))
def _normalized_x_to_pixel(self, value):
return math.floor(value * self.image_width)
def _normalized_y_to_pixel(self, value):
return math.floor(value * self.image_height)
def _landmark_to_x_y(self, landmark):
if isinstance(landmark, int):
landmark = self._landmark(landmark)
if not (self._is_valid_normalized_value(landmark.x) and self._is_valid_normalized_value(landmark.y)):
print(f'WARNING: {landmark.x} or {landmark.y} is not a valid normalized value')
return landmark.x, landmark.y
def _landmark_to_pixel_coordinates(self, landmark):
x, y = self._landmark_to_x_y(landmark)
return self._normalized_x_to_pixel(x), self._normalized_y_to_pixel(y)
def annotate_image(face_point_helper, image):
if not face_point_helper.results.multi_face_landmarks:
return image
image = cv2.rectangle(image, *face_point_helper.get_bounding_box(), (130, 0, 130))
for i in face_point_helper.get_bound_landmarks():
point = face_point_helper._landmark_to_pixel_coordinates(i)
image = cv2.circle(image, point, radius=1, color=(100,0,0), thickness=-1)
return image | _____no_output_____ | Apache-2.0 | 10a_viseme_tabular_identify_landmarks.ipynb | pete88b/expoco |
run the following cell to see the bounding box and the landmarks it encloses.press `ESC` to print all landmarks enclosed by the bounding box and stop capture | try: video_capture.release()
except: pass
video_capture = cv2.VideoCapture(0)
face_mesh = mp_face_mesh.FaceMesh(max_num_faces=1)
for vk in [win32con.VK_ESCAPE, ord('D')]: win32api.GetAsyncKeyState(vk)
retval, image = video_capture.read()
face_point_helper = FacePointHelper(*image.shape[:2], mouth_bounding_landmarks)
image_display_helper = ImageDisplayHelper(cv2.flip(image, 1), 'expoco: Dry Run')
while True:
retval, image = video_capture.read()
image = cv2.flip(image, 1)
results = face_point_helper.process(cv2.cvtColor(image, cv2.COLOR_BGR2RGB))
image_display_helper.show(annotate_image(face_point_helper, image))
if win32api.GetAsyncKeyState(win32con.VK_ESCAPE):
print(face_point_helper.get_bound_landmarks())
video_capture.release()
break
time.sleep(.05) | _____no_output_____ | Apache-2.0 | 10a_viseme_tabular_identify_landmarks.ipynb | pete88b/expoco |
General plotting tip: You can get pretty named colors from https://python-graph-gallery.com/196-select-one-color-with-matplotlib/ and unnamed colors from https://htmlcolorcodes.com/. | #Load data
#UPK 535
ra535,dec535,p535,pra535,pdec535,rv535a,G535,B535,R535,spt535,d535,binf535=opendat2(ddir,'UPK535_combined.dat',['#ra', 'dec', 'p','pra','pdec', 'rv', 'G', 'B', 'R', 'spt','d','binaryflag'])
p535err,pra535err,pdec535err,rv535aerr,G535err,B535err,R535err,spt535err,d535perr,d535merr=opendat2(ddir,'UPK535_combined.dat',['perr','praerr','pdecerr', 'rverr', 'Gerr', 'Berr', 'Rerr', 'spterr','dp','dm'])
#clean rv for simulation: already threw out my rverr>10 but gotta check new Gaia rvs.
rv535aa=[rv535a[i] if rv535aerr[i]<10. else np.float('nan') for i in range(len(ra535))]
rv535aaerr=[rv535aerr[i] if rv535aerr[i]<10. else np.float('nan') for i in range(len(ra535))]
#neutralize all binaries.
bindev=5.
rv535med=np.nanmedian(rv535aa)
binn535=[binf535[i] if binf535[i]!='nan' else 'SB1?' if abs(rv535aa[i]-rv535med)>bindev else 'nan' for i in range(len(ra535))]
#calculate cluster rv from nonbinary rvs and replace all nans with cluster rv.
rv535aaa=[rv535aa[i] if 'SB' not in binn535[i] else np.float('nan') for i in range(len(ra535))]
rv535aaaerr=[rv535aaerr[i] if 'SB' not in binn535[i] else np.float('nan') for i in range(len(ra535))]
rvcl535,rvcl535err=erm(rv535aaa,rv535aaaerr)[0],np.nanstd(rv535aaa)
print('UPK535:',rvcl535,'+/-',rvcl535err)
#clean rv:
rv535=[rvcl535 if np.isnan(r) else r for r in rv535aaa]
rv535err=[rvcl535err if np.isnan(rv535aaa[i]) else rv535aaaerr[i] for i in range(len(ra535))]
#Theia 120
ra120,dec120,p120,pra120,pdec120,rv120a,G120,B120,R120,spt120,d120,binf120=opendat2(ddir,'Theia120_combined.dat',['#ra', 'dec', 'p','pra','pdec', 'rv', 'G', 'B', 'R', 'spt','d','binaryflag'])
p120err,pra120err,pdec120err,rv120aerr,G120err,B120err,R120err,spt120err,d120perr,d120merr=opendat2(ddir,'Theia120_combined.dat',['perr','praerr','pdecerr', 'rverr', 'Gerr', 'Berr', 'Rerr', 'spterr','dp','dm'])
BR120=np.array(B120)-np.array(R120)
BR120err=np.sqrt(np.array(B120)**2.+np.array(R120)**2.)
#clean rv for simulation: already threw out my rverr>10 but gotta check new Gaia rvs.
rv120aa=[rv120a[i] if rv120aerr[i]<10. else np.float('nan') for i in range(len(ra120))]
rv120aaerr=[rv120aerr[i] if rv120aerr[i]<10. else np.float('nan') for i in range(len(ra120))]
#neutralize all binaries.
bindev=5.
rv120med=np.nanmedian(rv120aa)
binn120=[binf120[i] if binf120[i]!='nan' else 'SB1?' if abs(rv120aa[i]-rv120med)>bindev else 'nan' for i in range(len(ra120))]
#calculate cluster rv from nonbinary rvs and replace all nans with cluster rv.
rv120aaa=[rv120aa[i] if 'SB' not in binn120[i] else np.float('nan') for i in range(len(ra120))]
rv120aaaerr=[rv120aaerr[i] if 'SB' not in binn120[i] else np.float('nan') for i in range(len(ra120))]
rvcl120,rvcl120err=erm(rv120aaa,rv120aaaerr)[0],np.nanstd(rv120aaa)
print('Theia120:',rvcl120,'+/-',rvcl120err)
#clean rv:
rv120=[rvcl120 if np.isnan(r) else r for r in rv120aaa]
rv120err=[rvcl120err if np.isnan(rv120aaa[i]) else rv120aaaerr[i] for i in range(len(ra120))]
plt.rcParams.update({'font.size':22,'lines.linewidth':4, 'font.family':'serif','mathtext.fontset':'dejavuserif'})
# Histogram
f,((a11,a12),(a21,a22))=plt.subplots(2,2,figsize=(12,12),gridspec_kw = {'wspace':0.24,'hspace':0.28})
f.add_subplot(111, frameon=False,xticks=[],yticks=[]) #for tight layout
hpars1=[d535,[a for a in rv535aaa if np.isnan(a)==False],pra535,pdec535]
hpars2=[d120,[a for a in rv120aaa if np.isnan(a)==False],pra120,pdec120]
mins=[290,5,-15,1]
maxs=[380,22,-10,14]
ints=[10,1,0.5,1]
axs=[a11,a21,a12,a22]
labs=['Distance (pc)','$v_r$ (km s$^{-1}$)','$\mu_{\\alpha}$ (mas yr$^{-1}$)','$\mu_{\delta}$ (mas yr$^{-1}$)']
for i in range(4):
par1=hpars1[i]
par2=hpars2[i]
minn=mins[i]
maxx=maxs[i]
intt=ints[i]
ax=axs[i]
beans=np.arange(minn,maxx,intt)
h535,w535=np.histogram(par1,beans)
h120,w120=np.histogram(par2,beans)
#plt.figure(figsize=(10,10))
print(len(h535),len(w535))
ax.bar(w120[:-1],h120/len(par2),color='#008FFF',width=intt)
ax.bar(w535[:-1],h535/len(par1),color='blue',width=intt)
overlap=[np.min([h120[k]/len(par2),h535[k]/len(par1)]) for k in range(len(h120))]
ax.bar(w120[:-1],overlap,color='#00068D',width=intt)
ax.set_xlabel(labs[i])
plt.ylabel('Normalized Frequency\n\n')
plt.savefig(pdir+'CC_Histograms.png',bbox_inches='tight') | 8 9
16 17
9 10
12 13
| BSD-3-Clause | 2020_Workshop/Alex_Python/PrettyPlot.ipynb | imedan/AstroPAL_Coding_Workshop |
Color-Mapping! This basically gives you a 3rd dimension. Go wild! Color maps can be chosen from here: https://matplotlib.org/3.1.1/gallery/color/colormap_reference.html I personally like rainbow, warm, cool, and RdYlBu. You can also use lists for sizes to give different sizes to each data point. :) That's yet another dimension you can use! Be warned that the color bar can be a wily creature when you're adding it to plots with subplots. I often actually create a separate axis for it when that happens and calibrate its size by hand. D: With one plot though, it's easy. :) | #3-D position with ra, dec, AND distance:
plt.rcParams.update({'font.size':22,'lines.linewidth':4, 'font.family':'serif','mathtext.fontset':'dejavuserif'})
f=plt.figure(figsize=(10,10))
cm=plt.scatter(ra535,dec535,c=d535,cmap='rainbow',s=[d if d<300 else 300 for d in 10000./(d535-300)])
cbar = f.colorbar(cm,label='\nDistance (pc)')
plt.xlabel('RA ($^{o}$)')
plt.ylabel('Dec ($^{o}$)')
| _____no_output_____ | BSD-3-Clause | 2020_Workshop/Alex_Python/PrettyPlot.ipynb | imedan/AstroPAL_Coding_Workshop |
Lesson 04: Classification Performance ROCs- evaluating and comparing trained models is of extreme importance when deciding in favor/against + model architectures + hyperparameter sets - evaluating performance or quality of prediction is performed with a myriad of tests, figure-of-merits and even statistical hypothesis testing- in the following, the rather popular "Receiver Operating Characteristic" curve (spoken ROC curve)- the ROC was invented in WWII by radar engineers when seeking to detect enemy vessels and comparing different devices/techniques preface- two main ingredients to ROC: + TPR = True Positive Rate + FPR = False Positive Rate 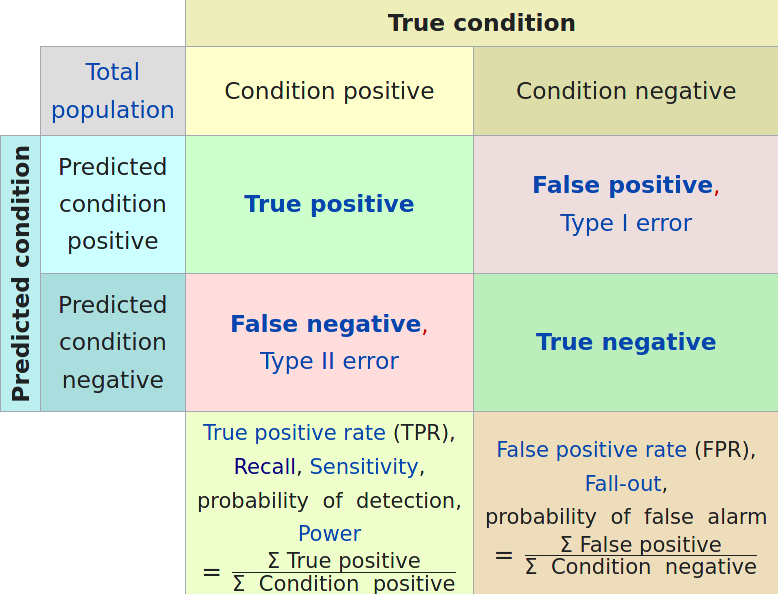- $TPR = \frac{TP}{TP+FN}$ also known as `recall`, always within $[0,1]$- $FPR = \frac{FP}{FP+TN}$ also known as `fall-out`, always within $[0,1]$ DataFor the following, I will rely (again) on the Palmer penguin dataset obtained from [this repo](https://github.com/allisonhorst/palmerpenguins). To quote the repo:> Data were collected and made available by [Dr. Kristen Gorman](https://www.uaf.edu/cfos/people/faculty/detail/kristen-gorman.php)> and the [Palmer Station, Antarctica LTER](https://pal.lternet.edu/), a member of the [Long Term Ecological Research Network](https://lternet.edu/). | import pandas as pd
import numpy as np
df = pd.read_csv("https://raw.githubusercontent.com/allisonhorst/palmerpenguins/master/inst/extdata/penguins.csv")
#let's remove the rows with NaN values
df = df[ df.bill_length_mm.notnull() ]
#convert species column to
df[["species_"]] = df[["species"]].astype("category")
print(df.shape)
print((df.species_.cat.codes < 1).shape)
#create binary column
df["is_adelie"] = (df.species_.cat.codes < 1).astype(np.int8)
print(df.head())
import matplotlib.pyplot as plt
plt.style.use('dark_background')
import seaborn as sns
print(f'seaborn version: {sns.__version__}')
from sklearn.neighbors import KNeighborsClassifier as knn
from sklearn.model_selection import train_test_split
kmeans = knn(n_neighbors=5)
#this time we train the knn algorithm, i.e. an unsupervised method is used in a supervised fashion
#prepare the data
X = np.stack((df.bill_length_mm, df.flipper_length_mm), axis=-1)
y = df.is_adelie
print(X.shape)
print(y.shape)
X_train, X_test, y_train, y_test = train_test_split(X, y,
test_size = .15,
random_state = 20210303)
print(X_train.shape)
print(y_train.shape)
print(X_test.shape)
print(y_test.shape)
kmeans = kmeans.fit(X_train, y_train)
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
y_test_hat = kmeans.predict(X_test)
cm = confusion_matrix( y_test, y_test_hat )
print(cm)
from sklearn.metrics import ConfusionMatrixDisplay
disp = ConfusionMatrixDisplay(confusion_matrix=cm)
disp.plot()
print(int(True)) | 1
| MIT | source/lesson04/script.ipynb | psteinb/deeplearning540.github.io |
Starting to ROC- let's take 4 samples of different size from our test set (as if we would conduct 4 experiments) | n_experiments = 4
X_test_exp = np.split(X_test[:32,...],n_experiments,axis=0)
y_test_exp = np.split(y_test.values[:32,...],n_experiments,axis=0)
print(X_test_exp[0].shape)
print(y_test_exp[0].shape)
y_test_exp
y_test_hat = kmeans.predict(X_test)
y_test_hat_exp = np.split(y_test_hat[:32,...],n_experiments,axis=0)
#let's compute tpr and fpr for each
from sklearn.metrics import recall_score as tpr
def fpr(y_true, y_pred):
""" compute the false positive rate using the confusion_matrix"""
cm = confusion_matrix(y_true, y_pred)
assert cm.shape == (2,2), f"{y_true.shape, y_pred.shape} => {cm,cm.shape}"
cond_negative = cm[:,1].sum()
value = cm[0,1] / cond_negative
return value
tpr_ = []
fpr_ = []
for i in range(len(y_test_exp)):
tpr_.append(tpr(y_test_exp[i], y_test_hat_exp[i]))
fpr_.append(fpr(y_test_exp[i], y_test_hat_exp[i]))
print(tpr_)
print(fpr_)
f, ax = plt.subplots(1)
ax.plot(fpr_, tpr_, 'ro', markersize=10)
ax.set_xlabel('False Positive Rate')
ax.set_ylabel('True Positive Rate')
ax.set_xlim(0,1)
ax.set_ylim(0,1)
| _____no_output_____ | MIT | source/lesson04/script.ipynb | psteinb/deeplearning540.github.io |
But how to get from single entries to a full curve?- in our case, we can employ the positive class prediction probabilities- for KNN, this is given by the amount of N(true label)/N in the neighborhood around a query point | kmeans.predict_proba(X_test[:10])
| _____no_output_____ | MIT | source/lesson04/script.ipynb | psteinb/deeplearning540.github.io |
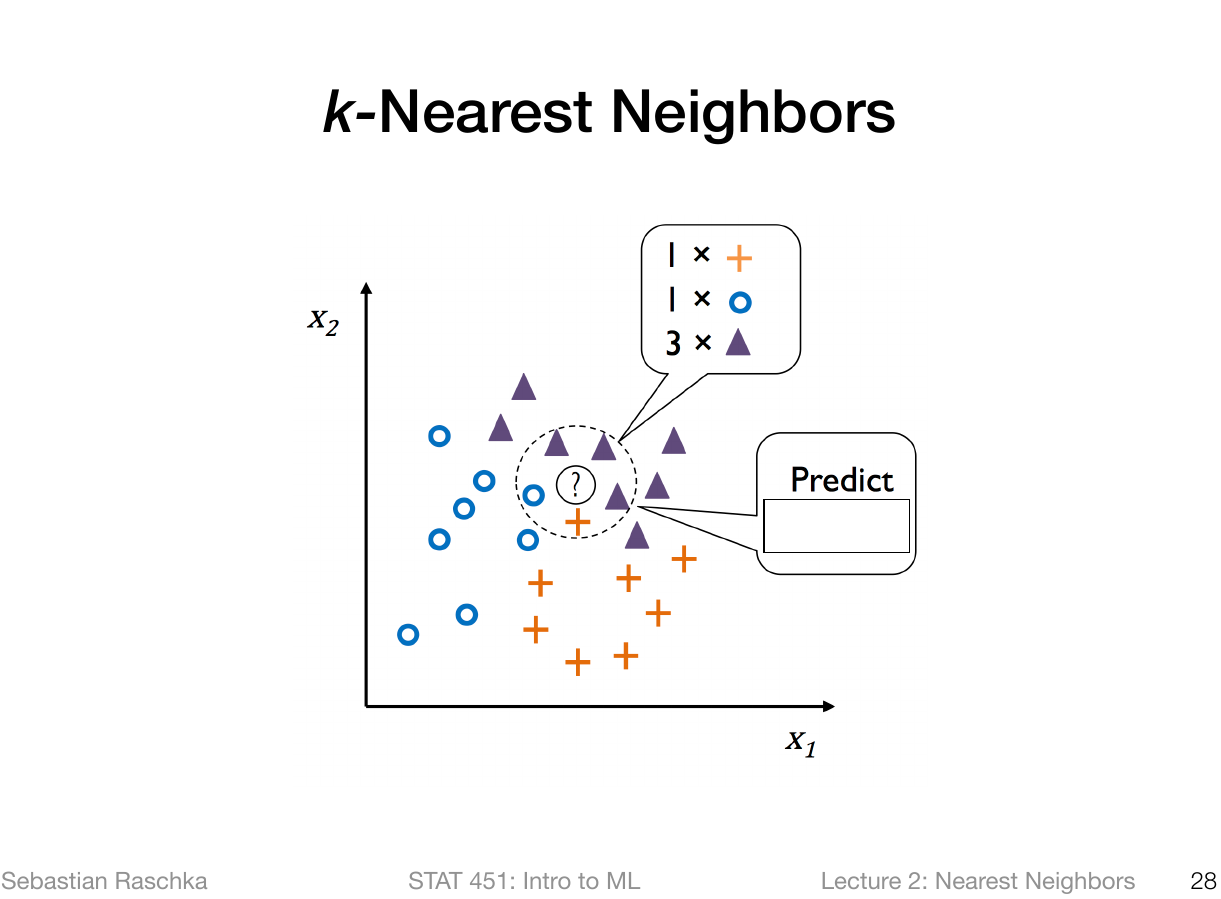- demonstrates how kNN classifyer is similar to `RandomForests`, `SVM`, ... : + spacial interpretation of the class prediction probability + the higher the probability for a sample, the more likely the sample belongs to `Adelie` in our case (i.e. the positive class in a binary classification setup) - relating this again to 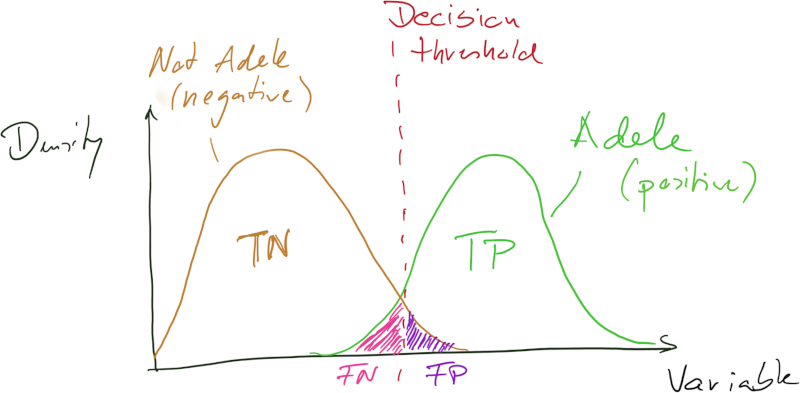the decision threshold for a `5`-neighborhood with a binary classification task is `0.6`, i.e. 3 of 5 neighbors have the positive class (then our query point will get the positive class assigned)- knowing these positive class prediction probabilities, I can now draw an envelope that gives me the ROC from the test set as with these probabilites and the theoretical threshold, we can compute FPR and TPR | from sklearn.metrics import roc_curve
probs = kmeans.predict_proba(X_test)
pos_pred_probs = probs[:,-1]
fpr, tpr, thr = roc_curve(y_test, pos_pred_probs)
print('false positive rate\n',fpr)
print('true positive rate\n',tpr)
print('thresholds\n',thr)
from sklearn.metrics import RocCurveDisplay
roc = RocCurveDisplay.from_estimator(kmeans, X_test, y_test)
| _____no_output_____ | MIT | source/lesson04/script.ipynb | psteinb/deeplearning540.github.io |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.