markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
Test out the RNN modelIt's always a good idea to run a few simple checks on our model to see that it behaves as expected. First, we can use the `Model.summary` function to print out a summary of our model's internal workings. Here we can check the layers in the model, the shape of the output of each of the layers, the batch size, etc. | model.summary() | Model: "sequential_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
embedding_1 (Embedding) (32, None, 256) 21248
_________________________________________________________________
lstm_1 (LSTM) (32, None, 1024) 5246976
_________________________________________________________________
dense_1 (Dense) (32, None, 83) 85075
=================================================================
Total params: 5,353,299
Trainable params: 5,353,299
Non-trainable params: 0
_________________________________________________________________
| MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
We can also quickly check the dimensionality of our output, using a sequence length of 100. Note that the model can be run on inputs of any length. | x, y = get_batch(vectorized_songs, seq_length=100, batch_size=32)
pred = model(x)
print("Input shape: ", x.shape, " # (batch_size, sequence_length)")
print("Prediction shape: ", pred.shape, "# (batch_size, sequence_length, vocab_size)")
x
y
pred | _____no_output_____ | MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
Predictions from the untrained modelLet's take a look at what our untrained model is predicting.To get actual predictions from the model, we sample from the output distribution, which is defined by a `softmax` over our character vocabulary. This will give us actual character indices. This means we are using a [categorical distribution](https://en.wikipedia.org/wiki/Categorical_distribution) to sample over the example prediction. This gives a prediction of the next character (specifically its index) at each timestep.Note here that we sample from this probability distribution, as opposed to simply taking the `argmax`, which can cause the model to get stuck in a loop.Let's try this sampling out for the first example in the batch. | # for batch 0, input sequence size: 100, so output sequence size: 100
sampled_indices = tf.random.categorical(pred[0], num_samples=1)
sampled_indices = tf.squeeze(sampled_indices,axis=-1).numpy()
sampled_indices
x[0] | _____no_output_____ | MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
We can now decode these to see the text predicted by the untrained model: | print("Input: \n", repr("".join(idx2char[x[0]])))
print()
print("Next Char Predictions: \n", repr("".join(idx2char[sampled_indices]))) | Input:
'AG|F2D DED|FEF GFG|!\nA3 cAG|AGA cde|fed cAG|Ad^c d2:|!\ne|f2d d^cd|f2a agf|e2c cBc|e2f gfe|!\nf2g agf|'
Next Char Predictions:
'^VIQXjybPEk-^_G/>#T9ZLYJ"CkYXBE\nUUDBU<AwqWFDa(]X09T)0GpF(5Q"k|\nKHU1fhFeuSM)s"i9F8hjYcj[Dl5\'KQzecQkKs'
| MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
As you can see, the text predicted by the untrained model is pretty nonsensical! How can we do better? We can train the network! 2.5 Training the model: loss and training operationsNow it's time to train the model!At this point, we can think of our next character prediction problem as a standard classification problem. Given the previous state of the RNN, as well as the input at a given time step, we want to predict the class of the next character -- that is, to actually predict the next character. To train our model on this classification task, we can use a form of the `crossentropy` loss (negative log likelihood loss). Specifically, we will use the [`sparse_categorical_crossentropy`](https://www.tensorflow.org/api_docs/python/tf/keras/losses/sparse_categorical_crossentropy) loss, as it utilizes integer targets for categorical classification tasks. We will want to compute the loss using the true targets -- the `labels` -- and the predicted targets -- the `logits`.Let's first compute the loss using our example predictions from the untrained model: | ### Defining the loss function ###
'''TODO: define the loss function to compute and return the loss between
the true labels and predictions (logits). Set the argument from_logits=True.'''
def compute_loss(labels, logits):
loss = tf.keras.losses.sparse_categorical_crossentropy(labels, logits, from_logits=True) # TODO
return loss
'''TODO: compute the loss using the true next characters from the example batch
and the predictions from the untrained model several cells above'''
example_batch_loss = compute_loss(y, pred) # TODO
print("Prediction shape: ", pred.shape, " # (batch_size, sequence_length, vocab_size)")
print("scalar_loss: ", example_batch_loss.numpy().mean())
y.shape
pred.shape
example_batch_loss.shape | _____no_output_____ | MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
Let's start by defining some hyperparameters for training the model. To start, we have provided some reasonable values for some of the parameters. It is up to you to use what we've learned in class to help optimize the parameter selection here! | ### Hyperparameter setting and optimization ###
# Optimization parameters:
num_training_iterations = 2000 # Increase this to train longer
batch_size = 4 # Experiment between 1 and 64
seq_length = 100 # Experiment between 50 and 500
learning_rate = 5e-3 # Experiment between 1e-5 and 1e-1
# Model parameters:
vocab_size = len(vocab)
embedding_dim = 256
rnn_units = 1024 # Experiment between 1 and 2048
# Checkpoint location:
checkpoint_dir = './training_checkpoints'
checkpoint_prefix = os.path.join(checkpoint_dir, "my_ckpt") | _____no_output_____ | MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
Now, we are ready to define our training operation -- the optimizer and duration of training -- and use this function to train the model. You will experiment with the choice of optimizer and the duration for which you train your models, and see how these changes affect the network's output. Some optimizers you may like to try are [`Adam`](https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/Adam?version=stable) and [`Adagrad`](https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/Adagrad?version=stable).First, we will instantiate a new model and an optimizer. Then, we will use the [`tf.GradientTape`](https://www.tensorflow.org/api_docs/python/tf/GradientTape) method to perform the backpropagation operations. We will also generate a print-out of the model's progress through training, which will help us easily visualize whether or not we are minimizing the loss. | ### Define optimizer and training operation ###
'''TODO: instantiate a new model for training using the `build_model`
function and the hyperparameters created above.'''
#model = build_model('''TODO: arguments''')
model = build_model(vocab_size, embedding_dim=embedding_dim, rnn_units=rnn_units, batch_size=batch_size)
'''TODO: instantiate an optimizer with its learning rate.
Checkout the tensorflow website for a list of supported optimizers.
https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/
Try using the Adam optimizer to start.'''
optimizer = tf.keras.optimizers.Adam(learning_rate=learning_rate)
@tf.function
def train_step(x, y):
# Use tf.GradientTape()
with tf.GradientTape() as tape:
'''TODO: feed the current input into the model and generate predictions'''
y_hat = model(x)
'''TODO: compute the loss!'''
loss = compute_loss(y, y_hat)
# Now, compute the gradients
'''TODO: complete the function call for gradient computation.
Remember that we want the gradient of the loss with respect all
of the model parameters.
HINT: use `model.trainable_variables` to get a list of all model
parameters.'''
grads = tape.gradient(loss, model.trainable_variables)
# Apply the gradients to the optimizer so it can update the model accordingly
optimizer.apply_gradients(zip(grads, model.trainable_variables))
return loss
##################
# Begin training!#
##################
history = []
plotter = mdl.util.PeriodicPlotter(sec=2, xlabel='Iterations', ylabel='Loss')
if hasattr(tqdm, '_instances'): tqdm._instances.clear() # clear if it exists
for iter in tqdm(range(num_training_iterations)):
# Grab a batch and propagate it through the network
x_batch, y_batch = get_batch(vectorized_songs, seq_length, batch_size)
loss = train_step(x_batch, y_batch)
# Update the progress bar
history.append(loss.numpy().mean())
plotter.plot(history)
# Update the model with the changed weights!
if iter % 100 == 0:
model.save_weights(checkpoint_prefix)
# Save the trained model and the weights
model.save_weights(checkpoint_prefix)
| _____no_output_____ | MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
2.6 Generate music using the RNN modelNow, we can use our trained RNN model to generate some music! When generating music, we'll have to feed the model some sort of seed to get it started (because it can't predict anything without something to start with!).Once we have a generated seed, we can then iteratively predict each successive character (remember, we are using the ABC representation for our music) using our trained RNN. More specifically, recall that our RNN outputs a `softmax` over possible successive characters. For inference, we iteratively sample from these distributions, and then use our samples to encode a generated song in the ABC format.Then, all we have to do is write it to a file and listen! Restore the latest checkpointTo keep this inference step simple, we will use a batch size of 1. Because of how the RNN state is passed from timestep to timestep, the model will only be able to accept a fixed batch size once it is built. To run the model with a different `batch_size`, we'll need to rebuild the model and restore the weights from the latest checkpoint, i.e., the weights after the last checkpoint during training: | '''TODO: Rebuild the model using a batch_size=1'''
model = build_model(vocab_size, embedding_dim=embedding_dim, rnn_units=rnn_units, batch_size=1)
# Restore the model weights for the last checkpoint after training
model.load_weights(tf.train.latest_checkpoint(checkpoint_dir))
model.build(tf.TensorShape([1, None]))
model.summary() | Model: "sequential_3"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
embedding_3 (Embedding) (1, None, 256) 21248
_________________________________________________________________
lstm_3 (LSTM) (1, None, 1024) 5246976
_________________________________________________________________
dense_3 (Dense) (1, None, 83) 85075
=================================================================
Total params: 5,353,299
Trainable params: 5,353,299
Non-trainable params: 0
_________________________________________________________________
| MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
Notice that we have fed in a fixed `batch_size` of 1 for inference. The prediction procedureNow, we're ready to write the code to generate text in the ABC music format:* Initialize a "seed" start string and the RNN state, and set the number of characters we want to generate.* Use the start string and the RNN state to obtain the probability distribution over the next predicted character.* Sample from multinomial distribution to calculate the index of the predicted character. This predicted character is then used as the next input to the model.* At each time step, the updated RNN state is fed back into the model, so that it now has more context in making the next prediction. After predicting the next character, the updated RNN states are again fed back into the model, which is how it learns sequence dependencies in the data, as it gets more information from the previous predictions.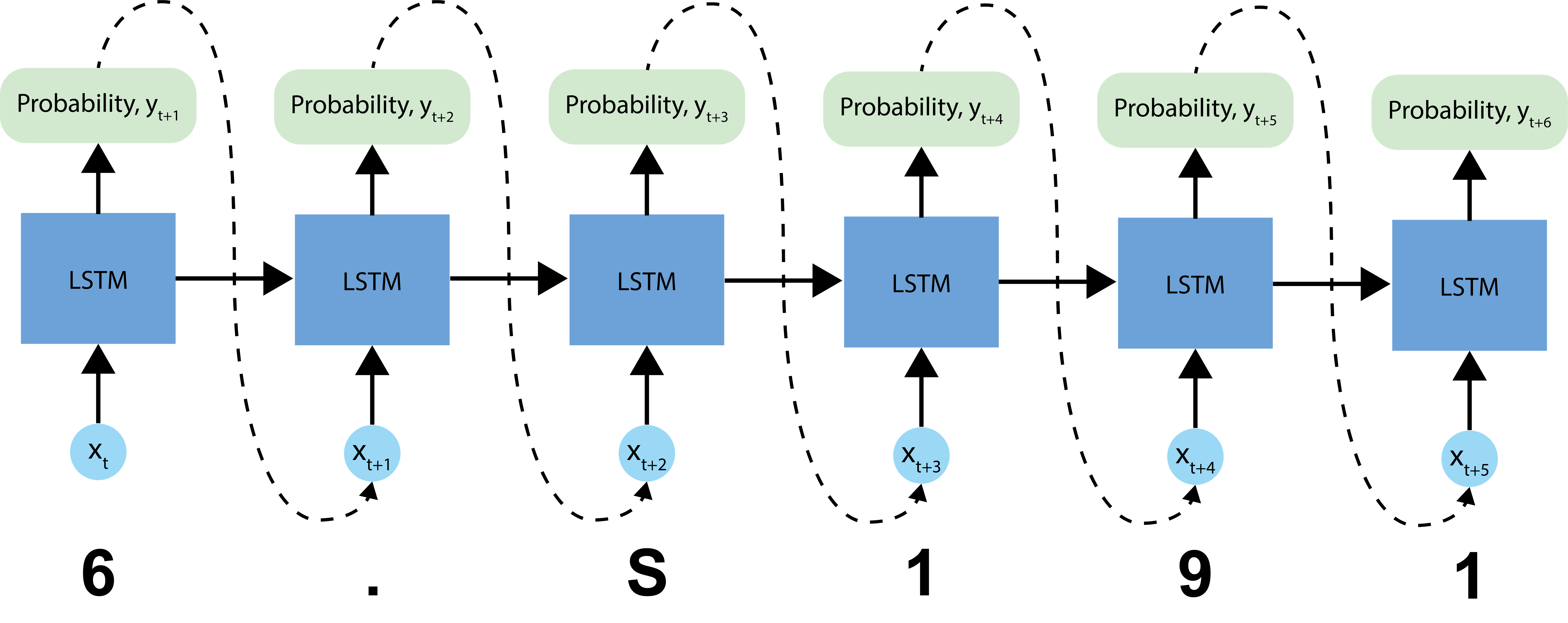Complete and experiment with this code block (as well as some of the aspects of network definition and training!), and see how the model performs. How do songs generated after training with a small number of epochs compare to those generated after a longer duration of training? | ### Prediction of a generated song ###
def generate_text(model, start_string, generation_length=1000):
# Evaluation step (generating ABC text using the learned RNN model)
'''TODO: convert the start string to numbers (vectorize)'''
input_eval = [char2idx[s] for s in start_string] # TODO
# input_eval = ['''TODO''']
input_eval = tf.expand_dims(input_eval, 0)
# Empty string to store our results
text_generated = []
# Here batch size == 1
model.reset_states()
tqdm._instances.clear()
for i in tqdm(range(generation_length)):
'''TODO: evaluate the inputs and generate the next character predictions'''
predictions = model(input_eval)
# predictions = model('''TODO''')
# Remove the batch dimension
predictions = tf.squeeze(predictions, 0)
'''TODO: use a multinomial distribution to sample'''
predicted_id = tf.random.categorical(predictions, num_samples=1)[-1,0].numpy()
# predicted_id = tf.random.categorical('''TODO''', num_samples=1)[-1,0].numpy()
# Pass the prediction along with the previous hidden state
# as the next inputs to the model
input_eval = tf.expand_dims([predicted_id], 0)
'''TODO: add the predicted character to the generated text!'''
# Hint: consider what format the prediction is in vs. the output
text_generated.append(idx2char[predicted_id]) # TODO
# text_generated.append('''TODO''')
return (start_string + ''.join(text_generated))
'''TODO: Use the model and the function defined above to generate ABC format text of length 1000!
As you may notice, ABC files start with "X" - this may be a good start string.'''
generated_text = generate_text(model, start_string="A", generation_length=1000) # TODO
# generated_text = generate_text('''TODO''', start_string="X", generation_length=1000)
generated_text | _____no_output_____ | MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
Play back the generated music!We can now call a function to convert the ABC format text to an audio file, and then play that back to check out our generated music! Try training longer if the resulting song is not long enough, or re-generating the song! | ### Play back generated songs ###
generated_songs = mdl.lab1.extract_song_snippet(generated_text)
for i, song in enumerate(generated_songs):
# Synthesize the waveform from a song
waveform = mdl.lab1.play_song(song)
# If its a valid song (correct syntax), lets play it!
if waveform:
print("Generated song", i)
ipythondisplay.display(waveform)
generated_songs | _____no_output_____ | MIT | lab1/Part2_Music_Generation.ipynb | mukesh5237/introtodeeplearning |
Working with Text Data Types of data represented as strings Example application: Sentiment analysis of movie reviews | ! wget -nc http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz -P data
! tar xzf data/aclImdb_v1.tar.gz --skip-old-files -C data
!tree -dL 2 data/aclImdb
!rm -r data/aclImdb/train/unsup
from sklearn.datasets import load_files
reviews_train = load_files("data/aclImdb/train/")
# load_files returns a bunch, containing training texts and training labels
text_train, y_train = reviews_train.data, reviews_train.target
print("type of text_train: {}".format(type(text_train)))
print("length of text_train: {}".format(len(text_train)))
print("text_train[6]:\n{}".format(text_train[6]))
text_train = [doc.replace(b"<br />", b" ") for doc in text_train]
np.unique(y_train)
print("Samples per class (training): {}".format(np.bincount(y_train)))
reviews_test = load_files("data/aclImdb/test/")
text_test, y_test = reviews_test.data, reviews_test.target
print("Number of documents in test data: {}".format(len(text_test)))
print("Samples per class (test): {}".format(np.bincount(y_test)))
text_test = [doc.replace(b"<br />", b" ") for doc in text_test] | Number of documents in test data: 25000
Samples per class (test): [12500 12500]
| MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Representing text data as Bag of Words  Applying bag-of-words to a toy dataset | bards_words =["The fool doth think he is wise,",
"but the wise man knows himself to be a fool"]
from sklearn.feature_extraction.text import CountVectorizer
vect = CountVectorizer()
vect.fit(bards_words)
print("Vocabulary size: {}".format(len(vect.vocabulary_)))
print("Vocabulary content:\n {}".format(vect.vocabulary_))
bag_of_words = vect.transform(bards_words)
print("bag_of_words: {}".format(repr(bag_of_words)))
print("Dense representation of bag_of_words:\n{}".format(
bag_of_words.toarray())) | Dense representation of bag_of_words:
[[0 0 1 1 1 0 1 0 0 1 1 0 1]
[1 1 0 1 0 1 0 1 1 1 0 1 1]]
| MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Bag-of-word for movie reviews | vect = CountVectorizer().fit(text_train)
X_train = vect.transform(text_train)
print("X_train:\n{}".format(repr(X_train)))
feature_names = vect.get_feature_names()
print("Number of features: {}".format(len(feature_names)))
print("First 20 features:\n{}".format(feature_names[:20]))
print("Features 20010 to 20030:\n{}".format(feature_names[20010:20030]))
print("Every 2000th feature:\n{}".format(feature_names[::2000]))
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
scores = cross_val_score(LogisticRegression(), X_train, y_train, cv=5)
print("Mean cross-validation accuracy: {:.2f}".format(np.mean(scores)))
from sklearn.model_selection import GridSearchCV
param_grid = {'C': [0.001, 0.01, 0.1, 1, 10]}
grid = GridSearchCV(LogisticRegression(), param_grid, cv=5)
grid.fit(X_train, y_train)
print("Best cross-validation score: {:.2f}".format(grid.best_score_))
print("Best parameters: ", grid.best_params_)
X_test = vect.transform(text_test)
print("Test score: {:.2f}".format(grid.score(X_test, y_test)))
vect = CountVectorizer(min_df=5).fit(text_train)
X_train = vect.transform(text_train)
print("X_train with min_df: {}".format(repr(X_train)))
feature_names = vect.get_feature_names()
print("First 50 features:\n{}".format(feature_names[:50]))
print("Features 20010 to 20030:\n{}".format(feature_names[20010:20030]))
print("Every 700th feature:\n{}".format(feature_names[::700]))
grid = GridSearchCV(LogisticRegression(), param_grid, cv=5)
grid.fit(X_train, y_train)
print("Best cross-validation score: {:.2f}".format(grid.best_score_)) | Best cross-validation score: 0.89
| MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Stop-words | from sklearn.feature_extraction.text import ENGLISH_STOP_WORDS
print("Number of stop words: {}".format(len(ENGLISH_STOP_WORDS)))
print("Every 10th stopword:\n{}".format(list(ENGLISH_STOP_WORDS)[::10]))
# Specifying stop_words="english" uses the built-in list.
# We could also augment it and pass our own.
vect = CountVectorizer(min_df=5, stop_words="english").fit(text_train)
X_train = vect.transform(text_train)
print("X_train with stop words:\n{}".format(repr(X_train)))
grid = GridSearchCV(LogisticRegression(), param_grid, cv=5)
grid.fit(X_train, y_train)
print("Best cross-validation score: {:.2f}".format(grid.best_score_)) | Best cross-validation score: 0.88
| MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Rescaling the Data with tf-idf\begin{equation*}\text{tfidf}(w, d) = \text{tf} \log\big(\frac{N + 1}{N_w + 1}\big) + 1\end{equation*} | from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.pipeline import make_pipeline
pipe = make_pipeline(TfidfVectorizer(min_df=5, norm=None),
LogisticRegression())
param_grid = {'logisticregression__C': [0.001, 0.01, 0.1, 1, 10]}
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(text_train, y_train)
print("Best cross-validation score: {:.2f}".format(grid.best_score_))
vectorizer = grid.best_estimator_.named_steps["tfidfvectorizer"]
# transform the training dataset:
X_train = vectorizer.transform(text_train)
# find maximum value for each of the features over dataset:
max_value = X_train.max(axis=0).toarray().ravel()
sorted_by_tfidf = max_value.argsort()
# get feature names
feature_names = np.array(vectorizer.get_feature_names())
print("Features with lowest tfidf:\n{}".format(
feature_names[sorted_by_tfidf[:20]]))
print("Features with highest tfidf: \n{}".format(
feature_names[sorted_by_tfidf[-20:]]))
sorted_by_idf = np.argsort(vectorizer.idf_)
print("Features with lowest idf:\n{}".format(
feature_names[sorted_by_idf[:100]])) | Features with lowest idf:
['the' 'and' 'of' 'to' 'this' 'is' 'it' 'in' 'that' 'but' 'for' 'with'
'was' 'as' 'on' 'movie' 'not' 'have' 'one' 'be' 'film' 'are' 'you' 'all'
'at' 'an' 'by' 'so' 'from' 'like' 'who' 'they' 'there' 'if' 'his' 'out'
'just' 'about' 'he' 'or' 'has' 'what' 'some' 'good' 'can' 'more' 'when'
'time' 'up' 'very' 'even' 'only' 'no' 'would' 'my' 'see' 'really' 'story'
'which' 'well' 'had' 'me' 'than' 'much' 'their' 'get' 'were' 'other'
'been' 'do' 'most' 'don' 'her' 'also' 'into' 'first' 'made' 'how' 'great'
'because' 'will' 'people' 'make' 'way' 'could' 'we' 'bad' 'after' 'any'
'too' 'then' 'them' 'she' 'watch' 'think' 'acting' 'movies' 'seen' 'its'
'him']
| MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Investigating model coefficients | mglearn.tools.visualize_coefficients(
grid.best_estimator_.named_steps["logisticregression"].coef_,
feature_names, n_top_features=40) | _____no_output_____ | MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Bag of words with more than one word (n-grams) | print("bards_words:\n{}".format(bards_words))
cv = CountVectorizer(ngram_range=(1, 1)).fit(bards_words)
print("Vocabulary size: {}".format(len(cv.vocabulary_)))
print("Vocabulary:\n{}".format(cv.get_feature_names()))
cv = CountVectorizer(ngram_range=(2, 2)).fit(bards_words)
print("Vocabulary size: {}".format(len(cv.vocabulary_)))
print("Vocabulary:\n{}".format(cv.get_feature_names()))
print("Transformed data (dense):\n{}".format(cv.transform(bards_words).toarray()))
cv = CountVectorizer(ngram_range=(1, 3)).fit(bards_words)
print("Vocabulary size: {}".format(len(cv.vocabulary_)))
print("Vocabulary:\n{}".format(cv.get_feature_names()))
pipe = make_pipeline(TfidfVectorizer(min_df=5), LogisticRegression())
# running the grid-search takes a long time because of the
# relatively large grid and the inclusion of trigrams
param_grid = {'logisticregression__C': [0.001, 0.01, 0.1, 1, 10, 100],
"tfidfvectorizer__ngram_range": [(1, 1), (1, 2), (1, 3)]}
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(text_train, y_train)
print("Best cross-validation score: {:.2f}".format(grid.best_score_))
print("Best parameters:\n{}".format(grid.best_params_))
# extract scores from grid_search
scores = grid.cv_results_['mean_test_score'].reshape(-1, 3).T
# visualize heat map
heatmap = mglearn.tools.heatmap(
scores, xlabel="C", ylabel="ngram_range", cmap="viridis", fmt="%.3f",
xticklabels=param_grid['logisticregression__C'],
yticklabels=param_grid['tfidfvectorizer__ngram_range'])
plt.colorbar(heatmap)
# extract feature names and coefficients
vect = grid.best_estimator_.named_steps['tfidfvectorizer']
feature_names = np.array(vect.get_feature_names())
coef = grid.best_estimator_.named_steps['logisticregression'].coef_
mglearn.tools.visualize_coefficients(coef, feature_names, n_top_features=40)
plt.ylim(-22, 22)
# find 3-gram features
mask = np.array([len(feature.split(" ")) for feature in feature_names]) == 3
# visualize only 3-gram features
mglearn.tools.visualize_coefficients(coef.ravel()[mask],
feature_names[mask], n_top_features=40)
plt.ylim(-22, 22) | _____no_output_____ | MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Advanced tokenization, stemming and lemmatization | import spacy
import nltk
# load spacy's English-language models
en_nlp = spacy.load('en')
# instantiate nltk's Porter stemmer
stemmer = nltk.stem.PorterStemmer()
# define function to compare lemmatization in spacy with stemming in nltk
def compare_normalization(doc):
# tokenize document in spacy
doc_spacy = en_nlp(doc)
# print lemmas found by spacy
print("Lemmatization:")
print([token.lemma_ for token in doc_spacy])
# print tokens found by Porter stemmer
print("Stemming:")
print([stemmer.stem(token.norm_.lower()) for token in doc_spacy])
compare_normalization(u"Our meeting today was worse than yesterday, "
"I'm scared of meeting the clients tomorrow.")
# Technicallity: we want to use the regexp based tokenizer
# that is used by CountVectorizer and only use the lemmatization
# from SpaCy. To this end, we replace en_nlp.tokenizer (the SpaCy tokenizer)
# with the regexp based tokenization
import re
# regexp used in CountVectorizer:
regexp = re.compile('(?u)\\b\\w\\w+\\b')
# load spacy language model
en_nlp = spacy.load('en', disable=['parser', 'ner'])
old_tokenizer = en_nlp.tokenizer
# replace the tokenizer with the preceding regexp
en_nlp.tokenizer = lambda string: old_tokenizer.tokens_from_list(
regexp.findall(string))
# create a custom tokenizer using the SpaCy document processing pipeline
# (now using our own tokenizer)
def custom_tokenizer(document):
doc_spacy = en_nlp(document)
return [token.lemma_ for token in doc_spacy]
# define a count vectorizer with the custom tokenizer
lemma_vect = CountVectorizer(tokenizer=custom_tokenizer, min_df=5)
# transform text_train using CountVectorizer with lemmatization
X_train_lemma = lemma_vect.fit_transform(text_train)
print("X_train_lemma.shape: {}".format(X_train_lemma.shape))
# standard CountVectorizer for reference
vect = CountVectorizer(min_df=5).fit(text_train)
X_train = vect.transform(text_train)
print("X_train.shape: {}".format(X_train.shape))
# build a grid-search using only 1% of the data as training set:
from sklearn.model_selection import StratifiedShuffleSplit
param_grid = {'C': [0.001, 0.01, 0.1, 1, 10]}
cv = StratifiedShuffleSplit(n_splits=5, test_size=0.99,
train_size=0.01, random_state=0)
grid = GridSearchCV(LogisticRegression(), param_grid, cv=cv)
# perform grid search with standard CountVectorizer
grid.fit(X_train, y_train)
print("Best cross-validation score "
"(standard CountVectorizer): {:.3f}".format(grid.best_score_))
# perform grid search with Lemmatization
grid.fit(X_train_lemma, y_train)
print("Best cross-validation score "
"(lemmatization): {:.3f}".format(grid.best_score_)) | Best cross-validation score (standard CountVectorizer): 0.721
Best cross-validation score (lemmatization): 0.731
| MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
Topic Modeling and Document Clustering Latent Dirichlet Allocation | vect = CountVectorizer(max_features=10000, max_df=.15)
X = vect.fit_transform(text_train)
from sklearn.decomposition import LatentDirichletAllocation
lda = LatentDirichletAllocation(n_topics=10, learning_method="batch",
max_iter=25, random_state=0)
# We build the model and transform the data in one step
# Computing transform takes some time,
# and we can save time by doing both at once
document_topics = lda.fit_transform(X)
print("lda.components_.shape: {}".format(lda.components_.shape))
# for each topic (a row in the components_), sort the features (ascending).
# Invert rows with [:, ::-1] to make sorting descending
sorting = np.argsort(lda.components_, axis=1)[:, ::-1]
# get the feature names from the vectorizer:
feature_names = np.array(vect.get_feature_names())
# Print out the 10 topics:
mglearn.tools.print_topics(topics=range(10), feature_names=feature_names,
sorting=sorting, topics_per_chunk=5, n_words=10)
lda100 = LatentDirichletAllocation(n_topics=100, learning_method="batch",
max_iter=25, random_state=0)
document_topics100 = lda100.fit_transform(X)
topics = np.array([7, 16, 24, 25, 28, 36, 37, 41, 45, 51, 53, 54, 63, 89, 97])
sorting = np.argsort(lda100.components_, axis=1)[:, ::-1]
feature_names = np.array(vect.get_feature_names())
mglearn.tools.print_topics(topics=topics, feature_names=feature_names,
sorting=sorting, topics_per_chunk=5, n_words=20)
# sort by weight of "music" topic 45
music = np.argsort(document_topics100[:, 45])[::-1]
# print the five documents where the topic is most important
for i in music[:10]:
# show first two sentences
print(b".".join(text_train[i].split(b".")[:2]) + b".\n")
fig, ax = plt.subplots(1, 2, figsize=(10, 10))
topic_names = ["{:>2} ".format(i) + " ".join(words)
for i, words in enumerate(feature_names[sorting[:, :2]])]
# two column bar chart:
for col in [0, 1]:
start = col * 50
end = (col + 1) * 50
ax[col].barh(np.arange(50), np.sum(document_topics100, axis=0)[start:end])
ax[col].set_yticks(np.arange(50))
ax[col].set_yticklabels(topic_names[start:end], ha="left", va="top")
ax[col].invert_yaxis()
ax[col].set_xlim(0, 2000)
yax = ax[col].get_yaxis()
yax.set_tick_params(pad=130)
plt.tight_layout() | _____no_output_____ | MIT | introduction_to_ml_with_python-master/07-working-with-text-data.ipynb | nosy0411/Programming_for_Data_Science |
CER100 - Configure Cluster with Self Signed Certificates========================================================This notebook will:1. Generate a new Root CA in the Big Data Cluster2. Create new certificates for each endpoint (Management, Gateway, App-Proxy and Controller)3. Sign each new certificate with the new generated Root CA, except the Controller cert (which is signed with the existing cluster Root CA)4. Install each certificate into the Big Data Cluster5. Download the new generated Root CA into this machine’s Trusted Root Cerification Authorities certificate store.All generated self-signed certificates will be stored in the controllerpod (at the `test_cert_store_root` location).**NOTE: When CER010 runs (the 3rd notebook), look for the ‘SecurityWarning’ dialog to pop up, and press ‘Yes’ to accept the installation ofthe new Root CA into this machine’s certificate store.**Upon completion of this notebook, all https:// access to the Big DataCluster from this machine (and any machine that installs the new RootCA) will show as being secure.The Notebook Runner chapter, will ensure CronJobs created (RUN003) torun App-Deploy will install the cluster Root CA to allow securelygetting JWT tokens and the swagger.json.Description----------- ParametersThe parameters set here will override the default parameters set in eachindividual notebook (`azdata notebook run` injects a `Parameters` cellat runtime with the values passed in from the `-a` arguement) | import getpass
common_name = "SQL Server Big Data Clusters Test CA"
country_name = "US"
state_or_province_name = "Illinois"
locality_name = "Chicago"
organization_name = "Contoso"
organizational_unit_name = "Finance"
email_address = f"{getpass.getuser()}@contoso.com"
days = "825" # the number of days to certify the certificates for
test_cert_store_root = "/var/opt/secrets/test-certificates" | _____no_output_____ | MIT | Big-Data-Clusters/CU3/Public/content/cert-management/cer100-create-root-ca-install-certs.ipynb | gantz-at-incomm/tigertoolbox |
Define notebooks and their arguments | import os
import copy
cer00_args = { "country_name": country_name, "state_or_province_name": state_or_province_name, "locality_name": locality_name, "organization_name": organization_name, "organizational_unit_name": organizational_unit_name, "common_name": common_name, "email_address": email_address, "days": days, "test_cert_store_root": test_cert_store_root }
cer02_args = copy.deepcopy(cer00_args)
cer02_args.pop("common_name") # no common_name (as this is the service name set per endpoint)
cer04_args = { "test_cert_store_root": test_cert_store_root }
notebooks = [
[ os.path.join("..", "common", "sop028-azdata-login.ipynb"), {} ],
[ os.path.join("..", "cert-management", "cer001-create-root-ca.ipynb"), cer00_args ],
[ os.path.join("..", "cert-management", "cer010-install-generated-root-ca-locally.ipynb"), cer04_args ],
[ os.path.join("..", "cert-management", "cer020-create-management-service-proxy-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer021-create-knox-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer022-create-app-proxy-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer023-create-controller-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer030-sign-service-proxy-generated-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer031-sign-knox-generated-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer032-sign-app-proxy-generated-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer033-sign-controller-generated-cert.ipynb"), cer02_args ],
[ os.path.join("..", "cert-management", "cer040-install-service-proxy-cert.ipynb"), cer04_args ],
[ os.path.join("..", "cert-management", "cer041-install-knox-cert.ipynb"), cer04_args ],
[ os.path.join("..", "cert-management", "cer042-install-app-proxy-cert.ipynb"), cer04_args ],
[ os.path.join("..", "cert-management", "cer043-install-controller-cert.ipynb"), cer04_args ],
[ os.path.join("..", "cert-management", "cer050-wait-cluster-healthly.ipynb"), {} ]
] | _____no_output_____ | MIT | Big-Data-Clusters/CU3/Public/content/cert-management/cer100-create-root-ca-install-certs.ipynb | gantz-at-incomm/tigertoolbox |
Common functionsDefine helper functions used in this notebook. | # Define `run` function for transient fault handling, hyperlinked suggestions, and scrolling updates on Windows
import sys
import os
import re
import json
import platform
import shlex
import shutil
import datetime
from subprocess import Popen, PIPE
from IPython.display import Markdown
retry_hints = {}
error_hints = {}
install_hint = {}
first_run = True
rules = None
def run(cmd, return_output=False, no_output=False, retry_count=0):
"""
Run shell command, stream stdout, print stderr and optionally return output
"""
MAX_RETRIES = 5
output = ""
retry = False
global first_run
global rules
if first_run:
first_run = False
rules = load_rules()
# shlex.split is required on bash and for Windows paths with spaces
#
cmd_actual = shlex.split(cmd)
# Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries
#
user_provided_exe_name = cmd_actual[0].lower()
# When running python, use the python in the ADS sandbox ({sys.executable})
#
if cmd.startswith("python "):
cmd_actual[0] = cmd_actual[0].replace("python", sys.executable)
# On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail
# with:
#
# UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)
#
# Setting it to a default value of "en_US.UTF-8" enables pip install to complete
#
if platform.system() == "Darwin" and "LC_ALL" not in os.environ:
os.environ["LC_ALL"] = "en_US.UTF-8"
# To aid supportabilty, determine which binary file will actually be executed on the machine
#
which_binary = None
# Special case for CURL on Windows. The version of CURL in Windows System32 does not work to
# get JWT tokens, it returns "(56) Failure when receiving data from the peer". If another instance
# of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost
# always the first curl.exe in the path, and it can't be uninstalled from System32, so here we
# look for the 2nd installation of CURL in the path)
if platform.system() == "Windows" and cmd.startswith("curl "):
path = os.getenv('PATH')
for p in path.split(os.path.pathsep):
p = os.path.join(p, "curl.exe")
if os.path.exists(p) and os.access(p, os.X_OK):
if p.lower().find("system32") == -1:
cmd_actual[0] = p
which_binary = p
break
# Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this
# seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound)
#
# NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.
#
if which_binary == None:
which_binary = shutil.which(cmd_actual[0])
if which_binary == None:
if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:
display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)")
else:
cmd_actual[0] = which_binary
start_time = datetime.datetime.now().replace(microsecond=0)
print(f"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)")
print(f" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})")
print(f" cwd: {os.getcwd()}")
# Command-line tools such as CURL and AZDATA HDFS commands output
# scrolling progress bars, which causes Jupyter to hang forever, to
# workaround this, use no_output=True
#
# Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait
#
wait = True
try:
if no_output:
p = Popen(cmd_actual)
else:
p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)
with p.stdout:
for line in iter(p.stdout.readline, b''):
line = line.decode()
if return_output:
output = output + line
else:
if cmd.startswith("azdata notebook run"): # Hyperlink the .ipynb file
regex = re.compile(' "(.*)"\: "(.*)"')
match = regex.match(line)
if match:
if match.group(1).find("HTML") != -1:
display(Markdown(f' - "{match.group(1)}": "{match.group(2)}"'))
else:
display(Markdown(f' - "{match.group(1)}": "[{match.group(2)}]({match.group(2)})"'))
wait = False
break # otherwise infinite hang, have not worked out why yet.
else:
print(line, end='')
if rules is not None:
apply_expert_rules(line)
if wait:
p.wait()
except FileNotFoundError as e:
if install_hint is not None:
display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)") from e
exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()
if not no_output:
for line in iter(p.stderr.readline, b''):
line_decoded = line.decode()
# azdata emits a single empty line to stderr when doing an hdfs cp, don't
# print this empty "ERR:" as it confuses.
#
if line_decoded == "":
continue
print(f"STDERR: {line_decoded}", end='')
if line_decoded.startswith("An exception has occurred") or line_decoded.startswith("ERROR: An error occurred while executing the following cell"):
exit_code_workaround = 1
if user_provided_exe_name in error_hints:
for error_hint in error_hints[user_provided_exe_name]:
if line_decoded.find(error_hint[0]) != -1:
display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))
if rules is not None:
apply_expert_rules(line_decoded)
if user_provided_exe_name in retry_hints:
for retry_hint in retry_hints[user_provided_exe_name]:
if line_decoded.find(retry_hint) != -1:
if retry_count < MAX_RETRIES:
print(f"RETRY: {retry_count} (due to: {retry_hint})")
retry_count = retry_count + 1
output = run(cmd, return_output=return_output, retry_count=retry_count)
if return_output:
return output
else:
return
elapsed = datetime.datetime.now().replace(microsecond=0) - start_time
# WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so
# don't wait here, if success known above
#
if wait:
if p.returncode != 0:
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(p.returncode)}.\n')
else:
if exit_code_workaround !=0 :
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(exit_code_workaround)}.\n')
print(f'\nSUCCESS: {elapsed}s elapsed.\n')
if return_output:
return output
def load_json(filename):
with open(filename, encoding="utf8") as json_file:
return json.load(json_file)
def load_rules():
try:
# Load this notebook as json to get access to the expert rules in the notebook metadata.
#
j = load_json("cer100-create-root-ca-install-certs.ipynb")
except:
pass # If the user has renamed the book, we can't load ourself. NOTE: Is there a way in Jupyter, to know your own filename?
else:
if "metadata" in j and \
"azdata" in j["metadata"] and \
"expert" in j["metadata"]["azdata"] and \
"rules" in j["metadata"]["azdata"]["expert"]:
rules = j["metadata"]["azdata"]["expert"]["rules"]
rules.sort() # Sort rules, so they run in priority order (the [0] element). Lowest value first.
# print (f"EXPERT: There are {len(rules)} rules to evaluate.")
return rules
def apply_expert_rules(line):
global rules
for rule in rules:
# rules that have 9 elements are the injected (output) rules (the ones we want). Rules
# with only 8 elements are the source (input) rules, which are not expanded (i.e. TSG029,
# not ../repair/tsg029-nb-name.ipynb)
if len(rule) == 9:
notebook = rule[1]
cell_type = rule[2]
output_type = rule[3] # i.e. stream or error
output_type_name = rule[4] # i.e. ename or name
output_type_value = rule[5] # i.e. SystemExit or stdout
details_name = rule[6] # i.e. evalue or text
expression = rule[7].replace("\\*", "*") # Something escaped *, and put a \ in front of it!
# print(f"EXPERT: If rule '{expression}' satisfied', run '{notebook}'.")
if re.match(expression, line, re.DOTALL):
# print("EXPERT: MATCH: name = value: '{0}' = '{1}' matched expression '{2}', therefore HINT '{4}'".format(output_type_name, output_type_value, expression, notebook))
match_found = True
display(Markdown(f'HINT: Use [{notebook}]({notebook}) to resolve this issue.'))
print('Common functions defined successfully.')
# Hints for binary (transient fault) retry, (known) error and install guide
#
retry_hints = {'azdata': ['Endpoint sql-server-master does not exist', 'Endpoint livy does not exist', 'Failed to get state for cluster', 'Endpoint webhdfs does not exist', 'Adaptive Server is unavailable or does not exist', 'Error: Address already in use']}
error_hints = {'azdata': [['azdata login', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['The token is expired', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Reason: Unauthorized', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Max retries exceeded with url: /api/v1/bdc/endpoints', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Look at the controller logs for more details', 'TSG027 - Observe cluster deployment', '../diagnose/tsg027-observe-bdc-create.ipynb'], ['provided port is already allocated', 'TSG062 - Get tail of all previous container logs for pods in BDC namespace', '../log-files/tsg062-tail-bdc-previous-container-logs.ipynb'], ['Create cluster failed since the existing namespace', 'SOP061 - Delete a big data cluster', '../install/sop061-delete-bdc.ipynb'], ['Failed to complete kube config setup', 'TSG067 - Failed to complete kube config setup', '../repair/tsg067-failed-to-complete-kube-config-setup.ipynb'], ['Error processing command: "ApiError', 'TSG110 - Azdata returns ApiError', '../repair/tsg110-azdata-returns-apierror.ipynb'], ['Error processing command: "ControllerError', 'TSG036 - Controller logs', '../log-analyzers/tsg036-get-controller-logs.ipynb'], ['ERROR: 500', 'TSG046 - Knox gateway logs', '../log-analyzers/tsg046-get-knox-logs.ipynb'], ['Data source name not found and no default driver specified', 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], ["Can't open lib 'ODBC Driver 17 for SQL Server", 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], ['Control plane upgrade failed. Failed to upgrade controller.', 'TSG108 - View the controller upgrade config map', '../diagnose/tsg108-controller-failed-to-upgrade.ipynb']]}
install_hint = {'azdata': ['SOP055 - Install azdata command line interface', '../install/sop055-install-azdata.ipynb']} | _____no_output_____ | MIT | Big-Data-Clusters/CU3/Public/content/cert-management/cer100-create-root-ca-install-certs.ipynb | gantz-at-incomm/tigertoolbox |
Create a temporary directory to stage files | # Create a temporary directory to hold configuration files
import tempfile
temp_dir = tempfile.mkdtemp()
print(f"Temporary directory created: {temp_dir}") | _____no_output_____ | MIT | Big-Data-Clusters/CU3/Public/content/cert-management/cer100-create-root-ca-install-certs.ipynb | gantz-at-incomm/tigertoolbox |
Helper function for running notebooks with `azdata notebook run`To pass ‘list’ types to `azdata notebook run --arguments`, flatten tostring | # Define helper function 'run_notebook'
def run_notebook(name, arguments):
for key, value in arguments.items():
if isinstance(value, list):
arguments[key] = str(value).replace("'", "")
# --arguments have to be passed as \" \" quoted strings on Windows cmd line
#
# `app create` and `app run` can take a long time, so pass in a 30 minute cell timeout
#
arguments = str(arguments).replace("'", '\\"')
run(f'azdata notebook run -p "{os.path.join("..", "notebook-runner", name)}" --arguments "{arguments}" --output-path "{os.getcwd()}" --output-html --timeout 1800')
print("Function 'run_notebook' defined") | _____no_output_____ | MIT | Big-Data-Clusters/CU3/Public/content/cert-management/cer100-create-root-ca-install-certs.ipynb | gantz-at-incomm/tigertoolbox |
Run the notebooks | for notebook in notebooks:
run_notebook(notebook[0], notebook[1])
print("Notebooks ran successfully.")
print('Notebook execution complete.') | _____no_output_____ | MIT | Big-Data-Clusters/CU3/Public/content/cert-management/cer100-create-root-ca-install-certs.ipynb | gantz-at-incomm/tigertoolbox |
Bernoulli Naive Bayes Classifier with Normalize This code template is facilitates to solve the problem of classification problem using Bernoulli Naive Bayes Algorithm using Normalize technique. Required Packages | !pip install imblearn
import warnings
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as se
from imblearn.over_sampling import RandomOverSampler
from sklearn.pipeline import make_pipeline
from sklearn.naive_bayes import BernoulliNB
from sklearn.preprocessing import LabelEncoder,Normalizer
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report,plot_confusion_matrix
warnings.filterwarnings('ignore') | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
InitializationFilepath of CSV file | #filepath
file_path= "" | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
List of features which are required for model training . | #x_values
features=[] | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Target feature for prediction. | #y_value
target='' | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Data FetchingPandas is an open-source, BSD-licensed library providing high-performance, easy-to-use data manipulation and data analysis tools.We will use panda's library to read the CSV file using its storage path.And we use the head function to display the initial row or entry. | df=pd.read_csv(file_path)
df.head() | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Feature SelectionsIt is the process of reducing the number of input variables when developing a predictive model. Used to reduce the number of input variables to both reduce the computational cost of modelling and, in some cases, to improve the performance of the model.We will assign all the required input features to X and target/outcome to Y. | X = df[features]
Y = df[target] | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Data PreprocessingSince the majority of the machine learning models in the Sklearn library doesn't handle string category data and Null value, we have to explicitly remove or replace null values. The below snippet have functions, which removes the null value if any exists. And convert the string classes data in the datasets by encoding them to integer classes. | def NullClearner(df):
if(isinstance(df, pd.Series) and (df.dtype in ["float64","int64"])):
df.fillna(df.mean(),inplace=True)
return df
elif(isinstance(df, pd.Series)):
df.fillna(df.mode()[0],inplace=True)
return df
else:return df
def EncodeX(df):
return pd.get_dummies(df)
def EncodeY(df):
if len(df.unique())<=2:
return df
else:
un_EncodedT=np.sort(pd.unique(df), axis=-1, kind='mergesort')
df=LabelEncoder().fit_transform(df)
EncodedT=[xi for xi in range(len(un_EncodedT))]
print("Encoded Target: {} to {}".format(un_EncodedT,EncodedT))
return df
x=X.columns.to_list()
for i in x:
X[i]=NullClearner(X[i])
X=EncodeX(X)
Y=EncodeY(NullClearner(Y))
X.head() | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Correlation MapIn order to check the correlation between the features, we will plot a correlation matrix. It is effective in summarizing a large amount of data where the goal is to see patterns. | f,ax = plt.subplots(figsize=(18, 18))
matrix = np.triu(X.corr())
se.heatmap(X.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax, mask=matrix)
plt.show() | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Distribution of Target Variable | plt.figure(figsize = (10,6))
se.countplot(Y) | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Data SplittingThe train-test split is a procedure for evaluating the performance of an algorithm. The procedure involves taking a dataset and dividing it into two subsets. The first subset is utilized to fit/train the model. The second subset is used for prediction. The main motive is to estimate the performance of the model on new data. | x_train,x_test,y_train,y_test=train_test_split(X,Y,test_size=0.2,random_state=123) | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Handling Target ImbalanceThe challenge of working with imbalanced datasets is that most machine learning techniques will ignore, and in turn have poor performance on, the minority class, although typically it is performance on the minority class that is most important.One approach to addressing imbalanced datasets is to oversample the minority class. The simplest approach involves duplicating examples in the minority class.We will perform overspampling using imblearn library. | x_train,y_train = RandomOverSampler(random_state=123).fit_resample(x_train, y_train) | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Data Rescalingsklearn.preprocessing.Normalizer()Normalize samples individually to unit norm.more details at [scikit-learn.org](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.Normalizer.htmlsklearn.preprocessing.Normalizer) | Scaler=Normalizer()
x_train=Scaler.fit_transform(x_train)
x_test=Scaler.transform(x_test) | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
ModelBernoulli Naive Bayes Classifier is used for discrete data and it works on Bernoulli distribution. The main feature of Bernoulli Naive Bayes is that it accepts features only as binary values like true or false, yes or no, success or failure, 0 or 1 and so on. So when the feature values are **binary** we know that we have to use Bernoulli Naive Bayes classifier. Model Tuning Parameters 1. alpha : float, default=1.0> Additive (Laplace/Lidstone) smoothing parameter (0 for no smoothing). 2. binarize : float or None, default=0.0> Threshold for binarizing (mapping to booleans) of sample features. If None, input is presumed to already consist of binary vectors. 3. fit_prior : bool, default=True> Whether to learn class prior probabilities or not. If false, a uniform prior will be used. 4. class_prior : array-like of shape (n_classes,), default=None> Prior probabilities of the classes. If specified the priors are not adjusted according to the data. | # BernoulliNB.
model = BernoulliNB()
model.fit(x_train, y_train) | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Model Accuracyscore() method return the mean accuracy on the given test data and labels.In multi-label classification, this is the subset accuracy which is a harsh metric since you require for each sample that each label set be correctly predicted. | print("Accuracy score {:.2f} %\n".format(model.score(x_test,y_test)*100)) | Accuracy score 41.25 %
| Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Confusion MatrixA confusion matrix is utilized to understand the performance of the classification model or algorithm in machine learning for a given test set where results are known. | plot_confusion_matrix(model,x_test,y_test,cmap=plt.cm.Blues) | _____no_output_____ | Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Classification ReportA Classification report is used to measure the quality of predictions from a classification algorithm. How many predictions are True, how many are False.* **where**: - Precision:- Accuracy of positive predictions. - Recall:- Fraction of positives that were correctly identified. - f1-score:- percent of positive predictions were correct - support:- Support is the number of actual occurrences of the class in the specified dataset. | print(classification_report(y_test,model.predict(x_test))) | precision recall f1-score support
0 0.54 0.44 0.48 50
1 0.28 0.37 0.32 30
accuracy 0.41 80
macro avg 0.41 0.40 0.40 80
weighted avg 0.44 0.41 0.42 80
| Apache-2.0 | Classification/Naive Bayes/BernoulliNB_Normalize.ipynb | shreepad-nade/ds-seed |
Tutorial on collocating a datafile with lagged data | import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
import pandas as pd
import warnings
import timeit
# filter some warning messages
warnings.filterwarnings("ignore")
#from geopy.distance import geodesic
####################you will need to change some paths here!#####################
#list of input files
filename_bird='~/Desktop/zoo_selgroups_HadSST_relabundance_5aug2019_plumchrusV_4regions_final.csv'
#output files
filename_bird_out='~/Desktop/zoo_selgroups_HadSST_relabundance_5aug2019_plumchrusV_4regions_final_satsst.csv'
filename_bird_out_netcdf='~/Desktop/zoo_selgroups_HadSST_relabundance_5aug2019_plumchrusV_4regions_final_satsst.nc'
#################################################################################
| _____no_output_____ | Apache-2.0 | Tutorials/Collocate_cloud_data_with_cvs_file.ipynb | caitlinkroeger/cloud_science |
Reading CSV datasets | #read in csv file in to panda dataframe & into xarray
df_bird = pd.read_csv(filename_bird)
# calculate time, it needs a datetime64[ns] format
df_bird.insert(3,'Year',df_bird['year'])
df_bird.insert(4,'Month',df_bird['month'])
df_bird.insert(5,'Day',df_bird['day'])
df_bird=df_bird.drop(columns={'day','month','year'})
df_bird['time'] = df_bird['time'].apply(lambda x: x.zfill(8))
df_bird.insert(6,'Hour',df_bird['time'].apply(lambda x: x[:2]))
df_bird.insert(7,'Min',df_bird['time'].apply(lambda x: x[3:5]))
df_bird.insert(3,'time64',pd.to_datetime(df_bird[list(df_bird)[3:7]]))
df_bird=df_bird.drop(columns={'Day','Month','Year','Hour','Min','time','Date'})
# transform to x array
ds_bird = df_bird.to_xarray()
#just check lat/lon & see looks okay
minlat,maxlat=ds_bird.lat.min(),ds_bird.lat.max()
minlon,maxlon=ds_bird.lon.min(),ds_bird.lon.max()
plt.scatter(ds_bird.lon,ds_bird.lat)
print(minlat,maxlat,minlon,maxlon)
#open cmc sst
ds = xr.open_zarr('F:/data/sat_data/sst/cmc/zarr').drop({'analysis_error','mask','sea_ice_fraction'})
ds
#average 0.6 deg in each direction to create mean
ds = ds.rolling(lat=3,center=True,keep_attrs=True).mean(keep_attrs=True)
ds = ds.rolling(lon=3,center=True,keep_attrs=True).mean(keep_attrs=True)
ds
ds_mon = ds.rolling(time=30, center=False,keep_attrs=True).mean(keep_attrs=True)
ds_15 = ds.rolling(time=15, center=False,keep_attrs=True).mean(keep_attrs=True)
ds['analysed_sst_1mon']=ds_mon['analysed_sst']
ds['analysed_sst_15dy']=ds_15['analysed_sst']
ds | _____no_output_____ | Apache-2.0 | Tutorials/Collocate_cloud_data_with_cvs_file.ipynb | caitlinkroeger/cloud_science |
Collocate all data with bird data | len(ds_bird.lat)
ds_data = ds
for var in ds_data:
var_tem=var
ds_bird[var_tem]=xr.DataArray(np.empty(ilen_bird, dtype=str(ds_data[var].dtype)), coords={'index': ds_bird.index}, dims=('index'))
ds_bird[var_tem].attrs=ds_data[var].attrs
print('var',var_tem)
for i in range(len(ds_bird.lat)):
# for i in range(len(ds_bird.lat)):
# if ds_bird.time[i]<ds_data.time.min():
# continue
# if ds_bird.time[i]>ds_data.time.max():
# continue
t1,t2 = ds_bird.time64[i]-np.timedelta64(24,'h'), ds_bird.time64[i]+np.timedelta64(24,'h')
lat1,lat2=ds_bird.lat[i]-.5,ds_bird.lat[i]+.5
lon1,lon2=ds_bird.lon[i]-.5,ds_bird.lon[i]+.5
tem = ds_data.sel(time=slice(t1,t2),lat=slice(lat1,lat2),lon=slice(lon1,lon2)).load()
tem = tem.interp(time=ds_bird.time64[i],lat=ds_bird.lat[i],lon=ds_bird.lon[i])
#tem = tem.load()
for var in ds_data:
var_tem=var
ds_bird[var_tem][i]=tem[var].data
if int(i/100)*100==i:
print(i,len(ds_bird.lat))
#output data
df_bird = ds_bird.to_dataframe()
df_bird.to_csv(filename_bird_out)
#ds_bird.to_netcdf(filename_bird_out_netcdf)
var2
#test rolling to check
print(da.data)
da = xr.DataArray(np.linspace(0, 11, num=12),coords=[pd.date_range( "15/12/1999", periods=12, freq=pd.DateOffset(months=1), )],dims="time",)
dar = da.rolling(time=3,center=False).mean() #before and up too
print(dar.data) | _____no_output_____ | Apache-2.0 | Tutorials/Collocate_cloud_data_with_cvs_file.ipynb | caitlinkroeger/cloud_science |
Plagiarism Detection, Feature EngineeringIn this project, you will be tasked with building a plagiarism detector that examines an answer text file and performs binary classification; labeling that file as either plagiarized or not, depending on how similar that text file is to a provided, source text. Your first task will be to create some features that can then be used to train a classification model. This task will be broken down into a few discrete steps:* Clean and pre-process the data.* Define features for comparing the similarity of an answer text and a source text, and extract similarity features.* Select "good" features, by analyzing the correlations between different features.* Create train/test `.csv` files that hold the relevant features and class labels for train/test data points.In the _next_ notebook, Notebook 3, you'll use the features and `.csv` files you create in _this_ notebook to train a binary classification model in a SageMaker notebook instance.You'll be defining a few different similarity features, as outlined in [this paper](https://s3.amazonaws.com/video.udacity-data.com/topher/2019/January/5c412841_developing-a-corpus-of-plagiarised-short-answers/developing-a-corpus-of-plagiarised-short-answers.pdf), which should help you build a robust plagiarism detector!To complete this notebook, you'll have to complete all given exercises and answer all the questions in this notebook.> All your tasks will be clearly labeled **EXERCISE** and questions as **QUESTION**.It will be up to you to decide on the features to include in your final training and test data.--- Read in the DataThe cell below will download the necessary, project data and extract the files into the folder `data/`.This data is a slightly modified version of a dataset created by Paul Clough (Information Studies) and Mark Stevenson (Computer Science), at the University of Sheffield. You can read all about the data collection and corpus, at [their university webpage](https://ir.shef.ac.uk/cloughie/resources/plagiarism_corpus.html). > **Citation for data**: Clough, P. and Stevenson, M. Developing A Corpus of Plagiarised Short Answers, Language Resources and Evaluation: Special Issue on Plagiarism and Authorship Analysis, In Press. [Download] | # NOTE:
# you only need to run this cell if you have not yet downloaded the data
# otherwise you may skip this cell or comment it out
!wget https://s3.amazonaws.com/video.udacity-data.com/topher/2019/January/5c4147f9_data/data.zip
!unzip data
# import libraries
import pandas as pd
import numpy as np
import os | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
This plagiarism dataset is made of multiple text files; each of these files has characteristics that are is summarized in a `.csv` file named `file_information.csv`, which we can read in using `pandas`. | csv_file = 'data/file_information.csv'
plagiarism_df = pd.read_csv(csv_file)
# print out the first few rows of data info
plagiarism_df.head() | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Types of PlagiarismEach text file is associated with one **Task** (task A-E) and one **Category** of plagiarism, which you can see in the above DataFrame. Tasks, A-EEach text file contains an answer to one short question; these questions are labeled as tasks A-E. For example, Task A asks the question: "What is inheritance in object oriented programming?" Categories of plagiarism Each text file has an associated plagiarism label/category:**1. Plagiarized categories: `cut`, `light`, and `heavy`.*** These categories represent different levels of plagiarized answer texts. `cut` answers copy directly from a source text, `light` answers are based on the source text but include some light rephrasing, and `heavy` answers are based on the source text, but *heavily* rephrased (and will likely be the most challenging kind of plagiarism to detect). **2. Non-plagiarized category: `non`.** * `non` indicates that an answer is not plagiarized; the Wikipedia source text is not used to create this answer. **3. Special, source text category: `orig`.*** This is a specific category for the original, Wikipedia source text. We will use these files only for comparison purposes. --- Pre-Process the DataIn the next few cells, you'll be tasked with creating a new DataFrame of desired information about all of the files in the `data/` directory. This will prepare the data for feature extraction and for training a binary, plagiarism classifier. EXERCISE: Convert categorical to numerical dataYou'll notice that the `Category` column in the data, contains string or categorical values, and to prepare these for feature extraction, we'll want to convert these into numerical values. Additionally, our goal is to create a binary classifier and so we'll need a binary class label that indicates whether an answer text is plagiarized (1) or not (0). Complete the below function `numerical_dataframe` that reads in a `file_information.csv` file by name, and returns a *new* DataFrame with a numerical `Category` column and a new `Class` column that labels each answer as plagiarized or not. Your function should return a new DataFrame with the following properties:* 4 columns: `File`, `Task`, `Category`, `Class`. The `File` and `Task` columns can remain unchanged from the original `.csv` file.* Convert all `Category` labels to numerical labels according to the following rules (a higher value indicates a higher degree of plagiarism): * 0 = `non` * 1 = `heavy` * 2 = `light` * 3 = `cut` * -1 = `orig`, this is a special value that indicates an original file.* For the new `Class` column * Any answer text that is not plagiarized (`non`) should have the class label `0`. * Any plagiarized answer texts should have the class label `1`. * And any `orig` texts will have a special label `-1`. Expected outputAfter running your function, you should get a DataFrame with rows that looks like the following: ``` File Task Category Class0 g0pA_taska.txt a 0 01 g0pA_taskb.txt b 3 12 g0pA_taskc.txt c 2 13 g0pA_taskd.txt d 1 14 g0pA_taske.txt e 0 0......99 orig_taske.txt e -1 -1``` | # Read in a csv file and return a transformed dataframe
def numerical_dataframe(csv_file='data/file_information.csv'):
'''Reads in a csv file which is assumed to have `File`, `Category` and `Task` columns.
This function does two things:
1) converts `Category` column values to numerical values
2) Adds a new, numerical `Class` label column.
The `Class` column will label plagiarized answers as 1 and non-plagiarized as 0.
Source texts have a special label, -1.
:param csv_file: The directory for the file_information.csv file
:return: A dataframe with numerical categories and a new `Class` label column'''
# your code here
df = pd.read_csv(csv_file) #read csv file and create a Dataframe
mapping_dict = {'Category':{'non': 0, 'heavy': 1, 'light': 2, 'cut': 3, 'orig': -1}} # Define numbers for each category
df.replace(mapping_dict, inplace=True) # replace string categories by the respective numbers
class_list = [(x if x < 1 else 1) for x in df['Category']] # this will be 0 and -1 for negatives and 1 for all toher categories
df['Class'] = class_list
return df
| _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Test cellsBelow are a couple of test cells. The first is an informal test where you can check that your code is working as expected by calling your function and printing out the returned result.The **second** cell below is a more rigorous test cell. The goal of a cell like this is to ensure that your code is working as expected, and to form any variables that might be used in _later_ tests/code, in this case, the data frame, `transformed_df`.> The cells in this notebook should be run in chronological order (the order they appear in the notebook). This is especially important for test cells.Often, later cells rely on the functions, imports, or variables defined in earlier cells. For example, some tests rely on previous tests to work.These tests do not test all cases, but they are a great way to check that you are on the right track! | # informal testing, print out the results of a called function
# create new `transformed_df`
transformed_df = numerical_dataframe(csv_file ='data/file_information.csv')
# check work
# check that all categories of plagiarism have a class label = 1
transformed_df.head(10)
# test cell that creates `transformed_df`, if tests are passed
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# importing tests
import problem_unittests as tests
# test numerical_dataframe function
tests.test_numerical_df(numerical_dataframe)
# if above test is passed, create NEW `transformed_df`
transformed_df = numerical_dataframe(csv_file ='data/file_information.csv')
# check work
print('\nExample data: ')
transformed_df.head() | Tests Passed!
Example data:
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Text Processing & Splitting DataRecall that the goal of this project is to build a plagiarism classifier. At it's heart, this task is a comparison text; one that looks at a given answer and a source text, compares them and predicts whether an answer has plagiarized from the source. To effectively do this comparison, and train a classifier we'll need to do a few more things: pre-process all of our text data and prepare the text files (in this case, the 95 answer files and 5 original source files) to be easily compared, and split our data into a `train` and `test` set that can be used to train a classifier and evaluate it, respectively. To this end, you've been provided code that adds additional information to your `transformed_df` from above. The next two cells need not be changed; they add two additional columns to the `transformed_df`:1. A `Text` column; this holds all the lowercase text for a `File`, with extraneous punctuation removed.2. A `Datatype` column; this is a string value `train`, `test`, or `orig` that labels a data point as part of our train or test setThe details of how these additional columns are created can be found in the `helpers.py` file in the project directory. You're encouraged to read through that file to see exactly how text is processed and how data is split.Run the cells below to get a `complete_df` that has all the information you need to proceed with plagiarism detection and feature engineering. | """
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
import helpers
# create a text column
text_df = helpers.create_text_column(transformed_df)
text_df.head()
# after running the cell above
# check out the processed text for a single file, by row index
row_idx = 9 # feel free to change this index
sample_text = text_df.iloc[row_idx]['Text']
print('Sample processed text:\n\n', sample_text) | Sample processed text:
dynamic programming is a method for solving mathematical programming problems that exhibit the properties of overlapping subproblems and optimal substructure this is a much quicker method than other more naive methods the word programming in dynamic programming relates optimization which is commonly referred to as mathematical programming richard bellman originally coined the term in the 1940s to describe a method for solving problems where one needs to find the best decisions one after another and by 1953 he refined his method to the current modern meaning optimal substructure means that by splitting the programming into optimal solutions of subproblems these can then be used to find the optimal solutions of the overall problem one example is the computing of the shortest path to a goal from a vertex in a graph first compute the shortest path to the goal from all adjacent vertices then using this the best overall path can be found thereby demonstrating the dynamic programming principle this general three step process can be used to solve a problem 1 break up the problem different smaller subproblems 2 recursively use this three step process to compute the optimal path in the subproblem 3 construct an optimal solution using the computed optimal subproblems for the original problem this process continues recursively working over the subproblems by dividing them into sub subproblems and so forth until a simple case is reached one that is easily solvable
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Split data into training and test setsThe next cell will add a `Datatype` column to a given DataFrame to indicate if the record is: * `train` - Training data, for model training.* `test` - Testing data, for model evaluation.* `orig` - The task's original answer from wikipedia. Stratified samplingThe given code uses a helper function which you can view in the `helpers.py` file in the main project directory. This implements [stratified random sampling](https://en.wikipedia.org/wiki/Stratified_sampling) to randomly split data by task & plagiarism amount. Stratified sampling ensures that we get training and test data that is fairly evenly distributed across task & plagiarism combinations. Approximately 26% of the data is held out for testing and 74% of the data is used for training.The function **train_test_dataframe** takes in a DataFrame that it assumes has `Task` and `Category` columns, and, returns a modified frame that indicates which `Datatype` (train, test, or orig) a file falls into. This sampling will change slightly based on a passed in *random_seed*. Due to a small sample size, this stratified random sampling will provide more stable results for a binary plagiarism classifier. Stability here is smaller *variance* in the accuracy of classifier, given a random seed. | random_seed = 1 # can change; set for reproducibility
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
import helpers
# create new df with Datatype (train, test, orig) column
# pass in `text_df` from above to create a complete dataframe, with all the information you need
complete_df = helpers.train_test_dataframe(text_df, random_seed=random_seed)
# check results
complete_df | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Determining PlagiarismNow that you've prepared this data and created a `complete_df` of information, including the text and class associated with each file, you can move on to the task of extracting similarity features that will be useful for plagiarism classification. > Note: The following code exercises, assume that the `complete_df` as it exists now, will **not** have its existing columns modified. The `complete_df` should always include the columns: `['File', 'Task', 'Category', 'Class', 'Text', 'Datatype']`. You can add additional columns, and you can create any new DataFrames you need by copying the parts of the `complete_df` as long as you do not modify the existing values, directly.--- Similarity Features One of the ways we might go about detecting plagiarism, is by computing **similarity features** that measure how similar a given answer text is as compared to the original wikipedia source text (for a specific task, a-e). The similarity features you will use are informed by [this paper on plagiarism detection](https://s3.amazonaws.com/video.udacity-data.com/topher/2019/January/5c412841_developing-a-corpus-of-plagiarised-short-answers/developing-a-corpus-of-plagiarised-short-answers.pdf). > In this paper, researchers created features called **containment** and **longest common subsequence**. Using these features as input, you will train a model to distinguish between plagiarized and not-plagiarized text files. Feature EngineeringLet's talk a bit more about the features we want to include in a plagiarism detection model and how to calculate such features. In the following explanations, I'll refer to a submitted text file as a **Student Answer Text (A)** and the original, wikipedia source file (that we want to compare that answer to) as the **Wikipedia Source Text (S)**. ContainmentYour first task will be to create **containment features**. To understand containment, let's first revisit a definition of [n-grams](https://en.wikipedia.org/wiki/N-gram). An *n-gram* is a sequential word grouping. For example, in a line like "bayes rule gives us a way to combine prior knowledge with new information," a 1-gram is just one word, like "bayes." A 2-gram might be "bayes rule" and a 3-gram might be "combine prior knowledge."> Containment is defined as the **intersection** of the n-gram word count of the Wikipedia Source Text (S) with the n-gram word count of the Student Answer Text (S) *divided* by the n-gram word count of the Student Answer Text.$$ \frac{\sum{count(\text{ngram}_{A}) \cap count(\text{ngram}_{S})}}{\sum{count(\text{ngram}_{A})}} $$If the two texts have no n-grams in common, the containment will be 0, but if _all_ their n-grams intersect then the containment will be 1. Intuitively, you can see how having longer n-gram's in common, might be an indication of cut-and-paste plagiarism. In this project, it will be up to you to decide on the appropriate `n` or several `n`'s to use in your final model. EXERCISE: Create containment featuresGiven the `complete_df` that you've created, you should have all the information you need to compare any Student Answer Text (A) with its appropriate Wikipedia Source Text (S). An answer for task A should be compared to the source text for task A, just as answers to tasks B, C, D, and E should be compared to the corresponding original source text.In this exercise, you'll complete the function, `calculate_containment` which calculates containment based upon the following parameters:* A given DataFrame, `df` (which is assumed to be the `complete_df` from above)* An `answer_filename`, such as 'g0pB_taskd.txt' * An n-gram length, `n` Containment calculationThe general steps to complete this function are as follows:1. From *all* of the text files in a given `df`, create an array of n-gram counts; it is suggested that you use a [CountVectorizer](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html) for this purpose.2. Get the processed answer and source texts for the given `answer_filename`.3. Calculate the containment between an answer and source text according to the following equation. >$$ \frac{\sum{count(\text{ngram}_{A}) \cap count(\text{ngram}_{S})}}{\sum{count(\text{ngram}_{A})}} $$ 4. Return that containment value.You are encouraged to write any helper functions that you need to complete the function below. | # Calculate the ngram containment for one answer file/source file pair in a df
from sklearn.feature_extraction.text import CountVectorizer
def calculate_containment(df, n, answer_filename):
'''Calculates the containment between a given answer text and its associated source text.
This function creates a count of ngrams (of a size, n) for each text file in our data.
Then calculates the containment by finding the ngram count for a given answer text,
and its associated source text, and calculating the normalized intersection of those counts.
:param df: A dataframe with columns,
'File', 'Task', 'Category', 'Class', 'Text', and 'Datatype'
:param n: An integer that defines the ngram size
:param answer_filename: A filename for an answer text in the df, ex. 'g0pB_taskd.txt'
:return: A single containment value that represents the similarity
between an answer text and its source text.
'''
# your code here
# print(f'calculate_containment(df=df, n={n}, answer_filename={answer_filename})')
# get the text from filename
answer_row = df.loc[df['File'] == answer_filename]
# print(f'answer_row =')
# print(answer_row)
# get the task, assuming no two files have the same name
# print(answer_row['Task'])
task = answer_row['Task'].tolist()[0]
#find appropriate original text
original_text_row = df.loc[(df['Task'] == task) & (df['Class'] == -1)]
original_text = original_text_row['Text']
# get answer text
answer_text = answer_row['Text']
# get one-hot-encoded matrix for our answer and get transformed strings
vectorizer = CountVectorizer(ngram_range=(n, n))
transformed_answer = vectorizer.fit_transform(answer_text).toarray()[0]
transformed_original = vectorizer.transform(original_text).toarray()[0]
# get the intersections between the two matrices
intersection = []
for i, element in enumerate(transformed_answer):
intersection.append(transformed_answer[i] if (transformed_answer[i] < transformed_original[i]) else transformed_original[i])
containment = sum(intersection) / sum(transformed_answer)
# print(f'containment = {containment}')
# print('------------------------------')
return containment | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Test cellsAfter you've implemented the containment function, you can test out its behavior. The cell below iterates through the first few files, and calculates the original category _and_ containment values for a specified n and file.>If you've implemented this correctly, you should see that the non-plagiarized have low or close to 0 containment values and that plagiarized examples have higher containment values, closer to 1.Note what happens when you change the value of n. I recommend applying your code to multiple files and comparing the resultant containment values. You should see that the highest containment values correspond to files with the highest category (`cut`) of plagiarism level. | # select a value for n
n = 1
# indices for first few files
test_indices = range(5)
# iterate through files and calculate containment
category_vals = []
containment_vals = []
for i in test_indices:
# get level of plagiarism for a given file index
category_vals.append(complete_df.loc[i, 'Category'])
# calculate containment for given file and n
filename = complete_df.loc[i, 'File']
c = calculate_containment(complete_df, n, filename)
containment_vals.append(c)
# print out result, does it make sense?
print('Original category values: \n', category_vals)
print()
print(str(n)+'-gram containment values: \n', containment_vals)
# run this test cell
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# test containment calculation
# params: complete_df from before, and containment function
tests.test_containment(complete_df, calculate_containment) | Tests Passed!
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
QUESTION 1: Why can we calculate containment features across *all* data (training & test), prior to splitting the DataFrame for modeling? That is, what about the containment calculation means that the test and training data do not influence each other? **Answer:**It's a property of a single entry (a feature) and not a property of the dataset or of a group of entries. --- Longest Common SubsequenceContainment a good way to find overlap in word usage between two documents; it may help identify cases of cut-and-paste as well as paraphrased levels of plagiarism. Since plagiarism is a fairly complex task with varying levels, it's often useful to include other measures of similarity. The paper also discusses a feature called **longest common subsequence**.> The longest common subsequence is the longest string of words (or letters) that are *the same* between the Wikipedia Source Text (S) and the Student Answer Text (A). This value is also normalized by dividing by the total number of words (or letters) in the Student Answer Text. In this exercise, we'll ask you to calculate the longest common subsequence of words between two texts. EXERCISE: Calculate the longest common subsequenceComplete the function `lcs_norm_word`; this should calculate the *longest common subsequence* of words between a Student Answer Text and corresponding Wikipedia Source Text. It may be helpful to think of this in a concrete example. A Longest Common Subsequence (LCS) problem may look as follows:* Given two texts: text A (answer text) of length n, and string S (original source text) of length m. Our goal is to produce their longest common subsequence of words: the longest sequence of words that appear left-to-right in both texts (though the words don't have to be in continuous order).* Consider: * A = "i think pagerank is a link analysis algorithm used by google that uses a system of weights attached to each element of a hyperlinked set of documents" * S = "pagerank is a link analysis algorithm used by the google internet search engine that assigns a numerical weighting to each element of a hyperlinked set of documents"* In this case, we can see that the start of each sentence of fairly similar, having overlap in the sequence of words, "pagerank is a link analysis algorithm used by" before diverging slightly. Then we **continue moving left -to-right along both texts** until we see the next common sequence; in this case it is only one word, "google". Next we find "that" and "a" and finally the same ending "to each element of a hyperlinked set of documents".* Below, is a clear visual of how these sequences were found, sequentially, in each text.* Now, those words appear in left-to-right order in each document, sequentially, and even though there are some words in between, we count this as the longest common subsequence between the two texts. * If I count up each word that I found in common I get the value 20. **So, LCS has length 20**. * Next, to normalize this value, divide by the total length of the student answer; in this example that length is only 27. **So, the function `lcs_norm_word` should return the value `20/27` or about `0.7408`.**In this way, LCS is a great indicator of cut-and-paste plagiarism or if someone has referenced the same source text multiple times in an answer. LCS, dynamic programmingIf you read through the scenario above, you can see that this algorithm depends on looking at two texts and comparing them word by word. You can solve this problem in multiple ways. First, it may be useful to `.split()` each text into lists of comma separated words to compare. Then, you can iterate through each word in the texts and compare them, adding to your value for LCS as you go. The method I recommend for implementing an efficient LCS algorithm is: using a matrix and dynamic programming. **Dynamic programming** is all about breaking a larger problem into a smaller set of subproblems, and building up a complete result without having to repeat any subproblems. This approach assumes that you can split up a large LCS task into a combination of smaller LCS tasks. Let's look at a simple example that compares letters:* A = "ABCD"* S = "BD"We can see right away that the longest subsequence of _letters_ here is 2 (B and D are in sequence in both strings). And we can calculate this by looking at relationships between each letter in the two strings, A and S.Here, I have a matrix with the letters of A on top and the letters of S on the left side:This starts out as a matrix that has as many columns and rows as letters in the strings S and O **+1** additional row and column, filled with zeros on the top and left sides. So, in this case, instead of a 2x4 matrix it is a 3x5.Now, we can fill this matrix up by breaking it into smaller LCS problems. For example, let's first look at the shortest substrings: the starting letter of A and S. We'll first ask, what is the Longest Common Subsequence between these two letters "A" and "B"? **Here, the answer is zero and we fill in the corresponding grid cell with that value.**Then, we ask the next question, what is the LCS between "AB" and "B"?**Here, we have a match, and can fill in the appropriate value 1**.If we continue, we get to a final matrix that looks as follows, with a **2** in the bottom right corner.The final LCS will be that value **2** *normalized* by the number of n-grams in A. So, our normalized value is 2/4 = **0.5**. The matrix rulesOne thing to notice here is that, you can efficiently fill up this matrix one cell at a time. Each grid cell only depends on the values in the grid cells that are directly on top and to the left of it, or on the diagonal/top-left. The rules are as follows:* Start with a matrix that has one extra row and column of zeros.* As you traverse your string: * If there is a match, fill that grid cell with the value to the top-left of that cell *plus* one. So, in our case, when we found a matching B-B, we added +1 to the value in the top-left of the matching cell, 0. * If there is not a match, take the *maximum* value from either directly to the left or the top cell, and carry that value over to the non-match cell.After completely filling the matrix, **the bottom-right cell will hold the non-normalized LCS value**.This matrix treatment can be applied to a set of words instead of letters. Your function should apply this to the words in two texts and return the normalized LCS value. | # Compute the normalized LCS given an answer text and a source text
def lcs_norm_word(answer_text, source_text):
'''Computes the longest common subsequence of words in two texts; returns a normalized value.
:param answer_text: The pre-processed text for an answer text
:param source_text: The pre-processed text for an answer's associated source text
:return: A normalized LCS value'''
# your code here
# split strings into words
answer = answer_text.lower().split()
source = source_text.lower().split()
# create a mtrix o zeros, with one additional row and one additional column
matrix = np.zeros((len(answer) + 1, len(source) + 1))
# iterate over all the word combinations
for row_idx, answer_word in enumerate(answer):
for col_idx, source_word in enumerate(source):
# define coordinates where I'll write a new value
# x and y are the column and row, respectively, where we'll write a new value
# col_idx and row_idx refer to the elements we are comparing
y = row_idx + 1
x = col_idx + 1
if answer_word == source_word:
# diagonal addition, as stated above
new_value = matrix[row_idx, col_idx] + 1
else:
# max of top/left values
value_north = matrix[row_idx, x]
value_west = matrix[y, col_idx]
new_value = max((value_north, value_west))
matrix[y, x] = new_value
return matrix[-1][-1] / len(answer) | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Test cellsLet's start by testing out your code on the example given in the initial description.In the below cell, we have specified strings A (answer text) and S (original source text). We know that these texts have 20 words in common and the submitted answer is 27 words long, so the normalized, longest common subsequence should be 20/27. | # Run the test scenario from above
# does your function return the expected value?
A = "i think pagerank is a link analysis algorithm used by google that uses a system of weights attached to each element of a hyperlinked set of documents"
S = "pagerank is a link analysis algorithm used by the google internet search engine that assigns a numerical weighting to each element of a hyperlinked set of documents"
# calculate LCS
lcs = lcs_norm_word(A, S)
print('LCS = ', lcs)
# expected value test
assert lcs==20/27., "Incorrect LCS value, expected about 0.7408, got "+str(lcs)
print('Test passed!') | LCS = 0.7407407407407407
Test passed!
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
This next cell runs a more rigorous test. | # run test cell
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# test lcs implementation
# params: complete_df from before, and lcs_norm_word function
tests.test_lcs(complete_df, lcs_norm_word) | Tests Passed!
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Finally, take a look at a few resultant values for `lcs_norm_word`. Just like before, you should see that higher values correspond to higher levels of plagiarism. | # test on your own
test_indices = range(5) # look at first few files
category_vals = []
lcs_norm_vals = []
# iterate through first few docs and calculate LCS
for i in test_indices:
category_vals.append(complete_df.loc[i, 'Category'])
# get texts to compare
answer_text = complete_df.loc[i, 'Text']
task = complete_df.loc[i, 'Task']
# we know that source texts have Class = -1
orig_rows = complete_df[(complete_df['Class'] == -1)]
orig_row = orig_rows[(orig_rows['Task'] == task)]
source_text = orig_row['Text'].values[0]
# calculate lcs
lcs_val = lcs_norm_word(answer_text, source_text)
lcs_norm_vals.append(lcs_val)
# print out result, does it make sense?
print('Original category values: \n', category_vals)
print()
print('Normalized LCS values: \n', lcs_norm_vals) | Original category values:
[0, 3, 2, 1, 0]
Normalized LCS values:
[0.1917808219178082, 0.8207547169811321, 0.8464912280701754, 0.3160621761658031, 0.24257425742574257]
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
--- Create All FeaturesNow that you've completed the feature calculation functions, it's time to actually create multiple features and decide on which ones to use in your final model! In the below cells, you're provided two helper functions to help you create multiple features and store those in a DataFrame, `features_df`. Creating multiple containment featuresYour completed `calculate_containment` function will be called in the next cell, which defines the helper function `create_containment_features`. > This function returns a list of containment features, calculated for a given `n` and for *all* files in a df (assumed to the the `complete_df`).For our original files, the containment value is set to a special value, -1.This function gives you the ability to easily create several containment features, of different n-gram lengths, for each of our text files. | """
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# Function returns a list of containment features, calculated for a given n
# Should return a list of length 100 for all files in a complete_df
def create_containment_features(df, n, column_name=None):
containment_values = []
if(column_name==None):
column_name = 'c_'+str(n) # c_1, c_2, .. c_n
# iterates through dataframe rows
for i in df.index:
file = df.loc[i, 'File']
# Computes features using calculate_containment function
if df.loc[i,'Category'] > -1:
c = calculate_containment(df, n, file)
containment_values.append(c)
# Sets value to -1 for original tasks
else:
containment_values.append(-1)
print(str(n)+'-gram containment features created!')
return containment_values
| _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Creating LCS featuresBelow, your complete `lcs_norm_word` function is used to create a list of LCS features for all the answer files in a given DataFrame (again, this assumes you are passing in the `complete_df`. It assigns a special value for our original, source files, -1. | """
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# Function creates lcs feature and add it to the dataframe
def create_lcs_features(df, column_name='lcs_word'):
lcs_values = []
# iterate through files in dataframe
for i in df.index:
# Computes LCS_norm words feature using function above for answer tasks
if df.loc[i,'Category'] > -1:
# get texts to compare
answer_text = df.loc[i, 'Text']
task = df.loc[i, 'Task']
# we know that source texts have Class = -1
orig_rows = df[(df['Class'] == -1)]
orig_row = orig_rows[(orig_rows['Task'] == task)]
source_text = orig_row['Text'].values[0]
# calculate lcs
lcs = lcs_norm_word(answer_text, source_text)
lcs_values.append(lcs)
# Sets to -1 for original tasks
else:
lcs_values.append(-1)
print('LCS features created!')
return lcs_values
| _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
EXERCISE: Create a features DataFrame by selecting an `ngram_range`The paper suggests calculating the following features: containment *1-gram to 5-gram* and *longest common subsequence*. > In this exercise, you can choose to create even more features, for example from *1-gram to 7-gram* containment features and *longest common subsequence*. You'll want to create at least 6 features to choose from as you think about which to give to your final, classification model. Defining and comparing at least 6 different features allows you to discard any features that seem redundant, and choose to use the best features for your final model!In the below cell **define an n-gram range**; these will be the n's you use to create n-gram containment features. The rest of the feature creation code is provided. | # Define an ngram range
ngram_range = range(1,10)
# The following code may take a minute to run, depending on your ngram_range
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
features_list = []
# Create features in a features_df
all_features = np.zeros((len(ngram_range)+1, len(complete_df)))
# Calculate features for containment for ngrams in range
i=0
for n in ngram_range:
column_name = 'c_'+str(n)
features_list.append(column_name)
# create containment features
all_features[i]=np.squeeze(create_containment_features(complete_df, n))
i+=1
# Calculate features for LCS_Norm Words
features_list.append('lcs_word')
all_features[i]= np.squeeze(create_lcs_features(complete_df))
# create a features dataframe
features_df = pd.DataFrame(np.transpose(all_features), columns=features_list)
# Print all features/columns
print()
print('Features: ', features_list)
print()
# print some results
features_df.head(10) | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Correlated FeaturesYou should use feature correlation across the *entire* dataset to determine which features are ***too*** **highly-correlated** with each other to include both features in a single model. For this analysis, you can use the *entire* dataset due to the small sample size we have. All of our features try to measure the similarity between two texts. Since our features are designed to measure similarity, it is expected that these features will be highly-correlated. Many classification models, for example a Naive Bayes classifier, rely on the assumption that features are *not* highly correlated; highly-correlated features may over-inflate the importance of a single feature. So, you'll want to choose your features based on which pairings have the lowest correlation. These correlation values range between 0 and 1; from low to high correlation, and are displayed in a [correlation matrix](https://www.displayr.com/what-is-a-correlation-matrix/), below. | """
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# Create correlation matrix for just Features to determine different models to test
corr_matrix = features_df.corr().abs().round(2)
# display shows all of a dataframe
display(corr_matrix) | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
EXERCISE: Create selected train/test dataComplete the `train_test_data` function below. This function should take in the following parameters:* `complete_df`: A DataFrame that contains all of our processed text data, file info, datatypes, and class labels* `features_df`: A DataFrame of all calculated features, such as containment for ngrams, n= 1-5, and lcs values for each text file listed in the `complete_df` (this was created in the above cells)* `selected_features`: A list of feature column names, ex. `['c_1', 'lcs_word']`, which will be used to select the final features in creating train/test sets of data.It should return two tuples:* `(train_x, train_y)`, selected training features and their corresponding class labels (0/1)* `(test_x, test_y)`, selected training features and their corresponding class labels (0/1)** Note: x and y should be arrays of feature values and numerical class labels, respectively; not DataFrames.**Looking at the above correlation matrix, you should decide on a **cutoff** correlation value, less than 1.0, to determine which sets of features are *too* highly-correlated to be included in the final training and test data. If you cannot find features that are less correlated than some cutoff value, it is suggested that you increase the number of features (longer n-grams) to choose from or use *only one or two* features in your final model to avoid introducing highly-correlated features.Recall that the `complete_df` has a `Datatype` column that indicates whether data should be `train` or `test` data; this should help you split the data appropriately. | # Takes in dataframes and a list of selected features (column names)
# and returns (train_x, train_y), (test_x, test_y)
def train_test_data(complete_df, features_df, selected_features):
'''Gets selected training and test features from given dataframes, and
returns tuples for training and test features and their corresponding class labels.
:param complete_df: A dataframe with all of our processed text data, datatypes, and labels
:param features_df: A dataframe of all computed, similarity features
:param selected_features: An array of selected features that correspond to certain columns in `features_df`
:return: training and test features and labels: (train_x, train_y), (test_x, test_y)'''
# assume all dataframes are in exactly the same order
# get indexes of training and testing features
train_df = complete_df.loc[(complete_df['Datatype'] == "train")]
test_df = complete_df.loc[(complete_df['Datatype'] == "test")]
train_indexes = train_df.index.tolist()
test_indexes = test_df.index.tolist()
# get the training features
train_x = features_df[selected_features].iloc[train_indexes].to_numpy()
# And training class labels (0 or 1)
train_y = complete_df.iloc[train_indexes]['Class'].to_numpy()
# get the test features and labels
test_x = features_df[selected_features].iloc[test_indexes].to_numpy()
test_y = complete_df.iloc[test_indexes]['Class'].to_numpy()
return (train_x, train_y), (test_x, test_y)
| _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Test cellsBelow, test out your implementation and create the final train/test data. | """
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
test_selection = list(features_df)[:2] # first couple columns as a test
# test that the correct train/test data is created
(train_x, train_y), (test_x, test_y) = train_test_data(complete_df, features_df, test_selection)
# params: generated train/test data
tests.test_data_split(train_x, train_y, test_x, test_y) | Tests Passed!
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
EXERCISE: Select "good" featuresIf you passed the test above, you can create your own train/test data, below. Define a list of features you'd like to include in your final mode, `selected_features`; this is a list of the features names you want to include. | # Select your list of features, this should be column names from features_df
# ex. ['c_1', 'lcs_word']
selected_features = ['c_1', 'c_9', 'lcs_word']
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
(train_x, train_y), (test_x, test_y) = train_test_data(complete_df, features_df, selected_features)
# check that division of samples seems correct
# these should add up to 95 (100 - 5 original files)
print('Training size: ', len(train_x))
print('Test size: ', len(test_x))
print()
print('Training df sample: \n', train_x[:10]) | Training size: 70
Test size: 25
Training df sample:
[[0.39814815 0. 0.19178082]
[0.86936937 0.21962617 0.84649123]
[0.59358289 0.01117318 0.31606218]
[0.54450262 0. 0.24257426]
[0.32950192 0. 0.16117216]
[0.59030837 0. 0.30165289]
[0.75977654 0.07017544 0.48430493]
[0.51612903 0. 0.27083333]
[0.44086022 0. 0.22395833]
[0.97945205 0.60869565 0.9 ]]
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Question 2: How did you decide on which features to include in your final model? **Answer:**By checking the correlation matrix we created, I selected two variables that are not strongly correlated (0.86), plus the wordLCS. --- Creating Final Data FilesNow, you are almost ready to move on to training a model in SageMaker!You'll want to access your train and test data in SageMaker and upload it to S3. In this project, SageMaker will expect the following format for your train/test data:* Training and test data should be saved in one `.csv` file each, ex `train.csv` and `test.csv`* These files should have class labels in the first column and features in the rest of the columnsThis format follows the practice, outlined in the [SageMaker documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/cdf-training.html), which reads: "Amazon SageMaker requires that a CSV file doesn't have a header record and that the target variable [class label] is in the first column." EXERCISE: Create csv filesDefine a function that takes in x (features) and y (labels) and saves them to one `.csv` file at the path `data_dir/filename`.It may be useful to use pandas to merge your features and labels into one DataFrame and then convert that into a csv file. You can make sure to get rid of any incomplete rows, in a DataFrame, by using `dropna`. | def make_csv(x, y, filename, data_dir):
'''Merges features and labels and converts them into one csv file with labels in the first column.
:param x: Data features
:param y: Data labels
:param file_name: Name of csv file, ex. 'train.csv'
:param data_dir: The directory where files will be saved
'''
# make data dir, if it does not exist
if not os.path.exists(data_dir):
os.makedirs(data_dir)
complete_path = os.path.join(data_dir, filename)
# your code here
df = pd.concat((pd.DataFrame(y),pd.DataFrame(x)), axis=1).dropna()
df.to_csv(path_or_buf=complete_path,
index=False,
header=False)
print(df)
# nothing is returned, but a print statement indicates that the function has run
print('Path created: '+str(data_dir)+'/'+str(filename)) | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Test cellsTest that your code produces the correct format for a `.csv` file, given some text features and labels. | """
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
fake_x = [ [0.39814815, 0.0001, 0.19178082],
[0.86936937, 0.44954128, 0.84649123],
[0.44086022, 0., 0.22395833] ]
fake_y = [0, 1, 1]
make_csv(fake_x, fake_y, filename='to_delete.csv', data_dir='test_csv')
# read in and test dimensions
fake_df = pd.read_csv('test_csv/to_delete.csv', header=None)
# check shape
assert fake_df.shape==(3, 4), \
'The file should have as many rows as data_points and as many columns as features+1 (for indices).'
# check that first column = labels
assert np.all(fake_df.iloc[:,0].values==fake_y), 'First column is not equal to the labels, fake_y.'
print('Tests passed!')
# delete the test csv file, generated above
! rm -rf test_csv | _____no_output_____ | MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
If you've passed the tests above, run the following cell to create `train.csv` and `test.csv` files in a directory that you specify! This will save the data in a local directory. Remember the name of this directory because you will reference it again when uploading this data to S3. | # can change directory, if you want
data_dir = 'plagiarism_data'
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
make_csv(train_x, train_y, filename='train.csv', data_dir=data_dir)
make_csv(test_x, test_y, filename='test.csv', data_dir=data_dir) | 0 0 1 2
0 0 0.398148 0.000000 0.191781
1 1 0.869369 0.219626 0.846491
2 1 0.593583 0.011173 0.316062
3 0 0.544503 0.000000 0.242574
4 0 0.329502 0.000000 0.161172
.. .. ... ... ...
65 1 0.845188 0.225108 0.643725
66 1 0.485000 0.000000 0.242718
67 1 0.950673 0.702326 0.839506
68 1 0.551220 0.187817 0.283019
69 0 0.361257 0.000000 0.161765
[70 rows x 4 columns]
Path created: plagiarism_data/train.csv
0 0 1 2
0 1 1.000000 0.835979 0.820755
1 1 0.765306 0.454545 0.621711
2 1 0.884444 0.064516 0.597458
3 1 0.619048 0.000000 0.427835
4 1 0.920000 0.179104 0.775000
5 1 0.992674 0.958491 0.993056
6 0 0.412698 0.000000 0.346667
7 0 0.462687 0.000000 0.189320
8 0 0.581152 0.000000 0.247423
9 0 0.584211 0.000000 0.294416
10 0 0.566372 0.000000 0.258333
11 0 0.481481 0.000000 0.278912
12 1 0.619792 0.005435 0.341584
13 1 0.921739 0.463964 0.929412
14 1 1.000000 0.842520 1.000000
15 1 0.861538 0.000000 0.504717
16 1 0.626168 0.067093 0.558559
17 1 1.000000 0.936759 0.996700
18 0 0.383838 0.000000 0.178744
19 1 1.000000 0.883895 0.854671
20 0 0.613924 0.000000 0.298343
21 1 0.972763 0.694779 0.927083
22 1 0.962810 0.495726 0.909804
23 0 0.415254 0.000000 0.177419
24 0 0.532189 0.000000 0.245833
Path created: plagiarism_data/test.csv
| MIT | Project_Plagiarism_Detection/2_Plagiarism_Feature_Engineering.ipynb | brunokiyoshi/ML_SageMaker_Studies |
Jugando con Probabilidades y Python La coincidencia de cumpleañosAquí vemos la solución de la paradija del cumpleaños que vimos en el apartado de proabilidad.La [paradoja del cumpleaños](https://es.wikipedia.org/wiki/Paradoja_del_cumplea%C3%B1os) es un problema muy conocido en el campo de la probabilidad. Plantea las siguientes interesantes preguntas: ¿Cuál es la probabilidad de que, en un grupo de personas elegidas al azar, al menos dos de ellas habrán nacido el mismo día del año? ¿Cuántas personas son necesarias para asegurar una probabilidad mayor al 50%?. Calcular esa probabilidad es complicado, así que vamos a calcular la probabilidad de que no coincidad, suponinedo que con eventos independietes (es decir las podemos multiplicar), y luego calcularemos la probabilidad de que coincidan como 1 menos esa probabilidad. Excluyendo el 29 de febrero de nuestros cálculos y asumiendo que los restantes 365 días de posibles cumpleaños son igualmente probables, vamos a calcular esas dós cuestiones. | # Ejemplo situación 2 La coincidencia de cumpleaños
prob = 1.0
asistentes = 50
# Calculamos el número de asistentes necesarios para asegurar
# que la probabilidad de coincidencia sea mayor del 50%
# asistentes = 50
asistentes = 1
prob = 1
while 1 - prob <= 0.5:
asistentes += 1
prob = prob * (365 - (asistentes - 1))/365
probabilidad_coincidir = 1 - prob
print(probabilidad_coincidir)
print("Para asegurar que la probabilidad es mayor del 50% necesitamos {0} asistentes".format(asistentes)) | 0.5072972343239855
Para asegurar que la probabilidad es mayor del 50% necesitamos 23 asistentes
| MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Variables aleatorias. Vamos a tirar un dadoVamos a trabajar con variables discretas, y en este caso vamos a vamos a reproducir un dado con la librería `random` que forma parte de la librería estandar de Python: | # importa la libreria random. puedes utilizar dir() para entender lo que ofrece
# utiliza help para obtener ayuda sobre el metodo randint
# utiliza randint() para simular un dado y haz una tirada
# ahora haz 20 tiradas, y crea una lista con las tiradas
# Vamos a calcular la media de las tiradas
# Calcula ahora la mediana
# Calcula la moda de las tiradas
# se te ocurre otra forma de calcularla?
a = [2, -9, -9, 2]
from scipy import stats
stats.mode(a).mode | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Viendo como evoluciona el número de 6 cuando sacamos más jugadasVamos a ver ahora como evoluciona el número de seises que obtenemos al lanzar el dado 10000 veces. Vamos a crear una lista en la que cada elemento sea el número de ocurrencias del número 6 dividido entre el número de lanzamientos. crea una lista llamadada ``frecuencia_seis[]`` que almacene estos valores | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
|
Vamos a tratar de hacerlo gráficamente¿Hacia que valor debería converger los números que forman la lista frecuencia_seis? Revisa la ley de los grandes números para la moneda, y aplica un poco de lógica para este caso. | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
|
Resolviendo el problema de Monty HallEste problema, más conocido con el nombre de [Monty Hall](https://es.wikipedia.org/wiki/Problema_de_Monty_Hall).En primer lugar trata de simular el problema de Monty Hall con Python, para ver cuantas veces gana el concursante y cuantas pierde. Realiza por ejemplo 10000 simulaciones del problema, en las que el usuario cambia siempre de puerta. Después puedes comparar con 10000 simulaciones en las que el usuario no cambie de puertas.Cuales son los resultados? Monty Hall sin Bayes - Simulación | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
|
Monthy Hall - una aproximación bayesianaTrata de resolver ahora el problema de Monthy Hall utilizando el teorema de Bayes.Puedes escribir la solución, o programar el código. Lo que prefieras. | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
|
El problema de las CookiesImagina que tienes 2 botes con galletas. El primero contiene 30 cookies de vainilla y 10 cookies de chocolate. El segundo bote tiene 20 cookies de chocolate y 20 cookies de vainilla.Ahora vamos a suponer que sacamos un cookie sin ver de que bote lo sacamos. El cookie es de vainilla. ¿Cuál es la probabilidad de que el cookie venga del primer bote? | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
|
El problema de los M&Ms En 1995 M&Ms lanzó los M&M’s azules. - Antes de ese año la distibución de una bolsa era: 30% Marrones, 20% Amarillos, 20% Rojos, 10% Verdes, 10% Naranjas, 10% Marron Claros. - Después de 1995 la distribución en una bolsa era la siguiente: 24% Azul , 20% Verde, 16% Naranjas, 14% Amarillos, 13% Rojos, 13% MarronesSin saber qué bolsa es cúal, sacas un M&Ms al azar de cada bolsa. Una es amarilla y otra es verde. ¿Cuál es la probabilidad de que la bolsa de la que salió el caramelo amarillo sea una bolsa de 1994?Pista: Para calcular la probabilidad a posteriori (likelihoods), tienes que multiplicar las probabilidades de sacar un amarillo de una bolsa y un verde de la otra, y viceversa.¿Cuál es la probabilidad de que el caramelo amarillo viniera de una bolsa de 1996? | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
|
Creando un clasificador basado en el teorema de Bayes Este es un problema extraido de la página web de Chris Albon, que ha replicado un ejemplo que puedes ver en la wikipedia. Trata de reproducirlo y entenderlo. Naive bayes is simple classifier known for doing well when only a small number of observations is available. In this tutorial we will create a gaussian naive bayes classifier from scratch and use it to predict the class of a previously unseen data point. This tutorial is based on an example on Wikipedia's [naive bayes classifier page](https://en.wikipedia.org/wiki/Naive_Bayes_classifier), I have implemented it in Python and tweaked some notation to improve explanation. Preliminaries | import pandas as pd
import numpy as np | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Create DataOur dataset is contains data on eight individuals. We will use the dataset to construct a classifier that takes in the height, weight, and foot size of an individual and outputs a prediction for their gender. | # Create an empty dataframe
data = pd.DataFrame()
# Create our target variable
data['Gender'] = ['male','male','male','male','female','female','female','female']
# Create our feature variables
data['Height'] = [6,5.92,5.58,5.92,5,5.5,5.42,5.75]
data['Weight'] = [180,190,170,165,100,150,130,150]
data['Foot_Size'] = [12,11,12,10,6,8,7,9]
# View the data
data | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
The dataset above is used to construct our classifier. Below we will create a new person for whom we know their feature values but not their gender. Our goal is to predict their gender. | # Create an empty dataframe
person = pd.DataFrame()
# Create some feature values for this single row
person['Height'] = [6]
person['Weight'] = [130]
person['Foot_Size'] = [8]
# View the data
person | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Bayes Theorem Bayes theorem is a famous equation that allows us to make predictions based on data. Here is the classic version of the Bayes theorem:$$\displaystyle P(A\mid B)={\frac {P(B\mid A)\,P(A)}{P(B)}}$$This might be too abstract, so let us replace some of the variables to make it more concrete. In a bayes classifier, we are interested in finding out the class (e.g. male or female, spam or ham) of an observation _given_ the data:$$p(\text{class} \mid \mathbf {\text{data}} )={\frac {p(\mathbf {\text{data}} \mid \text{class}) * p(\text{class})}{p(\mathbf {\text{data}} )}}$$where: - $\text{class}$ is a particular class (e.g. male)- $\mathbf {\text{data}}$ is an observation's data- $p(\text{class} \mid \mathbf {\text{data}} )$ is called the posterior- $p(\text{data|class})$ is called the likelihood- $p(\text{class})$ is called the prior- $p(\mathbf {\text{data}} )$ is called the marginal probabilityIn a bayes classifier, we calculate the posterior (technically we only calculate the numerator of the posterior, but ignore that for now) for every class for each observation. Then, classify the observation based on the class with the largest posterior value. In our example, we have one observation to predict and two possible classes (e.g. male and female), therefore we will calculate two posteriors: one for male and one for female.$$p(\text{person is male} \mid \mathbf {\text{person's data}} )={\frac {p(\mathbf {\text{person's data}} \mid \text{person is male}) * p(\text{person is male})}{p(\mathbf {\text{person's data}} )}}$$$$p(\text{person is female} \mid \mathbf {\text{person's data}} )={\frac {p(\mathbf {\text{person's data}} \mid \text{person is female}) * p(\text{person is female})}{p(\mathbf {\text{person's data}} )}}$$ Gaussian Naive Bayes Classifier A gaussian naive bayes is probably the most popular type of bayes classifier. To explain what the name means, let us look at what the bayes equations looks like when we apply our two classes (male and female) and three feature variables (height, weight, and footsize):$${\displaystyle {\text{posterior (male)}}={\frac {P({\text{male}})\,p({\text{height}}\mid{\text{male}})\,p({\text{weight}}\mid{\text{male}})\,p({\text{foot size}}\mid{\text{male}})}{\text{marginal probability}}}}$$$${\displaystyle {\text{posterior (female)}}={\frac {P({\text{female}})\,p({\text{height}}\mid{\text{female}})\,p({\text{weight}}\mid{\text{female}})\,p({\text{foot size}}\mid{\text{female}})}{\text{marginal probability}}}}$$Now let us unpack the top equation a bit:- $P({\text{male}})$ is the prior probabilities. It is, as you can see, simply the probability an observation is male. This is just the number of males in the dataset divided by the total number of people in the dataset.- $p({\text{height}}\mid{\text{female}})\,p({\text{weight}}\mid{\text{female}})\,p({\text{foot size}}\mid{\text{female}})$ is the likelihood. Notice that we have unpacked $\mathbf {\text{person's data}}$ so it is now every feature in the dataset. The "gaussian" and "naive" come from two assumptions present in this likelihood: 1. If you look each term in the likelihood you will notice that we assume each feature is uncorrelated from each other. That is, foot size is independent of weight or height etc.. This is obviously not true, and is a "naive" assumption - hence the name "naive bayes." 2. Second, we assume have that the value of the features (e.g. the height of women, the weight of women) are normally (gaussian) distributed. This means that $p(\text{height}\mid\text{female})$ is calculated by inputing the required parameters into the probability density function of the normal distribution: $$ p(\text{height}\mid\text{female})=\frac{1}{\sqrt{2\pi\text{variance of female height in the data}}}\,e^{ -\frac{(\text{observation's height}-\text{average height of females in the data})^2}{2\text{variance of female height in the data}} }$$- $\text{marginal probability}$ is probably one of the most confusing parts of bayesian approaches. In toy examples (including ours) it is completely possible to calculate the marginal probability. However, in many real-world cases, it is either extremely difficult or impossible to find the value of the marginal probability (explaining why is beyond the scope of this tutorial). This is not as much of a problem for our classifier as you might think. Why? Because we don't care what the true posterior value is, we only care which class has a the highest posterior value. And because the marginal probability is the same for all classes 1) we can ignore the denominator, 2) calculate only the posterior's numerator for each class, and 3) pick the largest numerator. That is, we can ignore the posterior's denominator and make a prediction solely on the relative values of the posterior's numerator.Okay! Theory over. Now let us start calculating all the different parts of the bayes equations. Calculate Priors Priors can be either constants or probability distributions. In our example, this is simply the probability of being a gender. Calculating this is simple: | # Number of males
n_male = data['Gender'][data['Gender'] == 'male'].count()
# Number of males
n_female = data['Gender'][data['Gender'] == 'female'].count()
# Total rows
total_ppl = data['Gender'].count()
# Number of males divided by the total rows
P_male = n_male/total_ppl
# Number of females divided by the total rows
P_female = n_female/total_ppl | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Calculate Likelihood Remember that each term (e.g. $p(\text{height}\mid\text{female})$) in our likelihood is assumed to be a normal pdf. For example:$$ p(\text{height}\mid\text{female})=\frac{1}{\sqrt{2\pi\text{variance of female height in the data}}}\,e^{ -\frac{(\text{observation's height}-\text{average height of females in the data})^2}{2\text{variance of female height in the data}} }$$This means that for each class (e.g. female) and feature (e.g. height) combination we need to calculate the variance and mean value from the data. Pandas makes this easy: | # Group the data by gender and calculate the means of each feature
data_means = data.groupby('Gender').mean()
# View the values
data_means
# Group the data by gender and calculate the variance of each feature
data_variance = data.groupby('Gender').var()
# View the values
data_variance | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Now we can create all the variables we need. The code below might look complex but all we are doing is creating a variable out of each cell in both of the tables above. | # Means for male
male_height_mean = data_means['Height'][data_variance.index == 'male'].values[0]
print(male_height_mean)
male_weight_mean = data_means['Weight'][data_variance.index == 'male'].values[0]
male_footsize_mean = data_means['Foot_Size'][data_variance.index == 'male'].values[0]
# Variance for male
male_height_variance = data_variance['Height'][data_variance.index == 'male'].values[0]
male_weight_variance = data_variance['Weight'][data_variance.index == 'male'].values[0]
male_footsize_variance = data_variance['Foot_Size'][data_variance.index == 'male'].values[0]
# Means for female
female_height_mean = data_means['Height'][data_variance.index == 'female'].values[0]
female_weight_mean = data_means['Weight'][data_variance.index == 'female'].values[0]
female_footsize_mean = data_means['Foot_Size'][data_variance.index == 'female'].values[0]
# Variance for female
female_height_variance = data_variance['Height'][data_variance.index == 'female'].values[0]
female_weight_variance = data_variance['Weight'][data_variance.index == 'female'].values[0]
female_footsize_variance = data_variance['Foot_Size'][data_variance.index == 'female'].values[0] | 5.855
| MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Finally, we need to create a function to calculate the probability density of each of the terms of the likelihood (e.g. $p(\text{height}\mid\text{female})$). | # Create a function that calculates p(x | y):
def p_x_given_y(x, mean_y, variance_y):
# Input the arguments into a probability density function
p = 1/(np.sqrt(2*np.pi*variance_y)) * np.exp((-(x-mean_y)**2)/(2*variance_y))
# return p
return p | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Apply Bayes Classifier To New Data Point Alright! Our bayes classifier is ready. Remember that since we can ignore the marginal probability (the demoninator), what we are actually calculating is this:$${\displaystyle {\text{numerator of the posterior}}={P({\text{female}})\,p({\text{height}}\mid{\text{female}})\,p({\text{weight}}\mid{\text{female}})\,p({\text{foot size}}\mid{\text{female}})}{}}$$To do this, we just need to plug in the values of the unclassified person (height = 6), the variables of the dataset (e.g. mean of female height), and the function (`p_x_given_y`) we made above: | # Numerator of the posterior if the unclassified observation is a male
P_male * \
p_x_given_y(person['Height'][0], male_height_mean, male_height_variance) * \
p_x_given_y(person['Weight'][0], male_weight_mean, male_weight_variance) * \
p_x_given_y(person['Foot_Size'][0], male_footsize_mean, male_footsize_variance)
# Numerator of the posterior if the unclassified observation is a female
P_female * \
p_x_given_y(person['Height'][0], female_height_mean, female_height_variance) * \
p_x_given_y(person['Weight'][0], female_weight_mean, female_weight_variance) * \
p_x_given_y(person['Foot_Size'][0], female_footsize_mean, female_footsize_variance) | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
Because the numerator of the posterior for female is greater than male, then we predict that the person is female. Crea un nuevo punto con tus datos y predice su resultado (fíjate en las unidades) | _____no_output_____ | MIT | Bloque 1 - Ramp-Up/06_Estadística/01_Probabilidad y Estadística descriptiva/06_Ejercicios.ipynb | franciscocanon-thebridge/bootcamp_thebridge_PTSep20 |
|
Data Set Preprocessing | import urllib.request, json
with urllib.request.urlopen("http://statistik.easycredit-bbl.de/XML/exchange/540/Schedule.php?type=json&saison=2017&fixedGamesOnly=0") as url:
games = json.loads(url.read().decode())
print(json.dumps(games, indent=4, sort_keys=True))
arena=[]
home_ids=[]
for i in range(0,len(games['competition'][0]['spiel'])):
if games['competition'][0]['spiel'][i]['home_id'] not in home_ids:
arena.append(games['competition'][0]['spiel'][i]['arenaName'])
home_ids.append(games['competition'][0]['spiel'][i]['home_id'])
print(arena)
print(len(arena))
print(home_ids)
from datetime import datetime
datetime_object = datetime.strptime(games['competition'][0]['spiel'][0]['datum']+" "+games['competition'][0]['spiel'][0]['uhrzeit'] , '%Y-%m-%d %H:%M:%S')
print(datetime_object)
print(datetime_object.strftime('%U'))
print(datetime_object.strftime('%w'))
#dictionary für die Hallenkapazitäten
arenakap = {486:6594,413:14500,433:4200,420:6150,415:6000,425:3300,430:6000,426:5002,540:3140,418:6200,421:4003,422:3603,483:3076,477:3447,428:3000,439:4200,517:3533,432:3132}
print(arenakap)
print(len(arenakap))
print(type(games['competition'][0]['spiel'][0]['home_id']))
#Die JSON Informationen sind als String angegeben | <class 'str'>
| MIT | ANN_moreFeatures.ipynb | F3rdixX/Basketball_BBL |
Deshalb parse (int) ich die Zuschauer und home_id für die weitere Berechnung | #Dataset zusammenstellen
dataset=[]
for i in range(0,len(games['competition'][0]['spiel'])):
datasetrow=[]
datasetrow.append(games['competition'][0]['spiel'][i]['home_id'])
datasetrow.append(games['competition'][0]['spiel'][i]['gast_id'])
datasetrow.append(int(games['competition'][0]['spiel'][i]['home_result']>games['competition'][0]['spiel'][i]['gast_result']))
datasetrow.append(int(games['competition'][0]['spiel'][i]['zuschauer']))
datasetrow.append(arenakap[int(games['competition'][0]['spiel'][i]['home_id'])])
dataset.append(datasetrow)
print(dataset)
# Umwandlung des Datasets in ein Numpy Array
import numpy as np
# : -> auslesen aller zeilen
dataset=np.asarray(dataset)
print(dataset[:,0])
print(len(dataset)) | ['540' '422' '483' '418' '477' '421' '432' '428' '413' '420' '426' '483'
'425' '486' '517' '415' '439' '413' '517' '477' '418' '428' '430' '540'
'432' '422' '421' '433' '426' '430' '477' '486' '483' '433' '432' '415'
'425' '418' '413' '422' '428' '486' '421' '540' '421' '413' '422' '477'
'415' '433' '430' '439' '426' '418' '540' '486' '428' '483' '420' '425'
'432' '517' '477' '422' '421' '432' '483' '439' '413' '426' '433' '420'
'428' '418' '430' '425' '439' '415' '486' '517' '420' '477' '426' '430'
'432' '421' '540' '415' '486' '439' '413' '486' '422' '425' '418' '433'
'483' '517' '420' '428' '426' '540' '486' '432' '420' '421' '415' '425'
'420' '439' '422' '418' '477' '433' '517' '430' '426' '483' '421' '418'
'425' '428' '413' '540' '432' '422' '426' '517' '477' '420' '433' '483'
'413' '540' '415' '418' '517' '439' '432' '428' '430' '425' '422' '477'
'439' '421' '433' '430' '426' '428' '420' '421' '415' '430' '428' '439'
'415' '517' '540' '413' '418' '421' '433' '432' '425' '415' '422' '428'
'439' '477' '486' '430' '413' '483' '426' '517' '422' '540' '418' '430'
'483' '425' '415' '428' '420' '413' '418' '425' '432' '433' '486' '483'
'540' '422' '433' '439' '477' '413' '433' '426' '483' '421' '425' '477'
'432' '439' '517' '418' '486' '415' '540' '483' '413' '418' '422' '421'
'420' '428' '430' '426' '477' '432' '540' '425' '433' '517' '439' '486'
'415' '477' '426' '517' '421' '430' '483' '517' '413' '418' '420' '422'
'426' '486' '425' '477' '486' '540' '433' '432' '428' '439' '415' '420'
'477' '413' '421' '483' '420' '430' '426' '422' '517' '540' '439' '420'
'425' '428' '415' '433' '486' '432' '486' '418' '430' '421' '426' '422'
'540' '425' '517' '439' '420' '413' '483' '428' '433' '415' '432' '430'
'486' '421' '517' '426' '477' '418' '420' '432' '425' '433' '439' '415'
'483' '422' '413' '430' '540' '428']
306
| MIT | ANN_moreFeatures.ipynb | F3rdixX/Basketball_BBL |
One hot encoding | from sklearn.preprocessing import LabelBinarizer
encoder = LabelBinarizer()
transformed_home_ids = encoder.fit_transform(dataset[:,0])
print(transformed_home_ids)
#ohne fit, damit die Teams eindeutig bleiben, nur transformation notwendig
transformed_gast_ids = encoder.transform(dataset[:,1])
print(transformed_gast_ids)
# Umformung der Zuschauer in eine Spalte (vorher war es nur eine Zeile)
print(np.reshape(dataset[:,3],(306,1)))
# Featurescaling der Zuschaueranzahl & Hallenkapazitäten
from sklearn.preprocessing import MinMaxScaler
arenaKap_scaler=MinMaxScaler()
arenaKap_scaler.fit([[0],[14500]]) #Maximum Berlin und 0 Minimum
#reshaping
transformed_zuschauer=arenaKap_scaler.transform(np.reshape(dataset[:,3],(306,1)))
transformed_kap=arenaKap_scaler.transform(np.reshape(dataset[:,4],(306,1)))
print(transformed_kap) | [[0.21655172]
[0.24848276]
[0.21213793]
[0.42758621]
[0.23772414]
[0.27606897]
[0.216 ]
[0.20689655]
[1. ]
[0.42413793]
[0.34496552]
[0.21213793]
[0.22758621]
[0.45475862]
[0.24365517]
[0.4137931 ]
[0.28965517]
[1. ]
[0.24365517]
[0.23772414]
[0.42758621]
[0.20689655]
[0.4137931 ]
[0.21655172]
[0.216 ]
[0.24848276]
[0.27606897]
[0.28965517]
[0.34496552]
[0.4137931 ]
[0.23772414]
[0.45475862]
[0.21213793]
[0.28965517]
[0.216 ]
[0.4137931 ]
[0.22758621]
[0.42758621]
[1. ]
[0.24848276]
[0.20689655]
[0.45475862]
[0.27606897]
[0.21655172]
[0.27606897]
[1. ]
[0.24848276]
[0.23772414]
[0.4137931 ]
[0.28965517]
[0.4137931 ]
[0.28965517]
[0.34496552]
[0.42758621]
[0.21655172]
[0.45475862]
[0.20689655]
[0.21213793]
[0.42413793]
[0.22758621]
[0.216 ]
[0.24365517]
[0.23772414]
[0.24848276]
[0.27606897]
[0.216 ]
[0.21213793]
[0.28965517]
[1. ]
[0.34496552]
[0.28965517]
[0.42413793]
[0.20689655]
[0.42758621]
[0.4137931 ]
[0.22758621]
[0.28965517]
[0.4137931 ]
[0.45475862]
[0.24365517]
[0.42413793]
[0.23772414]
[0.34496552]
[0.4137931 ]
[0.216 ]
[0.27606897]
[0.21655172]
[0.4137931 ]
[0.45475862]
[0.28965517]
[1. ]
[0.45475862]
[0.24848276]
[0.22758621]
[0.42758621]
[0.28965517]
[0.21213793]
[0.24365517]
[0.42413793]
[0.20689655]
[0.34496552]
[0.21655172]
[0.45475862]
[0.216 ]
[0.42413793]
[0.27606897]
[0.4137931 ]
[0.22758621]
[0.42413793]
[0.28965517]
[0.24848276]
[0.42758621]
[0.23772414]
[0.28965517]
[0.24365517]
[0.4137931 ]
[0.34496552]
[0.21213793]
[0.27606897]
[0.42758621]
[0.22758621]
[0.20689655]
[1. ]
[0.21655172]
[0.216 ]
[0.24848276]
[0.34496552]
[0.24365517]
[0.23772414]
[0.42413793]
[0.28965517]
[0.21213793]
[1. ]
[0.21655172]
[0.4137931 ]
[0.42758621]
[0.24365517]
[0.28965517]
[0.216 ]
[0.20689655]
[0.4137931 ]
[0.22758621]
[0.24848276]
[0.23772414]
[0.28965517]
[0.27606897]
[0.28965517]
[0.4137931 ]
[0.34496552]
[0.20689655]
[0.42413793]
[0.27606897]
[0.4137931 ]
[0.4137931 ]
[0.20689655]
[0.28965517]
[0.4137931 ]
[0.24365517]
[0.21655172]
[1. ]
[0.42758621]
[0.27606897]
[0.28965517]
[0.216 ]
[0.22758621]
[0.4137931 ]
[0.24848276]
[0.20689655]
[0.28965517]
[0.23772414]
[0.45475862]
[0.4137931 ]
[1. ]
[0.21213793]
[0.34496552]
[0.24365517]
[0.24848276]
[0.21655172]
[0.42758621]
[0.4137931 ]
[0.21213793]
[0.22758621]
[0.4137931 ]
[0.20689655]
[0.42413793]
[1. ]
[0.42758621]
[0.22758621]
[0.216 ]
[0.28965517]
[0.45475862]
[0.21213793]
[0.21655172]
[0.24848276]
[0.28965517]
[0.28965517]
[0.23772414]
[1. ]
[0.28965517]
[0.34496552]
[0.21213793]
[0.27606897]
[0.22758621]
[0.23772414]
[0.216 ]
[0.28965517]
[0.24365517]
[0.42758621]
[0.45475862]
[0.4137931 ]
[0.21655172]
[0.21213793]
[1. ]
[0.42758621]
[0.24848276]
[0.27606897]
[0.42413793]
[0.20689655]
[0.4137931 ]
[0.34496552]
[0.23772414]
[0.216 ]
[0.21655172]
[0.22758621]
[0.28965517]
[0.24365517]
[0.28965517]
[0.45475862]
[0.4137931 ]
[0.23772414]
[0.34496552]
[0.24365517]
[0.27606897]
[0.4137931 ]
[0.21213793]
[0.24365517]
[1. ]
[0.42758621]
[0.42413793]
[0.24848276]
[0.34496552]
[0.45475862]
[0.22758621]
[0.23772414]
[0.45475862]
[0.21655172]
[0.28965517]
[0.216 ]
[0.20689655]
[0.28965517]
[0.4137931 ]
[0.42413793]
[0.23772414]
[1. ]
[0.27606897]
[0.21213793]
[0.42413793]
[0.4137931 ]
[0.34496552]
[0.24848276]
[0.24365517]
[0.21655172]
[0.28965517]
[0.42413793]
[0.22758621]
[0.20689655]
[0.4137931 ]
[0.28965517]
[0.45475862]
[0.216 ]
[0.45475862]
[0.42758621]
[0.4137931 ]
[0.27606897]
[0.34496552]
[0.24848276]
[0.21655172]
[0.22758621]
[0.24365517]
[0.28965517]
[0.42413793]
[1. ]
[0.21213793]
[0.20689655]
[0.28965517]
[0.4137931 ]
[0.216 ]
[0.4137931 ]
[0.45475862]
[0.27606897]
[0.24365517]
[0.34496552]
[0.23772414]
[0.42758621]
[0.42413793]
[0.216 ]
[0.22758621]
[0.28965517]
[0.28965517]
[0.4137931 ]
[0.21213793]
[0.24848276]
[1. ]
[0.4137931 ]
[0.21655172]
[0.20689655]]
| MIT | ANN_moreFeatures.ipynb | F3rdixX/Basketball_BBL |
Data - Zusammenfügen der Spalten home_ids, gast_ids, zuschauer, Hallenkapazität, home_win | data=np.c_[transformed_home_ids,transformed_gast_ids,transformed_zuschauer,transformed_kap,dataset[:,2]]
print(data)
print(len(data[0])) | 39
| MIT | ANN_moreFeatures.ipynb | F3rdixX/Basketball_BBL |
Netz Modellierung | # Importing the Keras libraries and packages
from keras.models import Sequential
from keras.layers import Dense
# Initialising the ANN
regressor = Sequential()
# Adding the input layer and the first hidden layer
regressor.add(Dense(units = 38, kernel_initializer = 'uniform', activation = 'relu', input_shape = (38,)))
# Adding the second hidden layer
regressor.add(Dense(units = 18, kernel_initializer = 'uniform', activation = 'relu'))
# Adding the output layer
regressor.add(Dense(units = 1, kernel_initializer = 'uniform', activation = 'sigmoid'))
#Summary anzeigen
regressor.summary()
# Compiling the ANN - wie soll es lernen
regressor.compile(optimizer = 'adam', loss = 'mean_squared_error', metrics = ['accuracy'])#binary_crossentropy
# Fitting the ANN to the Training set
#input = data[:,0:4] output= (data[:,4]
history = regressor.fit(data[:,0:38], data[:,38], batch_size = 10, epochs = 100, validation_split = 0.1)
#
import matplotlib.pyplot as plt
handles = []
label, = plt.plot(history.history['acc'], label="acc")
handles.append(label)
label, = plt.plot(history.history['val_acc'], label="val_acc")
handles.append(label)
plt.title('Kostenfunktion')
plt.ylabel('Kosten')
plt.xlabel('Epochen')
plt.legend(handles=handles, loc='upper right')
figure = plt.gcf() # get current figure
figure.set_size_inches(8, 6) # um die größe des Plots anzupassen
#plt.savefig(pathpathpaht) # hiermit kannst das ding als auch als bild an dem angegebenen ort plus name ablegen
plt.show()
handles = []
label, = plt.plot(history.history['loss'], label="loss")
handles.append(label)
label, = plt.plot(history.history['val_loss'], label="val_loss")
handles.append(label)
plt.title('Kostenfunktion')
plt.ylabel('Kosten')
plt.xlabel('Epochen')
plt.legend(handles=handles, loc='upper right')
figure = plt.gcf() # get current figure
figure.set_size_inches(8, 6) # um die größe des Plots anzupassen
#plt.savefig(pathpathpaht) # hiermit kannst das ding als auch als bild an dem angegebenen ort plus name ablegen
plt.show()
import time as tm
import datetime
import pickle
def create_file_name():
ts = tm.time()
name = datetime.datetime.fromtimestamp(ts).strftime('%Y%m%d%H%M%S') + '_ann'
return name
path='./Netze/'
name_file= create_file_name()
with open(path + name_file + '.pkl', 'wb') as output:
ann_net = {'history_val_loss':history.history['val_loss'],'history_loss':history.history['loss']}
pickle.dump(ann_net, output) | _____no_output_____ | MIT | ANN_moreFeatures.ipynb | F3rdixX/Basketball_BBL |
This is a simple workbook used to generate the starting point for the weighting process. The tables in the workbook were in a strange format so I just did it manually here. | all_rows_post = pd.read_pickle('all_rows_post2.pkl')
all_rows_post = all_rows_post.groupby(['yearSale','Sector','Technology','HeatingEfficiency','CoolingEfficiency']).sum().reset_index()
all_rows_post = all_rows_post[all_rows_post['Sector'] == 'Residential']
key_techs = ['Heat Pump','Heat Pump - Ductless','Central Air Conditioning - Condenser','Gas Furnace']
all_rows_post = all_rows_post[all_rows_post['Technology'].isin(key_techs)]
all_rows_post = all_rows_post.drop(['Reported Quantity','Supplier','extrapped_qty','TOTAL'], axis=1)
all_rows_post = all_rows_post.replace('NA','UNDEFINED')
all_rows_post.to_pickle('sales_mix.pkl')
all_rows_post | _____no_output_____ | MIT | Generate Sales Mix Starting Point.ipynb | ischultz-cadeo/TO_43_Sales_Data |
Loading datas | datas = pd.read_csv('datasets/ISEAR.csv')
datas.head()
datas.columns
datas.drop('0', axis=1, inplace=True)
datas.size
datas.shape
column_name = datas.columns
datas = datas.rename(columns={column_name[0]: "Emotion",
column_name[1]: "Sentence"})
datas.head() | _____no_output_____ | FTL | notebooks/01.01_PL_sentiment_analysis_2020_05_18.ipynb | bhattbhuwan13/fuseai-training |
Adding $joy$ back to the dataset | missing_data = {"Emotion": column_name[0],
"Sentence": column_name[1]}
missing_data
datas = datas.append(missing_data, ignore_index=True) | _____no_output_____ | FTL | notebooks/01.01_PL_sentiment_analysis_2020_05_18.ipynb | bhattbhuwan13/fuseai-training |
Visualizing emotion ditribution | sns.catplot(kind='count', x='Emotion', data = datas)
plt.show()
datas.isna().sum()
datas.tail()
y = datas['Emotion']
y.head()
X = datas['Sentence']
X.head() | _____no_output_____ | FTL | notebooks/01.01_PL_sentiment_analysis_2020_05_18.ipynb | bhattbhuwan13/fuseai-training |
Converting all text to lovercase | Counter(y)
tfidf = TfidfVectorizer(tokenizer=nltk.word_tokenize, stop_words='english', min_df=3, ngram_range=(1, 2), lowercase=True)
tfidf.fit(X)
with open('tfidt_feature_vector.pkl', 'wb') as fp:
pickle.dump(tfidf, fp)
X = tfidf.transform(X)
tfidf.vocabulary_ | _____no_output_____ | FTL | notebooks/01.01_PL_sentiment_analysis_2020_05_18.ipynb | bhattbhuwan13/fuseai-training |
Making Models | bayes_classification = MultinomialNB()
dtree_classification = DecisionTreeClassifier()
Knn = KNeighborsClassifier()
def calculate_performance(test, pred, algorithm):
print(f'####For {algorithm}')
print(f'{classification_report(test, pred)}')
def train(X, y):
X_train, X_test, y_train, y_test = train_test_split(X, y)
bayes_classification.fit(X_train, y_train)
bayes_pred = bayes_classification.predict(X_test)
calculate_performance(y_test, bayes_pred, 'Naive Bayes')
pickle.dump(bayes_classification, open("Naive_bayes_model.pkl", 'wb'))
dtree_classification.fit(X_train, y_train)
dtree_pred = dtree_classification.predict(X_test)
calculate_performance(y_test, dtree_pred, 'Decision Tree')
Knn.fit(X_train, y_train)
knn_pred = Knn.predict(X_test)
print(knn_pred)
calculate_performance(y_test, dtree_pred, 'KNN')
train(X, y) | ####For Naive Bayes
precision recall f1-score support
anger 0.46 0.43 0.45 265
disgust 0.59 0.60 0.59 264
fear 0.62 0.66 0.64 260
guilt 0.51 0.48 0.50 262
joy 0.66 0.74 0.70 278
sadness 0.56 0.58 0.57 271
shame 0.53 0.47 0.49 262
accuracy 0.57 1862
macro avg 0.56 0.57 0.56 1862
weighted avg 0.56 0.57 0.56 1862
####For Decision Tree
precision recall f1-score support
anger 0.36 0.33 0.34 265
disgust 0.41 0.47 0.44 264
fear 0.56 0.56 0.56 260
guilt 0.39 0.36 0.37 262
joy 0.54 0.57 0.56 278
sadness 0.54 0.46 0.50 271
shame 0.35 0.39 0.37 262
accuracy 0.45 1862
macro avg 0.45 0.45 0.45 1862
weighted avg 0.45 0.45 0.45 1862
['guilt' 'anger' 'shame' ... 'anger' 'anger' 'sadness']
####For KNN
precision recall f1-score support
anger 0.36 0.33 0.34 265
disgust 0.41 0.47 0.44 264
fear 0.56 0.56 0.56 260
guilt 0.39 0.36 0.37 262
joy 0.54 0.57 0.56 278
sadness 0.54 0.46 0.50 271
shame 0.35 0.39 0.37 262
accuracy 0.45 1862
macro avg 0.45 0.45 0.45 1862
weighted avg 0.45 0.45 0.45 1862
| FTL | notebooks/01.01_PL_sentiment_analysis_2020_05_18.ipynb | bhattbhuwan13/fuseai-training |
Python 数据类型> Guido 对语言设计美学的深入理解让人震惊。我认识不少很不错的编程语言设计者,他们设计出来的东西确实很精彩,但是从来都不会有用户。Guido 知道如何在理论上做出一定妥协,设计出来的语言让使用者觉得如沐春风,这真是不可多得。 > ——Jim Hugunin > Jython 的作者,AspectJ 的作者之一,.NET DLR 架构师Python 最好的品质之一是**一致性**:你可以轻松理解 Python 语言,并通过 Python 的语言特性在类上定义**规范的接口**,来支持 Python 的核心语言特性,从而写出具有“Python 风格”的对象。 Python 解释器在碰到特殊的句法时,会使用特殊方法(我们称之为魔术方法)去激活一些基本的对象操作。> `__getitem__` 以双下划线开头的特殊方法,称为 dunder-getitem。特殊方法也称为双下方法(dunder-method)如 `my_c[key]` 语句执行时,就会调用 `my_c.__getitem__` 函数。这些特殊方法名能让你自己的对象实现和支持一下的语言构架,并与之交互:* 迭代* 集合类* 属性访问* 运算符重载* 函数和方法的调用* 对象的创建和销毁* 字符串表示形式和格式化* 管理上下文(即 `with` 块) 实现一个 Pythonic 的牌组 | # 通过实现魔术方法,来让内置函数支持你的自定义对象
# https://github.com/fluentpython/example-code/blob/master/01-data-model/frenchdeck.py
import collections
import random
Card = collections.namedtuple('Card', ['rank', 'suit'])
class FrenchDeck:
ranks = [str(n) for n in range(2, 11)] + list('JQKA')
suits = 'spades diamonds clubs hearts'.split()
def __init__(self):
self._cards = [Card(rank, suit) for suit in self.suits
for rank in self.ranks]
def __len__(self):
return len(self._cards)
def __getitem__(self, position):
return self._cards[position]
deck = FrenchDeck() | _____no_output_____ | MIT | 01-data-model/01-data-model.ipynb | yuechuanx/fluent-python-code-and-notes |
可以容易地获得一个纸牌对象 | beer_card = Card('7', 'diamonds')
print(beer_card) | _____no_output_____ | MIT | 01-data-model/01-data-model.ipynb | yuechuanx/fluent-python-code-and-notes |
和标准 Python 集合类型一样,使用 `len()` 查看一叠纸牌有多少张 | deck = FrenchDeck()
# 实现 __len__ 以支持下标操作
print(len(deck)) | _____no_output_____ | MIT | 01-data-model/01-data-model.ipynb | yuechuanx/fluent-python-code-and-notes |
可选取特定一张纸牌,这是由 `__getitem__` 方法提供的 | # 实现 __getitem__ 以支持下标操作
print(deck[1])
print(deck[5::13]) | _____no_output_____ | MIT | 01-data-model/01-data-model.ipynb | yuechuanx/fluent-python-code-and-notes |
随机抽取一张纸牌,使用 python 内置函数 `random.choice` | from random import choice
# 可以多运行几次观察
choice(deck) | _____no_output_____ | MIT | 01-data-model/01-data-model.ipynb | yuechuanx/fluent-python-code-and-notes |
实现特殊方法的两个好处:- 对于标准操作有固定命名- 更方便利用 Python 标准库 `__getitem__` 方法把 [] 操作交给了 `self._cards` 列表,deck 类自动支持切片操作 | deck[12::13]
deck[:3] | _____no_output_____ | MIT | 01-data-model/01-data-model.ipynb | yuechuanx/fluent-python-code-and-notes |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.