markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
__Exercise 2__ | import nltk
from nltk.corpus import names
import random
names = ([(name, 'male') for name in names.words('male.txt')] + [(name, 'female') for name in names.words('female.txt')])
random.shuffle(names)
test, devtest, training = names[:500], names[500:1000], names[1000:]
def gender_features1(name):
features = {}
features["firstletter"] = name[0].lower()
features["lastletter"] = name[-1].lower()
for letter in 'abcdefghijklmnopqrstuvwxyz':
features["count(%s)" % letter] = name.lower().count(letter)
features["has(%s)" % letter] = (letter in name.lower())
features["suffix2"] = name[-2:].lower()
return features
train_set = [(gender_features1(n), g) for (n,g) in training]
devtest_set = [(gender_features1(n), g) for (n,g) in devtest]
classifier = nltk.NaiveBayesClassifier.train(train_set)
print nltk.classify.accuracy(classifier, devtest_set)
def error_analysis(gender_features):
errors = []
for (name, tag) in devtest:
guess = classifier.classify(gender_features(name))
if guess != tag:
errors.append((tag, guess, name))
print 'no. of errors: ', len(errors)
for (tag, guess, name) in sorted(errors): # doctest: +ELLIPSIS +NORMALIZE_WHITESPACE
print 'correct=%-8s guess=%-8s name=%-30s' % (tag, guess, name)
error_analysis(gender_features1)
def gender_features2(name):
features = {}
features["firstletter"] = name[0].lower()
features["lastletter"] = name[-1].lower()
for letter in 'abcdefghijklmnopqrstuvwxyz':
features["count(%s)" % letter] = name.lower().count(letter)
features["has(%s)" % letter] = (letter in name.lower())
features["suffix2"] = name[-2:].lower()
features["suffix3"] = name[-3:].lower()
return features
train_set = [(gender_features2(n), g) for (n,g) in training]
devtest_set = [(gender_features2(n), g) for (n,g) in devtest]
classifier = nltk.NaiveBayesClassifier.train(train_set)
print nltk.classify.accuracy(classifier, devtest_set)
error_analysis(gender_features2)
def gender_features3(name):
features = {}
features["firstletter"] = name[0].lower()
features["lastletter"] = name[-1].lower()
for letter in 'abcdefghijklmnopqrstuvwxyz':
features["count(%s)" % letter] = name.lower().count(letter)
features["has(%s)" % letter] = (letter in name.lower())
features["suffix2"] = name[-2:].lower()
features["suffix3"] = name[-3:].lower()
features["prefix3"] = name[:3].lower()
return features
train_set = [(gender_features3(n), g) for (n,g) in training]
devtest_set = [(gender_features3(n), g) for (n,g) in devtest]
classifier = nltk.NaiveBayesClassifier.train(train_set)
print nltk.classify.accuracy(classifier, devtest_set)
error_analysis(gender_features3)
def gender_features4(name):
features = {}
features["firstletter"] = name[0].lower()
features["lastletter"] = name[-1].lower()
for letter in 'abcdefghijklmnopqrstuvwxyz':
features["count(%s)" % letter] = name.lower().count(letter)
features["has(%s)" % letter] = (letter in name.lower())
features["suffix2"] = name[-2:].lower()
features["suffix3"] = name[-3:].lower()
features["prefix3"] = name[:3].lower()
features["num_vowels"] = len([letter for letter in name if letter in 'aeiouy'])
return features
train_set = [(gender_features4(n), g) for (n,g) in training]
devtest_set = [(gender_features4(n), g) for (n,g) in devtest]
classifier = nltk.NaiveBayesClassifier.train(train_set)
print nltk.classify.accuracy(classifier, devtest_set)
# final performance test:
test_set = [(gender_features4(n), g) for (n,g) in test]
print nltk.classify.accuracy(classifier, test_set)
# performance slightly worse than in dev-test -> features reflect some idiosyncracies of dev-test | _____no_output_____ | MIT | chapter_6_exercises.ipynb | JuliaNeumann/nltk_book_exercises |
__Exercise 3)__ | from nltk.corpus import senseval
instances = senseval.instances('serve.pos')
size = int(len(instances) * 0.1)
training, test = instances[size:], instances[:size]
training[0]
def sense_features(instance):
features = {}
features["word-type"] = instance.word
features["word-tag"] = instance.context[instance.position][1]
features["prev-word"] = instance.context[instance.position-1][0]
features["prev-word-tag"] = instance.context[instance.position-1][1]
features["next-word"] = instance.context[instance.position+1][0]
features["next-word-tag"] = instance.context[instance.position+1][1]
return features
train_set = [(sense_features(instance), instance.senses) for instance in training]
test_set = [(sense_features(instance), instance.senses) for instance in test]
classifier = nltk.NaiveBayesClassifier.train(train_set)
print nltk.classify.accuracy(classifier, test_set) | 0.807780320366
| MIT | chapter_6_exercises.ipynb | JuliaNeumann/nltk_book_exercises |
__Exercise 4)__ | from nltk.corpus import movie_reviews
documents = [(list(movie_reviews.words(fileid)), category) for category in movie_reviews.categories() for fileid in movie_reviews.fileids(category)]
random.shuffle(documents)
all_words = nltk.FreqDist(w.lower() for w in movie_reviews.words())
word_features = all_words.keys()[:2000]
def document_features(document):
document_words = set(document)
features = {}
for word in word_features:
features['contains(%s)' % word] = (word in document_words)
return features
featuresets = [(document_features(d), c) for (d,c) in documents]
train_set, test_set = featuresets[100:], featuresets[:100]
classifier = nltk.NaiveBayesClassifier.train(train_set)
print nltk.classify.accuracy(classifier, test_set)
classifier.show_most_informative_features(30)
# most of them already indicate some judgment in themselves ('ugh', 'mediocrity') or belong to typical phrases that
# indicate one direction of judgement ('understands' -> '... understands how to create atmosphere' or something like that)
# some seem to be names of actors etc. which tend to be judged one direction or the other
# surprising -> '33', 'wires' | _____no_output_____ | MIT | chapter_6_exercises.ipynb | JuliaNeumann/nltk_book_exercises |
def isBalanced(S):
stack = []
brac = {"{": "}", "[": "]", "(" : ")"}
for i in S:
if i in brac.keys():
stack.append(i)
elif i in brac.values():
if i == brac.get(stack[-1]):
stack.pop()
else:
stack.append(i)
return "Success" if not stack else S.index(stack.pop()) + 1
S = input()
print(isBalanced(S)) | {[}
3
| Unlicense | Lab7_1.ipynb | hocthucv/bt |
|
Table of Contents1 Figure 1: Introduction2 Figure 2: Model Performance2.1 Load KaiABC model2.1.1 Estimate Errors2.2 Plot KaiABC model with $N_{\rm eff}$3 Figure 3: Plot all data together4 Figure 4: New Model5 Supplemental Plots | import numpy as np
import matplotlib.pyplot as plt
from decimal import Decimal
import pandas as pd
import pickle
from matplotlib.backends import backend_pdf as bpdf
from kaic_analysis.scripts import FirstPassage, RunModel, Current, StateData, FindParam, LoadExperiment, PlotExperiment, EntropyRate
from kaic_analysis.toymodel import SimulateClockKinetic
import os
from sklearn.decomposition import PCA
import seaborn as sns
import scipy.interpolate as interpolate
import bootstrapped.bootstrap as bs
import bootstrapped.stats_functions as bs_stats
import scipy.optimize as opt
%matplotlib inline
def var(values, axis=1):
'''Returns the variance of each row of a matrix'''
return np.var(np.asmatrix(values), axis=axis).A1
def compute_maxeig(A,C,N):
K0 = np.asarray([[-C*N*(np.sin(2*np.pi*(i-1-j)/N)-np.sin(2*np.pi*(i-j)/N))*(1-np.exp(-A/N)) for j in range(N)] for i in range (N)])
K1 = np.diag(np.ones(N-1),k=1)*N - np.eye(N)*N*(1+np.exp(-A/N)) + np.diag(np.ones(N-1),k=-1)*N*np.exp(-A/N)
K = K0 + K1
Keig = np.linalg.eig(K)[0]
max_ind = np.argmax(np.real(Keig))
return np.real(Keig[max_ind])
def compute_maxeig_imag(A,C,N):
K0 = np.asarray([[-C*N*(np.sin(2*np.pi*(i-1-j)/N)-np.sin(2*np.pi*(i-j)/N))*(1-np.exp(-A/N)) for j in range(N)] for i in range (N)])
K1 = np.diag(np.ones(N-1),k=1)*N - np.eye(N)*N*(1+np.exp(-A/N)) + np.diag(np.ones(N-1),k=-1)*N*np.exp(-A/N)
K = K0 + K1
Keig = np.linalg.eig(K)[0]
max_ind = np.argmax(np.real(Keig))
return np.imag(Keig[max_ind]) | _____no_output_____ | MIT | data/Generate Plots.ipynb | robertvsiii/kaic-analysis |
Figure 1: Introduction | fig,ax = plt.subplots(figsize=(3.25,1.75))
fig.subplots_adjust(left=0.17,bottom=0.25,right=0.95)
code_folder = '../KMC_KaiC_rev2'
data_low=RunModel(folder=code_folder,paramdict={'volume':1,'sample_cnt':1e4,'tequ':50,'rnd_seed':np.random.randint(1e6),'ATPfrac':0.45},name='data_low')
data_WT=RunModel(folder=code_folder,paramdict={'volume':1,'sample_cnt':3e5,'tequ':50,'rnd_seed':np.random.randint(1e6)},name='data_WT')
os.chdir('../data')
data_WT.index = data_WT.index-data_WT.index[0]
(data_WT['pT']/6).plot(ax=ax,color='k',legend=False)
ax.set_xlim((0,200))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.set_xlabel('Time (hours)')
ax.set_ylabel(r'$f_T$')
pdf = bpdf.PdfPages('Plots/timeseries_kaic.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
nT = data_WT['pT'].values*360 #volume = 1 corresponds to 360 hexamers
nS = data_WT['pS'].values*360
samp = np.arange(100000,300000,20,dtype=int)
fig,ax = plt.subplots(figsize=(3,3))
fig.subplots_adjust(left=0.17,bottom=0.25,right=0.95)
ax.plot(nT[samp],nS[samp])
ax.set_aspect('equal', 'box')
ax.set_xlabel('Number of phosphorylated threonines')
ax.set_ylabel('Number of phosphorylated serines')
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
pdf = bpdf.PdfPages('Plots/limit.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
fig,ax = plt.subplots(figsize=(3,3))
fig.subplots_adjust(left=0.17,bottom=0.25,right=0.9)
ax.plot(nT[100000:],nS[100000:])
ax.set_xlim((500,525))
ax.set_ylim((400,425))
ax.set_aspect('equal', 'box')
ax.set_xlabel('Number of phosphorylated threonines')
ax.set_ylabel('Number of phosphorylated serines')
ax.grid(True)
pdf = bpdf.PdfPages('Plots/zoom.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
param_name = 'ATPfrac'
run_nums = list(range(17,22))
fig,ax = plt.subplots(figsize=(3.25,1.75))
fig.subplots_adjust(left=0.17,bottom=0.25,right=0.95)
data = LoadExperiment(param_name,run_nums,date='2018-08-24',folder='kaic_data')
bins = np.linspace(0,150,150)
name = 'ATPfrac = 0.99999998477'
for Ncyc in range(1,6):
ax.hist(FirstPassage(data[2][name],Ncyc=Ncyc),bins=bins,density=True,alpha=0.5)
ax.set_xlim((0,150))
ax.set_xlabel('Time (hours)')
ax.set_ylabel('Fraction of runs')
pdf = bpdf.PdfPages('Plots/hist.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
Ncyclist = np.arange(1,30)
fig,ax = plt.subplots(figsize=(3.25,1.75))
fig.subplots_adjust(left=0.17,bottom=0.25,right=0.95)
ax.set_xlabel(r'$n$')
ax.set_ylabel(r'${\rm var}(\tau_n)$')
vartau = []
meantau = []
varw=[]
meanw=[]
for N in Ncyclist:
taus = np.asarray(FirstPassage(data[2][name],Ncyc=N))
bs_mean = bs.bootstrap(taus,stat_func=bs_stats.mean)
bs_var = bs.bootstrap(taus,stat_func=var)
vartau.append(bs_var.value)
meantau.append(bs_mean.value)
varw.append(2./(bs_var.upper_bound-bs_var.lower_bound))
meanw.append(2./(bs_mean.upper_bound-bs_mean.lower_bound))
varw = np.asarray(varw)
meanw=np.asarray(meanw)
vartau = np.asarray(vartau)
meantau = np.asarray(meantau)
[slope, intercept], cov = np.polyfit(Ncyclist,vartau,1,w=varw,cov=True)
ax.errorbar(Ncyclist,np.asarray(vartau),yerr=1/np.asarray(varw),color='k',alpha=0.5)
ax.plot(Ncyclist,intercept+slope*Ncyclist,color='k')
pdf = bpdf.PdfPages('Plots/D.pdf')
pdf.savefig(fig)
pdf.close()
plt.show() | _____no_output_____ | MIT | data/Generate Plots.ipynb | robertvsiii/kaic-analysis |
Figure 2: Model Performance Load KaiABC model | param_name = 'ATPfrac'
run_nums = list(range(1,13))
data = LoadExperiment(param_name,run_nums,date='2018-08-23',folder='kaic_data')[2]
run_nums = list(range(13,22))
data.update(LoadExperiment(param_name,run_nums,date='2018-08-24',folder='kaic_data')[2])
keylist = list(data.keys())
ATPfracs = [Decimal(keylist[j].split('=')[1]) for j in range(len(keylist))]
ATPfracs.sort()
namelist = [param_name+' = '+str(ATPfracs[j]) for j in range(len(ATPfracs))]
Ncyclist = np.arange(1,30)
D0 = []
D0_err = []
D0_2 = []
D0_2_err = []
T = []
vary = []
colors = sns.color_palette("RdBu_r",21)
fig,axs = plt.subplots(5,4,figsize=(6.5,9),sharex=True,sharey=True)
for k in range(4):
axs[4,k].set_xlabel('Number of Cycles')
axs[2,0].set_ylabel('Variance of Completion Time')
axs = axs.reshape(-1)
fig.subplots_adjust(hspace=0.4)
fig2,ax2 = plt.subplots(5,4,figsize=(6.5,9),sharex=True,sharey=True)
for k in range(4):
ax2[4,k].set_xlabel('Number of Cycles')
ax2[2,0].set_ylabel('Variance of Variance of Completion Time')
ax2 = ax2.reshape(-1)
fig2.subplots_adjust(hspace=0.4)
k = 0
for name in namelist[1:]:
vartau = []
meantau = []
varw=[]
meanw=[]
for N in Ncyclist:
taus = np.asarray(FirstPassage(data[name],Ncyc=N))
bs_mean = bs.bootstrap(taus,stat_func=bs_stats.mean,alpha=0.36)
bs_var = bs.bootstrap(taus,stat_func=var,alpha=0.36)
vartau.append(bs_var.value)
meantau.append(bs_mean.value)
varw.append(2./(bs_var.upper_bound-bs_var.lower_bound))
meanw.append(2./(bs_mean.upper_bound-bs_mean.lower_bound))
varw = np.asarray(varw)
meanw=np.asarray(meanw)
vartau = np.asarray(vartau)
meantau = np.asarray(meantau)
ax2[k].plot(Ncyclist,1./varw,color = colors[0])
slope, intercept = np.polyfit(Ncyclist,1./varw,1)
ax2[k].plot(Ncyclist,intercept+slope*Ncyclist,color=colors[-1])
vary.append(slope**2)
slope, intercept = np.polyfit(Ncyclist,meantau,1,w=meanw)
T.append(slope)
[slope, intercept], cov = np.polyfit(Ncyclist,vartau,1,w=varw,cov=True)
D0.append(slope)
D0_2.append(vartau[-1]/Ncyclist[-1])
cov = np.linalg.inv(np.asarray([[2*np.sum(varw**2*Ncyclist**2),2*np.sum(varw**2*Ncyclist)],
[2*np.sum(varw**2*Ncyclist),2*np.sum(varw**2)]]))
D0_err.append(np.sqrt(cov[0,0]))
D0_2_err.append(np.sqrt(1./varw[-1]))
axs[k].errorbar(Ncyclist,np.asarray(vartau),yerr=1/np.asarray(varw),color=colors[0])
axs[k].plot(Ncyclist,intercept+slope*Ncyclist,color=colors[-1])
axs[k].set_title(name[:14])
k+=1
pdf = bpdf.PdfPages('Plots/KaiC_fits.pdf')
pdf.savefig(fig)
pdf.close()
pdf = bpdf.PdfPages('Plots/KaiC_fits_var.pdf')
pdf.savefig(fig2)
pdf.close()
plt.show()
T = np.asarray(T)
D = np.asarray(D0)/T
D_err = np.asarray(D0_err)/T
D2 = np.asarray(D0_2)/T
D2_err = np.asarray(D0_2_err)/T
D3_err = np.sqrt(np.asarray(vary))/T
run_nums = list(range(1,13))
data = LoadExperiment(param_name,run_nums,date='2018-08-23',folder='kaic_data')[1]
run_nums = list(range(13,22))
data = data.join(LoadExperiment(param_name,run_nums,date='2018-08-24',folder='kaic_data')[1])
Scyc = data[namelist].values[0][1:]*T
with open('ModelData.dat', 'wb') as f:
pickle.dump([T, D, D_err, Scyc],f) | _____no_output_____ | MIT | data/Generate Plots.ipynb | robertvsiii/kaic-analysis |
Estimate Errors The graphs of the bootstrap error versus number of cycles indicate that the standard deviation of the completion time increases linearly with the number of cycles. This is what we would expect to happen if each run of the experiment (making the same number of trajectories and estimating the variance for each number of cycles) produces a slope $D_0+y$, where $y$ has mean 0 and variance $\sigma_y^2$, and is fixed for each iteration of the simulation.Specifically, we have\begin{align}{\rm var}(\tau_n) &= (D_0 + y)N_{\rm cyc}\\\sqrt{{\rm var}({\rm var}(\tau_n))} &= \sigma_y N_{\rm cyc}.\end{align}Under this noise model, the uncertainty in the slope is simply $\sigma_y$. Plot KaiABC model with $N_{\rm eff}$ | with open('ModelData.dat','rb') as f:
[T,D,D_err,Scyc] = pickle.load(f)
DelWmin = 2000
DelWmax = 3100
DelWvec = np.exp(np.linspace(np.log(DelWmin),np.log(DelWmax),5000))
M = 180*2
Neff = 1.1
Ncoh = T/D
Ncoh_err = (T/D**2)*D3_err
k=0
Scyc = Scyc[:-1]
Ncoh = Ncoh[:-1]
Ncoh_err = Ncoh_err[:-1]
fig,ax=plt.subplots(figsize=(3.5,3))
fig.subplots_adjust(left=0.2,bottom=0.2)
ax.plot(Scyc/M,Ncoh/M,label='KaiABC Model')
ax.errorbar(Scyc/M,Ncoh/M,yerr=Ncoh_err/M,linestyle='',capsize=2)
ax.plot([DelWmin,DelWmax],[Neff,Neff],'k--',label = r'$N_{\rm eff}/M = $'+str(Neff))
plt.legend(loc=4)
ax.set_xlabel(r'Entropy Production per Cycle $\Delta S/M$')
ax.set_ylabel(r'Number of Coherent Cycles $\mathcal{N}/M$')
pdf = bpdf.PdfPages('Plots/Figure2.pdf')
pdf.savefig(fig)
pdf.close()
plt.show() | _____no_output_____ | MIT | data/Generate Plots.ipynb | robertvsiii/kaic-analysis |
Figure 3: Plot all data together Empirical curve:\begin{align}\frac{V}{\mathcal{N}} - C &= W_0 (W-W_c)^\alpha\\\frac{\mathcal{N}}{V} &= \frac{1}{C+ W_0(W-W_c)^\alpha}\end{align} | with open('ModelData.dat','rb') as f:
[T,D,D_err,Scyc] = pickle.load(f)
def CaoN(DelW,params = {}):
N = (params['C']+params['W0']*(DelW-params['Wc'])**params['alpha'])**(-1)
N[np.where(DelW<params['Wc'])[0]] = np.nan
return N
#For comparing experiment data:
VKai = 3e13 #Convert volume to single hexamers (assuming 3.4 uM concentration of monomers, and 100 uL wells)
al = 1/(2*np.pi**2)
#Parameters from Cao2015
paramlist = {'Activator-Inhibitor':
{'C':0.6,
'W0':380,
'Wc':360,
'alpha':-0.99},
'AI Envelope':
{'C':0.36,
'W0':194,
'Wc':400,
'alpha':-1},
'Repressilator':
{'Wc':1.75,
'W0':25.9,
'alpha':-1.1,
'C':0.4},
'Brusselator':
{'Wc':100.4,
'W0':846,
'alpha':-1.0,
'C':0.5},
'Glycolysis':
{'Wc':80.5,
'W0':151.4,
'alpha':-1.1,
'C':0.5},
'KaiABC Experiment':
{'Wc':10.6*16*6,
'W0':0.28*16*6*VKai*al,
'alpha':-1.0,
'C':0.04*VKai*al}}
fig,ax=plt.subplots(figsize=(3.25,3))
fig.subplots_adjust(bottom=0.15,left=0.2,right=0.95,top=0.95)
del paramlist['AI Envelope']
del paramlist['KaiABC Experiment']
DelWmin = 1
DelWmax = 0.6*10000
DelWvec = np.exp(np.linspace(np.log(DelWmin),np.log(DelWmax),5000))
ax.plot(DelWvec,DelWvec/2,'k--',label='Thermodynamic Bound')
for item in ['Repressilator','Glycolysis','Activator-Inhibitor','Brusselator']:
ax.plot(DelWvec,CaoN(DelWvec,params=paramlist[item]),label=item)
N = 1
M = 180*2
Neff = 1.2*N*M
Ncoh = T/D
Ncoh_err = (T/D**2)*D_err
k=0
ax.plot(Scyc[:-1]/M,Ncoh[:-1]/M,label='KaiABC Model')
plt.legend(loc=1,fontsize=8)
ax.set_ylim((4e-1,2e1))
ax.set_xlim((10,3100))
ax.set_yscale('log')
ax.set_xscale('log')
ax.set_xlabel(r'$\Delta S/M$')
ax.set_ylabel(r'$\mathcal{N}/M$')
pdf = bpdf.PdfPages('Plots/Figure3.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
| /anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:5: RuntimeWarning: invalid value encountered in power
"""
| MIT | data/Generate Plots.ipynb | robertvsiii/kaic-analysis |
Figure 4: New Model | date = '2018-08-23'
date2 = '2018-08-24'
date3 = '2018-08-25'
D0 = []
D0_err = []
T = []
Scyc = []
sigy =[]
colors = sns.color_palette("RdBu_r",20)
k = 0
for expt_number in range(3):
low = 18*expt_number+1
high = 18*(expt_number+1)
if expt_number == 2: #Skip simulations that failed
low = 48
for run_number in range(low,high):
try:
t = pd.read_csv('toy_data/t_'+date+'_'+str(run_number)+'.csv',header=None)
t = t - t.loc[0]
data = pd.read_csv('toy_data/data_'+date+'_'+str(run_number)+'.csv',index_col=0)
except:
try:
t = pd.read_csv('toy_data/t_'+date2+'_'+str(run_number)+'.csv',header=None)
t = t - t.loc[0]
data = pd.read_csv('toy_data/data_'+date2+'_'+str(run_number)+'.csv',index_col=0)
except:
t = pd.read_csv('toy_data/t_'+date3+'_'+str(run_number)+'.csv',header=None)
t = t - t.loc[0]
data = pd.read_csv('toy_data/data_'+date3+'_'+str(run_number)+'.csv',index_col=0)
fig,axs = plt.subplots(2,figsize=(4,8))
fig2,axs2 = plt.subplots(2,figsize=(4,8))
vartau = []
meantau = []
varw=[]
meanw=[]
sigy=[]
for N in t.index:
taus = t.loc[N].values
bs_mean = bs.bootstrap(taus,stat_func=bs_stats.mean)
bs_var = bs.bootstrap(taus,stat_func=var)
vartau.append(bs_var.value)
meantau.append(bs_mean.value)
varw.append(2./(bs_var.upper_bound-bs_var.lower_bound))
meanw.append(2./(bs_mean.upper_bound-bs_mean.lower_bound))
varw = np.asarray(varw)
meanw=np.asarray(meanw)
vartau = np.asarray(vartau)
meantau = np.asarray(meantau)
usable = np.where(~np.isnan(meantau))[0]
usable = usable[1:]
try:
slope, intercept = np.polyfit(t.index.values[usable],meantau[usable],1,w=meanw[usable])
T.append(slope)
axs[0].set_title('A = '+str(data['A'].loc[0]))
axs[0].errorbar(t.index,meantau,yerr=1/meanw,color=colors[k])
axs[0].plot(t.index,intercept+slope*t.index,color=colors[k])
axs[0].set_xlabel('Number of Cycles')
axs[0].set_ylabel('Mean completion time')
axs2[0].plot(t.index,1./varw)
[slope, intercept], cov = np.polyfit(t.index.values[usable],1./varw[usable],1,cov=True)
sigy.append(slope)
[slope, intercept], cov = np.polyfit(t.index.values[usable],vartau[usable],1,w=varw[usable],cov=True)
D0.append(slope)
cov = np.linalg.inv(np.asarray([[np.nansum(varw[usable]**2*t.index.values[usable]**2),np.nansum(varw[usable]**2*t.index.values[usable])],
[np.nansum(varw[usable]**2*t.index.values[usable]),np.nansum(varw[usable]**2)]]))
D0_err.append(np.sqrt(cov[0,0]))
axs[1].errorbar(t.index,np.asarray(vartau),yerr=1/np.asarray(varw),color=colors[k])
axs[1].plot(t.index,intercept+slope*t.index,color=colors[k])
axs[1].set_xlabel('Number of Cycles')
axs[1].set_ylabel('Variance of completion time')
Scyc.append(data['Sdot'].mean()*T[-1])
k+=1
except:
print(str(run_number)+' failed!')
try:
del T[k]
except:
e = 1
try:
del D0[k]
except:
e = 1
plt.show()
T = np.asarray(T)
D = np.asarray(D0)/T
D_err = np.asarray(sigy)/T
Scyc = np.asarray(Scyc)
with open('ToyData_'+str(expt_number)+'_2.dat', 'wb') as f:
pickle.dump([T, D, D_err, Scyc],f)
fig,ax=plt.subplots(figsize=(3.25,3))
fig.subplots_adjust(bottom=0.15,left=0.21,right=0.95,top=0.95)
colors = sns.color_palette()
xvec = np.exp(np.linspace(-2,np.log(30),120))
Nlist = [3,10,50]
M = 100
colors = sns.color_palette()
k=0
for n in [2,1,0]:
with open('ToyData_'+str(n)+'_2.dat', 'rb') as f:
[T, D, D_err, Scyc] = pickle.load(f)
N = Nlist[n]
Neff = 1.3*N*M
Ncoh = T/D
Ncoh_err = 2*(T/D**2)*D_err
ax.errorbar(Scyc/M,Ncoh/M,yerr=Ncoh_err/M,color=colors[k],label='N = '+str(N))
k+=1
ax.plot(xvec,xvec/2,'--',color='k',label='Thermodynamic Bound')
plt.legend(loc=2,fontsize=10)
ax.set_xlim((0,30))
ax.set_ylim((0,20))
ax.set_xlabel(r'$\Delta S/M$',fontsize=14)
ax.set_ylabel(r'$\mathcal{N}/M$',fontsize=14)
pdf = bpdf.PdfPages('Plots/toymodel.pdf')
pdf.savefig(fig)
pdf.close()
plt.show() | _____no_output_____ | MIT | data/Generate Plots.ipynb | robertvsiii/kaic-analysis |
Supplemental Plots | data = []
N = 6
M = 100
C = 5
kwargs = {'tmax':4,'nsteps':1,'N':N,'M':M,'A':4.4,'C':C}
out1 = SimulateClockKinetic(**kwargs)
kwargs = {'tmax':4,'nsteps':1,'N':N,'M':M,'A':9.529,'C':C}
out2 = SimulateClockKinetic(**kwargs)
fig,ax=plt.subplots()
ax.plot(out1['t'],out1['f'][:,0], label=r'$\dot{S} = 160\,k_B/{\rm hr}$')
ax.plot(out2['t'],out2['f'][:,0], label=r'$\dot{S} = 880\,k_B/{\rm hr}$')
plt.legend()
ax.set_xlabel('Time (hours)')
ax.set_ylabel('Fraction in state 1')
pdf = bpdf.PdfPages('Plots/timeseries_new.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
fig,ax=plt.subplots()
N = 6
M = 100
C = 5
skip = 1
model = PCA(n_components=2).fit(out1['f'])
f1 = model.transform(out1['f'])
ax.plot(f1[np.arange(0,len(f1),skip),0],f1[np.arange(0,len(f1),skip),1],label=r'$\dot{S} = 160\,k_B/{\rm hr}$')
skip = 1
model = PCA(n_components=2).fit(out2['f'])
f2 = model.transform(out2['f'])
ax.plot(f2[np.arange(0,len(f2),skip),0],f2[np.arange(0,len(f2),skip),1],label=r'$\dot{S} = 880\,k_B/{\rm hr}$')
ax.plot([0],[0],'ko',markersize=8)
ax.plot([0,0],[0,0.6],'k',linewidth=2)
ax.set_ylim((-0.5,0.55))
plt.legend(loc=1)
ax.set_xlabel('PCA 1')
ax.set_ylabel('PCA 2')
pdf = bpdf.PdfPages('Plots/phase_new.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
Cvec = np.arange(2,7)
Acvec = {}
period = {}
Nvec = np.arange(3,50)
for C in Cvec:
Acvec.update({'C = '+str(C): []})
period.update({'C = '+str(C): []})
for N in Nvec:
try:
Acvec['C = '+str(C)].append(opt.brentq(compute_maxeig,0,2,args=(C,N)))
period['C = '+str(C)].append(2*np.pi/compute_maxeig_imag(Acvec['C = '+str(C)][-1],C,N))
except:
Acvec['C = '+str(C)].append(np.nan)
period['C = '+str(C)].append(np.nan)
with open('Ac2.dat','wb') as f:
pickle.dump([Nvec,Acvec,period],f)
with open('Ac2.dat','rb') as f:
Nvec,Acvec,period = pickle.load(f)
fig,ax=plt.subplots(figsize=(2.5,2.5))
fig.subplots_adjust(left=0.22,bottom=0.22)
for item in Acvec.keys():
ax.plot(Nvec,Acvec[item],label=item[-1])
plt.legend(title = r'$C$')
ax.set_xlabel(r'$N$')
ax.set_ylabel(r'$A_c$')
ax.set_ylim((0,1))
pdf = bpdf.PdfPages('Plots/Figure6b.pdf')
pdf.savefig(fig)
pdf.close()
plt.show()
fig,ax=plt.subplots(figsize=(3.5,3))
for item in Acvec.keys():
ax.plot(Nvec,period[item],label=item[-1])
plt.legend(title = r'$C$')
ax.set_xlabel(r'$N$')
ax.set_ylabel(r'Period at critical point')
pdf = bpdf.PdfPages('Plots/Figure5c.pdf')
pdf.savefig(fig)
pdf.close()
plt.show() | _____no_output_____ | MIT | data/Generate Plots.ipynb | robertvsiii/kaic-analysis |
Note* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps. | # Dependencies and Setup
import pandas as pd
# File to Load (Remember to Change These)
school_data_to_load = "Resources/schools_complete.csv"
student_data_to_load = "Resources/students_complete.csv"
# Read School and Student Data File and store into Pandas Data Frames
school_data = pd.read_csv(school_data_to_load)
student_data = pd.read_csv(student_data_to_load)
# Combine the data into a single dataset
school_data_complete = pd.merge(student_data, school_data, how="left", on=["school_name", "school_name"])
school_data_complete | _____no_output_____ | ADSL | PyCitySchools/Resources/PyCitySchools_starter_unsolved.ipynb | fkokro/Pandas---Challenge |
District Summary* Calculate the total number of schools* Calculate the total number of students* Calculate the total budget* Calculate the average math score * Calculate the average reading score* Calculate the overall passing rate (overall average score), i.e. (avg. math score + avg. reading score)/2* Calculate the percentage of students with a passing math score (70 or greater)* Calculate the percentage of students with a passing reading score (70 or greater)* Create a dataframe to hold the above results* Optional: give the displayed data cleaner formatting | #Calculates the total number of schools
arr_of_schools = school_data_complete["school_name"].unique()
total_schools = len(arr_of_schools)
#Calulates the number of students
arr_of_students = school_data_complete["student_name"]
total_students = len(arr_of_students)
#Calulates the total budget for all schools
arr_of_budgets = school_data_complete["budget"].unique()
total_budget = arr_of_budgets.sum()
#Calulates the average math score for all students
avg_math = school_data_complete["math_score"].mean()
#Calulates the average math score for all students
avg_read = school_data_complete["reading_score"].mean()
#Calculates the overall passing score
avg_overall = "{:.4f}".format((avg_read + avg_math)/2)
#Calculates the percentage of students with a passing math score (70 or greater)
percentage_math = "{:.6%}".format((len(school_data_complete.loc[(school_data_complete["math_score"]>= 70)])/total_students))
#Calculates the percentage of students with a passing reading score (70 or greater)
percentage_read = "{:.6%}".format((len(school_data_complete.loc[(school_data_complete["reading_score"]>= 70)])/total_students))
#Creates a dataframe to hold the above results
frame_df = pd.DataFrame({
"Total Schools":[total_schools],
"Total Students":["{:,}".format(total_students)],
"Total Budget":["${:,}".format(total_budget)],
"Average Math Score":[avg_math],
"Average Reading Score":[avg_read],
"% Passing Math":[percentage_math],
"% Passing Reading":[percentage_read],
"% Overall Passing Rate":[avg_overall]
})
frame_df | _____no_output_____ | ADSL | PyCitySchools/Resources/PyCitySchools_starter_unsolved.ipynb | fkokro/Pandas---Challenge |
School Summary * Create an overview table that summarizes key metrics about each school, including: * School Name * School Type * Total Students * Total School Budget * Per Student Budget * Average Math Score * Average Reading Score * % Passing Math * % Passing Reading * Overall Passing Rate (Average of the above two) * Create a dataframe to hold the above results Top Performing Schools (By Passing Rate) * Sort and display the top five schools in overall passing rate | #Sets index to school name
school_data_complete_name_index = school_data_complete.set_index("school_name") | _____no_output_____ | ADSL | PyCitySchools/Resources/PyCitySchools_starter_unsolved.ipynb | fkokro/Pandas---Challenge |
Bottom Performing Schools (By Passing Rate) * Sort and display the five worst-performing schools | _____no_output_____ | ADSL | PyCitySchools/Resources/PyCitySchools_starter_unsolved.ipynb | fkokro/Pandas---Challenge |
|
Math Scores by Grade * Create a table that lists the average Reading Score for students of each grade level (9th, 10th, 11th, 12th) at each school. * Create a pandas series for each grade. Hint: use a conditional statement. * Group each series by school * Combine the series into a dataframe * Optional: give the displayed data cleaner formatting Reading Score by Grade * Perform the same operations as above for reading scores Scores by School Spending * Create a table that breaks down school performances based on average Spending Ranges (Per Student). Use 4 reasonable bins to group school spending. Include in the table each of the following: * Average Math Score * Average Reading Score * % Passing Math * % Passing Reading * Overall Passing Rate (Average of the above two) | # Sample bins. Feel free to create your own bins.
spending_bins = [0, 585, 615, 645, 675]
group_names = ["<$585", "$585-615", "$615-645", "$645-675"] | _____no_output_____ | ADSL | PyCitySchools/Resources/PyCitySchools_starter_unsolved.ipynb | fkokro/Pandas---Challenge |
Scores by School Size * Perform the same operations as above, based on school size. | # Sample bins. Feel free to create your own bins.
size_bins = [0, 1000, 2000, 5000]
group_names = ["Small (<1000)", "Medium (1000-2000)", "Large (2000-5000)"] | _____no_output_____ | ADSL | PyCitySchools/Resources/PyCitySchools_starter_unsolved.ipynb | fkokro/Pandas---Challenge |
Copyright 2020 The TensorFlow Authors. | #@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License. | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
TensorFlow graph optimization with Grappler View on TensorFlow.org Run in Google Colab View source on GitHub Download notebook OverviewTensorFlow uses both graph and eager executions to execute computations. A `tf.Graph` contains a set of `tf.Operation` objects (ops) which represent units of computation and `tf.Tensor` objects which represent the units of data that flow between ops.Grappler is the default graph optimization system in the TensorFlow runtime. Grappler applies optimizations in graph mode (within `tf.function`) to improve the perormance of your TensorFlow computations through graph simplifications and other high-level optimizations such as inlining function bodies to enable inter-procedural optimizations. Optimizing the `tf.Graph` also reduces the device peak memory usage and improves hardware utilization by optimizing the mapping of graph nodes to compute resources. Use `tf.config.optimizer.set_experimental_options()` for finer control over your `tf.Graph` optimizations. Available graph optimizersGrappler performs graph optimizations through a top-level driver called the `MetaOptimizer`. The following graph optimizers are available with TensorFlow: * *Constant folding optimizer -* Statically infers the value of tensors when possible by folding constant nodes in the graph and materializes the result using constants.* *Arithmetic optimizer -* Simplifies arithmetic operations by eliminating common subexpressions and simplifying arithmetic statements. * *Layout optimizer -* Optimizes tensor layouts to execute data format dependent operations such as convolutions more efficiently.* *Remapper optimizer -* Remaps subgraphs onto more efficient implementations by replacing commonly occuring subgraphs with optimized fused monolithic kernels.* *Memory optimizer -* Analyzes the graph to inspect the peak memory usage for each operation and inserts CPU-GPU memory copy operations for swapping GPU memory to CPU to reduce the peak memory usage.* *Dependency optimizer -* Removes or rearranges control dependencies to shorten the critical path for a model step or enables otheroptimizations. Also removes nodes that are effectively no-ops such as Identity.* *Pruning optimizer -* Prunes nodes that have no effect on the output from the graph. It is usually run first to reduce the size of the graph and speed up processing in other Grappler passes.* *Function optimizer -* Optimizes the function library of a TensorFlow program and inlines function bodies to enable other inter-procedural optimizations.* *Shape optimizer -* Optimizes subgraphs that operate on shape and shape related information.* *Autoparallel optimizer -* Automatically parallelizes graphs by splitting along the batch dimension. This optimizer is turned OFF by default.* *Loop optimizer -* Optimizes the graph control flow by hoisting loop-invariant subgraphs out of loops and by removing redundant stack operations in loops. Also optimizes loops with statically known trip counts and removes statically known dead branches in conditionals.* *Scoped allocator optimizer -* Introduces scoped allocators to reduce data movement and to consolidate some operations.* *Pin to host optimizer -* Swaps small operations onto the CPU. This optimizer is turned OFF by default. * *Auto mixed precision optimizer -* Converts data types to float16 where applicable to improve performance. Currently applies only to GPUs.* *Debug stripper -* Strips nodes related to debugging operations such as `tf.debugging.Assert`, `tf.debugging.check_numerics`, and `tf.print` from the graph. This optimizer is turned OFF by default. Setup | from __future__ import absolute_import, division, print_function, unicode_literals
import numpy as np
import timeit
import traceback
import contextlib
try:
%tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
Create a context manager to easily toggle optimizer states. | @contextlib.contextmanager
def options(options):
old_opts = tf.config.optimizer.get_experimental_options()
tf.config.optimizer.set_experimental_options(options)
try:
yield
finally:
tf.config.optimizer.set_experimental_options(old_opts) | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
Compare execution performance with and without GrapplerTensorFlow 2 and beyond executes [eagerly](../eager.md) by default. Use `tf.function` to switch the default execution to Graph mode. Grappler runs automatically in the background to apply the graph optimizations above and improve execution performance. Constant folding optimizerAs a preliminary example, consider a function which performs operations on constants and returns an output. | def test_function_1():
@tf.function
def simple_function(input_arg):
print('Tracing!')
a = tf.constant(np.random.randn(2000,2000), dtype = tf.float32)
c = a
for n in range(50):
c = c@a
return tf.reduce_mean(c+input_arg)
return simple_function | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
Turn off the constant folding optimizer and execute the function: | with options({'constant_folding': False}):
print(tf.config.optimizer.get_experimental_options())
simple_function = test_function_1()
# Trace once
x = tf.constant(2.2)
simple_function(x)
print("Vanilla execution:", timeit.timeit(lambda: simple_function(x), number = 1), "s") | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
Enable the constant folding optimizer and execute the function again to observe a speed-up in function execution. | with options({'constant_folding': True}):
print(tf.config.optimizer.get_experimental_options())
simple_function = test_function_1()
# Trace once
x = tf.constant(2.2)
simple_function(x)
print("Constant folded execution:", timeit.timeit(lambda: simple_function(x), number = 1), "s") | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
Debug stripper optimizerConsider a simple function that checks the numeric value of its input argument and returns it. | def test_function_2():
@tf.function
def simple_func(input_arg):
output = input_arg
tf.debugging.check_numerics(output, "Bad!")
return output
return simple_func | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
First, execute the function with the debug stripper optimizer turned off. | test_func = test_function_2()
p1 = tf.constant(float('inf'))
try:
test_func(p1)
except tf.errors.InvalidArgumentError as e:
traceback.print_exc(limit=2) | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
`tf.debugging.check_numerics` raises an invalid argument error because of the `Inf` argument to `test_func`. Enable the debug stripper optimizer and execute the function again. | with options({'debug_stripper': True}):
test_func2 = test_function_2()
p1 = tf.constant(float('inf'))
try:
test_func2(p1)
except tf.errors.InvalidArgumentError as e:
traceback.print_exc(limit=2) | _____no_output_____ | Apache-2.0 | site/en/guide/graph_optimization.ipynb | atharva1503/docs |
Part I. ETL Pipeline for Pre-Processing the Files Import Python packages | # Import Python packages
import pandas as pd
import cassandra
import re
import os
import glob
import numpy as np
import json
import csv | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Creating list of filepaths to process original event csv data files | # checking your current working directory
print(os.getcwd())
# Get your current folder and subfolder event data
filepath = os.getcwd() + '/event_data'
# Create a for loop to create a list of files and collect each filepath
for root, dirs, files in os.walk(filepath):
# join the file path and roots with the subdirectories using glob
file_path_list = glob.glob(os.path.join(root,'*')) | /home/workspace
| MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Processing the files to create the data file csv that will be used for Apache Casssandra tables | # initiating an empty list of rows that will be generated from each file
full_data_rows_list = []
# for every filepath in the file path list
for f in file_path_list:
# reading csv file
with open(f, 'r', encoding = 'utf8', newline='') as csvfile:
# creating a csv reader object
csvreader = csv.reader(csvfile)
next(csvreader)
# extracting each data row one by one and append it
for line in csvreader:
#print(line)
full_data_rows_list.append(line)
# creating a smaller event data csv file called event_datafile_full csv that will be used to insert data into the \
# Apache Cassandra tables
csv.register_dialect('myDialect', quoting=csv.QUOTE_ALL, skipinitialspace=True)
with open('event_datafile_new.csv', 'w', encoding = 'utf8', newline='') as f:
writer = csv.writer(f, dialect='myDialect')
writer.writerow(['artist','firstName','gender','itemInSession','lastName','length',\
'level','location','sessionId','song','userId'])
for row in full_data_rows_list:
if (row[0] == ''):
continue
writer.writerow((row[0], row[2], row[3], row[4], row[5], row[6], row[7], row[8], row[12], row[13], row[16]))
# check the number of rows in your csv file
with open('event_datafile_new.csv', 'r', encoding = 'utf8') as f:
print(sum(1 for line in f)) | 6821
| MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
The event_datafile_new.csv now contains the following columns: - artist - firstName of user- gender of user- item number in session- last name of user- length of the song- level (paid or free song)- location of the user- sessionId- song title- userIdThe image below is a screenshot of what the denormalized data should appear like in the **event_datafile_new.csv** after the code above is run: Creating a Cluster | from cassandra.cluster import Cluster
try:
# This should make a connection to a Cassandra instance your local machine
# (127.0.0.1)
cluster = Cluster(['127.0.0.1'])
# To establish connection and begin executing queries, need a session
session = cluster.connect()
except Exception as e:
print(e) | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Create Keyspace | try:
session.execute("""
CREATE KEYSPACE IF NOT EXISTS sparkify
WITH REPLICATION =
{ 'class' : 'SimpleStrategy', 'replication_factor' : 1 }""")
except Exception as e:
print(e) | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Set Keyspace | try:
session.set_keyspace('sparkify')
except Exception as e:
print(e) | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Part II. Time to answer important query questions Query 1. Give me the artist, song title and song's length in the music app history that was heard during sessionId = 338, and itemInSession = 4This query focuses on session history, so the table will be named accordingly.Filtering needs to be done by session_id AND item_in_session. Looking at the data, it's safe to assume that both aforementioned columns would identify row uniquely, so both columns will be included for primary key. | session_history_create = """CREATE TABLE IF NOT EXISTS session_history(
session_id int,
item_in_session int,
artist_name text,
song_title text,
song_length float,
PRIMARY KEY(session_id, item_in_session));"""
try:
session.execute(session_history_create)
except Exception as e:
print(e)
file = 'event_datafile_new.csv'
with open(file, encoding = 'utf8') as f:
csvreader = csv.reader(f)
next(csvreader) # skip header
for line in csvreader:
query = "INSERT INTO session_history(session_id, item_in_session, artist_name, song_title, song_length)"
query = query + "VALUES(%s, %s, %s, %s, %s);"
session.execute(query, (int(line[8]), int(line[3]), line[0], line[9], float(line[5]))) | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Let's test query 1 with an example | query = """SELECT artist_name, song_title, song_length
FROM session_history
WHERE session_id=338 AND item_in_session=4;"""
try:
result = pd.DataFrame(list(session.execute(query)))
print(result.to_string())
except Exception as e:
print(e) | artist_name song_title song_length
0 Faithless Music Matters (Mark Knight Dub) 495.307312
| MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Query 2. Give me only the following: name of artist, song (sorted by itemInSession) and user (first and last name) for userid = 10, sessionid = 182This query focuses on specific user session history. We will be retrieving specific listening details by user_id and session_id. To support query, user_id, session_id and item_in_session will be used for primary key (where item_in_session is needed to give sorted output. | user_session_history_create = """
CREATE TABLE IF NOT EXISTS user_session_history(
user_id int,
session_id int,
item_in_session int,
artist_name text,
song_title text,
user_first_name text,
user_last_name text,
PRIMARY KEY(user_id, session_id, item_in_session));"""
try:
session.execute(user_session_history_create)
except Exception as e:
print(e)
file = 'event_datafile_new.csv'
with open(file, encoding = 'utf8') as f:
csvreader = csv.reader(f)
next(csvreader) # skip header
for line in csvreader:
query = "INSERT INTO user_session_history(user_id, session_id, item_in_session, artist_name, song_title, user_first_name, user_last_name)"
query = query + "VALUES(%s, %s, %s, %s, %s, %s, %s);"
session.execute(query, (int(line[10]), int(line[8]), int(line[3]), line[0], line[9], line[1], line[4])) | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Let's test query 2 with an example | query = """SELECT artist_name, song_title, user_first_name, user_last_name
FROM user_session_history
WHERE user_id=10 AND session_id=182;"""
try:
result = pd.DataFrame(list(session.execute(query)))
print(result.to_string())
except Exception as e:
print(e) | artist_name song_title user_first_name user_last_name
0 Down To The Bone Keep On Keepin' On Sylvie Cruz
1 Three Drives Greece 2000 Sylvie Cruz
2 Sebastien Tellier Kilometer Sylvie Cruz
3 Lonnie Gordon Catch You Baby (Steve Pitron & Max Sanna Radio... Sylvie Cruz
| MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Query 3. Give me every user name (first and last) in my music app history who listened to the song 'All Hands Against His Own'This query focuses on song history, highlighting which songs are being listened to by which users. We will be retrieving specific listening details by song_title. To support query, song_title and user_id will be used for primary key. Note: Song names might not be unique (e.g. different artists might have same song names). To remove ambiguity, in future it might makes sense to specify more details about this query. | song_user_history_create = """
CREATE TABLE IF NOT EXISTS song_user_history(
user_id int,
song_title text,
user_first_name text,
user_last_name text,
PRIMARY KEY(song_title, user_id));"""
try:
session.execute(song_user_history_create)
except Exception as e:
print(e)
file = 'event_datafile_new.csv'
with open(file, encoding = 'utf8') as f:
csvreader = csv.reader(f)
next(csvreader) # skip header
for line in csvreader:
query = "INSERT INTO song_user_history(user_id, song_title, user_first_name, user_last_name)"
query = query + "VALUES(%s, %s, %s, %s);"
session.execute(query, (int(line[10]), line[9], line[1], line[4])) | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Let's test query 3 with an example | query = """SELECT user_first_name, user_last_name
FROM song_user_history
WHERE song_title='All Hands Against His Own';"""
try:
result = pd.DataFrame(list(session.execute(query)))
print(result.to_string())
except Exception as e:
print(e) | user_first_name user_last_name
0 Jacqueline Lynch
1 Tegan Levine
2 Sara Johnson
| MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Dropping the tables before closing out the sessions | drop_session_history = "DROP TABLE IF EXISTS session_history;"
session.execute(drop_session_history)
drop_user_session_history = "DROP TABLE IF EXISTS user_session_history;"
session.execute(drop_user_session_history)
drop_song_user_history = "DROP TABLE IF EXISTS song_user_history;"
session.execute(drop_song_user_history) | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Don't forget to close the session and cluster connection¶ | session.shutdown()
cluster.shutdown() | _____no_output_____ | MIT | sparkify_cassandra_project.ipynb | iamfeniak/sparkify-etl-pipeline-cassandra |
Welcome! In finance momentum refers to the phenomenon of cross-sectional predictability of returns by past price data. A standard example would be the well documented tendency of stocks that have had high returns over the past one to twelve months to continue outperform stocks that have performed poorly over the same period. Positive returns from buying past winners and selling past losers is a long-standing market anomaly in financial research documented for basically every asset class and literally for hundreds of years. Note that since the stocks are compared to their peers we talk about cross-sectional predictability, in contrast to the time-series momentum, or trend following, where decision to buy or sell a stock depends on its own past performance only. Over the past quarter of a century the finance literature has proposed numerous ways to measure the momentum, e.g. in terms of lookback horizon, and identified a host of confounding variables, like market volatility, predicting its performance as an investment strategy. The emerging field of financial machine learning further finds past price data to be among the strongest predictors of future returns, dominating fundamental variables like book-to-market ratio. | import csv
import random
import matplotlib.pyplot as plt
import numpy as np
import scipy.stats as statfunc
import pandas as pd
import plotly.graph_objs as go
import mplfinance as mpf
from sklearn import mixture as mix
import seaborn as sns
from pandas_datareader import data as web
df= web.get_data_yahoo('RELIANCE.NS',start= '2018-04-08', end='2020-12-12')
df=df[['Open','High','Low','Close','Volume']]
df['open']=df['Open'].shift(1)
df['high']=df['High'].shift(1)
df['low']=df['Low'].shift(1)
df['close']=df['Close'].shift(1)
df['volume']=df['Volume'].shift(1)
df=df[['open','high','low','close','volume']]
df=df.dropna()
unsup = mix.GaussianMixture(n_components=4, covariance_type="spherical", n_init=100, random_state=42)
unsup.fit(np.reshape(df,(-1,df.shape[1])))
regime = unsup.predict(np.reshape(df,(-1,df.shape[1])))
df['Return']= np.log(df['close']/df['close'].shift(1))
Regimes=pd.DataFrame(regime,columns=['Regime'],index=df.index)\
.join(df, how='inner')\
.assign(market_cu_return=df.Return.cumsum())\
.reset_index(drop=False)\
.rename(columns={'index':'Date'})
order=[0,1,2,3]
fig = sns.FacetGrid(data=Regimes,hue='Regime',hue_order=order,aspect=2,size= 4)
fig.map(plt.scatter,'Date','market_cu_return', s=4).add_legend()
plt.show()
for i in order:
print('Mean for regime %i: '%i,unsup.means_[i][0])
print('Co-Variancefor regime %i: '%i,(unsup.covariances_[i])) | Mean for regime 0: 10784.766638714711
Co-Variancefor regime 0: 36141.444765725864
Mean for regime 1: 12047.572769668644
Co-Variancefor regime 1: 23143.320584827794
Mean for regime 2: 9447.270093875897
Co-Variancefor regime 2: 453141.17885501473
Mean for regime 3: 11509.5640340791
Co-Variancefor regime 3: 30112.327401162816
| MIT | FinanceProjectMain.ipynb | AmaaniGoose/FinanceProject |
Version 2: Using technical indicators | print(df)
def RSI(df, base="Close", period=21):
"""
Function to compute Relative Strength Index (RSI)
Args :
df : Pandas DataFrame which contains ['date', 'open', 'high', 'low', 'close', 'volume'] columns
base : String indicating the column name from which the MACD needs to be computed from (Default Close)
period : Integer indicates the period of computation in terms of number of candles
Returns :
df : Pandas DataFrame with new columns added for
Relative Strength Index (RSI_$period)
"""
delta = df[base].diff()
up, down = delta.copy(), delta.copy()
up[up < 0] = 0
down[down > 0] = 0
rUp = up.ewm(com=period - 1, adjust=False).mean()
rDown = down.ewm(com=period - 1, adjust=False).mean().abs()
df['RSI_' + str(period)] = 100 - 100 / (1 + rUp / rDown)
df['RSI_' + str(period)].fillna(0, inplace=True)
return df
def BBand(df, base='Close', period=20, multiplier=2):
"""
Function to compute Bollinger Band (BBand)
Args :
df : Pandas DataFrame which contains ['date', 'open', 'high', 'low', 'close', 'volume'] columns
base : String indicating the column name from which the MACD needs to be computed from (Default Close)
period : Integer indicates the period of computation in terms of number of candles
multiplier : Integer indicates value to multiply the SD
Returns :
df : Pandas DataFrame with new columns added for
Upper Band (UpperBB_$period_$multiplier)
Lower Band (LowerBB_$period_$multiplier)
"""
upper = 'UpperBB_' + str(period) + '_' + str(multiplier)
lower = 'LowerBB_' + str(period) + '_' + str(multiplier)
sma = df[base].rolling(window=period, min_periods=period - 1).mean()
sd = df[base].rolling(window=period).std()
df[upper] = sma + (multiplier * sd)
df[lower] = sma - (multiplier * sd)
df[upper].fillna(0, inplace=True)
df[lower].fillna(0, inplace=True)
return df
def MACD(df, fastEMA=12, slowEMA=26, signal=9, base='Close'):
"""
Function to compute Moving Average Convergence Divergence (MACD)
Args :
df : Pandas DataFrame which contains ['date', 'open', 'high', 'low', 'close', 'volume'] columns
fastEMA : Integer indicates faster EMA
slowEMA : Integer indicates slower EMA
signal : Integer indicates the signal generator for MACD
base : String indicating the column name from which the MACD needs to be computed from (Default Close)
Returns :
df : Pandas DataFrame with new columns added for
Fast EMA (ema_$fastEMA)
Slow EMA (ema_$slowEMA)
MACD (macd_$fastEMA_$slowEMA_$signal)
MACD Signal (signal_$fastEMA_$slowEMA_$signal)
MACD Histogram (MACD (hist_$fastEMA_$slowEMA_$signal))
"""
fE = "ema_" + str(fastEMA)
sE = "ema_" + str(slowEMA)
macd = "macd_" + str(fastEMA) + "_" + str(slowEMA) + "_" + str(signal)
sig = "signal_" + str(fastEMA) + "_" + str(slowEMA) + "_" + str(signal)
hist = "hist_" + str(fastEMA) + "_" + str(slowEMA) + "_" + str(signal)
# Compute fast and slow EMA
EMA(df, base, fE, fastEMA)
EMA(df, base, sE, slowEMA)
# Compute MACD
df[macd] = np.where(np.logical_and(np.logical_not(df[fE] == 0), np.logical_not(df[sE] == 0)), df[fE] - df[sE], 0)
# Compute MACD Signal
EMA(df, macd, sig, signal)
# Compute MACD Histogram
df[hist] = np.where(np.logical_and(np.logical_not(df[macd] == 0), np.logical_not(df[sig] == 0)), df[macd] - df[sig], 0)
return df
def ATR(df, period, ohlc=['Open', 'High', 'Low', 'Close']):
"""
Function to compute Average True Range (ATR)
Args :
df : Pandas DataFrame which contains ['date', 'open', 'high', 'low', 'close', 'volume'] columns
period : Integer indicates the period of computation in terms of number of candles
ohlc: List defining OHLC Column names (default ['Open', 'High', 'Low', 'Close'])
Returns :
df : Pandas DataFrame with new columns added for
True Range (TR)
ATR (ATR_$period)
"""
atr = 'ATR_' + str(period)
# Compute true range only if it is not computed and stored earlier in the df
if not 'TR' in df.columns:
df['h-l'] = df[ohlc[1]] - df[ohlc[2]]
df['h-yc'] = abs(df[ohlc[1]] - df[ohlc[3]].shift())
df['l-yc'] = abs(df[ohlc[2]] - df[ohlc[3]].shift())
df['TR'] = df[['h-l', 'h-yc', 'l-yc']].max(axis=1)
df.drop(['h-l', 'h-yc', 'l-yc'], inplace=True, axis=1)
# Compute EMA of true range using ATR formula after ignoring first row
EMA(df, 'TR', atr, period, alpha=True)
return df | _____no_output_____ | MIT | FinanceProjectMain.ipynb | AmaaniGoose/FinanceProject |
Objectives* Consider only two features of the dataset obtained after t-sne* Find out best performing value of perplexity for t-sne using cross-validation* Use linear model with no calibration vs non linear model to see how well these perform | def get_training_set(train):
X = train[train.columns.drop('Activity')]
y = train.Activity
return X, y
X, y = get_training_set(train)
Xtest = test
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, test_size=0.3, random_state=44) | _____no_output_____ | MIT | Kaggle-Competitions/Predict-Bio-Response/Predict-Bio-Response-Model-Building.ipynb | gopala-kr/ds-notebooks |
Cross validation | def get_cross_val_scores_by_perplexity(X, y, clf):
skf = StratifiedKFold(y, n_folds=3, shuffle=True, random_state=43)
perplexity = [50, 75, 100]
errors = []
for p in perplexity:
tsne = TSNE(n_components=2, perplexity=p)
Xhat = tsne.fit_transform(X)
cv_scores = cross_val_score(clf, Xhat, y, scoring='log_loss', cv=skf, n_jobs=-1)
errors.append(cv_scores.mean())
perplexity = np.array(perplexity)
errors = np.array(errors)
return perplexity, errors
perplexity, errors = get_cross_val_scores_by_perplexity(X, y, LogisticRegression(C=1.))
errors = map(abs, errors)
plt.scatter(perplexity, errors, color='b', alpha=0.5)
plt.xlabel('Perplexity')
plt.ylabel('Log Loss');
perplexity, errors = get_cross_val_scores_by_perplexity(X, y, RandomForestClassifier(n_jobs=-1))
errors = map(abs, errors)
plt.scatter(perplexity, errors, color='b', alpha=0.5)
plt.xlabel('Perplexity')
plt.ylabel('Log Loss'); | _____no_output_____ | MIT | Kaggle-Competitions/Predict-Bio-Response/Predict-Bio-Response-Model-Building.ipynb | gopala-kr/ds-notebooks |
Old way - csv | #js = json.load(open('ref_1554219251.json'))
#csv = open('ref_1554219251.csv').read()
csv = open('ref_1558361146.csv').read()
csv_data = []
for rec in [x.strip() for x in csv.split("\n")]:
p = rec.split(';')
if len(p) < 6:
continue
cur = collections.OrderedDict([
('method', p[0]),
('block', int(p[1])),
('deg', int(p[2])),
('comb_deg', int(p[3])),
('data_size', int(p[4])),
('zscores', [float(x.replace(',','.')) for x in p[6:]])
])
csv_data.append(cur)
print(json.dumps(csv_data[0]))
data = csv_data
data_filt = [x for x in data if x and len(x['zscores']) > 19000]
data_filt.sort(key=lambda x: (x['method'], x['block'], x['deg'], x['comb_deg'], x['data_size']))
np.random.seed(87655677)
print('#of inputs with data count over threshold: ', len(data_filt))
pval_db = []
for dix, val in enumerate(data_filt):
res = tabulate_pvals(val, abs_val=True)
pval_db.append(res)
print('Dump %s' % dix)
json.dump(pval_db, open('pval_db.json', 'w+'), indent=2)
nbins = 200
abs_val = True
for dix, val in enumerate(data_filt):
inp_iter = (val['zscores'])
if abs_val:
inp_iter = [abs(x) for x in inp_iter]
print('%s[%s:%s:%s:%s]: %s %s'
% (val['method'], val['block'], val['deg'], val['comb_deg'],
val['data_size'], len(val['zscores']),
'',#dst.ppf([1-0.0001, 1-0.001, 1-0.01, 1-0.05, 1-0.10, 1-0.5, 0, 1, 0.0001, 0.001, 0.1, 0.9])
#dst.stats(moments='mvsk')
))
bin_tup = get_bins(inp_iter, nbins=nbins, full=True)
bb = get_distrib_fbins(inp_iter, bin_tup)
bin_size = bin_tup[1]
minv, maxv = bin_tup[2], bin_tup[3]
bins = np.array([x[0] for x in bb])
dst = stats.rv_discrete(values=([x[0] for x in bb], [x[1] for x in bb]))
print(stats.rv_discrete)
x=np.array([bins[0], bins[1], bins[6]])
print(dst.pmf(x))
print(dst._pmf(x))
# Tabulate pvalues
build_integrator(bin_tup)
extremes = [
[minv, 1],
[0, -1],
[0, +1],
[maxv, -1]
] if not abs_val else [
[minv, 1],
[maxv, -1]
]
pvals = pvalue_comp(lambda x: binned_pmf(x, bin_tup), extremes,
dx=1./(nbins/10.), bin_tup=bin_tup, by_bins=True)
n_sample = 100
rvs = dst.rvs(size=n_sample)
f, l = np.histogram(rvs, bins=bins)
f = np.append(f, [0])
probs = np.array([x[1] for x in bb])
#print(bins, len(bins))
#print(probs, len(probs))
#print(f, len(f))
#sfreq = np.vstack([np.array([x[0] for x in bb]), f, probs*n_sample]).T
#print(sfreq)
print('%s[%s:%s:%s:%s]: %s %s'
% (val['method'], val['block'], val['deg'], val['comb_deg'],
val['data_size'], len(val['zscores']),
dst.ppf([1-0.0001, 1-0.001, 1-0.01, 1-0.05, 1-0.10, 1-0.5, 0, 1, 0.0001, 0.001, 0.1, 0.9])
#dst.stats(moments='mvsk')
))
x = np.linspace(min(bins),max(bins),1000)
plt.plot(x, dst.cdf(x))
plt.show()
cdf_dev = derivative(dst.cdf, x, dx=0.5)
plt.plot(x,cdf_dev)
sec_x = pvals[40] # 49
print('Plotting area under: ', sec_x)
for ix in range(len(sec_x[1])):
section = np.arange(sec_x[1][ix][0], sec_x[1][ix][1], 1/20.)
plt.fill_between(section, derivative(dst.cdf, section, dx=0.5))
plt.show()
#for pv in pvals:
# sec_x = pv
# for ix in range(len(sec_x[1])):
# section = np.arange(sec_x[1][ix][0], sec_x[1][ix][1], 1/20.)
# plt.fill_between(section, derivative(dst.cdf, section, dx=0.5))
# plt.show()
x = np.linspace(0,100,10000)
plt.plot(x,dst.ppf(x))
plt.show()
x = np.linspace(minv,maxv,10000)
plt.plot(bins, dst._pmf(bins))
plt.show()
x = np.linspace(minv,maxv,10000)
plt.plot(x, [binned_pmf(y, bin_tup) for y in x])
for ix in range(len(sec_x[1])):
section = np.linspace(sec_x[1][ix][0], sec_x[1][ix][1], 10000) #np.arange(sec_x[1][ix][0], sec_x[1][ix][1], 1/20.)
plt.fill_between(section, [binned_pmf(y, bin_tup)+0.0005 for y in section])
plt.show()
# Idea: pvalue function = pms of the distribution.
# If test returns z-score with p=0 then we reject the hypothesis as we didnt get such zscore
# If test returns with p=0.3 we dont reject as we have our alpha set somehow...
# Problem: number of bins. If too many, we have small probabilities -> some alphas not reachable.
#if dix > 3:
break
np.arange(7)
#np.zeros(np.shape(0.5),'d')
#print(dst.ppf([1-0.01, 1-0.05, 1-0.10, 0.5, 0.6]))
import itertools
def crit_regions(data, add_zscores=False):
iter_block = [128, 256, 384, 512]
iter_deg = [1, 2, 3]
iter_cdeg = [1, 2, 3]
iter = itertools.product(iter_block, iter_deg, iter_cdeg)
for ix, crec in enumerate(iter):
c = tuple(crec)
reldata = [x for x in data if (x['block'], x['deg'], x['comb_deg']) == c and len(x['zscores']) > 9999]
reldata.sort(key=lambda x: -len(x['zscores']))
relrecs = []
for crel in reldata:
zsz = [abs(x) for x in crel['zscores']]
relrecs.append((crel['method'], len(zsz), min(zsz), max(zsz), crel['zscores'] if add_zscores else None))
yield (crec, relrecs)
np.mean(list(range(10)))
# Prints markdown critical region table
def crit_regions_table(data):
dt = list(crit_regions(data, True))
max_num = 9
hdr = ['block', 'deg', 'comb-deg', 'samples', 'alpha', 'min', 'max', 'mean', 'stddev']
max_hdr = max([len(x) for x in hdr])
max_field = max(max_hdr, max_num) + 1
hdr_spaced = [x.rjust(max_field) for x in hdr]
print('|%s |' % ' |'.join(hdr_spaced))
print('|%s|' % '|'.join([''.rjust(max_field + 1, '-') for _ in hdr]))
for r in dt:
cr, cd = r[0], r[1][0]
zsz = [abs(x) for x in cd[4]]
elems = [*cr] + [cd[1], '%.1e' % (1./cd[1]),
'%9.6f' % cd[2], '%9.6f' % cd[3],
'%9.6f' % np.mean(zsz), '%9.6f' % np.std(zsz)]
elems = [str(x).rjust(max_field) for x in elems]
print('|%s |' % ' |'.join(elems))
crit_regions_table(data)
cregions = list(crit_regions(data, True))
colors = [
*sns.color_palette("ch:1.0,-.1,light=.7,dark=.4", 9),
*sns.color_palette("ch:2.0,-.1,light=.7,dark=.4", 9),
*sns.color_palette("ch:2.9,-.1,light=.7,dark=.4", 9),
*sns.color_palette("ch:0.5,-.1,light=.7,dark=.4", 9),
]
sns.palplot(colors)
# z-score dist plot
a4_dims = (10.7, 6.27) #None #(2*11.7, 8.27)
ylim = (0.0, 2.2740367425079864)
xlim = (1.1900278616886606, 11.30353802905374)
def distplots(cregions, colors, desc, fname, xlim=None, ylim=None):
fig, ax = pyplot.subplots(figsize=a4_dims)
for i in range(len(cregions)):
zs = [abs(x) for x in cregions[i][1][0][4]]
lbl = '-'.join([str(x) for x in cregions[i][0]])
r = sns.distplot(a=zs, ax=ax, hist=True, norm_hist=False, bins='auto', label=lbl, kde=True, color=colors[i])
lx = r.lines[-1].get_xdata()
ly = r.lines[-1].get_ydata()
maxi = np.argmax(ly)
ax.annotate(lbl, (lx[maxi], ly[maxi] + 0.1), xytext=(lx[maxi], 2.1),
rotation=90, ha='center', va='center',
arrowprops={'arrowstyle':'->', 'connectionstyle':'arc3',
"linestyle":"--", "linewidth":0.8, "color":'0.6'})
print(desc, i, len(zs), ly[maxi])
plt.legend(loc='upper right')
plt.title(desc if desc else 'z-score distributions')
ax.set_xlabel("z-score")
ax.set_ylabel("Hist")
if xlim: ax.set_xlim(xlim)
if ylim: ax.set_ylim(ylim)
plt.savefig(fname, bbox_inches='tight', dpi=400)
#plt.savefig(os.path.splitext(fname)[0] + '.pdf', bbox_inches='tight', dpi=400)
return fig, ax
# fig, ax = distplots(cregions, colors, 'z-score distributions', 'zscore-dists.png', xlim, ylim)
# print('ylim', ax.get_ylim())
# print('xlim', ax.get_xlim())
distplots(cregions[0*9:1*9], colors[0*9:1*9], 'z-score distributions, m=128', 'zscore-dists-128.png', xlim, ylim)
distplots(cregions[1*9:2*9], colors[1*9:2*9], 'z-score distributions, m=256', 'zscore-dists-256.png', xlim, ylim)
distplots(cregions[2*9:3*9], colors[2*9:3*9], 'z-score distributions, m=384', 'zscore-dists-384.png', xlim, ylim)
distplots(cregions[3*9:4*9], colors[3*9:4*9], 'z-score distributions, m=512', 'zscore-dists-512.png', xlim, ylim)
a4_dims = (2*11.7, 8.27)
fig, ax = pyplot.subplots(figsize=a4_dims)
zs = data_filt[1]['zscores']
for i in range(5):
zs = [(x) for x in data_filt[i]['zscores']]
print(len(zs))
sns.distplot(a=zs, ax=ax, hist=True, norm_hist=False, bins='auto') | _____no_output_____ | MIT | bool-pvalues.ipynb | sobuch/polynomial-distinguishers |
pvalues - pvalue = probability (in the null hypothesis distribution) to be observed as a value equal to or more extreme than the value observed computation - Derive CDF -> find 0 regions = extremes - Integrate from 0 regions towards region of increasing integral value. - Once sum of all integrations is alpha, stop. Integrated area is a critical region - Computation for x: integrate until the first integral boundary hits x. pvalue = sum of integrals - Tabulation: for each desired pvalue compute boundaries (4 values) where critical region starts. - pvalue(x): need to do the integration OR function table (\forall zscores: P(zscore) > 0). - In our case 4 extremes, integrate: - -\inf towards 0 - +\inf towards 0 - 0 towards +\inf - 0 towards -\inf - 10000 samples, pvalue = 0 -> 1/10000. - absolutize -> we have a new distribution -> 2x more datapoints, 2 tails. | np.arange(-1, 1, 1/20.)
counter = [0] * 8
MAXV = 2
def inc(counter):
global MAXV
ln = len(counter) - 1
while ln >= 0:
counter[ln] = (counter[ln] + 1) % MAXV
if (counter[ln] != 0):
return(counter)
ln-=1
raise ValueError('Overflow')
def dec(counter):
global MAXV
ln = len(counter) - 1
while ln >= 0:
counter[ln] = (counter[ln] - 1) % MAXV
if (counter[ln] != MAXV-1):
return counter
ln-=1
raise ValueError('Underflow')
for i in range(20):
print(inc(counter))
print('-'*80)
for i in range(20):
print(dec(counter))
from booltest import common
common.generate_seed(2)
import os
import time
tmp_files = os.scandir('/tmp')
for i in tmp_files:
print(i)
time.sleep(10)
3*3*3*10000*3
| _____no_output_____ | MIT | bool-pvalues.ipynb | sobuch/polynomial-distinguishers |
GradCAM (Gradient class activation mappinng)这个版本主要用来了解制作Grad_Cam的过程,并没有特意封装每行代码几乎都有注解---CNN卷积一直都是一个神秘的过程过去一直被以黑盒来形容能够窥探CNN是一件很有趣的事情, 特别是还能够帮助我们在进行一些任务的时候了解模型判别目标物的过程是不是出了什么问题而Grad-CAM就是一个很好的可视化选择因为能够生成高分辨率的图并叠加在原图上, 让我们在研究一些模型判别错误的时候, 有个更直观的了解那么具体是如何生成Grad CAM的呢?老规矩先上图, 说明流程一定要配图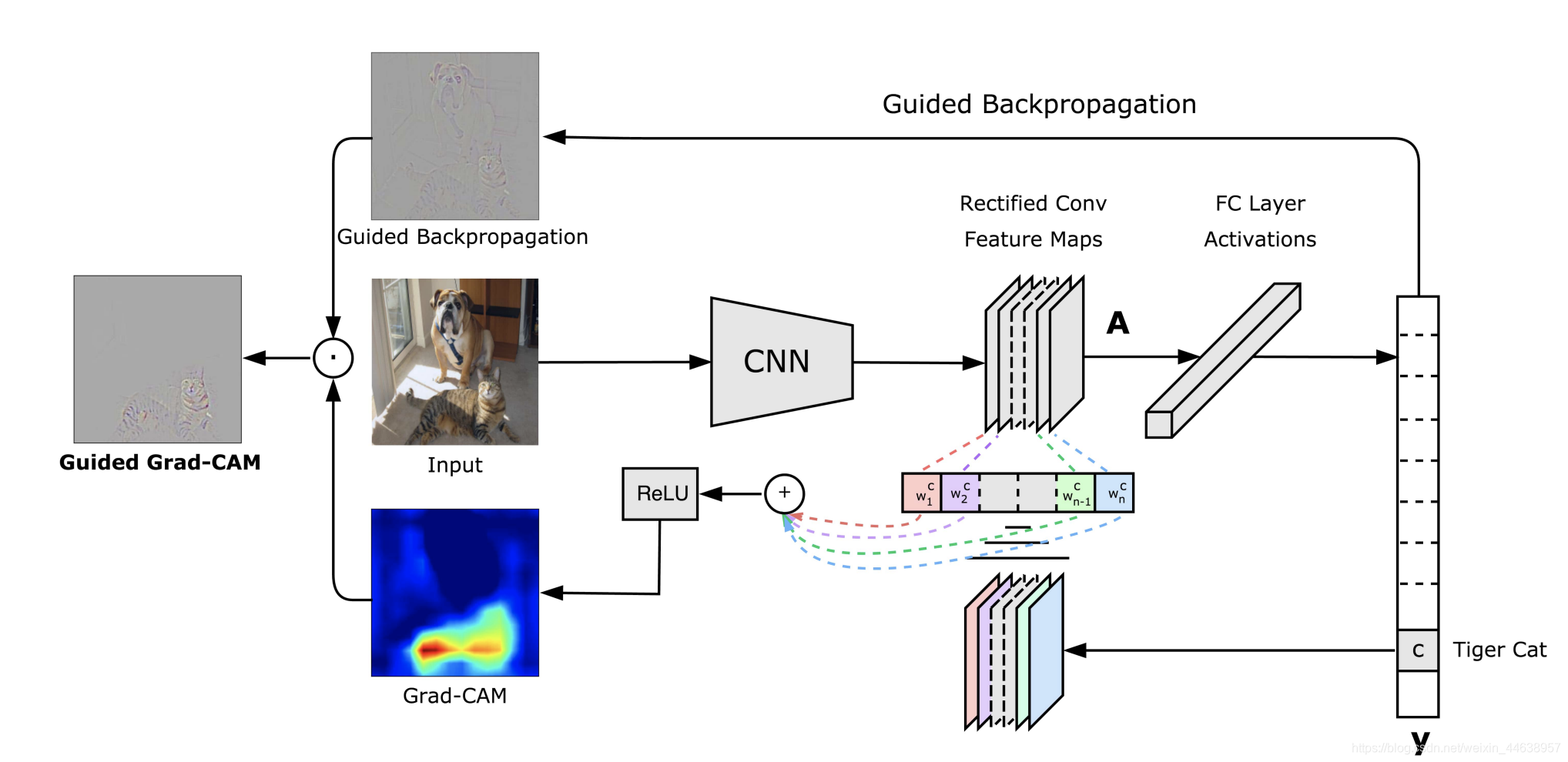借用一下论文作者的图我们能看到Input的地方就是我们要输入进网络进行判别的假设这里输入的input 类别是 Tiger cat 一如往常的进入CNN提取特征现在看到图中央的Rectified Conv Feature Maps 这个层就是我们要的目标层, 而这个层通常会是整个网路特征提取的最后一层, 为什么呢? 因为通常越深层的网络越能提取到越关键的特征, 这个是不变的如果你这边可以理解了就继续往下看呗我们能看到目标层下面有一排的 $w_1^c w_2^c....$等等的**这里的w就是权重, 也就是我们需要的目标层512维 每一维对类别Tiger cat的重要程度**>这里的权重不要理解为前向传播的 每个节点的权重, 两者是不同的而这些权重就是能够让模型判别为Tiger cat的最重要的依据我们要这些权重与目标层的feature map进行linear combination并提取出来那么该怎么求出这些权重呢?该是上公式的时候了, 公式由论文提出$L_{Grad-CAM}^c = ReLU(\sum_k\alpha^c_kA^k)$$\alpha_k^c$ 就是我们上面说的权重怎么求?摆出论文公式$\alpha_k^c = \frac{1}{Z} \sum_i \sum_j$ $\frac{\partial y^c}{\partial A^k_{ij}}$$\frac{1}{Z} \sum_i \sum_j$ 表示进行全局池化,也就是求feature map的平均值$\frac{\partial y^c}{\partial A^k_{ij}}$ 表示最终类别对我们要的目标层求梯度所有我们第一步先求出$A^k$ 的梯度之后, 在进行全局池化求得每一个feature map的平均梯度值就会是我们要的$\alpha_k^c$ , 整个池化过程可以是下图红框处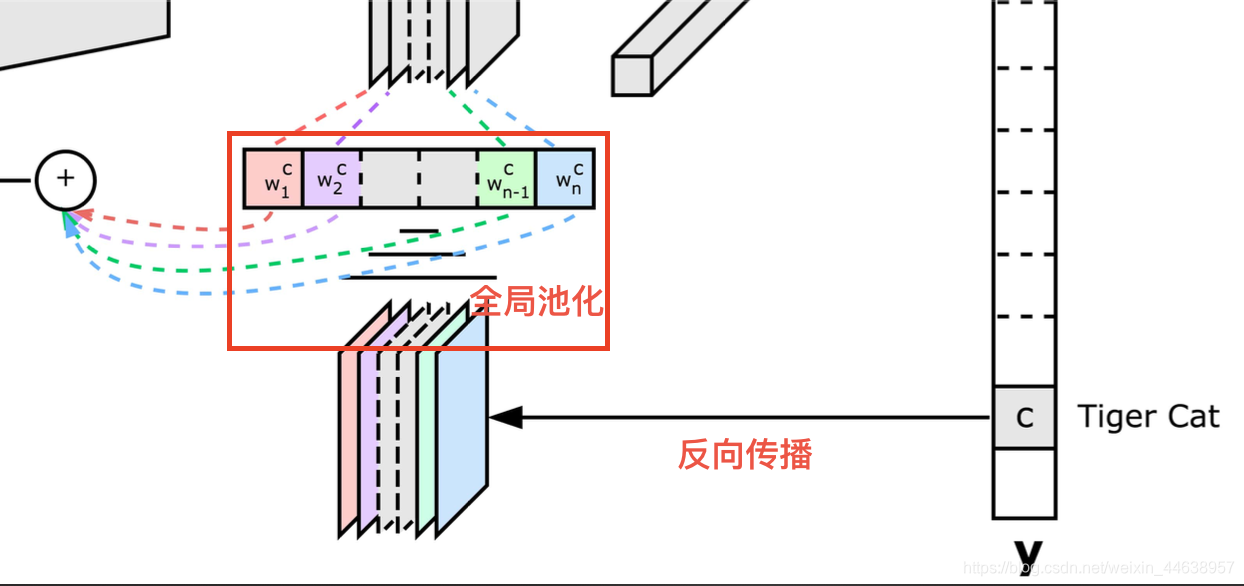这个$\alpha_k^c$ 代表的就是 经过全局池化求出梯度平均值$w_1^c w_2^c....$也就是我们前面所说的**目标层的512维 每一维对类别Tiger cat的重要程度**>这边很好理解, 就是把512层feature map分别取平均值, 就取出512个均值, 这512个均值就能分别表示每层的重要程度, 有的值高就显得重要, 有的低就不重要好的 回到$L_{Grad-CAM}^c = ReLU(\sum_k\alpha^c_kA^k)$现在$\alpha_k^c$ 我们有了$A^k$ 表示feature map A 然后深度是k, 如果网络是vgg16, 那么k就是512把我们求得的$\alpha_k^c$ 与 $A^k$两者进行相乘(这里做的就是线性相乘), k如果是512, 那么将512张feature map都与权重进行相乘然后加总$\sum_k$好,最终在经过Relu进行过滤我们回想一下Relu的作用是什么?是不是就是让大于0的参数原值输出,小于0的参数就直接等于0 相当于舍弃其实Relu在这里扮演很重要的角色, 因为我们真正感兴趣的就是哪些能够代表Tiger Cat这个类别的特征, 而那些小于0被舍弃的就是属于其他类别的, 在这里用不上, 所有经过relu的参数能有更好的表达能力于是 我们就提取出来这个Tiger Cat类别的CAM图啦那么要注意一下, 这个提取出来的CAM 大小以vgg16来说是14*14像素因为经过了很多层的卷积我们要将这个图进行放大到与原输入的尺寸一样大在进行叠加才能展现GradCAM容易分析的优势当然中间有很多实现的细节包含利用openCV将色彩空间转换就如下图是我做的一个范例那么是不是就很容易理解网络参数对于黑猩猩面部的权重被激活的比较多,白话的说就是网络是靠猩猩脸部来判别 哦 原来这是黑猩猩啊 !当然这样的效果是在预训练网络上(vgg16 imagenet 1000类 包含黑猩猩类)才会有的, 预训练好的参数早就可以轻易的判别黑猩猩了如果只是单纯的丢到不是预训练的网络会是下面这样子所以网络需要进行训练, 随着训练, 权重会越能突显这个类别的特征最后透过某些特定的纹路就能进行判别注意一下,代码中说的目标层就是我们锁定的VGG16中的第29层relu层论文连接: https://arxiv.org/pdf/1610.02391v1.pdf | import torch as t
from torchvision import models
import cv2
import sys
import numpy as np
import matplotlib.pyplot as plt
class FeatureExtractor():
"""
1. 提取目标层特征
2. register 目标层梯度
"""
def __init__(self, model, target_layers):
self.model = model
self.model_features = model.features
self.target_layers = target_layers
self.gradients = list()
def save_gradient(self, grad):
self.gradients.append(grad)
def get_gradients(self):
return self.gradients
def __call__(self, x):
target_activations = list()
self.gradients = list()
for name, module in self.model_features._modules.items(): #遍历的方式遍历网络的每一层
x = module(x) #input 会经过遍历的每一层
if name in self.target_layers: #设个条件,如果到了你指定的层, 则继续
x.register_hook(self.save_gradient) #利用hook来记录目标层的梯度
target_activations += [x] #这里只取得目标层的features
x = x.view(x.size(0), -1) #reshape成 全连接进入分类器
x = self.model.classifier(x)#进入分类器
return target_activations, x,
def preprocess_image(img):
"""
预处理层
将图像进行标准化处理
"""
mean = [0.485, 0.456, 0.406]
stds = [0.229, 0.224, 0.225]
preprocessed_img = img.copy()[:, :, ::-1] # BGR > RGB
#标准化处理, 将bgr三层都处理
for i in range(3):
preprocessed_img[:, :, i] = preprocessed_img[:, :, i] - mean[i]
preprocessed_img[:, :, i] = preprocessed_img[:, :, i] / stds[i]
preprocessed_img = \
np.ascontiguousarray(np.transpose(preprocessed_img, (2, 0, 1))) #transpose HWC > CHW
preprocessed_img = t.from_numpy(preprocessed_img) #totensor
preprocessed_img.unsqueeze_(0)
input = t.tensor(preprocessed_img, requires_grad=True)
return input
def show_cam_on_image(img, mask):
heatmap = cv2.applyColorMap(np.uint8(255*mask), cv2.COLORMAP_JET) #利用色彩空间转换将heatmap凸显
heatmap = np.float32(heatmap)/255 #归一化
cam = heatmap + np.float32(img) #将heatmap 叠加到原图
cam = cam / np.max(cam)
cv2.imwrite('GradCam_test.jpg', np.uint8(255 * cam))#生成图像
cam = cam[:, :, ::-1] #BGR > RGB
plt.figure(figsize=(10, 10))
plt.imshow(np.uint8(255*cam))
class GradCam():
"""
GradCam主要执行
1.提取特征(调用FeatureExtractor)
2.反向传播求目标层梯度
3.实现目标层的CAM图
"""
def __init__(self, model, target_layer_names):
self.model = model
self.extractor = FeatureExtractor(self.model, target_layer_names)
def forward(self, input):
return self.model(input)
def __call__(self, input):
features, output = self.extractor(input) #这里的feature 对应的就是目标层的输出, output是图像经过分类网络的输出
output.data
one_hot = output.max() #取1000个类中最大的值
self.model.features.zero_grad() #梯度清零
self.model.classifier.zero_grad() #梯度清零
one_hot.backward(retain_graph=True) #反向传播之后,为了取得目标层梯度
grad_val = self.extractor.get_gradients()[-1].data.numpy()
#调用函数get_gradients(), 得到目标层求得的梯
target = features[-1]
#features 目前是list 要把里面relu层的输出取出来, 也就是我们要的目标层 shape(1, 512, 14, 14)
target = target.data.numpy()[0, :] #(1, 512, 14, 14) > (512, 14, 14)
weights = np.mean(grad_val, axis = (2, 3))[0, :] #array shape (512, ) 求出relu梯度的 512层 每层权重
cam = np.zeros(target.shape[1:]) #做一个空白map,待会将值填上
#(14, 14) shape(512, 14, 14)tuple 索引[1:] 也就是从14开始开始
#for loop的方式将平均后的权重乘上目标层的每个feature map, 并且加到刚刚生成的空白map上
for i, w in enumerate(weights):
cam += w * target[i, :, :]
#w * target[i, :, :]
#target[i, :, :] = array:shape(14, 14)
#w = 512个的权重均值 shape(512, )
#每个均值分别乘上target的feature map
#在放到空白的14*14上(cam)
#最终 14*14的空白map 会被填满
cam = cv2.resize(cam, (224, 224)) #将14*14的featuremap 放大回224*224
cam = cam - np.min(cam)
cam = cam / np.max(cam)
return cam
grad_cam = GradCam(model = models.vgg16(pretrained=True), \
target_layer_names = ["29"])
#使用预训练vgg16
#我们的目标层取第29层relu, relu层只保留有用的结果, 所以取其层最能突显出特征
img = cv2.imread('./images/gorilla.jpg') #读取图像
img = np.float32(cv2.resize(img, (224, 224))) / 255 #为了丢到vgg16要求的224*224 先进行缩放并且归一化
input = preprocess_image(img)
mask = grad_cam(input)
show_cam_on_image(img, mask)
| /Library/Frameworks/Python.framework/Versions/3.7/lib/python3.7/site-packages/ipykernel_launcher.py:47: UserWarning: To copy construct from a tensor, it is recommended to use sourceTensor.clone().detach() or sourceTensor.clone().detach().requires_grad_(True), rather than torch.tensor(sourceTensor).
| OML | PaddleCare/Deep_learning/Grad_CAM_Pytorch-1.01-Stephenfang51-patch-1/Grad_CAM_Pytorch_1_tutorialv2.ipynb | momozi1996/Medical_AI_analysis |
1 ~ 9 (NO transistor) | GPIO.output(Pin.A.value, GPIO.LOW)
GPIO.output(Pin.B.value, GPIO.LOW)
GPIO.output(Pin.C.value,GPIO.LOW)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.HIGH)
GPIO.output(Pin.B.value,GPIO.LOW)
GPIO.output(Pin.C.value,GPIO.LOW)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.LOW)
GPIO.output(Pin.B.value,GPIO.HIGH)
GPIO.output(Pin.C.value,GPIO.LOW)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.HIGH)
GPIO.output(Pin.B.value,GPIO.HIGH)
GPIO.output(Pin.C.value,GPIO.LOW)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.LOW)
GPIO.output(Pin.B.value,GPIO.LOW)
GPIO.output(Pin.C.value,GPIO.HIGH)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.HIGH)
GPIO.output(Pin.B.value,GPIO.LOW)
GPIO.output(Pin.C.value,GPIO.HIGH)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.LOW)
GPIO.output(Pin.B.value,GPIO.HIGH)
GPIO.output(Pin.C.value,GPIO.HIGH)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.HIGH)
GPIO.output(Pin.B.value,GPIO.HIGH)
GPIO.output(Pin.C.value,GPIO.HIGH)
GPIO.output(Pin.D.value, GPIO.LOW)
GPIO.output(Pin.A.value,GPIO.LOW)
GPIO.output(Pin.B.value,GPIO.LOW)
GPIO.output(Pin.C.value,GPIO.LOW)
GPIO.output(Pin.D.value, GPIO.HIGH)
GPIO.output(Pin.A.value,GPIO.HIGH)
GPIO.output(Pin.B.value,GPIO.LOW)
GPIO.output(Pin.C.value,GPIO.LOW)
GPIO.output(Pin.D.value, GPIO.HIGH)
GPIO.output(Pin.A.value,GPIO.HIGH)
GPIO.output(Pin.B.value,GPIO.HIGH)
GPIO.output(Pin.C.value,GPIO.HIGH)
GPIO.output(Pin.D.value, GPIO.HIGH)
GPIO.cleanup() | _____no_output_____ | MIT | nixie/nixie.ipynb | tanico-rikudo/raspi4 |
(experimental) Channels Last Memory Format in PyTorch*********************************************************Author**: `Vitaly Fedyunin `_What is Channels Last---------------------Channels Last memory format is an alternative way of ordering NCHW tensors in memory preserving dimensions ordering. Channels Last tensors ordered in such a way that channels become the densest dimension (aka storing images pixel-per-pixel).For example, classic (contiguous) storage of NCHW tensor (in our case it is two 2x2 images with 3 color channels) look like this:.. figure:: /_static/img/classic_memory_format.png :alt: classic_memory_formatChannels Last memory format orders data differently:.. figure:: /_static/img/channels_last_memory_format.png :alt: channels_last_memory_formatPytorch supports memory formats (and provides back compatibility with existing models including eager, JIT, and TorchScript) by utilizing existing strides structure.For example, 10x3x16x16 batch in Channels Last format will have strides equal to (768, 1, 48, 3). Channels Last memory format is implemented for 4D NCWH Tensors only. | import torch
N, C, H, W = 10, 3, 32, 32 | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Memory Format API-----------------------Here is how to convert tensors between contiguous and channels last memory formats. Classic PyTorch contiguous tensor | x = torch.empty(N, C, H, W)
print(x.stride()) # Ouputs: (3072, 1024, 32, 1) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Conversion operator | x = x.contiguous(memory_format=torch.channels_last)
print(x.shape) # Outputs: (10, 3, 32, 32) as dimensions order preserved
print(x.stride()) # Outputs: (3072, 1, 96, 3) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Back to contiguous | x = x.contiguous(memory_format=torch.contiguous_format)
print(x.stride()) # Outputs: (3072, 1024, 32, 1) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Alternative option | x = x.to(memory_format=torch.channels_last)
print(x.stride()) # Ouputs: (3072, 1, 96, 3) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Format checks | print(x.is_contiguous(memory_format=torch.channels_last)) # Ouputs: True | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Create as Channels Last | x = torch.empty(N, C, H, W, memory_format=torch.channels_last)
print(x.stride()) # Ouputs: (3072, 1, 96, 3) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
``clone`` preserves memory format | y = x.clone()
print(y.stride()) # Ouputs: (3072, 1, 96, 3) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
``to``, ``cuda``, ``float`` ... preserves memory format | if torch.cuda.is_available():
y = x.cuda()
print(y.stride()) # Ouputs: (3072, 1, 96, 3) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
``empty_like``, ``*_like`` operators preserves memory format | y = torch.empty_like(x)
print(y.stride()) # Ouputs: (3072, 1, 96, 3) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Pointwise operators preserves memory format | z = x + y
print(z.stride()) # Ouputs: (3072, 1, 96, 3) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Conv, Batchnorm modules support Channels Last(only works for CudNN >= 7.6) | if torch.backends.cudnn.version() >= 7603:
input = torch.randint(1, 10, (2, 8, 4, 4), dtype=torch.float32, device="cuda", requires_grad=True)
model = torch.nn.Conv2d(8, 4, 3).cuda().float()
input = input.contiguous(memory_format=torch.channels_last)
model = model.to(memory_format=torch.channels_last) # Module parameters need to be Channels Last
out = model(input)
print(out.is_contiguous(memory_format=torch.channels_last)) # Ouputs: True | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Performance Gains-------------------------------------------------------------------------------------------The most significant performance gains are observed on NVidia's hardware with Tensor Cores support. We were able to archive over 22% perf gains while running 'AMP (Automated Mixed Precision) training scripts supplied by NVidia https://github.com/NVIDIA/apex.``python main_amp.py -a resnet50 --b 200 --workers 16 --opt-level O2 ./data`` | # opt_level = O2
# keep_batchnorm_fp32 = None <class 'NoneType'>
# loss_scale = None <class 'NoneType'>
# CUDNN VERSION: 7603
# => creating model 'resnet50'
# Selected optimization level O2: FP16 training with FP32 batchnorm and FP32 master weights.
# Defaults for this optimization level are:
# enabled : True
# opt_level : O2
# cast_model_type : torch.float16
# patch_torch_functions : False
# keep_batchnorm_fp32 : True
# master_weights : True
# loss_scale : dynamic
# Processing user overrides (additional kwargs that are not None)...
# After processing overrides, optimization options are:
# enabled : True
# opt_level : O2
# cast_model_type : torch.float16
# patch_torch_functions : False
# keep_batchnorm_fp32 : True
# master_weights : True
# loss_scale : dynamic
# Epoch: [0][10/125] Time 0.866 (0.866) Speed 230.949 (230.949) Loss 0.6735125184 (0.6735) Prec@1 61.000 (61.000) Prec@5 100.000 (100.000)
# Epoch: [0][20/125] Time 0.259 (0.562) Speed 773.481 (355.693) Loss 0.6968704462 (0.6852) Prec@1 55.000 (58.000) Prec@5 100.000 (100.000)
# Epoch: [0][30/125] Time 0.258 (0.461) Speed 775.089 (433.965) Loss 0.7877287269 (0.7194) Prec@1 51.500 (55.833) Prec@5 100.000 (100.000)
# Epoch: [0][40/125] Time 0.259 (0.410) Speed 771.710 (487.281) Loss 0.8285319805 (0.7467) Prec@1 48.500 (54.000) Prec@5 100.000 (100.000)
# Epoch: [0][50/125] Time 0.260 (0.380) Speed 770.090 (525.908) Loss 0.7370464802 (0.7447) Prec@1 56.500 (54.500) Prec@5 100.000 (100.000)
# Epoch: [0][60/125] Time 0.258 (0.360) Speed 775.623 (555.728) Loss 0.7592862844 (0.7472) Prec@1 51.000 (53.917) Prec@5 100.000 (100.000)
# Epoch: [0][70/125] Time 0.258 (0.345) Speed 774.746 (579.115) Loss 1.9698858261 (0.9218) Prec@1 49.500 (53.286) Prec@5 100.000 (100.000)
# Epoch: [0][80/125] Time 0.260 (0.335) Speed 770.324 (597.659) Loss 2.2505953312 (1.0879) Prec@1 50.500 (52.938) Prec@5 100.000 (100.000) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Passing ``--channels-last true`` allows running a model in Channels Last format with observed 22% perf gain.``python main_amp.py -a resnet50 --b 200 --workers 16 --opt-level O2 --channels-last true ./data`` | # opt_level = O2
# keep_batchnorm_fp32 = None <class 'NoneType'>
# loss_scale = None <class 'NoneType'>
#
# CUDNN VERSION: 7603
#
# => creating model 'resnet50'
# Selected optimization level O2: FP16 training with FP32 batchnorm and FP32 master weights.
#
# Defaults for this optimization level are:
# enabled : True
# opt_level : O2
# cast_model_type : torch.float16
# patch_torch_functions : False
# keep_batchnorm_fp32 : True
# master_weights : True
# loss_scale : dynamic
# Processing user overrides (additional kwargs that are not None)...
# After processing overrides, optimization options are:
# enabled : True
# opt_level : O2
# cast_model_type : torch.float16
# patch_torch_functions : False
# keep_batchnorm_fp32 : True
# master_weights : True
# loss_scale : dynamic
#
# Epoch: [0][10/125] Time 0.767 (0.767) Speed 260.785 (260.785) Loss 0.7579724789 (0.7580) Prec@1 53.500 (53.500) Prec@5 100.000 (100.000)
# Epoch: [0][20/125] Time 0.198 (0.482) Speed 1012.135 (414.716) Loss 0.7007197738 (0.7293) Prec@1 49.000 (51.250) Prec@5 100.000 (100.000)
# Epoch: [0][30/125] Time 0.198 (0.387) Speed 1010.977 (516.198) Loss 0.7113101482 (0.7233) Prec@1 55.500 (52.667) Prec@5 100.000 (100.000)
# Epoch: [0][40/125] Time 0.197 (0.340) Speed 1013.023 (588.333) Loss 0.8943189979 (0.7661) Prec@1 54.000 (53.000) Prec@5 100.000 (100.000)
# Epoch: [0][50/125] Time 0.198 (0.312) Speed 1010.541 (641.977) Loss 1.7113249302 (0.9551) Prec@1 51.000 (52.600) Prec@5 100.000 (100.000)
# Epoch: [0][60/125] Time 0.198 (0.293) Speed 1011.163 (683.574) Loss 5.8537774086 (1.7716) Prec@1 50.500 (52.250) Prec@5 100.000 (100.000)
# Epoch: [0][70/125] Time 0.198 (0.279) Speed 1011.453 (716.767) Loss 5.7595844269 (2.3413) Prec@1 46.500 (51.429) Prec@5 100.000 (100.000)
# Epoch: [0][80/125] Time 0.198 (0.269) Speed 1011.827 (743.883) Loss 2.8196096420 (2.4011) Prec@1 47.500 (50.938) Prec@5 100.000 (100.000) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
The following list of models has the full support of Channels Last and showing 8%-35% perf gains on Volta devices:``alexnet``, ``mnasnet0_5``, ``mnasnet0_75``, ``mnasnet1_0``, ``mnasnet1_3``, ``mobilenet_v2``, ``resnet101``, ``resnet152``, ``resnet18``, ``resnet34``, ``resnet50``, ``resnext50_32x4d``, ``shufflenet_v2_x0_5``, ``shufflenet_v2_x1_0``, ``shufflenet_v2_x1_5``, ``shufflenet_v2_x2_0``, ``squeezenet1_0``, ``squeezenet1_1``, ``vgg11``, ``vgg11_bn``, ``vgg13``, ``vgg13_bn``, ``vgg16``, ``vgg16_bn``, ``vgg19``, ``vgg19_bn``, ``wide_resnet101_2``, ``wide_resnet50_2`` Converting existing models--------------------------Channels Last support not limited by existing models, as any model can be converted to Channels Last and propagate format through the graph as soon as input formatted correctly. | # Need to be done once, after model initialization (or load)
model = model.to(memory_format=torch.channels_last) # Replace with your model
# Need to be done for every input
input = input.to(memory_format=torch.channels_last) # Replace with your input
output = model(input) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
However, not all operators fully converted to support Channels Last (usually returning contiguous output instead). That means you need to verify the list of used operators against supported operators list https://github.com/pytorch/pytorch/wiki/Operators-with-Channels-Last-support, or introduce memory format checks into eager execution mode and run your model.After running the code below, operators will raise an exception if the output of the operator doesn't match the memory format of the input. | def contains_cl(args):
for t in args:
if isinstance(t, torch.Tensor):
if t.is_contiguous(memory_format=torch.channels_last) and not t.is_contiguous():
return True
elif isinstance(t, list) or isinstance(t, tuple):
if contains_cl(list(t)):
return True
return False
def print_inputs(args, indent=''):
for t in args:
if isinstance(t, torch.Tensor):
print(indent, t.stride(), t.shape, t.device, t.dtype)
elif isinstance(t, list) or isinstance(t, tuple):
print(indent, type(t))
print_inputs(list(t), indent=indent + ' ')
else:
print(indent, t)
def check_wrapper(fn):
name = fn.__name__
def check_cl(*args, **kwargs):
was_cl = contains_cl(args)
try:
result = fn(*args, **kwargs)
except Exception as e:
print("`{}` inputs are:".format(name))
print_inputs(args)
print('-------------------')
raise e
failed = False
if was_cl:
if isinstance(result, torch.Tensor):
if result.dim() == 4 and not result.is_contiguous(memory_format=torch.channels_last):
print("`{}` got channels_last input, but output is not channels_last:".format(name),
result.shape, result.stride(), result.device, result.dtype)
failed = True
if failed and True:
print("`{}` inputs are:".format(name))
print_inputs(args)
raise Exception(
'Operator `{}` lost channels_last property'.format(name))
return result
return check_cl
def attribute(m):
for i in dir(m):
e = getattr(m, i)
exclude_functions = ['is_cuda', 'has_names', 'numel',
'stride', 'Tensor', 'is_contiguous', '__class__']
if i not in exclude_functions and not i.startswith('_') and '__call__' in dir(e):
try:
setattr(m, i, check_wrapper(e))
except Exception as e:
print(i)
print(e)
attribute(torch.Tensor)
attribute(torch.nn.functional)
attribute(torch) | _____no_output_____ | BSD-3-Clause | docs/_downloads/f11c58c36c9b8a5daf09d3f9a792ef84/memory_format_tutorial.ipynb | creduo/PyTorch-tutorials-kr |
Live in Lab**Neuro Video Series****By Neuromatch Academy****Content creators**: Anne Churchland, Chaoqun Yin, Alex Kostiuk, Lukas Oesch,Michael Ryan, Ashley Chen, Joao Couto**Content reviewers**: Tara van Viegen, Oluwatomisin Faniyan, Pooya Pakarian, Sirisha Sripada**Video editors, captioners, and translators**: Jeremy Forest, Tara van Viegen, Carolina Shimabuku, Shuze Liu **Our 2021 Sponsors, including Presenting Sponsor Facebook Reality Labs** Video | # @markdown
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = "https://player.bilibili.com/player.html?bvid={0}&page={1}".format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="BV1r5411T7pP", width=854, height=480, fs=1)
print("Video available at https://www.bilibili.com/video/{0}".format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="HCx-sOp9R7M", width=854, height=480, fs=1, rel=0)
print("Video available at https://youtube.com/watch?v=" + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out) | _____no_output_____ | CC-BY-4.0 | tutorials/W0D0_NeuroVideoSeries/student/W0D0_Tutorial4.ipynb | janeite/course-content |
**Chapter 2 – End-to-end Machine Learning project***Welcome to Machine Learning Housing Corp.! Your task is to predict median house values in Californian districts, given a number of features from these districts.**This notebook contains all the sample code and solutions to the exercices in chapter 2.* Setup First, let's import a few common modules, ensure MatplotLib plots figures inline and prepare a function to save the figures. We also check that Python 3.5 or later is installed (although Python 2.x may work, it is deprecated so we strongly recommend you use Python 3 instead), as well as Scikit-Learn ≥0.20. | # Python ≥3.5 is required
import sys
assert sys.version_info >= (3, 5)
# Scikit-Learn ≥0.20 is required
import sklearn
assert sklearn.__version__ >= "0.20"
# Common imports
import numpy as np
import os
# To plot pretty figures
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# Where to save the figures
PROJECT_ROOT_DIR = "."
CHAPTER_ID = "end_to_end_project"
IMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, "images", CHAPTER_ID)
os.makedirs(IMAGES_PATH, exist_ok=True)
def save_fig(fig_id, tight_layout=True, fig_extension="png", resolution=300):
path = os.path.join(IMAGES_PATH, fig_id + "." + fig_extension)
print("Saving figure", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
# Ignore useless warnings (see SciPy issue #5998)
import warnings
warnings.filterwarnings(action="ignore", message="^internal gelsd") | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Get the data | import os
import tarfile
import urllib
DOWNLOAD_ROOT = "https://raw.githubusercontent.com/ageron/handson-ml2/master/"
HOUSING_PATH = os.path.join("datasets", "housing")
HOUSING_URL = DOWNLOAD_ROOT + "datasets/housing/housing.tgz"
def fetch_housing_data(housing_url=HOUSING_URL, housing_path=HOUSING_PATH):
if not os.path.isdir(housing_path):
os.makedirs(housing_path)
tgz_path = os.path.join(housing_path, "housing.tgz")
urllib.request.urlretrieve(housing_url, tgz_path)
housing_tgz = tarfile.open(tgz_path)
housing_tgz.extractall(path=housing_path)
housing_tgz.close()
fetch_housing_data()
import pandas as pd
def load_housing_data(housing_path=HOUSING_PATH):
csv_path = os.path.join(housing_path, "housing.csv")
return pd.read_csv(csv_path)
housing = load_housing_data()
housing.head()
housing.info()
housing["ocean_proximity"].value_counts()
housing.describe()
%matplotlib inline
import matplotlib.pyplot as plt
housing.hist(bins=50, figsize=(20,15))
save_fig("attribute_histogram_plots")
plt.show()
# to make this notebook's output identical at every run
np.random.seed(42)
import numpy as np
# For illustration only. Sklearn has train_test_split()
def split_train_test(data, test_ratio):
shuffled_indices = np.random.permutation(len(data))
test_set_size = int(len(data) * test_ratio)
test_indices = shuffled_indices[:test_set_size]
train_indices = shuffled_indices[test_set_size:]
return data.iloc[train_indices], data.iloc[test_indices]
train_set, test_set = split_train_test(housing, 0.2)
len(train_set)
len(test_set)
from zlib import crc32
def test_set_check(identifier, test_ratio):
return crc32(np.int64(identifier)) & 0xffffffff < test_ratio * 2**32
def split_train_test_by_id(data, test_ratio, id_column):
ids = data[id_column]
in_test_set = ids.apply(lambda id_: test_set_check(id_, test_ratio))
return data.loc[~in_test_set], data.loc[in_test_set] | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
The implementation of `test_set_check()` above works fine in both Python 2 and Python 3. In earlier releases, the following implementation was proposed, which supported any hash function, but was much slower and did not support Python 2: | import hashlib
def test_set_check(identifier, test_ratio, hash=hashlib.md5):
return hash(np.int64(identifier)).digest()[-1] < 256 * test_ratio | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
If you want an implementation that supports any hash function and is compatible with both Python 2 and Python 3, here is one: | def test_set_check(identifier, test_ratio, hash=hashlib.md5):
return bytearray(hash(np.int64(identifier)).digest())[-1] < 256 * test_ratio
housing_with_id = housing.reset_index() # adds an `index` column
train_set, test_set = split_train_test_by_id(housing_with_id, 0.2, "index")
housing_with_id["id"] = housing["longitude"] * 1000 + housing["latitude"]
train_set, test_set = split_train_test_by_id(housing_with_id, 0.2, "id")
test_set.head()
from sklearn.model_selection import train_test_split
train_set, test_set = train_test_split(housing, test_size=0.2, random_state=42)
test_set.head()
housing["median_income"].hist()
housing["income_cat"] = pd.cut(housing["median_income"],
bins=[0., 1.5, 3.0, 4.5, 6., np.inf],
labels=[1, 2, 3, 4, 5])
housing["income_cat"].value_counts()
housing["income_cat"].hist()
from sklearn.model_selection import StratifiedShuffleSplit
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
for train_index, test_index in split.split(housing, housing["income_cat"]):
strat_train_set = housing.loc[train_index]
strat_test_set = housing.loc[test_index]
strat_test_set["income_cat"].value_counts() / len(strat_test_set)
housing["income_cat"].value_counts() / len(housing)
def income_cat_proportions(data):
return data["income_cat"].value_counts() / len(data)
train_set, test_set = train_test_split(housing, test_size=0.2, random_state=42)
compare_props = pd.DataFrame({
"Overall": income_cat_proportions(housing),
"Stratified": income_cat_proportions(strat_test_set),
"Random": income_cat_proportions(test_set),
}).sort_index()
compare_props["Rand. %error"] = 100 * compare_props["Random"] / compare_props["Overall"] - 100
compare_props["Strat. %error"] = 100 * compare_props["Stratified"] / compare_props["Overall"] - 100
compare_props
for set_ in (strat_train_set, strat_test_set):
set_.drop("income_cat", axis=1, inplace=True) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Discover and visualize the data to gain insights | housing = strat_train_set.copy()
housing.plot(kind="scatter", x="longitude", y="latitude")
save_fig("bad_visualization_plot")
housing.plot(kind="scatter", x="longitude", y="latitude", alpha=0.1)
save_fig("better_visualization_plot") | Saving figure better_visualization_plot
| Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
The argument `sharex=False` fixes a display bug (the x-axis values and legend were not displayed). This is a temporary fix (see: https://github.com/pandas-dev/pandas/issues/10611 ). Thanks to Wilmer Arellano for pointing it out. | housing.plot(kind="scatter", x="longitude", y="latitude", alpha=0.4,
s=housing["population"]/100, label="population", figsize=(10,7),
c="median_house_value", cmap=plt.get_cmap("jet"), colorbar=True,
sharex=False)
plt.legend()
save_fig("housing_prices_scatterplot")
import matplotlib.image as mpimg
california_img=mpimg.imread(PROJECT_ROOT_DIR + '/images/end_to_end_project/california.png')
ax = housing.plot(kind="scatter", x="longitude", y="latitude", figsize=(10,7),
s=housing['population']/100, label="Population",
c="median_house_value", cmap=plt.get_cmap("jet"),
colorbar=False, alpha=0.4,
)
plt.imshow(california_img, extent=[-124.55, -113.80, 32.45, 42.05], alpha=0.5,
cmap=plt.get_cmap("jet"))
plt.ylabel("Latitude", fontsize=14)
plt.xlabel("Longitude", fontsize=14)
prices = housing["median_house_value"]
tick_values = np.linspace(prices.min(), prices.max(), 11)
cbar = plt.colorbar()
cbar.ax.set_yticklabels(["$%dk"%(round(v/1000)) for v in tick_values], fontsize=14)
cbar.set_label('Median House Value', fontsize=16)
plt.legend(fontsize=16)
save_fig("california_housing_prices_plot")
plt.show()
corr_matrix = housing.corr()
corr_matrix["median_house_value"].sort_values(ascending=False)
# from pandas.tools.plotting import scatter_matrix # For older versions of Pandas
from pandas.plotting import scatter_matrix
attributes = ["median_house_value", "median_income", "total_rooms",
"housing_median_age"]
scatter_matrix(housing[attributes], figsize=(12, 8))
save_fig("scatter_matrix_plot")
housing.plot(kind="scatter", x="median_income", y="median_house_value",
alpha=0.1)
plt.axis([0, 16, 0, 550000])
save_fig("income_vs_house_value_scatterplot")
housing["rooms_per_household"] = housing["total_rooms"]/housing["households"]
housing["bedrooms_per_room"] = housing["total_bedrooms"]/housing["total_rooms"]
housing["population_per_household"]=housing["population"]/housing["households"]
corr_matrix = housing.corr()
corr_matrix["median_house_value"].sort_values(ascending=False)
housing.plot(kind="scatter", x="rooms_per_household", y="median_house_value",
alpha=0.2)
plt.axis([0, 5, 0, 520000])
plt.show()
housing.describe() | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Prepare the data for Machine Learning algorithms | housing = strat_train_set.drop("median_house_value", axis=1) # drop labels for training set
housing_labels = strat_train_set["median_house_value"].copy()
sample_incomplete_rows = housing[housing.isnull().any(axis=1)].head()
sample_incomplete_rows
sample_incomplete_rows.dropna(subset=["total_bedrooms"]) # option 1
sample_incomplete_rows.drop("total_bedrooms", axis=1) # option 2
median = housing["total_bedrooms"].median()
sample_incomplete_rows["total_bedrooms"].fillna(median, inplace=True) # option 3
sample_incomplete_rows
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(strategy="median") | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Remove the text attribute because median can only be calculated on numerical attributes: | housing_num = housing.drop("ocean_proximity", axis=1)
# alternatively: housing_num = housing.select_dtypes(include=[np.number])
imputer.fit(housing_num)
imputer.statistics_ | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Check that this is the same as manually computing the median of each attribute: | housing_num.median().values | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Transform the training set: | X = imputer.transform(housing_num)
housing_tr = pd.DataFrame(X, columns=housing_num.columns,
index=housing.index)
housing_tr.loc[sample_incomplete_rows.index.values]
imputer.strategy
housing_tr = pd.DataFrame(X, columns=housing_num.columns,
index=housing_num.index)
housing_tr.head() | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Now let's preprocess the categorical input feature, `ocean_proximity`: | housing_cat = housing[["ocean_proximity"]]
housing_cat.head(10)
from sklearn.preprocessing import OrdinalEncoder
ordinal_encoder = OrdinalEncoder()
housing_cat_encoded = ordinal_encoder.fit_transform(housing_cat)
housing_cat_encoded[:10]
ordinal_encoder.categories_
from sklearn.preprocessing import OneHotEncoder
cat_encoder = OneHotEncoder()
housing_cat_1hot = cat_encoder.fit_transform(housing_cat)
housing_cat_1hot | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
By default, the `OneHotEncoder` class returns a sparse array, but we can convert it to a dense array if needed by calling the `toarray()` method: | housing_cat_1hot.toarray() | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Alternatively, you can set `sparse=False` when creating the `OneHotEncoder`: | cat_encoder = OneHotEncoder(sparse=False)
housing_cat_1hot = cat_encoder.fit_transform(housing_cat)
housing_cat_1hot
cat_encoder.categories_ | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Let's create a custom transformer to add extra attributes: | from sklearn.base import BaseEstimator, TransformerMixin
# column index
rooms_ix, bedrooms_ix, population_ix, households_ix = 3, 4, 5, 6
class CombinedAttributesAdder(BaseEstimator, TransformerMixin):
def __init__(self, add_bedrooms_per_room = True): # no *args or **kargs
self.add_bedrooms_per_room = add_bedrooms_per_room
def fit(self, X, y=None):
return self # nothing else to do
def transform(self, X):
rooms_per_household = X[:, rooms_ix] / X[:, households_ix]
population_per_household = X[:, population_ix] / X[:, households_ix]
if self.add_bedrooms_per_room:
bedrooms_per_room = X[:, bedrooms_ix] / X[:, rooms_ix]
return np.c_[X, rooms_per_household, population_per_household,
bedrooms_per_room]
else:
return np.c_[X, rooms_per_household, population_per_household]
attr_adder = CombinedAttributesAdder(add_bedrooms_per_room=False)
housing_extra_attribs = attr_adder.transform(housing.values)
housing_extra_attribs = pd.DataFrame(
housing_extra_attribs,
columns=list(housing.columns)+["rooms_per_household", "population_per_household"],
index=housing.index)
housing_extra_attribs.head() | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Now let's build a pipeline for preprocessing the numerical attributes: | from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
num_pipeline = Pipeline([
('imputer', SimpleImputer(strategy="median")),
('attribs_adder', CombinedAttributesAdder()),
('std_scaler', StandardScaler()),
])
housing_num_tr = num_pipeline.fit_transform(housing_num)
housing_num_tr
from sklearn.compose import ColumnTransformer
num_attribs = list(housing_num)
cat_attribs = ["ocean_proximity"]
full_pipeline = ColumnTransformer([
("num", num_pipeline, num_attribs),
("cat", OneHotEncoder(), cat_attribs),
])
housing_prepared = full_pipeline.fit_transform(housing)
housing_prepared
housing_prepared.shape | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
For reference, here is the old solution based on a `DataFrameSelector` transformer (to just select a subset of the Pandas `DataFrame` columns), and a `FeatureUnion`: | from sklearn.base import BaseEstimator, TransformerMixin
# Create a class to select numerical or categorical columns
class OldDataFrameSelector(BaseEstimator, TransformerMixin):
def __init__(self, attribute_names):
self.attribute_names = attribute_names
def fit(self, X, y=None):
return self
def transform(self, X):
return X[self.attribute_names].values | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Now let's join all these components into a big pipeline that will preprocess both the numerical and the categorical features: | num_attribs = list(housing_num)
cat_attribs = ["ocean_proximity"]
old_num_pipeline = Pipeline([
('selector', OldDataFrameSelector(num_attribs)),
('imputer', SimpleImputer(strategy="median")),
('attribs_adder', CombinedAttributesAdder()),
('std_scaler', StandardScaler()),
])
old_cat_pipeline = Pipeline([
('selector', OldDataFrameSelector(cat_attribs)),
('cat_encoder', OneHotEncoder(sparse=False)),
])
from sklearn.pipeline import FeatureUnion
old_full_pipeline = FeatureUnion(transformer_list=[
("num_pipeline", old_num_pipeline),
("cat_pipeline", old_cat_pipeline),
])
old_housing_prepared = old_full_pipeline.fit_transform(housing)
old_housing_prepared | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
The result is the same as with the `ColumnTransformer`: | np.allclose(housing_prepared, old_housing_prepared) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Select and train a model | from sklearn.linear_model import LinearRegression
lin_reg = LinearRegression()
lin_reg.fit(housing_prepared, housing_labels)
# let's try the full preprocessing pipeline on a few training instances
some_data = housing.iloc[:5]
some_labels = housing_labels.iloc[:5]
some_data_prepared = full_pipeline.transform(some_data)
print("Predictions:", lin_reg.predict(some_data_prepared)) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Compare against the actual values: | print("Labels:", list(some_labels))
some_data_prepared
from sklearn.metrics import mean_squared_error
housing_predictions = lin_reg.predict(housing_prepared)
lin_mse = mean_squared_error(housing_labels, housing_predictions)
lin_rmse = np.sqrt(lin_mse)
lin_rmse
from sklearn.metrics import mean_absolute_error
lin_mae = mean_absolute_error(housing_labels, housing_predictions)
lin_mae
from sklearn.tree import DecisionTreeRegressor
tree_reg = DecisionTreeRegressor(random_state=42)
tree_reg.fit(housing_prepared, housing_labels)
housing_predictions = tree_reg.predict(housing_prepared)
tree_mse = mean_squared_error(housing_labels, housing_predictions)
tree_rmse = np.sqrt(tree_mse)
tree_rmse | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Fine-tune your model | from sklearn.model_selection import cross_val_score
scores = cross_val_score(tree_reg, housing_prepared, housing_labels,
scoring="neg_mean_squared_error", cv=10)
tree_rmse_scores = np.sqrt(-scores)
def display_scores(scores):
print("Scores:", scores)
print("Mean:", scores.mean())
print("Standard deviation:", scores.std())
display_scores(tree_rmse_scores)
lin_scores = cross_val_score(lin_reg, housing_prepared, housing_labels,
scoring="neg_mean_squared_error", cv=10)
lin_rmse_scores = np.sqrt(-lin_scores)
display_scores(lin_rmse_scores) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
**Note**: we specify `n_estimators=100` to be future-proof since the default value is going to change to 100 in Scikit-Learn 0.22 (for simplicity, this is not shown in the book). | from sklearn.ensemble import RandomForestRegressor
forest_reg = RandomForestRegressor(n_estimators=100, random_state=42)
forest_reg.fit(housing_prepared, housing_labels)
housing_predictions = forest_reg.predict(housing_prepared)
forest_mse = mean_squared_error(housing_labels, housing_predictions)
forest_rmse = np.sqrt(forest_mse)
forest_rmse
from sklearn.model_selection import cross_val_score
forest_scores = cross_val_score(forest_reg, housing_prepared, housing_labels,
scoring="neg_mean_squared_error", cv=10)
forest_rmse_scores = np.sqrt(-forest_scores)
display_scores(forest_rmse_scores)
scores = cross_val_score(lin_reg, housing_prepared, housing_labels, scoring="neg_mean_squared_error", cv=10)
pd.Series(np.sqrt(-scores)).describe()
from sklearn.svm import SVR
svm_reg = SVR(kernel="linear")
svm_reg.fit(housing_prepared, housing_labels)
housing_predictions = svm_reg.predict(housing_prepared)
svm_mse = mean_squared_error(housing_labels, housing_predictions)
svm_rmse = np.sqrt(svm_mse)
svm_rmse
from sklearn.model_selection import GridSearchCV
param_grid = [
# try 12 (3×4) combinations of hyperparameters
{'n_estimators': [3, 10, 30], 'max_features': [2, 4, 6, 8]},
# then try 6 (2×3) combinations with bootstrap set as False
{'bootstrap': [False], 'n_estimators': [3, 10], 'max_features': [2, 3, 4]},
]
forest_reg = RandomForestRegressor(random_state=42)
# train across 5 folds, that's a total of (12+6)*5=90 rounds of training
grid_search = GridSearchCV(forest_reg, param_grid, cv=5,
scoring='neg_mean_squared_error',
return_train_score=True)
grid_search.fit(housing_prepared, housing_labels) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
The best hyperparameter combination found: | grid_search.best_params_
grid_search.best_estimator_ | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Let's look at the score of each hyperparameter combination tested during the grid search: | cvres = grid_search.cv_results_
for mean_score, params in zip(cvres["mean_test_score"], cvres["params"]):
print(np.sqrt(-mean_score), params)
pd.DataFrame(grid_search.cv_results_)
from sklearn.model_selection import RandomizedSearchCV
from scipy.stats import randint
param_distribs = {
'n_estimators': randint(low=1, high=200),
'max_features': randint(low=1, high=8),
}
forest_reg = RandomForestRegressor(random_state=42)
rnd_search = RandomizedSearchCV(forest_reg, param_distributions=param_distribs,
n_iter=10, cv=5, scoring='neg_mean_squared_error', random_state=42)
rnd_search.fit(housing_prepared, housing_labels)
cvres = rnd_search.cv_results_
for mean_score, params in zip(cvres["mean_test_score"], cvres["params"]):
print(np.sqrt(-mean_score), params)
feature_importances = grid_search.best_estimator_.feature_importances_
feature_importances
extra_attribs = ["rooms_per_hhold", "pop_per_hhold", "bedrooms_per_room"]
#cat_encoder = cat_pipeline.named_steps["cat_encoder"] # old solution
cat_encoder = full_pipeline.named_transformers_["cat"]
cat_one_hot_attribs = list(cat_encoder.categories_[0])
attributes = num_attribs + extra_attribs + cat_one_hot_attribs
sorted(zip(feature_importances, attributes), reverse=True)
final_model = grid_search.best_estimator_
X_test = strat_test_set.drop("median_house_value", axis=1)
y_test = strat_test_set["median_house_value"].copy()
X_test_prepared = full_pipeline.transform(X_test)
final_predictions = final_model.predict(X_test_prepared)
final_mse = mean_squared_error(y_test, final_predictions)
final_rmse = np.sqrt(final_mse)
final_rmse | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
We can compute a 95% confidence interval for the test RMSE: | from scipy import stats
confidence = 0.95
squared_errors = (final_predictions - y_test) ** 2
np.sqrt(stats.t.interval(confidence, len(squared_errors) - 1,
loc=squared_errors.mean(),
scale=stats.sem(squared_errors))) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
We could compute the interval manually like this: | m = len(squared_errors)
mean = squared_errors.mean()
tscore = stats.t.ppf((1 + confidence) / 2, df=m - 1)
tmargin = tscore * squared_errors.std(ddof=1) / np.sqrt(m)
np.sqrt(mean - tmargin), np.sqrt(mean + tmargin) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Alternatively, we could use a z-scores rather than t-scores: | zscore = stats.norm.ppf((1 + confidence) / 2)
zmargin = zscore * squared_errors.std(ddof=1) / np.sqrt(m)
np.sqrt(mean - zmargin), np.sqrt(mean + zmargin) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Extra material A full pipeline with both preparation and prediction | full_pipeline_with_predictor = Pipeline([
("preparation", full_pipeline),
("linear", LinearRegression())
])
full_pipeline_with_predictor.fit(housing, housing_labels)
full_pipeline_with_predictor.predict(some_data) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Model persistence using joblib | my_model = full_pipeline_with_predictor
import joblib
joblib.dump(my_model, "my_model.pkl") # DIFF
#...
my_model_loaded = joblib.load("my_model.pkl") # DIFF | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Example SciPy distributions for `RandomizedSearchCV` | from scipy.stats import geom, expon
geom_distrib=geom(0.5).rvs(10000, random_state=42)
expon_distrib=expon(scale=1).rvs(10000, random_state=42)
plt.hist(geom_distrib, bins=50)
plt.show()
plt.hist(expon_distrib, bins=50)
plt.show() | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Exercise solutions 1. Question: Try a Support Vector Machine regressor (`sklearn.svm.SVR`), with various hyperparameters such as `kernel="linear"` (with various values for the `C` hyperparameter) or `kernel="rbf"` (with various values for the `C` and `gamma` hyperparameters). Don't worry about what these hyperparameters mean for now. How does the best `SVR` predictor perform? | from sklearn.model_selection import GridSearchCV
param_grid = [
{'kernel': ['linear'], 'C': [10., 30., 100., 300., 1000., 3000., 10000., 30000.0]},
{'kernel': ['rbf'], 'C': [1.0, 3.0, 10., 30., 100., 300., 1000.0],
'gamma': [0.01, 0.03, 0.1, 0.3, 1.0, 3.0]},
]
svm_reg = SVR()
grid_search = GridSearchCV(svm_reg, param_grid, cv=5, scoring='neg_mean_squared_error', verbose=2)
grid_search.fit(housing_prepared, housing_labels) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
The best model achieves the following score (evaluated using 5-fold cross validation): | negative_mse = grid_search.best_score_
rmse = np.sqrt(-negative_mse)
rmse | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
That's much worse than the `RandomForestRegressor`. Let's check the best hyperparameters found: | grid_search.best_params_ | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
The linear kernel seems better than the RBF kernel. Notice that the value of `C` is the maximum tested value. When this happens you definitely want to launch the grid search again with higher values for `C` (removing the smallest values), because it is likely that higher values of `C` will be better. 2. Question: Try replacing `GridSearchCV` with `RandomizedSearchCV`. | from sklearn.model_selection import RandomizedSearchCV
from scipy.stats import expon, reciprocal
# see https://docs.scipy.org/doc/scipy/reference/stats.html
# for `expon()` and `reciprocal()` documentation and more probability distribution functions.
# Note: gamma is ignored when kernel is "linear"
param_distribs = {
'kernel': ['linear', 'rbf'],
'C': reciprocal(20, 200000),
'gamma': expon(scale=1.0),
}
svm_reg = SVR()
rnd_search = RandomizedSearchCV(svm_reg, param_distributions=param_distribs,
n_iter=50, cv=5, scoring='neg_mean_squared_error',
verbose=2, random_state=42)
rnd_search.fit(housing_prepared, housing_labels) | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
The best model achieves the following score (evaluated using 5-fold cross validation): | negative_mse = rnd_search.best_score_
rmse = np.sqrt(-negative_mse)
rmse | _____no_output_____ | Apache-2.0 | 02_end_to_end_machine_learning_project.ipynb | Ruqyai/handson-ml2 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.