hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ec5e01101cfdf8c35654c0ff53783e5a768a73d8 | 7,150 | ipynb | Jupyter Notebook | notebooks/LR_SourceCode.ipynb | egmaziero/RefactoringML | 138138c37b919ef6885d647460cc19015573a53b | [
"Apache-2.0"
]
| null | null | null | notebooks/LR_SourceCode.ipynb | egmaziero/RefactoringML | 138138c37b919ef6885d647460cc19015573a53b | [
"Apache-2.0"
]
| null | null | null | notebooks/LR_SourceCode.ipynb | egmaziero/RefactoringML | 138138c37b919ef6885d647460cc19015573a53b | [
"Apache-2.0"
]
| null | null | null | 26.579926 | 245 | 0.541538 | [
[
[
"import pandas as pd\nimport csv\nimport sys\nimport re\nimport scipy\nimport numpy as np\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn import preprocessing\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.model_selection import RandomizedSearchCV\nfrom scipy.stats import randint as sp_randint\nfrom time import time\n\ncsv.field_size_limit(sys.maxsize)",
"_____no_output_____"
],
[
"df = pd.read_pickle('../data/instances.pkl')\nlabels = list(set(df['target'].values))",
"_____no_output_____"
],
[
"X = []\nY = []\n\nprint(\"Preparing lists...\")\nfor index, row in df.iterrows():\n X.append(row[\"source_code\"])\n Y.append(row[\"target\"])",
"Preparing lists...\n"
],
[
"le = preprocessing.LabelEncoder() # for use in logistic regression\nle.fit(labels)\nY = le.transform(Y)",
"_____no_output_____"
],
[
"print(\"Extracting features...\")\ncv = CountVectorizer(binary=True)\ncv.fit(X)\ninstances = cv.transform(X)",
"Extracting features...\n"
],
[
"X_train, X_test, y_train, y_test = train_test_split(instances, Y, train_size = 0.75, random_state=42)",
"/Users/erickmaziero/virtualenvs/refact_env/lib/python3.6/site-packages/sklearn/model_selection/_split.py:2179: FutureWarning: From version 0.21, test_size will always complement train_size unless both are specified.\n FutureWarning)\n"
]
],
[
[
"# Default parameters",
"_____no_output_____"
]
],
[
[
"lr_classifier = LogisticRegression(random_state=42, verbose=1)\nlr_classifier.fit(X_train, y_train)",
"/Users/erickmaziero/virtualenvs/refact_env/lib/python3.6/site-packages/sklearn/linear_model/logistic.py:433: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.\n FutureWarning)\n/Users/erickmaziero/virtualenvs/refact_env/lib/python3.6/site-packages/sklearn/linear_model/logistic.py:460: FutureWarning: Default multi_class will be changed to 'auto' in 0.22. Specify the multi_class option to silence this warning.\n \"this warning.\", FutureWarning)\n"
],
[
"print(\"============ EVALUATION on test set:\")\nprint(accuracy_score(y_test, lr_classifier.predict(X_test)))",
"============ EVALUATION on test set:\n0.752123379526151\n"
]
],
[
[
"# Hyperparametrization",
"_____no_output_____"
]
],
[
[
"def report(results, n_top=3):\n for i in range(1, n_top + 1):\n candidates = np.flatnonzero(results['rank_test_score'] == i)\n for candidate in candidates:\n print(\"Model with rank: {0}\".format(i))\n print(\"Mean validation score: {0:.3f} (std: {1:.3f})\".format(\n results['mean_test_score'][candidate],\n results['std_test_score'][candidate]))\n print(\"Parameters: {0}\".format(results['params'][candidate]))\n print(\"\")\n\nparam_dist = {'C': scipy.stats.expon(scale=100)}\n\nlr_classifier2 = LogisticRegression(random_state=42)\n\nn_iter_search = 20\nrandom_search = RandomizedSearchCV(lr_classifier2,\n param_distributions=param_dist,\n n_iter=n_iter_search,\n cv=5,\n n_jobs=-1)\nstart = time()\nprint(\"Hyperparameter tuning...\")\nrandom_search.fit(X_train, y_train)\nprint(\"RandomizedSearchCV took %.2f seconds for %d candidates\"\n \" parameter settings.\" % ((time() - start), n_iter_search))\nreport(random_search.cv_results_)\nprint(\"============ EVALUATION on test set:\")\nprint(accuracy_score(Y_test, random_search.best_estimator_.predict(test_instances)))",
"Hyperparameter tuning...\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec5e02260fc1a1b0fb3d64734bda73993d2153ec | 17,996 | ipynb | Jupyter Notebook | tutorials/Tutorial1_Basic_QA_Pipeline.ipynb | rsanjaykamath/haystack | 3434d5205d14e3f2b20ab483738a78a958e8708a | [
"Apache-2.0"
]
| 1 | 2021-03-26T05:29:54.000Z | 2021-03-26T05:29:54.000Z | tutorials/Tutorial1_Basic_QA_Pipeline.ipynb | rsanjaykamath/haystack | 3434d5205d14e3f2b20ab483738a78a958e8708a | [
"Apache-2.0"
]
| null | null | null | tutorials/Tutorial1_Basic_QA_Pipeline.ipynb | rsanjaykamath/haystack | 3434d5205d14e3f2b20ab483738a78a958e8708a | [
"Apache-2.0"
]
| null | null | null | 39.378556 | 765 | 0.614303 | [
[
[
"# Build Your First QA System\n\n<img style=\"float: right;\" src=\"https://upload.wikimedia.org/wikipedia/en/d/d8/Game_of_Thrones_title_card.jpg\">\n\nEXECUTABLE VERSION: [*colab*](https://colab.research.google.com/github/deepset-ai/haystack/blob/master/tutorials/Tutorial1_Basic_QA_Pipeline.ipynb)\n\nQuestion Answering can be used in a variety of use cases. A very common one: Using it to navigate through complex knowledge bases or long documents (\"search setting\").\n\nA \"knowledge base\" could for example be your website, an internal wiki or a collection of financial reports. \nIn this tutorial we will work on a slightly different domain: \"Game of Thrones\". \n\nLet's see how we can use a bunch of Wikipedia articles to answer a variety of questions about the \nmarvellous seven kingdoms... \n\n",
"_____no_output_____"
]
],
[
[
"# Install the latest release of Haystack in your own environment \n#! pip install farm-haystack\n\n# Install the latest master of Haystack\n!pip install git+https://github.com/deepset-ai/haystack.git",
"_____no_output_____"
],
[
"from haystack import Finder\nfrom haystack.preprocessor.cleaning import clean_wiki_text\nfrom haystack.preprocessor.utils import convert_files_to_dicts, fetch_archive_from_http\nfrom haystack.reader.farm import FARMReader\nfrom haystack.reader.transformers import TransformersReader\nfrom haystack.utils import print_answers",
"_____no_output_____"
]
],
[
[
"## Document Store\n\nHaystack finds answers to queries within the documents stored in a `DocumentStore`. The current implementations of `DocumentStore` include `ElasticsearchDocumentStore`, `FAISSDocumentStore`, `SQLDocumentStore`, and `InMemoryDocumentStore`.\n\n**Here:** We recommended Elasticsearch as it comes preloaded with features like [full-text queries](https://www.elastic.co/guide/en/elasticsearch/reference/current/full-text-queries.html), [BM25 retrieval](https://www.elastic.co/elasticon/conf/2016/sf/improved-text-scoring-with-bm25), and [vector storage for text embeddings](https://www.elastic.co/guide/en/elasticsearch/reference/7.6/dense-vector.html).\n\n**Alternatives:** If you are unable to setup an Elasticsearch instance, then follow the [Tutorial 3](https://github.com/deepset-ai/haystack/blob/master/tutorials/Tutorial3_Basic_QA_Pipeline_without_Elasticsearch.ipynb) for using SQL/InMemory document stores.\n\n**Hint**: This tutorial creates a new document store instance with Wikipedia articles on Game of Thrones. However, you can configure Haystack to work with your existing document stores.\n\n### Start an Elasticsearch server\nYou can start Elasticsearch on your local machine instance using Docker. If Docker is not readily available in your environment (eg., in Colab notebooks), then you can manually download and execute Elasticsearch from source.",
"_____no_output_____"
]
],
[
[
"# Recommended: Start Elasticsearch using Docker\n#! docker run -d -p 9200:9200 -e \"discovery.type=single-node\" elasticsearch:7.6.2",
"0ae423cd9c30d6f02ca2073e430d4e1f4403d88b8ec316411ec4c198bad3d416\r\n"
],
[
"# In Colab / No Docker environments: Start Elasticsearch from source\n! wget https://artifacts.elastic.co/downloads/elasticsearch/elasticsearch-7.6.2-linux-x86_64.tar.gz -q\n! tar -xzf elasticsearch-7.6.2-linux-x86_64.tar.gz\n! chown -R daemon:daemon elasticsearch-7.6.2\n\nimport os\nfrom subprocess import Popen, PIPE, STDOUT\nes_server = Popen(['elasticsearch-7.6.2/bin/elasticsearch'],\n stdout=PIPE, stderr=STDOUT,\n preexec_fn=lambda: os.setuid(1) # as daemon\n )\n# wait until ES has started\n! sleep 30",
"_____no_output_____"
],
[
"# Connect to Elasticsearch\n\nfrom haystack.document_store.elasticsearch import ElasticsearchDocumentStore\ndocument_store = ElasticsearchDocumentStore(host=\"localhost\", username=\"\", password=\"\", index=\"document\")",
"07/07/2020 10:41:47 - INFO - elasticsearch - PUT http://localhost:9200/document [status:200 request:0.364s]\n"
]
],
[
[
"## Preprocessing of documents\n\nHaystack provides a customizable pipeline for:\n - converting files into texts\n - cleaning texts\n - splitting texts\n - writing them to a Document Store\n\nIn this tutorial, we download Wikipedia articles about Game of Thrones, apply a basic cleaning function, and index them in Elasticsearch.",
"_____no_output_____"
]
],
[
[
"# Let's first fetch some documents that we want to query\n# Here: 517 Wikipedia articles for Game of Thrones\ndoc_dir = \"data/article_txt_got\"\ns3_url = \"https://s3.eu-central-1.amazonaws.com/deepset.ai-farm-qa/datasets/documents/wiki_gameofthrones_txt.zip\"\nfetch_archive_from_http(url=s3_url, output_dir=doc_dir)\n\n# Convert files to dicts\n# You can optionally supply a cleaning function that is applied to each doc (e.g. to remove footers)\n# It must take a str as input, and return a str.\ndicts = convert_files_to_dicts(dir_path=doc_dir, clean_func=clean_wiki_text, split_paragraphs=True)\n\n# We now have a list of dictionaries that we can write to our document store.\n# If your texts come from a different source (e.g. a DB), you can of course skip convert_files_to_dicts() and create the dictionaries yourself.\n# The default format here is:\n# {\n# 'text': \"<DOCUMENT_TEXT_HERE>\",\n# 'meta': {'name': \"<DOCUMENT_NAME_HERE>\", ...}\n#}\n# (Optionally: you can also add more key-value-pairs here, that will be indexed as fields in Elasticsearch and\n# can be accessed later for filtering or shown in the responses of the Finder)\n\n# Let's have a look at the first 3 entries:\nprint(dicts[:3])\n\n# Now, let's write the dicts containing documents to our DB.\ndocument_store.write_documents(dicts)",
"07/07/2020 10:41:48 - INFO - haystack.indexing.utils - Found data stored in `data/article_txt_got`. Delete this first if you really want to fetch new data.\n07/07/2020 10:41:48 - INFO - elasticsearch - POST http://localhost:9200/_bulk [status:200 request:0.461s]\n07/07/2020 10:41:49 - INFO - elasticsearch - POST http://localhost:9200/_bulk [status:200 request:0.259s]\n07/07/2020 10:41:49 - INFO - elasticsearch - POST http://localhost:9200/_bulk [status:200 request:0.205s]\n07/07/2020 10:41:49 - INFO - elasticsearch - POST http://localhost:9200/_bulk [status:200 request:0.158s]\n07/07/2020 10:41:49 - INFO - elasticsearch - POST http://localhost:9200/_bulk [status:200 request:0.126s]\n07/07/2020 10:41:49 - INFO - elasticsearch - POST http://localhost:9200/_bulk [status:200 request:0.095s]\n"
]
],
[
[
"## Initalize Retriever, Reader, & Finder\n\n### Retriever\n\nRetrievers help narrowing down the scope for the Reader to smaller units of text where a given question could be answered.\nThey use some simple but fast algorithm.\n\n**Here:** We use Elasticsearch's default BM25 algorithm\n\n**Alternatives:**\n\n- Customize the `ElasticsearchRetriever`with custom queries (e.g. boosting) and filters\n- Use `TfidfRetriever` in combination with a SQL or InMemory Document store for simple prototyping and debugging\n- Use `EmbeddingRetriever` to find candidate documents based on the similarity of embeddings (e.g. created via Sentence-BERT)\n- Use `DensePassageRetriever` to use different embedding models for passage and query (see Tutorial 6)",
"_____no_output_____"
]
],
[
[
"from haystack.retriever.sparse import ElasticsearchRetriever\nretriever = ElasticsearchRetriever(document_store=document_store)",
"_____no_output_____"
],
[
"# Alternative: An in-memory TfidfRetriever based on Pandas dataframes for building quick-prototypes with SQLite document store.\n\n# from haystack.retriever.sparse import TfidfRetriever\n# retriever = TfidfRetriever(document_store=document_store)",
"_____no_output_____"
]
],
[
[
"### Reader\n\nA Reader scans the texts returned by retrievers in detail and extracts the k best answers. They are based\non powerful, but slower deep learning models.\n\nHaystack currently supports Readers based on the frameworks FARM and Transformers.\nWith both you can either load a local model or one from Hugging Face's model hub (https://huggingface.co/models).\n\n**Here:** a medium sized RoBERTa QA model using a Reader based on FARM (https://huggingface.co/deepset/roberta-base-squad2)\n\n**Alternatives (Reader):** TransformersReader (leveraging the `pipeline` of the Transformers package)\n\n**Alternatives (Models):** e.g. \"distilbert-base-uncased-distilled-squad\" (fast) or \"deepset/bert-large-uncased-whole-word-masking-squad2\" (good accuracy)\n\n**Hint:** You can adjust the model to return \"no answer possible\" with the no_ans_boost. Higher values mean the model prefers \"no answer possible\"\n\n#### FARMReader",
"_____no_output_____"
]
],
[
[
"# Load a local model or any of the QA models on\n# Hugging Face's model hub (https://huggingface.co/models)\n\nreader = FARMReader(model_name_or_path=\"deepset/roberta-base-squad2\", use_gpu=False)",
"04/28/2020 12:29:45 - INFO - farm.utils - device: cpu n_gpu: 0, distributed training: False, automatic mixed precision training: None\n04/28/2020 12:29:45 - INFO - farm.infer - Could not find `deepset/roberta-base-squad2` locally. Try to download from model hub ...\n04/28/2020 12:29:49 - WARNING - farm.modeling.language_model - Could not automatically detect from language model name what language it is. \n\t We guess it's an *ENGLISH* model ... \n\t If not: Init the language model by supplying the 'language' param.\n04/28/2020 12:29:54 - WARNING - farm.modeling.prediction_head - Some unused parameters are passed to the QuestionAnsweringHead. Might not be a problem. Params: {\"loss_ignore_index\": -1}\n04/28/2020 12:29:58 - INFO - farm.utils - device: cpu n_gpu: 0, distributed training: False, automatic mixed precision training: None\n"
]
],
[
[
"#### TransformersReader",
"_____no_output_____"
]
],
[
[
"# Alternative:\n# reader = TransformersReader(model=\"distilbert-base-uncased-distilled-squad\", tokenizer=\"distilbert-base-uncased\", use_gpu=-1)",
"_____no_output_____"
]
],
[
[
"### Finder\n\nThe Finder sticks together reader and retriever in a pipeline to answer our actual questions. ",
"_____no_output_____"
]
],
[
[
"finder = Finder(reader, retriever)",
"_____no_output_____"
]
],
[
[
"## Voilà! Ask a question!",
"_____no_output_____"
]
],
[
[
"# You can configure how many candidates the reader and retriever shall return\n# The higher top_k_retriever, the better (but also the slower) your answers. \nprediction = finder.get_answers(question=\"Who is the father of Arya Stark?\", top_k_retriever=10, top_k_reader=5)",
"04/28/2020 12:27:53 - INFO - elasticsearch - GET http://localhost:9200/document/_search [status:200 request:0.113s]\n04/28/2020 12:27:53 - INFO - haystack.retriever.elasticsearch - Got 10 candidates from retriever\n04/28/2020 12:27:53 - INFO - haystack.finder - Reader is looking for detailed answer in 362347 chars ...\n"
],
[
"# prediction = finder.get_answers(question=\"Who created the Dothraki vocabulary?\", top_k_reader=5)\n# prediction = finder.get_answers(question=\"Who is the sister of Sansa?\", top_k_reader=5)",
"_____no_output_____"
],
[
"print_answers(prediction, details=\"minimal\")",
"[ { 'answer': 'Eddard',\n 'context': 's Nymeria after a legendary warrior queen. She travels '\n \"with her father, Eddard, to King's Landing when he is made \"\n 'Hand of the King. Before she leaves,'},\n { 'answer': 'Ned',\n 'context': 'girl disguised as a boy all along and is surprised to '\n \"learn she is Arya, Ned Stark's daughter. After the \"\n 'Goldcloaks get help from Ser Amory Lorch and '},\n { 'answer': 'Ned',\n 'context': 'in the television series.\\n'\n '\\n'\n '\\n'\n '====Season 1====\\n'\n 'Arya accompanies her father Ned and her sister Sansa to '\n \"King's Landing. Before their departure, Arya's ha\"},\n { 'answer': 'Balon Greyjoy',\n 'context': 'He sends Theon to the Iron Islands hoping to broker an '\n \"alliance with Balon Greyjoy, Theon's father. In exchange \"\n 'for Greyjoy support, Robb as the King '},\n { 'answer': 'Brynden Tully',\n 'context': 'o the weather. Sandor decides to instead take her to her '\n 'great-uncle Brynden Tully. On their way to Riverrun, they '\n \"encounter two men on Arya's death l\"}]\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec5e0d46e7797829b8beb2a12d002581f5a516dd | 72,937 | ipynb | Jupyter Notebook | IBM_DataScience/6_Data_Visualization_with_Python/labs/DV0101EN-2-3-1-Pie-Charts-Box-Plots-Scatter-Plots-and-Bubble-Plots-py-v2.0.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
]
| null | null | null | IBM_DataScience/6_Data_Visualization_with_Python/labs/DV0101EN-2-3-1-Pie-Charts-Box-Plots-Scatter-Plots-and-Bubble-Plots-py-v2.0.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
]
| null | null | null | IBM_DataScience/6_Data_Visualization_with_Python/labs/DV0101EN-2-3-1-Pie-Charts-Box-Plots-Scatter-Plots-and-Bubble-Plots-py-v2.0.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
]
| null | null | null | 30.479315 | 1,463 | 0.573934 | [
[
[
"<a href=\"https://cognitiveclass.ai\"><img src = \"https://ibm.box.com/shared/static/9gegpsmnsoo25ikkbl4qzlvlyjbgxs5x.png\" width = 400> </a>\n\n<h1 align=center><font size = 5>Pie Charts, Box Plots, Scatter Plots, and Bubble Plots</font></h1>",
"_____no_output_____"
],
[
"## Introduction\n\nIn this lab session, we continue exploring the Matplotlib library. More specificatlly, we will learn how to create pie charts, box plots, scatter plots, and bubble charts.",
"_____no_output_____"
],
[
"## Table of Contents\n\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n\n1. [Exploring Datasets with *p*andas](#0)<br>\n2. [Downloading and Prepping Data](#2)<br>\n3. [Visualizing Data using Matplotlib](#4) <br>\n4. [Pie Charts](#6) <br>\n5. [Box Plots](#8) <br>\n6. [Scatter Plots](#10) <br>\n7. [Bubble Plots](#12) <br> \n</div>\n<hr>",
"_____no_output_____"
],
[
"# Exploring Datasets with *pandas* and Matplotlib<a id=\"0\"></a>\n\nToolkits: The course heavily relies on [*pandas*](http://pandas.pydata.org/) and [**Numpy**](http://www.numpy.org/) for data wrangling, analysis, and visualization. The primary plotting library we will explore in the course is [Matplotlib](http://matplotlib.org/).\n\nDataset: Immigration to Canada from 1980 to 2013 - [International migration flows to and from selected countries - The 2015 revision](http://www.un.org/en/development/desa/population/migration/data/empirical2/migrationflows.shtml) from United Nation's website.\n\nThe dataset contains annual data on the flows of international migrants as recorded by the countries of destination. The data presents both inflows and outflows according to the place of birth, citizenship or place of previous / next residence both for foreigners and nationals. In this lab, we will focus on the Canadian Immigration data.",
"_____no_output_____"
],
[
"# Downloading and Prepping Data <a id=\"2\"></a>",
"_____no_output_____"
],
[
"Import primary modules.",
"_____no_output_____"
]
],
[
[
"import numpy as np # useful for many scientific computing in Python\nimport pandas as pd # primary data structure library",
"_____no_output_____"
]
],
[
[
"Let's download and import our primary Canadian Immigration dataset using *pandas* `read_excel()` method. Normally, before we can do that, we would need to download a module which *pandas* requires to read in excel files. This module is **xlrd**. For your convenience, we have pre-installed this module, so you would not have to worry about that. Otherwise, you would need to run the following line of code to install the **xlrd** module:\n```\n!conda install -c anaconda xlrd --yes\n```",
"_____no_output_____"
],
[
"Download the dataset and read it into a *pandas* dataframe.",
"_____no_output_____"
]
],
[
[
"df_can = pd.read_excel('https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DV0101EN/labs/Data_Files/Canada.xlsx',\n sheet_name='Canada by Citizenship',\n skiprows=range(20),\n skipfooter=2\n )\n\nprint('Data downloaded and read into a dataframe!')",
"_____no_output_____"
]
],
[
[
"Let's take a look at the first five items in our dataset.",
"_____no_output_____"
]
],
[
[
"df_can.head()",
"_____no_output_____"
]
],
[
[
"Let's find out how many entries there are in our dataset.",
"_____no_output_____"
]
],
[
[
"# print the dimensions of the dataframe\nprint(df_can.shape)",
"_____no_output_____"
]
],
[
[
"Clean up data. We will make some modifications to the original dataset to make it easier to create our visualizations. Refer to *Introduction to Matplotlib and Line Plots* and *Area Plots, Histograms, and Bar Plots* for a detailed description of this preprocessing.",
"_____no_output_____"
]
],
[
[
"# clean up the dataset to remove unnecessary columns (eg. REG) \ndf_can.drop(['AREA', 'REG', 'DEV', 'Type', 'Coverage'], axis=1, inplace=True)\n\n# let's rename the columns so that they make sense\ndf_can.rename(columns={'OdName':'Country', 'AreaName':'Continent','RegName':'Region'}, inplace=True)\n\n# for sake of consistency, let's also make all column labels of type string\ndf_can.columns = list(map(str, df_can.columns))\n\n# set the country name as index - useful for quickly looking up countries using .loc method\ndf_can.set_index('Country', inplace=True)\n\n# add total column\ndf_can['Total'] = df_can.sum(axis=1)\n\n# years that we will be using in this lesson - useful for plotting later on\nyears = list(map(str, range(1980, 2014)))\nprint('data dimensions:', df_can.shape)",
"_____no_output_____"
]
],
[
[
"# Visualizing Data using Matplotlib<a id=\"4\"></a>",
"_____no_output_____"
],
[
"Import `Matplotlib`.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\n\nmpl.style.use('ggplot') # optional: for ggplot-like style\n\n# check for latest version of Matplotlib\nprint('Matplotlib version: ', mpl.__version__) # >= 2.0.0",
"_____no_output_____"
]
],
[
[
"# Pie Charts <a id=\"6\"></a>\n\nA `pie chart` is a circualr graphic that displays numeric proportions by dividing a circle (or pie) into proportional slices. You are most likely already familiar with pie charts as it is widely used in business and media. We can create pie charts in Matplotlib by passing in the `kind=pie` keyword.\n\nLet's use a pie chart to explore the proportion (percentage) of new immigrants grouped by continents for the entire time period from 1980 to 2013. ",
"_____no_output_____"
],
[
"Step 1: Gather data. \n\nWe will use *pandas* `groupby` method to summarize the immigration data by `Continent`. The general process of `groupby` involves the following steps:\n\n1. **Split:** Splitting the data into groups based on some criteria.\n2. **Apply:** Applying a function to each group independently:\n .sum()\n .count()\n .mean() \n .std() \n .aggregate()\n .apply()\n .etc..\n3. **Combine:** Combining the results into a data structure.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DV0101EN/labs/Images/Mod3Fig4SplitApplyCombine.png\" height=400 align=\"center\">",
"_____no_output_____"
]
],
[
[
"# group countries by continents and apply sum() function \ndf_continents = df_can.groupby('Continent', axis=0).sum()\n\n# note: the output of the groupby method is a `groupby' object. \n# we can not use it further until we apply a function (eg .sum())\nprint(type(df_can.groupby('Continent', axis=0)))\n\ndf_continents.head()",
"_____no_output_____"
]
],
[
[
"Step 2: Plot the data. We will pass in `kind = 'pie'` keyword, along with the following additional parameters:\n- `autopct` - is a string or function used to label the wedges with their numeric value. The label will be placed inside the wedge. If it is a format string, the label will be `fmt%pct`.\n- `startangle` - rotates the start of the pie chart by angle degrees counterclockwise from the x-axis.\n- `shadow` - Draws a shadow beneath the pie (to give a 3D feel).",
"_____no_output_____"
]
],
[
[
"# autopct create %, start angle represent starting point\ndf_continents['Total'].plot(kind='pie',\n figsize=(5, 6),\n autopct='%1.1f%%', # add in percentages\n startangle=90, # start angle 90° (Africa)\n shadow=True, # add shadow \n )\n\nplt.title('Immigration to Canada by Continent [1980 - 2013]')\nplt.axis('equal') # Sets the pie chart to look like a circle.\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"The above visual is not very clear, the numbers and text overlap in some instances. Let's make a few modifications to improve the visuals:\n\n* Remove the text labels on the pie chart by passing in `legend` and add it as a seperate legend using `plt.legend()`.\n* Push out the percentages to sit just outside the pie chart by passing in `pctdistance` parameter.\n* Pass in a custom set of colors for continents by passing in `colors` parameter.\n* **Explode** the pie chart to emphasize the lowest three continents (Africa, North America, and Latin America and Carribbean) by pasing in `explode` parameter.\n",
"_____no_output_____"
]
],
[
[
"colors_list = ['gold', 'yellowgreen', 'lightcoral', 'lightskyblue', 'lightgreen', 'pink']\nexplode_list = [0.1, 0, 0, 0, 0.1, 0.1] # ratio for each continent with which to offset each wedge.\n\ndf_continents['Total'].plot(kind='pie',\n figsize=(15, 6),\n autopct='%1.1f%%', \n startangle=90, \n shadow=True, \n labels=None, # turn off labels on pie chart\n pctdistance=1.12, # the ratio between the center of each pie slice and the start of the text generated by autopct \n colors=colors_list, # add custom colors\n explode=explode_list # 'explode' lowest 3 continents\n )\n\n# scale the title up by 12% to match pctdistance\nplt.title('Immigration to Canada by Continent [1980 - 2013]', y=1.12) \n\nplt.axis('equal') \n\n# add legend\nplt.legend(labels=df_continents.index, loc='upper left') \n\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Question:** Using a pie chart, explore the proportion (percentage) of new immigrants grouped by continents in the year 2013.\n\n**Note**: You might need to play with the explore values in order to fix any overlapping slice values.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\nexplode_list = [0.1, 0, 0, 0, 0.1, 0.2] # ratio for each continent with which to offset each wedge.\n-->\n\n<!--\ndf_continents['2013'].plot(kind='pie',\n figsize=(15, 6),\n autopct='%1.1f%%', \n startangle=90, \n shadow=True, \n labels=None, # turn off labels on pie chart\n pctdistance=1.12, # the ratio between the pie center and start of text label\n explode=explode_list # 'explode' lowest 3 continents\n )\n-->\n\n<!--\n\\\\ # scale the title up by 12% to match pctdistance\nplt.title('Immigration to Canada by Continent in 2013', y=1.12) \nplt.axis('equal') \n-->\n\n<!--\n\\\\ # add legend\nplt.legend(labels=df_continents.index, loc='upper left') \n-->\n\n<!--\n\\\\ # show plot\nplt.show()\n-->",
"_____no_output_____"
],
[
"# Box Plots <a id=\"8\"></a>\n\nA `box plot` is a way of statistically representing the *distribution* of the data through five main dimensions: \n\n- **Minimun:** Smallest number in the dataset.\n- **First quartile:** Middle number between the `minimum` and the `median`.\n- **Second quartile (Median):** Middle number of the (sorted) dataset.\n- **Third quartile:** Middle number between `median` and `maximum`.\n- **Maximum:** Highest number in the dataset.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DV0101EN/labs/Images/boxplot_complete.png\" width=440, align=\"center\">",
"_____no_output_____"
],
[
"To make a `box plot`, we can use `kind=box` in `plot` method invoked on a *pandas* series or dataframe.\n\nLet's plot the box plot for the Japanese immigrants between 1980 - 2013.",
"_____no_output_____"
],
[
"Step 1: Get the dataset. Even though we are extracting the data for just one country, we will obtain it as a dataframe. This will help us with calling the `dataframe.describe()` method to view the percentiles.",
"_____no_output_____"
]
],
[
[
"# to get a dataframe, place extra square brackets around 'Japan'.\ndf_japan = df_can.loc[['Japan'], years].transpose()\ndf_japan.head()",
"_____no_output_____"
]
],
[
[
"Step 2: Plot by passing in `kind='box'`.",
"_____no_output_____"
]
],
[
[
"df_japan.plot(kind='box', figsize=(8, 6))\n\nplt.title('Box plot of Japanese Immigrants from 1980 - 2013')\nplt.ylabel('Number of Immigrants')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can immediately make a few key observations from the plot above:\n1. The minimum number of immigrants is around 200 (min), maximum number is around 1300 (max), and median number of immigrants is around 900 (median).\n2. 25% of the years for period 1980 - 2013 had an annual immigrant count of ~500 or fewer (First quartile).\n2. 75% of the years for period 1980 - 2013 had an annual immigrant count of ~1100 or fewer (Third quartile).\n\nWe can view the actual numbers by calling the `describe()` method on the dataframe.",
"_____no_output_____"
]
],
[
[
"df_japan.describe()",
"_____no_output_____"
]
],
[
[
"One of the key benefits of box plots is comparing the distribution of multiple datasets. In one of the previous labs, we observed that China and India had very similar immigration trends. Let's analyize these two countries further using box plots.\n\n**Question:** Compare the distribution of the number of new immigrants from India and China for the period 1980 - 2013.",
"_____no_output_____"
],
[
"Step 1: Get the dataset for China and India and call the dataframe **df_CI**.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\ndf_CI= df_can.loc[['China', 'India'], years].transpose()\ndf_CI.head()\n-->",
"_____no_output_____"
],
[
"Let's view the percentages associated with both countries using the `describe()` method.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\ndf_CI.describe()\n-->",
"_____no_output_____"
],
[
"Step 2: Plot data.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\ndf_CI.plot(kind='box', figsize=(10, 7))\n-->\n\n<!--\nplt.title('Box plots of Immigrants from China and India (1980 - 2013)')\nplt.xlabel('Number of Immigrants')\n-->\n\n<!--\nplt.show()\n-->",
"_____no_output_____"
],
[
"We can observe that, while both countries have around the same median immigrant population (~20,000), China's immigrant population range is more spread out than India's. The maximum population from India for any year (36,210) is around 15% lower than the maximum population from China (42,584).\n",
"_____no_output_____"
],
[
"If you prefer to create horizontal box plots, you can pass the `vert` parameter in the **plot** function and assign it to *False*. You can also specify a different color in case you are not a big fan of the default red color.",
"_____no_output_____"
]
],
[
[
"# horizontal box plots\ndf_CI.plot(kind='box', figsize=(10, 7), color='blue', vert=False)\n\nplt.title('Box plots of Immigrants from China and India (1980 - 2013)')\nplt.xlabel('Number of Immigrants')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Subplots**\n\nOften times we might want to plot multiple plots within the same figure. For example, we might want to perform a side by side comparison of the box plot with the line plot of China and India's immigration.\n\nTo visualize multiple plots together, we can create a **`figure`** (overall canvas) and divide it into **`subplots`**, each containing a plot. With **subplots**, we usually work with the **artist layer** instead of the **scripting layer**. \n\nTypical syntax is : <br>\n```python\n fig = plt.figure() # create figure\n ax = fig.add_subplot(nrows, ncols, plot_number) # create subplots\n```\nWhere\n- `nrows` and `ncols` are used to notionally split the figure into (`nrows` \\* `ncols`) sub-axes, \n- `plot_number` is used to identify the particular subplot that this function is to create within the notional grid. `plot_number` starts at 1, increments across rows first and has a maximum of `nrows` * `ncols` as shown below.\n\n<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DV0101EN/labs/Images/Mod3Fig5Subplots_V2.png\" width=500 align=\"center\">",
"_____no_output_____"
],
[
"We can then specify which subplot to place each plot by passing in the `ax` paramemter in `plot()` method as follows:",
"_____no_output_____"
]
],
[
[
"fig = plt.figure() # create figure\n\nax0 = fig.add_subplot(1, 2, 1) # add subplot 1 (1 row, 2 columns, first plot)\nax1 = fig.add_subplot(1, 2, 2) # add subplot 2 (1 row, 2 columns, second plot). See tip below**\n\n# Subplot 1: Box plot\ndf_CI.plot(kind='box', color='blue', vert=False, figsize=(20, 6), ax=ax0) # add to subplot 1\nax0.set_title('Box Plots of Immigrants from China and India (1980 - 2013)')\nax0.set_xlabel('Number of Immigrants')\nax0.set_ylabel('Countries')\n\n# Subplot 2: Line plot\ndf_CI.plot(kind='line', figsize=(20, 6), ax=ax1) # add to subplot 2\nax1.set_title ('Line Plots of Immigrants from China and India (1980 - 2013)')\nax1.set_ylabel('Number of Immigrants')\nax1.set_xlabel('Years')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"** * Tip regarding subplot convention **\n\nIn the case when `nrows`, `ncols`, and `plot_number` are all less than 10, a convenience exists such that the a 3 digit number can be given instead, where the hundreds represent `nrows`, the tens represent `ncols` and the units represent `plot_number`. For instance,\n```python\n subplot(211) == subplot(2, 1, 1) \n```\nproduces a subaxes in a figure which represents the top plot (i.e. the first) in a 2 rows by 1 column notional grid (no grid actually exists, but conceptually this is how the returned subplot has been positioned).",
"_____no_output_____"
],
[
"Let's try something a little more advanced. \n\nPreviously we identified the top 15 countries based on total immigration from 1980 - 2013.\n\n**Question:** Create a box plot to visualize the distribution of the top 15 countries (based on total immigration) grouped by the *decades* `1980s`, `1990s`, and `2000s`.",
"_____no_output_____"
],
[
"Step 1: Get the dataset. Get the top 15 countries based on Total immigrant population. Name the dataframe **df_top15**.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\ndf_top15 = df_can.sort_values(['Total'], ascending=False, axis=0).head(15)\ndf_top15\n-->",
"_____no_output_____"
],
[
"Step 2: Create a new dataframe which contains the aggregate for each decade. One way to do that:\n 1. Create a list of all years in decades 80's, 90's, and 00's.\n 2. Slice the original dataframe df_can to create a series for each decade and sum across all years for each country.\n 3. Merge the three series into a new data frame. Call your dataframe **new_df**.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\n\\\\ # create a list of all years in decades 80's, 90's, and 00's\nyears_80s = list(map(str, range(1980, 1990))) \nyears_90s = list(map(str, range(1990, 2000))) \nyears_00s = list(map(str, range(2000, 2010))) \n-->\n\n<!--\n\\\\ # slice the original dataframe df_can to create a series for each decade\ndf_80s = df_top15.loc[:, years_80s].sum(axis=1) \ndf_90s = df_top15.loc[:, years_90s].sum(axis=1) \ndf_00s = df_top15.loc[:, years_00s].sum(axis=1)\n-->\n\n<!--\n\\\\ # merge the three series into a new data frame\nnew_df = pd.DataFrame({'1980s': df_80s, '1990s': df_90s, '2000s':df_00s}) \n-->\n\n<!--\n\\\\ # display dataframe\nnew_df.head()\n-->",
"_____no_output_____"
],
[
"Let's learn more about the statistics associated with the dataframe using the `describe()` method.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\nnew_df.describe()\n-->",
"_____no_output_____"
],
[
"Step 3: Plot the box plots.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\nnew_df.plot(kind='box', figsize=(10, 6))\n-->\n\n<!--\nplt.title('Immigration from top 15 countries for decades 80s, 90s and 2000s')\n-->\n\n<!--\nplt.show()\n-->",
"_____no_output_____"
],
[
"Note how the box plot differs from the summary table created. The box plot scans the data and identifies the outliers. In order to be an outlier, the data value must be:<br>\n* larger than Q3 by at least 1.5 times the interquartile range (IQR), or,\n* smaller than Q1 by at least 1.5 times the IQR.\n\nLet's look at decade 2000s as an example: <br>\n* Q1 (25%) = 36,101.5 <br>\n* Q3 (75%) = 105,505.5 <br>\n* IQR = Q3 - Q1 = 69,404 <br>\n\nUsing the definition of outlier, any value that is greater than Q3 by 1.5 times IQR will be flagged as outlier.\n\nOutlier > 105,505.5 + (1.5 * 69,404) <br>\nOutlier > 209,611.5",
"_____no_output_____"
]
],
[
[
"# let's check how many entries fall above the outlier threshold \nnew_df[new_df['2000s']> 209611.5]",
"_____no_output_____"
]
],
[
[
"China and India are both considered as outliers since their population for the decade exceeds 209,611.5. \n\nThe box plot is an advanced visualizaiton tool, and there are many options and customizations that exceed the scope of this lab. Please refer to [Matplotlib documentation](http://matplotlib.org/api/pyplot_api.html#matplotlib.pyplot.boxplot) on box plots for more information.",
"_____no_output_____"
],
[
"# Scatter Plots <a id=\"10\"></a>\n\nA `scatter plot` (2D) is a useful method of comparing variables against each other. `Scatter` plots look similar to `line plots` in that they both map independent and dependent variables on a 2D graph. While the datapoints are connected together by a line in a line plot, they are not connected in a scatter plot. The data in a scatter plot is considered to express a trend. With further analysis using tools like regression, we can mathematically calculate this relationship and use it to predict trends outside the dataset.\n\nLet's start by exploring the following:\n\nUsing a `scatter plot`, let's visualize the trend of total immigrantion to Canada (all countries combined) for the years 1980 - 2013.",
"_____no_output_____"
],
[
"Step 1: Get the dataset. Since we are expecting to use the relationship betewen `years` and `total population`, we will convert `years` to `int` type.",
"_____no_output_____"
]
],
[
[
"# we can use the sum() method to get the total population per year\ndf_tot = pd.DataFrame(df_can[years].sum(axis=0))\n\n# change the years to type int (useful for regression later on)\ndf_tot.index = map(int, df_tot.index)\n\n# reset the index to put in back in as a column in the df_tot dataframe\ndf_tot.reset_index(inplace = True)\n\n# rename columns\ndf_tot.columns = ['year', 'total']\n\n# view the final dataframe\ndf_tot.head()",
"_____no_output_____"
]
],
[
[
"Step 2: Plot the data. In `Matplotlib`, we can create a `scatter` plot set by passing in `kind='scatter'` as plot argument. We will also need to pass in `x` and `y` keywords to specify the columns that go on the x- and the y-axis.",
"_____no_output_____"
]
],
[
[
"df_tot.plot(kind='scatter', x='year', y='total', figsize=(10, 6), color='darkblue')\n\nplt.title('Total Immigration to Canada from 1980 - 2013')\nplt.xlabel('Year')\nplt.ylabel('Number of Immigrants')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Notice how the scatter plot does not connect the datapoints together. We can clearly observe an upward trend in the data: as the years go by, the total number of immigrants increases. We can mathematically analyze this upward trend using a regression line (line of best fit). ",
"_____no_output_____"
],
[
"So let's try to plot a linear line of best fit, and use it to predict the number of immigrants in 2015.\n\nStep 1: Get the equation of line of best fit. We will use **Numpy**'s `polyfit()` method by passing in the following:\n- `x`: x-coordinates of the data. \n- `y`: y-coordinates of the data. \n- `deg`: Degree of fitting polynomial. 1 = linear, 2 = quadratic, and so on.",
"_____no_output_____"
]
],
[
[
"x = df_tot['year'] # year on x-axis\ny = df_tot['total'] # total on y-axis\nfit = np.polyfit(x, y, deg=1)\n\nfit",
"_____no_output_____"
]
],
[
[
"The output is an array with the polynomial coefficients, highest powers first. Since we are plotting a linear regression `y= a*x + b`, our output has 2 elements `[5.56709228e+03, -1.09261952e+07]` with the the slope in position 0 and intercept in position 1. \n\nStep 2: Plot the regression line on the `scatter plot`.",
"_____no_output_____"
]
],
[
[
"df_tot.plot(kind='scatter', x='year', y='total', figsize=(10, 6), color='darkblue')\n\nplt.title('Total Immigration to Canada from 1980 - 2013')\nplt.xlabel('Year')\nplt.ylabel('Number of Immigrants')\n\n# plot line of best fit\nplt.plot(x, fit[0] * x + fit[1], color='red') # recall that x is the Years\nplt.annotate('y={0:.0f} x + {1:.0f}'.format(fit[0], fit[1]), xy=(2000, 150000))\n\nplt.show()\n\n# print out the line of best fit\n'No. Immigrants = {0:.0f} * Year + {1:.0f}'.format(fit[0], fit[1]) ",
"_____no_output_____"
]
],
[
[
"Using the equation of line of best fit, we can estimate the number of immigrants in 2015:\n```python\nNo. Immigrants = 5567 * Year - 10926195\nNo. Immigrants = 5567 * 2015 - 10926195\nNo. Immigrants = 291,310\n```\nWhen compared to the actuals from Citizenship and Immigration Canada's (CIC) [2016 Annual Report](http://www.cic.gc.ca/english/resources/publications/annual-report-2016/index.asp), we see that Canada accepted 271,845 immigrants in 2015. Our estimated value of 291,310 is within 7% of the actual number, which is pretty good considering our original data came from United Nations (and might differ slightly from CIC data).\n\nAs a side note, we can observe that immigration took a dip around 1993 - 1997. Further analysis into the topic revealed that in 1993 Canada introcuded Bill C-86 which introduced revisions to the refugee determination system, mostly restrictive. Further amendments to the Immigration Regulations cancelled the sponsorship required for \"assisted relatives\" and reduced the points awarded to them, making it more difficult for family members (other than nuclear family) to immigrate to Canada. These restrictive measures had a direct impact on the immigration numbers for the next several years.",
"_____no_output_____"
],
[
"**Question**: Create a scatter plot of the total immigration from Denmark, Norway, and Sweden to Canada from 1980 to 2013?",
"_____no_output_____"
],
[
"Step 1: Get the data:\n 1. Create a dataframe the consists of the numbers associated with Denmark, Norway, and Sweden only. Name it **df_countries**.\n 2. Sum the immigration numbers across all three countries for each year and turn the result into a dataframe. Name this new dataframe **df_total**.\n 3. Reset the index in place.\n 4. Rename the columns to **year** and **total**.\n 5. Display the resulting dataframe.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\n\\\\ # create df_countries dataframe\ndf_countries = df_can.loc[['Denmark', 'Norway', 'Sweden'], years].transpose()\n-->\n\n<!--\n\\\\ # create df_total by summing across three countries for each year\ndf_total = pd.DataFrame(df_countries.sum(axis=1))\n-->\n\n<!--\n\\\\ # reset index in place\ndf_total.reset_index(inplace=True)\n-->\n\n<!--\n\\\\ # rename columns\ndf_total.columns = ['year', 'total']\n-->\n\n<!--\n\\\\ # change column year from string to int to create scatter plot\ndf_total['year'] = df_total['year'].astype(int)\n-->\n\n<!--\n\\\\ # show resulting dataframe\ndf_total.head()\n-->",
"_____no_output_____"
],
[
"Step 2: Generate the scatter plot by plotting the total versus year in **df_total**.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\n\\\\ # generate scatter plot\ndf_total.plot(kind='scatter', x='year', y='total', figsize=(10, 6), color='darkblue')\n-->\n\n<!--\n\\\\ # add title and label to axes\nplt.title('Immigration from Denmark, Norway, and Sweden to Canada from 1980 - 2013')\nplt.xlabel('Year')\nplt.ylabel('Number of Immigrants')\n-->\n\n<!--\n\\\\ # show plot\nplt.show()\n-->",
"_____no_output_____"
],
[
"# Bubble Plots <a id=\"12\"></a>\n\nA `bubble plot` is a variation of the `scatter plot` that displays three dimensions of data (x, y, z). The datapoints are replaced with bubbles, and the size of the bubble is determined by the third variable 'z', also known as the weight. In `maplotlib`, we can pass in an array or scalar to the keyword `s` to `plot()`, that contains the weight of each point.\n\n**Let's start by analyzing the effect of Argentina's great depression**.\n\nArgentina suffered a great depression from 1998 - 2002, which caused widespread unemployment, riots, the fall of the government, and a default on the country's foreign debt. In terms of income, over 50% of Argentines were poor, and seven out of ten Argentine children were poor at the depth of the crisis in 2002. \n\nLet's analyze the effect of this crisis, and compare Argentina's immigration to that of it's neighbour Brazil. Let's do that using a `bubble plot` of immigration from Brazil and Argentina for the years 1980 - 2013. We will set the weights for the bubble as the *normalized* value of the population for each year.",
"_____no_output_____"
],
[
"Step 1: Get the data for Brazil and Argentina. Like in the previous example, we will convert the `Years` to type int and bring it in the dataframe.",
"_____no_output_____"
]
],
[
[
"df_can_t = df_can[years].transpose() # transposed dataframe\n\n# cast the Years (the index) to type int\ndf_can_t.index = map(int, df_can_t.index)\n\n# let's label the index. This will automatically be the column name when we reset the index\ndf_can_t.index.name = 'Year'\n\n# reset index to bring the Year in as a column\ndf_can_t.reset_index(inplace=True)\n\n# view the changes\ndf_can_t.head()",
"_____no_output_____"
]
],
[
[
"Step 2: Create the normalized weights. \n\nThere are several methods of normalizations in statistics, each with its own use. In this case, we will use [feature scaling](https://en.wikipedia.org/wiki/Feature_scaling) to bring all values into the range [0,1]. The general formula is:\n\n<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DV0101EN/labs/Images/Mod3Fig3FeatureScaling.png\" align=\"center\">\n\nwhere *`X`* is an original value, *`X'`* is the normalized value. The formula sets the max value in the dataset to 1, and sets the min value to 0. The rest of the datapoints are scaled to a value between 0-1 accordingly.\n",
"_____no_output_____"
]
],
[
[
"# normalize Brazil data\nnorm_brazil = (df_can_t['Brazil'] - df_can_t['Brazil'].min()) / (df_can_t['Brazil'].max() - df_can_t['Brazil'].min())\n\n# normalize Argentina data\nnorm_argentina = (df_can_t['Argentina'] - df_can_t['Argentina'].min()) / (df_can_t['Argentina'].max() - df_can_t['Argentina'].min())",
"_____no_output_____"
]
],
[
[
"Step 3: Plot the data. \n- To plot two different scatter plots in one plot, we can include the axes one plot into the other by passing it via the `ax` parameter. \n- We will also pass in the weights using the `s` parameter. Given that the normalized weights are between 0-1, they won't be visible on the plot. Therefore we will:\n - multiply weights by 2000 to scale it up on the graph, and,\n - add 10 to compensate for the min value (which has a 0 weight and therefore scale with x2000).",
"_____no_output_____"
]
],
[
[
"# Brazil\nax0 = df_can_t.plot(kind='scatter',\n x='Year',\n y='Brazil',\n figsize=(14, 8),\n alpha=0.5, # transparency\n color='green',\n s=norm_brazil * 2000 + 10, # pass in weights \n xlim=(1975, 2015)\n )\n\n# Argentina\nax1 = df_can_t.plot(kind='scatter',\n x='Year',\n y='Argentina',\n alpha=0.5,\n color=\"blue\",\n s=norm_argentina * 2000 + 10,\n ax = ax0\n )\n\nax0.set_ylabel('Number of Immigrants')\nax0.set_title('Immigration from Brazil and Argentina from 1980 - 2013')\nax0.legend(['Brazil', 'Argentina'], loc='upper left', fontsize='x-large')",
"_____no_output_____"
]
],
[
[
"The size of the bubble corresponds to the magnitude of immigrating population for that year, compared to the 1980 - 2013 data. The larger the bubble, the more immigrants in that year.\n\nFrom the plot above, we can see a corresponding increase in immigration from Argentina during the 1998 - 2002 great depression. We can also observe a similar spike around 1985 to 1993. In fact, Argentina had suffered a great depression from 1974 - 1990, just before the onset of 1998 - 2002 great depression. \n\nOn a similar note, Brazil suffered the *Samba Effect* where the Brazilian real (currency) dropped nearly 35% in 1999. There was a fear of a South American financial crisis as many South American countries were heavily dependent on industrial exports from Brazil. The Brazilian government subsequently adopted an austerity program, and the economy slowly recovered over the years, culminating in a surge in 2010. The immigration data reflect these events.",
"_____no_output_____"
],
[
"**Question**: Previously in this lab, we created box plots to compare immigration from China and India to Canada. Create bubble plots of immigration from China and India to visualize any differences with time from 1980 to 2013. You can use **df_can_t** that we defined and used in the previous example.",
"_____no_output_____"
],
[
"Step 1: Normalize the data pertaining to China and India.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\n\\\\ # normalize China data\nnorm_china = (df_can_t['China'] - df_can_t['China'].min()) / (df_can_t['China'].max() - df_can_t['China'].min())\n-->\n\n<!--\n# normalize India data\nnorm_india = (df_can_t['India'] - df_can_t['India'].min()) / (df_can_t['India'].max() - df_can_t['India'].min())\n-->",
"_____no_output_____"
],
[
"Step 2: Generate the bubble plots.",
"_____no_output_____"
]
],
[
[
"### type your answer here\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n<!-- The correct answer is:\n\\\\ # China\nax0 = df_can_t.plot(kind='scatter',\n x='Year',\n y='China',\n figsize=(14, 8),\n alpha=0.5, # transparency\n color='green',\n s=norm_china * 2000 + 10, # pass in weights \n xlim=(1975, 2015)\n )\n-->\n\n<!--\n\\\\ # India\nax1 = df_can_t.plot(kind='scatter',\n x='Year',\n y='India',\n alpha=0.5,\n color=\"blue\",\n s=norm_india * 2000 + 10,\n ax = ax0\n )\n-->\n\n<!--\nax0.set_ylabel('Number of Immigrants')\nax0.set_title('Immigration from China and India from 1980 - 2013')\nax0.legend(['China', 'India'], loc='upper left', fontsize='x-large')\n-->",
"_____no_output_____"
],
[
"### Thank you for completing this lab!\n\nThis notebook was created by [Jay Rajasekharan](https://www.linkedin.com/in/jayrajasekharan) with contributions from [Ehsan M. Kermani](https://www.linkedin.com/in/ehsanmkermani), and [Slobodan Markovic](https://www.linkedin.com/in/slobodan-markovic).\n\nThis notebook was recently revamped by [Alex Aklson](https://www.linkedin.com/in/aklson/). I hope you found this lab session interesting. Feel free to contact me if you have any questions!",
"_____no_output_____"
],
[
"This notebook is part of a course on **Coursera** called *Data Visualization with Python*. If you accessed this notebook outside the course, you can take this course online by clicking [here](http://cocl.us/DV0101EN_Coursera_Week2_LAB2).",
"_____no_output_____"
],
[
"<hr>\n\nCopyright © 2019 [Cognitive Class](https://cognitiveclass.ai/?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec5e134b14686f77efd0df83acbeee6fbf2f11a5 | 18,850 | ipynb | Jupyter Notebook | NpOperations.ipynb | ABPande/MyPythonRepo | 51de39aee6c99b6ea2eb47fb199925ee63ca3750 | [
"Apache-2.0"
]
| null | null | null | NpOperations.ipynb | ABPande/MyPythonRepo | 51de39aee6c99b6ea2eb47fb199925ee63ca3750 | [
"Apache-2.0"
]
| null | null | null | NpOperations.ipynb | ABPande/MyPythonRepo | 51de39aee6c99b6ea2eb47fb199925ee63ca3750 | [
"Apache-2.0"
]
| null | null | null | 19.373073 | 86 | 0.415597 | [
[
[
"import numpy as np\nmylist = [1,2,3]\nnp.array(mylist)",
"_____no_output_____"
],
[
"np.array([[1,2,3],[4,5,6],[6,7,8]])",
"_____no_output_____"
],
[
"np.arange(0, 11, 2)",
"_____no_output_____"
],
[
"np.linspace(0,10,100).reshape(20,5)",
"_____no_output_____"
],
[
"np.zeros (3)",
"_____no_output_____"
],
[
"np.zeros((4,3))",
"_____no_output_____"
],
[
"np.ones(3)",
"_____no_output_____"
],
[
"np.ones((3,4))",
"_____no_output_____"
],
[
"np.eye (4)",
"_____no_output_____"
],
[
"np.eye(3)",
"_____no_output_____"
],
[
"np.random.rand(5,5)",
"_____no_output_____"
],
[
"np.random.randn(5,4)",
"_____no_output_____"
],
[
"np.random.randint(0,101,10).reshape(5,2)",
"_____no_output_____"
],
[
"arr = np.arange(25)",
"_____no_output_____"
],
[
"arr",
"_____no_output_____"
],
[
"arr.reshape(5,5)",
"_____no_output_____"
],
[
"arr.max()",
"_____no_output_____"
],
[
"arr.min()",
"_____no_output_____"
],
[
"arr.sum()",
"_____no_output_____"
],
[
"arr.std()",
"_____no_output_____"
],
[
"arr.shape",
"_____no_output_____"
],
[
"arr.reshape(5,5).shape",
"_____no_output_____"
],
[
"arr.dtype",
"_____no_output_____"
],
[
"from numpy.random import randint as rnd",
"_____no_output_____"
],
[
"rnd(0,5)",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
]
],
[
[
"arr",
"_____no_output_____"
],
[
"arr[1:4]",
"_____no_output_____"
],
[
"arr[5:9]",
"_____no_output_____"
],
[
"mat = arr.reshape(5,5)",
"_____no_output_____"
],
[
"mat",
"_____no_output_____"
],
[
"arr[5:] = 99",
"_____no_output_____"
],
[
"arr",
"_____no_output_____"
],
[
"arr = mat.reshape(25,)",
"_____no_output_____"
],
[
"arr",
"_____no_output_____"
],
[
"mat",
"_____no_output_____"
],
[
"arr = np.arange(25)",
"_____no_output_____"
],
[
"arr",
"_____no_output_____"
],
[
"mat",
"_____no_output_____"
],
[
"mat = arr.reshape(5,5)",
"_____no_output_____"
],
[
"mat",
"_____no_output_____"
],
[
"arr_2d = np.array([[5,10,15],[20,25,30],[35,40,45]])",
"_____no_output_____"
],
[
"arr_2d",
"_____no_output_____"
],
[
"arr_2d[1,]",
"_____no_output_____"
],
[
"arr_2d[:2,1:]",
"_____no_output_____"
],
[
"arr_2d[1:,:2]",
"_____no_output_____"
],
[
"arr_2d [arr_2d > 15]",
"_____no_output_____"
],
[
"arr_2d> 25",
"_____no_output_____"
]
]
]
| [
"code",
"raw",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5e14d8775301d36a66ffa679fa678152ac54c7 | 55,327 | ipynb | Jupyter Notebook | tamil word2vec and random encoding.ipynb | ankitmishra2232/Hope-Speech-Identification | 71c03cc61455315a52f6c79a86342eccdd7ed546 | [
"Apache-2.0"
]
| null | null | null | tamil word2vec and random encoding.ipynb | ankitmishra2232/Hope-Speech-Identification | 71c03cc61455315a52f6c79a86342eccdd7ed546 | [
"Apache-2.0"
]
| null | null | null | tamil word2vec and random encoding.ipynb | ankitmishra2232/Hope-Speech-Identification | 71c03cc61455315a52f6c79a86342eccdd7ed546 | [
"Apache-2.0"
]
| null | null | null | 55,327 | 55,327 | 0.575343 | [
[
[
"import pandas as pd\r\npd.set_option('display.max_colwidth',100)\r\nimport nltk",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"t_data=pd.read_csv('/content/drive/MyDrive/Ankit/EACL 2021 dec 2020/Hope speech/Tamil/tamil_hope_first_train.csv',names=[\"text\",\"label\",\"NaN\"], sep='\\t')\r\nt_data.pop('NaN')\r\nt_data",
"_____no_output_____"
],
[
"t_data['text']=t_data['text'].str.lower() #changing into lower case (remove and check acc too)",
"_____no_output_____"
],
[
"t_data['text']=t_data['text'].str.strip() #remove white spaces\r\nt_data['text']=t_data['text'].str.replace(r'\\d+','') #remove numbers\r\n#t_data['text']=t_data['text'].apply(lambda x: x.encode('ascii', 'ignore').decode('ascii')) #removing emoji\r\nt_data['text']=t_data['text'].str.replace('[^\\w\\s]','') #removing punct",
"_____no_output_____"
],
[
"#removing url if any\r\nimport re\r\ndef remove_URL(txt):\r\n url= re.compile(r\"https?://\\S+|www\\.\\S+\")\r\n return url.sub(r\"\",txt)\r\nt_data['text']=t_data['text'].apply(lambda x:remove_URL(x))",
"_____no_output_____"
],
[
"import numpy as np\r\nfrom keras.utils.np_utils import to_categorical\r\nfrom sklearn import preprocessing\r\nlabelEncode=preprocessing.LabelEncoder()\r\nlabelEncode.fit(t_data['label'])\r\nprint (labelEncode.classes_)\r\ntrain_labelEncode=labelEncode.transform(t_data['label'])\r\nlabel=to_categorical(np.asarray(train_labelEncode))\r\nlabel",
"['Hope_speech' 'Non_hope_speech' 'not-Tamil']\n"
],
[
"#counting labels \n \nt_data['label'].value_counts()\n#must use something to balance data",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"#words in each line\n \nt_data['totalwords'] = t_data['text'].str.count(' ') + 1\nt_data",
"_____no_output_____"
],
[
"import keras\r\nfrom keras.preprocessing.text import Tokenizer\r\ntok1 = Tokenizer(char_level=False, filters ='!\"$%&@()*+,-./:;”“<=>?[\\\\]^_`{|}~\\t\\n', lower = True)",
"_____no_output_____"
],
[
"tok2=Tokenizer(char_level=True,filters ='!\"$%&@()*+,-./:;”“<=>?[\\\\]^_`{|}~\\t\\n',lower=True)",
"_____no_output_____"
],
[
"tok1.fit_on_texts(t_data['text'])\r\nwords=len(tok1.word_counts)\r\nwords",
"_____no_output_____"
],
[
"tok2.fit_on_texts(t_data['text'])\r\nltr=len(tok2.word_counts)\r\nltr",
"_____no_output_____"
],
[
"Charlist=tok2.word_index\n#Charlist",
"_____no_output_____"
],
[
"wordlist=tok1.word_index\r\n#wordlist",
"_____no_output_____"
],
[
"\r\nencode=tok1.texts_to_sequences(t_data['text'])\r\nprint(t_data['text'][5555])\r\nencode[5555]",
"இநதய எலலப பகதயல சன சல அமபபத இநதய இரணவம அனமதகக கடத\n"
],
[
"## Padding encoded sequence of words\r\nfrom keras.preprocessing import sequence\r\nmax_length=30\r\npadd = sequence.pad_sequences(encode, maxlen=max_length, padding='post')\r\npadd",
"_____no_output_____"
],
[
"cencode=tok2.texts_to_sequences(t_data['text'])\r\ncpadd=sequence.pad_sequences(cencode,maxlen=70,padding='post')",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# importing libraries for creating model\r\nimport tensorflow as tf\r\nfrom tensorflow.keras.models import Sequential,Model\r\nfrom tensorflow.keras.layers import Dense, Dropout, Activation, Flatten, Embedding,MaxPool1D,Input\r\nfrom tensorflow.keras.layers import Conv1D, GlobalMaxPooling1D, MaxPooling1D,LSTM,Bidirectional,GlobalAveragePooling1D\r\nfrom keras.layers.merge import concatenate",
"_____no_output_____"
]
],
[
[
"#w2v Embedding",
"_____no_output_____"
]
],
[
[
"embeddings_index = {}\r\nf = open('/content/drive/MyDrive/Ankit/dec 2020 codalab/Hope speech/Tamil/tamil w2v.txt')\r\nfor line in f:\r\n values = line.split()\r\n word = values[0]\r\n coefs = np.asarray(values[1:])\r\n embeddings_index[word] = coefs\r\nf.close()\r\nprint('Loaded %s word vectors.' % len(embeddings_index))",
"Loaded 40997 word vectors.\n"
],
[
"embedding_matrix = np.zeros((words+1, 300))\r\nfor word, i in tok1.word_index.items():\r\n\tembedding_vector = embeddings_index.get(word)\r\n\tif embedding_vector is not None:\r\n\t\tembedding_matrix[i] = embedding_vector",
"_____no_output_____"
]
],
[
[
"#Embedding layer",
"_____no_output_____"
]
],
[
[
"Embedding_Layer = Embedding(words+1,300,weights=[embedding_matrix], input_length=max_length, trainable=False)\r\nEmbedding_Layer2=Embedding(words+1,300,input_length=max_length)\r\nEmbedding_Layer3=Embedding(ltr+1,300,input_length=70)",
"_____no_output_____"
]
],
[
[
"#CNN",
"_____no_output_____"
]
],
[
[
"# channel 1\r\ninputs0 = Input(shape=(70,))\r\nembedding0 = Embedding_Layer3(inputs0)\r\nconv0 = Bidirectional(LSTM(64))(embedding0)\r\ndrop0 = Dropout(0.5)(conv0)\r\n#pool0 = MaxPooling1D(pool_size=2)(drop0)\r\n#conv01=Conv1D(filters=64,kernel_size=4,activation='relu')(pool0)\r\n#drop01=Dropout(0.5)(conv01)\r\n#pool01=MaxPooling1D(pool_size=3,padding='same')(drop01)\r\n#flat0= Flatten()(pool01)",
"_____no_output_____"
],
[
"# channel 1\ninputs1 = Input(shape=(max_length,))\nembedding1 = Embedding_Layer2(inputs1)\nconv1 = Bidirectional(LSTM(32))(embedding1)\ndrop1 = Dropout(0.5)(conv1)\n#pool1 = MaxPooling1D(pool_size=2)(drop1)\n#conv12=Conv1D(filters=64,kernel_size=4,activation='relu')(pool1)\n#drop12=Dropout(0.5)(conv12)\n#pool12=MaxPooling1D(pool_size=3,padding='same')(drop12)\n#flat1 = (drop1)",
"_____no_output_____"
],
[
"# channel 2\r\ninputs2 = Input(shape=(max_length,))\r\nembedding2 =Embedding_Layer(inputs2)\r\nconv2 = Bidirectional(LSTM(32))(embedding2)\r\ndrop2 = Dropout(0.5)(conv2)\r\n#pool2 = MaxPooling1D(pool_size=2)(drop2)\r\n#flat2 =(drop2)",
"_____no_output_____"
],
[
"merged = concatenate([drop0,drop1,drop2])\r\ndense1 = Dense(64, activation='relu')(merged)\r\noutputs = Dense(3, activation='sigmoid')(dense1)\r\nmodel = Model(inputs=[inputs0,inputs1, inputs2], outputs=outputs)\r\nmodel.summary()",
"Model: \"model_3\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_11 (InputLayer) [(None, 70)] 0 \n__________________________________________________________________________________________________\ninput_12 (InputLayer) [(None, 30)] 0 \n__________________________________________________________________________________________________\ninput_13 (InputLayer) [(None, 30)] 0 \n__________________________________________________________________________________________________\nembedding_2 (Embedding) (None, 70, 300) 65700 input_11[0][0] \n__________________________________________________________________________________________________\nembedding_1 (Embedding) (None, 30, 300) 9612600 input_12[0][0] \n__________________________________________________________________________________________________\nembedding (Embedding) multiple 9612600 input_13[0][0] \n__________________________________________________________________________________________________\nbidirectional_9 (Bidirectional) (None, 128) 186880 embedding_2[2][0] \n__________________________________________________________________________________________________\nbidirectional_10 (Bidirectional (None, 64) 85248 embedding_1[2][0] \n__________________________________________________________________________________________________\nbidirectional_11 (Bidirectional (None, 64) 85248 embedding[5][0] \n__________________________________________________________________________________________________\ndropout_9 (Dropout) (None, 128) 0 bidirectional_9[0][0] \n__________________________________________________________________________________________________\ndropout_10 (Dropout) (None, 64) 0 bidirectional_10[0][0] \n__________________________________________________________________________________________________\ndropout_11 (Dropout) (None, 64) 0 bidirectional_11[0][0] \n__________________________________________________________________________________________________\nconcatenate_3 (Concatenate) (None, 256) 0 dropout_9[0][0] \n dropout_10[0][0] \n dropout_11[0][0] \n__________________________________________________________________________________________________\ndense_6 (Dense) (None, 64) 16448 concatenate_3[0][0] \n__________________________________________________________________________________________________\ndense_7 (Dense) (None, 3) 195 dense_6[0][0] \n==================================================================================================\nTotal params: 19,664,919\nTrainable params: 10,052,319\nNon-trainable params: 9,612,600\n__________________________________________________________________________________________________\n"
]
],
[
[
"#compile",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['acc'])\r\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"#Training models",
"_____no_output_____"
]
],
[
[
"model.fit([cpadd,padd,padd],label,epochs=50,verbose=1,batch_size=64)",
"Epoch 1/50\n253/253 [==============================] - 97s 348ms/step - loss: 0.5393 - acc: 0.5264\nEpoch 2/50\n253/253 [==============================] - 89s 351ms/step - loss: 0.4128 - acc: 0.6935\nEpoch 3/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.2508 - acc: 0.8498\nEpoch 4/50\n253/253 [==============================] - 88s 350ms/step - loss: 0.1756 - acc: 0.8922\nEpoch 5/50\n253/253 [==============================] - 88s 350ms/step - loss: 0.1348 - acc: 0.9120\nEpoch 6/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.1129 - acc: 0.9242\nEpoch 7/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0990 - acc: 0.9366\nEpoch 8/50\n253/253 [==============================] - 88s 346ms/step - loss: 0.0901 - acc: 0.9419\nEpoch 9/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0851 - acc: 0.9424\nEpoch 10/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.0751 - acc: 0.9496\nEpoch 11/50\n253/253 [==============================] - 89s 350ms/step - loss: 0.0738 - acc: 0.9520\nEpoch 12/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0699 - acc: 0.9536\nEpoch 13/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.0712 - acc: 0.9522\nEpoch 14/50\n253/253 [==============================] - 89s 350ms/step - loss: 0.0640 - acc: 0.9563\nEpoch 15/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0647 - acc: 0.9566\nEpoch 16/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0591 - acc: 0.9593\nEpoch 17/50\n253/253 [==============================] - 87s 346ms/step - loss: 0.0579 - acc: 0.9617\nEpoch 18/50\n253/253 [==============================] - 89s 350ms/step - loss: 0.0527 - acc: 0.9644\nEpoch 19/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0555 - acc: 0.9628\nEpoch 20/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0456 - acc: 0.9706\nEpoch 21/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0503 - acc: 0.9673\nEpoch 22/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.0517 - acc: 0.9692\nEpoch 23/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0498 - acc: 0.9655\nEpoch 24/50\n253/253 [==============================] - 88s 346ms/step - loss: 0.0454 - acc: 0.9710\nEpoch 25/50\n253/253 [==============================] - 88s 346ms/step - loss: 0.0499 - acc: 0.9664\nEpoch 26/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.0497 - acc: 0.9689\nEpoch 27/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0510 - acc: 0.9656\nEpoch 28/50\n253/253 [==============================] - 88s 346ms/step - loss: 0.0494 - acc: 0.9670\nEpoch 29/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0440 - acc: 0.9700\nEpoch 30/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.0428 - acc: 0.9723\nEpoch 31/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0429 - acc: 0.9713\nEpoch 32/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0413 - acc: 0.9719\nEpoch 33/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0415 - acc: 0.9727\nEpoch 34/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.0403 - acc: 0.9725\nEpoch 35/50\n253/253 [==============================] - 88s 347ms/step - loss: 0.0421 - acc: 0.9713\nEpoch 36/50\n253/253 [==============================] - 87s 344ms/step - loss: 0.0423 - acc: 0.9720\nEpoch 37/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0407 - acc: 0.9730\nEpoch 38/50\n253/253 [==============================] - 88s 346ms/step - loss: 0.0387 - acc: 0.9752\nEpoch 39/50\n253/253 [==============================] - 87s 346ms/step - loss: 0.0405 - acc: 0.9738\nEpoch 40/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0408 - acc: 0.9715\nEpoch 41/50\n253/253 [==============================] - 88s 346ms/step - loss: 0.0373 - acc: 0.9752\nEpoch 42/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0379 - acc: 0.9746\nEpoch 43/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0428 - acc: 0.9708\nEpoch 44/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0406 - acc: 0.9727\nEpoch 45/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0375 - acc: 0.9760\nEpoch 46/50\n253/253 [==============================] - 88s 349ms/step - loss: 0.0343 - acc: 0.9774\nEpoch 47/50\n253/253 [==============================] - 89s 351ms/step - loss: 0.0354 - acc: 0.9754\nEpoch 48/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0352 - acc: 0.9773\nEpoch 49/50\n253/253 [==============================] - 88s 348ms/step - loss: 0.0350 - acc: 0.9753\nEpoch 50/50\n253/253 [==============================] - 89s 350ms/step - loss: 0.0363 - acc: 0.9766\n"
]
],
[
[
"#Development data",
"_____no_output_____"
]
],
[
[
"d_data=pd.read_csv('/content/drive/MyDrive/Ankit/EACL 2021 dec 2020/Hope speech/Tamil/tamil_hope_first_dev.csv',names=['text','label','nan'],sep='\\t')",
"_____no_output_____"
],
[
"d_data=d_data[[\"text\",\"label\"]]\r\nd_data",
"_____no_output_____"
],
[
"d_data['label'].value_counts()",
"_____no_output_____"
],
[
"#processing as training set\r\n\r\nd_data['text']=d_data['text'].str.lower() #changing into lower case (remove and check acc too)\r\nd_data['text']=d_data['text'].str.strip() #remove white spaces\r\nd_data['text']=d_data['text'].str.replace(r'\\d+','') #remove numbers\r\n#d_data['text']=d_data['text'].apply(lambda x: x.encode('ascii', 'ignore').decode('ascii')) #removing emoji\r\nd_data['text']=d_data['text'].str.replace('[^\\w\\s]','') #removing punct\r\nd_data['text']=d_data['text'].apply(lambda x:remove_URL(x))\r\n#d_data['text'] = d_data['text'].apply(lambda x: remove_sw(x))\r\n",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"encoded1 =tok1.texts_to_sequences(d_data['text'])\r\nprint(d_data['text'][0])\r\nencoded1[0]",
"mg bro eve o clock video post pannuga\n"
],
[
"\r\npadded = sequence.pad_sequences(encoded1, maxlen=max_length, padding='post')\r\npadded[0]",
"_____no_output_____"
],
[
"cencoded1=tok2.texts_to_sequences(d_data['text'])\r\ncpadded=sequence.pad_sequences(cencoded1,maxlen=70,padding='post')",
"_____no_output_____"
],
[
"\r\nval_labelEncode=labelEncode.transform(d_data['label'])\r\nval_label=to_categorical(np.asarray(val_labelEncode))\r\nval_label[0]",
"_____no_output_____"
]
],
[
[
"#prediction",
"_____no_output_____"
]
],
[
[
"# devlopment prediction of CNN\r\n\r\ndev_predictions = model.predict([cpadded,padded,padded])\r\n# Finding out of two output neurons which one has highest probability\r\n# It will return us the predicted class\r\n# It will convert probability into final class 1 and 0\r\ndev_predictions1 = np.zeros_like(dev_predictions)\r\ndev_predictions1[np.arange(len(dev_predictions)), dev_predictions.argmax(1)] = 1\r\n#print(dev_predictions)\r\n#print(dev_predictions1)",
"_____no_output_____"
]
],
[
[
"#Acuracy report",
"_____no_output_____"
]
],
[
[
"#Accuracy CNN\r\nfrom sklearn.metrics import classification_report\r\nprint(classification_report(val_label,dev_predictions1))",
" precision recall f1-score support\n\n 0 0.49 0.52 0.50 757\n 1 0.63 0.58 0.61 998\n 2 0.53 0.56 0.55 263\n\n micro avg 0.56 0.56 0.56 2018\n macro avg 0.55 0.56 0.55 2018\nweighted avg 0.56 0.56 0.56 2018\n samples avg 0.56 0.56 0.56 2018\n\n"
]
],
[
[
"#TESTING",
"_____no_output_____"
]
],
[
[
"tx_data=pd.read_csv('/content/drive/MyDrive/Ankit/dec 2020 codalab/Hope speech/Tamil/tamil_hope_test.csv',names=['text'],sep=',')\r\ntx_data\r\n",
"_____no_output_____"
],
[
"#PREprocessing\r\n#processing as training set\r\n\r\ntx_data['text']=tx_data['text'].str.lower() #changing into lower case (remove and check acc too)\r\ntx_data['text']=tx_data['text'].str.strip() #remove white spaces\r\ntx_data['text']=tx_data['text'].str.replace(r'\\d+','') #remove numbers\r\n#t_data['text']=t_data['text'].apply(lambda x: x.encode('ascii', 'ignore').decode('ascii')) #removing emoji\r\ntx_data['text']=tx_data['text'].str.replace('[^\\w\\s]','') #removing punct\r\ntx_data['text']=tx_data['text'].apply(lambda x:remove_URL(x))\r\n#t_data['text'] = t_data['text'].apply(lambda x: remove_sw(x))\r\n",
"_____no_output_____"
],
[
"encoded2 =tok1.texts_to_sequences(tx_data['text'])\r\npadded2 = sequence.pad_sequences(encoded2, maxlen=max_length, padding='post')",
"_____no_output_____"
],
[
"cencoded2=tok2.texts_to_sequences(tx_data['text'])\r\ncpadded2=sequence.pad_sequences(cencoded2,maxlen=70,padding='post')",
"_____no_output_____"
],
[
"#prediction\r\n#prediction of CNN\r\n\r\npredictions = model.predict([cpadded2,padded2,padded2])\r\n# Finding out of two output neurons which one has highest probability\r\n# It will return us the predicted class\r\n# It will convert probability into final class 1 and 0\r\npredictions1 = np.zeros_like(predictions)\r\npredictions1[np.arange(len(predictions)), predictions.argmax(1)] = 1\r\n#print(dev_predictions)\r\nprint(predictions1)",
"[[1. 0. 0.]\n [0. 1. 0.]\n [1. 0. 0.]\n ...\n [1. 0. 0.]\n [1. 0. 0.]\n [0. 0. 1.]]\n"
],
[
"pred=np.argmax(predictions1,axis=1)",
"_____no_output_____"
],
[
"labelEncode.inverse_transform(pred)",
"_____no_output_____"
],
[
"test=pd.read_csv('/content/drive/MyDrive/Ankit/dec 2020 codalab/Hope speech/Tamil/tamil_hope_test.csv',names=['text'],sep=',')",
"_____no_output_____"
],
[
"test['label']=labelEncode.inverse_transform(pred)\r\ntest",
"_____no_output_____"
]
],
[
[
"#Into tSV",
"_____no_output_____"
]
],
[
[
"test.to_csv('iiit_dwd_Tamil.tsv',index=False)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec5e20a433620c4015093fcd03225ffcc9622857 | 358,899 | ipynb | Jupyter Notebook | lessons/CRISP_DM/How To Break Into the Field.ipynb | ZacksAmber/Udacity-Data-Scientist | b21595413f21a1200fe0b46f47e747ca9bff8d1f | [
"MIT"
]
| null | null | null | lessons/CRISP_DM/How To Break Into the Field.ipynb | ZacksAmber/Udacity-Data-Scientist | b21595413f21a1200fe0b46f47e747ca9bff8d1f | [
"MIT"
]
| null | null | null | lessons/CRISP_DM/How To Break Into the Field.ipynb | ZacksAmber/Udacity-Data-Scientist | b21595413f21a1200fe0b46f47e747ca9bff8d1f | [
"MIT"
]
| null | null | null | 383.849198 | 287,144 | 0.911404 | [
[
[
"### How To Break Into the Field\n\nNow you have had a closer look at the data, and you saw how I approached looking at how the survey respondents think you should break into the field. Let's recreate those results, as well as take a look at another question.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport HowToBreakIntoTheField as t\n%matplotlib inline\n\n# Load timing tool\ntry:\n %load_ext autotime\nexcept:\n !pip install autotime\n %load_ext autotime\n\ndf = pd.read_csv('./survey_results_public.csv')\nschema = pd.read_csv('./survey_results_schema.csv')\ndf.head()",
"_____no_output_____"
]
],
[
[
"#### Question 1\n\n**1.** In order to understand how to break into the field, we will look at the **CousinEducation** field. Use the **schema** dataset to answer this question. Write a function called **get_description** that takes the **schema dataframe** and the **column** as a string, and returns a string of the description for that column.",
"_____no_output_____"
]
],
[
[
"def get_description(column_name, schema=schema):\n '''\n INPUT - schema - pandas dataframe with the schema of the developers survey\n column_name - string - the name of the column you would like to know about\n OUTPUT - \n desc - string - the description of the column\n '''\n desc = list(schema[schema['Column'] == column_name]['Question'])[0]\n return desc\n\n#test your code\n#Check your function against solution - you shouldn't need to change any of the below code\nget_description(df.columns[0]) # This should return a string of the first column description",
"_____no_output_____"
],
[
"#Check your function against solution - you shouldn't need to change any of the below code\ndescrips = set(get_description(col) for col in df.columns)\nt.check_description(descrips)",
"Nice job it looks like your function works correctly!\ntime: 56.5 ms (started: 2021-08-08 11:27:14 -07:00)\n"
]
],
[
[
"The question we have been focused on has been around how to break into the field. Use your **get_description** function below to take a closer look at the **CousinEducation** column.",
"_____no_output_____"
]
],
[
[
"get_description('CousinEducation')",
"_____no_output_____"
]
],
[
[
"#### Question 2\n\n**2.** Provide a pandas series of the different **CousinEducation** status values in the dataset. Store this pandas series in **cous_ed_vals**. If you are correct, you should see a bar chart of the proportion of individuals in each status. If it looks terrible, and you get no information from it, then you followed directions. However, we should clean this up!",
"_____no_output_____"
]
],
[
[
"cous_ed_vals = df.CousinEducation.value_counts() #Provide a pandas series of the counts for each CousinEducation status\n\ncous_ed_vals # assure this looks right",
"_____no_output_____"
],
[
"# The below should be a bar chart of the proportion of individuals in your ed_vals\n# if it is set up correctly.\n\n(cous_ed_vals/df.shape[0]).plot(kind=\"bar\");\nplt.title(\"Formal Education\");",
"_____no_output_____"
]
],
[
[
"We definitely need to clean this. Above is an example of what happens when you do not clean your data. Below I am using the same code you saw in the earlier video to take a look at the data after it has been cleaned.",
"_____no_output_____"
]
],
[
[
"possible_vals = [\"Take online courses\", \"Buy books and work through the exercises\", \n \"None of these\", \"Part-time/evening courses\", \"Return to college\",\n \"Contribute to open source\", \"Conferences/meet-ups\", \"Bootcamp\",\n \"Get a job as a QA tester\", \"Participate in online coding competitions\",\n \"Master's degree\", \"Participate in hackathons\", \"Other\"]\n\ndef clean_and_plot(df, title='Method of Educating Suggested', plot=True):\n '''\n INPUT \n df - a dataframe holding the CousinEducation column\n title - string the title of your plot\n axis - axis object\n plot - bool providing whether or not you want a plot back\n \n OUTPUT\n study_df - a dataframe with the count of how many individuals\n Displays a plot of pretty things related to the CousinEducation column.\n '''\n study = df['CousinEducation'].value_counts().reset_index()\n study.rename(columns={'index': 'method', 'CousinEducation': 'count'}, inplace=True)\n study_df = t.total_count(study, 'method', 'count', possible_vals)\n\n study_df.set_index('method', inplace=True)\n if plot:\n (study_df/study_df.sum()).plot(kind='bar', legend=None);\n plt.title(title);\n plt.show()\n props_study_df = study_df/study_df.sum()\n return props_study_df\n \nprops_df = clean_and_plot(df)",
"_____no_output_____"
]
],
[
[
"#### Question 3\n\n**3.** I wonder if some of the individuals might have bias towards their own degrees. Complete the function below that will apply to the elements of the **FormalEducation** column in **df**. ",
"_____no_output_____"
]
],
[
[
"# Unique education levels from FormalEducation\ndf.FormalEducation.unique()",
"_____no_output_____"
],
[
"def higher_ed(formal_ed_str):\n '''\n INPUT\n formal_ed_str - a string of one of the values from the Formal Education column\n \n OUTPUT\n return 1 if the string is in (\"Master's degree\", \"Professional degree\")\n return 0 otherwise\n \n '''\n return 1 if formal_ed_str in [\"Master's degree\", \"Professional degree\"] else 0\n\ndf[\"FormalEducation\"].apply(higher_ed)[:5] #Test your function to assure it provides 1 and 0 values for the df",
"_____no_output_____"
],
[
"# Check your code here\ndf['HigherEd'] = df[\"FormalEducation\"].apply(higher_ed)\nhigher_ed_perc = df['HigherEd'].mean()\nt.higher_ed_test(higher_ed_perc)",
"Nice job! That's right. The percentage of individuals in these three groups is 0.2302376714480159.\ntime: 7.1 ms (started: 2021-08-08 11:27:33 -07:00)\n"
]
],
[
[
"#### Question 4\n\n**4.** Now we would like to find out if the proportion of individuals who completed one of these three programs feel differently than those that did not. Store a dataframe of only the individual's who had **HigherEd** equal to 1 in **ed_1**. Similarly, store a dataframe of only the **HigherEd** equal to 0 values in **ed_0**.\n\nNotice, you have already created the **HigherEd** column using the check code portion above, so here you only need to subset the dataframe using this newly created column.",
"_____no_output_____"
]
],
[
[
"ed_1 = df[df.HigherEd == 1] # Subset df to only those with HigherEd of 1\ned_0 = df[df.HigherEd == 0] # Subset df to only those with HigherEd of 0\n\n\nprint(ed_1['HigherEd'][:5]) #Assure it looks like what you would expect\nprint(ed_0['HigherEd'][:5]) #Assure it looks like what you would expect",
"4 1\n6 1\n7 1\n9 1\n14 1\nName: HigherEd, dtype: int64\n0 0\n1 0\n2 0\n3 0\n5 0\nName: HigherEd, dtype: int64\ntime: 28.9 ms (started: 2021-08-08 11:27:33 -07:00)\n"
],
[
"#Check your subset is correct - you should get a plot that was created using pandas styling\n#which you can learn more about here: https://pandas.pydata.org/pandas-docs/stable/style.html\n\ned_1_perc = clean_and_plot(ed_1, 'Higher Formal Education', plot=False)\ned_0_perc = clean_and_plot(ed_0, 'Max of Bachelors Higher Ed', plot=False)\n\ncomp_df = pd.merge(ed_1_perc, ed_0_perc, left_index=True, right_index=True)\ncomp_df.columns = ['ed_1_perc', 'ed_0_perc']\ncomp_df['Diff_HigherEd_Vals'] = comp_df['ed_1_perc'] - comp_df['ed_0_perc']\ncomp_df.style.bar(subset=['Diff_HigherEd_Vals'], align='mid', color=['#d65f5f', '#5fba7d']) # color=['rgb(214, 95, 94)', 'rgb(95, 186, 125)']",
"_____no_output_____"
]
],
[
[
"#### Question 5\n\n**5.** What can you conclude from the above plot? Change the dictionary to mark **True** for the keys of any statements you can conclude, and **False** for any of the statements you cannot conclude.",
"_____no_output_____"
]
],
[
[
"sol = {'Everyone should get a higher level of formal education': False, \n 'Regardless of formal education, online courses are the top suggested form of education': True,\n 'There is less than a 1% difference between suggestions of the two groups for all forms of education': False,\n 'Those with higher formal education suggest it more than those who do not have it': True}\n\nt.conclusions(sol)",
"Nice job that looks right!\ntime: 407 µs (started: 2021-08-08 11:27:33 -07:00)\n"
]
],
[
[
"This concludes another look at the way we could compare education methods by those currently writing code in industry.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec5e20fee0cfaf5e857a4d3623e256e3aab5a1c5 | 141,472 | ipynb | Jupyter Notebook | Vector Autoregression.ipynb | dilarakarabey/vector-autoregression | b7e687c00f3de6d4e5aee11c32fb6172a3c1220a | [
"MIT"
]
| null | null | null | Vector Autoregression.ipynb | dilarakarabey/vector-autoregression | b7e687c00f3de6d4e5aee11c32fb6172a3c1220a | [
"MIT"
]
| null | null | null | Vector Autoregression.ipynb | dilarakarabey/vector-autoregression | b7e687c00f3de6d4e5aee11c32fb6172a3c1220a | [
"MIT"
]
| null | null | null | 207.741557 | 69,580 | 0.877629 | [
[
[
"# Multivariate Time-Series Analysis with VAR",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport statsmodels.api as sm\nfrom statsmodels.tsa.stattools import adfuller\nfrom statsmodels.tsa.stattools import grangercausalitytests\nfrom statsmodels.tsa.api import VAR # vector autoregression\nfrom statsmodels.stats.stattools import durbin_watson\nfrom statsmodels.tsa.base.datetools import dates_from_str\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport warnings\n\nwarnings.filterwarnings(\"ignore\")\n\nsns.set(rc={'figure.figsize':(20, 4)})\n\n%matplotlib inline",
"_____no_output_____"
],
[
"data = pd.read_csv(\"beachwear.csv\")\ndata = data.set_index(\"Week\")\ndata.head()",
"_____no_output_____"
]
],
[
[
"We'll be using weekly search volume data from Google Trends for two Turkish search terms: \"mayo\" (meaning swimsuit) & \"sandalet\" (meaning sandals). Google scales search volume data between 0-100.",
"_____no_output_____"
],
[
"## Granger Causality Test\n\nWe will be using Granger Causality Test in order to see if these variables are useful in forecasting one another as well as themselves. The null hypothesis is that lagged x-values do not explain the variation in y. [Click here](https://www.statisticshowto.com/granger-causality/) to read more about it.",
"_____no_output_____"
]
],
[
[
"def grangers_causality_matrix(data, variables, test='ssr_chi2test', maxlag=2, verbose=True):\n\n dataset = pd.DataFrame(np.zeros((len(variables), len(variables))), columns=variables, index=variables)\n\n for c in dataset.columns:\n for r in dataset.index:\n test_result = grangercausalitytests(data[[r,c]], maxlag=maxlag, verbose=False)\n p_values = [round(test_result[i+1][0][test][1], 5) for i in range(maxlag)]\n if verbose:\n print(f'Y = {r}, X = {c}, P Values = {p_values}')\n\n min_p_value = np.min(p_values)\n dataset.loc[r,c] = min_p_value\n\n dataset.columns = [var + '_x' for var in variables]\n\n dataset.index = [var + '_y' for var in variables]\n\n return dataset\n\ngrangers_causality_matrix(data, variables=data.columns)",
"Y = mayo, X = mayo, P Values = [1.0, 1.0]\nY = sandalet, X = mayo, P Values = [0.0, 0.00211]\nY = mayo, X = sandalet, P Values = [0.0, 0.0]\nY = sandalet, X = sandalet, P Values = [1.0, 1.0]\n"
]
],
[
[
"#### As we can see, our p-values are smaller than 0.05 and therefore, the null hypothesis is rejected. That means these series can be used to predict each other.",
"_____no_output_____"
],
[
"## Augmented Dickey-Fuller Test\n\nWe will apply unit root testing to see whether the data is stationary or not. The null hypothesis is that the time series has a unit root and is therefore, non-stationary. If our data contains non-stationary series, we will be de-trending the data using log returns to make it more stationary.",
"_____no_output_____"
]
],
[
[
"mayo = data[\"mayo\"].values\nsandalet = data[\"sandalet\"].values\n\nmayo_result = adfuller(mayo)\nprint('Mayo - ADF Statistic: %f' % mayo_result[0])\nprint('Mayo - p-value: %f' % mayo_result[1])\n\nsandalet_result = adfuller(sandalet)\nprint('\\nSandalet - ADF Statistic: %f' % sandalet_result[0])\nprint('Sandalet - p-value: %f' % sandalet_result[1])",
"Mayo - ADF Statistic: -4.278713\nMayo - p-value: 0.000483\n\nSandalet - ADF Statistic: -4.343050\nSandalet - p-value: 0.000374\n"
]
],
[
[
"#### Since our p-values are smaller than 0.05, our null hypothesis is rejected and that means both these series are stationary.",
"_____no_output_____"
],
[
"## Training The Model\n\nWe will now be training the model. For the lag order selection process, we will be using the model's built-in method.",
"_____no_output_____"
]
],
[
[
"model = VAR(data)\nlag_orders = model.select_order(15)\nlag_orders.summary()",
"_____no_output_____"
]
],
[
[
"#### Let's think business for a moment. Say we are a retail company selling beachwear. Both false positives (BIC lead) & false negatives (AIC lead) may mislead us significantly & cripple our marketing efforts. In that case, we would fit two different models that have lag orders 3 and 8. However, since this is just an example case, we will be applying 8 (AIC lead) for now.",
"_____no_output_____"
]
],
[
[
"lag_order = 8\nresults = model.fit(lag_order, ic=\"aic\")\nresults.summary()",
"_____no_output_____"
]
],
[
[
"## Durbin-Watson Statistic\n\nWe will now check whether there is any correlation left in the residuals and if so, we will have to increase the order of our model. Values of this statistic varies between 0 & 4. Values closer to 2 mean that there is no significant serial correlation. Values close to 4 indicate strong negative correlation while those closer to 0 imply strong positive correlation. ",
"_____no_output_____"
]
],
[
[
"dw_r = durbin_watson(results.resid)\n\nfor col, val in zip(data.columns, dw_r):\n print(col, ':', round(val, 2))",
"mayo : 2.01\nsandalet : 2.02\n"
]
],
[
[
"#### As we can see, our values are close to 2. Therefore, we will be moving on to the next step.\n\n## Forecasting\n\nWe will now do forecasting for 5 observations.",
"_____no_output_____"
]
],
[
[
"fc = results.forecast(data.values[-lag_order:], steps=5)\nfc = pd.DataFrame(fc, columns=[\"mayo_forecast\", \"sandalet_forecast\"])\nfc",
"_____no_output_____"
],
[
"results.plot_forecast(5);",
"_____no_output_____"
]
],
[
[
"## Testing\n\nWe will begin testing with splitting the data into test and training datasets.",
"_____no_output_____"
]
],
[
[
"df_train, df_test = data[0:-5], data[-5:]",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=int(len(data.columns)/2), ncols=2, dpi=150, figsize=(10,3))\n\nfor i, (col, ax) in enumerate(zip(data.columns, axes.flatten())):\n fc[col + '_forecast'].plot(legend=True, ax=ax).autoscale(axis='x',tight=True)\n df_test[col][-5:].plot(legend=True, ax=ax);\n ax.set_title(col + \": Forecast vs Actuals\")\n ax.xaxis.set_ticks_position('none')\n ax.yaxis.set_ticks_position('none')\n ax.spines[\"top\"].set_alpha(0)\n ax.tick_params(labelsize=6)\n ax.set_ylim((0,100))",
"_____no_output_____"
]
],
[
[
"#### Our predictions seem to be pretty accurate.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
]
|
ec5e32ca40603738fb163b6da32634faae2fdf0b | 35,666 | ipynb | Jupyter Notebook | diabeticretinopathy_RF.ipynb | danhtaihoang/hyperparameters | 94f28bb02f87e02d234d8d9713a9c89861774a15 | [
"MIT"
]
| null | null | null | diabeticretinopathy_RF.ipynb | danhtaihoang/hyperparameters | 94f28bb02f87e02d234d8d9713a9c89861774a15 | [
"MIT"
]
| null | null | null | diabeticretinopathy_RF.ipynb | danhtaihoang/hyperparameters | 94f28bb02f87e02d234d8d9713a9c89861774a15 | [
"MIT"
]
| null | null | null | 38.023454 | 295 | 0.577889 | [
[
[
"import numpy as np\nimport pandas as pd\n\nfrom sklearn.model_selection import train_test_split,KFold,RandomizedSearchCV\nfrom sklearn.utils import shuffle\nfrom sklearn.metrics import accuracy_score\n\n#import ER_multiclass as ER\n\n#from sklearn.linear_model import LogisticRegression\n#from sklearn.naive_bayes import GaussianNB\n#from sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier\n\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"np.random.seed(1)",
"_____no_output_____"
],
[
"X = np.loadtxt('../diabeticretinopathy_X.txt')\ny = np.loadtxt('../diabeticretinopathy_y.txt')",
"_____no_output_____"
],
[
"def inference(X_train,y_train,X_test,y_test):\n \n ## Optimize hyper parameters by RandomizedSearchCV:\n model = RandomForestClassifier(random_state = 1)\n #model\n\n # Number of trees in random forest:\n n_estimators = [int(x) for x in np.linspace(start = 10, stop = 100, num = 10)]\n\n # Number of features to consider at every split:\n max_features = ['auto']\n\n # Maximum number of levels in tree:\n max_depth = [int(x) for x in np.linspace(1, 10, num = 10)]\n\n # Minimum number of samples required to split a node:\n min_samples_split = [5, 10, 15, 20]\n\n # Minimum number of samples required at each leaf node:\n min_samples_leaf = [int(x) for x in np.linspace(start = 1, stop = 5, num = 5)]\n\n # Method of selecting samples for training each tree:\n #bootstrap = [True, False]\n bootstrap = [False]\n \n # Create the random grid:\n random_grid = {'n_estimators': n_estimators,\n 'max_features': max_features,\n 'max_depth': max_depth,\n 'min_samples_split': min_samples_split,\n 'min_samples_leaf': min_samples_leaf,\n 'bootstrap': bootstrap}\n \n random_search = RandomizedSearchCV(estimator = model, param_distributions = random_grid, n_iter = 100, \n cv = 3, verbose=2, random_state=1, n_jobs = -1)\n\n random_search.fit(X_train, y_train)\n \n # best hyper parameters:\n print(random_search.best_params_)\n \n y_pred = random_search.best_estimator_.predict(X_test)\n accuracy = accuracy_score(y_test,y_pred)\n \n return accuracy",
"_____no_output_____"
],
[
"def compare_inference(X,y,train_size):\n npred = 10\n accuracy = np.zeros((len(list_methods),npred))\n precision = np.zeros((len(list_methods),npred))\n recall = np.zeros((len(list_methods),npred))\n accuracy_train = np.zeros((len(list_methods),npred))\n for ipred in range(npred):\n X_train0,X_test,y_train0,y_test = train_test_split(X,y,test_size=0.2,random_state = ipred)\n\n idx_train = np.random.choice(len(y_train0),size=int(train_size*len(y)),replace=False)\n X_train,y_train = X_train0[idx_train],y_train0[idx_train]\n\n for i,method in enumerate(list_methods):\n accuracy[i,ipred] = inference(X_train,y_train,X_test,y_test)\n \n return accuracy.mean(axis=1),accuracy.std(axis=1) ",
"_____no_output_____"
],
[
"list_train_size = [0.8,0.6,0.4,0.2]\n#list_methods=['logistic_regression','naive_bayes','random_forest','expectation_reflection']\n#list_train_size = [0.8,0.2]\nlist_methods=['random_forest']\nacc = np.zeros((len(list_train_size),len(list_methods)))\nacc_std = np.zeros((len(list_train_size),len(list_methods)))\nfor i,train_size in enumerate(list_train_size):\n acc[i,:],acc_std[i,:] = compare_inference(X,y,train_size)\n print(train_size,acc[i,:])",
"Fitting 3 folds for each of 100 candidates, totalling 300 fits\n"
],
[
"acc",
"_____no_output_____"
],
[
"acc_std",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5e369c25a300c80a4c57cbc7947f4e1ede72e2 | 263,879 | ipynb | Jupyter Notebook | courses/modsim2018/tasks/Desiree/Task12_MotorControl.ipynb | desireemiraldo/bmc | 5e50d806ea2e2d7494035ba120c3cd3a620a156a | [
"MIT"
]
| null | null | null | courses/modsim2018/tasks/Desiree/Task12_MotorControl.ipynb | desireemiraldo/bmc | 5e50d806ea2e2d7494035ba120c3cd3a620a156a | [
"MIT"
]
| null | null | null | courses/modsim2018/tasks/Desiree/Task12_MotorControl.ipynb | desireemiraldo/bmc | 5e50d806ea2e2d7494035ba120c3cd3a620a156a | [
"MIT"
]
| null | null | null | 71.862473 | 32,249 | 0.686728 | [
[
[
"# Task 12 - Motor Control\n### Introduction to modeling and simulation of human movement\nhttps://github.com/BMClab/bmc/blob/master/courses/ModSim2018.md",
"_____no_output_____"
],
[
"* Based on task for Lecture 11 (+ muscle activation dynamics):\n\nChange the derivative of the contractile element length function. The new function must compute the derivative according to the article from Thelen (2003) (Eqs. (1), (2), (6) and (7)):\n\n Thelen D; Adjustment of muscle mechanics model parameters to simulate dynamic contractions in older adults (2003)",
"_____no_output_____"
]
],
[
[
"import numpy as np\n#import pandas as pd\nimport matplotlib.pyplot as plt\nimport math\n\n%matplotlib notebook",
"_____no_output_____"
]
],
[
[
"### Muscle properties",
"_____no_output_____"
]
],
[
[
"Lslack = .223\nUmax = .04\nLce_o = .093 #optmal l\nwidth = .63#*Lce_o\nFmax = 3000\na = .01\nu = 0.5\n#b = .25*10#*Lce_o ",
"_____no_output_____"
]
],
[
[
"### Initial conditions",
"_____no_output_____"
]
],
[
[
"Lnorm_ce = .087/Lce_o #norm\nt0 = 0\ntf = 2.99\nh = 1e-3",
"_____no_output_____"
],
[
"t = np.arange(t0,tf,h)\nF = np.empty(t.shape)\nFkpe = np.empty(t.shape)\nFiberLen = np.empty(t.shape)\nTendonLen = np.empty(t.shape)\na_dynamics = np.empty(t.shape)",
"_____no_output_____"
]
],
[
[
"## Simulation - Series",
"_____no_output_____"
],
[
"for i in range (len(t)):\n #ramp\n if t[i]<=1:\n Lm = 0.31\n elif t[i]>1 and t[i]<2:\n Lm = .31 + .1*(t[i]-1)\n #print(Lm)\n \n #shortening at 4cm/s\n Lsee = Lm - Lce\n \n if Lsee<Lslack: \n F[i] = 0\n else: \n F[i] = Fmax*((Lsee-Lslack)/(Umax*Lslack))**2\n \n \n #isometric force at Lce from CE force length relationship\n F0 = max([0, Fmax*(1-((Lce-Lce_o)/width)**2)])\n \n #calculate CE velocity from Hill's equation\n if F[i]>F0: print('Error: cannot do eccentric contractions')\n \n Lcedot = -b*(F0-F[i])/(F[i]+a) #vel is negative for shortening\n \n # --- Euler integration step\n Lce += h*Lcedot\n\n ",
"_____no_output_____"
]
],
[
[
"def TendonForce (Lnorm_see,Lslack, Lce_o):\n '''\n Compute tendon force\n\n Inputs:\n Lnorm_see = normalized tendon length\n Lslack = slack length of the tendon (non-normalized)\n Lce_o = optimal length of the fiber\n \n Output:\n Fnorm_tendon = normalized tendon force\n \n '''\n Umax = .04\n \n if Lnorm_see<Lslack/Lce_o: \n Fnorm_tendon = 0\n else: \n Fnorm_tendon = ((Lnorm_see-Lslack/Lce_o)/(Umax*Lslack/Lce_o))**2\n \n return Fnorm_tendon",
"_____no_output_____"
],
[
"def ParallelElementForce (Lnorm_ce):\n '''\n Compute parallel element force\n \n Inputs:\n Lnorm_ce = normalized contractile element length\n \n Output:\n Fnorm_kpe = normalized parallel element force\n\n '''\n Umax = 1\n \n if Lnorm_ce< 1: \n Fnorm_kpe = 0\n else: \n Fnorm_kpe = ((Lnorm_ce-1)/(Umax*1))**2 \n \n return Fnorm_kpe",
"_____no_output_____"
],
[
"def ForceLengthCurve (Lnorm_ce,width):\n F0 = max([0, (1-((Lnorm_ce-1)/width)**2)])\n return F0",
"_____no_output_____"
]
],
[
[
"def ContractileElementDot(F0, Fnorm_CE, a, b):\n \n '''\n Compute Contractile Element Derivative\n\n Inputs:\n F0 = Force-Length Curve\n Fce = Contractile element force\n \n Output:\n Lnorm_cedot = normalized contractile element length derivative\n\n '''\n \n if Fnorm_CE>F0: print('Error: cannot do eccentric contractions')\n \n Lnorm_cedot = -b*(F0-Fnorm_CE)/(Fnorm_CE + a) #vel is negative for shortening\n \n return Lnorm_cedot",
"_____no_output_____"
]
],
[
[
"def ContractileElementDot(F0, Fnorm_CE, a):\n \n '''\n Compute Contractile Element Derivative\n\n Inputs:\n F0 = Force-Length Curve\n Fce = Contractile element force\n \n Output:\n Lnorm_cedot = normalized contractile element length derivative\n\n '''\n \n FMlen = 1.4 # young adults\n Vmax = 10 # young adults\n Af = 0.25 #force-velocity shape factor\n \n if Fnorm_CE > F0:\n \n b = ((2 + 2/Af)*(a*F0*FMlen - Fnorm_CE))/(FMlen-1)\n \n elif Fnorm_CE <= F0:\n \n b = a*F0 + Fnorm_CE/Af\n \n Lnorm_cedot = (.25 + .75*a)*Vmax*((Fnorm_CE - a*F0)/b)\n \n return Lnorm_cedot",
"_____no_output_____"
],
[
"def ContractileElementForce(Fnorm_tendon,Fnorm_kpe):\n '''\n Compute Contractile Element force\n\n Inputs:\n Fnorm_tendon = normalized tendon force\n Fnorm_kpe = normalized parallel element force\n \n Output:\n Fnorm_CE = normalized contractile element force\n '''\n Fnorm_CE = Fnorm_tendon - Fnorm_kpe\n return Fnorm_CE",
"_____no_output_____"
],
[
"def tendonLength(Lm,Lce_o,Lnorm_ce):\n '''\n Compute tendon length\n \n Inputs:\n Lm = \n Lce_o = optimal length of the fiber\n Lnorm_ce = normalized contractile element length\n \n Output:\n Lnorm_see = normalized tendon length \n '''\n Lnorm_see = Lm/Lce_o - Lnorm_ce\n return Lnorm_see",
"_____no_output_____"
],
[
"def activation(a,u,dt):\n '''\n Compute activation\n \n Inputs:\n u = idealized muscle excitation signal, 0 <= u <= 1\n a = muscular activation\n dt = time step\n \n Output:\n a = muscular activation \n '''\n \n tau_deact = 50e-3 #young adults\n tau_act = 15e-3\n \n if u>a:\n tau_a = tau_act*(0.5+1.5*a)\n elif u <=a:\n tau_a = tau_deact/(0.5+1.5*a)\n \n #-------\n dadt = (u-a)/tau_a # euler\n \n a += dadt*dt\n #-------\n return a",
"_____no_output_____"
]
],
[
[
"## Simulation - Parallel",
"_____no_output_____"
]
],
[
[
"#Normalizing\n\nfor i in range (len(t)):\n #ramp\n if t[i]<=1:\n Lm = 0.31\n elif t[i]>1 and t[i]<2:\n Lm = .31 - .04*(t[i]-1)\n #print(Lm)\n \n #shortening at 4cm/s\n \n Lnorm_see = tendonLength(Lm,Lce_o,Lnorm_ce)\n\n Fnorm_tendon = TendonForce (Lnorm_see,Lslack, Lce_o) \n \n Fnorm_kpe = ParallelElementForce (Lnorm_ce) \n \n #isometric force at Lce from CE force length relationship\n F0 = ForceLengthCurve (Lnorm_ce,width)\n \n Fnorm_CE = ContractileElementForce(Fnorm_tendon,Fnorm_kpe) #Fnorm_CE = ~Fm\n \n #computing activation\n a = activation(a,u,h)\n \n #calculate CE velocity from Hill's equation \n Lnorm_cedot = ContractileElementDot(F0, Fnorm_CE,a)\n \n # --- Euler integration step\n Lnorm_ce += h*Lnorm_cedot\n\n \n F[i] = Fnorm_tendon*Fmax\n Fkpe[i] = Fnorm_kpe*Fmax\n FiberLen[i] = Lnorm_ce*Lce_o\n TendonLen[i] = Lnorm_see*Lce_o\n a_dynamics[i] = a\n ",
"_____no_output_____"
]
],
[
[
"## Plots ",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(1, 1, figsize=(6,6), sharex=True)\n\nax.plot(t,a_dynamics,c='magenta')\nplt.grid()\nplt.xlabel('time (s)')\nplt.ylabel('Activation dynamics')\n\n\nax.legend()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6,6), sharex=True)\n\nax.plot(t,F,c='red')\nplt.grid()\nplt.xlabel('time (s)')\nplt.ylabel('Force (N)')\n\n\nax.legend()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6,6), sharex=True)\n\nax.plot(t,FiberLen, label = 'fiber')\nax.plot(t,TendonLen, label = 'tendon')\nplt.grid()\nplt.xlabel('time (s)')\nplt.ylabel('Length (m)')\nax.legend(loc='best')\n\n\nfig, ax = plt.subplots(1, 3, figsize=(12,4), sharex=True, sharey=True)\nax[0].plot(t,FiberLen, label = 'fiber')\nax[1].plot(t,TendonLen, label = 'tendon')\nax[2].plot(t,FiberLen + TendonLen, label = 'muscle (tendon + fiber)')\n\nax[1].set_xlabel('time (s)')\nax[0].set_ylabel('Length (m)')\n#plt.legend(loc='best')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec5e398e2f262912ead3cafeff5feb60c3522d09 | 10,091 | ipynb | Jupyter Notebook | Machine Learning/20_Classify_Kinematic_Data.ipynb | MukundAabha/machine_learning_projects | 28276d43235caf7d2b736285f242ee6d963060e7 | [
"MIT"
]
| null | null | null | Machine Learning/20_Classify_Kinematic_Data.ipynb | MukundAabha/machine_learning_projects | 28276d43235caf7d2b736285f242ee6d963060e7 | [
"MIT"
]
| null | null | null | Machine Learning/20_Classify_Kinematic_Data.ipynb | MukundAabha/machine_learning_projects | 28276d43235caf7d2b736285f242ee6d963060e7 | [
"MIT"
]
| null | null | null | 21.984749 | 305 | 0.51759 | [
[
[
"### Classify Kinematic Data\nDESCRIPTION\n\nYou are supposed to detect whether the person is running or walking based on the sensor data collected from iOS device. The dataset contains a single file which represents sensor data samples collected from accelerometer and gyroscope from iPhone 5c in 10 seconds interval and ~5.4/second frequency.",
"_____no_output_____"
],
[
"### Objective:\nPractice classification based on Naive Bayes algorithm. Identify the predictors that can be influential.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plot\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"###### Load the kinematics dataset as measured on mobile sensors from the file “run_or_walk.csv.”",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"run_or_walk.csv\")",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 88588 entries, 0 to 88587\nData columns (total 11 columns):\ndate 88588 non-null object\ntime 88588 non-null object\nusername 88588 non-null object\nwrist 88588 non-null int64\nactivity 88588 non-null int64\nacceleration_x 88588 non-null float64\nacceleration_y 88588 non-null float64\nacceleration_z 88588 non-null float64\ngyro_x 88588 non-null float64\ngyro_y 88588 non-null float64\ngyro_z 88588 non-null float64\ndtypes: float64(6), int64(2), object(3)\nmemory usage: 7.4+ MB\n"
]
],
[
[
"###### List the columns in the dataset.",
"_____no_output_____"
]
],
[
[
"df.columns",
"_____no_output_____"
]
],
[
[
"###### Let the target variable “y” be the activity, and assign all the columns after it to “x.”",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX, y = df.iloc[:, 5:].values,df.iloc[:, 4].values\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)",
"_____no_output_____"
],
[
"print(X_train.shape)\nprint(y_test[0:10])",
"(70870, 6)\n[1 0 0 1 1 1 0 1 1 1]\n"
]
],
[
[
"###### Using Scikit-learn, fit a Gaussian Naive Bayes model and observe the accuracy.",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import GaussianNB\nclassifier = GaussianNB()",
"_____no_output_____"
],
[
"classifier.fit(X_train,y_train)",
"_____no_output_____"
],
[
"y_predict = classifier.predict(X_test)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"accuracy = accuracy_score(y_predict,y_test)",
"_____no_output_____"
],
[
"print(accuracy)",
"0.955469014561\n"
],
[
"from sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"conf_mat =confusion_matrix(y_predict,y_test)",
"_____no_output_____"
],
[
"print(conf_mat)",
"[[8583 699]\n [ 90 8346]]\n"
],
[
"from sklearn.metrics import classification_report\ntarget_names = [\"Walk\",\"Run\"]",
"_____no_output_____"
]
],
[
[
"###### Generate a classification report using Scikit-learn.",
"_____no_output_____"
]
],
[
[
"print(classification_report(y_test, y_predict, target_names=target_names))",
" precision recall f1-score support\n\n Walk 0.92 0.99 0.96 8673\n Run 0.99 0.92 0.95 9045\n\navg / total 0.96 0.96 0.96 17718\n\n"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 88588 entries, 0 to 88587\nData columns (total 11 columns):\ndate 88588 non-null object\ntime 88588 non-null object\nusername 88588 non-null object\nwrist 88588 non-null int64\nactivity 88588 non-null int64\nacceleration_x 88588 non-null float64\nacceleration_y 88588 non-null float64\nacceleration_z 88588 non-null float64\ngyro_x 88588 non-null float64\ngyro_y 88588 non-null float64\ngyro_z 88588 non-null float64\ndtypes: float64(6), int64(2), object(3)\nmemory usage: 7.4+ MB\n"
]
],
[
[
"###### Repeat the model once using only the acceleration values as predictors and then using only the gyro values as predictors.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX, y = df.iloc[:, [5,6,7]].values,df.iloc[:, 4].values\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)",
"_____no_output_____"
],
[
"classifier.fit(X_train,y_train)\ny_predict = classifier.predict(X_test)\naccuracy_score(y_predict,y_test)",
"_____no_output_____"
]
],
[
[
"###### Repeat the model once using only the acceleration values as predictors and then using only the gyro values as predictors.",
"_____no_output_____"
]
],
[
[
"print(conf_mat)",
"[[8583 699]\n [ 90 8346]]\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec5e3f0b706c158e01b91684d2d6d0946b89ff08 | 124,971 | ipynb | Jupyter Notebook | complex-perturbation-test.ipynb | senderle/semantic-phasor-demo | fba8601829b38bbdb0313186b8d41f334b1ce20e | [
"MIT"
]
| 1 | 2019-11-07T13:59:06.000Z | 2019-11-07T13:59:06.000Z | complex-perturbation-test.ipynb | senderle/semantic-phasor-demo | fba8601829b38bbdb0313186b8d41f334b1ce20e | [
"MIT"
]
| null | null | null | complex-perturbation-test.ipynb | senderle/semantic-phasor-demo | fba8601829b38bbdb0313186b8d41f334b1ce20e | [
"MIT"
]
| null | null | null | 185.416914 | 29,760 | 0.881444 | [
[
[
"import numpy\nimport matplotlib.pyplot as plt\n\nplt.style.use('seaborn-darkgrid')",
"_____no_output_____"
],
[
"def complex_blob(n, scale=1.0, fixed_seed=False):\n # For a stable-but-random blob.\n if fixed_seed:\n numpy.random.seed(23)\n \n x, y = numpy.random.multivariate_normal(\n [0, 0], \n [[scale ** 2, 0], [0, scale ** 2]], \n n).T\n \n if fixed_seed:\n numpy.random.seed(None)\n\n return x + y * 1j\n\ndef plot_blob(c):\n plt.plot(c.real, c.imag, 'x')\n plt.axis('equal')\n plt.show()\n\n# Using 'seaborn-darkgrid':\nplot_blob(complex_blob(300, scale=0.1))\n\nwith plt.style.context('bmh'):\n plot_blob(complex_blob(300, scale=0.1))\n\nwith plt.style.context('ggplot'):\n plot_blob(complex_blob(300, scale=0.1))",
"_____no_output_____"
],
[
"# To generate a signature that will be very close to identical \n# when the starting blob is identical, we can start by picking \n# the three biggest outliers -- that is, the three phasors with \n# the greatest magnitudes. These outliers are likely to be the \n# same for blobs that are the same, even after different noise \n# is added.\n\n# We average the angles of the three outliers, and use the \n# result as a stable reference against which to measure the \n# angular positions of the remaining phasors. The calculation \n# for this amounts to dividing all the phasors by a unit-length \n# phasor at the given angle.\n\n# To denoise the phasors and reduce their dimension to k, we \n# multiply them by a fixed random unitary matrix, and drop all \n# but the first k dimensions. This denoises the phasors because \n# the added noise is i.i.d. in all directions, and so tends to \n# cancel itself out when summed up by the dot product. The \n# randomness of the projection matrix shouldn't change that \n# fact, though I'd like to work out a way to test that claim \n# empirically.\n\n# The matrix is \"random\" in the sense that we sample it \n# pseudo-randomly and uniformly from among all unitary \n# matrices. And it is \"fixed\" in the sense that the seed for \n# the random number generator is predetermined, so that we get\n# the same matrix every time. Otherwise, the signature for a \n# given document would change each time it was calculated, \n# making it useless as a signature!\n\n# NOTE: Since the above was written, I've found that reversing \n# the order of these steps and picking just one outlier gives \n# better results, at least on simulated data. In other words, \n# by performing the dimension reduction first, and then the \n# rotation, basing the rotation on a single outlier, we get a \n# more stable signature. It's possible that real data may \n# differ in this respect though.\n\n# Also, this way of doing things might limit the utility of \n# the signatures for tasks other than deduplication, because\n# the stabilizing rotation might be more arbitrary, whereas we\n# can hope that when the stabilizing rotation is based on \n# semantically marked outliers, it's reflecting *something* \n# about the text itself.",
"_____no_output_____"
],
[
"def normed_outliers(blobs, n_outliers):\n \n # Index first dimension\n blob_mags = numpy.abs(blobs)\n blob_mags_argsort = blob_mags.argsort(\n kind='stable', axis=0\n )[-n_outliers:, :]\n \n # Index second dimension\n arange_blob_mags = numpy.arange(blob_mags.shape[1])\n \n # Full index\n outliers = (blob_mags_argsort, arange_blob_mags)\n \n return blobs[outliers] / blob_mags[outliers]\n\ndef angle_offset(blobs, n_outliers):\n outlier_angles = numpy.angle(normed_outliers(blobs, n_outliers))\n offset_angle = outlier_angles.mean(axis=0)\n return numpy.cos(offset_angle) + 1j * numpy.sin(offset_angle)\n\ndef anchor_blob(blobs, n_anchors):\n anchors = angle_offset(blobs, n_anchors)\n return blobs / anchors\n\ndef haar_measure(size, fixed_seed=False):\n \"\"\"\n Sample a matrix from a uniform distribution over unitary matrices. \n \n Unitary matrices transform vectors of complex numbers without \n changing their magnitude, and transform pairs of complex vectors \n without changing the angle between them. Reflections and rotations\n are examples of unitary transformations, and in general you could \n think of unitary transformations as ones that preserve circular \n symmetries. For the purpose of random sampling, then, we should \n look for a definition of uniformity that does the same.\n \n It turns out that a way of measuring volume called the Haar \n measure does just that. When you distribute points on a sphere \n according to the Haar measure, for example, they are evenly\n distributed, so that no region of the sphere has more points than\n any other on average. The Haar measure is also provably the only\n measure with this property, so once you can show that your \n distribution preserves these symmetries, you have found the \n Haar measure, and vice versa. The `test_eigval_density` and \n `plot_haar_angles` functions below use that fact to test the\n correctness of the code here.\n\n\n `size` determines the edge size of the matrix. For example, \n passing `5` gives a 5x5 matrix.\n\n\n `fixed_seed` samples from the same pseudorandom sequence every \n time, in theory guaranteeing that the matrix returned will be \n identical. However, if numpy changes their RNG algorithm at all, \n the sequence will change, as will the matrix. So there may be \n some argument for creating a custom Mersenne twister. This\n point technically also applies to changes to the QR decomposition \n code. However, the numpy devs would have to change the LAPACK \n routines they use, or the LAPACK routines themselves would have\n to change, neither of which seems especially likely.\n \"\"\"\n\n # `z` is a square matrix full of random values sampled from a\n # complex normal distribution.\n z = complex_blob(\n size * size, \n fixed_seed=fixed_seed\n ).reshape((size, size)) / 2 ** 0.5\n \n # Decompose `z` into `q`, an orthogonal matrix, and `r`, an \n # upper-triangular matrix.\n \n q, r = numpy.linalg.qr(z)\n \n # In the `_haar_measure_wrong` variant of this code, we just \n # throw out `r`, and stop here, hoping that the orthogonal \n # component `q` is still random enough. In theory, it would\n # be, but it's generally not, because the LAPACK routines \n # use heuristics to reduce numerical error. Those heuristics\n # bias the code away from matrices that have small eigenvalues.\n \n # Ordinarily, that would make sense, since numerical error\n # is often caused by values that are very close to zero. But we\n # want to preserve circular symmetry, which means we want a\n # smooth distribution of eigenvalues across the range [-pi, pi).\n # The LAPACK heuristics create a gap in that distribution right\n # around `0`. If you plot the eigenvalues of the `wrong`\n # variant, using `plot_haar_angles(_haar_measure_wrong)`, you\n # can see the gap quite clearly.\n \n # So we have to do just a bit more work. The rationale behind\n # these additional steps is here:\n #\n # http://www.ams.org/notices/200705/fea-mezzadri-web.pdf\n #\n # Fortunately, in addition to supplying proofs, the paper \n # supplies pseudocode.\n\n # Take the diagonal of `r`, throwing out the rest of the\n # entries above the diagonal.\n rd = numpy.diagonal(r)\n \n # Normalize the diagonal, so that each entry is a complex\n # number somewhere on the unit circle.\n rd_abs = rd / numpy.abs(rd)\n \n # Rescale `q` with the result.\n return q * rd_abs\n\ndef stable_unitary_matrix(in_dim, out_dim):\n \"\"\"\n Use `haar_measure` to generate a stable matrix\n for dimension reduction. The output of `haar_measure`\n is a square matrix, which is not directly useful\n for dimension reduction, so this truncates the matrix,\n dropping all but the first `out_dim` dimensions.\n \n `in_dim` is the initial size of the matrix. We might\n save some time calculating just the first `out_dim`\n dimensions of the matrix, but it's not worth the extra\n complexity for now; the time saved turns out to be \n negligible compared to other overhead.\n \n Because this uses a `fixed_seed`, the output of this\n function for a given input should always be the same.\n \"\"\"\n return haar_measure(in_dim, fixed_seed=True)[:out_dim, :]\n\ndef signature(blobs, n_anchors=1, n_dims=5, reduce_first=True):\n \"\"\"\n `blobs` should be a 2D array containing one blob \n per column, and one feature dimension per row.\n \n `n_anchors` choses the number of outlier dimensions\n to use as anchors. `n_dims` determines the size of the\n signature, as measured in *complex* dimensions. (So\n the number of real parameters is twice this value.)\n \n `reduce_first` determines which step to start with.\n On synthetic data, reducing dimension first and then\n anchoring produces more stable signatures. This may\n not be true with real data, though.\n \"\"\"\n \n if reduce_first:\n projector = stable_unitary_matrix(blobs.shape[0], n_dims)\n reduced_blobs = projector @ blobs\n anchored_blobs = anchor_blob(reduced_blobs, n_anchors)\n return anchored_blobs\n else:\n anchored_blobs = anchor_blob(blobs, n_anchors)\n projector = stable_unitary_matrix(blobs.shape[0], n_dims)\n reduced_blobs = projector @ anchored_blobs\n return reduced_blobs\n",
"_____no_output_____"
],
[
"# Here, the next step is to create streamlined code paths \n# to the most useful portions of the basic phasor test \n# notebook. These don't have to be perfectly clean, but \n# they should not involve a lot of circling back through \n# states, the way slice_vec_bands does, for example.\n\n# Ideally, this will form the basis of a user-friendly \n# interface that lives in a new module, and that mostly \n# replaces the phasor module for everyday use.\n\n# To begin with, the API will be all functions. When \n# working with objects, like sklearn models, the model \n# should be passed in as a parameter if necessary, but only \n# if necessary, i.e, if we'd be creating a new object every \n# time in a tight loop. So we won't write anything that \n# manages state at first. Maybe by the end it will be clear \n# that we ahould manage some state but that will be a \n# decision we make later.\n\n# After thinking it through a bit more, this should all \n# be part of the update.py script. That script imports just \n# two functions from phasor.py, and although I think the \n# save function has some complicated dependencies, they \n# aren't so complicated that we can't simplify and \n# replicate them in update.py. From there we can do the \n# same for the signature save functions.\n#\n# There are three main dependencies:\n#\n# 1. load_one_sp_embedding\n# This loads the raw spacy vectors for each token. I \n# need to come up with better terms, because \"embedding\" \n# here refers to the underlying GloVe embeddings, but \n# elsewhere I talk about the fourier-transformed things \n# as embeddings. That's neither useful nor really all that \n# accurate.\n#\n# The main thing this uses is the VectorTable class, \n# which I think is actually a pretty good class. So this \n# part of the code doesn't need a lot of work.\n# \n# 2. embedding_fft \n# This runs through a series of utility functions, \n# mostly provided by numpy. The one non-numpy function is \n# the one that divides the volume into chunks and takes the \n# average GloVe vector for each chunk. I wish I had a line \n# that checks to see whether the chunks are larger than 200 \n# words or so, but I don't think I do. I will look at that \n# more carefully though. The rest uses standard numpy \n# functions to perform the actual fft and crunch and format\n# data.\n#\n# 3. save_compact_fft\n# The result of the last step is a 2-D complex array, \n# i.e. an array of blobs in the lingo I'm using here. Each \n# blob contains all the semantic dimensions of \n# one frequency band for one volume. I think. But, I think, \n# transposed relative to the blobs here in this notebook, \n# darnit! So a blob is a column instead of a row. Crumbs and \n# sausages. And, yes, a quick check confirms that the reduce \n# matrix in PhasorSignature is on the left, but the haar \n# projector here is on the right. So I gotta refactor the \n# code in this notebook so that blobs are columns. I think \n# this must be related to the shape of the fft output... \n# 300 rows 10 columns... = 10 blobs of 300 dims... vs... \n# yeah 10 rows of 300 dims = same but transposed\n\n\n# This function also decomposes the real and imaginary \n# parts because the numpy savez_compressed function doesnt \n# support complex arrays! It saves the two parts into \n# two arrays, `real` and `imag`. The end.\n",
"_____no_output_____"
],
[
"# The following portion of this notebook is dedicated to \n# testing the vectorized signature code above against a \n# simplified version of the same. There's nothing \n# particularly sophisticated about the underlying math here, \n# but correctness is critical in this part of the code.",
"_____no_output_____"
],
[
"def perturb_blob(blob, scale=0.05):\n \"\"\"\n Randomly rotate the starting blob and add a little bit of noise.\n \"\"\"\n # A single complex number.\n offset = complex_blob(1)[0] \n offset = offset / numpy.abs(offset)\n noise = complex_blob(len(blob), scale=scale)\n blob = blob * offset\n blob += noise\n return blob\n\ndef normed_outliers_basic(blob, n_outliers):\n blob_mags = numpy.abs(blob)\n outliers = blob_mags.argsort(kind='stable')[-n_outliers:]\n normed_outliers = blob[outliers] / blob_mags[outliers]\n return normed_outliers\n\ndef angle_offset_basic(blob, n_outliers):\n # numpy.angle works on 2d arrays.\n normed_outliers = normed_outliers_basic(blob, n_outliers)\n outlier_angles = numpy.angle(normed_outliers) \n offset_angle = outlier_angles.mean()\n return numpy.cos(offset_angle) + 1j * numpy.sin(offset_angle)\n\ndef anchor_blob_basic(blob, n_anchors):\n anchor = angle_offset_basic(blob, n_anchors)\n return blob / anchor\n\ndef signature_basic(blob, n_anchors=1, n_dims=5):\n projector = stable_unitary_matrix(len(blob), n_dims)\n blob = projector @ blob\n return anchor_blob_basic(blob, n_anchors)\n\n\ndef test_signature():\n # This uses too many transpositions. \n # It obscures the ideas behind the code. \n # Can we improve things?\n \n blob_1 = complex_blob(300)\n blob_2 = complex_blob(300)\n\n blob_1_perturbed = numpy.array(\n [perturb_blob(blob_1, scale=0.05) \n for i in range(10)]).T\n blob_2_perturbed = numpy.array(\n [perturb_blob(blob_2, scale=0.05) \n for i in range(10)]).T\n\n blob_1_signatures = signature(blob_1_perturbed)\n blob_1_signatures_basic = numpy.array(\n [signature_basic(p) \n for p in blob_1_perturbed.T]).T\n\n blob_2_signatures = signature(blob_2_perturbed)\n blob_2_signatures_basic = numpy.array(\n [signature_basic(p) \n for p in blob_2_perturbed.T]).T\n \n assert numpy.allclose(blob_1_signatures, \n blob_1_signatures_basic)\n assert numpy.allclose(blob_2_signatures, \n blob_2_signatures_basic)\n\ntest_signature()",
"_____no_output_____"
],
[
"# This portion of this notebook is devoted to testing \n# the sampling process, with particular emphasis on \n# 1) correctly generating unitary matrices, which \n# should preserve both the magnitudes of complex \n# vectors and the angles between them, and 2) sampling \n# from a uniform distribution over possible matrices, \n# as defined by the Haar measure, which ensures that \n# the eigenvalues of the matrices are smoothly distributed \n# between -pi and pi, i.e. symmetrically over the space of \n# a circle.\n\n# There are some subtle mathematical issues raised by \n# the second problem that I don't perfectly grasp. \n# Fortunately, the issues are described by the below \n# article in enough detail that I was able to write a \n# good test.\n\n# http://www.ams.org/notices/200705/fea-mezzadri-web.pdf \n\n# Unitary matrices are also used in quantum physics, \n# and it might be interesting to compare these two uses. \n# It could just be a sort of threefold coincidence -- \n# Fourier transforms + complex numbers + a need for \n# dimension reduction? But maybe that's unusual enough \n# to be notable anyway? I'll have to think about it \n# some more.",
"_____no_output_____"
],
[
"def _haar_measure_wrong(size):\n \"\"\"\n A demonstration of an incorrect random matrix generator.\n \n Pass this to plot_haar_angles to see what makes this incorrect.\n In brief, LAPACK heuristics designed to reduce numerical error\n bias the distribution away from small eigenvalues, creating a \n gap in the distribution rigth around zero.\n \"\"\"\n z = complex_blob(size * size).reshape((size, size)) / 2 ** 0.5\n q, r = numpy.linalg.qr(z)\n return q\n\ndef eigval_angles(u_mats):\n eig = numpy.linalg.eig\n return numpy.array([\n a \n for u_m in u_mats\n for e_vals, e_vecs in [eig(u_m)]\n for a in numpy.angle(e_vals)\n ])\n\ndef plot_haar_angles(haar_measure_func, size=50, \n n_mats=1000, n_bins=100):\n n, bins, patches = plt.hist(\n eigval_angles([haar_measure_func(size) \n for i in range(n_mats)]), \n bins=n_bins, \n density=True)\n plt.show()\n\ndef haar_angle_density(haar_measure_func, size=50, \n n_mats=1000, n_bins=100):\n angles = eigval_angles(\n [haar_measure_func(size) \n for i in range(n_mats)])\n hist, bins = numpy.histogram(\n angles, n_bins, density=True)\n return hist\n\ndef haar_angle_density_range(haar_measure_func, size=50, \n n_mats=1000, n_bins=100):\n density = haar_angle_density(\n haar_measure_func, size=size, \n n_mats=n_mats, n_bins=n_bins)\n return min(density), max(density)\n\ndef haar_angle_density_range_mean_std(\n haar_measure_func, size=50, \n n_mats=1000, n_bins=100, iters=10):\n \n ratios = [\n dmin / dmax \n for i in range(iters)\n for dmin, dmax in [\n haar_angle_density_range(\n haar_measure_func, \n size=size, \n n_mats=n_mats, \n n_bins=n_bins\n )]\n ]\n ratios = numpy.array(ratios)\n return ratios.mean(), ratios.std()\n\ndef test_angle(size=10, iters=1000):\n for i in range(iters):\n mat = haar_measure(size)\n v1 = complex_blob(size)[:, None]\n v2 = complex_blob(size)[:, None]\n assert numpy.allclose(\n v1.conj().T @ v2, \n (mat @ v1).conj().T @ (mat @ v2)\n )\n\ndef test_mag(size=10, iters=1000):\n for i in range(iters):\n mat = haar_measure(size)\n v1 = complex_blob(size)[:, None]\n assert numpy.allclose(\n v1.conj().T @ v1, \n (mat @ v1).conj().T @ (mat @ v1)\n )\n\ndef test_eigval_density():\n dmin, dmax = haar_angle_density_range(\n haar_measure, size=50, \n n_mats=1000, n_bins=100)\n \n # 0.5 is a reasonable requirement for the dmin / dmax ratio. \n # The average for this value is around 0.85 for what I believe \n # is a correct implementation, and the variance is very low at \n # the given settings.\n\n # In fact, it looks like the threshold lands at least 10 \n # standard deviations away from the mean, which I think \n # implies that the heat death of the universe will come \n # prior to a random test failure.\n\n # On the other hand, a modestly incorrect implementation \n # could fail pretty frequently -- at least one out of every \n # five tries, say -- and still be both tolerable as a solution,\n # and recognizable as a bad one. Worse solutions will fail \n # with greater regularity, and so on. A decent test, all \n # things considered.\n\n msg = (\"Minimum ratio tolerance: 0.5; \"\n f\"Actual ratio: {dmin / dmax:0.04f}\")\n assert (dmin / dmax) > 0.5, msg\n\ntest_mag()\ntest_angle()\ntest_eigval_density()\n\nplot_haar_angles(_haar_measure_wrong)\nplot_haar_angles(haar_measure)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5e4982e25c74da51bd227f3db3e3ecb43e8fec | 1,271 | ipynb | Jupyter Notebook | Capstone_Project_Notebook.ipynb | chaliuu/IBM_Data_Science_Capstone | 04819609b1bc6e476b9a304e42c2e8896efc32f9 | [
"MIT"
]
| null | null | null | Capstone_Project_Notebook.ipynb | chaliuu/IBM_Data_Science_Capstone | 04819609b1bc6e476b9a304e42c2e8896efc32f9 | [
"MIT"
]
| null | null | null | Capstone_Project_Notebook.ipynb | chaliuu/IBM_Data_Science_Capstone | 04819609b1bc6e476b9a304e42c2e8896efc32f9 | [
"MIT"
]
| null | null | null | 18.42029 | 83 | 0.522423 | [
[
[
"<b>This Notebook Will be used for the IBM Data Science Capstone Project! </b>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\npd.set_option(\"display.max_rows\", 19000)",
"_____no_output_____"
],
[
"print( 'Hello Capstone Project Course!')",
"Hello Capstone Project Course!\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
]
]
|
ec5e612210a49e64e902c1d9b889f3dec42f6359 | 21,353 | ipynb | Jupyter Notebook | _notebooks/2020-06-21-Model_Free_Prediction_Control.ipynb | dnlam/fastblog | bc06b33f617d78ef266f7bf618f815ad407cca6a | [
"Apache-2.0"
]
| null | null | null | _notebooks/2020-06-21-Model_Free_Prediction_Control.ipynb | dnlam/fastblog | bc06b33f617d78ef266f7bf618f815ad407cca6a | [
"Apache-2.0"
]
| null | null | null | _notebooks/2020-06-21-Model_Free_Prediction_Control.ipynb | dnlam/fastblog | bc06b33f617d78ef266f7bf618f815ad407cca6a | [
"Apache-2.0"
]
| null | null | null | 33.733017 | 529 | 0.602538 | [
[
[
"# Model Free Learning Method\n\n> A notebook that helps us to discover Reinforcement Learning\n- toc: true\n- branch: master\n- badges: true\n- comments: true\n- metadata_key1: metadata_value1\n- metadata_key2: metadata_value2\n- image: https://www.mdpi.com/symmetry/symmetry-12-01685/article_deploy/html/images/symmetry-12-01685-g002.png\n- description: Third in a series on understanding Reinforcement Learning.",
"_____no_output_____"
],
[
"# Introduction\nPreviously, we talked about using [Dynamic Programming](https://dnlam.github.io/fastblog/2020/06/14/MDP_DP.html), that have access to the full model, to solve optimal Bellman's Equation using MDPs. Thus, DP can learn directly from the interaction with the environment. \n\nIn this Notebook, we will study model free learning methods that do not require the full model's knowledge but only experience to attain optimal behaviour.\n",
"_____no_output_____"
],
[
"# Monte Carlo Algorithm",
"_____no_output_____"
],
[
"Monte Carlo (MC) methods are ways of solving the reinforcement learning problem based on averaging sample returns for episodic tasks. Only a completion of an episode, the value estimates and policies will be changed.\n\nHere, we talked about an episode which is defined as an trajectory of experience having some natural ending points. \n\nMC is a model free algorithm because it does not require any knowledge of MDP but only interaction returns or samples. As a concrete example, multi-armed bandit can be considered as a MC algorithm because the average reward samples (action values) are estimated for each action.",
"_____no_output_____"
],
[
"In the update form, the iterative true value of an action under multi-armed bandit is calculated as below:\n$$\nq_{t+1}(A_t) = q_t(A_t) + \\frac{1}{N(t)} (R_{t+1} - q_t(A_t))\n$$",
"_____no_output_____"
],
[
"Let's extend slightly the above bandit problem with different states. In MAB, the episodes are 1 step long as the bandit case before and it means that the actions do not affect the state transitions (you perform an action and you might see a new state that does not depend on your action.). Now, multiple states may appear and do not depend on the actions taken which means there is no long-term consequences. Then the goal is to estimate the action value that is conditioned on the action and state (contextual bandit).\n$$\nq(s,a) = \\mathbb{E} [R_{t+1} | S_t=s, A_t=a]\n$$",
"_____no_output_____"
],
[
"## Value Function Approximation\nSo far, we talked about the computation of values in MDP are based on the look up table. But it comes with a problem when a large MDP is introduced when it will consume a huge amount of memory to store every value entry.\n\nA possible solution for those problem is to use function approximation where the value function and action value function can be parameterized. Then, the parameter will be updated to obtain a value function instead of updating a huge entry during the learning process.\n$$\nv_w(s) \\approx v_\\pi(s) \\\\\\\\\nq_w(s,a) \\approx a_\\pi(s,a) \\\\\\\\\n$$",
"_____no_output_____"
],
[
"In model free learning, parameter $w$ can be updated as MC Algorithm or Temporal Difference Learning to generalize the unseen states. ",
"_____no_output_____"
],
[
"In case of the environment state is not fully observable, we will use agent state $S_t = u_w(S_{t-1},A_{t-1},O_t)$ parameterized by parameters $w$. Then, the current state is just a function of previous inputs.",
"_____no_output_____"
],
[
"### Linear Function Approximation",
"_____no_output_____"
],
[
"We will make a concrete example of a linear function approximation, where linear function can be represented a fixed feature vectors.\n\n$$\nx(s)=\\begin{pmatrix}\nx_1(s) \\\\\n... \\\\\nx_m(s) \\\\\n\\end{pmatrix}\n$$",
"_____no_output_____"
],
[
"By doing that, we can transfer the states into a bunch of vectors with elements. ",
"_____no_output_____"
],
[
"Then, the linear function approximation approach takes these features and defines our value function to be in the inner product.\n\n$$\nv_w(s) = w^Tx(s) = \\sum_{j=1}^n x_j(s)w_j\n$$ ",
"_____no_output_____"
],
[
"To update the our weights $w$, we need some sorts of objective. In our concern, the predictions will be the case so the objective will be the minimization of the loss between the true value and our estimate.\n$$\nL(w) = \\mathbb{E} [(v_\\pi(S) - w^T x(S))^2]\n$$",
"_____no_output_____"
],
[
"By apply Stochastic Gradient Descent for this objective, it will converge to global optimum of this loss function because the loss function is convex.\n\nThen, the update rule is relatively simple because the gradient of the value function with respect to our parameter $w$ is the feature $x$ and the stochastic gradient update if we would have the true value function $v_\\pi$ will be the step size times the error terms of prediction errors and the feature vector.\n$$\n\\Delta w = \\alpha (v_\\pi(S_t) - v_w(S_t))x_t \n$$",
"_____no_output_____"
],
[
"Basically, the table lookup is a special case of linear value function approximation. \nLet's n states be given by $\\mathcal{S} = \\left\\{s_1,...,s_n \\right\\}$\n\nOne hot feature represents the zeros on almost of the component except on the component that correspond the the state that we care about.",
"_____no_output_____"
],
[
"$$\nx(s)=\\begin{pmatrix}\n\\mathcal{I}(x_1(s)) \\\\\n... \\\\\n\\mathcal{I}(x_m(s)) \\\\\n\\end{pmatrix}\n$$",
"_____no_output_____"
],
[
"Then, parameter $w$ contains just value estimates for each state.\n\n$ v(s) = w^Tx(s) = \\sum _j w_j x_j(s) = w_s $",
"_____no_output_____"
],
[
"## Model-Free Prediction",
"_____no_output_____"
],
[
"Basically, q could be a parametric function (neural network) and we could use the following loss\n$$\nL(w) = \\frac{1}{2} \\mathbb{E} [(R_{t+1} - q_w(S_t,A_t))^2]\n$$",
"_____no_output_____"
],
[
"The gradient update will be\n\n$$\nw_{t+1} = w_t - \\alpha \\triangledown {w_t} L(w_t) \\\\\\\\\n= w_t - \\alpha \\triangledown _{w_t} \\frac{1}{2} \\mathbb{E} [(R_{t+1} - q_w(S_t,A_t))^2] \\\\\\\\\n= w_t + \\alpha \\mathbb{E} [(R_{t+1} - q_w(S_t,A_t)) \\triangledown _{w_t} q_{w_t}(S_t,A_t) ]\n$$",
"_____no_output_____"
],
[
"In case of a large continuous state space $\\mathcal{S}$ appears, it is just regression. ",
"_____no_output_____"
],
[
"If the action value function can be represented as a linear function, e.g. $q(s,a) = w^T x(s,a)$, then \n$$\nw_{t+1} = w_t + \\alpha (R_{t+1} - q_w(S_t,A_t)) x(s,a)\n$$",
"_____no_output_____"
],
[
"## MC Policy Evaluation\nNow, let's consider sequential decision problems where the objective is to learn $v_\\pi$ from episodes of experience under policy $\\pi$ under discouted factor $\\gamma$",
"_____no_output_____"
],
[
"$$\nS_1, A_1, R_2, ..., S_k \\sim \\pi\n$$",
"_____no_output_____"
],
[
"The discounted reward will be:\n$$\nG_t = R_{t+1} + ... + \\gamma ^{T-t-1} R_T\n$$",
"_____no_output_____"
],
[
"The expected return :\n$$\nv_\\pi(s) = \\mathbb{E}[G_t | S_t=s, \\pi]\n$$\nHere, we could use sample average return instead of expected return as the target of our updates and it is called Monte Carlo policy evaluation.",
"_____no_output_____"
],
[
"## Drawbacks of MC Learning\n- When episodes are long, learning can be quite slow because we need to wait until the end of the episode before start updating (The return is not well defined before we do.) and the return can have high variance. \n",
"_____no_output_____"
],
[
"# Temporal Difference Learning\n",
"_____no_output_____"
],
[
"Let's remind about the Bellman's equation relates the value of a state with the value of the next state and it is the definition of the value of a policy. \n$$\nv_\\pi(s) = \\mathbb{E} [R_{t+1} + \\gamma v_\\pi(S_{t+1}) | S_t=s, A_t \\sim \\pi(S_t)]\n$$",
"_____no_output_____"
],
[
"Before that, we learned that we can approximate these values by tuning this definition into an update where $v_\\pi$ is now replaced with the current estimate $v_k$. However, we should notice that the update can not happen at time step t because it is not known yet the return at time step t. \nThis iterative algorithm do learn and the do find the true value for the policy.\n$$\nv_{k+1}(s) = \\mathbb{E} [R_{t+1} + \\gamma v_k(S_{t+1}) | S_t=s, A_t \\sim \\pi(S_t)]\n$$",
"_____no_output_____"
],
[
"It suggested that we can sample this so that $v_{t+1}(S_t) = R_{t+1} + \\gamma v_t(S_{t+1}) $. then, it is better to take a small step with parameter $\\alpha$ than do all the way like that, so\n$$\nv_{t+1}(S_t) = v_t(S_t) + \\alpha _t (R_{t+1} + \\gamma v_t(S_{t+1}) - v_t(S_t))\n$$",
"_____no_output_____"
],
[
"It is very similar to MC algorithm but instead of updating towards a full return, we are going to update the target $R_{t+1} + \\gamma v_t(S_{t+1})$. So it uses our current value estimates instead of full return. It is called ```temporal difference``` learning",
"_____no_output_____"
],
[
"So, in temporal difference learning, the new target unrolls the experience one step and then uses our estimates to replace the rest of the return. \nThe ```temporal difference error ```($\\delta _t$) now is the difference in value from what we currently thing ($v_t(S_t)$) comparing to the one step into the future ($R_{t+1} + \\gamma v_t(S_{t+1})$)",
"_____no_output_____"
],
[
"## Bootstraping and Sampling\nAz we might notice now, Temporal Difference Learning combines the bootstraping approach of Dynamic Programming to derive the current estimate as part of the targer for the update and Sampling which does not require a model. ",
"_____no_output_____"
],
[
"Then we can apply the same idea to the action values where we have some action value function q and we do exactly as before.",
"_____no_output_____"
],
[
"$$\nq_{t+1} (S_t,A_t) \\leftarrow q_t(S_t,A_t) + \\alpha (R_{t+1} + \\gamma q_t(S_{t+1}, A_{t+1}) - q_t(S_t,A_t))\n$$",
"_____no_output_____"
],
[
"This algorithm is called ```SARSA``` because it uses a state action reward state and action $(S_t,A_t,R_{t+1}, S_{t+1},A_{t+1})$",
"_____no_output_____"
],
[
"## Temporal Difference Learning Properties",
"_____no_output_____"
],
[
"There are several properties of TD that we can take notice:\n- TD is model-free since it does not require the knowledge of MDP and it can learn directly from experience.\n- TD can learn from incomplete episodes bu bootstraping. It means that we dont have to wait all the way long until the end of episode before we can start learning. \n- TD can learn durning each episode. ",
"_____no_output_____"
],
[
"## Pros and Cons of MC and TD",
"_____no_output_____"
],
[
"- TD can learn before knowing the final outcomes because it can learn online after every step whereas MC must wait until the end of episode before return is known. \n- TD can learn without the final outcome, then TD can learn from incomplete sequences but MC can only learn from complete sequences. Additionally, TD works in continuing environment and MC works for episodic environments.\n\n- TD nees reasonable value estimates because the update is based on the estimation, so the bad estimate can lead to the low performance.",
"_____no_output_____"
],
[
"### Bias/Variance trade-off\nPractically speaking, MC return is an unbiased estimate of the true value but TD target is a biased estimate of the true value. In exchange, TD target has lower variance because the return in MC depends on many random actions, transitions, rewards but Td based merely on one random action, transition and reward.",
"_____no_output_____"
],
[
"## Batch mode of MC and TD\nWe have seen so far that tarbular MC and TD converge $v_t \\rightarrow v_\\pi$ as experience $\\rightarrow \\infty$ and $\\alpha_t \\rightarrow 0$. \nBut let's see what happen when we only have finite experience by considering fixed batch of experience\n$$\n\\text{episode 1:} \\quad S_1^1, A_1^1, R_2^1, ..., S^1_{T_1} \\\\\n. \\\\\n. \\\\\n. \\\\\n\\text{episode K:} \\quad S_1^K, A_1^K, R_2^K, ..., S^1_{T_K}\n\n$$",
"_____no_output_____"
],
[
"Here, instead of having a real model with every transitional probabilities, we only have observed frequencies of transitions (emperical model) ",
"_____no_output_____"
],
[
"By using MC learning, it will converge to best mean-squared fit for the observed returns $\\sum _{k=1}^K \\sum _{t=1} ^{T_k} (G_t^k - v(S_t^k))^2 $ by miniminzing this difference.",
"_____no_output_____"
],
[
"On the other hand, TD converges to solution of max likelihood Markov model, given the data samples. ",
"_____no_output_____"
],
[
"Therefore, TD exploits Markov property by means of helping in fully observable enioronments but MC does not.",
"_____no_output_____"
],
[
"# Unified view of Reinforcement Learning",
"_____no_output_____"
],
[
"Let's compare different RL algorithms where Dynamic Programming has been put on the top left corresponding to shallow update where we just look one step and full breadth into the future and we have full access on it (we need a model). \n\nBesides full breadth, if we can access on the full depth of the model, it would give us exhaustive search. \n\nIf we go all the way down, it exists an algorithm that only look at one trajectory and it is useful when we have only samples or we only deal with the interaction. TD appears on the bottom left and MC is on the bottom right. \nTD can be thought as having the breadth of one and depth of one so we can just take one step in the world and we can use that to update a value estimate whereas MC makes a very deep update when it rolls until the end of the episode and it use full trajectory to updates its value estimates.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"Following these principles, TD learning might not be accurate as it uses value estimates and additionally, information can propagate back quite slowly in some cases. MC information propagates faster as ot acknowledge all the state that have been visited before, but it comes with the cost of noisiy updates (high variance).",
"_____no_output_____"
],
[
"to go between MC and TD, we proposed ```Multi-step update```",
"_____no_output_____"
],
[
"### Multi-step prediction",
"_____no_output_____"
],
[
"Now, instead of looking exactly one step ahead as TD does, we could consider n-step ahead",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"In general, the n-step return is defined as\n$$\nG_t^{(n)} = R_{t+1} + \\gamma R_{t+2} + ... + \\gamma^{n-1} R_{t+n} + \\gamma ^n v(S_{t+n})\n$$",
"_____no_output_____"
],
[
"Then, the multi step TD learning can be transformed into the update as following:\n$$\nv(S_t) \\leftarrow v(S_t) + \\alpha (G_n^{(n)} - v(S_t))\n$$",
"_____no_output_____"
],
[
"### Mixed Multi-step return",
"_____no_output_____"
],
[
"Previously, if we have n-step return, we take one step $R_{t+1}$ and then we add $n-1$ one step return $G_t^{(n} = R_{t+1} +\\gamma G_{t+1}^{n-1} $.\n\nIt means that every step we lose one step until we have only one step left and then one step return will then bootstrap one our state value. \n\nIt means that we fully continue with random samples with trajectory in some steps but on other steps we fully stop and do bootstraping.",
"_____no_output_____"
],
[
"It is not necessarily applying the bootstrap at the end of the trajectory but we can do bootstraping a little bit with a parameter $\\lambda$",
"_____no_output_____"
],
[
"$$\nG_t^ \\lambda = R_{t+1} + \\gamma((1-\\lambda)v(S_{t+1}) + \\lambda G_{t+1}^\\lambda)\n$$",
"_____no_output_____"
],
[
"Now, instead of fully continuing or boostraping , we can do bootstraping a little bit that is controlled by $\\lambda$. This is a linear interpolation between our estimated matrix value and the rest of lambda return., Mathematically, it equals to the weighted sum of n-step return .\n$$\nG_t^\\lambda = \\sum _{n=1} ^\\infty (1-\\lambda) \\lambda ^{n-1} G_t ^{(n)} \\\\\n\\sum _{n=1} ^\\infty (1-\\lambda) \\lambda ^{n-1} = 1\n$$\n\n",
"_____no_output_____"
],
[
"If $\\lambda=0$ the return will be the same as TD case. If $\\lambda=1$, the update will follow MC algorithm.",
"_____no_output_____"
],
[
"### Benefits of multi-step returns\n- Multi-step return have both benefits of TD and MC\n- Bootstraping can have issues with bias\n- MC can have issues with variance\n- Then immediate values of n and $\\lambda$ are good.",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec5e70c756b52b5152e82fcb6c1df3876c16c825 | 4,373 | ipynb | Jupyter Notebook | tutorials/W1D5_DimensionalityReduction/student/W1D5_Outro.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| 2,294 | 2020-05-11T12:05:35.000Z | 2022-03-28T21:23:34.000Z | tutorials/W1D5_DimensionalityReduction/student/W1D5_Outro.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| 629 | 2020-05-11T15:42:26.000Z | 2022-03-29T12:23:35.000Z | tutorials/W1D5_DimensionalityReduction/student/W1D5_Outro.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| 917 | 2020-05-11T12:47:53.000Z | 2022-03-31T12:14:41.000Z | 25.87574 | 284 | 0.554539 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W1D5_DimensionalityReduction/student/W1D5_Outro.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Outro\n",
"_____no_output_____"
],
[
"**Our 2021 Sponsors, including Presenting Sponsor Facebook Reality Labs**\n\n<p align='center'><img src='https://github.com/NeuromatchAcademy/widgets/blob/master/sponsors.png?raw=True'/></p>",
"_____no_output_____"
],
[
"## Video",
"_____no_output_____"
]
],
[
[
"# @markdown\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1pi4y1V7tK\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"Vaut2BJlv10\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"## Daily survey\n\nDon't forget to complete your reflections and content check in the daily survey! Please be patient after logging in as there is\na small delay before you will be redirected to the survey.\n\n<a href=\"https://portal.neuromatchacademy.org/api/redirect/to/ec7e7e11-36cb-4ea1-b948-94332692674f\"><img src=\"https://github.com/NeuromatchAcademy/course-content/blob/master/tutorials/static/button.png?raw=1\" alt=\"button link to survey\" style=\"width:410px\"></a>",
"_____no_output_____"
],
[
"## Slides",
"_____no_output_____"
]
],
[
[
"# @markdown\nfrom IPython.display import IFrame\nIFrame(src=f\"https://mfr.ca-1.osf.io/render?url=https://osf.io/763p2/?direct%26mode=render%26action=download%26mode=render\", width=854, height=480)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec5e7c66cdc0be469420fd1eeeb73357db8f3acb | 30,747 | ipynb | Jupyter Notebook | Download Expression Matrix for Scanpy/Download SmartSeq2 Expression Matrix as Input to Scanpy.ipynb | HumanCellAtlas/data-consumer-vignettes | 1f682d945f5e2b7024874fd40ccc5b2c1e7a5f8c | [
"MIT"
]
| 18 | 2018-03-02T22:36:44.000Z | 2021-08-18T21:59:44.000Z | Download Expression Matrix for Scanpy/Download SmartSeq2 Expression Matrix as Input to Scanpy.ipynb | HumanCellAtlas/data-consumer-vignettes | 1f682d945f5e2b7024874fd40ccc5b2c1e7a5f8c | [
"MIT"
]
| 91 | 2018-03-02T00:04:25.000Z | 2020-04-30T18:29:26.000Z | Download Expression Matrix for Scanpy/Download SmartSeq2 Expression Matrix as Input to Scanpy.ipynb | HumanCellAtlas/data-consumer-vignettes | 1f682d945f5e2b7024874fd40ccc5b2c1e7a5f8c | [
"MIT"
]
| 7 | 2018-04-25T17:52:07.000Z | 2020-07-26T23:04:52.000Z | 65.141949 | 8,389 | 0.567372 | [
[
[
"# Download SmartSeq2 Expression Matrix as an Input to Scanpy",
"_____no_output_____"
],
[
"Suppose I want to get a SmartSeq2 expression matrix that I can analyze using scanpy. How can I go about finding something like this using the DSS API?",
"_____no_output_____"
]
],
[
[
"import hca.dss\nclient = hca.dss.DSSClient()",
"_____no_output_____"
]
],
[
[
"Well, first things first: We're going to need to search for a `.results` file. This file should contain what we need to put together an expression matrix.\n\nHere's our ElasticSearch query.",
"_____no_output_____"
]
],
[
[
"query = {\n \"query\": {\n \"bool\": {\n \"must\": [\n {\n \"wildcard\": {\n \"manifest.files.name\": {\n # We need a *.results file...\n \"value\": \"*.results\"\n }\n }\n },\n {\n \"range\": {\n \"manifest.version\": {\n # ...and preferably not too old, either.\n \"gte\": \"2018-07-12T100000.000000Z\"\n }\n }\n }\n ]\n }\n }\n}",
"_____no_output_____"
]
],
[
[
"This query looks for bundles that satisfy a *bool*ean condition consisting of two checks, both of which *must* be true. The *match* check looks for the `manifest.files.name` field and returns true if the name of a file listed in a bundle manifest ends with 'results'. The second check, *range*, returns true if the bundle's `manifest.version` has a value greater than or equal to 7/12/18. In short, __this query will find bundles that contain a `.results` file and are newer than July 12, 2018.__",
"_____no_output_____"
],
[
"Okay, now let's execute the search. Since the `files` section of the bundle is pretty long, I'll only print the portion containing a results file. If you want, you can always print the entire bundle to get a better picture of where the file is located.",
"_____no_output_____"
]
],
[
[
"import json\n\n# Print part of a recent analysis bundle with a results file\nbundles = client.post_search(es_query=query, replica='aws', output_format='raw')\nthe_first_bundle = bundles['results'][0]\nbundle_files = the_first_bundle['metadata']['manifest']['files']\nfor f in bundle_files:\n if f['name'].endswith('.results'):\n results_file_uuid, results_file_version = f['uuid'], f['version']\nprint(results_file_uuid, results_file_version)",
"ec727ae1-d47a-47a3-8c8e-b42d7a0e8cf4 2019-05-18T173116.989870Z\n"
]
],
[
[
"Okay! It looks like we've found a file uuid we can use. Let's retrieve that file and save it locally.",
"_____no_output_____"
]
],
[
[
"results = client.get_file(replica='aws', uuid=results_file_uuid, version=results_file_version)\nopen('matrix.results', 'w').write(results.decode(\"utf-8\"))",
"_____no_output_____"
]
],
[
[
"Here's what our file, `matrix.results`, looks like. I've truncated the output so it doesn't take up too much room.",
"_____no_output_____"
]
],
[
[
"print(open('matrix.results', 'r').read()[:852])",
"transcript_id\tgene_id\tlength\teffective_length\texpected_count\tTPM\tFPKM\tIsoPct\tposterior_mean_count\tposterior_standard_deviation_of_count\tpme_TPM\tpme_FPKM\tIsoPct_from_pme_TPM\nENST00000373020.8\tENSG00000000003.14\t2206\t2016.44\t0.00\t0.00\t0.00\t0.00\t0.00\t0.00\t0.12\t0.21\t9.99\nENST00000494424.1\tENSG00000000003.14\t820\t630.44\t0.00\t0.00\t0.00\t0.00\t0.00\t0.00\t0.38\t0.68\t31.95\nENST00000496771.5\tENSG00000000003.14\t1025\t835.44\t0.00\t0.00\t0.00\t0.00\t0.00\t0.00\t0.28\t0.51\t24.11\nENST00000612152.4\tENSG00000000003.14\t3796\t3606.44\t0.00\t0.00\t0.00\t0.00\t0.00\t0.00\t0.07\t0.12\t5.59\nENST00000614008.4\tENSG00000000003.14\t900\t710.44\t0.00\t0.00\t0.00\t0.00\t0.00\t0.00\t0.33\t0.60\t28.36\nENST00000373031.4\tENSG00000000005.5\t1339\t1149.44\t0.00\t0.00\t0.00\t0.00\t0.00\t0.00\t0.21\t0.37\t23.47\nENST00000485971.1\tENSG00000000005.5\t542\t352.44\t0.00\t0.00\t0.00\t0.00\t0.00\t0.00\t0.67\t1.22\t76.53\nENST00000371582.8\t\n"
]
],
[
[
"For our matrix, however, we might only want _some_ of these values. In my case, suppose I only want the `gene_id` and `TPM` values. We can extract these values easily using Python's `csv` module.",
"_____no_output_____"
]
],
[
[
"import csv\n\n# Take the data we want out of the results file and store it into a tsv file\n\nwith open('matrix.results', 'r') as infile, open('matrix.tsv', 'w', newline='') as outfile:\n reader = csv.DictReader(infile, delimiter='\\t')\n writer = csv.DictWriter(outfile, fieldnames=['gene_id', 'TPM'], delimiter='\\t')\n for row in reader:\n writer.writerow({'gene_id': row['gene_id'], 'TPM': row['TPM']})",
"_____no_output_____"
]
],
[
[
"Our new file, `matrix.tsv`, looks something like this:",
"_____no_output_____"
]
],
[
[
"print(open('matrix.tsv', 'r').read()[:214])",
"ENSG00000000003.14\t0.00\nENSG00000000003.14\t0.00\nENSG00000000003.14\t0.00\nENSG00000000003.14\t0.00\nENSG00000000003.14\t0.00\nENSG00000000005.5\t0.00\nENSG00000000005.5\t0.00\nENSG00000000419.12\t0.00\nENSG00000000419.12\t0.00\n\n"
]
],
[
[
"Now that we have a file containing what we want, we can transpose it and read it into scanpy.",
"_____no_output_____"
]
],
[
[
"import scanpy.api as sc\n\nadata = sc.read_csv(filename='matrix.tsv', delimiter='\\t').transpose()",
"Observation names are not unique. To make them unique, call `.obs_names_make_unique`.\nVariable names are not unique. To make them unique, call `.var_names_make_unique`.\n"
]
],
[
[
"But how do we know that everything worked? Let's print our AnnData object (truncating the output again).",
"_____no_output_____"
]
],
[
[
"print(adata)\nfor i in range(0, 153):\n print('{:<6}'.format('{:.1f}'.format(adata.X[i])), end='' if (i + 1) % 17 != 0 else '\\n' )",
"AnnData object with n_obs × n_vars = 1 × 200468 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 6.5 0.0 0.0 0.0 0.0 0.0 167.1 \n0.0 3.2 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 \n"
]
],
[
[
"And just to make it easier to see the relevant data in our matrix...",
"_____no_output_____"
]
],
[
[
"for i in range(len(adata.X)-1):\n if adata.X[i] != 0:\n print(adata.X[i], end=' ')",
"6.53 167.05 3.25 2.52 3.04 1.0 7.05 6.08 12.84 10.88 3.11 10.06 8.9 12.26 9.91 1.38 11.71 69.56 89.88 1.45 0.53 10.06 64.21 0.85 39.42 21.5 2.81 21.39 2.46 1.37 0.57 41.7 1.05 6.21 3.64 5.22 11.23 48.57 0.22 6.72 8.65 2.24 0.57 0.86 1.27 1.17 2.48 0.91 9.6 168.78 7.53 434.51 279.39 3.62 1.31 2.09 1.75 130.58 1.08 0.66 7.79 5.89 13.77 5.16 3.22 2.04 0.4 1.67 3.34 2.07 2.22 0.39 11.2 4.03 0.65 2.68 1.93 2.8 36.79 1.35 30.25 2.69 11.25 0.51 22.75 1.92 0.86 0.52 49.27 634.34 650.86 149.68 0.67 24.23 1.02 5.38 10.23 0.38 4.57 11.69 23.09 4.82 4.11 8.05 0.63 10.47 5.71 1.76 1.7 1.3 8.48 0.8 11.97 2.45 11.12 1.39 3.75 31.57 0.46 3.08 2.78 1.91 9.58 11.81 14.14 548.93 1033.31 2.02 11.71 1.48 6.2 1.87 2.8 1.62 2.94 4.99 37.99 0.92 8.03 3.22 65.9 6.83 47.61 2.16 0.44 15.74 6.07 1.0 0.42 2.15 0.88 2.23 5.08 1.11 4.08 4.42 2.3 1.11 19.06 0.62 4.89 1.44 9.8 1.55 1.64 6.58 0.81 1.24 2.85 2.9 17.77 25.15 0.49 4.73 0.04 0.78 2.04 2.26 2.43 0.44 1.34 24.36 2.94 6.62 6.65 21.71 20.38 4.72 0.83 10.44 5.54 90.53 1.73 1.36 3.57 3.13 0.35 1.03 97.68 2.18 11.05 2.25 12.1 3.28 0.85 131.76 2.93 3.52 1.42 7.53 16.79 11.52 0.85 2.19 12.8 1.43 4.72 14.51 2.35 30.29 3.62 10.23 4.01 1.81 124.66 0.78 2.98 0.36 2.94 1.71 1.8 1.44 2.19 59.13 182.93 23.63 0.43 1.3 98.35 11.2 2.77 2.47 41.67 2.33 1.04 9.27 5.48 1.41 6.83 48.74 7.78 2.14 2.78 0.73 22.63 0.53 8.67 0.46 4.95 3.39 22.52 1.47 76.77 3.88 2.22 3.0 13.9 2.89 12.11 2.49 7.58 11.23 5.36 0.74 0.93 5.24 8.51 5.14 0.53 1.03 2.83 3.24 6.0 2.22 0.96 7.05 1.54 12.07 2.25 0.5 50.93 1.14 5.8 1.85 0.84 7.31 3.26 22.54 1.47 10.48 11.36 7.27 3.64 2.95 1.59 32.68 1.0 28.59 0.47 1.19 0.68 27.62 2.6 33.37 2.1 0.95 1.06 23.9 10.48 2.57 0.71 2.23 6.01 1.18 4.22 6.14 1.22 0.83 2.09 1.8 0.7 3.95 7.84 3.79 4.68 6.24 1.64 1.92 8.9 4.25 1.98 4.91 2.32 55.25 3.11 0.92 11.21 0.87 1.08 1.49 1.18 16.12 24.25 326.34 8.45 12.06 2.22 24.31 10.8 7.74 1.02 25.23 296.06 4.24 1.04 3.36 1.34 2.18 4.24 3.67 0.8 14.28 4.82 8.14 2.9 88.49 1.11 6.28 8.02 3.32 22.09 1.9 1.29 9.45 1.04 0.76 2.39 11.39 2.79 1.26 7.54 0.63 12.68 1.5 1.82 1.04 3.27 0.85 2.07 1.14 27.14 8.23 3.48 1.8 23.09 0.56 2.68 1.4 5.36 4.24 0.63 1.38 8.29 1.01 0.43 1.58 0.51 0.96 2.44 5.32 13.1 4.12 4.75 2.68 6.47 5.72 1.5 11.24 0.89 10.41 13.64 4.66 0.64 36.29 22.94 2.24 4.72 6.46 1.44 19.38 2.98 3.17 7.23 137.56 10.51 1.02 47.85 195.22 2.37 106.51 39.35 0.48 2.25 8.32 4.72 9.92 1.54 3.14 5.68 61.23 0.86 0.52 0.95 17.92 4.37 0.77 5.71 15.94 0.81 1.19 1.14 9.21 3.22 60.52 0.66 10.42 0.61 2.31 1.53 31.52 14.38 2.06 2.39 4.38 0.75 30.6 50.04 183.7 2.09 3.96 0.96 0.63 1.45 2.13 0.6 1.66 0.92 35.13 2.72 10.57 23.61 0.73 0.5 0.8 1.21 0.51 11.93 0.9 1.02 27.74 10.38 32.67 22.91 1.4 0.95 4.0 1.34 2.42 1.84 11.2 2.84 10.53 2.57 4.03 0.59 30.72 0.63 0.97 5.56 0.74 7.17 51.1 14.43 1.47 7.07 0.07 1.98 8.87 1.22 1.61 0.45 6.62 0.74 5.12 7.33 1.85 14.68 3.52 8.83 6.23 1.19 9.39 16.43 5.62 2.02 1.19 4.84 0.68 4.97 0.77 2.99 1.48 2.24 1.6 3.92 5.06 1.75 32.17 2.85 6.55 311.55 13.32 29.74 2.21 1.04 1.59 3.96 4.11 0.54 9.32 9.48 3.39 8.69 0.72 17.86 1.44 0.81 3.69 0.81 0.92 5.05 1.87 0.74 0.35 113.35 25.65 2.52 1.0 2.16 1.36 30.22 101.08 2.24 4.65 12.77 2.27 37.87 17.82 8.23 0.76 54.69 2.34 0.31 1.87 3.37 0.62 1.03 28.82 1.03 17.35 0.78 1.7 7.52 8.37 30.66 1.68 1.68 1.34 2.91 17.09 2.79 5.21 2.71 2.21 1.06 1.29 67.59 1.69 2.36 1.04 13.41 0.84 1.0 4.73 0.87 2.07 0.92 1.21 1.98 9.61 4.83 6.83 39.35 0.85 1.91 2.9 10.69 1.42 2.19 1.29 56.65 0.56 5.73 0.27 0.46 11.56 3.17 4.82 0.98 1.11 3.31 7.09 1.8 3.03 0.64 1.94 5.84 2.34 3.31 14.07 10.77 22.58 2.1 0.54 4.17 0.81 123.82 3.47 35.76 0.62 0.43 150.55 32.87 8.36 2.76 1.52 8.48 4.72 17.71 2.05 2.84 0.73 1.21 4.81 2.82 2.0 3.36 1.17 2.45 0.56 3.57 1.56 14.08 3.93 4.18 233.27 0.22 8.56 2.05 1.24 0.53 3.36 49.84 5.35 4.1 2.64 2.17 28.82 7.7 3.02 0.44 7.6 21.66 24.81 4.24 19.68 7.78 3.81 3.73 13.4 10.14 6.06 3.18 2.35 99.53 0.77 3.8 4.12 13.42 12.03 1.47 1.05 6.45 69.98 504.94 1.81 3.0 14.76 7.06 2.28 1.14 4.35 0.84 2.14 1.08 4.62 115.54 3.31 1.64 1.76 2.91 148.09 94.85 0.98 1.79 2.78 2.31 0.93 0.28 0.8 3.61 1.41 9.15 1.27 6.24 0.86 8.15 2.23 0.4 4.5 0.89 9.79 14.32 19.39 0.78 1.02 2.71 45.18 177.82 3.33 8.36 0.7 0.69 3.84 203.35 2.17 0.69 1.22 0.63 21.12 2.25 2.19 3.82 29.15 4.78 4.67 10.53 7.93 0.98 6.84 3.07 45.21 3.54 22.56 10.4 50.91 2.57 68.99 39.33 7.52 8.28 2.9 0.7 9.24 9.38 2.2 1.59 0.55 2.36 22.37 5.32 0.31 5.74 3.94 1.16 5.2 12.16 0.45 11.83 1.63 1.6 76.23 432.41 2.88 1.88 0.51 15.99 0.61 0.74 70.8 4.5 1.42 1.96 109.96 1.35 2.99 1.59 3.33 2.6 9.05 0.72 1.26 9.5 2.75 0.56 0.42 1.16 2.73 23.54 1.57 1.6 1.08 12.09 0.54 138.51 0.78 9.29 11.57 11.02 1.01 1.4 0.53 9.36 0.72 1.15 0.56 2.69 12.15 1.97 10.44 23.45 0.73 0.63 589.71 1.17 1.18 0.78 23.45 37.21 1.56 5.34 3.2 0.35 12.6 4.65 3.23 2.48 3.27 24.27 0.63 13.02 1.56 11.82 2.88 21.33 3.55 11.21 1.36 1.32 3.77 3.08 3.08 0.84 5.93 1.82 4.26 2.55 2.07 5.14 3.21 9.21 2.5 35.12 0.62 5.03 1.16 0.58 1.18 1.98 0.4 0.32 18.73 2.36 1.88 6.97 1.89 227.08 1.7 2.87 3.47 0.59 1.46 2.19 0.64 2.06 2.41 0.36 1.54 0.48 17.43 1.05 1.46 1.53 12.26 2.51 1.66 99.07 5.6 5.83 4.5 2.22 1.86 87.19 33.95 5.85 10.51 1.48 2.9 2.21 74.29 0.88 5.77 2.22 6.78 1.65 2.33 3.41 18.54 1.32 2.62 0.95 1.97 1.98 6.47 0.75 9.14 2.31 0.76 1.72 4.18 0.39 32.35 4.1 17.52 0.84 6.63 8.03 1.14 3.49 1.23 11.27 92.66 0.96 1.1 3.42 0.6 1.25 2.13 0.76 0.99 2.56 91.74 0.83 0.38 11.11 38.32 2.16 3.19 411.22 29.43 4.37 6.58 10.77 3.17 4.15 2.12 2.18 2.34 6.97 0.89 0.62 26.5 0.91 20.04 1.9 2.88 2.11 1.33 8.72 1.09 2.39 1.04 1.75 2.71 14.68 3.66 6.67 15.25 6.47 1.72 7.86 7.36 9.6 5.23 2.02 1.82 2.93 12.19 6.89 2.07 65.18 4.26 1.77 0.94 82.34 242.68 0.35 0.89 10.36 2.15 12.72 27.82 6.68 11.08 6.23 9.97 0.62 35.65 6.12 1.35 3.06 12.26 1.56 2.33 22.61 522.42 964.06 5.81 2.6 0.38 23.04 25.42 0.64 25.21 3.61 3.31 0.76 3.6 4.22 2.86 5.02 2.11 4.31 1.57 4.11 1.77 7.78 15.67 332.41 9.47 112.63 11.47 1.61 4.95 2.05 1.19 1.43 1.51 4.93 2.29 0.46 5.2 1.25 2.45 0.79 18.62 19.04 1.64 74.67 17.11 1.64 12.19 3.92 2.82 1.58 1.18 7.68 2.0 6.03 2.07 11.0 9.13 1.88 0.16 1.71 1.86 0.4 0.83 59.97 0.87 1.38 4.63 7.45 1.31 0.71 1.45 17.11 9.59 1.14 1.93 1.91 1.65 3.8 0.45 3.68 13.79 2.09 39.22 8.1 56.76 1.47 2.85 0.75 3.85 4.3 6.89 1.72 5.17 79.28 2.42 2.72 2.41 0.29 0.82 0.66 0.88 3.34 12.78 13.26 1.18 7.27 5.04 0.72 2.26 1.03 1.07 0.85 1.81 6.51 0.97 12.52 4.36 7.49 6.42 10.32 5.85 1.89 198.26 1.73 7.93 9.59 1.58 2.33 59.19 2.81 3.8 2.61 8.0 2.39 1.92 2.39 8.12 2.1 1.75 12.09 5.54 0.72 2.17 7.09 83.98 5.38 9.79 1.09 2.19 2.31 8.05 2.39 54.41 9.54 2.05 4.18 6.36 2.97 5.61 1.56 2.19 1.06 2.74 2.29 11.42 1.37 10.65 29.03 1.53 1.11 0.98 1.86 2.09 0.65 0.81 18.24 2.68 10.46 8.0 14.53 3.61 7.06 0.95 0.79 1.15 4.32 1.72 6.83 7.26 1.4 1.49 11.17 0.94 144.56 28.11 0.74 3.88 3.52 4.89 4.62 8.8 0.71 10.18 5.5 0.81 19.98 1.54 11.42 4.77 14.25 1.77 3.3 3.59 2.51 5.57 2.09 1.55 98.8 1.93 22.5 1.16 0.51 4.16 5.65 6.93 7.04 8.96 1.87 132.96 7.7 7.88 54.35 3.39 9.36 3.52 2.37 1.12 0.32 2.03 21.2 0.2 12.45 1.91 1.02 15.42 1.8 2.97 307.46 7.12 0.46 1.75 4.31 1.57 24.92 6.14 2.95 0.33 13.73 0.62 0.74 3.97 0.2 15.72 1.51 1.82 146.29 0.65 14.94 10.72 2.76 28.63 10.91 2.49 9.59 0.23 1.18 2.16 3.09 3.33 0.98 198.18 2.23 1.01 0.9 2.5 22.71 0.85 118.79 0.46 4.05 4.79 1.9 1.9 4.04 1.72 1.35 1.68 7.49 0.96 98.32 26.67 8.43 10.86 0.87 9.16 58.09 1.71 0.92 3.33 2.0 2.49 2.11 7.13 202.57 8.83 2.49 14.62 0.78 2.89 0.61 409.27 561.15 11.72 0.64 1.29 31.7 2.09 12.85 2.11 1.58 1.7 1.8 2.72 679.33 1.55 8.83 132.02 220.41 265.84 0.85 0.8 3.31 9.92 0.9 2.4 11.8 1.83 11.08 2.68 1.25 317.51 1.07 4.6 1.27 11.61 3.64 5.43 0.77 73.39 12.8 6.01 11.69 73.39 1.28 0.47 1.1 11.0 2.5 134.02 1.02 1.64 8.45 23.5 1.05 1.69 26.39 2.19 1.1 0.69 1.62 1.19 1.24 1.35 1.0 1.32 10.3 53.66 4.06 33.99 1.29 1.18 1.1 5.14 1.67 1.11 0.55 19.02 8.66 1.36 1.09 1.96 3.02 12.39 7.26 6.65 22.52 1.3 4.47 1.94 2.9 0.71 0.35 1.7 1.61 5.34 125.14 1.57 1.53 3.35 0.51 0.81 11.98 1.37 2.77 11.15 3.95 1.09 36.25 1.25 2.81 2.92 6.35 3.54 6.8 0.95 1.24 20.95 4.0 1.21 2.54 6.19 1.76 2.91 7.27 74.44 2.14 10.36 2.16 28.23 1.74 88.37 11.63 1.41 4.16 10.52 1.62 26.27 8.27 7.66 97.92 21.15 20.62 0.23 0.55 1.97 69.96 7.85 8.58 2.06 4.07 0.77 10.83 0.78 54.02 0.68 3.81 0.34 1.49 2.03 0.53 0.9 3.3 0.37 2.19 1.97 2.06 2.64 88.83 2.77 4.48 0.95 0.99 1.92 1.31 11.93 2.68 2.36 13.81 21.34 1.17 2.66 0.12 7.34 4.89 1.74 4.98 0.65 4.53 13.76 17.66 3.22 0.66 "
]
],
[
[
"Okay, so we have one AnnData object we can use with scanpy. __But what if we want a second one?__ Let's go through the steps again, this time with a different `.results` file. We can use the same query to get a bunch of analysis bundles, but this time get our `.results` file from the _second_ bundle.",
"_____no_output_____"
]
],
[
[
"the_second_bundle = bundles['results'][1]\nbundle_files = the_second_bundle['metadata']['manifest']['files']\nfor f in bundle_files:\n if f['name'].endswith('.results'):\n results_file_uuid, results_file_version = f['uuid'], f['version']",
"_____no_output_____"
]
],
[
[
"With the new uuid, we can get the `.results` file itself.",
"_____no_output_____"
]
],
[
[
"results2 = client.get_file(replica='aws', uuid=results_file_uuid, version=results_file_version)\nopen('matrix2.results', 'w').write(results2.decode(\"utf-8\"))",
"Waiting 10s before redirect per Retry-After header\n"
]
],
[
[
"Again, let's take the data we want out of the results file and store it into a tsv file...",
"_____no_output_____"
]
],
[
[
"with open('matrix2.results', 'r') as infile, open('matrix2.tsv', 'w', newline='') as outfile:\n reader = csv.DictReader(infile, delimiter='\\t')\n writer = csv.DictWriter(outfile, fieldnames=['gene_id', 'TPM'], delimiter='\\t')\n for row in reader:\n writer.writerow({'gene_id': row['gene_id'], 'TPM': row['TPM']})",
"_____no_output_____"
]
],
[
[
"...and then create another AnnData object & print it.",
"_____no_output_____"
]
],
[
[
"adata2 = sc.read_csv( filename='matrix.tsv', delimiter='\\t' ).transpose()\n\nprint(adata2)\nfor i in range(0, 153):\n print( '{:<6}'.format('{:.1f}'.format(adata2.X[i])), end='' if (i + 1) % 17 != 0 else '\\n' )",
"Observation names are not unique. To make them unique, call `.obs_names_make_unique`.\nVariable names are not unique. To make them unique, call `.var_names_make_unique`.\n"
]
],
[
[
"Now that you've set up your two matrices in scanpy, you're ready to perform whatever analysis piques your interest.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec5e89987ae6779814e2a9222b132cf9ba01776d | 13,467 | ipynb | Jupyter Notebook | loanpy/data/examples.ipynb | martino-vic/loanpy | 5a5c1512470b4a9d5a451d252afc0f8613e714d1 | [
"AFL-3.0"
]
| 8 | 2020-11-18T20:13:51.000Z | 2022-02-07T20:12:27.000Z | loanpy/data/examples.ipynb | martino-vic/loanpy | 5a5c1512470b4a9d5a451d252afc0f8613e714d1 | [
"AFL-3.0"
]
| null | null | null | loanpy/data/examples.ipynb | martino-vic/loanpy | 5a5c1512470b4a9d5a451d252afc0f8613e714d1 | [
"AFL-3.0"
]
| null | null | null | 23.626316 | 132 | 0.541249 | [
[
[
"import pandas as pd\nfrom loanpy.helpers import filterdf\ndfin=pd.read_csv(\"dfhun_zaicz_backup.csv\", encoding=\"utf-8\")\ndffiltered = filterdf(dfin, \"L1_suffix\", occurs_or_bigger=False, term=\"\\+\")\nfilterdf(dffiltered, \"L1_year\", occurs_or_bigger=False, term=1600, write=True, name=\"example_dfhun_before1600.csv\")",
"_____no_output_____"
],
[
"import pandas as pd\nfrom loanpy.helpers import filterdf\ndfin=pd.read_csv(\"dfhun_zaicz_backup.csv\", encoding=\"utf-8\")\ndffiltered = filterdf(dfin, \"L1_suffix\", occurs_or_bigger=False, term=\"\\+\")\ndffiltered = filterdf(dffiltered, \"L1_year\", occurs_or_bigger=False, term=1600)\nfilterdf(dffiltered, \"L1_language\", occurs_or_bigger=True, term=\"unknown\", write=True, name=\"example_dfhun_unknown.csv\")",
"_____no_output_____"
],
[
"from loanpy import loanfinder as lf\nlf.adapt_or_reconstruct_col(\n inputcsv=\"dfhun_zaicz_before1600.csv\", inputcol=\"L1_ipa\",\n funcname=\"reconstruct\", howmany=float(\"inf\"), struc=False,\n vowelharmony=False, only_documented_clusters=False,\n write=True, outputcsv=\"example_dfhun_before1600.csv\",\n outputcol=\"example_rc\")",
"_____no_output_____"
],
[
"from loanpy import loanfinder as lf\nlf.adapt_or_reconstruct_col(\n inputcsv=\"dfgot_wikiling_backup.csv\", inputcol=\"L2_ipa\",\n funcname=\"adapt\", struc=True,howmany=1,\n only_documented_clusters=False, vowelharmony=True,\n write=True,\n outputcsv=\"example_dfgot.csv\", outputcol=\"example_ad\")",
"_____no_output_____"
],
[
"import pandas as pd\nfrom loanpy import loanfinder as lf\nlf.dfL2 = pd.read_csv(\"dfgot_wiktionary_backup.csv\",\n encoding=\"utf-8\")[\"L2_latin\"]\nlf.findphoneticmatches(\"^anna$\", 5)",
"_____no_output_____"
],
[
"import pandas as pd\nfrom loanpy import loanfinder as lf\nlf.dfL2 = pd.read_csv(\"dfgot_wiktionary_backup.csv\",\n encoding=\"utf-8\")[\"L2_latin\"]\nlf.findphoneticmatches(\"^abraham$|^anna$\", 123)",
"_____no_output_____"
],
[
"import pandas as pd\nfrom loanpy import loanfinder as lf\nlf.dfL2 = pd.read_csv(\"dfgot_wiktionary_backup.csv\",\n encoding=\"utf-8\")[\"L2_latin\"]\nlf.findphoneticmatches(\"^a(nn|br)a(ham)?$\", 5)",
"_____no_output_____"
],
[
"from loanpy import loanfinder as lf\nlf.findloans(\"example\", \"example_dfhun_before1600.csv\", \"example_dfgot.csv\", \"example_rc\", \"example_ad\")",
"_____no_output_____"
],
[
"from loanpy import adapter as ad\nad.launch()\nad.substidict",
"_____no_output_____"
],
[
"from loanpy import adapter as ad\nad.launch()\nad.allowedphonotactics",
"_____no_output_____"
],
[
"from loanpy import adapter as ad\nad.launch()\nad.scvalues",
"_____no_output_____"
],
[
"from loanpy import adapter as ad\nad.launch()\nad.adapt(\"wulɸɪla\", howmany=float(\"inf\"), struc=False,\n only_documented_clusters=False, vowelharmony=False)",
"_____no_output_____"
],
[
"from loanpy import adapter as ad\nad.launch()\nad.allowedphonotactics = [\"CVCVCVCV\",\"CVCV\"]\nad.adapt(\"wulɸɪla\", howmany=2, struc=True,\n vowelharmony=False, only_documented_clusters=False)",
"_____no_output_____"
],
[
"from loanpy import adapter as ad\nad.launch()\nad.adapt(\"wulɸɪla\", howmany=5, struc=False,\n vowelharmony=True, only_documented_clusters=False)",
"_____no_output_____"
],
[
"from loanpy import adapter as ad\nad.launch()\nad.adapt(\"wulɸɪla\", howmany=5, struc=False,\n vowelharmony=True, only_documented_clusters=True)",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch()\nrc.scdict",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch()\nrc.allowedphonotactics",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch()\nrc.nsedict",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch(se_or_edict=\"edict.txt\")\nrc.nsedict",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.getsoundchanges(\"ɟɒloɡ\", \"jɑlkɑ\")",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.dfetymology2dict()",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch()\nrc.getnse(\"ɟɒloɡ\",\"jɑlkɑ\")",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch(se_or_edict=\"edict.txt\")\nrc.getnse(\"ɟɒloɡ\", \"jɑlkɑ\", examples=True)",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch(se_or_edict=\"sedict.txt\")\nrc.getnse(\"ɟɒloɡ\", \"jɑlkɑ\", normalise=False)",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch(se_or_edict=\"sedict.txt\")\nrc.getnse(\"ɟɒloɡ\", \"jɑlkɑ\", examples=True)",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch()\nrc.reconstruct('mɒɟɒr', howmany=100)",
"_____no_output_____"
],
[
"from loanpy import reconstructor as rc\nrc.launch()\nrc.reconstruct(\"mɒɟɒr\", howmany=5, struc=True,\n vowelharmony=True, sort_by_nse=True)",
"_____no_output_____"
],
[
"from loanpy.helpers import phon2cv\nphon2cv(\"p\")",
"_____no_output_____"
],
[
"phon2cv(\"a\")",
"_____no_output_____"
],
[
"from loanpy.helpers import word2struc\nword2struc(\"mɒɟɒr\")",
"_____no_output_____"
],
[
"from loanpy.helpers import ipa2clusters\nipa2clusters(\"roflmao\")",
"_____no_output_____"
],
[
"from loanpy.helpers import list2regex\nlist2regex([\"b\", \"k\", \"v\"])",
"_____no_output_____"
],
[
"from loanpy.helpers import list2regex\nlist2regex([\"b\", \"k\", \"0\", \"v\"])",
"_____no_output_____"
],
[
"from loanpy.helpers import editdistancewith2ops\neditdistancewith2ops(\"hey\",\"hey\")",
"_____no_output_____"
],
[
"from loanpy.helpers import editdistancewith2ops\neditdistancewith2ops(\"hey\",\"he\")",
"_____no_output_____"
],
[
"from loanpy.helpers import editdistancewith2ops\neditdistancewith2ops(\"hey\",\"heyy\")",
"_____no_output_____"
],
[
"from loanpy.helpers import loadvectors, gensim_similarity\nloadvectors()\ngensim_similarity(\"house, sing, hello\", \"cottage, regrettable, car\",\n return_wordpair=False)",
"_____no_output_____"
],
[
"from loanpy.helpers import loadvectors, gensim_similarity\nloadvectors()\ngensim_similarity(\"house, sing, hello\", \"cottage, regrettable, car\",\n return_wordpair=True)",
"_____no_output_____"
],
[
"# import default vectors\nfrom loanpy.helpers import loadvectors\nloadvectors()",
"_____no_output_____"
],
[
"# now load some other vectors\nfrom loanpy import helpers\nhelpers.model=None\nhelpers.loadvectors(\"glove-twitter-25\")",
"_____no_output_____"
],
[
"from loanpy.helpers import vow2frontback\nvow2frontback(\"e\")",
"_____no_output_____"
],
[
"vow2frontback(\"o\")",
"_____no_output_____"
],
[
"from loanpy.helpers import harmony\nharmony(\"bot͡sibot͡si\")",
"_____no_output_____"
],
[
"from loanpy.helpers import harmony\nharmony([\"t\", \"ɒ\", \"r\", \"k\", \"ɒ\"])",
"_____no_output_____"
],
[
"from loanpy.helpers import harmony\nharmony(\"ʃɛfylɛʃɛ\")",
"_____no_output_____"
],
[
"from loanpy.helpers import adaptharmony\nadaptharmony('kɛsthɛj')",
"_____no_output_____"
],
[
"from loanpy.helpers import adaptharmony\nadaptharmony(['ɒ', 'l', 'ʃ', 'oː', 'ø', 'r', 'ʃ'])",
"_____no_output_____"
],
[
"from loanpy.helpers import adaptharmony\nadaptharmony('ʃioːfok')",
"_____no_output_____"
],
[
"from loanpy.helpers import adaptharmony\nadaptharmony(['b', 'ɒ', 'l', 'ɒ', 't', 'o', 'n', 'k', 'ɛ', 'n', 'ɛ', 'ʃ', 'ɛ'])",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5e8f1a2aa41be3b1f5bedda78f28f0940e3c91 | 37,412 | ipynb | Jupyter Notebook | notebooks/Scraper 2.0.ipynb | ActiveConclusion/COVID19_mobility | ca2579ab3206adc15c686a80dbe24cf0910a21b3 | [
"MIT"
]
| 239 | 2020-04-04T14:18:37.000Z | 2022-03-28T15:35:12.000Z | notebooks/Scraper 2.0.ipynb | ActiveConclusion/COVID19_mobility | ca2579ab3206adc15c686a80dbe24cf0910a21b3 | [
"MIT"
]
| 29 | 2020-04-06T14:20:15.000Z | 2022-03-29T19:24:09.000Z | notebooks/Scraper 2.0.ipynb | ActiveConclusion/COVID19_mobility | ca2579ab3206adc15c686a80dbe24cf0910a21b3 | [
"MIT"
]
| 90 | 2020-04-05T23:48:54.000Z | 2022-03-21T21:11:41.000Z | 45.074699 | 165 | 0.52542 | [
[
[
"# Scraper of Google, Apple and Waze Mobility reports",
"_____no_output_____"
],
[
"This notebook loads Google, Apple, Waze and TomTom Mobility reports, builds cleaned reports in different formats and builds merged files from these sources.\n\nOriginal data:\n - Google Community Mobility reports: https://www.google.com/covid19/mobility/\n - Apple Mobility Trends reports: https://www.apple.com/covid19/mobility\n - Waze COVID-19 local driving trends: https://www.waze.com/covid19\n - TomTom Traffic Index: https://www.tomtom.com/en_gb/traffic-index/ranking/",
"_____no_output_____"
]
],
[
[
"import os\nimport datetime\nimport time\n\nimport requests\nimport urllib.request\nfrom bs4 import BeautifulSoup\nimport re\nimport json\n\nimport pandas as pd\nimport zipfile as zp",
"_____no_output_____"
],
[
"def get_google_link():\n '''Get link of Google Community Mobility report file\n \n Returns:\n link (str): link of Google Community report file\n '''\n # get webpage source\n url = 'https://www.google.com/covid19/mobility/'\n response = requests.get(url)\n soup = BeautifulSoup(response.text, \"html.parser\")\n csv_tag = soup.find('a', {\"class\": \"icon-link\"})\n link = csv_tag['href']\n return link",
"_____no_output_____"
],
[
"def download_google_report(directory=\"google_reports\"):\n '''Download Google Community Mobility report in CSV format\n\n Args:\n directory: directory to which CSV report will be downloaded\n\n Returns:\n new_files (bool): flag indicating whether or not new files have been downloaded\n '''\n new_files = False\n\n # create directory if it don't exist\n if not os.path.exists(directory) and directory!='':\n os.makedirs(directory)\n\n # download CSV file\n link = get_google_link()\n file_name = \"Global_Mobility_Report.csv\"\n path = os.path.join(directory, file_name)\n old_size = os.path.getsize(path) if os.path.isfile(path) else 0\n urllib.request.urlretrieve(link, path)\n new_size = os.path.getsize(path)\n if old_size!=new_size:\n new_files = True\n\n if not new_files:\n print('Google: No updates')\n else:\n print('Google: Update available')\n \n return new_files",
"_____no_output_____"
],
[
"def build_google_report(\n source=os.path.join(\"google_reports\", \"Global_Mobility_Report.csv\"),\n report_type=\"regions\",\n countries=None,\n world_regions=None,\n country_regions_file = os.path.join(\"auxiliary_data\", \"country_worldregions.csv\")):\n '''Build cleaned Google report for the worldwide\n\n Args:\n source: location of the raw Google CSV report\n report_type: available options: \n 1) \"regions\" - basic report for the worldwide\n 2) \"US\" - report for the US\n 3) \"regions_detailed\" - detailed report for selected countries\n 4) \"world_regions_detailed\" - detailed report for selected regions (Europe, Asia etc)\n countries: list of countries for \"regions_detailed\" option. If None - all countries selected\n world_regions: list of regions for \"world_regions_detailed option. If None - all regions selected\n country_regions_file: path of the CSV file with matching table of countries and regions\n\n Returns:\n google (DataFrame): generated Google report\n '''\n # read the raw report\n google = pd.read_csv(source, low_memory=False)\n # shorten value column names\n google.columns = google.columns.str.replace(\n r'_percent_change_from_baseline', '')\n # remove underscores from column names\n google.columns = google.columns.str.replace(r'_', ' ')\n # rename country column\n google = google.rename(columns={'country region': 'country'})\n if report_type == \"regions\":\n # remove data of subregions of the second level\n google = google[google['sub region 2'].isnull()]\n # remove metropolitan data\n google = google[google['metro area'].isnull()]\n # rename region column\n google = google.rename(columns={'sub region 1': 'region'})\n google = google.loc[:,\n ['country',\n 'region',\n 'date',\n 'retail and recreation',\n 'grocery and pharmacy',\n 'parks',\n 'transit stations',\n 'workplaces',\n 'residential']]\n google['region'].fillna('Total', inplace=True)\n elif report_type == \"US\":\n google = google[(google['country'] == \"United States\")]\n google = google.rename(\n columns={\n 'sub region 1': 'state',\n 'sub region 2': 'county'})\n google = google.loc[:,\n ['state',\n 'county',\n 'date',\n 'retail and recreation',\n 'grocery and pharmacy',\n 'parks',\n 'transit stations',\n 'workplaces',\n 'residential']]\n google['state'].fillna('Total', inplace=True)\n google['county'].fillna('Total', inplace=True)\n elif report_type == \"regions_detailed\" or report_type == \"world_regions_detailed\":\n if countries is not None and report_type == \"regions_detailed\":\n google = google[google.country.isin(countries)]\n if report_type == \"world_regions_detailed\":\n if os.path.isfile(country_regions_file):\n country_regions = pd.read_csv(country_regions_file)\n google = pd.merge(google, country_regions, on='country')\n if world_regions is not None:\n google = google[google.world_region.isin(world_regions)]\n # metro area -> sub region 1 \n google['sub region 1'] = google.apply(lambda x: x['sub region 1'] if isinstance(x['sub region 1'],str)\n else x['metro area'], axis=1)\n column_list = ['world_region'] if report_type == \"world_regions_detailed\" else []\n column_list = column_list + ['country',\n 'sub region 1',\n 'sub region 2',\n 'date',\n 'retail and recreation',\n 'grocery and pharmacy',\n 'parks',\n 'transit stations',\n 'workplaces',\n 'residential']\n google = google.loc[:, column_list]\n google['sub region 1'].fillna('Total', inplace=True)\n google['sub region 2'].fillna('Total', inplace=True)\n return google",
"_____no_output_____"
],
[
"def get_apple_link():\n '''Get link of Apple Mobility Trends report file\n \n Returns:\n link (str): link of Apple Mobility Trends report file\n '''\n # get link via API\n json_link = \"https://covid19-static.cdn-apple.com/covid19-mobility-data/current/v3/index.json\"\n with urllib.request.urlopen(json_link) as url:\n json_data = json.loads(url.read().decode())\n link = \"https://covid19-static.cdn-apple.com\" + \\\n json_data['basePath'] + json_data['regions']['en-us']['csvPath']\n return link",
"_____no_output_____"
],
[
"def download_apple_report(directory=\"apple_reports\"):\n '''Download Apple Mobility Trends report in CSV\n\n Args:\n directory: directory to which CSV report will be downloaded\n\n Returns:\n new_files (bool): flag indicating whether or not a new file has been downloaded\n '''\n new_files = False\n \n # create directory if it don't exist\n if not os.path.exists(directory) and directory!='':\n os.makedirs(directory)\n \n link = get_apple_link()\n file_name = \"applemobilitytrends.csv\"\n path = os.path.join(directory, file_name)\n path = os.path.join(directory, file_name)\n old_size = os.path.getsize(path) if os.path.isfile(path) else 0\n urllib.request.urlretrieve(link, path)\n new_size = os.path.getsize(path)\n if old_size!=new_size:\n new_files = True\n\n if not new_files:\n print('Apple: No updates')\n else:\n print('Apple: Update available')\n \n return new_files",
"_____no_output_____"
],
[
"def build_apple_report(\n source=os.path.join(\n 'apple_reports',\n \"applemobilitytrends.csv\"),\n report_type=\"regions\"):\n '''Build cleaned Apple report (transform dates from columns to rows, add country names for subregions and cities)\n for the worldwide or for some country (currently only for the US)\n\n Args:\n source: location of the raw Apple CSV report\n destination: destination file path\n report_type: two options available: \"regions\" - report for the worldwide, \"US\" - report for the US\n\n Returns:\n apple (DataFrame): generated Apple report\n '''\n apple = pd.read_csv(source, low_memory=False)\n apple = apple.drop(columns=['alternative_name'])\n apple['country'] = apple.apply(\n lambda x: x['region'] if x['geo_type'] == 'country/region' else x['country'],\n axis=1)\n\n if report_type == 'regions':\n apple = apple[apple.geo_type != 'county']\n apple['sub-region'] = apple.apply(lambda x: 'Total' if x['geo_type'] == 'country/region' else (\n x['region'] if x['geo_type'] == 'sub-region' else x['sub-region']), axis=1)\n apple['subregion_and_city'] = apple.apply(\n lambda x: 'Total' if x['geo_type'] == 'country/region' else x['region'], axis=1)\n apple = apple.drop(columns=['region'])\n apple['sub-region'] = apple['sub-region'].fillna(\n apple['subregion_and_city'])\n\n apple = apple.melt(\n id_vars=[\n 'geo_type',\n 'subregion_and_city',\n 'sub-region',\n 'transportation_type',\n 'country'],\n var_name='date')\n apple['value'] = apple['value'] - 100\n\n apple = apple.pivot_table(\n index=[\n \"geo_type\",\n \"subregion_and_city\",\n \"sub-region\",\n \"date\",\n \"country\"],\n columns='transportation_type').reset_index()\n apple.columns = [t + (v if v != \"value\" else \"\")\n for v, t in apple.columns]\n apple = apple.loc[:,\n ['country',\n 'sub-region',\n 'subregion_and_city',\n 'geo_type',\n 'date',\n 'driving',\n 'transit',\n 'walking']]\n apple = apple.sort_values(by=['country',\n 'sub-region',\n 'subregion_and_city',\n 'date']).reset_index(drop=True)\n elif report_type == \"US\":\n apple = apple[apple.country == \"United States\"].drop(columns=[\n 'country'])\n apple['sub-region'] = apple['sub-region'].fillna(\n apple['region']).replace({\"United States\": \"Total\"})\n apple['region'] = apple.apply(lambda x: x['region'] if (\n x['geo_type'] == 'city' or x['geo_type'] == 'county') else 'Total', axis=1)\n apple = apple.rename(\n columns={\n 'sub-region': 'state',\n 'region': 'county_and_city'})\n\n apple = apple.melt(\n id_vars=[\n 'geo_type',\n 'state',\n 'county_and_city',\n 'transportation_type'],\n var_name='date')\n apple['value'] = apple['value'] - 100\n\n apple = apple.pivot_table(\n index=[\n 'geo_type',\n 'state',\n 'county_and_city',\n 'date'],\n columns='transportation_type').reset_index()\n apple.columns = [t + (v if v != \"value\" else \"\")\n for v, t in apple.columns]\n\n apple = apple.loc[:, ['state', 'county_and_city', 'geo_type',\n 'date', 'driving', 'transit', 'walking']]\n apple = apple.sort_values(\n by=['state', 'county_and_city', 'geo_type', 'date']).reset_index(drop=True)\n return apple",
"_____no_output_____"
],
[
"def get_waze_links():\n '''Get links of raw Waze local driving Trends report files\n \n Returns:\n link (list): links of Waze local driving Trends report files\n '''\n github_link = \"https://raw.githubusercontent.com/ActiveConclusion/waze_mobility_scraper/master/\"\n country_level_link = github_link + \"Waze_Country-Level_Data.csv\"\n city_level_link = github_link + \"Waze_City-Level_Data.csv\"\n return [country_level_link, city_level_link]",
"_____no_output_____"
],
[
"def download_waze_reports(directory=\"waze_reports\"):\n '''Download Waze local driving Trends reports in CSV\n\n Args:\n directory: directory to which CSV reports will be downloaded\n\n Returns:\n new_files (bool): flag indicating whether or not a new files has been downloaded\n '''\n new_files = False\n \n # create directory if it don't exist\n if not os.path.exists(directory) and directory!='':\n os.makedirs(directory)\n \n links = get_waze_links()\n file_names = [\"Waze_Country-Level_Data.csv\", \"Waze_City-Level_Data.csv\"]\n file_links = dict(zip(file_names, links))\n for file_name in file_names: \n path = os.path.join(directory, file_name)\n link = file_links[file_name]\n old_size = os.path.getsize(path) if os.path.isfile(path) else 0\n urllib.request.urlretrieve(link, path)\n new_size = os.path.getsize(path)\n if old_size!=new_size:\n new_files = True\n\n if not new_files:\n print('Waze: No updates')\n else:\n print('Waze: Update available')\n \n return new_files",
"_____no_output_____"
],
[
"def build_waze_report(countries_source=os.path.join(\"waze_reports\", \"Waze_Country-Level_Data.csv\"),\n cities_source=os.path.join(\"waze_reports\", \"Waze_City-Level_Data.csv\")):\n '''Build cleaned Waze report (transform dates from string to date format, merge country&city-level data,\n add geo_type column)\n\n Args:\n countries_source: location of the raw Waze country-level CSV report\n cities_source: location of the raw Waze city-level CSV report\n\n Returns:\n waze (DataFrame): generated Waze report\n '''\n waze_countries = pd.read_csv(countries_source, parse_dates=['Date'])\n waze_cities = pd.read_csv(cities_source, parse_dates=['Date'])\n waze_countries['City'] = 'Total'\n waze_countries['geo_type'] = 'country'\n waze_cities['geo_type'] = 'city'\n \n waze = waze_countries.append(waze_cities)\n waze = waze.rename(columns={'Country':'country', 'City':'city', \n 'Date':'date', '% Change In Waze Driven Miles/KMs':'driving_waze'})\n waze['driving_waze'] = waze['driving_waze'] * 100\n waze['date'] = waze['date'].dt.date\n waze = waze.loc[:,['country', 'city','geo_type','date', 'driving_waze']]\n waze = waze.sort_values(by=['country', 'city', 'geo_type', 'date']).reset_index(drop=True)\n return waze",
"_____no_output_____"
],
[
"def check_tomtom_update(\n tomtom_source=os.path.join(\"tomtom_reports\", \"tomtom_trafic_index.csv\"),\n api_key_check=\"JPN_tokyo\",\n):\n \"\"\"Check if new TomTom data available\n\n Args:\n tomtom_source: location of the TomTom report in CSV format (if exist)\n api_key_check: which city will be checked on the TomTom site\n Returns:\n new_files (bool): flag indicating whether or not new data available\n \"\"\"\n new_files = False\n # check if file available\n if not os.path.exists(tomtom_source):\n new_files = True\n else:\n # get max date from the CSV report\n tomtom = pd.read_csv(tomtom_source, low_memory=False)\n last_report_date = tomtom[\"date\"].max()\n # get last available date from API\n base_api_url = \"https://api.midway.tomtom.com/ranking/dailyStats/\"\n api_url = base_api_url + api_key_check\n response = requests.get(api_url)\n json_data = response.json()\n last_api_date = json_data[-1][\"date\"]\n if last_api_date != last_report_date:\n new_files = True\n if not new_files:\n print(\"TomTom: No updates\")\n else:\n print(\"TomTom: Update available\")\n\n return new_files",
"_____no_output_____"
],
[
"def download_tomtom_report(\n alpha_codes_filename=os.path.join(\"auxiliary_data\", \"country_alpha_codes.csv\")\n):\n \"\"\"Download TomTom Traffic Index\n\n Args:\n iso_codes_filename: path to alpha country codes file\n\n Returns:\n tomtom_data (DataFrame): scraped TomTom report\n \"\"\"\n # get all available cities\n json_link = (\n \"https://www.tomtom.com/en_gb/traffic-index/page-data/ranking/page-data.json\"\n )\n with urllib.request.urlopen(json_link) as url:\n json_data = json.loads(url.read().decode())\n # unpack data from json\n json_city_data = json_data[\"result\"][\"data\"][\"allCitiesJson\"][\"edges\"]\n # merge data and select necessary columns\n merged_data = {}\n for k in [\"name\", \"country\", \"countryName\", \"continent\", \"key\"]:\n merged_data[k] = tuple(merged_data[\"node\"][k] for merged_data in json_city_data)\n city_data = pd.DataFrame(merged_data)\n # define key exceptions\n key_exceptions = {\n \"birmingham-alabama\": \"birmingham\",\n \"hamilton-nz\": \"hamilton\",\n \"london-ontario\": \"london\",\n \"newcastle-au\": \"newcastle\",\n \"bengaluru\": \"bangalore\",\n }\n # replace exceptions in DataFrame\n city_data.replace({\"key\": key_exceptions}, inplace=True)\n # add Alpha3 country codes\n # read file with alpha codes\n alpha_codes = pd.read_csv(alpha_codes_filename)\n # match by alpha2 codes\n city_data = pd.merge(\n city_data, alpha_codes, left_on=\"country\", right_on=\"Alpha2\", how=\"left\"\n )\n city_data.drop(\"country\", axis=1, inplace=True)\n # create api key for scraping data\n city_data[\"api_key\"] = city_data[\"Alpha3\"] + \"_\" + city_data[\"key\"]\n # scrape data for each city\n base_api_url = \"https://api.midway.tomtom.com/ranking/dailyStats/\"\n city_df_list = []\n for _, row in city_data.iterrows():\n api_url = base_api_url + row[\"api_key\"]\n response = requests.get(api_url)\n city_df = pd.DataFrame(response.json())\n city_df[\"country\"] = row[\"countryName\"]\n city_df[\"city\"] = row[\"name\"]\n city_df_list.append(city_df)\n # merge all data\n tomtom_data = pd.concat(city_df_list, ignore_index=True)\n tomtom_data = tomtom_data.loc[\n :, [\"country\", \"city\", \"date\", \"congestion\", \"diffRatio\"]\n ]\n tomtom_data = tomtom_data.sort_values(by=[\"country\", \"city\", \"date\"]).reset_index(\n drop=True\n )\n return tomtom_data",
"_____no_output_____"
],
[
"def build_summary_report(apple_source, google_source, report_type=\"regions\"):\n '''Build a merged report from Google and Apple data\n\n Args:\n apple_source: location of the CSV report generated by build_apple_report function\n google_source: location of the CSV report generated by build_google_report function\n report_type: two options available: \"regions\" - report for the worldwide, \"US\" - report for the US\n\n Returns:\n summary (DataFrame): merged report from Google and Apple data\n '''\n apple = pd.read_csv(apple_source, low_memory=False)\n google = pd.read_csv(google_source, low_memory=False)\n summary = pd.DataFrame()\n # build report for regions\n if report_type == \"regions\":\n apple = apple.rename(columns={'subregion_and_city': 'region'})\n apple = apple.loc[:, ['country', 'region',\n 'date', 'driving', 'transit', 'walking']]\n # get matching table for converting Apple countries and subregions to\n # Google names\n country_AtoG_file = os.path.join(\n 'auxiliary_data', 'country_Apple_to_Google.csv')\n subregions_AtoG_file = os.path.join(\n 'auxiliary_data', 'subregions_Apple_to_Google.csv')\n\n if os.path.isfile(country_AtoG_file):\n country_AtoG = pd.read_csv(country_AtoG_file, index_col=0)\n else:\n country_AtoG = None\n if os.path.isfile(subregions_AtoG_file):\n subregions_AtoG = pd.read_csv(subregions_AtoG_file, index_col=0)\n else:\n subregions_AtoG = None\n # convert Apple countries and subregions to Google names\n apple['country'] = apple.apply(lambda x: country_AtoG.loc[x['country'], 'country_google'] if (\n country_AtoG is not None and x['country'] in country_AtoG.index) else x['country'], axis=1)\n apple['region'] = apple.apply(lambda x: subregions_AtoG.loc[x['region'], 'subregion_Google'] if (\n subregions_AtoG is not None and x['region'] in subregions_AtoG.index) else x['region'], axis=1)\n # merge reports\n apple = apple.set_index(['country', 'region', 'date'])\n google = google.set_index(['country', 'region', 'date'])\n summary = google.join(apple, how='outer')\n summary = summary.reset_index(level=['country', 'region', 'date'])\n elif report_type == \"US\":\n apple = apple.loc[:, ['state', 'county_and_city',\n 'date', 'driving', 'transit', 'walking']]\n apple.loc[apple.state == 'Washington DC',\n 'state'] = 'District of Columbia'\n apple.loc[apple.county_and_city ==\n 'Washington DC', 'county_and_city'] = 'Total'\n\n google = google.rename(columns={'county': 'county_and_city'})\n # merge reports\n apple = apple.set_index(['state', 'county_and_city', 'date'])\n google = google.set_index(['state', 'county_and_city', 'date'])\n summary = google.join(apple, how='outer')\n summary = summary.reset_index(\n level=['state', 'county_and_city', 'date'])\n return summary",
"_____no_output_____"
],
[
"os.chdir('..')",
"_____no_output_____"
],
[
"# process Google reports\nGOOGLE_ZIP_PATH = os.path.join(\"google_reports\", \"Global_Mobility_Report.zip\")\nGOOGLE_CSV_PATH = os.path.join(\"google_reports\", \"Global_Mobility_Report.csv\")\n# unzip existing report\nif os.path.exists(GOOGLE_ZIP_PATH):\n with zp.ZipFile(GOOGLE_ZIP_PATH, 'r') as zf:\n zf.extract('Global_Mobility_Report.csv', \"google_reports\")\n# download new report\nnew_files_status_google = download_google_report()\nif new_files_status_google:\n # build reports\n # build basic report for the worldwide\n google_world = build_google_report()\n # build report for the US\n google_US = build_google_report(report_type=\"US\")\n # build report for Brazil\n google_brazil = build_google_report( report_type=\"regions_detailed\", countries=[\"Brazil\"])\n # build detailed reports for world regions\n google_world_regions = build_google_report(report_type=\"world_regions_detailed\")\n google_europe = google_world_regions[google_world_regions.world_region.isin(['Europe'])]\n google_asia_africa = google_world_regions[google_world_regions.world_region.isin(['Asia', 'Africa'])]\n google_america_oceania = google_world_regions[\n google_world_regions.world_region.isin(['South America', 'North America', 'Oceania'])]\n # write reports to CSV and Excel\n google_world.to_csv(os.path.join(\"google_reports\", \"mobility_report_countries.csv\"), index=False)\n google_world.to_excel(os.path.join(\"google_reports\", \"mobility_report_countries.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n google_US.to_csv(os.path.join(\"google_reports\", \"mobility_report_US.csv\"), index=False)\n google_US.to_excel(os.path.join(\"google_reports\", \"mobility_report_US.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n google_brazil.to_csv(os.path.join(\"google_reports\", \"mobility_report_brazil.csv\"), index=False)\n google_brazil.to_excel(os.path.join(\"google_reports\", \"mobility_report_brazil.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n google_europe.to_csv(os.path.join(\"google_reports\", \"mobility_report_europe.csv\"), index=False)\n google_europe.to_excel(os.path.join(\"google_reports\", \"mobility_report_europe.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n google_asia_africa.to_csv(os.path.join(\"google_reports\", \"mobility_report_asia_africa.csv\"), index=False)\n google_asia_africa.to_excel(os.path.join(\"google_reports\", \"mobility_report_asia_africa.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n google_america_oceania.to_csv(os.path.join(\"google_reports\", \"mobility_report_america_oceania.csv\"), index=False)\n google_america_oceania.to_excel(os.path.join(\"google_reports\", \"mobility_report_america_oceania.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n # zip raw report\n with zp.ZipFile(GOOGLE_ZIP_PATH, 'w', zp.ZIP_DEFLATED) as zf:\n zf.write(GOOGLE_CSV_PATH,\"Global_Mobility_Report.csv\")\n# delete raw CSV report\nos.remove(GOOGLE_CSV_PATH)",
"_____no_output_____"
],
[
"# process Apple reports\n# download new report\nnew_files_status_apple = download_apple_report()\nif new_files_status_apple:\n # build reports\n apple_world = build_apple_report()\n apple_US = build_apple_report(report_type=\"US\")\n # write reports to CSV and Excel\n apple_world.to_csv(os.path.join(\"apple_reports\", \"apple_mobility_report.csv\"), index=False)\n apple_world.to_excel(os.path.join(\"apple_reports\", \"apple_mobility_report.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n apple_US.to_csv(os.path.join(\"apple_reports\", \"apple_mobility_report_US.csv\"), index=False)\n apple_US.to_excel(os.path.join(\"apple_reports\", \"apple_mobility_report_US.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')",
"_____no_output_____"
],
[
"# process Waze reports\n# download new report\nnew_files_status_waze = download_waze_reports()\nif new_files_status_waze:\n # build report\n waze = build_waze_report()\n # write report to CSV and Excel\n waze.to_csv(os.path.join(\"waze_reports\", \"waze_mobility.csv\"), index=False)\n waze.to_excel(os.path.join(\"waze_reports\", \"waze_mobility.xlsx\"),\n index=False, sheet_name='Data', engine='xlsxwriter')",
"_____no_output_____"
],
[
"# process TomTom reports\nnew_files_status_tomtom = check_tomtom_update()\nif new_files_status_tomtom:\n # scrape new data\n tomtom = download_tomtom_report()\n tomtom.to_csv(\n os.path.join(\"tomtom_reports\", \"tomtom_trafic_index.csv\"), index=False\n )\n tomtom.to_excel(\n os.path.join(\"tomtom_reports\", \"tomtom_trafic_index.xlsx\"),\n index=False,\n sheet_name=\"Data\",\n engine=\"xlsxwriter\",\n )",
"_____no_output_____"
],
[
"# build summary reports\nif new_files_status_apple or new_files_status_google:\n print(\"Merging reports...\")\n summary_regions = build_summary_report(os.path.join(\"apple_reports\",\"apple_mobility_report.csv\"),\n os.path.join(\"google_reports\", \"mobility_report_countries.csv\"))\n summary_US = build_summary_report(os.path.join(\"apple_reports\", \"apple_mobility_report_US.csv\"), \n os.path.join(\"google_reports\", \"mobility_report_US.csv\"), 'US')\n summary_countries = summary_regions[summary_regions['region']=='Total'].drop(columns=['region'])\n \n print('Writing merged reports to files...')\n summary_regions.to_csv(os.path.join(\"summary_reports\", \"summary_report_regions.csv\"), index=False)\n summary_regions.to_excel(os.path.join(\"summary_reports\", \"summary_report_regions.xlsx\"), \n index=False, sheet_name='Data', engine = 'xlsxwriter')\n summary_US.to_csv(os.path.join(\"summary_reports\", \"summary_report_US.csv\"), index=False)\n summary_US.to_excel(os.path.join(\"summary_reports\", \"summary_report_US.xlsx\"),\n index=False, sheet_name='Data', engine = 'xlsxwriter')\n summary_countries.to_csv(os.path.join(\"summary_reports\", \"summary_report_countries.csv\"), index=False)\n summary_countries.to_excel(os.path.join(\"summary_reports\", \"summary_report_countries.xlsx\"),\n index=False, sheet_name='Data', engine = 'xlsxwriter')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5e92f6963c3d9dd6f4751bf7084bb64dd3677a | 16,866 | ipynb | Jupyter Notebook | notebooks/TSP.ipynb | tail-island/pyqubo | 29974dbef0b14a4fcf26bc9b4669045eaf374186 | [
"Apache-2.0"
]
| 124 | 2018-09-21T06:50:17.000Z | 2022-03-16T07:56:49.000Z | notebooks/TSP.ipynb | tail-island/pyqubo | 29974dbef0b14a4fcf26bc9b4669045eaf374186 | [
"Apache-2.0"
]
| 107 | 2018-09-21T16:45:38.000Z | 2022-03-16T10:34:55.000Z | notebooks/TSP.ipynb | 29rou/pyqubo- | a71bcefd1a5fd3d989a38866c776ee0df4630cba | [
"Apache-2.0"
]
| 40 | 2018-09-21T19:51:55.000Z | 2022-03-22T20:45:31.000Z | 69.983402 | 7,420 | 0.802739 | [
[
[
"%matplotlib inline\nfrom pyqubo import Array, Placeholder, Constraint\nimport matplotlib.pyplot as plt\nimport networkx as nx\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## Traveling Salesman Problem (TSP)\n\nFind the shortest route that visits each city and returns to the origin city.",
"_____no_output_____"
]
],
[
[
"def plot_city(cities, sol=None):\n n_city = len(cities)\n cities_dict = dict(cities)\n G = nx.Graph()\n for city in cities_dict:\n G.add_node(city)\n \n # draw path\n if sol:\n city_order = []\n for i in range(n_city):\n for j in range(n_city):\n if sol.array('c', (i, j)) == 1:\n city_order.append(j)\n for i in range(n_city):\n city_index1 = city_order[i]\n city_index2 = city_order[(i+1) % n_city]\n G.add_edge(cities[city_index1][0], cities[city_index2][0])\n\n plt.figure(figsize=(3,3))\n pos = nx.spring_layout(G)\n nx.draw_networkx(G, cities_dict)\n plt.axis(\"off\")\n plt.show()\n\ndef dist(i, j, cities):\n pos_i = cities[i][1]\n pos_j = cities[j][1]\n return np.sqrt((pos_i[0] - pos_j[0])**2 + (pos_i[1] - pos_j[1])**2)",
"_____no_output_____"
],
[
"# City names and coordinates list[(\"name\", (x, y))]\ncities = [\n (\"a\", (0, 0)),\n (\"b\", (1, 3)),\n (\"c\", (3, 2)),\n (\"d\", (2, 1)),\n (\"e\", (0, 1))\n]\nplot_city(cities)",
"_____no_output_____"
]
],
[
[
"Prepare binary vector with bit $(i, j)$ representing to visit $j$ city at time $i$ ",
"_____no_output_____"
]
],
[
[
"n_city = len(cities)\nx = Array.create('c', (n_city, n_city), 'BINARY')",
"_____no_output_____"
],
[
"# Constraint not to visit more than two cities at the same time.\ntime_const = 0.0\nfor i in range(n_city):\n # If you wrap the hamiltonian by Const(...), this part is recognized as constraint\n time_const += Constraint((sum(x[i, j] for j in range(n_city)) - 1)**2, label=\"time{}\".format(i))\n\n# Constraint not to visit the same city more than twice.\ncity_const = 0.0\nfor j in range(n_city):\n city_const += Constraint((sum(x[i, j] for i in range(n_city)) - 1)**2, label=\"city{}\".format(j))",
"_____no_output_____"
],
[
"# distance of route\ndistance = 0.0\nfor i in range(n_city):\n for j in range(n_city):\n for k in range(n_city):\n d_ij = dist(i, j, cities)\n distance += d_ij * x[k, i] * x[(k+1)%n_city, j]",
"_____no_output_____"
],
[
"# Construct hamiltonian\nA = Placeholder(\"A\")\nH = distance + A * (time_const + city_const)",
"_____no_output_____"
],
[
"# Compile model\nmodel = H.compile()",
"_____no_output_____"
],
[
"# Generate QUBO\nfeed_dict = {'A': 4.0}\nbqm = model.to_bqm(feed_dict=feed_dict)",
"_____no_output_____"
],
[
"import neal\nsa = neal.SimulatedAnnealingSampler()\nsampleset = sa.sample(bqm, num_reads=100, num_sweeps=100)\n\n# Decode solution\ndecoded_samples = model.decode_sampleset(sampleset, feed_dict=feed_dict)\nbest_sample = min(decoded_samples, key=lambda x: x.energy)\nnum_broken = len(best_sample.constraints(only_broken=True))\nprint(\"number of broken constarint = {}\".format(num_broken))",
"number of broken constarint = 0\n"
],
[
"if num_broken == 0:\n plot_city(cities, best_sample)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5ea3439ba6785c066fe1090cb462b5f11a8386 | 108,054 | ipynb | Jupyter Notebook | notebooks/001-run-randomforest.ipynb | JiajunSong629/download-project | 83e31b3db18b83add4c7293a3375adb61b0c59c3 | [
"MIT"
]
| null | null | null | notebooks/001-run-randomforest.ipynb | JiajunSong629/download-project | 83e31b3db18b83add4c7293a3375adb61b0c59c3 | [
"MIT"
]
| null | null | null | notebooks/001-run-randomforest.ipynb | JiajunSong629/download-project | 83e31b3db18b83add4c7293a3375adb61b0c59c3 | [
"MIT"
]
| null | null | null | 188.575916 | 48,132 | 0.901614 | [
[
[
"import numpy as np\nimport pandas as pa\nimport matplotlib.pyplot as plt\nimport pickle\nfrom datetime import date\n\nfrom src.utils import get_framed_label, train_test_split\nfrom src.data import load_annotation\nfrom src.data import load_radar, load_water_distance, load_weight_sensor, load_audio\nfrom src import make_dataset\nimport config\n\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.metrics import classification_report, plot_roc_curve",
"_____no_output_____"
],
[
"def classification_result(model, testX, testY, threshold = 0.5):\n testYPredProb = model.predict_proba(testX)\n testYPred = (testYPredProb[:, 1] > threshold).astype(int)\n print (f\"threshold = {threshold}\", \"\\n\")\n print (classification_report(testY, testYPred))\n \ndef variable_importance(trainX, model, top=30):\n plt.figure(figsize=(20, 5))\n plt.bar(x = range(top), height = model.feature_importances_[:top])\n xticks_pos = np.arange(top)\n plt.xticks(xticks_pos, trainX.columns[:top], rotation=45, ha = 'right')\n pass",
"_____no_output_____"
]
],
[
[
"# Urination",
"_____no_output_____"
]
],
[
[
"CATEGORY = \"Urination\"\ndataset_config = config.DATASET_CONFIG[CATEGORY]\ncomplete_ids = load_annotation.get_complete_ids(\n category = CATEGORY\n)",
"_____no_output_____"
],
[
"selected_ids = complete_ids[:60]\nTRAIN_IDS, TEST_IDS = train_test_split(selected_ids, seed=1234)\n\nprint(f\"Category: {CATEGORY}\")\nprint(f\"Training {len(TRAIN_IDS)} use_ids: {TRAIN_IDS[:5]}...\")\nprint(f\"Testing {len(TEST_IDS)} use_ids: {TEST_IDS[:5]}...\")",
"Category: Urination\nTraining 48 use_ids: [1880, 1897, 1831, 1871, 1829]...\nTesting 12 use_ids: [1835, 1841, 1863, 1874, 1875]...\n"
],
[
"train_config = dataset_config.copy()\ntest_config = dataset_config.copy()\n\ntrain_config['USER_IDS'] = TRAIN_IDS\ntest_config['USER_IDS'] = TEST_IDS\n\ndataset = {}\ndataset['train'] = make_dataset.RandomForestExtended(train_config)\ndataset['test'] = make_dataset.RandomForestExtended(test_config)",
"_____no_output_____"
],
[
"# it may take around 20min to run\ntrain_x, train_y = dataset['train'].get_features_and_labels_from_users()\ntest_x, test_y = dataset['test'].get_features_and_labels_from_users()",
"_____no_output_____"
],
[
"print(f\"train_x.shape = {train_x.shape}, test_x.shape = {test_x.shape}\")\nprint(f\"#positive/#total train_y = {sum(train_y)}/{len(train_y)}\")\nprint(f\"#positive/#total test_y = {sum(test_y)}/{len(test_y)}\")",
"train_x.shape = (4292, 402), test_x.shape = (1376, 402)\n#positive/#total train_y = 708/4292\n#positive/#total test_y = 218/1376\n"
],
[
"rf = RandomForestClassifier(n_estimators = 30)\nrf.fit(train_x, train_y)",
"_____no_output_____"
],
[
"classification_result(\n rf,\n test_x, test_y,\n threshold = 0.3\n)",
"threshold = 0.3 \n\n precision recall f1-score support\n\n 0 0.96 0.99 0.97 1158\n 1 0.93 0.78 0.85 218\n\n accuracy 0.96 1376\n macro avg 0.95 0.89 0.91 1376\nweighted avg 0.96 0.96 0.96 1376\n\n"
],
[
"classification_result(\n rf,\n test_x, test_y,\n threshold = 0.2\n)",
"threshold = 0.2 \n\n precision recall f1-score support\n\n 0 0.97 0.97 0.97 1158\n 1 0.86 0.82 0.84 218\n\n accuracy 0.95 1376\n macro avg 0.91 0.90 0.90 1376\nweighted avg 0.95 0.95 0.95 1376\n\n"
],
[
"variable_importance(train_x, rf)",
"_____no_output_____"
],
[
"current_time = date.today().strftime(\"%Y-%m-%d\")\nmodel_name = f\"../models/urination-rf-extended-embedding-{current_time}.pkl\"\n\nwith open(model_name, \"wb\") as f:\n pickle.dump(rf, f)",
"_____no_output_____"
]
],
[
[
"# Defecation",
"_____no_output_____"
]
],
[
[
"CATEGORY = \"Defecation\"\ndataset_config = config.DATASET_CONFIG[CATEGORY]\ncomplete_ids = load_annotation.get_complete_ids(\n category = CATEGORY\n)",
"_____no_output_____"
],
[
"selected_ids = [idx for idx in complete_ids if idx <= 1950 and idx >= 1800]\nTRAIN_IDS, TEST_IDS = train_test_split(selected_ids)\n\nprint(f\"Category: {CATEGORY}\")\nprint(f\"Training {len(TRAIN_IDS)} use_ids: {TRAIN_IDS[:5]}...\")\nprint(f\"Testing {len(TEST_IDS)} use_ids: {TEST_IDS[:5]}...\")",
"Category: Defecation\nTraining 23 use_ids: [1898, 1930, 1919, 1926, 1941]...\nTesting 6 use_ids: [1854, 1870, 1875, 1882, 1890]...\n"
],
[
"train_config = dataset_config.copy()\ntest_config = dataset_config.copy()\n\ntrain_config['USER_IDS'] = TRAIN_IDS\ntest_config['USER_IDS'] = TEST_IDS\n\ndataset = {}\ndataset['train'] = make_dataset.RandomForestExtended(train_config)\ndataset['test'] = make_dataset.RandomForestExtended(test_config)",
"_____no_output_____"
],
[
"train_x, train_y = dataset['train'].get_features_and_labels_from_users()\ntest_x, test_y = dataset['test'].get_features_and_labels_from_users()",
"_____no_output_____"
],
[
"print(f'train_x.shape: {train_x.shape} test_x.shape: {test_x.shape}')\nprint(f'No. Positive in training {train_y.sum()}/{train_y.shape}')\nprint(f'No. Positive in testing {test_y.sum()}/{test_y.shape}')",
"train_x.shape: (3390, 402) test_x.shape: (1065, 402)\nNo. Positive in training 147/(3390,)\nNo. Positive in testing 37/(1065,)\n"
],
[
"rf = RandomForestClassifier(\n n_estimators = 10,\n class_weight = \"balanced\"\n)\nrf.fit(train_x, train_y)",
"_____no_output_____"
],
[
"classification_result(\n rf,\n test_x, test_y,\n threshold = 0.3\n)",
"threshold = 0.3 \n\n precision recall f1-score support\n\n 0 0.99 0.99 0.99 1028\n 1 0.62 0.65 0.63 37\n\n accuracy 0.97 1065\n macro avg 0.80 0.82 0.81 1065\nweighted avg 0.97 0.97 0.97 1065\n\n"
],
[
"classification_result(\n rf,\n test_x, test_y,\n threshold = 0.4\n)",
"threshold = 0.4 \n\n precision recall f1-score support\n\n 0 0.98 0.99 0.99 1028\n 1 0.61 0.54 0.57 37\n\n accuracy 0.97 1065\n macro avg 0.79 0.76 0.78 1065\nweighted avg 0.97 0.97 0.97 1065\n\n"
],
[
"variable_importance(train_x, rf)",
"_____no_output_____"
],
[
"current_time = date.today().strftime(\"%Y-%m-%d\")\nmodel_name = f\"../models/defecation-rf-extended-embedding-{current_time}.pkl\"\n\nwith open(model_name, \"wb\") as f:\n pickle.dump(rf, f)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5ea5e16b2f59d2e70bca6d484f9bf10f7959be | 4,266 | ipynb | Jupyter Notebook | extract-surface.ipynb | UttamBasu/ipygany_examples | 5b619951977a0e89f2c63a172d55a637519231d8 | [
"MIT"
]
| null | null | null | extract-surface.ipynb | UttamBasu/ipygany_examples | 5b619951977a0e89f2c63a172d55a637519231d8 | [
"MIT"
]
| null | null | null | extract-surface.ipynb | UttamBasu/ipygany_examples | 5b619951977a0e89f2c63a172d55a637519231d8 | [
"MIT"
]
| null | null | null | 47.4 | 1,621 | 0.5579 | [
[
[
"%matplotlib inline\nfrom pyvista import set_plot_theme\nset_plot_theme('document')",
"_____no_output_____"
]
],
[
[
"Extract Surface {#extract_surface_example}\n===============\n\nYou can extract the surface of nearly any object within `pyvista` using\nthe `extract_surface` filter.\n",
"_____no_output_____"
]
],
[
[
"# sphinx_gallery_thumbnail_number = 2\n\nimport numpy as np\nfrom vtk import VTK_QUADRATIC_HEXAHEDRON\n\nimport pyvista as pv",
"_____no_output_____"
]
],
[
[
"Create a quadratic cell and extract its surface\n===============================================\n\nHere we create a single quadratic hexahedral cell and then extract its\nsurface to demonstrate how to extract the surface of an\nUnstructuredGrid.\n",
"_____no_output_____"
]
],
[
[
"lin_pts = np.array([[-1, -1, -1], # point 0\n [ 1, -1, -1], # point 1\n [ 1, 1, -1], # point 2\n [-1, 1, -1], # point 3\n [-1, -1, 1], # point 4\n [ 1, -1, 1], # point 5\n [ 1, 1, 1], # point 6\n [-1, 1, 1]], np.double) # point 7\n\n# these are the \"midside\" points of a quad cell. See the definition of a\n# vtkQuadraticHexahedron at:\n# https://vtk.org/doc/nightly/html/classvtkQuadraticHexahedron.html\nquad_pts = np.array([\n (lin_pts[1] + lin_pts[0])/2, # between point 0 and 1\n (lin_pts[1] + lin_pts[2])/2, # between point 1 and 2\n (lin_pts[2] + lin_pts[3])/2, # and so on...\n (lin_pts[3] + lin_pts[0])/2,\n (lin_pts[4] + lin_pts[5])/2,\n (lin_pts[5] + lin_pts[6])/2,\n (lin_pts[6] + lin_pts[7])/2,\n (lin_pts[7] + lin_pts[4])/2,\n (lin_pts[0] + lin_pts[4])/2,\n (lin_pts[1] + lin_pts[5])/2,\n (lin_pts[2] + lin_pts[6])/2,\n (lin_pts[3] + lin_pts[7])/2])\n\n# introduce a minor variation to the location of the mid-side points\nquad_pts += np.random.random(quad_pts.shape)*0.3\npts = np.vstack((lin_pts, quad_pts))\n\n# create the grid\n\n# If you are using vtk>=9, you do not need the offset array\noffset = np.array([0])\ncells = np.hstack((20, np.arange(20))).astype(np.int64, copy=False)\ncelltypes = np.array([VTK_QUADRATIC_HEXAHEDRON])\ngrid = pv.UnstructuredGrid(offset, cells, celltypes, pts)\n\n# finally, extract the surface and plot it\nsurf = grid.extract_surface()\nsurf.plot(show_scalar_bar=False)",
"_____no_output_____"
]
],
[
[
"Nonlinear Surface Subdivision\n=============================\n\nShould your UnstructuredGrid contain quadratic cells, you can generate a\nsmooth surface based on the position of the \\\"mid-edge\\\" nodes. This\nallows the plotting of cells containing curvature. For additional\nreference, please see:\n<https://prod.sandia.gov/techlib-noauth/access-control.cgi/2004/041617.pdf>\n",
"_____no_output_____"
]
],
[
[
"surf_subdivided = grid.extract_surface(nonlinear_subdivision=5)\nsurf_subdivided.plot(show_scalar_bar=False)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec5eaa315f060464862261b98dff13dbf56f1de1 | 655,783 | ipynb | Jupyter Notebook | 1 - PCA.ipynb | wagglefoot/Target-Oxbridge-ML-workshop | d2b547169c45f985f8f6e501c837921950565dee | [
"CC0-1.0"
]
| null | null | null | 1 - PCA.ipynb | wagglefoot/Target-Oxbridge-ML-workshop | d2b547169c45f985f8f6e501c837921950565dee | [
"CC0-1.0"
]
| null | null | null | 1 - PCA.ipynb | wagglefoot/Target-Oxbridge-ML-workshop | d2b547169c45f985f8f6e501c837921950565dee | [
"CC0-1.0"
]
| null | null | null | 782.557279 | 139,827 | 0.941848 | [
[
[
"Generating Data\n===============\n\nWe first create 200 random two-dimensional data points.\nThe data points are sampled from a multinomial normal distribution.\n\nYou don't have to understand precisely how we do this. ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"Cov = np.array([[2.9, -2.2], [-2.2, 6.5]])\nX = np.random.multivariate_normal([1,2], Cov, size=200)",
"_____no_output_____"
]
],
[
[
"Let's have a look at the raw data first...",
"_____no_output_____"
]
],
[
[
"np.set_printoptions(4, suppress=True) # show only four decimals\nprint X[:10,:] # print the first 10 rows of X (from 0 to 9)",
"[[-1.8392 5.2679]\n [ 0.6284 3.0898]\n [-0.7239 3.1609]\n [ 1.5564 -2.3027]\n [ 0.7646 2.0468]\n [ 2.7401 2.4875]\n [ 2.1821 0.5568]\n [ 0.3088 -0.3838]\n [ 3.3336 -5.0069]\n [-1.2361 5.9021]]\n"
]
],
[
[
"Do you see a relationship between the two columns? Tricky, I'd say. However, since the data is two-dimensional, we can plot the data, which allows us to see the relationship.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(4,4))\nplt.scatter(X[:,0], X[:,1])\nplt.axis('equal') # equal scaling on both axis;\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can also have a look at the actual covariance matrix:",
"_____no_output_____"
]
],
[
[
"print np.cov(X,rowvar=False)",
"[[ 3.026 -2.3975]\n [-2.3975 8.3631]]\n"
]
],
[
[
"Running PCA\n===========\n\nWe would now like to analyze the directions in which the data varies most. For that, we \n\n1. place the point cloud in the center (0,0) and\n2. rotate it, such that the direction with most variance is parallel to the x-axis.\n\nBoth steps can be done using PCA, which is conveniently available in sklearn.\n\nWe start by loading the PCA class from the sklearn package and creating an instance of the class:",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\npca = PCA()",
"_____no_output_____"
]
],
[
[
"Now, `pca` is an object which has a function `pca.fit_transform(x)` which performs both steps from above to its argument `x`, and returns the centered and rotated version of `x`.",
"_____no_output_____"
]
],
[
[
"X_pca = pca.fit_transform(X)",
"_____no_output_____"
],
[
"pca.components_",
"_____no_output_____"
],
[
"pca.mean_",
"_____no_output_____"
],
[
"plt.figure(figsize=(4,4))\nplt.scatter(X_pca[:,0], X_pca[:,1])\nplt.axis('equal');",
"_____no_output_____"
]
],
[
[
"The covariances between different axes should be zero now. We can double-check by having a look at the non-diagonal entries of the covariance matrix:",
"_____no_output_____"
]
],
[
[
"print np.cov(X_pca, rowvar=False)",
"[[ 7.8372 -0. ]\n [-0. 1.7368]]\n"
]
],
[
[
"High-Dimensional Data\n=====================\n\nOur small example above was very easy, since we could get insight into the data by simply plotting it. This approach will not work once you have more than 3 dimensions, let's say we have the same data, but it is represented in four dimensions:",
"_____no_output_____"
]
],
[
[
"np.random.seed(1)\nX_HD = np.dot(X,np.random.uniform(0.2,3,(2,4))*(np.random.randint(0,2,(2,4))*2-1))",
"_____no_output_____"
]
],
[
[
"Lets look at the data again. First, the raw data:",
"_____no_output_____"
]
],
[
[
"print X_HD[:10]",
"[[-3.2623 4.9526 -0.8765 -2.9379]\n [-3.8924 6.2074 -0.6913 -3.1126]\n [-4.1499 5.8243 -1.6806 -4.3638]\n [-4.0622 3.9863 -3.6835 -6.5295]\n [ 0.5888 1.2708 2.7312 3.3803]\n [-1.9775 3.8727 0.5035 -0.6347]\n [-1.8163 1.7643 -1.6684 -2.9432]\n [-5.396 7.9641 -1.7204 -5.1592]\n [-0.0199 -2.6655 -3.2097 -3.5673]\n [-1.9595 3.206 -0.2517 -1.4603]]\n"
]
],
[
[
"That one is more tricky. See anything? We can also try plot a few two-dimensional projections:",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(8,8))\nfor i in xrange(4):\n for j in xrange(4):\n plt.subplot(4, 4, i * 4 + j + 1)\n plt.scatter(X_HD[:,i], X_HD[:,j])\n plt.axis('equal')\n plt.gca().set_aspect('equal')",
"_____no_output_____"
]
],
[
[
"It is not easy to see that this is still a two-dimensional dataset! \n\nHowever, if we now do PCA on it, you'll see that the last two dimensions do not matter at all:",
"_____no_output_____"
]
],
[
[
"X_HE = pca.fit_transform(X_HD)\nprint X_HE[:10,:]",
"[[-1.4996 1.3909 -0. 0. ]\n [-2.8628 1.813 -0. -0. ]\n [-3.161 0.1794 0. 0. ]\n [-2.4901 -3.232 0. -0. ]\n [ 5.679 6.8489 -0. -0. ]\n [ 0.8523 3.5109 0. 0. ]\n [ 1.718 -0.1997 -0. 0. ]\n [-5.7579 0.3254 0. 0. ]\n [ 5.8509 -3.1211 0. 0. ]\n [ 1.0737 2.2273 0. 0. ]]\n"
]
],
[
[
"Here it is easy to see, that the data is **still only two-dimensional**. Let's plot the two dimensions.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(4,4))\nplt.scatter(X_HE[:,0], X_HE[:,1])\nplt.axis('equal')\nplt.gca().set_aspect('equal')",
"_____no_output_____"
]
],
[
[
"Why does the result look differently than the two-dimensional data from which we generated it?",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(8,8))\nfor i in xrange(4):\n for j in xrange(4):\n plt.subplot(4, 4, i * 4 + j + 1)\n plt.scatter(X_HE[:,i], X_HE[:,j])\n plt.gca().set_xlim(-40,40)\n plt.gca().set_ylim(-40,40)\n plt.axis('equal')\n plt.gca().set_aspect('equal')",
"_____no_output_____"
]
],
[
[
"Dimension Reduction with PCA\n============================\n\nWe can see that there are actually only two dimensions in the dataset. \n\nLet's throw away even more data -- the second dimension -- and reconstruct the original data in `D`.",
"_____no_output_____"
]
],
[
[
"pca = PCA(1) # only keep one dimension!\nX_E = pca.fit_transform(X_HD)\nprint X_E[:10,:]",
"[[-1.4996]\n [-2.8628]\n [-3.161 ]\n [-2.4901]\n [ 5.679 ]\n [ 0.8523]\n [ 1.718 ]\n [-5.7579]\n [ 5.8509]\n [ 1.0737]]\n"
]
],
[
[
"Now lets plot the reconstructed data and compare to the original data D. We plot the original data in red, and the reconstruction with only one dimension in blue:",
"_____no_output_____"
]
],
[
[
"X_reconstructed = pca.inverse_transform(X_E)\nplt.figure(figsize=(8,8))\nfor i in xrange(4):\n for j in xrange(4):\n plt.subplot(4, 4, i * 4 + j + 1)\n plt.scatter(X_HD[:,i], X_HD[:,j],c='r')\n plt.scatter(X_reconstructed[:,i], X_reconstructed[:,j],c='b')\n plt.axis('equal')",
"_____no_output_____"
]
],
[
[
"## PCA on Images",
"_____no_output_____"
],
[
"In this final example, we use the $k$-Means algorithm on the classical MNIST dataset.\n\nThe MNIST dataset contains images of hand-written digits. \n\nLet's first fetch the dataset from the internet (which may take a while, note the asterisk [*]):",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import fetch_mldata\nfrom sklearn.cluster import KMeans\nfrom sklearn.utils import shuffle\nX_digits, _,_, Y_digits = fetch_mldata(\"MNIST Original\").values() # fetch dataset from internet\nX_digits, Y_digits = shuffle(X_digits,Y_digits) # shuffle dataset (which is ordered!)\nX_digits = X_digits[-5000:] # take only the last instances, to shorten runtime of PCA",
"_____no_output_____"
]
],
[
[
"Let's have a look at some of the instances in the dataset we just loaded:",
"_____no_output_____"
]
],
[
[
"plt.rc(\"image\", cmap=\"binary\")\nplt.figure(figsize=(8,4))\nfor i in xrange(10):\n plt.subplot(2,5,i+1)\n plt.imshow(X_digits[i].reshape(28,28))\n plt.xticks(())\n plt.yticks(())\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"**Warning**: This takes quite a few seconds, so be patient until the asterisk [*] disappears!",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\npca = PCA()\nX2_digits = pca.fit_transform(X_digits)",
"_____no_output_____"
]
],
[
[
"It does not make much sense to look at the transformed images, they will look like noise to us. \n\nInstead, let's have a look at the most important directions on which the dataset was projected:",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(8,6))\nW = pca.components_\n\nfor i in xrange(10): # loop over all means\n plt.subplot(2,5,i+1)\n plt.imshow(W[i].reshape(28,28))\n plt.xticks(())\n plt.yticks(())\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"The later directions (here, from the 100-th on) mainly show noise, small variations between different, but very similar training instances:",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(8,4))\nW = pca.components_\n\nfor i in xrange(10): # loop over all means\n plt.subplot(2,5,i+1)\n plt.imshow(W[100+i].reshape(28,28))\n plt.xticks(())\n plt.yticks(())\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"The number of \"interesting\" dimensions can be seen from the importance of the found directions. \n\nWe can simply plot them:",
"_____no_output_____"
]
],
[
[
"plt.plot(pca.explained_variance_);",
"_____no_output_____"
]
],
[
[
"We can see that the intrinsic dimensionality is not higher than maybe 100, even though the dataset has 784 dimensions!",
"_____no_output_____"
],
[
"Let's reconstruct the data again using only a handfull of the 784 dimensions:",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\npca = PCA(20)\nX2_few_digits = pca.fit_transform(X_digits)",
"_____no_output_____"
],
[
"plt.figure(figsize=(16,4))\nX_recons_digits = pca.inverse_transform(X2_few_digits)\nfor i in xrange(10):\n plt.subplot(2,10,i+1)\n plt.imshow(X_recons_digits[i].reshape(28,28))\n plt.xticks(())\n plt.yticks(())\nfor i in xrange(10):\n plt.subplot(2,10,11+i)\n plt.imshow(X_digits[i].reshape(28,28))\n plt.xticks(())\n plt.yticks(())\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"# Playing around with this Notebook",
"_____no_output_____"
],
[
"- What happens, when you multiply one of the data axis with a large (or small) number? \n\n e.g. using X[:,0] *= 100\n\n Does the result stay the same? Why/why not?\n\n-----\n\n- Try to explore the iris dataset below using PCA. \n\n What happens if you visualize the *last* components of PCA instead of the first ones?\n\n-----\n\n- Use PCA with 1, 2 or 3 axes and reconstruct the Iris data from each. How do the results change? Show plots!",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets\n_,data,target,_,_ = datasets.load_iris().values()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec5eb07c5cb448b5ce51d73ac36a783c46b27bef | 3,594 | ipynb | Jupyter Notebook | script/autotuning-compiler-flags-linux/start_pipeline_in_ipython_notebook.ipynb | ctuning/reproduce-pamela-project | de6509eb3ec6613c513a5176234af66c3c5ae20d | [
"BSD-3-Clause"
]
| 25 | 2015-12-30T11:26:22.000Z | 2019-05-17T21:10:15.000Z | script/autotuning-compiler-flags-linux/start_pipeline_in_ipython_notebook.ipynb | ctuning/reproduce-pamela-project | de6509eb3ec6613c513a5176234af66c3c5ae20d | [
"BSD-3-Clause"
]
| null | null | null | script/autotuning-compiler-flags-linux/start_pipeline_in_ipython_notebook.ipynb | ctuning/reproduce-pamela-project | de6509eb3ec6613c513a5176234af66c3c5ae20d | [
"BSD-3-Clause"
]
| null | null | null | 19.966667 | 85 | 0.484975 | [
[
[
"# Prepared by Grigori Fursin to demo CK",
"_____no_output_____"
],
[
"import ck.kernel as ck",
"_____no_output_____"
],
[
"print (ck.version({}))",
""
],
[
"# run from CMD\n# _clean_program_pipeline.bat\n# _clean_program_pipeline.bat\n# to clean and set up autotuning pipeline",
"_____no_output_____"
],
[
"# Turn off adaptive CPU frequency governor, \n# otherwise our pipeline will detect frequency change and will stop )",
"_____no_output_____"
],
[
"ii={'action':'run',\n 'module_uoa':'pipeline',\n 'data_uoa':'program',\n# 'save_to_file':'xyz.json',\n 'pipeline_from_file':'_setup_program_pipeline_tmp.json'}\nr=ck.access(ii)\nif r['return']>0:\n ck.err(r)",
""
],
[
"# check if pipeline failed\nlast_iteration_output=r['last_iteration_output']\nif last_iteration_output.get('fail','')=='yes':\n print ('Pipeline failed: '+last_iteration_output.get('fail_reason',''))\n exit(1)",
""
],
[
"# Get experiment desc\nexperiment_desc=r['experiment_desc']",
"_____no_output_____"
],
[
"# Get stat analysis and print available flat keys\nstat_analysis=r['last_stat_analysis']\ndict_flat=stat_analysis.get('dict_flat',{})\nfor k in dict_flat:\n print (k)",
""
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5eb663772e9b87aca5293354bf3239b4d06fcb | 529,203 | ipynb | Jupyter Notebook | Time Series Modelling/1.Forecasting_Solar_Irradiance_Model_VF.ipynb | eljimenezj/Team_51_DS4A_2021 | 6f8e1fca0962e1698e4b533fee6eabd36abea1cf | [
"MIT"
]
| null | null | null | Time Series Modelling/1.Forecasting_Solar_Irradiance_Model_VF.ipynb | eljimenezj/Team_51_DS4A_2021 | 6f8e1fca0962e1698e4b533fee6eabd36abea1cf | [
"MIT"
]
| null | null | null | Time Series Modelling/1.Forecasting_Solar_Irradiance_Model_VF.ipynb | eljimenezj/Team_51_DS4A_2021 | 6f8e1fca0962e1698e4b533fee6eabd36abea1cf | [
"MIT"
]
| null | null | null | 240.328338 | 133,784 | 0.894375 | [
[
[
"# Solar irradiance time series modeling and forecasting ",
"_____no_output_____"
],
[
"This notebook begins with the study of time series and then introduces the application of different forecasting techniques to obtain future values of solar irradiance in the different cities addressed in this project, which are mainly located in the Colombian Caribbean area.\n\nThis notebook is elaborated by Team 51 which is formed by:\n\n* Laura Milena Manrique\n* Edgar Leandro Jimenez \n* Juan Manuel Muskus \n* Arturo Quevedo\n* William Prieto\n* Juan Felipe Múnera\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport seaborn as sns\nimport numpy as np\nfrom prophet import Prophet\n\nfrom sqlalchemy import create_engine\n\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.linear_model import Lasso\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.metrics import mean_squared_error\n\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom skforecast.ForecasterAutoregCustom import ForecasterAutoregCustom\nfrom skforecast.ForecasterAutoregMultiOutput import ForecasterAutoregMultiOutput\nfrom skforecast.model_selection import grid_search_forecaster\nfrom skforecast.model_selection import time_series_spliter\nfrom skforecast.model_selection import cv_forecaster\nfrom skforecast.model_selection import backtesting_forecaster\nfrom skforecast.model_selection import backtesting_forecaster_intervals\n\nfrom sklearn.experimental import enable_hist_gradient_boosting # noqa\nfrom sklearn.ensemble import HistGradientBoostingRegressor\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.neural_network import MLPRegressor\nfrom sklearn.model_selection import train_test_split\n\nimport datetime\nfrom datetime import date\nimport joblib\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"## Load the data\n\nThe data provided by Dynamic Defense Solution was stored in a relational database that in Digital Ocean with the Postgres engine.\n\nThe following is the connection of Juyter Notebook to the instance in Digital Ocean where we have our data and application stored",
"_____no_output_____"
]
],
[
[
"# 1. Server Option \nhost = \"143.198.143.161\"\nport = \"5432\"\nuser = \"postgres\"\ndatabase = \"proyecto\"\npassword = \"sahagun21\"\n\nconnDB = create_engine(f'postgresql://{user}:{password}@{host}:{port}/{database}')\nconn = connDB.raw_connection()",
"_____no_output_____"
],
[
"%reload_ext sql\n%sql postgresql://postgres:[email protected]/proyecto",
"_____no_output_____"
],
[
"df = %sql select a.*, b.name, b.dpto, b.lat, b.lon, b.time_zone,b.elevation, b.local_time_zone, b.population from weather_sub as a , cities as b where a.id = b.id and a.id in ('1285504','1155322','1210398','1259275','1249863','1207119','1183916','1216586','1234322','1193453','1197836','1166758','1202369','1151781','1245138','1216434','1241377','1271225') ;\ndf = df.DataFrame()",
" * postgresql://postgres:***@143.198.143.161/proyecto\n6942624 rows affected.\n"
],
[
"# 2. CSV option --> used throughout the project locally as it is quicker to load \ndf = pd.read_csv('wheater_costa_vf.csv', sep = ',')",
"_____no_output_____"
]
],
[
[
"## Basic exploration and transformacion of dataset",
"_____no_output_____"
]
],
[
[
"## 1. Exploration\ndf.head(2)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.groupby('name')[['temperature','relative_humidity']].mean().reset_index().to_csv('cities_prom.csv',index=False)",
"_____no_output_____"
],
[
"df.groupby(['name','year'])['wind_speed'].max().reset_index()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"# Transformation\n\ndf['month'] = df['month'].map('{:0>2}'.format)\ndf['hour'] = df['hour'].map('{:0>2}'.format)\ndf['Date'] = pd.to_datetime(df[['year', 'month', 'day','hour']], \n errors='coerce')\n\n# Drop \ndf = df[df['minute']!=30]",
"_____no_output_____"
],
[
"# Boxplot DHI \nsns.boxplot(x=\"hour\", y=\"DHI\", data=df)",
"_____no_output_____"
],
[
"# Boxplot Win Spped \nsns.boxplot(x=\"hour\", y=\"wind_speed\", data=df)",
"_____no_output_____"
]
],
[
[
"## Data splitting by different cities",
"_____no_output_____"
],
[
"From the main dataframe we make a partition by city and obtain their respective data, then we store it in a dictionary where the key is the name of the city we are going to work with.",
"_____no_output_____"
]
],
[
[
"cities = {}\nnames = df['name'].unique()\n\nfor city in names:\n cities[city] = df[df['name']==city].groupby('Date')[['DHI','wind_speed']].mean().reset_index()",
"_____no_output_____"
],
[
"cities.keys()",
"_____no_output_____"
],
[
"cities['SAHAGÚN'].head(2)",
"_____no_output_____"
]
],
[
[
"## Modeling and forecasting\n",
"_____no_output_____"
],
[
"### Time Series modeling and Forecasting Using Prophet Library ",
"_____no_output_____"
]
],
[
[
"# Take cartegena data (Example)\ndf_cartegena = cities['CARTAGENA'].copy()\ndf_cartegena = df_cartegena[['Date', 'DHI']]\ndf_cartegena.columns = ['ds', 'y']\ndf_cartegena = df_cartegena[(df_cartegena['ds'].dt.hour>=6) & (df_cartegena['ds'].dt.hour<=18)]",
"_____no_output_____"
],
[
"# fit model\nm = Prophet().fit(df_cartegena)\nfuture = m.make_future_dataframe(periods=100, freq='H')\nfcst = m.predict(future)\nfig = m.plot(fcst)",
"_____no_output_____"
],
[
"# Lets check the descomposition\nfig = m.plot_components(fcst)",
"_____no_output_____"
]
],
[
[
"This model was tested for other cities and did not obtain good results, so no further modeling is presented. We will continue to explore other techniques",
"_____no_output_____"
],
[
"## Time series forecasting using recursive autoregressive modeling ",
"_____no_output_____"
]
],
[
[
"# Continue with Cartegena city\ndf_cartegena2 = df_cartegena.copy()\ndf_cartegena2 = df_cartegena2.set_index('ds')\ndf_cartegena2 = df_cartegena2.rename(columns={'x': 'y'})\ndf_cartegena2 = df_cartegena2.asfreq('H')\ndf_cartegena2 = df_cartegena2['y']\ndf_cartegena2 = df_cartegena2.sort_index()",
"_____no_output_____"
],
[
"# ==============================================================================\n(df_cartegena2.index == pd.date_range(start=df_cartegena2.index.min(),\n end=df_cartegena2.index.max(),\n freq=df_cartegena2.index.freq)).all()",
"_____no_output_____"
],
[
"# train-test split - 3 moths aprox\n# ==============================================================================\nsteps = 1170 \ndatos_train = df_cartegena2[:-steps]\ndatos_test = df_cartegena2[-steps:]\n\ndatos_train = datos_train.dropna()\ndatos_test = datos_test.dropna()\n\nfig, ax=plt.subplots(figsize=(9, 4))\ndatos_train.plot(ax=ax, label='train')\ndatos_test.plot(ax=ax, label='test')\nax.legend();",
"_____no_output_____"
],
[
"# Modelling \n# ==============================================================================\nforecaster_rf = ForecasterAutoreg(\n regressor=RandomForestRegressor(n_estimators=500,\n max_depth=10,\n random_state=123),\n lags=12\n )\n\nforecaster_rf.fit(y=datos_train)\n\nforecaster_rf",
"_____no_output_____"
],
[
"# Forecasting\n# ==============================================================================\nsteps = len(datos_test.index) \npredicciones = forecaster_rf.predict(steps=steps)\n\n# The time index is added to the predictions\npredicciones = pd.Series(data=predicciones, index=datos_test.index)\npredicciones.head()",
"_____no_output_____"
],
[
"# Plot\n# ==============================================================================\nfig, ax = plt.subplots(figsize=(9, 4))\ndatos_train.plot(ax=ax, label='train')\ndatos_test.plot(ax=ax, label='test')\npredicciones.plot(ax=ax, label='Predictions')\nax.legend();",
"_____no_output_____"
],
[
"# MSE Error\n# ==============================================================================\nerror_mse = mean_squared_error(\n y_true = np.nan_to_num(datos_test),\n y_pred = np.nan_to_num(predicciones)\n )\nprint(f\"Error de test (mse): {error_mse}\")",
"Error de test (mse): 15325.693476281564\n"
]
],
[
[
"### Hyperparameter adjustment ",
"_____no_output_____"
]
],
[
[
"# Grid search de hiperparámetros\n# ==============================================================================\nforecaster_rf = ForecasterAutoreg(\n regressor = RandomForestRegressor(random_state=123),\n lags = 12 \n )\n\n# Hyperparameters of the regressor\nparam_grid = {'n_estimators': [100, 200,500],\n 'max_depth': [3,5, 10]}\n\n# Lags \nlags_grid = [5, 20]\n\nresultados_grid = grid_search_forecaster(\n forecaster = forecaster_rf,\n y = datos_train,\n param_grid = param_grid,\n lags_grid = lags_grid,\n steps = 10,\n method = 'cv',\n metric = 'mean_squared_error',\n initial_train_size = int(len(datos_train)*0.5),\n allow_incomplete_fold = False,\n return_best = True,\n verbose = False\n )",
"2021-08-21 19:51:39,652 root INFO Number of models to fit: 18\nloop lags_grid: 0%| | 0/2 [00:00<?, ?it/s]\nloop param_grid: 0%| | 0/9 [00:00<?, ?it/s]\u001b[A\nloop param_grid: 11%|███████▎ | 1/9 [06:12<49:39, 372.45s/it]\u001b[A\nloop param_grid: 22%|██████████████▏ | 2/9 [18:08<1:07:03, 574.75s/it]\u001b[A\nloop param_grid: 33%|█████████████████████ | 3/9 [47:51<1:52:38, 1126.44s/it]\u001b[A\nloop param_grid: 44%|████████████████████████████▍ | 4/9 [56:52<1:14:36, 895.38s/it]\u001b[A\nloop param_grid: 56%|██████████████████████████████████▍ | 5/9 [1:14:23<1:03:25, 951.39s/it]\u001b[A\nloop param_grid: 67%|████████████████████████████████████████▋ | 6/9 [1:57:32<1:15:24, 1508.01s/it]\u001b[A\nloop param_grid: 78%|█████████████████████████████████████████████████ | 7/9 [2:12:59<43:56, 1318.05s/it]\u001b[A\nloop param_grid: 89%|████████████████████████████████████████████████████████ | 8/9 [2:43:33<24:42, 1482.40s/it]\u001b[A\nloop param_grid: 100%|███████████████████████████████████████████████████████████████| 9/9 [3:59:01<00:00, 2434.39s/it]\u001b[A\nloop lags_grid: 50%|██████████████████████████████▌ | 1/2 [3:59:01<3:59:01, 14341.03s/it]\u001b[A\nloop param_grid: 0%| | 0/9 [00:00<?, ?it/s]\u001b[A\nloop param_grid: 11%|███████ | 1/9 [17:45<2:22:02, 1065.30s/it]\u001b[A\nloop param_grid: 22%|██████████████ | 2/9 [52:50<3:15:38, 1676.97s/it]\u001b[A\nloop param_grid: 33%|████████████████████▎ | 3/9 [2:19:45<5:29:15, 3292.58s/it]\u001b[A\nloop param_grid: 44%|███████████████████████████ | 4/9 [2:48:07<3:42:03, 2664.65s/it]\u001b[A\nloop param_grid: 56%|█████████████████████████████████▉ | 5/9 [3:44:04<3:14:16, 2914.19s/it]\u001b[A\nloop param_grid: 67%|████████████████████████████████████████▋ | 6/9 [6:04:02<3:58:55, 4778.60s/it]\u001b[A\nloop param_grid: 78%|███████████████████████████████████████████████▍ | 7/9 [6:56:38<2:21:36, 4248.21s/it]\u001b[A\nloop param_grid: 89%|██████████████████████████████████████████████████████▏ | 8/9 [8:41:07<1:21:31, 4891.52s/it]\u001b[A\nloop param_grid: 100%|██████████████████████████████████████████████████████████████| 9/9 [13:02:19<00:00, 8261.81s/it]\u001b[A\nloop lags_grid: 100%|██████████████████████████████████████████████████████████████| 2/2 [17:01:20<00:00, 30640.36s/it]\u001b[A\n2021-08-22 12:53:00,378 root INFO Refitting `forecaster` using the best found parameters and the whole data set: \nlags: [ 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20] \nparams: {'max_depth': 10, 'n_estimators': 500}\n\n"
]
],
[
[
"### Direct multi-step forecasting using Lasso regression",
"_____no_output_____"
]
],
[
[
"# ==============================================================================\n\n\nforecaster_rf = ForecasterAutoregMultiOutput(\n regressor = Lasso(random_state=123, alpha=599.4842503189421),\n steps = len(datos_test.index),\n lags = 20\n )\n\n\nforecaster_rf.fit(y=datos_train)\n\nforecaster_rf\n",
"_____no_output_____"
],
[
"# Forecast\n# ==============================================================================\nsteps = len(datos_test.index) \npredicciones = forecaster_rf.predict(steps=steps)\npredicciones = pd.Series(data=predicciones, index=datos_test.index)\npredicciones.head()",
"_____no_output_____"
],
[
"# Plot\n# ==============================================================================\nfig, ax = plt.subplots(figsize=(9, 4))\ndatos_train.plot(ax=ax, label='train')\ndatos_test.plot(ax=ax, label='test')\npredicciones.plot(ax=ax, label='predicciones')\nax.legend();",
"_____no_output_____"
],
[
"# MSE Error\n# ==============================================================================\nerror_mse = mean_squared_error(y_true = datos_test, y_pred = predicciones)\nprint(f\"Error de test (mse) {error_mse}\")",
"Error de test (mse) 5813.363829269499\n"
],
[
"forecaster_rf = ForecasterAutoregMultiOutput(\n regressor = Lasso(random_state=123),\n steps = len(datos_test.index),\n lags = 12 # Este valor será remplazado en el grid search\n )\n\nparam_grid = {'alpha': np.logspace(-5, 5, 10)}\n\nlags_grid = [5, 12, 20]\n\nresultados_grid = grid_search_forecaster(\n forecaster = forecaster_rf,\n y = datos_train,\n param_grid = param_grid,\n lags_grid = lags_grid,\n steps = len(datos_test.index),\n method = 'cv',\n metric = 'mean_squared_error',\n initial_train_size = int(len(datos_train)*0.5),\n allow_incomplete_fold = False,\n return_best = True,\n verbose = False\n )",
"2021-08-22 12:53:49,891 root INFO Number of models to fit: 30\nloop lags_grid: 0%| | 0/3 [00:00<?, ?it/s]\nloop param_grid: 0%| | 0/10 [00:00<?, ?it/s]\u001b[A\nloop param_grid: 10%|██████▌ | 1/10 [00:37<05:37, 37.45s/it]\u001b[A\nloop param_grid: 20%|█████████████▏ | 2/10 [01:11<04:41, 35.18s/it]\u001b[A\nloop param_grid: 30%|███████████████████▊ | 3/10 [01:39<03:45, 32.23s/it]\u001b[A\nloop param_grid: 40%|██████████████████████████▍ | 4/10 [02:02<02:50, 28.34s/it]\u001b[A\nloop param_grid: 50%|█████████████████████████████████ | 5/10 [02:20<02:04, 24.90s/it]\u001b[A\nloop param_grid: 60%|███████████████████████████████████████▌ | 6/10 [02:37<01:27, 21.99s/it]\u001b[A\nloop param_grid: 70%|██████████████████████████████████████████████▏ | 7/10 [02:50<00:57, 19.25s/it]\u001b[A\nloop param_grid: 80%|████████████████████████████████████████████████████▊ | 8/10 [03:01<00:33, 16.53s/it]\u001b[A\nloop param_grid: 90%|███████████████████████████████████████████████████████████▍ | 9/10 [03:10<00:14, 14.04s/it]\u001b[A\nloop param_grid: 100%|█████████████████████████████████████████████████████████████████| 10/10 [03:18<00:00, 12.17s/it]\u001b[A\nloop lags_grid: 33%|██████████████████████▎ | 1/3 [03:18<06:36, 198.15s/it]\u001b[A\nloop param_grid: 0%| | 0/10 [00:00<?, ?it/s]\u001b[A\nloop param_grid: 10%|██████▌ | 1/10 [01:25<12:45, 85.04s/it]\u001b[A\nloop param_grid: 20%|█████████████▏ | 2/10 [02:39<10:32, 79.03s/it]\u001b[A\nloop param_grid: 30%|███████████████████▊ | 3/10 [03:43<08:25, 72.19s/it]\u001b[A\nloop param_grid: 40%|██████████████████████████▍ | 4/10 [04:37<06:29, 64.99s/it]\u001b[A\nloop param_grid: 50%|█████████████████████████████████ | 5/10 [05:19<04:43, 56.61s/it]\u001b[A\nloop param_grid: 60%|███████████████████████████████████████▌ | 6/10 [05:52<03:13, 48.49s/it]\u001b[A\nloop param_grid: 70%|██████████████████████████████████████████████▏ | 7/10 [06:15<02:00, 40.13s/it]\u001b[A\nloop param_grid: 80%|████████████████████████████████████████████████████▊ | 8/10 [06:33<01:06, 33.02s/it]\u001b[A\nloop param_grid: 90%|███████████████████████████████████████████████████████████▍ | 9/10 [06:43<00:25, 25.81s/it]\u001b[A\nloop param_grid: 100%|█████████████████████████████████████████████████████████████████| 10/10 [06:52<00:00, 20.74s/it]\u001b[A\nloop lags_grid: 67%|████████████████████████████████████████████▋ | 2/3 [10:10<05:24, 324.19s/it]\u001b[A\nloop param_grid: 0%| | 0/10 [00:00<?, ?it/s]\u001b[A\nloop param_grid: 10%|██████▌ | 1/10 [02:54<26:14, 174.94s/it]\u001b[A\nloop param_grid: 20%|█████████████ | 2/10 [05:26<21:27, 160.96s/it]\u001b[A\nloop param_grid: 30%|███████████████████▌ | 3/10 [07:34<17:02, 146.00s/it]\u001b[A\nloop param_grid: 40%|██████████████████████████ | 4/10 [09:24<13:11, 131.88s/it]\u001b[A\nloop param_grid: 50%|████████████████████████████████▌ | 5/10 [10:55<09:45, 117.02s/it]\u001b[A\nloop param_grid: 60%|███████████████████████████████████████▌ | 6/10 [12:00<06:37, 99.28s/it]\u001b[A\nloop param_grid: 70%|██████████████████████████████████████████████▏ | 7/10 [12:42<04:02, 80.70s/it]\u001b[A\nloop param_grid: 80%|████████████████████████████████████████████████████▊ | 8/10 [13:10<02:07, 63.78s/it]\u001b[A\nloop param_grid: 90%|███████████████████████████████████████████████████████████▍ | 9/10 [13:25<00:48, 48.81s/it]\u001b[A\nloop param_grid: 100%|█████████████████████████████████████████████████████████████████| 10/10 [13:39<00:00, 37.98s/it]\u001b[A\nloop lags_grid: 100%|███████████████████████████████████████████████████████████████████| 3/3 [23:50<00:00, 476.75s/it]\u001b[A\n2021-08-22 13:17:40,144 root INFO Refitting `forecaster` using the best found parameters and the whole data set: \nlags: [ 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20] \nparams: {'alpha': 599.4842503189421}\n\n"
]
],
[
[
"### Time series forecasting using Gradient Boosting Regressor ",
"_____no_output_____"
]
],
[
[
"# For this time lets check model with sahagun City\ndf2 = cities['SAHAGÚN'].copy()\ndf2 = df2[['Date', 'DHI']]\ndf2.columns = ['ds', 'y']\ndf2 = df2[(df2['ds'].dt.hour>=6) & (df2['ds'].dt.hour<=18)]\n#df2 = df2[df2['ds'].dt.year>1998] \n\n# Lag for the time: day, week, month, quarter, semester, annual\nserie =pd.concat([df2,df2.y.shift(91),df2.y.shift(104),df2.y.shift(117),df2.y.shift(130),df2.y.shift(143)\n ,df2.y.shift(156),df2.y.shift(169),df2.y.shift(182),df2.y.shift(390)\n ,df2.y.shift(403),df2.y.shift(1170), df2.y.shift(1183),df2.y.shift(1196)\n ,df2.y.shift(1209),df2.y.shift(2340), df2.y.shift(2353), df2.y.shift(2366)\n ,df2.y.shift(2379),df2.y.shift(3900),df2.y.shift(4745)],\n axis=1)\n\n# Columns \ncolumns_name = ['Date','y','t_7',\n 't_8','t_9','t_10','t_11','t_12','t_13','t_14',\n 't_30','t_31','t_90','t_91','t_92','t_93','t_180'\n ,'t_181','t_182','t_183','t_300','t_365']\n\nserie.columns = columns_name\n\n# Drops na\nserie = serie.dropna()",
"_____no_output_____"
],
[
"X = serie.drop(['Date', 'y'], axis=1)\ny = serie['y']\n\n# Train test split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.1, random_state = 42,shuffle=False)",
"_____no_output_____"
],
[
"est = HistGradientBoostingRegressor()\nest.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# Plotting\npred_gb = est.predict(X_test)\nforecast_gb = pd.DataFrame(y_test).reset_index()\nforecast_gb['pred'] = pd.DataFrame(pred_gb)\n\nfig, ax = plt.subplots(figsize=(9, 4))\nforecast_gb['y'].tail(91).plot(ax=ax, label='Real')\nforecast_gb['pred'].tail(91).plot(ax=ax, label='Forecast Gradient Boosting ')\nax.legend();",
"_____no_output_____"
],
[
"print(f\"Test R2 score: {est.score(X_test, y_test):.2f}\")",
"Test R2 score: 0.70\n"
],
[
"# MSE Error\n\nerror_mse_gb = mean_squared_error(y_test,pred_gb)\nprint(f\"Error de test (mse): {error_mse_gb}\")",
"Error de test (mse): 5909.477700698039\n"
]
],
[
[
"### Time series forecasting using Multi-layer Perceptron regressor.",
"_____no_output_____"
]
],
[
[
"## MLP without confg\nregr = MLPRegressor(random_state=1, max_iter=500).fit(X_train, y_train)\npred_MLP_1 = regr.predict(X_test)\n\nregr.score(X_test, y_test)",
"_____no_output_____"
],
[
"# Plotting\npred_MLP_1 = regr.predict(X_test)\nforecast_mlp1 = pd.DataFrame(y_test).reset_index()\nforecast_mlp1['pred'] = pd.DataFrame(pred_MLP_1)\n\nfig, ax = plt.subplots(figsize=(9, 4))\nforecast_mlp1['y'].tail(91).plot(ax=ax, label='Real')\nforecast_mlp1['pred'].tail(91).plot(ax=ax, label='Forecast MLP v1 ')\nax.legend();",
"_____no_output_____"
],
[
"## With more layers\n\nregr2 =MLPRegressor(hidden_layer_sizes=(20,50,50),learning_rate='adaptive',learning_rate_init=0.01,\n early_stopping=True).fit(X_train, y_train)\n\npred_MLP_2 = regr2.predict(X_test)\n\nregr2.score(X_test, y_test)",
"_____no_output_____"
],
[
"# Plotting\npred_MLP_2 = regr2.predict(X_test)\nforecast_mlp2 = pd.DataFrame(y_test).reset_index()\nforecast_mlp2['pred'] = pd.DataFrame(pred_MLP_2)\n\nfig, ax = plt.subplots(figsize=(9, 4))\nforecast_mlp2['y'].tail(91).plot(ax=ax, label='Real')\nforecast_mlp2['pred'].tail(91).plot(ax=ax, label='Forecast MLP v2 ')\nax.legend();",
"_____no_output_____"
],
[
"# MSE Error\n\nerror_mse_mlp = mean_squared_error(y_test,pred_MLP_2)\nprint(f\"Error de test (mse): {error_mse_mlp}\")",
"Error de test (mse): 6035.575493786521\n"
]
],
[
[
"## Save individual model",
"_____no_output_____"
]
],
[
[
"import joblib\n\n# save\njoblib.dump(model_sahagun, \"model_SAHAGUN2.pkl\") ",
"_____no_output_____"
],
[
"# load\nmodelo_prueba = joblib.load(\"model_SAHAGUN.pkl\")\n",
"_____no_output_____"
]
],
[
[
"## Create the models to other cities and forecasting function",
"_____no_output_____"
],
[
"To model the solar irradiance and make the forecasts for the different cities we will use the Multi-layer Perceptron regressor and Gradient Boosting.",
"_____no_output_____"
]
],
[
[
"# Model creation automation for multi layer perceptron\n\nfor key in cities.keys():\n\n df_city = cities[key].copy()\n df_city = df_city[df_city.Date < \"2021-09-01 01:00:00\"]\n df_city = df_city[(df_city['Date'].dt.hour>=6) & (df_city['Date'].dt.hour<=18)]\n df2 = df_city[['Date', 'DHI']]\n df2.columns = ['ds', 'y']\n\n # Lag for the time: day, week, month, quarter, semester, annual\n df_lags =pd.concat([df2,df2.y.shift(91),df2.y.shift(104),df2.y.shift(117),df2.y.shift(130),df2.y.shift(143)\n ,df2.y.shift(156),df2.y.shift(169),df2.y.shift(182),df2.y.shift(390)\n ,df2.y.shift(403),df2.y.shift(1170), df2.y.shift(1183),df2.y.shift(1196)\n ,df2.y.shift(1209),df2.y.shift(2340), df2.y.shift(2353), df2.y.shift(2366)\n ,df2.y.shift(2379),df2.y.shift(3900),df2.y.shift(4745)],\n axis=1)\n\n # Columns \n columns_name_2 = ['Date','y','t_7',\n 't_8','t_9','t_10','t_11','t_12','t_13','t_14',\n 't_30','t_31','t_90','t_91','t_92','t_93','t_180',\n 't_181','t_182','t_183','t_300','t_365']\n\n df_lags.columns = columns_name_2\n\n # Drops na\n df_lags = df_lags.dropna()\n\n X = df_lags.drop(['Date', 'y'], axis=1)\n y = df_lags['y']\n \n model = MLPRegressor(hidden_layer_sizes=(50,50,50)\n ,learning_rate='adaptive'\n ,learning_rate_init=0.01,\n early_stopping=True).fit(X, y)\n \n joblib.dump(model, str(key)+\"_MLP.pkl\") ",
"_____no_output_____"
],
[
"# Automation of model creation for gradient boosting\n\nfor key in cities.keys():\n\n df_city = cities[key].copy()\n df_city = df_city[df_city.Date < \"2021-09-01 01:00:00\"]\n df_city = df_city[(df_city['Date'].dt.hour>=6) & (df_city['Date'].dt.hour<=18)]\n df2 = df_city[['Date', 'DHI']]\n df2.columns = ['ds', 'y']\n\n # Lag for the time: day, week, month, quarter, semester, annual\n df_lags =pd.concat([df2,df2.y.shift(91),df2.y.shift(104),df2.y.shift(117),df2.y.shift(130),df2.y.shift(143)\n ,df2.y.shift(156),df2.y.shift(169),df2.y.shift(182),df2.y.shift(390)\n ,df2.y.shift(403),df2.y.shift(1170), df2.y.shift(1183),df2.y.shift(1196)\n ,df2.y.shift(1209),df2.y.shift(2340), df2.y.shift(2353), df2.y.shift(2366)\n ,df2.y.shift(2379),df2.y.shift(3900),df2.y.shift(4745)],\n axis=1)\n\n # Columns \n columns_name_2 = ['Date','y','t_7',\n 't_8','t_9','t_10','t_11','t_12','t_13','t_14',\n 't_30','t_31','t_90','t_91','t_92','t_93','t_180',\n 't_181','t_182','t_183','t_300','t_365']\n\n df_lags.columns = columns_name_2\n\n # Drops na\n df_lags = df_lags.dropna()\n\n X = df_lags.drop(['Date', 'y'], axis=1)\n y = df_lags['y']\n \n model = HistGradientBoostingRegressor().fit(X, y)\n \n joblib.dump(model, str(key)+\"_gb.pkl\") ",
"_____no_output_____"
]
],
[
[
"## Create the function to forecast data - BackEnd",
"_____no_output_____"
]
],
[
[
"def periodo(inicio, fin):\n d0 = date(*(int(s) for s in inicio.split('-')))\n d1 = date(*(int(s) for s in fin.split('-')))\n delta = d1 - d0\n \n if delta.days < 0:\n return print(\"Fecha inicio mayor que fecha fin\")\n neg = delta.days\n else:\n c = delta.days\n return c",
"_____no_output_____"
],
[
"#from datetime import datetime\ndef table_predict(city, inicio, fin):\n '''\n fecha : YYYY-MM-DD\n '''\n\n df2 = pd.read_csv(str(city)+'.csv')\n df2['Date'] = pd.to_datetime(df2['Date'],errors='coerce')\n df2 = df2[['Date', 'y']]\n df2.columns = ['ds', 'y']\n df2 = df2[(df2['ds'].dt.hour>=6) & (df2['ds'].dt.hour<=18)]\n \n # create datetime index passing the datetime series\n datetime_index = pd.DatetimeIndex(df2.ds.values)\n df2=df2.set_index(datetime_index)\n df2.drop('ds',axis=1,inplace=True)\n \n # Create future empty values\n c = periodo(inicio,fin)\n idx = pd.date_range(df2.index[-1] + pd.Timedelta(hours=7), periods=24*c, freq='h')[1:]\n \n table = df2.append(pd.DataFrame(pd.Series(np.repeat(0, len(idx)), index=idx), columns= ['y']))\n table = table[(table.index.hour>=6) & (table.index.hour<=18)]\n \n return table, c",
"_____no_output_____"
],
[
"## lags\ndef calculate_lags(df2):\n # Lag for the time: day, week, month, quarter, semester, annual\n serie2 =pd.concat([df2,df2.y.shift(91),df2.y.shift(104),df2.y.shift(117),df2.y.shift(130),df2.y.shift(143)\n ,df2.y.shift(156),df2.y.shift(169),df2.y.shift(182),df2.y.shift(390)\n ,df2.y.shift(403),df2.y.shift(1170), df2.y.shift(1183),df2.y.shift(1196)\n ,df2.y.shift(1209),df2.y.shift(2340), df2.y.shift(2353), df2.y.shift(2366)\n ,df2.y.shift(2379),df2.y.shift(3900),df2.y.shift(4745)],\n axis=1)\n\n # Columns \n columns_name2 = ['y','t_7','t_8','t_9','t_10','t_11','t_12','t_13','t_14',\n 't_30','t_31','t_90','t_91','t_92','t_93','t_180',\n 't_181','t_182','t_183','t_300','t_365']\n \n serie2.columns = columns_name2\n \n serie2 = serie2.dropna()\n\n return serie2",
"_____no_output_____"
],
[
"def forecast_values(serie, days):\n c = days * 13\n serie = serie[-c:]\n X_pred = serie.drop(['y'], axis=1)\n \n return X_pred",
"_____no_output_____"
],
[
"table, dias = table_predict('SAMPUES','2021-08-31', \"2021-09-02\")",
"_____no_output_____"
],
[
"table_lags = calculate_lags(table)",
"_____no_output_____"
],
[
"table_to_predict = forecast_values(table_lags, dias)",
"_____no_output_____"
],
[
"model_ipload = joblib.load(\"SAHAGÚN_MLP.pkl\")",
"_____no_output_____"
],
[
"pred_MLP_1 = model_ipload.predict(table_to_predict)",
"_____no_output_____"
],
[
"def table_show(table, inicio, forecast):\n \n inicio = date(*(int(s) for s in inicio.split('-')))\n inicio += datetime.timedelta(days=1)\n inicio = inicio.strftime('%Y/%m/%d')\n\n salida = table[table.index > inicio]\n salida['y'] = 0\n \n temp = pd.DataFrame(forecast)\n temp = round(temp,1)\n name = ['y']\n temp.columns= name\n \n salida = salida.assign(y=temp['y'].values)\n name2 = ['DHI_Forecast']\n salida.columns = name2\n \n return salida\n ",
"_____no_output_____"
],
[
"p_tabla = table_show(table, '2021-08-31', pred_MLP_1)",
"_____no_output_____"
],
[
"import modelos\nfrom modelos import densidad_aire, potencia_viento, potencia_viento_modificada, potencia_solar",
"_____no_output_____"
],
[
"ciudad_temp = pd.read_csv('cities_prom.csv')\n\ndef consumo_solar(city, irrad):\n if irrad <= 0:\n irrad = irrad + 0.1\n \n temp = ciudad_temp[ciudad_temp['name']==city]['temperature']\n pot_sol= potencia_solar(temp, irrad,36)\n if pot_sol < 0:\n pot_sol = 0\n return pot_sol",
"_____no_output_____"
],
[
"def final_table_solar(city,pred_MLP_1,p_tabla):\n column_generation = pd.DataFrame([consumo_solar(city,x) for x in pred_MLP_1])\n #name_cg = ['Generated Power (W)']\n name_temp = ['G2']\n column_generation.columns = name_temp\n \n p_tabla['G'] = 0\n final_table = p_tabla.assign(G=column_generation['G2'].values)\n name_cg = ['DHI_Forecast','Generated Power (W)']\n final_table.columns = name_cg\n return final_table",
"_____no_output_____"
],
[
"final_table_solar('SAMPUES',pred_MLP_1,p_tabla)",
"_____no_output_____"
]
],
[
[
"References\n\nhttps://www.cienciadedatos.net/documentos/py27-forecasting-series-temporales-python-scikitlearn.html\n\nhttps://towardsdatascience.com/playing-with-time-series-data-in-python-959e2485bff8\n\nhttps://setscholars.net/how-to-predict-a-time-series-using-xgboost-in-python/\n\nhttps://machinelearningmastery.com/random-forest-for-time-series-forecasting/\n\nhttps://facebook.github.io/prophet/docs/quick_start.html\n\nhttps://facebook.github.io/prophet/docs/non-daily_data.html#sub-daily-data\n\nhttps://www.cienciadedatos.net/documentos/py27-forecasting-series-temporales-python-scikitlearn.html\n\nhttps://stackoverflow.com/questions/61163759/tuning-mlpregressor-hyper-parameters\n\nhttps://scikit-learn.org/stable/auto_examples/inspection/plot_partial_dependence.html#sphx-glr-auto-examples-inspection-plot-partial-dependence-py\n\nhttps://scikit-learn.org/stable/modules/generated/sklearn.neural_network.MLPRegressor.html#sklearn.neural_network.MLPRegressor\n\nhttps://scikit-learn.org/stable/modules/neural_networks_supervised.html\n\nhttps://www.cienciadedatos.net/documentos/py27-forecasting-series-temporales-python-scikitlearn.html ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"raw",
"raw"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec5eb8dd2c6abb60fca0a422861e0b21581f2bc0 | 308,583 | ipynb | Jupyter Notebook | tutorials/notebook/mindinsight/mindinsight_dashboard.ipynb | Ming-blue/docs | 553d7b62a1eb4c445737e79a1873ac64a9a4a508 | [
"Apache-2.0",
"CC-BY-4.0"
]
| null | null | null | tutorials/notebook/mindinsight/mindinsight_dashboard.ipynb | Ming-blue/docs | 553d7b62a1eb4c445737e79a1873ac64a9a4a508 | [
"Apache-2.0",
"CC-BY-4.0"
]
| null | null | null | tutorials/notebook/mindinsight/mindinsight_dashboard.ipynb | Ming-blue/docs | 553d7b62a1eb4c445737e79a1873ac64a9a4a508 | [
"Apache-2.0",
"CC-BY-4.0"
]
| null | null | null | 365.619668 | 275,964 | 0.928995 | [
[
[
"# MindInsight训练看板\n\n通过MindSpore可以将训练过程中的标量、图像、参数分布直方图、张量、计算图和数据图记录到summary日志文件中,并通过MindInsight提供的可视化界面进行查看。\n\n- 通过查看特定的标量数值随着训练步骤的变化趋势,比如查看每个迭代的损失值、正确率、准确率这些标量的变化过程,追踪神经网络在整个训练过程中的信息,帮助用户了解模型是否过拟合,或者是否训练了过长时间。可以通过比较不同训练中的这些指标,以帮助调试和改善模型。\n\n- 通过查看训练过程中的图像数据,用户可以查看每个步骤所使用的数据集图像。\n\n- 参数分布直方图支持以直方图的形式呈现Tensor的变化趋势,用户可以查看训练过程中每个训练步骤的权重、bias和梯度参数变化信息。\n\n- 张量可视能够帮助用户直观查看训练过程中某个步骤的Tensor值,Tensor包括权重值、梯度值、激活值等。\n\n- 计算图的生成是通过将模型训练过程中的每个计算节点关联后所构成的,用户可以通过查看计算图,掌握整个模型的计算走向结构,数据流以及控制流的信息。对于高阶的使用人员,能够通过计算图验证计算节点的输入输出是否正确,并验证整个计算过程是否符合预期。\n\n- 数据图展示的是数据预处理的过程,在MindInsight可视化面板中可查看数据处理的图,能够更加直观地查看数据预处理的每一个环节,并帮助提升模型性能。\n\n接下来是本次流程的体验过程。\n\n## 整体流程\n\n1. 下载CIFAR-10二进制格式数据集。\n2. 对数据进行预处理。\n3. 定义AlexNet网络,在网络中使用summary算子记录数据。\n4. 训练网络,使用 `SummaryCollector` 记录损失值标量、权重梯度、计算图和数据图参数。同时启动MindInsight服务,实时查看损失值、参数直方图、输入图像、张量、计算图和数据图的变化。\n5. 完成训练后,查看MindInsight看板中记录到的损失值标量、直方图、图像信息、张量、计算图、数据图信息。\n6. 相关注意事项,关闭MindInsight服务。",
"_____no_output_____"
],
[
"## 准备环节\n\n### 下载数据集\n\n本次流程使用CIFAR-10二进制格式数据集,下载地址为:<https://www.cs.toronto.edu/~kriz/cifar-10-binary.tar.gz>。\n\nCIFAR-10二进制格式数据集包含10个类别的60000个32x32彩色图像。每个类别6000个图像,包含50000张训练图像和10000张测试图像。数据集分为5个训练批次和1个测试批次,每个批次具有10000张图像。测试批次包含每个类别中1000个随机选择的图像,训练批次按随机顺序包含剩余图像(某个训练批次包含的一类图像可能比另一类更多)。其中,每个训练批次精确地包含对应每个类别的5000张图像。\n\n执行下面一段代码下载CIFAR-10二进制格式数据集到当前工作目录,如果已经下载过数据集,则不重复下载。",
"_____no_output_____"
]
],
[
[
"import os, shutil\nimport urllib.request\nfrom urllib.parse import urlparse\n\n\ndef callbackfunc(blocknum, blocksize, totalsize):\n percent = 100.0 * blocknum * blocksize / totalsize\n if percent > 100:\n percent = 100\n print(\"downloaded {:.1f}\".format(percent), end=\"\\r\")\n\ndef _download_dataset():\n ds_url = \"https://www.cs.toronto.edu/~kriz/cifar-10-binary.tar.gz\"\n file_base_name = urlparse(ds_url).path.split(\"/\")[-1]\n file_name = os.path.join(\"./datasets\", file_base_name)\n if not os.path.exists(file_name):\n urllib.request.urlretrieve(ds_url, file_name, callbackfunc)\n print(\"{:*^40}\".format(\"DataSets Downloaded\"))\n shutil.unpack_archive(file_name, extract_dir=\"./datasets/cifar-10-binary\")\n\ndef _copy_dataset(ds_part, dest_path):\n data_source_path = \"./datasets/cifar-10-binary/cifar-10-batches-bin\"\n ds_part_source_path = os.path.join(data_source_path, ds_part)\n if not os.path.exists(ds_part_source_path):\n _download_dataset()\n shutil.copy(ds_part_source_path, dest_path)\n\ndef download_cifar10_dataset():\n ds_base_path = \"./datasets/cifar10\"\n train_path = os.path.join(ds_base_path, \"train\")\n test_path = os.path.join(ds_base_path, \"test\")\n print(\"{:*^40}\".format(\"Checking DataSets Path.\"))\n if not os.path.exists(train_path) and not os.path.exists(test_path):\n os.makedirs(train_path)\n os.makedirs(test_path)\n print(\"{:*^40}\".format(\"Downloading CIFAR-10 DataSets.\"))\n for i in range(1, 6):\n train_part = \"data_batch_{}.bin\".format(i)\n if not os.path.exists(os.path.join(train_path, train_part)):\n _copy_dataset(train_part, train_path)\n pops = train_part + \" is ok\"\n print(\"{:*^40}\".format(pops))\n test_part = \"test_batch.bin\"\n if not os.path.exists(os.path.join(test_path, test_part)):\n _copy_dataset(test_part, test_path)\n print(\"{:*^40}\".format(test_part+\" is ok\"))\n print(\"{:*^40}\".format(\"Downloaded CIFAR-10 DataSets Already.\"))\n\ndownload_cifar10_dataset()",
"********Checking DataSets Path.*********\n*****Downloading CIFAR-10 DataSets.*****\n*********data_batch_1.bin is ok*********\n*********data_batch_2.bin is ok*********\n*********data_batch_3.bin is ok*********\n*********data_batch_4.bin is ok*********\n*********data_batch_5.bin is ok*********\n**********test_batch.bin is ok**********\n*Downloaded CIFAR-10 DataSets Already.**\n"
]
],
[
[
"下载数据集后,CIFAR-10数据集目录(`datasets`)结构如下所示。\n\n```text\n $ tree datasets\n datasets\n └── cifar10\n ├── test\n │ └── test_batch.bin\n └── train\n ├── data_batch_1.bin\n ├── data_batch_2.bin\n ├── data_batch_3.bin\n ├── data_batch_4.bin\n └── data_batch_5.bin\n\n```\n\n其中:\n- `test_batch.bin`文件为测试数据集文件。\n- `data_batch_1.bin`文件为第1批次训练数据集文件。\n- `data_batch_2.bin`文件为第2批次训练数据集文件。\n- `data_batch_3.bin`文件为第3批次训练数据集文件。\n- `data_batch_4.bin`文件为第4批次训练数据集文件。\n- `data_batch_5.bin`文件为第5批次训练数据集文件。\n",
"_____no_output_____"
],
[
"## 数据处理\n\n好的数据集可以有效提高训练精度和效率,在加载数据集前,会进行一些处理,增加数据的可用性和随机性。下面一段代码定义函数`create_dataset_cifar10`来进行数据处理操作,并创建训练数据集(`ds_train`)和测试数据集(`ds_eval`)。\n",
"_____no_output_____"
]
],
[
[
"import mindspore.dataset as ds\nimport mindspore.dataset.transforms.c_transforms as C\nimport mindspore.dataset.vision.c_transforms as CV\nfrom mindspore import dtype as mstype\n\n\ndef create_dataset_cifar10(data_path, batch_size=32, repeat_size=1, status=\"train\"):\n \"\"\"\n create dataset for train or test\n \"\"\"\n cifar_ds = ds.Cifar10Dataset(data_path)\n rescale = 1.0 / 255.0\n shift = 0.0\n\n resize_op = CV.Resize(size=(227, 227))\n rescale_op = CV.Rescale(rescale, shift)\n normalize_op = CV.Normalize((0.4914, 0.4822, 0.4465), (0.2023, 0.1994, 0.2010))\n if status == \"train\":\n random_crop_op = CV.RandomCrop([32, 32], [4, 4, 4, 4])\n random_horizontal_op = CV.RandomHorizontalFlip()\n channel_swap_op = CV.HWC2CHW()\n typecast_op = C.TypeCast(mstype.int32)\n cifar_ds = cifar_ds.map(operations=typecast_op, input_columns=\"label\")\n if status == \"train\":\n cifar_ds = cifar_ds.map(operations=random_crop_op, input_columns=\"image\")\n cifar_ds = cifar_ds.map(operations=random_horizontal_op, input_columns=\"image\")\n cifar_ds = cifar_ds.map(operations=resize_op, input_columns=\"image\")\n cifar_ds = cifar_ds.map(operations=rescale_op, input_columns=\"image\")\n cifar_ds = cifar_ds.map(operations=normalize_op, input_columns=\"image\")\n cifar_ds = cifar_ds.map(operations=channel_swap_op, input_columns=\"image\")\n\n cifar_ds = cifar_ds.shuffle(buffer_size=1000)\n cifar_ds = cifar_ds.batch(batch_size, drop_remainder=True)\n cifar_ds = cifar_ds.repeat(repeat_size)\n return cifar_ds\n\nds_train = create_dataset_cifar10(data_path=\"./datasets/cifar10/train\")\nds_eval = create_dataset_cifar10(\"./datasets/cifar10/test\")",
"_____no_output_____"
]
],
[
[
"### 抽取数据集图像\n\n执行以下一段代码,抽取上步创建好的训练数据集`ds_train`中第一个`batch`的32张图像以及对应的类别名称进行展示。",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\nimport numpy as np\n\nlabel_list = [\"airplane\", \"automobile\", \"bird\", \"cat\", \"deer\", \"dog\", \"rog\", \"horse\", \"ship\", \"truck\"]\nprint(\"The 32 images with label of the first batch in ds_train are showed below:\")\nds_iterator = ds_train.create_dict_iterator()\nnext(ds_iterator)\nbatch_1 = next(ds_iterator)\nbatch_image = batch_1[\"image\"].asnumpy()\nbatch_label = batch_1[\"label\"].asnumpy()\n%matplotlib inline\nplt.figure(dpi=144)\nfor i,image in enumerate(batch_image):\n plt.subplot(4, 8, i+1)\n plt.subplots_adjust(wspace=0.2, hspace=0.2)\n image = image/np.amax(image)\n image = np.clip(image, 0, 1)\n image = np.transpose(image,(1,2,0))\n plt.imshow(image)\n num = batch_label[i]\n plt.title(f\"image {i+1}\\n{label_list[num]}\", y=-0.65, fontdict={\"fontsize\":8})\n plt.axis('off') \nplt.show()",
"The 32 images with label of the first batch in ds_train are showed below:\n"
]
],
[
[
"## 使用Summary算子记录数据\n\n在进行训练之前,需定义神经网络模型,本流程采用AlexNet网络。\n\nMindSpore提供了两种方法进行记录数据,分别为:\n\n- 通过Summary算子记录数据。\n- 通过 `SummaryCollector` 这个callback进行记录。\n\n下面为在AlexNet网络中使用Summary算子记录输入图像和张量数据的配置方法。\n\n- 使用 `ImageSummary` 记录输入图像数据。\n\n 1. 在 `__init__` 方法中初始化 `ImageSummary`。\n \n ```python\n # Init ImageSummary\n self.image_summary = ops.ImageSummary()\n ```\n \n 2. 在 `construct` 方法中使用 `ImageSummary` 算子记录输入图像。其中 \"Image\" 为该数据的名称,MindInsight在展示时,会将该名称展示出来以方便识别是哪个数据。\n \n ```python\n # Record image by Summary operator\n self.image_summary(\"Image\", x)\n ```\n \n- 使用 `TensorSummary` 记录张量数据。\n\n 1. 在 `__init__` 方法中初始化 `TensorSummary`。\n \n ```python\n # Init TensorSummary\n self.tensor_summary = ops.TensorSummary()\n ```\n \n 2. 在`construct`方法中使用`TensorSummary`算子记录张量数据。其中\"Tensor\"为该数据的名称。\n \n ```python\n # Record tensor by Summary operator\n self.tensor_summary(\"Tensor\", x)\n ```\n\n当前支持的Summary算子:\n\n- [ScalarSummary](https://www.mindspore.cn/doc/api_python/zh-CN/master/mindspore/ops/mindspore.ops.ScalarSummary.html): 记录标量数据\n- [TensorSummary](https://www.mindspore.cn/doc/api_python/zh-CN/master/mindspore/ops/mindspore.ops.TensorSummary.html): 记录张量数据\n- [ImageSummary](https://www.mindspore.cn/doc/api_python/zh-CN/master/mindspore/ops/mindspore.ops.ImageSummary.html): 记录图片数据\n- [HistogramSummary](https://www.mindspore.cn/doc/api_python/zh-CN/master/mindspore/ops/mindspore.ops.HistogramSummary.html): 将张量数据转为直方图数据记录\n\n以下一段代码中定义AlexNet网络结构。",
"_____no_output_____"
]
],
[
[
"import mindspore.nn as nn\nfrom mindspore.common.initializer import TruncatedNormal\nimport mindspore.ops as ops\n\ndef conv(in_channels, out_channels, kernel_size, stride=1, padding=0, pad_mode=\"valid\"):\n weight = weight_variable()\n return nn.Conv2d(in_channels, out_channels,\n kernel_size=kernel_size, stride=stride, padding=padding,\n weight_init=weight, has_bias=False, pad_mode=pad_mode)\n\ndef fc_with_initialize(input_channels, out_channels):\n weight = weight_variable()\n bias = weight_variable()\n return nn.Dense(input_channels, out_channels, weight, bias)\n\ndef weight_variable():\n return TruncatedNormal(0.02)\n\n\nclass AlexNet(nn.Cell):\n \"\"\"\n Alexnet\n \"\"\"\n def __init__(self, num_classes=10, channel=3):\n super(AlexNet, self).__init__()\n self.conv1 = conv(channel, 96, 11, stride=4)\n self.conv2 = conv(96, 256, 5, pad_mode=\"same\")\n self.conv3 = conv(256, 384, 3, pad_mode=\"same\")\n self.conv4 = conv(384, 384, 3, pad_mode=\"same\")\n self.conv5 = conv(384, 256, 3, pad_mode=\"same\")\n self.relu = nn.ReLU()\n self.max_pool2d = ops.MaxPool(ksize=3, strides=2)\n self.flatten = nn.Flatten()\n self.fc1 = fc_with_initialize(6*6*256, 4096)\n self.fc2 = fc_with_initialize(4096, 4096)\n self.fc3 = fc_with_initialize(4096, num_classes)\n # Init TensorSummary\n self.tensor_summary = ops.TensorSummary()\n # Init ImageSummary\n self.image_summary = ops.ImageSummary()\n\n def construct(self, x):\n # Record image by Summary operator\n self.image_summary(\"Image\", x)\n x = self.conv1(x)\n # Record tensor by Summary operator\n self.tensor_summary(\"Tensor\", x)\n x = self.relu(x)\n x = self.max_pool2d(x)\n x = self.conv2(x)\n x = self.relu(x)\n x = self.max_pool2d(x)\n x = self.conv3(x)\n x = self.relu(x)\n x = self.conv4(x)\n x = self.relu(x)\n x = self.conv5(x)\n x = self.relu(x)\n x = self.max_pool2d(x)\n x = self.flatten(x)\n x = self.fc1(x)\n x = self.relu(x)\n x = self.fc2(x)\n x = self.relu(x)\n x = self.fc3(x)\n return x",
"_____no_output_____"
]
],
[
[
"## 使用 `SummaryCollector` 记录数据\n\n下面展示使用`SummaryCollector`来记录标量、直方图信息。\n\n在MindSpore中通过`Callback`机制,提供支持快速简易地收集损失值、参数权重、梯度等信息的`Callback`, 叫做`SummaryCollector`(详细的用法可以参考API文档中[mindspore.train.callback.SummaryCollector](https://www.mindspore.cn/doc/api_python/zh-CN/master/mindspore/mindspore.train.html#mindspore.train.callback.SummaryCollector))。`SummaryCollector`使用方法如下: \n\n`SummaryCollector` 提供 `collect_specified_data` 参数,允许用户自定义想要收集的数据。\n\n下面的代码展示通过 `SummaryCollector` 收集损失值以及卷积层的参数值,参数值在MindInsight中以直方图展示。\n\n\n\n\n```python\nspecified={\"collect_metric\": True, \"histogram_regular\": \"^conv1.*|^conv2.*\",\"collect_graph\": True, \"collect_dataset_graph\": True}\nsummary_collector = SummaryCollector(summary_dir=\"./summary_dir/summary_01\", \n collect_specified_data=specified, \n collect_freq=1, \n keep_default_action=False, \n collect_tensor_freq=200)\n```\n\n- `summary_dir`:指定日志保存的路径。\n- `collect_specified_data`:指定需要记录的信息。\n- `collect_freq`:指定使用`SummaryCollector`记录数据的频率。\n- `keep_default_action`:指定是否除记录除指定信息外的其他数据信息。\n- `collect_tensor_freq`:指定记录张量信息的频率。\n- `\"collect_metric\"`为记录损失值标量信息。\n- `\"histogram_regular\"`为记录`conv1`层和`conv2`层直方图信息。\n- `\"collect_graph\"`为记录计算图信息。\n- `\"collect_dataset_graph\"`为记录数据图信息。\n\n  程序运行过程中将在本地`8080`端口自动启动MindInsight服务并自动遍历读取当前notebook目录下`summary_dir`子目录下所有日志文件、解析进行可视化展示。",
"_____no_output_____"
],
[
"### 导入模块",
"_____no_output_____"
]
],
[
[
"import os\nimport mindspore.nn as nn\nfrom mindspore.train.callback import ModelCheckpoint, CheckpointConfig, LossMonitor, TimeMonitor\nfrom mindspore.nn import Accuracy\nfrom mindspore.train.callback import SummaryCollector\nfrom mindspore import load_checkpoint, load_param_into_net\nfrom mindspore import Tensor, context, Model\n\ndevice_target = \"GPU\"\ncontext.set_context(mode=context.GRAPH_MODE, device_target=device_target)",
"_____no_output_____"
]
],
[
[
"### 定义学习率\n\n以下一段代码定义学习率。",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n\ndef get_lr(current_step, lr_max, total_epochs, steps_per_epoch):\n \"\"\"\n generate learning rate array\n\n Args:\n current_step(int): current steps of the training\n lr_max(float): max learning rate\n total_epochs(int): total epoch of training\n steps_per_epoch(int): steps of one epoch\n\n Returns:\n np.array, learning rate array\n \"\"\"\n lr_each_step = []\n total_steps = steps_per_epoch * total_epochs\n decay_epoch_index = [0.8 * total_steps]\n for i in range(total_steps):\n if i < decay_epoch_index[0]:\n lr = lr_max\n else:\n lr = lr_max * 0.1\n lr_each_step.append(lr)\n lr_each_step = np.array(lr_each_step).astype(np.float32)\n learning_rate = lr_each_step[current_step:]\n\n return learning_rate\n",
"_____no_output_____"
]
],
[
[
"### 执行训练",
"_____no_output_____"
]
],
[
[
"network = AlexNet(num_classes=10)\nnet_loss = nn.SoftmaxCrossEntropyWithLogits(sparse=True, reduction=\"mean\")\nlr = Tensor(get_lr(0, 0.002, 10, ds_train.get_dataset_size()))\nnet_opt = nn.Momentum(network.trainable_params(), learning_rate=lr, momentum=0.9)\ntime_cb = TimeMonitor(data_size=ds_train.get_dataset_size())\nconfig_ck = CheckpointConfig(save_checkpoint_steps=1562, keep_checkpoint_max=10)\nckpoint_cb = ModelCheckpoint(directory=\"./models/ckpt/mindinsight_dashboard\", prefix=\"checkpoint_alexnet\", config=config_ck)\nmodel = Model(network, net_loss, net_opt, metrics={\"Accuracy\": Accuracy()})\n\nsummary_base_dir = \"./summary_dir\"\nos.system(f\"mindinsight start --summary-base-dir {summary_base_dir} --port=8080\")\n\n# Init a SummaryCollector callback instance, and use it in model.train or model.eval\nspecified = {\"collect_metric\": True, \"histogram_regular\": \"^conv1.*|^conv2.*\", \"collect_graph\": True, \"collect_dataset_graph\": True}\nsummary_collector = SummaryCollector(summary_dir=\"./summary_dir/summary_01\", collect_specified_data=specified, collect_freq=1, keep_default_action=False, collect_tensor_freq=200)\n\nprint(\"============== Starting Training ==============\")\nmodel.train(epoch=10, train_dataset=ds_train, callbacks=[time_cb, ckpoint_cb, LossMonitor(), summary_collector], dataset_sink_mode=True)\n\nprint(\"============== Starting Testing ==============\")\nparam_dict = load_checkpoint(\"./models/ckpt/mindinsight_dashboard/checkpoint_alexnet-10_1562.ckpt\")\nload_param_into_net(network, param_dict)\nacc = model.eval(ds_eval, callbacks=summary_collector, dataset_sink_mode=True)\nprint(\"============== {} ==============\".format(acc))",
"============== Starting Training ==============\nepoch: 1 step: 1, loss is 2.3037791\nepoch: 1 step: 2, loss is 2.3127236\nepoch: 1 step: 3, loss is 2.3156757\nepoch: 1 step: 4, loss is 2.2910595\nepoch: 1 step: 5, loss is 2.3042145\nepoch: 1 step: 6, loss is 2.3150084\nepoch: 1 step: 7, loss is 2.2808924\nepoch: 1 step: 8, loss is 2.3073373\nepoch: 1 step: 9, loss is 2.308782\nepoch: 1 step: 10, loss is 2.2957213\n\n...\n\nepoch: 10 step: 1550, loss is 0.54039395\nepoch: 10 step: 1551, loss is 0.25690028\nepoch: 10 step: 1552, loss is 0.26572403\nepoch: 10 step: 1553, loss is 0.4429163\nepoch: 10 step: 1554, loss is 0.25716054\nepoch: 10 step: 1555, loss is 0.38538748\nepoch: 10 step: 1556, loss is 0.12103356\nepoch: 10 step: 1557, loss is 0.16565521\nepoch: 10 step: 1558, loss is 0.4364005\nepoch: 10 step: 1559, loss is 0.428179\nepoch: 10 step: 1560, loss is 0.42687342\nepoch: 10 step: 1561, loss is 0.6419081\nepoch: 10 step: 1562, loss is 0.5843237\nEpoch time: 115283.798, per step time: 73.805\n============== Starting Testing ==============\n============== {'Accuracy': 0.8302283653846154} ==============\n"
]
],
[
[
"## MindInsight看板\n\n在本地浏览器中打开地址:`127.0.0.1:8080`,进入到可视化面板。\n\n\n\n在上图所示面板中可以看到`summary_01`日志文件目录,点击**训练看板**进入到下图所示的训练数据展示面板,该面板展示了标量数据、直方图、图像和张量信息,并随着训练、测试的进行实时刷新数据,实时显示训练过程参数的变化情况。\n\n\n\n### 标量可视化\n\n标量可视化用于展示训练过程中标量的变化趋势,点击打开训练标量信息展示面板,该面板记录了迭代计算过程中的损失值标量信息,如下图展示了损失值标量趋势图。\n\n\n\n上图展示了神经网络在训练过程中损失值的变化过程。横坐标是训练步骤,纵坐标是损失值。\n\n图中右上角有几个按钮功能,从左到右功能分别是全屏展示,切换Y轴比例,开启/关闭框选,分步回退和还原图形。\n\n- 全屏展示即全屏展示该标量曲线,再点击一次即可恢复。\n- 切换Y轴比例是指可以将Y轴坐标进行对数转换。\n- 开启/关闭框选是指可以框选图中部分区域,并放大查看该区域,可以在已放大的图形上叠加框选。\n- 分步回退是指对同一个区域连续框选并放大查看时,可以逐步撤销操作。\n- 还原图形是指进行了多次框选后,点击此按钮可以将图还原回原始状态。\n\n\n\n上图展示的标量可视化的功能区,提供了根据选择不同标签,水平轴的不同维度和平滑度来查看标量信息的功能。\n\n- 标签选择:提供了对所有标签进行多项选择的功能,用户可以通过勾选所需的标签,查看对应的标量信息。\n- 水平轴:可以选择“步骤”、“相对时间”、“绝对时间”中的任意一项,来作为标量曲线的水平轴。\n- 平滑度:可以通过调整平滑度,对标量曲线进行平滑处理。\n- 标量合成:可以选中两条标量曲线进行合成并展示在一个图中,以方便对两条曲线进行对比或者查看合成后的图。\n 标量合成的功能区与标量可视化的功能区相似。其中与标量可视化功能区不一样的地方,在于标签选择时,标量合成功能最多只能同时选择两个标签,将其曲线合成并展示。\n\n### 直方图可视化\n\n\n直方图用于将用户所指定的张量以直方图的形式展示。点击打开直方图展示面板,以直方图的形式记录了在迭代过程中所有层参数分布信息。\n\n\n\n如下图为`conv1`层参数分布信息,点击图中右上角,可以将图放大。\n\n\n\n下图为直方图功能区。\n\n\n\n上图展示直方图的功能区,包含以下内容:\n\n- 标签选择:提供了对所有标签进行多项选择的功能,用户可以通过勾选所需的标签,查看对应的直方图。\n- 纵轴:可以选择步骤、相对时间、绝对时间中的任意一项,来作为直方图纵轴显示的数据。\n- 视角:可以选择正视和俯视中的一种。正视是指从正面的角度查看直方图,此时不同步骤之间的数据会覆盖在一起。俯视是指偏移以45度角俯视直方图区域,这时可以呈现不同步骤之间数据的差异。\n\n### 图像可视化\n\n图像可视化用于展示用户所指定的图片。点击数据抽样展示面板,展示了每个一步进行处理的图像信息。\n\n下图为展示`summary_01`记录的图像信息。\n\n\n\n通过滑动上图中的\"步骤\"滑条,查看不同步骤的图片。\n\n\n\n上图展示图像可视化的功能区,提供了选择查看不同标签,不同亮度和不同对比度来查看图片信息。\n\n- 标签:提供了对所有标签进行多项选择的功能,用户可以通过勾选所需的标签,查看对应的图片信息。\n- 亮度调整:可以调整所展示的所有图片亮度。\n- 对比度调整:可以调整所展示的所有图片对比度。\n\n### 张量可视化\n\n张量可视化用于将张量以表格以及直方图的形式进行展示。\n\n\n\n上图展示了张量可视化的功能区,包含以下内容:\n\n- 标签选择:提供了对所有标签进行多项选择的功能,用户可以通过勾选所需的标签,查看对应的表格数据或者直方图。\n- 视图:可以选择表格或者直方图来展示tensor数据。在直方图视图下存在纵轴和视角的功能选择。\n- 纵轴:可以选择步骤、相对时间、绝对时间中的任意一项,来作为直方图纵轴显示的数据。\n- 视角:可以选择正视和俯视中的一种。正视是指从正面的角度查看直方图,此时不同步骤之间的数据会覆盖在一起。俯视是指 偏移以45度角俯视直方图区域,这时可以呈现不同步骤之间数据的差异。\n\n\n\n上图中将用户所记录的张量以表格的形式展示,包含以下功能:\n\n- 点击表格右边小方框按钮,可以将表格放大。\n- 表格中白色方框显示当前展示的是哪个维度下的张量数据,其中冒号\":\"表示当前维度的所有值,可以在方框输入对应的索引或者:后按Enter键或者点击后边的打勾按钮来查询特定维度的张量数据。 假设某维度是32,则其索引范围是-32到31。注意:可以查询0维到2维的张量数据,不支持查询超过两维的张量数据,即不能设置超过两个冒号\":\"的查询条件。\n- 拖拽表格下方的空心圆圈可以查询特定步骤的张量数据。\n\n### 计算图可视化\n\n点击计算图可视化用于展示计算图的图结构,数据流以及控制流的走向,支持展示summary日志文件与通过`context`的`save_graphs`参数导出的`pb`文件。\n\n\n\n上展示了计算图的网络结构。如图中所展示的,在展示区中,选中其中一个算子(图中圈红算子),可以看到该算子有两个输入和一个输出(实线代表算子的数据流走向)。\n\n\n\n上图展示了计算图可视化的功能区,包含以下内容:\n\n- 文件选择框:可以选择查看不同文件的计算图。\n- 搜索框:可以对节点进行搜索,输入节点名称点击回车,即可展示该节点。\n- 缩略图:展示整个网络图结构的缩略图,在查看超大图结构时,方便查看当前浏览的区域。\n- 节点信息:展示选中的节点的基本信息,包括节点的名称、属性、输入节点、输出节点等信息。\n- 图例:展示的是计算图中各个图标的含义。\n\n### 数据图可视化\n\n数据图可视化用于展示单次模型训练的数据处理和数据增强信息。\n\n\n\n上图展示的数据图功能区包含以下内容:\n\n- 图例:展示数据溯源图中各个图标的含义。\n- 数据处理流水线:展示训练所使用的数据处理流水线,可以选择图中的单个节点查看详细信息。\n- 节点信息:展示选中的节点的基本信息,包括使用的数据处理和增强算子的名称、参数等。\n",
"_____no_output_____"
],
[
"### 单独记录损失值标量\n\n\n为了降低性能开销和日志文件大小,可以单独记录关心的数据。单独记录标量、参数分布直方图、计算图或数据图信息,可以通过配置`specified`参数为相应的值来单独记录。单独记录图像或张量信息,可以在AlexNet网络的`construct`方法中使用`ImageSummary`算子或`TensorSummary`算子来单独记录。\n",
"_____no_output_____"
],
[
"## 关闭MindInsight服务\n\n在终端命令行中执行以下代码关闭MindInsight服务。\n\n```bash\nmindinsight stop --port 8080\n```",
"_____no_output_____"
],
[
"## 注意事项和规格\n1. 为了控制列出summary文件目录的用时,MindInsight最多支持发现999个summary文件目录。\n2. 不能同时使用多个 `SummaryRecord` 实例 (`SummaryCollector` 中使用了 `SummaryRecord`)。\n\n 如果在 `model.train` 或者 `model.eval` 的callback列表中使用两个及以上的 `SummaryCollector` 实例,则视为同时使用 `SummaryRecord`,导致记录数据失败。\n\n 自定义callback中如果使用 `SummaryRecord`,则其不能和 `SummaryCollector` 同时使用。\n\n 正确代码:\n ```\n ...\n summary_collector = SummaryCollector('./summary_dir')\n model.train(2, train_dataset, callbacks=[summary_collector])\n\n ...\n model.eval(dataset, callbacks=[summary_collector])\n ```\n\n 错误代码:\n ```\n ...\n summary_collector1 = SummaryCollector('./summary_dir1')\n summary_collector2 = SummaryCollector('./summary_dir2')\n model.train(2, train_dataset, callbacks=[summary_collector1, summary_collector2])\n ```\n\n 错误代码:\n ```\n ...\n # Note: the 'ConfusionMatrixCallback' is user-defined, and it uses SummaryRecord to record data.\n confusion_callback = ConfusionMatrixCallback('./summary_dir1')\n summary_collector = SummaryCollector('./summary_dir2')\n model.train(2, train_dataset, callbacks=[confusion_callback, summary_collector])\n ```\n3. 每个summary日志文件目录中,应该只放置一次训练的数据。一个summary日志目录中如果存放了多次训练的summary数据,MindInsight在可视化数据时会将这些训练的summary数据进行叠加展示,可能会与预期可视化效果不相符。\n4. 当前 `SummaryCollector` 和 `SummaryRecord` 不支持GPU多卡运行的场景。\n5. 目前MindSpore仅支持在Ascend 910 AI处理器上导出算子融合后的计算图。\n6. 在训练中使用Summary算子收集数据时,`HistogramSummary` 算子会影响性能,所以请尽量少地使用。\n7. 为了控制内存占用,MindInsight对标签(tag)数目和步骤(step)数目进行了限制:\n - 每个训练看板的最大标签数量为300个标签。标量标签、图片标签、计算图标签、参数分布图(直方图)标签、张量标签的数量总和不得超过300个。特别地,每个训练看板最多有10个计算图标签、6个张量标签。当实际标签数量超过这一限制时,将依照MindInsight的处理顺序,保留最近处理的300个标签。\n - 每个训练看板的每个标量标签最多有1000个步骤的数据。当实际步骤的数目超过这一限制时,将对数据进行随机采样,以满足这一限制。\n - 每个训练看板的每个图片标签最多有10个步骤的数据。当实际步骤的数目超过这一限制时,将对数据进行随机采样,以满足这一限制。\n - 每个训练看板的每个参数分布图(直方图)标签最多有50个步骤的数据。当实际步骤的数目超过这一限制时,将对数据进行随机采样,以满足这一限制。\n - 每个训练看板的每个张量标签最多有20个步骤的数据。当实际步骤的数目超过这一限制时,将对数据进行随机采样,以满足这一限制。\n8. 由于`TensorSummary`会记录完整Tensor数据,数据量通常会比较大,为了控制内存占用和出于性能上的考虑,MindInsight对Tensor的大小以及返回前端展示的数值个数进行以下限制:\n - MindInsight最大支持加载含有1千万个数值的Tensor。\n - Tensor加载后,在张量可视的表格视图下,最大支持查看10万个数值,如果所选择的维度查询得到的数值超过这一限制,则无法显示。\n\n9. 由于张量可视(`TensorSummary`)会记录原始张量数据,需要的存储空间较大。使用`TensorSummary`前和训练过程中请注意检查系统存储空间充足。\n\n 通过以下方法可以降低张量可视功能的存储空间占用:\n\n 1)避免使用`TensorSummary`记录较大的Tensor。\n\n 2)减少网络中`TensorSummary`算子的使用个数。\n\n 功能使用完毕后,请及时清理不再需要的训练日志,以释放磁盘空间。\n\n 备注:估算`TensorSummary`空间使用量的方法如下:\n\n 一个`TensorSummary数据的大小 = Tensor中的数值个数 * 4 bytes`。假设使用`TensorSummary`记录的Tensor大小为`32 * 1 * 256 * 256`,则一个`TensorSummary`数据大约需要`32 * 1 * 256 * 256 * 4 bytes = 8,388,608 bytes = 8MiB`。`TensorSummary`默认会记录20个步骤的数据,则记录这20组数据需要的空间约为`20 * 8 MiB = 160MiB`。需要注意的是,由于数据结构等因素的开销,实际使用的存储空间会略大于160MiB。\n10. 当使用`TensorSummary`时,由于记录完整Tensor数据,训练日志文件较大,MindInsight需要更多时间解析训练日志文件,请耐心等待。",
"_____no_output_____"
],
[
"## 总结\n\n本次体验流程为完整的MindSpore深度学习及MindInsight可视化展示的过程,包括了下载数据集及预处理过程,构建网络、损失函数和优化器过程,生成模型并进行训练、验证的过程,以及启动MindInsight服务进行训练过程可视化展示。读者可以基于本次体验流程构建自己的网络模型进行训练,并使用`SummaryCollector`以及Summary算子记录关心的数据,然后在MindInsight服务看板中进行可视化展示,根据MindInsight服务中展示的结果调整相应的参数以提高训练精度。\n\n以上便完成了标量、直方图、图像和张量可视化的体验,我们通过本次体验全面了解了MindSpore执行训练的过程和MindInsight在标量、直方图、图像、张量、计算图和数据图可视化的应用,理解了如何使用`SummaryColletor`记录训练过程中的标量、直方图、图像、张量、计算图和数据图数据。",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec5ebb0d305203c97a82cef007583c511994ddf6 | 16,359 | ipynb | Jupyter Notebook | ML-Base-MOOC/chapt-3 Linear Regression/05-Gradient Descent in Linear Regression.ipynb | NovemberChopin/machine-learning | a87a2b8189b5a2990350083038c997fb22fb39ff | [
"MIT"
]
| 3 | 2019-10-28T19:46:36.000Z | 2020-02-25T06:59:19.000Z | ML-Base-MOOC/chapt-3 Linear Regression/05-Gradient Descent in Linear Regression.ipynb | NovemberChopin/machine-learning | a87a2b8189b5a2990350083038c997fb22fb39ff | [
"MIT"
]
| null | null | null | ML-Base-MOOC/chapt-3 Linear Regression/05-Gradient Descent in Linear Regression.ipynb | NovemberChopin/machine-learning | a87a2b8189b5a2990350083038c997fb22fb39ff | [
"MIT"
]
| 3 | 2019-10-05T16:52:11.000Z | 2021-06-17T02:16:53.000Z | 48.399408 | 9,892 | 0.768446 | [
[
[
"# Gradient Descent in Linear Regression",
"_____no_output_____"
],
[
"[](https://imgchr.com/i/8ZePW8)\n \n - $X_b$ 是在矩阵 $X$ 中加了一列,值为 1\n - 我们希望最后得出的梯度值和样本数量 m 无关,故变为如下形式\n\n[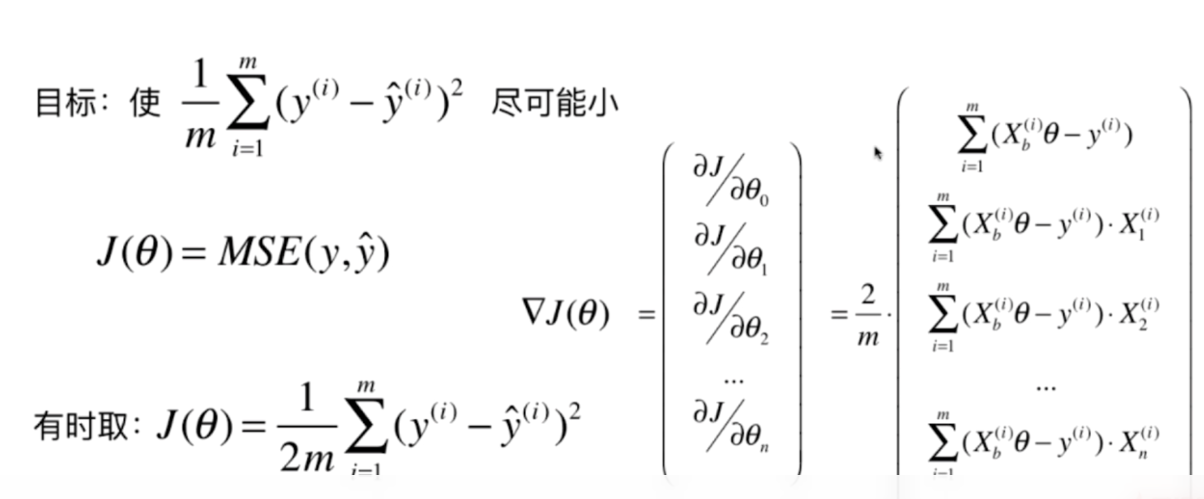](https://imgchr.com/i/8Znp28)\n",
"_____no_output_____"
],
[
"### 1. 编程实现",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"np.random.seed(333)\nx = 2 * np.random.random(size=100)\ny = x * 3. + 4. + np.random.normal(size = 100)",
"_____no_output_____"
],
[
"X = x.reshape(-1, 1)",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"y.shape",
"_____no_output_____"
],
[
"plt.scatter(x, y)",
"_____no_output_____"
]
],
[
[
"### 2. 使用梯度下降法训练模型",
"_____no_output_____"
]
],
[
[
"def J(theta, X_b, y):\n try:\n return np.sum((y - X_b.dot(theta))**2) / len(X_b)\n except:\n return float('inf')",
"_____no_output_____"
],
[
"def dJ(theta, X_b, y):\n res = np.empty(len(theta))\n res[0] = np.sum(X_b.dot(theta) - y)\n for i in range(1, len(theta)):\n res[i] = (X_b.dot(theta) - y).dot(X_b[:, i])\n return res * 2 / len(X_b)",
"_____no_output_____"
],
[
"def gradient_descent(X_b, y, initial_theta, eta, n_iters = 1e4, epsilon=1e-8):\n theta = initial_theta\n i_iter = 0\n \n while i_iter < n_iters:\n gradient = dJ(theta, X_b, y)\n last_theta = theta\n theta = theta - eta * gradient\n\n if np.abs(J(theta, X_b, y) - J(last_theta, X_b, y)) < epsilon:\n break\n \n i_iter += 1\n \n return theta",
"_____no_output_____"
],
[
"X_b = np.hstack([np.ones([len(X), 1]), X])\ninitial_theta = np.zeros(X_b.shape[1])\neta = 0.01\ntheta = gradient_descent(X_b, y, initial_theta, eta)",
"_____no_output_____"
],
[
"theta",
"_____no_output_____"
]
],
[
[
"### 3. 封装我们的线性回归梯度下降算法",
"_____no_output_____"
],
[
"- 其中梯度下降算法中的求导过程,可以转为向量化运算\n\n[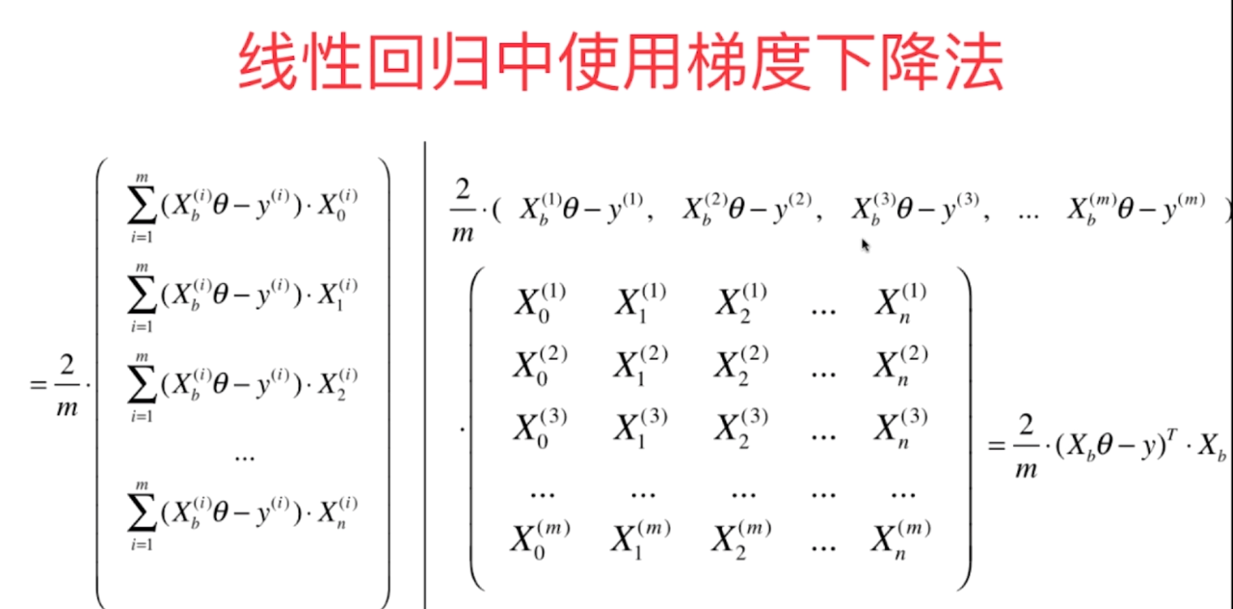](https://imgchr.com/i/8ZnS8f)\n\n- `X_b.T.dot(X_b.dot(theta) - y) * 2. / len(X_b)`",
"_____no_output_____"
]
],
[
[
"from LR.LinearRegression import LinearRegression",
"_____no_output_____"
],
[
"lin_reg = LinearRegression()\nlin_reg.fit_gd(X, y)",
"_____no_output_____"
],
[
"lin_reg.coef_",
"_____no_output_____"
],
[
"lin_reg.intercept_",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
ec5ebd7302df53423c6660772e88a2376bce9c96 | 209,426 | ipynb | Jupyter Notebook | Woof_128_ConvTwist_mix.ipynb | liuyao12/Ranger-Mish-ImageWoof-5 | 0504264b11e545b8d127ed8b139d9e200ded1f2f | [
"Apache-2.0"
]
| null | null | null | Woof_128_ConvTwist_mix.ipynb | liuyao12/Ranger-Mish-ImageWoof-5 | 0504264b11e545b8d127ed8b139d9e200ded1f2f | [
"Apache-2.0"
]
| null | null | null | Woof_128_ConvTwist_mix.ipynb | liuyao12/Ranger-Mish-ImageWoof-5 | 0504264b11e545b8d127ed8b139d9e200ded1f2f | [
"Apache-2.0"
]
| 1 | 2020-06-12T16:15:03.000Z | 2020-06-12T16:15:03.000Z | 84.994318 | 54,830 | 0.706951 | [
[
[
"<a href=\"https://colab.research.google.com/github/liuyao12/Ranger-Mish-ImageWoof-5/blob/master/Woof_128_ConvTwist_mix.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# ResNet with a Twist\n\n> ConvTwist + Ranger + Mish + MaxBlurPool + restrick",
"_____no_output_____"
],
[
"# setup and imports",
"_____no_output_____"
]
],
[
[
"# pip install git+https://github.com/kornia/kornia",
"_____no_output_____"
],
[
"pip install git+https://github.com/ayasyrev/model_constructor",
"Collecting git+https://github.com/ayasyrev/model_constructor\n Cloning https://github.com/ayasyrev/model_constructor to /tmp/pip-req-build-b3gs5d7j\n Running command git clone -q https://github.com/ayasyrev/model_constructor /tmp/pip-req-build-b3gs5d7j\nCollecting fastcore\n Downloading https://files.pythonhosted.org/packages/e2/6e/a18c0ff6cdca36915e65cf1690137134241a33d74ceef7882f4a63a6af55/fastcore-0.1.18-py3-none-any.whl\nRequirement already satisfied: torch in /usr/local/lib/python3.6/dist-packages (from model-constructor==0.1.1) (1.5.0+cu101)\nRequirement already satisfied: dataclasses>='0.7'; python_version < \"3.7\" in /usr/local/lib/python3.6/dist-packages (from fastcore->model-constructor==0.1.1) (0.7)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from fastcore->model-constructor==0.1.1) (1.18.5)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from torch->model-constructor==0.1.1) (0.16.0)\nBuilding wheels for collected packages: model-constructor\n Building wheel for model-constructor (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for model-constructor: filename=model_constructor-0.1.1-cp36-none-any.whl size=23583 sha256=3630ecc1cfbac3e6b6c29c693de0a206161c86ff8e12ad169af73ae46a9496be\n Stored in directory: /tmp/pip-ephem-wheel-cache-n_cwdche/wheels/5b/92/65/8093a307d5802f41f4a8776b40bc12b558e75a2a906ae8b683\nSuccessfully built model-constructor\nInstalling collected packages: fastcore, model-constructor\nSuccessfully installed fastcore-0.1.18 model-constructor-0.1.1\n"
],
[
"pip install git+https://github.com/ayasyrev/imagenette_experiments",
"Collecting git+https://github.com/ayasyrev/imagenette_experiments\n Cloning https://github.com/ayasyrev/imagenette_experiments to /tmp/pip-req-build-_ybqtee8\n Running command git clone -q https://github.com/ayasyrev/imagenette_experiments /tmp/pip-req-build-_ybqtee8\nRequirement already satisfied: fastai in /usr/local/lib/python3.6/dist-packages (from imagenette-experiments==0.0.1) (1.0.61)\nCollecting kornia\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/c2/60/f0c174c4a2a40b10b04b37c43f5afee3701cc145b48441a2dc5cf9286c3c/kornia-0.3.1-py2.py3-none-any.whl (158kB)\n\u001b[K |████████████████████████████████| 163kB 8.1MB/s \n\u001b[?25hRequirement already satisfied: model-constructor in /usr/local/lib/python3.6/dist-packages (from imagenette-experiments==0.0.1) (0.1.1)\nRequirement already satisfied: bottleneck in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (1.3.2)\nRequirement already satisfied: Pillow in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (7.0.0)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (2.23.0)\nRequirement already satisfied: matplotlib in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (3.2.1)\nRequirement already satisfied: nvidia-ml-py3 in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (7.352.0)\nRequirement already satisfied: torch>=1.0.0 in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (1.5.0+cu101)\nRequirement already satisfied: dataclasses; python_version < \"3.7\" in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (0.7)\nRequirement already satisfied: pandas in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (1.0.4)\nRequirement already satisfied: fastprogress>=0.2.1 in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (0.2.3)\nRequirement already satisfied: pyyaml in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (3.13)\nRequirement already satisfied: spacy>=2.0.18; python_version < \"3.8\" in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (2.2.4)\nRequirement already satisfied: numexpr in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (2.7.1)\nRequirement already satisfied: numpy>=1.15 in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (1.18.5)\nRequirement already satisfied: packaging in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (20.4)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (1.4.1)\nRequirement already satisfied: beautifulsoup4 in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (4.6.3)\nRequirement already satisfied: torchvision in /usr/local/lib/python3.6/dist-packages (from fastai->imagenette-experiments==0.0.1) (0.6.0+cu101)\nRequirement already satisfied: fastcore in /usr/local/lib/python3.6/dist-packages (from model-constructor->imagenette-experiments==0.0.1) (0.1.18)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->fastai->imagenette-experiments==0.0.1) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->fastai->imagenette-experiments==0.0.1) (2020.4.5.1)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->fastai->imagenette-experiments==0.0.1) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->fastai->imagenette-experiments==0.0.1) (2.9)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib->fastai->imagenette-experiments==0.0.1) (1.2.0)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib->fastai->imagenette-experiments==0.0.1) (2.8.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib->fastai->imagenette-experiments==0.0.1) (2.4.7)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib->fastai->imagenette-experiments==0.0.1) (0.10.0)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from torch>=1.0.0->fastai->imagenette-experiments==0.0.1) (0.16.0)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas->fastai->imagenette-experiments==0.0.1) (2018.9)\nRequirement already satisfied: preshed<3.1.0,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (3.0.2)\nRequirement already satisfied: srsly<1.1.0,>=1.0.2 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (1.0.2)\nRequirement already satisfied: catalogue<1.1.0,>=0.0.7 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (1.0.0)\nRequirement already satisfied: tqdm<5.0.0,>=4.38.0 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (4.41.1)\nRequirement already satisfied: cymem<2.1.0,>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (2.0.3)\nRequirement already satisfied: blis<0.5.0,>=0.4.0 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (0.4.1)\nRequirement already satisfied: murmurhash<1.1.0,>=0.28.0 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (1.0.2)\nRequirement already satisfied: thinc==7.4.0 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (7.4.0)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (47.1.1)\nRequirement already satisfied: wasabi<1.1.0,>=0.4.0 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (0.6.0)\nRequirement already satisfied: plac<1.2.0,>=0.9.6 in /usr/local/lib/python3.6/dist-packages (from spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (1.1.3)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from packaging->fastai->imagenette-experiments==0.0.1) (1.12.0)\nRequirement already satisfied: importlib-metadata>=0.20; python_version < \"3.8\" in /usr/local/lib/python3.6/dist-packages (from catalogue<1.1.0,>=0.0.7->spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (1.6.0)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.6/dist-packages (from importlib-metadata>=0.20; python_version < \"3.8\"->catalogue<1.1.0,>=0.0.7->spacy>=2.0.18; python_version < \"3.8\"->fastai->imagenette-experiments==0.0.1) (3.1.0)\nBuilding wheels for collected packages: imagenette-experiments\n Building wheel for imagenette-experiments (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for imagenette-experiments: filename=imagenette_experiments-0.0.1-cp36-none-any.whl size=15332 sha256=e292e396c4044928405e235e53aaa41efd8fd1bf5e3286f3394937c75bb54bfd\n Stored in directory: /tmp/pip-ephem-wheel-cache-3ykw6guo/wheels/af/99/98/2212941f45a18bf6d191f56c39e3569138414560c76defc0d4\nSuccessfully built imagenette-experiments\nInstalling collected packages: kornia, imagenette-experiments\nSuccessfully installed imagenette-experiments-0.0.1 kornia-0.3.1\n"
],
[
"from imagenette_experiments.train_utils import *",
"_____no_output_____"
],
[
"from kornia.contrib import MaxBlurPool2d",
"_____no_output_____"
],
[
"from fastai.basic_train import *\nfrom fastai.vision import *\n# from fastai.script import *\nfrom model_constructor.net import Net, act_fn\nfrom model_constructor.layers import SimpleSelfAttention, ConvLayer",
"_____no_output_____"
]
],
[
[
"# Twist",
"_____no_output_____"
]
],
[
[
"class ConvTwist(nn.Module): # replacing 3x3 Conv2d with in_channels=out_channels\n def __init__(self, ni, nf):\n super(ConvTwist, self).__init__()\n self.twist = True # and n>=128\n self.expansion = 1 # n//4\n nf *= self.expansion\n self.conv = nn.Conv2d(nf, nf, kernel_size=3, padding=1, bias=False, groups=nf)\n if self.twist:\n # self.conv_x = nn.Conv2d(nf, nf, kernel_size=3, padding=1, bias=False, groups=nf)\n # self.conv_y = nn.Conv2d(nf, nf, kernel_size=3, padding=1, bias=False, groups=nf)\n std = self.conv.weight.std().item()\n self.coeff_Ax = nn.Parameter(torch.empty((nf, 1)).normal_(0, std), requires_grad=True)\n self.coeff_Ay = nn.Parameter(torch.empty((nf, 1)).normal_(0, std), requires_grad=True)\n\n def DD(self, order=3):\n I = torch.Tensor([[0,0,0],[0,1,0],[0,0,0]]).view(1,1,3,3)\n D_x = torch.Tensor([[-1,0,1],[-2,0,2],[-1,0,1]]).view(1,1,3,3) / 10\n D_y = torch.Tensor([[1,2,1],[0,0,0],[-1,-2,-1]]).view(1,1,3,3) / 10\n def convolution(K1, K2):\n padding = K2.size()[2]//2+1\n return F.conv2d(K1, K2.flip(2).flip(3), padding=padding)\n D_xx = convolution(D_x, D_x).view(1,1,5,5)\n D_yy = convolution(D_y, D_y).view(1,1,5,5)\n D_xy = convolution(D_x, D_y).view(1,1,5,5)\n return {'x': D_x, 'y': D_y, 'xx': D_xx, 'yy': D_yy, 'xy': D_xy}\n\n def kernel(self, coeff_x, coeff_y):\n D_x = torch.Tensor([[-1,0,1],[-2,0,2],[-1,0,1]]).to(coeff_x.device)\n D_y = torch.Tensor([[1,2,1],[0,0,0],[-1,-2,-1]]).to(coeff_x.device)\n return coeff_x[:,:,None,None] * D_x + coeff_y[:,:,None,None] * D_y\n\n def _conv(self, x, kernel=None):\n if kernel is None:\n kernel = self.conv.weight\n groups = kernel.size()[0]//kernel.size()[1]\n return F.conv2d(x, kernel, padding=1, groups=groups)\n\n def symmetrize(self, conv_wt):\n conv_wt.data = (conv_wt-conv_wt.flip(2).flip(3))/2\n # if self.same:\n # n = conv_wt.size()[1]\n # for i in range(self.groups):\n # conv_wt.data[n*i:n*(i+1)] = (conv_wt[n*i:n*(i+1)] + torch.transpose(conv_wt[n*i:n*(i+1)],0,1)) / 2\n\n def forward(self, x):\n N,C,H,W = x.size()\n CC = C*self.expansion\n if self.expansion>1:\n x = torch.cat([x]*self.expansion, dim=1)\n x1 = x\n if self.twist:\n XX = torch.from_numpy(np.indices((1,1,H,W))[3]*2/W-1).type(x.dtype).to(x.device)\n YY = torch.from_numpy(np.indices((1,1,H,W))[2]*2/H-1).type(x.dtype).to(x.device)\n # mask = F.relu(x1)+0.01\n # mass = mask.sum((2,3))\n # Xbar = (XX*mask).sum((2,3))/mass\n # Ybar = (YY*mask).sum((2,3))/mass\n # XX = XX.expand((N,CC,H,W)) - Xbar[:,:,None,None]\n # YY = YY.expand((N,CC,H,W)) - Ybar[:,:,None,None]\n # self.symmetrize(self.conv_x.weight)\n # self.conv_y.weight.data = self.conv_x.weight.transpose(2,3).flip(3)\n kernel_x = self.kernel(self.coeff_Ax, self.coeff_Ay)\n kernel_y = kernel_x.transpose(2,3).flip(3)\n for _ in range(3):\n # x1 = x1 + self.conv(x1) + XX * self.conv_x(x1) + YY * self.conv_y(x1)\n x1 = x1 + self._conv(x1) + XX * self._conv(x1, kernel_x) + YY * self._conv(x1, kernel_y)\n else:\n for _ in range(3):\n x1 = x1 + self.conv(x1)\n x1 = x1 - x\n y = x1[:,:C]\n # y = torch.cat([y[:,1:], y[:,:1]], dim=1)\n y = y.view(N,C//4,4,H,W)\n y = torch.cat([y[:,:,1:], y[:,:,:1]], dim=2).view(N,C,H,W)\n for i in range(1,self.expansion):\n x2 = x1[:,C*i:C*(i+1)]\n # x2 = x2.view(N,C//4,4,H,W).sum((2))\n # x2 = x2.permute(0,2,1,3,4).reshape(N,C,H,W)\n x2 = torch.cat([x2[:,-4:], x2[:,:-4]], dim=1)\n # x2 = x2.view(N,4,C//4,H,W)\n # x2 = torch.cat([x2[:,1:], x2[:,:1]], dim=1).view(N,C,H,W)\n y[:,:] += x2\n return y",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nfig, axs = plt.subplots(2,2,figsize=(16,16))\naxs = axs.ravel()\ni=0\nfor n in [64,128,256,512]:\n model = ConvTwist(n)\n mask = model.make_mask(n).squeeze()\n axs[i].imshow(mask)\n i+=1",
"torch.Size([64, 64, 3, 3])\ntorch.Size([128, 128, 3, 3])\ntorch.Size([256, 256, 3, 3])\ntorch.Size([512, 512, 3, 3])\n"
]
],
[
[
"# ResBlock",
"_____no_output_____"
]
],
[
[
"class NewLayer(nn.Sequential):\n \"\"\"Basic conv layers block\"\"\"\n def __init__(self, ni, nf, ks=3, stride=1,\n act=True, act_fn=nn.ReLU(inplace=True),\n bn_layer=True, bn_1st=True, zero_bn=False,\n padding=None, bias=False, groups=1, **kwargs):\n\n if padding==None: padding = ks//2\n if ni%nf==0 and ks==3: layers = [('ConvTwist', ConvTwist(ni,nf))]\n else: layers = [('Conv{}x{}'.format(ks,ks), \n nn.Conv2d(ni, nf, ks, stride=stride, padding=padding, bias=bias, groups=groups))]\n\n act_bn = [('act_fn', act_fn)] if act else []\n if bn_layer:\n bn = nn.BatchNorm2d(nf)\n nn.init.constant_(bn.weight, 0. if zero_bn else 1.)\n act_bn += [('bn', bn)]\n if bn_1st: act_bn.reverse()\n layers += act_bn\n super().__init__(OrderedDict(layers))",
"_____no_output_____"
],
[
"class NewResBlock(Module):\n def __init__(self, expansion, ni, nh, stride=1,\n conv_layer=ConvLayer, act_fn=act_fn, bn_1st=True,\n pool=nn.AvgPool2d(2, ceil_mode=True), sa=False, sym=False, zero_bn=True):\n nf,ni = nh*expansion,ni*expansion\n conv_layer = NewLayer\n self.reduce = noop if stride==1 else pool\n layers = [(f\"conv_0\", conv_layer(ni, nh, 3, act_fn=act_fn, bn_1st=bn_1st)),\n (f\"conv_1\", conv_layer(nh, nf, 3, zero_bn=zero_bn, act=False, bn_1st=bn_1st))\n ] if expansion == 1 else [\n (f\"conv_0\", conv_layer(ni, nh, 1, act_fn=act_fn, bn_1st=bn_1st)),\n (f\"conv_1\", conv_layer(nh, nh, 3, act_fn=act_fn, bn_1st=bn_1st)),\n (f\"conv_2\", conv_layer(nh, nf, 1, zero_bn=zero_bn, act=False, bn_1st=bn_1st))\n ]\n if sa: layers.append(('sa', SimpleSelfAttention(nf,ks=1,sym=sym)))\n self.convs = nn.Sequential(OrderedDict(layers))\n self.idconv = noop if ni==nf else conv_layer(ni, nf, 1, act=False, bn_1st=bn_1st)\n self.merge = act_fn\n\n def forward(self, x):\n o = self.reduce(x)\n return self.merge(self.convs(o) + self.idconv(o))",
"_____no_output_____"
],
[
"A = torch.arange(15).view(3,5)\nprint(A)\nprint(A.sum(1))",
"tensor([[ 0, 1, 2, 3, 4],\n [ 5, 6, 7, 8, 9],\n [10, 11, 12, 13, 14]])\ntensor([10, 35, 60])\n"
]
],
[
[
"# Model Constructor",
"_____no_output_____"
]
],
[
[
"model = Net(c_out=10, layers=[3,6,8,3], expansion=4)\nmodel.block = NewResBlock\n# model.conv_layer = NewLayer # for the stem\npool = MaxBlurPool2d(3, True)\nmodel.pool = pool\nmodel.stem_pool = pool\nmodel.stem_sizes = [3,32,64,64]\nmodel.act_fn = Mish()\nmodel.sa = True\nres = []\n# epochs = [5]*3\nepochs = [20]",
"_____no_output_____"
]
],
[
[
"# Runs and results\n\n",
"_____no_output_____"
]
],
[
[
"for e in epochs:\n mixup=0 if e<=20 else 0.2\n learn = get_learn(model=model, size=192, bs=32, mixup=mixup)\n learn.fit_fc(e, lr=4e-3, moms=(0.95,0.95), start_pct=0.72)\n res += [learn.recorder.metrics[-1][0].item()]\nprint([round(x, 6) for x in res], sum(res)/len(res))",
"data path /root/.fastai/data/imagewoof2\n"
]
],
[
[
"Conv2d, full-connected ResNet50 \\\\\n`[0.777297, 0.783151, 0.793077] 0.7845083475112915`\n\nConvTwist, twist=False, depthwise, iters=1 \\\\\n`[0.807585, 0.81013, 0.811657] 0.8097904523213705`\n\nConvTwist, twist=False, depthwise, iters=3 \\\\\n`[0.813693, 0.822092, 0.804021] 0.8132688403129578` \\\\\n`0.874268`\n\nConvTwist, twist=True, depthwise, iters=1 \\\\\n`[0.806821, 0.797149, 0.805039] 0.8030033111572266`\n\nConvTwist, twist=True, depthwise, iters=3 \\\\\n`[0.803512, 0.805294, 0.798676] 0.8024942676226298` \\\\\n`0.87885`\n\n",
"_____no_output_____"
]
],
[
[
"# [4,6,8,10], size=192, epochs=80\nfor i in range(80):\n print('epoch {} {}'.format(i, learn.recorder.metrics[i][0].item()))",
"epoch 0 0.4026469886302948\nepoch 1 0.5433952808380127\nepoch 2 0.6449478268623352\nepoch 3 0.6805803179740906\nepoch 4 0.7218121886253357\nepoch 5 0.7612624168395996\nepoch 6 0.7750063538551331\nepoch 7 0.7683889269828796\nepoch 8 0.7907864451408386\nepoch 9 0.8139475584030151\nepoch 10 0.8106388449668884\nepoch 11 0.8220921158790588\nepoch 12 0.8307457566261292\nepoch 13 0.8452532291412354\nepoch 14 0.8294731378555298\nepoch 15 0.8388903141021729\nepoch 16 0.8460168242454529\nepoch 17 0.8348180055618286\nepoch 18 0.8442351818084717\nepoch 19 0.8388903141021729\nepoch 20 0.836090624332428\nepoch 21 0.840926468372345\nepoch 22 0.8439806699752808\nepoch 23 0.8572155833244324\nepoch 24 0.8597607612609863\nepoch 25 0.8439806699752808\nepoch 26 0.8442351818084717\nepoch 27 0.8467803597450256\nepoch 28 0.8500890731811523\nepoch 29 0.8498345613479614\nepoch 30 0.8541613817214966\nepoch 31 0.8424535393714905\nepoch 32 0.8567065596580505\nepoch 33 0.8401628732681274\nepoch 34 0.855942964553833\nepoch 35 0.8518707156181335\nepoch 36 0.8488165140151978\nepoch 37 0.8531432747840881\nepoch 38 0.8658691644668579\nepoch 39 0.86077880859375\nepoch 40 0.8597607612609863\nepoch 41 0.8511071801185608\nepoch 42 0.8508526086807251\nepoch 43 0.8544158935546875\nepoch 44 0.858488142490387\nepoch 45 0.8635784983634949\nepoch 46 0.8635784983634949\nepoch 47 0.8663781881332397\nepoch 48 0.8574700951576233\nepoch 49 0.8602697849273682\nepoch 50 0.855942964553833\nepoch 51 0.8526342511177063\nepoch 52 0.8638330101966858\nepoch 53 0.8503435850143433\nepoch 54 0.8623059391975403\nepoch 55 0.8635784983634949\nepoch 56 0.8638330101966858\nepoch 57 0.8551794290542603\nepoch 58 0.8610333204269409\nepoch 59 0.8600152730941772\nepoch 60 0.8600152730941772\nepoch 61 0.8579791188240051\nepoch 62 0.8653601408004761\nepoch 63 0.8615424036979675\nepoch 64 0.8668872714042664\nepoch 65 0.8707050085067749\nepoch 66 0.8729956746101379\nepoch 67 0.8796131610870361\nepoch 68 0.8747773170471191\nepoch 69 0.8773224949836731\nepoch 70 0.8806312084197998\nepoch 71 0.8821583390235901\nepoch 72 0.8811402320861816\nepoch 73 0.8831763863563538\nepoch 74 0.8826673626899719\nepoch 75 0.8862305879592896\nepoch 76 0.8844489455223083\nepoch 77 0.8869941234588623\nepoch 78 0.8854670524597168\nepoch 79 0.8847034573554993\n"
],
[
"# [4,6,8,10], size=192, epochs=80\nfor i in range(80):\n print('epoch {} {}'.format(i, learn.recorder.metrics[i][0].item()))",
"epoch 0 0.4059557020664215\nepoch 1 0.47213029861450195\nepoch 2 0.5886994004249573\nepoch 3 0.6602188944816589\nepoch 4 0.7177398800849915\nepoch 5 0.7317383289337158\nepoch 6 0.7602443099021912\nepoch 7 0.7795876860618591\nepoch 8 0.7912954688072205\nepoch 9 0.7887502908706665\nepoch 10 0.7793331742286682\nepoch 11 0.8027487993240356\nepoch 12 0.8149656653404236\nepoch 13 0.8045304417610168\nepoch 14 0.8139475584030151\nepoch 15 0.8180198669433594\nepoch 16 0.8022397756576538\nepoch 17 0.8337999582290649\nepoch 18 0.8340544700622559\nepoch 19 0.8315092921257019\nepoch 20 0.8404173851013184\nepoch 21 0.8449987173080444\nepoch 22 0.8434716463088989\nepoch 23 0.8460168242454529\nepoch 24 0.8327818512916565\nepoch 25 0.8567065596580505\nepoch 26 0.8623059391975403\nepoch 27 0.8483074307441711\nepoch 28 0.8589972257614136\nepoch 29 0.8577246069908142\nepoch 30 0.8551794290542603\nepoch 31 0.8531432747840881\nepoch 32 0.8500890731811523\nepoch 33 0.8569610714912415\nepoch 34 0.8628149628639221\nepoch 35 0.8648511171340942\nepoch 36 0.8521252274513245\nepoch 37 0.8666327595710754\nepoch 38 0.8554339408874512\nepoch 39 0.8531432747840881\nepoch 40 0.8488165140151978\nepoch 41 0.8505980968475342\nepoch 42 0.858488142490387\nepoch 43 0.8691779375076294\nepoch 44 0.8615424036979675\nepoch 45 0.86077880859375\nepoch 46 0.8549249172210693\nepoch 47 0.8686688542366028\nepoch 48 0.8460168242454529\nepoch 49 0.8645966053009033\nepoch 50 0.8689233660697937\nepoch 51 0.8623059391975403\nepoch 52 0.8500890731811523\nepoch 53 0.8595062494277954\nepoch 54 0.8612878322601318\nepoch 55 0.8612878322601318\nepoch 56 0.8628149628639221\nepoch 57 0.8707050085067749\nepoch 58 0.8750318288803101\nepoch 59 0.8750318288803101\nepoch 60 0.8709595203399658\nepoch 61 0.8714685440063477\nepoch 62 0.8663781881332397\nepoch 63 0.8740137219429016\nepoch 64 0.877577006816864\nepoch 65 0.8747773170471191\nepoch 66 0.8826673626899719\nepoch 67 0.8836854100227356\nepoch 68 0.8839399218559265\nepoch 69 0.8867396116256714\nepoch 70 0.8829218745231628\nepoch 71 0.8890302777290344\nepoch 72 0.8928480744361877\nepoch 73 0.8885212540626526\nepoch 74 0.8915754556655884\nepoch 75 0.8948841691017151\nepoch 76 0.8941206336021423\nepoch 77 0.8981929421424866\nepoch 78 0.8971748352050781\nepoch 79 0.8979384303092957\n"
],
[
"res = [0.762026, 0.753881, 0.763553, 0.772716, 0.758717]\nsum(res) / len(res)",
"_____no_output_____"
],
[
"res = [0.764826, 0.762789, 0.760244, 0.770425, 0.764826]\nsum(res) / len(res)",
"_____no_output_____"
],
[
"res = [0.775770, 0.762026, 0.768389, 0.748791, 0.763553]\nsum(res) / len(res)",
"_____no_output_____"
],
[
"res = [0.761262, 0.749809, 0.773988, 0.764571, 0.762026]\nsum(res) / len(res)",
"_____no_output_____"
],
[
"for i in range(80):\n print('epoch {} {}'.format(i, learn.recorder.metrics[i][0].item()))",
"_____no_output_____"
],
[
"for i in range(80):\n print('epoch {} {}'.format(i, learn.recorder.metrics[i][0].item()))",
"epoch 0 0.35607025027275085\nepoch 1 0.4662764072418213\nepoch 2 0.5454314351081848\nepoch 3 0.601679801940918\nepoch 4 0.6627640724182129\nepoch 5 0.6864342093467712\nepoch 6 0.7258844375610352\nepoch 7 0.7520997524261475\nepoch 8 0.7638075947761536\nepoch 9 0.7737337946891785\nepoch 10 0.7793331742286682\nepoch 11 0.7778060436248779\nepoch 12 0.7813693284988403\nepoch 13 0.7979129552841187\nepoch 14 0.8027487993240356\nepoch 15 0.8083481788635254\nepoch 16 0.8139475584030151\nepoch 17 0.8164927363395691\nepoch 18 0.8185288906097412\nepoch 19 0.8343089818954468\nepoch 20 0.8248918056488037\nepoch 21 0.828964114189148\nepoch 22 0.8304912447929382\nepoch 23 0.8307457566261292\nepoch 24 0.8373631834983826\nepoch 25 0.8195469379425049\nepoch 26 0.8279460668563843\nepoch 27 0.8376176953315735\nepoch 28 0.8439806699752808\nepoch 29 0.8332909345626831\nepoch 30 0.8411809802055359\nepoch 31 0.8358361124992371\nepoch 32 0.8282005786895752\nepoch 33 0.8297276496887207\nepoch 34 0.8475438952445984\nepoch 35 0.8421990275382996\nepoch 36 0.8353270292282104\nepoch 37 0.8442351818084717\nepoch 38 0.8355816006660461\nepoch 39 0.8447442054748535\nepoch 40 0.8439806699752808\nepoch 41 0.8493255376815796\nepoch 42 0.8434716463088989\nepoch 43 0.8505980968475342\nepoch 44 0.8396538496017456\nepoch 45 0.8546704053878784\nepoch 46 0.8495800495147705\nepoch 47 0.8500890731811523\nepoch 48 0.8294731378555298\nepoch 49 0.8414354920387268\nepoch 50 0.8432170748710632\nepoch 51 0.8518707156181335\nepoch 52 0.8493255376815796\nepoch 53 0.8386358022689819\nepoch 54 0.8483074307441711\nepoch 55 0.8406718969345093\nepoch 56 0.8444896936416626\nepoch 57 0.8511071801185608\nepoch 58 0.831254780292511\nepoch 59 0.8472893834114075\nepoch 60 0.8595062494277954\nepoch 61 0.8528887629508972\nepoch 62 0.8503435850143433\nepoch 63 0.8589972257614136\nepoch 64 0.8488165140151978\nepoch 65 0.8544158935546875\nepoch 66 0.8600152730941772\nepoch 67 0.8625604510307312\nepoch 68 0.8628149628639221\nepoch 69 0.8686688542366028\nepoch 70 0.870450496673584\nepoch 71 0.8740137219429016\nepoch 72 0.8724866509437561\nepoch 73 0.8737592101097107\nepoch 74 0.877577006816864\nepoch 75 0.875286340713501\nepoch 76 0.8780860304832458\nepoch 77 0.8750318288803101\nepoch 78 0.877577006816864\nepoch 79 0.8757953643798828\n"
],
[
"for i in range(80):\n print('epoch {} {}'.format(i, learn.recorder.metrics[i][0].item()))",
"epoch 0 0.39857470989227295\nepoch 1 0.4728938639163971\nepoch 2 0.5808093547821045\nepoch 3 0.6230593323707581\nepoch 4 0.6877067685127258\nepoch 5 0.7093408107757568\nepoch 6 0.7437006831169128\nepoch 7 0.7421735525131226\nepoch 8 0.7686434388160706\nepoch 9 0.7790786623954773\nepoch 10 0.7788241505622864\nepoch 11 0.7971494197845459\nepoch 12 0.7961313128471375\nepoch 13 0.7739883065223694\nepoch 14 0.8009671568870544\nepoch 15 0.8129295110702515\nepoch 16 0.8030033111572266\nepoch 17 0.8180198669433594\nepoch 18 0.8129295110702515\nepoch 19 0.8172562718391418\nepoch 20 0.8292186260223389\nepoch 21 0.828709602355957\nepoch 22 0.819037914276123\nepoch 23 0.8297276496887207\nepoch 24 0.8279460668563843\nepoch 25 0.8284550905227661\nepoch 26 0.8404173851013184\nepoch 27 0.8465258479118347\nepoch 28 0.821583092212677\nepoch 29 0.8343089818954468\nepoch 30 0.8256554007530212\nepoch 31 0.8388903141021729\nepoch 32 0.8437261581420898\nepoch 33 0.8404173851013184\nepoch 34 0.8404173851013184\nepoch 35 0.8399083614349365\nepoch 36 0.8388903141021729\nepoch 37 0.8355816006660461\nepoch 38 0.8449987173080444\nepoch 39 0.8411809802055359\nepoch 40 0.8365996479988098\nepoch 41 0.8460168242454529\nepoch 42 0.8467803597450256\nepoch 43 0.8386358022689819\nepoch 44 0.8345634937286377\nepoch 45 0.8427080512046814\nepoch 46 0.8381267786026001\nepoch 47 0.8424535393714905\nepoch 48 0.8493255376815796\nepoch 49 0.8457622528076172\nepoch 50 0.8488165140151978\nepoch 51 0.8404173851013184\nepoch 52 0.8485620021820068\nepoch 53 0.8439806699752808\nepoch 54 0.8470348715782166\nepoch 55 0.8485620021820068\nepoch 56 0.8528887629508972\nepoch 57 0.8452532291412354\nepoch 58 0.8483074307441711\nepoch 59 0.8447442054748535\nepoch 60 0.8447442054748535\nepoch 61 0.8432170748710632\nepoch 62 0.8508526086807251\nepoch 63 0.8551794290542603\nepoch 64 0.8539068698883057\nepoch 65 0.8513616919517517\nepoch 66 0.8551794290542603\nepoch 67 0.8615424036979675\nepoch 68 0.8663781881332397\nepoch 69 0.868159830570221\nepoch 70 0.8732501864433289\nepoch 71 0.8676508069038391\nepoch 72 0.8719776272773743\nepoch 73 0.8760498762130737\nepoch 74 0.8729956746101379\nepoch 75 0.8768134117126465\nepoch 76 0.879867672920227\nepoch 77 0.8788495659828186\nepoch 78 0.880122184753418\nepoch 79 0.8803766965866089\n"
],
[
"for _ in range(1):\n learn = get_learn(model=model, size=128, bs=32, mixup=0.5)\n learn.fit_fc(80, lr=4e-3, moms=(0.95,0.95), start_pct=0.72)",
"Downloading https://s3.amazonaws.com/fast-ai-imageclas/imagewoof2\n"
],
[
"learn.recorder.plot_losses()",
"_____no_output_____"
],
[
"for _ in range(3):\n learn = get_learn(model=model, size=128, bs=32, mixup=0)\n learn.fit_fc(5, lr=4e-3, moms=(0.95,0.95), start_pct=0.72)",
"data path /root/.fastai/data/imagewoof2\nLearn path /root/.fastai/data/imagewoof2\n"
],
[
"for name, param in learn.model.body.named_parameters():\n if 'ConvTwist' in name:\n print(name, param.size())",
"l_0.bl_0.convs.conv_1.ConvTwist.conv.weight torch.Size([64, 8, 3, 3])\nl_0.bl_0.convs.conv_1.ConvTwist.conv_x.weight torch.Size([64, 8, 3, 3])\nl_0.bl_0.convs.conv_1.ConvTwist.conv_y.weight torch.Size([64, 8, 3, 3])\nl_0.bl_1.convs.conv_1.ConvTwist.conv.weight torch.Size([64, 8, 3, 3])\nl_0.bl_1.convs.conv_1.ConvTwist.conv_x.weight torch.Size([64, 8, 3, 3])\nl_0.bl_1.convs.conv_1.ConvTwist.conv_y.weight torch.Size([64, 8, 3, 3])\nl_0.bl_2.convs.conv_1.ConvTwist.conv.weight torch.Size([64, 8, 3, 3])\nl_0.bl_2.convs.conv_1.ConvTwist.conv_x.weight torch.Size([64, 8, 3, 3])\nl_0.bl_2.convs.conv_1.ConvTwist.conv_y.weight torch.Size([64, 8, 3, 3])\nl_1.bl_0.convs.conv_1.ConvTwist.conv.weight torch.Size([128, 8, 3, 3])\nl_1.bl_0.convs.conv_1.ConvTwist.conv_x.weight torch.Size([128, 8, 3, 3])\nl_1.bl_0.convs.conv_1.ConvTwist.conv_y.weight torch.Size([128, 8, 3, 3])\nl_1.bl_1.convs.conv_1.ConvTwist.conv.weight torch.Size([128, 8, 3, 3])\nl_1.bl_1.convs.conv_1.ConvTwist.conv_x.weight torch.Size([128, 8, 3, 3])\nl_1.bl_1.convs.conv_1.ConvTwist.conv_y.weight torch.Size([128, 8, 3, 3])\nl_1.bl_2.convs.conv_1.ConvTwist.conv.weight torch.Size([128, 8, 3, 3])\nl_1.bl_2.convs.conv_1.ConvTwist.conv_x.weight torch.Size([128, 8, 3, 3])\nl_1.bl_2.convs.conv_1.ConvTwist.conv_y.weight torch.Size([128, 8, 3, 3])\nl_1.bl_3.convs.conv_1.ConvTwist.conv.weight torch.Size([128, 8, 3, 3])\nl_1.bl_3.convs.conv_1.ConvTwist.conv_x.weight torch.Size([128, 8, 3, 3])\nl_1.bl_3.convs.conv_1.ConvTwist.conv_y.weight torch.Size([128, 8, 3, 3])\nl_2.bl_0.convs.conv_1.ConvTwist.conv.weight torch.Size([256, 8, 3, 3])\nl_2.bl_0.convs.conv_1.ConvTwist.conv_x.weight torch.Size([256, 8, 3, 3])\nl_2.bl_0.convs.conv_1.ConvTwist.conv_y.weight torch.Size([256, 8, 3, 3])\nl_2.bl_1.convs.conv_1.ConvTwist.conv.weight torch.Size([256, 8, 3, 3])\nl_2.bl_1.convs.conv_1.ConvTwist.conv_x.weight torch.Size([256, 8, 3, 3])\nl_2.bl_1.convs.conv_1.ConvTwist.conv_y.weight torch.Size([256, 8, 3, 3])\nl_2.bl_2.convs.conv_1.ConvTwist.conv.weight torch.Size([256, 8, 3, 3])\nl_2.bl_2.convs.conv_1.ConvTwist.conv_x.weight torch.Size([256, 8, 3, 3])\nl_2.bl_2.convs.conv_1.ConvTwist.conv_y.weight torch.Size([256, 8, 3, 3])\nl_2.bl_3.convs.conv_1.ConvTwist.conv.weight torch.Size([256, 8, 3, 3])\nl_2.bl_3.convs.conv_1.ConvTwist.conv_x.weight torch.Size([256, 8, 3, 3])\nl_2.bl_3.convs.conv_1.ConvTwist.conv_y.weight torch.Size([256, 8, 3, 3])\nl_2.bl_4.convs.conv_1.ConvTwist.conv.weight torch.Size([256, 8, 3, 3])\nl_2.bl_4.convs.conv_1.ConvTwist.conv_x.weight torch.Size([256, 8, 3, 3])\nl_2.bl_4.convs.conv_1.ConvTwist.conv_y.weight torch.Size([256, 8, 3, 3])\nl_2.bl_5.convs.conv_1.ConvTwist.conv.weight torch.Size([256, 8, 3, 3])\nl_2.bl_5.convs.conv_1.ConvTwist.conv_x.weight torch.Size([256, 8, 3, 3])\nl_2.bl_5.convs.conv_1.ConvTwist.conv_y.weight torch.Size([256, 8, 3, 3])\nl_3.bl_0.convs.conv_1.ConvTwist.conv.weight torch.Size([512, 8, 3, 3])\nl_3.bl_0.convs.conv_1.ConvTwist.conv_x.weight torch.Size([512, 8, 3, 3])\nl_3.bl_0.convs.conv_1.ConvTwist.conv_y.weight torch.Size([512, 8, 3, 3])\nl_3.bl_1.convs.conv_1.ConvTwist.conv.weight torch.Size([512, 8, 3, 3])\nl_3.bl_1.convs.conv_1.ConvTwist.conv_x.weight torch.Size([512, 8, 3, 3])\nl_3.bl_1.convs.conv_1.ConvTwist.conv_y.weight torch.Size([512, 8, 3, 3])\nl_3.bl_2.convs.conv_1.ConvTwist.conv.weight torch.Size([512, 8, 3, 3])\nl_3.bl_2.convs.conv_1.ConvTwist.conv_x.weight torch.Size([512, 8, 3, 3])\nl_3.bl_2.convs.conv_1.ConvTwist.conv_y.weight torch.Size([512, 8, 3, 3])\n"
],
[
"for i in range(80):\n print('epoch {} {}'.format(i, learn.recorder.metrics[i][0].item()))",
"epoch 0 0.3866123557090759\nepoch 1 0.4499872624874115\nepoch 2 0.5008907914161682\nepoch 3 0.5970984697341919\nepoch 4 0.6304402947425842\nepoch 5 0.6688724756240845\nepoch 6 0.6986510753631592\nepoch 7 0.7327564358711243\nepoch 8 0.7401374578475952\nepoch 9 0.768134355545044\nepoch 10 0.7643166184425354\nepoch 11 0.7688979506492615\nepoch 12 0.7673708200454712\nepoch 13 0.7956222891807556\nepoch 14 0.7930771112442017\nepoch 15 0.814202070236206\nepoch 16 0.8058030009269714\nepoch 17 0.8180198669433594\nepoch 18 0.8024942874908447\nepoch 19 0.8170017600059509\nepoch 20 0.8088572025299072\nepoch 21 0.8198014497756958\nepoch 22 0.8213285803794861\nepoch 23 0.8187834024429321\nepoch 24 0.8282005786895752\nepoch 25 0.828964114189148\nepoch 26 0.8261644244194031\nepoch 27 0.8139475584030151\nepoch 28 0.836090624332428\nepoch 29 0.8332909345626831\nepoch 30 0.8340544700622559\nepoch 31 0.8345634937286377\nepoch 32 0.833545446395874\nepoch 33 0.8368541598320007\nepoch 34 0.8381267786026001\nepoch 35 0.8294731378555298\nepoch 36 0.8294731378555298\nepoch 37 0.8452532291412354\nepoch 38 0.833545446395874\nepoch 39 0.8315092921257019\nepoch 40 0.8429625630378723\nepoch 41 0.8432170748710632\nepoch 42 0.8299821615219116\nepoch 43 0.8526342511177063\nepoch 44 0.8388903141021729\nepoch 45 0.836090624332428\nepoch 46 0.8457622528076172\nepoch 47 0.8401628732681274\nepoch 48 0.8505980968475342\nepoch 49 0.8421990275382996\nepoch 50 0.8404173851013184\nepoch 51 0.8414354920387268\nepoch 52 0.838381290435791\nepoch 53 0.8513616919517517\nepoch 54 0.838381290435791\nepoch 55 0.8505980968475342\nepoch 56 0.8444896936416626\nepoch 57 0.8419445157051086\nepoch 58 0.8477984070777893\nepoch 59 0.8526342511177063\nepoch 60 0.8442351818084717\nepoch 61 0.8455077409744263\nepoch 62 0.8480529189109802\nepoch 63 0.8414354920387268\nepoch 64 0.8546704053878784\nepoch 65 0.8541613817214966\nepoch 66 0.8505980968475342\nepoch 67 0.8651056289672852\nepoch 68 0.8635784983634949\nepoch 69 0.868159830570221\nepoch 70 0.8653601408004761\nepoch 71 0.8673962950706482\nepoch 72 0.8750318288803101\nepoch 73 0.8757953643798828\nepoch 74 0.8793585896492004\nepoch 75 0.8791040778160095\nepoch 76 0.8785950541496277\nepoch 77 0.8796131610870361\nepoch 78 0.8803766965866089\nepoch 79 0.8788495659828186\n"
],
[
"learn.recorder.plot_metrics()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5ecb06b96868e43cb81e1d0721c48361cce8da | 193,932 | ipynb | Jupyter Notebook | LinearRegression.ipynb | carbonlkg/Kaggle_HousingPrices | 7e00ee3f4a8280f9ba049a38785922a8e10b1d92 | [
"MIT"
]
| null | null | null | LinearRegression.ipynb | carbonlkg/Kaggle_HousingPrices | 7e00ee3f4a8280f9ba049a38785922a8e10b1d92 | [
"MIT"
]
| null | null | null | LinearRegression.ipynb | carbonlkg/Kaggle_HousingPrices | 7e00ee3f4a8280f9ba049a38785922a8e10b1d92 | [
"MIT"
]
| null | null | null | 172.384 | 50,348 | 0.870511 | [
[
[
"# Linear Regression analysis of the Kaggle Housing Prices dataset\n\nIn this anlaysis the [Kaggle Housing prices dataset](https://www.kaggle.com/c/house-prices-advanced-regression-techniques/overview) is analysed using Linear Regression methods.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.metrics import mean_squared_error, mean_squared_log_error",
"_____no_output_____"
],
[
"import math\n\n#A function to calculate Root Mean Squared Logarithmic Error (RMSLE)\ndef rmsle(y, y_pred):\n assert len(y) == len(y_pred)\n terms_to_sum = [(math.log(y_pred[i] + 1) - math.log(y[i] + 1)) ** 2.0 for i,pred in enumerate(y_pred)]\n return (sum(terms_to_sum) * (1.0/len(y))) ** 0.5",
"_____no_output_____"
]
],
[
[
"## Importing data and cleaning",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('Data/train.csv')\ndata.head()",
"_____no_output_____"
]
],
[
[
"### Extract Numerical columns \n\nIn the first iteration, only the numerical columns are considered for the linear regression.",
"_____no_output_____"
]
],
[
[
"num_df = data.select_dtypes(include='number').copy()\n\nnum_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1460 entries, 0 to 1459\nData columns (total 38 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Id 1460 non-null int64 \n 1 MSSubClass 1460 non-null int64 \n 2 LotFrontage 1201 non-null float64\n 3 LotArea 1460 non-null int64 \n 4 OverallQual 1460 non-null int64 \n 5 OverallCond 1460 non-null int64 \n 6 YearBuilt 1460 non-null int64 \n 7 YearRemodAdd 1460 non-null int64 \n 8 MasVnrArea 1452 non-null float64\n 9 BsmtFinSF1 1460 non-null int64 \n 10 BsmtFinSF2 1460 non-null int64 \n 11 BsmtUnfSF 1460 non-null int64 \n 12 TotalBsmtSF 1460 non-null int64 \n 13 1stFlrSF 1460 non-null int64 \n 14 2ndFlrSF 1460 non-null int64 \n 15 LowQualFinSF 1460 non-null int64 \n 16 GrLivArea 1460 non-null int64 \n 17 BsmtFullBath 1460 non-null int64 \n 18 BsmtHalfBath 1460 non-null int64 \n 19 FullBath 1460 non-null int64 \n 20 HalfBath 1460 non-null int64 \n 21 BedroomAbvGr 1460 non-null int64 \n 22 KitchenAbvGr 1460 non-null int64 \n 23 TotRmsAbvGrd 1460 non-null int64 \n 24 Fireplaces 1460 non-null int64 \n 25 GarageYrBlt 1379 non-null float64\n 26 GarageCars 1460 non-null int64 \n 27 GarageArea 1460 non-null int64 \n 28 WoodDeckSF 1460 non-null int64 \n 29 OpenPorchSF 1460 non-null int64 \n 30 EnclosedPorch 1460 non-null int64 \n 31 3SsnPorch 1460 non-null int64 \n 32 ScreenPorch 1460 non-null int64 \n 33 PoolArea 1460 non-null int64 \n 34 MiscVal 1460 non-null int64 \n 35 MoSold 1460 non-null int64 \n 36 YrSold 1460 non-null int64 \n 37 SalePrice 1460 non-null int64 \ndtypes: float64(3), int64(35)\nmemory usage: 433.6 KB\n"
]
],
[
[
">**Calculating the age of the building since initial construction (Age_built) and the number of years since last remodelling (Age_remod).**",
"_____no_output_____"
]
],
[
[
"num_df['Age_built'] = num_df['YrSold'] - num_df['YearBuilt']\nnum_df['Age_remod'] = num_df['YrSold'] - num_df['YearRemodAdd']\nnum_df.drop(['YrSold','YearBuilt', 'YearRemodAdd'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"num_df.nunique().sort_values()",
"_____no_output_____"
]
],
[
[
">**The following columns are removed, since they have very low unique values and the information they provide is covered in other variables.**",
"_____no_output_____"
]
],
[
[
"drop_cols = ['HalfBath', 'BsmtHalfBath','BsmtFullBath','KitchenAbvGr','Fireplaces','FullBath','GarageCars','PoolArea','BedroomAbvGr', 'MoSold', 'GarageYrBlt', 'MasVnrArea', 'Id']",
"_____no_output_____"
],
[
"num_df.drop(drop_cols, axis=1, inplace=True)",
"_____no_output_____"
],
[
"num_df.describe()",
"_____no_output_____"
],
[
"corr_df = num_df.corr().abs()\nplt.subplots(figsize=(10,10))\nsns.heatmap(corr_df[corr_df>0.5], cmap='Blues')",
"_____no_output_____"
],
[
"corr_df['SalePrice'].sort_values(ascending=False)",
"_____no_output_____"
]
],
[
[
">**The following columns represent categorical variables and are hence converetd to dummies.**",
"_____no_output_____"
]
],
[
[
"categ_cols = ['OverallCond', 'OverallQual', 'MSSubClass']",
"_____no_output_____"
],
[
"num_df[categ_cols] = num_df[categ_cols].astype('category')",
"_____no_output_____"
],
[
"dummies = pd.get_dummies(num_df[categ_cols])",
"_____no_output_____"
],
[
"num_df = pd.concat([num_df, dummies], axis=1)\nnum_df.drop(categ_cols, axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"### Missing data handling\n\nCalculate the percentage of missing values in all the columns. Fill missing values with the mean of the relevant column.",
"_____no_output_____"
]
],
[
[
"def pct_mv(data):\n '''\n Function calculates the percentage of missing values in all columns of dataset. \n \n ------\n Inputs\n ------\n data (required in pd.DataFrame format)\n The dataframe for which the missing values have to be calculated\n \n ------\n Outputs\n ------\n Dataframe containing the column names and percentage missing values, sorted ascendingly.\n '''\n data_pct_mv = data.isna().sum()/len(data)\n data_pct_mv.sort_values(ascending=False, inplace=True)\n return data_pct_mv",
"_____no_output_____"
],
[
"pct_mv(num_df).plot.barh(figsize = (5,10))",
"_____no_output_____"
],
[
"num_df['LotFrontage'].fillna(num_df['LotFrontage'].mean(), inplace=True)",
"_____no_output_____"
],
[
"pct_mv(num_df).plot.barh(figsize = (5,10))",
"_____no_output_____"
]
],
[
[
"## Linear Regression - Iteration 1",
"_____no_output_____"
]
],
[
[
"def train_test(data, target):\n train_rows = round(0.75* len(data))\n train = data[:train_rows]\n test = data[train_rows:]\n \n features = data.columns.drop([target])\n \n lr = LinearRegression()\n lr.fit(train[features], train[target])\n predictions = lr.predict(train[features])\n predictions[predictions<0] = 0\n train_rmse = mean_squared_error(predictions, train[target])**0.5\n train_rmsle= mean_squared_log_error(train[target], predictions)\n predictions = lr.predict(test[features])\n predictions[predictions<0] = 0\n test_rmse = mean_squared_error(predictions, test[target])**0.5\n test_rmsle= mean_squared_log_error(test[target], predictions)\n \n return lr, train_rmse, test_rmse, train_rmsle, test_rmsle",
"_____no_output_____"
],
[
"corr_df= num_df.corr().abs()['SalePrice'].sort_values(ascending=False)",
"_____no_output_____"
],
[
"train_rmses = []\ntest_rmses = []\ntrain_rmsles = []\ntest_rmsles = []\ncols = range(2,35)\n\nfor i in cols:\n lr, train_rmse, test_rmse, train_rmsle, test_rmsle = train_test(num_df[list(corr_df.index[:i])], 'SalePrice')\n train_rmses.append(train_rmse)\n test_rmses.append(test_rmse)\n train_rmsles.append(train_rmsle)\n test_rmsles.append(test_rmsle)\n\nfig, ax = plt.subplots(2,1, figsize = (7,7))\nax[0].plot(cols, train_rmses, c='blue', label='Train RMSE')\nax[0].plot(cols,test_rmses, c='red', label='Test RMSE')\nax[0].legend()\n\nax[1].plot(cols, train_rmsles, c='blue', label='Train RMSLE')\nax[1].plot(cols,test_rmsles, c='red', label='Test RMSLE')\nax[1].legend()\n\n\nplt.show()",
"_____no_output_____"
],
[
"num_df['SalePrice'].plot.hist()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
ec5ed0a88cb27d2680591848d36efacb9db9c77e | 46,392 | ipynb | Jupyter Notebook | pytorch-implementation/model-training.ipynb | rcharan/phutball | b0ae375d06d1b4e27b511e5b6729aeb278c7886f | [
"MIT"
]
| 4 | 2020-06-04T00:38:00.000Z | 2020-11-26T08:18:30.000Z | pytorch-implementation/model-training.ipynb | rcharan/phutball | b0ae375d06d1b4e27b511e5b6729aeb278c7886f | [
"MIT"
]
| 8 | 2021-03-30T13:18:08.000Z | 2022-02-27T10:59:45.000Z | pytorch-implementation/model-training.ipynb | rcharan/phutball | b0ae375d06d1b4e27b511e5b6729aeb278c7886f | [
"MIT"
]
| 1 | 2020-09-07T15:52:27.000Z | 2020-09-07T15:52:27.000Z | 54.006985 | 16,810 | 0.640046 | [
[
[
"# Setup",
"_____no_output_____"
],
[
"## Create Filesystem\nThis notebook is primarily meant to be executed in Colab as a computational backend. If you want to run on your own hardware with data, you need to set `data_dir` and `ALLOW_IO`\n\nThis notebook viewable directly on Colab from [https://colab.research.google.com/github/rcharan/phutball/blob/master/pytorch-implementation/model-training.ipynb](https://colab.research.google.com/github/rcharan/phutball/blob/master/pytorch-implementation/model-training.ipynb) (it is a mirror of github). But if it has moved branches or you are looking at a past commit, look at the [Google instructions](https://colab.research.google.com/github/googlecolab/colabtools/blob/master/notebooks/colab-github-demo.ipynb) on where to find this file.\n\nThe workflow is:\n - Data stored in (my personal/private) Google Drive\n - Utilities/library files (for importing) on github, edited on local hardware and pushed to github.\n - Notebook hosted on github, edited both in Colab or locally (depending on the relative value of having a GPU attached versus being able to use regular Jupyter keyboard shortcuts/a superior interface)",
"_____no_output_____"
]
],
[
[
"# Attempt Colab setup if on Colab\ntry:\n import google.colab\nexcept:\n ALLOW_IO = False\nelse:\n # Mount Google Drive at data_dir\n # (for data)\n from google.colab import drive\n from os.path import join\n ROOT = '/content/drive'\n DATA = 'My Drive/phutball'\n drive.mount(ROOT)\n ALLOW_IO = True\n data_dir = join(ROOT, DATA)\n !mkdir \"{data_dir}\" # in case we haven't created it already \n\n # Pull in code from github\n %cd /content\n github_repo = 'https://github.com/rcharan/phutball'\n !git clone -b master {github_repo}\n %cd /content/phutball\n \n # Point python to code base\n import sys\n sys.path.append('/content/phutball/pytorch-implementation')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive\nmkdir: cannot create directory ‘/content/drive/My Drive/phutball’: File exists\n/content\nCloning into 'phutball'...\nremote: Enumerating objects: 259, done.\u001b[K\nremote: Counting objects: 100% (259/259), done.\u001b[K\nremote: Compressing objects: 100% (165/165), done.\u001b[K\nremote: Total 2243 (delta 175), reused 151 (delta 92), pack-reused 1984\u001b[K\nReceiving objects: 100% (2243/2243), 38.58 MiB | 39.51 MiB/s, done.\nResolving deltas: 100% (1426/1426), done.\n/content/phutball\n"
]
],
[
[
"## Imports",
"_____no_output_____"
]
],
[
[
"# !git pull",
"Already up to date.\n"
],
[
"%%capture\n\n%load_ext autoreload\n%autoreload 2\n\nimport os\nimport gc\nimport numpy as np\n\n# Codebase\nfrom lib.models.model_v3 import TDConway\nfrom lib.off_policy import EpsilonGreedy\nfrom lib.optim import AlternatingTDLambda\n\nfrom lib.training import training_loop\n\nfrom lib.utilities import config, lfilter, Timer, product\n# from lib.testing_utilities import create_state, visualize_state, boards\n\nfrom lib.move_selection import get_next_move_training\n\nfrom lib.pretraining.fit_one_cycle import fit_one_cycle\nfrom lib.pretraining.pre_training import pre_train\nfrom torch.optim import SGD\n\nfrom lib.arena import Player, Battle, RandoTron\n\n# Time Zone management utilities\nfrom datetime import datetime\nfrom pytz import timezone\neastern = timezone('US/Eastern')\n\n# Graphics for visualization\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\n%matplotlib inline\nplt.ioff()",
"_____no_output_____"
]
],
[
[
"## Device Management Utilities\nSetup for GPU, CPU, or (not working well/fully implemented) TPU",
"_____no_output_____"
]
],
[
[
"import os\n\ndef install_tpu():\n VERSION = \"1.5\"\n !curl https://raw.githubusercontent.com/pytorch/xla/master/contrib/scripts/env-setup.py -o pytorch-xla-env-setup.py\n !python pytorch-xla-env-setup.py --version $VERSION\n \nuse_tpu = 'COLAB_TPU_ADDR' in os.environ",
"_____no_output_____"
],
[
"import torch\n\nif use_tpu:\n # Install PyTorch/XLA\n install_tpu()\n import torch_xla\n import torch_xla.core.xla_model as xm\n \n # Set the device\n device = xm.xla_device()\n \n # Memory inspection\n def print_memory_usage():\n print('TPU memory inspection not implemented')\n def print_max_memory_usage():\n print('TPU memory inspection not implemented')\n def garbage_collect():\n gc.collect() # No TPU specific implementation yet\n \nelif torch.cuda.is_available():\n # Set the device\n device = torch.device('cuda')\n \n # Echo GPU info\n gpu_info = !nvidia-smi\n gpu_info = '\\n'.join(gpu_info)\n print(gpu_info)\n \n # Memory inspection and management\n from lib.memory import (\n print_memory_usage_cuda as print_memory_usage,\n print_max_memory_usage_cuda as print_max_memory_usage,\n garbage_collect_cuda as garbage_collect\n )\n\nelse:\n # Set the device to CPU\n device = torch.device('cpu')\n \n # Echo RAM info\n from psutil import virtual_memory\n from lib.memory import format_bytes\n ram = virtual_memory().total\n print(format_bytes(ram), 'available memory on CPU-based runtime')\n \n # Memory inspection and management\n from lib.memory import (\n print_memory_usage, \n print_max_memory_usage,\n garbage_collect\n )",
"Sat Jul 18 18:55:54 2020 \n+-----------------------------------------------------------------------------+\n| NVIDIA-SMI 450.51.05 Driver Version: 418.67 CUDA Version: 10.1 |\n|-------------------------------+----------------------+----------------------+\n| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |\n| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |\n| | | MIG M. |\n|===============================+======================+======================|\n| 0 Tesla P100-PCIE... Off | 00000000:00:04.0 Off | 0 |\n| N/A 34C P0 25W / 250W | 10MiB / 16280MiB | 0% Default |\n| | | ERR! |\n+-------------------------------+----------------------+----------------------+\n \n+-----------------------------------------------------------------------------+\n| Processes: |\n| GPU GI CI PID Type Process name GPU Memory |\n| ID ID Usage |\n|=============================================================================|\n| No running processes found |\n+-----------------------------------------------------------------------------+\n"
]
],
[
[
"# Utilities",
"_____no_output_____"
]
],
[
[
"def save(fname, model):\n state_dict = {\n 'model' : model.state_dict(),\n }\n torch.save(state_dict, f'{data_dir}/{fname}.pt')\n\ndef fname(version, game_num):\n return f'v{version}-{game_num}'\n \ndef load(version, game_num, model):\n sd = torch.load(f'{data_dir}/{fname(version, game_num)}.pt')\n model.load_state_dict(sd['model'])\n return model",
"_____no_output_____"
]
],
[
[
"# Training",
"_____no_output_____"
],
[
"## Fit one cycle",
"_____no_output_____"
]
],
[
[
"model = TDConway(config).to(device)\ndata = fit_one_cycle(model)",
"6942/6942 [==============================] - 72s 10ms/step\n"
],
[
"df = pd.DataFrame(data, columns = ['lr', 'loss'])\ndf['loss'] = df.loss.apply(lambda l : l.item())\ndf = df.groupby('lr').mean().reset_index()\n\nfig, ax = plt.subplots()\nsns.lineplot(x = 'lr', y='loss', data = df, ax = ax)\nax.set_xscale('log', basex = 10)\nax.set_yscale('log', basey = 10)\nfig",
"_____no_output_____"
]
],
[
[
"## Pretraining",
"_____no_output_____"
]
],
[
[
"model = TDConway(config).to(device)\noptimizer = SGD(model.parameters(), lr = 0.05)\n\npre_train(model, optimizer, loops = 40000, batch_size = 300)",
"40000/40000 [==============================] - 517s 13ms/step - loss: 0.0011\n"
],
[
"version = '0.5.1'\ngame_num = 'pre40000x300'\n\nsave(fname(version, game_num), model)",
"_____no_output_____"
]
],
[
[
"## Profile",
"_____no_output_____"
]
],
[
[
"initial_state = create_state('H10').to(device)\nepsilon_greedy = EpsilonGreedy(0.01)\n\ndef num_params(model):\n return sum(\n product(t.shape) for t in model.parameters()\n )",
"_____no_output_____"
],
[
"# version = '0.3.1'\n# game_num = '70000'\nmodel = TDConway(config, temperature = 0.1).to(device)\nmodel = load(version, game_num, model)\n\noptimizer = AlternatingTDLambda(model.parameters(), alpha = 0.05, lamda = 0.9)\n\nprint(f'{num_params(model):,d} parameters')\n%timeit get_next_move_training(initial_state, model, device)\n%timeit training_loop(model, optimizer, 1, device, off_policy = epsilon_greedy)\n# %prun training_loop(model, optimizer, 1, device, off_policy = epsilon_greedy)",
"1,935,105 parameters\n100 loops, best of 3: 3.28 ms per loop\n1/1 [==============================] - 0s 622us/step\n1/1 [==============================] - 0s 188us/step\n1/1 [==============================] - 0s 192us/step\n1/1 [==============================] - 0s 172us/step\n1 loop, best of 3: 473 ms per loop\n"
]
],
[
[
"## Train",
"_____no_output_____"
]
],
[
[
"epsilon_greedy = EpsilonGreedy(0.05)\nmodel = TDConway(config, temperature = 0.1).to(device)\noptimizer = AlternatingTDLambda(model.parameters(), 0.05, 0.9)\n\nversion = '0.5.1'\ngame_num = 'pre40000x300'\n\nload(version, game_num, model);",
"_____no_output_____"
],
[
"game_num = 0\n\nbatch_size = 500\n\nwhile True: # Until Colab or User disconnects out of boredom\n try:\n training_loop(model, optimizer, batch_size, device, \n off_policy = epsilon_greedy, \n verbose = 1,\n initial = 0.1)\n game_num += batch_size\n\n save(fname(version, game_num), model)\n\n print(f'Finished {game_num} games at', datetime.now(eastern).strftime('%I:%M%p %Z'))\n except KeyboardInterrupt:\n break",
" 13/500 [..............................] - ETA: 7s - game-length: 2.0769"
],
[
"# Versioning:\n# Major versions - major change in approach\n# Minor versions - incompatible architecture tweaks\n# Build - retraining or changes in training parameters\n# Game number - number of games trained or pre{E}x{B} where E is the the\n# number of batches and B is the batch size for pre-training\n# Example: v0.1.2 @400 is the second attempt at training the v0.1 architecture\n# and was trained for 400 games\n\n# Performance benchmarks.\n# GPU benchmarks are on a P100 unless otherwise stated\n# per move : training-relevant\n# forward pass: evaluation (arena mode) relevant\n# CPU benchmarks are for inference on a fixed set of 300 randomly\n# generated boards on an Intel i5 chipset. (deployment-relevant)\n# Memory consumption has not been an issue\n\n# v0.1: architecture from model_v1. Training: Alternating TD(λ)\n# ~60M params (59,943,809)\n# GPU: 100–110ms/move with 50-60ms for forward pass\n# CPU: 8.1s ± 0.5s \n# alpha = 0.01, lambda = 0.9, epsilon = 0.1\n#\n# - v0.1.1 - don't use, bug in training\n# - v0.1.2 - Use. available @400. Win rate v RandoTron 51% (😢)\n\n# v0.2: architecture from model_v2. Smaller Residual ConvNet (17 layers)\n# Training: Alternating TD(λ) WITH pretraining to prefer the ball to the\n# right on randomly generated boards\n# ~4.4M params (4,381,505)\n# GPU: 30-35ms/move with ~12ms for forward pass\n# CPU: 1.1s ± 0.02s\n#\n# - v0.2.1 - Available @pre10000x300, @400, @1500, @3500\n# - Hyperparameters same as v0.1\n# Win rate v RandoTron \n# - @pre-trained: 75.4% ±1.4% (!?)\n# - @400: 49%\n# - @1500: 56.9% ±1.8%\n# - @3500: 54.8% ±1.7%\n# - In further increments of 500 as [4000, 4500, ..., 20500]\n# - @10500: 60.3% ±1.6%\n# - @17500: 59.5% ±1.2%\n# - v0.2.2 - Increased pretraining, epsilon = 0.01\n# - Available @pre30000x300 (71.2% ± 2.3%)\n# - And in increments of 500 [500, 1000, ..., 82000]\n# - @17500: 71.5% ±1.1%\n# - @82000: 99.8% ±0.1%\n\n# v0.3: architecture from model_v3. Much smaller (7 layers), no residuals\n# ~1.9M params (1,935,105)\n# alpha = 0.05, epsilon = 0.1, lambda = 0.9\n#\n# -v0.3.1 - Available at @pre40000x300, increments of 1000\n# - @27000: 71.3% ±1.1%\n# - @73000: 58.8% \n# -v0.3.2 - With ε = 0.05",
"_____no_output_____"
]
],
[
[
"# Evaluate",
"_____no_output_____"
]
],
[
[
"# Player 1\nmodel = TDConway(config).to(device)\nversion = '0.5.1'\ngame_num = 4000\nsd = torch.load(f'{data_dir}/{fname(version, game_num)}.pt', map_location = device)\nmodel.load_state_dict(sd['model'])\n\ntd_conway = Player(model, name = f'TD Conway v{version} @{game_num}')\ntd_conway.eval()\nrandotron = RandoTron()\nbattle = Battle(td_conway, randotron, verbose = 1)",
"_____no_output_____"
],
[
"battle.play_match(1600, device)",
"1600/1600 [==============================] - 262s 164ms/step\n1600 games were played between TD Conway v0.5.1 @4000 and RandoTron with 0 draws.\nThe winner was TD Conway v0.5.1 @4000 with a 61.6% win rate!\nTD Conway v0.5.1 @4000 on average won in a game of length 69.1.\nRandoTron on average won in a game of length 77.3\nOverall average length of game was 72.26125\nTotal time taken: 4:21 at\n - 164ms per finished game.\n - 2ms per move in a finished game\n"
],
[
"# Player 1 @3500\nmodel = TDConway(config).to(device)\nversion = '0.2.1'\ngame_num = 3500\nsd = torch.load(f'{data_dir}/{fname(version, game_num)}.pt', map_location = device)\nmodel.load_state_dict(sd['model'])\ntd_conway_1 = Player(model, name = f'TD Conway v{version} @{game_num}')\ntd_conway_1.eval()\n\n# Player 2 @ 10500\nmodel = TDConway(config).to(device)\nversion = '0.2.1'\ngame_num = 10500\nsd = torch.load(f'{data_dir}/{fname(version, game_num)}.pt', map_location = device)\nmodel.load_state_dict(sd['model'])\ntd_conway_2 = Player(model, name = f'TD Conway v{version} @{game_num}')\ntd_conway_2.eval()\n\nbattle = Battle(td_conway_1, td_conway_2, verbose = 1)",
"_____no_output_____"
],
[
"battle.play_match(900, device)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
ec5ed46fbb5573e79bdd073da74acff20a5f2979 | 4,658 | ipynb | Jupyter Notebook | ch08/02_DifferentTokenizers.ipynb | oreilly-japan/practical-nlp-ja | 2491adbc24f5df15e1ea03ccd27dfad8572f4ad7 | [
"MIT"
]
| 29 | 2021-12-14T10:22:09.000Z | 2022-03-29T01:33:40.000Z | ch08/02_DifferentTokenizers.ipynb | oreilly-japan/practical-nlp-ja | 2491adbc24f5df15e1ea03ccd27dfad8572f4ad7 | [
"MIT"
]
| null | null | null | ch08/02_DifferentTokenizers.ipynb | oreilly-japan/practical-nlp-ja | 2491adbc24f5df15e1ea03ccd27dfad8572f4ad7 | [
"MIT"
]
| 5 | 2021-11-16T00:56:26.000Z | 2022-03-28T02:32:33.000Z | 33.035461 | 308 | 0.464148 | [
[
[
"!pip install twikenizer==1.0 emoji==1.5.0",
"Collecting twikenizer\n Downloading twikenizer-1.0.tar.gz (4.4 kB)\nCollecting emoji\n Downloading emoji-1.5.0.tar.gz (185 kB)\n\u001b[K |████████████████████████████████| 185 kB 2.7 MB/s \n\u001b[?25hBuilding wheels for collected packages: twikenizer, emoji\n Building wheel for twikenizer (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for twikenizer: filename=twikenizer-1.0-py3-none-any.whl size=4851 sha256=10dfd57c7175eea6cf30c3416bc82d9077fe60008c175e163f9faede6a2977b2\n Stored in directory: /root/.cache/pip/wheels/b2/6e/1e/8e16444d4adf9e5bb49cf3089ebbb5d81e7e51d216430afcc5\n Building wheel for emoji (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for emoji: filename=emoji-1.5.0-py3-none-any.whl size=187457 sha256=f8d16035f46b97b8c04834735a9a108ec5789e49418336f6ca4564378fc49a37\n Stored in directory: /root/.cache/pip/wheels/db/b5/f6/b39abf14e94b3d6640613bbe630a66c10ccf7a12882d064fb5\nSuccessfully built twikenizer emoji\nInstalling collected packages: twikenizer, emoji\nSuccessfully installed emoji-1.5.0 twikenizer-1.0\n"
],
[
"tweet3 = \"Tw33t a_!aa&!a?b #%lol # @dude_really #b3st_day $ad (b@e) (beep#d) @dude. 😀😀 !😀abc %😀lol #loveit #love.it $%&/ d*ck-\"",
"_____no_output_____"
],
[
"import twikenizer as twk\n\ntwk = twk.Twikenizer()\nprint(twk.tokenize(tweet3))",
"['Tw33t', 'a', '_', '!', 'aa', '&', '!', 'a', '?', 'b', '#%lol', '#', '@dude_really', '#b3st_day', '$ad', '(', 'b', '@', 'e', ')', '(', 'beep', '#', 'd', ')', '@dude', '.', '😀', '😀', '!', '😀', 'abc', '%', '😀', 'lol', '#loveit', '#love', '.', 'it', '$', '%', '&', '/', 'd*ck', '-']\n"
],
[
"from nltk.tokenize import TweetTokenizer\n\ntknzr = TweetTokenizer()\nprint(tknzr.tokenize(tweet3))",
"['Tw33t', 'a_', '!', 'aa', '&', '!', 'a', '?', 'b', '#', '%', 'lol', '#', '@dude_really', '#b3st_day', '$', 'ad', '(', 'b', '@e', ')', '(', 'beep', '#', 'd', ')', '@dude', '.', '😀', '😀', '!', '😀', 'abc', '%', '😀', 'lol', '#loveit', '#love', '.', 'it', '$', '%', '&', '/', 'd', '*', 'ck', '-']\n"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5f0e3300bbd2e827fbafb870df1f0613499d96 | 5,849 | ipynb | Jupyter Notebook | 05_objectDetection/Faster_rcnn_resnet/04_share_map.ipynb | fioletflying/tensorflowcook | 10b9feaf0db780f94ace4a9e019b163999b6c38e | [
"MIT"
]
| null | null | null | 05_objectDetection/Faster_rcnn_resnet/04_share_map.ipynb | fioletflying/tensorflowcook | 10b9feaf0db780f94ace4a9e019b163999b6c38e | [
"MIT"
]
| null | null | null | 05_objectDetection/Faster_rcnn_resnet/04_share_map.ipynb | fioletflying/tensorflowcook | 10b9feaf0db780f94ace4a9e019b163999b6c38e | [
"MIT"
]
| null | null | null | 31.788043 | 117 | 0.520944 | [
[
[
"import keras.layers as KL\nfrom keras.models import Model\nimport keras.backend as K\nimport keras\nimport tensorflow as tf\nfrom keras.utils import plot_model\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom PIL import Image\n%matplotlib inline",
"_____no_output_____"
],
[
"def building_block(filters,block):\n \n if block != 0:\n stride = 1\n else:\n stride = 2\n \n def f(x):\n \n #主通路的网络结构\n y = KL.Conv2D(filters=filters,kernel_size=(1,1),strides=stride)(x)\n y = KL.BatchNormalization(axis=3)(y)\n y = KL.Activation('relu')(y)\n \n y = KL.Conv2D(filters=filters,kernel_size=(3,3),padding='same')(y)\n y = KL.BatchNormalization(axis=3)(y)\n y = KL.Activation('relu')(y)\n \n #主通路的输出\n y = KL.Conv2D(filters=4*filters,kernel_size=(1,1))(y)\n y = KL.BatchNormalization(axis=3)(y)\n \n #判断block的类型\n if block == 0:\n shortcut = KL.Conv2D(filters = 4*filters,kernel_size=(1,1),strides=stride)(x)\n shortcut = KL.BatchNormalization(axis=3)(shortcut)\n else:\n shortcut = x\n \n #主通路与shortcut 相加 \n y = KL.Add()([y,shortcut])\n import random\n y = KL.Activation('relu',name='last'+ str((random.randint(100,300))))(y)\n #y = KL.Activation('relu')(y)\n \n return y\n return f\n ",
"_____no_output_____"
],
[
"def ResNet_Extractor(inputs):\n \n # reanet 预处理 heading\n x = KL.Conv2D(filters=64,kernel_size=(3,3),padding='same')(inputs)\n x = KL.BatchNormalization(axis=3)(x)\n x = KL.Activation('relu')(x)\n \n filters = 64\n \n block=[2,2,2]\n for i,block_num in enumerate(block):\n for block_id in range(block_num):\n x = building_block(filters=filters,block=block_id)(x)\n filters *=2\n \n return x\n ",
"_____no_output_____"
],
[
"def RpnNet(inputs,k=9):\n \n #共享层输出\n shareMap = KL.Conv2D(filters=256,kernel_size=(3,3),padding='same',name='sharemap')(inputs)\n shareMap = KL.Activation('linear')(shareMap) #使用linear\n \n #RPN 分类 前后景\n rpn_classfication = KL.Conv2D(filters=2*k,kernel_size=(1,1))(shareMap)\n # 为了保证与原始分类输入一致,所以需要reshape\n rpn_classfication = KL.Lambda(lambda x:tf.reshape(x,[tf.shape(x)[0],-1,2]))(rpn_classfication)\n rpn_classfication = KL.Activation('linear',name='rpn_classfication')(rpn_classfication)\n \n rpn_probability = KL.Activation('softmax',name='rpn_prob')(rpn_classfication)\n \n # 计算回归修正\n rpn_position = KL.Conv2D(filters=4*k,kernel_size=(1,1))(shareMap)\n rpn_position = KL.Activation('linear')(rpn_position)\n # -l 表示anchor数量不确定\n rpn_BoundingBox = KL.Lambda(lambda x:tf.reshape(x,[tf.shape(x)[0],-1,4]),name='rpn_POS')(rpn_position)\n \n return rpn_classfication,rpn_probability,rpn_BoundingBox\n",
"_____no_output_____"
],
[
"x = KL.Input((64,64,3))\nfeatureMap = ResNet_Extractor(x)\nrpn_classfication,rpn_probability,rpn_BoundingBox = RpnNet(featureMap,k=9)\nmodel = Model(inputs=[x],outputs=[rpn_classfication,rpn_probability,rpn_BoundingBox])\nplot_model(model=model,to_file='sharemap-64.png',show_shapes=True)",
"_____no_output_____"
],
[
"def RPNClassLoss(rpn_match,rpn_Cal):\n rpn_match = tf.squeeze(rpn_match,axis=-1)\n indices = tf.where(K.not_equal(x=rpn_match,y=0))\n #1=1,0,-1 = 0\n anchor_class = K.cast(K.equal(rpn_match,1),tf.int32)\n \n # 原始样本结果\n anchor_class = tf.gather_nd(params=anchor_class, indices=indices)\n #这个是rpn计算结果\n rpn_cal_class = tf.gather_nd(params=rpn_Cal,indices=indices)\n \n # one hot \n loss = K.sparse_categorical_crossentropy(target=anchor_class,output=rpn_cal_class,\n from_logits=True)\n \n loss = K.switch(condition=tf.size(loss)>0,then_expression=K.mean(loss),else_expression=tf.constant(0.0))\n \n return loss\n ",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5f0fe43f52f76f3ef7fbf61f647ae3733aa1c2 | 1,019 | ipynb | Jupyter Notebook | baseline/Merge-chunks.ipynb | EBI-Metagenomics/iseq-prof-analysis | 4ac27f2d5ee935c5c87f9238141978945ee81142 | [
"MIT"
]
| null | null | null | baseline/Merge-chunks.ipynb | EBI-Metagenomics/iseq-prof-analysis | 4ac27f2d5ee935c5c87f9238141978945ee81142 | [
"MIT"
]
| null | null | null | baseline/Merge-chunks.ipynb | EBI-Metagenomics/iseq-prof-analysis | 4ac27f2d5ee935c5c87f9238141978945ee81142 | [
"MIT"
]
| null | null | null | 18.196429 | 49 | 0.534838 | [
[
[
"import config\nimport iseq_prof\nfrom tqdm import tqdm",
"_____no_output_____"
],
[
"prof = iseq_prof.Profiling(config.root_dir)",
"_____no_output_____"
],
[
"for organism in tqdm(prof.organisms):\n prof.merge_chunks(organism, force=True)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
ec5f1c0dab98652b322f0a7526738c9277103fed | 9,030 | ipynb | Jupyter Notebook | test/QuadrotorILQRTest.ipynb | MinRegret/deluca-igpc | a060c5e505bf253d19649e456f7df4fd7c75bcac | [
"CC0-1.0"
]
| 4 | 2021-06-15T14:28:23.000Z | 2022-01-02T21:34:30.000Z | test/QuadrotorILQRTest.ipynb | MinRegret/deluca-igpc | a060c5e505bf253d19649e456f7df4fd7c75bcac | [
"CC0-1.0"
]
| 1 | 2021-07-25T20:30:10.000Z | 2021-07-25T20:30:10.000Z | test/QuadrotorILQRTest.ipynb | MinRegret/deluca-igpc | a060c5e505bf253d19649e456f7df4fd7c75bcac | [
"CC0-1.0"
]
| 2 | 2021-10-16T09:29:16.000Z | 2022-02-26T21:24:26.000Z | 41.612903 | 125 | 0.554153 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import jax\nimport jax.numpy as jnp\nimport tqdm\nimport flax\nimport deluca.core\nimport matplotlib.pyplot as plt\nfrom deluca.igpc.ilqr import iLQR, iLQR_open, iLQR_closed\nfrom deluca.envs import PlanarQuadrotor",
"WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)\n"
],
[
"## Testing ILQR\nenv = PlanarQuadrotor.create()\ndef cost(x, u, sim):\n return 0.1 * (u - sim.goal_action) @ (u - sim.goal_action) + (x.arr - sim.goal_state) @ (x.arr - sim.goal_state)\n\nU_initial = jnp.tile(jnp.zeros_like(env.goal_action), (env.H, 1))\nX, U, k, K, c = iLQR(env, cost, U_initial, 10, verbose=True)",
"iLQR (true): t = -1, r = 1, c = 357221.34375\niLQR (true): t = 0, r = 2, alphac = 0.550000011920929, cost = 89396.578125\niLQR (true): t = 0, r = 2, c = 89396.578125\niLQR (true): t = 1, r = 3, alphac = 0.6050000190734863, cost = 18147.2578125\niLQR (true): t = 1, r = 3, c = 18147.2578125\niLQR (true): t = 2, r = 4, alphac = 0.6655000448226929, cost = 2876.22216796875\niLQR (true): t = 2, r = 4, c = 2876.22216796875\niLQR (true): t = 3, r = 5, alphac = 0.7320500612258911, cost = 368.6937255859375\niLQR (true): t = 3, r = 5, c = 368.6937255859375\niLQR (true): t = 4, r = 6, alphac = 0.8052550554275513, cost = 75.40985870361328\niLQR (true): t = 4, r = 6, c = 75.40985870361328\niLQR (true): t = 5, r = 7, alphac = 0.8857805728912354, cost = 53.57438278198242\niLQR (true): t = 5, r = 7, c = 53.57438278198242\niLQR (true): t = 6, r = 8, alphac = 0.9743586182594299, cost = 52.72297668457031\niLQR (true): t = 6, r = 8, c = 52.72297668457031\niLQR (true): t = 7, r = 9, alphac = 1.0717945098876953, cost = 52.71146774291992\niLQR (true): t = 7, r = 9, c = 52.71146774291992\niLQR (true): t = 8, r = 10, alphac = 1.178973913192749, cost = 52.7114372253418\niLQR (true): t = 8, r = 10, c = 52.7114372253418\niLQR (true): t = 9, r = 11, alphac = 1.296871304512024, cost = 52.71142578125\niLQR (true): t = 9, r = 11, c = 52.71142578125\n"
],
[
"## Testing ILQR Open\n\ndef cost(x, u, sim):\n return 0.1 * (u - sim.goal_action) @ (u - sim.goal_action) + (x.arr - sim.goal_state) @ (x.arr - sim.goal_state)\n\n\nenv_true, env_sim = PlanarQuadrotor.create(wind=0.1), PlanarQuadrotor.create()\nU_initial = jnp.tile(env_sim.goal_action, (env_sim.H, 1))\nX, U, _, _, c = iLQR_open(env_true, env_sim, cost, U_initial, 10)\nassert c < 72000",
"iLQR (sim): t = -1, r = 1, c = 199.999267578125\niLQR (sim): t = 0, r = 2, alphac = 0.550000011920929, cost = 89.76972961425781\niLQR (sim): t = 0, r = 2, c = 89.76972961425781\niLQR (sim): t = 1, r = 3, alphac = 0.6050000190734863, cost = 60.33021545410156\niLQR (sim): t = 1, r = 3, c = 60.33021545410156\niLQR (sim): t = 2, r = 4, alphac = 0.6655000448226929, cost = 53.93872833251953\niLQR (sim): t = 2, r = 4, c = 53.93872833251953\niLQR (sim): t = 3, r = 5, alphac = 0.7320500612258911, cost = 52.85933303833008\niLQR (sim): t = 3, r = 5, c = 52.85933303833008\niLQR (sim): t = 4, r = 6, alphac = 0.8052550554275513, cost = 52.72452926635742\niLQR (sim): t = 4, r = 6, c = 52.72452926635742\niLQR (sim): t = 5, r = 7, alphac = 0.8857805728912354, cost = 52.7124137878418\niLQR (sim): t = 5, r = 7, c = 52.7124137878418\niLQR (sim): t = 6, r = 8, alphac = 0.9743586182594299, cost = 52.71151351928711\niLQR (sim): t = 6, r = 8, c = 52.71151351928711\niLQR (sim): t = 7, r = 9, alphac = 1.0717945098876953, cost = 52.711429595947266\niLQR (sim): t = 7, r = 9, c = 52.711429595947266\niLQR (sim): t = 8, r = 10, alphac = 1.178973913192749, cost = 52.7114372253418\niLQR (sim): t = 8, r = 11, alphac = 1.0717943906784058, cost = 52.7114372253418\niLQR (sim): t = 8, r = 12, alphac = 0.8052549362182617, cost = 52.7114372253418\niLQR (sim): t = 8, r = 13, alphac = 0.4999999403953552, cost = 52.7114372253418\niLQR (sim): t = 8, r = 14, alphac = 0.2565789818763733, cost = 52.7114372253418\niLQR (sim): t = 8, r = 15, alphac = 0.10881450772285461, cost = 52.7114372253418\niLQR (sim): t = 8, r = 16, alphac = 0.03813881427049637, cost = 52.7114372253418\niLQR (sim): t = 8, r = 17, alphac = 0.011047453619539738, cost = 52.7114372253418\niLQR (sim): t = 8, r = 18, alphac = 0.002644671592861414, cost = 52.7114372253418\niLQR (sim): t = 8, r = 19, alphac = 0.0005232339608483016, cost = 52.7114372253418\niLQR (sim): t = 9, r = 20, alphac = 1.178973913192749, cost = 52.7114372253418\niLQR (sim): t = 9, r = 21, alphac = 1.0717943906784058, cost = 52.7114372253418\niLQR (sim): t = 9, r = 22, alphac = 0.8052549362182617, cost = 52.7114372253418\niLQR (sim): t = 9, r = 23, alphac = 0.4999999403953552, cost = 52.7114372253418\niLQR (sim): t = 9, r = 24, alphac = 0.2565789818763733, cost = 52.7114372253418\niLQR (sim): t = 9, r = 25, alphac = 0.10881450772285461, cost = 52.7114372253418\niLQR (sim): t = 9, r = 26, alphac = 0.03813881427049637, cost = 52.7114372253418\niLQR (sim): t = 9, r = 27, alphac = 0.011047453619539738, cost = 52.7114372253418\niLQR (sim): t = 9, r = 28, alphac = 0.002644671592861414, cost = 52.7114372253418\niLQR (sim): t = 9, r = 29, alphac = 0.0005232339608483016, cost = 52.7114372253418\niLQR (open): t = 1, r = 1, c = 64758.3671875\n"
],
[
"## Testing ILQR Closed\n\ndef cost(x, u, sim):\n return 0.1 * (u - sim.goal_action) @ (u - sim.goal_action) + (x.arr - sim.goal_state) @ (x.arr - sim.goal_state)\n\n\nenv_true, env_sim = PlanarQuadrotor.create(wind=0.1), PlanarQuadrotor.create()\nU_initial = jnp.tile(env_sim.goal_action, (env_sim.H, 1))\nX, U, _, _, c = iLQR_closed(env_true, env_sim, cost, U_initial, 10)\nassert c < 90",
"iLQR (sim): t = -1, r = 1, c = 199.999267578125\niLQR (sim): t = 0, r = 2, alphac = 0.550000011920929, cost = 89.76972961425781\niLQR (sim): t = 0, r = 2, c = 89.76972961425781\niLQR (sim): t = 1, r = 3, alphac = 0.6050000190734863, cost = 60.33021545410156\niLQR (sim): t = 1, r = 3, c = 60.33021545410156\niLQR (sim): t = 2, r = 4, alphac = 0.6655000448226929, cost = 53.93872833251953\niLQR (sim): t = 2, r = 4, c = 53.93872833251953\niLQR (sim): t = 3, r = 5, alphac = 0.7320500612258911, cost = 52.85933303833008\niLQR (sim): t = 3, r = 5, c = 52.85933303833008\niLQR (sim): t = 4, r = 6, alphac = 0.8052550554275513, cost = 52.72452926635742\niLQR (sim): t = 4, r = 6, c = 52.72452926635742\niLQR (sim): t = 5, r = 7, alphac = 0.8857805728912354, cost = 52.7124137878418\niLQR (sim): t = 5, r = 7, c = 52.7124137878418\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5f234ae1ea62555438774a5c1cfa819e712b6a | 211,255 | ipynb | Jupyter Notebook | case.ipynb | Akash-tirupati/pandas | 9661f8ae218755fa4f3d9f8c3b6f8f529cdb6f75 | [
"Apache-2.0"
]
| null | null | null | case.ipynb | Akash-tirupati/pandas | 9661f8ae218755fa4f3d9f8c3b6f8f529cdb6f75 | [
"Apache-2.0"
]
| null | null | null | case.ipynb | Akash-tirupati/pandas | 9661f8ae218755fa4f3d9f8c3b6f8f529cdb6f75 | [
"Apache-2.0"
]
| null | null | null | 131.869538 | 42,148 | 0.844998 | [
[
[
"\\# Various plot for Visualization",
"_____no_output_____"
],
[
"### Quantitative:\n\n1. Kernel Density plot\n2. Histogram\n3. Box plot \n",
"_____no_output_____"
],
[
"### Categorical: \n\n1. Pie chart",
"_____no_output_____"
],
[
"### Quantitative vs. Quantitative\n 1. Scatterplot\n 2. Line plot",
"_____no_output_____"
],
[
"### Categorical vs. Quantitative\n 1. Bar chart (on summary statistics)\n 2. Grouped kernel density plots\n 3. Box plots\n 4. Violin plots\n",
"_____no_output_____"
],
[
"### Categorical vs. Categorical\n 1. Stacked bar chart\n 2. Grouped bar chart\n 3. Segmented bar chart\n",
"_____no_output_____"
],
[
"## Case study 1",
"_____no_output_____"
],
[
"\n### Problem Feature:(Heart Disease)\n\n### Data Set: \n\n * age - age in years \n * sex - (1 = male; 0 = female) \n * cp - chest pain type \n * trestbps - resting blood pressure (in mm Hg on admission to the hospital) \n * chol - serum cholestoral in mg/dl \n * fbs - (fasting blood sugar > 120 mg/dl) (1 = true; 0 = false) \n * restecg - resting electrocardiographic results \n * exang - exercise induced angina (1 = yes; 0 = no) \n * oldpeak - ST depression induced by exercise relative to rest \n * slope - the slope of the peak exercise ST segment \n * ca - number of major vessels (0-3) colored by flourosopy \n * thal - 3 = normal; 6 = fixed defect; 7 = reversable defect \n * target - have disease or not (1=yes, 0=no)\n\n",
"_____no_output_____"
],
[
"### 1.1 Loading the libraries ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"### 1.2 import the dataset ? (1 mark)",
"_____no_output_____"
]
],
[
[
"data=pd.read_csv(\"heart.csv\")\ndata",
"_____no_output_____"
]
],
[
[
"### 1.3 How to see the size of your data? (1 mark)",
"_____no_output_____"
]
],
[
[
"data.size",
"_____no_output_____"
]
],
[
[
"### 1.4 How to view the statistical characteristics of the data? (1 mark)",
"_____no_output_____"
]
],
[
[
"data.describe()",
"_____no_output_____"
]
],
[
[
"### 1.5 a How to see just one column?",
"_____no_output_____"
]
],
[
[
"data[\"sex\"]",
"_____no_output_____"
]
],
[
[
"### 1.5 b How to check the column names? (1 mark)",
"_____no_output_____"
]
],
[
[
"data.columns",
"_____no_output_____"
]
],
[
[
"### 1.5 c Find the Numerical Features,Categorical Features, Alphanumeric Features? (1 mark)",
"_____no_output_____"
]
],
[
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 303 entries, 0 to 302\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 age 303 non-null int64 \n 1 sex 303 non-null object \n 2 cp 303 non-null int64 \n 3 trestbps 303 non-null int64 \n 4 chol 303 non-null int64 \n 5 fbs 303 non-null int64 \n 6 restecg 303 non-null int64 \n 7 thalach 303 non-null int64 \n 8 exang 303 non-null int64 \n 9 oldpeak 303 non-null float64\n 10 slope 303 non-null int64 \n 11 ca 303 non-null int64 \n 12 thal 303 non-null int64 \n 13 target 303 non-null int64 \ndtypes: float64(1), int64(12), object(1)\nmemory usage: 33.3+ KB\n"
]
],
[
[
"#### Change the column names?",
"_____no_output_____"
]
],
[
[
"#Change the sex(0,1)=(female,male)\ndata[\"sex\"].replace(0,\"female\",inplace=True)\ndata[\"sex\"].replace(1,\"male\",inplace=True)\ndata",
"_____no_output_____"
]
],
[
[
"## 1.6 Quantitative: ( 5 marks)",
"_____no_output_____"
],
[
"### 1.6 a) Create kdeplot for showing age",
"_____no_output_____"
]
],
[
[
"sns.countplot(x='age',data=data)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 1.6 b) Create histogram for showing cholestrol with Number of bins 5",
"_____no_output_____"
]
],
[
[
"data[\"chol\"].hist(bins=5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 1.6 c) Create Boxplot for showing trestbps and comment what the dark spot indicate",
"_____no_output_____"
]
],
[
[
"sns.catplot(x='trestbps',\n y='thalach',\n data=data,\n kind='box')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"dark spot indicate outlier",
"_____no_output_____"
]
],
[
[
"## Categorical vs. Quantitative (10 Marks)",
"_____no_output_____"
]
],
[
[
" * Categorical: chest_pain_type, Sex, fasting_blood_sugar,rest_ecg,exercise_induced_angina,st_slope,thalassemia\n * Numerical Features: Age (Continuous), resting_blood_pressure ,cholesterol,max_heart_rate_achieved,st_depression, num_major_vessels,target",
"_____no_output_____"
],
[
"### 1.8a) Create bar plot for showing Gender and target. And your Observation:",
"_____no_output_____"
]
],
[
[
"\n#draw a bar plot of target by sex\nsns.catplot(x='sex',\n y='target',\n data=data,\n kind='bar')\nplt.show()\n\n#print percentages of females vs. males Heart Disease\n",
"_____no_output_____"
]
],
[
[
"### Observation:\n\n",
"_____no_output_____"
],
[
"### 1.8b) Create Bar plot for checking the both target vs Sex.",
"_____no_output_____"
]
],
[
[
"#create a subplot\nsns.relplot(x='target',y='sex',data=data,kind='scatter')\nplt.show()\n# create bar plot using groupby\nsns.catplot(x='target',y='cp',data=data,kind='bar')\nplt.show()\n# create count plot\nsns.countplot(x=\"target\",data=data,hue=\"sex\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 1.8d) Create Bar plot for checking the both Number Of people having chest_pain_type vs chest_pain_type:Heart Disease or Not",
"_____no_output_____"
]
],
[
[
"# create subplot plot\nsns.relplot(x='target',\n y='cp',\n data=data,\n kind='scatter')\nplt.show()\n# create bar plot using groupby\nsns.catplot(x='target',\n y='cp',\n data=data,\n kind='bar')\nplt.show()\n\n# create count plot\nsns.countplot(x=\"cp\",data=data,hue=\"target\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 1.8c) Create violinplot plot for checking the fasting_blood_sugar and Age vs target'",
"_____no_output_____"
]
],
[
[
"# create subplot plot\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.relplot(x='target',y='age',data=data,kind='scatter',col='fbs')\nplt.show()\n\n# create violinplot plot using groupby\n\nsns.catplot(x='target',y='age',data=data,kind='violin',col='fbs')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Box Plot\na box plot or boxplot is a method for graphically depicting groups of numerical data through their quartiles. Box plots may also have lines extending vertically from the boxes (whiskers) indicating variability outside the upper and lower quartiles, ",
"_____no_output_____"
],
[
"### 1.8 d)create a box plot for Sex & Age ",
"_____no_output_____"
]
],
[
[
"# create boxplot\nsns.catplot(x='sex',\n y='age',\n data=data,\n kind='box')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# 1.9 Categorical vs. Categorical (3 marks)\n\n* Categorical: target, Sex.",
"_____no_output_____"
],
[
"### Stacked bar chart",
"_____no_output_____"
],
[
"A stacked bar chart, also known as a stacked bar graph, is a graph that is used to break down and compare parts of a whole. Each bar in the chart represents a whole, and segments in the bar represent different parts or categories of that whole. Different colors are used to illustrate the different categories in the bar.\n\n",
"_____no_output_____"
],
[
"### 1.9a) Create a Stacked bar chart for showing target & Sex",
"_____no_output_____"
]
],
[
[
"#create crosstab\ndf=data.groupby(['sex','target'])['sex'].count().unstack('target')\ndf.plot(kind='bar',stacked=True,color=['green','blue'])\nplt.show()",
"_____no_output_____"
]
],
[
[
"# 1.10 Quantitative vs. Quantitative ( 2 marks)",
"_____no_output_____"
],
[
"### 1.10a) Create a scatter plot for showing age & st_depression",
"_____no_output_____"
]
],
[
[
"sns.scatterplot(x=\"age\",y=\"oldpeak\",data=data)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Overall Observation ( 5 marks):\n ",
"_____no_output_____"
],
[
"I have observed the following things:\n1.Females are targeted more than males.\n2.People of the age group between 50-60 are more targeted.\n3.People doing exercise affects are less affected.\n4. exercise induced angina are less affected.",
"_____no_output_____"
],
[
"# THE END",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec5f27b8e49464cf5328e9c070ae7a44e3fbd2c3 | 9,935 | ipynb | Jupyter Notebook | RF_notebooks/Cross_validation.ipynb | haddocking/MD-scoring | f744cf4abf4eed8c6b742332042d473d99107941 | [
"Apache-2.0"
]
| null | null | null | RF_notebooks/Cross_validation.ipynb | haddocking/MD-scoring | f744cf4abf4eed8c6b742332042d473d99107941 | [
"Apache-2.0"
]
| null | null | null | RF_notebooks/Cross_validation.ipynb | haddocking/MD-scoring | f744cf4abf4eed8c6b742332042d473d99107941 | [
"Apache-2.0"
]
| 1 | 2021-11-23T10:24:36.000Z | 2021-11-23T10:24:36.000Z | 30.289634 | 166 | 0.557524 | [
[
[
"# Cross validation of training set ",
"_____no_output_____"
],
[
"## Open features file",
"_____no_output_____"
]
],
[
[
"\n# Pandas is used for data manipulation\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\ntime='80_100'\n# Read in data as pandas dataframe and display first 5 rows\nfeatures = pd.read_csv('../features/features_training1//features_{}.csv'.format(time))\nfeatures_num=features.to_numpy()\nfeatures[:] = np.nan_to_num(features_num)\nnp.where(pd.isnull(features_num))\nRSEED=50\n#Check the start of the file\nfeatures.head(7)\n#features.describe(include='all')",
"_____no_output_____"
]
],
[
[
"## Plot the distribution of features for native and non-native models",
"_____no_output_____"
]
],
[
[
"# sklearn provides the iris species as integer values since this is required for classification\n# here we're just adding a column with the species names to the dataframe for visualisation\n\n#features['quality'] = np.array([features[i] for i in features)\ncolors = ['#91174A','#0072B2']\nsns.set(font_scale=3, context='notebook') \nsns.pairplot(features, hue='quality', palette=colors, height=4)\ng = sns.PairGrid(features, hue='quality', height=4)\ng.map_diag(sns.kdeplot)\ng.map_offdiag(sns.kdeplot, n_levels=6);\n\n",
"_____no_output_____"
]
],
[
[
"### Define Features and Labels and Convert Data to Arrays",
"_____no_output_____"
]
],
[
[
"# Use numpy to convert to arrays\nimport numpy as np\n\n\nlabels = features['quality']\n\n# Remove the labels from the features\n# axis 1 refers to the columns\nfeatures= features.drop('quality',axis = 1)\n\n\n# Saving feature names for later use\nfeature_list = list(features.columns)\nfeatures[features==np.inf]=np.nan\nfeatures.fillna(features.mean(), inplace=True)\ny = labels.map({'native':1,\"non-native\":0})\nx = features.values",
"_____no_output_____"
]
],
[
[
"## Specify the Random Forest classifier",
"_____no_output_____"
]
],
[
[
"# Using Skicit-learn to split data into training and testing sets\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestClassifier\n\n\nrf = RandomForestClassifier(bootstrap= True, max_depth=50, max_features='auto', min_samples_leaf=1, min_samples_split=2, n_estimators = 1000,oob_score= True,\n random_state = 42)\n\n# Split the data into training and testing sets\ntrain_features, test_features, train_labels, test_labels = train_test_split(x, y, test_size = 0.25,\n random_state = 42)",
"_____no_output_____"
]
],
[
[
"### Define plotting of ROC curve",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.metrics import roc_curve, auc\n# Plot an ROC. pred - the predictions, y - the expected output.\ndef plot_roc(pred,y):\n fpr, tpr, _ = roc_curve(y, pred)\n roc_auc = auc(fpr, tpr)\n plt.figure()\n plt.plot(fpr, tpr, label='ROC curve (area = %0.2f)' % roc_auc)\n plt.plot([0, 1], [0, 1], 'k--')\n plt.xlim([0.0, 1.0])\n plt.ylim([0.0, 1.05])\n plt.xlabel('False Positive Rate')\n plt.ylabel('True Positive Rate')\n plt.title('Receiver Operating Characteristic (ROC)')\n plt.legend(loc=\"lower right\")\n plt.show()\n\n\n",
"_____no_output_____"
]
],
[
[
"## Run the Random Forest classifier and plot the ROC curve",
"_____no_output_____"
]
],
[
[
"print(__doc__)\nsns.set(style=\"white\" )\n\n\nfrom sklearn.model_selection import RepeatedKFold\n\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy import interp\nfrom sklearn.model_selection import KFold\n\nfrom sklearn.model_selection import LeavePOut\nfrom sklearn import svm, datasets\nfrom sklearn.metrics import auc\nfrom sklearn.metrics import roc_curve\nfrom sklearn.model_selection import StratifiedKFold\n\n\n# #############################################################################\n# Classification and ROC analysis\n\n# Run classifier with cross-validation and plot ROC curves\ncv = StratifiedKFold(n_splits=100, random_state=26124)\n\n\n\nfig, ax = plt.subplots()\ntprs = []\naucs = []\nmean_fpr = np.linspace(0, 1, 100)\n\ni = 0\nfor train, test in cv.split(x, y):\n \n probas_ = rf.fit(x[train], y[train]).predict_proba(x[test])\n # Compute ROC curve and area the curve\n fpr, tpr, thresholds = roc_curve(y[test], probas_[:, 1])\n tprs.append(interp(mean_fpr, fpr, tpr))\n tprs[-1][0] = 0.0\n roc_auc = auc(fpr, tpr)\n aucs.append(roc_auc)\n plt.plot(fpr, tpr, lw=1, alpha=0.1,\n )\n\n i += 1\nplt.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',\n label='Luck', alpha=1)\n\nmean_tpr = np.mean(tprs, axis=0)\nmean_tpr[-1] = 1.0\nmean_auc = auc(mean_fpr, mean_tpr)\nstd_auc = np.std(aucs)\nplt.plot(mean_fpr, mean_tpr, color='b',\n label=r'Mean ROC (AUC = %0.2f $\\pm$ %0.2f)' % (mean_auc, std_auc),\n lw=2, alpha=1)\n\nstd_tpr = np.std(tprs, axis=0)\ntprs_upper = np.minimum(mean_tpr + std_tpr, 1)\ntprs_lower = np.maximum(mean_tpr - std_tpr, 0)\nplt.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.1,\n label=r'$\\pm$ 1 std. dev.')\n\nplt.xlim([-0.05, 1.05])\nplt.ylim([-0.05, 1.05])\nplt.xlabel('False Positive Rate', fontsize=14)\nplt.ylabel('True Positive Rate', fontsize=14)\nplt.title('Receiver operating characteristic example', fontsize=14)\nplt.legend(loc=\"lower right\", fontsize=14)\nplt.xticks(fontsize=14)\nplt.yticks(fontsize=14)\n\n",
"_____no_output_____"
]
],
[
[
"## Extract Feature Importances",
"_____no_output_____"
]
],
[
[
"\n\n# Get numerical feature importances\nimportances = list(rf.feature_importances_)\n\n# List of tuples with variable and importance\nfeature_importances = [(feature, round(importance, 2)) for feature, importance in zip(feature_list, importances)]\n\n# Sort the feature importances by most important first\nfeature_importances = sorted(feature_importances, key = lambda x: x[1], reverse = True)\n\n# Print out the feature and importances \n[print('Variable: {:20} Importance: {}'.format(*pair)) for pair in feature_importances];\n\n# list of x locations for plotting\nx_values = list(range(len(importances)))\n\n# Make a bar chart\nplt.bar(x_values, importances, orientation = 'vertical')\n\n# Tick labels for x axis\nplt.xticks(x_values, feature_list, rotation='vertical',fontsize=15)\nplt.yticks(fontsize=15)\n\n# Axis labels and title\nplt.ylabel('Importance', fontsize=20); plt.xlabel('Variable', fontsize=20); plt.title('Variable Importances time = {}ns'.format(time), fontsize=25); \n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec5f3358c5583b1c66430fc860d9a5decf90dfaa | 384,659 | ipynb | Jupyter Notebook | exercises/04-BikesRent_solution.ipynb | S7evenrg/PracticalMachineLearningClass | 2795827dd3e90124b663e7566a143bf70e6e8358 | [
"MIT"
]
| 2 | 2019-01-23T02:10:02.000Z | 2020-09-19T20:58:21.000Z | exercises/04-BikesRent_solution.ipynb | S7evenrg/PracticalMachineLearningClass | 2795827dd3e90124b663e7566a143bf70e6e8358 | [
"MIT"
]
| null | null | null | exercises/04-BikesRent_solution.ipynb | S7evenrg/PracticalMachineLearningClass | 2795827dd3e90124b663e7566a143bf70e6e8358 | [
"MIT"
]
| 3 | 2017-12-11T01:56:57.000Z | 2019-01-15T03:55:36.000Z | 327.648211 | 88,290 | 0.911631 | [
[
[
"# Exercise 04\n\nEstimate a regression using the Capital Bikeshare data\n\n\n## Forecast use of a city bikeshare system\n\nWe'll be working with a dataset from Capital Bikeshare that was used in a Kaggle competition ([data dictionary](https://www.kaggle.com/c/bike-sharing-demand/data)).\n\nGet started on this competition through Kaggle Scripts\n\nBike sharing systems are a means of renting bicycles where the process of obtaining membership, rental, and bike return is automated via a network of kiosk locations throughout a city. Using these systems, people are able rent a bike from a one location and return it to a different place on an as-needed basis. Currently, there are over 500 bike-sharing programs around the world.\n\nThe data generated by these systems makes them attractive for researchers because the duration of travel, departure location, arrival location, and time elapsed is explicitly recorded. Bike sharing systems therefore function as a sensor network, which can be used for studying mobility in a city. In this competition, participants are asked to combine historical usage patterns with weather data in order to forecast bike rental demand in the Capital Bikeshare program in Washington, D.C.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\n%matplotlib inline\nimport matplotlib.pyplot as plt\n\n# read the data and set the datetime as the index\nurl = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/bikeshare.csv'\nbikes = pd.read_csv(url, index_col='datetime', parse_dates=True)\n\n# \"count\" is a method, so it's best to name that column something else\nbikes.rename(columns={'count':'total'}, inplace=True)\n\nbikes.head()",
"_____no_output_____"
]
],
[
[
"* datetime - hourly date + timestamp \n* season - \n * 1 = spring\n * 2 = summer \n * 3 = fall \n * 4 = winter \n* holiday - whether the day is considered a holiday\n* workingday - whether the day is neither a weekend nor holiday\n* weather - \n * 1: Clear, Few clouds, Partly cloudy, Partly cloudy \n * 2: Mist + Cloudy, Mist + Broken clouds, Mist + Few clouds, Mist \n * 3: Light Snow, Light Rain + Thunderstorm + Scattered clouds, Light Rain + Scattered clouds \n * 4: Heavy Rain + Ice Pallets + Thunderstorm + Mist, Snow + Fog \n* temp - temperature in Celsius\n* atemp - \"feels like\" temperature in Celsius\n* humidity - relative humidity\n* windspeed - wind speed\n* casual - number of non-registered user rentals initiated\n* registered - number of registered user rentals initiated\n* total - number of total rentals",
"_____no_output_____"
]
],
[
[
"bikes.shape",
"_____no_output_____"
]
],
[
[
"# Exercise 4.1 \n\nWhat is the relation between the temperature and total?\n\nFor a one percent increase in temperature how much the bikes shares increases?\n\nUsing sklearn estimate a linear regression and predict the total bikes share when the temperature is 31 degrees ",
"_____no_output_____"
]
],
[
[
"# Pandas scatter plot\nbikes.plot(kind='scatter', x='temp', y='total', alpha=0.2)",
"_____no_output_____"
],
[
"# create X and y\nfeature_cols = ['temp']\nX = bikes[feature_cols]\ny = bikes.total\n\n# import, instantiate, fit\nfrom sklearn.linear_model import LinearRegression\nlinreg = LinearRegression()\nlinreg.fit(X, y)",
"_____no_output_____"
],
[
"# Pandas scatter plot\nbikes.plot(kind='scatter', x='temp', y='total', alpha=0.2)\n\n# Plot the linear regression\nx_plot = np.arange(bikes['temp'].min(), bikes['temp'].max())\ny_plot = linreg.intercept_ + linreg.coef_ * x_plot\nplt.plot(x_plot, y_plot, 'r', lw=5)",
"_____no_output_____"
],
[
"linreg.coef_",
"_____no_output_____"
]
],
[
[
"Interpreting the **\"temp\" coefficient** ($\\beta_1$):\n\n- It is the change in $y$ divided by change in $x$, or the \"slope\".\n- Thus, a temperature increase of 1 degree Celsius is **associated with** a rental increase of 9.17 bikes.\n- This is not a statement of causation.\n- $\\beta_1$ would be **negative** if an increase in temperature was associated with a **decrease** in rentals.",
"_____no_output_____"
]
],
[
[
"linreg.predict(31)",
"_____no_output_____"
]
],
[
[
"# Exercise 04.2\n\nEvaluate the model using the MSE",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\n\ny_pred = linreg.predict(X)\nmetrics.mean_squared_error(y, y_pred)",
"_____no_output_____"
]
],
[
[
"# Exercise 04.3\n\nDoes the scale of the features matter?\n\nLet's say that temperature was measured in Fahrenheit, rather than Celsius. How would that affect the model?",
"_____no_output_____"
]
],
[
[
"# create a new column for Fahrenheit temperature\nbikes['temp_F'] = bikes.temp * 9 / 5 + 32\nbikes.head()",
"_____no_output_____"
],
[
"# Seaborn scatter plot with regression line\nbikes.plot(kind='scatter', x='temp_F', y='total', alpha=0.2)",
"_____no_output_____"
],
[
"# create X and y\nfeature_cols = ['temp_F']\nX = bikes[feature_cols]\ny = bikes.total\n\n# instantiate and fit\nlinreg = LinearRegression()\nlinreg.fit(X, y)\n\n# print the coefficients\nprint(linreg.intercept_)\nprint(linreg.coef_)",
"-156.985617821\n[ 5.09474471]\n"
],
[
"# convert 31 degrees Celsius to Fahrenheit\n31. * 1.8 + 32",
"_____no_output_____"
],
[
"# predict rentals for 77 degrees Fahrenheit\nlinreg.predict(87.8)",
"_____no_output_____"
]
],
[
[
"**Conclusion:** The scale of the features is **irrelevant** for linear regression models. When changing the scale, we simply change our **interpretation** of the coefficients.",
"_____no_output_____"
],
[
"\n# Exercise 04.4\n\nRun a regression model using as features the temperature and temperature$^2$ using the OLS equations",
"_____no_output_____"
]
],
[
[
"# create X and y\nX = bikes['temp']\ny = bikes.total\n\nX = np.c_[np.ones(X.shape[0]), X, X**2 ]\n \nbetas = np.dot(np.linalg.inv(np.dot(X.T, X)),np.dot(X.T, y))",
"_____no_output_____"
],
[
"betas",
"_____no_output_____"
]
],
[
[
"# Exercise 04.5\n\nData visualization.\n\nWhat behavior is unexpected?",
"_____no_output_____"
]
],
[
[
"# explore more features\nfeature_cols = ['temp', 'season', 'weather', 'humidity']",
"_____no_output_____"
],
[
"# multiple scatter plots in Pandas\nfig, axs = plt.subplots(1, len(feature_cols), sharey=True)\nfor index, feature in enumerate(feature_cols):\n bikes.plot(kind='scatter', x=feature, y='total', ax=axs[index], figsize=(16, 3))",
"_____no_output_____"
]
],
[
[
"Are you seeing anything that you did not expect?",
"_____no_output_____"
],
[
" seasons: \n * 1 = spring\n * 2 = summer \n * 3 = fall \n * 4 = winter ",
"_____no_output_____"
]
],
[
[
"# pivot table of season and month\nmonth = bikes.index.month\npd.pivot_table(bikes, index='season', columns=month, values='temp', aggfunc=np.count_nonzero).fillna(0)",
"_____no_output_____"
],
[
"# box plot of rentals, grouped by season\nbikes.boxplot(column='total', by='season')",
"_____no_output_____"
],
[
"# line plot of rentals\nbikes.total.plot()",
"_____no_output_____"
]
],
[
[
"What does this tell us?\n\nThere are more rentals in the winter than the spring, but only because the system is experiencing **overall growth** and the winter months happen to come after the spring months.",
"_____no_output_____"
],
[
"# Exercise 04.6\n\nEstimate a regression using more features ['temp', 'season', 'weather', 'humidity'].\n\nHow is the performance compared to using only the temperature?",
"_____no_output_____"
]
],
[
[
"# create a list of features\nfeature_cols = ['temp', 'season', 'weather', 'humidity']\n\n# create X and y\nX = bikes[feature_cols]\ny = bikes.total\n\n# instantiate and fit\nlinreg = LinearRegression()\nlinreg.fit(X, y)\n\n# print the coefficients\nprint(linreg.intercept_)\nprint(linreg.coef_)",
"159.520687861\n[ 7.86482499 22.53875753 6.67030204 -3.11887338]\n"
],
[
"y_pred = linreg.predict(X)\nmetrics.mean_squared_error(y, y_pred)",
"_____no_output_____"
]
],
[
[
"# Exercise 04.7 (3 points)\n\nSplit the data in train and test\n\nWhich of the following models is the best in the testing set?\n* ['temp', 'season', 'weather', 'humidity']\n* ['temp', 'season', 'weather']\n* ['temp', 'season', 'humidity']\n",
"_____no_output_____"
]
],
[
[
"from sklearn.cross_validation import train_test_split\n\nfeature_cols = [\n ['temp', 'season', 'weather', 'humidity'],\n ['temp', 'season', 'weather'],\n ['temp', 'season', 'humidity']\n]\n\nfor features in feature_cols:\n X = bikes[features]\n y = bikes.total\n X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=123)\n\n linreg = LinearRegression()\n linreg.fit(X_train, y_train)\n\n y_pred = linreg.predict(X_test)\n print(features, metrics.mean_squared_error(y_test, y_pred))",
"['temp', 'season', 'weather', 'humidity'] 24226.7541277\n['temp', 'season', 'weather'] 26950.2784793\n['temp', 'season', 'humidity'] 24210.7965343\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec5f3643132b090bfdd8aa47bc5673c938cddc10 | 10,887 | ipynb | Jupyter Notebook | examples/pointcloud_tr.ipynb | dewloosh/dewloosh-geom | 5c97fbab4b68f4748bf4309184b9e0e877f94cd6 | [
"MIT"
]
| 2 | 2021-12-11T17:25:51.000Z | 2022-01-06T15:36:27.000Z | examples/pointcloud_tr.ipynb | dewloosh/dewloosh-geom | 5c97fbab4b68f4748bf4309184b9e0e877f94cd6 | [
"MIT"
]
| null | null | null | examples/pointcloud_tr.ipynb | dewloosh/dewloosh-geom | 5c97fbab4b68f4748bf4309184b9e0e877f94cd6 | [
"MIT"
]
| null | null | null | 20.936538 | 93 | 0.439423 | [
[
[
"from dewloosh.geom.space import StandardFrame, PointCloud\nfrom dewloosh.math.linalg.vector import Vector\nfrom dewloosh.math.linalg import linspace\nfrom dewloosh.geom.utils import center_of_points\nimport numpy as np",
"_____no_output_____"
],
[
"Lx, Ly, Lz = 300, 300, 300\npoints_per_edge = 3\nmesh_size = Lx / (points_per_edge-1)",
"_____no_output_____"
],
[
"points = []\nnTotalPoints = 0 # node counter\n\n# corners\ncorner_coords = [\n [-Lx/2, -Ly/2, -Lz/2],\n [Lx/2, -Ly/2, -Lz/2],\n [Lx/2, Ly/2, -Lz/2],\n [-Lx/2, Ly/2, -Lz/2],\n [-Lx/2, -Ly/2, Lz/2],\n [Lx/2, -Ly/2, Lz/2],\n [Lx/2, Ly/2, Lz/2],\n [-Lx/2, Ly/2, Lz/2]\n]\ncorner_coords = np.array(corner_coords)\npoints.append(corner_coords)\nnTotalPoints += len(corner_coords)\n\n# populate edges\nnodes_of_edges = [\n [0, 1], [1, 2], [2, 3], [3, 0],\n [4, 5], [5, 6], [6, 7], [7, 4],\n [0, 4], [1, 5], [2, 6], [3, 7]\n ]\nedge_coords = []\nN = points_per_edge + 2\nfor nodes in nodes_of_edges:\n p0 = corner_coords[nodes[0]]\n p1 = corner_coords[nodes[1]]\n edge_coords.append(linspace(p0, p1, N)[1:-1])\nedge_coords = np.vstack(edge_coords)\npoints.append(edge_coords)\nnTotalPoints += len(edge_coords)\n\n# faces\ncorners_of_faces = {\n 'front' : [1, 2, 6, 5], \n 'back' : [0, 3, 7, 4], \n 'left' : [2, 3, 7, 6], \n 'right' : [0, 1, 5, 4],\n 'bottom' : [0, 1, 2, 3], \n 'top' : [4, 5, 6, 7], \n}\nedges_of_faces = {\n 'front' : [1, 5, 9, 10], \n 'back' : [3, 7, 8, 11], \n 'right' : [0, 9, 4, 8], \n 'left' : [2, 6, 10, 11],\n 'bottom' : [0, 1, 2, 3], \n 'top' : [4, 5, 6, 7], \n}\n\n# center of face\ndef cof(id) : return center_of_points(corner_coords[corners_of_faces[id]])\n\n# face frames\nframes = {}\nframes['front'] = StandardFrame(dim=3, origo=cof('front'))\nrot90z = 'Body', [0, 0, np.pi/2], 'XYZ'\nframes['left'] = frames['front'].fork(*rot90z).move(cof('left') - cof('front'))\nframes['back'] = frames['left'].fork(*rot90z).move(cof('back') - cof('left'))\nframes['right'] = frames['back'].fork(*rot90z).move(cof('right') - cof('back'))\nrot_front_top = 'Body', [0, -np.pi/2, 0], 'XYZ'\nframes['top'] = frames['front'].fork(*rot_front_top).move(cof('top') - cof('front'))\nrot180y = 'Body', [0, np.pi, 0], 'XYZ'\nframes['bottom'] = frames['top'].fork(*rot180y).move(cof('bottom') - cof('top'))",
"_____no_output_____"
],
[
"frames['left'].origo()",
"_____no_output_____"
],
[
"coords = np.array([[0, -Lx/2, 0], [0, Lx/2, -Ly/2], [0, Lx/2, 0], [0, Lx/2, Ly/2]])\ncoords",
"_____no_output_____"
],
[
"pc = PointCloud(coords, frame=frames['left'])\npc",
"_____no_output_____"
],
[
"pc.show()",
"_____no_output_____"
],
[
"pc.center()",
"_____no_output_____"
]
],
[
[
"## **Single Vector Test**",
"_____no_output_____"
]
],
[
[
"A = StandardFrame(dim=3)",
"_____no_output_____"
],
[
"v1 = Vector([Lx/2, Ly/2, 0.0], frame=A)\nv1",
"_____no_output_____"
],
[
"v1.show()",
"_____no_output_____"
],
[
"arr2 = v1.show(frames['left'])\narr2",
"_____no_output_____"
],
[
"v2 = Vector(arr2, frame=frames['left'])\nv2",
"_____no_output_____"
],
[
"v2.show()",
"_____no_output_____"
],
[
"coords = np.array([v2.array]) - np.array([150., 0., 0.])\ncoords",
"_____no_output_____"
],
[
"v = Vector(coords[0], frame=frames['left'])\nv.show()",
"_____no_output_____"
],
[
"pc = PointCloud(coords, frame=frames['left'])\npc.show()",
"_____no_output_____"
],
[
"v = Vector(coords[0], frame=frames['left'])\nv.show()",
"_____no_output_____"
],
[
"pc = PointCloud(coords, frame=frames['left'])\npc.show()",
"_____no_output_____"
],
[
"frames['left'].origo()",
"_____no_output_____"
],
[
"frames['left'].origo(frames['left'])",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5f48b126f59091be41634993fab324e0f18271 | 734,340 | ipynb | Jupyter Notebook | Part 1 - questionnaire scales - 2014-11.ipynb | coej/Timing-study-data-processing | 4443d2c19d55077a3809930a70cfbfe63e3a5834 | [
"MIT"
]
| null | null | null | Part 1 - questionnaire scales - 2014-11.ipynb | coej/Timing-study-data-processing | 4443d2c19d55077a3809930a70cfbfe63e3a5834 | [
"MIT"
]
| null | null | null | Part 1 - questionnaire scales - 2014-11.ipynb | coej/Timing-study-data-processing | 4443d2c19d55077a3809930a70cfbfe63e3a5834 | [
"MIT"
]
| null | null | null | 58.709626 | 124 | 0.664783 | [
[
[
"from __future__ import print_function\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pylab as plt\nfrom collections import OrderedDict\n%matplotlib inline\n\n#pd.options.display.mpl_style = 'default'\n#pd.options.display.max_columns = 30\n#pd.options.display.max_rows = 40\n\ninput_scales_csv = \"RS2_QD_20141028a.csv\"",
"_____no_output_____"
]
],
[
[
"### Search & display functions",
"_____no_output_____"
]
],
[
[
"# using this for inline documentation so that it's clear\n# that the printing statement isn't part of the necessary\n# transformation code.\ndef html_print(df):\n from IPython.display import HTML\n try:\n out = df.to_html()\n except AttributeError:\n out = pd.DataFrame(df).to_html()\n return HTML(out)\n\n\ndef htmljoin(df_list, delimiter=''):\n from IPython.display import HTML\n return HTML(delimiter.join([x.to_html() for x in df_list]))\n\n\ndef col_matches(df, regex): \n import re \n cols = list(enumerate(df.columns)) \n matches = [c for (i, c) in cols \n if re.findall(regex, c)] \n return matches\n\n\ndef concat_matches(df, *args): \n assert all([len(r) for r in args])\n import re \n col_match_lists = [col_matches(df, regex) for regex in args] \n col_set = [df[matches] for matches in col_match_lists] \n if len(col_set) == 0:\n return None \n elif len(col_set) == 1:\n return col_set[0] \n else:\n return pd.concat(col_set, axis=1)\n\n \ndef show_frames(frame_list, delimiter=''): \n from IPython.display import HTML \n if len(frame_list) == len(delimiter): \n html_out = \"\"\n item_template = '<p><strong>{}</strong></p>{}<br>' \n for i, tup in enumerate(zip(frame_list, delimiter)):\n frame = tup[0]\n tag = tup[1]\n html_out += item_template.format(tag, frame.to_html()) \n return HTML(html_out) \n else: \n html_out = [df.to_html() for df in frame_list] \n return HTML(delimiter.join(html_out))\n \n \ndef compare_transformations(df, columns, functions, **kwargs): \n print('raw')\n df[columns].hist(**kwargs)\n plt.show() \n for name, func in functions.items(): \n print(name) \n df[columns].apply(func).hist(**kwargs)\n plt.show()\n \n \ndef quickcompare(r, size=(15,7)): \n return compare_transformations(dfo, col_matches(dfo, r),\n {'log1p': np.log1p,\n 'sqrt': np.sqrt, },\n figsize=size)\n",
"_____no_output_____"
]
],
[
[
"### Load data and set participant ID index",
"_____no_output_____"
]
],
[
[
"dfo = pd.read_csv(input_scales_csv,\n index_col='pid',\n )\n\ndef add_leading_zeroes(pid):\n no_leading_zero = str(pid)\n with_leading_zeroes = '000'[len(no_leading_zero):] + no_leading_zero\n return with_leading_zeroes\n\ndfo.index = dfo.reset_index('pid').pid.apply(add_leading_zeroes)\n\ndfo[::12].T.head()",
"_____no_output_____"
]
],
[
[
"## Missing values",
"_____no_output_____"
]
],
[
[
"#show missing values from original CSV entry ('77777')\n\nhtml_print(dfo.loc['053'].head(10)) #example P with missing WASI subtest",
"_____no_output_____"
],
[
"# Replace NaNs in note-taking columns with blanks for readability\n\ndef note_columns():\n note_columns = col_matches(dfo, \"notes\")\n assert len(note_columns) == 25\n \n more_note_columns = ['qbasic_ethnicityother',\n 'qmusic_dancestyle',\n 'qmusic_drumstyles',\n 'qmusic_instrumentlist',\n 'qmusic_gamenames']\n\n assert set(more_note_columns).issubset(dfo.columns) \n note_columns += more_note_columns \n assert len(note_columns) == 30 \n return note_columns\n\ndfo.loc[:, note_columns()] = dfo.loc[:, note_columns()].fillna(\"\")\n\n# Replace missing data (coded \"77777\") with NaNs\n# (but will change this back before exporting for external analyses)\ndfo.replace('77777', np.nan, inplace=True)\n\nhtml_print(dfo[30:40].T[18:22])",
"_____no_output_____"
],
[
"# View missing values\nhtmljoin([pd.DataFrame(row[row.isnull()])[:7].T \n for i, row in dfo.iterrows()\n if len(row[row.isnull()]) > 0])",
"_____no_output_____"
]
],
[
[
"## Transforming questionnaire items",
"_____no_output_____"
],
[
"### Quantiles and dichotomizing",
"_____no_output_____"
]
],
[
[
"simple_hour_items = col_matches(dfo, 'hours$') # '$' --> matches only end of name\nsimple_hour_items",
"_____no_output_____"
],
[
"# Extreme floor effect for \"practice hours\" items when viewing\n# the overall sample, so:\n# Zero or nonzero monthly practice hours?\n\nhour_counts_A = ['qmusic_singinghours',\n 'qmusic_singingtimes',\n 'qmusic_dancehours',\n 'qmusic_instrumenthours',\n 'qmusic_drumhours',\n 'qmusic_behaviors_09_danceprv', \n 'qmusic_behaviors_10_dancepub',\n 'qmusic_gamehoursall',\n 'qmusic_gamehoursdrumsticks',\n ]\n\nfor varname in hour_counts_A:\n s = dfo[varname]\n is_nonzero = (s > 0) # --> False, True\n dfo[varname + '_nonzero'] = is_nonzero.astype(int) # --> 0, 1 ",
"_____no_output_____"
],
[
"#pleasant matplotlib style set by pandas library\npd.options.display.mpl_style = 'default'\n\nprint('raw distributions:')\ndfo[hour_counts_A].hist(figsize=(10,10))\nplt.show()\n\nprint(concat_matches(dfo, '_nonzero$').head(4).T)",
"raw distributions:\n"
],
[
"pos_skewed_vars = ['qmusic_behaviors_07_yourself',\n 'qmusic_behaviors_08_otherprs',\n 'qmusic_behaviors_09_danceprv',\n 'qmusic_dancelevel']",
"_____no_output_____"
],
[
"compare_transformations(dfo, pos_skewed_vars, \n {'log1p': np.log1p, \n 'sqrt': np.sqrt,}, \n figsize=(15,7))",
"raw\n"
],
[
"for varname in pos_skewed_vars:\n s = dfo[varname] \n dfo[varname + '_ln1p'] = np.log1p(s) #ln of (x + 1)",
"_____no_output_____"
],
[
"from functools import partial\nmedsplit = partial(pd.qcut, q=2, labels=False)\n\ncompare_transformations(dfo, \n ['qmusic_dancelevel'], \n {'medsplit': medsplit}, \n figsize=(10,3))",
"raw\n"
],
[
"dfo['qmusic_dancelevel_tophalf'] = medsplit(dfo['qmusic_dancelevel'])\n # same as recoding as (0-1) --> 0\n # (2-7) --> 1",
"_____no_output_____"
],
[
"compare_transformations(dfo, \n col_matches(dfo, 'rel_totalmonths'), \n {'log1p': np.log1p,\n 'sqrt': np.sqrt, },\n figsize=(10,3))",
"raw\n"
],
[
"quickcompare('times$', size=(5,3))",
"raw\n"
],
[
"#zero-indexed order of tasks of this ISI among the two types\ndfo['orders_500'] = 1 - dfo.order_500ms_first\ndfo['orders_800'] = 0 + dfo.order_500ms_first\n\n# entered as strings in session_taskorder:\n# 1. Iso, Lin, Phase\n# 2. Iso, Phase, Lin\n# 3. Lin, Iso, Phase\n# 4. Lin, Phase, Iso\n# 5. Phase, Iso, Lin\n# 6. Phase, Lin, Iso\n\n# first character (number indicating overall order)\norder_n = dfo['session_taskorder'].apply(lambda x: int(x[0]))\n\n#zero-indexed order of tasks of this type among the three types\nisochronous_placement = {1: 0, 2: 0, 3: 1,\n 4: 2, 5: 1, 6: 2,}\n\nphaseshift_placement = {1: 2, 2: 1, 3: 2,\n 4: 1, 5: 0, 6: 0,}\n\nlinearchange_placement = {1: 1, 2: 2, 3: 0,\n 4: 0, 5: 2, 6: 1,}\n\ndfo['orders_iso'] = order_n.apply(lambda x: isochronous_placement[x])\ndfo['orders_phase'] = order_n.apply(lambda x: phaseshift_placement[x])\ndfo['orders_linear'] = order_n.apply(lambda x: linearchange_placement[x])\n\ndfo['orderc_iso_before_lin'] = (dfo.orders_iso < dfo.orders_linear).astype(int)\ndfo['orderc_iso_before_phase'] = (dfo.orders_iso < dfo.orders_phase).astype(int)\ndfo['orderc_phase_before_lin'] = (dfo.orders_phase < dfo.orders_linear).astype(int)\n\n\n# Set up precice ordering of each task in the set (1-indexed)\n# practice trials of isochronous/single-stimulus\ndfo['order_iso5t1'] = dfo.orders_500 + 1\ndfo['order_iso8t1'] = dfo.orders_800 + 1\n\n# +1 to set from zero-indexed to one-indexed,\n# then +2 because the the first two have passed (iso5t1 and iso5t2)\ndfo['order_iso5t2'] = 2 + (2 * dfo.orders_iso) + dfo.orders_500 + 1\ndfo['order_iso8t2'] = 2 + (2 * dfo.orders_iso) + dfo.orders_800 + 1\ndfo['order_psh5t'] = 2 + (2 * dfo.orders_phase) + dfo.orders_500 + 1\ndfo['order_psh8t'] = 2 + (2 * dfo.orders_phase) + dfo.orders_800 + 1\ndfo['order_lin5t'] = 2 + (2 * dfo.orders_linear) + dfo.orders_500 + 1\ndfo['order_lin8t'] = 2 + (2 * dfo.orders_linear) + dfo.orders_800 + 1\n\ndfo['order_iso5j'] = 6 + dfo['order_iso5t2'] #already adjusted to 1-index\ndfo['order_iso8j'] = 6 + dfo['order_iso8t2']\ndfo['order_psh5j'] = 6 + dfo['order_psh5t']\ndfo['order_psh8j'] = 6 + dfo['order_psh8t']\ndfo['order_lin5j'] = 6 + dfo['order_lin5t']\ndfo['order_lin8j'] = 6 + dfo['order_lin8t']\n\ndfo['order_isip5'] = dfo['order_iso5t1'] + 2 + 6 + 6\ndfo['order_isip8'] = dfo['order_iso8t1'] + 2 + 6 + 6\n\nhtml_print(concat_matches(dfo, '^order|taskorder')[::20].T.sort())",
"_____no_output_____"
],
[
"#Looking at high-end outliers, no transformations done here\n\nhours_col_names = col_matches(dfo, 'hours')\nhours_col_names += col_matches(dfo, 'behaviors_07')\nhours_col_names += col_matches(dfo, 'behaviors_08')\nhours_col_names += col_matches(dfo, 'behaviors_09')\nhours_col_names += col_matches(dfo, 'behaviors_10')\n\nhours_cols = dfo[hours_col_names]\n\nframes = [hours_cols.sort(c, ascending=False).head(8).T \n for c in hours_cols.columns]\n\nshow_frames(frames, hours_col_names)\n\n#log_hours_columns = np.log1p(hours_cols)\n#log_hours_columns.sort(col)",
"_____no_output_____"
],
[
"quickcompare('^calc')",
"raw\n"
],
[
"concat_matches(dfo, 'sumhours', 'qmusic_dance', 'iq').T",
"_____no_output_____"
],
[
"#Finally: set NaNs back to '77777'\ndfo.replace(np.nan, '77777', inplace=True)\n\nscales_output_updated = '2014-10-29a'\nprefix = \"c:/db_pickles/pickle - dfo-scales - \"\n\nimport cPickle as pickle\noutput_file= prefix + scales_output_updated + '.pickle'\npickle.dump(dfo, open(output_file, \"wb\"))\n\n# Proceed with pickle to Part 5",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5f4da5d44a1d25da9bb20634234bc440bd351d | 8,397 | ipynb | Jupyter Notebook | LEGALST-123/lab4/lab4.ipynb | jdmarshl/LS190 | 2b79b02c31006bd60b75b82e0bf8fbc2be3130d6 | [
"MIT"
]
| null | null | null | LEGALST-123/lab4/lab4.ipynb | jdmarshl/LS190 | 2b79b02c31006bd60b75b82e0bf8fbc2be3130d6 | [
"MIT"
]
| null | null | null | LEGALST-123/lab4/lab4.ipynb | jdmarshl/LS190 | 2b79b02c31006bd60b75b82e0bf8fbc2be3130d6 | [
"MIT"
]
| null | null | null | 22.155673 | 231 | 0.552816 | [
[
[
"from datascience import *\nfrom collections import Counter\nimport numpy as np\nimport pandas as pd\nfrom scipy import stats\n%matplotlib inline\nimport matplotlib.pyplot as plot\nplot.style.use('fivethirtyeight')",
"_____no_output_____"
]
],
[
[
"## Data",
"_____no_output_____"
],
[
"We'll continue using the ANES data for this lab!",
"_____no_output_____"
]
],
[
[
"incidents = pd.read_csv('../data/anes/ANES_legalst123_cleaned.csv')\nincidents.head()",
"_____no_output_____"
]
],
[
[
"## Empirical Distributions",
"_____no_output_____"
],
[
"### Data Manipulation and Plotting Review",
"_____no_output_____"
],
[
"Write code that saves the \"post_liberal_rating\" column in the NCVS data to a Series variable, and only includes values below 150.",
"_____no_output_____"
],
[
"Plot a histogram of the data:",
"_____no_output_____"
],
[
"### Question 1",
"_____no_output_____"
],
[
"What patterns do you notice about the histogram? Is the distribution left or right skewed? What does the skew suggest about what cash values are usually implicated in crime?",
"_____no_output_____"
],
[
"### Law of Averages",
"_____no_output_____"
],
[
"Write a function, \"empirical_hist_crime\" that takes a Series and a sample size as its argument, and then draws a histogram based on the results. Consult the book for help!",
"_____no_output_____"
]
],
[
[
"# empirical_hist_crime function",
"_____no_output_____"
]
],
[
[
"Check how many rows are in the table with the \"size\" method, and then use your self-defined function to plot histograms taking sample sizes 10, 100, 1000, and the total number of rows.",
"_____no_output_____"
]
],
[
[
"# Check number of rows",
"_____no_output_____"
]
],
[
[
"### Question 2",
"_____no_output_____"
],
[
"What happens to the histograms (compared to the original frin Q1) as you increase the sample size? How does this relate to the Law of Averages? What is the relationship between sample size and population parameter estimation?",
"_____no_output_____"
],
[
"## Hypothesis Testing",
"_____no_output_____"
],
[
"In this section, we'll cover the basic tools for hypothesis testing!",
"_____no_output_____"
],
[
"### Jury Selection",
"_____no_output_____"
],
[
"First, we'll use the jury selection example from the book. Here, we are concerned with making sure that the racial composition of a jury is not statistically different from the racial composition of the population.",
"_____no_output_____"
]
],
[
[
"# Create the table\njury = pd.DataFrame(data = {'Ethnicity': ['Asian', 'Black', 'Latino', 'White', 'Other'],\n 'Eligible': [0.15, 0.18, 0.12, 0.54, 0.01],\n 'Panels': [0.26, 0.08, 0.08, 0.54, 0.04]}\n)\n\njury",
"_____no_output_____"
],
[
"# Horizontal Bar Chart\njury.barh('Ethnicity')",
"_____no_output_____"
],
[
"# Augment with the difference between the \"panels\" columns and \"eligible\" column\njury_with_diffs = jury.assign(Difference = jury.loc[:, 'Panels'] - jury.loc[:, 'Eligible'])\njury_with_diffs",
"_____no_output_____"
]
],
[
[
"Write code that does a t-test between the \"Eligible\" and \"Panels\" columns. Hint: https://docs.scipy.org/doc/scipy-0.15.1/reference/generated/scipy.stats.ttest_ind.html#scipy.stats.ttest_ind",
"_____no_output_____"
]
],
[
[
"# scipy t-test",
"_____no_output_____"
]
],
[
[
"### Hypothesis Testing on NCVS Data",
"_____no_output_____"
],
[
"Now let's try with the NCVS data! Write code that creates a new DataFrame with the \"post_liberal_rating\" and \"post_conservative_rating\" as columns, and only includes values below 150.",
"_____no_output_____"
]
],
[
[
"value_lost = incidents.loc[:, [\"post_liberal_rating\", \"post_conservative_rating\"]]\nvalue_lost = value_lost.where(value_lost[\"post_liberal_rating\"] < 150)\nvalue_lost = value_lost.where(value_lost[\"post_conservative_rating\"] < 150)\n\nvalue_lost.head()",
"_____no_output_____"
]
],
[
[
"### Question 3",
"_____no_output_____"
],
[
"Plot a histogram of both the post liberal rating and post conservating rating side by side. Experiment with different bin widths. Visually, what can you infer about the shape of each data?",
"_____no_output_____"
]
],
[
[
"value_lost.hist(sharex=True, sharey=True)",
"_____no_output_____"
]
],
[
[
"### Question 4",
"_____no_output_____"
],
[
"Now write code to do a t-test between liberl and conservative. For the T-Test to work, you have to remove NaN values first (Check pandas documentation to find out how to do so).",
"_____no_output_____"
]
],
[
[
"# Drop NaN",
"_____no_output_____"
],
[
"# t-test",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
ec5f55854d7a0f8d7e436512894e6ab7e59c7398 | 326,831 | ipynb | Jupyter Notebook | case_study_1_and_2.ipynb | diphosphane/MLinQCbook22-delta | 1d1acb1b79a7e81d7818497cb5437e1a0791debc | [
"MIT"
]
| null | null | null | case_study_1_and_2.ipynb | diphosphane/MLinQCbook22-delta | 1d1acb1b79a7e81d7818497cb5437e1a0791debc | [
"MIT"
]
| null | null | null | case_study_1_and_2.ipynb | diphosphane/MLinQCbook22-delta | 1d1acb1b79a7e81d7818497cb5437e1a0791debc | [
"MIT"
]
| 1 | 2022-02-23T03:51:36.000Z | 2022-02-23T03:51:36.000Z | 791.358354 | 135,396 | 0.945599 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom typing import Dict, List, Any\nfrom itertools import product\nfrom copy import deepcopy\nfrom abc import abstractmethod, ABC",
"_____no_output_____"
]
],
[
[
"# define KRR class",
"_____no_output_____"
]
],
[
[
"class BaseEstimator(ABC):\n def fit(self, X, y): \n if len(X)[0] != len(y)[0]:\n raise Error('Error, size of X and y is not equal.')\n \n @abstractmethod\n def predict(self, X): pass\n \n @staticmethod\n def mse(y_ref, y_pred) -> float:\n tmp = (y_ref - y_pred) ** 2\n return tmp.sum() / len(tmp)\n \n @abstractmethod\n def set_kw(self, kw: Dict[str, Any]): pass\n \n\nclass KRR(BaseEstimator):\n def __init__(self, kernel='Linear', lambda_=0.1, **kw):\n self.kernel = getattr(self, kernel)\n self.params = {'lambda': lambda_, 'sigma': 0.01}\n self.set_kw(kw)\n self.train_x = None\n self.alpha = None\n \n def set_kw(self, kw: Dict[str, Any]):\n for k, v in kw.items():\n self.params[k] = v\n \n def Linear(self, x1, x2):\n return x1 @ x2.T\n \n def Gaussian(self, x1, x2):\n dist = np.sum(x1**2, 1).reshape(-1, 1) \\\n + np.sum(x2**2, 1) \\\n - 2*np.dot(x1, x2.T)\n return np.exp(-0.5 * dist / (self.params['sigma']**2))\n \n def fit(self, X, y):\n self.train_x = np.asarray(X)\n self.train_y = np.asarray(y).reshape(-1, 1)\n K = self.kernel(self.train_x, self.train_x)\n self.alpha = np.linalg.inv(K + self.params['lambda'] * np.eye(len(self.train_x))) @ self.train_y\n \n def predict(self, X):\n self.pred_x = np.asarray(X)\n K = self.kernel(self.pred_x, self.train_x)\n return K @ self.alpha",
"_____no_output_____"
]
],
[
[
"# define hyperparameter optimization class",
"_____no_output_____"
]
],
[
[
"class GridSearch(BaseEstimator):\n def __init__(self, estimator: BaseEstimator, param_grid: Dict[str, Any], cv: int=0) -> None:\n super().__init__()\n self.estimator = estimator\n self.param_grid = param_grid\n self.cv = cv\n self.best_param: Dict[str, Any] = dict()\n \n def fit(self, X, y):\n train_idx_list, val_idx_list = self._cv_split(X.shape[0])\n best_mse = 1e50\n # best_model = None\n self.best_param: Dict[str, Any] = dict()\n keys, values = zip(*self.param_grid.items())\n for params_value in product(*values):\n params = dict(zip(keys, params_value))\n self.estimator.set_kw(params)\n mse = self._evaluate_model(self.estimator, X, y, train_idx_list, val_idx_list)\n if mse < best_mse:\n best_mse = mse\n self.best_param = params\n # best_model = deepcopy(self.estimator)\n self.estimator.set_kw(self.best_param)\n self.estimator.fit(X, y)\n \n def predict(self, X):\n return self.estimator.predict(X)\n \n def _cv_split(self, cnt: int):\n train_cnt = int(cnt * 0.8); # val_cnt = cnt - train_cnt\n train_idx_list = []; val_idx_list = []\n if self.cv in [0, 1]:\n train_idx_list.append(np.arange(train_cnt))\n val_idx_list.append(np.arange(train_cnt, cnt))\n else:\n batch_num = cnt // self.cv\n for i in range(self.cv):\n flg1, flg2 = batch_num * i, batch_num * (i+1)\n train_idx_list.append(np.r_[np.arange(flg1), np.arange(flg2, cnt)])\n val_idx_list.append(np.arange(flg1, flg2))\n return train_idx_list, val_idx_list\n \n @classmethod\n def _evaluate_model(cls, estimator: BaseEstimator, X, y, train_idx_list: List[Any], val_idx_list: List[Any]):\n mse_list = []\n for train_idx, val_idx in zip(train_idx_list, val_idx_list):\n estimator.fit(X[train_idx], y[train_idx])\n y_pred = estimator.predict(X[val_idx])\n mse = cls.mse(y[val_idx], y_pred)\n mse_list.append(mse)\n return np.array(mse_list).mean()\n \n def set_kw(self, kw: Dict[str, Any]): pass",
"_____no_output_____"
]
],
[
[
"# load data",
"_____no_output_____"
]
],
[
[
"x = np.loadtxt('R_451.dat')\nyt = np.loadtxt('E_FCI_451.dat')\nyb = np.loadtxt('E_UHF_451.dat')\ndelta_y = yt - yb\ntrain_idx = np.loadtxt('itrain.dat', dtype=np.int64) - 1\ntrain_idx = train_idx[3:]\ntest_idx = np.loadtxt('itest.dat', dtype=np.int64) - 1",
"_____no_output_____"
]
],
[
[
"## Fig 1",
"_____no_output_____"
]
],
[
[
"krr = KRR(kernel='Gaussian')\ngs = GridSearch(krr, param_grid={'lambda': np.logspace(-40, -2, 20),\n 'sigma': np.logspace(-10, 8, 20)})\nplt.figure(figsize=(15, 5), dpi=120)\nplt.subplot(121)\nplt.plot(x, yt, 'k', lw=4, label='FCI')\nplt.plot(x, yb, 'c', lw=2, label='UHF')\ntrain_x, train_y = x[train_idx], yt[train_idx]\ntest_x, test_y = x[test_idx], yt[test_idx]\ngs.fit(train_x.reshape(-1, 1), train_y.reshape(-1, 1))\npred_y = gs.predict(test_x.reshape(-1, 1))\nplt.plot(test_x, pred_y.ravel(), 'r', label='ML')\nprint(f'ML param: {gs.best_param}')\n\ntrain_x, train_y = x[train_idx], delta_y[train_idx]\ntest_x, test_y = x[test_idx], delta_y[test_idx]\ngs.fit(train_x.reshape(-1, 1), train_y.reshape(-1, 1))\npred_y = gs.predict(test_x.reshape(-1, 1))\nplt.plot(test_x, pred_y.ravel() + yb[test_idx], 'b', label='∆-ML')\nprint(f'∆-ML param: {gs.best_param}')\n\nplt.plot(train_x, yt[train_idx], 'ko', markerfacecolor='none', markersize=10, label='FCI training data')\nplt.xlabel('R, $\\mathrm{\\AA}$'); plt.ylabel('Energy, Hartree')\nplt.legend()\nplt.text(-0.1, 1.0, 'a', transform=plt.gca().transAxes, fontsize=18, va='top', ha='right')\n\nplt.subplot(122)\nplt.plot(x, yt-yb, 'k', label='difference between FCI and UHF')\nplt.plot(x[test_idx], pred_y, 'r', label='ML prediction of the difference')\nplt.plot(train_x, train_y, 'ko', markerfacecolor='none', markersize=10, label='training data of difference between FCI and UHF')\nplt.xlabel('R, $\\mathrm{\\AA}$'); plt.ylabel('Energy, Hartree')\nplt.legend()\nplt.text(-0.125, 1.0, 'b', transform=plt.gca().transAxes, fontsize=18, va='top', ha='right')\n\nplt.gcf().savefig('fig1.jpg')",
"ML param: {'lambda': 1e-08, 'sigma': 2.6366508987303554}\n∆-ML param: {'lambda': 1e-10, 'sigma': 2.6366508987303554}\n"
]
],
[
[
"## Case Study 1 & Fig 3",
"_____no_output_____"
]
],
[
[
"krr = KRR(kernel='Gaussian')\ngs = GridSearch(krr, param_grid={'lambda': np.logspace(-40, -2, 20),\n 'sigma': np.logspace(-10, 8, 20)}, cv=5)\nplt.figure(figsize=(7, 4), dpi=150)\nplt.plot(x, yt, lw=8, alpha=0.3, label='FCI ground truth')\ntrain_x, train_y = x[train_idx], yt[train_idx]\ntest_x, test_y = x[test_idx], yt[test_idx]\ngs.fit(train_x.reshape(-1, 1), train_y.reshape(-1, 1))\nprint(f'FCI direct learning parameter: {gs.best_param}')\npred_y = gs.predict(test_x.reshape(-1, 1))\nplt.plot(test_x, pred_y.ravel(), label='FCI direct learning')\nplt.plot(train_x, train_y, 'ro', markersize=5, label='FCI training data')\nplt.xlabel('R, $\\mathrm{\\AA}$'); plt.ylabel('Energy, Hartree')\n\ntrain_x, train_y = x[train_idx], delta_y[train_idx]\ntest_x, test_y = x[test_idx], delta_y[test_idx]\ngs.fit(train_x.reshape(-1, 1), train_y.reshape(-1, 1))\npred_y = gs.predict(test_x.reshape(-1, 1))\nplt.plot(test_x, pred_y.ravel() + yb[test_idx], alpha=0.7, label='∆-learning prediction')\nplt.legend()\n\nplt.gcf().savefig('fig4.jpg')",
"FCI direct learning parameter: {'lambda': 1e-08, 'sigma': 2.6366508987303554}\n"
]
],
[
[
"## Case Study 2 & Fig 4",
"_____no_output_____"
]
],
[
[
"krr = KRR(kernel='Gaussian')\ngs = GridSearch(krr, param_grid={'lambda': np.logspace(-40, -2, 20),\n 'sigma': np.logspace(-10, 8, 20)}, cv=5)\nplt.figure(figsize=(15, 5), dpi=120)\nplt.subplot(121)\ntrain_x, train_y = x[train_idx], yb[train_idx]\ntest_x, test_y = x[test_idx], yb[test_idx]\ngs.fit(train_x.reshape(-1, 1), train_y.reshape(-1, 1))\npred_uhf = gs.predict(test_x.reshape(-1, 1))\ntrain_x, train_y = x[train_idx], (yt - yb)[train_idx]\ntest_x, test_y = x[test_idx], (yt - yb)[test_idx]\ngs.fit(train_x.reshape(-1, 1), train_y.reshape(-1, 1))\npred_delta = gs.predict(test_x.reshape(-1, 1))\npred_hML = pred_uhf + pred_delta\n\nplt.plot(test_x, pred_hML.ravel(), label='hML prediction')\nplt.plot(x, yt, alpha=0.3, lw=5, label='FCI ground truth')\nplt.plot(x, yb, alpha=0.3, lw=5, label='UHF ground truth')\n# plt.plot(x[train_idx], yt[train_idx], 'ro', label='training data')\nplt.plot(x[train_idx], yt[train_idx], 'ro', markerfacecolor='none', markersize=8, label='training data of target value')\nplt.plot(x[train_idx], yb[train_idx], 'go', markerfacecolor='none', markersize=8, label='training data of base line')\nplt.xlabel('R, $\\mathrm{\\AA}$'); plt.ylabel('Energy, Hartree')\nplt.legend()\nplt.text(-0.1, 1.0, 'a', transform=plt.gca().transAxes, fontsize=18, va='top', ha='right')\n\nplt.subplot(122)\ntest_x, test_y = x[test_idx], yb[test_idx]\ngs.fit(x.reshape(-1, 1), yb.reshape(-1, 1))\n# plt.plot(x, yb, 'co', markerfacecolor='none', markersize=10, label='training data of base line')\npred_uhf = gs.predict(test_x.reshape(-1, 1))\npred_hML = pred_uhf + pred_delta\nplt.plot(test_x, pred_hML.ravel(), label='hML prediction')\nplt.plot(test_x, pred_uhf.ravel(), label='UHF prediction')\nplt.plot(x, yt, lw=5, alpha=0.3, label='FCI ground truth')\nplt.plot(x, yb, lw=5, alpha=0.3, label='UHF ground truth')\nplt.plot(x[train_idx], yt[train_idx], 'ko', markerfacecolor='none', markersize=8, label='training data of target value')\nplt.xlabel('R, $\\mathrm{\\AA}$'); plt.ylabel('Energy, Hartree')\nplt.legend()\nplt.text(-0.1, 1.0, 'b', transform=plt.gca().transAxes, fontsize=18, va='top', ha='right')\n\nplt.gcf().savefig('fig5.jpg')",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec5f5bf0427c4b539808187279eaa0251cbb7cb5 | 129,707 | ipynb | Jupyter Notebook | Louvain_tutorial.ipynb | qlinhta/graph-clustering-louvain | 53bb2ea990f2fffe867feaf5700bb7d521c5943d | [
"MIT"
]
| null | null | null | Louvain_tutorial.ipynb | qlinhta/graph-clustering-louvain | 53bb2ea990f2fffe867feaf5700bb7d521c5943d | [
"MIT"
]
| null | null | null | Louvain_tutorial.ipynb | qlinhta/graph-clustering-louvain | 53bb2ea990f2fffe867feaf5700bb7d521c5943d | [
"MIT"
]
| null | null | null | 128.55005 | 47,491 | 0.865088 | [
[
[
"graph={('a','b'):1,('a','c'):2,('b','c'):2,('c','d'):1,('d','e'):4,('e','g'):1,('f','g'):4,('f','d'):1,('f','f'):1}",
"_____no_output_____"
],
[
"import networkx as nx\nimport matplotlib.pyplot as plt\n\nplt.figure()\nG=nx.Graph()\nfor edge in graph:\n G.add_edge(edge[0],edge[1],weight=graph[edge])\n \npos=nx.spring_layout(G)\nnx.draw_networkx_nodes(G,pos)\nnx.draw_networkx_edges(G,pos)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Calculating modularity\nThe Louvain algorithm maximizes a property known as modularity. If each node has a color then the modularity measures the relative likelihood that edges in the network will connect nodes of the same color. For example we can asign the following colors:",
"_____no_output_____"
]
],
[
[
"color= {'a': 'c0', 'b':'c1', 'c':'c2', 'd':'c1', 'e':'c1', 'f':'c0','g':'c2'} ",
"_____no_output_____"
]
],
[
[
"to get",
"_____no_output_____"
]
],
[
[
"plt.figure()\n\ncolor_list=['r','b','g','c','m','orange','yellow','k','pink']\nn=0\nfor c in set(color.values()):\n nodes=[node for node in color if color[node]==c]\n nx.draw_networkx_nodes(G,pos,nodelist=nodes, node_color=color_list[n],alpha=0.8)\n n=n+1\n nx.draw_networkx_edges(G,pos)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Modularity was priginally defined in Newman's 2004 paper to be\n$$\nQ=\\sum_{c}\\left(e_{cc}-a_{c}^{2}\\right)\n$$\nwhere $e_{cc}$ denotes the fraction of the total weight of edges that both begin and end at nodes of color $c$, and $a_{c}$ is the fraction of weight the total weight of edges that end (or begin) at nodes of color $c$. \n\n### A note of the definition of modularity\nThe Louvain paper refers to \nThe definition given in the louvain paper is expressed differently as\n$$\nQ=\\frac{1}{S}\\sum_{i,j}\\left[w_{ij}-\\frac{s_{i}s_{j}}{S}\\right]\\delta(c_{i},c_{j})\n$$\nwhere $w_{ij}$ is the weight of the edge between $i$ and $j$, $s_{i}=\\sum_{j}w_{ij}$, and $S=\\sum_{i}s_{i}$. The $\\delta$ used here is the Kronecker delta function; $\\delta(x,y)=1$ if $x=y$, and 0 otherwise. The notation $\\sum_{ij}$ means the sum over all elements in a matrix, in this case it is the matrix whose $i,j$th element is $w_{ij}-s_{i}s_{j}/S$. Note that this is not the same as the sum over all pairs of nodes since it counts each pair of nodes twice (with the exception of pairs that consist of the same node twice). \n\nHere I will explain why these two definitions are not the same. First, if $G$ is a graph and $G_{c}$ is the subgraph of nodes of color $c$ then \n $e_{cc}$ is defined as\n$$\ne_{cc}=\\frac{1}{W}\\sum_{(i,j)\\in G_{c}}w_{ij}\n$$\nwhere $W$ is the total weight of the graph, and \n$$\nW=\\sum_{(i,j)\\in G}w_{ij}=\\frac{1}{2}\\left[S+\\sum_{i}w_{ii}\\right].\n$$\nTo make it look more like the first term in the Louvain equation we note that \n$$\n\\sum_{(i,j)\\in G_{c}}w_{ij}=\\frac{1}{2}\\left[\\sum_{i,j}w_{ij}+\\sum_{i}w_{ii}\\right]\\delta(c,c_{i})\\delta(c,c_{j})\n$$\nso \n$$\ne_{cc}=\\frac{1}{2W}\\left[\\sum_{i,j}w_{ij}+\\sum_{i}w_{ii}\\right]\\delta(c,c_{i})\\delta(c,c_{j}).\n$$\n\nNext, the Newman paper defines $a_{c}$ as fraction of the weight of edges that connect to nodes of color $c$. It then goes on to say that in a randomly colored graph the expectation is $e_{c1c2}=a_{c1}a_{c2}$ but this is incorrect. Consider the following graph:",
"_____no_output_____"
]
],
[
[
"plt.figure()\ngraph2={('a','b'):1,('b','c'):1,('c','d'):1,('d','a'):1}\nG2=nx.Graph()\nfor edge in graph2:\n G2.add_edge(edge[0],edge[1],weight=graph2[edge])\npos2=nx.spring_layout(G2)\ncolor2={'a': 'c0', 'b':'c0', 'c':'c1', 'd':'c1'} \nn=0\nfor c in set(color2.values()):\n nodes2=[node for node in color2 if color2[node]==c]\n nx.draw_networkx_nodes(G2,pos2,nodelist=nodes2, node_color=color_list[n],alpha=0.8)\n n=n+1\n nx.draw_networkx_edges(G2,pos2)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Here $a_{red}=3/4$ and $a_{blue}=3/4$ which means the null expectation for the fraction of weight between red and red is $9/16$, the same for edges between blue and blue, and the same for edges between red and blue. Adding these together we ought to get $1$ but instead we get $27/16$. A better definition (that appears to have been used in the Louvain definition of modularity) is that $a_{c}$ is the fraction of ends of edges that are attached to nodes of color $c$,\n$$\na_{c}=\\frac{1}{S}\\sum_{i\\in G_{c}}s_{i}.\n$$\nNow the expectation for the weight of edges between red and red is $(1/2)\\times(1/2)$ and its the same for blue and blue. For edges that go from one color to the other, the expectation is $2\\times(1/2)\\times(1/2)$ since there are effectively two ways that the edge can be placed, either the red stub could be selected then the blue one, or the blue one is selected first then the red. From this definition the sum of these expected fractions is $1$, as it should be.\n\nFor Newman's formula we need to know $a_{c}^{2}$. Noting that\n$$\n\\begin{split}\na_{c}^{2}&=\\frac{1}{S^{2}}\\left[\\sum_{i\\in G_{c}}s_{i}\\right]^{2}\\\\\n&=\\frac{1}{S^{2}}\\sum_{i,j}s_{i}s_{j}\\delta(c,c_{i})\\delta(c,c_{j})\n\\end{split}\n$$\nwe have \n$$\nQ=\\sum_{c}\\left[\\sum_{i,j}w_{ij}+\\sum_{i}w_{ii}\\right]\\delta(c,c_{i})\\delta(c,c_{j})-\\sum_{i,j}s_{i}s_{j}\\delta(c,c_{i})\\delta(c,c_{j})\n$$\nwhich can be rewritten\n$$\nQ=\\sum_{i,j}\\left[\\frac{w_{ij}}{2W}-\\frac{s_{i}s_{j}}{S^{2}}\\right]\\delta(c_{i},c_{j})+\\sum_{i}\\frac{w_{ii}}{2W}.\n$$\nComparing this to the definition given in the Louvain paper we see two differences: an extra term that depends on the weight of the self-loops, and that the two terms within the sum have different denominators. If the network contains no self-loops then $S=2W$ and the two definitions become equivalent to each other. In other words, Newman's definition of modularity and the version used in the Louvain paper are only the same if there are no self-loops.\n\nTo make the calculation of modularity as fast as possible we can reduce the number of operations by summing over each distinct pair of nodes in the network rather than summing over every matrix element. First lets get a list of the nodes,",
"_____no_output_____"
]
],
[
[
"nodes=sorted(list(set([edge[0] for edge in graph]+[edge[1] for edge in graph])))\nN=len(nodes)\nprint('nodes:',nodes)",
"nodes: ['a', 'b', 'c', 'd', 'e', 'f', 'g']\n"
]
],
[
[
"Now we need to know the weight of edges between each distinct pair:",
"_____no_output_____"
]
],
[
[
"Weight={}\nfor i in nodes:\n for j in nodes:\n edge=tuple(sorted([i,j]))\n if (i,j) in graph:\n Weight[edge]=graph[(i,j)]\n elif (j,i) in graph:\n Weight[edge]=graph[(j,i)]\n else:\n Weight[edge]=0\nprint(Weight)",
"{('a', 'a'): 0, ('a', 'b'): 1, ('a', 'c'): 2, ('a', 'd'): 0, ('a', 'e'): 0, ('a', 'f'): 0, ('a', 'g'): 0, ('b', 'b'): 0, ('b', 'c'): 2, ('b', 'd'): 0, ('b', 'e'): 0, ('b', 'f'): 0, ('b', 'g'): 0, ('c', 'c'): 0, ('c', 'd'): 1, ('c', 'e'): 0, ('c', 'f'): 0, ('c', 'g'): 0, ('d', 'd'): 0, ('d', 'e'): 4, ('d', 'f'): 1, ('d', 'g'): 0, ('e', 'e'): 0, ('e', 'f'): 0, ('e', 'g'): 1, ('f', 'f'): 1, ('f', 'g'): 4, ('g', 'g'): 0}\n"
]
],
[
[
"Notice that when we read the edge from the network we sort it. This prevents the same edge form appearing twice in the dictionary.\n\nThe sum over matrix elements ($\\sum_{i,j}$) relates to the sum over distinct pairs of nodes ($\\sum_{(i,j)}$) in the following way:\n$$\n\\sum_{(i,j)}w_{ij}=\\frac{1}{2}\\left[\\sum_{i,j}w_{ij}+\\sum_{i}w_{ii}\\right].\n$$\nand so the modularity equation can be expressed as \n$$\nQ=\\sum_{i}\\left(\\frac{s_{i}}{S}\\right)^{2}+\\sum_{(i,j)}\\left[\\frac{w_{ij}}{W}-\\frac{2s_{i}s_{j}}{S^{2}}\\right]\\delta(c_{i},c_{j}).\n$$\n\n### Computing modularity\n\nThe formula above turns out to be quite easy to write as code. First, we will want to know the strength of every node ($s_{i}$ for all nodes $i$).",
"_____no_output_____"
]
],
[
[
"strength={}\nfor i in nodes:\n strength[i]=sum([graph[edge] for edge in graph if edge[0]==i or edge[1]==i])\nprint('strength:',strength)",
"strength: {'a': 3, 'b': 3, 'c': 5, 'd': 6, 'e': 5, 'f': 6, 'g': 5}\n"
]
],
[
[
"and the total strength (S)",
"_____no_output_____"
]
],
[
[
"total_strength=sum([strength[i] for i in nodes])\nprint('total strength:',total_strength)",
"total strength: 33\n"
]
],
[
[
"and the total weight (W)",
"_____no_output_____"
]
],
[
[
"total_weight=sum([graph[edge] for edge in graph])\nprint('total weightt:',total_weight)",
"total weightt: 17\n"
]
],
[
[
"Now we can put it all together to find the modularity",
"_____no_output_____"
]
],
[
[
"Q=sum([(strength[i]/total_strength)**2 for i in nodes])\nfor edge in Weight:\n i=edge[0]\n j=edge[1]\n if color[i]==color[j]: \n Q=Q+Weight[edge]/(total_weight)-2*strength[i]*strength[j]/(total_strength**2)\n\nprint('Modularity of this coloring =',Q)",
"Modularity of this coloring = -0.05207151731215902\n"
]
],
[
[
"This code is included in the Louvain module and can be accessed as follows:",
"_____no_output_____"
]
],
[
[
"import Louvain\nprint('Q =',Louvain.modularity(Weight,color))",
"Q = -0.05207151731215902\n"
]
],
[
[
"For comparison, I also copied the code from the python community library. ",
"_____no_output_____"
]
],
[
[
"print('Q =',Louvain.net_x_modularity(color,G))",
"Q = -0.04844290657439447\n"
]
],
[
[
"The small difference in the result is because of the existence of self-loop in the graph; in $\\verb|net_x_modularity|$, self-loops are considered to attach to the same node twice the weight of a self-loop is counted twice when calculating the strength of a node (which I believe is incorrect). Additionally, the denominator for the second term is $2W$ when it should be $S$. ",
"_____no_output_____"
],
[
"## Finding an optimal coloring\n\nThis is where the Louvain algorithm begins. Actually, to make things easier later on we first want to create an adjacency list for every node. We start with an empty dictionary and add each node as a neighbor of itself",
"_____no_output_____"
]
],
[
[
"neighbors={}\nfor i in nodes:\n neighbors[i]=list(set([edge[0] for edge in graph if edge[1]==i]+[edge[1] for edge in graph if edge[0]==i])) \n\nprint(neighbors)",
"{'a': ['b', 'c'], 'b': ['a', 'c'], 'c': ['b', 'a', 'd'], 'd': ['f', 'c', 'e'], 'e': ['g', 'd'], 'f': ['g', 'f', 'd'], 'g': ['f', 'e']}\n"
]
],
[
[
"The procedure starts by asigning a unique color to every node. We are going to keep track of the node colors using a dictionary",
"_____no_output_____"
]
],
[
[
"# color tells us the color of each node\ncolor={}\nfor n in range(N):\n i=nodes[n]\n # this will be the label for the color\n c='c'+str(n)\n color[i]=c \nprint(color)",
"{'a': 'c0', 'b': 'c1', 'c': 'c2', 'd': 'c3', 'e': 'c4', 'f': 'c5', 'g': 'c6'}\n"
]
],
[
[
"The first part of the procedure is to compute the change in modularity that occurs when the color of a node changes. For a node, $i$ that changes color from $c1$ to $c2$ the magnitude of this change is \n$$\n\\Delta Q=\\frac{w_{i,c2}}{W}-\\frac{2s_{i}(s_{i}+w_{c2})}{S^{2}}-\\frac{w_{i,c1}}{W}+\\frac{2s_{i}w_{c1}}{S^{2}}\n$$\n\nWhere $w_{i,c}$ denotes the weight of edges adjacent to $i$ that link to nodes of color $c$, initially this is ",
"_____no_output_____"
]
],
[
[
"weight_in_color={} \nfor j in nodes:\n for i in nodes:\n if i in neighbors[j]:\n weight_in_color[(i,color[j])]=Weight[tuple(sorted((i,j)))]\n else:\n weight_in_color[(i,color[j])]=0",
"_____no_output_____"
]
],
[
[
"and we need to know $w_{c}$ the total weight of edges of edges attached to nodes of color $c$",
"_____no_output_____"
]
],
[
[
"total_weight_of_color={}\nfor c in set(color.values()):\n total_weight_of_color[c]=sum([strength[i] for i in nodes if color[i]==c])",
"_____no_output_____"
]
],
[
[
"When each node is considered, the source is the color that it currently is and the target is the color that it may change to. T1 - T4 are the four terms in the equation for $\\Delta Q$.",
"_____no_output_____"
]
],
[
[
"i='a'\nsource='c0'\ntarget='c2'\n\nT1 =(weight_in_color[(i,target)]+Weight[(i,i)])/total_weight\n\nT2 =2*strength[i]*(strength[i]+total_weight_of_color[target])/(total_strength**2)\n \nT3=weight_in_color[(i,source)]/total_weight\n\nT4=2*strength[i]*total_weight_of_color[source]/total_strength**2\n \n# compute the total change if ID moved from source community to target community\ndelta_Q=T1-T2-T3+T4",
"_____no_output_____"
]
],
[
[
"Just to prove that the formula is correct we can calculate the modularity befor and after making the change",
"_____no_output_____"
]
],
[
[
"Q_before=Louvain.modularity(Weight,color)\n# make the change\ncolor[i]=target\nQ_after=Louvain.modularity(Weight,color)\nprint(Q_after-Q_before)\nprint(delta_Q)\n\n# now reset it to how it was before so we don't mess things up!\ncolor[i]=source",
"0.09009884945713824\n0.09009884945713822\n"
]
],
[
[
"Now we have finally reached the fun part! The following is a loop that sequentially chooses nodes. It calculates, for every color, the change in Q that would occur if the node changed to that color, if the biggest change it finds is positive (and not tiny) then it makes that change.",
"_____no_output_____"
]
],
[
[
"# if no improvements occur in N iterations then this number reaches N and the loop ends\nunsuccessful_iterations=0\n# n is the index of the node we want to check\nn=0\n# the loop terminates when we iterate through every node and find that none of them yield an improvement\nwhile unsuccessful_iterations<N:\n i=nodes[n]\n n=(n+1) % N\n # source is the community ID is currently in\n source=color[i]\n # compute the weight of edges between ID and nodes in source community (including itself)\n T3=weight_in_color[(i,source)]/total_weight \n # compute the expectation \n T4=2*strength[i]*total_weight_of_color[source]/total_strength**2\n\n best_delta=0\n # instead of choosing all possible target colors, rule out the ones ID has no connection to\n for target in [x for x in set(color.values()) if x!=source and weight_in_color[(i,x)]>0]: \n # compute the weight of edges between ID and nodes in target community (including itself) \n T1 =(weight_in_color[(i,target)]+Weight[(i,i)])/total_weight\n # compute the expectation\n T2 =2*strength[i]*(strength[i]+total_weight_of_color[target])/(total_strength**2)\n # compute the total change if ID moved from source community to target community\n delta_Q=T1-T2-T3+T4\n\n # keep track of the largest\n if delta_Q>best_delta: \n best_delta=delta_Q\n best_target=target\n\n if best_delta>0.00000001:\n #print(modularity(graph,color)+best_delta)\n color[i]=best_target\n #print(modularity(graph,color))\n #print()\n # update the total_similarity of source\n total_weight_of_color[source]=total_weight_of_color[source]-strength[i]\n # update the total_similarity of target\n total_weight_of_color[best_target]=total_weight_of_color[best_target]+strength[i]\n # update similarity to the source/target community of every node\n for j in neighbors[i]:\n weight_in_color[(j,source)]=weight_in_color[(j,source)]-Weight[tuple(sorted((i,j)))]\n weight_in_color[(j,best_target)]=weight_in_color[(j,best_target)]+Weight[tuple(sorted((i,j)))]\n\n # best delta is large so the iteration was successful. Reset the counter\n unsuccessful_iterations=0\n else:\n # if no improvements occur in N iterations then this number reaches N and the loop ends\n unsuccessful_iterations=unsuccessful_iterations+1 \n \nprint(color)",
"{'a': 'c2', 'b': 'c2', 'c': 'c2', 'd': 'c4', 'e': 'c4', 'f': 'c6', 'g': 'c6'}\n"
]
],
[
[
"Lets plot the result to see if it worked!",
"_____no_output_____"
]
],
[
[
"plt.figure()\nn=0\nfor c in set(color.values()):\n node_group=[node for node in color if color[node]==c]\n nx.draw_networkx_nodes(G,pos,nodelist=node_group, node_color=color_list[n])\n n=n+1\n nx.draw_networkx_edges(G,pos)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Recomputing the modularity we have ",
"_____no_output_____"
]
],
[
[
"print('Q =',Louvain.modularity(Weight,color))",
"Q = 0.4901960784313726\n"
]
],
[
[
"So that seems to have worked, however, there may still be room for improvement. The next stage of the algorithm looks to see if an improvement can be made by merging any pair of communities. \n\n## Merging communities\nWe make a new network with each color as a community ",
"_____no_output_____"
]
],
[
[
"# start by creating a Weight dictionary where all the pairs have weight 0\nnew_weight={}\n\nfor c_i in set(color.values()):\n for c_j in set(color.values()):\n new_edge=tuple(sorted((c_i,c_j)))\n if new_edge not in new_weight:\n new_weight[new_edge]=0\n# for each of the old edges, add its weight to the weight of the appropriate new edge\nfor edge in Weight:\n # new-edge is the edge between two colors \n new_edge=tuple(sorted((color[edge[0]],color[edge[1]])))\n # once we know which new edge to add to, we can add it\n new_weight[new_edge]=new_weight[new_edge]+Weight[edge]\n\nprint('color:',color)\nprint('new weight',new_weight)",
"color: {'a': 'c2', 'b': 'c2', 'c': 'c2', 'd': 'c4', 'e': 'c4', 'f': 'c6', 'g': 'c6'}\nnew weight {('c2', 'c2'): 5, ('c2', 'c4'): 1, ('c2', 'c6'): 0, ('c4', 'c4'): 4, ('c4', 'c6'): 2, ('c6', 'c6'): 5}\n"
]
],
[
[
"Everything we have done up to here is combined into one function in the Louvain module. The function $\\verb|modulize|$ takes the dictionary Weight as input and gives a new graph as output. There next stage of the algorithm simply involves putting the the new network through the $\\verb|modulize|$ process repeatedly until no more changes occur.",
"_____no_output_____"
]
],
[
[
"new_weight,new_color=Louvain.modulize(new_weight)\n\nprint('color:',new_color)",
"color: {'c2': 'c0', 'c4': 'c1', 'c6': 'c2'}\n"
]
],
[
[
"Since the new color dictionary only tells us how the old colors have changed, the color of the nodes is now the color of the color it became during the first iteration. To get a dictionary that maps the actual node to its new color we do",
"_____no_output_____"
]
],
[
[
"final_color={}\nfor i in nodes:\n final_color[i]=new_color[color[i]] \nprint('Final color',final_color)",
"Final color {'a': 'c0', 'b': 'c0', 'c': 'c0', 'd': 'c1', 'e': 'c1', 'f': 'c2', 'g': 'c2'}\n"
]
],
[
[
"Lets take a look, ",
"_____no_output_____"
]
],
[
[
"plt.figure()\nn=0\nfor c in set(color.values()):\n node_group=[node for node in color if color[node]==c]\n nx.draw_networkx_nodes(G,pos,nodelist=node_group, node_color=color_list[n])\n n=n+1\n nx.draw_networkx_edges(G,pos)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"The only thing that has changed is the names of the colors; that means we have reached a local optimum!\n\nWe can automate this process in a 'while' loop that stops when there is no change between two iterations. Note that each time we iterate we need to keep track of the colors of each node. The following code performs this on our original graph (named 'graph') and outputs the final coloring:",
"_____no_output_____"
]
],
[
[
"end_loop=False\nnumber_of_modules=len(nodes)\nfinal_color={}\nfor i in nodes:\n final_color[i]=i\n#stopping criteria: no change has occured\nwhile end_loop==False: \n #partition produces 0) the partition 1) a dictionary of similarities between groups partition \n graph,color=Louvain.modulize(graph)\n for i in nodes:\n final_color[i]=color[final_color[i]] \n\n # if number of modules is the same then end the loop\n if number_of_modules==len(set(color.values())):\n end_loop=True\n else:\n number_of_modules=len(set(color.values())) \nprint(final_color)",
"{'a': 'c0', 'b': 'c0', 'c': 'c0', 'd': 'c1', 'e': 'c1', 'f': 'c2', 'g': 'c2'}\n"
]
],
[
[
"## Real networks\n\nFirst we need to read the files and put them in the desired format",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# choose a network from shark_0, parakeet_2_2, stumptailed_macaque, Howler_monkeys, Macaques_Massen\nnetwork='stumptailed_macaque'\ndf=pd.read_csv('Data/'+network+'_edgelist.csv')\n\ngraph={}\nfor i,row in df.iterrows():\n graph[(row['ID1'],row['ID2'])]=row['Weight']\n \ncolor=Louvain.get_colors(graph)",
"_____no_output_____"
]
],
[
[
"Often it is better to have the result in the form of a partition",
"_____no_output_____"
]
],
[
[
"plt.figure()\nG=nx.Graph()\nfor edge in graph:\n G.add_edge(edge[0],edge[1],weight=graph[edge])\npos=nx.spring_layout(G)\n\nn=0\nfor c in set(color.values()):\n node_group=[node for node in color if color[node]==c]\n nx.draw_networkx_nodes(G,pos,nodelist=node_group, node_color=color_list[n])\n n=n+1\n nx.draw_networkx_edges(G,pos)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
],
[
"Q=Louvain.modularity(graph,color)\nprint(Q)",
"0.27721936051031304\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec5f7e1b70ec725a7550fcd04c5edcce3ea8330c | 6,400 | ipynb | Jupyter Notebook | src/FileTransformer.ipynb | Pristor/Thoth | 83e46a4ae728dd30a8f657c17eadd232661c8695 | [
"MIT"
]
| null | null | null | src/FileTransformer.ipynb | Pristor/Thoth | 83e46a4ae728dd30a8f657c17eadd232661c8695 | [
"MIT"
]
| 3 | 2020-11-13T18:41:04.000Z | 2022-02-10T01:37:01.000Z | src/FileTransformer.ipynb | Pristor/Thoth | 83e46a4ae728dd30a8f657c17eadd232661c8695 | [
"MIT"
]
| null | null | null | 38.787879 | 123 | 0.431094 | [
[
[
"import os\nfrom data_source import preproc as pp\nfrom multiprocessing import Pool\nfrom functools import partial\nimport string\nimport h5py",
"_____no_output_____"
],
[
"class Dataset():\n \"\"\"\n Dataset class functions:\n read_iam: reads the lines from iam Dataset saves test, valid and train set in a dict\n dataset structure : {train:{dt:[], gt:[]}, valid:{dt:[], gt:[]}, test:{dt:[], gt:[]}}\n \n preprocess_partitions preprocesses the data\n \"\"\"\n \n def __init__(self, iam_path):\n \n self.iam_path = iam_path\n self.dataset = None\n self.partitions = ['train', 'valid', 'test']\n \n \n def read_iam(self):\n \"\"\"reads iam dataset\"\"\"\n pt_path = os.path.join(self.iam_path, \"partitions\")\n paths = {\"train\": open(os.path.join(pt_path, \"trainset.txt\")).read().splitlines(),\n \"valid\": open(os.path.join(pt_path, \"validationset1.txt\")).read().splitlines(),\n \"test\": open(os.path.join(pt_path, \"testset.txt\")).read().splitlines()}\n \n lines = open(os.path.join(self.iam_path, \"lines.txt\")).read().splitlines()\n gt_dict = dict()\n \n for line in lines:\n if (not line or line[0]== \"#\"):\n continue\n \n splitted = line.split()\n \n if splitted[1] == \"ok\":\n gt_dict[splitted[0]] = \" \".join(splitted[8::]).replace(\"|\",\" \")\n \n dataset = dict()\n \n for i in self.partitions:\n dataset[i] = {\"dt\": [], \"gt\": []}\n \n for line in paths[i]:\n try:\n split = line.split(\"-\")\n \n folder = f\"{split[0]}-{split[1]}\"\n image = f\"{split[0]}-{split[1]}-{split[2]}.png\"\n \n image_path = os.path.join(self.iam_path, \"lines\", split[0], folder, image)\n \n dataset[i]['gt'].append(gt_dict[line])\n dataset[i]['dt'].append(image_path)\n \n except KeyError:\n pass\n \n self.dataset = dataset\n \n \n def preprocess_partitions(self, input_size):\n \"\"\" function to preprocess the data, removes bad samples, preprocesses images\"\"\"\n \n print(\"Partitions will be preprocessed...\")\n \n for i in self.partitions:\n \n arange = range(len(self.dataset[i]['gt']))\n \n for j in reversed(arange):\n #handles spaces around punctations\n text = pp.text_standardize(self.dataset[i]['gt'][j])\n \n if not self.check_text(text):\n #remove if the example has more punctations than letters\n self.dataset[i]['gt'].pop(j)\n self.dataset[i]['dt'].pop(j)\n continue\n \n self.dataset[i]['gt'][j] = text.encode()\n \n pool = Pool()\n #multiprocess: apllies pp.preprocess on each value of the array\n #partial -> changes args of function\n self.dataset[i]['dt'] = pool.map(partial(pp.preprocess, input_size=input_size), self.dataset[i]['dt'])\n pool.close()\n pool.join()\n \n print(\"Partitions preprocessing finished\")\n \n def save(self, source_path):\n \"saves partitions as hdf5\"\n os.makedirs(os.path.dirname(source_path), exist_ok=True)\n \n for i in self.partitions:\n with h5py.File(source_path, \"a\") as hf:\n hf.create_dataset(f\"{i}/dt\", data=ds.dataset[i]['dt'], compression=\"gzip\", compression_opts=9)\n hf.create_dataset(f\"{i}/gt\", data=ds.dataset[i]['gt'], compression=\"gzip\", compression_opts=9)\n print(f\"[OK] {i} partition.\")\n\n print(f\"Transformation finished.\")\n \n \n \n \n @staticmethod\n def check_text(text):\n \"\"\"Make sure text has more characters instead of punctuation marks\"\"\"\n\n strip_punc = text.strip(string.punctuation).strip()\n no_punc = text.translate(str.maketrans(\"\", \"\", string.punctuation)).strip()\n\n if len(text) == 0 or len(strip_punc) == 0 or len(no_punc) == 0:\n return False\n\n punc_percent = (len(strip_punc) - len(no_punc)) / len(strip_punc)\n\n return len(no_punc) >= 2 and punc_percent <= 0.1\n ",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
ec5f84d578af57d0950fe15f8d33ebf7b5d760e3 | 1,909 | ipynb | Jupyter Notebook | tests/nb_builds/nb_alice/10.00-The_Lobster_Quadrille.ipynb | rmsrosa/nbbinder | abaa46a256d8f02944c3383d6c1f8caf3d185a10 | [
"MIT"
]
| 2 | 2020-02-26T21:26:03.000Z | 2020-07-06T21:29:37.000Z | tests/nb_builds/nb_alice/10.00-The_Lobster_Quadrille.ipynb | rmsrosa/nbbinder | abaa46a256d8f02944c3383d6c1f8caf3d185a10 | [
"MIT"
]
| 6 | 2019-12-10T11:58:38.000Z | 2020-07-17T14:30:39.000Z | tests/nb_builds/nb_alice/10.00-The_Lobster_Quadrille.ipynb | rmsrosa/nbbinder | abaa46a256d8f02944c3383d6c1f8caf3d185a10 | [
"MIT"
]
| null | null | null | 21.449438 | 193 | 0.55736 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec5f8b72705649332295f527a839abbb0df5f540 | 63,892 | ipynb | Jupyter Notebook | demo.ipynb | sinjax/micrograd | 35b7ca4ac12a7fca9b14bc147a88882729f40baf | [
"MIT"
]
| null | null | null | demo.ipynb | sinjax/micrograd | 35b7ca4ac12a7fca9b14bc147a88882729f40baf | [
"MIT"
]
| null | null | null | demo.ipynb | sinjax/micrograd | 35b7ca4ac12a7fca9b14bc147a88882729f40baf | [
"MIT"
]
| null | null | null | 138.895652 | 23,880 | 0.86053 | [
[
[
"### MicroGrad demo",
"_____no_output_____"
]
],
[
[
"import random\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"from micrograd.engine import Value\nfrom micrograd.nn import Neuron, Layer, MLP",
"_____no_output_____"
],
[
"np.random.seed(1337)\nrandom.seed(1337)",
"_____no_output_____"
],
[
"# make up a dataset\n\nfrom sklearn.datasets import make_moons, make_blobs\nX, y = make_moons(n_samples=100, noise=0.1)\n\ny = y*2 - 1 # make y be -1 or 1\n# visualize in 2D\nplt.figure(figsize=(5,5))\nplt.scatter(X[:,0], X[:,1], c=y, s=20, cmap='jet')",
"_____no_output_____"
],
[
"# initialize a model \nmodel = MLP(2, [16, 16, 1]) # 2-layer neural network\nprint(model)\nprint(\"number of parameters\", len(model.parameters()))",
"MLP of [Layer of [ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2), ReLUNeuron(2)], Layer of [ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16), ReLUNeuron(16)], Layer of [LinearNeuron(16)]]\nnumber of parameters 337\n"
],
[
"# loss function\ndef loss(batch_size=None):\n \n # inline DataLoader :)\n if batch_size is None:\n Xb, yb = X, y\n else:\n ri = np.random.permutation(X.shape[0])[:batch_size]\n Xb, yb = X[ri], y[ri]\n inputs = [list(map(Value, xrow)) for xrow in Xb]\n \n # forward the model to get scores\n scores = list(map(model, inputs))\n \n # svm \"max-margin\" loss\n losses = [(1 + -yi * scorei).relu() for yi, scorei in zip(yb, scores)]\n data_loss = sum(losses) * (1.0 / len(losses))\n # L2 regularization\n alpha = 1e-4\n reg_loss = alpha * sum((p*p for p in model.parameters()))\n total_loss = data_loss + reg_loss\n \n # also get accuracy\n accuracy = [(yi > 0) == (scorei.data > 0) for yi, scorei in zip(yb, scores)]\n return total_loss, sum(accuracy) / len(accuracy)\n\ntotal_loss, acc = loss()\nprint(total_loss, acc)",
"Value(data=0.8958441028683222, grad=0) 0.5\n"
],
[
"# optimization\nlearning_rate = 0.001\n\nfor k in range(200):\n \n # forward\n total_loss, acc = loss()\n \n # backward\n model.zero_grad()\n total_loss.grad = 1\n total_loss.backward()\n \n # update (sgd)\n for p in model.parameters():\n p.data -= learning_rate * p.grad\n \n if k % 1 == 0:\n print(f\"step {k} loss {total_loss.data}, accuracy {acc*100}%\")\n",
"step 0 loss 0.8958441028683222, accuracy 50.0%\nstep 1 loss 1.1385269605233466, accuracy 76.0%\nstep 2 loss 0.6492743386774888, accuracy 78.0%\nstep 3 loss 0.360925921530329, accuracy 83.0%\nstep 4 loss 0.8907324403996303, accuracy 77.0%\nstep 5 loss 0.5646826452300904, accuracy 79.0%\nstep 6 loss 0.3156801109748185, accuracy 85.0%\nstep 7 loss 0.4157825430989441, accuracy 78.0%\nstep 8 loss 0.6213067306738933, accuracy 79.0%\nstep 9 loss 0.4180567496592728, accuracy 83.0%\nstep 10 loss 0.28219041839006803, accuracy 86.0%\nstep 11 loss 0.2743239585624415, accuracy 86.0%\nstep 12 loss 0.29492786796191506, accuracy 84.0%\nstep 13 loss 0.36221204595037265, accuracy 85.0%\nstep 14 loss 0.3129397150958227, accuracy 82.0%\nstep 15 loss 0.39887106321336463, accuracy 85.0%\nstep 16 loss 0.26381378000762695, accuracy 88.0%\nstep 17 loss 0.2611445705079117, accuracy 89.0%\nstep 18 loss 0.2752681553368485, accuracy 87.0%\nstep 19 loss 0.28804160476940854, accuracy 87.0%\nstep 20 loss 0.3197969880189577, accuracy 84.0%\nstep 21 loss 0.3449379623363138, accuracy 86.0%\nstep 22 loss 0.2756387236601572, accuracy 89.0%\nstep 23 loss 0.27820808925472196, accuracy 89.0%\nstep 24 loss 0.3085868094404226, accuracy 88.0%\nstep 25 loss 0.31010112931687156, accuracy 89.0%\nstep 26 loss 0.28038435283369106, accuracy 88.0%\nstep 27 loss 0.2997974224235103, accuracy 88.0%\nstep 28 loss 0.27001777994095405, accuracy 91.0%\nstep 29 loss 0.27067772677770013, accuracy 89.0%\nstep 30 loss 0.2747605031172743, accuracy 89.0%\nstep 31 loss 0.3035014820642558, accuracy 89.0%\nstep 32 loss 0.2725855361876734, accuracy 91.0%\nstep 33 loss 0.28687238814026, accuracy 90.0%\nstep 34 loss 0.26764861548783747, accuracy 91.0%\nstep 35 loss 0.2852017826044789, accuracy 90.0%\nstep 36 loss 0.2578400625465807, accuracy 92.0%\nstep 37 loss 0.2554404590646637, accuracy 92.0%\nstep 38 loss 0.25550183383522723, accuracy 92.0%\nstep 39 loss 0.25029321021808515, accuracy 92.0%\nstep 40 loss 0.25192826275312213, accuracy 91.0%\nstep 41 loss 0.24987624192733948, accuracy 92.0%\nstep 42 loss 0.2680397351117543, accuracy 89.0%\nstep 43 loss 0.2745380022217276, accuracy 90.0%\nstep 44 loss 0.2477204796558657, accuracy 91.0%\nstep 45 loss 0.24282540173487477, accuracy 92.0%\nstep 46 loss 0.2562948834124049, accuracy 90.0%\nstep 47 loss 0.25213740846499766, accuracy 91.0%\nstep 48 loss 0.2638952006101125, accuracy 90.0%\nstep 49 loss 0.2778945587919835, accuracy 90.0%\nstep 50 loss 0.2436047530471408, accuracy 91.0%\nstep 51 loss 0.23063827402272657, accuracy 93.0%\nstep 52 loss 0.23542341678734385, accuracy 92.0%\nstep 53 loss 0.2219834172350498, accuracy 92.0%\nstep 54 loss 0.22176346609417014, accuracy 91.0%\nstep 55 loss 0.21920879536035612, accuracy 93.0%\nstep 56 loss 0.22446965828988596, accuracy 93.0%\nstep 57 loss 0.2084693846294397, accuracy 92.0%\nstep 58 loss 0.206263334077088, accuracy 96.0%\nstep 59 loss 0.20786962471400225, accuracy 94.0%\nstep 60 loss 0.1988647997815982, accuracy 94.0%\nstep 61 loss 0.2016031698218395, accuracy 94.0%\nstep 62 loss 0.19666227291358532, accuracy 94.0%\nstep 63 loss 0.1944721856566291, accuracy 94.0%\nstep 64 loss 0.19236768609906907, accuracy 96.0%\nstep 65 loss 0.2032755234506043, accuracy 95.0%\nstep 66 loss 0.22538748467144445, accuracy 92.0%\nstep 67 loss 0.25042015120374894, accuracy 93.0%\nstep 68 loss 0.18123011875165812, accuracy 97.0%\nstep 69 loss 0.18273680027356018, accuracy 95.0%\nstep 70 loss 0.18423354022975694, accuracy 95.0%\nstep 71 loss 0.17888393396821947, accuracy 95.0%\nstep 72 loss 0.18556825263698637, accuracy 94.0%\nstep 73 loss 0.20908171358324087, accuracy 95.0%\nstep 74 loss 0.1709706278890858, accuracy 95.0%\nstep 75 loss 0.17090834162914526, accuracy 95.0%\nstep 76 loss 0.1745509344182436, accuracy 94.0%\nstep 77 loss 0.18130787961990374, accuracy 95.0%\nstep 78 loss 0.17773293491721473, accuracy 94.0%\nstep 79 loss 0.18341075061379242, accuracy 95.0%\nstep 80 loss 0.14949546295725546, accuracy 95.0%\nstep 81 loss 0.15014263640927955, accuracy 95.0%\nstep 82 loss 0.15182412297913686, accuracy 94.0%\nstep 83 loss 0.14348549612688635, accuracy 95.0%\nstep 84 loss 0.1563821936790777, accuracy 94.0%\nstep 85 loss 0.16125698219655338, accuracy 95.0%\nstep 86 loss 0.152525171751533, accuracy 94.0%\nstep 87 loss 0.15162638747164667, accuracy 95.0%\nstep 88 loss 0.14396671682895215, accuracy 95.0%\nstep 89 loss 0.1385541131587785, accuracy 95.0%\nstep 90 loss 0.14706217508404354, accuracy 95.0%\nstep 91 loss 0.13884892497809492, accuracy 95.0%\nstep 92 loss 0.13986800002428076, accuracy 95.0%\nstep 93 loss 0.12292058123909481, accuracy 96.0%\nstep 94 loss 0.12278580131606984, accuracy 97.0%\nstep 95 loss 0.13011906763950176, accuracy 95.0%\nstep 96 loss 0.1299448575848511, accuracy 95.0%\nstep 97 loss 0.12280725339498925, accuracy 96.0%\nstep 98 loss 0.1296829351238715, accuracy 96.0%\nstep 99 loss 0.11515452948465027, accuracy 96.0%\nstep 100 loss 0.11680576492300558, accuracy 97.0%\nstep 101 loss 0.1275530486641907, accuracy 95.0%\nstep 102 loss 0.11858138147679602, accuracy 96.0%\nstep 103 loss 0.11945506060480983, accuracy 96.0%\nstep 104 loss 0.11925739263187832, accuracy 96.0%\nstep 105 loss 0.11163385762683546, accuracy 96.0%\nstep 106 loss 0.12130756357145137, accuracy 96.0%\nstep 107 loss 0.10539507969359126, accuracy 97.0%\nstep 108 loss 0.10607042452822817, accuracy 97.0%\nstep 109 loss 0.11843703016931181, accuracy 96.0%\nstep 110 loss 0.1112830252690315, accuracy 97.0%\nstep 111 loss 0.10555242555247156, accuracy 97.0%\nstep 112 loss 0.11162802205102697, accuracy 97.0%\nstep 113 loss 0.10103415557576888, accuracy 97.0%\nstep 114 loss 0.10718859834558549, accuracy 98.0%\nstep 115 loss 0.10130682167954483, accuracy 97.0%\nstep 116 loss 0.10468012310808926, accuracy 98.0%\nstep 117 loss 0.09980474990087512, accuracy 97.0%\nstep 118 loss 0.10170948184913117, accuracy 98.0%\nstep 119 loss 0.10495815750128004, accuracy 97.0%\nstep 120 loss 0.092576498815773, accuracy 98.0%\nstep 121 loss 0.0901964221489017, accuracy 97.0%\nstep 122 loss 0.09041792528797574, accuracy 98.0%\nstep 123 loss 0.08804854434935536, accuracy 97.0%\nstep 124 loss 0.0870586305109394, accuracy 98.0%\nstep 125 loss 0.08797611555460179, accuracy 97.0%\nstep 126 loss 0.09941611918364339, accuracy 98.0%\nstep 127 loss 0.10709685404791819, accuracy 96.0%\nstep 128 loss 0.08000252569363407, accuracy 98.0%\nstep 129 loss 0.08113262441545457, accuracy 97.0%\nstep 130 loss 0.08547340772828471, accuracy 99.0%\nstep 131 loss 0.10516048590823834, accuracy 96.0%\nstep 132 loss 0.07559220445754242, accuracy 98.0%\nstep 133 loss 0.07845938939984295, accuracy 97.0%\nstep 134 loss 0.08242277084218921, accuracy 99.0%\nstep 135 loss 0.08694419037295546, accuracy 97.0%\nstep 136 loss 0.08445075888982714, accuracy 99.0%\nstep 137 loss 0.10322381966156025, accuracy 97.0%\nstep 138 loss 0.0710055214746157, accuracy 98.0%\nstep 139 loss 0.07500647400975607, accuracy 97.0%\nstep 140 loss 0.09587144248198697, accuracy 98.0%\nstep 141 loss 0.09121096305530267, accuracy 97.0%\nstep 142 loss 0.07069210435052661, accuracy 99.0%\nstep 143 loss 0.07647040769217672, accuracy 97.0%\nstep 144 loss 0.08818150167078187, accuracy 99.0%\nstep 145 loss 0.09890129125191513, accuracy 97.0%\nstep 146 loss 0.07138802455102707, accuracy 99.0%\nstep 147 loss 0.09010668266439284, accuracy 97.0%\nstep 148 loss 0.06791853696826573, accuracy 99.0%\nstep 149 loss 0.09220739636954929, accuracy 97.0%\nstep 150 loss 0.0643692446312098, accuracy 99.0%\nstep 151 loss 0.07273292738879687, accuracy 97.0%\nstep 152 loss 0.07228146815870656, accuracy 99.0%\nstep 153 loss 0.07895800533159007, accuracy 97.0%\nstep 154 loss 0.06799893233839355, accuracy 99.0%\nstep 155 loss 0.08056580902623867, accuracy 97.0%\nstep 156 loss 0.06078100204949933, accuracy 99.0%\nstep 157 loss 0.07195113186485842, accuracy 97.0%\nstep 158 loss 0.06844854411312444, accuracy 100.0%\nstep 159 loss 0.07294675650433287, accuracy 97.0%\nstep 160 loss 0.06437460834512392, accuracy 100.0%\nstep 161 loss 0.07428710521108998, accuracy 97.0%\nstep 162 loss 0.0622937526916013, accuracy 100.0%\nstep 163 loss 0.07541556054232305, accuracy 97.0%\nstep 164 loss 0.06394637269093684, accuracy 100.0%\nstep 165 loss 0.08293375527558466, accuracy 97.0%\nstep 166 loss 0.06082765981653514, accuracy 100.0%\nstep 167 loss 0.07681538895696388, accuracy 97.0%\nstep 168 loss 0.057111325548537456, accuracy 100.0%\nstep 169 loss 0.07328121721595349, accuracy 97.0%\nstep 170 loss 0.053724377496977636, accuracy 100.0%\nstep 171 loss 0.08099515571591993, accuracy 97.0%\nstep 172 loss 0.049734038623435346, accuracy 99.0%\nstep 173 loss 0.049361536270227824, accuracy 99.0%\nstep 174 loss 0.04863448536082203, accuracy 99.0%\nstep 175 loss 0.04810623611904974, accuracy 99.0%\nstep 176 loss 0.048250428982465086, accuracy 99.0%\nstep 177 loss 0.07497522614399739, accuracy 99.0%\nstep 178 loss 0.08495355376855729, accuracy 97.0%\nstep 179 loss 0.04709437019581432, accuracy 99.0%\nstep 180 loss 0.056402990231506994, accuracy 100.0%\nstep 181 loss 0.06663074328132418, accuracy 97.0%\nstep 182 loss 0.053012274049893525, accuracy 100.0%\nstep 183 loss 0.06753366094209322, accuracy 97.0%\nstep 184 loss 0.04831724542217604, accuracy 100.0%\nstep 185 loss 0.05596167821372465, accuracy 98.0%\nstep 186 loss 0.05564894748430812, accuracy 100.0%\nstep 187 loss 0.06927325334812592, accuracy 97.0%\nstep 188 loss 0.0445937745452226, accuracy 100.0%\nstep 189 loss 0.047753064634323675, accuracy 99.0%\nstep 190 loss 0.06181955161874489, accuracy 100.0%\nstep 191 loss 0.07755593228094528, accuracy 97.0%\nstep 192 loss 0.043917282444800435, accuracy 100.0%\nstep 193 loss 0.04270600293452227, accuracy 100.0%\nstep 194 loss 0.041682039328595474, accuracy 100.0%\nstep 195 loss 0.041534050249874416, accuracy 100.0%\nstep 196 loss 0.0525323370939684, accuracy 100.0%\nstep 197 loss 0.06391991521093969, accuracy 98.0%\nstep 198 loss 0.04066538736980109, accuracy 100.0%\nstep 199 loss 0.0401602488304608, accuracy 100.0%\n"
],
[
"# visualize decision boundary\n\nh = 0.25\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nxx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\nXmesh = np.c_[xx.ravel(), yy.ravel()]\ninputs = [list(map(Value, xrow)) for xrow in Xmesh]\nscores = list(map(model, inputs))\nZ = np.array([s.data > 0 for s in scores])\nZ = Z.reshape(xx.shape)\n\nfig = plt.figure()\nplt.contourf(xx, yy, Z, cmap=plt.cm.Spectral, alpha=0.8)\nplt.scatter(X[:, 0], X[:, 1], c=y, s=40, cmap=plt.cm.Spectral)\nplt.xlim(xx.min(), xx.max())\nplt.ylim(yy.min(), yy.max())\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5f9e20352bb0a94f1b2e8791e049f71d7bd5ee | 12,564 | ipynb | Jupyter Notebook | site/en/tutorials/eager/automatic_differentiation.ipynb | bananabowl/docs | 3d8f77242fa1f492e28e2739582db08e8bd3849c | [
"Apache-2.0"
]
| null | null | null | site/en/tutorials/eager/automatic_differentiation.ipynb | bananabowl/docs | 3d8f77242fa1f492e28e2739582db08e8bd3849c | [
"Apache-2.0"
]
| null | null | null | site/en/tutorials/eager/automatic_differentiation.ipynb | bananabowl/docs | 3d8f77242fa1f492e28e2739582db08e8bd3849c | [
"Apache-2.0"
]
| 2 | 2019-11-13T20:05:03.000Z | 2019-12-15T08:32:27.000Z | 34.61157 | 644 | 0.531439 | [
[
[
"##### Copyright 2018 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Automatic differentiation and gradient tape",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\"><td>\n<a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/tensorflow/blob/master/tensorflow/contrib/eager/python/examples/notebooks/automatic_differentiation.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n</td><td>\n<a target=\"_blank\" href=\"https://github.com/tensorflow/tensorflow/blob/master/tensorflow/contrib/eager/python/examples/notebooks/automatic_differentiation.ipynb\"><img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a></td></table>",
"_____no_output_____"
],
[
"In the previous tutorial we introduced `Tensor`s and operations on them. In this tutorial we will cover [automatic differentiation](https://en.wikipedia.org/wiki/Automatic_differentiation), a key technique for optimizing machine learning models.",
"_____no_output_____"
],
[
"## Setup\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\ntf.enable_eager_execution()\n\ntfe = tf.contrib.eager # Shorthand for some symbols",
"_____no_output_____"
]
],
[
[
"## Derivatives of a function\n\nTensorFlow provides APIs for automatic differentiation - computing the derivative of a function. The way that more closely mimics the math is to encapsulate the computation in a Python function, say `f`, and use `tfe.gradients_function` to create a function that computes the derivatives of `f` with respect to its arguments. If you're familiar with [autograd](https://github.com/HIPS/autograd) for differentiating numpy functions, this will be familiar. For example: ",
"_____no_output_____"
]
],
[
[
"from math import pi\n\ndef f(x):\n return tf.square(tf.sin(x))\n\nassert f(pi/2).numpy() == 1.0\n\n\n# grad_f will return a list of derivatives of f\n# with respect to its arguments. Since f() has a single argument,\n# grad_f will return a list with a single element.\ngrad_f = tfe.gradients_function(f)\nassert tf.abs(grad_f(pi/2)[0]).numpy() < 1e-7",
"_____no_output_____"
]
],
[
[
"### Higher-order gradients\n\nThe same API can be used to differentiate as many times as you like:\n",
"_____no_output_____"
]
],
[
[
"def f(x):\n return tf.square(tf.sin(x))\n\ndef grad(f):\n return lambda x: tfe.gradients_function(f)(x)[0]\n\nx = tf.lin_space(-2*pi, 2*pi, 100) # 100 points between -2π and +2π\n\nimport matplotlib.pyplot as plt\n\nplt.plot(x, f(x), label=\"f\")\nplt.plot(x, grad(f)(x), label=\"first derivative\")\nplt.plot(x, grad(grad(f))(x), label=\"second derivative\")\nplt.plot(x, grad(grad(grad(f)))(x), label=\"third derivative\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Gradient tapes\n\nEvery differentiable TensorFlow operation has an associated gradient function. For example, the gradient function of `tf.square(x)` would be a function that returns `2.0 * x`. To compute the gradient of a user-defined function (like `f(x)` in the example above), TensorFlow first \"records\" all the operations applied to compute the output of the function. We call this record a \"tape\". It then uses that tape and the gradients functions associated with each primitive operation to compute the gradients of the user-defined function using [reverse mode differentiation](https://en.wikipedia.org/wiki/Automatic_differentiation).\n\nSince operations are recorded as they are executed, Python control flow (using `if`s and `while`s for example) is naturally handled:\n\n",
"_____no_output_____"
]
],
[
[
"def f(x, y):\n output = 1\n # Must use range(int(y)) instead of range(y) in Python 3 when\n # using TensorFlow 1.10 and earlier. Can use range(y) in 1.11+\n for i in range(int(y)):\n output = tf.multiply(output, x)\n return output\n\ndef g(x, y):\n # Return the gradient of `f` with respect to it's first parameter\n return tfe.gradients_function(f)(x, y)[0]\n\nassert f(3.0, 2).numpy() == 9.0 # f(x, 2) is essentially x * x\nassert g(3.0, 2).numpy() == 6.0 # And its gradient will be 2 * x\nassert f(4.0, 3).numpy() == 64.0 # f(x, 3) is essentially x * x * x\nassert g(4.0, 3).numpy() == 48.0 # And its gradient will be 3 * x * x",
"_____no_output_____"
]
],
[
[
"At times it may be inconvenient to encapsulate computation of interest into a function. For example, if you want the gradient of the output with respect to intermediate values computed in the function. In such cases, the slightly more verbose but explicit [tf.GradientTape](https://www.tensorflow.org/api_docs/python/tf/GradientTape) context is useful. All computation inside the context of a `tf.GradientTape` is \"recorded\".\n\nFor example:",
"_____no_output_____"
]
],
[
[
"x = tf.ones((2, 2))\n \n# a single t.gradient() call when the bug is resolved.\nwith tf.GradientTape(persistent=True) as t:\n t.watch(x)\n y = tf.reduce_sum(x)\n z = tf.multiply(y, y)\n\n# Use the same tape to compute the derivative of z with respect to the\n# intermediate value y.\ndz_dy = t.gradient(z, y)\nassert dz_dy.numpy() == 8.0\n\n# Derivative of z with respect to the original input tensor x\ndz_dx = t.gradient(z, x)\nfor i in [0, 1]:\n for j in [0, 1]:\n assert dz_dx[i][j].numpy() == 8.0",
"_____no_output_____"
]
],
[
[
"### Higher-order gradients\n\nOperations inside of the `GradientTape` context manager are recorded for automatic differentiation. If gradients are computed in that context, then the gradient computation is recorded as well. As a result, the exact same API works for higher-order gradients as well. For example:",
"_____no_output_____"
]
],
[
[
"x = tf.constant(1.0) # Convert the Python 1.0 to a Tensor object\n\nwith tf.GradientTape() as t:\n with tf.GradientTape() as t2:\n t2.watch(x)\n y = x * x * x\n # Compute the gradient inside the 't' context manager\n # which means the gradient computation is differentiable as well.\n dy_dx = t2.gradient(y, x)\nd2y_dx2 = t.gradient(dy_dx, x)\n\nassert dy_dx.numpy() == 3.0\nassert d2y_dx2.numpy() == 6.0",
"_____no_output_____"
]
],
[
[
"## Next Steps\n\nIn this tutorial we covered gradient computation in TensorFlow. With that we have enough of the primitives required to build an train neural networks, which we will cover in the [next tutorial](https://github.com/tensorflow/models/tree/master/official/contrib/eager/python/examples/notebooks/3_neural_networks.ipynb).",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec5fa1168f5104431234bf2b3260d01f325a0d5d | 10,300 | ipynb | Jupyter Notebook | notebooks/punishment.ipynb | statesman/daycare-data-unwatched | a1ff4a731d3561cbd4e58ef3e92f1c311415d475 | [
"CC-BY-4.0",
"MIT"
]
| 1 | 2018-12-06T05:35:51.000Z | 2018-12-06T05:35:51.000Z | notebooks/punishment.ipynb | statesman/daycare-data-unwatched | a1ff4a731d3561cbd4e58ef3e92f1c311415d475 | [
"CC-BY-4.0",
"MIT"
]
| null | null | null | notebooks/punishment.ipynb | statesman/daycare-data-unwatched | a1ff4a731d3561cbd4e58ef3e92f1c311415d475 | [
"CC-BY-4.0",
"MIT"
]
| null | null | null | 31.402439 | 488 | 0.468155 | [
[
[
"# Prohibited punishment violations\n\nQueries [violation](https://data.texas.gov/Social-Services/DFPS-CCL-Non-Compliance-Data/tqgd-mf4x), [operation](https://data.texas.gov/Social-Services/DFPS-CCL-Daycare-and-Residential-Operations-Data/bc5r-88dy) and [assessment](https://data.texas.gov/Social-Services/DFPS-CCL-Inspection-Investigation-Assessment-Data/m5q4-3y3d) datasets downloaded from the state data portal on Feb. 1, 2018, against a spreadsheet of fines provided by the Texas Health and Human Services Commission.",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"non_compliance = pd.read_csv('../src/dfps-2018-02-01/non_compliance.csv').drop('Unnamed: 0', axis=1)\noperations = pd.read_csv('../src/dfps-2018-02-01/operations.csv').drop('Unnamed: 0', axis=1)\ninspections = pd.read_csv('../src/dfps-2018-02-01/assessment.csv').drop('Unnamed: 0', axis=1)",
"_____no_output_____"
],
[
"punishment = non_compliance.assign(\n cruel = lambda x: x.STANDARD_NUMBER_DESCRIPTION.str.lower().isin(\n pd.Series(list(filter(\n lambda x: 'harsh, cruel' in x,\n [a for a in non_compliance.STANDARD_NUMBER_DESCRIPTION.str.lower().unique()]\n )))\n ) | x.STANDARD_NUMBER_DESCRIPTION.isin([\n '746.2805(4) - Prohibited Punishments - Hitting with Hand or Instrument',\n '746.2805(3) - Prohibited Punishments - Pinching, Shaking, or Biting',\n '746.2805(8) - Prohibited Punishments -Placing Child in Locked, Dark Room'\n ])\n).merge(\n operations,\n on='OPERATION_ID'\n).query(\n 'OPERATION_TYPE != \"Child Placing Agency\" & OPERATION_TYPE != \"General Residential Operation\"'\n).query('cruel == True').append(\n non_compliance.assign(\n cruel = lambda x: x.STANDARD_NUMBER_DESCRIPTION.str.lower().isin(\n pd.Series(list(filter(\n lambda x: 'neglected' in x,\n [a for a in non_compliance.STANDARD_NUMBER_DESCRIPTION.str.lower().unique()]\n )))\n )\n ).merge(\n operations,\n on='OPERATION_ID'\n ).query(\n 'OPERATION_TYPE != \"Child Placing Agency\" & OPERATION_TYPE != \"General Residential Operation\"'\n ).query('cruel == True').set_index(\n 'ACTIVITY_ID'\n )\n)[[\n 'OPERATION_ID', 'OPERATION_NAME', 'LOCATION_ADDRESS', 'COUNTY',\n 'STANDARD_NUMBER_DESCRIPTION',\n 'CORRECTED_DATE',\n 'NARRATIVE', 'TECHNICAL_ASSISTANCE_GIVEN', 'OPERATION_TYPE',\n]].assign(\n in_timeframe = lambda x: pd.to_datetime(x.CORRECTED_DATE).apply(\n lambda x: x >= pd.to_datetime('2016-02-01')\n )\n).query('in_timeframe').drop(\n 'in_timeframe', axis=1\n)",
"_____no_output_____"
],
[
"# Export for database\npunishment.to_csv(\n '../database/punishment.csv',\n index=False\n)",
"_____no_output_____"
],
[
"fines = pd.read_excel(\n '../Fines.xlsx',\n skiprows=3,\n skip_footer=1,\n converters={'Operation Number': int}\n).rename(\n columns = lambda x: x.replace(' ', '')\n)",
"_____no_output_____"
],
[
"fines.head()",
"_____no_output_____"
],
[
"# Search operations cited for prohibited punishments in fines spreadsheet\n\nfines.assign(\n name_number = lambda x: x.OperationName.astype(str) + x.OperationNumber.astype(str)\n).merge(\n punishment.assign(\n name_number = lambda x: x.OPERATION_NAME.astype(str) + x.OPERATION_ID.astype(str)\n ),\n on='name_number'\n)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5fadf1082ef5bfc76f8b5273ac60f402c99d46 | 165,811 | ipynb | Jupyter Notebook | 0902_training_template_ResNeSt_1D.ipynb | root4kaido/Cornell-Birdcall-Identification | 186008965ad7e8797fc181a2836bb63aacb324e4 | [
"MIT"
]
| 1 | 2020-11-21T12:03:07.000Z | 2020-11-21T12:03:07.000Z | 0902_training_template_ResNeSt_1D.ipynb | root4kaido/Cornell-Birdcall-Identification | 186008965ad7e8797fc181a2836bb63aacb324e4 | [
"MIT"
]
| null | null | null | 0902_training_template_ResNeSt_1D.ipynb | root4kaido/Cornell-Birdcall-Identification | 186008965ad7e8797fc181a2836bb63aacb324e4 | [
"MIT"
]
| null | null | null | 36.797825 | 769 | 0.354759 | [
[
[
"!pip install /home/knikaido/work/Cornell-Birdcall-Identification/data/resnest50-fast-package/resnest-0.0.6b20200701/resnest/\n!pip install torch==1.4.0\n!pip install opencv-python\n!pip install slackweb\n!pip install torchvision==0.2.2\n!pip install torch_summary",
"Defaulting to user installation because normal site-packages is not writeable\nProcessing /home/knikaido/work/Cornell-Birdcall-Identification/data/resnest50-fast-package/resnest-0.0.6b20200701/resnest\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200902) (1.17.4)\nRequirement already satisfied: tqdm in /home/user/.local/lib/python3.6/site-packages (from resnest==0.0.6b20200902) (4.19.9)\nRequirement already satisfied: nose in /home/user/.local/lib/python3.6/site-packages (from resnest==0.0.6b20200902) (1.3.7)\nRequirement already satisfied: torch>=1.0.0 in /home/user/.local/lib/python3.6/site-packages (from resnest==0.0.6b20200902) (1.4.0)\nRequirement already satisfied: Pillow in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200902) (7.1.2)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200902) (1.1.0)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200902) (2.24.0)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->resnest==0.0.6b20200902) (2.9)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->resnest==0.0.6b20200902) (2020.4.5.2)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /home/user/.local/lib/python3.6/site-packages (from requests->resnest==0.0.6b20200902) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->resnest==0.0.6b20200902) (3.0.4)\nBuilding wheels for collected packages: resnest\n Building wheel for resnest (setup.py) ... \u001b[?25ldone\n\u001b[?25h Created wheel for resnest: filename=resnest-0.0.6b20200902-py3-none-any.whl size=30752 sha256=a9841597b999e692de5591aee068e08f91c441fd7ec76fb4de6529698aa76a10\n Stored in directory: /tmp/pip-ephem-wheel-cache-dmqmhdfq/wheels/98/b8/20/14b175a058326076510265be935570257f33b40bafba7255a9\nSuccessfully built resnest\nInstalling collected packages: resnest\n Attempting uninstall: resnest\n Found existing installation: resnest 0.0.6b20200902\n Uninstalling resnest-0.0.6b20200902:\n Successfully uninstalled resnest-0.0.6b20200902\nSuccessfully installed resnest-0.0.6b20200902\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: torch==1.4.0 in /home/user/.local/lib/python3.6/site-packages (1.4.0)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: opencv-python in /home/user/.local/lib/python3.6/site-packages (4.4.0.42)\nRequirement already satisfied: numpy>=1.13.3 in /usr/local/lib/python3.6/dist-packages (from opencv-python) (1.17.4)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: slackweb in /home/user/.local/lib/python3.6/site-packages (1.0.5)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: torchvision==0.2.2 in /home/user/.local/lib/python3.6/site-packages (0.2.2)\nRequirement already satisfied: torch in /home/user/.local/lib/python3.6/site-packages (from torchvision==0.2.2) (1.4.0)\nRequirement already satisfied: pillow>=4.1.1 in /usr/local/lib/python3.6/dist-packages (from torchvision==0.2.2) (7.1.2)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torchvision==0.2.2) (1.17.4)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from torchvision==0.2.2) (1.11.0)\nRequirement already satisfied: tqdm==4.19.9 in /home/user/.local/lib/python3.6/site-packages (from torchvision==0.2.2) (4.19.9)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: torch_summary in /home/user/.local/lib/python3.6/site-packages (1.4.2)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\n"
],
[
"from pathlib import Path\nimport numpy as np\nimport pandas as pd\nimport typing as tp\nimport yaml\nimport random\nimport os\nimport sys\nimport soundfile as sf\nimport librosa\nimport cv2\nimport matplotlib.pyplot as plt\nimport time\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.utils.data as data\nimport resnest.torch as resnest_torch\n\nfrom torchvision import models\n\nfrom sklearn.model_selection import StratifiedKFold\nfrom sklearn.metrics import f1_score\nfrom radam import RAdam\n\n\npd.options.display.max_rows = 500\npd.options.display.max_columns = 500",
"_____no_output_____"
],
[
"with open('config.yml', 'r') as yml:\n settings = yaml.safe_load(yml)",
"_____no_output_____"
],
[
"def set_seed(seed: int = 42):\n random.seed(seed)\n np.random.seed(seed)\n os.environ[\"PYTHONHASHSEED\"] = str(seed)\n torch.manual_seed(seed)\n torch.cuda.manual_seed(seed) # type: ignore\n# torch.backends.cudnn.deterministic = True # type: ignore\n# torch.backends.cudnn.benchmark = True # type: ignore\n ",
"_____no_output_____"
],
[
"# def progress_bar(i):\n# pro_bar = ('=' * i) + (' ' * (pro_size - i))\n# print('\\r[{0}] {1}%'.format(pro_bar, i / pro_size * 100.), end='')",
"_____no_output_____"
],
[
"# ROOT = Path.cwd().parent\n# INPUT_ROOT = ROOT / \"input\"\nINPUT_ROOT = Path(\"/home/knikaido/work/Cornell-Birdcall-Identification/data\")\nRAW_DATA = INPUT_ROOT / \"birdsong-recognition\"\nTRAIN_AUDIO_DIR = RAW_DATA / \"train_audio\"\nTRAIN_RESAMPLED_AUDIO_DIRS = [\n INPUT_ROOT / \"birdsong-resampled-train-audio-{:0>2}\".format(i) for i in range(5)\n]\nTEST_AUDIO_DIR = RAW_DATA / \"test_audio\"",
"_____no_output_____"
],
[
"# train = pd.read_csv(RAW_DATA / \"train.csv\")\ntrain = pd.read_csv(TRAIN_RESAMPLED_AUDIO_DIRS[0] / \"train_mod.csv\")\ntrain.head().T",
"_____no_output_____"
],
[
"if not TEST_AUDIO_DIR.exists():\n TEST_AUDIO_DIR = INPUT_ROOT / \"birdcall-check\" / \"test_audio\"\n test = pd.read_csv(INPUT_ROOT / \"birdcall-check\" / \"test.csv\")\nelse:\n test = pd.read_csv(RAW_DATA / \"test.csv\")\ntest.head().T",
"_____no_output_____"
],
[
"BIRD_CODE = {\n 'aldfly': 0, 'ameavo': 1, 'amebit': 2, 'amecro': 3, 'amegfi': 4,\n 'amekes': 5, 'amepip': 6, 'amered': 7, 'amerob': 8, 'amewig': 9,\n 'amewoo': 10, 'amtspa': 11, 'annhum': 12, 'astfly': 13, 'baisan': 14,\n 'baleag': 15, 'balori': 16, 'banswa': 17, 'barswa': 18, 'bawwar': 19,\n 'belkin1': 20, 'belspa2': 21, 'bewwre': 22, 'bkbcuc': 23, 'bkbmag1': 24,\n 'bkbwar': 25, 'bkcchi': 26, 'bkchum': 27, 'bkhgro': 28, 'bkpwar': 29,\n 'bktspa': 30, 'blkpho': 31, 'blugrb1': 32, 'blujay': 33, 'bnhcow': 34,\n 'boboli': 35, 'bongul': 36, 'brdowl': 37, 'brebla': 38, 'brespa': 39,\n 'brncre': 40, 'brnthr': 41, 'brthum': 42, 'brwhaw': 43, 'btbwar': 44,\n 'btnwar': 45, 'btywar': 46, 'buffle': 47, 'buggna': 48, 'buhvir': 49,\n 'bulori': 50, 'bushti': 51, 'buwtea': 52, 'buwwar': 53, 'cacwre': 54,\n 'calgul': 55, 'calqua': 56, 'camwar': 57, 'cangoo': 58, 'canwar': 59,\n 'canwre': 60, 'carwre': 61, 'casfin': 62, 'caster1': 63, 'casvir': 64,\n 'cedwax': 65, 'chispa': 66, 'chiswi': 67, 'chswar': 68, 'chukar': 69,\n 'clanut': 70, 'cliswa': 71, 'comgol': 72, 'comgra': 73, 'comloo': 74,\n 'commer': 75, 'comnig': 76, 'comrav': 77, 'comred': 78, 'comter': 79,\n 'comyel': 80, 'coohaw': 81, 'coshum': 82, 'cowscj1': 83, 'daejun': 84,\n 'doccor': 85, 'dowwoo': 86, 'dusfly': 87, 'eargre': 88, 'easblu': 89,\n 'easkin': 90, 'easmea': 91, 'easpho': 92, 'eastow': 93, 'eawpew': 94,\n 'eucdov': 95, 'eursta': 96, 'evegro': 97, 'fiespa': 98, 'fiscro': 99,\n 'foxspa': 100, 'gadwal': 101, 'gcrfin': 102, 'gnttow': 103, 'gnwtea': 104,\n 'gockin': 105, 'gocspa': 106, 'goleag': 107, 'grbher3': 108, 'grcfly': 109,\n 'greegr': 110, 'greroa': 111, 'greyel': 112, 'grhowl': 113, 'grnher': 114,\n 'grtgra': 115, 'grycat': 116, 'gryfly': 117, 'haiwoo': 118, 'hamfly': 119,\n 'hergul': 120, 'herthr': 121, 'hoomer': 122, 'hoowar': 123, 'horgre': 124,\n 'horlar': 125, 'houfin': 126, 'houspa': 127, 'houwre': 128, 'indbun': 129,\n 'juntit1': 130, 'killde': 131, 'labwoo': 132, 'larspa': 133, 'lazbun': 134,\n 'leabit': 135, 'leafly': 136, 'leasan': 137, 'lecthr': 138, 'lesgol': 139,\n 'lesnig': 140, 'lesyel': 141, 'lewwoo': 142, 'linspa': 143, 'lobcur': 144,\n 'lobdow': 145, 'logshr': 146, 'lotduc': 147, 'louwat': 148, 'macwar': 149,\n 'magwar': 150, 'mallar3': 151, 'marwre': 152, 'merlin': 153, 'moublu': 154,\n 'mouchi': 155, 'moudov': 156, 'norcar': 157, 'norfli': 158, 'norhar2': 159,\n 'normoc': 160, 'norpar': 161, 'norpin': 162, 'norsho': 163, 'norwat': 164,\n 'nrwswa': 165, 'nutwoo': 166, 'olsfly': 167, 'orcwar': 168, 'osprey': 169,\n 'ovenbi1': 170, 'palwar': 171, 'pasfly': 172, 'pecsan': 173, 'perfal': 174,\n 'phaino': 175, 'pibgre': 176, 'pilwoo': 177, 'pingro': 178, 'pinjay': 179,\n 'pinsis': 180, 'pinwar': 181, 'plsvir': 182, 'prawar': 183, 'purfin': 184,\n 'pygnut': 185, 'rebmer': 186, 'rebnut': 187, 'rebsap': 188, 'rebwoo': 189,\n 'redcro': 190, 'redhea': 191, 'reevir1': 192, 'renpha': 193, 'reshaw': 194,\n 'rethaw': 195, 'rewbla': 196, 'ribgul': 197, 'rinduc': 198, 'robgro': 199,\n 'rocpig': 200, 'rocwre': 201, 'rthhum': 202, 'ruckin': 203, 'rudduc': 204,\n 'rufgro': 205, 'rufhum': 206, 'rusbla': 207, 'sagspa1': 208, 'sagthr': 209,\n 'savspa': 210, 'saypho': 211, 'scatan': 212, 'scoori': 213, 'semplo': 214,\n 'semsan': 215, 'sheowl': 216, 'shshaw': 217, 'snobun': 218, 'snogoo': 219,\n 'solsan': 220, 'sonspa': 221, 'sora': 222, 'sposan': 223, 'spotow': 224,\n 'stejay': 225, 'swahaw': 226, 'swaspa': 227, 'swathr': 228, 'treswa': 229,\n 'truswa': 230, 'tuftit': 231, 'tunswa': 232, 'veery': 233, 'vesspa': 234,\n 'vigswa': 235, 'warvir': 236, 'wesblu': 237, 'wesgre': 238, 'weskin': 239,\n 'wesmea': 240, 'wessan': 241, 'westan': 242, 'wewpew': 243, 'whbnut': 244,\n 'whcspa': 245, 'whfibi': 246, 'whtspa': 247, 'whtswi': 248, 'wilfly': 249,\n 'wilsni1': 250, 'wiltur': 251, 'winwre3': 252, 'wlswar': 253, 'wooduc': 254,\n 'wooscj2': 255, 'woothr': 256, 'y00475': 257, 'yebfly': 258, 'yebsap': 259,\n 'yehbla': 260, 'yelwar': 261, 'yerwar': 262, 'yetvir': 263\n}\n\nINV_BIRD_CODE = {v: k for k, v in BIRD_CODE.items()}",
"_____no_output_____"
],
[
"PERIOD = 5\n\ndef mono_to_color(\n X: np.ndarray, mean=None, std=None,\n norm_max=None, norm_min=None, eps=1e-6\n):\n # Stack X as [X,X,X]\n X = np.stack([X, X, X], axis=-1)\n\n # Standardize\n mean = mean or X.mean()\n X = X - mean\n std = std or X.std()\n Xstd = X / (std + eps)\n _min, _max = Xstd.min(), Xstd.max()\n norm_max = norm_max or _max\n norm_min = norm_min or _min\n if (_max - _min) > eps:\n # Normalize to [0, 255]\n V = Xstd\n V[V < norm_min] = norm_min\n V[V > norm_max] = norm_max\n V = 255 * (V - norm_min) / (norm_max - norm_min)\n V = V.astype(np.uint8)\n else:\n # Just zero\n V = np.zeros_like(Xstd, dtype=np.uint8)\n return V\n\nclass SpectrogramDataset(data.Dataset):\n def __init__(\n self,\n file_list: tp.List[tp.List[str]], img_size=224,\n waveform_transforms=None, spectrogram_transforms=None, melspectrogram_parameters={}\n ):\n self.file_list = file_list # list of list: [file_path, ebird_code]\n self.img_size = img_size\n self.waveform_transforms = waveform_transforms\n self.spectrogram_transforms = spectrogram_transforms\n self.melspectrogram_parameters = melspectrogram_parameters\n\n def __len__(self):\n return len(self.file_list)\n\n def __getitem__(self, idx: int):\n wav_path, ebird_code = self.file_list[idx]\n\n y, sr = sf.read(wav_path)\n\n if self.waveform_transforms:\n y = self.waveform_transforms(y)\n else:\n len_y = len(y)\n effective_length = sr * PERIOD\n if len_y < effective_length:\n new_y = np.zeros(effective_length, dtype=y.dtype)\n start = np.random.randint(effective_length - len_y)\n new_y[start:start + len_y] = y\n y = new_y.astype(np.float32)\n elif len_y > effective_length:\n start = np.random.randint(len_y - effective_length)\n y = y[start:start + effective_length].astype(np.float32)\n else:\n y = y.astype(np.float32)\n\n melspec = librosa.feature.melspectrogram(y, sr=sr, **self.melspectrogram_parameters)\n melspec = librosa.power_to_db(melspec).astype(np.float32)\n\n if self.spectrogram_transforms:\n melspec = self.spectrogram_transforms(melspec)\n else:\n pass\n melspec = np.reshape(melspec, (1, 128, -1))\n\n# image = mono_to_color(melspec)\n# height, width, _ = image.shape\n# image = cv2.resize(image, (int(width * self.img_size / height), self.img_size))\n# image = np.moveaxis(image, 2, 0)\n# image = (image / 255.0).astype(np.float32)\n\n# labels = np.zeros(len(BIRD_CODE), dtype=\"i\")\n labels = np.zeros(len(BIRD_CODE), dtype=\"f\")\n labels[BIRD_CODE[ebird_code]] = 1\n\n return melspec, labels",
"_____no_output_____"
],
[
"def get_loaders_for_training(\n args_dataset: tp.Dict, args_loader: tp.Dict,\n train_file_list: tp.List[str], val_file_list: tp.List[str]\n):\n # # make dataset\n train_dataset = SpectrogramDataset(train_file_list, **args_dataset)\n val_dataset = SpectrogramDataset(val_file_list, **args_dataset)\n # # make dataloader\n train_loader = data.DataLoader(train_dataset, **args_loader[\"train\"])\n val_loader = data.DataLoader(val_dataset, **args_loader[\"val\"])\n \n return train_loader, val_loader",
"_____no_output_____"
],
[
"class ResNeSt(nn.Module):\n def __init__(self, base_model_name: str, pretrained=True,\n num_classes=264):\n super().__init__()\n torch.hub.list('zhanghang1989/ResNeSt', force_reload=True)\n # load pretrained models, using ResNeSt-50 as an example\n base_model = torch.hub.load('zhanghang1989/ResNeSt', 'resnest50', pretrained=True)\n layers = list(base_model.children())[:-2]\n# layers.append(nn.AdaptiveMaxPool2d(1))\n self.encoder = nn.Sequential(*layers)\n\n in_features = base_model.fc.in_features\n\n self.classifier = nn.Sequential(\n nn.Linear(in_features, 1024), nn.ReLU(), nn.Dropout(p=0.2),\n nn.Linear(1024, 1024), nn.ReLU(), nn.Dropout(p=0.2),\n nn.Linear(1024, num_classes))\n\n def forward(self, x):\n batch_size = x.size(0)\n x = self.encoder(x).view(batch_size, -1)\n x = self.classifier(x)\n return x\n# multiclass_proba = F.softmax(x, dim=1)\n# multilabel_proba = torch.sigmoid(x)\n# return {\n# \"logits\": x,\n# \"multiclass_proba\": multiclass_proba,\n# \"multilabel_proba\": multilabel_proba\n# }",
"_____no_output_____"
],
[
"# def get_model(args: tp.Dict):\n# model = ResNeSt('resnet50')\n \n# return model",
"_____no_output_____"
],
[
"def get_model():\n model =getattr(resnest_torch, 'resnest50_fast_1s1x64d')(pretrained=True)\n del model.fc\n # # use the same head as the baseline notebook.\n model.fc = nn.Sequential(\n nn.Linear(2048, 1024), nn.ReLU(), nn.Dropout(p=0.2),\n nn.Linear(1024, 1024), nn.ReLU(), nn.Dropout(p=0.2),\n nn.Linear(1024, 264))\n \n conv1 = nn.Sequential(nn.Conv2d(1, 32, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False),\n nn.BatchNorm2d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True),\n nn.ReLU(inplace=True),\n nn.Conv2d(32, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False),\n nn.BatchNorm2d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True),\n nn.ReLU(inplace=True),\n nn.Conv2d(32, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)) \n \n layers = list(model.children())[1:]\n# layers.append(nn.AdaptiveMaxPool2d(1))\n model = nn.Sequential(conv1,*layers)\n \n return model",
"_____no_output_____"
]
],
[
[
"https://nonbiri-tereka.hatenablog.com/entry/2020/05/15/082637",
"_____no_output_____"
]
],
[
[
"tmp_list = []\nfor audio_d in TRAIN_RESAMPLED_AUDIO_DIRS:\n if not audio_d.exists():\n continue\n for ebird_d in audio_d.iterdir():\n if ebird_d.is_file():\n continue\n for wav_f in ebird_d.iterdir():\n tmp_list.append([ebird_d.name, wav_f.name, wav_f.as_posix()])\n \ntrain_wav_path_exist = pd.DataFrame(\n tmp_list, columns=[\"ebird_code\", \"resampled_filename\", \"file_path\"])\n\ndel tmp_list\n\ntrain_all = pd.merge(\n train, train_wav_path_exist, on=[\"ebird_code\", \"resampled_filename\"], how=\"inner\")\n\nprint(train.shape)\nprint(train_wav_path_exist.shape)\nprint(train_all.shape)\ntrain_all.head()",
"(21375, 38)\n(21375, 3)\n(21375, 39)\n"
],
[
"skf = StratifiedKFold(settings[\"split\"][\"params\"][\"n_splits\"])\n\ntrain_all[\"fold\"] = -1\nfor fold_id, (train_index, val_index) in enumerate(skf.split(train_all, train_all[\"ebird_code\"])):\n train_all.iloc[val_index, -1] = fold_id\n \n# # check the propotion\nfold_proportion = pd.pivot_table(train_all, index=\"ebird_code\", columns=\"fold\", values=\"xc_id\", aggfunc=len)\nprint(fold_proportion.shape)\nfold_proportion",
"(264, 5)\n"
],
[
"use_fold = settings[\"globals\"][\"use_fold\"]\ntrain_file_list = train_all.query(\"fold != @use_fold\")[[\"file_path\", \"ebird_code\"]].values.tolist()\nval_file_list = train_all.query(\"fold == @use_fold\")[[\"file_path\", \"ebird_code\"]].values.tolist()\n\nprint(\"[fold {}] train: {}, val: {}\".format(use_fold, len(train_file_list), len(val_file_list)))",
"[fold 0] train: 17057, val: 4318\n"
],
[
"set_seed(settings[\"globals\"][\"seed\"])\ndevice = torch.device(settings[\"globals\"][\"device\"])\noutput_dir = Path(settings[\"globals\"][\"output_dir\"])\nepoch = settings[\"globals\"][\"num_epochs\"]\n\n# # # get loader\ntrain_loader, val_loader = get_loaders_for_training(\n settings[\"dataset\"][\"params\"], settings[\"loader\"], train_file_list, val_file_list)\n\n# # # get model\n# model = get_model(settings[\"model\"])\nmodel = get_model()\nmodel = model.to(device)\n\n\n\n# # # get optimizer\n# optimizer = getattr(\n# torch.optim, settings[\"optimizer\"][\"name\"]\n# )(model.parameters(), **settings[\"optimizer\"][\"params\"])\noptimizer = RAdam(model.parameters(),lr=0.001)\n\n# # # get scheduler\nscheduler = getattr(\n torch.optim.lr_scheduler, settings[\"scheduler\"][\"name\"]\n)(optimizer, **settings[\"scheduler\"][\"params\"])\n\n# # # get loss\nloss_func = getattr(nn, settings[\"loss\"][\"name\"])(**settings[\"loss\"][\"params\"])\n",
"_____no_output_____"
],
[
"def log(message):\n print(message)\n with open(output_dir / 'log.txt', 'a+') as logger:\n logger.write(f'{message}\\n')",
"_____no_output_____"
],
[
"import cloudpickle\ndef save_model(model):\n with open('model_ResNeSt_1ch_best.pkl', 'wb') as f:\n cloudpickle.dump(model, f)",
"_____no_output_____"
],
[
"def _train_loop(\n manager, args, model, device,\n train_loader, val_loader, optimizer, scheduler, loss_func\n):\n \"\"\"Run minibatch training loop\"\"\"\n while not manager.stop_trigger:\n model.train()\n for batch_idx, (data, target) in enumerate(train_loader):\n with manager.run_iteration():\n data, target = data.to(device), target.to(device)\n optimizer.zero_grad()\n output = model(data)\n loss = loss_func(output, target)\n ppe.reporting.report({'train/loss': loss.item()})\n loss.backward()\n optimizer.step()\n scheduler.step()\n\ndef train_loop(\n args, model, device,\n train_loader, val_loader, optimizer, scheduler, loss_func\n):\n \"\"\"Run minibatch training loop\"\"\"\n train_losses = []\n valid_losses = []\n best_f1_micro = 0\n f1_macros = []\n f1_micros = []\n threshold = 0.8\n start = time.time()\n \n for i in range(epoch):\n epoch_start = time.time()\n model.train()\n train_loss = 0\n valid_loss = 0\n for batch_idx, (data, target) in enumerate(train_loader):\n # with manager.run_iteration():\n data, target = data.to(device), target.to(device)\n optimizer.zero_grad()\n output = model(data)\n loss = loss_func(output, target)\n # ppe.reporting.report({'train/loss': loss.item()})\n loss.backward()\n optimizer.step()\n scheduler.step()\n train_loss += loss.item()\n# train_loss += loss.item() * data.size(0)\n print(\"\\r\"+'train_roop...'+str(batch_idx),end=\"\")\n \n# break\n\n print('')\n epoch_train_loss = train_loss / (batch_idx + 1)\n train_losses.append(epoch_train_loss)\n \n outputs = []\n targets = []\n model.eval()\n for batch_idx, (data, target) in enumerate(val_loader):\n with torch.no_grad():\n data, target = data.to(device), target.to(device)\n output = model(data)\n loss = loss_func(output, target)\n valid_loss += loss.item()\n# valid_loss += loss.item() * data.size(0)\n outputs.extend(np.argmax(torch.sigmoid(output).to('cpu').detach().numpy().copy(), axis = 1))\n targets.extend(np.argmax(target.to('cpu').detach().numpy().copy(), axis = 1))\n print(\"\\r\"+'valid_roop...'+str(batch_idx),end=\"\")\n \n# break\n print('')\n epoch_valid_loss = valid_loss / (batch_idx + 1)\n valid_losses.append(epoch_valid_loss)\n\n f1_macro = f1_score(np.array(targets), np.array(outputs), average='macro')\n f1_micro = f1_score(np.array(targets), np.array(outputs), average='micro')\n log(f'epoch [{i+1}/{epoch}] train_loss = {epoch_train_loss}, valid_loss = {epoch_valid_loss}')\n log(f'epoch [{i+1}/{epoch}] f1macro = {f1_macro}, f1micro = {f1_micro}')\n epoch_end = time.time() - epoch_start\n log(\"epoch_time:{0}\".format(epoch_end) + \"[sec]\")\n log('\\n')\n f1_micros.append(f1_micro)\n f1_macros.append(f1_macro)\n \n if(f1_micro > best_f1_micro):\n print('save_model')\n save_model(model)\n best_f1_micro = f1_micro\n\n whole_time = time.time() - start\n log(\"elapsed_time:{0}\".format(whole_time) + \"[sec]\")\n# break\n\n return model, train_losses, valid_losses, f1_micros, f1_macros\n \n ",
"_____no_output_____"
],
[
"%%time\n# # runtraining\nmodel_, train_losses, valid_losses, f1_micros, f1_macros = train_loop(\n settings, model, device,\n train_loader, val_loader, optimizer, scheduler, loss_func)",
"train_roop...425\nvalid_roop...431\nepoch [1/45] train_loss = 0.07864445356975698, valid_loss = 0.025222051535146655\nepoch [1/45] f1macro = 0.00011386354081929753, f1micro = 0.005094951366373321\nepoch_time:557.373229265213[sec]\n\n\nsave_model\n"
],
[
"import cloudpickle",
"_____no_output_____"
],
[
"with open(output_dir / 'model_ResNeSt_1ch.pkl', 'wb') as f:\n cloudpickle.dump(model_, f)",
"_____no_output_____"
],
[
"import slackweb\nslack = slackweb.Slack(url=\"https://hooks.slack.com/services/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\")\nslack.notify(text=\"おわた\")",
"_____no_output_____"
],
[
"with open(output_dir / 'train_losses.pkl', 'wb') as f:\n cloudpickle.dump(train_losses, f)\nwith open(output_dir / 'valid_losses.pkl', 'wb') as f:\n cloudpickle.dump(valid_losses, f)\nwith open(output_dir / 'f1_micros.pkl', 'wb') as f:\n cloudpickle.dump(f1_micros, f)\nwith open(output_dir / 'f1_macros.pkl', 'wb') as f:\n cloudpickle.dump(f1_macros, f)",
"_____no_output_____"
],
[
"with open(output_dir / 'model_ResNeSt.pkl', 'rb') as f:\n net = cloudpickle.load(f)",
"_____no_output_____"
],
[
"plt.figure(figsize=(5,5), dpi= 80)\nplt.plot(train_losses, color='tab:red', label='train')\nplt.plot(valid_losses, color='tab:blue', label='valid')\nplt.grid(which='minor',color='black',linestyle='-')\nplt.legend()",
"_____no_output_____"
],
[
"plt.figure(figsize=(5,5), dpi= 80)\nplt.plot(f1_micros, color='tab:red', label='micro')\nplt.plot(f1_macros, color='tab:blue', label='macro')\nplt.grid(which='minor',color='black',linestyle='-')\nplt.legend()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5faed36b5ac591cedcefef52851de3ff028264 | 2,908 | ipynb | Jupyter Notebook | Chapter6_ObjectOriented/iterator_and_generators2.ipynb | Hasideluxe/UdemyPythonPro | 67f3b55ebdacc9024ffbcf4fd4d3268a9d3bd760 | [
"MIT"
]
| 4 | 2020-12-28T23:43:35.000Z | 2022-01-01T18:34:18.000Z | Chapter6_ObjectOriented/iterator_and_generators2.ipynb | Hasideluxe/UdemyPythonPro | 67f3b55ebdacc9024ffbcf4fd4d3268a9d3bd760 | [
"MIT"
]
| null | null | null | Chapter6_ObjectOriented/iterator_and_generators2.ipynb | Hasideluxe/UdemyPythonPro | 67f3b55ebdacc9024ffbcf4fd4d3268a9d3bd760 | [
"MIT"
]
| 9 | 2020-09-26T19:29:28.000Z | 2022-02-07T06:41:00.000Z | 22.897638 | 72 | 0.517538 | [
[
[
"class NamesIterator:\n def __init__(self, names):\n self.names = names\n self.num_names = len(self.names)\n self.current_n = 0\n\n def __iter__(self):\n self.current_n = 0\n return self\n\n def __next__(self):\n if self.current_n < self.num_names:\n current_result = self.names[self.current_n]\n self.current_n += 1\n return current_result\n else:\n self.current_n = 0\n current_result = self.names[self.current_n]\n self.current_n += 1\n return current_result",
"_____no_output_____"
],
[
"names = [\"Jan\", \"Peter\", \"Dennis\"]\n\nmy_iterator = NameIterator(names)\n\nfor i in range(7):\n print(next(my_iterator))",
"Jan\nPeter\nDennis\nJan\nPeter\nDennis\nJan\n"
],
[
"import itertools\n\nnames2 = [\"Jan\", \"Peter\", \"Dennis\"]\n\nmy_iterator2 = itertools.cycle(names)\n\nfor i in range(7):\n print(next(my_iterator2))",
"Jan\nPeter\nDennis\nJan\nPeter\nDennis\nJan\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
ec5fb6afdf5e57e5ebd3454af06690b28d49890e | 60,109 | ipynb | Jupyter Notebook | ML0101EN-Clas-K-Nearest-neighbors-CustCat-py-v1.ipynb | SimranAnand1/Machine-Learning-Tasks | 4ec9e359582144af740e3c9a3ea7b05e4bbb88b7 | [
"MIT"
]
| 4 | 2021-04-08T20:48:05.000Z | 2021-07-22T11:16:20.000Z | ML0101EN-Clas-K-Nearest-neighbors-CustCat-py-v1.ipynb | SimranAnand1/Machine-Learning-Tasks | 4ec9e359582144af740e3c9a3ea7b05e4bbb88b7 | [
"MIT"
]
| null | null | null | ML0101EN-Clas-K-Nearest-neighbors-CustCat-py-v1.ipynb | SimranAnand1/Machine-Learning-Tasks | 4ec9e359582144af740e3c9a3ea7b05e4bbb88b7 | [
"MIT"
]
| null | null | null | 50.682125 | 21,740 | 0.7249 | [
[
[
"<a href=\"https://www.bigdatauniversity.com\"><img src=\"https://ibm.box.com/shared/static/cw2c7r3o20w9zn8gkecaeyjhgw3xdgbj.png\" width=\"400\" align=\"center\"></a>\n\n<h1><center>K-Nearest Neighbors</center></h1>\n",
"_____no_output_____"
],
[
"In this Lab you will load a customer dataset, fit the data, and use K-Nearest Neighbors to predict a data point. But what is **K-Nearest Neighbors**?\n",
"_____no_output_____"
],
[
"**K-Nearest Neighbors** is an algorithm for supervised learning. Where the data is 'trained' with data points corresponding to their classification. Once a point is to be predicted, it takes into account the 'K' nearest points to it to determine it's classification.\n",
"_____no_output_____"
],
[
"### Here's an visualization of the K-Nearest Neighbors algorithm.\n\n<img src=\"https://ibm.box.com/shared/static/mgkn92xck0z05v7yjq8pqziukxvc2461.png\">\n",
"_____no_output_____"
],
[
"In this case, we have data points of Class A and B. We want to predict what the star (test data point) is. If we consider a k value of 3 (3 nearest data points) we will obtain a prediction of Class B. Yet if we consider a k value of 6, we will obtain a prediction of Class A.\n",
"_____no_output_____"
],
[
"In this sense, it is important to consider the value of k. But hopefully from this diagram, you should get a sense of what the K-Nearest Neighbors algorithm is. It considers the 'K' Nearest Neighbors (points) when it predicts the classification of the test point.\n",
"_____no_output_____"
],
[
"<h1>Table of contents</h1>\n\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ol>\n <li><a href=\"#about_dataset\">About the dataset</a></li>\n <li><a href=\"#visualization_analysis\">Data Visualization and Analysis</a></li>\n <li><a href=\"#classification\">Classification</a></li>\n </ol>\n</div>\n<br>\n<hr>\n",
"_____no_output_____"
],
[
"Lets load required libraries\n",
"_____no_output_____"
]
],
[
[
"import itertools\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom matplotlib.ticker import NullFormatter\nimport pandas as pd\nimport numpy as np\nimport matplotlib.ticker as ticker\nfrom sklearn import preprocessing\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<div id=\"about_dataset\">\n <h2>About the dataset</h2>\n</div>\n",
"_____no_output_____"
],
[
"Imagine a telecommunications provider has segmented its customer base by service usage patterns, categorizing the customers into four groups. If demographic data can be used to predict group membership, the company can customize offers for individual prospective customers. It is a classification problem. That is, given the dataset, with predefined labels, we need to build a model to be used to predict class of a new or unknown case. \n\nThe example focuses on using demographic data, such as region, age, and marital, to predict usage patterns. \n\nThe target field, called **custcat**, has four possible values that correspond to the four customer groups, as follows:\n 1- Basic Service\n 2- E-Service\n 3- Plus Service\n 4- Total Service\n\nOur objective is to build a classifier, to predict the class of unknown cases. We will use a specific type of classification called K nearest neighbour.\n",
"_____no_output_____"
],
[
"Lets download the dataset. To download the data, we will use !wget to download it from IBM Object Storage.\n",
"_____no_output_____"
]
],
[
[
"!wget -O teleCust1000t.csv https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-ML0101EN-Coursera/labs/Data_files/teleCust1000t.csv",
"--2021-02-08 13:26:14-- https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-ML0101EN-Coursera/labs/Data_files/teleCust1000t.csv\nResolving cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud (cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud)... 169.63.118.104\nConnecting to cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud (cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud)|169.63.118.104|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 36047 (35K) [text/csv]\nSaving to: ‘teleCust1000t.csv’\n\nteleCust1000t.csv 100%[===================>] 35.20K --.-KB/s in 0.02s \n\n2021-02-08 13:26:15 (1.88 MB/s) - ‘teleCust1000t.csv’ saved [36047/36047]\n\n"
]
],
[
[
"**Did you know?** When it comes to Machine Learning, you will likely be working with large datasets. As a business, where can you host your data? IBM is offering a unique opportunity for businesses, with 10 Tb of IBM Cloud Object Storage: [Sign up now for free](http://cocl.us/ML0101EN-IBM-Offer-CC)\n",
"_____no_output_____"
],
[
"### Load Data From CSV File\n",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('teleCust1000t.csv')\ndf.head()",
"_____no_output_____"
]
],
[
[
"<div id=\"visualization_analysis\">\n <h2>Data Visualization and Analysis</h2> \n</div>\n",
"_____no_output_____"
],
[
"#### Let’s see how many of each class is in our data set\n",
"_____no_output_____"
]
],
[
[
"df['custcat'].value_counts()",
"_____no_output_____"
]
],
[
[
"#### 281 Plus Service, 266 Basic-service, 236 Total Service, and 217 E-Service customers\n",
"_____no_output_____"
],
[
"You can easily explore your data using visualization techniques:\n",
"_____no_output_____"
]
],
[
[
"df.hist(column='income', bins=50)",
"_____no_output_____"
]
],
[
[
"### Feature set\n",
"_____no_output_____"
],
[
"Lets define feature sets, X:\n",
"_____no_output_____"
]
],
[
[
"df.columns",
"_____no_output_____"
]
],
[
[
"To use scikit-learn library, we have to convert the Pandas data frame to a Numpy array:\n",
"_____no_output_____"
]
],
[
[
"X = df[['region', 'tenure','age', 'marital', 'address', 'income', 'ed', 'employ','retire', 'gender', 'reside']] .values #.astype(float)\nX[0:5]\n",
"_____no_output_____"
]
],
[
[
"What are our labels?\n",
"_____no_output_____"
]
],
[
[
"y = df['custcat'].values\ny[0:5]",
"_____no_output_____"
]
],
[
[
"## Normalize Data\n",
"_____no_output_____"
],
[
"Data Standardization give data zero mean and unit variance, it is good practice, especially for algorithms such as KNN which is based on distance of cases:\n",
"_____no_output_____"
]
],
[
[
"X = preprocessing.StandardScaler().fit(X).transform(X.astype(float))\nX[0:5]",
"_____no_output_____"
]
],
[
[
"### Train Test Split\n\nOut of Sample Accuracy is the percentage of correct predictions that the model makes on data that that the model has NOT been trained on. Doing a train and test on the same dataset will most likely have low out-of-sample accuracy, due to the likelihood of being over-fit.\n\nIt is important that our models have a high, out-of-sample accuracy, because the purpose of any model, of course, is to make correct predictions on unknown data. So how can we improve out-of-sample accuracy? One way is to use an evaluation approach called Train/Test Split.\nTrain/Test Split involves splitting the dataset into training and testing sets respectively, which are mutually exclusive. After which, you train with the training set and test with the testing set. \n\nThis will provide a more accurate evaluation on out-of-sample accuracy because the testing dataset is not part of the dataset that have been used to train the data. It is more realistic for real world problems.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split( X, y, test_size=0.2, random_state=4)\nprint ('Train set:', X_train.shape, y_train.shape)\nprint ('Test set:', X_test.shape, y_test.shape)",
"Train set: (800, 11) (800,)\nTest set: (200, 11) (200,)\n"
]
],
[
[
"<div id=\"classification\">\n <h2>Classification</h2>\n</div>\n",
"_____no_output_____"
],
[
"<h3>K nearest neighbor (KNN)</h3>\n",
"_____no_output_____"
],
[
"#### Import library\n",
"_____no_output_____"
],
[
"Classifier implementing the k-nearest neighbors vote.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsClassifier",
"_____no_output_____"
]
],
[
[
"### Training\n\nLets start the algorithm with k=4 for now:\n",
"_____no_output_____"
]
],
[
[
"k = 4\n#Train Model and Predict \nneigh = KNeighborsClassifier(n_neighbors = k).fit(X_train,y_train)\nneigh",
"_____no_output_____"
]
],
[
[
"### Predicting\n\nwe can use the model to predict the test set:\n",
"_____no_output_____"
]
],
[
[
"yhat = neigh.predict(X_test)\nyhat[0:5]",
"_____no_output_____"
]
],
[
[
"### Accuracy evaluation\n\nIn multilabel classification, **accuracy classification score** is a function that computes subset accuracy. This function is equal to the jaccard_similarity_score function. Essentially, it calculates how closely the actual labels and predicted labels are matched in the test set.\n",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\nprint(\"Train set Accuracy: \", metrics.accuracy_score(y_train, neigh.predict(X_train)))\nprint(\"Test set Accuracy: \", metrics.accuracy_score(y_test, yhat))",
"Train set Accuracy: 0.5475\nTest set Accuracy: 0.32\n"
]
],
[
[
"## Practice\n\nCan you build the model again, but this time with k=6?\n",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsClassifier\nk=6\nneigh6=KNeighborsClassifier(n_neighbors=k).fit(X_train,y_train)\nyhat6=neigh6.predict(X_test)\nprint(\"Train set Accuracy: \", metrics.accuracy_score(y_train, neigh6.predict(X_train)))\nprint(\"Test set Accuracy: \", metrics.accuracy_score(y_test, yhat6))\n\n",
"Train set Accuracy: 0.51625\nTest set Accuracy: 0.31\n"
]
],
[
[
"Double-click **here** for the solution.\n\n<!-- Your answer is below:\n \n \nk = 6\nneigh6 = KNeighborsClassifier(n_neighbors = k).fit(X_train,y_train)\nyhat6 = neigh6.predict(X_test)\nprint(\"Train set Accuracy: \", metrics.accuracy_score(y_train, neigh6.predict(X_train)))\nprint(\"Test set Accuracy: \", metrics.accuracy_score(y_test, yhat6))\n\n-->\n",
"_____no_output_____"
],
[
"#### What about other K?\n\nK in KNN, is the number of nearest neighbors to examine. It is supposed to be specified by the User. So, how can we choose right value for K?\nThe general solution is to reserve a part of your data for testing the accuracy of the model. Then chose k =1, use the training part for modeling, and calculate the accuracy of prediction using all samples in your test set. Repeat this process, increasing the k, and see which k is the best for your model.\n\nWe can calculate the accuracy of KNN for different Ks.\n",
"_____no_output_____"
]
],
[
[
"Ks = 10\nmean_acc = np.zeros((Ks-1))\nstd_acc = np.zeros((Ks-1))\nConfustionMx = [];\nfor n in range(1,Ks):\n \n #Train Model and Predict \n neigh = KNeighborsClassifier(n_neighbors = n).fit(X_train,y_train)\n yhat=neigh.predict(X_test)\n mean_acc[n-1] = metrics.accuracy_score(y_test, yhat)\n\n \n std_acc[n-1]=np.std(yhat==y_test)/np.sqrt(yhat.shape[0])\n\nmean_acc",
"_____no_output_____"
]
],
[
[
"#### Plot model accuracy for Different number of Neighbors\n",
"_____no_output_____"
]
],
[
[
"plt.plot(range(1,Ks),mean_acc,'g')\nplt.fill_between(range(1,Ks),mean_acc - 1 * std_acc,mean_acc + 1 * std_acc, alpha=0.10)\nplt.legend(('Accuracy ', '+/- 3xstd'))\nplt.ylabel('Accuracy ')\nplt.xlabel('Number of Nabors (K)')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"print( \"The best accuracy was with\", mean_acc.max(), \"with k=\", mean_acc.argmax()+1) ",
"The best accuracy was with 0.34 with k= 9\n"
]
],
[
[
"<h2>Want to learn more?</h2>\n\nIBM SPSS Modeler is a comprehensive analytics platform that has many machine learning algorithms. It has been designed to bring predictive intelligence to decisions made by individuals, by groups, by systems – by your enterprise as a whole. A free trial is available through this course, available here: <a href=\"http://cocl.us/ML0101EN-SPSSModeler\">SPSS Modeler</a>\n\nAlso, you can use Watson Studio to run these notebooks faster with bigger datasets. Watson Studio is IBM's leading cloud solution for data scientists, built by data scientists. With Jupyter notebooks, RStudio, Apache Spark and popular libraries pre-packaged in the cloud, Watson Studio enables data scientists to collaborate on their projects without having to install anything. Join the fast-growing community of Watson Studio users today with a free account at <a href=\"https://cocl.us/ML0101EN_DSX\">Watson Studio</a>\n\n<h3>Thanks for completing this lesson!</h3>\n\n<h4>Author: <a href=\"https://ca.linkedin.com/in/saeedaghabozorgi\">Saeed Aghabozorgi</a></h4>\n<p><a href=\"https://ca.linkedin.com/in/saeedaghabozorgi\">Saeed Aghabozorgi</a>, PhD is a Data Scientist in IBM with a track record of developing enterprise level applications that substantially increases clients’ ability to turn data into actionable knowledge. He is a researcher in data mining field and expert in developing advanced analytic methods like machine learning and statistical modelling on large datasets.</p>\n",
"_____no_output_____"
],
[
"| Date (YYYY-MM-DD) | Version | Changed By | Change Description |\n| ----------------- | ------- | ---------- | --------------------- |\n| 2020-08-04 | 0 | Nayef | Upload file to Gitlab |\n| | | | |\n",
"_____no_output_____"
],
[
"<hr>\n\n<p>Copyright © 2018 <a href=\"https://cocl.us/DX0108EN_CC\">Cognitive Class</a>. This notebook and its source code are released under the terms of the <a href=\"https://bigdatauniversity.com/mit-license/\">MIT License</a>.</p>\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec5fc487aef8bec4f23bf6147a55a85099b58ac7 | 4,621 | ipynb | Jupyter Notebook | ipynb/Madagascar.ipynb | skirienko/oscovida.github.io | eda5412d02365a8a000239be5480512c53bee8c2 | [
"CC-BY-4.0"
]
| null | null | null | ipynb/Madagascar.ipynb | skirienko/oscovida.github.io | eda5412d02365a8a000239be5480512c53bee8c2 | [
"CC-BY-4.0"
]
| null | null | null | ipynb/Madagascar.ipynb | skirienko/oscovida.github.io | eda5412d02365a8a000239be5480512c53bee8c2 | [
"CC-BY-4.0"
]
| null | null | null | 28.349693 | 164 | 0.509197 | [
[
[
"# Madagascar\n\n* Homepage of project: https://oscovida.github.io\n* Plots are explained at http://oscovida.github.io/plots.html\n* [Execute this Jupyter Notebook using myBinder](https://mybinder.org/v2/gh/oscovida/binder/master?filepath=ipynb/Madagascar.ipynb)",
"_____no_output_____"
]
],
[
[
"import datetime\nimport time\n\nstart = datetime.datetime.now()\nprint(f\"Notebook executed on: {start.strftime('%d/%m/%Y %H:%M:%S%Z')} {time.tzname[time.daylight]}\")",
"_____no_output_____"
],
[
"%config InlineBackend.figure_formats = ['svg']\nfrom oscovida import *",
"_____no_output_____"
],
[
"overview(\"Madagascar\", weeks=5);",
"_____no_output_____"
],
[
"overview(\"Madagascar\");",
"_____no_output_____"
],
[
"compare_plot(\"Madagascar\", normalise=True);\n",
"_____no_output_____"
],
[
"# load the data\ncases, deaths = get_country_data(\"Madagascar\")\n\n# compose into one table\ntable = compose_dataframe_summary(cases, deaths)\n\n# show tables with up to 500 rows\npd.set_option(\"max_rows\", 500)\n\n# display the table\ntable",
"_____no_output_____"
]
],
[
[
"# Explore the data in your web browser\n\n- If you want to execute this notebook, [click here to use myBinder](https://mybinder.org/v2/gh/oscovida/binder/master?filepath=ipynb/Madagascar.ipynb)\n- and wait (~1 to 2 minutes)\n- Then press SHIFT+RETURN to advance code cell to code cell\n- See http://jupyter.org for more details on how to use Jupyter Notebook",
"_____no_output_____"
],
[
"# Acknowledgements:\n\n- Johns Hopkins University provides data for countries\n- Robert Koch Institute provides data for within Germany\n- Atlo Team for gathering and providing data from Hungary (https://atlo.team/koronamonitor/)\n- Open source and scientific computing community for the data tools\n- Github for hosting repository and html files\n- Project Jupyter for the Notebook and binder service\n- The H2020 project Photon and Neutron Open Science Cloud ([PaNOSC](https://www.panosc.eu/))\n\n--------------------",
"_____no_output_____"
]
],
[
[
"print(f\"Download of data from Johns Hopkins university: cases at {fetch_cases_last_execution()} and \"\n f\"deaths at {fetch_deaths_last_execution()}.\")",
"_____no_output_____"
],
[
"# to force a fresh download of data, run \"clear_cache()\"",
"_____no_output_____"
],
[
"print(f\"Notebook execution took: {datetime.datetime.now()-start}\")\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec5fc970d7c88584ffdee67c1731d9798aea52c2 | 611,124 | ipynb | Jupyter Notebook | Final_Project_Balancing_SMOTE_(Drop_Size).ipynb | heriswn/LatihanDTS | 033074044980748e4c152a82b0b7f1bfa1c3d997 | [
"MIT"
]
| null | null | null | Final_Project_Balancing_SMOTE_(Drop_Size).ipynb | heriswn/LatihanDTS | 033074044980748e4c152a82b0b7f1bfa1c3d997 | [
"MIT"
]
| null | null | null | Final_Project_Balancing_SMOTE_(Drop_Size).ipynb | heriswn/LatihanDTS | 033074044980748e4c152a82b0b7f1bfa1c3d997 | [
"MIT"
]
| null | null | null | 42.934101 | 31,122 | 0.362961 | [
[
[
"<a href=\"https://colab.research.google.com/github/heriswn/LatihanDTS/blob/master/Final_Project_Balancing_SMOTE_(Drop_Size).ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# <center> FINAL PROJECT",
"_____no_output_____"
],
[
"## <center>Invalid Syntax(]",
"_____no_output_____"
],
[
"# Importing Data",
"_____no_output_____"
]
],
[
[
"import warnings\nimport numpy as np\nimport pandas as pd\nimport scipy as sp\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"warnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"data=pd.read_csv('googleplaystore.csv')",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
]
],
[
[
"# Data Preparation",
"_____no_output_____"
]
],
[
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10841 entries, 0 to 10840\nData columns (total 13 columns):\nApp 10841 non-null object\nCategory 10841 non-null object\nRating 9367 non-null float64\nReviews 10841 non-null object\nSize 10841 non-null object\nInstalls 10841 non-null object\nType 10840 non-null object\nPrice 10841 non-null object\nContent Rating 10840 non-null object\nGenres 10841 non-null object\nLast Updated 10841 non-null object\nCurrent Ver 10833 non-null object\nAndroid Ver 10838 non-null object\ndtypes: float64(1), object(12)\nmemory usage: 1.1+ MB\n"
]
],
[
[
"## Checking Data",
"_____no_output_____"
],
[
"### Check Data Unique",
"_____no_output_____"
]
],
[
[
"for colum in data.columns:\n print(colum,':\\n',data[colum].unique())\n print()",
"App :\n ['Photo Editor & Candy Camera & Grid & ScrapBook' 'Coloring book moana'\n 'U Launcher Lite – FREE Live Cool Themes, Hide Apps' ...\n 'Parkinson Exercices FR' 'The SCP Foundation DB fr nn5n'\n 'iHoroscope - 2018 Daily Horoscope & Astrology']\n\nCategory :\n ['ART_AND_DESIGN' 'AUTO_AND_VEHICLES' 'BEAUTY' 'BOOKS_AND_REFERENCE'\n 'BUSINESS' 'COMICS' 'COMMUNICATION' 'DATING' 'EDUCATION' 'ENTERTAINMENT'\n 'EVENTS' 'FINANCE' 'FOOD_AND_DRINK' 'HEALTH_AND_FITNESS' 'HOUSE_AND_HOME'\n 'LIBRARIES_AND_DEMO' 'LIFESTYLE' 'GAME' 'FAMILY' 'MEDICAL' 'SOCIAL'\n 'SHOPPING' 'PHOTOGRAPHY' 'SPORTS' 'TRAVEL_AND_LOCAL' 'TOOLS'\n 'PERSONALIZATION' 'PRODUCTIVITY' 'PARENTING' 'WEATHER' 'VIDEO_PLAYERS'\n 'NEWS_AND_MAGAZINES' 'MAPS_AND_NAVIGATION' '1.9']\n\nRating :\n [ 4.1 3.9 4.7 4.5 4.3 4.4 3.8 4.2 4.6 3.2 4. nan 4.8 4.9\n 3.6 3.7 3.3 3.4 3.5 3.1 5. 2.6 3. 1.9 2.5 2.8 2.7 1.\n 2.9 2.3 2.2 1.7 2. 1.8 2.4 1.6 2.1 1.4 1.5 1.2 19. ]\n\nReviews :\n ['159' '967' '87510' ... '603' '1195' '398307']\n\nSize :\n ['19M' '14M' '8.7M' '25M' '2.8M' '5.6M' '29M' '33M' '3.1M' '28M' '12M'\n '20M' '21M' '37M' '2.7M' '5.5M' '17M' '39M' '31M' '4.2M' '7.0M' '23M'\n '6.0M' '6.1M' '4.6M' '9.2M' '5.2M' '11M' '24M' 'Varies with device'\n '9.4M' '15M' '10M' '1.2M' '26M' '8.0M' '7.9M' '56M' '57M' '35M' '54M'\n '201k' '3.6M' '5.7M' '8.6M' '2.4M' '27M' '2.5M' '16M' '3.4M' '8.9M'\n '3.9M' '2.9M' '38M' '32M' '5.4M' '18M' '1.1M' '2.2M' '4.5M' '9.8M' '52M'\n '9.0M' '6.7M' '30M' '2.6M' '7.1M' '3.7M' '22M' '7.4M' '6.4M' '3.2M'\n '8.2M' '9.9M' '4.9M' '9.5M' '5.0M' '5.9M' '13M' '73M' '6.8M' '3.5M'\n '4.0M' '2.3M' '7.2M' '2.1M' '42M' '7.3M' '9.1M' '55M' '23k' '6.5M' '1.5M'\n '7.5M' '51M' '41M' '48M' '8.5M' '46M' '8.3M' '4.3M' '4.7M' '3.3M' '40M'\n '7.8M' '8.8M' '6.6M' '5.1M' '61M' '66M' '79k' '8.4M' '118k' '44M' '695k'\n '1.6M' '6.2M' '18k' '53M' '1.4M' '3.0M' '5.8M' '3.8M' '9.6M' '45M' '63M'\n '49M' '77M' '4.4M' '4.8M' '70M' '6.9M' '9.3M' '10.0M' '8.1M' '36M' '84M'\n '97M' '2.0M' '1.9M' '1.8M' '5.3M' '47M' '556k' '526k' '76M' '7.6M' '59M'\n '9.7M' '78M' '72M' '43M' '7.7M' '6.3M' '334k' '34M' '93M' '65M' '79M'\n '100M' '58M' '50M' '68M' '64M' '67M' '60M' '94M' '232k' '99M' '624k'\n '95M' '8.5k' '41k' '292k' '11k' '80M' '1.7M' '74M' '62M' '69M' '75M'\n '98M' '85M' '82M' '96M' '87M' '71M' '86M' '91M' '81M' '92M' '83M' '88M'\n '704k' '862k' '899k' '378k' '266k' '375k' '1.3M' '975k' '980k' '4.1M'\n '89M' '696k' '544k' '525k' '920k' '779k' '853k' '720k' '713k' '772k'\n '318k' '58k' '241k' '196k' '857k' '51k' '953k' '865k' '251k' '930k'\n '540k' '313k' '746k' '203k' '26k' '314k' '239k' '371k' '220k' '730k'\n '756k' '91k' '293k' '17k' '74k' '14k' '317k' '78k' '924k' '902k' '818k'\n '81k' '939k' '169k' '45k' '475k' '965k' '90M' '545k' '61k' '283k' '655k'\n '714k' '93k' '872k' '121k' '322k' '1.0M' '976k' '172k' '238k' '549k'\n '206k' '954k' '444k' '717k' '210k' '609k' '308k' '705k' '306k' '904k'\n '473k' '175k' '350k' '383k' '454k' '421k' '70k' '812k' '442k' '842k'\n '417k' '412k' '459k' '478k' '335k' '782k' '721k' '430k' '429k' '192k'\n '200k' '460k' '728k' '496k' '816k' '414k' '506k' '887k' '613k' '243k'\n '569k' '778k' '683k' '592k' '319k' '186k' '840k' '647k' '191k' '373k'\n '437k' '598k' '716k' '585k' '982k' '222k' '219k' '55k' '948k' '323k'\n '691k' '511k' '951k' '963k' '25k' '554k' '351k' '27k' '82k' '208k' '913k'\n '514k' '551k' '29k' '103k' '898k' '743k' '116k' '153k' '209k' '353k'\n '499k' '173k' '597k' '809k' '122k' '411k' '400k' '801k' '787k' '237k'\n '50k' '643k' '986k' '97k' '516k' '837k' '780k' '961k' '269k' '20k' '498k'\n '600k' '749k' '642k' '881k' '72k' '656k' '601k' '221k' '228k' '108k'\n '940k' '176k' '33k' '663k' '34k' '942k' '259k' '164k' '458k' '245k'\n '629k' '28k' '288k' '775k' '785k' '636k' '916k' '994k' '309k' '485k'\n '914k' '903k' '608k' '500k' '54k' '562k' '847k' '957k' '688k' '811k'\n '270k' '48k' '329k' '523k' '921k' '874k' '981k' '784k' '280k' '24k'\n '518k' '754k' '892k' '154k' '860k' '364k' '387k' '626k' '161k' '879k'\n '39k' '970k' '170k' '141k' '160k' '144k' '143k' '190k' '376k' '193k'\n '246k' '73k' '658k' '992k' '253k' '420k' '404k' '1,000+' '470k' '226k'\n '240k' '89k' '234k' '257k' '861k' '467k' '157k' '44k' '676k' '67k' '552k'\n '885k' '1020k' '582k' '619k']\n\nInstalls :\n ['10,000+' '500,000+' '5,000,000+' '50,000,000+' '100,000+' '50,000+'\n '1,000,000+' '10,000,000+' '5,000+' '100,000,000+' '1,000,000,000+'\n '1,000+' '500,000,000+' '50+' '100+' '500+' '10+' '1+' '5+' '0+' '0'\n 'Free']\n\nType :\n ['Free' 'Paid' nan '0']\n\nPrice :\n ['0' '$4.99' '$3.99' '$6.99' '$1.49' '$2.99' '$7.99' '$5.99' '$3.49'\n '$1.99' '$9.99' '$7.49' '$0.99' '$9.00' '$5.49' '$10.00' '$24.99'\n '$11.99' '$79.99' '$16.99' '$14.99' '$1.00' '$29.99' '$12.99' '$2.49'\n '$10.99' '$1.50' '$19.99' '$15.99' '$33.99' '$74.99' '$39.99' '$3.95'\n '$4.49' '$1.70' '$8.99' '$2.00' '$3.88' '$25.99' '$399.99' '$17.99'\n '$400.00' '$3.02' '$1.76' '$4.84' '$4.77' '$1.61' '$2.50' '$1.59' '$6.49'\n '$1.29' '$5.00' '$13.99' '$299.99' '$379.99' '$37.99' '$18.99' '$389.99'\n '$19.90' '$8.49' '$1.75' '$14.00' '$4.85' '$46.99' '$109.99' '$154.99'\n '$3.08' '$2.59' '$4.80' '$1.96' '$19.40' '$3.90' '$4.59' '$15.46' '$3.04'\n '$4.29' '$2.60' '$3.28' '$4.60' '$28.99' '$2.95' '$2.90' '$1.97'\n '$200.00' '$89.99' '$2.56' '$30.99' '$3.61' '$394.99' '$1.26' 'Everyone'\n '$1.20' '$1.04']\n\nContent Rating :\n ['Everyone' 'Teen' 'Everyone 10+' 'Mature 17+' 'Adults only 18+' 'Unrated'\n nan]\n\nGenres :\n ['Art & Design' 'Art & Design;Pretend Play' 'Art & Design;Creativity'\n 'Art & Design;Action & Adventure' 'Auto & Vehicles' 'Beauty'\n 'Books & Reference' 'Business' 'Comics' 'Comics;Creativity'\n 'Communication' 'Dating' 'Education;Education' 'Education'\n 'Education;Creativity' 'Education;Music & Video'\n 'Education;Action & Adventure' 'Education;Pretend Play'\n 'Education;Brain Games' 'Entertainment' 'Entertainment;Music & Video'\n 'Entertainment;Brain Games' 'Entertainment;Creativity' 'Events' 'Finance'\n 'Food & Drink' 'Health & Fitness' 'House & Home' 'Libraries & Demo'\n 'Lifestyle' 'Lifestyle;Pretend Play' 'Adventure;Action & Adventure'\n 'Arcade' 'Casual' 'Card' 'Casual;Pretend Play' 'Action' 'Strategy'\n 'Puzzle' 'Sports' 'Music' 'Word' 'Racing' 'Casual;Creativity'\n 'Casual;Action & Adventure' 'Simulation' 'Adventure' 'Board' 'Trivia'\n 'Role Playing' 'Simulation;Education' 'Action;Action & Adventure'\n 'Casual;Brain Games' 'Simulation;Action & Adventure'\n 'Educational;Creativity' 'Puzzle;Brain Games' 'Educational;Education'\n 'Card;Brain Games' 'Educational;Brain Games' 'Educational;Pretend Play'\n 'Entertainment;Education' 'Casual;Education' 'Music;Music & Video'\n 'Racing;Action & Adventure' 'Arcade;Pretend Play'\n 'Role Playing;Action & Adventure' 'Simulation;Pretend Play'\n 'Puzzle;Creativity' 'Sports;Action & Adventure'\n 'Educational;Action & Adventure' 'Arcade;Action & Adventure'\n 'Entertainment;Action & Adventure' 'Puzzle;Action & Adventure'\n 'Strategy;Action & Adventure' 'Music & Audio;Music & Video'\n 'Health & Fitness;Education' 'Adventure;Education' 'Board;Brain Games'\n 'Board;Action & Adventure' 'Board;Pretend Play' 'Casual;Music & Video'\n 'Role Playing;Pretend Play' 'Entertainment;Pretend Play'\n 'Video Players & Editors;Creativity' 'Card;Action & Adventure' 'Medical'\n 'Social' 'Shopping' 'Photography' 'Travel & Local'\n 'Travel & Local;Action & Adventure' 'Tools' 'Tools;Education'\n 'Personalization' 'Productivity' 'Parenting' 'Parenting;Music & Video'\n 'Parenting;Education' 'Parenting;Brain Games' 'Weather'\n 'Video Players & Editors' 'Video Players & Editors;Music & Video'\n 'News & Magazines' 'Maps & Navigation'\n 'Health & Fitness;Action & Adventure' 'Educational' 'Casino'\n 'Adventure;Brain Games' 'Trivia;Education' 'Lifestyle;Education'\n 'Books & Reference;Creativity' 'Books & Reference;Education'\n 'Puzzle;Education' 'Role Playing;Education' 'Role Playing;Brain Games'\n 'Strategy;Education' 'Racing;Pretend Play' 'Communication;Creativity'\n 'February 11, 2018' 'Strategy;Creativity']\n\nLast Updated :\n ['January 7, 2018' 'January 15, 2018' 'August 1, 2018' ...\n 'January 20, 2014' 'February 16, 2014' 'March 23, 2014']\n\nCurrent Ver :\n ['1.0.0' '2.0.0' '1.2.4' ... '1.0.612928' '0.3.4' '2.0.148.0']\n\nAndroid Ver :\n ['4.0.3 and up' '4.2 and up' '4.4 and up' '2.3 and up' '3.0 and up'\n '4.1 and up' '4.0 and up' '2.3.3 and up' 'Varies with device'\n '2.2 and up' '5.0 and up' '6.0 and up' '1.6 and up' '1.5 and up'\n '2.1 and up' '7.0 and up' '5.1 and up' '4.3 and up' '4.0.3 - 7.1.1'\n '2.0 and up' '3.2 and up' '4.4W and up' '7.1 and up' '7.0 - 7.1.1'\n '8.0 and up' '5.0 - 8.0' '3.1 and up' '2.0.1 and up' '4.1 - 7.1.1' nan\n '5.0 - 6.0' '1.0 and up' '2.2 - 7.1.1' '5.0 - 7.1.1']\n\n"
],
[
"data.Category.unique()",
"_____no_output_____"
],
[
"data[data.Category=='1.9']",
"_____no_output_____"
]
],
[
[
"### Droping Unclear Data",
"_____no_output_____"
]
],
[
[
"data=data.drop(10472,axis=0)\ndata",
"_____no_output_____"
]
],
[
[
"## Missing Value",
"_____no_output_____"
],
[
"### Finding Missing Value",
"_____no_output_____"
]
],
[
[
"count=0\nfor i in data.isnull().sum(axis=1):\n if i>0:\n count+=1\n\nif count>0:\n print(count,'Rows have Missing Value')",
"1480 Rows have Missing Value\n"
],
[
"for colum in data.columns:\n missed=data[data[colum].isnull()==True].shape[0]\n if missed>0:\n print('Attribute ',colum,': ',missed)\n else:\n print('Attribute ',colum,': No Missing Value')",
"Attribute App : No Missing Value\nAttribute Category : No Missing Value\nAttribute Rating : 1474\nAttribute Reviews : No Missing Value\nAttribute Size : No Missing Value\nAttribute Installs : No Missing Value\nAttribute Type : 1\nAttribute Price : No Missing Value\nAttribute Content Rating : No Missing Value\nAttribute Genres : No Missing Value\nAttribute Last Updated : No Missing Value\nAttribute Current Ver : 8\nAttribute Android Ver : 2\n"
]
],
[
[
"### Replace the Missing Value",
"_____no_output_____"
]
],
[
[
"for colum in data.columns:\n if (data[data[colum].isnull()].shape[0]>0):\n print('\\nAttribute-',colum,' (before) :',data[data[colum].isnull()].shape[0])\n if (data[colum].dtypes in ['int64','float64']):\n data[colum].fillna(data[data[colum].notnull()][colum].mean(), inplace=True)\n else:\n data[colum].fillna(data[data[colum].notnull()][colum].mode(), inplace=True)\n print('\\nAttribute-',colum,' (after) :',data[data[colum].isnull()].shape[0])",
"\nAttribute- Rating (before) : 1474\n\nAttribute- Rating (after) : 0\n\nAttribute- Type (before) : 1\n\nAttribute- Type (after) : 1\n\nAttribute- Current Ver (before) : 8\n\nAttribute- Current Ver (after) : 8\n\nAttribute- Android Ver (before) : 2\n\nAttribute- Android Ver (after) : 2\n"
],
[
"data",
"_____no_output_____"
]
],
[
[
"## Data Type",
"_____no_output_____"
],
[
"### Rating",
"_____no_output_____"
]
],
[
[
"data.Rating.dtype",
"_____no_output_____"
]
],
[
[
"### Reviews",
"_____no_output_____"
]
],
[
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 10840 entries, 0 to 10840\nData columns (total 13 columns):\nApp 10840 non-null object\nCategory 10840 non-null object\nRating 10840 non-null float64\nReviews 10840 non-null object\nSize 10840 non-null object\nInstalls 10840 non-null object\nType 10839 non-null object\nPrice 10840 non-null object\nContent Rating 10840 non-null object\nGenres 10840 non-null object\nLast Updated 10840 non-null object\nCurrent Ver 10832 non-null object\nAndroid Ver 10838 non-null object\ndtypes: float64(1), object(12)\nmemory usage: 1.2+ MB\n"
],
[
"data.Reviews.dtype",
"_____no_output_____"
],
[
"data.Reviews",
"_____no_output_____"
],
[
"data.Reviews=data.Reviews.astype('int64')",
"_____no_output_____"
]
],
[
[
"### Size",
"_____no_output_____"
]
],
[
[
"data.Size.dtype",
"_____no_output_____"
],
[
"data.Size",
"_____no_output_____"
],
[
"k_indices=data.Size.loc[data.Size.str.contains('k')].index.tolist()\nconverter=pd.DataFrame(data.loc[k_indices,'Size'].apply(lambda x: x.strip('k')).astype(float).apply(lambda x: x / 1024).apply(lambda x: round(x, 3)).astype(str))\ndata.loc[k_indices,'Size'] = converter",
"_____no_output_____"
],
[
"data.Size=data.Size.apply(lambda x: x.strip('M'))\ndata.Size=data.Size.apply(lambda x: x.strip('Varies with device'))\ndata.Size=data.Size.replace('','NaN')\ndata.Size=data.Size.astype('float64')\ndata.Size",
"_____no_output_____"
],
[
"data.Size.value_counts()",
"_____no_output_____"
],
[
"data=data.dropna()\ndata",
"_____no_output_____"
]
],
[
[
"### Installs",
"_____no_output_____"
]
],
[
[
"data.Installs.dtype",
"_____no_output_____"
],
[
"data.Installs",
"_____no_output_____"
],
[
"data.Installs=data.Installs.apply(lambda x: x.strip('+'))\ndata.Installs=data.Installs.apply(lambda x: x.replace(',',''))\ndata.Installs=data.Installs.astype('int64')\ndata.Installs",
"_____no_output_____"
]
],
[
[
"### Price",
"_____no_output_____"
]
],
[
[
"data.Price.dtype",
"_____no_output_____"
],
[
"data.Price",
"_____no_output_____"
],
[
"data.Price=data.Price.apply(lambda x: x.strip('$'))\ndata.Price=data.Price.astype('float64')\ndata.Price",
"_____no_output_____"
],
[
"data.Installs.value_counts().sort_index()",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 9135 entries, 0 to 10840\nData columns (total 13 columns):\nApp 9135 non-null object\nCategory 9135 non-null object\nRating 9135 non-null float64\nReviews 9135 non-null int64\nSize 9135 non-null float64\nInstalls 9135 non-null int64\nType 9135 non-null object\nPrice 9135 non-null float64\nContent Rating 9135 non-null object\nGenres 9135 non-null object\nLast Updated 9135 non-null object\nCurrent Ver 9135 non-null object\nAndroid Ver 9135 non-null object\ndtypes: float64(3), int64(2), object(8)\nmemory usage: 999.1+ KB\n"
]
],
[
[
"### Labeling Popularity Apps",
"_____no_output_____"
]
],
[
[
"data['Popularity']=data.apply(lambda row:'Very Not Popular' if (row['Installs']<50) \n else('Not Popular' if ((row['Installs']>=50) and (row['Installs']<5000))\n else('Ordinary' if ((row['Installs']>=5000) and (row['Installs']<500000))\n else('Popular' if ((row['Installs']>=500000) and (row['Installs']<50000000))\n else 'Very popular'))),axis=1)",
"_____no_output_____"
],
[
"data.Popularity.value_counts()",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 9135 entries, 0 to 10840\nData columns (total 14 columns):\nApp 9135 non-null object\nCategory 9135 non-null object\nRating 9135 non-null float64\nReviews 9135 non-null int64\nSize 9135 non-null float64\nInstalls 9135 non-null int64\nType 9135 non-null object\nPrice 9135 non-null float64\nContent Rating 9135 non-null object\nGenres 9135 non-null object\nLast Updated 9135 non-null object\nCurrent Ver 9135 non-null object\nAndroid Ver 9135 non-null object\nPopularity 9135 non-null object\ndtypes: float64(3), int64(2), object(9)\nmemory usage: 1.0+ MB\n"
]
],
[
[
"### Dropping Columns Less Effect",
"_____no_output_____"
]
],
[
[
"data",
"_____no_output_____"
],
[
"data=data.drop(columns=['App','Category'],axis=1)\ndata=data.drop(columns=['Type','Installs'],axis=1)\ndata=data.drop(columns=['Last Updated','Current Ver','Android Ver'],axis=1)\ndata",
"_____no_output_____"
]
],
[
[
"## Encoding",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\nfrom sklearn.preprocessing import LabelEncoder",
"_____no_output_____"
]
],
[
[
"### Content Rating",
"_____no_output_____"
]
],
[
[
"data['Content Rating']=LabelEncoder().fit_transform(data['Content Rating'])\ndata",
"_____no_output_____"
]
],
[
[
"### Genres",
"_____no_output_____"
]
],
[
[
"data['Genres']=LabelEncoder().fit_transform(data['Genres'])\ndata",
"_____no_output_____"
]
],
[
[
"### Correlation",
"_____no_output_____"
]
],
[
[
"corr=data.corr()\nsns.heatmap(corr,annot=True, fmt='.2f')",
"_____no_output_____"
]
],
[
[
"# Modelling",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.model_selection import KFold\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import f1_score\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"from imblearn.over_sampling import SMOTE\nfrom imblearn.over_sampling import RandomOverSampler",
"_____no_output_____"
]
],
[
[
"### Data Target",
"_____no_output_____"
]
],
[
[
"X=data.iloc[:,:-1].values\nY=data.iloc[:,-1].values\n\nx_train,x_test,y_train,y_test=train_test_split(X,Y,test_size=0.2,random_state=123,stratify=Y)",
"_____no_output_____"
],
[
"pd.value_counts(pd.Series(Y))",
"_____no_output_____"
],
[
"sns.countplot(data.Popularity).set(xlabel='Popularity', ylabel='Frequency')",
"_____no_output_____"
]
],
[
[
"### Balancing Data",
"_____no_output_____"
]
],
[
[
"y_train.shape",
"_____no_output_____"
],
[
"x_train.shape",
"_____no_output_____"
],
[
"sm=SMOTE(random_state=123)\nx_train_re,y_train_re=sm.fit_resample(x_train,y_train)",
"_____no_output_____"
],
[
"pd.value_counts(pd.Series(y_train))",
"_____no_output_____"
],
[
"pd.value_counts(pd.Series(y_train_re))",
"_____no_output_____"
],
[
"f,axes=plt.subplots(1,2,figsize=(15,10),sharex=True,sharey=True)\n\nsns.countplot(y_train,ax=axes[0]).set(xlabel='Popularity (Before Balancing)', ylabel='Frequency')\nsns.countplot(y_train_re,ax=axes[1]).set(xlabel='Popularity (After Balancing)', ylabel='Frequency')",
"_____no_output_____"
]
],
[
[
"# Experiment",
"_____no_output_____"
],
[
"## KNN Models",
"_____no_output_____"
],
[
"### Training and Testing Model",
"_____no_output_____"
],
[
"#### Find the Best Parameter for KNN Model",
"_____no_output_____"
]
],
[
[
"paramknn={\n 'n_neighbors':[5,6,7,8,9,10],\n 'leaf_size':[1,2,3,5],\n 'weights':['uniform', 'distance'],\n 'algorithm':['auto', 'ball_tree','kd_tree','brute'],\n 'n_jobs':[-1]\n}",
"_____no_output_____"
],
[
"knn_parameters=[]",
"_____no_output_____"
],
[
"modelknn=KNeighborsClassifier(n_jobs=-1)",
"_____no_output_____"
],
[
"modelknn_best=GridSearchCV(modelknn,param_grid=paramknn,n_jobs=-1)\nmodelknn_best.fit(x_train,y_train)\nknn_parameters.append(modelknn_best.best_params_)\nknn_parameters[0]",
"_____no_output_____"
],
[
"modelknn_re_best=GridSearchCV(modelknn,param_grid=paramknn,n_jobs=-1)\nmodelknn_re_best.fit(x_train_re,y_train_re)\nknn_parameters.append(modelknn_re_best.best_params_)\nknn_parameters[1]",
"_____no_output_____"
],
[
"# def modelknn(parameter,i):\n# modell=KNeighborsClassifier(n_jobs=-1)\n# parameters=parameter[i].values\n# model=GridSearchCV(modell,param_grid=parameters,n_jobs=-1)\n# return model\n\ndef modelknn(n,l,w,a,j):\n model=KNeighborsClassifier(n_neighbors=n,leaf_size=l,weights=w,algorithm=a,n_jobs=j)\n return model",
"_____no_output_____"
],
[
"model_knn=[]",
"_____no_output_____"
],
[
"# model_knn.append(modelknn(knn_parameters,0))\n\nmodel_knn.append(modelknn(10,2,'distance','auto',-1))",
"_____no_output_____"
],
[
"train=model_knn[0].fit(x_train,y_train)\npredicted=train.predict(x_test)\nprint(\"KNN (Before Balancing) :\\n\", classification_report(y_test,predicted))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted))",
"KNN (Before Balancing) :\n precision recall f1-score support\n\n Not Popular 0.76 0.79 0.77 419\n Ordinary 0.82 0.83 0.82 594\n Popular 0.89 0.89 0.89 630\nVery Not Popular 0.55 0.40 0.46 106\n Very popular 0.85 0.85 0.85 78\n\n accuracy 0.82 1827\n macro avg 0.77 0.75 0.76 1827\n weighted avg 0.81 0.82 0.81 1827\n\n\n\nConfusion Matrix:\n\n[[329 55 0 35 0]\n [ 41 492 61 0 0]\n [ 0 55 563 0 12]\n [ 64 0 0 42 0]\n [ 0 1 11 0 66]]\n"
],
[
"train_re=model_knn[0].fit(x_train_re,y_train_re)\npredicted_re=train_re.predict(x_test)\nprint(\"KNN (After Balancing) :\\n\", classification_report(y_test,predicted_re))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_re))",
"KNN (After Balancing) :\n precision recall f1-score support\n\n Not Popular 0.74 0.68 0.70 419\n Ordinary 0.82 0.79 0.81 594\n Popular 0.90 0.84 0.87 630\nVery Not Popular 0.43 0.64 0.51 106\n Very popular 0.62 0.94 0.75 78\n\n accuracy 0.78 1827\n macro avg 0.70 0.78 0.73 1827\n weighted avg 0.80 0.78 0.78 1827\n\n\n\nConfusion Matrix:\n\n[[283 46 0 90 0]\n [ 64 472 57 1 0]\n [ 0 58 528 0 44]\n [ 38 0 0 68 0]\n [ 0 1 4 0 73]]\n"
],
[
"model_knn.append(modelknn(7,1,'distance','brute',-1))",
"_____no_output_____"
],
[
"train_2=model_knn[1].fit(x_train,y_train)\npredicted_2=train_2.predict(x_test)\nprint(\"KNN (Before Balancing) :\\n\", classification_report(y_test,predicted_2))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_2))",
"KNN (Before Balancing) :\n precision recall f1-score support\n\n Not Popular 0.75 0.77 0.76 419\n Ordinary 0.81 0.82 0.82 594\n Popular 0.88 0.89 0.88 630\nVery Not Popular 0.51 0.38 0.43 106\n Very popular 0.84 0.85 0.84 78\n\n accuracy 0.81 1827\n macro avg 0.76 0.74 0.75 1827\n weighted avg 0.80 0.81 0.81 1827\n\n\n\nConfusion Matrix:\n\n[[324 56 0 39 0]\n [ 41 487 66 0 0]\n [ 0 57 560 0 13]\n [ 66 0 0 40 0]\n [ 0 1 11 0 66]]\n"
],
[
"train_re_2=model_knn[1].fit(x_train_re,y_train_re)\npredicted_re_2=train_re_2.predict(x_test)\nprint(\"KNN (After Balancing) :\\n\", classification_report(y_test,predicted_re_2))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_re_2))",
"KNN (After Balancing) :\n precision recall f1-score support\n\n Not Popular 0.75 0.66 0.70 419\n Ordinary 0.81 0.80 0.80 594\n Popular 0.89 0.83 0.86 630\nVery Not Popular 0.43 0.66 0.52 106\n Very popular 0.62 0.94 0.74 78\n\n accuracy 0.78 1827\n macro avg 0.70 0.78 0.73 1827\n weighted avg 0.79 0.78 0.78 1827\n\n\n\nConfusion Matrix:\n\n[[278 51 0 90 0]\n [ 59 473 61 1 0]\n [ 0 59 526 0 45]\n [ 36 0 0 70 0]\n [ 0 1 4 0 73]]\n"
]
],
[
[
"### KFold",
"_____no_output_____"
]
],
[
[
"def kfoldknnclass(x,y,n):\n for i in range(len(model_knn)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_knn[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Not balanced')\n print('=================')\n print('Parameter:',knn_parameters[i],'\\n')\n print('Hasil dari model KNN:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n model.fit(X_train,Y_train)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('Subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfoldknnclass(X,Y,5)",
"Not balanced\n=================\nParameter: {'algorithm': 'auto', 'leaf_size': 2, 'n_jobs': -1, 'n_neighbors': 10, 'weights': 'distance'} \n\nHasil dari model KNN:\n\nSubset 1 accuracy= 0.8445539135194308\nSubset 1 f1 score= 0.8461557398973909\nSubset 2 accuracy= 0.8319649698960043\nSubset 2 f1 score= 0.8263070876379154\nSubset 3 accuracy= 0.7586206896551724\nSubset 3 f1 score= 0.7591120993687734\nSubset 4 accuracy= 0.7848932676518884\nSubset 4 f1 score= 0.780589182981329\nSubset 5 accuracy= 0.789272030651341\nSubset 5 f1 score= 0.7842931327982124\n\n\nAverage Accuracy= 0.8018609742747674\nAverage F1 Score= 0.7992914485367242\n\n\nConfusion Matrix:\n [[467 70 0 42 0]\n [ 58 521 56 0 1]\n [ 0 41 369 0 8]\n [104 0 0 70 0]\n [ 0 0 5 0 15]]\n\n\n\nNot balanced\n=================\nParameter: {'algorithm': 'brute', 'leaf_size': 1, 'n_jobs': -1, 'n_neighbors': 7, 'weights': 'distance'} \n\nHasil dari model KNN:\n\nSubset 1 accuracy= 0.8418171866447729\nSubset 1 f1 score= 0.8435532821682452\nSubset 2 accuracy= 0.8286808976464148\nSubset 2 f1 score= 0.8240712574548607\nSubset 3 accuracy= 0.7624521072796935\nSubset 3 f1 score= 0.7634235301437723\nSubset 4 accuracy= 0.7788724685276409\nSubset 4 f1 score= 0.7750162135873271\nSubset 5 accuracy= 0.7876299945265463\nSubset 5 f1 score= 0.7807348624607738\n\n\nAverage Accuracy= 0.7998905309250136\nAverage F1 Score= 0.7973598291629959\n\n\nConfusion Matrix:\n [[464 67 0 48 0]\n [ 51 530 54 0 1]\n [ 0 39 372 0 7]\n [116 0 0 58 0]\n [ 0 0 5 0 15]]\n\n\n\n"
],
[
"def kfoldknnclass_re(x,y,n):\n for i in range(len(model_knn)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_knn[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Balanced')\n print('=================')\n print('Parameter:',knn_parameters[i],'\\n')\n print('Hasil dari model KNN:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n sm=SMOTE(random_state=123)\n X_train_re,Y_train_re=sm.fit_resample(X_train,Y_train)\n \n model.fit(X_train_re,Y_train_re)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfoldknnclass_re(X,Y,5)",
"Balanced\n=================\nParameter: {'algorithm': 'auto', 'leaf_size': 2, 'n_jobs': -1, 'n_neighbors': 10, 'weights': 'distance'} \n\nHasil dari model KNN:\n\nSubset 1 accuracy= 0.8237547892720306\nsubset 1 f1 score= 0.8267353643425251\nSubset 2 accuracy= 0.8056923918992884\nsubset 2 f1 score= 0.8079135421447345\nSubset 3 accuracy= 0.7323481116584565\nsubset 3 f1 score= 0.7425524711167146\nSubset 4 accuracy= 0.7542419266557198\nsubset 4 f1 score= 0.7609052302768805\nSubset 5 accuracy= 0.7629994526546251\nsubset 5 f1 score= 0.7688162314966768\n\n\nAverage Accuracy= 0.7758073344280241\nAverage F1 Score= 0.7813845678755063\n\n\nConfusion Matrix:\n [[392 49 0 138 0]\n [ 80 508 47 0 1]\n [ 0 46 350 0 22]\n [ 48 0 0 126 0]\n [ 0 0 2 0 18]]\n\n\n\nBalanced\n=================\nParameter: {'algorithm': 'brute', 'leaf_size': 1, 'n_jobs': -1, 'n_neighbors': 7, 'weights': 'distance'} \n\nHasil dari model KNN:\n\nSubset 1 accuracy= 0.8111658456486043\nsubset 1 f1 score= 0.8146905361062797\nSubset 2 accuracy= 0.8029556650246306\nsubset 2 f1 score= 0.8048042857074877\nSubset 3 accuracy= 0.7345374931581828\nsubset 3 f1 score= 0.7443646519804836\nSubset 4 accuracy= 0.7416529830322933\nsubset 4 f1 score= 0.7482579316554809\nSubset 5 accuracy= 0.7608100711548987\nsubset 5 f1 score= 0.7658368058427554\n\n\nAverage Accuracy= 0.7702244116037218\nAverage F1 Score= 0.7755908422584975\n\n\nConfusion Matrix:\n [[392 54 0 133 0]\n [ 84 501 50 0 1]\n [ 0 45 353 0 20]\n [ 48 0 0 126 0]\n [ 0 0 2 0 18]]\n\n\n\n"
]
],
[
[
"## Decision Tree Models",
"_____no_output_____"
],
[
"### Training and Testing Model",
"_____no_output_____"
],
[
"#### Find the Best Parameter for Decision Tree Model",
"_____no_output_____"
]
],
[
[
"paramdtree={\n 'max_features': ['auto', 'sqrt', 'log2'],\n 'min_samples_split': [2,3,4,5,6,7,8,9,10,11,12,13,14,15], \n 'min_samples_leaf':[1,2,3,4,5,6,7,8,9,10,11],\n 'random_state':[123]\n}",
"_____no_output_____"
],
[
"dtree_parameters=[]",
"_____no_output_____"
],
[
"modeldtree=DecisionTreeClassifier(random_state=1234)",
"_____no_output_____"
],
[
"modeldtree_best=GridSearchCV(modeldtree,param_grid=paramdtree,n_jobs=-1)\nmodeldtree_best.fit(x_train,y_train)\ndtree_parameters.append(modeldtree_best.best_params_)\ndtree_parameters[0]",
"_____no_output_____"
],
[
"modeldtree_re_best=GridSearchCV(modeldtree,param_grid=paramdtree,n_jobs=-1)\nmodeldtree_re_best.fit(x_train_re,y_train_re)\ndtree_parameters.append(modeldtree_re_best.best_params_)\ndtree_parameters[1]",
"_____no_output_____"
],
[
"# def modelknn(parameter,i):\n# model=KNeighborsClassifier(n_jobs=-1)\n# model=GridSearchCV(model,param_grid=parameters[i],n_jobs=-1)\n# return model\n\ndef modeldtree(f,s,l,r):\n model=DecisionTreeClassifier(max_features=f,min_samples_split=s,min_samples_leaf=l,random_state=r)\n return model",
"_____no_output_____"
],
[
"model_dtree=[]",
"_____no_output_____"
],
[
"model_dtree.append(modeldtree('auto',2,5,123))",
"_____no_output_____"
],
[
"train=model_dtree[0].fit(x_train,y_train)\npredicted=train.predict(x_test)\nprint(\"Decision Tree (Before Balancing) :\\n\", classification_report(y_test,predicted))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted))",
"Decision Tree (Before Balancing) :\n precision recall f1-score support\n\n Not Popular 0.76 0.78 0.77 419\n Ordinary 0.81 0.81 0.81 594\n Popular 0.89 0.89 0.89 630\nVery Not Popular 0.60 0.50 0.54 106\n Very popular 0.84 0.83 0.84 78\n\n accuracy 0.82 1827\n macro avg 0.78 0.76 0.77 1827\n weighted avg 0.81 0.82 0.81 1827\n\n\n\nConfusion Matrix:\n\n[[328 56 0 35 0]\n [ 52 483 58 1 0]\n [ 0 56 562 0 12]\n [ 53 0 0 53 0]\n [ 0 0 13 0 65]]\n"
],
[
"train_re=model_dtree[0].fit(x_train_re,y_train_re)\npredicted_re=train_re.predict(x_test)\nprint(\"Decision Tree (After Balancing) :\\n\", classification_report(y_test,predicted_re))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_re))",
"Decision Tree (After Balancing) :\n precision recall f1-score support\n\n Not Popular 0.73 0.70 0.72 419\n Ordinary 0.79 0.76 0.78 594\n Popular 0.88 0.87 0.87 630\nVery Not Popular 0.51 0.68 0.58 106\n Very popular 0.74 0.88 0.81 78\n\n accuracy 0.78 1827\n macro avg 0.73 0.78 0.75 1827\n weighted avg 0.79 0.78 0.78 1827\n\n\n\nConfusion Matrix:\n\n[[293 61 4 61 0]\n [ 69 452 64 9 0]\n [ 3 58 545 0 24]\n [ 34 0 0 72 0]\n [ 0 0 9 0 69]]\n"
],
[
"model_dtree.append(modeldtree('auto',10,1,123))",
"_____no_output_____"
],
[
"train_2=model_dtree[1].fit(x_train,y_train)\npredicted_2=train_2.predict(x_test)\nprint(\"Decision Tree (Before Balancing) :\\n\", classification_report(y_test,predicted_2))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_2))",
"Decision Tree (Before Balancing) :\n precision recall f1-score support\n\n Not Popular 0.71 0.75 0.73 419\n Ordinary 0.77 0.77 0.77 594\n Popular 0.88 0.88 0.88 630\nVery Not Popular 0.57 0.42 0.49 106\n Very popular 0.84 0.85 0.84 78\n\n accuracy 0.79 1827\n macro avg 0.75 0.74 0.74 1827\n weighted avg 0.79 0.79 0.79 1827\n\n\n\nConfusion Matrix:\n\n[[316 69 0 34 0]\n [ 73 460 61 0 0]\n [ 1 61 555 0 13]\n [ 55 6 0 45 0]\n [ 0 0 12 0 66]]\n"
],
[
"train_re_2=model_dtree[1].fit(x_train_re,y_train_re)\npredicted_re_2=train_re_2.predict(x_test)\nprint(\"Decision Tree (After Balancing) :\\n\", classification_report(y_test,predicted_re_2))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_re_2))",
"Decision Tree (After Balancing) :\n precision recall f1-score support\n\n Not Popular 0.71 0.70 0.71 419\n Ordinary 0.78 0.79 0.79 594\n Popular 0.87 0.84 0.85 630\nVery Not Popular 0.50 0.52 0.51 106\n Very popular 0.63 0.79 0.70 78\n\n accuracy 0.77 1827\n macro avg 0.70 0.73 0.71 1827\n weighted avg 0.77 0.77 0.77 1827\n\n\n\nConfusion Matrix:\n\n[[294 72 2 51 0]\n [ 61 468 61 4 0]\n [ 9 58 527 0 36]\n [ 50 0 1 55 0]\n [ 0 0 16 0 62]]\n"
]
],
[
[
"### KFold",
"_____no_output_____"
]
],
[
[
"def kfolddtreeclass(x,y,n):\n for i in range(len(model_dtree)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_dtree[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Not Balanced')\n print('=================')\n print('Parameter:',dtree_parameters[i],'\\n')\n print('Hasil dari model Decision Tree:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n model.fit(X_train,Y_train)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('Subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfolddtreeclass(X,Y,5)",
"Not Balanced\n=================\nParameter: {'max_features': 'auto', 'min_samples_leaf': 5, 'min_samples_split': 2, 'random_state': 123} \n\nHasil dari model Decision Tree:\n\nSubset 1 accuracy= 0.8434592227695676\nSubset 1 f1 score= 0.8453978213931522\nSubset 2 accuracy= 0.8182813355227149\nSubset 2 f1 score= 0.8183024815899147\nSubset 3 accuracy= 0.7690202517788725\nSubset 3 f1 score= 0.7685101052544145\nSubset 4 accuracy= 0.7870826491516146\nSubset 4 f1 score= 0.7830343259917703\nSubset 5 accuracy= 0.7837985769020251\nSubset 5 f1 score= 0.7748065333610413\n\n\nAverage Accuracy= 0.800328407224959\nAverage F1 Score= 0.7980102535180587\n\n\nConfusion Matrix:\n [[477 71 0 31 0]\n [ 72 526 38 0 0]\n [ 4 45 365 0 4]\n [123 0 0 51 0]\n [ 0 0 7 0 13]]\n\n\n\nNot Balanced\n=================\nParameter: {'max_features': 'auto', 'min_samples_leaf': 1, 'min_samples_split': 10, 'random_state': 123} \n\nHasil dari model Decision Tree:\n\nSubset 1 accuracy= 0.8407224958949097\nSubset 1 f1 score= 0.8438384809641608\nSubset 2 accuracy= 0.8013136288998358\nSubset 2 f1 score= 0.800538674692416\nSubset 3 accuracy= 0.7777777777777778\nSubset 3 f1 score= 0.7781201870896611\nSubset 4 accuracy= 0.7723043240284619\nSubset 4 f1 score= 0.7698468936929426\nSubset 5 accuracy= 0.7794198139025725\nSubset 5 f1 score= 0.7775645886561883\n\n\nAverage Accuracy= 0.7943076081007115\nAverage F1 Score= 0.7939817650190737\n\n\nConfusion Matrix:\n [[451 73 3 51 1]\n [ 69 507 58 1 1]\n [ 0 39 367 0 12]\n [ 86 1 0 87 0]\n [ 0 0 8 0 12]]\n\n\n\n"
],
[
"def kfolddtreeclass_re(x,y,n):\n for i in range(len(model_dtree)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_dtree[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Balanced')\n print('=================')\n print('Parameter:',dtree_parameters[i],'\\n')\n print('Hasil dari model Decision Tree:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n sm=SMOTE(random_state=123)\n X_train_re,Y_train_re=sm.fit_resample(X_train,Y_train)\n \n model.fit(X_train_re,Y_train_re)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfolddtreeclass_re(X,Y,5)",
"Balanced\n=================\nParameter: {'max_features': 'auto', 'min_samples_leaf': 5, 'min_samples_split': 2, 'random_state': 123} \n\nHasil dari model Decision Tree:\n\nSubset 1 accuracy= 0.7427476737821566\nsubset 1 f1 score= 0.7488023985877542\nSubset 2 accuracy= 0.80623973727422\nsubset 2 f1 score= 0.8088531478847452\nSubset 3 accuracy= 0.7733990147783252\nsubset 3 f1 score= 0.7791454016052648\nSubset 4 accuracy= 0.7416529830322933\nsubset 4 f1 score= 0.7445132686764745\nSubset 5 accuracy= 0.7843459222769568\nsubset 5 f1 score= 0.7878993878466167\n\n\nAverage Accuracy= 0.7696770662287903\nAverage F1 Score= 0.773842720920171\n\n\nConfusion Matrix:\n [[431 46 2 99 1]\n [ 82 507 43 3 1]\n [ 2 40 361 1 14]\n [ 56 0 0 118 0]\n [ 0 0 4 0 16]]\n\n\n\nBalanced\n=================\nParameter: {'max_features': 'auto', 'min_samples_leaf': 1, 'min_samples_split': 10, 'random_state': 123} \n\nHasil dari model Decision Tree:\n\nSubset 1 accuracy= 0.8325123152709359\nsubset 1 f1 score= 0.8371772290157722\nSubset 2 accuracy= 0.8024083196496989\nsubset 2 f1 score= 0.8051776749580094\nSubset 3 accuracy= 0.7619047619047619\nsubset 3 f1 score= 0.7655163407498617\nSubset 4 accuracy= 0.7640941434044882\nsubset 4 f1 score= 0.7668990285497664\nSubset 5 accuracy= 0.7646414887794198\nsubset 5 f1 score= 0.7641243524822754\n\n\nAverage Accuracy= 0.7851122058018609\nAverage F1 Score= 0.787778925151137\n\n\nConfusion Matrix:\n [[453 63 2 61 0]\n [ 83 495 54 3 1]\n [ 0 61 345 0 12]\n [ 84 1 0 89 0]\n [ 0 0 5 0 15]]\n\n\n\n"
]
],
[
[
"## Random Forest Models",
"_____no_output_____"
],
[
"### Training and Testing Model",
"_____no_output_____"
],
[
"#### Find the Best Parameter for Random Forest Model",
"_____no_output_____"
]
],
[
[
"paramrf={\n 'criterion':['gini','entropy'],\n 'n_estimators':[10,15,20,25,30],\n 'min_samples_leaf':[1,2,3],\n 'min_samples_split':[3,4,5,6,7], \n 'random_state':[123],\n 'n_jobs':[-1]\n}",
"_____no_output_____"
],
[
"rf_parameters=[]",
"_____no_output_____"
],
[
"modelrf=RandomForestClassifier()",
"_____no_output_____"
],
[
"modelrf_best=GridSearchCV(modelrf,param_grid=paramrf,n_jobs=-1)\nmodelrf_best.fit(x_train,y_train)\nrf_parameters.append(modelrf_best.best_params_)\nrf_parameters[0]",
"_____no_output_____"
],
[
"modelrf_re_best=GridSearchCV(modelrf,param_grid=paramrf,n_jobs=-1)\nmodelrf_re_best.fit(x_train_re,y_train_re)\nrf_parameters.append(modelrf_re_best.best_params_)\nrf_parameters[1]",
"_____no_output_____"
],
[
"# def modelknn(parameter,i):\n# modell=KNeighborsClassifier(n_jobs=-1)\n# parameters=parameter[i].values\n# model=GridSearchCV(modell,param_grid=parameters,n_jobs=-1)\n# return model\n\ndef modelrf(c,e,l,s,r,j):\n model=RandomForestClassifier(criterion=c,n_estimators=e,min_samples_leaf=l,\n min_samples_split=s,random_state=r,n_jobs=j)\n return model",
"_____no_output_____"
],
[
"model_rf=[]",
"_____no_output_____"
],
[
"# model_knn.append(modelknn(knn_parameters,0))\n\nmodel_rf.append(modelrf('entropy',15,3,3,123,-1))",
"_____no_output_____"
],
[
"train=model_rf[0].fit(x_train,y_train)\npredicted=train.predict(x_test)\nprint(\"Random Forest (Before Balancing) :\\n\", classification_report(y_test,predicted))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted))",
"Random Forest (Before Balancing) :\n precision recall f1-score support\n\n Not Popular 0.78 0.81 0.80 419\n Ordinary 0.86 0.86 0.86 594\n Popular 0.92 0.92 0.92 630\nVery Not Popular 0.63 0.52 0.57 106\n Very popular 0.88 0.87 0.88 78\n\n accuracy 0.85 1827\n macro avg 0.81 0.80 0.80 1827\n weighted avg 0.85 0.85 0.85 1827\n\n\n\nConfusion Matrix:\n\n[[340 47 0 32 0]\n [ 43 510 41 0 0]\n [ 0 39 582 0 9]\n [ 51 0 0 55 0]\n [ 0 0 10 0 68]]\n"
],
[
"train_re=model_rf[0].fit(x_train_re,y_train_re)\npredicted_re=train_re.predict(x_test)\nprint(\"Random Forest (After Balancing) :\\n\", classification_report(y_test,predicted_re))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_re))",
"Random Forest (After Balancing) :\n precision recall f1-score support\n\n Not Popular 0.78 0.76 0.77 419\n Ordinary 0.85 0.85 0.85 594\n Popular 0.93 0.90 0.92 630\nVery Not Popular 0.56 0.66 0.60 106\n Very popular 0.81 0.94 0.87 78\n\n accuracy 0.84 1827\n macro avg 0.79 0.82 0.80 1827\n weighted avg 0.84 0.84 0.84 1827\n\n\n\nConfusion Matrix:\n\n[[320 44 0 55 0]\n [ 52 502 39 1 0]\n [ 0 43 570 0 17]\n [ 36 0 0 70 0]\n [ 0 0 5 0 73]]\n"
],
[
"model_rf.append(modelrf('gini',30,1,4,123,-1))",
"_____no_output_____"
],
[
"train_2=model_rf[1].fit(x_train,y_train)\npredicted_2=train_2.predict(x_test)\nprint(\"Random Forest (Before Balancing) :\\n\", classification_report(y_test,predicted_2))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_2))",
"Random Forest (Before Balancing) :\n precision recall f1-score support\n\n Not Popular 0.78 0.80 0.79 419\n Ordinary 0.85 0.87 0.86 594\n Popular 0.92 0.92 0.92 630\nVery Not Popular 0.59 0.49 0.54 106\n Very popular 0.89 0.85 0.87 78\n\n accuracy 0.85 1827\n macro avg 0.81 0.78 0.79 1827\n weighted avg 0.84 0.85 0.85 1827\n\n\n\nConfusion Matrix:\n\n[[334 50 0 35 0]\n [ 38 515 40 1 0]\n [ 0 42 580 0 8]\n [ 54 0 0 52 0]\n [ 0 0 12 0 66]]\n"
],
[
"train_re_2=model_rf[1].fit(x_train_re,y_train_re)\npredicted_re_2=train_re_2.predict(x_test)\nprint(\"Random Forest (After Balancing) :\\n\", classification_report(y_test,predicted_re_2))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_re_2))",
"Random Forest (After Balancing) :\n precision recall f1-score support\n\n Not Popular 0.78 0.78 0.78 419\n Ordinary 0.85 0.86 0.86 594\n Popular 0.94 0.92 0.93 630\nVery Not Popular 0.58 0.56 0.57 106\n Very popular 0.87 0.94 0.90 78\n\n accuracy 0.85 1827\n macro avg 0.80 0.81 0.81 1827\n weighted avg 0.85 0.85 0.85 1827\n\n\n\nConfusion Matrix:\n\n[[327 50 0 42 0]\n [ 47 513 33 1 0]\n [ 0 39 580 0 11]\n [ 47 0 0 59 0]\n [ 0 0 5 0 73]]\n"
]
],
[
[
"### KFold",
"_____no_output_____"
]
],
[
[
"def kfoldrfclass(x,y,n):\n for i in range(len(model_rf)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_rf[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Not balanced')\n print('=================')\n print('Parameter:',rf_parameters[i],'\\n')\n print('Hasil dari model Random Forest:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n model.fit(X_train,Y_train)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('Subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfoldrfclass(X,Y,5)",
"Not balanced\n=================\nParameter: {'criterion': 'entropy', 'min_samples_leaf': 3, 'min_samples_split': 3, 'n_estimators': 15, 'n_jobs': -1, 'random_state': 123} \n\nHasil dari model Random Forest:\n\nSubset 1 accuracy= 0.8735632183908046\nSubset 1 f1 score= 0.8753098125924427\nSubset 2 accuracy= 0.8659003831417624\nSubset 2 f1 score= 0.8645566685934465\nSubset 3 accuracy= 0.8155446086480569\nSubset 3 f1 score= 0.8168406751779548\nSubset 4 accuracy= 0.8264915161466886\nSubset 4 f1 score= 0.8237275186685532\nSubset 5 accuracy= 0.8325123152709359\nSubset 5 f1 score= 0.8268868425748483\n\n\nAverage Accuracy= 0.8428024083196497\nAverage F1 Score= 0.8414643035214491\n\n\nConfusion Matrix:\n [[498 49 0 32 0]\n [ 47 551 38 0 0]\n [ 0 30 384 0 4]\n [101 0 0 73 0]\n [ 0 0 5 0 15]]\n\n\n\nNot balanced\n=================\nParameter: {'criterion': 'gini', 'min_samples_leaf': 1, 'min_samples_split': 4, 'n_estimators': 30, 'n_jobs': -1, 'random_state': 123} \n\nHasil dari model Random Forest:\n\nSubset 1 accuracy= 0.879584017515052\nSubset 1 f1 score= 0.8816825036368711\nSubset 2 accuracy= 0.8642583470169677\nSubset 2 f1 score= 0.8631651414424691\nSubset 3 accuracy= 0.8139025725232621\nSubset 3 f1 score= 0.8153814589540481\nSubset 4 accuracy= 0.8210180623973727\nSubset 4 f1 score= 0.818312488177958\nSubset 5 accuracy= 0.8385331143951834\nSubset 5 f1 score= 0.835018355566771\n\n\nAverage Accuracy= 0.8434592227695676\nAverage F1 Score= 0.8427119895556234\n\n\nConfusion Matrix:\n [[498 46 0 35 0]\n [ 46 552 37 1 0]\n [ 0 34 381 0 3]\n [ 88 0 0 86 0]\n [ 0 0 5 0 15]]\n\n\n\n"
],
[
"def kfoldrfclass_re(x,y,n):\n for i in range(len(model_rf)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_rf[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Balanced')\n print('=================')\n print('Parameter:',rf_parameters[i],'\\n')\n print('Hasil dari model Random Forest:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n sm=SMOTE(random_state=123)\n X_train_re,Y_train_re=sm.fit_resample(X_train,Y_train)\n \n model.fit(X_train_re,Y_train_re)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfoldrfclass_re(X,Y,5)",
"Balanced\n=================\nParameter: {'criterion': 'entropy', 'min_samples_leaf': 3, 'min_samples_split': 3, 'n_estimators': 15, 'n_jobs': -1, 'random_state': 123} \n\nHasil dari model Random Forest:\n\nSubset 1 accuracy= 0.8768472906403941\nsubset 1 f1 score= 0.8790413738831654\nSubset 2 accuracy= 0.8631636562671046\nsubset 2 f1 score= 0.8644758568405776\nSubset 3 accuracy= 0.8051450465243569\nsubset 3 f1 score= 0.810180599380624\nSubset 4 accuracy= 0.819376026272578\nsubset 4 f1 score= 0.821408228913684\nSubset 5 accuracy= 0.8352490421455939\nsubset 5 f1 score= 0.8354784985815905\n\n\nAverage Accuracy= 0.8399562123700056\nAverage F1 Score= 0.8421169115199284\n\n\nConfusion Matrix:\n [[483 40 0 56 0]\n [ 57 542 36 1 0]\n [ 0 35 373 0 10]\n [ 64 0 0 110 0]\n [ 0 0 2 0 18]]\n\n\n\nBalanced\n=================\nParameter: {'criterion': 'gini', 'min_samples_leaf': 1, 'min_samples_split': 4, 'n_estimators': 30, 'n_jobs': -1, 'random_state': 123} \n\nHasil dari model Random Forest:\n\nSubset 1 accuracy= 0.8762999452654625\nsubset 1 f1 score= 0.8787133176986556\nSubset 2 accuracy= 0.8653530377668309\nsubset 2 f1 score= 0.8663549391845903\nSubset 3 accuracy= 0.8111658456486043\nsubset 3 f1 score= 0.8151253467269255\nSubset 4 accuracy= 0.8199233716475096\nsubset 4 f1 score= 0.8203500090994248\nSubset 5 accuracy= 0.8264915161466886\nsubset 5 f1 score= 0.8258538851417931\n\n\nAverage Accuracy= 0.8398467432950193\nAverage F1 Score= 0.8412794995702779\n\n\nConfusion Matrix:\n [[481 44 0 54 0]\n [ 59 546 30 1 0]\n [ 0 41 368 0 9]\n [ 77 0 0 97 0]\n [ 0 0 2 0 18]]\n\n\n\n"
]
],
[
[
"## Naive Bayes",
"_____no_output_____"
],
[
"### Training dan Testing",
"_____no_output_____"
],
[
"#### Find the Best Parameter for Naive Bayes Model",
"_____no_output_____"
]
],
[
[
"paramnaive={\n 'priors':[None],\n 'var_smoothing':[1e-09,1e-05,2e-09,3e-09,5e-09]\n}",
"_____no_output_____"
],
[
"naive_parameters=[]",
"_____no_output_____"
],
[
"modelnaive=GaussianNB()",
"_____no_output_____"
],
[
"modelnaive_best=GridSearchCV(modelnaive,param_grid=paramnaive,n_jobs=-1)\nmodelnaive_best.fit(x_train,y_train)\nnaive_parameters.append(modelnaive_best.best_params_)\nnaive_parameters[0]",
"_____no_output_____"
],
[
"modelnaive_re_best=GridSearchCV(modelnaive,param_grid=paramnaive,n_jobs=-1)\nmodelnaive_re_best.fit(x_train_re,y_train_re)\nnaive_parameters.append(modelnaive_re_best.best_params_)\nnaive_parameters[1]",
"_____no_output_____"
],
[
"def modelnaivebayes(prior,var_smooth):\n model=GaussianNB(priors=prior,var_smoothing=var_smooth)\n return model",
"_____no_output_____"
],
[
"model_naive=[]",
"_____no_output_____"
],
[
"model_naive.append(modelnaivebayes(None,1e-09))",
"_____no_output_____"
],
[
"train=model_naive[0].fit(x_train,y_train)\npredicted=train.predict(x_test)\nprint(\"Naive Bayes (Before Balancing) :\\n\", classification_report(y_test,predicted))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted))",
"Naive Bayes (Before Balancing) :\n precision recall f1-score support\n\n Not Popular 0.54 0.99 0.70 419\n Ordinary 0.72 0.57 0.63 594\n Popular 0.95 0.77 0.85 630\nVery Not Popular 0.00 0.00 0.00 106\n Very popular 0.82 0.86 0.84 78\n\n accuracy 0.71 1827\n macro avg 0.61 0.64 0.60 1827\n weighted avg 0.72 0.71 0.69 1827\n\n\n\nConfusion Matrix:\n\n[[415 1 0 3 0]\n [242 336 16 0 0]\n [ 1 131 483 0 15]\n [106 0 0 0 0]\n [ 0 1 10 0 67]]\n"
],
[
"train_re=model_naive[0].fit(x_train_re,y_train_re)\npredicted_re=train_re.predict(x_test)\nprint(\"Naive Bayes (After Balancing) :\\n\", classification_report(y_test,predicted_re))\nprint(\"\\n\")\nprint(\"Confusion Matrix:\\n\")\nprint(confusion_matrix(y_test,predicted_re))",
"Naive Bayes (After Balancing) :\n precision recall f1-score support\n\n Not Popular 0.12 0.07 0.09 419\n Ordinary 0.68 0.48 0.56 594\n Popular 0.96 0.75 0.84 630\nVery Not Popular 0.18 1.00 0.31 106\n Very popular 0.75 0.90 0.82 78\n\n accuracy 0.53 1827\n macro avg 0.54 0.64 0.52 1827\n weighted avg 0.62 0.53 0.55 1827\n\n\n\nConfusion Matrix:\n\n[[ 29 1 0 389 0]\n [217 284 15 78 0]\n [ 2 131 474 0 23]\n [ 0 0 0 106 0]\n [ 0 1 7 0 70]]\n"
]
],
[
[
"### KFold",
"_____no_output_____"
]
],
[
[
"def kfoldnaiveclass(x,y,n):\n for i in range(len(model_naive)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_naive[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Not Balanced')\n print('=================')\n print('Parameter:',naive_parameters[i],'\\n')\n print('Hasil dari model Naive Bayes:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n model.fit(X_train,Y_train)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('Subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfoldnaiveclass(X,Y,5)",
"Not Balanced\n=================\nParameter: {'priors': None, 'var_smoothing': 1e-09} \n\nHasil dari model Naive Bayes:\n\nSubset 1 accuracy= 0.7536945812807881\nSubset 1 f1 score= 0.7637672843177205\nSubset 2 accuracy= 0.7263273125342091\nSubset 2 f1 score= 0.7311088602873101\nSubset 3 accuracy= 0.6978653530377669\nSubset 3 f1 score= 0.6690208948165416\nSubset 4 accuracy= 0.6995073891625616\nSubset 4 f1 score= 0.666383763538018\nSubset 5 accuracy= 0.6694033935413246\nSubset 5 f1 score= 0.6335658755670062\n\n\nAverage Accuracy= 0.7093596059113301\nAverage F1 Score= 0.6927693357053192\n\n\nConfusion Matrix:\n [[579 0 0 0 0]\n [296 333 7 0 0]\n [ 0 111 299 0 8]\n [174 0 0 0 0]\n [ 0 0 8 0 12]]\n\n\n\n"
],
[
"def kfoldnaiveclass_re(x,y,n):\n for i in range(len(model_naive)):\n kf=KFold(n_splits=n)\n kf.get_n_splits(x)\n model=model_naive[i]\n j=0\n accuracy=[]\n f1_scores=[]\n print('Balanced')\n print('=================')\n print('Parameter:',naive_parameters[i],'\\n')\n print('Hasil dari model Naive Bayes:\\n')\n for train_index,test_index in kf.split(x):\n X_train,X_test=x[train_index],x[test_index]\n Y_train,Y_test=y[train_index],y[test_index]\n \n sm=SMOTE(random_state=123)\n X_train_re,Y_train_re=sm.fit_resample(X_train,Y_train)\n \n model.fit(X_train_re,Y_train_re)\n Y_pred=model.predict(X_test)\n \n accuracy.append(accuracy_score(Y_test,Y_pred))\n f1_scores.append(f1_score(Y_test,Y_pred,average='weighted'))\n print('Subset',j+1,'accuracy= ',accuracy[j])\n print('subset',j+1,'f1 score=',f1_scores[j])\n j+=1\n \n print('\\n')\n print('Average Accuracy= ',np.mean(accuracy))\n print('Average F1 Score= ',np.mean(f1_scores))\n print('\\n')\n print('Confusion Matrix:\\n',confusion_matrix(Y_test,Y_pred))\n print('\\n\\n')",
"_____no_output_____"
],
[
"kfoldnaiveclass_re(X,Y,5)",
"Balanced\n=================\nParameter: {'priors': None, 'var_smoothing': 1e-09} \n\nHasil dari model Naive Bayes:\n\nSubset 1 accuracy= 0.6869184455391352\nsubset 1 f1 score= 0.7191083861697086\nSubset 2 accuracy= 0.6595511767925561\nsubset 2 f1 score= 0.6882823928082262\nSubset 3 accuracy= 0.4428024083196497\nsubset 3 f1 score= 0.46366778621916377\nSubset 4 accuracy= 0.4482758620689655\nsubset 4 f1 score= 0.46030406857878237\nSubset 5 accuracy= 0.4318555008210181\nsubset 5 f1 score= 0.4346993033788781\n\n\nAverage Accuracy= 0.5338806787082649\nAverage F1 Score= 0.5532123874309518\n\n\nConfusion Matrix:\n [[ 23 0 0 556 0]\n [234 283 7 112 0]\n [ 0 111 296 0 11]\n [ 2 0 0 172 0]\n [ 0 0 5 0 15]]\n\n\n\n"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5fca40d75da0ec9ba432b3be807b09e9365a07 | 11,387 | ipynb | Jupyter Notebook | notebooks/BertEmbedding.ipynb | CS-savvy/Essay-scoring | eaad7c974e59e3eb75669d2a2a64b1ce16cfbeb5 | [
"MIT"
]
| null | null | null | notebooks/BertEmbedding.ipynb | CS-savvy/Essay-scoring | eaad7c974e59e3eb75669d2a2a64b1ce16cfbeb5 | [
"MIT"
]
| null | null | null | notebooks/BertEmbedding.ipynb | CS-savvy/Essay-scoring | eaad7c974e59e3eb75669d2a2a64b1ce16cfbeb5 | [
"MIT"
]
| null | null | null | 26.983412 | 402 | 0.536752 | [
[
[
"import torch\nimport numpy as np\nfrom transformers import BertTokenizer, BertModel\nimport logging\nlogging.basicConfig(level=logging.INFO)\n\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"# Load pre-trained model tokenizer (vocabulary)\ntokenizer = BertTokenizer.from_pretrained('bert-base-uncased')",
"_____no_output_____"
],
[
"text = \"Here is the sentence I want embeddings for.\"\nmarked_text = \"[CLS] \" + text + \" [SEP]\"",
"_____no_output_____"
],
[
"tokenized_text = tokenizer.tokenize(marked_text)",
"_____no_output_____"
],
[
"print (tokenized_text)",
"['[CLS]', 'here', 'is', 'the', 'sentence', 'i', 'want', 'em', '##bed', '##ding', '##s', 'for', '.', '[SEP]']\n"
],
[
"tokens = tokenizer.encode_plus(text, add_special_tokens=True)\ntokens",
"_____no_output_____"
],
[
"len(list(tokenizer.vocab))",
"_____no_output_____"
],
[
"model = BertModel.from_pretrained('bert-base-uncased', output_hidden_states = True,)",
"Some weights of the model checkpoint at bert-base-uncased were not used when initializing BertModel: ['cls.predictions.transform.dense.bias', 'cls.predictions.transform.LayerNorm.weight', 'cls.seq_relationship.bias', 'cls.seq_relationship.weight', 'cls.predictions.decoder.weight', 'cls.predictions.bias', 'cls.predictions.transform.dense.weight', 'cls.predictions.transform.LayerNorm.bias']\n- This IS expected if you are initializing BertModel from the checkpoint of a model trained on another task or with another architecture (e.g. initializing a BertForSequenceClassification model from a BertForPreTraining model).\n- This IS NOT expected if you are initializing BertModel from the checkpoint of a model that you expect to be exactly identical (initializing a BertForSequenceClassification model from a BertForSequenceClassification model).\n"
],
[
"def bert_text_preparation(text, tokenizer):\n \"\"\"Preparing the input for BERT\n \n Takes a string argument and performs\n pre-processing like adding special tokens,\n tokenization, tokens to ids, and tokens to\n segment ids. All tokens are mapped to seg-\n ment id = 1.\n \n Args:\n text (str): Text to be converted\n tokenizer (obj): Tokenizer object\n to convert text into BERT-re-\n adable tokens and ids\n \n Returns:\n list: List of BERT-readable tokens\n obj: Torch tensor with token ids\n obj: Torch tensor segment ids\n \n \n \"\"\"\n marked_text = \"[CLS] \" + text + \" [SEP]\"\n tokenized_text = tokenizer.tokenize(marked_text)\n indexed_tokens = tokenizer.convert_tokens_to_ids(tokenized_text)\n segments_ids = [1]*len(indexed_tokens)\n\n # Convert inputs to PyTorch tensors\n tokens_tensor = torch.tensor([indexed_tokens])\n segments_tensors = torch.tensor([segments_ids])\n\n return tokenized_text, tokens_tensor, segments_tensors",
"_____no_output_____"
],
[
"def get_bert_embeddings(tokens_tensor, segments_tensors, model):\n \"\"\"Get embeddings from an embedding model\n \n Args:\n tokens_tensor (obj): Torch tensor size [n_tokens]\n with token ids for each token in text\n segments_tensors (obj): Torch tensor size [n_tokens]\n with segment ids for each token in text\n model (obj): Embedding model to generate embeddings\n from token and segment ids\n \n Returns:\n list: List of list of floats of size\n [n_tokens, n_embedding_dimensions]\n containing embeddings for each token\n \n \"\"\"\n \n # Gradient calculation id disabled\n # Model is in inference mode\n with torch.no_grad():\n outputs = model(tokens_tensor, segments_tensors)\n # Removing the first hidden state\n # The first state is the input state\n hidden_states = outputs[2][1:]\n\n # Getting embeddings from the final BERT layer\n token_embeddings = hidden_states[-1]\n # Collapsing the tensor into 1-dimension\n token_embeddings = torch.squeeze(token_embeddings, dim=0)\n # Converting torchtensors to lists\n list_token_embeddings = [token_embed.tolist() for token_embed in token_embeddings]\n\n return list_token_embeddings",
"_____no_output_____"
],
[
"texts = [\"Ram and Shayam are playing.\"]\n \n # \"The bank vault was robust.\",\n # \"He had to bank on her for support.\",\n # \"The bank was out of money.\",\n # \"The bank teller was a man.\"]",
"_____no_output_____"
],
[
"target_word_embeddings = []\nfor text in texts:\n tokenized_text, tokens_tensor, segments_tensors = bert_text_preparation(text, tokenizer)\n list_token_embeddings = get_bert_embeddings(tokens_tensor, segments_tensors, model)\n \n # Find the position 'bank' in list of tokens\n # word_index = tokenized_text.index('bank')\n # Get the embedding for bank\n # word_embedding = list_token_embeddings[word_index]\n break",
"_____no_output_____"
],
[
"len(list_token_embeddings), tokenized_text",
"_____no_output_____"
],
[
"len(list_token_embeddings[0])",
"_____no_output_____"
],
[
"list_token_embeddings[1][:10] #ram",
"_____no_output_____"
],
[
"list_token_embeddings[3][:10] #shay",
"_____no_output_____"
],
[
"list_token_embeddings[4][:10] #shay",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec5fd6bb311d6250fa513a1aaf8d8d33413a7fbe | 188,202 | ipynb | Jupyter Notebook | 18. dcgan-svhn/DCGAN_Solution.ipynb | josancamon19/DeepLearningNanodegree | 35a2bdfe874a8e4ff301af1ed365b1ed3272dff1 | [
"MIT"
]
| null | null | null | 18. dcgan-svhn/DCGAN_Solution.ipynb | josancamon19/DeepLearningNanodegree | 35a2bdfe874a8e4ff301af1ed365b1ed3272dff1 | [
"MIT"
]
| null | null | null | 18. dcgan-svhn/DCGAN_Solution.ipynb | josancamon19/DeepLearningNanodegree | 35a2bdfe874a8e4ff301af1ed365b1ed3272dff1 | [
"MIT"
]
| null | null | null | 158.686341 | 81,568 | 0.84065 | [
[
[
"# Deep Convolutional GANs\n\nIn this notebook, you'll build a GAN using convolutional layers in the generator and discriminator. This is called a Deep Convolutional GAN, or DCGAN for short. The DCGAN architecture was first explored in 2016 and has seen impressive results in generating new images; you can read the [original paper, here](https://arxiv.org/pdf/1511.06434.pdf).\n\nYou'll be training DCGAN on the [Street View House Numbers](http://ufldl.stanford.edu/housenumbers/) (SVHN) dataset. These are color images of house numbers collected from Google street view. SVHN images are in color and much more variable than MNIST. \n\n<img src='assets/svhn_dcgan.png' width=80% />\n\nSo, our goal is to create a DCGAN that can generate new, realistic-looking images of house numbers. We'll go through the following steps to do this:\n* Load in and pre-process the house numbers dataset\n* Define discriminator and generator networks\n* Train these adversarial networks\n* Visualize the loss over time and some sample, generated images\n\n#### Deeper Convolutional Networks\n\nSince this dataset is more complex than our MNIST data, we'll need a deeper network to accurately identify patterns in these images and be able to generate new ones. Specifically, we'll use a series of convolutional or transpose convolutional layers in the discriminator and generator. It's also necessary to use batch normalization to get these convolutional networks to train. \n\nBesides these changes in network structure, training the discriminator and generator networks should be the same as before. That is, the discriminator will alternate training on real and fake (generated) images, and the generator will aim to trick the discriminator into thinking that its generated images are real!",
"_____no_output_____"
]
],
[
[
"# import libraries\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pickle as pkl\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Getting the data\n\nHere you can download the SVHN dataset. It's a dataset built-in to the PyTorch datasets library. We can load in training data, transform it into Tensor datatypes, then create dataloaders to batch our data into a desired size.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision import datasets\nfrom torchvision import transforms\n\n# Tensor transform\ntransform = transforms.ToTensor()\n\n# SVHN training datasets\nsvhn_train = datasets.SVHN(root='data/', split='train', download=True, transform=transform)\n\nbatch_size = 128\nnum_workers = 0\n\n# build DataLoaders for SVHN dataset\ntrain_loader = torch.utils.data.DataLoader(dataset=svhn_train,\n batch_size=batch_size,\n shuffle=True,\n num_workers=num_workers)\n",
"Using downloaded and verified file: data/train_32x32.mat\n"
]
],
[
[
"### Visualize the Data\n\nHere I'm showing a small sample of the images. Each of these is 32x32 with 3 color channels (RGB). These are the real, training images that we'll pass to the discriminator. Notice that each image has _one_ associated, numerical label.",
"_____no_output_____"
]
],
[
[
"# obtain one batch of training images\ndataiter = iter(train_loader)\nimages, labels = dataiter.next()\n\n# plot the images in the batch, along with the corresponding labels\nfig = plt.figure(figsize=(25, 4))\nplot_size=20\nfor idx in np.arange(plot_size):\n ax = fig.add_subplot(2, plot_size/2, idx+1, xticks=[], yticks=[])\n ax.imshow(np.transpose(images[idx], (1, 2, 0)))\n # print out the correct label for each image\n # .item() gets the value contained in a Tensor\n ax.set_title(str(labels[idx].item()))",
"_____no_output_____"
]
],
[
[
"### Pre-processing: scaling from -1 to 1\n\nWe need to do a bit of pre-processing; we know that the output of our `tanh` activated generator will contain pixel values in a range from -1 to 1, and so, we need to rescale our training images to a range of -1 to 1. (Right now, they are in a range from 0-1.)",
"_____no_output_____"
]
],
[
[
"# current range\nimg = images[0]\n\nprint('Min: ', img.min())\nprint('Max: ', img.max())",
"Min: tensor(0.3255)\nMax: tensor(0.7882)\n"
],
[
"# helper scale function\ndef scale(x, feature_range=(-1, 1)):\n ''' Scale takes in an image x and returns that image, scaled\n with a feature_range of pixel values from -1 to 1. \n This function assumes that the input x is already scaled from 0-1.'''\n # assume x is scaled to (0, 1)\n # scale to feature_range and return scaled x\n min, max = feature_range\n x = x * (max - min) + min\n return x\n",
"_____no_output_____"
],
[
"# scaled range\nscaled_img = scale(img)\n\nprint('Scaled min: ', scaled_img.min())\nprint('Scaled max: ', scaled_img.max())",
"Scaled min: tensor(-0.3490)\nScaled max: tensor(0.5765)\n"
]
],
[
[
"---\n# Define the Model\n\nA GAN is comprised of two adversarial networks, a discriminator and a generator.",
"_____no_output_____"
],
[
"## Discriminator\n\nHere you'll build the discriminator. This is a convolutional classifier like you've built before, only without any maxpooling layers. \n* The inputs to the discriminator are 32x32x3 tensor images\n* You'll want a few convolutional, hidden layers\n* Then a fully connected layer for the output; as before, we want a sigmoid output, but we'll add that in the loss function, [BCEWithLogitsLoss](https://pytorch.org/docs/stable/nn.html#bcewithlogitsloss), later\n\n<img src='assets/conv_discriminator.png' width=80%/>\n\nFor the depths of the convolutional layers I suggest starting with 32 filters in the first layer, then double that depth as you add layers (to 64, 128, etc.). Note that in the DCGAN paper, they did all the downsampling using only strided convolutional layers with no maxpooling layers.\n\nYou'll also want to use batch normalization with [nn.BatchNorm2d](https://pytorch.org/docs/stable/nn.html#batchnorm2d) on each layer **except** the first convolutional layer and final, linear output layer. \n\n#### Helper `conv` function \n\nIn general, each layer should look something like convolution > batch norm > leaky ReLU, and so we'll define a function to put these layers together. This function will create a sequential series of a convolutional + an optional batch norm layer. We'll create these using PyTorch's [Sequential container](https://pytorch.org/docs/stable/nn.html#sequential), which takes in a list of layers and creates layers according to the order that they are passed in to the Sequential constructor.\n\nNote: It is also suggested that you use a **kernel_size of 4** and a **stride of 2** for strided convolutions.",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn\nimport torch.nn.functional as F\n\n# helper conv function\ndef conv(in_channels, out_channels, kernel_size, stride=2, padding=1, batch_norm=True):\n \"\"\"Creates a convolutional layer, with optional batch normalization.\n \"\"\"\n layers = []\n conv_layer = nn.Conv2d(in_channels, out_channels, \n kernel_size, stride, padding, bias=False)\n \n # append conv layer\n layers.append(conv_layer)\n\n if batch_norm:\n # append batchnorm layer\n layers.append(nn.BatchNorm2d(out_channels))\n \n # using Sequential container\n return nn.Sequential(*layers)\n",
"_____no_output_____"
],
[
"class Discriminator(nn.Module):\n\n def __init__(self, conv_dim=32):\n super(Discriminator, self).__init__()\n\n # complete init function\n self.conv_dim = conv_dim\n\n # 32x32 input\n self.conv1 = conv(3, conv_dim, 4, batch_norm=False) # first layer, no batch_norm\n # 16x16 out\n self.conv2 = conv(conv_dim, conv_dim*2, 4)\n # 8x8 out\n self.conv3 = conv(conv_dim*2, conv_dim*4, 4)\n # 4x4 out\n \n # final, fully-connected layer\n self.fc = nn.Linear(conv_dim*4*4*4, 1)\n\n def forward(self, x):\n # all hidden layers + leaky relu activation\n out = F.leaky_relu(self.conv1(x), 0.2)\n out = F.leaky_relu(self.conv2(out), 0.2)\n out = F.leaky_relu(self.conv3(out), 0.2)\n \n # flatten\n out = out.view(-1, self.conv_dim*4*4*4)\n \n # final output layer\n out = self.fc(out) \n return out\n ",
"_____no_output_____"
]
],
[
[
"## Generator\n\nNext, you'll build the generator network. The input will be our noise vector `z`, as before. And, the output will be a $tanh$ output, but this time with size 32x32 which is the size of our SVHN images.\n\n<img src='assets/conv_generator.png' width=80% />\n\nWhat's new here is we'll use transpose convolutional layers to create our new images. \n* The first layer is a fully connected layer which is reshaped into a deep and narrow layer, something like 4x4x512. \n* Then, we use batch normalization and a leaky ReLU activation. \n* Next is a series of [transpose convolutional layers](https://pytorch.org/docs/stable/nn.html#convtranspose2d), where you typically halve the depth and double the width and height of the previous layer. \n* And, we'll apply batch normalization and ReLU to all but the last of these hidden layers. Where we will just apply a `tanh` activation.\n\n#### Helper `deconv` function\n\nFor each of these layers, the general scheme is transpose convolution > batch norm > ReLU, and so we'll define a function to put these layers together. This function will create a sequential series of a transpose convolutional + an optional batch norm layer. We'll create these using PyTorch's Sequential container, which takes in a list of layers and creates layers according to the order that they are passed in to the Sequential constructor.\n\nNote: It is also suggested that you use a **kernel_size of 4** and a **stride of 2** for transpose convolutions.",
"_____no_output_____"
]
],
[
[
"# helper deconv function\ndef deconv(in_channels, out_channels, kernel_size, stride=2, padding=1, batch_norm=True):\n \"\"\"Creates a transposed-convolutional layer, with optional batch normalization.\n \"\"\"\n # create a sequence of transpose + optional batch norm layers\n layers = []\n transpose_conv_layer = nn.ConvTranspose2d(in_channels, out_channels, \n kernel_size, stride, padding, bias=False)\n # append transpose convolutional layer\n layers.append(transpose_conv_layer)\n \n if batch_norm:\n # append batchnorm layer\n layers.append(nn.BatchNorm2d(out_channels))\n \n return nn.Sequential(*layers)\n",
"_____no_output_____"
],
[
"class Generator(nn.Module):\n \n def __init__(self, z_size, conv_dim=32):\n super(Generator, self).__init__()\n\n # complete init function\n \n self.conv_dim = conv_dim\n \n # first, fully-connected layer\n self.fc = nn.Linear(z_size, conv_dim*4*4*4)\n\n # transpose conv layers\n self.t_conv1 = deconv(conv_dim*4, conv_dim*2, 4)\n self.t_conv2 = deconv(conv_dim*2, conv_dim, 4)\n self.t_conv3 = deconv(conv_dim, 3, 4, batch_norm=False)\n \n\n def forward(self, x):\n # fully-connected + reshape \n out = self.fc(x)\n out = out.view(-1, self.conv_dim*4, 4, 4) # (batch_size, depth, 4, 4)\n \n # hidden transpose conv layers + relu\n out = F.relu(self.t_conv1(out))\n out = F.relu(self.t_conv2(out))\n \n # last layer + tanh activation\n out = self.t_conv3(out)\n out = F.tanh(out)\n \n return out\n ",
"_____no_output_____"
]
],
[
[
"## Build complete network\n\nDefine your models' hyperparameters and instantiate the discriminator and generator from the classes defined above. Make sure you've passed in the correct input arguments.",
"_____no_output_____"
]
],
[
[
"# define hyperparams\nconv_dim = 32\nz_size = 100\n\n# define discriminator and generator\nD = Discriminator(conv_dim)\nG = Generator(z_size=z_size, conv_dim=conv_dim)\n\nprint(D)\nprint()\nprint(G)",
"Discriminator(\n (conv1): Sequential(\n (0): Conv2d(3, 32, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n (conv2): Sequential(\n (0): Conv2d(32, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv3): Sequential(\n (0): Conv2d(64, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (fc): Linear(in_features=2048, out_features=1, bias=True)\n)\n\nGenerator(\n (fc): Linear(in_features=100, out_features=2048, bias=True)\n (t_conv1): Sequential(\n (0): ConvTranspose2d(128, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (t_conv2): Sequential(\n (0): ConvTranspose2d(64, 32, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (t_conv3): Sequential(\n (0): ConvTranspose2d(32, 3, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n)\n"
]
],
[
[
"### Training on GPU\n\nCheck if you can train on GPU. If you can, set this as a variable and move your models to GPU. \n> Later, we'll also move any inputs our models and loss functions see (real_images, z, and ground truth labels) to GPU as well.",
"_____no_output_____"
]
],
[
[
"train_on_gpu = torch.cuda.is_available()\n\nif train_on_gpu:\n # move models to GPU\n G.cuda()\n D.cuda()\n print('GPU available for training. Models moved to GPU')\nelse:\n print('Training on CPU.')\n ",
"Training on CPU.\n"
]
],
[
[
"---\n## Discriminator and Generator Losses\n\nNow we need to calculate the losses. And this will be exactly the same as before.\n\n### Discriminator Losses\n\n> * For the discriminator, the total loss is the sum of the losses for real and fake images, `d_loss = d_real_loss + d_fake_loss`. \n* Remember that we want the discriminator to output 1 for real images and 0 for fake images, so we need to set up the losses to reflect that.\n\nThe losses will by binary cross entropy loss with logits, which we can get with [BCEWithLogitsLoss](https://pytorch.org/docs/stable/nn.html#bcewithlogitsloss). This combines a `sigmoid` activation function **and** and binary cross entropy loss in one function.\n\nFor the real images, we want `D(real_images) = 1`. That is, we want the discriminator to classify the the real images with a label = 1, indicating that these are real. The discriminator loss for the fake data is similar. We want `D(fake_images) = 0`, where the fake images are the _generator output_, `fake_images = G(z)`. \n\n### Generator Loss\n\nThe generator loss will look similar only with flipped labels. The generator's goal is to get `D(fake_images) = 1`. In this case, the labels are **flipped** to represent that the generator is trying to fool the discriminator into thinking that the images it generates (fakes) are real!",
"_____no_output_____"
]
],
[
[
"def real_loss(D_out, smooth=False):\n batch_size = D_out.size(0)\n # label smoothing\n if smooth:\n # smooth, real labels = 0.9\n labels = torch.ones(batch_size)*0.9\n else:\n labels = torch.ones(batch_size) # real labels = 1\n # move labels to GPU if available \n if train_on_gpu:\n labels = labels.cuda()\n # binary cross entropy with logits loss\n criterion = nn.BCEWithLogitsLoss()\n # calculate loss\n loss = criterion(D_out.squeeze(), labels)\n return loss\n\ndef fake_loss(D_out):\n batch_size = D_out.size(0)\n labels = torch.zeros(batch_size) # fake labels = 0\n if train_on_gpu:\n labels = labels.cuda()\n criterion = nn.BCEWithLogitsLoss()\n # calculate loss\n loss = criterion(D_out.squeeze(), labels)\n return loss",
"_____no_output_____"
]
],
[
[
"## Optimizers\n\nNot much new here, but notice how I am using a small learning rate and custom parameters for the Adam optimizers, This is based on some research into DCGAN model convergence.\n\n### Hyperparameters\n\nGANs are very sensitive to hyperparameters. A lot of experimentation goes into finding the best hyperparameters such that the generator and discriminator don't overpower each other. Try out your own hyperparameters or read [the DCGAN paper](https://arxiv.org/pdf/1511.06434.pdf) to see what worked for them.",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\n\n# params\nlr = 0.0002\nbeta1=0.5\nbeta2=0.999 # default value\n\n# Create optimizers for the discriminator and generator\nd_optimizer = optim.Adam(D.parameters(), lr, [beta1, beta2])\ng_optimizer = optim.Adam(G.parameters(), lr, [beta1, beta2])",
"_____no_output_____"
]
],
[
[
"---\n## Training\n\nTraining will involve alternating between training the discriminator and the generator. We'll use our functions `real_loss` and `fake_loss` to help us calculate the discriminator losses in all of the following cases.\n\n### Discriminator training\n1. Compute the discriminator loss on real, training images \n2. Generate fake images\n3. Compute the discriminator loss on fake, generated images \n4. Add up real and fake loss\n5. Perform backpropagation + an optimization step to update the discriminator's weights\n\n### Generator training\n1. Generate fake images\n2. Compute the discriminator loss on fake images, using **flipped** labels!\n3. Perform backpropagation + an optimization step to update the generator's weights\n\n#### Saving Samples\n\nAs we train, we'll also print out some loss statistics and save some generated \"fake\" samples.\n\n**Evaluation mode**\n\nNotice that, when we call our generator to create the samples to display, we set our model to evaluation mode: `G.eval()`. That's so the batch normalization layers will use the population statistics rather than the batch statistics (as they do during training), *and* so dropout layers will operate in eval() mode; not turning off any nodes for generating samples.",
"_____no_output_____"
]
],
[
[
"import pickle as pkl\n\n# training hyperparams\nnum_epochs = 50\n\n# keep track of loss and generated, \"fake\" samples\nsamples = []\nlosses = []\n\nprint_every = 300\n\n# Get some fixed data for sampling. These are images that are held\n# constant throughout training, and allow us to inspect the model's performance\nsample_size=16\nfixed_z = np.random.uniform(-1, 1, size=(sample_size, z_size))\nfixed_z = torch.from_numpy(fixed_z).float()\n\n# train the network\nfor epoch in range(num_epochs):\n \n for batch_i, (real_images, _) in enumerate(train_loader):\n \n batch_size = real_images.size(0)\n \n # important rescaling step\n real_images = scale(real_images)\n \n # ============================================\n # TRAIN THE DISCRIMINATOR\n # ============================================\n \n d_optimizer.zero_grad()\n \n # 1. Train with real images\n if train_on_gpu:\n real_images = real_images.cuda()\n D_real = D(real_images) \n \n # Compute the discriminator losses on real images \n d_real_loss = real_loss(D_real)\n \n # 2. Train with fake images\n \n # Generate fake images\n z = np.random.uniform(-1, 1, size=(batch_size, z_size))\n z = torch.from_numpy(z).float()\n # move x to GPU, if available\n if train_on_gpu:\n z = z.cuda()\n fake_images = G(z)\n \n # Compute the discriminator losses on fake images \n D_fake = D(fake_images)\n d_fake_loss = fake_loss(D_fake)\n \n # add up loss and perform backprop\n d_loss = d_real_loss + d_fake_loss\n d_loss.backward()\n d_optimizer.step()\n \n \n # =========================================\n # TRAIN THE GENERATOR\n # =========================================\n g_optimizer.zero_grad()\n \n # 1. Train with fake images and flipped labels\n \n # Generate fake images\n z = np.random.uniform(-1, 1, size=(batch_size, z_size))\n z = torch.from_numpy(z).float()\n if train_on_gpu:\n z = z.cuda()\n fake_images = G(z)\n \n # Compute the discriminator losses on fake images \n # using flipped labels!\n D_fake = D(fake_images)\n g_loss = real_loss(D_fake) # use real loss to flip labels\n \n # perform backprop\n g_loss.backward()\n g_optimizer.step()\n\n # Print some loss stats\n if batch_i % print_every == 0:\n # append discriminator loss and generator loss\n losses.append((d_loss.item(), g_loss.item()))\n # print discriminator and generator loss\n print('Epoch [{:5d}/{:5d}] | d_loss: {:6.4f} | g_loss: {:6.4f}'.format(\n epoch+1, num_epochs, d_loss.item(), g_loss.item()))\n\n \n ## AFTER EACH EPOCH## \n # generate and save sample, fake images\n G.eval() # for generating samples\n if train_on_gpu:\n fixed_z = fixed_z.cuda()\n samples_z = G(fixed_z)\n samples.append(samples_z)\n G.train() # back to training mode\n\n\n# Save training generator samples\nwith open('train_samples.pkl', 'wb') as f:\n pkl.dump(samples, f)",
"_____no_output_____"
],
[
"# set number of epochs \nn_epochs = 5\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\n# call training function\nlosses = train(D, G, n_epochs=n_epochs)",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\torch\\nn\\functional.py:1320: UserWarning: nn.functional.tanh is deprecated. Use torch.tanh instead.\n warnings.warn(\"nn.functional.tanh is deprecated. Use torch.tanh instead.\")\n"
]
],
[
[
"## Training loss\n\nHere we'll plot the training losses for the generator and discriminator, recorded after each epoch.",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\nlosses = np.array(losses)\nplt.plot(losses.T[0], label='Discriminator', alpha=0.5)\nplt.plot(losses.T[1], label='Generator', alpha=0.5)\nplt.title(\"Training Losses\")\nplt.legend()",
"_____no_output_____"
]
],
[
[
"## Generator samples from training\n\nHere we can view samples of images from the generator. We'll look at the images we saved during training.",
"_____no_output_____"
]
],
[
[
"# helper function for viewing a list of passed in sample images\ndef view_samples(epoch, samples):\n fig, axes = plt.subplots(figsize=(16,4), nrows=2, ncols=8, sharey=True, sharex=True)\n for ax, img in zip(axes.flatten(), samples[epoch]):\n img = img.detach().cpu().numpy()\n img = np.transpose(img, (1, 2, 0))\n img = ((img +1)*255 / (2)).astype(np.uint8) # rescale to pixel range (0-255)\n ax.xaxis.set_visible(False)\n ax.yaxis.set_visible(False)\n im = ax.imshow(img.reshape((32,32,3)))",
"_____no_output_____"
],
[
"_ = view_samples(-1, samples)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec5fd859866a3f2ef2a59a91e811f27fd33ebc3a | 271,631 | ipynb | Jupyter Notebook | 2 Coursera - Improving Deep Neural Networks - Hyperparameter Tuning, Regularization and Optimization/Programming Assignments/Week 1 - Practical Aspects of Deep Learning/W1A2/Regularization.ipynb | ishaan-narula/coursera-deep-learning-specialisation | ee33cd3cb5b0b72beadd73d9ec51acd97f1398ff | [
"MIT"
]
| null | null | null | 2 Coursera - Improving Deep Neural Networks - Hyperparameter Tuning, Regularization and Optimization/Programming Assignments/Week 1 - Practical Aspects of Deep Learning/W1A2/Regularization.ipynb | ishaan-narula/coursera-deep-learning-specialisation | ee33cd3cb5b0b72beadd73d9ec51acd97f1398ff | [
"MIT"
]
| null | null | null | 2 Coursera - Improving Deep Neural Networks - Hyperparameter Tuning, Regularization and Optimization/Programming Assignments/Week 1 - Practical Aspects of Deep Learning/W1A2/Regularization.ipynb | ishaan-narula/coursera-deep-learning-specialisation | ee33cd3cb5b0b72beadd73d9ec51acd97f1398ff | [
"MIT"
]
| null | null | null | 216.439044 | 55,056 | 0.896816 | [
[
[
"# Regularization\n\nWelcome to the second assignment of this week. Deep Learning models have so much flexibility and capacity that **overfitting can be a serious problem**, if the training dataset is not big enough. Sure it does well on the training set, but the learned network **doesn't generalize to new examples** that it has never seen!\n\n**You will learn to:** Use regularization in your deep learning models.\n\nLet's get started!",
"_____no_output_____"
],
[
"## Table of Contents\n- [1 - Packages](#1)\n- [2 - Problem Statement](#2)\n- [3 - Loading the Dataset](#3)\n- [4 - Non-Regularized Model](#4)\n- [5 - L2 Regularization](#5)\n - [Exercise 1 - compute_cost_with_regularization](#ex-1)\n - [Exercise 2 - backward_propagation_with_regularization](#ex-2)\n- [6 - Dropout](#6)\n - [6.1 - Forward Propagation with Dropout](#6-1)\n - [Exercise 3 - forward_propagation_with_dropout](#ex-3)\n - [6.2 - Backward Propagation with Dropout](#6-2)\n - [Exercise 4 - backward_propagation_with_dropout](#ex-4)\n- [7 - Conclusions](#7)",
"_____no_output_____"
],
[
"<a name='1'></a>\n## 1 - Packages",
"_____no_output_____"
]
],
[
[
"# import packages\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport sklearn\nimport sklearn.datasets\nimport scipy.io\nfrom reg_utils import sigmoid, relu, plot_decision_boundary, initialize_parameters, load_2D_dataset, predict_dec\nfrom reg_utils import compute_cost, predict, forward_propagation, backward_propagation, update_parameters\nfrom testCases import *\nfrom public_tests import *\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (7.0, 4.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"<a name='2'></a>\n## 2 - Problem Statement",
"_____no_output_____"
],
[
"You have just been hired as an AI expert by the French Football Corporation. They would like you to recommend positions where France's goal keeper should kick the ball so that the French team's players can then hit it with their head. \n\n<img src=\"images/field_kiank.png\" style=\"width:600px;height:350px;\">\n\n<caption><center><font color='purple'><b>Figure 1</b>: Football field. The goal keeper kicks the ball in the air, the players of each team are fighting to hit the ball with their head </font></center></caption>\n\n\nThey give you the following 2D dataset from France's past 10 games.",
"_____no_output_____"
],
[
"<a name='3'></a>\n## 3 - Loading the Dataset",
"_____no_output_____"
]
],
[
[
"train_X, train_Y, test_X, test_Y = load_2D_dataset()",
"_____no_output_____"
]
],
[
[
"Each dot corresponds to a position on the football field where a football player has hit the ball with his/her head after the French goal keeper has shot the ball from the left side of the football field.\n- If the dot is blue, it means the French player managed to hit the ball with his/her head\n- If the dot is red, it means the other team's player hit the ball with their head\n\n**Your goal**: Use a deep learning model to find the positions on the field where the goalkeeper should kick the ball.",
"_____no_output_____"
],
[
"**Analysis of the dataset**: This dataset is a little noisy, but it looks like a diagonal line separating the upper left half (blue) from the lower right half (red) would work well. \n\nYou will first try a non-regularized model. Then you'll learn how to regularize it and decide which model you will choose to solve the French Football Corporation's problem. ",
"_____no_output_____"
],
[
"<a name='4'></a>\n## 4 - Non-Regularized Model\n\nYou will use the following neural network (already implemented for you below). This model can be used:\n- in *regularization mode* -- by setting the `lambd` input to a non-zero value. We use \"`lambd`\" instead of \"`lambda`\" because \"`lambda`\" is a reserved keyword in Python. \n- in *dropout mode* -- by setting the `keep_prob` to a value less than one\n\nYou will first try the model without any regularization. Then, you will implement:\n- *L2 regularization* -- functions: \"`compute_cost_with_regularization()`\" and \"`backward_propagation_with_regularization()`\"\n- *Dropout* -- functions: \"`forward_propagation_with_dropout()`\" and \"`backward_propagation_with_dropout()`\"\n\nIn each part, you will run this model with the correct inputs so that it calls the functions you've implemented. Take a look at the code below to familiarize yourself with the model.",
"_____no_output_____"
]
],
[
[
"def model(X, Y, learning_rate = 0.3, num_iterations = 30000, print_cost = True, lambd = 0, keep_prob = 1):\n \"\"\"\n Implements a three-layer neural network: LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID.\n \n Arguments:\n X -- input data, of shape (input size, number of examples)\n Y -- true \"label\" vector (1 for blue dot / 0 for red dot), of shape (output size, number of examples)\n learning_rate -- learning rate of the optimization\n num_iterations -- number of iterations of the optimization loop\n print_cost -- If True, print the cost every 10000 iterations\n lambd -- regularization hyperparameter, scalar\n keep_prob - probability of keeping a neuron active during drop-out, scalar.\n \n Returns:\n parameters -- parameters learned by the model. They can then be used to predict.\n \"\"\"\n \n grads = {}\n costs = [] # to keep track of the cost\n m = X.shape[1] # number of examples\n layers_dims = [X.shape[0], 20, 3, 1]\n \n # Initialize parameters dictionary.\n parameters = initialize_parameters(layers_dims)\n\n # Loop (gradient descent)\n\n for i in range(0, num_iterations):\n\n # Forward propagation: LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID.\n if keep_prob == 1:\n a3, cache = forward_propagation(X, parameters)\n elif keep_prob < 1:\n a3, cache = forward_propagation_with_dropout(X, parameters, keep_prob)\n \n # Cost function\n if lambd == 0:\n cost = compute_cost(a3, Y)\n else:\n cost = compute_cost_with_regularization(a3, Y, parameters, lambd)\n \n # Backward propagation.\n assert (lambd == 0 or keep_prob == 1) # it is possible to use both L2 regularization and dropout, \n # but this assignment will only explore one at a time\n if lambd == 0 and keep_prob == 1:\n grads = backward_propagation(X, Y, cache)\n elif lambd != 0:\n grads = backward_propagation_with_regularization(X, Y, cache, lambd)\n elif keep_prob < 1:\n grads = backward_propagation_with_dropout(X, Y, cache, keep_prob)\n \n # Update parameters.\n parameters = update_parameters(parameters, grads, learning_rate)\n \n # Print the loss every 10000 iterations\n if print_cost and i % 10000 == 0:\n print(\"Cost after iteration {}: {}\".format(i, cost))\n if print_cost and i % 1000 == 0:\n costs.append(cost)\n \n # plot the cost\n plt.plot(costs)\n plt.ylabel('cost')\n plt.xlabel('iterations (x1,000)')\n plt.title(\"Learning rate =\" + str(learning_rate))\n plt.show()\n \n return parameters",
"_____no_output_____"
]
],
[
[
"Let's train the model without any regularization, and observe the accuracy on the train/test sets.",
"_____no_output_____"
]
],
[
[
"parameters = model(train_X, train_Y)\nprint (\"On the training set:\")\npredictions_train = predict(train_X, train_Y, parameters)\nprint (\"On the test set:\")\npredictions_test = predict(test_X, test_Y, parameters)",
"Cost after iteration 0: 0.6557412523481002\nCost after iteration 10000: 0.16329987525724204\nCost after iteration 20000: 0.13851642423234922\n"
]
],
[
[
"The train accuracy is 94.8% while the test accuracy is 91.5%. This is the **baseline model** (you will observe the impact of regularization on this model). Run the following code to plot the decision boundary of your model.",
"_____no_output_____"
]
],
[
[
"plt.title(\"Model without regularization\")\naxes = plt.gca()\naxes.set_xlim([-0.75,0.40])\naxes.set_ylim([-0.75,0.65])\nplot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)",
"_____no_output_____"
]
],
[
[
"The non-regularized model is obviously overfitting the training set. It is fitting the noisy points! Lets now look at two techniques to reduce overfitting.",
"_____no_output_____"
],
[
"<a name='5'></a>\n## 5 - L2 Regularization\n\nThe standard way to avoid overfitting is called **L2 regularization**. It consists of appropriately modifying your cost function, from:\n$$J = -\\frac{1}{m} \\sum\\limits_{i = 1}^{m} \\large{(}\\small y^{(i)}\\log\\left(a^{[L](i)}\\right) + (1-y^{(i)})\\log\\left(1- a^{[L](i)}\\right) \\large{)} \\tag{1}$$\nTo:\n$$J_{regularized} = \\small \\underbrace{-\\frac{1}{m} \\sum\\limits_{i = 1}^{m} \\large{(}\\small y^{(i)}\\log\\left(a^{[L](i)}\\right) + (1-y^{(i)})\\log\\left(1- a^{[L](i)}\\right) \\large{)} }_\\text{cross-entropy cost} + \\underbrace{\\frac{1}{m} \\frac{\\lambda}{2} \\sum\\limits_l\\sum\\limits_k\\sum\\limits_j W_{k,j}^{[l]2} }_\\text{L2 regularization cost} \\tag{2}$$\n\nLet's modify your cost and observe the consequences.\n\n<a name='ex-1'></a>\n### Exercise 1 - compute_cost_with_regularization\nImplement `compute_cost_with_regularization()` which computes the cost given by formula (2). To calculate $\\sum\\limits_k\\sum\\limits_j W_{k,j}^{[l]2}$ , use :\n```python\nnp.sum(np.square(Wl))\n```\nNote that you have to do this for $W^{[1]}$, $W^{[2]}$ and $W^{[3]}$, then sum the three terms and multiply by $ \\frac{1}{m} \\frac{\\lambda}{2} $.",
"_____no_output_____"
]
],
[
[
"W1 = parameters[\"W1\"]\nW2 = parameters[\"W2\"]\nW3 = parameters[\"W3\"]\n(np.sum(np.square(W1)) + np.sum(np.square(W2)) + np.sum(np.square(W3)))",
"_____no_output_____"
],
[
"# GRADED FUNCTION: compute_cost_with_regularization\n\ndef compute_cost_with_regularization(A3, Y, parameters, lambd):\n \"\"\"\n Implement the cost function with L2 regularization. See formula (2) above.\n \n Arguments:\n A3 -- post-activation, output of forward propagation, of shape (output size, number of examples)\n Y -- \"true\" labels vector, of shape (output size, number of examples)\n parameters -- python dictionary containing parameters of the model\n \n Returns:\n cost - value of the regularized loss function (formula (2))\n \"\"\"\n m = Y.shape[1]\n W1 = parameters[\"W1\"]\n W2 = parameters[\"W2\"]\n W3 = parameters[\"W3\"]\n \n cross_entropy_cost = compute_cost(A3, Y) # This gives you the cross-entropy part of the cost\n \n #(≈ 1 lines of code)\n # L2_regularization_cost = \n # YOUR CODE STARTS HERE\n L2_regularization_cost = (np.sum(np.square(W1)) + np.sum(np.square(W2)) + np.sum(np.square(W3))) * (lambd/(2*m))\n \n # YOUR CODE ENDS HERE\n \n cost = cross_entropy_cost + L2_regularization_cost\n \n return cost",
"_____no_output_____"
],
[
"A3, t_Y, parameters = compute_cost_with_regularization_test_case()\ncost = compute_cost_with_regularization(A3, t_Y, parameters, lambd=0.1)\nprint(\"cost = \" + str(cost))\n\ncompute_cost_with_regularization_test(compute_cost_with_regularization)",
"cost = 1.7864859451590758\n\u001b[92m All tests passed.\n"
]
],
[
[
"Of course, because you changed the cost, you have to change backward propagation as well! All the gradients have to be computed with respect to this new cost. \n\n<a name='ex-2'></a>\n### Exercise 2 - backward_propagation_with_regularization\nImplement the changes needed in backward propagation to take into account regularization. The changes only concern dW1, dW2 and dW3. For each, you have to add the regularization term's gradient ($\\frac{d}{dW} ( \\frac{1}{2}\\frac{\\lambda}{m} W^2) = \\frac{\\lambda}{m} W$).",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: backward_propagation_with_regularization\n\ndef backward_propagation_with_regularization(X, Y, cache, lambd):\n \"\"\"\n Implements the backward propagation of our baseline model to which we added an L2 regularization.\n \n Arguments:\n X -- input dataset, of shape (input size, number of examples)\n Y -- \"true\" labels vector, of shape (output size, number of examples)\n cache -- cache output from forward_propagation()\n lambd -- regularization hyperparameter, scalar\n \n Returns:\n gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables\n \"\"\"\n \n m = X.shape[1]\n (Z1, A1, W1, b1, Z2, A2, W2, b2, Z3, A3, W3, b3) = cache\n \n dZ3 = A3 - Y\n #(≈ 1 lines of code)\n # dW3 = 1./m * np.dot(dZ3, A2.T) + None\n # YOUR CODE STARTS HERE\n dW3 = 1./m * np.dot(dZ3, A2.T) + (lambd/m) * W3\n \n # YOUR CODE ENDS HERE\n db3 = 1. / m * np.sum(dZ3, axis=1, keepdims=True)\n \n dA2 = np.dot(W3.T, dZ3)\n dZ2 = np.multiply(dA2, np.int64(A2 > 0))\n #(≈ 1 lines of code)\n # dW2 = 1./m * np.dot(dZ2, A1.T) + None\n # YOUR CODE STARTS HERE\n dW2 = 1./m * np.dot(dZ2, A1.T) + (lambd/m) * W2\n \n # YOUR CODE ENDS HERE\n db2 = 1. / m * np.sum(dZ2, axis=1, keepdims=True)\n \n dA1 = np.dot(W2.T, dZ2)\n dZ1 = np.multiply(dA1, np.int64(A1 > 0))\n #(≈ 1 lines of code)\n # dW1 = 1./m * np.dot(dZ1, X.T) + None\n # YOUR CODE STARTS HERE\n dW1 = 1./m * np.dot(dZ1, X.T) + (lambd/m) * W1\n \n # YOUR CODE ENDS HERE\n db1 = 1. / m * np.sum(dZ1, axis=1, keepdims=True)\n \n gradients = {\"dZ3\": dZ3, \"dW3\": dW3, \"db3\": db3,\"dA2\": dA2,\n \"dZ2\": dZ2, \"dW2\": dW2, \"db2\": db2, \"dA1\": dA1, \n \"dZ1\": dZ1, \"dW1\": dW1, \"db1\": db1}\n \n return gradients",
"_____no_output_____"
],
[
"t_X, t_Y, cache = backward_propagation_with_regularization_test_case()\n\ngrads = backward_propagation_with_regularization(t_X, t_Y, cache, lambd = 0.7)\nprint (\"dW1 = \\n\"+ str(grads[\"dW1\"]))\nprint (\"dW2 = \\n\"+ str(grads[\"dW2\"]))\nprint (\"dW3 = \\n\"+ str(grads[\"dW3\"]))\nbackward_propagation_with_regularization_test(backward_propagation_with_regularization)",
"dW1 = \n[[-0.25604646 0.12298827 -0.28297129]\n [-0.17706303 0.34536094 -0.4410571 ]]\ndW2 = \n[[ 0.79276486 0.85133918]\n [-0.0957219 -0.01720463]\n [-0.13100772 -0.03750433]]\ndW3 = \n[[-1.77691347 -0.11832879 -0.09397446]]\n\u001b[92m All tests passed.\n"
]
],
[
[
"Let's now run the model with L2 regularization $(\\lambda = 0.7)$. The `model()` function will call: \n- `compute_cost_with_regularization` instead of `compute_cost`\n- `backward_propagation_with_regularization` instead of `backward_propagation`",
"_____no_output_____"
]
],
[
[
"parameters = model(train_X, train_Y, lambd = 0.7)\nprint (\"On the train set:\")\npredictions_train = predict(train_X, train_Y, parameters)\nprint (\"On the test set:\")\npredictions_test = predict(test_X, test_Y, parameters)",
"Cost after iteration 0: 0.6974484493131264\nCost after iteration 10000: 0.2684918873282238\nCost after iteration 20000: 0.26809163371273004\n"
]
],
[
[
"Congrats, the test set accuracy increased to 93%. You have saved the French football team!\n\nYou are not overfitting the training data anymore. Let's plot the decision boundary.",
"_____no_output_____"
]
],
[
[
"plt.title(\"Model with L2-regularization\")\naxes = plt.gca()\naxes.set_xlim([-0.75,0.40])\naxes.set_ylim([-0.75,0.65])\nplot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)",
"_____no_output_____"
]
],
[
[
"**Observations**:\n- The value of $\\lambda$ is a hyperparameter that you can tune using a dev set.\n- L2 regularization makes your decision boundary smoother. If $\\lambda$ is too large, it is also possible to \"oversmooth\", resulting in a model with high bias.\n\n**What is L2-regularization actually doing?**:\n\nL2-regularization relies on the assumption that a model with small weights is simpler than a model with large weights. Thus, by penalizing the square values of the weights in the cost function you drive all the weights to smaller values. It becomes too costly for the cost to have large weights! This leads to a smoother model in which the output changes more slowly as the input changes. \n\n<br>\n<font color='blue'>\n \n**What you should remember:** the implications of L2-regularization on:\n- The cost computation:\n - A regularization term is added to the cost.\n- The backpropagation function:\n - There are extra terms in the gradients with respect to weight matrices.\n- Weights end up smaller (\"weight decay\"): \n - Weights are pushed to smaller values.",
"_____no_output_____"
],
[
"<a name='6'></a>\n## 6 - Dropout\n\nFinally, **dropout** is a widely used regularization technique that is specific to deep learning. \n**It randomly shuts down some neurons in each iteration.** Watch these two videos to see what this means!\n\n<!--\nTo understand drop-out, consider this conversation with a friend:\n- Friend: \"Why do you need all these neurons to train your network and classify images?\". \n- You: \"Because each neuron contains a weight and can learn specific features/details/shape of an image. The more neurons I have, the more featurse my model learns!\"\n- Friend: \"I see, but are you sure that your neurons are learning different features and not all the same features?\"\n- You: \"Good point... Neurons in the same layer actually don't talk to each other. It should be definitly possible that they learn the same image features/shapes/forms/details... which would be redundant. There should be a solution.\"\n!--> \n\n\n<center>\n<video width=\"620\" height=\"440\" src=\"images/dropout1_kiank.mp4\" type=\"video/mp4\" controls>\n</video>\n</center>\n<br>\n<caption><center><font color='purple'><b>Figure 2 </b>: <b>Drop-out on the second hidden layer.</b> <br> At each iteration, you shut down (= set to zero) each neuron of a layer with probability $1 - keep\\_prob$ or keep it with probability $keep\\_prob$ (50% here). The dropped neurons don't contribute to the training in both the forward and backward propagations of the iteration. </font></center></caption>\n\n<center>\n<video width=\"620\" height=\"440\" src=\"images/dropout2_kiank.mp4\" type=\"video/mp4\" controls>\n</video>\n</center>\n\n<caption><center><font color='purple'><b>Figure 3</b>:<b> Drop-out on the first and third hidden layers. </b><br> $1^{st}$ layer: we shut down on average 40% of the neurons. $3^{rd}$ layer: we shut down on average 20% of the neurons. </font></center></caption>\n\n\nWhen you shut some neurons down, you actually modify your model. The idea behind drop-out is that at each iteration, you train a different model that uses only a subset of your neurons. With dropout, your neurons thus become less sensitive to the activation of one other specific neuron, because that other neuron might be shut down at any time. \n\n<a name='6-1'></a>\n### 6.1 - Forward Propagation with Dropout\n\n<a name='ex-3'></a>\n### Exercise 3 - forward_propagation_with_dropout\n\nImplement the forward propagation with dropout. You are using a 3 layer neural network, and will add dropout to the first and second hidden layers. We will not apply dropout to the input layer or output layer. \n\n**Instructions**:\nYou would like to shut down some neurons in the first and second layers. To do that, you are going to carry out 4 Steps:\n1. In lecture, we dicussed creating a variable $d^{[1]}$ with the same shape as $a^{[1]}$ using `np.random.rand()` to randomly get numbers between 0 and 1. Here, you will use a vectorized implementation, so create a random matrix $D^{[1]} = [d^{[1](1)} d^{[1](2)} ... d^{[1](m)}] $ of the same dimension as $A^{[1]}$.\n2. Set each entry of $D^{[1]}$ to be 1 with probability (`keep_prob`), and 0 otherwise.\n\n**Hint:** Let's say that keep_prob = 0.8, which means that we want to keep about 80% of the neurons and drop out about 20% of them. We want to generate a vector that has 1's and 0's, where about 80% of them are 1 and about 20% are 0.\nThis python statement: \n`X = (X < keep_prob).astype(int)` \n\nis conceptually the same as this if-else statement (for the simple case of a one-dimensional array) :\n\n```\nfor i,v in enumerate(x):\n if v < keep_prob:\n x[i] = 1\n else: # v >= keep_prob\n x[i] = 0\n```\nNote that the `X = (X < keep_prob).astype(int)` works with multi-dimensional arrays, and the resulting output preserves the dimensions of the input array.\n\nAlso note that without using `.astype(int)`, the result is an array of booleans `True` and `False`, which Python automatically converts to 1 and 0 if we multiply it with numbers. (However, it's better practice to convert data into the data type that we intend, so try using `.astype(int)`.)\n\n3. Set $A^{[1]}$ to $A^{[1]} * D^{[1]}$. (You are shutting down some neurons). You can think of $D^{[1]}$ as a mask, so that when it is multiplied with another matrix, it shuts down some of the values.\n4. Divide $A^{[1]}$ by `keep_prob`. By doing this you are assuring that the result of the cost will still have the same expected value as without drop-out. (This technique is also called inverted dropout.)",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: forward_propagation_with_dropout\n\ndef forward_propagation_with_dropout(X, parameters, keep_prob = 0.5):\n \"\"\"\n Implements the forward propagation: LINEAR -> RELU + DROPOUT -> LINEAR -> RELU + DROPOUT -> LINEAR -> SIGMOID.\n \n Arguments:\n X -- input dataset, of shape (2, number of examples)\n parameters -- python dictionary containing your parameters \"W1\", \"b1\", \"W2\", \"b2\", \"W3\", \"b3\":\n W1 -- weight matrix of shape (20, 2)\n b1 -- bias vector of shape (20, 1)\n W2 -- weight matrix of shape (3, 20)\n b2 -- bias vector of shape (3, 1)\n W3 -- weight matrix of shape (1, 3)\n b3 -- bias vector of shape (1, 1)\n keep_prob - probability of keeping a neuron active during drop-out, scalar\n \n Returns:\n A3 -- last activation value, output of the forward propagation, of shape (1,1)\n cache -- tuple, information stored for computing the backward propagation\n \"\"\"\n \n np.random.seed(1)\n \n # retrieve parameters\n W1 = parameters[\"W1\"]\n b1 = parameters[\"b1\"]\n W2 = parameters[\"W2\"]\n b2 = parameters[\"b2\"]\n W3 = parameters[\"W3\"]\n b3 = parameters[\"b3\"]\n \n # LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID\n Z1 = np.dot(W1, X) + b1\n A1 = relu(Z1)\n #(≈ 4 lines of code) # Steps 1-4 below correspond to the Steps 1-4 described above. \n # D1 = # Step 1: initialize matrix D1 = np.random.rand(..., ...)\n # D1 = # Step 2: convert entries of D1 to 0 or 1 (using keep_prob as the threshold)\n # A1 = # Step 3: shut down some neurons of A1\n # A1 = # Step 4: scale the value of neurons that haven't been shut down\n # YOUR CODE STARTS HERE\n D1 = np.random.rand(A1.shape[0],A1.shape[1])\n D1 = (D1 < keep_prob).astype(int)\n A1 = A1*D1\n A1 = A1/keep_prob\n \n # YOUR CODE ENDS HERE\n Z2 = np.dot(W2, A1) + b2\n A2 = relu(Z2)\n #(≈ 4 lines of code)\n # D2 = # Step 1: initialize matrix D2 = np.random.rand(..., ...)\n # D2 = # Step 2: convert entries of D2 to 0 or 1 (using keep_prob as the threshold)\n # A2 = # Step 3: shut down some neurons of A2\n # A2 = # Step 4: scale the value of neurons that haven't been shut down\n # YOUR CODE STARTS HERE\n D2 = np.random.rand(A2.shape[0],A2.shape[1])\n D2 = (D2 < keep_prob).astype(int)\n A2 = A2*D2\n A2 = A2/keep_prob\n \n \n # YOUR CODE ENDS HERE\n Z3 = np.dot(W3, A2) + b3\n A3 = sigmoid(Z3)\n \n cache = (Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3)\n \n return A3, cache",
"_____no_output_____"
],
[
"t_X, parameters = forward_propagation_with_dropout_test_case()\n\nA3, cache = forward_propagation_with_dropout(t_X, parameters, keep_prob=0.7)\nprint (\"A3 = \" + str(A3))\n\nforward_propagation_with_dropout_test(forward_propagation_with_dropout)",
"A3 = [[0.36974721 0.00305176 0.04565099 0.49683389 0.36974721]]\n\u001b[92m All tests passed.\n"
]
],
[
[
"<a name='6-2'></a>\n### 6.2 - Backward Propagation with Dropout\n\n<a name='ex-4'></a>\n### Exercise 4 - backward_propagation_with_dropout\nImplement the backward propagation with dropout. As before, you are training a 3 layer network. Add dropout to the first and second hidden layers, using the masks $D^{[1]}$ and $D^{[2]}$ stored in the cache. \n\n**Instruction**:\nBackpropagation with dropout is actually quite easy. You will have to carry out 2 Steps:\n1. You had previously shut down some neurons during forward propagation, by applying a mask $D^{[1]}$ to `A1`. In backpropagation, you will have to shut down the same neurons, by reapplying the same mask $D^{[1]}$ to `dA1`. \n2. During forward propagation, you had divided `A1` by `keep_prob`. In backpropagation, you'll therefore have to divide `dA1` by `keep_prob` again (the calculus interpretation is that if $A^{[1]}$ is scaled by `keep_prob`, then its derivative $dA^{[1]}$ is also scaled by the same `keep_prob`).\n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: backward_propagation_with_dropout\n\ndef backward_propagation_with_dropout(X, Y, cache, keep_prob):\n \"\"\"\n Implements the backward propagation of our baseline model to which we added dropout.\n \n Arguments:\n X -- input dataset, of shape (2, number of examples)\n Y -- \"true\" labels vector, of shape (output size, number of examples)\n cache -- cache output from forward_propagation_with_dropout()\n keep_prob - probability of keeping a neuron active during drop-out, scalar\n \n Returns:\n gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables\n \"\"\"\n \n m = X.shape[1]\n (Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3) = cache\n \n dZ3 = A3 - Y\n dW3 = 1./m * np.dot(dZ3, A2.T)\n db3 = 1./m * np.sum(dZ3, axis=1, keepdims=True)\n dA2 = np.dot(W3.T, dZ3)\n #(≈ 2 lines of code)\n # dA2 = # Step 1: Apply mask D2 to shut down the same neurons as during the forward propagation\n # dA2 = # Step 2: Scale the value of neurons that haven't been shut down\n # YOUR CODE STARTS HERE\n dA2 = dA2*D2\n dA2 = dA2/keep_prob\n \n # YOUR CODE ENDS HERE\n dZ2 = np.multiply(dA2, np.int64(A2 > 0))\n dW2 = 1./m * np.dot(dZ2, A1.T)\n db2 = 1./m * np.sum(dZ2, axis=1, keepdims=True)\n \n dA1 = np.dot(W2.T, dZ2)\n #(≈ 2 lines of code)\n # dA1 = # Step 1: Apply mask D1 to shut down the same neurons as during the forward propagation\n # dA1 = # Step 2: Scale the value of neurons that haven't been shut down\n # YOUR CODE STARTS HERE\n dA1 = dA1*D1\n dA1 = dA1/keep_prob\n \n # YOUR CODE ENDS HERE\n dZ1 = np.multiply(dA1, np.int64(A1 > 0))\n dW1 = 1./m * np.dot(dZ1, X.T)\n db1 = 1./m * np.sum(dZ1, axis=1, keepdims=True)\n \n gradients = {\"dZ3\": dZ3, \"dW3\": dW3, \"db3\": db3,\"dA2\": dA2,\n \"dZ2\": dZ2, \"dW2\": dW2, \"db2\": db2, \"dA1\": dA1, \n \"dZ1\": dZ1, \"dW1\": dW1, \"db1\": db1}\n \n return gradients",
"_____no_output_____"
],
[
"t_X, t_Y, cache = backward_propagation_with_dropout_test_case()\n\ngradients = backward_propagation_with_dropout(t_X, t_Y, cache, keep_prob=0.8)\n\nprint (\"dA1 = \\n\" + str(gradients[\"dA1\"]))\nprint (\"dA2 = \\n\" + str(gradients[\"dA2\"]))\n\nbackward_propagation_with_dropout_test(backward_propagation_with_dropout)",
"dA1 = \n[[ 0.36544439 0. -0.00188233 0. -0.17408748]\n [ 0.65515713 0. -0.00337459 0. -0. ]]\ndA2 = \n[[ 0.58180856 0. -0.00299679 0. -0.27715731]\n [ 0. 0.53159854 -0. 0.53159854 -0.34089673]\n [ 0. 0. -0.00292733 0. -0. ]]\n\u001b[92m All tests passed.\n"
]
],
[
[
"Let's now run the model with dropout (`keep_prob = 0.86`). It means at every iteration you shut down each neurons of layer 1 and 2 with 14% probability. The function `model()` will now call:\n- `forward_propagation_with_dropout` instead of `forward_propagation`.\n- `backward_propagation_with_dropout` instead of `backward_propagation`.",
"_____no_output_____"
]
],
[
[
"parameters = model(train_X, train_Y, keep_prob = 0.86, learning_rate = 0.3)\n\nprint (\"On the train set:\")\npredictions_train = predict(train_X, train_Y, parameters)\nprint (\"On the test set:\")\npredictions_test = predict(test_X, test_Y, parameters)",
"Cost after iteration 0: 0.6543912405149825\nCost after iteration 10000: 0.0610169865749056\nCost after iteration 20000: 0.060582435798513114\n"
]
],
[
[
"Dropout works great! The test accuracy has increased again (to 95%)! Your model is not overfitting the training set and does a great job on the test set. The French football team will be forever grateful to you! \n\nRun the code below to plot the decision boundary.",
"_____no_output_____"
]
],
[
[
"plt.title(\"Model with dropout\")\naxes = plt.gca()\naxes.set_xlim([-0.75,0.40])\naxes.set_ylim([-0.75,0.65])\nplot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)",
"_____no_output_____"
]
],
[
[
"**Note**:\n- A **common mistake** when using dropout is to use it both in training and testing. You should use dropout (randomly eliminate nodes) only in training. \n- Deep learning frameworks like [tensorflow](https://www.tensorflow.org/api_docs/python/tf/nn/dropout), [PaddlePaddle](http://doc.paddlepaddle.org/release_doc/0.9.0/doc/ui/api/trainer_config_helpers/attrs.html), [keras](https://keras.io/layers/core/#dropout) or [caffe](http://caffe.berkeleyvision.org/tutorial/layers/dropout.html) come with a dropout layer implementation. Don't stress - you will soon learn some of these frameworks.\n\n<font color='blue'>\n \n**What you should remember about dropout:**\n- Dropout is a regularization technique.\n- You only use dropout during training. Don't use dropout (randomly eliminate nodes) during test time.\n- Apply dropout both during forward and backward propagation.\n- During training time, divide each dropout layer by keep_prob to keep the same expected value for the activations. For example, if keep_prob is 0.5, then we will on average shut down half the nodes, so the output will be scaled by 0.5 since only the remaining half are contributing to the solution. Dividing by 0.5 is equivalent to multiplying by 2. Hence, the output now has the same expected value. You can check that this works even when keep_prob is other values than 0.5. ",
"_____no_output_____"
],
[
"<a name='7'></a>\n## 7 - Conclusions",
"_____no_output_____"
],
[
"**Here are the results of our three models**: \n\n<table> \n <tr>\n <td>\n <b>model</b>\n </td>\n <td>\n <b>train accuracy</b>\n </td>\n <td>\n <b>test accuracy</b>\n </td>\n </tr>\n <td>\n 3-layer NN without regularization\n </td>\n <td>\n 95%\n </td>\n <td>\n 91.5%\n </td>\n <tr>\n <td>\n 3-layer NN with L2-regularization\n </td>\n <td>\n 94%\n </td>\n <td>\n 93%\n </td>\n </tr>\n <tr>\n <td>\n 3-layer NN with dropout\n </td>\n <td>\n 93%\n </td>\n <td>\n 95%\n </td>\n </tr>\n</table> ",
"_____no_output_____"
],
[
"Note that regularization hurts training set performance! This is because it limits the ability of the network to overfit to the training set. But since it ultimately gives better test accuracy, it is helping your system. ",
"_____no_output_____"
],
[
"Congratulations for finishing this assignment! And also for revolutionizing French football. :-) ",
"_____no_output_____"
],
[
"<font color='blue'>\n \n**What we want you to remember from this notebook**:\n- Regularization will help you reduce overfitting.\n- Regularization will drive your weights to lower values.\n- L2 regularization and Dropout are two very effective regularization techniques.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec5fdc2d17de9ed2833e85310d17f1c862e71f34 | 697,274 | ipynb | Jupyter Notebook | 07_train/wip/debugger-monitor/02_deploy_and_monitor/deploy_and_monitor.ipynb | NRauschmayr/workshop | c890e38a5f4a339540697206ebdea479e66534e5 | [
"Apache-2.0"
]
| 1 | 2021-02-20T14:55:02.000Z | 2021-02-20T14:55:02.000Z | 07_train/wip/debugger-monitor/02_deploy_and_monitor/deploy_and_monitor.ipynb | NRauschmayr/workshop | c890e38a5f4a339540697206ebdea479e66534e5 | [
"Apache-2.0"
]
| null | null | null | 07_train/wip/debugger-monitor/02_deploy_and_monitor/deploy_and_monitor.ipynb | NRauschmayr/workshop | c890e38a5f4a339540697206ebdea479e66534e5 | [
"Apache-2.0"
]
| null | null | null | 118.56385 | 16,414 | 0.584099 | [
[
[
"# Deploying and Monitoring\n\nIn this notebook we will deploy the network traffic classification model that we have trained in the previous steps to Amazon SageMaker hosting, which will expose a fully-managed real-time endpoint to execute inferences.\n\nAmazon SageMaker is adding new capabilities that monitor ML models while in production and detect deviations in data quality in comparison to a baseline dataset (e.g. training data set). They enable you to capture the metadata and the input and output for invocations of the models that you deploy with Amazon SageMaker. They also enable you to analyze the data and monitor its quality. \n\nWe will deploy the model to a real-time endpoint with data capture enabled and start collecting some inference inputs/outputs. Then, we will create a baseline and finally enable model monitoring to compare inference data with respect to the baseline and analyze the quality.",
"_____no_output_____"
],
[
"First, we set some variables, including the AWS region we are working in, the IAM execution role of the notebook instance and the Amazon S3 bucket where we will store data and outputs.",
"_____no_output_____"
]
],
[
[
"import os\nimport boto3\nimport sagemaker\n\nregion = boto3.Session().region_name\nrole = sagemaker.get_execution_role()\nsagemaker_session = sagemaker.Session()\nbucket_name = sagemaker_session.default_bucket()\nprefix = 'aim362'\n\nprint(region)\nprint(role)\nprint(bucket_name)",
"us-east-1\narn:aws:iam::806570384721:role/service-role/AmazonSageMaker-ExecutionRole-20191201T115647\nsagemaker-us-east-1-806570384721\n"
]
],
[
[
"## Deployment with Data Capture\n\nWe are going to deploy the latest network traffic classification model that we have trained. To deploy a model using the SM Python SDK, we need to make sure we have the Amazon S3 URI where the model artifacts are stored and the URI of the Docker container that will be used for hosting this model.\n\nFirst, let's determine the Amazon S3 URI of the model artifacts by using a couple of utility functions which query Amazon SageMaker service to get the latest training job whose name starts with 'nw-traffic-classification-xgb' and then describing the training job.",
"_____no_output_____"
]
],
[
[
"import boto3\n\ndef get_latest_training_job_name(base_job_name):\n client = boto3.client('sagemaker')\n response = client.list_training_jobs(NameContains=base_job_name, SortBy='CreationTime', \n SortOrder='Descending', StatusEquals='Completed')\n if len(response['TrainingJobSummaries']) > 0 :\n return response['TrainingJobSummaries'][0]['TrainingJobName']\n else:\n raise Exception('Training job not found.')\n\ndef get_training_job_s3_model_artifacts(job_name):\n client = boto3.client('sagemaker')\n response = client.describe_training_job(TrainingJobName=job_name)\n s3_model_artifacts = response['ModelArtifacts']['S3ModelArtifacts']\n return s3_model_artifacts\n\nlatest_training_job_name = get_latest_training_job_name('nw-traffic-classification-xgb')\nprint(latest_training_job_name)\nmodel_path = get_training_job_s3_model_artifacts(latest_training_job_name)\nprint(model_path)",
"nw-traffic-classification-xgb-2020-02-04-19-56-23-672\ns3://sagemaker-us-east-1-806570384721/aim362/output/nw-traffic-classification-xgb-2020-02-04-19-56-23-672/output/model.tar.gz\n"
]
],
[
[
"For this model, we are going to use the same XGBoost Docker container we used for training, which also offers inference capabilities. As a consequence, we can just create the XGBoostModel object of the Amazon SageMaker Python SDK and then invoke its .deploy() method to execute deployment.",
"_____no_output_____"
],
[
"We will also provide an entrypoint script to be invoked at deployment/inference time. The purpose of this code is deserializing and loading the XGB model. In addition, we are re-defining the output functions as we want to extract the class value from the default array output. For example, for class 3 the XGB container would output [3.] but we want to extract only the 3 value.",
"_____no_output_____"
]
],
[
[
"!pygmentize source_dir/deploy_xgboost.py",
"\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mos\u001b[39;49;00m\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mpickle\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mpkl\u001b[39;49;00m\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mnumpy\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mnp\u001b[39;49;00m\n\n\u001b[34mfrom\u001b[39;49;00m \u001b[04m\u001b[36msagemaker_containers.beta.framework\u001b[39;49;00m \u001b[34mimport\u001b[39;49;00m (\n content_types, encoders, env, modules, transformer, worker)\n\n\u001b[34mdef\u001b[39;49;00m \u001b[32mmodel_fn\u001b[39;49;00m(model_dir):\n model_file = model_dir + \u001b[33m'\u001b[39;49;00m\u001b[33m/model.bin\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\n model = pkl.load(\u001b[36mopen\u001b[39;49;00m(model_file, \u001b[33m'\u001b[39;49;00m\u001b[33mrb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m))\n \u001b[34mreturn\u001b[39;49;00m model\n\n\u001b[34mdef\u001b[39;49;00m \u001b[32moutput_fn\u001b[39;49;00m(prediction, accept):\n \n pred_array_value = np.array(prediction)\n pred_value = \u001b[36mint\u001b[39;49;00m(pred_array_value[\u001b[34m0\u001b[39;49;00m])\n \n \u001b[34mreturn\u001b[39;49;00m worker.Response(\u001b[36mstr\u001b[39;49;00m(pred_value), accept, mimetype=accept)\n"
]
],
[
[
"Now we are ready to create the XGBoostModel object.",
"_____no_output_____"
]
],
[
[
"from time import gmtime, strftime\nfrom sagemaker.xgboost import XGBoostModel\n\nmodel_name = 'nw-traffic-classification-xgb-model-' + strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime())\n\ncode_location = 's3://{0}/{1}/code'.format(bucket_name, prefix)\nxgboost_model = XGBoostModel(model_data=model_path,\n entry_point='deploy_xgboost.py',\n source_dir='source_dir/',\n name=model_name,\n code_location=code_location,\n framework_version='0.90-2',\n role=role, \n sagemaker_session=sagemaker_session)",
"_____no_output_____"
]
],
[
[
"Finally we create an endpoint with data capture enabled, for monitoring the model data quality.\nData capture is enabled at enpoint configuration level for the Amazon SageMaker real-time endpoint. You can choose to capture the request payload, the response payload or both and captured data is stored in JSON format.",
"_____no_output_____"
]
],
[
[
"from time import gmtime, strftime\nfrom sagemaker.model_monitor import DataCaptureConfig\n\ns3_capture_upload_path = 's3://{}/{}/monitoring/datacapture'.format(bucket_name, prefix)\nprint(s3_capture_upload_path)\n\nendpoint_name = 'nw-traffic-classification-xgb-ep-' + strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime())\nprint(endpoint_name)\n\npred = xgboost_model.deploy(initial_instance_count=1,\n instance_type='ml.m5.xlarge',\n endpoint_name=endpoint_name,\n data_capture_config=DataCaptureConfig(\n enable_capture=True,\n sampling_percentage=100,\n destination_s3_uri=s3_capture_upload_path))",
"s3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture\nnw-traffic-classification-xgb-ep-2020-02-04-20-24-20\n-----------------!"
]
],
[
[
"After the deployment has been completed, we can leverage on the RealTimePredictor object to execute HTTPs requests against the deployed endpoint and get inference results.",
"_____no_output_____"
]
],
[
[
"from sagemaker.predictor import RealTimePredictor\n\npred = RealTimePredictor(endpoint_name)\npred.content_type = 'text/csv'\npred.accept = 'text/csv'\n\n# Expecting class 4\ntest_values = \"80,1056736,3,4,20,964,20,0,6.666666667,11.54700538,964,0,241.0,482.0,931.1691850999999,6.6241710320000005,176122.6667,\\\n431204.4454,1056315,2,394,197.0,275.77164469999997,392,2,1056733,352244.3333,609743.1115,1056315,24,0,0,0,0,72,92,\\\n2.8389304419999997,3.78524059,0,964,123.0,339.8873763,115523.4286,0,0,1,1,0,0,0,1,1.0,140.5714286,6.666666667,\\\n241.0,0.0,0.0,0.0,0.0,0.0,0.0,3,20,4,964,8192,211,1,20,0.0,0.0,0,0,0.0,0.0,0,0,20,2,2018,1,0,1,0\"\n\nresult = pred.predict(test_values)\nprint(result)\n\n# Expecting class 7\ntest_values = \"80,10151,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,197.0249237,10151.0,0.0,10151,10151,10151,10151.0,0.0,10151,10151,0,0.0,\\\n0.0,0,0,0,0,0,0,40,0,197.0249237,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,2,0,0,0,32738,\\\n-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,21,2,2018,2,0,1,0\"\n\nresult = pred.predict(test_values)\nprint(result)\n\n# Expecting class 0\ntest_values = \"80,54322832,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,0.0368169318,54322832.0,0.0,54322832,54322832,54322832,54322832.0,0.0,\\\n54322832,54322832,0,0.0,0.0,0,0,0,0,0,0,40,0,0.0368169318,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,\\\n0.0,0.0,0.0,0.0,2,0,0,0,279,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,23,2,2018,4,0,1,0\"\n\nresult = pred.predict(test_values)\nprint(result)",
"b'4'\nb'7'\nb'0'\n"
]
],
[
[
"Now let's list the data capture files stored in S3. You should expect to see different files from different time periods organized based on the hour in which the invocation occurred.\n\n**Note that the delivery of capture data to Amazon S3 can require a couple of minutes so next cell might error. If this happens, please retry after a minute.**",
"_____no_output_____"
]
],
[
[
"s3_client = boto3.Session().client('s3')\ncurrent_endpoint_capture_prefix = '{}/monitoring/datacapture/{}'.format(prefix, endpoint_name)\n\nresult = s3_client.list_objects(Bucket=bucket_name, Prefix=current_endpoint_capture_prefix)\ncapture_files = ['s3://{0}/{1}'.format(bucket_name, capture_file.get(\"Key\")) for capture_file in result.get('Contents')]\n\nprint(\"Capture Files: \")\nprint(\"\\n \".join(capture_files))",
"Capture Files: \ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/33-57-898-0815a2e8-c235-4891-bd5d-91a59c41b6ca.jsonl\n"
]
],
[
[
"We can also read the contents of one of these files and see how capture records are organized in JSON lines format.",
"_____no_output_____"
]
],
[
[
"!aws s3 cp {capture_files[0]} datacapture/captured_data_example.jsonl\n!head datacapture/captured_data_example.jsonl",
"download: s3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/33-57-898-0815a2e8-c235-4891-bd5d-91a59c41b6ca.jsonl to datacapture/captured_data_example.jsonl\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,1056736,3,4,20,964,20,0,6.666666667,11.54700538,964,0,241.0,482.0,931.1691850999999,6.6241710320000005,176122.6667,431204.4454,1056315,2,394,197.0,275.77164469999997,392,2,1056733,352244.3333,609743.1115,1056315,24,0,0,0,0,72,92,2.8389304419999997,3.78524059,0,964,123.0,339.8873763,115523.4286,0,0,1,1,0,0,0,1,1.0,140.5714286,6.666666667,241.0,0.0,0.0,0.0,0.0,0.0,0.0,3,20,4,964,8192,211,1,20,0.0,0.0,0,0,0.0,0.0,0,0,20,2,2018,1,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"4\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"1e4cf25e-310d-4d6e-b9fa-5aea6b9a1f82\",\"inferenceTime\":\"2020-02-04T20:33:57Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,10151,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,197.0249237,10151.0,0.0,10151,10151,10151,10151.0,0.0,10151,10151,0,0.0,0.0,0,0,0,0,0,0,40,0,197.0249237,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,2,0,0,0,32738,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,21,2,2018,2,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"7\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"ce71debf-9537-4250-b970-869d9ff811da\",\"inferenceTime\":\"2020-02-04T20:33:57Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,54322832,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,0.0368169318,54322832.0,0.0,54322832,54322832,54322832,54322832.0,0.0,54322832,54322832,0,0.0,0.0,0,0,0,0,0,0,40,0,0.0368169318,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,2,0,0,0,279,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,23,2,2018,4,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"0\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"42f9be4d-3a0d-4630-9965-7102a8dcdf7b\",\"inferenceTime\":\"2020-02-04T20:33:58Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,1056736,3,4,20,964,20,0,6.666666667,11.54700538,964,0,241.0,482.0,931.1691850999999,6.6241710320000005,176122.6667,431204.4454,1056315,2,394,197.0,275.77164469999997,392,2,1056733,352244.3333,609743.1115,1056315,24,0,0,0,0,72,92,2.8389304419999997,3.78524059,0,964,123.0,339.8873763,115523.4286,0,0,1,1,0,0,0,1,1.0,140.5714286,6.666666667,241.0,0.0,0.0,0.0,0.0,0.0,0.0,3,20,4,964,8192,211,1,20,0.0,0.0,0,0,0.0,0.0,0,0,20,2,2018,1,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"4\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"14796b3c-a92c-4349-9260-50fe6fa8c329\",\"inferenceTime\":\"2020-02-04T20:34:37Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,10151,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,197.0249237,10151.0,0.0,10151,10151,10151,10151.0,0.0,10151,10151,0,0.0,0.0,0,0,0,0,0,0,40,0,197.0249237,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,2,0,0,0,32738,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,21,2,2018,2,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"7\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"fe6be844-6f06-46d3-8ace-49774af6d36a\",\"inferenceTime\":\"2020-02-04T20:34:37Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,54322832,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,0.0368169318,54322832.0,0.0,54322832,54322832,54322832,54322832.0,0.0,54322832,54322832,0,0.0,0.0,0,0,0,0,0,0,40,0,0.0368169318,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,2,0,0,0,279,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,23,2,2018,4,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"0\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"5cc6507e-08f7-45e0-a48e-dd7344faa979\",\"inferenceTime\":\"2020-02-04T20:34:37Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,1056736,3,4,20,964,20,0,6.666666667,11.54700538,964,0,241.0,482.0,931.1691850999999,6.6241710320000005,176122.6667,431204.4454,1056315,2,394,197.0,275.77164469999997,392,2,1056733,352244.3333,609743.1115,1056315,24,0,0,0,0,72,92,2.8389304419999997,3.78524059,0,964,123.0,339.8873763,115523.4286,0,0,1,1,0,0,0,1,1.0,140.5714286,6.666666667,241.0,0.0,0.0,0.0,0.0,0.0,0.0,3,20,4,964,8192,211,1,20,0.0,0.0,0,0,0.0,0.0,0,0,20,2,2018,1,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"4\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"4fd42f24-77cf-4caa-bee8-6884b7a296bf\",\"inferenceTime\":\"2020-02-04T20:34:44Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,10151,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,197.0249237,10151.0,0.0,10151,10151,10151,10151.0,0.0,10151,10151,0,0.0,0.0,0,0,0,0,0,0,40,0,197.0249237,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,2,0,0,0,32738,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,21,2,2018,2,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"7\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"f3815389-2dc2-4c43-b901-cc0e7301ace7\",\"inferenceTime\":\"2020-02-04T20:34:44Z\"},\"eventVersion\":\"0\"}\n{\"captureData\":{\"endpointInput\":{\"observedContentType\":\"text/csv\",\"mode\":\"INPUT\",\"data\":\"80,54322832,2,0,0,0,0,0,0.0,0.0,0,0,0.0,0.0,0.0,0.0368169318,54322832.0,0.0,54322832,54322832,54322832,54322832.0,0.0,54322832,54322832,0,0.0,0.0,0,0,0,0,0,0,40,0,0.0368169318,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,2,0,0,0,279,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,23,2,2018,4,0,1,0\",\"encoding\":\"CSV\"},\"endpointOutput\":{\"observedContentType\":\"text/csv; charset=utf-8\",\"mode\":\"OUTPUT\",\"data\":\"0\",\"encoding\":\"CSV\"}},\"eventMetadata\":{\"eventId\":\"2e1d543d-661a-4ec1-8dab-6a7e06f75fd5\",\"inferenceTime\":\"2020-02-04T20:34:44Z\"},\"eventVersion\":\"0\"}\n"
]
],
[
[
"In addition, we can better understand the content of each JSON line like follows:",
"_____no_output_____"
]
],
[
[
"import json\nwith open (\"datacapture/captured_data_example.jsonl\", \"r\") as myfile:\n data=myfile.read()\n\nprint(json.dumps(json.loads(data.split('\\n')[0]), indent=2))",
"{\n \"captureData\": {\n \"endpointInput\": {\n \"observedContentType\": \"text/csv\",\n \"mode\": \"INPUT\",\n \"data\": \"80,1056736,3,4,20,964,20,0,6.666666667,11.54700538,964,0,241.0,482.0,931.1691850999999,6.6241710320000005,176122.6667,431204.4454,1056315,2,394,197.0,275.77164469999997,392,2,1056733,352244.3333,609743.1115,1056315,24,0,0,0,0,72,92,2.8389304419999997,3.78524059,0,964,123.0,339.8873763,115523.4286,0,0,1,1,0,0,0,1,1.0,140.5714286,6.666666667,241.0,0.0,0.0,0.0,0.0,0.0,0.0,3,20,4,964,8192,211,1,20,0.0,0.0,0,0,0.0,0.0,0,0,20,2,2018,1,0,1,0\",\n \"encoding\": \"CSV\"\n },\n \"endpointOutput\": {\n \"observedContentType\": \"text/csv; charset=utf-8\",\n \"mode\": \"OUTPUT\",\n \"data\": \"4\",\n \"encoding\": \"CSV\"\n }\n },\n \"eventMetadata\": {\n \"eventId\": \"1e4cf25e-310d-4d6e-b9fa-5aea6b9a1f82\",\n \"inferenceTime\": \"2020-02-04T20:33:57Z\"\n },\n \"eventVersion\": \"0\"\n}\n"
]
],
[
[
"For each inference request, we get input data, output data and some metadata like the inference time captured and saved.",
"_____no_output_____"
],
[
"## Baselining",
"_____no_output_____"
],
[
"From our validation dataset let's ask Amazon SageMaker to suggest a set of baseline constraints and generate descriptive statistics for our features. Note that we are using the validation dataset for this workshop to make sure baselining time is short, and that file extension needs to be changed since the baselining jobs require .CSV file extension as default.\nIn reality, you might be willing to use a larger dataset as baseline.",
"_____no_output_____"
]
],
[
[
"import boto3\n\ns3 = boto3.resource('s3')\n\nbucket_key_prefix = \"aim362/data/val/\"\nbucket = s3.Bucket(bucket_name)\n\nfor s3_object in bucket.objects.filter(Prefix=bucket_key_prefix):\n target_key = s3_object.key.replace('data/val/', 'monitoring/baselining/data/').replace('.part', '.csv')\n print('Copying {0} to {1} ...'.format(s3_object.key, target_key))\n \n copy_source = {\n 'Bucket': bucket_name,\n 'Key': s3_object.key\n }\n s3.Bucket(bucket_name).copy(copy_source, target_key)",
"Copying aim362/data/val/0.part to aim362/monitoring/baselining/data/0.csv ...\nCopying aim362/data/val/1.part to aim362/monitoring/baselining/data/1.csv ...\nCopying aim362/data/val/2.part to aim362/monitoring/baselining/data/2.csv ...\nCopying aim362/data/val/3.part to aim362/monitoring/baselining/data/3.csv ...\nCopying aim362/data/val/4.part to aim362/monitoring/baselining/data/4.csv ...\nCopying aim362/data/val/5.part to aim362/monitoring/baselining/data/5.csv ...\nCopying aim362/data/val/6.part to aim362/monitoring/baselining/data/6.csv ...\nCopying aim362/data/val/7.part to aim362/monitoring/baselining/data/7.csv ...\nCopying aim362/data/val/8.part to aim362/monitoring/baselining/data/8.csv ...\nCopying aim362/data/val/9.part to aim362/monitoring/baselining/data/9.csv ...\n"
],
[
"baseline_data_path = 's3://{0}/{1}/monitoring/baselining/data'.format(bucket_name, prefix)\nbaseline_results_path = 's3://{0}/{1}/monitoring/baselining/results'.format(bucket_name, prefix)\n\nprint(baseline_data_path)\nprint(baseline_results_path)",
"s3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/data\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results\n"
]
],
[
[
"Please note that running the baselining job will require 8-10 minutes. In the meantime, you can take a look at the Deequ library, used to execute these analyses with the default Model Monitor container: https://github.com/awslabs/deequ",
"_____no_output_____"
]
],
[
[
"from sagemaker.model_monitor import DefaultModelMonitor\nfrom sagemaker.model_monitor.dataset_format import DatasetFormat\n\nmy_default_monitor = DefaultModelMonitor(\n role=role,\n instance_count=1,\n instance_type='ml.c5.4xlarge',\n volume_size_in_gb=20,\n max_runtime_in_seconds=3600,\n)",
"_____no_output_____"
],
[
"my_default_monitor.suggest_baseline(\n baseline_dataset=baseline_data_path,\n dataset_format=DatasetFormat.csv(header=True),\n output_s3_uri=baseline_results_path,\n wait=True\n)",
"\nJob Name: baseline-suggestion-job-2020-02-04-20-36-40-816\nInputs: [{'InputName': 'baseline_dataset_input', 'S3Input': {'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/data', 'LocalPath': '/opt/ml/processing/input/baseline_dataset_input', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3DataDistributionType': 'FullyReplicated', 'S3CompressionType': 'None'}}]\nOutputs: [{'OutputName': 'monitoring_output', 'S3Output': {'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results', 'LocalPath': '/opt/ml/processing/output', 'S3UploadMode': 'EndOfJob'}}]\n...................\u001b[34m2020-02-04 20:39:44,924 - __main__ - INFO - All params:{'ProcessingJobArn': 'arn:aws:sagemaker:us-east-1:806570384721:processing-job/baseline-suggestion-job-2020-02-04-20-36-40-816', 'ProcessingJobName': 'baseline-suggestion-job-2020-02-04-20-36-40-816', 'Environment': {'dataset_format': '{\"csv\": {\"header\": true, \"output_columns_position\": \"START\"}}', 'dataset_source': '/opt/ml/processing/input/baseline_dataset_input', 'output_path': '/opt/ml/processing/output', 'publish_cloudwatch_metrics': 'Disabled'}, 'AppSpecification': {'ImageUri': '156813124566.dkr.ecr.us-east-1.amazonaws.com/sagemaker-model-monitor-analyzer', 'ContainerEntrypoint': None, 'ContainerArguments': None}, 'ProcessingInputs': [{'InputName': 'baseline_dataset_input', 'S3Input': {'LocalPath': '/opt/ml/processing/input/baseline_dataset_input', 'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/data', 'S3DataDistributionType': 'FullyReplicated', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3CompressionType': 'None', 'S3DownloadMode': 'StartOfJob'}}], 'ProcessingOutputConfig': {'Outputs': [{'OutputName': 'monitoring_output', 'S3Output': {'LocalPath': '/opt/ml/processing/output', 'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results', 'S3UploadMode': 'EndOfJob'}}], 'KmsKeyId': None}, 'ProcessingResources': {'ClusterConfig': {'InstanceCount': 1, 'InstanceType': 'ml.c5.4xlarge', 'VolumeSizeInGB': 20, 'VolumeKmsKeyId': None}}, 'RoleArn': 'arn:aws:iam::806570384721:role/service-role/AmazonSageMaker-ExecutionRole-20191201T115647', 'StoppingCondition': {'MaxRuntimeInSeconds': 3600}}\u001b[0m\n\u001b[34m2020-02-04 20:39:44,924 - __main__ - INFO - Current Environment:{'dataset_format': '{\"csv\": {\"header\": true, \"output_columns_position\": \"START\"}}', 'dataset_source': '/opt/ml/processing/input/baseline_dataset_input', 'output_path': '/opt/ml/processing/output', 'publish_cloudwatch_metrics': 'Disabled'}\u001b[0m\n\u001b[34m2020-02-04 20:39:44,924 - DefaultDataAnalyzer - INFO - Performing analysis with input: {\"dataset_source\": \"/opt/ml/processing/input/baseline_dataset_input\", \"dataset_format\": {\"csv\": {\"header\": true, \"output_columns_position\": \"START\"}}, \"record_preprocessor_script\": null, \"post_analytics_processor_script\": null, \"baseline_constraints\": null, \"baseline_statistics\": null, \"output_path\": \"/opt/ml/processing/output\", \"start_time\": null, \"end_time\": null, \"cloudwatch_metrics_directory\": \"/opt/ml/output/metrics/cloudwatch\", \"publish_cloudwatch_metrics\": \"Disabled\", \"sagemaker_endpoint_name\": null, \"sagemaker_monitoring_schedule_name\": null, \"output_message_file\": \"/opt/ml/output/message\"}\u001b[0m\n\u001b[34m2020-02-04 20:39:44,924 - DefaultDataAnalyzer - INFO - Bootstrapping yarn\u001b[0m\n\u001b[34m2020-02-04 20:39:44,924 - bootstrap - INFO - Copy aws jars\u001b[0m\n\u001b[34m2020-02-04 20:39:44,975 - bootstrap - INFO - Copy cluster config\u001b[0m\n\u001b[34m2020-02-04 20:39:44,976 - bootstrap - INFO - Write runtime cluster config\u001b[0m\n\u001b[34m2020-02-04 20:39:44,976 - bootstrap - INFO - Resource Config is: {'current_host': 'algo-1', 'hosts': ['algo-1']}\u001b[0m\n\u001b[34m2020-02-04 20:39:44,985 - bootstrap - INFO - Finished Yarn configuration files setup.\u001b[0m\n\u001b[34m2020-02-04 20:39:44,985 - bootstrap - INFO - Starting spark process for master node algo-1\u001b[0m\n\u001b[34m2020-02-04 20:39:44,985 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/hdfs namenode -format -force\u001b[0m\n\u001b[34mWARNING: /usr/hadoop-3.0.0/logs does not exist. Creating.\u001b[0m\n\u001b[34m2020-02-04 20:39:45,365 INFO namenode.NameNode: STARTUP_MSG: \u001b[0m\n\u001b[34m/************************************************************\u001b[0m\n\u001b[34mSTARTUP_MSG: Starting NameNode\u001b[0m\n\u001b[34mSTARTUP_MSG: host = algo-1/10.0.188.93\u001b[0m\n\u001b[34mSTARTUP_MSG: args = [-format, -force]\u001b[0m\n\u001b[34mSTARTUP_MSG: version = 3.0.0\u001b[0m\n\u001b[34mSTARTUP_MSG: classpath = /usr/hadoop-3.0.0/etc/hadoop:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-cli-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jul-to-slf4j-1.7.25.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-io-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-server-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/avro-1.7.7.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-servlet-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-config-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/junit-4.11.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-json-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-common-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/htrace-core4-4.1.0-incubating.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-lang-2.6.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/accessors-smart-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsr311-api-1.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/hadoop-annotations-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/woodstox-core-5.0.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-crypto-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-logging-1.1.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jcip-annotations-1.0-1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-configuration2-2.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-security-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-asn1-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsch-0.1.54.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/paranamer-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/curator-client-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-servlet-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-compress-1.4.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-xml-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-lang3-3.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-http-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/metrics-core-3.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-codec-1.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/javax.servlet-api-3.1.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-io-2.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-server-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jaxb-impl-2.2.3-1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/asm-5.0.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-databind-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-jaxrs-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-core-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsp-api-2.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-core-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-core-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/hadoop-auth-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-server-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-xc-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-net-3.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-simplekdc-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/protobuf-java-2.5.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-annotations-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-collections-3.2.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jaxb-api-2.2.11.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/slf4j-api-1.7.25.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-identity-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-math3-3.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/httpcore-4.4.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/nimbus-jose-jwt-4.41.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/slf4j-log4j12-1.7.25.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/token-provider-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-pkix-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/re2j-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/json-smart-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-xdr-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/mockito-all-1.8.5.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-client-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/curator-recipes-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/snappy-java-1.0.5.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/xz-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/zookeeper-3.4.9.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/netty-3.10.5.Final.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-util-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsr305-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/hamcrest-core-1.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/httpclient-4.5.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-admin-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-core-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/guava-11.0.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jettison-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/stax2-api-3.1.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/gson-2.2.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/curator-framework-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-beanutils-1.9.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-mapper-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/log4j-1.2.17.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-webapp-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/hadoop-aws-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/aws-java-sdk-bundle-1.11.199.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-nfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-kms-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-common-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-util-ajax-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-cli-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-io-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-server-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/avro-1.7.7.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-servlet-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-config-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-json-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-common-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/htrace-core4-4.1.0-incubating.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-lang-2.6.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/accessors-smart-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jsr311-api-1.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/hadoop-annotations-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/woodstox-core-5.0.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-crypto-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-logging-1.1.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jcip-annotations-1.0-1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-configuration2-2.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-security-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-asn1-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jsch-0.1.54.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/paranamer-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/curator-client-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-servlet-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-compress-1.4.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-xml-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-lang3-3.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-http-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-codec-1.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/javax.servlet-api-3.1.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/okio-1.4.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-io-2.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-server-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jaxb-impl-2.2.3-1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/asm-5.0.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-databind-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-jaxrs-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-core-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-core-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-core-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/hadoop-auth-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-server-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-xc-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-net-3.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-simplekdc-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/protobuf-java-2.5.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/netty-all-4.0.23.Final.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-annotations-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-collections-3.2.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jaxb-api-2.2.11.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-identity-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/okhttp-2.4.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/leveldbjni-all-1.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-math3-3.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/httpcore-4.4.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/nimbus-jose-jwt-4.41.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/token-provider-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-pkix-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/re2j-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/json-smart-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-xdr-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-client-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/curator-recipes-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/snappy-java-1.0.5.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/xz-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/zookeeper-3.4.9.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/netty-3.10.5.Final.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-daemon-1.0.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-util-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jsr305-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/httpclient-4.5.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-admin-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/json-simple-1.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-core-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/guava-11.0.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jettison-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/stax2-api-3.1.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/gson-2.2.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/curator-framework-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-beanutils-1.9.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-mapper-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/log4j-1.2.17.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-webapp-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-client-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-httpfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-native-client-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-nfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-native-client-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-client-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-app-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-examples-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-hs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-jobclient-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-shuffle-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-core-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-jobclient-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-nativetask-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-hs-plugins-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/HikariCP-java7-2.4.12.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/java-util-1.9.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/aopalliance-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-math-2.2.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/javax.inject-1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/metrics-core-2.2.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-server-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-prefix-tree-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-hadoop-compat-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/htrace-core-3.1.0-incubating.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-el-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-procedure-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-annotations-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/disruptor-3.3.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jackson-jaxrs-base-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-protocol-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-client-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/guice-4.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jasper-runtime-5.5.23.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jackson-jaxrs-json-provider-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jcodings-1.0.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/metrics-core-3.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-csv-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jasper-compiler-5.5.23.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-common-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/json-io-2.5.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jersey-guice-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-httpclient-3.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/findbugs-annotations-1.3.9-1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/fst-2.50.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jsp-2.1-6.1.14.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/guice-servlet-4.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-hadoop2-compat-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/ehcache-3.3.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/mssql-jdbc-6.2.1.jre7.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jersey-client-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/geronimo-jcache_1.0_spec-1.0-alpha-1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jsp-api-2.1-6.1.14.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jamon-runtime-2.4.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/servlet-api-2.5-6.1.14.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jackson-module-jaxb-annotations-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/joni-2.1.2.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-applicationhistoryservice-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-tests-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-nodemanager-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-sharedcachemanager-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-api-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-applications-unmanaged-am-launcher-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-resourcemanager-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yar\u001b[0m\n\u001b[34mn/hadoop-yarn-applications-distributedshell-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-web-proxy-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-router-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-registry-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-timelineservice-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-timelineservice-hbase-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-client-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-timelineservice-hbase-tests-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-timeline-pluginstorage-3.0.0.jar\u001b[0m\n\u001b[34mSTARTUP_MSG: build = https://git-wip-us.apache.org/repos/asf/hadoop.git -r c25427ceca461ee979d30edd7a4b0f50718e6533; compiled by 'andrew' on 2017-12-08T19:16Z\u001b[0m\n\u001b[34mSTARTUP_MSG: java = 1.8.0_222\u001b[0m\n\u001b[34m************************************************************/\u001b[0m\n\u001b[34m2020-02-04 20:39:45,371 INFO namenode.NameNode: registered UNIX signal handlers for [TERM, HUP, INT]\u001b[0m\n\u001b[34m2020-02-04 20:39:45,374 INFO namenode.NameNode: createNameNode [-format, -force]\u001b[0m\n\u001b[34mFormatting using clusterid: CID-36b037a8-86ad-49f6-a83a-2b0f3bd75e46\u001b[0m\n\u001b[34m2020-02-04 20:39:45,740 INFO namenode.FSEditLog: Edit logging is async:true\u001b[0m\n\u001b[34m2020-02-04 20:39:45,751 INFO namenode.FSNamesystem: KeyProvider: null\u001b[0m\n\u001b[34m2020-02-04 20:39:45,752 INFO namenode.FSNamesystem: fsLock is fair: true\u001b[0m\n\u001b[34m2020-02-04 20:39:45,754 INFO namenode.FSNamesystem: Detailed lock hold time metrics enabled: false\u001b[0m\n\u001b[34m2020-02-04 20:39:45,758 INFO namenode.FSNamesystem: fsOwner = root (auth:SIMPLE)\u001b[0m\n\u001b[34m2020-02-04 20:39:45,758 INFO namenode.FSNamesystem: supergroup = supergroup\u001b[0m\n\u001b[34m2020-02-04 20:39:45,758 INFO namenode.FSNamesystem: isPermissionEnabled = true\u001b[0m\n\u001b[34m2020-02-04 20:39:45,758 INFO namenode.FSNamesystem: HA Enabled: false\u001b[0m\n\u001b[34m2020-02-04 20:39:45,789 INFO common.Util: dfs.datanode.fileio.profiling.sampling.percentage set to 0. Disabling file IO profiling\u001b[0m\n\u001b[34m2020-02-04 20:39:45,801 INFO blockmanagement.DatanodeManager: dfs.block.invalidate.limit: configured=1000, counted=60, effected=1000\u001b[0m\n\u001b[34m2020-02-04 20:39:45,801 INFO blockmanagement.DatanodeManager: dfs.namenode.datanode.registration.ip-hostname-check=true\u001b[0m\n\u001b[34m2020-02-04 20:39:45,804 INFO blockmanagement.BlockManager: dfs.namenode.startup.delay.block.deletion.sec is set to 000:00:00:00.000\u001b[0m\n\u001b[34m2020-02-04 20:39:45,805 INFO blockmanagement.BlockManager: The block deletion will start around 2020 Feb 04 20:39:45\u001b[0m\n\u001b[34m2020-02-04 20:39:45,806 INFO util.GSet: Computing capacity for map BlocksMap\u001b[0m\n\u001b[34m2020-02-04 20:39:45,806 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 20:39:45,807 INFO util.GSet: 2.0% max memory 6.5 GB = 133.7 MB\u001b[0m\n\u001b[34m2020-02-04 20:39:45,807 INFO util.GSet: capacity = 2^24 = 16777216 entries\u001b[0m\n\u001b[34m2020-02-04 20:39:45,858 INFO blockmanagement.BlockManager: dfs.block.access.token.enable = false\u001b[0m\n\u001b[34m2020-02-04 20:39:45,861 INFO Configuration.deprecation: No unit for dfs.namenode.safemode.extension(30000) assuming MILLISECONDS\u001b[0m\n\u001b[34m2020-02-04 20:39:45,861 INFO blockmanagement.BlockManagerSafeMode: dfs.namenode.safemode.threshold-pct = 0.9990000128746033\u001b[0m\n\u001b[34m2020-02-04 20:39:45,861 INFO blockmanagement.BlockManagerSafeMode: dfs.namenode.safemode.min.datanodes = 0\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManagerSafeMode: dfs.namenode.safemode.extension = 30000\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManager: defaultReplication = 3\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManager: maxReplication = 512\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManager: minReplication = 1\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManager: maxReplicationStreams = 2\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManager: redundancyRecheckInterval = 3000ms\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManager: encryptDataTransfer = false\u001b[0m\n\u001b[34m2020-02-04 20:39:45,862 INFO blockmanagement.BlockManager: maxNumBlocksToLog = 1000\u001b[0m\n\u001b[34m2020-02-04 20:39:45,891 INFO util.GSet: Computing capacity for map INodeMap\u001b[0m\n\u001b[34m2020-02-04 20:39:45,891 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 20:39:45,891 INFO util.GSet: 1.0% max memory 6.5 GB = 66.8 MB\u001b[0m\n\u001b[34m2020-02-04 20:39:45,891 INFO util.GSet: capacity = 2^23 = 8388608 entries\u001b[0m\n\u001b[34m2020-02-04 20:39:45,909 INFO namenode.FSDirectory: ACLs enabled? false\u001b[0m\n\u001b[34m2020-02-04 20:39:45,909 INFO namenode.FSDirectory: POSIX ACL inheritance enabled? true\u001b[0m\n\u001b[34m2020-02-04 20:39:45,909 INFO namenode.FSDirectory: XAttrs enabled? true\u001b[0m\n\u001b[34m2020-02-04 20:39:45,909 INFO namenode.NameNode: Caching file names occurring more than 10 times\u001b[0m\n\u001b[34m2020-02-04 20:39:45,914 INFO snapshot.SnapshotManager: Loaded config captureOpenFiles: false, skipCaptureAccessTimeOnlyChange: false, snapshotDiffAllowSnapRootDescendant: true\u001b[0m\n\u001b[34m2020-02-04 20:39:45,917 INFO util.GSet: Computing capacity for map cachedBlocks\u001b[0m\n\u001b[34m2020-02-04 20:39:45,917 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 20:39:45,918 INFO util.GSet: 0.25% max memory 6.5 GB = 16.7 MB\u001b[0m\n\u001b[34m2020-02-04 20:39:45,918 INFO util.GSet: capacity = 2^21 = 2097152 entries\u001b[0m\n\u001b[34m2020-02-04 20:39:45,924 INFO metrics.TopMetrics: NNTop conf: dfs.namenode.top.window.num.buckets = 10\u001b[0m\n\u001b[34m2020-02-04 20:39:45,924 INFO metrics.TopMetrics: NNTop conf: dfs.namenode.top.num.users = 10\u001b[0m\n\u001b[34m2020-02-04 20:39:45,924 INFO metrics.TopMetrics: NNTop conf: dfs.namenode.top.windows.minutes = 1,5,25\u001b[0m\n\u001b[34m2020-02-04 20:39:45,928 INFO namenode.FSNamesystem: Retry cache on namenode is enabled\u001b[0m\n\u001b[34m2020-02-04 20:39:45,928 INFO namenode.FSNamesystem: Retry cache will use 0.03 of total heap and retry cache entry expiry time is 600000 millis\u001b[0m\n\u001b[34m2020-02-04 20:39:45,929 INFO util.GSet: Computing capacity for map NameNodeRetryCache\u001b[0m\n\u001b[34m2020-02-04 20:39:45,929 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 20:39:45,929 INFO util.GSet: 0.029999999329447746% max memory 6.5 GB = 2.0 MB\u001b[0m\n\u001b[34m2020-02-04 20:39:45,929 INFO util.GSet: capacity = 2^18 = 262144 entries\u001b[0m\n\u001b[34m2020-02-04 20:39:45,948 INFO namenode.FSImage: Allocated new BlockPoolId: BP-68915925-10.0.188.93-1580848785943\u001b[0m\n\u001b[34m2020-02-04 20:39:45,960 INFO common.Storage: Storage directory /opt/amazon/hadoop/hdfs/namenode has been successfully formatted.\u001b[0m\n\u001b[34m2020-02-04 20:39:45,967 INFO namenode.FSImageFormatProtobuf: Saving image file /opt/amazon/hadoop/hdfs/namenode/current/fsimage.ckpt_0000000000000000000 using no compression\u001b[0m\n\u001b[34m2020-02-04 20:39:46,044 INFO namenode.FSImageFormatProtobuf: Image file /opt/amazon/hadoop/hdfs/namenode/current/fsimage.ckpt_0000000000000000000 of size 386 bytes saved in 0 seconds.\u001b[0m\n\u001b[34m2020-02-04 20:39:46,054 INFO namenode.NNStorageRetentionManager: Going to retain 1 images with txid >= 0\u001b[0m\n\u001b[34m2020-02-04 20:39:46,058 INFO namenode.NameNode: SHUTDOWN_MSG: \u001b[0m\n\u001b[34m/************************************************************\u001b[0m\n\u001b[34mSHUTDOWN_MSG: Shutting down NameNode at algo-1/10.0.188.93\u001b[0m\n\u001b[34m************************************************************/\u001b[0m\n\u001b[34m2020-02-04 20:39:46,070 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/hdfs --daemon start namenode\u001b[0m\n\u001b[34m2020-02-04 20:39:48,111 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/hdfs --daemon start namenode, return code 1\u001b[0m\n\u001b[34m2020-02-04 20:39:48,111 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/hdfs --daemon start datanode\u001b[0m\n\u001b[34m2020-02-04 20:39:50,152 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/hdfs --daemon start datanode, return code 1\u001b[0m\n\u001b[34m2020-02-04 20:39:50,153 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/yarn --daemon start resourcemanager\u001b[0m\n\u001b[34mWARNING: YARN_LOG_DIR has been replaced by HADOOP_LOG_DIR. Using value of YARN_LOG_DIR.\u001b[0m\n\u001b[34mWARNING: /var/log/yarn/ does not exist. Creating.\u001b[0m\n\u001b[34m2020-02-04 20:39:52,195 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/yarn --daemon start resourcemanager, return code 1\u001b[0m\n\u001b[34m2020-02-04 20:39:52,195 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/yarn --daemon start nodemanager\u001b[0m\n\u001b[34mWARNING: YARN_LOG_DIR has been replaced by HADOOP_LOG_DIR. Using value of YARN_LOG_DIR.\u001b[0m\n\u001b[34m2020-02-04 20:39:54,237 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/yarn --daemon start nodemanager, return code 1\u001b[0m\n\u001b[34m2020-02-04 20:39:54,237 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/yarn --daemon start proxyserver\u001b[0m\n\u001b[34mWARNING: YARN_LOG_DIR has been replaced by HADOOP_LOG_DIR. Using value of YARN_LOG_DIR.\u001b[0m\n\u001b[34mERROR: Cannot set priority of proxyserver process 808\u001b[0m\n\u001b[34m2020-02-04 20:39:56,299 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/yarn --daemon start proxyserver, return code 1\u001b[0m\n\u001b[34m2020-02-04 20:39:56,300 - DefaultDataAnalyzer - INFO - Total number of hosts in the cluster: 1\u001b[0m\n\u001b[34m2020-02-04 20:40:06,304 - DefaultDataAnalyzer - INFO - Running command: bin/spark-submit --master yarn --deploy-mode client --conf spark.hadoop.fs.s3a.aws.credentials.provider=org.apache.hadoop.fs.s3a.SimpleAWSCredentialsProvider --conf spark.serializer=org.apache.spark.serializer.KryoSerializer /opt/amazon/sagemaker-data-analyzer-1.0-jar-with-dependencies.jar --analytics_input /tmp/spark_job_config.json\u001b[0m\n\u001b[34m2020-02-04 20:40:07 WARN NativeCodeLoader:60 - Unable to load native-hadoop library for your platform... using builtin-java classes where applicable\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO Main:28 - Start analyzing with args: --analytics_input /tmp/spark_job_config.json\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO Main:31 - Analytics input path: DataAnalyzerParams(/tmp/spark_job_config.json)\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO FileUtil:66 - Read file from path /tmp/spark_job_config.json.\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO DataAnalyzer:30 - Analytics input: AnalyticsInput(/opt/ml/processing/input/baseline_dataset_input,DataSetFormat(Some(CSV(Some(true),None,None,None,None)),None,None),None,None,None,None,/opt/ml/processing/output,None,None,/opt/ml/output/metrics/cloudwatch,Some(Disabled),None,None,/opt/ml/output/message)\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SparkContext:54 - Running Spark version 2.3.1\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SparkContext:54 - Submitted application: SageMakerDataAnalyzer\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SecurityManager:54 - Changing view acls to: root\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SecurityManager:54 - Changing modify acls to: root\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SecurityManager:54 - Changing view acls groups to: \u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SecurityManager:54 - Changing modify acls groups to: \u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SecurityManager:54 - SecurityManager: authentication disabled; ui acls disabled; users with view permissions: Set(root); groups with view permissions: Set(); users with modify permissions: Set(root); groups with modify permissions: Set()\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO Utils:54 - Successfully started service 'sparkDriver' on port 42557.\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SparkEnv:54 - Registering MapOutputTracker\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SparkEnv:54 - Registering BlockManagerMaster\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO BlockManagerMasterEndpoint:54 - Using org.apache.spark.storage.DefaultTopologyMapper for getting topology information\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO BlockManagerMasterEndpoint:54 - BlockManagerMasterEndpoint up\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO DiskBlockManager:54 - Created local directory at /tmp/blockmgr-23ddd2aa-20a2-4f5e-9411-eaa31167537d\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO MemoryStore:54 - MemoryStore started with capacity 1458.6 MB\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SparkEnv:54 - Registering OutputCommitCoordinator\u001b[0m\n\u001b[34m2020-02-04 20:40:07 INFO SparkContext:54 - Added JAR file:/opt/amazon/sagemaker-data-analyzer-1.0-jar-with-dependencies.jar at spark://10.0.188.93:42557/jars/sagemaker-data-analyzer-1.0-jar-with-dependencies.jar with timestamp 1580848807936\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO RMProxy:133 - Connecting to ResourceManager at /10.0.188.93:8032\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO Client:54 - Requesting a new application from cluster with 1 NodeManagers\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO Configuration:2636 - resource-types.xml not found\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO ResourceUtils:427 - Unable to find 'resource-types.xml'.\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO Client:54 - Verifying our application has not requested more than the maximum memory capability of the cluster (31144 MB per container)\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO Client:54 - Will allocate AM container, with 896 MB memory including 384 MB overhead\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO Client:54 - Setting up container launch context for our AM\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO Client:54 - Setting up the launch environment for our AM container\u001b[0m\n\u001b[34m2020-02-04 20:40:08 INFO Client:54 - Preparing resources for our AM container\u001b[0m\n\u001b[34m2020-02-04 20:40:09 WARN Client:66 - Neither spark.yarn.jars nor spark.yarn.archive is set, falling back to uploading libraries under SPARK_HOME.\u001b[0m\n\u001b[34m2020-02-04 20:40:10 INFO Client:54 - Uploading resource file:/tmp/spark-0d5ac611-dffa-4128-8a53-56c642d1822e/__spark_libs__7496089848976976159.zip -> hdfs://10.0.188.93/user/root/.sparkStaging/application_1580848791166_0001/__spark_libs__7496089848976976159.zip\u001b[0m\n\u001b[34m2020-02-04 20:40:11 INFO Client:54 - Uploading resource file:/tmp/spark-0d5ac611-dffa-4128-8a53-56c642d1822e/__spark_conf__2601539848326300604.zip -> hdfs://10.0.188.93/user/root/.sparkStaging/application_1580848791166_0001/__spark_conf__.zip\u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO SecurityManager:54 - Changing view acls to: root\u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO SecurityManager:54 - Changing modify acls to: root\u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO SecurityManager:54 - Changing view acls groups to: \u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO SecurityManager:54 - Changing modify acls groups to: \u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO SecurityManager:54 - SecurityManager: authentication disabled; ui acls disabled; users with view permissions: Set(root); groups with view permissions: Set(); users with modify permissions: Set(root); groups with modify permissions: Set()\u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO Client:54 - Submitting application application_1580848791166_0001 to ResourceManager\u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO YarnClientImpl:310 - Submitted application application_1580848791166_0001\u001b[0m\n\u001b[34m2020-02-04 20:40:12 INFO SchedulerExtensionServices:54 - Starting Yarn extension services with app application_1580848791166_0001 and attemptId None\u001b[0m\n\u001b[34m2020-02-04 20:40:13 INFO Client:54 - Application report for application_1580848791166_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 20:40:13 INFO Client:54 - \u001b[0m\n\u001b[34m#011 client token: N/A\u001b[0m\n\u001b[34m#011 diagnostics: AM container is launched, waiting for AM container to Register with RM\u001b[0m\n\u001b[34m#011 ApplicationMaster host: N/A\u001b[0m\n\u001b[34m#011 ApplicationMaster RPC port: -1\u001b[0m\n\u001b[34m#011 queue: default\u001b[0m\n\u001b[34m#011 start time: 1580848812134\u001b[0m\n\u001b[34m#011 final status: UNDEFINED\u001b[0m\n\u001b[34m#011 tracking URL: http://algo-1:8088/proxy/application_1580848791166_0001/\u001b[0m\n\u001b[34m#011 user: root\u001b[0m\n\u001b[34m2020-02-04 20:40:14 INFO Client:54 - Application report for application_1580848791166_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 20:40:15 INFO Client:54 - Application report for application_1580848791166_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 20:40:16 INFO YarnClientSchedulerBackend:54 - Add WebUI Filter. org.apache.hadoop.yarn.server.webproxy.amfilter.AmIpFilter, Map(PROXY_HOSTS -> algo-1, PROXY_URI_BASES -> http://algo-1:8088/proxy/application_1580848791166_0001), /proxy/application_1580848791166_0001\u001b[0m\n\u001b[34m2020-02-04 20:40:16 INFO Client:54 - Application report for application_1580848791166_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 20:40:16 INFO YarnSchedulerBackend$YarnSchedulerEndpoint:54 - ApplicationMaster registered as NettyRpcEndpointRef(spark-client://YarnAM)\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO Client:54 - Application report for application_1580848791166_0001 (state: RUNNING)\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO Client:54 - \u001b[0m\n\u001b[34m#011 client token: N/A\u001b[0m\n\u001b[34m#011 diagnostics: N/A\u001b[0m\n\u001b[34m#011 ApplicationMaster host: 10.0.188.93\u001b[0m\n\u001b[34m#011 ApplicationMaster RPC port: 0\u001b[0m\n\u001b[34m#011 queue: default\u001b[0m\n\u001b[34m#011 start time: 1580848812134\u001b[0m\n\u001b[34m#011 final status: UNDEFINED\u001b[0m\n\u001b[34m#011 tracking URL: http://algo-1:8088/proxy/application_1580848791166_0001/\u001b[0m\n\u001b[34m#011 user: root\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO YarnClientSchedulerBackend:54 - Application application_1580848791166_0001 has started running.\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO Utils:54 - Successfully started service 'org.apache.spark.network.netty.NettyBlockTransferService' on port 46703.\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO NettyBlockTransferService:54 - Server created on 10.0.188.93:46703\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO BlockManager:54 - Using org.apache.spark.storage.RandomBlockReplicationPolicy for block replication policy\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO BlockManagerMaster:54 - Registering BlockManager BlockManagerId(driver, 10.0.188.93, 46703, None)\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO BlockManagerMasterEndpoint:54 - Registering block manager 10.0.188.93:46703 with 1458.6 MB RAM, BlockManagerId(driver, 10.0.188.93, 46703, None)\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO BlockManagerMaster:54 - Registered BlockManager BlockManagerId(driver, 10.0.188.93, 46703, None)\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO BlockManager:54 - Initialized BlockManager: BlockManagerId(driver, 10.0.188.93, 46703, None)\u001b[0m\n\u001b[34m2020-02-04 20:40:17 INFO log:192 - Logging initialized @10946ms\u001b[0m\n\u001b[34m2020-02-04 20:40:18 INFO YarnSchedulerBackend$YarnDriverEndpoint:54 - Registered executor NettyRpcEndpointRef(spark-client://Executor) (10.0.188.93:45832) with ID 1\u001b[0m\n\u001b[34m2020-02-04 20:40:18 INFO BlockManagerMasterEndpoint:54 - Registering block manager algo-1:34693 with 11.7 GB RAM, BlockManagerId(1, algo-1, 34693, None)\u001b[0m\n\u001b[34m2020-02-04 20:40:37 INFO YarnClientSchedulerBackend:54 - SchedulerBackend is ready for scheduling beginning after waiting maxRegisteredResourcesWaitingTime: 30000(ms)\u001b[0m\n\u001b[34m2020-02-04 20:40:38 WARN SparkContext:66 - Spark is not running in local mode, therefore the checkpoint directory must not be on the local filesystem. Directory '/tmp' appears to be on the local filesystem.\u001b[0m\n\u001b[34m2020-02-04 20:40:38 INFO SharedState:54 - Setting hive.metastore.warehouse.dir ('null') to the value of spark.sql.warehouse.dir ('file:/usr/spark-2.3.1/spark-warehouse').\u001b[0m\n\u001b[34m2020-02-04 20:40:38 INFO SharedState:54 - Warehouse path is 'file:/usr/spark-2.3.1/spark-warehouse'.\u001b[0m\n\u001b[34m2020-02-04 20:40:38 INFO StateStoreCoordinatorRef:54 - Registered StateStoreCoordinator endpoint\u001b[0m\n\u001b[34m2020-02-04 20:40:38 INFO DatasetReader:90 - Files to process:List(file:///opt/ml/processing/input/baseline_dataset_input/6.csv, file:///opt/ml/processing/input/baseline_dataset_input/0.csv, file:///opt/ml/processing/input/baseline_dataset_input/2.csv, file:///opt/ml/processing/input/baseline_dataset_input/3.csv, file:///opt/ml/processing/input/baseline_dataset_input/5.csv, file:///opt/ml/processing/input/baseline_dataset_input/7.csv, file:///opt/ml/processing/input/baseline_dataset_input/8.csv, file:///opt/ml/processing/input/baseline_dataset_input/1.csv, file:///opt/ml/processing/input/baseline_dataset_input/9.csv, file:///opt/ml/processing/input/baseline_dataset_input/4.csv)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO FileSourceStrategy:54 - Post-Scan Filters: (length(trim(value#0, None)) > 0)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO FileSourceStrategy:54 - Output Data Schema: struct<value: string>\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO CodeGenerator:54 - Code generated in 142.597842 ms\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO CodeGenerator:54 - Code generated in 19.170156 ms\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO MemoryStore:54 - Block broadcast_0 stored as values in memory (estimated size 429.5 KB, free 1458.2 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO MemoryStore:54 - Block broadcast_0_piece0 stored as bytes in memory (estimated size 38.2 KB, free 1458.1 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO BlockManagerInfo:54 - Added broadcast_0_piece0 in memory on 10.0.188.93:46703 (size: 38.2 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO SparkContext:54 - Created broadcast 0 from csv at DatasetReader.scala:51\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 47316495 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO SparkContext:54 - Starting job: csv at DatasetReader.scala:51\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO DAGScheduler:54 - Got job 0 (csv at DatasetReader.scala:51) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO DAGScheduler:54 - Final stage: ResultStage 0 (csv at DatasetReader.scala:51)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO DAGScheduler:54 - Submitting ResultStage 0 (MapPartitionsRDD[5] at csv at DatasetReader.scala:51), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO MemoryStore:54 - Block broadcast_1 stored as values in memory (estimated size 9.4 KB, free 1458.1 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO MemoryStore:54 - Block broadcast_1_piece0 stored as bytes in memory (estimated size 4.5 KB, free 1458.1 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO BlockManagerInfo:54 - Added broadcast_1_piece0 in memory on 10.0.188.93:46703 (size: 4.5 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO SparkContext:54 - Created broadcast 1 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 0 (MapPartitionsRDD[5] at csv at DatasetReader.scala:51) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO YarnScheduler:54 - Adding task set 0.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 20:40:39 INFO TaskSetManager:54 - Starting task 0.0 in stage 0.0 (TID 0, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8426 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:40:40 INFO BlockManagerInfo:54 - Added broadcast_1_piece0 in memory on algo-1:34693 (size: 4.5 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:40 INFO BlockManagerInfo:54 - Added broadcast_0_piece0 in memory on algo-1:34693 (size: 38.2 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO TaskSetManager:54 - Finished task 0.0 in stage 0.0 (TID 0) in 1443 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO YarnScheduler:54 - Removed TaskSet 0.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - ResultStage 0 (csv at DatasetReader.scala:51) finished in 1.502 s\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - Job 0 finished: csv at DatasetReader.scala:51, took 1.534367 s\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceStrategy:54 - Post-Scan Filters: \u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceStrategy:54 - Output Data Schema: struct<value: string>\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO CodeGenerator:54 - Code generated in 5.886757 ms\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO MemoryStore:54 - Block broadcast_2 stored as values in memory (estimated size 429.5 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO MemoryStore:54 - Block broadcast_2_piece0 stored as bytes in memory (estimated size 38.2 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO BlockManagerInfo:54 - Added broadcast_2_piece0 in memory on 10.0.188.93:46703 (size: 38.2 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO SparkContext:54 - Created broadcast 2 from csv at DatasetReader.scala:51\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 47316495 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceStrategy:54 - Post-Scan Filters: \u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceStrategy:54 - Output Data Schema: struct<Target: string, Dst Port: string, Flow Duration: string, Tot Fwd Pkts: string, Tot Bwd Pkts: string ... 83 more fields>\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO MemoryStore:54 - Block broadcast_3 stored as values in memory (estimated size 429.5 KB, free 1457.3 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO MemoryStore:54 - Block broadcast_3_piece0 stored as bytes in memory (estimated size 38.2 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO BlockManagerInfo:54 - Added broadcast_3_piece0 in memory on 10.0.188.93:46703 (size: 38.2 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO SparkContext:54 - Created broadcast 3 from cache at DataAnalyzer.scala:90\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 47316495 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 20:40:41 WARN Utils:66 - Truncated the string representation of a plan since it was too large. This behavior can be adjusted by setting 'spark.debug.maxToStringFields' in SparkEnv.conf.\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO CodeGenerator:54 - Code generated in 80.700884 ms\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO CodeGenerator:54 - Code generated in 45.486266 ms\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO SparkContext:54 - Starting job: head at DataAnalyzer.scala:93\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - Got job 1 (head at DataAnalyzer.scala:93) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - Final stage: ResultStage 1 (head at DataAnalyzer.scala:93)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - Submitting ResultStage 1 (MapPartitionsRDD[19] at head at DataAnalyzer.scala:93), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO MemoryStore:54 - Block broadcast_4 stored as values in memory (estimated size 101.8 KB, free 1457.1 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO MemoryStore:54 - Block broadcast_4_piece0 stored as bytes in memory (estimated size 32.5 KB, free 1457.1 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO BlockManagerInfo:54 - Added broadcast_4_piece0 in memory on 10.0.188.93:46703 (size: 32.5 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO SparkContext:54 - Created broadcast 4 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 1 (MapPartitionsRDD[19] at head at DataAnalyzer.scala:93) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO YarnScheduler:54 - Adding task set 1.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO TaskSetManager:54 - Starting task 0.0 in stage 1.0 (TID 1, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8426 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO BlockManagerInfo:54 - Added broadcast_4_piece0 in memory on algo-1:34693 (size: 32.5 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:41 INFO BlockManagerInfo:54 - Added broadcast_3_piece0 in memory on algo-1:34693 (size: 38.2 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:46 INFO BlockManagerInfo:54 - Added rdd_13_0 in memory on algo-1:34693 (size: 30.9 MB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO TaskSetManager:54 - Finished task 0.0 in stage 1.0 (TID 1) in 5311 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO YarnScheduler:54 - Removed TaskSet 1.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO DAGScheduler:54 - ResultStage 1 (head at DataAnalyzer.scala:93) finished in 5.347 s\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO DAGScheduler:54 - Job 1 finished: head at DataAnalyzer.scala:93, took 5.352143 s\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 45\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 11\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 33\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 0\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 57\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 19\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 32\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 50\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 64\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 63\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 24\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 49\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 43\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 28\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 26\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 53\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 10\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 4\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 60\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 12\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO BlockManagerInfo:54 - Removed broadcast_0_piece0 on 10.0.188.93:46703 in memory (size: 38.2 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO BlockManagerInfo:54 - Removed broadcast_0_piece0 on algo-1:34693 in memory (size: 38.2 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 34\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 48\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 27\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 62\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 59\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 9\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO BlockManagerInfo:54 - Removed broadcast_4_piece0 on 10.0.188.93:46703 in memory (size: 32.5 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO BlockManagerInfo:54 - Removed broadcast_4_piece0 on algo-1:34693 in memory (size: 32.5 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 47\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 67\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 17\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 42\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 5\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 35\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 46\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 6\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 21\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 58\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 65\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 1\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 69\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 30\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 44\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 7\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO BlockManagerInfo:54 - Removed broadcast_2_piece0 on 10.0.188.93:46703 in memory (size: 38.2 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 52\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 66\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 3\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 8\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 56\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 14\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 31\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 22\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 2\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 29\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 55\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 16\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 15\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 23\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 18\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 13\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 25\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO BlockManagerInfo:54 - Removed broadcast_1_piece0 on 10.0.188.93:46703 in memory (size: 4.5 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO BlockManagerInfo:54 - Removed broadcast_1_piece0 on algo-1:34693 in memory (size: 4.5 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 54\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 51\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 68\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 61\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO ContextCleaner:54 - Cleaned accumulator 20\u001b[0m\n\u001b[34m2020-02-04 20:40:47 INFO CodeGenerator:54 - Code generated in 61.21319 ms\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO SparkContext:54 - Starting job: collect at AnalysisRunner.scala:313\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO DAGScheduler:54 - Registering RDD 24 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO DAGScheduler:54 - Got job 2 (collect at AnalysisRunner.scala:313) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO DAGScheduler:54 - Final stage: ResultStage 3 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO DAGScheduler:54 - Parents of final stage: List(ShuffleMapStage 2)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO DAGScheduler:54 - Missing parents: List(ShuffleMapStage 2)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO DAGScheduler:54 - Submitting ShuffleMapStage 2 (MapPartitionsRDD[24] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO MemoryStore:54 - Block broadcast_5 stored as values in memory (estimated size 790.6 KB, free 1457.4 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO MemoryStore:54 - Block broadcast_5_piece0 stored as bytes in memory (estimated size 240.3 KB, free 1457.1 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO BlockManagerInfo:54 - Added broadcast_5_piece0 in memory on 10.0.188.93:46703 (size: 240.3 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO SparkContext:54 - Created broadcast 5 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO DAGScheduler:54 - Submitting 5 missing tasks from ShuffleMapStage 2 (MapPartitionsRDD[24] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0, 1, 2, 3, 4))\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO YarnScheduler:54 - Adding task set 2.0 with 5 tasks\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO TaskSetManager:54 - Starting task 0.0 in stage 2.0 (TID 2, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO TaskSetManager:54 - Starting task 1.0 in stage 2.0 (TID 3, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO TaskSetManager:54 - Starting task 2.0 in stage 2.0 (TID 4, algo-1, executor 1, partition 2, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO TaskSetManager:54 - Starting task 3.0 in stage 2.0 (TID 5, algo-1, executor 1, partition 3, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO TaskSetManager:54 - Starting task 4.0 in stage 2.0 (TID 6, algo-1, executor 1, partition 4, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:40:48 INFO BlockManagerInfo:54 - Added broadcast_5_piece0 in memory on algo-1:34693 (size: 240.3 KB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:54 INFO BlockManagerInfo:54 - Added rdd_13_3 in memory on algo-1:34693 (size: 30.7 MB, free: 11.7 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:54 INFO BlockManagerInfo:54 - Added rdd_13_1 in memory on algo-1:34693 (size: 30.8 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:54 INFO BlockManagerInfo:54 - Added rdd_13_4 in memory on algo-1:34693 (size: 30.6 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:54 INFO BlockManagerInfo:54 - Added rdd_13_2 in memory on algo-1:34693 (size: 30.7 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:40:58 INFO TaskSetManager:54 - Finished task 0.0 in stage 2.0 (TID 2) in 10732 ms on algo-1 (executor 1) (1/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO TaskSetManager:54 - Finished task 2.0 in stage 2.0 (TID 4) in 14459 ms on algo-1 (executor 1) (2/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO TaskSetManager:54 - Finished task 4.0 in stage 2.0 (TID 6) in 14496 ms on algo-1 (executor 1) (3/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO TaskSetManager:54 - Finished task 1.0 in stage 2.0 (TID 3) in 14541 ms on algo-1 (executor 1) (4/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO TaskSetManager:54 - Finished task 3.0 in stage 2.0 (TID 5) in 14570 ms on algo-1 (executor 1) (5/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO YarnScheduler:54 - Removed TaskSet 2.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO DAGScheduler:54 - ShuffleMapStage 2 (collect at AnalysisRunner.scala:313) finished in 14.608 s\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO DAGScheduler:54 - looking for newly runnable stages\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO DAGScheduler:54 - running: Set()\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO DAGScheduler:54 - waiting: Set(ResultStage 3)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO DAGScheduler:54 - failed: Set()\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO DAGScheduler:54 - Submitting ResultStage 3 (MapPartitionsRDD[27] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO MemoryStore:54 - Block broadcast_6 stored as values in memory (estimated size 971.6 KB, free 1456.2 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO MemoryStore:54 - Block broadcast_6_piece0 stored as bytes in memory (estimated size 292.8 KB, free 1455.9 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO BlockManagerInfo:54 - Added broadcast_6_piece0 in memory on 10.0.188.93:46703 (size: 292.8 KB, free: 1458.0 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO SparkContext:54 - Created broadcast 6 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 3 (MapPartitionsRDD[27] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO YarnScheduler:54 - Adding task set 3.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO TaskSetManager:54 - Starting task 0.0 in stage 3.0 (TID 7, algo-1, executor 1, partition 0, NODE_LOCAL, 7765 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO BlockManagerInfo:54 - Added broadcast_6_piece0 in memory on algo-1:34693 (size: 292.8 KB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:02 INFO MapOutputTrackerMasterEndpoint:54 - Asked to send map output locations for shuffle 0 to 10.0.188.93:45832\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO TaskSetManager:54 - Finished task 0.0 in stage 3.0 (TID 7) in 1533 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO YarnScheduler:54 - Removed TaskSet 3.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO DAGScheduler:54 - ResultStage 3 (collect at AnalysisRunner.scala:313) finished in 1.561 s\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO DAGScheduler:54 - Job 2 finished: collect at AnalysisRunner.scala:313, took 16.184545 s\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 71\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO BlockManagerInfo:54 - Removed broadcast_6_piece0 on 10.0.188.93:46703 in memory (size: 292.8 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO BlockManagerInfo:54 - Removed broadcast_6_piece0 on algo-1:34693 in memory (size: 292.8 KB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 70\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 95\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 74\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 118\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 123\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 94\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 96\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 93\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 128\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 89\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 101\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 73\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 82\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 103\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 104\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 78\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 77\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 99\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 72\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 120\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 85\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 98\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 110\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 97\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 107\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 125\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 126\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 130\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 81\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 105\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 84\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 92\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 109\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 108\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 106\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 91\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 86\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 87\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 80\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 116\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 113\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 114\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 83\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 100\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 112\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 90\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 79\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 75\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 122\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 111\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 129\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 119\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 132\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 102\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 115\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 127\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO BlockManagerInfo:54 - Removed broadcast_5_piece0 on 10.0.188.93:46703 in memory (size: 240.3 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO BlockManagerInfo:54 - Removed broadcast_5_piece0 on algo-1:34693 in memory (size: 240.3 KB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 117\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 131\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 76\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 124\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned shuffle 0\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 88\u001b[0m\n\u001b[34m2020-02-04 20:41:04 INFO ContextCleaner:54 - Cleaned accumulator 121\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO CodeGenerator:54 - Code generated in 79.496618 ms\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO SparkContext:54 - Starting job: treeReduce at KLLRunner.scala:107\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO DAGScheduler:54 - Got job 3 (treeReduce at KLLRunner.scala:107) with 5 output partitions\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO DAGScheduler:54 - Final stage: ResultStage 4 (treeReduce at KLLRunner.scala:107)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO DAGScheduler:54 - Submitting ResultStage 4 (MapPartitionsRDD[36] at treeReduce at KLLRunner.scala:107), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO MemoryStore:54 - Block broadcast_7 stored as values in memory (estimated size 144.0 KB, free 1458.0 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO MemoryStore:54 - Block broadcast_7_piece0 stored as bytes in memory (estimated size 43.9 KB, free 1458.0 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO BlockManagerInfo:54 - Added broadcast_7_piece0 in memory on 10.0.188.93:46703 (size: 43.9 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO SparkContext:54 - Created broadcast 7 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO DAGScheduler:54 - Submitting 5 missing tasks from ResultStage 4 (MapPartitionsRDD[36] at treeReduce at KLLRunner.scala:107) (first 15 tasks are for partitions Vector(0, 1, 2, 3, 4))\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO YarnScheduler:54 - Adding task set 4.0 with 5 tasks\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO TaskSetManager:54 - Starting task 0.0 in stage 4.0 (TID 8, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8426 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO TaskSetManager:54 - Starting task 1.0 in stage 4.0 (TID 9, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8426 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO TaskSetManager:54 - Starting task 2.0 in stage 4.0 (TID 10, algo-1, executor 1, partition 2, PROCESS_LOCAL, 8426 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO TaskSetManager:54 - Starting task 3.0 in stage 4.0 (TID 11, algo-1, executor 1, partition 3, PROCESS_LOCAL, 8426 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO TaskSetManager:54 - Starting task 4.0 in stage 4.0 (TID 12, algo-1, executor 1, partition 4, PROCESS_LOCAL, 8426 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:06 INFO BlockManagerInfo:54 - Added broadcast_7_piece0 in memory on algo-1:34693 (size: 43.9 KB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO BlockManagerInfo:54 - Added taskresult_11 in memory on algo-1:34693 (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO TransportClientFactory:267 - Successfully created connection to algo-1/10.0.188.93:34693 after 5 ms (0 ms spent in bootstraps)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO BlockManagerInfo:54 - Added taskresult_12 in memory on algo-1:34693 (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO BlockManagerInfo:54 - Added taskresult_9 in memory on algo-1:34693 (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO TaskSetManager:54 - Finished task 3.0 in stage 4.0 (TID 11) in 28815 ms on algo-1 (executor 1) (1/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO BlockManagerInfo:54 - Removed taskresult_11 on algo-1:34693 in memory (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO TaskSetManager:54 - Finished task 4.0 in stage 4.0 (TID 12) in 28824 ms on algo-1 (executor 1) (2/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO BlockManagerInfo:54 - Removed taskresult_12 on algo-1:34693 in memory (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO TaskSetManager:54 - Finished task 1.0 in stage 4.0 (TID 9) in 28853 ms on algo-1 (executor 1) (3/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:34 INFO BlockManagerInfo:54 - Removed taskresult_9 on algo-1:34693 in memory (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:35 INFO BlockManagerInfo:54 - Added taskresult_8 in memory on algo-1:34693 (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:35 INFO BlockManagerInfo:54 - Added taskresult_10 in memory on algo-1:34693 (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:35 INFO TaskSetManager:54 - Finished task 2.0 in stage 4.0 (TID 10) in 29027 ms on algo-1 (executor 1) (4/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:35 INFO TaskSetManager:54 - Finished task 0.0 in stage 4.0 (TID 8) in 29029 ms on algo-1 (executor 1) (5/5)\u001b[0m\n\u001b[34m2020-02-04 20:41:35 INFO YarnScheduler:54 - Removed TaskSet 4.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 20:41:35 INFO BlockManagerInfo:54 - Removed taskresult_10 on algo-1:34693 in memory (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:35 INFO BlockManagerInfo:54 - Removed taskresult_8 on algo-1:34693 in memory (size: 3.9 MB, free: 11.6 GB)\u001b[0m\n\u001b[34m2020-02-04 20:41:38 INFO DAGScheduler:54 - ResultStage 4 (treeReduce at KLLRunner.scala:107) finished in 32.139 s\u001b[0m\n\u001b[34m2020-02-04 20:41:38 INFO DAGScheduler:54 - Job 3 finished: treeReduce at KLLRunner.scala:107, took 32.503410 s\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO CodeGenerator:54 - Code generated in 217.926996 ms\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO SparkContext:54 - Starting job: collect at AnalysisRunner.scala:313\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO DAGScheduler:54 - Registering RDD 42 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO DAGScheduler:54 - Got job 4 (collect at AnalysisRunner.scala:313) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO DAGScheduler:54 - Final stage: ResultStage 6 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO DAGScheduler:54 - Parents of final stage: List(ShuffleMapStage 5)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO DAGScheduler:54 - Missing parents: List(ShuffleMapStage 5)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO DAGScheduler:54 - Submitting ShuffleMapStage 5 (MapPartitionsRDD[42] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO MemoryStore:54 - Block broadcast_8 stored as values in memory (estimated size 322.5 KB, free 1457.6 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO MemoryStore:54 - Block broadcast_8_piece0 stored as bytes in memory (estimated size 98.6 KB, free 1457.5 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO BlockManagerInfo:54 - Added broadcast_8_piece0 in memory on 10.0.188.93:46703 (size: 98.6 KB, free: 1458.4 MB)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO SparkContext:54 - Created broadcast 8 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO DAGScheduler:54 - Submitting 5 missing tasks from ShuffleMapStage 5 (MapPartitionsRDD[42] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0, 1, 2, 3, 4))\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO YarnScheduler:54 - Adding task set 5.0 with 5 tasks\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO TaskSetManager:54 - Starting task 0.0 in stage 5.0 (TID 13, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO TaskSetManager:54 - Starting task 1.0 in stage 5.0 (TID 14, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO TaskSetManager:54 - Starting task 2.0 in stage 5.0 (TID 15, algo-1, executor 1, partition 2, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO TaskSetManager:54 - Starting task 3.0 in stage 5.0 (TID 16, algo-1, executor 1, partition 3, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO TaskSetManager:54 - Starting task 4.0 in stage 5.0 (TID 17, algo-1, executor 1, partition 4, PROCESS_LOCAL, 8415 bytes)\u001b[0m\n\u001b[34m2020-02-04 20:41:40 INFO BlockManagerInfo:54 - Added broadcast_8_piece0 in memory on algo-1:34693 (size: 98.6 KB, free: 11.6 GB)\u001b[0m\n"
]
],
[
[
"Let's display the statistics that were generated by the baselining job.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nbaseline_job = my_default_monitor.latest_baselining_job\nschema_df = pd.io.json.json_normalize(baseline_job.baseline_statistics().body_dict[\"features\"])\nschema_df.head(10)",
"_____no_output_____"
]
],
[
[
"Then, we can also visualize the constraints.",
"_____no_output_____"
]
],
[
[
"constraints_df = pd.io.json.json_normalize(baseline_job.suggested_constraints().body_dict[\"features\"])\nconstraints_df.head(10)",
"_____no_output_____"
]
],
[
[
"#### Results\n\nThe baselining job has inspected the validation dataset and generated constraints and statistics, that will be used to monitor our endpoint.",
"_____no_output_____"
],
[
"## Generating violations artificially",
"_____no_output_____"
],
[
"In order to get some result relevant to monitoring analysis, we are going to generate artificially some inferences with feature values causing specific violations, and then invoke the endpoint with this data.\n\nThis requires about 2 minutes for 1000 inferences.",
"_____no_output_____"
]
],
[
[
"import time\nimport numpy as np\ndist_values = np.random.normal(1, 0.2, 1000)\n\n# Tot Fwd Pkts -> set to float (expected integer) [second feature]\n# Flow Duration -> set to empty (missing value) [third feature]\n# Fwd Pkt Len Mean -> sampled from random normal distribution [nineth feature]\n\nartificial_values = \"22,,40.3,0,0,0,0,0,{0},0.0,0,0,0.0,0.0,0.0,0.0368169318,54322832.0,0.0,54322832,54322832,54322832,54322832.0,0.0,\\\n54322832,54322832,0,0.0,0.0,0,0,0,0,0,0,40,0,0.0368169318,0.0,0,0,0.0,0.0,0.0,0,0,0,0,1,0,0,0,0.0,0.0,0.0,0.0,0.0,0.0,\\\n0.0,0.0,0.0,0.0,2,0,0,0,279,-1,0,20,0.0,0.0,0,0,0.0,0.0,0,0,23,2,2018,4,0,1,0\"\n\nfor i in range(1000):\n pred.predict(artificial_values.format(str(dist_values[i])))\n time.sleep(0.15)\n if i > 0 and i % 100 == 0 :\n print('Executed {0} inferences.'.format(i))",
"Executed 100 inferences.\nExecuted 200 inferences.\nExecuted 300 inferences.\nExecuted 400 inferences.\nExecuted 500 inferences.\nExecuted 600 inferences.\nExecuted 700 inferences.\nExecuted 800 inferences.\nExecuted 900 inferences.\n"
]
],
[
[
"## Monitoring",
"_____no_output_____"
],
[
"Once we have built the baseline for our data, we can enable endpoint monitoring by creating a monitoring schedule.\nWhen the schedule fires, a monitoring job will be kicked-off and will inspect the data captured at the endpoint with respect to the baseline; then it will generate some report files that can be used to analyze monitoring results.",
"_____no_output_____"
],
[
"### Create Monitoring Schedule",
"_____no_output_____"
],
[
"Let's create the monitoring schedule for the previously created endpoint. When we create the schedule, we can also specify two scripts that will preprocess the records before the analysis takes place and execute post-processing at the end.\nFor this example, we are not going to use a record preprocessor, and we are just specifying a post-processor that outputs some text for demo purposes.",
"_____no_output_____"
]
],
[
[
"!pygmentize postprocessor.py",
"\u001b[34mdef\u001b[39;49;00m \u001b[32mpostprocess_handler\u001b[39;49;00m():\n \u001b[34mprint\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mHello from post-proc script!\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\n"
]
],
[
[
"We copy the script to Amazon S3 and specify the path where the monitoring reports will be saved to.",
"_____no_output_____"
]
],
[
[
"import boto3\n\nmonitoring_code_prefix = '{0}/monitoring/code'.format(prefix)\nprint(monitoring_code_prefix)\n\nboto3.Session().resource('s3').Bucket(bucket_name).Object(monitoring_code_prefix + '/postprocessor.py').upload_file('postprocessor.py')\npostprocessor_path = 's3://{0}/{1}/monitoring/code/postprocessor.py'.format(bucket_name, prefix)\nprint(postprocessor_path)\n\nreports_path = 's3://{0}/{1}/monitoring/reports'.format(bucket_name, prefix)\nprint(reports_path)",
"aim362/monitoring/code\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/code/postprocessor.py\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports\n"
]
],
[
[
"Finally, we create the monitoring schedule with hourly schedule execution.",
"_____no_output_____"
]
],
[
[
"from sagemaker.model_monitor import CronExpressionGenerator\nfrom time import gmtime, strftime\n\nendpoint_name = pred.endpoint\n\nmon_schedule_name = 'nw-traffic-classification-xgb-mon-sch-' + strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime())\nmy_default_monitor.create_monitoring_schedule(\n monitor_schedule_name=mon_schedule_name,\n endpoint_input=endpoint_name,\n post_analytics_processor_script=postprocessor_path,\n output_s3_uri=reports_path,\n statistics=my_default_monitor.baseline_statistics(),\n constraints=my_default_monitor.suggested_constraints(),\n schedule_cron_expression=CronExpressionGenerator.hourly(),\n enable_cloudwatch_metrics=True\n)",
"\nCreating Monitoring Schedule with name: nw-traffic-classification-xgb-mon-sch-2020-02-04-20-59-51\n"
]
],
[
[
"### Describe Monitoring Schedule",
"_____no_output_____"
]
],
[
[
"desc_schedule_result = my_default_monitor.describe_schedule()\ndesc_schedule_result",
"_____no_output_____"
]
],
[
[
"### Delete Monitoring Schedule\n\nOnce the schedule is created, it will kick of jobs at specified intervals. Note that if you are kicking this off after creating the hourly schedule, you might find the executions empty. \nYou might have to wait till you cross the hour boundary (in UTC) to see executions kick off. Since we don't want to wait for the hour in this example we can delete the schedule and use the code in next steps to simulate what will happen when a schedule is triggered, by running an Amazon SageMaker Processing Job.",
"_____no_output_____"
]
],
[
[
"# Note: this is just for the purpose of running this example.\nmy_default_monitor.delete_monitoring_schedule()",
"\nDeleting Monitoring Schedule with name: nw-traffic-classification-xgb-mon-sch-2020-02-04-20-59-51\n"
]
],
[
[
"### Triggering execution manually\n\nIn oder to trigger the execution manually, we first get all paths to data capture, baseline statistics, baseline constraints, etc.\nThen, we use a utility fuction, defined in <a href=\"./monitoringjob_utils.py\">monitoringjob_utils.py</a>, to run the processing job.",
"_____no_output_____"
]
],
[
[
"result = s3_client.list_objects(Bucket=bucket_name, Prefix=current_endpoint_capture_prefix)\ncapture_files = ['s3://{0}/{1}'.format(bucket_name, capture_file.get(\"Key\")) for capture_file in result.get('Contents')]\n\nprint(\"Capture Files: \")\nprint(\"\\n \".join(capture_files))\n\ndata_capture_path = capture_files[len(capture_files) - 1][: capture_files[len(capture_files) - 1].rfind('/')]\nstatistics_path = baseline_results_path + '/statistics.json'\nconstraints_path = baseline_results_path + '/constraints.json'\n\nprint(data_capture_path)\nprint(postprocessor_path)\nprint(statistics_path)\nprint(constraints_path)\nprint(reports_path)",
"Capture Files: \ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/33-57-898-0815a2e8-c235-4891-bd5d-91a59c41b6ca.jsonl\n s3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/51-28-839-f722c428-f4a3-48a1-a6ab-a5f695d3c99a.jsonl\n s3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/52-28-908-839decf0-a61a-48d0-9b34-f8d50576eea6.jsonl\n s3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/53-28-991-e0c9ae91-1350-405e-9929-df8a0c8eda9b.jsonl\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/code/postprocessor.py\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/statistics.json\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/constraints.json\ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports\n"
],
[
"from monitoringjob_utils import run_model_monitor_job_processor\n\nrun_model_monitor_job_processor(region, 'ml.m5.xlarge', role, data_capture_path, statistics_path, constraints_path, reports_path,\n postprocessor_path=postprocessor_path)",
"\nJob Name: sagemaker-model-monitor-analyzer-2020-02-04-21-00-30-633\nInputs: [{'InputName': 'input_1', 'S3Input': {'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20', 'LocalPath': '/opt/ml/processing/input/endpoint/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3DataDistributionType': 'FullyReplicated', 'S3CompressionType': 'None'}}, {'InputName': 'baseline', 'S3Input': {'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/statistics.json', 'LocalPath': '/opt/ml/processing/baseline/stats', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3DataDistributionType': 'FullyReplicated', 'S3CompressionType': 'None'}}, {'InputName': 'constraints', 'S3Input': {'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/constraints.json', 'LocalPath': '/opt/ml/processing/baseline/constraints', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3DataDistributionType': 'FullyReplicated', 'S3CompressionType': 'None'}}, {'InputName': 'post_processor_script', 'S3Input': {'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/code/postprocessor.py', 'LocalPath': '/opt/ml/processing/code/postprocessing', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3DataDistributionType': 'FullyReplicated', 'S3CompressionType': 'None'}}]\nOutputs: [{'OutputName': 'result', 'S3Output': {'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20', 'LocalPath': '/opt/ml/processing/output', 'S3UploadMode': 'Continuous'}}]\n........................\u001b[34m2020-02-04 21:04:13,950 - __main__ - INFO - All params:{'ProcessingJobArn': 'arn:aws:sagemaker:us-east-1:806570384721:processing-job/sagemaker-model-monitor-analyzer-2020-02-04-21-00-30-633', 'ProcessingJobName': 'sagemaker-model-monitor-analyzer-2020-02-04-21-00-30-633', 'Environment': {'baseline_constraints': '/opt/ml/processing/baseline/constraints/constraints.json', 'baseline_statistics': '/opt/ml/processing/baseline/stats/statistics.json', 'dataset_format': '{\"sagemakerCaptureJson\":{\"captureIndexNames\":[\"endpointInput\",\"endpointOutput\"]}}', 'dataset_source': '/opt/ml/processing/input/endpoint', 'output_path': '/opt/ml/processing/output', 'post_analytics_processor_script': '/opt/ml/processing/code/postprocessing/postprocessor.py', 'publish_cloudwatch_metrics': 'Disabled'}, 'AppSpecification': {'ImageUri': '156813124566.dkr.ecr.us-east-1.amazonaws.com/sagemaker-model-monitor-analyzer', 'ContainerEntrypoint': None, 'ContainerArguments': None}, 'ProcessingInputs': [{'InputName': 'input_1', 'S3Input': {'LocalPath': '/opt/ml/processing/input/endpoint/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20', 'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/datacapture/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20', 'S3DataDistributionType': 'FullyReplicated', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3CompressionType': 'None', 'S3DownloadMode': 'StartOfJob'}}, {'InputName': 'baseline', 'S3Input': {'LocalPath': '/opt/ml/processing/baseline/stats', 'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/statistics.json', 'S3DataDistributionType': 'FullyReplicated', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3CompressionType': 'None', 'S3DownloadMode': 'StartOfJob'}}, {'InputName': 'constraints', 'S3Input': {'LocalPath': '/opt/ml/processing/baseline/constraints', 'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/constraints.json', 'S3DataDistributionType': 'FullyReplicated', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3CompressionType': 'None', 'S3DownloadMode': 'StartOfJob'}}, {'InputName': 'post_processor_script', 'S3Input': {'LocalPath': '/opt/ml/processing/code/postprocessing', 'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/code/postprocessor.py', 'S3DataDistributionType': 'FullyReplicated', 'S3DataType': 'S3Prefix', 'S3InputMode': 'File', 'S3CompressionType': 'None', 'S3DownloadMode': 'StartOfJob'}}], 'ProcessingOutputConfig': {'Outputs': [{'OutputName': 'result', 'S3Output': {'LocalPath': '/opt/ml/processing/output', 'S3Uri': 's3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20', 'S3UploadMode': 'Continuous'}}], 'KmsKeyId': None}, 'ProcessingResources': {'ClusterConfig': {'InstanceCount': 1, 'InstanceType': 'ml.m5.xlarge', 'VolumeSizeInGB': 30, 'VolumeKmsKeyId': None}}, 'RoleArn': 'arn:aws:iam::806570384721:role/service-role/AmazonSageMaker-ExecutionRole-20191201T115647', 'StoppingCondition': {'MaxRuntimeInSeconds': 86400}}\u001b[0m\n\u001b[34m2020-02-04 21:04:13,950 - __main__ - INFO - Current Environment:{'baseline_constraints': '/opt/ml/processing/baseline/constraints/constraints.json', 'baseline_statistics': '/opt/ml/processing/baseline/stats/statistics.json', 'dataset_format': '{\"sagemakerCaptureJson\":{\"captureIndexNames\":[\"endpointInput\",\"endpointOutput\"]}}', 'dataset_source': '/opt/ml/processing/input/endpoint', 'output_path': '/opt/ml/processing/output', 'post_analytics_processor_script': '/opt/ml/processing/code/postprocessing/postprocessor.py', 'publish_cloudwatch_metrics': 'Disabled'}\u001b[0m\n\u001b[34m2020-02-04 21:04:13,950 - DefaultDataAnalyzer - INFO - Performing analysis with input: {\"dataset_source\": \"/opt/ml/processing/input/endpoint\", \"dataset_format\": {\"sagemakerCaptureJson\": {\"captureIndexNames\": [\"endpointInput\", \"endpointOutput\"]}}, \"record_preprocessor_script\": null, \"post_analytics_processor_script\": \"/opt/ml/processing/code/postprocessing/postprocessor.py\", \"baseline_constraints\": \"/opt/ml/processing/baseline/constraints/constraints.json\", \"baseline_statistics\": \"/opt/ml/processing/baseline/stats/statistics.json\", \"output_path\": \"/opt/ml/processing/output\", \"start_time\": null, \"end_time\": null, \"cloudwatch_metrics_directory\": \"/opt/ml/output/metrics/cloudwatch\", \"publish_cloudwatch_metrics\": \"Disabled\", \"sagemaker_endpoint_name\": null, \"sagemaker_monitoring_schedule_name\": null, \"output_message_file\": \"/opt/ml/output/message\"}\u001b[0m\n\u001b[34m2020-02-04 21:04:13,951 - DefaultDataAnalyzer - INFO - Bootstrapping yarn\u001b[0m\n\u001b[34m2020-02-04 21:04:13,951 - bootstrap - INFO - Copy aws jars\u001b[0m\n\u001b[34m2020-02-04 21:04:14,004 - bootstrap - INFO - Copy cluster config\u001b[0m\n\u001b[34m2020-02-04 21:04:14,005 - bootstrap - INFO - Write runtime cluster config\u001b[0m\n\u001b[34m2020-02-04 21:04:14,005 - bootstrap - INFO - Resource Config is: {'current_host': 'algo-1', 'hosts': ['algo-1']}\u001b[0m\n\u001b[34m2020-02-04 21:04:14,012 - bootstrap - INFO - Finished Yarn configuration files setup.\u001b[0m\n\u001b[34m2020-02-04 21:04:14,013 - bootstrap - INFO - Starting spark process for master node algo-1\u001b[0m\n\u001b[34m2020-02-04 21:04:14,013 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/hdfs namenode -format -force\u001b[0m\n\u001b[34mWARNING: /usr/hadoop-3.0.0/logs does not exist. Creating.\u001b[0m\n\u001b[34m2020-02-04 21:04:14,442 INFO namenode.NameNode: STARTUP_MSG: \u001b[0m\n\u001b[34m/************************************************************\u001b[0m\n\u001b[34mSTARTUP_MSG: Starting NameNode\u001b[0m\n\u001b[34mSTARTUP_MSG: host = algo-1/10.0.106.106\u001b[0m\n\u001b[34mSTARTUP_MSG: args = [-format, -force]\u001b[0m\n\u001b[34mSTARTUP_MSG: version = 3.0.0\u001b[0m\n\u001b[34mSTARTUP_MSG: classpath = /usr/hadoop-3.0.0/etc/hadoop:/usr/hadoop-3.0.0/share/hadoop/common/lib/hadoop-annotations-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-asn1-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/woodstox-core-5.0.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/avro-1.7.7.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-jaxrs-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/snappy-java-1.0.5.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/curator-framework-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-io-2.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/hadoop-auth-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-client-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-admin-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-collections-3.2.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-xml-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/mockito-all-1.8.5.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-lang3-3.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/accessors-smart-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/log4j-1.2.17.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/protobuf-java-2.5.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-webapp-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/stax2-api-3.1.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-servlet-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-beanutils-1.9.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/curator-recipes-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-core-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-cli-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/gson-2.2.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-math3-3.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-crypto-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-annotations-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-pkix-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-net-3.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/slf4j-api-1.7.25.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-mapper-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/xz-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-io-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsch-0.1.54.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/guava-11.0.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jaxb-api-2.2.11.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/htrace-core4-4.1.0-incubating.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-lang-2.6.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/junit-4.11.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/netty-3.10.5.Final.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/token-provider-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/zookeeper-3.4.9.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-util-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-configuration2-2.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/re2j-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-server-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-xc-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-security-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/nimbus-jose-jwt-4.41.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-core-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-core-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/hamcrest-core-1.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/javax.servlet-api-3.1.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/asm-5.0.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsr311-api-1.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/json-smart-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-servlet-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/httpclient-4.5.2.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-logging-1.1.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsr305-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-http-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jaxb-impl-2.2.3-1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/curator-client-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/slf4j-log4j12-1.7.25.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-core-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/httpcore-4.4.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-simplekdc-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-config-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-codec-1.4.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jul-to-slf4j-1.7.25.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-identity-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerby-xdr-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-server-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/kerb-common-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jersey-json-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jcip-annotations-1.0-1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/paranamer-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jsp-api-2.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jetty-server-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jackson-databind-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/jettison-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/metrics-core-3.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/commons-compress-1.4.1.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/hadoop-aws-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/lib/aws-java-sdk-bundle-1.11.199.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-kms-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-nfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/common/hadoop-common-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/hadoop-annotations-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-asn1-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/woodstox-core-5.0.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/netty-all-4.0.23.Final.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/avro-1.7.7.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-jaxrs-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/snappy-java-1.0.5.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/curator-framework-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-io-2.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/hadoop-auth-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-client-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-util-ajax-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-admin-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-collections-3.2.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/okhttp-2.4.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-util-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-xml-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-lang3-3.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/accessors-smart-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/log4j-1.2.17.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/protobuf-java-2.5.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-webapp-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/stax2-api-3.1.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-servlet-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-beanutils-1.9.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/curator-recipes-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-core-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-cli-1.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/gson-2.2.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-math3-3.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-crypto-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-annotations-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-pkix-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-net-3.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-mapper-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/xz-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-io-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jsch-0.1.54.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/guava-11.0.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jaxb-api-2.2.11.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/htrace-core4-4.1.0-incubating.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-lang-2.6.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/netty-3.10.5.Final.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/token-provider-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/zookeeper-3.4.9.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-util-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-configuration2-2.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/re2j-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/json-simple-1.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-server-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-xc-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-daemon-1.0.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-security-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/nimbus-jose-jwt-4.41.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-core-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-core-asl-1.9.13.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/javax.servlet-api-3.1.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/asm-5.0.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jsr311-api-1.1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/json-smart-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-servlet-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/okio-1.4.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/httpclient-4.5.2.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-logging-1.1.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jsr305-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-http-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jaxb-impl-2.2.3-1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/curator-client-2.12.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-core-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/httpcore-4.4.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-simplekdc-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-config-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-codec-1.4.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-identity-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerby-xdr-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-server-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/kerb-common-1.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jersey-json-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jcip-annotations-1.0-1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/paranamer-2.3.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jetty-server-9.3.19.v20170502.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/leveldbjni-all-1.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jackson-databind-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/jettison-1.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/lib/commons-compress-1.4.1.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-native-client-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-client-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-nfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-httpfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-client-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-native-client-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/hdfs/hadoop-hdfs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-jobclient-3.0.0-tests.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-jobclient-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-app-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-nativetask-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-hs-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-core-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-examples-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-hs-plugins-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/mapreduce/hadoop-mapreduce-client-shuffle-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jackson-module-jaxb-annotations-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/fst-2.50.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jasper-compiler-5.5.23.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-procedure-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/findbugs-annotations-1.3.9-1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/mssql-jdbc-6.2.1.jre7.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jamon-runtime-2.4.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/ehcache-3.3.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jackson-jaxrs-json-provider-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-common-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-hadoop2-compat-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/metrics-core-2.2.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-math-2.2.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/javax.inject-1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jcodings-1.0.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/geronimo-jcache_1.0_spec-1.0-alpha-1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-el-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-protocol-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-annotations-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-httpclient-3.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/aopalliance-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/guice-servlet-4.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jsp-api-2.1-6.1.14.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jersey-client-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-hadoop-compat-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/joni-2.1.2.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/commons-csv-1.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/HikariCP-java7-2.4.12.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/json-io-2.5.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/disruptor-3.3.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/htrace-core-3.1.0-incubating.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-server-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/guice-4.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jackson-jaxrs-base-2.7.8.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jsp-2.1-6.1.14.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/java-util-1.9.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jersey-guice-1.19.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-client-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/jasper-runtime-5.5.23.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/servlet-api-2.5-6.1.14.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/metrics-core-3.0.1.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/lib/hbase-prefix-tree-1.2.6.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-resourcemanager-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-router-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-applications-unmanaged-am-launcher-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-applications-distributedshell-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-tests-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-nodemanager-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-common-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/had\u001b[0m\n\u001b[34moop-yarn-server-timelineservice-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-timelineservice-hbase-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-registry-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-sharedcachemanager-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-api-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-web-proxy-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-applicationhistoryservice-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-timelineservice-hbase-tests-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-client-3.0.0.jar:/usr/hadoop-3.0.0/share/hadoop/yarn/hadoop-yarn-server-timeline-pluginstorage-3.0.0.jar\u001b[0m\n\u001b[34mSTARTUP_MSG: build = https://git-wip-us.apache.org/repos/asf/hadoop.git -r c25427ceca461ee979d30edd7a4b0f50718e6533; compiled by 'andrew' on 2017-12-08T19:16Z\u001b[0m\n\u001b[34mSTARTUP_MSG: java = 1.8.0_222\u001b[0m\n\u001b[34m************************************************************/\u001b[0m\n\u001b[34m2020-02-04 21:04:14,449 INFO namenode.NameNode: registered UNIX signal handlers for [TERM, HUP, INT]\u001b[0m\n\u001b[34m2020-02-04 21:04:14,452 INFO namenode.NameNode: createNameNode [-format, -force]\u001b[0m\n\u001b[34mFormatting using clusterid: CID-df9009fc-e4ca-47c8-9d1e-4867588df247\u001b[0m\n\u001b[34m2020-02-04 21:04:14,920 INFO namenode.FSEditLog: Edit logging is async:true\u001b[0m\n\u001b[34m2020-02-04 21:04:14,935 INFO namenode.FSNamesystem: KeyProvider: null\u001b[0m\n\u001b[34m2020-02-04 21:04:14,936 INFO namenode.FSNamesystem: fsLock is fair: true\u001b[0m\n\u001b[34m2020-02-04 21:04:14,939 INFO namenode.FSNamesystem: Detailed lock hold time metrics enabled: false\u001b[0m\n\u001b[34m2020-02-04 21:04:14,944 INFO namenode.FSNamesystem: fsOwner = root (auth:SIMPLE)\u001b[0m\n\u001b[34m2020-02-04 21:04:14,944 INFO namenode.FSNamesystem: supergroup = supergroup\u001b[0m\n\u001b[34m2020-02-04 21:04:14,944 INFO namenode.FSNamesystem: isPermissionEnabled = true\u001b[0m\n\u001b[34m2020-02-04 21:04:14,944 INFO namenode.FSNamesystem: HA Enabled: false\u001b[0m\n\u001b[34m2020-02-04 21:04:14,976 INFO common.Util: dfs.datanode.fileio.profiling.sampling.percentage set to 0. Disabling file IO profiling\u001b[0m\n\u001b[34m2020-02-04 21:04:14,986 INFO blockmanagement.DatanodeManager: dfs.block.invalidate.limit: configured=1000, counted=60, effected=1000\u001b[0m\n\u001b[34m2020-02-04 21:04:14,987 INFO blockmanagement.DatanodeManager: dfs.namenode.datanode.registration.ip-hostname-check=true\u001b[0m\n\u001b[34m2020-02-04 21:04:14,990 INFO blockmanagement.BlockManager: dfs.namenode.startup.delay.block.deletion.sec is set to 000:00:00:00.000\u001b[0m\n\u001b[34m2020-02-04 21:04:14,991 INFO blockmanagement.BlockManager: The block deletion will start around 2020 Feb 04 21:04:14\u001b[0m\n\u001b[34m2020-02-04 21:04:14,992 INFO util.GSet: Computing capacity for map BlocksMap\u001b[0m\n\u001b[34m2020-02-04 21:04:14,992 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 21:04:14,993 INFO util.GSet: 2.0% max memory 3.1 GB = 63.3 MB\u001b[0m\n\u001b[34m2020-02-04 21:04:14,993 INFO util.GSet: capacity = 2^23 = 8388608 entries\u001b[0m\n\u001b[34m2020-02-04 21:04:15,014 INFO blockmanagement.BlockManager: dfs.block.access.token.enable = false\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO Configuration.deprecation: No unit for dfs.namenode.safemode.extension(30000) assuming MILLISECONDS\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManagerSafeMode: dfs.namenode.safemode.threshold-pct = 0.9990000128746033\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManagerSafeMode: dfs.namenode.safemode.min.datanodes = 0\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManagerSafeMode: dfs.namenode.safemode.extension = 30000\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManager: defaultReplication = 3\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManager: maxReplication = 512\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManager: minReplication = 1\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManager: maxReplicationStreams = 2\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManager: redundancyRecheckInterval = 3000ms\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManager: encryptDataTransfer = false\u001b[0m\n\u001b[34m2020-02-04 21:04:15,066 INFO blockmanagement.BlockManager: maxNumBlocksToLog = 1000\u001b[0m\n\u001b[34m2020-02-04 21:04:15,094 INFO util.GSet: Computing capacity for map INodeMap\u001b[0m\n\u001b[34m2020-02-04 21:04:15,095 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 21:04:15,095 INFO util.GSet: 1.0% max memory 3.1 GB = 31.6 MB\u001b[0m\n\u001b[34m2020-02-04 21:04:15,095 INFO util.GSet: capacity = 2^22 = 4194304 entries\u001b[0m\n\u001b[34m2020-02-04 21:04:15,097 INFO namenode.FSDirectory: ACLs enabled? false\u001b[0m\n\u001b[34m2020-02-04 21:04:15,097 INFO namenode.FSDirectory: POSIX ACL inheritance enabled? true\u001b[0m\n\u001b[34m2020-02-04 21:04:15,097 INFO namenode.FSDirectory: XAttrs enabled? true\u001b[0m\n\u001b[34m2020-02-04 21:04:15,097 INFO namenode.NameNode: Caching file names occurring more than 10 times\u001b[0m\n\u001b[34m2020-02-04 21:04:15,101 INFO snapshot.SnapshotManager: Loaded config captureOpenFiles: false, skipCaptureAccessTimeOnlyChange: false, snapshotDiffAllowSnapRootDescendant: true\u001b[0m\n\u001b[34m2020-02-04 21:04:15,104 INFO util.GSet: Computing capacity for map cachedBlocks\u001b[0m\n\u001b[34m2020-02-04 21:04:15,105 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 21:04:15,105 INFO util.GSet: 0.25% max memory 3.1 GB = 7.9 MB\u001b[0m\n\u001b[34m2020-02-04 21:04:15,105 INFO util.GSet: capacity = 2^20 = 1048576 entries\u001b[0m\n\u001b[34m2020-02-04 21:04:15,111 INFO metrics.TopMetrics: NNTop conf: dfs.namenode.top.window.num.buckets = 10\u001b[0m\n\u001b[34m2020-02-04 21:04:15,111 INFO metrics.TopMetrics: NNTop conf: dfs.namenode.top.num.users = 10\u001b[0m\n\u001b[34m2020-02-04 21:04:15,111 INFO metrics.TopMetrics: NNTop conf: dfs.namenode.top.windows.minutes = 1,5,25\u001b[0m\n\u001b[34m2020-02-04 21:04:15,115 INFO namenode.FSNamesystem: Retry cache on namenode is enabled\u001b[0m\n\u001b[34m2020-02-04 21:04:15,115 INFO namenode.FSNamesystem: Retry cache will use 0.03 of total heap and retry cache entry expiry time is 600000 millis\u001b[0m\n\u001b[34m2020-02-04 21:04:15,116 INFO util.GSet: Computing capacity for map NameNodeRetryCache\u001b[0m\n\u001b[34m2020-02-04 21:04:15,116 INFO util.GSet: VM type = 64-bit\u001b[0m\n\u001b[34m2020-02-04 21:04:15,116 INFO util.GSet: 0.029999999329447746% max memory 3.1 GB = 972.1 KB\u001b[0m\n\u001b[34m2020-02-04 21:04:15,117 INFO util.GSet: capacity = 2^17 = 131072 entries\u001b[0m\n\u001b[34m2020-02-04 21:04:15,136 INFO namenode.FSImage: Allocated new BlockPoolId: BP-498651783-10.0.106.106-1580850255131\u001b[0m\n\u001b[34m2020-02-04 21:04:15,147 INFO common.Storage: Storage directory /opt/amazon/hadoop/hdfs/namenode has been successfully formatted.\u001b[0m\n\u001b[34m2020-02-04 21:04:15,153 INFO namenode.FSImageFormatProtobuf: Saving image file /opt/amazon/hadoop/hdfs/namenode/current/fsimage.ckpt_0000000000000000000 using no compression\u001b[0m\n\u001b[34m2020-02-04 21:04:15,229 INFO namenode.FSImageFormatProtobuf: Image file /opt/amazon/hadoop/hdfs/namenode/current/fsimage.ckpt_0000000000000000000 of size 389 bytes saved in 0 seconds.\u001b[0m\n\u001b[34m2020-02-04 21:04:15,240 INFO namenode.NNStorageRetentionManager: Going to retain 1 images with txid >= 0\u001b[0m\n\u001b[34m2020-02-04 21:04:15,243 INFO namenode.NameNode: SHUTDOWN_MSG: \u001b[0m\n\u001b[34m/************************************************************\u001b[0m\n\u001b[34mSHUTDOWN_MSG: Shutting down NameNode at algo-1/10.0.106.106\u001b[0m\n\u001b[34m************************************************************/\u001b[0m\n\u001b[34m2020-02-04 21:04:15,253 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/hdfs --daemon start namenode\u001b[0m\n\u001b[34m2020-02-04 21:04:17,297 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/hdfs --daemon start namenode, return code 1\u001b[0m\n\u001b[34m2020-02-04 21:04:17,298 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/hdfs --daemon start datanode\u001b[0m\n\u001b[34m2020-02-04 21:04:19,356 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/hdfs --daemon start datanode, return code 1\u001b[0m\n\u001b[34m2020-02-04 21:04:19,356 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/yarn --daemon start resourcemanager\u001b[0m\n\u001b[34mWARNING: YARN_LOG_DIR has been replaced by HADOOP_LOG_DIR. Using value of YARN_LOG_DIR.\u001b[0m\n\u001b[34mWARNING: /var/log/yarn/ does not exist. Creating.\u001b[0m\n\u001b[34m2020-02-04 21:04:21,426 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/yarn --daemon start resourcemanager, return code 1\u001b[0m\n\u001b[34m2020-02-04 21:04:21,427 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/yarn --daemon start nodemanager\u001b[0m\n\u001b[34mWARNING: YARN_LOG_DIR has been replaced by HADOOP_LOG_DIR. Using value of YARN_LOG_DIR.\u001b[0m\n\u001b[34m2020-02-04 21:04:23,518 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/yarn --daemon start nodemanager, return code 1\u001b[0m\n\u001b[34m2020-02-04 21:04:23,518 - bootstrap - INFO - Running command: /usr/hadoop-3.0.0/bin/yarn --daemon start proxyserver\u001b[0m\n\u001b[34mWARNING: YARN_LOG_DIR has been replaced by HADOOP_LOG_DIR. Using value of YARN_LOG_DIR.\u001b[0m\n\u001b[34m2020-02-04 21:04:25,590 - bootstrap - INFO - Failed to run /usr/hadoop-3.0.0/bin/yarn --daemon start proxyserver, return code 1\u001b[0m\n\u001b[34m2020-02-04 21:04:25,590 - DefaultDataAnalyzer - INFO - Total number of hosts in the cluster: 1\u001b[0m\n\u001b[34m2020-02-04 21:04:35,600 - DefaultDataAnalyzer - INFO - Running command: bin/spark-submit --master yarn --deploy-mode client --conf spark.hadoop.fs.s3a.aws.credentials.provider=org.apache.hadoop.fs.s3a.SimpleAWSCredentialsProvider --conf spark.serializer=org.apache.spark.serializer.KryoSerializer /opt/amazon/sagemaker-data-analyzer-1.0-jar-with-dependencies.jar --analytics_input /tmp/spark_job_config.json\u001b[0m\n\u001b[34m2020-02-04 21:04:36 WARN NativeCodeLoader:60 - Unable to load native-hadoop library for your platform... using builtin-java classes where applicable\u001b[0m\n\u001b[34m2020-02-04 21:04:36 INFO Main:28 - Start analyzing with args: --analytics_input /tmp/spark_job_config.json\u001b[0m\n\u001b[34m2020-02-04 21:04:36 INFO Main:31 - Analytics input path: DataAnalyzerParams(/tmp/spark_job_config.json)\u001b[0m\n\u001b[34m2020-02-04 21:04:36 INFO FileUtil:66 - Read file from path /tmp/spark_job_config.json.\u001b[0m\n\u001b[34m2020-02-04 21:04:36 INFO DataAnalyzer:30 - Analytics input: AnalyticsInput(/opt/ml/processing/input/endpoint,DataSetFormat(None,None,Some(SageMakerCaptureJson(List(endpointInput, endpointOutput),None))),None,Some(/opt/ml/processing/code/postprocessing/postprocessor.py),Some(/opt/ml/processing/baseline/constraints/constraints.json),Some(/opt/ml/processing/baseline/stats/statistics.json),/opt/ml/processing/output,None,None,/opt/ml/output/metrics/cloudwatch,Some(Disabled),None,None,/opt/ml/output/message)\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SparkContext:54 - Running Spark version 2.3.1\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SparkContext:54 - Submitted application: SageMakerDataAnalyzer\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SecurityManager:54 - Changing view acls to: root\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SecurityManager:54 - Changing modify acls to: root\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SecurityManager:54 - Changing view acls groups to: \u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SecurityManager:54 - Changing modify acls groups to: \u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SecurityManager:54 - SecurityManager: authentication disabled; ui acls disabled; users with view permissions: Set(root); groups with view permissions: Set(); users with modify permissions: Set(root); groups with modify permissions: Set()\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO Utils:54 - Successfully started service 'sparkDriver' on port 45119.\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SparkEnv:54 - Registering MapOutputTracker\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SparkEnv:54 - Registering BlockManagerMaster\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO BlockManagerMasterEndpoint:54 - Using org.apache.spark.storage.DefaultTopologyMapper for getting topology information\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO BlockManagerMasterEndpoint:54 - BlockManagerMasterEndpoint up\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO DiskBlockManager:54 - Created local directory at /tmp/blockmgr-46cb2f4d-b039-4ccc-8f03-8ae3050bdb81\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO MemoryStore:54 - MemoryStore started with capacity 1458.6 MB\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SparkEnv:54 - Registering OutputCommitCoordinator\u001b[0m\n\u001b[34m2020-02-04 21:04:37 INFO SparkContext:54 - Added JAR file:/opt/amazon/sagemaker-data-analyzer-1.0-jar-with-dependencies.jar at spark://10.0.106.106:45119/jars/sagemaker-data-analyzer-1.0-jar-with-dependencies.jar with timestamp 1580850277424\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO RMProxy:133 - Connecting to ResourceManager at /10.0.106.106:8032\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO Client:54 - Requesting a new application from cluster with 1 NodeManagers\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO Configuration:2636 - resource-types.xml not found\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO ResourceUtils:427 - Unable to find 'resource-types.xml'.\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO Client:54 - Verifying our application has not requested more than the maximum memory capability of the cluster (15578 MB per container)\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO Client:54 - Will allocate AM container, with 896 MB memory including 384 MB overhead\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO Client:54 - Setting up container launch context for our AM\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO Client:54 - Setting up the launch environment for our AM container\u001b[0m\n\u001b[34m2020-02-04 21:04:38 INFO Client:54 - Preparing resources for our AM container\u001b[0m\n\u001b[34m2020-02-04 21:04:39 WARN Client:66 - Neither spark.yarn.jars nor spark.yarn.archive is set, falling back to uploading libraries under SPARK_HOME.\u001b[0m\n\u001b[34m2020-02-04 21:04:40 INFO Client:54 - Uploading resource file:/tmp/spark-eb9f0aad-a906-4aa7-a0a2-503e2073af1a/__spark_libs__633775288156079250.zip -> hdfs://10.0.106.106/user/root/.sparkStaging/application_1580850260617_0001/__spark_libs__633775288156079250.zip\u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO Client:54 - Uploading resource file:/tmp/spark-eb9f0aad-a906-4aa7-a0a2-503e2073af1a/__spark_conf__5695000870469261038.zip -> hdfs://10.0.106.106/user/root/.sparkStaging/application_1580850260617_0001/__spark_conf__.zip\u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO SecurityManager:54 - Changing view acls to: root\u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO SecurityManager:54 - Changing modify acls to: root\u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO SecurityManager:54 - Changing view acls groups to: \u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO SecurityManager:54 - Changing modify acls groups to: \u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO SecurityManager:54 - SecurityManager: authentication disabled; ui acls disabled; users with view permissions: Set(root); groups with view permissions: Set(); users with modify permissions: Set(root); groups with modify permissions: Set()\u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO Client:54 - Submitting application application_1580850260617_0001 to ResourceManager\u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO YarnClientImpl:310 - Submitted application application_1580850260617_0001\u001b[0m\n\u001b[34m2020-02-04 21:04:42 INFO SchedulerExtensionServices:54 - Starting Yarn extension services with app application_1580850260617_0001 and attemptId None\u001b[0m\n\u001b[34m2020-02-04 21:04:43 INFO Client:54 - Application report for application_1580850260617_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 21:04:43 INFO Client:54 - \u001b[0m\n\u001b[34m#011 client token: N/A\u001b[0m\n\u001b[34m#011 diagnostics: [Tue Feb 04 21:04:43 +0000 2020] Scheduler has assigned a container for AM, waiting for AM container to be launched\u001b[0m\n\u001b[34m#011 ApplicationMaster host: N/A\u001b[0m\n\u001b[34m#011 ApplicationMaster RPC port: -1\u001b[0m\n\u001b[34m#011 queue: default\u001b[0m\n\u001b[34m#011 start time: 1580850282667\u001b[0m\n\u001b[34m#011 final status: UNDEFINED\u001b[0m\n\u001b[34m#011 tracking URL: http://algo-1:8088/proxy/application_1580850260617_0001/\u001b[0m\n\u001b[34m#011 user: root\u001b[0m\n\u001b[34m2020-02-04 21:04:44 INFO Client:54 - Application report for application_1580850260617_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 21:04:45 INFO Client:54 - Application report for application_1580850260617_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 21:04:46 INFO Client:54 - Application report for application_1580850260617_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 21:04:47 INFO Client:54 - Application report for application_1580850260617_0001 (state: ACCEPTED)\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO YarnClientSchedulerBackend:54 - Add WebUI Filter. org.apache.hadoop.yarn.server.webproxy.amfilter.AmIpFilter, Map(PROXY_HOSTS -> algo-1, PROXY_URI_BASES -> http://algo-1:8088/proxy/application_1580850260617_0001), /proxy/application_1580850260617_0001\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO YarnSchedulerBackend$YarnSchedulerEndpoint:54 - ApplicationMaster registered as NettyRpcEndpointRef(spark-client://YarnAM)\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO Client:54 - Application report for application_1580850260617_0001 (state: RUNNING)\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO Client:54 - \u001b[0m\n\u001b[34m#011 client token: N/A\u001b[0m\n\u001b[34m#011 diagnostics: N/A\u001b[0m\n\u001b[34m#011 ApplicationMaster host: 10.0.106.106\u001b[0m\n\u001b[34m#011 ApplicationMaster RPC port: 0\u001b[0m\n\u001b[34m#011 queue: default\u001b[0m\n\u001b[34m#011 start time: 1580850282667\u001b[0m\n\u001b[34m#011 final status: UNDEFINED\u001b[0m\n\u001b[34m#011 tracking URL: http://algo-1:8088/proxy/application_1580850260617_0001/\u001b[0m\n\u001b[34m#011 user: root\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO YarnClientSchedulerBackend:54 - Application application_1580850260617_0001 has started running.\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO Utils:54 - Successfully started service 'org.apache.spark.network.netty.NettyBlockTransferService' on port 39767.\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO NettyBlockTransferService:54 - Server created on 10.0.106.106:39767\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO BlockManager:54 - Using org.apache.spark.storage.RandomBlockReplicationPolicy for block replication policy\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO BlockManagerMaster:54 - Registering BlockManager BlockManagerId(driver, 10.0.106.106, 39767, None)\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO BlockManagerMasterEndpoint:54 - Registering block manager 10.0.106.106:39767 with 1458.6 MB RAM, BlockManagerId(driver, 10.0.106.106, 39767, None)\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO BlockManagerMaster:54 - Registered BlockManager BlockManagerId(driver, 10.0.106.106, 39767, None)\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO BlockManager:54 - Initialized BlockManager: BlockManagerId(driver, 10.0.106.106, 39767, None)\u001b[0m\n\u001b[34m2020-02-04 21:04:48 INFO log:192 - Logging initialized @13199ms\u001b[0m\n\u001b[34m2020-02-04 21:04:50 INFO YarnSchedulerBackend$YarnDriverEndpoint:54 - Registered executor NettyRpcEndpointRef(spark-client://Executor) (10.0.106.106:36964) with ID 1\u001b[0m\n\u001b[34m2020-02-04 21:04:50 INFO BlockManagerMasterEndpoint:54 - Registering block manager algo-1:39511 with 5.8 GB RAM, BlockManagerId(1, algo-1, 39511, None)\u001b[0m\n\u001b[34m2020-02-04 21:05:07 INFO YarnClientSchedulerBackend:54 - SchedulerBackend is ready for scheduling beginning after waiting maxRegisteredResourcesWaitingTime: 30000(ms)\u001b[0m\n\u001b[34m2020-02-04 21:05:07 WARN SparkContext:66 - Spark is not running in local mode, therefore the checkpoint directory must not be on the local filesystem. Directory '/tmp' appears to be on the local filesystem.\u001b[0m\n\u001b[34m2020-02-04 21:05:07 INFO SharedState:54 - Setting hive.metastore.warehouse.dir ('null') to the value of spark.sql.warehouse.dir ('file:/usr/spark-2.3.1/spark-warehouse').\u001b[0m\n\u001b[34m2020-02-04 21:05:07 INFO SharedState:54 - Warehouse path is 'file:/usr/spark-2.3.1/spark-warehouse'.\u001b[0m\n\u001b[34m2020-02-04 21:05:08 INFO StateStoreCoordinatorRef:54 - Registered StateStoreCoordinator endpoint\u001b[0m\n\u001b[34m2020-02-04 21:05:08 INFO DatasetReader:90 - Files to process:List(file:///opt/ml/processing/input/endpoint/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/33-57-898-0815a2e8-c235-4891-bd5d-91a59c41b6ca.jsonl, file:///opt/ml/processing/input/endpoint/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/51-28-839-f722c428-f4a3-48a1-a6ab-a5f695d3c99a.jsonl, file:///opt/ml/processing/input/endpoint/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/53-28-991-e0c9ae91-1350-405e-9929-df8a0c8eda9b.jsonl, file:///opt/ml/processing/input/endpoint/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/52-28-908-839decf0-a61a-48d0-9b34-f8d50576eea6.jsonl)\u001b[0m\n\u001b[34m2020-02-04 21:05:08 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 21:05:08 INFO FileSourceStrategy:54 - Post-Scan Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:08 INFO FileSourceStrategy:54 - Output Data Schema: struct<value: string>\u001b[0m\n\u001b[34m2020-02-04 21:05:08 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO CodeGenerator:54 - Code generated in 187.547994 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO MemoryStore:54 - Block broadcast_0 stored as values in memory (estimated size 429.4 KB, free 1458.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO MemoryStore:54 - Block broadcast_0_piece0 stored as bytes in memory (estimated size 38.3 KB, free 1458.1 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO BlockManagerInfo:54 - Added broadcast_0_piece0 in memory on 10.0.106.106:39767 (size: 38.3 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO SparkContext:54 - Created broadcast 0 from json at DatasetReader.scala:57\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 5817759 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO SparkContext:54 - Starting job: json at DatasetReader.scala:57\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO DAGScheduler:54 - Got job 0 (json at DatasetReader.scala:57) with 2 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO DAGScheduler:54 - Final stage: ResultStage 0 (json at DatasetReader.scala:57)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO DAGScheduler:54 - Submitting ResultStage 0 (MapPartitionsRDD[5] at json at DatasetReader.scala:57), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO MemoryStore:54 - Block broadcast_1 stored as values in memory (estimated size 9.4 KB, free 1458.1 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO MemoryStore:54 - Block broadcast_1_piece0 stored as bytes in memory (estimated size 5.1 KB, free 1458.1 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO BlockManagerInfo:54 - Added broadcast_1_piece0 in memory on 10.0.106.106:39767 (size: 5.1 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO SparkContext:54 - Created broadcast 1 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO DAGScheduler:54 - Submitting 2 missing tasks from ResultStage 0 (MapPartitionsRDD[5] at json at DatasetReader.scala:57) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO YarnScheduler:54 - Adding task set 0.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO TaskSetManager:54 - Starting task 0.0 in stage 0.0 (TID 0, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8648 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:09 INFO TaskSetManager:54 - Starting task 1.0 in stage 0.0 (TID 1, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8648 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:10 INFO BlockManagerInfo:54 - Added broadcast_1_piece0 in memory on algo-1:39511 (size: 5.1 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO BlockManagerInfo:54 - Added broadcast_0_piece0 in memory on algo-1:39511 (size: 38.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO TaskSetManager:54 - Finished task 0.0 in stage 0.0 (TID 0) in 1847 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO TaskSetManager:54 - Finished task 1.0 in stage 0.0 (TID 1) in 1839 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO YarnScheduler:54 - Removed TaskSet 0.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - ResultStage 0 (json at DatasetReader.scala:57) finished in 1.937 s\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - Job 0 finished: json at DatasetReader.scala:57, took 1.989476 s\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO FileSourceStrategy:54 - Post-Scan Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO FileSourceStrategy:54 - Output Data Schema: struct<captureData: struct<endpointInput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>, endpointOutput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>>>\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO CodeGenerator:54 - Code generated in 20.276509 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO CodeGenerator:54 - Code generated in 31.6196 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO MemoryStore:54 - Block broadcast_2 stored as values in memory (estimated size 429.4 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO MemoryStore:54 - Block broadcast_2_piece0 stored as bytes in memory (estimated size 38.3 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO BlockManagerInfo:54 - Added broadcast_2_piece0 in memory on 10.0.106.106:39767 (size: 38.3 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO SparkContext:54 - Created broadcast 2 from head at CapturedDataReader.scala:49\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 5817759 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO SparkContext:54 - Starting job: head at CapturedDataReader.scala:49\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - Got job 1 (head at CapturedDataReader.scala:49) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - Final stage: ResultStage 1 (head at CapturedDataReader.scala:49)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - Submitting ResultStage 1 (MapPartitionsRDD[9] at head at CapturedDataReader.scala:49), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO MemoryStore:54 - Block broadcast_3 stored as values in memory (estimated size 13.4 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO MemoryStore:54 - Block broadcast_3_piece0 stored as bytes in memory (estimated size 6.2 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO BlockManagerInfo:54 - Added broadcast_3_piece0 in memory on 10.0.106.106:39767 (size: 6.2 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO SparkContext:54 - Created broadcast 3 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 1 (MapPartitionsRDD[9] at head at CapturedDataReader.scala:49) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO YarnScheduler:54 - Adding task set 1.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO TaskSetManager:54 - Starting task 0.0 in stage 1.0 (TID 2, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8648 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:11 INFO BlockManagerInfo:54 - Added broadcast_3_piece0 in memory on algo-1:39511 (size: 6.2 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Added broadcast_2_piece0 in memory on algo-1:39511 (size: 38.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO TaskSetManager:54 - Finished task 0.0 in stage 1.0 (TID 2) in 247 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO YarnScheduler:54 - Removed TaskSet 1.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - ResultStage 1 (head at CapturedDataReader.scala:49) finished in 0.268 s\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Job 1 finished: head at CapturedDataReader.scala:49, took 0.274701 s\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Post-Scan Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Output Data Schema: struct<captureData: struct<endpointInput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>, endpointOutput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>>>\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO CodeGenerator:54 - Code generated in 9.494394 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO CodeGenerator:54 - Code generated in 45.201424 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_4 stored as values in memory (estimated size 429.4 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_4_piece0 stored as bytes in memory (estimated size 38.3 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Added broadcast_4_piece0 in memory on 10.0.106.106:39767 (size: 38.3 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO SparkContext:54 - Created broadcast 4 from csv at CapturedDataReader.scala:148\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 5817759 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 31\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO SparkContext:54 - Starting job: csv at CapturedDataReader.scala:148\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Got job 2 (csv at CapturedDataReader.scala:148) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Final stage: ResultStage 2 (csv at CapturedDataReader.scala:148)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Submitting ResultStage 2 (MapPartitionsRDD[13] at csv at CapturedDataReader.scala:148), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_5 stored as values in memory (estimated size 22.2 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_5_piece0 stored as bytes in memory (estimated size 8.7 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_0_piece0 on 10.0.106.106:39767 in memory (size: 38.3 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_0_piece0 on algo-1:39511 in memory (size: 38.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Added broadcast_5_piece0 in memory on 10.0.106.106:39767 (size: 8.7 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO SparkContext:54 - Created broadcast 5 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 2 (MapPartitionsRDD[13] at csv at CapturedDataReader.scala:148) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO YarnScheduler:54 - Adding task set 2.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO TaskSetManager:54 - Starting task 0.0 in stage 2.0 (TID 3, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8648 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 19\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 3\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 52\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 48\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 46\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 51\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 15\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 18\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 0\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 1\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 30\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 49\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 25\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 23\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 47\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 24\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_2_piece0 on 10.0.106.106:39767 in memory (size: 38.3 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_2_piece0 on algo-1:39511 in memory (size: 38.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Added broadcast_5_piece0 in memory on algo-1:39511 (size: 8.7 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 5\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 44\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 34\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 38\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 39\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 22\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 9\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 45\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 28\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 6\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 17\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 27\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 32\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 13\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 2\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 20\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 33\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 37\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 10\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 58\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 11\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 53\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_1_piece0 on 10.0.106.106:39767 in memory (size: 5.1 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_1_piece0 on algo-1:39511 in memory (size: 5.1 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 50\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 35\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 41\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 55\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 26\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 57\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 16\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 56\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 12\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 36\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 14\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_3_piece0 on 10.0.106.106:39767 in memory (size: 6.2 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Removed broadcast_3_piece0 on algo-1:39511 in memory (size: 6.2 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 40\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 7\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 59\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 4\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 8\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 43\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 29\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 21\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 42\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO ContextCleaner:54 - Cleaned accumulator 54\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Added broadcast_4_piece0 in memory on algo-1:39511 (size: 38.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO TaskSetManager:54 - Finished task 0.0 in stage 2.0 (TID 3) in 247 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - ResultStage 2 (csv at CapturedDataReader.scala:148) finished in 0.263 s\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO DAGScheduler:54 - Job 2 finished: csv at CapturedDataReader.scala:148, took 0.275215 s\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO YarnScheduler:54 - Removed TaskSet 2.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Post-Scan Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Output Data Schema: struct<captureData: struct<endpointInput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>, endpointOutput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>>>\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO CodeGenerator:54 - Code generated in 33.927824 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_6 stored as values in memory (estimated size 429.4 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_6_piece0 stored as bytes in memory (estimated size 38.3 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Added broadcast_6_piece0 in memory on 10.0.106.106:39767 (size: 38.3 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO SparkContext:54 - Created broadcast 6 from csv at CapturedDataReader.scala:148\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 5817759 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Pruning directories with: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Post-Scan Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceStrategy:54 - Output Data Schema: struct<captureData: struct<endpointInput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>, endpointOutput: struct<data: string, encoding: string, mode: string, observedContentType: string ... 2 more fields>>>\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceScanExec:54 - Pushed Filters: \u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO CodeGenerator:54 - Code generated in 31.692123 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_7 stored as values in memory (estimated size 429.4 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO MemoryStore:54 - Block broadcast_7_piece0 stored as bytes in memory (estimated size 38.3 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO BlockManagerInfo:54 - Added broadcast_7_piece0 in memory on 10.0.106.106:39767 (size: 38.3 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO SparkContext:54 - Created broadcast 7 from csv at CapturedDataReader.scala:148\u001b[0m\n\u001b[34m2020-02-04 21:05:12 INFO FileSourceScanExec:54 - Planning scan with bin packing, max size: 5817759 bytes, open cost is considered as scanning 4194304 bytes.\u001b[0m\n\u001b[34m2020-02-04 21:05:13 INFO FileUtil:66 - Read file from path /opt/ml/processing/baseline/constraints/constraints.json.\u001b[0m\n\u001b[34m2020-02-04 21:05:13 INFO FileUtil:66 - Read file from path /opt/ml/processing/baseline/stats/statistics.json.\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO DataAnalyzer:79 - Using baseline columns: Target, Dst Port, Flow Duration, Tot Fwd Pkts, Tot Bwd Pkts, TotLen Fwd Pkts, TotLen Bwd Pkts, Fwd Pkt Len Max, Fwd Pkt Len Min, Fwd Pkt Len Mean, Fwd Pkt Len Std, Bwd Pkt Len Max, Bwd Pkt Len Min, Bwd Pkt Len Mean, Bwd Pkt Len Std, Flow Byts/s, Flow Pkts/s, Flow IAT Mean, Flow IAT Std, Flow IAT Max, Flow IAT Min, Fwd IAT Tot, Fwd IAT Mean, Fwd IAT Std, Fwd IAT Max, Fwd IAT Min, Bwd IAT Tot, Bwd IAT Mean, Bwd IAT Std, Bwd IAT Max, Bwd IAT Min, Fwd PSH Flags, Bwd PSH Flags, Fwd URG Flags, Bwd URG Flags, Fwd Header Len, Bwd Header Len, Fwd Pkts/s, Bwd Pkts/s, Pkt Len Min, Pkt Len Max, Pkt Len Mean, Pkt Len Std, Pkt Len Var, FIN Flag Cnt, SYN Flag Cnt, RST Flag Cnt, PSH Flag Cnt, ACK Flag Cnt, URG Flag Cnt, CWE Flag Count, ECE Flag Cnt, Down/Up Ratio, Pkt Size Avg, Fwd Seg Size Avg, Bwd Seg Size Avg, Fwd Byts/b Avg, Fwd Pkts/b Avg, Fwd Blk Rate Avg, Bwd Byts/b Avg, Bwd Pkts/b Avg, Bwd Blk Rate Avg, Subflow Fwd Pkts, Subflow Fwd Byts, Subflow Bwd Pkts, Subflow Bwd Byts, Init Fwd Win Byts, Init Bwd Win Byts, Fwd Act Data Pkts, Fwd Seg Size Min, Active Mean, Active Std, Active Max, Active Min, Idle Mean, Idle Std, Idle Max, Idle Min, day, month, year, dayofweek, prot_0, prot_6, prot_17\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO CodeGenerator:54 - Code generated in 72.221163 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:14 WARN Utils:66 - Truncated the string representation of a plan since it was too large. This behavior can be adjusted by setting 'spark.debug.maxToStringFields' in SparkEnv.conf.\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO CodeGenerator:54 - Code generated in 61.809453 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 70\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 92\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 68\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO BlockManagerInfo:54 - Removed broadcast_6_piece0 on 10.0.106.106:39767 in memory (size: 38.3 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO BlockManagerInfo:54 - Removed broadcast_5_piece0 on 10.0.106.106:39767 in memory (size: 8.7 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO BlockManagerInfo:54 - Removed broadcast_5_piece0 on algo-1:39511 in memory (size: 8.7 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 89\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 84\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 72\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 66\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 62\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 63\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 65\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 79\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 77\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO BlockManagerInfo:54 - Removed broadcast_4_piece0 on 10.0.106.106:39767 in memory (size: 38.3 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO BlockManagerInfo:54 - Removed broadcast_4_piece0 on algo-1:39511 in memory (size: 38.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 67\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 69\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 94\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 83\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 95\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 88\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 74\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 78\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 86\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 93\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 90\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 64\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 73\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 87\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 75\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 82\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 76\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 71\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 81\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 60\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 85\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 91\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 80\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO ContextCleaner:54 - Cleaned accumulator 61\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO CodeGenerator:54 - Code generated in 49.096628 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO SparkContext:54 - Starting job: head at DataAnalyzer.scala:93\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO DAGScheduler:54 - Got job 3 (head at DataAnalyzer.scala:93) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO DAGScheduler:54 - Final stage: ResultStage 3 (head at DataAnalyzer.scala:93)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO DAGScheduler:54 - Submitting ResultStage 3 (MapPartitionsRDD[34] at head at DataAnalyzer.scala:93), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO MemoryStore:54 - Block broadcast_8 stored as values in memory (estimated size 155.8 KB, free 1458.0 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO MemoryStore:54 - Block broadcast_8_piece0 stored as bytes in memory (estimated size 46.4 KB, free 1457.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO BlockManagerInfo:54 - Added broadcast_8_piece0 in memory on 10.0.106.106:39767 (size: 46.4 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO SparkContext:54 - Created broadcast 8 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 3 (MapPartitionsRDD[34] at head at DataAnalyzer.scala:93) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO YarnScheduler:54 - Adding task set 3.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO TaskSetManager:54 - Starting task 0.0 in stage 3.0 (TID 4, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8648 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:14 INFO BlockManagerInfo:54 - Added broadcast_8_piece0 in memory on algo-1:39511 (size: 46.4 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:15 INFO BlockManagerInfo:54 - Added broadcast_7_piece0 in memory on algo-1:39511 (size: 38.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:16 INFO BlockManagerInfo:54 - Added rdd_28_0 in memory on algo-1:39511 (size: 35.6 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:16 INFO TaskSetManager:54 - Finished task 0.0 in stage 3.0 (TID 4) in 1721 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 21:05:16 INFO YarnScheduler:54 - Removed TaskSet 3.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:16 INFO DAGScheduler:54 - ResultStage 3 (head at DataAnalyzer.scala:93) finished in 1.788 s\u001b[0m\n\u001b[34m2020-02-04 21:05:16 INFO DAGScheduler:54 - Job 3 finished: head at DataAnalyzer.scala:93, took 1.800666 s\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO CodeGenerator:54 - Code generated in 53.644593 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO SparkContext:54 - Starting job: collect at AnalysisRunner.scala:313\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO DAGScheduler:54 - Registering RDD 39 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO DAGScheduler:54 - Got job 4 (collect at AnalysisRunner.scala:313) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO DAGScheduler:54 - Final stage: ResultStage 5 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO DAGScheduler:54 - Parents of final stage: List(ShuffleMapStage 4)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO DAGScheduler:54 - Missing parents: List(ShuffleMapStage 4)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO DAGScheduler:54 - Submitting ShuffleMapStage 4 (MapPartitionsRDD[39] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO MemoryStore:54 - Block broadcast_9 stored as values in memory (estimated size 851.0 KB, free 1457.1 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO MemoryStore:54 - Block broadcast_9_piece0 stored as bytes in memory (estimated size 257.5 KB, free 1456.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO BlockManagerInfo:54 - Added broadcast_9_piece0 in memory on 10.0.106.106:39767 (size: 257.5 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO SparkContext:54 - Created broadcast 9 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO DAGScheduler:54 - Submitting 2 missing tasks from ShuffleMapStage 4 (MapPartitionsRDD[39] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO YarnScheduler:54 - Adding task set 4.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO TaskSetManager:54 - Starting task 0.0 in stage 4.0 (TID 5, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO TaskSetManager:54 - Starting task 1.0 in stage 4.0 (TID 6, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:17 INFO BlockManagerInfo:54 - Added broadcast_9_piece0 in memory on algo-1:39511 (size: 257.5 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:18 INFO BlockManagerInfo:54 - Added rdd_28_1 in memory on algo-1:39511 (size: 29.3 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO TaskSetManager:54 - Finished task 1.0 in stage 4.0 (TID 6) in 4152 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO TaskSetManager:54 - Finished task 0.0 in stage 4.0 (TID 5) in 4187 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO YarnScheduler:54 - Removed TaskSet 4.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO DAGScheduler:54 - ShuffleMapStage 4 (collect at AnalysisRunner.scala:313) finished in 4.228 s\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO DAGScheduler:54 - looking for newly runnable stages\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO DAGScheduler:54 - running: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO DAGScheduler:54 - waiting: Set(ResultStage 5)\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO DAGScheduler:54 - failed: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO DAGScheduler:54 - Submitting ResultStage 5 (MapPartitionsRDD[42] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:21 INFO MemoryStore:54 - Block broadcast_10 stored as values in memory (estimated size 1032.3 KB, free 1455.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO MemoryStore:54 - Block broadcast_10_piece0 stored as bytes in memory (estimated size 310.6 KB, free 1455.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO BlockManagerInfo:54 - Added broadcast_10_piece0 in memory on 10.0.106.106:39767 (size: 310.6 KB, free: 1458.0 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO SparkContext:54 - Created broadcast 10 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 5 (MapPartitionsRDD[42] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO YarnScheduler:54 - Adding task set 5.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO TaskSetManager:54 - Starting task 0.0 in stage 5.0 (TID 7, algo-1, executor 1, partition 0, NODE_LOCAL, 7765 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO BlockManagerInfo:54 - Added broadcast_10_piece0 in memory on algo-1:39511 (size: 310.6 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:22 INFO MapOutputTrackerMasterEndpoint:54 - Asked to send map output locations for shuffle 0 to 10.0.106.106:36964\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO TaskSetManager:54 - Finished task 0.0 in stage 5.0 (TID 7) in 2083 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO DAGScheduler:54 - ResultStage 5 (collect at AnalysisRunner.scala:313) finished in 2.130 s\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO YarnScheduler:54 - Removed TaskSet 5.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO DAGScheduler:54 - Job 4 finished: collect at AnalysisRunner.scala:313, took 6.391461 s\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 172\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 152\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 144\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 147\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 137\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 129\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 150\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 122\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 135\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 166\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 167\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 118\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 184\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 188\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 127\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 121\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 115\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 153\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned shuffle 0\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 165\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 111\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 125\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 181\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 160\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 138\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 136\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 168\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 148\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 116\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 162\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 187\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 174\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 126\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 164\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 186\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 192\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 108\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 189\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 110\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO BlockManagerInfo:54 - Removed broadcast_10_piece0 on 10.0.106.106:39767 in memory (size: 310.6 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO BlockManagerInfo:54 - Removed broadcast_10_piece0 on algo-1:39511 in memory (size: 310.6 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 191\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 106\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 117\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 155\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 123\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO BlockManagerInfo:54 - Removed broadcast_8_piece0 on 10.0.106.106:39767 in memory (size: 46.4 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO BlockManagerInfo:54 - Removed broadcast_8_piece0 on algo-1:39511 in memory (size: 46.4 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 180\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 120\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 130\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 163\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 190\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 132\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 133\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 134\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 145\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO BlockManagerInfo:54 - Removed broadcast_9_piece0 on 10.0.106.106:39767 in memory (size: 257.5 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO BlockManagerInfo:54 - Removed broadcast_9_piece0 on algo-1:39511 in memory (size: 257.5 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 161\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 170\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 182\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 149\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 169\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 107\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 178\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 114\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 183\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 171\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 194\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 113\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 193\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 142\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 154\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 112\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 128\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 185\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 143\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 119\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 176\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 109\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 140\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 179\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 177\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 157\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 146\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 131\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 105\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 141\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 156\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 159\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 151\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 124\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 139\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 158\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 173\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 195\u001b[0m\n\u001b[34m2020-02-04 21:05:24 INFO ContextCleaner:54 - Cleaned accumulator 175\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO CodeGenerator:54 - Code generated in 115.987207 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO SparkContext:54 - Starting job: treeReduce at KLLRunner.scala:107\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - Got job 5 (treeReduce at KLLRunner.scala:107) with 2 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - Final stage: ResultStage 6 (treeReduce at KLLRunner.scala:107)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - Parents of final stage: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - Missing parents: List()\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - Submitting ResultStage 6 (MapPartitionsRDD[51] at treeReduce at KLLRunner.scala:107), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO MemoryStore:54 - Block broadcast_11 stored as values in memory (estimated size 198.8 KB, free 1457.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO MemoryStore:54 - Block broadcast_11_piece0 stored as bytes in memory (estimated size 58.8 KB, free 1457.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO BlockManagerInfo:54 - Added broadcast_11_piece0 in memory on 10.0.106.106:39767 (size: 58.8 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO SparkContext:54 - Created broadcast 11 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - Submitting 2 missing tasks from ResultStage 6 (MapPartitionsRDD[51] at treeReduce at KLLRunner.scala:107) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO YarnScheduler:54 - Adding task set 6.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO TaskSetManager:54 - Starting task 0.0 in stage 6.0 (TID 8, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8648 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO TaskSetManager:54 - Starting task 1.0 in stage 6.0 (TID 9, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8648 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO BlockManagerInfo:54 - Added broadcast_11_piece0 in memory on algo-1:39511 (size: 58.8 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO TaskSetManager:54 - Finished task 1.0 in stage 6.0 (TID 9) in 583 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO TaskSetManager:54 - Finished task 0.0 in stage 6.0 (TID 8) in 592 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - ResultStage 6 (treeReduce at KLLRunner.scala:107) finished in 0.606 s\u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO YarnScheduler:54 - Removed TaskSet 6.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:26 INFO DAGScheduler:54 - Job 5 finished: treeReduce at KLLRunner.scala:107, took 0.613939 s\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 220\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 219\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 222\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 210\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 216\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 211\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 200\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 201\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 217\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 206\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 213\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 208\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 223\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 204\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 209\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 221\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 212\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 215\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 207\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 214\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 218\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 199\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO BlockManagerInfo:54 - Removed broadcast_11_piece0 on 10.0.106.106:39767 in memory (size: 58.8 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO BlockManagerInfo:54 - Removed broadcast_11_piece0 on algo-1:39511 in memory (size: 58.8 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 205\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 203\u001b[0m\n\u001b[34m2020-02-04 21:05:27 INFO ContextCleaner:54 - Cleaned accumulator 202\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO CodeGenerator:54 - Code generated in 240.525064 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO ContextCleaner:54 - Cleaned accumulator 224\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO SparkContext:54 - Starting job: collect at AnalysisRunner.scala:313\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO DAGScheduler:54 - Registering RDD 57 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO DAGScheduler:54 - Got job 6 (collect at AnalysisRunner.scala:313) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO DAGScheduler:54 - Final stage: ResultStage 8 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO DAGScheduler:54 - Parents of final stage: List(ShuffleMapStage 7)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO DAGScheduler:54 - Missing parents: List(ShuffleMapStage 7)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO DAGScheduler:54 - Submitting ShuffleMapStage 7 (MapPartitionsRDD[57] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO MemoryStore:54 - Block broadcast_12 stored as values in memory (estimated size 377.7 KB, free 1457.8 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO MemoryStore:54 - Block broadcast_12_piece0 stored as bytes in memory (estimated size 112.9 KB, free 1457.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO BlockManagerInfo:54 - Added broadcast_12_piece0 in memory on 10.0.106.106:39767 (size: 112.9 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO SparkContext:54 - Created broadcast 12 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO DAGScheduler:54 - Submitting 2 missing tasks from ShuffleMapStage 7 (MapPartitionsRDD[57] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO YarnScheduler:54 - Adding task set 7.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO TaskSetManager:54 - Starting task 0.0 in stage 7.0 (TID 10, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO TaskSetManager:54 - Starting task 1.0 in stage 7.0 (TID 11, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:28 INFO BlockManagerInfo:54 - Added broadcast_12_piece0 in memory on algo-1:39511 (size: 112.9 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO TaskSetManager:54 - Finished task 1.0 in stage 7.0 (TID 11) in 1415 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO TaskSetManager:54 - Finished task 0.0 in stage 7.0 (TID 10) in 1600 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO YarnScheduler:54 - Removed TaskSet 7.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO DAGScheduler:54 - ShuffleMapStage 7 (collect at AnalysisRunner.scala:313) finished in 1.619 s\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO DAGScheduler:54 - looking for newly runnable stages\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO DAGScheduler:54 - running: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO DAGScheduler:54 - waiting: Set(ResultStage 8)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO DAGScheduler:54 - failed: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO DAGScheduler:54 - Submitting ResultStage 8 (MapPartitionsRDD[60] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO MemoryStore:54 - Block broadcast_13 stored as values in memory (estimated size 459.3 KB, free 1457.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO MemoryStore:54 - Block broadcast_13_piece0 stored as bytes in memory (estimated size 132.8 KB, free 1457.1 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO BlockManagerInfo:54 - Added broadcast_13_piece0 in memory on 10.0.106.106:39767 (size: 132.8 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO SparkContext:54 - Created broadcast 13 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 8 (MapPartitionsRDD[60] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO YarnScheduler:54 - Adding task set 8.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO TaskSetManager:54 - Starting task 0.0 in stage 8.0 (TID 12, algo-1, executor 1, partition 0, NODE_LOCAL, 7765 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO BlockManagerInfo:54 - Added broadcast_13_piece0 in memory on algo-1:39511 (size: 132.8 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:30 INFO MapOutputTrackerMasterEndpoint:54 - Asked to send map output locations for shuffle 1 to 10.0.106.106:36964\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO TaskSetManager:54 - Finished task 0.0 in stage 8.0 (TID 12) in 1231 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO YarnScheduler:54 - Removed TaskSet 8.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - ResultStage 8 (collect at AnalysisRunner.scala:313) finished in 1.269 s\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Job 6 finished: collect at AnalysisRunner.scala:313, took 2.899361 s\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO SparkContext:54 - Starting job: countByKey at ColumnProfiler.scala:566\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Registering RDD 67 (countByKey at ColumnProfiler.scala:566)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Got job 7 (countByKey at ColumnProfiler.scala:566) with 2 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Final stage: ResultStage 10 (countByKey at ColumnProfiler.scala:566)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Parents of final stage: List(ShuffleMapStage 9)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Missing parents: List(ShuffleMapStage 9)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Submitting ShuffleMapStage 9 (MapPartitionsRDD[67] at countByKey at ColumnProfiler.scala:566), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO MemoryStore:54 - Block broadcast_14 stored as values in memory (estimated size 136.2 KB, free 1457.0 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO MemoryStore:54 - Block broadcast_14_piece0 stored as bytes in memory (estimated size 44.9 KB, free 1456.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO BlockManagerInfo:54 - Added broadcast_14_piece0 in memory on 10.0.106.106:39767 (size: 44.9 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO SparkContext:54 - Created broadcast 14 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO DAGScheduler:54 - Submitting 2 missing tasks from ShuffleMapStage 9 (MapPartitionsRDD[67] at countByKey at ColumnProfiler.scala:566) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO YarnScheduler:54 - Adding task set 9.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO TaskSetManager:54 - Starting task 0.0 in stage 9.0 (TID 13, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO TaskSetManager:54 - Starting task 1.0 in stage 9.0 (TID 14, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:31 INFO BlockManagerInfo:54 - Added broadcast_14_piece0 in memory on algo-1:39511 (size: 44.9 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Finished task 1.0 in stage 9.0 (TID 14) in 282 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Finished task 0.0 in stage 9.0 (TID 13) in 347 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO YarnScheduler:54 - Removed TaskSet 9.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - ShuffleMapStage 9 (countByKey at ColumnProfiler.scala:566) finished in 0.376 s\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - looking for newly runnable stages\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - running: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - waiting: Set(ResultStage 10)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - failed: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Submitting ResultStage 10 (ShuffledRDD[68] at countByKey at ColumnProfiler.scala:566), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MemoryStore:54 - Block broadcast_15 stored as values in memory (estimated size 3.2 KB, free 1456.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MemoryStore:54 - Block broadcast_15_piece0 stored as bytes in memory (estimated size 1921.0 B, free 1456.9 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO BlockManagerInfo:54 - Added broadcast_15_piece0 in memory on 10.0.106.106:39767 (size: 1921.0 B, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO SparkContext:54 - Created broadcast 15 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Submitting 2 missing tasks from ResultStage 10 (ShuffledRDD[68] at countByKey at ColumnProfiler.scala:566) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO YarnScheduler:54 - Adding task set 10.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Starting task 0.0 in stage 10.0 (TID 15, algo-1, executor 1, partition 0, NODE_LOCAL, 7660 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Starting task 1.0 in stage 10.0 (TID 16, algo-1, executor 1, partition 1, NODE_LOCAL, 7660 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO BlockManagerInfo:54 - Added broadcast_15_piece0 in memory on algo-1:39511 (size: 1921.0 B, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MapOutputTrackerMasterEndpoint:54 - Asked to send map output locations for shuffle 2 to 10.0.106.106:36964\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Finished task 1.0 in stage 10.0 (TID 16) in 93 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Finished task 0.0 in stage 10.0 (TID 15) in 101 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO YarnScheduler:54 - Removed TaskSet 10.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - ResultStage 10 (countByKey at ColumnProfiler.scala:566) finished in 0.112 s\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Job 7 finished: countByKey at ColumnProfiler.scala:566, took 0.500770 s\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO ConstraintGenerator:46 - Generating Constraints:\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO ConstraintGenerator:51 - Constraints: {\n \"version\" : 0.0,\n \"features\" : [ {\n \"name\" : \"Target\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Dst Port\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Flow Duration\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 0.008919722497522299,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Tot Fwd Pkts\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Tot Bwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"TotLen Fwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"TotLen Bwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Pkt Len Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Pkt Len Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Pkt Len Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Pkt Len Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Pkt Len Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Pkt Len Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Pkt Len Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Pkt Len Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Flow Byts/s\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Flow Pkts/s\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Flow IAT Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Flow IAT Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Flow IAT Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Flow IAT Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd IAT Tot\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd IAT Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd IAT Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd IAT Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd IAT Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd IAT Tot\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd IAT Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd IAT Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd IAT Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd IAT Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd PSH Flags\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd PSH Flags\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd URG Flags\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd URG Flags\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Header Len\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Header Len\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Pkts/s\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Pkts/s\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Pkt Len Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Pkt Len Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Pkt Len Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Pkt Len Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Pkt Len Var\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"FIN Flag Cnt\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"SYN Flag Cnt\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"RST Flag Cnt\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"PSH Flag Cnt\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"ACK Flag Cnt\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"URG Flag Cnt\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"CWE Flag Count\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"ECE Flag Cnt\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Down/Up Ratio\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Pkt Size Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Seg Size Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Seg Size Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Byts/b Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Pkts/b Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Blk Rate Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Byts/b Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Pkts/b Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Bwd Blk Rate Avg\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Subflow Fwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Subflow Fwd Byts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Subflow Bwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Subflow Bwd Byts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Init Fwd Win Byts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Init Bwd Win Byts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : false\n }\n }, {\n \"name\" : \"Fwd Act Data Pkts\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Fwd Seg Size Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Active Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Active Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Active Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Active Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Idle Mean\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Idle Std\",\n \"inferred_type\" : \"Fractional\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Idle Max\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"Idle Min\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"day\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"month\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"year\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"dayofweek\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"prot_0\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"prot_6\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n }, {\n \"name\" : \"prot_17\",\n \"inferred_type\" : \"Integral\",\n \"completeness\" : 1.0,\n \"num_constraints\" : {\n \"is_non_negative\" : true\n }\n } ],\n \"monitoring_config\" : {\n \"evaluate_constraints\" : \"Enabled\",\n \"emit_metrics\" : \"Enabled\",\n \"datatype_check_threshold\" : 1.0,\n \"domain_content_threshold\" : 1.0,\n \"distribution_constraints\" : {\n \"perform_comparison\" : \"Enabled\",\n \"comparison_threshold\" : 0.1,\n \"comparison_method\" : \"Robust\"\n }\n }\u001b[0m\n\u001b[34m}\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO FileUtil:29 - Write to file constraints.json at path /opt/ml/processing/output.\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO StatsGenerator:67 - Generating Stats:\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO CodeGenerator:54 - Code generated in 10.954084 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO CodeGenerator:54 - Code generated in 7.471544 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO SparkContext:54 - Starting job: count at StatsGenerator.scala:69\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Registering RDD 73 (count at StatsGenerator.scala:69)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Got job 8 (count at StatsGenerator.scala:69) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Final stage: ResultStage 12 (count at StatsGenerator.scala:69)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Parents of final stage: List(ShuffleMapStage 11)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Missing parents: List(ShuffleMapStage 11)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Submitting ShuffleMapStage 11 (MapPartitionsRDD[73] at count at StatsGenerator.scala:69), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MemoryStore:54 - Block broadcast_16 stored as values in memory (estimated size 129.8 KB, free 1456.8 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MemoryStore:54 - Block broadcast_16_piece0 stored as bytes in memory (estimated size 43.9 KB, free 1456.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO BlockManagerInfo:54 - Added broadcast_16_piece0 in memory on 10.0.106.106:39767 (size: 43.9 KB, free: 1458.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO SparkContext:54 - Created broadcast 16 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Submitting 2 missing tasks from ShuffleMapStage 11 (MapPartitionsRDD[73] at count at StatsGenerator.scala:69) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO YarnScheduler:54 - Adding task set 11.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Starting task 0.0 in stage 11.0 (TID 17, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Starting task 1.0 in stage 11.0 (TID 18, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO BlockManagerInfo:54 - Added broadcast_16_piece0 in memory on algo-1:39511 (size: 43.9 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Finished task 1.0 in stage 11.0 (TID 18) in 84 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Finished task 0.0 in stage 11.0 (TID 17) in 89 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO YarnScheduler:54 - Removed TaskSet 11.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - ShuffleMapStage 11 (count at StatsGenerator.scala:69) finished in 0.116 s\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - looking for newly runnable stages\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - running: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - waiting: Set(ResultStage 12)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - failed: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Submitting ResultStage 12 (MapPartitionsRDD[76] at count at StatsGenerator.scala:69), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MemoryStore:54 - Block broadcast_17 stored as values in memory (estimated size 7.4 KB, free 1456.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MemoryStore:54 - Block broadcast_17_piece0 stored as bytes in memory (estimated size 3.8 KB, free 1456.7 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO BlockManagerInfo:54 - Added broadcast_17_piece0 in memory on 10.0.106.106:39767 (size: 3.8 KB, free: 1458.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO SparkContext:54 - Created broadcast 17 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 12 (MapPartitionsRDD[76] at count at StatsGenerator.scala:69) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO YarnScheduler:54 - Adding task set 12.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Starting task 0.0 in stage 12.0 (TID 19, algo-1, executor 1, partition 0, NODE_LOCAL, 7765 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO BlockManagerInfo:54 - Added broadcast_17_piece0 in memory on algo-1:39511 (size: 3.8 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO MapOutputTrackerMasterEndpoint:54 - Asked to send map output locations for shuffle 3 to 10.0.106.106:36964\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO TaskSetManager:54 - Finished task 0.0 in stage 12.0 (TID 19) in 36 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO YarnScheduler:54 - Removed TaskSet 12.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - ResultStage 12 (count at StatsGenerator.scala:69) finished in 0.055 s\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO DAGScheduler:54 - Job 8 finished: count at StatsGenerator.scala:69, took 0.176341 s\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO StatsGenerator:72 - Stats: {\n \"version\" : 0.0,\n \"dataset\" : {\n \"item_count\" : 1009\n },\n \"features\" : [ {\n \"name\" : \"Target\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.03270564915758176,\n \"sum\" : 33.0,\n \"std_dev\" : 0.43839593363324475,\n \"min\" : 0.0,\n \"max\" : 7.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.7,\n \"count\" : 1003.0\n }, {\n \"lower_bound\" : 0.7,\n \"upper_bound\" : 1.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.4,\n \"upper_bound\" : 2.1,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.1,\n \"upper_bound\" : 2.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.8,\n \"upper_bound\" : 3.5,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.5,\n \"upper_bound\" : 4.2,\n \"count\" : 3.0\n }, {\n \"lower_bound\" : 4.2,\n \"upper_bound\" : 4.9,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.9,\n \"upper_bound\" : 5.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 5.6,\n \"upper_bound\" : 6.3,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 6.3,\n \"upper_bound\" : 7.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 4.0, 7.0, 0.0, 4.0, 7.0, 0.0, 4.0, 7.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Dst Port\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 22.517343904856293,\n \"sum\" : 22720.0,\n \"std_dev\" : 5.45328357650444,\n \"min\" : 22.0,\n \"max\" : 80.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 22.0,\n \"upper_bound\" : 27.8,\n \"count\" : 1000.0\n }, {\n \"lower_bound\" : 27.8,\n \"upper_bound\" : 33.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 33.6,\n \"upper_bound\" : 39.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 39.4,\n \"upper_bound\" : 45.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 45.2,\n \"upper_bound\" : 51.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 51.0,\n \"upper_bound\" : 56.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 56.8,\n \"upper_bound\" : 62.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 62.6,\n \"upper_bound\" : 68.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 68.4,\n \"upper_bound\" : 74.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 74.2,\n \"upper_bound\" : 80.0,\n \"count\" : 9.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 80.0, 80.0, 80.0, 80.0, 80.0, 80.0, 80.0, 80.0, 80.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0, 22.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Flow Duration\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 9,\n \"num_missing\" : 1000\n },\n \"mean\" : 1.8463239666666668E7,\n \"sum\" : 1.66169157E8,\n \"std_dev\" : 2.5360160446064483E7,\n \"min\" : 10151.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 10151.0,\n \"upper_bound\" : 5441419.1,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5441419.1,\n \"upper_bound\" : 1.08726872E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.08726872E7,\n \"upper_bound\" : 1.63039553E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.63039553E7,\n \"upper_bound\" : 2.17352234E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.17352234E7,\n \"upper_bound\" : 2.71664915E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.71664915E7,\n \"upper_bound\" : 3.25977596E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.25977596E7,\n \"upper_bound\" : 3.80290277E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.80290277E7,\n \"upper_bound\" : 4.34602958E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.34602958E7,\n \"upper_bound\" : 4.88915639E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.88915639E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 1056736.0, 10151.0, 5.4322832E7, 1056736.0, 10151.0, 5.4322832E7, 1056736.0, 10151.0, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Tot Fwd Pkts\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 39.961347869177025,\n \"sum\" : 40320.99999999962,\n \"std_dev\" : 3.5699845184177255,\n \"min\" : 2.0,\n \"max\" : 40.3,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 2.0,\n \"upper_bound\" : 5.83,\n \"count\" : 9.0\n }, {\n \"lower_bound\" : 5.83,\n \"upper_bound\" : 9.66,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 9.66,\n \"upper_bound\" : 13.489999999999998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 13.489999999999998,\n \"upper_bound\" : 17.32,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 17.32,\n \"upper_bound\" : 21.15,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 21.15,\n \"upper_bound\" : 24.979999999999997,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 24.979999999999997,\n \"upper_bound\" : 28.809999999999995,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 28.809999999999995,\n \"upper_bound\" : 32.64,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 32.64,\n \"upper_bound\" : 36.47,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 36.47,\n \"upper_bound\" : 40.3,\n \"count\" : 1000.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 3.0, 2.0, 2.0, 3.0, 2.0, 2.0, 3.0, 2.0, 2.0, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3, 40.3 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Tot Bwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.011892963330029732,\n \"sum\" : 12.0,\n \"std_dev\" : 0.21778523995750837,\n \"min\" : 0.0,\n \"max\" : 4.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.4,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 0.4,\n \"upper_bound\" : 0.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.8,\n \"upper_bound\" : 1.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.2,\n \"upper_bound\" : 1.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.6,\n \"upper_bound\" : 2.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.0,\n \"upper_bound\" : 2.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.4,\n \"upper_bound\" : 2.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.8,\n \"upper_bound\" : 3.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.2,\n \"upper_bound\" : 3.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.6,\n \"upper_bound\" : 4.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 4.0, 0.0, 0.0, 4.0, 0.0, 0.0, 4.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"TotLen Fwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.05946481665014866,\n \"sum\" : 60.0,\n \"std_dev\" : 1.0889261997875417,\n \"min\" : 0.0,\n \"max\" : 20.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 2.0,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 2.0,\n \"upper_bound\" : 4.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.0,\n \"upper_bound\" : 6.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 6.0,\n \"upper_bound\" : 8.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 8.0,\n \"upper_bound\" : 10.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 10.0,\n \"upper_bound\" : 12.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 12.0,\n \"upper_bound\" : 14.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 14.0,\n \"upper_bound\" : 16.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 16.0,\n \"upper_bound\" : 18.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 18.0,\n \"upper_bound\" : 20.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 20.0, 0.0, 0.0, 20.0, 0.0, 0.0, 20.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"TotLen Bwd Pkts\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 2.8662041625371657,\n \"sum\" : 2892.0,\n \"std_dev\" : 52.48624282975952,\n \"min\" : 0.0,\n \"max\" : 964.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 96.4,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 96.4,\n \"upper_bound\" : 192.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 192.8,\n \"upper_bound\" : 289.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 289.2,\n \"upper_bound\" : 385.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 385.6,\n \"upper_bound\" : 482.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 482.0,\n \"upper_bound\" : 578.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 578.4,\n \"upper_bound\" : 674.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 674.8,\n \"upper_bound\" : 771.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 771.2,\n \"upper_bound\" : 867.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 867.6,\n \"upper_bound\" : 964.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 964.0, 0.0, 0.0, 964.0, 0.0, 0.0, 964.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd Pkt Len Max\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.05946481665014866,\n \"sum\" : 60.0,\n \"std_dev\" : 1.0889261997875417,\n \"min\" : 0.0,\n \"max\" : 20.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 2.0,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 2.0,\n \"upper_bound\" : 4.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.0,\n \"upper_bound\" : 6.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 6.0,\n \"upper_bound\" : 8.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 8.0,\n \"upper_bound\" : 10.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 10.0,\n \"upper_bound\" : 12.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 12.0,\n \"upper_bound\" : 14.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 14.0,\n \"upper_bound\" : 16.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 16.0,\n \"upper_bound\" : 18.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 18.0,\n \"upper_bound\" : 20.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 20.0, 0.0, 0.0, 20.0, 0.0, 0.0, 20.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd Pkt Len Min\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.0,\n \"sum\" : 0.0,\n \"std_dev\" : 0.0,\n \"min\" : 0.0,\n \"max\" : 0.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 1009.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd Pkt Len Mean\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 1.0042306382560349,\n \"sum\" : 1013.2687140003393,\n \"std_dev\" : 0.37442435793978285,\n \"min\" : 0.0,\n \"max\" : 6.666666667,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.6666666667000001,\n \"count\" : 57.0\n }, {\n \"lower_bound\" : 0.6666666667000001,\n \"upper_bound\" : 1.3333333334000002,\n \"count\" : 916.0\n }, {\n \"lower_bound\" : 1.3333333334000002,\n \"upper_bound\" : 2.0000000001,\n \"count\" : 33.0\n }, {\n \"lower_bound\" : 2.0000000001,\n \"upper_bound\" : 2.6666666668000003,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.6666666668000003,\n \"upper_bound\" : 3.3333333334999997,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.3333333334999997,\n \"upper_bound\" : 4.0000000002,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.0000000002,\n \"upper_bound\" : 4.6666666669,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.6666666669,\n \"upper_bound\" : 5.333333333600001,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 5.333333333600001,\n \"upper_bound\" : 6.0000000003,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 6.0000000003,\n \"upper_bound\" : 6.666666666999999,\n \"count\" : 0.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.746989504947279, 1.1206173085600735, 0.9730594629751137, 1.0291816138757108, 1.0607422959504458, 1.0813093307970258, 1.0805533941096637, 1.2858366832722126, 0.9966644818977114, 0.7327669693602721, 1.2122157395912665, 1.2317624204319026, 1.2107324617493656, 1.183505444115929, 1.1457478565198456, 0.9361195693920595, 0.8422083137703985, 1.0547009877396447, 0.5822236232250695, 0.8079067402053163, 1.190194629375473, 0.8876829830580462, 1.2040891610487179, 1.2762425828914723, 1.0451182585308558, 1.0264488272964307, 0.9149583491546008, 0.7572046705194118, 0.7263427006736167, 1.273578770805067, 0.8196471495077503, 1.070553852459601, 0.8810946392604552, 0.7583703906031055, 1.370011438682092, 0.7772067064289138, 0.6494812985571745, 1.1203899850838006, 0.7632099539677769, 0.7317305132491743, 1.0037697316605554, 0.7003056247427717, 0.9637908997659987, 0.7936947132936578, 0.9539793900701655, 0.6576768480794484, 1.0190049001460337, 1.0461820595606077, 1.1800462511922913, 0.6689914476263483, 0.8872277329379729, 0.8444201929104964, 1.0858374843669194, 1.081333950329977, 0.668894323671694, 1.1786467157598053, 1.1500745903670562, 0.5721272988708768, 1.1947696962009193, 1.1917243019139343, 0.9530016162124333, 1.1252951262017703, 0.8864598140967683, 1.0112923764620907, 1.1919061743295853, 1.0338016120338533, 1.0652865491449157, 0.6030798863330207, 0.825014866319617, 0.9683647650054076, 1.180867033067194, 1.3044335399271079, 1.185471163964403, 1.3958513100334673, 1.0698033503341304, 0.7160843166595559, 0.7763446809240415, 1.265583644893016, 0.8201747400780215, 0.8529414621092158, 0.943452929804391, 0.7347022820074054, 1.0223232053870912, 0.9628693361920359, 1.0752860978654932, 1.0336631670302039, 1.152859172580336, 1.116467256755739, 1.0140790700690148, 0.8134098689479979, 0.9560138365429587, 0.9426729785744657, 0.9110016778192674, 0.9156380543683037, 1.004140621790672, 0.7570066170057141, 0.7518912129104973, 0.8344029723155516, 1.2837568475921735, 0.5504108586653458, 0.9791371981708832, 0.9260546732842723, 1.1164518279161713, 0.7878096474947904, 1.0911075984195893, 1.1124797361512178, 1.1462207245572884, 0.6002202096952811, 0.8146912325069238, 0.7255142119330584, 1.1436232495550054, 1.176760933793268, 1.0489008965503943, 1.4628397369198556, 0.6914924487987464, 0.9663092153461219, 1.1535488089919093, 0.694573782126682, 1.1402616764387734, 1.1070631009746028, 1.2853790924567419, 1.1768907229882943, 0.8218885296912729, 0.8249673729648839, 0.7867333821931503, 1.1227731823981446, 1.36411729986408, 0.9523012747908641, 0.8474342721769501, 1.0056590834474706, 0.8333243400280026, 1.1097869667596905, 0.7009100331463161, 0.9629420171902994, 1.1469643355542254, 1.0278118524611966, 1.1705871475155398, 0.9811631982990563, 0.9685590044376828, 0.5061470085656521, 1.029228484065922, 1.2421740789605047, 0.6552091107390262, 1.2046002550329442, 1.0013068215297314, 0.6992032632915217, 0.7938846532088816, 0.9699946432124363, 0.9492909890063963, 1.068516739792617, 0.7840383797952823, 0.9232714089286357, 0.9228975638269051, 0.7726594114656525, 1.1932279998978235, 0.8819743058589906, 0.8647039846136844, 0.9990929560022017, 0.7702093964514649, 0.6905512215282786, 0.9093723222298753, 1.1849069727507588, 1.2718863804610059, 0.7757573108975876, 0.6049427000954644, 1.0443936361850958, 0.6954302784518092, 1.2925292637837673, 1.0159114802808296, 1.041378684387264, 0.839863850235302, 1.158629354775706, 1.2526493134621988, 1.2323454547013455, 1.0870965828944876, 0.882143565263157, 0.9090169955687706, 0.9857909525627042, 0.7103825874874776, 0.9861048483225235, 1.2818139938589637, 0.9667170968842635, 0.8528240831348102, 1.149358483254821, 0.7203883769967876, 1.0204853780948928, 1.237343048398173, 0.7302518889139413, 1.0748651555107842, 0.7199523300671928, 0.8339218123917678, 1.0343276099255052, 1.1841209718255286, 1.0888856636956232, 1.1810281040126398, 0.6232689565895433, 0.7015533650541501, 0.8163273506578514, 1.0042819199461364, 1.0544133990855804, 0.7437613302543777, 1.1538802459313215, 1.0953235842454214, 1.2504848841196023, 0.7361599877329638, 1.3039372646694, 0.9856514918931002, 1.0095949447907206, 1.348374387605678, 1.1482559181792857, 0.8345407393742542, 1.3564614874593155, 1.1211442337888347, 0.8884822840439399, 0.8864361352721659, 1.0493027616806667, 1.2669821745591563, 1.0893885399549836, 1.32219894063308, 1.0591235364817622, 0.6336053859016975, 0.6006831468866451, 1.0285467224924898, 0.7950330724164048, 0.4141283862922178, 0.8946219096962027, 0.681446687439032, 0.9898781132073049, 0.6958853934790925, 0.8968294905491392, 1.151416975870089, 0.954922211520799, 1.1925370776288857, 0.9313405878927886, 1.5362994649593897, 0.9449085646991524, 1.1195091406954423, 0.9018423060976579, 1.2119353070708492, 0.7994734997410435, 1.00607142482175, 1.1761834655442913, 1.2494255639972032, 0.626706980371827, 1.5546671695022347, 0.7563864389612198, 1.141077041093937, 1.0389041432467705, 0.8017883281624356, 1.0251073257856482, 1.1921695630741191, 0.8171588167652867, 0.8511019665120947, 0.980546184666291, 1.116219324328978, 0.8261535553496357, 1.3347591796218679, 1.161875165116222, 0.9551030030834977, 0.7210569371600827, 1.170721559504421, 0.4907725788599664, 1.099957097956701, 0.6486020747713978, 0.9300837518323442, 1.0458106178031459, 1.2590987301697611, 1.1792220194830416, 1.0287194135374653, 1.1289693594239416, 1.1526894160591814, 1.1730085001718467, 0.7635870505876335, 0.9516725557815677, 1.1787226151925916, 1.3044192037346118, 1.0758904120382977, 0.9212317959440584, 0.8086611959549803, 1.2085891843858336, 1.0769610442693498, 0.848003064234272, 1.230831387977605, 1.1051737378330797, 1.1246947834759566, 0.8890666443628923, 0.8974792244358064, 0.541595160967971, 1.2456245164902524, 1.1249293836307892, 1.2014779876469481, 6.666666667, 0.0, 0.0, 6.666666667, 0.0, 0.0, 6.666666667, 0.0, 0.0, 0.9549042444517206, 1.021146879077744, 1.2291549256210057, 1.066023843819341, 0.7205516693354455, 1.136349594936645, 0.9626766696481165, 1.1283977989087648, 0.8699720162112676, 0.9046420625709664, 1.1900726922847875, 1.1881299491647608, 1.0142383122103902, 0.7902393093269163, 1.0718014303532093, 0.9857897334163683, 0.9301176542556939, 0.9305857164368769, 0.8398629953001184, 0.8141025293651463, 0.991419584799469, 0.963957546888709, 1.354038618096421, 1.084687835749879, 1.2741305904756586, 0.5844801214530218, 0.879874626761135, 1.0948586976534398, 0.8766583775665289, 0.8316280342813751, 0.6631284005203597, 0.7165295223767443, 1.1251898462275933, 0.9673332669074467, 0.9581901986768958, 1.338278307864329, 0.954800015064386, 0.7178457376787503, 1.1226996092428847, 1.3465376925318937, 1.1012305346070592, 1.0519586205042437, 1.0598613219038469, 0.37936166228238255, 1.3438182202548663, 0.9756545391452229, 0.8676672379859097, 1.0212574216895578, 0.6945121282613353, 0.9438777069467013, 0.7717517907296425, 0.693793197279486, 0.8464806994204871, 0.7571531065691475, 0.836541377992738, 0.5999674194055746, 0.884727007356806, 1.2677415582695986, 0.9526232584343306, 1.2539832293656117, 0.43221320226091076, 1.1524464168085489, 1.0219571013956321, 0.9966163397202419, 0.8560178398585117, 1.0970046771810378, 1.1037058157132207, 0.9206640727455211, 0.8581070613519233, 0.9604166681676353, 1.2838709728936022, 1.0739186748035703, 0.8896988165554519, 1.0589165480731044, 1.0596283719675745, 0.9232055369879504, 0.8993669464017714, 1.095413695332944, 1.287456023859114, 0.9743114931112279, 1.0775425525423352, 1.2181400016870518, 0.6381638613651801, 0.9954384265426709, 1.175674106253768, 0.6550172372647836, 0.8892982360185623, 1.2582872408979828, 1.0663645144319236, 1.7265820539972294, 0.4480164035676426, 0.9376351383290458, 1.0337074240084776, 0.9341052787872842, 1.0886703940330957, 1.2784025285847602, 0.7388611275649652, 1.3079343557734813, 1.2020566403621968, 1.0398921862304078, 1.311748968495464, 0.9712230496036361, 0.9701957713276953, 0.7557843531522143, 0.9395837406927494, 0.8949896236933099, 1.0504078073951844, 1.1603491447550596, 0.9831483526003711, 0.9674119867601022, 0.7974172436451074, 1.04267099907387, 1.1273065195794945, 1.1786677661652192, 0.8438002662406144, 1.1553617671175924, 0.6198054970528849, 0.8258230159700956, 0.8266035689257014, 0.9088117496012279, 1.1096862870666164, 1.1127036660612517, 0.6566664831498946, 1.3363266695063878, 0.8118434424011123, 0.9505410434854852, 0.9201117239103014, 1.1092098304112754, 0.7077665294177348, 1.1121980285249908, 0.7045126234279421, 0.8266359723025863, 0.845112042515736, 0.9102168138043977, 1.0006027916348674, 1.2308272785165575, 1.048981007596588, 0.8702226202984925, 1.0692327893884566, 0.6233621120807713, 1.2307648409462404, 1.1008867212788422, 0.933101210867308, 0.7077491093140844, 0.6367048030725186, 1.1584559030354222, 1.3536153655418817, 1.1678819931191866, 0.9980582822846842, 1.0379862062019978, 0.905682339142586, 1.0901404434096655, 1.1166174381442944, 0.8717410402172414, 1.2452810529966092, 0.7710125728252615, 0.8267521652857953, 0.6528760345899669, 0.8839511740238222, 0.7168064907529785, 0.7320714383181028, 0.8651030853472726, 0.7660085203035476, 0.9957541151708679, 0.8254908024334451, 0.813974806646658, 1.2051881045253974, 1.1430224737149743, 0.7279546339969487, 1.0677775406963508, 1.275443163306547, 0.9405745994714173, 0.8578017409470206, 1.1186668202800245, 1.0111685963478159, 1.1978278621195337, 0.8456286220127904, 0.799642771265285, 0.9349078007340373, 1.172234843108231, 0.6524166100955398, 0.7555476536827208, 0.8747154319039241, 1.042954808395808, 1.1699383221982063, 0.9651893741074853, 1.0171733571193187, 1.0134038745102103, 1.0887038956481023, 1.1542224234724399, 1.1619282392005439, 1.0352590163064828, 1.213151984270153, 1.8069978687657762, 1.26290206516018, 0.9962156864763718, 0.7356264640780504, 1.0553492139921499, 0.688517664141061, 1.0729261244389083, 1.1797469228505353, 1.1979691885621848, 1.0587380537415614, 1.0294700001358381, 0.9557707652506899, 1.0993380393517882, 0.7517862438070704, 0.48448522963965635, 1.1957855973433267, 0.6947468783794677, 1.0392907511944245, 0.8124552268492531, 1.2432237472181418, 1.2947072812360017, 1.2164863202906855, 0.7526816951699422, 0.9186501460211952, 0.7553260063515775, 1.128246030464562, 1.0186809017207765, 1.0433086162845144, 1.0823198472258997, 0.8954701589678304, 1.0424245080115218, 1.0010192608569235, 1.01795726767169, 1.0174237192432696, 1.1747208027372393, 1.0830458604793527, 0.9018020455526403, 0.8267367558126845, 0.839196576018584, 0.730376381252068, 0.9038986635143595, 0.9092975801007572, 0.9290097134561156, 1.1219572174576764, 1.176817373559033, 0.8971796845404897, 0.8911270657032315, 0.7662183769863188, 0.9774460615999389, 1.0983432660340338, 1.1760562809503923, 1.0179259062529913, 1.002584007858695, 1.1077055471083384, 0.9134120049577396, 0.8436153934936658, 0.9941213563172353, 1.3468884310692175, 1.0133611812843961, 0.9138789453366346, 0.9614007269549749, 1.0829165436315502, 1.2422069443343662, 0.8849555197009118, 0.999551856036739, 0.886197423689973, 1.039546705607713, 0.9956031566760756, 0.8724284875900482, 1.1319490873589442, 0.6780635920942291, 1.0527047653563424, 1.2648542995820564, 0.9312774108868106, 1.3284979289960073, 0.9385670948627178, 0.7615149552328557, 0.6746903228164782, 1.436310915290467, 0.8377376618101431, 0.5936150485571752, 0.9100522938139354, 0.716584291326829, 1.2128980696696514, 1.0222462200057263, 0.8936062056217653, 0.7146184565457256, 1.2687757398105215, 0.7099584642538307, 1.0485864351124305, 0.9459212502715644, 0.8804479862136904, 0.8244653391048391, 0.973374540474125, 0.6211671656176467, 0.8866700114182117, 0.7214027296362917, 0.6949939271669578, 0.978074341030571, 0.8241150149595017, 1.099131810860252, 0.9387708213502862, 1.2411245814471723, 0.7235615499218031, 1.2109779878358196, 0.872896468155921, 1.435549325670597, 0.9833787252708307, 0.5439486238175031, 1.0606405296884864, 1.1761499729225984, 0.8294323835744306, 1.1336745241820854, 0.8845311281575404, 0.994943205945073, 1.4154047968657453, 0.5791927893555318, 0.745472356998592, 0.975463250198956, 1.003995578114161, 1.00131273083247, 1.1427245818616187, 0.9532070837212552, 1.0079523718282446, 1.0871413009841695, 1.001310849956808, 0.656982453539481, 1.13031149570234, 0.6452078862814863, 1.0052982624708517, 0.7425129501184381, 1.0256782557282087, 0.800446276213219, 1.209213295591672, 1.1214934622450254, 1.081515486709797, 1.0809671571451989, 0.4625943195365707, 1.2285240558013626, 1.1124677492562658, 0.9116829146667514, 1.1638468490875067, 1.114485416012853, 0.7997553289714877, 1.4257096371220048, 0.8883267845549389, 0.95626198339674, 1.2615411390396727, 1.0392332539665508, 0.9938999632092876, 1.2804408177036528, 1.0106873679296902, 1.0105829052085578, 0.9758517460177468, 0.8219072436297736, 0.6517032428922935, 1.241215083557, 0.8802846708369974, 1.077869990871475, 1.066711092317819, 0.6405751623073217, 1.3704139303063754, 1.232485230091298, 0.8057033278004779, 1.2530940054526185, 1.3572346737954069, 1.3168690411433883, 1.0053626985225272, 0.9613741744509201, 0.9071371508308228, 1.0599025631631669, 1.1047680377521283, 1.0490157105367701, 1.2413681059086858, 1.0353562094357573, 1.1288323780946175, 0.8401860704696195, 1.0416404441743214, 0.9706161931486506, 0.7389403519176263, 0.8972796714723661, 0.9523068566236232, 1.2557323713844044, 1.1624450126638202, 1.0514530626858538, 0.8736349396155798, 1.1340862684403576, 1.0414012210246733, 1.2261001465433328, 0.9610744391490793, 1.1120413636540454, 0.7394149168167761, 0.8191524875386132, 0.7246314406277754, 1.2332540105911822, 1.0365316215387756, 1.0963489818939087, 1.1163377834012982, 1.2726331235141826, 1.0923237699618782, 0.7851332124479763, 0.6456429127466101, 0.672723652219553, 1.1689503973001205, 1.01201583471957, 1.0653641236909244, 1.2145505177831557, 0.9196983273265977, 0.8601681123961351, 1.2235735724644594, 1.276183977956995, 0.994552742469622, 1.0481713404632451, 0.7789670725236839, 1.2725241122579114, 0.9494650937314321, 1.0897173715943396, 1.1075923753123726, 1.1403739372010528, 1.0068571600711311, 0.9174475892609426, 0.7329677183931471, 1.015950881841398, 0.7448518879882857, 1.3026096288707145, 0.8425788079657135, 0.9580722130512884, 0.836652785766265, 0.9760274805356115, 1.0665917681337276, 1.2316725076297808, 1.2677435879769385, 0.8381364482447372, 0.7894521696860757, 1.3059877055056766, 0.9021172765273175, 0.8213866035646299, 0.9741779338113692, 0.8168734405417992, 0.6159540000916219, 0.7857553492661554, 1.3177940238067816, 0.8600469959587664, 0.9857222652203034, 1.1046594884560192, 0.9546862749976899, 1.1566611300740064, 1.4137409167582473, 1.1068555126062345, 1.1462953233630095, 0.93864298034088, 1.2888619231328575, 1.1443078212019768, 1.2155907514062143, 0.8902675319480035, 0.7612959202957101, 1.2579938702889284, 0.9762677895895445, 1.0288193554385772, 0.9090129957105797, 0.8904935269860623, 0.969904908284857, 1.018787146809621, 0.9391290630409672, 0.9297990026898153, 1.0551632243400528, 0.9324287754228945, 1.1248449759857542, 0.98462812437094, 0.8599171444575757, 0.7580716998867434, 1.005720215257612, 0.5805692653782208, 0.6680424574378498, 1.221504860603636, 1.2412158797860104, 0.7401618710206519, 1.3315111676429312, 0.8000221363472884, 0.8867319912827121, 1.0713286582336463, 1.2445495329241167, 0.9646995797042279, 0.7650967720005917, 1.2119554493282496, 0.9973008210969153, 1.4677069986318352, 1.249476968710374, 0.9423608415711633, 0.5929885008094276, 0.9514830862896041, 1.0089158244092915, 1.0778548280332654, 0.9235614879277807, 1.2221033932563838, 1.080082073269146, 1.04614602623584, 1.0931121429982995, 1.1962100521067205, 0.6831492729569502, 0.7538548527180635, 0.9132018695956048, 0.764338794312407, 1.0141945881837748, 1.1660350084577251, 1.1675819355533297, 0.9199626943084409, 1.0649800598781978, 0.9604429323501039, 1.1604091403099572, 0.9993805184363517, 1.037487348904696, 1.2589290725976987, 1.0852738376137696, 1.1138498202072649, 0.4551182779898235, 1.1049729866172098, 0.9002981918544, 0.8458239884423921, 1.0870165998738393, 1.3310536475759422, 1.0397026457872063, 0.7655060926137767, 0.7352733017939586, 0.8367006594200368, 1.1759955471844212, 1.1687119558837522, 1.146957766603728, 1.1365703128891846, 0.9779557787957938, 0.82411026958505, 0.8629138025756916, 1.176383119631406, 0.6868944267416046, 0.9645918452896298, 0.945323201983036, 1.34565698824138, 1.0337384303992085,\n 0.9982429575463955, 0.8387900805027508, 0.7792255473461621, 1.0659026614433829, 1.3525413689421026, 0.7921659484750309, 0.9384808721014868, 1.0536829197828048, 1.0607325340129812, 1.0777731294349697, 1.0411575377969335, 0.7923610044290794, 0.8293330267894333, 1.0258706767049819, 1.2638420781178488, 1.012539786572193, 0.9517576476123244, 0.8998453338330956, 1.0797757114741187, 1.4102335519060307, 0.941965410934329, 0.8993509733157297, 1.021274010289744, 0.9007181037142952, 0.8283758243039211, 0.9315388101505266, 1.205324639072545, 0.8796313217360741, 1.1439445867388311, 1.1608601641365457, 1.0973177702880983, 1.1909919910805975, 1.1471855715910293, 1.1320241099962156, 0.8997084102379451, 1.219635271795571, 0.8332425393157287, 0.85426445774827, 1.1034825586703867, 0.9265319497823384, 0.9563904213096308, 1.1195914800613203, 1.2041995342094676, 0.9168539169418289, 1.0435728381078, 1.3689864866186374, 0.8719691398949099, 1.0250875113553475, 1.0914820579206377, 1.1802451007988872, 1.1271304155371122, 0.6172708311185815, 1.2928753100028474, 1.1461658170385327, 0.7778952480169814, 1.1701445605206924, 1.1448538420217522, 0.741885935672676, 1.1583717499426642, 0.8433896717829775, 1.0622401691385388, 0.8722703540179189, 0.9879227497028529, 0.883897764924859, 0.7870187346494247, 1.0812374296367993, 0.9947947775781496, 1.1106838291722632, 1.0671721499085218, 0.9288927470967313, 1.1198380133972825, 0.8761902188857753, 0.9729095968586889, 0.9552665512444917, 0.9347066328024073, 1.076676373183516, 0.8147585825580762, 1.2349631497929634, 0.3886946596595924, 0.8986309986525198, 0.8107746176003945, 0.8678567423073906, 1.119137614432499, 0.7400471514618203, 1.118162071645473, 0.919219640375075, 0.7661965324815206, 1.150222563474162, 1.0962309792271407, 0.6849759627249339, 0.9180502468321823, 1.0470258235081182, 1.0067646021955405, 1.2243765506056687, 0.9993752616711851, 0.6001542239936344, 1.2222703815952913, 0.796487027924132, 0.727480129732668, 1.039367327559225, 1.3200526465233744, 0.9576796313989059, 1.037507352657514, 1.1092763535533086, 0.9710570949622043, 1.1326409100536807, 0.9168047515796545, 0.9863347783736117, 1.1957460212462454, 1.092388167635826, 1.0603206658908748, 0.9499797059370189, 0.6458377612669117, 1.0431512074600093, 1.1544191097502676, 0.9341677552675542, 1.0736727464858251, 1.0094466906409456, 1.1174116715864437, 1.1130109465830331, 1.2713664158140932, 0.80071488699671, 1.198218383552184, 1.0046583089567316, 0.7260998815157471, 1.124769212233923, 0.7294812053826416, 1.0920585535059864, 1.1028935465346517, 0.9045979620058824, 1.2497023184200224, 1.0317098569545988, 1.324105508891703, 1.1899569507642025, 1.4629203547911143, 1.2317942897017211, 0.8852307865431468, 1.1247762575708191, 0.6824218894146927, 0.895836137848413, 0.7946502074025527, 0.9507337480602653, 1.1283211803722073, 1.0485925626227866, 0.8969621050329869, 0.8206376188339286, 1.0767339473244728, 0.8960027536311557, 1.0371913620990012, 0.9079604795533623, 0.8627434367628857, 1.1164140330180587, 1.26638362016188, 0.9934215486985182, 1.4936421200247956, 0.9810021577964947, 1.0641235483507792, 0.9841851802666446, 0.7512142150142374, 1.2687601932601755, 1.1274429793765428, 1.0668757796857125, 0.9093164443387418, 1.1681046444729437, 1.1482665173566102, 0.9137304961695388, 0.9945983700997858, 1.3962756733830406, 0.9801795400752518, 1.1898030104222757, 1.1771058499066467, 1.0464949606011962, 0.858616131007625, 0.849962439766064, 1.2821864434451677, 0.8736394943447566, 0.899810475299067, 1.1856744922350082 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd Pkt Len Std\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.03433202788899901,\n \"sum\" : 34.64101614,\n \"std_dev\" : 0.628691834368485,\n \"min\" : 0.0,\n \"max\" : 11.54700538,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 1.154700538,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 1.154700538,\n \"upper_bound\" : 2.309401076,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.309401076,\n \"upper_bound\" : 3.4641016139999996,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.4641016139999996,\n \"upper_bound\" : 4.618802152,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.618802152,\n \"upper_bound\" : 5.77350269,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 5.77350269,\n \"upper_bound\" : 6.928203227999999,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 6.928203227999999,\n \"upper_bound\" : 8.082903766,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 8.082903766,\n \"upper_bound\" : 9.237604304,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 9.237604304,\n \"upper_bound\" : 10.392304842,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 10.392304842,\n \"upper_bound\" : 11.54700538,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 11.54700538, 0.0, 0.0, 11.54700538, 0.0, 0.0, 11.54700538, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Bwd Pkt Len Max\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 2.8662041625371657,\n \"sum\" : 2892.0,\n \"std_dev\" : 52.48624282975952,\n \"min\" : 0.0,\n \"max\" : 964.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 96.4,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 96.4,\n \"upper_bound\" : 192.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 192.8,\n \"upper_bound\" : 289.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 289.2,\n \"upper_bound\" : 385.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 385.6,\n \"upper_bound\" : 482.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 482.0,\n \"upper_bound\" : 578.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 578.4,\n \"upper_bound\" : 674.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 674.8,\n \"upper_bound\" : 771.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 771.2,\n \"upper_bound\" : 867.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 867.6,\n \"upper_bound\" : 964.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 964.0, 0.0, 0.0, 964.0, 0.0, 0.0, 964.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Bwd Pkt Len Min\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.0,\n \"sum\" : 0.0,\n \"std_dev\" : 0.0,\n \"min\" : 0.0,\n \"max\" : 0.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 0.0,\n \"count\" : 1009.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Bwd Pkt Len Mean\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.7165510406342914,\n \"sum\" : 723.0,\n \"std_dev\" : 13.12156070743988,\n \"min\" : 0.0,\n \"max\" : 241.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 24.1,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 24.1,\n \"upper_bound\" : 48.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 48.2,\n \"upper_bound\" : 72.3,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 72.3,\n \"upper_bound\" : 96.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 96.4,\n \"upper_bound\" : 120.5,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 120.5,\n \"upper_bound\" : 144.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 144.6,\n \"upper_bound\" : 168.7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 168.7,\n \"upper_bound\" : 192.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 192.8,\n \"upper_bound\" : 216.9,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 216.9,\n \"upper_bound\" : 241.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 241.0, 0.0, 0.0, 241.0, 0.0, 0.0, 241.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Bwd Pkt Len Std\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 1.4331020812685829,\n \"sum\" : 1446.0,\n \"std_dev\" : 26.24312141487976,\n \"min\" : 0.0,\n \"max\" : 482.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 48.2,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 48.2,\n \"upper_bound\" : 96.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 96.4,\n \"upper_bound\" : 144.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 144.6,\n \"upper_bound\" : 192.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 192.8,\n \"upper_bound\" : 241.0,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 241.0,\n \"upper_bound\" : 289.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 289.2,\n \"upper_bound\" : 337.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 337.4,\n \"upper_bound\" : 385.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 385.6,\n \"upper_bound\" : 433.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 433.8,\n \"upper_bound\" : 482.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 482.0, 0.0, 0.0, 482.0, 0.0, 0.0, 482.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Flow Byts/s\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 2.768590243111992,\n \"sum\" : 2793.5075552999997,\n \"std_dev\" : 50.69872610451025,\n \"min\" : 0.0,\n \"max\" : 931.1691850999999,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 93.11691850999999,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 93.11691850999999,\n \"upper_bound\" : 186.23383701999998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 186.23383701999998,\n \"upper_bound\" : 279.35075552999996,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 279.35075552999996,\n \"upper_bound\" : 372.46767403999996,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 372.46767403999996,\n \"upper_bound\" : 465.58459254999997,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 465.58459254999997,\n \"upper_bound\" : 558.7015110599999,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 558.7015110599999,\n \"upper_bound\" : 651.81842957,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 651.81842957,\n \"upper_bound\" : 744.9353480799999,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 744.9353480799999,\n \"upper_bound\" : 838.05226659,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 838.05226659,\n \"upper_bound\" : 931.1691850999999,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 931.1691850999999, 0.0, 0.0, 931.1691850999999, 0.0, 0.0, 931.1691850999999, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Flow Pkts/s\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.6420958045504462,\n \"sum\" : 647.8746667914003,\n \"std_dev\" : 10.730201652463874,\n \"min\" : 0.0368169318,\n \"max\" : 197.0249237,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0368169318,\n \"upper_bound\" : 19.73562760862,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 19.73562760862,\n \"upper_bound\" : 39.434438285439995,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 39.434438285439995,\n \"upper_bound\" : 59.133248962260005,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 59.133248962260005,\n \"upper_bound\" : 78.83205963908,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 78.83205963908,\n \"upper_bound\" : 98.5308703159,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 98.5308703159,\n \"upper_bound\" : 118.22968099272002,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 118.22968099272002,\n \"upper_bound\" : 137.92849166954,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 137.92849166954,\n \"upper_bound\" : 157.62730234635998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 157.62730234635998,\n \"upper_bound\" : 177.32611302317997,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 177.32611302317997,\n \"upper_bound\" : 197.0249237,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 6.6241710320000005, 197.0249237, 0.0368169318, 6.6241710320000005, 197.0249237, 0.0368169318, 6.6241710320000005, 197.0249237, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318, 0.0368169318 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Flow IAT Mean\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 5.400035611199217E7,\n \"sum\" : 5.44863593170001E10,\n \"std_dev\" : 4169390.7658364405,\n \"min\" : 10151.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 10151.0,\n \"upper_bound\" : 5441419.1,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5441419.1,\n \"upper_bound\" : 1.08726872E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.08726872E7,\n \"upper_bound\" : 1.63039553E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.63039553E7,\n \"upper_bound\" : 2.17352234E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.17352234E7,\n \"upper_bound\" : 2.71664915E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.71664915E7,\n \"upper_bound\" : 3.25977596E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.25977596E7,\n \"upper_bound\" : 3.80290277E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.80290277E7,\n \"upper_bound\" : 4.34602958E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.34602958E7,\n \"upper_bound\" : 4.88915639E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.88915639E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 1003.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 176122.6667, 10151.0, 5.4322832E7, 176122.6667, 10151.0, 5.4322832E7, 176122.6667, 10151.0, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Flow IAT Std\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 1282.074664222002,\n \"sum\" : 1293613.3362,\n \"std_dev\" : 23477.49090304583,\n \"min\" : 0.0,\n \"max\" : 431204.4454,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 43120.444540000004,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 43120.444540000004,\n \"upper_bound\" : 86240.88908000001,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 86240.88908000001,\n \"upper_bound\" : 129361.33362,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 129361.33362,\n \"upper_bound\" : 172481.77816000002,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 172481.77816000002,\n \"upper_bound\" : 215602.22269999998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 215602.22269999998,\n \"upper_bound\" : 258722.66724,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 258722.66724,\n \"upper_bound\" : 301843.11178000004,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 301843.11178000004,\n \"upper_bound\" : 344963.55632000003,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 344963.55632000003,\n \"upper_bound\" : 388084.00086000003,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 388084.00086000003,\n \"upper_bound\" : 431204.44539999997,\n \"count\" : 0.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 431204.4454, 0.0, 0.0, 431204.4454, 0.0, 0.0, 431204.4454, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Flow IAT Max\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 5.4002973135777995E7,\n \"sum\" : 5.4488999894E10,\n \"std_dev\" : 4135746.2924290346,\n \"min\" : 10151.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 10151.0,\n \"upper_bound\" : 5441419.1,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5441419.1,\n \"upper_bound\" : 1.08726872E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.08726872E7,\n \"upper_bound\" : 1.63039553E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.63039553E7,\n \"upper_bound\" : 2.17352234E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.17352234E7,\n \"upper_bound\" : 2.71664915E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.71664915E7,\n \"upper_bound\" : 3.25977596E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.25977596E7,\n \"upper_bound\" : 3.80290277E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.80290277E7,\n \"upper_bound\" : 4.34602958E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.34602958E7,\n \"upper_bound\" : 4.88915639E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.88915639E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 1003.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 1056315.0, 10151.0, 5.4322832E7, 1056315.0, 10151.0, 5.4322832E7, 1056315.0, 10151.0, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Flow IAT Min\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 5.399983246283449E7,\n \"sum\" : 5.4485830955E10,\n \"std_dev\" : 4176156.287602919,\n \"min\" : 2.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 2.0,\n \"upper_bound\" : 5432285.0,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5432285.0,\n \"upper_bound\" : 1.0864568E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.0864568E7,\n \"upper_bound\" : 1.6296851E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.6296851E7,\n \"upper_bound\" : 2.1729134E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.1729134E7,\n \"upper_bound\" : 2.7161417E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.7161417E7,\n \"upper_bound\" : 3.25937E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.25937E7,\n \"upper_bound\" : 3.8025983E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.8025983E7,\n \"upper_bound\" : 4.3458266E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.3458266E7,\n \"upper_bound\" : 4.8890549E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.8890549E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 1003.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 2.0, 10151.0, 5.4322832E7, 2.0, 10151.0, 5.4322832E7, 2.0, 10151.0, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd IAT Tot\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 5.399983362834489E7,\n \"sum\" : 5.4485832131E10,\n \"std_dev\" : 4176141.21698635,\n \"min\" : 394.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 394.0,\n \"upper_bound\" : 5432637.8,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5432637.8,\n \"upper_bound\" : 1.08648816E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.08648816E7,\n \"upper_bound\" : 1.62971254E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.62971254E7,\n \"upper_bound\" : 2.17293692E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.17293692E7,\n \"upper_bound\" : 2.7161613E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.7161613E7,\n \"upper_bound\" : 3.25938568E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.25938568E7,\n \"upper_bound\" : 3.80261006E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.80261006E7,\n \"upper_bound\" : 4.34583444E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.34583444E7,\n \"upper_bound\" : 4.88905882E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.88905882E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 1003.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 394.0, 10151.0, 5.4322832E7, 394.0, 10151.0, 5.4322832E7, 394.0, 10151.0, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd IAT Mean\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 5.399983304261645E7,\n \"sum\" : 5.448583154E10,\n \"std_dev\" : 4176148.7907332485,\n \"min\" : 197.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 197.0,\n \"upper_bound\" : 5432460.5,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5432460.5,\n \"upper_bound\" : 1.0864724E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.0864724E7,\n \"upper_bound\" : 1.62969875E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.62969875E7,\n \"upper_bound\" : 2.1729251E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.1729251E7,\n \"upper_bound\" : 2.71615145E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.71615145E7,\n \"upper_bound\" : 3.2593778E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.2593778E7,\n \"upper_bound\" : 3.80260415E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.80260415E7,\n \"upper_bound\" : 4.3458305E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.3458305E7,\n \"upper_bound\" : 4.88905685E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.88905685E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 1003.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 197.0, 10151.0, 5.4322832E7, 197.0, 10151.0, 5.4322832E7, 197.0, 10151.0, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd IAT Std\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 0.8199355144697719,\n \"sum\" : 827.3149340999998,\n \"std_dev\" : 15.014748453616557,\n \"min\" : 0.0,\n \"max\" : 275.77164469999997,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 27.577164469999996,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 27.577164469999996,\n \"upper_bound\" : 55.15432893999999,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 55.15432893999999,\n \"upper_bound\" : 82.73149340999998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 82.73149340999998,\n \"upper_bound\" : 110.30865787999998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 110.30865787999998,\n \"upper_bound\" : 137.88582234999998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 137.88582234999998,\n \"upper_bound\" : 165.46298681999997,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 165.46298681999997,\n \"upper_bound\" : 193.04015128999998,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 193.04015128999998,\n \"upper_bound\" : 220.61731575999997,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 220.61731575999997,\n \"upper_bound\" : 248.19448022999995,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 248.19448022999995,\n \"upper_bound\" : 275.77164469999997,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 275.77164469999997, 0.0, 0.0, 275.77164469999997, 0.0, 0.0, 275.77164469999997, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd IAT Max\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 5.399983362239841E7,\n \"sum\" : 5.4485832125E10,\n \"std_dev\" : 4176141.293877112,\n \"min\" : 392.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 392.0,\n \"upper_bound\" : 5432636.0,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5432636.0,\n \"upper_bound\" : 1.086488E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.086488E7,\n \"upper_bound\" : 1.6297124E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.6297124E7,\n \"upper_bound\" : 2.1729368E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.1729368E7,\n \"upper_bound\" : 2.7161612E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.7161612E7,\n \"upper_bound\" : 3.2593856E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.2593856E7,\n \"upper_bound\" : 3.80261E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.80261E7,\n \"upper_bound\" : 4.3458344E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.3458344E7,\n \"upper_bound\" : 4.8890588E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.8890588E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 1003.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 392.0, 10151.0, 5.4322832E7, 392.0, 10151.0, 5.4322832E7, 392.0, 10151.0, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Fwd IAT Min\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 5.399983246283449E7,\n \"sum\" : 5.4485830955E10,\n \"std_dev\" : 4176156.287602919,\n \"min\" : 2.0,\n \"max\" : 5.4322832E7,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 2.0,\n \"upper_bound\" : 5432285.0,\n \"count\" : 6.0\n }, {\n \"lower_bound\" : 5432285.0,\n \"upper_bound\" : 1.0864568E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.0864568E7,\n \"upper_bound\" : 1.6296851E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 1.6296851E7,\n \"upper_bound\" : 2.1729134E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.1729134E7,\n \"upper_bound\" : 2.7161417E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 2.7161417E7,\n \"upper_bound\" : 3.25937E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.25937E7,\n \"upper_bound\" : 3.8025983E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 3.8025983E7,\n \"upper_bound\" : 4.3458266E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.3458266E7,\n \"upper_bound\" : 4.8890549E7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 4.8890549E7,\n \"upper_bound\" : 5.4322832E7,\n \"count\" : 1003.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 2.0, 10151.0, 5.4322832E7, 2.0, 10151.0, 5.4322832E7, 2.0, 10151.0, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7, 5.4322832E7 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Bwd IAT Tot\",\n \"inferred_type\" : \"Integral\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 3141.9217046580775,\n \"sum\" : 3170199.0,\n \"std_dev\" : 57535.21249400442,\n \"min\" : 0.0,\n \"max\" : 1056733.0,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 105673.3,\n \"count\" : 1006.0\n }, {\n \"lower_bound\" : 105673.3,\n \"upper_bound\" : 211346.6,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 211346.6,\n \"upper_bound\" : 317019.9,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 317019.9,\n \"upper_bound\" : 422693.2,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 422693.2,\n \"upper_bound\" : 528366.5,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 528366.5,\n \"upper_bound\" : 634039.8,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 634039.8,\n \"upper_bound\" : 739713.1,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 739713.1,\n \"upper_bound\" : 845386.4,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 845386.4,\n \"upper_bound\" : 951059.7,\n \"count\" : 0.0\n }, {\n \"lower_bound\" : 951059.7,\n \"upper_bound\" : 1056733.0,\n \"count\" : 3.0\n } ],\n \"sketch\" : {\n \"parameters\" : {\n \"c\" : 0.64,\n \"k\" : 2048.0\n },\n \"data\" : [ [ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1056733.0, 0.0, 0.0, 1056733.0, 0.0, 0.0, 1056733.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 ] ]\n }\n }\n }\n }\n }, {\n \"name\" : \"Bwd IAT Mean\",\n \"inferred_type\" : \"Fractional\",\n \"numerical_statistics\" : {\n \"common\" : {\n \"num_present\" : 1009,\n \"num_missing\" : 0\n },\n \"mean\" : 1047.3072347869177,\n \"sum\" : 1056732.9999,\n \"std_dev\" : 19178.40416285326,\n \"min\" : 0.0,\n \"max\" : 352244.3333,\n \"distribution\" : {\n \"kll\" : {\n \"buckets\" : [ {\n \"lower_bound\" : 0.0,\n \"upper_bound\" : 35224.43333,\n }, {\n }\u001b[0m\n\u001b[34m}\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO FileUtil:29 - Write to file statistics.json at path /opt/ml/processing/output.\u001b[0m\n\u001b[34m2020-02-04 21:05:32 INFO ConstraintGenerator:61 - Evaluating constraints\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO CodeGenerator:54 - Code generated in 29.440889 ms\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO SparkContext:54 - Starting job: collect at AnalysisRunner.scala:313\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO DAGScheduler:54 - Registering RDD 81 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO DAGScheduler:54 - Got job 9 (collect at AnalysisRunner.scala:313) with 1 output partitions\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO DAGScheduler:54 - Final stage: ResultStage 14 (collect at AnalysisRunner.scala:313)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO DAGScheduler:54 - Parents of final stage: List(ShuffleMapStage 13)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO DAGScheduler:54 - Missing parents: List(ShuffleMapStage 13)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO DAGScheduler:54 - Submitting ShuffleMapStage 13 (MapPartitionsRDD[81] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO MemoryStore:54 - Block broadcast_18 stored as values in memory (estimated size 247.3 KB, free 1456.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO MemoryStore:54 - Block broadcast_18_piece0 stored as bytes in memory (estimated size 66.2 KB, free 1456.4 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO BlockManagerInfo:54 - Added broadcast_18_piece0 in memory on 10.0.106.106:39767 (size: 66.2 KB, free: 1458.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO SparkContext:54 - Created broadcast 18 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO DAGScheduler:54 - Submitting 2 missing tasks from ShuffleMapStage 13 (MapPartitionsRDD[81] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0, 1))\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO YarnScheduler:54 - Adding task set 13.0 with 2 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO TaskSetManager:54 - Starting task 0.0 in stage 13.0 (TID 20, algo-1, executor 1, partition 0, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO TaskSetManager:54 - Starting task 1.0 in stage 13.0 (TID 21, algo-1, executor 1, partition 1, PROCESS_LOCAL, 8637 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:33 INFO BlockManagerInfo:54 - Added broadcast_18_piece0 in memory on algo-1:39511 (size: 66.2 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO TaskSetManager:54 - Finished task 1.0 in stage 13.0 (TID 21) in 1133 ms on algo-1 (executor 1) (1/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO TaskSetManager:54 - Finished task 0.0 in stage 13.0 (TID 20) in 1216 ms on algo-1 (executor 1) (2/2)\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO YarnScheduler:54 - Removed TaskSet 13.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO DAGScheduler:54 - ShuffleMapStage 13 (collect at AnalysisRunner.scala:313) finished in 1.243 s\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO DAGScheduler:54 - looking for newly runnable stages\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO DAGScheduler:54 - running: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO DAGScheduler:54 - waiting: Set(ResultStage 14)\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO DAGScheduler:54 - failed: Set()\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO DAGScheduler:54 - Submitting ResultStage 14 (MapPartitionsRDD[84] at collect at AnalysisRunner.scala:313), which has no missing parents\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO MemoryStore:54 - Block broadcast_19 stored as values in memory (estimated size 309.7 KB, free 1456.1 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO MemoryStore:54 - Block broadcast_19_piece0 stored as bytes in memory (estimated size 80.4 KB, free 1456.0 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO BlockManagerInfo:54 - Added broadcast_19_piece0 in memory on 10.0.106.106:39767 (size: 80.4 KB, free: 1458.1 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO SparkContext:54 - Created broadcast 19 from broadcast at DAGScheduler.scala:1039\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO DAGScheduler:54 - Submitting 1 missing tasks from ResultStage 14 (MapPartitionsRDD[84] at collect at AnalysisRunner.scala:313) (first 15 tasks are for partitions Vector(0))\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO YarnScheduler:54 - Adding task set 14.0 with 1 tasks\u001b[0m\n\u001b[34m2020-02-04 21:05:34 INFO TaskSetManager:54 - Starting task 0.0 in stage 14.0 (TID 22, algo-1, executor 1, partition 0, NODE_LOCAL, 7765 bytes)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Added broadcast_19_piece0 in memory on algo-1:39511 (size: 80.4 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO MapOutputTrackerMasterEndpoint:54 - Asked to send map output locations for shuffle 4 to 10.0.106.106:36964\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO TaskSetManager:54 - Finished task 0.0 in stage 14.0 (TID 22) in 226 ms on algo-1 (executor 1) (1/1)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO DAGScheduler:54 - ResultStage 14 (collect at AnalysisRunner.scala:313) finished in 0.252 s\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO YarnScheduler:54 - Removed TaskSet 14.0, whose tasks have all completed, from pool \u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO DAGScheduler:54 - Job 9 finished: collect at AnalysisRunner.scala:313, took 1.503842 s\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 256\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 237\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 254\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 420\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 283\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 360\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 246\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 385\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 407\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 379\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 336\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 405\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 234\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 279\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 294\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 316\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 460\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 377\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 301\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 406\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 422\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 394\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 302\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 308\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 271\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 231\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 435\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 299\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 331\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 353\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 226\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 441\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 247\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 322\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 439\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 249\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 446\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_13_piece0 on 10.0.106.106:39767 in memory (size: 132.8 KB, free: 1458.2 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_13_piece0 on algo-1:39511 in memory (size: 132.8 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 266\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 438\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 432\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 382\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 324\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned shuffle 1\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_19_piece0 on 10.0.106.106:39767 in memory (size: 80.4 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_19_piece0 on algo-1:39511 in memory (size: 80.4 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 282\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 426\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 362\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 399\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 343\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 370\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 416\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 374\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 459\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 371\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 325\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 368\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 409\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 298\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 251\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned shuffle 2\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 402\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 293\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 235\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 372\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 348\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 387\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 337\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 332\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 276\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 230\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 410\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 447\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 424\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 280\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 425\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_17_piece0 on 10.0.106.106:39767 in memory (size: 3.8 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_17_piece0 on algo-1:39511 in memory (size: 3.8 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 361\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 267\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 367\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 241\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 306\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 314\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 462\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 376\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 296\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 321\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 436\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 373\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 440\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 434\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 344\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 328\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 339\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 341\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 334\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 275\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 310\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 342\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 421\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 259\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 401\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 378\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 239\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 228\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 304\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 375\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 253\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 464\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 358\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 404\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 335\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 442\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 386\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 388\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 381\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 366\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 389\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 445\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 408\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_16_piece0 on 10.0.106.106:39767 in memory (size: 43.9 KB, free: 1458.3 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_16_piece0 on algo-1:39511 in memory (size: 43.9 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 414\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 469\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 273\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_14_piece0 on 10.0.106.106:39767 in memory (size: 44.9 KB, free: 1458.4 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_14_piece0 on algo-1:39511 in memory (size: 44.9 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 443\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 227\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 248\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 257\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 463\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 268\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 323\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 319\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 397\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 352\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 419\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 450\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 390\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 260\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 285\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned shuffle 4\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 262\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 456\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 309\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 245\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 229\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 307\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 417\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 278\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_12_piece0 on 10.0.106.106:39767 in memory (size: 112.9 KB, free: 1458.5 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_12_piece0 on algo-1:39511 in memory (size: 112.9 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 284\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 429\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 265\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 287\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 392\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 449\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 451\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 458\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 461\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 327\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 356\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 295\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 258\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 240\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned shuffle 3\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 277\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 357\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 398\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 431\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_18_piece0 on 10.0.106.106:39767 in memory (size: 66.2 KB, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_18_piece0 on algo-1:39511 in memory (size: 66.2 KB, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 264\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 313\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 338\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 225\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 393\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 444\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 454\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 418\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 433\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 430\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 467\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 351\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 236\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 250\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 380\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 318\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 297\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 448\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 363\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 317\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 453\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 455\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 330\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 369\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 255\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 300\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 364\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 269\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 232\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_15_piece0 on 10.0.106.106:39767 in memory (size: 1921.0 B, free: 1458.6 MB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO BlockManagerInfo:54 - Removed broadcast_15_piece0 on algo-1:39511 in memory (size: 1921.0 B, free: 5.8 GB)\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 333\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 354\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 270\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 395\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 391\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 303\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 305\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 359\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 423\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 286\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 468\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 349\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 238\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 242\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 291\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 412\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 196\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 312\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 427\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 383\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 396\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 400\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 244\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 315\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 320\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 345\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 252\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 350\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 457\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 198\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 355\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 326\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 311\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 233\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 274\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 452\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 403\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 411\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 466\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 261\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 365\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 329\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 437\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 272\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 347\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 263\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 292\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 384\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 340\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 281\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 197\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 288\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 413\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 428\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 465\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 346\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 415\u001b[0m\n\u001b[34m2020-02-04 21:05:35 INFO ContextCleaner:54 - Cleaned accumulator 243\u001b[0m\n\u001b[34m2020-02-04 21:05:36 INFO DistanceGenerator:63 - Inferred data type don't match for column Tot Fwd Pkts. Inferred data type in baseline statistics: Integral. Inferred data type in current dataset: Fractional\u001b[0m\n\n\u001b[34m2020-02-04 21:05:48 INFO StatsGenerator:86 - Evaluating stats\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO DataAnalyzer:148 - Constraint and Stats violations: {\n \"violations\" : [ {\n \"feature_name\" : \"Tot Fwd Pkts\",\n \"constraint_check_type\" : \"data_type_check\",\n \"description\" : \"Data type match requirement is not met. Expected data type: Integral, Expected match: 100.0%. Observed: Only 0.8919722497522299% of data is Integral.\"\n }, {\n \"feature_name\" : \"Flow Duration\",\n \"constraint_check_type\" : \"completeness_check\",\n \"description\" : \"Data completeness requirement is not met. Expected: 100.0%. Observed: Only 0.8919722497522299%.\"\n }, {\n \"feature_name\" : \"Fwd Pkt Len Mean\",\n \"constraint_check_type\" : \"baseline_drift_check\",\n \"description\" : \"Baseline drift distance: 0.9293200129316993 exceeds threshold: 0.1\"\n } ]\u001b[0m\n\u001b[34m}\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO FileUtil:29 - Write to file constraint_violations.json at path /opt/ml/processing/output.\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO YarnClientSchedulerBackend:54 - Interrupting monitor thread\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO YarnClientSchedulerBackend:54 - Shutting down all executors\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO YarnSchedulerBackend$YarnDriverEndpoint:54 - Asking each executor to shut down\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO SchedulerExtensionServices:54 - Stopping SchedulerExtensionServices\u001b[0m\n\u001b[34m(serviceOption=None,\n services=List(),\n started=false)\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO YarnClientSchedulerBackend:54 - Stopped\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO MapOutputTrackerMasterEndpoint:54 - MapOutputTrackerMasterEndpoint stopped!\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO MemoryStore:54 - MemoryStore cleared\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO BlockManager:54 - BlockManager stopped\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO BlockManagerMaster:54 - BlockManagerMaster stopped\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO OutputCommitCoordinator$OutputCommitCoordinatorEndpoint:54 - OutputCommitCoordinator stopped!\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO SparkContext:54 - Successfully stopped SparkContext\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO Main:53 - CompletedWithViolations: Job completed successfully with 3 violations.\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO Main:115 - Write to file /opt/ml/output/message.\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO ShutdownHookManager:54 - Shutdown hook called\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO ShutdownHookManager:54 - Deleting directory /tmp/spark-eb9f0aad-a906-4aa7-a0a2-503e2073af1a\u001b[0m\n\u001b[34m2020-02-04 21:05:48 INFO ShutdownHookManager:54 - Deleting directory /tmp/spark-ddf5a4d2-3150-4bb4-aca8-4bbb91237c44\u001b[0m\n\u001b[34m2020-02-04 21:05:49,195 - DefaultDataAnalyzer - INFO - Completed spark-submit with return code : 0\u001b[0m\n\u001b[34m2020-02-04 21:05:49,195 - DefaultDataAnalyzer - INFO - Invoking customer's post-processing script...\u001b[0m\n\u001b[34m2020-02-04 21:05:49,196 - DefaultDataAnalyzer - INFO - Spark job completed.\u001b[0m\n\u001b[34mHello from post-proc script!\u001b[0m\n"
]
],
[
[
"### Analysis",
"_____no_output_____"
],
[
"When the monitoring job completes, monitoring reports are saved to Amazon S3. Let's list the generated reports.",
"_____no_output_____"
]
],
[
[
"s3_client = boto3.Session().client('s3')\nmonitoring_reports_prefix = '{}/monitoring/reports/{}'.format(prefix, pred.endpoint)\n\nresult = s3_client.list_objects(Bucket=bucket_name, Prefix=monitoring_reports_prefix)\ntry:\n monitoring_reports = ['s3://{0}/{1}'.format(bucket_name, capture_file.get(\"Key\")) for capture_file in result.get('Contents')]\n print(\"Monitoring Reports Files: \")\n print(\"\\n \".join(monitoring_reports))\nexcept:\n print('No monitoring reports found.')",
"Monitoring Reports Files: \ns3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/constraint_violations.json\n s3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/constraints.json\n s3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/statistics.json\n"
]
],
[
[
"We then copy monitoring reports locally.",
"_____no_output_____"
]
],
[
[
"!aws s3 cp {monitoring_reports[0]} monitoring/\n!aws s3 cp {monitoring_reports[1]} monitoring/\n!aws s3 cp {monitoring_reports[2]} monitoring/",
"download: s3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/constraint_violations.json to monitoring/constraint_violations.json\ndownload: s3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/constraints.json to monitoring/constraints.json\ndownload: s3://sagemaker-us-east-1-806570384721/aim362/monitoring/reports/nw-traffic-classification-xgb-ep-2020-02-04-20-24-20/AllTraffic/2020/02/04/20/statistics.json to monitoring/statistics.json\n"
]
],
[
[
"Let's display the violations identified by the monitoring execution.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\npd.set_option('display.max_colwidth', -1)\n\nfile = open('monitoring/constraint_violations.json', 'r')\ndata = file.read()\n\nviolations_df = pd.io.json.json_normalize(json.loads(data)['violations'])\nviolations_df.head(10)",
"_____no_output_____"
]
],
[
[
"We can see that the violations identified correspond to the ones that we artificially generated and that there is a feature that is generating some drift from the baseline.",
"_____no_output_____"
],
[
"### Advanced Hints",
"_____no_output_____"
],
[
"You might be asking yourself what are the type of violations that are monitored and how drift from the baseline is computed.\n\nThe types of violations monitored are listed here: https://docs.aws.amazon.com/sagemaker/latest/dg/model-monitor-interpreting-violations.html. Most of them use configurable thresholds, that are specified in the monitoring configuration section of the baseline constraints JSON. Let's take a look at this configuration from the baseline constraints file:",
"_____no_output_____"
]
],
[
[
"!aws s3 cp {statistics_path} baseline/\n!aws s3 cp {constraints_path} baseline/",
"download: s3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/statistics.json to baseline/statistics.json\ndownload: s3://sagemaker-us-east-1-806570384721/aim362/monitoring/baselining/results/constraints.json to baseline/constraints.json\n"
],
[
"import json\nwith open (\"baseline/constraints.json\", \"r\") as myfile:\n data=myfile.read()\n\nprint(json.dumps(json.loads(data)['monitoring_config'], indent=2))",
"{\n \"evaluate_constraints\": \"Enabled\",\n \"emit_metrics\": \"Enabled\",\n \"datatype_check_threshold\": 1.0,\n \"domain_content_threshold\": 1.0,\n \"distribution_constraints\": {\n \"perform_comparison\": \"Enabled\",\n \"comparison_threshold\": 0.1,\n \"comparison_method\": \"Robust\"\n }\n}\n"
]
],
[
[
"This configuration is intepreted when the monitoring job is executed and used to compare captured data to the baseline. If you want to customize this section, you will have to update the **constraints.json** file and upload it back to Amazon S3 before launching the monitoring job.\n\nWhen data distributions are compared to detect potential drift, you can choose to use either a _Simple_ or _Robust_ comparison method, where the latter has to be preferred when dealing with small datasets. Additional info: https://docs.aws.amazon.com/sagemaker/latest/dg/model-monitor-byoc-constraints.html.",
"_____no_output_____"
],
[
"## Delete Endpoint",
"_____no_output_____"
],
[
"Finally we can delete the endpoint to free-up resources.",
"_____no_output_____"
]
],
[
[
"pred.delete_endpoint()\npred.delete_model()",
"_____no_output_____"
]
],
[
[
"## References\n\nA Realistic Cyber Defense Dataset (CSE-CIC-IDS2018) https://registry.opendata.aws/cse-cic-ids2018/",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec601108efb488b0b1d26ca8468d67dbef7c14db | 237,349 | ipynb | Jupyter Notebook | PCA.ipynb | researcher2312/machine-learning | fb13332045c08a38d6fdf56e17671a529ec4bc96 | [
"Unlicense"
]
| null | null | null | PCA.ipynb | researcher2312/machine-learning | fb13332045c08a38d6fdf56e17671a529ec4bc96 | [
"Unlicense"
]
| null | null | null | PCA.ipynb | researcher2312/machine-learning | fb13332045c08a38d6fdf56e17671a529ec4bc96 | [
"Unlicense"
]
| null | null | null | 262.844961 | 75,282 | 0.914758 | [
[
[
"# <center>PCA</center>",
"_____no_output_____"
],
[
"## Calculate the number of principal components",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib as mpl\nfrom matplotlib import pyplot as plt\nfrom sklearn.datasets import load_wine\nfrom sklearn.preprocessing import StandardScaler",
"_____no_output_____"
]
],
[
[
"Data import and scaling",
"_____no_output_____"
]
],
[
[
"wineData = load_wine().data\nwineData = StandardScaler().fit_transform(wineData)",
"_____no_output_____"
]
],
[
[
"Covariance matrix calculation",
"_____no_output_____"
]
],
[
[
"features = wineData.T\ncov_matrix = np.cov(features)",
"_____no_output_____"
],
[
"values, vectors = np.linalg.eig(cov_matrix)\nvalueSum = np.sum(values)\n\nvalues = sorted(((v, i) for i, v in enumerate(values)), reverse=True)\nvariancesSum = []\npartSum = 0\nfor i, j in values:\n variancesSum.append(i/valueSum+partSum)\n partSum += i/valueSum\n\nplt.title(\"Variances sum\")\nplt.scatter(range(1,14), variancesSum)\nplt.axhline(y=0.9)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Based on this plot, 7 vectors are needed. The vectors are as following: ",
"_____no_output_____"
]
],
[
[
"for i, num in values[:7]:\n print(vectors[num])",
"[-0.1443294 0.48365155 -0.20738262 0.0178563 -0.26566365 0.21353865\n 0.05639636 -0.01496997 0.39613926 -0.26628645 -0.50861912 -0.22591696\n 0.21160473]\n[ 0.24518758 0.22493093 0.08901289 -0.53689028 0.03521363 0.53681385\n -0.42052391 -0.02596375 0.06582674 0.12169604 0.07528304 0.07648554\n -0.30907994]\n[ 0.00205106 0.31606881 0.6262239 0.21417556 -0.14302547 0.15447466\n 0.14917061 0.14121803 -0.17026002 -0.04962237 0.30769445 -0.49869142\n -0.02712539]\n[ 0.23932041 -0.0105905 0.61208035 -0.06085941 0.06610294 -0.10082451\n 0.28696914 -0.09168285 0.42797018 -0.05574287 -0.20044931 0.47931378\n 0.05279942]\n[-0.14199204 0.299634 0.13075693 0.35179658 0.72704851 0.03814394\n -0.3228833 -0.05677422 -0.15636143 0.06222011 -0.27140257 0.07128891\n 0.06787022]\n[-0.39466085 0.06503951 0.14617896 -0.19806835 -0.14931841 -0.0841223\n 0.02792498 0.46390791 -0.40593409 -0.30388245 -0.28603452 0.30434119\n -0.32013135]\n[-0.4229343 -0.00335981 0.1506819 -0.15229479 -0.10902584 -0.01892002\n 0.06068521 -0.83225706 -0.18724536 -0.04289883 -0.04957849 -0.02569409\n -0.16315051]\n"
]
],
[
[
"## US Arrest dataset analysis",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('usarrests.csv', index_col=0)\ndf.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nIndex: 50 entries, Alabama to Wyoming\nData columns (total 4 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Murder 50 non-null float64\n 1 Assault 50 non-null int64 \n 2 UrbanPop 50 non-null int64 \n 3 Rape 50 non-null float64\ndtypes: float64(2), int64(2)\nmemory usage: 2.0+ KB\n"
],
[
"df.mean()",
"_____no_output_____"
],
[
"df.var()",
"_____no_output_____"
],
[
"from sklearn.preprocessing import scale\nX = pd.DataFrame(scale(df), index=df.index, columns=df.columns)",
"_____no_output_____"
],
[
"from sklearn.decomposition import PCA\npca_loadings = pd.DataFrame(PCA().fit(X).components_.T, index=df.columns, columns=['V1', 'V2', 'V3', 'V4'])\npca_loadings",
"_____no_output_____"
],
[
"# Fit the PCA model and transform X to get the principal components\npca = PCA()\ndf_plot = pd.DataFrame(pca.fit_transform(X), columns=['PC1', 'PC2', 'PC3', 'PC4'], index=X.index)\ndf_plot.head()",
"_____no_output_____"
],
[
"fig , ax1 = plt.subplots(figsize=(9,7))\nax1.set_xlim(-3.5,3.5)\nax1.set_ylim(-3.5,3.5)\n# Plot Principal Components 1 and 2\nfor i in df_plot.index:\n ax1.annotate(i, (-df_plot.PC1.loc[i], -df_plot.PC2.loc[i]), ha='center')\n# Plot reference lines\nax1.hlines(0,-3.5,3.5, linestyles='dotted', colors='grey')\nax1.vlines(0,-3.5,3.5, linestyles='dotted', colors='grey')\nax1.set_xlabel('First Principal Component')\nax1.set_ylabel('Second Principal Component')\n# Plot Principal Component loading vectors, using a second y-axis.\nax2 = ax1.twinx().twiny()\nax2.set_ylim(-1,1)\nax2.set_xlim(-1,1)\nax2.set_xlabel('Principal Component loading vectors', color='red')\n# Plot labels for vectors. Variable 'a' is a small offset parameter to separate arrow tip and text.\na = 1.07\nfor i in pca_loadings[['V1', 'V2']].index:\n ax2.annotate(i, (-pca_loadings.V1.loc[i]*a, -pca_loadings.V2.loc[i]*a), color='red')\n# Plot vectors\nax2.arrow(0,0,-pca_loadings.V1[0], -pca_loadings.V2[0])\nax2.arrow(0,0,-pca_loadings.V1[1], -pca_loadings.V2[1])\nax2.arrow(0,0,-pca_loadings.V1[2], -pca_loadings.V2[2])\nax2.arrow(0,0,-pca_loadings.V1[3], -pca_loadings.V2[3])\nplt.show()",
"_____no_output_____"
],
[
"pca.explained_variance_",
"_____no_output_____"
],
[
"pca.explained_variance_ratio_",
"_____no_output_____"
],
[
"plt.figure(figsize=(7,5))\nplt.plot([1,2,3,4], pca.explained_variance_ratio_, '-o')\nplt.ylabel('Proportion of Variance Explained')\nplt.xlabel('Principal Component')\nplt.xlim(0.75,4.25)\nplt.ylim(0,1.05)\nplt.xticks([1,2,3,4])\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(7,5))\nplt.plot([1,2,3,4], np.cumsum(pca.explained_variance_ratio_), '-s')\nplt.ylabel('Proportion of Variance Explained')\nplt.xlabel('Principal Component')\nplt.xlim(0.75,4.25)\nplt.ylim(0,1.05)\nplt.xticks([1,2,3,4])\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Cancer cell data analysis",
"_____no_output_____"
]
],
[
[
"df2 = pd.read_csv('nci60.csv').drop('Unnamed: 0', axis=1)\ndf2.columns = np.arange(df2.columns.size)\ny = pd.read_csv('nci60_y.csv', usecols=[1], skiprows=1, names=['type'])\ndf2.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 64 entries, 0 to 63\nColumns: 6830 entries, 0 to 6829\ndtypes: float64(6830)\nmemory usage: 3.3 MB\n"
],
[
"# Scale the data\nX = pd.DataFrame(scale(df2))\nX.shape\n# Fit the PCA model and transform X to get the principal components\npca2 = PCA()\ndf2_plot = pd.DataFrame(pca2.fit_transform(X))",
"_____no_output_____"
],
[
"fig, (ax1, ax2) = plt.subplots(1,2, figsize=(15,6))\ncolor_idx = pd.factorize(y.type)[0]\ncmap = mpl.cm.hsv\n# Left plot\nax1.scatter(df2_plot.iloc[:,0], df2_plot.iloc[:,1], c=color_idx, cmap=cmap, alpha=0.5, s=50)\nax1.set_ylabel('Principal Component 2')\n# Right plot\nax2.scatter(df2_plot.iloc[:,0], df2_plot.iloc[:,2], c=color_idx, cmap=cmap, alpha=0.5, s=50)\nax2.set_ylabel('Principal Component 3')\n# Custom legend for the classes (y) since we do not create scatter plots per class (which could have their own labels).\nhandles = []\nlabels = pd.factorize(y.type.unique())\nnorm = mpl.colors.Normalize(vmin=0.0, vmax=14.0)\nfor i, v in zip(labels[0], labels[1]):\n handles.append(mpl.patches.Patch(color=cmap(norm(i)), label=v, alpha=0.5))\nax2.legend(handles=handles, bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\n# xlabel for both plots\nfor ax in fig.axes:\n ax.set_xlabel('Principal Component 1')\n plt.show()",
"_____no_output_____"
],
[
"pd.DataFrame([df2_plot.iloc[:,:5].std(axis=0, ddof=0).to_numpy(),\npca2.explained_variance_ratio_[:5],\nnp.cumsum(pca2.explained_variance_ratio_[:5])],\nindex=['Standard Deviation', 'Proportion of Variance', 'Cumulative Proportion'],\ncolumns=['PC1', 'PC2', 'PC3', 'PC4', 'PC5'])",
"_____no_output_____"
],
[
"df2_plot.iloc[:,:10].var(axis=0, ddof=0).plot(kind='bar', rot=0)\nplt.ylabel('Variances')\nplt.show()",
"_____no_output_____"
],
[
"fig , (ax1,ax2) = plt.subplots(1,2, figsize=(15,5))\n# Left plot\nax1.plot(pca2.explained_variance_ratio_, '-o')\nax1.set_ylabel('Proportion of Variance Explained')\nax1.set_ylim(ymin=-0.01)\n# Right plot\nax2.plot(np.cumsum(pca2.explained_variance_ratio_), '-ro')\nax2.set_ylabel('Cumulative Proportion of Variance Explained')\nax2.set_ylim(ymax=1.05)\nfor ax in fig.axes:\n ax.set_xlabel('Principal Component')\nax.set_xlim(-1,65)\nplt.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6020f6d719bfea443f13e5f5b70d45e4c2cea9 | 45,849 | ipynb | Jupyter Notebook | dev/40_tabular_core.ipynb | BenjaminDev/fastai_dev | 89e7957ff18883bd2e840d93968025dabe9b3563 | [
"Apache-2.0"
]
| null | null | null | dev/40_tabular_core.ipynb | BenjaminDev/fastai_dev | 89e7957ff18883bd2e840d93968025dabe9b3563 | [
"Apache-2.0"
]
| 5 | 2019-10-28T23:03:36.000Z | 2019-11-04T23:41:04.000Z | dev/40_tabular_core.ipynb | BenjaminDev/fastai_dev | 89e7957ff18883bd2e840d93968025dabe9b3563 | [
"Apache-2.0"
]
| null | null | null | 32.129643 | 179 | 0.448581 | [
[
[
"#default_exp tabular.core",
"_____no_output_____"
],
[
"#export\nfrom local.torch_basics import *\nfrom local.test import *\nfrom local.core import *\nfrom local.data.all import *",
"_____no_output_____"
],
[
"from local.notebook.showdoc import *",
"_____no_output_____"
],
[
"#export\npd.set_option('mode.chained_assignment','raise')",
"_____no_output_____"
]
],
[
[
"# Tabular core\n\n> Basic function to preprocess tabular data before assembling it in a `DataBunch`.",
"_____no_output_____"
],
[
"## Tabular -",
"_____no_output_____"
]
],
[
[
"#export\nclass _TabIloc:\n \"Get/set rows by iloc and cols by name\"\n def __init__(self,to): self.to = to\n def __getitem__(self, idxs):\n df = self.to.items\n if isinstance(idxs,tuple):\n rows,cols = idxs\n cols = df.columns.isin(cols) if is_listy(cols) else df.columns.get_loc(cols)\n else: rows,cols = idxs,slice(None)\n return self.to.new(df.iloc[rows, cols])",
"_____no_output_____"
],
[
"#export\nclass Tabular(CollBase, GetAttr, FilteredBase):\n \"A `DataFrame` wrapper that knows which cols are cont/cat/y, and returns rows in `__getitem__`\"\n _default='items'\n def __init__(self, df, procs=None, cat_names=None, cont_names=None, y_names=None, is_y_cat=True, splits=None, do_setup=True):\n if splits is None: splits=[range_of(df)]\n df = df.iloc[sum(splits, [])].copy()\n super().__init__(df)\n\n store_attr(self, 'y_names,is_y_cat')\n self.cat_names,self.cont_names,self.procs = L(cat_names),L(cont_names),Pipeline(procs, as_item=True)\n self.cat_y = None if not is_y_cat else y_names\n self.cont_y = None if is_y_cat else y_names\n self.split = len(splits[0])\n if do_setup: self.procs.setup(self)\n\n def subset(self, i): return self.new(self.items[slice(0,self.split) if i==0 else slice(self.split,len(self))])\n def copy(self): self.items = self.items.copy(); return self\n def new(self, df): return type(self)(df, do_setup=False, **attrdict(self, 'procs','cat_names','cont_names','y_names','is_y_cat'))\n def show(self, max_n=10, **kwargs): display_df(self.all_cols[:max_n])\n def setup(self): self.procs.setup(self)\n def process(self): self.procs(self)\n def iloc(self): return _TabIloc(self)\n def targ(self): return self.items[self.y_names]\n def all_cont_names(self): return self.cont_names + self.cont_y\n def all_cat_names (self): return self.cat_names + self.cat_y\n def all_col_names (self): return self.all_cont_names + self.all_cat_names\n def n_subsets(self): return 2\n\nproperties(Tabular,'iloc','targ','all_cont_names','all_cat_names','all_col_names','n_subsets')",
"_____no_output_____"
],
[
"#export\nclass TabularPandas(Tabular):\n def transform(self, cols, f): self[cols] = self[cols].transform(f)",
"_____no_output_____"
],
[
"#export\ndef _add_prop(cls, nm):\n @property\n def f(o): return o[list(getattr(o,nm+'_names'))]\n @f.setter\n def fset(o, v): o[getattr(o,nm+'_names')] = v\n setattr(cls, nm+'s', f)\n setattr(cls, nm+'s', fset)\n\n_add_prop(Tabular, 'cat')\n_add_prop(Tabular, 'all_cat')\n_add_prop(Tabular, 'cont')\n_add_prop(Tabular, 'all_cont')\n_add_prop(Tabular, 'all_col')",
"_____no_output_____"
],
[
"df = pd.DataFrame({'a':[0,1,2,0,2], 'b':[0,0,0,0,1]})\nto = TabularPandas(df, cat_names='a')\nt = pickle.loads(pickle.dumps(to))\ntest_eq(t.items,to.items)\ntest_eq(to.all_cols,to[['a']])\nto.show() # only shows 'a' since that's the only col in `TabularPandas`",
"_____no_output_____"
],
[
"#export\nclass TabularProc(InplaceTransform):\n \"Base class to write a non-lazy tabular processor for dataframes\"\n def setup(self, items=None):\n super().setup(getattr(items,'train',items))\n # Procs are called as soon as data is available\n return self(items.items if isinstance(items,DataSource) else items)",
"_____no_output_____"
],
[
"#export\nclass Categorify(TabularProc):\n \"Transform the categorical variables to that type.\"\n order = 1\n def setups(self, to):\n self.classes = {n:CategoryMap(to.iloc[:,n].items, add_na=(n in to.cat_names)) for n in to.all_cat_names}\n def _apply_cats (self, add, c): return c.cat.codes+add if is_categorical_dtype(c) else c.map(self[c.name].o2i)\n def _decode_cats(self, c): return c.map(dict(enumerate(self[c.name].items)))\n def encodes(self, to):\n to.transform(to.cat_names, partial(self._apply_cats,1))\n to.transform(L(to.cat_y), partial(self._apply_cats,0))\n def decodes(self, to): to.transform(to.all_cat_names, self._decode_cats)\n def __getitem__(self,k): return self.classes[k]",
"_____no_output_____"
],
[
"show_doc(Categorify, title_level=3)",
"_____no_output_____"
],
[
"df = pd.DataFrame({'a':[0,1,2,0,2]})\nto = TabularPandas(df, Categorify, 'a')\ncat = to.procs.categorify\ntest_eq(cat['a'], ['#na#',0,1,2])\ntest_eq(to.a, [1,2,3,1,3])",
"_____no_output_____"
],
[
"df1 = pd.DataFrame({'a':[1,0,3,-1,2]})\nto1 = to.new(df1)\nto1.process()\n#Values that weren't in the training df are sent to 0 (na)\ntest_eq(to1.a, [2,1,0,0,3])\nto2 = cat.decode(to1)\ntest_eq(to2.a, [1,0,'#na#','#na#',2])",
"_____no_output_____"
],
[
"to1.procs",
"_____no_output_____"
],
[
"#test with splits\ncat = Categorify()\ndf = pd.DataFrame({'a':[0,1,2,3,2]})\nto = TabularPandas(df, cat, 'a', splits=[[0,1,2],[3,4]])\ntest_eq(cat['a'], ['#na#',0,1,2])\ntest_eq(to['a'], [1,2,3,0,3])",
"_____no_output_____"
],
[
"df = pd.DataFrame({'a':pd.Categorical(['M','H','L','M'], categories=['H','M','L'], ordered=True)})\nto = TabularPandas(df, Categorify, 'a')\ncat = to.procs.categorify\ntest_eq(cat['a'], ['#na#','H','M','L'])\ntest_eq(to.a, [2,1,3,2])\nto2 = cat.decode(to)\ntest_eq(to2.a, ['M','H','L','M'])",
"_____no_output_____"
],
[
"#export\nclass Normalize(TabularProc):\n \"Normalize the continuous variables.\"\n order = 2\n def setups(self, dsrc): self.means,self.stds = dsrc.conts.mean(),dsrc.conts.std(ddof=0)+1e-7\n def encodes(self, to): to.conts = (to.conts-self.means) / self.stds\n def decodes(self, to): to.conts = (to.conts*self.stds ) + self.means",
"_____no_output_____"
],
[
"show_doc(Normalize, title_level=3)",
"_____no_output_____"
],
[
"norm = Normalize()\ndf = pd.DataFrame({'a':[0,1,2,3,4]})\nto = TabularPandas(df, norm, cont_names='a')\nx = np.array([0,1,2,3,4])\nm,s = x.mean(),x.std()\ntest_eq(norm.means['a'], m)\ntest_close(norm.stds['a'], s)\ntest_close(to.a.values, (x-m)/s)",
"_____no_output_____"
],
[
"df1 = pd.DataFrame({'a':[5,6,7]})\nto1 = to.new(df1)\nto1.process()\ntest_close(to1['a'].values, (np.array([5,6,7])-m)/s)\nto2 = norm.decode(to1)\ntest_close(to2.a.values, [5,6,7])",
"_____no_output_____"
],
[
"norm = Normalize()\ndf = pd.DataFrame({'a':[0,1,2,3,4]})\nto = TabularPandas(df, norm, cont_names='a', splits=[[0,1,2],[3,4]])\nx = np.array([0,1,2])\nm,s = x.mean(),x.std()\ntest_eq(norm.means['a'], m)\ntest_close(norm.stds['a'], s)\ntest_close(to['a'].values, (np.array([0,1,2,3,4])-m)/s)",
"_____no_output_____"
],
[
"#export\nclass FillStrategy:\n \"Namespace containing the various filling strategies.\"\n def median (c,fill): return c.median()\n def constant(c,fill): return fill\n def mode (c,fill): return c.dropna().value_counts().idxmax()",
"_____no_output_____"
],
[
"#export\nclass FillMissing(TabularProc):\n \"Fill the missing values in continuous columns.\"\n def __init__(self, fill_strategy=FillStrategy.median, add_col=True, fill_vals=None):\n if fill_vals is None: fill_vals = defaultdict(int)\n store_attr(self, 'fill_strategy,add_col,fill_vals')\n\n def setups(self, dsrc):\n self.na_dict = {n:self.fill_strategy(dsrc[n], self.fill_vals[n])\n for n in pd.isnull(dsrc.conts).any().keys()}\n\n def encodes(self, to):\n missing = pd.isnull(to.conts)\n for n in missing.any().keys():\n assert n in self.na_dict, f\"nan values in `{n}` but not in setup training set\"\n to[n].fillna(self.na_dict[n], inplace=True)\n if self.add_col:\n to.loc[:,n+'_na'] = missing[n]\n if n+'_na' not in to.cat_names: to.cat_names.append(n+'_na')",
"_____no_output_____"
],
[
"show_doc(FillMissing, title_level=3)",
"_____no_output_____"
],
[
"fill1,fill2,fill3 = (FillMissing(fill_strategy=s) \n for s in [FillStrategy.median, FillStrategy.constant, FillStrategy.mode])\ndf = pd.DataFrame({'a':[0,1,np.nan,1,2,3,4]})\ndf1 = df.copy(); df2 = df.copy()\ntos = TabularPandas(df, fill1, cont_names='a'),TabularPandas(df1, fill2, cont_names='a'),TabularPandas(df2, fill3, cont_names='a')\ntest_eq(fill1.na_dict, {'a': 1.5})\ntest_eq(fill2.na_dict, {'a': 0})\ntest_eq(fill3.na_dict, {'a': 1.0})\n\nfor t in tos: test_eq(t.cat_names, ['a_na'])\n\nfor to_,v in zip(tos, [1.5, 0., 1.]):\n test_eq(to_.a.values, np.array([0, 1, v, 1, 2, 3, 4]))\n test_eq(to_.a_na.values, np.array([0, 0, 1, 0, 0, 0, 0]))",
"_____no_output_____"
],
[
"dfa = pd.DataFrame({'a':[np.nan,0,np.nan]})\ntos = [t.new(o) for t,o in zip(tos,(dfa,dfa.copy(),dfa.copy()))]\nfor t in tos: t.process()\nfor to_,v in zip(tos, [1.5, 0., 1.]):\n test_eq(to_.a.values, np.array([v, 0, v]))\n test_eq(to_.a_na.values, np.array([1, 0, 1]))",
"_____no_output_____"
]
],
[
[
"## TabularPandas Pipelines -",
"_____no_output_____"
]
],
[
[
"procs = [Normalize, Categorify, FillMissing, noop]\ndf = pd.DataFrame({'a':[0,1,2,1,1,2,0], 'b':[0,1,np.nan,1,2,3,4]})\nto = TabularPandas(df, procs, cat_names='a', cont_names='b')\n\n#Test setup and apply on df_main\ntest_eq(to.cat_names, ['a', 'b_na'])\ntest_eq(to.a, [1,2,3,2,2,3,1])\ntest_eq(to.b_na, [1,1,2,1,1,1,1])\nx = np.array([0,1,1.5,1,2,3,4])\nm,s = x.mean(),x.std()\ntest_close(to.b.values, (x-m)/s)\ntest_eq(to.procs.classes, {'a': ['#na#',0,1,2], 'b_na': ['#na#',False,True]})",
"_____no_output_____"
],
[
"#Test apply on y_names\ndf = pd.DataFrame({'a':[0,1,2,1,1,2,0], 'b':[0,1,np.nan,1,2,3,4], 'c': ['b','a','b','a','a','b','a']})\nto = TabularPandas(df, procs, 'a', 'b', y_names='c')\n\ntest_eq(to.cat_names, ['a', 'b_na'])\ntest_eq(to.a, [1,2,3,2,2,3,1])\ntest_eq(to.b_na, [1,1,2,1,1,1,1])\ntest_eq(to.c, [1,0,1,0,0,1,0])\nx = np.array([0,1,1.5,1,2,3,4])\nm,s = x.mean(),x.std()\ntest_close(to.b.values, (x-m)/s)\ntest_eq(to.procs.classes, {'a': ['#na#',0,1,2], 'b_na': ['#na#',False,True], 'c': ['a','b']})",
"_____no_output_____"
],
[
"df = pd.DataFrame({'a':[0,1,2,1,1,2,0], 'b':[0,1,np.nan,1,2,3,4], 'c': ['b','a','b','a','a','b','a']})\nto = TabularPandas(df, procs, 'a', 'b', y_names='c')\n\ntest_eq(to.cat_names, ['a', 'b_na'])\ntest_eq(to.a, [1,2,3,2,2,3,1])\ntest_eq(df.a.dtype,int)\ntest_eq(to.b_na, [1,1,2,1,1,1,1])\ntest_eq(to.c, [1,0,1,0,0,1,0])",
"_____no_output_____"
],
[
"df = pd.DataFrame({'a':[0,1,2,1,1,2,0], 'b':[0,np.nan,1,1,2,3,4], 'c': ['b','a','b','a','a','b','a']})\nto = TabularPandas(df, procs, cat_names='a', cont_names='b', y_names='c', splits=[[0,1,4,6], [2,3,5]])\n\ntest_eq(to.cat_names, ['a', 'b_na'])\ntest_eq(to.a, [1,2,2,1,0,2,0])\ntest_eq(df.a.dtype,int)\ntest_eq(to.b_na, [1,2,1,1,1,1,1])\ntest_eq(to.c, [1,0,0,0,1,0,1])",
"_____no_output_____"
],
[
"#export\nclass ReadTabBatch(ItemTransform):\n def __init__(self, to): self.to = to\n # TODO: use float for cont targ\n def encodes(self, to): return tensor(to.cats).long(),tensor(to.conts).float(), tensor(to.targ).long()\n\n def decodes(self, o):\n cats,conts,targs = to_np(o)\n vals = np.concatenate([cats,conts,targs[:,None]], axis=1)\n df = pd.DataFrame(vals, columns=self.to.cat_names+self.to.cont_names+self.to.y_names)\n to = self.to.new(df)\n to = self.to.procs.decode(to)\n return to",
"_____no_output_____"
],
[
"#export\n@typedispatch\ndef show_batch(x: Tabular, y, samples, max_n=10, ctxs=None):\n x.show()",
"_____no_output_____"
],
[
"#export\n@delegates()\nclass TabDataLoader(TfmdDL):\n do_item = noops\n def __init__(self, dataset, bs=16, shuffle=False, after_batch=None, num_workers=0, **kwargs):\n after_batch = L(after_batch)+ReadTabBatch(dataset)\n super().__init__(dataset, bs=bs, shuffle=shuffle, after_batch=after_batch, num_workers=num_workers, **kwargs)\n\n def create_batch(self, b): return self.dataset.iloc[b]\n\nTabularPandas._dl_type = TabDataLoader",
"_____no_output_____"
]
],
[
[
"## Integration example",
"_____no_output_____"
]
],
[
[
"path = untar_data(URLs.ADULT_SAMPLE)\ndf = pd.read_csv(path/'adult.csv')\ndf_main,df_test = df.iloc[:10000].copy(),df.iloc[10000:].copy()\ndf_main.head()",
"_____no_output_____"
],
[
"cat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race']\ncont_names = ['age', 'fnlwgt', 'education-num']\nprocs = [Categorify, FillMissing, Normalize]\nsplits = RandomSplitter()(range_of(df_main))",
"_____no_output_____"
],
[
"%time to = TabularPandas(df_main, procs, cat_names, cont_names, y_names=\"salary\", splits=splits)",
"CPU times: user 194 ms, sys: 4.32 ms, total: 198 ms\nWall time: 197 ms\n"
],
[
"dbch = to.databunch()\ndbch.valid_dl.show_batch()",
"_____no_output_____"
],
[
"to.procs[-1].means",
"_____no_output_____"
],
[
"to_tst = to.new(df_test)\nto_tst.process()\nto_tst.all_cols.head()",
"_____no_output_____"
]
],
[
[
"## Not being used now - for multi-modal",
"_____no_output_____"
]
],
[
[
"class TensorTabular(Tuple):\n def get_ctxs(self, max_n=10, **kwargs):\n n_samples = min(self[0].shape[0], max_n)\n df = pd.DataFrame(index = range(n_samples))\n return [df.iloc[i] for i in range(n_samples)]\n\n def display(self, ctxs): display_df(pd.DataFrame(ctxs))\n\nclass TabularLine(pd.Series):\n \"A line of a dataframe that knows how to show itself\"\n def show(self, ctx=None, **kwargs): return self if ctx is None else ctx.append(self)\n\nclass ReadTabLine(ItemTransform):\n def __init__(self, proc): self.proc = proc\n\n def encodes(self, row):\n cats,conts = (o.map(row.__getitem__) for o in (self.proc.cat_names,self.proc.cont_names))\n return TensorTabular(tensor(cats).long(),tensor(conts).float())\n\n def decodes(self, o):\n to = TabularPandas(o, self.proc.cat_names, self.proc.cont_names, self.proc.y_names)\n to = self.proc.decode(to)\n return TabularLine(pd.Series({c: v for v,c in zip(to.items[0]+to.items[1], self.proc.cat_names+self.proc.cont_names)}))\n\nclass ReadTabTarget(ItemTransform):\n def __init__(self, proc): self.proc = proc\n def encodes(self, row): return row[self.proc.y_names].astype(np.int64)\n def decodes(self, o): return Category(self.proc.classes[self.proc.y_names][o])",
"_____no_output_____"
],
[
"# tds = TfmdDS(to.items, tfms=[[ReadTabLine(proc)], ReadTabTarget(proc)])\n# enc = tds[1]\n# test_eq(enc[0][0], tensor([2,1]))\n# test_close(enc[0][1], tensor([-0.628828]))\n# test_eq(enc[1], 1)\n\n# dec = tds.decode(enc)\n# assert isinstance(dec[0], TabularLine)\n# test_close(dec[0], pd.Series({'a': 1, 'b_na': False, 'b': 1}))\n# test_eq(dec[1], 'a')\n\n# test_stdout(lambda: print(show_at(tds, 1)), \"\"\"a 1\n# b_na False\n# b 1\n# category a\n# dtype: object\"\"\")",
"_____no_output_____"
]
],
[
[
"## Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom local.notebook.export import notebook2script\nnotebook2script(all_fs=True)",
"Converted 00_test.ipynb.\nConverted 01_core.ipynb.\nConverted 01a_utils.ipynb.\nConverted 01b_dispatch.ipynb.\nConverted 01c_transform.ipynb.\nConverted 02_script.ipynb.\nConverted 03_torch_core.ipynb.\nConverted 03a_layers.ipynb.\nConverted 04_dataloader.ipynb.\nConverted 05_data_core.ipynb.\nConverted 06_data_transforms.ipynb.\nConverted 07_data_block.ipynb.\nConverted 08_vision_core.ipynb.\nConverted 09_vision_augment.ipynb.\nConverted 10_pets_tutorial.ipynb.\nConverted 11_vision_models_xresnet.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_learner.ipynb.\nConverted 13a_metrics.ipynb.\nConverted 14_callback_schedule.ipynb.\nConverted 14a_callback_data.ipynb.\nConverted 15_callback_hook.ipynb.\nConverted 15a_vision_models_unet.ipynb.\nConverted 16_callback_progress.ipynb.\nConverted 17_callback_tracker.ipynb.\nConverted 18_callback_fp16.ipynb.\nConverted 19_callback_mixup.ipynb.\nConverted 21_vision_learner.ipynb.\nConverted 22_tutorial_imagenette.ipynb.\nConverted 23_tutorial_transfer_learning.ipynb.\nConverted 30_text_core.ipynb.\nConverted 31_text_data.ipynb.\nConverted 32_text_models_awdlstm.ipynb.\nConverted 33_text_models_core.ipynb.\nConverted 34_callback_rnn.ipynb.\nConverted 35_tutorial_wikitext.ipynb.\nConverted 36_text_models_qrnn.ipynb.\nConverted 37_text_learner.ipynb.\nConverted 38_tutorial_ulmfit.ipynb.\nConverted 40_tabular_core.ipynb.\nConverted 41_tabular_model.ipynb.\nConverted 42_tabular_rapids.ipynb.\nConverted 50_data_block_examples.ipynb.\nConverted 60_medical_imaging.ipynb.\nConverted 65_medical_text.ipynb.\nConverted 90_notebook_core.ipynb.\nConverted 91_notebook_export.ipynb.\nConverted 92_notebook_showdoc.ipynb.\nConverted 93_notebook_export2html.ipynb.\nConverted 94_notebook_test.ipynb.\nConverted 95_index.ipynb.\nConverted 96_data_external.ipynb.\nConverted 97_utils_test.ipynb.\nConverted notebook2jekyll.ipynb.\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6021c3cca3dac7a69625b8c6463508ed73d903 | 7,634 | ipynb | Jupyter Notebook | docs/samples/custom/kfserving-custom-model/kfserving_sdk_custom_image.ipynb | waylayio/kfserving | a4fafea70fb07ca4739a605bfdb3ae4df7a50ac8 | [
"Apache-2.0"
]
| 2 | 2020-12-03T22:45:44.000Z | 2020-12-28T03:52:28.000Z | docs/samples/custom/kfserving-custom-model/kfserving_sdk_custom_image.ipynb | doguaraci/kfserving | 2c1f66898b2cdfe2f580c1b455ef8ee57f73bc03 | [
"Apache-2.0"
]
| 1 | 2020-04-28T18:53:40.000Z | 2020-04-28T18:53:40.000Z | docs/samples/custom/kfserving-custom-model/kfserving_sdk_custom_image.ipynb | doguaraci/kfserving | 2c1f66898b2cdfe2f580c1b455ef8ee57f73bc03 | [
"Apache-2.0"
]
| null | null | null | 28.274074 | 347 | 0.582395 | [
[
[
"# Sample for KFServing SDK with a custom image\n\nThis is a sample for KFServing SDK using a custom image.\n\nThe notebook shows how to use KFServing SDK to create, get and delete InferenceService with a custom image.\n\n### Setup\n- Your `~/.kube/config` should point to a cluster with KFServing installed.\n- Your cluster's Istio Ingress gateway must be network accessible.",
"_____no_output_____"
],
[
"### Build the docker image we will be using.\n\nThe goal of custom image support is to allow users to bring their own wrapped model inside a container and serve it with KFServing. Please note that you will need to ensure that your container is also running a web server e.g. Flask to expose your model endpoints. This example extends kfserving.KFModel which uses the tornado web server.\n\n\nTo build and push with Docker Hub set the `DOCKER_HUB_USERNAME` variable below with your Docker Hub username",
"_____no_output_____"
]
],
[
[
"# Set this to be your dockerhub username\n# It will be used when building your image and when creating the InferenceService for your image\nDOCKER_HUB_USERNAME = \"your_docker_username\"",
"_____no_output_____"
],
[
"%%bash -s \"$DOCKER_HUB_USERNAME\"\ndocker build -t $1/kfserving-custom-model ./model-server",
"_____no_output_____"
],
[
"%%bash -s \"$DOCKER_HUB_USERNAME\"\ndocker push $1/kfserving-custom-model",
"_____no_output_____"
]
],
[
[
"### KFServing Client SDK\n\nWe will use the [KFServing client SDK](https://github.com/kubeflow/kfserving/blob/master/python/kfserving/README.md#kfserving-client) to create the InferenceService and deploy our custom image.",
"_____no_output_____"
]
],
[
[
"from kubernetes import client\nfrom kubernetes.client import V1Container\n\nfrom kfserving import KFServingClient\nfrom kfserving import constants\nfrom kfserving import utils\nfrom kfserving import V1alpha2EndpointSpec\nfrom kfserving import V1alpha2PredictorSpec\nfrom kfserving import V1alpha2InferenceServiceSpec\nfrom kfserving import V1alpha2InferenceService\nfrom kfserving import V1alpha2CustomSpec",
"_____no_output_____"
],
[
"namespace = utils.get_default_target_namespace()\nprint(namespace)",
"_____no_output_____"
]
],
[
[
"### Define InferenceService\n\nFirstly define default endpoint spec, and then define the inferenceservice using the endpoint spec.\n\nTo use a custom image we need to use V1alphaCustomSpec which takes a [V1Container](https://github.com/kubernetes-client/python/blob/master/kubernetes/docs/V1Container.md)\n from the kuberenetes library\n",
"_____no_output_____"
]
],
[
[
"api_version = constants.KFSERVING_GROUP + '/' + constants.KFSERVING_VERSION\n\ndefault_endpoint_spec = V1alpha2EndpointSpec(\n predictor=V1alpha2PredictorSpec(\n custom=V1alpha2CustomSpec(\n container=V1Container(\n name=\"kfserving-custom-model\",\n image=f\"{DOCKER_HUB_USERNAME}/kfserving-custom-model\"))))\n\nisvc = V1alpha2InferenceService(api_version=api_version,\n kind=constants.KFSERVING_KIND,\n metadata=client.V1ObjectMeta(\n name='kfserving-custom-model', namespace=namespace),\n spec=V1alpha2InferenceServiceSpec(default=default_endpoint_spec))",
"_____no_output_____"
]
],
[
[
"### Create the InferenceService\n\nCall KFServingClient to create InferenceService.",
"_____no_output_____"
]
],
[
[
"KFServing = KFServingClient()\nKFServing.create(isvc)",
"_____no_output_____"
]
],
[
[
"### Check the InferenceService",
"_____no_output_____"
]
],
[
[
"KFServing.get('kfserving-custom-model', namespace=namespace, watch=True, timeout_seconds=120)",
"_____no_output_____"
]
],
[
[
"### Run a prediction ",
"_____no_output_____"
]
],
[
[
"MODEL_NAME = \"kfserving-custom-model\"",
"_____no_output_____"
],
[
"%%bash --out CLUSTER_IP\n# Use \"kfserving-ingressgateway\" as your `INGRESS_GATEWAY` if you are deploying KFServing\n# as part of Kubeflow install, and not independently.\nINGRESS_GATEWAY=\"istio-ingressgateway\"\necho \"$(kubectl -n istio-system get service $INGRESS_GATEWAY -o jsonpath='{.status.loadBalancer.ingress[0].ip}')\"",
"_____no_output_____"
],
[
"%%bash -s \"$MODEL_NAME\" --out SERVICE_HOSTNAME\necho \"$(kubectl get inferenceservice $1 -o jsonpath='{.status.url}' | cut -d \"/\" -f 3)\"",
"_____no_output_____"
],
[
"import requests\nimport json\n\nwith open('input.json') as json_file:\n data = json.load(json_file)\n url = f\"http://{CLUSTER_IP.strip()}/v1/models/{MODEL_NAME}:predict\"\n headers = {\"Host\": SERVICE_HOSTNAME.strip()}\n result = requests.post(url, data=json.dumps(data), headers=headers)\n print(result.content)",
"_____no_output_____"
]
],
[
[
"### Delete the InferenceService",
"_____no_output_____"
]
],
[
[
"KFServing.delete(MODEL_NAME, namespace=namespace)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec60241d60a1f69141ae0a9b8e376d8d164cab23 | 109,948 | ipynb | Jupyter Notebook | Module4/Module4 - Lab2.ipynb | marselag/DAT210x | 68e1743485a52a9c4623e3e7b478786dd434c452 | [
"MIT"
]
| null | null | null | Module4/Module4 - Lab2.ipynb | marselag/DAT210x | 68e1743485a52a9c4623e3e7b478786dd434c452 | [
"MIT"
]
| null | null | null | Module4/Module4 - Lab2.ipynb | marselag/DAT210x | 68e1743485a52a9c4623e3e7b478786dd434c452 | [
"MIT"
]
| null | null | null | 49.79529 | 33,400 | 0.538846 | [
[
[
"# DAT210x - Programming with Python for DS",
"_____no_output_____"
],
[
"## Module4- Lab2",
"_____no_output_____"
]
],
[
[
"import math\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport matplotlib\n\nfrom sklearn import preprocessing",
"_____no_output_____"
],
[
"# Look pretty...\n\n# matplotlib.style.use('ggplot')\nplt.style.use('ggplot')",
"_____no_output_____"
]
],
[
[
"### Some Boilerplate Code",
"_____no_output_____"
],
[
"For your convenience, we've included some boilerplate code here which will help you out. You aren't expected to know how to write this code on your own at this point, but it'll assist with your visualizations. We've added some notes to the code in case you're interested in knowing what it's doing:",
"_____no_output_____"
],
[
"### A Note on SKLearn's `.transform()` calls:",
"_____no_output_____"
],
[
"Any time you perform a transformation on your data, you lose the column header names because the output of SciKit-Learn's `.transform()` method is an NDArray and not a daraframe.\n\nThis actually makes a lot of sense because there are essentially two types of transformations:\n- Those that adjust the scale of your features, and\n- Those that change alter the number of features, perhaps even changing their values entirely.\n\nAn example of adjusting the scale of a feature would be changing centimeters to inches. Changing the feature entirely would be like using PCA to reduce 300 columns to 30. In either case, the original column's units have either been altered or no longer exist at all, so it's up to you to assign names to your columns after any transformation, if you'd like to store the resulting NDArray back into a dataframe.",
"_____no_output_____"
]
],
[
[
"def scaleFeaturesDF(df):\n # Feature scaling is a type of transformation that only changes the\n # scale, but not number of features. Because of this, we can still\n # use the original dataset's column names... so long as we keep in\n # mind that the _units_ have been altered:\n\n scaled = preprocessing.StandardScaler().fit_transform(df)\n scaled = pd.DataFrame(scaled, columns=df.columns)\n \n print(\"New Variances:\\n\", scaled.var())\n print(\"New Describe:\\n\", scaled.describe())\n return scaled",
"_____no_output_____"
]
],
[
[
"SKLearn contains many methods for transforming your features by scaling them, a type of pre-processing):\n - `RobustScaler`\n - `Normalizer`\n - `MinMaxScaler`\n - `MaxAbsScaler`\n - `StandardScaler`\n - ...\n\nhttp://scikit-learn.org/stable/modules/classes.html#module-sklearn.preprocessing\n\nHowever in order to be effective at PCA, there are a few requirements that must be met, and which will drive the selection of your scaler. PCA requires your data is standardized -- in other words, it's _mean_ should equal 0, and it should have unit variance.\n\nSKLearn's regular `Normalizer()` doesn't zero out the mean of your data, it only clamps it, so it could be inappropriate to use depending on your data. `MinMaxScaler` and `MaxAbsScaler` both fail to set a unit variance, so you won't be using them here either. `RobustScaler` can work, again depending on your data (watch for outliers!). So for this assignment, you're going to use the `StandardScaler`. Get familiar with it by visiting these two websites:\n\n- http://scikit-learn.org/stable/modules/preprocessing.html#preprocessing-scaler\n- http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html#sklearn.preprocessing.StandardScaler",
"_____no_output_____"
],
[
"Lastly, some code to help with visualizations:",
"_____no_output_____"
]
],
[
[
"def drawVectors(transformed_features, components_, columns, plt, scaled):\n if not scaled:\n return plt.axes() # No cheating ;-)\n\n num_columns = len(columns)\n\n # This funtion will project your *original* feature (columns)\n # onto your principal component feature-space, so that you can\n # visualize how \"important\" each one was in the\n # multi-dimensional scaling\n\n # Scale the principal components by the max value in\n # the transformed set belonging to that component\n xvector = components_[0] * max(transformed_features[:,0])\n yvector = components_[1] * max(transformed_features[:,1])\n\n ## visualize projections\n\n # Sort each column by it's length. These are your *original*\n # columns, not the principal components.\n important_features = { columns[i] : math.sqrt(xvector[i]**2 + yvector[i]**2) for i in range(num_columns) }\n important_features = sorted(zip(important_features.values(), important_features.keys()), reverse=True)\n print(\"Features by importance:\\n\", important_features)\n\n ax = plt.axes()\n\n for i in range(num_columns):\n # Use an arrow to project each original feature as a\n # labeled vector on your principal component axes\n plt.arrow(0, 0, xvector[i], yvector[i], color='b', width=0.0005, head_width=0.02, alpha=0.75)\n plt.text(xvector[i]*1.2, yvector[i]*1.2, list(columns)[i], color='b', alpha=0.75)\n\n return ax",
"_____no_output_____"
]
],
[
[
"### And Now, The Assignment",
"_____no_output_____"
]
],
[
[
"# Do * NOT * alter this line, until instructed!\n# scaleFeatures = False\nscaleFeatures = True",
"_____no_output_____"
]
],
[
[
"Load up the dataset specified on the lab instructions page and remove any and all _rows_ that have a NaN in them. You should be a pro at this by now ;-)\n\n**QUESTION**: Should the `id` column be included in your dataset as a feature?",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\ndf = pd.read_csv('C:/Users/mgavrilova/Desktop/DAT210x/Module4/Datasets/kidney_disease.csv')\ndf.dropna(axis = 0, how = 'any', inplace = True)\ndf = df.dropna(axis=0, thresh=1)\ndf = df.reset_index(drop=True)\ndf",
"_____no_output_____"
]
],
[
[
"Let's build some color-coded labels; the actual label feature will be removed prior to executing PCA, since it's unsupervised. You're only labeling by color so you can see the effects of PCA:",
"_____no_output_____"
]
],
[
[
"labels = ['red' if i=='ckd' else 'green' for i in df.classification]",
"_____no_output_____"
]
],
[
[
"Use an indexer to select only the following columns: `['bgr','wc','rc']`",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\ndf = df[['bgr', 'wc', 'rc']]",
"_____no_output_____"
]
],
[
[
"Either take a look at the dataset's webpage in the attribute info section of UCI's [Chronic Kidney Disease]() page,: https://archive.ics.uci.edu/ml/datasets/Chronic_Kidney_Disease or alternatively, you can actually look at the first few rows of your dataframe using `.head()`. What kind of data type should these three columns be? Compare what you see with the results when you print out your dataframe's `dtypes`.\n\nIf Pandas did not properly detect and convert your columns to the data types you expected, use an appropriate command to coerce these features to the right type.",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\ndf.wc = pd.to_numeric(df.wc)\ndf.rc = pd.to_numeric(df.rc)\ndf.dtypes",
"C:\\Users\\mgavrilova\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\pandas\\core\\generic.py:4401: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self[name] = value\n"
]
],
[
[
"PCA Operates based on variance. The variable with the greatest variance will dominate. Examine your data using a command that will check the variance of every feature in your dataset, and then print out the results. Also print out the results of running `.describe` on your dataset.\n\n_Hint:_ If you do not see all three variables: `'bgr'`, `'wc'`, and `'rc'`, then it's likely you probably did not complete the previous step properly.",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nprint(df.var())\nprint(df.describe())",
"bgr 4.217182e+03\nwc 9.777380e+06\nrc 1.039104e+00\ndtype: float64\n bgr wc rc\ncount 158.000000 158.000000 158.000000\nmean 131.341772 8475.949367 4.891772\nstd 64.939832 3126.880181 1.019364\nmin 70.000000 3800.000000 2.100000\n25% 97.000000 6525.000000 4.500000\n50% 115.500000 7800.000000 4.950000\n75% 131.750000 9775.000000 5.600000\nmax 490.000000 26400.000000 8.000000\n"
]
],
[
[
"Below, we assume your dataframe's variable is named `df`. If it isn't, make the appropriate changes. But do not alter the code in `scaleFeaturesDF()` just yet!",
"_____no_output_____"
]
],
[
[
"# .. your (possible) code adjustment here ..\nif scaleFeatures: df = scaleFeaturesDF(df)",
"New Variances:\n bgr 1.006369\nwc 1.006369\nrc 1.006369\ndtype: float64\nNew Describe:\n bgr wc rc\ncount 1.580000e+02 1.580000e+02 1.580000e+02\nmean -9.755075e-17 9.345548e-17 1.068063e-16\nstd 1.003180e+00 1.003180e+00 1.003180e+00\nmin -9.475974e-01 -1.500159e+00 -2.747446e+00\n25% -5.305059e-01 -6.259123e-01 -3.855519e-01\n50% -2.447210e-01 -2.168611e-01 5.730335e-02\n75% 6.306235e-03 4.167672e-01 6.969831e-01\nmax 5.540492e+00 5.750474e+00 3.058878e+00\n"
]
],
[
[
"Run PCA on your dataset, reducing it to 2 principal components. Make sure your PCA model is saved in a variable called `'pca'`, and that the results of your transformation are saved in another variable `'T'`:",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nfrom sklearn.decomposition import PCA\n\npca = PCA(n_components = 2)\n\npca.fit(df)\n\nT = pca.transform(df)",
"_____no_output_____"
]
],
[
[
"Now, plot the transformed data as a scatter plot. Recall that transforming the data will result in a NumPy NDArray. You can either use MatPlotLib to graph it directly, or you can convert it back to DataFrame and have Pandas do it for you.\n\nSince we've already demonstrated how to plot directly with MatPlotLib in `Module4/assignment1.ipynb`, this time we'll show you how to convert your transformed data back into to a Pandas Dataframe and have Pandas plot it from there.",
"_____no_output_____"
]
],
[
[
"# Since we transformed via PCA, we no longer have column names; but we know we\n# are in `principal-component` space, so we'll just define the coordinates accordingly:\nax = drawVectors(T, pca.components_, df.columns.values, plt, scaleFeatures)\nT = pd.DataFrame(T)\n\nT.columns = ['component1', 'component2']\nT.plot.scatter(x='component1', y='component2', marker='o', c=labels, alpha=0.75, ax=ax)\n\nplt.show()",
"Features by importance:\n [(3.9998071556884853, 'wc'), (3.2588876641210898, 'bgr'), (3.0097527529983648, 'rc')]\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec605c80e4c88649df29d6d54d52c9bf6affe471 | 2,152 | ipynb | Jupyter Notebook | notebooks/emo_predict.ipynb | tanchangsheng/publicpolicy_sentiment_analysis | 5a52a40c97d8f9ef8b402e6b9e1c3058b68250c0 | [
"MIT"
]
| null | null | null | notebooks/emo_predict.ipynb | tanchangsheng/publicpolicy_sentiment_analysis | 5a52a40c97d8f9ef8b402e6b9e1c3058b68250c0 | [
"MIT"
]
| null | null | null | notebooks/emo_predict.ipynb | tanchangsheng/publicpolicy_sentiment_analysis | 5a52a40c97d8f9ef8b402e6b9e1c3058b68250c0 | [
"MIT"
]
| null | null | null | 23.391304 | 180 | 0.533922 | [
[
[
"import os\nimport sys\n\n# add the 'src' directory as one where we can import modules\n# os.pardir refers to parent directory. \n# src_dir = os.path.join(os.getcwd(), '../src') => works the same\nsrc_dir = os.path.join(os.getcwd(), os.pardir, 'src')\nsys.path.append(src_dir)\n\n# Load the \"autoreload\" extension\n%load_ext autoreload\n# always reload modules marked with \"%aimport\"\n%autoreload 2",
"_____no_output_____"
],
[
"%run ../src/models/predict_model.py --data ../data/train/emotion-large.2-label.tsv.gz --vtr ../models/exp6_vtr_dict --clf ../models/exp6_clf_lr --output ../data/results/emo_1",
"data : ../data/train/emotion-large.2-label.tsv.gz\nid_index : 0\ntext_index : 1\nvtr : ../models/exp6_vtr_dict\nclf : ../models/exp6_clf_lr\noutput : ../data/results/emo_1\npreprocess : True\nstop_words : False\nfeat : count\nemotion_words : None\nlsd_words : None\nemoticons : None\nword_feature : False\nngrams : None\nfiveWoneH : None\npos : False\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
ec60976817df4ea9ce4b3de8637f9cee916e1d79 | 18,739 | ipynb | Jupyter Notebook | Final .ipynb | rahulrzende/Restoration-and-colorization-of-grayscale-memories-using-CNNs | db1a4e9001bb7a041bb2a9ddc9210b6e3cddf5fb | [
"MIT"
]
| null | null | null | Final .ipynb | rahulrzende/Restoration-and-colorization-of-grayscale-memories-using-CNNs | db1a4e9001bb7a041bb2a9ddc9210b6e3cddf5fb | [
"MIT"
]
| 8 | 2020-04-16T05:27:56.000Z | 2020-05-26T09:13:31.000Z | Final .ipynb | rahulrzende/Restoration-and-colorization-of-grayscale-memories-using-CNNs | db1a4e9001bb7a041bb2a9ddc9210b6e3cddf5fb | [
"MIT"
]
| 1 | 2020-04-16T05:22:31.000Z | 2020-04-16T05:22:31.000Z | 36.599609 | 162 | 0.582048 | [
[
[
"# Load all required libraries\n\n#### Summary - \n\n- Numpy for stacking images in an array\n- OpenCV for image restoration\n- Matplotlib for general plotting tasks\n- Skimage for image restoration\n- Pandas for general python data manipulations\n- Random for selecting random images from dataset for testing colorization feature\n- PIL for Image manipulation\n- OS for file manipulation, etc.\n- TensorFlow for foundational support with creation of deep learning model\n- Keras for high-level experimentation with deep learning model",
"_____no_output_____"
]
],
[
[
"#import libraries\n\nimport numpy as np\nimport cv2 as cv\nfrom matplotlib import pyplot as plt\nimport skimage \nfrom skimage import io\nimport pandas as pd\nimport random\nfrom PIL import Image\nimport skimage \nfrom skimage.transform import resize\nfrom skimage.color import rgb2lab\nimport os\nimport tensorflow as tf\nfrom tensorflow.keras.applications import MobileNetV2\nfrom tensorflow.keras.models import Model, Sequential, load_model\nfrom tensorflow.keras.layers import Conv2D, Conv2DTranspose, Reshape, Dropout, LeakyReLU, BatchNormalization, Input, Concatenate, Activation, concatenate\nfrom tensorflow.keras.initializers import RandomNormal\nfrom tensorflow.keras.optimizers import Adam\nfrom tensorflow.keras.utils import plot_model",
"_____no_output_____"
]
],
[
[
"# Define structure for a convolutional encoder-decoder deep learning model that colorizes grayscale images\n\nThe main inspiration for this approach was this paper published in 2017 - \nhttps://arxiv.org/abs/1712.03400\n\n#### The below code block does following actions -\n\n- Randomly initialize the weights for our neural network (to satisfy the expectation of SGD i.e. Stochastic Gradient Descent)\n- Initialize an input layer that takes in images of the size 224 by 224 pixels\n- Use the pretrained (imagenet data) neural network MobileNetV2 for high-level feature extraction\n- Dropouts are implemented to prevent overfitting (this is done by dropping neurons at certain stages to train different neural networks each time)\n- The output layer is configured for A and B channels (intended to be merged with the L channel) of our input images",
"_____no_output_____"
]
],
[
[
"# build a network for training on our dataset, use the pretrained MobileNet for deep layers\n\n# prepare the kernel initializer values\nweight_init = RandomNormal(stddev=0.02)\n\n# prepare the Input layer\nnet_input = Input((224,224,3))\n\n# download mobile net, and use it as the base.\nmobile_net_base = MobileNetV2(include_top=False, input_shape=(224,224,3), weights='imagenet')\nmobilenet = mobile_net_base(net_input)\n\n# encoder block #\n\n# 224x224\nconv1 = Conv2D(64, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(net_input)\nconv1 = LeakyReLU(alpha=0.2)(conv1)\n\n# 112x112\nconv2 = Conv2D(128, (3, 3), strides=(1, 1), padding='same', kernel_initializer=weight_init)(conv1)\nconv2 = LeakyReLU(alpha=0.2)(conv2)\n\n# 112x112\nconv3 = Conv2D(128, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(conv2)\nconv3 = Activation('relu')(conv3)\n\n# 56x56\nconv4 = Conv2D(256, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(conv3)\nconv4 = Activation('relu')(conv4)\n\n# 28x28\nconv4_ = Conv2D(256, (3, 3), strides=(1, 1), padding='same', kernel_initializer=weight_init)(conv4)\nconv4_ = Activation('relu')(conv4_)\n\n# 28x28\nconv5 = Conv2D(512, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(conv4_)\nconv5 = Activation('relu')(conv5)\n\n# 14x14\nconv5_ = Conv2D(256, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(conv5)\nconv5_ = Activation('relu')(conv5_)\n\n#7x7\n# fusion layer - connect MobileNet with our encoder\nconc = concatenate([mobilenet, conv5_])\nfusion = Conv2D(512, (1, 1), padding='same', kernel_initializer=weight_init)(conc)\nfusion = Activation('relu')(fusion)\n\n# skip fusion layer\nskip_fusion = concatenate([fusion, conv5_])\n\n# decoder block #\n\n# 7x7\ndecoder = Conv2DTranspose(512, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(skip_fusion)\ndecoder = Activation('relu')(decoder)\ndecoder = Dropout(0.25)(decoder)\n\n# skip layer from conv5 (with added dropout)\nskip_4_drop = Dropout(0.25)(conv5)\nskip_4 = concatenate([decoder, skip_4_drop])\n\n# 14x14\ndecoder = Conv2DTranspose(256, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(skip_4)\ndecoder = Activation('relu')(decoder)\ndecoder = Dropout(0.25)(decoder)\n\n# skip layer from conv4_ (with added dropout)\nskip_3_drop = Dropout(0.25)(conv4_)\nskip_3 = concatenate([decoder, skip_3_drop])\n\n# 28x28\ndecoder = Conv2DTranspose(128, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(skip_3)\ndecoder = Activation('relu')(decoder)\ndecoder = Dropout(0.25)(decoder)\n\n# 56x56\ndecoder = Conv2DTranspose(64, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(decoder)\ndecoder = Activation('relu')(decoder)\ndecoder = Dropout(0.25)(decoder)\n\n# 112x112\ndecoder = Conv2DTranspose(64, (3, 3), strides=(1, 1), padding='same', kernel_initializer=weight_init)(decoder)\ndecoder = Activation('relu')(decoder)\n\n# 112x112\ndecoder = Conv2DTranspose(32, (3, 3), strides=(2, 2), padding='same', kernel_initializer=weight_init)(decoder)\ndecoder = Activation('relu')(decoder)\n\n# 224x224\n# output layer, with 2 channels (a and b)\noutput_layer = Conv2D(2, (1, 1), activation='tanh')(decoder)",
"_____no_output_____"
]
],
[
[
"# Configure model to be optimized using Adam optimizer, specified learning rate and mean-squared error loss\n\n- This step loads the weights derived upon training our deep learning model for 100 epochs on the entire MIRFLICKR25k dataset (in batches of 3000 each)",
"_____no_output_____"
]
],
[
[
"# configure model\nmodel = Model(net_input, output_layer)\nmodel.compile(Adam(lr=0.0002), loss='mse', metrics=['accuracy'])\n\n# load weights\nmodel.load_weights('trained_on_all_is_for_100_es.h5')",
"_____no_output_____"
]
],
[
[
"# Define all functions for getting predictions and constructing complete colorized RGB image based on LAB inputs",
"_____no_output_____"
]
],
[
[
"def get_pred(model, image_l):\n \"\"\"\n Summary -\n This function generates the predicted A and B channels of the colorized image\n\n :param model: Trained model\n :type model: tensorflow.python.keras.engine.training.Model\n :param image_l: The L channel of the input grayscale image\n :type image_l: numpy.ndarray\n :return: Predicted A and B channels of the input grayscale image\n :rtype: numpy.ndarray\n \n \"\"\"\n # repeat the L value to match input shape\n image_l_R = np.repeat(image_l[..., np.newaxis], 3, -1)\n image_l_R = image_l_R.reshape((1, 224, 224, 3))\n # normalize the input\n image_l_R = (image_l_R.astype('float32') - 127.5) / 127.5\n # make prediction\n prediction = model.predict(image_l_R)\n # normalize the output\n pred = (prediction[0].astype('float32') * 127.5) + 127.5\n return pred\n\ndef get_LAB(image_l, image_ab):\n \"\"\"\n Summary - \n This function generates the compiled RGB equivalent of the inputted L plus the predicted A and B channels\n\n :param image_l: Grayscale image (1 layer, L)\n :type image_l: numpy.ndarray\n :param image_ab: Predicted image (2 layers, A and B)\n :type image_ab: numpy.ndarray\n :return: Compiled image (3 layers, RGB)\n :rtype: numpy.ndarray\n \n \"\"\"\n image_l = image_l.reshape((224, 224, 1))\n image_lab = np.concatenate((image_l, image_ab), axis=2)\n image_lab = image_lab.astype(\"uint8\")\n image_rgb = cv.cvtColor(image_lab, cv.COLOR_LAB2RGB)\n image_rgb = Image.fromarray(image_rgb)\n return image_rgb\n\ndef create_sample(model, image_gray):\n \"\"\"\n Summary - \n This function creates the output we need from colorization\n\n :param model: Trained model\n :type model: tensorflow.python.keras.engine.training.Model\n :param image_gray: Grayscale image (1 layer, L)\n :type image_gray: numpy.ndarray\n :return: Result image\n :rtype: numpy.ndarray\n \n \"\"\"\n # get the model's prediction\n pred = get_pred(model, image_gray)\n # combine input and output to LAB image\n image = get_LAB(image_gray, pred)\n # create new combined image, save it\n new_image = Image.new('RGB', (224, 224))\n new_image.paste(image, (0, 0))\n return new_image",
"_____no_output_____"
],
[
"# Define function for ",
"_____no_output_____"
],
[
"def show_image(Selection,l,i):\n \"\"\"\n Summary -\n Selects the image based on user choice and sends it to the deep learning model for processing\n \n :param Selection: Image label chosen by user\n :type model: String\n :param l: list of labels\n :type l: list\n :param i: list of processed images\n :type i: list\n \"\"\"\n index = l.index(Selection)\n img = i[index]\n mod_img = resize(rgb2lab(img.copy())[:,:,0], (224,224), anti_aliasing=True)\n sample = create_sample(model, mod_img)\n sample.save('output/output.jpg')\n \n fig, axes = plt.subplots(ncols = 2, figsize = (15,5))\n axes[0].imshow(img)\n axes[0].axis('off')\n axes[0].set_title(Selection)\n\n axes[1].imshow(sample)\n axes[1].axis('off')\n axes[1].set_title('Auto-Colored')",
"_____no_output_____"
],
[
"def img_restoration(img):\n \"\"\"\n Summary -\n This function is created to carry out image restoration process in steps\n \n :param img: Image that needs to be restored\n :type img: Imageio class array \n :return: list of images and labels \n :rtype: String and imageio array list\n \"\"\"\n #perform denoising of the image\n denoised = cv.fastNlMeansDenoisingColored(img,None,7,10,6,21)\n #canny edge detection\n edges = cv.Canny(denoised,200,250)\n #filter for image processing\n kernel = np.ones((5,5),np.uint8)\n #image dilation\n dilation = cv.morphologyEx(edges, cv.MORPH_DILATE, kernel)\n closing = cv.morphologyEx(dilation, cv.MORPH_CLOSE, kernel)\n erode = cv.morphologyEx(closing,cv.MORPH_ERODE, kernel)\n #fill in missing gaps \n inpainted = cv.inpaint(denoised,erode,5,cv.INPAINT_TELEA)\n #overlay denoised image over smoothed out image\n alpha = 0.5\n overlaid = inpainted.copy()\n cv.addWeighted(denoised, alpha, overlaid, 1 - alpha,0, overlaid)\n \n #convert image to gray\n img2gray = cv.cvtColor(denoised,cv.COLOR_BGR2GRAY)\n #create mask and inverse mask based on threshold\n ret, mask = cv.threshold(img2gray, 100, 255, cv.THRESH_BINARY_INV)\n #combine via bit addition denoised image human and smoothed out background of inpainted image\n bg1 = cv.bitwise_and(denoised,denoised,mask = mask)\n mask_inv = cv.bitwise_not(mask)\n bg2 = cv.bitwise_and(overlaid,overlaid,mask = mask_inv)\n combined = cv.add(bg1,bg2)\n \n #store the various processed images\n images = [img,denoised,inpainted,overlaid,combined]\n labels = ['Original','Choice 1','Choice 2', 'Choice 3', 'Choice 4']\n return images, labels",
"_____no_output_____"
],
[
"def display_images(images,labels):\n \"\"\"\n Summary - \n This function displays the various processed images and labels associated with them\n :param images: list of processed images\n :type images: list\n :param labels: list of labels\n :type labels: list \n \"\"\"\n fig, axes = plt.subplots(ncols = 5, figsize = (15,5))\n axes[0].imshow(images[0])\n axes[0].axis('off')\n axes[0].set_title(labels[0])\n\n axes[1].imshow(images[1])\n axes[1].axis('off')\n axes[1].set_title(labels[1])\n \n axes[2].imshow(images[2])\n axes[2].axis('off')\n axes[2].set_title(labels[2])\n \n axes[3].imshow(images[3])\n axes[3].axis('off')\n axes[3].set_title(labels[3])\n \n axes[4].imshow(images[4])\n axes[4].axis('off')\n axes[4].set_title(labels[4])",
"_____no_output_____"
],
[
"!jupyter nbextension enable --py widgetsnbextension",
"_____no_output_____"
],
[
"import ipywidgets as widgets\nfrom IPython.display import display, clear_output\nfrom ipywidgets import interact, interact_manual, fixed, Output\n\n#create button for widget\nbutton = widgets.Button(description=\"Begin Restoration\")\nsave = widgets.Button(description=\"Save Result\")\n#create text box for widget\nfilename = widgets.Text(value='<filename>.jpg',placeholder='Type something',disabled=False)\n#create output area for widget\noutput = widgets.Output()\n\n#layout setting piece for code\nvertical = widgets.VBox([widgets.Label(value=\"Enter the name of the picture you want to restore:\"),widgets.HBox([filename, button])])\ndisplay(vertical)\ndisplay(output)\n\n\ndef on_button_clicked(b):\n \"\"\"\n Summary -\n This function handles the button clicked event\n :param b: passes a reference to the button itself\n :type b: Widgets Button \n \"\"\"\n with output:\n clear_output()\n #read the image\n img = io.imread(\"images/\"+filename.value)\n #gets 4 different kinds of processed images\n images, labels = img_restoration(img)\n #displays the various options\n display_images(images,labels)\n display(widgets.Label(value=\"Select the picture you want to color:\"))\n #code that calls the model based on user selected image\n interact(show_image, Selection = labels, l=fixed(labels), i=fixed(images), description = 'Choose image to color')\n display(widgets.Label(value=\"Output saved to the output folder\"))\n\nbutton.on_click(on_button_clicked)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec60a2857948a73765f79aebd792ceb86027437d | 1,254 | ipynb | Jupyter Notebook | notebooks/Untitled.ipynb | jhconning/simpleTeach | d89d29465e97cbb3bcb99c82f20af68bb2361ace | [
"MIT"
]
| 5 | 2020-02-03T06:04:54.000Z | 2021-12-25T07:14:28.000Z | notebooks/Untitled.ipynb | jhconning/simpleTeach | d89d29465e97cbb3bcb99c82f20af68bb2361ace | [
"MIT"
]
| 1 | 2020-02-01T01:34:52.000Z | 2020-02-21T16:43:02.000Z | notebooks/Untitled.ipynb | jhconning/simpleTeach | d89d29465e97cbb3bcb99c82f20af68bb2361ace | [
"MIT"
]
| 3 | 2019-09-02T15:19:32.000Z | 2020-05-18T16:31:37.000Z | 18.173913 | 45 | 0.519139 | [
[
[
"## Read FRED",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown"
]
]
|
ec60ab6997822f834c378cf4cf6fe3a3504d50bf | 1,489 | ipynb | Jupyter Notebook | .ipynb_checkpoints/11 偏最小二乘回归分析-checkpoint.ipynb | ZDDWLIG/math-model | ec64b7856c7bc43988b1b7e4e68e389cc9c9a8af | [
"MIT"
]
| 47 | 2020-12-02T02:06:56.000Z | 2022-03-28T02:51:43.000Z | .ipynb_checkpoints/11 偏最小二乘回归分析-checkpoint.ipynb | RabbitWhite1/Mathematical_Modeling_in_Python | ec64b7856c7bc43988b1b7e4e68e389cc9c9a8af | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/11 偏最小二乘回归分析-checkpoint.ipynb | RabbitWhite1/Mathematical_Modeling_in_Python | ec64b7856c7bc43988b1b7e4e68e389cc9c9a8af | [
"MIT"
]
| 15 | 2021-01-02T15:38:51.000Z | 2022-03-02T13:37:47.000Z | 23.634921 | 149 | 0.535259 | [
[
[
"# 偏最小二乘回归分析\n[参考网站](https://www.cnblogs.com/duye/p/9031511.html)\n- 当数据量小,甚至比变量维数还小,而相关性又比较大时使用,这个方法甚至优于主成分回归。 \n\n[sklearn.cross_decomposition.PLSRegression文档](https://scikit-learn.org/stable/modules/generated/sklearn.cross_decomposition.PLSRegression.html)",
"_____no_output_____"
]
],
[
[
"from sklearn.cross_decomposition import PLSRegression\nX = [[0., 0., 1.], [1.,0.,0.], [2.,2.,2.], [2.,5.,4.]]\nY = [[0.1, -0.2], [0.9, 1.1], [6.2, 5.9], [11.9, 12.3]]\npls2 = PLSRegression(n_components=2)\npls2.fit(X, Y)\nY_pred = pls2.predict(X)\nprint(Y_pred)",
"[[ 0.26087869 0.15302213]\n [ 0.60667302 0.45634164]\n [ 6.46856199 6.48931562]\n [11.7638863 12.00132061]]\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
]
]
|
ec60c5f3fca3cea423637ef5ee67e9e795da90cd | 262,457 | ipynb | Jupyter Notebook | requests_practice.ipynb | myusernameisuseless/web_services_creation_python | a2cddc95b06741e9d32365c2b08f0e33c8cc5123 | [
"Apache-2.0"
]
| null | null | null | requests_practice.ipynb | myusernameisuseless/web_services_creation_python | a2cddc95b06741e9d32365c2b08f0e33c8cc5123 | [
"Apache-2.0"
]
| null | null | null | requests_practice.ipynb | myusernameisuseless/web_services_creation_python | a2cddc95b06741e9d32365c2b08f0e33c8cc5123 | [
"Apache-2.0"
]
| null | null | null | 52.157591 | 526 | 0.561612 | [
[
[
"import requests\nimport json\n\n\nACCESS_TOKEN = '17da724517da724517da72458517b8abce117da17da72454d235c274f1a2be5f45ee711'\n\nuser = requests.get('https://api.vk.com/method/users.get?v=5.71&access_token=17da724517da724517da72458517b8abce117da17da72454d235c274f1a2be5f45ee711&user_ids=kipah')\n\nprint(user.text)",
"{\"response\":[{\"first_name\":\"Egar\",\"id\":54927323,\"last_name\":\"Khvoynitsky\"}]}\n"
],
[
"friends = requests.get('https://api.vk.com/method/friends.get?v=5.71&access_token=17da724517da724517da72458517b8abce117da17da72454d235c274f1a2be5f45ee711&user_id=54927323&fields=bdate')\n\nfriends = friends.json()",
"_____no_output_____"
],
[
"friends",
"_____no_output_____"
],
[
"ages = []\n\nfor friend in friends['response']['items']:\n try:\n if len(friend['bdate']) > 6:\n bdate = int(friend['bdate'][-1:-5:-1][::-1])\n age = 2021 - bdate\n ages.append(age)\n else:\n continue\n except:\n pass",
"_____no_output_____"
],
[
"from collections import Counter\n\n\n#print(Counter(ages).keys(), Counter(ages).values())\n\nunique_ages = list(set(ages))\nanswer = []\n\nfor age in unique_ages:\n answer.append((age, ages.count(age)))\n \nprint(answer)",
"[(16, 1), (17, 3), (18, 5), (19, 17), (20, 46), (21, 60), (22, 68), (23, 62), (24, 58), (25, 60), (26, 57), (27, 51), (28, 40), (29, 45), (30, 43), (31, 53), (32, 47), (33, 40), (34, 31), (35, 26), (36, 10), (37, 18), (38, 8), (39, 6), (40, 2), (41, 5), (42, 3), (43, 1), (47, 2), (49, 1), (50, 1), (51, 1), (52, 2), (55, 2), (58, 1), (59, 1), (61, 2), (67, 1), (72, 1), (88, 1), (89, 1), (90, 1), (91, 1), (100, 1), (101, 1), (104, 2), (110, 1), (111, 3), (113, 1), (114, 2), (116, 3), (117, 1), (118, 2), (119, 7)]\n"
],
[
"answer = sorted(answer, key=lambda x: x[0])\nanswer = sorted(answer, key=lambda x: x[1], reverse=True)\nanswer",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec60c904b1cf50ed16774a0f9d8ae561e31080ee | 699,900 | ipynb | Jupyter Notebook | solution/p2_full_solution.ipynb | ASU-CompMethodsPhysics-PHY494/project-snowwhite-7dwarves | 1b6308901b0599a7bb0ae5e26aec9ac536984d12 | [
"MIT"
]
| null | null | null | solution/p2_full_solution.ipynb | ASU-CompMethodsPhysics-PHY494/project-snowwhite-7dwarves | 1b6308901b0599a7bb0ae5e26aec9ac536984d12 | [
"MIT"
]
| null | null | null | solution/p2_full_solution.ipynb | ASU-CompMethodsPhysics-PHY494/project-snowwhite-7dwarves | 1b6308901b0599a7bb0ae5e26aec9ac536984d12 | [
"MIT"
]
| null | null | null | 587.657431 | 93,512 | 0.936741 | [
[
[
"# Project 2: Solution \nCopyright © 2017 Oliver Beckstein. All Rights Reserved.",
"_____no_output_____"
],
[
"## Goal and Objectives\n\nSimulate the movements of planets near the star TRAPPIST-1\n\n1. Simulate movement of the 6 closest planets (b, c, d, e, f, g) for 1000 earth days\n2. From which planets can an observer see the most other planets so close that one could resolve features of size 1000 km by naked eye?\n\nThe Space Tourism Bureau suggests TRAPPIST-1e (see poster below). Are they right?",
"_____no_output_____"
],
[
"\n(From [NASA Exoplanet Exploration: Galleries: TRAPPIST-1](https://exoplanets.nasa.gov/resources/2159/?linkId=34784370).)",
"_____no_output_____"
],
[
"## Outline of a Solution",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt\n%matplotlib inline\nmatplotlib.style.use('ggplot')",
"_____no_output_____"
]
],
[
[
"Most of the code is in the module `simulation`. First load system parameters from `parameters`.",
"_____no_output_____"
]
],
[
[
"import parameters\nimport simulation\nimport analysis\n\nfrom parameters import planets, star",
"_____no_output_____"
],
[
"from importlib import reload\nreload(simulation)\nreload(analysis)",
"_____no_output_____"
]
],
[
[
"## Orbits with star fixed ",
"_____no_output_____"
],
[
"### Initial conditions",
"_____no_output_____"
]
],
[
[
"planets_x = simulation.rmax(planets['semi-major'], planets['eccentricity'])\nplanets_vy = simulation.v_aphelion(planets['semi-major'], planets['eccentricity'], M=star['mass'])\n\nr0 = np.array([planets_x, np.zeros_like(planets_x)]).T\nv0 = np.array([np.zeros_like(planets_vy), planets_vy]).T\nplanet_masses = planets['mass']\nmasses = np.concatenate(([star['mass']], planet_masses))",
"_____no_output_____"
]
],
[
[
"### Simulation ",
"_____no_output_____"
],
[
"Test that the integration is (likely) correct and reproduces the period for the innermost planet: Check visually that after 1.5 d, TRAPPIST-1b has almost completed one revolution (period 1.51 d).",
"_____no_output_____"
]
],
[
[
"t, r, v = simulation.orbits(r0, v0, planet_masses, t_max=1.5, dt=0.01)\nanalysis.plot_orbits(r)",
"_____no_output_____"
],
[
"t, r, v = simulation.orbits(r0, v0, planet_masses, t_max=5, dt=0.01)\nanalysis.plot_orbits(r)",
"_____no_output_____"
]
],
[
[
"Run the simulation for 1000 d.",
"_____no_output_____"
]
],
[
[
"t, r, v = simulation.orbits(r0, v0, planet_masses, t_max=1000, dt=0.01)\nanalysis.plot_orbits(r)",
"_____no_output_____"
]
],
[
[
"### Energy conservation\nTest energy conservation\n\n$$\nE = T + U = \\text{const}\n$$\n\nfor the 1000-d run.",
"_____no_output_____"
],
[
"The `kinetic_energy()` function compute the kinetic energy for each planet and time step. To get the system energies, we have to sum over all bodies (`axis=-1`) but we also need to compute the potential energy for *all* $N$-body interactions\n\n$$\nU = \\frac{1}{2} \\sum_{i=1}^N \\sum_{j=1, j\\neq i}^N \\left( - \\frac{G m_i m_j}{r_{ij}} \\right)\n$$\n\n(as implemented in `simulation.U_tot()`). The total energy as a function of time\n\n$$\nE(t) = T(t) + U(t)\n$$\n\nor in code:",
"_____no_output_____"
],
[
"```python\nT = np.sum(analysis.kinetic_energy(v, masses), axis=-1)\nU = np.array([simulation.U_tot(rt, masses) for rt in r])\nE_tot = T + U\n```\n\n(`U_tot()` can only operate on individual frames of the trajectory, so we need to loop and put the results into an array.)",
"_____no_output_____"
],
[
"Note that for the analysis we provide all masses, not just the planet masses:",
"_____no_output_____"
]
],
[
[
"analysis.analyze_energies(t, r, v, simulation.U_tot, m=masses)",
"_____no_output_____"
]
],
[
[
"Energy conservation is acceptable, given the time step of `dt = 0.01`. (The unit of time is 1 so for the step of $10^{-2}$ we expect an error on the order of $\\Delta t^2$, i.e., $10^{-4}$ as seen.)",
"_____no_output_____"
],
[
"## BONUS: Star wobble ",
"_____no_output_____"
],
[
"Allow the central star to move, too, but set initial conditions so that the total momentum is 0. (Total momentum is conserved by velocity Verlet, and thus the center of mass of our simulation will remain fixed during the simulation.)",
"_____no_output_____"
]
],
[
[
"# planets \nr0 = np.array([planets_x, np.zeros_like(planets_x)]).T\nv0 = np.array([np.zeros_like(planets_vy), planets_vy]).T\n\n# prepend star\nr0_system = np.vstack((np.array([[0, 0]]), r0))\n\n# choose star velocity to make total momentum 0\np_planets = np.sum(planet_masses[:, np.newaxis] * v0, axis=0)\nv0_star = -p_planets / star['mass']\nv0_system = np.vstack((v0_star, v0))",
"_____no_output_____"
]
],
[
[
"NOTE: perhaps we should put the center of mass at 0 and move the star to\n\n\n\\begin{equation}\n\\mathbf{r}_{star} = - \\frac{\\sum_{i=1}^N m_i \\mathbf{r}_i}{M_{star}},\n\\end{equation}\n",
"_____no_output_____"
],
[
"### Trajectories with star wobbling ",
"_____no_output_____"
],
[
"1000 d trajectory with very fine time step (as a base line); later we go back to 0.01d time step.",
"_____no_output_____"
]
],
[
[
"t, r, v = simulation.orbits2(r0_system, v0_system, masses, t_max=1000, dt=0.001)\nanalysis.plot_orbits2(r)",
"_____no_output_____"
],
[
"analysis.analyze_energies(t, r, v, simulation.U_tot, m=masses)",
"_____no_output_____"
]
],
[
[
"Simulate for 1000 d with standard $\\Delta t = 0.01\\,\\text{d}$:",
"_____no_output_____"
]
],
[
[
"t, r, v = simulation.orbits2(r0_system, v0_system, masses, t_max=1000, dt=0.01)\nanalysis.plot_orbits2(r)",
"_____no_output_____"
]
],
[
[
"Note that the larger time step of 0.01 d introduces some inaccuracies in the inner orbits, compared to the trajectories for the 0.001 time step.",
"_____no_output_____"
],
[
"### Energy conservation with wobble",
"_____no_output_____"
]
],
[
[
"analysis.analyze_energies(t, r, v, simulation.U_tot, m=masses)",
"_____no_output_____"
]
],
[
[
"Qualitatively, the movement of the central star does not seem to affect the observed orbits and it also does not negatively affect the stability of the integrator. The relative accuracy is about $10^{-4}$, as expected for velocity Verlet's accuracy $\\mathcal{O}(\\Delta t^2/T)$ (where $T$ is the shortest time scale for an orbit, namely the orbital period of TRAPPIST-1b, 1.51 d) with a time step of $\\Delta t/T \\approx 10^{-2}$ and hence $\\epsilon \\approx (10^{-2})^2 = 10^{-4}$.",
"_____no_output_____"
],
[
"### Total momentum conservation\nAnalyze conservation of total system momentum:\n\n$$\n\\mathbf{P}(t) = \\sum_{i=1}^N m_i \\mathbf{v}_i(t)\n$$\n\nBecause we start with $P = 0$ we expect $P(t)$ to be close to 0 during the whole trajectory.",
"_____no_output_____"
]
],
[
[
"P = np.sum(v * masses[np.newaxis, :, np.newaxis], axis=1) # total momentum vector P(t)\np = np.sqrt(np.sum(P*P, axis=-1)) # length of momentum vector\nplt.plot(t, p)\nplt.xlabel(\"time (d)\")\nplt.ylabel(r\"total momentum $|\\mathbf{P}(t)|$\")",
"_____no_output_____"
],
[
"p.max()",
"_____no_output_____"
]
],
[
[
"The largest deviation is on the order of the machine precision. Therefore, momentum *is conserved* in our simulation to an excellent degree.",
"_____no_output_____"
],
[
"### Movement of the star ",
"_____no_output_____"
],
[
"Plot the movement of the center of the star for the first 5 days; the star's size is shown by an orange circle (which is much bigger than the area covered by the movement).",
"_____no_output_____"
]
],
[
[
"r_star = r[:int(5/0.01), 0][:, np.newaxis, :]\nax = analysis.plot_orbits_fancy2(r_star)\nax.set_xlabel(\"x (mAU)\")\nax.set_ylabel(\"y (mAU)\")",
"_____no_output_____"
]
],
[
[
"Movement of the star over 1000 d:",
"_____no_output_____"
]
],
[
[
"r_star = r[:, 0][:, np.newaxis, :]\nax = analysis.plot_orbits_fancy2(r_star)\nax.set_xlabel(\"x (mAU)\")\nax.set_ylabel(\"y (mAU)\")",
"_____no_output_____"
]
],
[
[
"Compare the extend of the movement to the size of the star itself (in mAU):",
"_____no_output_____"
]
],
[
[
"parameters.star_radius_localunits",
"_____no_output_____"
]
],
[
[
"The orbit of the star TRAPPIST-1a is inside the star itself. Thus the common center of mass of the system is inside the star itself and probably no clear wobble is observable. But we will check.",
"_____no_output_____"
],
[
"### Velocity analysis of TRAPPIST-1: Is the wobble detectable with Dopple spectroscopy? ",
"_____no_output_____"
],
[
"Could the wobble be observed by [Doppler spectroscopy](https://en.wikipedia.org/wiki/Doppler_spectroscopy) measurements? Third generation instruments are sensitive to 0.1 m/s.\n\nAssume ideal observation geometry (namely, observation side on). We approximate the [radial velocity](https://en.wikipedia.org/wiki/Radial_velocity) by the y-component of the velocity, $v_y$. (In principle it is the projection of the velocity on the observation direction but the change in angle is going to be very small and not much different from 0 because the observation distances is so much larger than the extend of the star motion. We could have also picked $v_x$ or any other viewing direction to get a sense for the magnitudes involved.)\n\nCompute the time series of star velocities and look at the maximum differences.",
"_____no_output_____"
],
[
"Transform from center-of-mass coordinates (simulation) to lab frame (observer) coordinates:\n\n$$\n\\mathbf{v}_{*} = -\\frac{\\sum_{i=1}^{N_{\\text{planets}}}\n m_{i}}{M_{\\text{star}} + \\sum_{i=1}^{N_{\\text{planets}}} m_{i}}\\mathbf{v}\n$$",
"_____no_output_____"
]
],
[
[
"def v_observer(v_com, M_star, masses_planets):\n return - v_com * masses_planets.sum() / (M_star + masses_planets.sum())",
"_____no_output_____"
],
[
"v_star_com = v[:, 0]\nv_star = v_observer(v_star_com, star['mass'], planet_masses)\nvy_star = v_star[:, 1] # in 1e-3 au/d\nvy_star_ms = analysis.au2km(vy_star * 1e-3) * 1e3 / (24*3600) # in m/s\ndt = 0.01\ntime = dt * np.arange(0, len(vy_star))",
"_____no_output_____"
],
[
"ax1 = plt.subplot(121)\nax1.plot(time, vy_star_ms)\nax1.set_xlabel(r\"time $t$ (d)\")\nax1.set_ylabel(r\"radial $v_y$ (m/s)\")\nax2 = plt.subplot(122)\nax2.plot(time, vy_star_ms)\nax2.set_xlabel(r\"time $t$ (d)\")\nax2.set_xlim(250, 280)\nax1.figure.tight_layout()",
"_____no_output_____"
],
[
"vy_max, vy_min = vy_star_ms.max(), vy_star_ms.min()\nprint(\"Delta v_radial = {0:.3f} m/s\\nmax v_r = {1:.3f} m/s\\nmin v_r = {2:.3f} m/s\".format(vy_max-vy_min, vy_max, vy_min))",
"Delta v_radial = 0.004 m/s\nmax v_r = 0.002 m/s\nmin v_r = -0.002 m/s\n"
]
],
[
[
"Current Doppler spectroscopy can resolve velocities on the order of 1 m/s (and latest generation instruments can do perhaps 0.1 m/s) but the radial velocities for TRAPPIST-1 are still 1-2 orders of magnitude smaller. Therefore, the wobble cannot be detected with Doppler spectroscopy. (Instead, light curver measurements were used.)",
"_____no_output_____"
],
[
"## Analyze close planets\n\nWe want to count only planets as *nearby* by if we can resolve features of size `feature_size` with the nake eye:",
"_____no_output_____"
]
],
[
[
"feature_size = 1000 # km",
"_____no_output_____"
]
],
[
[
"Given the angular resolution of the naked eye ($\\theta_\\text{eye} = 1'$), we look for planets within a cutoff distance $d$ where features of size $y$ are still resolved:\n\n$$\nd(y) ≤ \\frac{y}{\\theta_\\text{eye}}\n$$\n",
"_____no_output_____"
],
[
"The critical distance is",
"_____no_output_____"
]
],
[
[
"d_max = analysis.dmax(feature_size)\nprint(\"{0:g} km\".format(d_max))\nprint(\"{0:.1f} mAU\".format(analysis.km2au(d_max) * 1e3))",
"3.43775e+06 km\n23.0 mAU\n"
]
],
[
[
"Analyze the whole trajectory (here we are looking at the trajectory with the star allowed to wobble and so we remove the star from the trajectory with `r[:, 1:]`; qualitatively similar results are obtained when the star is fixed).",
"_____no_output_____"
]
],
[
[
"counts = analysis.find_nearby_planets(r[:, 1:], ymin=feature_size)",
"_____no_output_____"
]
],
[
[
"Plot the time series for TRAPPIST-1e (index 3 in the `counts` time series):",
"_____no_output_____"
]
],
[
[
"plt.plot(t, counts[:, 3], color=\"gray\")\nplt.xlabel(\"time (days)\")\nplt.ylabel(r\"number of close planets\")",
"_____no_output_____"
]
],
[
[
"Probability distribution for observing close planets:",
"_____no_output_____"
]
],
[
[
"plt.hist(counts, bins=6, range=(-0.5, 5.5), normed=True, label=planets['name'])\nplt.legend(loc=\"best\")\nplt.xlabel(\"number of other nearby planets\")\nplt.ylabel(\"probability\")\nplt.title(\"Features of size $y ≥ {}$ km are visible\".format(feature_size))\nplt.xlim(-0.5, 5.5);",
"_____no_output_____"
]
],
[
[
"In general, the inner planets (b, c, d) provide more opportunities to see other planets up close. However TRAPPIST-1e is the planet where one is most likely to see *four* other planets up close and it still has the third highest probability of any planet to have a close up view on *three* planets.",
"_____no_output_____"
],
[
"Only TRAPPIST-1d and TRAPPIST-1e see *five* planets at all:",
"_____no_output_____"
]
],
[
[
"np.sum(counts >= 5, axis=0)",
"_____no_output_____"
]
],
[
[
"although it happens rarely. Still TRAPPIST-1e is ",
"_____no_output_____"
]
],
[
[
"266/28",
"_____no_output_____"
]
],
[
[
"about 9 times more likely to see five planets than the only other contender.",
"_____no_output_____"
],
[
"The probability of observing *five* planets is small, but non-zero: the table shows the probability that one can see $N$ other planets from a given planet:",
"_____no_output_____"
]
],
[
[
"# make probability density (histogram) from the time series of each planet separately\nhistograms = [np.histogram(counts[:, i], bins=6, range=(-0.5, 5.5), density=True) for i in range(counts.shape[1])]\n_, edges = histograms[0]\nn = 0.5*(edges[1:] + edges[:-1]) # number of planets\n\n# formatting as a table (using only python)\nrow_format = \"{:6s}\" + \"{:>8}\" * len(n)\nrule = \"-\" * len(row_format.format(\"x\", *n))\nprint(rule + \"\\n\" +\n row_format.format(\"planet\", *n.astype(int)) + \"\\n\" +\n rule)\nfor name, (probability, _) in zip(planets['name'], histograms):\n print(row_format.format(name, *np.round(probability * 100, decimals=2)))\nprint(rule)\nprint(\"Probability in % to observe N planets.\")",
"------------------------------------------------------\nplanet 0 1 2 3 4 5\n------------------------------------------------------\nb 12.85 41.22 36.88 9.06 0.0 0.0\nc 13.11 38.77 35.93 11.51 0.68 0.0\nd 16.67 40.62 33.39 8.2 1.09 0.03\ne 26.6 38.64 23.1 9.49 1.91 0.27\nf 50.24 35.98 12.65 0.87 0.26 0.0\ng 72.11 25.1 2.8 0.0 0.0 0.0\n------------------------------------------------------\nProbability in % to observe N planets.\n"
]
],
[
[
"Only about 0.27% of the time are 5 planets visible from TRAPPIST-1e, but this is still more than any other; and the probability to observe 4 planets 1.9% is the highest for all planets.",
"_____no_output_____"
],
[
"Thus, the poster advertising TRAPPIST-1e speaks the truth: TRAPPIST-1e **is the best destination for planet watching**.",
"_____no_output_____"
],
[
"(even though the poster depicts a very rare sight)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec61036011f7768509a771ad116784acbde24f64 | 407,599 | ipynb | Jupyter Notebook | notebooks/archive/augmentation.ipynb | Neuralwood-Net/face-recognizer-9000 | b7804355927540bf07ce70cfe44dac6988a9b8cc | [
"MIT"
]
| null | null | null | notebooks/archive/augmentation.ipynb | Neuralwood-Net/face-recognizer-9000 | b7804355927540bf07ce70cfe44dac6988a9b8cc | [
"MIT"
]
| null | null | null | notebooks/archive/augmentation.ipynb | Neuralwood-Net/face-recognizer-9000 | b7804355927540bf07ce70cfe44dac6988a9b8cc | [
"MIT"
]
| null | null | null | 3,111.442748 | 402,648 | 0.962063 | [
[
[
"!pip install imgaug",
"Collecting imgaug\n Downloading imgaug-0.4.0-py2.py3-none-any.whl (948 kB)\n\u001b[K |████████████████████████████████| 948 kB 4.9 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: Pillow in /opt/conda/lib/python3.7/site-packages (from imgaug) (7.2.0)\nCollecting opencv-python\n Downloading opencv_python-4.4.0.46-cp37-cp37m-manylinux2014_x86_64.whl (49.5 MB)\n\u001b[K |████████████████████████████████| 49.5 MB 29.3 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: imageio in /opt/conda/lib/python3.7/site-packages (from imgaug) (2.9.0)\nRequirement already satisfied: scikit-image>=0.14.2 in /opt/conda/lib/python3.7/site-packages (from imgaug) (0.17.2)\nCollecting Shapely\n Downloading Shapely-1.7.1-cp37-cp37m-manylinux1_x86_64.whl (1.0 MB)\n\u001b[K |████████████████████████████████| 1.0 MB 93.8 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: matplotlib in /opt/conda/lib/python3.7/site-packages (from imgaug) (3.3.2)\nRequirement already satisfied: six in /opt/conda/lib/python3.7/site-packages (from imgaug) (1.15.0)\nRequirement already satisfied: numpy>=1.15 in /opt/conda/lib/python3.7/site-packages (from imgaug) (1.18.5)\nRequirement already satisfied: scipy in /opt/conda/lib/python3.7/site-packages (from imgaug) (1.5.2)\nRequirement already satisfied: networkx>=2.0 in /opt/conda/lib/python3.7/site-packages (from scikit-image>=0.14.2->imgaug) (2.5)\nRequirement already satisfied: tifffile>=2019.7.26 in /opt/conda/lib/python3.7/site-packages (from scikit-image>=0.14.2->imgaug) (2020.10.1)\nRequirement already satisfied: PyWavelets>=1.1.1 in /opt/conda/lib/python3.7/site-packages (from scikit-image>=0.14.2->imgaug) (1.1.1)\nRequirement already satisfied: kiwisolver>=1.0.1 in /opt/conda/lib/python3.7/site-packages (from matplotlib->imgaug) (1.2.0)\nRequirement already satisfied: cycler>=0.10 in /opt/conda/lib/python3.7/site-packages (from matplotlib->imgaug) (0.10.0)\nRequirement already satisfied: certifi>=2020.06.20 in /opt/conda/lib/python3.7/site-packages (from matplotlib->imgaug) (2020.6.20)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.3 in /opt/conda/lib/python3.7/site-packages (from matplotlib->imgaug) (2.4.7)\nRequirement already satisfied: python-dateutil>=2.1 in /opt/conda/lib/python3.7/site-packages (from matplotlib->imgaug) (2.8.1)\nRequirement already satisfied: decorator>=4.3.0 in /opt/conda/lib/python3.7/site-packages (from networkx>=2.0->scikit-image>=0.14.2->imgaug) (4.4.2)\nInstalling collected packages: opencv-python, Shapely, imgaug\nSuccessfully installed Shapely-1.7.1 imgaug-0.4.0 opencv-python-4.4.0.46\n"
],
[
"import imageio\nfrom skimage import io\nimport imgaug as ia\n%matplotlib inline\n\nimage_sk = io.imread(\"https://upload.wikimedia.org/wikipedia/en/7/7d/Lenna_%28test_image%29.png\")\nprint(type(image))\nprint(type(image_sk))\n\nprint(\"Original:\")\nia.imshow(image_sk)",
"<class 'imageio.core.util.Array'>\n<class 'numpy.ndarray'>\nOriginal:\n"
],
[
"from imgaug import augmenters as iaa\nia.seed(4)\n\nrotate = iaa.Affine(rotate=(-25, 25))\nimage_aug = rotate(image=image)\n\nprint(\"Augmented:\")\nia.imshow(image_aug)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
ec61075fb6995c9e82f1a14ba51e0ce90750840f | 251,159 | ipynb | Jupyter Notebook | python/projects/chess/Basic Chess.ipynb | BharathC15/NielitChennai | c817aaf63b741eb7a8e4c1df16b5038a0b4f0df7 | [
"MIT"
]
| null | null | null | python/projects/chess/Basic Chess.ipynb | BharathC15/NielitChennai | c817aaf63b741eb7a8e4c1df16b5038a0b4f0df7 | [
"MIT"
]
| null | null | null | python/projects/chess/Basic Chess.ipynb | BharathC15/NielitChennai | c817aaf63b741eb7a8e4c1df16b5038a0b4f0df7 | [
"MIT"
]
| 1 | 2020-06-11T08:04:43.000Z | 2020-06-11T08:04:43.000Z | 164.586501 | 44,072 | 0.868223 | [
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()",
"_____no_output_____"
],
[
"df=pd.read_csv(\"games.csv\")\ndf.head(3)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"plt.figure(dpi=120)\nsns.distplot(df[\"white_rating\"])",
"D:\\bharath\\Anaconda3\\lib\\site-packages\\scipy\\stats\\stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval\n"
],
[
"plt.figure(dpi=120)\nsns.distplot(df[\"black_rating\"])",
"D:\\bharath\\Anaconda3\\lib\\site-packages\\scipy\\stats\\stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval\n"
],
[
"print(\"Mean Rating of White \",df[\"white_rating\"].mean())\nprint(\"Mean Rating of Black \",df[\"black_rating\"].mean())",
"Mean Rating of White 1596.6318675840064\nMean Rating of Black 1588.8319872370128\n"
],
[
"df.groupby(\"winner\")[\"id\"].count()",
"_____no_output_____"
],
[
"df[\"winnerRating\"]=np.nan",
"_____no_output_____"
],
[
"df.head(3)",
"_____no_output_____"
],
[
"df.loc[df[\"winner\"]==\"white\",\"winnerRating\"]=df[\"white_rating\"]\ndf.loc[df[\"winner\"]==\"black\",\"winnerRating\"]=df[\"black_rating\"]\ndf.loc[df[\"winner\"]==\"draw\",\"winnerRating\"]=df[[\"black_rating\",\"white_rating\"]].mean(axis=1)",
"_____no_output_____"
],
[
"df[\"moves\"].head()",
"_____no_output_____"
],
[
"df[\"moves\"][0]",
"_____no_output_____"
]
],
[
[
"white_first,black_first=[],[]\nfor i in df[\"moves\"]:\n try:\n white_first.append(i.split(' ')[0])\n except:\n white_first.append(np.nan)\n try:\n black_first.append(i.split(' ')[1])\n except:\n black_first.append(np.nan)\ndf[\"blackFirst\"]=black_first\ndf[\"whiteFirst\"]=white_first",
"_____no_output_____"
]
],
[
[
"### Alternate method",
"_____no_output_____"
]
],
[
[
"df[\"whiteFirst\"]=df[\"moves\"].str.split(' ').str[0]\ndf[\"blackFirst\"]=df[\"moves\"].str.split(' ').str[1]",
"_____no_output_____"
]
],
[
[
"# Most Common moves by White and Black",
"_____no_output_____"
]
],
[
[
"df[\"whiteFirst\"].value_counts()",
"_____no_output_____"
],
[
"df[\"blackFirst\"].value_counts()",
"_____no_output_____"
],
[
"df[\"openingMove\"]=df[\"opening_name\"].str.replace(\"|\",\",\").str.split(\":\").str[0]",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df1=df[['rated','turns','victory_status','winner','white_rating',\n 'black_rating','openingMove','whiteFirst', 'blackFirst','winnerRating']]\ndf1.head()",
"_____no_output_____"
],
[
"df1[\"openingMove\"].value_counts().head(20)",
"_____no_output_____"
],
[
"plt.figure(dpi=120)\nprint(\"Mean Rating of the winners\",df[\"winnerRating\"].mean())\nsns.distplot(df1[\"winnerRating\"])\nplt.show()",
"Mean Rating of the winners 1636.692466846146\n"
],
[
"df1.describe()[\"winnerRating\"]",
"_____no_output_____"
],
[
"winnerRating_min=df1[\"winnerRating\"].min()\nwinnerRating_max=df1[\"winnerRating\"].max()",
"_____no_output_____"
],
[
"winnerRating_cut=pd.cut(df1.winnerRating,np.linspace(winnerRating_min,winnerRating_max,9))\nwinnerRating_cut.head()",
"_____no_output_____"
],
[
"df.pivot_table(values=\"turns\",index=winnerRating_cut,columns=\"winner\")",
"_____no_output_____"
],
[
"df2=df.pivot_table(values=\"turns\",index=winnerRating_cut,columns=\"winner\").reset_index()\ndf2",
"_____no_output_____"
],
[
"df2[\"meanMoves\"]=df2[[\"black\",\"draw\",\"white\"]].mean(axis=1)\ndf2",
"_____no_output_____"
],
[
"plt.figure(dpi=120)\nsns.barplot(x=\"winnerRating\",y=\"meanMoves\",data=df2)\nplt.xticks(rotation=90)\nplt.show()",
"_____no_output_____"
],
[
"df1.columns",
"_____no_output_____"
],
[
"df3=df.pivot_table(values=\"turns\",index=winnerRating_cut,columns=\"victory_status\").unstack().reset_index()\ndf3.head()",
"_____no_output_____"
],
[
"df3.columns=[\"victory_status\",\"winnerRating\",\"meanTurns\"]",
"_____no_output_____"
],
[
"plt.figure(dpi=120)\nsns.barplot(x=\"winnerRating\",y=\"meanTurns\",data=df3,palette='gist_rainbow_r')\nplt.xticks(rotation=90)\nplt.show()",
"D:\\bharath\\Anaconda3\\lib\\site-packages\\scipy\\stats\\stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval\n"
],
[
"plt.figure(dpi=120)\nsns.barplot(x=\"winnerRating\",y=\"meanTurns\",data=df3,hue=\"victory_status\",palette='deep')\nplt.xticks(rotation=90)\nplt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"raw",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6116bf6969d0e17aaea3fa644405f673608bd9 | 2,770 | ipynb | Jupyter Notebook | All about Python/Python IS Keyword.ipynb | SuperSaiyan-God/Machine-Learning | cb6c3859c8091a43d8a7ffd8f8d8f3437ba7a172 | [
"MIT"
]
| 1 | 2022-03-24T21:57:26.000Z | 2022-03-24T21:57:26.000Z | All about Python/Python IS Keyword.ipynb | SuperSaiyan-God/Machine-Learning | cb6c3859c8091a43d8a7ffd8f8d8f3437ba7a172 | [
"MIT"
]
| null | null | null | All about Python/Python IS Keyword.ipynb | SuperSaiyan-God/Machine-Learning | cb6c3859c8091a43d8a7ffd8f8d8f3437ba7a172 | [
"MIT"
]
| null | null | null | 16.390533 | 47 | 0.441155 | [
[
[
"#is vs ==\nlst1=[\"Krish\",\"Krish1\",\"Krish2\"]\nlst2=[\"Krish\",\"Krish1\",\"Krish2\"]\nlst1==lst2",
"_____no_output_____"
],
[
"a=\"Krish\"\nb=\"Krish\"\na==b",
"_____no_output_____"
],
[
"lst1=[\"Krish\",\"Krish1\",\"Krish2\"]\nlst2=lst1\nlst1 is lst2",
"_____no_output_____"
],
[
"lst2 is lst1",
"_____no_output_____"
],
[
"lst2[0]=\"Krish2\"",
"_____no_output_____"
],
[
"lst2",
"_____no_output_____"
],
[
"lst1",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6129fc3961d91ae81f4e7d39cce7eb2ad7b8a4 | 79,453 | ipynb | Jupyter Notebook | EmployeeSQL/code_pandas/.ipynb_checkpoints/HP_employee_db_pandas_bonus_analysis_backup-checkpoint.ipynb | robgauer/sql-challenge | e3259f38c1823d9e80adf33c91394312a15ceec4 | [
"ADSL"
]
| null | null | null | EmployeeSQL/code_pandas/.ipynb_checkpoints/HP_employee_db_pandas_bonus_analysis_backup-checkpoint.ipynb | robgauer/sql-challenge | e3259f38c1823d9e80adf33c91394312a15ceec4 | [
"ADSL"
]
| null | null | null | EmployeeSQL/code_pandas/.ipynb_checkpoints/HP_employee_db_pandas_bonus_analysis_backup-checkpoint.ipynb | robgauer/sql-challenge | e3259f38c1823d9e80adf33c91394312a15ceec4 | [
"ADSL"
]
| null | null | null | 71.838156 | 27,572 | 0.712774 | [
[
[
"# SQL Homework - Employee Database: A Mystery in Two Parts\n\n\n## File Information and Author\n#### ----------------------------------------------\n#### HEADER COMMENTS\n#### DATE May 8, 2020\n#### AUTHOR Rob Gauer\n#### FILE NAME 'HP_employee_db_pandas_bonus_analysis.ipynb'\n#### ----------------------------------------------\n\n## Bonus (Optional)\n##### As you examine the data, you are overcome with a creeping suspicion that the dataset is fake. \n##### You surmise that your boss handed you spurious data in order to test the data engineering skills of a new employee. \n##### To confirm your hunch, you decide to take the following steps to generate a visualization of the data, \n##### with which you will confront your boss:\n\n##### 1. Import the SQL database into Pandas. (Yes, you could read the CSVs directly in Pandas, but you are, after all, trying to prove your technical mettle.) This step may require some research. Feel free to use the code below to get started. Be sure to make any necessary modifications for your username, password, host, port, and database name:\n # from sqlalchemy import create_engine\n # engine = create_engine('postgresql://localhost:5432/<your_db_name>')\n # connection = engine.connect()\n \n##### Consult SQLAlchemy documentation for more information.\n##### If using a password, do not upload your password to your GitHub repository. \n##### See https://www.youtube.com/watch?v=2uaTPmNvH0I and https://martin-thoma.com/configuration-files-in-python/ \n##### for more information.\n\n##### 2. Create a histogram to visualize the most common salary ranges for employees.\n##### 3. Create a bar chart of average salary by title.\n#####",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\n#%matplotlib notebook\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport os\nimport sys\nimport psycopg2\n\n# define current date string to append to the names of output files...\nimport datetime \ndef _getToday(): return datetime.date.today().strftime(\"%Y%m%d\") ",
"_____no_output_____"
]
],
[
[
"# Secure Connectivity",
"_____no_output_____"
]
],
[
[
"# Secure database connectivity\n# Import path to permissions file for database\nsys.path.insert(0, \"/Users/rober/Desktop/ClassHomework/keys\")\n\n# Methon 1 - from conndb.py file import username, password, host, for secure connectivity\nfrom conndb import (username, password, host)\n\n# Method 2 - from conndb import username, password, host, port, for secure connectivity\n#from conndb import host\n#from conndb import port\n#from conndb import username\n#from conndb import password\n#print(username)\n#print(password)\n#print(host)\n#print(port)",
"_____no_output_____"
]
],
[
[
"# Database Connectivity",
"_____no_output_____"
]
],
[
[
"# Connect to local database\nfrom sqlalchemy import create_engine\nengine = create_engine(f\"postgresql://{username}:{password}@{host}:5432/employees_db\")\n#engine = create_engine(f\"postgresql://{username}:{password}@{host}/employees_db\")\nconnection = engine.connect()",
"_____no_output_____"
],
[
"# Review tables present in database\nengine.table_names()",
"_____no_output_____"
]
],
[
[
"# Create DataFrames",
"_____no_output_____"
]
],
[
[
"# Query database and create pandas dataframe\nemployees_df = pd.read_sql_query(\"\\\nSELECT * FROM salaries \\\nINNER JOIN employees ON employees.emp_no = salaries.emp_no \\\nINNER JOIN titles ON titles.title_id = employees.emp_title_id\",connection)",
"_____no_output_____"
],
[
"# Display column headers - validated\nemployees_df.columns",
"_____no_output_____"
],
[
"# Display dataframe\nemployees_df.head()",
"_____no_output_____"
],
[
"# Clean up dataframe - rename columns from dataframe \n# NOTE two instances of column header called 'emp_no' - both will be renamed to 'Employee No.'.\nemployees_df=employees_df.rename(columns={'emp_no':'Employee No.', 'salary':'Salary', 'emp_title_id':'Employee Title ID', 'birth_date':'Birth Date', 'first_name':'First Name', 'last_name':'Last Name', 'sex':'Sex', 'hire_date':'Hire Date', 'title_id':'Title ID','title':'Title'})\nemployees_df.head()",
"_____no_output_____"
],
[
"# Display column headers - validated\nemployees_df.columns",
"_____no_output_____"
],
[
"# Display dataframe\nemployees_df=employees_df[['Employee No.','Last Name','First Name','Sex','Birth Date','Hire Date','Title ID','Title','Salary']]\nemployees_df",
"_____no_output_____"
],
[
"# Display dataframe count \nemployees_df.count()",
"_____no_output_____"
]
],
[
[
"# CHART: Histogram - Common Salary Ranges for Employees",
"_____no_output_____"
]
],
[
[
"## CHART: Histogram to visualize the most common salary ranges for employees.\n\n# Set a Title and labels\ntitle=('Employee Salary Distribution')\n#plt.ylabel('Freqency (Number of Employees)')\n#plt.ylabel(\"Frequency (Number of Employees)\") \nplt.xlabel(\"Salary - Annual (US Dollars)\")\nxticks = np.arange(40000,130000,10000)\nplt.xticks(xticks,rotation=45)\n\n# Define chart\nemployees_df['Salary'].plot.hist(title=title, color='blue',figsize=(12, 7))\n# option for readibility - add grid=True\n\n# Save chart to file \nplt.savefig(\"../output_images/Histogram_Common_Salary_Ranges_for_Employees.png\") \n\n# Display figure\nplt.show()\nplt.tight_layout()",
"_____no_output_____"
],
[
"# Create dataframe to determine average salary by title.\nsalary_df = employees_df[['Salary','Title']].groupby('Title').mean()\nsalary_df",
"_____no_output_____"
]
],
[
[
"# CHART: Bar Chart - Average (mean) Salary by Title",
"_____no_output_____"
]
],
[
[
"## CHART: Bar Chart to visualize the average (mean) salary by title.\n\n# Define chart\ntitle='Average (Mean) Salary by Title'\nsalary_df.plot.bar(title=title, color='green',figsize=(12, 7))\n# option for readibility - add grid=True\n\n# Set a Title and labels\nplt.xticks(rotation=45)\nplt.ylabel(\"Salary - Annual (US Dollars)\")\nplt.xlabel(\"Employee Title\")\n\n# Save chart to file \nplt.savefig(\"../output_images/Bar_Chart_Average_Salary_by_Title.png\") \n\n# Display figure\nplt.show()\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"# Epilogue - Display your Employee Information (499942)",
"_____no_output_____"
]
],
[
[
"## Display your employee information.\n\n# Evidence in hand, you march into your boss's office and present the visualization. \n# With a sly grin, your boss thanks you for your work. On your way out of the office, \n# you hear the words, \"Search your ID number.\" \n# You look down at your badge to see that your employee ID number is 499942.\n\n#employees_df\nemployees_df.query('emp_no == 499942')",
"_____no_output_____"
]
],
[
[
"# Create & View all Dataframes from Database (optional)",
"_____no_output_____"
]
],
[
[
"# Review tables present in database\nengine.table_names()",
"_____no_output_____"
],
[
"# Display the database table - Salaries (salaries).\nsalaries_df=pd.read_sql('SELECT * FROM salaries', connection)\nsalaries_df.head()",
"_____no_output_____"
],
[
"# Display the database table - Titles (titles).\ntitles_df=pd.read_sql('SELECT * FROM titles', connection)\ntitles_df.head()",
"_____no_output_____"
],
[
"# Display the database table - Employees (employees)\nemployees2_df=pd.read_sql('SELECT * FROM employees', connection)\nemployees2_df.head()",
"_____no_output_____"
],
[
"# Display the database table - Departments (departments).\ndepartments_df=pd.read_sql('SELECT * FROM departments', connection)\ndepartments_df.head()",
"_____no_output_____"
],
[
"# Display the database table - Department Manager (dept_manager).\ndept_manager_df=pd.read_sql('SELECT * FROM dept_manager', connection)\ndept_manager_df.head()",
"_____no_output_____"
],
[
"# Display the database table - Department Employees (dept_emp).\ndept_emp_df=pd.read_sql('SELECT * FROM dept_emp', connection)\ndept_emp_df.head()",
"_____no_output_____"
],
[
"## EOF ##\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec613360f0266414986ad800c9d5b661939703ba | 57,843 | ipynb | Jupyter Notebook | 04-early-stopping.ipynb | edwinb-ai/deep-workshop | ed9f60a429dbac0b136bc270051d08e882cee8f3 | [
"MIT"
]
| null | null | null | 04-early-stopping.ipynb | edwinb-ai/deep-workshop | ed9f60a429dbac0b136bc270051d08e882cee8f3 | [
"MIT"
]
| 10 | 2019-08-11T17:34:11.000Z | 2019-10-11T02:37:45.000Z | 04-early-stopping.ipynb | edwinb-ai/deep-workshop | ed9f60a429dbac0b136bc270051d08e882cee8f3 | [
"MIT"
]
| null | null | null | 132.363844 | 32,296 | 0.864426 | [
[
[
"# Entrenamiento automático (Early Stopping)\n\nLa técnica de _Early Stopping_ es de suma importancia para el entrenamiento de redes neuronales. Consiste en monitorear una métrica o valor correspondiente que está siendo evaluado por el modelo en cuestión, tal que cuando este valor empeore el entrenamiento se detenga automáticamente.\n\nEs una técnica de suma importancia para evitar los problemas de sobreajuste en redes neuronales, esto se logra cuando se genera un conocimiento de lo que está sucediendo en el entrenamiento de las redes neuronales.\n\nEn este documento se verá un ejemplo aplicado a una CNN que clasifica el conjunto de datos CIFAR-10, con algunas de las técnicas conocidas como _Batch Normalization_ y _Early Stopping_ para ver su efecto.",
"_____no_output_____"
]
],
[
[
"from keras.datasets import cifar10\nfrom keras.models import Sequential, load_model\nfrom keras.layers import Dense, BatchNormalization, Activation\nfrom keras.layers import Conv2D, MaxPooling2D, Flatten\nfrom keras.utils import to_categorical\nfrom keras.callbacks import EarlyStopping, ModelCheckpoint\nfrom keras.optimizers import Nadam\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"Como siempre, se inicializan los hiperparámetros necesarios para el modelo, en este caso se emplean 100 épocas, pero no se asegura que se utilicen todas.",
"_____no_output_____"
]
],
[
[
"batch_size = 32\nnum_classes = 10\nepocas = 100",
"_____no_output_____"
]
],
[
[
"## Preprocesamiento de datos\n\nComo se acostumbra a hacer, se cargan los datos, se hace una exploración general del conjunto y se codifican las clases utilizando _One Hot_ para poder emplear la función de pérdida usual que es la entropía cruzada.",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (x_test, y_test) = cifar10.load_data()",
"_____no_output_____"
],
[
"print(\"Tamaño de x_train:\", x_train.shape)\nprint(x_train.shape[0], \"Ejemplos de entrenamiento\")\nprint(x_test.shape[0], \"Ejemplos de prueba\")",
"Tamaño de x_train: (50000, 32, 32, 3)\n50000 Ejemplos de entrenamiento\n10000 Ejemplos de prueba\n"
],
[
"plt.imshow(x_train[10, :, :, :])",
"_____no_output_____"
],
[
"x_train = x_train.astype(\"float32\")\nx_test = x_test.astype(\"float32\")\n# Siempre se deben normalizar los datos\nx_train /= 255\nx_test /= 255",
"_____no_output_____"
],
[
"# Codificar a One Hot cada clase del conjunto de datos\ny_train = to_categorical(y_train, num_classes)\ny_test = to_categorical(y_test, num_classes)",
"_____no_output_____"
]
],
[
[
"## Arquitectura de la CNN\n\nEn este caso se va a aplicar la siguiente arquitectura:\n\n- La **primera** capa es de convolución con 32 unidades, con relleno de $3 \\times 3$, y función de activación ReLU.\n- La **segunda** capa también es de convolución con 32 unidades, sin relleno, y función de activación ReLU.\n- A estas dos capas le sigue una capa de _max pooling_ de $2 \\times 2$.\n- La **tercera** capa es de convolución con 64 unidades, con relleno de $3 \\times 3$, y función de activación ReLU.\n- La **cuarta** capa también es de convolución con 64 unidades, sin relleno, y función de activación ReLU.\n- A estas dos capas le sigue una nueva capa de _max pooling_ de $2 \\times 2$; aquí termina la etapa de características\n- Con la etapa de clasificación se **aplanan** las imágenes y se ingresan a una red neuronal totalmente conectada de 512 unidades con función de activación ReLU.\n- La **capa de salida** tiene tantas unidades como clases y función de activación softmax.\n\nEntre cada capa se aplica _batch normalization_ y se quita el uso de sesgos para acelerar el entrenamiento.",
"_____no_output_____"
]
],
[
[
"# Todas las capas normalizadas, excpeto la última\narquitectura_norm = [\n # Capa de entrada, primera capa de características\n Conv2D(32, kernel_size=(3, 3),\n padding=\"same\",\n input_shape=x_train.shape[1:],\n use_bias=False),\n BatchNormalization(scale=False),\n Activation(\"relu\"),\n Conv2D(32, (3, 3), use_bias=False),\n BatchNormalization(scale=False),\n Activation(\"relu\"),\n MaxPooling2D(pool_size=(2, 2)),\n # Segunda capa de características\n Conv2D(64, kernel_size=(3, 3),\n padding=\"same\",\n use_bias=False),\n BatchNormalization(scale=False),\n Activation(\"relu\"),\n Conv2D(64, (3, 3), use_bias=False),\n BatchNormalization(scale=False),\n Activation(\"relu\"),\n MaxPooling2D(pool_size=(2, 2)),\n # Capa totalmente conectada\n Flatten(),\n Dense(512, use_bias=False),\n BatchNormalization(scale=False),\n Activation(\"relu\"),\n # Capa de salida\n Dense(num_classes, activation=\"softmax\")\n]",
"_____no_output_____"
],
[
"# Se construye el modelo con esta arquitectura\nmodel_normalizado = Sequential(arquitectura_norm)",
"_____no_output_____"
]
],
[
[
"## Optimizador y entrenamiento\n\nPara el optimizador se emplea _Nadam_ con los [valores default](https://keras.io/optimizers/) de `keras`.",
"_____no_output_____"
]
],
[
[
"model_normalizado.compile(loss=\"categorical_crossentropy\",\n optimizer=Nadam(),\n metrics=[\"accuracy\"])",
"_____no_output_____"
]
],
[
[
"## Llamadas al modelo\n\nAhora se va a implementar _Early stopping_ como una llamada al modelo. Afortunadamente `keras` ya cuenta con una implementación de esta metodología. En este caso se monitereará la **pérdida del conjunto de validación**, y se estará observando su cambio a lo largo de 5 épocas.\n\nAdicionalmente se va a implementar el guardado del mejor modelo cada que se evalua. A este se le conoce como _Model checkpoint_ en `keras`, lo que permite guardar el modelo que tenga el mejor valor monitoreado. En este caso se va a monitorear la **precisión en el conjunto de validación** y solamente se guardará el mejor modelo encontrado.",
"_____no_output_____"
]
],
[
[
"# Aplicar Early Stopping\nes = EarlyStopping(monitor=\"val_loss\", mode=\"min\", verbose=1, patience=5)\n# Guardar siempre el mejor modelo encontrado en base a la precisión de validación\nmc = ModelCheckpoint(\"mejor_modelo.h5\", monitor=\"val_acc\", mode=\"max\", verbose=1, save_best_only=True)",
"_____no_output_____"
],
[
"# Se procede a entrenar el modelo\nhistoria = model_normalizado.fit(x_train, y_train,\n validation_split=0.2,\n epochs=epocas,\n verbose=1,\n callbacks=[es, mc])",
"Train on 40000 samples, validate on 10000 samples\nEpoch 1/100\n40000/40000 [==============================] - 126s 3ms/step - loss: 1.1688 - acc: 0.5898 - val_loss: 1.1700 - val_acc: 0.6021\n\nEpoch 00001: val_acc improved from -inf to 0.60210, saving model to mejor_modelo.h5\nEpoch 2/100\n40000/40000 [==============================] - 120s 3ms/step - loss: 0.7645 - acc: 0.7328 - val_loss: 1.0196 - val_acc: 0.6518\n\nEpoch 00002: val_acc improved from 0.60210 to 0.65180, saving model to mejor_modelo.h5\nEpoch 3/100\n40000/40000 [==============================] - 124s 3ms/step - loss: 0.5962 - acc: 0.7924 - val_loss: 0.7436 - val_acc: 0.7483\n\nEpoch 00003: val_acc improved from 0.65180 to 0.74830, saving model to mejor_modelo.h5\nEpoch 4/100\n40000/40000 [==============================] - 125s 3ms/step - loss: 0.4569 - acc: 0.8398 - val_loss: 0.8535 - val_acc: 0.7280\n\nEpoch 00004: val_acc did not improve from 0.74830\nEpoch 5/100\n40000/40000 [==============================] - 126s 3ms/step - loss: 0.3435 - acc: 0.8795 - val_loss: 0.8401 - val_acc: 0.7474\n\nEpoch 00005: val_acc did not improve from 0.74830\nEpoch 6/100\n40000/40000 [==============================] - 121s 3ms/step - loss: 0.2414 - acc: 0.9166 - val_loss: 0.9202 - val_acc: 0.7379\n\nEpoch 00006: val_acc did not improve from 0.74830\nEpoch 7/100\n40000/40000 [==============================] - 127s 3ms/step - loss: 0.1821 - acc: 0.9371 - val_loss: 0.9331 - val_acc: 0.7487\n\nEpoch 00007: val_acc improved from 0.74830 to 0.74870, saving model to mejor_modelo.h5\nEpoch 8/100\n40000/40000 [==============================] - 128s 3ms/step - loss: 0.1359 - acc: 0.9536 - val_loss: 0.9541 - val_acc: 0.7612\n\nEpoch 00008: val_acc improved from 0.74870 to 0.76120, saving model to mejor_modelo.h5\nEpoch 00008: early stopping\n"
]
],
[
[
"Como se puede ver, el modelo dejó de entrenar abruptamente. Esto se debe a que el valor de pérdida de la última época de entrenamiento empezó a aumentar. Vamos a analizar esto con más detalle.\n\nEn la época 7 el **valor de la pérdida en el conjunto de validación** fue de 0.9331, mientras que en el época 8 fue de 09541. Al ver que este valor aumentó se activa el _Early stopping_ deteniendo por completo el entrenamiento.\n\nSe debe notar que el monitoreo es cada 5 épocas. Con este se puede ver el siguiente gráfica para analizar visualmente el entrenamiento.",
"_____no_output_____"
]
],
[
[
"# Graficar los valores de pérdida en el entrenamiento\nplt.figure(figsize=(13, 9))\nplt.plot(historia.history['loss'])\nplt.plot(historia.history['val_loss'])\nplt.legend(['Train', 'Test'], loc='upper left')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Claramente se presenta sobreajuste, pero más importante que eso, el valor de la pérdida el conjunto de entrenamiento y el conjunto de validación están muy dispersos.\n\nEste es el propósito de _Early stopping._ Cuando no existe una mejora sustancial en la clasificación del conjunto de validación es mejor detener el entrenamiento y afinar los hiperparámetros necesarios para mejorar el rendimiento del modelo.",
"_____no_output_____"
]
],
[
[
"# Cargar el mejor modelo encontrado\nmejor_modelo = load_model(\"mejor_modelo.h5\")",
"_____no_output_____"
]
],
[
[
"Con esto se carga el mejor modelo encontrado en el proceso de entrenamiento, que fue guardado mientras se evaluaba cada época. La ventaja de _Early stopping_ es que siempre se puede tener un modelo guardado y emplearlo para tener el conjunto de datos de prueba al respecto.\n\nEn el siguiente pedazo de código se hace justamente esto. Se cargó el modelo guardado y ahora se evalúa con un conjunto de datos que nunca fue visto por el modelo.",
"_____no_output_____"
]
],
[
[
"resultado = mejor_modelo.evaluate(x_test, y_test)\nprint(f\"Pérdida: {resultado[0]}\\nPrecisión: {resultado[1]}\")",
"10000/10000 [==============================] - 10s 960us/step\nPérdida: 0.9576307853221894\nPrecisión: 0.7606\n"
]
],
[
[
"Como se puede ver, la pérdida y la precisión no son tan malos. Claramente no es un resultado _estado del arte_ pero al emplear _Early stopping_ el entrenamiento y cálculos innecesarios fueron evitados para obtener el mejor modelo posible.",
"_____no_output_____"
],
[
"## Ejercicios\n\n1. Probar con diferentes valores para `patience` en el _Early stopping_. ¿Qué sucede si se aumentan? ¿Qué pasa cuando disminuyen?",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
ec61548f57bb02008a56e1e1c3cceb4239ce1f0e | 104,106 | ipynb | Jupyter Notebook | 1.3 R Data Frames/de-DE/1.3.18 R - Data Frame Basics.ipynb | softfactories/r_train_2 | ffa0530ba13c7b43f5f33a4141b1690647603141 | [
"MIT"
]
| null | null | null | 1.3 R Data Frames/de-DE/1.3.18 R - Data Frame Basics.ipynb | softfactories/r_train_2 | ffa0530ba13c7b43f5f33a4141b1690647603141 | [
"MIT"
]
| null | null | null | 1.3 R Data Frames/de-DE/1.3.18 R - Data Frame Basics.ipynb | softfactories/r_train_2 | ffa0530ba13c7b43f5f33a4141b1690647603141 | [
"MIT"
]
| null | null | null | 61.059238 | 439 | 0.350585 | [
[
[
"# Tag 1. Kapitel 3. Data Frames\n\n## Lektion 18. Data Frame Grundlagen\n\nWir haben bereits Vektoren und deren zweidimensionales Gegenstück, die Matrizen, kennengelernt. Bevor wir uns abschließend mit Listen befassen schauen wir uns jetzt *Data Frames* an. Diese bilden das Hauptwerkzeug zur Datenanalyse in R! Matrizen sind dazu etwas zu beschränkt, da alle Elemente in ihnen nur einen Datentyp haben dürfen. Durch Data Frames können wir jetzt verschiedene Daten Typen zusammen speichern und bearbeiten.\n\nR bietet seinerseits einige vorinstallierte Data Frames, die gut sind, um die Funktionalität kennenzulernen und etwas herumzuspielen. Es folgen einige Beispiele:",
"_____no_output_____"
]
],
[
[
"# Data Frame der US Staaten\nstate.x77",
"_____no_output_____"
],
[
"# US persönlich Ausgaben\nUSPersonalExpenditure",
"_____no_output_____"
],
[
"# Frauen \nwomen",
"_____no_output_____"
]
],
[
[
"Um eine Liste aller verfügbaren Data Frames zu erhalten können wir `data()` nutzen.",
"_____no_output_____"
]
],
[
[
"data() # aktuelle geladene Datensätze anzeigen",
"_____no_output_____"
],
[
"sleep",
"_____no_output_____"
]
],
[
[
"Wir können auch Daten aus externen Datenpaketn laden. Z.B. so kann man bio-medizinische Daten aus dem \"survival\"-Paket laden:",
"_____no_output_____"
]
],
[
[
"install.packages('survival')",
"package 'survival' successfully unpacked and MD5 sums checked\n\nThe downloaded binary packages are in\n\tC:\\Users\\dm_78\\AppData\\Local\\Temp\\Rtmp6bbZNZ\\downloaded_packages\n"
],
[
"library(survival)\ndata()",
"_____no_output_____"
],
[
"# Daten zur Stanford Herz-Transplantation \nheart",
"_____no_output_____"
],
[
"ovarian",
"_____no_output_____"
]
],
[
[
"## Mit Data Frames arbeiten\n\nBestimmt ist euch aufgefallen, dass manche Daten sehr umfangreich sind. Wir können die Funktionen `head()` bzw. `tail()` verwenden, um die ersten bzw. letzten 6 Zeilen anzuzeigen. Schauen wir uns das an:",
"_____no_output_____"
]
],
[
[
"# Kurze Variablenzuweisung, um etwas Tippen zu sparen\nht <- heart\nclass(ht)",
"_____no_output_____"
],
[
"head(ht)",
"_____no_output_____"
],
[
"tail(ht)",
"_____no_output_____"
]
],
[
[
"### Data Frames - Informationsübersicht\n\nWir können die `str()` Funktion nutzen, um mehr über die Struktur eines Data Frame zu erhalten. Wir erhalten bspw. Informationen über die Variablennamen und Datentypen, die der Data Frame beinhaltet. Zusätzlich können wir `summary()` nutzen, um eine schnelle statistische Auswertung zu erhalten. Für einen ersten Eindruck ist dies außerordentlich nützlich.",
"_____no_output_____"
]
],
[
[
"# Statistische Zusammenfassung\nst <- state.x77\nsummary(st)",
"_____no_output_____"
],
[
"summary(iris)\nhead(iris)",
"_____no_output_____"
],
[
"summary(ht)",
"_____no_output_____"
],
[
"# Struktur der Daten\nstr(st)",
" num [1:50, 1:8] 3615 365 2212 2110 21198 ...\n - attr(*, \"dimnames\")=List of 2\n ..$ : chr [1:50] \"Alabama\" \"Alaska\" \"Arizona\" \"Arkansas\" ...\n ..$ : chr [1:8] \"Population\" \"Income\" \"Illiteracy\" \"Life Exp\" ...\n"
],
[
"str(ht)",
"'data.frame':\t172 obs. of 8 variables:\n $ start : num 0 0 0 1 0 36 0 0 0 51 ...\n $ stop : num 50 6 1 16 36 39 18 3 51 675 ...\n $ event : num 1 1 0 1 0 1 1 1 0 1 ...\n $ age : num -17.16 3.84 6.3 6.3 -7.74 ...\n $ year : num 0.123 0.255 0.266 0.266 0.49 ...\n $ surgery : num 0 0 0 0 0 0 0 0 0 0 ...\n $ transplant: Factor w/ 2 levels \"0\",\"1\": 1 1 1 2 1 2 1 1 1 2 ...\n $ id : num 1 2 3 3 4 4 5 6 7 7 ...\n"
]
],
[
[
"# Data Frames erstellen\n\n*Hinweis: Teilweise verwenden Leute Dataframe als ein Wort. In R ist es jedoch üblich von zwei Worten \"Data Frame\" zu sprechen. Kein wirklich großer Unterschied, doch kann es sein, dass DataFrame sich auf Python/Pandas bezieht, wenn es jemand dementsprechend schreibt.*\n\nWir können Data Frames erstellen, indem wir die `data.frame()` Funktion verwenden und Vektoren als Argumente übergeben. Diese Vektoren werden dann in Spalten des Data Frame umgewandelt. Hier ist ein Beispiel:",
"_____no_output_____"
]
],
[
[
"# Einige erfunden Wetterdaten\ntage <- c('Mo','Di','Mi','Do','Fr')\ntemp <- c(22.2,NA,NA,24.3,25)\nregen <- c(TRUE, TRUE, FALSE, FALSE, TRUE)",
"_____no_output_____"
],
[
"# Die Vektoren übergeben\ndf <- data.frame(tage,temp,regen)",
"_____no_output_____"
],
[
"tage\ntemp\nregen\n\ndf",
"_____no_output_____"
],
[
"# Struktur überprüfen\nstr(df)",
"'data.frame':\t5 obs. of 3 variables:\n $ tage : Factor w/ 5 levels \"Di\",\"Do\",\"Fr\",..: 5 1 4 2 3\n $ temp : num 22.2 NA NA 24.3 25\n $ regen: logi TRUE TRUE FALSE FALSE TRUE\n"
],
[
"summary(df)",
"_____no_output_____"
]
],
[
[
"Herzlichen Glückwunsch! Sie sind mit Lektion 18. fertig!",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec615867a2ffa819de761bc324f24deb01753383 | 133,942 | ipynb | Jupyter Notebook | test.ipynb | Ryan-Amaral/cifar-10-100-tpg | 08a08577e921ce802764005c50eebd766e896f77 | [
"MIT"
]
| null | null | null | test.ipynb | Ryan-Amaral/cifar-10-100-tpg | 08a08577e921ce802764005c50eebd766e896f77 | [
"MIT"
]
| null | null | null | test.ipynb | Ryan-Amaral/cifar-10-100-tpg | 08a08577e921ce802764005c50eebd766e896f77 | [
"MIT"
]
| null | null | null | 131.187071 | 11,136 | 0.836056 | [
[
[
"import pickle\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport random",
"_____no_output_____"
],
[
"labels = []\ndata = []",
"_____no_output_____"
],
[
"with open(\"cifar-10-batches-py/data_batch_1\", \"rb\") as f:\n mdict = pickle.load(f, encoding=\"bytes\")\n \nprint(mdict.keys())\nmdict[b\"data\"]",
"dict_keys([b'batch_label', b'labels', b'data', b'filenames'])\n"
],
[
"for i in range(5):\n with open(\"cifar-10-batches-py/data_batch_\" + str(i+1), \"rb\") as f:\n mdict = pickle.load(f, encoding=\"bytes\")\n for i in range(len(mdict[b\"data\"])):\n data.append(mdict[b\"data\"][i])\n labels.append(mdict[b\"labels\"][i])",
"_____no_output_____"
],
[
"labelNames = ['airplane', 'automobile', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']\n\nfor i in range(10):\n print(labelNames[labels[i]])\n plt.imshow(np.transpose(data[i].reshape((3,32,32)), \n (1,2,0)))\n plt.show()",
"frog\n"
]
],
[
[
"## Experiment Settings:\n\n- With and without memory.\n - 0: Without memory.\n - 1: With memory.\n- Different generational data sampling methods to try:\n - 0: All examples every generation.\n - 1: Random stratified subsample per generation.\n - 2: Single class per generation.\n- Different ordering in generation:\n - 0: Single individual guesses all samples, then do next individual.\n - 1: All individual guess single sample, then do next sample.\n- Different Fitness methods:\n - 0: Mean score across classes.\n - 1: Min score across classes.\n - 2: Static Lexicographic.\n - 3: Dynamic Lexicographic.\n - 4: Score of only current class (only with data sampling #2).\n - 5: Score of only current class + diminishing previous score (only with data sampling #2).\n \n40 Experiments to run.\n\n## Experiment Process:\n- Separate environments for memory or not.\n- First get list of all labels and list of all datas.\n- Setup TPG\n- run stuff etc.\n\n## Reporting:\n- Generational log:\n - Accuracy per class per generation.\n - Cumulative time per generation (hours).\n - Size of best agent per generation.\n- Confusion matrix per generation of best individual.\n- Final scores on test data.\n- Full population and best individual after last generation.",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(labels).add_prefix(\"y\").join(pd.DataFrame(data).add_prefix(\"x\"))\ndf.head()",
"_____no_output_____"
],
[
"classes = random.sample(range(10), 3)\nprint(classes)\nsampleDf = df[df[\"y0\"].isin(classes)].groupby(\"y0\", group_keys=False).apply(lambda x: x.sample(min(len(x), 2)))\nsampleDf.head(7)",
"[7, 9, 4]\n"
],
[
"sampleDf = df[:5].groupby(\"y0\", group_keys=False)#.apply(lambda x: x.sample(min(len(x), 2)))\nsampleDf.count()\npd.concat([sampleDf.get_group(i) for i in [1,9]])",
"_____no_output_____"
],
[
"data = np.array(sampleDf, dtype=float)[:,1:]\nlabels = np.array(sampleDf)[:,0]",
"_____no_output_____"
],
[
"data/255",
"_____no_output_____"
],
[
"pd.DataFrame(labels)[0].value_counts()[0]",
"_____no_output_____"
],
[
"guess = 1\nlabel = 1\n\nscore = int(guess==label)\nscore",
"_____no_output_____"
],
[
"0==False",
"_____no_output_____"
],
[
"data[0]",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec616d39dc94dc9d07b587f5724c8310b9ed9f05 | 71,450 | ipynb | Jupyter Notebook | docs/tutorials/tfx/components_keras.ipynb | AseiSugiyama/tfx | d7057809198d17dcd3216537677e6af0fcd02202 | [
"Apache-2.0"
]
| null | null | null | docs/tutorials/tfx/components_keras.ipynb | AseiSugiyama/tfx | d7057809198d17dcd3216537677e6af0fcd02202 | [
"Apache-2.0"
]
| null | null | null | docs/tutorials/tfx/components_keras.ipynb | AseiSugiyama/tfx | d7057809198d17dcd3216537677e6af0fcd02202 | [
"Apache-2.0"
]
| null | null | null | 36.39837 | 538 | 0.550595 | [
[
[
"##### Copyright © 2020 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# TFX Component Tutorial\n\n***A Component-by-Component Introduction to TensorFlow Extended (TFX)***",
"_____no_output_____"
],
[
"Note: We recommend running this tutorial in a Colab notebook, with no setup required! Just click \"Run in Google Colab\".\n\n<div class=\"devsite-table-wrapper\"><table class=\"tfo-notebook-buttons\" align=\"left\">\n<td><a target=\"_blank\" href=\"https://www.tensorflow.org/tfx/tutorials/tfx/components_keras\">\n<img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a></td>\n<td><a target=\"_blank\" href=\"https://colab.sandbox.google.com/github/tensorflow/tfx/blob/master/docs/tutorials/tfx/components_keras.ipynb\">\n<img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\">Run in Google Colab</a></td>\n<td><a target=\"_blank\" href=\"https://github.com/tensorflow/tfx/tree/master/docs/tutorials/tfx/components_keras.ipynb\">\n<img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\">View source on GitHub</a></td>\n</table></div>",
"_____no_output_____"
],
[
"\nThis Colab-based tutorial will interactively walk through each built-in component of TensorFlow Extended (TFX).\n\nIt covers every step in an end-to-end machine learning pipeline, from data ingestion to pushing a model to serving.\n\nWhen you're done, the contents of this notebook can be automatically exported as TFX pipeline source code, which you can orchestrate with Apache Airflow and Apache Beam.\n\nNote: This notebook demonstrates the use of native Keras models in TFX pipelines. **TFX only supports the TensorFlow 2 version of Keras**.\n\nNote: This notebook and its associated APIs are **experimental** and are\nin active development. Major changes in functionality, behavior, and\npresentation are expected.",
"_____no_output_____"
],
[
"## Background\nThis notebook demonstrates how to use TFX in a Jupyter/Colab environment. Here, we walk through the Chicago Taxi example in an interactive notebook.\n\nWorking in an interactive notebook is a useful way to become familiar with the structure of a TFX pipeline. It's also useful when doing development of your own pipelines as a lightweight development environment, but you should be aware that there are differences in the way interactive notebooks are orchestrated, and how they access metadata artifacts.\n\n### Orchestration\n\nIn a production deployment of TFX, you will use an orchestrator such as Apache Airflow, Kubeflow Pipelines, or Apache Beam to orchestrate a pre-defined pipeline graph of TFX components. In an interactive notebook, the notebook itself is the orchestrator, running each TFX component as you execute the notebook cells.\n\n### Metadata\n\nIn a production deployment of TFX, you will access metadata through the ML Metadata (MLMD) API. MLMD stores metadata properties in a database such as MySQL or SQLite, and stores the metadata payloads in a persistent store such as on your filesystem. In an interactive notebook, both properties and payloads are stored in an ephemeral SQLite database in the `/tmp` directory on the Jupyter notebook or Colab server.",
"_____no_output_____"
],
[
"## Setup\nFirst, we install and import the necessary packages, set up paths, and download data.",
"_____no_output_____"
],
[
"### Install TFX\n\n**Note: Because of package updates, the first time you run this cell you must then restart the runtime (Runtime > Restart runtime ...).**",
"_____no_output_____"
]
],
[
[
"!pip install -q \"tfx>=0.21.1,<0.22\" \"tensorflow>=2.1,<2.2\" \"tensorboard>=2.1,<2.2\"",
"_____no_output_____"
]
],
[
[
"## Did you restart the runtime?\n\nThe first time that you run the cell above, you must restart the runtime (Runtime > Restart runtime ...). This is because of the way that Colab loads packages.",
"_____no_output_____"
],
[
"### Import packages\nWe import necessary packages, including standard TFX component classes.",
"_____no_output_____"
]
],
[
[
"import os\nimport pprint\nimport tempfile\nimport urllib\n\nimport absl\nimport tensorflow as tf\nimport tensorflow_model_analysis as tfma\ntf.get_logger().propagate = False\npp = pprint.PrettyPrinter()\n\nimport tfx\nfrom tfx.components import CsvExampleGen\nfrom tfx.components import Evaluator\nfrom tfx.components import ExampleValidator\nfrom tfx.components import Pusher\nfrom tfx.components import ResolverNode\nfrom tfx.components import SchemaGen\nfrom tfx.components import StatisticsGen\nfrom tfx.components import Trainer\nfrom tfx.components import Transform\nfrom tfx.components.base import executor_spec\nfrom tfx.components.trainer.executor import GenericExecutor\nfrom tfx.dsl.experimental import latest_blessed_model_resolver\nfrom tfx.orchestration import metadata\nfrom tfx.orchestration import pipeline\nfrom tfx.orchestration.experimental.interactive.interactive_context import InteractiveContext\nfrom tfx.proto import pusher_pb2\nfrom tfx.proto import trainer_pb2\nfrom tfx.types import Channel\nfrom tfx.types.standard_artifacts import Model\nfrom tfx.types.standard_artifacts import ModelBlessing\nfrom tfx.utils.dsl_utils import external_input\n\n\n%load_ext tfx.orchestration.experimental.interactive.notebook_extensions.skip",
"_____no_output_____"
]
],
[
[
"Let's check the library versions.",
"_____no_output_____"
]
],
[
[
"print('TensorFlow version: {}'.format(tf.__version__))\nprint('TFX version: {}'.format(tfx.__version__))",
"_____no_output_____"
]
],
[
[
"### Set up pipeline paths",
"_____no_output_____"
]
],
[
[
"# This is the root directory for your TFX pip package installation.\n_tfx_root = tfx.__path__[0]\n\n# This is the directory containing the TFX Chicago Taxi Pipeline example.\n_taxi_root = os.path.join(_tfx_root, 'examples/chicago_taxi_pipeline')\n\n# This is the path where your model will be pushed for serving.\n_serving_model_dir = os.path.join(\n tempfile.mkdtemp(), 'serving_model/taxi_simple')\n\n# Set up logging.\nabsl.logging.set_verbosity(absl.logging.INFO)",
"_____no_output_____"
]
],
[
[
"### Download example data\nWe download the example dataset for use in our TFX pipeline.\n\nThe dataset we're using is the [Taxi Trips dataset](https://data.cityofchicago.org/Transportation/Taxi-Trips/wrvz-psew) released by the City of Chicago. The columns in this dataset are:\n\n<table>\n<tr><td>pickup_community_area</td><td>fare</td><td>trip_start_month</td></tr>\n<tr><td>trip_start_hour</td><td>trip_start_day</td><td>trip_start_timestamp</td></tr>\n<tr><td>pickup_latitude</td><td>pickup_longitude</td><td>dropoff_latitude</td></tr>\n<tr><td>dropoff_longitude</td><td>trip_miles</td><td>pickup_census_tract</td></tr>\n<tr><td>dropoff_census_tract</td><td>payment_type</td><td>company</td></tr>\n<tr><td>trip_seconds</td><td>dropoff_community_area</td><td>tips</td></tr>\n</table>\n\nWith this dataset, we will build a model that predicts the `tips` of a trip.",
"_____no_output_____"
]
],
[
[
"_data_root = tempfile.mkdtemp(prefix='tfx-data')\nDATA_PATH = 'https://raw.githubusercontent.com/tensorflow/tfx/master/tfx/examples/chicago_taxi_pipeline/data/simple/data.csv'\n_data_filepath = os.path.join(_data_root, \"data.csv\")\nurllib.request.urlretrieve(DATA_PATH, _data_filepath)",
"_____no_output_____"
]
],
[
[
"Take a quick look at the CSV file.",
"_____no_output_____"
]
],
[
[
"!head {_data_filepath}",
"_____no_output_____"
]
],
[
[
"*Disclaimer: This site provides applications using data that has been modified for use from its original source, www.cityofchicago.org, the official website of the City of Chicago. The City of Chicago makes no claims as to the content, accuracy, timeliness, or completeness of any of the data provided at this site. The data provided at this site is subject to change at any time. It is understood that the data provided at this site is being used at one’s own risk.*",
"_____no_output_____"
],
[
"### Create the InteractiveContext\nLast, we create an InteractiveContext, which will allow us to run TFX components interactively in this notebook.",
"_____no_output_____"
]
],
[
[
"# Here, we create an InteractiveContext using default parameters. This will\n# use a temporary directory with an ephemeral ML Metadata database instance.\n# To use your own pipeline root or database, the optional properties\n# `pipeline_root` and `metadata_connection_config` may be passed to\n# InteractiveContext. Calls to InteractiveContext are no-ops outside of the\n# notebook.\ncontext = InteractiveContext()",
"_____no_output_____"
]
],
[
[
"## Run TFX components interactively\nIn the cells that follow, we create TFX components one-by-one, run each of them, and visualize their output artifacts.",
"_____no_output_____"
],
[
"### ExampleGen\n\nThe `ExampleGen` component is usually at the start of a TFX pipeline. It will:\n\n1. Split data into training and evaluation sets (by default, 2/3 training + 1/3 eval)\n2. Convert data into the `tf.Example` format\n3. Copy data into the `_tfx_root` directory for other components to access\n\n`ExampleGen` takes as input the path to your data source. In our case, this is the `_data_root` path that contains the downloaded CSV.\n\nNote: In this notebook, we can instantiate components one-by-one and run them with `InteractiveContext.run()`. By contrast, in a production setting, we would specify all the components upfront in a `Pipeline` to pass to the orchestrator (see the \"Export to Pipeline\" section).",
"_____no_output_____"
]
],
[
[
"example_gen = CsvExampleGen(input=external_input(_data_root))\ncontext.run(example_gen)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `ExampleGen`. This component produces two artifacts, training examples and evaluation examples:\n\nNote: The `%%skip_for_export` cell magic will omit the contents of this cell in the exported pipeline file (see the \"Export to pipeline\" section). This is useful for notebook-specific code that you don't want to run in an orchestrated pipeline.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nartifact = example_gen.outputs['examples'].get()[0]\nprint(artifact.split_names, artifact.uri)",
"_____no_output_____"
]
],
[
[
"We can also take a look at the first three training examples:",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\n# Get the URI of the output artifact representing the training examples, which is a directory\ntrain_uri = os.path.join(example_gen.outputs['examples'].get()[0].uri, 'train')\n\n# Get the list of files in this directory (all compressed TFRecord files)\ntfrecord_filenames = [os.path.join(train_uri, name)\n for name in os.listdir(train_uri)]\n\n# Create a `TFRecordDataset` to read these files\ndataset = tf.data.TFRecordDataset(tfrecord_filenames, compression_type=\"GZIP\")\n\n# Iterate over the first 3 records and decode them.\nfor tfrecord in dataset.take(3):\n serialized_example = tfrecord.numpy()\n example = tf.train.Example()\n example.ParseFromString(serialized_example)\n pp.pprint(example)",
"_____no_output_____"
]
],
[
[
"Now that `ExampleGen` has finished ingesting the data, the next step is data analysis.",
"_____no_output_____"
],
[
"### StatisticsGen\nThe `StatisticsGen` component computes statistics over your dataset for data analysis, as well as for use in downstream components. It uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`StatisticsGen` takes as input the dataset we just ingested using `ExampleGen`.",
"_____no_output_____"
]
],
[
[
"statistics_gen = StatisticsGen(\n examples=example_gen.outputs['examples'])\ncontext.run(statistics_gen)",
"_____no_output_____"
]
],
[
[
"After `StatisticsGen` finishes running, we can visualize the outputted statistics. Try playing with the different plots!",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(statistics_gen.outputs['statistics'])",
"_____no_output_____"
]
],
[
[
"### SchemaGen\n\nThe `SchemaGen` component generates a schema based on your data statistics. (A schema defines the expected bounds, types, and properties of the features in your dataset.) It also uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`SchemaGen` will take as input the statistics that we generated with `StatisticsGen`, looking at the training split by default.",
"_____no_output_____"
]
],
[
[
"schema_gen = SchemaGen(\n statistics=statistics_gen.outputs['statistics'],\n infer_feature_shape=False)\ncontext.run(schema_gen)",
"_____no_output_____"
]
],
[
[
"After `SchemaGen` finishes running, we can visualize the generated schema as a table.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(schema_gen.outputs['schema'])",
"_____no_output_____"
]
],
[
[
"Each feature in your dataset shows up as a row in the schema table, alongside its properties. The schema also captures all the values that a categorical feature takes on, denoted as its domain.\n\nTo learn more about schemas, see [the SchemaGen documentation](https://www.tensorflow.org/tfx/guide/schemagen).",
"_____no_output_____"
],
[
"### ExampleValidator\nThe `ExampleValidator` component detects anomalies in your data, based on the expectations defined by the schema. It also uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`ExampleValidator` will take as input the statistics from `StatisticsGen`, and the schema from `SchemaGen`.\n\nBy default, it compares the statistics from the evaluation split to the schema from the training split.",
"_____no_output_____"
]
],
[
[
"example_validator = ExampleValidator(\n statistics=statistics_gen.outputs['statistics'],\n schema=schema_gen.outputs['schema'])\ncontext.run(example_validator)",
"_____no_output_____"
]
],
[
[
"After `ExampleValidator` finishes running, we can visualize the anomalies as a table.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(example_validator.outputs['anomalies'])",
"_____no_output_____"
]
],
[
[
"In the anomalies table, we can see that the `company` feature takes on new values that were not in the training split. This information can be used to debug model performance, understand how your data evolves over time, and identify data errors.\n\nIn our case, this `company` anomaly is innocuous, but the `payment_type` could be fixed. For now we move on to the next step of transforming the data.",
"_____no_output_____"
],
[
"### Transform\nThe `Transform` component performs feature engineering for both training and serving. It uses the [TensorFlow Transform](https://www.tensorflow.org/tfx/transform/get_started) library.\n\n`Transform` will take as input the data from `ExampleGen`, the schema from `SchemaGen`, as well as a module that contains user-defined Transform code.\n\nLet's see an example of user-defined Transform code below (for an introduction to the TensorFlow Transform APIs, [see the tutorial](https://www.tensorflow.org/tfx/tutorials/transform/simple)). First, we define a few constants for feature engineering:\n\nNote: The `%%writefile` cell magic will save the contents of the cell as a `.py` file on disk. This allows the `Transform` component to load your code as a module.\n\n",
"_____no_output_____"
]
],
[
[
"_taxi_constants_module_file = 'taxi_constants.py'",
"_____no_output_____"
],
[
"%%skip_for_export\n%%writefile {_taxi_constants_module_file}\n\n# Categorical features are assumed to each have a maximum value in the dataset.\nMAX_CATEGORICAL_FEATURE_VALUES = [24, 31, 12]\n\nCATEGORICAL_FEATURE_KEYS = [\n 'trip_start_hour', 'trip_start_day', 'trip_start_month',\n 'pickup_census_tract', 'dropoff_census_tract', 'pickup_community_area',\n 'dropoff_community_area'\n]\n\nDENSE_FLOAT_FEATURE_KEYS = ['trip_miles', 'fare', 'trip_seconds']\n\n# Number of buckets used by tf.transform for encoding each feature.\nFEATURE_BUCKET_COUNT = 10\n\nBUCKET_FEATURE_KEYS = [\n 'pickup_latitude', 'pickup_longitude', 'dropoff_latitude',\n 'dropoff_longitude'\n]\n\n# Number of vocabulary terms used for encoding VOCAB_FEATURES by tf.transform\nVOCAB_SIZE = 1000\n\n# Count of out-of-vocab buckets in which unrecognized VOCAB_FEATURES are hashed.\nOOV_SIZE = 10\n\nVOCAB_FEATURE_KEYS = [\n 'payment_type',\n 'company',\n]\n\n# Keys\nLABEL_KEY = 'tips'\nFARE_KEY = 'fare'\n\ndef transformed_name(key):\n return key + '_xf'",
"_____no_output_____"
]
],
[
[
"Next, we write a `preprocessing_fn` that takes in raw data as input, and returns transformed features that our model can train on:",
"_____no_output_____"
]
],
[
[
"_taxi_transform_module_file = 'taxi_transform.py'",
"_____no_output_____"
],
[
"%%skip_for_export\n%%writefile {_taxi_transform_module_file}\n\nimport tensorflow as tf\nimport tensorflow_transform as tft\n\nimport taxi_constants\n\n_DENSE_FLOAT_FEATURE_KEYS = taxi_constants.DENSE_FLOAT_FEATURE_KEYS\n_VOCAB_FEATURE_KEYS = taxi_constants.VOCAB_FEATURE_KEYS\n_VOCAB_SIZE = taxi_constants.VOCAB_SIZE\n_OOV_SIZE = taxi_constants.OOV_SIZE\n_FEATURE_BUCKET_COUNT = taxi_constants.FEATURE_BUCKET_COUNT\n_BUCKET_FEATURE_KEYS = taxi_constants.BUCKET_FEATURE_KEYS\n_CATEGORICAL_FEATURE_KEYS = taxi_constants.CATEGORICAL_FEATURE_KEYS\n_FARE_KEY = taxi_constants.FARE_KEY\n_LABEL_KEY = taxi_constants.LABEL_KEY\n_transformed_name = taxi_constants.transformed_name\n\n\ndef preprocessing_fn(inputs):\n \"\"\"tf.transform's callback function for preprocessing inputs.\n Args:\n inputs: map from feature keys to raw not-yet-transformed features.\n Returns:\n Map from string feature key to transformed feature operations.\n \"\"\"\n outputs = {}\n for key in _DENSE_FLOAT_FEATURE_KEYS:\n # Preserve this feature as a dense float, setting nan's to the mean.\n outputs[_transformed_name(key)] = tft.scale_to_z_score(\n _fill_in_missing(inputs[key]))\n\n for key in _VOCAB_FEATURE_KEYS:\n # Build a vocabulary for this feature.\n outputs[_transformed_name(key)] = tft.compute_and_apply_vocabulary(\n _fill_in_missing(inputs[key]),\n top_k=_VOCAB_SIZE,\n num_oov_buckets=_OOV_SIZE)\n\n for key in _BUCKET_FEATURE_KEYS:\n outputs[_transformed_name(key)] = tft.bucketize(\n _fill_in_missing(inputs[key]), _FEATURE_BUCKET_COUNT,\n always_return_num_quantiles=False)\n\n for key in _CATEGORICAL_FEATURE_KEYS:\n outputs[_transformed_name(key)] = _fill_in_missing(inputs[key])\n\n # Was this passenger a big tipper?\n taxi_fare = _fill_in_missing(inputs[_FARE_KEY])\n tips = _fill_in_missing(inputs[_LABEL_KEY])\n outputs[_transformed_name(_LABEL_KEY)] = tf.where(\n tf.math.is_nan(taxi_fare),\n tf.cast(tf.zeros_like(taxi_fare), tf.int64),\n # Test if the tip was > 20% of the fare.\n tf.cast(\n tf.greater(tips, tf.multiply(taxi_fare, tf.constant(0.2))), tf.int64))\n\n return outputs\n\n\ndef _fill_in_missing(x):\n \"\"\"Replace missing values in a SparseTensor.\n Fills in missing values of `x` with '' or 0, and converts to a dense tensor.\n Args:\n x: A `SparseTensor` of rank 2. Its dense shape should have size at most 1\n in the second dimension.\n Returns:\n A rank 1 tensor where missing values of `x` have been filled in.\n \"\"\"\n default_value = '' if x.dtype == tf.string else 0\n return tf.squeeze(\n tf.sparse.to_dense(\n tf.SparseTensor(x.indices, x.values, [x.dense_shape[0], 1]),\n default_value),\n axis=1)",
"_____no_output_____"
]
],
[
[
"Now, we pass in this feature engineering code to the `Transform` component and run it to transform your data.",
"_____no_output_____"
]
],
[
[
"transform = Transform(\n examples=example_gen.outputs['examples'],\n schema=schema_gen.outputs['schema'],\n module_file=os.path.abspath(_taxi_transform_module_file))\ncontext.run(transform)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `Transform`. This component produces two types of outputs:\n\n* `transform_graph` is the graph that can perform the preprocessing operations (this graph will be included in the serving and evaluation models).\n* `transformed_examples` represents the preprocessed training and evaluation data.",
"_____no_output_____"
]
],
[
[
"transform.outputs",
"_____no_output_____"
]
],
[
[
"Take a peek at the `transform_graph` artifact. It points to a directory containing three subdirectories.",
"_____no_output_____"
]
],
[
[
"train_uri = transform.outputs['transform_graph'].get()[0].uri\nos.listdir(train_uri)",
"_____no_output_____"
]
],
[
[
"The `transformed_metadata` subdirectory contains the schema of the preprocessed data. The `transform_fn` subdirectory contains the actual preprocessing graph. The `metadata` subdirectory contains the schema of the original data.\n\nWe can also take a look at the first three transformed examples:",
"_____no_output_____"
]
],
[
[
"# Get the URI of the output artifact representing the transformed examples, which is a directory\ntrain_uri = os.path.join(transform.outputs['transformed_examples'].get()[0].uri, 'train')\n\n# Get the list of files in this directory (all compressed TFRecord files)\ntfrecord_filenames = [os.path.join(train_uri, name)\n for name in os.listdir(train_uri)]\n\n# Create a `TFRecordDataset` to read these files\ndataset = tf.data.TFRecordDataset(tfrecord_filenames, compression_type=\"GZIP\")\n\n# Iterate over the first 3 records and decode them.\nfor tfrecord in dataset.take(3):\n serialized_example = tfrecord.numpy()\n example = tf.train.Example()\n example.ParseFromString(serialized_example)\n pp.pprint(example)",
"_____no_output_____"
]
],
[
[
"After the `Transform` component has transformed your data into features, and the next step is to train a model.",
"_____no_output_____"
],
[
"### Trainer\nThe `Trainer` component will train a model that you define in TensorFlow. Default Trainer support Estimator API, to use Keras API, you need to specify [Generic Trainer](https://github.com/tensorflow/community/blob/master/rfcs/20200117-tfx-generic-trainer.md) by setup `custom_executor_spec=executor_spec.ExecutorClassSpec(GenericExecutor)` in Trainer's contructor.\n\n`Trainer` takes as input the schema from `SchemaGen`, the transformed data and graph from `Transform`, training parameters, as well as a module that contains user-defined model code.\n\nLet's see an example of user-defined model code below (for an introduction to the TensorFlow Keras APIs, [see the tutorial](https://www.tensorflow.org/guide/keras)):",
"_____no_output_____"
]
],
[
[
"_taxi_trainer_module_file = 'taxi_trainer.py'",
"_____no_output_____"
],
[
"%%skip_for_export\n%%writefile {_taxi_trainer_module_file}\n\nfrom typing import List, Text\n\nimport os\nimport absl\nimport datetime\nimport tensorflow as tf\nimport tensorflow_transform as tft\n\nfrom tfx.components.trainer.executor import TrainerFnArgs\n\nimport taxi_constants\n\n_DENSE_FLOAT_FEATURE_KEYS = taxi_constants.DENSE_FLOAT_FEATURE_KEYS\n_VOCAB_FEATURE_KEYS = taxi_constants.VOCAB_FEATURE_KEYS\n_VOCAB_SIZE = taxi_constants.VOCAB_SIZE\n_OOV_SIZE = taxi_constants.OOV_SIZE\n_FEATURE_BUCKET_COUNT = taxi_constants.FEATURE_BUCKET_COUNT\n_BUCKET_FEATURE_KEYS = taxi_constants.BUCKET_FEATURE_KEYS\n_CATEGORICAL_FEATURE_KEYS = taxi_constants.CATEGORICAL_FEATURE_KEYS\n_MAX_CATEGORICAL_FEATURE_VALUES = taxi_constants.MAX_CATEGORICAL_FEATURE_VALUES\n_LABEL_KEY = taxi_constants.LABEL_KEY\n_transformed_name = taxi_constants.transformed_name\n\n\ndef _transformed_names(keys):\n return [_transformed_name(key) for key in keys]\n\n\ndef _gzip_reader_fn(filenames):\n \"\"\"Small utility returning a record reader that can read gzip'ed files.\"\"\"\n return tf.data.TFRecordDataset(\n filenames,\n compression_type='GZIP')\n\n\ndef _get_serve_tf_examples_fn(model, tf_transform_output):\n \"\"\"Returns a function that parses a serialized tf.Example and applies TFT.\"\"\"\n\n model.tft_layer = tf_transform_output.transform_features_layer()\n\n @tf.function\n def serve_tf_examples_fn(serialized_tf_examples):\n \"\"\"Returns the output to be used in the serving signature.\"\"\"\n feature_spec = tf_transform_output.raw_feature_spec()\n feature_spec.pop(_LABEL_KEY)\n parsed_features = tf.io.parse_example(serialized_tf_examples, feature_spec)\n\n transformed_features = model.tft_layer(parsed_features)\n transformed_features.pop(_transformed_name(_LABEL_KEY))\n\n return model(transformed_features)\n\n return serve_tf_examples_fn\n\n\ndef _input_fn(file_pattern: Text,\n tf_transform_output: tft.TFTransformOutput,\n batch_size: int = 200) -> tf.data.Dataset:\n \"\"\"Generates features and label for tuning/training.\n\n Args:\n file_pattern: input tfrecord file pattern.\n tf_transform_output: A TFTransformOutput.\n batch_size: representing the number of consecutive elements of returned\n dataset to combine in a single batch\n\n Returns:\n A dataset that contains (features, indices) tuple where features is a\n dictionary of Tensors, and indices is a single Tensor of label indices.\n \"\"\"\n transformed_feature_spec = (\n tf_transform_output.transformed_feature_spec().copy())\n\n dataset = tf.data.experimental.make_batched_features_dataset(\n file_pattern=file_pattern,\n batch_size=batch_size,\n features=transformed_feature_spec,\n reader=_gzip_reader_fn,\n label_key=_transformed_name(_LABEL_KEY))\n\n return dataset\n\n\ndef _build_keras_model(hidden_units: List[int] = None) -> tf.keras.Model:\n \"\"\"Creates a DNN Keras model for classifying taxi data.\n\n Args:\n hidden_units: [int], the layer sizes of the DNN (input layer first).\n\n Returns:\n A keras Model.\n \"\"\"\n real_valued_columns = [\n tf.feature_column.numeric_column(key, shape=())\n for key in _transformed_names(_DENSE_FLOAT_FEATURE_KEYS)\n ]\n categorical_columns = [\n tf.feature_column.categorical_column_with_identity(\n key, num_buckets=_VOCAB_SIZE + _OOV_SIZE, default_value=0)\n for key in _transformed_names(_VOCAB_FEATURE_KEYS)\n ]\n categorical_columns += [\n tf.feature_column.categorical_column_with_identity(\n key, num_buckets=_FEATURE_BUCKET_COUNT, default_value=0)\n for key in _transformed_names(_BUCKET_FEATURE_KEYS)\n ]\n categorical_columns += [\n tf.feature_column.categorical_column_with_identity( # pylint: disable=g-complex-comprehension\n key,\n num_buckets=num_buckets,\n default_value=0) for key, num_buckets in zip(\n _transformed_names(_CATEGORICAL_FEATURE_KEYS),\n _MAX_CATEGORICAL_FEATURE_VALUES)\n ]\n indicator_column = [\n tf.feature_column.indicator_column(categorical_column)\n for categorical_column in categorical_columns\n ]\n\n model = _wide_and_deep_classifier(\n # TODO(b/139668410) replace with premade wide_and_deep keras model\n wide_columns=indicator_column,\n deep_columns=real_valued_columns,\n dnn_hidden_units=hidden_units or [100, 70, 50, 25])\n return model\n\n\ndef _wide_and_deep_classifier(wide_columns, deep_columns, dnn_hidden_units):\n \"\"\"Build a simple keras wide and deep model.\n\n Args:\n wide_columns: Feature columns wrapped in indicator_column for wide (linear)\n part of the model.\n deep_columns: Feature columns for deep part of the model.\n dnn_hidden_units: [int], the layer sizes of the hidden DNN.\n\n Returns:\n A Wide and Deep Keras model\n \"\"\"\n # Following values are hard coded for simplicity in this example,\n # However prefarably they should be passsed in as hparams.\n\n # Keras needs the feature definitions at compile time.\n # TODO(b/139081439): Automate generation of input layers from FeatureColumn.\n input_layers = {\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype=tf.float32)\n for colname in _transformed_names(_DENSE_FLOAT_FEATURE_KEYS)\n }\n input_layers.update({\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype='int32')\n for colname in _transformed_names(_VOCAB_FEATURE_KEYS)\n })\n input_layers.update({\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype='int32')\n for colname in _transformed_names(_BUCKET_FEATURE_KEYS)\n })\n input_layers.update({\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype='int32')\n for colname in _transformed_names(_CATEGORICAL_FEATURE_KEYS)\n })\n\n # TODO(b/144500510): SparseFeatures for feature columns + Keras.\n deep = tf.keras.layers.DenseFeatures(deep_columns)(input_layers)\n for numnodes in dnn_hidden_units:\n deep = tf.keras.layers.Dense(numnodes)(deep)\n wide = tf.keras.layers.DenseFeatures(wide_columns)(input_layers)\n\n output = tf.keras.layers.Dense(\n 1, activation='sigmoid')(\n tf.keras.layers.concatenate([deep, wide]))\n\n model = tf.keras.Model(input_layers, output)\n model.compile(\n loss='binary_crossentropy',\n optimizer=tf.keras.optimizers.Adam(lr=0.001),\n metrics=[tf.keras.metrics.BinaryAccuracy()])\n model.summary(print_fn=absl.logging.info)\n return model\n\n\n# TFX Trainer will call this function.\ndef run_fn(fn_args: TrainerFnArgs):\n \"\"\"Train the model based on given args.\n\n Args:\n fn_args: Holds args used to train the model as name/value pairs.\n \"\"\"\n # Number of nodes in the first layer of the DNN\n first_dnn_layer_size = 100\n num_dnn_layers = 4\n dnn_decay_factor = 0.7\n\n tf_transform_output = tft.TFTransformOutput(fn_args.transform_output)\n\n train_dataset = _input_fn(fn_args.train_files, tf_transform_output, 40)\n eval_dataset = _input_fn(fn_args.eval_files, tf_transform_output, 40)\n\n model = _build_keras_model(\n # Construct layers sizes with exponetial decay\n hidden_units=[\n max(2, int(first_dnn_layer_size * dnn_decay_factor**i))\n for i in range(num_dnn_layers)\n ])\n\n # This log path might change in the future.\n log_dir = os.path.join(os.path.dirname(fn_args.serving_model_dir), 'logs')\n tensorboard_callback = tf.keras.callbacks.TensorBoard(\n log_dir=log_dir, update_freq='batch')\n model.fit(\n train_dataset,\n steps_per_epoch=fn_args.train_steps,\n validation_data=eval_dataset,\n validation_steps=fn_args.eval_steps,\n callbacks=[tensorboard_callback])\n\n signatures = {\n 'serving_default':\n _get_serve_tf_examples_fn(model,\n tf_transform_output).get_concrete_function(\n tf.TensorSpec(\n shape=[None],\n dtype=tf.string,\n name='examples')),\n }\n model.save(fn_args.serving_model_dir, save_format='tf', signatures=signatures)",
"_____no_output_____"
]
],
[
[
"Now, we pass in this model code to the `Trainer` component and run it to train the model.",
"_____no_output_____"
]
],
[
[
"trainer = Trainer(\n module_file=os.path.abspath(_taxi_trainer_module_file),\n custom_executor_spec=executor_spec.ExecutorClassSpec(GenericExecutor),\n examples=transform.outputs['transformed_examples'],\n transform_graph=transform.outputs['transform_graph'],\n schema=schema_gen.outputs['schema'],\n train_args=trainer_pb2.TrainArgs(num_steps=10000),\n eval_args=trainer_pb2.EvalArgs(num_steps=5000))\ncontext.run(trainer)",
"_____no_output_____"
]
],
[
[
"#### Analyze Training with TensorBoard\nTake a peek at the trainer artifact. It points to a directory containing the model subdirectories.",
"_____no_output_____"
]
],
[
[
"model_artifact_dir = trainer.outputs['model'].get()[0].uri\npp.pprint(os.listdir(model_artifact_dir))\nmodel_dir = os.path.join(model_artifact_dir, 'serving_model_dir')\npp.pprint(os.listdir(model_dir))",
"_____no_output_____"
]
],
[
[
"Optionally, we can connect TensorBoard to the Trainer to analyze our model's training curves.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nlog_dir = os.path.join(model_artifact_dir, 'logs')\n\n%load_ext tensorboard\n%tensorboard --logdir {log_dir}",
"_____no_output_____"
]
],
[
[
"### Evaluator\nThe `Evaluator` component computes model performance metrics over the evaluation set. It uses the [TensorFlow Model Analysis](https://www.tensorflow.org/tfx/model_analysis/get_started) library. The `Evaluator` can also optionally validate that a newly trained model is better than the previous model. This is useful in a production pipeline setting where you may automatically train and validate a model every day. In this notebook, we only train one model, so the `Evaluator` automatically will label the model as \"good\". \n\n`Evaluator` will take as input the data from `ExampleGen`, the trained model from `Trainer`, and slicing configuration. The slicing configuration allows you to slice your metrics on feature values (e.g. how does your model perform on taxi trips that start at 8am versus 8pm?). See an example of this configuration below:",
"_____no_output_____"
]
],
[
[
"eval_config = tfma.EvalConfig(\n model_specs=[\n # This assumes a serving model with signature 'serving_default'. If\n # using estimator based EvalSavedModel, add signature_name: 'eval' and \n # remove the label_key.\n tfma.ModelSpec(label_key='tips')\n ],\n metrics_specs=[\n tfma.MetricsSpec(\n # The metrics added here are in addition to those saved with the\n # model (assuming either a keras model or EvalSavedModel is used).\n # Any metrics added into the saved model (for example using\n # model.compile(..., metrics=[...]), etc) will be computed\n # automatically.\n metrics=[\n tfma.MetricConfig(class_name='ExampleCount')\n ],\n # To add validation thresholds for metrics saved with the model,\n # add them keyed by metric name to the thresholds map.\n thresholds = {\n 'binary_accuracy': tfma.MetricThreshold(\n value_threshold=tfma.GenericValueThreshold(\n lower_bound={'value': 0.5}),\n change_threshold=tfma.GenericChangeThreshold(\n direction=tfma.MetricDirection.HIGHER_IS_BETTER,\n absolute={'value': -1e-10}))\n }\n )\n ],\n slicing_specs=[\n # An empty slice spec means the overall slice, i.e. the whole dataset.\n tfma.SlicingSpec(),\n # Data can be sliced along a feature column. In this case, data is\n # sliced along feature column trip_start_hour.\n tfma.SlicingSpec(feature_keys=['trip_start_hour'])\n ])",
"_____no_output_____"
]
],
[
[
"Next, we give this configuration to `Evaluator` and run it.",
"_____no_output_____"
]
],
[
[
"# Use TFMA to compute a evaluation statistics over features of a model and\n# validate them against a baseline.\n\n# The model resolver is only required if performing model validation in addition\n# to evaluation. In this case we validate against the latest blessed model. If\n# no model has been blessed before (as in this case) the evaluator will make our\n# candidate the first blessed model.\nmodel_resolver = ResolverNode(\n instance_name='latest_blessed_model_resolver',\n resolver_class=latest_blessed_model_resolver.LatestBlessedModelResolver,\n model=Channel(type=Model),\n model_blessing=Channel(type=ModelBlessing))\ncontext.run(model_resolver)\n\nevaluator = Evaluator(\n examples=example_gen.outputs['examples'],\n model=trainer.outputs['model'],\n baseline_model=model_resolver.outputs['model'],\n # Change threshold will be ignored if there is no baseline (first run).\n eval_config=eval_config)\ncontext.run(evaluator)",
"_____no_output_____"
]
],
[
[
"Now let's examine the output artifacts of `Evaluator`. ",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nevaluator.outputs",
"_____no_output_____"
]
],
[
[
"Using the `evaluation` output we can show the default visualization of global metrics on the entire evaluation set.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(evaluator.outputs['evaluation'])",
"_____no_output_____"
]
],
[
[
"To see the visualization for sliced evaluation metrics, we can directly call the TensorFlow Model Analysis library.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nimport tensorflow_model_analysis as tfma\n\n# Get the TFMA output result path and load the result.\nPATH_TO_RESULT = evaluator.outputs['evaluation'].get()[0].uri\ntfma_result = tfma.load_eval_result(PATH_TO_RESULT)\n\n# Show data sliced along feature column trip_start_hour.\ntfma.view.render_slicing_metrics(\n tfma_result, slicing_column='trip_start_hour')",
"_____no_output_____"
]
],
[
[
"This visualization shows the same metrics, but computed at every feature value of `trip_start_hour` instead of on the entire evaluation set.\n\nTensorFlow Model Analysis supports many other visualizations, such as Fairness Indicators and plotting a time series of model performance. To learn more, see [the tutorial](https://www.tensorflow.org/tfx/tutorials/model_analysis/tfma_basic).",
"_____no_output_____"
],
[
"Since we added thresholds to our config, validation output is also available. The precence of a `blessing` artifact indicates that our model passed validation. Since this is the first validation being performed the candidate is automatically blessed.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nblessing_uri = evaluator.outputs.blessing.get()[0].uri\n!ls -l {blessing_uri}",
"_____no_output_____"
]
],
[
[
"Now can also verify the success by loading the validation result record:",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nPATH_TO_RESULT = evaluator.outputs['evaluation'].get()[0].uri\nprint(tfma.load_validation_result(PATH_TO_RESULT))",
"_____no_output_____"
]
],
[
[
"### Pusher\nThe `Pusher` component is usually at the end of a TFX pipeline. It checks whether a model has passed validation, and if so, exports the model to `_serving_model_dir`.",
"_____no_output_____"
]
],
[
[
"pusher = Pusher(\n model=trainer.outputs['model'],\n model_blessing=evaluator.outputs['blessing'],\n push_destination=pusher_pb2.PushDestination(\n filesystem=pusher_pb2.PushDestination.Filesystem(\n base_directory=_serving_model_dir)))\ncontext.run(pusher)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `Pusher`. ",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\npusher.outputs",
"_____no_output_____"
]
],
[
[
"In particular, the Pusher will export your model in the SavedModel format, which looks like this:",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\npush_uri = pusher.outputs.model_push.get()[0].uri\nlatest_version = max(os.listdir(push_uri))\nlatest_version_path = os.path.join(push_uri, latest_version)\nmodel = tf.saved_model.load(latest_version_path)\n\nfor item in model.signatures.items():\n pp.pprint(item)",
"_____no_output_____"
]
],
[
[
"We're finished our tour of built-in TFX components!\n\nAfter you're happy with experimenting with TFX components and code in this notebook, you may want to export it as a pipeline to be orchestrated with Apache Airflow or Apache Beam. See the final section.",
"_____no_output_____"
],
[
"## Export to pipeline\n\nTo export the contents of this notebook as a pipeline to be orchestrated with Airflow or Beam, follow the instructions below.\n\nIf you're using Colab, make sure to **save this notebook to Google Drive** (`File` → `Save a Copy in Drive`) before exporting.",
"_____no_output_____"
],
[
"### 1. Mount Google Drive (Colab-only)\n\nIf you're using Colab, this notebook needs to mount your Google Drive to be able to access its own `.ipynb` file.",
"_____no_output_____"
]
],
[
[
"#docs_infra: no_execute\n%%skip_for_export\n\n#@markdown Run this cell and enter the authorization code to mount Google Drive.\n\nimport sys\n\nif 'google.colab' in sys.modules:\n # Colab.\n from google.colab import drive\n\n drive.mount('/content/drive')",
"_____no_output_____"
]
],
[
[
"### 2. Select an orchestrator",
"_____no_output_____"
]
],
[
[
"_runner_type = 'beam' #@param [\"beam\", \"airflow\"]\n_pipeline_name = 'chicago_taxi_%s' % _runner_type",
"_____no_output_____"
]
],
[
[
"### 3. Set up paths for the pipeline",
"_____no_output_____"
]
],
[
[
"#docs_infra: no_execute\n# For Colab notebooks only.\n# TODO(USER): Fill out the path to this notebook.\n_notebook_filepath = (\n '/content/drive/My Drive/Colab Notebooks/taxi_pipeline_interactive.ipynb')\n\n# For Jupyter notebooks only.\n# _notebook_filepath = os.path.join(os.getcwd(),\n# 'taxi_pipeline_interactive.ipynb')\n\n# TODO(USER): Fill out the paths for the exported pipeline.\n_tfx_root = os.path.join(os.environ['HOME'], 'tfx')\n_taxi_root = os.path.join(os.environ['HOME'], 'taxi')\n_serving_model_dir = os.path.join(_taxi_root, 'serving_model')\n_data_root = os.path.join(_taxi_root, 'data', 'simple')\n_pipeline_root = os.path.join(_tfx_root, 'pipelines', _pipeline_name)\n_metadata_path = os.path.join(_tfx_root, 'metadata', _pipeline_name,\n 'metadata.db')",
"_____no_output_____"
]
],
[
[
"### 4. Choose components to include in the pipeline",
"_____no_output_____"
]
],
[
[
"#docs_infra: no_execute\n# TODO(USER): Specify components to be included in the exported pipeline.\ncomponents = [\n example_gen, statistics_gen, schema_gen, example_validator, transform,\n trainer, evaluator, pusher\n]",
"_____no_output_____"
]
],
[
[
"### 5. Generate pipeline files",
"_____no_output_____"
]
],
[
[
"#docs_infra: no_execute\n%%skip_for_export\n\n#@markdown Run this cell to generate the pipeline files.\n\nif get_ipython().magics_manager.auto_magic:\n print('Warning: %automagic is ON. Line magics specified without the % prefix '\n 'will not be scrubbed during export to pipeline.')\n\n_pipeline_export_filepath = 'export_%s.py' % _pipeline_name\ncontext.export_to_pipeline(notebook_filepath=_notebook_filepath,\n export_filepath=_pipeline_export_filepath,\n runner_type=_runner_type)",
"_____no_output_____"
]
],
[
[
"### 6. Download pipeline files",
"_____no_output_____"
]
],
[
[
"#docs_infra: no_execute\n%%skip_for_export\n\n#@markdown Run this cell to download the pipeline files as a `.zip`.\n\nif 'google.colab' in sys.modules:\n from google.colab import files\n import zipfile\n\n zip_export_path = os.path.join(\n tempfile.mkdtemp(), 'export.zip')\n with zipfile.ZipFile(zip_export_path, mode='w') as export_zip:\n export_zip.write(_pipeline_export_filepath)\n export_zip.write(_taxi_constants_module_file)\n export_zip.write(_taxi_transform_module_file)\n export_zip.write(_taxi_trainer_module_file)\n\n files.download(zip_export_path)",
"_____no_output_____"
]
],
[
[
"To learn how to run the orchestrated pipeline with Apache Airflow, please refer to the [TFX Orchestration Tutorial](https://www.tensorflow.org/tfx/tutorials/tfx/airflow_workshop).",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec616e119685deec4befb25474689f24f07d7f33 | 10,415 | ipynb | Jupyter Notebook | AnEconomistPlease.ipynb | eniktab/MoE_nlp | 3adf81410d0c91c568ae9c3b6df8e7dfe57b15e9 | [
"MIT"
]
| null | null | null | AnEconomistPlease.ipynb | eniktab/MoE_nlp | 3adf81410d0c91c568ae9c3b6df8e7dfe57b15e9 | [
"MIT"
]
| null | null | null | AnEconomistPlease.ipynb | eniktab/MoE_nlp | 3adf81410d0c91c568ae9c3b6df8e7dfe57b15e9 | [
"MIT"
]
| null | null | null | 44.131356 | 489 | 0.545943 | [
[
[
"<a href=\"https://colab.research.google.com/github/eniktab/MoE_nlp/blob/master/AnEconomistPlease.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport numpy as np\nimport os\nimport time\nimport random\n\n\ntrained_model_path = [\"https://storage.googleapis.com/bucket-1-free/%20philip/ArthurGrimes.txt\", \n \"https://storage.googleapis.com/bucket-1-free/%20philip/ShamubeelEaqubs.txt\"]\n!rm -r /content/training_checkpoints\nfor i in trained_model_path:\n tf.keras.utils.get_file(\n i.split(\"/\")[-1], i, cache_subdir=os.path.abspath(\"/content\"))\n\necolist = ['/content/ArthurGrimes.txt', '/content/ShamubeelEaqubs.txt']\ntemtxt= random.choice(ecolist) \n# Read, then decode for py2 compat.\ntext = open(temtxt, 'rb').read().decode(encoding='utf-8')\nprint('Length of text: {} characters'.format(len(text)))\n# The unique characters in the file\nvocab = sorted(set(text))\nchar2idx = {u:i for i, u in enumerate(vocab)}\nidx2char = np.array(vocab)\ntext_as_int = np.array([char2idx[c] for c in text])\n# The maximum length sentence you want for a single input in characters\nseq_length = 100\nexamples_per_epoch = len(text)//(seq_length+1)\n# Create training examples / targets\nchar_dataset = tf.data.Dataset.from_tensor_slices(text_as_int)\nsequences = char_dataset.batch(seq_length+1, drop_remainder=True)\ndef split_input_target(chunk):\n input_text = chunk[:-1]\n target_text = chunk[1:]\n return input_text, target_text\ndataset = sequences.map(split_input_target)\n# Batch size\nBATCH_SIZE = 64\nBUFFER_SIZE = 10000\ndataset = dataset.shuffle(BUFFER_SIZE).batch(BATCH_SIZE, drop_remainder=True)\nvocab_size = len(vocab)\nembedding_dim = 256\nrnn_units = 1024\ndef build_model(vocab_size, embedding_dim, rnn_units, batch_size):\n model = tf.keras.Sequential([\n tf.keras.layers.Embedding(vocab_size, embedding_dim,\n batch_input_shape=[batch_size, None]),\n tf.keras.layers.GRU(rnn_units,\n return_sequences=True,\n stateful=True,\n recurrent_initializer='glorot_uniform'),\n tf.keras.layers.Dense(vocab_size)\n ])\n return model\nmodel = build_model(\n vocab_size=len(vocab),\n embedding_dim=embedding_dim,\n rnn_units=rnn_units,\n batch_size=BATCH_SIZE)\ndef loss(labels, logits):\n return tf.keras.losses.sparse_categorical_crossentropy(labels, logits, from_logits=True)\nmodel.compile(optimizer='adam', loss=loss)\n# Directory where the checkpoints will be saved\ncheckpoint_dir = './training_checkpoints/ArthurGrimes'\n# Name of the checkpoint files\ncheckpoint_prefix = os.path.join(checkpoint_dir, \"ckpt_{epoch}\")\n\ncheckpoint_callback = tf.keras.callbacks.ModelCheckpoint(\n filepath=checkpoint_prefix,\n save_weights_only=True\n )\n\nmodel = build_model(\n vocab_size=len(vocab),\n embedding_dim=embedding_dim,\n rnn_units=rnn_units,\n batch_size=BATCH_SIZE)\n\noptimizer = tf.keras.optimizers.Adam()\n\[email protected]\ndef train_step(inp, target):\n with tf.GradientTape() as tape:\n predictions = model(inp)\n loss = tf.reduce_mean(\n tf.keras.losses.sparse_categorical_crossentropy(\n target, predictions, from_logits=True))\n grads = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n\n return loss\n\ndef generate_text(model, start_string):\n # Evaluation step (generating text using the learned model)\n\n # Number of characters to generate\n num_generate = 1000\n\n # Converting our start string to numbers (vectorizing)\n input_eval = [char2idx[s] for s in start_string]\n input_eval = tf.expand_dims(input_eval, 0)\n\n # Empty string to store our results\n text_generated = []\n\n # Low temperature results in more predictable text.\n # Higher temperature results in more surprising text.\n # Experiment to find the best setting.\n temperature = 0.5\n\n model.reset_states()\n for i in range(num_generate):\n predictions = model(input_eval)\n # remove the batch dimension\n predictions = tf.squeeze(predictions, 0)\n\n # using a categorical distribution to predict the character returned by the model\n predictions = predictions / temperature\n predicted_id = tf.random.categorical(predictions, num_samples=1)[-1,0].numpy()\n\n # Pass the predicted character as the next input to the model\n # along with the previous hidden state\n input_eval = tf.expand_dims([predicted_id], 0)\n\n text_generated.append(idx2char[predicted_id])\n\n return (start_string + ''.join(text_generated))\n \ntf.train.latest_checkpoint(checkpoint_dir)\n# Training step\nEPOCHS = 1500\n\nfor epoch in range(EPOCHS):\n start = time.time()\n\n # resetting the hidden state at the start of every epoch\n model.reset_states()\n\n for (batch_n, (inp, target)) in enumerate(dataset):\n loss = train_step(inp, target)\n\n if batch_n % 100 == 0:\n template = 'Epoch {} Batch {} Loss {}'\n print(template.format(epoch + 1, batch_n, loss))\n\n # saving (checkpoint) the model every 5 epochs\n if (epoch + 1) % 5 == 0:\n model.save_weights(checkpoint_prefix.format(epoch=epoch))\n\n print('Epoch {} Loss {:}'.format(epoch + 1, loss))\n print('Time taken for 1 epoch {} sec\\n'.format(time.time() - start))\n\nmodel.save_weights(checkpoint_prefix.format(epoch=epoch))\n\ntf.train.latest_checkpoint(checkpoint_dir)\nmodel = build_model(vocab_size, embedding_dim, rnn_units, batch_size=1)\nmodel.load_weights(tf.train.latest_checkpoint(checkpoint_dir))\nmodel.build(tf.TensorShape([1, None]))\n\n",
"_____no_output_____"
],
[
"add = temtxt.split(r\"/\")[-1].replace(\".txt\", \" says: \")\r\nprint(add)\r\nprint(generate_text(model, start_string=u\"life \"))\r\n\r\n",
"ArthurGrimes Says: \nlife risk that the prevalence of drug use increases more than what has been seen internationally following deliver social benefits of $19m. This is based on various meta-studies of drug treatment and prevention programmes. There may also be benefits, such as reduced use of other drugs and connecontinues, then the housing market will have a downturn.\nBut its hard to tell if it this tightening for organised crime. It also generates tax revenues of $185m-$240m per year.\nIf these taxes are put have been cut in the first place.\nIncreases in infrastructure spending is welcome, but we need to increase infrastructure spending by around half again to close our ing drug use away – by banning it, rather than accepting many people use drugs regardless – doesn’t w Zealand Drug Foundation, the New Zealand Needle Exchange Programme and Matua Raki. This health-based for sale, and who it is sold to. There is no reliable data on the effect of standardising cannabis would also be allow Zealand Drug Foundat\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
ec6179e2015eb6c61e4a3a90f63a0d4b645443f3 | 545,179 | ipynb | Jupyter Notebook | Pneumonia_Detection.ipynb | ReJackTion/cat304 | b961d22e6353a9fbe0edf4bf7ad7bbde3268fe2c | [
"MIT"
]
| null | null | null | Pneumonia_Detection.ipynb | ReJackTion/cat304 | b961d22e6353a9fbe0edf4bf7ad7bbde3268fe2c | [
"MIT"
]
| null | null | null | Pneumonia_Detection.ipynb | ReJackTion/cat304 | b961d22e6353a9fbe0edf4bf7ad7bbde3268fe2c | [
"MIT"
]
| null | null | null | 87.578956 | 24,070 | 0.69942 | [
[
[
"<a href=\"https://colab.research.google.com/github/ReJackTion/cat304/blob/main/Pneumonia_Detection.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Getting the dataset",
"_____no_output_____"
]
],
[
[
"!pip install -q kaggle",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nfrom google.colab import files\nfrom pathlib import Path\nimport os.path\n\nfrom sklearn.model_selection import train_test_split\n\nimport tensorflow as tf\n\nfrom sklearn.metrics import confusion_matrix, classification_report\n\nimport warnings\nwarnings.filterwarnings('ignore') #ignore warning to imporve readability",
"_____no_output_____"
]
],
[
[
"To upload Kaggle's API to download dataset from Kaggle",
"_____no_output_____"
]
],
[
[
"files.upload()",
"_____no_output_____"
],
[
"!mkdir ~/.kaggle #make a new folder\n!cp /content/kaggle.json ~/.kaggle/ #copy file to that folder\n!chmod 600 ~/.kaggle/kaggle.json #to allow read and write the file",
"_____no_output_____"
]
],
[
[
"dataset source:\nhttps://www.kaggle.com/paultimothymooney/chest-xray-pneumonia",
"_____no_output_____"
]
],
[
[
"!kaggle datasets download -d paultimothymooney/chest-xray-pneumonia",
"Downloading chest-xray-pneumonia.zip to /content\n100% 2.29G/2.29G [00:42<00:00, 121MB/s]\n100% 2.29G/2.29G [00:42<00:00, 58.1MB/s]\n"
],
[
"!unzip chest-xray-pneumonia.zip",
"\u001b[1;30;43mStreaming output truncated to the last 5000 lines.\u001b[0m\n inflating: chest_xray/train/NORMAL/IM-0435-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0435-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0437-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0437-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0437-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0438-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0439-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0439-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0439-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0440-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0441-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0442-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0444-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0445-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0446-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0447-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0448-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0449-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0450-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0451-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0452-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0453-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0453-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0455-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0456-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0457-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0458-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0459-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0460-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0461-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0463-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0464-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0465-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0466-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0467-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0467-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0467-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0469-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0471-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0472-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0473-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0474-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0475-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0476-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0477-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0478-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0479-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0480-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0481-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0482-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0483-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0484-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0485-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0486-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0487-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0488-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0489-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0490-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0491-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0491-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0491-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0492-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0493-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0494-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0495-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0496-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0497-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0497-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0497-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0499-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0499-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0499-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0500-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0501-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0501-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0501-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0502-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0503-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0504-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0505-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0505-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0505-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0506-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0507-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0508-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0509-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0509-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0509-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0510-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0511-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0511-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0511-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0512-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0513-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0514-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0515-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0516-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0517-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0517-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0519-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0519-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0519-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0520-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0521-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0522-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0523-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0523-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0523-0001-0003.jpeg \n inflating: chest_xray/train/NORMAL/IM-0523-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0524-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0525-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0525-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0525-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0526-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0527-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0528-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0529-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0530-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0531-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0531-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0532-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0533-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0533-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0533-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0534-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0535-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0536-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0537-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0538-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0539-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0539-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0539-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0540-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0541-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0542-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0543-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0543-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0544-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0545-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0545-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0545-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0546-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0547-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0548-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0549-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0549-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0549-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0551-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0551-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0551-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0552-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0553-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0553-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0553-0001-0003.jpeg \n inflating: chest_xray/train/NORMAL/IM-0553-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0554-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0555-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0555-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0555-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0556-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0557-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0559-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0560-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0561-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0562-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0563-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0564-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0565-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0566-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0568-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0569-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0570-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0571-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0574-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0575-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0577-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0578-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0579-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0580-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0581-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0582-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0583-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0584-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0586-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0588-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0590-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0591-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0592-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0593-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0595-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0596-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0598-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0599-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0600-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0601-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0602-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0604-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0605-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0606-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0607-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0608-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0608-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0608-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0609-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0612-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0612-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0612-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0613-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0614-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0615-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0616-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0617-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0618-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0618-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0618-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0619-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0620-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0620-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0620-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0621-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0622-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0622-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0622-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0623-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0624-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0624-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0625-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0626-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0626-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0627-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0628-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0629-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0629-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0629-0001-0003.jpeg \n inflating: chest_xray/train/NORMAL/IM-0629-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0630-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0631-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0631-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0631-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0632-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0633-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0634-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0635-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0636-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0637-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0640-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0640-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0640-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0641-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0642-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0643-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0644-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0644-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0644-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0645-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0646-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0647-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0648-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0649-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0650-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0650-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0650-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0651-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0652-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0652-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0654-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0655-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0656-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0656-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0656-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0657-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0658-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0659-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0660-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0660-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0660-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0661-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0662-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0663-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0664-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0665-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0666-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0666-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/IM-0666-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0667-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0668-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0669-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0670-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0671-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0672-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0673-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0674-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0675-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0676-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0677-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0678-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0679-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0680-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0681-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0682-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0683-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0684-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0685-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0686-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0687-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0688-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0689-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0691-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0692-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0693-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0694-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0695-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0696-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0697-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0698-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0700-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0701-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0702-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0703-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0704-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0705-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0706-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0707-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0709-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0710-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0711-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0712-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0713-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0714-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0715-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0716-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0717-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0718-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0719-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0721-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0722-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0724-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0727-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0728-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0729-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0730-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0732-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0733-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0734-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0735-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0736-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0737-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0738-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0739-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0740-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0741-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0742-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0746-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0747-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0748-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0750-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0751-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0752-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0753-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0754-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0755-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0757-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0761-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0764-0001.jpeg \n inflating: chest_xray/train/NORMAL/IM-0766-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0383-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0384-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0385-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0386-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0388-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0389-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0390-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0391-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0392-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0393-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0394-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0395-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0395-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0395-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0396-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0397-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0399-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0401-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0402-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0403-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0404-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0406-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0407-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0408-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0409-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0410-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0412-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0413-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0414-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0415-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0416-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0416-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0416-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0417-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0418-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0419-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0421-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0423-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0424-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0425-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0427-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0428-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0429-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0433-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0435-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0437-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0439-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0440-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0441-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0443-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0445-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0447-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0448-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0449-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0450-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0451-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0452-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0453-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0454-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0455-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0456-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0458-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0460-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0462-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0463-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0464-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0465-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0466-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0468-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0472-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0473-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0474-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0475-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0476-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0478-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0479-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0480-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0481-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0482-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0485-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0486-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0487-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0488-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0489-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0490-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0491-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0493-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0496-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0497-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0499-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0500-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0501-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0502-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0503-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0506-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0507-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0508-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0509-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0511-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0512-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0513-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0515-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0516-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0517-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0518-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0520-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0521-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0522-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0523-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0525-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0526-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0528-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0529-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0530-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0531-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0533-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0535-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0535-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0536-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0537-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0539-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0540-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0541-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0543-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0545-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0547-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0550-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0551-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0552-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0553-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0554-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0555-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0555-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0555-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0557-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0558-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0559-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0561-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0563-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0564-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0566-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0567-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0568-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0569-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0571-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0572-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0573-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0575-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0576-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0577-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0578-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0579-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0580-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0582-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0583-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0585-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0587-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0587-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0587-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0588-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0589-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0592-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0594-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0595-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0596-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0599-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0600-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0601-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0602-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0603-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0604-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0609-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0611-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0616-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0617-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0618-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0619-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0620-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0621-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0622-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0623-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0626-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0627-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0629-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0630-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0633-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0634-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0635-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0636-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0637-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0640-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0641-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0642-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0643-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0645-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0647-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0648-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0649-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0650-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0651-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0651-0004.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0652-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0653-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0654-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0655-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0657-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0659-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0660-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0661-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0662-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0663-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0664-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0665-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0666-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0667-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0668-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0669-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0671-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0672-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0673-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0675-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0678-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0680-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0682-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0683-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0684-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0686-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0687-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0689-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0690-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0692-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0693-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0694-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0695-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0696-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0698-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0699-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0700-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0702-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0705-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0707-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0718-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0719-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0723-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0725-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0727-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0730-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0736-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0741-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0744-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0746-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0749-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0753-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0757-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0765-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0771-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0772-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0774-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0775-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0776-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0777-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0780-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0781-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0790-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0793-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0796-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0797-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0798-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0799-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0803-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0804-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0806-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0807-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0808-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0809-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0810-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0811-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0812-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0814-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0815-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0816-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0818-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0818-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0819-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0820-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0821-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0822-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0824-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0825-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0826-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0827-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0828-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0829-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0830-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0831-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0832-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0832-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0832-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0833-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0834-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0836-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0837-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0838-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0839-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0840-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0841-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0842-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0843-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0845-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0846-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0847-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0848-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0849-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0851-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0851-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0851-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0852-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0853-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0854-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0855-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0856-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0857-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0858-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0859-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0860-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0862-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0863-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0865-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0866-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0867-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0868-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0869-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0870-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0871-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0872-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0873-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0874-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0875-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0876-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0877-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0879-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0880-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0881-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0882-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0885-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0886-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0887-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0888-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0890-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0892-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0893-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0894-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0895-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0896-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0897-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0898-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0899-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0900-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0903-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0904-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0905-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0906-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0907-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0908-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0909-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0910-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0911-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0912-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0913-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0914-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0915-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0917-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0918-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0919-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0922-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0923-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0924-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0925-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0926-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0927-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0929-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0930-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0931-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0932-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0933-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0934-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0935-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0936-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0937-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0939-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0941-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0942-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0944-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0945-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0946-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0947-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0948-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0949-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0950-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0951-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0952-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0954-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0955-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0956-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0957-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0959-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0960-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0961-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0962-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0965-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0966-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0967-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0969-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0970-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0971-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0971-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0974-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0975-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0976-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0977-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0978-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0979-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0980-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0981-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0983-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0983-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0983-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0986-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0987-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0988-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0989-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0992-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0993-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0994-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0995-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0995-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0995-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0997-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0998-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-0999-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1002-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1004-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1005-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1006-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1008-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1010-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1011-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1014-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1015-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1016-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1017-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1018-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1019-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1020-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1020-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1020-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1022-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1023-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1024-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1025-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1026-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1027-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1028-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1030-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1033-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1035-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1037-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1038-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1039-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1040-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1041-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1043-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1044-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1045-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1046-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1047-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1048-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1049-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1050-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1051-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1052-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1053-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1054-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1055-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1056-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1058-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1059-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1060-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1062-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1064-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1067-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1067-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1073-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1084-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1086-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1088-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1089-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1090-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1091-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1093-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1094-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1094-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1094-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1096-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1096-0001-0003.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1096-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1098-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1100-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1102-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1102-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1102-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1103-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1104-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1105-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1106-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1108-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1109-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1110-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1111-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1112-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1113-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1114-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1116-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1116-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1116-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1117-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1118-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1120-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1122-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1123-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1124-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1125-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1126-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1127-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1128-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1128-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1128-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1130-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1131-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1132-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1134-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1135-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1136-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1138-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1141-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1142-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1142-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1142-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1144-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1145-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1147-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1148-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1149-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1150-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1151-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1152-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1152-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1152-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1153-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1154-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1154-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1154-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1155-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1156-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1157-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1158-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1160-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1161-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1162-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1163-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1164-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1167-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1168-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1169-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1170-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1171-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1173-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1174-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1175-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1176-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1177-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1178-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1179-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1180-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1181-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1182-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1183-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1184-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1185-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1187-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1188-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1189-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1190-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1191-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1192-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1194-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1196-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1197-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1198-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1200-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1201-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1202-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1203-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1204-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1205-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1206-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1209-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1214-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1218-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1219-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1220-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1221-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1222-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1223-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1224-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1225-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1226-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1227-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1228-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1231-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1232-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1234-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1236-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1237-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1240-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1241-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1242-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1243-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1244-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1245-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1247-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1250-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1252-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1253-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1254-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1256-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1257-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1258-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1258-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1258-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1260-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1261-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1262-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1264-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1266-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1266-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1266-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1267-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1269-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1269-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1269-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1270-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1271-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1272-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1273-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1274-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1275-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1276-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1277-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1277-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1277-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1278-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1279-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1280-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1281-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1282-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1285-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1286-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1287-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1288-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1289-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1290-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1291-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1292-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1293-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1294-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1294-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1294-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1295-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1296-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1300-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1301-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1302-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1303-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1304-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1305-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1306-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1307-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1308-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1310-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1311-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1314-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1315-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1316-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1317-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1318-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1319-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1320-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1321-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1322-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1323-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1326-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1327-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1328-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1329-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1330-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1332-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1333-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1334-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1335-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1336-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1337-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1338-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1341-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1342-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1343-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1344-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1345-0001-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1345-0001-0002.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1345-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1346-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1347-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1348-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1349-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1350-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1351-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1356-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1357-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1360-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1362-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1365-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1371-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1376-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1379-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1385-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1396-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1400-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1401-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1406-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1412-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1419-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1422-0001.jpeg \n inflating: chest_xray/train/NORMAL/NORMAL2-IM-1423-0001.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1000_bacteria_2931.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1000_virus_1681.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1001_bacteria_2932.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1002_bacteria_2933.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1003_bacteria_2934.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1003_virus_1685.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1004_bacteria_2935.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1004_virus_1686.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1005_bacteria_2936.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1005_virus_1688.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1006_bacteria_2937.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1007_bacteria_2938.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1007_virus_1690.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1008_bacteria_2939.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1008_virus_1691.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1009_virus_1694.jpeg \n inflating: chest_xray/train/PNEUMONIA/person100_virus_184.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1010_bacteria_2941.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1010_virus_1695.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1011_bacteria_2942.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1012_bacteria_2943.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1014_bacteria_2945.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1015_virus_1701.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1015_virus_1702.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1016_bacteria_2947.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1016_virus_1704.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1017_bacteria_2948.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1018_bacteria_2949.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1018_virus_1706.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1019_bacteria_2950.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1019_virus_1707.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1019_virus_1708.jpeg \n inflating: chest_xray/train/PNEUMONIA/person101_virus_187.jpeg \n inflating: chest_xray/train/PNEUMONIA/person101_virus_188.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1020_bacteria_2951.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1020_virus_1710.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1021_virus_1711.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1022_bacteria_2953.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1022_virus_1712.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1023_bacteria_2954.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1023_virus_1714.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1024_bacteria_2955.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1024_virus_1716.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1026_bacteria_2957.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1026_virus_1718.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1028_bacteria_2959.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1028_bacteria_2960.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1029_bacteria_2961.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1029_virus_1721.jpeg \n inflating: chest_xray/train/PNEUMONIA/person102_virus_189.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1030_virus_1722.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1031_bacteria_2963.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1031_bacteria_2964.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1031_virus_1723.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1033_bacteria_2966.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1034_bacteria_2968.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1034_virus_1728.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1035_bacteria_2969.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1035_virus_1729.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1036_bacteria_2970.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1036_virus_1730.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1037_bacteria_2971.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1038_bacteria_2972.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1038_virus_1733.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1039_bacteria_2973.jpeg \n inflating: chest_xray/train/PNEUMONIA/person103_virus_190.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1040_bacteria_2974.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1040_virus_1735.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1041_bacteria_2975.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1041_virus_1736.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1042_virus_1737.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1043_bacteria_2977.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1043_virus_1738.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1044_bacteria_2978.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1044_virus_1740.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1045_bacteria_2979.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1045_virus_1741.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1046_bacteria_2980.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1046_virus_1742.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1048_bacteria_2982.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1048_virus_1744.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1049_bacteria_2983.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1049_virus_1746.jpeg \n inflating: chest_xray/train/PNEUMONIA/person104_virus_191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1050_bacteria_2984.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1051_bacteria_2985.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1051_virus_1750.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1052_bacteria_2986.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1052_virus_1751.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1053_bacteria_2987.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1054_bacteria_2988.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1055_bacteria_2989.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1056_bacteria_2990.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1056_virus_1755.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1057_bacteria_2991.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1057_virus_1756.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1058_bacteria_2992.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1058_virus_1757.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1059_bacteria_2993.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1059_virus_1758.jpeg \n inflating: chest_xray/train/PNEUMONIA/person105_virus_192.jpeg \n inflating: chest_xray/train/PNEUMONIA/person105_virus_193.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1060_virus_1760.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1062_bacteria_2996.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1062_virus_1762.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1063_bacteria_2997.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1063_virus_1765.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1065_bacteria_2999.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1065_virus_1768.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1066_bacteria_3000.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1066_virus_1769.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1067_bacteria_3001.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1067_virus_1770.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1068_bacteria_3002.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1068_virus_1771.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1069_bacteria_3003.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1069_virus_1772.jpeg \n inflating: chest_xray/train/PNEUMONIA/person106_virus_194.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1070_virus_1773.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1071_bacteria_3005.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1071_virus_1774.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1072_bacteria_3006.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1072_bacteria_3007.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1072_virus_1775.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1073_bacteria_3008.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1073_bacteria_3011.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1073_virus_1776.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1074_bacteria_3012.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1074_bacteria_3014.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1075_bacteria_3015.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1076_bacteria_3016.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1077_bacteria_3017.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1077_virus_1787.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1078_bacteria_3018.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1078_virus_1788.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1079_bacteria_3019.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1079_virus_1789.jpeg \n inflating: chest_xray/train/PNEUMONIA/person107_virus_197.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1080_bacteria_3020.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1080_virus_1791.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1081_bacteria_3021.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1081_virus_1793.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1082_bacteria_3022.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1082_virus_1794.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1083_bacteria_3023.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1083_virus_1795.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1084_bacteria_3024.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1084_virus_1796.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1085_bacteria_3025.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1085_virus_1797.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1086_bacteria_3026.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1086_virus_1798.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1087_bacteria_3027.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1087_virus_1799.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1088_bacteria_3028.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1088_virus_1800.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1089_bacteria_3029.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1089_virus_1808.jpeg \n inflating: chest_xray/train/PNEUMONIA/person108_virus_199.jpeg \n inflating: chest_xray/train/PNEUMONIA/person108_virus_200.jpeg \n inflating: chest_xray/train/PNEUMONIA/person108_virus_201.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1090_virus_1809.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1091_bacteria_3031.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1091_virus_1810.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1092_bacteria_3032.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1092_virus_1811.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1093_bacteria_3033.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1094_virus_1814.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1095_virus_1815.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1096_bacteria_3037.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1096_virus_1816.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1097_bacteria_3038.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1097_virus_1817.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1098_bacteria_3039.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1098_virus_1818.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1099_bacteria_3040.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1099_virus_1819.jpeg \n inflating: chest_xray/train/PNEUMONIA/person109_virus_203.jpeg \n inflating: chest_xray/train/PNEUMONIA/person10_bacteria_43.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1100_bacteria_3041.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1100_virus_1820.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1101_bacteria_3042.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1102_bacteria_3043.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1102_virus_1822.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1103_bacteria_3044.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1103_virus_1825.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1104_virus_1826.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1105_bacteria_3046.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1106_virus_1829.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1107_bacteria_3048.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1107_virus_1831.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1107_virus_1832.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1108_bacteria_3049.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1108_virus_1833.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1109_bacteria_3050.jpeg \n inflating: chest_xray/train/PNEUMONIA/person110_virus_205.jpeg \n inflating: chest_xray/train/PNEUMONIA/person110_virus_206.jpeg \n inflating: chest_xray/train/PNEUMONIA/person110_virus_207.jpeg \n inflating: chest_xray/train/PNEUMONIA/person110_virus_208.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1110_bacteria_3051.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1110_virus_1835.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1111_bacteria_3052.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1111_virus_1836.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1112_bacteria_3053.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1112_virus_1837.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1113_virus_1838.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1114_bacteria_3055.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1115_bacteria_3056.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1115_virus_1840.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1116_virus_1841.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1118_bacteria_3059.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1119_virus_1844.jpeg \n inflating: chest_xray/train/PNEUMONIA/person111_virus_209.jpeg \n inflating: chest_xray/train/PNEUMONIA/person111_virus_210.jpeg \n inflating: chest_xray/train/PNEUMONIA/person111_virus_212.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1120_virus_1845.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1121_virus_1846.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1122_bacteria_3063.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1122_virus_1847.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1123_virus_1848.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1124_bacteria_3065.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1124_virus_1851.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1125_bacteria_3066.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1125_virus_1852.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1126_virus_1853.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1127_bacteria_3068.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1127_virus_1854.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1128_bacteria_3069.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1129_bacteria_3070.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1129_virus_1857.jpeg \n inflating: chest_xray/train/PNEUMONIA/person112_virus_213.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1130_bacteria_3072.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1130_virus_1860.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1131_bacteria_3073.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1132_virus_1863.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1133_bacteria_3075.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1133_virus_1865.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1134_bacteria_3076.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1135_bacteria_3077.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1136_bacteria_3078.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1137_virus_1876.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1138_bacteria_3080.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1138_virus_1877.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1138_virus_1879.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1139_bacteria_3081.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1139_bacteria_3082.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1139_virus_1882.jpeg \n inflating: chest_xray/train/PNEUMONIA/person113_virus_215.jpeg \n inflating: chest_xray/train/PNEUMONIA/person113_virus_216.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1140_bacteria_3083.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1140_virus_1885.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1141_bacteria_3084.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1141_bacteria_3085.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1141_virus_1886.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1141_virus_1890.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1142_bacteria_3086.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1142_virus_1892.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1143_virus_1896.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1143_virus_1897.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1144_bacteria_3089.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1145_bacteria_3090.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1145_virus_1902.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1145_virus_1905.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1145_virus_1906.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1146_bacteria_3091.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1147_virus_1917.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1147_virus_1919.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1147_virus_1920.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1149_bacteria_3094.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1149_virus_1924.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1149_virus_1925.jpeg \n inflating: chest_xray/train/PNEUMONIA/person114_virus_217.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1150_bacteria_3095.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1151_virus_1928.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1152_virus_1930.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1153_virus_1932.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1154_bacteria_3099.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1154_virus_1933.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1155_bacteria_3100.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1155_virus_1934.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1156_bacteria_3101.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1156_virus_1935.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1156_virus_1936.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1157_bacteria_3102.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1157_virus_1937.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1158_bacteria_3103.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1158_virus_1938.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1158_virus_1940.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1158_virus_1941.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1158_virus_1942.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1158_virus_1943.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1159_bacteria_3104.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1159_virus_1944.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1159_virus_1945.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1159_virus_1946.jpeg \n inflating: chest_xray/train/PNEUMONIA/person115_virus_218.jpeg \n inflating: chest_xray/train/PNEUMONIA/person115_virus_219.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1160_bacteria_3105.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1160_virus_1947.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1161_virus_1948.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1162_bacteria_3107.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1162_virus_1949.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1162_virus_1950.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1163_virus_1951.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1164_bacteria_3110.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1164_virus_1952.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1164_virus_1955.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1164_virus_1956.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1164_virus_1957.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1164_virus_1958.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1165_bacteria_3111.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1165_virus_1959.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1167_bacteria_3113.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1168_bacteria_3114.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1168_bacteria_3115.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1168_virus_1965.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1168_virus_1966.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1169_virus_1968.jpeg \n inflating: chest_xray/train/PNEUMONIA/person116_virus_221.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1170_bacteria_3117.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1170_virus_1969.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1170_virus_1970.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1171_bacteria_3118.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1172_bacteria_3119.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1172_virus_1977.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1173_virus_1978.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1174_virus_1980.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1175_bacteria_3122.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1175_virus_1981.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1176_bacteria_3123.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1176_bacteria_3124.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1176_virus_1996.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1176_virus_1997.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1176_virus_1998.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1177_bacteria_3125.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1177_virus_1999.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1177_virus_2000.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1177_virus_2001.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1177_virus_2002.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1178_bacteria_3126.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1178_virus_2004.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1179_bacteria_3127.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1179_virus_2006.jpeg \n inflating: chest_xray/train/PNEUMONIA/person117_virus_223.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_bacteria_3128.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2007.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2008.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2009.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2010.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2011.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2012.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2013.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2014.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1180_virus_2015.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1181_bacteria_3129.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1181_virus_2016.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1182_virus_2017.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1183_bacteria_3131.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1183_virus_2018.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1184_bacteria_3132.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1184_virus_2019.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1185_bacteria_3133.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1186_bacteria_3134.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1186_bacteria_3135.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1186_virus_2021.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1186_virus_2022.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1187_bacteria_3136.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1187_virus_2023.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1188_bacteria_3137.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1188_virus_2024.jpeg \n inflating: chest_xray/train/PNEUMONIA/person118_virus_224.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1190_virus_2031.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1191_virus_2032.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1192_bacteria_3141.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1193_virus_2034.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1194_bacteria_3143.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1195_bacteria_3144.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1196_bacteria_3146.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1197_bacteria_3147.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1197_virus_2039.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1198_bacteria_3148.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1199_bacteria_3149.jpeg \n inflating: chest_xray/train/PNEUMONIA/person119_virus_225.jpeg \n inflating: chest_xray/train/PNEUMONIA/person11_bacteria_45.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1200_virus_2042.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1201_bacteria_3151.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1202_bacteria_3152.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1202_bacteria_3153.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1202_virus_2045.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1203_bacteria_3154.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1203_bacteria_3155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1204_bacteria_3156.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1205_bacteria_3157.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1206_bacteria_3158.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1206_virus_2051.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1207_bacteria_3159.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1208_bacteria_3160.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1209_bacteria_3161.jpeg \n inflating: chest_xray/train/PNEUMONIA/person120_virus_226.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1211_bacteria_3163.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1211_virus_2056.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1212_bacteria_3164.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1212_virus_2057.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1213_virus_2058.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1214_bacteria_3166.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1214_virus_2059.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1215_bacteria_3167.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1216_bacteria_3168.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1216_virus_2062.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1217_bacteria_3169.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1217_virus_2063.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1218_bacteria_3171.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1218_virus_2066.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1219_bacteria_3172.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1219_virus_2067.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1220_bacteria_3173.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1220_bacteria_3174.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1220_virus_2068.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1222_bacteria_3177.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1222_virus_2071.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1223_bacteria_3178.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1223_virus_2073.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1224_virus_2074.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1225_bacteria_3180.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1225_virus_2076.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1226_virus_2077.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1227_bacteria_3182.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1227_virus_2078.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1228_bacteria_3183.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1228_virus_2079.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1229_virus_2080.jpeg \n inflating: chest_xray/train/PNEUMONIA/person122_virus_229.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1230_bacteria_3185.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1230_virus_2081.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1231_bacteria_3186.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1231_virus_2088.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1232_virus_2089.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1233_bacteria_3188.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1233_virus_2090.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1234_bacteria_3189.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1234_bacteria_3190.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1234_virus_2093.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1235_bacteria_3191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1235_virus_2095.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1236_bacteria_3192.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1236_virus_2096.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1237_bacteria_3193.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1237_virus_2097.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1238_bacteria_3194.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1238_virus_2098.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1239_bacteria_3195.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1239_virus_2099.jpeg \n inflating: chest_xray/train/PNEUMONIA/person123_virus_230.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1240_bacteria_3196.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1241_bacteria_3197.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1241_virus_2106.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1242_bacteria_3198.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1242_virus_2108.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1242_virus_2109.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1243_bacteria_3199.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1243_virus_2110.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1244_bacteria_3200.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1244_virus_2111.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1246_bacteria_3202.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1247_bacteria_3203.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1247_virus_2115.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1248_bacteria_3204.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1248_virus_2117.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1249_bacteria_3205.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1249_virus_2118.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_231.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_233.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_234.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_236.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_237.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_238.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_239.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_242.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_244.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_245.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_246.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_247.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_249.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_250.jpeg \n inflating: chest_xray/train/PNEUMONIA/person124_virus_251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1250_bacteria_3207.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1251_bacteria_3208.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1252_bacteria_3209.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1252_virus_2124.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1253_bacteria_3211.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1253_virus_2129.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1254_virus_2130.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1255_virus_2132.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1256_bacteria_3214.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1257_bacteria_3215.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1258_bacteria_3216.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1258_virus_2138.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1259_bacteria_3217.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1259_virus_2139.jpeg \n inflating: chest_xray/train/PNEUMONIA/person125_virus_254.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1260_bacteria_3218.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1260_virus_2140.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1261_bacteria_3219.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1261_virus_2145.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1261_virus_2147.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1261_virus_2148.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1262_bacteria_3220.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1263_bacteria_3221.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1264_bacteria_3222.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1264_virus_2155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1265_bacteria_3223.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1265_virus_2156.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1266_bacteria_3224.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1266_bacteria_3225.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1266_virus_2158.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1267_bacteria_3226.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1267_virus_2160.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1268_bacteria_3227.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1268_bacteria_3228.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1268_virus_2161.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1269_bacteria_3229.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1269_virus_2162.jpeg \n inflating: chest_xray/train/PNEUMONIA/person126_virus_255.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1270_bacteria_3230.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1270_virus_2163.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1271_bacteria_3231.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1271_virus_2164.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1272_bacteria_3232.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1272_virus_2190.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1273_bacteria_3233.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1273_bacteria_3234.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1273_virus_2191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1274_bacteria_3235.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1274_bacteria_3236.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1274_virus_2193.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1275_bacteria_3237.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1276_bacteria_3239.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1276_virus_2198.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1277_bacteria_3240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1278_virus_2201.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1279_bacteria_3242.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1280_bacteria_3243.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1281_bacteria_3244.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1281_virus_2204.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1282_bacteria_3245.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1283_bacteria_3246.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1283_virus_2206.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1284_bacteria_3247.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1284_virus_2207.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1285_virus_2208.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1286_bacteria_3249.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1286_virus_2209.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1287_bacteria_3250.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1287_virus_2210.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1288_bacteria_3251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1288_virus_2211.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1289_bacteria_3252.jpeg \n inflating: chest_xray/train/PNEUMONIA/person128_virus_261.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1290_bacteria_3253.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1290_virus_2215.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1290_virus_2216.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1291_virus_2217.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1292_bacteria_3255.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1292_virus_2218.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1293_virus_2219.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1294_bacteria_3257.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1294_virus_2221.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1294_virus_2222.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1295_bacteria_3258.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1295_virus_2223.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1296_virus_2224.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1297_bacteria_3260.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1298_bacteria_3261.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1298_virus_2226.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1298_virus_2228.jpeg \n inflating: chest_xray/train/PNEUMONIA/person12_bacteria_46.jpeg \n inflating: chest_xray/train/PNEUMONIA/person12_bacteria_47.jpeg \n inflating: chest_xray/train/PNEUMONIA/person12_bacteria_48.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1300_bacteria_3264.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1300_virus_2240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1301_virus_2241.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1302_bacteria_3266.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1303_bacteria_3267.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1303_virus_2243.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1304_bacteria_3269.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1305_bacteria_3271.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1306_bacteria_3272.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1306_bacteria_3274.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1306_bacteria_3275.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1306_bacteria_3276.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1306_bacteria_3277.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1306_virus_2249.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1307_bacteria_3278.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1307_virus_2251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_bacteria_3280.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_bacteria_3283.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_bacteria_3285.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_bacteria_3286.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_bacteria_3288.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_bacteria_3290.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_bacteria_3292.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_virus_2252.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1308_virus_2253.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1309_bacteria_3294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1309_virus_2254.jpeg \n inflating: chest_xray/train/PNEUMONIA/person130_virus_263.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1310_bacteria_3295.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1310_bacteria_3297.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1310_bacteria_3300.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1310_bacteria_3301.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1310_bacteria_3302.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1310_bacteria_3304.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1310_virus_2255.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1311_bacteria_3312.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1311_virus_2257.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1311_virus_2259.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1312_bacteria_3313.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1312_bacteria_3314.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1312_bacteria_3316.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1312_bacteria_3317.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1312_bacteria_3318.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1312_bacteria_3319.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1312_virus_2261.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1313_bacteria_3320.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1313_virus_2264.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1314_virus_2266.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1314_virus_2268.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1314_virus_2269.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1315_bacteria_3322.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1315_virus_2270.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1316_bacteria_3326.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1316_virus_2271.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1317_bacteria_3332.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1317_virus_2273.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1318_bacteria_3334.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1318_bacteria_3335.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1318_virus_2274.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1319_virus_2276.jpeg \n inflating: chest_xray/train/PNEUMONIA/person131_virus_265.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3339.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3340.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3342.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3344.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3345.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3346.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3347.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3348.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3350.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3351.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3352.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3353.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_bacteria_3355.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1320_virus_2277.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1321_bacteria_3358.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1321_bacteria_3359.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1321_virus_2279.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1322_bacteria_3360.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1323_bacteria_3361.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1323_bacteria_3362.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1323_bacteria_3363.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1323_virus_2282.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1323_virus_2283.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1324_virus_2284.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1324_virus_2285.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1325_bacteria_3366.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1325_virus_2287.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1326_bacteria_3372.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1327_bacteria_3373.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1327_bacteria_3374.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1328_bacteria_3376.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1328_virus_2293.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1328_virus_2294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1328_virus_2295.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1329_bacteria_3377.jpeg \n inflating: chest_xray/train/PNEUMONIA/person132_virus_266.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1331_bacteria_3380.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1331_virus_2299.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1332_virus_2300.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1333_bacteria_3383.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1333_bacteria_3384.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1333_bacteria_3385.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1333_bacteria_3386.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1333_virus_2301.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1336_virus_2306.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1337_virus_2307.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1338_bacteria_3394.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1338_bacteria_3395.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1338_bacteria_3397.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1338_virus_2308.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1339_bacteria_3399.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1339_bacteria_3402.jpeg \n inflating: chest_xray/train/PNEUMONIA/person133_virus_267.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1340_bacteria_3405.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1340_virus_2311.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1340_virus_2312.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1341_bacteria_3406.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1341_virus_2313.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1342_bacteria_3407.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1342_virus_2315.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3409.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3411.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3413.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3414.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3415.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3416.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3417.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3418.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_bacteria_3419.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_virus_2316.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1343_virus_2317.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1344_bacteria_3421.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1344_virus_2319.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1344_virus_2320.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1345_bacteria_3422.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1345_bacteria_3424.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1345_bacteria_3425.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1345_bacteria_3426.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1345_bacteria_3427.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1345_bacteria_3428.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1345_virus_2321.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1346_bacteria_3430.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1346_virus_2322.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1347_virus_2323.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1348_virus_2324.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1348_virus_2326.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1349_bacteria_3434.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1349_bacteria_3436.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1349_bacteria_3437.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1349_bacteria_3438.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1349_bacteria_3439.jpeg \n inflating: chest_xray/train/PNEUMONIA/person134_virus_268.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1350_virus_2329.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1351_bacteria_3441.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1351_virus_2330.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1352_bacteria_3442.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1352_bacteria_3443.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1352_bacteria_3444.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1352_bacteria_3445.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1353_bacteria_3446.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1353_virus_2333.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1354_bacteria_3448.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1355_bacteria_3449.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1355_bacteria_3452.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1355_virus_2336.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1356_virus_2337.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1357_virus_2338.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1358_bacteria_3463.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1358_bacteria_3465.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1358_virus_2339.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1359_virus_2340.jpeg \n inflating: chest_xray/train/PNEUMONIA/person135_virus_270.jpeg \n inflating: chest_xray/train/PNEUMONIA/person135_virus_271.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1360_virus_2341.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1361_bacteria_3476.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1361_bacteria_3477.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1361_virus_2342.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1361_virus_2344.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1362_virus_2345.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1363_bacteria_3483.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1363_bacteria_3484.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1363_virus_2346.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1365_bacteria_3489.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1365_virus_2348.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1366_bacteria_3490.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1366_virus_2349.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1367_virus_2351.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1368_virus_2352.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1368_virus_2353.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1368_virus_2354.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1369_virus_2355.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1369_virus_2356.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1371_virus_2361.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1371_virus_2362.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1372_bacteria_3498.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1372_bacteria_3499.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1372_bacteria_3500.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1372_bacteria_3501.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1372_bacteria_3502.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1372_bacteria_3503.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1374_bacteria_3506.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1374_bacteria_3507.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1374_virus_2365.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1375_bacteria_3509.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1375_bacteria_3510.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1375_virus_2366.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1376_bacteria_3511.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1376_virus_2367.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1377_bacteria_3512.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1377_virus_2369.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1378_bacteria_3513.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1379_bacteria_3514.jpeg \n inflating: chest_xray/train/PNEUMONIA/person137_virus_281.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1380_bacteria_3515.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1381_bacteria_3516.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1381_bacteria_3517.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1381_virus_2375.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1383_bacteria_3521.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1383_virus_2377.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1384_bacteria_3522.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1385_bacteria_3524.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1385_virus_2380.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1387_virus_2382.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1388_bacteria_3529.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1389_bacteria_3531.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1389_virus_2387.jpeg \n inflating: chest_xray/train/PNEUMONIA/person138_virus_282.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1390_bacteria_3534.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1390_bacteria_3535.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1391_bacteria_3536.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1391_bacteria_3537.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1392_bacteria_3538.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1393_virus_2396.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1394_virus_2397.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1395_bacteria_3544.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1395_virus_2398.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1396_bacteria_3545.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1396_virus_2399.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1397_virus_2400.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1398_bacteria_3548.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1398_virus_2401.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1399_bacteria_3549.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1399_virus_2402.jpeg \n inflating: chest_xray/train/PNEUMONIA/person139_virus_283.jpeg \n inflating: chest_xray/train/PNEUMONIA/person13_bacteria_49.jpeg \n inflating: chest_xray/train/PNEUMONIA/person13_bacteria_50.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1400_bacteria_3550.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1400_bacteria_3551.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1400_bacteria_3553.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1400_bacteria_3554.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1401_bacteria_3555.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1402_virus_2405.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1403_bacteria_3557.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1403_bacteria_3559.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1403_virus_2406.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1404_bacteria_3561.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1405_bacteria_3564.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1405_bacteria_3566.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1405_bacteria_3567.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1405_bacteria_3571.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1405_bacteria_3573.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1405_virus_2408.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1406_bacteria_3574.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1406_bacteria_3575.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1406_virus_2409.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1407_virus_2410.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1408_bacteria_3579.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1408_bacteria_3581.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1408_virus_2411.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1409_bacteria_3583.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1409_bacteria_3585.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1409_virus_2413.jpeg \n inflating: chest_xray/train/PNEUMONIA/person140_virus_284.jpeg \n inflating: chest_xray/train/PNEUMONIA/person140_virus_285.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3591.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3593.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3598.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3599.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3601.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3602.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3603.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3604.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3607.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3609.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_bacteria_3610.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1411_virus_2415.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1412_bacteria_3612.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1413_bacteria_3613.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1413_bacteria_3615.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1413_bacteria_3617.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1413_bacteria_3620.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1413_virus_2422.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1413_virus_2423.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1414_bacteria_3627.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1414_bacteria_3628.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1414_virus_2424.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1415_bacteria_3629.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1415_virus_2425.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1416_virus_2427.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1417_bacteria_3635.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1418_bacteria_3636.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1418_bacteria_3637.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1418_bacteria_3638.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1418_bacteria_3639.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1418_bacteria_3643.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1418_virus_2429.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1419_bacteria_3645.jpeg \n inflating: chest_xray/train/PNEUMONIA/person141_virus_287.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1420_bacteria_3647.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1420_virus_2431.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1422_bacteria_3649.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1422_virus_2434.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1423_bacteria_3650.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1424_bacteria_3651.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1424_virus_2437.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1425_virus_2438.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1426_bacteria_3667.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1426_bacteria_3668.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1426_virus_2439.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1427_virus_2441.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1428_virus_2442.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1429_bacteria_3688.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1429_bacteria_3690.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1429_bacteria_3691.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1429_virus_2443.jpeg \n inflating: chest_xray/train/PNEUMONIA/person142_virus_288.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1430_bacteria_3693.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1430_bacteria_3694.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1430_bacteria_3695.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1430_bacteria_3696.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1430_bacteria_3697.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1430_virus_2444.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1431_bacteria_3698.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1432_bacteria_3699.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1433_bacteria_3701.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1433_bacteria_3704.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1433_bacteria_3705.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1433_virus_2447.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1436_bacteria_3711.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1436_bacteria_3712.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1438_bacteria_3715.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1438_bacteria_3718.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1438_bacteria_3721.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1438_virus_2452.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1439_bacteria_3722.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1439_virus_2453.jpeg \n inflating: chest_xray/train/PNEUMONIA/person143_virus_289.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1440_bacteria_3723.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1441_bacteria_3724.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1441_virus_2454.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1441_virus_2457.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1442_bacteria_3726.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1444_bacteria_3732.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1445_bacteria_3734.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1445_bacteria_3735.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1446_bacteria_3737.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1446_bacteria_3739.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1446_bacteria_3740.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1447_bacteria_3741.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1448_virus_2468.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1449_bacteria_3743.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1449_bacteria_3745.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1449_bacteria_3746.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1449_bacteria_3747.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1449_virus_2474.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1449_virus_2476.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1450_bacteria_3753.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1451_virus_2479.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1451_virus_2480.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1451_virus_2482.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1452_virus_2484.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1453_bacteria_3770.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1453_bacteria_3771.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1453_bacteria_3772.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1453_virus_2485.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1454_bacteria_3774.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1454_bacteria_3778.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1454_bacteria_3779.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1454_bacteria_3780.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1454_bacteria_3781.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1454_bacteria_3782.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1454_virus_2486.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1455_bacteria_3784.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1455_virus_2487.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1455_virus_2488.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1455_virus_2489.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1455_virus_2490.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1455_virus_2492.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1455_virus_2496.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1457_virus_2498.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1458_virus_2501.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1458_virus_2502.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1458_virus_2503.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1459_bacteria_3796.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1459_bacteria_3797.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1459_virus_2506.jpeg \n inflating: chest_xray/train/PNEUMONIA/person145_virus_294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person145_virus_295.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1460_bacteria_3801.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1460_bacteria_3805.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1460_virus_2507.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1460_virus_2509.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1461_virus_2510.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1462_virus_2512.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1463_bacteria_3808.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1463_bacteria_3809.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1463_bacteria_3811.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1463_virus_2516.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1465_virus_2530.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1465_virus_2531.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1465_virus_2532.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1465_virus_2537.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1466_virus_2541.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1466_virus_2542.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1466_virus_2543.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1467_virus_2544.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1468_bacteria_3822.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1468_virus_2545.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1468_virus_2546.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1469_bacteria_3824.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1469_bacteria_3827.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1469_virus_2547.jpeg \n inflating: chest_xray/train/PNEUMONIA/person146_virus_296.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1470_bacteria_3829.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1470_bacteria_3830.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1471_bacteria_3831.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1471_virus_2549.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1472_bacteria_3833.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1472_bacteria_3834.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1472_virus_2550.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1473_bacteria_3836.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1473_virus_2551.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1473_virus_2553.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1474_bacteria_3837.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1474_virus_2556.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1474_virus_2557.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1475_virus_2558.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1476_bacteria_3842.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1476_bacteria_3843.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1476_virus_2560.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1477_virus_2561.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1478_bacteria_3848.jpeg \n inflating: chest_xray/train/PNEUMONIA/person147_virus_297.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1480_bacteria_3858.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1480_bacteria_3859.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1480_virus_2566.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_bacteria_3862.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_bacteria_3863.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_bacteria_3864.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_bacteria_3865.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_bacteria_3866.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_bacteria_3867.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_bacteria_3868.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1481_virus_2567.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1482_bacteria_3870.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1482_bacteria_3874.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1482_virus_2569.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1482_virus_2570.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1482_virus_2571.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1482_virus_2572.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1482_virus_2573.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1483_bacteria_3876.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1483_virus_2574.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1484_bacteria_3878.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1484_virus_2576.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1484_virus_2577.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1486_bacteria_3881.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1486_bacteria_3883.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1486_bacteria_3884.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1486_bacteria_3885.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1486_virus_2580.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1488_bacteria_3887.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1488_virus_2585.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1488_virus_2587.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1488_virus_2589.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1488_virus_2592.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1488_virus_2593.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1489_bacteria_3889.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1489_virus_2594.jpeg \n inflating: chest_xray/train/PNEUMONIA/person148_virus_298.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1490_bacteria_3891.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1490_virus_2596.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1491_bacteria_3892.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1491_bacteria_3893.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1491_virus_2597.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1492_bacteria_3894.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1492_virus_2599.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1493_bacteria_3895.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1493_bacteria_3896.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1493_bacteria_3897.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1493_bacteria_3898.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1493_bacteria_3899.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1494_bacteria_3901.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1494_virus_2601.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1495_virus_2603.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_bacteria_3905.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_bacteria_3906.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_bacteria_3907.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_bacteria_3908.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_bacteria_3909.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_bacteria_3910.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_bacteria_3911.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_virus_2605.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1496_virus_2606.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1497_bacteria_3912.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1497_virus_2607.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1499_bacteria_3915.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1499_virus_2609.jpeg \n inflating: chest_xray/train/PNEUMONIA/person149_virus_299.jpeg \n inflating: chest_xray/train/PNEUMONIA/person14_bacteria_51.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1500_bacteria_3916.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1500_virus_2610.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1501_virus_2611.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_bacteria_3922.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_bacteria_3923.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_bacteria_3924.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_bacteria_3925.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_bacteria_3927.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_bacteria_3928.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_bacteria_3929.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1502_virus_2612.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1503_virus_2613.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1504_bacteria_3931.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1504_virus_2614.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1505_virus_2615.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1506_bacteria_3933.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1506_virus_2616.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3935.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3942.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3943.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3944.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3945.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3946.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3947.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1507_bacteria_3948.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1508_bacteria_3949.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1509_bacteria_3951.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1509_virus_2621.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1510_virus_2628.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1510_virus_2629.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1511_bacteria_3955.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1512_bacteria_3958.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1512_virus_2631.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1513_bacteria_3962.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1513_virus_2632.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1514_bacteria_3964.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1514_virus_2633.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1516_virus_2643.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1517_bacteria_3968.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1517_virus_2644.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1518_bacteria_3969.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1518_virus_2645.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1519_bacteria_3970.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1519_virus_2646.jpeg \n inflating: chest_xray/train/PNEUMONIA/person151_virus_301.jpeg \n inflating: chest_xray/train/PNEUMONIA/person151_virus_302.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1520_bacteria_3971.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1520_virus_2647.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1521_virus_2649.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1522_bacteria_3977.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1522_virus_2651.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1523_bacteria_3979.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1523_bacteria_3980.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1524_bacteria_3983.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1524_bacteria_3984.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1524_virus_2658.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1525_bacteria_3985.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1525_virus_2659.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1526_bacteria_3986.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1526_virus_2660.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1527_bacteria_3988.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1527_bacteria_3989.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1527_bacteria_3990.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1527_virus_2661.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1528_bacteria_3991.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1528_bacteria_3996.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1528_virus_2662.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1529_virus_2663.jpeg \n inflating: chest_xray/train/PNEUMONIA/person152_virus_303.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1530_bacteria_4000.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1530_virus_2664.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1531_bacteria_4003.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1531_virus_2666.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1532_virus_2667.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1534_virus_2670.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1535_bacteria_4015.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1535_bacteria_4016.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1535_bacteria_4017.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1535_virus_2672.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1536_bacteria_4018.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1536_virus_2673.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1537_bacteria_4019.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1537_bacteria_4020.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1537_virus_2674.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1538_bacteria_4021.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1539_bacteria_4022.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1539_virus_2678.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1539_virus_2679.jpeg \n inflating: chest_xray/train/PNEUMONIA/person153_virus_304.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1540_bacteria_4023.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1540_virus_2680.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1541_virus_2681.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1542_bacteria_4029.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1542_virus_2683.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1543_virus_2684.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1544_bacteria_4033.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1544_bacteria_4035.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1544_bacteria_4037.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1544_bacteria_4038.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1544_virus_2685.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1545_bacteria_4041.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1545_bacteria_4042.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1546_bacteria_4044.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1546_bacteria_4045.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1546_virus_2687.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1547_virus_2688.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1548_bacteria_4048.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1548_virus_2689.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1549_bacteria_4050.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1549_virus_2690.jpeg \n inflating: chest_xray/train/PNEUMONIA/person154_virus_305.jpeg \n inflating: chest_xray/train/PNEUMONIA/person154_virus_306.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1550_bacteria_4051.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1550_virus_2691.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1551_bacteria_4053.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1551_bacteria_4054.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1551_virus_2692.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1552_bacteria_4055.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1554_bacteria_4057.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1554_virus_2696.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1555_bacteria_4058.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1555_bacteria_4059.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1555_bacteria_4060.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1556_bacteria_4061.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1556_bacteria_4062.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1556_virus_2699.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1557_bacteria_4063.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1557_bacteria_4065.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1558_bacteria_4066.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1559_bacteria_4067.jpeg \n inflating: chest_xray/train/PNEUMONIA/person155_virus_307.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1561_bacteria_4077.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1562_bacteria_4078.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1562_bacteria_4081.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1562_bacteria_4087.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1562_bacteria_4089.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1563_bacteria_4092.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1564_bacteria_4094.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1564_virus_2719.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1565_bacteria_4095.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1566_bacteria_4099.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1567_bacteria_4100.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1567_virus_2722.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1568_virus_2723.jpeg \n inflating: chest_xray/train/PNEUMONIA/person156_virus_308.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1571_bacteria_4108.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1571_bacteria_4110.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1571_virus_2728.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1574_bacteria_4118.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1575_bacteria_4119.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4120.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4121.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4122.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4124.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4126.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4127.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4128.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_bacteria_4129.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1576_virus_2734.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1577_virus_2735.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1579_bacteria_4133.jpeg \n inflating: chest_xray/train/PNEUMONIA/person157_virus_311.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1580_virus_2739.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1581_bacteria_4135.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1581_virus_2741.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1582_bacteria_4136.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1582_bacteria_4137.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1582_bacteria_4140.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1582_bacteria_4142.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1582_bacteria_4143.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1583_bacteria_4144.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1584_bacteria_4146.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1584_bacteria_4148.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1585_bacteria_4149.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1585_bacteria_4151.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1585_bacteria_4155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1588_virus_2762.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1589_bacteria_4171.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1589_bacteria_4172.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1589_virus_2763.jpeg \n inflating: chest_xray/train/PNEUMONIA/person158_virus_312.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1590_bacteria_4174.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1590_bacteria_4175.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1590_bacteria_4176.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1590_virus_2764.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1591_bacteria_4177.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1591_virus_2765.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1592_bacteria_4178.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1592_virus_2766.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1593_virus_2767.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1594_bacteria_4182.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1594_virus_2768.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1595_bacteria_4183.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1595_virus_2771.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1596_bacteria_4184.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4187.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4188.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4189.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4190.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4192.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4193.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1597_bacteria_4194.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1598_bacteria_4195.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1598_bacteria_4197.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1598_bacteria_4198.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1599_bacteria_4200.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1599_bacteria_4201.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1599_virus_2775.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1599_virus_2776.jpeg \n inflating: chest_xray/train/PNEUMONIA/person15_bacteria_52.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1600_bacteria_4202.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1600_virus_2777.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1601_bacteria_4209.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1601_bacteria_4212.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1602_bacteria_4218.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1602_virus_2780.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1603_virus_2781.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1604_virus_2782.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1605_bacteria_4226.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1607_bacteria_4232.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1607_virus_2785.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1608_bacteria_4235.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1609_bacteria_4236.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1609_bacteria_4237.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1609_bacteria_4239.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1609_virus_2790.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1609_virus_2791.jpeg \n inflating: chest_xray/train/PNEUMONIA/person160_virus_316.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1610_bacteria_4240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1611_bacteria_4241.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1613_bacteria_4247.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1614_bacteria_4248.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1615_bacteria_4249.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1616_bacteria_4251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1617_bacteria_4254.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1617_bacteria_4255.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1617_bacteria_4256.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1618_bacteria_4258.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4261.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4262.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4266.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4267.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4268.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4269.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4270.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1619_bacteria_4271.jpeg \n inflating: chest_xray/train/PNEUMONIA/person161_virus_317.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1620_bacteria_4272.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1625_bacteria_4290.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1626_bacteria_4291.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1628_bacteria_4293.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1628_bacteria_4294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1628_bacteria_4295.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1628_bacteria_4296.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1628_bacteria_4297.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1628_bacteria_4298.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1629_bacteria_4299.jpeg \n inflating: chest_xray/train/PNEUMONIA/person162_virus_319.jpeg \n inflating: chest_xray/train/PNEUMONIA/person162_virus_320.jpeg \n inflating: chest_xray/train/PNEUMONIA/person162_virus_321.jpeg \n inflating: chest_xray/train/PNEUMONIA/person162_virus_322.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1630_bacteria_4303.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1630_bacteria_4304.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1634_bacteria_4326.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1634_bacteria_4331.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1634_bacteria_4334.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1635_bacteria_4335.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1636_bacteria_4337.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1636_bacteria_4338.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1637_bacteria_4339.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1638_bacteria_4340.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1638_bacteria_4341.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1638_bacteria_4342.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1639_bacteria_4343.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1639_bacteria_4344.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1639_bacteria_4345.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1639_bacteria_4347.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1640_bacteria_4348.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1641_bacteria_4350.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1642_bacteria_4352.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1642_bacteria_4353.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1643_bacteria_4354.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1644_bacteria_4356.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1644_bacteria_4357.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1644_bacteria_4358.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1644_bacteria_4360.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1644_bacteria_4361.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1644_bacteria_4362.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1645_bacteria_4363.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1646_bacteria_4368.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1647_bacteria_4372.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1648_bacteria_4373.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1648_bacteria_4375.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1648_bacteria_4376.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1649_bacteria_4377.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1649_bacteria_4378.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1649_bacteria_4379.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1651_bacteria_4381.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1652_bacteria_4383.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1657_bacteria_4398.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1657_bacteria_4399.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1657_bacteria_4400.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1658_bacteria_4402.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1660_bacteria_4404.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1661_bacteria_4406.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1663_bacteria_4411.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1663_bacteria_4412.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1665_bacteria_4415.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1666_bacteria_4416.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1667_bacteria_4417.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1667_bacteria_4418.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1668_bacteria_4420.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1668_bacteria_4421.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1669_bacteria_4422.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1669_bacteria_4423.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1669_bacteria_4424.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1670_bacteria_4425.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1670_bacteria_4426.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1670_bacteria_4427.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1670_bacteria_4428.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1670_bacteria_4429.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1670_bacteria_4430.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1670_bacteria_4431.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1671_bacteria_4432.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1673_bacteria_4434.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1674_bacteria_4437.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1676_bacteria_4441.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1677_bacteria_4443.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1677_bacteria_4444.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1678_bacteria_4446.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1679_bacteria_4448.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1679_bacteria_4449.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1679_bacteria_4450.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1679_bacteria_4452.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1679_bacteria_4453.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1680_bacteria_4455.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1682_bacteria_4459.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1683_bacteria_4460.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1684_bacteria_4461.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1684_bacteria_4462.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1684_bacteria_4463.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1685_bacteria_4465.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1686_bacteria_4466.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1687_bacteria_4468.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1689_bacteria_4472.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1689_bacteria_4473.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1689_bacteria_4474.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1690_bacteria_4475.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1691_bacteria_4479.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1691_bacteria_4481.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1693_bacteria_4485.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1695_bacteria_4492.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1696_bacteria_4495.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1697_bacteria_4496.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1698_bacteria_4497.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1699_bacteria_4498.jpeg \n inflating: chest_xray/train/PNEUMONIA/person16_bacteria_53.jpeg \n inflating: chest_xray/train/PNEUMONIA/person16_bacteria_54.jpeg \n inflating: chest_xray/train/PNEUMONIA/person16_bacteria_55.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1700_bacteria_4500.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1700_bacteria_4502.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1701_bacteria_4504.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1702_bacteria_4506.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1702_bacteria_4508.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1702_bacteria_4509.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1702_bacteria_4510.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1702_bacteria_4511.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1705_bacteria_4515.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1706_bacteria_4516.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1707_bacteria_4520.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1708_bacteria_4521.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1709_bacteria_4522.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1709_bacteria_4523.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1709_bacteria_4524.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1710_bacteria_4525.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1710_bacteria_4526.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1711_bacteria_4527.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1712_bacteria_4529.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1713_bacteria_4530.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1715_bacteria_4532.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1716_bacteria_4533.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1717_bacteria_4534.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1717_bacteria_4536.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1718_bacteria_4538.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1718_bacteria_4540.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1719_bacteria_4541.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1719_bacteria_4542.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1719_bacteria_4544.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1720_bacteria_4545.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1721_bacteria_4546.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1722_bacteria_4547.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1723_bacteria_4548.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1724_bacteria_4549.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1725_bacteria_4550.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1725_bacteria_4551.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1726_bacteria_4552.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1728_bacteria_4555.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1728_bacteria_4556.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1729_bacteria_4557.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1730_bacteria_4558.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1730_bacteria_4559.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1731_bacteria_4563.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1732_bacteria_4564.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1733_bacteria_4566.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1735_bacteria_4570.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1737_bacteria_4573.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1739_bacteria_4576.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1740_bacteria_4579.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1744_bacteria_4583.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1745_bacteria_4584.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1746_bacteria_4585.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1748_bacteria_4588.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1749_bacteria_4590.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1751_bacteria_4592.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1753_bacteria_4594.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1756_bacteria_4598.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1757_bacteria_4599.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1758_bacteria_4600.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1760_bacteria_4602.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1761_bacteria_4603.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1763_bacteria_4606.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1764_bacteria_4607.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1765_bacteria_4608.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1770_bacteria_4613.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1770_bacteria_4614.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1771_bacteria_4615.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1777_bacteria_4622.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1779_bacteria_4626.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1784_bacteria_4631.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1787_bacteria_4634.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1790_bacteria_4638.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1796_bacteria_4644.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1799_bacteria_4647.jpeg \n inflating: chest_xray/train/PNEUMONIA/person17_bacteria_56.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1803_bacteria_4651.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1803_bacteria_4652.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1810_bacteria_4664.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1812_bacteria_4667.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1814_bacteria_4669.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1816_bacteria_4673.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1816_bacteria_4674.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1817_bacteria_4675.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1818_bacteria_4676.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1819_bacteria_4677.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1823_bacteria_4682.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1830_bacteria_4693.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1835_bacteria_4699.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1838_bacteria_4703.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1839_bacteria_4705.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1841_bacteria_4708.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1843_bacteria_4710.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1847_bacteria_4716.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1848_bacteria_4719.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1850_bacteria_4721.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1851_bacteria_4722.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1852_bacteria_4724.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1855_bacteria_4727.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1857_bacteria_4729.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1858_bacteria_4730.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1859_bacteria_4731.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1860_bacteria_4732.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1863_bacteria_4735.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1864_bacteria_4736.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1865_bacteria_4737.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1865_bacteria_4739.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1866_bacteria_4740.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1867_bacteria_4741.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1868_bacteria_4743.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1869_bacteria_4745.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1872_bacteria_4750.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1872_bacteria_4751.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1875_bacteria_4756.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1876_bacteria_4760.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1877_bacteria_4761.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1879_bacteria_4764.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1880_bacteria_4765.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1881_bacteria_4767.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1883_bacteria_4769.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1884_bacteria_4771.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1885_bacteria_4772.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1886_bacteria_4773.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1888_bacteria_4775.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1893_bacteria_4781.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1896_bacteria_4788.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1897_bacteria_4789.jpeg \n inflating: chest_xray/train/PNEUMONIA/person18_bacteria_57.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1901_bacteria_4795.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1903_bacteria_4797.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1904_bacteria_4798.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1905_bacteria_4801.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1906_bacteria_4803.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1907_bacteria_4806.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1908_bacteria_4811.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1910_bacteria_4814.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1911_bacteria_4815.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1912_bacteria_4816.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1912_bacteria_4817.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1916_bacteria_4821.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1917_bacteria_4823.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1918_bacteria_4825.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1921_bacteria_4828.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1922_bacteria_4830.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1923_bacteria_4831.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1924_bacteria_4832.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1924_bacteria_4833.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1926_bacteria_4835.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1927_bacteria_4836.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1927_bacteria_4837.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1929_bacteria_4839.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1930_bacteria_4841.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1931_bacteria_4842.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1932_bacteria_4843.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1933_bacteria_4844.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1934_bacteria_4846.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1935_bacteria_4847.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1935_bacteria_4848.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1935_bacteria_4849.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1935_bacteria_4850.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1936_bacteria_4852.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1937_bacteria_4853.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1938_bacteria_4854.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1940_bacteria_4859.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1940_bacteria_4861.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1940_bacteria_4862.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1941_bacteria_4863.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1942_bacteria_4865.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1943_bacteria_4868.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1944_bacteria_4869.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1945_bacteria_4872.jpeg \n inflating: chest_xray/train/PNEUMONIA/person19_bacteria_58.jpeg \n inflating: chest_xray/train/PNEUMONIA/person19_bacteria_59.jpeg \n inflating: chest_xray/train/PNEUMONIA/person19_bacteria_60.jpeg \n inflating: chest_xray/train/PNEUMONIA/person19_bacteria_61.jpeg \n inflating: chest_xray/train/PNEUMONIA/person19_bacteria_62.jpeg \n inflating: chest_xray/train/PNEUMONIA/person19_bacteria_63.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1_bacteria_1.jpeg \n inflating: chest_xray/train/PNEUMONIA/person1_bacteria_2.jpeg \n inflating: chest_xray/train/PNEUMONIA/person20_bacteria_64.jpeg \n inflating: chest_xray/train/PNEUMONIA/person20_bacteria_66.jpeg \n inflating: chest_xray/train/PNEUMONIA/person20_bacteria_67.jpeg \n inflating: chest_xray/train/PNEUMONIA/person20_bacteria_69.jpeg \n inflating: chest_xray/train/PNEUMONIA/person20_bacteria_70.jpeg \n inflating: chest_xray/train/PNEUMONIA/person21_bacteria_72.jpeg \n inflating: chest_xray/train/PNEUMONIA/person21_bacteria_73.jpeg \n inflating: chest_xray/train/PNEUMONIA/person22_bacteria_74.jpeg \n inflating: chest_xray/train/PNEUMONIA/person22_bacteria_76.jpeg \n inflating: chest_xray/train/PNEUMONIA/person22_bacteria_77.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_100.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_101.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_102.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_103.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_104.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_105.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_106.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_107.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_78.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_79.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_80.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_81.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_82.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_83.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_84.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_85.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_86.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_87.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_88.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_89.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_90.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_91.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_92.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_93.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_94.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_95.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_96.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_97.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_98.jpeg \n inflating: chest_xray/train/PNEUMONIA/person23_bacteria_99.jpeg \n inflating: chest_xray/train/PNEUMONIA/person24_bacteria_108.jpeg \n inflating: chest_xray/train/PNEUMONIA/person24_bacteria_109.jpeg \n inflating: chest_xray/train/PNEUMONIA/person24_bacteria_110.jpeg \n inflating: chest_xray/train/PNEUMONIA/person24_bacteria_111.jpeg \n inflating: chest_xray/train/PNEUMONIA/person24_bacteria_112.jpeg \n inflating: chest_xray/train/PNEUMONIA/person253_bacteria_1152.jpeg \n inflating: chest_xray/train/PNEUMONIA/person253_bacteria_1153.jpeg \n inflating: chest_xray/train/PNEUMONIA/person253_bacteria_1154.jpeg \n inflating: chest_xray/train/PNEUMONIA/person253_bacteria_1155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person253_bacteria_1156.jpeg \n inflating: chest_xray/train/PNEUMONIA/person253_bacteria_1157.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_bacteria_1160.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_bacteria_1161.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_bacteria_1162.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_bacteria_1165.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_bacteria_1175.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_bacteria_1182.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_bacteria_1188.jpeg \n inflating: chest_xray/train/PNEUMONIA/person255_virus_531.jpeg \n inflating: chest_xray/train/PNEUMONIA/person256_bacteria_1189.jpeg \n inflating: chest_xray/train/PNEUMONIA/person256_virus_537.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1193.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1194.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1195.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1196.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1197.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1199.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_bacteria_1200.jpeg \n inflating: chest_xray/train/PNEUMONIA/person257_virus_538.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1205.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1206.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1207.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1208.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1209.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1210.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1212.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1214.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1215.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_bacteria_1216.jpeg \n inflating: chest_xray/train/PNEUMONIA/person258_virus_539.jpeg \n inflating: chest_xray/train/PNEUMONIA/person259_bacteria_1217.jpeg \n inflating: chest_xray/train/PNEUMONIA/person259_bacteria_1219.jpeg \n inflating: chest_xray/train/PNEUMONIA/person259_bacteria_1220.jpeg \n inflating: chest_xray/train/PNEUMONIA/person259_virus_540.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_113.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_114.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_115.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_116.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_117.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_118.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_119.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_120.jpeg \n inflating: chest_xray/train/PNEUMONIA/person25_bacteria_121.jpeg \n inflating: chest_xray/train/PNEUMONIA/person260_bacteria_1222.jpeg \n inflating: chest_xray/train/PNEUMONIA/person260_bacteria_1223.jpeg \n inflating: chest_xray/train/PNEUMONIA/person260_bacteria_1224.jpeg \n inflating: chest_xray/train/PNEUMONIA/person260_virus_541.jpeg \n inflating: chest_xray/train/PNEUMONIA/person261_bacteria_1225.jpeg \n inflating: chest_xray/train/PNEUMONIA/person261_virus_543.jpeg \n inflating: chest_xray/train/PNEUMONIA/person262_bacteria_1226.jpeg \n inflating: chest_xray/train/PNEUMONIA/person262_virus_544.jpeg \n inflating: chest_xray/train/PNEUMONIA/person262_virus_545.jpeg \n inflating: chest_xray/train/PNEUMONIA/person263_bacteria_1227.jpeg \n inflating: chest_xray/train/PNEUMONIA/person263_virus_546.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_bacteria_1228.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_bacteria_1229.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_bacteria_1230.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_bacteria_1231.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_bacteria_1232.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_bacteria_1233.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_bacteria_1234.jpeg \n inflating: chest_xray/train/PNEUMONIA/person264_virus_547.jpeg \n inflating: chest_xray/train/PNEUMONIA/person265_bacteria_1235.jpeg \n inflating: chest_xray/train/PNEUMONIA/person265_bacteria_1236.jpeg \n inflating: chest_xray/train/PNEUMONIA/person265_virus_548.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1237.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1238.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1239.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1241.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1242.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1244.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1245.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1247.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1248.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_bacteria_1249.jpeg \n inflating: chest_xray/train/PNEUMONIA/person266_virus_549.jpeg \n inflating: chest_xray/train/PNEUMONIA/person267_bacteria_1250.jpeg \n inflating: chest_xray/train/PNEUMONIA/person267_bacteria_1251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person267_bacteria_1252.jpeg \n inflating: chest_xray/train/PNEUMONIA/person267_bacteria_1253.jpeg \n inflating: chest_xray/train/PNEUMONIA/person267_virus_552.jpeg \n inflating: chest_xray/train/PNEUMONIA/person268_virus_553.jpeg \n inflating: chest_xray/train/PNEUMONIA/person269_virus_554.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_122.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_123.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_124.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_126.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_127.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_128.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_129.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_130.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_131.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_132.jpeg \n inflating: chest_xray/train/PNEUMONIA/person26_bacteria_133.jpeg \n inflating: chest_xray/train/PNEUMONIA/person270_virus_555.jpeg \n inflating: chest_xray/train/PNEUMONIA/person271_virus_556.jpeg \n inflating: chest_xray/train/PNEUMONIA/person272_virus_559.jpeg \n inflating: chest_xray/train/PNEUMONIA/person273_virus_561.jpeg \n inflating: chest_xray/train/PNEUMONIA/person273_virus_562.jpeg \n inflating: chest_xray/train/PNEUMONIA/person274_bacteria_1288.jpeg \n inflating: chest_xray/train/PNEUMONIA/person274_bacteria_1289.jpeg \n inflating: chest_xray/train/PNEUMONIA/person274_bacteria_1290.jpeg \n inflating: chest_xray/train/PNEUMONIA/person274_virus_563.jpeg \n inflating: chest_xray/train/PNEUMONIA/person275_bacteria_1291.jpeg \n inflating: chest_xray/train/PNEUMONIA/person275_bacteria_1293.jpeg \n inflating: chest_xray/train/PNEUMONIA/person275_bacteria_1294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person275_virus_565.jpeg \n inflating: chest_xray/train/PNEUMONIA/person276_bacteria_1295.jpeg \n inflating: chest_xray/train/PNEUMONIA/person276_bacteria_1296.jpeg \n inflating: chest_xray/train/PNEUMONIA/person276_bacteria_1297.jpeg \n inflating: chest_xray/train/PNEUMONIA/person276_bacteria_1298.jpeg \n inflating: chest_xray/train/PNEUMONIA/person276_bacteria_1299.jpeg \n inflating: chest_xray/train/PNEUMONIA/person276_virus_569.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1300.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1301.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1302.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1303.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1304.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1305.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1306.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_bacteria_1307.jpeg \n inflating: chest_xray/train/PNEUMONIA/person277_virus_571.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_bacteria_1309.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_bacteria_1311.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_bacteria_1313.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_bacteria_1314.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_virus_572.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_virus_573.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_virus_574.jpeg \n inflating: chest_xray/train/PNEUMONIA/person278_virus_575.jpeg \n inflating: chest_xray/train/PNEUMONIA/person279_bacteria_1315.jpeg \n inflating: chest_xray/train/PNEUMONIA/person279_bacteria_1316.jpeg \n inflating: chest_xray/train/PNEUMONIA/person279_virus_576.jpeg \n inflating: chest_xray/train/PNEUMONIA/person27_bacteria_135.jpeg \n inflating: chest_xray/train/PNEUMONIA/person27_bacteria_136.jpeg \n inflating: chest_xray/train/PNEUMONIA/person27_bacteria_137.jpeg \n inflating: chest_xray/train/PNEUMONIA/person27_bacteria_138.jpeg \n inflating: chest_xray/train/PNEUMONIA/person280_bacteria_1318.jpeg \n inflating: chest_xray/train/PNEUMONIA/person280_bacteria_1320.jpeg \n inflating: chest_xray/train/PNEUMONIA/person280_bacteria_1321.jpeg \n inflating: chest_xray/train/PNEUMONIA/person280_bacteria_1322.jpeg \n inflating: chest_xray/train/PNEUMONIA/person280_virus_577.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1323.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1324.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1325.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1326.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1327.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1328.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1329.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1330.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1331.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1332.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_bacteria_1333.jpeg \n inflating: chest_xray/train/PNEUMONIA/person281_virus_578.jpeg \n inflating: chest_xray/train/PNEUMONIA/person282_virus_579.jpeg \n inflating: chest_xray/train/PNEUMONIA/person284_virus_582.jpeg \n inflating: chest_xray/train/PNEUMONIA/person286_virus_585.jpeg \n inflating: chest_xray/train/PNEUMONIA/person287_bacteria_1354.jpeg \n inflating: chest_xray/train/PNEUMONIA/person287_bacteria_1355.jpeg \n inflating: chest_xray/train/PNEUMONIA/person287_virus_586.jpeg \n inflating: chest_xray/train/PNEUMONIA/person288_virus_587.jpeg \n inflating: chest_xray/train/PNEUMONIA/person289_virus_593.jpeg \n inflating: chest_xray/train/PNEUMONIA/person28_bacteria_139.jpeg \n inflating: chest_xray/train/PNEUMONIA/person28_bacteria_141.jpeg \n inflating: chest_xray/train/PNEUMONIA/person28_bacteria_142.jpeg \n inflating: chest_xray/train/PNEUMONIA/person28_bacteria_143.jpeg \n inflating: chest_xray/train/PNEUMONIA/person290_bacteria_1372.jpeg \n inflating: chest_xray/train/PNEUMONIA/person290_virus_594.jpeg \n inflating: chest_xray/train/PNEUMONIA/person291_bacteria_1374.jpeg \n inflating: chest_xray/train/PNEUMONIA/person291_bacteria_1376.jpeg \n inflating: chest_xray/train/PNEUMONIA/person291_bacteria_1377.jpeg \n inflating: chest_xray/train/PNEUMONIA/person291_virus_596.jpeg \n inflating: chest_xray/train/PNEUMONIA/person292_bacteria_1378.jpeg \n inflating: chest_xray/train/PNEUMONIA/person292_virus_597.jpeg \n inflating: chest_xray/train/PNEUMONIA/person292_virus_598.jpeg \n inflating: chest_xray/train/PNEUMONIA/person292_virus_599.jpeg \n inflating: chest_xray/train/PNEUMONIA/person292_virus_600.jpeg \n inflating: chest_xray/train/PNEUMONIA/person292_virus_602.jpeg \n inflating: chest_xray/train/PNEUMONIA/person293_bacteria_1379.jpeg \n inflating: chest_xray/train/PNEUMONIA/person293_virus_604.jpeg \n inflating: chest_xray/train/PNEUMONIA/person293_virus_605.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1380.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1381.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1382.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1383.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1384.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1385.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1386.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_bacteria_1388.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_virus_606.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_virus_610.jpeg \n inflating: chest_xray/train/PNEUMONIA/person294_virus_611.jpeg \n inflating: chest_xray/train/PNEUMONIA/person295_bacteria_1389.jpeg \n inflating: chest_xray/train/PNEUMONIA/person295_bacteria_1390.jpeg \n inflating: chest_xray/train/PNEUMONIA/person295_virus_612.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_bacteria_1391.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_bacteria_1392.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_bacteria_1393.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_bacteria_1394.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_bacteria_1395.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_bacteria_1396.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_bacteria_1397.jpeg \n inflating: chest_xray/train/PNEUMONIA/person296_virus_613.jpeg \n inflating: chest_xray/train/PNEUMONIA/person297_bacteria_1400.jpeg \n inflating: chest_xray/train/PNEUMONIA/person297_bacteria_1404.jpeg \n inflating: chest_xray/train/PNEUMONIA/person297_virus_614.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_bacteria_1408.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_bacteria_1409.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_bacteria_1410.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_bacteria_1411.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_bacteria_1412.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_bacteria_1413.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_virus_617.jpeg \n inflating: chest_xray/train/PNEUMONIA/person298_virus_618.jpeg \n inflating: chest_xray/train/PNEUMONIA/person299_bacteria_1414.jpeg \n inflating: chest_xray/train/PNEUMONIA/person299_bacteria_1416.jpeg \n inflating: chest_xray/train/PNEUMONIA/person299_bacteria_1417.jpeg \n inflating: chest_xray/train/PNEUMONIA/person299_bacteria_1418.jpeg \n inflating: chest_xray/train/PNEUMONIA/person299_bacteria_1419.jpeg \n inflating: chest_xray/train/PNEUMONIA/person299_virus_620.jpeg \n inflating: chest_xray/train/PNEUMONIA/person29_bacteria_144.jpeg \n inflating: chest_xray/train/PNEUMONIA/person2_bacteria_3.jpeg \n inflating: chest_xray/train/PNEUMONIA/person2_bacteria_4.jpeg \n inflating: chest_xray/train/PNEUMONIA/person300_bacteria_1421.jpeg \n inflating: chest_xray/train/PNEUMONIA/person300_bacteria_1422.jpeg \n inflating: chest_xray/train/PNEUMONIA/person300_bacteria_1423.jpeg \n inflating: chest_xray/train/PNEUMONIA/person300_virus_621.jpeg \n inflating: chest_xray/train/PNEUMONIA/person301_bacteria_1424.jpeg \n inflating: chest_xray/train/PNEUMONIA/person301_bacteria_1427.jpeg \n inflating: chest_xray/train/PNEUMONIA/person301_bacteria_1428.jpeg \n inflating: chest_xray/train/PNEUMONIA/person301_bacteria_1429.jpeg \n inflating: chest_xray/train/PNEUMONIA/person301_virus_622.jpeg \n inflating: chest_xray/train/PNEUMONIA/person302_bacteria_1430.jpeg \n inflating: chest_xray/train/PNEUMONIA/person302_virus_623.jpeg \n inflating: chest_xray/train/PNEUMONIA/person303_bacteria_1431.jpeg \n inflating: chest_xray/train/PNEUMONIA/person303_virus_624.jpeg \n inflating: chest_xray/train/PNEUMONIA/person304_virus_625.jpeg \n inflating: chest_xray/train/PNEUMONIA/person305_bacteria_1435.jpeg \n inflating: chest_xray/train/PNEUMONIA/person305_bacteria_1436.jpeg \n inflating: chest_xray/train/PNEUMONIA/person305_bacteria_1437.jpeg \n inflating: chest_xray/train/PNEUMONIA/person305_virus_627.jpeg \n inflating: chest_xray/train/PNEUMONIA/person306_bacteria_1439.jpeg \n inflating: chest_xray/train/PNEUMONIA/person306_bacteria_1440.jpeg \n inflating: chest_xray/train/PNEUMONIA/person306_virus_628.jpeg \n inflating: chest_xray/train/PNEUMONIA/person307_bacteria_1441.jpeg \n inflating: chest_xray/train/PNEUMONIA/person307_bacteria_1442.jpeg \n inflating: chest_xray/train/PNEUMONIA/person307_virus_629.jpeg \n inflating: chest_xray/train/PNEUMONIA/person308_bacteria_1443.jpeg \n inflating: chest_xray/train/PNEUMONIA/person308_bacteria_1445.jpeg \n inflating: chest_xray/train/PNEUMONIA/person308_virus_630.jpeg \n inflating: chest_xray/train/PNEUMONIA/person309_bacteria_1447.jpeg \n inflating: chest_xray/train/PNEUMONIA/person309_bacteria_1449.jpeg \n inflating: chest_xray/train/PNEUMONIA/person309_virus_631.jpeg \n inflating: chest_xray/train/PNEUMONIA/person309_virus_632.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_145.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_146.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_147.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_148.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_149.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_150.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_151.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_152.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_153.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_154.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_156.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_157.jpeg \n inflating: chest_xray/train/PNEUMONIA/person30_bacteria_158.jpeg \n inflating: chest_xray/train/PNEUMONIA/person310_bacteria_1450.jpeg \n inflating: chest_xray/train/PNEUMONIA/person310_bacteria_1451.jpeg \n inflating: chest_xray/train/PNEUMONIA/person310_virus_633.jpeg \n inflating: chest_xray/train/PNEUMONIA/person311_bacteria_1452.jpeg \n inflating: chest_xray/train/PNEUMONIA/person311_bacteria_1453.jpeg \n inflating: chest_xray/train/PNEUMONIA/person311_virus_634.jpeg \n inflating: chest_xray/train/PNEUMONIA/person312_bacteria_1454.jpeg \n inflating: chest_xray/train/PNEUMONIA/person312_bacteria_1455.jpeg \n inflating: chest_xray/train/PNEUMONIA/person312_bacteria_1456.jpeg \n inflating: chest_xray/train/PNEUMONIA/person312_virus_635.jpeg \n inflating: chest_xray/train/PNEUMONIA/person313_bacteria_1457.jpeg \n inflating: chest_xray/train/PNEUMONIA/person313_bacteria_1458.jpeg \n inflating: chest_xray/train/PNEUMONIA/person313_bacteria_1459.jpeg \n inflating: chest_xray/train/PNEUMONIA/person313_bacteria_1460.jpeg \n inflating: chest_xray/train/PNEUMONIA/person313_virus_637.jpeg \n inflating: chest_xray/train/PNEUMONIA/person314_bacteria_1461.jpeg \n inflating: chest_xray/train/PNEUMONIA/person314_bacteria_1462.jpeg \n inflating: chest_xray/train/PNEUMONIA/person314_virus_639.jpeg \n inflating: chest_xray/train/PNEUMONIA/person315_bacteria_1464.jpeg \n inflating: chest_xray/train/PNEUMONIA/person315_bacteria_1465.jpeg \n inflating: chest_xray/train/PNEUMONIA/person315_bacteria_1466.jpeg \n inflating: chest_xray/train/PNEUMONIA/person315_bacteria_1467.jpeg \n inflating: chest_xray/train/PNEUMONIA/person315_bacteria_1468.jpeg \n inflating: chest_xray/train/PNEUMONIA/person316_bacteria_1469.jpeg \n inflating: chest_xray/train/PNEUMONIA/person316_bacteria_1470.jpeg \n inflating: chest_xray/train/PNEUMONIA/person316_virus_641.jpeg \n inflating: chest_xray/train/PNEUMONIA/person317_bacteria_1471.jpeg \n inflating: chest_xray/train/PNEUMONIA/person317_bacteria_1473.jpeg \n inflating: chest_xray/train/PNEUMONIA/person317_virus_643.jpeg \n inflating: chest_xray/train/PNEUMONIA/person318_bacteria_1474.jpeg \n inflating: chest_xray/train/PNEUMONIA/person318_virus_644.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_bacteria_1475.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_bacteria_1476.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_bacteria_1477.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_bacteria_1478.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_bacteria_1479.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_bacteria_1480.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_virus_645.jpeg \n inflating: chest_xray/train/PNEUMONIA/person319_virus_646.jpeg \n inflating: chest_xray/train/PNEUMONIA/person31_bacteria_160.jpeg \n inflating: chest_xray/train/PNEUMONIA/person31_bacteria_161.jpeg \n inflating: chest_xray/train/PNEUMONIA/person31_bacteria_162.jpeg \n inflating: chest_xray/train/PNEUMONIA/person31_bacteria_163.jpeg \n inflating: chest_xray/train/PNEUMONIA/person31_bacteria_164.jpeg \n inflating: chest_xray/train/PNEUMONIA/person320_virus_647.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_bacteria_1483.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_bacteria_1484.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_bacteria_1485.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_bacteria_1486.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_bacteria_1487.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_bacteria_1488.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_bacteria_1489.jpeg \n inflating: chest_xray/train/PNEUMONIA/person321_virus_648.jpeg \n inflating: chest_xray/train/PNEUMONIA/person322_bacteria_1494.jpeg \n inflating: chest_xray/train/PNEUMONIA/person322_virus_655.jpeg \n inflating: chest_xray/train/PNEUMONIA/person323_virus_656.jpeg \n inflating: chest_xray/train/PNEUMONIA/person324_virus_658.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_bacteria_1497.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_bacteria_1498.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_bacteria_1500.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_bacteria_1501.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_bacteria_1502.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_virus_659.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_virus_660.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_virus_661.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_virus_664.jpeg \n inflating: chest_xray/train/PNEUMONIA/person325_virus_665.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_bacteria_1503.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_bacteria_1504.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_bacteria_1505.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_bacteria_1506.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_bacteria_1507.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_virus_666.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_virus_668.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_virus_670.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_virus_672.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_virus_673.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_virus_675.jpeg \n inflating: chest_xray/train/PNEUMONIA/person326_virus_677.jpeg \n inflating: chest_xray/train/PNEUMONIA/person327_bacteria_1508.jpeg \n inflating: chest_xray/train/PNEUMONIA/person327_bacteria_1509.jpeg \n inflating: chest_xray/train/PNEUMONIA/person327_virus_679.jpeg \n inflating: chest_xray/train/PNEUMONIA/person328_bacteria_1510.jpeg \n inflating: chest_xray/train/PNEUMONIA/person328_bacteria_1511.jpeg \n inflating: chest_xray/train/PNEUMONIA/person328_bacteria_1512.jpeg \n inflating: chest_xray/train/PNEUMONIA/person328_bacteria_1513.jpeg \n inflating: chest_xray/train/PNEUMONIA/person328_bacteria_1514.jpeg \n inflating: chest_xray/train/PNEUMONIA/person328_bacteria_1515.jpeg \n inflating: chest_xray/train/PNEUMONIA/person328_virus_681.jpeg \n inflating: chest_xray/train/PNEUMONIA/person329_virus_682.jpeg \n inflating: chest_xray/train/PNEUMONIA/person32_bacteria_165.jpeg \n inflating: chest_xray/train/PNEUMONIA/person32_bacteria_166.jpeg \n inflating: chest_xray/train/PNEUMONIA/person330_virus_683.jpeg \n inflating: chest_xray/train/PNEUMONIA/person331_bacteria_1526.jpeg \n inflating: chest_xray/train/PNEUMONIA/person331_bacteria_1527.jpeg \n inflating: chest_xray/train/PNEUMONIA/person331_bacteria_1528.jpeg \n inflating: chest_xray/train/PNEUMONIA/person331_bacteria_1529.jpeg \n inflating: chest_xray/train/PNEUMONIA/person331_bacteria_1530.jpeg \n inflating: chest_xray/train/PNEUMONIA/person331_virus_684.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_bacteria_1531.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_bacteria_1533.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_bacteria_1534.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_bacteria_1535.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_bacteria_1536.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_bacteria_1537.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_bacteria_1538.jpeg \n inflating: chest_xray/train/PNEUMONIA/person332_virus_685.jpeg \n inflating: chest_xray/train/PNEUMONIA/person333_bacteria_1539.jpeg \n inflating: chest_xray/train/PNEUMONIA/person333_bacteria_1540.jpeg \n inflating: chest_xray/train/PNEUMONIA/person333_virus_688.jpeg \n inflating: chest_xray/train/PNEUMONIA/person334_bacteria_1541.jpeg \n inflating: chest_xray/train/PNEUMONIA/person334_bacteria_1542.jpeg \n inflating: chest_xray/train/PNEUMONIA/person334_bacteria_1544.jpeg \n inflating: chest_xray/train/PNEUMONIA/person334_bacteria_1545.jpeg \n inflating: chest_xray/train/PNEUMONIA/person334_virus_689.jpeg \n inflating: chest_xray/train/PNEUMONIA/person335_virus_690.jpeg \n inflating: chest_xray/train/PNEUMONIA/person336_bacteria_1548.jpeg \n inflating: chest_xray/train/PNEUMONIA/person336_bacteria_1549.jpeg \n inflating: chest_xray/train/PNEUMONIA/person336_bacteria_1550.jpeg \n inflating: chest_xray/train/PNEUMONIA/person336_bacteria_1551.jpeg \n inflating: chest_xray/train/PNEUMONIA/person336_bacteria_1552.jpeg \n inflating: chest_xray/train/PNEUMONIA/person336_bacteria_1553.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1554.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1557.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1558.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1560.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1561.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1562.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1563.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1564.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1565.jpeg \n inflating: chest_xray/train/PNEUMONIA/person337_bacteria_1566.jpeg \n inflating: chest_xray/train/PNEUMONIA/person338_bacteria_1567.jpeg \n inflating: chest_xray/train/PNEUMONIA/person338_bacteria_1568.jpeg \n inflating: chest_xray/train/PNEUMONIA/person338_virus_694.jpeg \n inflating: chest_xray/train/PNEUMONIA/person339_bacteria_1572.jpeg \n inflating: chest_xray/train/PNEUMONIA/person339_bacteria_1573.jpeg \n inflating: chest_xray/train/PNEUMONIA/person339_bacteria_1574.jpeg \n inflating: chest_xray/train/PNEUMONIA/person339_virus_695.jpeg \n inflating: chest_xray/train/PNEUMONIA/person33_bacteria_169.jpeg \n inflating: chest_xray/train/PNEUMONIA/person33_bacteria_172.jpeg \n inflating: chest_xray/train/PNEUMONIA/person33_bacteria_173.jpeg \n inflating: chest_xray/train/PNEUMONIA/person33_bacteria_174.jpeg \n inflating: chest_xray/train/PNEUMONIA/person33_bacteria_175.jpeg \n inflating: chest_xray/train/PNEUMONIA/person340_bacteria_1575.jpeg \n inflating: chest_xray/train/PNEUMONIA/person340_virus_698.jpeg \n inflating: chest_xray/train/PNEUMONIA/person341_bacteria_1577.jpeg \n inflating: chest_xray/train/PNEUMONIA/person341_virus_699.jpeg \n inflating: chest_xray/train/PNEUMONIA/person342_virus_701.jpeg \n inflating: chest_xray/train/PNEUMONIA/person342_virus_702.jpeg \n inflating: chest_xray/train/PNEUMONIA/person343_bacteria_1583.jpeg \n inflating: chest_xray/train/PNEUMONIA/person343_bacteria_1584.jpeg \n inflating: chest_xray/train/PNEUMONIA/person343_virus_704.jpeg \n inflating: chest_xray/train/PNEUMONIA/person344_bacteria_1585.jpeg \n inflating: chest_xray/train/PNEUMONIA/person344_virus_705.jpeg \n inflating: chest_xray/train/PNEUMONIA/person346_bacteria_1590.jpeg \n inflating: chest_xray/train/PNEUMONIA/person346_virus_708.jpeg \n inflating: chest_xray/train/PNEUMONIA/person346_virus_709.jpeg \n inflating: chest_xray/train/PNEUMONIA/person347_bacteria_1594.jpeg \n inflating: chest_xray/train/PNEUMONIA/person347_bacteria_1595.jpeg \n inflating: chest_xray/train/PNEUMONIA/person347_bacteria_1597.jpeg \n inflating: chest_xray/train/PNEUMONIA/person347_bacteria_1599.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_bacteria_1601.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_bacteria_1602.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_bacteria_1603.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_bacteria_1604.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_711.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_714.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_715.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_716.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_717.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_719.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_720.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_721.jpeg \n inflating: chest_xray/train/PNEUMONIA/person348_virus_723.jpeg \n inflating: chest_xray/train/PNEUMONIA/person349_bacteria_1605.jpeg \n inflating: chest_xray/train/PNEUMONIA/person349_bacteria_1606.jpeg \n inflating: chest_xray/train/PNEUMONIA/person349_bacteria_1607.jpeg \n inflating: chest_xray/train/PNEUMONIA/person349_virus_724.jpeg \n inflating: chest_xray/train/PNEUMONIA/person34_bacteria_176.jpeg \n inflating: chest_xray/train/PNEUMONIA/person350_virus_725.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_bacteria_1617.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_bacteria_1619.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_bacteria_1620.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_bacteria_1621.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_bacteria_1622.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_bacteria_1623.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_bacteria_1624.jpeg \n inflating: chest_xray/train/PNEUMONIA/person351_virus_726.jpeg \n inflating: chest_xray/train/PNEUMONIA/person352_bacteria_1625.jpeg \n inflating: chest_xray/train/PNEUMONIA/person353_bacteria_1626.jpeg \n inflating: chest_xray/train/PNEUMONIA/person353_bacteria_1628.jpeg \n inflating: chest_xray/train/PNEUMONIA/person353_virus_728.jpeg \n inflating: chest_xray/train/PNEUMONIA/person354_bacteria_1632.jpeg \n inflating: chest_xray/train/PNEUMONIA/person354_bacteria_1633.jpeg \n inflating: chest_xray/train/PNEUMONIA/person354_bacteria_1634.jpeg \n inflating: chest_xray/train/PNEUMONIA/person354_bacteria_1635.jpeg \n inflating: chest_xray/train/PNEUMONIA/person354_virus_729.jpeg \n inflating: chest_xray/train/PNEUMONIA/person355_bacteria_1637.jpeg \n inflating: chest_xray/train/PNEUMONIA/person355_virus_730.jpeg \n inflating: chest_xray/train/PNEUMONIA/person355_virus_731.jpeg \n inflating: chest_xray/train/PNEUMONIA/person356_bacteria_1638.jpeg \n inflating: chest_xray/train/PNEUMONIA/person356_virus_733.jpeg \n inflating: chest_xray/train/PNEUMONIA/person357_bacteria_1639.jpeg \n inflating: chest_xray/train/PNEUMONIA/person357_bacteria_1640.jpeg \n inflating: chest_xray/train/PNEUMONIA/person357_virus_734.jpeg \n inflating: chest_xray/train/PNEUMONIA/person357_virus_735.jpeg \n inflating: chest_xray/train/PNEUMONIA/person357_virus_736.jpeg \n inflating: chest_xray/train/PNEUMONIA/person358_virus_737.jpeg \n inflating: chest_xray/train/PNEUMONIA/person359_bacteria_1642.jpeg \n inflating: chest_xray/train/PNEUMONIA/person359_bacteria_1643.jpeg \n inflating: chest_xray/train/PNEUMONIA/person359_bacteria_1644.jpeg \n inflating: chest_xray/train/PNEUMONIA/person359_bacteria_1645.jpeg \n inflating: chest_xray/train/PNEUMONIA/person359_bacteria_1646.jpeg \n inflating: chest_xray/train/PNEUMONIA/person359_virus_738.jpeg \n inflating: chest_xray/train/PNEUMONIA/person35_bacteria_178.jpeg \n inflating: chest_xray/train/PNEUMONIA/person35_bacteria_180.jpeg \n inflating: chest_xray/train/PNEUMONIA/person35_bacteria_181.jpeg \n inflating: chest_xray/train/PNEUMONIA/person360_bacteria_1647.jpeg \n inflating: chest_xray/train/PNEUMONIA/person360_virus_739.jpeg \n inflating: chest_xray/train/PNEUMONIA/person361_bacteria_1651.jpeg \n inflating: chest_xray/train/PNEUMONIA/person361_virus_740.jpeg \n inflating: chest_xray/train/PNEUMONIA/person362_bacteria_1652.jpeg \n inflating: chest_xray/train/PNEUMONIA/person362_virus_741.jpeg \n inflating: chest_xray/train/PNEUMONIA/person363_bacteria_1653.jpeg \n inflating: chest_xray/train/PNEUMONIA/person363_bacteria_1654.jpeg \n inflating: chest_xray/train/PNEUMONIA/person363_bacteria_1655.jpeg \n inflating: chest_xray/train/PNEUMONIA/person363_virus_742.jpeg \n inflating: chest_xray/train/PNEUMONIA/person364_bacteria_1656.jpeg \n inflating: chest_xray/train/PNEUMONIA/person364_bacteria_1657.jpeg \n inflating: chest_xray/train/PNEUMONIA/person364_bacteria_1658.jpeg \n inflating: chest_xray/train/PNEUMONIA/person364_bacteria_1659.jpeg \n inflating: chest_xray/train/PNEUMONIA/person364_bacteria_1660.jpeg \n inflating: chest_xray/train/PNEUMONIA/person364_virus_743.jpeg \n inflating: chest_xray/train/PNEUMONIA/person365_virus_745.jpeg \n inflating: chest_xray/train/PNEUMONIA/person366_bacteria_1664.jpeg \n inflating: chest_xray/train/PNEUMONIA/person366_virus_746.jpeg \n inflating: chest_xray/train/PNEUMONIA/person367_bacteria_1665.jpeg \n inflating: chest_xray/train/PNEUMONIA/person367_virus_747.jpeg \n inflating: chest_xray/train/PNEUMONIA/person368_bacteria_1666.jpeg \n inflating: chest_xray/train/PNEUMONIA/person368_bacteria_1667.jpeg \n inflating: chest_xray/train/PNEUMONIA/person368_bacteria_1668.jpeg \n inflating: chest_xray/train/PNEUMONIA/person368_bacteria_1672.jpeg \n inflating: chest_xray/train/PNEUMONIA/person368_bacteria_1678.jpeg \n inflating: chest_xray/train/PNEUMONIA/person368_virus_748.jpeg \n inflating: chest_xray/train/PNEUMONIA/person369_bacteria_1680.jpeg \n inflating: chest_xray/train/PNEUMONIA/person369_virus_750.jpeg \n inflating: chest_xray/train/PNEUMONIA/person36_bacteria_182.jpeg \n inflating: chest_xray/train/PNEUMONIA/person36_bacteria_183.jpeg \n inflating: chest_xray/train/PNEUMONIA/person36_bacteria_184.jpeg \n inflating: chest_xray/train/PNEUMONIA/person36_bacteria_185.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_bacteria_1687.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_bacteria_1688.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_bacteria_1689.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_bacteria_1690.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_bacteria_1691.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_bacteria_1692.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_virus_752.jpeg \n inflating: chest_xray/train/PNEUMONIA/person370_virus_753.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1694.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1695.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1696.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1698.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1699.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1700.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1701.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1702.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_bacteria_1703.jpeg \n inflating: chest_xray/train/PNEUMONIA/person371_virus_754.jpeg \n inflating: chest_xray/train/PNEUMONIA/person372_bacteria_1704.jpeg \n inflating: chest_xray/train/PNEUMONIA/person372_bacteria_1705.jpeg \n inflating: chest_xray/train/PNEUMONIA/person372_bacteria_1706.jpeg \n inflating: chest_xray/train/PNEUMONIA/person372_virus_755.jpeg \n inflating: chest_xray/train/PNEUMONIA/person373_bacteria_1707.jpeg \n inflating: chest_xray/train/PNEUMONIA/person373_bacteria_1708.jpeg \n inflating: chest_xray/train/PNEUMONIA/person373_bacteria_1709.jpeg \n inflating: chest_xray/train/PNEUMONIA/person373_virus_756.jpeg \n inflating: chest_xray/train/PNEUMONIA/person374_bacteria_1710.jpeg \n inflating: chest_xray/train/PNEUMONIA/person374_bacteria_1711.jpeg \n inflating: chest_xray/train/PNEUMONIA/person374_bacteria_1712.jpeg \n inflating: chest_xray/train/PNEUMONIA/person374_virus_757.jpeg \n inflating: chest_xray/train/PNEUMONIA/person375_bacteria_1713.jpeg \n inflating: chest_xray/train/PNEUMONIA/person375_virus_758.jpeg \n inflating: chest_xray/train/PNEUMONIA/person376_bacteria_1715.jpeg \n inflating: chest_xray/train/PNEUMONIA/person376_bacteria_1716.jpeg \n inflating: chest_xray/train/PNEUMONIA/person376_virus_759.jpeg \n inflating: chest_xray/train/PNEUMONIA/person377_bacteria_1717.jpeg \n inflating: chest_xray/train/PNEUMONIA/person377_bacteria_1718.jpeg \n inflating: chest_xray/train/PNEUMONIA/person377_virus_760.jpeg \n inflating: chest_xray/train/PNEUMONIA/person378_virus_761.jpeg \n inflating: chest_xray/train/PNEUMONIA/person379_bacteria_1721.jpeg \n inflating: chest_xray/train/PNEUMONIA/person379_bacteria_1722.jpeg \n inflating: chest_xray/train/PNEUMONIA/person379_virus_762.jpeg \n inflating: chest_xray/train/PNEUMONIA/person37_bacteria_186.jpeg \n inflating: chest_xray/train/PNEUMONIA/person37_bacteria_187.jpeg \n inflating: chest_xray/train/PNEUMONIA/person37_bacteria_188.jpeg \n inflating: chest_xray/train/PNEUMONIA/person37_bacteria_189.jpeg \n inflating: chest_xray/train/PNEUMONIA/person380_virus_763.jpeg \n inflating: chest_xray/train/PNEUMONIA/person381_bacteria_1730.jpeg \n inflating: chest_xray/train/PNEUMONIA/person381_bacteria_1731.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1737.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1738.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1739.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1740.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1741.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1742.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1745.jpeg \n inflating: chest_xray/train/PNEUMONIA/person382_bacteria_1746.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1747.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1748.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1749.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1750.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1751.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1752.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1753.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_bacteria_1754.jpeg \n inflating: chest_xray/train/PNEUMONIA/person383_virus_767.jpeg \n inflating: chest_xray/train/PNEUMONIA/person384_bacteria_1755.jpeg \n inflating: chest_xray/train/PNEUMONIA/person384_virus_769.jpeg \n inflating: chest_xray/train/PNEUMONIA/person385_bacteria_1765.jpeg \n inflating: chest_xray/train/PNEUMONIA/person385_bacteria_1766.jpeg \n inflating: chest_xray/train/PNEUMONIA/person385_virus_770.jpeg \n inflating: chest_xray/train/PNEUMONIA/person386_virus_771.jpeg \n inflating: chest_xray/train/PNEUMONIA/person387_bacteria_1769.jpeg \n inflating: chest_xray/train/PNEUMONIA/person387_bacteria_1770.jpeg \n inflating: chest_xray/train/PNEUMONIA/person387_bacteria_1772.jpeg \n inflating: chest_xray/train/PNEUMONIA/person387_virus_772.jpeg \n inflating: chest_xray/train/PNEUMONIA/person388_virus_775.jpeg \n inflating: chest_xray/train/PNEUMONIA/person388_virus_777.jpeg \n inflating: chest_xray/train/PNEUMONIA/person389_bacteria_1778.jpeg \n inflating: chest_xray/train/PNEUMONIA/person389_bacteria_1780.jpeg \n inflating: chest_xray/train/PNEUMONIA/person389_virus_778.jpeg \n inflating: chest_xray/train/PNEUMONIA/person38_bacteria_190.jpeg \n inflating: chest_xray/train/PNEUMONIA/person38_bacteria_191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person38_bacteria_192.jpeg \n inflating: chest_xray/train/PNEUMONIA/person38_bacteria_193.jpeg \n inflating: chest_xray/train/PNEUMONIA/person38_bacteria_194.jpeg \n inflating: chest_xray/train/PNEUMONIA/person38_bacteria_195.jpeg \n inflating: chest_xray/train/PNEUMONIA/person38_bacteria_196.jpeg \n inflating: chest_xray/train/PNEUMONIA/person390_bacteria_1781.jpeg \n inflating: chest_xray/train/PNEUMONIA/person391_bacteria_1782.jpeg \n inflating: chest_xray/train/PNEUMONIA/person391_virus_781.jpeg \n inflating: chest_xray/train/PNEUMONIA/person392_bacteria_1783.jpeg \n inflating: chest_xray/train/PNEUMONIA/person392_bacteria_1784.jpeg \n inflating: chest_xray/train/PNEUMONIA/person392_bacteria_1785.jpeg \n inflating: chest_xray/train/PNEUMONIA/person392_bacteria_1786.jpeg \n inflating: chest_xray/train/PNEUMONIA/person392_bacteria_1787.jpeg \n inflating: chest_xray/train/PNEUMONIA/person392_virus_782.jpeg \n inflating: chest_xray/train/PNEUMONIA/person393_bacteria_1789.jpeg \n inflating: chest_xray/train/PNEUMONIA/person393_virus_784.jpeg \n inflating: chest_xray/train/PNEUMONIA/person394_bacteria_1791.jpeg \n inflating: chest_xray/train/PNEUMONIA/person394_bacteria_1792.jpeg \n inflating: chest_xray/train/PNEUMONIA/person394_virus_786.jpeg \n inflating: chest_xray/train/PNEUMONIA/person395_bacteria_1794.jpeg \n inflating: chest_xray/train/PNEUMONIA/person395_bacteria_1795.jpeg \n inflating: chest_xray/train/PNEUMONIA/person395_virus_788.jpeg \n inflating: chest_xray/train/PNEUMONIA/person396_bacteria_1796.jpeg \n inflating: chest_xray/train/PNEUMONIA/person396_virus_789.jpeg \n inflating: chest_xray/train/PNEUMONIA/person397_bacteria_1797.jpeg \n inflating: chest_xray/train/PNEUMONIA/person397_virus_790.jpeg \n inflating: chest_xray/train/PNEUMONIA/person398_bacteria_1799.jpeg \n inflating: chest_xray/train/PNEUMONIA/person398_bacteria_1801.jpeg \n inflating: chest_xray/train/PNEUMONIA/person399_bacteria_1804.jpeg \n inflating: chest_xray/train/PNEUMONIA/person399_bacteria_1805.jpeg \n inflating: chest_xray/train/PNEUMONIA/person399_bacteria_1806.jpeg \n inflating: chest_xray/train/PNEUMONIA/person399_virus_793.jpeg \n inflating: chest_xray/train/PNEUMONIA/person39_bacteria_198.jpeg \n inflating: chest_xray/train/PNEUMONIA/person39_bacteria_200.jpeg \n inflating: chest_xray/train/PNEUMONIA/person3_bacteria_10.jpeg \n inflating: chest_xray/train/PNEUMONIA/person3_bacteria_11.jpeg \n inflating: chest_xray/train/PNEUMONIA/person3_bacteria_12.jpeg \n inflating: chest_xray/train/PNEUMONIA/person3_bacteria_13.jpeg \n inflating: chest_xray/train/PNEUMONIA/person400_bacteria_1807.jpeg \n inflating: chest_xray/train/PNEUMONIA/person400_virus_794.jpeg \n inflating: chest_xray/train/PNEUMONIA/person401_bacteria_1808.jpeg \n inflating: chest_xray/train/PNEUMONIA/person401_virus_795.jpeg \n inflating: chest_xray/train/PNEUMONIA/person401_virus_797.jpeg \n inflating: chest_xray/train/PNEUMONIA/person401_virus_798.jpeg \n inflating: chest_xray/train/PNEUMONIA/person402_bacteria_1809.jpeg \n inflating: chest_xray/train/PNEUMONIA/person402_bacteria_1810.jpeg \n inflating: chest_xray/train/PNEUMONIA/person402_bacteria_1811.jpeg \n inflating: chest_xray/train/PNEUMONIA/person402_bacteria_1812.jpeg \n inflating: chest_xray/train/PNEUMONIA/person402_bacteria_1813.jpeg \n inflating: chest_xray/train/PNEUMONIA/person402_virus_799.jpeg \n inflating: chest_xray/train/PNEUMONIA/person402_virus_801.jpeg \n inflating: chest_xray/train/PNEUMONIA/person403_bacteria_1814.jpeg \n inflating: chest_xray/train/PNEUMONIA/person403_virus_803.jpeg \n inflating: chest_xray/train/PNEUMONIA/person405_bacteria_1817.jpeg \n inflating: chest_xray/train/PNEUMONIA/person405_virus_805.jpeg \n inflating: chest_xray/train/PNEUMONIA/person406_bacteria_1818.jpeg \n inflating: chest_xray/train/PNEUMONIA/person406_bacteria_1819.jpeg \n inflating: chest_xray/train/PNEUMONIA/person406_bacteria_1820.jpeg \n inflating: chest_xray/train/PNEUMONIA/person407_bacteria_1822.jpeg \n inflating: chest_xray/train/PNEUMONIA/person407_virus_811.jpeg \n inflating: chest_xray/train/PNEUMONIA/person407_virus_812.jpeg \n inflating: chest_xray/train/PNEUMONIA/person407_virus_814.jpeg \n inflating: chest_xray/train/PNEUMONIA/person408_bacteria_1823.jpeg \n inflating: chest_xray/train/PNEUMONIA/person408_virus_815.jpeg \n inflating: chest_xray/train/PNEUMONIA/person409_bacteria_1824.jpeg \n inflating: chest_xray/train/PNEUMONIA/person409_virus_816.jpeg \n inflating: chest_xray/train/PNEUMONIA/person409_virus_818.jpeg \n inflating: chest_xray/train/PNEUMONIA/person409_virus_820.jpeg \n inflating: chest_xray/train/PNEUMONIA/person40_bacteria_202.jpeg \n inflating: chest_xray/train/PNEUMONIA/person40_bacteria_203.jpeg \n inflating: chest_xray/train/PNEUMONIA/person40_bacteria_204.jpeg \n inflating: chest_xray/train/PNEUMONIA/person40_bacteria_205.jpeg \n inflating: chest_xray/train/PNEUMONIA/person410_bacteria_1825.jpeg \n inflating: chest_xray/train/PNEUMONIA/person410_virus_821.jpeg \n inflating: chest_xray/train/PNEUMONIA/person411_bacteria_1826.jpeg \n inflating: chest_xray/train/PNEUMONIA/person412_bacteria_1827.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_bacteria_1828.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_bacteria_1829.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_bacteria_1830.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_bacteria_1831.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_bacteria_1832.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_bacteria_1833.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_bacteria_1834.jpeg \n inflating: chest_xray/train/PNEUMONIA/person413_virus_844.jpeg \n inflating: chest_xray/train/PNEUMONIA/person414_bacteria_1835.jpeg \n inflating: chest_xray/train/PNEUMONIA/person414_virus_845.jpeg \n inflating: chest_xray/train/PNEUMONIA/person415_bacteria_1837.jpeg \n inflating: chest_xray/train/PNEUMONIA/person415_bacteria_1838.jpeg \n inflating: chest_xray/train/PNEUMONIA/person415_bacteria_1839.jpeg \n inflating: chest_xray/train/PNEUMONIA/person415_virus_847.jpeg \n inflating: chest_xray/train/PNEUMONIA/person416_bacteria_1840.jpeg \n inflating: chest_xray/train/PNEUMONIA/person416_virus_849.jpeg \n inflating: chest_xray/train/PNEUMONIA/person417_bacteria_1841.jpeg \n inflating: chest_xray/train/PNEUMONIA/person417_bacteria_1842.jpeg \n inflating: chest_xray/train/PNEUMONIA/person417_virus_850.jpeg \n inflating: chest_xray/train/PNEUMONIA/person418_bacteria_1843.jpeg \n inflating: chest_xray/train/PNEUMONIA/person418_virus_852.jpeg \n inflating: chest_xray/train/PNEUMONIA/person419_bacteria_1844.jpeg \n inflating: chest_xray/train/PNEUMONIA/person419_bacteria_1845.jpeg \n inflating: chest_xray/train/PNEUMONIA/person419_virus_855.jpeg \n inflating: chest_xray/train/PNEUMONIA/person419_virus_857.jpeg \n inflating: chest_xray/train/PNEUMONIA/person419_virus_859.jpeg \n inflating: chest_xray/train/PNEUMONIA/person419_virus_861.jpeg \n inflating: chest_xray/train/PNEUMONIA/person41_bacteria_206.jpeg \n inflating: chest_xray/train/PNEUMONIA/person41_bacteria_207.jpeg \n inflating: chest_xray/train/PNEUMONIA/person41_bacteria_208.jpeg \n inflating: chest_xray/train/PNEUMONIA/person41_bacteria_209.jpeg \n inflating: chest_xray/train/PNEUMONIA/person41_bacteria_210.jpeg \n inflating: chest_xray/train/PNEUMONIA/person41_bacteria_211.jpeg \n inflating: chest_xray/train/PNEUMONIA/person420_bacteria_1847.jpeg \n inflating: chest_xray/train/PNEUMONIA/person420_bacteria_1848.jpeg \n inflating: chest_xray/train/PNEUMONIA/person420_bacteria_1849.jpeg \n inflating: chest_xray/train/PNEUMONIA/person420_bacteria_1850.jpeg \n inflating: chest_xray/train/PNEUMONIA/person420_bacteria_1851.jpeg \n inflating: chest_xray/train/PNEUMONIA/person421_bacteria_1852.jpeg \n inflating: chest_xray/train/PNEUMONIA/person421_virus_866.jpeg \n inflating: chest_xray/train/PNEUMONIA/person422_bacteria_1853.jpeg \n inflating: chest_xray/train/PNEUMONIA/person422_virus_867.jpeg \n inflating: chest_xray/train/PNEUMONIA/person422_virus_868.jpeg \n inflating: chest_xray/train/PNEUMONIA/person423_bacteria_1854.jpeg \n inflating: chest_xray/train/PNEUMONIA/person423_bacteria_1855.jpeg \n inflating: chest_xray/train/PNEUMONIA/person423_bacteria_1856.jpeg \n inflating: chest_xray/train/PNEUMONIA/person423_bacteria_1857.jpeg \n inflating: chest_xray/train/PNEUMONIA/person423_bacteria_1858.jpeg \n inflating: chest_xray/train/PNEUMONIA/person423_virus_869.jpeg \n inflating: chest_xray/train/PNEUMONIA/person424_bacteria_1859.jpeg \n inflating: chest_xray/train/PNEUMONIA/person425_bacteria_1860.jpeg \n inflating: chest_xray/train/PNEUMONIA/person425_virus_871.jpeg \n inflating: chest_xray/train/PNEUMONIA/person426_bacteria_1861.jpeg \n inflating: chest_xray/train/PNEUMONIA/person426_bacteria_1862.jpeg \n inflating: chest_xray/train/PNEUMONIA/person426_bacteria_1863.jpeg \n inflating: chest_xray/train/PNEUMONIA/person426_virus_873.jpeg \n inflating: chest_xray/train/PNEUMONIA/person427_bacteria_1864.jpeg \n inflating: chest_xray/train/PNEUMONIA/person427_bacteria_1865.jpeg \n inflating: chest_xray/train/PNEUMONIA/person427_bacteria_1866.jpeg \n inflating: chest_xray/train/PNEUMONIA/person427_bacteria_1867.jpeg \n inflating: chest_xray/train/PNEUMONIA/person427_bacteria_1868.jpeg \n inflating: chest_xray/train/PNEUMONIA/person427_virus_875.jpeg \n inflating: chest_xray/train/PNEUMONIA/person428_bacteria_1869.jpeg \n inflating: chest_xray/train/PNEUMONIA/person428_virus_876.jpeg \n inflating: chest_xray/train/PNEUMONIA/person429_bacteria_1870.jpeg \n inflating: chest_xray/train/PNEUMONIA/person429_virus_877.jpeg \n inflating: chest_xray/train/PNEUMONIA/person430_bacteria_1871.jpeg \n inflating: chest_xray/train/PNEUMONIA/person430_virus_879.jpeg \n inflating: chest_xray/train/PNEUMONIA/person431_bacteria_1872.jpeg \n inflating: chest_xray/train/PNEUMONIA/person431_virus_880.jpeg \n inflating: chest_xray/train/PNEUMONIA/person432_virus_881.jpeg \n inflating: chest_xray/train/PNEUMONIA/person433_bacteria_1874.jpeg \n inflating: chest_xray/train/PNEUMONIA/person433_bacteria_1875.jpeg \n inflating: chest_xray/train/PNEUMONIA/person433_bacteria_1876.jpeg \n inflating: chest_xray/train/PNEUMONIA/person433_virus_882.jpeg \n inflating: chest_xray/train/PNEUMONIA/person434_bacteria_1877.jpeg \n inflating: chest_xray/train/PNEUMONIA/person434_virus_883.jpeg \n inflating: chest_xray/train/PNEUMONIA/person434_virus_884.jpeg \n inflating: chest_xray/train/PNEUMONIA/person435_bacteria_1879.jpeg \n inflating: chest_xray/train/PNEUMONIA/person435_virus_885.jpeg \n inflating: chest_xray/train/PNEUMONIA/person436_bacteria_1883.jpeg \n inflating: chest_xray/train/PNEUMONIA/person436_virus_886.jpeg \n inflating: chest_xray/train/PNEUMONIA/person437_bacteria_1884.jpeg \n inflating: chest_xray/train/PNEUMONIA/person437_bacteria_1885.jpeg \n inflating: chest_xray/train/PNEUMONIA/person437_bacteria_1886.jpeg \n inflating: chest_xray/train/PNEUMONIA/person437_bacteria_1887.jpeg \n inflating: chest_xray/train/PNEUMONIA/person437_bacteria_1888.jpeg \n inflating: chest_xray/train/PNEUMONIA/person437_virus_888.jpeg \n inflating: chest_xray/train/PNEUMONIA/person438_bacteria_1889.jpeg \n inflating: chest_xray/train/PNEUMONIA/person438_bacteria_1890.jpeg \n inflating: chest_xray/train/PNEUMONIA/person438_bacteria_1891.jpeg \n inflating: chest_xray/train/PNEUMONIA/person438_bacteria_1892.jpeg \n inflating: chest_xray/train/PNEUMONIA/person438_bacteria_1893.jpeg \n inflating: chest_xray/train/PNEUMONIA/person438_virus_889.jpeg \n inflating: chest_xray/train/PNEUMONIA/person439_bacteria_1895.jpeg \n inflating: chest_xray/train/PNEUMONIA/person439_virus_890.jpeg \n inflating: chest_xray/train/PNEUMONIA/person439_virus_891.jpeg \n inflating: chest_xray/train/PNEUMONIA/person43_bacteria_213.jpeg \n inflating: chest_xray/train/PNEUMONIA/person43_bacteria_216.jpeg \n inflating: chest_xray/train/PNEUMONIA/person440_bacteria_1897.jpeg \n inflating: chest_xray/train/PNEUMONIA/person440_bacteria_1898.jpeg \n inflating: chest_xray/train/PNEUMONIA/person440_virus_893.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1900.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1902.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1903.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1904.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1905.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1907.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1910.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1911.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1912.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1914.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1915.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1916.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1917.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_bacteria_1918.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_virus_894.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_virus_895.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_virus_896.jpeg \n inflating: chest_xray/train/PNEUMONIA/person441_virus_897.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_898.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_899.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_900.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_901.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_902.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_903.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_904.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_905.jpeg \n inflating: chest_xray/train/PNEUMONIA/person442_virus_906.jpeg \n inflating: chest_xray/train/PNEUMONIA/person443_bacteria_1923.jpeg \n inflating: chest_xray/train/PNEUMONIA/person443_bacteria_1924.jpeg \n inflating: chest_xray/train/PNEUMONIA/person443_bacteria_1926.jpeg \n inflating: chest_xray/train/PNEUMONIA/person443_virus_908.jpeg \n inflating: chest_xray/train/PNEUMONIA/person444_bacteria_1927.jpeg \n inflating: chest_xray/train/PNEUMONIA/person444_virus_911.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_bacteria_1928.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_bacteria_1929.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_bacteria_1930.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_912.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_913.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_914.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_915.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_916.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_917.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_918.jpeg \n inflating: chest_xray/train/PNEUMONIA/person445_virus_919.jpeg \n inflating: chest_xray/train/PNEUMONIA/person446_bacteria_1931.jpeg \n inflating: chest_xray/train/PNEUMONIA/person446_virus_920.jpeg \n inflating: chest_xray/train/PNEUMONIA/person447_bacteria_1932.jpeg \n inflating: chest_xray/train/PNEUMONIA/person447_virus_921.jpeg \n inflating: chest_xray/train/PNEUMONIA/person447_virus_921_1.jpeg \n inflating: chest_xray/train/PNEUMONIA/person448_bacteria_1933.jpeg \n inflating: chest_xray/train/PNEUMONIA/person448_bacteria_1934.jpeg \n inflating: chest_xray/train/PNEUMONIA/person448_bacteria_1935.jpeg \n inflating: chest_xray/train/PNEUMONIA/person448_bacteria_1936.jpeg \n inflating: chest_xray/train/PNEUMONIA/person448_bacteria_1937.jpeg \n inflating: chest_xray/train/PNEUMONIA/person448_virus_922.jpeg \n inflating: chest_xray/train/PNEUMONIA/person449_bacteria_1938.jpeg \n inflating: chest_xray/train/PNEUMONIA/person449_bacteria_1939.jpeg \n inflating: chest_xray/train/PNEUMONIA/person449_bacteria_1940.jpeg \n inflating: chest_xray/train/PNEUMONIA/person44_bacteria_218.jpeg \n inflating: chest_xray/train/PNEUMONIA/person44_bacteria_219.jpeg \n inflating: chest_xray/train/PNEUMONIA/person450_bacteria_1941.jpeg \n inflating: chest_xray/train/PNEUMONIA/person450_virus_931.jpeg \n inflating: chest_xray/train/PNEUMONIA/person451_bacteria_1942.jpeg \n inflating: chest_xray/train/PNEUMONIA/person451_virus_932.jpeg \n inflating: chest_xray/train/PNEUMONIA/person452_bacteria_1943.jpeg \n inflating: chest_xray/train/PNEUMONIA/person453_virus_935.jpeg \n inflating: chest_xray/train/PNEUMONIA/person453_virus_936.jpeg \n inflating: chest_xray/train/PNEUMONIA/person454_bacteria_1945.jpeg \n inflating: chest_xray/train/PNEUMONIA/person454_virus_938.jpeg \n inflating: chest_xray/train/PNEUMONIA/person455_bacteria_1947.jpeg \n inflating: chest_xray/train/PNEUMONIA/person456_bacteria_1948.jpeg \n inflating: chest_xray/train/PNEUMONIA/person456_virus_943.jpeg \n inflating: chest_xray/train/PNEUMONIA/person457_bacteria_1949.jpeg \n inflating: chest_xray/train/PNEUMONIA/person457_virus_944.jpeg \n inflating: chest_xray/train/PNEUMONIA/person458_bacteria_1950.jpeg \n inflating: chest_xray/train/PNEUMONIA/person458_bacteria_1951.jpeg \n inflating: chest_xray/train/PNEUMONIA/person458_bacteria_1952.jpeg \n inflating: chest_xray/train/PNEUMONIA/person458_bacteria_1953.jpeg \n inflating: chest_xray/train/PNEUMONIA/person458_bacteria_1954.jpeg \n inflating: chest_xray/train/PNEUMONIA/person458_bacteria_1955.jpeg \n inflating: chest_xray/train/PNEUMONIA/person458_virus_945.jpeg \n inflating: chest_xray/train/PNEUMONIA/person459_bacteria_1956.jpeg \n inflating: chest_xray/train/PNEUMONIA/person459_bacteria_1957.jpeg \n inflating: chest_xray/train/PNEUMONIA/person459_virus_947.jpeg \n inflating: chest_xray/train/PNEUMONIA/person45_bacteria_220.jpeg \n inflating: chest_xray/train/PNEUMONIA/person45_bacteria_221.jpeg \n inflating: chest_xray/train/PNEUMONIA/person45_bacteria_222.jpeg \n inflating: chest_xray/train/PNEUMONIA/person460_bacteria_1958.jpeg \n inflating: chest_xray/train/PNEUMONIA/person460_virus_948.jpeg \n inflating: chest_xray/train/PNEUMONIA/person461_bacteria_1960.jpeg \n inflating: chest_xray/train/PNEUMONIA/person461_virus_949.jpeg \n inflating: chest_xray/train/PNEUMONIA/person461_virus_950.jpeg \n inflating: chest_xray/train/PNEUMONIA/person462_bacteria_1961.jpeg \n inflating: chest_xray/train/PNEUMONIA/person462_bacteria_1963.jpeg \n inflating: chest_xray/train/PNEUMONIA/person462_bacteria_1967.jpeg \n inflating: chest_xray/train/PNEUMONIA/person462_bacteria_1968.jpeg \n inflating: chest_xray/train/PNEUMONIA/person462_virus_951.jpeg \n inflating: chest_xray/train/PNEUMONIA/person463_bacteria_1971.jpeg \n inflating: chest_xray/train/PNEUMONIA/person463_virus_952.jpeg \n inflating: chest_xray/train/PNEUMONIA/person463_virus_953.jpeg \n inflating: chest_xray/train/PNEUMONIA/person464_bacteria_1974.jpeg \n inflating: chest_xray/train/PNEUMONIA/person464_bacteria_1975.jpeg \n inflating: chest_xray/train/PNEUMONIA/person464_virus_954.jpeg \n inflating: chest_xray/train/PNEUMONIA/person464_virus_956.jpeg \n inflating: chest_xray/train/PNEUMONIA/person465_bacteria_1976.jpeg \n inflating: chest_xray/train/PNEUMONIA/person465_bacteria_1977.jpeg \n inflating: chest_xray/train/PNEUMONIA/person465_bacteria_1979.jpeg \n inflating: chest_xray/train/PNEUMONIA/person465_bacteria_1980.jpeg \n inflating: chest_xray/train/PNEUMONIA/person465_bacteria_1981.jpeg \n inflating: chest_xray/train/PNEUMONIA/person465_bacteria_1982.jpeg \n inflating: chest_xray/train/PNEUMONIA/person465_virus_957.jpeg \n inflating: chest_xray/train/PNEUMONIA/person466_bacteria_1983.jpeg \n inflating: chest_xray/train/PNEUMONIA/person466_bacteria_1984.jpeg \n inflating: chest_xray/train/PNEUMONIA/person466_bacteria_1986.jpeg \n inflating: chest_xray/train/PNEUMONIA/person466_bacteria_1987.jpeg \n inflating: chest_xray/train/PNEUMONIA/person466_virus_960.jpeg \n inflating: chest_xray/train/PNEUMONIA/person467_bacteria_1988.jpeg \n inflating: chest_xray/train/PNEUMONIA/person467_bacteria_1989.jpeg \n inflating: chest_xray/train/PNEUMONIA/person467_virus_961.jpeg \n inflating: chest_xray/train/PNEUMONIA/person468_bacteria_1990.jpeg \n inflating: chest_xray/train/PNEUMONIA/person468_bacteria_1991.jpeg \n inflating: chest_xray/train/PNEUMONIA/person468_virus_963.jpeg \n inflating: chest_xray/train/PNEUMONIA/person469_bacteria_1992.jpeg \n inflating: chest_xray/train/PNEUMONIA/person469_bacteria_1993.jpeg \n inflating: chest_xray/train/PNEUMONIA/person469_bacteria_1994.jpeg \n inflating: chest_xray/train/PNEUMONIA/person469_bacteria_1995.jpeg \n inflating: chest_xray/train/PNEUMONIA/person469_virus_965.jpeg \n inflating: chest_xray/train/PNEUMONIA/person46_bacteria_224.jpeg \n inflating: chest_xray/train/PNEUMONIA/person46_bacteria_225.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_bacteria_1996.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_bacteria_1998.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_bacteria_1999.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_bacteria_2000.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_bacteria_2001.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_bacteria_2002.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_bacteria_2003.jpeg \n inflating: chest_xray/train/PNEUMONIA/person470_virus_966.jpeg \n inflating: chest_xray/train/PNEUMONIA/person471_bacteria_2004.jpeg \n inflating: chest_xray/train/PNEUMONIA/person471_bacteria_2005.jpeg \n inflating: chest_xray/train/PNEUMONIA/person471_bacteria_2006.jpeg \n inflating: chest_xray/train/PNEUMONIA/person471_virus_967.jpeg \n inflating: chest_xray/train/PNEUMONIA/person471_virus_968.jpeg \n inflating: chest_xray/train/PNEUMONIA/person472_bacteria_2007.jpeg \n inflating: chest_xray/train/PNEUMONIA/person472_bacteria_2008.jpeg \n inflating: chest_xray/train/PNEUMONIA/person472_bacteria_2010.jpeg \n inflating: chest_xray/train/PNEUMONIA/person472_bacteria_2014.jpeg \n inflating: chest_xray/train/PNEUMONIA/person472_bacteria_2015.jpeg \n inflating: chest_xray/train/PNEUMONIA/person472_virus_969.jpeg \n inflating: chest_xray/train/PNEUMONIA/person473_bacteria_2018.jpeg \n inflating: chest_xray/train/PNEUMONIA/person474_virus_971.jpeg \n inflating: chest_xray/train/PNEUMONIA/person475_bacteria_2020.jpeg \n inflating: chest_xray/train/PNEUMONIA/person475_bacteria_2021.jpeg \n inflating: chest_xray/train/PNEUMONIA/person475_bacteria_2022.jpeg \n inflating: chest_xray/train/PNEUMONIA/person475_bacteria_2023.jpeg \n inflating: chest_xray/train/PNEUMONIA/person475_bacteria_2024.jpeg \n inflating: chest_xray/train/PNEUMONIA/person475_bacteria_2025.jpeg \n inflating: chest_xray/train/PNEUMONIA/person475_virus_972.jpeg \n inflating: chest_xray/train/PNEUMONIA/person476_bacteria_2026.jpeg \n inflating: chest_xray/train/PNEUMONIA/person476_virus_973.jpeg \n inflating: chest_xray/train/PNEUMONIA/person477_bacteria_2028.jpeg \n inflating: chest_xray/train/PNEUMONIA/person477_bacteria_2029.jpeg \n inflating: chest_xray/train/PNEUMONIA/person477_bacteria_2030.jpeg \n inflating: chest_xray/train/PNEUMONIA/person477_bacteria_2031.jpeg \n inflating: chest_xray/train/PNEUMONIA/person478_bacteria_2032.jpeg \n inflating: chest_xray/train/PNEUMONIA/person478_bacteria_2035.jpeg \n inflating: chest_xray/train/PNEUMONIA/person478_virus_975.jpeg \n inflating: chest_xray/train/PNEUMONIA/person479_virus_978.jpeg \n inflating: chest_xray/train/PNEUMONIA/person47_bacteria_229.jpeg \n inflating: chest_xray/train/PNEUMONIA/person480_bacteria_2038.jpeg \n inflating: chest_xray/train/PNEUMONIA/person480_bacteria_2039.jpeg \n inflating: chest_xray/train/PNEUMONIA/person480_bacteria_2040.jpeg \n inflating: chest_xray/train/PNEUMONIA/person480_virus_981.jpeg \n inflating: chest_xray/train/PNEUMONIA/person480_virus_982.jpeg \n inflating: chest_xray/train/PNEUMONIA/person481_bacteria_2041.jpeg \n inflating: chest_xray/train/PNEUMONIA/person481_bacteria_2042.jpeg \n inflating: chest_xray/train/PNEUMONIA/person481_virus_983.jpeg \n inflating: chest_xray/train/PNEUMONIA/person482_bacteria_2043.jpeg \n inflating: chest_xray/train/PNEUMONIA/person482_bacteria_2044.jpeg \n inflating: chest_xray/train/PNEUMONIA/person482_bacteria_2045.jpeg \n inflating: chest_xray/train/PNEUMONIA/person482_virus_984.jpeg \n inflating: chest_xray/train/PNEUMONIA/person483_bacteria_2046.jpeg \n inflating: chest_xray/train/PNEUMONIA/person483_virus_985.jpeg \n inflating: chest_xray/train/PNEUMONIA/person484_virus_986.jpeg \n inflating: chest_xray/train/PNEUMONIA/person485_bacteria_2049.jpeg \n inflating: chest_xray/train/PNEUMONIA/person485_virus_988.jpeg \n inflating: chest_xray/train/PNEUMONIA/person486_bacteria_2051.jpeg \n inflating: chest_xray/train/PNEUMONIA/person486_bacteria_2052.jpeg \n inflating: chest_xray/train/PNEUMONIA/person486_bacteria_2053.jpeg \n inflating: chest_xray/train/PNEUMONIA/person486_bacteria_2054.jpeg \n inflating: chest_xray/train/PNEUMONIA/person486_virus_990.jpeg \n inflating: chest_xray/train/PNEUMONIA/person487_bacteria_2055.jpeg \n inflating: chest_xray/train/PNEUMONIA/person487_bacteria_2056.jpeg \n inflating: chest_xray/train/PNEUMONIA/person487_bacteria_2057.jpeg \n inflating: chest_xray/train/PNEUMONIA/person487_bacteria_2058.jpeg \n inflating: chest_xray/train/PNEUMONIA/person487_bacteria_2059.jpeg \n inflating: chest_xray/train/PNEUMONIA/person487_bacteria_2060.jpeg \n inflating: chest_xray/train/PNEUMONIA/person487_virus_991.jpeg \n inflating: chest_xray/train/PNEUMONIA/person488_bacteria_2061.jpeg \n inflating: chest_xray/train/PNEUMONIA/person488_bacteria_2062.jpeg \n inflating: chest_xray/train/PNEUMONIA/person488_virus_992.jpeg \n inflating: chest_xray/train/PNEUMONIA/person489_bacteria_2063.jpeg \n inflating: chest_xray/train/PNEUMONIA/person489_bacteria_2064.jpeg \n inflating: chest_xray/train/PNEUMONIA/person489_bacteria_2065.jpeg \n inflating: chest_xray/train/PNEUMONIA/person489_bacteria_2066.jpeg \n inflating: chest_xray/train/PNEUMONIA/person489_bacteria_2067.jpeg \n inflating: chest_xray/train/PNEUMONIA/person489_virus_994.jpeg \n inflating: chest_xray/train/PNEUMONIA/person489_virus_995.jpeg \n inflating: chest_xray/train/PNEUMONIA/person48_bacteria_230.jpeg \n inflating: chest_xray/train/PNEUMONIA/person48_bacteria_231.jpeg \n inflating: chest_xray/train/PNEUMONIA/person48_bacteria_232.jpeg \n inflating: chest_xray/train/PNEUMONIA/person48_bacteria_233.jpeg \n inflating: chest_xray/train/PNEUMONIA/person490_bacteria_2068.jpeg \n inflating: chest_xray/train/PNEUMONIA/person490_bacteria_2069.jpeg \n inflating: chest_xray/train/PNEUMONIA/person490_bacteria_2070.jpeg \n inflating: chest_xray/train/PNEUMONIA/person490_virus_996.jpeg \n inflating: chest_xray/train/PNEUMONIA/person491_bacteria_2071.jpeg \n inflating: chest_xray/train/PNEUMONIA/person491_bacteria_2073.jpeg \n inflating: chest_xray/train/PNEUMONIA/person491_bacteria_2075.jpeg \n inflating: chest_xray/train/PNEUMONIA/person491_bacteria_2080.jpeg \n inflating: chest_xray/train/PNEUMONIA/person491_bacteria_2081.jpeg \n inflating: chest_xray/train/PNEUMONIA/person491_bacteria_2082.jpeg \n inflating: chest_xray/train/PNEUMONIA/person491_virus_997.jpeg \n inflating: chest_xray/train/PNEUMONIA/person492_bacteria_2083.jpeg \n inflating: chest_xray/train/PNEUMONIA/person492_bacteria_2084.jpeg \n inflating: chest_xray/train/PNEUMONIA/person492_bacteria_2085.jpeg \n inflating: chest_xray/train/PNEUMONIA/person492_virus_998.jpeg \n inflating: chest_xray/train/PNEUMONIA/person493_bacteria_2086.jpeg \n inflating: chest_xray/train/PNEUMONIA/person493_bacteria_2087.jpeg \n inflating: chest_xray/train/PNEUMONIA/person493_virus_999.jpeg \n inflating: chest_xray/train/PNEUMONIA/person494_bacteria_2088.jpeg \n inflating: chest_xray/train/PNEUMONIA/person494_bacteria_2089.jpeg \n inflating: chest_xray/train/PNEUMONIA/person494_bacteria_2090.jpeg \n inflating: chest_xray/train/PNEUMONIA/person494_virus_1000.jpeg \n inflating: chest_xray/train/PNEUMONIA/person495_bacteria_2094.jpeg \n inflating: chest_xray/train/PNEUMONIA/person495_virus_1001.jpeg \n inflating: chest_xray/train/PNEUMONIA/person496_bacteria_2095.jpeg \n inflating: chest_xray/train/PNEUMONIA/person496_virus_1003.jpeg \n inflating: chest_xray/train/PNEUMONIA/person497_virus_1005.jpeg \n inflating: chest_xray/train/PNEUMONIA/person498_bacteria_2100.jpeg \n inflating: chest_xray/train/PNEUMONIA/person498_bacteria_2101.jpeg \n inflating: chest_xray/train/PNEUMONIA/person498_bacteria_2102.jpeg \n inflating: chest_xray/train/PNEUMONIA/person498_virus_1007.jpeg \n inflating: chest_xray/train/PNEUMONIA/person499_bacteria_2103.jpeg \n inflating: chest_xray/train/PNEUMONIA/person499_bacteria_2104.jpeg \n inflating: chest_xray/train/PNEUMONIA/person499_virus_1008.jpeg \n inflating: chest_xray/train/PNEUMONIA/person49_bacteria_235.jpeg \n inflating: chest_xray/train/PNEUMONIA/person49_bacteria_236.jpeg \n inflating: chest_xray/train/PNEUMONIA/person49_bacteria_237.jpeg \n inflating: chest_xray/train/PNEUMONIA/person4_bacteria_14.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_bacteria_2105.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_bacteria_2106.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_bacteria_2107.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_bacteria_2108.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_bacteria_2109.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_bacteria_2110.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_bacteria_2111.jpeg \n inflating: chest_xray/train/PNEUMONIA/person500_virus_1009.jpeg \n inflating: chest_xray/train/PNEUMONIA/person501_bacteria_2112.jpeg \n inflating: chest_xray/train/PNEUMONIA/person501_bacteria_2113.jpeg \n inflating: chest_xray/train/PNEUMONIA/person501_bacteria_2114.jpeg \n inflating: chest_xray/train/PNEUMONIA/person501_bacteria_2115.jpeg \n inflating: chest_xray/train/PNEUMONIA/person501_bacteria_2116.jpeg \n inflating: chest_xray/train/PNEUMONIA/person501_virus_1010.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_bacteria_2117.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_bacteria_2118.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_bacteria_2119.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_bacteria_2120.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_bacteria_2121.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_bacteria_2122.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_bacteria_2123.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_virus_1011.jpeg \n inflating: chest_xray/train/PNEUMONIA/person502_virus_1012.jpeg \n inflating: chest_xray/train/PNEUMONIA/person503_bacteria_2125.jpeg \n inflating: chest_xray/train/PNEUMONIA/person503_bacteria_2126.jpeg \n inflating: chest_xray/train/PNEUMONIA/person503_virus_1013.jpeg \n inflating: chest_xray/train/PNEUMONIA/person504_bacteria_2127.jpeg \n inflating: chest_xray/train/PNEUMONIA/person504_bacteria_2129.jpeg \n inflating: chest_xray/train/PNEUMONIA/person504_bacteria_2130.jpeg \n inflating: chest_xray/train/PNEUMONIA/person504_bacteria_2132.jpeg \n inflating: chest_xray/train/PNEUMONIA/person504_bacteria_2133.jpeg \n inflating: chest_xray/train/PNEUMONIA/person505_bacteria_2135.jpeg \n inflating: chest_xray/train/PNEUMONIA/person505_virus_1017.jpeg \n inflating: chest_xray/train/PNEUMONIA/person506_bacteria_2136.jpeg \n inflating: chest_xray/train/PNEUMONIA/person506_bacteria_2138.jpeg \n inflating: chest_xray/train/PNEUMONIA/person506_virus_1018.jpeg \n inflating: chest_xray/train/PNEUMONIA/person507_bacteria_2139.jpeg \n inflating: chest_xray/train/PNEUMONIA/person507_bacteria_2140.jpeg \n inflating: chest_xray/train/PNEUMONIA/person507_bacteria_2141.jpeg \n inflating: chest_xray/train/PNEUMONIA/person507_virus_1019.jpeg \n inflating: chest_xray/train/PNEUMONIA/person508_bacteria_2142.jpeg \n inflating: chest_xray/train/PNEUMONIA/person508_bacteria_2143.jpeg \n inflating: chest_xray/train/PNEUMONIA/person508_bacteria_2144.jpeg \n inflating: chest_xray/train/PNEUMONIA/person508_virus_1020.jpeg \n inflating: chest_xray/train/PNEUMONIA/person508_virus_1021.jpeg \n inflating: chest_xray/train/PNEUMONIA/person509_bacteria_2145.jpeg \n inflating: chest_xray/train/PNEUMONIA/person509_bacteria_2146.jpeg \n inflating: chest_xray/train/PNEUMONIA/person509_virus_1024.jpeg \n inflating: chest_xray/train/PNEUMONIA/person509_virus_1025.jpeg \n inflating: chest_xray/train/PNEUMONIA/person50_bacteria_238.jpeg \n inflating: chest_xray/train/PNEUMONIA/person510_bacteria_2147.jpeg \n inflating: chest_xray/train/PNEUMONIA/person510_bacteria_2148.jpeg \n inflating: chest_xray/train/PNEUMONIA/person510_bacteria_2149.jpeg \n inflating: chest_xray/train/PNEUMONIA/person510_bacteria_2150.jpeg \n inflating: chest_xray/train/PNEUMONIA/person510_virus_1026.jpeg \n inflating: chest_xray/train/PNEUMONIA/person511_bacteria_2152.jpeg \n inflating: chest_xray/train/PNEUMONIA/person511_bacteria_2153.jpeg \n inflating: chest_xray/train/PNEUMONIA/person511_virus_1027.jpeg \n inflating: chest_xray/train/PNEUMONIA/person512_bacteria_2154.jpeg \n inflating: chest_xray/train/PNEUMONIA/person512_bacteria_2155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person512_virus_1029.jpeg \n inflating: chest_xray/train/PNEUMONIA/person513_virus_1030.jpeg \n inflating: chest_xray/train/PNEUMONIA/person514_bacteria_2184.jpeg \n inflating: chest_xray/train/PNEUMONIA/person514_virus_1031.jpeg \n inflating: chest_xray/train/PNEUMONIA/person515_bacteria_2185.jpeg \n inflating: chest_xray/train/PNEUMONIA/person515_bacteria_2186.jpeg \n inflating: chest_xray/train/PNEUMONIA/person515_bacteria_2187.jpeg \n inflating: chest_xray/train/PNEUMONIA/person515_bacteria_2188.jpeg \n inflating: chest_xray/train/PNEUMONIA/person515_bacteria_2189.jpeg \n inflating: chest_xray/train/PNEUMONIA/person515_bacteria_2190.jpeg \n inflating: chest_xray/train/PNEUMONIA/person515_virus_1032.jpeg \n inflating: chest_xray/train/PNEUMONIA/person516_bacteria_2191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person516_bacteria_2192.jpeg \n inflating: chest_xray/train/PNEUMONIA/person516_virus_1033.jpeg \n inflating: chest_xray/train/PNEUMONIA/person517_bacteria_2196.jpeg \n inflating: chest_xray/train/PNEUMONIA/person517_virus_1034.jpeg \n inflating: chest_xray/train/PNEUMONIA/person517_virus_1035.jpeg \n inflating: chest_xray/train/PNEUMONIA/person518_bacteria_2197.jpeg \n inflating: chest_xray/train/PNEUMONIA/person518_bacteria_2198.jpeg \n inflating: chest_xray/train/PNEUMONIA/person518_bacteria_2199.jpeg \n inflating: chest_xray/train/PNEUMONIA/person518_bacteria_2200.jpeg \n inflating: chest_xray/train/PNEUMONIA/person518_virus_1036.jpeg \n inflating: chest_xray/train/PNEUMONIA/person519_virus_1038.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_239.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_241.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_242.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_243.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_244.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_245.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_246.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_247.jpeg \n inflating: chest_xray/train/PNEUMONIA/person51_bacteria_248.jpeg \n inflating: chest_xray/train/PNEUMONIA/person520_bacteria_2203.jpeg \n inflating: chest_xray/train/PNEUMONIA/person520_bacteria_2204.jpeg \n inflating: chest_xray/train/PNEUMONIA/person520_bacteria_2205.jpeg \n inflating: chest_xray/train/PNEUMONIA/person520_virus_1039.jpeg \n inflating: chest_xray/train/PNEUMONIA/person521_virus_1040.jpeg \n inflating: chest_xray/train/PNEUMONIA/person522_bacteria_2210.jpeg \n inflating: chest_xray/train/PNEUMONIA/person522_bacteria_2211.jpeg \n inflating: chest_xray/train/PNEUMONIA/person522_virus_1041.jpeg \n inflating: chest_xray/train/PNEUMONIA/person523_virus_1043.jpeg \n inflating: chest_xray/train/PNEUMONIA/person524_virus_1045.jpeg \n inflating: chest_xray/train/PNEUMONIA/person525_bacteria_2216.jpeg \n inflating: chest_xray/train/PNEUMONIA/person525_bacteria_2217.jpeg \n inflating: chest_xray/train/PNEUMONIA/person525_bacteria_2218.jpeg \n inflating: chest_xray/train/PNEUMONIA/person525_bacteria_2220.jpeg \n inflating: chest_xray/train/PNEUMONIA/person525_virus_1046.jpeg \n inflating: chest_xray/train/PNEUMONIA/person526_bacteria_2221.jpeg \n inflating: chest_xray/train/PNEUMONIA/person527_bacteria_2225.jpeg \n inflating: chest_xray/train/PNEUMONIA/person527_bacteria_2226.jpeg \n inflating: chest_xray/train/PNEUMONIA/person527_virus_1048.jpeg \n inflating: chest_xray/train/PNEUMONIA/person528_bacteria_2227.jpeg \n inflating: chest_xray/train/PNEUMONIA/person528_virus_1049.jpeg \n inflating: chest_xray/train/PNEUMONIA/person529_bacteria_2228.jpeg \n inflating: chest_xray/train/PNEUMONIA/person529_bacteria_2229.jpeg \n inflating: chest_xray/train/PNEUMONIA/person529_bacteria_2230.jpeg \n inflating: chest_xray/train/PNEUMONIA/person529_virus_1050.jpeg \n inflating: chest_xray/train/PNEUMONIA/person52_bacteria_249.jpeg \n inflating: chest_xray/train/PNEUMONIA/person52_bacteria_251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person530_bacteria_2231.jpeg \n inflating: chest_xray/train/PNEUMONIA/person530_bacteria_2233.jpeg \n inflating: chest_xray/train/PNEUMONIA/person530_virus_1052.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2235.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2236.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2237.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2238.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2239.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2241.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_bacteria_2242.jpeg \n inflating: chest_xray/train/PNEUMONIA/person531_virus_1053.jpeg \n inflating: chest_xray/train/PNEUMONIA/person532_virus_1054.jpeg \n inflating: chest_xray/train/PNEUMONIA/person533_bacteria_2245.jpeg \n inflating: chest_xray/train/PNEUMONIA/person533_bacteria_2250.jpeg \n inflating: chest_xray/train/PNEUMONIA/person533_virus_1055.jpeg \n inflating: chest_xray/train/PNEUMONIA/person534_bacteria_2251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person534_bacteria_2252.jpeg \n inflating: chest_xray/train/PNEUMONIA/person534_bacteria_2253.jpeg \n inflating: chest_xray/train/PNEUMONIA/person534_bacteria_2254.jpeg \n inflating: chest_xray/train/PNEUMONIA/person534_virus_1061.jpeg \n inflating: chest_xray/train/PNEUMONIA/person535_bacteria_2255.jpeg \n inflating: chest_xray/train/PNEUMONIA/person535_bacteria_2256.jpeg \n inflating: chest_xray/train/PNEUMONIA/person535_virus_1062.jpeg \n inflating: chest_xray/train/PNEUMONIA/person536_bacteria_2257.jpeg \n inflating: chest_xray/train/PNEUMONIA/person536_bacteria_2258.jpeg \n inflating: chest_xray/train/PNEUMONIA/person536_bacteria_2259.jpeg \n inflating: chest_xray/train/PNEUMONIA/person536_bacteria_2260.jpeg \n inflating: chest_xray/train/PNEUMONIA/person536_virus_1064.jpeg \n inflating: chest_xray/train/PNEUMONIA/person536_virus_1065.jpeg \n inflating: chest_xray/train/PNEUMONIA/person537_bacteria_2261.jpeg \n inflating: chest_xray/train/PNEUMONIA/person537_bacteria_2262.jpeg \n inflating: chest_xray/train/PNEUMONIA/person537_bacteria_2263.jpeg \n inflating: chest_xray/train/PNEUMONIA/person537_bacteria_2264.jpeg \n inflating: chest_xray/train/PNEUMONIA/person537_bacteria_2265.jpeg \n inflating: chest_xray/train/PNEUMONIA/person537_bacteria_2266.jpeg \n inflating: chest_xray/train/PNEUMONIA/person537_virus_1067.jpeg \n inflating: chest_xray/train/PNEUMONIA/person538_bacteria_2268.jpeg \n inflating: chest_xray/train/PNEUMONIA/person538_virus_1068.jpeg \n inflating: chest_xray/train/PNEUMONIA/person539_bacteria_2269.jpeg \n inflating: chest_xray/train/PNEUMONIA/person539_bacteria_2270.jpeg \n inflating: chest_xray/train/PNEUMONIA/person539_virus_1069.jpeg \n inflating: chest_xray/train/PNEUMONIA/person53_bacteria_252.jpeg \n inflating: chest_xray/train/PNEUMONIA/person53_bacteria_253.jpeg \n inflating: chest_xray/train/PNEUMONIA/person53_bacteria_254.jpeg \n inflating: chest_xray/train/PNEUMONIA/person53_bacteria_255.jpeg \n inflating: chest_xray/train/PNEUMONIA/person540_bacteria_2271.jpeg \n inflating: chest_xray/train/PNEUMONIA/person540_bacteria_2272.jpeg \n inflating: chest_xray/train/PNEUMONIA/person540_bacteria_2273.jpeg \n inflating: chest_xray/train/PNEUMONIA/person540_virus_1070.jpeg \n inflating: chest_xray/train/PNEUMONIA/person541_bacteria_2274.jpeg \n inflating: chest_xray/train/PNEUMONIA/person541_bacteria_2275.jpeg \n inflating: chest_xray/train/PNEUMONIA/person541_virus_1071.jpeg \n inflating: chest_xray/train/PNEUMONIA/person542_bacteria_2276.jpeg \n inflating: chest_xray/train/PNEUMONIA/person542_virus_1072.jpeg \n inflating: chest_xray/train/PNEUMONIA/person543_bacteria_2279.jpeg \n inflating: chest_xray/train/PNEUMONIA/person543_bacteria_2280.jpeg \n inflating: chest_xray/train/PNEUMONIA/person543_bacteria_2281.jpeg \n inflating: chest_xray/train/PNEUMONIA/person543_bacteria_2282.jpeg \n inflating: chest_xray/train/PNEUMONIA/person543_bacteria_2283.jpeg \n inflating: chest_xray/train/PNEUMONIA/person543_bacteria_2284.jpeg \n inflating: chest_xray/train/PNEUMONIA/person543_virus_1073.jpeg \n inflating: chest_xray/train/PNEUMONIA/person544_bacteria_2286.jpeg \n inflating: chest_xray/train/PNEUMONIA/person544_virus_1074.jpeg \n inflating: chest_xray/train/PNEUMONIA/person544_virus_1075.jpeg \n inflating: chest_xray/train/PNEUMONIA/person544_virus_1076.jpeg \n inflating: chest_xray/train/PNEUMONIA/person544_virus_1078.jpeg \n inflating: chest_xray/train/PNEUMONIA/person544_virus_1079.jpeg \n inflating: chest_xray/train/PNEUMONIA/person544_virus_1080.jpeg \n inflating: chest_xray/train/PNEUMONIA/person545_bacteria_2287.jpeg \n inflating: chest_xray/train/PNEUMONIA/person545_bacteria_2288.jpeg \n inflating: chest_xray/train/PNEUMONIA/person545_bacteria_2289.jpeg \n inflating: chest_xray/train/PNEUMONIA/person545_bacteria_2290.jpeg \n inflating: chest_xray/train/PNEUMONIA/person545_virus_1081.jpeg \n inflating: chest_xray/train/PNEUMONIA/person546_virus_1085.jpeg \n inflating: chest_xray/train/PNEUMONIA/person547_bacteria_2292.jpeg \n inflating: chest_xray/train/PNEUMONIA/person547_bacteria_2294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person547_bacteria_2296.jpeg \n inflating: chest_xray/train/PNEUMONIA/person547_virus_1086.jpeg \n inflating: chest_xray/train/PNEUMONIA/person548_bacteria_2297.jpeg \n inflating: chest_xray/train/PNEUMONIA/person548_bacteria_2298.jpeg \n inflating: chest_xray/train/PNEUMONIA/person548_bacteria_2299.jpeg \n inflating: chest_xray/train/PNEUMONIA/person548_bacteria_2300.jpeg \n inflating: chest_xray/train/PNEUMONIA/person548_bacteria_2301.jpeg \n inflating: chest_xray/train/PNEUMONIA/person548_bacteria_2302.jpeg \n inflating: chest_xray/train/PNEUMONIA/person548_virus_1088.jpeg \n inflating: chest_xray/train/PNEUMONIA/person549_bacteria_2303.jpeg \n inflating: chest_xray/train/PNEUMONIA/person549_bacteria_2304.jpeg \n inflating: chest_xray/train/PNEUMONIA/person549_bacteria_2305.jpeg \n inflating: chest_xray/train/PNEUMONIA/person549_bacteria_2306.jpeg \n inflating: chest_xray/train/PNEUMONIA/person549_bacteria_2307.jpeg \n inflating: chest_xray/train/PNEUMONIA/person549_virus_1089.jpeg \n inflating: chest_xray/train/PNEUMONIA/person54_bacteria_257.jpeg \n inflating: chest_xray/train/PNEUMONIA/person54_bacteria_258.jpeg \n inflating: chest_xray/train/PNEUMONIA/person550_bacteria_2308.jpeg \n inflating: chest_xray/train/PNEUMONIA/person550_bacteria_2309.jpeg \n inflating: chest_xray/train/PNEUMONIA/person550_virus_1090.jpeg \n inflating: chest_xray/train/PNEUMONIA/person551_bacteria_2310.jpeg \n inflating: chest_xray/train/PNEUMONIA/person551_bacteria_2311.jpeg \n inflating: chest_xray/train/PNEUMONIA/person551_virus_1091.jpeg \n inflating: chest_xray/train/PNEUMONIA/person552_bacteria_2313.jpeg \n inflating: chest_xray/train/PNEUMONIA/person552_bacteria_2315.jpeg \n inflating: chest_xray/train/PNEUMONIA/person552_virus_1092.jpeg \n inflating: chest_xray/train/PNEUMONIA/person553_bacteria_2316.jpeg \n inflating: chest_xray/train/PNEUMONIA/person553_bacteria_2317.jpeg \n inflating: chest_xray/train/PNEUMONIA/person553_virus_1093.jpeg \n inflating: chest_xray/train/PNEUMONIA/person554_bacteria_2320.jpeg \n inflating: chest_xray/train/PNEUMONIA/person554_bacteria_2321.jpeg \n inflating: chest_xray/train/PNEUMONIA/person554_bacteria_2322.jpeg \n inflating: chest_xray/train/PNEUMONIA/person554_bacteria_2323.jpeg \n inflating: chest_xray/train/PNEUMONIA/person554_virus_1094.jpeg \n inflating: chest_xray/train/PNEUMONIA/person555_bacteria_2325.jpeg \n inflating: chest_xray/train/PNEUMONIA/person556_bacteria_2326.jpeg \n inflating: chest_xray/train/PNEUMONIA/person556_virus_1096.jpeg \n inflating: chest_xray/train/PNEUMONIA/person557_bacteria_2327.jpeg \n inflating: chest_xray/train/PNEUMONIA/person557_virus_1097.jpeg \n inflating: chest_xray/train/PNEUMONIA/person558_bacteria_2328.jpeg \n inflating: chest_xray/train/PNEUMONIA/person558_virus_1098.jpeg \n inflating: chest_xray/train/PNEUMONIA/person559_bacteria_2329.jpeg \n inflating: chest_xray/train/PNEUMONIA/person559_virus_1099.jpeg \n inflating: chest_xray/train/PNEUMONIA/person55_bacteria_260.jpeg \n inflating: chest_xray/train/PNEUMONIA/person55_bacteria_261.jpeg \n inflating: chest_xray/train/PNEUMONIA/person55_bacteria_262.jpeg \n inflating: chest_xray/train/PNEUMONIA/person55_bacteria_263.jpeg \n inflating: chest_xray/train/PNEUMONIA/person55_bacteria_264.jpeg \n inflating: chest_xray/train/PNEUMONIA/person55_bacteria_265.jpeg \n inflating: chest_xray/train/PNEUMONIA/person55_bacteria_266.jpeg \n inflating: chest_xray/train/PNEUMONIA/person560_bacteria_2330.jpeg \n inflating: chest_xray/train/PNEUMONIA/person561_bacteria_2331.jpeg \n inflating: chest_xray/train/PNEUMONIA/person562_bacteria_2332.jpeg \n inflating: chest_xray/train/PNEUMONIA/person562_virus_1102.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2333.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2334.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2335.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2336.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2337.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2338.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2339.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_bacteria_2340.jpeg \n inflating: chest_xray/train/PNEUMONIA/person563_virus_1103.jpeg \n inflating: chest_xray/train/PNEUMONIA/person564_bacteria_2342.jpeg \n inflating: chest_xray/train/PNEUMONIA/person564_bacteria_2343.jpeg \n inflating: chest_xray/train/PNEUMONIA/person564_bacteria_2344.jpeg \n inflating: chest_xray/train/PNEUMONIA/person564_bacteria_2345.jpeg \n inflating: chest_xray/train/PNEUMONIA/person564_bacteria_2346.jpeg \n inflating: chest_xray/train/PNEUMONIA/person564_bacteria_2347.jpeg \n inflating: chest_xray/train/PNEUMONIA/person564_virus_1104.jpeg \n inflating: chest_xray/train/PNEUMONIA/person565_bacteria_2348.jpeg \n inflating: chest_xray/train/PNEUMONIA/person565_virus_1105.jpeg \n inflating: chest_xray/train/PNEUMONIA/person566_bacteria_2351.jpeg \n inflating: chest_xray/train/PNEUMONIA/person566_virus_1106.jpeg \n inflating: chest_xray/train/PNEUMONIA/person567_bacteria_2352.jpeg \n inflating: chest_xray/train/PNEUMONIA/person567_bacteria_2353.jpeg \n inflating: chest_xray/train/PNEUMONIA/person567_bacteria_2354.jpeg \n inflating: chest_xray/train/PNEUMONIA/person567_virus_1107.jpeg \n inflating: chest_xray/train/PNEUMONIA/person568_bacteria_2358.jpeg \n inflating: chest_xray/train/PNEUMONIA/person568_bacteria_2359.jpeg \n inflating: chest_xray/train/PNEUMONIA/person569_bacteria_2360.jpeg \n inflating: chest_xray/train/PNEUMONIA/person569_bacteria_2362.jpeg \n inflating: chest_xray/train/PNEUMONIA/person569_bacteria_2363.jpeg \n inflating: chest_xray/train/PNEUMONIA/person569_bacteria_2364.jpeg \n inflating: chest_xray/train/PNEUMONIA/person569_virus_1110.jpeg \n inflating: chest_xray/train/PNEUMONIA/person56_bacteria_267.jpeg \n inflating: chest_xray/train/PNEUMONIA/person56_bacteria_268.jpeg \n inflating: chest_xray/train/PNEUMONIA/person56_bacteria_269.jpeg \n inflating: chest_xray/train/PNEUMONIA/person570_bacteria_2365.jpeg \n inflating: chest_xray/train/PNEUMONIA/person570_virus_1112.jpeg \n inflating: chest_xray/train/PNEUMONIA/person571_bacteria_2367.jpeg \n inflating: chest_xray/train/PNEUMONIA/person571_virus_1114.jpeg \n inflating: chest_xray/train/PNEUMONIA/person572_bacteria_2368.jpeg \n inflating: chest_xray/train/PNEUMONIA/person573_bacteria_2369.jpeg \n inflating: chest_xray/train/PNEUMONIA/person573_virus_1116.jpeg \n inflating: chest_xray/train/PNEUMONIA/person574_bacteria_2370.jpeg \n inflating: chest_xray/train/PNEUMONIA/person574_bacteria_2371.jpeg \n inflating: chest_xray/train/PNEUMONIA/person574_bacteria_2372.jpeg \n inflating: chest_xray/train/PNEUMONIA/person574_bacteria_2373.jpeg \n inflating: chest_xray/train/PNEUMONIA/person574_virus_1118.jpeg \n inflating: chest_xray/train/PNEUMONIA/person575_bacteria_2374.jpeg \n inflating: chest_xray/train/PNEUMONIA/person575_virus_1119.jpeg \n inflating: chest_xray/train/PNEUMONIA/person576_bacteria_2375.jpeg \n inflating: chest_xray/train/PNEUMONIA/person576_bacteria_2376.jpeg \n inflating: chest_xray/train/PNEUMONIA/person576_virus_1120.jpeg \n inflating: chest_xray/train/PNEUMONIA/person577_bacteria_2378.jpeg \n inflating: chest_xray/train/PNEUMONIA/person577_virus_1121.jpeg \n inflating: chest_xray/train/PNEUMONIA/person578_bacteria_2379.jpeg \n inflating: chest_xray/train/PNEUMONIA/person578_virus_1122.jpeg \n inflating: chest_xray/train/PNEUMONIA/person579_bacteria_2381.jpeg \n inflating: chest_xray/train/PNEUMONIA/person579_bacteria_2382.jpeg \n inflating: chest_xray/train/PNEUMONIA/person579_bacteria_2383.jpeg \n inflating: chest_xray/train/PNEUMONIA/person579_bacteria_2384.jpeg \n inflating: chest_xray/train/PNEUMONIA/person579_bacteria_2386.jpeg \n inflating: chest_xray/train/PNEUMONIA/person579_virus_1123.jpeg \n inflating: chest_xray/train/PNEUMONIA/person57_bacteria_270.jpeg \n inflating: chest_xray/train/PNEUMONIA/person57_bacteria_271.jpeg \n inflating: chest_xray/train/PNEUMONIA/person580_bacteria_2387.jpeg \n inflating: chest_xray/train/PNEUMONIA/person580_bacteria_2388.jpeg \n inflating: chest_xray/train/PNEUMONIA/person580_bacteria_2389.jpeg \n inflating: chest_xray/train/PNEUMONIA/person581_bacteria_2390.jpeg \n inflating: chest_xray/train/PNEUMONIA/person581_bacteria_2392.jpeg \n inflating: chest_xray/train/PNEUMONIA/person581_bacteria_2393.jpeg \n inflating: chest_xray/train/PNEUMONIA/person581_bacteria_2394.jpeg \n inflating: chest_xray/train/PNEUMONIA/person581_bacteria_2395.jpeg \n inflating: chest_xray/train/PNEUMONIA/person581_bacteria_2400.jpeg \n inflating: chest_xray/train/PNEUMONIA/person581_virus_1125.jpeg \n inflating: chest_xray/train/PNEUMONIA/person582_bacteria_2403.jpeg \n inflating: chest_xray/train/PNEUMONIA/person582_bacteria_2404.jpeg \n inflating: chest_xray/train/PNEUMONIA/person582_bacteria_2405.jpeg \n inflating: chest_xray/train/PNEUMONIA/person583_bacteria_2406.jpeg \n inflating: chest_xray/train/PNEUMONIA/person583_bacteria_2408.jpeg \n inflating: chest_xray/train/PNEUMONIA/person583_bacteria_2409.jpeg \n inflating: chest_xray/train/PNEUMONIA/person583_virus_1127.jpeg \n inflating: chest_xray/train/PNEUMONIA/person584_virus_1128.jpeg \n inflating: chest_xray/train/PNEUMONIA/person585_bacteria_2411.jpeg \n inflating: chest_xray/train/PNEUMONIA/person585_bacteria_2412.jpeg \n inflating: chest_xray/train/PNEUMONIA/person585_bacteria_2413.jpeg \n inflating: chest_xray/train/PNEUMONIA/person585_bacteria_2414.jpeg \n inflating: chest_xray/train/PNEUMONIA/person585_bacteria_2415.jpeg \n inflating: chest_xray/train/PNEUMONIA/person585_bacteria_2416.jpeg \n inflating: chest_xray/train/PNEUMONIA/person585_virus_1129.jpeg \n inflating: chest_xray/train/PNEUMONIA/person586_bacteria_2417.jpeg \n inflating: chest_xray/train/PNEUMONIA/person586_bacteria_2418.jpeg \n inflating: chest_xray/train/PNEUMONIA/person586_bacteria_2420.jpeg \n inflating: chest_xray/train/PNEUMONIA/person586_virus_1130.jpeg \n inflating: chest_xray/train/PNEUMONIA/person587_bacteria_2421.jpeg \n inflating: chest_xray/train/PNEUMONIA/person588_bacteria_2422.jpeg \n inflating: chest_xray/train/PNEUMONIA/person588_bacteria_2423.jpeg \n inflating: chest_xray/train/PNEUMONIA/person588_virus_1134.jpeg \n inflating: chest_xray/train/PNEUMONIA/person588_virus_1135.jpeg \n inflating: chest_xray/train/PNEUMONIA/person589_bacteria_2424.jpeg \n inflating: chest_xray/train/PNEUMONIA/person589_bacteria_2425.jpeg \n inflating: chest_xray/train/PNEUMONIA/person58_bacteria_272.jpeg \n inflating: chest_xray/train/PNEUMONIA/person58_bacteria_273.jpeg \n inflating: chest_xray/train/PNEUMONIA/person58_bacteria_274.jpeg \n inflating: chest_xray/train/PNEUMONIA/person58_bacteria_275.jpeg \n inflating: chest_xray/train/PNEUMONIA/person58_bacteria_276.jpeg \n inflating: chest_xray/train/PNEUMONIA/person58_bacteria_277.jpeg \n inflating: chest_xray/train/PNEUMONIA/person58_bacteria_278.jpeg \n inflating: chest_xray/train/PNEUMONIA/person590_bacteria_2428.jpeg \n inflating: chest_xray/train/PNEUMONIA/person590_virus_1138.jpeg \n inflating: chest_xray/train/PNEUMONIA/person591_bacteria_2429.jpeg \n inflating: chest_xray/train/PNEUMONIA/person591_virus_1139.jpeg \n inflating: chest_xray/train/PNEUMONIA/person592_bacteria_2431.jpeg \n inflating: chest_xray/train/PNEUMONIA/person592_bacteria_2434.jpeg \n inflating: chest_xray/train/PNEUMONIA/person592_virus_1141.jpeg \n inflating: chest_xray/train/PNEUMONIA/person593_bacteria_2435.jpeg \n inflating: chest_xray/train/PNEUMONIA/person593_virus_1142.jpeg \n inflating: chest_xray/train/PNEUMONIA/person594_bacteria_2436.jpeg \n inflating: chest_xray/train/PNEUMONIA/person594_virus_1145.jpeg \n inflating: chest_xray/train/PNEUMONIA/person595_bacteria_2438.jpeg \n inflating: chest_xray/train/PNEUMONIA/person595_virus_1147.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2440.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2441.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2443.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2444.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2445.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2446.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2447.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_bacteria_2449.jpeg \n inflating: chest_xray/train/PNEUMONIA/person596_virus_1149.jpeg \n inflating: chest_xray/train/PNEUMONIA/person597_bacteria_2450.jpeg \n inflating: chest_xray/train/PNEUMONIA/person597_bacteria_2451.jpeg \n inflating: chest_xray/train/PNEUMONIA/person597_virus_1150.jpeg \n inflating: chest_xray/train/PNEUMONIA/person598_bacteria_2453.jpeg \n inflating: chest_xray/train/PNEUMONIA/person598_bacteria_2454.jpeg \n inflating: chest_xray/train/PNEUMONIA/person598_virus_1151.jpeg \n inflating: chest_xray/train/PNEUMONIA/person598_virus_1153.jpeg \n inflating: chest_xray/train/PNEUMONIA/person598_virus_1154.jpeg \n inflating: chest_xray/train/PNEUMONIA/person599_bacteria_2455.jpeg \n inflating: chest_xray/train/PNEUMONIA/person599_virus_1155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person59_bacteria_279.jpeg \n inflating: chest_xray/train/PNEUMONIA/person59_bacteria_280.jpeg \n inflating: chest_xray/train/PNEUMONIA/person59_bacteria_281.jpeg \n inflating: chest_xray/train/PNEUMONIA/person59_bacteria_282.jpeg \n inflating: chest_xray/train/PNEUMONIA/person59_bacteria_283.jpeg \n inflating: chest_xray/train/PNEUMONIA/person59_bacteria_284.jpeg \n inflating: chest_xray/train/PNEUMONIA/person5_bacteria_15.jpeg \n inflating: chest_xray/train/PNEUMONIA/person5_bacteria_16.jpeg \n inflating: chest_xray/train/PNEUMONIA/person5_bacteria_17.jpeg \n inflating: chest_xray/train/PNEUMONIA/person5_bacteria_19.jpeg \n inflating: chest_xray/train/PNEUMONIA/person600_bacteria_2456.jpeg \n inflating: chest_xray/train/PNEUMONIA/person600_bacteria_2457.jpeg \n inflating: chest_xray/train/PNEUMONIA/person600_bacteria_2458.jpeg \n inflating: chest_xray/train/PNEUMONIA/person600_virus_1156.jpeg \n inflating: chest_xray/train/PNEUMONIA/person601_bacteria_2459.jpeg \n inflating: chest_xray/train/PNEUMONIA/person602_bacteria_2460.jpeg \n inflating: chest_xray/train/PNEUMONIA/person603_bacteria_2461.jpeg \n inflating: chest_xray/train/PNEUMONIA/person603_virus_1164.jpeg \n inflating: chest_xray/train/PNEUMONIA/person604_bacteria_2462.jpeg \n inflating: chest_xray/train/PNEUMONIA/person604_bacteria_2463.jpeg \n inflating: chest_xray/train/PNEUMONIA/person604_virus_1165.jpeg \n inflating: chest_xray/train/PNEUMONIA/person605_bacteria_2464.jpeg \n inflating: chest_xray/train/PNEUMONIA/person605_bacteria_2465.jpeg \n inflating: chest_xray/train/PNEUMONIA/person605_bacteria_2466.jpeg \n inflating: chest_xray/train/PNEUMONIA/person605_bacteria_2467.jpeg \n inflating: chest_xray/train/PNEUMONIA/person605_bacteria_2468.jpeg \n inflating: chest_xray/train/PNEUMONIA/person605_virus_1166.jpeg \n inflating: chest_xray/train/PNEUMONIA/person605_virus_1169.jpeg \n inflating: chest_xray/train/PNEUMONIA/person606_bacteria_2469.jpeg \n inflating: chest_xray/train/PNEUMONIA/person607_bacteria_2470.jpeg \n inflating: chest_xray/train/PNEUMONIA/person607_virus_1173.jpeg \n inflating: chest_xray/train/PNEUMONIA/person608_bacteria_2471.jpeg \n inflating: chest_xray/train/PNEUMONIA/person608_bacteria_2472.jpeg \n inflating: chest_xray/train/PNEUMONIA/person608_bacteria_2473.jpeg \n inflating: chest_xray/train/PNEUMONIA/person608_virus_1175.jpeg \n inflating: chest_xray/train/PNEUMONIA/person609_bacteria_2474.jpeg \n inflating: chest_xray/train/PNEUMONIA/person609_virus_1176.jpeg \n inflating: chest_xray/train/PNEUMONIA/person60_bacteria_285.jpeg \n inflating: chest_xray/train/PNEUMONIA/person60_bacteria_286.jpeg \n inflating: chest_xray/train/PNEUMONIA/person60_bacteria_287.jpeg \n inflating: chest_xray/train/PNEUMONIA/person610_bacteria_2475.jpeg \n inflating: chest_xray/train/PNEUMONIA/person610_virus_1177.jpeg \n inflating: chest_xray/train/PNEUMONIA/person611_bacteria_2476.jpeg \n inflating: chest_xray/train/PNEUMONIA/person612_bacteria_2477.jpeg \n inflating: chest_xray/train/PNEUMONIA/person612_bacteria_2478.jpeg \n inflating: chest_xray/train/PNEUMONIA/person612_virus_1179.jpeg \n inflating: chest_xray/train/PNEUMONIA/person613_bacteria_2479.jpeg \n inflating: chest_xray/train/PNEUMONIA/person613_virus_1181.jpeg \n inflating: chest_xray/train/PNEUMONIA/person614_bacteria_2480.jpeg \n inflating: chest_xray/train/PNEUMONIA/person614_bacteria_2481.jpeg \n inflating: chest_xray/train/PNEUMONIA/person614_bacteria_2483.jpeg \n inflating: chest_xray/train/PNEUMONIA/person614_virus_1183.jpeg \n inflating: chest_xray/train/PNEUMONIA/person615_virus_1184.jpeg \n inflating: chest_xray/train/PNEUMONIA/person616_bacteria_2487.jpeg \n inflating: chest_xray/train/PNEUMONIA/person616_virus_1186.jpeg \n inflating: chest_xray/train/PNEUMONIA/person617_bacteria_2488.jpeg \n inflating: chest_xray/train/PNEUMONIA/person617_virus_1187.jpeg \n inflating: chest_xray/train/PNEUMONIA/person618_bacteria_2489.jpeg \n inflating: chest_xray/train/PNEUMONIA/person618_virus_1189.jpeg \n inflating: chest_xray/train/PNEUMONIA/person619_bacteria_2490.jpeg \n inflating: chest_xray/train/PNEUMONIA/person619_bacteria_2491.jpeg \n inflating: chest_xray/train/PNEUMONIA/person619_virus_1190.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_288.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_289.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_290.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_291.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_292.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_293.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_295.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_296.jpeg \n inflating: chest_xray/train/PNEUMONIA/person61_bacteria_297.jpeg \n inflating: chest_xray/train/PNEUMONIA/person620_bacteria_2492.jpeg \n inflating: chest_xray/train/PNEUMONIA/person620_virus_1191.jpeg \n inflating: chest_xray/train/PNEUMONIA/person620_virus_1192.jpeg \n inflating: chest_xray/train/PNEUMONIA/person621_virus_1194.jpeg \n inflating: chest_xray/train/PNEUMONIA/person621_virus_1195.jpeg \n inflating: chest_xray/train/PNEUMONIA/person622_bacteria_2494.jpeg \n inflating: chest_xray/train/PNEUMONIA/person622_virus_1196.jpeg \n inflating: chest_xray/train/PNEUMONIA/person623_bacteria_2495.jpeg \n inflating: chest_xray/train/PNEUMONIA/person623_bacteria_2496.jpeg \n inflating: chest_xray/train/PNEUMONIA/person623_virus_1197.jpeg \n inflating: chest_xray/train/PNEUMONIA/person624_bacteria_2497.jpeg \n inflating: chest_xray/train/PNEUMONIA/person624_virus_1198.jpeg \n inflating: chest_xray/train/PNEUMONIA/person625_bacteria_2499.jpeg \n inflating: chest_xray/train/PNEUMONIA/person625_bacteria_2500.jpeg \n inflating: chest_xray/train/PNEUMONIA/person625_virus_1199.jpeg \n inflating: chest_xray/train/PNEUMONIA/person626_bacteria_2502.jpeg \n inflating: chest_xray/train/PNEUMONIA/person626_virus_1202.jpeg \n inflating: chest_xray/train/PNEUMONIA/person627_virus_1204.jpeg \n inflating: chest_xray/train/PNEUMONIA/person628_bacteria_2505.jpeg \n inflating: chest_xray/train/PNEUMONIA/person628_virus_1206.jpeg \n inflating: chest_xray/train/PNEUMONIA/person629_bacteria_2506.jpeg \n inflating: chest_xray/train/PNEUMONIA/person629_bacteria_2507.jpeg \n inflating: chest_xray/train/PNEUMONIA/person629_bacteria_2508.jpeg \n inflating: chest_xray/train/PNEUMONIA/person629_bacteria_2509.jpeg \n inflating: chest_xray/train/PNEUMONIA/person629_bacteria_2510.jpeg \n inflating: chest_xray/train/PNEUMONIA/person629_virus_1207.jpeg \n inflating: chest_xray/train/PNEUMONIA/person62_bacteria_298.jpeg \n inflating: chest_xray/train/PNEUMONIA/person62_bacteria_299.jpeg \n inflating: chest_xray/train/PNEUMONIA/person62_bacteria_300.jpeg \n inflating: chest_xray/train/PNEUMONIA/person62_bacteria_301.jpeg \n inflating: chest_xray/train/PNEUMONIA/person62_bacteria_302.jpeg \n inflating: chest_xray/train/PNEUMONIA/person62_bacteria_303.jpeg \n inflating: chest_xray/train/PNEUMONIA/person630_bacteria_2512.jpeg \n inflating: chest_xray/train/PNEUMONIA/person630_bacteria_2513.jpeg \n inflating: chest_xray/train/PNEUMONIA/person630_bacteria_2514.jpeg \n inflating: chest_xray/train/PNEUMONIA/person630_bacteria_2515.jpeg \n inflating: chest_xray/train/PNEUMONIA/person630_bacteria_2516.jpeg \n inflating: chest_xray/train/PNEUMONIA/person630_virus_1209.jpeg \n inflating: chest_xray/train/PNEUMONIA/person632_bacteria_2520.jpeg \n inflating: chest_xray/train/PNEUMONIA/person632_bacteria_2521.jpeg \n inflating: chest_xray/train/PNEUMONIA/person632_virus_1211.jpeg \n inflating: chest_xray/train/PNEUMONIA/person633_bacteria_2522.jpeg \n inflating: chest_xray/train/PNEUMONIA/person633_virus_1213.jpeg \n inflating: chest_xray/train/PNEUMONIA/person634_bacteria_2525.jpeg \n inflating: chest_xray/train/PNEUMONIA/person635_bacteria_2526.jpeg \n inflating: chest_xray/train/PNEUMONIA/person636_bacteria_2527.jpeg \n inflating: chest_xray/train/PNEUMONIA/person636_virus_1217.jpeg \n inflating: chest_xray/train/PNEUMONIA/person637_bacteria_2528.jpeg \n inflating: chest_xray/train/PNEUMONIA/person637_bacteria_2529.jpeg \n inflating: chest_xray/train/PNEUMONIA/person637_virus_1218.jpeg \n inflating: chest_xray/train/PNEUMONIA/person639_virus_1220.jpeg \n inflating: chest_xray/train/PNEUMONIA/person63_bacteria_306.jpeg \n inflating: chest_xray/train/PNEUMONIA/person640_bacteria_2532.jpeg \n inflating: chest_xray/train/PNEUMONIA/person640_virus_1221.jpeg \n inflating: chest_xray/train/PNEUMONIA/person641_bacteria_2533.jpeg \n inflating: chest_xray/train/PNEUMONIA/person641_virus_1222.jpeg \n inflating: chest_xray/train/PNEUMONIA/person642_virus_1223.jpeg \n inflating: chest_xray/train/PNEUMONIA/person643_bacteria_2534.jpeg \n inflating: chest_xray/train/PNEUMONIA/person644_bacteria_2536.jpeg \n inflating: chest_xray/train/PNEUMONIA/person644_virus_1225.jpeg \n inflating: chest_xray/train/PNEUMONIA/person645_bacteria_2537.jpeg \n inflating: chest_xray/train/PNEUMONIA/person645_virus_1226.jpeg \n inflating: chest_xray/train/PNEUMONIA/person646_bacteria_2538.jpeg \n inflating: chest_xray/train/PNEUMONIA/person646_virus_1227.jpeg \n inflating: chest_xray/train/PNEUMONIA/person647_bacteria_2539.jpeg \n inflating: chest_xray/train/PNEUMONIA/person647_virus_1228.jpeg \n inflating: chest_xray/train/PNEUMONIA/person647_virus_1229.jpeg \n inflating: chest_xray/train/PNEUMONIA/person648_bacteria_2540.jpeg \n inflating: chest_xray/train/PNEUMONIA/person648_virus_1230.jpeg \n inflating: chest_xray/train/PNEUMONIA/person649_bacteria_2541.jpeg \n inflating: chest_xray/train/PNEUMONIA/person649_virus_1231.jpeg \n inflating: chest_xray/train/PNEUMONIA/person64_bacteria_310.jpeg \n inflating: chest_xray/train/PNEUMONIA/person64_bacteria_312.jpeg \n inflating: chest_xray/train/PNEUMONIA/person64_bacteria_316.jpeg \n inflating: chest_xray/train/PNEUMONIA/person64_bacteria_317.jpeg \n inflating: chest_xray/train/PNEUMONIA/person64_bacteria_318.jpeg \n inflating: chest_xray/train/PNEUMONIA/person64_bacteria_319.jpeg \n inflating: chest_xray/train/PNEUMONIA/person64_bacteria_320.jpeg \n inflating: chest_xray/train/PNEUMONIA/person650_bacteria_2542.jpeg \n inflating: chest_xray/train/PNEUMONIA/person650_virus_1232.jpeg \n inflating: chest_xray/train/PNEUMONIA/person651_bacteria_2543.jpeg \n inflating: chest_xray/train/PNEUMONIA/person652_bacteria_2544.jpeg \n inflating: chest_xray/train/PNEUMONIA/person652_virus_1234.jpeg \n inflating: chest_xray/train/PNEUMONIA/person653_bacteria_2545.jpeg \n inflating: chest_xray/train/PNEUMONIA/person653_virus_1235.jpeg \n inflating: chest_xray/train/PNEUMONIA/person654_bacteria_2546.jpeg \n inflating: chest_xray/train/PNEUMONIA/person655_bacteria_2547.jpeg \n inflating: chest_xray/train/PNEUMONIA/person656_bacteria_2548.jpeg \n inflating: chest_xray/train/PNEUMONIA/person656_virus_1238.jpeg \n inflating: chest_xray/train/PNEUMONIA/person657_bacteria_2549.jpeg \n inflating: chest_xray/train/PNEUMONIA/person657_virus_1240.jpeg \n inflating: chest_xray/train/PNEUMONIA/person658_bacteria_2550.jpeg \n inflating: chest_xray/train/PNEUMONIA/person658_virus_1241.jpeg \n inflating: chest_xray/train/PNEUMONIA/person659_bacteria_2551.jpeg \n inflating: chest_xray/train/PNEUMONIA/person659_virus_1243.jpeg \n inflating: chest_xray/train/PNEUMONIA/person65_bacteria_322.jpeg \n inflating: chest_xray/train/PNEUMONIA/person660_bacteria_2552.jpeg \n inflating: chest_xray/train/PNEUMONIA/person660_virus_1244.jpeg \n inflating: chest_xray/train/PNEUMONIA/person661_bacteria_2553.jpeg \n inflating: chest_xray/train/PNEUMONIA/person661_virus_1245.jpeg \n inflating: chest_xray/train/PNEUMONIA/person662_bacteria_2554.jpeg \n inflating: chest_xray/train/PNEUMONIA/person662_virus_1246.jpeg \n inflating: chest_xray/train/PNEUMONIA/person663_bacteria_2555.jpeg \n inflating: chest_xray/train/PNEUMONIA/person663_virus_1247.jpeg \n inflating: chest_xray/train/PNEUMONIA/person663_virus_1248.jpeg \n inflating: chest_xray/train/PNEUMONIA/person664_virus_1249.jpeg \n inflating: chest_xray/train/PNEUMONIA/person665_bacteria_2557.jpeg \n inflating: chest_xray/train/PNEUMONIA/person665_virus_1250.jpeg \n inflating: chest_xray/train/PNEUMONIA/person666_bacteria_2558.jpeg \n inflating: chest_xray/train/PNEUMONIA/person666_virus_1251.jpeg \n inflating: chest_xray/train/PNEUMONIA/person667_virus_1252.jpeg \n inflating: chest_xray/train/PNEUMONIA/person667_virus_1253.jpeg \n inflating: chest_xray/train/PNEUMONIA/person669_bacteria_2561.jpeg \n inflating: chest_xray/train/PNEUMONIA/person669_bacteria_2562.jpeg \n inflating: chest_xray/train/PNEUMONIA/person669_virus_1255.jpeg \n inflating: chest_xray/train/PNEUMONIA/person66_bacteria_323.jpeg \n inflating: chest_xray/train/PNEUMONIA/person66_bacteria_324.jpeg \n inflating: chest_xray/train/PNEUMONIA/person66_bacteria_325.jpeg \n inflating: chest_xray/train/PNEUMONIA/person66_bacteria_326.jpeg \n inflating: chest_xray/train/PNEUMONIA/person670_bacteria_2563.jpeg \n inflating: chest_xray/train/PNEUMONIA/person670_virus_1256.jpeg \n inflating: chest_xray/train/PNEUMONIA/person670_virus_1259.jpeg \n inflating: chest_xray/train/PNEUMONIA/person671_bacteria_2564.jpeg \n inflating: chest_xray/train/PNEUMONIA/person671_virus_1260.jpeg \n inflating: chest_xray/train/PNEUMONIA/person672_bacteria_2565.jpeg \n inflating: chest_xray/train/PNEUMONIA/person672_virus_1261.jpeg \n inflating: chest_xray/train/PNEUMONIA/person673_bacteria_2566.jpeg \n inflating: chest_xray/train/PNEUMONIA/person673_virus_1263.jpeg \n inflating: chest_xray/train/PNEUMONIA/person674_bacteria_2568.jpeg \n inflating: chest_xray/train/PNEUMONIA/person675_bacteria_2569.jpeg \n inflating: chest_xray/train/PNEUMONIA/person677_bacteria_2571.jpeg \n inflating: chest_xray/train/PNEUMONIA/person677_virus_1268.jpeg \n inflating: chest_xray/train/PNEUMONIA/person678_bacteria_2572.jpeg \n inflating: chest_xray/train/PNEUMONIA/person679_virus_1270.jpeg \n inflating: chest_xray/train/PNEUMONIA/person67_bacteria_328.jpeg \n inflating: chest_xray/train/PNEUMONIA/person67_bacteria_329.jpeg \n inflating: chest_xray/train/PNEUMONIA/person67_bacteria_330.jpeg \n inflating: chest_xray/train/PNEUMONIA/person67_bacteria_331.jpeg \n inflating: chest_xray/train/PNEUMONIA/person67_bacteria_332.jpeg \n inflating: chest_xray/train/PNEUMONIA/person67_bacteria_333.jpeg \n inflating: chest_xray/train/PNEUMONIA/person67_bacteria_334.jpeg \n inflating: chest_xray/train/PNEUMONIA/person680_bacteria_2575.jpeg \n inflating: chest_xray/train/PNEUMONIA/person681_bacteria_2576.jpeg \n inflating: chest_xray/train/PNEUMONIA/person681_virus_1272.jpeg \n inflating: chest_xray/train/PNEUMONIA/person682_virus_1273.jpeg \n inflating: chest_xray/train/PNEUMONIA/person683_bacteria_2578.jpeg \n inflating: chest_xray/train/PNEUMONIA/person684_bacteria_2580.jpeg \n inflating: chest_xray/train/PNEUMONIA/person684_virus_1275.jpeg \n inflating: chest_xray/train/PNEUMONIA/person685_bacteria_2581.jpeg \n inflating: chest_xray/train/PNEUMONIA/person687_bacteria_2583.jpeg \n inflating: chest_xray/train/PNEUMONIA/person688_bacteria_2584.jpeg \n inflating: chest_xray/train/PNEUMONIA/person688_virus_1281.jpeg \n inflating: chest_xray/train/PNEUMONIA/person688_virus_1282.jpeg \n inflating: chest_xray/train/PNEUMONIA/person689_bacteria_2585.jpeg \n inflating: chest_xray/train/PNEUMONIA/person689_bacteria_2586.jpeg \n inflating: chest_xray/train/PNEUMONIA/person68_bacteria_335.jpeg \n inflating: chest_xray/train/PNEUMONIA/person68_bacteria_336.jpeg \n inflating: chest_xray/train/PNEUMONIA/person68_bacteria_337.jpeg \n inflating: chest_xray/train/PNEUMONIA/person690_bacteria_2587.jpeg \n inflating: chest_xray/train/PNEUMONIA/person691_bacteria_2588.jpeg \n inflating: chest_xray/train/PNEUMONIA/person692_bacteria_2589.jpeg \n inflating: chest_xray/train/PNEUMONIA/person692_virus_1286.jpeg \n inflating: chest_xray/train/PNEUMONIA/person693_bacteria_2590.jpeg \n inflating: chest_xray/train/PNEUMONIA/person696_bacteria_2594.jpeg \n inflating: chest_xray/train/PNEUMONIA/person698_virus_1294.jpeg \n inflating: chest_xray/train/PNEUMONIA/person699_bacteria_2598.jpeg \n inflating: chest_xray/train/PNEUMONIA/person699_virus_1295.jpeg \n inflating: chest_xray/train/PNEUMONIA/person69_bacteria_338.jpeg \n inflating: chest_xray/train/PNEUMONIA/person6_bacteria_22.jpeg \n inflating: chest_xray/train/PNEUMONIA/person700_bacteria_2599.jpeg \n inflating: chest_xray/train/PNEUMONIA/person701_bacteria_2600.jpeg \n inflating: chest_xray/train/PNEUMONIA/person701_virus_1297.jpeg \n inflating: chest_xray/train/PNEUMONIA/person702_bacteria_2601.jpeg \n inflating: chest_xray/train/PNEUMONIA/person702_virus_1299.jpeg \n inflating: chest_xray/train/PNEUMONIA/person703_bacteria_2602.jpeg \n inflating: chest_xray/train/PNEUMONIA/person703_virus_1300.jpeg \n inflating: chest_xray/train/PNEUMONIA/person704_bacteria_2603.jpeg \n inflating: chest_xray/train/PNEUMONIA/person704_virus_1301.jpeg \n inflating: chest_xray/train/PNEUMONIA/person705_virus_1302.jpeg \n inflating: chest_xray/train/PNEUMONIA/person705_virus_1303.jpeg \n inflating: chest_xray/train/PNEUMONIA/person706_virus_1304.jpeg \n inflating: chest_xray/train/PNEUMONIA/person707_bacteria_2606.jpeg \n inflating: chest_xray/train/PNEUMONIA/person707_virus_1305.jpeg \n inflating: chest_xray/train/PNEUMONIA/person709_bacteria_2608.jpeg \n inflating: chest_xray/train/PNEUMONIA/person70_bacteria_341.jpeg \n inflating: chest_xray/train/PNEUMONIA/person70_bacteria_342.jpeg \n inflating: chest_xray/train/PNEUMONIA/person70_bacteria_343.jpeg \n inflating: chest_xray/train/PNEUMONIA/person70_bacteria_344.jpeg \n inflating: chest_xray/train/PNEUMONIA/person70_bacteria_345.jpeg \n inflating: chest_xray/train/PNEUMONIA/person70_bacteria_346.jpeg \n inflating: chest_xray/train/PNEUMONIA/person710_bacteria_2611.jpeg \n inflating: chest_xray/train/PNEUMONIA/person710_virus_1308.jpeg \n inflating: chest_xray/train/PNEUMONIA/person711_bacteria_2612.jpeg \n inflating: chest_xray/train/PNEUMONIA/person711_virus_1309.jpeg \n inflating: chest_xray/train/PNEUMONIA/person712_bacteria_2613.jpeg \n inflating: chest_xray/train/PNEUMONIA/person712_virus_1310.jpeg \n inflating: chest_xray/train/PNEUMONIA/person713_bacteria_2614.jpeg \n inflating: chest_xray/train/PNEUMONIA/person714_bacteria_2615.jpeg \n inflating: chest_xray/train/PNEUMONIA/person716_bacteria_2617.jpeg \n inflating: chest_xray/train/PNEUMONIA/person716_virus_1314.jpeg \n inflating: chest_xray/train/PNEUMONIA/person717_bacteria_2618.jpeg \n inflating: chest_xray/train/PNEUMONIA/person718_bacteria_2620.jpeg \n inflating: chest_xray/train/PNEUMONIA/person718_virus_1316.jpeg \n inflating: chest_xray/train/PNEUMONIA/person719_bacteria_2621.jpeg \n inflating: chest_xray/train/PNEUMONIA/person719_virus_1338.jpeg \n inflating: chest_xray/train/PNEUMONIA/person71_bacteria_347.jpeg \n inflating: chest_xray/train/PNEUMONIA/person71_bacteria_348.jpeg \n inflating: chest_xray/train/PNEUMONIA/person71_bacteria_349.jpeg \n inflating: chest_xray/train/PNEUMONIA/person71_bacteria_350.jpeg \n inflating: chest_xray/train/PNEUMONIA/person71_bacteria_351.jpeg \n inflating: chest_xray/train/PNEUMONIA/person720_bacteria_2622.jpeg \n inflating: chest_xray/train/PNEUMONIA/person720_virus_1339.jpeg \n inflating: chest_xray/train/PNEUMONIA/person721_bacteria_2623.jpeg \n inflating: chest_xray/train/PNEUMONIA/person721_virus_1340.jpeg \n inflating: chest_xray/train/PNEUMONIA/person722_virus_1341.jpeg \n inflating: chest_xray/train/PNEUMONIA/person723_bacteria_2625.jpeg \n inflating: chest_xray/train/PNEUMONIA/person723_virus_1342.jpeg \n inflating: chest_xray/train/PNEUMONIA/person724_bacteria_2626.jpeg \n inflating: chest_xray/train/PNEUMONIA/person724_virus_1343.jpeg \n inflating: chest_xray/train/PNEUMONIA/person724_virus_1344.jpeg \n inflating: chest_xray/train/PNEUMONIA/person725_bacteria_2627.jpeg \n inflating: chest_xray/train/PNEUMONIA/person726_bacteria_2628.jpeg \n inflating: chest_xray/train/PNEUMONIA/person727_bacteria_2629.jpeg \n inflating: chest_xray/train/PNEUMONIA/person727_virus_1347.jpeg \n inflating: chest_xray/train/PNEUMONIA/person728_bacteria_2630.jpeg \n inflating: chest_xray/train/PNEUMONIA/person72_bacteria_352.jpeg \n inflating: chest_xray/train/PNEUMONIA/person72_bacteria_353.jpeg \n inflating: chest_xray/train/PNEUMONIA/person72_bacteria_354.jpeg \n inflating: chest_xray/train/PNEUMONIA/person730_bacteria_2632.jpeg \n inflating: chest_xray/train/PNEUMONIA/person730_virus_1351.jpeg \n inflating: chest_xray/train/PNEUMONIA/person731_bacteria_2633.jpeg \n inflating: chest_xray/train/PNEUMONIA/person731_virus_1352.jpeg \n inflating: chest_xray/train/PNEUMONIA/person732_bacteria_2634.jpeg \n inflating: chest_xray/train/PNEUMONIA/person732_virus_1353.jpeg \n inflating: chest_xray/train/PNEUMONIA/person733_bacteria_2635.jpeg \n inflating: chest_xray/train/PNEUMONIA/person734_bacteria_2637.jpeg \n inflating: chest_xray/train/PNEUMONIA/person734_virus_1355.jpeg \n inflating: chest_xray/train/PNEUMONIA/person735_bacteria_2638.jpeg \n inflating: chest_xray/train/PNEUMONIA/person735_virus_1356.jpeg \n inflating: chest_xray/train/PNEUMONIA/person736_bacteria_2639.jpeg \n inflating: chest_xray/train/PNEUMONIA/person736_virus_1358.jpeg \n inflating: chest_xray/train/PNEUMONIA/person737_bacteria_2640.jpeg \n inflating: chest_xray/train/PNEUMONIA/person738_bacteria_2641.jpeg \n inflating: chest_xray/train/PNEUMONIA/person738_virus_1360.jpeg \n inflating: chest_xray/train/PNEUMONIA/person739_bacteria_2642.jpeg \n inflating: chest_xray/train/PNEUMONIA/person739_virus_1361.jpeg \n inflating: chest_xray/train/PNEUMONIA/person73_bacteria_355.jpeg \n inflating: chest_xray/train/PNEUMONIA/person73_bacteria_356.jpeg \n inflating: chest_xray/train/PNEUMONIA/person73_bacteria_357.jpeg \n inflating: chest_xray/train/PNEUMONIA/person73_bacteria_358.jpeg \n inflating: chest_xray/train/PNEUMONIA/person73_bacteria_359.jpeg \n inflating: chest_xray/train/PNEUMONIA/person73_bacteria_360.jpeg \n inflating: chest_xray/train/PNEUMONIA/person740_bacteria_2643.jpeg \n inflating: chest_xray/train/PNEUMONIA/person740_virus_1362.jpeg \n inflating: chest_xray/train/PNEUMONIA/person740_virus_1363.jpeg \n inflating: chest_xray/train/PNEUMONIA/person741_bacteria_2644.jpeg \n inflating: chest_xray/train/PNEUMONIA/person741_virus_1364.jpeg \n inflating: chest_xray/train/PNEUMONIA/person742_virus_1365.jpeg \n inflating: chest_xray/train/PNEUMONIA/person743_bacteria_2646.jpeg \n inflating: chest_xray/train/PNEUMONIA/person743_virus_1366.jpeg \n inflating: chest_xray/train/PNEUMONIA/person744_bacteria_2647.jpeg \n inflating: chest_xray/train/PNEUMONIA/person744_virus_1367.jpeg \n inflating: chest_xray/train/PNEUMONIA/person745_bacteria_2648.jpeg \n inflating: chest_xray/train/PNEUMONIA/person745_virus_1368.jpeg \n inflating: chest_xray/train/PNEUMONIA/person746_bacteria_2649.jpeg \n inflating: chest_xray/train/PNEUMONIA/person746_virus_1369.jpeg \n inflating: chest_xray/train/PNEUMONIA/person747_bacteria_2650.jpeg \n inflating: chest_xray/train/PNEUMONIA/person747_virus_1370.jpeg \n inflating: chest_xray/train/PNEUMONIA/person747_virus_1372.jpeg \n inflating: chest_xray/train/PNEUMONIA/person748_virus_1373.jpeg \n inflating: chest_xray/train/PNEUMONIA/person749_bacteria_2652.jpeg \n inflating: chest_xray/train/PNEUMONIA/person749_virus_1374.jpeg \n inflating: chest_xray/train/PNEUMONIA/person74_bacteria_361.jpeg \n inflating: chest_xray/train/PNEUMONIA/person74_bacteria_362.jpeg \n inflating: chest_xray/train/PNEUMONIA/person74_bacteria_363.jpeg \n inflating: chest_xray/train/PNEUMONIA/person750_bacteria_2653.jpeg \n inflating: chest_xray/train/PNEUMONIA/person751_bacteria_2654.jpeg \n inflating: chest_xray/train/PNEUMONIA/person752_virus_1377.jpeg \n inflating: chest_xray/train/PNEUMONIA/person753_bacteria_2656.jpeg \n inflating: chest_xray/train/PNEUMONIA/person753_virus_1378.jpeg \n inflating: chest_xray/train/PNEUMONIA/person754_virus_1379.jpeg \n inflating: chest_xray/train/PNEUMONIA/person755_bacteria_2659.jpeg \n inflating: chest_xray/train/PNEUMONIA/person755_virus_1380.jpeg \n inflating: chest_xray/train/PNEUMONIA/person755_virus_1382.jpeg \n inflating: chest_xray/train/PNEUMONIA/person756_bacteria_2660.jpeg \n inflating: chest_xray/train/PNEUMONIA/person757_bacteria_2661.jpeg \n inflating: chest_xray/train/PNEUMONIA/person757_virus_1385.jpeg \n inflating: chest_xray/train/PNEUMONIA/person758_bacteria_2662.jpeg \n inflating: chest_xray/train/PNEUMONIA/person759_bacteria_2663.jpeg \n inflating: chest_xray/train/PNEUMONIA/person759_virus_1387.jpeg \n inflating: chest_xray/train/PNEUMONIA/person75_bacteria_364.jpeg \n inflating: chest_xray/train/PNEUMONIA/person75_bacteria_365.jpeg \n inflating: chest_xray/train/PNEUMONIA/person75_bacteria_366.jpeg \n inflating: chest_xray/train/PNEUMONIA/person75_bacteria_367.jpeg \n inflating: chest_xray/train/PNEUMONIA/person75_bacteria_368.jpeg \n inflating: chest_xray/train/PNEUMONIA/person75_bacteria_369.jpeg \n inflating: chest_xray/train/PNEUMONIA/person760_bacteria_2664.jpeg \n inflating: chest_xray/train/PNEUMONIA/person760_virus_1388.jpeg \n inflating: chest_xray/train/PNEUMONIA/person761_bacteria_2665.jpeg \n inflating: chest_xray/train/PNEUMONIA/person761_virus_1389.jpeg \n inflating: chest_xray/train/PNEUMONIA/person762_virus_1390.jpeg \n inflating: chest_xray/train/PNEUMONIA/person763_bacteria_2667.jpeg \n inflating: chest_xray/train/PNEUMONIA/person763_virus_1391.jpeg \n inflating: chest_xray/train/PNEUMONIA/person764_bacteria_2668.jpeg \n inflating: chest_xray/train/PNEUMONIA/person764_virus_1392.jpeg \n inflating: chest_xray/train/PNEUMONIA/person765_bacteria_2669.jpeg \n inflating: chest_xray/train/PNEUMONIA/person765_virus_1393.jpeg \n inflating: chest_xray/train/PNEUMONIA/person766_bacteria_2670.jpeg \n inflating: chest_xray/train/PNEUMONIA/person767_bacteria_2671.jpeg \n inflating: chest_xray/train/PNEUMONIA/person768_bacteria_2672.jpeg \n inflating: chest_xray/train/PNEUMONIA/person768_virus_1396.jpeg \n inflating: chest_xray/train/PNEUMONIA/person769_bacteria_2673.jpeg \n inflating: chest_xray/train/PNEUMONIA/person76_bacteria_370.jpeg \n inflating: chest_xray/train/PNEUMONIA/person76_bacteria_371.jpeg \n inflating: chest_xray/train/PNEUMONIA/person76_bacteria_372.jpeg \n inflating: chest_xray/train/PNEUMONIA/person770_bacteria_2674.jpeg \n inflating: chest_xray/train/PNEUMONIA/person770_virus_1398.jpeg \n inflating: chest_xray/train/PNEUMONIA/person771_bacteria_2675.jpeg \n inflating: chest_xray/train/PNEUMONIA/person771_virus_1399.jpeg \n inflating: chest_xray/train/PNEUMONIA/person772_virus_1401.jpeg \n inflating: chest_xray/train/PNEUMONIA/person773_virus_1402.jpeg \n inflating: chest_xray/train/PNEUMONIA/person774_bacteria_2678.jpeg \n inflating: chest_xray/train/PNEUMONIA/person774_virus_1403.jpeg \n inflating: chest_xray/train/PNEUMONIA/person775_bacteria_2679.jpeg \n inflating: chest_xray/train/PNEUMONIA/person775_virus_1404.jpeg \n inflating: chest_xray/train/PNEUMONIA/person776_bacteria_2680.jpeg \n inflating: chest_xray/train/PNEUMONIA/person776_virus_1405.jpeg \n inflating: chest_xray/train/PNEUMONIA/person778_bacteria_2682.jpeg \n inflating: chest_xray/train/PNEUMONIA/person778_virus_1408.jpeg \n inflating: chest_xray/train/PNEUMONIA/person779_bacteria_2683.jpeg \n inflating: chest_xray/train/PNEUMONIA/person779_virus_1409.jpeg \n inflating: chest_xray/train/PNEUMONIA/person779_virus_1410.jpeg \n inflating: chest_xray/train/PNEUMONIA/person77_bacteria_374.jpeg \n inflating: chest_xray/train/PNEUMONIA/person77_bacteria_375.jpeg \n inflating: chest_xray/train/PNEUMONIA/person77_bacteria_376.jpeg \n inflating: chest_xray/train/PNEUMONIA/person77_bacteria_377.jpeg \n inflating: chest_xray/train/PNEUMONIA/person780_bacteria_2684.jpeg \n inflating: chest_xray/train/PNEUMONIA/person780_virus_1411.jpeg \n inflating: chest_xray/train/PNEUMONIA/person781_virus_1412.jpeg \n inflating: chest_xray/train/PNEUMONIA/person782_bacteria_2686.jpeg \n inflating: chest_xray/train/PNEUMONIA/person783_bacteria_2687.jpeg \n inflating: chest_xray/train/PNEUMONIA/person783_virus_1414.jpeg \n inflating: chest_xray/train/PNEUMONIA/person785_bacteria_2689.jpeg \n inflating: chest_xray/train/PNEUMONIA/person786_bacteria_2690.jpeg \n inflating: chest_xray/train/PNEUMONIA/person787_bacteria_2691.jpeg \n inflating: chest_xray/train/PNEUMONIA/person788_virus_1419.jpeg \n inflating: chest_xray/train/PNEUMONIA/person789_bacteria_2694.jpeg \n inflating: chest_xray/train/PNEUMONIA/person789_virus_1420.jpeg \n inflating: chest_xray/train/PNEUMONIA/person790_virus_1421.jpeg \n inflating: chest_xray/train/PNEUMONIA/person791_virus_1422.jpeg \n inflating: chest_xray/train/PNEUMONIA/person793_virus_1424.jpeg \n inflating: chest_xray/train/PNEUMONIA/person794_bacteria_2700.jpeg \n inflating: chest_xray/train/PNEUMONIA/person795_virus_1427.jpeg \n inflating: chest_xray/train/PNEUMONIA/person796_bacteria_2702.jpeg \n inflating: chest_xray/train/PNEUMONIA/person796_virus_1428.jpeg \n inflating: chest_xray/train/PNEUMONIA/person797_virus_1429.jpeg \n inflating: chest_xray/train/PNEUMONIA/person798_virus_1430.jpeg \n inflating: chest_xray/train/PNEUMONIA/person799_bacteria_2705.jpeg \n inflating: chest_xray/train/PNEUMONIA/person799_virus_1431.jpeg \n inflating: chest_xray/train/PNEUMONIA/person7_bacteria_24.jpeg \n inflating: chest_xray/train/PNEUMONIA/person7_bacteria_25.jpeg \n inflating: chest_xray/train/PNEUMONIA/person7_bacteria_28.jpeg \n inflating: chest_xray/train/PNEUMONIA/person7_bacteria_29.jpeg \n inflating: chest_xray/train/PNEUMONIA/person800_bacteria_2706.jpeg \n inflating: chest_xray/train/PNEUMONIA/person801_virus_1434.jpeg \n inflating: chest_xray/train/PNEUMONIA/person802_bacteria_2708.jpeg \n inflating: chest_xray/train/PNEUMONIA/person803_bacteria_2710.jpeg \n inflating: chest_xray/train/PNEUMONIA/person803_virus_1436.jpeg \n inflating: chest_xray/train/PNEUMONIA/person804_bacteria_2711.jpeg \n inflating: chest_xray/train/PNEUMONIA/person805_bacteria_2712.jpeg \n inflating: chest_xray/train/PNEUMONIA/person806_virus_1439.jpeg \n inflating: chest_xray/train/PNEUMONIA/person806_virus_1440.jpeg \n inflating: chest_xray/train/PNEUMONIA/person807_virus_1441.jpeg \n inflating: chest_xray/train/PNEUMONIA/person808_bacteria_2716.jpeg \n inflating: chest_xray/train/PNEUMONIA/person808_virus_1442.jpeg \n inflating: chest_xray/train/PNEUMONIA/person809_bacteria_2717.jpeg \n inflating: chest_xray/train/PNEUMONIA/person809_bacteria_2718.jpeg \n inflating: chest_xray/train/PNEUMONIA/person80_virus_150.jpeg \n inflating: chest_xray/train/PNEUMONIA/person810_bacteria_2719.jpeg \n inflating: chest_xray/train/PNEUMONIA/person810_virus_1445.jpeg \n inflating: chest_xray/train/PNEUMONIA/person810_virus_1446.jpeg \n inflating: chest_xray/train/PNEUMONIA/person811_bacteria_2721.jpeg \n inflating: chest_xray/train/PNEUMONIA/person811_virus_1447.jpeg \n inflating: chest_xray/train/PNEUMONIA/person813_bacteria_2723.jpeg \n inflating: chest_xray/train/PNEUMONIA/person813_bacteria_2724.jpeg \n inflating: chest_xray/train/PNEUMONIA/person813_virus_1449.jpeg \n inflating: chest_xray/train/PNEUMONIA/person814_bacteria_2725.jpeg \n inflating: chest_xray/train/PNEUMONIA/person815_bacteria_2726.jpeg \n inflating: chest_xray/train/PNEUMONIA/person816_bacteria_2727.jpeg \n inflating: chest_xray/train/PNEUMONIA/person817_bacteria_2728.jpeg \n inflating: chest_xray/train/PNEUMONIA/person818_bacteria_2729.jpeg \n inflating: chest_xray/train/PNEUMONIA/person819_bacteria_2730.jpeg \n inflating: chest_xray/train/PNEUMONIA/person819_virus_1455.jpeg \n inflating: chest_xray/train/PNEUMONIA/person81_virus_152.jpeg \n inflating: chest_xray/train/PNEUMONIA/person81_virus_153.jpeg \n inflating: chest_xray/train/PNEUMONIA/person820_bacteria_2731.jpeg \n inflating: chest_xray/train/PNEUMONIA/person820_virus_1456.jpeg \n inflating: chest_xray/train/PNEUMONIA/person823_virus_1459.jpeg \n inflating: chest_xray/train/PNEUMONIA/person825_bacteria_2736.jpeg \n inflating: chest_xray/train/PNEUMONIA/person826_bacteria_2737.jpeg \n inflating: chest_xray/train/PNEUMONIA/person826_virus_1462.jpeg \n inflating: chest_xray/train/PNEUMONIA/person827_bacteria_2738.jpeg \n inflating: chest_xray/train/PNEUMONIA/person829_bacteria_2740.jpeg \n inflating: chest_xray/train/PNEUMONIA/person82_virus_154.jpeg \n inflating: chest_xray/train/PNEUMONIA/person82_virus_155.jpeg \n inflating: chest_xray/train/PNEUMONIA/person830_bacteria_2741.jpeg \n inflating: chest_xray/train/PNEUMONIA/person830_virus_1466.jpeg \n inflating: chest_xray/train/PNEUMONIA/person831_bacteria_2742.jpeg \n inflating: chest_xray/train/PNEUMONIA/person832_bacteria_2743.jpeg \n inflating: chest_xray/train/PNEUMONIA/person832_virus_1468.jpeg \n inflating: chest_xray/train/PNEUMONIA/person833_virus_1469.jpeg \n inflating: chest_xray/train/PNEUMONIA/person834_bacteria_2747.jpeg \n inflating: chest_xray/train/PNEUMONIA/person834_bacteria_2748.jpeg \n inflating: chest_xray/train/PNEUMONIA/person835_bacteria_2749.jpeg \n inflating: chest_xray/train/PNEUMONIA/person835_bacteria_2750.jpeg \n inflating: chest_xray/train/PNEUMONIA/person835_virus_1472.jpeg \n inflating: chest_xray/train/PNEUMONIA/person836_bacteria_2752.jpeg \n inflating: chest_xray/train/PNEUMONIA/person836_virus_1473.jpeg \n inflating: chest_xray/train/PNEUMONIA/person837_bacteria_2753.jpeg \n inflating: chest_xray/train/PNEUMONIA/person837_bacteria_2754.jpeg \n inflating: chest_xray/train/PNEUMONIA/person837_virus_1475.jpeg \n inflating: chest_xray/train/PNEUMONIA/person838_virus_1476.jpeg \n inflating: chest_xray/train/PNEUMONIA/person839_bacteria_2757.jpeg \n inflating: chest_xray/train/PNEUMONIA/person83_virus_156.jpeg \n inflating: chest_xray/train/PNEUMONIA/person840_bacteria_2758.jpeg \n inflating: chest_xray/train/PNEUMONIA/person840_bacteria_2759.jpeg \n inflating: chest_xray/train/PNEUMONIA/person841_bacteria_2760.jpeg \n inflating: chest_xray/train/PNEUMONIA/person841_bacteria_2761.jpeg \n inflating: chest_xray/train/PNEUMONIA/person841_virus_1481.jpeg \n inflating: chest_xray/train/PNEUMONIA/person842_bacteria_2762.jpeg \n inflating: chest_xray/train/PNEUMONIA/person842_virus_1483.jpeg \n inflating: chest_xray/train/PNEUMONIA/person843_bacteria_2763.jpeg \n inflating: chest_xray/train/PNEUMONIA/person843_virus_1485.jpeg \n inflating: chest_xray/train/PNEUMONIA/person844_virus_1487.jpeg \n inflating: chest_xray/train/PNEUMONIA/person845_virus_1489.jpeg \n inflating: chest_xray/train/PNEUMONIA/person846_bacteria_2766.jpeg \n inflating: chest_xray/train/PNEUMONIA/person846_virus_1491.jpeg \n inflating: chest_xray/train/PNEUMONIA/person847_bacteria_2767.jpeg \n inflating: chest_xray/train/PNEUMONIA/person847_virus_1492.jpeg \n inflating: chest_xray/train/PNEUMONIA/person848_bacteria_2769.jpeg \n inflating: chest_xray/train/PNEUMONIA/person848_virus_1493.jpeg \n inflating: chest_xray/train/PNEUMONIA/person849_bacteria_2770.jpeg \n inflating: chest_xray/train/PNEUMONIA/person849_virus_1494.jpeg \n inflating: chest_xray/train/PNEUMONIA/person84_virus_157.jpeg \n inflating: chest_xray/train/PNEUMONIA/person850_bacteria_2771.jpeg \n inflating: chest_xray/train/PNEUMONIA/person851_virus_1496.jpeg \n inflating: chest_xray/train/PNEUMONIA/person852_virus_1497.jpeg \n inflating: chest_xray/train/PNEUMONIA/person853_bacteria_2774.jpeg \n inflating: chest_xray/train/PNEUMONIA/person853_bacteria_2775.jpeg \n inflating: chest_xray/train/PNEUMONIA/person853_virus_1498.jpeg \n inflating: chest_xray/train/PNEUMONIA/person854_bacteria_2776.jpeg \n inflating: chest_xray/train/PNEUMONIA/person855_bacteria_2777.jpeg \n inflating: chest_xray/train/PNEUMONIA/person855_virus_1500.jpeg \n inflating: chest_xray/train/PNEUMONIA/person858_bacteria_2780.jpeg \n inflating: chest_xray/train/PNEUMONIA/person859_virus_1504.jpeg \n inflating: chest_xray/train/PNEUMONIA/person860_virus_1505.jpeg \n inflating: chest_xray/train/PNEUMONIA/person861_virus_1506.jpeg \n inflating: chest_xray/train/PNEUMONIA/person862_bacteria_2784.jpeg \n inflating: chest_xray/train/PNEUMONIA/person862_virus_1507.jpeg \n inflating: chest_xray/train/PNEUMONIA/person863_bacteria_2785.jpeg \n inflating: chest_xray/train/PNEUMONIA/person863_virus_1508.jpeg \n inflating: chest_xray/train/PNEUMONIA/person864_virus_1509.jpeg \n inflating: chest_xray/train/PNEUMONIA/person866_bacteria_2788.jpeg \n inflating: chest_xray/train/PNEUMONIA/person866_virus_1511.jpeg \n inflating: chest_xray/train/PNEUMONIA/person867_bacteria_2789.jpeg \n inflating: chest_xray/train/PNEUMONIA/person867_virus_1512.jpeg \n inflating: chest_xray/train/PNEUMONIA/person868_virus_1513.jpeg \n inflating: chest_xray/train/PNEUMONIA/person868_virus_1514.jpeg \n inflating: chest_xray/train/PNEUMONIA/person86_virus_159.jpeg \n inflating: chest_xray/train/PNEUMONIA/person870_bacteria_2792.jpeg \n inflating: chest_xray/train/PNEUMONIA/person870_virus_1516.jpeg \n inflating: chest_xray/train/PNEUMONIA/person871_bacteria_2793.jpeg \n inflating: chest_xray/train/PNEUMONIA/person871_virus_1517.jpeg \n inflating: chest_xray/train/PNEUMONIA/person872_bacteria_2795.jpeg \n inflating: chest_xray/train/PNEUMONIA/person873_bacteria_2796.jpeg \n inflating: chest_xray/train/PNEUMONIA/person874_bacteria_2797.jpeg \n inflating: chest_xray/train/PNEUMONIA/person875_bacteria_2798.jpeg \n inflating: chest_xray/train/PNEUMONIA/person876_bacteria_2799.jpeg \n inflating: chest_xray/train/PNEUMONIA/person877_bacteria_2800.jpeg \n inflating: chest_xray/train/PNEUMONIA/person877_virus_1525.jpeg \n inflating: chest_xray/train/PNEUMONIA/person878_bacteria_2801.jpeg \n inflating: chest_xray/train/PNEUMONIA/person878_virus_1526.jpeg \n inflating: chest_xray/train/PNEUMONIA/person87_virus_160.jpeg \n inflating: chest_xray/train/PNEUMONIA/person880_bacteria_2804.jpeg \n inflating: chest_xray/train/PNEUMONIA/person880_virus_1529.jpeg \n inflating: chest_xray/train/PNEUMONIA/person881_bacteria_2805.jpeg \n inflating: chest_xray/train/PNEUMONIA/person881_virus_1531.jpeg \n inflating: chest_xray/train/PNEUMONIA/person882_bacteria_2806.jpeg \n inflating: chest_xray/train/PNEUMONIA/person883_bacteria_2807.jpeg \n inflating: chest_xray/train/PNEUMONIA/person883_virus_1533.jpeg \n inflating: chest_xray/train/PNEUMONIA/person884_bacteria_2808.jpeg \n inflating: chest_xray/train/PNEUMONIA/person884_virus_1534.jpeg \n inflating: chest_xray/train/PNEUMONIA/person885_bacteria_2809.jpeg \n inflating: chest_xray/train/PNEUMONIA/person886_bacteria_2810.jpeg \n inflating: chest_xray/train/PNEUMONIA/person886_virus_1536.jpeg \n inflating: chest_xray/train/PNEUMONIA/person887_bacteria_2811.jpeg \n inflating: chest_xray/train/PNEUMONIA/person888_bacteria_2812.jpeg \n inflating: chest_xray/train/PNEUMONIA/person888_virus_1538.jpeg \n inflating: chest_xray/train/PNEUMONIA/person889_bacteria_2813.jpeg \n inflating: chest_xray/train/PNEUMONIA/person88_virus_161.jpeg \n inflating: chest_xray/train/PNEUMONIA/person88_virus_163.jpeg \n inflating: chest_xray/train/PNEUMONIA/person88_virus_164.jpeg \n inflating: chest_xray/train/PNEUMONIA/person88_virus_165.jpeg \n inflating: chest_xray/train/PNEUMONIA/person88_virus_166.jpeg \n inflating: chest_xray/train/PNEUMONIA/person88_virus_167.jpeg \n inflating: chest_xray/train/PNEUMONIA/person890_bacteria_2814.jpeg \n inflating: chest_xray/train/PNEUMONIA/person890_virus_1540.jpeg \n inflating: chest_xray/train/PNEUMONIA/person891_virus_1541.jpeg \n inflating: chest_xray/train/PNEUMONIA/person892_bacteria_2817.jpeg \n inflating: chest_xray/train/PNEUMONIA/person893_bacteria_2818.jpeg \n inflating: chest_xray/train/PNEUMONIA/person894_bacteria_2819.jpeg \n inflating: chest_xray/train/PNEUMONIA/person894_virus_1546.jpeg \n inflating: chest_xray/train/PNEUMONIA/person895_bacteria_2820.jpeg \n inflating: chest_xray/train/PNEUMONIA/person895_virus_1547.jpeg \n inflating: chest_xray/train/PNEUMONIA/person896_bacteria_2821.jpeg \n inflating: chest_xray/train/PNEUMONIA/person896_virus_1548.jpeg \n inflating: chest_xray/train/PNEUMONIA/person897_bacteria_2822.jpeg \n inflating: chest_xray/train/PNEUMONIA/person898_bacteria_2823.jpeg \n inflating: chest_xray/train/PNEUMONIA/person898_virus_1552.jpeg \n inflating: chest_xray/train/PNEUMONIA/person899_virus_1553.jpeg \n inflating: chest_xray/train/PNEUMONIA/person89_virus_168.jpeg \n inflating: chest_xray/train/PNEUMONIA/person8_bacteria_37.jpeg \n inflating: chest_xray/train/PNEUMONIA/person900_virus_1554.jpeg \n inflating: chest_xray/train/PNEUMONIA/person901_virus_1555.jpeg \n inflating: chest_xray/train/PNEUMONIA/person902_bacteria_2827.jpeg \n inflating: chest_xray/train/PNEUMONIA/person903_bacteria_2828.jpeg \n inflating: chest_xray/train/PNEUMONIA/person904_bacteria_2829.jpeg \n inflating: chest_xray/train/PNEUMONIA/person905_bacteria_2830.jpeg \n inflating: chest_xray/train/PNEUMONIA/person905_virus_1561.jpeg \n inflating: chest_xray/train/PNEUMONIA/person906_bacteria_2831.jpeg \n inflating: chest_xray/train/PNEUMONIA/person906_virus_1562.jpeg \n inflating: chest_xray/train/PNEUMONIA/person907_bacteria_2832.jpeg \n inflating: chest_xray/train/PNEUMONIA/person907_virus_1563.jpeg \n inflating: chest_xray/train/PNEUMONIA/person908_virus_1564.jpeg \n inflating: chest_xray/train/PNEUMONIA/person909_bacteria_2834.jpeg \n inflating: chest_xray/train/PNEUMONIA/person909_virus_1565.jpeg \n inflating: chest_xray/train/PNEUMONIA/person90_virus_169.jpeg \n inflating: chest_xray/train/PNEUMONIA/person90_virus_170.jpeg \n inflating: chest_xray/train/PNEUMONIA/person911_bacteria_2836.jpeg \n inflating: chest_xray/train/PNEUMONIA/person911_virus_1567.jpeg \n inflating: chest_xray/train/PNEUMONIA/person912_bacteria_2837.jpeg \n inflating: chest_xray/train/PNEUMONIA/person913_bacteria_2838.jpeg \n inflating: chest_xray/train/PNEUMONIA/person913_virus_1570.jpeg \n inflating: chest_xray/train/PNEUMONIA/person914_bacteria_2839.jpeg \n inflating: chest_xray/train/PNEUMONIA/person914_virus_1571.jpeg \n inflating: chest_xray/train/PNEUMONIA/person915_virus_1572.jpeg \n inflating: chest_xray/train/PNEUMONIA/person916_bacteria_2841.jpeg \n inflating: chest_xray/train/PNEUMONIA/person917_bacteria_2842.jpeg \n inflating: chest_xray/train/PNEUMONIA/person918_bacteria_2843.jpeg \n inflating: chest_xray/train/PNEUMONIA/person918_virus_1575.jpeg \n inflating: chest_xray/train/PNEUMONIA/person919_bacteria_2844.jpeg \n inflating: chest_xray/train/PNEUMONIA/person919_virus_1576.jpeg \n inflating: chest_xray/train/PNEUMONIA/person920_bacteria_2845.jpeg \n inflating: chest_xray/train/PNEUMONIA/person920_virus_1577.jpeg \n inflating: chest_xray/train/PNEUMONIA/person921_bacteria_2846.jpeg \n inflating: chest_xray/train/PNEUMONIA/person921_virus_1578.jpeg \n inflating: chest_xray/train/PNEUMONIA/person922_bacteria_2847.jpeg \n inflating: chest_xray/train/PNEUMONIA/person923_bacteria_2848.jpeg \n inflating: chest_xray/train/PNEUMONIA/person924_bacteria_2849.jpeg \n inflating: chest_xray/train/PNEUMONIA/person924_virus_1581.jpeg \n inflating: chest_xray/train/PNEUMONIA/person925_bacteria_2850.jpeg \n inflating: chest_xray/train/PNEUMONIA/person925_virus_1582.jpeg \n inflating: chest_xray/train/PNEUMONIA/person926_virus_1583.jpeg \n inflating: chest_xray/train/PNEUMONIA/person927_bacteria_2852.jpeg \n inflating: chest_xray/train/PNEUMONIA/person927_virus_1584.jpeg \n inflating: chest_xray/train/PNEUMONIA/person928_virus_1586.jpeg \n inflating: chest_xray/train/PNEUMONIA/person929_bacteria_2854.jpeg \n inflating: chest_xray/train/PNEUMONIA/person929_virus_1588.jpeg \n inflating: chest_xray/train/PNEUMONIA/person929_virus_1589.jpeg \n inflating: chest_xray/train/PNEUMONIA/person92_virus_174.jpeg \n inflating: chest_xray/train/PNEUMONIA/person930_bacteria_2855.jpeg \n inflating: chest_xray/train/PNEUMONIA/person931_bacteria_2856.jpeg \n inflating: chest_xray/train/PNEUMONIA/person931_virus_1592.jpeg \n inflating: chest_xray/train/PNEUMONIA/person932_virus_1593.jpeg \n inflating: chest_xray/train/PNEUMONIA/person933_bacteria_2858.jpeg \n inflating: chest_xray/train/PNEUMONIA/person933_virus_1594.jpeg \n inflating: chest_xray/train/PNEUMONIA/person934_bacteria_2859.jpeg \n inflating: chest_xray/train/PNEUMONIA/person934_virus_1595.jpeg \n inflating: chest_xray/train/PNEUMONIA/person935_virus_1597.jpeg \n inflating: chest_xray/train/PNEUMONIA/person936_bacteria_2861.jpeg \n inflating: chest_xray/train/PNEUMONIA/person936_virus_1598.jpeg \n inflating: chest_xray/train/PNEUMONIA/person937_bacteria_2862.jpeg \n inflating: chest_xray/train/PNEUMONIA/person937_virus_1599.jpeg \n inflating: chest_xray/train/PNEUMONIA/person938_bacteria_2863.jpeg \n inflating: chest_xray/train/PNEUMONIA/person938_virus_1600.jpeg \n inflating: chest_xray/train/PNEUMONIA/person939_bacteria_2864.jpeg \n inflating: chest_xray/train/PNEUMONIA/person93_virus_175.jpeg \n inflating: chest_xray/train/PNEUMONIA/person940_bacteria_2865.jpeg \n inflating: chest_xray/train/PNEUMONIA/person940_virus_1602.jpeg \n inflating: chest_xray/train/PNEUMONIA/person940_virus_1604.jpeg \n inflating: chest_xray/train/PNEUMONIA/person940_virus_1605.jpeg \n inflating: chest_xray/train/PNEUMONIA/person940_virus_1607.jpeg \n inflating: chest_xray/train/PNEUMONIA/person940_virus_1609.jpeg \n inflating: chest_xray/train/PNEUMONIA/person941_virus_1610.jpeg \n inflating: chest_xray/train/PNEUMONIA/person942_bacteria_2867.jpeg \n inflating: chest_xray/train/PNEUMONIA/person942_virus_1611.jpeg \n inflating: chest_xray/train/PNEUMONIA/person943_bacteria_2868.jpeg \n inflating: chest_xray/train/PNEUMONIA/person944_bacteria_2869.jpeg \n inflating: chest_xray/train/PNEUMONIA/person945_bacteria_2870.jpeg \n inflating: chest_xray/train/PNEUMONIA/person945_virus_1616.jpeg \n inflating: chest_xray/train/PNEUMONIA/person946_bacteria_2871.jpeg \n inflating: chest_xray/train/PNEUMONIA/person947_bacteria_2872.jpeg \n inflating: chest_xray/train/PNEUMONIA/person947_virus_1618.jpeg \n inflating: chest_xray/train/PNEUMONIA/person949_bacteria_2874.jpeg \n inflating: chest_xray/train/PNEUMONIA/person949_virus_1620.jpeg \n inflating: chest_xray/train/PNEUMONIA/person94_virus_176.jpeg \n inflating: chest_xray/train/PNEUMONIA/person950_virus_1621.jpeg \n inflating: chest_xray/train/PNEUMONIA/person951_bacteria_2876.jpeg \n inflating: chest_xray/train/PNEUMONIA/person951_virus_1622.jpeg \n inflating: chest_xray/train/PNEUMONIA/person952_bacteria_2877.jpeg \n inflating: chest_xray/train/PNEUMONIA/person952_virus_1623.jpeg \n inflating: chest_xray/train/PNEUMONIA/person953_bacteria_2878.jpeg \n inflating: chest_xray/train/PNEUMONIA/person954_bacteria_2879.jpeg \n inflating: chest_xray/train/PNEUMONIA/person954_virus_1626.jpeg \n inflating: chest_xray/train/PNEUMONIA/person955_bacteria_2880.jpeg \n inflating: chest_xray/train/PNEUMONIA/person955_virus_1627.jpeg \n inflating: chest_xray/train/PNEUMONIA/person956_bacteria_2881.jpeg \n inflating: chest_xray/train/PNEUMONIA/person956_virus_1628.jpeg \n inflating: chest_xray/train/PNEUMONIA/person957_bacteria_2882.jpeg \n inflating: chest_xray/train/PNEUMONIA/person957_virus_1629.jpeg \n inflating: chest_xray/train/PNEUMONIA/person958_bacteria_2883.jpeg \n inflating: chest_xray/train/PNEUMONIA/person958_virus_1630.jpeg \n inflating: chest_xray/train/PNEUMONIA/person959_bacteria_2884.jpeg \n inflating: chest_xray/train/PNEUMONIA/person95_virus_177.jpeg \n inflating: chest_xray/train/PNEUMONIA/person960_bacteria_2885.jpeg \n inflating: chest_xray/train/PNEUMONIA/person960_virus_1633.jpeg \n inflating: chest_xray/train/PNEUMONIA/person961_bacteria_2886.jpeg \n inflating: chest_xray/train/PNEUMONIA/person961_virus_1634.jpeg \n inflating: chest_xray/train/PNEUMONIA/person962_bacteria_2887.jpeg \n inflating: chest_xray/train/PNEUMONIA/person962_virus_1635.jpeg \n inflating: chest_xray/train/PNEUMONIA/person963_bacteria_2888.jpeg \n inflating: chest_xray/train/PNEUMONIA/person963_virus_1636.jpeg \n inflating: chest_xray/train/PNEUMONIA/person964_bacteria_2889.jpeg \n inflating: chest_xray/train/PNEUMONIA/person964_virus_1637.jpeg \n inflating: chest_xray/train/PNEUMONIA/person965_bacteria_2890.jpeg \n inflating: chest_xray/train/PNEUMONIA/person965_virus_1638.jpeg \n inflating: chest_xray/train/PNEUMONIA/person966_bacteria_2891.jpeg \n inflating: chest_xray/train/PNEUMONIA/person966_virus_1639.jpeg \n inflating: chest_xray/train/PNEUMONIA/person967_bacteria_2892.jpeg \n inflating: chest_xray/train/PNEUMONIA/person967_virus_1640.jpeg \n inflating: chest_xray/train/PNEUMONIA/person968_bacteria_2893.jpeg \n inflating: chest_xray/train/PNEUMONIA/person968_virus_1642.jpeg \n inflating: chest_xray/train/PNEUMONIA/person969_bacteria_2894.jpeg \n inflating: chest_xray/train/PNEUMONIA/person969_virus_1643.jpeg \n inflating: chest_xray/train/PNEUMONIA/person96_virus_178.jpeg \n inflating: chest_xray/train/PNEUMONIA/person96_virus_179.jpeg \n inflating: chest_xray/train/PNEUMONIA/person970_bacteria_2895.jpeg \n inflating: chest_xray/train/PNEUMONIA/person970_virus_1644.jpeg \n inflating: chest_xray/train/PNEUMONIA/person971_bacteria_2896.jpeg \n inflating: chest_xray/train/PNEUMONIA/person972_bacteria_2897.jpeg \n inflating: chest_xray/train/PNEUMONIA/person972_virus_1646.jpeg \n inflating: chest_xray/train/PNEUMONIA/person973_virus_1647.jpeg \n inflating: chest_xray/train/PNEUMONIA/person974_bacteria_2899.jpeg \n inflating: chest_xray/train/PNEUMONIA/person974_virus_1649.jpeg \n inflating: chest_xray/train/PNEUMONIA/person975_virus_1650.jpeg \n inflating: chest_xray/train/PNEUMONIA/person976_bacteria_2901.jpeg \n inflating: chest_xray/train/PNEUMONIA/person976_virus_1651.jpeg \n inflating: chest_xray/train/PNEUMONIA/person977_bacteria_2902.jpeg \n inflating: chest_xray/train/PNEUMONIA/person977_virus_1652.jpeg \n inflating: chest_xray/train/PNEUMONIA/person978_bacteria_2904.jpeg \n inflating: chest_xray/train/PNEUMONIA/person978_virus_1653.jpeg \n inflating: chest_xray/train/PNEUMONIA/person979_bacteria_2905.jpeg \n inflating: chest_xray/train/PNEUMONIA/person979_virus_1654.jpeg \n inflating: chest_xray/train/PNEUMONIA/person97_virus_180.jpeg \n inflating: chest_xray/train/PNEUMONIA/person97_virus_181.jpeg \n inflating: chest_xray/train/PNEUMONIA/person980_bacteria_2906.jpeg \n inflating: chest_xray/train/PNEUMONIA/person980_virus_1655.jpeg \n inflating: chest_xray/train/PNEUMONIA/person981_bacteria_2907.jpeg \n inflating: chest_xray/train/PNEUMONIA/person981_bacteria_2908.jpeg \n inflating: chest_xray/train/PNEUMONIA/person981_virus_1657.jpeg \n inflating: chest_xray/train/PNEUMONIA/person982_bacteria_2909.jpeg \n inflating: chest_xray/train/PNEUMONIA/person982_virus_1658.jpeg \n inflating: chest_xray/train/PNEUMONIA/person983_bacteria_2910.jpeg \n inflating: chest_xray/train/PNEUMONIA/person983_virus_1660.jpeg \n inflating: chest_xray/train/PNEUMONIA/person984_bacteria_2911.jpeg \n inflating: chest_xray/train/PNEUMONIA/person985_bacteria_2912.jpeg \n inflating: chest_xray/train/PNEUMONIA/person986_bacteria_2913.jpeg \n inflating: chest_xray/train/PNEUMONIA/person987_bacteria_2914.jpeg \n inflating: chest_xray/train/PNEUMONIA/person988_bacteria_2915.jpeg \n inflating: chest_xray/train/PNEUMONIA/person988_virus_1666.jpeg \n inflating: chest_xray/train/PNEUMONIA/person989_virus_1667.jpeg \n inflating: chest_xray/train/PNEUMONIA/person98_virus_182.jpeg \n inflating: chest_xray/train/PNEUMONIA/person990_bacteria_2917.jpeg \n inflating: chest_xray/train/PNEUMONIA/person991_bacteria_2918.jpeg \n inflating: chest_xray/train/PNEUMONIA/person991_virus_1669.jpeg \n inflating: chest_xray/train/PNEUMONIA/person992_bacteria_2919.jpeg \n inflating: chest_xray/train/PNEUMONIA/person992_bacteria_2920.jpeg \n inflating: chest_xray/train/PNEUMONIA/person992_virus_1670.jpeg \n inflating: chest_xray/train/PNEUMONIA/person993_bacteria_2921.jpeg \n inflating: chest_xray/train/PNEUMONIA/person993_virus_1671.jpeg \n inflating: chest_xray/train/PNEUMONIA/person994_bacteria_2922.jpeg \n inflating: chest_xray/train/PNEUMONIA/person994_virus_1672.jpeg \n inflating: chest_xray/train/PNEUMONIA/person995_bacteria_2923.jpeg \n inflating: chest_xray/train/PNEUMONIA/person995_virus_1676.jpeg \n inflating: chest_xray/train/PNEUMONIA/person996_bacteria_2924.jpeg \n inflating: chest_xray/train/PNEUMONIA/person996_virus_1677.jpeg \n inflating: chest_xray/train/PNEUMONIA/person997_bacteria_2926.jpeg \n inflating: chest_xray/train/PNEUMONIA/person997_virus_1678.jpeg \n inflating: chest_xray/train/PNEUMONIA/person998_bacteria_2927.jpeg \n inflating: chest_xray/train/PNEUMONIA/person998_bacteria_2928.jpeg \n inflating: chest_xray/train/PNEUMONIA/person99_virus_183.jpeg \n inflating: chest_xray/train/PNEUMONIA/person9_bacteria_38.jpeg \n inflating: chest_xray/train/PNEUMONIA/person9_bacteria_39.jpeg \n inflating: chest_xray/train/PNEUMONIA/person9_bacteria_40.jpeg \n inflating: chest_xray/train/PNEUMONIA/person9_bacteria_41.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1427-0001.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1430-0001.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1431-0001.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1436-0001.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1437-0001.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1438-0001.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1440-0001.jpeg \n inflating: chest_xray/val/NORMAL/NORMAL2-IM-1442-0001.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1946_bacteria_4874.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1946_bacteria_4875.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1947_bacteria_4876.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1949_bacteria_4880.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1950_bacteria_4881.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1951_bacteria_4882.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1952_bacteria_4883.jpeg \n inflating: chest_xray/val/PNEUMONIA/person1954_bacteria_4886.jpeg \n"
]
],
[
[
"# Creating Data Generators",
"_____no_output_____"
]
],
[
[
"train_dir = '/content/chest_xray/train'\nval_dir = '/content/chest_xray/val'\ntest_dir = '/content/chest_xray/test'",
"_____no_output_____"
],
[
"IMG_HEIGHT = 224\nIMG_WIDTH = 224\n\nBATCH_SIZE = 32",
"_____no_output_____"
],
[
"train_datagen = tf.keras.preprocessing.image.ImageDataGenerator(\n rescale=1./255,\n rotation_range=20,\n brightness_range=(1.2, 1.5),\n horizontal_flip=True\n)\n\nval_datagen = tf.keras.preprocessing.image.ImageDataGenerator(\n rescale=1./255\n)\n\ntest_datagen = tf.keras.preprocessing.image.ImageDataGenerator(\n rescale=1./255\n)",
"_____no_output_____"
],
[
"train_data = train_datagen.flow_from_directory(\n train_dir,\n target_size=(IMG_HEIGHT, IMG_WIDTH),\n class_mode='binary',\n batch_size=BATCH_SIZE\n)\n\nval_data = train_datagen.flow_from_directory(\n val_dir,\n target_size=(IMG_HEIGHT, IMG_WIDTH),\n class_mode='binary',\n batch_size=BATCH_SIZE\n)\n\ntest_data = train_datagen.flow_from_directory(\n test_dir,\n target_size=(IMG_HEIGHT, IMG_WIDTH),\n class_mode='binary',\n batch_size=BATCH_SIZE\n)",
"Found 5216 images belonging to 2 classes.\nFound 16 images belonging to 2 classes.\nFound 624 images belonging to 2 classes.\n"
]
],
[
[
"# Model Building",
"_____no_output_____"
],
[
"## Model 1 (MobileNetV2)",
"_____no_output_____"
]
],
[
[
"mobilenet = tf.keras.applications.MobileNetV2(\n input_shape=(IMG_HEIGHT, IMG_WIDTH, 3),\n include_top=False,\n weights='imagenet',\n pooling='avg'\n)\n\nmobilenet.trainable = False",
"Downloading data from https://storage.googleapis.com/tensorflow/keras-applications/mobilenet_v2/mobilenet_v2_weights_tf_dim_ordering_tf_kernels_1.0_224_no_top.h5\n9412608/9406464 [==============================] - 0s 0us/step\n9420800/9406464 [==============================] - 0s 0us/step\n"
],
[
"model_1 = tf.keras.models.Sequential([\n mobilenet,\n\n tf.keras.layers.Dense(64, activation='leaky_relu', kernel_regularizer='l2', name='fc1'),\n tf.keras.layers.Dense(32, activation='leaky_relu', kernel_regularizer='l2', name='fc2'),\n tf.keras.layers.Dense(16, activation='leaky_relu', kernel_regularizer='l2', name='fc3'),\n\n tf.keras.layers.Dense(1, activation='sigmoid', name='output')\n ])\n\n\nprint(model_1.summary())\ntf.keras.utils.plot_model(model_1)",
"Model: \"sequential\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n mobilenetv2_1.00_224 (Funct (None, 1280) 2257984 \n ional) \n \n fc1 (Dense) (None, 64) 81984 \n \n fc2 (Dense) (None, 32) 2080 \n \n fc3 (Dense) (None, 16) 528 \n \n output (Dense) (None, 1) 17 \n \n=================================================================\nTotal params: 2,342,593\nTrainable params: 84,609\nNon-trainable params: 2,257,984\n_________________________________________________________________\nNone\n"
],
[
"EPOCHS = 10\n\nmodel_1.compile(\n optimizer='adam',\n loss='binary_crossentropy',\n metrics=[\n 'accuracy',\n tf.keras.metrics.AUC(name='auc')\n ]\n)\n\nhistory_1 = model_1.fit(\n train_data,\n validation_data=val_data,\n batch_size=BATCH_SIZE,\n epochs=EPOCHS,\n callbacks=[\n tf.keras.callbacks.EarlyStopping(\n monitor='val_loss',\n patience=3,\n restore_best_weights=True\n )\n ]\n)",
"Epoch 1/10\n163/163 [==============================] - 129s 770ms/step - loss: 1.0477 - accuracy: 0.9227 - auc: 0.9727 - val_loss: 0.6392 - val_accuracy: 0.9375 - val_auc: 0.9844\nEpoch 2/10\n163/163 [==============================] - 122s 745ms/step - loss: 0.4816 - accuracy: 0.9477 - auc: 0.9863 - val_loss: 0.6502 - val_accuracy: 0.8125 - val_auc: 0.9375\nEpoch 3/10\n163/163 [==============================] - 121s 742ms/step - loss: 0.3325 - accuracy: 0.9536 - auc: 0.9886 - val_loss: 0.9734 - val_accuracy: 0.6250 - val_auc: 0.9219\nEpoch 4/10\n163/163 [==============================] - 121s 743ms/step - loss: 0.2801 - accuracy: 0.9519 - auc: 0.9871 - val_loss: 0.5849 - val_accuracy: 0.7500 - val_auc: 1.0000\nEpoch 5/10\n163/163 [==============================] - 125s 767ms/step - loss: 0.2293 - accuracy: 0.9578 - auc: 0.9909 - val_loss: 0.3756 - val_accuracy: 0.9375 - val_auc: 0.9844\nEpoch 6/10\n163/163 [==============================] - 126s 775ms/step - loss: 0.2155 - accuracy: 0.9582 - auc: 0.9898 - val_loss: 0.4949 - val_accuracy: 0.8125 - val_auc: 0.8906\nEpoch 7/10\n163/163 [==============================] - 126s 770ms/step - loss: 0.2049 - accuracy: 0.9559 - auc: 0.9894 - val_loss: 0.4525 - val_accuracy: 0.8125 - val_auc: 0.9688\nEpoch 8/10\n163/163 [==============================] - 127s 778ms/step - loss: 0.2015 - accuracy: 0.9526 - auc: 0.9887 - val_loss: 0.7513 - val_accuracy: 0.7500 - val_auc: 0.9844\n"
],
[
"def evaluate(model): \n results = model.evaluate(test_data, verbose=0)\n\n accuracy = results[1]\n auc = results[2]\n\n true_labels = test_data.labels\n pred_labels = np.squeeze(np.array(model.predict(test_data) >= 0.5, dtype=np.int))\n cm = confusion_matrix(true_labels, pred_labels)\n\n tn, fp, fn, tp = cm.ravel()\n\n precision = tp / (tp + fp)\n recall = tp / (tp + fn)\n\n print(\"Accuracy: {:.2f}\".format(accuracy))\n print(\"AUC: {:.2f}\".format(auc))\n print(\"Precision: {:.2f}\".format(precision))\n print(\"Recall: {:.2f}\".format(recall))",
"_____no_output_____"
],
[
"evaluate(model_1)",
"Accuracy: 0.86\nAUC: 0.94\nPrecision: 0.62\nRecall: 0.71\n"
]
],
[
[
"## Model 2 (ResNet50V2)",
"_____no_output_____"
]
],
[
[
"resnet = tf.keras.applications.resnet_v2.ResNet50V2(\n input_shape=(IMG_HEIGHT, IMG_WIDTH, 3),\n include_top=False,\n weights='imagenet',\n pooling='avg'\n)\n\nresnet.trainable = False",
"_____no_output_____"
],
[
"model_2 = tf.keras.models.Sequential([\n resnet,\n\n tf.keras.layers.Dense(64, activation='leaky_relu', kernel_regularizer='l2', name='fc1'),\n tf.keras.layers.Dense(32, activation='leaky_relu', kernel_regularizer='l2', name='fc2'),\n tf.keras.layers.Dense(16, activation='leaky_relu', kernel_regularizer='l2', name='fc3'),\n\n tf.keras.layers.Dense(1, activation='sigmoid', name='output')\n ])\n\n\nprint(model_2.summary())\ntf.keras.utils.plot_model(model_2)",
"Model: \"sequential_6\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n resnet50v2 (Functional) (None, 2048) 23564800 \n \n fc1 (Dense) (None, 64) 131136 \n \n fc2 (Dense) (None, 32) 2080 \n \n fc3 (Dense) (None, 16) 528 \n \n output (Dense) (None, 1) 17 \n \n=================================================================\nTotal params: 23,698,561\nTrainable params: 133,761\nNon-trainable params: 23,564,800\n_________________________________________________________________\nNone\n"
],
[
"EPOCHS = 10\n\nmodel_2.compile(\n optimizer='adam',\n loss='binary_crossentropy',\n metrics=[\n 'accuracy',\n tf.keras.metrics.AUC(name='auc')\n ]\n)\n\nhistory_2 = model_2.fit(\n train_data,\n validation_data=val_data,\n batch_size=BATCH_SIZE,\n epochs=EPOCHS,\n callbacks=[\n tf.keras.callbacks.EarlyStopping(\n monitor='val_loss',\n patience=3,\n restore_best_weights=True\n )\n ]\n)",
"Epoch 1/10\n163/163 [==============================] - 181s 820ms/step - loss: 0.9415 - accuracy: 0.9317 - auc: 0.9772 - val_loss: 1.0034 - val_accuracy: 0.7500 - val_auc: 0.9844\nEpoch 2/10\n163/163 [==============================] - 133s 813ms/step - loss: 0.4437 - accuracy: 0.9434 - auc: 0.9822 - val_loss: 0.9529 - val_accuracy: 0.7500 - val_auc: 0.9453\nEpoch 3/10\n163/163 [==============================] - 132s 811ms/step - loss: 0.2977 - accuracy: 0.9601 - auc: 0.9886 - val_loss: 0.7499 - val_accuracy: 0.7500 - val_auc: 0.9531\nEpoch 4/10\n163/163 [==============================] - 133s 817ms/step - loss: 0.2532 - accuracy: 0.9546 - auc: 0.9880 - val_loss: 0.4708 - val_accuracy: 0.8750 - val_auc: 1.0000\nEpoch 5/10\n163/163 [==============================] - 133s 814ms/step - loss: 0.2149 - accuracy: 0.9584 - auc: 0.9896 - val_loss: 0.5044 - val_accuracy: 0.7500 - val_auc: 0.9219\nEpoch 6/10\n163/163 [==============================] - 132s 808ms/step - loss: 0.1977 - accuracy: 0.9590 - auc: 0.9904 - val_loss: 0.2456 - val_accuracy: 0.9375 - val_auc: 1.0000\nEpoch 7/10\n163/163 [==============================] - 132s 807ms/step - loss: 0.1895 - accuracy: 0.9582 - auc: 0.9907 - val_loss: 0.6586 - val_accuracy: 0.7500 - val_auc: 0.9844\nEpoch 8/10\n163/163 [==============================] - 133s 815ms/step - loss: 0.1803 - accuracy: 0.9601 - auc: 0.9899 - val_loss: 0.6218 - val_accuracy: 0.7500 - val_auc: 0.9688\nEpoch 9/10\n163/163 [==============================] - 133s 812ms/step - loss: 0.1853 - accuracy: 0.9525 - auc: 0.9889 - val_loss: 0.3314 - val_accuracy: 0.8125 - val_auc: 0.9688\n"
],
[
"evaluate(model_2)",
"Accuracy: 0.88\nAUC: 0.95\nPrecision: 0.63\nRecall: 0.72\n"
]
],
[
[
"## Model 3",
"_____no_output_____"
]
],
[
[
"# Unfreeze all of the layers in the base model\nresnet.trainable = True\n\n# Refreeze every layer except for the last 3\nfor layer in resnet.layers[:-37]:\n layer.trainable = False",
"_____no_output_____"
],
[
"for layer_number, layer in enumerate(resnet.layers):\n print(layer_number, layer.name, layer.trainable)",
"0 input_5 False\n1 conv1_pad False\n2 conv1_conv False\n3 pool1_pad False\n4 pool1_pool False\n5 conv2_block1_preact_bn False\n6 conv2_block1_preact_relu False\n7 conv2_block1_1_conv False\n8 conv2_block1_1_bn False\n9 conv2_block1_1_relu False\n10 conv2_block1_2_pad False\n11 conv2_block1_2_conv False\n12 conv2_block1_2_bn False\n13 conv2_block1_2_relu False\n14 conv2_block1_0_conv False\n15 conv2_block1_3_conv False\n16 conv2_block1_out False\n17 conv2_block2_preact_bn False\n18 conv2_block2_preact_relu False\n19 conv2_block2_1_conv False\n20 conv2_block2_1_bn False\n21 conv2_block2_1_relu False\n22 conv2_block2_2_pad False\n23 conv2_block2_2_conv False\n24 conv2_block2_2_bn False\n25 conv2_block2_2_relu False\n26 conv2_block2_3_conv False\n27 conv2_block2_out False\n28 conv2_block3_preact_bn False\n29 conv2_block3_preact_relu False\n30 conv2_block3_1_conv False\n31 conv2_block3_1_bn False\n32 conv2_block3_1_relu False\n33 conv2_block3_2_pad False\n34 conv2_block3_2_conv False\n35 conv2_block3_2_bn False\n36 conv2_block3_2_relu False\n37 max_pooling2d_6 False\n38 conv2_block3_3_conv False\n39 conv2_block3_out False\n40 conv3_block1_preact_bn False\n41 conv3_block1_preact_relu False\n42 conv3_block1_1_conv False\n43 conv3_block1_1_bn False\n44 conv3_block1_1_relu False\n45 conv3_block1_2_pad False\n46 conv3_block1_2_conv False\n47 conv3_block1_2_bn False\n48 conv3_block1_2_relu False\n49 conv3_block1_0_conv False\n50 conv3_block1_3_conv False\n51 conv3_block1_out False\n52 conv3_block2_preact_bn False\n53 conv3_block2_preact_relu False\n54 conv3_block2_1_conv False\n55 conv3_block2_1_bn False\n56 conv3_block2_1_relu False\n57 conv3_block2_2_pad False\n58 conv3_block2_2_conv False\n59 conv3_block2_2_bn False\n60 conv3_block2_2_relu False\n61 conv3_block2_3_conv False\n62 conv3_block2_out False\n63 conv3_block3_preact_bn False\n64 conv3_block3_preact_relu False\n65 conv3_block3_1_conv False\n66 conv3_block3_1_bn False\n67 conv3_block3_1_relu False\n68 conv3_block3_2_pad False\n69 conv3_block3_2_conv False\n70 conv3_block3_2_bn False\n71 conv3_block3_2_relu False\n72 conv3_block3_3_conv False\n73 conv3_block3_out False\n74 conv3_block4_preact_bn False\n75 conv3_block4_preact_relu False\n76 conv3_block4_1_conv False\n77 conv3_block4_1_bn False\n78 conv3_block4_1_relu False\n79 conv3_block4_2_pad False\n80 conv3_block4_2_conv False\n81 conv3_block4_2_bn False\n82 conv3_block4_2_relu False\n83 max_pooling2d_7 False\n84 conv3_block4_3_conv False\n85 conv3_block4_out False\n86 conv4_block1_preact_bn False\n87 conv4_block1_preact_relu False\n88 conv4_block1_1_conv False\n89 conv4_block1_1_bn False\n90 conv4_block1_1_relu False\n91 conv4_block1_2_pad False\n92 conv4_block1_2_conv False\n93 conv4_block1_2_bn False\n94 conv4_block1_2_relu False\n95 conv4_block1_0_conv False\n96 conv4_block1_3_conv False\n97 conv4_block1_out False\n98 conv4_block2_preact_bn False\n99 conv4_block2_preact_relu False\n100 conv4_block2_1_conv False\n101 conv4_block2_1_bn False\n102 conv4_block2_1_relu False\n103 conv4_block2_2_pad False\n104 conv4_block2_2_conv False\n105 conv4_block2_2_bn False\n106 conv4_block2_2_relu False\n107 conv4_block2_3_conv False\n108 conv4_block2_out False\n109 conv4_block3_preact_bn False\n110 conv4_block3_preact_relu False\n111 conv4_block3_1_conv False\n112 conv4_block3_1_bn False\n113 conv4_block3_1_relu False\n114 conv4_block3_2_pad False\n115 conv4_block3_2_conv False\n116 conv4_block3_2_bn False\n117 conv4_block3_2_relu False\n118 conv4_block3_3_conv False\n119 conv4_block3_out False\n120 conv4_block4_preact_bn False\n121 conv4_block4_preact_relu False\n122 conv4_block4_1_conv False\n123 conv4_block4_1_bn False\n124 conv4_block4_1_relu False\n125 conv4_block4_2_pad False\n126 conv4_block4_2_conv False\n127 conv4_block4_2_bn False\n128 conv4_block4_2_relu False\n129 conv4_block4_3_conv False\n130 conv4_block4_out False\n131 conv4_block5_preact_bn False\n132 conv4_block5_preact_relu False\n133 conv4_block5_1_conv False\n134 conv4_block5_1_bn False\n135 conv4_block5_1_relu False\n136 conv4_block5_2_pad False\n137 conv4_block5_2_conv False\n138 conv4_block5_2_bn False\n139 conv4_block5_2_relu False\n140 conv4_block5_3_conv False\n141 conv4_block5_out False\n142 conv4_block6_preact_bn False\n143 conv4_block6_preact_relu False\n144 conv4_block6_1_conv False\n145 conv4_block6_1_bn False\n146 conv4_block6_1_relu False\n147 conv4_block6_2_pad False\n148 conv4_block6_2_conv False\n149 conv4_block6_2_bn False\n150 conv4_block6_2_relu False\n151 max_pooling2d_8 False\n152 conv4_block6_3_conv False\n153 conv4_block6_out False\n154 conv5_block1_preact_bn True\n155 conv5_block1_preact_relu True\n156 conv5_block1_1_conv True\n157 conv5_block1_1_bn True\n158 conv5_block1_1_relu True\n159 conv5_block1_2_pad True\n160 conv5_block1_2_conv True\n161 conv5_block1_2_bn True\n162 conv5_block1_2_relu True\n163 conv5_block1_0_conv True\n164 conv5_block1_3_conv True\n165 conv5_block1_out True\n166 conv5_block2_preact_bn True\n167 conv5_block2_preact_relu True\n168 conv5_block2_1_conv True\n169 conv5_block2_1_bn True\n170 conv5_block2_1_relu True\n171 conv5_block2_2_pad True\n172 conv5_block2_2_conv True\n173 conv5_block2_2_bn True\n174 conv5_block2_2_relu True\n175 conv5_block2_3_conv True\n176 conv5_block2_out True\n177 conv5_block3_preact_bn True\n178 conv5_block3_preact_relu True\n179 conv5_block3_1_conv True\n180 conv5_block3_1_bn True\n181 conv5_block3_1_relu True\n182 conv5_block3_2_pad True\n183 conv5_block3_2_conv True\n184 conv5_block3_2_bn True\n185 conv5_block3_2_relu True\n186 conv5_block3_3_conv True\n187 conv5_block3_out True\n188 post_bn True\n189 post_relu True\n190 avg_pool True\n"
],
[
"model_3 = tf.keras.models.Sequential([\n resnet,\n\n tf.keras.layers.Dense(64, activation='leaky_relu', kernel_regularizer='l2', name='fc2'),\n tf.keras.layers.Dense(32, activation='leaky_relu', kernel_regularizer='l2', name='fc3'),\n tf.keras.layers.Dense(16, activation='leaky_relu', kernel_regularizer='l2', name='fc4'),\n\n tf.keras.layers.Dense(1, activation='sigmoid', name='output')\n ])\n\n\nprint(model_3.summary())\ntf.keras.utils.plot_model(model_3)",
"Model: \"sequential_9\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n resnet50v2 (Functional) (None, 2048) 23564800 \n \n fc2 (Dense) (None, 64) 131136 \n \n fc3 (Dense) (None, 32) 2080 \n \n fc4 (Dense) (None, 16) 528 \n \n output (Dense) (None, 1) 17 \n \n=================================================================\nTotal params: 23,698,561\nTrainable params: 15,104,641\nNon-trainable params: 8,593,920\n_________________________________________________________________\nNone\n"
],
[
"EPOCHS = 10\n\nmodel_3.compile(\n optimizer = tf.keras.optimizers.Adam(5e-4), # 2x lower learning rate than default,\n loss='binary_crossentropy',\n metrics=[\n 'accuracy',\n tf.keras.metrics.AUC(name='auc')\n ]\n)\n\nhistory_3 = model_3.fit(\n train_data,\n validation_data=val_data,\n batch_size=BATCH_SIZE,\n epochs=EPOCHS,\n callbacks=[\n tf.keras.callbacks.EarlyStopping(\n monitor='val_loss',\n patience=3,\n restore_best_weights=True\n )\n ]\n)",
"Epoch 1/10\n163/163 [==============================] - 144s 850ms/step - loss: 1.0704 - accuracy: 0.9551 - auc: 0.9861 - val_loss: 3.8306 - val_accuracy: 0.6250 - val_auc: 0.6875\nEpoch 2/10\n163/163 [==============================] - 138s 847ms/step - loss: 0.3732 - accuracy: 0.9714 - auc: 0.9943 - val_loss: 0.5758 - val_accuracy: 0.8125 - val_auc: 1.0000\nEpoch 3/10\n163/163 [==============================] - 138s 843ms/step - loss: 0.1941 - accuracy: 0.9822 - auc: 0.9980 - val_loss: 0.2591 - val_accuracy: 0.8750 - val_auc: 0.9844\nEpoch 4/10\n163/163 [==============================] - 137s 839ms/step - loss: 0.1288 - accuracy: 0.9841 - auc: 0.9982 - val_loss: 0.3659 - val_accuracy: 0.8750 - val_auc: 1.0000\nEpoch 5/10\n163/163 [==============================] - 136s 837ms/step - loss: 0.1011 - accuracy: 0.9839 - auc: 0.9979 - val_loss: 0.3914 - val_accuracy: 0.8750 - val_auc: 0.9844\nEpoch 6/10\n163/163 [==============================] - 137s 838ms/step - loss: 0.0805 - accuracy: 0.9860 - auc: 0.9986 - val_loss: 0.0601 - val_accuracy: 1.0000 - val_auc: 1.0000\nEpoch 7/10\n163/163 [==============================] - 137s 838ms/step - loss: 0.0679 - accuracy: 0.9875 - auc: 0.9987 - val_loss: 0.1111 - val_accuracy: 1.0000 - val_auc: 1.0000\nEpoch 8/10\n163/163 [==============================] - 136s 837ms/step - loss: 0.0584 - accuracy: 0.9908 - auc: 0.9988 - val_loss: 0.8631 - val_accuracy: 0.7500 - val_auc: 1.0000\nEpoch 9/10\n163/163 [==============================] - 137s 837ms/step - loss: 0.0554 - accuracy: 0.9910 - auc: 0.9992 - val_loss: 0.0736 - val_accuracy: 1.0000 - val_auc: 1.0000\n"
],
[
"evaluate(model_3)",
"Accuracy: 0.89\nAUC: 0.97\nPrecision: 0.63\nRecall: 0.74\n"
]
],
[
[
"# Model Selection",
"_____no_output_____"
]
],
[
[
"print(\"Model 1:--\")\nevaluate(model_1)\nprint(\"\\nModel 2:--\")\nevaluate(model_2)\nprint(\"\\nModel 3:--\")\nevaluate(model_3)",
"Model 1:--\nAccuracy: 0.85\nAUC: 0.95\nPrecision: 0.64\nRecall: 0.75\n\nModel 2:--\nAccuracy: 0.89\nAUC: 0.96\nPrecision: 0.61\nRecall: 0.67\n\nModel 3:--\nAccuracy: 0.89\nAUC: 0.97\nPrecision: 0.64\nRecall: 0.74\n"
]
],
[
[
"\nAs we can see, model 3 performs the best. Therefore, model 3 is chosen.",
"_____no_output_____"
]
],
[
[
"model_3.save_weights('pneumonia_prediction_model', save_format='h5')",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec617f332aec5adee78eb957597f3a7e3bab226b | 4,777 | ipynb | Jupyter Notebook | 01/file_2.ipynb | yoonkt200/learning-spoons-nano-degree | 1732d08be0b17957f82b8bf876450fe3a95e989c | [
"MIT"
]
| null | null | null | 01/file_2.ipynb | yoonkt200/learning-spoons-nano-degree | 1732d08be0b17957f82b8bf876450fe3a95e989c | [
"MIT"
]
| null | null | null | 01/file_2.ipynb | yoonkt200/learning-spoons-nano-degree | 1732d08be0b17957f82b8bf876450fe3a95e989c | [
"MIT"
]
| 1 | 2021-07-19T10:18:55.000Z | 2021-07-19T10:18:55.000Z | 30.234177 | 85 | 0.452376 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv(\"../dataset/olist_customers_dataset.csv\")",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
ec618d9d8fc9287cdb61be92bef1311c6bd8d600 | 35,305 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Teste-checkpoint.ipynb | daviseemann/bancada-didatica-de-conversores-INEP | 77673e48da75244cb492c27714f9307461058270 | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Teste-checkpoint.ipynb | daviseemann/bancada-didatica-de-conversores-INEP | 77673e48da75244cb492c27714f9307461058270 | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Teste-checkpoint.ipynb | daviseemann/bancada-didatica-de-conversores-INEP | 77673e48da75244cb492c27714f9307461058270 | [
"MIT"
]
| null | null | null | 60.042517 | 22,836 | 0.797932 | [
[
[
"Buck Converter\n=========\n#### Conversor abaixador\n\nO conversor de buck tem uma estrutura simples e de operação direta, uma ótima opção de conversor CC-CC PWM. A tensão de saída de um conversor buck é sempre menor que a tensão de entrada, por isso ele é conhecido também como conversor abaixador.\n\nO conversor buck é composto por uma fonte de tensão contínua, um transistor que funciona como uma chave, um diodo, um indutor, um capacitor e a carga. A chave irá controlar em qual estado estará o circuito, on-time ou off-time. Durante o período on-time, a chave é fechada e o diodo está no estado desligado. Neste intervalo a fonte de tensão transfere energia para o indutor e a corrente sobre ele aumenta. Já no período off-time, a chave estará aberta e o diodo ligado. Assim, a energia armazenada no indutor é liberada para a carga e a corrente no indutor diminui. \n\nO estado estacionário no conversor é atingido quando a transferência de energia dentro do tempo torna-se a mesma que a liberação de energia fora do tempo. Nesse estado a corrente no indutor estabele uma forma de onda triangular periódica e a tensão de saída torna-se quase constante com um pequeno componente de ondulação. ",
"_____no_output_____"
],
[
"#### Esquemático de um conversor buck\n\n",
"_____no_output_____"
],
[
"#### Ganho estático do conversor \n\n",
"_____no_output_____"
],
[
"#### Etapas do projeto:\n\n 1. Definir parâmetros de projeto (Vin, Vout, Po, fs, variação de IL e de Vc)\n 2. Calcular a razão cíclica (D=Vout/Vin)\n 3. Calcular indutância (L=(Vin - Vout).D/fs.(variação de IL)\n 4. Calcular a capacitância (C=variação de IL/8.fs.variação de Vc)\n 5. Calcular os esforços nos semicondutores (Ismd, Isef, Ismax, Idmds, Idef, Idmax, Vsmax e Vdmax)",
"_____no_output_____"
],
[
"#### Tensão de Entrada:",
"_____no_output_____"
]
],
[
[
"Vin = 75",
"_____no_output_____"
]
],
[
[
"#### Tensão de Saída: ",
"_____no_output_____"
]
],
[
[
"Vout = 30",
"_____no_output_____"
]
],
[
[
"#### Potência máxima de saída:",
"_____no_output_____"
]
],
[
[
"Po = 20",
"_____no_output_____"
]
],
[
[
"#### Frequência de comutação: ",
"_____no_output_____"
]
],
[
[
"fs = 20000",
"_____no_output_____"
]
],
[
[
"#### Razão ciclíca:",
"_____no_output_____"
]
],
[
[
"D = Vout/Vin\nprint(\"A razão ciclíca é\", D)",
"A razão ciclíca é 0.4\n"
]
],
[
[
"#### Corrente de saída:",
"_____no_output_____"
]
],
[
[
"Io = Po/Vout\nprint(\"A corrente de saída é\", Io, \"A\")",
"A corrente de saída é 0.6666666666666666 A\n"
]
],
[
[
"#### Ondulação de corrente no indutor:",
"_____no_output_____"
]
],
[
[
"delta_Il = 0.1*Io\nprint(\"A ondulação de corrente no indutor é\", delta_Il, \"A\")",
"A ondulação de corrente no indutor é 0.06666666666666667 A\n"
]
],
[
[
"#### Ondulação de tensão no capacitor:",
"_____no_output_____"
]
],
[
[
"delta_Vc = 0.01*Vout\nprint(\"A ondulação de tensão no capacitor é\", delta_Vc, \"V\")",
"A ondulação de tensão no capacitor é 0.3 V\n"
]
],
[
[
"#### Resistência de carga:",
"_____no_output_____"
]
],
[
[
"Ro = (Vout**2)/Po\nprint(\"A resistência de carga é\", Ro, \"ohms\")",
"A resistência de carga é 45.0 ohms\n"
]
],
[
[
"#### Indutor de Saída:",
"_____no_output_____"
]
],
[
[
"Lo = (Vin-Vout)*D/(fs*delta_Il)\nprint(\"O indutor de saída é\", Lo, \"H\")",
"O indutor de saída é 0.013500000000000002 H\n"
]
],
[
[
"#### Capacitor de Saída:",
"_____no_output_____"
]
],
[
[
"Co = delta_Il/(8*fs*delta_Vc)\nprint(\"O capacitor de saída é\", Co, \"F\")",
"O capacitor de saída é 1.388888888888889e-06 F\n"
]
],
[
[
"### Esforços na chave:\n\n#### Valor médio da corrente na chave:",
"_____no_output_____"
]
],
[
[
"Is_md = D*Io \nprint(\"O valor médio da corrente na chave é\", Is_md, \"A\")",
"O valor médio da corrente na chave é 0.26666666666666666 A\n"
]
],
[
[
"#### Valor eficaz da corrente na chave:",
"_____no_output_____"
]
],
[
[
"Is_ef = (D**0.5)*Io \nprint(\"O valor eficaz da corrente na chave é\", Is_ef, \"A\")",
"O valor eficaz da corrente na chave é 0.4216370213557839 A\n"
]
],
[
[
"#### Valor máximo da corrente na chave:",
"_____no_output_____"
]
],
[
[
"Is_max = Io + delta_Il/2 \nprint(\"O valor máximo da corrente na chave é\", Is_max, \"A\")",
"O valor máximo da corrente na chave é 0.7 A\n"
]
],
[
[
"#### Valor máximo da tensão na chave:",
"_____no_output_____"
]
],
[
[
"Vs_max = Vin\nprint(\"O valor máximo da tensão na chave é\", Vs_max, \"V\")",
"O valor máximo da tensão na chave é 75 V\n"
]
],
[
[
"### Esforços no diodo:\n\n#### Valor médio da corrente no diodo:",
"_____no_output_____"
]
],
[
[
"Id_md = (1-D)*Io \nprint(\"O valor médio da corrente no diodo é\", Id_md, \"A\")",
"O valor médio da corrente no diodo é 0.39999999999999997 A\n"
]
],
[
[
"#### Valor eficaz da corrente no diodo:",
"_____no_output_____"
]
],
[
[
"Id_ef = ((1-D)**0.5)*Io \nprint(\"O valor eficaz da corrente no diodo é\", Id_ef, \"A\")",
"O valor eficaz da corrente no diodo é 0.5163977794943222 A\n"
]
],
[
[
"#### Valor máximo da corrente no diodo:",
"_____no_output_____"
]
],
[
[
"Id_max = Io + delta_Il/2 \nprint(\"O valor máximo da corrente no diodo é\", Id_max, \"A\")",
"O valor máximo da corrente no diodo é 0.7 A\n"
]
],
[
[
"#### Valor máximo da tensão no diodo:",
"_____no_output_____"
]
],
[
[
"Vd_max = Vin\nprint(\"O valor máximo da tensão no diodo é\", Vd_max, \"V\")",
"O valor máximo da tensão no diodo é 75 V\n"
]
],
[
[
"#### Resistência crítica:",
"_____no_output_____"
]
],
[
[
"Rcrit = 2*Lo*fs/(1-D)\nprint(\"A resistência crítica é\", Rcrit, \"ohms\")",
"A resistência crítica é 900.0000000000002 ohms\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6192b6cae88f7d1502fea401eb35c8802c7fbe | 14,977 | ipynb | Jupyter Notebook | notebooks/lab-01.ipynb | 18lejoh/mm-labs | 9cc81a388034c661a1de18921110146424cf41e2 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/lab-01.ipynb | 18lejoh/mm-labs | 9cc81a388034c661a1de18921110146424cf41e2 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/lab-01.ipynb | 18lejoh/mm-labs | 9cc81a388034c661a1de18921110146424cf41e2 | [
"BSD-3-Clause"
]
| null | null | null | 26.840502 | 478 | 0.568271 | [
[
[
"# Lab 1\n## Introduction\nIn this lab, we will begin to learn how to use Python to solve mathematical problems.\n\nThe goal of this lab is to teach you how to write basic, everyday mathematical expressions in Python. This will give you the basic tools you’ll need for future labs, covering quiver plots, Euler’s method and the numerical solution of differential equations, and for subsequent topics in second- and third-year Mathematics.",
"_____no_output_____"
],
[
"### Executing simple mathematical expressions\nIn Jupyter notebooks like this one you can execute Python in cells by pressing Ctrl-Enter (or Shift-Enter).\n\nLet's start simple! Execute the cell below and observe the output.",
"_____no_output_____"
]
],
[
[
"1 + 1",
"_____no_output_____"
]
],
[
[
"Python uses standard symbols for the basic algebraic operations of addition, subtraction, multiplication and division. Here are some examples to try.",
"_____no_output_____"
]
],
[
[
"17 - 2.5",
"_____no_output_____"
],
[
"7 * 4",
"_____no_output_____"
],
[
"1 / 9",
"_____no_output_____"
]
],
[
[
"Exponents are represents in Python by double-star `**`. For example, to compute the number googol, which is ten raised to the one-hundredth power, do the following.",
"_____no_output_____"
]
],
[
[
"10**100",
"_____no_output_____"
]
],
[
[
"Note that in Jupyter, only the output of the final line will be displayed. To suppress that output, terminate the final line with `;`.",
"_____no_output_____"
]
],
[
[
"1 + 1\n2 + 3",
"_____no_output_____"
],
[
"1 + 1\n2 + 3;",
"_____no_output_____"
]
],
[
[
"If you would like to view a result from a line that is not the final line, use the `print` function.",
"_____no_output_____"
]
],
[
[
"print('one plus one is', 1 + 1)\n2 + 3",
"_____no_output_____"
]
],
[
[
"## Using numpy mathematical functions\n\nYou are already familiar with a wide array of mathematical functions, such as trig functions, logs and square roots. Here, we see how to use these functions in Python.\n\nPython is a general-purpose programming language, so you have to `import` the mathematical functions that you want from a _module_ called `numpy` (aka NumPy).",
"_____no_output_____"
]
],
[
[
"from numpy import log, log10, sqrt, sin, cos, tan",
"_____no_output_____"
]
],
[
[
"Names of the functions in NumPy are all more or less the same as the names you use when writing them down or on your graphics calculator. \n\nFor example, NumPy’s sine function is sin, its cosine function cos and its tangent function tan. Therefore, to compute the cosine of 3.1415926, you proceed as follows.",
"_____no_output_____"
]
],
[
[
"cos(3.1415926)",
"_____no_output_____"
]
],
[
[
"Note that arguments passed to functions are always enclosed in brackets. Note also that numpy assumes that all angles are in radians.\n\nSome function names in NumPy are a little surprising. For instance, the natural logarithm function is `log`, not `ln`.\n",
"_____no_output_____"
]
],
[
[
"log(2.7182818)",
"_____no_output_____"
]
],
[
[
"You can compute the base-10 logarithm using `log10`.",
"_____no_output_____"
]
],
[
[
"log10(100)",
"_____no_output_____"
]
],
[
[
"Other functions that you would usually write with symbols require functions calls. For instance $\\sqrt{4}$ becomes",
"_____no_output_____"
]
],
[
[
"sqrt(4)",
"_____no_output_____"
]
],
[
[
"## Exercise 1\nCalculate `tan(3.14159)` in the cell below.",
"_____no_output_____"
]
],
[
[
"#Hello",
"_____no_output_____"
]
],
[
[
"You can execute the same cell as many times as you like. Go back and change your answer above to calculate `tan(3.14159/4)`. You can also copy and paste like you ordinarily would, and by tapping or click next a cell you can select the whole cell. You can move, copy, or paste cells. Exactly how you do this depends on your browser, so experiment a little bit.",
"_____no_output_____"
],
[
"## Names for common constants in NumPy\nThree constants that are ubiquitous in mathematics are $\\pi$, $\\mathrm{e}$, and the imaginary number $i$. We can import the first two from NumPy.",
"_____no_output_____"
]
],
[
[
"from numpy import pi, e",
"_____no_output_____"
]
],
[
[
"We know that $\\pi$ and $\\mathrm{e}$ are constants, but the objects we have just imported from NumPy are _variables_, not functions like we imported from NumPy last time. That means we don't put brackets after them.",
"_____no_output_____"
]
],
[
[
"cos(pi/4)",
"_____no_output_____"
]
],
[
[
"We don't import $i$ because Python already knows about it. Electrical engineers use $j$ instead of $i$ (because $i$ is current), and so does python.",
"_____no_output_____"
]
],
[
[
"1j * 1j",
"_____no_output_____"
]
],
[
[
"Note that `1j` is a complex number, so the output of the above multiplication is also a complex number (whose imaginary part is zero).",
"_____no_output_____"
],
[
"If you really want $\\mathrm{e}$ you can use the constant imported above, but most of the time we will use NumPy's `exp` function (which calculates $\\mathrm{e}^x$).",
"_____no_output_____"
]
],
[
[
"from numpy import exp\nexp(1)",
"_____no_output_____"
]
],
[
[
"## Exercise 2\nCalculate $\\mathrm{e}^{i\\pi}$.\n\nUnless you import another specialised module, all arithmetic in Python will be _floating point_ arithmetic, where irrational numbers are truncated to have finite precision. For that reason, you may observe a very small _floating point error_ in your results.",
"_____no_output_____"
],
[
"## Plotting functions\nWhen we plot a function given by a formula, say $f(x) = \\sin(x)$, we first must define the range of $x$ values we want to use, the domain of the plot.\n### Example 1\nPlot the curve $y = \\sin(x)$ in the range $−5 \\leq x \\leq 5$. We define the range of $x$ by typing the following command.",
"_____no_output_____"
]
],
[
[
"from numpy import linspace\nx = linspace(-5, 5, 101)",
"_____no_output_____"
]
],
[
[
"Note that if you don't know what a function does (or can't remember the order of its arguments), help is only a question mark away.",
"_____no_output_____"
]
],
[
[
"linspace?",
"_____no_output_____"
]
],
[
[
"The above call to `linspace` creates a $1\\times 101$ NumPy array whose elements are from -5 to 5. We chose the steps to be small enough to make the plot smooth.\n\nThere are several good visualisation modules available in Python. We will use Plotly.",
"_____no_output_____"
]
],
[
[
"from plotly import graph_objs as go",
"_____no_output_____"
]
],
[
[
"First calculate the $y$ values that correspond to our $x$ values.",
"_____no_output_____"
]
],
[
[
"y = sin(x)",
"_____no_output_____"
]
],
[
[
"Now draw a line through the points defined by the $x$ and $y$ arrays. This may be the first time that you have encountered object-oriented programming. The `.` in `go.Figure` means that `Figure` belongs to `go`. `go.Figure()` returns a new `Figure` object, which you can then manipulate with its methods. The `.` in `fig.add_trace` means that the `add_trace` function belongs to `fig`. When you call it, it changes `fig`. Finally, `fig.show()` tells fig to display itself.",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\nfig.add_trace(go.Scatter(x=x, y=y))\nfig.show('png')",
"_____no_output_____"
]
],
[
[
"Decorating your plot uses more `.` operators. `fig.layout.update` updates the layout that belongs to fig. Don't worry if you don't get the object-oriented paradigm, we will mostly just use it in the course to generate plots. If you copy this syntax, Jupyter will draw plots for you.",
"_____no_output_____"
]
],
[
[
"fig.layout.update(dict(title='A Plot',\n xaxis=dict(title='x'),\n yaxis=dict(title='y=sin(y)')))\nfig.show('png')",
"_____no_output_____"
]
],
[
[
"### Steps for basic plotting\n1. define the range: `x = linspace(`start`,`stop`, `num points`)`\n2. define the function you want to plot: `y = `function`(x)`\n3. type the plot command:\n```\nfig = go.Figure()\nfig.add_trace(go.Scatter(x=x, y=y))\nfig.show()\n```\n\n### Example 2\nPlot the function\n$$\ny = x^2 - x\\sin(3x) - x\\mathrm{e}^x,\\qquad -3\\leq x \\leq 3\n$$",
"_____no_output_____"
]
],
[
[
"x = linspace(-3, 3, 61)\ny = x**2 - x*sin(3*x) - x*exp(x)\nfig = go.Figure()\nfig.add_trace(go.Scatter(x=x, y=y))\nfig.show('png')",
"_____no_output_____"
]
],
[
[
"### Example 3\nSometimes we have a set of measurements that we want to plot. Essentially we do what we did above, but the variables now contain the data, rather than values of an independent variable $x$ and the corresponding function values $y$.\n\nHere are some actual data on influenza cases collected during the 2003-2004 flu season.",
"_____no_output_____"
]
],
[
[
"data = {'Weeks': \n [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, \n 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26],\n '% Flu Cases': \n [0.9, 1.0, 1.3, 1.3, 1.8, 2.4, 2.7, 3.7, 4.5, \n 5.4, 7.4, 7.5, 7.6, 5.2, 2.9, 2.1, 1.9, 1.8,\n 1.5, 1.4, 1.3, 1.1, 1.0, 1.0, 1.1, 0.9]}",
"_____no_output_____"
]
],
[
[
"Let's visualise it using a `DataFrame` (a type of table) from the `pandas` module.",
"_____no_output_____"
]
],
[
[
"from pandas import DataFrame\nfluCases = DataFrame(data).set_index('Weeks')\nfluCases",
"_____no_output_____"
]
],
[
[
"Week 1 is the first full week of October 2003. _% Flu Cases_ is the percentage of all patients seen by doctors in that week who had flu-like symptoms.\n\n`pandas` is already friends with `plotly`, so if you ask our new DataFrame to plot using the Plotly backend, it can create a figure for you. You can then change the attributes of that figure just as you did above for the basic figures.",
"_____no_output_____"
]
],
[
[
"fig = fluCases.plot(backend='plotly')\nfig.layout.update(dict(title='The 2003-2004 Flu Season',\n xaxis=dict(title='weeks'),\n yaxis=dict(title='% patients with flu-like symptoms')))\nfig.show('png')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6193f259f2f8dff4a065f70b86b8668239cf01 | 174,950 | ipynb | Jupyter Notebook | Intro.ipynb | sheejong/desihigh | 370d97b356df89dc0afddf1c5d0993143bbba6d4 | [
"BSD-3-Clause"
]
| null | null | null | Intro.ipynb | sheejong/desihigh | 370d97b356df89dc0afddf1c5d0993143bbba6d4 | [
"BSD-3-Clause"
]
| null | null | null | Intro.ipynb | sheejong/desihigh | 370d97b356df89dc0afddf1c5d0993143bbba6d4 | [
"BSD-3-Clause"
]
| null | null | null | 140.97502 | 29,764 | 0.870106 | [
[
[
"from IPython.display import IFrame",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# *Hubble and the origins of DESI*",
"_____no_output_____"
],
[
"The year 1929 brought us the Oscars, the first car radio and Edwin Hubble's unexpected observation that all galaxies are moving away from us!",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"Let's take a quick look at some of the galaxies he was looking at, Triangulum and the Large Magellanic Cloud.",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"In total, Edwin studied the distances of 24 galaxies from us, and their observed `redshifts'. What does that mean?",
"_____no_output_____"
],
[
"Maybe you already know that the energy levels of Hydrogen are __quantized__, with electrons habitating a series of shells with __discrete__ energies. When an electron transitions between any two levels, light is emitted with a wavelength neatly given by the \"Rydberg\" formula: \n\n$$\n\\frac{1}{\\lambda_{\\rm vac}} = 1.096 \\times 10^{7} \\left ( \\frac{1}{n^2} - \\frac{1}{m^2} \\right ) \n$$\n\nwhere $n$ and $m$ (any one of $[0, 1, 2, ... \\infty]$) label the two energy levels. ",
"_____no_output_____"
]
],
[
[
"# First, let's import some useful packages:\nimport os\nimport sys\nimport astropy\nimport pylab as pl\nimport pandas as pd\nimport numpy as np\n\n\nfrom matplotlib import pyplot as plt\nfrom scipy import stats\nfrom IPython.display import Image\nfrom pkg_resources import resource_filename",
"_____no_output_____"
],
[
"# For our friends running on Google Colab - remember to \n# mount your drive for all notebooks other than Colab.ipynb\n# \n# Safely ignore otherwise!\nsys.path.append('/content/drive/MyDrive/desihigh/')",
"_____no_output_____"
],
[
"from tools import pubplot\n\nfrom tools.wave2rgb import wavelength_to_rgb",
"_____no_output_____"
],
[
"def Rydberg(n, m):\n # Vacuum wavelengths [nanometres]\n result = 1.096e-2 * (1. / n / n - 1. / m / m)\n return 1. / result",
"_____no_output_____"
]
],
[
[
"Let's workout the wavelengths of light that Hydrogen can emit:",
"_____no_output_____"
]
],
[
[
"waves = []\n\nprint('n \\t m \\t Wavelength [nm]')\n\nfor n in np.arange(1, 10, 1):\n for m in np.arange(n+1, 10, 1):\n wave = Rydberg(n, m) \n waves.append(wave)\n \n print('{:d} \\t {:d} \\t {:.3f}'.format(n, m, wave))",
"n \t m \t Wavelength [nm]\n1 \t 2 \t 121.655\n1 \t 3 \t 102.646\n1 \t 4 \t 97.324\n1 \t 5 \t 95.043\n1 \t 6 \t 93.848\n1 \t 7 \t 93.142\n1 \t 8 \t 92.689\n1 \t 9 \t 92.381\n2 \t 3 \t 656.934\n2 \t 4 \t 486.618\n2 \t 5 \t 434.480\n2 \t 6 \t 410.584\n2 \t 7 \t 397.405\n2 \t 8 \t 389.294\n2 \t 9 \t 383.923\n3 \t 4 \t 1876.955\n3 \t 5 \t 1283.075\n3 \t 6 \t 1094.891\n3 \t 7 \t 1005.931\n3 \t 8 \t 955.541\n3 \t 9 \t 923.814\n4 \t 5 \t 4055.150\n4 \t 6 \t 2627.737\n4 \t 7 \t 2167.662\n4 \t 8 \t 1946.472\n4 \t 9 \t 1819.203\n5 \t 6 \t 7465.163\n5 \t 7 \t 4657.086\n5 \t 8 \t 3743.215\n5 \t 9 \t 3299.335\n6 \t 7 \t 12380.685\n6 \t 8 \t 7507.821\n6 \t 9 \t 5912.409\n7 \t 8 \t 19075.426\n7 \t 9 \t 11316.720\n8 \t 9 \t 27823.100\n"
]
],
[
[
"Now let's plot the wavelengths and see the color of these lines. If we were to look at a emitting Hydrogen atom, you'd see this:",
"_____no_output_____"
]
],
[
[
"for wave in waves:\n # color = [r, g, b]\n color = wavelength_to_rgb(wave) \n pl.axvline(x=wave, c=color)\n\npl.xlabel('Vacuum wavelength [nanometers]')\npl.xlim(380., 780.)",
"_____no_output_____"
]
],
[
[
"If the hydrogen exists in a galaxy that is moving, we see the lines Doppler shifted. We will call this the \"redshift\" of the galaxy, often denote as $z$ (https://en.wikipedia.org/wiki/Redshift). Let's say the galaxy is moving at 1% the speed of light (v = 0.1*c), we can calculate the redshift with the following equation:\n$$\n1 + z = \\sqrt{\\frac{1 + v/c}{1 - v/c}}\n$$",
"_____no_output_____"
]
],
[
[
"def redshift(v):\n # v [speed of light].\n result = (1. + v) / (1. - v) \n result = np.sqrt(result) - 1.\n \n return result",
"_____no_output_____"
],
[
"zz = redshift(0.01) \n\nfor restwave in waves:\n obswave = (1. + zz) * restwave \n\n color = wavelength_to_rgb(restwave) \n pl.axvline(x=restwave, c=color, alpha=0.25)\n\n color = wavelength_to_rgb(obswave) \n pl.axvline(x=obswave, c=color)\n\npl.xlabel('Vacuum wavelength [nanometers]')\npl.xlim(380., 780.)",
"_____no_output_____"
]
],
[
[
"Here you see the original line (faint) and the line shifted if the galaxy with the emitting Hydrogen is moving. https://en.wikipedia.org/wiki/Doppler_effect will tell you all the details.",
"_____no_output_____"
],
[
"Hubble knew the lines of Hydrogen, and for many other elements. By reversing above, he was able to calculate the velocity for many galaxies. He found out how far away there were (from how bright some special stars in the galaxy were - https://en.wikipedia.org/wiki/Cepheid_variable) and how fast they were moving (from their redshift, as above):",
"_____no_output_____"
]
],
[
[
"hub = resource_filename('desihigh','dat/hubble.dat')",
"_____no_output_____"
],
[
"dat = pd.read_csv(hub, sep='\\s+', comment='#', names=['Galaxy name', 'Distance [Mpc]', 'Velocity [km/s]'])\ndat",
"_____no_output_____"
]
],
[
[
"Let's plot them.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10, 7.5))\nax = fig.add_subplot(1, 1, 1)\nplt.close()",
"_____no_output_____"
],
[
"label_style = {'fontname': 'Georgia', 'fontsize': 16} ",
"_____no_output_____"
],
[
"ax.plot(dat['Distance [Mpc]'], dat['Velocity [km/s]'], '-', c='k', marker='*', lw=0)\n\nax.set_xlabel('Distance from us [Megaparsecs]', **label_style)\nax.set_ylabel('Recession velocity [km/s]', **label_style)\n\nplt.tight_layout()",
"_____no_output_____"
],
[
"fig",
"_____no_output_____"
]
],
[
[
"Edwin saw a clear trend, but the measurements seemed pretty noisy. Let's figure out our best guess at the true relationship between the two. We'll look at a linear relationship (regression) using the scipy stats package:",
"_____no_output_____"
]
],
[
[
"slope, intercept, r_value, p_value, std_err = stats.linregress(dat['Distance [Mpc]'],dat['Velocity [km/s]'])",
"_____no_output_____"
],
[
"print('The gradient to this trend is known as the Hubble constant: {:.3f} [km/s/Mpc]'.format(slope))",
"The gradient to this trend is known as the Hubble constant: 454.158 [km/s/Mpc]\n"
]
],
[
[
"Let's see what that looks like. ",
"_____no_output_____"
]
],
[
[
"distances = np.linspace(-0.5, 2.5, 10)\nvelocities = slope * distances\n\nax.plot(distances, velocities, lw=0.25, c='k')\nax.set_xlim(0.0, 2.5)",
"_____no_output_____"
],
[
"fig",
"_____no_output_____"
]
],
[
[
"Seems a pretty good fit! ",
"_____no_output_____"
],
[
"Now it's your turn, can you figure out a good estimate of the error on this measurement of the Hubble costant. How accurately can we predict the recession of a galaxy at a given distance, i.e. how fast or slow could it be moving? ",
"_____no_output_____"
],
[
"So in conclusion, every galaxy is likely to be moving away from us! We find this to be true of all galaxies - we are not at center or special in any way. Every galaxy is moving away from every other. The fact that the Universe was expanding came as a shock to many in 1929, but an even greater surprise was in store. ",
"_____no_output_____"
],
[
"# *Dark Energy*",
"_____no_output_____"
],
[
"In 1998, the world would change forever. Larry Page and Sergey Brin founded Google, the American Unity node and Russian Zarya module would be brought together to form the [International Space Station](https://en.wikipedia.org/wiki/International_Space_Station), and Lawrence Berkeley Lab's\nvery own Saul Perlmutter, Brian Schmidt and Adam Reiss irrefutably confirmed the existence of _Dark Energy_. Here's Saul impressing some young Berkeley researchers with these results at the time:",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"So what was everyone looking at? Let's breakdown the data.",
"_____no_output_____"
],
[
"Saul and his team measured the redshift ($z$) and the effective magnitude for several Type Ia Supernovae (https://en.wikipedia.org/wiki/Type_Ia_supernova)",
"_____no_output_____"
]
],
[
[
"perl = resource_filename('desihigh', 'dat/perlmutter.txt')",
"_____no_output_____"
],
[
"dat = pd.read_csv(perl, names=['z', 'Effective magnitude'], comment='#', sep='\\s+')\ntoprint = dat[:10]\ntoprint",
"_____no_output_____"
]
],
[
[
"A plot would show this a lot more clearly:",
"_____no_output_____"
]
],
[
[
"pl.plot(dat['z'], dat['Effective magnitude'], marker='.', lw=0.0)\n\npl.xlabel('z')\npl.ylabel('Effective magnitude')",
"_____no_output_____"
]
],
[
[
"Saul has good reason to believe (really, he had to tweak them a bit first) that every [type Ia supernovae](https://en.wikipedia.org/wiki/Type_Ia_supernova) shown here was equally bright intrinsically, but those at high redshift appeared relatively faint compared to those at low redshift, as they were simply further away. This explains the trend shown, given that 'effective magnitude' is the awkward way in which astronomers typically express how bright something appears.",
"_____no_output_____"
],
[
"The useful thing about this measurement is that how far away a supernovae or galaxy is for a given redshift depends on a few parameters, one of which is how much Dark Energy there might be in the Universe. Almost everyone expected this data to prove there was _no_ _Dark Energy_ when Saul made it, but a few guessed otherwise. \n\nWhen Hubble discovered the expansion, a natural consequence was that the amount of (rest mass) energy contained within a cubic meter would dilute with time. Dark Energy would be special, as the amount of energy per cubic meter would instead be constant with time and would suggest that spooky effects of [quantum mechanics](https://en.wikipedia.org/wiki/Quantum_mechanics) would be causing the galaxies to separate. ",
"_____no_output_____"
],
[
"So let's use Saul's data to figure out how much Dark Energy is in the Universe. First, we need a model for the (luminosity) distance of a supernovae at a given redshift, given some amount of Dark Energy. We use $\\Omega_\\Lambda$ to denote the _fraction_ of all matter that behaves like Dark Energy. ",
"_____no_output_____"
]
],
[
[
"from astropy.cosmology import FlatLambdaCDM\n\ndef lumdist(z, olambda):\n cosmo = FlatLambdaCDM(H0=70, Om0=1. - olambda, Tcmb0=2.725)\n \n return cosmo.luminosity_distance(z) ",
"_____no_output_____"
]
],
[
[
"We then need to convert this distance into how astronomers measure brightness:",
"_____no_output_____"
]
],
[
[
"def effmag(z, olambda, MB):\n DL = lumdist(z, olambda) \n\n return MB + 5. * np.log10(DL.value) ",
"_____no_output_____"
],
[
"zs = np.arange(0.01, 0.85, 0.01)\n\npl.plot(dat['z'], dat['Effective magnitude'], marker='.', lw=0.0)\n\npl.plot(zs, effmag(zs, 0.0, 6.), c='k', label='No Dark Energy', alpha=0.5)\npl.plot(zs, effmag(zs, 0.5, 6.), c='k', label='Dark Energy!')\n\npl.xlabel('z')\npl.ylabel('Effective magnitude')\n\npl.legend(loc=4, frameon=False)",
"_____no_output_____"
]
],
[
[
"Even by eye, the data looks to prefer some Dark Energy. But there's not a huge amount in it. Let's figure out what exactly the data prefers. To do this, we'll assume that minimising the distance between each point and the line is the best measure of how well the theory fits the data (see https://en.wikipedia.org/wiki/Least_squares). Together with the fraction of Dark Energy, we also don't know how bright every supernovae is intrinsically so we'll fit for that simultaneously. ",
"_____no_output_____"
]
],
[
[
"from scipy.optimize import minimize",
"_____no_output_____"
],
[
"def chi2(x):\n olambda = x[0]\n MB = x[1] \n \n model = effmag(dat['z'], olambda, MB) \n \n return np.sum((dat['Effective magnitude'] - model)**2.) ",
"_____no_output_____"
],
[
"res = minimize(chi2, x0=[0.5, 5.0], options={'disp': True})",
"Optimization terminated successfully.\n Current function value: 6.018565\n Iterations: 10\n Function evaluations: 39\n Gradient evaluations: 13\n"
],
[
"res.x",
"_____no_output_____"
],
[
"zs = np.arange(0.01, 0.85, 0.01)\n\npl.plot(dat['z'], dat['Effective magnitude'], marker='.', lw=0.0)\n\npl.plot(zs, effmag(zs, 0.0, 6.), c='k', label='No Dark Energy', alpha=0.5)\npl.plot(zs, effmag(zs, 0.5, 6.), c='k', label='50% Dark Energy!')\npl.plot(zs, effmag(zs, 0.75, 6.), c='c', label='75% Dark Energy!')\n\npl.xlabel('z')\npl.ylabel('Effective magnitude')\n\npl.legend(loc=4, frameon=False)",
"_____no_output_____"
]
],
[
[
"So there's something like 75% dark energy in the Universe! As the first people to make this measurement, Saul, together with Brian Schmidt and Adam Reiss, would be awarded the 2011 Nobel Prize for their work.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"You can find all the details of his work here: https://arxiv.org/pdf/astro-ph/9812133.pdf. Warning, this is for the pros so don't worry if you don't understand too much!",
"_____no_output_____"
],
[
"As honorary principal at DESI High, Saul has a special opening address to all the students, including you! ",
"_____no_output_____"
],
[
"[A welcome to DESI High, Prof. Saul Perlmutter](https://github.com/michaelJwilson/desihigh/blob/main/desihigh/perlmutter/letter.pdf)",
"_____no_output_____"
],
[
"The primary motivation for DESI is to repeat similar distant-redshift measurements much more precisely and learn much more about this spooky Dark Energy! ",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec61a3ddece12c6a317b414342fc7187903b5f44 | 330,888 | ipynb | Jupyter Notebook | UGRID/ESTOFS_water_levels.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
]
| 32 | 2015-01-07T01:48:05.000Z | 2022-03-02T07:07:42.000Z | UGRID/ESTOFS_water_levels.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
]
| 1 | 2015-04-13T21:00:18.000Z | 2015-04-13T21:00:18.000Z | UGRID/ESTOFS_water_levels.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
]
| 30 | 2015-01-28T09:31:29.000Z | 2022-03-07T03:08:28.000Z | 327.611881 | 213,463 | 0.899253 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec61a71091162595154677db85f5e6b7e9a56b14 | 27,644 | ipynb | Jupyter Notebook | adventofcode/2017/.ipynb_checkpoints/day1_9-checkpoint.ipynb | bicepjai/mypuzzles | 1918700562b38320bc30f61ee2fdcf9b8f404b8d | [
"BSD-3-Clause"
]
| null | null | null | adventofcode/2017/.ipynb_checkpoints/day1_9-checkpoint.ipynb | bicepjai/mypuzzles | 1918700562b38320bc30f61ee2fdcf9b8f404b8d | [
"BSD-3-Clause"
]
| null | null | null | adventofcode/2017/.ipynb_checkpoints/day1_9-checkpoint.ipynb | bicepjai/mypuzzles | 1918700562b38320bc30f61ee2fdcf9b8f404b8d | [
"BSD-3-Clause"
]
| null | null | null | 27.75502 | 588 | 0.537115 | [
[
[
"# Setup",
"_____no_output_____"
]
],
[
[
"import sys\nimport os\n\nimport re\nimport collections\nimport itertools\nimport bcolz\nimport pickle\n\nimport numpy as np\nimport pandas as pd\nimport gc\nimport random\nimport smart_open\nimport h5py\nimport csv\n\nimport tensorflow as tf\nimport gensim\nimport string\n\nimport datetime as dt\nfrom tqdm import tqdm_notebook as tqdm\n\nimport numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nrandom_state_number = 967898",
"_____no_output_____"
]
],
[
[
"# Code",
"_____no_output_____"
],
[
"## Day 1: Inverse Captcha",
"_____no_output_____"
],
[
"The captcha requires you to review a sequence of digits (your puzzle input) and \n\nfind the sum of all digits that match the next digit in the list. The list is circular, \n\nso the digit after the last digit is the first digit in the list.",
"_____no_output_____"
]
],
[
[
"! cat day1_input.txt",
"R4, R5, L5, L5, L3, R2, R1, R1, L5, R5, R2, L1, L3, L4, R3, L1, L1, R2, R3, R3, R1, L3, L5, R3, R1, L1, R1, R2, L1, L4, L5, R4, R2, L192, R5, L2, R53, R1, L5, R73, R5, L5, R186, L3, L2, R1, R3, L3, L3, R1, L4, L2, R3, L5, R4, R3, R1, L1, R5, R2, R1, R1, R1, R3, R2, L1, R5, R1, L5, R2, L2, L4, R3, L1, R4, L5, R4, R3, L5, L3, R4, R2, L5, L5, R2, R3, R5, R4, R2, R1, L1, L5, L2, L3, L4, L5, L4, L5, L1, R3, R4, R5, R3, L5, L4, L3, L1, L4, R2, R5, R5, R4, L2, L4, R3, R1, L2, R5, L5, R1, R1, L1, L5, L5, L2, L1, R5, R2, L4, L1, R4, R3, L3, R1, R5, L1, L4, R2, L3, R5, R3, R1, L3\r\n"
],
[
"input_data = None\nwith open(\"day1_input.txt\") as f:\n input_data = f.read().strip().split()\n input_data = [w.strip(\",\") for w in input_data ]",
"_____no_output_____"
]
],
[
[
"We will form the direction map since they are finite.",
"_____no_output_____"
]
],
[
[
"directions = {\n (\"N\",\"R\") : (\"E\",0,1),\n (\"N\",\"L\") : (\"W\",0,-1),\n \n (\"W\",\"R\") : (\"N\",1,1),\n (\"W\",\"L\") : (\"S\",1,-1),\n \n (\"E\",\"R\") : (\"S\",1,-1),\n (\"E\",\"L\") : (\"N\",1,1),\n \n (\"S\",\"R\") : (\"W\",0,-1),\n (\"S\",\"L\") : (\"E\",0,1)\n}",
"_____no_output_____"
],
[
"def get_distance(data):\n d,pos = \"N\",[0,0]\n for code in data:\n d1,v = code[0], int(code[1:])\n d,i,m = directions[(d, code[0])]\n pos[i] += m*v\n #print(code,d,v,pos)\n return sum([abs(n) for n in pos]) ",
"_____no_output_____"
],
[
"data = [\"R2\", \"R2\", \"R2\"]\nget_distance(data)",
"_____no_output_____"
],
[
"data = [\"R5\", \"L5\", \"R5\", \"R3\"]\nget_distance(data)",
"_____no_output_____"
],
[
"get_distance(input_data)",
"_____no_output_____"
]
],
[
[
"## Day 2: Bathroom Security \n",
"_____no_output_____"
],
[
"### part1",
"_____no_output_____"
],
[
"You arrive at Easter Bunny Headquarters under cover of darkness. However, you left in such a rush that you forgot to use the bathroom! Fancy office buildings like this one usually have keypad locks on their bathrooms, so you search the front desk for the code.\n\n\"In order to improve security,\" the document you find says, \"bathroom codes will no longer be written down. Instead, please memorize and follow the procedure below to access the bathrooms.\"\n\nThe document goes on to explain that each button to be pressed can be found by starting on the previous button and moving to adjacent buttons on the keypad: U moves up, D moves down, L moves left, and R moves right. Each line of instructions corresponds to one button, starting at the previous button (or, for the first line, the \"5\" button); press whatever button you're on at the end of each line. If a move doesn't lead to a button, ignore it.\n\nYou can't hold it much longer, so you decide to figure out the code as you walk to the bathroom. You picture a keypad like this:\n\n1 2 3\n4 5 6\n7 8 9\nSuppose your instructions are:\n\nULL\nRRDDD\nLURDL\nUUUUD\nYou start at \"5\" and move up (to \"2\"), left (to \"1\"), and left (you can't, and stay on \"1\"), so the first button is 1.\nStarting from the previous button (\"1\"), you move right twice (to \"3\") and then down three times (stopping at \"9\" after two moves and ignoring the third), ending up with 9.\nContinuing from \"9\", you move left, up, right, down, and left, ending with 8.\nFinally, you move up four times (stopping at \"2\"), then down once, ending with 5.\nSo, in this example, the bathroom code is 1985.\n\nYour puzzle input is the instructions from the document you found at the front desk. What is the bathroom code?",
"_____no_output_____"
]
],
[
[
"input_data = None\nwith open(\"day2_input.txt\") as f:\n input_data = f.read().strip().split()",
"_____no_output_____"
],
[
"def get_codes(data, keypad, keypad_max_size, start_index=(1,1), verbose=False):\n r,c = start_index\n digit = \"\"\n for codes in data:\n if verbose: print(\" \",codes)\n for code in codes:\n if verbose: print(\" before\",r,c,keypad[r][c])\n if code == 'R' and c+1 < keypad_max_size and keypad[r][c+1] is not None:\n c += 1\n elif code == 'L' and c-1 >= 0 and keypad[r][c-1] is not None:\n c -= 1\n elif code == 'U' and r-1 >= 0 and keypad[r-1][c] is not None:\n r -= 1\n elif code == 'D' and r+1 < keypad_max_size and keypad[r+1][c] is not None:\n r += 1\n if verbose: print(\" after\",code,r,c,keypad[r][c])\n digit += str(keypad[r][c])\n return digit\n ",
"_____no_output_____"
],
[
"sample = [\"ULL\",\n\"RRDDD\",\n\"LURDL\",\n\"UUUUD\"]",
"_____no_output_____"
],
[
"keypad = [[1,2,3],[4,5,6],[7,8,9]]\nget_codes(sample, keypad, keypad_max_size=3)",
"_____no_output_____"
],
[
"keypad = [[1,2,3],[4,5,6],[7,8,9]]\nget_codes(input_data, keypad, keypad_max_size=3)",
"_____no_output_____"
]
],
[
[
"### part2",
"_____no_output_____"
],
[
"You finally arrive at the bathroom (it's a several minute walk from the lobby so visitors can behold the many fancy conference rooms and water coolers on this floor) and go to punch in the code. Much to your bladder's dismay, the keypad is not at all like you imagined it. Instead, you are confronted with the result of hundreds of man-hours of bathroom-keypad-design meetings:\n\n 1\n 2 3 4\n5 6 7 8 9\n A B C\n D\nYou still start at \"5\" and stop when you're at an edge, but given the same instructions as above, the outcome is very different:\n\nYou start at \"5\" and don't move at all (up and left are both edges), ending at 5.\nContinuing from \"5\", you move right twice and down three times (through \"6\", \"7\", \"B\", \"D\", \"D\"), ending at D.\nThen, from \"D\", you move five more times (through \"D\", \"B\", \"C\", \"C\", \"B\"), ending at B.\nFinally, after five more moves, you end at 3.\nSo, given the actual keypad layout, the code would be 5DB3.\n\nUsing the same instructions in your puzzle input, what is the correct bathroom code?\n\nAlthough it hasn't changed, you can still get your puzzle input.\n",
"_____no_output_____"
]
],
[
[
"input_data = None\nwith open(\"day21_input.txt\") as f:\n input_data = f.read().strip().split()",
"_____no_output_____"
],
[
"keypad = [[None, None, 1, None, None],\n [None, 2, 3, 4, None],\n [ 5, 6, 7, 8, None],\n [None, 'A', 'B', 'C', None],\n [None, None, 'D', None, None]]",
"_____no_output_____"
],
[
"sample = [\"ULL\",\n\"RRDDD\",\n\"LURDL\",\n\"UUUUD\"]\nget_codes(sample, keypad, keypad_max_size=5, start_index=(2,0), verbose=False)",
"_____no_output_____"
],
[
"get_codes(input_data, keypad, keypad_max_size=5, start_index=(2,0), verbose=False)",
"_____no_output_____"
]
],
[
[
"## Day3 squares With Three Sides",
"_____no_output_____"
],
[
"### part1",
"_____no_output_____"
],
[
"Now that you can think clearly, you move deeper into the labyrinth of hallways and office furniture that makes up this part of Easter Bunny HQ. This must be a graphic design department; the walls are covered in specifications for triangles.\n\nOr are they?\n\nThe design document gives the side lengths of each triangle it describes, but... 5 10 25? Some of these aren't triangles. You can't help but mark the impossible ones.\n\nIn a valid triangle, the sum of any two sides must be larger than the remaining side. For example, the \"triangle\" given above is impossible, because 5 + 10 is not larger than 25.\n\nIn your puzzle input, how many of the listed triangles are possible?",
"_____no_output_____"
]
],
[
[
"input_data = None\nwith open(\"day3_input.txt\") as f:\n input_data = f.read().strip().split(\"\\n\")",
"_____no_output_____"
],
[
"input_data = [list(map(int, l.strip().split())) for l in input_data]",
"_____no_output_____"
],
[
"result = [ (sides[0]+sides[1] > sides[2]) and (sides[2]+sides[1] > sides[0]) and (sides[0]+sides[2] > sides[1]) for sides in input_data]",
"_____no_output_____"
],
[
"sum(result)",
"_____no_output_____"
]
],
[
[
"### part2",
"_____no_output_____"
],
[
"Now that you've helpfully marked up their design documents, it occurs to you that triangles are specified in groups of three vertically. Each set of three numbers in a column specifies a triangle. Rows are unrelated.\n\nFor example, given the following specification, numbers with the same hundreds digit would be part of the same triangle:\n\n101 301 501\n102 302 502\n103 303 503\n201 401 601\n202 402 602\n203 403 603\nIn your puzzle input, and instead reading by columns, how many of the listed triangles are possible?",
"_____no_output_____"
]
],
[
[
"input_data = None\nwith open(\"day31_input.txt\") as f:\n input_data = f.read().strip().split(\"\\n\")",
"_____no_output_____"
],
[
"input_data = [list(map(int, l.strip().split())) for l in input_data]\ninput_data[:5]",
"_____no_output_____"
],
[
"def chunks(l, n):\n \"\"\"Yield successive n-sized chunks from l.\"\"\"\n for i in range(0, len(l), n):\n yield l[i:i + n]\n \nsingle_list = [input_data[r][c] for c in [0,1,2] for r in range(len(input_data))]\nresult = [ (sides[0]+sides[1] > sides[2]) and (sides[2]+sides[1] > sides[0]) and (sides[0]+sides[2] > sides[1]) for sides in chunks(single_list, 3)]\nsum(result)",
"_____no_output_____"
]
],
[
[
"## Day4",
"_____no_output_____"
],
[
"### part1: Security Through Obscurity ",
"_____no_output_____"
],
[
"Finally, you come across an information kiosk with a list of rooms. Of course, the list is encrypted and full of decoy data, but the instructions to decode the list are barely hidden nearby. Better remove the decoy data first.\n\nEach room consists of an encrypted name (lowercase letters separated by dashes) followed by a dash, a sector ID, and a checksum in square brackets.\n\nA room is real (not a decoy) if the checksum is the five most common letters in the encrypted name, in order, with ties broken by alphabetization. For example:\n\naaaaa-bbb-z-y-x-123[abxyz] is a real room because the most common letters are a (5), b (3), and then a tie between x, y, and z, which are listed alphabetically.\na-b-c-d-e-f-g-h-987[abcde] is a real room because although the letters are all tied (1 of each), the first five are listed alphabetically.\nnot-a-real-room-404[oarel] is a real room.\ntotally-real-room-200[decoy] is not.\nOf the real rooms from the list above, the sum of their sector IDs is 1514.\n\nWhat is the sum of the sector IDs of the real rooms?",
"_____no_output_____"
]
],
[
[
"input_data = None\nwith open(\"day4_input.txt\") as f:\n input_data = f.read().strip().split(\"\\n\")\nlen(input_data), input_data[:5]",
"_____no_output_____"
],
[
"answer = 0\nfor code in input_data:\n m = re.match(r'(.+)-(\\d+)\\[([a-z]*)\\]', code)\n code, sector, checksum = m.groups()\n code = code.replace(\"-\",\"\")\n counts = collections.Counter(code).most_common()\n counts.sort(key=lambda k: (-k[1], k[0]))\n if ''.join([ch for ch,_ in counts[:5]]) == checksum:\n answer += int(sector)\nanswer",
"_____no_output_____"
]
],
[
[
"### part2",
"_____no_output_____"
],
[
"With all the decoy data out of the way, it's time to decrypt this list and get moving.\n\nThe room names are encrypted by a state-of-the-art shift cipher, which is nearly unbreakable without the right software. However, the information kiosk designers at Easter Bunny HQ were not expecting to deal with a master cryptographer like yourself.\n\nTo decrypt a room name, rotate each letter forward through the alphabet a number of times equal to the room's sector ID. A becomes B, B becomes C, Z becomes A, and so on. Dashes become spaces.\n\nFor example, the real name for qzmt-zixmtkozy-ivhz-343 is very encrypted name.\n\nWhat is the sector ID of the room where North Pole objects are stored?\n\n",
"_____no_output_____"
]
],
[
[
"for code in input_data:\n m = re.match(r'(.+)-(\\d+)\\[([a-z]*)\\]', code)\n code, sector, checksum = m.groups()\n sector = int(sector)\n code = code.replace(\"-\",\"\")\n counts = collections.Counter(code).most_common()\n counts.sort(key=lambda k: (-k[1], k[0]))\n string_maps = string.ascii_lowercase\n cipher_table = str.maketrans(string_maps, string_maps[sector%26:] + string_maps[:sector%26])\n if ''.join([ch for ch,_ in counts[:5]]) == checksum:\n if \"north\" in code.translate(cipher_table):\n print(code.translate(cipher_table))\n print(\"sector\",sector)\n",
"northpoleobjectstorage\nsector 991\n"
]
],
[
[
"## Day5 How About a Nice Game of Chess?",
"_____no_output_____"
],
[
"### part1",
"_____no_output_____"
],
[
"You are faced with a security door designed by Easter Bunny engineers that seem to have acquired most of their security knowledge by watching hacking movies.\n\nThe eight-character password for the door is generated one character at a time by finding the MD5 hash of some Door ID (your puzzle input) and an increasing integer index (starting with 0).\n\nA hash indicates the next character in the password if its hexadecimal representation starts with five zeroes. If it does, the sixth character in the hash is the next character of the password.\n\nFor example, if the Door ID is abc:\n\nThe first index which produces a hash that starts with five zeroes is 3231929, which we find by hashing abc3231929; the sixth character of the hash, and thus the first character of the password, is 1.\n5017308 produces the next interesting hash, which starts with 000008f82..., so the second character of the password is 8.\nThe third time a hash starts with five zeroes is for abc5278568, discovering the character f.\nIn this example, after continuing this search a total of eight times, the password is 18f47a30.\n\nGiven the actual Door ID, what is the password?",
"_____no_output_____"
],
[
"### part2",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec61af5d5e5117132608ae8bc5bccb3ac028e5cd | 7,687 | ipynb | Jupyter Notebook | nmbs-analytics/Untitled.ipynb | datamindedbe/nmbs-realtime-serverless | d4250f3ab02f5209574e3025c64fb8a553c57274 | [
"Apache-2.0"
]
| null | null | null | nmbs-analytics/Untitled.ipynb | datamindedbe/nmbs-realtime-serverless | d4250f3ab02f5209574e3025c64fb8a553c57274 | [
"Apache-2.0"
]
| null | null | null | nmbs-analytics/Untitled.ipynb | datamindedbe/nmbs-realtime-serverless | d4250f3ab02f5209574e3025c64fb8a553c57274 | [
"Apache-2.0"
]
| null | null | null | 71.841121 | 1,309 | 0.605698 | [
[
[
"\nimport plotly.plotly as py\nimport pandas as pd\n\ndf = pd.read_csv('https://raw.githubusercontent.com/plotly/datasets/master/2011_february_us_airport_traffic.csv')\ndf.head()\n\ndf['text'] = df['airport'] + '' + df['city'] + ', ' + df['state'] + '' + 'Arrivals: ' + df['cnt'].astype(str)\n\nscl = [ [0,\"rgb(5, 10, 172)\"],[0.35,\"rgb(40, 60, 190)\"],[0.5,\"rgb(70, 100, 245)\"],\\\n [0.6,\"rgb(90, 120, 245)\"],[0.7,\"rgb(106, 137, 247)\"],[1,\"rgb(220, 220, 220)\"] ]\n\ndata = [ dict(\n type = 'scattergeo',\n locationmode = 'USA-states',\n lon = df['long'],\n lat = df['lat'],\n text = df['text'],\n mode = 'markers',\n marker = dict(\n size = 8,\n opacity = 0.8,\n reversescale = True,\n autocolorscale = False,\n symbol = 'square',\n line = dict(\n width=1,\n color='rgba(102, 102, 102)'\n ),\n colorscale = scl,\n cmin = 0,\n color = df['cnt'],\n cmax = df['cnt'].max(),\n colorbar=dict(\n title=\"Incoming flightsFebruary 2011\"\n )\n ))]\n\nlayout = dict(\n title = 'Most trafficked US airports<br>(Hover for airport names)',\n colorbar = True,\n geo = dict(\n scope='usa',\n projection=dict( type='albers usa' ),\n showland = True,\n landcolor = \"rgb(250, 250, 250)\",\n subunitcolor = \"rgb(217, 217, 217)\",\n countrycolor = \"rgb(217, 217, 217)\",\n countrywidth = 0.5,\n subunitwidth = 0.5\n ),\n )\n\nfig = dict( data=data, layout=layout )\npy.iplot( fig, validate=False, filename='d3-airports' )",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
ec61e034e40e511d4059c94aca8f1952acdd25e6 | 141,931 | ipynb | Jupyter Notebook | module3-make-explanatory-visualizations/LS_DS_123_Make_Explanatory_Visualizations_Assignment.ipynb | Lrizika/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | 480b7497a665d57335e61406d09cf52d9be46daa | [
"MIT"
]
| null | null | null | module3-make-explanatory-visualizations/LS_DS_123_Make_Explanatory_Visualizations_Assignment.ipynb | Lrizika/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | 480b7497a665d57335e61406d09cf52d9be46daa | [
"MIT"
]
| null | null | null | module3-make-explanatory-visualizations/LS_DS_123_Make_Explanatory_Visualizations_Assignment.ipynb | Lrizika/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | 480b7497a665d57335e61406d09cf52d9be46daa | [
"MIT"
]
| null | null | null | 141,931 | 141,931 | 0.943909 | [
[
[
"# Change directory to VSCode workspace root so that relative path loads work correctly. Turn this addition off with the DataScience.changeDirOnImportExport setting\nimport os\ntry:\n\tos.chdir(os.path.join(os.getcwd(), '..'))\n\tprint(os.getcwd())\nexcept:\n\tpass\n",
"_____no_output_____"
],
[
"import os\ntry:\n\tos.chdir(os.path.join(os.getcwd(), 'module3-make-explanatory-visualizations'))\n\t#print(os.getcwd())\nexcept:\n\tpass",
"_____no_output_____"
]
],
[
[
" # ASSIGNMENT\n\n ### 1) Replicate the lesson code. I recommend that you [do not copy-paste](https://docs.google.com/document/d/1ubOw9B3Hfip27hF2ZFnW3a3z9xAgrUDRReOEo-FHCVs/edit).\n\n Get caught up to where we got our example in class and then try and take things further. How close to \"pixel perfect\" can you make the lecture graph?\n\n Once you have something that you're proud of, share your graph in the cohort channel and move on to the second exercise.\n\n ### 2) Reproduce another example from [FiveThityEight's shared data repository](https://data.fivethirtyeight.com/).\n\n **WARNING**: There are a lot of very custom graphs and tables at the above link. I **highly** recommend not trying to reproduce any that look like a table of values or something really different from the graph types that we are already familiar with. Search through the posts until you find a graph type that you are more or less familiar with: histogram, bar chart, stacked bar chart, line chart, [seaborn relplot](https://seaborn.pydata.org/generated/seaborn.relplot.html), etc. Recreating some of the graphics that 538 uses would be a lot easier in Adobe photoshop/illustrator than with matplotlib.\n\n - If you put in some time to find a graph that looks \"easy\" to replicate you'll probably find that it's not as easy as you thought.\n\n - If you start with a graph that looks hard to replicate you'll probably run up against a brick wall and be disappointed with your afternoon.\n\n\n\n\n\n\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Image\nimport pandas\n\ndf = pandas.read_csv('https://raw.githubusercontent.com/fivethirtyeight/data/master/inconvenient-sequel/ratings.csv')\n\nurl = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'\nexample = Image(url=url, width=400)\n\ndisplay(example)\n",
"_____no_output_____"
],
[
"votes = df.tail(1).loc[80052,'1_votes':'10_votes']\nvotes_percent = votes/votes.sum()*100\nvotes_percent\n",
"_____no_output_____"
],
[
"from matplotlib import pyplot\n\nsize_px = (1150,960)\ndpi = 300\nsize_in = [i/dpi for i in size_px]\nbar_color = '#ED713A'#(237, 113, 58)\ntick_color = '#999999'#(153, 153, 153)\ngrid_color = '#CDCDCD'#(205, 205, 205)\ntitle_color = '#222222'\nbar_width_px = 85\nbar_width_in = bar_width_px/dpi\nbar_divide_px = 10\nbar_divide_in = bar_divide_px/dpi\nbar_width_rel = bar_width_px/(bar_width_px+bar_divide_px)\nbar_divide_rel = bar_divide_px/(bar_width_px+bar_divide_px)\nsubplot_width_px = 960\nsubplot_width_in = subplot_width_px/dpi\ndpimult = 100/dpi\n\nfigure = pyplot.figure(figsize=size_in, dpi=dpi)\nfigure.patch.set(facecolor='None')\nsubplot = figure.add_subplot(1, 1, 1)\nfigure.subplots_adjust(top=0.85, bottom=0.1,left=0.15,right=1.0)\nsubplot.patch.set(facecolor='None')\nfor spine in subplot.spines:\n\tsubplot.spines[spine].set_visible(False)\n\nsubplot.grid(color=grid_color)\n\nsubplot.bar(range(1,11),votes_percent,width=bar_width_rel,color=bar_color, zorder=20)\n\ntitle_font = {'color': title_color,\n\t\t\t'weight': 700,\n\t\t\t'size': int(26*dpimult)}\nsubtitle_font = {'color': title_color,\n\t\t\t'size': int(24*dpimult)}\nlabel_font = {'color': 'black',\n\t\t\t'weight': 700,\n\t\t\t'size': int(20*dpimult)}\ntick_font = {'color': tick_color,\n\t\t\t'size': int(20*dpimult)}\nsubplot.text(-1.5,45,'\\'An Inconvenient Sequel: Truth to Power\\' is divisive', fontdict=title_font)\nsubplot.text(-1.5,42.5,'IMDb ratings for the film as of Aug. 29', fontdict=subtitle_font)\n\nxaxis = subplot.xaxis\nxaxis.set_label_text('Rating', fontdict=label_font)\nyaxis = subplot.yaxis\nyaxis.set_label_text('Percent of total values', fontdict=label_font)\n\nsubplot.set_xticks(range(1,11))\nsubplot.set_xticklabels(range(1,11), fontdict=tick_font)\nsubplot.set_yticks(range(0,50,10))\nsubplot.set_yticklabels(list(range(0,40,10))+['40%'], fontdict=tick_font)\n\nfigure.savefig('rebuilt.png', transparent=True)\npyplot.show()\n",
"_____no_output_____"
]
],
[
[
" # Scrabble score histograms\n From https://fivethirtyeight.com/features/how-qi-and-za-changed-scrabble/",
"_____no_output_____"
]
],
[
[
"df_scrabble = pandas.read_csv('https://github.com/fivethirtyeight/data/blob/master/scrabble-games/scrabble_games.csv?raw=true')\n\nurl = 'https://fivethirtyeight.com/wp-content/uploads/2017/04/roeder-scrabble-1.png'\nexample = Image(url=url, width=400)\n\ndisplay(example)\n",
"_____no_output_____"
],
[
"# Drop zero values in score columns\n# While it is, technically, possible to score zero\n# It's certainly not the case for > 500,000 games\nimport numpy\ndf_scrabble['winnerscore'].replace(0,numpy.NaN,inplace=True)\ndf_scrabble['loserscore'].replace(0,numpy.NaN,inplace=True)\ndf_scrabble.dropna(inplace=True)\n\n",
"_____no_output_____"
],
[
"import matplotlib.transforms as transforms\n#df_scrabble.describe()\n\nsize_px = (1150,1090)\ndpi = 300\nsize_in = [i/dpi for i in size_px]\n#bar_color = '#ED713A'#(237, 113, 58)\ntick_color = '#999999'#(153, 153, 153)\ngrid_color = '#CDCDCD'#(205, 205, 205)\ntitle_color = '#222222'\naxis_color = title_color\nloser_color = '#FA5347'\nloser_color_t = '#FF5448FA'\nwinner_color = '#6AB86B'\nwinner_color_t = '#6DBD6DFA'\n# bar_width_px = 10\n# bar_width_in = bar_width_px/dpi\n# bar_divide_px = 10\n# bar_divide_in = bar_divide_px/dpi\n# bar_width_rel = bar_width_px/(bar_width_px+bar_divide_px)\n# bar_divide_rel = bar_divide_px/(bar_width_px+bar_divide_px)\nsubplot_width_px = 960\nsubplot_width_in = subplot_width_px/dpi\ndpimult = 100/dpi\n\nfigure = pyplot.figure(figsize=size_in, dpi=dpi)\nfigure.patch.set(facecolor='None')\nsubplot = figure.add_subplot(1, 1, 1)\nfigure.subplots_adjust(top=0.85, bottom=0.1,left=0.1,right=0.9)\nsubplot.patch.set(facecolor='None')\nfor spine in subplot.spines:\n\tsubplot.spines[spine].set_visible(False)\n# subplot.spines['right'].set_visible(False)\n# subplot.spines['top'].set_visible(False)\n\nsubplot.grid(color=grid_color)\n\nbins = range(800)\nsubplot.hist(df_scrabble['loserscore'].append(df_scrabble['winnerscore']),bins=bins,color=loser_color, zorder=25)\nsubplot.hist(df_scrabble['winnerscore'],bins=bins,color=winner_color, zorder=30)\n#subplot.bar(range(1,11),votes_percent,width=bar_width_rel,color=bar_color)\n\ntitle_font = {'color': title_color,\n\t\t\t'weight': 700,\n\t\t\t'size': int(40*dpimult)}\nsubtitle_font = {'color': title_color,\n\t\t\t'size': int(32*dpimult)}\nlabel_font = {'color': 'black',\n\t\t\t'weight': 700,\n\t\t\t'size': int(20*dpimult)}\ntick_font = {'color': tick_color,\n\t\t\t'size': int(20*dpimult)}\ninline_label_font = {'color': 'white',\n\t\t\t'weight': 700,\n\t\t\t'horizontalalignment': 'center',\n\t\t\t'size': int(20*dpimult)}\nsubplot.text(-110, 12000, '700,000 games of Scrabble', fontdict=title_font)\nsubplot.text(-110, 11250, 'All tournament Scrabble games on cross-tables.com', fontdict=subtitle_font)\n\nsubplot.xaxis.set_label_text('Rating', fontdict=label_font)\n\nsubplot.set_xlim(left=-40, right=840)\nsubplot.set_xticks(range(0,1000,200))\n# subplot.set_xticklabels(range(1,11), fontdict=tick_font)\nsubplot.set_xticklabels(['0 points']+list(range(200,1000,200)), fontdict=tick_font)\nzeroxTick = subplot.get_xticklabels()[0]\nzeroxTickTransform = transforms.ScaledTranslation(31/72*dpimult,0,figure.dpi_scale_trans)\nzeroxTick.set_transform(zeroxTick.get_transform()+zeroxTickTransform)\n\nsubplot.set_yticks(range(0,12500,2500))\nsubplot.set_ylim(bottom=-500, top=10500)\nsubplot.set_yticklabels([i/4 for i in range(0,40,10)]+['10.0k scores'], fontdict=tick_font)\ntenkyTick = subplot.get_yticklabels()[-1]\ntenkyTickTransform = transforms.ScaledTranslation(72/72*dpimult,0,figure.dpi_scale_trans)\ntenkyTick.set_transform(tenkyTick.get_transform()+tenkyTickTransform)\n\nsubplot.text(370,6600,'Losing\\nscores', fontdict=inline_label_font, zorder=67)\nsubplot.text(410,1500,'Winning\\nscores', fontdict=inline_label_font, zorder=67)\n\nsubplot.axhline(y=0, color=axis_color, lw=1)\nsubplot.axvline(x=0, color=axis_color, lw=1)\n\nfigure.savefig('rebuilt_scrabble.png', transparent=True)\npyplot.show()\n",
"_____no_output_____"
]
],
[
[
" # STRETCH OPTIONS\n\n ### 1) Reproduce one of the following using the matplotlib or seaborn libraries:\n\n - [thanksgiving-2015](https://fivethirtyeight.com/features/heres-what-your-part-of-america-eats-on-thanksgiving/)\n - [candy-power-ranking](https://fivethirtyeight.com/features/the-ultimate-halloween-candy-power-ranking/)\n - or another example of your choice!\n\n ### 2) Make more charts!\n\n Choose a chart you want to make, from [Visual Vocabulary - Vega Edition](http://ft.com/vocabulary).\n\n Find the chart in an example gallery of a Python data visualization library:\n - [Seaborn](http://seaborn.pydata.org/examples/index.html)\n - [Altair](https://altair-viz.github.io/gallery/index.html)\n - [Matplotlib](https://matplotlib.org/gallery.html)\n - [Pandas](https://pandas.pydata.org/pandas-docs/stable/visualization.html)\n\n Reproduce the chart. [Optionally, try the \"Ben Franklin Method.\"](https://docs.google.com/document/d/1ubOw9B3Hfip27hF2ZFnW3a3z9xAgrUDRReOEo-FHCVs/edit) If you want, experiment and make changes.\n\n Take notes. Consider sharing your work with your cohort!",
"_____no_output_____"
]
],
[
[
"# More Work Here\n\n\n\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec61eaea878f52a4c56ae54f7422bc8e8e2f00cd | 2,396 | ipynb | Jupyter Notebook | index.ipynb | ecervera/perceptrons | 9a7e3234b4ca68ca2ef433d335900d20cf3e73b9 | [
"MIT"
]
| null | null | null | index.ipynb | ecervera/perceptrons | 9a7e3234b4ca68ca2ef433d335900d20cf3e73b9 | [
"MIT"
]
| null | null | null | index.ipynb | ecervera/perceptrons | 9a7e3234b4ca68ca2ef433d335900d20cf3e73b9 | [
"MIT"
]
| null | null | null | 38.031746 | 540 | 0.65818 | [
[
[
"# Perceptrons\n\n[<img src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/3/31/Perceptron.svg/320px-Perceptron.svg.png\" align=\"right\">](https://commons.wikimedia.org/wiki/File:Perceptron.svg)\n*From Wikipedia, the free encyclopedia*\n\nThe [perceptron](https://en.wikipedia.org/wiki/Perceptron) is an algorithm for supervised learning of binary classifiers (functions that can decide whether an input, represented by a vector of numbers, belongs to some specific class or not). It is a type of linear classifier, i.e. a classification algorithm that makes its predictions based on a linear predictor function combining a set of weights with the feature vector. The algorithm allows for online learning, in that it processes elements in the training set one at a time.\n\nThe perceptron algorithm dates back to the late 1950s; its first implementation, in custom hardware, was one of the first artificial neural networks to be produced.",
"_____no_output_____"
],
[
"## Activities\n\nYou should run first the example of the AND function, then complete with a similar code the exercises of the OR function and the linearly separable classes.\n\n* [Example: the AND function](AND.ipynb)\n* [Exercise: the OR function](OR_Exercise.ipynb)\n* [Exercise: linearly separable classes](Classes_Exercise.ipynb)\n\n<!--\nFinally, you should experiment the limitations of the perceptron.\n\n* [Linear Separability and the XOR Problem](XOR.ipynb)\n* [Linearly non-separable classes](non-separable.ipynb)\n-->\n\nFor the submission of this task, please follow the instructions of the corresponding workshop in Aula Virtual.",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown"
]
]
|
ec61f5372afa1f0d0a9ed6578cd8191acfd7b405 | 24,600 | ipynb | Jupyter Notebook | torch-knowledge-competitions/experiments/digit_classifier.ipynb | Charl-AI/Kaggle-Knowledge-Competitions | 509dff4146dbc3015749744a08d2a06005dc4259 | [
"MIT"
]
| null | null | null | torch-knowledge-competitions/experiments/digit_classifier.ipynb | Charl-AI/Kaggle-Knowledge-Competitions | 509dff4146dbc3015749744a08d2a06005dc4259 | [
"MIT"
]
| 6 | 2021-12-04T10:46:40.000Z | 2022-03-24T10:36:53.000Z | torch-knowledge-competitions/experiments/digit_classifier.ipynb | Charl-AI/Kaggle-Knowledge-Competitions | 509dff4146dbc3015749744a08d2a06005dc4259 | [
"MIT"
]
| null | null | null | 76.875 | 15,222 | 0.798821 | [
[
[
"# MNIST Digit Classifier",
"_____no_output_____"
]
],
[
[
"# This cell assumes a project structure of: project-root/src/experiments/this_notebook.ipynb\n# We append the parent directory to the system path, so now we can import modules from src\n# We also create a variable named path which points to the project root.\n\nimport sys\nfrom pathlib import Path\n\nsys.path.append(\"../\") # go to parent dir\npath = str(Path().resolve().parent.parent)\n\nprint(path)\n",
"/workspaces/Kaggle-Knowledge-Competitions\n"
]
],
[
[
"Class of configurations (consider using Hydra for heavier workloads in future):",
"_____no_output_____"
]
],
[
[
"from dataclasses import dataclass\n\n@dataclass\nclass Configurations:\n data_dir: str = path + \"/data/kaggle_mnist\"\n batch_size: int = 128\n num_workers: int = 4\n\n lr: float = 0.01\n\n num_epochs: int = 5\n\n device: str = \"cuda\"\n log_dir: str = path + \"/logs/torch-digit-classifier\"\n log_every_n_steps: int = 50\n\ncfg = Configurations()",
"_____no_output_____"
],
[
"import torch\nimport os\nfrom torch.utils.data import DataLoader, random_split\nfrom torch.utils.tensorboard.writer import SummaryWriter\n\nfrom trainer.digit_classifier_trainer import train_digit_classifier\nfrom datasets.kaggle_mnist import KaggleMNIST\nfrom models.digit_classifier import ResNet18\n\nimport socket\nfrom datetime import datetime\ncurrent_time = datetime.now().strftime('%b%d_%H-%M-%S')\nlog_dir = os.path.join(\n cfg.log_dir, current_time + '_' + socket.gethostname())\n\ndevice = torch.device(cfg.device)\nmodel = ResNet18(in_channels=1, out_classes=10)\ndata = KaggleMNIST(data_dir=cfg.data_dir, train=True, transform=None)\noptimizer = torch.optim.SGD(model.parameters(), lr=cfg.lr)\nlogger = SummaryWriter(log_dir=log_dir)\nprofiler = torch.profiler.profile(\n schedule=torch.profiler.schedule(wait=1, warmup=1, active=3, repeat=2),\n on_trace_ready=torch.profiler.tensorboard_trace_handler(log_dir),\n record_shapes=True,\n profile_memory=True,\n with_flops=True,\n with_stack=True)\n\nn_val = int(len(data) * 0.2)\nn_train = len(data) - n_val\ntrain_data, val_data = random_split(data, [n_train, n_val])\n\ntrain_loader = DataLoader(\n train_data,\n shuffle=True,\n batch_size=cfg.batch_size,\n num_workers=cfg.num_workers,\n)\n\nval_loader = DataLoader(\n val_data,\n shuffle=False,\n batch_size=cfg.batch_size,\n num_workers=cfg.num_workers,\n)\n\ntrain_digit_classifier(\n model,\n train_loader,\n val_loader,\n num_epochs=cfg.num_epochs,\n device=device,\n optimizer=optimizer,\n logger=logger,\n log_every_n_steps=cfg.log_every_n_steps,\n profiler=profiler,\n)",
"Epoch 0: train: 100%|██████████| 263/263 [00:09<00:00, 28.05it/s]\nEpoch 0: val: 100%|██████████| 66/66 [00:00<00:00, 78.75it/s] \nEpoch 1: train: 100%|██████████| 263/263 [00:08<00:00, 31.97it/s]\nEpoch 1: val: 100%|██████████| 66/66 [00:00<00:00, 88.99it/s] \nEpoch 2: train: 100%|██████████| 263/263 [00:07<00:00, 33.02it/s]\nEpoch 2: val: 100%|██████████| 66/66 [00:00<00:00, 101.61it/s]\nEpoch 3: train: 100%|██████████| 263/263 [00:07<00:00, 33.80it/s]\nEpoch 3: val: 100%|██████████| 66/66 [00:00<00:00, 100.63it/s]\nEpoch 4: train: 100%|██████████| 263/263 [00:08<00:00, 32.08it/s]\nEpoch 4: val: 100%|██████████| 66/66 [00:00<00:00, 92.16it/s] \n"
],
[
"from trainer.digit_classifier_trainer import test_digit_classifier\n\ndata = KaggleMNIST(data_dir=cfg.data_dir, train=False, transform=None)\ntest_loader = DataLoader(\n data,\n shuffle=False,\n batch_size=cfg.batch_size,\n num_workers=cfg.num_workers,\n)\n\nraw_preds = test_digit_classifier(model, test_loader, device)",
"Generating predictions on test set: 100%|██████████| 219/219 [00:01<00:00, 201.79it/s]\n"
]
],
[
[
"Our raw prediction is currently a list of tuples representing the predictions for each batch. We want to turn this into two tensors, each the length of the test set.",
"_____no_output_____"
]
],
[
[
"def unpack_predictions(predictions):\n \"\"\"Takes the output of trainer.predict and unpacks it into a tuple of two tensors\n over the data set:\n (imgs, predictions)\n \"\"\"\n # predictions start as list of tuples (img tensor, pred_tensor), of length num_batches.\n # each tensor is 1D with length batch_size.\n # we want to convert this to two tensors which are the length of the val/test set.\n unpacked_imgs = torch.Tensor().to(predictions[0][0].device)\n unpacked_predictions = torch.Tensor().to(predictions[0][0].device)\n for batch in predictions:\n imgs, preds = batch\n unpacked_predictions = torch.cat([unpacked_predictions, preds], dim=0)\n unpacked_imgs = torch.cat([unpacked_imgs, imgs], dim=0)\n\n return unpacked_imgs, unpacked_predictions\n\npredictions = unpack_predictions(raw_preds)",
"_____no_output_____"
]
],
[
[
"Now, lets sanity check our predictions to see if they're reasonable:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport random\n\nimgs, preds = predictions\nimgs = imgs.squeeze().detach().cpu().numpy()\npreds = preds.int().squeeze().detach().cpu().numpy()\n\nnrows=2\nncols=5\nfig, axs = plt.subplots(nrows=nrows, ncols=ncols,figsize=(10,5))\n\nfor i in range(nrows):\n for j in range(ncols):\n k = random.randint(0,28000)\n axs[i,j].imshow(imgs[k],cmap=\"gray\")\n print(preds[k])\n",
"1\n8\n5\n4\n9\n2\n9\n2\n0\n5\n"
]
],
[
[
"Finally, save our predictions in the format for Kaggle submission. You can submit by running the following line:\n```bash\n# submits preds.csv to the mnist classification competition\nkaggle competitions submit -c digit-recognizer -f data/kaggle_mnist/preds.csv --message first_submission_with_api\n```",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.DataFrame({\"ImageId\" : list(range(1, 28001)),\"Label\" : preds})\ndf.to_csv(path+\"/data/kaggle_mnist/preds.csv\", index=False)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6204a5289eca99c398ba0bcc573e4088a27c6c | 11,363 | ipynb | Jupyter Notebook | notebooks/Simulated population.ipynb | adrn/TidalCircularization | 53d785db210720fbd8993dd58d6e3f620bae4fa6 | [
"MIT"
]
| null | null | null | notebooks/Simulated population.ipynb | adrn/TidalCircularization | 53d785db210720fbd8993dd58d6e3f620bae4fa6 | [
"MIT"
]
| null | null | null | notebooks/Simulated population.ipynb | adrn/TidalCircularization | 53d785db210720fbd8993dd58d6e3f620bae4fa6 | [
"MIT"
]
| null | null | null | 27.647202 | 129 | 0.493444 | [
[
[
"import glob\nfrom os import path\nimport re\n\nfrom astropy.table import QTable\nfrom astropy.constants import G\nimport astropy.units as u\nimport numpy as np\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom scipy.integrate import simps\nfrom tqdm import tqdm\n\nfrom twoface.mass import period_at_surface, stellar_radius\n\nimport mesa_reader as mr\n\nfrom helpers import (MESAHelper, MatchedSimulatedSample, \n compute_dlne, solve_final_ea)",
"_____no_output_____"
]
],
[
[
"### Load the MESA models",
"_____no_output_____"
]
],
[
[
"mesa = MESAHelper('../mesa/')",
"_____no_output_____"
]
],
[
[
"### Load the data",
"_____no_output_____"
]
],
[
[
"unimodal = QTable.read('../../twoface/paper/1-catalog/tables/highK-unimodal.fits', \n character_as_bytes=False)\nclean_uni = unimodal[ (unimodal['clean_flag'] == 0)]\nhigh_logg = clean_uni[clean_uni['LOGG'] > 2]",
"_____no_output_____"
],
[
"(np.nanmedian(high_logg['M1']), \n np.nanmedian(high_logg['M2_min']), \n np.median(high_logg['LOGG'][high_logg['LOGG'] > -999]))",
"_____no_output_____"
]
],
[
[
"### Plot the stellar evolution curves",
"_____no_output_____"
]
],
[
[
"cmap = plt.get_cmap('rainbow_r')\nnorm = mpl.colors.Normalize(vmin=0.8, vmax=3)\n\nfig, ax = plt.subplots(1, 1, figsize=(5, 5))\nfor k in sorted(mesa.h.keys()):\n h = mesa.h[k]\n min_idx = h.log_R.argmin()\n slc = slice(min_idx, None)\n ax.plot(h.Teff[slc], h.log_g[slc], \n marker='', label=r'${0}\\,{{\\rm M}}_\\odot$'.format(k),\n linewidth=2, alpha=0.8, \n color=cmap(norm(float(k))))\n \nax.set_ylim(5, -0.1)\nax.set_xlim(13000, 3000)\nax.legend(loc='upper left', fontsize=12, borderaxespad=1.)\n\nax.set_xlabel(r'$T_{\\rm eff}$ [K]')\nax.set_ylabel(r'$\\log g$')\n\nfig.tight_layout()\n\nfig.savefig('../paper/figures/mesa.pdf')",
"_____no_output_____"
]
],
[
[
"## Generate a fake population of binaries",
"_____no_output_____"
]
],
[
[
"s = MatchedSimulatedSample(logg=high_logg['LOGG'],\n M1=high_logg['M1'],\n M2=high_logg['M2_min'],\n mesa_helper=mesa,\n seed=42)",
"_____no_output_____"
],
[
"t = s.generate_sample(size=1024)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(1, 4, figsize=(15, 5))\n\nax = axes[0]\nax.scatter(t['P'], t['e'], marker='.')\nax.scatter(high_logg['P'], high_logg['e'], marker='.')\n\nax.set_xscale('log')\nax.set_ylim(0, 1)\nax.set_xlabel('$P$')\nax.set_ylabel('$e$')\n\n_, bins, _ = axes[1].hist(t['M1_orig'], bins='auto', alpha=0.5, normed=True);\naxes[1].hist(high_logg['M1'][np.isfinite(high_logg['M1'])], bins='auto', alpha=0.5, normed=True);\naxes[1].set_xlabel('$M_1$')\n\nbins = np.linspace(0.01, 1.5, 10)\naxes[2].hist(t['M2']/t['M1_orig'], bins=bins, normed=True);\naxes[2].hist(high_logg['M2_min'][np.isfinite(high_logg['M2_min'])] / high_logg['M1'][np.isfinite(high_logg['M2_min'])], \n bins='auto', alpha=0.5, normed=True);\naxes[2].set_xlabel('$q$')\n\n_, bins, _ = axes[3].hist(t['logg'], bins='auto', alpha=0.5, normed=True);\naxes[3].hist(high_logg['LOGG'], bins='auto', alpha=0.5, normed=True);\naxes[3].set_xlabel(r'$\\log g$')\n\nfig.tight_layout()",
"_____no_output_____"
],
[
"_, bins, _ = plt.hist(t['M1_orig'].value, bins='auto', normed=True) #, h_Mstr[np.abs(h_M - t['M1'].value[i]).argmin()]\nplt.hist(high_logg['M1'][np.isfinite(high_logg['M1'])], bins='auto', normed=True);\nplt.hist(t['M1'], bins=bins, normed=True);",
"_____no_output_____"
]
],
[
[
"## Now simulate circularization:",
"_____no_output_____"
]
],
[
[
"dlnes = []\nfor row in tqdm(t):\n dlne = compute_dlne(row['logg'], M1=row['M1'], M2=row['M2'], a=row['a'], \n mesa_helper=mesa)\n dlnes.append(dlne)\n \nt['dlne'] = dlnes\nt['e_f'] = np.exp(np.log(t['e']) + t['dlne'])\n\n# mask = (t['a'] * (1 - t['e_f'])) < t['R1']\n# t['e_f'][mask] = np.nan",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6, 5))\nax.scatter(-t['dlne'], t['e_f'])\nax.set_xscale('log')\nax.set_xlim(1E6, 1E-12)",
"_____no_output_____"
],
[
"def plot_simulated_P_e(t, e_col='e_f', P_col='P'):\n fig, axes = plt.subplots(1, 3, figsize=(14, 5.5), sharey=True)\n \n cmap = plt.get_cmap('inferno')\n style = dict(marker='o', edgecolor='#555555', linewidth=0.5,\n alpha=0.5, vmin=2, vmax=4, cmap=cmap,\n s=30, c=t['logg'], rasterized=True)\n \n P_surf = period_at_surface(t['M1'], t['logg'], t[e_col], t['M2'])\n \n # Actually plot the markers\n cs = axes[0].scatter(t['P'], t['e'], **style)\n axes[1].scatter(t[P_col], t[e_col], **style)\n axes[2].scatter(t[P_col]/P_surf, t[e_col], **style)\n \n # Label all the things\n axes[0].set_ylabel(r'$e$')\n axes[0].set_xlabel(r'$P$ [day]')\n axes[1].set_xlabel(r'$P$ [day]')\n axes[2].set_xlabel(r'$P/P_{\\rm surface}$')\n \n axes[0].set_title('initial')\n axes[1].set_title('final')\n axes[2].set_title('final')\n \n # Scales, lims, ticks:\n for ax in axes:\n ax.set_xscale('log')\n \n axes[0].set_ylim(-0.05, 1)\n loc = mpl.ticker.LogLocator(numticks=10)\n for ax in axes[:2]:\n ax.xaxis.set_ticks(10**np.arange(-1, 4+0.1))\n ax.xaxis.set_ticks(np.concatenate([x*np.arange(1, 10+1) for x in ax.get_xticks()[:-1]]), minor=True)\n ax.set_xlim(8E-1, 1E4)\n\n axes[2].xaxis.set_ticks(10**np.arange(-1, 4+0.1))\n axes[2].xaxis.set_ticks(np.concatenate([x*np.arange(1, 10+1) for x in ax.get_xticks()[:-1]]), minor=True)\n axes[2].set_xlim(8E-1, 1.5E3)\n\n # Colorbar\n cax = fig.add_axes([0.865, 0.165, 0.02, 0.615])\n cb = fig.colorbar(cs, cax=cax)\n cb.ax.xaxis.set_ticks_position('top')\n cb.ax.xaxis.set_label_position('top')\n cb.set_label(r'$\\log g$', labelpad=10)\n cb.solids.set_rasterized(True) \n cb.solids.set_edgecolor('face')\n cb.set_ticks(np.arange(2, 4+0.1, 0.5))\n cb.ax.invert_yaxis()\n\n fig.tight_layout()\n fig.subplots_adjust(top=0.78, right=0.85, wspace=0.1)\n fig.set_facecolor('w')\n \n fig.suptitle(r'${\\bf Simulated\\,\\,binaries}$', y=0.94, x=0.45, fontsize=26)\n \n return fig",
"_____no_output_____"
],
[
"fig = plot_simulated_P_e(t)\nfig.savefig('../paper/figures/simulated.pdf', rasterized=True, dpi=250)",
"_____no_output_____"
]
],
[
[
"---\n\n## Try solving de/dt and da/dt simultaneously",
"_____no_output_____"
]
],
[
[
"t2 = t.copy()",
"_____no_output_____"
],
[
"efs = []\nafs = []\nfor row in tqdm(t2):\n ef, af = solve_final_ea(row['e'], row['a'], row['logg'], \n row['M1'], row['M2'], mesa)\n efs.append(ef)\n afs.append(af)\n\nefs = np.array(efs)\nafs = u.Quantity(afs).to(u.au)\nt2['e_f'] = efs\nt2['a_f'] = afs\nt2['P_f'] = 2*np.pi * np.sqrt(t2['a_f']**3 / (G * (t2['M1'] + t2['M2']))).to(u.day)",
"_____no_output_____"
],
[
"fig = plot_simulated_P_e(t2, P_col='P_f')",
"_____no_output_____"
]
],
[
[
"Conclusion: solving both $a$ and $e$ looks almost the same as the simpler case!",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(5,5))\nplt.scatter(t['P'], t2['P_f'])\nplt.xscale('log')\nplt.yscale('log')\nplt.xlim(1E0, 1E4)\nplt.ylim(1E0, 1E4)",
"_____no_output_____"
],
[
"plt.figure(figsize=(5,5))\nplt.scatter(t['e_f'], t2['e_f'])",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6206dc8742e9aecc1a70248cc1eb81f2a0894b | 47,720 | ipynb | Jupyter Notebook | _labs/Lab02/Lab02-Summarizing Variables.ipynb | M-Sender/cmps3160 | 54546d307f913b35caa45efe6c5528dadb8055f2 | [
"MIT"
]
| null | null | null | _labs/Lab02/Lab02-Summarizing Variables.ipynb | M-Sender/cmps3160 | 54546d307f913b35caa45efe6c5528dadb8055f2 | [
"MIT"
]
| null | null | null | _labs/Lab02/Lab02-Summarizing Variables.ipynb | M-Sender/cmps3160 | 54546d307f913b35caa45efe6c5528dadb8055f2 | [
"MIT"
]
| null | null | null | 30.826873 | 590 | 0.519111 | [
[
[
"# Lab 02: Suummarizing Variables\n\nThis lab is presented with some revisions from [Dennis Sun at Cal Poly](https://web.calpoly.edu/~dsun09/index.html) and his [Data301 Course](http://users.csc.calpoly.edu/~dsun09/data301/lectures.html)\n\n### When you have filled out all the questions, submit via [Tulane Canvas](https://tulane.instructure.com/)",
"_____no_output_____"
],
[
"In the previous section, we emphasized the difference between quantitative and categorical variables. The distinction is not merely pedantic; `pandas` will actually behave differently depending on whether it thinks a variable is quantitative or categorical.\n\nIt is not easy for a human to make sense of _all_ the values of a variable. In this section, we focus on ways to reduce the values to just a handful of summary statistics. Our working example will again be the Titanic data set, which contains both quantitative and categorical variables.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\npd.options.display.max_rows = 8\n\ndf = pd.read_csv(\"../data/titanic.csv\")\ndf.head()",
"_____no_output_____"
]
],
[
[
"To get a quick summary of a variable, we can use the `.describe()` function. Let's see what happens when we call `.describe()` on a quantitative variable, like `age`.",
"_____no_output_____"
]
],
[
[
"df.age.describe()",
"_____no_output_____"
]
],
[
[
"It returns the count (the number of observations with non-missing values), the mean, the standard deviation (`std`), and various percentiles (`min`, `25%`, `50%`, `75%`, `max`).\n\nNow, what if we call `.describe()` on a categorical variable, like `embarked`? This is a variable that takes on the values `C`, `Q`, or `S`, depending on whether the passenger embarked at Cherbourg, Queenstown, or Southampton.",
"_____no_output_____"
]
],
[
[
"df.embarked.describe()",
"_____no_output_____"
]
],
[
[
"The description of this variable is very different. We still get the count (of non-missing values). But instead of the mean and standard deviation (how would you calculate the mean of `Q` and `S`, anyway?), we get the number of unique values (`unique`), the value that appeared most often (`top`), and how often it appeared (`freq`). These are more natural summaries for a categorical variable, which only take on a limited set of values, where the values are often not even numeric.",
"_____no_output_____"
],
[
"The `.describe()` function only provides a handful of the many summary statistics that are available in `pandas`. We extract additional summary statistics below.",
"_____no_output_____"
],
[
"## Summary Statistics for Quantitative Variables\n\nWhat statistics should we use to summarize a quantitative variable? The most salient features of a quantitative variable are its **center** and **spread**.\n\n### Measures of Center\n\nSome statistics measure the **center** of a variable. Two commonly used measures of the center are:\n\n- the **mean** (a.k.a. average): the sum of the values divided by the count\n- the **median**: the middle value when you sort the values (i.e., a value such that 50% of the values lie below and 50% of the values lie above)\n\nA measure of center gives us information about the \"typical\" value of a variable. For example, you might not know whether a typical fare on the Titanic was £1, £10, or £100. But if we calculate the mean:",
"_____no_output_____"
]
],
[
[
"df.fare.mean()",
"_____no_output_____"
]
],
[
[
"we see that a typical fare is around £30.\n\nLet's see what the median says about the \"typical\" fare:",
"_____no_output_____"
]
],
[
[
"df.fare.median()",
"_____no_output_____"
]
],
[
[
"The median is quite different from the mean! It says that about 50% of the passengers paid less than £15 and about 50% paid more, so another reasonable value for the \"typical\" fare is £15.\n\nThe mean was twice the median! What explains this discrepancy? The reason is that the mean is very sensitive to extreme values. To see this, let's look at the highest fare that any passenger paid.",
"_____no_output_____"
]
],
[
[
"df.fare.max()",
"_____no_output_____"
]
],
[
[
"The highest fare paid was over £500! Even if most passengers paid less than £15, extreme values like this one will drag the mean upward. On the other hand, since the median is always the middle value, it is not affected by the extreme values, as long as the ordering of the values is not changed.\n\nTo drive this point home, let's see what would happen to the mean and median if that maximum fare were actually £10,000.",
"_____no_output_____"
]
],
[
[
"fare_10k = df.fare.replace(df.fare.max(), 10000)\nfare_10k.mean(), fare_10k.median()",
"_____no_output_____"
]
],
[
[
"Notice how the mean is now over £60, but the median is unchanged.",
"_____no_output_____"
],
[
"Just to satisfy our curiosity, let's learn more about this passenger who paid the maximum fare. To do this, we have to find the row that achieved this maximum value. Fortunately, there is a convenient `pandas` function, `.idxmax()`, that returns the _row index_ of the maximum fare. (A mathematician might call this the [\"arg max\"](https://en.wikipedia.org/wiki/Arg_max).)",
"_____no_output_____"
]
],
[
[
"df.fare.idxmax()",
"_____no_output_____"
]
],
[
[
"Now we can select the row corresponding to this index using `.loc`, as we learned in the previous section.",
"_____no_output_____"
]
],
[
[
"df.loc[df.fare.idxmax()]",
"_____no_output_____"
]
],
[
[
"The median is a number below which 50% of the values fall. What if we want to know some other percentile? We can use the `.quantile()` function, which takes a percentile rank (between 0 and 1) as input and returns the corresponding percentile.\n\nFor example, the 75th percentile is:",
"_____no_output_____"
]
],
[
[
"df.fare.quantile(.75)",
"_____no_output_____"
]
],
[
[
"which is pretty close to the mean. So only about 25% of the passengers paid more than the mean! The mean is not a great measure of center when there are extreme values, as in this data set.",
"_____no_output_____"
],
[
"To summarize, we have encountered several `pandas` functions that can be used to summarize a quantitative variable:\n\n- `.mean()` calculates the mean or average.\n- `.median()` calculates the median.\n- `.quantile(q)` returns a value such that a fraction `q` of the values fall below that value (in other words, the (100q)th percentile).\n- `.max()` calculates the maximum value.\n- `.idxmax()` returns the index of the row with the maximum value. If there are multiple rows that achieve this value, then it will only return the index of the first occurrence.\n\nThe corresponding functions for the _minimum_ value exist as well:\n\n- `.min()` calculates the minimum value.\n- `.idxmin()` returns the index of the row with the minimum value. If there are multiple rows that achieve this value, then it will only return the index of the first occurrence.",
"_____no_output_____"
]
],
[
[
"df.fare.min()",
"_____no_output_____"
]
],
[
[
"Some passengers boarded the Titanic for free, apparently.",
"_____no_output_____"
],
[
"### Measures of Spread\n\nThe center of a quantitative variable only tells part of the story. For one, it tells us nothing about how spread out the values are. Therefore, it is important to also report a measure of **spread**.\n\nLet's investigate a few measures of spread that are built into `pandas`. For completeness, the formulas for these statistics are provided, where $x_1, ..., x_n$ represent the values and $\\bar x$ their mean.\n\nThe first statistic that might come to mind is the **mean absolute deviation**, or MAD. To calculate the MAD, you first calculate the difference between each observation and the mean. Values below the mean will have a negative difference, while values above the mean will have a positive difference. We don't want the negative differences to cancel out the positive differences, since _all_ of them contribute to the spread. So we take the absolute value of all the differences and then average.\n\n$$\n\\begin{align*}\n\\textrm{MAD} &= \\textrm{mean of } |x_i - \\bar x| \\\\\n&= \\frac{1}{n} \\sum_{i=1}^n |x_i - \\bar x|\n\\end{align*}\n$$\n\nWe can implement the MAD ourselves using the `.mean()` and `.abs()` functions.",
"_____no_output_____"
]
],
[
[
"# STEP 1: Calculate the difference between each fare and the mean.\n(df.fare - df.fare.mean())",
"_____no_output_____"
],
[
"# STEP 2: Calculate the absolute value of each difference.\n(df.fare - df.fare.mean()).abs()",
"_____no_output_____"
],
[
"# STEP 3: Take the mean of these absolute differences.\n(df.fare - df.fare.mean()).abs().mean()",
"_____no_output_____"
]
],
[
[
"Notice that in Step 1, we subtracted a single value (`df.fare.mean()`) from a `pandas` `Series` (`df.fare`). A `Series` is like an array, and in most programming languages, subtracting a number from an array is a type mismatch. But `pandas` automatically **broadcasted** the subtraction over each number in the `Series`.\n\nThe `.abs()` function in Step 2 is another example of broadcasting. The absolute value function is applied to each element of the `Series`.",
"_____no_output_____"
],
[
"The MAD is actually built into `pandas`, so there really is no reason to implement it from scratch, as we did above. Let's check that we get the same answer when we call the built-in function.",
"_____no_output_____"
]
],
[
[
"df.fare.mad()",
"_____no_output_____"
]
],
[
[
"Since the MAD is a mean of the absolute differences and the mean represents the \"typical\" value, we can interpret the MAD as saying that the \"typical\" fare is about £30 away from the average.",
"_____no_output_____"
],
[
"Another way to ensure that the negative and positive differences don't cancel is to square all the differences before averaging. This leads to the definition of **variance**.\n\n$$\\textrm{Variance} = \\textrm{mean of } (x_i - \\bar x)^2$$\n\nWe can implement the variance ourselves using the .mean() and power (`**`) functions. Again, notice how the subtraction and the power function are broadcast over the elements of the `Series`.",
"_____no_output_____"
]
],
[
[
"((df.fare - df.fare.mean()) ** 2).mean()",
"_____no_output_____"
]
],
[
[
"Alternatively, we can simply call the `.var()` function in `pandas`.",
"_____no_output_____"
]
],
[
[
"df.fare.var()",
"_____no_output_____"
]
],
[
[
"You might be surprised that `.var()` produces a slightly different number. This is because `pandas` divides by $n-1$ in calculating the mean of the squared differences, rather than $n$. That is, the formula that `pandas` uses is \n\n$$\\text{Variance} = \\frac{1}{n-1} \\sum_{i=1}^n (x_i - \\bar x)^2.$$\n\nTo force Pandas to divide by $n$, you can set `ddof=0`.",
"_____no_output_____"
]
],
[
[
"df.fare.var(ddof=0)",
"_____no_output_____"
]
],
[
[
"Now the value returned by `pandas` matches the value we obtained manually.",
"_____no_output_____"
],
[
"#### Why We Divide By $n-1$ \n\nData is often a sample from some population. The point of calculating the variance of a sample is to be able to say something about the spread of the population.\n\nTo see why we divide by $n-1$ to measure the spread of a population, consider the extreme case where we have a sample of size $n=1$. What can we say about the spread of the population based on this one observation? Absolutely nothing! We need a sample of size at least $n=2$ to be able to say anything about the _spread_. Therefore, the variance is undefined when $n=1$. In order to make the variance not defined for $n=1$, we divide by $n-1$ so that we have $0/0$ when $n=1$. The variance formula above is only defined when $n \\geq 2$.",
"_____no_output_____"
],
[
"The trouble with variance is that its units are wrong. If the original values $x_1, ..., x_n$ were in pounds, the variance would be in pounds _squared_. This is obvious if you simply look at the magnitude of the variance in the example above; the variance is in the _thousands_, even though the largest fare is just over £500. To correct the units of variance, we take the square root to obtain a more interpretable measure of spread, called the **standard deviation**:\n\n$$\\textrm{SD} = \\sqrt{\\textrm{Variance}}.$$",
"_____no_output_____"
]
],
[
[
"df.fare.std()",
"_____no_output_____"
]
],
[
[
"We can interpret this standard deviation as saying: the \"typical\" fare is about £50 away from the average.",
"_____no_output_____"
],
[
"The standard deviation is the most widely used measure of spread, more common than the MAD. At first, this might seem odd. To calculate the standard deviation, we squared the differences from the mean, only to take a square root in the end. Why bother with this rigamarole, when we could just calculate absolute values instead?\n\nThe reasons for preferring the standard deviation are complicated. But the short answer is that the variance (which is the square of the standard deviation) is much nicer mathematically. If you know calculus, you might remember that the absolute value function does not have a derivative at 0. Therefore, the MAD is not _differentiable_, which makes it inconvenient mathematically (especially for various machine learning applications). That doesn't necessarily mean that it's any worse as a measure of spread.",
"_____no_output_____"
],
[
"## Summary Statistics for Categorical Variables\n\nAlthough there are many ways to summarize a quantitative variable, there is really only one way to summarize a categorical variable. Since a categorical variable can only take on a limited set of values, we can completely summarize the variable by reporting the frequencies of the different categories. The `pandas` function that produces this summary is `.value_counts()`.",
"_____no_output_____"
]
],
[
[
"embarked_counts = df.embarked.value_counts()\nembarked_counts",
"_____no_output_____"
]
],
[
[
"Note that the counts are sorted in decreasing order by default, so the first element corresponds to `top` in the summary produced by `.describe()`. Southampton was the most common point of embarkation. \n\nSince the counts are stored in a `pandas` `Series` indexed by category, we can extract a particular count using either label-based or position-based selection:",
"_____no_output_____"
]
],
[
[
"embarked_counts.loc[\"C\"], embarked_counts.iloc[1]",
"_____no_output_____"
]
],
[
[
"Instead of the _number_ of passengers embarking at each location, we might instead want to know the _percentage_ of passengers. To do this, divide the `Series` by the sum to turn the counts into **proportions**. (The term _proportion_ refers to a percentage when it is expressed as a number between 0 and 1, instead of between 0% and 100%.) Proportions must add up to 1, just as percentages must add up to 100%.",
"_____no_output_____"
]
],
[
[
"embarked_counts / embarked_counts.sum()",
"_____no_output_____"
]
],
[
[
"Notice the use of _broadcasting_ again; `embarked_counts` is a `Series`, but `embarked_counts.sum()` is a number. When a `Series` is divided by a number, the division is automatically applied to each element of the `Series`, producing another `Series`.",
"_____no_output_____"
],
[
"### Binary Categorical Variables\n\nA binary categorical variable (i.e., a categorical variable with two categories) can be represented as a quantitative variable by coding one category as 1 and the other as 0.\n\nIn the Titanic data set, the `survived` variable has been coded this way. Each passenger either survived (1) or didn't (0).",
"_____no_output_____"
]
],
[
[
"df.survived",
"_____no_output_____"
]
],
[
[
"Although we can use `.value_counts()` to determine how many passengers survived:",
"_____no_output_____"
]
],
[
[
"df.survived.value_counts()",
"_____no_output_____"
]
],
[
[
"we can also call `.sum()` and `.mean()` on this variable because the values are numeric.\n\nWhat does `.sum()` do?",
"_____no_output_____"
]
],
[
[
"df.survived.sum()",
"_____no_output_____"
]
],
[
[
"`.sum()` returns the _number_ of ones. To see why, remember that this `Series` only 0s and 1s. Each 1 we encounter increments the sum by one, and each 0 contributes nothing to the sum. So when we add up all the numbers, we end up with the number of ones---or, in this example, the number of survivors.\n\nWhat about `.mean()`?",
"_____no_output_____"
]
],
[
[
"df.survived.mean()",
"_____no_output_____"
]
],
[
[
"`.mean()` returns the _proportion_ of ones. To see why, remember that the mean is the sum divided by the number of observations. The sum, as we have just discussed, is the number of 1s. Dividing this by the number of observations gives us the proportion of 1s---or, in this example, the proportion of survivors.\n\n$$ \\text{mean} = \\frac{\\text{sum}}{n} = \\frac{\\text{number of survivors}}{\\text{number of passengers}} = \\text{proportion of passengers who survived}.$$",
"_____no_output_____"
],
[
"## Summary Statistics for Other Variables?\n\nIn the last section, we noted that `name` is not a categorical variable because it does not take on a limited set of values. Hopefully, you now see why it was important to make this distinction. It does not make sense to analyze `name` like we analyzed `embarked` above. For example, if we calculate the frequency of each unique value in `name`, we don't learn much, since names generally do not repeat. ",
"_____no_output_____"
]
],
[
[
"df.name.value_counts()",
"_____no_output_____"
]
],
[
[
"That is why `name` was classified as an \"other\" variable. \"Other\" variables require additional processing before they can be summarized and analyzed. For example, if we extracted just the surnames from the `name` variable, then it might make sense to analyze this new variable as a categorical variable. The following case study shows how.\n\n### Case Study: Extracting the Surname from the Names\n\nWe can extract the surnames from the names using the [built-in string processing functions](https://pandas.pydata.org/pandas-docs/stable/text.html), all of which are preceded by `.str`. The string processing function that will be most useful to us is `.str.split()`, which allows us to split each string in a `Series` by some sequence of characters. (In other words, the `split()` function is _broadcast_ over the strings in the `Series`.) Since the surname and other names are separated by `\", \"`, we will split by `\", \"` to obtain two chunks, the first of which is the surname.",
"_____no_output_____"
]
],
[
[
"df.name.str.split(\", \")",
"_____no_output_____"
]
],
[
[
"We can specify the option `expand=True` to get a `DataFrame` where each chunk is a separate column. The surnames are now in the first column.",
"_____no_output_____"
]
],
[
[
"df.name.str.split(\", \", expand=True)",
"_____no_output_____"
]
],
[
[
"Now we can select the surnames column (the column is named `0` in the `DataFrame`).",
"_____no_output_____"
]
],
[
[
"surnames = df.name.str.split(\", \", expand=True)[0]\nsurnames",
"_____no_output_____"
]
],
[
[
"Since there are multiple passengers with the same surname, this is a categorical variable. We can use `.value_counts()` to find out which surnames were most common.",
"_____no_output_____"
]
],
[
[
"surnames.value_counts()",
"_____no_output_____"
]
],
[
[
"## Summary\n\n- Quantitative and categorical variables are summarized differently.\n- For quantitative variables, we typically report a measure of center (e.g., mean, median, quantiles) and a measure of spread (e.g., variance, standard deviation, MAD).\n- For categorical variables, we typically report the frequencies of the various categories, either as counts or as proportions.\n- Other variables require additional processing before they can be analyzed.",
"_____no_output_____"
],
[
"# Exercises\n\nAll of the following exercises use the Tips data set (`../data/tips.csv`).",
"_____no_output_____"
],
[
"**Exercise 1.** How many people were in the largest party served by the waiter? The smallest?",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"../data/tips.csv\")\nprint(df.head())\nprint(\"Largest was \",df['size'].max(),\". And smallest was \",df['size'].min(),\".\",sep=\"\")",
" total_bill tip sex smoker day time size\n0 16.99 1.01 Female No Sun Dinner 2\n1 10.34 1.66 Male No Sun Dinner 3\n2 21.01 3.50 Male No Sun Dinner 3\n3 23.68 3.31 Male No Sun Dinner 2\n4 24.59 3.61 Female No Sun Dinner 4\nLargest was 6. And smallest was 1.\n"
]
],
[
[
"**Exercise 2.** How could you use the `.quantile()` function to calculate the median? Check that your method works on an appropriate variable from the Tips data set.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE AND ANSWERS HERE (AS COMMENTS)\n#if you were to get the 50th percentile, that is equal to the median.\nprint(df['size'].quantile(0.5))\nprint(df['size'].quantile(0.5)==df['size'].median())\n",
"2.0\nTrue\n"
]
],
[
[
"**Exercise 3.** Another measure of spread is the **interquartile range**, or IQR, defined as:\n\n$$ \\textrm{IQR} = \\textrm{75th percentile} - \\textrm{25th percentile}. $$\n\nMeasure the spread in the total bills by reporting the IQR.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE AND ANSWERS HERE (AS COMMENTS)\nIQR = df['size'].quantile(0.75) - df['size'].quantile(0.25)\nprint(IQR) #The IQR is 1.0",
"1.0\n"
]
],
[
[
"**Exercise 4.** Some people use MAD to refer to the **median absolute deviation**. The median absolute deviation is the same as the mean absolute deviation, but it uses the median instead of the mean:\n\n$$\\textrm{M(edian)AD} = \\textrm{median of } |x_i - \\textrm{median}|. $$\n\nCalculate the median absolute deviation of the total bills. (The median absolute deviation is not built into Pandas, so you will have to implement it from scratch.)",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE AND ANSWERS HERE (AS COMMENTS)\nstep1 = (df.total_bill - df.total_bill.median())\nstep2 = step1.abs()\nstep3 = step2.median()\nprint(\"M(edian)AD =\",step3) #The M(edian)AD = 5.03",
"M(edian)AD = 5.03\n"
]
],
[
[
"**Exercise 5.** Who pays the bill more often: men or women?",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE AND ANSWERS HERE (AS COMMENTS)\nprint(df.sex.value_counts())\n#Men pay the bill more often as 157 (amount of men who paid) is greater than 87( amount of women who paid).",
"157\nMale 157\nFemale 87\nName: sex, dtype: int64\n"
]
],
[
[
"### When you have filled out all the questions, submit via [Tulane Canvas](https://tulane.instructure.com/)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec62098e2b65fe797abf84c394b1052ac43e1e3d | 44,196 | ipynb | Jupyter Notebook | 06_Functions__Primer.ipynb | hphilamore/ILAS_PyEv2019 | b3c8ebe00d6795f67879a50ce6ef517353b069c1 | [
"MIT"
]
| null | null | null | 06_Functions__Primer.ipynb | hphilamore/ILAS_PyEv2019 | b3c8ebe00d6795f67879a50ce6ef517353b069c1 | [
"MIT"
]
| null | null | null | 06_Functions__Primer.ipynb | hphilamore/ILAS_PyEv2019 | b3c8ebe00d6795f67879a50ce6ef517353b069c1 | [
"MIT"
]
| null | null | null | 25.283753 | 3,057 | 0.515974 | [
[
[
"# 06 Functions\n## PRIMER\n\n\n<br> <a href='#AnatomyFunction'>1. The Anatomy of a Function</a> \n<br> <a href='#Scope'>2. Scope</a>",
"_____no_output_____"
],
[
"<a id='AnatomyFunction'></a>\n## 1. The Anatomy of a Function\n\n\n<br> <a href='#FunctionChecklist'>1.1 Function Checklist</a> \n<a href='#DocumentationString'>1.2 The Documentation String</a>\n<br> <a href='#FunctionArguments'>1.3 Function Arguments</a> \n<a href='#return'>1.4 `return`</a>\n\n\nA function is just a piece of code that we can use in our program by typing its name. \n\n\n\nHere is a python function:\n```python\ndef sum_and_increment(a, b): \n c = a + b + 1\n return c\n```\n\nIts name is `sum_and_increment`.\n \n \n\n",
"_____no_output_____"
],
[
"<a id='FunctionChecklist'></a>\n## 1.1 Function Checklist\n\nA custom function is __declared__ using:\n1. The definition keyword, __`def`__.\n1. A __function name__ of your choice.\n1. __() parentheses__ which optionally contain __arguments__ (the *inputs* to the function)\n1. __: a colon__ character\n1. A __documentation string__ that says what the function does.\n1. The __body code__ to be executed when the function is *called*.\n1. An optional __return__ statement (the *output* of the function)\n\n<img src=\"img/function_anotated.png\" alt=\"Drawing\" style=\"width: 600px;\"/>\n",
"_____no_output_____"
]
],
[
[
"def sum_and_increment(a, b): \n c = a + b + 1\n return c\n\nd = sum_and_increment(2, 3)\nprint(d)\n\nprint(sum_and_increment(2, 3))",
"6\n6\n"
]
],
[
[
"__Function name:__ `sum_and_increment`\n\n__Arguments:__ \n<br>`a` and `b`\n<br> Function inputs are placed within () parentheses.\n<br> Function inputs are variables to be used within (\"parsed to\") the function.\n\n ```python\n def sum_and_increment(a, b): \n \n ```\n \n\n",
"_____no_output_____"
],
[
"__Body:__ \n<br>The code to be executed when the function is called. \n<br>Indented (typically four spaces, automatic). \n\n ```python\n def sum_and_increment(a, b): \n c = a + b + 1\n\n ```",
"_____no_output_____"
],
[
"__`return`__ statement: \n<br>Defines the output of the function.\n<br>Often placed at the end of a function.\n\n\n ```python\n def sum_and_increment(a, b): \n c = a + b + 1\n return c\n \n ```",
"_____no_output_____"
],
[
"To execute (*call*) the function, type:\n - a variable to store the output if the function `return`s a value\n - the function name\n - any arguments ",
"_____no_output_____"
]
],
[
[
"m = sum_and_increment(3, 4)\nprint(m) # Expect 8",
"8\n"
],
[
"m = 10\nn = sum_and_increment(m, m)\nprint(n) # Expect 21",
"21\n"
],
[
"l = 5\nm = 6\nn = sum_and_increment(m, l)\nprint(n) ",
"12\n"
]
],
[
[
"<a id='DocumentationString'></a>\n## 1.2 The Documentation String\nIt is best practise to include a *documentation string* (\"docstring\").\n - Describes __in words__ what the function does.\n - Begins and end with `\"\"\"`.\n - *Optional* - however it makes your code much more understandadble. ",
"_____no_output_____"
]
],
[
[
"def sum_and_increment(a, b):\n \"\"\"\n Return the sum of a and b, plus 1\n \"\"\"\n c = a + b + 1\n return c\n",
"_____no_output_____"
]
],
[
[
"A function does not necessarily:\n- take input arguments\n- return output variables\n\n__Example__\n<br>A function with:\n- no input arguments - empty () parentheses\n- no output variables - no `return` statement ",
"_____no_output_____"
]
],
[
[
"def print_message():\n print(\"The function 'print_message' has been called.\")\n\nprint_message()",
"The function 'print_message' has been called.\n"
]
],
[
[
"<a id='FunctionArguments'></a>\n\n## 1.3 Function Arguments\n",
"_____no_output_____"
],
[
"### What can be passed as a function argument?\n\n*Object* types that can be passed as arguments to functions include:\n- single variables (`int`, `float`...)\n- data structures (`list`, `array`...)\n- other functions \n\n",
"_____no_output_____"
],
[
"<a id='SingleVariablesFunctionArguments'></a>\n### Single Variables as Function Arguments. \nWe can define the position of a tree by how far it is offset from an initial set of x,y coordinates.\n",
"_____no_output_____"
]
],
[
[
"import pygame\n\ndef tree(x, y):\n \"Draws a tree\"\n pygame.draw.rect(window, black, [160+x, 300+y, 30, 45])\n pygame.draw.polygon(window, green, [[250+x, 300+y], [175+x, 150+y], [100+x, 300+y]])\n pygame.draw.polygon(window, green, [[240+x, 250+y], [175+x, 130+y], [110+x, 250+y]])",
"_____no_output_____"
],
[
"# Draw a tree at original position\ntree(0, 0)\n \n\n# Draw tree at offset (x, y) = (100, -100)\ntree(100, -100)",
"_____no_output_____"
]
],
[
[
"\nIf we want to draw multiple objects, it can be convenient to express their coordinates usign a data structure.\n\n trees = [[0, 0], [200, -150], [350, -150]]\n \n for t in trees:\n tree(t[0], t[1])\n ",
"_____no_output_____"
],
[
"<a id='DataStructuresFunctionArguments'></a>\n### Data Structures as Function Arguments. \n\nWe can also write a function that accepts a data structure as an argument\n\n<a id='ExampleTrees'></a>\n#### Example: Trees",
"_____no_output_____"
]
],
[
[
"def draw_trees(trees):\n \"Draws trees\"\n \n for t in trees:\n \n x = t[0]\n y = t[1]\n \n pygame.draw.rect(window, black, [160+x, 300+y, 30, 45])\n pygame.draw.polygon(window, green, [[250+x, 300+y], [175+x, 150+y], [100+x, 300+y]])\n pygame.draw.polygon(window, green, [[240+x, 250+y], [175+x, 130+y], [110+x, 250+y]])",
"_____no_output_____"
]
],
[
[
"<a id='FunctionsFunctionArguments'></a>\n### Functions as Function Arguments. \nConsider the two functions.\n<br>The docstring of each function explains what it does.",
"_____no_output_____"
]
],
[
[
"# Function A\ndef f0(y):\n \"Computes y^2 - 10\"\n return y*y - 10\n\n# Function B\ndef is_positive(x):\n \"Checks if x is positive\"\n return x > 0\n\nprint(f0(3))\nprint(is_positive(3))",
"_____no_output_____"
]
],
[
[
"Let's say we want to test if $y^2 - 1$ (*Function A*) is positive (*Function B*).\n\nWe can nest one function within another function:",
"_____no_output_____"
]
],
[
[
"print(is_positive( f0(3) ))",
"_____no_output_____"
]
],
[
[
"Alternatively we can re-write Function B to take a function and a variable as *seperate* input arguments: ",
"_____no_output_____"
]
],
[
[
"# Function A\ndef f0(y):\n \"Computes y^2 - 1\"\n return y*y - 10\n\n# Function B\n\n# def is_positive(x):\n# \"Checks if x is positive\"\n# return x > 0\n\ndef is_positive(f, x):\n \"Checks if the function value f(x) is positive\"\n return f(x) > 0\n\nprint(is_positive(f0, 3))\n \n",
"_____no_output_____"
]
],
[
[
"This is useful, for example, where the use of the function depends on the input value.\n\nThis time *Function B* includes `if-else`:",
"_____no_output_____"
]
],
[
[
"# Function B\ndef is_positive(f, x):\n \"Checks if the function value f(x) is positive\"\n # odd\n if x%2:\n return f(x) > 0\n # even\n else:\n return x > 0\n\nprint(is_positive(f0, 2))\nprint(is_positive(f0, 3))",
"_____no_output_____"
]
],
[
[
"Multiple functions can be input as arguments.",
"_____no_output_____"
]
],
[
[
"# Function A\ndef f0(y):\n \"Computes y^2 - 1\"\n return y*y - 10\n\n\n# Function A'\ndef f1(y):\n \"Computes y^2 - 1\"\n return y*y*y - 10\n\n\n# Function B\ndef is_positive(x, f_0, f_1):\n \"Checks if the function value f(x) is positive\"\n if x%2:\n return f_0(x) > 0\n else:\n return f_1(x) > 0\n \n \nprint(is_positive(2, f0, f1))\nprint(is_positive(3, f0, f1))",
"_____no_output_____"
]
],
[
[
"### Rules for Inputting Arguments\n<a id='RulesInputtingArguments'></a>",
"_____no_output_____"
],
[
"It is important input arguments in the correct order. ",
"_____no_output_____"
]
],
[
[
"def sum_and_increment(a, b):\n \"\"\"\"\n Return the sum of a and b, plus 1\n \"\"\"\n c = a + b + 1\n return c",
"_____no_output_____"
]
],
[
[
"The function `sum_and_increment` finds the sum of:\n - the first argument, `a`\n - the second argument `b`\n - 1\n \nIf the order of a and b is switched, the result is the same.\n",
"_____no_output_____"
]
],
[
[
"print(sum_and_increment(3,4))\nprint(sum_and_increment(4,3))",
"_____no_output_____"
]
],
[
[
"However, if we subtract one argument from the other, the result depends on the input order: ",
"_____no_output_____"
]
],
[
[
"def subtract_and_increment(a, b):\n \"\"\"\"\n Return a minus b, plus 1\n \"\"\"\n c = a - b + 1\n return c\n\nprint(subtract_and_increment(3,4))\nprint(subtract_and_increment(4,3))",
"_____no_output_____"
]
],
[
[
"### Named Arguments\n<a id='NamedArguments'></a>\nIt can be easy to make a mistake in the input order, leading to incorrect output. \n\nWe can reduce this risk by giving inputs as *named* arguments. \n\nWhen we use named arguments, the order of input does not matter. ",
"_____no_output_____"
]
],
[
[
"def subtract_and_increment(a, b):\n \"Return a minus b, plus 1\"\n c = a - b + 1\n return c\n\nalpha = 3\nbeta = 4\n\nprint(subtract_and_increment(a=alpha, b=beta))\nprint(subtract_and_increment(b=beta, a=alpha)) ",
"_____no_output_____"
]
],
[
[
"<a id='DefaultKeywordArguments'></a>\n### Default / Keyword Arguments\n\n'Default' or 'keyword' arguments have a default initial value.\n\nThe default value can be overridden when the function is called. \n\nIn some cases it just saves the programmer effort - they can write less code. \n\nIn other cases default arguments allow a function to be applied to a wider range of problems. \n\n",
"_____no_output_____"
],
[
"#### Example: Blue Trees\n<a id='ExampleBlueTrees'></a>\nEvery time we have drawn a tree it has been green, with a black tree-trunk.\n\nWe can therefore write the function `tree`:",
"_____no_output_____"
]
],
[
[
"# original code\n\ndef tree(x, y):\n \"Draws a tree\"\n pygame.draw.rect(window, black, [160+x, 300+y, 30, 45])\n pygame.draw.polygon(window, green, [[250+x, 300+y], [175+x, 150+y], [100+x, 300+y]])\n pygame.draw.polygon(window, green, [[240+x, 250+y], [175+x, 130+y], [110+x, 250+y]])\n\n\n# function with keyword arguments\n\ndef tree(x, y, tree_colour=green, trunk_colour=black):\n \"Draws a tree\"\n pygame.draw.rect(window, trunk_colour, [160+x, 300+y, 30, 45])\n pygame.draw.polygon(window, tree_colour, [[250+x, 300+y], [175+x, 150+y], [100+x, 300+y]])\n pygame.draw.polygon(window, tree_colour, [[240+x, 250+y], [175+x, 130+y], [110+x, 250+y]])",
"_____no_output_____"
]
],
[
[
"The function can now take *up to* four arguments.\n\nHowever, we only need to enter the *first two arguments* (`x, y`), if using the default values (`tree_colour=green, trunk_colour=black`). \n",
"_____no_output_____"
]
],
[
[
"tree(0, 0)\ntree(100, -100)",
"_____no_output_____"
]
],
[
[
"Sometimes we want to draw blue tree trunks. \n\nIn this case, we simply override the default value for `trunk_colour`: ",
"_____no_output_____"
]
],
[
[
"tree(100, -100, green, blue)",
"_____no_output_____"
]
],
[
[
"__Note__ that we have *also* entered the `tree_colour`.\n\nAs the value to overide is the 4th argument, the 3rd argument must also be input. \n\nThe function interprets:\n\n tree(100, -100, blue)\n \nas\n\n tree(100, -100, blue, black)\n \n\n",
"_____no_output_____"
],
[
"Manually inputting an argument, `tree_colour` when we want to use its default is a potential source of error. \n\nWe may accidentally input the default value of `tree_colour` incorrectly, causing a bug in our program. \n\nA more robust solution is to specify the colour by using a __named argument__. ",
"_____no_output_____"
]
],
[
[
"tree(100, -100, trunk_colour=black)",
"_____no_output_____"
]
],
[
[
"The program overwrites the correct default value.\n\nWe do not have to specify `tree_colour`. ",
"_____no_output_____"
],
[
"### Forcing Default Arguments\n<a id='ForcingDefaultArguments'></a>\nAs an additional safety measure, you can force arguments to be entered as named arguments by preceding them with a * star in the function definition.\n\nAll arguments after the star must be entered as named arguments.\n\nBelow is an example:",
"_____no_output_____"
]
],
[
[
"# redefine tree function, forcing keyword arguments\n\ndef tree(x, y, *, tree_colour=green, trunk_colour=black):\n \"Draws a tree\"\n pygame.draw.rect(window, trunk_colour, [160+x, 300+y, 30, 45])\n pygame.draw.polygon(window, tree_colour, [[250+x, 300+y], [175+x, 150+y], [100+x, 300+y]])\n pygame.draw.polygon(window, tree_colour, [[240+x, 250+y], [175+x, 130+y], [110+x, 250+y]])\n\n \ntree(x, y, tree_colour=blue)\n#tree(x, y, blue)",
"_____no_output_____"
]
],
[
[
"<a id='return'></a>\n## 1.4 `return` \n\nThe `return` keyword defines the outputs of the function.\n\nA __single__ Python function can return:\n- no values\n- a single value \n- multiple return values",
"_____no_output_____"
],
[
"For example, we could have a function that:\n - takes three values (`x0, x1, x2`)\n - returns the maximum, the minimum and the mean",
"_____no_output_____"
]
],
[
[
"def compute_max_min_mean(x0, x1, x2):\n \"Return maximum, minimum and mean values\"\n \n x_min = x0\n if x1 < x_min:\n x_min = x1\n if x2 < x_min:\n x_min = x2\n\n x_max = x0\n if x1 > x_max:\n x_max = x1\n if x2 > x_max:\n x_max = x2\n\n x_mean = (x0 + x1 + x2)/3 \n \n return x_min, x_max, x_mean\n\n\nXmin, Xmax, Xmean = compute_max_min_mean(0.5, 0.1, -20)\n\nX = compute_max_min_mean(0.5, 0.1, -20)\n\n#print(Xmin, Xmax, Xmean)\nprint(X)\nprint(X[2])",
"_____no_output_____"
]
],
[
[
"The __`return`__ keyword works a bit like the __`break`__ statement does in a loop.\n\nIt returns the value and then exits the function before running the rest of the code.",
"_____no_output_____"
],
[
"Any code following the `return` statement will not be run.\n\nIn this example, the code to increase x by 1 comes after the return statement. ",
"_____no_output_____"
]
],
[
[
"x = 1\n\ndef process_value(X):\n \"Returns a value that depends on the input value x \"\n \n if X > 10:\n return str(X) + \" > 10\"\n elif X > 5:\n return str(X) + \" > 5\"\n elif X > 0:\n return str(X) + \" > 0\"\n else:\n return str(X)\n \n # Increment global x by +1\n global x\n x = X + 1 \n \nprint(process_value(x))\nprint(process_value(x))\nprint(process_value(x))",
"_____no_output_____"
]
],
[
[
"The return statement must come last.",
"_____no_output_____"
]
],
[
[
"x = 1\n\ndef process_value(X):\n \"Returns a value that depends on the input value x \"\n \n #Increment global x by +1 \n global x\n x = X + 1 \n \n if x > 10:\n return str(X) + \" > 10\"\n elif x > 5:\n return str(X) + \" > 5\"\n elif x > 0:\n return str(X) + \" > 0\"\n else:\n return str(X) \n \nprint(process_value(x))\nprint(process_value(x))\nprint(process_value(x))",
"_____no_output_____"
]
],
[
[
"It may be more appropriate to store the return item as a varable if multiple items are to be returned...\n<br> ",
"_____no_output_____"
]
],
[
[
"x = -3\n\ndef process_value(X): \n \"Returns two values that depend on the input value x \"\n if X > 10:\n i = (str(X) + \" > 10\")\n elif X > 0:\n i = (str(X) + \" > 0\")\n else:\n i = None\n \n if X < 0:\n j = (str(X) + \" < 0\")\n elif X < 10:\n j = (str(X) + \" < 10\")\n else:\n j = None\n \n global x\n x = X + 1 \n \n return i, j\n\n\n# if i and j: \n# return i, j \n# elif i:\n# return (i,)\n# else:\n# return (j,)\n\n\nfor k in range(14):\n print(process_value(x))",
"_____no_output_____"
]
],
[
[
"<a id='Scope'></a>\n# 2. Scope\n\n__Global variables:__ Variable that are *declared* __outside__ of a function *can* be used __inside__ of the function. <br>\nThey have *global scope*. \n\n__Local variables:__ Variables that are *declared* __inside__ of a function *can not* be used __outside__ of the function. \n<br>\nThey have *local scope*. ",
"_____no_output_____"
],
[
"#### Example: Global Variables\nGlobal variables are accessible anywhere",
"_____no_output_____"
]
],
[
[
"# global variable\nglobal_var = \"Global variable\"\n\n# define function\ndef my_func():\n \"\"\"\n Prints a global variable \n \"\"\"\n # the function can access the global variable\n print(global_var) \n \n\n\n# call function\nmy_func() ",
"_____no_output_____"
]
],
[
[
"The global variable may be created *after* the function is __defined__,\n<br>*but must* be created *before* the function is __called__.",
"_____no_output_____"
]
],
[
[
"# define function\ndef my_func():\n \"\"\"\n Prints a global variable \n \"\"\"\n # the function can access the global variable\n print(global_var) \n \n \n \n# global variable\nglobal_var = \"Global variable\"\n\n\n# call function\nmy_func() ",
"_____no_output_____"
]
],
[
[
"A global variable can be __created__ *inside* a function using the `global` keyword:",
"_____no_output_____"
]
],
[
[
"def my_func():\n \n # Locally assigned global variable\n global var\n var = \"Locally assigned global variable\"\n \n\n# global variable does not exit before function call\n# print(var)\n\nmy_func()\n\nprint(var)",
"_____no_output_____"
]
],
[
[
"#### Example: Local Variables\nLocal variables only accessible within the function in which they are defined",
"_____no_output_____"
]
],
[
[
"# define function\ndef my_func():\n \"\"\"\n Prints a local variable \n \"\"\" \n \n # global variable\n local_var = \"Local variable\"\n print(local_var)\n \n \n# call function\nmy_func()\n\n\n# try to print local variable\n# print(local_var)",
"_____no_output_____"
]
],
[
[
"__Readability:__ \n\nThe limited scope of local variables can be useful。\n\nFor example, some variable names can be useful for different tasks in our program. \n\nWe may not want to \"use them up\" on a single task.",
"_____no_output_____"
],
[
"Due to scope, variables with the *same name* can appear globally and locally without conflict. \n\nThis prevents variables declared inside a function from unexpectedly affecting other parts of a program. \n\n",
"_____no_output_____"
],
[
"Where a local and global variable have the same name, the program will use the __local__ version.\n\nLet's modify our function `my_func` so now both the local and global varibale have the same name...\n\nThis time the first `print(var)` raises an error.\n\nThe local variable overrides the global variable, \n<br>however the local variable has not yet been assigned a value.",
"_____no_output_____"
]
],
[
[
"# global variable\nvar = \"Global variable\"\n\n\ndef my_func():\n # notice what happens this time if we try to access the global variable within the function\n print(var) \n \n # local variable of the same name\n var = \"Local variable\"\n print(var)\n\n \n# Call the function.\n# print(my_func())",
"_____no_output_____"
]
],
[
[
"\n\nThe global variable `var` is unaffected by the local variable `var`.",
"_____no_output_____"
]
],
[
[
"# global variable\nvar = \"Global variable\"\n\ndef my_func():\n \n # local variable of the same name\n var = \"Local variable\"\n return var\n\n\n# Call the function.\nprint(my_func())\n\n\n# The global variable is unaffected by the local variable\nprint(var)\n\n\n# We can overwrite the global varibale with the returned value\nvar = my_func()\nprint(var)",
"_____no_output_____"
]
],
[
[
"If we *really* want to use a global variable and a local variable:\n- with the same name \n- within the same function\n\nwe can input use the global variable as a __function argument__. \n\nBy inputting it as an argument we rename the global variable for use within the function....",
"_____no_output_____"
]
],
[
[
"# Global \nvar = \"Global variable\"\n\n\ndef my_func(input_var):\n # The global variable is renameed for use within a function with a local variable with the same name\n print(input_var) \n \n # Local\n var = \"Local variable\"\n print(var)\n \n return (input_var + \" \" + var)\n\n\n# Run the function, giving the global variable as an argument\nprint(my_func(var))",
"_____no_output_____"
]
],
[
[
"The global variable is unaffected by the function",
"_____no_output_____"
]
],
[
[
"print(var)",
"_____no_output_____"
]
],
[
[
"...unless we overwrite the value of the global variable.",
"_____no_output_____"
]
],
[
[
"print(var)\n\nvar = my_func(var)\n\nprint(var)",
"_____no_output_____"
]
],
[
[
"__Try it yourself__\nIn the cell below:\n1. Create a global variable called `my_var`, with a numeric value\n1. Create a function, called `my_func`, that:\n - takes a single argument, `input_var` \n - creates a local variable called `my_var` (same name as global variable).\n - returns the sum of the function argument and the local variable: `input_var + my_var`.<br><br>\n1. Print the output when the function `my_func` is called, giving the global varable `my_var` as the input agument.\n1. print the global variable `my_var`.\n1. Add a docstring to say what your function does",
"_____no_output_____"
],
[
"A global variable can be __modified__ from *inside* a function using the `global` keyword:\n1. Use Python `global` keyword. Give the variable a name.\n```python\nglobal var\n```\n1. Assign the variable a value.\n```python\nvar = 10\n```",
"_____no_output_____"
]
],
[
[
"# global variable\nvar = \"Global variable\"\n\n\ndef my_func():\n \n # Locally assigned global variable\n global var\n var = \"Locally assigned global variable\"\n \n\n \nprint(\"Before function call, var =\", var)\n\n\n# Call the function.\nmy_func()\n\n\nprint(\"After function call, var =\", var)",
"_____no_output_____"
]
],
[
[
"__Try it yourself__\n\nIn the cell below:\n1. Copy and paste your code from the previous exercise.\n1. Edit your code so that:\n - The function `my_func` takes no input arguments. \n - The global variable `my_var` is overwritten within the function using the prefix `global`. \n1. Print the global variable before and after calling the function to check your code. \n1. Modify the docstring as necessary.",
"_____no_output_____"
]
],
[
[
"# Copy and paste code here:",
"_____no_output_____"
],
[
"# Global and local scope",
"_____no_output_____"
]
],
[
[
"As we have seen, a *local variable* can be accessed from outside the function by *returning* it. ",
"_____no_output_____"
],
[
"# Summary\n<a id='Summary'></a>\n - Functions are defined using the `def` keyword.\n - Functions contain indented statements to execute when the function is called.\n - Global variables can be used eveywhere.\n - Local variables can be used within the function in which they are defined.\n - Function arguments (inputs) are declared between () parentheses, seperated by commas.\n - Function arguments muct be specified each time a function is called. \n - Default arguments do not need to be specified when a function is called unless different values are to be used. \n - The keyword used to define the function outputs is `return`\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
]
|
ec621bef121035e18686f7dfb43e0c7df0ec195b | 89,434 | ipynb | Jupyter Notebook | Daniel_Ogechi_PythonNotebook.ipynb | Dogechi/dataAnalysis | b8a752d59695b0c5c60d82e14693b94492533e96 | [
"MIT"
]
| null | null | null | Daniel_Ogechi_PythonNotebook.ipynb | Dogechi/dataAnalysis | b8a752d59695b0c5c60d82e14693b94492533e96 | [
"MIT"
]
| null | null | null | Daniel_Ogechi_PythonNotebook.ipynb | Dogechi/dataAnalysis | b8a752d59695b0c5c60d82e14693b94492533e96 | [
"MIT"
]
| null | null | null | 38.532529 | 109 | 0.316781 | [
[
[
"#import libraries\nimport pandas as pd\nimport numpy as np\n",
"_____no_output_____"
],
[
"#loading datasets\n\ntelcom1 = pd.read_csv('Telcom_dataset.csv')\ntelcom1.head(10)\n\n\n\na = ['PRODUCT','VALUE','DATE_TIME','CELL_ON_SITE',\n'DW_A_NUMBER_INT',\n'DW_B_NUMBER_INT',\n'COUNTRY_A',\n'COUNTRY_B',\n'CELL_ID',\n'SITE_ID']\n\n\ntelcom1.columns.values[:] = [i for i in a ]\n\ntelcom1",
"_____no_output_____"
],
[
"#Getting rid of unwanted columns for the first data set\n# uwanted_columns = ['COUNTRY_A','COUNTRY_B','CELL_ON_SITE']\n\ntelcom_1_clean = telcom1.drop(columns=(['COUNTRY_A','COUNTRY_B','CELL_ON_SITE']))\ntelcom_1_clean",
"_____no_output_____"
],
[
"#reading and renaming columns for the second dataset provided\n\ntelcom2 = pd.read_csv('Telcom_dataset2.csv')\ntelcom2.head(10)\n\ntelcom2.columns = ['PRODUCT','VALUE','DATE_TIME','CELL_ON_SITE',\n'DW_A_NUMBER_INT',\n'DW_B_NUMBER_INT',\n'COUNTRY_A',\n'COUNTRY_B',\n'CELL_ID',\n'SITE_ID']",
"_____no_output_____"
],
[
"#Droping the uncessary columns forthe second dataset\n\ntelcom_2_clean = telcom2.drop(columns=(['COUNTRY_A','COUNTRY_B','CELL_ON_SITE']))\ntelcom_2_clean",
"_____no_output_____"
],
[
"#read the third dataset\ntelcom3 = pd.read_csv('Telcom_dataset3.csv')\ntelcom3.head(10)\n\ntelcom3.columns = ['PRODUCT','VALUE','DATE_TIME','CELL_ON_SITE',\n'DW_A_NUMBER_INT',\n'DW_B_NUMBER_INT',\n'COUNTRY_A',\n'COUNTRY_B',\n'CELL_ID',\n'SITE_ID']",
"_____no_output_____"
],
[
"#Drop the unwanted columns)\n\ntelcom_3_clean = telcom3.drop(columns=(['COUNTRY_A','COUNTRY_B','CELL_ON_SITE']))\ntelcom_3_clean",
"_____no_output_____"
],
[
"# columns=['VILLES',\n# 'STATUS',\n# 'LOCALISATION',\n# 'DECOUPZONE',\n# 'ZONENAME',\n# 'LONGITUDE',\n# 'LATITUDE',\n# 'REGION',\n# 'AREA',\n# 'CELL_ID',\n# 'SITE_CODE']\ndeli = [',',' ',',']\ncells = pd.read_csv('/content/cells_geo.csv', delimiter=';')\ncells_df = pd.DataFrame(cells)\ncells_df.head(10)",
"_____no_output_____"
],
[
"#CONCATNATE ALL THE TABLES TO BE ABLE TO DO SOME OPERATIONS ON THE DATA\n\ndt1 = pd.concat([telcom_1_clean,telcom_2_clean,telcom_3_clean],axis = 0,sort = False)\ndt1",
"_____no_output_____"
],
[
"#check for null values and delete rows with no data and value is equal to 0\n\ndt1.dropna(how='all')\ndt1 = dt1[dt1.VALUE != 0]\ndt1\ndt1 = dt1.drop_duplicates()\ndt1\n",
"_____no_output_____"
],
[
"#replacing null values with 0 and converting it to lowercase\ndt1 = dt1.fillna('.')\ndt1 = dt1.astype(str).apply(lambda x: x.str.lower())\ndt1",
"_____no_output_____"
],
[
"#conveting the geo dataset to lowercase\n\n\n# cells_df = cells_df.fillna(0)\n\n# cells_df = cells_df.dropna()\ncells_df = cells_df.astype(str).apply(lambda x: x.str.lower())\ncells_df\ncells_df = cells_df.drop_duplicates()\ncells_df.describe()",
"_____no_output_____"
],
[
"# Which ones were the most used city for the three days?\n#We merger our dataset with the geographical data on the common column which is cell_ID where\ncombined = dt1.merge(cells_df, left_on= 'SITE_ID', right_on='CELL_ID')\ncombined.head(10)\n\ncombined = combined.drop_duplicates()\ncombined.head(10)",
"_____no_output_____"
],
[
"# merged = dt1.merge(cells_df, how='inner', on='CELL_ID')\n# most_city = merged.groupby('VILLES')['VALUE'].sum()\n# most_city.sort_values(ascending=False)\n# # merged\n# most_city\n\ncombined_count = combined.groupby('VILLES')['VILLES'].count()\ncombined_count.sort_values(ascending=False)",
"_____no_output_____"
],
[
"# Question 2",
"_____no_output_____"
],
[
"# Which cities were the most used during business and home hours?\n#split date and time columns\n\ndt1_no_date = dt1.drop(columns='DATE_TIME')\ndt1_no_date\n\n#time column\ndt2 = dt1['DATE_TIME'].apply(lambda x: pd.Series([i for i in reversed(x.lower().split(' '))]))\ndt2.columns =['DATE','TIME']\ndt2_time = dt2.drop(columns='DATE')\ndt2_time\n\n#date column\ndt2 = dt2['DATE'].apply(lambda x: pd.Series([i for i in reversed(x.lower().split('.'))]))\ndt2.columns = ['index', 'DATE']\ndt3_date = dt2.drop('index',axis=1)\ndt3_date\n\n\ndt4 = pd.concat([dt1_no_date, dt2_time, dt3_date],axis = 1,sort = False)\ndt4.head()\ndt4.rename(columns= {'TIME':'DATE','DATE':'TIME'},inplace =True)\ndt4\n\n#using the cleaned date/time dateset and the geo data we can find the cities \n\nbusiness_hours = (dt4['TIME'] < '08:00:00') \nbusiness_hours.describe()",
"_____no_output_____"
],
[
"combined_s = combined.groupby('PRODUCT')['VALUE'].sum()\ncombined_s",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec621f41c4c1b4dca16f9d49da632578f7375ba4 | 7,958 | ipynb | Jupyter Notebook | notebooks/2.1-am-PolicyGradients_Pong.ipynb | adityamangal410/nlp_with_pytorch | 81919102339ee483210f366aeaec0dd30273a846 | [
"MIT"
]
| 2 | 2020-12-26T07:32:42.000Z | 2021-01-16T19:07:13.000Z | notebooks/2.1-am-PolicyGradients_Pong.ipynb | adityamangal410/nlp_with_pytorch | 81919102339ee483210f366aeaec0dd30273a846 | [
"MIT"
]
| null | null | null | notebooks/2.1-am-PolicyGradients_Pong.ipynb | adityamangal410/nlp_with_pytorch | 81919102339ee483210f366aeaec0dd30273a846 | [
"MIT"
]
| null | null | null | 36.172727 | 164 | 0.560945 | [
[
[
"# Using Policy Gradients to play Pong - Reinforcement Learning",
"_____no_output_____"
],
[
"https://gist.github.com/karpathy/a4166c7fe253700972fcbc77e4ea32c5",
"_____no_output_____"
]
],
[
[
"# !pip install gym[atari]",
"_____no_output_____"
],
[
"\"\"\" Trains an agent with (stochastic) Policy Gradients on Pong. Uses OpenAI Gym. \"\"\"\nimport numpy as np\n# import cPickle as pickle\nimport gym\nimport pickle",
"_____no_output_____"
],
[
"\n# hyperparameters\nH = 200 # number of hidden layer neurons\nbatch_size = 10 # every how many episodes to do a param update?\nlearning_rate = 1e-4\ngamma = 0.99 # discount factor for reward\ndecay_rate = 0.99 # decay factor for RMSProp leaky sum of grad^2\nresume = True # resume from previous checkpoint?\nrender = True\n\n# model initialization\nD = 80 * 80 # input dimensionality: 80x80 grid\nif resume:\n model = pickle.load(open('save.p', 'rb'))\nelse:\n model = {}\n model['W1'] = np.random.randn(H,D) / np.sqrt(D) # \"Xavier\" initialization\n model['W2'] = np.random.randn(H) / np.sqrt(H)\n \ngrad_buffer = { k : np.zeros_like(v) for k,v in model.items() } # update buffers that add up gradients over a batch\nrmsprop_cache = { k : np.zeros_like(v) for k,v in model.items() } # rmsprop memory",
"_____no_output_____"
],
[
"def sigmoid(x): \n return 1.0 / (1.0 + np.exp(-x)) # sigmoid \"squashing\" function to interval [0,1]\n\n\ndef prepro(I):\n \"\"\" prepro 210x160x3 uint8 frame into 6400 (80x80) 1D float vector \"\"\"\n I = I[35:195] # crop\n I = I[::2,::2,0] # downsample by factor of 2\n I[I == 144] = 0 # erase background (background type 1)\n I[I == 109] = 0 # erase background (background type 2)\n I[I != 0] = 1 # everything else (paddles, ball) just set to 1\n return I.astype(np.float).ravel()\n\ndef discount_rewards(r):\n \"\"\" take 1D float array of rewards and compute discounted reward \"\"\"\n discounted_r = np.zeros_like(r)\n running_add = 0\n for t in reversed(range(0, r.size)):\n if r[t] != 0: running_add = 0 # reset the sum, since this was a game boundary (pong specific!)\n running_add = running_add * gamma + r[t]\n discounted_r[t] = running_add\n return discounted_r\n\ndef policy_forward(x):\n h = np.dot(model['W1'], x)\n h[h<0] = 0 # ReLU nonlinearity\n logp = np.dot(model['W2'], h)\n p = sigmoid(logp)\n return p, h # return probability of taking action 2, and hidden state\n\ndef policy_backward(eph, epdlogp):\n \"\"\" backward pass. (eph is array of intermediate hidden states) \"\"\"\n dW2 = np.dot(eph.T, epdlogp).ravel()\n dh = np.outer(epdlogp, model['W2'])\n dh[eph <= 0] = 0 # backpro prelu\n dW1 = np.dot(dh.T, epx)\n return {'W1':dW1, 'W2':dW2}",
"_____no_output_____"
],
[
"\nenv = gym.make(\"Pong-v0\")\nobservation = env.reset()\nprev_x = None # used in computing the difference frame\nxs,hs,dlogps,drs = [],[],[],[]\nrunning_reward = None\nreward_sum = 0\nepisode_number = 0\nwhile True:\n if render: env.render()\n\n # preprocess the observation, set input to network to be difference image\n cur_x = prepro(observation)\n x = cur_x - prev_x if prev_x is not None else np.zeros(D)\n prev_x = cur_x\n\n # forward the policy network and sample an action from the returned probability\n aprob, h = policy_forward(x)\n action = 2 if np.random.uniform() < aprob else 3 # roll the dice!\n\n # record various intermediates (needed later for backprop)\n xs.append(x) # observation\n hs.append(h) # hidden state\n y = 1 if action == 2 else 0 # a \"fake label\"\n dlogps.append(y - aprob) # grad that encourages the action that was taken to be taken (see http://cs231n.github.io/neural-networks-2/#losses if confused)\n\n # step the environment and get new measurements\n observation, reward, done, info = env.step(action)\n reward_sum += reward\n\n drs.append(reward) # record reward (has to be done after we call step() to get reward for previous action)\n\n if done: # an episode finished\n episode_number += 1\n\n # stack together all inputs, hidden states, action gradients, and rewards for this episode\n epx = np.vstack(xs)\n eph = np.vstack(hs)\n epdlogp = np.vstack(dlogps)\n epr = np.vstack(drs)\n xs,hs,dlogps,drs = [],[],[],[] # reset array memory\n\n # compute the discounted reward backwards through time\n discounted_epr = discount_rewards(epr)\n # standardize the rewards to be unit normal (helps control the gradient estimator variance)\n discounted_epr -= np.mean(discounted_epr)\n discounted_epr /= np.std(discounted_epr)\n\n epdlogp *= discounted_epr # modulate the gradient with advantage (PG magic happens right here.)\n grad = policy_backward(eph, epdlogp)\n for k in model: grad_buffer[k] += grad[k] # accumulate grad over batch\n\n # perform rmsprop parameter update every batch_size episodes\n if episode_number % batch_size == 0:\n for k,v in model.items():\n g = grad_buffer[k] # gradient\n rmsprop_cache[k] = decay_rate * rmsprop_cache[k] + (1 - decay_rate) * g**2\n model[k] += learning_rate * g / (np.sqrt(rmsprop_cache[k]) + 1e-5)\n grad_buffer[k] = np.zeros_like(v) # reset batch gradient buffer\n\n # boring book-keeping\n running_reward = reward_sum if running_reward is None else running_reward * 0.99 + reward_sum * 0.01\n print('resetting env. episode reward total was %f. running mean: %f' % (reward_sum, running_reward))\n if episode_number % 100 == 0: pickle.dump(model, open('save.p', 'wb'))\n reward_sum = 0\n observation = env.reset() # reset env\n prev_x = None\n\n if reward != 0: # Pong has either +1 or -1 reward exactly when game ends.\n print (('ep %d: game finished, reward: %f' % (episode_number, reward)) + ('' if reward == -1 else ' !!!!!!!!'))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6230ebeaee2e4093d7d20cfd0d20397c78a3bc | 1,541 | ipynb | Jupyter Notebook | Chapter07/unit_tests/.ipynb_checkpoints/Exercise 81-checkpoint.ipynb | arifmudi/The-Data-Wrangling-Workshop | c325f6fa1c6daf8dd22e9705df48ce2644217a73 | [
"MIT"
]
| 22 | 2020-06-27T04:21:49.000Z | 2022-03-08T04:39:44.000Z | Chapter07/unit_tests/.ipynb_checkpoints/Exercise 81-checkpoint.ipynb | arifmudi/The-Data-Wrangling-Workshop | c325f6fa1c6daf8dd22e9705df48ce2644217a73 | [
"MIT"
]
| 2 | 2021-02-02T22:49:16.000Z | 2021-06-02T02:09:21.000Z | Chapter07/unit_tests/.ipynb_checkpoints/Exercise 81-checkpoint.ipynb | Hubertus444/The-Data-Wrangling-Workshop | ddad20f8676602ac6624e72e802769fcaff45b0f | [
"MIT"
]
| 46 | 2020-04-20T13:04:11.000Z | 2022-03-22T05:23:52.000Z | 21.109589 | 68 | 0.519143 | [
[
[
"def test_exercise_81__1(x) -> bool:\n import requests\n if \"requests\" not in sys.modules:\n return False\n else:\n return True",
"_____no_output_____"
],
[
"def test_exercise_81__2(x) -> bool:\n return \"https://en.wikipedia.org/wiki/Main_Page\" == x",
"_____no_output_____"
],
[
"def test_exercise_81__3(x) -> bool:\n import requests \n wiki_home = \"https://en.wikipedia.org/wiki/Main_Page\"\n response = requests.get(wiki_home)\n return response == x",
"_____no_output_____"
],
[
"def test_exercise_81__4(x) -> bool:\n return \"requests.models.Response\" == x",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
ec623a2bb60945f85c9372119b257b685654fe0b | 20,270 | ipynb | Jupyter Notebook | 05_Merge/Auto_MPG/Exercises.ipynb | pidodo92/pandas_exercises | 3563081adc9a6e1601eb8f01ccbca330d65715d5 | [
"BSD-3-Clause"
]
| null | null | null | 05_Merge/Auto_MPG/Exercises.ipynb | pidodo92/pandas_exercises | 3563081adc9a6e1601eb8f01ccbca330d65715d5 | [
"BSD-3-Clause"
]
| null | null | null | 05_Merge/Auto_MPG/Exercises.ipynb | pidodo92/pandas_exercises | 3563081adc9a6e1601eb8f01ccbca330d65715d5 | [
"BSD-3-Clause"
]
| null | null | null | 29.548105 | 261 | 0.343661 | [
[
[
"# MPG Cars",
"_____no_output_____"
],
[
"### Introduction:\n\nThe following exercise utilizes data from [UC Irvine Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Auto+MPG)\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the first dataset [cars1](https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/05_Merge/Auto_MPG/cars1.csv) and [cars2](https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/05_Merge/Auto_MPG/cars2.csv). ",
"_____no_output_____"
],
[
" ### Step 3. Assign each to a variable called cars1 and cars2",
"_____no_output_____"
]
],
[
[
"cars1 = pd.read_csv('https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/05_Merge/Auto_MPG/cars1.csv')\ncars2 = pd.read_csv('https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/05_Merge/Auto_MPG/cars2.csv')",
"_____no_output_____"
]
],
[
[
"### Step 4. Oops, it seems our first dataset has some unnamed blank columns, fix cars1",
"_____no_output_____"
]
],
[
[
"cars1\n\ncars1 = cars1.loc[:,'mpg':'car']",
"_____no_output_____"
]
],
[
[
"### Step 5. What is the number of observations in each dataset?",
"_____no_output_____"
]
],
[
[
"cars1.shape[0]\ncars1.head()",
"_____no_output_____"
],
[
"cars2.shape[0]\ncars2.head()",
"_____no_output_____"
]
],
[
[
"### Step 6. Join cars1 and cars2 into a single DataFrame called cars",
"_____no_output_____"
]
],
[
[
"cars =cars1.append(cars2)",
"_____no_output_____"
]
],
[
[
"### Step 7. Oops, there is a column missing, called owners. Create a random number Series from 15,000 to 73,000.",
"_____no_output_____"
]
],
[
[
"owners = np.random.randint(15000,73000, size = 398, dtype='I')\nlen(owners)",
"_____no_output_____"
]
],
[
[
"### Step 8. Add the column owners to cars",
"_____no_output_____"
]
],
[
[
"cars['owners']=owners",
"_____no_output_____"
],
[
"cars",
"_____no_output_____"
],
[
"cars.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 398 entries, 0 to 199\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 mpg 398 non-null float64\n 1 cylinders 398 non-null int64 \n 2 displacement 398 non-null int64 \n 3 horsepower 398 non-null object \n 4 weight 398 non-null int64 \n 5 acceleration 398 non-null float64\n 6 model 398 non-null int64 \n 7 origin 398 non-null int64 \n 8 car 398 non-null object \n 9 owners 398 non-null uint32 \ndtypes: float64(2), int64(5), object(2), uint32(1)\nmemory usage: 32.6+ KB\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec623cb7185dd7d3e90f5977086bfbe959c947d8 | 15,513 | ipynb | Jupyter Notebook | web/home/ipython/examples/r.ipynb | joequant/bitquant-local | 0271a2b1b0f4159612f3cbb165721a5182afed05 | [
"BSD-2-Clause"
]
| null | null | null | web/home/ipython/examples/r.ipynb | joequant/bitquant-local | 0271a2b1b0f4159612f3cbb165721a5182afed05 | [
"BSD-2-Clause"
]
| null | null | null | web/home/ipython/examples/r.ipynb | joequant/bitquant-local | 0271a2b1b0f4159612f3cbb165721a5182afed05 | [
"BSD-2-Clause"
]
| 1 | 2020-06-07T11:58:15.000Z | 2020-06-07T11:58:15.000Z | 23.39819 | 505 | 0.556243 | [
[
[
"# Rmagic Functions Extension",
"_____no_output_____"
]
],
[
[
"%pylab inline",
"_____no_output_____"
]
],
[
[
"## Line magics",
"_____no_output_____"
],
[
"IPython has an `rmagic` extension that contains a some magic functions for working with R via rpy2. This extension can be loaded using the `%load_ext` magic as follows:",
"_____no_output_____"
]
],
[
[
"%load_ext rpy2.ipython ",
"_____no_output_____"
]
],
[
[
"A typical use case one imagines is having some numpy arrays, wanting to compute some statistics of interest on these\n arrays and return the result back to python. Let's suppose we just want to fit a simple linear model to a scatterplot.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pylab\nX = np.array([0,1,2,3,4])\nY = np.array([3,5,4,6,7])\npylab.scatter(X, Y)",
"_____no_output_____"
]
],
[
[
"We can accomplish this by first pushing variables to R, fitting a model and returning the results. The line magic %Rpush copies its arguments to variables of the same name in rpy2. The %R line magic evaluates the string in rpy2 and returns the results. In this case, the coefficients of a linear model.",
"_____no_output_____"
]
],
[
[
"%Rpush X Y\n%R lm(Y~X)$coef",
"_____no_output_____"
]
],
[
[
"We can check that this is correct fairly easily:",
"_____no_output_____"
]
],
[
[
"Xr = X - X.mean(); Yr = Y - Y.mean()\nslope = (Xr*Yr).sum() / (Xr**2).sum()\nintercept = Y.mean() - X.mean() * slope\n(intercept, slope)",
"_____no_output_____"
]
],
[
[
"It is also possible to return more than one value with %R.",
"_____no_output_____"
]
],
[
[
"%R resid(lm(Y~X)); coef(lm(X~Y))\n",
"_____no_output_____"
]
],
[
[
"One can also easily capture the results of %R into python objects. Like R, the return value of this multiline expression (multiline in the sense that it is separated by ';') is the final value, which is \nthe *coef(lm(X~Y))*. To pull other variables from R, there is one more magic.",
"_____no_output_____"
],
[
"There are two more line magics, %Rpull and %Rget. Both are useful after some R code has been executed and there are variables\nin the rpy2 namespace that one would like to retrieve. The main difference is that one\n returns the value (%Rget), while the other pulls it to self.shell.user_ns (%Rpull). Imagine we've stored the results\nof some calculation in the variable \"a\" in rpy2's namespace. By using the %R magic, we can obtain these results and\nstore them in b. We can also pull them directly to user_ns with %Rpull. They are both views on the same data.",
"_____no_output_____"
],
[
"%Rpull is equivalent to calling %R with just -o\n",
"_____no_output_____"
]
],
[
[
"b = %R a=resid(lm(Y~X))\n%Rpull a\nprint a\n%R -o a",
"_____no_output_____"
],
[
"%R d=resid(lm(Y~X)); e=coef(lm(Y~X))\n%R -o d -o e\n%Rpull e\nprint d\nprint e\nimport numpy as np\nnp.testing.assert_almost_equal(d, a)",
"_____no_output_____"
]
],
[
[
"On the other hand %Rpush is equivalent to calling %R with just -i and no trailing code.",
"_____no_output_____"
]
],
[
[
"A = np.arange(20)\n%R -i A\n%R mean(A)\n",
"_____no_output_____"
]
],
[
[
"The magic %Rget retrieves one variable from R.",
"_____no_output_____"
]
],
[
[
"%Rget A",
"_____no_output_____"
]
],
[
[
"## Plotting and capturing output",
"_____no_output_____"
],
[
"R's console (i.e. its stdout() connection) is captured by ipython, as are any plots which are published as PNG files like the notebook with arguments --pylab inline. As a call to %R may produce a return value (see above) we must ask what happens to a magic like the one below. The R code specifies that something is published to the notebook. If anything is published to the notebook, that call to %R returns None.",
"_____no_output_____"
]
],
[
[
"v1 = %R plot(X,Y); print(summary(lm(Y~X))); vv=mean(X)*mean(Y)\nprint 'v1 is:', v1\nv2 = %R mean(X)*mean(Y)\nprint 'v2 is:', v2",
"_____no_output_____"
]
],
[
[
"## What value is returned from %R?",
"_____no_output_____"
],
[
"Some calls have no particularly interesting return value, the magic %R will not return anything in this case. The return value in rpy2 is actually NULL so %R returns None.",
"_____no_output_____"
]
],
[
[
"v = %R plot(X,Y)\nassert v == None",
"_____no_output_____"
]
],
[
[
"Also, if the return value of a call to %R (in line mode) has just been printed to the console, then its value is also not returned.",
"_____no_output_____"
]
],
[
[
"v = %R print(X)\nassert v == None",
"_____no_output_____"
]
],
[
[
"But, if the last value did not print anything to console, the value is returned:\n",
"_____no_output_____"
]
],
[
[
"v = %R print(summary(X)); X\nprint 'v:', v",
"_____no_output_____"
]
],
[
[
"The return value can be suppressed by a trailing ';' or an -n argument.\n",
"_____no_output_____"
]
],
[
[
"%R -n X",
"_____no_output_____"
],
[
"%R X; ",
"_____no_output_____"
]
],
[
[
"## Cell level magic",
"_____no_output_____"
],
[
"Often, we will want to do more than a simple linear regression model. There may be several lines of R code that we want to \nuse before returning to python. This is the cell-level magic.\n\n\nFor the cell level magic, inputs can be passed via the -i or --inputs argument in the line. These variables are copied \nfrom the shell namespace to R's namespace using rpy2.robjects.r.assign. It would be nice not to have to copy these into R: rnumpy ( http://bitbucket.org/njs/rnumpy/wiki/API ) has done some work to limit or at least make transparent the number of copies of an array. This seems like a natural thing to try to build on. Arrays can be output from R via the -o or --outputs argument in the line. All other arguments are sent to R's png function, which is the graphics device used to create the plots.\n\nWe can redo the above calculations in one ipython cell. We might also want to add some output such as a summary\n from R or perhaps the standard plotting diagnostics of the lm.",
"_____no_output_____"
]
],
[
[
"%%R -i X,Y -o XYcoef\nXYlm = lm(Y~X)\nXYcoef = coef(XYlm)\nprint(summary(XYlm))\npar(mfrow=c(2,2))\nplot(XYlm)",
"_____no_output_____"
]
],
[
[
"## Passing data back and forth",
"_____no_output_____"
],
[
"Currently, data is passed through RMagics.pyconverter when going from python to R and RMagics.Rconverter when \ngoing from R to python. These currently default to numpy.ndarray. Future work will involve writing better converters, most likely involving integration with http://pandas.sourceforge.net.\n\nPassing ndarrays into R seems to require a copy, though once an object is returned to python, this object is NOT copied, and it is possible to change its values.\n",
"_____no_output_____"
]
],
[
[
"seq1 = np.arange(10)",
"_____no_output_____"
],
[
"%%R -i seq1 -o seq2\nseq2 = rep(seq1, 2)\nprint(seq2)",
"_____no_output_____"
],
[
"seq2[::2] = 0\nseq2",
"_____no_output_____"
],
[
"%%R\nprint(seq2)",
"_____no_output_____"
]
],
[
[
"Once the array data has been passed to R, modifring its contents does not modify R's copy of the data.",
"_____no_output_____"
]
],
[
[
"seq1[0] = 200\n%R print(seq1)",
"_____no_output_____"
]
],
[
[
"But, if we pass data as both input and output, then the value of \"data\" in user_ns will be overwritten and the\nnew array will be a view of the data in R's copy.",
"_____no_output_____"
]
],
[
[
"print seq1\n%R -i seq1 -o seq1\nprint seq1\nseq1[0] = 200\n%R print(seq1)\nseq1_view = %R seq1\nassert(id(seq1_view.data) == id(seq1.data))",
"_____no_output_____"
]
],
[
[
"## Exception handling",
"_____no_output_____"
],
[
"Exceptions are handled by passing back rpy2's exception and the line that triggered it.",
"_____no_output_____"
]
],
[
[
"try:\n %R -n nosuchvar\nexcept Exception as e:\n print e.message\n pass",
"_____no_output_____"
]
],
[
[
"## Structured arrays and data frames",
"_____no_output_____"
],
[
"In R, data frames play an important role as they allow array-like objects of mixed type with column names (and row names). In bumpy, the closest analogy is a structured array with named fields. In future work, it would be nice to use pandas to return full-fledged DataFrames from rpy2. In the mean time, structured arrays can be passed back and forth with the -d flag to %R, %Rpull, and %Rget",
"_____no_output_____"
]
],
[
[
"datapy= np.array([(1, 2.9, 'a'), (2, 3.5, 'b'), (3, 2.1, 'c')],\n dtype=[('x', '<i4'), ('y', '<f8'), ('z', '|S1')])\n",
"_____no_output_____"
],
[
"%%R -i datapy -d datar\ndatar = datapy",
"_____no_output_____"
],
[
"datar",
"_____no_output_____"
],
[
"%R datar2 = datapy\n%Rpull -d datar2\ndatar2",
"_____no_output_____"
],
[
"%Rget -d datar2",
"_____no_output_____"
]
],
[
[
"For arrays without names, the -d argument has no effect because the R object has no colnames or names.",
"_____no_output_____"
]
],
[
[
"Z = np.arange(6)\n%R -i Z\n%Rget -d Z",
"_____no_output_____"
]
],
[
[
"For mixed-type data frames in R, if the -d flag is not used, then an array of a single type is returned and\nits value is transposed. This would be nice to fix, but it seems something that should be fixed at the rpy2 level (See: https://bitbucket.org/lgautier/rpy2/issue/44/numpyrecarray-as-dataframe)",
"_____no_output_____"
]
],
[
[
"%Rget datar2",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec623eb9efe1a88f01f7da786589bcf7ae33bd42 | 140,220 | ipynb | Jupyter Notebook | analysis/data/SI_FigS1_code.ipynb | Peyara/Evolution-Counterdiabatic-Driving | e695fad703b2d339bed0013e5b4254ba2365c105 | [
"MIT"
]
| 3 | 2020-08-24T20:24:41.000Z | 2020-08-26T02:16:16.000Z | analysis/data/SI_FigS1_code.ipynb | Peyara/Evolution-Counterdiabatic-Driving | e695fad703b2d339bed0013e5b4254ba2365c105 | [
"MIT"
]
| null | null | null | analysis/data/SI_FigS1_code.ipynb | Peyara/Evolution-Counterdiabatic-Driving | e695fad703b2d339bed0013e5b4254ba2365c105 | [
"MIT"
]
| null | null | null | 141.350806 | 98,163 | 0.814834 | [
[
[
"Code to generate SI Fig. S1",
"_____no_output_____"
]
],
[
[
"from pylab import *\nfrom matplotlib import rc\nimport matplotlib.gridspec as gridspec\nimport numpy as np\n\nfrom matplotlib.font_manager import FontProperties\nimport matplotlib.colors as colors\n\nimport matplotlib.cm as cm\nfrom matplotlib import patches\n\nfrom matplotlib.collections import LineCollection\nfrom matplotlib.colors import ListedColormap, BoundaryNorm\n\nfrom mpl_toolkits.axes_grid1.inset_locator import inset_axes",
"_____no_output_____"
],
[
"%matplotlib notebook\nclf()\n\nxoff=0.2\nyoff=0.2\nwidth=0.85-xoff\nheight=0.85-yoff\n\ndashes=[3,2]\nlabelx=-0.11\n\n##################\nfig_width_pt = 246.0\ninches_per_pt = 1.0/72.27\ngolden_mean = (sqrt(5)-1.0)/2.0\nfig_width = fig_width_pt*inches_per_pt\nfig_height = fig_width_pt*inches_per_pt*golden_mean\nfig_size = [1.5*fig_width,3.5*fig_height]\nparams = {'backend': 'pdf',\n 'axes.labelsize': 9,\n #'text.fontsize': 9,\n 'xtick.labelsize': 9,\n 'ytick.labelsize': 9,\n 'text.usetex': False,\n 'figure.figsize': fig_size,\n 'figure.subplot.left': xoff,\n 'figure.subplot.bottom': yoff,\n 'figure.subplot.right': xoff+width,\n 'figure.subplot.top': yoff+height,\n 'mathtext.fontset': 'stixsans'}\n\nrcParams.update(params)\nrc('font',**{'family':'sans-serif','sans-serif':['Tex Gyre Heros']})\n \nfigure(1)\n\ngs1=gridspec.GridSpec(1,1)\ngs1.update(hspace=0.0,wspace=0,bottom=0.6)\n\ngs2=gridspec.GridSpec(1,1)\ngs2.update(hspace=0.0,wspace=0.0,top=0.6)\n\ncols={\"teal\": '#469990',\"cyan\" : '#42d4f4',\"green\" : '#3cb44b', \"red\" : '#e6194B', \"yellow\" : '#ffe119', \"blue\" : '#4363d8', \"orange\" : '#f58231', \"pink\" : '#fabebe', \"lavender\" : '#e6beff', \"maroon\" : '#800000', \"navy\" : '#000075', \"grey\" : '#a9a9a9', \"black\" : '#000000'}\nylpos=-0.12\n\n############################################3\n \nsubplot(gs1[0,0])\n\nnval=np.array(np.loadtxt('Ntab.txt',delimiter='\\t'))\n\nplot([e[0] for e in nval],[e[1] for e in nval],lw=1.25,color=cols[\"red\"])\nplot([e[0] for e in nval],[e[2] for e in nval],lw=1.25,color=cols[\"orange\"])\nplot([e[0] for e in nval],[e[3] for e in nval],lw=1.25,color=cols[\"green\"])\nplot([e[0] for e in nval],[e[4] for e in nval],lw=1.25,color=cols[\"blue\"])\nplot([e[0] for e in nval],[e[5] for e in nval],lw=1.25,color='Purple')\n\n\ngca().set_yscale('log')\nsetp(gca(),xlim=[0,300],ylim=[1e3,1.5e5],xticklabels=[])\nylabel(r'Population $N(t\\,)$')\ngca().yaxis.set_label_coords(ylpos,0.5)\ngca().tick_params(axis='x',direction='in')\n\n#leg=legend([lorig,hs,leq],[r'$p\\,(x_1,t\\,)$ theory',r'$p\\,(x_1,t\\,)$ simulations',r'$\\rho\\,(x_1;\\lambda(t))$'],handlelength=2.5,loc=(0.02,0.4),numpoints=3,prop = FontProperties(size=9),ncol=1,labelspacing=0.35,handletextpad=0.5)\n#leg.draw_frame(0)\n\n############################################3\n \nsubplot(gs2[0,0])\n\nBm40=np.array(np.loadtxt('klcdBm40.txt',delimiter='\\t'))\nBm40no=np.array(np.loadtxt('klnocdBm40.txt',delimiter='\\t'))\n\nBm20=np.array(np.loadtxt('klcdBm20.txt',delimiter='\\t'))\nBm20no=np.array(np.loadtxt('klnocdBm20.txt',delimiter='\\t'))\n\nBm0=np.array(np.loadtxt('klcd.txt',delimiter='\\t'))\nBm0no=np.array(np.loadtxt('klnocd.txt',delimiter='\\t'))\n\nB200=np.array(np.loadtxt('klcdB200.txt',delimiter='\\t'))\nB200no=np.array(np.loadtxt('klnocdB200.txt',delimiter='\\t'))\n\nB450=np.array(np.loadtxt('klcdB450.txt',delimiter='\\t'))\nB450no=np.array(np.loadtxt('klnocdB450.txt',delimiter='\\t'))\n\nplot([e[0] for e in Bm40],[e[1] for e in Bm40],lw=1.25,color=cols[\"red\"])\nll,=plot([e[0] for e in Bm40no],[e[1] for e in Bm40no],lw=1.25,color=cols[\"red\"])\nll.set_dashes([2,2])\n\nplot([e[0] for e in Bm20],[e[1] for e in Bm20],lw=1.25,color=cols[\"orange\"])\nll,=plot([e[0] for e in Bm20no],[e[1] for e in Bm20no],lw=1.25,color=cols[\"orange\"])\nll.set_dashes([2,2])\n\nplot([e[0] for e in Bm0],[e[1] for e in Bm0],lw=1.25,color=cols[\"green\"])\nll,=plot([e[0] for e in Bm0no],[e[1] for e in Bm0no],lw=1.25,color=cols[\"green\"])\nll.set_dashes([2,2])\n\nplot([e[0] for e in B200],[e[1] for e in B200],lw=1.25,color=cols[\"blue\"])\nll,=plot([e[0] for e in B200no],[e[1] for e in B200no],lw=1.25,color=cols[\"blue\"])\nll.set_dashes([2,2])\n\nplot([e[0] for e in B450],[e[1] for e in B450],lw=1.25,color='Purple')\nll,=plot([e[0] for e in B450no],[e[1] for e in B450no],lw=1.25,color='Purple')\nll.set_dashes([2,2])\n\n\ngca().set_yscale('log')\nsetp(gca(),xlim=[0,300],ylim=[1e-4,95])\nxlabel(r'Time [generations]')\nylabel(r'$D_{\\rm KL}(\\rho\\,||\\,p)$ [bits]')\ngca().yaxis.set_label_coords(ylpos,0.5)\n\n\n\n############################################3\n\nfigtext(0.22,0.82,'A',fontsize=12,fontweight='bold')\nfigtext(0.22,0.57,'B',fontsize=12,fontweight='bold')\n\nfigtext(0.645,0.613,r'$\\zeta = -40$',fontsize=9,color=cols[\"red\"])\nfigtext(0.645,0.668,r'$\\zeta = -20$',fontsize=9,color=cols[\"orange\"])\nfigtext(0.645,0.724,r'$\\zeta = 0$',fontsize=9,color=cols[\"green\"])\nfigtext(0.645,0.773,r'$\\zeta = 200$',fontsize=9,color=cols[\"blue\"])\nfigtext(0.645,0.808,r'$\\zeta = 450$',fontsize=9,color='Purple')\n\n\nshow()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
]
]
|
ec625289a23e7d6abc92b758c49955f1ff0e392e | 94,209 | ipynb | Jupyter Notebook | Chapter5_Exercise2_Stock_Trading.ipynb | packkkk/data-science-and-machine-learning-fundamentals | 801cdbf226749fa02b49bc6ff843ee8d06760c69 | [
"MIT"
]
| null | null | null | Chapter5_Exercise2_Stock_Trading.ipynb | packkkk/data-science-and-machine-learning-fundamentals | 801cdbf226749fa02b49bc6ff843ee8d06760c69 | [
"MIT"
]
| null | null | null | Chapter5_Exercise2_Stock_Trading.ipynb | packkkk/data-science-and-machine-learning-fundamentals | 801cdbf226749fa02b49bc6ff843ee8d06760c69 | [
"MIT"
]
| null | null | null | 29.175906 | 162 | 0.360634 | [
[
[
"# Chapter 5 - Exercise 2: Giao dịch chứng khoán",
"_____no_output_____"
],
[
"##### Cho 3 file .csv sau:\n* **stocks1.csv :** *date*, *symbol*, *open*, *high*, *low*, *close*, *volume* : chứa thông tin giao dịch chứng khoán các công ty khác nhau\n* **stocks2.csv :** *date*, *symbol*, *open*, *high*, *low*, *close*, *volume* : chứa thông tin giao dịch chứng khoán các công ty khác nhau\n* **companies.csv :** *name*, *employees*, *headquarters_city*, *headquarters_state* : chứa thông tin về trụ sở và số lượng nhân viên cho một công ty cụ thể",
"_____no_output_____"
],
[
"#### Thực hiện các yêu cầu sau, và đối chiếu với kết quả cho trước:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Câu 1a: Đọc file stocks1.csv => đưa dữ liệu vào stocks1\nstocks1 = pd.read_csv('stock_trading_data/stocks1.csv')\n# Hiển thị 5 dòng dữ liệu đầu của stocks1\nstocks1.head(5)",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>0</th>\n <td>01-03-19</td>\n <td>AMZN</td>\n <td>1655.13</td>\n <td>1674.26</td>\n <td>1651.00</td>\n <td>1671.73</td>\n <td>4974877</td>\n </tr>\n <tr>\n <th>1</th>\n <td>04-03-19</td>\n <td>AMZN</td>\n <td>1685.00</td>\n <td>1709.43</td>\n <td>1674.36</td>\n <td>1696.17</td>\n <td>6167358</td>\n </tr>\n <tr>\n <th>2</th>\n <td>05-03-19</td>\n <td>AMZN</td>\n <td>1702.95</td>\n <td>1707.80</td>\n <td>1689.01</td>\n <td>1692.43</td>\n <td>3681522</td>\n </tr>\n <tr>\n <th>3</th>\n <td>06-03-19</td>\n <td>AMZN</td>\n <td>1695.97</td>\n <td>NaN</td>\n <td>NaN</td>\n <td>1668.95</td>\n <td>3996001</td>\n </tr>\n <tr>\n <th>4</th>\n <td>07-03-19</td>\n <td>AMZN</td>\n <td>1667.37</td>\n <td>1669.75</td>\n <td>1620.51</td>\n <td>1625.95</td>\n <td>4957017</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Hiển thị 5 dòng dữ liệu cuối của stocks1\nstocks1.tail(5)",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>10</th>\n <td>01-03-19</td>\n <td>GOOG</td>\n <td>1124.90</td>\n <td>1142.97</td>\n <td>1124.75</td>\n <td>1140.99</td>\n <td>1450316</td>\n </tr>\n <tr>\n <th>11</th>\n <td>04-03-19</td>\n <td>GOOG</td>\n <td>1146.99</td>\n <td>1158.28</td>\n <td>1130.69</td>\n <td>1147.80</td>\n <td>1446047</td>\n </tr>\n <tr>\n <th>12</th>\n <td>05-03-19</td>\n <td>GOOG</td>\n <td>1150.06</td>\n <td>NaN</td>\n <td>NaN</td>\n <td>1162.03</td>\n <td>1443174</td>\n </tr>\n <tr>\n <th>13</th>\n <td>06-03-19</td>\n <td>GOOG</td>\n <td>1162.49</td>\n <td>1167.57</td>\n <td>1155.49</td>\n <td>1157.86</td>\n <td>1099289</td>\n </tr>\n <tr>\n <th>14</th>\n <td>07-03-19</td>\n <td>GOOG</td>\n <td>1155.72</td>\n <td>1156.76</td>\n <td>1134.91</td>\n <td>1143.30</td>\n <td>1166559</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Cho biết kiểu dữ liệu (dtype) của các cột của stocks1\nprint(stocks1.dtypes)",
"date object\nsymbol object\nopen float64\nhigh float64\nlow float64\nclose float64\nvolume int64\ndtype: object\n"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <pre>date object\nsymbol object\nopen float64\nhigh float64\nlow float64\nclose float64\nvolume int64\ndtype: object</pre>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Xem thông tin (info) của stocks1\nprint(stocks1.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 15 entries, 0 to 14\nData columns (total 7 columns):\ndate 15 non-null object\nsymbol 15 non-null object\nopen 15 non-null float64\nhigh 13 non-null float64\nlow 13 non-null float64\nclose 15 non-null float64\nvolume 15 non-null int64\ndtypes: float64(4), int64(1), object(2)\nmemory usage: 920.0+ bytes\nNone\n"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <pre><class 'pandas.core.frame.DataFrame'>\nRangeIndex: 15 entries, 0 to 14\nData columns (total 7 columns):\ndate 15 non-null object\nsymbol 15 non-null object\nopen 15 non-null float64\nhigh 13 non-null float64\nlow 13 non-null float64\nclose 15 non-null float64\nvolume 15 non-null int64\ndtypes: float64(4), int64(1), object(2)\nmemory usage: 968.0+ bytes\n</pre>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 1b: Đọc file stocks2.csv => đưa dữ liệu vào stocks2\nstocks2 = pd.read_csv('stock_trading_data/stocks2.csv')\n# Hiển thị 5 dòng dữ liệu đầu của stocks2\nstocks2.head(5)",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>0</th>\n <td>01-03-19</td>\n <td>FB</td>\n <td>162.60</td>\n <td>163.132</td>\n <td>161.69</td>\n <td>162.28</td>\n <td>11097770</td>\n </tr>\n <tr>\n <th>1</th>\n <td>04-03-19</td>\n <td>FB</td>\n <td>163.90</td>\n <td>167.500</td>\n <td>163.83</td>\n <td>167.37</td>\n <td>18894689</td>\n </tr>\n <tr>\n <th>2</th>\n <td>05-03-19</td>\n <td>FB</td>\n <td>167.37</td>\n <td>171.880</td>\n <td>166.55</td>\n <td>171.26</td>\n <td>28187890</td>\n </tr>\n <tr>\n <th>3</th>\n <td>06-03-19</td>\n <td>FB</td>\n <td>172.90</td>\n <td>173.570</td>\n <td>171.27</td>\n <td>172.51</td>\n <td>21531723</td>\n </tr>\n <tr>\n <th>4</th>\n <td>07-03-19</td>\n <td>FB</td>\n <td>171.50</td>\n <td>171.740</td>\n <td>167.61</td>\n <td>169.13</td>\n <td>18306504</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Hiển thị 5 dòng dữ liệu cuối của stocks2\nstocks2.tail(5)",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>5</th>\n <td>01-03-19</td>\n <td>TSLA</td>\n <td>306.94</td>\n <td>307.1300</td>\n <td>291.90</td>\n <td>294.79</td>\n <td>22911375</td>\n </tr>\n <tr>\n <th>6</th>\n <td>04-03-19</td>\n <td>TSLA</td>\n <td>298.12</td>\n <td>299.0000</td>\n <td>282.78</td>\n <td>285.36</td>\n <td>17096818</td>\n </tr>\n <tr>\n <th>7</th>\n <td>05-03-19</td>\n <td>TSLA</td>\n <td>282.00</td>\n <td>284.0000</td>\n <td>270.10</td>\n <td>276.54</td>\n <td>18764740</td>\n </tr>\n <tr>\n <th>8</th>\n <td>06-03-19</td>\n <td>TSLA</td>\n <td>276.48</td>\n <td>281.5058</td>\n <td>274.39</td>\n <td>276.24</td>\n <td>10335485</td>\n </tr>\n <tr>\n <th>9</th>\n <td>07-03-19</td>\n <td>TSLA</td>\n <td>278.84</td>\n <td>284.7000</td>\n <td>274.25</td>\n <td>276.59</td>\n <td>9442483</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Cho biết kiểu dữ liệu (dtype) của các cột của stocks2\nprint(stocks2.dtypes)",
"date object\nsymbol object\nopen float64\nhigh float64\nlow float64\nclose float64\nvolume int64\ndtype: object\n"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <pre>date object\nsymbol object\nopen float64\nhigh float64\nlow float64\nclose float64\nvolume int64\ndtype: object</pre>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Xem thông tin (info) của stocks2\nprint(stocks2.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10 entries, 0 to 9\nData columns (total 7 columns):\ndate 10 non-null object\nsymbol 10 non-null object\nopen 10 non-null float64\nhigh 10 non-null float64\nlow 10 non-null float64\nclose 10 non-null float64\nvolume 10 non-null int64\ndtypes: float64(4), int64(1), object(2)\nmemory usage: 640.0+ bytes\nNone\n"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <pre><class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10 entries, 0 to 9\nData columns (total 7 columns):\ndate 10 non-null object\nsymbol 10 non-null object\nopen 10 non-null float64\nhigh 10 non-null float64\nlow 10 non-null float64\nclose 10 non-null float64\nvolume 10 non-null int64\ndtypes: float64(4), int64(1), object(2)\nmemory usage: 688.0+ bytes\n</pre>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 1c: Đọc file companies.csv => đưa dữ liệu vào companies\ncompanies = pd.read_csv('stock_trading_data/companies.csv')\n# Xem dữ liệu của companies\ncompanies",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>name</th>\n <th>employees</th>\n <th>headquarters_city</th>\n <th>headquarters_state</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>0</th>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n </tr>\n <tr>\n <th>1</th>\n <td>GOOG</td>\n <td>98771</td>\n <td>Mountain View</td>\n <td>CA</td>\n </tr>\n <tr>\n <th>2</th>\n <td>AAPL</td>\n <td>132000</td>\n <td>Cupertino</td>\n <td>CA</td>\n </tr>\n <tr>\n <th>3</th>\n <td>FB</td>\n <td>48268</td>\n <td>Menlo Park</td>\n <td>CA</td>\n </tr>\n <tr>\n <th>4</th>\n <td>TSLA</td>\n <td>48016</td>\n <td>Palo Alto</td>\n <td>CA</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Cho biết kiểu dữ liệu (dtype) của các cột của companies\nprint(companies.dtypes)",
"name object\nemployees int64\nheadquarters_city object\nheadquarters_state object\ndtype: object\n"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <pre>name object\nemployees int64\nheadquarters_city object\nheadquarters_state object\ndtype: object</pre>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Xem thông tin (info) của companies\nprint(companies.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5 entries, 0 to 4\nData columns (total 4 columns):\nname 5 non-null object\nemployees 5 non-null int64\nheadquarters_city 5 non-null object\nheadquarters_state 5 non-null object\ndtypes: int64(1), object(3)\nmemory usage: 240.0+ bytes\nNone\n"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <pre><class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5 entries, 0 to 4\nData columns (total 4 columns):\nname 5 non-null object\nemployees 5 non-null int64\nheadquarters_city 5 non-null object\nheadquarters_state 5 non-null object\ndtypes: int64(1), object(3)\nmemory usage: 288.0+ bytes\n</pre>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 2: Cho biết trong stocks1 có dữ liệu Null hay không? \nstocks1.isnull().any()",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <pre>date False\nsymbol False\nopen False\nhigh True\nlow True\nclose False\nvolume False\ndtype: bool</pre>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Nếu có, hãy thay thế với quy tắc sau:\n# Nếu Null cột 'high' thì thay bằng giá trị max trên cột 'high' của mã chứng khoán đó\nstocks1.high.fillna(stocks1.high.max(), inplace=True)\n# Nếu Null cột 'low' thì thay bằng giá trị min trên cột 'low' của mã chứng khoán đó\nstocks1.low.fillna(stocks1.low.min(), inplace=True)\nstocks1",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>0</th>\n <td>01-03-19</td>\n <td>AMZN</td>\n <td>1655.13</td>\n <td>1674.26</td>\n <td>1651.00</td>\n <td>1671.73</td>\n <td>4974877</td>\n </tr>\n <tr>\n <th>1</th>\n <td>04-03-19</td>\n <td>AMZN</td>\n <td>1685.00</td>\n <td>1709.43</td>\n <td>1674.36</td>\n <td>1696.17</td>\n <td>6167358</td>\n </tr>\n <tr>\n <th>2</th>\n <td>05-03-19</td>\n <td>AMZN</td>\n <td>1702.95</td>\n <td>1707.80</td>\n <td>1689.01</td>\n <td>1692.43</td>\n <td>3681522</td>\n </tr>\n <tr>\n <th>3</th>\n <td>06-03-19</td>\n <td>AMZN</td>\n <td>1695.97</td>\n <td>1709.43</td>\n <td>1620.51</td>\n <td>1668.95</td>\n <td>3996001</td>\n </tr>\n <tr>\n <th>4</th>\n <td>07-03-19</td>\n <td>AMZN</td>\n <td>1667.37</td>\n <td>1669.75</td>\n <td>1620.51</td>\n <td>1625.95</td>\n <td>4957017</td>\n </tr>\n <tr>\n <th>5</th>\n <td>01-03-19</td>\n <td>AAPL</td>\n <td>174.28</td>\n <td>175.15</td>\n <td>172.89</td>\n <td>174.97</td>\n <td>25886167</td>\n </tr>\n <tr>\n <th>6</th>\n <td>04-03-19</td>\n <td>AAPL</td>\n <td>175.69</td>\n <td>177.75</td>\n <td>173.97</td>\n <td>175.85</td>\n <td>27436203</td>\n </tr>\n <tr>\n <th>7</th>\n <td>05-03-19</td>\n <td>AAPL</td>\n <td>175.94</td>\n <td>176.00</td>\n <td>174.54</td>\n <td>175.53</td>\n <td>19737419</td>\n </tr>\n <tr>\n <th>8</th>\n <td>06-03-19</td>\n <td>AAPL</td>\n <td>174.67</td>\n <td>175.49</td>\n <td>173.94</td>\n <td>174.52</td>\n <td>20810384</td>\n </tr>\n <tr>\n <th>9</th>\n <td>07-03-19</td>\n <td>AAPL</td>\n <td>173.87</td>\n <td>174.44</td>\n <td>172.02</td>\n <td>172.50</td>\n <td>24796374</td>\n </tr>\n <tr>\n <th>10</th>\n <td>01-03-19</td>\n <td>GOOG</td>\n <td>1124.90</td>\n <td>1142.97</td>\n <td>1124.75</td>\n <td>1140.99</td>\n <td>1450316</td>\n </tr>\n <tr>\n <th>11</th>\n <td>04-03-19</td>\n <td>GOOG</td>\n <td>1146.99</td>\n <td>1158.28</td>\n <td>1130.69</td>\n <td>1147.80</td>\n <td>1446047</td>\n </tr>\n <tr>\n <th>12</th>\n <td>05-03-19</td>\n <td>GOOG</td>\n <td>1150.06</td>\n <td>1167.57</td>\n <td>1124.75</td>\n <td>1162.03</td>\n <td>1443174</td>\n </tr>\n <tr>\n <th>13</th>\n <td>06-03-19</td>\n <td>GOOG</td>\n <td>1162.49</td>\n <td>1167.57</td>\n <td>1155.49</td>\n <td>1157.86</td>\n <td>1099289</td>\n </tr>\n <tr>\n <th>14</th>\n <td>07-03-19</td>\n <td>GOOG</td>\n <td>1155.72</td>\n <td>1156.76</td>\n <td>1134.91</td>\n <td>1143.30</td>\n <td>1166559</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 3: Tạo dataframe stocks bằng cách gộp stocks1 và stocks2 theo dòng\n# stocks = pd.merge(stocks1, stocks2, how='inner')\nstocks = pd.merge(left=stocks1, right=stocks2, how='outer')\n# Xem 15 dòng dữ liệu cuối của stocks\nstocks.tail(15)",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>10</th>\n <td>01-03-19</td>\n <td>GOOG</td>\n <td>1124.90</td>\n <td>1142.9700</td>\n <td>1124.75</td>\n <td>1140.99</td>\n <td>1450316</td>\n </tr>\n <tr>\n <th>11</th>\n <td>04-03-19</td>\n <td>GOOG</td>\n <td>1146.99</td>\n <td>1158.2800</td>\n <td>1130.69</td>\n <td>1147.80</td>\n <td>1446047</td>\n </tr>\n <tr>\n <th>12</th>\n <td>05-03-19</td>\n <td>GOOG</td>\n <td>1150.06</td>\n <td>1167.5700</td>\n <td>1124.75</td>\n <td>1162.03</td>\n <td>1443174</td>\n </tr>\n <tr>\n <th>13</th>\n <td>06-03-19</td>\n <td>GOOG</td>\n <td>1162.49</td>\n <td>1167.5700</td>\n <td>1155.49</td>\n <td>1157.86</td>\n <td>1099289</td>\n </tr>\n <tr>\n <th>14</th>\n <td>07-03-19</td>\n <td>GOOG</td>\n <td>1155.72</td>\n <td>1156.7600</td>\n <td>1134.91</td>\n <td>1143.30</td>\n <td>1166559</td>\n </tr>\n <tr>\n <th>15</th>\n <td>01-03-19</td>\n <td>FB</td>\n <td>162.60</td>\n <td>163.1320</td>\n <td>161.69</td>\n <td>162.28</td>\n <td>11097770</td>\n </tr>\n <tr>\n <th>16</th>\n <td>04-03-19</td>\n <td>FB</td>\n <td>163.90</td>\n <td>167.5000</td>\n <td>163.83</td>\n <td>167.37</td>\n <td>18894689</td>\n </tr>\n <tr>\n <th>17</th>\n <td>05-03-19</td>\n <td>FB</td>\n <td>167.37</td>\n <td>171.8800</td>\n <td>166.55</td>\n <td>171.26</td>\n <td>28187890</td>\n </tr>\n <tr>\n <th>18</th>\n <td>06-03-19</td>\n <td>FB</td>\n <td>172.90</td>\n <td>173.5700</td>\n <td>171.27</td>\n <td>172.51</td>\n <td>21531723</td>\n </tr>\n <tr>\n <th>19</th>\n <td>07-03-19</td>\n <td>FB</td>\n <td>171.50</td>\n <td>171.7400</td>\n <td>167.61</td>\n <td>169.13</td>\n <td>18306504</td>\n </tr>\n <tr>\n <th>20</th>\n <td>01-03-19</td>\n <td>TSLA</td>\n <td>306.94</td>\n <td>307.1300</td>\n <td>291.90</td>\n <td>294.79</td>\n <td>22911375</td>\n </tr>\n <tr>\n <th>21</th>\n <td>04-03-19</td>\n <td>TSLA</td>\n <td>298.12</td>\n <td>299.0000</td>\n <td>282.78</td>\n <td>285.36</td>\n <td>17096818</td>\n </tr>\n <tr>\n <th>22</th>\n <td>05-03-19</td>\n <td>TSLA</td>\n <td>282.00</td>\n <td>284.0000</td>\n <td>270.10</td>\n <td>276.54</td>\n <td>18764740</td>\n </tr>\n <tr>\n <th>23</th>\n <td>06-03-19</td>\n <td>TSLA</td>\n <td>276.48</td>\n <td>281.5058</td>\n <td>274.39</td>\n <td>276.24</td>\n <td>10335485</td>\n </tr>\n <tr>\n <th>24</th>\n <td>07-03-19</td>\n <td>TSLA</td>\n <td>278.84</td>\n <td>284.7000</td>\n <td>274.25</td>\n <td>276.59</td>\n <td>9442483</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 4: Tạo dataframe stocks_companies bằng cách gộp stocks và companies \nstock_companies = pd.merge(stocks, companies, left_on='symbol', right_on='name')\n# Xem 5 dòng dữ liệu đầu của stocks_companies\nstock_companies.head(5)",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n <th>name</th>\n <th>employees</th>\n <th>headquarters_city</th>\n <th>headquarters_state</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>0</th>\n <td>01-03-19</td>\n <td>AMZN</td>\n <td>1655.13</td>\n <td>1674.26</td>\n <td>1651.00</td>\n <td>1671.73</td>\n <td>4974877</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n </tr>\n <tr>\n <th>1</th>\n <td>04-03-19</td>\n <td>AMZN</td>\n <td>1685.00</td>\n <td>1709.43</td>\n <td>1674.36</td>\n <td>1696.17</td>\n <td>6167358</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n </tr>\n <tr>\n <th>2</th>\n <td>05-03-19</td>\n <td>AMZN</td>\n <td>1702.95</td>\n <td>1707.80</td>\n <td>1689.01</td>\n <td>1692.43</td>\n <td>3681522</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n </tr>\n <tr>\n <th>3</th>\n <td>06-03-19</td>\n <td>AMZN</td>\n <td>1695.97</td>\n <td>1709.43</td>\n <td>1620.51</td>\n <td>1668.95</td>\n <td>3996001</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n </tr>\n <tr>\n <th>4</th>\n <td>07-03-19</td>\n <td>AMZN</td>\n <td>1667.37</td>\n <td>1669.75</td>\n <td>1620.51</td>\n <td>1625.95</td>\n <td>4957017</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 5: Cho biết giá (open, high, low, close) trung bình và volume trung bình của mỗi công ty\nstock_companies.groupby('symbol').agg(['mean'])",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n </tr>\n <tr>\n <th>symbol</th>\n <th></th>\n <th></th>\n <th></th>\n <th></th>\n <th></th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>AAPL</th>\n <td>174.890</td>\n <td>175.76600</td>\n <td>173.472</td>\n <td>174.674</td>\n <td>23733309.4</td>\n </tr>\n <tr>\n <th>AMZN</th>\n <td>1681.284</td>\n <td>1694.13400</td>\n <td>1651.078</td>\n <td>1671.046</td>\n <td>4755355.0</td>\n </tr>\n <tr>\n <th>FB</th>\n <td>167.654</td>\n <td>169.56440</td>\n <td>166.190</td>\n <td>168.510</td>\n <td>19603715.2</td>\n </tr>\n <tr>\n <th>GOOG</th>\n <td>1148.032</td>\n <td>1158.63000</td>\n <td>1134.118</td>\n <td>1150.396</td>\n <td>1321077.0</td>\n </tr>\n <tr>\n <th>TSLA</th>\n <td>288.476</td>\n <td>291.26716</td>\n <td>278.684</td>\n <td>281.904</td>\n <td>15710180.2</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 6: Cho biết giá đóng cửa (close) trung bình, lớn nhất và nhỏ nhất ở mỗi công ty\nstock_companies.groupby('symbol').close.agg(['mean', 'max', 'min'])",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>mean</th>\n <th>min</th>\n <th>max</th>\n </tr>\n <tr>\n <th>symbol</th>\n <th></th>\n <th></th>\n <th></th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>AAPL</th>\n <td>174.674</td>\n <td>172.50</td>\n <td>175.85</td>\n </tr>\n <tr>\n <th>AMZN</th>\n <td>1671.046</td>\n <td>1625.95</td>\n <td>1696.17</td>\n </tr>\n <tr>\n <th>FB</th>\n <td>168.510</td>\n <td>162.28</td>\n <td>172.51</td>\n </tr>\n <tr>\n <th>GOOG</th>\n <td>1150.396</td>\n <td>1140.99</td>\n <td>1162.03</td>\n </tr>\n <tr>\n <th>TSLA</th>\n <td>281.904</td>\n <td>276.24</td>\n <td>294.79</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 7: Tạo cột parsed_time trong stocks_companies bằng cách đổi thời gian sang định dạng DateTime\nstock_companies['parsed_time'] = pd.to_datetime(stock_companies['date'])\n# Cho biết kiểu dữ liệu của cột parsed_time\nprint(stock_companies.parsed_time.dtype)\n# Hiển thị 5 dòng dữ liệu đầu của stocks_companies\nstock_companies.head(5)",
"datetime64[ns]\n"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n <th>name</th>\n <th>employees</th>\n <th>headquarters_city</th>\n <th>headquarters_state</th>\n <th>parsed_time</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>0</th>\n <td>01-03-19</td>\n <td>AMZN</td>\n <td>1655.13</td>\n <td>1674.26</td>\n <td>1651.00</td>\n <td>1671.73</td>\n <td>4974877</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-01-03</td>\n </tr>\n <tr>\n <th>1</th>\n <td>04-03-19</td>\n <td>AMZN</td>\n <td>1685.00</td>\n <td>1709.43</td>\n <td>1674.36</td>\n <td>1696.17</td>\n <td>6167358</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-04-03</td>\n </tr>\n <tr>\n <th>2</th>\n <td>05-03-19</td>\n <td>AMZN</td>\n <td>1702.95</td>\n <td>1707.80</td>\n <td>1689.01</td>\n <td>1692.43</td>\n <td>3681522</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-05-03</td>\n </tr>\n <tr>\n <th>3</th>\n <td>06-03-19</td>\n <td>AMZN</td>\n <td>1695.97</td>\n <td>1709.43</td>\n <td>1620.51</td>\n <td>1668.95</td>\n <td>3996001</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-06-03</td>\n </tr>\n <tr>\n <th>4</th>\n <td>07-03-19</td>\n <td>AMZN</td>\n <td>1667.37</td>\n <td>1669.75</td>\n <td>1620.51</td>\n <td>1625.95</td>\n <td>4957017</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-07-03</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
],
[
[
"# Câu 8: Thêm cột result, nếu giá 'close' > 'open' thì cột result có giá trị 'up', ngược lại 'down'\n",
"_____no_output_____"
]
],
[
[
"<details>\n <summary>Nhấn vào đây để xem kết quả!</summary>\n <div>\n<style scoped=\"\">\n .dataframe tbody tr th:only-of-type {\n vertical-align: middle;\n }\n\n .dataframe tbody tr th {\n vertical-align: top;\n }\n\n .dataframe thead th {\n text-align: right;\n }\n</style>\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>date</th>\n <th>symbol</th>\n <th>open</th>\n <th>high</th>\n <th>low</th>\n <th>close</th>\n <th>volume</th>\n <th>name</th>\n <th>employees</th>\n <th>headquarters_city</th>\n <th>headquarters_state</th>\n <th>parsed_time</th>\n <th>result</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>0</th>\n <td>01-03-19</td>\n <td>AMZN</td>\n <td>1655.13</td>\n <td>1674.26</td>\n <td>1651.00</td>\n <td>1671.73</td>\n <td>4974877</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-01-03</td>\n <td>up</td>\n </tr>\n <tr>\n <th>1</th>\n <td>04-03-19</td>\n <td>AMZN</td>\n <td>1685.00</td>\n <td>1709.43</td>\n <td>1674.36</td>\n <td>1696.17</td>\n <td>6167358</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-04-03</td>\n <td>up</td>\n </tr>\n <tr>\n <th>2</th>\n <td>05-03-19</td>\n <td>AMZN</td>\n <td>1702.95</td>\n <td>1707.80</td>\n <td>1689.01</td>\n <td>1692.43</td>\n <td>3681522</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-05-03</td>\n <td>down</td>\n </tr>\n <tr>\n <th>3</th>\n <td>06-03-19</td>\n <td>AMZN</td>\n <td>1695.97</td>\n <td>1709.43</td>\n <td>1620.51</td>\n <td>1668.95</td>\n <td>3996001</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-06-03</td>\n <td>down</td>\n </tr>\n <tr>\n <th>4</th>\n <td>07-03-19</td>\n <td>AMZN</td>\n <td>1667.37</td>\n <td>1669.75</td>\n <td>1620.51</td>\n <td>1625.95</td>\n <td>4957017</td>\n <td>AMZN</td>\n <td>613300</td>\n <td>Seattle</td>\n <td>WA</td>\n <td>2019-07-03</td>\n <td>down</td>\n </tr>\n </tbody>\n</table>\n</div>\n \n</details>",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec625de3e485716365814ab095692af6edde3a57 | 19,171 | ipynb | Jupyter Notebook | posts/post-9-adversarial-policies-attacking-deep-reinforcement-learning.ipynb | deepboltzer/brAIn | 7c8f6c2e162ae21027b227bc4c685c283ed3d8a0 | [
"MIT"
]
| 2 | 2021-11-24T18:58:05.000Z | 2021-12-03T12:43:32.000Z | posts/post-9-adversarial-policies-attacking-deep-reinforcement-learning.ipynb | deepboltzer/brAIn | 7c8f6c2e162ae21027b227bc4c685c283ed3d8a0 | [
"MIT"
]
| null | null | null | posts/post-9-adversarial-policies-attacking-deep-reinforcement-learning.ipynb | deepboltzer/brAIn | 7c8f6c2e162ae21027b227bc4c685c283ed3d8a0 | [
"MIT"
]
| 4 | 2021-11-14T10:18:22.000Z | 2022-03-22T21:09:58.000Z | 92.168269 | 1,269 | 0.72135 | [
[
[
"# Adversarial Policies: Attacking Deep Reinforcement Learning\n\n## Introduction\n\nAfter introducing adversarial attacks in a previous post (TODO: hier hyperlink) we will now showcase adversarial policy attacks in greater detail. To this end we will utilize the work of Adam Gleaves et. al. from their 2019 paper \"Adversarial Policies: Attacking Deep Reinforcement Learning\". This showcase will illustrate the theory behind adversarial policies in various environment and will also include the results of experiments we conducted ourselves. ",
"_____no_output_____"
],
[
"## What is an Adversarial Policy?\n\nAdversarial policies are a type of adversarial attack which can be utiliize in multiagent environments. In a multiagent environments multiple agents act together on the same enviornment. This can be cooperative but also competetive. For this post we will focus on competetive tasks, but the same type of attack could be used in cooperative environments.\n\nAn adversarial policy attack can occure if an attacker gains control of one or more agents in the environment. The attacker can then train the controlled agent with the explicit goal of minimizing the reward of other agents. This can be very effective as the deployed agents in an environment will ",
"_____no_output_____"
],
[
"## Setup for the Berkley Repository\n\nTo implement and see the effectiveness of Adversarial Policies we used this Berkeley [GitHub](https://github.com/HumanCompatibleAI/adversarial-policies). The easiest way to work with the repository is by utilizing [Docker](https://www.docker.com/). This will allow you to use the repository in an isolated environment, preventing potential complications with various dependencies. \n\nWe will give a detailed guide on how to setup the repository on Windows yet the process is almost identical on Linux systems.\n\n### Reproducing results on Windows\n\n\n#### Setting up Docker\nOn Windows, you first need to install WSL. This allows Windows to run a Linux environment directly and is required to use Docker. To do this just follow along this guide by [Microsoft](https://docs.microsoft.com/en-us/windows/wsl/install).\n\nAfterwards download and install Docker Desktop according to the [tutorial](https://docs.docker.com/desktop/windows/install/) provided by Docker. On startup the Desktop application will start a tutorial which can be very helpful to familiarize yourself with Docker. \n\n\n#### Preparing the Repositroy\nThe Berkley repository can simply be cloned or downloaded from [GitHub](https://github.com/HumanCompatibleAI/adversarial-policies). Since the Repository utilizes Mujoco we need to download a [Mujoco activation key](https://www.roboti.us/license.html). So that the key can be accessed later on simply move the key file directly into the repository in your filesystem.\n\nNow we can utilize Docker to work with the repositroy. Create a terminal and navigate to the repository. Then build a docker image from the repository with ```docker build -t rl_adversarial```. \n\nAfter succesfully building the image start a docker container with the Mujoco key by calling ```docker run -it --name rl_adv --env MUJOCO_PY_MJKEY_PATH=/adversarial-policies/mjkey.txt rl_adversarial /bin/bash```. When you get an error similar to ```ERROR [python-req 6/6] RUN touch /root/.mujoco/mjkey.txt && ci/build_venv.sh /venv && rm -rf HOME/.cache/ ``` while building the image consider running ``` git config --global core.autocrlf false ``` and repeating previous steps can help. \n\nIf everything went smoothly a Linux command line should appear in the terminal. You are now able to train the Adversarial Policies using the implementation from the 2019 paper.\n\n\n## Getting started with the repository\n\nNow you have several options you can follow. We would suggest that you first run ``` python -m aprl.train```. This will come in handy when searching for the trained models. If an error like ``` multi_train is not in list``` occurs simply restarting docker would fix it for us.\n\n### aprl.train\n```python -m aprl.train``` lets you train a policy. To get a better understanding of the different settings you can run, head to ```aprl/train``` and take a look at different parameters under ```train_config()```. The environment supports a total of 6 games. A summary is provided under ```Games```. To simply recreate the results in the game SumoHumans use ``` python -m aprl.train with env_name=multicomp/SumoHumans-v0 paper```. This will train a policy for a total of 20 Million time steps. After the training is finished you can test the policy by using ```aprl.score_agent```\n\n### aprl.score_agent\n``` python -m aprl.score_agent``` allows us to test the quality of our trained policy. Just like ```aprl.train```, there are lot of different paramters. You can find these at ```aprl/score_agent default_score_config()```. To evaluate the quality of our trained Policy from before run the following command```python -m aprl.score_agent with env_name=multicomp/SumoHumans-v0 agent_b_type=ppo2 agent_b_path=/adversarial-policies/data/baselines/20220322_162856-default/final_model/ episodes=100 ```. You need to change ```20220322_162856-default``` to the actual name of the folder the policy is stored in. Simply follow along the ``` Save location``` part below to find the folders. ```aprl.score_agent``` has the option to creat videos aswell. To create videos of the policies simply add ```videos=True```. In our case we had to set the ```annotated``` parameter to False under ```video_params``` or we would recieve an error. Occasionally other errors while creating videos can occur. Most of the time restarting ```WSL```would fix these for us. The videos are stored in the same folder as the logs of the score session if the path is not changed. To find these folders a small guide to locate them is provided below.\n\n### Save location\nTo find the directory in which the trained policies and the scores are safed head to ``` \\\\wsl$ ``` -> ```docker-desktop-data``` -> ``` version-pack-data ``` -> ``` community``` -> ```docker```-> ```overlay2``` -> At this point there should be several folders with weird names. Simply sort by last edited and open the last edited folder(to make sure this works atleast one Policy should´ve been trained already). -> ```diff```-> ```adversarial-policies``` -> ```data```. The trained policies are stored in ```baselines```. The logs of the training sessions and the scoring sessions are stored in ```sacred```.\n\n### Games\n\nA total of 6 games are provided by ```gym_compete``` in which two agents compete against each other in the ```MuJoCo robotics simulator```. There are ``` KickAndDefend-v0, RunToGoalHumans-v0, SumoHumans-v0, YouShallNotPassHumans-v0 SumoAnts-v0 and RunToGoalAnts-v0```. To see the games in action simply run ```aprl.score_agent``` with the specific game as ```env_name``` and create a few Videos. \n\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\KickAndDefend-gif\\vid.gif\" style=\"width: 300px;\">\n<figcaption>Example of KickAndDefend</figcaption>\n</figure>\n</center>\n\n### Mistakes to avoid\n\nThe most time consuming mistake we encountered was training the wrong agent in ```YouShallNotPassHumans-v0```. While it doesn´t really matter which Agent you select to train in the Sumo games its important to select the correct agent here. If you simply select the game and run ``` aprl.train``` you will train the attacking agent. To make sure you select the defending agent use ```python -m aprl.train with env_name=multicomp/YouShallNotPassHumans-v0 embed_index=1 paper```. \n",
"_____no_output_____"
],
[
"## Results\n\nTo test the effectiveness of adversarial policies we trained several agents in different games and let them compete against each other. One advantage prior to the acutal results of adversarial policies is their fast learning. Compared to training a policy from scratch one to play a game it took the adversarial policy only a fraction of this time to learn an efficient attack.\n\n### SumoHumans\n\nIn this game you have two agents fighting each other in a small arena. The goal is to push the opposing agent over or out of the arena(Fig.1 left). A total of 3 different baseline agents are provided by gym_compete and we trained one adversary for each. In all three cases the adversarial policy had similiar approaches. The adversary simply choose to kneel down thus confusing the zoo agent and forcing him to make wrong decisions(Fig.1 middle) while aditionally avoiding the losing condition of falling over. This form of attack was especially effective against the first zoo policy(vic_v1 in Fig.2) with a 74% win rate and only a 7% loss rate while performing significant worse against vic_v2. That is mostly due to the fact that vic_v2 uses the same strategy and fights in a kneeling position, thus most games end in a tie. Another interesting observation is the strong difference in performance between attacking the victim with a adversarial policy that was traineed based on that victim and with a policy that was trained against another victim version. In the first case the adversary wins way more then he loses(except against vic_v2) but the moment he plays against another victim version he loses almost 50% or even more of the games played(Fig.2).\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\Sumo_Humans_1v1_tourney\\gifs\\1v1_norm.gif\" style=\"width: 300px;\">\n<img src=\"..\\workspace\\adv_policy_training\\Sumo_Humans_1v1_tourney\\gifs\\1v1_adv(v1).gif\" style=\"width: 300px;\">\n<img src=\"..\\workspace\\adv_policy_training\\Sumo_Humans_1v1_tourney\\gifs\\1v1_adv(v2).gif\" style=\"width: 300px;\">\n<figcaption>(Fig.1)Left: 1 vs 1 between two zoo agents. Middle: 1 vs 1 between vic_v1(red) and adv_v1(green)- Right: 1 vs 1 between vic_v2(red) and adv_v2(green)</figcaption>\n</figure>\n</center>\n\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\Sumo_humans_1v1_tourney/Übersicht.png\" style=\"width: 1000px;\">\n<figcaption>(Fig.2)From left to right: Adversary wins, victim wins and ties</figcaption>\n</figure>\n</center>\n\n### SumoAnts\n\nSumoAnts uses the same setup and rules as SumoHumans. The only difference is the agent. While the human version has more dimensions and thus more room for errors the ant version has fewer dimensions(Fig.3). The comparison between the two games makes a weakness of adversarial policies obvious. The less complex the environment the less effective an adversarial attack is. In the SumoAnt game the adversary didn´t manage to get a higher winrate then 9%(Fig.4) even after training for 20 million timesteps.\n\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\Sumo_ants_1v1_tourney/gif_1vs1.gif\" style=\"width: 300px;\">\n<figcaption>(Fig.3)1 vs 1 between two zoo agents.</figcaption>\n</figure>\n</center>\n\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\Sumo_ants_1v1_tourney/Übersicht2.png\" style=\"width: 1000px;\">\n<figcaption>(Fig.4)From left to right: Adversary wins, victim wins and ties</figcaption>\n</figure>\n</center>\n\n\n### YouShallNotPass\n\nThe two prior expamples had one thing in common. Both agents had the same goal. To make the other agent fall over or fall out of the arena. In YouShallNotPass the agent has a different task(Fig.4 left). The attacking agent has to run past the defending agent. The defending agent has to stop the other agent from reaching the finish line somehow. We choose the defending agent as the adversary. Just like in the two sumo games the adversary throws himself on the ground to confuse the attacking agent(Fig.5 middle). This tactic seems to be consistent as he wins 75% of the matches(Fig.6 right).\n\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\YouShallNotPass_1v1_tourney+Videos\\gif\\normal1v1.gif\" style=\"width: 300px;\">\n<img src=\"..\\workspace\\adv_policy_training\\YouShallNotPass_1v1_tourney+Videos\\gif\\advvsvic.gif\" style=\"width: 300px;\">\n<figcaption>(Fig.5)Left: 1 vs 1 between two zoo agents(red is defending, green is attacking). Middle: 1 vs 1 between adversary and a zoo agent.</figcaption>\n</figure>\n</center>\n\nFurthermore we decided to mask the attacking agent. This means the attacking agent doesn´t observe his opponent and decides what actions to take without taking the opponents actions in account(Fig.6 middle). This counters the approach the adversary takes completely, resulting in a 97% win rate(Fig.6) A second approach we took was training a zoo agent to play against the adversary and defend himself from the attack. We started the training with an estimation of 20 million time steps but it took the trained agent way less to learn the adversary attack and counter it(Fig.6 middle). After less then 1 million timesteps the defending victim already achieved a 88% win rate(Fig.6 right). But this robustness against attacks is not without drawbacks. The moment the defended or the masked agent has to play against a normal zoo opponent he gets beaten severly and only manages to win 1% of the games, thus making these two approaches of defence strong against the adversary but poor if the agent is meant to perform as well as possible against normaly trained agents. \n\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\YouShallNotPass_1v1_tourney+Videos\\gif\\advvsmas.gif\" style=\"width: 300px;\">\n<img src=\"..\\workspace\\adv_policy_training\\YouShallNotPass_1v1_tourney+Videos\\gif\\advvsdef.gif\" style=\"width: 300px;\">\n<img src=\"..\\workspace\\adv_policy_training\\YouShallNotPass_1v1_tourney+Videos\\Übersicht4.png\" style=\"width: 500px;\">\n<figcaption>(Fig.6) Left: 1 vs 1 between adv(red) and vic_Masked(green). Middle: 1 vs 1 between adv(red) and vic_Ddefended(green). Right: 1 vs 1 results between the adversary and a zoo agent(vic_v1), a masked zoo agent(Vic_M) and a defended zoo agent(Vic_D)</figcaption>\n</figure>\n</center>\n\nOne solution for the overfitting is training the victim to defend itself against the Adversary and the zoo agent at the same time. By simply playing against the adversary in one episode and against the zoo agent in the next episode the victim is robust against the adversary while also being able to achieve the original results he had versus the zoo agent(Fig.7). With this method it is possible to create a victim that appears safe at first glance. And it is safe as long as the adversary stays the same and doesn´t evolve himself. But as mentioned before the training of the adversary takes little to no time compared to victim hardening and thus it took us only a few hours to create a second version of the adversary that isn´t just very effective against the modified victim but also against the original zoo agent(Fig.7).\n<center>\n<figure>\n<img src=\"..\\workspace\\adv_policy_training\\YouShallNotPass_1v1_tourney+Videos\\Übersicht3.png\" style=\"width: 500px;\">\n<figcaption>(Fig.7)Test results of hardened victim and Adv2</figcaption>\n</figure>\n</center>\n\n\n",
"_____no_output_____"
],
[
"## Conclusion\n\nThese results give us a good overview of the strength and weaknesses of adversarial policies. One of the big strength of adversarial policies lies in their fast training with little computational power and their adaptability to different problems. Like mentioned above if you have access to a black box with the victim it doesn´t take much time to find their weakness if possible.\n\nYet adversarial policies are not withour drawbacks. First, the victim need to be static as the victim would otherwise adapt to the adversary's strategy, nullifying the effect of the attack.\n\nSecond, the more dimensions of the victim's observation the adversary can impact, the easier it becomes to discover a vulnerability for the adversarial agent to exploit. This was evident in the difference between SumoAnts and SumoHumans. In SumoAnts the adversary was unable to find an exploit while in SumoHumans the adversarial attack was successful under the same conditions. This might remind one of the curse of dimesnionality. The more complex the state space, the more sparse the states of the agent's collected episodes. Thus the adversary can create a state for the victim which the victim was not properly trained on, thus creating an exploitable weakness.\n\nThis also explains, why simple countermeasures like masking the victim's observation, thereby removing the adversary's influence over the next state, made the adversary useless and even with the slow learning speed of the victim it developed a strategy to counter the adversary in little to no time. Both of thes defensive strategies are, however, not without cost. They require addittonal training and weaken the performance of the agent against non-adversarial agents.\n\nThere does not seem to be a singular best strategy for an agent to follow. For each agent there also exists an adversary capable exploiting or outperfroming the agents strategy. Thus we also need to balance the robustness and the perfomance of an agent as we can not create an agent that is both the best performing and robust against all attacks.",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec627bfce2a2f1d5dd019dc2695206d9ecfc7da9 | 867,416 | ipynb | Jupyter Notebook | Managing big data with MySQL (Duke)/MySQL_Exercise_12_Queries_that_Test_Relationships_Between_Test_Completion_and_Test_Circumstances.ipynb | yunhaojohn/MOOC | 83864010b4548866fc0c64f02769f67b5d8369c5 | [
"MIT"
]
| null | null | null | Managing big data with MySQL (Duke)/MySQL_Exercise_12_Queries_that_Test_Relationships_Between_Test_Completion_and_Test_Circumstances.ipynb | yunhaojohn/MOOC | 83864010b4548866fc0c64f02769f67b5d8369c5 | [
"MIT"
]
| null | null | null | Managing big data with MySQL (Duke)/MySQL_Exercise_12_Queries_that_Test_Relationships_Between_Test_Completion_and_Test_Circumstances.ipynb | yunhaojohn/MOOC | 83864010b4548866fc0c64f02769f67b5d8369c5 | [
"MIT"
]
| null | null | null | 41.999516 | 1,023 | 0.437909 | [
[
[
"Copyright Jana Schaich Borg/Attribution-NonCommercial 4.0 International (CC BY-NC 4.0)",
"_____no_output_____"
],
[
"# MySQL Exercise 12: Queries that Test Relationships Between Test Completion and Testing Circumstances \n\nIn this lesson, we are going to practice integrating more of the concepts we learned over the past few weeks to address whether issues in our Dognition sPAP are related to the number of tests dogs complete. We are going to focus on a subset of the issues listed in the \"Features of Testing Circumstances\" branch of our sPAP. You will need to look up new functions several times and the final queries at which we will arrive by the end of this lesson will be quite complex, but we will work up to them step-by-step. \n\nTo begin, load the sql library and database, and make the Dognition database your default database:",
"_____no_output_____"
]
],
[
[
"%load_ext sql\n%sql mysql://studentuser:studentpw@mysqlserver/dognitiondb\n%sql USE dognitiondb",
"0 rows affected.\n"
]
],
[
[
"<img src=\"https://duke.box.com/shared/static/p2eucjdttai08eeo7davbpfgqi3zrew0.jpg\" width=600 alt=\"SELECT FROM WHERE\" />\n\n## 1. During which weekdays do Dognition users complete the most tests?\n\nThe first question we are going to address is whether there is a certain day of the week when users are more or less likely to complete Dognition tests. If so, targeting promotions or reminder emails to those times of the week might increase the number of tests users complete.\n\nAt first, the query we need to address this question might seem a bit intimidating, but once you can describe what the query needs to do in words, writing the query won't seem so challenging. \n\nUltimately, we want a count of the number of tests completed on each day of the week, with all of the dog_guids and user_guids the Dognition team flagged in their exclude column excluded. To achieve this, we are going to have to use the GROUP BY clause to break up counts of the records in the completed_tests table according to days of the week. We will also have to join the completed_tests table with the dogs and users table in order to exclude completed_tests records that are associated with dog_guids or user_guids that should be excluded. First, though, we need a method for extracting the day of the week from a time stamp. In MySQL Exercise 2 we used a function called \"DAYNAME\". That is the most efficient function to use for this purpose, but not all database systems have this function, so let's try using a different method for the queries in this lesson. Search these sites to find a function that will output a number from 1-7 for time stamps where 1 = Sunday, 2 = Monday, …, 7 = Saturday:\n\nhttp://dev.mysql.com/doc/refman/5.7/en/func-op-summary-ref.html \nhttp://www.w3resource.com/mysql/mysql-functions-and-operators.php\n\n**Question 1: Using the function you found in the websites above, write a query that will output one column with the original created_at time stamp from each row in the completed_tests table, and another column with a number that represents the day of the week associated with each of those time stamps. Limit your output to 200 rows starting at row 50.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT created_at,DAYOFWEEK(created_at)\nFROM complete_tests\nLIMIT 200 OFFSET 49",
"200 rows affected.\n"
]
],
[
[
"Of course, the results of the query in Question 1 would be much easier to interpret if the output included the name of the day of the week (or a relevant abbreviation) associated with each time stamp rather than a number index.\n\n**Question 2: Include a CASE statement in the query you wrote in Question 1 to output a third column that provides the weekday name (or an appropriate abbreviation) associated with each created_at time stamp.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT created_at,\n CASE DAYOFWEEK(created_at)\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name\nFROM complete_tests\nLIMIT 200 OFFSET 49",
"200 rows affected.\n"
]
],
[
[
"Now that we are confident we have the correct syntax for extracting weekday labels from the created_at time stamps, we can start building our larger query that examines the number of tests completed on each weekday.\n\n**Question 3: Adapt the query you wrote in Question 2 to report the total number of tests completed on each weekday. Sort the results by the total number of tests completed in descending order. You should get a total of 33,190 tests in the Sunday row of your output.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT CASE DAYOFWEEK(created_at)\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name, COUNT(created_at) AS num_tests\nFROM complete_tests\nGROUP BY weekday_name\nORDER BY num_tests DESC",
"7 rows affected.\n"
]
],
[
[
"So far these results suggest that users complete the most tests on Sunday night and the fewest tests on Friday night. We need to determine if this trend remains after flagged dog_guids and user_guids are excluded. Let's start by removing the dog_guids that have an exclude flag. We'll exclude user_guids with an exclude flag in later queries.\n\n**Question 4: Rewrite the query in Question 3 to exclude the dog_guids that have a value of \"1\" in the exclude column (Hint: this query will require a join.) This time you should get a total of 31,092 tests in the Sunday row of your output.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT CASE DAYOFWEEK(c.created_at)\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name, COUNT(c.created_at) AS num_tests\nFROM complete_tests c, dogs d\nWHERE c.dog_guid=d.dog_guid AND (d.exclude IS NULL OR d.exclude=0)\nGROUP BY weekday_name\nORDER BY num_tests DESC",
"7 rows affected.\n"
]
],
[
[
"Now we need to exclude the user_guids that have a value of \"1\" in the exclude column as well. One way to do this would be to join the completed_tests, dogs, and users table with a sequence of inner joins. However, we've seen in previous lessons that there are duplicate rows in the users table. These duplicates will get passed through the join and will affect the count calculations. To illustrate this, compare the following two queries.\n\n**Question 5: Write a query to retrieve all the dog_guids for users common to the dogs and users table using the traditional inner join syntax (your output will have 950,331 rows).**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT d.dog_guid\nFROM dogs d JOIN users u\n ON d.user_guid=u.user_guid",
"950331 rows affected.\n"
]
],
[
[
"**Question 6: Write a query to retrieve all the *distinct* dog_guids common to the dogs and users table using the traditional inner join syntax (your output will have 35,048 rows).**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT DISTINCT d.dog_guid\nFROM dogs d JOIN users u\n ON d.user_guid=u.user_guid",
"35048 rows affected.\n"
]
],
[
[
"The strategy we will use to handle duplicate rows in the users table will be to, first, write a subquery that retrieves the distinct dog_guids from an inner join between the dogs and users table with the appropriate records excluded. Then, second, we will join the result of this subquery to the complete_tests table and group the results according to the day of the week.\n\n**Question 7: Start by writing a query that retrieves distinct dog_guids common to the dogs and users table, excuding dog_guids and user_guids with a \"1\" in their respective exclude columns (your output will have 34,121 rows).**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT DISTINCT d.dog_guid\nFROM dogs d JOIN users u\n ON d.user_guid=u.user_guid\nWHERE (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0)",
"34121 rows affected.\n"
]
],
[
[
"**Question 8: Now adapt your query from Question 4 so that it inner joins on the result of the subquery you wrote in Question 7 instead of the dogs table. This will give you a count of the number of tests completed on each day of the week, excluding all of the dog_guids and user_guids that the Dognition team flagged in the exclude columns.** ",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT CASE DAYOFWEEK(c.created_at)\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name, COUNT(c.created_at) AS num_tests\nFROM complete_tests c JOIN (SELECT DISTINCT d.dog_guid\n FROM dogs d JOIN users u\n ON d.user_guid=u.user_guid\n WHERE (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0)) AS Distinct_dogs\n ON c.dog_guid=Distinct_dogs.dog_guid\nGROUP BY weekday_name\nORDER BY num_tests DESC",
"7 rows affected.\n"
]
],
[
[
"These results still suggest that Sunday is the day when the most tests are completed and Friday is the day when the fewest tests are completed. However, our first query suggested that more tests were completed on Tuesday than Saturday; our current query suggests that slightly more tests are completed on Saturday than Tuesday, now that flagged dog_guids and user_guids are excluded.\n\nIt's always a good idea to see if a data pattern replicates before you interpret it too strongly. The ideal way to do this would be to have a completely separate and independent data set to analyze. We don't have such a data set, but we can assess the reliability of the day of the week patterns in a different way. We can test whether the day of the week patterns are the same in all years of our data set.\n\n**Question 9: Adapt your query from Question 8 to provide a count of the number of tests completed on each weekday of each year in the Dognition data set. Exclude all dog_guids and user_guids with a value of \"1\" in their exclude columns. Sort the output by year in ascending order, and then by the total number of tests completed in descending order. HINT: you will need a function described in one of these references to retrieve the year of each time stamp in the created_at field:**\n\nhttp://dev.mysql.com/doc/refman/5.7/en/func-op-summary-ref.html \nhttp://www.w3resource.com/mysql/mysql-functions-and-operators.php",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT YEAR(c.created_at) AS year,\n CASE DAYOFWEEK(c.created_at)\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name, COUNT(c.created_at) AS num_tests\nFROM complete_tests c JOIN (SELECT DISTINCT d.dog_guid\n FROM dogs d JOIN users u\n ON d.user_guid=u.user_guid\n WHERE (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0)) AS Distinct_dogs\n ON c.dog_guid=Distinct_dogs.dog_guid\nGROUP BY year,weekday_name\nORDER BY year ASC,num_tests DESC",
"21 rows affected.\n"
]
],
[
[
"These results suggest that although the precise order of the weekdays with the most to fewest completed tests changes slightly from year to year, Sundays always have a lot of completed tests, and Fridays always have the fewest or close to the fewest completed tests. So far, it seems like it might be a good idea for Dognition to target reminder or encouragement messages to customers on Sundays. However, there is one more issue our analysis does not address. All of the time stamps in the created_at column are in Coordinated Universal Time (abbreviated UTC). This is a time convention that is constant around the globe. Nonetheless, as the picture below illustrates, countries and states have different time zones. The same UTC time can correspond with local times in different countries that are as much as 24 hours apart:\n\n<img src=\"https://duke.box.com/shared/static/0p8ee9az908soq1m0o4jst94vqlh2oh7.jpg\" width=600 alt=\"TIME_ZONE_MAP\" />\n\n\nTherefore, the weekdays we have extracted so far may not accurately reflect the weekdays in the local times of different countries. The only way to correct the time stamps for time zone differences is to obtain a table with the time zones of every city, state, or country. Such a table was not available to us in this course, but we can run some analyses that approximate a time zone correction for United States customers.\n\n**Question 10: First, adapt your query from Question 9 so that you only examine customers located in the United States, with Hawaii and Alaska residents excluded. HINTS: In this data set, the abbreviation for the United States is \"US\", the abbreviation for Hawaii is \"HI\" and the abbreviation for Alaska is \"AK\". You should have 5,860 tests completed on Sunday of 2013.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT YEAR(c.created_at) AS year,\n CASE DAYOFWEEK(c.created_at)\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name, COUNT(c.created_at) AS num_tests\nFROM complete_tests c JOIN (SELECT DISTINCT d.dog_guid\n FROM dogs d JOIN users u\n ON d.user_guid=u.user_guid\n WHERE (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0) AND u.country='US'\n AND u.state NOT IN ('HI','AK')) AS Distinct_dogs\n ON c.dog_guid=Distinct_dogs.dog_guid\nGROUP BY year,weekday_name\nORDER BY year ASC,num_tests DESC",
"21 rows affected.\n"
]
],
[
[
"The next step is to adjust the created_at times for differences in time zone. Most United States states (excluding Hawaii and Alaska) have a time zone of UTC time -5 hours (in the eastern-most regions) to -8 hours (in the western-most regions). To get a general idea for how much our weekday analysis is likely to change based on time zone, we will subtract 6 hours from every time stamp in the complete_tests table. Although this means our time stamps can be inaccurate by 1 or 2 hours, people are not likely to be playing Dognition games at midnight, so 1-2 hours should not affect the weekdays extracted from each time stamp too much. \n\nThe functions used to subtract time differ across database systems, so you should double-check which function you need to use every time you are working with a new database. We will use the date_sub function:\n\nhttp://www.w3schools.com/sql/func_date_sub.asp\n\n**Question 11: Write a query that extracts the original created_at time stamps for rows in the complete_tests table in one column, and the created_at time stamps with 6 hours subtracted in another column. Limit your output to 100 rows.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT created_at, DATE_SUB(created_at, INTERVAL 6 HOUR)\nFROM complete_tests",
"193246 rows affected.\n"
]
],
[
[
"**Question 12: Use your query from Question 11 to adapt your query from Question 10 in order to provide a count of the number of tests completed on each day of the week, with approximate time zones taken into account, in each year in the Dognition data set. Exclude all dog_guids and user_guids with a value of \"1\" in their exclude columns. Sort the output by year in ascending order, and then by the total number of tests completed in descending order. HINT: Don't forget to adjust for the time zone in your DAYOFWEEK statement and your CASE statement.** ",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT YEAR(DATE_SUB(c.created_at, INTERVAL 6 HOUR)) AS year,\n CASE DAYOFWEEK(DATE_SUB(created_at, INTERVAL 6 HOUR))\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name, COUNT(DATE_SUB(c.created_at, INTERVAL 6 HOUR)) AS num_tests\nFROM complete_tests c JOIN (SELECT DISTINCT d.dog_guid\n FROM dogs d JOIN users u\n ON d.user_guid=u.user_guid\n WHERE (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0) AND u.country='US'\n AND u.state NOT IN ('HI','AK')) AS Distinct_dogs\n ON c.dog_guid=Distinct_dogs.dog_guid\nGROUP BY year,weekday_name\nORDER BY year ASC,num_tests DESC",
"21 rows affected.\n"
]
],
[
[
"You can try re-running the query with time-zone corrections of 5, 7, or 8 hours, and the results remain essentially the same. All of these analyses suggest that customers are most likely to complete tests around Sunday and Monday, and least likely to complete tests around the end of the work week, on Thursday and Friday. This is certainly valuable information for Dognition to take advantage of.\n\nIf you were presenting this information to the Dognition team, you might want to present the information in the form of a graph that you make in another program. The graph would be easier to read if the output was ordered according to the days of the week shown in standard calendars, with Monday being the first day and Sunday being the last day. MySQL provides an easy way to do this using the FIELD function in the ORDER BY statement:\n\nhttps://www.virendrachandak.com/techtalk/mysql-ordering-results-by-specific-field-values/\n\n**Question 13: Adapt your query from Question 12 so that the results are sorted by year in ascending order, and then by the day of the week in the following order: Monday, Tuesday, Wednesday, Thursday, Friday, Saturday, Sunday.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT YEAR(DATE_SUB(c.created_at, INTERVAL 6 HOUR)) AS year,\n CASE DAYOFWEEK(DATE_SUB(created_at, INTERVAL 6 HOUR))\n WHEN 1 THEN 'SUN'\n WHEN 2 THEN 'MON'\n WHEN 3 THEN 'TUE'\n WHEN 4 THEN 'WED'\n WHEN 5 THEN 'THU'\n WHEN 6 THEN 'FRI'\n ELSE 'SAT'\n END AS weekday_name, COUNT(DATE_SUB(c.created_at, INTERVAL 6 HOUR)) AS num_tests\nFROM complete_tests c JOIN (SELECT DISTINCT d.dog_guid\n FROM dogs d JOIN users u\n ON d.user_guid=u.user_guid\n WHERE (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0) AND u.country='US'\n AND u.state NOT IN ('HI','AK')) AS Distinct_dogs\n ON c.dog_guid=Distinct_dogs.dog_guid\nGROUP BY year,weekday_name\nORDER BY year ASC,FIELD(weekday_name,'MON','TUE','WED','THU','FRI','SAT','SUN') ASC",
"21 rows affected.\n"
]
],
[
[
"Unfortunately other database platforms do not have the ORDER BY FIELD functionality. To achieve the same result in other platforms, you would have to use a CASE statement or a more advanced solution:\n\nhttp://stackoverflow.com/questions/1309624/simulating-mysqls-order-by-field-in-postgresql\n\nThe link provided above is to a discussion on stackoverflow.com. Stackoverflow is a great website that, in their words, \"is a community of 4.7 million programmers, just like you, helping each other.\" You can ask questions about SQL queries and get help from other experts, or search through questions posted previously to see if somebody else has already asked a question that is relevant to the problem you are trying to solve. It's a great resource to use whenever you run into trouble with your queries.\n\n## 2. Which states and countries have the most Dognition users?\n\nYou ended up with a pretty long and complex query in the questions above that you tested step-by-step. Many people save these types of queries so that they can be adapted for similar queries in the future without having to redesign and retest the entire query. \n \nIn the next two questions, we will practice repurposing previously-designed queries for new questions. Both questions can be answered through relatively minor modifications of the queries you wrote above.\n\n**Question 14: Which 5 states within the United States have the most Dognition customers, once all dog_guids and user_guids with a value of \"1\" in their exclude columns are removed? Try using the following general strategy: count how many unique user_guids are associated with dogs in the complete_tests table, break up the counts according to state, sort the results by counts of unique user_guids in descending order, and then limit your output to 5 rows. California (\"CA\") and New York (\"NY\") should be at the top of your list.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT state,COUNT(DISTINCT u.user_guid) AS num_users\nFROM dogs d,complete_tests c,users u\nWHERE d.user_guid=u.user_guid AND c.dog_guid=d.dog_guid AND country='US'\n AND (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0)\nGROUP BY state\nORDER BY num_users DESC\nLIMIT 5",
"5 rows affected.\n"
]
],
[
[
"The number of unique Dognition users in California is more than two times greater than any other state. This information could be very helpful to Dognition. Useful follow-up questions would be: were special promotions run in California that weren't run in other states? Did Dognition use advertising channels that are particularly effective in California? If not, what traits differentiate California users from other users? Can these traits be taken advantage of in future marketing efforts or product developments?\n\nLet's try one more analysis that examines testing circumstances from a different angle.\n\n**Question 15: Which 10 countries have the most Dognition customers, once all dog_guids and user_guids with a value of \"1\" in their exclude columns are removed? HINT: don't forget to remove the u.country=\"US\" statement from your WHERE clause.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT country,COUNT(DISTINCT u.user_guid) AS num_users\nFROM dogs d,complete_tests c,users u\nWHERE d.user_guid=u.user_guid AND c.dog_guid=d.dog_guid\n AND (d.exclude IS NULL OR d.exclude=0) AND (u.exclude IS NULL OR u.exclude=0)\nGROUP BY country\nORDER BY num_users DESC\nLIMIT 10",
"10 rows affected.\n"
]
],
[
[
"The United States, Canada, Australia, and Great Britain are the countries with the most Dognition users. N/A refers to \"not applicable\" which essentially means we have no usable country data from those rows. After Great Britain, the number of Dognition users drops quite a lot. This analysis suggests that Dognition is most likely to be used by English-speaking countries. One question Dognition might want to consider is whether there are any countries whose participation would dramatically increase if a translated website were available.\n\n## 3. Congratulations!\n\nYou have now written many complex queries on your own that address real analysis questions about a real business problem. You know how to look up new functions, you know how to troubleshoot your queries by isolating each piece of the query until you are sure the syntax is correct, and you know where to look for help if you get stuck. You are ready to start using SQL in your own business ventures. Keep learning, keep trying new things, and keep asking questions. Congratulations for taking your career to the next level!\n\nThere is another video to watch, and of course, more exercises to work through using the Dillard's data set. \n \n**In the meantime, enjoy practicing any other queries you want to try here:**",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec62864275f0584ca32603d3c2d082ba229c8ae0 | 129,467 | ipynb | Jupyter Notebook | Applied AI Study Group #2 - February 2020/week3/2- Anomaly Detection Model.ipynb | cerob/Applied-AI-Study-Group | 976a9db54e71bc2408586fcd69c0ccb288d5d5b6 | [
"MIT"
]
| 40 | 2020-11-23T11:35:15.000Z | 2022-02-11T21:09:47.000Z | Applied AI Study Group #2 - February 2020/week3/2- Anomaly Detection Model.ipynb | cerob/Applied-AI-Study-Group | 976a9db54e71bc2408586fcd69c0ccb288d5d5b6 | [
"MIT"
]
| 3 | 2021-07-29T08:40:20.000Z | 2022-02-26T10:04:18.000Z | Applied AI Study Group #2 - February 2020/week3/2- Anomaly Detection Model.ipynb | cerob/Applied-AI-Study-Group | 976a9db54e71bc2408586fcd69c0ccb288d5d5b6 | [
"MIT"
]
| 34 | 2020-12-27T13:13:41.000Z | 2022-03-29T15:09:46.000Z | 129,467 | 129,467 | 0.895487 | [
[
[
"###Load Data",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive/', force_remount=False)\n%cd /content/drive/My Drive\n\n",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive/\n/content/drive/My Drive\n"
],
[
"import pandas as pd\ndf_healthy = pd.read_csv('result_healthy_pandas.csv', engine='python', header=None)\ndf_healthy.head()",
"_____no_output_____"
],
[
"import numpy\ndf_faulty = pd.read_csv('result_faulty_pandas.csv', engine='python', header=None)\nprint(numpy.shape(df_faulty))\ndf_faulty.head()",
"(488309, 4)\n"
]
],
[
[
"Check normalization",
"_____no_output_____"
]
],
[
[
"import numpy as np\nprint(np.amax(df_healthy.round(1).loc[:]))\nprint(np.amin(df_healthy.round(1).loc[:]))",
"0 1213483.0\n1 100.0\n2 0.3\n3 0.4\ndtype: float64\n0 0.0\n1 97.0\n2 -0.3\n3 -0.3\ndtype: float64\n"
],
[
"print(np.amax(df_faulty.round(1).loc[:]))\nprint(np.amin(df_faulty.round(1).loc[:]))",
"0 488308.0\n1 108.0\n2 1.7\n3 1.0\ndtype: float64\n0 0.0\n1 105.0\n2 -1.5\n3 -1.2\ndtype: float64\n"
]
],
[
[
"### Prepare Sequential",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom numpy import concatenate\nfrom matplotlib import pyplot\nfrom pandas import read_csv\nfrom pandas import DataFrame\nfrom pandas import concat\n",
"_____no_output_____"
],
[
"def get_recording(df,file_id):\n return np.array(df.sort_values(by=0, ascending=True).loc[df[1] == file_id].drop(columns = [0,1]))\n\ndef create_trimmed_recording(df,file_id,timesteps,dim):\n recording = get_recording(df,file_id) \n samples = len(recording)\n trim = samples % 100\n trimmed = samples-trim\n recording_trimmed = recording[:trimmed]\n recording_trimmed.shape = (int((trimmed)/timesteps),timesteps,dim)\n return recording_trimmed\n",
"_____no_output_____"
],
[
"#pd.unique()\n#df_healthy.drop(0,1).drop(2,1).drop(3,1)\npd.unique(df_healthy.iloc[:,1])",
"_____no_output_____"
]
],
[
[
"### Model",
"_____no_output_____"
]
],
[
[
"import sklearn\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.metrics import mean_squared_error\n\nfrom keras.models import Sequential\nfrom keras.layers import LSTM, Dense, Activation\nfrom keras.callbacks import Callback\n\n\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\n\nimport time\n\n%matplotlib inline",
"Using TensorFlow backend.\n"
],
[
"class LossHistory(Callback):\n def on_train_begin(self, logs={}):\n self.losses = []\n\n def on_batch_end(self, batch, logs={}):\n self.losses.append(logs.get('loss'))",
"_____no_output_____"
],
[
"timesteps = 100\ndim = 2\nlossHistory = LossHistory()\n# design network\n\nmodel = Sequential()\nmodel.add(LSTM(50,input_shape=(timesteps,dim),return_sequences=True))\nmodel.add(Dense(2))\nmodel.compile(loss='mae', optimizer='adam')\n\ndef train(data):\n model.fit(data, data, epochs=5, batch_size=72, validation_data=(data, data), verbose=1, shuffle=False,callbacks=[lossHistory])\n\ndef score(data):\n yhat = model.predict(data)\n return yhat",
"_____no_output_____"
],
[
"file_ids = pd.unique(df_healthy.iloc[:,1])\nstart = time.time()\nfor file_id in file_ids:\n recording_trimmed = create_trimmed_recording(df_healthy,file_id,timesteps,dim)\n print(\"Staring training on %s\" % (file_id))\n #train(recording_trimmed)\n model.fit(recording_trimmed, recording_trimmed, epochs=5, batch_size=72, validation_data=(recording_trimmed, recording_trimmed), verbose=1, shuffle=False,callbacks=[lossHistory])\n print(\"Finished training on %s after %s seconds\" % (file_id,time.time()-start))\n\nprint(\"Finished job on after %s seconds\" % (time.time()-start))\nhealthy_losses = lossHistory.losses\n",
"Staring training on 97\nTrain on 2439 samples, validate on 2439 samples\nEpoch 1/5\n2439/2439 [==============================] - 7s 3ms/step - loss: 0.0569 - val_loss: 0.0505\nEpoch 2/5\n2439/2439 [==============================] - 7s 3ms/step - loss: 0.0441 - val_loss: 0.0354\nEpoch 3/5\n2439/2439 [==============================] - 7s 3ms/step - loss: 0.0269 - val_loss: 0.0205\nEpoch 4/5\n2439/2439 [==============================] - 7s 3ms/step - loss: 0.0159 - val_loss: 0.0113\nEpoch 5/5\n2439/2439 [==============================] - 6s 3ms/step - loss: 0.0094 - val_loss: 0.0078\nFinished training on 97 after 35.24387216567993 seconds\nStaring training on 98\nTrain on 4839 samples, validate on 4839 samples\nEpoch 1/5\n4839/4839 [==============================] - 13s 3ms/step - loss: 0.0061 - val_loss: 0.0039\nEpoch 2/5\n4839/4839 [==============================] - 14s 3ms/step - loss: 0.0039 - val_loss: 0.0035\nEpoch 3/5\n4839/4839 [==============================] - 13s 3ms/step - loss: 0.0031 - val_loss: 0.0030\nEpoch 4/5\n4839/4839 [==============================] - 14s 3ms/step - loss: 0.0027 - val_loss: 0.0023\nEpoch 5/5\n4839/4839 [==============================] - 13s 3ms/step - loss: 0.0032 - val_loss: 0.0034\nFinished training on 98 after 102.27773571014404 seconds\nStaring training on 100\nTrain on 4856 samples, validate on 4856 samples\nEpoch 1/5\n4856/4856 [==============================] - 13s 3ms/step - loss: 0.0033 - val_loss: 0.0031\nEpoch 2/5\n4856/4856 [==============================] - 13s 3ms/step - loss: 0.0027 - val_loss: 0.0022\nEpoch 3/5\n4856/4856 [==============================] - 14s 3ms/step - loss: 0.0023 - val_loss: 0.0025\nEpoch 4/5\n4856/4856 [==============================] - 13s 3ms/step - loss: 0.0020 - val_loss: 0.0019\nEpoch 5/5\n4856/4856 [==============================] - 13s 3ms/step - loss: 0.0018 - val_loss: 0.0016\nFinished training on 100 after 168.73098278045654 seconds\nFinished job on after 168.73126482963562 seconds\n"
],
[
"healthy_losses = lossHistory.losses",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(num=None, figsize=(14, 6), dpi=80, facecolor='w', edgecolor='k')\nsize = len(healthy_losses)\nplt.ylim(0,0.008)\nax.plot(range(0,size), healthy_losses, '-', color='blue', animated = True, linewidth=1)",
"_____no_output_____"
],
[
"file_ids = pd.unique(df_faulty.iloc[:,1])\nstart = time.time()\nfor file_id in file_ids:\n recording_trimmed = create_trimmed_recording(df_faulty,file_id,timesteps,dim)\n print(\"Staring training on %s\" % (file_id))\n model.fit(recording_trimmed, recording_trimmed, epochs=5, batch_size=72, validation_data=(recording_trimmed, recording_trimmed), verbose=1, shuffle=False,callbacks=[lossHistory])\n print(\"Finished training on %s after %s seconds\" % (file_id,time.time()-start))\n\nprint(\"Finished job on after %s seconds\" % (time.time()-start))\nfaulty_losses = lossHistory.losses\n",
"Staring training on 105\nTrain on 1212 samples, validate on 1212 samples\nEpoch 1/5\n1212/1212 [==============================] - 4s 3ms/step - loss: 0.0125 - val_loss: 0.0090\nEpoch 2/5\n1212/1212 [==============================] - 3s 3ms/step - loss: 0.0078 - val_loss: 0.0065\nEpoch 3/5\n1212/1212 [==============================] - 3s 3ms/step - loss: 0.0059 - val_loss: 0.0054\nEpoch 4/5\n1212/1212 [==============================] - 3s 3ms/step - loss: 0.0051 - val_loss: 0.0048\nEpoch 5/5\n1212/1212 [==============================] - 3s 3ms/step - loss: 0.0046 - val_loss: 0.0044\nFinished training on 105 after 16.649536609649658 seconds\nStaring training on 106\nTrain on 1219 samples, validate on 1219 samples\nEpoch 1/5\n1219/1219 [==============================] - 3s 3ms/step - loss: 0.0043 - val_loss: 0.0041\nEpoch 2/5\n1219/1219 [==============================] - 3s 3ms/step - loss: 0.0040 - val_loss: 0.0039\nEpoch 3/5\n1219/1219 [==============================] - 3s 3ms/step - loss: 0.0038 - val_loss: 0.0037\nEpoch 4/5\n1219/1219 [==============================] - 4s 3ms/step - loss: 0.0036 - val_loss: 0.0035\nEpoch 5/5\n1219/1219 [==============================] - 3s 3ms/step - loss: 0.0034 - val_loss: 0.0033\nFinished training on 106 after 33.45457649230957 seconds\nStaring training on 107\nTrain on 1221 samples, validate on 1221 samples\nEpoch 1/5\n1221/1221 [==============================] - 3s 3ms/step - loss: 0.0032 - val_loss: 0.0032\nEpoch 2/5\n1221/1221 [==============================] - 3s 3ms/step - loss: 0.0031 - val_loss: 0.0030\nEpoch 3/5\n1221/1221 [==============================] - 3s 3ms/step - loss: 0.0030 - val_loss: 0.0029\nEpoch 4/5\n1221/1221 [==============================] - 3s 3ms/step - loss: 0.0028 - val_loss: 0.0028\nEpoch 5/5\n1221/1221 [==============================] - 3s 3ms/step - loss: 0.0027 - val_loss: 0.0027\nFinished training on 107 after 50.136409521102905 seconds\nStaring training on 108\nTrain on 1229 samples, validate on 1229 samples\nEpoch 1/5\n1229/1229 [==============================] - 3s 3ms/step - loss: 0.0028 - val_loss: 0.0027\nEpoch 2/5\n1229/1229 [==============================] - 3s 3ms/step - loss: 0.0026 - val_loss: 0.0026\nEpoch 3/5\n1229/1229 [==============================] - 3s 3ms/step - loss: 0.0026 - val_loss: 0.0025\nEpoch 4/5\n1229/1229 [==============================] - 4s 3ms/step - loss: 0.0024 - val_loss: 0.0024\nEpoch 5/5\n1229/1229 [==============================] - 4s 3ms/step - loss: 0.0024 - val_loss: 0.0023\nFinished training on 108 after 67.63817548751831 seconds\nFinished job on after 67.63835382461548 seconds\n"
],
[
"fig, ax = plt.subplots(num=None, figsize=(14, 6), dpi=80, facecolor='w', edgecolor='k')\nsize = len(healthy_losses+faulty_losses)\nplt.ylim(0,0.008)\nax.plot(range(0,size), healthy_losses+faulty_losses, '-', color='blue', animated = True, linewidth=1)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec62961a188d32ba200ce3aed0dd17ad5708fb88 | 31,157 | ipynb | Jupyter Notebook | Surrey/sourcecode/surrey_avg_ensemble_A.ipynb | leonlha/e2e-3m | 6f3f8a7aee8e74388e7e09d3fdf0df6bbf72526c | [
"MIT"
]
| 8 | 2020-08-05T16:19:31.000Z | 2021-06-23T05:56:12.000Z | Surrey/sourcecode/surrey_avg_ensemble_A.ipynb | leonlha/e2e-3m | 6f3f8a7aee8e74388e7e09d3fdf0df6bbf72526c | [
"MIT"
]
| null | null | null | Surrey/sourcecode/surrey_avg_ensemble_A.ipynb | leonlha/e2e-3m | 6f3f8a7aee8e74388e7e09d3fdf0df6bbf72526c | [
"MIT"
]
| 3 | 2020-08-05T22:36:28.000Z | 2021-06-02T09:50:49.000Z | 31.250752 | 121 | 0.511667 | [
[
[
"cd /media/datastorage/Phong/",
"/media/datastorage/Phong\n"
],
[
"# #G4\n# from google.colab import drive\n# drive.mount('/content/gdrive')",
"Mounted at /content/gdrive\n"
],
[
"# cp gdrive/My\\ Drive/fingerspelling5.tar.bz2 fingerspelling5.tar.bz2",
"_____no_output_____"
],
[
"# !tar xjf fingerspelling5.tar.bz2",
"_____no_output_____"
],
[
"# cd dataset5",
"/content/dataset5\n"
],
[
"# #remove depth files\n# import glob\n# import os\n# import shutil\n\n# # get parts of image's path\n# def get_image_parts(image_path):\n# \"\"\"Given a full path to an image, return its parts.\"\"\"\n# parts = image_path.split(os.path.sep)\n# #print(parts)\n# filename = parts[2]\n# filename_no_ext = filename.split('.')[0]\n# classname = parts[1]\n# train_or_test = parts[0]\n\n# return train_or_test, classname, filename_no_ext, filename\n\n\n# #del_folders = ['A','B','C','D','E'] \n# move_folders_1 = ['A','B','C','D'] \n# move_folders_2 = ['E']\n\n# # look for all images in sub-folders\n# for folder in move_folders_1:\n# class_folders = glob.glob(os.path.join(folder, '*'))\n \n# for iid_class in class_folders:\n# #move depth files\n# class_files = glob.glob(os.path.join(iid_class, 'depth*.png'))\n \n# print('copying %d files' %(len(class_files)))\n# for idx in range(len(class_files)): \n# src = class_files[idx]\n# if \"0001\" not in src: \n# train_or_test, classname, _, filename = get_image_parts(src)\n\n# dst = os.path.join('train_depth', classname, train_or_test+'_'+ filename)\n\n# # image directory\n# img_directory = os.path.join('train_depth', classname)\n\n# # create folder if not existed\n# if not os.path.exists(img_directory):\n# os.makedirs(img_directory)\n\n# #copying\n# shutil.copy(src, dst)\n# else:\n# print('ignor: %s' %src)\n \n# #move color files \n# for iid_class in class_folders:\n# #move depth files\n# class_files = glob.glob(os.path.join(iid_class, 'color*.png'))\n \n# print('copying %d files' %(len(class_files)))\n# for idx in range(len(class_files)): \n# src = class_files[idx]\n# train_or_test, classname, _, filename = get_image_parts(src)\n\n# dst = os.path.join('train_color', classname, train_or_test+'_'+ filename)\n \n# # image directory\n# img_directory = os.path.join('train_color', classname)\n\n# # create folder if not existed\n# if not os.path.exists(img_directory):\n# os.makedirs(img_directory)\n \n# #copying\n# shutil.copy(src, dst)\n \n# # look for all images in sub-folders\n# for folder in move_folders_2:\n# class_folders = glob.glob(os.path.join(folder, '*'))\n \n# for iid_class in class_folders:\n# #move depth files\n# class_files = glob.glob(os.path.join(iid_class, 'depth*.png'))\n \n# print('copying %d files' %(len(class_files)))\n# for idx in range(len(class_files)): \n# src = class_files[idx]\n# if \"0001\" not in src: \n# train_or_test, classname, _, filename = get_image_parts(src)\n\n# dst = os.path.join('test_depth', classname, train_or_test+'_'+ filename)\n\n# # image directory\n# img_directory = os.path.join('test_depth', classname)\n\n# # create folder if not existed\n# if not os.path.exists(img_directory):\n# os.makedirs(img_directory)\n\n# #copying\n# shutil.copy(src, dst)\n# else:\n# print('ignor: %s' %src)\n \n# #move color files \n# for iid_class in class_folders:\n# #move depth files\n# class_files = glob.glob(os.path.join(iid_class, 'color*.png'))\n \n# print('copying %d files' %(len(class_files)))\n# for idx in range(len(class_files)): \n# src = class_files[idx]\n# train_or_test, classname, _, filename = get_image_parts(src)\n\n# dst = os.path.join('test_color', classname, train_or_test+'_'+ filename)\n \n# # image directory\n# img_directory = os.path.join('test_color', classname)\n\n# # create folder if not existed\n# if not os.path.exists(img_directory):\n# os.makedirs(img_directory)\n \n# #copying\n# shutil.copy(src, dst) ",
"copying 528 files\nignor: A/r/depth_17_0001.png\ncopying 524 files\nignor: A/t/depth_19_0001.png\ncopying 536 files\nignor: A/d/depth_3_0001.png\ncopying 523 files\nignor: A/h/depth_7_0001.png\ncopying 572 files\nignor: A/p/depth_15_0001.png\ncopying 533 files\nignor: A/y/depth_24_0001.png\ncopying 524 files\nignor: A/u/depth_20_0001.png\ncopying 512 files\nignor: A/o/depth_14_0001.png\ncopying 522 files\nignor: A/x/depth_23_0001.png\ncopying 519 files\nignor: A/f/depth_5_0001.png\ncopying 530 files\nignor: A/n/depth_13_0001.png\ncopying 516 files\nignor: A/l/depth_11_0001.png\ncopying 471 files\ncopying 524 files\nignor: A/e/depth_4_0001.png\ncopying 528 files\nignor: A/g/depth_6_0001.png\ncopying 528 files\nignor: A/a/depth_0_0001.png\ncopying 525 files\nignor: A/w/depth_22_0001.png\ncopying 557 files\nignor: A/c/depth_2_0001.png\ncopying 516 files\nignor: A/b/depth_1_0001.png\ncopying 507 files\nignor: A/v/depth_21_0001.png\ncopying 527 files\nignor: A/m/depth_12_0001.png\ncopying 518 files\nignor: A/k/depth_10_0001.png\ncopying 516 files\nignor: A/q/depth_16_0001.png\ncopying 515 files\nignor: A/i/depth_8_0001.png\ncopying 527 files\ncopying 523 files\ncopying 535 files\ncopying 522 files\ncopying 571 files\ncopying 532 files\ncopying 523 files\ncopying 511 files\ncopying 521 files\ncopying 518 files\ncopying 529 files\ncopying 515 files\ncopying 470 files\ncopying 523 files\ncopying 527 files\ncopying 527 files\ncopying 524 files\ncopying 556 files\ncopying 515 files\ncopying 506 files\ncopying 526 files\ncopying 517 files\ncopying 515 files\ncopying 514 files\ncopying 550 files\nignor: B/r/depth_17_0001.png\ncopying 514 files\nignor: B/t/depth_19_0001.png\ncopying 552 files\nignor: B/d/depth_3_0001.png\ncopying 545 files\nignor: B/h/depth_7_0001.png\ncopying 642 files\nignor: B/p/depth_15_0001.png\ncopying 537 files\nignor: B/y/depth_24_0001.png\ncopying 542 files\nignor: B/u/depth_20_0001.png\ncopying 529 files\nignor: B/o/depth_14_0001.png\ncopying 622 files\nignor: B/x/depth_23_0001.png\ncopying 530 files\nignor: B/f/depth_5_0001.png\ncopying 544 files\nignor: B/n/depth_13_0001.png\ncopying 577 files\nignor: B/l/depth_11_0001.png\ncopying 749 files\nignor: B/s/depth_18_0001.png\ncopying 565 files\nignor: B/e/depth_4_0001.png\ncopying 544 files\nignor: B/g/depth_6_0001.png\ncopying 536 files\nignor: B/a/depth_0_0001.png\ncopying 648 files\nignor: B/w/depth_22_0001.png\ncopying 549 files\nignor: B/c/depth_2_0001.png\ncopying 577 files\nignor: B/b/depth_1_0001.png\ncopying 628 files\nignor: B/v/depth_21_0001.png\ncopying 582 files\nignor: B/m/depth_12_0001.png\ncopying 777 files\nignor: B/k/depth_10_0001.png\ncopying 540 files\nignor: B/q/depth_16_0001.png\ncopying 543 files\nignor: B/i/depth_8_0001.png\ncopying 549 files\ncopying 513 files\ncopying 551 files\ncopying 544 files\ncopying 641 files\ncopying 536 files\ncopying 541 files\ncopying 528 files\ncopying 621 files\ncopying 529 files\ncopying 543 files\ncopying 576 files\ncopying 748 files\ncopying 564 files\ncopying 543 files\ncopying 535 files\ncopying 647 files\ncopying 548 files\ncopying 576 files\ncopying 627 files\ncopying 581 files\ncopying 776 files\ncopying 539 files\ncopying 542 files\ncopying 530 files\nignor: C/r/depth_17_0001.png\ncopying 529 files\nignor: C/t/depth_19_0001.png\ncopying 526 files\nignor: C/d/depth_3_0001.png\ncopying 536 files\nignor: C/h/depth_7_0001.png\ncopying 534 files\nignor: C/p/depth_15_0001.png\ncopying 543 files\nignor: C/y/depth_24_0001.png\ncopying 530 files\nignor: C/u/depth_20_0001.png\ncopying 548 files\nignor: C/o/depth_14_0001.png\ncopying 531 files\nignor: C/x/depth_23_0001.png\ncopying 518 files\nignor: C/f/depth_5_0001.png\ncopying 564 files\nignor: C/n/depth_13_0001.png\ncopying 586 files\nignor: C/l/depth_11_0001.png\ncopying 507 files\nignor: C/s/depth_18_0001.png\ncopying 529 files\nignor: C/e/depth_4_0001.png\ncopying 532 files\nignor: C/g/depth_6_0001.png\ncopying 524 files\nignor: C/a/depth_0_0001.png\ncopying 890 files\nignor: C/w/depth_22_0001.png\ncopying 743 files\nignor: C/c/depth_2_0001.png\ncopying 533 files\nignor: C/b/depth_1_0001.png\ncopying 536 files\nignor: C/v/depth_21_0001.png\ncopying 538 files\nignor: C/m/depth_12_0001.png\ncopying 539 files\nignor: C/k/depth_10_0001.png\ncopying 541 files\nignor: C/q/depth_16_0001.png\ncopying 530 files\nignor: C/i/depth_8_0001.png\ncopying 529 files\ncopying 528 files\ncopying 525 files\ncopying 535 files\ncopying 533 files\ncopying 542 files\ncopying 529 files\ncopying 547 files\ncopying 530 files\ncopying 517 files\ncopying 563 files\ncopying 585 files\ncopying 506 files\ncopying 528 files\ncopying 531 files\ncopying 523 files\ncopying 889 files\ncopying 742 files\ncopying 532 files\ncopying 535 files\ncopying 537 files\ncopying 538 files\ncopying 540 files\ncopying 529 files\ncopying 774 files\nignor: D/r/depth_17_0001.png\ncopying 530 files\nignor: D/t/depth_19_0001.png\ncopying 526 files\nignor: D/d/depth_3_0001.png\ncopying 562 files\nignor: D/h/depth_7_0001.png\ncopying 532 files\nignor: D/p/depth_15_0001.png\ncopying 536 files\nignor: D/y/depth_24_0001.png\ncopying 532 files\nignor: D/u/depth_20_0001.png\ncopying 539 files\nignor: D/o/depth_14_0001.png\ncopying 538 files\nignor: D/x/depth_23_0001.png\ncopying 525 files\nignor: D/f/depth_5_0001.png\ncopying 530 files\nignor: D/n/depth_13_0001.png\ncopying 565 files\nignor: D/l/depth_11_0001.png\ncopying 535 files\nignor: D/s/depth_18_0001.png\ncopying 528 files\nignor: D/e/depth_4_0001.png\ncopying 542 files\nignor: D/g/depth_6_0001.png\ncopying 547 files\nignor: D/a/depth_0_0001.png\ncopying 536 files\nignor: D/w/depth_22_0001.png\ncopying 531 files\nignor: D/c/depth_2_0001.png\ncopying 572 files\nignor: D/b/depth_1_0001.png\ncopying 544 files\nignor: D/v/depth_21_0001.png\ncopying 542 files\nignor: D/m/depth_12_0001.png\ncopying 540 files\nignor: D/k/depth_10_0001.png\ncopying 550 files\nignor: D/q/depth_16_0001.png\ncopying 522 files\nignor: D/i/depth_8_0001.png\ncopying 773 files\ncopying 529 files\ncopying 525 files\ncopying 561 files\ncopying 531 files\ncopying 535 files\ncopying 531 files\ncopying 538 files\ncopying 537 files\ncopying 524 files\ncopying 529 files\ncopying 564 files\ncopying 534 files\ncopying 527 files\ncopying 541 files\ncopying 546 files\ncopying 535 files\ncopying 530 files\ncopying 571 files\ncopying 543 files\ncopying 541 files\ncopying 539 files\ncopying 549 files\ncopying 521 files\ncopying 563 files\nignor: E/r/depth_17_0001.png\ncopying 532 files\nignor: E/t/depth_19_0001.png\ncopying 545 files\nignor: E/d/depth_3_0001.png\ncopying 535 files\nignor: E/h/depth_7_0001.png\ncopying 528 files\nignor: E/p/depth_15_0001.png\ncopying 522 files\nignor: E/y/depth_24_0001.png\ncopying 530 files\nignor: E/u/depth_20_0001.png\ncopying 533 files\nignor: E/o/depth_14_0001.png\ncopying 523 files\nignor: E/x/depth_23_0001.png\ncopying 528 files\nignor: E/f/depth_5_0001.png\ncopying 531 files\nignor: E/n/depth_13_0001.png\ncopying 515 files\nignor: E/l/depth_11_0001.png\ncopying 527 files\nignor: E/s/depth_18_0001.png\ncopying 540 files\nignor: E/e/depth_4_0001.png\ncopying 538 files\nignor: E/g/depth_6_0001.png\ncopying 546 files\nignor: E/a/depth_0_0001.png\ncopying 514 files\nignor: E/w/depth_22_0001.png\ncopying 541 files\nignor: E/c/depth_2_0001.png\ncopying 535 files\nignor: E/b/depth_1_0001.png\ncopying 528 files\nignor: E/v/depth_21_0001.png\ncopying 526 files\nignor: E/m/depth_12_0001.png\ncopying 570 files\nignor: E/k/depth_10_0001.png\ncopying 530 files\nignor: E/q/depth_16_0001.png\ncopying 526 files\nignor: E/i/depth_8_0001.png\ncopying 562 files\ncopying 531 files\ncopying 544 files\ncopying 534 files\ncopying 527 files\ncopying 521 files\ncopying 529 files\ncopying 532 files\ncopying 522 files\ncopying 527 files\ncopying 530 files\ncopying 514 files\ncopying 526 files\ncopying 539 files\ncopying 537 files\ncopying 545 files\ncopying 513 files\ncopying 540 files\ncopying 534 files\ncopying 527 files\ncopying 525 files\ncopying 569 files\ncopying 529 files\ncopying 525 files\n"
],
[
"cd ..",
"/content\n"
],
[
"# mkdir surrey",
"_____no_output_____"
],
[
"# mkdir surrey/E",
"_____no_output_____"
],
[
"# mv dataset5/* surrey/E/",
"_____no_output_____"
],
[
"# cp /content/gdrive/MyDrive/Surrey_ASL/npy/4_Surrey_MobileNet_E_L3.hdf5.npy 4_Surrey_MobileNet_E_L3.hdf5.npy",
"_____no_output_____"
],
[
"# cp /content/gdrive/MyDrive/Surrey_ASL/npy/4_Surrey_MobileNet_E_L1.hdf5.npy 4_Surrey_MobileNet_E_L1.hdf5.npy",
"_____no_output_____"
],
[
"# cp /content/gdrive/MyDrive/Surrey_ASL/npy/4_Surrey_MobileNet_E_L2.hdf5.npy 4_Surrey_MobileNet_E_L2.hdf5.npy",
"_____no_output_____"
],
[
"# cp /content/gdrive/MyDrive/Surrey_ASL/npy/9_Surrey_InceptionV3_E_L2.hdf5.npy 9_Surrey_InceptionV3_E_L2.hdf5.npy",
"_____no_output_____"
],
[
"# cp /content/gdrive/MyDrive/Surrey_ASL/npy/9_Surrey_InceptionV3_E_L3.hdf5.npy 9_Surrey_InceptionV3_E_L3.hdf5.npy",
"_____no_output_____"
],
[
"# cp /content/gdrive/MyDrive/Surrey_ASL/npy/9_Surrey_InceptionV3_E_L1.hdf5.npy 9_Surrey_InceptionV3_E_L1.hdf5.npy",
"_____no_output_____"
],
[
"import numpy as np\nimport os\n\nmean_pred3_1 = np.load(os.path.join('surrey', 'A', 'npy', 'Surrey_MobileNet_A_0902.npy'))\nmean_pred3_2 = np.load(os.path.join('surrey', 'A', 'npy', 'Surrey_MobileNet_A_L2_0902.npy'))\nmean_pred3_3 = np.load(os.path.join('surrey', 'A', 'npy', 'Surrey_MobileNet_A_L3_0902.npy'))\n\nmean_pred3_4 = np.load(os.path.join('surrey', 'A', 'npy', 'Surrey_InceptionV3_A_0902.npy'))\nmean_pred3_5 = np.load(os.path.join('surrey', 'A', 'npy', 'Surrey_InceptionV3_A_L2_0902.npy'))\nmean_pred3_6 = np.load(os.path.join('surrey', 'A', 'npy', 'Surrey_InceptionV3_A_L3_0902.npy'))\n",
"_____no_output_____"
],
[
"# a1 = mean_pred3_1\n# a2 = mean_pred3_2\n# a3 = mean_pred3_3\n# a4 = mean_pred3_4",
"_____no_output_____"
],
[
"ls",
"\u001b[0m\u001b[01;34mcassava\u001b[0m/ \u001b[01;34mNat19\u001b[0m/\r\n\u001b[01;34mcifar10\u001b[0m/ \u001b[01;34mNat19_2\u001b[0m/\r\n\u001b[01;34mcifar100_png\u001b[0m/ nohup.out\r\n\u001b[01;31mcifar-100-python.tar.gz\u001b[0m \u001b[01;34msiim_isic_melanoma\u001b[0m/\r\n\u001b[01;34mdataset5\u001b[0m/ \u001b[01;34mSTFDG\u001b[0m/\r\n\u001b[01;34mDogAge\u001b[0m/ \u001b[01;34msurrey\u001b[0m/\r\n\u001b[01;34mfashion_mnist\u001b[0m/ Surrey_InceptionV3_D_L1_0902.csv\r\n\u001b[01;31mfingerspelling5.tar.bz2\u001b[0m Surrey_InceptionV3_D_L2_0902.csv\r\n\u001b[01;34mImageNet\u001b[0m/ Surrey_InceptionV3_D_L3_0902.csv\r\n\u001b[01;31mimages.tar\u001b[0m \u001b[01;34msvhn\u001b[0m/\r\n\u001b[01;31mlibcudnn7_7.6.4.38-1+cuda10.1_amd64.deb\u001b[0m \u001b[01;34msvhn_v2\u001b[0m/\r\n\u001b[01;31mlibcudnn7-dev_7.6.4.38-1+cuda10.1_amd64.deb\u001b[0m \u001b[01;31mtest.zip\u001b[0m\r\n"
],
[
"mean_a14 = (mean_pred3_1+mean_pred3_4)/2\nmean_a25 = (mean_pred3_2+mean_pred3_5)/2\nmean_a36 = (mean_pred3_3+mean_pred3_6)/2\nmean_a34 = (mean_pred3_2+mean_pred3_4)/2\n",
"_____no_output_____"
],
[
"from keras.preprocessing.image import ImageDataGenerator\nfrom math import ceil\nimport numpy as np\n\nbatch_size = 20\n\n#Crop-Official Test\ndef random_crop(img, random_crop_size):\n # Note: image_data_format is 'channel_last'\n assert img.shape[2] == 3\n height, width = img.shape[0], img.shape[1]\n dy, dx = random_crop_size\n x = np.random.randint(0, width - dx + 1)\n y = np.random.randint(0, height - dy + 1)\n return img[y:(y+dy), x:(x+dx), :]\n\ndef crop_generator(batches, crop_length):\n \"\"\"Generate random crops from the image batches\"\"\"\n while True:\n batch_x, batch_y = next(batches)\n batch_crops = np.zeros((batch_x.shape[0], crop_length, crop_length, 3))\n for i in range(batch_x.shape[0]):\n batch_crops[i] = random_crop(batch_x[i], (crop_length, crop_length))\n yield (batch_crops, batch_y)\n\ntrain_datagen = ImageDataGenerator(\n\n)\n\ntrain_set = train_datagen.flow_from_directory('surrey/A/train_color',\n target_size = (299, 299),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=True,\n seed=7,\n# subset=\"training\"\n )\n \ntest_datagen_crop = ImageDataGenerator(\n)\n\ntesting_set_crop = test_datagen_crop.flow_from_directory('surrey/A/test_color',\n target_size = (299, 299),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=False,\n seed=7,\n# subset=\"training\"\n )\n#customized generator\ntest_crops = crop_generator(testing_set_crop, 299)\n\nstep_size_test_crop = ceil(testing_set_crop.n/testing_set_crop.batch_size)\n\npredicted_class_indices_mean=np.argmax(mean_a34,axis=1)\nlabels = (train_set.class_indices)\nlabels = dict((v,k) for k,v in labels.items())\nfinalpre = [labels[k] for k in predicted_class_indices_mean]\n\nimport pandas as pd\nfilenames=testing_set_crop.filenames\nresults=pd.DataFrame({\"id\":filenames,\n \"predicted\":finalpre,\n })\nresults.to_csv('surrey_A_Ensemble_0902_meana34.csv')\nresults.head(5)",
"Found 53227 images belonging to 24 classes.\nFound 12547 images belonging to 24 classes.\n"
],
[
"cp surrey_A_Ensemble_0902_meana34.csv /home/bribeiro/Phong/Nat19/surrey_A_Ensemble_0902_meana34.csv",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec629de972df38952043c8d4c5289f495e4d8944 | 1,034,397 | ipynb | Jupyter Notebook | notebooks/rv-uncert.ipynb | adrn/one-datum | f879f23e5ec256e871d03f008199f1866ac53e5c | [
"MIT"
]
| 2 | 2021-01-20T01:36:48.000Z | 2021-01-20T14:33:15.000Z | notebooks/rv-uncert.ipynb | adrn/one-datum | f879f23e5ec256e871d03f008199f1866ac53e5c | [
"MIT"
]
| 11 | 2021-02-03T17:07:49.000Z | 2021-04-07T17:08:17.000Z | notebooks/rv-uncert.ipynb | adrn/one-datum | f879f23e5ec256e871d03f008199f1866ac53e5c | [
"MIT"
]
| 1 | 2021-02-03T17:09:47.000Z | 2021-02-03T17:09:47.000Z | 3,554.628866 | 404,784 | 0.961644 | [
[
[
"%matplotlib inline",
"Matplotlib created a temporary config/cache directory at /tmp/matplotlib-1o491zv4 because the default path (/home/dforeman/.cache/matplotlib) is not a writable directory; it is highly recommended to set the MPLCONFIGDIR environment variable to a writable directory, in particular to speed up the import of Matplotlib and to better support multiprocessing.\n"
],
[
"import numpy as np\nfrom scipy.ndimage import gaussian_filter\n\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport matplotlib.colors as colors\n\nfrom astropy.io import fits",
"_____no_output_____"
],
[
"plt.style.use(\"default\")\nplt.rcParams[\"savefig.dpi\"] = 100\nplt.rcParams[\"figure.dpi\"] = 100\nplt.rcParams[\"font.size\"] = 16\nplt.rcParams[\"font.family\"] = \"sans-serif\"\nplt.rcParams[\"font.sans-serif\"] = [\"Liberation Sans\"]\nplt.rcParams[\"font.cursive\"] = [\"Liberation Sans\"]\nplt.rcParams[\"mathtext.fontset\"] = \"custom\"\nget_ipython().magic('config InlineBackend.figure_format = \"retina\"')",
"_____no_output_____"
],
[
"with fits.open(\"/data/rv_uncertainty_grid.fits\") as f:\n hdr = f[0].header\n mu = f[1].data\n sigma = f[2].data\n \ncolor_bins = np.linspace(hdr[\"MIN_COL\"], hdr[\"MAX_COL\"], hdr[\"NUM_COL\"] + 1)\nmag_bins = np.linspace(hdr[\"MIN_MAG\"], hdr[\"MAX_MAG\"], hdr[\"NUM_MAG\"] + 1)\nivar = 1.0 / sigma ** 2",
"_____no_output_____"
],
[
"mu_smooth = gaussian_filter(np.mean(mu, axis=-1), (1, 0.8))",
"_____no_output_____"
],
[
"plt.figure(figsize=(7, 5))\n\n# plt.pcolor(color_bins, mag_bins, np.mean(mu, axis=-1))\nc = plt.contourf(\n 0.5 * (color_bins[:-1] + color_bins[1:]),\n 0.5 * (mag_bins[:-1] + mag_bins[1:]),\n np.mean(mu, axis=-1),\n levels=15\n)\nplt.contour(\n 0.5 * (color_bins[:-1] + color_bins[1:]),\n 0.5 * (mag_bins[:-1] + mag_bins[1:]),\n mu_smooth,\n colors=\"k\",\n linestyles=\"solid\",\n linewidths=0.5,\n levels=15\n)\n\nplt.colorbar(c, label=r\"$\\ln(\\sigma_\\mathrm{rv})$ [km/s]\")\nplt.ylim(plt.ylim()[::-1])\nplt.ylabel(\"$m_\\mathrm{G}$\")\nplt.xlabel(\"$G_\\mathrm{BP}-G_\\mathrm{RP}$\");",
"_____no_output_____"
],
[
"for m in range(len(color_bins) - 1):\n plt.plot(\n 0.5 * (mag_bins[1:] + mag_bins[:-1]),\n np.mean(mu[:, m], axis=-1),\n \":\",\n lw=0.5,\n color=plt.cm.viridis(m / (len(color_bins) - 1)))\n plt.errorbar(\n 0.5 * (mag_bins[1:] + mag_bins[:-1]),\n mu_smooth[:, m], \n yerr=np.std(mu[:, m], axis=-1),\n fmt=\".-\",\n color=plt.cm.viridis(m / (len(color_bins) - 1)),\n label=\"BP$-$RP = {0:.3f}\".format(0.5*(color_bins[m] + color_bins[m + 1])))\nplt.ylabel(r\"$\\ln (\\sigma_\\mathrm{rv})$ [km/s]\")\nplt.xlabel(\"$m_\\mathrm{G}$\")\nylim = plt.ylim()\n\nax2 = plt.gca().twinx()\nax2.set_ylim(np.exp(ylim))\nax2.set_yscale(\"log\")\nax2.yaxis.set_major_formatter(mpl.ticker.ScalarFormatter())\nax2.yaxis.set_minor_formatter(mpl.ticker.ScalarFormatter())\nax2.tick_params(axis='both', which='major', labelsize=10)\nax2.tick_params(axis='both', which='minor', labelsize=8)\nax2.set_ylabel(r\"$\\sigma_\\mathrm{rv}$ [km/s]\");",
"_____no_output_____"
],
[
"for n in range(len(mag_bins) - 1):\n plt.plot(\n 0.5 * (color_bins[1:] + color_bins[:-1]),\n np.mean(mu[n], axis=-1),\n \":\",\n lw=0.5,\n color=plt.cm.viridis(n / (len(mag_bins) - 1)))\n plt.errorbar(\n 0.5 * (color_bins[1:] + color_bins[:-1]),\n mu_smooth[n], \n yerr=np.std(mu[n], axis=-1),\n fmt=\".-\",\n color=plt.cm.viridis(n / (len(mag_bins) - 1)),\n label=\"$m_G$ = {0:.3f}\".format(0.5*(mag_bins[n] + mag_bins[n + 1])))\nplt.ylabel(r\"$\\ln(\\sigma_\\mathrm{rv})$ [km/s]\")\nplt.xlabel(\"$G_\\mathrm{BP}-G_\\mathrm{RP}$\")\n# plt.legend(fontsize=10)\n\nax2 = plt.gca().twinx()\nax2.set_ylim(np.exp(ylim))\nax2.set_yscale(\"log\")\nax2.yaxis.set_major_formatter(mpl.ticker.ScalarFormatter())\nax2.yaxis.set_minor_formatter(mpl.ticker.ScalarFormatter())\nax2.tick_params(axis='both', which='major', labelsize=10)\nax2.tick_params(axis='both', which='minor', labelsize=8)\nax2.set_ylabel(r\"$\\sigma_\\mathrm{rv}$ [km/s]\");",
"_____no_output_____"
],
[
"plt.hist(np.std(mu, axis=-1).flatten())\nplt.xlabel(\"scatter on the RV uncertainty within a bin\");",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec629e342dc245a9a1699bd0d821f107246e0b9b | 484,999 | ipynb | Jupyter Notebook | notebooks/02_second_model.ipynb | kylercape/GameRecon | f68dd14d7a5227d2501fad6348936fdf47b77bb6 | [
"Apache-2.0"
]
| 1 | 2020-10-21T15:43:43.000Z | 2020-10-21T15:43:43.000Z | notebooks/02_second_model.ipynb | kylercape/GameRecon | f68dd14d7a5227d2501fad6348936fdf47b77bb6 | [
"Apache-2.0"
]
| null | null | null | notebooks/02_second_model.ipynb | kylercape/GameRecon | f68dd14d7a5227d2501fad6348936fdf47b77bb6 | [
"Apache-2.0"
]
| 1 | 2020-10-20T20:03:16.000Z | 2020-10-20T20:03:16.000Z | 1,837.117424 | 477,876 | 0.958548 | [
[
[
"# First model\n\nTrain a small CNN to recognize game screenshots",
"_____no_output_____"
]
],
[
[
"from fastai.vision.all import *\nfrom pathlib import Path",
"_____no_output_____"
],
[
"data_path = Path(\"../Data\")\n\ndata_path.ls()",
"_____no_output_____"
],
[
"files = get_image_files(data_path)",
"_____no_output_____"
],
[
"files",
"_____no_output_____"
],
[
"dls = ImageDataLoaders.from_folder(data_path, item_tfms=Resize(224*3), batch_tfms=aug_transforms(size=224))",
"_____no_output_____"
],
[
"dls.show_batch()",
"_____no_output_____"
],
[
"learn = cnn_learner(dls, resnet34, metrics=error_rate)\nlearn.fine_tune(epochs=10, base_lr=3e-3, freeze_epochs=1)",
"_____no_output_____"
],
[
"learn.show_results(max_n=16)",
"_____no_output_____"
],
[
"interp = Interpretation.from_learner(learn)",
"_____no_output_____"
],
[
"# this will show the images that caused the most confusion from the network\ninterp.plot_top_losses(9, figsize=(15,10))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.