hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ec86df12ae035f0adbcca17423e5d93df52f031a | 190,836 | ipynb | Jupyter Notebook | notebooks/08.3-Fig2v2.ipynb | elijahc/vae | 5cd80518f876d4ca9e97de2ece7c266e3df09cb7 | [
"MIT"
] | null | null | null | notebooks/08.3-Fig2v2.ipynb | elijahc/vae | 5cd80518f876d4ca9e97de2ece7c266e3df09cb7 | [
"MIT"
] | null | null | null | notebooks/08.3-Fig2v2.ipynb | elijahc/vae | 5cd80518f876d4ca9e97de2ece7c266e3df09cb7 | [
"MIT"
] | null | null | null | 269.1622 | 58,308 | 0.90529 | [
[
[
"import os\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib as mpl\n\nimport neptune\n\nfrom src.results.neptune import get_model_files, load_models, load_assemblies, load_params, load_properties,prep_assemblies,NeptuneExperimentRun\n\ndef set_style():\n # This sets reasonable defaults for font size for\n # a figure that will go in a paper\n sns.set_context(\"paper\")\n \n # Set the font to be serif, rather than sans\n# sns.set(font='serif')\n \n # Make the background white, and specify the\n # specific font family\n sns.set_style(\"white\", {\n \"font.family\": \"sans-serif\",\n \"font.serif\": [\"Helvetica\",\"serif\"]\n })",
"_____no_output_____"
]
],
[
[
"## Load dicarlo single unit selectivities",
"_____no_output_____"
]
],
[
[
"import xarray\npix_da = xarray.open_dataarray(os.path.join(proj_root,'data','dicarlo_images','hi_pix.nc'))\npix_da = pix_da.rename(ty='tx',tz='ty')\npix_da = pix_da.set_index({\n 'neuroid':['neuroid_id','region','subregion','layer'],\n 'presentation':['image_id','object_name','category_name','tx','ty','rxy']\n })",
"_____no_output_____"
],
[
"next(runs[0].load_assemblies(['dicarlo.DPX-64.nc'])).sel(layer=0)",
"_____no_output_____"
],
[
"proj_root = '/home/elijahc/projects/vae'",
"_____no_output_____"
],
[
"fp = os.path.join(proj_root,'data','su_selectivity_dicarlo_hi_var.pqt')\nhda = pd.read_parquet(fp).dropna()[['neuroid_id','layer','region','tx','ty','rxy','category_name']]\n\nhda = pd.melt(hda,id_vars=['neuroid_id','layer','region'],value_vars=['tx','ty','rxy','category_name'],var_name='attribute',value_name='selectivity')\nhda['stimulus']='dicarlo'\nhda['model']='macaque'\n\nhda.head()",
"_____no_output_____"
],
[
"hda.groupby(['region','attribute','stimulus']).count()",
"_____no_output_____"
],
[
"'NEPTUNE_API_KEY' in os.environ.keys()",
"_____no_output_____"
],
[
"neptune.init('elijahc/DuplexAE',api_token=os.environ['NEPTUNE_API_KEY'])\nneptune.set_project('elijahc/DuplexAE')",
"WARNING: It is not secure to place API token in your source code. You should treat it as a password to your account. It is strongly recommended to use NEPTUNE_API_TOKEN environment variable instead. Remember not to upload source file with API token to any public repository.\n"
],
[
"exps = neptune.project.get_experiments(id=['DPX-64','DPX-65','DPX-66'])\nruns = [NeptuneExperimentRun(proj_root=proj_root,neptune_exp=e) for e in exps]",
"_____no_output_____"
],
[
"su_select = [pd.read_parquet('../data/DPX6465_su_selectivities.parquet').query('model == \"{}\"'.format(m)) for m in ['full recon', 'no recon']]\nsu_select.append(pd.read_parquet(os.path.join(runs[-1].experiment_dir,'su_selectivity.DPX-66.parquet')))",
"_____no_output_____"
],
[
"dat = pd.concat([pd.concat(su_select).query('stimulus == \"fashion_mnist\"'),hda],sort=True)\ndat.head()",
"_____no_output_____"
],
[
"dat.model = dat.model.replace({'full recon':'combined','no recon':'classify'})",
"_____no_output_____"
],
[
"set_style()\nsns.set_context('talk')\ng = sns.catplot(x='model',y='selectivity',col='attribute', col_wrap=2,hue='layer',hue_order=[0,1,2,3,4,7], order=['classify', 'recon','combined','macaque'],\n kind='bar',data=dat,aspect=1.75,palette='magma',height=4,sharey=False,sharex=True,dodge=True)\n\ng.set(ylim=(0, 0.4))\n# g.axes[-1].set_ylim(0,1)\n\nfor ax,sub_title in zip(g.axes,['Horizontal Translation', 'Vertical Translation', 'Rotation', 'Category Name']):\n ax.get_yaxis().set_minor_locator(mpl.ticker.AutoMinorLocator())\n ax.grid(b=True, which='major', axis='y', color='gray', linewidth=0.5)\n\n ax.get_children()[-4].set_text(sub_title)\n# g.fig\nsns.despine(g.fig)\n# mpl.pyplot.tight_layout()",
"_____no_output_____"
],
[
"g.savefig(os.path.join(proj_root,'figures','pub','neural_networks_revision','su_selectivity0.4max.png'),dpi=200)\ng.savefig(os.path.join(proj_root,'figures','pub','neural_networks_revision','su_selectivity_0.4max.pdf'),dpi=200)",
"_____no_output_____"
],
[
"ax = g.axes[-1]",
"_____no_output_____"
],
[
"ax.figure",
"_____no_output_____"
],
[
"ax.set_ylim(0,0.55)\nax.figure",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec86e785277b10aae362542267c72e5c9fd5732b | 21,816 | ipynb | Jupyter Notebook | notebooks/pca.ipynb | SJern/quest_for_ml | 0ac0d62e9b3e4082d9fabb0c0b15998376bcb40f | [
"MIT"
] | null | null | null | notebooks/pca.ipynb | SJern/quest_for_ml | 0ac0d62e9b3e4082d9fabb0c0b15998376bcb40f | [
"MIT"
] | null | null | null | notebooks/pca.ipynb | SJern/quest_for_ml | 0ac0d62e9b3e4082d9fabb0c0b15998376bcb40f | [
"MIT"
] | null | null | null | 34.738854 | 478 | 0.486432 | [
[
[
"# What do Principal Components Actually do Mathematically?\nI have recently taken an interest in PCA after watching Professor Gilbert Strang’s [PCA lecture](https://www.youtube.com/watch?v=Y4f7K9XF04k). I must have watched at least 15 other videos and read 7 different blog posts on PCA since. They are all very excellent resources, but I found myself somewhat unsatisfied. What they do a lot is teaching us the following:\n- What the PCA promise is;\n- Why that promise is very useful in Data Science; and\n- How to extract these principal components. (Although I don't agree with how some of them do it by applying SVD on the covariance matrix, that can be saved for another post.)\n\nSome of them go the extra mile to show how the promise is being fulfilled graphically. For example, a transformed vector can be shown to be still clustered with its original group in a plot.\n\n## Objective\nTo me, the plot does not provide a visual effect that is striking enough. The components extraction part, on the other hand, mostly talks about how only. Therefore, the objective of this post is to shift our focus onto these 2 areas - to establish a more precise goal before we dive into the components extraction part, and to bring an end to this post with a more striking visual.\n\n## Prerequisites\nThis post is for you if:\n- You have already seen the aforementioned plot - just a bonus actually;\n- You have a decent understanding of what the covariance matrix is about;\n- You have a good foundation in linear algebra; and\n- Your heart is longing to discover the principal components, instead of being told what they are!\n\n## How to Choose P?\nAfter hearing my dissatisfaction, my friend [Calvin](https://calvinfeng.github.io/) recommended this paper by Jonathon Shlens - [A Tutorial on Principal Component Analysis](https://arxiv.org/pdf/1404.1100.pdf) to me. It is by far the best resource I have come across on PCA. However, it's also a bit lengthier than your typical blog post, so the remainder of this post will focus on section 5 of the paper. In there, Jonathon immediately establishes the following goal:\n> The [original] dataset is $X$, an $m × n$ matrix.<br />\n> Find some orthonormal matrix $P$ in $Y = PX$ such that $C_Y \\equiv \\frac{1}{n}YY^T$ is a diagonal matrix.[1]<br />\n> The rows of $P$ shall be principal components of $X$.\n\nAs you might have noticed, $C_Y$ here is the covariance matrix of our rotated dataset $Y$. Why do we want $C_Y$ to be diagonal? Before we address this question, let’s generate a dataset $X$ consisting of 4 features with some random values.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nMostly helper functions. \nSkip ahead unles you would like to follow the steps on your local machine.\n\"\"\"\nfrom IPython.display import Latex, display\nfrom string import ascii_lowercase\nimport numpy as np\nimport pandas as pd\n\nFEAT_NUM, SAMPLE_NUM = 4, 4\n\ndef covariance_matrix(dataset):\n return dataset @ dataset.transpose() / SAMPLE_NUM\n\ndef tabulate(dataset, rotated=False):\n '''\n Label row(s) and column(s) of a matrix by wrapping it in a dataframe.\n '''\n if rotated:\n prefix = 'new_'\n feats = ascii_lowercase[FEAT_NUM:2 * FEAT_NUM]\n else:\n prefix = ''\n feats = ascii_lowercase[0:FEAT_NUM]\n return pd.DataFrame.from_records(dataset, \n columns=['sample{}'.format(num) for num in range(SAMPLE_NUM)], \n index=['{}feat_{}'.format(prefix, feat) for feat in feats])\n\ndef display_df(dataset, latex=False):\n rounded = dataset.round(15)\n if latex:\n display(Latex(rounded.to_latex()))\n else:\n display(rounded)",
"_____no_output_____"
],
[
"x = tabulate(np.random.rand(FEAT_NUM, SAMPLE_NUM))\ndisplay_df(x)",
"_____no_output_____"
]
],
[
[
"$X$ above just looks like a normal dataset. Nothing special. What about its covariance matrix?",
"_____no_output_____"
]
],
[
[
"c_x = covariance_matrix(x)\ndisplay_df(c_x)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"Its covariance matrix $C_X$ doesn't look that intersting either. However, let us recall that the covariance matrix is always a symmetric matrix with the variances on its diagonal and the covariances off-diagonal, i.e., having the following form:\n$\n\\large\n\\begin{vmatrix}\nvar(a, a) & cov(a, b) & cov(a, c) \\\\\ncov(b, a) & var(b, b) & cov(b, c) \\\\\ncov(c, a) & cov(c, b) & var(c, c) \\\\\n\\end{vmatrix}\n$\nLet's also recall that $cov(x, y)$ is zero if and only if feature x and y are uncorrelated. The non-zero convariances in $C_X$ is an indication that there are quite some redundant features in $X$. What we are going to do here is feature extraction. We would like to rotate our dataset in a way such that the change of basis will bring us features that are uncorrelated to each other, i.e., having a new covariance matrix that is diagonal.\n\n### Time to Choose\nWith a clearer goal now, let's figure out how we can achieve it.\n$\n\\begin{array}{ccc}\n\\text{Givens} & \\text{Goal} & \\text{Unknown} \\\\\n\\hline\n\\begin{gathered}Y = PX \\\\ C_X \\equiv \\frac{1}{n}XX^T \\\\ C_Y \\equiv \\frac{1}{n}YY^T\\end{gathered}\n& C_Y\\text{ to be diagonal}\n& \\text{How to Choose }P\\text{?}\n\\end{array}\n$\nFrom the givens above, we are able to derive the relationship between $C_Y$ and $C_X$ in terms of $P$:\n$\nC_Y = \\frac{1}{n}YY^T = \\frac{1}{n}(PX)(PX)^T = \\frac{1}{n}PXX^TP^T\n$\n$\nC_Y = PC_XP^T\n$\nLet's recall one more time that all covariance matrices are symmetric, and any symmetric matrix can be \"Eigendecomposed\" as\n$\nQ{\\Lambda}Q^T\n$\nwhere $Q$ is an orthogonal matrix whose columns are the eigenvectors of the symmetric matrix, and $\\Lambda$ is a diagonal matrix whose entries are the eigenvalues. There is usally more than one way to choose $P$, but Eigendecomposing $C_X$ will prove to make our life much easier. Let's see what we can do with it:\n$\nC_Y = PQ{\\Lambda}Q^TP^T\n$\nSince we know $\\Lambda$ is diagonal and $Q^TQ \\equiv I$, what if we choose $P$ to be $Q^T$?\n$\nC_Y = Q^TQ{\\Lambda}Q^TQ = I{\\Lambda}I\n$\n$\nC_Y = \\Lambda\n$\nVoilà, by choosing $P$ to be eigenvectors of $C_X$, we are able to transform $X$ into $Y$ whose features are uncorrelated to each other!\n\n### Test it\nWell, that was quite convenient, wasn't it? What's even better is that we can demonstrate it in a few lines of code:",
"_____no_output_____"
]
],
[
[
"_, q = np.linalg.eig(c_x) # Eigendecomposition\np = q.transpose()\ny = tabulate(p @ x, rotated=True)\ndisplay_df(y)",
"_____no_output_____"
]
],
[
[
"The transformed dataset $Y$ with the newly extracted features $e$ to $h$, doesn't look like anything either. What about its convariance matrix??",
"_____no_output_____"
]
],
[
[
"c_y = covariance_matrix(y)\ndisplay_df(c_y)",
"_____no_output_____"
]
],
[
[
"Holy moly, isn't this exactly what we were aiming for from the beginning, with just a few lines of code? From a dataset with some redundant and less interesting fetures, we have extracted new features that are much more meaningful to look at, simply by diagonalizing its convariance matrix. Let's wrap this up with some side-by-side comparisons.",
"_____no_output_____"
]
],
[
[
"display_df(x, latex=True)\ndisplay_df(c_x, latex=True)\ndisplay_df(y, latex=True)\ndisplay_df(c_y, latex=True)",
"_____no_output_____"
]
],
[
[
"Look at this. Isn't it just beautiful?",
"_____no_output_____"
],
[
"[1]: The reason orthonormality is part of the goal is that we do not want to do anything more than rotations. We do not want to modify $X$. We only want to re-express $X$ by carefully choosing a change of basis.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ec86eb1c17141d6474b6282823178442eb5dd8fe | 12,504 | ipynb | Jupyter Notebook | Hands-on-ML(Scikit-Learn&TF)/src/Labs/07_Ensemble_Learning.ipynb | Akhilj786/Books | 6d837f6fcc06c1b2ee5a600984f0de9c9f371f18 | [
"Apache-2.0"
] | null | null | null | Hands-on-ML(Scikit-Learn&TF)/src/Labs/07_Ensemble_Learning.ipynb | Akhilj786/Books | 6d837f6fcc06c1b2ee5a600984f0de9c9f371f18 | [
"Apache-2.0"
] | null | null | null | Hands-on-ML(Scikit-Learn&TF)/src/Labs/07_Ensemble_Learning.ipynb | Akhilj786/Books | 6d837f6fcc06c1b2ee5a600984f0de9c9f371f18 | [
"Apache-2.0"
] | null | null | null | 23.637051 | 182 | 0.515675 | [
[
[
"from __future__ import division, print_function, unicode_literals\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
],
[
"try:\n from sklearn.datasets import fetch_openml\n mnist = fetch_openml('mnist_784', version=1)\n mnist.target = mnist.target.astype(np.int64)\nexcept ImportError:\n from sklearn.datasets import fetch_mldata\n mnist = fetch_mldata('MNIST original')",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test=train_test_split(mnist['data'],mnist['target'], test_size=0.2,random_state=42)",
"_____no_output_____"
],
[
"from sklearn.ensemble import RandomForestClassifier,ExtraTreesClassifier\nfrom sklearn.svm import LinearSVC\nfrom sklearn.neural_network import MLPClassifier",
"_____no_output_____"
],
[
"## Problem :#8\n\nlin_clf=LinearSVC(random_state=42)\nrf_clf=RandomForestClassifier(n_estimators=10,random_state=42)\nextra_clf=ExtraTreesClassifier(n_estimators=10,random_state=42)\nmlp_clf=MLPClassifier(random_state=42)\n",
"_____no_output_____"
],
[
"estimators = [lin_clf, rf_clf, extra_clf, mlp_clf]\nfor estimator in estimators:\n print(\"Training the\", estimator)\n estimator.fit(X_train, y_train)",
"Training the LinearSVC(random_state=42)\n"
],
[
"[estimator.score(X_test, y_test) for estimator in estimators]\n",
"_____no_output_____"
],
[
"from sklearn.ensemble import VotingClassifier\nnamed_estimator=[\n ('lr',lin_clf),\n ('rf',rf_clf),\n ('extra',extra_clf),\n ('mlp',mlp_clf)\n]\nvoting_clf=VotingClassifier(estimators=named_estimator,voting='hard')\nvoting_clf.fit(X_train,y_train)",
"/opt/anaconda3/envs/AI-Notebooks/lib/python3.7/site-packages/sklearn/svm/_base.py:977: ConvergenceWarning: Liblinear failed to converge, increase the number of iterations.\n \"the number of iterations.\", ConvergenceWarning)\n"
],
[
"voting_clf.score(X_test,y_test)",
"_____no_output_____"
],
[
"[estimator.score(X_test, y_test) for estimator in voting_clf.estimators_]\n",
"_____no_output_____"
],
[
"voting_clf.estimators",
"_____no_output_____"
],
[
"voting_clf.set_params(lr=None)",
"_____no_output_____"
],
[
"voting_clf.estimators_",
"_____no_output_____"
],
[
"del voting_clf.estimators_[0]",
"_____no_output_____"
],
[
"voting_clf.score(X_test,y_test)",
"_____no_output_____"
],
[
"[estimator.score(X_test, y_test) for estimator in voting_clf.estimators_]\n",
"_____no_output_____"
],
[
"voting_clf.voting='soft'\nprint(voting_clf.score(X_test,y_test))\n[estimator.score(X_test, y_test) for estimator in voting_clf.estimators_]\n",
"0.9693571428571428\n"
]
],
[
[
"# Problem 9",
"_____no_output_____"
]
],
[
[
"X_val_predictions = np.empty((len(X_val), len(estimators)), dtype=np.float32)\n\nfor index, estimator in enumerate(estimators):\n X_val_predictions[:, index] = estimator.predict(X_val)",
"_____no_output_____"
],
[
"X_val_predictions\n",
"_____no_output_____"
],
[
"rnd_forest_blender = RandomForestClassifier(n_estimators=200, oob_score=True, random_state=42)\nrnd_forest_blender.fit(X_val_predictions, y_test)",
"_____no_output_____"
],
[
"rnd_forest_blender.oob_score_\n",
"_____no_output_____"
],
[
"X_test_predictions = np.empty((len(X_test), len(estimators)), dtype=np.float32)\n\nfor index, estimator in enumerate(estimators):\n X_test_predictions[:, index] = estimator.predict(X_test)",
"_____no_output_____"
],
[
"y_pred = rnd_forest_blender.predict(X_test_predictions)\n",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test, y_pred)\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec86f8c8aaf1c398360cd9d69901e5d0b2d388c9 | 30,100 | ipynb | Jupyter Notebook | Machine_learning_template_1.ipynb | arjencupido/METSIM | 1d75405c97d913861ac2906f0fc72edc4d37d07c | [
"Apache-2.0"
] | null | null | null | Machine_learning_template_1.ipynb | arjencupido/METSIM | 1d75405c97d913861ac2906f0fc72edc4d37d07c | [
"Apache-2.0"
] | null | null | null | Machine_learning_template_1.ipynb | arjencupido/METSIM | 1d75405c97d913861ac2906f0fc72edc4d37d07c | [
"Apache-2.0"
] | null | null | null | 430 | 28,290 | 0.938339 | [
[
[
"<a href=\"https://colab.research.google.com/github/arjencupido/METSIM/blob/main/Machine_learning_template_1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom matplotlib import pyplot as plt\n\nys = 200 + np.random.randn(100)\nx = [x for x in range(len(ys))]\n\nplt.plot(x, ys, '-')\nplt.fill_between(x, ys, 195, where=(ys > 195), facecolor='g', alpha=0.6)\n\nplt.title(\"Sample Visualization\")\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
ec87013e5a713cdb59ce41d5d63ca3692e04cc6f | 40,317 | ipynb | Jupyter Notebook | docs/tutorials/pythontutorial.ipynb | phoenixdong/nrn | 211b737e274cf7980d87510d1692adb92ebbd385 | [
"BSD-3-Clause"
] | null | null | null | docs/tutorials/pythontutorial.ipynb | phoenixdong/nrn | 211b737e274cf7980d87510d1692adb92ebbd385 | [
"BSD-3-Clause"
] | null | null | null | docs/tutorials/pythontutorial.ipynb | phoenixdong/nrn | 211b737e274cf7980d87510d1692adb92ebbd385 | [
"BSD-3-Clause"
] | null | null | null | 25.198125 | 1,412 | 0.550835 | [
[
[
"# Introduction to Python",
"_____no_output_____"
],
[
"This page demonstrates some basic Python concepts and essentials. See the <a class=\"reference external\" href=\"http://docs.python.org/tutorial/index.html\">Python Tutorial</a> and <a class=\"reference external\" href=\"http://docs.python.org/library/index.html\">Reference</a> for more exhaustive resources.\n<p>This page provides a brief introduction to:</p>\n<ul class=\"simple\">\n<li>Python syntax</li>\n<li>Variables</li>\n<li>Lists and Dicts</li>\n<li>For loops and iterators</li>\n<li>Functions</li>\n<li>Classes</li>\n<li>Importing modules</li>\n<li>Writing and reading files with Pickling.</li>\n</ul>",
"_____no_output_____"
],
[
"## Displaying results",
"_____no_output_____"
],
[
"The following command simply prints \"Hello world!\". Run it, and then re-evaluate with a different string.",
"_____no_output_____"
]
],
[
[
"print(\"Hello world!\")",
"_____no_output_____"
]
],
[
[
"Here <tt>print</tt> is a function that displays its input on the screen.",
"_____no_output_____"
],
[
"We can use a string's <tt>format</tt> method with <tt>{}</tt> placeholders to substitute calculated values into the output in specific locations; for example:",
"_____no_output_____"
]
],
[
[
"print(\"3 + 4 = {}. Amazing!\".format(3 + 4))",
"_____no_output_____"
]
],
[
[
"Multiple values can be substituted:",
"_____no_output_____"
]
],
[
[
"print(\"The length of the {} is {} microns.\".format(\"apical dendrite\", 100))",
"_____no_output_____"
]
],
[
[
"There are more sophisticated ways of using <tt>format</tt>\n(see <a class=\"reference external\" href=\"https://docs.python.org/3/library/string.html#format-examples\">examples</a> on the Python website).",
"_____no_output_____"
],
[
"## Variables: Strings, numbers, and dynamic type casting",
"_____no_output_____"
],
[
"Variables are easily assigned:",
"_____no_output_____"
]
],
[
[
"my_name = \"Tom\"\nmy_age = 45",
"_____no_output_____"
]
],
[
[
"Let’s work with these variables.",
"_____no_output_____"
]
],
[
[
"print(my_name)",
"_____no_output_____"
],
[
"print(my_age)",
"_____no_output_____"
]
],
[
[
"Strings can be combined with the + operator.",
"_____no_output_____"
]
],
[
[
"greeting = \"Hello, \" + my_name\nprint(greeting)",
"_____no_output_____"
]
],
[
[
"Let’s move on to numbers.",
"_____no_output_____"
]
],
[
[
"print(my_age)",
"_____no_output_____"
]
],
[
[
"If you try using the + operator on my_name and my_age:",
"_____no_output_____"
],
[
"```python\nprint(my_name + my_age)\n```",
"_____no_output_____"
],
[
"<p>You will get a <a class=\"reference external\" href=\"https://docs.python.org/3/library/exceptions.html#TypeError\" title=\"(in Python v3.6)\"><code class=\"xref py py-class docutils literal\"><span class=\"pre\">TypeError</span></code></a>. What is wrong?</p>\n<p>my_name is a string and my_age is a number. Adding in this context does not make any sense.</p>\n<p>We can determine an object’s type with the <code class=\"xref py py-func docutils literal\"><span class=\"pre\">type()</span></code> function.</p>",
"_____no_output_____"
]
],
[
[
"type(my_name)",
"_____no_output_____"
],
[
"type(my_age)",
"_____no_output_____"
]
],
[
[
"<p>The function <a class=\"reference external\" href=\"https://docs.python.org/3/library/functions.html#isinstance\" title=\"(in Python v3.6)\"><code class=\"xref py py-func docutils literal\"><span class=\"pre\">isinstance()</span></code></a> is typically more useful than comparing variable types as it allows handling subclasses:</p>",
"_____no_output_____"
]
],
[
[
"isinstance(my_name, str)",
"_____no_output_____"
],
[
"my_valid_var = None\nif my_valid_var is not None:\n print(my_valid_var)\nelse:\n print(\"The variable is None!\")",
"_____no_output_____"
]
],
[
[
"## Arithmetic: <tt>+</tt>, <tt>-</tt>, <tt>*</tt>, <tt>/</tt>, <tt>**</tt>, <tt>%</tt> and comparisons",
"_____no_output_____"
]
],
[
[
"2 * 6",
"_____no_output_____"
]
],
[
[
"Check for equality using two equal signs:",
"_____no_output_____"
]
],
[
[
"2 * 6 == 4 * 3",
"_____no_output_____"
],
[
"5 < 2",
"_____no_output_____"
],
[
"5 < 2 or 3 < 5",
"_____no_output_____"
],
[
"5 < 2 and 3 < 5",
"_____no_output_____"
],
[
"2 * 3 != 5",
"_____no_output_____"
]
],
[
[
"<tt>%</tt> is the modulus operator. It returns the remainder from doing a division.",
"_____no_output_____"
]
],
[
[
"5 % 3",
"_____no_output_____"
]
],
[
[
"The above is because 5 / 3 is 1 with a remainder of 2.",
"_____no_output_____"
],
[
"In decimal, that is:",
"_____no_output_____"
]
],
[
[
"5 / 3",
"_____no_output_____"
]
],
[
[
"<div style='background-color:pink'>\n<p class=\"first admonition-title\"><b>Warning</b></p>\n<p class=\"last\">In older versions of Python (prior to 3.0), the <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">/</span></code> operator when used on integers performed integer division; i.e. <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">3/2</span></code> returned <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">1</span></code>, but <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">3/2.0</span></code> returned <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">1.5</span></code>. Beginning with Python 3.0, the <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">/</span></code> operator returns a float if integers do not divide evenly; i.e. <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">3/2</span></code> returns <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">1.5</span></code>. Integer division is still available using the <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">//</span></code> operator, i.e. <code class=\"docutils literal\" style='background-color:pink'><span class=\"pre\">3</span> <span class=\"pre\">//</span> <span class=\"pre\">2</span></code> evaluates to 1.</p></div>",
"_____no_output_____"
],
[
"## Making choices: if, else",
"_____no_output_____"
]
],
[
[
"section = 'soma'\nif section == 'soma':\n print('working on the soma')\nelse:\n print('not working on the soma')",
"_____no_output_____"
],
[
"p = 0.06\nif p < 0.05:\n print('statistically significant')\nelse:\n print('not statistically significant')",
"_____no_output_____"
]
],
[
[
"Note that here we used a single quote instead of a double quote to indicate the beginning and end of a string. Either way is fine, as long as the beginning and end of a string match.",
"_____no_output_____"
],
[
"Python also has a special object called <tt>None</tt>. This is one way you can specify whether or not an object is valid. When doing comparisons with <tt>None</tt>, it is generally recommended to use <tt>is</tt> and <tt>is not</tt>:",
"_____no_output_____"
]
],
[
[
"postsynaptic_cell = None\nif postsynaptic_cell is not None:\n print(\"Connecting to postsynaptic cell\")\nelse:\n print(\"No postsynaptic cell to connect to\")",
"_____no_output_____"
]
],
[
[
"## Lists",
"_____no_output_____"
],
[
"Lists are comma-separated values surrounded by square brackets:",
"_____no_output_____"
]
],
[
[
"my_list = [1, 3, 5, 8, 13]\nprint(my_list)",
"_____no_output_____"
]
],
[
[
"Lists are zero-indexed. That is, the first element is 0.",
"_____no_output_____"
]
],
[
[
"my_list[0]",
"_____no_output_____"
]
],
[
[
"You may often find yourself wanting to know how many items are in a list.",
"_____no_output_____"
]
],
[
[
"len(my_list)",
"_____no_output_____"
]
],
[
[
"Python interprets negative indices as counting backwards from the end of the list. That is, the -1 index refers to the last item, the -2 index refers to the second-to-last item, etc.",
"_____no_output_____"
]
],
[
[
"print(my_list)\nprint(my_list[-1])",
"_____no_output_____"
]
],
[
[
"“Slicing” is extracting particular sub-elements from the list in a particular range. However, notice that the right-side is excluded, and the left is included.",
"_____no_output_____"
]
],
[
[
"print(my_list)\nprint(my_list[2:4]) # Includes the range from index 2 to 3\nprint(my_list[2:-1]) # Includes the range from index 2 to the element before -1\nprint(my_list[:2]) # Includes everything before index 2\nprint(my_list[2:]) # Includes everything from index 2",
"_____no_output_____"
]
],
[
[
"We can check if our list contains a given value using the <tt>in</tt> operator:",
"_____no_output_____"
]
],
[
[
"42 in my_list",
"_____no_output_____"
],
[
"5 in my_list",
"_____no_output_____"
]
],
[
[
"We can append an element to a list using the <tt>append</tt> method:",
"_____no_output_____"
]
],
[
[
"my_list.append(42)\nprint(my_list)",
"_____no_output_____"
]
],
[
[
"To make a variable equal to a copy of a list, set it equal to <tt>list(the_old_list)</tt>. For example:",
"_____no_output_____"
]
],
[
[
"list_a = [1, 3, 5, 8, 13]\nlist_b = list(list_a)\nlist_b.reverse()\nprint(\"list_a = \" + str(list_a))\nprint(\"list_b = \" + str(list_b))",
"_____no_output_____"
]
],
[
[
"In particular, note that assigning one list to another variable <i>does not</i> make a copy. Instead, it just gives another way of accessing the same list.",
"_____no_output_____"
]
],
[
[
"print('initial: list_a[0] = %g' % list_a[0])\nfoo = list_a\nfoo[0] = 42\nprint('final: list_a[0] = %g' % list_a[0])",
"_____no_output_____"
]
],
[
[
"<p>If the second line in the previous example was replaced with <tt>list_b = list_a</tt>, then what would happen?</p>\n<p>In that case, <tt>list_b</tt> is the same list as <tt>list_a</tt> (as opposed to a copy), so when <tt>list_b</tt> was reversed so is <tt>list_a</a> (since <tt>list_b<tt> <i>is</i> <tt>list_a</tt>).</p>",
"_____no_output_____"
],
[
"We can sort a list and get a new list using the <tt>sorted</tt> function. e.g.",
"_____no_output_____"
]
],
[
[
"sorted(['soma', 'basal', 'apical', 'axon', 'obliques'])",
"_____no_output_____"
]
],
[
[
"Here we have sorted by alphabetical order. If our list had only numbers, it would by default sort by numerical order:",
"_____no_output_____"
]
],
[
[
"sorted([6, 1, 165, 1.51, 4])",
"_____no_output_____"
]
],
[
[
"If we wanted to sort by another attribute, we can specify a function that returns that attribute value as the optional <tt>key</tt> keyword argument. For example, to sort our list of neuron parts by the length of the name (returned by the function <tt>len</tt>):",
"_____no_output_____"
]
],
[
[
"sorted(['soma', 'basal', 'apical', 'axon', 'obliques'], key=len)",
"_____no_output_____"
]
],
[
[
"<p>Lists can contain arbitrary data types, but if you find yourself doing this, you should probably consider making classes or dictionaries (described below).</p>",
"_____no_output_____"
]
],
[
[
"confusing_list = ['abc', 1.0, 2, \"another string\"]\nprint(confusing_list)\nprint(confusing_list[3])",
"_____no_output_____"
]
],
[
[
"It is sometimes convenient to assign the elements of a list with a known length to an equivalent number of variables:",
"_____no_output_____"
]
],
[
[
"first, second, third = [42, 35, 25]\nprint(second)",
"_____no_output_____"
]
],
[
[
"The <tt>range</tt> function can be used to create a list-like object (prior to Python 3.0, it generated lists; beginning with 3.0, it generates a more memory efficient structure) of evenly spaced integers; a list can be produced from this using the <tt>list</tt> function. With one argument, range produces integers from 0 to the argument, with two integer arguments, it produces integers between the two values, and with three arguments, the third specifies the interval between the first two. The ending value is not included.",
"_____no_output_____"
]
],
[
[
"print(list(range(10))) # [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]",
"_____no_output_____"
],
[
"print(list(range(0, 10))) # [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]",
"_____no_output_____"
],
[
"print(list(range(3, 10))) # [3, 4, 5, 6, 7, 8, 9]",
"_____no_output_____"
],
[
"print(list(range(0, 10, 2))) # [0, 2, 4, 6, 8]",
"_____no_output_____"
],
[
"print(list(range(0, -10))) # []",
"_____no_output_____"
],
[
"print(list(range(0, -10, -1))) # [0, -1, -2, -3, -4, -5, -6, -7, -8, -9]",
"_____no_output_____"
],
[
"print(list(range(0, -10, -2))) # [0, -2, -4, -6, -8]",
"_____no_output_____"
]
],
[
[
"For non-integer ranges, use <tt><a href=\"https://docs.scipy.org/doc/numpy/reference/generated/numpy.arange.html\">numpy.arange</a></tt> from the <tt><a href=\"http://www.numpy.org/\">numpy</a></tt> module.",
"_____no_output_____"
],
[
"### List comprehensions (set theory)",
"_____no_output_____"
],
[
"List comprehensions provide a rule for building a list from another list (or any other Python iterable).\n\nFor example, the list of all integers from 0 to 9 inclusive is <tt>range(10)</tt> as shown above. We can get a list of the squares of all those integers via:",
"_____no_output_____"
]
],
[
[
"[x ** 2 for x in range(10)]",
"_____no_output_____"
]
],
[
[
"Including an <tt>if</tt> condition allows us to filter the list:\n\nWhat are all the integers <tt>x</tt> between 0 and 29 inclusive that satisfy <tt>(x - 3) * (x - 10) == 0</tt>?",
"_____no_output_____"
]
],
[
[
"[x for x in range(30) if (x - 3) * (x - 10) == 0]",
"_____no_output_____"
]
],
[
[
"## For loops and iterators",
"_____no_output_____"
],
[
"We can iterate over elements in a list by following the format: <tt>for element in list:</tt> Notice that indentation is important in Python! After a colon, the block needs to be indented. (Any consistent indentation will work, but the Python standard is 4 spaces).",
"_____no_output_____"
]
],
[
[
"cell_parts = ['soma', 'axon', 'dendrites']\nfor part in cell_parts:\n print(part)",
"_____no_output_____"
]
],
[
[
"Note that we are iterating over the elements of a list; much of the time, the index of the items is irrelevant. If, on the other hand, we need to know the index as well, we can use <tt>enumerate</tt>:",
"_____no_output_____"
]
],
[
[
"cell_parts = ['soma', 'axon', 'dendrites']\nfor part_num, part in enumerate(cell_parts):\n print('%d %s' % (part_num, part))",
"_____no_output_____"
]
],
[
[
"Multiple aligned lists (such as occurs with time series data) can be looped over simultaneously using <tt>zip</tt>:",
"_____no_output_____"
]
],
[
[
"cell_parts = ['soma', 'axon', 'dendrites']\ndiams = [20, 2, 3]\nfor diam, part in zip(diams, cell_parts):\n print('%10s %g' % (part, diam))",
"_____no_output_____"
]
],
[
[
"Another example:",
"_____no_output_____"
]
],
[
[
"y = ['a', 'b', 'c', 'd', 'e']\nx = list(range(len(y)))\nprint(\"x = {}\".format(x))\nprint(\"y = {}\".format(y))\nprint(list(zip(x, y)))",
"_____no_output_____"
]
],
[
[
"This is a list of tuples. Given a list of tuples, then we iterate with each tuple.",
"_____no_output_____"
]
],
[
[
"for x_val, y_val in zip(x, y):\n print(\"index {}: {}\".format(x_val, y_val))",
"_____no_output_____"
]
],
[
[
"Tuples are similar to lists, except they are immutable (cannot be changed). You can retrieve individual elements of a tuple, but once they are set upon creation, you cannot change them. Also, you cannot add or remove elements of a tuple.",
"_____no_output_____"
]
],
[
[
"my_tuple = (1, 'two', 3)\nprint(my_tuple)\nprint(my_tuple[1])",
"_____no_output_____"
]
],
[
[
"Attempting to modify a tuple, e.g.",
"_____no_output_____"
],
[
"```python\nmy_tuple[1] = 2\n```",
"_____no_output_____"
],
[
"will cause a <a class=\"reference external\" href=\"https://docs.python.org/3/library/exceptions.html#TypeError\" title=\"(in Python v3.6)\"><code class=\"xref py py-class docutils literal\"><span class=\"pre\">TypeError</span></code></a>.",
"_____no_output_____"
],
[
"Because you cannot modify an element in a tuple, or add or remove individual elements of it, it can operate in Python more efficiently than a list. A tuple can even serve as a key to a dictionary.",
"_____no_output_____"
],
[
"## Dictionaries",
"_____no_output_____"
],
[
"A dictionary (also called a dict or hash table) is a set of (key, value) pairs, denoted by curly brackets:",
"_____no_output_____"
]
],
[
[
"about_me = {'name': my_name, 'age': my_age, 'height': \"5'8\"}\nprint(about_me)",
"_____no_output_____"
]
],
[
[
"You can obtain values by referencing the key:",
"_____no_output_____"
]
],
[
[
"print(about_me['height'])",
"_____no_output_____"
]
],
[
[
"Similarly, we can modify existing values by referencing the key.",
"_____no_output_____"
]
],
[
[
"about_me['name'] = \"Thomas\"\nprint(about_me)",
"_____no_output_____"
]
],
[
[
"We can even add new values.",
"_____no_output_____"
]
],
[
[
"about_me['eye_color'] = \"brown\"\nprint(about_me)",
"_____no_output_____"
]
],
[
[
"We can use curly braces with keys to indicate dictionary fields when using <tt>format</tt>. e.g.",
"_____no_output_____"
]
],
[
[
"print('I am a {age} year old person named {name}. Again, my name is {name}.'.format(**about_me))",
"_____no_output_____"
]
],
[
[
"Important: note the use of the <tt>**</tt> inside the <tt>format</tt> call.",
"_____no_output_____"
],
[
"We can iterate keys (<tt>.keys()</tt>), values (<tt>.values()</tt>) or key-value value pairs (<tt>.items()</tt>)in the dict. Here is an example of key-value pairs.",
"_____no_output_____"
]
],
[
[
"for k, v in about_me.items():\n print('key = {:10s} val = {}'.format(k, v))",
"_____no_output_____"
]
],
[
[
"To test for the presence of a key in a dict, we just ask:",
"_____no_output_____"
]
],
[
[
"if 'hair_color' in about_me:\n print(\"Yes. 'hair_color' is a key in the dict\")\nelse:\n print(\"No. 'hair_color' is NOT a key in the dict\")",
"_____no_output_____"
]
],
[
[
"Dictionaries can be nested, e.g.",
"_____no_output_____"
]
],
[
[
"neurons = {\n 'purkinje cells': {\n 'location': 'cerebellum',\n 'role': 'motor movement'\n },\n 'ca1 pyramidal cells': {\n 'location': 'hippocampus',\n 'role': 'learning and memory'\n }\n}\nprint(neurons['purkinje cells']['location'])",
"_____no_output_____"
]
],
[
[
"## Functions",
"_____no_output_____"
],
[
"Functions are defined with a “def” keyword in front of them, end with a colon, and the next line is indented. Indentation of 4-spaces (again, any non-zero consistent amount will do) demarcates functional blocks.",
"_____no_output_____"
]
],
[
[
"def print_hello():\n print(\"Hello\")",
"_____no_output_____"
]
],
[
[
"Now let's call our function.",
"_____no_output_____"
]
],
[
[
"print_hello()",
"_____no_output_____"
]
],
[
[
"We can also pass in an argument.",
"_____no_output_____"
]
],
[
[
"def my_print(the_arg):\n print(the_arg)",
"_____no_output_____"
]
],
[
[
"Now try passing various things to the <tt>my_print()</tt> function.",
"_____no_output_____"
]
],
[
[
"my_print(\"Hello\")",
"_____no_output_____"
]
],
[
[
"We can even make default arguments.",
"_____no_output_____"
]
],
[
[
"def my_print(message=\"Hello\"):\n print(message)\n\nmy_print()\nmy_print(list(range(4)))",
"_____no_output_____"
]
],
[
[
"And we can also return values.",
"_____no_output_____"
]
],
[
[
"def fib(n=5):\n \"\"\"Get a Fibonacci series up to n.\"\"\"\n a, b = 0, 1\n series = [a]\n while b < n:\n a, b = b, a + b\n series.append(a)\n return series\n\nprint(fib())",
"_____no_output_____"
]
],
[
[
"Note the assignment line for a and b inside the while loop. That line says that a becomes the old value of b and that b becomes the old value of a plus the old value of b. The ability to calculate multiple values before assigning them allows Python to do things like swapping the values of two variables in one line while many other programming languages would require the introduction of a temporary variable.\n\nWhen a function begins with a string as in the above, that string is known as a doc string, and is shown whenever <tt>help</tt> is invoked on the function (this, by the way, is a way to learn more about Python's many functions):",
"_____no_output_____"
]
],
[
[
"help(fib)",
"_____no_output_____"
]
],
[
[
"You may have noticed the string beginning the <tt>fib</tt> function was triple-quoted. This enables a string to span multiple lines.",
"_____no_output_____"
]
],
[
[
"multi_line_str = \"\"\"This is the first line\nThis is the second,\nand a third.\"\"\"\n\nprint(multi_line_str)",
"_____no_output_____"
]
],
[
[
"## Classes",
"_____no_output_____"
],
[
"Objects are instances of a <tt>class</tt>. They are useful for encapsulating ideas, and mostly for having multiple instances of a structure. (In NEURON, for example, one might use a class to represent a neuron type and create many instances.) Usually you will have an <tt>__init__()</tt> method. Also note that every method of the class will have <tt>self</tt> as the first argument. While <tt>self</tt> has to be listed in the argument list of a class's method, you do not pass a <tt>self</tt> argument when calling any of the class’s methods; instead, you refer to those methods as <tt>self.method_name</tt>.",
"_____no_output_____"
]
],
[
[
"class Contact(object):\n \"\"\"A given person for my database of friends.\"\"\"\n\n def __init__(self, first_name=None, last_name=None, email=None, phone=None):\n self.first_name = first_name\n self.last_name = last_name\n self.email = email\n self.phone = phone\n\n def print_info(self):\n \"\"\"Print all of the information of this contact.\"\"\"\n my_str = \"Contact info:\"\n if self.first_name:\n my_str += \" \" + self.first_name\n if self.last_name:\n my_str += \" \" + self.last_name\n if self.email:\n my_str += \" \" + self.email\n if self.phone:\n my_str += \" \" + self.phone\n print(my_str)",
"_____no_output_____"
]
],
[
[
"By convention, the first letter of a class name is capitalized. Notice in the class definition above that the object can contain fields, which are used within the class as <tt>self.field</tt>. This field can be another method in the class, or another object of another class.",
"_____no_output_____"
],
[
"Let's make a couple instances of Contact.",
"_____no_output_____"
]
],
[
[
"bob = Contact('Bob','Smith')\njoe = Contact(email='[email protected]')",
"_____no_output_____"
]
],
[
[
"Notice that in the first case, if we are filling each argument, we do not need to explicitly denote “first_name” and “last_name”. However, in the second case, since “first” and “last” are omitted, the first parameter passed in would be assigned to the first_name field so we have to explicitly set it to “email”.",
"_____no_output_____"
],
[
"Let’s set a field.",
"_____no_output_____"
]
],
[
[
"joe.first_name = \"Joe\"",
"_____no_output_____"
]
],
[
[
"Similarly, we can retrieve fields from the object.",
"_____no_output_____"
]
],
[
[
"the_name = joe.first_name\nprint(the_name)",
"_____no_output_____"
]
],
[
[
"And we call methods of the object using the format instance.method().",
"_____no_output_____"
]
],
[
[
"joe.print_info()",
"_____no_output_____"
]
],
[
[
"Remember the importance of docstrings!",
"_____no_output_____"
]
],
[
[
"help(Contact)",
"_____no_output_____"
]
],
[
[
"## Importing modules",
"_____no_output_____"
],
[
"Extensions to core Python are made by importing modules, which may contain more variables, objects, methods, and functions. Many modules come with Python, but are not part of its core. Other packages and modules have to be installed.\n\nThe <tt>numpy</tt> module contains a function called <tt>arange()</tt> that is similar to Python’s <tt>range()</tt> function, but permits non-integer steps.",
"_____no_output_____"
]
],
[
[
"import numpy\nmy_vec = numpy.arange(0, 1, 0.1)\nprint(my_vec)",
"_____no_output_____"
]
],
[
[
"<b>Note</b>: <tt>numpy</tt> is available in many distributions of Python, but it is not part of Python itself. If the <tt>import numpy</tt> line gave an error message, you either do not have numpy installed or Python cannot find it for some reason. You should resolve this issue before proceeding because we will use numpy in some of the examples in other parts of the tutorial. The standard tool for installing Python modules is called pip; other options may be available depending on your platform.",
"_____no_output_____"
],
[
"We can get 20 evenly spaced values between 0 and $\\pi$ using <tt>numpy.linspace</tt>:",
"_____no_output_____"
]
],
[
[
"x = numpy.linspace(0, numpy.pi, 20)",
"_____no_output_____"
],
[
"print(x)",
"_____no_output_____"
]
],
[
[
"NumPy provides vectorized trig (and other) functions. For example, we can get another array with the sines of all those x values via:",
"_____no_output_____"
]
],
[
[
"y = numpy.sin(x)",
"_____no_output_____"
],
[
"print(y)",
"_____no_output_____"
]
],
[
[
"The <tt>bokeh</tt> module is one way to plot graphics that works especially well in a Jupyter notebook environment. To use this library in a Jupyter notebook, we first load it and tell it to display in the notebook:",
"_____no_output_____"
]
],
[
[
"from bokeh.io import output_notebook\nimport bokeh.plotting as plt\noutput_notebook() # skip this line if not working in Jupyter",
"_____no_output_____"
]
],
[
[
"Here we plot y = sin(x) vs x:",
"_____no_output_____"
]
],
[
[
"f = plt.figure(x_axis_label='x', y_axis_label='sin(x)')\nf.line(x, y, line_width=2)\nplt.show(f)",
"_____no_output_____"
]
],
[
[
"## Pickling objects",
"_____no_output_____"
],
[
"There are various file io operations in Python, but one of the easiest is “<a class=\"reference external\" href=\"https://docs.python.org/3/library/pickle.html#module-pickle\" title=\"(in Python v3.6)\"><code class=\"xref py py-mod docutils literal\"><span class=\"pre\">Pickling</span></code></a>”, which attempts to save a Python object to a file for later restoration with the load command.",
"_____no_output_____"
]
],
[
[
"import pickle\ncontacts = [joe, bob] # Make a list of contacts\n\nwith open('contacts.p', 'wb') as pickle_file: # Make a new file\n pickle.dump(contacts, pickle_file) # Write contact list\n\nwith open('contacts.p', 'rb') as pickle_file: # Open the file for reading\n contacts2 = pickle.load(pickle_file) # Load the pickled contents\n\nfor elem in contacts2:\n elem.print_info()",
"_____no_output_____"
]
],
[
[
"The next part of this tutorial introduces basic NEURON commands.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec8710ccd35ea33a3e08873d4f70318d2b1392a0 | 12,883 | ipynb | Jupyter Notebook | notebook/4-2-asyncio.ipynb | xuanyeleyou/easy-scraping-tutorial | e52c45d27ffce90a7bfe12ea1da09f049e052099 | [
"MIT"
] | 2 | 2020-05-16T07:47:45.000Z | 2020-05-16T07:48:48.000Z | notebook/4-2-asyncio.ipynb | yubo105139/easy-scraping-tutorial | e52c45d27ffce90a7bfe12ea1da09f049e052099 | [
"MIT"
] | null | null | null | notebook/4-2-asyncio.ipynb | yubo105139/easy-scraping-tutorial | e52c45d27ffce90a7bfe12ea1da09f049e052099 | [
"MIT"
] | 1 | 2018-04-06T07:05:29.000Z | 2018-04-06T07:05:29.000Z | 27.825054 | 184 | 0.480168 | [
[
[
"# Asyncio tutorial\n\n## A normal way in python\n**Firstly, let's see a running time in a normal way.**",
"_____no_output_____"
]
],
[
[
"import time\n\n\ndef job(t):\n print('Start job ', t)\n time.sleep(t) # wait for \"t\" seconds\n print('Job ', t, ' takes ', t, ' s')\n \n\ndef main():\n [job(t) for t in range(1, 3)]\n \n \nt1 = time.time()\nmain()\nprint(\"NO async total time : \", time.time() - t1)",
"Start job 1\n"
]
],
[
[
"## Translate above to async\n**Now, let's see the running time using asyncio**",
"_____no_output_____"
]
],
[
[
"import asyncio\n\n\nasync def job(t):\n print('Start job ', t)\n await asyncio.sleep(t) # wait for \"t\" seconds, it will look for another job while await\n print('Job ', t, ' takes ', t, ' s')\n \n\nasync def main(loop):\n tasks = [loop.create_task(job(t)) for t in range(1, 3)] # just create, not run job\n await asyncio.wait(tasks) # run jobs and wait for all tasks done\n\nt1 = time.time()\nloop = asyncio.get_event_loop()\nloop.run_until_complete(main(loop))\n# loop.close() # Ipython notebook gives error if close loop\nprint(\"Async total time : \", time.time() - t1)\n",
"Start job 1\nStart job 2\n"
]
],
[
[
"## A normal way in crawling webpage\n**We can use this machanism in requesting for a website. Await for download a page and switch to do another job.**",
"_____no_output_____"
]
],
[
[
"import requests\n\nURL = 'https://morvanzhou.github.io/'\n\n\ndef normal(): \n for i in range(2):\n r = requests.get(URL)\n url = r.url\n print(url)\n \nt1 = time.time()\nnormal()\nprint(\"Normal total time:\", time.time()-t1)",
"https://morvanzhou.github.io/\nhttps://morvanzhou.github.io/\nNormal total time: 0.3869960308074951\n"
]
],
[
[
"## Translate above to async using aiohttp\n**We have to install another useful package called [aiohttp](https://aiohttp.readthedocs.io/en/stable/index.html). You can simply run \"pip3 install aiohttp\" in your terminal.**",
"_____no_output_____"
]
],
[
[
"import aiohttp\n\n\nasync def job(session):\n response = await session.get(URL)\n return str(response.url)\n\n\nasync def main(loop):\n async with aiohttp.ClientSession() as session:\n tasks = [loop.create_task(job(session)) for _ in range(2)]\n finished, unfinished = await asyncio.wait(tasks)\n all_results = [r.result() for r in finished] # get return from job\n print(all_results)\n \nt1 = time.time()\nloop = asyncio.get_event_loop()\nloop.run_until_complete(main(loop))\n# loop.close() # Ipython notebook gives error if close loop\nprint(\"Async total time:\", time.time() - t1)",
"['https://morvanzhou.github.io/', 'https://morvanzhou.github.io/']\nAsync total time: 0.11447715759277344\n"
]
],
[
[
"## Compare async with multiprocessing\n**The following code scrape my website with async**",
"_____no_output_____"
]
],
[
[
"import aiohttp\nimport asyncio\nimport time\nfrom bs4 import BeautifulSoup\nfrom urllib.request import urljoin\nimport re\nimport multiprocessing as mp\n\n# base_url = \"https://morvanzhou.github.io/\"\nbase_url = \"http://127.0.0.1:4000/\"\n\n# DON'T OVER CRAWL THE WEBSITE OR YOU MAY NEVER VISIT AGAIN\nif base_url != \"http://127.0.0.1:4000/\":\n restricted_crawl = True\nelse:\n restricted_crawl = False\n \n \nseen = set()\nunseen = set([base_url])\n\n\ndef parse(html):\n soup = BeautifulSoup(html, 'lxml')\n urls = soup.find_all('a', {\"href\": re.compile('^/.+?/$')})\n title = soup.find('h1').get_text().strip()\n page_urls = set([urljoin(base_url, url['href']) for url in urls])\n url = soup.find('meta', {'property': \"og:url\"})['content']\n return title, page_urls, url\n\n\nasync def crawl(url, session):\n r = await session.get(url)\n html = await r.text()\n await asyncio.sleep(0.1) # slightly delay for downloading\n return html\n\n\nasync def main(loop):\n pool = mp.Pool(8) # slightly affected\n async with aiohttp.ClientSession() as session:\n count = 1\n while len(unseen) != 0:\n print('\\nAsync Crawling...')\n tasks = [loop.create_task(crawl(url, session)) for url in unseen]\n finished, unfinished = await asyncio.wait(tasks)\n htmls = [f.result() for f in finished]\n \n print('\\nDistributed Parsing...')\n parse_jobs = [pool.apply_async(parse, args=(html,)) for html in htmls]\n results = [j.get() for j in parse_jobs]\n \n print('\\nAnalysing...')\n seen.update(unseen)\n unseen.clear()\n for title, page_urls, url in results:\n # print(count, title, url)\n unseen.update(page_urls - seen)\n count += 1\n\nif __name__ == \"__main__\":\n t1 = time.time()\n loop = asyncio.get_event_loop()\n loop.run_until_complete(main(loop))\n # loop.close()\n print(\"Async total time: \", time.time() - t1)",
"\nAsync Crawling...\n\nDistributed Parsing...\n\nAnalysing...\n\nAsync Crawling...\n"
]
],
[
[
"**Here we try multiprocessing and test the speed**",
"_____no_output_____"
]
],
[
[
"from urllib.request import urlopen, urljoin\nfrom bs4 import BeautifulSoup\nimport multiprocessing as mp\nimport re\nimport time\n\n\ndef crawl(url):\n response = urlopen(url)\n time.sleep(0.1) # slightly delay for downloading\n return response.read().decode()\n\n\ndef parse(html):\n soup = BeautifulSoup(html, 'lxml')\n urls = soup.find_all('a', {\"href\": re.compile('^/.+?/$')})\n title = soup.find('h1').get_text().strip()\n page_urls = set([urljoin(base_url, url['href']) for url in urls])\n url = soup.find('meta', {'property': \"og:url\"})['content']\n return title, page_urls, url\n\n\nif __name__ == '__main__':\n # base_url = 'https://morvanzhou.github.io/'\n base_url = \"http://127.0.0.1:4000/\"\n \n # DON'T OVER CRAWL THE WEBSITE OR YOU MAY NEVER VISIT AGAIN\n if base_url != \"http://127.0.0.1:4000/\":\n restricted_crawl = True\n else:\n restricted_crawl = False\n \n unseen = set([base_url,])\n seen = set()\n\n pool = mp.Pool(8) # number strongly affected\n count, t1 = 1, time.time()\n while len(unseen) != 0: # still get some url to visit\n if restricted_crawl and len(seen) > 20:\n break\n print('\\nDistributed Crawling...')\n crawl_jobs = [pool.apply_async(crawl, args=(url,)) for url in unseen]\n htmls = [j.get() for j in crawl_jobs] # request connection\n htmls = [h for h in htmls if h is not None] # remove None\n\n print('\\nDistributed Parsing...')\n parse_jobs = [pool.apply_async(parse, args=(html,)) for html in htmls]\n results = [j.get() for j in parse_jobs] # parse html\n\n print('\\nAnalysing...')\n seen.update(unseen)\n unseen.clear()\n\n for title, page_urls, url in results:\n # print(count, title, url)\n count += 1\n unseen.update(page_urls - seen)\n\n print('Total time: %.1f s' % (time.time()-t1, ))\n\n",
"\nDistributed Crawling...\n\nDistributed Parsing...\n\nAnalysing...\n\nDistributed Crawling...\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec871945e20a2fcb337e6c57dd269e6e9418b561 | 349,550 | ipynb | Jupyter Notebook | temp/extractor_Rockaway2014.ipynb | esturdivant-usgs/geomorph-working-files | bd8d5391714ad0d8243580b6278c21ba419d83c5 | [
"CC0-1.0"
] | null | null | null | temp/extractor_Rockaway2014.ipynb | esturdivant-usgs/geomorph-working-files | bd8d5391714ad0d8243580b6278c21ba419d83c5 | [
"CC0-1.0"
] | 1 | 2018-12-26T18:11:51.000Z | 2018-12-26T18:12:02.000Z | temp/extractor_Rockaway2014.ipynb | esturdivant-usgs/geomorph-working-files | bd8d5391714ad0d8243580b6278c21ba419d83c5 | [
"CC0-1.0"
] | null | null | null | 103.356002 | 70,954 | 0.768906 | [
[
[
"# Extract barrier island metrics along transects\n\nAuthor: Emily Sturdivant, [email protected]\n\n***\n\nExtract barrier island metrics along transects for Bayesian Network Deep Dive\n\n\n## Pre-requisites:\n- All the input layers (transects, shoreline, etc.) must be ready. This is performed with the notebook file prepper.ipynb.\n- The files servars.py and configmap.py may need to be updated for the current dataset.\n\n## Notes:\n- This code contains some in-line quality checking during the processing, which requires the user's attention. For thorough QC'ing, we recommend displaying the layers in ArcGIS, especially to confirm the integrity of values for variables such as distance to inlet (__Dist2Inlet__) and widths of the landmass (__WidthPart__, etc.). \n\n\n***\n\n## Import modules",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nimport pandas as pd\nimport numpy as np\nimport arcpy\nimport matplotlib.pyplot as plt\nimport matplotlib\nmatplotlib.style.use('ggplot')\nimport core.functions_warcpy as fwa\nimport core.functions as fun",
"_____no_output_____"
]
],
[
[
"### Initialize variables\n\nBased on the project directory, and the site and year you have input, setvars.py will set a bunch of variables as the names of folders, files, and fields. Set-up the project folder and paths: ",
"_____no_output_____"
]
],
[
[
"from core.setvars import *\n\norig_trans = os.path.join(home, 'origTrans')\nextendedTrans = os.path.join(home, 'extTrans')\nextTrans_tidy = os.path.join(home, 'extTrans')\n\ninletLines = os.path.join(home, 'BP_inletLines_2014')\nShorelinePts = os.path.join(home, 'BreezyPt2014_shoreline')\ndlPts = os.path.join(home, 'BP2014_DLpts')\ndhPts = os.path.join(home, 'BP2014_DHpts')\n\narmorLines = os.path.join(home, 'BP_armorshoreward_2014')\nbarrierBoundary = os.path.join(home, 'bndpoly_2sl') \nshoreline = os.path.join(home, 'ShoreBetweenInlets')\n\nelevGrid = os.path.join(home, 'BP_DEM_2014')\nelevGrid_5m = os.path.join(home, 'BP_DEM_2014_5m')\nslopeGrid = 'slope_5m' # Slope in 5 m grids\n\nSubType = os.path.join(home, 'BP14_SubType')\nVegType = os.path.join(home, 'BP14_VegType')\nVegDens = os.path.join(home, 'BP14_VegDen')\nGeoSet = os.path.join(home, 'BP14_GeoSet')\nDisMOSH = os.path.join(home, 'BP14_DisMOSH')\n\ntr_w_anthro = os.path.join(home, 'extTrans_wAnthro')\nSA_bounds = os.path.join(home, 'SA_bounds')",
"site: Rockaway\nyear: 2014\nPath to project directory (e.g. \\\\Mac\u000bolume\\dir\\FireIsland2014): \\\\Mac\\stor\\Projects\\TransectExtraction\\Rockaway2014\nsetvars.py initialized variables.\n"
]
],
[
[
"## Transect-averaged values\nWe work with the shapefile/feature class as a Pandas Dataframe as much as possible to speed processing and minimize reliance on the ArcGIS GUI display.\n\n1. Create a pandas dataframe from the transects feature class. In the process, we remove some of the unnecessary fields. The resulting dataframe is indexed by __sort_ID__ with columns corresponding to the attribute fields in the transects feature class. ",
"_____no_output_____"
]
],
[
[
"# Copy feature class to dataframe.\ntrans_df = fwa.FCtoDF(extendedTrans, id_fld=tID_fld, extra_fields=extra_fields)\ntrans_df['DD_ID'] = trans_df[tID_fld] + sitevals['id_init_val']\n\n# Get anthro fields and join to DF\ntrdf_anthro = fwa.FCtoDF(tr_w_anthro, id_fld=tID_fld, dffields=['Development', 'Nourishment','Construction'])\ntrans_df = fun.join_columns(trans_df, trdf_anthro) \n\n# Save\ntrans_df.to_pickle(os.path.join(scratch_dir, 'trans_df.pkl'))\n\n# Display\nprint(\"\\nHeader of transects dataframe (rows 1-5 out of {}): \".format(len(trans_df)))\ntrans_df.head()",
"Converting feature class to array...\nConverting array to dataframe...\nConverting feature class to array...\nConverting array to dataframe...\n\nHeader of transects dataframe (rows 1-5 out of 376): \n"
]
],
[
[
"### Add XY and Z/slope from DH, DL, SL points within 25 m of transects\nAdd to each transect row the positions of the nearest pre-created beach geomorphic features (shoreline, dune toe, and dune crest).\n\n#### Shoreline\n\nThe MHW shoreline easting and northing (__SL_x__, __SL_y__) are the coordinates of the intersection of the oceanside shoreline with the transect. Each transect is assigned the foreshore slope (__Bslope__) from the nearest shoreline point within 25 m. These values are populated for each transect as follows: \n1. get __SL_x__ and __SL_y__ at the point where the transect crosses the oceanside shoreline; \n2. find the closest shoreline point to the intersection point (must be within 25 m) and copy the slope value from the point to the transect in the field __Bslope__.",
"_____no_output_____"
]
],
[
[
"if not arcpy.Exists(shoreline):\n shoreline = fwa.CreateShoreBetweenInlets(barrierBoundary, inletLines, shoreline, ShorelinePts, proj_code, SA_bounds)\n\n# Get the XY position where transect crosses the oceanside shoreline\nsl2trans_df = fwa.add_shorelinePts2Trans(extendedTrans, ShorelinePts, shoreline, \n tID_fld, proximity=pt2trans_disttolerance)\n\n# Save as pickle\nsl2trans_df.to_pickle(os.path.join(scratch_dir, 'sl2trans.pkl'))",
"\nMatching shoreline points to transects...\n...duration at transect 100: 0:0:31.7 seconds\n...duration at transect 200: 0:1:1.1 seconds\n...duration at transect 300: 0:1:30.5 seconds\nDuration: 0:1:52.6 seconds\n"
],
[
"sl2trans_df.sample(10)",
"_____no_output_____"
]
],
[
[
"#### Dune positions along transects\n\n__DL_x__, __DL_y__, and __DL_zMHW__ are the easting, northing, and height above MHW, respectively, of the nearest dune toe point within 25 meters of the transect. __DH_x__, __DH_y__, and __DH_zMHW__ are the easting, northing, and height above MHW, respectively, of the nearest dune crest point within 25 meters. \n\n__DL_snapX__, __DL_snapY__, __DH_snapX__, and __DH_snapY__ are the eastings and northings of the points 'snapped' to the transect. \"Snapping\" finds the position along the transect nearest to the point, i.e. orthogonal to the transect. These values are used to find the beach width.\n\nThese values are populated as follows: \n\n1. Find the nearest dune crest/toe point to the transect and proceed if the distance is less than 25 m. \n2. Get the X, Y, and Z values of the point and the XY position 'snapped' to the transect. The 'snapped' XY position is calculated using the arcpy geometry method. If there are no points within 25 m of the transect, populate the row with Null values; \n3. Convert the elevations to the MHW datum by applying the MHW offset.",
"_____no_output_____"
]
],
[
[
"# Create dataframe for both dune crest and dune toe positions\ndune2trans_df = fwa.find_ClosestPt2Trans_snap(extendedTrans, dhPts, dlPts, trans_df, \n tID_fld, proximity=pt2trans_disttolerance)\n\n# Save\ndune2trans_df.to_pickle(os.path.join(scratch_dir, 'dune2trans.pkl'))",
"\nMatching dune points with transects:\nGetting name of DH Z field...\nGetting name of DL Z field...\nLooping through transects and dune points to find nearest point within 25 m...\n...duration at transect 100: 0:0:40.6 seconds\n...duration at transect 200: 0:1:18.2 seconds\n...duration at transect 300: 0:1:58.0 seconds\nDuration: 0:2:27.2 seconds\n"
],
[
"dune2trans_df.sample(10)",
"_____no_output_____"
]
],
[
[
"#### Armoring\n__Arm_x__, __Arm_y__, and __Arm_zMHW__ are the easting, northing, and height above MHW, respectively, where an artificial structure crosses the transect in the vicinity of the beach. These features are meant to supplement the dune toe data set by providing an upper limit to the beach in areas where dune toe extraction was confounded by the presence of an artificial structure. Values are populated for each transect as follows: \n\n1. Get the positions of intersection between the digitized armoring lines and the transects (Intersect tool from the Overlay toolset); \n2. Extract the elevation value at each intersection point from the DEM (Extract Multi Values to Points tool from Spatial Analyst); \n4. Convert the elevations to the MHW datum by applying the MHW offset.",
"_____no_output_____"
]
],
[
[
"# Armoring line\narm2trans_df = fwa.ArmorLineToTrans_PD(extendedTrans, armorLines, sl2trans_df, tID_fld, proj_code, elevGrid_5m)\n\n# Save\narm2trans_df.to_pickle(os.path.join(scratch_dir, 'arm2trans.pkl'))",
"Converting feature class to array with X and Y...\nConverting array to dataframe...\nLooks like these transects [375] are intersected by armoring lines multiple times. We will select the more seaward of the points.\n"
]
],
[
[
"### Add all the positions to the trans_df\nJoin the new dataframes to the transect dataframe. Before it performs the join, `join_columns_id_check()` checks the index and the ID field for potential errors such as whether they are the equal and whether there are duplicated IDs or null values in either.",
"_____no_output_____"
]
],
[
[
"trans_df = pd.read_pickle(os.path.join(scratch_dir, 'trans_df.pkl'))\nsl2trans_df = pd.read_pickle(os.path.join(scratch_dir, 'sl2trans.pkl'))\ndune2trans_df = pd.read_pickle(os.path.join(scratch_dir, 'dune2trans.pkl'))\narm2trans_df = pd.read_pickle(os.path.join(scratch_dir, 'arm2trans.pkl'))",
"_____no_output_____"
],
[
"# Join positions of shoreline, dune crest, dune toe, armoring\ntrans_df = fun.join_columns_id_check(trans_df, sl2trans_df, tID_fld)\ntrans_df = fun.join_columns_id_check(trans_df, dune2trans_df, tID_fld)\ntrans_df = fun.join_columns_id_check(trans_df, arm2trans_df, tID_fld)\n\n# Save\ntrans_df.to_pickle(os.path.join(scratch_dir, 'trans_df_beachmetrics.pkl'))\n\n# Display\ntrans_df.sample(10)",
"_____no_output_____"
]
],
[
[
"### Check for errors: display summary stats / histograms and create feature classes\n*Optional*\n\nThe feature classes will display the locations that will be used to calculate beach width. Output files for validation:\n\n- pts2trans_SL\n- ptSnap2trans_DL\n- ptSnap2trans_DH\n- arm2trans",
"_____no_output_____"
]
],
[
[
"print(trans_df.describe([]))\n\ntrans_df.hist(['DH_z', 'DL_z', 'Arm_z'])\nplt.show()\nplt.close()",
" AZIMUTH LRR TRANSECTID TRANSORDER DD_ID \\\ncount 197.000000 197.000000 197.000000 197.000000 376.000000 \nmean 339.940457 0.633401 108.604061 108.604061 20188.500000 \nstd 4.852957 0.602143 58.625111 58.625111 108.686092 \nmin 338.070000 -0.490000 3.000000 3.000000 20001.000000 \n50% 338.070000 0.610000 108.000000 108.000000 20188.500000 \nmax 355.040000 2.190000 211.000000 211.000000 20376.000000 \n\n Construction Development Nourishment SL_x SL_y \\\ncount 376.000000 376.000000 376.0 344.000000 3.440000e+02 \nmean 386.433511 395.880319 222.0 597631.436474 4.491836e+06 \nstd 122.920618 96.655065 0.0 4590.687315 1.740392e+03 \nmin 111.000000 111.000000 222.0 589758.151996 4.488519e+06 \n50% 444.000000 444.000000 222.0 597625.860710 4.492016e+06 \nmax 444.000000 444.000000 222.0 605581.041702 4.494117e+06 \n\n ... DH_snapX DH_snapY DL_x DL_y \\\ncount ... 306.000000 3.060000e+02 287.000000 2.870000e+02 \nmean ... 597559.069703 4.491926e+06 597444.233065 4.491881e+06 \nstd ... 4486.760543 1.690052e+03 4420.261656 1.684453e+03 \nmin ... 589912.646650 4.488761e+06 589912.633805 4.488747e+06 \n50% ... 597605.908987 4.492066e+06 597585.970543 4.492045e+06 \nmax ... 605527.626472 4.494258e+06 605375.307618 4.494249e+06 \n\n DL_z DL_snapX DL_snapY Arm_x Arm_y \\\ncount 135.000000 287.000000 2.870000e+02 296.000000 2.960000e+02 \nmean 2.791535 597444.490373 4.491881e+06 599854.140524 4.492737e+06 \nstd 0.679758 4420.766211 1.684713e+03 3975.967972 1.379072e+03 \nmin 1.603063 589918.592368 4.488750e+06 592765.671846 4.490166e+06 \n50% 2.733410 597587.185143 4.492045e+06 599860.474373 4.493026e+06 \nmax 4.738086 605372.509579 4.494249e+06 606752.575422 4.494597e+06 \n\n Arm_z \ncount 290.000000 \nmean 2.939457 \nstd 0.636446 \nmin 0.555400 \n50% 2.982320 \nmax 4.345000 \n\n[6 rows x 24 columns]\n"
],
[
"# Convert dataframe to feature class - shoreline points with slope\nfwa.DFtoFC(sl2trans_df, os.path.join(arcpy.env.scratchGDB, 'pts2trans_SL'), \n spatial_ref=utmSR, id_fld=tID_fld, xy=[\"SL_x\", \"SL_y\"], keep_fields=['Bslope'])\n\n# Dune crests\ntry:\n fwa.DFtoFC(dune2trans_df, os.path.join(arcpy.env.scratchGDB, 'ptSnap2trans_DH'), \n spatial_ref=utmSR, id_fld=tID_fld, xy=[\"DH_snapX\", \"DH_snapY\"], keep_fields=['DH_z'])\nexcept Exception as err:\n print(err)\n pass\n\n# Dune toes\ntry:\n fwa.DFtoFC(dune2trans_df, os.path.join(arcpy.env.scratchGDB, 'ptSnap2trans_DL'), \n spatial_ref=utmSR, id_fld=tID_fld, xy=[\"DL_snapX\", \"DL_snapY\"], keep_fields=['DL_z'])\nexcept Exception as err:\n print(err)\n pass",
"_____no_output_____"
]
],
[
[
"### Calculate upper beach width and height\nUpper beach width (__uBW__) and upper beach height (__uBH__) are calculated based on the difference in position between two points: the position of MHW along the transect (__SL_x__, __SL_y__) and the dune toe position or equivalent (usually __DL_x__, __DL_y__). In some cases, the dune toe is not appropriate to designate the \"top of beach\" so beach width and height are calculated from either the position of the dune toe, the dune crest, or the base of an armoring structure. The dune crest was only considered a possibility if the dune crest elevation (__DH_zMHW__) was less than or equal to `maxDH`. \n\nThey are calculated as follows: \n1. Find the position along the transect of an orthogonal line drawn to the dune point (__DL_x__, __DL_y__ and __DH_x__, __DH_y__)\n2. Calculate distances from MHW to the position along the transect of the dune toe (__DistDL__), dune crest (__DistDH__), and armoring (__DistArm__). \n3. Conditionally select the appropriate feature to represent \"top of beach.\" Dune toe is prioritized. If it is not available and __DH_zMHW__ is less than or equal to maxDH, use dune crest. If neither of the dune positions satisfy the conditions and an armoring feature intersects with the transect, use the armoring position. If none of the three are possible, __uBW__ and __uBH__ will be null. \n4. Copy the distance to shoreline and height above MHW (__Dist--__, __---zMHW__) to __uBW__ and __uBH__, respectively. ",
"_____no_output_____"
]
],
[
[
"trans_df = pd.read_pickle(os.path.join(scratch_dir, 'trans_df_beachmetrics.pkl'))",
"_____no_output_____"
],
[
"#%% Calculate distances from shore to dunes, etc.\ntrans_df = fwa.calc_BeachWidth_fill(extendedTrans, trans_df, maxDH, tID_fld, sitevals['MHW'], fill, skip_missing_z=True)",
"_____no_output_____"
]
],
[
[
"### Dist2Inlet\n\n\nDistance to nearest tidal inlet (__Dist2Inlet__) is computed as alongshore distance of each sampling transect from the nearest tidal inlet. This distance includes changes in the path of the shoreline instead of simply a Euclidean distance and reflects sediment transport pathways. It is measured using the oceanside shoreline between inlets (ShoreBetweenInlets). \n\nNote that the ShoreBetweenInlets feature class must be both 'dissolved' and 'singlepart' so that each feature represents one-and-only-one shoreline that runs the entire distance between two inlets or equivalent. If the shoreline is bounded on both sides by an inlet, measure the distance to both and assign the minimum distance of the two. If the shoreline meets only one inlet (meaning the study area ends before the island ends), use the distance to the only inlet. \n\nThe process uses the cut, disjoint, and length geometry methods and properties in ArcPy data access module. The function measure_Dist2Inlet() prints a warning when the difference in Dist2Inlet between two consecutive transects is greater than 300. ",
"_____no_output_____"
]
],
[
[
"# Calc Dist2Inlet in new dataframe \ndist_df = fwa.measure_Dist2Inlet(shoreline, extendedTrans, inletLines, tID_fld)\n\n# Save\ndist_df.to_pickle(os.path.join(scratch_dir, 'dist2inlet_df.pkl'))\n\n# Join\ntrans_df = fun.join_columns_id_check(trans_df, dist_df, tID_fld, fill=fill)",
"Duration: 0:0:4.6 seconds\n"
],
[
"dist_df.tail(10)",
"_____no_output_____"
]
],
[
[
"### Clip transects, get barrier widths\nCalculates __WidthLand__, __WidthFull__, and __WidthPart__, which measure different flavors of the cross-shore width of the barrier island. __WidthLand__ is the above-water distance between the back-barrier and seaward MHW shorelines. __WidthLand__ only includes regions of the barrier within the shoreline polygon (bndpoly_2sl) and does not extend into any of the sinuous or intervening back-barrier waterways and islands. __WidthFull__ is the total distance between the back-barrier and seaward MHW shorelines (including space occupied by waterways). __WidthPart__ is the width of only the most seaward portion of land within the shoreline. \n\nThese are calculated as follows: \n\n1. Clip the transect to the full island shoreline (Clip in the Analysis toolbox); \n2. For __WidthLand__, get the length of the multipart line segment from "SHAPE@LENGTH" feature class attribute. When the feature is multipart, this will include only the remaining portions of the transect; \n3. For __WidthPart__, convert the clipped transect from multipart to singlepart and get the length of the first line segment, which should be the most seaward; \n4. For __WidthFull__, calculate the distance between the first vertex and the last vertex of the clipped transect (Feature Class to NumPy Array with explode to points, pandas groupby, numpy hypot).\n\nTemporary layers (assuming default names):\n\n- clip2island\n- clip2islandSingle_temp'",
"_____no_output_____"
]
],
[
[
"# Clip transects, get barrier widths *SPATIAL*\nwidths_df = fwa.calc_IslandWidths(extendedTrans, barrierBoundary, tID_fld=tID_fld)\n\n# # Save\nwidths_df.to_pickle(os.path.join(scratch_dir, 'widths_df.pkl'))\n# widths_df = pd.read_pickle(os.path.join(scratch_dir, 'widths_df.pkl'))\n\n# Join\ntrans_df = fun.join_columns_id_check(trans_df, widths_df, tID_fld, fill=fill)\n\n# Save\ntrans_df.to_pickle(os.path.join(scratch_dir, trans_name+'_null_prePts.pkl'))\n# trans_df = pd.read_pickle(os.path.join(scratch_dir, trans_name+'_null_prePts.pkl'))",
"Getting the width along each transect of the oceanside land (WidthPart)...\nConverting feature class to array...\nConverting array to dataframe...\nGetting the width along each transect of the entire barrier (WidthFull)...\nConverting feature class vertices to array with X and Y...\nConverting array to dataframe...\nGetting the width along each transect of above water portion of the barrier (WidthLand)...\n"
],
[
"trans_df.sample(10)",
"_____no_output_____"
]
],
[
[
"## 5-m Points\nThe point dataset samples the land every 5 m along each shore-normal transect. \n\n### Split transects into points at 5-m intervals. \n\nIt is created from the tidied transects (tidyTrans, created during pre-processing) as follows: \n\n1. Clip the tidied transects (tidyTrans) to the shoreline polygon (bndpoly_2sl) , retaining only those portions of the transects that represent land.\n2. Produce a dataframe of point positions along each transect every 5 m starting from the ocean-side shoreline. This uses the positionAlongLine geometry method accessed witha Search Cursor and saves the outputs in a new dataframe. \n3. Create a point feature class from the dataframe. \n\nTemporary files: \n- tran5mPts_unsorted",
"_____no_output_____"
]
],
[
[
"# if os.path.exists(os.path.join(scratch_dir, pts_name+'_null.pkl')):\n# pts_df = pd.read_pickle(os.path.join(scratch_dir, pts_name+'_null.pkl'))\n# trans_df = pd.read_pickle(os.path.join(scratch_dir, trans_name+'_null_prePts.pkl'))\nextTrans_tidy = os.path.join(home, 'extTrans')\npts_df, pts_presort = fwa.TransectsToPointsDF(extTrans_tidy, barrierBoundary, fc_out=os.path.join(arcpy.env.scratchGDB, 'transPts_unsorted')) # 5 minutes for FireIsland",
"Getting points every 5m along each transect and saving in both dataframe and feature class...\nConverting new dataframe to feature class...\nDuration: 0:4:24.3 seconds\n"
]
],
[
[
"### Add Elevation and Slope to points\n\n__ptZmhw__ and __ptSlp__ are the elevation and slope at the 5-m cell corresponding to the point. \n1. Create the slope and DEM rasters if they don't already exist. We use the 5-m DEM to generate a slope surface (Slope tool in 3D Analyst). \n2. Use Extract Multi Values to Points tool in Spatial Analyst. \n3. Convert the feature class back to a dataframe",
"_____no_output_____"
]
],
[
[
"# Create elevation and slope rasters at 5-m resolution\nif not arcpy.Exists(elevGrid_5m):\n fwa.ProcessDEM(elevGrid, elevGrid_5m, utmSR)\nif not arcpy.Exists(slopeGrid):\n arcpy.Slope_3d(elevGrid_5m, slopeGrid, 'PERCENT_RISE')\n \n# Add elevation and slope values at points.\narcpy.sa.ExtractMultiValuesToPoints(pts_presort, \n [[elevGrid_5m, 'ptZ'], [slopeGrid, 'ptSlp']])",
"_____no_output_____"
],
[
"# Add substrate type, geomorphic setting, veg type, veg density values at points.\narcpy.sa.ExtractMultiValuesToPoints(pts_presort, [[SubType, 'SubType'], [VegType, 'VegType'], \n [VegDens, 'VegDens'], [GeoSet, 'GeoSet'],\n [DisMOSH, 'DisMOSH']])\n\n# Convert to dataframe\npts_df = fwa.FCtoDF(pts_presort, xy=True, dffields=[tID_fld,'ptZ', 'ptSlp', 'SubType', \n 'VegType', 'VegDens', 'GeoSet','DisMOSH'])\n# Recode fill values\npts_df.replace({'GeoSet': {9999:np.nan}, 'SubType': {9999:np.nan}, 'VegType': {9999:np.nan},\n 'VegDens': {9999:np.nan}, 'DisMOSH': {9999:np.nan}}, inplace=True)\n\n# Save\npts_df.to_pickle(os.path.join(scratch_dir, 'pts_extractedvalues_presort.pkl'))\n\n# View 10 randomly selected rows \npts_df.sample(10)",
"Converting feature class to array with X and Y...\nConverting array to dataframe...\n"
],
[
"pts_df.hist('ptZ')\nplt.show()\nplt.close()",
"_____no_output_____"
]
],
[
[
"### Calculate distances and sort points\n\n__SplitSort__ is a unique numeric identifier of the 5-m points at the study site, sorted by order along shoreline and by distance from oceanside. __SplitSort__ values are populated by sorting the points by __sort_ID__ and __Dist_Seg__ (see below). \n\n__Dist_Seg__ is the Euclidean distance between the point and the seaward shoreline (__SL_x__, __SL_y__). __Dist_MHWbay__ is the distance between the point and the bayside shoreline and is calculated by subtracting the __Dist_Seg__ value from the __WidthPart__ value of the transect. \n\n__DistSegDH__, __DistSegDL__, and __DistSegArm__ measure the distance of each 5-m point from the dune crest and dune toe position along a particular transect. They are calculated as the Euclidean distance between the 5-m point and the given feature. ",
"_____no_output_____"
]
],
[
[
"pts_df = pd.read_pickle(os.path.join(scratch_dir, 'pts_extractedvalues_presort.pkl'))\ntrans_df = pd.read_pickle(os.path.join(scratch_dir, trans_name+'_null_prePts.pkl'))\n\n# Calculate DistSeg, Dist_MHWbay, DistSegDH, DistSegDL, DistSegArm, and sort points (SplitSort)\npts_df = fun.join_columns(pts_df, trans_df, tID_fld)\npts_df = fun.prep_points(pts_df, tID_fld, pID_fld, sitevals['MHW'], fill)\n\n# Aggregate ptZmhw to max and mean and join to transects\npts_df, zmhw = fun.aggregate_z(pts_df, sitevals['MHW'], tID_fld, 'ptZ', fill)\ntrans_df = fun.join_columns(trans_df, zmhw) \n\n# Join transect values to pts\npts_df = fun.join_columns(pts_df, trans_df, tID_fld)\n\n# pID_fld needs to be among the columns\nif not pID_fld in pts_df.columns:\n pts_df.reset_index(drop=False, inplace=True)\n\n# Drop extra fields and sort columns\ntrans_df.drop(extra_fields, axis=1, inplace=True, errors='ignore')\npts_df = pts_df.reindex_axis(sorted_pt_flds, axis=1)\n\n# Save dataframes to open elsewhere or later\ntrans_df.to_pickle(os.path.join(scratch_dir, trans_name+'_null.pkl'))\npts_df.to_pickle(os.path.join(scratch_dir, pts_name+'_null.pkl'))\n\n# View 10 randomly selected rows \npts_df.sample(5)",
"_____no_output_____"
]
],
[
[
"### Recode the values for CSV output and model running",
"_____no_output_____"
]
],
[
[
"# Recode\npts_df4csv = pts_df.replace({'SubType': {7777:'{1111, 2222}', 1000:'{1111, 3333}'}, \n 'VegType': {77:'{11, 22}', 88:'{22, 33}', 99:'{33, 44}'},\n 'VegDens': {666: '{111, 222}', 777: '{222, 333}', \n 888: '{333, 444}', 999: '{222, 333, 444}'}})\n\n# Fill NAs\npts_df4csv.fillna(fill, inplace=True) \n\n# Save as pickle\npts_df4csv.to_pickle(os.path.join(scratch_dir, pts_name+'_csv.pkl'))\n\n# Take a look at some rows\npts_df4csv.sample(5)",
"_____no_output_____"
],
[
"pts_df = pd.read_pickle(os.path.join(scratch_dir, pts_name+'_null.pkl'))\ntrans_df = pd.read_pickle(os.path.join(scratch_dir, trans_name+'_null.pkl'))\npts_df.sample(5)",
"_____no_output_____"
]
],
[
[
"## Quality checking\nLook at extracted profiles from around the island. Entered the transect ID within the available range when prompted. Evaluated the plots for consistency among variables. Repeat various times until you can be satisfied that the variables are consistent with each other and appear to represent reality.",
"_____no_output_____"
]
],
[
[
"desccols = ['DL_zmhw', 'DH_zmhw', 'Arm_zmhw', 'uBW', 'uBH', 'Dist2Inlet', \n 'WidthPart', 'WidthLand', 'WidthFull', 'mean_Zmhw', 'max_Zmhw']\n\n# Histograms\ntrans_df.hist(desccols, sharey=True, figsize=[15, 10], bins=20)\nplt.show()\nplt.close('all')",
"_____no_output_____"
],
[
"flds_dist = ['SplitSort', 'Dist_Seg', 'Dist_MHWbay', 'DistSegDH', 'DistSegDL', 'DistSegArm']\nflds_z = ['ptZmhw', 'ptZ', 'ptSlp']\npts_df.loc[:,flds_dist+flds_z].describe()\npts_df.hist(flds_dist, sharey=True, figsize=[15, 8], layout=(2,3))\npts_df.hist(flds_z, sharey=True, figsize=[15, 4], layout=(1,3))\nplt.show()\nplt.close('all')",
"_____no_output_____"
],
[
"# Prompt for transect identifier (sort_ID) and get all points from that transect.\ntrans_in = int(input('Transect ID (\"sort_ID\" {:d}-{:d}): '.format(int(pts_df[tID_fld].head(1)), int(pts_df[tID_fld].tail(1)))))\npts_set = pts_df[pts_df[tID_fld] == trans_in]\n\n# Plot\nfig = plt.figure(figsize=(13,10))\n\n# Plot the width of the island.\nax1 = fig.add_subplot(211)\nfun.plot_island_profile(ax1, pts_set, sitevals['MHW'], sitevals['MTL'])\n\n# Zoom in on the upper beach.\nax2 = fig.add_subplot(212)\nfun.plot_beach_profile(ax2, pts_set, sitevals['MHW'], sitevals['MTL'], maxDH)\n\n# Display\nplt.show()\nplt.close('all')",
"Transect ID (\"sort_ID\" 1-376): 250\n"
]
],
[
[
"## Outputs\n\n### Transect-averaged\n#### Vector format",
"_____no_output_____"
]
],
[
[
"trans_df = pd.read_pickle(os.path.join(scratch_dir, trans_name+'_null.pkl'))\npts_df = pd.read_pickle(os.path.join(scratch_dir, pts_name+'_null.pkl'))",
"_____no_output_____"
],
[
"# Create transect FC with fill values - Join values from trans_df to the transect FC as a new file.\ntrans_fc = fwa.JoinDFtoFC(trans_df, extendedTrans, tID_fld, out_fc=trans_name+'_fill')\n\n# Create transect FC with null values\nfwa.CopyFCandReplaceValues(trans_fc, fill, None, out_fc=trans_name+'_null', out_dir=home)\n\n# Save final transect SHP with fill values\nout_shp = arcpy.FeatureClassToFeatureClass_conversion(trans_fc, scratch_dir, trans_name+'_shp.shp')\nprint(\"OUTPUT: {} in specified scratch_dir.\".format(os.path.basename(str(out_shp))))",
"Created rock14_trans_fill from input dataframe and extTrans file.\nOUTPUT: rock14_trans_null\nOUTPUT: rock14_trans_shp.shp in specified scratch_dir.\n"
]
],
[
[
"#### Raster - beach width",
"_____no_output_____"
]
],
[
[
"# Create a template raster corresponding to the transects. \nif not arcpy.Exists(rst_transID):\n print(\"{} was not found so we will create the base raster.\".format(os.path.basename(rst_transID)))\n outEucAll = arcpy.sa.EucAllocation(extTrans_tidy, maximum_distance=50, cell_size=cell_size, source_field=tID_fld)\n outEucAll.save(os.path.basename(rst_transID))\n\n# Create raster of uBW values by joining trans_df to the template raster.\nout_rst = fwa.JoinDFtoRaster(trans_df, os.path.basename(rst_transID), bw_rst, fill, tID_fld, 'uBW')",
"OUTPUT: rock14_ubw. Field \"Value\" is ID and \"uBW\" is beachwidth.\n"
]
],
[
[
"### 5-m points",
"_____no_output_____"
],
[
"#### Tabular format",
"_____no_output_____"
],
[
"Save final pts with fill values as CSV",
"_____no_output_____"
]
],
[
[
"trans_df = pd.read_pickle(os.path.join(scratch_dir, trans_name+'_null.pkl'))\npts_df4csv = pd.read_pickle(os.path.join(scratch_dir, pts_name+'_csv.pkl'))",
"_____no_output_____"
],
[
"# Save CSV in scratch_dir\ncsv_fname = os.path.join(scratch_dir, pts_name +'.csv')\npts_df4csv.to_csv(csv_fname, na_rep=fill, index=False)\nprint(\"OUTPUT: {} in specified scratch_dir.\".format(os.path.basename(csv_fname)))",
"OUTPUT: rock14_pts.csv in specified scratch_dir.\n"
]
],
[
[
"#### Vector format",
"_____no_output_____"
]
],
[
[
"# Convert pts_df to FC - automatically converts NaNs to fills (default fill is -99999)\npts_fc = fwa.DFtoFC_large(pts_df, out_fc=os.path.join(arcpy.env.workspace, pts_name+'_fill'), \n spatial_ref=utmSR, df_id=pID_fld, xy=[\"seg_x\", \"seg_y\"])\n\n# Save final FCs with null values\nfwa.CopyFCandReplaceValues(pts_fc, fill, None, out_fc=pts_name+'_null', out_dir=home)\n\n# Save final points as SHP and XLS with fill values\nout_pts_shp = arcpy.FeatureClassToFeatureClass_conversion(pts_fc, scratch_dir, pts_name+'_shp.shp')\nprint(\"OUTPUT: {} in specified scratch_dir.\".format(os.path.basename(str(out_pts_shp))))",
"Converting points DF to FC...\nOUTPUT: rock14_pts_fill\nDuration: 0:6:58.6 seconds\nOUTPUT: rock14_pts_null\nOUTPUT: rock14_pts_shp.shp in specified scratch_dir.\n"
]
],
[
[
"### Report values for Entity and Attribute Information ",
"_____no_output_____"
]
],
[
[
"print('{}{} -- Number of points in dataset: {}'.format(site, year, pts_df4csv.shape))\nfor fld in pts_df4csv.columns:\n if fld in ['GeoSet', 'SubType', 'VegDens', 'VegType', 'Construction', 'Development', 'Nourishment', 'ub_feat']:\n print('{} values: {}'.format(fld, ' | '.join(str(x) for x in pts_df4csv.loc[:,fld].unique())))\n else:\n if not any(pd.isnull(pts_df4csv.loc[:,fld])):\n mn = min(pts_df4csv.loc[:,fld])\n else:\n mn = -99999\n mx = np.max(pts_df4csv.loc[:,fld])\n print('{} min|max: {} | {}'.format(fld, mn, mx))",
"Rockaway2014 -- Number of points in dataset: (85936, 54)\nSplitSort min|max: 0 | 85935\nseg_x min|max: -99999.0 | 606857.4244118808\nseg_y min|max: -99999.0 | 4496663.707794331\nDist_Seg min|max: -99999.0 | 2564.999972378326\nDist_MHWbay min|max: -99999.0 | 2566.1516249920132\nDistSegDH min|max: -99999.0 | 2523.0856470807003\nDistSegDL min|max: -99999.0 | 2531.797153888941\nDistSegArm min|max: -99999.0 | 2533.2068455604176\nptZ min|max: -99999.0 | 20.68008041381836\nptSlp min|max: -99999.0 | 128.31585693359375\nptZmhw min|max: -99999.0 | 20.220081329345703\nGeoSet values: -99999.0 | 1.0 | 2.0 | 5.0 | 3.0 | 4.0\nSubType values: -99999 | 4444.0 | {1111, 2222} | 6666.0 | 2222.0 | 1111.0\nVegDens values: -99999 | 111.0 | {111, 222} | 555.0 | 222.0 | 333.0 | 444.0 | {333, 444}\nVegType values: -99999 | 11.0 | {11, 22} | 55.0 | 22.0 | 33.0 | {33, 44} | {22, 33}\nsort_ID min|max: 1.0 | 376.0\nTRANSORDER min|max: -99999.0 | 211.0\nTRANSECTID min|max: -99999.0 | 211.0\nDD_ID min|max: 20001 | 20376\nLRR min|max: -99999.0 | 2.19\nSL_x min|max: -99999.0 | 605581.0417019129\nSL_y min|max: -99999.0 | 4494116.911421059\nBslope min|max: -99999.0 | -0.013101\nDL_x min|max: -99999.0 | 605375.3076182976\nDL_y min|max: -99999.0 | 4494249.472522093\nDL_z min|max: -99999.0 | 4.738086\nDL_zmhw min|max: -99999.0 | 4.278086\nDL_snapX min|max: -99999.0 | 605372.5095792771\nDL_snapY min|max: -99999.0 | 4494249.21319848\nDH_x min|max: -99999.0 | 605522.6227301983\nDH_y min|max: -99999.0 | 4494258.164789788\nDH_z min|max: -99999.0 | 8.156864\nDH_zmhw min|max: -99999.0 | 7.696864000000001\nDH_snapX min|max: -99999.0 | 605527.626472122\nDH_snapY min|max: -99999.0 | 4494257.739110547\nArm_x min|max: -99999.0 | 606752.5754216788\nArm_y min|max: -99999.0 | 4494597.309792919\nArm_z min|max: -99999.0 | 4.345000267028809\nArm_zmhw min|max: -99999.0 | 3.885000228881836\nDistDH min|max: -99999.0 | 255.10942271195344\nDistDL min|max: -99999.0 | 246.38538445644008\nDistArm min|max: -99999.0 | 239.7291582507877\nDist2Inlet min|max: -99999.0 | 8566.102089233871\nWidthPart min|max: -99999.0 | 2566.15165261419\nWidthLand min|max: -99999.0 | 2566.15165261419\nWidthFull min|max: -99999.0 | 2566.15165261419\nuBW min|max: -99999.0 | 246.38538445644008\nuBH min|max: -99999.0 | 4.278086\nub_feat values: -99999 | DL | DH | Arm\nmean_Zmhw min|max: -99999.0 | 7.125663757324219\nmax_Zmhw min|max: -99999.0 | 20.220081329345703\nConstruction values: 333.0 | 111.0 | 444.0\nDevelopment values: 111.0 | 222.0 | 333.0 | 444.0\nNourishment values: 222.0\n"
],
[
"trans_flds = ['TRANSECTID', 'TRANSORDER', 'DD_ID']\ntrans_4pub = fwa.JoinDFtoFC(trans_df.loc[:,trans_flds], extendedTrans, tID_fld, out_fc=sitevals['code']+'_trans')\nout_shp = arcpy.FeatureClassToFeatureClass_conversion(trans_4pub, scratch_dir, sitevals['code']+'_trans.shp')\nprint(\"OUTPUT: {} in specified scratch_dir.\".format(os.path.basename(str(out_shp))))",
"Created rock_trans from input dataframe and extTrans file.\nOUTPUT: rock_trans.shp in specified scratch_dir.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec8725a8738872f4443c3935efdffe532a98f919 | 701,739 | ipynb | Jupyter Notebook | 12-Scraping-APIs/lecture-12-apis.ipynb | uwsd/2018-datascience-lectures | 4ddaa512d3f618e3fcd36b1f8151687e9ea1facf | [
"MIT"
] | 36 | 2018-01-09T18:25:45.000Z | 2021-08-08T02:15:43.000Z | 12-Scraping-APIs/lecture-12-apis.ipynb | uwsd/2018-datascience-lectures | 4ddaa512d3f618e3fcd36b1f8151687e9ea1facf | [
"MIT"
] | 1 | 2018-11-16T10:49:32.000Z | 2018-11-17T12:35:17.000Z | 12-Scraping-APIs/lecture-12-apis.ipynb | uwsd/2018-datascience-lectures | 4ddaa512d3f618e3fcd36b1f8151687e9ea1facf | [
"MIT"
] | 41 | 2018-01-10T20:40:25.000Z | 2020-08-20T03:25:15.000Z | 53.847376 | 28,230 | 0.562195 | [
[
[
"# Introduction to Data Science – Lecture 12 - APIs\n*COMP 5360 / MATH 4100, University of Utah, http://datasciencecourse.net/*\n\nIn this lecture we will explore how we can extract data from web-APIs. The first part of this tutorial is based on [this blog](https://www.dataquest.io/blog/python-api-tutorial/).\n\nAPI stands for **A**pplication **P**rogramming **I**nterface. It is supposed to be a **well defined interface** for consuming data (in contrast to web-scraping), or, more generally, for the **interaction with a remote program** on a server. APIs aren't only generated so that you can read data; they're often designed so that you could write custom software to interact with the API. You could, for example, write your own twitter client! \n\nAPIs are usually well documented, as the organization that provides APIs wants you to use them. \n\nHere are a couple of examples of sites that have documented APIs:\n\n * [StackExchange](https://api.stackexchange.com/docs) \n * [Twitter](https://dev.twitter.com/rest/public)\n * [Facebook](https://developers.facebook.com/docs/graph-api)\n * [KEGG](http://www.genome.jp/kegg/rest/keggapi.html) \n * [Google Maps](https://developers.google.com/maps/) \n * and many many more. \n \nMost APIs require some form of authentication and have rules on what you're allowed to do and what not. Follow those rules, or you will be blocked faster than you can say `GET`. \n \nAn API works a lot like working with a website, but instead of a human in front of the screen, it's computers talking to each other. \n\nWe'll be using the dominant form of APIs, REST (REpresentational State Transfer), which are now the dominant way of exposing and API on the web. REST APIs use an [URI](https://en.wikipedia.org/wiki/Uniform_Resource_Identifier) (a Uniform Resource Identifier; URLs are one specific form of URIs) to specify what you want to do with and API.\n\nAll API methods in REST are relative to a base URL. E.g., for twitter this would be\n```\nhttps://api.twitter.com/1.1/\n```\n\nA query would look like this: \n\n```\nhttps://api.twitter.com/1.1/search/tweets.json?q=%40twitterapi\n```\n\nREST APIs can return data in different forms, the most common ones are JSON and XML, and of those two, JSON is now dominant. \n\nJSON stands for JavaScript Object Notation, and is a pretty convenient format, as we'll see. \n \n\nLet's start with a very simple example, retrieving the current position of the ISS. We will be using the [requests library](http://www.python-requests.org/en/latest/), which will handle our communication with the server.",
"_____no_output_____"
]
],
[
[
"import requests \n# Make a get request to get the latest position of the international space station from the opennotify api.\nresponse = requests.get(\"http://api.open-notify.org/iss-now.json\")\n\nresponse",
"_____no_output_____"
]
],
[
[
"That looks good, we've received a response and it has status 200. What does the 200 mean? It's a status code - you've probably seen the \"Error 404\" on the internet. \n\nHere are a couple of codes:\n\n * **200** – everything went okay, and the result has been returned (if any)\n * **301** – the server is redirecting you to a different endpoint. This can happen when a company switches domain names, or an endpoint name is changed.\n * **401** – the server thinks you’re not authenticated. This happens when you don’t send the right credentials to access an API (we’ll talk about authentication later).\n * **400** – the server thinks you made a bad request. This can happen when you don’t send along the right data, among other things.\n * **403** – the resource you’re trying to access is forbidden – you don’t have the right permissions to see it.\n * **404** – the resource you tried to access wasn’t found on the server.\n\nLet's try to get a response with a wrong URL:",
"_____no_output_____"
]
],
[
[
"response_failed = requests.get(\"http://api.open-notify.org/iss\")\n\nresponse_failed",
"_____no_output_____"
]
],
[
[
"We used the get methods to put in a GET request. There are four different types of requests: \n\n\n[Source](https://en.wikipedia.org/wiki/Representational_state_transfer)",
"_____no_output_____"
],
[
"Let's look at the payload of our previous, successful response:",
"_____no_output_____"
]
],
[
[
"response.content",
"_____no_output_____"
]
],
[
[
"We can already see that this is JSON (though it is stored a `bytes` object), but we can check formally:",
"_____no_output_____"
]
],
[
[
"response.headers['content-type']",
"_____no_output_____"
]
],
[
[
"We can decode this byte object, then the JSON will be readable. ",
"_____no_output_____"
]
],
[
[
"response_j = response.content.decode(\"utf-8\")\nprint(response_j)",
"{\"timestamp\": 1518671239, \"iss_position\": {\"longitude\": \"14.9127\", \"latitude\": \"-32.1697\"}, \"message\": \"success\"}\n"
]
],
[
[
"Let's take a look at the JSON here:\n\n```JSON\n{\n \"iss_position\": {\n \"latitude\": -30.005751854107206, \n \"longitude\": -104.20085371352678\n }, \n \"message\": \"success\", \n \"timestamp\": 1475240215\n}\n```\n\nThis looks a lot like a dictionary! We have key-value pairs. \n\nWe can use the [json library](https://docs.python.org/3/library/json.html) to convert JSON into objects:",
"_____no_output_____"
]
],
[
[
"import json\nresponse_d = json.loads(response_j)\nprint(type(response_d))\nprint(response_d)\nresponse_d[\"iss_position\"]",
"<class 'dict'>\n{'timestamp': 1518671239, 'iss_position': {'longitude': '14.9127', 'latitude': '-32.1697'}, 'message': 'success'}\n"
]
],
[
[
"Or, not surprisingly, pandas can also load a jsong object:",
"_____no_output_____"
]
],
[
[
"import pandas as pd \n\ndf = pd.read_json(response_j)\ndf",
"_____no_output_____"
]
],
[
[
"This isn't quite what we want - we probably want one row per timestamp and longitude and latitude as columns:",
"_____no_output_____"
]
],
[
[
"def flatten(response_d):\n response_d[\"latitude\"] = response_d[\"iss_position\"][\"latitude\"]\n response_d[\"longitude\"] = response_d[\"iss_position\"][\"longitude\"]\n del(response_d[\"iss_position\"])\n return response_d\nflatten(response_d)",
"_____no_output_____"
]
],
[
[
"That looks better. Let's get a couple of positions of the ISS over time and save it as an array:",
"_____no_output_____"
]
],
[
[
"import time\n\ndef pull_position():\n \"\"\"Retreives the position of the ISS and returns it as a flat dictionary\"\"\"\n response = requests.get(\"http://api.open-notify.org/iss-now.json\")\n response_j = response.content.decode(\"utf-8\")\n response_d = json.loads(response_j)\n flat_response = flatten(response_d)\n return flat_response \n\niss_position = []\n \n# calls pull_position 10 times with 3 seconds break\nfor i in range(10):\n flat_response = pull_position()\n iss_position.append(flat_response)\n print(flat_response)\n time.sleep(3)\n \nlen(iss_position)",
"{'timestamp': 1518671240, 'message': 'success', 'latitude': '-32.1256', 'longitude': '14.9646'}\n{'timestamp': 1518671243, 'message': 'success', 'latitude': '-31.9932', 'longitude': '15.1201'}\n{'timestamp': 1518671246, 'message': 'success', 'latitude': '-31.8607', 'longitude': '15.2749'}\n{'timestamp': 1518671250, 'message': 'success', 'latitude': '-31.7057', 'longitude': '15.4551'}\n{'timestamp': 1518671253, 'message': 'success', 'latitude': '-31.5726', 'longitude': '15.6090'}\n{'timestamp': 1518671256, 'message': 'success', 'latitude': '-31.4393', 'longitude': '15.7624'}\n{'timestamp': 1518671259, 'message': 'success', 'latitude': '-31.3057', 'longitude': '15.9153'}\n{'timestamp': 1518671262, 'message': 'success', 'latitude': '-31.1720', 'longitude': '16.0678'}\n{'timestamp': 1518671265, 'message': 'success', 'latitude': '-31.0380', 'longitude': '16.2198'}\n{'timestamp': 1518671268, 'message': 'success', 'latitude': '-30.8815', 'longitude': '16.3966'}\n"
]
],
[
[
"Now we can convert this into a nice dataframe:",
"_____no_output_____"
]
],
[
[
"iss_position_df = pd.DataFrame(iss_position)\niss_position_df['timestamp'] = pd.to_datetime(iss_position_df['timestamp'], unit=\"s\")\n\niss_position_df = iss_position_df.set_index(pd.DatetimeIndex(iss_position_df['timestamp']))\niss_position_df[\"latitude\"] = iss_position_df[\"latitude\"].map(float)\niss_position_df[\"longitude\"] = iss_position_df[\"longitude\"].map(float)",
"_____no_output_____"
]
],
[
[
"Let's see how the ISS moves",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n# This next line tells jupyter to render the images inline\n%matplotlib inline\nplt.style.use('ggplot')\niss_position_df.plot(kind=\"scatter\", x=\"latitude\", y=\"longitude\")",
"_____no_output_____"
]
],
[
[
"### Querying with Parameters\n\nRequests can be parametrized. You can search for tweets of a specific user, for example, or [retrieve the time the ISS is over SLC](http://open-notify.org/Open-Notify-API/ISS-Pass-Times/)!\n\nThe way to query with a get request for the ISS is this:\n\n`http://api.open-notify.org/iss-pass.json?lat=40.758701&lon=-111.876183`\n\nWe, of course, could generate that URL ourselves, but the requests library is helpful here. Since JSON is similar to dictionaries, the requests library takes dictionaries for parameters.",
"_____no_output_____"
]
],
[
[
"url = \"http://api.open-notify.org/iss-pass.json\"\ncoordinates = {\"lat\": 40.758701, \"lon\": -111.876183, \"n\":5}\n\nr = requests.get(url, params=coordinates)\ndata = r.json()\ndata",
"_____no_output_____"
],
[
"timestamp = data[\"response\"][0][\"risetime\"]\npd.to_datetime(timestamp, unit=\"s\")",
"_____no_output_____"
]
],
[
[
"So, the ISS will be visible at 4:13:34!",
"_____no_output_____"
],
[
"### Exercise: How many Astronauts? \n\nUse the [Open Notify API](http://open-notify.org/Open-Notify-API/People-In-Space/) to find out how many people are in space right now.",
"_____no_output_____"
],
[
"## Libraries and Authentication\n\nWhile we now have the skills to directly talk to an API, it's sometimes a little tedious. Popular APIs often have existing Python libraries that wrap around them. [Here](https://github.com/realpython/list-of-python-api-wrappers) is a long list of wrappers! \n\nNow we'll explore the Twitter API using the [twython library](https://github.com/ryanmcgrath/twython). Check out the [documentation](https://twython.readthedocs.io/en/latest/).\n\nUnfortunately, most professional APIs will require you to authenticate and will limit you in what you can do – mostly they limit how much data you can retreive at a certain time. To run the following code, you'll have to put in your own credentials (sorry – I can't share mine). \n\nInstall twython:\n`pip install twython`\n\n* Go to https://apps.twitter.com/ and create a new app\n* Save your consumer key and consumer secret in a file `credentials.py` in the format:\n```python\nCONSUMER_KEY = \"KEY\"\nCONSUMER_SECRET = \"KEY\"\n```\n\nBut before we get started, let's check out what [a tweet looks like](https://dev.twitter.com/overview/api/tweets):",
"_____no_output_____"
],
[
"```JSON\n{'contributors': None,\n 'coordinates': None,\n 'created_at': 'Fri Sep 30 18:37:48 +0000 2016',\n 'entities': {'hashtags': [], 'symbols': [], 'urls': [], 'user_mentions': []},\n 'favorite_count': 2960,\n 'favorited': False,\n 'geo': None,\n 'id': 781926033159249920,\n 'id_str': '781926033159249920',\n 'in_reply_to_screen_name': None,\n 'in_reply_to_status_id': None,\n 'in_reply_to_status_id_str': None,\n 'in_reply_to_user_id': None,\n 'in_reply_to_user_id_str': None,\n 'is_quote_status': False,\n 'lang': 'en',\n 'place': None,\n 'retweet_count': 1283,\n 'retweeted': False,\n 'source': '<a href=\"http://twitter.com/download/android\" rel=\"nofollow\">Twitter for Android</a>',\n 'text': \"For those few people knocking me for tweeting at three o'clock in the morning, at least you know I will be there, awake, to answer the call!\",\n 'truncated': False,\n 'user': {'contributors_enabled': False,\n 'created_at': 'Wed Mar 18 13:46:38 +0000 2009',\n 'default_profile': False,\n 'default_profile_image': False,\n 'description': '',\n 'entities': {'description': {'urls': []},\n 'url': {'urls': [{'display_url': 'DonaldJTrump.com',\n 'expanded_url': 'http://www.DonaldJTrump.com',\n 'indices': [0, 23],\n 'url': 'https://t.co/mZB2hymxC9'}]}},\n 'favourites_count': 39,\n 'follow_request_sent': None,\n 'followers_count': 11929387,\n 'following': None,\n 'friends_count': 41,\n 'geo_enabled': True,\n 'has_extended_profile': False,\n 'id': 25073877,\n 'id_str': '25073877',\n 'is_translation_enabled': True,\n 'is_translator': False,\n 'lang': 'en',\n 'listed_count': 39225,\n 'location': 'New York, NY',\n 'name': 'Donald J. Trump',\n 'notifications': None,\n 'profile_background_color': '6D5C18',\n 'profile_background_image_url': 'http://pbs.twimg.com/profile_background_images/530021613/trump_scotland__43_of_70_cc.jpg',\n 'profile_background_image_url_https': 'https://pbs.twimg.com/profile_background_images/530021613/trump_scotland__43_of_70_cc.jpg',\n 'profile_background_tile': True,\n 'profile_banner_url': 'https://pbs.twimg.com/profile_banners/25073877/1468988952',\n 'profile_image_url': 'http://pbs.twimg.com/profile_images/1980294624/DJT_Headshot_V2_normal.jpg',\n 'profile_image_url_https': 'https://pbs.twimg.com/profile_images/1980294624/DJT_Headshot_V2_normal.jpg',\n 'profile_link_color': '0D5B73',\n 'profile_sidebar_border_color': 'BDDCAD',\n 'profile_sidebar_fill_color': 'C5CEC0',\n 'profile_text_color': '333333',\n 'profile_use_background_image': True,\n 'protected': False,\n 'screen_name': 'realDonaldTrump',\n 'statuses_count': 33344,\n 'time_zone': 'Eastern Time (US & Canada)',\n 'url': 'https://t.co/mZB2hymxC9',\n 'utc_offset': -14400,\n 'verified': True}\n```\n",
"_____no_output_____"
]
],
[
[
"from twython import Twython\n# credentials is a local file with your own credentials. It defines CONSUMER_KEY and CONSUMER_SECRET\nfrom credentials_alex import * \ntwitter = Twython(CONSUMER_KEY, CONSUMER_SECRET)",
"_____no_output_____"
]
],
[
[
"Here, we have created the Twython library object and authenticated against our user.\n\nNow let's search for a hashtag:",
"_____no_output_____"
]
],
[
[
"tag = \"#resist\"\nresult = twitter.search(q=tag, tweet_mode=\"extended\")\nresult",
"_____no_output_____"
]
],
[
[
"The result is an array of tweets, we can look at specific text:",
"_____no_output_____"
]
],
[
[
"result[\"statuses\"][0][\"full_text\"]",
"_____no_output_____"
]
],
[
[
"Or print all the tweets:",
"_____no_output_____"
]
],
[
[
"for status in result[\"statuses\"]:\n print(status[\"full_text\"])\n print(\"----\")",
"RT @24CarlyS119: @NBCNews THE GOP & NRA AREN'T ALL THAT'S WRONG. PEOPLE WHO DO NOTHING ARE NOW WORSE. #VOLUNTEER #DONATE #RECRUIT #RESIST…\n----\nWe are all wrestling with these compounding tragedies & the emotional strain as they tear at our souls\nIn these times as in the dark days before we need the voices that calm educate & lift us. You @sherilynch & yours are those voices.\n#Resist\n----\nAt 19, Nikolas is an adult who could purchase his weapons legally. Where he got the money to do so has yet to be disclosed. One rumor states that he was bullied in school. Aha!\n#trump #maga #potus #gop #dems #IngrahamAngle #wattersworld #resist #TuckerCarlsonTonight #SeanHannity\n----\nRT @progresslabsorg: Now Just Five Men Own Almost as Much Wealth as Half the World's Population https://t.co/1Jn0V1YYwo #resist #fbr\n----\n'I invite him to get off his ass': Dem Congressman slams Trump in wake of another school shooting \nhttps://t.co/xI96RDZk1B\n#GOPCorruptAF #LGBT #LGBTQ #Resist #TheResistance\n----\nThis is the #GrandInquisitor of the\n#BenghaziCircus 8 million and 2.5\nyears and they couldn't find Jesus\nAnother caveat is 150K paid out to\na Air Force Veteran he tried to black\nball because he wouldn't lie about\nfindings in the case against Clinton!\n#Gowdy #RemoveGowdy #resist\n----\nRT @ALTUSNPS: We keep talking about #resisting. To #resist successfully we must #participate.\n\nVote\nTweet\nWrite\nSpeak\nVolunteer\nPersist\nDon…\n----\n@outer_spaeth Finding people willing to listen is HARD! I wished I could vote, but still have roughly 2 years until I can apply for citizenship. In the meantime, #resist!\n----\nRT @ALTUSNPS: Ok, new motto for the rest of the week- PARTICIPATE\n\nDon't just #resist, but get involved, take action & make a difference by…\n----\nhttps://t.co/DJmqZfIoaD #resist\n----\n@realDonaldTrump Do not give *pass* for suspicious activity, whether it be student showing kids cigarette lighter resembling a pistol, pop can labeled \"fake bomb\", aggressive notes on textbook cover, or serious taunt. Stop violence at the beginning. Use the facts and keep repeating them. #resist\n----\nRT @22Avengers: RT If You're Mueller's Big Fan ! #TheResistance #Resist https://t.co/EmOj8J34tx\n----\nRT @ALTUSNPS: We keep talking about #resisting. To #resist successfully we must #participate.\n\nVote\nTweet\nWrite\nSpeak\nVolunteer\nPersist\nDon…\n----\nRT @iRhysTay: I voted for Hillary Clinton! \n#ThePeoplesPresident\n#ImStillWithHer\n#ShePersisted\n#HillaryWarnedUs\n#WhatHappened\n#Resist #Insi…\n----\nYes. You. Are. #resist https://t.co/Ro6xeDgUIN\n----\n"
]
],
[
[
"We can also search for tweets based on usernames:",
"_____no_output_____"
]
],
[
[
"result = twitter.search(q=\"@realDonaldTrump\", tweet_mode=\"extended\")\nfor status in result[\"statuses\"]:\n print(status[\"full_text\"])\n print(\"----\")",
"RT @realDonaldTrump: As we come together to celebrate the extraordinary contributions of African-Americans to our nation, our thoughts turn…\n----\nRT @BGibbles: @realDonaldTrump Your prayers and condolences won’t do shit to prevent this from happening more in the future. Use your power…\n----\nRT @SarahBoothHenry: @realDonaldTrump ....then do something about it! 18 worldwide in 2 decades. 18 in the US in 35 days. DO SOMETHING!\n----\nRT @MaggieJordanACN: . @realDonaldTrump received $31,194,646 from the NRA in 2016 https://t.co/qSBSp9U9MD\n----\n@icegov Excellent. Keep up God's Work. He too has a wall that only certain people can enter. \n@FrantzRadio @WayneDupreeShow @realDonaldTrump @Scavino45 @PressSec @WhiteHouse @mike_pence @VP @SpeakerRyan @SenMajLdr https://t.co/ltkg9Syz0o\n----\n@FalconsKingdom @christy_doyon @Tnlady56King @realDonaldTrump In special cases, security can carry one around.\n----\nRT @timmYTiggerG13: @realDonaldTrump U r not doing anyx. What Abt law enforcement?? The kid is in custody. Your job is nra getting away w…\n----\nRT @ANOMALY1: Thank You Sir🤝#GodBless🙏 \n\n🦅🇺🇸@POTUS @realDonaldTrump\n\n“After spending trillions of dollars overseas rebuilding other countri…\n----\n@OliverMcGee @realDonaldTrump He's not watching at all..he's working hard deeply with his own Infidels 2keep harming our GREAT PRESIDENT! Obama was born to destroy USA! It's in Koran! But TRUMP WAS SENT BY GOD TO SAVE USA & THE ENTIRE WESTERN WORLD 🌎! God bless us all! Evil is 💪&has ☝️mission destruction\n----\n@realDonaldTrump I wish you would also work on not taking our rights and freedoms away.\n----\nRT @brandovader: @Crayzcapper @JaketheSerling @BGibbles @realDonaldTrump Just because its impossible to always stop it doesnt mean we cant…\n----\n@StacyLStiles @realDonaldTrump @Hoosiers1986 @AMErikaNGIRLBOT @JrcheneyJohn @ClintonM614 @mikandynothem @IsraelUSAforevr @GrizzleMeister @SandraTXAS @SKYRIDER4538 @TrumpGirlStrong I think it’s an ugly painting even if the palest white man ever were sitting in that chair with the tacky background. Or any sentient being\n----\n@realDonaldTrump Look at the global stats. Japan has banned ALL guns and stunningly has NO gun violence. Deaths per year by guns is under 10. Under TEN! Think about that!\n----\n@realDonaldTrump but us students fucking do. We are legitimately scared to show up to class tomorrow because of the sole fact anyone can obtain a gun. no matter of their intention a gun should not be able to reach a child’s hand and should never enter a school.\n----\n@JaketheSerling @BGibbles @realDonaldTrump Yes it freaking does stop people from getting guns!\n----\n"
]
],
[
[
"This returns all tweets that are mentioning a username. We can also explicitly get the tweets of a person. Let's download The Donald's last 50 tweets. [Here](https://dev.twitter.com/rest/reference/get/statuses/user_timeline) is the relevant API documentation, [here](https://github.com/ryanmcgrath/twython/blob/master/twython/endpoints.py) are the definitions for twython.",
"_____no_output_____"
]
],
[
[
"#twitter = Twython(CONSUMER_KEY, CONSUMER_SECRET, ACCESS_TOKEN, ACCESS_TOKEN_SECRET)\n\n# count is limited to the last 3200 tweets of a user, a max of 200 per request\nthe_donald_result = twitter.get_user_timeline(screen_name=\"realDonaldTrump\", count=50)\nthe_donald_result",
"_____no_output_____"
]
],
[
[
"Let's count the numbers for The Donald's favorites and retweets.",
"_____no_output_____"
]
],
[
[
"the_donald_favorites = []\nthe_donald_retweets = []\nfor status in the_donald_result:\n print(status[\"text\"])\n print(status[\"favorite_count\"])\n print(\"----\")\n the_donald_favorites.append(status[\"favorite_count\"])\n the_donald_retweets.append(status[\"retweet_count\"])",
"Just spoke to Governor Rick Scott. We are working closely with law enforcement on the terrible Florida school shooting.\n76290\n----\nMy prayers and condolences to the families of the victims of the terrible Florida shooting. No child, teacher or an… https://t.co/RFJxgM1jhm\n116218\n----\nToday, I was honored to be joined by Republicans and Democrats from both the House and Senate, as well as members o… https://t.co/38kUIIV8Cp\n38193\n----\nAs we come together to celebrate the extraordinary contributions of African-Americans to our nation, our thoughts t… https://t.co/SwAJxO88zC\n47068\n----\nNegotiations on DACA have begun. Republicans want to make a deal and Democrats say they want to make a deal. Wouldn… https://t.co/YW4mCpHr0R\n89413\n----\nOur infrastructure plan has been put forward and has received great reviews by everyone except, of course, the Demo… https://t.co/2EfB9sObdN\n98910\n----\nThe journey to #MAGA began @CPAC 2011 and the opportunity to reconnect with friends and supporters is something I l… https://t.co/DSz3Szp4xE\n63652\n----\nThank you to Sue Kruczek, who lost her wonderful and talented son Nick to the Opioid scourge, for your kind words w… https://t.co/0kIdepXBdi\n83215\n----\nThis will be a big week for Infrastructure. After so stupidly spending $7 trillion in the Middle East, it is now ti… https://t.co/OcX7UN18Er\n134070\n----\nJust spoke to @JohnKasich to express condolences and prayers to all for the horrible shooting of two great police o… https://t.co/X7AhuEYwHI\n74595\n----\nRep. Lou Barletta, a Great Republican from Pennsylvania who was one of my very earliest supporters, will make a FAN… https://t.co/dA2lK3WHFU\n94180\n----\n4.2 million hard working Americans have already received a large Bonus and/or Pay Increase because of our recently… https://t.co/oJl5AiCPZs\n121340\n----\nSo many positive things going on for the U.S.A. and the Fake News Media just doesn’t want to go there. Same negativ… https://t.co/LBnzRfAkJx\n120700\n----\nMy Administration has identified three major priorities for creating a safe, modern and lawful immigration system:… https://t.co/DrjXUak41S\n116405\n----\nMy thoughts and prayers are with the two police officers, their families, and everybody at the @WestervillePD. https://t.co/AoingY77Ky\n83053\n----\nRepublicans want to fix DACA far more than the Democrats do. The Dems had all three branches of government back in… https://t.co/0Or3U2CFZR\n116164\n----\n....agencies, not just the FBI & DOJ, now the State Department to dig up dirt on him in the days leading up to the… https://t.co/Of9d7fmsA3\n74093\n----\n“My view is that not only has Trump been vindicated in the last several weeks about the mishandling of the Dossier… https://t.co/dWJuKECfwn\n99572\n----\nPeoples lives are being shattered and destroyed by a mere allegation. Some are true and some are false. Some are ol… https://t.co/6Zz5RAeoQe\n135075\n----\nAccording to the @nytimes, a Russian sold phony secrets on “Trump” to the U.S. Asking price was $10 million, brough… https://t.co/CBMaAX5VMt\n104089\n----\nThe Democrats sent a very political and long response memo which they knew, because of sources and methods (and mor… https://t.co/iR1265rmh3\n116231\n----\nJobless claims have dropped to a 45 year low!\n63319\n----\nCosts on non-military lines will never come down if we do not elect more Republicans in the 2018 Election, and beyo… https://t.co/WgvQYGEaMu\n88462\n----\nWithout more Republicans in Congress, we were forced to increase spending on things we do not like or want in order… https://t.co/7dfsJYWjRY\n109408\n----\nJust signed Bill. Our Military will now be stronger than ever before. We love and need our Military and gave them e… https://t.co/9MtCabhzj8\n128809\n----\nWow! -Senator Mark Warner got caught having extensive contact with a lobbyist for a Russian oligarch. Warner did no… https://t.co/N5SoaLnllm\n126812\n----\nTime to end the visa lottery. Congress must secure the immigration system and protect Americans. https://t.co/yukxm48x9X\n96792\n----\nAs long as we open our eyes to God’s grace - and open our hearts to God’s love - then America will forever be the l… https://t.co/wGUIaD1JX6\n108945\n----\nI will be meeting with Henry Kissinger at 1:45pm. Will be discussing North Korea, China and the Middle East.\n78979\n----\nOur founders invoked our Creator four times in the Declaration of Independence. Our currency declares “IN GOD WE TR… https://t.co/q8ib233y1M\n105822\n----\nWill be heading over shortly to make remarks at The National Prayer Breakfast in Washington. Great religious and po… https://t.co/bEReTLNLo4\n105609\n----\nThe Budget Agreement today is so important for our great Military. It ends the dangerous sequester and gives Secret… https://t.co/VRkKsNDZGC\n108721\n----\nCongratulations to the Republic of Korea on what will be a MAGNIFICENT Winter Olympics! What the South Korean peopl… https://t.co/gGaZUnZBMa\n69734\n----\nBest wishes to the Republic of Korea on hosting the @Olympics! What a wonderful opportunity to show everyone that y… https://t.co/NL6cIfHZgN\n64716\n----\nNEW FBI TEXTS ARE BOMBSHELLS!\n137712\n----\nIn the “old days,” when good news was reported, the Stock Market would go up. Today, when good news is reported, th… https://t.co/mW5vEWhZYY\n111978\n----\nCongratulations @ElonMusk and @SpaceX on the successful #FalconHeavy launch. This achievement, along with @NASA’s c… https://t.co/MaQocjQKcI\n166694\n----\nHAPPY BIRTHDAY to our 40th President of the United States of America, Ronald Reagan! https://t.co/JtEglBhm4c\n180752\n----\nToday, we heard the experiences of law enforcement professionals and community leaders working to combat the threat… https://t.co/BAHG9Ajkio\n75521\n----\nWe need a 21st century MERIT-BASED immigration system. Chain migration and the visa lottery are outdated programs… https://t.co/5SzmbuLXuY\n92211\n----\nPolling shows nearly 7 in 10 Americans support an immigration reform package that includes DACA, fully secures the… https://t.co/OD93mZebu7\n89159\n----\nMy prayers and best wishes are with the family of Edwin Jackson, a wonderful young man whose life was so senselessly taken. @Colts\n104179\n----\nSo disgraceful that a person illegally in our country killed @Colts linebacker Edwin Jackson. This is just one of m… https://t.co/r8vqtb4Tjb\n142531\n----\nThanks to the historic TAX CUTS that I signed into law, your paychecks are going way UP, your taxes are going way D… https://t.co/DKVuox1qa6\n105514\n----\nRepresentative Devin Nunes, a man of tremendous courage and grit, may someday be recognized as a Great American Her… https://t.co/p7egMdnqQs\n133513\n----\nAny deal on DACA that does not include STRONG border security and the desperately needed WALL is a total waste of t… https://t.co/BuGrQqbWfs\n123043\n----\nLittle Adam Schiff, who is desperate to run for higher office, is one of the biggest liars and leakers in Washingto… https://t.co/7csaXTJLoO\n142100\n----\nThank you to @foxandfriends for exposing the truth. Perhaps that’s why your ratings are soooo much better than your untruthful competition!\n107357\n----\nThe Democrats are pushing for Universal HealthCare while thousands of people are marching in the UK because their U… https://t.co/9JK8HHq2bb\n120698\n----\nCongratulations to the Philadelphia Eagles on a great Super Bowl victory!\n212218\n----\n"
]
],
[
[
"Now let's do the same for Hillary.",
"_____no_output_____"
]
],
[
[
"hillary_results = twitter.get_user_timeline(screen_name=\"HillaryClinton\", count=50)\nhillary_results",
"_____no_output_____"
],
[
"hillary_favorites = []\nhillary_retweets = []\nfor status in hillary_results:\n print(status[\"text\"])\n print(status[\"favorite_count\"])\n print(\"----\")\n hillary_favorites.append(status[\"favorite_count\"])\n hillary_retweets.append(status[\"retweet_count\"])",
"RT @GiffordsCourage: Read @GabbyGiffords' statement about today's school shooting in Parkland, Florida: https://t.co/78PglvHbTO\n0\n----\nLove this! https://t.co/8wMSuoQa34\n27800\n----\nTune in today at 2:30pm ET/11:30am PT! https://t.co/rnrRfjlsAI\n9421\n----\nA new book is out today that picks up where I left off in What Happened in explaining “Those Damn Emails.” Terrific… https://t.co/gWO6S0OXuO\n22572\n----\n25 years ago, @billclinton signed the Family and Medical Leave Act, which remains a success all these years later.… https://t.co/hYGHBixDNB\n38978\n----\nLooking forward to being a part of #MAKERSConference2018 and talking about how we can all raise our voices. Watch… https://t.co/ep1R2ldDHU\n9468\n----\nTune in! https://t.co/8OP5wp7gwa\n11524\n----\n@PhilippeReines @RWPUSA @ErinBurnett @CNN 🙏\n1638\n----\nI wrote a Facebook post about a decision I made 10 years ago, what’s changed, & on an issue you didn’t hear a singl… https://t.co/g26PP5U8qJ\n51886\n----\nI called her today to tell her how proud I am of her and to make sure she knows what all women should: we deserve to be heard.\n32336\n----\nA story appeared today about something that happened in 2008. I was dismayed when it occurred, but was heartened th… https://t.co/opzKyXCYR7\n41815\n----\nThank you, @CecileRichards, for your tireless advocacy on behalf of women and girls, and for your grace under press… https://t.co/fsvHCeqD92\n57891\n----\nFor those of you who don’t know @aminatou, she’s a supremely talented young woman with a terrific podcast,… https://t.co/vhTSaNOroT\n28455\n----\nIn 2017, the Women’s March was a beacon of hope and defiance. In 2018, it is a testament to the power and resilienc… https://t.co/i7O6nwg3Qh\n247169\n----\nRT @civilrights_MS: Reps. John Lewis, Bennie Thompson to Attend Grand Celebration of Mississippi Civil Rights Museum https://t.co/t4cnfVXixy\n0\n----\nI’m so heartened by all of you. Onward! https://t.co/vkRj7M3kBS\n129479\n----\nThese words from Dr. King also come to mind today: https://t.co/0qFK3RxBAF\n85872\n----\nBeautifully said, @BerniceKing. An important message today and every day. https://t.co/eYJAGc6i2b\n44619\n----\nThe anniversary of the devastating earthquake 8 years ago is a day to remember the tragedy, honor the resilient peo… https://t.co/58mOD3wtR8\n250146\n----\n@NancyEMcFadden Nancy has a record of beating the odds - from DC to CA - where she helped Gov Brown do a fantastic… https://t.co/itsUB8QgbS\n781\n----\nRT @BillKristol: Two weeks ago a 26-year old soldier raced repeatedly into a burning Bronx apartment building, saving four people before he…\n0\n----\nRT @melvillehouse: Today, the amazing @SusanBordo's THE DESTRUCTION OF HILLARY CLINTON is out in paperback! We're celebrating with this exc…\n0\n----\nFamilies across America had to start 2018 worried that their kids wouldn’t have health care. Failing to act now sho… https://t.co/t08bih6PPb\n29792\n----\nTime to bring CHIP to the Senate floor as promised. This alleged extension until March doesn’t cut it as states fre… https://t.co/vCiOaewaNL\n45523\n----\nThe Iranian people, especially the young, are protesting for the freedom and future they deserve. I hope their gove… https://t.co/lEf6RfM7qj\n115521\n----\nRT @tribelaw: Retweet if you agree it’s totally crazy to suggest that the FBI — having helped sink Hillary’s campaign by revealing that she…\n0\n----\nRT @Jorge_Silva: Must read: Former Clinton deputies: How Rex Tillerson can right the State Department. #statedepartment https://t.co/G2mx…\n0\n----\nThank you to everyone who has donated to Onward Together in our first year -- we’re only able to support these grea… https://t.co/ntArAtXI7Z\n15107\n----\nAlong with @IndivisibleTeam, @ColorOfChange, @EmergeAmerica, @swingleft, and @runforsomething, I know these groups… https://t.co/Uay6VOKdUZ\n11641\n----\nAnd with the help of @collectivepac, 23 African-American candidates have been elected to local, state, and federal… https://t.co/eycWtjE0xt\n6039\n----\nMore than 400 students and leaders met last month at the @votolatino Power Summit to share resources and tools for national advocacy.\n4974\n----\nThe team at @latinovictoryus is working to elect more Latino candidates at every level of government.\n5399\n----\nAt @alliance4youth, young people are using local organizing to address economic inequality, attacks on voting rights, and more.\n5163\n----\n450 brand-new civic leaders and activists attended the very first @ArenaSummit -- and they're on track to train 5,0… https://t.co/s4ZJAcnGRH\n5704\n----\nThe organizers at @iVoteFund worked with partners on the ground to get automatic voter registration on the ballot in Nevada.\n7180\n----\nOnward Together is ending 2017 by supporting six more incredible organizations fighting to protect voting rights an… https://t.co/mE17T8Mffd\n51624\n----\nRT @NickMerrill: What Happened by @HillaryClinton was named a best book of the year by:\n✔️ The New York Times\n✔️ The Washington Post\n✔️ NPR…\n0\n----\nSomething productive to do with your outrage today: https://t.co/XEUb0xLsSl\n51879\n----\nI guest-edited this month’s issue of @TeenVogue. It was a wonderful experience, with lots of terrific contributions… https://t.co/6VQUojh1rj\n42357\n----\nA little girl power in Pasadena...\nhttps://t.co/nqNAMl16yS https://t.co/jdWhdXHL4Y\n10460\n----\nSome photos from the book tour. I met so many incredible people, including some little heros, & the occasional supe… https://t.co/4mHrVcDoPM\n25905\n----\nReturning home after the final week of the book tour for 2017. This was the last copy of What Happened I signed at… https://t.co/UOJUYESAqc\n38166\n----\nThank you to the @lapromisefund & @msjwilly for a terrific discussion, a wonderful group of young people, & a truly… https://t.co/QK39ERYvmz\n10102\n----\nToday is the final day to sign up for 2018 health coverage. Go to https://t.co/3Ll40zSLQe and get covered! https://t.co/PiNnum27J0\n17580\n----\n@ELaffGarb @DHochsprung @ELaffGarb, thinking of you, @nelba_MG, and all the Sandy Hook families this week. Thank you for your courage.\n1923\n----\nYes!\nElections matter. https://t.co/xU8dpew1Cn\n85484\n----\nTonight, Alabama voters elected a senator who'll make them proud. \n\nAnd if Democrats can win in Alabama, we can --… https://t.co/m18Cuh5Iuc\n455044\n----\nMayor Ed Lee’s death is a terrible loss for the people of San Francisco. He was a good friend & a vocal advocate fo… https://t.co/5AsOrwby8h\n43707\n----\nIf you don't live in an area that could be affected, read this to see how you can help those who do:\nhttps://t.co/QTJROuLU1k\n9618\n----\nThinking about all affected & those who could be affected by the California wildfires. If you live near a fire, mak… https://t.co/zL8LOj8AZs\n21424\n----\n"
]
],
[
[
"And let's create DataFrames for both of them and explore their stats:",
"_____no_output_____"
]
],
[
[
"donald_stats = pd.DataFrame({\n \"The Donald Fav\":the_donald_favorites,\n \"The Donald RT\":the_donald_retweets\n })\n\nhillary_stats = pd.DataFrame({\n \"Hillary Fav\":hillary_favorites,\n \"Hillary RT\":hillary_retweets\n })",
"_____no_output_____"
],
[
"donald_stats.head()",
"_____no_output_____"
],
[
"donald_stats.describe()",
"_____no_output_____"
],
[
"hillary_stats.describe()",
"_____no_output_____"
]
],
[
[
"Now we're plotting the tweets; but we have to sort them first so that they make sense. ",
"_____no_output_____"
]
],
[
[
"donald_stats = donald_stats.sort_values(\"The Donald Fav\")\ndonald_stats = donald_stats.reset_index(drop=True)\ndonald_stats.head()",
"_____no_output_____"
],
[
"hillary_stats = hillary_stats.sort_values(\"Hillary Fav\")\nhillary_stats = hillary_stats.reset_index(drop=True)\nhillary_stats.head(15)",
"_____no_output_____"
],
[
"combined = hillary_stats.copy()\ncombined[\"The Donald Fav\"] = donald_stats[\"The Donald Fav\"]\ncombined[\"The Donald RT\"] = donald_stats[\"The Donald RT\"]",
"_____no_output_____"
],
[
"combined.plot()",
"_____no_output_____"
]
],
[
[
"### Exercise\n",
"_____no_output_____"
],
[
"Retreive the last 50 tweets with the hashtag #datavis in the SLC area, print the text and the username.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ec8732a032842a1a9ce3f94e6fe38490f0a640ea | 243,049 | ipynb | Jupyter Notebook | study_roadmaps/4_image_classification_zoo/Classifier - Friedburg Groceries Dataset.ipynb | take2rohit/monk_v1 | 9c567bf2c8b571021b120d879ba9edf7751b9f92 | [
"Apache-2.0"
] | 542 | 2019-11-10T12:09:31.000Z | 2022-03-28T11:39:07.000Z | study_roadmaps/4_image_classification_zoo/Classifier - Friedburg Groceries Dataset.ipynb | take2rohit/monk_v1 | 9c567bf2c8b571021b120d879ba9edf7751b9f92 | [
"Apache-2.0"
] | 117 | 2019-11-12T09:39:24.000Z | 2022-03-12T00:20:41.000Z | study_roadmaps/4_image_classification_zoo/Classifier - Friedburg Groceries Dataset.ipynb | take2rohit/monk_v1 | 9c567bf2c8b571021b120d879ba9edf7751b9f92 | [
"Apache-2.0"
] | 246 | 2019-11-09T21:53:24.000Z | 2022-03-29T00:57:07.000Z | 411.947458 | 120,984 | 0.936786 | [
[
[
"<a href=\"https://colab.research.google.com/github/Tessellate-Imaging/monk_v1/blob/master/study_roadmaps/4_image_classification_zoo/Classifier%20-%20Friedburg%20Groceries%20Dataset.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Table of contents\n\n\n## Install Monk\n\n\n## Using pretrained model for classifying Monkey specie types\n\n\n## Training a classifier from scratch",
"_____no_output_____"
],
[
"<a id='0'></a>\n# Install Monk",
"_____no_output_____"
],
[
"## Using pip (Recommended)\n\n - colab (gpu) \n - All bakcends: `pip install -U monk-colab`\n \n\n - kaggle (gpu) \n - All backends: `pip install -U monk-kaggle`\n \n\n - cuda 10.2\t\n - All backends: `pip install -U monk-cuda102`\n - Gluon bakcned: `pip install -U monk-gluon-cuda102`\n\t - Pytorch backend: `pip install -U monk-pytorch-cuda102`\n - Keras backend: `pip install -U monk-keras-cuda102`\n \n\n - cuda 10.1\t\n - All backend: `pip install -U monk-cuda101`\n\t - Gluon bakcned: `pip install -U monk-gluon-cuda101`\n\t - Pytorch backend: `pip install -U monk-pytorch-cuda101`\n\t - Keras backend: `pip install -U monk-keras-cuda101`\n \n\n - cuda 10.0\t\n - All backend: `pip install -U monk-cuda100`\n\t - Gluon bakcned: `pip install -U monk-gluon-cuda100`\n\t - Pytorch backend: `pip install -U monk-pytorch-cuda100`\n\t - Keras backend: `pip install -U monk-keras-cuda100`\n \n\n - cuda 9.2\t\n - All backend: `pip install -U monk-cuda92`\n\t - Gluon bakcned: `pip install -U monk-gluon-cuda92`\n\t - Pytorch backend: `pip install -U monk-pytorch-cuda92`\n\t - Keras backend: `pip install -U monk-keras-cuda92`\n \n\n - cuda 9.0\t\n - All backend: `pip install -U monk-cuda90`\n\t - Gluon bakcned: `pip install -U monk-gluon-cuda90`\n\t - Pytorch backend: `pip install -U monk-pytorch-cuda90`\n\t - Keras backend: `pip install -U monk-keras-cuda90`\n \n\n - cpu \t\t\n - All backend: `pip install -U monk-cpu`\n\t - Gluon bakcned: `pip install -U monk-gluon-cpu`\n\t - Pytorch backend: `pip install -U monk-pytorch-cpu`\n\t - Keras backend: `pip install -U monk-keras-cpu`",
"_____no_output_____"
],
[
"## Install Monk Manually (Not recommended)\n \n### Step 1: Clone the library\n - git clone https://github.com/Tessellate-Imaging/monk_v1.git\n \n \n \n \n### Step 2: Install requirements \n - Linux\n - Cuda 9.0\n - `cd monk_v1/installation/Linux && pip install -r requirements_cu90.txt`\n - Cuda 9.2\n - `cd monk_v1/installation/Linux && pip install -r requirements_cu92.txt`\n - Cuda 10.0\n - `cd monk_v1/installation/Linux && pip install -r requirements_cu100.txt`\n - Cuda 10.1\n - `cd monk_v1/installation/Linux && pip install -r requirements_cu101.txt`\n - Cuda 10.2\n - `cd monk_v1/installation/Linux && pip install -r requirements_cu102.txt`\n - CPU (Non gpu system)\n - `cd monk_v1/installation/Linux && pip install -r requirements_cpu.txt`\n \n \n - Windows\n - Cuda 9.0 (Experimental support)\n - `cd monk_v1/installation/Windows && pip install -r requirements_cu90.txt`\n - Cuda 9.2 (Experimental support)\n - `cd monk_v1/installation/Windows && pip install -r requirements_cu92.txt`\n - Cuda 10.0 (Experimental support)\n - `cd monk_v1/installation/Windows && pip install -r requirements_cu100.txt`\n - Cuda 10.1 (Experimental support)\n - `cd monk_v1/installation/Windows && pip install -r requirements_cu101.txt`\n - Cuda 10.2 (Experimental support)\n - `cd monk_v1/installation/Windows && pip install -r requirements_cu102.txt`\n - CPU (Non gpu system)\n - `cd monk_v1/installation/Windows && pip install -r requirements_cpu.txt`\n \n \n - Mac\n - CPU (Non gpu system)\n - `cd monk_v1/installation/Mac && pip install -r requirements_cpu.txt`\n \n \n - Misc\n - Colab (GPU)\n - `cd monk_v1/installation/Misc && pip install -r requirements_colab.txt`\n - Kaggle (GPU)\n - `cd monk_v1/installation/Misc && pip install -r requirements_kaggle.txt`\n \n \n \n### Step 3: Add to system path (Required for every terminal or kernel run)\n - `import sys`\n - `sys.path.append(\"monk_v1/\");`",
"_____no_output_____"
],
[
"# Used trained classifier for demo",
"_____no_output_____"
]
],
[
[
"#Using mxnet-gluon backend \n\n# When installed using pip\nfrom monk.gluon_prototype import prototype\n\n\n# When installed manually (Uncomment the following)\n#import os\n#import sys\n#sys.path.append(\"monk_v1/\");\n#sys.path.append(\"monk_v1/monk/\");\n#from monk.gluon_prototype import prototype",
"_____no_output_____"
],
[
"# Download trained weights",
"_____no_output_____"
],
[
"! wget --load-cookies /tmp/cookies.txt \"https://docs.google.com/uc?export=download&confirm=$(wget --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://docs.google.com/uc?export=download&id=1VW9ExZXHmBKwjqxCZ07rATxLnbXEqWuW' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\\1\\n/p')&id=1VW9ExZXHmBKwjqxCZ07rATxLnbXEqWuW\" -O cls_freiburg_trained.zip && rm -rf /tmp/cookies.txt",
"_____no_output_____"
],
[
"! unzip -qq cls_freiburg_trained.zip ",
"_____no_output_____"
],
[
"ls workspace/Project-Groceries",
"\u001b[0m\u001b[01;34mGluon-mobilenetv3_large\u001b[0m/ \u001b[01;34mGluon-mobilenetv3_small\u001b[0m/\r\n"
],
[
"# Load project in inference mode\n\ngtf = prototype(verbose=1);\ngtf.Prototype(\"Project-Groceries\", \"Gluon-mobilenetv3_large\", eval_infer=True);\n\n#Other trained models - uncomment \n#gtf.Prototype(\"Project-Groceries\", \"Gluon-mobilenetv3_small\", eval_infer=True);",
"Mxnet Version: 1.5.1\n\nModel Details\n Loading model - workspace/Project-Groceries/Gluon-mobilenetv3_large/output/models/final-symbol.json\n Model loaded!\n\nExperiment Details\n Project: Project-Groceries\n Experiment: Gluon-mobilenetv3_large\n Dir: /home/ubuntu/Desktop/cls/workspace/Project-Groceries/Gluon-mobilenetv3_large/\n\n"
],
[
"#Infer",
"_____no_output_____"
],
[
"ls -a workspace/test/",
"\u001b[0m\u001b[01;34m.\u001b[0m/ \u001b[01;35mBEANS0001.png\u001b[0m \u001b[01;35mCEREAL0001.png\u001b[0m \u001b[01;35mCHOCOLATE0002.png\u001b[0m \u001b[01;35mFISH0000.png\u001b[0m\r\n\u001b[01;34m..\u001b[0m/ \u001b[01;35mCAKE0000.png\u001b[0m \u001b[01;35mCHIPS0001.png\u001b[0m \u001b[01;35mCOFFEE0001.png\u001b[0m \u001b[01;35mJUICE0002.png\u001b[0m\r\n"
],
[
"img_name = \"workspace/test/BEANS0001.png\"\npredictions = gtf.Infer(img_name=img_name);\nfrom IPython.display import Image\nImage(filename=img_name)",
"Prediction\n Image name: workspace/test/BEANS0001.png\n Predicted class: BEANS\n Predicted score: 0.9995871782302856\n\n"
],
[
"img_name = \"workspace/test/CHOCOLATE0002.png\"\npredictions = gtf.Infer(img_name=img_name);\nfrom IPython.display import Image\nImage(filename=img_name)",
"Prediction\n Image name: workspace/test/CHOCOLATE0002.png\n Predicted class: CHOCOLATE\n Predicted score: 0.8742499947547913\n\n"
]
],
[
[
"# Training custom classifier from scratch",
"_____no_output_____"
],
[
"## Dataset\n - Credits: https://github.com/PhilJd/freiburg_groceries_dataset",
"_____no_output_____"
],
[
"## Download",
"_____no_output_____"
]
],
[
[
"! wget --load-cookies /tmp/cookies.txt \"https://docs.google.com/uc?export=download&confirm=$(wget --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://docs.google.com/uc?export=download&id=19tdHgRNitTOdMT8kSuvGh6p17qxOB34s' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\\1\\n/p')&id=19tdHgRNitTOdMT8kSuvGh6p17qxOB34s\" -O groceries_dataset.zip && rm -rf /tmp/cookies.txt",
"_____no_output_____"
],
[
"! unzip -qq groceries_dataset.zip",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"# Using mxnet-gluon backend \nfrom monk.gluon_prototype import prototype\n\n# For pytorch backend\n#from monk.pytorch_prototype import prototype\n\n# For Keras backend\n#from monk.keras_prototype import prototype",
"_____no_output_____"
],
[
"# Create Project and Experiment\n\ngtf = prototype(verbose=1);\ngtf.Prototype(\"Project-Groceries\", \"Gluon-mobilenetv3_small\");",
"_____no_output_____"
],
[
"gtf.Default(dataset_path=\"groceries_dataset/train\",\n model_name=\"mobilenetv3_small\", \n freeze_base_network=False,\n num_epochs=2);",
"_____no_output_____"
]
],
[
[
"### How to change hyper parameters and models \n - Docs - https://github.com/Tessellate-Imaging/monk_v1#4\n - Examples - https://github.com/Tessellate-Imaging/monk_v1/tree/master/study_roadmaps/1_getting_started_roadmap",
"_____no_output_____"
]
],
[
[
"#Start Training\ngtf.Train();\n\n#Read the training summary generated once you run the cell and training is completed",
"_____no_output_____"
]
],
[
[
"## Validating on the same dataset",
"_____no_output_____"
]
],
[
[
"# Using mxnet-gluon backend \nfrom monk.gluon_prototype import prototype\n\n# For pytorch backend\n#from monk.pytorch_prototype import prototype\n\n# For Keras backend\n#from monk.keras_prototype import prototype",
"_____no_output_____"
],
[
"# Create Project and Experiment\n\ngtf = prototype(verbose=1);\ngtf.Prototype(\"Project-Groceries\", \"Gluon-mobilenetv3_small\", eval_infer=True);",
"_____no_output_____"
],
[
"# Load dataset for validaion\ngtf.Dataset_Params(dataset_path=\"groceries_dataset/train\");\ngtf.Dataset();",
"_____no_output_____"
],
[
"# Run validation\naccuracy, class_based_accuracy = gtf.Evaluate();",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ec8732bba4f951c8bbb64caa848976094fbc0790 | 3,576 | ipynb | Jupyter Notebook | examples/example_parametric_components/make_plasmas.ipynb | zmarkan/paramak | ecf9a46394adb4d6bb5744000ec6e2f74c30f2ba | [
"MIT"
] | 25 | 2021-06-22T07:29:32.000Z | 2022-03-28T13:30:53.000Z | examples/example_parametric_components/make_plasmas.ipynb | zmarkan/paramak | ecf9a46394adb4d6bb5744000ec6e2f74c30f2ba | [
"MIT"
] | 181 | 2021-06-22T00:56:30.000Z | 2022-03-31T22:36:54.000Z | examples/example_parametric_components/make_plasmas.ipynb | zmarkan/paramak | ecf9a46394adb4d6bb5744000ec6e2f74c30f2ba | [
"MIT"
] | 8 | 2021-07-05T12:29:43.000Z | 2022-03-24T13:17:37.000Z | 26.488889 | 113 | 0.510347 | [
[
[
"\"\"\"\nThis python script demonstrates the creation of plasmas\n\"\"\"\n\nimport paramak\n\n \nITER_simple = paramak.Plasma(\n major_radius=620,\n minor_radius=210,\n triangularity=0.33,\n elongation=1.85,\n name=\"ITER_plasma\",\n rotation_angle=1,\n color=(0,1,0)\n)\n\nITER = paramak.PlasmaBoundaries(\n A=-0.155,\n major_radius=620,\n minor_radius=210,\n triangularity=0.33,\n elongation=1.85,\n name=\"ITER_plasma_plasmaboundaries\",\n rotation_angle=1,\n color=(1,0,0),\n azimuth_placement_angle=(360/7)\n)\n\nEU_DEMO = paramak.Plasma(\n name=\"EU_DEMO_plasma\",\n major_radius=910,\n minor_radius=290,\n triangularity=0.33,\n elongation=1.59,\n rotation_angle=1,\n color=(0.9, 0.1, 0.1),\n azimuth_placement_angle=(360/7)*2\n)\n\nST = paramak.Plasma(\n name=\"ST_plasma\",\n major_radius=170,\n minor_radius=129,\n triangularity=0.55,\n elongation=2.3,\n rotation_angle=1,\n color=(0.2, 0.6, 0.2),\n azimuth_placement_angle=(360/7)*3\n)\n\nAST = paramak.Plasma(\n name=\"AST_plasma\",\n major_radius=170,\n minor_radius=129,\n triangularity=-0.55,\n elongation=2.3,\n rotation_angle=1,\n color=(0, 0, 0),\n azimuth_placement_angle=(360/7)*4\n)\n\nNSTX_double_null = paramak.PlasmaBoundaries(\n name=\"NSTX_double_null_plasma_plasmaboundaries\",\n A=0,\n major_radius=850,\n minor_radius=680,\n triangularity=0.35,\n elongation=2,\n color=(1, 1, 0),\n rotation_angle=1,\n configuration=\"double-null\",\n azimuth_placement_angle=(360/7)*5\n)\n\nNSTX_single_null = paramak.PlasmaBoundaries(\n name=\"NSTX_single_null_plasma_plasmaboundaries\",\n A=-0.05,\n major_radius=850,\n minor_radius=680,\n triangularity=0.35,\n elongation=2,\n color=(0.6, 0.3, 0.6),\n rotation_angle=1,\n configuration=\"single-null\",\n azimuth_placement_angle=(360/7)*6\n)\n\n# combines all the plasma into a single object\nall_plasmas = paramak.Reactor([ITER_simple, ITER, EU_DEMO, ST, AST, NSTX_double_null, NSTX_single_null])\n\nall_plasmas.show() ",
"_____no_output_____"
],
[
"all_plasmas.export_html()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ec873f7376997095c0833d0139acc8afacb5983b | 94,612 | ipynb | Jupyter Notebook | NSL_Analysis/NSL_NeuralNetwork.ipynb | suchetsapre/intrusion-dataset-comparison | 2ba607219394bea6f2cb08eecdca74b7047c6b3a | [
"MIT"
] | null | null | null | NSL_Analysis/NSL_NeuralNetwork.ipynb | suchetsapre/intrusion-dataset-comparison | 2ba607219394bea6f2cb08eecdca74b7047c6b3a | [
"MIT"
] | null | null | null | NSL_Analysis/NSL_NeuralNetwork.ipynb | suchetsapre/intrusion-dataset-comparison | 2ba607219394bea6f2cb08eecdca74b7047c6b3a | [
"MIT"
] | null | null | null | 255.708108 | 48,612 | 0.90739 | [
[
[
"import pandas as pd\nimport tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.preprocessing import normalize\nfrom tensorflow.keras.callbacks import EarlyStopping\nimport seaborn as sns\nimport Standard_Functions as sf",
"_____no_output_____"
],
[
"x_train = pd.read_pickle('NSL_x_train_dummy.pkl').values\nx_test = pd.read_pickle('NSL_x_test_dummy.pkl').values\nx_test_21 = pd.read_pickle('NSL_x_test_21_dummy.pkl').values\ny_train_type = pd.read_pickle('NSL_y_train_col_5.pkl').values\ny_test_type = pd.read_pickle('NSL_y_test_col_5.pkl').values\ny_test_21_type = pd.read_pickle('NSL_y_test_21_col_5.pkl').values\ny_train_binary = pd.read_pickle('NSL_y_train_col_1.pkl').values\ny_test_binary = pd.read_pickle('NSL_y_test_col_1.pkl').values\ny_test_21_binary = pd.read_pickle('NSL_y_test_21_col_1.pkl').values\ny_train_intrusion = pd.read_pickle('NSL_y_train_col_40.pkl').values\ny_test_intrusion = pd.read_pickle('NSL_y_test_col_40.pkl').values\ny_test_21_intrusion = pd.read_pickle('NSL_y_test_21_col_40.pkl').values",
"_____no_output_____"
],
[
"x_train_norm= normalize(x_train, axis=1, norm='l2')\nx_test_norm= normalize(x_test, axis=1, norm='l2')\nx_test_21_norm= normalize(x_test_21, axis=1, norm='l2')\ny_train_type_norm= normalize(y_train_type, axis=1, norm='l2')\ny_test_type_norm= normalize(y_test_type, axis=1, norm='l2')\ny_test_21_type_norm= normalize(y_test_21_type, axis=1, norm='l2')\ny_train_binary_norm= normalize(y_train_binary, axis=1, norm='l2')\ny_test_binary_norm= normalize(y_test_binary, axis=1, norm='l2')\ny_test_21_binary_norm= normalize(y_test_21_binary, axis=1, norm='l2')\ny_train_intrusion_norm= normalize(y_train_intrusion, axis=1, norm='l2')\ny_test_intrusion_norm= normalize(y_test_intrusion, axis=1, norm='l2')\ny_test_21_intrusion_norm= normalize(y_test_21_intrusion, axis=1, norm='l2')",
"_____no_output_____"
],
[
"model = tf.keras.models.Sequential()\n# model.add(tf.keras.layers.Flatten())\nmodel.add(tf.keras.layers.Dense(100, activation = tf.nn.relu)) \nmodel.add(tf.keras.layers.Dense(100, activation = tf.nn.relu))\n# model.add(tf.keras.layers.Dense(200, activation = tf.nn.relu))\nmodel.add(tf.keras.layers.Dense(1, activation = tf.nn.sigmoid))\n\nmodel.compile(optimizer = 'adam', loss = 'binary_crossentropy', metrics=['accuracy']) #categorical_crossentropy\n\nearly_stopping_monitor = EarlyStopping(patience=20)",
"_____no_output_____"
],
[
"history = model.fit(x_train_norm, y_train_binary, batch_size=256, epochs=20, validation_data=(x_test_norm, y_test_binary), callbacks=[early_stopping_monitor])",
"Train on 125972 samples, validate on 22543 samples\nEpoch 1/20\n125972/125972 [==============================] - 3s 24us/sample - loss: 0.1678 - acc: 0.9415 - val_loss: 0.8528 - val_acc: 0.7687\nEpoch 2/20\n125972/125972 [==============================] - 3s 20us/sample - loss: 0.0837 - acc: 0.9683 - val_loss: 1.0476 - val_acc: 0.7485\nEpoch 3/20\n125972/125972 [==============================] - 3s 20us/sample - loss: 0.0681 - acc: 0.9738 - val_loss: 1.0636 - val_acc: 0.7680\nEpoch 4/20\n125972/125972 [==============================] - 3s 20us/sample - loss: 0.0593 - acc: 0.9791 - val_loss: 1.1999 - val_acc: 0.7709\nEpoch 5/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0524 - acc: 0.9828 - val_loss: 1.2650 - val_acc: 0.7664\nEpoch 6/20\n125972/125972 [==============================] - 3s 20us/sample - loss: 0.0475 - acc: 0.9846 - val_loss: 1.4104 - val_acc: 0.7584\nEpoch 7/20\n125972/125972 [==============================] - 3s 22us/sample - loss: 0.0432 - acc: 0.9866 - val_loss: 1.1908 - val_acc: 0.7810\nEpoch 8/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0391 - acc: 0.9877 - val_loss: 1.0843 - val_acc: 0.7931\nEpoch 9/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0365 - acc: 0.9884 - val_loss: 1.2811 - val_acc: 0.7779\nEpoch 10/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0336 - acc: 0.9895 - val_loss: 1.1624 - val_acc: 0.7816\nEpoch 11/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0309 - acc: 0.9905 - val_loss: 1.1255 - val_acc: 0.7851\nEpoch 12/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0287 - acc: 0.9912 - val_loss: 1.0765 - val_acc: 0.7870\nEpoch 13/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0268 - acc: 0.9918 - val_loss: 1.3106 - val_acc: 0.7672\nEpoch 14/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0249 - acc: 0.9926 - val_loss: 1.0077 - val_acc: 0.7939\nEpoch 15/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0242 - acc: 0.9928 - val_loss: 1.6499 - val_acc: 0.7678\nEpoch 16/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0229 - acc: 0.9931 - val_loss: 1.1857 - val_acc: 0.7786\nEpoch 17/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0216 - acc: 0.9937 - val_loss: 1.4131 - val_acc: 0.7823\nEpoch 18/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0216 - acc: 0.9936 - val_loss: 1.4192 - val_acc: 0.7747\nEpoch 19/20\n125972/125972 [==============================] - 3s 21us/sample - loss: 0.0209 - acc: 0.9940 - val_loss: 1.3083 - val_acc: 0.7821\nEpoch 20/20\n125972/125972 [==============================] - 3s 22us/sample - loss: 0.0196 - acc: 0.9945 - val_loss: 1.5234 - val_acc: 0.7716\n"
],
[
"sf.plot_history(history)\n ",
"_____no_output_____"
],
[
"y_pred = model.predict(x_test_norm)",
"_____no_output_____"
],
[
"#accuracy_per_class(model.predict(x_test_21), y_test_21_intrusion, list(pd.read_pickle('NSL_y_train_col_40.pkl').columns)) #[\"normal\", \"dos\", \"r2l\", \"u2r\", \"probe\"]\nsf.accuracy_per_class_binary(model.predict(x_test_21), y_test_21_binary)",
"_____no_output_____"
],
[
"binary_classification_accuracy_per_class = sf.accuracy_per_class_binary(model.predict(x_test_norm), y_test_binary)\n\ntp, tn, fp, fn = sf.tp_tn_fp_fn(binary_classification_accuracy_per_class)\n\nprecision = tp/(tp+fp)\nrecall = tp/(tp+fn)\nf1_score = 2*(recall * precision) / (recall + precision)\n\nprecision, recall, f1_score",
"_____no_output_____"
]
],
[
[
"precision, recall, f1_score \n(0.9665514858327574, 0.544965710723192, 0.6969651666915833)",
"_____no_output_____"
],
[
" index_to_type_dict = {\n 0 : 'normal',\n 1 : 'dos',\n 2 : 'r2l',\n 3 : 'u2r',\n 4 : 'probe'\n }",
"_____no_output_____"
]
],
[
[
"sf.confusion_matrix_type(y_pred, y_test_type)",
"_____no_output_____"
],
[
"cm = sf.confusion_matrix_type(y_pred, y_test_type)",
"_____no_output_____"
],
[
"def show_confusion_matrix(confusion_matrix_dict):\n SMALL_SIZE = 14\n MEDIUM_SIZE = 17\n BIGGER_SIZE = 50\n\n plt.rc('font', size=SMALL_SIZE) # controls default text sizes\n plt.rc('axes', titlesize=SMALL_SIZE) # fontsize of the axes title\n plt.rc('axes', labelsize=MEDIUM_SIZE) # fontsize of the x and y labels\n plt.rc('xtick', labelsize=SMALL_SIZE) # fontsize of the tick labels\n plt.rc('ytick', labelsize=SMALL_SIZE) # fontsize of the tick labels\n plt.rc('legend', fontsize=SMALL_SIZE) # legend fontsize\n plt.rc('figure', titlesize=BIGGER_SIZE) # fontsize of the figure title\n \n index_to_type_dict = {\n 0 : 'normal',\n 1 : 'dos',\n 2 : 'r2l',\n 3 : 'u2r',\n 4 : 'probe'\n }\n ax = plt.subplot()\n confusion_matrix_array = []\n for i in range(5):\n the_type = index_to_type_dict[i]\n confusion_matrix_array.append(confusion_matrix_dict[the_type])\n confusion_matrix_array = np.asarray(confusion_matrix_array)\n confusion_matrix_array = normalize(confusion_matrix_array, axis=1, norm='l2')\n sns.heatmap(confusion_matrix_array, annot=True, ax = ax, cmap='Reds')\n ax.set_xlabel('Predicted Labels')\n ax.set_ylabel('True Labels')\n ax.set_title('Confusion Matrix', fontsize=23)\n ax.xaxis.set_ticklabels(['normal', 'dos', 'r2l', 'u2r', 'probe'])\n ax.yaxis.set_ticklabels(['probe', 'u2r', 'r2l', 'dos', 'normal'])\n #plt.matshow(confusion_matrix_array)\n #plt.colorbar()\n plt.show()",
"_____no_output_____"
],
[
"show_confusion_matrix(cm)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ec874f8fb1475790ce5d1bac80f3a98b7b2f5c88 | 208,744 | ipynb | Jupyter Notebook | examples/notebooks/module_02_01_matplotlib_pyplot.ipynb | yngtodd/eleven | aaf2777f0e89d919d992dd9c65dc78e9845fbd48 | [
"BSD-3-Clause"
] | 1 | 2021-09-06T22:52:16.000Z | 2021-09-06T22:52:16.000Z | examples/notebooks/module_02_01_matplotlib_pyplot.ipynb | yngtodd/eleven | aaf2777f0e89d919d992dd9c65dc78e9845fbd48 | [
"BSD-3-Clause"
] | 5 | 2020-08-10T18:28:56.000Z | 2020-10-28T03:09:50.000Z | examples/notebooks/module_02_01_matplotlib_pyplot.ipynb | yngtodd/eleven | aaf2777f0e89d919d992dd9c65dc78e9845fbd48 | [
"BSD-3-Clause"
] | 7 | 2020-08-05T20:56:43.000Z | 2021-12-16T15:35:56.000Z | 372.757143 | 37,636 | 0.93995 | [
[
[
"# Matplotlib Pyplot\n\nThe `matplotlib.pyplot` module is a collection of functions that let us get up and running with visualization quickly. Interacting with `pyplot` directly is referred to as the state-machine interface of Matplotlib. This is the first of two approaches that we will take. \n\nThere are two things that we want to keep in mind with `pyplot`: one, `pyplot` is stateful; and two, `pyplot` tries to set reasonable defaults for us. These two things together can make `pyplot` feel a bit like magic. If you find yourself thinking, \"Wait a minute, what is Matplotlib doing here?\", then hang tight. When we introduce the next Matplotlib interface, then things may seem a bit more clear.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n%matplotlib inline",
"_____no_output_____"
],
[
"# Let's make our runs consistent\nnp.random.seed(13)",
"_____no_output_____"
]
],
[
[
"## Data\n\nLet's start by creating some simple curves to plot.",
"_____no_output_____"
]
],
[
[
"squares = [x**2 for x in range(11)]\ncubes = [x**3 for x in range(11)]\nquads = [x**4 for x in range(11)]",
"_____no_output_____"
],
[
"squares",
"_____no_output_____"
]
],
[
[
"## A Simple Plot",
"_____no_output_____"
]
],
[
[
"plt.plot(squares)\nplt.show()",
"_____no_output_____"
]
],
[
[
"This is about as simple as a Matplotlib figure can get. Given `squares`, a Python list of the first 10 squares, Matplotlib gives us a line plot. The `pyplot` module is inferring a lot here. Notice that we did not specify our x and y axes. We didn’t even tell it Matplotlib that we were looking to plot a curve here!\n\nAlso note that we never instantiated an object from the pyplot module. Instead, we are calling the module directly. First, we feed our data into `plt.plt(squares)`, then we call `plt.show()` to render our figure. If we were to call `plt.show()` again without passing the data back into `plt.plot()`, Matplotlib will not render the figure a second time. That is because Matploglib clears the state of our plot as soon as we call `show`.",
"_____no_output_____"
]
],
[
[
"# Where did our state go?\nplt.show()",
"_____no_output_____"
]
],
[
[
"`pyplot` gives us some functions to add titles, axis labels, and more",
"_____no_output_____"
]
],
[
[
"plt.plot(squares)\nplt.title('Squares Plot')\nplt.ylabel(r'$X^{2}$') # note the LaTeX\nplt.xlabel('X')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Plotting Multiple Curves",
"_____no_output_____"
]
],
[
[
"plt.plot(squares, label='b = 2')\nplt.plot(cubes, label='b = 3')\nplt.title('Curves')\nplt.ylabel(r'$X^{b}$')\nplt.xlabel('X')\nplt.legend(loc=\"upper left\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"Note how the scale of the y-axis changed in this figure. Scale is something that is important to consider whenever we are working with visualizations. Since Matplotlib is keeping the overall dimensions of the figure the same across each of these figures, the curves can look deceptive. Take a close look at the orange curve, $X^{b}$ where $b = 3$. If you were to quickly glance at this figure compared with the previous figure, you might think that the only change we made was the color of the curve! \n\nIt is always a good idea to think about how relative scale effects our perceptions of visualization. This is also true of color choices, and a host of other design features. Time to take some studio art classes! ",
"_____no_output_____"
],
[
"## Plotting Multiple Subplots\n\nMaybe our plot will be more clear if we separate the two plots into two subplots.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(18, 5))\nplt.subplot(131)\nplt.plot(cubes, label='b = 3')\nplt.ylabel(r'$X^{b}$')\nplt.xlabel('X')\nplt.subplot(132)\nplt.plot(squares, label='b = 2')\nplt.title('Curves')\nplt.xlabel('X')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Oh no, this is worse! Look at those y-axis scales! And why is the title associated with the second subplot. And we no longer have a legend since these plots don't belong to the `Axes`. Which one of these curves corresponds to $X^{2}$ again?",
"_____no_output_____"
],
[
"## What happens when we keep adding on?",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(18, 5))\nplt.subplot(131)\nplt.plot(cubes, label='b = 3')\nplt.ylabel(r'$X^{b}$')\nplt.xlabel('X')\nplt.subplot(132)\nplt.plot(squares, label='b = 2')\nplt.title('Curves')\nplt.xlabel('X')\nplt.subplot(133)\nplt.plot(quads, label='b = 4')\nplt.xlabel('X')\nplt.show()",
"_____no_output_____"
]
],
[
[
"This is getting out of hand. Look at all of that repetition! You know how I created this figure? I copied and pasted snippets from the previous figure, and then modified some of the variables. The horror. What if I messed up when editing, or accidentally copied the wrong bit of the previous code? But hey, that title is now centered in the figure.\n\nSince this is confusing, let's go back the single plot:",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(20, 10))\nplt.plot(squares, label='b = 2')\nplt.plot(cubes, label='b = 3')\nplt.plot(quads, label='b = 4')\nplt.ylabel(r'$X^{b}$')\nplt.xlabel('X')\nplt.title('Curves')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Not All Plots Are Line Plots, Todd\n\nthe default plot in `pyplot` is a line plot, but are other functions in the module that we can use.",
"_____no_output_____"
],
[
"### Scatter Plot",
"_____no_output_____"
]
],
[
[
"num_points = 30\nx = np.random.rand(num_points)\ny = np.random.rand(num_points)\ncolors = np.random.rand(num_points)\nsize = (50 * np.random.rand(num_points))**2\n\nprint(f'First five entries of size:\\n\\n{size[:5]}')",
"First five entries of size:\n\n[1783.19778834 1120.26145061 29.86890733 19.47171832 2335.12219308]\n"
],
[
"plt.figure(figsize=(20, 10))\nplt.scatter(x, y, s=size, c=colors, alpha=0.5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"By using an `alpha=0.5`, we can some transparency. This can be helpful when some of the visual information contained in your plot overlaps.",
"_____no_output_____"
],
[
"### Bar Plot",
"_____no_output_____"
]
],
[
[
"labels = ['Vanilla', 'Chocolate', 'Pistachio', 'Mint']\nyoung_todd = [40, 40, 5, 5]\nless_young_todd = [25, 25, 25, 25]\n\nlabel_loc = np.arange(len(labels))\n# What happens if we try to use a list?\n#label_loc = [x for x in range(len(labels))]\nwidth = 0.3\n\nplt.figure(figsize=(20, 10))\nplt.bar(label_loc - width/2, young_todd, width, label=\"Baby Todd\")\nplt.bar(label_loc + width/2, less_young_todd, width, label=\"Current Todd\")\n\nplt.title(\"Todd's Ice Cream Preferences by Age\")\nplt.ylabel('Todd Score')\nplt.xticks(ticks=label_loc, labels=labels)\nplt.legend()",
"_____no_output_____"
]
],
[
[
"We have only scratched the surface with these plots. You can really fine tune your visualizations to meet your needs. If you are like me, remembering all of details can be a bit cumbersome. How do I set my x-axis labels, What was that thing about offsetting bars in a bar plot? What are the parameters of `plt.xticks`? Luckily, the docs have our back.",
"_____no_output_____"
]
],
[
[
"# Check a function's documentation\n# plt.xticks?",
"_____no_output_____"
]
],
[
[
"### Histograms",
"_____no_output_____"
]
],
[
[
"x1 = np.random.normal(0, 0.8, 1000)\nx2 = np.random.normal(-2, 1, 1000)\nx3 = np.random.normal(3, 2, 1000)\n\nkwargs = dict(density='stepfilled', alpha=0.3, bins=50)\n\nplt.figure(figsize=(20, 10))\n_ = plt.hist(x1, **kwargs)\n_ = plt.hist(x2, **kwargs)\n_ = plt.hist(x3, **kwargs)\n\nplt.title(\"Three Gaussian Distributions\")",
"_____no_output_____"
]
],
[
[
"We could get into a lot more detail here. But, before we do, this might be a good time to introduce the second interface for Matplotlib. Let's switch over to our next notebook, `module_02_02_matplotlib_object_oriented.ipynb`. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec8759e209e30053fc0a05093bdf3b4d7cb17735 | 8,397 | ipynb | Jupyter Notebook | ipynb/_a1-starting.ipynb | geocompr/pytest | fea416c99ddd47961ae36c3f8cfecfaa792eb778 | [
"CC0-1.0"
] | 1 | 2022-02-17T13:32:58.000Z | 2022-02-17T13:32:58.000Z | ipynb/_a1-starting.ipynb | geocompr/pytest | fea416c99ddd47961ae36c3f8cfecfaa792eb778 | [
"CC0-1.0"
] | 6 | 2022-01-13T21:20:06.000Z | 2022-02-17T19:45:31.000Z | ipynb/a1-starting.ipynb | anitagraser/py | 84f4102b96380a3acbe533ed4820427676ba226e | [
"CC0-1.0"
] | 1 | 2022-02-17T08:24:23.000Z | 2022-02-17T08:24:23.000Z | 87.46875 | 702 | 0.677861 | [
[
[
"# Installing R and RStudio {#sec-appendix-starting}\n\nTo get started with R, you need to acquire your own copy. This appendix will show you how to download R as well as RStudio, a software application that makes R easier to use. You'll go from downloading R to opening your first R session.\n\nBoth R and RStudio are free and easy to download.\n\n## How to Download and Install R\n\nR is maintained by an international team of developers who make the language available through the web page of [The Comprehensive R Archive Network](http://cran.r-project.org). The top of the web page provides three links for downloading R. Follow the link that describes your operating system: Windows, Mac, or Linux.\n\n### Windows\n\nTo install R on Windows, click the \"Download R for Windows\" link. Then click the \"base\" link. Next, click the first link at the top of the new page. This link should say something like \"Download R 3.0.3 for Windows,\" except the 3.0.3 will be replaced by the most current version of R. The link downloads an installer program, which installs the most up-to-date version of R for Windows. Run this program and step through the installation wizard that appears. The wizard will install R into your program files folders and place a shortcut in your Start menu. Note that you'll need to have all of the appropriate administration privileges to install new software on your machine.\n\n### Mac\n\nTo install R on a Mac, click the \"Download R for Mac\" link. Next, click on the `R-3.0.3` package link (or the package link for the most current release of R). An installer will download to guide you through the installation process, which is very easy. The installer lets you customize your installation, but the defaults will be suitable for most users. I've never found a reason to change them. If your computer requires a password before installing new progams, you'll need it here.\n\n::: callout-note\n## Binaries Versus Source\n\nR can be installed from precompiled binaries or built from source on any operating system. For Windows and Mac machines, installing R from binaries is extremely easy. The binary comes preloaded in its own installer. Although you can build R from source on these platforms, the process is much more complicated and won't provide much benefit for most users. For Linux systems, the opposite is true. Precompiled binaries can be found for some systems, but it is much more common to build R from source files when installing on Linux. The download pages on [CRAN's website](http://cran.r-project.org) provide information about building R from source for the Windows, Mac, and Linux platforms.\n:::\n\n### Linux\n\nR comes preinstalled on many Linux systems, but you'll want the newest version of R if yours is out of date. [The CRAN website](http://cran.r-project.org) provides files to build R from source on Debian, Redhat, SUSE, and Ubuntu systems under the link \"Download R for Linux.\" Click the link and then follow the directory trail to the version of Linux you wish to install on. The exact installation procedure will vary depending on the Linux system you use. CRAN guides the process by grouping each set of source files with documentation or README files that explain how to install on your system.\n\n::: callout-note\n## 32-bit Versus 64-bit\n\nR comes in both 32-bit and 64-bit versions. Which should you use? In most cases, it won't matter. Both versions use 32-bit integers, which means they compute numbers to the same numerical precision. The difference occurs in the way each version manages memory. 64-bit R uses 64-bit memory pointers, and 32-bit R uses 32-bit memory pointers. This means 64-bit R has a larger memory space to use (and search through).\n\nAs a rule of thumb, 32-bit builds of R are faster than 64-bit builds, though not always. On the other hand, 64-bit builds can handle larger files and data sets with fewer memory management problems. In either version, the maximum allowable vector size tops out at around 2 billion elements. If your operating system doesn't support 64-bit programs, or your RAM is less than 4 GB, 32-bit R is for you. The Windows and Mac installers will automatically install both versions if your system supports 64-bit R.\n:::\n\n## Using R\n\nR isn't a program that you can open and start using, like Microsoft Word or Internet Explorer. Instead, R is a computer language, like C, C++, or UNIX. You use R by writing commands in the R language and asking your computer to interpret them. In the old days, people ran R code in a UNIX terminal window---as if they were hackers in a movie from the 1980s. Now almost everyone uses R with an application called RStudio, and I recommend that you do, too.\n\n::: callout-tip\n## R and UNIX\n\nYou can still run R in a UNIX or BASH window by typing the command:\n\n``` bash\n$ R\n```\n\nwhich opens an R interpreter. You can then do your work and close the interpreter by running `q()` when you are finished.\n:::\n\n## RStudio\n\nRStudio *is* an application like Microsoft Word---except that instead of helping you write in English, RStudio helps you write in R. I use RStudio throughout the book because it makes using R much easier. Also, the RStudio interface looks the same for Windows, Mac OS, and Linux. That will help me match the book to your personal experience.\n\nYou can [download RStudio](http://www.rstudio.com/ide) for free. Just click the \"Download RStudio\" button and follow the simple instructions that follow. Once you've installed RStudio, you can open it like any other program on your computer---usually by clicking an icon on your desktop.\n\n::: callout-tip\n## The R GUIs\n\nWindows and Mac users usually do not program from a terminal window, so the Windows and Mac downloads for R come with a simple program that opens a terminal-like window for you to run R code in. This is what opens when you click the R icon on your Windows or Mac computer. These programs do a little more than the basic terminal window, but not much. You may hear people refer to them as the Windows or Mac R GUIs.\n:::\n\nWhen you open RStudio, a window appears with three panes in it, as in @fig-layout. The largest pane is a console window. This is where you'll run your R code and see results. The console window is exactly what you'd see if you ran R from a UNIX console or the Windows or Mac GUIs. Everything else you see is unique to RStudio. Hidden in the other panes are a text editor, a graphics window, a debugger, a file manager, and much more. You'll learn about these panes as they become useful throughout the course of this book.\n\n{#fig-layout}\n\n::: callout-tip\n## Do I still need to download R?\n\nEven if you use RStudio, you'll still need to download R to your computer. RStudio helps you use the version of R that lives on your computer, but it doesn't come with a version of R on its own.\n:::\n\n## Opening R\n\nNow that you have both R and RStudio on your computer, you can begin using R by opening the RStudio program. Open RStudio just as you would any program, by clicking on its icon or by typing \"RStudio\" at the Windows Run prompt.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ec876433e9444e0abeb567693f827cfa80227519 | 140,865 | ipynb | Jupyter Notebook | tests/scikit-learn/plot_out_of_core_classification.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 1 | 2019-05-10T09:16:23.000Z | 2019-05-10T09:16:23.000Z | tests/scikit-learn/plot_out_of_core_classification.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | null | null | null | tests/scikit-learn/plot_out_of_core_classification.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 1 | 2019-05-10T09:17:28.000Z | 2019-05-10T09:17:28.000Z | 214.406393 | 42,028 | 0.887247 | [
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"#environment setup with watermark\n%load_ext watermark\n%watermark -a 'Gopala KR' -u -d -v -p watermark,numpy,pandas,matplotlib,nltk,sklearn,tensorflow,theano,mxnet,chainer",
"WARNING (theano.tensor.blas): Using NumPy C-API based implementation for BLAS functions.\n"
]
],
[
[
"\n# Out-of-core classification of text documents\n\n\nThis is an example showing how scikit-learn can be used for classification\nusing an out-of-core approach: learning from data that doesn't fit into main\nmemory. We make use of an online classifier, i.e., one that supports the\npartial_fit method, that will be fed with batches of examples. To guarantee\nthat the features space remains the same over time we leverage a\nHashingVectorizer that will project each example into the same feature space.\nThis is especially useful in the case of text classification where new\nfeatures (words) may appear in each batch.\n\nThe dataset used in this example is Reuters-21578 as provided by the UCI ML\nrepository. It will be automatically downloaded and uncompressed on first run.\n\nThe plot represents the learning curve of the classifier: the evolution\nof classification accuracy over the course of the mini-batches. Accuracy is\nmeasured on the first 1000 samples, held out as a validation set.\n\nTo limit the memory consumption, we queue examples up to a fixed amount before\nfeeding them to the learner.\n\n",
"_____no_output_____"
]
],
[
[
"# Authors: Eustache Diemert <[email protected]>\n# @FedericoV <https://github.com/FedericoV/>\n# License: BSD 3 clause\n\nfrom __future__ import print_function\n\nfrom glob import glob\nimport itertools\nimport os.path\nimport re\nimport tarfile\nimport time\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom matplotlib import rcParams\n\nfrom sklearn.externals.six.moves import html_parser\nfrom sklearn.externals.six.moves.urllib.request import urlretrieve\nfrom sklearn.datasets import get_data_home\nfrom sklearn.feature_extraction.text import HashingVectorizer\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.linear_model import PassiveAggressiveClassifier\nfrom sklearn.linear_model import Perceptron\nfrom sklearn.naive_bayes import MultinomialNB\n\n\ndef _not_in_sphinx():\n # Hack to detect whether we are running by the sphinx builder\n return '__file__' in globals()",
"_____no_output_____"
]
],
[
[
"Reuters Dataset related routines\n--------------------------------\n\n\n",
"_____no_output_____"
]
],
[
[
"class ReutersParser(html_parser.HTMLParser):\n \"\"\"Utility class to parse a SGML file and yield documents one at a time.\"\"\"\n\n def __init__(self, encoding='latin-1'):\n html_parser.HTMLParser.__init__(self)\n self._reset()\n self.encoding = encoding\n\n def handle_starttag(self, tag, attrs):\n method = 'start_' + tag\n getattr(self, method, lambda x: None)(attrs)\n\n def handle_endtag(self, tag):\n method = 'end_' + tag\n getattr(self, method, lambda: None)()\n\n def _reset(self):\n self.in_title = 0\n self.in_body = 0\n self.in_topics = 0\n self.in_topic_d = 0\n self.title = \"\"\n self.body = \"\"\n self.topics = []\n self.topic_d = \"\"\n\n def parse(self, fd):\n self.docs = []\n for chunk in fd:\n self.feed(chunk.decode(self.encoding))\n for doc in self.docs:\n yield doc\n self.docs = []\n self.close()\n\n def handle_data(self, data):\n if self.in_body:\n self.body += data\n elif self.in_title:\n self.title += data\n elif self.in_topic_d:\n self.topic_d += data\n\n def start_reuters(self, attributes):\n pass\n\n def end_reuters(self):\n self.body = re.sub(r'\\s+', r' ', self.body)\n self.docs.append({'title': self.title,\n 'body': self.body,\n 'topics': self.topics})\n self._reset()\n\n def start_title(self, attributes):\n self.in_title = 1\n\n def end_title(self):\n self.in_title = 0\n\n def start_body(self, attributes):\n self.in_body = 1\n\n def end_body(self):\n self.in_body = 0\n\n def start_topics(self, attributes):\n self.in_topics = 1\n\n def end_topics(self):\n self.in_topics = 0\n\n def start_d(self, attributes):\n self.in_topic_d = 1\n\n def end_d(self):\n self.in_topic_d = 0\n self.topics.append(self.topic_d)\n self.topic_d = \"\"\n\n\ndef stream_reuters_documents(data_path=None):\n \"\"\"Iterate over documents of the Reuters dataset.\n\n The Reuters archive will automatically be downloaded and uncompressed if\n the `data_path` directory does not exist.\n\n Documents are represented as dictionaries with 'body' (str),\n 'title' (str), 'topics' (list(str)) keys.\n\n \"\"\"\n\n DOWNLOAD_URL = ('http://archive.ics.uci.edu/ml/machine-learning-databases/'\n 'reuters21578-mld/reuters21578.tar.gz')\n ARCHIVE_FILENAME = 'reuters21578.tar.gz'\n\n if data_path is None:\n data_path = os.path.join(get_data_home(), \"reuters\")\n if not os.path.exists(data_path):\n \"\"\"Download the dataset.\"\"\"\n print(\"downloading dataset (once and for all) into %s\" %\n data_path)\n os.mkdir(data_path)\n\n def progress(blocknum, bs, size):\n total_sz_mb = '%.2f MB' % (size / 1e6)\n current_sz_mb = '%.2f MB' % ((blocknum * bs) / 1e6)\n if _not_in_sphinx():\n print('\\rdownloaded %s / %s' % (current_sz_mb, total_sz_mb),\n end='')\n\n archive_path = os.path.join(data_path, ARCHIVE_FILENAME)\n urlretrieve(DOWNLOAD_URL, filename=archive_path,\n reporthook=progress)\n if _not_in_sphinx():\n print('\\r', end='')\n print(\"untarring Reuters dataset...\")\n tarfile.open(archive_path, 'r:gz').extractall(data_path)\n print(\"done.\")\n\n parser = ReutersParser()\n for filename in glob(os.path.join(data_path, \"*.sgm\")):\n for doc in parser.parse(open(filename, 'rb')):\n yield doc",
"_____no_output_____"
]
],
[
[
"Main\n----\n\nCreate the vectorizer and limit the number of features to a reasonable\nmaximum\n\n",
"_____no_output_____"
]
],
[
[
"vectorizer = HashingVectorizer(decode_error='ignore', n_features=2 ** 18,\n alternate_sign=False)\n\n\n# Iterator over parsed Reuters SGML files.\ndata_stream = stream_reuters_documents()\n\n# We learn a binary classification between the \"acq\" class and all the others.\n# \"acq\" was chosen as it is more or less evenly distributed in the Reuters\n# files. For other datasets, one should take care of creating a test set with\n# a realistic portion of positive instances.\nall_classes = np.array([0, 1])\npositive_class = 'acq'\n\n# Here are some classifiers that support the `partial_fit` method\npartial_fit_classifiers = {\n 'SGD': SGDClassifier(),\n 'Perceptron': Perceptron(),\n 'NB Multinomial': MultinomialNB(alpha=0.01),\n 'Passive-Aggressive': PassiveAggressiveClassifier(),\n}\n\n\ndef get_minibatch(doc_iter, size, pos_class=positive_class):\n \"\"\"Extract a minibatch of examples, return a tuple X_text, y.\n\n Note: size is before excluding invalid docs with no topics assigned.\n\n \"\"\"\n data = [(u'{title}\\n\\n{body}'.format(**doc), pos_class in doc['topics'])\n for doc in itertools.islice(doc_iter, size)\n if doc['topics']]\n if not len(data):\n return np.asarray([], dtype=int), np.asarray([], dtype=int)\n X_text, y = zip(*data)\n return X_text, np.asarray(y, dtype=int)\n\n\ndef iter_minibatches(doc_iter, minibatch_size):\n \"\"\"Generator of minibatches.\"\"\"\n X_text, y = get_minibatch(doc_iter, minibatch_size)\n while len(X_text):\n yield X_text, y\n X_text, y = get_minibatch(doc_iter, minibatch_size)\n\n\n# test data statistics\ntest_stats = {'n_test': 0, 'n_test_pos': 0}\n\n# First we hold out a number of examples to estimate accuracy\nn_test_documents = 1000\ntick = time.time()\nX_test_text, y_test = get_minibatch(data_stream, 1000)\nparsing_time = time.time() - tick\ntick = time.time()\nX_test = vectorizer.transform(X_test_text)\nvectorizing_time = time.time() - tick\ntest_stats['n_test'] += len(y_test)\ntest_stats['n_test_pos'] += sum(y_test)\nprint(\"Test set is %d documents (%d positive)\" % (len(y_test), sum(y_test)))\n\n\ndef progress(cls_name, stats):\n \"\"\"Report progress information, return a string.\"\"\"\n duration = time.time() - stats['t0']\n s = \"%20s classifier : \\t\" % cls_name\n s += \"%(n_train)6d train docs (%(n_train_pos)6d positive) \" % stats\n s += \"%(n_test)6d test docs (%(n_test_pos)6d positive) \" % test_stats\n s += \"accuracy: %(accuracy).3f \" % stats\n s += \"in %.2fs (%5d docs/s)\" % (duration, stats['n_train'] / duration)\n return s\n\n\ncls_stats = {}\n\nfor cls_name in partial_fit_classifiers:\n stats = {'n_train': 0, 'n_train_pos': 0,\n 'accuracy': 0.0, 'accuracy_history': [(0, 0)], 't0': time.time(),\n 'runtime_history': [(0, 0)], 'total_fit_time': 0.0}\n cls_stats[cls_name] = stats\n\nget_minibatch(data_stream, n_test_documents)\n# Discard test set\n\n# We will feed the classifier with mini-batches of 1000 documents; this means\n# we have at most 1000 docs in memory at any time. The smaller the document\n# batch, the bigger the relative overhead of the partial fit methods.\nminibatch_size = 1000\n\n# Create the data_stream that parses Reuters SGML files and iterates on\n# documents as a stream.\nminibatch_iterators = iter_minibatches(data_stream, minibatch_size)\ntotal_vect_time = 0.0\n\n# Main loop : iterate on mini-batches of examples\nfor i, (X_train_text, y_train) in enumerate(minibatch_iterators):\n\n tick = time.time()\n X_train = vectorizer.transform(X_train_text)\n total_vect_time += time.time() - tick\n\n for cls_name, cls in partial_fit_classifiers.items():\n tick = time.time()\n # update estimator with examples in the current mini-batch\n cls.partial_fit(X_train, y_train, classes=all_classes)\n\n # accumulate test accuracy stats\n cls_stats[cls_name]['total_fit_time'] += time.time() - tick\n cls_stats[cls_name]['n_train'] += X_train.shape[0]\n cls_stats[cls_name]['n_train_pos'] += sum(y_train)\n tick = time.time()\n cls_stats[cls_name]['accuracy'] = cls.score(X_test, y_test)\n cls_stats[cls_name]['prediction_time'] = time.time() - tick\n acc_history = (cls_stats[cls_name]['accuracy'],\n cls_stats[cls_name]['n_train'])\n cls_stats[cls_name]['accuracy_history'].append(acc_history)\n run_history = (cls_stats[cls_name]['accuracy'],\n total_vect_time + cls_stats[cls_name]['total_fit_time'])\n cls_stats[cls_name]['runtime_history'].append(run_history)\n\n if i % 3 == 0:\n print(progress(cls_name, cls_stats[cls_name]))\n if i % 3 == 0:\n print('\\n')",
"/srv/venv/lib/python3.6/site-packages/sklearn/linear_model/stochastic_gradient.py:84: FutureWarning: max_iter and tol parameters have been added in <class 'sklearn.linear_model.stochastic_gradient.SGDClassifier'> in 0.19. If both are left unset, they default to max_iter=5 and tol=None. If tol is not None, max_iter defaults to max_iter=1000. From 0.21, default max_iter will be 1000, and default tol will be 1e-3.\n \"and default tol will be 1e-3.\" % type(self), FutureWarning)\n/srv/venv/lib/python3.6/site-packages/sklearn/linear_model/stochastic_gradient.py:84: FutureWarning: max_iter and tol parameters have been added in <class 'sklearn.linear_model.perceptron.Perceptron'> in 0.19. If both are left unset, they default to max_iter=5 and tol=None. If tol is not None, max_iter defaults to max_iter=1000. From 0.21, default max_iter will be 1000, and default tol will be 1e-3.\n \"and default tol will be 1e-3.\" % type(self), FutureWarning)\n/srv/venv/lib/python3.6/site-packages/sklearn/linear_model/stochastic_gradient.py:84: FutureWarning: max_iter and tol parameters have been added in <class 'sklearn.linear_model.passive_aggressive.PassiveAggressiveClassifier'> in 0.19. If both are left unset, they default to max_iter=5 and tol=None. If tol is not None, max_iter defaults to max_iter=1000. From 0.21, default max_iter will be 1000, and default tol will be 1e-3.\n \"and default tol will be 1e-3.\" % type(self), FutureWarning)\n"
]
],
[
[
"Plot results\n------------\n\n",
"_____no_output_____"
]
],
[
[
"def plot_accuracy(x, y, x_legend):\n \"\"\"Plot accuracy as a function of x.\"\"\"\n x = np.array(x)\n y = np.array(y)\n plt.title('Classification accuracy as a function of %s' % x_legend)\n plt.xlabel('%s' % x_legend)\n plt.ylabel('Accuracy')\n plt.grid(True)\n plt.plot(x, y)\n\nrcParams['legend.fontsize'] = 10\ncls_names = list(sorted(cls_stats.keys()))\n\n# Plot accuracy evolution\nplt.figure()\nfor _, stats in sorted(cls_stats.items()):\n # Plot accuracy evolution with #examples\n accuracy, n_examples = zip(*stats['accuracy_history'])\n plot_accuracy(n_examples, accuracy, \"training examples (#)\")\n ax = plt.gca()\n ax.set_ylim((0.8, 1))\nplt.legend(cls_names, loc='best')\n\nplt.figure()\nfor _, stats in sorted(cls_stats.items()):\n # Plot accuracy evolution with runtime\n accuracy, runtime = zip(*stats['runtime_history'])\n plot_accuracy(runtime, accuracy, 'runtime (s)')\n ax = plt.gca()\n ax.set_ylim((0.8, 1))\nplt.legend(cls_names, loc='best')\n\n# Plot fitting times\nplt.figure()\nfig = plt.gcf()\ncls_runtime = []\nfor cls_name, stats in sorted(cls_stats.items()):\n cls_runtime.append(stats['total_fit_time'])\n\ncls_runtime.append(total_vect_time)\ncls_names.append('Vectorization')\nbar_colors = ['b', 'g', 'r', 'c', 'm', 'y']\n\nax = plt.subplot(111)\nrectangles = plt.bar(range(len(cls_names)), cls_runtime, width=0.5,\n color=bar_colors)\n\nax.set_xticks(np.linspace(0.25, len(cls_names) - 0.75, len(cls_names)))\nax.set_xticklabels(cls_names, fontsize=10)\nymax = max(cls_runtime) * 1.2\nax.set_ylim((0, ymax))\nax.set_ylabel('runtime (s)')\nax.set_title('Training Times')\n\n\ndef autolabel(rectangles):\n \"\"\"attach some text vi autolabel on rectangles.\"\"\"\n for rect in rectangles:\n height = rect.get_height()\n ax.text(rect.get_x() + rect.get_width() / 2.,\n 1.05 * height, '%.4f' % height,\n ha='center', va='bottom')\n\nautolabel(rectangles)\nplt.show()\n\n# Plot prediction times\nplt.figure()\ncls_runtime = []\ncls_names = list(sorted(cls_stats.keys()))\nfor cls_name, stats in sorted(cls_stats.items()):\n cls_runtime.append(stats['prediction_time'])\ncls_runtime.append(parsing_time)\ncls_names.append('Read/Parse\\n+Feat.Extr.')\ncls_runtime.append(vectorizing_time)\ncls_names.append('Hashing\\n+Vect.')\n\nax = plt.subplot(111)\nrectangles = plt.bar(range(len(cls_names)), cls_runtime, width=0.5,\n color=bar_colors)\n\nax.set_xticks(np.linspace(0.25, len(cls_names) - 0.75, len(cls_names)))\nax.set_xticklabels(cls_names, fontsize=8)\nplt.setp(plt.xticks()[1], rotation=30)\nymax = max(cls_runtime) * 1.2\nax.set_ylim((0, ymax))\nax.set_ylabel('runtime (s)')\nax.set_title('Prediction Times (%d instances)' % n_test_documents)\nautolabel(rectangles)\nplt.show()",
"_____no_output_____"
],
[
"test complete; Gopal",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec8764ff4197c37e0f655666aa372e4409d217ac | 22,914 | ipynb | Jupyter Notebook | analysis/EDA/add_tuning_decision_boosted.ipynb | natmourajr/CERN-ATLAS-Qualify | 9b40106df97df5f75ba09a7acbbc763a9fdbb8b9 | [
"MIT"
] | null | null | null | analysis/EDA/add_tuning_decision_boosted.ipynb | natmourajr/CERN-ATLAS-Qualify | 9b40106df97df5f75ba09a7acbbc763a9fdbb8b9 | [
"MIT"
] | null | null | null | analysis/EDA/add_tuning_decision_boosted.ipynb | natmourajr/CERN-ATLAS-Qualify | 9b40106df97df5f75ba09a7acbbc763a9fdbb8b9 | [
"MIT"
] | 1 | 2022-03-17T15:29:27.000Z | 2022-03-17T15:29:27.000Z | 37.198052 | 311 | 0.389544 | [
[
[
"import os\nimport numpy as np\nimport pandas as pd\nimport joblib\nimport json\nimport tensorflow as tf\nimport gc",
"_____no_output_____"
],
[
"tuning_path = '../tunings/TrigL2_20180125_v8'\ntuning_file = \"ElectronRingerLooseTriggerConfig.json\"\nwith open(os.path.join(tuning_path,tuning_file)) as f:\n conf = json.load(f)",
"_____no_output_____"
],
[
"#conf",
"_____no_output_____"
],
[
"def open_boosted(path= \"\"):\n '''\n This function will get a .npz file and transform into a pandas DataFrame. \n The .npz has three types of data: float, int and bool this function will concatenate these features and reorder them.\n\n Arguments:\n path (str) - the full path to .npz file\n '''\n # open the file\n d = dict(np.load(path, allow_pickle=True)) \n #print(d.keys()) \n # create a list of temporary dataframes that should be concateneted into a final one\n df_list = []\n for itype in ['float', 'int', 'bool', 'object']:\n df_list.append(pd.DataFrame(data=d['data_%s' %itype], columns=d['features_%s' %itype]))\n # concat the list\n df = pd.concat(df_list, axis=1)\n # return the DataFrame with ordered features.\n df = df[d['ordered_features']]\n # add the target information\n df['target'] = d['target']\n df['et_bin'] = d['etBinIdx']\n df['eta_bin'] = d['etaBinIdx']\n # remove the list of DataFrame and collect into garbage collector\n del df_list, d\n gc.collect()\n return df",
"_____no_output_____"
],
[
"# my pc path\n#data_path = '/media/natmourajr/Backup/Work/CERN/Qualify/cern_data/Zee_boosted/mc16_13TeV.302236_309995_341330.sgn.boosted_probes.WZ_llqq_plus_radion_ZZ_llqq_plus_ggH3000.merge.25bins/mc16_13TeV.302236_309995_341330.sgn.boosted_probes.WZ_llqq_plus_radion_ZZ_llqq_plus_ggH3000.merge.25bins_et4_eta0.npz'\n# LPS path\ndata_path = '/home/natmourajr/Workspace/CERN/Qualify/data/Zee_boosted/mc16_13TeV.302236_309995_341330.sgn.boosted_probes.WZ_llqq_plus_radion_ZZ_llqq_plus_ggH3000.merge.25bins/mc16_13TeV.302236_309995_341330.sgn.boosted_probes.WZ_llqq_plus_radion_ZZ_llqq_plus_ggH3000.merge.25bins_et4_eta0.npz'\ndf = open_boosted(data_path)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"def add_tuning_decision(df, tuning_path, config_dict):\n #find et and eta bin\n idx = (df['et_bin']+df['eta_bin']).unique()[0]\n print(idx)\n\n m_path = tuning_path\n m_model_path = os.path.join(m_path,config_dict[\"Model__path\"][idx].replace('.onnx',''))\n with open(m_model_path + '.json', 'r') as f:\n sequential = json.load(f) \n model = tf.keras.models.model_from_json(json.dumps(sequential, separators=(',', ':')))\n model.load_weights(m_model_path + '.h5')\n slope, offset = config_dict[\"Threshold__slope\"][idx],config_dict[\"Threshold__offset\"][idx]\n\n # ring list\n rings = ['trig_L2_cl_ring_%i' %(iring) for iring in range(100)] \n\n def norm1( data ): \n norms = np.abs( data.sum(axis=1) )\n norms[norms==0] = 1\n return data/norms[:,None] \n df['nn_output_%s_%s'%(config_dict['__operation__'].lower(),config_dict['__version__'])] = model.predict(norm1(df[rings].values)) \n df['thr_%s_%s' %(config_dict['__operation__'].lower(),config_dict['__version__'])] = df['avgmu']*slope + offset\n df['nn_decision_%s_%s' %(config_dict['__operation__'].lower(),config_dict['__version__'])] = 0\n df.loc[df['nn_output_%s_%s' %(config_dict['__operation__'].lower(),config_dict['__version__'])] > df['thr_%s_%s' %(config_dict['__operation__'].lower(),config_dict['__version__'])],'nn_decision_%s_%s' %(config_dict['__operation__'].lower(),config_dict['__version__'])] = 1 \n",
"_____no_output_____"
],
[
"tuning_path = '../tunings/TrigL2_20180125_v8'\nadd_tuning_decision(df=df, tuning_path=tuning_path, config_dict=conf)",
"4\n"
],
[
"df.head()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec87743133b34fb7bb137125cc73047e8c66286e | 187,360 | ipynb | Jupyter Notebook | lab_gan/runGAN-1.ipynb | Nicco20/ArtificialFlyingObjects | d8a5156c514604caa949004ca1c3f33a71357c38 | [
"MIT"
] | null | null | null | lab_gan/runGAN-1.ipynb | Nicco20/ArtificialFlyingObjects | d8a5156c514604caa949004ca1c3f33a71357c38 | [
"MIT"
] | null | null | null | lab_gan/runGAN-1.ipynb | Nicco20/ArtificialFlyingObjects | d8a5156c514604caa949004ca1c3f33a71357c38 | [
"MIT"
] | null | null | null | 50.149893 | 331 | 0.532093 | [
[
[
"<h1 style=\"font-size:40px;\"><center>Exercise V:<br> GANs\n</center></h1>\n\n## Short summary\nIn this exercise, we will design a generative network to generate the last rgb image given the first image. These folder has **three files**: \n- **configGAN.py:** this involves definitions of all parameters and data paths\n- **utilsGAN.py:** includes utility functions required to grab and visualize data \n- **runGAN.ipynb:** contains the script to design, train and test the network \n\nMake sure that before running this script, you created an environment and **installed all required libraries** such \nas keras.\n\n## The data\nThere exists also a subfolder called **data** which contains the traning, validation, and testing data each has both RGB input images together with the corresponding ground truth images.\n\n\n## The exercises\nAs for the previous lab all exercises are found below.\n\n\n## The different 'Cells'\nThis notebook contains several cells with python code, together with the markdown cells (like this one) with only text. Each of the cells with python code has a \"header\" markdown cell with information about the code. The table below provides a short overview of the code cells. \n\n| # | CellName | CellType | Comment |\n| :--- | :-------- | :-------- | :------- |\n| 1 | Init | Needed | Sets up the environment|\n| 2 | Ex | Exercise 1| A class definition of a network model |\n| 3 | Loading | Needed | Loading parameters and initializing the model |\n| 4 | Stats | Needed | Show data distribution | \n| 5 | Data | Needed | Generating the data batches |\n| 6 | Debug | Needed | Debugging the data |\n| 7 | Device | Needed | Selecting CPU/GPU |\n| 8 | Init | Needed | Sets up the timer and other neccessary components |\n| 9 | Training | Exercise 1-2 | Training the model |\n| 10 | Testing | Exercise 1-2| Testing the method | \n\n\nIn order for you to start with the exercise you need to run all cells. It is important that you do this in the correct order, starting from the top and continuing with the next cells. Later when you have started to work with the notebook it may be easier to use the command \"Run All\" found in the \"Cell\" dropdown menu.\n\n## Writing the report\n\nThere is no need to provide any report. However, implemented network architecuture and observed experimental results must be presented as a short presentation in the last lecture, May 28.",
"_____no_output_____"
],
[
"1) We first start with importing all required modules",
"_____no_output_____"
]
],
[
[
"import os\nfrom configGAN import *\ncfg = flying_objects_config()\nif cfg.GPU >=0:\n print(\"creating network model using gpu \" + str(cfg.GPU))\n os.environ['CUDA_VISIBLE_DEVICES'] = str(cfg.GPU)\nelif cfg.GPU >=-1:\n print(\"creating network model using cpu \") \n os.environ[\"CUDA_DEVICE_ORDER\"] = \"PCI_BUS_ID\" # see issue #152\n os.environ[\"CUDA_VISIBLE_DEVICES\"] = \"\"\n\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom utilsGAN import *\nfrom sklearn.metrics import confusion_matrix\n# import seaborn as sns\nfrom datetime import datetime\nimport imageio\nfrom skimage import img_as_ubyte\n\nimport pprint\n# import the necessary packages\nfrom keras.models import Sequential\nfrom keras.layers.normalization import BatchNormalization\nfrom keras.layers.convolutional import Conv3D, Conv2D, Conv1D, Convolution2D, Deconvolution2D, Cropping2D, UpSampling2D\nfrom keras.layers import Input, Conv2DTranspose, ConvLSTM2D, TimeDistributed\nfrom keras.layers.convolutional import MaxPooling2D\nfrom keras.layers.core import Activation\nfrom keras.layers import Concatenate, concatenate, Reshape\nfrom keras.layers.core import Flatten\nfrom keras.layers.core import Dropout\nfrom keras.layers.core import Dense\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.optimizers import Adam\nfrom keras.models import Model\nfrom keras.callbacks import TensorBoard\nfrom keras.applications.vgg16 import VGG16, preprocess_input, decode_predictions\nfrom keras.layers import Input, merge\nfrom keras.regularizers import l2\nfrom keras.layers import Input, merge, Convolution2D, MaxPooling2D, UpSampling2D, Reshape, core, Dropout, LeakyReLU\nimport keras.backend as kb\n",
"creating network model using gpu 0\n"
]
],
[
[
"2) Here, we have the network model class definition. In this class, the most important functions are **build_generator()** and **build_discriminator()**. As defined in the exercises section, your task is to update the both network architectures defined in these functions.",
"_____no_output_____"
]
],
[
[
"class GANModel():\n def __init__(self, batch_size=32, inputShape=(64, 64, 3), dropout_prob=0.25): \n self.batch_size = batch_size\n self.inputShape = inputShape\n self.dropout_prob = dropout_prob\n\n # Calculate the shape of patches\n patch = int(self.inputShape[0] / 2**4)\n self.disc_patch = (patch, patch, 1)\n \n # Build and compile the discriminator\n self.discriminator = self.build_discriminator()\n self.discriminator.compile(loss='mse', optimizer=Adam(0.0002, 0.5),metrics=['accuracy'])\n \n # Build the generator\n self.generator = self.build_generator()\n\n # Input images and their conditioning images\n first_frame = Input(shape=self.inputShape)\n last_frame = Input(shape=self.inputShape)\n\n # By conditioning on the first frame generate a fake version of the last frame\n fake_last_frame = self.generator(first_frame)\n\n # For the combined model we will only train the generator\n self.discriminator.trainable = False\n \n # Discriminators determines validity of fake and condition first image pairs\n valid = self.discriminator([fake_last_frame, first_frame])\n\n self.combined = Model(inputs=[last_frame, first_frame], outputs=[valid, fake_last_frame])\n self.combined.compile(loss=['mse', 'mae'], # mean squared and mean absolute errors\n loss_weights=[1, 100],\n optimizer=Adam(0.0002, 0.5))\n\n def build_generator(self):\n inputs = Input(shape=self.inputShape)\n \n conv1= Conv2D(filters=32,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(inputs)\n conv1 = BatchNormalization(momentum=0.8)(conv1)\n conv1= Conv2D(filters=32,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv1)\n conv1 = BatchNormalization(momentum=0.8)(conv1)\n pool1 = MaxPooling2D(pool_size=(2, 2))(conv1)\n \n conv2= Conv2D(filters=64,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(pool1)\n conv2 = BatchNormalization(momentum=0.8)(conv2)\n conv2= Conv2D(filters=64,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv2)\n conv2 = BatchNormalization(momentum=0.8)(conv2)\n pool2 = MaxPooling2D(pool_size=(2, 2))(conv2)\n \n conv3= Conv2D(filters=128,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(pool2)\n conv3 = BatchNormalization(momentum=0.8)(conv3)\n conv3= Conv2D(filters=128,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv3)\n conv3 = BatchNormalization(momentum=0.8)(conv3)\n pool3 = MaxPooling2D(pool_size=(2, 2))(conv3)\n \n conv4= Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(pool3)\n conv4 = BatchNormalization(momentum=0.8)(conv4)\n conv4= Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv4)\n conv4 = BatchNormalization(momentum=0.8)(conv4)\n drop4 = Dropout(0.5)(conv4)\n '''pool4 = MaxPooling2D(pool_size=(2, 2))(drop4)\n \n conv5= Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(pool4)\n conv5= Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv5)\n #pool2 = MaxPooling2D(pool_size=(2, 2))(conv2)\n drop5 = Dropout(0.5)(conv5)\n \n \n up6 = UpSampling2D(size=(2, 2))(drop5)\n up6 = Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(up6)\n up6 = Concatenate(axis=3)([drop4, up6])\n conv6 = Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(up6)\n conv6 = Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv6)'''\n\n up7 = UpSampling2D(size=(2, 2))(conv4)\n up7 = Conv2D(filters=256,kernel_size=2,activation='relu',padding='same',kernel_initializer='he_normal')(up7)\n up7 = BatchNormalization(momentum=0.8)(up7)\n up7 = Concatenate(axis=3)([conv3, up7])\n conv7 = Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(up7)\n conv7 = BatchNormalization(momentum=0.8)(conv7)\n conv7 = Conv2D(filters=256,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv7)\n conv7 = BatchNormalization(momentum=0.8)(conv7)\n\n up8 = UpSampling2D(size=(2, 2))(conv7)\n up8 = Conv2D(filters=128,kernel_size=2,activation='relu',padding='same',kernel_initializer='he_normal')(up8)\n up8 = BatchNormalization(momentum=0.8)(up8)\n up8 = Concatenate(axis=3)([conv2, up8])\n conv8 = Conv2D(filters=128,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(up8)\n conv8 = BatchNormalization(momentum=0.8)(conv8)\n conv8 = Conv2D(filters=128,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv8)\n\n up9 = UpSampling2D(size=(2, 2))(conv8)\n up9 = Conv2D(filters=64,kernel_size=2,activation='relu',padding='same',kernel_initializer='he_normal')(up9)\n up9 = BatchNormalization(momentum=0.8)(up9)\n up9 = Concatenate(axis=3)([conv1, up9])\n conv9 = Conv2D(filters=64,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(up9)\n conv9 = BatchNormalization(momentum=0.8)(conv9)\n conv9 = Conv2D(filters=64,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv9)\n conv9 = BatchNormalization(momentum=0.8)(conv9)\n conv9 = Conv2D(filters=32,kernel_size=3,activation='relu',padding='same',kernel_initializer='he_normal')(conv9)\n conv9 = BatchNormalization(momentum=0.8)(conv9)\n \n nbr_img_channels = self.inputShape[2]\n\n outputs = Conv2D(nbr_img_channels, (1, 1), activation='sigmoid')(conv9)\n\n model = Model(inputs=inputs, outputs=outputs, name='Generator')\n model.summary()\n \n '''inputs = Input(shape=self.inputShape)\n print(inputs.shape)\n \n down1 = Conv2D(32, (3, 3),padding='same')(inputs)\n down1 = Activation('relu')(down1) \n down1_pool = MaxPooling2D((2, 2), strides=(2, 2))(down1)\n \n down2 = Conv2D(64, (3, 3), padding='same')(down1_pool)\n down2 = Activation('relu')(down2) \n \n\n up1 = UpSampling2D((2, 2))(down2)\n up1 = concatenate([down1, up1], axis=3)\n up1 = Conv2D(256, (3, 3), padding='same')(up1) \n up1 = Activation('relu')(up1) \n \n \n up2 = Conv2D(256, (3, 3), padding='same')(up1) \n up2 = Activation('relu')(up2) \n \n nbr_img_channels = self.inputShape[2]\n outputs = Conv2D(nbr_img_channels, (1, 1), activation='sigmoid')(up2)\n\n model = Model(inputs=inputs, outputs=outputs, name='Generator')\n model.summary()'''\n \n return model\n\n def build_discriminator(self):\n \n last_img = Input(shape=self.inputShape)\n first_img = Input(shape=self.inputShape)\n\n # Concatenate image and conditioning image by channels to produce input\n combined_imgs = Concatenate(axis=-1)([last_img, first_img])\n \n d1 = Conv2D(32, (3, 3), strides=2, padding='same')(combined_imgs) \n d1 = Activation('relu')(d1) \n d2 = Conv2D(64, (3, 3), strides=2, padding='same')(d1)\n d2 = Activation('relu')(d2) \n d3 = Conv2D(128, (3, 3), strides=2, padding='same')(d2)\n d3 = Activation('relu')(d3) \n \n validity = Conv2D(1, (3, 3), strides=2, padding='same')(d3)\n\n model = Model([last_img, first_img], validity)\n model.summary()\n\n return model",
"_____no_output_____"
]
],
[
[
"3) We import the network **hyperparameters** and build a simple network by calling the class introduced in the previous step. Please note that to change the hyperparameters, you just need to change the values in the file called **configPredictor.py.**",
"_____no_output_____"
]
],
[
[
"image_shape = (cfg.IMAGE_HEIGHT, cfg.IMAGE_WIDTH, cfg.IMAGE_CHANNEL)\nmodelObj = GANModel(batch_size=cfg.BATCH_SIZE, inputShape=image_shape,\n dropout_prob=cfg.DROPOUT_PROB)",
"Model: \"model\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 64, 64, 3)] 0 \n__________________________________________________________________________________________________\ninput_2 (InputLayer) [(None, 64, 64, 3)] 0 \n__________________________________________________________________________________________________\nconcatenate (Concatenate) (None, 64, 64, 6) 0 input_1[0][0] \n input_2[0][0] \n__________________________________________________________________________________________________\nconv2d (Conv2D) (None, 32, 32, 32) 1760 concatenate[0][0] \n__________________________________________________________________________________________________\nactivation (Activation) (None, 32, 32, 32) 0 conv2d[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 16, 16, 64) 18496 activation[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 16, 16, 64) 0 conv2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 8, 8, 128) 73856 activation_1[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 8, 8, 128) 0 conv2d_2[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 4, 4, 1) 1153 activation_2[0][0] \n==================================================================================================\nTotal params: 95,265\nTrainable params: 95,265\nNon-trainable params: 0\n__________________________________________________________________________________________________\nModel: \"Generator\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_3 (InputLayer) [(None, 64, 64, 3)] 0 \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 64, 64, 32) 896 input_3[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 64, 64, 32) 9248 conv2d_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization (BatchNorma (None, 64, 64, 32) 128 conv2d_5[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 32, 32, 32) 0 batch_normalization[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 32, 32, 64) 18496 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_1 (BatchNor (None, 32, 32, 64) 256 conv2d_6[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 32, 32, 64) 36928 batch_normalization_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_2 (BatchNor (None, 32, 32, 64) 256 conv2d_7[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_1 (MaxPooling2D) (None, 16, 16, 64) 0 batch_normalization_2[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 16, 16, 128) 73856 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_3 (BatchNor (None, 16, 16, 128) 512 conv2d_8[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 16, 16, 128) 147584 batch_normalization_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_4 (BatchNor (None, 16, 16, 128) 512 conv2d_9[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_2 (MaxPooling2D) (None, 8, 8, 128) 0 batch_normalization_4[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 8, 8, 256) 295168 max_pooling2d_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_5 (BatchNor (None, 8, 8, 256) 1024 conv2d_10[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 8, 8, 256) 590080 batch_normalization_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_6 (BatchNor (None, 8, 8, 256) 1024 conv2d_11[0][0] \n__________________________________________________________________________________________________\nup_sampling2d (UpSampling2D) (None, 16, 16, 256) 0 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 16, 16, 256) 262400 up_sampling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_7 (BatchNor (None, 16, 16, 256) 1024 conv2d_12[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 16, 16, 384) 0 batch_normalization_4[0][0] \n batch_normalization_7[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 16, 16, 256) 884992 concatenate_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_8 (BatchNor (None, 16, 16, 256) 1024 conv2d_13[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 16, 16, 256) 590080 batch_normalization_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_9 (BatchNor (None, 16, 16, 256) 1024 conv2d_14[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_1 (UpSampling2D) (None, 32, 32, 256) 0 batch_normalization_9[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 32, 32, 128) 131200 up_sampling2d_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 32, 32, 128) 512 conv2d_15[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 32, 32, 192) 0 batch_normalization_2[0][0] \n batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 32, 32, 128) 221312 concatenate_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 32, 32, 128) 512 conv2d_16[0][0] \n__________________________________________________________________________________________________\nconv2d_17 (Conv2D) (None, 32, 32, 128) 147584 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_2 (UpSampling2D) (None, 64, 64, 128) 0 conv2d_17[0][0] \n__________________________________________________________________________________________________\nconv2d_18 (Conv2D) (None, 64, 64, 64) 32832 up_sampling2d_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 64, 64, 64) 256 conv2d_18[0][0] \n__________________________________________________________________________________________________\nconcatenate_3 (Concatenate) (None, 64, 64, 96) 0 batch_normalization[0][0] \n batch_normalization_12[0][0] \n__________________________________________________________________________________________________\nconv2d_19 (Conv2D) (None, 64, 64, 64) 55360 concatenate_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 64, 64, 64) 256 conv2d_19[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 64, 64, 64) 36928 batch_normalization_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 64, 64, 64) 256 conv2d_20[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 64, 64, 32) 18464 batch_normalization_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_15 (BatchNo (None, 64, 64, 32) 128 conv2d_21[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 64, 64, 3) 99 batch_normalization_15[0][0] \n==================================================================================================\nTotal params: 3,562,211\nTrainable params: 3,557,859\nNon-trainable params: 4,352\n__________________________________________________________________________________________________\n"
]
],
[
[
"4) We call the utility function **show_statistics** to display the data distribution. This is just for debugging purpose.",
"_____no_output_____"
]
],
[
[
"#### show how the data looks like\nshow_statistics(cfg.training_data_dir, fineGrained=False, title=\" Training Data Statistics \")\nshow_statistics(cfg.validation_data_dir, fineGrained=False, title=\" Validation Data Statistics \")\nshow_statistics(cfg.testing_data_dir, fineGrained=False, title=\" Testing Data Statistics \")",
"\n######################################################################\n##################### Training Data Statistics #####################\n######################################################################\ntotal image number \t 10817\ntotal class number \t 3\nclass square \t 3488 images\nclass circular \t 3626 images\nclass triangle \t 3703 images\n######################################################################\n\n######################################################################\n##################### Validation Data Statistics #####################\n######################################################################\ntotal image number \t 2241\ntotal class number \t 3\nclass triangle \t 745 images\nclass square \t 783 images\nclass circular \t 713 images\n######################################################################\n\n######################################################################\n##################### Testing Data Statistics #####################\n######################################################################\ntotal image number \t 2220\ntotal class number \t 3\nclass triangle \t 733 images\nclass square \t 765 images\nclass circular \t 722 images\n######################################################################\n"
]
],
[
[
"5) We now create batch generators to get small batches from the entire dataset. There is no need to change these functions as they already return **normalized inputs as batches**.",
"_____no_output_____"
]
],
[
[
"nbr_train_data = get_dataset_size(cfg.training_data_dir)\nnbr_valid_data = get_dataset_size(cfg.validation_data_dir)\nnbr_test_data = get_dataset_size(cfg.testing_data_dir)\ntrain_batch_generator = generate_lastframepredictor_batches(cfg.training_data_dir, image_shape, cfg.BATCH_SIZE)\nvalid_batch_generator = generate_lastframepredictor_batches(cfg.validation_data_dir, image_shape, cfg.BATCH_SIZE)\ntest_batch_generator = generate_lastframepredictor_batches(cfg.testing_data_dir, image_shape, cfg.BATCH_SIZE)\nprint(\"Data batch generators are created!\")",
"Data batch generators are created!\n"
]
],
[
[
"6) We can visualize how the data looks like for debugging purpose",
"_____no_output_____"
]
],
[
[
"if cfg.DEBUG_MODE:\n t_x, t_y = next(train_batch_generator)\n print('train_x', t_x.shape, t_x.dtype, t_x.min(), t_x.max())\n print('train_y', t_y.shape, t_y.dtype, t_y.min(), t_y.max()) \n #plot_sample_lastframepredictor_data_with_groundtruth(t_x, t_y, t_y)\n pprint.pprint (cfg)",
"train_x (30, 64, 64, 3) float32 0.0 1.0\ntrain_y (30, 64, 64, 3) float32 0.0 1.0\n{'BATCH_SIZE': 30,\n 'DATA_AUGMENTATION': True,\n 'DEBUG_MODE': True,\n 'DROPOUT_PROB': 0.5,\n 'GPU': 0,\n 'IMAGE_CHANNEL': 3,\n 'IMAGE_HEIGHT': 64,\n 'IMAGE_WIDTH': 64,\n 'LEARNING_RATE': 0.01,\n 'LR_DECAY_FACTOR': 0.1,\n 'NUM_EPOCHS': 5,\n 'PRINT_EVERY': 20,\n 'SAVE_EVERY': 1,\n 'SEQUENCE_LENGTH': 10,\n 'testing_data_dir': '../data/FlyingObjectDataset_10K/testing',\n 'training_data_dir': '../data/FlyingObjectDataset_10K/training',\n 'validation_data_dir': '../data/FlyingObjectDataset_10K/validation'}\n"
]
],
[
[
"7) Start timer and init matrices",
"_____no_output_____"
]
],
[
[
"start_time = datetime.now()\n# Adversarial loss ground truths\nvalid = np.ones((cfg.BATCH_SIZE,) + modelObj.disc_patch)\nfake = np.zeros((cfg.BATCH_SIZE,) + modelObj.disc_patch)\n# log file\noutput_log_dir = \"./logs/{}\".format(datetime.now().strftime(\"%Y%m%d-%H%M%S\"))\nif not os.path.exists(output_log_dir):\n os.makedirs(output_log_dir)",
"_____no_output_____"
]
],
[
[
"8) We can now feed the training and validation data to the network. This will train the network for **some epochs**. Note that the epoch number is also predefined in the file called **configGAN.py.**",
"_____no_output_____"
]
],
[
[
"import imageio\nimport matplotlib.pyplot as plt\nfrom skimage import img_as_ubyte\nimport numpy as np \n\n%matplotlib inline\n\n\ntest_first_imgs, test_last_imgs = next(test_batch_generator)\n\nfor epoch in range(cfg.NUM_EPOCHS):\n steps_per_epoch = (nbr_train_data // cfg.BATCH_SIZE) \n for batch_i in range(steps_per_epoch):\n first_frames, last_frames= next(train_batch_generator)\n if first_frames.shape[0] == cfg.BATCH_SIZE: \n \n # Condition on the first frame and generate the last frame\n fake_last_frames = modelObj.generator.predict(first_frames)\n #plt.imshow(fake_last_frames[1])\n print(fake_last_frames.shape)\n #print(tf.keras.backend.mean(fake_last_frames[0]))\n print(np.mean(fake_last_frames[0]))\n\n # Train the discriminator with combined loss \n d_loss_real = modelObj.discriminator.train_on_batch([last_frames, first_frames], valid)\n d_loss_fake = modelObj.discriminator.train_on_batch([fake_last_frames, first_frames], fake)\n d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)\n \n # Train the generator\n g_loss = modelObj.combined.train_on_batch([last_frames, first_frames], [valid, last_frames])\n\n elapsed_time = datetime.now() - start_time \n print (\"[Epoch %d/%d] [Batch %d/%d] [D loss: %f] [G loss: %f] time: %s\" % (epoch, cfg.NUM_EPOCHS,\n batch_i,\n steps_per_epoch,\n d_loss[0], \n g_loss[0],\n elapsed_time))\n # run some tests to check how the generated images evolve during training\n test_fake_last_imgs = modelObj.generator.predict(test_first_imgs)\n test_img_name = output_log_dir + \"/gen_img_epoc_\" + str(epoch) + \".png\"\n merged_img = np.vstack((first_frames[0],last_frames[0],fake_last_frames[0]))\n imageio.imwrite(test_img_name, img_as_ubyte(merged_img)) #scipy.misc.imsave(test_img_name, merged_img)\n ",
"(30, 64, 64, 3)\n0.5123859\n[Epoch 0/5] [Batch 0/360] [D loss: 0.521882] [G loss: 51.729744] time: 0:01:55.399744\n(30, 64, 64, 3)\n0.46975246\n[Epoch 0/5] [Batch 1/360] [D loss: 0.382826] [G loss: 49.014347] time: 0:01:55.661834\n(30, 64, 64, 3)\n0.5017248\n[Epoch 0/5] [Batch 2/360] [D loss: 0.308842] [G loss: 47.192730] time: 0:01:55.842164\n(30, 64, 64, 3)\n0.5099958\n[Epoch 0/5] [Batch 3/360] [D loss: 0.263464] [G loss: 45.672604] time: 0:01:56.036658\n(30, 64, 64, 3)\n0.51462656\n[Epoch 0/5] [Batch 4/360] [D loss: 0.232008] [G loss: 44.569897] time: 0:01:56.244516\n(30, 64, 64, 3)\n0.52320975\n[Epoch 0/5] [Batch 5/360] [D loss: 0.214573] [G loss: 42.836452] time: 0:01:56.460845\n(30, 64, 64, 3)\n0.5407674\n[Epoch 0/5] [Batch 6/360] [D loss: 0.204692] [G loss: 41.815075] time: 0:01:56.650737\n(30, 64, 64, 3)\n0.56274575\n[Epoch 0/5] [Batch 7/360] [D loss: 0.199852] [G loss: 40.513672] time: 0:01:56.869258\n(30, 64, 64, 3)\n0.558219\n[Epoch 0/5] [Batch 8/360] [D loss: 0.197937] [G loss: 39.192722] time: 0:01:57.100405\n(30, 64, 64, 3)\n0.57522684\n[Epoch 0/5] [Batch 9/360] [D loss: 0.195689] [G loss: 38.770912] time: 0:01:57.297703\n(30, 64, 64, 3)\n0.61223525\n[Epoch 0/5] [Batch 10/360] [D loss: 0.194392] [G loss: 37.923569] time: 0:01:57.506539\n(30, 64, 64, 3)\n0.64503044\n[Epoch 0/5] [Batch 11/360] [D loss: 0.194248] [G loss: 37.632103] time: 0:01:57.685478\n(30, 64, 64, 3)\n0.6177899\n[Epoch 0/5] [Batch 12/360] [D loss: 0.192824] [G loss: 36.585800] time: 0:01:57.897593\n(30, 64, 64, 3)\n0.6607342\n[Epoch 0/5] [Batch 13/360] [D loss: 0.195086] [G loss: 35.837852] time: 0:01:58.086765\n(30, 64, 64, 3)\n0.7082332\n[Epoch 0/5] [Batch 14/360] [D loss: 0.192257] [G loss: 35.931820] time: 0:01:58.288584\n(30, 64, 64, 3)\n0.6127825\n[Epoch 0/5] [Batch 15/360] [D loss: 0.195523] [G loss: 34.343960] time: 0:01:58.464667\n(30, 64, 64, 3)\n0.59361637\n[Epoch 0/5] [Batch 16/360] [D loss: 0.189859] [G loss: 34.356030] time: 0:01:58.624539\n(30, 64, 64, 3)\n0.5967233\n[Epoch 0/5] [Batch 17/360] [D loss: 0.187571] [G loss: 33.540260] time: 0:01:58.801689\n(30, 64, 64, 3)\n0.6924434\n[Epoch 0/5] [Batch 18/360] [D loss: 0.182821] [G loss: 32.614262] time: 0:01:58.980561\n(30, 64, 64, 3)\n0.69658685\n[Epoch 0/5] [Batch 19/360] [D loss: 0.180001] [G loss: 32.091885] time: 0:01:59.148898\n(30, 64, 64, 3)\n0.67934245\n[Epoch 0/5] [Batch 20/360] [D loss: 0.179236] [G loss: 32.982826] time: 0:01:59.342928\n(30, 64, 64, 3)\n0.6189347\n[Epoch 0/5] [Batch 21/360] [D loss: 0.175368] [G loss: 32.358463] time: 0:01:59.536235\n(30, 64, 64, 3)\n0.6695819\n[Epoch 0/5] [Batch 22/360] [D loss: 0.171176] [G loss: 31.839575] time: 0:01:59.711505\n(30, 64, 64, 3)\n0.6538427\n[Epoch 0/5] [Batch 23/360] [D loss: 0.166772] [G loss: 31.374453] time: 0:01:59.898445\n(30, 64, 64, 3)\n0.67038816\n[Epoch 0/5] [Batch 24/360] [D loss: 0.159651] [G loss: 31.912554] time: 0:02:00.080578\n(30, 64, 64, 3)\n0.64601374\n[Epoch 0/5] [Batch 25/360] [D loss: 0.152984] [G loss: 31.202366] time: 0:02:00.257917\n(30, 64, 64, 3)\n0.6769021\n[Epoch 0/5] [Batch 26/360] [D loss: 0.149574] [G loss: 30.488167] time: 0:02:00.472089\n(30, 64, 64, 3)\n0.564105\n[Epoch 0/5] [Batch 27/360] [D loss: 0.146421] [G loss: 30.339027] time: 0:02:00.669467\n(30, 64, 64, 3)\n0.56238407\n[Epoch 0/5] [Batch 28/360] [D loss: 0.135006] [G loss: 30.110241] time: 0:02:00.885116\n(30, 64, 64, 3)\n0.72594863\n[Epoch 0/5] [Batch 29/360] [D loss: 0.126961] [G loss: 30.413858] time: 0:02:01.087997\n(30, 64, 64, 3)\n0.5908888\n[Epoch 0/5] [Batch 30/360] [D loss: 0.125321] [G loss: 30.480207] time: 0:02:01.283374\n(30, 64, 64, 3)\n0.61987364\n[Epoch 0/5] [Batch 31/360] [D loss: 0.117734] [G loss: 29.801392] time: 0:02:01.499260\n(30, 64, 64, 3)\n0.5751477\n[Epoch 0/5] [Batch 32/360] [D loss: 0.103989] [G loss: 30.570192] time: 0:02:01.673665\n(30, 64, 64, 3)\n0.69060975\n[Epoch 0/5] [Batch 33/360] [D loss: 0.090282] [G loss: 29.562237] time: 0:02:01.887374\n(30, 64, 64, 3)\n0.6775036\n[Epoch 0/5] [Batch 34/360] [D loss: 0.085323] [G loss: 28.962849] time: 0:02:02.090860\n(30, 64, 64, 3)\n0.61252403\n[Epoch 0/5] [Batch 35/360] [D loss: 0.073937] [G loss: 29.094028] time: 0:02:02.242036\n(30, 64, 64, 3)\n0.6485998\n[Epoch 0/5] [Batch 36/360] [D loss: 0.073477] [G loss: 30.136679] time: 0:02:02.471435\n(30, 64, 64, 3)\n0.6894965\n[Epoch 0/5] [Batch 37/360] [D loss: 0.060577] [G loss: 29.561184] time: 0:02:02.690959\n(30, 64, 64, 3)\n0.6365028\n[Epoch 0/5] [Batch 38/360] [D loss: 0.053819] [G loss: 30.129847] time: 0:02:02.896402\n(30, 64, 64, 3)\n0.70599204\n[Epoch 0/5] [Batch 39/360] [D loss: 0.045669] [G loss: 29.035303] time: 0:02:03.095090\n(30, 64, 64, 3)\n0.633985\n[Epoch 0/5] [Batch 40/360] [D loss: 0.035719] [G loss: 29.182255] time: 0:02:03.312937\n(30, 64, 64, 3)\n0.6192128\n[Epoch 0/5] [Batch 41/360] [D loss: 0.032138] [G loss: 29.022223] time: 0:02:03.498836\n(30, 64, 64, 3)\n0.67283374\n[Epoch 0/5] [Batch 42/360] [D loss: 0.031922] [G loss: 28.749182] time: 0:02:03.738325\n(30, 64, 64, 3)\n0.687185\n[Epoch 0/5] [Batch 43/360] [D loss: 0.026213] [G loss: 28.719744] time: 0:02:03.926111\n(30, 64, 64, 3)\n0.6240085\n[Epoch 0/5] [Batch 44/360] [D loss: 0.026381] [G loss: 29.246387] time: 0:02:04.107141\n(30, 64, 64, 3)\n0.5748121\n[Epoch 0/5] [Batch 45/360] [D loss: 0.019812] [G loss: 28.288385] time: 0:02:04.305959\n(30, 64, 64, 3)\n0.6181051\n[Epoch 0/5] [Batch 46/360] [D loss: 0.022145] [G loss: 27.961037] time: 0:02:04.473115\n(30, 64, 64, 3)\n0.6150862\n[Epoch 0/5] [Batch 47/360] [D loss: 0.018966] [G loss: 27.876186] time: 0:02:04.663238\n(30, 64, 64, 3)\n0.7814126\n[Epoch 0/5] [Batch 48/360] [D loss: 0.020166] [G loss: 28.679783] time: 0:02:04.850780\n(30, 64, 64, 3)\n0.58656424\n[Epoch 0/5] [Batch 49/360] [D loss: 0.019588] [G loss: 28.503212] time: 0:02:05.040777\n(30, 64, 64, 3)\n0.62443537\n[Epoch 0/5] [Batch 50/360] [D loss: 0.016852] [G loss: 27.472406] time: 0:02:05.236798\n(30, 64, 64, 3)\n0.6721311\n[Epoch 0/5] [Batch 51/360] [D loss: 0.019973] [G loss: 27.602173] time: 0:02:05.401295\n(30, 64, 64, 3)\n0.67418146\n[Epoch 0/5] [Batch 52/360] [D loss: 0.018032] [G loss: 27.539967] time: 0:02:05.584270\n(30, 64, 64, 3)\n0.6508574\n[Epoch 0/5] [Batch 53/360] [D loss: 0.019671] [G loss: 27.158953] time: 0:02:05.793990\n(30, 64, 64, 3)\n0.66510963\n[Epoch 0/5] [Batch 54/360] [D loss: 0.014121] [G loss: 27.624315] time: 0:02:05.978783\n(30, 64, 64, 3)\n0.6940932\n[Epoch 0/5] [Batch 55/360] [D loss: 0.020137] [G loss: 27.056953] time: 0:02:06.166308\n(30, 64, 64, 3)\n0.5655505\n[Epoch 0/5] [Batch 56/360] [D loss: 0.013460] [G loss: 28.618711] time: 0:02:06.375954\n(30, 64, 64, 3)\n0.6261441\n[Epoch 0/5] [Batch 57/360] [D loss: 0.021789] [G loss: 27.897959] time: 0:02:06.592447\n(30, 64, 64, 3)\n0.6623556\n[Epoch 0/5] [Batch 58/360] [D loss: 0.015561] [G loss: 27.271538] time: 0:02:06.820122\n(30, 64, 64, 3)\n0.76060796\n[Epoch 0/5] [Batch 59/360] [D loss: 0.017480] [G loss: 27.193972] time: 0:02:07.054230\n(30, 64, 64, 3)\n0.76513594\n[Epoch 0/5] [Batch 60/360] [D loss: 0.022780] [G loss: 27.114626] time: 0:02:07.270998\n(30, 64, 64, 3)\n0.7328604\n[Epoch 0/5] [Batch 61/360] [D loss: 0.012830] [G loss: 26.944971] time: 0:02:07.457541\n(30, 64, 64, 3)\n0.70357823\n[Epoch 0/5] [Batch 62/360] [D loss: 0.014118] [G loss: 27.393007] time: 0:02:07.640281\n(30, 64, 64, 3)\n0.6130781\n[Epoch 0/5] [Batch 63/360] [D loss: 0.013361] [G loss: 27.670343] time: 0:02:07.834291\n(30, 64, 64, 3)\n0.7073018\n[Epoch 0/5] [Batch 64/360] [D loss: 0.012796] [G loss: 26.774895] time: 0:02:08.048192\n(30, 64, 64, 3)\n0.6648252\n[Epoch 0/5] [Batch 65/360] [D loss: 0.014944] [G loss: 26.484747] time: 0:02:08.232434\n(30, 64, 64, 3)\n0.60865253\n[Epoch 0/5] [Batch 66/360] [D loss: 0.012262] [G loss: 26.813086] time: 0:02:08.425397\n(30, 64, 64, 3)\n0.69256145\n[Epoch 0/5] [Batch 67/360] [D loss: 0.017052] [G loss: 27.779707] time: 0:02:08.634811\n(30, 64, 64, 3)\n0.6764321\n[Epoch 0/5] [Batch 68/360] [D loss: 0.017146] [G loss: 26.449053] time: 0:02:08.830335\n(30, 64, 64, 3)\n0.6417978\n[Epoch 0/5] [Batch 69/360] [D loss: 0.011392] [G loss: 26.968487] time: 0:02:09.020873\n(30, 64, 64, 3)\n0.70213556\n[Epoch 0/5] [Batch 70/360] [D loss: 0.013112] [G loss: 26.744894] time: 0:02:09.219532\n(30, 64, 64, 3)\n0.61458963\n[Epoch 0/5] [Batch 71/360] [D loss: 0.010658] [G loss: 27.126829] time: 0:02:09.421085\n(30, 64, 64, 3)\n0.6639778\n[Epoch 0/5] [Batch 72/360] [D loss: 0.013123] [G loss: 26.011003] time: 0:02:09.615508\n(30, 64, 64, 3)\n0.73892146\n[Epoch 0/5] [Batch 73/360] [D loss: 0.017413] [G loss: 25.999971] time: 0:02:09.816683\n(30, 64, 64, 3)\n0.62463135\n[Epoch 0/5] [Batch 74/360] [D loss: 0.014285] [G loss: 26.254631] time: 0:02:09.982513\n(30, 64, 64, 3)\n0.551145\n[Epoch 0/5] [Batch 75/360] [D loss: 0.009854] [G loss: 26.141327] time: 0:02:10.202803\n(30, 64, 64, 3)\n0.69273335\n[Epoch 0/5] [Batch 76/360] [D loss: 0.010801] [G loss: 26.133678] time: 0:02:10.399207\n(30, 64, 64, 3)\n0.67151076\n[Epoch 0/5] [Batch 77/360] [D loss: 0.012695] [G loss: 26.353382] time: 0:02:10.603044\n(30, 64, 64, 3)\n0.7605972\n[Epoch 0/5] [Batch 78/360] [D loss: 0.033067] [G loss: 26.016432] time: 0:02:10.808345\n(30, 64, 64, 3)\n0.61671793\n[Epoch 0/5] [Batch 79/360] [D loss: 0.015674] [G loss: 25.587311] time: 0:02:11.029863\n(30, 64, 64, 3)\n0.65884143\n[Epoch 0/5] [Batch 80/360] [D loss: 0.016044] [G loss: 26.009743] time: 0:02:11.236985\n(30, 64, 64, 3)\n0.7099996\n[Epoch 0/5] [Batch 81/360] [D loss: 0.023153] [G loss: 26.390219] time: 0:02:11.446770\n(30, 64, 64, 3)\n0.70928806\n[Epoch 0/5] [Batch 82/360] [D loss: 0.010415] [G loss: 25.734440] time: 0:02:11.629822\n(30, 64, 64, 3)\n0.73869795\n[Epoch 0/5] [Batch 83/360] [D loss: 0.008567] [G loss: 26.441698] time: 0:02:11.854180\n(30, 64, 64, 3)\n0.69918174\n[Epoch 0/5] [Batch 84/360] [D loss: 0.013909] [G loss: 25.602587] time: 0:02:12.078478\n(30, 64, 64, 3)\n0.6936342\n[Epoch 0/5] [Batch 85/360] [D loss: 0.009918] [G loss: 25.968977] time: 0:02:12.271849\n(30, 64, 64, 3)\n0.73177224\n[Epoch 0/5] [Batch 86/360] [D loss: 0.034223] [G loss: 25.534449] time: 0:02:12.486856\n(30, 64, 64, 3)\n0.70246094\n[Epoch 0/5] [Batch 87/360] [D loss: 0.023187] [G loss: 25.110731] time: 0:02:12.672182\n(30, 64, 64, 3)\n0.7385576\n[Epoch 0/5] [Batch 88/360] [D loss: 0.019185] [G loss: 25.306429] time: 0:02:12.854894\n(30, 64, 64, 3)\n0.6412718\n[Epoch 0/5] [Batch 89/360] [D loss: 0.035861] [G loss: 25.137922] time: 0:02:13.049954\n(30, 64, 64, 3)\n0.6813607\n[Epoch 0/5] [Batch 90/360] [D loss: 0.012891] [G loss: 24.508562] time: 0:02:13.255216\n(30, 64, 64, 3)\n0.6634572\n[Epoch 0/5] [Batch 91/360] [D loss: 0.009231] [G loss: 24.908018] time: 0:02:13.457549\n(30, 64, 64, 3)\n0.69351107\n[Epoch 0/5] [Batch 92/360] [D loss: 0.006080] [G loss: 25.189596] time: 0:02:13.678964\n(30, 64, 64, 3)\n0.6998659\n[Epoch 0/5] [Batch 93/360] [D loss: 0.009230] [G loss: 24.548727] time: 0:02:13.904714\n(30, 64, 64, 3)\n0.58556676\n[Epoch 0/5] [Batch 94/360] [D loss: 0.008092] [G loss: 24.257200] time: 0:02:14.103211\n(30, 64, 64, 3)\n0.7079932\n[Epoch 0/5] [Batch 95/360] [D loss: 0.009584] [G loss: 24.801746] time: 0:02:14.296893\n(30, 64, 64, 3)\n0.690011\n[Epoch 0/5] [Batch 96/360] [D loss: 0.008696] [G loss: 24.527821] time: 0:02:14.486698\n(30, 64, 64, 3)\n0.7087186\n[Epoch 0/5] [Batch 97/360] [D loss: 0.006676] [G loss: 24.731321] time: 0:02:14.661069\n(30, 64, 64, 3)\n0.7287164\n[Epoch 0/5] [Batch 98/360] [D loss: 0.005972] [G loss: 24.745256] time: 0:02:14.851819\n(30, 64, 64, 3)\n0.66922766\n[Epoch 0/5] [Batch 99/360] [D loss: 0.008528] [G loss: 25.037413] time: 0:02:15.063134\n(30, 64, 64, 3)\n0.7472017\n[Epoch 0/5] [Batch 100/360] [D loss: 0.012495] [G loss: 24.762697] time: 0:02:15.260561\n(30, 64, 64, 3)\n0.71304697\n[Epoch 0/5] [Batch 101/360] [D loss: 0.008148] [G loss: 25.731260] time: 0:02:15.454503\n(30, 64, 64, 3)\n0.68221974\n[Epoch 0/5] [Batch 102/360] [D loss: 0.012907] [G loss: 24.966658] time: 0:02:15.672023\n(30, 64, 64, 3)\n0.7281988\n[Epoch 0/5] [Batch 103/360] [D loss: 0.022593] [G loss: 24.280764] time: 0:02:15.882281\n(30, 64, 64, 3)\n0.64991343\n[Epoch 0/5] [Batch 104/360] [D loss: 0.070364] [G loss: 25.121197] time: 0:02:16.064847\n(30, 64, 64, 3)\n0.67631644\n[Epoch 0/5] [Batch 105/360] [D loss: 0.023815] [G loss: 24.440727] time: 0:02:16.246119\n(30, 64, 64, 3)\n0.6250861\n[Epoch 0/5] [Batch 106/360] [D loss: 0.032124] [G loss: 23.901295] time: 0:02:16.458922\n(30, 64, 64, 3)\n0.73868245\n[Epoch 0/5] [Batch 107/360] [D loss: 0.017329] [G loss: 24.527252] time: 0:02:16.618698\n(30, 64, 64, 3)\n0.74258184\n[Epoch 0/5] [Batch 108/360] [D loss: 0.006957] [G loss: 23.865784] time: 0:02:16.821819\n(30, 64, 64, 3)\n0.76660115\n[Epoch 0/5] [Batch 109/360] [D loss: 0.016305] [G loss: 24.532391] time: 0:02:17.030180\n(30, 64, 64, 3)\n0.6812201\n[Epoch 0/5] [Batch 110/360] [D loss: 0.008858] [G loss: 24.008121] time: 0:02:17.247370\n(30, 64, 64, 3)\n0.64364845\n[Epoch 0/5] [Batch 111/360] [D loss: 0.008039] [G loss: 24.187166] time: 0:02:17.448796\n(30, 64, 64, 3)\n0.64303195\n[Epoch 0/5] [Batch 112/360] [D loss: 0.007085] [G loss: 24.132818] time: 0:02:17.611415\n(30, 64, 64, 3)\n0.64014673\n[Epoch 0/5] [Batch 113/360] [D loss: 0.012323] [G loss: 23.520546] time: 0:02:17.813337\n(30, 64, 64, 3)\n0.7419877\n[Epoch 0/5] [Batch 114/360] [D loss: 0.005913] [G loss: 24.074360] time: 0:02:17.994331\n(30, 64, 64, 3)\n0.75602454\n[Epoch 0/5] [Batch 115/360] [D loss: 0.005182] [G loss: 24.339649] time: 0:02:18.188097\n(30, 64, 64, 3)\n0.6787753\n[Epoch 0/5] [Batch 116/360] [D loss: 0.005764] [G loss: 23.839758] time: 0:02:18.359874\n(30, 64, 64, 3)\n0.68748456\n[Epoch 0/5] [Batch 117/360] [D loss: 0.007252] [G loss: 23.761801] time: 0:02:18.548806\n(30, 64, 64, 3)\n0.6385508\n[Epoch 0/5] [Batch 118/360] [D loss: 0.005017] [G loss: 23.868656] time: 0:02:18.721177\n(30, 64, 64, 3)\n0.67944765\n[Epoch 0/5] [Batch 119/360] [D loss: 0.005809] [G loss: 23.634714] time: 0:02:18.913284\n(30, 64, 64, 3)\n0.6728402\n[Epoch 0/5] [Batch 120/360] [D loss: 0.008137] [G loss: 23.056444] time: 0:02:19.120369\n(30, 64, 64, 3)\n0.67592734\n[Epoch 0/5] [Batch 121/360] [D loss: 0.005750] [G loss: 22.956636] time: 0:02:19.274556\n(30, 64, 64, 3)\n0.6424126\n[Epoch 0/5] [Batch 122/360] [D loss: 0.008223] [G loss: 23.469046] time: 0:02:19.469693\n(30, 64, 64, 3)\n0.755826\n[Epoch 0/5] [Batch 123/360] [D loss: 0.008272] [G loss: 23.768263] time: 0:02:19.631727\n(30, 64, 64, 3)\n0.7222281\n[Epoch 0/5] [Batch 124/360] [D loss: 0.007127] [G loss: 23.324516] time: 0:02:19.828478\n(30, 64, 64, 3)\n0.69405895\n[Epoch 0/5] [Batch 125/360] [D loss: 0.007152] [G loss: 23.154234] time: 0:02:20.037243\n(30, 64, 64, 3)\n0.6082084\n[Epoch 0/5] [Batch 126/360] [D loss: 0.005690] [G loss: 22.531027] time: 0:02:20.254180\n(30, 64, 64, 3)\n0.7152688\n[Epoch 0/5] [Batch 127/360] [D loss: 0.008566] [G loss: 23.138641] time: 0:02:20.420693\n(30, 64, 64, 3)\n0.7267654\n[Epoch 0/5] [Batch 128/360] [D loss: 0.010448] [G loss: 23.540173] time: 0:02:20.598980\n(30, 64, 64, 3)\n0.6752288\n[Epoch 0/5] [Batch 129/360] [D loss: 0.012454] [G loss: 23.148726] time: 0:02:20.777900\n(30, 64, 64, 3)\n0.65454847\n[Epoch 0/5] [Batch 130/360] [D loss: 0.006169] [G loss: 23.189882] time: 0:02:20.950432\n(30, 64, 64, 3)\n0.6445578\n[Epoch 0/5] [Batch 131/360] [D loss: 0.007619] [G loss: 23.240803] time: 0:02:21.147889\n(30, 64, 64, 3)\n0.7329698\n[Epoch 0/5] [Batch 132/360] [D loss: 0.005943] [G loss: 22.891701] time: 0:02:21.361962\n(30, 64, 64, 3)\n0.66356117\n[Epoch 0/5] [Batch 133/360] [D loss: 0.006622] [G loss: 22.347902] time: 0:02:21.575184\n(30, 64, 64, 3)\n0.69151855\n[Epoch 0/5] [Batch 134/360] [D loss: 0.005138] [G loss: 23.378637] time: 0:02:21.745576\n(30, 64, 64, 3)\n0.7207988\n[Epoch 0/5] [Batch 135/360] [D loss: 0.003746] [G loss: 23.070902] time: 0:02:21.925797\n(30, 64, 64, 3)\n0.7411954\n[Epoch 0/5] [Batch 136/360] [D loss: 0.006114] [G loss: 22.446808] time: 0:02:22.160562\n(30, 64, 64, 3)\n0.7770726\n[Epoch 0/5] [Batch 137/360] [D loss: 0.005328] [G loss: 22.372046] time: 0:02:22.361691\n(30, 64, 64, 3)\n0.74007136\n[Epoch 0/5] [Batch 138/360] [D loss: 0.012290] [G loss: 23.044474] time: 0:02:22.535979\n(30, 64, 64, 3)\n0.7214894\n[Epoch 0/5] [Batch 139/360] [D loss: 0.011149] [G loss: 21.998549] time: 0:02:22.741966\n(30, 64, 64, 3)\n0.6857675\n[Epoch 0/5] [Batch 140/360] [D loss: 0.016424] [G loss: 23.522943] time: 0:02:22.974254\n(30, 64, 64, 3)\n0.74278235\n[Epoch 0/5] [Batch 141/360] [D loss: 0.037606] [G loss: 22.222425] time: 0:02:23.184872\n(30, 64, 64, 3)\n0.806804\n[Epoch 0/5] [Batch 142/360] [D loss: 0.042214] [G loss: 22.760725] time: 0:02:23.348227\n(30, 64, 64, 3)\n0.70603395\n[Epoch 0/5] [Batch 143/360] [D loss: 0.011101] [G loss: 22.855669] time: 0:02:23.563317\n(30, 64, 64, 3)\n0.7706942\n[Epoch 0/5] [Batch 144/360] [D loss: 0.010405] [G loss: 23.382397] time: 0:02:23.739807\n(30, 64, 64, 3)\n0.7319676\n[Epoch 0/5] [Batch 145/360] [D loss: 0.022122] [G loss: 23.107281] time: 0:02:23.967049\n(30, 64, 64, 3)\n0.70898753\n[Epoch 0/5] [Batch 146/360] [D loss: 0.008602] [G loss: 22.270201] time: 0:02:24.175300\n(30, 64, 64, 3)\n0.7408007\n[Epoch 0/5] [Batch 147/360] [D loss: 0.025250] [G loss: 22.361868] time: 0:02:24.345953\n(30, 64, 64, 3)\n0.7056327\n[Epoch 0/5] [Batch 148/360] [D loss: 0.007189] [G loss: 21.525280] time: 0:02:24.558343\n(30, 64, 64, 3)\n0.76971716\n[Epoch 0/5] [Batch 149/360] [D loss: 0.004897] [G loss: 23.377596] time: 0:02:24.777261\n(30, 64, 64, 3)\n0.69629306\n[Epoch 0/5] [Batch 150/360] [D loss: 0.004924] [G loss: 21.839725] time: 0:02:24.989387\n(30, 64, 64, 3)\n0.74316835\n[Epoch 0/5] [Batch 151/360] [D loss: 0.005720] [G loss: 22.504208] time: 0:02:25.230811\n(30, 64, 64, 3)\n0.73087174\n[Epoch 0/5] [Batch 152/360] [D loss: 0.003755] [G loss: 22.195042] time: 0:02:25.418355\n(30, 64, 64, 3)\n0.7044309\n[Epoch 0/5] [Batch 153/360] [D loss: 0.004024] [G loss: 22.438469] time: 0:02:25.639092\n(30, 64, 64, 3)\n0.7239897\n[Epoch 0/5] [Batch 154/360] [D loss: 0.004323] [G loss: 22.264221] time: 0:02:25.826474\n(30, 64, 64, 3)\n0.7163673\n[Epoch 0/5] [Batch 155/360] [D loss: 0.006011] [G loss: 21.950752] time: 0:02:26.017825\n(30, 64, 64, 3)\n0.69925827\n[Epoch 0/5] [Batch 156/360] [D loss: 0.008188] [G loss: 21.918806] time: 0:02:26.227096\n(30, 64, 64, 3)\n0.7647745\n[Epoch 0/5] [Batch 157/360] [D loss: 0.005339] [G loss: 21.450489] time: 0:02:26.427937\n(30, 64, 64, 3)\n0.73845124\n[Epoch 0/5] [Batch 158/360] [D loss: 0.006963] [G loss: 22.356817] time: 0:02:26.665287\n(30, 64, 64, 3)\n0.75178546\n[Epoch 0/5] [Batch 159/360] [D loss: 0.005848] [G loss: 22.046598] time: 0:02:26.862330\n(30, 64, 64, 3)\n0.73805386\n[Epoch 0/5] [Batch 160/360] [D loss: 0.005226] [G loss: 21.832022] time: 0:02:27.025186\n(30, 64, 64, 3)\n0.6899292\n[Epoch 0/5] [Batch 161/360] [D loss: 0.004878] [G loss: 21.663408] time: 0:02:27.175592\n(30, 64, 64, 3)\n0.76498264\n[Epoch 0/5] [Batch 162/360] [D loss: 0.006013] [G loss: 21.220213] time: 0:02:27.371171\n(30, 64, 64, 3)\n0.6920287\n[Epoch 0/5] [Batch 163/360] [D loss: 0.004594] [G loss: 21.797070] time: 0:02:27.532501\n(30, 64, 64, 3)\n0.7175813\n[Epoch 0/5] [Batch 164/360] [D loss: 0.005165] [G loss: 21.304726] time: 0:02:27.760474\n(30, 64, 64, 3)\n0.7663326\n[Epoch 0/5] [Batch 165/360] [D loss: 0.006865] [G loss: 21.747213] time: 0:02:27.970623\n(30, 64, 64, 3)\n0.73790497\n[Epoch 0/5] [Batch 166/360] [D loss: 0.004575] [G loss: 22.035582] time: 0:02:28.156013\n(30, 64, 64, 3)\n0.7131322\n[Epoch 0/5] [Batch 167/360] [D loss: 0.004622] [G loss: 22.134373] time: 0:02:28.361364\n(30, 64, 64, 3)\n0.7467156\n[Epoch 0/5] [Batch 168/360] [D loss: 0.007694] [G loss: 21.620100] time: 0:02:28.570253\n(30, 64, 64, 3)\n0.73543745\n[Epoch 0/5] [Batch 169/360] [D loss: 0.003515] [G loss: 21.844011] time: 0:02:28.768712\n(30, 64, 64, 3)\n0.70486075\n[Epoch 0/5] [Batch 170/360] [D loss: 0.007560] [G loss: 20.810282] time: 0:02:28.962992\n(30, 64, 64, 3)\n0.699122\n[Epoch 0/5] [Batch 171/360] [D loss: 0.006291] [G loss: 20.982769] time: 0:02:29.169731\n(30, 64, 64, 3)\n0.7302776\n[Epoch 0/5] [Batch 172/360] [D loss: 0.003952] [G loss: 21.165592] time: 0:02:29.377341\n(30, 64, 64, 3)\n0.8237994\n[Epoch 0/5] [Batch 173/360] [D loss: 0.004635] [G loss: 21.029091] time: 0:02:29.564688\n(30, 64, 64, 3)\n0.76005083\n[Epoch 0/5] [Batch 174/360] [D loss: 0.004064] [G loss: 21.273434] time: 0:02:29.722152\n(30, 64, 64, 3)\n0.7100854\n[Epoch 0/5] [Batch 175/360] [D loss: 0.004049] [G loss: 21.325537] time: 0:02:29.954246\n(30, 64, 64, 3)\n0.650436\n[Epoch 0/5] [Batch 176/360] [D loss: 0.003777] [G loss: 21.981535] time: 0:02:30.131844\n(30, 64, 64, 3)\n0.7709503\n[Epoch 0/5] [Batch 177/360] [D loss: 0.004060] [G loss: 21.284750] time: 0:02:30.328180\n(30, 64, 64, 3)\n0.6850965\n[Epoch 0/5] [Batch 178/360] [D loss: 0.005197] [G loss: 20.829712] time: 0:02:30.531291\n(30, 64, 64, 3)\n0.72811633\n[Epoch 0/5] [Batch 179/360] [D loss: 0.010180] [G loss: 20.942110] time: 0:02:30.710241\n(30, 64, 64, 3)\n0.7118683\n[Epoch 0/5] [Batch 180/360] [D loss: 0.021682] [G loss: 21.113873] time: 0:02:30.880754\n(30, 64, 64, 3)\n0.7253241\n[Epoch 0/5] [Batch 181/360] [D loss: 0.059086] [G loss: 21.499578] time: 0:02:31.092551\n(30, 64, 64, 3)\n0.6986349\n[Epoch 0/5] [Batch 182/360] [D loss: 0.095216] [G loss: 21.278425] time: 0:02:31.308518\n(30, 64, 64, 3)\n0.71511966\n[Epoch 0/5] [Batch 183/360] [D loss: 0.022174] [G loss: 21.169138] time: 0:02:31.496421\n(30, 64, 64, 3)\n0.75717276\n[Epoch 0/5] [Batch 184/360] [D loss: 0.087520] [G loss: 20.297392] time: 0:02:31.720815\n(30, 64, 64, 3)\n0.6840756\n[Epoch 0/5] [Batch 185/360] [D loss: 0.127885] [G loss: 20.764269] time: 0:02:31.900544\n(30, 64, 64, 3)\n0.6988674\n[Epoch 0/5] [Batch 186/360] [D loss: 0.012839] [G loss: 21.051197] time: 0:02:32.080871\n(30, 64, 64, 3)\n0.73776346\n[Epoch 0/5] [Batch 187/360] [D loss: 0.050421] [G loss: 20.827662] time: 0:02:32.279411\n(30, 64, 64, 3)\n0.7458343\n[Epoch 0/5] [Batch 188/360] [D loss: 0.007357] [G loss: 21.707973] time: 0:02:32.491689\n(30, 64, 64, 3)\n0.7339032\n[Epoch 0/5] [Batch 189/360] [D loss: 0.011071] [G loss: 20.549242] time: 0:02:32.656950\n(30, 64, 64, 3)\n0.67983675\n[Epoch 0/5] [Batch 190/360] [D loss: 0.008143] [G loss: 21.205452] time: 0:02:32.849666\n(30, 64, 64, 3)\n0.78614336\n[Epoch 0/5] [Batch 191/360] [D loss: 0.010262] [G loss: 20.567020] time: 0:02:33.056202\n(30, 64, 64, 3)\n0.782983\n[Epoch 0/5] [Batch 192/360] [D loss: 0.006908] [G loss: 20.546278] time: 0:02:33.230574\n(30, 64, 64, 3)\n0.744054\n[Epoch 0/5] [Batch 193/360] [D loss: 0.006658] [G loss: 20.348690] time: 0:02:33.575364\n(30, 64, 64, 3)\n0.73216987\n[Epoch 0/5] [Batch 194/360] [D loss: 0.005795] [G loss: 20.371367] time: 0:02:33.765192\n(30, 64, 64, 3)\n0.6899789\n[Epoch 0/5] [Batch 195/360] [D loss: 0.005112] [G loss: 20.475201] time: 0:02:33.955516\n(30, 64, 64, 3)\n0.76003176\n[Epoch 0/5] [Batch 196/360] [D loss: 0.004330] [G loss: 20.777777] time: 0:02:34.160832\n(30, 64, 64, 3)\n0.75065786\n[Epoch 0/5] [Batch 197/360] [D loss: 0.004265] [G loss: 20.088335] time: 0:02:34.334657\n(30, 64, 64, 3)\n0.6991523\n[Epoch 0/5] [Batch 198/360] [D loss: 0.005516] [G loss: 20.413252] time: 0:02:34.551964\n(30, 64, 64, 3)\n0.7521227\n[Epoch 0/5] [Batch 199/360] [D loss: 0.005314] [G loss: 20.006470] time: 0:02:34.771183\n(30, 64, 64, 3)\n0.70553964\n[Epoch 0/5] [Batch 200/360] [D loss: 0.005383] [G loss: 19.672842] time: 0:02:34.990388\n(30, 64, 64, 3)\n0.6816532\n[Epoch 0/5] [Batch 201/360] [D loss: 0.004710] [G loss: 20.265093] time: 0:02:35.205106\n(30, 64, 64, 3)\n0.7203037\n[Epoch 0/5] [Batch 202/360] [D loss: 0.004179] [G loss: 20.814396] time: 0:02:35.416318\n(30, 64, 64, 3)\n0.75785303\n[Epoch 0/5] [Batch 203/360] [D loss: 0.004689] [G loss: 19.860996] time: 0:02:35.618609\n(30, 64, 64, 3)\n0.795481\n[Epoch 0/5] [Batch 204/360] [D loss: 0.003884] [G loss: 19.778522] time: 0:02:35.836265\n(30, 64, 64, 3)\n0.73262787\n[Epoch 0/5] [Batch 205/360] [D loss: 0.005224] [G loss: 19.440760] time: 0:02:36.010362\n(30, 64, 64, 3)\n0.7809368\n[Epoch 0/5] [Batch 206/360] [D loss: 0.003521] [G loss: 20.517738] time: 0:02:36.221970\n(30, 64, 64, 3)\n0.7800346\n[Epoch 0/5] [Batch 207/360] [D loss: 0.003502] [G loss: 20.439070] time: 0:02:36.435053\n(30, 64, 64, 3)\n0.6912895\n[Epoch 0/5] [Batch 208/360] [D loss: 0.004378] [G loss: 19.731628] time: 0:02:36.619829\n(30, 64, 64, 3)\n0.70280856\n[Epoch 0/5] [Batch 209/360] [D loss: 0.004405] [G loss: 20.169025] time: 0:02:36.809548\n(30, 64, 64, 3)\n0.7533956\n[Epoch 0/5] [Batch 210/360] [D loss: 0.006647] [G loss: 19.819710] time: 0:02:37.023074\n(30, 64, 64, 3)\n0.71523637\n[Epoch 0/5] [Batch 211/360] [D loss: 0.005328] [G loss: 19.291948] time: 0:02:37.191139\n(30, 64, 64, 3)\n0.74072295\n[Epoch 0/5] [Batch 212/360] [D loss: 0.005985] [G loss: 19.830008] time: 0:02:37.407459\n(30, 64, 64, 3)\n0.7858626\n[Epoch 0/5] [Batch 213/360] [D loss: 0.007253] [G loss: 19.324644] time: 0:02:37.617386\n(30, 64, 64, 3)\n0.703465\n[Epoch 0/5] [Batch 214/360] [D loss: 0.005554] [G loss: 19.557528] time: 0:02:37.859644\n(30, 64, 64, 3)\n0.8108079\n[Epoch 0/5] [Batch 215/360] [D loss: 0.004131] [G loss: 19.872610] time: 0:02:38.047562\n(30, 64, 64, 3)\n0.75132996\n[Epoch 0/5] [Batch 216/360] [D loss: 0.005182] [G loss: 19.305256] time: 0:02:38.258020\n(30, 64, 64, 3)\n0.79804105\n[Epoch 0/5] [Batch 217/360] [D loss: 0.005223] [G loss: 20.080688] time: 0:02:38.498996\n(30, 64, 64, 3)\n0.80678064\n[Epoch 0/5] [Batch 218/360] [D loss: 0.004115] [G loss: 19.488400] time: 0:02:38.695675\n(30, 64, 64, 3)\n0.7783349\n[Epoch 0/5] [Batch 219/360] [D loss: 0.004585] [G loss: 19.255194] time: 0:02:38.885851\n(30, 64, 64, 3)\n0.7522025\n[Epoch 0/5] [Batch 220/360] [D loss: 0.007821] [G loss: 18.870420] time: 0:02:39.053346\n(30, 64, 64, 3)\n0.76253086\n[Epoch 0/5] [Batch 221/360] [D loss: 0.004228] [G loss: 19.799822] time: 0:02:39.272501\n(30, 64, 64, 3)\n0.7748749\n[Epoch 0/5] [Batch 222/360] [D loss: 0.006159] [G loss: 18.506105] time: 0:02:39.495637\n(30, 64, 64, 3)\n0.8422127\n[Epoch 0/5] [Batch 223/360] [D loss: 0.007221] [G loss: 19.082033] time: 0:02:39.730303\n(30, 64, 64, 3)\n0.7691856\n[Epoch 0/5] [Batch 224/360] [D loss: 0.004567] [G loss: 19.180817] time: 0:02:39.935002\n(30, 64, 64, 3)\n0.748116\n[Epoch 0/5] [Batch 225/360] [D loss: 0.005905] [G loss: 19.381313] time: 0:02:40.152566\n(30, 64, 64, 3)\n0.72433424\n[Epoch 0/5] [Batch 226/360] [D loss: 0.005013] [G loss: 19.144453] time: 0:02:40.360105\n(30, 64, 64, 3)\n0.7556529\n[Epoch 0/5] [Batch 227/360] [D loss: 0.004973] [G loss: 18.896889] time: 0:02:40.546224\n(30, 64, 64, 3)\n0.7045899\n[Epoch 0/5] [Batch 228/360] [D loss: 0.004035] [G loss: 19.005075] time: 0:02:40.728157\n(30, 64, 64, 3)\n0.74115855\n[Epoch 0/5] [Batch 229/360] [D loss: 0.004116] [G loss: 18.913330] time: 0:02:40.925297\n(30, 64, 64, 3)\n0.74564236\n[Epoch 0/5] [Batch 230/360] [D loss: 0.004032] [G loss: 19.405169] time: 0:02:41.154102\n(30, 64, 64, 3)\n0.7702465\n[Epoch 0/5] [Batch 231/360] [D loss: 0.008475] [G loss: 19.216290] time: 0:02:41.343630\n(30, 64, 64, 3)\n0.71171755\n[Epoch 0/5] [Batch 232/360] [D loss: 0.004077] [G loss: 19.304739] time: 0:02:41.545326\n(30, 64, 64, 3)\n0.79918057\n[Epoch 0/5] [Batch 233/360] [D loss: 0.004201] [G loss: 18.802656] time: 0:02:41.791112\n(30, 64, 64, 3)\n0.80554277\n[Epoch 0/5] [Batch 234/360] [D loss: 0.002706] [G loss: 19.773149] time: 0:02:41.966748\n(30, 64, 64, 3)\n0.7766482\n[Epoch 0/5] [Batch 235/360] [D loss: 0.004276] [G loss: 18.380888] time: 0:02:42.189822\n(30, 64, 64, 3)\n0.7248693\n[Epoch 0/5] [Batch 236/360] [D loss: 0.004389] [G loss: 18.652290] time: 0:02:42.360844\n(30, 64, 64, 3)\n0.7681573\n[Epoch 0/5] [Batch 237/360] [D loss: 0.012657] [G loss: 19.025425] time: 0:02:42.586182\n(30, 64, 64, 3)\n0.7860141\n[Epoch 0/5] [Batch 238/360] [D loss: 0.014505] [G loss: 19.102051] time: 0:02:42.801190\n(30, 64, 64, 3)\n0.7604416\n[Epoch 0/5] [Batch 239/360] [D loss: 0.010991] [G loss: 18.594877] time: 0:02:42.995728\n(30, 64, 64, 3)\n0.7246766\n[Epoch 0/5] [Batch 240/360] [D loss: 0.009801] [G loss: 18.698719] time: 0:02:43.224375\n(30, 64, 64, 3)\n0.8070337\n[Epoch 0/5] [Batch 241/360] [D loss: 0.004403] [G loss: 18.619230] time: 0:02:43.410490\n(30, 64, 64, 3)\n0.7917769\n[Epoch 0/5] [Batch 242/360] [D loss: 0.004294] [G loss: 18.256693] time: 0:02:43.640869\n(30, 64, 64, 3)\n0.7631939\n[Epoch 0/5] [Batch 243/360] [D loss: 0.004599] [G loss: 18.555643] time: 0:02:43.882276\n(30, 64, 64, 3)\n0.7376079\n[Epoch 0/5] [Batch 244/360] [D loss: 0.003923] [G loss: 19.008333] time: 0:02:44.064883\n(30, 64, 64, 3)\n0.810134\n[Epoch 0/5] [Batch 245/360] [D loss: 0.003805] [G loss: 18.720051] time: 0:02:44.318861\n(30, 64, 64, 3)\n0.7173231\n[Epoch 0/5] [Batch 246/360] [D loss: 0.004901] [G loss: 18.427313] time: 0:02:44.510467\n(30, 64, 64, 3)\n0.73673564\n[Epoch 0/5] [Batch 247/360] [D loss: 0.003404] [G loss: 18.357851] time: 0:02:44.683320\n(30, 64, 64, 3)\n0.7802279\n[Epoch 0/5] [Batch 248/360] [D loss: 0.003408] [G loss: 19.031725] time: 0:02:44.859590\n(30, 64, 64, 3)\n0.748012\n[Epoch 0/5] [Batch 249/360] [D loss: 0.003558] [G loss: 18.577662] time: 0:02:45.019388\n(30, 64, 64, 3)\n0.64493006\n[Epoch 0/5] [Batch 250/360] [D loss: 0.005923] [G loss: 18.572920] time: 0:02:45.228140\n(30, 64, 64, 3)\n0.7659469\n[Epoch 0/5] [Batch 251/360] [D loss: 0.005220] [G loss: 18.207027] time: 0:02:45.452348\n(30, 64, 64, 3)\n0.68739015\n[Epoch 0/5] [Batch 252/360] [D loss: 0.005511] [G loss: 18.857718] time: 0:02:45.655667\n(30, 64, 64, 3)\n0.66253847\n[Epoch 0/5] [Batch 253/360] [D loss: 0.019234] [G loss: 17.984903] time: 0:02:45.843050\n(30, 64, 64, 3)\n0.8169797\n[Epoch 0/5] [Batch 254/360] [D loss: 0.060318] [G loss: 19.476921] time: 0:02:46.066988\n(30, 64, 64, 3)\n0.74439985\n[Epoch 0/5] [Batch 255/360] [D loss: 0.316534] [G loss: 18.670830] time: 0:02:46.255811\n(30, 64, 64, 3)\n0.7276368\n[Epoch 0/5] [Batch 256/360] [D loss: 0.122206] [G loss: 17.910076] time: 0:02:46.438010\n(30, 64, 64, 3)\n0.7160016\n[Epoch 0/5] [Batch 257/360] [D loss: 0.013852] [G loss: 18.701939] time: 0:02:46.641616\n(30, 64, 64, 3)\n0.72477347\n[Epoch 0/5] [Batch 258/360] [D loss: 0.025299] [G loss: 17.705444] time: 0:02:46.848418\n(30, 64, 64, 3)\n0.7858483\n[Epoch 0/5] [Batch 259/360] [D loss: 0.004691] [G loss: 17.753174] time: 0:02:47.040410\n(30, 64, 64, 3)\n0.7875426\n[Epoch 0/5] [Batch 260/360] [D loss: 0.005393] [G loss: 17.914730] time: 0:02:47.244979\n(30, 64, 64, 3)\n0.7443164\n[Epoch 0/5] [Batch 261/360] [D loss: 0.005549] [G loss: 18.249554] time: 0:02:47.459558\n(30, 64, 64, 3)\n0.7898237\n[Epoch 0/5] [Batch 262/360] [D loss: 0.005168] [G loss: 18.069458] time: 0:02:47.643819\n(30, 64, 64, 3)\n0.7427611\n[Epoch 0/5] [Batch 263/360] [D loss: 0.004750] [G loss: 18.596827] time: 0:02:47.852154\n(30, 64, 64, 3)\n0.7519443\n[Epoch 0/5] [Batch 264/360] [D loss: 0.004650] [G loss: 18.862680] time: 0:02:48.021787\n(30, 64, 64, 3)\n0.7515319\n[Epoch 0/5] [Batch 265/360] [D loss: 0.007191] [G loss: 18.061451] time: 0:02:48.230982\n(30, 64, 64, 3)\n0.70239186\n[Epoch 0/5] [Batch 266/360] [D loss: 0.005591] [G loss: 18.198130] time: 0:02:48.401649\n(30, 64, 64, 3)\n0.7778442\n[Epoch 0/5] [Batch 267/360] [D loss: 0.004962] [G loss: 18.167566] time: 0:02:48.604095\n(30, 64, 64, 3)\n0.6960225\n[Epoch 0/5] [Batch 268/360] [D loss: 0.003630] [G loss: 18.095066] time: 0:02:48.819697\n(30, 64, 64, 3)\n0.7206407\n[Epoch 0/5] [Batch 269/360] [D loss: 0.004348] [G loss: 17.378683] time: 0:02:49.007888\n(30, 64, 64, 3)\n0.73921585\n[Epoch 0/5] [Batch 270/360] [D loss: 0.003414] [G loss: 18.661472] time: 0:02:49.235886\n(30, 64, 64, 3)\n0.77003986\n[Epoch 0/5] [Batch 271/360] [D loss: 0.005022] [G loss: 17.219669] time: 0:02:49.412988\n(30, 64, 64, 3)\n0.76250166\n[Epoch 0/5] [Batch 272/360] [D loss: 0.003373] [G loss: 18.290649] time: 0:02:49.594365\n(30, 64, 64, 3)\n0.75990087\n[Epoch 0/5] [Batch 273/360] [D loss: 0.004982] [G loss: 18.162367] time: 0:02:49.826567\n(30, 64, 64, 3)\n0.6851118\n[Epoch 0/5] [Batch 274/360] [D loss: 0.004677] [G loss: 17.733223] time: 0:02:50.040787\n(30, 64, 64, 3)\n0.71609014\n[Epoch 0/5] [Batch 275/360] [D loss: 0.005353] [G loss: 17.092268] time: 0:02:50.213849\n(30, 64, 64, 3)\n0.742565\n[Epoch 0/5] [Batch 276/360] [D loss: 0.004504] [G loss: 16.978333] time: 0:02:50.423394\n(30, 64, 64, 3)\n0.741396\n[Epoch 0/5] [Batch 277/360] [D loss: 0.004736] [G loss: 17.439989] time: 0:02:50.649370\n(30, 64, 64, 3)\n0.82205796\n[Epoch 0/5] [Batch 278/360] [D loss: 0.004357] [G loss: 18.328596] time: 0:02:50.856246\n(30, 64, 64, 3)\n0.78237873\n[Epoch 0/5] [Batch 279/360] [D loss: 0.003912] [G loss: 16.894800] time: 0:02:51.036791\n(30, 64, 64, 3)\n0.75504524\n[Epoch 0/5] [Batch 280/360] [D loss: 0.003122] [G loss: 17.951532] time: 0:02:51.246392\n(30, 64, 64, 3)\n0.7524951\n[Epoch 0/5] [Batch 281/360] [D loss: 0.006059] [G loss: 17.594898] time: 0:02:51.442967\n(30, 64, 64, 3)\n0.78371996\n[Epoch 0/5] [Batch 282/360] [D loss: 0.003504] [G loss: 17.182140] time: 0:02:51.676877\n(30, 64, 64, 3)\n0.8190525\n[Epoch 0/5] [Batch 283/360] [D loss: 0.003640] [G loss: 18.624531] time: 0:02:51.881930\n(30, 64, 64, 3)\n0.7171524\n[Epoch 0/5] [Batch 284/360] [D loss: 0.003519] [G loss: 17.075802] time: 0:02:52.072631\n(30, 64, 64, 3)\n0.7366927\n[Epoch 0/5] [Batch 285/360] [D loss: 0.004717] [G loss: 16.652967] time: 0:02:52.297063\n(30, 64, 64, 3)\n0.7381428\n[Epoch 0/5] [Batch 286/360] [D loss: 0.003548] [G loss: 17.802803] time: 0:02:52.487824\n(30, 64, 64, 3)\n0.739322\n[Epoch 0/5] [Batch 287/360] [D loss: 0.004231] [G loss: 16.945568] time: 0:02:52.701750\n(30, 64, 64, 3)\n0.8017549\n[Epoch 0/5] [Batch 288/360] [D loss: 0.003775] [G loss: 17.469614] time: 0:02:52.905990\n(30, 64, 64, 3)\n0.830808\n[Epoch 0/5] [Batch 289/360] [D loss: 0.004408] [G loss: 17.096733] time: 0:02:53.101436\n(30, 64, 64, 3)\n0.78838235\n[Epoch 0/5] [Batch 290/360] [D loss: 0.003555] [G loss: 16.738586] time: 0:02:53.316298\n(30, 64, 64, 3)\n0.79905605\n[Epoch 0/5] [Batch 291/360] [D loss: 0.004168] [G loss: 17.105068] time: 0:02:53.481274\n(30, 64, 64, 3)\n0.75910425\n[Epoch 0/5] [Batch 292/360] [D loss: 0.003656] [G loss: 17.214102] time: 0:02:53.677975\n(30, 64, 64, 3)\n0.75241905\n[Epoch 0/5] [Batch 293/360] [D loss: 0.003262] [G loss: 17.045849] time: 0:02:53.875391\n(30, 64, 64, 3)\n0.8022451\n[Epoch 0/5] [Batch 294/360] [D loss: 0.003704] [G loss: 16.973854] time: 0:02:54.050135\n(30, 64, 64, 3)\n0.7688973\n[Epoch 0/5] [Batch 295/360] [D loss: 0.004257] [G loss: 16.660614] time: 0:02:54.238336\n(30, 64, 64, 3)\n0.7554781\n[Epoch 0/5] [Batch 296/360] [D loss: 0.004640] [G loss: 16.379541] time: 0:02:54.403736\n(30, 64, 64, 3)\n0.6888897\n[Epoch 0/5] [Batch 297/360] [D loss: 0.006695] [G loss: 16.966276] time: 0:02:54.617790\n(30, 64, 64, 3)\n0.7799003\n[Epoch 0/5] [Batch 298/360] [D loss: 0.004136] [G loss: 16.690910] time: 0:02:54.821837\n(30, 64, 64, 3)\n0.68228406\n[Epoch 0/5] [Batch 299/360] [D loss: 0.003990] [G loss: 16.099737] time: 0:02:55.050871\n(30, 64, 64, 3)\n0.79642755\n[Epoch 0/5] [Batch 300/360] [D loss: 0.004544] [G loss: 16.636393] time: 0:02:55.235398\n(30, 64, 64, 3)\n0.7825256\n[Epoch 0/5] [Batch 301/360] [D loss: 0.003586] [G loss: 17.608622] time: 0:02:55.413149\n(30, 64, 64, 3)\n0.78830177\n[Epoch 0/5] [Batch 302/360] [D loss: 0.004057] [G loss: 16.693066] time: 0:02:55.573620\n(30, 64, 64, 3)\n0.7357879\n[Epoch 0/5] [Batch 303/360] [D loss: 0.004395] [G loss: 16.326082] time: 0:02:55.792326\n(30, 64, 64, 3)\n0.8062961\n[Epoch 0/5] [Batch 304/360] [D loss: 0.005540] [G loss: 16.638878] time: 0:02:56.001375\n(30, 64, 64, 3)\n0.7135234\n[Epoch 0/5] [Batch 305/360] [D loss: 0.003224] [G loss: 16.870844] time: 0:02:56.208120\n(30, 64, 64, 3)\n0.78462344\n[Epoch 0/5] [Batch 306/360] [D loss: 0.003998] [G loss: 16.453732] time: 0:02:56.405715\n(30, 64, 64, 3)\n0.7701834\n[Epoch 0/5] [Batch 307/360] [D loss: 0.003674] [G loss: 16.507215] time: 0:02:56.616685\n(30, 64, 64, 3)\n0.7568538\n[Epoch 0/5] [Batch 308/360] [D loss: 0.004044] [G loss: 17.109625] time: 0:02:56.827164\n(30, 64, 64, 3)\n0.8089984\n[Epoch 0/5] [Batch 309/360] [D loss: 0.003565] [G loss: 16.722094] time: 0:02:57.040905\n(30, 64, 64, 3)\n0.77272224\n[Epoch 0/5] [Batch 310/360] [D loss: 0.003871] [G loss: 16.226736] time: 0:02:57.232503\n(30, 64, 64, 3)\n0.8011615\n[Epoch 0/5] [Batch 311/360] [D loss: 0.005058] [G loss: 16.839121] time: 0:02:57.462585\n(30, 64, 64, 3)\n0.84341496\n[Epoch 0/5] [Batch 312/360] [D loss: 0.003348] [G loss: 16.636749] time: 0:02:57.676246\n(30, 64, 64, 3)\n0.80351686\n[Epoch 0/5] [Batch 313/360] [D loss: 0.004095] [G loss: 16.732443] time: 0:02:57.905173\n(30, 64, 64, 3)\n0.78452826\n[Epoch 0/5] [Batch 314/360] [D loss: 0.007152] [G loss: 16.960217] time: 0:02:58.104304\n(30, 64, 64, 3)\n0.7586022\n[Epoch 0/5] [Batch 315/360] [D loss: 0.003730] [G loss: 17.010891] time: 0:02:58.303769\n(30, 64, 64, 3)\n0.81850624\n[Epoch 0/5] [Batch 316/360] [D loss: 0.003288] [G loss: 16.363802] time: 0:02:58.476422\n(30, 64, 64, 3)\n0.7612114\n[Epoch 0/5] [Batch 317/360] [D loss: 0.003396] [G loss: 16.599684] time: 0:02:58.690200\n(30, 64, 64, 3)\n0.7575524\n[Epoch 0/5] [Batch 318/360] [D loss: 0.004398] [G loss: 16.465334] time: 0:02:58.916450\n(30, 64, 64, 3)\n0.76719594\n[Epoch 0/5] [Batch 319/360] [D loss: 0.002829] [G loss: 17.142086] time: 0:02:59.127094\n(30, 64, 64, 3)\n0.82172227\n[Epoch 0/5] [Batch 320/360] [D loss: 0.003686] [G loss: 16.429943] time: 0:02:59.332431\n(30, 64, 64, 3)\n0.7533719\n[Epoch 0/5] [Batch 321/360] [D loss: 0.004500] [G loss: 15.882351] time: 0:02:59.531692\n(30, 64, 64, 3)\n0.8233974\n[Epoch 0/5] [Batch 322/360] [D loss: 0.002654] [G loss: 16.432064] time: 0:02:59.707847\n(30, 64, 64, 3)\n0.82469076\n[Epoch 0/5] [Batch 323/360] [D loss: 0.003618] [G loss: 16.786449] time: 0:02:59.894578\n(30, 64, 64, 3)\n0.75797534\n[Epoch 0/5] [Batch 324/360] [D loss: 0.003187] [G loss: 15.964214] time: 0:03:00.070952\n(30, 64, 64, 3)\n0.8079795\n[Epoch 0/5] [Batch 325/360] [D loss: 0.004036] [G loss: 16.085417] time: 0:03:00.262069\n(30, 64, 64, 3)\n0.7662625\n[Epoch 0/5] [Batch 326/360] [D loss: 0.004866] [G loss: 15.724848] time: 0:03:00.431144\n(30, 64, 64, 3)\n0.763128\n[Epoch 0/5] [Batch 327/360] [D loss: 0.002600] [G loss: 16.264114] time: 0:03:00.662637\n(30, 64, 64, 3)\n0.8187248\n[Epoch 0/5] [Batch 328/360] [D loss: 0.004295] [G loss: 15.980284] time: 0:03:00.851484\n(30, 64, 64, 3)\n0.7467795\n[Epoch 0/5] [Batch 329/360] [D loss: 0.004297] [G loss: 15.697162] time: 0:03:01.077218\n(30, 64, 64, 3)\n0.7700519\n[Epoch 0/5] [Batch 330/360] [D loss: 0.003725] [G loss: 15.950444] time: 0:03:01.283667\n(30, 64, 64, 3)\n0.7656393\n[Epoch 0/5] [Batch 331/360] [D loss: 0.010147] [G loss: 15.816264] time: 0:03:01.449153\n(30, 64, 64, 3)\n0.7842393\n[Epoch 0/5] [Batch 332/360] [D loss: 0.024345] [G loss: 16.031410] time: 0:03:01.631975\n(30, 64, 64, 3)\n0.7501275\n[Epoch 0/5] [Batch 333/360] [D loss: 0.006968] [G loss: 16.070568] time: 0:03:01.850466\n(30, 64, 64, 3)\n0.79534405\n[Epoch 0/5] [Batch 334/360] [D loss: 0.023009] [G loss: 16.120764] time: 0:03:02.085796\n(30, 64, 64, 3)\n0.7935074\n[Epoch 0/5] [Batch 335/360] [D loss: 0.046940] [G loss: 15.680136] time: 0:03:02.271128\n(30, 64, 64, 3)\n0.78418905\n[Epoch 0/5] [Batch 336/360] [D loss: 0.146115] [G loss: 16.457993] time: 0:03:02.478614\n(30, 64, 64, 3)\n0.75877565\n[Epoch 0/5] [Batch 337/360] [D loss: 0.408503] [G loss: 15.649289] time: 0:03:02.659942\n(30, 64, 64, 3)\n0.80507296\n[Epoch 0/5] [Batch 338/360] [D loss: 0.014646] [G loss: 16.067209] time: 0:03:02.851853\n(30, 64, 64, 3)\n0.7455309\n[Epoch 0/5] [Batch 339/360] [D loss: 0.072509] [G loss: 15.743466] time: 0:03:03.116942\n(30, 64, 64, 3)\n0.82345134\n[Epoch 0/5] [Batch 340/360] [D loss: 0.006045] [G loss: 16.497253] time: 0:03:03.395416\n(30, 64, 64, 3)\n0.82011145\n[Epoch 0/5] [Batch 341/360] [D loss: 0.013923] [G loss: 15.787183] time: 0:03:03.601769\n(30, 64, 64, 3)\n0.79009753\n[Epoch 0/5] [Batch 342/360] [D loss: 0.005562] [G loss: 15.571603] time: 0:03:03.785243\n(30, 64, 64, 3)\n0.78429586\n[Epoch 0/5] [Batch 343/360] [D loss: 0.008527] [G loss: 16.075327] time: 0:03:03.975952\n(30, 64, 64, 3)\n0.75202674\n[Epoch 0/5] [Batch 344/360] [D loss: 0.005125] [G loss: 15.743366] time: 0:03:04.175536\n(30, 64, 64, 3)\n0.751126\n[Epoch 0/5] [Batch 345/360] [D loss: 0.005091] [G loss: 15.811646] time: 0:03:04.394855\n(30, 64, 64, 3)\n0.7255834\n[Epoch 0/5] [Batch 346/360] [D loss: 0.005434] [G loss: 15.410718] time: 0:03:04.585811\n(30, 64, 64, 3)\n0.8165625\n[Epoch 0/5] [Batch 347/360] [D loss: 0.004901] [G loss: 15.417832] time: 0:03:04.780412\n(30, 64, 64, 3)\n0.70489883\n[Epoch 0/5] [Batch 348/360] [D loss: 0.005700] [G loss: 15.594007] time: 0:03:04.995897\n(30, 64, 64, 3)\n0.8001995\n[Epoch 0/5] [Batch 349/360] [D loss: 0.002961] [G loss: 15.784444] time: 0:03:05.203673\n(30, 64, 64, 3)\n0.8131946\n[Epoch 0/5] [Batch 350/360] [D loss: 0.004939] [G loss: 16.341028] time: 0:03:05.426149\n(30, 64, 64, 3)\n0.7513499\n[Epoch 0/5] [Batch 351/360] [D loss: 0.004512] [G loss: 15.510555] time: 0:03:05.650536\n(30, 64, 64, 3)\n0.7728014\n[Epoch 0/5] [Batch 352/360] [D loss: 0.003747] [G loss: 15.811953] time: 0:03:05.879590\n(30, 64, 64, 3)\n0.7573573\n[Epoch 0/5] [Batch 353/360] [D loss: 0.003094] [G loss: 16.124603] time: 0:03:06.088536\n(30, 64, 64, 3)\n0.8004399\n[Epoch 0/5] [Batch 354/360] [D loss: 0.003011] [G loss: 16.114723] time: 0:03:06.284225\n(30, 64, 64, 3)\n0.7504163\n[Epoch 0/5] [Batch 355/360] [D loss: 0.004778] [G loss: 15.627082] time: 0:03:06.482415\n(30, 64, 64, 3)\n0.8086811\n[Epoch 0/5] [Batch 356/360] [D loss: 0.003027] [G loss: 16.591644] time: 0:03:06.685640\n(30, 64, 64, 3)\n0.7988686\n[Epoch 0/5] [Batch 357/360] [D loss: 0.003426] [G loss: 15.432536] time: 0:03:06.892288\n(30, 64, 64, 3)\n0.8262453\n[Epoch 0/5] [Batch 358/360] [D loss: 0.004113] [G loss: 15.442795] time: 0:03:07.099523\n(30, 64, 64, 3)\n0.8303111\n[Epoch 1/5] [Batch 0/360] [D loss: 0.004771] [G loss: 14.632837] time: 0:03:07.374464\n(30, 64, 64, 3)\n0.8217961\n[Epoch 1/5] [Batch 1/360] [D loss: 0.004438] [G loss: 14.547250] time: 0:03:07.611936\n(30, 64, 64, 3)\n0.8533416\n[Epoch 1/5] [Batch 2/360] [D loss: 0.003604] [G loss: 15.166718] time: 0:03:07.833744\n(30, 64, 64, 3)\n0.7928872\n[Epoch 1/5] [Batch 3/360] [D loss: 0.005319] [G loss: 15.491902] time: 0:03:08.062976\n(30, 64, 64, 3)\n0.7809291\n[Epoch 1/5] [Batch 4/360] [D loss: 0.003738] [G loss: 15.332727] time: 0:03:08.239839\n(30, 64, 64, 3)\n0.7452016\n[Epoch 1/5] [Batch 5/360] [D loss: 0.003992] [G loss: 15.228599] time: 0:03:08.424529\n(30, 64, 64, 3)\n0.7698111\n[Epoch 1/5] [Batch 6/360] [D loss: 0.003316] [G loss: 15.488581] time: 0:03:08.595465\n(30, 64, 64, 3)\n0.7734607\n[Epoch 1/5] [Batch 7/360] [D loss: 0.003235] [G loss: 15.321564] time: 0:03:08.786767\n(30, 64, 64, 3)\n0.80780506\n[Epoch 1/5] [Batch 8/360] [D loss: 0.004980] [G loss: 14.625796] time: 0:03:08.968209\n(30, 64, 64, 3)\n0.75939614\n[Epoch 1/5] [Batch 9/360] [D loss: 0.003094] [G loss: 15.481733] time: 0:03:09.169812\n(30, 64, 64, 3)\n0.80304956\n[Epoch 1/5] [Batch 10/360] [D loss: 0.004602] [G loss: 15.873953] time: 0:03:09.375545\n(30, 64, 64, 3)\n0.7913478\n[Epoch 1/5] [Batch 11/360] [D loss: 0.003544] [G loss: 15.598336] time: 0:03:09.555094\n(30, 64, 64, 3)\n0.83034325\n[Epoch 1/5] [Batch 12/360] [D loss: 0.003167] [G loss: 15.001308] time: 0:03:09.741551\n(30, 64, 64, 3)\n0.81987286\n[Epoch 1/5] [Batch 13/360] [D loss: 0.003653] [G loss: 15.489408] time: 0:03:09.913469\n(30, 64, 64, 3)\n0.7411945\n[Epoch 1/5] [Batch 14/360] [D loss: 0.007628] [G loss: 15.143878] time: 0:03:10.125657\n(30, 64, 64, 3)\n0.79013914\n[Epoch 1/5] [Batch 15/360] [D loss: 0.004641] [G loss: 15.125076] time: 0:03:10.313339\n(30, 64, 64, 3)\n0.70341426\n[Epoch 1/5] [Batch 16/360] [D loss: 0.005779] [G loss: 14.396868] time: 0:03:10.503061\n(30, 64, 64, 3)\n0.8267915\n[Epoch 1/5] [Batch 17/360] [D loss: 0.003526] [G loss: 15.110131] time: 0:03:10.719823\n(30, 64, 64, 3)\n0.7462489\n[Epoch 1/5] [Batch 18/360] [D loss: 0.015293] [G loss: 14.218528] time: 0:03:10.930522\n(30, 64, 64, 3)\n0.7729786\n[Epoch 1/5] [Batch 19/360] [D loss: 0.004900] [G loss: 15.701384] time: 0:03:11.109634\n(30, 64, 64, 3)\n0.83290577\n[Epoch 1/5] [Batch 20/360] [D loss: 0.003141] [G loss: 14.922021] time: 0:03:11.358717\n(30, 64, 64, 3)\n0.82011765\n[Epoch 1/5] [Batch 21/360] [D loss: 0.002467] [G loss: 14.906857] time: 0:03:11.579783\n(30, 64, 64, 3)\n0.7468934\n[Epoch 1/5] [Batch 22/360] [D loss: 0.003990] [G loss: 14.678727] time: 0:03:11.755791\n(30, 64, 64, 3)\n0.82097197\n[Epoch 1/5] [Batch 23/360] [D loss: 0.003866] [G loss: 14.519544] time: 0:03:11.954516\n(30, 64, 64, 3)\n0.80668706\n[Epoch 1/5] [Batch 24/360] [D loss: 0.003382] [G loss: 14.515939] time: 0:03:12.191878\n(30, 64, 64, 3)\n0.8068137\n[Epoch 1/5] [Batch 25/360] [D loss: 0.004757] [G loss: 14.655063] time: 0:03:12.386699\n(30, 64, 64, 3)\n0.8215949\n[Epoch 1/5] [Batch 26/360] [D loss: 0.002925] [G loss: 15.312774] time: 0:03:12.610449\n(30, 64, 64, 3)\n0.7921641\n[Epoch 1/5] [Batch 27/360] [D loss: 0.003847] [G loss: 14.920681] time: 0:03:12.866251\n(30, 64, 64, 3)\n0.81752616\n[Epoch 1/5] [Batch 28/360] [D loss: 0.003039] [G loss: 14.443041] time: 0:03:13.043950\n(30, 64, 64, 3)\n0.85590315\n[Epoch 1/5] [Batch 29/360] [D loss: 0.007251] [G loss: 14.770486] time: 0:03:13.254534\n(30, 64, 64, 3)\n0.75610095\n[Epoch 1/5] [Batch 30/360] [D loss: 0.004729] [G loss: 14.493303] time: 0:03:13.462822\n(30, 64, 64, 3)\n0.8272709\n[Epoch 1/5] [Batch 31/360] [D loss: 0.002818] [G loss: 15.159875] time: 0:03:13.639682\n(30, 64, 64, 3)\n0.8082762\n[Epoch 1/5] [Batch 32/360] [D loss: 0.003692] [G loss: 14.608438] time: 0:03:13.847034\n(30, 64, 64, 3)\n0.787191\n[Epoch 1/5] [Batch 33/360] [D loss: 0.003533] [G loss: 14.084882] time: 0:03:14.056449\n(30, 64, 64, 3)\n0.82802534\n[Epoch 1/5] [Batch 34/360] [D loss: 0.017298] [G loss: 14.568651] time: 0:03:14.251362\n(30, 64, 64, 3)\n0.70508677\n[Epoch 1/5] [Batch 35/360] [D loss: 0.044722] [G loss: 14.580846] time: 0:03:14.437566\n(30, 64, 64, 3)\n0.786622\n[Epoch 1/5] [Batch 36/360] [D loss: 0.015652] [G loss: 14.104079] time: 0:03:14.655669\n(30, 64, 64, 3)\n0.763658\n[Epoch 1/5] [Batch 37/360] [D loss: 0.021089] [G loss: 14.344448] time: 0:03:14.857326\n(30, 64, 64, 3)\n0.8102062\n[Epoch 1/5] [Batch 38/360] [D loss: 0.013440] [G loss: 13.903037] time: 0:03:15.087063\n(30, 64, 64, 3)\n0.833981\n[Epoch 1/5] [Batch 39/360] [D loss: 0.003758] [G loss: 14.476164] time: 0:03:15.296131\n(30, 64, 64, 3)\n0.8403697\n[Epoch 1/5] [Batch 40/360] [D loss: 0.008181] [G loss: 14.180944] time: 0:03:15.477447\n(30, 64, 64, 3)\n0.8622529\n[Epoch 1/5] [Batch 41/360] [D loss: 0.004244] [G loss: 14.424094] time: 0:03:15.690831\n(30, 64, 64, 3)\n0.8044092\n[Epoch 1/5] [Batch 42/360] [D loss: 0.017650] [G loss: 14.773619] time: 0:03:15.868828\n(30, 64, 64, 3)\n0.7117174\n[Epoch 1/5] [Batch 43/360] [D loss: 0.006236] [G loss: 14.754608] time: 0:03:16.095143\n(30, 64, 64, 3)\n0.8272183\n[Epoch 1/5] [Batch 44/360] [D loss: 0.005216] [G loss: 14.443670] time: 0:03:16.274626\n(30, 64, 64, 3)\n0.75021935\n[Epoch 1/5] [Batch 45/360] [D loss: 0.004869] [G loss: 15.088010] time: 0:03:16.443441\n(30, 64, 64, 3)\n0.80698663\n[Epoch 1/5] [Batch 46/360] [D loss: 0.016735] [G loss: 14.350901] time: 0:03:16.661033\n(30, 64, 64, 3)\n0.7590025\n[Epoch 1/5] [Batch 47/360] [D loss: 0.004288] [G loss: 14.435834] time: 0:03:16.832437\n(30, 64, 64, 3)\n0.7900465\n[Epoch 1/5] [Batch 48/360] [D loss: 0.002816] [G loss: 14.297620] time: 0:03:17.028850\n(30, 64, 64, 3)\n0.75772566\n[Epoch 1/5] [Batch 49/360] [D loss: 0.003639] [G loss: 14.751752] time: 0:03:17.282160\n(30, 64, 64, 3)\n0.8202713\n[Epoch 1/5] [Batch 50/360] [D loss: 0.014154] [G loss: 14.416879] time: 0:03:17.510367\n(30, 64, 64, 3)\n0.8275261\n[Epoch 1/5] [Batch 51/360] [D loss: 0.009426] [G loss: 14.842789] time: 0:03:17.700357\n(30, 64, 64, 3)\n0.7923145\n[Epoch 1/5] [Batch 52/360] [D loss: 0.020862] [G loss: 14.212986] time: 0:03:17.888938\n(30, 64, 64, 3)\n0.8345575\n[Epoch 1/5] [Batch 53/360] [D loss: 0.114074] [G loss: 14.038408] time: 0:03:18.093695\n(30, 64, 64, 3)\n0.742614\n[Epoch 1/5] [Batch 54/360] [D loss: 0.330283] [G loss: 14.286210] time: 0:03:18.316985\n(30, 64, 64, 3)\n0.79336387\n[Epoch 1/5] [Batch 55/360] [D loss: 0.013716] [G loss: 14.211987] time: 0:03:18.521862\n(30, 64, 64, 3)\n0.7868187\n[Epoch 1/5] [Batch 56/360] [D loss: 0.066605] [G loss: 14.101266] time: 0:03:18.683236\n(30, 64, 64, 3)\n0.80629474\n[Epoch 1/5] [Batch 57/360] [D loss: 0.006612] [G loss: 13.956211] time: 0:03:18.879988\n(30, 64, 64, 3)\n0.81854135\n[Epoch 1/5] [Batch 58/360] [D loss: 0.018337] [G loss: 14.030013] time: 0:03:19.038687\n(30, 64, 64, 3)\n0.8216744\n[Epoch 1/5] [Batch 59/360] [D loss: 0.006150] [G loss: 13.954692] time: 0:03:19.249735\n(30, 64, 64, 3)\n0.76838106\n[Epoch 1/5] [Batch 60/360] [D loss: 0.005701] [G loss: 14.046259] time: 0:03:19.418304\n(30, 64, 64, 3)\n0.7313548\n[Epoch 1/5] [Batch 61/360] [D loss: 0.005480] [G loss: 13.754184] time: 0:03:19.616824\n(30, 64, 64, 3)\n0.8168335\n[Epoch 1/5] [Batch 62/360] [D loss: 0.005812] [G loss: 13.839202] time: 0:03:19.778477\n(30, 64, 64, 3)\n0.741385\n[Epoch 1/5] [Batch 63/360] [D loss: 0.004708] [G loss: 13.653640] time: 0:03:20.013266\n(30, 64, 64, 3)\n0.81051064\n[Epoch 1/5] [Batch 64/360] [D loss: 0.006105] [G loss: 14.182746] time: 0:03:20.207590\n(30, 64, 64, 3)\n0.7466337\n[Epoch 1/5] [Batch 65/360] [D loss: 0.006972] [G loss: 13.594400] time: 0:03:20.410255\n(30, 64, 64, 3)\n0.87644124\n[Epoch 1/5] [Batch 66/360] [D loss: 0.005716] [G loss: 13.359414] time: 0:03:20.637071\n(30, 64, 64, 3)\n0.7799192\n[Epoch 1/5] [Batch 67/360] [D loss: 0.006883] [G loss: 13.616696] time: 0:03:20.804424\n(30, 64, 64, 3)\n0.81083935\n[Epoch 1/5] [Batch 68/360] [D loss: 0.003727] [G loss: 14.271995] time: 0:03:21.008397\n(30, 64, 64, 3)\n0.87601906\n[Epoch 1/5] [Batch 69/360] [D loss: 0.007026] [G loss: 13.863400] time: 0:03:21.233357\n(30, 64, 64, 3)\n0.7976043\n[Epoch 1/5] [Batch 70/360] [D loss: 0.004613] [G loss: 13.490416] time: 0:03:21.429723\n(30, 64, 64, 3)\n0.8078081\n[Epoch 1/5] [Batch 71/360] [D loss: 0.003443] [G loss: 14.608051] time: 0:03:21.592887\n(30, 64, 64, 3)\n0.80752164\n[Epoch 1/5] [Batch 72/360] [D loss: 0.004625] [G loss: 13.739341] time: 0:03:21.779655\n(30, 64, 64, 3)\n0.8336752\n[Epoch 1/5] [Batch 73/360] [D loss: 0.003536] [G loss: 14.148263] time: 0:03:21.989420\n(30, 64, 64, 3)\n0.8286495\n[Epoch 1/5] [Batch 74/360] [D loss: 0.007663] [G loss: 13.853058] time: 0:03:22.209041\n(30, 64, 64, 3)\n0.7999466\n[Epoch 1/5] [Batch 75/360] [D loss: 0.003670] [G loss: 13.524625] time: 0:03:22.407223\n(30, 64, 64, 3)\n0.7856643\n[Epoch 1/5] [Batch 76/360] [D loss: 0.004282] [G loss: 13.565998] time: 0:03:22.605992\n(30, 64, 64, 3)\n0.77128506\n[Epoch 1/5] [Batch 77/360] [D loss: 0.003955] [G loss: 13.443272] time: 0:03:22.827869\n(30, 64, 64, 3)\n0.7544527\n[Epoch 1/5] [Batch 78/360] [D loss: 0.011798] [G loss: 12.824796] time: 0:03:23.070845\n(30, 64, 64, 3)\n0.7871955\n[Epoch 1/5] [Batch 79/360] [D loss: 0.005184] [G loss: 13.538568] time: 0:03:23.300259\n(30, 64, 64, 3)\n0.79778117\n[Epoch 1/5] [Batch 80/360] [D loss: 0.003580] [G loss: 13.597935] time: 0:03:23.482715\n(30, 64, 64, 3)\n0.78380233\n[Epoch 1/5] [Batch 81/360] [D loss: 0.003095] [G loss: 13.284327] time: 0:03:23.664167\n(30, 64, 64, 3)\n0.80416924\n[Epoch 1/5] [Batch 82/360] [D loss: 0.004561] [G loss: 13.097714] time: 0:03:23.864188\n(30, 64, 64, 3)\n0.8022144\n[Epoch 1/5] [Batch 83/360] [D loss: 0.004483] [G loss: 13.580516] time: 0:03:24.060822\n(30, 64, 64, 3)\n0.8059404\n[Epoch 1/5] [Batch 84/360] [D loss: 0.006900] [G loss: 13.409391] time: 0:03:24.243552\n(30, 64, 64, 3)\n0.82303524\n[Epoch 1/5] [Batch 85/360] [D loss: 0.003064] [G loss: 13.746194] time: 0:03:24.402623\n(30, 64, 64, 3)\n0.8435111\n[Epoch 1/5] [Batch 86/360] [D loss: 0.004421] [G loss: 13.000741] time: 0:03:24.576635\n(30, 64, 64, 3)\n0.8780031\n[Epoch 1/5] [Batch 87/360] [D loss: 0.004733] [G loss: 13.790207] time: 0:03:24.756209\n(30, 64, 64, 3)\n0.7966905\n[Epoch 1/5] [Batch 88/360] [D loss: 0.011769] [G loss: 13.269823] time: 0:03:24.949712\n(30, 64, 64, 3)\n0.80486673\n[Epoch 1/5] [Batch 89/360] [D loss: 0.005848] [G loss: 12.871856] time: 0:03:25.160502\n(30, 64, 64, 3)\n0.7741828\n[Epoch 1/5] [Batch 90/360] [D loss: 0.041028] [G loss: 13.566634] time: 0:03:25.350634\n(30, 64, 64, 3)\n0.82941943\n[Epoch 1/5] [Batch 91/360] [D loss: 0.092614] [G loss: 14.344952] time: 0:03:25.541789\n(30, 64, 64, 3)\n0.82741326\n[Epoch 1/5] [Batch 92/360] [D loss: 0.026318] [G loss: 13.154767] time: 0:03:25.736517\n(30, 64, 64, 3)\n0.81286985\n[Epoch 1/5] [Batch 93/360] [D loss: 0.026691] [G loss: 13.452489] time: 0:03:25.914552\n(30, 64, 64, 3)\n0.7749383\n[Epoch 1/5] [Batch 94/360] [D loss: 0.109876] [G loss: 13.732473] time: 0:03:26.142057\n(30, 64, 64, 3)\n0.8169725\n[Epoch 1/5] [Batch 95/360] [D loss: 0.027104] [G loss: 13.572575] time: 0:03:26.315476\n(30, 64, 64, 3)\n0.81709915\n[Epoch 1/5] [Batch 96/360] [D loss: 0.007089] [G loss: 12.980503] time: 0:03:26.494691\n(30, 64, 64, 3)\n0.8281421\n[Epoch 1/5] [Batch 97/360] [D loss: 0.009861] [G loss: 12.668849] time: 0:03:26.687145\n(30, 64, 64, 3)\n0.8463332\n[Epoch 1/5] [Batch 98/360] [D loss: 0.003876] [G loss: 13.420447] time: 0:03:26.865030\n(30, 64, 64, 3)\n0.81432706\n[Epoch 1/5] [Batch 99/360] [D loss: 0.004154] [G loss: 13.449734] time: 0:03:27.059086\n(30, 64, 64, 3)\n0.8068023\n[Epoch 1/5] [Batch 100/360] [D loss: 0.005605] [G loss: 13.575326] time: 0:03:27.256697\n(30, 64, 64, 3)\n0.7978513\n[Epoch 1/5] [Batch 101/360] [D loss: 0.018861] [G loss: 13.027775] time: 0:03:27.444445\n(30, 64, 64, 3)\n0.8193671\n[Epoch 1/5] [Batch 102/360] [D loss: 0.005463] [G loss: 13.085851] time: 0:03:27.620095\n(30, 64, 64, 3)\n0.7766083\n[Epoch 1/5] [Batch 103/360] [D loss: 0.005597] [G loss: 13.204276] time: 0:03:27.831305\n(30, 64, 64, 3)\n0.81013554\n[Epoch 1/5] [Batch 104/360] [D loss: 0.005772] [G loss: 13.052438] time: 0:03:28.035119\n(30, 64, 64, 3)\n0.8282826\n[Epoch 1/5] [Batch 105/360] [D loss: 0.003583] [G loss: 13.110657] time: 0:03:28.210148\n(30, 64, 64, 3)\n0.7649177\n[Epoch 1/5] [Batch 106/360] [D loss: 0.004761] [G loss: 12.839192] time: 0:03:28.401815\n(30, 64, 64, 3)\n0.78966594\n[Epoch 1/5] [Batch 107/360] [D loss: 0.003348] [G loss: 12.967223] time: 0:03:28.635429\n(30, 64, 64, 3)\n0.8359496\n[Epoch 1/5] [Batch 108/360] [D loss: 0.006995] [G loss: 13.339203] time: 0:03:28.847655\n(30, 64, 64, 3)\n0.81503254\n[Epoch 1/5] [Batch 109/360] [D loss: 0.005099] [G loss: 13.093711] time: 0:03:29.091178\n(30, 64, 64, 3)\n0.8498362\n[Epoch 1/5] [Batch 110/360] [D loss: 0.005878] [G loss: 13.218973] time: 0:03:29.250071\n(30, 64, 64, 3)\n0.85968584\n[Epoch 1/5] [Batch 111/360] [D loss: 0.003662] [G loss: 13.013436] time: 0:03:29.441423\n(30, 64, 64, 3)\n0.8327169\n[Epoch 1/5] [Batch 112/360] [D loss: 0.008424] [G loss: 12.242448] time: 0:03:29.617363\n(30, 64, 64, 3)\n0.8164441\n[Epoch 1/5] [Batch 113/360] [D loss: 0.004292] [G loss: 12.919320] time: 0:03:29.842995\n(30, 64, 64, 3)\n0.8868975\n[Epoch 1/5] [Batch 114/360] [D loss: 0.029329] [G loss: 13.125769] time: 0:03:30.017304\n(30, 64, 64, 3)\n0.8795096\n[Epoch 1/5] [Batch 115/360] [D loss: 0.009961] [G loss: 12.830725] time: 0:03:30.239023\n(30, 64, 64, 3)\n0.82591933\n[Epoch 1/5] [Batch 116/360] [D loss: 0.016524] [G loss: 13.459532] time: 0:03:30.432339\n(30, 64, 64, 3)\n0.7775688\n[Epoch 1/5] [Batch 117/360] [D loss: 0.040664] [G loss: 12.609166] time: 0:03:30.625711\n(30, 64, 64, 3)\n0.80332136\n[Epoch 1/5] [Batch 118/360] [D loss: 0.005713] [G loss: 12.914264] time: 0:03:30.874351\n(30, 64, 64, 3)\n0.78897816\n[Epoch 1/5] [Batch 119/360] [D loss: 0.010015] [G loss: 13.704987] time: 0:03:31.090423\n(30, 64, 64, 3)\n0.8020775\n[Epoch 1/5] [Batch 120/360] [D loss: 0.005027] [G loss: 12.394193] time: 0:03:31.282883\n(30, 64, 64, 3)\n0.7988329\n[Epoch 1/5] [Batch 121/360] [D loss: 0.004193] [G loss: 12.785321] time: 0:03:31.511078\n(30, 64, 64, 3)\n0.8049826\n[Epoch 1/5] [Batch 122/360] [D loss: 0.005529] [G loss: 12.258134] time: 0:03:31.742861\n(30, 64, 64, 3)\n0.79788584\n[Epoch 1/5] [Batch 123/360] [D loss: 0.021729] [G loss: 12.585679] time: 0:03:31.938157\n(30, 64, 64, 3)\n0.7902813\n[Epoch 1/5] [Batch 124/360] [D loss: 0.004348] [G loss: 13.551263] time: 0:03:32.104004\n(30, 64, 64, 3)\n0.7887203\n[Epoch 1/5] [Batch 125/360] [D loss: 0.005539] [G loss: 12.384624] time: 0:03:32.291556\n(30, 64, 64, 3)\n0.87036353\n[Epoch 1/5] [Batch 126/360] [D loss: 0.010274] [G loss: 12.779136] time: 0:03:32.499670\n(30, 64, 64, 3)\n0.8299389\n[Epoch 1/5] [Batch 127/360] [D loss: 0.025036] [G loss: 12.663925] time: 0:03:32.669564\n(30, 64, 64, 3)\n0.8395443\n[Epoch 1/5] [Batch 128/360] [D loss: 0.008092] [G loss: 12.148825] time: 0:03:32.859135\n(30, 64, 64, 3)\n0.83413213\n[Epoch 1/5] [Batch 129/360] [D loss: 0.011177] [G loss: 12.419131] time: 0:03:33.054433\n(30, 64, 64, 3)\n0.8517696\n[Epoch 1/5] [Batch 130/360] [D loss: 0.082613] [G loss: 12.921801] time: 0:03:33.264316\n(30, 64, 64, 3)\n0.7970083\n[Epoch 1/5] [Batch 131/360] [D loss: 0.272662] [G loss: 12.158792] time: 0:03:33.463867\n(30, 64, 64, 3)\n0.8089649\n[Epoch 1/5] [Batch 132/360] [D loss: 0.015730] [G loss: 12.329244] time: 0:03:33.674036\n(30, 64, 64, 3)\n0.81961125\n[Epoch 1/5] [Batch 133/360] [D loss: 0.059893] [G loss: 12.844100] time: 0:03:33.837209\n(30, 64, 64, 3)\n0.84358805\n[Epoch 1/5] [Batch 134/360] [D loss: 0.010483] [G loss: 12.661746] time: 0:03:34.000835\n(30, 64, 64, 3)\n0.82798356\n[Epoch 1/5] [Batch 135/360] [D loss: 0.007264] [G loss: 12.226956] time: 0:03:34.159942\n(30, 64, 64, 3)\n0.72692823\n[Epoch 1/5] [Batch 136/360] [D loss: 0.005123] [G loss: 12.478705] time: 0:03:34.364017\n(30, 64, 64, 3)\n0.8521328\n[Epoch 1/5] [Batch 137/360] [D loss: 0.007339] [G loss: 12.309030] time: 0:03:34.564308\n(30, 64, 64, 3)\n0.83993655\n[Epoch 1/5] [Batch 138/360] [D loss: 0.004879] [G loss: 12.473965] time: 0:03:34.758656\n(30, 64, 64, 3)\n0.8485617\n[Epoch 1/5] [Batch 139/360] [D loss: 0.004191] [G loss: 12.751367] time: 0:03:34.946554\n(30, 64, 64, 3)\n0.88536674\n[Epoch 1/5] [Batch 140/360] [D loss: 0.002597] [G loss: 12.589905] time: 0:03:35.161658\n(30, 64, 64, 3)\n0.80859536\n[Epoch 1/5] [Batch 141/360] [D loss: 0.005576] [G loss: 12.557035] time: 0:03:35.358315\n(30, 64, 64, 3)\n0.810749\n[Epoch 1/5] [Batch 142/360] [D loss: 0.003215] [G loss: 12.639303] time: 0:03:35.513778\n(30, 64, 64, 3)\n0.8462177\n[Epoch 1/5] [Batch 143/360] [D loss: 0.004461] [G loss: 12.636027] time: 0:03:35.678791\n(30, 64, 64, 3)\n0.8186379\n[Epoch 1/5] [Batch 144/360] [D loss: 0.003779] [G loss: 12.117444] time: 0:03:35.903724\n(30, 64, 64, 3)\n0.8032016\n[Epoch 1/5] [Batch 145/360] [D loss: 0.005458] [G loss: 12.096737] time: 0:03:36.118955\n(30, 64, 64, 3)\n0.8755428\n[Epoch 1/5] [Batch 146/360] [D loss: 0.006160] [G loss: 12.398019] time: 0:03:36.381322\n(30, 64, 64, 3)\n0.8024311\n[Epoch 1/5] [Batch 147/360] [D loss: 0.004877] [G loss: 12.139889] time: 0:03:36.571440\n(30, 64, 64, 3)\n0.86419415\n[Epoch 1/5] [Batch 148/360] [D loss: 0.003585] [G loss: 12.029099] time: 0:03:36.788537\n(30, 64, 64, 3)\n0.8151594\n[Epoch 1/5] [Batch 149/360] [D loss: 0.005298] [G loss: 11.747252] time: 0:03:36.987547\n(30, 64, 64, 3)\n0.8240795\n[Epoch 1/5] [Batch 150/360] [D loss: 0.005510] [G loss: 12.533697] time: 0:03:37.184866\n(30, 64, 64, 3)\n0.81155235\n[Epoch 1/5] [Batch 151/360] [D loss: 0.003612] [G loss: 12.752323] time: 0:03:37.375959\n(30, 64, 64, 3)\n0.7487747\n[Epoch 1/5] [Batch 152/360] [D loss: 0.005545] [G loss: 11.824879] time: 0:03:37.588824\n(30, 64, 64, 3)\n0.856425\n[Epoch 1/5] [Batch 153/360] [D loss: 0.007183] [G loss: 11.867030] time: 0:03:37.755706\n(30, 64, 64, 3)\n0.77885675\n[Epoch 1/5] [Batch 154/360] [D loss: 0.022936] [G loss: 11.735703] time: 0:03:37.938701\n(30, 64, 64, 3)\n0.7546541\n[Epoch 1/5] [Batch 155/360] [D loss: 0.007459] [G loss: 11.953355] time: 0:03:38.126665\n(30, 64, 64, 3)\n0.84018975\n[Epoch 1/5] [Batch 156/360] [D loss: 0.005025] [G loss: 12.023451] time: 0:03:38.380711\n(30, 64, 64, 3)\n0.784975\n[Epoch 1/5] [Batch 157/360] [D loss: 0.007578] [G loss: 11.177072] time: 0:03:38.587433\n(30, 64, 64, 3)\n0.84245163\n[Epoch 1/5] [Batch 158/360] [D loss: 0.022623] [G loss: 11.924679] time: 0:03:38.833305\n(30, 64, 64, 3)\n0.8204338\n[Epoch 1/5] [Batch 159/360] [D loss: 0.017541] [G loss: 11.896173] time: 0:03:39.020855\n(30, 64, 64, 3)\n0.80628514\n[Epoch 1/5] [Batch 160/360] [D loss: 0.011790] [G loss: 11.786216] time: 0:03:39.185592\n(30, 64, 64, 3)\n0.80450726\n[Epoch 1/5] [Batch 161/360] [D loss: 0.011067] [G loss: 11.482999] time: 0:03:39.380856\n(30, 64, 64, 3)\n0.8426876\n[Epoch 1/5] [Batch 162/360] [D loss: 0.004603] [G loss: 12.173666] time: 0:03:39.577559\n(30, 64, 64, 3)\n0.8018195\n[Epoch 1/5] [Batch 163/360] [D loss: 0.027077] [G loss: 12.004983] time: 0:03:39.736731\n(30, 64, 64, 3)\n0.8074749\n[Epoch 1/5] [Batch 164/360] [D loss: 0.031395] [G loss: 11.739624] time: 0:03:39.919980\n(30, 64, 64, 3)\n0.8538169\n[Epoch 1/5] [Batch 165/360] [D loss: 0.010873] [G loss: 12.017669] time: 0:03:40.102936\n(30, 64, 64, 3)\n0.80034447\n[Epoch 1/5] [Batch 166/360] [D loss: 0.055070] [G loss: 11.403998] time: 0:03:40.314285\n(30, 64, 64, 3)\n0.8348201\n[Epoch 1/5] [Batch 167/360] [D loss: 0.106557] [G loss: 11.488236] time: 0:03:40.518544\n(30, 64, 64, 3)\n0.8493441\n[Epoch 1/5] [Batch 168/360] [D loss: 0.191730] [G loss: 11.947191] time: 0:03:40.751757\n(30, 64, 64, 3)\n0.777194\n[Epoch 1/5] [Batch 169/360] [D loss: 0.036443] [G loss: 11.516825] time: 0:03:40.950967\n(30, 64, 64, 3)\n0.84170836\n[Epoch 1/5] [Batch 170/360] [D loss: 0.027564] [G loss: 12.264881] time: 0:03:41.146103\n(30, 64, 64, 3)\n0.85101455\n[Epoch 1/5] [Batch 171/360] [D loss: 0.031120] [G loss: 12.236696] time: 0:03:41.349052\n(30, 64, 64, 3)\n0.7862005\n[Epoch 1/5] [Batch 172/360] [D loss: 0.006348] [G loss: 11.596332] time: 0:03:41.548064\n(30, 64, 64, 3)\n0.8310895\n[Epoch 1/5] [Batch 173/360] [D loss: 0.006552] [G loss: 11.326175] time: 0:03:41.770921\n(30, 64, 64, 3)\n0.847408\n[Epoch 1/5] [Batch 174/360] [D loss: 0.008666] [G loss: 11.908268] time: 0:03:41.942708\n(30, 64, 64, 3)\n0.7845004\n[Epoch 1/5] [Batch 175/360] [D loss: 0.013693] [G loss: 11.351649] time: 0:03:42.122863\n(30, 64, 64, 3)\n0.78914434\n[Epoch 1/5] [Batch 176/360] [D loss: 0.007149] [G loss: 11.575963] time: 0:03:42.312298\n(30, 64, 64, 3)\n0.87648195\n[Epoch 1/5] [Batch 177/360] [D loss: 0.006843] [G loss: 12.065296] time: 0:03:42.532926\n(30, 64, 64, 3)\n0.8715666\n[Epoch 1/5] [Batch 178/360] [D loss: 0.004949] [G loss: 11.219635] time: 0:03:42.775080\n(30, 64, 64, 3)\n0.8053913\n[Epoch 1/5] [Batch 179/360] [D loss: 0.005005] [G loss: 10.989188] time: 0:03:43.016202\n(30, 64, 64, 3)\n0.8402841\n[Epoch 1/5] [Batch 180/360] [D loss: 0.006250] [G loss: 10.872471] time: 0:03:43.223109\n(30, 64, 64, 3)\n0.8658843\n[Epoch 1/5] [Batch 181/360] [D loss: 0.006382] [G loss: 12.428500] time: 0:03:43.427041\n(30, 64, 64, 3)\n0.8146672\n[Epoch 1/5] [Batch 182/360] [D loss: 0.004607] [G loss: 11.920047] time: 0:03:43.614169\n(30, 64, 64, 3)\n0.7631375\n[Epoch 1/5] [Batch 183/360] [D loss: 0.003333] [G loss: 12.043550] time: 0:03:43.807979\n(30, 64, 64, 3)\n0.84589654\n[Epoch 1/5] [Batch 184/360] [D loss: 0.005835] [G loss: 11.728181] time: 0:03:44.030058\n(30, 64, 64, 3)\n0.83128256\n[Epoch 1/5] [Batch 185/360] [D loss: 0.004324] [G loss: 11.040371] time: 0:03:44.216090\n(30, 64, 64, 3)\n0.84970146\n[Epoch 1/5] [Batch 186/360] [D loss: 0.004548] [G loss: 11.129046] time: 0:03:44.386784\n(30, 64, 64, 3)\n0.8357101\n[Epoch 1/5] [Batch 187/360] [D loss: 0.006041] [G loss: 10.575469] time: 0:03:44.566056\n(30, 64, 64, 3)\n0.81274134\n[Epoch 1/5] [Batch 188/360] [D loss: 0.005158] [G loss: 12.169154] time: 0:03:44.756912\n(30, 64, 64, 3)\n0.81307936\n[Epoch 1/5] [Batch 189/360] [D loss: 0.003359] [G loss: 11.402562] time: 0:03:44.963995\n(30, 64, 64, 3)\n0.81068945\n[Epoch 1/5] [Batch 190/360] [D loss: 0.002968] [G loss: 11.759844] time: 0:03:45.197799\n(30, 64, 64, 3)\n0.8497974\n[Epoch 1/5] [Batch 191/360] [D loss: 0.004849] [G loss: 11.505380] time: 0:03:45.431693\n(30, 64, 64, 3)\n0.81417066\n[Epoch 1/5] [Batch 192/360] [D loss: 0.003829] [G loss: 11.191339] time: 0:03:45.614126\n(30, 64, 64, 3)\n0.8293869\n[Epoch 1/5] [Batch 193/360] [D loss: 0.004327] [G loss: 11.464327] time: 0:03:45.832049\n(30, 64, 64, 3)\n0.76964664\n[Epoch 1/5] [Batch 194/360] [D loss: 0.003730] [G loss: 11.659084] time: 0:03:46.017316\n(30, 64, 64, 3)\n0.8349616\n[Epoch 1/5] [Batch 195/360] [D loss: 0.004563] [G loss: 11.362738] time: 0:03:46.204148\n(30, 64, 64, 3)\n0.7931688\n[Epoch 1/5] [Batch 196/360] [D loss: 0.003988] [G loss: 11.232528] time: 0:03:46.441038\n(30, 64, 64, 3)\n0.8221705\n[Epoch 1/5] [Batch 197/360] [D loss: 0.004602] [G loss: 11.483027] time: 0:03:46.669040\n(30, 64, 64, 3)\n0.85056895\n[Epoch 1/5] [Batch 198/360] [D loss: 0.003855] [G loss: 11.080771] time: 0:03:46.871756\n(30, 64, 64, 3)\n0.83693314\n[Epoch 1/5] [Batch 199/360] [D loss: 0.003965] [G loss: 11.217344] time: 0:03:47.101848\n(30, 64, 64, 3)\n0.84461397\n[Epoch 1/5] [Batch 200/360] [D loss: 0.004671] [G loss: 10.845703] time: 0:03:47.309942\n(30, 64, 64, 3)\n0.7938381\n[Epoch 1/5] [Batch 201/360] [D loss: 0.003704] [G loss: 11.446568] time: 0:03:47.536925\n(30, 64, 64, 3)\n0.8164611\n[Epoch 1/5] [Batch 202/360] [D loss: 0.004154] [G loss: 11.407430] time: 0:03:47.710379\n(30, 64, 64, 3)\n0.820045\n[Epoch 1/5] [Batch 203/360] [D loss: 0.005704] [G loss: 11.272894] time: 0:03:47.885960\n(30, 64, 64, 3)\n0.8114879\n[Epoch 1/5] [Batch 204/360] [D loss: 0.004627] [G loss: 11.242739] time: 0:03:48.075707\n(30, 64, 64, 3)\n0.79184026\n[Epoch 1/5] [Batch 205/360] [D loss: 0.006308] [G loss: 11.096220] time: 0:03:48.256584\n(30, 64, 64, 3)\n0.82559896\n[Epoch 1/5] [Batch 206/360] [D loss: 0.005762] [G loss: 10.710133] time: 0:03:48.432607\n(30, 64, 64, 3)\n0.8584368\n[Epoch 1/5] [Batch 207/360] [D loss: 0.005619] [G loss: 10.934218] time: 0:03:48.642900\n(30, 64, 64, 3)\n0.8083852\n[Epoch 1/5] [Batch 208/360] [D loss: 0.012089] [G loss: 11.061778] time: 0:03:48.832738\n(30, 64, 64, 3)\n0.77709883\n[Epoch 1/5] [Batch 209/360] [D loss: 0.028082] [G loss: 11.983307] time: 0:03:49.034816\n(30, 64, 64, 3)\n0.88237077\n[Epoch 1/5] [Batch 210/360] [D loss: 0.019804] [G loss: 10.954106] time: 0:03:49.234560\n(30, 64, 64, 3)\n0.8284592\n[Epoch 1/5] [Batch 211/360] [D loss: 0.016734] [G loss: 11.177935] time: 0:03:49.449783\n(30, 64, 64, 3)\n0.844021\n[Epoch 1/5] [Batch 212/360] [D loss: 0.065604] [G loss: 10.751602] time: 0:03:49.621768\n(30, 64, 64, 3)\n0.8414693\n[Epoch 1/5] [Batch 213/360] [D loss: 0.390641] [G loss: 11.188334] time: 0:03:49.796626\n(30, 64, 64, 3)\n0.81867486\n[Epoch 1/5] [Batch 214/360] [D loss: 0.062467] [G loss: 10.786057] time: 0:03:50.020891\n(30, 64, 64, 3)\n0.8164211\n[Epoch 1/5] [Batch 215/360] [D loss: 0.108971] [G loss: 11.018533] time: 0:03:50.222546\n(30, 64, 64, 3)\n0.82326984\n[Epoch 1/5] [Batch 216/360] [D loss: 0.045483] [G loss: 11.569798] time: 0:03:50.450981\n(30, 64, 64, 3)\n0.7634349\n[Epoch 1/5] [Batch 217/360] [D loss: 0.037591] [G loss: 11.794821] time: 0:03:50.629910\n(30, 64, 64, 3)\n0.8727929\n[Epoch 1/5] [Batch 218/360] [D loss: 0.016657] [G loss: 11.732834] time: 0:03:50.821008\n(30, 64, 64, 3)\n0.8244829\n[Epoch 1/5] [Batch 219/360] [D loss: 0.013979] [G loss: 10.612988] time: 0:03:51.037461\n(30, 64, 64, 3)\n0.81053406\n[Epoch 1/5] [Batch 220/360] [D loss: 0.012340] [G loss: 10.836810] time: 0:03:51.274388\n(30, 64, 64, 3)\n0.8523636\n[Epoch 1/5] [Batch 221/360] [D loss: 0.015582] [G loss: 10.668518] time: 0:03:51.504229\n(30, 64, 64, 3)\n0.8421056\n[Epoch 1/5] [Batch 222/360] [D loss: 0.010505] [G loss: 10.451117] time: 0:03:51.691085\n(30, 64, 64, 3)\n0.8411746\n[Epoch 1/5] [Batch 223/360] [D loss: 0.015488] [G loss: 10.693519] time: 0:03:51.900670\n(30, 64, 64, 3)\n0.8371642\n[Epoch 1/5] [Batch 224/360] [D loss: 0.008518] [G loss: 11.573159] time: 0:03:52.280648\n(30, 64, 64, 3)\n0.86913425\n[Epoch 1/5] [Batch 225/360] [D loss: 0.007682] [G loss: 10.918625] time: 0:03:52.471404\n(30, 64, 64, 3)\n0.79702264\n[Epoch 1/5] [Batch 226/360] [D loss: 0.005203] [G loss: 11.366698] time: 0:03:52.628329\n(30, 64, 64, 3)\n0.85421515\n[Epoch 1/5] [Batch 227/360] [D loss: 0.007682] [G loss: 10.632730] time: 0:03:52.804370\n(30, 64, 64, 3)\n0.8942564\n[Epoch 1/5] [Batch 228/360] [D loss: 0.006209] [G loss: 10.712003] time: 0:03:53.032907\n(30, 64, 64, 3)\n0.8331427\n[Epoch 1/5] [Batch 229/360] [D loss: 0.010599] [G loss: 11.293878] time: 0:03:53.246709\n(30, 64, 64, 3)\n0.8448376\n[Epoch 1/5] [Batch 230/360] [D loss: 0.006971] [G loss: 10.480639] time: 0:03:53.408665\n(30, 64, 64, 3)\n0.8104933\n[Epoch 1/5] [Batch 231/360] [D loss: 0.004738] [G loss: 11.335039] time: 0:03:53.581124\n(30, 64, 64, 3)\n0.82414657\n[Epoch 1/5] [Batch 232/360] [D loss: 0.004594] [G loss: 11.025922] time: 0:03:53.758494\n(30, 64, 64, 3)\n0.76632386\n[Epoch 1/5] [Batch 233/360] [D loss: 0.004605] [G loss: 10.548059] time: 0:03:53.953034\n(30, 64, 64, 3)\n0.80033606\n[Epoch 1/5] [Batch 234/360] [D loss: 0.007422] [G loss: 10.734185] time: 0:03:54.189057\n(30, 64, 64, 3)\n0.8526041\n[Epoch 1/5] [Batch 235/360] [D loss: 0.008650] [G loss: 10.790258] time: 0:03:54.345609\n(30, 64, 64, 3)\n0.85357594\n[Epoch 1/5] [Batch 236/360] [D loss: 0.004689] [G loss: 10.591612] time: 0:03:54.503752\n(30, 64, 64, 3)\n0.848012\n[Epoch 1/5] [Batch 237/360] [D loss: 0.004162] [G loss: 11.245341] time: 0:03:54.690497\n(30, 64, 64, 3)\n0.7842874\n[Epoch 1/5] [Batch 238/360] [D loss: 0.007006] [G loss: 10.824711] time: 0:03:54.858046\n(30, 64, 64, 3)\n0.76968\n[Epoch 1/5] [Batch 239/360] [D loss: 0.006981] [G loss: 10.106206] time: 0:03:55.076678\n(30, 64, 64, 3)\n0.8241403\n[Epoch 1/5] [Batch 240/360] [D loss: 0.006017] [G loss: 10.926955] time: 0:03:55.304229\n(30, 64, 64, 3)\n0.86601895\n[Epoch 1/5] [Batch 241/360] [D loss: 0.005336] [G loss: 10.669475] time: 0:03:55.528409\n(30, 64, 64, 3)\n0.8543172\n[Epoch 1/5] [Batch 242/360] [D loss: 0.008515] [G loss: 10.154014] time: 0:03:55.709349\n(30, 64, 64, 3)\n0.8390899\n[Epoch 1/5] [Batch 243/360] [D loss: 0.005094] [G loss: 10.414115] time: 0:03:55.924999\n(30, 64, 64, 3)\n0.8346613\n[Epoch 1/5] [Batch 244/360] [D loss: 0.006838] [G loss: 10.534036] time: 0:03:56.105217\n(30, 64, 64, 3)\n0.8545585\n[Epoch 1/5] [Batch 245/360] [D loss: 0.006723] [G loss: 10.238903] time: 0:03:56.271507\n(30, 64, 64, 3)\n0.8360246\n[Epoch 1/5] [Batch 246/360] [D loss: 0.007962] [G loss: 10.099887] time: 0:03:56.439467\n(30, 64, 64, 3)\n0.86775285\n[Epoch 1/5] [Batch 247/360] [D loss: 0.006587] [G loss: 10.893668] time: 0:03:56.600820\n(30, 64, 64, 3)\n0.8490713\n[Epoch 1/5] [Batch 248/360] [D loss: 0.006673] [G loss: 10.679172] time: 0:03:56.795770\n(30, 64, 64, 3)\n0.7946493\n[Epoch 1/5] [Batch 249/360] [D loss: 0.007295] [G loss: 10.585209] time: 0:03:57.000760\n(30, 64, 64, 3)\n0.82809037\n[Epoch 1/5] [Batch 250/360] [D loss: 0.007543] [G loss: 10.720749] time: 0:03:57.193791\n(30, 64, 64, 3)\n0.8158018\n[Epoch 1/5] [Batch 251/360] [D loss: 0.008437] [G loss: 10.717311] time: 0:03:57.397187\n(30, 64, 64, 3)\n0.7849365\n[Epoch 1/5] [Batch 252/360] [D loss: 0.006327] [G loss: 10.436175] time: 0:03:57.571625\n(30, 64, 64, 3)\n0.8828075\n[Epoch 1/5] [Batch 253/360] [D loss: 0.009658] [G loss: 11.037832] time: 0:03:57.758599\n(30, 64, 64, 3)\n0.8304696\n[Epoch 1/5] [Batch 254/360] [D loss: 0.123655] [G loss: 10.831046] time: 0:03:57.950985\n(30, 64, 64, 3)\n0.8968115\n[Epoch 1/5] [Batch 255/360] [D loss: 0.587557] [G loss: 10.395963] time: 0:03:58.131762\n(30, 64, 64, 3)\n0.86463475\n[Epoch 1/5] [Batch 256/360] [D loss: 0.079988] [G loss: 9.902197] time: 0:03:58.365270\n(30, 64, 64, 3)\n0.7433856\n[Epoch 1/5] [Batch 257/360] [D loss: 0.167815] [G loss: 10.330790] time: 0:03:58.610742\n(30, 64, 64, 3)\n0.8385064\n[Epoch 1/5] [Batch 258/360] [D loss: 0.097172] [G loss: 10.326024] time: 0:03:58.805057\n(30, 64, 64, 3)\n0.8190561\n[Epoch 1/5] [Batch 259/360] [D loss: 0.073077] [G loss: 10.030618] time: 0:03:59.010293\n(30, 64, 64, 3)\n0.77344567\n[Epoch 1/5] [Batch 260/360] [D loss: 0.066750] [G loss: 9.717032] time: 0:03:59.212223\n(30, 64, 64, 3)\n0.8265193\n[Epoch 1/5] [Batch 261/360] [D loss: 0.062516] [G loss: 11.342500] time: 0:03:59.395908\n(30, 64, 64, 3)\n0.82663316\n[Epoch 1/5] [Batch 262/360] [D loss: 0.053634] [G loss: 10.420810] time: 0:03:59.583146\n(30, 64, 64, 3)\n0.87607145\n[Epoch 1/5] [Batch 263/360] [D loss: 0.042036] [G loss: 10.106339] time: 0:03:59.791533\n(30, 64, 64, 3)\n0.84933424\n[Epoch 1/5] [Batch 264/360] [D loss: 0.057798] [G loss: 10.872731] time: 0:03:59.956915\n(30, 64, 64, 3)\n0.8504629\n[Epoch 1/5] [Batch 265/360] [D loss: 0.044103] [G loss: 10.395633] time: 0:04:00.140990\n(30, 64, 64, 3)\n0.8827295\n[Epoch 1/5] [Batch 266/360] [D loss: 0.060067] [G loss: 10.078870] time: 0:04:00.366136\n(30, 64, 64, 3)\n0.77808255\n[Epoch 1/5] [Batch 267/360] [D loss: 0.103961] [G loss: 10.553190] time: 0:04:00.542981\n(30, 64, 64, 3)\n0.79479367\n[Epoch 1/5] [Batch 268/360] [D loss: 0.069263] [G loss: 10.224995] time: 0:04:00.709393\n(30, 64, 64, 3)\n0.8409979\n[Epoch 1/5] [Batch 269/360] [D loss: 0.021120] [G loss: 10.481665] time: 0:04:00.864397\n(30, 64, 64, 3)\n0.7630046\n[Epoch 1/5] [Batch 270/360] [D loss: 0.033381] [G loss: 10.427011] time: 0:04:01.045442\n(30, 64, 64, 3)\n0.8542405\n[Epoch 1/5] [Batch 271/360] [D loss: 0.061728] [G loss: 10.948812] time: 0:04:01.228900\n(30, 64, 64, 3)\n0.850346\n[Epoch 1/5] [Batch 272/360] [D loss: 0.031743] [G loss: 9.926161] time: 0:04:01.426848\n(30, 64, 64, 3)\n0.8548939\n[Epoch 1/5] [Batch 273/360] [D loss: 0.014210] [G loss: 10.155290] time: 0:04:01.636487\n(30, 64, 64, 3)\n0.86145073\n[Epoch 1/5] [Batch 274/360] [D loss: 0.019682] [G loss: 9.969718] time: 0:04:01.820167\n(30, 64, 64, 3)\n0.85327315\n[Epoch 1/5] [Batch 275/360] [D loss: 0.014902] [G loss: 10.588321] time: 0:04:02.004280\n(30, 64, 64, 3)\n0.7583801\n[Epoch 1/5] [Batch 276/360] [D loss: 0.011988] [G loss: 10.240867] time: 0:04:02.200818\n(30, 64, 64, 3)\n0.8232886\n[Epoch 1/5] [Batch 277/360] [D loss: 0.026031] [G loss: 10.796651] time: 0:04:02.390134\n(30, 64, 64, 3)\n0.80368835\n[Epoch 1/5] [Batch 278/360] [D loss: 0.229851] [G loss: 10.460450] time: 0:04:02.594466\n(30, 64, 64, 3)\n0.86693764\n[Epoch 1/5] [Batch 279/360] [D loss: 0.032417] [G loss: 10.208077] time: 0:04:02.759544\n(30, 64, 64, 3)\n0.76625586\n[Epoch 1/5] [Batch 280/360] [D loss: 0.030090] [G loss: 10.444667] time: 0:04:02.972930\n(30, 64, 64, 3)\n0.85453826\n[Epoch 1/5] [Batch 281/360] [D loss: 0.091154] [G loss: 10.533751] time: 0:04:03.198317\n(30, 64, 64, 3)\n0.86513454\n[Epoch 1/5] [Batch 282/360] [D loss: 0.338101] [G loss: 9.724272] time: 0:04:03.431808\n(30, 64, 64, 3)\n0.86318064\n[Epoch 1/5] [Batch 283/360] [D loss: 0.035633] [G loss: 9.609518] time: 0:04:03.620294\n(30, 64, 64, 3)\n0.8698616\n[Epoch 1/5] [Batch 284/360] [D loss: 0.142910] [G loss: 10.232991] time: 0:04:03.838968\n(30, 64, 64, 3)\n0.8265195\n[Epoch 1/5] [Batch 285/360] [D loss: 0.060264] [G loss: 9.849954] time: 0:04:04.016373\n(30, 64, 64, 3)\n0.86203694\n[Epoch 1/5] [Batch 286/360] [D loss: 0.072731] [G loss: 9.530831] time: 0:04:04.248425\n(30, 64, 64, 3)\n0.8459889\n[Epoch 1/5] [Batch 287/360] [D loss: 0.025982] [G loss: 10.008951] time: 0:04:04.504029\n(30, 64, 64, 3)\n0.8367157\n[Epoch 1/5] [Batch 288/360] [D loss: 0.016407] [G loss: 10.102016] time: 0:04:04.722146\n(30, 64, 64, 3)\n0.81571674\n[Epoch 1/5] [Batch 289/360] [D loss: 0.012911] [G loss: 9.846527] time: 0:04:04.921231\n(30, 64, 64, 3)\n0.8448979\n[Epoch 1/5] [Batch 290/360] [D loss: 0.016509] [G loss: 9.175504] time: 0:04:05.145568\n(30, 64, 64, 3)\n0.86925656\n[Epoch 1/5] [Batch 291/360] [D loss: 0.017136] [G loss: 10.269197] time: 0:04:05.336007\n(30, 64, 64, 3)\n0.77197117\n[Epoch 1/5] [Batch 292/360] [D loss: 0.011845] [G loss: 10.141925] time: 0:04:05.588030\n(30, 64, 64, 3)\n0.8749241\n[Epoch 1/5] [Batch 293/360] [D loss: 0.015178] [G loss: 10.168641] time: 0:04:05.795908\n(30, 64, 64, 3)\n0.8586147\n[Epoch 1/5] [Batch 294/360] [D loss: 0.015166] [G loss: 9.684262] time: 0:04:05.971017\n(30, 64, 64, 3)\n0.8606779\n[Epoch 1/5] [Batch 295/360] [D loss: 0.010259] [G loss: 9.961617] time: 0:04:06.164514\n(30, 64, 64, 3)\n0.85695505\n[Epoch 1/5] [Batch 296/360] [D loss: 0.017478] [G loss: 9.787992] time: 0:04:06.367534\n(30, 64, 64, 3)\n0.8134103\n[Epoch 1/5] [Batch 297/360] [D loss: 0.010546] [G loss: 9.570606] time: 0:04:06.545068\n(30, 64, 64, 3)\n0.8790104\n[Epoch 1/5] [Batch 298/360] [D loss: 0.009935] [G loss: 9.601557] time: 0:04:06.750606\n(30, 64, 64, 3)\n0.8570957\n[Epoch 1/5] [Batch 299/360] [D loss: 0.011854] [G loss: 9.318510] time: 0:04:06.931975\n(30, 64, 64, 3)\n0.8490071\n[Epoch 1/5] [Batch 300/360] [D loss: 0.008965] [G loss: 9.423849] time: 0:04:07.133634\n(30, 64, 64, 3)\n0.82834965\n[Epoch 1/5] [Batch 301/360] [D loss: 0.010441] [G loss: 9.954537] time: 0:04:07.327693\n(30, 64, 64, 3)\n0.8177387\n[Epoch 1/5] [Batch 302/360] [D loss: 0.016110] [G loss: 9.893490] time: 0:04:07.520030\n(30, 64, 64, 3)\n0.86400527\n[Epoch 1/5] [Batch 303/360] [D loss: 0.010248] [G loss: 10.005666] time: 0:04:07.719081\n(30, 64, 64, 3)\n0.84561723\n[Epoch 1/5] [Batch 304/360] [D loss: 0.014111] [G loss: 9.567921] time: 0:04:07.886906\n(30, 64, 64, 3)\n0.8657382\n[Epoch 1/5] [Batch 305/360] [D loss: 0.009621] [G loss: 9.948355] time: 0:04:08.079149\n(30, 64, 64, 3)\n0.8916456\n[Epoch 1/5] [Batch 306/360] [D loss: 0.010650] [G loss: 9.731984] time: 0:04:08.251759\n(30, 64, 64, 3)\n0.8005437\n[Epoch 1/5] [Batch 307/360] [D loss: 0.010421] [G loss: 9.114942] time: 0:04:08.417150\n(30, 64, 64, 3)\n0.8771486\n[Epoch 1/5] [Batch 308/360] [D loss: 0.008919] [G loss: 10.200412] time: 0:04:08.624181\n(30, 64, 64, 3)\n0.78744227\n[Epoch 1/5] [Batch 309/360] [D loss: 0.014289] [G loss: 9.884434] time: 0:04:08.811586\n(30, 64, 64, 3)\n0.8820637\n[Epoch 1/5] [Batch 310/360] [D loss: 0.009872] [G loss: 9.688173] time: 0:04:08.994218\n(30, 64, 64, 3)\n0.8635289\n[Epoch 1/5] [Batch 311/360] [D loss: 0.010631] [G loss: 9.653303] time: 0:04:09.174247\n(30, 64, 64, 3)\n0.8739145\n[Epoch 1/5] [Batch 312/360] [D loss: 0.008761] [G loss: 10.195224] time: 0:04:09.349457\n(30, 64, 64, 3)\n0.85810536\n[Epoch 1/5] [Batch 313/360] [D loss: 0.018708] [G loss: 9.133027] time: 0:04:09.560804\n(30, 64, 64, 3)\n0.8252112\n[Epoch 1/5] [Batch 314/360] [D loss: 0.013856] [G loss: 9.663775] time: 0:04:09.791802\n(30, 64, 64, 3)\n0.8081855\n[Epoch 1/5] [Batch 315/360] [D loss: 0.010746] [G loss: 9.803483] time: 0:04:09.986347\n(30, 64, 64, 3)\n0.8662526\n[Epoch 1/5] [Batch 316/360] [D loss: 0.011182] [G loss: 8.969952] time: 0:04:10.195983\n(30, 64, 64, 3)\n0.84285235\n[Epoch 1/5] [Batch 317/360] [D loss: 0.014532] [G loss: 9.111957] time: 0:04:10.396044\n(30, 64, 64, 3)\n0.871247\n[Epoch 1/5] [Batch 318/360] [D loss: 0.024013] [G loss: 9.273561] time: 0:04:10.619569\n(30, 64, 64, 3)\n0.8339582\n[Epoch 1/5] [Batch 319/360] [D loss: 0.036596] [G loss: 10.199162] time: 0:04:10.809547\n(30, 64, 64, 3)\n0.8593158\n[Epoch 1/5] [Batch 320/360] [D loss: 0.358910] [G loss: 9.479290] time: 0:04:11.015764\n(30, 64, 64, 3)\n0.868112\n[Epoch 1/5] [Batch 321/360] [D loss: 0.015463] [G loss: 10.061769] time: 0:04:11.215445\n(30, 64, 64, 3)\n0.89918685\n[Epoch 1/5] [Batch 322/360] [D loss: 0.145030] [G loss: 9.553054] time: 0:04:11.405140\n(30, 64, 64, 3)\n0.8261234\n[Epoch 1/5] [Batch 323/360] [D loss: 0.044098] [G loss: 9.681189] time: 0:04:11.564693\n(30, 64, 64, 3)\n0.77894616\n[Epoch 1/5] [Batch 324/360] [D loss: 0.032809] [G loss: 9.236493] time: 0:04:11.733951\n(30, 64, 64, 3)\n0.7983187\n[Epoch 1/5] [Batch 325/360] [D loss: 0.054276] [G loss: 10.177660] time: 0:04:11.938785\n(30, 64, 64, 3)\n0.85794187\n[Epoch 1/5] [Batch 326/360] [D loss: 0.263344] [G loss: 8.969471] time: 0:04:12.086988\n(30, 64, 64, 3)\n0.7628643\n[Epoch 1/5] [Batch 327/360] [D loss: 0.018382] [G loss: 9.018636] time: 0:04:12.278133\n(30, 64, 64, 3)\n0.8405602\n[Epoch 1/5] [Batch 328/360] [D loss: 0.057176] [G loss: 9.870684] time: 0:04:12.458885\n(30, 64, 64, 3)\n0.88723445\n[Epoch 1/5] [Batch 329/360] [D loss: 0.052193] [G loss: 9.416790] time: 0:04:12.649658\n(30, 64, 64, 3)\n0.8258731\n[Epoch 1/5] [Batch 330/360] [D loss: 0.023146] [G loss: 9.685861] time: 0:04:12.853154\n(30, 64, 64, 3)\n0.86963934\n[Epoch 1/5] [Batch 331/360] [D loss: 0.016217] [G loss: 9.994110] time: 0:04:13.038078\n(30, 64, 64, 3)\n0.87890834\n[Epoch 1/5] [Batch 332/360] [D loss: 0.021493] [G loss: 9.786473] time: 0:04:13.292944\n(30, 64, 64, 3)\n0.87018746\n[Epoch 1/5] [Batch 333/360] [D loss: 0.011359] [G loss: 9.711921] time: 0:04:13.474227\n(30, 64, 64, 3)\n0.85005695\n[Epoch 1/5] [Batch 334/360] [D loss: 0.016996] [G loss: 9.207345] time: 0:04:13.640864\n(30, 64, 64, 3)\n0.8903656\n[Epoch 1/5] [Batch 335/360] [D loss: 0.029114] [G loss: 9.582791] time: 0:04:13.855188\n(30, 64, 64, 3)\n0.8578474\n[Epoch 1/5] [Batch 336/360] [D loss: 0.011942] [G loss: 9.537892] time: 0:04:14.052074\n(30, 64, 64, 3)\n0.81200266\n[Epoch 1/5] [Batch 337/360] [D loss: 0.021977] [G loss: 9.290660] time: 0:04:14.272451\n(30, 64, 64, 3)\n0.91141015\n[Epoch 1/5] [Batch 338/360] [D loss: 0.088601] [G loss: 9.943697] time: 0:04:14.477989\n(30, 64, 64, 3)\n0.887764\n[Epoch 1/5] [Batch 339/360] [D loss: 0.756370] [G loss: 8.479629] time: 0:04:14.673202\n(30, 64, 64, 3)\n0.8622052\n[Epoch 1/5] [Batch 340/360] [D loss: 0.100634] [G loss: 9.203760] time: 0:04:14.870793\n(30, 64, 64, 3)\n0.8026719\n[Epoch 1/5] [Batch 341/360] [D loss: 0.120592] [G loss: 9.414018] time: 0:04:15.085895\n(30, 64, 64, 3)\n0.8497017\n[Epoch 1/5] [Batch 342/360] [D loss: 0.133374] [G loss: 9.566379] time: 0:04:15.264247\n(30, 64, 64, 3)\n0.8314788\n[Epoch 1/5] [Batch 343/360] [D loss: 0.097669] [G loss: 9.339572] time: 0:04:15.427606\n(30, 64, 64, 3)\n0.83715105\n[Epoch 1/5] [Batch 344/360] [D loss: 0.082967] [G loss: 9.395182] time: 0:04:15.600867\n(30, 64, 64, 3)\n0.80178404\n[Epoch 1/5] [Batch 345/360] [D loss: 0.066095] [G loss: 9.608442] time: 0:04:15.795252\n(30, 64, 64, 3)\n0.7916255\n[Epoch 1/5] [Batch 346/360] [D loss: 0.073346] [G loss: 9.741716] time: 0:04:15.976388\n(30, 64, 64, 3)\n0.86604214\n[Epoch 1/5] [Batch 347/360] [D loss: 0.095986] [G loss: 9.383099] time: 0:04:16.162498\n(30, 64, 64, 3)\n0.8932646\n[Epoch 1/5] [Batch 348/360] [D loss: 0.065657] [G loss: 9.468138] time: 0:04:16.348149\n(30, 64, 64, 3)\n0.83184963\n[Epoch 1/5] [Batch 349/360] [D loss: 0.053187] [G loss: 9.602016] time: 0:04:16.548460\n(30, 64, 64, 3)\n0.8535962\n[Epoch 1/5] [Batch 350/360] [D loss: 0.125609] [G loss: 10.279267] time: 0:04:16.718065\n(30, 64, 64, 3)\n0.84717625\n[Epoch 1/5] [Batch 351/360] [D loss: 0.060853] [G loss: 8.818589] time: 0:04:16.885317\n(30, 64, 64, 3)\n0.8443915\n[Epoch 1/5] [Batch 352/360] [D loss: 0.022951] [G loss: 9.313452] time: 0:04:17.050921\n(30, 64, 64, 3)\n0.8836729\n[Epoch 1/5] [Batch 353/360] [D loss: 0.035671] [G loss: 9.431663] time: 0:04:17.262164\n(30, 64, 64, 3)\n0.895371\n[Epoch 1/5] [Batch 354/360] [D loss: 0.038570] [G loss: 9.382573] time: 0:04:17.456217\n(30, 64, 64, 3)\n0.8812229\n[Epoch 1/5] [Batch 355/360] [D loss: 0.019787] [G loss: 9.328539] time: 0:04:17.645044\n(30, 64, 64, 3)\n0.8426308\n[Epoch 1/5] [Batch 356/360] [D loss: 0.073235] [G loss: 9.290811] time: 0:04:17.827089\n(30, 64, 64, 3)\n0.91351205\n[Epoch 1/5] [Batch 357/360] [D loss: 0.331272] [G loss: 9.241428] time: 0:04:17.997274\n(30, 64, 64, 3)\n0.8684637\n[Epoch 1/5] [Batch 358/360] [D loss: 0.050683] [G loss: 9.578181] time: 0:04:18.205641\n(30, 64, 64, 3)\n0.8668199\n[Epoch 1/5] [Batch 359/360] [D loss: 0.142009] [G loss: 9.391651] time: 0:04:18.421070\n(30, 64, 64, 3)\n0.8294371\n[Epoch 2/5] [Batch 1/360] [D loss: 0.061276] [G loss: 9.861098] time: 0:04:18.704457\n(30, 64, 64, 3)\n0.83730143\n[Epoch 2/5] [Batch 2/360] [D loss: 0.080587] [G loss: 8.937701] time: 0:04:18.948023\n(30, 64, 64, 3)\n0.8337353\n[Epoch 2/5] [Batch 3/360] [D loss: 0.031199] [G loss: 8.686855] time: 0:04:19.129862\n(30, 64, 64, 3)\n0.9096491\n[Epoch 2/5] [Batch 4/360] [D loss: 0.059155] [G loss: 9.667027] time: 0:04:19.318870\n(30, 64, 64, 3)\n0.85804003\n[Epoch 2/5] [Batch 5/360] [D loss: 0.185219] [G loss: 9.123701] time: 0:04:19.495983\n(30, 64, 64, 3)\n0.8227399\n[Epoch 2/5] [Batch 6/360] [D loss: 0.097509] [G loss: 9.491867] time: 0:04:19.695662\n(30, 64, 64, 3)\n0.82282\n[Epoch 2/5] [Batch 7/360] [D loss: 0.038111] [G loss: 8.957864] time: 0:04:19.924630\n(30, 64, 64, 3)\n0.803953\n[Epoch 2/5] [Batch 8/360] [D loss: 0.032373] [G loss: 9.594410] time: 0:04:20.132094\n(30, 64, 64, 3)\n0.8145438\n[Epoch 2/5] [Batch 9/360] [D loss: 0.093652] [G loss: 9.467355] time: 0:04:20.328311\n(30, 64, 64, 3)\n0.86114645\n[Epoch 2/5] [Batch 10/360] [D loss: 0.062372] [G loss: 9.118885] time: 0:04:20.487447\n(30, 64, 64, 3)\n0.86137944\n[Epoch 2/5] [Batch 11/360] [D loss: 0.024191] [G loss: 8.738434] time: 0:04:20.677588\n(30, 64, 64, 3)\n0.87145025\n[Epoch 2/5] [Batch 12/360] [D loss: 0.041738] [G loss: 9.508257] time: 0:04:20.885163\n(30, 64, 64, 3)\n0.8279557\n[Epoch 2/5] [Batch 13/360] [D loss: 0.384288] [G loss: 8.192307] time: 0:04:21.118258\n(30, 64, 64, 3)\n0.8775689\n[Epoch 2/5] [Batch 14/360] [D loss: 0.108918] [G loss: 9.077845] time: 0:04:21.330480\n(30, 64, 64, 3)\n0.8736539\n[Epoch 2/5] [Batch 15/360] [D loss: 0.081844] [G loss: 8.806483] time: 0:04:21.506580\n(30, 64, 64, 3)\n0.85935575\n[Epoch 2/5] [Batch 16/360] [D loss: 0.103853] [G loss: 8.969625] time: 0:04:21.701719\n(30, 64, 64, 3)\n0.83506036\n[Epoch 2/5] [Batch 17/360] [D loss: 0.063200] [G loss: 9.286945] time: 0:04:21.937248\n(30, 64, 64, 3)\n0.9071718\n[Epoch 2/5] [Batch 18/360] [D loss: 0.048177] [G loss: 9.060016] time: 0:04:22.140566\n(30, 64, 64, 3)\n0.828572\n[Epoch 2/5] [Batch 19/360] [D loss: 0.046040] [G loss: 9.080580] time: 0:04:22.326751\n(30, 64, 64, 3)\n0.8288958\n[Epoch 2/5] [Batch 20/360] [D loss: 0.030770] [G loss: 8.615175] time: 0:04:22.531240\n(30, 64, 64, 3)\n0.7948206\n[Epoch 2/5] [Batch 21/360] [D loss: 0.035842] [G loss: 8.750318] time: 0:04:22.694772\n(30, 64, 64, 3)\n0.827998\n[Epoch 2/5] [Batch 22/360] [D loss: 0.040382] [G loss: 9.056130] time: 0:04:22.913691\n(30, 64, 64, 3)\n0.87607694\n[Epoch 2/5] [Batch 23/360] [D loss: 0.098341] [G loss: 9.541148] time: 0:04:23.150447\n(30, 64, 64, 3)\n0.81524295\n[Epoch 2/5] [Batch 24/360] [D loss: 0.108545] [G loss: 9.222827] time: 0:04:23.391655\n(30, 64, 64, 3)\n0.8741928\n[Epoch 2/5] [Batch 25/360] [D loss: 0.026729] [G loss: 8.765711] time: 0:04:23.595719\n(30, 64, 64, 3)\n0.85616106\n[Epoch 2/5] [Batch 26/360] [D loss: 0.036417] [G loss: 8.667024] time: 0:04:23.827624\n(30, 64, 64, 3)\n0.86431545\n[Epoch 2/5] [Batch 27/360] [D loss: 0.185494] [G loss: 9.737656] time: 0:04:23.994096\n(30, 64, 64, 3)\n0.83272004\n[Epoch 2/5] [Batch 28/360] [D loss: 0.151712] [G loss: 9.094715] time: 0:04:24.177777\n(30, 64, 64, 3)\n0.8879122\n[Epoch 2/5] [Batch 29/360] [D loss: 0.230292] [G loss: 8.728302] time: 0:04:24.391762\n(30, 64, 64, 3)\n0.84081906\n[Epoch 2/5] [Batch 30/360] [D loss: 0.264609] [G loss: 8.596226] time: 0:04:24.657625\n(30, 64, 64, 3)\n0.8684618\n[Epoch 2/5] [Batch 31/360] [D loss: 0.026105] [G loss: 8.608868] time: 0:04:24.861753\n(30, 64, 64, 3)\n0.8063553\n[Epoch 2/5] [Batch 32/360] [D loss: 0.049213] [G loss: 8.651999] time: 0:04:25.069543\n(30, 64, 64, 3)\n0.8624727\n[Epoch 2/5] [Batch 33/360] [D loss: 0.041897] [G loss: 8.769027] time: 0:04:25.272497\n(30, 64, 64, 3)\n0.83221406\n[Epoch 2/5] [Batch 34/360] [D loss: 0.024828] [G loss: 8.940800] time: 0:04:25.433860\n(30, 64, 64, 3)\n0.9134926\n[Epoch 2/5] [Batch 35/360] [D loss: 0.059735] [G loss: 8.464597] time: 0:04:25.635729\n(30, 64, 64, 3)\n0.8234108\n[Epoch 2/5] [Batch 36/360] [D loss: 0.088191] [G loss: 8.941156] time: 0:04:25.847151\n(30, 64, 64, 3)\n0.87437105\n[Epoch 2/5] [Batch 37/360] [D loss: 0.065803] [G loss: 8.625784] time: 0:04:26.044641\n(30, 64, 64, 3)\n0.79462034\n[Epoch 2/5] [Batch 38/360] [D loss: 0.023929] [G loss: 8.942017] time: 0:04:26.253228\n(30, 64, 64, 3)\n0.83828515\n[Epoch 2/5] [Batch 39/360] [D loss: 0.027190] [G loss: 8.392560] time: 0:04:26.459959\n(30, 64, 64, 3)\n0.83005947\n[Epoch 2/5] [Batch 40/360] [D loss: 0.037951] [G loss: 8.259995] time: 0:04:26.638218\n(30, 64, 64, 3)\n0.863347\n[Epoch 2/5] [Batch 41/360] [D loss: 0.018593] [G loss: 8.589247] time: 0:04:26.841852\n(30, 64, 64, 3)\n0.87389404\n[Epoch 2/5] [Batch 42/360] [D loss: 0.019721] [G loss: 9.009238] time: 0:04:27.038382\n(30, 64, 64, 3)\n0.83862686\n[Epoch 2/5] [Batch 43/360] [D loss: 0.122321] [G loss: 8.555246] time: 0:04:27.260180\n(30, 64, 64, 3)\n0.8123782\n[Epoch 2/5] [Batch 44/360] [D loss: 0.142779] [G loss: 8.698080] time: 0:04:27.432731\n(30, 64, 64, 3)\n0.8772841\n[Epoch 2/5] [Batch 45/360] [D loss: 0.122218] [G loss: 8.980123] time: 0:04:27.622237\n(30, 64, 64, 3)\n0.8551037\n[Epoch 2/5] [Batch 46/360] [D loss: 0.202212] [G loss: 8.785493] time: 0:04:27.822136\n(30, 64, 64, 3)\n0.81231433\n[Epoch 2/5] [Batch 47/360] [D loss: 0.038023] [G loss: 9.039211] time: 0:04:27.999455\n(30, 64, 64, 3)\n0.83084744\n[Epoch 2/5] [Batch 48/360] [D loss: 0.032479] [G loss: 9.176832] time: 0:04:28.177754\n(30, 64, 64, 3)\n0.8563331\n[Epoch 2/5] [Batch 49/360] [D loss: 0.028325] [G loss: 8.784700] time: 0:04:28.359779\n(30, 64, 64, 3)\n0.84865546\n[Epoch 2/5] [Batch 50/360] [D loss: 0.031620] [G loss: 8.918521] time: 0:04:28.587699\n(30, 64, 64, 3)\n0.85436773\n[Epoch 2/5] [Batch 51/360] [D loss: 0.118482] [G loss: 8.596505] time: 0:04:28.755832\n(30, 64, 64, 3)\n0.87928486\n[Epoch 2/5] [Batch 52/360] [D loss: 0.102048] [G loss: 7.996586] time: 0:04:28.939820\n(30, 64, 64, 3)\n0.82382745\n[Epoch 2/5] [Batch 53/360] [D loss: 0.028829] [G loss: 8.676611] time: 0:04:29.168952\n(30, 64, 64, 3)\n0.84727424\n[Epoch 2/5] [Batch 54/360] [D loss: 0.023605] [G loss: 9.285073] time: 0:04:29.388966\n(30, 64, 64, 3)\n0.854795\n[Epoch 2/5] [Batch 55/360] [D loss: 0.039181] [G loss: 7.959604] time: 0:04:29.575410\n(30, 64, 64, 3)\n0.8268151\n[Epoch 2/5] [Batch 56/360] [D loss: 0.019777] [G loss: 8.003026] time: 0:04:29.756693\n(30, 64, 64, 3)\n0.8718768\n[Epoch 2/5] [Batch 57/360] [D loss: 0.017651] [G loss: 8.558772] time: 0:04:29.992209\n(30, 64, 64, 3)\n0.8783547\n[Epoch 2/5] [Batch 58/360] [D loss: 0.013886] [G loss: 8.752698] time: 0:04:30.198418\n(30, 64, 64, 3)\n0.86127955\n[Epoch 2/5] [Batch 59/360] [D loss: 0.015672] [G loss: 8.273173] time: 0:04:30.391169\n(30, 64, 64, 3)\n0.87124205\n[Epoch 2/5] [Batch 60/360] [D loss: 0.013632] [G loss: 8.739828] time: 0:04:30.591809\n(30, 64, 64, 3)\n0.8463784\n[Epoch 2/5] [Batch 61/360] [D loss: 0.017148] [G loss: 8.582269] time: 0:04:30.772708\n(30, 64, 64, 3)\n0.8740074\n[Epoch 2/5] [Batch 62/360] [D loss: 0.024886] [G loss: 8.333685] time: 0:04:30.962075\n(30, 64, 64, 3)\n0.82229567\n[Epoch 2/5] [Batch 63/360] [D loss: 0.017092] [G loss: 8.569901] time: 0:04:31.143992\n(30, 64, 64, 3)\n0.8298368\n[Epoch 2/5] [Batch 64/360] [D loss: 0.020055] [G loss: 7.812581] time: 0:04:31.363736\n(30, 64, 64, 3)\n0.83626026\n[Epoch 2/5] [Batch 65/360] [D loss: 0.019750] [G loss: 8.160746] time: 0:04:31.571871\n(30, 64, 64, 3)\n0.82567215\n[Epoch 2/5] [Batch 66/360] [D loss: 0.011548] [G loss: 8.319510] time: 0:04:31.775177\n(30, 64, 64, 3)\n0.86699325\n[Epoch 2/5] [Batch 67/360] [D loss: 0.014667] [G loss: 7.807200] time: 0:04:31.968717\n(30, 64, 64, 3)\n0.8357032\n[Epoch 2/5] [Batch 68/360] [D loss: 0.017873] [G loss: 7.814360] time: 0:04:32.140591\n(30, 64, 64, 3)\n0.88638407\n[Epoch 2/5] [Batch 69/360] [D loss: 0.041268] [G loss: 8.074452] time: 0:04:32.393861\n(30, 64, 64, 3)\n0.8213458\n[Epoch 2/5] [Batch 70/360] [D loss: 0.112125] [G loss: 8.824152] time: 0:04:32.614249\n(30, 64, 64, 3)\n0.9060438\n[Epoch 2/5] [Batch 71/360] [D loss: 0.279975] [G loss: 8.601279] time: 0:04:32.837820\n(30, 64, 64, 3)\n0.86965466\n[Epoch 2/5] [Batch 72/360] [D loss: 0.470416] [G loss: 8.093515] time: 0:04:33.041484\n(30, 64, 64, 3)\n0.911088\n[Epoch 2/5] [Batch 73/360] [D loss: 0.026531] [G loss: 8.244648] time: 0:04:33.244698\n(30, 64, 64, 3)\n0.8982093\n[Epoch 2/5] [Batch 74/360] [D loss: 0.095450] [G loss: 8.184966] time: 0:04:33.453221\n(30, 64, 64, 3)\n0.82437104\n[Epoch 2/5] [Batch 75/360] [D loss: 0.029930] [G loss: 8.052133] time: 0:04:33.678241\n(30, 64, 64, 3)\n0.8637211\n[Epoch 2/5] [Batch 76/360] [D loss: 0.034493] [G loss: 8.102020] time: 0:04:33.874970\n(30, 64, 64, 3)\n0.88046616\n[Epoch 2/5] [Batch 77/360] [D loss: 0.034916] [G loss: 8.209344] time: 0:04:34.041855\n(30, 64, 64, 3)\n0.8104079\n[Epoch 2/5] [Batch 78/360] [D loss: 0.033981] [G loss: 8.522060] time: 0:04:34.234197\n(30, 64, 64, 3)\n0.8021083\n[Epoch 2/5] [Batch 79/360] [D loss: 0.019435] [G loss: 8.504379] time: 0:04:34.402176\n(30, 64, 64, 3)\n0.8565903\n[Epoch 2/5] [Batch 80/360] [D loss: 0.026692] [G loss: 8.019991] time: 0:04:34.603708\n(30, 64, 64, 3)\n0.905528\n[Epoch 2/5] [Batch 81/360] [D loss: 0.028049] [G loss: 8.670983] time: 0:04:34.806683\n(30, 64, 64, 3)\n0.8645107\n[Epoch 2/5] [Batch 82/360] [D loss: 0.019737] [G loss: 8.211490] time: 0:04:34.994737\n(30, 64, 64, 3)\n0.8126216\n[Epoch 2/5] [Batch 83/360] [D loss: 0.018376] [G loss: 8.558345] time: 0:04:35.220049\n(30, 64, 64, 3)\n0.8671157\n[Epoch 2/5] [Batch 84/360] [D loss: 0.020416] [G loss: 8.281677] time: 0:04:35.394125\n(30, 64, 64, 3)\n0.80764514\n[Epoch 2/5] [Batch 85/360] [D loss: 0.017598] [G loss: 7.964083] time: 0:04:35.575289\n(30, 64, 64, 3)\n0.8633423\n[Epoch 2/5] [Batch 86/360] [D loss: 0.031299] [G loss: 8.195501] time: 0:04:35.772742\n(30, 64, 64, 3)\n0.9173441\n[Epoch 2/5] [Batch 87/360] [D loss: 0.045679] [G loss: 8.686682] time: 0:04:35.992058\n(30, 64, 64, 3)\n0.90097827\n[Epoch 2/5] [Batch 88/360] [D loss: 0.063157] [G loss: 8.317509] time: 0:04:36.170713\n(30, 64, 64, 3)\n0.91806465\n[Epoch 2/5] [Batch 89/360] [D loss: 0.026445] [G loss: 8.057409] time: 0:04:36.369269\n(30, 64, 64, 3)\n0.8308328\n[Epoch 2/5] [Batch 90/360] [D loss: 0.018093] [G loss: 8.597827] time: 0:04:36.571562\n(30, 64, 64, 3)\n0.85440975\n[Epoch 2/5] [Batch 91/360] [D loss: 0.041015] [G loss: 8.205815] time: 0:04:36.799661\n(30, 64, 64, 3)\n0.8770712\n[Epoch 2/5] [Batch 92/360] [D loss: 0.066663] [G loss: 8.017258] time: 0:04:37.009404\n(30, 64, 64, 3)\n0.85368663\n[Epoch 2/5] [Batch 93/360] [D loss: 0.075796] [G loss: 7.783727] time: 0:04:37.201414\n(30, 64, 64, 3)\n0.80948\n[Epoch 2/5] [Batch 94/360] [D loss: 0.037665] [G loss: 8.116616] time: 0:04:37.451984\n(30, 64, 64, 3)\n0.8252127\n[Epoch 2/5] [Batch 95/360] [D loss: 0.018470] [G loss: 8.087255] time: 0:04:37.674791\n(30, 64, 64, 3)\n0.85213965\n[Epoch 2/5] [Batch 96/360] [D loss: 0.020403] [G loss: 8.713223] time: 0:04:37.861370\n(30, 64, 64, 3)\n0.84823674\n[Epoch 2/5] [Batch 97/360] [D loss: 0.030459] [G loss: 7.988657] time: 0:04:38.070175\n(30, 64, 64, 3)\n0.83792514\n[Epoch 2/5] [Batch 98/360] [D loss: 0.019041] [G loss: 7.867125] time: 0:04:38.291885\n(30, 64, 64, 3)\n0.8247053\n[Epoch 2/5] [Batch 99/360] [D loss: 0.026908] [G loss: 7.747796] time: 0:04:38.484760\n(30, 64, 64, 3)\n0.88085383\n[Epoch 2/5] [Batch 100/360] [D loss: 0.039353] [G loss: 8.311252] time: 0:04:38.696520\n(30, 64, 64, 3)\n0.91833705\n[Epoch 2/5] [Batch 101/360] [D loss: 0.073396] [G loss: 7.941414] time: 0:04:38.912136\n(30, 64, 64, 3)\n0.8298783\n[Epoch 2/5] [Batch 102/360] [D loss: 0.117036] [G loss: 8.071662] time: 0:04:39.155623\n(30, 64, 64, 3)\n0.8244327\n[Epoch 2/5] [Batch 103/360] [D loss: 0.497948] [G loss: 7.180910] time: 0:04:39.329252\n(30, 64, 64, 3)\n0.87057585\n[Epoch 2/5] [Batch 104/360] [D loss: 0.028174] [G loss: 7.889644] time: 0:04:39.498126\n(30, 64, 64, 3)\n0.86954236\n[Epoch 2/5] [Batch 105/360] [D loss: 0.219228] [G loss: 7.760309] time: 0:04:39.747655\n(30, 64, 64, 3)\n0.9014004\n[Epoch 2/5] [Batch 106/360] [D loss: 0.084501] [G loss: 7.318175] time: 0:04:39.920430\n(30, 64, 64, 3)\n0.8883131\n[Epoch 2/5] [Batch 107/360] [D loss: 0.056590] [G loss: 7.906036] time: 0:04:40.098853\n(30, 64, 64, 3)\n0.87138224\n[Epoch 2/5] [Batch 108/360] [D loss: 0.083923] [G loss: 8.070314] time: 0:04:40.285440\n(30, 64, 64, 3)\n0.84345865\n[Epoch 2/5] [Batch 109/360] [D loss: 0.092952] [G loss: 7.267732] time: 0:04:40.472162\n(30, 64, 64, 3)\n0.8847668\n[Epoch 2/5] [Batch 110/360] [D loss: 0.035258] [G loss: 7.635301] time: 0:04:40.668774\n(30, 64, 64, 3)\n0.8569835\n[Epoch 2/5] [Batch 111/360] [D loss: 0.031521] [G loss: 8.128624] time: 0:04:40.843742\n(30, 64, 64, 3)\n0.92863846\n[Epoch 2/5] [Batch 112/360] [D loss: 0.034099] [G loss: 8.209901] time: 0:04:41.062322\n(30, 64, 64, 3)\n0.8874466\n[Epoch 2/5] [Batch 113/360] [D loss: 0.035507] [G loss: 7.901157] time: 0:04:41.254446\n(30, 64, 64, 3)\n0.7769943\n[Epoch 2/5] [Batch 114/360] [D loss: 0.020904] [G loss: 7.889304] time: 0:04:41.451500\n(30, 64, 64, 3)\n0.82685477\n[Epoch 2/5] [Batch 115/360] [D loss: 0.017803] [G loss: 8.122330] time: 0:04:41.645122\n(30, 64, 64, 3)\n0.8492196\n[Epoch 2/5] [Batch 116/360] [D loss: 0.018271] [G loss: 7.419212] time: 0:04:41.814243\n(30, 64, 64, 3)\n0.7971919\n[Epoch 2/5] [Batch 117/360] [D loss: 0.017901] [G loss: 7.835560] time: 0:04:42.010633\n(30, 64, 64, 3)\n0.84625524\n[Epoch 2/5] [Batch 118/360] [D loss: 0.020640] [G loss: 7.823187] time: 0:04:42.202010\n(30, 64, 64, 3)\n0.85285586\n[Epoch 2/5] [Batch 119/360] [D loss: 0.015705] [G loss: 7.489946] time: 0:04:42.416801\n(30, 64, 64, 3)\n0.8988684\n[Epoch 2/5] [Batch 120/360] [D loss: 0.011822] [G loss: 8.218699] time: 0:04:42.624684\n(30, 64, 64, 3)\n0.89731413\n[Epoch 2/5] [Batch 121/360] [D loss: 0.026416] [G loss: 7.453041] time: 0:04:42.845119\n(30, 64, 64, 3)\n0.85965544\n[Epoch 2/5] [Batch 122/360] [D loss: 0.021117] [G loss: 7.768237] time: 0:04:43.042097\n(30, 64, 64, 3)\n0.8826852\n[Epoch 2/5] [Batch 123/360] [D loss: 0.020932] [G loss: 7.426774] time: 0:04:43.262170\n(30, 64, 64, 3)\n0.8511303\n[Epoch 2/5] [Batch 124/360] [D loss: 0.038393] [G loss: 7.547175] time: 0:04:43.443377\n(30, 64, 64, 3)\n0.84673613\n[Epoch 2/5] [Batch 125/360] [D loss: 0.086005] [G loss: 7.861975] time: 0:04:43.664502\n(30, 64, 64, 3)\n0.8948348\n[Epoch 2/5] [Batch 126/360] [D loss: 0.378372] [G loss: 7.846583] time: 0:04:43.856382\n(30, 64, 64, 3)\n0.835969\n[Epoch 2/5] [Batch 127/360] [D loss: 0.019718] [G loss: 7.881302] time: 0:04:44.093134\n(30, 64, 64, 3)\n0.89058524\n[Epoch 2/5] [Batch 128/360] [D loss: 0.132774] [G loss: 8.006011] time: 0:04:44.291053\n(30, 64, 64, 3)\n0.8730231\n[Epoch 2/5] [Batch 129/360] [D loss: 0.044770] [G loss: 7.601029] time: 0:04:44.453418\n(30, 64, 64, 3)\n0.883262\n[Epoch 2/5] [Batch 130/360] [D loss: 0.042901] [G loss: 7.572725] time: 0:04:44.624768\n(30, 64, 64, 3)\n0.8860381\n[Epoch 2/5] [Batch 131/360] [D loss: 0.398797] [G loss: 7.333388] time: 0:04:44.805264\n(30, 64, 64, 3)\n0.85509473\n[Epoch 2/5] [Batch 132/360] [D loss: 0.070601] [G loss: 7.732456] time: 0:04:45.017821\n(30, 64, 64, 3)\n0.8181853\n[Epoch 2/5] [Batch 133/360] [D loss: 0.132033] [G loss: 7.658635] time: 0:04:45.201366\n(30, 64, 64, 3)\n0.88615435\n[Epoch 2/5] [Batch 134/360] [D loss: 0.060943] [G loss: 7.864050] time: 0:04:45.404843\n(30, 64, 64, 3)\n0.8955144\n[Epoch 2/5] [Batch 135/360] [D loss: 0.049307] [G loss: 7.281221] time: 0:04:45.615636\n(30, 64, 64, 3)\n0.8707824\n[Epoch 2/5] [Batch 136/360] [D loss: 0.036198] [G loss: 7.197370] time: 0:04:45.805400\n(30, 64, 64, 3)\n0.86407095\n[Epoch 2/5] [Batch 137/360] [D loss: 0.031770] [G loss: 8.074692] time: 0:04:46.000699\n(30, 64, 64, 3)\n0.894064\n[Epoch 2/5] [Batch 138/360] [D loss: 0.037280] [G loss: 7.628061] time: 0:04:46.182917\n(30, 64, 64, 3)\n0.884248\n[Epoch 2/5] [Batch 139/360] [D loss: 0.035826] [G loss: 7.742716] time: 0:04:46.423172\n(30, 64, 64, 3)\n0.82958835\n[Epoch 2/5] [Batch 140/360] [D loss: 0.032243] [G loss: 7.545616] time: 0:04:46.622503\n(30, 64, 64, 3)\n0.8209071\n[Epoch 2/5] [Batch 141/360] [D loss: 0.045569] [G loss: 7.860268] time: 0:04:46.835982\n(30, 64, 64, 3)\n0.916929\n[Epoch 2/5] [Batch 142/360] [D loss: 0.108207] [G loss: 7.598845] time: 0:04:47.020799\n(30, 64, 64, 3)\n0.87901324\n[Epoch 2/5] [Batch 143/360] [D loss: 0.409806] [G loss: 7.465102] time: 0:04:47.256724\n(30, 64, 64, 3)\n0.8705295\n[Epoch 2/5] [Batch 144/360] [D loss: 0.076579] [G loss: 7.961498] time: 0:04:47.481117\n(30, 64, 64, 3)\n0.82445353\n[Epoch 2/5] [Batch 145/360] [D loss: 0.080313] [G loss: 7.184301] time: 0:04:47.721677\n(30, 64, 64, 3)\n0.84126455\n[Epoch 2/5] [Batch 146/360] [D loss: 0.098047] [G loss: 7.517327] time: 0:04:47.930230\n(30, 64, 64, 3)\n0.78222543\n[Epoch 2/5] [Batch 147/360] [D loss: 0.080329] [G loss: 7.353908] time: 0:04:48.110267\n(30, 64, 64, 3)\n0.8881884\n[Epoch 2/5] [Batch 148/360] [D loss: 0.047009] [G loss: 7.594724] time: 0:04:48.319497\n(30, 64, 64, 3)\n0.8912077\n[Epoch 2/5] [Batch 149/360] [D loss: 0.053652] [G loss: 7.080897] time: 0:04:48.507117\n(30, 64, 64, 3)\n0.8576812\n[Epoch 2/5] [Batch 150/360] [D loss: 0.044802] [G loss: 7.368513] time: 0:04:48.731978\n(30, 64, 64, 3)\n0.87642556\n[Epoch 2/5] [Batch 151/360] [D loss: 0.028555] [G loss: 7.503169] time: 0:04:48.969553\n(30, 64, 64, 3)\n0.84407735\n[Epoch 2/5] [Batch 152/360] [D loss: 0.025001] [G loss: 7.562111] time: 0:04:49.151835\n(30, 64, 64, 3)\n0.8380553\n[Epoch 2/5] [Batch 153/360] [D loss: 0.025086] [G loss: 7.654046] time: 0:04:49.329840\n(30, 64, 64, 3)\n0.8454096\n[Epoch 2/5] [Batch 154/360] [D loss: 0.048379] [G loss: 7.455315] time: 0:04:49.538317\n(30, 64, 64, 3)\n0.9096144\n[Epoch 2/5] [Batch 155/360] [D loss: 0.167919] [G loss: 7.889795] time: 0:04:49.753239\n(30, 64, 64, 3)\n0.81631273\n[Epoch 2/5] [Batch 156/360] [D loss: 0.465243] [G loss: 6.701303] time: 0:04:49.952368\n(30, 64, 64, 3)\n0.9101365\n[Epoch 2/5] [Batch 157/360] [D loss: 0.104978] [G loss: 7.371297] time: 0:04:50.158848\n(30, 64, 64, 3)\n0.81986856\n[Epoch 2/5] [Batch 158/360] [D loss: 0.082504] [G loss: 7.359524] time: 0:04:50.342258\n(30, 64, 64, 3)\n0.8912678\n[Epoch 2/5] [Batch 159/360] [D loss: 0.093935] [G loss: 7.584564] time: 0:04:50.562921\n(30, 64, 64, 3)\n0.910346\n[Epoch 2/5] [Batch 160/360] [D loss: 0.113584] [G loss: 7.370079] time: 0:04:50.738036\n(30, 64, 64, 3)\n0.8921368\n[Epoch 2/5] [Batch 161/360] [D loss: 0.035008] [G loss: 7.290982] time: 0:04:50.913456\n(30, 64, 64, 3)\n0.8836892\n[Epoch 2/5] [Batch 162/360] [D loss: 0.043814] [G loss: 7.023370] time: 0:04:51.086773\n(30, 64, 64, 3)\n0.92156225\n[Epoch 2/5] [Batch 163/360] [D loss: 0.028288] [G loss: 7.731320] time: 0:04:51.322730\n(30, 64, 64, 3)\n0.8621404\n[Epoch 2/5] [Batch 164/360] [D loss: 0.030102] [G loss: 7.369248] time: 0:04:51.518139\n(30, 64, 64, 3)\n0.8493285\n[Epoch 2/5] [Batch 165/360] [D loss: 0.028900] [G loss: 7.022588] time: 0:04:51.693450\n(30, 64, 64, 3)\n0.8530577\n[Epoch 2/5] [Batch 166/360] [D loss: 0.027852] [G loss: 7.345344] time: 0:04:51.902084\n(30, 64, 64, 3)\n0.81625485\n[Epoch 2/5] [Batch 167/360] [D loss: 0.075898] [G loss: 7.533359] time: 0:04:52.139715\n(30, 64, 64, 3)\n0.885516\n[Epoch 2/5] [Batch 168/360] [D loss: 0.092414] [G loss: 7.901878] time: 0:04:52.357901\n(30, 64, 64, 3)\n0.84806895\n[Epoch 2/5] [Batch 169/360] [D loss: 0.051813] [G loss: 7.172698] time: 0:04:52.593530\n(30, 64, 64, 3)\n0.9305434\n[Epoch 2/5] [Batch 170/360] [D loss: 0.020888] [G loss: 7.667854] time: 0:04:52.804846\n(30, 64, 64, 3)\n0.8481843\n[Epoch 2/5] [Batch 171/360] [D loss: 0.033410] [G loss: 6.853996] time: 0:04:53.004400\n(30, 64, 64, 3)\n0.8789675\n[Epoch 2/5] [Batch 172/360] [D loss: 0.038934] [G loss: 7.236163] time: 0:04:53.213521\n(30, 64, 64, 3)\n0.856528\n[Epoch 2/5] [Batch 173/360] [D loss: 0.078139] [G loss: 7.406964] time: 0:04:53.426488\n(30, 64, 64, 3)\n0.81847745\n[Epoch 2/5] [Batch 174/360] [D loss: 0.238712] [G loss: 7.395112] time: 0:04:53.656044\n(30, 64, 64, 3)\n0.89218384\n[Epoch 2/5] [Batch 175/360] [D loss: 0.076891] [G loss: 7.622932] time: 0:04:53.817175\n(30, 64, 64, 3)\n0.8884743\n[Epoch 2/5] [Batch 176/360] [D loss: 0.026630] [G loss: 7.129149] time: 0:04:54.010356\n(30, 64, 64, 3)\n0.91068\n[Epoch 2/5] [Batch 177/360] [D loss: 0.041243] [G loss: 7.127751] time: 0:04:54.192717\n(30, 64, 64, 3)\n0.8771736\n[Epoch 2/5] [Batch 178/360] [D loss: 0.204335] [G loss: 7.246322] time: 0:04:54.397564\n(30, 64, 64, 3)\n0.85753375\n[Epoch 2/5] [Batch 179/360] [D loss: 0.361046] [G loss: 6.766628] time: 0:04:54.608834\n(30, 64, 64, 3)\n0.8681874\n[Epoch 2/5] [Batch 180/360] [D loss: 0.029016] [G loss: 7.688205] time: 0:04:54.836451\n(30, 64, 64, 3)\n0.8290207\n[Epoch 2/5] [Batch 181/360] [D loss: 0.241346] [G loss: 6.667188] time: 0:04:55.048200\n(30, 64, 64, 3)\n0.8521549\n[Epoch 2/5] [Batch 182/360] [D loss: 0.088477] [G loss: 8.024444] time: 0:04:55.243150\n(30, 64, 64, 3)\n0.89035577\n[Epoch 2/5] [Batch 183/360] [D loss: 0.087399] [G loss: 7.243837] time: 0:04:55.440038\n(30, 64, 64, 3)\n0.8543518\n[Epoch 2/5] [Batch 184/360] [D loss: 0.083781] [G loss: 8.267483] time: 0:04:55.619926\n(30, 64, 64, 3)\n0.8397121\n[Epoch 2/5] [Batch 185/360] [D loss: 0.166873] [G loss: 7.348469] time: 0:04:55.804080\n(30, 64, 64, 3)\n0.843241\n[Epoch 2/5] [Batch 186/360] [D loss: 0.038669] [G loss: 7.216058] time: 0:04:55.978991\n(30, 64, 64, 3)\n0.8551695\n[Epoch 2/5] [Batch 187/360] [D loss: 0.051752] [G loss: 7.107588] time: 0:04:56.149712\n(30, 64, 64, 3)\n0.8408992\n[Epoch 2/5] [Batch 188/360] [D loss: 0.040534] [G loss: 7.074183] time: 0:04:56.365127\n(30, 64, 64, 3)\n0.8564852\n[Epoch 2/5] [Batch 189/360] [D loss: 0.030816] [G loss: 6.949321] time: 0:04:56.589554\n(30, 64, 64, 3)\n0.8808708\n[Epoch 2/5] [Batch 190/360] [D loss: 0.048668] [G loss: 7.038843] time: 0:04:56.825760\n(30, 64, 64, 3)\n0.9066059\n[Epoch 2/5] [Batch 191/360] [D loss: 0.032414] [G loss: 6.718074] time: 0:04:57.006596\n(30, 64, 64, 3)\n0.86376387\n[Epoch 2/5] [Batch 192/360] [D loss: 0.023859] [G loss: 7.152301] time: 0:04:57.231552\n(30, 64, 64, 3)\n0.8789455\n[Epoch 2/5] [Batch 193/360] [D loss: 0.027525] [G loss: 7.368412] time: 0:04:57.414645\n(30, 64, 64, 3)\n0.8750667\n[Epoch 2/5] [Batch 194/360] [D loss: 0.025302] [G loss: 6.725982] time: 0:04:57.588488\n(30, 64, 64, 3)\n0.84113437\n[Epoch 2/5] [Batch 195/360] [D loss: 0.029409] [G loss: 7.054451] time: 0:04:57.771508\n(30, 64, 64, 3)\n0.84558994\n[Epoch 2/5] [Batch 196/360] [D loss: 0.021422] [G loss: 7.441612] time: 0:04:57.962794\n(30, 64, 64, 3)\n0.8311201\n[Epoch 2/5] [Batch 197/360] [D loss: 0.017656] [G loss: 7.414119] time: 0:04:58.118803\n(30, 64, 64, 3)\n0.8916761\n[Epoch 2/5] [Batch 198/360] [D loss: 0.016202] [G loss: 7.260391] time: 0:04:58.297775\n(30, 64, 64, 3)\n0.8885026\n[Epoch 2/5] [Batch 199/360] [D loss: 0.023141] [G loss: 6.768197] time: 0:04:58.458479\n(30, 64, 64, 3)\n0.8380385\n[Epoch 2/5] [Batch 200/360] [D loss: 0.035858] [G loss: 7.439940] time: 0:04:58.649802\n(30, 64, 64, 3)\n0.8250577\n[Epoch 2/5] [Batch 201/360] [D loss: 0.028790] [G loss: 6.935070] time: 0:04:58.856381\n(30, 64, 64, 3)\n0.9245265\n[Epoch 2/5] [Batch 202/360] [D loss: 0.019436] [G loss: 7.271949] time: 0:04:59.095734\n(30, 64, 64, 3)\n0.87991714\n[Epoch 2/5] [Batch 203/360] [D loss: 0.054723] [G loss: 7.296504] time: 0:04:59.276869\n(30, 64, 64, 3)\n0.8568986\n[Epoch 2/5] [Batch 204/360] [D loss: 0.137438] [G loss: 7.271648] time: 0:04:59.486213\n(30, 64, 64, 3)\n0.9053251\n[Epoch 2/5] [Batch 205/360] [D loss: 0.074515] [G loss: 7.497812] time: 0:04:59.690105\n(30, 64, 64, 3)\n0.8746233\n[Epoch 2/5] [Batch 206/360] [D loss: 0.015603] [G loss: 7.170533] time: 0:04:59.844482\n(30, 64, 64, 3)\n0.8896411\n[Epoch 2/5] [Batch 207/360] [D loss: 0.016689] [G loss: 7.350577] time: 0:05:00.038404\n(30, 64, 64, 3)\n0.8386452\n[Epoch 2/5] [Batch 208/360] [D loss: 0.020508] [G loss: 7.374006] time: 0:05:00.245741\n(30, 64, 64, 3)\n0.8432633\n[Epoch 2/5] [Batch 209/360] [D loss: 0.021996] [G loss: 7.407105] time: 0:05:00.454768\n(30, 64, 64, 3)\n0.8540407\n[Epoch 2/5] [Batch 210/360] [D loss: 0.015416] [G loss: 6.555439] time: 0:05:00.635349\n(30, 64, 64, 3)\n0.87246084\n[Epoch 2/5] [Batch 211/360] [D loss: 0.023274] [G loss: 7.583427] time: 0:05:00.797851\n(30, 64, 64, 3)\n0.89635324\n[Epoch 2/5] [Batch 212/360] [D loss: 0.027763] [G loss: 7.105660] time: 0:05:01.033084\n(30, 64, 64, 3)\n0.8610514\n[Epoch 2/5] [Batch 213/360] [D loss: 0.097539] [G loss: 7.476156] time: 0:05:01.240919\n(30, 64, 64, 3)\n0.8706843\n[Epoch 2/5] [Batch 214/360] [D loss: 0.442292] [G loss: 6.532024] time: 0:05:01.460802\n(30, 64, 64, 3)\n0.8467373\n[Epoch 2/5] [Batch 215/360] [D loss: 0.069036] [G loss: 7.346165] time: 0:05:01.657232\n(30, 64, 64, 3)\n0.8687787\n[Epoch 2/5] [Batch 216/360] [D loss: 0.473522] [G loss: 5.697380] time: 0:05:01.855209\n(30, 64, 64, 3)\n0.91437435\n[Epoch 2/5] [Batch 217/360] [D loss: 0.159520] [G loss: 6.115507] time: 0:05:02.035170\n(30, 64, 64, 3)\n0.89443177\n[Epoch 2/5] [Batch 218/360] [D loss: 0.102388] [G loss: 6.485472] time: 0:05:02.225939\n(30, 64, 64, 3)\n0.8467925\n[Epoch 2/5] [Batch 219/360] [D loss: 0.122477] [G loss: 6.575648] time: 0:05:02.433448\n(30, 64, 64, 3)\n0.87768775\n[Epoch 2/5] [Batch 220/360] [D loss: 0.105584] [G loss: 6.661679] time: 0:05:02.629400\n(30, 64, 64, 3)\n0.9223562\n[Epoch 2/5] [Batch 221/360] [D loss: 0.092934] [G loss: 6.599540] time: 0:05:02.846932\n(30, 64, 64, 3)\n0.9131322\n[Epoch 2/5] [Batch 222/360] [D loss: 0.080542] [G loss: 6.760729] time: 0:05:03.074707\n(30, 64, 64, 3)\n0.83985823\n[Epoch 2/5] [Batch 223/360] [D loss: 0.092663] [G loss: 6.458893] time: 0:05:03.295382\n(30, 64, 64, 3)\n0.8799613\n[Epoch 2/5] [Batch 224/360] [D loss: 0.095030] [G loss: 6.498758] time: 0:05:03.499892\n(30, 64, 64, 3)\n0.8728852\n[Epoch 2/5] [Batch 225/360] [D loss: 0.075467] [G loss: 7.061043] time: 0:05:03.736012\n(30, 64, 64, 3)\n0.8599172\n[Epoch 2/5] [Batch 226/360] [D loss: 0.072323] [G loss: 6.576604] time: 0:05:03.913326\n(30, 64, 64, 3)\n0.90056294\n[Epoch 2/5] [Batch 227/360] [D loss: 0.057184] [G loss: 6.844611] time: 0:05:04.097414\n(30, 64, 64, 3)\n0.8930149\n[Epoch 2/5] [Batch 228/360] [D loss: 0.153181] [G loss: 6.945486] time: 0:05:04.318682\n(30, 64, 64, 3)\n0.87596816\n[Epoch 2/5] [Batch 229/360] [D loss: 0.112620] [G loss: 6.694433] time: 0:05:04.521697\n(30, 64, 64, 3)\n0.88564175\n[Epoch 2/5] [Batch 230/360] [D loss: 0.039425] [G loss: 6.838265] time: 0:05:04.676776\n(30, 64, 64, 3)\n0.80888253\n[Epoch 2/5] [Batch 231/360] [D loss: 0.049020] [G loss: 6.875766] time: 0:05:04.902261\n(30, 64, 64, 3)\n0.8808675\n[Epoch 2/5] [Batch 232/360] [D loss: 0.079610] [G loss: 6.483555] time: 0:05:05.063063\n(30, 64, 64, 3)\n0.88647693\n[Epoch 2/5] [Batch 233/360] [D loss: 0.032529] [G loss: 6.144383] time: 0:05:05.225849\n(30, 64, 64, 3)\n0.90699434\n[Epoch 2/5] [Batch 234/360] [D loss: 0.054147] [G loss: 6.446342] time: 0:05:05.426404\n(30, 64, 64, 3)\n0.8729966\n[Epoch 2/5] [Batch 235/360] [D loss: 0.284693] [G loss: 6.627099] time: 0:05:05.646121\n(30, 64, 64, 3)\n0.8665512\n[Epoch 2/5] [Batch 236/360] [D loss: 0.070905] [G loss: 7.087224] time: 0:05:05.835770\n(30, 64, 64, 3)\n0.9017076\n[Epoch 2/5] [Batch 237/360] [D loss: 0.047733] [G loss: 6.864686] time: 0:05:06.029177\n(30, 64, 64, 3)\n0.89956886\n[Epoch 2/5] [Batch 238/360] [D loss: 0.044155] [G loss: 7.233632] time: 0:05:06.235062\n(30, 64, 64, 3)\n0.8865535\n[Epoch 2/5] [Batch 239/360] [D loss: 0.069988] [G loss: 7.290987] time: 0:05:06.438135\n(30, 64, 64, 3)\n0.89099246\n[Epoch 2/5] [Batch 240/360] [D loss: 0.089538] [G loss: 6.739630] time: 0:05:06.697350\n(30, 64, 64, 3)\n0.83790874\n[Epoch 2/5] [Batch 241/360] [D loss: 0.088194] [G loss: 7.110896] time: 0:05:06.899980\n(30, 64, 64, 3)\n0.8554514\n[Epoch 2/5] [Batch 242/360] [D loss: 0.178687] [G loss: 7.095422] time: 0:05:07.093914\n(30, 64, 64, 3)\n0.8794908\n[Epoch 2/5] [Batch 243/360] [D loss: 0.089550] [G loss: 6.952402] time: 0:05:07.306095\n(30, 64, 64, 3)\n0.8880527\n[Epoch 2/5] [Batch 244/360] [D loss: 0.027895] [G loss: 6.582946] time: 0:05:07.502845\n(30, 64, 64, 3)\n0.8795716\n[Epoch 2/5] [Batch 245/360] [D loss: 0.033482] [G loss: 6.857618] time: 0:05:07.689577\n(30, 64, 64, 3)\n0.8803196\n[Epoch 2/5] [Batch 246/360] [D loss: 0.056184] [G loss: 7.015878] time: 0:05:07.882695\n(30, 64, 64, 3)\n0.8560193\n[Epoch 2/5] [Batch 247/360] [D loss: 0.301317] [G loss: 7.114709] time: 0:05:08.088207\n(30, 64, 64, 3)\n0.8453768\n[Epoch 2/5] [Batch 248/360] [D loss: 0.052877] [G loss: 7.006164] time: 0:05:08.325756\n(30, 64, 64, 3)\n0.82666683\n[Epoch 2/5] [Batch 249/360] [D loss: 0.042320] [G loss: 6.700946] time: 0:05:08.557432\n(30, 64, 64, 3)\n0.9032743\n[Epoch 2/5] [Batch 250/360] [D loss: 0.127687] [G loss: 7.327629] time: 0:05:08.744446\n(30, 64, 64, 3)\n0.88498306\n[Epoch 2/5] [Batch 251/360] [D loss: 0.292539] [G loss: 6.961227] time: 0:05:08.916859\n(30, 64, 64, 3)\n0.83508563\n[Epoch 2/5] [Batch 252/360] [D loss: 0.027173] [G loss: 7.031213] time: 0:05:09.102106\n(30, 64, 64, 3)\n0.91075546\n[Epoch 2/5] [Batch 253/360] [D loss: 0.085582] [G loss: 7.114581] time: 0:05:09.272426\n(30, 64, 64, 3)\n0.8849716\n[Epoch 2/5] [Batch 254/360] [D loss: 0.094885] [G loss: 6.581625] time: 0:05:09.450726\n(30, 64, 64, 3)\n0.86092085\n[Epoch 2/5] [Batch 255/360] [D loss: 0.028070] [G loss: 6.676363] time: 0:05:09.803270\n(30, 64, 64, 3)\n0.91992015\n[Epoch 2/5] [Batch 256/360] [D loss: 0.051342] [G loss: 6.175388] time: 0:05:10.019730\n(30, 64, 64, 3)\n0.8891471\n[Epoch 2/5] [Batch 257/360] [D loss: 0.122257] [G loss: 6.667206] time: 0:05:10.230819\n(30, 64, 64, 3)\n0.89023966\n[Epoch 2/5] [Batch 258/360] [D loss: 0.310854] [G loss: 6.099066] time: 0:05:10.408805\n(30, 64, 64, 3)\n0.870771\n[Epoch 2/5] [Batch 259/360] [D loss: 0.093431] [G loss: 6.629173] time: 0:05:10.604447\n(30, 64, 64, 3)\n0.8631625\n[Epoch 2/5] [Batch 260/360] [D loss: 0.127122] [G loss: 6.319006] time: 0:05:10.795808\n(30, 64, 64, 3)\n0.90120405\n[Epoch 2/5] [Batch 261/360] [D loss: 0.104734] [G loss: 6.262616] time: 0:05:10.966543\n(30, 64, 64, 3)\n0.9086149\n[Epoch 2/5] [Batch 262/360] [D loss: 0.086483] [G loss: 6.813387] time: 0:05:11.114150\n(30, 64, 64, 3)\n0.89258283\n[Epoch 2/5] [Batch 263/360] [D loss: 0.098247] [G loss: 6.579489] time: 0:05:11.306908\n(30, 64, 64, 3)\n0.8629046\n[Epoch 2/5] [Batch 264/360] [D loss: 0.086302] [G loss: 6.526937] time: 0:05:11.481167\n(30, 64, 64, 3)\n0.83301306\n[Epoch 2/5] [Batch 265/360] [D loss: 0.097698] [G loss: 6.488541] time: 0:05:11.700408\n(30, 64, 64, 3)\n0.8788207\n[Epoch 2/5] [Batch 266/360] [D loss: 0.096064] [G loss: 6.488579] time: 0:05:11.867251\n(30, 64, 64, 3)\n0.8856616\n[Epoch 2/5] [Batch 267/360] [D loss: 0.067195] [G loss: 6.353868] time: 0:05:12.055351\n(30, 64, 64, 3)\n0.8904526\n[Epoch 2/5] [Batch 268/360] [D loss: 0.024059] [G loss: 6.060432] time: 0:05:12.285249\n(30, 64, 64, 3)\n0.8914214\n[Epoch 2/5] [Batch 269/360] [D loss: 0.035911] [G loss: 6.942231] time: 0:05:12.456016\n(30, 64, 64, 3)\n0.841106\n[Epoch 2/5] [Batch 270/360] [D loss: 0.039083] [G loss: 6.662906] time: 0:05:12.649374\n(30, 64, 64, 3)\n0.92304534\n[Epoch 2/5] [Batch 271/360] [D loss: 0.027574] [G loss: 6.137290] time: 0:05:12.859432\n(30, 64, 64, 3)\n0.8762476\n[Epoch 2/5] [Batch 272/360] [D loss: 0.031818] [G loss: 7.190288] time: 0:05:13.097565\n(30, 64, 64, 3)\n0.886982\n[Epoch 2/5] [Batch 273/360] [D loss: 0.046799] [G loss: 6.866181] time: 0:05:13.343770\n(30, 64, 64, 3)\n0.9193086\n[Epoch 2/5] [Batch 274/360] [D loss: 0.022471] [G loss: 6.665140] time: 0:05:13.603528\n(30, 64, 64, 3)\n0.9103861\n[Epoch 2/5] [Batch 275/360] [D loss: 0.026589] [G loss: 6.101126] time: 0:05:13.842002\n(30, 64, 64, 3)\n0.8518569\n[Epoch 2/5] [Batch 276/360] [D loss: 0.030230] [G loss: 6.028717] time: 0:05:14.021495\n(30, 64, 64, 3)\n0.8740187\n[Epoch 2/5] [Batch 277/360] [D loss: 0.042512] [G loss: 5.573484] time: 0:05:14.231692\n(30, 64, 64, 3)\n0.89371556\n[Epoch 2/5] [Batch 278/360] [D loss: 0.034667] [G loss: 6.002675] time: 0:05:14.418045\n(30, 64, 64, 3)\n0.8791509\n[Epoch 2/5] [Batch 279/360] [D loss: 0.069288] [G loss: 6.398297] time: 0:05:14.601104\n(30, 64, 64, 3)\n0.9435794\n[Epoch 2/5] [Batch 280/360] [D loss: 0.501878] [G loss: 6.120923] time: 0:05:14.818897\n"
]
],
[
[
"9) We can test the model with 100 test data which will be saved as images",
"_____no_output_____"
]
],
[
[
"for batch_i in range(100):\n test_first_imgs, test_last_imgs = next(test_batch_generator)\n test_fake_last_imgs = modelObj.generator.predict(test_first_imgs) \n\n test_img_name = output_log_dir + \"/gen_img_test_\" + str(batch_i) + \".png\"\n merged_img = np.vstack((test_first_imgs[0],test_last_imgs[0],test_fake_last_imgs[0]))\n imageio.imwrite(test_img_name, img_as_ubyte(merged_img))",
"_____no_output_____"
]
],
[
[
"## EXERCISES",
"_____no_output_____"
],
[
"#### Exercise 1)\nUpdate the network architecture given in **build_generator** and **build_discriminator** of the class GANModel. Please note that the current image resolution is set to 32x32 (i.e. IMAGE_WIDTH and IMAGE_HEIGHT values) in the file configGAN.py. \nThis way initial experiements can run faster. Once you implement the inital version of the network, please set the resolution values back to 128x128. Experimental results should be provided for this high resolution images. \n\n**Hint:** As a generator model, you can use the segmentation model implemented in lab03. Do not forget to adapt the input and output shapes of the generator model in this case.",
"_____no_output_____"
],
[
"#### Exercise 2) \nUse different **optimization** (e.g. ADAM, SGD, etc) and **regularization** (e.g. data augmentation, dropout) methods to increase the network accuracy. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ec877dfdbbbb5bf69d9a58b9b7fd20e44f89d21b | 14,959 | ipynb | Jupyter Notebook | Tensorflow chat-bot model.ipynb | bsivavenu/Google_Colab_Notebooks | 30e8f25c5ac1410b7e34c32879c391780ebc7048 | [
"Apache-2.0"
] | null | null | null | Tensorflow chat-bot model.ipynb | bsivavenu/Google_Colab_Notebooks | 30e8f25c5ac1410b7e34c32879c391780ebc7048 | [
"Apache-2.0"
] | null | null | null | Tensorflow chat-bot model.ipynb | bsivavenu/Google_Colab_Notebooks | 30e8f25c5ac1410b7e34c32879c391780ebc7048 | [
"Apache-2.0"
] | null | null | null | 14,959 | 14,959 | 0.665686 | [
[
[
"# things we need for NLP\nimport nltk\nfrom nltk.stem.lancaster import LancasterStemmer\nstemmer = LancasterStemmer()\n\n# things we need for Tensorflow\nimport numpy as np\nimport tflearn\nimport tensorflow as tf\nimport random",
"_____no_output_____"
],
[
"!pip install tflearn",
"Collecting tflearn\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/16/ec/e9ce1b52e71f6dff3bd944f020cef7140779e783ab27512ea7c7275ddee5/tflearn-0.3.2.tar.gz (98kB)\n\r\u001b[K |███▎ | 10kB 18.5MB/s eta 0:00:01\r\u001b[K |██████▋ | 20kB 1.6MB/s eta 0:00:01\r\u001b[K |██████████ | 30kB 2.2MB/s eta 0:00:01\r\u001b[K |█████████████▎ | 40kB 2.4MB/s eta 0:00:01\r\u001b[K |████████████████▋ | 51kB 1.9MB/s eta 0:00:01\r\u001b[K |████████████████████ | 61kB 2.2MB/s eta 0:00:01\r\u001b[K |███████████████████████▎ | 71kB 2.4MB/s eta 0:00:01\r\u001b[K |██████████████████████████▋ | 81kB 2.6MB/s eta 0:00:01\r\u001b[K |██████████████████████████████ | 92kB 2.8MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 102kB 2.2MB/s \n\u001b[?25hRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from tflearn) (1.18.5)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from tflearn) (1.15.0)\nRequirement already satisfied: Pillow in /usr/local/lib/python3.6/dist-packages (from tflearn) (7.0.0)\nBuilding wheels for collected packages: tflearn\n Building wheel for tflearn (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for tflearn: filename=tflearn-0.3.2-cp36-none-any.whl size=128208 sha256=1b3bcce937d4d20bdf6ed288a655ce7ae97b5895b45f3742530c73a6f682767e\n Stored in directory: /root/.cache/pip/wheels/d0/f6/69/0ef3ee395aac2e5d15d89efd29a9a216f3c27767b43b72c006\nSuccessfully built tflearn\nInstalling collected packages: tflearn\nSuccessfully installed tflearn-0.3.2\n"
],
[
"# import our chat-bot intents file\nimport json\nwith open('intents.json') as json_data:\n intents = json.load(json_data)",
"_____no_output_____"
],
[
"words = []\nclasses = []\ndocuments = []\nignore_words = ['?']\n# loop through each sentence in our intents patterns\nfor intent in intents['intents']:\n for pattern in intent['patterns']:\n # tokenize each word in the sentence\n w = nltk.word_tokenize(pattern)\n # add to our words list\n words.extend(w)\n # add to documents in our corpus\n documents.append((w, intent['tag']))\n # add to our classes list\n if intent['tag'] not in classes:\n classes.append(intent['tag'])\n\n# stem and lower each word and remove duplicates\nwords = [stemmer.stem(w.lower()) for w in words if w not in ignore_words]\nwords = sorted(list(set(words)))\n\n# remove duplicates\nclasses = sorted(list(set(classes)))\n\nprint (len(documents), \"documents\")\nprint (len(classes), \"classes\", classes)\nprint (len(words), \"unique stemmed words\", words)",
"27 documents\n9 classes ['goodbye', 'greeting', 'hours', 'mopeds', 'opentoday', 'payments', 'rental', 'thanks', 'today']\n48 unique stemmed words [\"'d\", \"'s\", 'a', 'acceiv', 'anyon', 'ar', 'bye', 'can', 'card', 'cash', 'credit', 'day', 'do', 'doe', 'good', 'goodby', 'hav', 'hello', 'help', 'hi', 'hour', 'how', 'i', 'is', 'kind', 'lat', 'lik', 'mastercard', 'mop', 'of', 'on', 'op', 'rent', 'see', 'tak', 'thank', 'that', 'ther', 'thi', 'to', 'today', 'we', 'what', 'when', 'which', 'work', 'yo', 'you']\n"
],
[
"# create our training data\ntraining = []\noutput = []\n# create an empty array for our output\noutput_empty = [0] * len(classes)\n\n# training set, bag of words for each sentence\nfor doc in documents:\n # initialize our bag of words\n bag = []\n # list of tokenized words for the pattern\n pattern_words = doc[0]\n # stem each word\n pattern_words = [stemmer.stem(word.lower()) for word in pattern_words]\n # create our bag of words array\n for w in words:\n bag.append(1) if w in pattern_words else bag.append(0)\n\n # output is a '0' for each tag and '1' for current tag\n output_row = list(output_empty)\n output_row[classes.index(doc[1])] = 1\n\n training.append([bag, output_row])\n\n# shuffle our features and turn into np.array\nrandom.shuffle(training)\ntraining = np.array(training)\n\n# create train and test lists\ntrain_x = list(training[:,0])\ntrain_y = list(training[:,1])",
"_____no_output_____"
],
[
"# reset underlying graph data\ntf.reset_default_graph()\n# Build neural network\nnet = tflearn.input_data(shape=[None, len(train_x[0])])\nnet = tflearn.fully_connected(net, 8)\nnet = tflearn.fully_connected(net, 8)\nnet = tflearn.fully_connected(net, len(train_y[0]), activation='softmax')\nnet = tflearn.regression(net)\n\n# Define model and setup tensorboard\nmodel = tflearn.DNN(net, tensorboard_dir='tflearn_logs')\n# Start training (apply gradient descent algorithm)\nmodel.fit(train_x, train_y, n_epoch=1000, batch_size=8, show_metric=True)\nmodel.save('model.tflearn')",
"Training Step: 3999 | total loss: \u001b[1m\u001b[32m0.54986\u001b[0m\u001b[0m | time: 0.008s\n| Adam | epoch: 1000 | loss: 0.54986 - acc: 0.9616 -- iter: 24/27\nTraining Step: 4000 | total loss: \u001b[1m\u001b[32m0.50465\u001b[0m\u001b[0m | time: 0.011s\n| Adam | epoch: 1000 | loss: 0.50465 - acc: 0.9654 -- iter: 27/27\n--\nINFO:tensorflow:/home/gk/gensim/notebooks/model.tflearn is not in all_model_checkpoint_paths. Manually adding it.\n"
],
[
"def clean_up_sentence(sentence):\n # tokenize the pattern\n sentence_words = nltk.word_tokenize(sentence)\n # stem each word\n sentence_words = [stemmer.stem(word.lower()) for word in sentence_words]\n return sentence_words\n\n# return bag of words array: 0 or 1 for each word in the bag that exists in the sentence\ndef bow(sentence, words, show_details=False):\n # tokenize the pattern\n sentence_words = clean_up_sentence(sentence)\n # bag of words\n bag = [0]*len(words) \n for s in sentence_words:\n for i,w in enumerate(words):\n if w == s: \n bag[i] = 1\n if show_details:\n print (\"found in bag: %s\" % w)\n\n return(np.array(bag))",
"_____no_output_____"
],
[
"p = bow(\"is your shop open today?\", words)\nprint (p)\nprint (classes)",
"[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0\n 0 0 0 1 0 0 0 0 0 1 0]\n['goodbye', 'greeting', 'hours', 'mopeds', 'opentoday', 'payments', 'rental', 'thanks', 'today']\n"
],
[
"print(model.predict([p]))",
"[[5.111963474746517e-08, 0.00029038049979135394, 0.19395901262760162, 4.018096966262874e-10, 0.7987914085388184, 0.0005724101793020964, 8.153344310812827e-08, 5.96670907127006e-11, 0.0063865589909255505]]\n"
],
[
"# save all of our data structures\nimport pickle\npickle.dump( {'words':words, 'classes':classes, 'train_x':train_x, 'train_y':train_y}, open( \"training_data\", \"wb\" ) )",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec87920e215fb3843d205e4fa81f9a075945ae7d | 4,066 | ipynb | Jupyter Notebook | day17/day17a.ipynb | kannix68/advent_of_code_2019 | b02a86e1f8e83111973cc2bc8c7f4d5dcf1c10aa | [
"MIT"
] | null | null | null | day17/day17a.ipynb | kannix68/advent_of_code_2019 | b02a86e1f8e83111973cc2bc8c7f4d5dcf1c10aa | [
"MIT"
] | null | null | null | day17/day17a.ipynb | kannix68/advent_of_code_2019 | b02a86e1f8e83111973cc2bc8c7f4d5dcf1c10aa | [
"MIT"
] | null | null | null | 24.792683 | 89 | 0.506394 | [
[
[
"##\n# Advent of code 2019, AoC day 17 puzzle 1\n# This solution (python3.7 jupyter notebook) by kannix68, @ 2020-01-05.\n\nimport sys\nsys.path.insert(0, '..') # allow import from parent dir\nimport lib.aochelper as aoc",
"_____no_output_____"
],
[
"import logging\nlogging.basicConfig(stream=sys.stdout, level=logging.INFO)\nlog = logging.getLogger(__name__)",
"_____no_output_____"
],
[
"## PROBLEM DOMAIN code\nfrom lib.intcode_interpreter import *",
"_____no_output_____"
],
[
"def get_mazelst_intersect_prodsum(maze_lst):\n result = 0\n for y in range(len(maze_lst)):\n for x in range(len(maze_lst[0])):\n if maze_lst[y][x] == '#': # crossings only if own cell is on a rail\n nbrs = aoc.get_neighbors(maze_lst, x, y)\n if ''.join(nbrs) == '####':\n log.info(f\"intersection found at ({x,y})\")\n result += x*y\n return result",
"_____no_output_____"
],
[
"def solve(program: list, base_combi: list) -> int:\n return 0",
"_____no_output_____"
],
[
"## MAIN",
"_____no_output_____"
],
[
"### tests\nlog.setLevel(logging.DEBUG)",
"_____no_output_____"
],
[
"# example \"1\"\nmaze_str = \"\"\"\n..#..........\n..#..........\n#######...###\n#.#...#...#.#\n#############\n..#...#...#..\n..#####...^..\n\"\"\".strip()\nmaze_lst = maze_str.split(\"\\n\")\nlog.info(f\"maze_lst=\\n{aoc.represent_strlist(maze_lst)}\")\nresult = get_mazelst_intersect_prodsum(maze_lst)\nprint(f\"result={result}\") ",
"_____no_output_____"
],
[
"### personal input solution, puzzle 1\nlog.setLevel(logging.INFO)\ndata = aoc.read_file_firstline_to_str('day17.in')\ndata = list(map(int, data.split(',')))\nlog.debug(f\"data-type={type(data)} data=\\n{aoc.cl(data)}\")\n\ncomputer = IntcodeInterpreter(data, mem_growable=True)\ncomputer.interpret_program()\noutputs = computer.pseudo_stdout\nlog.debug(f\"outputs={outputs}\")\nmaze_str = \"\"\nfor i in outputs:\n if i < 32:\n if i == 10:\n maze_str += \"\\n\"\n else:\n raise(RuntimeError(f\"input ASCII non-printable char #{i} not handled\")) \n else:\n maze_str += chr(i)\n\nmaze_lst = maze_str.strip().split(\"\\n\")\n#log.info(f\"mazestr=\\n{maze_str}\")\nlog.info(f\"maze_lst=\\n{aoc.represent_strlist(maze_lst)}\")\nresult = get_mazelst_intersect_prodsum(maze_lst)\nprint(f\"result={result}\") ",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8792204a3264320ffca67876841e8e670d41bf | 11,622 | ipynb | Jupyter Notebook | hackathons/hackathon2/problem2.ipynb | wd15/chimad-phase-field | b8ead2ef666201b500033052d0a4efb55796c2da | [
"MIT"
] | 11 | 2016-04-19T13:51:02.000Z | 2021-05-30T07:52:21.000Z | hackathons/hackathon2/problem2.ipynb | usnistgov/chimad-phase-field | 7f07e5ab046b917dfa32d84a68421ed94ec03a3b | [
"MIT"
] | 417 | 2015-03-20T16:39:11.000Z | 2018-01-16T16:33:53.000Z | hackathons/hackathon2/problem2.ipynb | wd15/chimad-phase-field | b8ead2ef666201b500033052d0a4efb55796c2da | [
"MIT"
] | 15 | 2015-03-20T21:44:06.000Z | 2017-12-05T23:39:36.000Z | 46.119048 | 751 | 0.5746 | [
[
[
"from IPython.display import HTML\n\nHTML('''<script>\ncode_show=true; \nfunction code_toggle() {\n if (code_show){\n $('div.input').hide();\n $('div.prompt').hide();\n } else {\n $('div.input').show();\n$('div.prompt').show();\n }\n code_show = !code_show\n} \n$( document ).ready(code_toggle);\n</script>\n<form action=\"javascript:code_toggle()\"><input type=\"submit\" value=\"Code Toggle\"></form>''')",
"_____no_output_____"
],
[
"from IPython.display import HTML\n\nHTML('''\n<a href=\"{{ site.links.github }}/raw/nist-pages/hackathons/hackathon2/problem2.ipynb\"\n download>\n<button type=\"submit\">Download Notebook</button>\n</a>\n''')",
"_____no_output_____"
],
[
"from IPython.display import HTML\n\nHTML('''{% include jupyter_benchmark_table.html num=\"[4]\" revision=0 %}''')",
"_____no_output_____"
]
],
[
[
"# Benchmark Problem 4: Linear Elasticity in 3D\n\nThe linear elastic energy of a body is\n\n\\begin{equation}\nE_{\\rm elastic}=\\frac{1}{2}\\int \\sigma_{ij}\\epsilon_{ij}\\,dV=\\frac{1}{2}\\int C_{ijkl}\\epsilon_{ij}\\epsilon_{kl}\\,dV,\n\\end{equation}\n\nwhere $\\sigma_{ij}=C_{ijkl}\\epsilon_{kl}$ is the stress, $C_{ijkl}$ is the elastic tensor, and $\\epsilon_{ij}$ is the strain,\n\\begin{equation}\n\\epsilon_{ij}=\\frac{1}{2}\\left[\\frac{\\partial u_i}{\\partial x_j}+\\frac{\\partial u_j}{\\partial x_i}\\right],\n\\end{equation}\nwith $u_i$ the displacement field. The indices $i,j,k,l$ run from 1 to 3, and $x_i,\\,i=1,2,3$, are Cartesian coordinates; we are using Einstein summation convention so repeated indices are summed over.\n\nThe elastic tensor obeys symmetries $C_{ijkl}=C_{jikl}=C_{ijlk}=C_{jilk}$ and $C_{ijkl}=C_{klij}$. These symmetries imply that there are only 21 independent entries in the elastic tensor. Usually Voigt notation is used, in which the four indices $ijkl$ are replaced by two indices $IJ$. The mapping for each pair $ij$ (or $kl$) is $11\\to1$, $22\\to2$, $33\\to3$, $23\\to4$ (and $32\\to4$), $13\\to5$ (and $31\\to5$), and $12\\to6$ (and $21\\to6$). The crystal symmetry may further reduce the number of independent entries. In an orthorombic crystal, there are only nine independent entries, and they are (in Voigt notation) $C_{11}, C_{22}, C_{33}, C_{44}, C_{55}, C_{66}, C_{12}, C_{13}$, and $C_{23}$. The tensor $C_{IJ}$ thus has the form\n\n\\begin{equation}\n\\left(\n\\begin{matrix}\nC_{11} & C_{12} & C_{13} & 0 & 0 & 0\\\\\nC_{12} & C_{22} & C_{23} & 0 & 0 & 0 \\\\\nC_{13} & C_{23} & C_{33} & 0 & 0 & 0 \\\\\n0 & 0 & 0 & C_{44} & 0 & 0\\\\\n0 & 0 & 0 & 0 & C_{55} & 0\\\\\n0 & 0 & 0 & 0 & 0 & C_{66}\n\\end{matrix}\n\\right).\n\\end{equation}\n\nFor tetragonal symmetry, there are six independent entries, $C_{11}, C_{33}, C_{44}, C_{66}, C_{12}$, and $C_{13}$. \n\nAluminum silicate, Al$_2$SiO$_5$ is a crystal with orthorombic symmetry and unit cell parameters $a=7.738\\;\\unicode{x212B}$ , $b=7.857\\;\\unicode{x212B}$, and $c=5.534\\;\\unicode{x212B}$. The elastic tensor is given by $C_{11}=233.4$, $C_{22}=289.0$, $C_{33}=380.1$, $C_{44}=99.5$, $C_{55}=87.8$, $C_{66}=112.3$; $C_{11}+C_{22}-2C_{12}=233.4$, $C_{11}+C_{33}-2C_{13}=380.9$, and $C_{22}+C_{33}-2C_{23}=506.3$, all in units of GPa.\n\n(a) (Potato in space) What is the equilibrium shape of a 0.0042~$\\mu$m$^3$ volume of Al$_2$SiO$_5$ in free space (stress-free boundaries)? Take the surface energy, $\\gamma$, to be equal to 200 mJ/m$^2$. The crystalline axes $a$, $b$, and $c$ are aligned with the $x$, $y$, and $z$-axes of a Cartesian lab coordinate system.\n\nThis problem can be cast as a phase field problem, where the phase field $\\varphi\\in[0,1]$ takes the value of 1 in one phase (the \"potato\"), and a value of 0 in the other (the surrounding vacuum). Thus, the total free energy can be written\n\\begin{equation}\n{\\mathcal F}=\\int \\left[f_{\\rm elastic}+\\frac{\\kappa}{2}|\\nabla\\varphi|^2+h_0f(\\varphi)\\right]\\,dV,\n\\end{equation}\nwith the integral extended over all space,\nwhere\n$f(\\varphi)$ is a (dimensionless) double-well function\n\\begin{equation}\nf(\\varphi)=\\varphi^2\\left[\\varphi-1\\right]^2,\n\\end{equation}\nand $h_0$ has the dimension of energy per unit volume. The interface width $W$ between approximately $\\varphi=0.1$ and $\\varphi=0.9$ in this model is given by $W=2\\sqrt{2\\kappa/h_0}$, while $\\gamma=\\sqrt{\\kappa h_0/18}$, $\\kappa=1.5\\gamma W$, and $h_0=12\\gamma/W$ (ignoring modification of the phase field order parameter $\\varphi$ by the elastic interactions through the interface). Use $\\kappa=3\\times10^{-9}$~J/m, and\n$h_0=2.4\\times10^{8}$~J/m$^3$. \n\nWe use a simple interpolation of the elastic constants, \n\n\\begin{equation}\nC_{ijkl}=h(\\varphi)C_{ijkl}^{\\rm potato},%+\\left[1-h(\\varphi)\\right]C_{ijkl}^{\\rm matrix},\n\\end{equation}\n\nwhere $h(\\varphi)$ is a smooth interpolation function,\n\\begin{equation}\nh(\\varphi)=\\varphi^3\\left[6\\varphi^2-15\\varphi+10\\right],\n\\end{equation}\nthat interpolates between $h(\\varphi=0)=0$ and $h(\\varphi=1)=1$.\n\nHint: find time-evolution equations for $\\varphi$ that monotonically drive the total energy to a minimum while preserving the volume. One way to do this is to set up a Cahn-Hilliard equation for $\\varphi$.\n\n(b) (Compressed potato in space) What is the equilibrium shape of a 0.0042 $\\mu$m$^3$ volume Al$_2$SiO$_5$ with uniaxial compressive stress of 500~MPa applied in the $z$-direction? Note that the strain must go to zero far from the \"potato.\"\n\n(c) (Potato in a stew) A volume of Al$_2$SiO$_5$ is embedded coherently in a matrix of tetragonal symmetry with unit cell parameters $a_m=b_m=7.6918\\;\\unicode{x212B}$ and $c_m=5.5674\\;\\unicode{x212B}$ such that $a$ and $a_m$, $b$ and $b_m$ and $c$ and $c_m$ are pairwise aligned. The elastic tensor of the matrix is given by $C_{11}=C_{22}=269.0$, $C_{12}=177.0$, $C_{13}=146.0$, $C_{33}=480.0$, $C_{44}=124.0$, and $C_{66}=192.0$, all in units of GPa.\n\nThis problem can also be cast as a phase field problem, where now the phase field $\\varphi\\in[0,1]$ takes the value of 1 in one phase (the inclusion), and a value of 0 in the other (the matrix). The total free energy is again\n\\begin{equation}\n{\\mathcal F}=\\int \\left[f_{\\rm elastic}+\\frac{\\kappa}{2}|\\nabla\\varphi|^2+h_0f(\\varphi)\\right]\\,dV,\n\\end{equation}\nwhere \n$f(\\varphi)$ is given by above.\n\nThe interpolation of the elastic constants is now, \n\n\\begin{equation}\nC_{ijkl}=h(\\varphi)C_{ijkl}^{\\rm inclusion}+\\left[1-h(\\varphi)\\right]C_{ijkl}^{\\rm matrix},\n\\end{equation}\n\nwhere again $h(\\varphi)$ is a smooth interpolation function given above.\n\nIn the elastic energy, the relevant strain is the total strain $\\epsilon_{ij}$ minus the local misfit strain $\\epsilon^0_{ij}$, so\n\n\\begin{equation}\nf_{\\rm elastic}=\\frac{1}{2}C_{ijkl}\\left(\\epsilon_{ij}-\\epsilon^0_{ij}\\right)\\left(\\epsilon_{kl}-\\epsilon^0_{kl}\\right).\n\\end{equation}\n\nWe will use Vegard's law to determine the local misfit strain, for which we just interpolate the crystallographic misfit strain tensor, $\\epsilon^T_{ij}$:\n\n\\begin{equation}\n\\epsilon^0_{ij}=h(\\varphi)\\epsilon^T_{ij},\n\\end{equation}\n\nwhere $h(\\varphi)$ is the interpolation function, and $\\epsilon^T_{ij}$ is\n\\begin{equation}\n\\epsilon^T_{ij}=\\left(\n\\begin{matrix}\n\\frac{a-a_m}{a_m} & 0 & 0\\\\\n0 & \\frac{b-b_m}{b_m} & 0\\\\\n0& 0 & \\frac{c-c_c}{c_m}\n\\end{matrix}\n\\right).\n\\end{equation}\n\nFind the equilibrium shape of an isolated inclusion with a volume of 0.0042 $\\mu$m$^3$ for $\\kappa=3\\times10^{-9}$ J/m, and $h_0=2.4\\times10^{8}$ J/m$^3$. Use as an initial condition a prolate ellipsoid with $a=c=0.155$ $\\mu$m and $b=0.042$ $\\mu$m. Because of elastic strain energy and interfacial energy within the system, you will need to increase the intial values of the phase field variable in the matrix and the precipitate (e.g., $\\varphi$ in the precipitate $\\approx$ 1.05; $\\varphi$ in the matrix $\\approx 0.05$). Note that the strain must go to zero far from the \"potato\".\n\nFor parts (a) - (c), present a rendering of the final shape as well as the lengths of the \"potato\" along the principal axes, and also track the total free energy as function of \"time\" and plot it.\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ec87a22923cfc792e5511f060204596c95f8fbcf | 18,546 | ipynb | Jupyter Notebook | exercises/segmentation/label_generation.ipynb | avalenzu/manipulation | 13cacd5731557c3f2d844e7d72d8c40af2762821 | [
"BSD-3-Clause"
] | null | null | null | exercises/segmentation/label_generation.ipynb | avalenzu/manipulation | 13cacd5731557c3f2d844e7d72d8c40af2762821 | [
"BSD-3-Clause"
] | null | null | null | exercises/segmentation/label_generation.ipynb | avalenzu/manipulation | 13cacd5731557c3f2d844e7d72d8c40af2762821 | [
"BSD-3-Clause"
] | null | null | null | 37.771894 | 633 | 0.612855 | [
[
[
"# Mask-RCNN Label Generation\n\n## Notebook Setup \nThe following cell will install Drake, checkout the manipulation repository, and set up the path (only if necessary).\n- On Google's Colaboratory, this **will take approximately two minutes** on the first time it runs (to provision the machine), but should only need to reinstall once every 12 hours. \n\nMore details are available [here](http://manipulation.mit.edu/drake.html).",
"_____no_output_____"
]
],
[
[
"import importlib\nimport os, sys\nfrom urllib.request import urlretrieve\n\nif 'google.colab' in sys.modules and importlib.util.find_spec('manipulation') is None:\n urlretrieve(f\"http://manipulation.csail.mit.edu/scripts/setup/setup_manipulation_colab.py\",\n \"setup_manipulation_colab.py\")\n from setup_manipulation_colab import setup_manipulation\n setup_manipulation(manipulation_sha='c1bdae733682f8a390f848bc6cb0dbbf9ea98602', drake_version='0.25.0', drake_build='releases')\n\nfrom manipulation import running_as_notebook\n\n# Setup rendering (with xvfb), if necessary:\nimport os\nif 'google.colab' in sys.modules and os.getenv(\"DISPLAY\") is None:\n from pyvirtualdisplay import Display\n display = Display(visible=0, size=(1400, 900))\n display.start()\n\n# setup ngrok server\nserver_args = []\nif 'google.colab' in sys.modules:\n server_args = ['--ngrok_http_tunnel']\n\nfrom meshcat.servers.zmqserver import start_zmq_server_as_subprocess\nproc, zmq_url, web_url = start_zmq_server_as_subprocess(server_args=server_args)\n\nimport numpy as np\n\nfrom pydrake.all import ( \n AddMultibodyPlantSceneGraph, ConnectMeshcatVisualizer, \n DiagramBuilder, RigidTransform, RotationMatrix, Parser, Simulator,\n FindResourceOrThrow,Quaternion, AddTriad\n)\n\nimport open3d as o3d\nimport meshcat\nimport meshcat.geometry as g\nimport meshcat.transformations as tf\n\nfrom manipulation.meshcat_utils import draw_open3d_point_cloud, draw_points\nfrom manipulation.mustard_depth_camera_example import MustardExampleSystem\nfrom manipulation.utils import FindResource\nfrom manipulation.open3d_utils import create_open3d_point_cloud\n\nimport matplotlib.pyplot as plt\nfrom matplotlib.patches import Rectangle\nfrom copy import deepcopy\nimport pydot\nfrom ipywidgets import Dropdown, Layout\nfrom IPython.display import display, HTML, SVG",
"_____no_output_____"
],
[
"class SimpleCameraSystem:\n def __init__(self):\n diagram = MustardExampleSystem()\n context = diagram.CreateDefaultContext()\n \n # setup\n self.meshcat_vis = meshcat.Visualizer(zmq_url=zmq_url)\n self.meshcat_vis[\"/Background\"].set_property(\"visible\", False)\n \n # getting data\n self.point_cloud = diagram.GetOutputPort(\"camera0_point_cloud\").Eval(context)\n self.depth_im_read = diagram.GetOutputPort('camera0_depth_image').Eval(context).data.squeeze()\n self.depth_im = deepcopy(self.depth_im_read)\n self.depth_im[self.depth_im == np.inf] = 10.0\n label_im = diagram.GetOutputPort('camera0_label_image').Eval(context).data.squeeze()\n self.rgb_im = diagram.GetOutputPort('camera0_rgb_image').Eval(context).data\n self.mask = label_im == 1\n\n # draw visualization\n pcd = create_open3d_point_cloud(self.point_cloud)\n draw_open3d_point_cloud(self.meshcat_vis[\"self.point_cloud\"], pcd)\n \n # camera specs\n cam0 = diagram.GetSubsystemByName('camera0')\n cam0_context = cam0.GetMyMutableContextFromRoot(context)\n cam0_pose = cam0.GetOutputPort('X_WB').Eval(cam0_context)\n self.cam_info = cam0.depth_camera_info()\n self.X_WC = RigidTransform(quaternion=Quaternion(cam0_pose[3:]), p=cam0_pose[0:3])\n\n # get points for mustard bottle\n depth_mustard = self.mask * self.depth_im\n u_range = np.arange(depth_mustard.shape[0])\n v_range = np.arange(depth_mustard.shape[1])\n depth_v, depth_u = np.meshgrid(v_range, u_range)\n depth_pnts = np.dstack([depth_u, depth_v, depth_mustard])\n depth_pnts = depth_pnts.reshape([depth_pnts.shape[0]*depth_pnts.shape[1], 3])\n pC =self.project_depth_to_pC(depth_pnts)\n p_C_mustard = pC[pC[:,2] > 0]\n self.p_W_mustard = self.X_WC.multiply(p_C_mustard.T).T\n \n def get_color_image(self):\n return deepcopy(self.rgb_im[:,:,0:3])\n \n def get_intrinsics(self):\n # read camera intrinsics\n cx = self.cam_info.center_x()\n cy = self.cam_info.center_y()\n fx = self.cam_info.focal_x()\n fy = self.cam_info.focal_y()\n return cx, cy, fx, fy\n def project_depth_to_pC(self, depth_pixel):\n \"\"\"\n project depth pixels to points in camera frame\n using pinhole camera model\n Input:\n depth_pixels: numpy array of (nx3) or (3,)\n Output:\n pC: 3D point in camera frame, numpy array of (nx3)\n \"\"\"\n # switch u,v due to python convention\n v = depth_pixel[:,0]\n u = depth_pixel[:,1]\n Z = depth_pixel[:,2]\n cx, cy, fx, fy = self.get_intrinsics()\n X = (u-cx) * Z/fx\n Y = (v-cy) * Z/fy\n pC = np.c_[X,Y,Z]\n return pC\n \ndef bbox(img):\n a = np.where(img != 0)\n bbox = ([np.min(a[0]), np.max(a[0])], [np.min(a[1]), np.max(a[1])])\n return bbox\n \nenv = SimpleCameraSystem()\nX_WC = env.X_WC\np_W_mustard = env.p_W_mustard\nK = env.cam_info.intrinsic_matrix()\nrgb_im = env.get_color_image()",
"_____no_output_____"
]
],
[
[
"# Generate Mask Labels\n\nIn the lecture, you have learned about Mask-RCNN. A major difficulty in training/fine-tuning Mask-RCNN is to obtain high-quality real training data, especially the mask labels for the objects of interest. Although you can get training labels from [Amazon Mechanical Turk](https://www.mturk.com/), it is a paid service and you will have to wait for some time until you get your data labeled. An alternative method is to design clever pipelines to generate labeled masks automatically without requiring manual labor. \n\nConsider a setup where an object of interest is placed on a planar surface, and an RGBD camera is mounted at a fixed location pointing to the object. From the RGBD camera, you should be able to generate the corresponding point clouds of the desired object and the surrounding environment (e.g. planar surface). You can easily remove the points associated with the planar surface (recall RANSAC exercise in the problem set 2). The remaining points then should all belong to the desired object. To generate mask labels, all you need to do is to project the points back to the camera image plane using the pinhole camera model!\n\nLet's quickly review the pinhole camera model!\n\nIn the last problem set, you have played with [pinhole camera model](https://docs.opencv.org/3.4/d9/d0c/group__calib3d.html). In particular, you used the pinhole camera model to map the depth pixels to 3D points. You may review the `SimpleCameraSystem` class from the last problem set.\n\nThe mathematical description of the pinhole camera model is written below (you may also use the intrinsics matrix by `env.cam_info.intrinsic_matrix()`).\n\nThe camera intrinsics are:\n\\begin{equation}\nX_c = (u-c_x)\\frac{Z_c}{f_x}\n\\end{equation}\n\n\\begin{equation}\nY_c = (v-c_y)\\frac{Z_c}{f_y}\n\\end{equation}\n\nNotations:\n- $f_x$: focal length in x direction\n- $f_y$: focal length in y direction\n- $c_x$: principal point in x direction (pixels)\n- $c_y$: principal point in y direction (pixels)\n- $(X_C, Y_C, Z_C)$: points in camera frame\n\nwhere $f_x$, $f_y$, $c_x$, $c_y$ specify the intrinsics of the camera.\n\nThe diagram of the pinhole camera model below is found from [OpenCV documentation](https://docs.opencv.org/3.4/d9/d0c/group__calib3d.html). Note that the $u$ and $v$ directions are reversed in Python due to the difference in the convention.",
"_____no_output_____"
],
[
"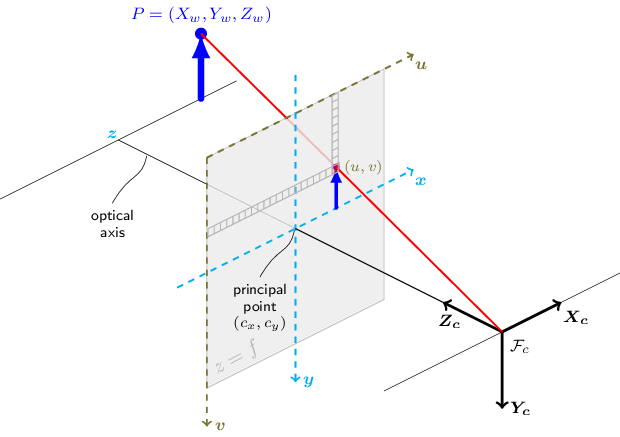",
"_____no_output_____"
],
[
"## Generate Mask from Point Clouds\n\nNow given the points of the mustard bottle in the world frame p_W_mustard, can you re-project these points back to the image plane to construct the mask of the mustard bottle? Note that you may need to reverse u,v indices to get the mask of the mustard bottle upright. Your mask should be of the same size as the original depth image, which is (480, 640)",
"_____no_output_____"
]
],
[
[
"def deproject_pW_to_image(p_W_mustard, cx, cy, fx, fy, X_WC):\n \"\"\"\n convert points in the world frame to camera pixels\n Input:\n - p_W_mustard: points of the mustard bottle in world frame (nx3)\n - fx, fy, cx, cy: camera intrinsics\n - X_WC: camera pose in the world frame \n Output:\n - mask: numpy array of size 480x640\n \"\"\"\n\n mask = np.zeros([480, 640])\n return mask",
"_____no_output_____"
],
[
"cx, cy, fx, fy = env.get_intrinsics()\nmask = deproject_pW_to_image(p_W_mustard, cx, cy, fx, fy, X_WC)\nplt.imshow(mask)",
"_____no_output_____"
]
],
[
[
"You should be able to visually verify that the generated mask perfectly align with the mustard bottle!",
"_____no_output_____"
]
],
[
[
"plt.imshow(rgb_im)",
"_____no_output_____"
],
[
"masked_rgb = rgb_im * mask[:,:,np.newaxis].astype(int) \nplt.imshow(masked_rgb)",
"_____no_output_____"
]
],
[
[
"# Analysis for Cluttered Scenes\n**Now you have a working pipeline for generating mask labels for single objects on a planar surface. Now consider besides a mustard bottle, you also have a Cheez-It box in the scene. Both objects appear in your RGBD image. Answer the following questions.**\n\n**6.1.b** Assume the Cheez-It box is not occluding the mustard bottle, without any modification to the pipeline above, can you get the separate masks of the mustard bottle and Cheez-It box? Explain your reasoning.\n\n**6.1.c** Assume the Cheez-It box is occluding the mustard bottle, without any modification to the pipeline above, can you get the separate masks of the mustard bottle and Cheez-It box? Explain your reasoning.",
"_____no_output_____"
],
[
"# Generate Training Images and Masks via Data Augmentation",
"_____no_output_____"
],
[
"[Data augmentation](https://en.wikipedia.org/wiki/Data_augmentation) is commonly used to generate more training data from the existing data. For example, a common trick to generate training images and masks for occluded scenes is to randomly insert rendered objects on top of the real image. Similarly, you can randomly scale, flip, rotate, duplicate, and crop to \"simulate\" more complex scenes. ",
"_____no_output_____"
],
[
"<figure>\n<center>\n<img src='https://developers.google.com/machine-learning/practica/image-classification/images/data_augmentation.png' />\n<figcaption>Example Image Data Augmentation (credit: developers.google.com)</figcaption></center>\n</figure>",
"_____no_output_____"
],
[
"In this exercise, we ask you to explore different ways to augment from our existing mustard bottle image:\n- flipping\n- translating\n- duplication \n- cropping\n- adding noise \n\n**Please complete the function below to generate 1 more pair of a color image and mask label using at least 2 tricks above to augment your data. You may use Numpy only!** \n\n**Note: make sure you display both of the your new image and mask below in your notebook submission. Your results will be visually graded**",
"_____no_output_____"
]
],
[
[
"def augment_mustard_bottle(rgb_im, mask):\n \"\"\"\n perform random rotation, scaling, and duplication to generate\n more training images and labels\n rgb_im: original rgb image of the mustard bottle\n mask: binay mask of the mustard bottle\n \"\"\"\n augmented_rgb = np.zeros((480, 640, 3))\n augmented_mask = np.zeros((480, 640))\n return augmented_rgb, augmented_mask",
"_____no_output_____"
],
[
"new_img, new_mask = augment_mustard_bottle(rgb_im, mask)",
"_____no_output_____"
],
[
"plt.imshow(new_img)",
"_____no_output_____"
],
[
"plt.imshow(new_mask)",
"_____no_output_____"
]
],
[
[
"## How will this notebook be Graded?\n\nIf you are enrolled in the class, this notebook will be graded using [Gradescope](www.gradescope.com). You should have gotten the enrollement code on our announcement in Piazza. \n\nFor submission of this assignment, you must do two things. \n- Download and submit the notebook `label_generation.ipynb` to Gradescope's notebook submission section, along with your notebook for the other problems.\n- Write down your answers to 6.1.b and 6.1.c to a separately pdf file and submit it to Gradescope's written submission section. \n\nWe will evaluate the local functions in the notebook to see if the function behaves as we have expected. For this exercise, the rubric is as follows:\n- [3 pts] Correct Implementation of `deproject_pW_to_image` method.\n- [2 pts] Analysis for Cluttered Scenes: reasonable answers and explanations. \n- [2 pts] Visually reasonable output from `augment_mustard_bottle`.\n",
"_____no_output_____"
]
],
[
[
"from manipulation.exercises.segmentation.test_mask import TestMask\nfrom manipulation.exercises.grader import Grader \n\nGrader.grade_output([TestMask], [locals()], 'results.json')\nGrader.print_test_results('results.json')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec87a27e2651e0d55e7bec93243a29d2067c4bb1 | 14,480 | ipynb | Jupyter Notebook | aata/exercises-homomorph.ipynb | johnperry-math/cocalc-examples | 394479e972dc2b74211113bbb43bc1ec4ec9978c | [
"Apache-2.0",
"CC-BY-4.0"
] | 13 | 2017-09-06T23:04:59.000Z | 2021-04-05T11:08:51.000Z | aata/exercises-homomorph.ipynb | johnperry-math/cocalc-examples | 394479e972dc2b74211113bbb43bc1ec4ec9978c | [
"Apache-2.0",
"CC-BY-4.0"
] | 9 | 2018-02-01T15:58:28.000Z | 2021-07-14T15:18:35.000Z | aata/exercises-homomorph.ipynb | johnperry-math/cocalc-examples | 394479e972dc2b74211113bbb43bc1ec4ec9978c | [
"Apache-2.0",
"CC-BY-4.0"
] | 10 | 2017-10-26T17:30:03.000Z | 2021-12-11T07:25:28.000Z | 482.666667 | 2,212 | 0.63163 | [
[
[
"%%html\n<link href=\"http://mathbook.pugetsound.edu/beta/mathbook-content.css\" rel=\"stylesheet\" type=\"text/css\" />\n<link href=\"https://aimath.org/mathbook/mathbook-add-on.css\" rel=\"stylesheet\" type=\"text/css\" />\n<style>.subtitle {font-size:medium; display:block}</style>\n<link href=\"https://fonts.googleapis.com/css?family=Open+Sans:400,400italic,600,600italic\" rel=\"stylesheet\" type=\"text/css\" />\n<link href=\"https://fonts.googleapis.com/css?family=Inconsolata:400,700&subset=latin,latin-ext\" rel=\"stylesheet\" type=\"text/css\" /><!-- Hide this cell. -->\n<script>\nvar cell = $(\".container .cell\").eq(0), ia = cell.find(\".input_area\")\nif (cell.find(\".toggle-button\").length == 0) {\nia.after(\n $('<button class=\"toggle-button\">Toggle hidden code</button>').click(\n function (){ ia.toggle() }\n )\n )\nia.hide()\n}\n</script>\n",
"_____no_output_____"
]
],
[
[
"**Important:** to view this notebook properly you will need to execute the cell above, which assumes you have an Internet connection. It should already be selected, or place your cursor anywhere above to select. Then press the \"Run\" button in the menu bar above (the right-pointing arrowhead), or press Shift-Enter on your keyboard.",
"_____no_output_____"
],
[
"$\\newcommand{\\identity}{\\mathrm{id}}\n\\newcommand{\\notdivide}{\\nmid}\n\\newcommand{\\notsubset}{\\not\\subset}\n\\newcommand{\\lcm}{\\operatorname{lcm}}\n\\newcommand{\\gf}{\\operatorname{GF}}\n\\newcommand{\\inn}{\\operatorname{Inn}}\n\\newcommand{\\aut}{\\operatorname{Aut}}\n\\newcommand{\\Hom}{\\operatorname{Hom}}\n\\newcommand{\\cis}{\\operatorname{cis}}\n\\newcommand{\\chr}{\\operatorname{char}}\n\\newcommand{\\Null}{\\operatorname{Null}}\n\\newcommand{\\lt}{<}\n\\newcommand{\\gt}{>}\n\\newcommand{\\amp}{&}\n$",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><h2 class=\"heading hide-type\" alt=\"Exercises 11.3 Exercises\"><span class=\"type\">Section</span><span class=\"codenumber\">11.3</span><span class=\"title\">Exercises</span></h2><a href=\"exercises-homomorph.ipynb\" class=\"permalink\">¶</a></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-387\"><h6 class=\"heading\"><span class=\"codenumber\">1</span></h6><p id=\"p-1749\">Prove that $\\det( AB) = \\det(A) \\det(B)$ for $A, B \\in GL_2( {\\mathbb R} )\\text{.}$ This shows that the determinant is a homomorphism from $GL_2( {\\mathbb R} )$ to ${\\mathbb R}^*\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-388\"><h6 class=\"heading\"><span class=\"codenumber\">2</span></h6><p id=\"p-1750\">Which of the following maps are homomorphisms? If the map is a homomorphism, what is the kernel? </p><ol class=\"lower-alpha\"><li id=\"li-409\"><p id=\"p-1751\">$\\phi : {\\mathbb R}^\\ast \\rightarrow GL_2 ( {\\mathbb R})$ defined by</p><div class=\"displaymath\">\n\\begin{equation*}\n\\phi( a ) =\n\\begin{pmatrix}\n1 & 0 \\\\\n0 & a\n\\end{pmatrix}\n\\end{equation*}\n</div></li><li id=\"li-410\"><p id=\"p-1752\">$\\phi : {\\mathbb R} \\rightarrow GL_2 ( {\\mathbb R})$ defined by</p><div class=\"displaymath\">\n\\begin{equation*}\n\\phi( a ) =\n\\begin{pmatrix}\n1 & 0 \\\\\na & 1\n\\end{pmatrix}\n\\end{equation*}\n</div></li><li id=\"li-411\"><p id=\"p-1753\">$\\phi : GL_2 ({\\mathbb R}) \\rightarrow {\\mathbb R}$ defined by</p><div class=\"displaymath\">\n\\begin{equation*}\n\\phi\n\\left(\n\\begin{pmatrix}\na & b \\\\\nc & d\n\\end{pmatrix}\n\\right)\n= a + d\n\\end{equation*}\n</div></li><li id=\"li-412\"><p id=\"p-1754\">$\\phi : GL_2 ( {\\mathbb R}) \\rightarrow {\\mathbb R}^\\ast$ defined by</p><div class=\"displaymath\">\n\\begin{equation*}\n\\phi\n\\left(\n\\begin{pmatrix}\na & b \\\\\nc & d\n\\end{pmatrix}\n\\right)\n= ad - bc\n\\end{equation*}\n</div></li><li id=\"li-413\"><p id=\"p-1755\">$\\phi : {\\mathbb M}_2( {\\mathbb R}) \\rightarrow {\\mathbb R}$ defined by</p><div class=\"displaymath\">\n\\begin{equation*}\n\\phi\n\\left(\n\\begin{pmatrix}\na & b \\\\\nc & d\n\\end{pmatrix}\n\\right)\n= b,\n\\end{equation*}\n</div><p>where ${\\mathbb M}_2( {\\mathbb R})$ is the additive group of $2 \\times 2$ matrices with entries in ${\\mathbb R}\\text{.}$</p></li></ol><div class=\"solutions\"><span class=\"solution\"><a knowl=\"\" class=\"id-ref\" refid=\"hk-hint-105\" id=\"hint-105\"><span class=\"type\">Hint</span></a></span><div id=\"hk-hint-105\" class=\"hidden-content tex2jax_ignore\"><span class=\"solution\"><p id=\"p-1756\">(a) is a homomorphism with kernel $\\{ 1 \\}\\text{;}$ (c) is not a homomorphism.</p></span></div></div></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-389\"><h6 class=\"heading\"><span class=\"codenumber\">3</span></h6><p id=\"p-1757\">Let $A$ be an $m \\times n$ matrix. Show that matrix multiplication, $x \\mapsto Ax\\text{,}$ defines a homomorphism $\\phi : {\\mathbb R}^n \\rightarrow {\\mathbb R}^m\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-390\"><h6 class=\"heading\"><span class=\"codenumber\">4</span></h6><p id=\"p-1758\">Let $\\phi : {\\mathbb Z} \\rightarrow {\\mathbb Z}$ be given by $\\phi(n) = 7n\\text{.}$ Prove that $\\phi$ is a group homomorphism. Find the kernel and the image of $\\phi\\text{.}$</p><div class=\"solutions\"><span class=\"solution\"><a knowl=\"\" class=\"id-ref\" refid=\"hk-hint-106\" id=\"hint-106\"><span class=\"type\">Hint</span></a></span><div id=\"hk-hint-106\" class=\"hidden-content tex2jax_ignore\"><span class=\"solution\"><p id=\"p-1759\">Since $\\phi(m + n) = 7(m+n) = 7m + 7n = \\phi(m) + \\phi(n)\\text{,}$ $\\phi$ is a homomorphism.</p></span></div></div></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-391\"><h6 class=\"heading\"><span class=\"codenumber\">5</span></h6><p id=\"p-1760\">Describe all of the homomorphisms from ${\\mathbb Z}_{24}$ to ${\\mathbb Z}_{18}\\text{.}$</p><div class=\"solutions\"><span class=\"solution\"><a knowl=\"\" class=\"id-ref\" refid=\"hk-hint-107\" id=\"hint-107\"><span class=\"type\">Hint</span></a></span><div id=\"hk-hint-107\" class=\"hidden-content tex2jax_ignore\"><span class=\"solution\"><p id=\"p-1761\">For any homomorphism $\\phi : {\\mathbb Z}_{24} \\rightarrow {\\mathbb Z}_{18}\\text{,}$ the kernel of $\\phi$ must be a subgroup of ${\\mathbb Z}_{24}$ and the image of $\\phi$ must be a subgroup of ${\\mathbb Z}_{18}\\text{.}$ Now use the fact that a generator must map to a generator.</p></span></div></div></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-392\"><h6 class=\"heading\"><span class=\"codenumber\">6</span></h6><p id=\"p-1762\">Describe all of the homomorphisms from ${\\mathbb Z}$ to ${\\mathbb Z}_{12}\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-393\"><h6 class=\"heading\"><span class=\"codenumber\">7</span></h6><p id=\"p-1763\">In the group ${\\mathbb Z}_{24}\\text{,}$ let $H = \\langle 4 \\rangle$ and $N = \\langle 6 \\rangle\\text{.}$ </p><ol class=\"lower-alpha\"><li id=\"li-414\"><p id=\"p-1764\">List the elements in $HN$ (we usually write $H + N$ for these additive groups) and $H \\cap N\\text{.}$</p></li><li id=\"li-415\"><p id=\"p-1765\">List the cosets in $HN/N\\text{,}$ showing the elements in each coset.</p></li><li id=\"li-416\"><p id=\"p-1766\">List the cosets in $H/(H \\cap N)\\text{,}$ showing the elements in each coset.</p></li><li id=\"li-417\"><p id=\"p-1767\">Give the correspondence between $HN/N$ and $H/(H \\cap N)$ described in the proof of the Second Isomorphism Theorem.</p></li></ol></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-394\"><h6 class=\"heading\"><span class=\"codenumber\">8</span></h6><p id=\"p-1768\">If $G$ is an abelian group and $n \\in {\\mathbb N}\\text{,}$ show that $\\phi : G \\rightarrow G$ defined by $g \\mapsto g^n$ is a group homomorphism.</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-395\"><h6 class=\"heading\"><span class=\"codenumber\">9</span></h6><p id=\"p-1769\">If $\\phi : G \\rightarrow H$ is a group homomorphism and $G$ is abelian, prove that $\\phi(G)$ is also abelian.</p><div class=\"solutions\"><span class=\"solution\"><a knowl=\"\" class=\"id-ref\" refid=\"hk-hint-108\" id=\"hint-108\"><span class=\"type\">Hint</span></a></span><div id=\"hk-hint-108\" class=\"hidden-content tex2jax_ignore\"><span class=\"solution\"><p id=\"p-1770\">Let $a, b \\in G\\text{.}$ Then $\\phi(a) \\phi(b) = \\phi(ab) = \\phi(ba) = \\phi(b)\\phi(a)\\text{.}$</p></span></div></div></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-396\"><h6 class=\"heading\"><span class=\"codenumber\">10</span></h6><p id=\"p-1771\">If $\\phi : G \\rightarrow H$ is a group homomorphism and $G$ is cyclic, prove that $\\phi(G)$ is also cyclic.</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-397\"><h6 class=\"heading\"><span class=\"codenumber\">11</span></h6><p id=\"p-1772\">Show that a homomorphism defined on a cyclic group is completely determined by its action on the generator of the group.</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-398\"><h6 class=\"heading\"><span class=\"codenumber\">12</span></h6><p id=\"p-1773\">If a group $G$ has exactly one subgroup $H$ of order $k\\text{,}$ prove that $H$ is normal in $G\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-399\"><h6 class=\"heading\"><span class=\"codenumber\">13</span></h6><p id=\"p-1774\">Prove or disprove: ${\\mathbb Q} / {\\mathbb Z} \\cong {\\mathbb Q}\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-400\"><h6 class=\"heading\"><span class=\"codenumber\">14</span></h6><p id=\"p-1775\">Let $G$ be a finite group and $N$ a normal subgroup of $G\\text{.}$ If $H$ is a subgroup of $G/N\\text{,}$ prove that $\\phi^{-1}(H)$ is a subgroup in $G$ of order $|H| \\cdot |N|\\text{,}$ where $\\phi : G \\rightarrow G/N$ is the canonical homomorphism.</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-401\"><h6 class=\"heading\"><span class=\"codenumber\">15</span></h6><p id=\"p-1776\">Let $G_1$ and $G_2$ be groups, and let $H_1$ and $H_2$ be normal subgroups of $G_1$ and $G_2$ respectively. Let $\\phi : G_1 \\rightarrow G_2$ be a homomorphism. Show that $\\phi$ induces a natural homomorphism $\\overline{\\phi} : (G_1/H_1) \\rightarrow (G_2/H_2)$ if $\\phi(H_1) \\subset H_2\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-402\"><h6 class=\"heading\"><span class=\"codenumber\">16</span></h6><p id=\"p-1777\">If $H$ and $K$ are normal subgroups of $G$ and $H \\cap K = \\{ e \\}\\text{,}$ prove that $G$ is isomorphic to a subgroup of $G/H \\times G/K\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-403\"><h6 class=\"heading\"><span class=\"codenumber\">17</span></h6><p id=\"p-1778\">Let $\\phi : G_1 \\rightarrow G_2$ be a surjective group homomorphism. Let $H_1$ be a normal subgroup of $G_1$ and suppose that $\\phi(H_1) = H_2\\text{.}$ Prove or disprove that $G_1/H_1 \\cong G_2/H_2\\text{.}$</p><div class=\"solutions\"><span class=\"solution\"><a knowl=\"\" class=\"id-ref\" refid=\"hk-hint-109\" id=\"hint-109\"><span class=\"type\">Hint</span></a></span><div id=\"hk-hint-109\" class=\"hidden-content tex2jax_ignore\"><span class=\"solution\"><p id=\"p-1779\">Find a counterexample.</p></span></div></div></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-trivial-kernel\"><h6 class=\"heading\"><span class=\"codenumber\">18</span></h6><p id=\"p-1780\">Let $\\phi : G \\rightarrow H$ be a group homomorphism. Show that $\\phi$ is one-to-one if and only if $\\phi^{-1}(e) = \\{ e \\}\\text{.}$</p></article></div>",
"_____no_output_____"
],
[
"<div class=\"mathbook-content\"><article class=\"exercise-like\" id=\"exercise-405\"><h6 class=\"heading\"><span class=\"codenumber\">19</span></h6><p id=\"p-1781\">Given a homomorphism $\\phi :G \\rightarrow H$ define a relation $\\sim$ on $G$ by $a \\sim b$ if $\\phi(a) = \\phi(b)$ for $a, b \\in G\\text{.}$ Show this relation is an equivalence relation and describe the equivalence classes.</p></article></div>",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ec87b38bd710e001f94901e9478b898d981f1f3a | 47,987 | ipynb | Jupyter Notebook | tools/annotation_tools/text_to_tree_tool/postprocessing_notebooks/convert_old_dict_to_new.ipynb | bharadwaj1098/droidlet | c2b6059cdf9631731b3dd94afa43243721c54d3f | [
"MIT"
] | 669 | 2020-11-21T01:20:20.000Z | 2021-09-13T13:25:16.000Z | tools/annotation_tools/text_to_tree_tool/postprocessing_notebooks/convert_old_dict_to_new.ipynb | bharadwaj1098/droidlet | c2b6059cdf9631731b3dd94afa43243721c54d3f | [
"MIT"
] | 324 | 2020-12-07T18:20:34.000Z | 2021-09-14T17:17:18.000Z | tools/annotation_tools/text_to_tree_tool/postprocessing_notebooks/convert_old_dict_to_new.ipynb | bharadwaj1098/droidlet | c2b6059cdf9631731b3dd94afa43243721c54d3f | [
"MIT"
] | 56 | 2021-01-04T19:57:40.000Z | 2021-09-13T21:20:08.000Z | 47.231299 | 359 | 0.532498 | [
[
[
"import json\nfrom pprint import pprint\nimport copy\nimport re",
"_____no_output_____"
],
[
"with open('../old_data/all_data_newdct.json') as f:\n data = json.load(f)\n",
"_____no_output_____"
],
[
"import spacy\nfrom spacy.lang.en import English\ntokenizer = English().Defaults.create_tokenizer()\n\ndef tokenize(st):\n return ' '.join([str(x) for x in tokenizer(st)])",
"_____no_output_____"
],
[
"print(data.keys())\n# do also for templated\ntrain_data, valid_data, test_data = data['train'], data['valid'], data['test']\nprint(train_data.keys())\nprint(valid_data.keys())\nprint(test_data.keys())\ntotal = {}\nfor data_type in [train_data, valid_data, test_data]:\n for key in ['templated', 'rephrased', 'prompts', 'humanbot']:\n if key in data_type:\n total[key] = total.get(key, 0) + len(data_type[key])\nprint(total)",
"[['kindly proceed forty six paces to the left of the yard'], {'dialogue_type': 'HUMAN_GIVE_COMMAND', 'action_sequence': [{'location': {'reference_object': {'has_name': [0, [10, 10]]}, 'steps': [0, [2, 3]], 'relative_direction': 'LEFT', 'location_type': 'REFERENCE_OBJECT'}, 'action_type': 'MOVE'}]}]\n"
],
[
"def to_snake_case(word, case=\"lower\"):\n \"\"\"convert a given word to snake case\"\"\"\n s1 = re.sub(\"(.)([A-Z][a-z]+)\", r\"\\1_\\2\", word)\n snake_case = re.sub(\"([a-z0-9])([A-Z])\", r\"\\1_\\2\", s1)\n if case == \"upper\":\n return snake_case.upper()\n return snake_case.lower()",
"_____no_output_____"
],
[
"def fix_keys(d):\n for key, val in d.items():\n if key in ['action_reference_object', 'location_reference_object']:\n d['reference_object'] = d[key]\n del d[key]\n elif key.endswith('_'):\n d[key[:-1]] = d[key]\n del d[key]\n if type(val) == dict:\n fix_keys(val)\n \n return d",
"_____no_output_____"
],
[
"def fix_vals(d):\n for key, val in d.items():\n if type(val) == dict:\n fix_vals(val)\n # fix spans\n elif type(val) == list:\n d[key] = [0, val]\n else:\n d[key] = to_snake_case(val, \"upper\")\n return d",
"_____no_output_____"
],
[
"def check_loc_type(d):\n for key, val in d.items():\n if key == \"location_type\" and val in ['SpeakerLook', 'AgentPos', 'SpeakerPos']:\n return True\n if type(val) == dict:\n if check_loc_type(val):\n return True\n return False",
"_____no_output_____"
],
[
"COLOURS = [\"red\", \"blue\", \"green\", \"yellow\", \"purple\", \"grey\", \"brown\", \"black\", \"orange\"]\nABSTRACT_SIZE = [\n \"really tiny\",\n \"very tiny\",\n \"tiny\",\n \"really small\",\n \"very small\",\n \"really little\",\n \"very little\",\n \"small\",\n \"little\",\n \"medium\",\n \"medium sized\",\n \"big\",\n \"large\",\n \"really big\",\n \"very big\",\n \"really large\",\n \"very large\",\n \"huge\",\n \"gigantic\",\n \"really huge\",\n \"very huge\",\n \"really gigantic\",\n \"very gigantic\",\n]\n",
"_____no_output_____"
],
[
"def make_new_data(datalist):\n new_data = []\n\n skip_data = []\n\n for data_point in datalist:\n text = data_point[0]\n text = tokenize(text)\n re_dict = data_point[1]\n \n # skip the ones that need re-annotation with coref-type\n if check_loc_type(re_dict):\n skip_data += [([text], re_dict)]\n else:\n x = copy.deepcopy(re_dict)\n new_dict = fix_keys(x)\n n_dict = fix_vals(new_dict)\n updated_dict = {}\n\n if n_dict['action_type'] == 'TAG':\n updated_dict['dialogue_type'] = 'PUT_MEMORY'\n if 'reference_object' in n_dict:\n updated_dict['filters'] = {'reference_object' : n_dict['reference_object']}\n if 'tag' in n_dict:\n updated_dict['upsert'] = {'memory_data' : {\"memory_type\": 'TRIPLE'}}\n tag_range = n_dict['tag'][1]\n words = \" \".join(text.split()[tag_range[0]: tag_range[1]+1])\n if words.strip() in COLOURS:\n updated_dict['upsert']['memory_data']['has_colour'] = n_dict['tag']\n elif words.strip() in ABSTRACT_SIZE:\n updated_dict['upsert']['memory_data']['has_size'] = n_dict['tag']\n else:\n updated_dict['upsert']['memory_data']['has_tag'] = n_dict['tag']\n elif n_dict['action_type'] == 'ANSWER':\n updated_dict['dialogue_type'] = 'GET_MEMORY'\n if 'reference_object' not in n_dict or 'answer_type' not in n_dict:\n skip_data.append([[text], re_dict])\n continue\n updated_dict['filters'] = {'reference_object' : n_dict['reference_object'], 'type' : 'REFERENCE_OBJECT'}\n if n_dict['answer_type'] == 'QUERY_TAG':\n if 'query_tag_name' in n_dict and 'query_tag_val' not in n_dict:\n updated_dict['answer_type'] = 'TAG'\n updated_dict['tag_name'] = 'has_' + n_dict['query_tag_name'].lower()\n elif 'query_tag_name' in n_dict and 'query_tag_val' in n_dict:\n updated_dict['answer_type'] = 'EXISTS'\n val = n_dict['query_tag_val']\n key = 'has_' + n_dict['query_tag_name'].lower()\n updated_dict['filters']['reference_object'][key] = val\n else:\n skip_data.append([[text], re_dict])\n continue\n elif n_dict['answer_type'] == 'QUERY_ALL_TAGS':\n updated_dict['answer_type'] = 'TAG'\n updated_dict['tag_name'] = 'has_name'\n elif n_dict['action_type'] == 'NOOP':\n updated_dict['dialogue_type'] = 'NOOP'\n else:\n updated_dict[\"dialogue_type\"] = \"HUMAN_GIVE_COMMAND\"\n updated_dict[\"action\"] = n_dict\n \n new_data+= [([text], updated_dict)]\n\n return new_data, skip_data",
"_____no_output_____"
],
[
"# for rephrases\nrephrases = []\nnew_rephrases, skip_rephrases = [], []\nrephrases = train_data['rephrased']\nrephrases.extend(valid_data['rephrased'])\nrephrases.extend(test_data['rephrased'])\n\n# for x in rephrases:\n# if 'Name each very large item in front of the orange airport with big' in x[0]:\n# print(x)\nnew_rephrases, skip_rephrases = make_new_data(rephrases)\nprint(len(rephrases))\nprint(len(new_rephrases))\nprint(len(skip_rephrases))",
"35200\n25402\n9798\n"
],
[
"print(skip_rephrases[700])",
"(['what are the dimensions of the structure'], {'reference_object': {'location': {'location_type': 'SpeakerLook'}}, 'answer_type': 'query_tag', 'query_tag_name': 'size', 'action_type': 'Answer'})\n"
],
[
"# write things that need to be annotated\n\nwith open('/checkpoint/kavyasrinet/release_data_dialogue/rephrases.json', 'w') as f:\n json.dump(new_rephrases, f)\n\nwith open('/checkpoint/kavyasrinet/reannotate_data/rephrases.json', 'w') as f:\n json.dump(skip_rephrases, f)\n",
"_____no_output_____"
],
[
"prompts = []\nprompts = valid_data['prompts']\nprompts.extend(test_data['prompts'])\n\nnew_prompts, skip_prompts = make_new_data(prompts)\nprint(len(prompts))\nprint(len(new_prompts))\nprint(len(skip_prompts))\n\nwith open('/checkpoint/kavyasrinet/release_data_dialogue/prompts.json', 'w') as f:\n json.dump(new_prompts, f)\n\nwith open('/checkpoint/kavyasrinet/reannotate_data/prompts.json', 'w') as f:\n json.dump(skip_prompts, f)\n",
"3065\n2513\n552\n"
],
[
"# humanbot\nhumanbot = []\nhumanbot = valid_data['humanbot']\nhumanbot.extend(test_data['humanbot'])\nnew_humanbot, skip_humanbot = make_new_data(humanbot)\nprint(len(humanbot), len(new_humanbot), len(skip_humanbot))\nwith open('/checkpoint/kavyasrinet/release_data_dialogue/humanbot.json', 'w') as f:\n json.dump(new_humanbot, f)\n\nwith open('/checkpoint/kavyasrinet/reannotate_data/humanbot.json', 'w') as f:\n json.dump(skip_humanbot, f)\n",
"817 708 109\n"
],
[
"with open('dialogue_data.json') as f:\n dialogue_data = json.load(f)",
"_____no_output_____"
],
[
"templated = []\nfor key in ['train', 'valid', 'test']:\n data = dialogue_data[key]['templated']\n templated.extend(data)\nprint(len(templated))\nprint(templated[17])\n\nwith open('/checkpoint/kavyasrinet/release_data_dialogue/generated_dialogues.json')",
"800000\n[['please reproduce 90 red porches over here'], {'dialogue_type': 'HUMAN_GIVE_COMMAND', 'action': {'reference_object': {'repeat': {'repeat_key': 'FOR', 'repeat_count': [0, [2, 2]]}, 'has_colour': [0, [3, 3]], 'has_name': [0, [4, 4]], 'has_attribute': [[0, [3, 3]], [0, [4, 4]]]}, 'location': {'coref_resolve': [0, [6, 6]]}, 'action_type': 'BUILD'}}]\n"
],
[
"# print(dialogue_data['test']['templated'][20])\nprint(len(dialogue_data['test']['templated']))\nprint(len(dialogue_data['train']['templated']))\nfor d_type in ['train', 'test', 'valid']:\n for example in dialogue_data[d_type]['templated']:\n if 'what are you doing' in example[0]:\n print(example)",
"5000\n790000\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'NOOP'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n[['what are you doing'], {'dialogue_type': 'GET_MEMORY', 'filters': {'temporal': 'CURRENT', 'type': 'ACTION'}, 'answer_type': 'TAG', 'tag_name': 'action_name'}]\n"
],
[
"print(dialogue_data['train'].keys())\n\nfor x, y in zip(['rephrased'], [new_rephrases]): # add rephrases only\n# for x, y in zip(['rephrased', 'prompts', 'humanbot'], [new_rephrases, new_prompts, new_humanbot]): \n n_train = (9 * len(y)) // 10\n n_valid = len(y) // 20\n dialogue_data['train'][x] = y[ : n_train]\n dialogue_data['valid'][x] = y[n_train : n_train + n_valid]\n dialogue_data['test'][x] = y[n_train + n_valid : ]\n #print(x, len(dialogue_data['train'][x]), len(dialogue_data['valid'][x]), len(dialogue_data['test'][x]))\n# write as train, valid, test and each is a dict -> rephrased, prompt, humanbot",
"dict_keys(['templated'])\n"
],
[
"print(dialogue_data['test']['rephrased'][29])",
"(['Complete the construction of a couple tiny stairways please'], {'dialogue_type': 'HUMAN_GIVE_COMMAND', 'action': {'reference_object': {'repeat': {'repeat_key': 'FOR', 'repeat_count': [0, [4, 5]]}, 'has_size': [0, [6, 6]], 'has_name': [0, [7, 7]]}, 'action_type': 'FREE_BUILD'}})\n"
],
[
"print(dialogue_data.keys())\n# do also for templated\ntrain_data, valid_data, test_data = dialogue_data['train'], dialogue_data['valid'], dialogue_data['test']\nprint(train_data.keys())\nprint(valid_data.keys())\nprint(test_data.keys())\ntotal = {}\nprint(len(new_rephrases))\nprint(len(new_prompts))\nprint(len(new_humanbot))\nfor data_type in [train_data, valid_data, test_data]:\n for key in ['templated', 'rephrased']:\n \n# for key in ['templated', 'rephrased', 'prompts', 'humanbot']:\n print(key, len(data_type[key]))\n print(\"-----------------------------\")\n# if key in data_type:\n# print(key, data_type[key])\n# total[key] = total.get(key, 0) + len(data_type[key])\n",
"dict_keys(['train', 'valid', 'test'])\ndict_keys(['templated', 'rephrased'])\ndict_keys(['templated', 'rephrased'])\ndict_keys(['templated', 'rephrased'])\n25402\n2513\n708\ntemplated 790000\nrephrased 22861\n-----------------------------\ntemplated 5000\nrephrased 1270\n-----------------------------\ntemplated 5000\nrephrased 1271\n-----------------------------\n"
],
[
"with open('combined_dialogue_data.json', 'w') as f:\n json.dump(dialogue_data, f)",
"_____no_output_____"
],
[
"import json\nwith open('combined_dialogue_data.json') as f:\n data = json.load(f)",
"_____no_output_____"
],
[
"for data_type in ['train', 'valid', 'test']:\n for example in data[data_type]['rephrased']:\n if 'good job' in example[0]:\n print(example)",
"_____no_output_____"
],
[
"with open('/Users/kavyasrinet/rephrases.json') as f:\n data_r = json.load(f)",
"_____no_output_____"
],
[
"print(type(data))",
"<class 'list'>\n"
],
[
"def check_coref(d):\n for key, val in d.items():\n if key == \"coref_resolve\":\n return True\n if type(val) == dict:\n if check_coref(val):\n return True\n if type(val) == list and type(val[0]) == dict:\n for v in val:\n if check_coref(v):\n return True\n return False",
"_____no_output_____"
],
[
"def location_type(d):\n \n for key, val in d.items():\n #print(key, val)\n if key == \"location_type\" and val in ['SPEAKER_LOOK', 'AGENT_POS', 'SPEAKER_POS']:\n return True\n if type(val) == dict:\n if location_type(val):\n return True\n if type(val) == list and type(val[0]) == dict:\n for v in val:\n if location_type(v):\n return True\n return False",
"_____no_output_____"
],
[
"print(len(data))",
"35192\n"
],
[
"def update_coref(d):\n updated_d = {}\n for key, val in d.items():\n if key == \"coref_resolve\":\n updated_d['contains_coreference'] = \"yes\"\n elif type(val) == dict:\n updated_d[key] = update_coref(val)\n elif type(val) == list and type(val[0]) == dict:\n updated_d[key] = []\n for v in val:\n updated_d[key].append(update_coref(v))\n else:\n updated_d[key] = val\n return updated_d",
"_____no_output_____"
],
[
"def fix_freebuild(d):\n updated_d = {}\n for key, val in d.items():\n if key == \"action_type\" and val == 'FREE_BUILD':\n updated_d[key] = \"FREEBUILD\"\n elif type(val) == dict:\n updated_d[key] = fix_freebuild(val)\n elif type(val) == list and type(val[0]) == dict:\n updated_d[key] = []\n for v in val:\n updated_d[key].append(fix_freebuild(v))\n else:\n updated_d[key] = val\n return updated_d",
"_____no_output_____"
],
[
"def get_action_type(d):\n if 'action_sequence' in ad:\n return ad['action_sequence'][0]['action_type']\n return None",
"_____no_output_____"
],
[
"action_types = set()\nfor sentence, ad in data:\n if 'action_sequence' in ad:\n val = ad['action_sequence']\n if len(val)> 1:\n print(\"YESSSSS\")\n for v in val:\n# if v['action_type'] == 'FREE_BUILD':\n# print(sentence, ad)\n action_types.add(v['action_type'])\nprint(action_types)",
"{'FREE_BUILD', 'DESTROY', 'RESUME', 'BUILD', 'DIG', 'MOVE', 'UNDO', 'STOP', 'FILL'}\n"
],
[
"def fix_dig(ad):\n new_dict = {}\n action_dict = ad['action_sequence'][0]\n for key, val in action_dict.items():\n if key.startswith('has_'):\n if 'schematic' not in new_dict:\n new_dict['schematic'] = {}\n new_dict['schematic'][key] = val\n else:\n new_dict[key] = val\n out= {}\n out['dialogue_type'] = 'HUMAN_GIVE_COMMAND'\n out['action_sequence'] = [new_dict]\n return out",
"_____no_output_____"
],
[
"def check_target_action_type(d):\n for k, v in d.items():\n if k =='target_action_type':\n return True\n if type(v) == dict:\n if check_target_action_type(v):\n return True\n if type(v) == list and type(v[0]) == dict:\n for val in v:\n if check_target_action_type(val):\n return True\n return False",
"_____no_output_____"
],
[
"i = 0\nj = 0\ntarget = 0\nupdated_data = []\ndig, fill, freebuild = 0, 0, 0\nfor sentence, ad in data:\n \n text = sentence\n new_d = ad\n \n if check_target_action_type(ad):\n target += 1\n if get_action_type(ad) == 'DIG':\n dig += 1\n new_d = fix_dig(ad)\n# if get_action_type(ad) == 'FILL':\n# fill += 1\n# print(sentence, ad)\n# continue\n\n if get_action_type(new_d) == 'FREE_BUILD':\n freebuild += 1\n new_d = fix_freebuild(new_d)\n \n# if location_type(ad):\n# i +=1\n \n # contains coreference\n if check_coref(ad):\n new_d = update_coref(new_d)\n \n updated_data+= [(text, new_d)]\n\nprint(i, dig, fill, freebuild)\nprint(target)",
"0 2624 0 2779\n0\n"
],
[
"#print(updated_data[0])\nwith open('/Users/kavyasrinet/Github/minecraft/python/craftassist/ttad/rephrases_data/rephrases.json', 'w') as f:\n json.dump(updated_data, f)",
"(['kindly proceed forty six paces to the left of the yard'], {'dialogue_type': 'HUMAN_GIVE_COMMAND', 'action_sequence': [{'location': {'reference_object': {'has_name': [0, [10, 10]]}, 'steps': [0, [2, 3]], 'relative_direction': 'LEFT', 'location_type': 'REFERENCE_OBJECT'}, 'action_type': 'MOVE'}]})\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec87bc0dd8b6aa431e84b7b9976e5bd7937d74fb | 46,994 | ipynb | Jupyter Notebook | trash/garbage/flat_arch__GGG.ipynb | redjerdai/financial_news_re | d7950da28d77ade29628f3b2fa266f5059527f52 | [
"MIT"
] | null | null | null | trash/garbage/flat_arch__GGG.ipynb | redjerdai/financial_news_re | d7950da28d77ade29628f3b2fa266f5059527f52 | [
"MIT"
] | null | null | null | trash/garbage/flat_arch__GGG.ipynb | redjerdai/financial_news_re | d7950da28d77ade29628f3b2fa266f5059527f52 | [
"MIT"
] | null | null | null | 49.156904 | 3,937 | 0.421181 | [
[
[
"#",
"_____no_output_____"
],
[
"import json\nimport time\nimport numpy\nimport pandas\nimport datetime",
"_____no_output_____"
],
[
"import torch\nfrom torch import nn",
"_____no_output_____"
],
[
"from premodels.prenet import Generale as GGG",
"_____no_output_____"
],
[
"from thats_the_stuff import gonet as golags",
"_____no_output_____"
],
[
"from mpydge.holy.data_keeper.keeper import Conductor\ndef gonet(data_set, y_names, removes, categoricals, net, net_init_params, net_fit_params):\n\n exclude = y_names + removes\n x_names = [x for x in data_set.columns.values if (x not in exclude and 'LAG0' not in x)]\n used_names = x_names + y_names\n data_set = data_set[used_names]\n\n categorical_columns = categoricals\n numerical_columns = x_names\n\n outputs = y_names\n\n if categorical_columns is not None:\n for category in categorical_columns:\n data_set[category] = data_set[category].astype('category')\n if numerical_columns is not None:\n for numeric in numerical_columns:\n data_set[numeric] = data_set[numeric].astype('float64')\n\n data_set[outputs[0]] = data_set[outputs[0]].astype('category')\n\n data = Conductor(data_frame=data_set, target=outputs)\n\n net = net(data=data, **net_init_params)\n print(net)\n\n net_fit_params['optimiser'] = net_fit_params['optimiser'](net.parameters(), lr=0.001)\n\n net.fit(**net_fit_params)\n net.fit_plot()\n\n from sklearn.metrics import mean_squared_error as MSE, r2_score as R2\n\n summary_params = {'on': 'train',\n 'loss_function': MSE,\n 'score': R2}\n net.summary(**summary_params)\n\n summary_params = {'on': 'validation',\n 'loss_function': MSE,\n 'score': R2}\n net.summary(**summary_params)\n\n summary_params = {'on': 'test',\n 'loss_function': MSE,\n 'score': R2}\n net.summary(**summary_params)\n\n return None\n\ngolags = gonet",
"_____no_output_____"
],
[
"d = './dataset.csv'\ndata = pandas.read_csv(d)\ndata = data.rename(columns={'lag': 'news_horizon'})\ndata = data.set_index(['ticker', 'time', 'news_horizon'], drop=False)\ndata = data.sort_index()\ndata",
"_____no_output_____"
],
[
"tsi_names = ['news_time']\ny_names = ['open_LAG0']\nremoves = ['ticker', 'time', 'id', 'title', 'news_time']\ncategoricals = None\n\nnet_init_params = {'layers': [nn.Linear], \n 'layers_dimensions': [data.shape[0]],\n 'activators': [nn.Tanh], \n 'preprocessor': nn.BatchNorm1d, \n 'embeddingdrop': 0.0, \n 'activators_args': {}, \n 'interprocessors': None,\n 'interdrops': None, \n 'postlayer': nn.Linear}\nnet_fit_params = {'loss_function': nn.MSELoss(),\n 'optimiser': torch.optim.Adam,\n 'epochs': 300}\n\n\ngolags(data, y_names, removes, categoricals, GGG, net_init_params, net_fit_params)",
"<ipython-input-23-3ccc25d3ef2d>:19: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n data_set[numeric] = data_set[numeric].astype('float64')\n<ipython-input-23-3ccc25d3ef2d>:21: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n data_set[outputs[0]] = data_set[outputs[0]].astype('category')\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec87c15407335d7990dde505c0cd8c8e056a05a5 | 143,593 | ipynb | Jupyter Notebook | WeatherDataCollection/Excel Clean-up.ipynb | mhnam/weather-conditions-COVID19 | 8b4fe5f554ee62fdf0a92f4f7941ede23ee02bbd | [
"MIT"
] | null | null | null | WeatherDataCollection/Excel Clean-up.ipynb | mhnam/weather-conditions-COVID19 | 8b4fe5f554ee62fdf0a92f4f7941ede23ee02bbd | [
"MIT"
] | null | null | null | WeatherDataCollection/Excel Clean-up.ipynb | mhnam/weather-conditions-COVID19 | 8b4fe5f554ee62fdf0a92f4f7941ede23ee02bbd | [
"MIT"
] | null | null | null | 64.594242 | 126 | 0.263502 | [
[
[
"## Scale data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport pickle\n\ndef scale_dfs(dfs, start_column_index):\n for var in dfs:\n dfs[var].iloc[:,start_column_index:] = dfs[var].iloc[:,start_column_index:].mul(scales[var])",
"_____no_output_____"
],
[
"pm_scale = 1000000000 #to micrograms\no3_scale = 603447629\nno2_scale = 629585593\nso2_scale = 452118045 #to ppbv",
"_____no_output_____"
],
[
"scales = {\n \"pm2p5\":pm_scale,\n \"o3\":o3_scale,\n \"no2\":no2_scale,\n \"so2\":so2_scale\n }",
"_____no_output_____"
],
[
"#pollution_sheet_names = [\"pm2p5\", \"o3\", \"no2\", \"so2\"]\npollution_sheet_names = [\"o3\", \"no2\", \"so2\"]\n\nmain_var_sheet_names = [\"maxtempC\",\"mintempC\",\"avgtempC\",\"cloudcover\",\"humidity\",\"precipMM\",\"pressure\",\n \"windspeedKmph\",\"totalSnow_cm\",'sunHour','moon_illumination','moonrise',\n 'moonset','sunrise','sunset','DewPointC','FeelsLikeC',#'HeatIndexC',\n 'WindChillC','WindGustKmph','visibility','winddirDegree','uvIndex'\n ]\n\nvars_not_to_weight = ['moonrise','moonset','sunrise','sunset']",
"_____no_output_____"
],
[
"file_name = \"US_pollution_gases\"",
"_____no_output_____"
],
[
"dfs = pd.read_excel(file_name + \".xlsx\", index_col = 0, sheet_name = pollution_sheet_names)",
"_____no_output_____"
],
[
"start_column_index = list(dfs[\"o3\"].columns).index(\"2020-04-01\")\nstart_column_index",
"_____no_output_____"
],
[
"dfs[\"o3\"].head()",
"_____no_output_____"
],
[
"scale_dfs(dfs, start_column_index)",
"_____no_output_____"
],
[
"#dfs[\"pm2p5\"].head()\ndfs",
"_____no_output_____"
],
[
"writer = pd.ExcelWriter('scaled_'+ file_name +'.xlsx', engine='xlsxwriter')\nfor var in dfs:\n print(\"Saving\", var)\n dfs[var].to_excel(writer, sheet_name = var, index = True)\n \nwriter.save()",
"Saving o3\nSaving no2\nSaving so2\n"
]
],
[
[
"## Replacing Missing Data with avg of neighboring values",
"_____no_output_____"
]
],
[
[
"def flag_pollution_value(row, index):\n #After Scaling, threshold chosen after inspection (correct values always more than 10^-3, avg is ~\n return row[index] < 10**(-5)",
"_____no_output_____"
],
[
"def flag_temperature_value(row, index):\n return row[index]==0 and abs(row[index-1]) > 8 and abs(row[index+1]) > 8",
"_____no_output_____"
],
[
"def flag_UV_value(row, index):\n return row[index]==0",
"_____no_output_____"
],
[
"def avg_neighbors(row, index):\n return (row[index-2]+ row[index-1] + row[index+1]+row[index+2])/4",
"_____no_output_____"
],
[
"def interpolate_pollution_data(dfs, start_column_index):\n for var in dfs:\n num_corrections = 0\n arr = dfs[var].to_numpy()\n n = len(arr[0])\n for row in arr:\n for j in range(start_column_index+2,n-2): #avoid first 2 and last 2 vals for avging\n if flag_pollution_value(row, j):\n row[j] = avg_neighbors(row, j)\n num_corrections += 1\n dfs[var] = pd.DataFrame(arr, columns = dfs[var].columns, index = dfs[var].index)\n \n print(num_corrections,\"corrections made for variable\",var)",
"_____no_output_____"
],
[
"def interpolate_temperature_data(dfs, start_column_index):\n num_corrections = 0\n maxTemp = dfs[\"maxtempC\"].to_numpy()\n minTemp = dfs[\"mintempC\"].to_numpy()\n avgTemp = dfs[\"avgtempC\"].to_numpy()\n n = len(maxTemp[0])\n for i, row in enumerate(maxTemp):\n for j in range(start_column_index+2,n-2): #avoid first 2 and last 2 vals for avging\n if flag_temperature_value(row, j):\n row[j] = avg_neighbors(row, j)\n minTemp[i][j] = avg_neighbors(minTemp[i], j)\n avgTemp[i][j] = avg_neighbors(avgTemp[i], j)\n num_corrections+=1\n \n dfs[\"maxtempC\"] = pd.DataFrame(maxTemp, columns = dfs[\"maxtempC\"].columns, index = dfs[\"maxtempC\"].index)\n dfs[\"mintempC\"] = pd.DataFrame(minTemp, columns = dfs[\"mintempC\"].columns, index = dfs[\"mintempC\"].index)\n dfs[\"avgtempC\"] = pd.DataFrame(avgTemp, columns = dfs[\"avgtempC\"].columns, index = dfs[\"avgtempC\"].index)\n \n print(num_corrections,\"corrections made for temperature\")",
"_____no_output_____"
],
[
"def interpolate_UV_data(dfs, start_column_index):\n num_corrections = 0\n arr = dfs[\"uvIndex\"].to_numpy()\n n = len(arr[0])\n for row in arr:\n for j in range(start_column_index+2,n-2): #avoid first 2 and last 2 vals for avging\n if flag_UV_value(row, j):\n row[j] = avg_neighbors(row, j)\n num_corrections += 1\n dfs[\"uvIndex\"] = pd.DataFrame(arr, columns = dfs[\"uvIndex\"].columns, index = dfs[\"uvIndex\"].index)\n \n print(num_corrections,\"corrections made for uvIndex\")",
"_____no_output_____"
],
[
"pollution_sheet_names = [\"pm2p5\", \"o3\", \"no2\", \"so2\"]\n\nmain_var_sheet_names = [\"maxtempC\",\"mintempC\",\"avgtempC\",\"cloudcover\",\"humidity\",\"precipMM\",\"pressure\",\n \"windspeedKmph\",\"totalSnow_cm\",'sunHour','moon_illumination','moonrise',\n 'moonset','sunrise','sunset','DewPointC','FeelsLikeC',#'HeatIndexC',\n 'WindChillC','WindGustKmph','visibility','winddirDegree','uvIndex'\n ]\n\ntemperature_vars = [\"maxtempC\",\"mintempC\",\"avgtempC\"]",
"_____no_output_____"
]
],
[
[
"#### Load Pollution Files Here",
"_____no_output_____"
]
],
[
[
"file_name = \"\"",
"_____no_output_____"
],
[
"dfs = pd.read_excel(file_name + \".xlsx\", index_col = 0, sheet_name = pollution_sheet_names)",
"_____no_output_____"
],
[
"start_column_index = list(dfs[\"pm2p5\"].columns).index(\"2019-01-01\")\nstart_column_index",
"_____no_output_____"
],
[
"interpolate_pollution_data(dfs, start_column_index)",
"_____no_output_____"
],
[
"writer = pd.ExcelWriter('interpolated_'+ file_name +'.xlsx', engine='xlsxwriter')\nfor var in dfs:\n print(\"Saving\", var)\n dfs[var].to_excel(writer, sheet_name = var, index = True)\n \nwriter.save()",
"_____no_output_____"
]
],
[
[
"### Load Temperature Files Here",
"_____no_output_____"
]
],
[
[
"file_name = \"\"",
"_____no_output_____"
],
[
"dfs = pd.read_excel(file_name + \".xlsx\", index_col = 0, sheet_name = main_var_sheet_names)",
"_____no_output_____"
],
[
"start_column_index = list(dfs[\"maxtempC\"].columns).index(\"2019-01-01\")\nstart_column_index",
"_____no_output_____"
],
[
"interpolate_temperature_data(dfs, start_column_index)",
"_____no_output_____"
],
[
"interpolate_UV_data(dfs, start_column_index)",
"_____no_output_____"
],
[
"writer = pd.ExcelWriter('interpolated_'+ file_name +'.xlsx', engine='xlsxwriter')\nfor var in dfs:\n print(\"Saving\", var)\n dfs[var].to_excel(writer, sheet_name = var, index = True)\n \nwriter.save()",
"_____no_output_____"
]
],
[
[
"## Weighted Avg of Weather Variables",
"_____no_output_____"
],
[
"Weights by population",
"_____no_output_____"
]
],
[
[
"pollution_sheet_names = [\"pm2p5\", \"o3\", \"no2\", \"so2\"]\nmain_var_sheet_names = [\"maxtempC\",\"mintempC\",\"avgtempC\",\"cloudcover\",\"humidity\",\"precipMM\",\"pressure\",\n \"windspeedKmph\",\"totalSnow_cm\",'sunHour','moon_illumination','moonrise',\n 'moonset','sunrise','sunset','DewPointC','FeelsLikeC',#'HeatIndexC',\n 'WindChillC','WindGustKmph','visibility','winddirDegree','uvIndex'\n ]\n\nvars_not_to_weight = ['moonrise','moonset','sunrise','sunset']\n\nsheet_names = pollution_sheet_names",
"_____no_output_____"
],
[
"dfs = pd.read_excel(\"\", index_col = 0, \n sheet_name = sheet_names)\n\n#dfs = dict: sheet_names --> dataframes",
"_____no_output_____"
],
[
"for var in dfs:\n df = dfs[var]\n df.drop(columns = [\"Lat\", \"Long\"], inplace = True)\n",
"_____no_output_____"
],
[
"start_column_index = list(dfs[sheet_names[0]].columns).index(\"2019-01-01\")\nprint(start_column_index)\ndate_columns = list(dfs[sheet_names[0]].columns[start_column_index:])",
"_____no_output_____"
],
[
"countries = list(dict.fromkeys(dfs[sheet_names[0]].index))",
"_____no_output_____"
],
[
"print(countries)",
"_____no_output_____"
],
[
"dfs[sheet_names[0]].loc[countries[0]].iloc[0][:5]",
"_____no_output_____"
],
[
"def weighted_weather_for_country(country, df):\n country_df = df.loc[country]\n weights = np.array(country_df[\"Population\"])\n weights = weights/np.sum(weights)\n row = [country] #output row\n for date in date_columns:\n row.append(np.average(country_df[date], weights = weights))\n \n return row",
"_____no_output_____"
],
[
"output_frames = []\nfor var in sheet_names:\n data = []\n df = dfs[var]\n for country in countries:\n if var in vars_not_to_weight:\n row = [country] + list(df.iloc[0][start_column_index:])\n else:\n try:\n row = weighted_weather_for_country(country, df)\n except:\n print(country, var)\n break\n data.append(row)\n \n df = pd.DataFrame(data, columns = [\"Country\"] + date_columns).set_index(\"Country\")\n output_frames.append(df)\n ",
"_____no_output_____"
],
[
"writer = pd.ExcelWriter('Weights Applied.xlsx', engine='xlsxwriter') #rename!",
"_____no_output_____"
],
[
"for i, sheet in enumerate(output_frames):\n sheet.to_excel(writer, sheet_name = sheet_names[i], index = True)",
"_____no_output_____"
],
[
"writer.save()",
"_____no_output_____"
]
],
[
[
"## Reorder Global Rows based on canonical ordering",
"_____no_output_____"
]
],
[
[
"import pickle\nimport pandas as pd",
"_____no_output_____"
],
[
"canonical_order = pickle.load(open(\"canonical_order.p\", \"rb\"))",
"_____no_output_____"
],
[
"canonical_order",
"_____no_output_____"
],
[
"weighted_countries = pickle.load(open(\"weighted_countries_list.p\", \"rb\"))",
"_____no_output_____"
],
[
"weighted_countries",
"_____no_output_____"
],
[
"pollution_sheet_names = [\"pm2p5\", \"o3\", \"no2\", \"so2\"]\n\nmain_var_sheet_names = [\"maxtempC\",\"mintempC\",\"avgtempC\",\"cloudcover\",\"humidity\",\"precipMM\",\"pressure\",\n \"windspeedKmph\",\"totalSnow_cm\",'sunHour','moon_illumination','moonrise',\n 'moonset','sunrise','sunset','DewPointC','FeelsLikeC',#'HeatIndexC',\n 'WindChillC','WindGustKmph','visibility','winddirDegree','uvIndex'\n ]",
"_____no_output_____"
],
[
"weighted_filename = \"\"\nsimple_filename = \"\"",
"_____no_output_____"
],
[
"weighted_dfs = pd.read_excel(weighted_filename + \".xlsx\", index_col = 0, sheet_name = pollution_sheet_names)",
"_____no_output_____"
],
[
"simple_dfs = pd.read_excel(simple_filename + \".xlsx\", index_col = 0, sheet_name = pollution_sheet_names)",
"_____no_output_____"
],
[
"simple_dfs[\"pm2p5\"]",
"_____no_output_____"
],
[
"for var in simple_dfs:\n simple_dfs[var].drop(weighted_countries, inplace = True)",
"_____no_output_____"
],
[
"final_dfs = {var:simple_dfs[var].append(weighted_dfs[var]) for var in simple_dfs}",
"_____no_output_____"
],
[
"columns = [\"lat\",\"long\"] + list(weighted_dfs[\"pm2p5\"].columns)",
"_____no_output_____"
],
[
"columns",
"_____no_output_____"
],
[
"for var in final_dfs:\n final_dfs[var] = final_dfs[var].reindex(canonical_order)\n final_dfs[var] = final_dfs[var][columns]",
"_____no_output_____"
],
[
"final_dfs[\"pm2p5\"]",
"_____no_output_____"
],
[
"final_dfs.keys()",
"_____no_output_____"
],
[
"writer = pd.ExcelWriter('Merged Global Pollution.xlsx', engine='xlsxwriter')",
"_____no_output_____"
],
[
"for var in final_dfs:\n final_dfs[var].to_excel(writer, sheet_name = var, index = True)",
"_____no_output_____"
],
[
"writer.save()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec87cfb23781d6fa5a98a0689b2d4d6aea416a99 | 244,438 | ipynb | Jupyter Notebook | site/en-snapshot/probability/examples/FFJORD_Demo.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
] | 491 | 2020-01-27T19:05:32.000Z | 2022-03-31T08:50:44.000Z | site/en-snapshot/probability/examples/FFJORD_Demo.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
] | 511 | 2020-01-27T22:40:05.000Z | 2022-03-21T08:40:55.000Z | site/en-snapshot/probability/examples/FFJORD_Demo.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
] | 627 | 2020-01-27T21:49:52.000Z | 2022-03-28T18:11:50.000Z | 389.232484 | 94,262 | 0.928673 | [
[
[
"##### Copyright 2018 The TensorFlow Probability Authors.\n\nLicensed under the Apache License, Version 2.0 (the \"License\");",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\"); { display-mode: \"form\" }\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# FFJORD\n\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/probability/examples/FFJORD_Demo\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/probability/blob/main/tensorflow_probability/examples/jupyter_notebooks/FFJORD_Demo.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/probability/blob/main/tensorflow_probability/examples/jupyter_notebooks/FFJORD_Demo.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/probability/tensorflow_probability/examples/jupyter_notebooks/FFJORD_Demo.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Setup\n\nFirst install packages used in this demo.",
"_____no_output_____"
]
],
[
[
"!pip install -q dm-sonnet",
"_____no_output_____"
],
[
"#@title Imports (tf, tfp with adjoint trick, etc)\n\nimport numpy as np\nimport tqdm as tqdm\nimport sklearn.datasets as skd\n\n# visualization\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom scipy.stats import kde\n\n# tf and friends\nimport tensorflow.compat.v2 as tf\nimport tensorflow_probability as tfp\nimport sonnet as snt\ntf.enable_v2_behavior()\n\ntfb = tfp.bijectors\ntfd = tfp.distributions\n\ndef make_grid(xmin, xmax, ymin, ymax, gridlines, pts):\n xpts = np.linspace(xmin, xmax, pts)\n ypts = np.linspace(ymin, ymax, pts)\n xgrid = np.linspace(xmin, xmax, gridlines)\n ygrid = np.linspace(ymin, ymax, gridlines)\n xlines = np.stack([a.ravel() for a in np.meshgrid(xpts, ygrid)])\n ylines = np.stack([a.ravel() for a in np.meshgrid(xgrid, ypts)])\n return np.concatenate([xlines, ylines], 1).T\n\ngrid = make_grid(-3, 3, -3, 3, 4, 100)",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"#@title Helper functions for visualization\ndef plot_density(data, axis):\n x, y = np.squeeze(np.split(data, 2, axis=1))\n levels = np.linspace(0.0, 0.75, 10)\n kwargs = {'levels': levels}\n return sns.kdeplot(x, y, cmap=\"viridis\", shade=True, \n shade_lowest=True, ax=axis, **kwargs)\n\n\ndef plot_points(data, axis, s=10, color='b', label=''):\n x, y = np.squeeze(np.split(data, 2, axis=1))\n axis.scatter(x, y, c=color, s=s, label=label)\n\n\ndef plot_panel(\n grid, samples, transformed_grid, transformed_samples,\n dataset, axarray, limits=True):\n if len(axarray) != 4:\n raise ValueError('Expected 4 axes for the panel')\n ax1, ax2, ax3, ax4 = axarray\n plot_points(data=grid, axis=ax1, s=20, color='black', label='grid')\n plot_points(samples, ax1, s=30, color='blue', label='samples')\n plot_points(transformed_grid, ax2, s=20, color='black', label='ode(grid)')\n plot_points(transformed_samples, ax2, s=30, color='blue', label='ode(samples)')\n ax3 = plot_density(transformed_samples, ax3)\n ax4 = plot_density(dataset, ax4)\n if limits:\n set_limits([ax1], -3.0, 3.0, -3.0, 3.0)\n set_limits([ax2], -2.0, 3.0, -2.0, 3.0)\n set_limits([ax3, ax4], -1.5, 2.5, -0.75, 1.25)\n\n\ndef set_limits(axes, min_x, max_x, min_y, max_y):\n if isinstance(axes, list):\n for axis in axes:\n set_limits(axis, min_x, max_x, min_y, max_y)\n else:\n axes.set_xlim(min_x, max_x)\n axes.set_ylim(min_y, max_y)",
"_____no_output_____"
]
],
[
[
"# FFJORD bijector\nIn this colab we demonstrate FFJORD bijector, originally proposed in the paper by Grathwohl, Will, et al. [arxiv link](https://arxiv.org/pdf/1810.01367.pdf).\n\nIn the nutshell the idea behind such approach is to establish a correspondence between a known **base distribution** and the **data distribution**.\n\nTo establish this connection, we need to\n\n 1. Define a bijective map $\\mathcal{T}_{\\theta}:\\mathbf{x} \\rightarrow \\mathbf{y}$, $\\mathcal{T}_{\\theta}^{1}:\\mathbf{y} \\rightarrow \\mathbf{x}$ between the space $\\mathcal{Y}$ on which **base distribution** is defined and space $\\mathcal{X}$ of the data domain.\n 2. Efficiently keep track of the deformations we perform to transfer the notion of probability onto $\\mathcal{X}$.\n\nThe second condition is formalized in the following expression for probability\ndistribution defined on $\\mathcal{X}$:\n\n$$\n\\log p_{\\mathbf{x}}(\\mathbf{x})=\\log p_{\\mathbf{y}}(\\mathbf{y})-\\log \\operatorname{det}\\left|\\frac{\\partial \\mathcal{T}_{\\theta}(\\mathbf{y})}{\\partial \\mathbf{y}}\\right|\n$$\n\nFFJORD bijector accomplishes this by defining a transformation \n$$\n\\mathcal{T_{\\theta}}: \\mathbf{x} = \\mathbf{z}(t_{0}) \\rightarrow \\mathbf{y} = \\mathbf{z}(t_{1}) \\quad : \\quad \\frac{d \\mathbf{z}}{dt} = \\mathbf{f}(t, \\mathbf{z}, \\theta)\n$$\n\nThis transformation is invertible, as long as function $\\mathbf{f}$ describing the evolution of the state $\\mathbf{z}$ is well behaved and the `log_det_jacobian` can be calculated by integrating the following expression.\n\n$$\n\\log \\operatorname{det}\\left|\\frac{\\partial \\mathcal{T}_{\\theta}(\\mathbf{y})}{\\partial \\mathbf{y}}\\right| = \n-\\int_{t_{0}}^{t_{1}} \\operatorname{Tr}\\left(\\frac{\\partial \\mathbf{f}(t, \\mathbf{z}, \\theta)}{\\partial \\mathbf{z}(t)}\\right) d t\n$$\n\nIn this demo we will train a FFJORD bijector to warp a gaussian distribution onto the distribution defined by `moons` dataset. This will be done in 3 steps:\n * Define **base distribution**\n * Define FFJORD bijector\n * Minimize exact log-likelihood of the dataset",
"_____no_output_____"
],
[
"First, we load the data",
"_____no_output_____"
]
],
[
[
"#@title Dataset\nDATASET_SIZE = 1024 * 8 #@param\nBATCH_SIZE = 256 #@param\nSAMPLE_SIZE = DATASET_SIZE\n\nmoons = skd.make_moons(n_samples=DATASET_SIZE, noise=.06)[0]\n\nmoons_ds = tf.data.Dataset.from_tensor_slices(moons.astype(np.float32))\nmoons_ds = moons_ds.prefetch(tf.data.experimental.AUTOTUNE)\nmoons_ds = moons_ds.cache()\nmoons_ds = moons_ds.shuffle(DATASET_SIZE)\nmoons_ds = moons_ds.batch(BATCH_SIZE)\n\nplt.figure(figsize=[8, 8])\nplt.scatter(moons[:, 0], moons[:, 1])\nplt.show()",
"_____no_output_____"
]
],
[
[
"Next, we instantiate a base distribution",
"_____no_output_____"
]
],
[
[
"base_loc = np.array([0.0, 0.0]).astype(np.float32)\nbase_sigma = np.array([0.8, 0.8]).astype(np.float32)\nbase_distribution = tfd.MultivariateNormalDiag(base_loc, base_sigma)",
"_____no_output_____"
]
],
[
[
"We use a multi-layer perceptron to model `state_derivative_fn`.\n\nWhile not necessary for this dataset, it is often benefitial to make `state_derivative_fn` dependent on time. Here we achieve this by concatenating `t` to inputs of our network.",
"_____no_output_____"
]
],
[
[
"class MLP_ODE(snt.Module):\n \"\"\"Multi-layer NN ode_fn.\"\"\"\n def __init__(self, num_hidden, num_layers, num_output, name='mlp_ode'):\n super(MLP_ODE, self).__init__(name=name)\n self._num_hidden = num_hidden\n self._num_output = num_output\n self._num_layers = num_layers\n self._modules = []\n for _ in range(self._num_layers - 1):\n self._modules.append(snt.Linear(self._num_hidden))\n self._modules.append(tf.math.tanh)\n self._modules.append(snt.Linear(self._num_output))\n self._model = snt.Sequential(self._modules)\n\n def __call__(self, t, inputs):\n inputs = tf.concat([tf.broadcast_to(t, inputs.shape), inputs], -1)\n return self._model(inputs)",
"_____no_output_____"
],
[
"#@title Model and training parameters\nLR = 1e-2 #@param\nNUM_EPOCHS = 80 #@param\nSTACKED_FFJORDS = 4 #@param\nNUM_HIDDEN = 8 #@param\nNUM_LAYERS = 3 #@param\nNUM_OUTPUT = 2",
"_____no_output_____"
]
],
[
[
"Now we construct a stack of FFJORD bijectors. Each bijector is provided with `ode_solve_fn` and `trace_augmentation_fn` and it's own `state_derivative_fn` model, so that they represent a sequence of different transformations.",
"_____no_output_____"
]
],
[
[
"#@title Building bijector\nsolver = tfp.math.ode.DormandPrince(atol=1e-5)\node_solve_fn = solver.solve\ntrace_augmentation_fn = tfb.ffjord.trace_jacobian_exact\n\nbijectors = []\nfor _ in range(STACKED_FFJORDS):\n mlp_model = MLP_ODE(NUM_HIDDEN, NUM_LAYERS, NUM_OUTPUT)\n next_ffjord = tfb.FFJORD(\n state_time_derivative_fn=mlp_model,ode_solve_fn=ode_solve_fn,\n trace_augmentation_fn=trace_augmentation_fn)\n bijectors.append(next_ffjord)\n\nstacked_ffjord = tfb.Chain(bijectors[::-1])",
"_____no_output_____"
]
],
[
[
"Now we can use `TransformedDistribution` which is the result of warping `base_distribution` with `stacked_ffjord` bijector.",
"_____no_output_____"
]
],
[
[
"transformed_distribution = tfd.TransformedDistribution(\n distribution=base_distribution, bijector=stacked_ffjord)",
"_____no_output_____"
]
],
[
[
"Now we define our training procedure. We simply minimize negative log-likelihood of the data.",
"_____no_output_____"
]
],
[
[
"#@title Training\[email protected]\ndef train_step(optimizer, target_sample):\n with tf.GradientTape() as tape:\n loss = -tf.reduce_mean(transformed_distribution.log_prob(target_sample))\n variables = tape.watched_variables()\n gradients = tape.gradient(loss, variables)\n optimizer.apply(gradients, variables)\n return loss",
"_____no_output_____"
],
[
"#@title Samples\[email protected]\ndef get_samples():\n base_distribution_samples = base_distribution.sample(SAMPLE_SIZE)\n transformed_samples = transformed_distribution.sample(SAMPLE_SIZE)\n return base_distribution_samples, transformed_samples\n\n\[email protected]\ndef get_transformed_grid():\n transformed_grid = stacked_ffjord.forward(grid)\n return transformed_grid",
"_____no_output_____"
]
],
[
[
"Plot samples from base and transformed distributions.",
"_____no_output_____"
]
],
[
[
"evaluation_samples = []\nbase_samples, transformed_samples = get_samples()\ntransformed_grid = get_transformed_grid()\nevaluation_samples.append((base_samples, transformed_samples, transformed_grid))",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/resource_variable_ops.py:1817: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\n"
],
[
"panel_id = 0\npanel_data = evaluation_samples[panel_id]\nfig, axarray = plt.subplots(\n 1, 4, figsize=(16, 6))\nplot_panel(\n grid, panel_data[0], panel_data[2], panel_data[1], moons, axarray, False)\nplt.tight_layout()",
"_____no_output_____"
],
[
"learning_rate = tf.Variable(LR, trainable=False)\noptimizer = snt.optimizers.Adam(learning_rate)\n\nfor epoch in tqdm.trange(NUM_EPOCHS // 2):\n base_samples, transformed_samples = get_samples()\n transformed_grid = get_transformed_grid()\n evaluation_samples.append(\n (base_samples, transformed_samples, transformed_grid))\n for batch in moons_ds:\n _ = train_step(optimizer, batch)",
"\r 0%| | 0/40 [00:00<?, ?it/s]"
],
[
"panel_id = -1\npanel_data = evaluation_samples[panel_id]\nfig, axarray = plt.subplots(\n 1, 4, figsize=(16, 6))\nplot_panel(grid, panel_data[0], panel_data[2], panel_data[1], moons, axarray)\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"Training it for longer with learning rate results in further improvements.",
"_____no_output_____"
],
[
"Not convered in this example, FFJORD bijector supports hutchinson's stochastic trace estimation. The particular estimator can be provided via `trace_augmentation_fn`. Similarly alternative integrators can be used by defining custom `ode_solve_fn`. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ec87e282ce4abecd1b94bbc33131e4957f56a032 | 13,096 | ipynb | Jupyter Notebook | training.ipynb | kipgparker/RLFromHumanPrefrences | 8f40902708a4f0c55295da519df35d160a9671fd | [
"MIT"
] | 1 | 2021-07-06T05:47:58.000Z | 2021-07-06T05:47:58.000Z | training.ipynb | kipgparker/RLFromHumanPrefrences | 8f40902708a4f0c55295da519df35d160a9671fd | [
"MIT"
] | null | null | null | training.ipynb | kipgparker/RLFromHumanPrefrences | 8f40902708a4f0c55295da519df35d160a9671fd | [
"MIT"
] | null | null | null | 25.184615 | 914 | 0.484575 | [
[
[
"from reward_predictor import Reward_Predictor\nfrom pref_db import PrefBuffer, PrefDB, Segment\n\nimport numpy as np\nimport torch",
"_____no_output_____"
],
[
"db_val = PrefDB(maxlen=100)\nfor i in range(10):\n db_val.append(torch.rand((32, 1 ,84, 84), dtype=torch.float), torch.rand((32, 1 ,84, 84), dtype=torch.float), 1)\n \ndb_train = PrefDB(maxlen=100)\nfor i in range(10):\n db_train.append(torch.rand((32, 1 ,84, 84), dtype=torch.float), torch.rand((32, 1 ,84, 84), dtype=torch.float), 0)",
"_____no_output_____"
],
[
"device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\nfrom a2c_ppo_acktr.envs import make_vec_envs\nenvs = make_vec_envs('PongNoFrameskip-v4', 42, 8,\n 0.99, '/tmp/gym/', device, False)\n\nreward_predictor = Reward_Predictor(\n envs.observation_space.shape\n)\nreward_predictor.to(device)\nprint('cudaa')\n#reward_predictor(s1s.cuda(), s2s.cuda())",
"Logging to /tmp/openai-2020-11-21-16-49-47-344020\nCreating dummy env object to get spaces\ncudaa\n"
],
[
"for i in range(100):\n reward_predictor.train(db_train, db_val, device)",
"_____no_output_____"
],
[
"o",
"_____no_output_____"
],
[
"#def train(self, pref_buffer):\ndef train(train_db, val_db, device):\n #train_db, val_db = pref_buffer.get_dbs()\n train_loader = torch.utils.data.DataLoader(\n train_db,\n batch_size=32,\n shuffle=False,\n num_workers=8\n )\n val_loader = torch.utils.data.DataLoader(\n val_db,\n batch_size=32,\n shuffle=False,\n num_workers=8\n )\n\n for batch in train_loader:\n s1s, s2s, prefs = batch\n\n reward_predictor.optimizer.zero_grad()\n\n preds = reward_predictor(s1s.to(device), s2s.to(device))\n\n loss = reward_predictor.criterion(preds, prefs.to(device))\n loss.backward()\n reward_predictor.optimizer.step()\n\n correct = 0\n total = 0\n with torch.no_grad():\n for batch in val_loader:\n s1s, s2s, prefs = batch\n preds = reward_predictor(s1s.to(device), s2s.to(device))\n\n predicted = torch.max(preds.data, 1)[1]\n\n total += predicted.size(0)\n correct += (predicted == prefs.to(device)).sum().item()\n\n print(f'accuracy of reward on {total} trajectories: {(100 * correct / total)}')\n \ntrain(db_train, db_val, device)",
"accuracy of reward on 10 trajectories: 60.0\n"
],
[
"train(db_train, db_val, device)",
"accuracy of reward on 10 trajectories: 50.0\n"
],
[
"prefs = torch.rand(4,2)\npreds.data",
"_____no_output_____"
],
[
"_, predicted = torch.max(preds.data, 1)\npredicted",
"_____no_output_____"
],
[
"_, predicted = torch.max(preds.data, 1)\npredicted",
"_____no_output_____"
],
[
"torch.equal(torch.max(preds.data, 1)[1])",
"_____no_output_____"
],
[
"labels = torch.max(prefs.data, 1)[1]\npredicted = torch.max(preds.data, 1)[1]\n\ntotal += predicted.size(0)\ncorrect += (predicted == labels.cuda()).sum().item()",
"_____no_output_____"
],
[
"torch.equal(torch.argmax(prefs, 1), torch.argmax(preds, 1))\n\nfrom a2c_ppo_acktr.envs import make_vec_envs\nimport garner as g\n\ng.login('[email protected]')\ng.select_pool('Deep reinforcement learning from human prefrences')",
"_____no_output_____"
],
[
"pref_db_train = PrefDB(maxlen=20000)\npref_db_val = PrefDB(maxlen=5000)\npref_buffer = PrefBuffer(garner = g, db_train=pref_db_train,\n db_val=pref_db_val, maxlen=30)",
"_____no_output_____"
],
[
"envs.observation_space.shape",
"_____no_output_____"
],
[
"s1s.shape",
"_____no_output_____"
],
[
"b = envs.observation_space.shape",
"_____no_output_____"
],
[
"b = torch.zeros((4,12,84,84))",
"_____no_output_____"
],
[
"a = (1, b[1:])\na",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec87e68156d5188d97f9f8d47701d0a8d53c70ba | 50,897 | ipynb | Jupyter Notebook | chinese_ocr/.ipynb_checkpoints/understand_detect-checkpoint.ipynb | bing1zhi2/chinese_ocr | d5b70457079e9ae5c965588c940020da742ea9c1 | [
"MIT"
] | 43 | 2019-07-23T11:28:18.000Z | 2022-01-17T12:51:18.000Z | chinese_ocr/.ipynb_checkpoints/understand_detect-checkpoint.ipynb | dun933/chinese_ocr-1 | d5b70457079e9ae5c965588c940020da742ea9c1 | [
"MIT"
] | 4 | 2019-07-25T05:04:40.000Z | 2020-01-06T08:01:36.000Z | chinese_ocr/.ipynb_checkpoints/understand_detect-checkpoint.ipynb | dun933/chinese_ocr-1 | d5b70457079e9ae5c965588c940020da742ea9c1 | [
"MIT"
] | 13 | 2019-09-07T03:34:56.000Z | 2020-10-14T22:12:01.000Z | 126.925187 | 37,720 | 0.833684 | [
[
[
"import time\n\nfrom PIL import Image\nimport cv2\nimport numpy as np\n\nimport model\nfrom utils.image import union_rbox, adjust_box_to_origin\nfrom utils.image import get_boxes, letterbox_image\nfrom global_obj import ocr_predict\nfrom config.config import IMGSIZE\nfrom utils import visualize\n",
"Using TensorFlow backend.\n"
]
],
[
[
"# 1 test on an image",
"_____no_output_____"
]
],
[
[
"detectAngle = True\npath = \"test_images/shi.png\"\n\nimg = cv2.imread(path) ##GBR\nH, W = img.shape[:2]\ntimeTake = time.time()\n",
"_____no_output_____"
],
[
"\n# 多行识别 multi line\n_, result, angle = model.model(img,\n detectAngle=detectAngle, ##是否进行文字方向检测,通过web传参控制\n config=dict(MAX_HORIZONTAL_GAP=50, ##字符之间的最大间隔,用于文本行的合并\n MIN_V_OVERLAPS=0.6,\n MIN_SIZE_SIM=0.6,\n TEXT_PROPOSALS_MIN_SCORE=0.1,\n TEXT_PROPOSALS_NMS_THRESH=0.3,\n TEXT_LINE_NMS_THRESH=0.7, ##文本行之间测iou值\n ),\n leftAdjust=True, ##对检测的文本行进行向左延伸\n rightAdjust=True, ##对检测的文本行进行向右延伸\n alph=0.1, ##对检测的文本行进行向右、左延伸的倍数\n )\n\nprint(result, angle)\nresult = union_rbox(result, 0.2)\nprint(result, angle)\n\n\n",
"text detect complete, it took 4.687s\n[{'cx': 78.5, 'cy': 13.5, 'text': '|1.《千年的愿望》', 'w': 155.05, 'h': 23.0, 'degree': 0.0}, {'cx': 32.75, 'cy': 70.25, 'text': '总希望', 'w': 65.57721759772025, 'h': 23.510864433221343, 'degree': 0.8052535989639354}, {'cx': 94.75, 'cy': 128.0, 'text': '二十岁的那个月夜', 'w': 161.14999999999998, 'h': 24.0104121494643, 'degree': -0.06638731531194668}, {'cx': 42.5, 'cy': 184.5, 'text': '能再回来', 'w': 86.05, 'h': 23.0, 'degree': 0.0}, {'cx': 85.0, 'cy': 243.0, 'text': '再重新活那么一次', 'w': 175.3, 'h': 24.0, 'degree': 0.0}, {'cx': 23.0, 'cy': 300.5, 'text': '然而', 'w': 44.55670631985086, 'h': 24.020824298928574, 'degree': -1.663921062330226}, {'cx': 32.5, 'cy': 357.5, 'text': '商时风', 'w': 65.05, 'h': 23.0, 'degree': 0.0}, {'cx': 32.75, 'cy': 415.0, 'text': '唐时雨', 'w': 65.57947154465685, 'h': 23.010864433221286, 'degree': -0.823569106588903}, {'cx': 42.75, 'cy': 473.5, 'text': '多少枝花', 'w': 86.57837423311449, 'h': 24.010412149464287, 'degree': -0.6524109307870573}, {'cx': 85.0, 'cy': 530.25, 'text': '多少个闲情的少女', 'w': 168.7008928477311, 'h': 23.5, 'degree': -0.36349282343671985}, {'cx': 115.0, 'cy': 588.0, 'text': '想她们在玉阶上转回以后', 'w': 238.3, 'h': 24.0, 'degree': 0.0}, {'cx': 102.25, 'cy': 645.0, 'text': '也只能枉然地剪下玫瑰', 'w': 211.5373435255704, 'h': 23.010864433221286, 'degree': 0.8437489635904356}, {'cx': 42.5, 'cy': 703.0, 'text': '插入瓶中', 'w': 86.05, 'h': 24.0, 'degree': 0.0}] 0\n[{'text': '|1.《千年的愿望》', 'cx': 78.5, 'cy': 13.5, 'w': 155.05, 'h': 23.0, 'degree': 0.0}, {'text': '总希望', 'cx': 32.75, 'cy': 70.25, 'w': 65.57721759772025, 'h': 23.510864433221347, 'degree': 0.8052535989639396}, {'text': '二十岁的那个月夜', 'cx': 94.75, 'cy': 128.0, 'w': 161.15, 'h': 24.010412149464294, 'degree': -0.06638731531194593}, {'text': '能再回来', 'cx': 42.5, 'cy': 184.5, 'w': 86.05000000000001, 'h': 23.0, 'degree': 3.2841453741284767e-15}, {'text': '再重新活那么一次', 'cx': 85.0, 'cy': 243.0, 'w': 175.3, 'h': 24.0, 'degree': 0.0}, {'text': '然而', 'cx': 23.0, 'cy': 300.5, 'w': 44.55670631985086, 'h': 24.02082429892859, 'degree': -1.6639210623302045}, {'text': '商时风', 'cx': 32.5, 'cy': 357.5, 'w': 65.05000000000001, 'h': 23.0, 'degree': 5.47318354533983e-15}, {'text': '唐时雨', 'cx': 32.75, 'cy': 415.0, 'w': 65.57947154465685, 'h': 23.010864433221325, 'degree': -0.8235691065889122}, {'text': '多少枝花', 'cx': 42.75, 'cy': 473.5, 'w': 86.57837423311449, 'h': 24.01041214946426, 'degree': -0.6524109307870586}, {'text': '多少个闲情的少女', 'cx': 85.0, 'cy': 530.25, 'w': 168.7008928477311, 'h': 23.499999999999922, 'degree': -0.3634928234367317}, {'text': '想她们在玉阶上转回以后', 'cx': 115.0, 'cy': 588.0, 'w': 238.3, 'h': 24.0, 'degree': 0.0}, {'text': '也只能枉然地剪下玫瑰', 'cx': 102.25, 'cy': 645.0, 'w': 211.53734352557044, 'h': 23.010864433221233, 'degree': 0.8437489635904399}, {'text': '插入瓶中', 'cx': 42.5, 'cy': 703.0, 'w': 86.05000000000001, 'h': 24.0, 'degree': 3.264804123567002e-15}] 0\n"
],
[
"print(\"result str: \\n\", )\nfor x in result:\n print(x['text'])\n\nprint(\"done\")",
"result str: \n\n|1.《千年的愿望》\n总希望\n二十岁的那个月夜\n能再回来\n再重新活那么一次\n然而\n商时风\n唐时雨\n多少枝花\n多少个闲情的少女\n想她们在玉阶上转回以后\n也只能枉然地剪下玫瑰\n插入瓶中\ndone\n"
],
[
"res = [{'text': x['text'],\n 'name': str(i),\n 'box': {'cx': x['cx'],\n 'cy': x['cy'],\n 'w': x['w'],\n 'h': x['h'],\n 'angle': x['degree']\n \n }\n } for i, x in enumerate(result)]\nres = adjust_box_to_origin(img, angle, res) ##修正box\nres",
"_____no_output_____"
]
],
[
[
"## show box",
"_____no_output_____"
]
],
[
[
"# resize the box to (m,n,2)\nboxes=[]\nfor d in res:\n x1 = d['box'][0]\n y1 = d['box'][1]\n x2 = d['box'][2]\n y2 = d['box'][3]\n x3 = d['box'][4]\n y3 = d['box'][5]\n x4 = d['box'][6]\n y4 = d['box'][7]\n xy = [[x1,y1],\n [x2,y2],\n [x3,y3],\n [x4,y4]]\n boxes.append(xy)\n\nd_boxes = np.array(boxes)\nd_boxes.shape",
"_____no_output_____"
],
[
"visualize.drow_polygon(img,boxes=d_boxes,title='result')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec87ed95e8d19e370c7bbdc94e9183d177aba44e | 833,746 | ipynb | Jupyter Notebook | TheEffectOfPsychology_TR.ipynb | iremdereli/the-effect-of-psychology-on-b12 | 7bd0796a54aafc6d79ee251739c00c03ddf19fdb | [
"MIT"
] | 4 | 2020-07-10T07:27:10.000Z | 2021-02-26T18:21:22.000Z | TheEffectOfPsychology_TR.ipynb | iremdereli/the-effect-of-psychology-on-b12 | 7bd0796a54aafc6d79ee251739c00c03ddf19fdb | [
"MIT"
] | null | null | null | TheEffectOfPsychology_TR.ipynb | iremdereli/the-effect-of-psychology-on-b12 | 7bd0796a54aafc6d79ee251739c00c03ddf19fdb | [
"MIT"
] | null | null | null | 351.791561 | 129,662 | 0.690106 | [
[
[
"# **Psikolojinin B12'ye Etkisi**\n\n#### İrem Dereli\n\nProje, bir kişinin B12 seviyesini psikoloji ve zihinsel sağlığına bakarak tespit etmeyi amaçlamaktadır.",
"_____no_output_____"
],
[
"### **İçerik**\n\n[Problem](#problem) \n[Verinin Anlaşılması](#data_understanding) \n[Verilerin Hazırlanması](#data_preparation) \n[Modelleme](#modeling) \n[Değerlendirme](#evaluation) \n[Referanslar](#references) \n",
"_____no_output_____"
],
[
"## **Problem** <a class=\"anchor\" id=\"problem\"></a>",
"_____no_output_____"
],
[
"Psikoloji ve zihinsel sağlık sorunları, bu yüzyılda çok yaygın olan sorunlardandır. Ve B12 vitamini eksikliği, anemi, unutkanlık, nörolojik, gastrointestinal problemler vb. gibi büyük sağlık sorunlarına sebep olmaktadır. Psikolojik problemler, çocukluk tramvaları, özel hayat, ailesel sorunlar, iş hayatı ve bunlar gibi sebeplerden ortaya çıkabilmektedir.<br>\nBir kişinin B12 seviyesi düşük, normal veya yüksek olabilir. Tıbbi makalelere bağlı olarak, B12'nin alt limiti 200pg/ml'dir ve 300pg/ml'den düşük olmamalıdır. 300pg/ml'den az değerler B12 için çok düşük seviyelerdir ve bir kişinin günlerce hastanede kalmasına sebep olabilir. Ancak doktorlar, B12 seviyeniz 500pg/ml'den düşük ise oral takviye kullanmanızı önerir.\n\n\n---\n\nYukarıda bahsedildiği üzere, B12 seviyesinin sonucu düşük, normal veya yüksek olacaktır. Ve bu, üç kategorili bir sınıflandırma problemidir.",
"_____no_output_____"
],
[
"## **Verinin Anlaşılması**<a class=\"anchor\" id=\"data_understanding\"></a>",
"_____no_output_____"
],
[
"### Veri İncelemesi",
"_____no_output_____"
],
[
"Ulusal Sağlık ve Beslenme İnceleme Anketi (NHANES), www.cdc.gov adresinde çevrimiçi veri kümesi deposudur. 1999 yılından başlayarak, her iki yıl için bir tane olmak üzere toplamda 11 veri kümesi bulunmaktadır. Bu projede seçilen veri kümesi, B12 verisinin fazlalığından dolayı 2013-2014 yıllarındaki veri kümesidir. Ankete katılan toplam kişi sayısı 14,332'dir.\n\nVeri kümesi 5 bölüm içerir. Bunlar demografi, inceleme sonuçları, laboratuvar sonuçları, diyet verileri ve anket sorularıdır. Bu projede demografi, laboratuvar sonuçları ve anket bölümü kullanılacaktır.\n\nDemografi kısmı kişinin kişisel, ailesel ve evsel bilgilerini içermektedir. Cinsiyet ve yaş özellikleri bu kısımdan alınacaktır.\n\nLaboratuvar sonuçları kişinin kan değerlerini içermektedir ve yalnızca B12 değeri kullanılacaktır. Laboratuvar verileri sadece 20 ile 80 yaş aralığındaki insanlar için mevcuttur.\n\nVe son olarak, anket bölümü zihinsel sağlık taraması, ilişki, aile ve örnek kişi sorularını içerir. Psikoloji, zihinsel sağlık ve ev geliri, uyku bozuklukları gibi psikolojiyle alakalı sorular modelde kullanılacaktır.",
"_____no_output_____"
]
],
[
[
"# pandas kütüphanesini yükle\nimport pandas as pd\n\n# veri kümesini yükle\ndemografi = pd.read_csv('dataset/demographic.csv', index_col=False)\nlab = pd.read_csv('dataset/labs.csv', index_col=False)\nanket = pd.read_csv('dataset/questionnaire.csv', index_col=False)",
"_____no_output_____"
]
],
[
[
"### Demografi verisinin incelemesi",
"_____no_output_____"
]
],
[
[
"demografi.shape",
"_____no_output_____"
]
],
[
[
"Demografi veri kümesinde 47 özellik ve 10175 kayıt bulunmaktadır. Model, bu veri kümesinden yalnızca yaş ve cinsiyeti kullanacaktır.<br>RIAGENDR cinsiyeti, RIDAGEYR yaşı temsil etmektedir.\n",
"_____no_output_____"
]
],
[
[
"demografi.head()",
"_____no_output_____"
],
[
"# id, cinsiyet ve yaş sütununu al\ndemografi = demografi[['SEQN', 'RIDAGEYR', 'RIAGENDR']]\n\n# sütunların isimlerini değiştir\ndemografi.rename(columns={'SEQN': 'ID', 'RIDAGEYR': 'Yas', 'RIAGENDR': 'Cinsiyet'}, inplace=True)\n\ndemografi.head()",
"_____no_output_____"
]
],
[
[
"Cinsiyet sütununda, 1 erkeği, 2 kadını temsil etmektedir.<br>\nCinsiyet dağılımının görüntülemesi\n",
"_____no_output_____"
]
],
[
[
"# görüntüleme kütüphanelerini yükle\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\nsns.countplot(demografi['Cinsiyet'], palette='husl')\nplt.ylabel('Sayı')\nplt.title('Cinsiyet Dağılımı')\nplt.show()",
"_____no_output_____"
],
[
"erkek = (demografi['Cinsiyet']==1).sum()\nkadın = (demografi['Cinsiyet']==2).sum()\n\nprint(\"Toplam {} erkek katılımcı vardır\".format(erkek))\nprint(\"Toplam {} kadın katılımcı vardır\".format(kadın))",
"Toplam 5003 erkek katılımcı vardır\nToplam 5172 kadın katılımcı vardır\n"
],
[
"print(\"Toplam {} farklı yaş vardır\".format(len(demografi['Yas'].unique())))",
"Toplam 81 farklı yaş vardır\n"
]
],
[
[
"### Laboratuvar verilerinin incelenmesi",
"_____no_output_____"
]
],
[
[
"lab.shape",
"_____no_output_____"
],
[
"lab.head()",
"_____no_output_____"
]
],
[
[
"Laboratuvar verisinde 424 özellik ve 9813 kayıt bulunmaktadır. Bu veri kümesinden yalnızca B12 sütunu kullanılacaktır.<br>LBDB12 sütunu B12'yi temsil etmektedir.",
"_____no_output_____"
]
],
[
[
"# id ve b12 sütunlarını al\nlab = lab[['SEQN', 'LBDB12']]\n\n# sütunların isimlerini değiştir\nlab.rename(columns={'SEQN': 'ID', 'LBDB12': 'B12'}, inplace=True)\n\nlab.head()",
"_____no_output_____"
],
[
"lab['B12'].dtype",
"_____no_output_____"
]
],
[
[
"B12 verileri ondalıklı sayı olarak tutulmaktadır.",
"_____no_output_____"
]
],
[
[
"lab['B12'].isnull().sum()",
"_____no_output_____"
]
],
[
[
"4497 kayıt B12 verisi içermemektedir.",
"_____no_output_____"
]
],
[
[
"lab['B12'].describe()",
"_____no_output_____"
]
],
[
[
"Veri kümesindeki maksimum B12 değeri 26801, minimum ise 18'dir. İnsanların ortalama olarak normal B12 değerine sahip olduğunu söyleyebiliriz.\n\n---\n\n",
"_____no_output_____"
],
[
"### Anket verilerinin incelenmesi",
"_____no_output_____"
]
],
[
[
"anket.shape",
"_____no_output_____"
]
],
[
[
"Bu veri kümesi, diğerleriyle karşılaştırıldığında daha fazla özellik içermekte. Fakat yukarıda bahsedildiği üzere, sadece zihinsel sağlık ve psikoloji ile alakalı sütunlar seçilecektir.",
"_____no_output_____"
]
],
[
[
"anket.head()",
"_____no_output_____"
]
],
[
[
"Anket veri kümesinden seçilecek sütunlar aşağıda listelenmiştir.\n\n* **DPQ010:** Bir şeyler yapma isteğinin az olması\n* **DPQ020:** Keyifsiz, depresif veya umutsuz hissetmek\n* **DPQ030:** Uyumakta veya fazla uyumakta sorun yaşamak\n* **DPQ040:** Yorgun veya az enerjik hissetmek\n* **DPQ050:** İştahsızlık veya aşırı yemek\n* **DPQ060:** Kendin hakkında kötü hissetmek\n* **DPQ070:** Bir şeylere odaklanmakta sorun yaşamak\n* **DPQ080:** Hareket ederken veya konuşurken yavaş olmak veya çok hızlı olmak\n* **DPQ090:** Ölmenin kendi için daha iyi olacağını düşünmek\n* **DPQ100:** İşteki görevlerini yapmakta, evdeki işleri idare etmekte veya insanlarla geçimde ne kadar zorluk yaşıyorsun?\n<br><br>\n* **SLQ050:** Hiç doktoruna uyumakta zorlandığını söyledin mi?\n* **SLQ060:** Hiç doktorun uyku bozukluğun olduğunu söyledi mi?\n<br><br>\n* **IND235:** Aylık aile geliri\n\n",
"_____no_output_____"
]
],
[
[
"# sütunları al\nanket = anket[['SEQN', 'DPQ010', 'DPQ020', 'DPQ030', 'DPQ040', 'DPQ050', 'DPQ060', 'DPQ070', 'DPQ080', 'DPQ090', 'DPQ100', 'SLQ050', 'SLQ060', 'IND235']]\n\n# id sütununun ismini değiştir\nanket.rename(columns={'SEQN': 'ID'}, inplace=True)\n\nanket.head()",
"_____no_output_____"
]
],
[
[
"Seçilecek sütunlardan biri DPQ020'dir. Katılımcılara 'Keyifsiz, depresif veya umutsuz hissetmek' sorusunu sorar. Bu sütunu inceleyelim. Mental sağlık ile alakalı diğer tüm sütunlar da aynıdır (DPQ ile başlayan sütunlar).",
"_____no_output_____"
]
],
[
[
"anket['DPQ020'].unique()",
"_____no_output_____"
]
],
[
[
"\n\n* 0: Hiç\n* 1: Birkaç gün ara ile\n* 2: Günlerin yarısından fazla\n* 3: Neredeyse her gün\n* 7: Reddedildi\n* 9: Bilmiyor\n* Eksik değerler\n\n",
"_____no_output_____"
],
[
"Bir diğer seçilecek sütun uyku bozukluklarıyla alakalı ve bu sütun da kategorik değerler içermektedir.",
"_____no_output_____"
]
],
[
[
"anket['SLQ050'].unique()",
"_____no_output_____"
]
],
[
[
"\n\n* 1: Evet\n* 2: Hayır\n* 9: Bilmiyor\n\n",
"_____no_output_____"
],
[
"## **Verilerin Hazırlanması** <a class=\"anchor\" id=\"data_preparation\"></a>",
"_____no_output_____"
],
[
"Kategorik değer içermeyen yaş sütunu, modele pek bir anlam ifade etmeyecektir. Değerlerin kategorize edilmesi gerekmektedir ve aşağıda listelenmiştir.",
"_____no_output_____"
],
[
"\n\n* 1: 20 - 29\n* 2: 30 - 39\n* 3: 40 - 49\n* 4: 50 - 59\n* 5: 60 - 69\n* 6: 70 - 80\n",
"_____no_output_____"
]
],
[
[
"# veriyi kategorize et\ndemografi['Yas'] = pd.cut(demografi['Yas'], bins=[20, 30, 40, 50, 60, 70, 80], labels=[1, 2, 3, 4, 5, 6], include_lowest=True)\n\ndemografi.head()",
"_____no_output_____"
]
],
[
[
"Veri kümesinin boş değer içermesinin sebebi yukarıda bahsedildiği üzere, laboratuvar verisinin yalnızca 20 ile 80 yaş aralığındaki insanları kapsamasından kaynaklanmaktadır. 20 ile 80 yaş aralığının dışındaki insanlar boş değere sahip olacaktır.\n\n\n",
"_____no_output_____"
]
],
[
[
"# yaş dağılımının gösterimi\nsns.countplot(demografi['Yas'], palette='husl')\nplt.ylabel('Sayı')\nplt.title('Yaş Dağılımı Grafiği')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Yaşların dağılımı genellikle birbirine yakındır. En fazla katılımın 20-29 yaş aralığından olduğunu söyleyebiliriz. En az katılım da 70-80 yaş aralığındandır.\n\n---\n\n",
"_____no_output_____"
],
[
"B12 değeri boş olan kayıtlar veri kümesinden atılacaktır. Çünkü model B12 değerini tahmin etme üzerine kurulmuştur.",
"_____no_output_____"
]
],
[
[
"# B12 sütunundaki değeri boş olan kayıtları at\nlab.dropna(axis=0, inplace=True)\n\nlab.shape",
"_____no_output_____"
]
],
[
[
"Boş olan kayıtları attıktan sonra elimizde 5316 kayıt kalmıştır.",
"_____no_output_____"
],
[
"Laboratuvar veri kümesinde 424 özellik var. İhtiyacımız olan sadece B12 değeridir. Ve B12 değerleri de aşağıda gösterildiği gibi kategorize edilecektir.\n\n\n* Düşük = 1: 0 - 300\n* Normal = 2: 301 - 950\n* Yüksek = 3: 950 - 27000\n\n",
"_____no_output_____"
]
],
[
[
"# veriyi kategorize et\nlab['B12'] = pd.cut(lab['B12'], bins=[0, 300, 950, 27000], labels=[1, 2, 3])\n\nlab.head()",
"_____no_output_____"
],
[
"# B12 dağılımını görüntüle\nsns.countplot(lab['B12'], label='Toplam', palette='husl')\nplt.title('B12\\'nin dağılımı')\nplt.show()",
"_____no_output_____"
],
[
"lab['B12'].value_counts()",
"_____no_output_____"
]
],
[
[
"Grafikten görebileceğimiz üzere, genellikle insanlar normal B12 seviyesine sahiptir. Ancak bu durum, verinin dengeli olmadığını da gösteriyor. Verinin böyle olması, modelin 2. kategoriye meyilli olmasına sebep olacaktır.",
"_____no_output_____"
],
[
"Demografi, laboratuvar ve anket bölümlerini birleştirelim.",
"_____no_output_____"
]
],
[
[
"veri = lab.merge(demografi, on=\"ID\").merge(anket, on=\"ID\")\n\nveri.head()",
"_____no_output_____"
]
],
[
[
"Id sütunu artık gereksizdir. Veri kümesinden atacağız.",
"_____no_output_____"
]
],
[
[
"veri.drop('ID', axis=1, inplace=True)\n\nveri.head()",
"_____no_output_____"
]
],
[
[
"Sütunların değerlerini inceleyelim.",
"_____no_output_____"
]
],
[
[
"veri.hist(figsize=(15, 10), grid=False)\nplt.show()",
"_____no_output_____"
]
],
[
[
"İnsanların sorulara genelde 'hiç (0)' cevabı verdiğini söyleyebiliriz.",
"_____no_output_____"
]
],
[
[
"veri.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 5316 entries, 0 to 5315\nData columns (total 16 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 B12 5316 non-null category\n 1 Yas 5316 non-null category\n 2 Cinsiyet 5316 non-null int64 \n 3 DPQ010 4875 non-null float64 \n 4 DPQ020 4873 non-null float64 \n 5 DPQ030 4872 non-null float64 \n 6 DPQ040 4872 non-null float64 \n 7 DPQ050 4872 non-null float64 \n 8 DPQ060 4871 non-null float64 \n 9 DPQ070 4871 non-null float64 \n 10 DPQ080 4871 non-null float64 \n 11 DPQ090 4870 non-null float64 \n 12 DPQ100 3315 non-null float64 \n 13 SLQ050 5316 non-null float64 \n 14 SLQ060 5316 non-null float64 \n 15 IND235 5113 non-null float64 \ndtypes: category(2), float64(13), int64(1)\nmemory usage: 633.7 KB\n"
]
],
[
[
"Veri kümesinde toplamda 5316 kayıt bulunmakta. Fakat bazı sütunlar boş değerler içermekte. Özellikle, DPQ100 sütunu, diğerleriyle karşılaştırıldığında çok fazla boş değer bulundurmakta. Ortalama değer ile boş değerleri değiştirmek modelin daha kötü çalışmasına sebep olmakta, bu yüzden bu sütunu veri kümesinden atacağız.",
"_____no_output_____"
]
],
[
[
"veri.drop('DPQ100', axis=1, inplace=True)\nveri.head()",
"_____no_output_____"
],
[
"veri.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 5316 entries, 0 to 5315\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 B12 5316 non-null category\n 1 Yas 5316 non-null category\n 2 Cinsiyet 5316 non-null int64 \n 3 DPQ010 4875 non-null float64 \n 4 DPQ020 4873 non-null float64 \n 5 DPQ030 4872 non-null float64 \n 6 DPQ040 4872 non-null float64 \n 7 DPQ050 4872 non-null float64 \n 8 DPQ060 4871 non-null float64 \n 9 DPQ070 4871 non-null float64 \n 10 DPQ080 4871 non-null float64 \n 11 DPQ090 4870 non-null float64 \n 12 SLQ050 5316 non-null float64 \n 13 SLQ060 5316 non-null float64 \n 14 IND235 5113 non-null float64 \ndtypes: category(2), float64(12), int64(1)\nmemory usage: 592.1 KB\n"
]
],
[
[
"Sütunu veri kümesinden attıktan sonra, 13 özelliğimiz ve hala boş değerlerimiz bulunmakta. Zihinsel sağlık ile alakalı (DPQ ile başlayan sütunlar) özellikler, diğer özellikler ile karşılaştırıldığında daha önemlidir. Bu yüzden, bu özellikler boş değer içeremez. Zihinsel sağlık özelliklerinde boş değer içeren kayıtlar veri kümesinden atılacaktır.",
"_____no_output_____"
]
],
[
[
"veri.dropna(subset=['DPQ010', 'DPQ020', 'DPQ030', 'DPQ040', 'DPQ050', 'DPQ060', 'DPQ070', 'DPQ080', 'DPQ090'], axis=0, inplace=True)\nveri.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 4870 entries, 0 to 5314\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 B12 4870 non-null category\n 1 Yas 4870 non-null category\n 2 Cinsiyet 4870 non-null int64 \n 3 DPQ010 4870 non-null float64 \n 4 DPQ020 4870 non-null float64 \n 5 DPQ030 4870 non-null float64 \n 6 DPQ040 4870 non-null float64 \n 7 DPQ050 4870 non-null float64 \n 8 DPQ060 4870 non-null float64 \n 9 DPQ070 4870 non-null float64 \n 10 DPQ080 4870 non-null float64 \n 11 DPQ090 4870 non-null float64 \n 12 SLQ050 4870 non-null float64 \n 13 SLQ060 4870 non-null float64 \n 14 IND235 4702 non-null float64 \ndtypes: category(2), float64(12), int64(1)\nmemory usage: 542.5 KB\n"
]
],
[
[
"Şimdi, 4870 kaydımız ve hala IND235 özelliğinde boş değerlerimiz var. DPQ özellikleri önemli olduğundan dolayı, bu kayıtları veri kümesinden atmayıp, boş değerleri ortalama değer ile değiştireceğiz.",
"_____no_output_____"
]
],
[
[
"veri['IND235'].describe()",
"_____no_output_____"
]
],
[
[
"Ortalama değer 6.95 ~ 7'dir. Ayrıca veri kümesinde 77 'reddedildi' ve 99 'bilmiyor' değerlerini temsil etmektedir. Bu değerleri de 7 ile değiştireceğiz.",
"_____no_output_____"
]
],
[
[
"veri['IND235'].replace({77: 7, 99: 7, None: 7}, inplace=True)",
"_____no_output_____"
]
],
[
[
"DPQ ve SLQ özelliklerinde de ayrıca 7 reddedildiyi, 9 bilmiyoru temsil etmekte. Bunları da en çok görülen değer (0 = hiç) ile değiştireceğiz.",
"_____no_output_____"
]
],
[
[
"veri[['DPQ010', 'DPQ020', 'DPQ030', 'DPQ040', 'DPQ050', 'DPQ060', 'DPQ070', 'DPQ080', 'DPQ090', 'SLQ050', 'SLQ060']].replace({7: 0, 9: 0})\nveri.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 4870 entries, 0 to 5314\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 B12 4870 non-null category\n 1 Yas 4870 non-null category\n 2 Cinsiyet 4870 non-null int64 \n 3 DPQ010 4870 non-null float64 \n 4 DPQ020 4870 non-null float64 \n 5 DPQ030 4870 non-null float64 \n 6 DPQ040 4870 non-null float64 \n 7 DPQ050 4870 non-null float64 \n 8 DPQ060 4870 non-null float64 \n 9 DPQ070 4870 non-null float64 \n 10 DPQ080 4870 non-null float64 \n 11 DPQ090 4870 non-null float64 \n 12 SLQ050 4870 non-null float64 \n 13 SLQ060 4870 non-null float64 \n 14 IND235 4870 non-null float64 \ndtypes: category(2), float64(12), int64(1)\nmemory usage: 542.5 KB\n"
]
],
[
[
"Kategori şeklinde tutulan yaş ve B12 sütunlarını, tam sayı şekline çevirelim",
"_____no_output_____"
]
],
[
[
"veri['B12'] = veri['B12'].astype(int)\nveri['Yas'] = veri['Yas'].astype(int)",
"_____no_output_____"
]
],
[
[
"Şimdi veri kümemiz hazır.",
"_____no_output_____"
]
],
[
[
"# numpy kütüphanesini yükle\nimport numpy as np\n\n# grafik büyüklüğü\nplt.figure(figsize=(15, 10))\n\n# korelasyon grafiğinin yarısını alabilmek için numpy kütüphanesi ile alt üçgeni alıyoruz\nmask = np.triu(veri.iloc[:, 1:].corr())\n\nsns.heatmap(veri.iloc[:, 1:].corr(), annot=True, mask=mask)\n\n# y eksenindeki yazıları yatay yap\nplt.yticks(rotation=0)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"\n\n* DPQ ile başlayan sütunların birbirleriyle pozitif ve diğer sütunlarla kıyaslandığında yüksek korelasyona sahip olduğunu söyleyebiliriz.\n* SLQ ile başlayan sütunların yüksek korelasyona sahip olmadığını ve pozitif korelasyona sahip olduğunu ancak cinsiyet hariç diğer tüm sütunlarla negatif korelasyona sahip olduğunu görebiliyoruz (SLQ050 sütunu cinsiyetle de negatif korelasyona sahip).\n* Toplam aylık gelirin diğer sütunlarla bir korelasyona sahip olmadığını görebiliyoruz.\n",
"_____no_output_____"
],
[
"## **Modelleme** <a class=\"anchor\" id=\"modeling\"></a>",
"_____no_output_____"
],
[
"### Dengesiz Veri İçin",
"_____no_output_____"
]
],
[
[
"# model kütüphanelerini yükle\nfrom sklearn.model_selection import train_test_split\n\nfrom sklearn import tree\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn import svm\n\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.utils.extmath import density\n\nfrom sklearn import metrics\n\n# veriyi özellikler ve hedef olarak ayır\nX = veri.iloc[:, 1:15]\ny = veri.iloc[:, 0]\n\n# test ve eğitim verisi olarak böl\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42, shuffle=True) # 70% eğitim ve 30% test",
"_____no_output_____"
]
],
[
[
"Benchmark sınıflandırması, farklı algoritmaların sonuçlarını karşılaştırmamızı sağlayan bir fonksiyondur.",
"_____no_output_____"
]
],
[
[
"from time import time\n# Benchmark sınıflandırması\ndef benchmark(clf, X_train, y_train, X_test, y_test):\n print('_' * 80)\n print(\"Training: \")\n print(clf)\n t0 = time()\n clf.fit(X_train, y_train)\n train_time = time() - t0\n print(\"train time: %0.3fs\" % train_time)\n\n t0 = time()\n pred = clf.predict(X_test)\n test_time = time() - t0\n print(\"test time: %0.3fs\" % test_time)\n\n score = metrics.accuracy_score(y_test, pred)\n print(\"accuracy: %0.3f\" % score)\n\n if hasattr(clf, 'coef_'):\n print(\"dimensionality: %d\" % clf.coef_.shape[1])\n print(\"density: %f\" % density(clf.coef_))\n\n print(\"classification report:\")\n print(metrics.classification_report(y_test, pred))\n\n print(\"confusion matrix:\")\n print(metrics.confusion_matrix(y_test, pred))\n\n clf_descr = str(clf).split('(')[0]\n return clf_descr, score, train_time, test_time",
"_____no_output_____"
],
[
"# Farklı algoritmalar\nresults = []\nfor clf, name in (\n (tree.DecisionTreeClassifier(), \"Decision Tree\"),\n (RandomForestClassifier(n_estimators=100), \"Random Forest\"),\n (GaussianNB(), \"Gauissian Naive Bayes\"),\n (KNeighborsClassifier(n_neighbors=10), \"kNN\"),\n (LogisticRegression(), \"Logistic Regression\"),\n (svm.SVC(), \"Support Vector Machines\"),\n ):\n print('=' * 80)\n print(name)\n results.append(benchmark(clf, X_train, y_train, X_test, y_test))\n",
"================================================================================\nDecision Tree\n________________________________________________________________________________\nTraining: \nDecisionTreeClassifier(ccp_alpha=0.0, class_weight=None, criterion='gini',\n max_depth=None, max_features=None, max_leaf_nodes=None,\n min_impurity_decrease=0.0, min_impurity_split=None,\n min_samples_leaf=1, min_samples_split=2,\n min_weight_fraction_leaf=0.0, presort='deprecated',\n random_state=None, splitter='best')\ntrain time: 0.019s\ntest time: 0.002s\naccuracy: 0.641\nclassification report:\n precision recall f1-score support\n\n 1 0.10 0.10 0.10 169\n 2 0.78 0.80 0.79 1116\n 3 0.19 0.15 0.16 176\n\n accuracy 0.64 1461\n macro avg 0.35 0.35 0.35 1461\nweighted avg 0.63 0.64 0.63 1461\n\nconfusion matrix:\n[[ 17 133 19]\n [127 894 95]\n [ 28 122 26]]\n================================================================================\nRandom Forest\n________________________________________________________________________________\nTraining: \nRandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,\n criterion='gini', max_depth=None, max_features='auto',\n max_leaf_nodes=None, max_samples=None,\n min_impurity_decrease=0.0, min_impurity_split=None,\n min_samples_leaf=1, min_samples_split=2,\n min_weight_fraction_leaf=0.0, n_estimators=100,\n n_jobs=None, oob_score=False, random_state=None,\n verbose=0, warm_start=False)\ntrain time: 0.350s\ntest time: 0.034s\naccuracy: 0.736\nclassification report:\n precision recall f1-score support\n\n 1 0.17 0.04 0.06 169\n 2 0.77 0.95 0.85 1116\n 3 0.16 0.03 0.06 176\n\n accuracy 0.74 1461\n macro avg 0.37 0.34 0.32 1461\nweighted avg 0.62 0.74 0.66 1461\n\nconfusion matrix:\n[[ 6 157 6]\n [ 27 1064 25]\n [ 2 168 6]]\n================================================================================\nGauissian Naive Bayes\n________________________________________________________________________________\nTraining: \nGaussianNB(priors=None, var_smoothing=1e-09)\ntrain time: 0.003s\ntest time: 0.001s\naccuracy: 0.721\nclassification report:\n precision recall f1-score support\n\n 1 0.12 0.04 0.05 169\n 2 0.77 0.93 0.84 1116\n 3 0.19 0.07 0.10 176\n\n accuracy 0.72 1461\n macro avg 0.36 0.34 0.33 1461\nweighted avg 0.62 0.72 0.66 1461\n\nconfusion matrix:\n[[ 6 156 7]\n [ 37 1036 43]\n [ 9 155 12]]\n================================================================================\nkNN\n________________________________________________________________________________\nTraining: \nKNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',\n metric_params=None, n_jobs=None, n_neighbors=10, p=2,\n weights='uniform')\ntrain time: 0.009s\ntest time: 0.099s\naccuracy: 0.760\nclassification report:\n precision recall f1-score support\n\n 1 0.00 0.00 0.00 169\n 2 0.76 1.00 0.86 1116\n 3 0.00 0.00 0.00 176\n\n accuracy 0.76 1461\n macro avg 0.25 0.33 0.29 1461\nweighted avg 0.58 0.76 0.66 1461\n\nconfusion matrix:\n[[ 0 167 2]\n [ 2 1111 3]\n [ 1 175 0]]\n================================================================================\nLogistic Regression\n________________________________________________________________________________\nTraining: \nLogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,\n intercept_scaling=1, l1_ratio=None, max_iter=100,\n multi_class='auto', n_jobs=None, penalty='l2',\n random_state=None, solver='lbfgs', tol=0.0001, verbose=0,\n warm_start=False)\ntrain time: 0.106s\ntest time: 0.001s\naccuracy: 0.763\ndimensionality: 14\ndensity: 1.000000\nclassification report:\n precision recall f1-score support\n\n 1 0.00 0.00 0.00 169\n 2 0.76 1.00 0.87 1116\n 3 0.00 0.00 0.00 176\n\n accuracy 0.76 1461\n macro avg 0.25 0.33 0.29 1461\nweighted avg 0.58 0.76 0.66 1461\n\nconfusion matrix:\n[[ 0 169 0]\n [ 1 1115 0]\n [ 0 176 0]]\n================================================================================\nSupport Vector Machines\n________________________________________________________________________________\nTraining: \nSVC(C=1.0, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,\n decision_function_shape='ovr', degree=3, gamma='scale', kernel='rbf',\n max_iter=-1, probability=False, random_state=None, shrinking=True,\n tol=0.001, verbose=False)\ntrain time: 0.649s\ntest time: 0.066s\naccuracy: 0.764\nclassification report:\n precision recall f1-score support\n\n 1 0.00 0.00 0.00 169\n 2 0.76 1.00 0.87 1116\n 3 0.00 0.00 0.00 176\n\n accuracy 0.76 1461\n macro avg 0.25 0.33 0.29 1461\nweighted avg 0.58 0.76 0.66 1461\n\nconfusion matrix:\n[[ 0 169 0]\n [ 0 1116 0]\n [ 0 176 0]]\n"
],
[
"# grafikler\n\nindices = np.arange(len(results))\n\nresults = [[x[i] for x in results] for i in range(4)]\n\nclf_names, score, training_time, test_time = results\ntraining_time = np.array(training_time) / np.max(training_time)\ntest_time = np.array(test_time) / np.max(test_time)\n\nplt.figure(figsize=(12, 8))\nplt.title(\"Score\")\nplt.barh(indices, score, .2, label=\"score\", color='navy')\nplt.barh(indices + .3, training_time, .2, label=\"training time\",\n color='c')\nplt.barh(indices + .6, test_time, .2, label=\"test time\", color='darkorange')\nplt.yticks(())\nplt.legend(loc='best')\nplt.subplots_adjust(left=.25)\nplt.subplots_adjust(top=.95)\nplt.subplots_adjust(bottom=.05)\n\nfor i, c in zip(indices, clf_names):\n plt.text(-.3, i, c)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Modellerin doğruluk oranı karşılaştırıldığında, en yüksek doğruluk oranına sahip iki algoritma Logistic Regression ve Support Vector Classifier olduğunu, fakat 'precision', 'recall', 'f1score' değerlerini göz önüne aldığımız zaman, çok başarılı modeller olmadığını, yalnızca 'Normal' değerli B12 tahmini yaptığını görebiliyoruz. Bu yüzden, bu iki modelle karşılaştırdığımızda doğruluk oranı çok da farklı olmayan, ancak diğer kategorilere de tahmin yapan Gaussian Naive Bayes algoritmasını projemizde kullanacağız.",
"_____no_output_____"
],
[
"Gaussian Naive Bayes algoritmasında 'hyperparameters' yapılamadığı için Grid Search kullanamıyoruz. Bunun yerine, Naive Bayes'ten sonra gelen en iyi model olan Random Forest'ta daha iyi sonuç alıp alamayacağımızı görmek açısından Grid Search algoritmasını deniyoruz.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\n\nforest = RandomForestClassifier(random_state=1, n_estimators = 10, min_samples_split = 1)\n\nn_estimators = [100, 300, 500, 800, 1200]\nmax_depth = [5, 8, 15, 25, 30]\nmin_samples_split = [2, 5, 10, 15, 100]\nmin_samples_leaf = [1, 2, 5, 10] \n\nhyperF = dict(n_estimators = n_estimators, max_depth = max_depth, \n min_samples_split = min_samples_split, \n min_samples_leaf = min_samples_leaf)\n\ngridF = GridSearchCV(forest, hyperF, cv = 3, verbose = 1, \n n_jobs = -1)\nbestF = gridF.fit(X_train, y_train)",
"Fitting 3 folds for each of 500 candidates, totalling 1500 fits\n[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 34 tasks | elapsed: 12.0s\n[Parallel(n_jobs=-1)]: Done 184 tasks | elapsed: 54.1s\n[Parallel(n_jobs=-1)]: Done 434 tasks | elapsed: 2.1min\n[Parallel(n_jobs=-1)]: Done 784 tasks | elapsed: 3.8min\n[Parallel(n_jobs=-1)]: Done 1234 tasks | elapsed: 26.8min\n[Parallel(n_jobs=-1)]: Done 1500 out of 1500 | elapsed: 28.4min finished\n"
],
[
"# modelin en iyi çalıştığı parametreler\ngridF.best_params_",
"_____no_output_____"
],
[
"gridF.refit\npreds = gridF.predict(X_test)\nprobs = gridF.predict_proba(X_test)\n\n# doğruluk oranı\nnp.mean(preds == y_test)",
"_____no_output_____"
],
[
"print(metrics.classification_report(y_test, preds, target_names=['Düşük', 'Normal', 'Yüksek']))",
"precision recall f1-score support\n\n Düşük 0.00 0.00 0.00 169\n Normal 0.76 1.00 0.87 1116\n Yüksek 0.00 0.00 0.00 176\n\n accuracy 0.76 1461\n macro avg 0.25 0.33 0.29 1461\nweighted avg 0.58 0.76 0.66 1461\n\n"
]
],
[
[
"Grid Search sonucunda, doğruluk oranının %3 arttığını, ancak sınıflandırma raporunda negatif bir etkiye sebep olduğunu görüyoruz. Düşük ve yüksek B12 kategorilerini (1 ve 3) tahmin edemediğini görüyoruz. Bu yüzden, Naive Bayes algoritmasıyla devam ediyoruz.",
"_____no_output_____"
],
[
"### Dengeli Veri İçin",
"_____no_output_____"
],
[
"Verimizde \"Normal\" B12 değerine sahip olan katılımcıların çok daha fazla olduğunu yukarıda görmüştük. Bu yüzden, kategorileri daha dengeli hale getirip, modelimizi tekrar oluşturacağız. Diğer kategorilerde yaklaşık 530 veri bulunmakta. Normal kategoriden 1000 tane veri seçiyoruz. Bunun sebebi de, çok fazla veri kaybetmek istemememiz.",
"_____no_output_____"
]
],
[
[
"dengeli_veri = veri.copy()\n\ndengeli_veri[dengeli_veri['B12'] == 2] = dengeli_veri[dengeli_veri['B12'] == 2].sample(1000, random_state=42)\ndengeli_veri = dengeli_veri[dengeli_veri['B12'].notnull()]\ndengeli_veri['B12'].value_counts()\n",
"_____no_output_____"
],
[
"# veriyi özellikler ve hedef olarak ayır\nX_dengeli = dengeli_veri.iloc[:, 1:15]\ny_dengeli = dengeli_veri.iloc[:, 0]\n\n# test ve eğitim verisi olarak böl\nX_train_dengeli, X_test_dengeli, y_train_dengeli, y_test_dengeli = train_test_split(X_dengeli, y_dengeli, test_size=0.3, random_state=42, shuffle=True) # 70% training and 30% test",
"_____no_output_____"
],
[
"results_dengeli = []\nfor clf, name in (\n (tree.DecisionTreeClassifier(), \"Decision Tree\"),\n (RandomForestClassifier(n_estimators=100), \"Random Forest\"),\n (GaussianNB(), \"Gauissian Naive Bayes\"),\n (KNeighborsClassifier(n_neighbors=10), \"kNN\"),\n (LogisticRegression(), \"Logistic Regression\"),\n (svm.SVC(), \"Support Vector Machines\"),\n ):\n print('=' * 80)\n print(name)\n results_dengeli.append(benchmark(clf, X_train_dengeli, y_train_dengeli, X_test_dengeli, y_test_dengeli))",
"================================================================================\nDecision Tree\n________________________________________________________________________________\nTraining: \nDecisionTreeClassifier(ccp_alpha=0.0, class_weight=None, criterion='gini',\n max_depth=None, max_features=None, max_leaf_nodes=None,\n min_impurity_decrease=0.0, min_impurity_split=None,\n min_samples_leaf=1, min_samples_split=2,\n min_weight_fraction_leaf=0.0, presort='deprecated',\n random_state=None, splitter='best')\ntrain time: 0.011s\ntest time: 0.002s\naccuracy: 0.371\nclassification report:\n precision recall f1-score support\n\n 1.0 0.22 0.27 0.24 145\n 2.0 0.48 0.49 0.48 288\n 3.0 0.35 0.26 0.30 187\n\n accuracy 0.37 620\n macro avg 0.35 0.34 0.34 620\nweighted avg 0.38 0.37 0.37 620\n\nconfusion matrix:\n[[ 39 74 32]\n [ 85 142 61]\n [ 56 82 49]]\n================================================================================\nRandom Forest\n________________________________________________________________________________\nTraining: \nRandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,\n criterion='gini', max_depth=None, max_features='auto',\n max_leaf_nodes=None, max_samples=None,\n min_impurity_decrease=0.0, min_impurity_split=None,\n min_samples_leaf=1, min_samples_split=2,\n min_weight_fraction_leaf=0.0, n_estimators=100,\n n_jobs=None, oob_score=False, random_state=None,\n verbose=0, warm_start=False)\ntrain time: 0.236s\ntest time: 0.020s\naccuracy: 0.416\nclassification report:\n precision recall f1-score support\n\n 1.0 0.21 0.17 0.19 145\n 2.0 0.48 0.67 0.56 288\n 3.0 0.38 0.22 0.28 187\n\n accuracy 0.42 620\n macro avg 0.36 0.35 0.34 620\nweighted avg 0.39 0.42 0.39 620\n\nconfusion matrix:\n[[ 24 93 28]\n [ 56 192 40]\n [ 32 113 42]]\n================================================================================\nGauissian Naive Bayes\n________________________________________________________________________________\nTraining: \nGaussianNB(priors=None, var_smoothing=1e-09)\ntrain time: 0.002s\ntest time: 0.001s\naccuracy: 0.434\nclassification report:\n precision recall f1-score support\n\n 1.0 0.24 0.12 0.16 145\n 2.0 0.52 0.58 0.55 288\n 3.0 0.38 0.45 0.41 187\n\n accuracy 0.43 620\n macro avg 0.38 0.38 0.37 620\nweighted avg 0.41 0.43 0.42 620\n\nconfusion matrix:\n[[ 18 78 49]\n [ 33 167 88]\n [ 25 78 84]]\n================================================================================\nkNN\n________________________________________________________________________________\nTraining: \nKNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',\n metric_params=None, n_jobs=None, n_neighbors=10, p=2,\n weights='uniform')\ntrain time: 0.004s\ntest time: 0.032s\naccuracy: 0.400\nclassification report:\n precision recall f1-score support\n\n 1.0 0.21 0.20 0.20 145\n 2.0 0.47 0.70 0.56 288\n 3.0 0.32 0.10 0.15 187\n\n accuracy 0.40 620\n macro avg 0.33 0.33 0.31 620\nweighted avg 0.37 0.40 0.35 620\n\nconfusion matrix:\n[[ 29 107 9]\n [ 58 201 29]\n [ 51 118 18]]\n================================================================================\nLogistic Regression\n________________________________________________________________________________\nTraining: \nLogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,\n intercept_scaling=1, l1_ratio=None, max_iter=100,\n multi_class='auto', n_jobs=None, penalty='l2',\n random_state=None, solver='lbfgs', tol=0.0001, verbose=0,\n warm_start=False)\ntrain time: 0.049s\ntest time: 0.001s\naccuracy: 0.476\ndimensionality: 14\ndensity: 1.000000\nclassification report:\n precision recall f1-score support\n\n 1.0 0.12 0.01 0.02 145\n 2.0 0.48 0.93 0.63 288\n 3.0 0.58 0.14 0.22 187\n\n accuracy 0.48 620\n macro avg 0.39 0.36 0.29 620\nweighted avg 0.43 0.48 0.37 620\n\nconfusion matrix:\n[[ 2 136 7]\n [ 9 267 12]\n [ 5 156 26]]\n================================================================================\nSupport Vector Machines\n________________________________________________________________________________\nTraining: \nSVC(C=1.0, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,\n decision_function_shape='ovr', degree=3, gamma='scale', kernel='rbf',\n max_iter=-1, probability=False, random_state=None, shrinking=True,\n tol=0.001, verbose=False)\ntrain time: 0.107s\ntest time: 0.020s\naccuracy: 0.465\nclassification report:\n precision recall f1-score support\n\n 1.0 0.00 0.00 0.00 145\n 2.0 0.46 1.00 0.63 288\n 3.0 0.00 0.00 0.00 187\n\n accuracy 0.46 620\n macro avg 0.15 0.33 0.21 620\nweighted avg 0.22 0.46 0.29 620\n\nconfusion matrix:\n[[ 0 145 0]\n [ 0 288 0]\n [ 0 187 0]]\n"
],
[
"# grafikleri çıkar\n\nindices = np.arange(len(results_dengeli))\n\nresults_dengeli = [[x[i] for x in results_dengeli] for i in range(4)]\n\nclf_names, score, training_time, test_time = results_dengeli\ntraining_time = np.array(training_time) / np.max(training_time)\ntest_time = np.array(test_time) / np.max(test_time)\n\nplt.figure(figsize=(12, 8))\nplt.title(\"Score\")\nplt.barh(indices, score, .2, label=\"score\", color='navy')\nplt.barh(indices + .3, training_time, .2, label=\"training time\",\n color='c')\nplt.barh(indices + .6, test_time, .2, label=\"test time\", color='darkorange')\nplt.yticks(())\nplt.legend(loc='best')\nplt.subplots_adjust(left=.25)\nplt.subplots_adjust(top=.95)\nplt.subplots_adjust(bottom=.05)\n\nfor i, c in zip(indices, clf_names):\n plt.text(-.3, i, c)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Doğruluk oranı, dengesiz veriye göre daha düşüktür. Yine doğruluk oranı, f1score, recall ve precision değerlerini göz önüne aldığımızda Gaussian Naive Bayes algoritmasının daha iyi çalıştığını görebiliyoruz.",
"_____no_output_____"
]
],
[
[
"gridF_dengeli = GridSearchCV(forest, hyperF, cv = 3, verbose = 1, \n n_jobs = -1)\nbestF_dengeli = gridF_dengeli.fit(X_train_dengeli, y_train_dengeli)",
"Fitting 3 folds for each of 500 candidates, totalling 1500 fits\n[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 34 tasks | elapsed: 8.6s\n[Parallel(n_jobs=-1)]: Done 184 tasks | elapsed: 41.9s\n[Parallel(n_jobs=-1)]: Done 434 tasks | elapsed: 1.7min\n[Parallel(n_jobs=-1)]: Done 784 tasks | elapsed: 3.1min\n[Parallel(n_jobs=-1)]: Done 1234 tasks | elapsed: 4.9min\n[Parallel(n_jobs=-1)]: Done 1500 out of 1500 | elapsed: 5.9min finished\n"
],
[
"gridF_dengeli.best_params_",
"_____no_output_____"
],
[
"gridF_dengeli.refit\npreds = gridF_dengeli.predict(X_test_dengeli)\nprobs = gridF_dengeli.predict_proba(X_test_dengeli)\n\n# doğruluk oranı\nnp.mean(preds == y_test_dengeli)",
"_____no_output_____"
],
[
"print(metrics.classification_report(y_test_dengeli, preds, target_names=['Düşük', 'Normal', 'Yüksek']))",
"precision recall f1-score support\n\n Düşük 0.00 0.00 0.00 145\n Normal 0.47 0.98 0.64 288\n Yüksek 0.61 0.07 0.13 187\n\n accuracy 0.48 620\n macro avg 0.36 0.35 0.26 620\nweighted avg 0.40 0.48 0.34 620\n\n"
]
],
[
[
"Dengesiz veri setinde olduğu gibi, doğruluk oranımız yaklaşık %6 arttırış gösterdi ancak sınıflandırma raporumuza negatif bir etkisi oldu. ",
"_____no_output_____"
],
[
"## **Değerlendirme** <a class=\"anchor\" id=\"evaluation\"></a>",
"_____no_output_____"
]
],
[
[
"model = GaussianNB()\nmodel.fit(X_train, y_train)\npred = model.predict(X_test)\n\nprint(\"Doğruluk oranımız {}\".format(metrics.accuracy_score(y_test, pred)))\nprint(metrics.classification_report(y_test, pred, target_names=['Düşük', 'Normal', 'Yüksek']))",
"Doğruluk oranımız 0.7214236824093087\n precision recall f1-score support\n\n Düşük 0.12 0.04 0.05 169\n Normal 0.77 0.93 0.84 1116\n Yüksek 0.19 0.07 0.10 176\n\n accuracy 0.72 1461\n macro avg 0.36 0.34 0.33 1461\nweighted avg 0.62 0.72 0.66 1461\n\n"
]
],
[
[
"Hatırlama, bir sınıflandırma modelinin ilgili tüm örnekleri belirleme yeteneğidir. Hatırlamanın sadece normal B12 değerleri için iyi olduğunu görebiliriz. Model düşük ve yüksek değerler için çok başarılı değildir.\n\nHassasiyet, bir sınıflandırma modelinin yalnızca ilgili örnekleri döndürme becerisidir. Modelimiz normal değerlerde çok başarılı, ancak düşük ve yüksek değerlerde çok başarılı değildir.",
"_____no_output_____"
]
],
[
[
"rslt = metrics.confusion_matrix(y_test, pred)\nsns.heatmap(pd.DataFrame(rslt, index=['Tahmin Edilen Düşük', 'Tahmin Edilen Normal', 'Tahmin Edilen Yüksek'], columns=['Aslında Düşük Olan', 'Aslında Normal Olan', 'Aslında Yüksek Olan']), annot=True, fmt='d', cmap='Blues')\nplt.title('Dengeli Olmayan Confision Matrix')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Konfeksiyon matrisi, modelin normal değerleri daha fazla tahmin etme eğiliminde olduğunu göstermektedir. Bunun nedeni verilerin dengesiz olması ve normal değerlerin diğerlerinden çok daha fazla olmasıdır. Gördüğümüz gibi, model aslında düşük veya yüksek olan 80 değeri normal olarak tahmin ediyor.",
"_____no_output_____"
]
],
[
[
"model.fit(X_train_dengeli, y_train_dengeli)\npred_dengeli = model.predict(X_test_dengeli)\n\nprint(\"Dengeli veri seti için doğruluk oranımız {}\".format(metrics.accuracy_score(y_test_dengeli, pred_dengeli)))\nprint(metrics.classification_report(y_test_dengeli, pred_dengeli, target_names=['Düşük', 'Normal', 'Yüksek']))",
"Dengeli veri seti için doğruluk oranımız 0.4338709677419355\n precision recall f1-score support\n\n Düşük 0.24 0.12 0.16 145\n Normal 0.52 0.58 0.55 288\n Yüksek 0.38 0.45 0.41 187\n\n accuracy 0.43 620\n macro avg 0.38 0.38 0.37 620\nweighted avg 0.41 0.43 0.42 620\n\n"
]
],
[
[
"Hatırlama, bir sınıflandırma modelinin ilgili tüm örnekleri belirleme yeteneğidir. Dengeli verilerde hatırlama, dengesiz verilerin hatırlamasından daha iyi görünüyor. Yüksek değerler şimdi daha iyi hatırlama yüzdesine sahip, normal hala iyi. Düşük daha iyi olabilirdi.\n\nHassasiyet, bir sınıflandırma modelinin yalnızca ilgili örnekleri döndürme becerisidir. Dengeli verilerin hassasiyeti, dengesiz verilerin kesinliğinden çok daha iyidir. Tüm kategoriler artık kabul edilebilir hassasiyetlere sahiptir.",
"_____no_output_____"
]
],
[
[
"rslt_dengeli = metrics.confusion_matrix(y_test_dengeli, pred_dengeli)\nsns.heatmap(pd.DataFrame(rslt_dengeli, index=['Tahmin Edilen Düşük', 'Tahmin Edilen Normal', 'Tahmin Edilen Yüksek'], columns=['Aslında Düşük Olan', 'Aslında Normal Olan', 'Aslında Yüksek Olan']), annot=True, fmt='d', cmap='Blues')\nplt.title('Dengeli Olan Confision Matrix')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Dengeli verilerle elde edilen sonuç, modelin normal değerleri daha fazla tahmin etme eğiliminde olmadığını göstermektedir. Tüm değerleri tahmin eder, ancak doğruluk oranı kötü olduğu için daha fazla yanlış tahminde bulunur.\n\n---\n",
"_____no_output_____"
],
[
"İki sonucu karşılaştırdığımız zaman, doğruluk oranının dengesiz veri seti için daha iyi olduğunu, ancak sınıflandırma raporuna baktığımız zaman dengeli veri setinin daha iyi olduğunu görebiliyoruz. ",
"_____no_output_____"
],
[
"## **Referanslar**<a class=\"anchor\" id=\"references\"></a>",
"_____no_output_____"
],
[
"\n\n* https://github.com/Jean-njoroge/Breast-cancer-risk-prediction\n* https://dizziness-and-balance.com/disorders/central/b12.html\n",
"_____no_output_____"
],
[
"\n**Disclaimer!** <font color='grey'>This notebook was prepared by İrem Dereli as a term project for the *BBM469 - Data Intensive Applications Laboratory* class. The notebook is available for educational purposes only. There is no guarantee on the correctness of the content provided as it is a student work.\n\nIf you think there is any copyright violation, please let us [know](https://forms.gle/BNNRB2kR8ZHVEREq8). \n</font>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ec883725e3046d10efe7c391c7cf8629e78dc195 | 239,577 | ipynb | Jupyter Notebook | ontology/ontol_sim_terms.ipynb | ehbeam/neuro-knowledge-engine | 9dc56ade0bbbd8d14f0660774f787c3f46d7e632 | [
"MIT"
] | 15 | 2020-07-17T07:10:26.000Z | 2022-02-18T05:51:45.000Z | ontology/ontol_sim_terms.ipynb | YifeiCAO/neuro-knowledge-engine | 9dc56ade0bbbd8d14f0660774f787c3f46d7e632 | [
"MIT"
] | 2 | 2022-01-14T09:10:12.000Z | 2022-01-28T17:32:42.000Z | ontology/ontol_sim_terms.ipynb | YifeiCAO/neuro-knowledge-engine | 9dc56ade0bbbd8d14f0660774f787c3f46d7e632 | [
"MIT"
] | 4 | 2021-12-22T13:27:32.000Z | 2022-02-18T05:51:47.000Z | 96.72063 | 42,000 | 0.792251 | [
[
[
"import pandas as pd\nimport numpy as np\nnp.random.seed(42)\n\nfrom collections import OrderedDict\n\nimport sys\nsys.path.append(\"..\")\nimport utilities, ontology",
"_____no_output_____"
]
],
[
[
"# Load brain and text data",
"_____no_output_____"
]
],
[
[
"act_bin = utilities.load_coordinates().astype(float)\nprint(\"Document N={}, Structure N={}\".format(act_bin.shape[0], act_bin.shape[1]))",
"Document N=18155, Structure N=118\n"
],
[
"version = 190325\ndtm_bin = utilities.load_doc_term_matrix(version=version, binarize=True)\nprint(\"Document N={}, Term N={}\".format(dtm_bin.shape[0], dtm_bin.shape[1]))",
"Document N=18155, Term N=4107\n"
],
[
"frameworks = [\"data-driven\", \"data-driven\", \"rdoc\", \"dsm\"]\nlist_suffixes = [\"\", \"\", \"_opsim\", \"_opsim\"]\ncircuit_suffixes = [\"_terms\", \"_lr\", \"\", \"\"]\nlists, circuits = {}, {}\nfor fw, list_suffix, circuit_suffix in zip(frameworks, list_suffixes, circuit_suffixes):\n lists[fw+circuit_suffix], circuits[fw+circuit_suffix] = utilities.load_framework(fw, suffix=list_suffix, clf=circuit_suffix)",
"_____no_output_____"
],
[
"frameworks = [fw+circuit_suffix for fw, circuit_suffix in zip(frameworks, circuit_suffixes)]",
"_____no_output_____"
],
[
"scores = {fw: utilities.score_lists(lists[fw], dtm_bin, label_var=\"DOMAIN\") for fw in frameworks}",
"_____no_output_____"
],
[
"words = []\nfor fw in frameworks:\n words += list(lists[fw][\"TOKEN\"])\nwords = sorted(list(set(words)))\nstructures = list(act_bin.columns)\nprint(\"Term N={}, Structure N={}\".format(len(words), len(structures)))",
"Term N=349, Structure N=118\n"
],
[
"domains = {fw: list(OrderedDict.fromkeys(lists[fw][\"DOMAIN\"])) for fw in frameworks}",
"_____no_output_____"
],
[
"pmids = act_bin.index.intersection(scores[\"rdoc\"].index).intersection(scores[\"dsm\"].index)\nlen(pmids)",
"_____no_output_____"
],
[
"for fw in frameworks:\n scores[fw] = scores[fw].loc[pmids]",
"_____no_output_____"
],
[
"dtm_bin = dtm_bin.loc[pmids, words]\nact_bin = act_bin.loc[pmids, structures]",
"_____no_output_____"
]
],
[
[
"# Load frameworks",
"_____no_output_____"
]
],
[
[
"fw2names = {\n \"data-driven_lr\": [\"Memory\", \"Reward\", \"Cognition\", \"Vision\", \"Manipulation\", \"Language\"],\n \"data-driven_terms\": [\"Memory\", \"Emotion\", \"Inference\", \"Cognition\", \"Vision\", \"Language\"],\n \"rdoc\": [\"Negative\", \"Positive\", \"Arousal & Reg.\", \"Social\", \"Cognitive\", \"Sensorimotor\"],\n \"dsm\": [\"Depressive\", \"Anxiety\", \"Trauma & Stressor\", \"Obsess.-Compuls.\", \n \"Disruptive\", \"Substance\", \"Developmental\", \"Psychotic\", \"Bipolar\"]\n}",
"_____no_output_____"
],
[
"systems = {}\nfor fw in frameworks:\n fw_df = pd.DataFrame(0.0, index=words+structures, columns=domains[fw])\n for dom in domains[fw]:\n for word in lists[fw].loc[lists[fw][\"DOMAIN\"] == dom, \"TOKEN\"]:\n fw_df.loc[word, dom] = 1.0\n for struct in structures:\n fw_df.loc[struct, dom] = circuits[fw].loc[struct, dom]\n fw_df[fw_df > 0.0] = 1.0\n fw_df.columns = fw2names[fw]\n systems[fw] = fw_df",
"_____no_output_____"
],
[
"domains = fw2names",
"_____no_output_____"
]
],
[
[
"# Similarity of RDoC and data-driven terms\n\n## Observed values",
"_____no_output_____"
]
],
[
[
"from scipy.spatial.distance import dice, cdist\nimport matplotlib.pyplot as plt\nfrom matplotlib import font_manager, rcParams\nfrom style import style\n\nfont_prop = font_manager.FontProperties(fname=style.font, size=20)\nrcParams[\"axes.linewidth\"] = 1.5\n\n%matplotlib inline",
"_____no_output_____"
],
[
"def compute_sim_obs(fw, fw_base=\"data-driven_terms\"):\n sims = pd.DataFrame(index=domains[fw_base], columns=domains[fw])\n for k in domains[fw_base]:\n for r in domains[fw]:\n sims.loc[k,r] = 1.0 - dice(systems[fw_base][k], systems[fw][r])\n return sims",
"_____no_output_____"
],
[
"sims = compute_sim_obs(\"rdoc\")\nsims",
"_____no_output_____"
],
[
"def plot_sims(sims, outfile, figsize=(2.8,2.8), plot_cbar=False):\n \n fig = plt.figure(figsize=figsize)\n ax = fig.add_axes([0,0,1,1])\n font_prop = font_manager.FontProperties(fname=style.font, size=20)\n\n im = plt.imshow(sims.values.astype(float), cmap=plt.cm.gray)\n\n plt.xticks(range(0,sims.shape[1]), fontproperties=font_prop, rotation=60, ha=\"right\")\n plt.yticks(range(0,sims.shape[0]), fontproperties=font_prop)\n ax.set_xticklabels(sims.columns)\n ax.set_yticklabels(sims.index)\n ax.xaxis.set_tick_params(width=1.5, length=7, top=False)\n ax.yaxis.set_tick_params(width=1.5, length=7)\n plt.clim(0, 1)\n\n if plot_cbar:\n cbar = plt.colorbar()\n cbar.solids.set_edgecolor(\"face\")\n \n plt.savefig(outfile, dpi=250, bbox_inches=\"tight\")\n plt.show()\n plt.close()",
"_____no_output_____"
],
[
"plot_sims(sims, \"figures/sim_dd-terms_rdoc.png\", plot_cbar=False)",
"_____no_output_____"
]
],
[
[
"## Null distribution",
"_____no_output_____"
]
],
[
[
"def compute_sim_null(fw, n_iter=10000, fw_base=\"data-driven_terms\"):\n sims_null = np.empty((len(domains[fw_base]), len(domains[fw]), n_iter))\n for n in range(n_iter):\n null = np.random.choice(words+structures, \n size=len(words+structures), replace=False)\n sims_null[:,:,n] = 1.0 - cdist(systems[fw_base].loc[null].values.T, \n systems[fw].values.T, metric=\"dice\")\n if n % (float(n_iter) / 10.0) == 0:\n print(\"Iteration {}\".format(n))\n return sims_null",
"_____no_output_____"
],
[
"sims_null = compute_sim_null(\"rdoc\")",
"Iteration 0\nIteration 1000\nIteration 2000\nIteration 3000\nIteration 4000\nIteration 5000\nIteration 6000\nIteration 7000\nIteration 8000\nIteration 9000\n"
]
],
[
[
"## False discovery rate",
"_____no_output_____"
]
],
[
[
"from statsmodels.stats.multitest import multipletests",
"_____no_output_____"
],
[
"def compute_sim_fdr(fw, sims, sims_null, fw_base=\"data-driven_terms\"):\n n_iter = sims_null.shape[2]\n pvals = pd.DataFrame(index=domains[fw_base], columns=domains[fw])\n for i, k in enumerate(domains[fw_base]):\n for j, r in enumerate(domains[fw]):\n pvals.loc[k,r] = np.sum(sims_null[i,j,:] > sims.loc[k,r]) / float(n_iter)\n fdrs = multipletests(pvals.values.ravel(), method=\"fdr_bh\")[1]\n fdrs = np.reshape(fdrs, pvals.shape)\n fdrs = pd.DataFrame(fdrs, index=domains[fw_base], columns=domains[fw])\n return fdrs",
"_____no_output_____"
],
[
"fdrs = compute_sim_fdr(\"rdoc\", sims, sims_null)\nfdrs",
"_____no_output_____"
],
[
"def compute_sim_star(fw, fdrs, fw_base=\"data-driven_terms\"):\n stars = pd.DataFrame(\"\", index=domains[fw_base], columns=domains[fw])\n for k in domains[fw_base]:\n for r in domains[fw]:\n fdr = fdrs.loc[k,r]\n if fdr < 0.05:\n stars.loc[k,r] = \"*\"\n if fdr < 0.01:\n stars.loc[k,r] = \"**\"\n if fdr < 0.001:\n stars.loc[k,r] = \"***\"\n return stars",
"_____no_output_____"
],
[
"stars = compute_sim_star(\"rdoc\", fdrs)\nstars",
"_____no_output_____"
]
],
[
[
"## Weights for figure",
"_____no_output_____"
]
],
[
[
"sims[fdrs < 0.05] * 50",
"_____no_output_____"
]
],
[
[
"# Similarity of RDoC and data-driven LR",
"_____no_output_____"
]
],
[
[
"sims = compute_sim_obs(\"rdoc\", fw_base=\"data-driven_lr\")\nplot_sims(sims, \"figures/sim_dd-lr_rdoc.png\")",
"_____no_output_____"
]
],
[
[
"# Similarity of DSM and data-driven terms\n\n## Observed values",
"_____no_output_____"
]
],
[
[
"sims = compute_sim_obs(\"dsm\")\nsims",
"_____no_output_____"
],
[
"plot_sims(sims, \"figures/sim_dd-terms_dsm.png\", figsize=(5,2.8))",
"_____no_output_____"
]
],
[
[
"## Null distribution",
"_____no_output_____"
]
],
[
[
"sims_null = compute_sim_null(\"dsm\")",
"Iteration 0\nIteration 1000\nIteration 2000\nIteration 3000\nIteration 4000\nIteration 5000\nIteration 6000\nIteration 7000\nIteration 8000\nIteration 9000\n"
]
],
[
[
"## False discovery rate",
"_____no_output_____"
]
],
[
[
"fdrs = compute_sim_fdr(\"dsm\", sims, sims_null)\nfdrs",
"_____no_output_____"
],
[
"stars = compute_sim_star(\"dsm\", fdrs)\nstars",
"_____no_output_____"
]
],
[
[
"## Weights for figure",
"_____no_output_____"
]
],
[
[
"sims[fdrs < 0.05] * 50",
"_____no_output_____"
]
],
[
[
"# Similarity of DSM and data-driven LR",
"_____no_output_____"
]
],
[
[
"sims = compute_sim_obs(\"dsm\", fw_base=\"data-driven_lr\")\nplot_sims(sims, \"figures/sim_dd-lr_dsm.png\", figsize=(5,2.8))",
"_____no_output_____"
]
],
[
[
"# Similarity of data-driven system versions\n\n## Observed values",
"_____no_output_____"
]
],
[
[
"sims = compute_sim_obs(\"data-driven_lr\")\nsims",
"_____no_output_____"
]
],
[
[
"## Null distribution",
"_____no_output_____"
]
],
[
[
"sims_null = compute_sim_null(\"data-driven_lr\")",
"Iteration 0\nIteration 1000\nIteration 2000\nIteration 3000\nIteration 4000\nIteration 5000\nIteration 6000\nIteration 7000\nIteration 8000\nIteration 9000\n"
]
],
[
[
"## False discovery rate",
"_____no_output_____"
]
],
[
[
"fdrs = compute_sim_fdr(\"data-driven_lr\", sims, sims_null)\nfdrs",
"_____no_output_____"
],
[
"stars = compute_sim_star(\"data-driven_lr\", fdrs)\nstars",
"_____no_output_____"
]
],
[
[
"## Weights for figure",
"_____no_output_____"
]
],
[
[
"sims[fdrs < 0.05] * 50",
"_____no_output_____"
]
],
[
[
"# Compare RDoC similarity between data-driven versions",
"_____no_output_____"
]
],
[
[
"sims_terms = compute_sim_obs(\"rdoc\", fw_base=\"data-driven_terms\")\nsims_lr = compute_sim_obs(\"rdoc\", fw_base=\"data-driven_lr\")\nsims = {\"lr\": sims_lr, \"terms\": sims_terms}",
"_____no_output_____"
],
[
"# null_terms = compute_sim_null(\"rdoc\", fw_base=\"data-driven_terms\")\n# null_lr = compute_sim_null(\"rdoc\", fw_base=\"data-driven_lr\")",
"_____no_output_____"
],
[
"def compute_sim_boot(fw, n_iter=10000, fw_base=\"data-driven_terms\"):\n sims_boot = np.empty((len(domains[fw_base]), len(domains[fw]), n_iter))\n for n in range(n_iter):\n boot = np.random.choice(words+structures, \n size=len(words+structures), replace=True)\n sims_boot[:,:,n] = 1.0 - cdist(systems[fw_base].loc[boot].values.T, \n systems[fw].loc[boot].values.T, metric=\"dice\")\n if n % (float(n_iter) / 10.0) == 0:\n print(\"Iteration {}\".format(n))\n return sims_boot",
"_____no_output_____"
],
[
"boot_terms = compute_sim_boot(\"rdoc\", fw_base=\"data-driven_terms\")\nboot_lr = compute_sim_boot(\"rdoc\", fw_base=\"data-driven_lr\")",
"Iteration 0\nIteration 1000\nIteration 2000\nIteration 3000\nIteration 4000\nIteration 5000\nIteration 6000\nIteration 7000\nIteration 8000\nIteration 9000\nIteration 0\nIteration 1000\nIteration 2000\nIteration 3000\nIteration 4000\nIteration 5000\nIteration 6000\nIteration 7000\nIteration 8000\nIteration 9000\n"
],
[
"mean_boot_terms = np.mean(np.mean(boot_terms, axis=0), axis=0)\nmean_boot_lr = np.mean(np.mean(boot_lr, axis=0), axis=0)\nboot = {\"lr\": list(mean_boot_lr), \"terms\": list(mean_boot_terms)}",
"_____no_output_____"
],
[
"n_iter = boot_terms.shape[2]\npval_dif = np.sum(np.less(mean_boot_terms - mean_boot_lr, 0.0)) / n_iter\npval_dif",
"_____no_output_____"
],
[
"def plot_comparison(boot, sims, filename, path=\"\", n_iter=1000, print_fig=True, \n font=style.font, dx=0.38, ylim=[0,0.25], yticks=[0,0.0625,0.125,0.1875,0.25]):\n\n font_lg = font_manager.FontProperties(fname=font, size=20)\n rcParams[\"axes.linewidth\"] = 1.5\n\n fig = plt.figure(figsize=(1.25, 4.25))\n ax = fig.add_axes([0,0,1,1])\n\n i = 0\n labels = []\n for fw, dist in boot.items():\n labels.append(fw)\n dist_avg = np.mean(dist, axis=0)\n macro_avg = np.mean(np.mean(sims[fw], axis=0), axis=0)\n plt.plot([i-dx, i+dx], [macro_avg, macro_avg], \n c=\"gray\", alpha=1, lw=2, zorder=-1)\n v = ax.violinplot(sorted(dist), positions=[i], \n showmeans=False, showmedians=False, widths=0.85)\n for pc in v[\"bodies\"]:\n pc.set_facecolor(\"gray\")\n pc.set_edgecolor(\"gray\")\n pc.set_linewidth(2)\n pc.set_alpha(0.5)\n for line in [\"cmaxes\", \"cmins\", \"cbars\"]:\n v[line].set_edgecolor(\"none\")\n i += 1\n\n ax.set_xticks(range(len(boot.keys())))\n ax.set_xticklabels([], rotation=60, ha=\"right\")\n plt.xticks(fontproperties=font_lg)\n plt.yticks(yticks, fontproperties=font_lg)\n plt.xlim([-0.75, len(boot.keys())-0.25])\n plt.ylim(ylim)\n \n for side in [\"right\", \"top\"]:\n ax.spines[side].set_visible(False)\n ax.xaxis.set_tick_params(width=1.5, length=7)\n ax.yaxis.set_tick_params(width=1.5, length=7)\n \n plt.savefig(\"{}figures/{}\".format(path, filename), dpi=250, bbox_inches=\"tight\")\n\n if print_fig:\n plt.show()\n plt.close()",
"_____no_output_____"
],
[
"plot_comparison(boot, sims, \"sim_dd_rdoc.png\")",
"_____no_output_____"
]
],
[
[
"# Compare DSM similarity between data-driven versions",
"_____no_output_____"
]
],
[
[
"sims_terms = compute_sim_obs(\"dsm\", fw_base=\"data-driven_terms\")\nsims_lr = compute_sim_obs(\"dsm\", fw_base=\"data-driven_lr\")\nsims = {\"lr\": sims_lr, \"terms\": sims_terms}",
"_____no_output_____"
],
[
"# null_terms = compute_sim_null(\"dsm\", fw_base=\"data-driven_terms\")\n# null_lr = compute_sim_null(\"dsm\", fw_base=\"data-driven_lr\")",
"_____no_output_____"
],
[
"boot_terms = compute_sim_boot(\"dsm\", fw_base=\"data-driven_terms\")\nboot_lr = compute_sim_boot(\"dsm\", fw_base=\"data-driven_lr\")",
"Iteration 0\nIteration 1000\nIteration 2000\nIteration 3000\nIteration 4000\nIteration 5000\nIteration 6000\nIteration 7000\nIteration 8000\nIteration 9000\nIteration 0\nIteration 1000\nIteration 2000\nIteration 3000\nIteration 4000\nIteration 5000\nIteration 6000\nIteration 7000\nIteration 8000\nIteration 9000\n"
],
[
"mean_boot_terms = np.mean(np.mean(boot_terms, axis=0), axis=0)\nmean_boot_lr = np.mean(np.mean(boot_lr, axis=0), axis=0)\nboot = {\"lr\": list(mean_boot_lr), \"terms\": list(mean_boot_terms)}",
"_____no_output_____"
],
[
"np.mean(np.mean(sims[\"lr\"], axis=0), axis=0)",
"_____no_output_____"
],
[
"np.mean(np.mean(sims[\"terms\"], axis=0), axis=0)",
"_____no_output_____"
],
[
"n_iter = boot_terms.shape[2]\npval_dif = np.sum(np.less(mean_boot_terms - mean_boot_lr, 0.0)) / n_iter\npval_dif",
"_____no_output_____"
],
[
"plot_comparison(boot, sims, \"sim_dd_dsm.png\")",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec883bb91bc6375985bac418c98a7da314a727e0 | 855 | ipynb | Jupyter Notebook | pset_pandas_ext/101problems/solutions/nb/p61.ipynb | mottaquikarim/pydev-psets | 9749e0d216ee0a5c586d0d3013ef481cc21dee27 | [
"MIT"
] | 5 | 2019-04-08T20:05:37.000Z | 2019-12-04T20:48:45.000Z | pset_pandas_ext/101problems/solutions/nb/p61.ipynb | mottaquikarim/pydev-psets | 9749e0d216ee0a5c586d0d3013ef481cc21dee27 | [
"MIT"
] | 8 | 2019-04-15T15:16:05.000Z | 2022-02-12T10:33:32.000Z | pset_pandas_ext/101problems/solutions/nb/p61.ipynb | mottaquikarim/pydev-psets | 9749e0d216ee0a5c586d0d3013ef481cc21dee27 | [
"MIT"
] | 2 | 2019-04-10T00:14:42.000Z | 2020-02-26T20:35:21.000Z | 23.108108 | 82 | 0.479532 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ec8844512247c77e678d4a0ce73e34012106e5e4 | 10,200 | ipynb | Jupyter Notebook | 005_Python_Keywords_and_Identifiers.ipynb | Alana951/01_Python_Introduction | 3835e250d1e6131410585525a3e665d511a347cc | [
"Unlicense"
] | 3 | 2021-06-29T10:27:31.000Z | 2021-06-29T10:29:23.000Z | 005_Python_Keywords_and_Identifiers.ipynb | zhengyongjian123/01_Python_Introduction | 49e02d7a5b93fb239a106701d4bd780d777bec5d | [
"Unlicense"
] | null | null | null | 005_Python_Keywords_and_Identifiers.ipynb | zhengyongjian123/01_Python_Introduction | 49e02d7a5b93fb239a106701d4bd780d777bec5d | [
"Unlicense"
] | null | null | null | 29.142857 | 350 | 0.545392 | [
[
[
"<small><small><i>\nAll the IPython Notebooks in this lecture series by Dr. Milan Parmar are available @ **[GitHub](https://github.com/milaan9/01_Python_Introduction)**\n</i></small></small>",
"_____no_output_____"
],
[
"# Python Keywords and Identifiers\n\nIn this class, you will learn about keywords (reserved words in Python) and identifiers (names given to variables, functions, etc.).",
"_____no_output_____"
],
[
"# 1. Python Keywords\n\nKeywords are the reserved words in Python.\n\nWe cannot use a keyword as a **[variable](https://github.com/milaan9/01_Python_Introduction/blob/main/009_Python_Data_Types.ipynb)** name, **[function](https://github.com/milaan9/04_Python_Functions/blob/main/001_Python_Functions.ipynb)** name or any other identifier. They are used to define the syntax and structure of the Python language.\n\nIn Python, keywords are **case sensitive**.\n\nThere are **33** keywords in Python 3.9. This number can vary slightly over the course of time.\n\nAll the keywords except **`True`**, **`False`** and **`None`** are in lowercase and they must be written as they are. The **[list of all the keywords](https://github.com/milaan9/01_Python_Introduction/blob/main/Python_Keywords_List.ipynb)** is given below.\n\n**Keywords in Python**\n\n| | | | | |\n|:----|:----|:----|:----|:----|\n| **`False`** | **`await`** | **`else`** | **`import`** | **`pass`** |\n| **`None`** | **`break`** | **`except`** | **`in`** | **`raise`** |\n| **`True`** | **`class`** | **`finally`** | **`is`** | **`return`** |\n| **`and`** | **`continue`** | **`for`** | **`lambda`** | **`try`** |\n| **`as`** | **`def`** | **`from`** | **`nonlocal`** | **`while`** |\n| **`assert`** | **`del`** | **`global`** | **`not`** | **`with`** |\n| **`async`** | **`elif`** | **`if`** | **`or`** | **`yield`** |\n\nYou can see this list any time by typing help **`keywords`** to the Python interpreter. \n\nTrying to create a variable with the same name as any reserved word results in an **error**:\n\n```python\n>>>for = 6\n\nFile \"<ipython-input-1-50b154750974>\", line 1\nfor = 6 # It will give error becasue \"for\" is keyword and we cannot use as a variable name.\n ^\nSyntaxError: invalid syntax\n```",
"_____no_output_____"
]
],
[
[
"for = 6 # It will give error becasue \"for\" is keyword and we cannot use as a variable name.",
"_____no_output_____"
],
[
"For = 6 # \"for\" is keyword but \"For\" is not keyword so we can use it as variable name\nFor",
"_____no_output_____"
]
],
[
[
"# 2. Python Identifiers\n\nAn **identifier** is a name given to entities like **class, functions, variables, etc**. It helps to differentiate one entity from another.",
"_____no_output_____"
],
[
"### Rules for writing identifiers\n\n1. **Identifiers** can be a combination of letters in lowercase **(a to z)** or uppercase **(A to Z)** or digits **(0 to 9)** or an underscore **`_`**. Names like **`myClass`**, **`var_1`** and **`print_this_to_screen`**, all are valid example. \n\n2. An identifier cannot start with a digit. **`1variable`** is invalid, but **`variable1`** is perfectly fine. \n\n3. Keywords cannot be used as identifiers\n\n```python\n>>>global = 3\n\nFile \"<ipython-input-2-43186c7d3555>\", line 1\n global = 3 # because \"global\" is a keyword\n ^\nSyntaxError: invalid syntax\n```",
"_____no_output_____"
]
],
[
[
"global = 3 # because \"global\" is a keyword",
"_____no_output_____"
]
],
[
[
"4. We cannot use special symbols like **!**, **@**, **#**,<b> $, % </b>, etc. in our identifier.\n\n```python\n>>>m@ = 3\n\nFile \"<ipython-input-3-4d4a0e714c73>\", line 1\n m@ = 3\n ^\nSyntaxError: invalid syntax\n```",
"_____no_output_____"
]
],
[
[
"m@ = 3",
"_____no_output_____"
]
],
[
[
"## Things to Remember\n\nPython is a case-sensitive language. This means, **`Variable`** and **`variable`** are not the same.\n\nAlways give the identifiers a name that makes sense. While **`c = 10`** is a valid name, writing **`count = 10`** would make more sense, and it would be easier to figure out what it represents when you look at your code after a long gap.\n\nMultiple words can be separated using an underscore, like **`this_is_a_long_variable`**.",
"_____no_output_____"
]
],
[
[
"this_is_a_long_variable = 6+3\nthis_is_a_long_variable",
"_____no_output_____"
],
[
"add_5_and_3 = 6+3\nadd_5_and_3",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec8845c20f6b7099629d0c78c3769e448fd9cab6 | 34,899 | ipynb | Jupyter Notebook | IntroToEnums.ipynb | akubera/COhPy-Presentations | ab29b9fcb2d3de02cb3e3ef681f1779af14d4f3a | [
"Unlicense"
] | 1 | 2016-01-28T05:19:56.000Z | 2016-01-28T05:19:56.000Z | IntroToEnums.ipynb | akubera/COhPy-Presentations | ab29b9fcb2d3de02cb3e3ef681f1779af14d4f3a | [
"Unlicense"
] | null | null | null | IntroToEnums.ipynb | akubera/COhPy-Presentations | ab29b9fcb2d3de02cb3e3ef681f1779af14d4f3a | [
"Unlicense"
] | null | null | null | 17.124141 | 133 | 0.454483 | [
[
[
"# Intro to Enums\n\n> Andrew Kubera\n\n> COhPy - Feb 26, 2018",
"_____no_output_____"
],
[
"### What is an Enumerated Type?\n\n* Simply a type comprising a finite-set of named values",
"_____no_output_____"
],
[
"* Standard example is a card suit:\n```\n Hearts, Spades, Diamonds, Clubs\n```",
"_____no_output_____"
],
[
"* Most languages store these internally as as simple integers\n * Compilers often typecheck, providing some safety\n * Easily copiable ",
"_____no_output_____"
],
[
"Enumerated types are a common tool in programming languages:\n\n* C-based\n* Java\n* Rust\n* Swift\n* TypeScript\n* Haskell\n* Lisp",
"_____no_output_____"
],
[
"- Missing : JavaScript, Ruby, *Python???*\n - Unnecessary in dynamically-typed languages?",
"_____no_output_____"
],
[
"`enum` module added to Python standard library in version *3.4*",
"_____no_output_____"
],
[
"Here's how to use it:",
"_____no_output_____"
]
],
[
[
"from enum import Enum",
"_____no_output_____"
],
[
"class A(Enum):\n X = 1\n Y = 2\n Z = 3",
"_____no_output_____"
]
],
[
[
"Access members as usual:",
"_____no_output_____"
]
],
[
[
"A.X",
"_____no_output_____"
],
[
"A.X == 1",
"_____no_output_____"
],
[
"A.X == A.X",
"_____no_output_____"
],
[
"A.Y == A.X",
"_____no_output_____"
],
[
"a = A.X\nb = A.X\n\na is b",
"_____no_output_____"
]
],
[
[
"Values may be looked up by \"name\" using `[ ]` notation",
"_____no_output_____"
]
],
[
[
"key = \"Y\"\n\n...\n\nA[key]",
"_____no_output_____"
]
],
[
[
"Enums have dict-like access!",
"_____no_output_____"
]
],
[
[
"A[key] is A.Y",
"_____no_output_____"
]
],
[
[
"Also supports \"backwards\" lookup via `( )`:",
"_____no_output_____"
]
],
[
[
"A(1)",
"_____no_output_____"
]
],
[
[
"(note: this would *usually* be creating a new object of type `A`)",
"_____no_output_____"
],
[
"**Bidirectional mapping** between a name (string) and some value!",
"_____no_output_____"
],
[
"### \"Standard\" errors when doing lookups",
"_____no_output_____"
]
],
[
[
"A.W # AttributeError",
"_____no_output_____"
],
[
"A['W'] # KeyError",
"_____no_output_____"
],
[
"A[1] # KeyError",
"_____no_output_____"
],
[
"A(9) # ValueError",
"_____no_output_____"
]
],
[
[
"## Iteration over members\n\nEnums are *iterable* over all values (order is preserved)",
"_____no_output_____"
]
],
[
[
"list(A)",
"_____no_output_____"
]
],
[
[
"### Alternative means of construction",
"_____no_output_____"
]
],
[
[
"B = Enum(\"B\", \"X Y Z\")",
"_____no_output_____"
],
[
"B.X",
"_____no_output_____"
],
[
"B(3)",
"_____no_output_____"
],
[
"B = Enum(\"B\", [('X', 90), ('Y', 45)])",
"_____no_output_____"
],
[
"B.X",
"_____no_output_____"
]
],
[
[
"### Lets look more at the values...",
"_____no_output_____"
]
],
[
[
"# Values can be mixed types\nclass B(Enum):\n X = 1\n Y = \"some string.\"\n Z = 0.3\n L = [] # note the list!",
"_____no_output_____"
],
[
"B(\"some string.\")",
"_____no_output_____"
],
[
"B([])",
"_____no_output_____"
],
[
"B(0.3)",
"_____no_output_____"
]
],
[
[
"<small>BTW, you should **NOT** use floating points to do indexing</small>",
"_____no_output_____"
]
],
[
[
"key = 0.1 + 0.2",
"_____no_output_____"
],
[
"B(key) # B(0.3)",
"_____no_output_____"
]
],
[
[
"# Namedtuple like behavior",
"_____no_output_____"
]
],
[
[
"# Cannot reassign values\nB.X = 2",
"_____no_output_____"
],
[
"B.L.append(1)",
"_____no_output_____"
]
],
[
[
"We will have to start thinking differently.....",
"_____no_output_____"
],
[
"## Look deeper at the values",
"_____no_output_____"
]
],
[
[
"class A(Enum):\n X = 1\n Y = 2\n Z = 3",
"_____no_output_____"
],
[
"A.X",
"_____no_output_____"
],
[
"isinstance(A.X, int)",
"_____no_output_____"
],
[
"isinstance(A.X, A)",
"_____no_output_____"
],
[
"type(A.X)",
"_____no_output_____"
]
],
[
[
"When creating the class:\n\n* The members are \"intercepted\" and wrapped with instances of the class.\n* After that, the class is \"frozen\" and *no further instances* may be created",
"_____no_output_____"
],
[
"One little \"goof\":",
"_____no_output_____"
]
],
[
[
"A.X.X.Y",
"_____no_output_____"
]
],
[
[
"^ Because instances of objects have access to members of their class, all enums have each-other as attributes (Don't use this!)",
"_____no_output_____"
],
[
"So where are `1, 2, 3` in A?",
"_____no_output_____"
]
],
[
[
"A.X.value",
"_____no_output_____"
],
[
"A.X.name",
"_____no_output_____"
]
],
[
[
"The `value` & `name` attributes are ***the*** important things to know about python Enums ",
"_____no_output_____"
]
],
[
[
"B.L",
"_____no_output_____"
],
[
"B.L.value.append(7)",
"_____no_output_____"
],
[
"B.L",
"_____no_output_____"
]
],
[
[
"So they are *semi-*immutable (like tuples)",
"_____no_output_____"
],
[
"### Methods",
"_____no_output_____"
]
],
[
[
"class A(Enum):\n X = 1\n Y = 2\n \n def add_ten(self):\n return self.value + 10",
"_____no_output_____"
],
[
"list(A)",
"_____no_output_____"
]
],
[
[
"`add_ten` **not** included as enum member -- behaves as typical python method",
"_____no_output_____"
]
],
[
[
"A.add_ten",
"_____no_output_____"
],
[
"A.X.add_ten()",
"_____no_output_____"
],
[
"class C(Enum):\n X = 1\n Y = lambda a: print('Y(a=%r)' % a)",
"_____no_output_____"
],
[
"list(C)",
"_____no_output_____"
],
[
"C.X.Y() ",
"_____no_output_____"
]
],
[
[
"So lambdas are interpreted as methods (as usual)",
"_____no_output_____"
],
[
"Can I get a little crazy now?",
"_____no_output_____"
]
],
[
[
"class A(Enum):\n \n class J:\n def __init__(self):\n print(\"constructing J\")\n \n class M:\n def __init__(self):\n print(\"constructing M\")\n",
"_____no_output_____"
],
[
"list(A)",
"_____no_output_____"
],
[
"m = A.M.value()",
"_____no_output_____"
],
[
"type(m)",
"_____no_output_____"
],
[
"class A(Enum):\n \n class J:\n def __init__(self):\n print(\"constructing J\")\n \n class M:\n def __init__(self):\n print(\"constructing M\")\n\n def New(self):\n return self.value()\n \n @classmethod\n def From(cls, foo):\n return cls[foo.upper()].New()",
"_____no_output_____"
],
[
"classname = 'J'\nA[classname].New()",
"_____no_output_____"
],
[
"A.From('m')",
"_____no_output_____"
]
],
[
[
"Back to basics...",
"_____no_output_____"
],
[
"## Cannot subclass Enums",
"_____no_output_____"
]
],
[
[
"class A(Enum):\n X = 1\n Y = 2",
"_____no_output_____"
],
[
"# TypeError: Cannot extend enumerations\nclass B(A):\n Z = 3",
"_____no_output_____"
],
[
"[(a.name, a.value) for a in A] ",
"_____no_output_____"
],
[
"B = Enum(\"B\", [(a.name, a.value) for a in A] + [('Z', 3)])\nlist(B)",
"_____no_output_____"
]
],
[
[
"## Other Enums",
"_____no_output_____"
],
[
"### IntEnum\n\n* The values must be integers\n * Or rather, they are `int()` compatible\n* The type inherits from `int`, so comparisons works \n * no need for `.value`",
"_____no_output_____"
]
],
[
[
"from enum import IntEnum",
"_____no_output_____"
],
[
"class A(IntEnum):\n X = 1\n Y = 2",
"_____no_output_____"
],
[
"A.X",
"_____no_output_____"
],
[
"A.X == 1",
"_____no_output_____"
],
[
"class B(IntEnum):\n X = 1\n Y = '2'",
"_____no_output_____"
],
[
"B.Y == 2",
"_____no_output_____"
],
[
"B('2')",
"_____no_output_____"
]
],
[
[
"### Flag & IntFlag\n\n* New in py3.6\n* Flags support bitwise `&`, `|` operations to *combine* enumerated values\n* Bitflags should be kept to powers of two",
"_____no_output_____"
]
],
[
[
"from enum import Flag, IntFlag",
"_____no_output_____"
],
[
"class A(Flag):\n X = 1 # 1 << 0\n Y = 2 # 1 << 1\n Z = 4 # 1 << 2",
"_____no_output_____"
],
[
"a = A.X | A.Z\na",
"_____no_output_____"
],
[
"isinstance(a, A)",
"_____no_output_____"
],
[
"a & A.X",
"_____no_output_____"
],
[
"a & A.Y",
"_____no_output_____"
],
[
"a & A.Z",
"_____no_output_____"
]
],
[
[
"auto-enumeration works as expected",
"_____no_output_____"
]
],
[
[
"A = IntFlag(\"A\", \"W X Y Z\")\nlist(A)",
"_____no_output_____"
],
[
"A(13)",
"_____no_output_____"
],
[
"for a in A:\n print(f'{a.name} = 0b{a:05b}')",
"_____no_output_____"
]
],
[
[
"### When to use?\n\n* When you have finite set of things and want name & value lookup + iteration for free\n * \"Finite\" means \"Nameable\"\n\n* Examples:\n * Finite states:\n * On/Off\n * Low, Medium, High\n * String or Byte \"validation\" (IntFlag)",
"_____no_output_____"
],
[
"## Example: Suit Of Cards",
"_____no_output_____"
]
],
[
[
"class Suit(Enum):\n Hearts = 1\n Clubs = 2\n Spades = 3\n Diamonds = 4\n \n def is_red(self):\n return self in (Suit.Hearts, Suit.Diamonds)",
"_____no_output_____"
],
[
"list(Suit)",
"_____no_output_____"
],
[
"Suit.Diamonds.is_red()",
"_____no_output_____"
],
[
"from itertools import product\ndeck = list(product(Suit, range(1, 14)))",
"_____no_output_____"
],
[
"deck[:5]",
"_____no_output_____"
],
[
"from random import shuffle\nshuffle(deck)\ndeck[:6]",
"_____no_output_____"
],
[
"class Card:\n def __init__(self, suit, face):\n if isinstance(suit, str):\n suit = Suit[suit]\n self.suit = suit\n self.face = face\n\n def __repr__(self):\n if 1 < self.face < 11:\n f = self.face\n else:\n f = self.FaceToLetter(self.face).name\n return \"<Card %s %s>\" % (f, self.suit.name)\n\n class FaceToLetter(Enum):\n A = 1\n J = 11\n Q = 12\n K = 13",
"_____no_output_____"
],
[
"from itertools import starmap\ndeck = list(starmap(Card, product(Suit, range(1, 14))))",
"_____no_output_____"
],
[
"deck[::5]",
"_____no_output_____"
]
],
[
[
"## Example: Unix File Permissions",
"_____no_output_____"
]
],
[
[
"class Perm(IntFlag):\n READ = 1\n WRITE = 2\n EXEC = 4\n \n def __str__(self):\n return (('r' if self & self.READ else '-') +\n ('w' if self & self.WRITE else '-') +\n ('x' if self & self.EXEC else '-'))",
"_____no_output_____"
],
[
"'%s' % (Perm.READ | Perm.EXEC)",
"_____no_output_____"
],
[
"Perm(6)",
"_____no_output_____"
]
],
[
[
"* Full UNIX file permission has a \"Perm\" for `user`, `group`, `all`.\n* NOT a good case for Enum\n - Many (3 × 2^3) states to choose from\n - You do not want to name them all!",
"_____no_output_____"
]
],
[
[
"class UnixPermission:\n def __init__(self, user: Perm, group: Perm, all: Perm):\n self.user = user\n self.group = group\n self.all = all\n \n @classmethod\n def from_int(cls, num):\n return cls(Perm((num >> 6) & 7),\n Perm((num >> 3) & 7),\n Perm((num >> 0) & 7))\n\n def __str__(self):\n return '%s%s%s' % (self.user, self.group, self.all)",
"_____no_output_____"
],
[
"str(UnixPermission.from_int(0o755))",
"_____no_output_____"
]
],
[
[
"## Example : HTTP Methods & Status Codes\n\n\n* [HTTP Methods](https://en.wikipedia.org/wiki/Hypertext_Transfer_Protocol#Summary_table)\n* [HTTP Status Codes](https://en.wikipedia.org/wiki/List_of_HTTP_status_codes)\n",
"_____no_output_____"
],
[
"Famous Status Codes:\n * 200 - OK\n * 404 - Not Found\n * 500 - Internal Server Error",
"_____no_output_____"
]
],
[
[
"HttpMethod = Enum(\"HttpMethod\", \"GET POST PUT DELETE\")",
"_____no_output_____"
],
[
"try:\n method = HttpMethod[method_str] \nexcept KeyError:\n raise UnknownHttpMethodException(method_str)",
"_____no_output_____"
],
[
"class HttpStatus(IntEnum):\n OK = 200\n NOT_FOUND = 404\n INTERNAL_SERVER_ERROR = 500",
"_____no_output_____"
],
[
"class HttpError(Exception):\n pass\n\nclass HttpErrorNotFound(HttpError):\n status = HttpStatus.NOT_FOUND\n\nclass HttpErrorISE(HttpError):\n status = HttpStatus.INTERNAL_SERVER_ERROR\n",
"_____no_output_____"
],
[
"from http import HTTPStatus\nHTTPStatus",
"_____no_output_____"
],
[
"HTTPStatus['OK']",
"_____no_output_____"
],
[
"HTTPStatus[\"OK\"].description",
"_____no_output_____"
],
[
"HTTPStatus(404)",
"_____no_output_____"
],
[
"list(HTTPStatus)",
"_____no_output_____"
]
],
[
[
"## Summary\n\n* Enums are a useful tool in your programming toolbox\n * Should be aware of them and when to use them\n* Nothing truely new (you could implement with dictionaries), but get automatic features\n* For more information, check the documentation https://docs.python.org/3/library/enum.html\n",
"_____no_output_____"
],
[
"# END",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ec8853bd7c47a5b8a4cf224010b4ada37cb0e39b | 28,425 | ipynb | Jupyter Notebook | 06_neural_network.ipynb | Chanonsersa/Artificial-Intelligence | bfbdfcf4204048565323060044267bff9b040348 | [
"MIT"
] | null | null | null | 06_neural_network.ipynb | Chanonsersa/Artificial-Intelligence | bfbdfcf4204048565323060044267bff9b040348 | [
"MIT"
] | null | null | null | 06_neural_network.ipynb | Chanonsersa/Artificial-Intelligence | bfbdfcf4204048565323060044267bff9b040348 | [
"MIT"
] | null | null | null | 50.578292 | 8,682 | 0.676482 | [
[
[
"# 6. Deep Learning Neural Network",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers import Conv2D, MaxPooling2D, Flatten, Dense\nfrom keras import Input\nfrom keras.utils import np_utils\nfrom keras.datasets import mnist\nimport visualkeras\n\nimport numpy as np\n",
"_____no_output_____"
]
],
[
[
"## Neuron Network",
"_____no_output_____"
],
[
"### 1. Create a model\n\n- Create a model as a sequential model\n- Defining the number of neuron in each layer and activation function",
"_____no_output_____"
]
],
[
[
"# Model / data parameter\nnum_classes = 10\ninput_shape = (784)\n\nmodel = Sequential(\n [\n Input(shape=input_shape),\n Dense(32, activation='relu'),\n Dense(10, activation='softmax')\n ]\n)\nmodel.summary()\n",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 32) 25120 \n_________________________________________________________________\ndense_1 (Dense) (None, 10) 330 \n=================================================================\nTotal params: 25,450\nTrainable params: 25,450\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"visualkeras.layered_view(model, legend=True, draw_volume=True, spacing=10)",
"_____no_output_____"
]
],
[
[
"#### Model parameters\n\n- 1<sup>st</sup> layer: (784 + 1) * 32 = 25,120\n- 2<sup>nd</sup> layer: (32 + 1) * 10 = 330\n- Total parameter: 25,450\n",
"_____no_output_____"
],
[
"### 2. Compile a model\n\n- Setting optimizer\n - `adam`, `SGD`, `rmsprop`, etc\n- Setting loss function\n - `mean_squared_error`, `mean_absolute_error`, `categorical_crossentropy`, etc\n- Setting evaluation metric",
"_____no_output_____"
]
],
[
[
"model.compile(loss=\"categorical_crossentropy\",\n optimizer=\"adam\", metrics=[\"accuracy\"])\n",
"_____no_output_____"
]
],
[
[
"### 3. Training a model\n\n- Load MNIST data\n- Traning data (60k) + Testing data (10k)\n- Convert from 28x28 pixel to 784 one dimensional vector",
"_____no_output_____"
]
],
[
[
"# the data, split between train and test sets\n(X_train, y_train), (X_test, y_test) = mnist.load_data()\n\n# Scale images to the [0, 1] range\nX_train = X_train.astype(\"float32\") / 255\nX_test = X_test.astype(\"float32\") / 255\n\n# Convert from (28, 28) to (784)\nX_train = X_train.reshape(60000, 784)\nX_test = X_test.reshape(10000, 784)\n\n# Make sure images have shape (784)\nprint(\"x_train shape:\", X_train.shape)\nprint(X_train.shape[0], \"train samples\")\nprint(X_test.shape[0], \"test samples\")\n\n\n# Convert class vectors to binary class matrices\ny_train = np_utils.to_categorical(y_train, num_classes)\ny_test = np_utils.to_categorical(y_test, num_classes)\n",
"x_train shape: (60000, 784)\n60000 train samples\n10000 test samples\n"
],
[
"batch_size = 128\nepochs = 15\n\nmodel.fit(X_train, y_train, batch_size=batch_size,\n epochs=epochs, verbose=1, validation_split=.1)\n",
"2022-02-08 00:01:54.993679: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:185] None of the MLIR Optimization Passes are enabled (registered 2)\n"
]
],
[
[
"### 4. Evaluate the trained model",
"_____no_output_____"
]
],
[
[
"score = model.evaluate(X_test, y_test, verbose=0)\nprint(\"Test loss:\", score[0])\nprint(\"Test accuracy:\", score[1])\n",
"Test loss: 0.10482573509216309\nTest accuracy: 0.9681000113487244\n"
]
],
[
[
"## Convolutional Neuron Network (CNN)",
"_____no_output_____"
],
[
"### 1. Create a model",
"_____no_output_____"
]
],
[
[
"# Model / data parameters\nnum_classes = 10\ninput_shape = (28, 28, 1)\n\n\ncnn_model = Sequential(\n [\n Input(shape=input_shape),\n Conv2D(32, kernel_size=(3, 3), activation=\"relu\"),\n MaxPooling2D(pool_size=(2, 2)),\n Conv2D(32, kernel_size=(3, 3), activation=\"relu\"),\n MaxPooling2D(pool_size=(2, 2)),\n Flatten(),\n Dense(64, activation='relu'),\n Dense(num_classes, activation=\"softmax\"),\n ]\n)\n\ncnn_model.summary()\n",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d (Conv2D) (None, 26, 26, 32) 320 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 13, 13, 32) 0 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 11, 11, 32) 9248 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 5, 5, 32) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 800) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 64) 51264 \n_________________________________________________________________\ndense_3 (Dense) (None, 10) 650 \n=================================================================\nTotal params: 61,482\nTrainable params: 61,482\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"visualkeras.layered_view(cnn_model, legend=True, draw_volume=True, spacing=10)\n",
"_____no_output_____"
]
],
[
[
"### 2. Complie a model",
"_____no_output_____"
]
],
[
[
"cnn_model.compile(loss=\"categorical_crossentropy\",\n optimizer=\"adam\", metrics=[\"accuracy\"])",
"_____no_output_____"
]
],
[
[
"### 3. Training a model\n\n- Load MNIST data\n- Traning data (60k) + Testing data (10k)",
"_____no_output_____"
]
],
[
[
"# the data, split between train and test sets\n(X_train, y_train), (X_test, y_test) = mnist.load_data()\n\n# Scale images to the [0, 1] range\nX_train = X_train.astype(\"float32\") / 255\nX_test = X_test.astype(\"float32\") / 255\n\n# Make sure images have shape (28, 28, 1)\nX_train = np.expand_dims(X_train, -1)\nX_test = np.expand_dims(X_test, -1)\nprint(\"x_train shape:\", X_train.shape)\nprint(X_train.shape[0], \"train samples\")\nprint(X_train.shape[0], \"test samples\")\n\n\n# Convert class vectors to binary class matrices\ny_train = np_utils.to_categorical(y_train, num_classes)\ny_test = np_utils.to_categorical(y_test, num_classes)\n",
"x_train shape: (60000, 28, 28, 1)\n60000 train samples\n60000 test samples\n"
],
[
"batch_size = 128\nepochs = 15\n\ncnn_model.fit(X_train, y_train, batch_size=batch_size,\n epochs=epochs, verbose=1, validation_split=.1)\n",
"Epoch 1/15\n422/422 [==============================] - 17s 40ms/step - loss: 0.3123 - accuracy: 0.9106 - val_loss: 0.0941 - val_accuracy: 0.9738\nEpoch 2/15\n422/422 [==============================] - 17s 40ms/step - loss: 0.0858 - accuracy: 0.9733 - val_loss: 0.0608 - val_accuracy: 0.9842\nEpoch 3/15\n422/422 [==============================] - 18s 42ms/step - loss: 0.0617 - accuracy: 0.9806 - val_loss: 0.0492 - val_accuracy: 0.9867\nEpoch 4/15\n422/422 [==============================] - 17s 41ms/step - loss: 0.0503 - accuracy: 0.9844 - val_loss: 0.0457 - val_accuracy: 0.9877\nEpoch 5/15\n422/422 [==============================] - 17s 41ms/step - loss: 0.0416 - accuracy: 0.9869 - val_loss: 0.0426 - val_accuracy: 0.9863\nEpoch 6/15\n422/422 [==============================] - 18s 41ms/step - loss: 0.0357 - accuracy: 0.9890 - val_loss: 0.0396 - val_accuracy: 0.9887\nEpoch 7/15\n422/422 [==============================] - 19s 45ms/step - loss: 0.0308 - accuracy: 0.9904 - val_loss: 0.0344 - val_accuracy: 0.9905\nEpoch 8/15\n422/422 [==============================] - 17s 41ms/step - loss: 0.0264 - accuracy: 0.9918 - val_loss: 0.0394 - val_accuracy: 0.9890\nEpoch 9/15\n422/422 [==============================] - 16s 38ms/step - loss: 0.0237 - accuracy: 0.9929 - val_loss: 0.0413 - val_accuracy: 0.9880\nEpoch 10/15\n422/422 [==============================] - 17s 39ms/step - loss: 0.0210 - accuracy: 0.9934 - val_loss: 0.0410 - val_accuracy: 0.9892\nEpoch 11/15\n422/422 [==============================] - 16s 38ms/step - loss: 0.0174 - accuracy: 0.9944 - val_loss: 0.0393 - val_accuracy: 0.9898\nEpoch 12/15\n422/422 [==============================] - 18s 42ms/step - loss: 0.0150 - accuracy: 0.9956 - val_loss: 0.0408 - val_accuracy: 0.9897\nEpoch 13/15\n422/422 [==============================] - 17s 41ms/step - loss: 0.0141 - accuracy: 0.9953 - val_loss: 0.0339 - val_accuracy: 0.9910\nEpoch 14/15\n422/422 [==============================] - 18s 42ms/step - loss: 0.0117 - accuracy: 0.9958 - val_loss: 0.0381 - val_accuracy: 0.9905\nEpoch 15/15\n422/422 [==============================] - 15s 36ms/step - loss: 0.0102 - accuracy: 0.9963 - val_loss: 0.0506 - val_accuracy: 0.9882\n"
]
],
[
[
"### 4. Evaluate the trained model",
"_____no_output_____"
]
],
[
[
"score = cnn_model.evaluate(X_test, y_test, verbose=0)\nprint(\"Test loss:\", score[0])\nprint(\"Test accuracy:\", score[1])\n",
"Test loss: 0.04123755171895027\nTest accuracy: 0.9883999824523926\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec885a3375e4f4bd2610ff994ad13555e6ead5c3 | 9,201 | ipynb | Jupyter Notebook | WordCount.ipynb | mesc08/movie-reviews-sentiment-analysis | beb06e096853c245230f665082bd681ae91728fd | [
"MIT"
] | null | null | null | WordCount.ipynb | mesc08/movie-reviews-sentiment-analysis | beb06e096853c245230f665082bd681ae91728fd | [
"MIT"
] | null | null | null | WordCount.ipynb | mesc08/movie-reviews-sentiment-analysis | beb06e096853c245230f665082bd681ae91728fd | [
"MIT"
] | null | null | null | 27.061765 | 264 | 0.461146 | [
[
[
"## IMPORT LIBRARIES",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport os \nimport re\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.metrics import accuracy_score, confusion_matrix, classification_report\nfrom sklearn.linear_model import LogisticRegression",
"C:\\Users\\srija\\Anaconda3\\lib\\site-packages\\sklearn\\utils\\__init__.py:4: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated since Python 3.3,and in 3.9 it will stop working\n from collections import Sequence\nC:\\Users\\srija\\Anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:18: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated since Python 3.3,and in 3.9 it will stop working\n from collections import Iterable\nC:\\Users\\srija\\Anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_search.py:16: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated since Python 3.3,and in 3.9 it will stop working\n from collections import Mapping, namedtuple, defaultdict, Sequence\n"
]
],
[
[
"## READFILE",
"_____no_output_____"
]
],
[
[
"train_review =[]\nfor line in open('../movie-sentiment-analysis/aclImdb/movie_data/full_train.txt','r',encoding='utf-8'):\n train_review.append(line.strip())\n",
"_____no_output_____"
],
[
"test_review=[]\nfor line in open('../movie-sentiment-analysis/aclImdb/movie_data/full_test.txt','r',encoding='utf-8'):\n test_review.append(line.strip())",
"_____no_output_____"
]
],
[
[
"## CLEANING AND PROCESSING",
"_____no_output_____"
]
],
[
[
"no_space = re.compile(\"(\\.)|(\\;)|(\\:)|(\\!)|(\\')|(\\?)|(\\,)|(\\\")|(\\()|(\\))|(\\[)|(\\])|(\\d+)\")\nwith_space = re.compile(\"(<br\\s*/><br\\s*/>)|(\\-)|(\\/)\")\ndef cleaningandprocessing(reviews):\n reviews = [no_space.sub(\"\",line.lower()) for line in reviews]\n reviews = [with_space.sub(\" \",line) for line in reviews]\n return reviews\nclean_train_reviews = cleaningandprocessing(train_review)\nclean_test_reviews = cleaningandprocessing(test_review)",
"_____no_output_____"
]
],
[
[
"## USING WORD COUNT ANALYSIS",
"_____no_output_____"
]
],
[
[
"cv = CountVectorizer(binary=False)\ncv.fit(clean_train_reviews)\nX_train = cv.transform(clean_train_reviews)\nX_test = cv.transform(clean_test_reviews)",
"_____no_output_____"
],
[
"target = [1 if i <12500 else 0 for i in range(25000)]\nx_train, x_test, y_train, y_test = train_test_split(X_train, target, test_size=0.8, random_state=42)",
"_____no_output_____"
],
[
"for c in [0.01,0.05,0.1,0.25,0.5,1]:\n lr = LogisticRegression(C=c)\n lr.fit(x_train, y_train)\n print(\"For: \", c)\n print(\"\\n\")\n print(accuracy_score(y_test, lr.predict(x_test)))\n print(\"\\n\")\n print(confusion_matrix(y_test, lr.predict(x_test)))\n print(\"\\n\")\n print(classification_report(y_test, lr.predict(x_test)))\n print(\"\\n\\n\")",
"For: 0.01\n\n\n0.8462\n\n\n[[8371 1591]\n [1485 8553]]\n\n\n precision recall f1-score support\n\n 0 0.85 0.84 0.84 9962\n 1 0.84 0.85 0.85 10038\n\navg / total 0.85 0.85 0.85 20000\n\n\n\n\nFor: 0.05\n\n\n0.85785\n\n\n[[8496 1466]\n [1377 8661]]\n\n\n precision recall f1-score support\n\n 0 0.86 0.85 0.86 9962\n 1 0.86 0.86 0.86 10038\n\navg / total 0.86 0.86 0.86 20000\n\n\n\n\nFor: 0.1\n\n\n0.857\n\n\n[[8490 1472]\n [1388 8650]]\n\n\n precision recall f1-score support\n\n 0 0.86 0.85 0.86 9962\n 1 0.85 0.86 0.86 10038\n\navg / total 0.86 0.86 0.86 20000\n\n\n\n\nFor: 0.25\n\n\n0.85645\n\n\n[[8469 1493]\n [1378 8660]]\n\n\n precision recall f1-score support\n\n 0 0.86 0.85 0.86 9962\n 1 0.85 0.86 0.86 10038\n\navg / total 0.86 0.86 0.86 20000\n\n\n\n\nFor: 0.5\n\n\n0.85425\n\n\n[[8446 1516]\n [1399 8639]]\n\n\n precision recall f1-score support\n\n 0 0.86 0.85 0.85 9962\n 1 0.85 0.86 0.86 10038\n\navg / total 0.85 0.85 0.85 20000\n\n\n\n\nFor: 1\n\n\n0.85235\n\n\n[[8427 1535]\n [1418 8620]]\n\n\n precision recall f1-score support\n\n 0 0.86 0.85 0.85 9962\n 1 0.85 0.86 0.85 10038\n\navg / total 0.85 0.85 0.85 20000\n\n\n\n\n"
]
],
[
[
"### FOR C = 0.05 ACCURACY SCORE = 85.875",
"_____no_output_____"
]
],
[
[
"final_model = LogisticRegression(C=0.05)\nfinal_model.fit(X_train, target)\npredictions1 = final_model.predict(X_test)\nprint(\"Accuracy score is: \",accuracy_score(target,predictions1))\nprint(\"\\n\")\nprint(\"Confusion matrix is: \", confusion_matrix(target, predictions1))\nprint(\"\\n\")\nprint(\"Classification matrix is: \",classification_report(target, predictions1))",
"Accuracy score is: 0.88196\n\n\nConfusion matrix is: [[11033 1467]\n [ 1484 11016]]\n\n\nClassification matrix is: precision recall f1-score support\n\n 0 0.88 0.88 0.88 12500\n 1 0.88 0.88 0.88 12500\n\navg / total 0.88 0.88 0.88 25000\n\n"
]
],
[
[
"### FINAL MODEL ACCURACY SCORE IS 88.196",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec885de200a88cae0db7f4329f6ccca5f35c74c0 | 17,058 | ipynb | Jupyter Notebook | ANTARES/lsst_broker_workshop_2021.ipynb | dmitryduev/tutorials | 4c234c7327e77f997e06c7f4010f2bed303d4e25 | [
"MIT"
] | 5 | 2021-04-04T21:03:19.000Z | 2021-04-24T20:45:10.000Z | ANTARES/lsst_broker_workshop_2021.ipynb | dmitryduev/tutorials | 4c234c7327e77f997e06c7f4010f2bed303d4e25 | [
"MIT"
] | 1 | 2021-04-09T05:36:46.000Z | 2021-04-09T05:36:46.000Z | ANTARES/lsst_broker_workshop_2021.ipynb | dmitryduev/tutorials | 4c234c7327e77f997e06c7f4010f2bed303d4e25 | [
"MIT"
] | 9 | 2021-04-06T01:46:46.000Z | 2021-08-28T13:24:35.000Z | 33.51277 | 306 | 0.511666 | [
[
[
"## Before you begin:\n- Clone the repo, navigate to correct directory.\n- Install all package requirements.",
"_____no_output_____"
]
],
[
[
"! git clone https://github.com/broker-workshop/tutorials.git",
"fatal: destination path 'tutorials' already exists and is not an empty directory.\n"
],
[
"%cd tutorials/ANTARES/data",
"/content/tutorials/ANTARES/data\n"
],
[
"%%capture\n! pip install matplotlib pandas astropy astroquery ligo.skymap healpy pickle5 keras-tcn\n! pip install git+https://github.com/deepchatterjeeligo/astrorapid.git@broker-workshop",
"_____no_output_____"
]
],
[
[
"The typical scenario considered here is:\n* A binary neutron star coalescence is observed by LIGO/Virgo/KAGRA (LVK) gravitational-wave detectors\n* The LVK alert contains a sky-localization, which is a probability distribution in sky coordinates.\n \n > GW data can only provide an sky-localization based on the strength of the signal, the bandwidth of the signal, and participating detectors. These can be order $\\mathcal{O}(10)$ to $\\mathcal{O}(1000)$ sq. deg. in sky\n\n* There is an associated counterpart: the _kilonova_, like GW170817 \n* There may be several contaminants in the field of view of the kilonova, specially as we get more sensitive telescopes. We need to down select potential objects.\n* Communicate to scheduling facilities. For example [Treasure Map](http://treasuremap.space/).\n\nWe will consider the situation with simulated lightcurves.\n- We have seen just one kilonova\n- Data is proprietary at the time of GW discovery (at least from high probability region of skymap)\n- Need simulated objects for mock tests. (see talk + demo on PLAsTiCC)",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"try:\n lightcurves = pd.read_pickle('lightcurves.pickle')\nexcept ValueError:\n import pickle5\n with open('lightcurves.pickle', 'rb') as f:\n lightcurves = pickle5.load(f)",
"_____no_output_____"
]
],
[
[
"## Here are 10 lightcurves (9 contaminants and 1 kilonova)\n\nThese were simulated considering an observing cadence of ZTF. The last one is the Kilonova (KN).\n\n(For those interested, the data (SED models) are publicly available as a part of [SNANA package data](https://zenodo.org/record/4015340#.YHZ3ehJOlcA))",
"_____no_output_____"
]
],
[
[
"lightcurves.SIM_MODEL_NAME",
"_____no_output_____"
]
],
[
[
"## Plot the KN",
"_____no_output_____"
]
],
[
[
"kn = lightcurves.iloc[-1] # the last one is the KN",
"_____no_output_____"
],
[
"plt.figure(figsize=(14, 6))\n\npkmjd_kn = kn.pkmjd\nmjd_r_kn = kn.mjd_r - pkmjd_kn\nmjd_g_kn = kn.mjd_g - pkmjd_kn\n\nplt.errorbar(mjd_r_kn, kn.mag_r, yerr=kn.magerr_r, color='red',\n marker='*', fmt='o', label='R')\nplt.errorbar(mjd_g_kn, kn.mag_g, yerr=kn.magerr_g, color='green',\n marker='^', fmt='o', label='g')\nplt.ylim(plt.ylim()[::-1])\nplt.legend(fontsize=14)\nplt.xlabel('Time (days)', fontsize=14)\nplt.title(\"Kilonova\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"Kilonovae evolve rapidly. Given LSST cadence, discovery will be hard. Need rapid turnaround time (look at the light curve in comparison to SN below)",
"_____no_output_____"
],
[
"## Plot a few contaminant objects",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(14, 6))\n\nfor idx, row in lightcurves.iterrows():\n if 'SALT' not in row.SIM_MODEL_NAME: # plot SALT2 Ia; try a few others from list above\n continue\n pkmjd = row.pkmjd\n mjd_r = row.mjd_r - pkmjd\n mjd_g = row.mjd_g - pkmjd\n plt.errorbar(mjd_r, row.mag_r, yerr=row.magerr_r, color='red',\n marker='*', fmt='o', label='R')\n plt.errorbar(mjd_g, row.mag_g, yerr=row.magerr_g, color='green',\n marker='^', fmt='o', label='g')\nplt.ylim(plt.ylim()[::-1])\nplt.legend(fontsize=14)\nplt.xlabel('Time (days)', fontsize=14)\nplt.title(\"Contaminant\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Associated skymap",
"_____no_output_____"
]
],
[
[
"import healpy as hp\nfrom ligo.skymap.io import read_sky_map",
"_____no_output_____"
],
[
"skymap, *h = read_sky_map('skymap.fits')\nplt.figure(figsize=(15, 10))\nhp.mollview(skymap, fig=0, cmap='YlOrBr', cbar=False, hold=True,\n title=\"Mock LVK skymap associated with KN\")\n\n# plot locations of all objects\nfor idx, row in lightcurves.iterrows():\n hp.visufunc.projplot(\n row.SIM_RA, row.SIM_DEC, lonlat=True, marker='x',\n c='r' if row.true_label else 'b',\n markersize=15 if row.true_label else 5\n )\nhp.graticule()",
"_____no_output_____"
]
],
[
[
"From the skymap, we can see that there are several supernovae which coincide with the kilonova in sky location as \"contaminants\". It is imperative to single out the kilonovae from the other events.",
"_____no_output_____"
],
[
"A simple filter\n- check temporal coincidence\n- check consistency with skymap\n- crossmatch with a galaxy catalog",
"_____no_output_____"
]
],
[
[
"from astropy import coordinates, units as u\nfrom astroquery.vizier import Vizier\nfrom ligo.skymap.postprocess import crossmatch\n\n\ngw_trigger_mjd = 58347\nskymap_filename = 'skymap.fits'\nskymap = read_sky_map(skymap_filename, moc=True)\n\n# load a catalog\ncatalog, = Vizier.query_constraints(\n catalog='VII/281/glade2',\n)",
"_____no_output_____"
],
[
"def simple_kn_filter(obj):\n \"\"\"Simple filter to downselect an associated KN\"\"\"\n # check if there are any detections before GW trigger time\n mjd = np.hstack((obj.mjd_g, obj.mjd_r, obj.mjd_i))\n print(f\"\\nChecking for object {obj.SIM_MODEL_NAME.strip()}\")\n if np.any(mjd < gw_trigger_mjd):\n print(\"Found detections before GW trigger time\")\n return False\n \n # check consistency with skymap\n obj_location = coordinates.SkyCoord(\n obj.SIM_RA * u.deg, # in reality this will be part of the alert\n obj.SIM_DEC * u.deg\n )\n # get a ligo.skymap crossmatch result\n crossmatch_result = crossmatch(\n skymap, coordinates=(obj_location,),\n contours=(0.5, 0.95)\n )\n # get line of sight p-value\n p_val = crossmatch_result.probdensity\n # get angular offset from mode\n offset, *_ = crossmatch_result.offset * u.deg\n # get sky areas of skymap\n area_fifty, area_ninety = crossmatch_result.contour_areas * u.deg**2\n # get searched area from posterior mode to target\n searched_area, *_ = crossmatch_result.searched_area * u.deg**2\n # put a threshold on searched area\n if searched_area > area_ninety:\n print(f\"Searched area for {obj.SIM_MODEL_NAME.strip()} is {searched_area:.2f} > \"\n f\"90% area of {area_ninety:.2f}\")\n return False\n \n # print objects matched to a galaxy catalog\n catalog_object_locations = coordinates.SkyCoord(catalog['RAJ2000'], catalog['DEJ2000'])\n idx, sep2d, dist3d = obj_location.match_to_catalog_sky(catalog_object_locations)\n closest_galaxy = catalog[idx]\n print(\n f\"Found matching galaxy @ RA: {closest_galaxy['RAJ2000']:.3f} / \"\n f\"DEC: {closest_galaxy['DEJ2000']:.3f}\"\n )\n print(\n f\"Transient @ RA:{obj.SIM_RA:.3f} \"\n f\"DEC: {obj.SIM_DEC:.3f} / \"\n f\"Angular sep = {sep2d[0]:.3f}\"\n )\n return True",
"_____no_output_____"
],
[
"[simple_kn_filter(row) for idx, row in lightcurves.iterrows()]",
"_____no_output_____"
]
],
[
[
"Thus simple temporal and spatial selection cuts can help us downselect. A more realistic situation may have an associated galaxy (it also may not since the galaxy catalog may be incomplete). These temporal information and spatial cuts based on the skymap was used during O3 operations in ANTARES.",
"_____no_output_____"
],
[
"## Classifying the lightcurve (WIP)\n\nEarly epoch classification code [RAPID](https://astrorapid.readthedocs.io/en/latest/) is a part of ANTARES.\n\nBut classification may be challenging just from the lightcurve. Since there may not be enough data. Hence we want to use the available contextual information available. Luckily, we get some contextual info for free - LVK skymap itself.",
"_____no_output_____"
]
],
[
[
"from astrorapid import Classify\n\nclassification = Classify(\n model_filepath='trained_model.hdf5',\n known_redshift=False,\n passbands=('g', 'r', 'i'),\n class_names = ('Pre-explosion', 'Kilonova', 'Other'),\n mintime=-5,\n timestep=3\n)",
"_____no_output_____"
],
[
"def get_data_to_classify(obj):\n mjd = np.hstack((obj.mjd_g, obj.mjd_r, obj.mjd_i))\n sort_mask = np.argsort(mjd)\n\n flux = np.hstack((obj.fluxcal_g, obj.fluxcal_r, obj.fluxcal_i))\n fluxerr = np.hstack((obj.fluxcalerr_g, obj.fluxcalerr_r, obj.fluxcalerr_i))\n photflag = np.hstack((obj.photflag_g, obj.photflag_r, obj.photflag_i))\n passbands = np.array(obj.mjd_g.size*['g'] + obj.mjd_r.size*['r'] + obj.mjd_i.size*['i'])\n \n objid = obj.SIM_MODEL_NAME.strip()\n ra = obj.SIM_RA\n dec = obj.SIM_DEC\n redshift = obj.z\n mwebv = obj.SIM_MWEBV\n return (\n mjd[sort_mask], flux[sort_mask], fluxerr[sort_mask],\n passbands[sort_mask], photflag[sort_mask], ra, dec,\n objid, redshift, mwebv\n )",
"_____no_output_____"
],
[
"for idx, row in lightcurves.iterrows():\n print(f\"\\n#### True model: {row.SIM_MODEL_NAME.strip()} ####\")\n lightcurve_data = get_data_to_classify(row)\n other_meta_data = dict(offset=row.offset, logprob=row.logprob)\n\n predictions, time_steps = classification.get_predictions(\n [lightcurve_data], return_predictions_at_obstime=False,\n other_meta_data=[other_meta_data]\n )\n if not predictions:\n continue\n kn_prediction = predictions[0].T[1]\n print(\"Time:\", time_steps[0] - gw_trigger_mjd)\n print(\"KN class probabilities:\", kn_prediction)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
ec885e6b49ca0007480abd6de8af7942638f43e5 | 829,199 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Kaggle House Prices-checkpoint.ipynb | bmaelum/kaggle_houseprices | 774765a1408e836b7e370dd2da64197d0acc832c | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Kaggle House Prices-checkpoint.ipynb | bmaelum/kaggle_houseprices | 774765a1408e836b7e370dd2da64197d0acc832c | [
"MIT"
] | 23 | 2020-03-24T17:07:39.000Z | 2022-03-29T22:15:49.000Z | .ipynb_checkpoints/Kaggle House Prices-checkpoint.ipynb | bmaelum/kaggle_houseprices | 774765a1408e836b7e370dd2da64197d0acc832c | [
"MIT"
] | null | null | null | 76.02448 | 331,140 | 0.727898 | [
[
[
"# House Prices: Advanced Regression Techniques\n### [Link to Kaggle Challenge](https://www.kaggle.com/c/house-prices-advanced-regression-techniques/data)",
"_____no_output_____"
],
[
"## Workflow\n1. Question or problem definition.\n2. Acquire training and testing data.\n3. Wrangle, prepare, cleanse the data.\n4. Analyze, identify patterns, and explore the data.\n5. Model, predict and solve the problem.\n6. Visualize, report, and present the problem solving steps and final solution.\n7. Supply or submit the results.",
"_____no_output_____"
],
[
"### Workflow goals\n\nThe data science solutions workflow solves for seven major goals.\n\n**Classifying:** We may want to classify or categorize our samples. We may also want to understand the implications or correlation of different classes with our solution goal.\n\n**Correlating:** One can approach the problem based on available features within the training dataset. Which features within the dataset contribute significantly to our solution goal? Statistically speaking is there a correlation among a feature and solution goal? As the feature values change does the solution state change as well, and visa-versa? This can be tested both for numerical and categorical features in the given dataset. We may also want to determine correlation among features other than survival for subsequent goals and workflow stages. Correlating certain features may help in creating, completing, or correcting features.\n\n**Converting:** For modeling stage, one needs to prepare the data. Depending on the choice of model algorithm one may require all features to be converted to numerical equivalent values. So for instance converting text categorical values to numeric values.\n\n**Completing.** Data preparation may also require us to estimate any missing values within a feature. Model algorithms may work best when there are no missing values.\n\n**Correcting:** We may also analyze the given training dataset for errors or possibly innacurate values within features and try to corrent these values or exclude the samples containing the errors. One way to do this is to detect any outliers among our samples or features. We may also completely discard a feature if it is not contribting to the analysis or may significantly skew the results.\n\n**Creating:** Can we create new features based on an existing feature or a set of features, such that the new feature follows the correlation, conversion, completeness goals.\n\n**Charting:** How to select the right visualization plots and charts depending on nature of the data and the solution goals.",
"_____no_output_____"
],
[
"# 1. Competition Description\nAsk a home buyer to describe their dream house, and they probably won't begin with the height of the basement ceiling or the proximity to an east-west railroad. But this playground competition's dataset proves that much more influences price negotiations than the number of bedrooms or a white-picket fence.\n\nWith 79 explanatory variables describing (almost) every aspect of residential homes in Ames, Iowa, this competition challenges you to predict the final price of each home.",
"_____no_output_____"
],
[
"# 2. Acquire training and test data",
"_____no_output_____"
]
],
[
[
"# data analysis and wrangling\nimport pandas as pd\nimport numpy as np\nimport random as rnd\n\n# visualization\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# machine learning\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.svm import SVC, LinearSVC\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.linear_model import Perceptron\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\n\nimport xgboost\nfrom xgboost import XGBRegressor\n\n# Feature selection\nfrom boruta import BorutaPy\n\n",
"_____no_output_____"
],
[
"!pip freeze > requirements.txt",
"_____no_output_____"
],
[
"df_train = pd.read_csv('data/train.csv') \ndf_test = pd.read_csv('data/test.csv')\ncombine = [df_train, df_test]",
"_____no_output_____"
],
[
"df_train.shape",
"_____no_output_____"
],
[
"df_train.head(3)",
"_____no_output_____"
],
[
"df_test.shape",
"_____no_output_____"
],
[
"df_test.sort_values(by='Id').head(3)",
"_____no_output_____"
],
[
"df_test.interpolate().isna().sum()",
"_____no_output_____"
],
[
"f = open(\"data/data_description.txt\", \"r\")\nprint(f.read())",
"MSSubClass: Identifies the type of dwelling involved in the sale.\t\n\n 20\t1-STORY 1946 & NEWER ALL STYLES\n 30\t1-STORY 1945 & OLDER\n 40\t1-STORY W/FINISHED ATTIC ALL AGES\n 45\t1-1/2 STORY - UNFINISHED ALL AGES\n 50\t1-1/2 STORY FINISHED ALL AGES\n 60\t2-STORY 1946 & NEWER\n 70\t2-STORY 1945 & OLDER\n 75\t2-1/2 STORY ALL AGES\n 80\tSPLIT OR MULTI-LEVEL\n 85\tSPLIT FOYER\n 90\tDUPLEX - ALL STYLES AND AGES\n 120\t1-STORY PUD (Planned Unit Development) - 1946 & NEWER\n 150\t1-1/2 STORY PUD - ALL AGES\n 160\t2-STORY PUD - 1946 & NEWER\n 180\tPUD - MULTILEVEL - INCL SPLIT LEV/FOYER\n 190\t2 FAMILY CONVERSION - ALL STYLES AND AGES\n\nMSZoning: Identifies the general zoning classification of the sale.\n\t\t\n A\tAgriculture\n C\tCommercial\n FV\tFloating Village Residential\n I\tIndustrial\n RH\tResidential High Density\n RL\tResidential Low Density\n RP\tResidential Low Density Park \n RM\tResidential Medium Density\n\t\nLotFrontage: Linear feet of street connected to property\n\nLotArea: Lot size in square feet\n\nStreet: Type of road access to property\n\n Grvl\tGravel\t\n Pave\tPaved\n \t\nAlley: Type of alley access to property\n\n Grvl\tGravel\n Pave\tPaved\n NA \tNo alley access\n\t\t\nLotShape: General shape of property\n\n Reg\tRegular\t\n IR1\tSlightly irregular\n IR2\tModerately Irregular\n IR3\tIrregular\n \nLandContour: Flatness of the property\n\n Lvl\tNear Flat/Level\t\n Bnk\tBanked - Quick and significant rise from street grade to building\n HLS\tHillside - Significant slope from side to side\n Low\tDepression\n\t\t\nUtilities: Type of utilities available\n\t\t\n AllPub\tAll public Utilities (E,G,W,& S)\t\n NoSewr\tElectricity, Gas, and Water (Septic Tank)\n NoSeWa\tElectricity and Gas Only\n ELO\tElectricity only\t\n\t\nLotConfig: Lot configuration\n\n Inside\tInside lot\n Corner\tCorner lot\n CulDSac\tCul-de-sac\n FR2\tFrontage on 2 sides of property\n FR3\tFrontage on 3 sides of property\n\t\nLandSlope: Slope of property\n\t\t\n Gtl\tGentle slope\n Mod\tModerate Slope\t\n Sev\tSevere Slope\n\t\nNeighborhood: Physical locations within Ames city limits\n\n Blmngtn\tBloomington Heights\n Blueste\tBluestem\n BrDale\tBriardale\n BrkSide\tBrookside\n ClearCr\tClear Creek\n CollgCr\tCollege Creek\n Crawfor\tCrawford\n Edwards\tEdwards\n Gilbert\tGilbert\n IDOTRR\tIowa DOT and Rail Road\n MeadowV\tMeadow Village\n Mitchel\tMitchell\n Names\tNorth Ames\n NoRidge\tNorthridge\n NPkVill\tNorthpark Villa\n NridgHt\tNorthridge Heights\n NWAmes\tNorthwest Ames\n OldTown\tOld Town\n SWISU\tSouth & West of Iowa State University\n Sawyer\tSawyer\n SawyerW\tSawyer West\n Somerst\tSomerset\n StoneBr\tStone Brook\n Timber\tTimberland\n Veenker\tVeenker\n\t\t\t\nCondition1: Proximity to various conditions\n\t\n Artery\tAdjacent to arterial street\n Feedr\tAdjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad\n\t\nCondition2: Proximity to various conditions (if more than one is present)\n\t\t\n Artery\tAdjacent to arterial street\n Feedr\tAdjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad\n\t\nBldgType: Type of dwelling\n\t\t\n 1Fam\tSingle-family Detached\t\n 2FmCon\tTwo-family Conversion; originally built as one-family dwelling\n Duplx\tDuplex\n TwnhsE\tTownhouse End Unit\n TwnhsI\tTownhouse Inside Unit\n\t\nHouseStyle: Style of dwelling\n\t\n 1Story\tOne story\n 1.5Fin\tOne and one-half story: 2nd level finished\n 1.5Unf\tOne and one-half story: 2nd level unfinished\n 2Story\tTwo story\n 2.5Fin\tTwo and one-half story: 2nd level finished\n 2.5Unf\tTwo and one-half story: 2nd level unfinished\n SFoyer\tSplit Foyer\n SLvl\tSplit Level\n\t\nOverallQual: Rates the overall material and finish of the house\n\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\n 5\tAverage\n 4\tBelow Average\n 3\tFair\n 2\tPoor\n 1\tVery Poor\n\t\nOverallCond: Rates the overall condition of the house\n\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\t\n 5\tAverage\n 4\tBelow Average\t\n 3\tFair\n 2\tPoor\n 1\tVery Poor\n\t\t\nYearBuilt: Original construction date\n\nYearRemodAdd: Remodel date (same as construction date if no remodeling or additions)\n\nRoofStyle: Type of roof\n\n Flat\tFlat\n Gable\tGable\n Gambrel\tGabrel (Barn)\n Hip\tHip\n Mansard\tMansard\n Shed\tShed\n\t\t\nRoofMatl: Roof material\n\n ClyTile\tClay or Tile\n CompShg\tStandard (Composite) Shingle\n Membran\tMembrane\n Metal\tMetal\n Roll\tRoll\n Tar&Grv\tGravel & Tar\n WdShake\tWood Shakes\n WdShngl\tWood Shingles\n\t\t\nExterior1st: Exterior covering on house\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\t\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles\n\t\nExterior2nd: Exterior covering on house (if more than one material)\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles\n\t\nMasVnrType: Masonry veneer type\n\n BrkCmn\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n None\tNone\n Stone\tStone\n\t\nMasVnrArea: Masonry veneer area in square feet\n\nExterQual: Evaluates the quality of the material on the exterior \n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t\nExterCond: Evaluates the present condition of the material on the exterior\n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t\nFoundation: Type of foundation\n\t\t\n BrkTil\tBrick & Tile\n CBlock\tCinder Block\n PConc\tPoured Contrete\t\n Slab\tSlab\n Stone\tStone\n Wood\tWood\n\t\t\nBsmtQual: Evaluates the height of the basement\n\n Ex\tExcellent (100+ inches)\t\n Gd\tGood (90-99 inches)\n TA\tTypical (80-89 inches)\n Fa\tFair (70-79 inches)\n Po\tPoor (<70 inches\n NA\tNo Basement\n\t\t\nBsmtCond: Evaluates the general condition of the basement\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical - slight dampness allowed\n Fa\tFair - dampness or some cracking or settling\n Po\tPoor - Severe cracking, settling, or wetness\n NA\tNo Basement\n\t\nBsmtExposure: Refers to walkout or garden level walls\n\n Gd\tGood Exposure\n Av\tAverage Exposure (split levels or foyers typically score average or above)\t\n Mn\tMimimum Exposure\n No\tNo Exposure\n NA\tNo Basement\n\t\nBsmtFinType1: Rating of basement finished area\n\n GLQ\tGood Living Quarters\n ALQ\tAverage Living Quarters\n BLQ\tBelow Average Living Quarters\t\n Rec\tAverage Rec Room\n LwQ\tLow Quality\n Unf\tUnfinshed\n NA\tNo Basement\n\t\t\nBsmtFinSF1: Type 1 finished square feet\n\nBsmtFinType2: Rating of basement finished area (if multiple types)\n\n GLQ\tGood Living Quarters\n ALQ\tAverage Living Quarters\n BLQ\tBelow Average Living Quarters\t\n Rec\tAverage Rec Room\n LwQ\tLow Quality\n Unf\tUnfinshed\n NA\tNo Basement\n\nBsmtFinSF2: Type 2 finished square feet\n\nBsmtUnfSF: Unfinished square feet of basement area\n\nTotalBsmtSF: Total square feet of basement area\n\nHeating: Type of heating\n\t\t\n Floor\tFloor Furnace\n GasA\tGas forced warm air furnace\n GasW\tGas hot water or steam heat\n Grav\tGravity furnace\t\n OthW\tHot water or steam heat other than gas\n Wall\tWall furnace\n\t\t\nHeatingQC: Heating quality and condition\n\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t\nCentralAir: Central air conditioning\n\n N\tNo\n Y\tYes\n\t\t\nElectrical: Electrical system\n\n SBrkr\tStandard Circuit Breakers & Romex\n FuseA\tFuse Box over 60 AMP and all Romex wiring (Average)\t\n FuseF\t60 AMP Fuse Box and mostly Romex wiring (Fair)\n FuseP\t60 AMP Fuse Box and mostly knob & tube wiring (poor)\n Mix\tMixed\n\t\t\n1stFlrSF: First Floor square feet\n \n2ndFlrSF: Second floor square feet\n\nLowQualFinSF: Low quality finished square feet (all floors)\n\nGrLivArea: Above grade (ground) living area square feet\n\nBsmtFullBath: Basement full bathrooms\n\nBsmtHalfBath: Basement half bathrooms\n\nFullBath: Full bathrooms above grade\n\nHalfBath: Half baths above grade\n\nBedroom: Bedrooms above grade (does NOT include basement bedrooms)\n\nKitchen: Kitchens above grade\n\nKitchenQual: Kitchen quality\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical/Average\n Fa\tFair\n Po\tPoor\n \t\nTotRmsAbvGrd: Total rooms above grade (does not include bathrooms)\n\nFunctional: Home functionality (Assume typical unless deductions are warranted)\n\n Typ\tTypical Functionality\n Min1\tMinor Deductions 1\n Min2\tMinor Deductions 2\n Mod\tModerate Deductions\n Maj1\tMajor Deductions 1\n Maj2\tMajor Deductions 2\n Sev\tSeverely Damaged\n Sal\tSalvage only\n\t\t\nFireplaces: Number of fireplaces\n\nFireplaceQu: Fireplace quality\n\n Ex\tExcellent - Exceptional Masonry Fireplace\n Gd\tGood - Masonry Fireplace in main level\n TA\tAverage - Prefabricated Fireplace in main living area or Masonry Fireplace in basement\n Fa\tFair - Prefabricated Fireplace in basement\n Po\tPoor - Ben Franklin Stove\n NA\tNo Fireplace\n\t\t\nGarageType: Garage location\n\t\t\n 2Types\tMore than one type of garage\n Attchd\tAttached to home\n Basment\tBasement Garage\n BuiltIn\tBuilt-In (Garage part of house - typically has room above garage)\n CarPort\tCar Port\n Detchd\tDetached from home\n NA\tNo Garage\n\t\t\nGarageYrBlt: Year garage was built\n\t\t\nGarageFinish: Interior finish of the garage\n\n Fin\tFinished\n RFn\tRough Finished\t\n Unf\tUnfinished\n NA\tNo Garage\n\t\t\nGarageCars: Size of garage in car capacity\n\nGarageArea: Size of garage in square feet\n\nGarageQual: Garage quality\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical/Average\n Fa\tFair\n Po\tPoor\n NA\tNo Garage\n\t\t\nGarageCond: Garage condition\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical/Average\n Fa\tFair\n Po\tPoor\n NA\tNo Garage\n\t\t\nPavedDrive: Paved driveway\n\n Y\tPaved \n P\tPartial Pavement\n N\tDirt/Gravel\n\t\t\nWoodDeckSF: Wood deck area in square feet\n\nOpenPorchSF: Open porch area in square feet\n\nEnclosedPorch: Enclosed porch area in square feet\n\n3SsnPorch: Three season porch area in square feet\n\nScreenPorch: Screen porch area in square feet\n\nPoolArea: Pool area in square feet\n\nPoolQC: Pool quality\n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n NA\tNo Pool\n\t\t\nFence: Fence quality\n\t\t\n GdPrv\tGood Privacy\n MnPrv\tMinimum Privacy\n GdWo\tGood Wood\n MnWw\tMinimum Wood/Wire\n NA\tNo Fence\n\t\nMiscFeature: Miscellaneous feature not covered in other categories\n\t\t\n Elev\tElevator\n Gar2\t2nd Garage (if not described in garage section)\n Othr\tOther\n Shed\tShed (over 100 SF)\n TenC\tTennis Court\n NA\tNone\n\t\t\nMiscVal: $Value of miscellaneous feature\n\nMoSold: Month Sold (MM)\n\nYrSold: Year Sold (YYYY)\n\nSaleType: Type of sale\n\t\t\n WD \tWarranty Deed - Conventional\n CWD\tWarranty Deed - Cash\n VWD\tWarranty Deed - VA Loan\n New\tHome just constructed and sold\n COD\tCourt Officer Deed/Estate\n Con\tContract 15% Down payment regular terms\n ConLw\tContract Low Down payment and low interest\n ConLI\tContract Low Interest\n ConLD\tContract Low Down\n Oth\tOther\n\t\t\nSaleCondition: Condition of sale\n\n Normal\tNormal Sale\n Abnorml\tAbnormal Sale - trade, foreclosure, short sale\n AdjLand\tAdjoining Land Purchase\n Alloca\tAllocation - two linked properties with separate deeds, typically condo with a garage unit\t\n Family\tSale between family members\n Partial\tHome was not completed when last assessed (associated with New Homes)\n\n"
]
],
[
[
"# 3. Feature Engineering",
"_____no_output_____"
]
],
[
[
"df_train.describe()",
"_____no_output_____"
]
],
[
[
"**Function to create ranges for continuous feature**",
"_____no_output_____"
]
],
[
[
"def create_ranges(max_value, min_value, value):\n #print(value)\n shifted_max_value = max_value + abs(min_value)\n shifted_value = value + abs(min_value)\n #print(type(shifted_max_value))\n #print(type(shifted_value))\n percentage = np.divide(shifted_value, shifted_max_value)\n #print(type(percentage))\n #print(percentage)\n output_range = int(round(percentage*10))\n #print(output_range)\n\n return output_range",
"_____no_output_____"
]
],
[
[
"## 3.1 MSSubClass \nChange feature values to be represented by number from 1 and up.",
"_____no_output_____"
],
[
"**Feature Description:** \nMSSubClass: Identifies the type of dwelling involved in the sale.\t\n\n 20\t1-STORY 1946 & NEWER ALL STYLES\n 30\t1-STORY 1945 & OLDER\n 40\t1-STORY W/FINISHED ATTIC ALL AGES\n 45\t1-1/2 STORY - UNFINISHED ALL AGES\n 50\t1-1/2 STORY FINISHED ALL AGES\n 60\t2-STORY 1946 & NEWER\n 70\t2-STORY 1945 & OLDER\n 75\t2-1/2 STORY ALL AGES\n 80\tSPLIT OR MULTI-LEVEL\n 85\tSPLIT FOYER\n 90\tDUPLEX - ALL STYLES AND AGES\n 120\t1-STORY PUD (Planned Unit Development) - 1946 & NEWER\n 150\t1-1/2 STORY PUD - ALL AGES\n 160\t2-STORY PUD - 1946 & NEWER\n 180\tPUD - MULTILEVEL - INCL SPLIT LEV/FOYER\n 190\t2 FAMILY CONVERSION - ALL STYLES AND AGES\n \n**Feature Engineering:**\nCreate a binary feature where 1 is 1 story and 0 is more than 1 story.",
"_____no_output_____"
]
],
[
[
"df_train.MSSubClass.isna().sum()",
"_____no_output_____"
],
[
"df_train.MSSubClass.value_counts()",
"_____no_output_____"
],
[
"one_story_nums = [20, 30, 40, 120]\n\ndef mssubclass_feateng(input):\n if input in one_story_nums:\n return 1\n else:\n return 0",
"_____no_output_____"
],
[
"df_train['MSSubClass_1story'] = df_train.apply(lambda x: mssubclass_feateng(x.MSSubClass), axis=1)\ndf_test['MSSubClass_1story'] = df_test.apply(lambda x: mssubclass_feateng(x.MSSubClass), axis=1)",
"_____no_output_____"
],
[
"df_train.MSSubClass_1story.value_counts()",
"_____no_output_____"
],
[
"df_train.MSSubClass_1story.value_counts()",
"_____no_output_____"
],
[
"df_train['MSSubClass_1story'] = df_train['MSSubClass_1story'].astype('category')\ndf_test['MSSubClass_1story'] = df_test['MSSubClass_1story'].astype('category')",
"_____no_output_____"
],
[
"df_train.drop('MSSubClass', axis=1, inplace=True)\ndf_test.drop('MSSubClass', axis=1, inplace=True)\n\ndf_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.2 MSZoning",
"_____no_output_____"
],
[
"**Feature Description:** \nMSZoning: Identifies the general zoning classification of the sale.\n\t\t\n A\tAgriculture\n C\tCommercial\n FV\tFloating Village Residential\n I\tIndustrial\n RH\tResidential High Density\n RL\tResidential Low Density\n RP\tResidential Low Density Park \n RM\tResidential Medium Density",
"_____no_output_____"
]
],
[
[
"df_train.MSZoning.value_counts()",
"_____no_output_____"
],
[
"df_test.MSZoning.value_counts()",
"_____no_output_____"
],
[
"### Create dummies to handle categorical feature",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train.drop('MSZoning', axis=1), pd.get_dummies(df_train['MSZoning'], prefix='MSZoning')], axis=1)\ndf_test = pd.concat([df_test.drop('MSZoning', axis=1), pd.get_dummies(df_test['MSZoning'], prefix='MSZoning')], axis=1)\n",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.3 LotFrontage",
"_____no_output_____"
]
],
[
[
"df_train.LotFrontage.describe()",
"_____no_output_____"
],
[
"df_train.LotFrontage.isna().sum()",
"_____no_output_____"
],
[
"df_test.LotFrontage.isna().sum()",
"_____no_output_____"
]
],
[
[
"### Drop LotFrontage\nDrop this feature as there are 259 NaNs which is close to 20% of the samples.",
"_____no_output_____"
],
[
"An alternative would be to estimate the LotFrontage based on the LotArea.",
"_____no_output_____"
]
],
[
[
"sns.scatterplot(x='LotFrontage', y='LotArea', data=df_train)",
"_____no_output_____"
],
[
"df_train.drop('LotFrontage', axis=1, inplace=True)\ndf_test.drop('LotFrontage', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.4 Lot Area",
"_____no_output_____"
]
],
[
[
"df_train.LotArea.describe()",
"_____no_output_____"
]
],
[
[
"**Remove outliers**",
"_____no_output_____"
]
],
[
[
"df_train = df_train[df_train['LotArea'] < 150000]\ndf_test = df_test[df_test['LotArea'] < 150000]",
"_____no_output_____"
],
[
"df_train.LotArea.describe()",
"_____no_output_____"
]
],
[
[
"## 3.5 Street\nDrop this feature as 99.7% has the same value.",
"_____no_output_____"
]
],
[
[
"df_train.Street.value_counts()",
"_____no_output_____"
],
[
"df_train.drop('Street', axis=1, inplace=True)\ndf_test.drop('Street', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.6 Alley",
"_____no_output_____"
]
],
[
[
"df_train.Alley.value_counts()",
"_____no_output_____"
],
[
"df_train.Alley.isna().sum()",
"_____no_output_____"
]
],
[
[
"**Drop Alley feature as 1366 of 1452 are NaNs**",
"_____no_output_____"
]
],
[
[
"df_train.drop('Alley', axis=1, inplace=True)\ndf_test.drop('Alley', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.7 LotShape",
"_____no_output_____"
],
[
"**Feature Description** \nLotShape: General shape of property\n\n Reg\tRegular\t\n IR1\tSlightly irregular\n IR2\tModerately Irregular\n IR3\tIrregular",
"_____no_output_____"
]
],
[
[
"df_train.LotShape.value_counts()",
"_____no_output_____"
],
[
"df_train.LotShape.isna().sum()",
"_____no_output_____"
]
],
[
[
"**Replace LotShape with binary feature LotShape_regular**",
"_____no_output_____"
]
],
[
[
"df_train['LotShape_regular'] = df_train.apply(lambda x: 1 if x.LotShape == \"Reg\" else 0, axis=1)\ndf_test['LotShape_regular'] = df_test.apply(lambda x: 1 if x.LotShape == \"Reg\" else 0, axis=1)",
"_____no_output_____"
],
[
"df_train.LotShape_regular.value_counts()",
"_____no_output_____"
],
[
"df_train.drop('LotShape', axis=1, inplace=True)\ndf_test.drop('LotShape', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.8 Land Contour\n**Feature Description** \nLandContour: Flatness of the property\n\n Lvl\tNear Flat/Level\t\n Bnk\tBanked - Quick and significant rise from street grade to building\n HLS\tHillside - Significant slope from side to side\n Low\tDepression",
"_____no_output_____"
]
],
[
[
"df_train.LandContour.value_counts()",
"_____no_output_____"
],
[
"df_train.LandContour.isna().sum()",
"_____no_output_____"
],
[
"df_train['LandContour_level'] = df_train.apply(lambda x: 1 if \"Lvl\" in x.LandContour else 0, axis=1)\ndf_test['LandContour_level'] = df_test.apply(lambda x: 1 if \"Lvl\" in x.LandContour else 0, axis=1)\n",
"_____no_output_____"
],
[
"df_train.LandContour_level.value_counts()",
"_____no_output_____"
],
[
"df_train.drop('LandContour', axis=1, inplace=True)\ndf_test.drop('LandContour', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.9 Utilities\nUtilities: Type of utilities available\n\t\t\n AllPub\tAll public Utilities (E,G,W,& S)\t\n NoSewr\tElectricity, Gas, and Water (Septic Tank)\n NoSeWa\tElectricity and Gas Only\n ELO\tElectricity only",
"_____no_output_____"
]
],
[
[
"df_train.Utilities.value_counts()",
"_____no_output_____"
]
],
[
[
"**Drop this feature as all but one sample are the same**",
"_____no_output_____"
]
],
[
[
"df_train.drop('Utilities', axis=1, inplace=True)\ndf_test.drop('Utilities', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.10 LotConfig\nLotConfig: Lot configuration\n\n Inside\tInside lot\n Corner\tCorner lot\n CulDSac\tCul-de-sac\n FR2\tFrontage on 2 sides of property\n FR3\tFrontage on 3 sides of property",
"_____no_output_____"
]
],
[
[
"df_train.LotConfig.value_counts()",
"_____no_output_____"
],
[
"df_train.LotConfig.isna().sum()",
"_____no_output_____"
],
[
"df_train['LotConfig'] = df_train.apply(lambda x: x.LotConfig[:2] if \"FR\" in x.LotConfig else x.LotConfig, axis=1)\ndf_test['LotConfig'] = df_test.apply(lambda x: x.LotConfig[:2] if \"FR\" in x.LotConfig else x.LotConfig, axis=1)",
"_____no_output_____"
],
[
"df_train.LotConfig.value_counts()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train.drop('LotConfig', axis=1), pd.get_dummies(df_train['LotConfig'], prefix='LotConfig')], axis=1)\ndf_test = pd.concat([df_test.drop('LotConfig', axis=1), pd.get_dummies(df_test['LotConfig'], prefix='LotConfig')], axis=1)",
"_____no_output_____"
]
],
[
[
"## 3.11 LandSlope\nLandSlope: Slope of property\n\t\t\n Gtl\tGentle slope\n Mod\tModerate Slope\t\n Sev\tSevere Slope",
"_____no_output_____"
]
],
[
[
"df_train.LandSlope.value_counts()",
"_____no_output_____"
],
[
"df_train.LandSlope.isna().sum()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train.drop('LandSlope', axis=1), pd.get_dummies(df_train['LandSlope'], prefix='LandSlope')], axis=1)\ndf_test = pd.concat([df_test.drop('LandSlope', axis=1), pd.get_dummies(df_test['LandSlope'], prefix='LandSlope')], axis=1)\n",
"_____no_output_____"
]
],
[
[
"## 3.12 Neighborhood\nNeighborhood: Physical locations within Ames city limits\n\n Blmngtn\tBloomington Heights\n Blueste\tBluestem\n BrDale\tBriardale\n BrkSide\tBrookside\n ClearCr\tClear Creek\n CollgCr\tCollege Creek\n Crawfor\tCrawford\n Edwards\tEdwards\n Gilbert\tGilbert\n IDOTRR\tIowa DOT and Rail Road\n MeadowV\tMeadow Village\n Mitchel\tMitchell\n Names\tNorth Ames\n NoRidge\tNorthridge\n NPkVill\tNorthpark Villa\n NridgHt\tNorthridge Heights\n NWAmes\tNorthwest Ames\n OldTown\tOld Town\n SWISU\tSouth & West of Iowa State University\n Sawyer\tSawyer\n SawyerW\tSawyer West\n Somerst\tSomerset\n StoneBr\tStone Brook\n Timber\tTimberland\n Veenker\tVeenker",
"_____no_output_____"
]
],
[
[
"df_train.Neighborhood.value_counts()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train.drop('Neighborhood', axis=1), pd.get_dummies(df_train['Neighborhood'], prefix='Neighborhood')], axis=1)\ndf_test = pd.concat([df_test.drop('Neighborhood', axis=1), pd.get_dummies(df_test['Neighborhood'], prefix='Neighborhood')], axis=1)",
"_____no_output_____"
]
],
[
[
"## 3.13 MasVnrArea\nMasVnrArea: Masonry veneer area in square feet",
"_____no_output_____"
]
],
[
[
"df_train.MasVnrArea.isna().sum()",
"_____no_output_____"
]
],
[
[
"**Set all NaN values to mean**",
"_____no_output_____"
]
],
[
[
"MasVnrArea_mean = df_train.MasVnrArea.mean()\nMasVnrArea_mean",
"_____no_output_____"
],
[
"df_train['MasVnrArea'].fillna(MasVnrArea_mean, inplace=True)\ndf_test['MasVnrArea'].fillna(MasVnrArea_mean, inplace=True)",
"_____no_output_____"
],
[
"df_train.MasVnrArea.isna().sum()",
"_____no_output_____"
]
],
[
[
"## 3.14 Condition1: Proximity to various conditions\n Artery Adjacent to arterial street\n Feedr Adjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad",
"_____no_output_____"
]
],
[
[
"df_train.Condition1.value_counts()",
"_____no_output_____"
],
[
"df_train['Condition1_Norm'] = df_train.apply(lambda x: 1 if x.Condition1 =='Norm' else 0, axis=1)\ndf_test['Condition1_Norm'] = df_test.apply(lambda x: 1 if x.Condition1 =='Norm' else 0, axis=1)",
"_____no_output_____"
],
[
"df_train[['Condition1', 'Condition1_Norm']];",
"_____no_output_____"
],
[
"df_train.drop('Condition1', axis=1, inplace=True)\ndf_test.drop('Condition1', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.15 Condition2: Proximity to various conditions (if more than one is present)\n\n Artery Adjacent to arterial street\n Feedr Adjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad",
"_____no_output_____"
]
],
[
[
"df_train.Condition2.value_counts()",
"_____no_output_____"
],
[
"df_train.drop('Condition2', axis=1, inplace=True)\ndf_test.drop('Condition2', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.16 BldgType: Type of dwelling\n 1Fam\t Single-family Detached\t\n 2FmCon\tTwo-family Conversion; originally built as one-family dwelling\n Duplx\t Duplex\n TwnhsE\tTownhouse End Unit\n TwnhsI\tTownhouse Inside Unit",
"_____no_output_____"
]
],
[
[
"df_train.BldgType.isna().sum()",
"_____no_output_____"
],
[
"df_train.BldgType.value_counts()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.BldgType, prefix='BldgType')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.BldgType, prefix='BldgType')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BldgType', axis=1, inplace=True)\ndf_test.drop('BldgType', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.17 HouseStyle: Style of dwelling\n 1Story\tOne story\n 1.5Fin\tOne and one-half story: 2nd level finished\n 1.5Unf\tOne and one-half story: 2nd level unfinished\n 2Story\tTwo story\n 2.5Fin\tTwo and one-half story: 2nd level finished\n 2.5Unf\tTwo and one-half story: 2nd level unfinished\n SFoyer\tSplit Foyer\n SLvl\t Split Level",
"_____no_output_____"
]
],
[
[
"df_train.HouseStyle.value_counts()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.HouseStyle, prefix='HouseStyle')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.HouseStyle, prefix='HouseStyle')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('HouseStyle', axis=1, inplace=True)\ndf_test.drop('HouseStyle', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.18 OverallQual: Rates the overall material and finish of the house\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\n 5\tAverage\n 4\tBelow Average\n 3\tFair\n 2\tPoor\n 1\tVery Poor",
"_____no_output_____"
]
],
[
[
"df_train.OverallQual.value_counts()",
"_____no_output_____"
],
[
"df_train.OverallQual.isna().sum()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.OverallQual, prefix='OverallQual')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.OverallQual, prefix='OverallQual')], axis=1)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.19 OverallCond: Rates the overall condition of the house\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\t\n 5\tAverage\n 4\tBelow Average\t\n 3\tFair\n 2\tPoor\n 1\tVery Poor",
"_____no_output_____"
]
],
[
[
"df_train.OverallCond.value_counts()",
"_____no_output_____"
],
[
"df_train.OverallCond.isna().sum()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.OverallCond, prefix='OverallCond')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.OverallCond, prefix='OverallCond')], axis=1)",
"_____no_output_____"
]
],
[
[
"## 3.20 YearBuilt\nOriginal construction date",
"_____no_output_____"
]
],
[
[
"def YearBuilt_ranges(year):\n if year <= 1900:\n built_range = 'pre_1900'\n elif year > 1900 and year <= 1925:\n built_range = '1900_1925'\n elif year > 1925 and year <= 1950:\n built_range = '1925_1950'\n elif year > 1950 and year <= 1975:\n built_range = '1950_1975'\n elif year > 1975 and year <= 2000:\n built_range = '1975_2000'\n elif year > 2000:\n built_range = 'post_2000'\n \n return built_range",
"_____no_output_____"
],
[
"df_train['YearBuilt_range'] = df_train.apply(lambda x: YearBuilt_ranges(x.YearBuilt), axis=1)\ndf_test['YearBuilt_range'] = df_test.apply(lambda x: YearBuilt_ranges(x.YearBuilt), axis=1)",
"_____no_output_____"
],
[
"df_train[['YearBuilt_range']];",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.YearBuilt_range, prefix='YearBuilt_range')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.YearBuilt_range, prefix='YearBuilt_range')], axis=1)",
"_____no_output_____"
],
[
"df_train['Age'] = df_train.apply(lambda x: 2010 - x.YearBuilt, axis=1)\ndf_test['Age'] = df_test.apply(lambda x: 2010 - x.YearBuilt, axis=1)",
"_____no_output_____"
],
[
"df_train.drop('YearBuilt', axis=1, inplace=True)\ndf_test.drop('YearBuilt', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.21 YearRemodAdd\nRemodel date (same as construction date if no remodeling or additions)",
"_____no_output_____"
]
],
[
[
"df_train.YearRemodAdd.value_counts().head()",
"_____no_output_____"
],
[
"df_train['YearsSinceRemod'] = df_train.apply(lambda x: 2010 - x.YearRemodAdd, axis=1)\ndf_test['YearsSinceRemod'] = df_test.apply(lambda x: 2010 - x.YearRemodAdd, axis=1)",
"_____no_output_____"
],
[
"df_train.drop('YearRemodAdd', axis=1, inplace=True)\ndf_test.drop('YearRemodAdd', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.22 RoofStyle: Type of roof\n\n Flat\t Flat\n Gable\t Gable\n Gambrel\t Gabrel (Barn)\n Hip\t Hip\n Mansard\t Mansard\n Shed\t Shed",
"_____no_output_____"
]
],
[
[
"df_train.RoofStyle.value_counts()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.RoofStyle, prefix='RoofStyle')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.RoofStyle, prefix='RoofStyle')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('RoofStyle', axis=1, inplace=True)\ndf_test.drop('RoofStyle', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.23 RoofMatl: Roof material\n\n ClyTile\tClay or Tile\n CompShg\tStandard (Composite) Shingle\n Membran\tMembrane\n Metal\tMetal\n Roll\tRoll\n Tar&Grv\tGravel & Tar\n WdShake\tWood Shakes\n WdShngl\tWood Shingles",
"_____no_output_____"
]
],
[
[
"df_train.RoofMatl.value_counts()",
"_____no_output_____"
]
],
[
[
"**Remove feature as 98% of the samples are one category**",
"_____no_output_____"
]
],
[
[
"df_train.drop('RoofMatl', axis=1, inplace=True)\ndf_test.drop('RoofMatl', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.24 Exterior1st: Exterior covering on house\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\t\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles",
"_____no_output_____"
]
],
[
[
"df_train.Exterior1st.value_counts()",
"_____no_output_____"
],
[
"df_train.Exterior1st.isna().sum()",
"_____no_output_____"
],
[
"def transform_Exterior1st(category):\n list_above100 = ['VinylSd', 'HdBoard', 'MetalSd', 'Wd Sdng', 'Plywood']\n if category not in list_above100:\n return 'Other'\n else:\n return category",
"_____no_output_____"
],
[
"df_train['Exterior1st'] = df_train.apply(lambda x: transform_Exterior1st(x.Exterior1st), axis=1)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.Exterior1st, prefix='Exterior1st')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.Exterior1st, prefix='Exterior1st')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('Exterior1st', axis=1, inplace=True)\ndf_test.drop('Exterior1st', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.25 Exterior2nd: Exterior covering on house (if more than one material)\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles",
"_____no_output_____"
]
],
[
[
"df_train.Exterior2nd.value_counts()",
"_____no_output_____"
],
[
"df_train.Exterior2nd.isna().sum()",
"_____no_output_____"
],
[
"def transform_Exterior2nd(category):\n list_above100 = ['VinylSd', 'HdBoard', 'MetalSd', 'Wd Sdng', 'Plywood']\n if category not in list_above100:\n return 'Other'\n else:\n return category",
"_____no_output_____"
],
[
"df_train['Exterior2nd'] = df_train.apply(lambda x: transform_Exterior2nd(x.Exterior2nd), axis=1)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.Exterior2nd, prefix='Exterior2nd')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.Exterior2nd, prefix='Exterior2nd')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('Exterior2nd', axis=1, inplace=True)\ndf_test.drop('Exterior2nd', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.26 MasVnrType: Masonry veneer type\n\n BrkCmn\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n None\tNone\n Stone\tStone",
"_____no_output_____"
]
],
[
[
"df_train.MasVnrType.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.MasVnrType, prefix='MasVnrType')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.MasVnrType, prefix='MasVnrType')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('MasVnrType', axis=1, inplace=True)\ndf_test.drop('MasVnrType', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.27 MasVnrArea\n Masonry veneer area in square feet\n",
"_____no_output_____"
]
],
[
[
"df_train.MasVnrArea.describe()",
"_____no_output_____"
],
[
"df_train.MasVnrArea.plot()",
"_____no_output_____"
]
],
[
[
"## 3.28 ExterQual: Evaluates the quality of the material on the exterior \n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor",
"_____no_output_____"
]
],
[
[
"df_train.ExterQual.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.ExterQual, prefix='ExterQual')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.ExterQual, prefix='ExterQual')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('ExterQual', axis=1, inplace=True)\ndf_test.drop('ExterQual', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.29 ExterCond: Evaluates the present condition of the material on the exterior\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor",
"_____no_output_____"
]
],
[
[
"df_train.ExterCond.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.ExterCond, prefix='ExterCond')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.ExterCond, prefix='ExterCond')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('ExterCond', axis=1, inplace=True)\ndf_test.drop('ExterCond', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.30 Foundation: Type of foundation\n BrkTil\tBrick & Tile\n CBlock\tCinder Block\n PConc\t Poured Contrete\t\n Slab\t Slab\n Stone\t Stone\n Wood\t Wood\n",
"_____no_output_____"
]
],
[
[
"df_train.Foundation.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.Foundation, prefix='Foundation')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.Foundation, prefix='Foundation')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('Foundation', axis=1, inplace=True)\ndf_test.drop('Foundation', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.31 BsmtQual: Evaluates the height of the basement\n Ex\tExcellent (100+ inches)\t\n Gd\tGood (90-99 inches)\n TA\tTypical (80-89 inches)\n Fa\tFair (70-79 inches)\n Po\tPoor (<70 inches\n NA\tNo Basement",
"_____no_output_____"
]
],
[
[
"df_train.BsmtQual.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.BsmtQual, prefix='BsmtQual')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.BsmtQual, prefix='BsmtQual')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BsmtQual', axis=1, inplace=True)\ndf_test.drop('BsmtQual', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.32 BsmtCond: Evaluates the general condition of the basement\n Ex\tExcellent\n Gd\tGood\n TA\tTypical - slight dampness allowed\n Fa\tFair - dampness or some cracking or settling\n Po\tPoor - Severe cracking, settling, or wetness\n NA\tNo Basement",
"_____no_output_____"
]
],
[
[
"df_train.BsmtCond.value_counts(dropna=False)",
"_____no_output_____"
]
],
[
[
"**Drop as there are a big majority of TA and 37 NaN**",
"_____no_output_____"
]
],
[
[
"df_train.drop('BsmtCond', axis=1, inplace=True)\ndf_test.drop('BsmtCond', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.33 BsmtExposure: Refers to walkout or garden level walls\n Gd\tGood Exposure\n Av\tAverage Exposure (split levels or foyers typically score average or above)\t\n Mn\tMimimum Exposure\n No\tNo Exposure\n NA\tNo Basement",
"_____no_output_____"
]
],
[
[
"df_train.BsmtExposure.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.BsmtExposure, prefix='BsmtExposure')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.BsmtExposure, prefix='BsmtExposure')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BsmtExposure', axis=1, inplace=True)\ndf_test.drop('BsmtExposure', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.34 BsmtFinType1: Rating of basement finished area\n GLQ\tGood Living Quarters\n ALQ\tAverage Living Quarters\n BLQ\tBelow Average Living Quarters\t\n Rec\tAverage Rec Room\n LwQ\tLow Quality\n Unf\tUnfinshed\n NA\tNo Basement",
"_____no_output_____"
]
],
[
[
"df_train.BsmtFinType1.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.BsmtFinType1, prefix='BsmtFinType1')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.BsmtFinType1, prefix='BsmtFinType1')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BsmtFinType1', axis=1, inplace=True)\ndf_test.drop('BsmtFinType1', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.35 BsmtFinSF1: Type 1 finished square feet",
"_____no_output_____"
]
],
[
[
"df_train.BsmtFinSF1.describe()",
"_____no_output_____"
],
[
"df_train.BsmtFinSF1.isna().sum()",
"_____no_output_____"
],
[
"df_test.BsmtFinSF1.isna().sum()",
"_____no_output_____"
],
[
"feature = 'BsmtFinSF1'\nmax_value = df_train[[feature]].max()[0]\nmin_value = df_train[[feature]].min()[0]\n\ndf_train[feature+'_range'] = df_train.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
]
],
[
[
"**Fill NaN in test set**",
"_____no_output_____"
]
],
[
[
"df_test.BsmtFinSF1.isna().sum()",
"_____no_output_____"
],
[
"df_test.BsmtFinSF1.fillna(df_test.BsmtFinSF1.mean(), inplace=True)",
"_____no_output_____"
],
[
"feature = 'BsmtFinSF1'\nmax_value = df_test[[feature]].max()[0]\nmin_value = df_test[[feature]].min()[0]\n\ndf_test[feature+'_range'] = df_test.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BsmtFinSF1', axis=1, inplace=True)\ndf_test.drop('BsmtFinSF1', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.36 BsmtFinType2: Rating of basement finished area (if multiple types)\n GLQ\tGood Living Quarters\n ALQ\tAverage Living Quarters\n BLQ\tBelow Average Living Quarters\t\n Rec\tAverage Rec Room\n LwQ\tLow Quality\n Unf\tUnfinshed\n NA\tNo Basement",
"_____no_output_____"
]
],
[
[
"df_train.BsmtFinType2.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.BsmtFinType2, prefix='BsmtFinType2')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.BsmtFinType2, prefix='BsmtFinType2')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BsmtFinType2', axis=1, inplace=True)\ndf_test.drop('BsmtFinType2', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"## 3.37 BsmtFinSF2: Type 2 finished square feet",
"_____no_output_____"
]
],
[
[
"df_train.BsmtFinSF2.describe()",
"_____no_output_____"
],
[
"df_train.BsmtFinSF2.plot()",
"_____no_output_____"
],
[
"feature = 'BsmtFinSF2'\nmax_value = df_train[[feature]].max()[0]\nmin_value = df_train[[feature]].min()[0]\n\ndf_train[feature+'_range'] = df_train.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
]
],
[
[
"**Fill NaN in test set**",
"_____no_output_____"
]
],
[
[
"df_test.BsmtFinSF2.isna().sum()",
"_____no_output_____"
],
[
"df_test.BsmtFinSF2.fillna(df_test.BsmtFinSF2.mean(), inplace=True)",
"_____no_output_____"
],
[
"feature = 'BsmtFinSF2'\nmax_value = df_test[[feature]].max()[0]\nmin_value = df_test[[feature]].min()[0]\n\ndf_test[feature+'_range'] = df_test.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BsmtFinSF2', axis=1, inplace=True)\ndf_test.drop('BsmtFinSF2', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.38 BsmtUnfSF: Unfinished square feet of basement area",
"_____no_output_____"
]
],
[
[
"df_train.BsmtUnfSF.describe()",
"_____no_output_____"
],
[
"df_train.BsmtUnfSF.plot()",
"_____no_output_____"
],
[
"feature = 'BsmtUnfSF'\nmax_value = df_train[[feature]].max()[0]\nmin_value = df_train[[feature]].min()[0]\n\ndf_train[feature+'_range'] = df_train.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
]
],
[
[
"**Fill NaN in test set**",
"_____no_output_____"
]
],
[
[
"df_test.BsmtUnfSF.isna().sum()",
"_____no_output_____"
],
[
"df_test.BsmtUnfSF.fillna(df_test.BsmtUnfSF.mean(), inplace=True)",
"_____no_output_____"
],
[
"feature = 'BsmtUnfSF'\nmax_value = df_test[[feature]].max()[0]\nmin_value = df_test[[feature]].min()[0]\n\ndf_test[feature+'_range'] = df_test.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
],
[
"df_train.drop('BsmtUnfSF', axis=1, inplace=True)\ndf_test.drop('BsmtUnfSF', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.39 TotalBsmtSF: Total square feet of basement area",
"_____no_output_____"
]
],
[
[
"df_train.TotalBsmtSF.describe()",
"_____no_output_____"
],
[
"df_train.TotalBsmtSF.plot()",
"_____no_output_____"
],
[
"TotalBsmtSF_mean = df_train.TotalBsmtSF.mean()\nTotalBsmtSF_mean",
"_____no_output_____"
]
],
[
[
"**Remove outliers**",
"_____no_output_____"
]
],
[
[
"df_train['TotalBsmtSF_range'] = df_train.apply(lambda x: TotalBsmtSF_mean if x.TotalBsmtSF > 4000 else x.TotalBsmtSF, axis=1)\ndf_test['TotalBsmtSF_range'] = df_test.apply(lambda x: TotalBsmtSF_mean if x.TotalBsmtSF > 4000 else x.TotalBsmtSF, axis=1)",
"_____no_output_____"
],
[
"df_train.TotalBsmtSF_range.plot()",
"_____no_output_____"
],
[
"feature = 'TotalBsmtSF'\nmax_value = df_train[[feature]].max()[0]\nmin_value = df_train[[feature]].min()[0]\n\ndf_train[feature+'_range'] = df_train.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
]
],
[
[
"**Fill NaN in test set**",
"_____no_output_____"
]
],
[
[
"df_test.TotalBsmtSF.isna().sum()",
"_____no_output_____"
],
[
"df_test.TotalBsmtSF.fillna(df_test.TotalBsmtSF.mean(), inplace=True)",
"_____no_output_____"
],
[
"feature = 'TotalBsmtSF'\nmax_value = df_test[[feature]].max()[0]\nmin_value = df_test[[feature]].min()[0]\n\ndf_test[feature+'_range'] = df_test.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
],
[
"df_train.drop('TotalBsmtSF', axis=1, inplace=True)\ndf_test.drop('TotalBsmtSF', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.40 Heating: Type of heating\n\n Floor\tFloor Furnace\n GasA\tGas forced warm air furnace\n GasW\tGas hot water or steam heat\n Grav\tGravity furnace\t\n OthW\tHot water or steam heat other than gas\n Wall\tWall furnace",
"_____no_output_____"
]
],
[
[
"df_train.Heating.value_counts(dropna=False)",
"_____no_output_____"
]
],
[
[
"**Remove as it gives no new information**",
"_____no_output_____"
]
],
[
[
"df_train.drop('Heating', axis=1, inplace=True)\ndf_test.drop('Heating', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.41 HeatingQC: Heating quality and condition\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t",
"_____no_output_____"
]
],
[
[
"df_train.HeatingQC.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.HeatingQC, prefix='HeatingQC')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.HeatingQC, prefix='HeatingQC')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('HeatingQC', axis=1, inplace=True)\ndf_test.drop('HeatingQC', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.42 CentralAir: Central air conditioning\n\n N\tNo\n Y\tYes",
"_____no_output_____"
]
],
[
[
"df_train.CentralAir.value_counts(dropna=False)",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.CentralAir, prefix='CentralAir')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.CentralAir, prefix='CentralAir')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('CentralAir', axis=1, inplace=True)\ndf_test.drop('CentralAir', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.43 Electrical: Electrical system\n\n SBrkr\tStandard Circuit Breakers & Romex\n FuseA\tFuse Box over 60 AMP and all Romex wiring (Average)\t\n FuseF\t60 AMP Fuse Box and mostly Romex wiring (Fair)\n FuseP\t60 AMP Fuse Box and mostly knob & tube wiring (poor)\n Mix\t Mixed",
"_____no_output_____"
]
],
[
[
"df_train.Electrical.value_counts(dropna=False)",
"_____no_output_____"
]
],
[
[
"**Remove as it gives no new information**",
"_____no_output_____"
]
],
[
[
"df_train.drop('Electrical', axis=1, inplace=True)\ndf_test.drop('Electrical', axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## 3.44 1stFlrSF: First Floor square feet",
"_____no_output_____"
]
],
[
[
"df_train.rename({'1stFlrSF':'FirstFlrSF'}, axis=1, inplace=True)\ndf_test.rename({'1stFlrSF':'FirstFlrSF'}, axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train[['FirstFlrSF']].isna().sum()",
"_____no_output_____"
],
[
"df_train[['FirstFlrSF']].plot()",
"_____no_output_____"
]
],
[
[
"**Remove outliers**",
"_____no_output_____"
]
],
[
[
"FirststFlrSF_mean = df_train[['FirstFlrSF']].mean()[0]\nFirststFlrSF_mean",
"_____no_output_____"
],
[
"df_train['FirstFlrSF'] = df_train.apply(lambda x: FirststFlrSF_mean if x.FirstFlrSF > 4000 else x.FirstFlrSF, axis=1)\ndf_test['FirstFlrSF'] = df_test.apply(lambda x: FirststFlrSF_mean if x.FirstFlrSF > 4000 else x.FirstFlrSF, axis=1)",
"_____no_output_____"
]
],
[
[
"**Create and implement ranges**",
"_____no_output_____"
]
],
[
[
"feature = 'FirstFlrSF'\nmax_value = df_train[[feature]].max()[0]\nmin_value = df_train[[feature]].min()[0]\n\ndf_train[feature+'_range'] = df_train.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
],
[
"feature = 'FirstFlrSF'\nmax_value = df_test[[feature]].max()[0]\nmin_value = df_test[[feature]].min()[0]\n\ndf_test[feature+'_range'] = df_test.apply(lambda x: create_ranges(max_value, min_value, x[[feature]][0]), axis=1)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
],
[
"df_train = pd.concat([df_train, pd.get_dummies(df_train.FirstFlrSF, prefix='FirstFlrSF')], axis=1)\ndf_test = pd.concat([df_test, pd.get_dummies(df_test.FirstFlrSF, prefix='FirstFlrSF')], axis=1)",
"_____no_output_____"
],
[
"df_train.drop('FirstFlrSF', axis=1, inplace=True)\ndf_test.drop('FirstFlrSF', axis=1, inplace=True)",
"_____no_output_____"
],
[
"#df_train.drop('FirstFlrSF_range', axis=1, inplace=True)\n#df_test.drop('FirstFlrSF_range', axis=1, inplace=True)",
"_____no_output_____"
],
[
"f = open(\"data/data_description.txt\", \"r\")\nprint(f.read())",
"MSSubClass: Identifies the type of dwelling involved in the sale.\t\n\n 20\t1-STORY 1946 & NEWER ALL STYLES\n 30\t1-STORY 1945 & OLDER\n 40\t1-STORY W/FINISHED ATTIC ALL AGES\n 45\t1-1/2 STORY - UNFINISHED ALL AGES\n 50\t1-1/2 STORY FINISHED ALL AGES\n 60\t2-STORY 1946 & NEWER\n 70\t2-STORY 1945 & OLDER\n 75\t2-1/2 STORY ALL AGES\n 80\tSPLIT OR MULTI-LEVEL\n 85\tSPLIT FOYER\n 90\tDUPLEX - ALL STYLES AND AGES\n 120\t1-STORY PUD (Planned Unit Development) - 1946 & NEWER\n 150\t1-1/2 STORY PUD - ALL AGES\n 160\t2-STORY PUD - 1946 & NEWER\n 180\tPUD - MULTILEVEL - INCL SPLIT LEV/FOYER\n 190\t2 FAMILY CONVERSION - ALL STYLES AND AGES\n\nMSZoning: Identifies the general zoning classification of the sale.\n\t\t\n A\tAgriculture\n C\tCommercial\n FV\tFloating Village Residential\n I\tIndustrial\n RH\tResidential High Density\n RL\tResidential Low Density\n RP\tResidential Low Density Park \n RM\tResidential Medium Density\n\t\nLotFrontage: Linear feet of street connected to property\n\nLotArea: Lot size in square feet\n\nStreet: Type of road access to property\n\n Grvl\tGravel\t\n Pave\tPaved\n \t\nAlley: Type of alley access to property\n\n Grvl\tGravel\n Pave\tPaved\n NA \tNo alley access\n\t\t\nLotShape: General shape of property\n\n Reg\tRegular\t\n IR1\tSlightly irregular\n IR2\tModerately Irregular\n IR3\tIrregular\n \nLandContour: Flatness of the property\n\n Lvl\tNear Flat/Level\t\n Bnk\tBanked - Quick and significant rise from street grade to building\n HLS\tHillside - Significant slope from side to side\n Low\tDepression\n\t\t\nUtilities: Type of utilities available\n\t\t\n AllPub\tAll public Utilities (E,G,W,& S)\t\n NoSewr\tElectricity, Gas, and Water (Septic Tank)\n NoSeWa\tElectricity and Gas Only\n ELO\tElectricity only\t\n\t\nLotConfig: Lot configuration\n\n Inside\tInside lot\n Corner\tCorner lot\n CulDSac\tCul-de-sac\n FR2\tFrontage on 2 sides of property\n FR3\tFrontage on 3 sides of property\n\t\nLandSlope: Slope of property\n\t\t\n Gtl\tGentle slope\n Mod\tModerate Slope\t\n Sev\tSevere Slope\n\t\nNeighborhood: Physical locations within Ames city limits\n\n Blmngtn\tBloomington Heights\n Blueste\tBluestem\n BrDale\tBriardale\n BrkSide\tBrookside\n ClearCr\tClear Creek\n CollgCr\tCollege Creek\n Crawfor\tCrawford\n Edwards\tEdwards\n Gilbert\tGilbert\n IDOTRR\tIowa DOT and Rail Road\n MeadowV\tMeadow Village\n Mitchel\tMitchell\n Names\tNorth Ames\n NoRidge\tNorthridge\n NPkVill\tNorthpark Villa\n NridgHt\tNorthridge Heights\n NWAmes\tNorthwest Ames\n OldTown\tOld Town\n SWISU\tSouth & West of Iowa State University\n Sawyer\tSawyer\n SawyerW\tSawyer West\n Somerst\tSomerset\n StoneBr\tStone Brook\n Timber\tTimberland\n Veenker\tVeenker\n\t\t\t\nCondition1: Proximity to various conditions\n\t\n Artery\tAdjacent to arterial street\n Feedr\tAdjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad\n\t\nCondition2: Proximity to various conditions (if more than one is present)\n\t\t\n Artery\tAdjacent to arterial street\n Feedr\tAdjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad\n\t\nBldgType: Type of dwelling\n\t\t\n 1Fam\tSingle-family Detached\t\n 2FmCon\tTwo-family Conversion; originally built as one-family dwelling\n Duplx\tDuplex\n TwnhsE\tTownhouse End Unit\n TwnhsI\tTownhouse Inside Unit\n\t\nHouseStyle: Style of dwelling\n\t\n 1Story\tOne story\n 1.5Fin\tOne and one-half story: 2nd level finished\n 1.5Unf\tOne and one-half story: 2nd level unfinished\n 2Story\tTwo story\n 2.5Fin\tTwo and one-half story: 2nd level finished\n 2.5Unf\tTwo and one-half story: 2nd level unfinished\n SFoyer\tSplit Foyer\n SLvl\tSplit Level\n\t\nOverallQual: Rates the overall material and finish of the house\n\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\n 5\tAverage\n 4\tBelow Average\n 3\tFair\n 2\tPoor\n 1\tVery Poor\n\t\nOverallCond: Rates the overall condition of the house\n\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\t\n 5\tAverage\n 4\tBelow Average\t\n 3\tFair\n 2\tPoor\n 1\tVery Poor\n\t\t\nYearBuilt: Original construction date\n\nYearRemodAdd: Remodel date (same as construction date if no remodeling or additions)\n\nRoofStyle: Type of roof\n\n Flat\tFlat\n Gable\tGable\n Gambrel\tGabrel (Barn)\n Hip\tHip\n Mansard\tMansard\n Shed\tShed\n\t\t\nRoofMatl: Roof material\n\n ClyTile\tClay or Tile\n CompShg\tStandard (Composite) Shingle\n Membran\tMembrane\n Metal\tMetal\n Roll\tRoll\n Tar&Grv\tGravel & Tar\n WdShake\tWood Shakes\n WdShngl\tWood Shingles\n\t\t\nExterior1st: Exterior covering on house\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\t\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles\n\t\nExterior2nd: Exterior covering on house (if more than one material)\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles\n\t\nMasVnrType: Masonry veneer type\n\n BrkCmn\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n None\tNone\n Stone\tStone\n\t\nMasVnrArea: Masonry veneer area in square feet\n\nExterQual: Evaluates the quality of the material on the exterior \n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t\nExterCond: Evaluates the present condition of the material on the exterior\n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t\nFoundation: Type of foundation\n\t\t\n BrkTil\tBrick & Tile\n CBlock\tCinder Block\n PConc\tPoured Contrete\t\n Slab\tSlab\n Stone\tStone\n Wood\tWood\n\t\t\nBsmtQual: Evaluates the height of the basement\n\n Ex\tExcellent (100+ inches)\t\n Gd\tGood (90-99 inches)\n TA\tTypical (80-89 inches)\n Fa\tFair (70-79 inches)\n Po\tPoor (<70 inches\n NA\tNo Basement\n\t\t\nBsmtCond: Evaluates the general condition of the basement\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical - slight dampness allowed\n Fa\tFair - dampness or some cracking or settling\n Po\tPoor - Severe cracking, settling, or wetness\n NA\tNo Basement\n\t\nBsmtExposure: Refers to walkout or garden level walls\n\n Gd\tGood Exposure\n Av\tAverage Exposure (split levels or foyers typically score average or above)\t\n Mn\tMimimum Exposure\n No\tNo Exposure\n NA\tNo Basement\n\t\nBsmtFinType1: Rating of basement finished area\n\n GLQ\tGood Living Quarters\n ALQ\tAverage Living Quarters\n BLQ\tBelow Average Living Quarters\t\n Rec\tAverage Rec Room\n LwQ\tLow Quality\n Unf\tUnfinshed\n NA\tNo Basement\n\t\t\nBsmtFinSF1: Type 1 finished square feet\n\nBsmtFinType2: Rating of basement finished area (if multiple types)\n\n GLQ\tGood Living Quarters\n ALQ\tAverage Living Quarters\n BLQ\tBelow Average Living Quarters\t\n Rec\tAverage Rec Room\n LwQ\tLow Quality\n Unf\tUnfinshed\n NA\tNo Basement\n\nBsmtFinSF2: Type 2 finished square feet\n\nBsmtUnfSF: Unfinished square feet of basement area\n\nTotalBsmtSF: Total square feet of basement area\n\nHeating: Type of heating\n\t\t\n Floor\tFloor Furnace\n GasA\tGas forced warm air furnace\n GasW\tGas hot water or steam heat\n Grav\tGravity furnace\t\n OthW\tHot water or steam heat other than gas\n Wall\tWall furnace\n\t\t\nHeatingQC: Heating quality and condition\n\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t\nCentralAir: Central air conditioning\n\n N\tNo\n Y\tYes\n\t\t\nElectrical: Electrical system\n\n SBrkr\tStandard Circuit Breakers & Romex\n FuseA\tFuse Box over 60 AMP and all Romex wiring (Average)\t\n FuseF\t60 AMP Fuse Box and mostly Romex wiring (Fair)\n FuseP\t60 AMP Fuse Box and mostly knob & tube wiring (poor)\n Mix\tMixed\n\t\t\n1stFlrSF: First Floor square feet\n \n2ndFlrSF: Second floor square feet\n\nLowQualFinSF: Low quality finished square feet (all floors)\n\nGrLivArea: Above grade (ground) living area square feet\n\nBsmtFullBath: Basement full bathrooms\n\nBsmtHalfBath: Basement half bathrooms\n\nFullBath: Full bathrooms above grade\n\nHalfBath: Half baths above grade\n\nBedroom: Bedrooms above grade (does NOT include basement bedrooms)\n\nKitchen: Kitchens above grade\n\nKitchenQual: Kitchen quality\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical/Average\n Fa\tFair\n Po\tPoor\n \t\nTotRmsAbvGrd: Total rooms above grade (does not include bathrooms)\n\nFunctional: Home functionality (Assume typical unless deductions are warranted)\n\n Typ\tTypical Functionality\n Min1\tMinor Deductions 1\n Min2\tMinor Deductions 2\n Mod\tModerate Deductions\n Maj1\tMajor Deductions 1\n Maj2\tMajor Deductions 2\n Sev\tSeverely Damaged\n Sal\tSalvage only\n\t\t\nFireplaces: Number of fireplaces\n\nFireplaceQu: Fireplace quality\n\n Ex\tExcellent - Exceptional Masonry Fireplace\n Gd\tGood - Masonry Fireplace in main level\n TA\tAverage - Prefabricated Fireplace in main living area or Masonry Fireplace in basement\n Fa\tFair - Prefabricated Fireplace in basement\n Po\tPoor - Ben Franklin Stove\n NA\tNo Fireplace\n\t\t\nGarageType: Garage location\n\t\t\n 2Types\tMore than one type of garage\n Attchd\tAttached to home\n Basment\tBasement Garage\n BuiltIn\tBuilt-In (Garage part of house - typically has room above garage)\n CarPort\tCar Port\n Detchd\tDetached from home\n NA\tNo Garage\n\t\t\nGarageYrBlt: Year garage was built\n\t\t\nGarageFinish: Interior finish of the garage\n\n Fin\tFinished\n RFn\tRough Finished\t\n Unf\tUnfinished\n NA\tNo Garage\n\t\t\nGarageCars: Size of garage in car capacity\n\nGarageArea: Size of garage in square feet\n\nGarageQual: Garage quality\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical/Average\n Fa\tFair\n Po\tPoor\n NA\tNo Garage\n\t\t\nGarageCond: Garage condition\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical/Average\n Fa\tFair\n Po\tPoor\n NA\tNo Garage\n\t\t\nPavedDrive: Paved driveway\n\n Y\tPaved \n P\tPartial Pavement\n N\tDirt/Gravel\n\t\t\nWoodDeckSF: Wood deck area in square feet\n\nOpenPorchSF: Open porch area in square feet\n\nEnclosedPorch: Enclosed porch area in square feet\n\n3SsnPorch: Three season porch area in square feet\n\nScreenPorch: Screen porch area in square feet\n\nPoolArea: Pool area in square feet\n\nPoolQC: Pool quality\n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n NA\tNo Pool\n\t\t\nFence: Fence quality\n\t\t\n GdPrv\tGood Privacy\n MnPrv\tMinimum Privacy\n GdWo\tGood Wood\n MnWw\tMinimum Wood/Wire\n NA\tNo Fence\n\t\nMiscFeature: Miscellaneous feature not covered in other categories\n\t\t\n Elev\tElevator\n Gar2\t2nd Garage (if not described in garage section)\n Othr\tOther\n Shed\tShed (over 100 SF)\n TenC\tTennis Court\n NA\tNone\n\t\t\nMiscVal: $Value of miscellaneous feature\n\nMoSold: Month Sold (MM)\n\nYrSold: Year Sold (YYYY)\n\nSaleType: Type of sale\n\t\t\n WD \tWarranty Deed - Conventional\n CWD\tWarranty Deed - Cash\n VWD\tWarranty Deed - VA Loan\n New\tHome just constructed and sold\n COD\tCourt Officer Deed/Estate\n Con\tContract 15% Down payment regular terms\n ConLw\tContract Low Down payment and low interest\n ConLI\tContract Low Interest\n ConLD\tContract Low Down\n Oth\tOther\n\t\t\nSaleCondition: Condition of sale\n\n Normal\tNormal Sale\n Abnorml\tAbnormal Sale - trade, foreclosure, short sale\n AdjLand\tAdjoining Land Purchase\n Alloca\tAllocation - two linked properties with separate deeds, typically condo with a garage unit\t\n Family\tSale between family members\n Partial\tHome was not completed when last assessed (associated with New Homes)\n\n"
]
],
[
[
"# 4. Analysis",
"_____no_output_____"
],
[
"## 4.1 Correlation Matrix",
"_____no_output_____"
]
],
[
[
"cols = df_train.columns.tolist()\nn = int(cols.index('SalePrice'))\ncols = cols[:n] + cols[n+1:] + [cols[n]]\ndf_train = df_train[cols]",
"_____no_output_____"
],
[
"corr_mat = df_train.corr()",
"_____no_output_____"
],
[
"corr_mat.columns",
"_____no_output_____"
],
[
"corr_mat[corr_mat['SalePrice'].abs().sort_values(ascending=False) > 0.4]",
"/Users/bjornar/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: UserWarning: Boolean Series key will be reindexed to match DataFrame index.\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"correlated_cols = list(corr_mat[corr_mat.SalePrice.abs()>0.5].index)\n#correlated_cols.append('Id')\ncorrelated_cols",
"_____no_output_____"
],
[
"corr_mat[correlated_cols].loc[correlated_cols]",
"_____no_output_____"
],
[
"corr_mat_plot = pd.merge(corr_mat[correlated_cols].loc[correlated_cols].reset_index(), corr_mat['SalePrice'][correlated_cols].reset_index(), on='index').set_index('index')",
"_____no_output_____"
],
[
"corr_mat_plot",
"_____no_output_____"
],
[
"sns.set(rc={'figure.figsize':(25,25)})\nsns.set_context(\"notebook\", font_scale=1.5, rc={\"lines.linewidth\": 2.5})\n\nax = sns.heatmap(abs(corr_mat_plot), annot=True)\n",
"_____no_output_____"
]
],
[
[
"## 4.2 Boruta Feature Selection",
"_____no_output_____"
]
],
[
[
"X_train = df_train.copy()[correlated_cols].drop(\"SalePrice\", axis=1).interpolate(method='ffill')\nY_train = df_train.copy()[\"SalePrice\"]",
"_____no_output_____"
],
[
"def run_boruta():\n # load X and y\n # NOTE BorutaPy accepts numpy arrays only, hence the .values attribute\n X = X_train.values\n y = Y_train.ravel()\n\n # define random forest classifier, with utilising all cores and\n # sampling in proportion to y labels\n rf = RandomForestClassifier(n_jobs=-1, class_weight='balanced', max_depth=5)\n\n # define Boruta feature selection method\n feat_selector = BorutaPy(rf, n_estimators='auto', verbose=10, random_state=1)\n\n # find all relevant features - 5 features should be selected\n feat_selector.fit(X, y)\n\n # check selected features - first 5 features are selected\n feat_selector.support_\n\n # check ranking of features\n feat_selector.ranking_\n\n # call transform() on X to filter it down to selected features\n X_filtered = feat_selector.transform(X)\n \n#run_boruta()",
"_____no_output_____"
]
],
[
[
"feat_selector.support_",
"_____no_output_____"
],
[
"X_train.columns[feat_selector.support_]",
"_____no_output_____"
],
[
"df_boruta = df_train[[X_train.columns[feat_selector.support_][0], 'SalePrice']]",
"_____no_output_____"
],
[
"df_boruta.head()",
"_____no_output_____"
],
[
"# 5. Prediction",
"_____no_output_____"
],
[
"## Random Forest",
"_____no_output_____"
]
],
[
[
"X_train = df_train.copy().interpolate(method='ffill')[correlated_cols].drop(\"SalePrice\", axis=1)\nY_train = df_train[\"SalePrice\"]\ncorrelated_cols.remove('SalePrice')\nX_test = df_test.copy()[correlated_cols].interpolate()\nX_train.shape, Y_train.shape, X_test.shape",
"_____no_output_____"
],
[
"X_train.isna().interpolate().sum()",
"_____no_output_____"
],
[
"X_train.isna().sum()",
"_____no_output_____"
],
[
"random_forest = RandomForestClassifier(n_estimators=100)\nrandom_forest.fit(X_train, Y_train)\nY_pred = random_forest.predict(X_test)",
"_____no_output_____"
],
[
"random_forest.score(X_train, Y_train)\nacc_random_forest = round(random_forest.score(X_train, Y_train) * 100, 2)\nacc_random_forest",
"_____no_output_____"
]
],
[
[
"# XGBoost",
"_____no_output_____"
],
[
"model = xgboost.train({\"learning_rate\": 0.5}, xgboost.DMatrix(X_train, label=Y_train), 10)",
"_____no_output_____"
],
[
"fit model no training data\nmodel = XGBRegressor(objective ='reg:linear', colsample_bytree = 0.3, learning_rate = 0.5,\n max_depth = 5, alpha = 10, n_estimators = 10)\nmodel.fit(X_train[['OverallQual', 'YearBuilt']], Y_train, verbose=True)",
"_____no_output_____"
],
[
"print(model)",
"_____no_output_____"
],
[
"xgb_model = XGBRegressor().fit(X_train[['OverallQual', 'YearBuilt']], Y_train)\npredictions = xgb_model.predict(X_test)\nactuals = Y_test\nprint(mean_squared_error(actuals, predictions))",
"_____no_output_____"
],
[
"# 7. Submit results",
"_____no_output_____"
]
],
[
[
"submission = pd.DataFrame({ \"Id\": df_test[\"Id\"], \"SalePrice\": Y_pred\n }).set_index('Id')",
"_____no_output_____"
],
[
"submission.to_csv('submission.csv')",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
]
],
[
[
"# 8. Explain model",
"_____no_output_____"
]
],
[
[
"import shap\n\n# load JS visualization code to notebook\nshap.initjs()",
"_____no_output_____"
],
[
"# explain the model's predictions using SHAP values\n# (same syntax works for LightGBM, CatBoost, and scikit-learn models)\nexplainer = shap.TreeExplainer(random_forest)\nshap_values = explainer.shap_values(X_train)",
"_____no_output_____"
],
[
"# visualize the first prediction's explanation (use matplotlib=True to avoid Javascript)\nshap.force_plot(explainer.expected_value, shap_values)",
"_____no_output_____"
],
[
"shap.dependence_plot(0, shap_values, X_train)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ec8862b5ebfff8a6aefdee97048c479a062f18a7 | 23,003 | ipynb | Jupyter Notebook | workshop_sections/wide_n_deep/wide_n_deep_flow3.ipynb | thesmarthomeninja/Tensorflow_Census_Example | 09de7d23e665996d21db3e13eb8f7abe752db018 | [
"Apache-2.0"
] | 1 | 2017-12-05T07:57:13.000Z | 2017-12-05T07:57:13.000Z | workshop_sections/wide_n_deep/wide_n_deep_flow3.ipynb | thesmarthomeninja/Tensorflow_Census_Example | 09de7d23e665996d21db3e13eb8f7abe752db018 | [
"Apache-2.0"
] | null | null | null | workshop_sections/wide_n_deep/wide_n_deep_flow3.ipynb | thesmarthomeninja/Tensorflow_Census_Example | 09de7d23e665996d21db3e13eb8f7abe752db018 | [
"Apache-2.0"
] | 1 | 2020-01-20T08:29:10.000Z | 2020-01-20T08:29:10.000Z | 38.856419 | 533 | 0.60253 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ec886a7912fff08dd0690c088a0de0c4bc2a0ff1 | 409,151 | ipynb | Jupyter Notebook | token_prices/Untitled.ipynb | howardpen9/eth_analytics | cf9f35f867c88cd3ec11836253861908a9bc3d92 | [
"MIT"
] | 2 | 2021-08-09T21:37:34.000Z | 2021-08-09T21:37:35.000Z | token_prices/Untitled.ipynb | howardpen9/eth_analytics | cf9f35f867c88cd3ec11836253861908a9bc3d92 | [
"MIT"
] | null | null | null | token_prices/Untitled.ipynb | howardpen9/eth_analytics | cf9f35f867c88cd3ec11836253861908a9bc3d92 | [
"MIT"
] | null | null | null | 2,922.507143 | 405,450 | 0.702442 | [
[
[
"import quantstats as qs\nfrom pycoingecko import CoinGeckoAPI\nimport pandas as pd \nfrom datetime import datetime\nimport statistics\ncg = CoinGeckoAPI()\ndate = []\neth_price = []\n\neth_price_history = cg.get_coin_market_chart_by_id(id='defipulse-index', vs_currency='usd', days='165')\neth_price_dict = eth_price_history['prices']\n\nfor x in eth_price_dict: \n date.append(datetime.fromtimestamp(x[0]/1000))\n eth_price.append(x[1])\ntotal_yield = .28\ndaily_reduction = .00105\nstarting_value = 10000\ndays = 165\nvalues = []\nwhile days > 0: \n #print(starting_value)\n starting_value = starting_value + (starting_value*(total_yield/365))\n total_yield = total_yield - daily_reduction\n values.append(starting_value)\n days = days - 1 \n\nlist_returns_percent_a = [100 * (b - a) / a for a, b in zip(eth_price[::1], eth_price[1::1])]\n#print(list_returns_percent_a)\n#percents_list = statistics.pstdev(list_returns_percent_a)\n\n# fetch the daily returns for a stock\n#stock = qs.utils.download_returns('FB')\n#print(stock)\n#print(type(stock))\n# show sharpe ratio\nx = pd.Series(list_returns_percent_a)\nfrom datetime import datetime\n\ndate_range = pd.date_range(end = datetime.today(), periods = 165).to_pydatetime().tolist()\n\nx.index = date_range\nx = x/100\nprint(x)\nqs.reports.html(x)\n\n'''\nx.index = pd.to_datetime(x.index)\nprint(x)\nqs.reports.full(x)\n'''",
"2021-01-01 17:17:30.801950 0.015137\n2021-01-02 17:17:30.801950 0.004440\n2021-01-03 17:17:30.801950 0.142626\n2021-01-04 17:17:30.801950 0.040895\n2021-01-05 17:17:30.801950 0.163195\n ... \n2021-06-10 17:17:30.801950 -0.061763\n2021-06-11 17:17:30.801950 -0.089739\n2021-06-12 17:17:30.801950 -0.019045\n2021-06-13 17:17:30.801950 0.096384\n2021-06-14 17:17:30.801950 0.016649\nLength: 165, dtype: float64\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ec886dfa905ca232f735424438e6d14499589733 | 700,007 | ipynb | Jupyter Notebook | Titanic_ds&ml.ipynb | SaadMuhammad/Titanic_Feature_Engineering_guide | fe9097f175b76d86f855e302d4299b9d57307424 | [
"MIT"
] | null | null | null | Titanic_ds&ml.ipynb | SaadMuhammad/Titanic_Feature_Engineering_guide | fe9097f175b76d86f855e302d4299b9d57307424 | [
"MIT"
] | null | null | null | Titanic_ds&ml.ipynb | SaadMuhammad/Titanic_Feature_Engineering_guide | fe9097f175b76d86f855e302d4299b9d57307424 | [
"MIT"
] | null | null | null | 700,007 | 700,007 | 0.894114 | [
[
[
"# An Example of Data Science work on Titanic Dataset",
"_____no_output_____"
],
[
"## A Guide on how to perform different feature engineering tasks\n ",
"_____no_output_____"
],
[
" \n \n##### We will start from exploring the data to see what we have to do clean the provided data set. \"Always consider in any data science problem we have to perform some exploratory data analysis as first steps\" (if data is readily available, if its not like for data mining strategy will be a bit different)\n\n",
"_____no_output_____"
],
[
"##### it is important to have a clear understanding of what libs you will be using and import/install them beforehand ",
"_____no_output_____"
]
],
[
[
"#important libraries\nimport numpy as np \nimport pandas as pd\n\n%matplotlib inline \nfrom matplotlib import pyplot as plt\nfrom matplotlib import style\nimport seaborn as sns\n\nimport string\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"#### Explanations:\n##### %matplotlib inline:\n##### is specifically used here for jupyter notebook to store plots in notebook document \n\n##### warnings: \n##### has been imported to avoid raise of warning when a function is deprecated",
"_____no_output_____"
],
[
"##### The main reason to have seaborn apart from matplot lib is It is used to create more attractive and informative statistical graphics",
"_____no_output_____"
]
],
[
[
"# Importing provided dataset one for prediction and one for training and testing\ntest_frame = pd.read_csv(\"test.csv\")\ntrain_frame = pd.read_csv(\"train.csv\")",
"_____no_output_____"
],
[
"train_frame.head()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Note:\n\n##### shape function provide number of rows and column in a given dataset like (rows, cols)",
"_____no_output_____"
]
],
[
[
"print(train_frame.shape)\nprint(test_frame.shape)",
"(891, 12)\n(418, 11)\n"
]
],
[
[
" ",
"_____no_output_____"
],
[
"\n\n##### so our train and pred data have 891 and 418 rows and 12 and 11 cols respectively, df_pred has obviously 1 col less because that is the target col which we have to predict",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"# to see what types of data each col contains\ntrain_frame.dtypes",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Explanation:\n##### Dataframe.Describe()\n\n##### provide a good EDA understanding of the dataset in hand, it provide mean, std and fragments of each 25% and is good to have a glimpse of outliers in data prior to visualzations",
"_____no_output_____"
]
],
[
[
"train_frame.describe()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Dataframe.describe(include='all') is used to provide information about categorical data. This helps us to identify what categories we are seeing the most ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"#include all provide an understanding of category in form of unique values in each col and freq of most common value\ntrain_frame.describe(include='all')",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Lets check how many null/missing values we have in both dataset so we can define our strategy of how to treat them",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"train_null = train_frame.isnull().sum().sort_values(ascending=False)\ntrain_null",
"_____no_output_____"
],
[
"null_test = test_frame.isnull().sum().sort_values(ascending=False)\nnull_test",
"_____no_output_____"
],
[
"#performing EDA \ntrain_frame['Parch'].value_counts()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"#### Note:\n##### The main reason behind employing visualization is because it is helpful in understanding data/distributions by placing it in a visual context so that patterns, trends and correlations that might not otherwise be detected can be worked with",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"#initial visualization, droping missing rows to avoid errors in visuals\nmale = train_frame[train_frame['Sex']=='male']\nfemale = train_frame[train_frame['Sex']=='female']\nx = male[male['Survived']==1].Age.dropna()\nx1 = male[male['Survived']==0].Age.dropna()\ny = female[female['Survived']==1].Age.dropna()\ny1 = female[female['Survived']==0].Age.dropna()",
"_____no_output_____"
],
[
"#lets have some visualization on sex and survial ratio\n\nfig, axes = plt.subplots(nrows=1, ncols=2,figsize=(10, 4))\nax = sns.distplot(x, bins=15, label = 'survived', ax = axes[0], kde = False, color = 'g')\nax = sns.distplot(x1, bins=30, label = 'not survived', ax = axes[0], kde = False, color = 'b')\nax.legend()\nax.set_title('Male')\nax = sns.distplot(y, bins=15, label = 'survived', ax = axes[1], kde = False, color = 'y')\nax = sns.distplot(y1, bins=30, label = 'not survived', ax = axes[1], kde = False, color = 'r')\nax.legend()\nax.set_title('Female')\nplt.show()",
"_____no_output_____"
],
[
"#survival ration w.r.t to class\nsns.barplot(x='Pclass', y='Survived', data=train_frame)",
"_____no_output_____"
],
[
"sns.barplot(x='Parch', y='Survived', data=train_frame)",
"_____no_output_____"
],
[
"dft1 = train_frame.copy()\ncat_cols = ['Sex', 'Embarked', 'Survived']\n\ndft1[cat_cols]= dft1[cat_cols].astype('category')",
"_____no_output_____"
],
[
"f,ax =plt.subplots(len(cat_cols),1,figsize=(5,10))\nfor idx,col in enumerate(cat_cols):\n if col!='Survived':\n sns.countplot(x=col,data=dft1[cat_cols],hue='Survived', ax=ax[idx])",
"_____no_output_____"
],
[
"#Advance Vis\na = sns.FacetGrid(train_frame, hue = 'Survived', aspect=4, palette=\"Set1\" )\na.map(sns.kdeplot, 'Age', shade= True )\na.set(xlim=(0 , train_frame['Age'].max()))\na.add_legend()",
"_____no_output_____"
],
[
"b = sns.FacetGrid(train_frame, row = 'Sex', col = 'Pclass', hue = 'Survived', palette=\"Set2\")\nb.map(plt.hist, 'Age', alpha = .75)\nb.add_legend()",
"_____no_output_____"
],
[
"add_1 = sns.FacetGrid(train_frame, col = 'Embarked')\nadd_1.map(sns.pointplot, 'Pclass', 'Survived', 'Sex', ci=95.0, palette = 'deep')\nadd_1.add_legend()",
"_____no_output_____"
],
[
"sns.scatterplot(x=\"Age\", y=\"Fare\",\n hue=\"Survived\", palette=\"Set2\",\n sizes=(20, 200), hue_norm=(0, 7),\n legend=\"full\", data=train_frame)",
"_____no_output_____"
],
[
"sns.scatterplot(x=\"Age\", y=\"Fare\", hue=\"Survived\", size=\"Sex\", palette=\"Set2\",\n sizes=(20, 200), hue_norm=(0, 7),\n legend=\"full\", data=train_frame)",
"_____no_output_____"
],
[
"sns.scatterplot(x=\"Pclass\", y=\"Parch\",\n hue=\"Survived\", palette=\"Set1\",\n sizes=(20, 200), hue_norm=(0, 7),\n legend=\"full\", data=train_frame)",
"_____no_output_____"
],
[
"cols = ['Survived', 'Pclass', 'Sex', 'Parch', 'Fare', 'Embarked']\nsns.pairplot(train_frame[cols], diag_kind=\"hist\", hue = 'Survived', palette=\"Set2\")",
"_____no_output_____"
],
[
"sns.jointplot(train_frame['Age'],train_frame['Survived'], kind=\"hex\")\nplt.title('Age Vs Survived')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"sns.jointplot(train_frame['Age'],train_frame['Fare'], kind=\"reg\")\nplt.title('Age Vs fare')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"#### Note: \n##### We now have some understanding of what we are expecting from different features like for 'Parch' we have decision boundires for survial indications of which classes are going with 100% survival and which are with 0% and so on.",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"## Working on Mising data",
"_____no_output_____"
],
[
"#### Explanation:\n##### Filling missing values in dataset is of utmost importance because the fate and accuracy of your model rely heavily on your strategy. There are numerous ways for filling your missing data with most easy is to replace it by mean/median or most common value.",
"_____no_output_____"
],
[
"#### What we will do\n##### Here i have taken 3 dif strategies just for knowledge sharing as how we can fill nan/missing values. \n\n",
"_____no_output_____"
],
[
"#### 1. Filling NaN with Mean and Standard Deviation\n\n#### 2. Imputations using simple impute/KNN\n\n#### 3. Using statistics to fill NaN ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"#### Note: \n##### sometime it is wise to drop missing data rows/cols mainly when there are too much nan values or if filling them not make any sense. This is part where our understanding of statistics and visualization skills provide us the insights about how to deal with your datset",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"df_train1 = train_frame.copy()\ndf_test1 = test_frame.copy()",
"_____no_output_____"
]
],
[
[
"### 1. Taking mean & std ",
"_____no_output_____"
],
[
"##### Since Age feature has the most num of NaN, easiest way is to take mean and fill is by +,_ of its std ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"Age = [df_train1, df_test1]\n\nfor dataset in Age:\n #making use of both test and train frame\n mean = train_frame[\"Age\"].mean()\n std = test_frame[\"Age\"].std()\n is_null = dataset[\"Age\"].isnull().sum()\n # computing random numbers between the mean, std and is_null\n rand_age = np.random.randint(mean - std, mean + std, size = is_null)\n # fill NaN values in Age column with random values generated in range of mean +/- std\n age_slice = dataset[\"Age\"].copy()\n age_slice[np.isnan(age_slice)] = rand_age\n dataset[\"Age\"] = age_slice\n dataset[\"Age\"] = df_train1[\"Age\"].astype(int)\ndf_train1[\"Age\"].isnull().sum()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"#### Note:\n##### This is not the most intelligent or accurate approach since there are different groups w.r.t to pclass, embarked and etc so the model understanding will be biased. So we will not be continuing with this approach",
"_____no_output_____"
]
],
[
[
"#re-checking if any nulls of Age feautes \ntrain_null = df_train1.isnull().sum().sort_values(ascending=False)\ntrain_null",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"### 2. Imputations\n",
"_____no_output_____"
],
[
"##### Imputation is the process of replacing missing data with substituted values\n\n#### Imputations can be performed in many ways as describe below:",
"_____no_output_____"
],
[
"##### SimpleFill: Replaces missing entries with the mean or median of each column.\n\n##### •KNN: Nearest neighbor imputations which weights samples using the mean squared difference on features for which two rows both have observed data.\n\n##### •SoftImpute: Matrix completion by iterative soft thresholding of SVD decompositions. Inspired by the softImpute package for R, which is based on Spectral Regularization Algorithms for Learning Large Incomplete Matrices by Mazumder et. al.\n\n##### •IterativeSVD: Matrix completion by iterative low-rank SVD decomposition. Should be similar to SVDimpute from Missing value estimation methods for DNA microarrays by Troyanskaya et. al.\n\n##### •MICE: Reimplementation of Multiple Imputation by Chained Equations.\n\n##### •MatrixFactorization: Direct factorization of the incomplete matrix into low-rank U and V, with an L1 sparsity penalty on the elements of U and an L2 penalty on the elements of V. Solved by gradient descent.\n\n##### •NuclearNormMinimization: Simple implementation of Exact Matrix Completion via Convex Optimization by Emmanuel Candes and Benjamin Recht using cvxpy. Too slow for large matrices.\n\n##### •BiScaler: Iterative estimation of row/column means and standard deviations to get doubly normalized matrix. Not guaranteed to converge but works well in practice. Taken from Matrix Completion and Low-Rank SVD via Fast Alternating Least Squares.",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"##### For learning purpose will use 2 imputations Simple Impute and KNN since others are out of context for this task\n##### (You are free to experiment with others as this improve your understanding of different process)",
"_____no_output_____"
],
[
"#### 2.1 Imputing Using Simple Impute",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"df1 = train_frame['Age'].isnull()\ndf2 = pd.DataFrame(df1)\ndf2.columns = [\"new\"]\ndf2.head()",
"_____no_output_____"
],
[
"#using boolean expressions to divide Nan rows\ndf3 = train_frame[df2['new']==True] #for nan val of age\ndf4 = train_frame[df2['new']==False]\ndf3.head()",
"_____no_output_____"
],
[
"df4.head()",
"_____no_output_____"
],
[
"import datawig",
"_____no_output_____"
],
[
"#Using a SimpleImputer model\nimputer = datawig.SimpleImputer(\n input_columns = ['Sex','Pclass', 'Parch', 'Embarked', 'Survived'], output_column = 'Age',\n output_path = 'imputer_model')\n#input cols serves as data on whose basis imputation is performed\n#output col is one on which imputation will be performed",
"_____no_output_____"
],
[
"#Fit an imputer model on the train data\nimputer.fit(train_df=df4, num_epochs=50)",
"2020-07-08 11:21:52,682 [INFO] \n========== start: fit model\n2020-07-08 11:21:52,683 [WARNING] Already bound, ignoring bind()\n2020-07-08 11:21:52,684 [WARNING] optimizer already initialized, ignoring...\n2020-07-08 11:21:53,451 [INFO] Epoch[0] Batch [0-21]\tSpeed: 466.26 samples/sec\tcross-entropy=12.126804\tAge-accuracy=0.000000\n2020-07-08 11:21:54,146 [INFO] Epoch[0] Train-cross-entropy=11.686669\n2020-07-08 11:21:54,148 [INFO] Epoch[0] Train-Age-accuracy=0.000000\n2020-07-08 11:21:54,149 [INFO] Epoch[0] Time cost=1.464\n2020-07-08 11:21:54,229 [INFO] Saved checkpoint to \"imputer_model/model-0000.params\"\n2020-07-08 11:21:54,335 [INFO] Epoch[0] Validation-cross-entropy=15.185138\n2020-07-08 11:21:54,336 [INFO] Epoch[0] Validation-Age-accuracy=0.000000\n2020-07-08 11:21:55,049 [INFO] Epoch[1] Batch [0-21]\tSpeed: 477.16 samples/sec\tcross-entropy=12.035890\tAge-accuracy=0.000000\n2020-07-08 11:21:55,743 [INFO] Epoch[1] Train-cross-entropy=11.626136\n2020-07-08 11:21:55,745 [INFO] Epoch[1] Train-Age-accuracy=0.000000\n2020-07-08 11:21:55,746 [INFO] Epoch[1] Time cost=1.409\n2020-07-08 11:21:55,750 [INFO] Saved checkpoint to \"imputer_model/model-0001.params\"\n2020-07-08 11:21:55,940 [INFO] Epoch[1] Validation-cross-entropy=15.303293\n2020-07-08 11:21:55,941 [INFO] Epoch[1] Validation-Age-accuracy=0.000000\n2020-07-08 11:21:56,738 [INFO] Epoch[2] Batch [0-21]\tSpeed: 473.90 samples/sec\tcross-entropy=11.969215\tAge-accuracy=0.000000\n2020-07-08 11:21:57,334 [INFO] Epoch[2] Train-cross-entropy=11.580678\n2020-07-08 11:21:57,336 [INFO] Epoch[2] Train-Age-accuracy=0.000000\n2020-07-08 11:21:57,337 [INFO] Epoch[2] Time cost=1.394\n2020-07-08 11:21:57,340 [INFO] Saved checkpoint to \"imputer_model/model-0002.params\"\n2020-07-08 11:21:57,528 [INFO] Epoch[2] Validation-cross-entropy=15.400109\n2020-07-08 11:21:57,530 [INFO] Epoch[2] Validation-Age-accuracy=0.000000\n2020-07-08 11:21:58,331 [INFO] Epoch[3] Batch [0-21]\tSpeed: 424.27 samples/sec\tcross-entropy=11.911020\tAge-accuracy=0.000000\n2020-07-08 11:21:58,949 [INFO] Epoch[3] Train-cross-entropy=11.539261\n2020-07-08 11:21:58,950 [INFO] Epoch[3] Train-Age-accuracy=0.000000\n2020-07-08 11:21:59,027 [INFO] Epoch[3] Time cost=1.497\n2020-07-08 11:21:59,032 [INFO] Saved checkpoint to \"imputer_model/model-0003.params\"\n2020-07-08 11:21:59,147 [INFO] Epoch[3] Validation-cross-entropy=15.468325\n2020-07-08 11:21:59,148 [INFO] Epoch[3] Validation-Age-accuracy=0.000000\n2020-07-08 11:22:00,048 [INFO] Epoch[4] Batch [0-21]\tSpeed: 414.13 samples/sec\tcross-entropy=11.864084\tAge-accuracy=0.000000\n2020-07-08 11:22:00,931 [INFO] Epoch[4] Train-cross-entropy=11.505636\n2020-07-08 11:22:00,933 [INFO] Epoch[4] Train-Age-accuracy=0.000000\n2020-07-08 11:22:00,935 [INFO] Epoch[4] Time cost=1.785\n2020-07-08 11:22:00,940 [INFO] Saved checkpoint to \"imputer_model/model-0004.params\"\n2020-07-08 11:22:01,134 [INFO] Epoch[4] Validation-cross-entropy=15.500107\n2020-07-08 11:22:01,136 [INFO] Epoch[4] Validation-Age-accuracy=0.000000\n2020-07-08 11:22:02,232 [INFO] Epoch[5] Batch [0-21]\tSpeed: 309.88 samples/sec\tcross-entropy=11.827355\tAge-accuracy=0.000000\n2020-07-08 11:22:03,129 [INFO] Epoch[5] Train-cross-entropy=11.478054\n2020-07-08 11:22:03,131 [INFO] Epoch[5] Train-Age-accuracy=0.000000\n2020-07-08 11:22:03,133 [INFO] Epoch[5] Time cost=1.996\n2020-07-08 11:22:03,137 [INFO] Saved checkpoint to \"imputer_model/model-0005.params\"\n2020-07-08 11:22:03,329 [INFO] No improvement detected for 5 epochs compared to 15.185137844085693 last error obtained: 15.557070255279541, stopping here\n2020-07-08 11:22:03,331 [INFO] \n========== done (10.649701595306396 s) fit model\n"
],
[
"#Impute missing values and return original dataframe with predictions\nimputed = imputer.predict(df3)",
"_____no_output_____"
],
[
"imputed[\"Age_imputed\"].isnull().sum()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### As we now have 2 features of 'Age', 1 is orignal with Nan and second with imputated values so droping NaN col.",
"_____no_output_____"
]
],
[
[
"del imputed['Age']",
"_____no_output_____"
],
[
"imputed = imputed.rename(columns={'Age_imputed' : 'Age'})",
"_____no_output_____"
],
[
"imputed.head()",
"_____no_output_____"
],
[
"imputed = imputed[['PassengerId','Survived', 'Pclass','Name','Sex','Age','SibSp','Parch','Ticket','Fare','Cabin',\n 'Embarked']]",
"_____no_output_____"
],
[
"df6 = imputed.append(df4)\ndf6.sort_values(by=['PassengerId'])",
"_____no_output_____"
],
[
"df6[\"Age\"].isnull().sum()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"#### 2.2 Imputing Using KNN",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"##### This part is just to understand how KNN can be useful for filling NaN values, we'll just cover how its done and move forward to next strategy",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"from fancyimpute import KNN ",
"Using TensorFlow backend.\n"
],
[
"cols = ['Survived', 'Pclass', 'SibSp', 'Parch', 'Age']",
"_____no_output_____"
],
[
"k_n = KNN(k=7).fit_transform(df[cols]) ",
"Imputing row 1/891 with 0 missing, elapsed time: 0.211\nImputing row 101/891 with 0 missing, elapsed time: 0.212\nImputing row 201/891 with 0 missing, elapsed time: 0.213\nImputing row 301/891 with 1 missing, elapsed time: 0.214\nImputing row 401/891 with 0 missing, elapsed time: 0.215\nImputing row 501/891 with 0 missing, elapsed time: 0.216\nImputing row 601/891 with 0 missing, elapsed time: 0.217\nImputing row 701/891 with 0 missing, elapsed time: 0.218\nImputing row 801/891 with 0 missing, elapsed time: 0.219\n"
],
[
"k_n",
"_____no_output_____"
],
[
"df1 = pd.DataFrame(KNN(k=5).fit_transform(df[cols]) )",
"Imputing row 1/891 with 0 missing, elapsed time: 0.148\nImputing row 101/891 with 0 missing, elapsed time: 0.151\nImputing row 201/891 with 0 missing, elapsed time: 0.152\nImputing row 301/891 with 1 missing, elapsed time: 0.153\nImputing row 401/891 with 0 missing, elapsed time: 0.154\nImputing row 501/891 with 0 missing, elapsed time: 0.156\nImputing row 601/891 with 0 missing, elapsed time: 0.157\nImputing row 701/891 with 0 missing, elapsed time: 0.158\nImputing row 801/891 with 0 missing, elapsed time: 0.159\n"
],
[
"df1.head()",
"_____no_output_____"
]
],
[
[
"##### Now this can be re-added to orignal data frame and processed as rest",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"### 3. Using The power of Statistics",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"##### Let's think more logically and find certain features which have significant impact on missing data, lets start corelations and find which features are similar to Age",
"_____no_output_____"
]
],
[
[
"df_train2 = train_frame.copy()\ndf_test2 = test_frame.copy()",
"_____no_output_____"
],
[
"df_corr = df_train2.corr().abs().unstack().sort_values(kind=\"quicksort\", ascending=False).reset_index()\ndf_corr.rename(columns={\"level_0\": \"Feature 1\", \"level_1\": \"Feature 2\", 0: 'Correlation Coefficient'}, inplace=True)\ndf_corr[df_corr['Feature 1'] == 'Age']",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### As evident age is mostly correlated with Pclass, this will help us. This time i will use median age by Pclass feature and fill missing value by this correlation",
"_____no_output_____"
]
],
[
[
"age_by_pclass_sex = df_train2.groupby(['Sex', 'Pclass']).median()['Age']\n\nfor pclass in range(1, 4):\n for sex in ['female', 'male']:\n print('Median age of Pclass {} {}s: {}'.format(pclass, sex, age_by_pclass_sex[sex][pclass]))\nprint('Median age of all passengers: {}'.format(df_train1['Age'].median()))",
"Median age of Pclass 1 females: 35.0\nMedian age of Pclass 1 males: 40.0\nMedian age of Pclass 2 females: 28.0\nMedian age of Pclass 2 males: 30.0\nMedian age of Pclass 3 females: 21.5\nMedian age of Pclass 3 males: 25.0\nMedian age of all passengers: 28.0\n"
],
[
"df_train2['Age'] = df_train2.groupby(['Sex', 'Pclass'])['Age'].apply(lambda x: x.fillna(x.median()))",
"_____no_output_____"
],
[
"# Filling the missing values in test frame as well for Age with the medians of Sex and Pclass groups\nage_by_pclass_sex = df_test2.groupby(['Sex', 'Pclass']).median()['Age']\n\nfor pclass in range(1, 4):\n for sex in ['female', 'male']:\n print('Median age of Pclass {} {}s: {}'.format(pclass, sex, age_by_pclass_sex[sex][pclass]))\nprint('Median age of all passengers: {}'.format(df_test2['Age'].median()))",
"Median age of Pclass 1 females: 41.0\nMedian age of Pclass 1 males: 42.0\nMedian age of Pclass 2 females: 24.0\nMedian age of Pclass 2 males: 28.0\nMedian age of Pclass 3 females: 22.0\nMedian age of Pclass 3 males: 24.0\nMedian age of all passengers: 27.0\n"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Notice the difference of Medians for train and test data",
"_____no_output_____"
]
],
[
[
"# Filling the missing values in Age with the medians of Sex and Pclass groups\ndf_test2['Age'] = df_test2.groupby(['Sex', 'Pclass'])['Age'].apply(lambda x: x.fillna(x.median()))",
"_____no_output_____"
]
],
[
[
"##### This seems much better as we used some statistical understanding to fill missing data",
"_____no_output_____"
],
[
"##### now check if any pending nan left in age feature",
"_____no_output_____"
]
],
[
[
"train_null = df_train2.isnull().sum().sort_values(ascending=False)\ntrain_null",
"_____no_output_____"
],
[
"test_null = df_test2.isnull().sum().sort_values(ascending=False)\ntest_null",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### As Embarked has only few missing values we can use the most frequent occuring strategy here",
"_____no_output_____"
]
],
[
[
"#filling embarked with most frquent value\ncommon_value = 'S'\nembark = [df_train2, df_test2]\n\nfor dataset in embark:\n dataset['Embarked'] = dataset['Embarked'].fillna(common_value)",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Using Median approach for Fare as it is also have few occuring",
"_____no_output_____"
]
],
[
[
"med_fare = df_test2.groupby(['Pclass', 'Parch', 'SibSp']).Fare.median()[3][0][0]\nmed_fare",
"_____no_output_____"
],
[
"# Filling the missing value in Fare with the median Fare of 3rd class alone passenger\nfare = [df_test2]\n\nfor dfall in fare:\n dfall['Fare'] = dfall['Fare'].fillna(med_fare)",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"## Feature Creation",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"##### It is a part of Feature Engineering and it is process of creating features that don't already exist in the dataset or creating meaning out of not so important features",
"_____no_output_____"
],
[
"##### We'll start with Name Feature which doesn't hold much information for a model but a have deeper look and you'll find we can actually extract title from it like 'Mr.' , 'Mrs' etc which can be made useful",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"#take title out of name\nFeature = [df_train2, df_test2]\nmin_feature = 10\n\nfor dataset in Feature:\n dataset['Title'] = dataset['Name'].str.split(\", \", expand=True)[1].str.split(\".\", expand=True)[0]\n \ntitle = (df_train2['Title'].value_counts() < min_feature)\ndf_train2['Title'] = df_train2['Title'].apply(lambda x: 'Misc' if title.loc[x] == True else x)\n\ndf_train2['Title'].value_counts()",
"_____no_output_____"
],
[
"Survived_female = df_train2[df_train2.Sex=='female'].groupby(['Sex','Title'])['Survived'].mean()\nSurvived_female",
"_____no_output_____"
],
[
"Survived_male = df_train2[df_train2.Sex=='male'].groupby(['Sex','Title'])['Survived'].mean()\nSurvived_male",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"### Some fun with waffle chart",
"_____no_output_____"
],
[
"##### lets visualize our newly created feature in waffle",
"_____no_output_____"
]
],
[
[
"from pywaffle import Waffle",
"_____no_output_____"
],
[
"waf = {'Mr':517, 'Miss':182, 'Mrs':125, 'Master':40, 'Misc':27}\nwaf1 = pd.DataFrame(waf.items(), columns=['Title', 'Value'])",
"_____no_output_____"
],
[
"total_values = sum(waf1['Value'])\ncategory_proportions = [(float(value) / total_values) for value in waf1['Value']]\n\n# print out proportions\nfor i, proportion in enumerate(category_proportions):\n print (waf1.Title.values[i] + ': ' + str(proportion))",
"Mr: 0.5802469135802469\nMiss: 0.20426487093153758\nMrs: 0.14029180695847362\nMaster: 0.04489337822671156\nMisc: 0.030303030303030304\n"
],
[
"#add waffle for title\nwidth = 40 # width of chart\nheight = 10 # height of chart\n\ntotal_num_tiles = width * height # total number of tiles\n\n# compute the number of tiles for each catagory\ntiles_per_category = [round(proportion * total_num_tiles) for proportion in category_proportions]\n\n# print out number of tiles per category\nfor i, tiles in enumerate(tiles_per_category):\n print (waf1.Title.values[i] + ': ' + str(tiles))",
"Mr: 232\nMiss: 82\nMrs: 56\nMaster: 18\nMisc: 12\n"
],
[
"data = {'Mr': 232, 'Miss': 82, 'Mrs': 56, 'Master': 18, 'Misc': 12}\nfig = plt.figure(\n FigureClass=Waffle, \n rows=10, \n columns=40,\n values=data, \n cmap_name=\"tab10\",\n legend={'loc': 'upper left', 'bbox_to_anchor': (1, 1)},\n icons='male', icon_size=18, \n icon_legend=True,\n figsize=(14, 18)\n)",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"Encoding the feature manually, we can employ One Hot or Label Encoder for this task but this is done to show it can be mapped manually as well if you are not sure if encoders will perform differently on validations/test/train set.",
"_____no_output_____"
]
],
[
[
"#encoding\nFeature1 = [df_train2, df_test2]\ntitles = {\"Mr\": 1, \"Miss\": 2, \"Mrs\": 3, \"Master\": 4, \"Misc\": 5}\n\nfor dataset in Feature1:\n dataset['Title'] = dataset['Title'].map(titles)\n # filling NaN with 0, to get safe\n dataset['Title'] = dataset['Title'].fillna(0)\n \ndf_train2['Title'].value_counts()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Note: The major distinction between 'pd.cut & pd.qcut' is that 'qcut' will calculate the size of each bin in order to make sure the distribution of data in the bins is equal. In other words, all bins will have (roughly) the same number of observations. While 'cut' will create bins on average and each bin may have different vaulues(unequal samples).",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"pd.qcut(df_train2['Age'], 5).value_counts()",
"_____no_output_____"
],
[
"pd.qcut(df_train2['Fare'], 5).value_counts()",
"_____no_output_____"
],
[
"age_bin = [df_train2, df_test2]\nfor dataset in age_bin:\n dataset['Age'] = dataset['Age'].astype(int)\n dataset.loc[ dataset['Age'] <= 20, 'Age'] = 0\n dataset.loc[(dataset['Age'] > 20) & (dataset['Age'] <= 25), 'Age'] = 1\n dataset.loc[(dataset['Age'] > 25) & (dataset['Age'] <= 30), 'Age'] = 2\n dataset.loc[(dataset['Age'] > 30) & (dataset['Age'] <= 40), 'Age'] = 3\n dataset.loc[(dataset['Age'] > 40) & (dataset['Age'] <= 80), 'Age'] = 4\n",
"_____no_output_____"
],
[
"df_test2['Age'].value_counts()",
"_____no_output_____"
],
[
"Fare_bin = [df_train2, df_test2]\nfor dataset in Fare_bin:\n dataset['Fare'] = dataset['Fare'].astype(int)\n dataset.loc[ dataset['Fare'] <= 7.854, 'Fare'] = 0\n dataset.loc[(dataset['Fare'] > 7.854) & (dataset['Fare'] <= 10.5), 'Fare'] = 1\n dataset.loc[(dataset['Fare'] > 10.5) & (dataset['Fare'] <= 21.679), 'Fare'] = 2\n dataset.loc[(dataset['Fare'] > 21.679) & (dataset['Fare'] <= 39.688), 'Fare'] = 3\n dataset.loc[(dataset['Fare'] > 39.688) & (dataset['Fare'] <= 513), 'Fare'] = 4",
"_____no_output_____"
],
[
"df_train2['Fare'].value_counts()",
"_____no_output_____"
],
[
"Fig = sns.FacetGrid(df_train2, col = 'Fare', col_wrap=3)\nFig.map(sns.pointplot, 'Age', 'Survived', 'Sex', ci=95.0, palette = 'deep')\nFig.add_legend()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### See how a combination of multiple feature tell us details about target. We can actually create our own probabalistic model with these ratios.",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"## some learning\nfemale_mean = df_train2[df_train2.Sex=='female'].groupby(['Sex','Pclass', 'Embarked','Fare'])['Survived'].mean()\nfemale_mean",
"_____no_output_____"
],
[
"male_mean = df_train2[df_train2.Sex=='male'].groupby(['Sex','Pclass', 'Embarked','Fare'])['Survived'].mean()\nmale_mean",
"_____no_output_____"
],
[
"# Creating Deck column from the first letter of the Cabin column, For Missing using M\ndf_train2['Deck'] = df_train2['Cabin'].apply(lambda s: s[0] if pd.notnull(s) else 'M')\n\ndf_decks = df_train2.groupby(['Deck', 'Pclass']).count().drop(columns=['Survived', 'Sex', 'Age', 'SibSp',\n 'Parch','Fare', 'Embarked', 'Cabin', 'PassengerId', \n 'Ticket', 'Title']).rename(columns={'Name': 'Count'}).transpose()",
"_____no_output_____"
],
[
"df_decks",
"_____no_output_____"
]
],
[
[
"##### Since Deck T is negligible assigning it to frequent deck value",
"_____no_output_____"
]
],
[
[
"deck = df_train2[df_train2['Deck'] == 'T'].index\ndf_train2.loc[deck, 'Deck'] = 'M'",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"#### waffle visual for deck",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"waf2 = {'A':15, 'B':47, 'C':59, 'D':33, 'E':32, 'F':13, 'G':4, 'M':647, 'T':1}\nwaf2 = pd.DataFrame(waf2.items(), columns=['Deck', 'Value'])",
"_____no_output_____"
],
[
"total_values = sum(waf2['Value'])\ncategory_proportions = [(float(value) / total_values) for value in waf2['Value']]\n\nwidth = 30 # width of chart\nheight = 10 # height of chart\n\ntotal_num_tiles = width * height # total number of tiles\n\n# compute the number of tiles for each catagory\ntiles_per_category = [round(proportion * total_num_tiles) for proportion in category_proportions]\n\n# print out number of tiles per category\nfor i, tiles in enumerate(tiles_per_category):\n print (waf2.Deck.values[i] + ': ' + str(tiles))\n",
"A: 5\nB: 17\nC: 21\nD: 12\nE: 11\nF: 5\nG: 1\nM: 228\nT: 0\n"
],
[
"data = {'A':5, 'B':17, 'C':21, 'D':12, 'E':11, 'F':5, 'G':1, 'M':228, 'T':0}\nfig = plt.figure(\n FigureClass=Waffle, \n rows=10, \n columns=40,\n values=data, \n cmap_name=\"tab10\",\n legend={'loc': 'upper left', 'bbox_to_anchor': (1, 1)},\n icons='ship', icon_size=18, \n icon_legend=True,\n figsize=(14, 18)\n)",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Bining Deck feature ",
"_____no_output_____"
]
],
[
[
"df_train2['Deck'] = df_train2['Deck'].replace(['A', 'B', 'C'], 'ABC')\ndf_train2['Deck'] = df_train2['Deck'].replace(['D', 'E'], 'DE')\ndf_train2['Deck'] = df_train2['Deck'].replace(['F', 'G'], 'FG')\n\ndf_train2['Deck'].value_counts()",
"_____no_output_____"
]
],
[
[
"Same for Test dataset",
"_____no_output_____"
]
],
[
[
"df_test2['Deck'] = df_test2['Cabin'].apply(lambda s: s[0] if pd.notnull(s) else 'M')\n\ndft_decks = df_test2.groupby(['Deck', 'Pclass']).count().drop(columns=['Sex', 'Age', 'SibSp',\n 'Parch','Fare', 'Embarked', 'Cabin', 'PassengerId', \n 'Ticket', 'Title']).rename(columns={'Name': 'Count'}).transpose()",
"_____no_output_____"
],
[
"df_test2['Deck'] = df_test2['Deck'].replace(['A', 'B', 'C'], 'ABC')\ndf_test2['Deck'] = df_test2['Deck'].replace(['D', 'E'], 'DE')\ndf_test2['Deck'] = df_test2['Deck'].replace(['F', 'G'], 'FG')\n\ndf_test2['Deck'].value_counts()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"##### Note: Here we are going to check correlation or odds of surviving with respect to all features. This will give us a boost in understanding which charcteristics of features have more importance.",
"_____no_output_____"
]
],
[
[
"###ckeck corr between difernt class\nTarget = ['Survived']\ncorr_cols = ['Sex','Pclass', 'Embarked', 'Title','SibSp', 'Parch', 'Age', 'Fare']\n\nfor corr in corr_cols:\n if df_train2[corr].dtype != 'float64' :\n print('Surviving Correlation by:',corr)\n print(df_train2[[corr, Target[0]]].groupby(corr, as_index=False).mean())\n print('-'*10, '\\n')",
"Surviving Correlation by: Sex\n Sex Survived\n0 female 0.742038\n1 male 0.188908\n---------- \n\nSurviving Correlation by: Pclass\n Pclass Survived\n0 1 0.629630\n1 2 0.472826\n2 3 0.242363\n---------- \n\nSurviving Correlation by: Embarked\n Embarked Survived\n0 C 0.553571\n1 Q 0.389610\n2 S 0.339009\n---------- \n\nSurviving Correlation by: Title\n Title Survived\n0 1 0.156673\n1 2 0.697802\n2 3 0.792000\n3 4 0.575000\n4 5 0.444444\n---------- \n\nSurviving Correlation by: SibSp\n SibSp Survived\n0 0 0.345395\n1 1 0.535885\n2 2 0.464286\n3 3 0.250000\n4 4 0.166667\n5 5 0.000000\n6 8 0.000000\n---------- \n\nSurviving Correlation by: Parch\n Parch Survived\n0 0 0.343658\n1 1 0.550847\n2 2 0.500000\n3 3 0.600000\n4 4 0.000000\n5 5 0.200000\n6 6 0.000000\n---------- \n\nSurviving Correlation by: Age\n Age Survived\n0 0 0.455556\n1 1 0.295720\n2 2 0.380165\n3 3 0.448649\n4 4 0.371622\n---------- \n\nSurviving Correlation by: Fare\n Fare Survived\n0 0 0.215768\n1 1 0.195122\n2 2 0.426901\n3 3 0.444444\n4 4 0.642045\n---------- \n\n"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Note: Apart from heatmaps which are used further in this notebook we can also sortout corr between different features for feature selections as highly correlated feature sometime exibit no extra information than their counterparts",
"_____no_output_____"
]
],
[
[
"corr_train = df_train2.drop(['PassengerId'], axis=1).corr().abs().unstack().sort_values(kind=\"quicksort\", ascending=False).reset_index()\ncorr_train.rename(columns={\"level_0\": \"Attribute 1\", \"level_1\": \"Attribute 2\", 0: 'Correlation Coefficient'}, inplace=True)\ncorr_train.drop(corr_train.iloc[1::2].index, inplace=True)\ncorr_train1 = corr_train.drop(corr_train[corr_train['Correlation Coefficient'] == 1.0].index)",
"_____no_output_____"
],
[
"#Train frame correlations check\ncorr = corr_train1['Correlation Coefficient'] > 0.3\ncorr_train1[corr]",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### The only downside is we cannot track if the correlation is negative (if you want to do it, remove abs() from the code)",
"_____no_output_____"
]
],
[
[
"corr_test = df_test2.corr().abs().unstack().sort_values(kind=\"quicksort\", ascending=False).reset_index()\ncorr_test.rename(columns={\"level_0\": \"Feature 1\", \"level_1\": \"Feature 2\", 0: 'Correlation Coefficient'}, inplace=True)\ncorr_test.drop(corr_test.iloc[1::2].index, inplace=True)\ncorr_test1 = corr_test.drop(corr_test[corr_test['Correlation Coefficient'] == 1.0].index)",
"_____no_output_____"
],
[
"# Test frame correlations check\ncorr1 = corr_test1['Correlation Coefficient'] > 0.3\ncorr_test1[corr1]",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Same as above but with heatmaps",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(nrows=2, figsize=(25, 20))\n\nsns.heatmap(df_train2.drop(['PassengerId'], axis=1).corr(), ax=axs[0], annot=True, square=True, cmap=\"YlGnBu\", annot_kws={'size': 14})\nsns.heatmap(df_test2.drop(['PassengerId'], axis=1).corr(), ax=axs[1], annot=True, square=True, cmap='coolwarm', annot_kws={'size': 14})\n\nfor i in range(2): \n axs[i].tick_params(axis='x', labelsize=14)\n axs[i].tick_params(axis='y', labelsize=14)\n \naxs[0].set_title('Correlations for Training data features', size=15)\naxs[1].set_title('Correlations for Test data features', size=15)\n\n\naxs[0].set_ylim(6.0, 0)\naxs[1].set_ylim(6.0, 0)\nplt.show()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"## Model Training and Evaluation",
"_____no_output_____"
],
[
"##### We'll use multiple model and see they behave w.r.t to training data and choose the best model to submit for the competetion",
"_____no_output_____"
]
],
[
[
"#Models for checking, might not use all\n\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.linear_model import Perceptron\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.svm import SVC, LinearSVC\nfrom sklearn.naive_bayes import GaussianNB\nfrom xgboost import XGBClassifier",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Dropping Features not relevant for performace",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"df_train2.drop(['PassengerId', 'Name', 'Ticket', 'Cabin'], axis=1, inplace=True)\ndf_test2.drop(['PassengerId', 'Name', 'Ticket', 'Cabin'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_train2.head()",
"_____no_output_____"
],
[
"df_test2.head()",
"_____no_output_____"
],
[
"from sklearn.preprocessing import LabelEncoder",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### LabelEncoder: \n##### can turn [dog,cat,dog,mouse,cat] into [1,2,1,3,2], but then the imposed ordinality means that the average of dog and mouse is cat. Still there are algorithms like decision trees and random forests that can work with categorical variables just fine and LabelEncoder can be used to store values using less disk space.\n\n##### One-Hot-Encoding: \n##### has the advantage that the result is binary rather than ordinal and that everything sits in an orthogonal vector space. The disadvantage is that for high cardinality, the feature space can really blow up quickly and you start fighting with the curse of dimensionality. In these cases, I typically employ one-hot-encoding followed by PCA for dimensionality reduction. I find that the judicious combination of one-hot plus PCA can seldom be beat by other encoding schemes. PCA finds the linear overlap, so will naturally tend to group similar features into the same feature.",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"class FeatureEncoder:\n def __init__(self,columns = None):\n self.columns = columns # array of column names to encode\n \n def fit(self,X,y=None):\n return self \n\n def transform(self,X):\n '''\n Transforms columns of X specified in self.columns using\n LabelEncoder(). If no columns specified, transforms all\n columns in X.\n '''\n output = X.copy()\n if self.columns is not None:\n for col in self.columns:\n output[col] = LabelEncoder().fit_transform(output[col])\n else:\n for colname,col in output.iteritems():\n output[colname] = LabelEncoder().fit_transform(col)\n return output\n\n def fit_transform(self,X,y=None):\n return self.fit(X,y).transform(X)",
"_____no_output_____"
]
],
[
[
"To avoid encoding feature by feautre we employ above method which is written With help of : https://stackoverflow.com/questions/24458645/label-encoding-across-multiple-columns-in-scikit-learn",
"_____no_output_____"
]
],
[
[
"df_tr = df_train2.copy()\ndf_tr = FeatureEncoder(columns = ['Sex','Embarked', 'Deck']).fit_transform(df_tr)",
"_____no_output_____"
],
[
"df_te = df_test2.copy()\ndf_te = FeatureEncoder(columns = ['Sex','Embarked', 'Deck']).fit_transform(df_te)",
"_____no_output_____"
],
[
"df_te.head()",
"_____no_output_____"
],
[
"df_tr.dtypes",
"_____no_output_____"
],
[
"df_te = df_te.astype(int)\ndf_te.dtypes",
"_____no_output_____"
]
],
[
[
"##### Splitting target and features",
"_____no_output_____"
],
[
"##### iLoc : iloc returns a Pandas Series when one row is selected, and a Pandas DataFrame when multiple rows are selected, or if any column in full is selected \n\nwhereas, \n\n##### Loc : loc is label-based, which means that you have to specify rows and columns based on their row and column labels\n",
"_____no_output_____"
]
],
[
[
"x_train = df_tr.iloc[:,1:]\ny_train = df_tr.iloc[:,:1]",
"_____no_output_____"
],
[
"x_test = df_te.copy()",
"_____no_output_____"
],
[
"from sklearn.preprocessing import PolynomialFeatures\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import PowerTransformer",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### pipelines are set up with the fit/transform/predict functionality, so that we can fit the whole pipeline to the training data and transform to the test data without having to do it individually for everything",
"_____no_output_____"
],
[
"##### PowerTransformer provides non-linear transformations in which data is mapped to a normal distribution to stabilize variance and minimize skewness.",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"#start with PF of degree 3 then we will check till 5 if efficieny increase else we will leave it\npr=PolynomialFeatures(degree=3)\nz = pr.fit_transform(x_train)\nz.shape",
"_____no_output_____"
]
],
[
[
"##### Notice how applying Polynomial featuring have increase dimensions/variables to the equation",
"_____no_output_____"
],
[
"##### One more thing to make note of is the higher in terms of 'degree' you go in Polynomial the the more feature it will create and you might end up breaking your code, so it is better to add a break for this model",
"_____no_output_____"
]
],
[
[
"Input=[('scale',PowerTransformer()), ('polynomial', PolynomialFeatures(include_bias=False)), ('model',LogisticRegression())]\npipe=Pipeline(Input)\npipe.fit(x_train,y_train)\nlr_score = round(pipe.score(x_train, y_train) * 100, 2)\nprint(\"score:\", lr_score, \"%\")",
"score: 84.06 %\n"
],
[
"decision_tree = DecisionTreeClassifier(criterion='entropy', splitter='random') \ndecision_tree.fit(x_train, y_train) \ndt_pred = decision_tree.predict(x_test) \ndt_score = round(decision_tree.score(x_train, y_train) * 100, 2)\nprint(\"score:\", dt_score, \"%\")",
"score: 91.69 %\n"
],
[
"knn = KNeighborsClassifier(n_neighbors = 5) \nknn.fit(x_train, y_train) \nknn_pred = knn.predict(x_test) \nknn_score1 = round(knn.score(x_train, y_train) * 100, 2)\nprint(\"score:\", knn_score1, \"%\")",
"score: 86.08 %\n"
],
[
"knn = KNeighborsClassifier(n_neighbors = 3) \nknn.fit(x_train, y_train) \nknn_pred = knn.predict(x_test) \nknn_score = round(knn.score(x_train, y_train) * 100, 2)\nprint(\"score:\", knn_score, \"%\")",
"score: 87.54 %\n"
],
[
"# Random Forest\nrandom_forest = RandomForestClassifier(criterion = \"gini\", \n min_samples_leaf = 1, \n min_samples_split = 10, \n n_estimators=100, \n max_features='auto', \n oob_score=True, \n random_state=1, \n n_jobs=-1)\n\nrandom_forest.fit(x_train, y_train)\nrf_pred = random_forest.predict(x_test)\n\nrf_score = round(random_forest.score(x_train, y_train) * 100, 2)\nprint(\"score:\", rf_score, \"%\")",
"score: 88.22 %\n"
],
[
"from sklearn import svm\nfrom sklearn.svm import SVC\n\nmodel = svm.SVC(C=1, kernel='poly', random_state=0, gamma = 'auto', degree = 5)\nmodel.fit(x_train, y_train)\nsvm_pred = model.predict(x_test)\nsvm_score = round(model.score(x_train, y_train) * 100, 2)\nprint(\"score:\", svm_score, \"%\")",
"score: 89.56 %\n"
],
[
"clf = XGBClassifier()\nclf.fit(x_train, y_train, eval_metric='auc', verbose=True)\nxgb_pred = clf.predict(x_test)\nxgb_score = round(clf.score(x_train, y_train) * 100, 2)\nprint(\"score:\", xgb_score, \"%\")",
"score: 91.13 %\n"
],
[
"clf1 = XGBClassifier(booster='dart', min_split_loss = 1, max_depth= 7)\nclf1.fit(x_train, y_train, eval_metric='auc', verbose=True)\nxgb_pred = clf1.predict(x_test)\nxgb1_score = round(clf.score(x_train, y_train) * 100, 2)\nprint(\"score:\", xgb1_score, \"%\")",
"score: 91.13 %\n"
],
[
"perceptron = Perceptron(max_iter=5)\nperceptron.fit(x_train, y_train)\n\npercep_pred = perceptron.predict(x_test)\n\npercep_score = round(perceptron.score(x_train, y_train) * 100, 2)\nprint(\"score:\", percep_score, \"%\")",
"score: 77.55 %\n"
],
[
"gaussian = GaussianNB() \ngaussian.fit(x_train, y_train) \nnb_pred = gaussian.predict(x_test) \nnb_score = round(gaussian.score(x_train, y_train) * 100, 2)\nprint(\"score:\", nb_score, \"%\")",
"score: 77.78 %\n"
]
],
[
[
" ",
"_____no_output_____"
],
[
"##### Adding all model performance in a single df",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"#area for Model Score visual\nresults = pd.DataFrame({\n 'Model': ['Support Vector Machines', 'KNN_3 ', 'KNN_5', 'Logistic Regression', \n 'Random Forest', 'Naive Bayes', 'Perceptron', 'XGB Classifier',\n 'Decision Tree'],\n 'Score': [svm_score, knn_score, knn_score1, lr_score, \n rf_score, nb_score, percep_score, xgb_score, dt_score]})\nresult_df = results.sort_values(by='Score', ascending=False)\n#result_df = result_df.set_index('Score')\nresult_df.head(9)",
"_____no_output_____"
],
[
"ax = sns.barplot(x=\"Model\", y=\"Score\", data=result_df, palette=\"Set2\")\nax.set_xticklabels(ax.get_xticklabels(), rotation=40, ha=\"right\")\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"#area for feature importance visual\nimportances = pd.DataFrame({'feature':x_train.columns,'importance':np.round(random_forest.feature_importances_,3)})\nimportances = importances.sort_values('importance',ascending=False).set_index('feature')\nimportances.head(9)",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.barplot(x='Importance', y=importances.index, data=importances)\n\nplt.tick_params(axis='x', labelsize=10)\nplt.tick_params(axis='y', labelsize=10)\nplt.title('Random Forest Classifier', size=15)\n\nplt.show()",
"_____no_output_____"
],
[
"importances['importance'].plot.bar()",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
],
[
"### Predict probabilty via NaiveBayes and Decision Tree",
"_____no_output_____"
],
[
"##### predict_proba gives you the probabilities for the target (0 and 1 in our case) in array form. The number of probabilities for each row is equal to the number of categories in target variable",
"_____no_output_____"
],
[
"##### This is a very powerful function of NaiveBayes and Decision Tree (also employed in Random Forest, XGB and so on). Having a strong understanding of probablities and distribution functions combine with with this may beat some of the top models. If you can somehow work on probablities that are in range of 0.4-0.6 which are the real problem for model and create your function/layer between models say: Naive Bayes -> Your Function -> New features -> XGB/LightGBM might actually perform exceptionally well. \n##### If you want to get going with this i recommed read about probablities and distributions functions",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
],
[
[
"nb_p = gaussian.predict_proba(x_train)",
"_____no_output_____"
],
[
"nb_p[0]",
"_____no_output_____"
],
[
"prob_nb = pd.DataFrame(nb_p, columns=['Prob_NotSurvived', 'Prob_Survived']) ",
"_____no_output_____"
],
[
"prob_nb.head()",
"_____no_output_____"
],
[
"pd.cut(prob_nb['Prob_NotSurvived'], 5).value_counts()",
"_____no_output_____"
],
[
"## lets probability which may be cuasing issue for classifier ie .41 to 0.59\nprob_issue = prob_nb[prob_nb['Prob_NotSurvived']>=0.41]",
"_____no_output_____"
],
[
"prob_issue = prob_issue[prob_issue['Prob_NotSurvived']<=0.59]",
"_____no_output_____"
],
[
"prob_issue.head()",
"_____no_output_____"
]
],
[
[
"#### see how this will effect model performace, even humans will have issue with such odds :D",
"_____no_output_____"
]
],
[
[
"dfp = decision_tree.predict_proba(x_train)",
"_____no_output_____"
],
[
"prob_dt = pd.DataFrame(dfp, columns=['Prob_NotSurvived', 'Prob_Survived'])",
"_____no_output_____"
],
[
"pd.cut(prob_dt['Prob_NotSurvived'], 5).value_counts()",
"_____no_output_____"
]
],
[
[
"##### we can further join index with orignal dataframe to check where on which features our Naive Bayes and decision tree model is having issue and with further analysis we can tune our model better but this will be out of scope for this notebook.",
"_____no_output_____"
]
],
[
[
"#then end at df test pred\n\ndf_submit = pd.DataFrame(columns=['PassengerId', 'Survived'])\ndf_submit['PassengerId'] = test_frame['PassengerId']\ndf_submit['Survived'] = dt_pred\ndf_submit.to_csv('submit01.csv', header=True, index=False)\n",
"_____no_output_____"
]
],
[
[
"##### Lastly, there are tonnes of options I didn't discuss here, some of the prominent are Cross Validations, Grid Seacrh for paramters tuning, PCA. But remember that they are also essential part of any ML project.",
"_____no_output_____"
],
[
"##### Feel free to contact me here or at : https://www.linkedin.com/in/muhammad-saad-31740060/",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ec889541efa439c3a767fb5c721844bed9680f70 | 6,495 | ipynb | Jupyter Notebook | local_optimization/FOAS/migration/migration_e04dg_e04kf.ipynb | numericalalgorithmsgroup/NAGPythonExamples | 4bd5beb75e7dec2baf8d147d93427989bac9105c | [
"BSD-3-Clause"
] | 40 | 2018-12-06T20:20:01.000Z | 2022-03-05T23:09:31.000Z | local_optimization/FOAS/migration/migration_e04dg_e04kf.ipynb | kelly1208/NAGPythonExamples | bd20f719c176bbbbc878fea7d0962e5fa10d9d3e | [
"BSD-3-Clause"
] | 11 | 2019-03-25T11:52:51.000Z | 2021-04-12T14:08:31.000Z | local_optimization/FOAS/migration/migration_e04dg_e04kf.ipynb | kelly1208/NAGPythonExamples | bd20f719c176bbbbc878fea7d0962e5fa10d9d3e | [
"BSD-3-Clause"
] | 21 | 2019-01-22T13:30:57.000Z | 2021-12-15T13:05:14.000Z | 29.930876 | 202 | 0.554888 | [
[
[
"# Migrating from `uncon_conjgrd_comp` to `handle_solve_bounds_foas`\n\nThis notebook illustrates the steps required to upgrade from the solver `uncon_conjgrd_comp` (`E04DG`) to `handle_solve_bounds_foas` (`E04KF`) introduced at Mark 27 of the NAG Library.\n\nFrom the usage perspective, the main difference between the solvers is the user call-backs,\n`uncon_conjgrd_comp` has a single user call-back that can return *objective \nfunction* and *gradient* evaluations, while `handle_solve_bounds_foas` has two separate user call-backs, \none for the *objective funtion* and one for the *objective gradient*.\n\nIn this notebook the 2d Rosenbrock problem is solved with both solvers and illustrates the changes necessary for the migration to `handle_solve_bounds_foas`. The solution to the problem is (1, 1).",
"_____no_output_____"
]
],
[
[
"# NAG Copyright 2020.\nfrom naginterfaces.base import utils\nfrom naginterfaces.library import opt\nimport numpy as np",
"_____no_output_____"
],
[
"# Define E04DG user call-back\ndef objfun_e04dg(mode, x, _nstate, _data=None):\n objf = (1. - x[0])**2 + 100.*(x[1] - x[0]**2)**2\n if mode == 2:\n fdx = [\n 2.*x[0] - 400.*x[0]*(x[1]-x[0]**2) - 2.,\n 200.*(x[1]-x[0]**2),\n ]\n return objf, fdx\n return objf, np.zeros(len(x))",
"_____no_output_____"
],
[
"# Define user call-backs for E04KF\ndef objfun_e04kf(x, inform, _data=None): \n \"\"\"Return the objective function value\"\"\"\n objf = (1. - x[0])**2 + 100.*(x[1] - x[0]**2)**2\n return objf, inform\n\ndef objgrd_e04kf(x, fdx, inform, _data=None):\n \"\"\"The objective's gradient. Note that fdx has to be updated IN-PLACE\"\"\"\n fdx[:] = [\n 2.*x[0] - 400.*x[0]*(x[1]-x[0]**2) - 2.,\n 200.*(x[1]-x[0]**2),\n ]\n return inform",
"_____no_output_____"
],
[
"# The initial guess\nx = [-1.5, 1.9]\n\n# Use an explicit I/O manager for abbreviated iteration output\niom = utils.FileObjManager(locus_in_output=False)",
"_____no_output_____"
]
],
[
[
"## Solve the problem with `uncon_conjgrd_comp`",
"_____no_output_____"
]
],
[
[
"comm = opt.nlp1_init('uncon_conjgrd_comp')\nslv = opt.uncon_conjgrd_comp(objfun_e04dg, x, comm, io_manager=iom)\nprint('Solution: \\n', slv.x)",
"<ipython-input-5-a8bdc418615c>:2: NagDeprecatedWarning: (NAG Python function naginterfaces.library.opt.uncon_conjgrd_comp)\nThis function is deprecated.\nThe following advice is given for making a replacement:\nPlease use handle_solve_bounds_foas instead.\nSee also https://www.nag.com/numeric/py/nagdoc_latest/replace.html\n slv = opt.uncon_conjgrd_comp(objfun_e04dg, x, comm, io_manager=iom)\n"
]
],
[
[
"## Now solve with the new solver `handle_solve_bounds_foas`",
"_____no_output_____"
]
],
[
[
"# Create an empty handle for the problem\nnvar = len(x)\nhandle = opt.handle_init(nvar)\n\n# Define the nonlinear objective in the handle\n# Setup a gradient vector of length nvar\nopt.handle_set_nlnobj(handle, idxfd=list(range(1, nvar+1)))\n\n# Set some algorithmic options\nfor option in [\n 'Print Options = No', # print Options?\n 'Print Solution = Yes', # print on the screen the solution point X\n 'Print Level = 1', # print details of each iteration (screen)\n]:\n opt.handle_opt_set(handle, option)\n \n# Solve the problem and print the solution\nopt.handle_solve_bounds_foas(handle, x, objfun=objfun_e04kf, objgrd=objgrd_e04kf,\n io_manager=iom)\n\n# Destroy the handle and free allocated memory\nopt.handle_free(handle)",
" E04KF, First order method for bound-constrained problems\n\n Status: converged, an optimal solution was found\n Value of the objective 2.12807E-15\n Norm of gradient 3.67342E-08\n\n Primal variables:\n idx Lower bound Value Upper bound\n 1 -inf 1.00000E+00 inf\n 2 -inf 1.00000E+00 inf\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec88cbe3b75c7768f39af110180df2354895904e | 15,274 | ipynb | Jupyter Notebook | src/examples/tutorial/ascent_intro/notebooks/03_conduit_blueprint_mesh_examples.ipynb | ax3l/ascent | 2c70bac3b9ee21e9472c4725841bd096e9da653d | [
"BSD-3-Clause"
] | null | null | null | src/examples/tutorial/ascent_intro/notebooks/03_conduit_blueprint_mesh_examples.ipynb | ax3l/ascent | 2c70bac3b9ee21e9472c4725841bd096e9da653d | [
"BSD-3-Clause"
] | null | null | null | src/examples/tutorial/ascent_intro/notebooks/03_conduit_blueprint_mesh_examples.ipynb | ax3l/ascent | 2c70bac3b9ee21e9472c4725841bd096e9da653d | [
"BSD-3-Clause"
] | null | null | null | 31.688797 | 252 | 0.539217 | [
[
[
"# Conduit Mesh Blueprint Examples\nSimulation mesh data is passed to Ascent using a shared set of conventions called the\n[Mesh Blueprint] (https://llnl-conduit.readthedocs.io/en/latest/blueprint_mesh.html).\n\nSimply described, these conventions outline a structure to follow to create Conduit trees \nthat can represent a wide range of simulation mesh types (uniform grids, unstructured meshes, etc). Conduit's dynamic tree and zero-copy support make it easy to adapt existing data to conform to the Mesh Blueprint for use in tools like Ascent.\n\nThese examples outline how to create Conduit Nodes that describe simple single domain meshes and review\nsome of Conduits built-in mesh examples. More Mesh Blueprint examples are also detailed in Conduit's [Mesh Blueprint Examples Docs](https://llnl-conduit.readthedocs.io/en/latest/blueprint_mesh.html#examples).",
"_____no_output_____"
]
],
[
[
"# cleanup any old results\n#!./cleanup.sh\n# conduit + ascent imports\nimport conduit\nimport conduit.blueprint\nimport ascent\n\nimport math\nimport numpy as np\n\n# Jupyter imports\nfrom IPython.display import Image\n# helpers we use when displaying results in the notebook\nfrom ascent_tutorial_jupyter_utils import img_display_width\nfrom ascent_tutorial_jupyter_utils import ImageSeqViewer",
"_____no_output_____"
]
],
[
[
"## Mesh Blueprint Example 1\n### Creating a uniform grid with a single field",
"_____no_output_____"
]
],
[
[
"#\n# Create a 3D mesh defined on a uniform grid of points\n# with a single vertex associated field named `alternating`\n#\n\nmesh = conduit.Node()\n\n# create the coordinate set\nnum_per_dim = 9\nmesh[\"coordsets/coords/type\"] = \"uniform\";\nmesh[\"coordsets/coords/dims/i\"] = num_per_dim\nmesh[\"coordsets/coords/dims/j\"] = num_per_dim\nmesh[\"coordsets/coords/dims/k\"] = num_per_dim\n\n# add origin and spacing to the coordset (optional)\nmesh[\"coordsets/coords/origin/x\"] = -10.0\nmesh[\"coordsets/coords/origin/y\"] = -10.0\nmesh[\"coordsets/coords/origin/z\"] = -10.0\n\ndistance_per_step = 20.0/(num_per_dim-1)\n\nmesh[\"coordsets/coords/spacing/dx\"] = distance_per_step\nmesh[\"coordsets/coords/spacing/dy\"] = distance_per_step\nmesh[\"coordsets/coords/spacing/dz\"] = distance_per_step\n\n# add the topology\n# this case is simple b/c it's implicitly derived from the coordinate set\nmesh[\"topologies/topo/type\"] = \"uniform\";\n# reference the coordinate set by name\nmesh[\"topologies/topo/coordset\"] = \"coords\";\n\n# create a vertex associated field named alternating\nnum_vertices = num_per_dim * num_per_dim * num_per_dim\nvals = np.zeros(num_vertices,dtype=np.float32)\nfor i in range(num_vertices):\n if i%2:\n vals[i] = 0.0\n else:\n vals[i] = 1.0\nmesh[\"fields/alternating/association\"] = \"vertex\";\nmesh[\"fields/alternating/topology\"] = \"topo\";\nmesh[\"fields/alternating/values\"].set_external(vals)",
"_____no_output_____"
],
[
"# print the mesh we created\nprint(mesh.to_yaml())",
"_____no_output_____"
],
[
"# make sure the mesh we created conforms to the blueprint\nverify_info = conduit.Node()\nif not conduit.blueprint.mesh.verify(mesh,verify_info):\n print(\"Mesh Verify failed!\")\n print(verify_info.to_yaml())\nelse:\n print(\"Mesh verify success!\")",
"_____no_output_____"
],
[
"# now lets look at the mesh with Ascent\na = ascent.Ascent()\na.open()\n\n# publish mesh to ascent\na.publish(mesh)\n\n# setup actions\nactions = conduit.Node()\nadd_act = actions.append();\nadd_act[\"action\"] = \"add_scenes\";\n\n# declare a scene (s1) with one plot (p1) \n# to render the dataset\nscenes = add_act[\"scenes\"]\nscenes[\"s1/plots/p1/type\"] = \"pseudocolor\"\nscenes[\"s1/plots/p1/field\"] = \"alternating\"\n# Set the output file name (ascent will add \".png\")\nscenes[\"s1/image_name\"] = \"out_ascent_render_uniform\"\n\n# print our full actions tree\nprint(actions.to_yaml())\n\n# execute the actions\na.execute(actions)\n\n# close ascent\na.close()",
"_____no_output_____"
],
[
"# show the resulting image\nImage(\"out_ascent_render_uniform.png\",width=img_display_width())",
"_____no_output_____"
]
],
[
[
"## Mesh Blueprint Example 2\n### Creating an unstructured tet mesh with fields",
"_____no_output_____"
]
],
[
[
"#\n# Create a 3D mesh defined on an explicit set of points,\n# composed of two tets, with two element associated fields\n# (`var1` and `var2`)\n#\n\nmesh = conduit.Node()\n\n# create an explicit coordinate set\nx = np.array( [-1.0, 0.0, 0.0, 0.0, 1.0 ], dtype=np.float64 )\ny = np.array( [0.0, -1.0, 0.0, 1.0, 0.0 ], dtype=np.float64 )\nz = np.array( [ 0.0, 0.0, 1.0, 0.0, 0.0 ], dtype=np.float64 )\n\nmesh[\"coordsets/coords/type\"] = \"explicit\";\nmesh[\"coordsets/coords/values/x\"].set_external(x)\nmesh[\"coordsets/coords/values/y\"].set_external(y)\nmesh[\"coordsets/coords/values/z\"].set_external(z)\n\n# add an unstructured topology\nmesh[\"topologies/mesh/type\"] = \"unstructured\"\n# reference the coordinate set by name\nmesh[\"topologies/mesh/coordset\"] = \"coords\"\n# set topology shape type\nmesh[\"topologies/mesh/elements/shape\"] = \"tet\"\n# add a connectivity array for the tets\nconnectivity = np.array([0, 1, 3, 2, 4, 3, 1, 2 ],dtype=np.int64)\nmesh[\"topologies/mesh/elements/connectivity\"].set_external(connectivity)\n \nvar1 = np.array([0,1],dtype=np.float32)\nvar2 = np.array([1,0],dtype=np.float32)\n\n# create a field named var1\nmesh[\"fields/var1/association\"] = \"element\"\nmesh[\"fields/var1/topology\"] = \"mesh\"\nmesh[\"fields/var1/values\"].set_external(var1)\n\n# create a field named var2\nmesh[\"fields/var2/association\"] = \"element\"\nmesh[\"fields/var2/topology\"] = \"mesh\"\nmesh[\"fields/var2/values\"].set_external(var2)",
"_____no_output_____"
],
[
"# print the mesh we created\nprint(mesh.to_yaml())",
"_____no_output_____"
],
[
"# make sure the mesh we created conforms to the blueprint\nverify_info = conduit.Node()\nif not conduit.blueprint.mesh.verify(mesh,verify_info):\n print(\"Mesh Verify failed!\")\n print(verify_info.to_yaml())\nelse:\n print(\"Mesh verify success!\")",
"_____no_output_____"
],
[
"# now lets look at the mesh with Ascent\na = ascent.Ascent()\na.open()\n\n# publish mesh to ascent\na.publish(mesh)\n\n# setup actions\nactions = conduit.Node()\nadd_act = actions.append();\nadd_act[\"action\"] = \"add_scenes\"\n\n# declare a scene (s1) with one plot (p1) \n# to render the dataset\nscenes = add_act[\"scenes\"]\nscenes[\"s1/plots/p1/type\"] = \"pseudocolor\"\nscenes[\"s1/plots/p1/field\"] = \"var1\"\n# Set the output file name (ascent will add \".png\")\nscenes[\"s1/image_name\"] = \"out_ascent_render_tets\"\n\n# print our full actions tree\nprint(actions.to_yaml())\n\n# execute the actions\na.execute(actions)\n\n# close ascent\na.close()",
"_____no_output_____"
],
[
"# show the resulting image\nImage(\"out_ascent_render_tets.png\",width=img_display_width())",
"_____no_output_____"
]
],
[
[
"## Mesh Blueprint Example 3\n### Experimenting with the built-in braid example\nRelated docs: [Braid Example Docs](https://llnl-conduit.readthedocs.io/en/latest/blueprint_mesh.html#braid)",
"_____no_output_____"
]
],
[
[
"# The conduit blueprint library provides several \n# simple builtin examples that cover the range of\n# supported coordinate sets, topologies, field etc\n# \n# Here we create a mesh using the braid example\n# (https://llnl-conduit.readthedocs.io/en/latest/blueprint_mesh.html#braid)\n# and modify one of its fields to create a time-varying\n# example",
"_____no_output_____"
],
[
"# Define a function that will calcualte a time varying field\ndef braid_time_varying(npts_x, npts_y, npts_z, interp, res):\n if npts_z < 1:\n npts_z = 1\n\n npts = npts_x * npts_y * npts_z\n \n res[\"association\"] = \"vertex\"\n res[\"topology\"] = \"mesh\"\n vals = res[\"values\"]\n \n dx_seed_start = 0.0\n dx_seed_end = 5.0\n dx_seed = interp * (dx_seed_end - dx_seed_start) + dx_seed_start\n \n dy_seed_start = 0.0\n dy_seed_end = 2.0\n dy_seed = interp * (dy_seed_end - dy_seed_start) + dy_seed_start\n \n dz_seed = 3.0\n\n dx = (float) (dx_seed * math.pi) / float(npts_x - 1)\n dy = (float) (dy_seed * math.pi) / float(npts_y-1)\n dz = (float) (dz_seed * math.pi) / float(npts_z-1)\n \n idx = 0\n for k in range(npts_z):\n cz = (k * dz) - (1.5 * math.pi)\n for j in range(npts_y):\n cy = (j * dy) - (math.pi)\n for i in range(npts_x):\n cx = (i * dx) + (2.0 * math.pi)\n cv = math.sin( cx ) + \\\n math.sin( cy ) + \\\n 2.0 * math.cos(math.sqrt( (cx*cx)/2.0 +cy*cy) / .75) + \\\n 4.0 * math.cos( cx*cy / 4.0)\n \n if npts_z > 1:\n cv += math.sin( cz ) + \\\n 1.5 * math.cos(math.sqrt(cx*cx + cy*cy + cz*cz) / .75)\n vals[idx] = cv\n idx+=1",
"_____no_output_____"
],
[
"# create conduit node with an example mesh using conduit blueprint's braid function\n# ref: https://llnl-conduit.readthedocs.io/en/latest/blueprint_mesh.html#braid\nmesh = conduit.Node()\nconduit.blueprint.mesh.examples.braid(\"hexs\",\n 50,\n 50,\n 50,\n mesh)",
"_____no_output_____"
],
[
"a = ascent.Ascent()\n# open ascent\na.open()\n\n# create our actions\nactions = conduit.Node()\nadd_act =actions.append()\nadd_act[\"action\"] = \"add_scenes\"\n\n# declare a scene (s1) and plot (p1)\n# to render braid field \nscenes = add_act[\"scenes\"] \nscenes[\"s1/plots/p1/type\"] = \"pseudocolor\"\nscenes[\"s1/plots/p1/field\"] = \"braid\"\n\n# print our actions tree\nprint(actions.to_yaml())\n\n# loop over a set of steps and \n# render a time varying version of the braid field\n\nnsteps = 20\ninterps = np.linspace(0.0, 1.0, num=nsteps)\ni = 0\n\nfor interp in interps:\n print(\"{}: interp = {}\".format(i,interp))\n # update the braid field\n braid_time_varying(50,50,50,interp,mesh[\"fields/braid\"])\n # update the mesh cycle\n mesh[\"state/cycle\"] = i\n # Set the output file name (ascent will add \".png\")\n scenes[\"s1/renders/r1/image_name\"] = \"out_ascent_render_braid_tv_%04d\" % i\n scenes[\"s1/renders/r1/camera/azimuth\"] = 25.0\n \n # publish mesh to ascent\n a.publish(mesh)\n\n # execute the actions\n a.execute(actions)\n \n i+=1\n\n# close ascent\na.close()",
"_____no_output_____"
],
[
"result_image_files = [\"out_ascent_render_braid_tv_%04d.png\" % i for i in range(nsteps)]",
"_____no_output_____"
],
[
"ImageSeqViewer(result_image_files).show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec88cd38e9d70298f31c6876085e1751c5b32a96 | 5,651 | ipynb | Jupyter Notebook | jupyter/notebooks/Use case 1 - Setup vars - Process files.ipynb | samarthdd/cdr-plugin-folder-to-folder | def780f5590f63066194ff7b348fd256d7f74a10 | [
"Apache-2.0"
] | null | null | null | jupyter/notebooks/Use case 1 - Setup vars - Process files.ipynb | samarthdd/cdr-plugin-folder-to-folder | def780f5590f63066194ff7b348fd256d7f74a10 | [
"Apache-2.0"
] | null | null | null | jupyter/notebooks/Use case 1 - Setup vars - Process files.ipynb | samarthdd/cdr-plugin-folder-to-folder | def780f5590f63066194ff7b348fd256d7f74a10 | [
"Apache-2.0"
] | 1 | 2021-07-03T13:15:14.000Z | 2021-07-03T13:15:14.000Z | 23.644351 | 141 | 0.482216 | [
[
[
"# Use case 1 - Setup vars - Process files",
"_____no_output_____"
],
[
"### Step 1 : configure server and check health",
"_____no_output_____"
]
],
[
[
"from jupyter_apis.API_Client import API_Client\nimport json\n\nurl_server='http://api:8880' # api dns resolution is provided by docker-compose or kubernetes\napi = API_Client(url_server)\n\nassert api.health() =={'status': 'ok'} ",
"_____no_output_____"
]
],
[
[
"### Step 2 : Configure hard discs\n\n- Configure hd1_path, hd2_path and hd3_path in \"data\"",
"_____no_output_____"
]
],
[
[
"\ndata = { \"hd1_path\" : \"./test_data/scenario-2/hd1\", \n \"hd2_path\" : \"./test_data/scenario-2/hd2\", \n \"hd3_path\" : \"./test_data/scenario-2/hd3\"} \n\nresponse=api.configure_environment(data=data)\nassert response.status_code == 200 \nassert response.json() == data\n\n\"Hard discs are configured\"\n",
"_____no_output_____"
]
],
[
[
"### Step 3 : Configure GW SDK Endpoints\n\n- Configure gw sdk IP and Port\n- Edit IP and Port in varibale \"data\"\n- You can pass multiple endpoints\n- Example : data = '{ \"Endpoints\": [{ \"IP\": \"92.109.25.70\", \"Port\": \"8080\" },{ \"IP\": \"127.0.0.1\", \"Port\": \"8080\" }]}'",
"_____no_output_____"
]
],
[
[
"data = data = { \"Endpoints\": [ { \"IP\": \"34.245.221.234\", \"Port\": \"8080\" },\n { \"IP\": \"34.240.183.4\" , \"Port\": \"8080\" },\n { \"IP\": \"84.16.229.232\" , \"Port\": \"8080\" }]}\n\nresponse=api.set_gw_sdk_endpoints(data=data)\nassert response.status_code == 200\nassert response.json() == data\n\n\"SDK Endpoints are set\"\n",
"_____no_output_____"
]
],
[
[
"### Step 4 : Process files\n\nThese calls will:\n\n- copy all files from HD1 into HD2 (based on hash)\n- process all files from HD2 using Glasswall Cloud SDK\n- copy all processed files into HD3\n",
"_____no_output_____"
]
],
[
[
"#api.clear_data_and_status()\n#assert api.pre_process() == '[\"Processing is done\"]'\nassert api.start_process() == '\"Loop completed\"'\n\"all files processed \"",
"_____no_output_____"
]
],
[
[
"# Step 5 : Folder structure visualization",
"_____no_output_____"
]
],
[
[
"from jupyter_apis.Display_Path import display_path\n\ndisplay_path('../test_data/scenario-1')",
"scenario-1/\n├── .DS_Store\n├── hd1/\n│ └── image2.jpg\n├── hd2/\n│ ├── .DS_Store\n│ ├── data/\n│ │ └── 86df34018436a99e1e98ff51346591e189cd76c8518f7288bb1ea8336396259b/\n│ │ ├── events.json\n│ │ ├── metadata.json\n│ │ └── source\n│ └── status/\n│ ├── events.json\n│ ├── hash.json\n│ └── status.json\n└── hd3/\n ├── image2.jpg\n └── image3.jpg\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec88d689cac43a60e905b8c6d0f0720f93caf4ac | 34,867 | ipynb | Jupyter Notebook | reinforce/REINFORCE.ipynb | arpradha/deep-reinforcement-learning | 01cfc7ab19453285886900d9c6332c8cb435df51 | [
"MIT"
] | null | null | null | reinforce/REINFORCE.ipynb | arpradha/deep-reinforcement-learning | 01cfc7ab19453285886900d9c6332c8cb435df51 | [
"MIT"
] | null | null | null | reinforce/REINFORCE.ipynb | arpradha/deep-reinforcement-learning | 01cfc7ab19453285886900d9c6332c8cb435df51 | [
"MIT"
] | null | null | null | 142.314286 | 28,372 | 0.883558 | [
[
[
"# REINFORCE\n\n---\n\nIn this notebook, we will train REINFORCE with OpenAI Gym's Cartpole environment.",
"_____no_output_____"
],
[
"### 1. Import the Necessary Packages",
"_____no_output_____"
]
],
[
[
"import gym\ngym.logger.set_level(40) # suppress warnings (please remove if gives error)\nimport numpy as np\nfrom collections import deque\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport torch\ntorch.manual_seed(0) # set random seed\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nfrom torch.distributions import Categorical",
"_____no_output_____"
]
],
[
[
"### 2. Define the Architecture of the Policy",
"_____no_output_____"
]
],
[
[
"env = gym.make('CartPole-v0')\nenv.seed(0)\nprint('observation space:', env.observation_space)\nprint('action space:', env.action_space)\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\nclass Policy(nn.Module):\n def __init__(self, s_size=4, h_size=16, a_size=2):\n super(Policy, self).__init__()\n self.fc1 = nn.Linear(s_size, h_size)\n self.fc2 = nn.Linear(h_size, a_size)\n\n def forward(self, x):\n x = F.relu(self.fc1(x))\n x = self.fc2(x)\n return F.softmax(x, dim=1)\n \n def act(self, state):\n state = torch.from_numpy(state).float().unsqueeze(0).to(device)\n probs = self.forward(state).cpu()\n m = Categorical(probs)\n action = m.sample()\n return action.item(), m.log_prob(action)",
"observation space: Box(4,)\naction space: Discrete(2)\n"
]
],
[
[
"### 3. Train the Agent with REINFORCE",
"_____no_output_____"
]
],
[
[
"policy = Policy().to(device)\noptimizer = optim.Adam(policy.parameters(), lr=1e-2)\n\ndef reinforce(n_episodes=1000, max_t=1000, gamma=1.0, print_every=100):\n scores_deque = deque(maxlen=100)\n scores = []\n for i_episode in range(1, n_episodes+1):\n saved_log_probs = []\n rewards = []\n state = env.reset()\n for t in range(max_t):\n action, log_prob = policy.act(state)\n saved_log_probs.append(log_prob)\n state, reward, done, _ = env.step(action)\n rewards.append(reward)\n if done:\n break \n scores_deque.append(sum(rewards))\n scores.append(sum(rewards))\n \n discounts = [gamma**i for i in range(len(rewards)+1)]\n R = sum([a*b for a,b in zip(discounts, rewards)])\n \n policy_loss = []\n for log_prob in saved_log_probs:\n policy_loss.append(-log_prob * R)\n policy_loss = torch.cat(policy_loss).sum()\n \n optimizer.zero_grad()\n policy_loss.backward()\n optimizer.step()\n \n if i_episode % print_every == 0:\n print('Episode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))\n if np.mean(scores_deque)>=195.0:\n print('Environment solved in {:d} episodes!\\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_deque)))\n break\n \n return scores\n \nscores = reinforce()",
"Episode 100\tAverage Score: 34.47\nEpisode 200\tAverage Score: 66.26\nEpisode 300\tAverage Score: 87.82\nEpisode 400\tAverage Score: 72.83\nEpisode 500\tAverage Score: 172.00\nEpisode 600\tAverage Score: 160.65\nEpisode 700\tAverage Score: 167.15\nEnvironment solved in 691 episodes!\tAverage Score: 196.69\n"
]
],
[
[
"### 4. Plot the Scores",
"_____no_output_____"
]
],
[
[
"fig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(1, len(scores)+1), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 5. Watch a Smart Agent!",
"_____no_output_____"
]
],
[
[
"env = gym.make('CartPole-v0')\n\nstate = env.reset()\nfor t in range(1000):\n action, _ = policy.act(state)\n env.render()\n state, reward, done, _ = env.step(action)\n if done:\n break \n\nenv.close()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec88e70984ddb95f426b2f70ae6765387251bd2b | 26,530 | ipynb | Jupyter Notebook | 4 Tuples.ipynb | mzamanpk/Python-IBM | c85b7c191caacdd3a07c7a65406ee81dbf71db74 | [
"MIT"
] | null | null | null | 4 Tuples.ipynb | mzamanpk/Python-IBM | c85b7c191caacdd3a07c7a65406ee81dbf71db74 | [
"MIT"
] | null | null | null | 4 Tuples.ipynb | mzamanpk/Python-IBM | c85b7c191caacdd3a07c7a65406ee81dbf71db74 | [
"MIT"
] | null | null | null | 21.49919 | 716 | 0.510931 | [
[
[
"<h1>Tuples in Python</h1>",
"_____no_output_____"
],
[
"<p><strong>Welcome!</strong> This notebook will teach you about the tuples in the Python Programming Language. By the end of this lab, you'll know the basics tuple operations in Python, including indexing, slicing and sorting.</p> ",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <a href=\"https://cocl.us/topNotebooksPython101Coursera\">\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/TopAd.png\" width=\"750\" align=\"center\">\n </a>\n</div>",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ul>\n <li>\n <a href=\"#dataset\">About the Dataset</a>\n </li>\n <li>\n <a href=\"#tuple\">Tuples</a>\n <ul>\n <li><a href=\"index\">Indexing</a></li>\n <li><a href=\"slice\">Slicing</a></li>\n <li><a href=\"sort\">Sorting</a></li>\n </ul>\n </li>\n <li>\n <a href=\"#escape\">Quiz on Tuples</a>\n </li>\n </ul>\n <p>\n Estimated time needed: <strong>15 min</strong>\n </p>\n</div>\n\n<hr>",
"_____no_output_____"
],
[
"<h2 id=\"tuple\">Tuples</h2>",
"_____no_output_____"
],
[
"In Python, there are different data types: string, integer and float. These data types can all be contained in a tuple as follows:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesType.png\" width=\"750\" align=\"center\" />",
"_____no_output_____"
],
[
"Now, let us create your first tuple with string, integer and float.",
"_____no_output_____"
]
],
[
[
"# Create your first tuple\n\ntuple1 = (\"disco\",10,1.2 )\ntuple1",
"_____no_output_____"
]
],
[
[
"The type of variable is a **tuple**. ",
"_____no_output_____"
]
],
[
[
"# Print the type of the tuple you created\n\ntype(tuple1)",
"_____no_output_____"
]
],
[
[
"<h3 id=\"index\">Indexing</h3>",
"_____no_output_____"
],
[
" Each element of a tuple can be accessed via an index. The following table represents the relationship between the index and the items in the tuple. Each element can be obtained by the name of the tuple followed by a square bracket with the index number:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesIndex.gif\" width=\"750\" align=\"center\">",
"_____no_output_____"
],
[
"We can print out each value in the tuple:",
"_____no_output_____"
]
],
[
[
"# Print the variable on each index\n\nprint(tuple1[0])\nprint(tuple1[1])\nprint(tuple1[2])",
"disco\n10\n1.2\n"
]
],
[
[
"We can print out the **type** of each value in the tuple:\n",
"_____no_output_____"
]
],
[
[
"# Print the type of value on each index\n\nprint(type(tuple1[0]))\nprint(type(tuple1[1]))\nprint(type(tuple1[2]))",
"<class 'str'>\n<class 'int'>\n<class 'float'>\n"
]
],
[
[
"We can also use negative indexing. We use the same table above with corresponding negative values:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesNeg.png\" width=\"750\" align=\"center\">",
"_____no_output_____"
],
[
"We can obtain the last element as follows (this time we will not use the print statement to display the values):",
"_____no_output_____"
]
],
[
[
"# Use negative index to get the value of the last element\n\ntuple1[-1]",
"_____no_output_____"
]
],
[
[
"We can display the next two elements as follows:",
"_____no_output_____"
]
],
[
[
"# Use negative index to get the value of the second last element\n\ntuple1[-2]",
"_____no_output_____"
],
[
"# Use negative index to get the value of the third last element\n\ntuple1[-3]",
"_____no_output_____"
]
],
[
[
"<h3 id=\"concate\">Concatenate Tuples</h3>",
"_____no_output_____"
],
[
"We can concatenate or combine tuples by using the **+** sign:",
"_____no_output_____"
]
],
[
[
"# Concatenate two tuples\n\ntuple2 = tuple1 + (\"hard rock\", 10)\ntuple2",
"_____no_output_____"
]
],
[
[
"We can slice tuples obtaining multiple values as demonstrated by the figure below:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesSlice.gif\" width=\"750\" align=\"center\">",
"_____no_output_____"
],
[
"<h3 id=\"slice\">Slicing</h3>",
"_____no_output_____"
],
[
"We can slice tuples, obtaining new tuples with the corresponding elements: ",
"_____no_output_____"
]
],
[
[
"# Slice from index 0 to index 2\n\ntuple2[0:3]",
"_____no_output_____"
]
],
[
[
"We can obtain the last two elements of the tuple:",
"_____no_output_____"
]
],
[
[
"# Slice from index 3 to index 4\n\ntuple2[3:5]",
"_____no_output_____"
]
],
[
[
"We can obtain the length of a tuple using the length command: ",
"_____no_output_____"
]
],
[
[
"# Get the length of tuple\n\nlen(tuple2)",
"_____no_output_____"
]
],
[
[
"This figure shows the number of elements:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesElement.png\" width=\"750\" align=\"center\">",
"_____no_output_____"
],
[
"<h3 id=\"sort\">Sorting</h3>",
"_____no_output_____"
],
[
" Consider the following tuple:",
"_____no_output_____"
]
],
[
[
"# A sample tuple\n\nRatings = (0, 9, 6, 5, 10, 8, 9, 6, 2)",
"_____no_output_____"
]
],
[
[
"We can sort the values in a tuple and save it to a new tuple: ",
"_____no_output_____"
]
],
[
[
"# Sort the tuple\n\nRatingsSorted = sorted(Ratings)\nRatingsSorted",
"_____no_output_____"
]
],
[
[
"<h3 id=\"nest\">Nested Tuple</h3>",
"_____no_output_____"
],
[
"A tuple can contain another tuple as well as other more complex data types. This process is called 'nesting'. Consider the following tuple with several elements: ",
"_____no_output_____"
]
],
[
[
"# Create a nest tuple\n\nNestedT =(1, 2, (\"pop\", \"rock\") ,(3,4),(\"disco\",(1,2)))",
"_____no_output_____"
]
],
[
[
"Each element in the tuple including other tuples can be obtained via an index as shown in the figure:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesNestOne.png\" width=\"750\" align=\"center\">",
"_____no_output_____"
]
],
[
[
"# Print element on each index\n\nprint(\"Element 0 of Tuple: \", NestedT[0])\nprint(\"Element 1 of Tuple: \", NestedT[1])\nprint(\"Element 2 of Tuple: \", NestedT[2])\nprint(\"Element 3 of Tuple: \", NestedT[3])\nprint(\"Element 4 of Tuple: \", NestedT[4])",
"Element 0 of Tuple: 1\nElement 1 of Tuple: 2\nElement 2 of Tuple: ('pop', 'rock')\nElement 3 of Tuple: (3, 4)\nElement 4 of Tuple: ('disco', (1, 2))\n"
]
],
[
[
"We can use the second index to access other tuples as demonstrated in the figure:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesNestTwo.png\" width=\"750\" align=\"center\">",
"_____no_output_____"
],
[
" We can access the nested tuples :",
"_____no_output_____"
]
],
[
[
"# Print element on each index, including nest indexes\n\nprint(\"Element 2, 0 of Tuple: \", NestedT[2][0])\nprint(\"Element 2, 1 of Tuple: \", NestedT[2][1])\nprint(\"Element 3, 0 of Tuple: \", NestedT[3][0])\nprint(\"Element 3, 1 of Tuple: \", NestedT[3][1])\nprint(\"Element 4, 0 of Tuple: \", NestedT[4][0])\nprint(\"Element 4, 1 of Tuple: \", NestedT[4][1])",
"Element 2, 0 of Tuple: pop\nElement 2, 1 of Tuple: rock\nElement 3, 0 of Tuple: 3\nElement 3, 1 of Tuple: 4\nElement 4, 0 of Tuple: disco\nElement 4, 1 of Tuple: (1, 2)\n"
]
],
[
[
"We can access strings in the second nested tuples using a third index:",
"_____no_output_____"
]
],
[
[
"# Print the first element in the second nested tuples\n\nNestedT[2][1][0]",
"_____no_output_____"
],
[
"# Print the second element in the second nested tuples\n\nNestedT[2][1][1]",
"_____no_output_____"
]
],
[
[
" We can use a tree to visualise the process. Each new index corresponds to a deeper level in the tree:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesNestThree.gif\" width=\"750\" align=\"center\">",
"_____no_output_____"
],
[
"Similarly, we can access elements nested deeper in the tree with a fourth index:",
"_____no_output_____"
]
],
[
[
"# Print the first element in the second nested tuples\n\nNestedT[4][1][0]",
"_____no_output_____"
],
[
"# Print the second element in the second nested tuples\n\nNestedT[4][1][1]",
"_____no_output_____"
]
],
[
[
"The following figure shows the relationship of the tree and the element <code>NestedT[4][1][1]</code>:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesNestFour.gif\" width=\"750\" align=\"center\">",
"_____no_output_____"
],
[
"<h2 id=\"quiz\">Quiz on Tuples</h2>",
"_____no_output_____"
],
[
"Consider the following tuple:",
"_____no_output_____"
]
],
[
[
"# sample tuple\n\ngenres_tuple = (\"pop\", \"rock\", \"soul\", \"hard rock\", \"soft rock\", \\\n \"R&B\", \"progressive rock\", \"disco\") \ngenres_tuple",
"_____no_output_____"
]
],
[
[
"Find the length of the tuple, <code>genres_tuple</code>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\nlen(genres_tuple)",
"_____no_output_____"
]
],
[
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/TuplesQuiz.png\" width=\"1100\" align=\"center\">",
"_____no_output_____"
],
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nlen(genres_tuple)\n-->",
"_____no_output_____"
],
[
"Access the element, with respect to index 3: ",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\ngenres_tuple[3]",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\ngenres_tuple[3]\n-->",
"_____no_output_____"
],
[
"Use slicing to obtain indexes 3, 4 and 5:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\ngenres_tuple[3:6]",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\ngenres_tuple[3:6]\n-->",
"_____no_output_____"
],
[
"Find the first two elements of the tuple <code>genres_tuple</code>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\ngenres_tuple[0:2]",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\ngenres_tuple[0:2]\n-->",
"_____no_output_____"
],
[
"Find the first index of <code>\"disco\"</code>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\ngenres_tuple.index(\"disco\")\ngenres_tuple[7][0]",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\ngenres_tuple.index(\"disco\")\n-->",
"_____no_output_____"
],
[
"Generate a sorted List from the Tuple <code>C_tuple=(-5, 1, -3)</code>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nC_tuple=(-5, 1, -3)\nc_tuple=sorted(C_tuple)\nprint(C_tuple)\nprint(c_tuple)",
"(-5, 1, -3)\n[-5, -3, 1]\n"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nC_tuple = (-5, 1, -3)\nC_list = sorted(C_tuple)\nC_list\n-->",
"_____no_output_____"
],
[
"<hr>\n<h2>The last exercise!</h2>\n<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href=\"https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/\" target=\"_blank\">this article</a> to learn how to share your work.\n<hr>",
"_____no_output_____"
],
[
"<h3>About the Authors:</h3> \n<p><a href=\"https://www.linkedin.com/in/joseph-s-50398b136/\" target=\"_blank\">Joseph Santarcangelo</a> is a Data Scientist at IBM, and holds a PhD in Electrical Engineering. His research focused on using Machine Learning, Signal Processing, and Computer Vision to determine how videos impact human cognition. Joseph has been working for IBM since he completed his PhD.</p>",
"_____no_output_____"
],
[
"Other contributors: <a href=\"www.linkedin.com/in/jiahui-mavis-zhou-a4537814a\">Mavis Zhou</a>",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<p>Copyright © 2018 IBM Developer Skills Network. This notebook and its source code are released under the terms of the <a href=\"https://cognitiveclass.ai/mit-license/\">MIT License</a>.</p>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ec88e8f56bea591f4ecc2e0f0116af165510d5c1 | 800 | ipynb | Jupyter Notebook | content/lessons/01/Watch-Me-Code/WMC-walkthrough.ipynb | MahopacHS/spring2019-rizzenM | 2860b3338c8f452e45aac04f04388a417b2cf506 | [
"MIT"
] | 1 | 2019-04-03T00:43:15.000Z | 2019-04-03T00:43:15.000Z | content/lessons/01/Watch-Me-Code/WMC-walkthrough.ipynb | MahopacHS/spring2019-rizzenM | 2860b3338c8f452e45aac04f04388a417b2cf506 | [
"MIT"
] | null | null | null | content/lessons/01/Watch-Me-Code/WMC-walkthrough.ipynb | MahopacHS/spring2019-rizzenM | 2860b3338c8f452e45aac04f04388a417b2cf506 | [
"MIT"
] | 5 | 2018-09-17T03:54:06.000Z | 2019-10-17T02:47:20.000Z | 17.777778 | 62 | 0.53 | [
[
[
"# Watch Me Code: Walkthrough\n\nWatch me codes are the code examples from lecture days. ",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ec88ead85dea3bfd7746ea0813e27e56636c73e0 | 724,174 | ipynb | Jupyter Notebook | my_working_set.ipynb | dtn067/cms-git-set | df7a810225e717a20324556ab48e566f8a59be63 | [
"MIT"
] | null | null | null | my_working_set.ipynb | dtn067/cms-git-set | df7a810225e717a20324556ab48e566f8a59be63 | [
"MIT"
] | null | null | null | my_working_set.ipynb | dtn067/cms-git-set | df7a810225e717a20324556ab48e566f8a59be63 | [
"MIT"
] | 2 | 2019-04-08T16:27:15.000Z | 2019-04-08T16:29:47.000Z | 96.556533 | 145,023 | 0.745441 | [
[
[
"from __future__ import print_function\nimport datetime\nfrom functools import reduce\nimport os\n\nimport pandas as pd\nimport numpy as np\n%matplotlib nbagg\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"#import psutil\n#from multiprocessing import cpu_count\n\n#p = psutil.Process(os.getpid())\n#p.set_cpu_affinity(range(cpu_count()))\n#print p.get_cpu_affinity()\nos.system(\"taskset -p 0xff %d\" % os.getpid())",
"_____no_output_____"
],
[
"startTime = datetime.datetime.now()",
"_____no_output_____"
],
[
"# Data collected from a spark query at CERN, in pandas pickle format\n# CRAB jobs only have data after Oct. 2017\nws = pd.read_pickle(\"data/working_set_day.pkl.gz\")\n# spark returns lists, we want to use sets\nws['working_set_blocks'] = ws.apply(lambda x: set(x.working_set_blocks), 'columns')\nws['working_set'] = ws.apply(lambda x: set(x.working_set), 'columns')",
"_____no_output_____"
],
[
"# DBS BLOCKS table schema:\n# BLOCK_ID NOT NULL NUMBER(38)\n# BLOCK_NAME NOT NULL VARCHAR2(500)\n# DATASET_ID NOT NULL NUMBER(38)\n# OPEN_FOR_WRITING NOT NULL NUMBER(38)\n# ORIGIN_SITE_NAME NOT NULL VARCHAR2(100)\n# BLOCK_SIZE NUMBER(38)\n# FILE_COUNT NUMBER(38)\n# CREATION_DATE NUMBER(38)\n# CREATE_BY VARCHAR2(500)\n# LAST_MODIFICATION_DATE NUMBER(38)\n# LAST_MODIFIED_BY VARCHAR2(500)\nif not os.path.exists('data/block_size.npy'):\n blocksize = pd.read_csv(\"data/dbs_blocks.csv.gz\", dtype='i8', usecols=(0,5), names=['block_id', 'block_size'])\n np.save('data/block_size.npy', blocksize.values)\n blocksize = blocksize.values\nelse:\n blocksize = np.load('data/block_size.npy')\n\n# We'll be accessing randomly, make a dictionary\nblocksize = {v[0]:v[1] for v in blocksize}",
"_____no_output_____"
],
[
"# join the data tier definitions\ndatatiers = pd.read_csv('data/dbs_datatiers.csv').set_index('id')\nws['data_tier'] = datatiers.loc[ws.d_data_tier_id].data_tier.values",
"_____no_output_____"
],
[
"date_index = np.arange(np.min(ws.day.values//86400), np.max(ws.day.values//86400)+1)\ndate_index_ts = np.array(list(datetime.date.fromtimestamp(day*86400) for day in date_index))",
"_____no_output_____"
],
[
"ws_filtered = ws[(ws.crab_job==True) & (ws.data_tier.str.contains('MINIAOD'))]\n\nblocks_day = []\nfor i, day in enumerate(date_index):\n today = (ws_filtered.day==day*86400)\n blocks_day.append(reduce(lambda a,b: a.union(b), ws_filtered[today].working_set_blocks, set()))\n\nprint(\"Done assembling blocklists\")\n\nnrecords = np.zeros_like(date_index)\nlifetimes = {\n '1w': 7,\n '1m': 30,\n '3m': 90,\n '6m': 120,\n}\nws_size = {k: np.zeros_like(date_index) for k in lifetimes}\nnrecalls = {k: np.zeros_like(date_index) for k in lifetimes}\nrecall_size = {k: np.zeros_like(date_index) for k in lifetimes}\nprevious = {k: set() for k in lifetimes}\n\nfor i, day in enumerate(date_index):\n nrecords[i] = ws_filtered[(ws_filtered.day==day*86400)].size\n for key in lifetimes:\n current = reduce(lambda a,b: a.union(b), blocks_day[max(0,i-lifetimes[key]):i+1], set())\n recall = current - previous[key]\n nrecalls[key][i] = len(recall)\n ws_size[key][i] = sum(blocksize[bid] for bid in current)\n recall_size[key][i] = sum(blocksize[bid] for bid in recall)\n previous[key] = current\n if i%30==0:\n print(\"Day \", i)\n\nprint(\"Done\")",
"Done assembling blocklists\nDay 0\nDay 30\nDay 60\nDay 90\nDay 120\nDay 150\nDay 180\nDay 210\nDay 240\nDay 270\nDay 300\nDay 330\nDay 360\nDay 390\nDay 420\nDay 450\nDay 480\nDay 510\nDay 540\nDay 570\nDay 600\nDay 630\nDay 660\nDay 690\nDay 720\nDay 750\nDone\n"
],
[
"print(list(blocksize.keys())[0:10])\nprint(list(blocksize.values())[0:10])",
"[16777216, 16777217, 16777218, 16777219, 16777220, 16777221, 16777222, 16777223, 16777224, 16777225]\n[20329608129, 16116886675, 12668970442, 12449380625, 1270370651, 2724755002, 18445001, 18137333201, 16618896308, 20835210196]\n"
],
[
"fig, ax = plt.subplots(1,1)\nax.plot(date_index_ts, recall_size['1w']/1e15, label='1 week')\nax.plot(date_index_ts, recall_size['1m']/1e15, label='1 month')\nax.plot(date_index_ts, recall_size['3m']/1e15, label='3 months')\nax.legend(title='Block lifetime')\nax.set_title('Simulated block recalls for CRAB users')\nax.set_ylabel('Recall rate [PB/day]')\nax.set_xlabel('Date')\nax.set_ylim(0, None)\nax.set_xlim(datetime.date(2017,10,1), None)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,1)\nax.plot(date_index_ts, ws_size['1w']/1e15, label='1 week')\nax.plot(date_index_ts, ws_size['1m']/1e15, label='1 month')\nax.plot(date_index_ts, ws_size['3m']/1e15, label='3 months')\nax.legend(title='Block lifetime')\nax.set_title('Working set for CRAB users, MINIAOD*')\nax.set_ylabel('Working set size [PB]')\nax.set_xlabel('Date')\nax.set_ylim(0, None)\nax.set_xlim(datetime.date(2017,10,1), None)",
"_____no_output_____"
],
[
"recall_size['3m'].mean()/1e12",
"_____no_output_____"
],
[
"print(ws_filtered)",
" day d_data_tier_id crab_job input_campaign \\\n29 1505779200 31223 True Run2017C \n36 1506556800 31224 True PhaseIFall16MiniAOD \n41 1508025600 31223 True Run2016G \n43 1508630400 31223 True Run2016B \n51 1510099200 31223 True Run2016D \n53 1511136000 31223 True Run2017B \n59 1512777600 31223 True Run2017C \n64 1513900800 31224 True RunIISpring16MiniAODv2 \n65 1514505600 31223 True Run2017E \n69 1514937600 31224 True RunIISummer17MiniAOD \n75 1515801600 31223 True Run2017F \n78 1516060800 31224 True RunIISummer16MiniAODv2 \n80 1516665600 31224 True RunIISpring16MiniAODv2 \n88 1518307200 31223 True Run2016H \n92 1519257600 31223 True Run2017C \n96 1519603200 31223 True Run2016H \n104 1520553600 31223 True Run2016D \n124 1524614400 31223 True Run2016F \n139 1527811200 31223 True Run2016G \n150 1529539200 31223 True Run2015D \n151 1529539200 31224 True PhaseIITDRFall17MiniAOD \n152 1529798400 31223 True Run2016C \n154 1530057600 31224 True RunIISummer16MiniAODv2 \n163 1530662400 31223 True Run2016B \n166 1531699200 31223 True Run2018A \n167 1532044800 31223 True Run2017B \n168 1532131200 31223 True Run2016G \n170 1532304000 31224 True RunIISummer16MiniAODv2 \n180 1534291200 31223 True Run2018C \n187 1534809600 31223 True Run2017B \n... ... ... ... ... \n56077 1526515200 31223 True Run2016E \n56087 1528934400 31223 True Run2017F \n56091 1529366400 31223 True Run2016E \n56092 1529625600 31223 True Run2017B \n56093 1529712000 31224 True PhaseIITDRFall17MiniAOD \n56100 1531008000 31223 True Run2017D \n56108 1532131200 31223 True Run2016D \n56111 1532563200 31223 True Run2017D \n56112 1532822400 31223 True Run2018A \n56119 1533427200 31223 True Run2016E \n56123 1533859200 31223 True Run2017C \n56128 1534809600 31224 True RunIIFall17MiniAOD \n56133 1536192000 31223 True Run2017D \n56134 1536192000 31223 True Run2018D \n56135 1536364800 31223 True Run2017F \n56136 1536537600 31223 True Run2017B \n56138 1536883200 31223 True Run2016E \n56148 1538524800 31223 True Run2016H \n56153 1539302400 31223 True Run2017E \n56163 1540857600 31224 True RunIIFall18MiniAOD \n56165 1541030400 31223 True Run2017D \n56167 1541808000 31224 True RunIIFall17MiniAODv2 \n56168 1542412800 31223 True Run2016F \n56169 1542931200 31223 True Run2016F \n56188 1544659200 31223 True Run2018B \n56191 1545091200 31223 True Run2018B \n56198 1546560000 31223 True Run2017C \n56200 1546992000 31223 True Run2016D \n56203 1547769600 31224 True Spring14miniaod \n56205 1547942400 31223 True Run2017B \n\n working_set_blocks \\\n29 {17821696, 17788929, 17735690, 17795096, 17850... \n36 {17459591, 17460747, 17459596, 17459597, 17459... \n41 {18001926, 18046986, 17258508, 17252365, 17899... \n43 {17836038, 18030599, 17838091, 17836046, 17264... \n51 {17256962, 17249797, 17253894, 17253897, 18029... \n53 {17670659, 17655300, 18125316, 17679875, 18123... \n59 {18074625, 17863681, 17742342, 17734153, 17735... \n64 {15582576, 15578996, 15573181} \n65 {18071057, 18108440, 18102812, 18060317, 18108... \n69 {17984444, 17979358, 17984448, 17951584, 17984... \n75 {18124807, 18329609, 18141194, 18124820, 18302... \n78 {17072137, 17072141, 17956879, 16875546, 16875... \n80 {16010763, 16005133, 16008215, 16014629, 15873... \n88 {17281026, 17256464, 17254426, 17256485, 17260... \n92 {18282496, 18397186, 18282503, 18247691, 18290... \n96 {17829889, 17829380, 17249286, 17248776, 17261... \n104 {18002945, 18002949, 17253894, 17249797, 17253... \n124 {16664065, 17255941, 16664585, 16664075, 17251... \n139 {16654340, 16721927, 16717831, 16621579, 17258... \n150 {14839968, 14840100, 14840101, 14840136, 14839... \n151 {18196480, 18196487, 18225166, 18180118, 18217... \n152 {16635917, 17250324, 17260565, 17254421, 18026... \n154 {17006592, 17481728, 17006595, 17006596, 17680... \n163 {17266692, 17264655, 17266705, 17250327, 17260... \n166 {18799618, 18718723, 18785284, 18805764, 18789... \n167 {18280448, 18268161, 18272259, 18259978, 18651... \n168 {17250824, 17251850, 17251857, 17249300, 17248... \n170 {17073553, 17063942, 17063944, 17063945, 17063... \n180 {19081740, 19032589, 19103760, 19037203, 19078... \n187 {18227713, 18651146, 18238477, 18237458, 18237... \n... ... \n56077 {18011136, 17256965, 17248774, 17984518, 17248... \n56087 {18632711, 18302998, 18630685, 18630686, 18630... \n56091 {17256965, 17248774, 17248773, 17251339, 17250... \n56092 {18312196, 18318341, 18317319, 18327564, 18318... \n56093 {18224640, 18193412, 18266118, 18198034, 18176... \n56100 {18324997, 18286086, 18342918, 18304520, 18633... \n56108 {17250323, 17250326, 17252892, 17254940, 17250... \n56111 {18304513, 18324997, 18342918, 18297351, 18286... \n56112 {18842448, 18850593, 18847995, 18853585, 18853... \n56119 {17248774, 17250828, 17970189, 17248269, 17248... \n56123 {18629760, 18636289, 18634626, 18632835, 18627... \n56128 {18317323, 18309132, 18374673, 18366483, 18341... \n56133 {18324997, 18342918, 18297351, 18633225, 18299... \n56134 {19165700, 19231238, 19170826, 19226130, 19213... \n56135 {18632193, 18632194, 18329092, 18323978, 18629... \n56136 {18275840, 18275841, 18280448, 18272259, 18280... \n56138 {17255425, 17248773, 17248774, 17262089, 17250... \n56148 {17829380, 17249286, 17248776, 17248777, 17249... \n56153 {18259968, 18276353, 18319362, 18259971, 18298... \n56163 {19440964, 19456421, 19440965, 19432583, 19356... \n56165 {18324997, 18342918, 18297351, 18633225, 18299... \n56167 {19259393, 18677767, 19243018, 19251216, 18677... \n56168 {19102209, 17903107, 17257988, 17957893, 17946... \n56169 {17826817, 17266690, 17258499, 17257988, 17255... \n56188 {18980879, 18941970, 18855965, 18907172, 19437... \n56191 {19499784, 19512206, 18898447, 18892304, 19502... \n56198 {18629760, 18636289, 18634626, 18632835, 18627... \n56200 {16758792, 17250323, 17963042, 17250342, 19077... \n56203 {12379168, 12379137, 12379213} \n56205 {18635908, 18642183, 18647816, 18651146, 18636... \n\n working_set sum_throughput \\\n29 {13433184, 13414272, 13428515, 13428612, 13433... 5.907146e+05 \n36 {13375987, 13384516, 13384517} 1.519105e+00 \n41 {13333474, 13333442, 13444164, 13444197, 13334... 7.141626e+05 \n43 {13333764, 13453981, 13268166, 13434055, 13335... 2.062321e+05 \n51 {13333477, 13333479, 13338280, 13445225, 13441... 3.024643e+05 \n53 {13404738, 13411331, 13455240, 13404528, 13411... 9.594011e+04 \n59 {13428515, 13433316, 13428549, 13428612, 13433... 1.494795e+04 \n64 {12911666} 9.514780e+00 \n65 {13454773, 13454749} 5.483359e+04 \n69 {13441587, 13444932} 1.761178e+04 \n75 {13470851, 13470663, 13470699, 13470828, 13470... 5.623874e+05 \n78 {13297665, 13358594, 13409795, 13306886, 13301... 1.078694e+06 \n80 {13381242, 12984923, 13004818, 12986117} 1.615160e+02 \n88 {13333443, 13335491, 13333510, 13333383, 13333... 7.441337e+05 \n92 {13481472, 13481154, 13495718, 13481095, 13480... 1.536124e+05 \n96 {13333510, 13334599, 13333415, 13333383, 13333... 3.715777e+04 \n104 {13333426, 13333460, 13333741, 13443790, 13333... 3.697534e+04 \n124 {13333640, 13333617, 13264891, 13333380, 13334... 1.379823e+05 \n139 {13264768, 13333474, 13334020, 13333348, 13263... 1.488345e+05 \n150 {12554332} 1.191977e+04 \n151 {13475845, 13484678, 13476232, 13476489, 13475... 7.731586e+01 \n152 {13334565, 13334696, 13263722, 13445707, 13334... 1.990327e+05 \n154 {13292032, 13297665, 13405188, 13301254, 13295... 4.335670e+05 \n163 {13432728, 13267399, 13335979, 13333764, 13333... 4.222341e+05 \n166 {13554434, 13673731, 13600413, 13554439, 13605... 8.100208e+05 \n167 {13541633, 13485162, 13541612, 13481465, 13541... 8.420172e+05 \n168 {13333348, 13333535} 2.031260e+04 \n170 {13303299, 13295620, 13312519, 13315595, 13297... 3.277416e+05 \n180 {13690179, 13686887, 13686536, 13686889, 13686... 5.533044e+04 \n187 {13484989, 13479291, 13541612, 13541900, 13543... 3.683596e+04 \n... ... ... \n56077 {13334601, 13445389, 13394000, 13333332, 13333... 3.350703e+05 \n56087 {13485215, 13484481, 13487202, 13470851, 13541... 2.316855e+05 \n56091 {13337934, 13333614, 13333332, 13437941, 13333... 4.816278e+05 \n56092 {13541600, 13541900, 13481234, 13541627, 13484... 6.268421e+03 \n56093 {13475845, 13484678, 13476232, 13476489, 13478... 6.170478e+00 \n56100 {13481752, 13485040, 13542526, 13481839} 4.121466e+04 \n56108 {13333426, 13333479} 1.325424e+04 \n56111 {13543587, 13541638, 13481839, 13481752, 13541... 6.074144e+04 \n56112 {13673753} 5.182276e+01 \n56119 {13444045, 13690254, 13437905, 13333332, 13690... 8.720875e+04 \n56123 {13541568, 13541664, 13543107, 13543301, 13542... 1.189269e+06 \n56128 {13491202, 13504517, 13505032, 13502474, 13490... 2.063884e+06 \n56133 {13541624, 13543587, 13541638, 13485012, 13543... 7.072433e+05 \n56134 {13694256, 13694359, 13694399} 6.097633e+05 \n56135 {13543088, 13541650, 13541701, 13542750, 13485... 8.669431e+04 \n56136 {13481465, 13541900, 13484861, 13543423} 1.938581e+04 \n56138 {13690397, 13433605, 13334601, 13337934, 13334... 2.405789e+05 \n56148 {13333510, 13333734, 13333383, 13333358, 13690... 2.819607e+05 \n56153 {13481988, 13485285, 13481126, 13543561, 13542... 6.435625e+05 \n56163 {13713238} 2.821496e+03 \n56165 {13541730, 13543587, 13542629, 13541638, 13541... 4.766528e+05 \n56167 {13605894, 13711372, 13671437, 13675022, 13548... 2.938925e+05 \n56168 {13432611, 13333380, 13439622, 13690855, 13333... 8.277549e+04 \n56169 {13432611, 13333380, 13690474, 13334576, 13333... 1.108705e+05 \n56188 {13720832, 13674081, 13720424, 13722730, 13684... 3.627144e+05 \n56191 {13678020, 13678087, 13722730, 13678027, 13725... 1.586768e+03 \n56198 {13541568, 13541664, 13543107, 13543095, 13542... 1.381626e+05 \n56200 {13690498, 13689827, 13443558, 13333479, 13338... 4.012125e+05 \n56203 {9543033} NaN \n56205 {13541547, 13541612, 13541900, 13541649, 13543... 1.781885e+04 \n\n sum_walltime njobs data_tier \n29 5.587856e+06 6877 MINIAOD \n36 7.820499e+04 310 MINIAODSIM \n41 3.321386e+07 21808 MINIAOD \n43 4.769794e+07 10422 MINIAOD \n51 5.938120e+07 19222 MINIAOD \n53 5.617182e+06 3078 MINIAOD \n59 1.605389e+07 3470 MINIAOD \n64 1.129383e+05 5 MINIAODSIM \n65 7.179609e+06 3380 MINIAOD \n69 3.151977e+06 1212 MINIAODSIM \n75 2.742331e+07 8361 MINIAOD \n78 1.203028e+08 52845 MINIAODSIM \n80 2.644691e+05 46 MINIAODSIM \n88 1.194136e+08 33981 MINIAOD \n92 6.104821e+07 13756 MINIAOD \n96 7.080728e+06 2699 MINIAOD \n104 1.718378e+07 3295 MINIAOD \n124 8.651919e+06 5529 MINIAOD \n139 1.964368e+07 10514 MINIAOD \n150 4.846800e+04 286 MINIAOD \n151 1.143785e+06 703 MINIAODSIM \n152 1.090482e+07 6945 MINIAOD \n154 2.905792e+08 53409 MINIAODSIM \n163 6.351877e+07 16783 MINIAOD \n166 1.622244e+07 14482 MINIAOD \n167 4.504520e+07 17198 MINIAOD \n168 9.470104e+06 1189 MINIAOD \n170 4.080411e+07 17555 MINIAODSIM \n180 7.356446e+06 4664 MINIAOD \n187 3.494917e+07 7770 MINIAOD \n... ... ... ... \n56077 5.796609e+07 12900 MINIAOD \n56087 3.836791e+07 13785 MINIAOD \n56091 4.547205e+07 10456 MINIAOD \n56092 4.850918e+06 1087 MINIAOD \n56093 4.014404e+05 174 MINIAODSIM \n56100 2.245850e+06 1842 MINIAOD \n56108 6.046667e+06 770 MINIAOD \n56111 1.665589e+07 2924 MINIAOD \n56112 7.164783e+05 30 MINIAOD \n56119 3.167241e+07 7814 MINIAOD \n56123 4.831500e+06 13455 MINIAOD \n56128 2.364470e+08 52805 MINIAODSIM \n56133 2.287123e+07 10503 MINIAOD \n56134 1.080126e+07 8310 MINIAOD \n56135 2.416709e+07 7359 MINIAOD \n56136 1.827936e+06 1005 MINIAOD \n56138 6.061037e+07 10344 MINIAOD \n56148 2.201857e+07 7860 MINIAOD \n56153 2.977756e+07 13779 MINIAOD \n56163 2.194266e+05 299 MINIAODSIM \n56165 1.556698e+07 5757 MINIAOD \n56167 2.415294e+08 38916 MINIAODSIM \n56168 2.843550e+07 3793 MINIAOD \n56169 2.686420e+07 5160 MINIAOD \n56188 2.942816e+07 7111 MINIAOD \n56191 1.897340e+06 197 MINIAOD \n56198 4.489844e+07 8055 MINIAOD \n56200 3.475360e+07 14922 MINIAOD \n56203 5.725263e+03 0 MINIAODSIM \n56205 1.360171e+07 3277 MINIAOD \n\n[10321 rows x 10 columns]\n"
],
[
"# block_dict is a dictionary that holds the lists of blocks\n# for all of the days for which the lists are nonzero\n# Keys: Day\n# Values: List of Blocks that were accessed on that Day\nblock_dict = {}\ni=0\nfor el in blocks_day:\n i=i+1\n if len(el)>0:\n block_dict[i] = el\n\nprint(\"Merging daily block lists into one block set\")\nblock_list = []\nfor i in range(len(blocks_day)):\n block_list += blocks_day[i]\n# block_set is a set of all unique blocks.\n# This can be used to isolate properties of individual blocks\n# (e.g. how many times a block is accessed)\nblock_set = set(block_list)\nprint(\"Block Set Created\")",
"Merging daily block lists into one block set\nBlock Set Created\n"
],
[
"# Rounds down the number to the multiple specified by the divisor\ndef round_down(num, divisor):\n return num - (num % divisor)\n\n# Counts the frequencies of the quantities of blocks reused in an\n# n day period (over all given time) where n = threshold\n# Parameters: threshold - integer that determines the range of days (counted back from the given day)\n# over which the blocks are counted\n# bucket - integer that determines \"tolerance\" for quantity of blocks such that\n# the blocks are counted together (e.g. a bucket of 1000 means that a\n# quantity of blocks less than 2000 would be counted as part of the\n# 1000 bucket because it is being rounded down)\ndef countBlockReuseFreq(threshold, bucket):\n block_reuse_dict = {}\n for day in block_dict:\n try:\n b = block_dict[day]\n i = day\n for i in range(day+1, day+threshold+1):\n b = b.union(block_dict[i])\n # Rounds down to the bin and checks to see if such a bin exists in the dictionary\n if round_down(len(b), bucket) in block_reuse_dict:\n block_reuse_dict[round_down(len(b), bucket)] += 1\n else:\n block_reuse_dict[round_down(len(b), bucket)] = 1\n except Exception as e:\n print(\"Empty Key: \", e)\n \n # Returns block_reuse_dict\n # Keys: Number of Blocks reused within the given threshold\n # (organized by the given buckets)\n # Values: Frequencies of occurrence\n return block_reuse_dict\n\n# Counts the frequencies of the total number of bytes reused in an\n# n day period (over all given time) where n = threshold\n# Parameters: threshold - integer that determines the range of days (counted back from the given day)\n# over which the blocks are counted\n# bucket - integer that determines \"tolerance\" for bytes such that\n# the bytes are counted together (e.g. a bucket of 1000 means that a\n# size of less than 2000 bytes would be counted as part of the\n# 1000 byte bucket because it is being rounded down)\ndef countByteReuseFreq(threshold, bucket):\n byte_reuse_dict = {}\n for day in block_dict:\n try:\n b = block_dict[day]\n i = day\n for i in range(day+1, day+threshold+1):\n b = b.union(block_dict[i])\n # Counting the number of bytes in b\n bSize = 0\n for block in b:\n bSize += blocksize[block]\n # Rounds down to the bin and checks to see if such a bin exists in the dictionary\n if round_down(bSize, bucket) in byte_reuse_dict:\n # Adding the number of bytes into the given block into the current bin\n byte_reuse_dict[round_down(bSize, bucket)] += 1\n else:\n byte_reuse_dict[round_down(bSize, bucket)] = 1\n except Exception as e:\n print(\"Empty Key: \", e)\n \n # Returns byte_reuse_dict\n # Keys: Bytes reused within the given threshold (organized by the given buckets)\n # Values: Frequencies of occurrence\n return byte_reuse_dict\n\n# Counts the frequencies of the total number of bytes reused in an\n# n day period (over all given time) where n = threshold\n# Returns them as a fraction: (Reused Bytes for a given day) / (Bytes accessed for a given day) \n# Parameters: threshold - integer that determines the range of days (counted back from the given day)\n# over which the blocks are counted\n# bucket - integer that determines \"tolerance\" for percentage of byte usage \n# such that the percentage of byte usage are counted together (e.g. \n# a bucket of 0.05 means that a size of less than 0.10 would be counted as\n# part of the 0.05 percentage bucket because it is being rounded down)\ndef countByteFracReuseFreq(threshold, bucket):\n byte_reuse_dict = {}\n for day in block_dict:\n try:\n b = block_dict[day]\n i = day\n tSize = 0\n # Populating tSize with the bytes accessed in the given day\n for block in block_dict[day]:\n tSize += blocksize[block]\n for i in range(day+1, day+threshold+1):\n b = b.union(block_dict[i])\n # Counting the number of bytes accessed in the days within the threshold\n for block in block_dict[i]:\n tSize += blocksize[block]\n # Counting the number of bytes in b\n bSize = 0\n for block in b:\n bSize += blocksize[block]\n percent = float(bSize)/float(tSize)\n # Rounds down to the bin and checks to see if such a bin exists in the dictionary\n if round_down(percent, bucket) in byte_reuse_dict:\n # Adding the number of bytes into the given block into the current bin\n byte_reuse_dict[round_down(percent, bucket)] += 1\n else:\n byte_reuse_dict[round_down(percent, bucket)] = 1\n except Exception as e:\n print(\"Empty Key: \", e)\n \n # Returns byte_reuse_dict\n # Keys: Bytes reused within the given threshold (organized by the given buckets)\n # Values: Frequencies of occurrence\n return byte_reuse_dict",
"_____no_output_____"
],
[
"print(list(countByteFracReuseFreq(1, 0.05).keys()))\nprint(list(countByteFracReuseFreq(1, 0.05).values()))\nprint(list(countByteFracReuseFreq(14, 0.05).keys()))\nprint(list(countByteFracReuseFreq(14, 0.05).values()))",
"Empty Key: 245\nEmpty Key: 762\n[0.75, 0.9, 0.8, 0.6000000000000001, 0.55, 0.8500000000000001, 0.7000000000000001, 0.65]\nEmpty Key: 245\nEmpty Key: 762\n[35, 2, 9, 111, 3, 3, 103, 247]\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\n[0.2, 0.15000000000000002, 0.25]\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 245\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\nEmpty Key: 762\n[196, 285, 12]\n"
],
[
"fig, ax = plt.subplots(1,1)\nbucket = 5000\nblock_reuse_dict1 = countBlockReuseFreq(1, bucket)\nax.bar(list(block_reuse_dict1.keys()), list(block_reuse_dict1.values()), width=bucket)\nax.set_title('Frequency Vs. Number of Blocks Reused, Threshold: 1 Day')\nax.legend(title='Bucket ' + str(bucket))\nax.set_ylabel('Frequency')\nax.set_xlabel('Number of Blocks Reused')\nax.set_ylim(0, None)\nax.set_xlim(0, None)\nplt.savefig(\"BlockReuseThreshold1.png\")",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,1)\nbucket = 5000\nblock_reuse_dict14 = countBlockReuseFreq(14, bucket)\nax.bar(list(block_reuse_dict14.keys()), list(block_reuse_dict14.values()), width=bucket)\nax.set_title('Frequency Vs. Number of Blocks Reused, Threshold: 14 Day')\nax.legend(title='Bucket ' + str(bucket))\nax.set_ylabel('Frequency')\nax.set_xlabel('Number of Blocks Reused')\nax.set_ylim(0, None)\nax.set_xlim(0, None)\nplt.savefig(\"BlockReuseThreshold14.png\")",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,1)\nbucket = 8*1e13\nblock_reuse_dict1 = countByteReuseFreq(1, bucket)\nax.bar(list(block_reuse_dict1.keys()), list(block_reuse_dict1.values()), width=bucket)\nax.set_title('Frequency Vs. Pedabytes Reused, Threshold: 1 Day')\nax.legend(title='Bucket ' + str(format(bucket, \"10.2E\")))\nax.set_ylabel('Frequency')\nax.set_xlabel('Pedabytes Reused')\nax.set_ylim(0, None)\nax.set_xlim(0, None)\nplt.savefig(\"ByteReuseThreshold1.png\")",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,1)\nbucket = 12*1e13\nblock_reuse_dict14 = countByteReuseFreq(14, bucket)\nax.bar(list(block_reuse_dict14.keys()), list(block_reuse_dict14.values()), width=bucket)\nax.set_title('Frequency Vs. Pedabytes Reused, Threshold: 14 Day')\nax.legend(title='Bucket ' + str(format(bucket, \"10.2E\")))\nax.set_ylabel('Frequency')\nax.set_xlabel('Pedabytes Reused')\nax.set_ylim(0, None)\nax.set_xlim(0, None)\nplt.savefig(\"ByteReuseThreshold14.png\")",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,1)\nbucket = 0.05\nblock_reuse_dict1 = countByteFracReuseFreq(1, bucket)\nax.bar(list(block_reuse_dict1.keys()), list(block_reuse_dict1.values()), width=bucket)\nax.set_title('Frequency Vs. Fraction of Bytes Reused, Threshold: 1 Day')\nax.legend(title='Bucket ' + str(bucket))\nax.set_ylabel('Frequency')\nax.set_xlabel('Fraction of Pedabytes Reused')\nax.set_ylim(0, None)\nax.set_xlim(0, None)\nplt.savefig(\"ByteFracReuseThreshold1.png\")",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,1)\nbucket = 0.05\nblock_reuse_dict14 = countByteFracReuseFreq(14, bucket)\nax.bar(list(block_reuse_dict14.keys()), list(block_reuse_dict14.values()), width=bucket)\nax.set_title('Frequency Vs. Fraction of Bytes Reused, Threshold: 14 Day')\nax.legend(title='Bucket ' + str(bucket))\nax.set_ylabel('Frequency')\nax.set_xlabel('Fraction of Pedabytes Reused')\nax.set_ylim(0, None)\nax.set_xlim(0, None)\nplt.savefig(\"ByteFracReuseThreshold14.png\")",
"_____no_output_____"
],
[
"endTime = datetime.datetime.now()",
"_____no_output_____"
],
[
"timeDifference = endTime-startTime\nprint(\"Total Runtime:\")\nprint(timeDifference)",
"Total Runtime:\n0:20:25.865028\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec88f00b4dbf18d331b1aae2f1187681d5d450a2 | 197,435 | ipynb | Jupyter Notebook | model/trading_model_with_comission.ipynb | BGTCapital/pairstrade-fyp-2019 | 032ccf3c6151f3c2179237a771abadb8d9c98424 | [
"MIT"
] | 1 | 2021-11-26T12:28:03.000Z | 2021-11-26T12:28:03.000Z | model/trading_model_with_comission.ipynb | BGTCapital/pairstrade-fyp-2019 | 032ccf3c6151f3c2179237a771abadb8d9c98424 | [
"MIT"
] | null | null | null | model/trading_model_with_comission.ipynb | BGTCapital/pairstrade-fyp-2019 | 032ccf3c6151f3c2179237a771abadb8d9c98424 | [
"MIT"
] | null | null | null | 109.686111 | 59,340 | 0.802487 | [
[
[
"import tensorflow as tf\nimport numpy as np\nimport pandas as pd\nimport math\nimport os\nfrom os.path import isfile, join, splitext\nimport random\nfrom datetime import datetime, timedelta\nimport time\nimport matplotlib.pyplot as plt\n%matplotlib inline\nplt.rcParams[\"patch.force_edgecolor\"] = True\n\nfrom process_raw_prices import *\n\n\n# Some more magic so that the notebook will reload external python modules;\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"# # make sure the jupyter notebook run in this working directory\n# %cd ~/fyp/code/model",
"_____no_output_____"
],
[
"tf.enable_eager_execution()",
"_____no_output_____"
],
[
"num_of_pair = 6670",
"_____no_output_____"
],
[
"# processed dataset folder path\ndataset_folder_path = '../../dataset/nyse-daily-transformed-1'\nos.makedirs(dataset_folder_path, exist_ok=True)\n\n# raw dataset files pattern\nraw_files_path_pattern = \"../../dataset/nyse-daily-trimmed-same-length/*.csv\"\n\ndf_columns = ['close1', 'close2', 'normalizedLogClose1', 'normalizedLogClose2', 'spread', 'alpha', 'beta']\nind = {'y_close': 0, 'x_close': 1, 'spread': 4}\n\n# compute dataset\nall_pairs_slices = [splitext(f)[0] for f in os.listdir(dataset_folder_path) if isfile(join(dataset_folder_path, f))]\nif len(all_pairs_slices) == 0:\n generate_pairs_data(raw_files_path_pattern, result_path=dataset_folder_path)\n all_pairs_slices = [splitext(f)[0] for f in os.listdir(dataset_folder_path) if isfile(join(dataset_folder_path, f))]\nprint(\"Total number of pair slices: %d\" % len(all_pairs_slices))\n\n# split for training and testing\nall_pairs = sorted(list(set(['-'.join(p.split('-')[0:2]) for p in all_pairs_slices])))[:num_of_pair]\n# all_pairs = [\"VMW-WUBA\"]\n# all_pairs = [\"TWTR-UIS\"]\nall_pairs_slices_train = []\nall_pairs_slices_valid = []\n# all_pairs_slices_test = []\nfor p in all_pairs:\n all_pairs_slices_train += [p+'-0']\n all_pairs_slices_valid += [p+'-1']\n# all_pairs_slices_test += [p+'-2']\n # note that we also have 3 that can use\nprint(\"Total number of pair slices for training: %d\" % len(all_pairs_slices_train))\nprint(\"Total number of pair slices for validation: %d\" % len(all_pairs_slices_valid))\n# print(\"Total number of pair slices for testing: %d\" % len(all_pairs_slices_test))\n\nstart_t = time.time()\nall_pairs_df = {}\nfor s in all_pairs_slices_train:\n all_pairs_df[s] = pd.read_csv(join(dataset_folder_path, s+\".csv\"))\nfor s in all_pairs_slices_valid:\n all_pairs_df[s] = pd.read_csv(join(dataset_folder_path, s+\".csv\"))\n# for s in all_pairs_slices_test:\n# all_pairs_df[s] = pd.read_csv(join(dataset_folder_path, s+\".csv\"))\nprint('time spent for loading df = {}s'.format(time.time()-start_t))",
"Total number of pair slices: 13340\nTotal number of pair slices for training: 6670\nTotal number of pair slices for validation: 6670\ntime spent for loading df = 50.1102294921875s\n"
],
[
"# update batch size\nbatch_size = 300\n\n# number of batch in training\nnum_of_epoch = 80\nnum_of_batch = 2*num_of_pair*num_of_epoch//batch_size\n\n# fixed number of time steps in one episode (not used)\ntrading_period = 200\n\n# # 1 is zscore\n# num_features = 1\n\n# 0 is no position. 1 is long the spread. 2 is short the spread.\na_num = position_num = 3\n\n# RNN hidden state dimension\nh_dim = 30\n\n# number of RNN layer\nnum_layers = 1\n\n# number of layer1 output\nlayer1_out_num = 20\n\n# learning rate\nlr = 5e-4\n\nreg = 0.00001\n\n# discount factor in reinforcement learning\ngamma = 1\n\n# random action probability lower is better...\nrand_action_prob = 0.0\n\nbatches_per_print = 10*5\n\n# dummy initial cash\ninitial_cash = 10000\n\n# checkpoint folder\ncheckpoint_dir = '../../model_checkpoint/'\nos.makedirs(checkpoint_dir, exist_ok=True)",
"_____no_output_____"
],
[
"# functions\n\n# glob_mode should be assigned to None if an epoch finished\nglob_mode = None\nsample_start_index = None\ncurr_pairs = None\ndef get_random_history(batch_size, mode):\n \"\"\"Sample some pairs and get the history of those pairs. The history should have\n three dimension. The first dimension is for time. The second dimension is indexed\n by features name. The third dimension is the index of training instance.\n \"\"\"\n global glob_mode\n global sample_start_index\n global curr_pairs\n \n if mode == 'train': # user intended to use training data\n # first batch of training data\n # shuffle the data first before sampling\n if glob_mode != 'train':\n glob_mode = 'train'\n random.shuffle(all_pairs_slices_train)\n sample_start_index = 0\n sample_end_index = sample_start_index+batch_size\n sample_pair_slices = all_pairs_slices_train[sample_start_index:sample_end_index]\n\n # end of one epoch\n if sample_end_index >= len(all_pairs_slices_train):\n glob_mode = None\n\n elif mode == 'valid': # user intended to use validation data\n # first batch of validation data\n # shuffle the data first before sampling\n if glob_mode != 'valid':\n glob_mode = 'valid'\n random.shuffle(all_pairs_slices_valid)\n sample_start_index = 0\n sample_end_index = sample_start_index+batch_size\n sample_pair_slices = all_pairs_slices_valid[sample_start_index:sample_end_index]\n\n # end of one epoch\n if sample_end_index >= len(all_pairs_slices_valid):\n glob_mode = None\n elif mode == 'test': # user intended to use validation data\n # first batch of test data\n # shuffle the data first before sampling\n if glob_mode != 'test':\n glob_mode = 'test'\n random.shuffle(all_pairs_slices_test)\n sample_start_index = 0\n sample_end_index = sample_start_index+batch_size\n sample_pair_slices = all_pairs_slices_test[sample_start_index:sample_end_index]\n\n # end of one epoch\n if sample_end_index >= len(all_pairs_slices_test):\n glob_mode = None\n else:\n raise Exception(\"mode should be in ['train', 'valid', 'test'].\")\n \n curr_pairs = sample_pair_slices\n \n # update index for next batch\n sample_start_index += batch_size\n \n # return to the environment. this should be no greater than batch_size\n actual_batch_size = len(sample_pair_slices)\n \n history = []\n for s in sample_pair_slices:\n# df = pd.read_csv(join(dataset_folder_path, s+\".csv\"))\n df = all_pairs_df[s]\n df_val = df[df_columns].values\n history.append(df_val)\n \n history = np.array(history)\n return np.transpose(history, (1, 2, 0)), actual_batch_size\n\ndef compute_input_history(history):\n \"\"\"Slicing history in its second dimension.\"\"\"\n # no slicing for now\n return history[:,2:5]\n\ndef sample_action(logits, random=False, batch_size=batch_size):\n if random:\n dist = tf.distributions.Categorical(logits=tf.zeros([batch_size, a_num]))\n else:\n dist = tf.distributions.Categorical(logits=logits)\n \n # 1-D Tensor where the i-th element correspond to a sample from\n # the i-th categorical distribution\n return dist.sample()\n\ndef long_portfolio_value(q, p):\n return q*p\n\ndef short_portfolio_value(q, p, init_p):\n return q*(3.0*init_p/2 - p)\n\ndef incur_commission(price, qty):\n return min(max(1, 0.005*qty), 0.01*price*qty)\n\n# def discount_rewards(r, all_actions):\n# \"\"\"\n# r is a numpy array in the shape of (n, batch_size).\n# all_actions is a numpy array in the same shape as r.\n \n# return the discounted and cumulative rewards\"\"\"\n \n# result = np.zeros_like(r, dtype=float)\n# n = r.shape[0]\n# sum_ = np.zeros_like(r[0], dtype=float)\n# pre_action = all_actions[n-1]\n# for i in range(n-1,-1,-1):\n# sum_ *= gamma\n \n# # when the previous action(position) not equal to the current one,\n# # set the previous sum of reward to be zero.\n# sum_ = sum_*(all_actions[i]==pre_action) + r[i]\n# result[i] = sum_\n \n# # update pre_action\n# pre_action = all_actions[i]\n \n# return result\n\ndef discount_rewards(r):\n \"\"\"\n r is a numpy array in the shape of (n, batch_size).\n \n return the discounted and cumulative rewards\"\"\"\n \n result = np.zeros_like(r, dtype=float)\n n = r.shape[0]\n sum_ = np.zeros_like(r[0], dtype=float)\n for i in range(n-1,-1,-1):\n sum_ *= gamma\n sum_ += r[i]\n result[i] = sum_\n \n return result\n\ndef loss(all_logits, all_actions, all_advantages):\n neg_log_select_prob = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=all_logits, labels=all_actions)\n \n # 0 axis is the time axis. 1 axis is the batch axis\n return tf.reduce_mean(neg_log_select_prob * all_advantages, 0)\n\ndef extract_pair_name(s):\n return '_'.join(s.split('-')[:2])\n\ndef extract_pair_index(s):\n return int(s.split('-')[-1])\n\ndef save_model():\n hkg_time = datetime.now() + timedelta(hours=16)\n checkpoint_name = hkg_time.strftime(\"%Y%m%d_%H%M%S\")\n checkpoint_prefix = os.path.join(checkpoint_dir, checkpoint_name)\n root.save(checkpoint_prefix)\n tf.train.latest_checkpoint(checkpoint_dir)\n \ndef restore_model(checkpoint_name):\n root.restore(join(checkpoint_dir, checkpoint_name))\n\n\nmyLeakyReLU = tf.keras.layers.LeakyReLU()\nmyLeakyReLU.__name__ = \"myLeakyReLU\"\n\n# classes\nclass TradingPolicyModel(tf.keras.Model):\n def __init__(self):\n super(TradingPolicyModel, self).__init__()\n self.dense1 = tf.layers.Dense(units=layer1_out_num,\n activation=myLeakyReLU,\n kernel_initializer=tf.contrib.layers.xavier_initializer(),\n# kernel_regularizer=tf.contrib.layers.l2_regularizer(reg)\n )\n self.dense2 = tf.layers.Dense(units=layer1_out_num,\n activation=myLeakyReLU,\n kernel_initializer=tf.contrib.layers.xavier_initializer(),\n# kernel_regularizer=tf.contrib.layers.l2_regularizer(reg)\n )\n# self.dense3 = tf.layers.Dense(units=layer1_out_num,\n# activation=myLeakyReLU,\n# kernel_initializer=tf.contrib.layers.xavier_initializer(),\n# kernel_regularizer=tf.contrib.layers.l2_regularizer(reg)\n# )\n# self.dense4 = tf.layers.Dense(units=layer1_out_num,\n# activation=tf.keras.layers.LeakyReLU(),\n# kernel_initializer=tf.contrib.layers.xavier_initializer()\n# )\n self.logits = tf.layers.Dense(units=a_num,\n activation=myLeakyReLU,\n kernel_initializer=tf.contrib.layers.xavier_initializer(),\n# kernel_regularizer=tf.contrib.layers.l2_regularizer(reg)\n )\n\n def call(self, inputs):\n # Forward pass\n inputs = self.dense1(inputs)\n inputs = self.dense2(inputs)\n# inputs = self.dense3(inputs)\n# inputs = self.dense4(inputs)\n logits = self.logits(inputs)\n return logits\n\n\nclass StateEncodingModel(tf.keras.Model):\n def __init__(self, batch_size=batch_size):\n super(StateEncodingModel, self).__init__()\n self.cell = tf.contrib.rnn.MultiRNNCell([tf.contrib.rnn.LSTMCell(h_dim) for i in range(num_layers)])\n self.reset_state(batch_size)\n \n def call(self, inputs):\n output, self.state = self.cell(inputs, self.state)\n return output\n \n def reset_state(self, batch_size=batch_size):\n self.state = self.cell.zero_state(batch_size, tf.float32)\n\n\nclass TradingEnvironment():\n \"\"\"Trading environment for reinforcement learning training.\n \n NOTE: Call reset first before calling step!\n \n Arguments:\n state_encoding_model: the model that encode past input_history data into a state\n vector which will be fed as input to the policy network.\n \"\"\"\n def __init__(self, state_encoding_model):\n # do some initialization\n self.state_encoding_model = state_encoding_model\n \n def _reset_env(self, mode, batch_size=batch_size):\n \n # prepare a batch of history and input_history\n # actual batch_size depends on the dataset\n self.history, curr_batch_size = get_random_history(batch_size, mode)\n batch_size = curr_batch_size\n self.input_history = compute_input_history(self.history)\n \n self.t = 0\n self.state_encoding_model.reset_state(batch_size)\n\n # 0 is no position. 1 is long the spread. 2 is short the spread\n self.position = np.zeros(batch_size, dtype=int)\n \n # initialize the cash each agent has\n self.cash = np.ones(batch_size)*initial_cash\n self.port_val = np.ones(batch_size)*initial_cash\n self.port_val_minus_com = np.ones(batch_size)*initial_cash\n \n # only useful when there is a postion on the spread\n self.quantity = {'x': np.zeros(batch_size), 'y': np.zeros(batch_size)}\n \n # for compute current portfolio value of the short side\n self.short_side_init_price = np.zeros(batch_size)\n \n # create or update self.state variable\n self.update_state()\n \n def reset(self, mode):\n \"\"\"Return an initial state for the trading environment\"\"\"\n \n # determine what dataset to use\n self._reset_env(mode)\n return self.state\n \n def compute_reward(self, action):\n \"\"\"Compute the reward at time t which is the change in total portfolio value\n from time t to t+1. It also update the position for time t+1. Exit trade when\n the short side portfolio value <= 0.\"\"\"\n \n r = np.zeros_like(action, dtype=float)\n cur_his = self.history[self.t]\n nex_his = self.history[self.t+1]\n \n # compute for each training instance in a batch\n for i, a in enumerate(action):\n y_p = cur_his[ind[\"y_close\"], i]\n x_p = cur_his[ind[\"x_close\"], i]\n nex_y_p = nex_his[ind[\"y_close\"], i]\n nex_x_p = nex_his[ind[\"x_close\"], i]\n \n if a == 0: # take no position on the spread at time t (current time step)\n if self.position[i] != 0:\n # need to exit at current time step\n self.cash[i] = self.port_val_minus_com[i]\n self.port_val[i] = self.port_val_minus_com[i]\n \n # compute reward (no change since no position on the spread)\n r[i] = 0\n \n # record the current situation\n self.position[i] = 0\n self.quantity['y'][i] = 0\n self.quantity['x'][i] = 0\n elif a == 1: # long the spread: long Y and short X\n if self.position[i] == 2:\n # need to exit at current time step\n self.cash[i] = self.port_val_minus_com[i]\n \n # quantity of each stock will change when the current position is not previous position\n if self.position[i] != 1:\n # compute quantity from cash\n self.quantity['y'][i] = int(2.0*self.cash[i]/3.0/y_p)\n self.quantity['x'][i] = int(2.0*self.cash[i]/3.0/x_p)\n self.short_side_init_price[i] = x_p\n \n # compute entering commission\n enter_commission = (incur_commission(y_p, self.quantity['y'][i])\n +incur_commission(x_p, self.quantity['x'][i]))\n \n # cash remaining after entering a position\n # initial cash - investment amount and commission\n self.cash[i] -= (0.5*self.quantity['x'][i]*x_p + self.quantity['y'][i]*y_p\n + enter_commission)\n \n lpv = long_portfolio_value(self.quantity['y'][i], y_p)\n spv = short_portfolio_value(self.quantity['x'][i], x_p, self.short_side_init_price[i])\n current_port_val = self.cash[i] + lpv + spv\n\n lpv_nex = long_portfolio_value(self.quantity['y'][i], nex_y_p)\n spv_nex = short_portfolio_value(self.quantity['x'][i], nex_x_p, self.short_side_init_price[i])\n \n # the zero here can be changed to other positive threshold ...\n if spv_nex <= 0:\n # we loss all the money in the short side\n # so need to exit the long side\n self.port_val_minus_com[i] = (\n self.cash[i] + lpv_nex - incur_commission(nex_y_p, self.quantity['y'][i])\n )\n \n # forced to take position 0. this mean all the assets transformed into cash\n self.position[i] = 0\n self.quantity['y'][i] = 0\n self.quantity['x'][i] = 0\n self.cash[i] = self.port_val_minus_com[i]\n self.port_val[i] = self.port_val_minus_com[i]\n else:\n exit_commission = (incur_commission(nex_y_p, self.quantity['y'][i])\n +incur_commission(nex_x_p, self.quantity['x'][i]))\n self.port_val[i] = self.cash[i] + lpv_nex + spv_nex\n self.port_val_minus_com[i] = self.cash[i] + lpv_nex + spv_nex - exit_commission\n self.position[i] = 1\n \n r[i] = self.port_val_minus_com[i] - current_port_val\n \n elif a == 2: # short the spread: short Y and long X\n if self.position[i] == 1:\n # need to exit at current time step\n self.cash[i] = self.port_val_minus_com[i]\n \n # quantity will change when the current position is not previous position\n if self.position[i] != 2:\n # compute quantity from cash\n self.quantity['y'][i] = int(2.0*self.cash[i]/3.0/y_p)\n self.quantity['x'][i] = int(2.0*self.cash[i]/3.0/x_p)\n self.short_side_init_price[i] = y_p\n \n # compute entering commission\n enter_commission = (incur_commission(y_p, self.quantity['y'][i])\n +incur_commission(x_p, self.quantity['x'][i]))\n \n # cash remaining after entering a position\n # initial cash - investment amount and commission\n self.cash[i] -= (self.quantity['x'][i]*x_p + 0.5*self.quantity['y'][i]*y_p\n + enter_commission)\n \n lpv = long_portfolio_value(self.quantity['x'][i], x_p)\n spv = short_portfolio_value(self.quantity['y'][i], y_p, self.short_side_init_price[i])\n current_port_val = self.cash[i] + lpv + spv\n\n lpv_nex = long_portfolio_value(self.quantity['x'][i], nex_x_p)\n spv_nex = short_portfolio_value(self.quantity['y'][i], nex_y_p, self.short_side_init_price[i])\n \n if spv_nex <= 0:\n # we loss all the money in the short side\n # so need to exit the long side\n self.port_val_minus_com[i] = (\n self.cash[i] + lpv_nex - incur_commission(nex_x_p, self.quantity['x'][i])\n )\n \n # forced to take position 0. this mean all the assets transformed into cash\n self.position[i] = 0\n self.quantity['y'][i] = 0\n self.quantity['x'][i] = 0\n self.cash[i] = self.port_val_minus_com[i]\n self.port_val[i] = self.port_val_minus_com[i]\n else:\n exit_commission = (incur_commission(nex_y_p, self.quantity['y'][i])\n +incur_commission(nex_x_p, self.quantity['x'][i]))\n self.port_val[i] = self.cash[i] + lpv_nex + spv_nex\n self.port_val_minus_com[i] = self.cash[i] + lpv_nex + spv_nex - exit_commission\n self.position[i] = 2\n \n r[i] = self.port_val_minus_com[i] - current_port_val\n\n return r\n \n def update_state(self):\n# # concate next_input_history and next position to form next partial state\n# partial_state = tf.concat([self.input_history[self.t].T, tf.one_hot(self.position, position_num)], 1)\n \n# # update state\n# self.state = self.state_encoding_model(partial_state)\n\n action_observation = tf.concat([\n tf.one_hot(self.position, position_num),\n tf.convert_to_tensor(self.input_history[self.t].T, dtype=tf.float32)\n ], 1)\n \n # use rnn to encode observationans and current stock state into next stock state\n stock_state = self.state_encoding_model(action_observation)\n \n# # do normalization for total_portfolio_value\n# # this is extremely important. if not normalized, the action will be highly biased.\n# portfolio_state = np.array([\n# self.total_portfolio_value/initial_cash,\n# # self.quantity['y'],\n# # self.quantity['x']\n# ]).T\n \n# # stock state and portfolio state together form the whole environment state\n# self.state = tf.concat([\n# stock_state,\n# tf.one_hot(self.position, position_num)\n# ], 1)\n self.state = stock_state\n \n def step(self, action):\n \"\"\"Given the current state and action, return the reward, next state and done.\n This function should be called after reset.\n \n reward is of type numpy array. state is of type tensor. done is of type boolean.\n \n \n Arguments:\n action: a numpy array containing the current action for each training pair.\n\n Note that we follow the convention where the trajectory is indexed as s_0, a_0, r_0,\n s_1, ... . Therefore t is updated just after computing the reward is computed and\n before computing next state.\n \"\"\"\n # r_t\n r = self.compute_reward(action) # also update the position for time t+1\n\n # t = t+1\n self.t += 1\n \n # compute s_(t+1)\n self.update_state()\n\n return r, self.state, (self.t+1) == trading_period",
"_____no_output_____"
],
[
"# create objects\npi = TradingPolicyModel()\nstate_encoding_model = StateEncodingModel()\nenv = TradingEnvironment(state_encoding_model)\noptimizer = tf.train.AdamOptimizer(learning_rate=lr)\n\n# create checkpoint object\nroot = tf.train.Checkpoint(pi=pi, state_encoding_model=state_encoding_model, optimizer=optimizer)",
"_____no_output_____"
],
[
"def run_batch_for_evaluate_performance(reward_list, mode):\n done = False\n s = env.reset(mode)\n# print('portfolio val:', env.port_val[0])\n\n # for accumalting episode statistics\n act_batch_size = tf.shape(s).numpy()[0]\n total_r = np.zeros(act_batch_size)\n\n # internally the episode length is fixed by trading_period\n while not done:\n logits = pi(s)\n a = sample_action(logits, batch_size=act_batch_size)\n\n # get immediate reward, update state, and get done\n r, s, done = env.step(a.numpy())\n \n\n# # for debugging\n# print('logits:', logits)\n# print('a:', a.numpy())\n# print('r:', r)\n# print('s:', s)\n# print('portfolio val:', env.port_val[0])\n\n total_r += r\n reward_list += total_r.tolist()\n return {extract_pair_name(curr_pairs[i]): total_r[i] for i in range(act_batch_size)}\n\n\ndef run_epoch_for_evaluate_performance(rs, total_r_dict, mode):\n counter = 0\n temp_dict = run_batch_for_evaluate_performance(rs, mode)\n total_r_dict.update(temp_dict)\n print('{}, '.format(counter), end='')\n counter += 1\n while glob_mode != None:\n temp_dict = run_batch_for_evaluate_performance(rs, mode)\n total_r_dict.update(temp_dict)\n print('{}, '.format(counter), end='')\n counter += 1\n print()\n\n\ndef plot_rs_dist(rs):\n print(len(rs))\n print(np.mean(rs))\n stat = plt.hist(rs, bins=30)\n plt.gcf().set_size_inches(10, 5)\n stat",
"_____no_output_____"
],
[
"# evaluate performance on train dataset\ntrain_rs = []\ntrain_total_r_dict = {}\nrun_epoch_for_evaluate_performance(train_rs, train_total_r_dict, 'train')\n\nplot_rs_dist(train_rs)",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, \n6670\n-511.15507467766105\n"
],
[
"# evaluate performance on valid dataset\nvalid_rs = []\nvalid_total_r_dict = {}\nrun_epoch_for_evaluate_performance(valid_rs, valid_total_r_dict, 'valid')\n\nplot_rs_dist(valid_rs)",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, \n6670\n-581.3031622188907\n"
],
[
"# pick best K pairs from train result and see how good those pairs are in valid\nK = 20\ntrain_total_r_ordered = sorted(list(train_total_r_dict.items()), key=lambda x: x[1], reverse=True)\ntrain_total_r_ordered_sliced = train_total_r_ordered[:K]\nvalid_total_r_sliced = [(x[0], valid_total_r_dict[x[0]]) for x in train_total_r_ordered_sliced]\nvalid_total_r_corresponding_value = [x[1] for x in valid_total_r_sliced]\ntrain_total_r_sliced_value = [x[1] for x in train_total_r_ordered_sliced]\n\n# see if the model overfits a lot by checking the performace in valid of best K pairs in train\nbins = np.linspace(-10000, 30000, 30)\nplt.hist(train_total_r_sliced_value, bins, alpha=0.3, label='train')\nplt.hist(valid_total_r_corresponding_value, bins, alpha=0.3, label='valid')\nplt.legend(loc='upper right')\nplt.gcf().set_size_inches(10, 5)",
"_____no_output_____"
],
[
"# global batch_id to keep track of the progress\nbatch_no = 0\ndef train(num_of_batch=num_of_batch):\n global batch_no\n global glob_mode\n # print parameters\n print('num_of_pair =', num_of_pair)\n print('batch_size =', batch_size)\n print('num_of_batch = {}, estimated epoch = {}'.format(num_of_batch, num_of_batch*batch_size/2/num_of_pair))\n print('rand_action_prob =', rand_action_prob)\n print('lr =', lr)\n glob_mode = None # reset the dataset\n\n # for training reference only\n average_total_r = 0.0\n epoch_average_total_r = 0.0\n num_eps_over = 0\n total_r_dict = {}\n\n start_time = time.time()\n for batch in range(num_of_batch):\n\n with tf.GradientTape() as gt:\n # saving for update\n all_logits = []\n all_actions = []\n all_rewards = []\n\n # episode starts here~\n done = False\n s = env.reset('train')\n\n # for accumalting episode statistics\n act_batch_size = tf.shape(s).numpy()[0]\n num_eps_over += act_batch_size\n total_r = np.zeros(act_batch_size)\n\n # internally the episode length is fixed by trading_period\n while not done:\n logits = pi(s)\n a = sample_action(logits, random=np.random.rand() <= rand_action_prob,\n batch_size=act_batch_size)\n r, s, done = env.step(a.numpy())\n\n # save the episode\n all_logits.append(logits)\n all_actions.append(a)\n all_rewards.append(r)\n\n r_sum = np.sum(r)\n average_total_r += r_sum\n epoch_average_total_r += r_sum\n total_r += r\n\n # # debugging\n # print(env.t)\n # print(env.t+1==200)\n # print(r[0])\n # print('a:', a.numpy())\n # print(done)\n # print(logits)\n\n # keep track of the pair performance (of course this is not totally fair for all pairs\n # as there are parameters update).\n total_r_dict.update({curr_pairs[i]: total_r[i] for i in range(act_batch_size)})\n\n all_logits_stack = tf.stack(all_logits)\n all_actions_stack = tf.stack(all_actions)\n all_rewards_stack = np.array(all_rewards)\n\n # compute cummulative rewards for each action\n all_cum_rewards = discount_rewards(all_rewards_stack)\n all_cum_rewards -= np.mean(all_cum_rewards)\n # all_cum_rewards /= np.std(all_cum_rewards)\n # all_cum_rewards /= np.mean(np.abs(all_cum_rewards))\n all_cum_rewards /= initial_cash\n all_cum_rewards = tf.convert_to_tensor(all_cum_rewards, dtype=tf.float32)\n\n loss_value = loss(all_logits_stack, all_actions_stack, all_cum_rewards)\n\n grads = gt.gradient(loss_value, state_encoding_model.variables + pi.variables)\n optimizer.apply_gradients(zip(grads, state_encoding_model.variables + pi.variables))\n\n if (batch_no+1) % batches_per_print == 0:\n end_time = time.time()\n print((\"batch_id: {}, num_eps_over: {}, average_total_r_per_ep: {:.2f}, \"+\n \"time_spent: {:.1f}s\").format(\n batch_no, num_eps_over, average_total_r/num_eps_over, end_time-start_time))\n\n # reset\n average_total_r = 0.0\n num_eps_over = 0\n start_time = time.time()\n\n # print epoch summary\n if glob_mode == None:\n # compute average total reward in one epoch to evaluate agent performance\n print(\"average total_r over one epoch: {:.2f}\".format(\n epoch_average_total_r/len(total_r_dict)))\n\n # reset\n epoch_average_total_r = 0.0\n total_r_dict = {}\n\n batch_no += 1\n\n print('Finished training~')",
"_____no_output_____"
],
[
"train()",
"num_of_pair = 6670\nbatch_size = 300\nnum_of_batch = 3557, estimated epoch = 79.99250374812594\nrand_action_prob = 0.0\nlr = 0.0005\naverage total_r over one epoch: -513.14\naverage total_r over one epoch: -368.25\nbatch_id: 49, num_eps_over: 14540, average_total_r_per_ep: -407.01, time_spent: 75.4s\naverage total_r over one epoch: -129.28\naverage total_r over one epoch: 138.04\nbatch_id: 99, num_eps_over: 14540, average_total_r_per_ep: 74.79, time_spent: 78.2s\naverage total_r over one epoch: 491.62\naverage total_r over one epoch: 793.90\nbatch_id: 149, num_eps_over: 14540, average_total_r_per_ep: 790.62, time_spent: 83.5s\naverage total_r over one epoch: 1126.15\naverage total_r over one epoch: 1288.20\nbatch_id: 199, num_eps_over: 14540, average_total_r_per_ep: 1287.64, time_spent: 83.5s\naverage total_r over one epoch: 1334.88\naverage total_r over one epoch: 1427.53\nbatch_id: 249, num_eps_over: 14540, average_total_r_per_ep: 1402.51, time_spent: 79.3s\naverage total_r over one epoch: 1409.61\naverage total_r over one epoch: 1422.56\naverage total_r over one epoch: 1425.06\nbatch_id: 299, num_eps_over: 14310, average_total_r_per_ep: 1427.23, time_spent: 78.2s\naverage total_r over one epoch: 1452.47\naverage total_r over one epoch: 1458.46\nbatch_id: 349, num_eps_over: 14540, average_total_r_per_ep: 1442.73, time_spent: 77.0s\naverage total_r over one epoch: 1416.12\naverage total_r over one epoch: 1488.70\nbatch_id: 399, num_eps_over: 14540, average_total_r_per_ep: 1470.62, time_spent: 79.5s\naverage total_r over one epoch: 1503.79\naverage total_r over one epoch: 1465.44\nbatch_id: 449, num_eps_over: 14540, average_total_r_per_ep: 1481.58, time_spent: 82.8s\naverage total_r over one epoch: 1481.56\naverage total_r over one epoch: 1429.82\nbatch_id: 499, num_eps_over: 14540, average_total_r_per_ep: 1430.54, time_spent: 82.5s\naverage total_r over one epoch: 1449.74\naverage total_r over one epoch: 1419.13\nbatch_id: 549, num_eps_over: 14540, average_total_r_per_ep: 1455.12, time_spent: 77.4s\naverage total_r over one epoch: 1464.64\naverage total_r over one epoch: 1487.86\naverage total_r over one epoch: 1452.93\nbatch_id: 599, num_eps_over: 14310, average_total_r_per_ep: 1472.89, time_spent: 75.7s\naverage total_r over one epoch: 1384.61\naverage total_r over one epoch: 1353.78\nbatch_id: 649, num_eps_over: 14540, average_total_r_per_ep: 1344.56, time_spent: 76.1s\naverage total_r over one epoch: 1367.06\naverage total_r over one epoch: 1429.41\nbatch_id: 699, num_eps_over: 14540, average_total_r_per_ep: 1413.86, time_spent: 76.0s\naverage total_r over one epoch: 1381.78\naverage total_r over one epoch: 1447.73\nbatch_id: 749, num_eps_over: 14540, average_total_r_per_ep: 1458.56, time_spent: 77.2s\naverage total_r over one epoch: 1491.03\naverage total_r over one epoch: 1504.21\nbatch_id: 799, num_eps_over: 14540, average_total_r_per_ep: 1497.04, time_spent: 75.5s\naverage total_r over one epoch: 1531.72\naverage total_r over one epoch: 1472.15\nbatch_id: 849, num_eps_over: 14540, average_total_r_per_ep: 1508.98, time_spent: 76.8s\naverage total_r over one epoch: 1537.21\naverage total_r over one epoch: 1494.65\naverage total_r over one epoch: 1507.82\nbatch_id: 899, num_eps_over: 14310, average_total_r_per_ep: 1493.47, time_spent: 74.9s\naverage total_r over one epoch: 1529.32\naverage total_r over one epoch: 1535.24\nbatch_id: 949, num_eps_over: 14540, average_total_r_per_ep: 1565.44, time_spent: 75.4s\naverage total_r over one epoch: 1597.96\naverage total_r over one epoch: 1599.31\nbatch_id: 999, num_eps_over: 14540, average_total_r_per_ep: 1585.01, time_spent: 75.2s\naverage total_r over one epoch: 1568.43\naverage total_r over one epoch: 1590.03\nbatch_id: 1049, num_eps_over: 14540, average_total_r_per_ep: 1598.19, time_spent: 74.6s\naverage total_r over one epoch: 1622.18\naverage total_r over one epoch: 1581.71\nbatch_id: 1099, num_eps_over: 14540, average_total_r_per_ep: 1540.72, time_spent: 75.7s\naverage total_r over one epoch: 1484.33\naverage total_r over one epoch: 1502.01\nbatch_id: 1149, num_eps_over: 14310, average_total_r_per_ep: 1502.33, time_spent: 74.1s\naverage total_r over one epoch: 1508.31\naverage total_r over one epoch: 1532.07\naverage total_r over one epoch: 1519.08\nbatch_id: 1199, num_eps_over: 14540, average_total_r_per_ep: 1524.89, time_spent: 74.5s\naverage total_r over one epoch: 1527.19\naverage total_r over one epoch: 1573.20\nbatch_id: 1249, num_eps_over: 14540, average_total_r_per_ep: 1551.58, time_spent: 75.6s\naverage total_r over one epoch: 1566.45\naverage total_r over one epoch: 1658.72\nbatch_id: 1299, num_eps_over: 14540, average_total_r_per_ep: 1633.41, time_spent: 74.5s\naverage total_r over one epoch: 1662.81\naverage total_r over one epoch: 1646.39\nbatch_id: 1349, num_eps_over: 14540, average_total_r_per_ep: 1675.24, time_spent: 73.7s\naverage total_r over one epoch: 1676.54\naverage total_r over one epoch: 1676.84\nbatch_id: 1399, num_eps_over: 14540, average_total_r_per_ep: 1668.75, time_spent: 73.2s\naverage total_r over one epoch: 1674.22\naverage total_r over one epoch: 1641.16\naverage total_r over one epoch: 1597.89\nbatch_id: 1449, num_eps_over: 14310, average_total_r_per_ep: 1621.76, time_spent: 72.8s\naverage total_r over one epoch: 1646.23\naverage total_r over one epoch: 1625.50\nbatch_id: 1499, num_eps_over: 14540, average_total_r_per_ep: 1632.83, time_spent: 75.7s\naverage total_r over one epoch: 1613.57\naverage total_r over one epoch: 1681.78\nbatch_id: 1549, num_eps_over: 14540, average_total_r_per_ep: 1655.39, time_spent: 72.8s\naverage total_r over one epoch: 1690.21\naverage total_r over one epoch: 1738.02\nbatch_id: 1599, num_eps_over: 14540, average_total_r_per_ep: 1714.41, time_spent: 72.6s\naverage total_r over one epoch: 1731.78\naverage total_r over one epoch: 1746.48\nbatch_id: 1649, num_eps_over: 14540, average_total_r_per_ep: 1740.42, time_spent: 73.8s\naverage total_r over one epoch: 1720.14\naverage total_r over one epoch: 1766.12\nbatch_id: 1699, num_eps_over: 14540, average_total_r_per_ep: 1756.30, time_spent: 73.2s\naverage total_r over one epoch: 1754.45\naverage total_r over one epoch: 1738.04\naverage total_r over one epoch: 1782.17\nbatch_id: 1749, num_eps_over: 14310, average_total_r_per_ep: 1771.23, time_spent: 71.9s\naverage total_r over one epoch: 1786.39\naverage total_r over one epoch: 1725.69\nbatch_id: 1799, num_eps_over: 14540, average_total_r_per_ep: 1754.09, time_spent: 72.1s\naverage total_r over one epoch: 1711.57\naverage total_r over one epoch: 1695.72\nbatch_id: 1849, num_eps_over: 14540, average_total_r_per_ep: 1701.55, time_spent: 72.0s\naverage total_r over one epoch: 1769.25\naverage total_r over one epoch: 1805.68\nbatch_id: 1899, num_eps_over: 14540, average_total_r_per_ep: 1799.78, time_spent: 72.4s\naverage total_r over one epoch: 1791.15\naverage total_r over one epoch: 1824.37\nbatch_id: 1949, num_eps_over: 14540, average_total_r_per_ep: 1795.17, time_spent: 71.7s\naverage total_r over one epoch: 1783.14\naverage total_r over one epoch: 1782.02\nbatch_id: 1999, num_eps_over: 14540, average_total_r_per_ep: 1782.30, time_spent: 72.2s\naverage total_r over one epoch: 1775.72\naverage total_r over one epoch: 1724.44\naverage total_r over one epoch: 1762.20\nbatch_id: 2049, num_eps_over: 14310, average_total_r_per_ep: 1745.38, time_spent: 70.5s\naverage total_r over one epoch: 1780.36\naverage total_r over one epoch: 1776.83\nbatch_id: 2099, num_eps_over: 14540, average_total_r_per_ep: 1773.75, time_spent: 71.2s\naverage total_r over one epoch: 1797.77\naverage total_r over one epoch: 1744.33\nbatch_id: 2149, num_eps_over: 14540, average_total_r_per_ep: 1755.27, time_spent: 76.4s\naverage total_r over one epoch: 1684.86\naverage total_r over one epoch: 1712.17\nbatch_id: 2199, num_eps_over: 14540, average_total_r_per_ep: 1717.44, time_spent: 70.2s\naverage total_r over one epoch: 1736.57\naverage total_r over one epoch: 1775.72\nbatch_id: 2249, num_eps_over: 14540, average_total_r_per_ep: 1786.79, time_spent: 71.2s\naverage total_r over one epoch: 1812.85\naverage total_r over one epoch: 1839.00\nbatch_id: 2299, num_eps_over: 14310, average_total_r_per_ep: 1832.33, time_spent: 75.8s\naverage total_r over one epoch: 1841.45\naverage total_r over one epoch: 1828.32\naverage total_r over one epoch: 1865.55\n"
],
[
"def save_model():\n hkg_time = datetime.now() + timedelta(hours=16)\n checkpoint_name = hkg_time.strftime(\"%Y%m%d_%H%M%S\")\n print(checkpoint_name)\n checkpoint_prefix = os.path.join(checkpoint_dir, checkpoint_name)\n root.save(checkpoint_prefix)\n tf.train.latest_checkpoint(checkpoint_dir)",
"_____no_output_____"
],
[
"save_model()",
"20190213_014123\n"
],
[
"train()",
"num_of_pair = 6670\nbatch_size = 300\nnum_of_batch = 3557, estimated epoch = 79.99250374812594\nrand_action_prob = 0.0\nlr = 0.0005\naverage total_r over one epoch: 1916.15\nbatch_id: 3599, num_eps_over: 12670, average_total_r_per_ep: 1908.59, time_spent: 60.5s\naverage total_r over one epoch: 1897.89\naverage total_r over one epoch: 1877.67\naverage total_r over one epoch: 1849.71\nbatch_id: 3649, num_eps_over: 14310, average_total_r_per_ep: 1862.63, time_spent: 72.5s\naverage total_r over one epoch: 1839.02\naverage total_r over one epoch: 1879.64\nbatch_id: 3699, num_eps_over: 14540, average_total_r_per_ep: 1859.49, time_spent: 68.8s\naverage total_r over one epoch: 1906.49\naverage total_r over one epoch: 1916.48\nbatch_id: 3749, num_eps_over: 14540, average_total_r_per_ep: 1937.64, time_spent: 69.2s\naverage total_r over one epoch: 1985.88\naverage total_r over one epoch: 1998.85\nbatch_id: 3799, num_eps_over: 14540, average_total_r_per_ep: 1991.26, time_spent: 74.3s\naverage total_r over one epoch: 1982.44\naverage total_r over one epoch: 2025.10\nbatch_id: 3849, num_eps_over: 14540, average_total_r_per_ep: 2000.94, time_spent: 70.7s\naverage total_r over one epoch: 1981.86\naverage total_r over one epoch: 2000.41\nbatch_id: 3899, num_eps_over: 14540, average_total_r_per_ep: 2010.22, time_spent: 70.3s\naverage total_r over one epoch: 2039.67\naverage total_r over one epoch: 2029.53\naverage total_r over one epoch: 2049.88\nbatch_id: 3949, num_eps_over: 14310, average_total_r_per_ep: 2035.06, time_spent: 69.9s\naverage total_r over one epoch: 1975.69\naverage total_r over one epoch: 1971.95\nbatch_id: 3999, num_eps_over: 14540, average_total_r_per_ep: 1977.74, time_spent: 69.1s\naverage total_r over one epoch: 1970.55\naverage total_r over one epoch: 1989.92\nbatch_id: 4049, num_eps_over: 14540, average_total_r_per_ep: 1995.25, time_spent: 68.8s\naverage total_r over one epoch: 2008.65\naverage total_r over one epoch: 1967.17\nbatch_id: 4099, num_eps_over: 14540, average_total_r_per_ep: 1976.40, time_spent: 69.4s\naverage total_r over one epoch: 1960.86\naverage total_r over one epoch: 1985.95\nbatch_id: 4149, num_eps_over: 14540, average_total_r_per_ep: 1985.75, time_spent: 69.8s\naverage total_r over one epoch: 2032.51\naverage total_r over one epoch: 1995.21\nbatch_id: 4199, num_eps_over: 14540, average_total_r_per_ep: 2021.23, time_spent: 71.1s\naverage total_r over one epoch: 2041.94\naverage total_r over one epoch: 2016.24\naverage total_r over one epoch: 2022.04\nbatch_id: 4249, num_eps_over: 14310, average_total_r_per_ep: 2024.30, time_spent: 72.2s\naverage total_r over one epoch: 2014.35\naverage total_r over one epoch: 2000.70\nbatch_id: 4299, num_eps_over: 14540, average_total_r_per_ep: 1990.69, time_spent: 71.8s\naverage total_r over one epoch: 1997.64\naverage total_r over one epoch: 1998.15\nbatch_id: 4349, num_eps_over: 14540, average_total_r_per_ep: 2014.42, time_spent: 71.1s\naverage total_r over one epoch: 2029.87\naverage total_r over one epoch: 2026.87\nbatch_id: 4399, num_eps_over: 14540, average_total_r_per_ep: 2021.19, time_spent: 73.3s\naverage total_r over one epoch: 2023.27\naverage total_r over one epoch: 1974.82\nbatch_id: 4449, num_eps_over: 14540, average_total_r_per_ep: 1995.01, time_spent: 77.2s\naverage total_r over one epoch: 1980.71\naverage total_r over one epoch: 1908.88\nbatch_id: 4499, num_eps_over: 14310, average_total_r_per_ep: 1886.92, time_spent: 74.9s\naverage total_r over one epoch: 1860.12\naverage total_r over one epoch: 1897.05\naverage total_r over one epoch: 1918.48\nbatch_id: 4549, num_eps_over: 14540, average_total_r_per_ep: 1907.47, time_spent: 74.4s\naverage total_r over one epoch: 1911.92\naverage total_r over one epoch: 1955.11\nbatch_id: 4599, num_eps_over: 14540, average_total_r_per_ep: 1939.78, time_spent: 71.1s\naverage total_r over one epoch: 1960.07\naverage total_r over one epoch: 1920.29\nbatch_id: 4649, num_eps_over: 14540, average_total_r_per_ep: 1934.14, time_spent: 71.5s\naverage total_r over one epoch: 1906.66\naverage total_r over one epoch: 1907.07\nbatch_id: 4699, num_eps_over: 14540, average_total_r_per_ep: 1892.55, time_spent: 72.9s\naverage total_r over one epoch: 1878.69\naverage total_r over one epoch: 1914.39\nbatch_id: 4749, num_eps_over: 14540, average_total_r_per_ep: 1906.81, time_spent: 75.6s\naverage total_r over one epoch: 1907.09\naverage total_r over one epoch: 1912.88\naverage total_r over one epoch: 1917.58\nbatch_id: 4799, num_eps_over: 14310, average_total_r_per_ep: 1915.73, time_spent: 70.2s\naverage total_r over one epoch: 1932.56\naverage total_r over one epoch: 1914.00\nbatch_id: 4849, num_eps_over: 14540, average_total_r_per_ep: 1931.66, time_spent: 72.8s\naverage total_r over one epoch: 1916.85\naverage total_r over one epoch: 1889.60\nbatch_id: 4899, num_eps_over: 14540, average_total_r_per_ep: 1880.56, time_spent: 72.9s\naverage total_r over one epoch: 1870.21\naverage total_r over one epoch: 1935.94\nbatch_id: 4949, num_eps_over: 14540, average_total_r_per_ep: 1905.58, time_spent: 71.3s\naverage total_r over one epoch: 1882.93\naverage total_r over one epoch: 1838.63\nbatch_id: 4999, num_eps_over: 14540, average_total_r_per_ep: 1810.56, time_spent: 72.8s\naverage total_r over one epoch: 1727.17\naverage total_r over one epoch: 1743.37\nbatch_id: 5049, num_eps_over: 14540, average_total_r_per_ep: 1738.16, time_spent: 70.3s\naverage total_r over one epoch: 1733.77\naverage total_r over one epoch: 1787.36\naverage total_r over one epoch: 1757.17\nbatch_id: 5099, num_eps_over: 14310, average_total_r_per_ep: 1769.28, time_spent: 71.7s\naverage total_r over one epoch: 1727.50\naverage total_r over one epoch: 1751.55\nbatch_id: 5149, num_eps_over: 14540, average_total_r_per_ep: 1753.00, time_spent: 77.6s\naverage total_r over one epoch: 1762.36\naverage total_r over one epoch: 1775.35\nbatch_id: 5199, num_eps_over: 14540, average_total_r_per_ep: 1753.52, time_spent: 71.4s\naverage total_r over one epoch: 1731.81\naverage total_r over one epoch: 1600.97\nbatch_id: 5249, num_eps_over: 14540, average_total_r_per_ep: 1601.82, time_spent: 71.5s\naverage total_r over one epoch: 1461.93\naverage total_r over one epoch: 1321.16\nbatch_id: 5299, num_eps_over: 14540, average_total_r_per_ep: 1346.82, time_spent: 72.6s\naverage total_r over one epoch: 1361.94\naverage total_r over one epoch: 1454.94\nbatch_id: 5349, num_eps_over: 14540, average_total_r_per_ep: 1463.03, time_spent: 72.0s\naverage total_r over one epoch: 1485.27\naverage total_r over one epoch: 1528.52\naverage total_r over one epoch: 1582.71\nbatch_id: 5399, num_eps_over: 14310, average_total_r_per_ep: 1552.67, time_spent: 74.3s\naverage total_r over one epoch: 1646.91\naverage total_r over one epoch: 1644.45\nbatch_id: 5449, num_eps_over: 14540, average_total_r_per_ep: 1665.76, time_spent: 71.4s\naverage total_r over one epoch: 1634.41\naverage total_r over one epoch: 1667.02\nbatch_id: 5499, num_eps_over: 14540, average_total_r_per_ep: 1625.43, time_spent: 77.1s\naverage total_r over one epoch: 1651.96\naverage total_r over one epoch: 1684.88\nbatch_id: 5549, num_eps_over: 14540, average_total_r_per_ep: 1684.73, time_spent: 73.7s\naverage total_r over one epoch: 1668.97\naverage total_r over one epoch: 1682.80\nbatch_id: 5599, num_eps_over: 14540, average_total_r_per_ep: 1692.62, time_spent: 72.3s\naverage total_r over one epoch: 1705.15\naverage total_r over one epoch: 1722.13\nbatch_id: 5649, num_eps_over: 14310, average_total_r_per_ep: 1718.56, time_spent: 70.7s\naverage total_r over one epoch: 1715.65\naverage total_r over one epoch: 1717.86\naverage total_r over one epoch: 1690.40\nbatch_id: 5699, num_eps_over: 14540, average_total_r_per_ep: 1697.83, time_spent: 72.3s\naverage total_r over one epoch: 1682.01\naverage total_r over one epoch: 1685.01\nbatch_id: 5749, num_eps_over: 14540, average_total_r_per_ep: 1685.48, time_spent: 71.3s\naverage total_r over one epoch: 1700.65\naverage total_r over one epoch: 1709.28\nbatch_id: 5799, num_eps_over: 14540, average_total_r_per_ep: 1710.94, time_spent: 72.1s\naverage total_r over one epoch: 1722.10\naverage total_r over one epoch: 1725.63\nbatch_id: 5849, num_eps_over: 14540, average_total_r_per_ep: 1737.50, time_spent: 74.8s\naverage total_r over one epoch: 1738.72\naverage total_r over one epoch: 1732.42\n"
],
[
"save_model()",
"20190213_030613\n"
],
[
"# evaluate performance on train dataset\nglob_mode = None # reset the dataset\ntrain_rs = []\ntrain_total_r_dict = {}\nrun_epoch_for_evaluate_performance(train_rs, train_total_r_dict, 'train')\n\nplot_rs_dist(train_rs)",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, \n6670\n1836.8113364767623\n"
],
[
"# evaluate performance on valid dataset\nvalid_rs = []\nvalid_total_r_dict = {}\nrun_epoch_for_evaluate_performance(valid_rs, valid_total_r_dict, 'valid')\n\nplot_rs_dist(valid_rs)",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, \n6670\n1015.2148457496246\n"
],
[
"# # evaluate performance on test dataset\n# test_rs = []\n# test_total_r_dict = {}\n# run_epoch_for_evaluate_performance(test_rs, test_total_r_dict, 'test')\n\n# plot_rs_dist(test_rs)",
"_____no_output_____"
],
[
"# pick best K pairs from train result and see how good those pairs are in valid\nK = 5\ntrain_total_r_ordered = sorted(list(train_total_r_dict.items()), key=lambda x: x[1], reverse=True)\ntrain_total_r_ordered_sliced = train_total_r_ordered[:K]\nvalid_total_r_sliced = [(x[0], valid_total_r_dict[x[0]]) for x in train_total_r_ordered_sliced]\nvalid_total_r_corresponding_value = [x[1] for x in valid_total_r_sliced]\ntrain_total_r_sliced_value = [x[1] for x in train_total_r_ordered_sliced]\n\n# see if the model overfits a lot by checking the performace in valid of best K pairs in train\nbins = np.linspace(-10000, 30000, 30)\nplt.hist(train_total_r_sliced_value, bins, alpha=0.3, label='train')\nplt.hist(valid_total_r_corresponding_value, bins, alpha=0.3, label='valid')\nplt.legend(loc='upper right')\nplt.gcf().set_size_inches(10, 5)",
"_____no_output_____"
],
[
"# # pick best K pairs from valid result and see how good those pairs are in test\n# K = 5\n# valid_total_r_ordered = sorted(list(valid_total_r_dict.items()), key=lambda x: x[1], reverse=True)\n# valid_total_r_ordered_sliced = valid_total_r_ordered[:K]\n# test_total_r_sliced = [(x[0], test_total_r_dict[x[0]]) for x in valid_total_r_ordered_sliced]\n# test_total_r_corresponding_value = [x[1] for x in test_total_r_sliced]\n# valid_total_r_sliced_value = [x[1] for x in valid_total_r_ordered_sliced]\n\n# # see if the model overfits a lot by checking the performace in valid of best K pairs in train\n# bins = np.linspace(-10000, 30000, 30)\n# plt.hist(valid_total_r_sliced_value, bins, alpha=0.3, label='valid')\n# plt.hist(test_total_r_corresponding_value, bins, alpha=0.3, label='test')\n# plt.legend(loc='upper right')\n# plt.gcf().set_size_inches(10, 5)",
"_____no_output_____"
],
[
"# pick best K pairs from train result and see how good those pairs are in valid\nK = 20\ntrain_total_r_ordered = sorted(list(train_total_r_dict.items()), key=lambda x: x[1], reverse=True)\ntrain_total_r_ordered_sliced = train_total_r_ordered[:K]\nvalid_total_r_sliced = [(x[0], valid_total_r_dict[x[0]]) for x in train_total_r_ordered_sliced]\nvalid_total_r_corresponding_value = [x[1] for x in valid_total_r_sliced]\ntrain_total_r_sliced_value = [x[1] for x in train_total_r_ordered_sliced]\n\n# see if the model overfits a lot by checking the performace in valid of best K pairs in train\nbins = np.linspace(-10000, 30000, 30)\nplt.hist(train_total_r_sliced_value, bins, alpha=0.3, label='train')\nplt.hist(valid_total_r_corresponding_value, bins, alpha=0.3, label='valid')\nplt.legend(loc='upper right')\nplt.gcf().set_size_inches(10, 5)",
"_____no_output_____"
],
[
"# # pick best K pairs from valid result and see how good those pairs are in test\n# K = 20\n# valid_total_r_ordered = sorted(list(valid_total_r_dict.items()), key=lambda x: x[1], reverse=True)\n# valid_total_r_ordered_sliced = valid_total_r_ordered[:K]\n# test_total_r_sliced = [(x[0], test_total_r_dict[x[0]]) for x in valid_total_r_ordered_sliced]\n# test_total_r_corresponding_value = [x[1] for x in test_total_r_sliced]\n# valid_total_r_sliced_value = [x[1] for x in valid_total_r_ordered_sliced]\n\n# # see if the model overfits a lot by checking the performace in valid of best K pairs in train\n# bins = np.linspace(-10000, 30000, 30)\n# plt.hist(valid_total_r_sliced_value, bins, alpha=0.3, label='valid')\n# plt.hist(test_total_r_corresponding_value, bins, alpha=0.3, label='test')\n# plt.legend(loc='upper right')\n# plt.gcf().set_size_inches(10, 5)",
"_____no_output_____"
],
[
"train_total_r_ordered_by_name = sorted(list(train_total_r_dict.items()), key=lambda x: x[0])\nvalid_total_r_ordered_by_name = sorted(list(valid_total_r_dict.items()), key=lambda x: x[0])\ntrain_total_r_ordered_val = [x[1] for x in train_total_r_ordered_by_name]\nvalid_total_r_ordered_val = [x[1] for x in valid_total_r_ordered_by_name]\n\nplt.scatter(train_total_r_ordered_val, valid_total_r_ordered_val, s=15, alpha=0.3, label='train')\nplt.xlabel('train')\nplt.ylabel('valid')\nplt.xlim(-10000, 40000)\nplt.ylim(-10000, 40000)\nplt.plot(np.linspace(-10000, 40000, 30), np.linspace(-10000, 40000, 30), color='r')\nplt.gca().set_aspect('equal', adjustable='box')\nplt.gcf().set_size_inches(10, 10)",
"_____no_output_____"
],
[
"# valid_total_r_ordered_by_name = sorted(list(valid_total_r_dict.items()), key=lambda x: x[0])\n# test_total_r_ordered_by_name = sorted(list(test_total_r_dict.items()), key=lambda x: x[0])\n# valid_total_r_ordered_val = [x[1] for x in valid_total_r_ordered_by_name]\n# test_total_r_ordered_val = [x[1] for x in test_total_r_ordered_by_name]\n\n# plt.scatter(valid_total_r_ordered_val, test_total_r_ordered_val, s=15, alpha=0.3, label='train')\n# plt.xlabel('valid')\n# plt.ylabel('test')\n# plt.xlim(-10000, 40000)\n# plt.ylim(-10000, 40000)\n# plt.plot(np.linspace(-10000, 40000, 30), np.linspace(-10000, 40000, 30), color='r')\n# plt.gca().set_aspect('equal', adjustable='box')\n# plt.gcf().set_size_inches(10, 10)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec890377a0fc7bd42845cd783d0fd16093b8b327 | 6,129 | ipynb | Jupyter Notebook | experiments/breakout/AMN_ens_SNR.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | experiments/breakout/AMN_ens_SNR.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | experiments/breakout/AMN_ens_SNR.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | 30.954545 | 141 | 0.58574 | [
[
[
"from tqdm.notebook import tqdm\nimport math\nimport gym\nimport torch\nimport torch.optim as optim \nfrom torch.utils.tensorboard import SummaryWriter\nfrom collections import deque\n\nimport active_rl.networks as networks\nfrom active_rl.networks.dqn_atari import ENS_DQN\nfrom active_rl.utils.memory import RankedReplayMemory, LabeledReplayMemory\nfrom active_rl.utils.optimization import AMN_optimization_ensemble\nfrom active_rl.environments.atari_wrappers import make_atari, wrap_deepmind\nfrom active_rl.utils.atari_utils import fp, ActionSelector, evaluate\nfrom active_rl.utils.acquisition_functions import ens_SNR",
"_____no_output_____"
],
[
"env_name = 'Breakout'\nenv_raw = make_atari('{}NoFrameskip-v4'.format(env_name))\nenv = wrap_deepmind(env_raw, frame_stack=False, episode_life=False, clip_rewards=True)\nc,h,w = c,h,w = fp(env.reset()).shape\nn_actions = env.action_space.n",
"_____no_output_____"
],
[
"BATCH_SIZE = 64\nLR = 0.0000625\nGAMMA = 0.99\nEPS = 0.05\nNUM_STEPS = 10000000\nNOT_LABELLED_CAPACITY = 10000\nLABELLED_CAPACITY = 200000\nINITIAL_STEPS=NOT_LABELLED_CAPACITY\nTRAINING_PER_LABEL = 80.\nPERCENTAGE = 0.005\nTRAINING_ITER = int(TRAINING_PER_LABEL * NOT_LABELLED_CAPACITY * PERCENTAGE)\n\nNAME = 'AMN_Ensemble_SNR_05_more_training'",
"_____no_output_____"
],
[
"device = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\") # if gpu is to be used\n# AMN_net = MC_DQN(n_actions).to(device)\nAMN_net = ENS_DQN(n_actions).to(device)\nexpert_net = torch.load(\"models/dqn_expert_breakout_model\").to(device)\nAMN_net.apply(AMN_net.init_weights)\nexpert_net.eval()\n# optimizer = optim.Adam(AMN_net.parameters(), lr=LR, eps=1.5e-4)\noptimizer = optim.Adam(AMN_net.parameters(), lr=LR, eps=1.5e-4)",
"_____no_output_____"
],
[
"memory = LabeledReplayMemory(NOT_LABELLED_CAPACITY, LABELLED_CAPACITY, [5,h,w], n_actions, ens_SNR, AMN_net, \n device=device)\naction_selector = ActionSelector(EPS, EPS, AMN_net, 1, n_actions, device)",
"_____no_output_____"
],
[
"steps_done = 0\nwriter = SummaryWriter(f'runs/{NAME}')",
"_____no_output_____"
],
[
"q = deque(maxlen=5)\ndone=True\neps = 0\nepisode_len = 0\nnum_labels = 0",
"_____no_output_____"
],
[
"progressive = tqdm(range(NUM_STEPS), total=NUM_STEPS, ncols=400, leave=False, unit='b')\nfor step in progressive:\n if done:\n env.reset()\n sum_reward = 0\n episode_len = 0\n img, _, _, _ = env.step(1) # BREAKOUT specific !!!\n for i in range(10): # no-op\n n_frame, _, _, _ = env.step(0)\n n_frame = fp(n_frame)\n q.append(n_frame)\n \n # Select and perform an action\n state = torch.cat(list(q))[1:].unsqueeze(0)\n action, eps = action_selector.select_action(state)\n n_frame, reward, done, info = env.step(action)\n n_frame = fp(n_frame)\n\n # 5 frame as memory\n q.append(n_frame)\n memory.push(torch.cat(list(q)).unsqueeze(0), action, reward, done) # here the n_frame means next frame from the previous time step\n episode_len += 1\n\n # Perform one step of the optimization (on the target network)\n if step % NOT_LABELLED_CAPACITY == 0 and step >= INITIAL_STEPS:\n num_labels += memory.label_sample(percentage=PERCENTAGE)\n loss = 0\n for _ in range(TRAINING_ITER):\n loss += AMN_optimization_ensemble(AMN_net, expert_net, optimizer, memory, batch_size=BATCH_SIZE, \n device=device)\n loss /= TRAINING_ITER\n if loss is not None:\n writer.add_scalar('loss_vs_#labels', loss, num_labels)\n evaluated_reward = evaluate(step, AMN_net, device, env_raw, n_actions, eps=0.05, num_episode=15)\n writer.add_scalar('reward_vs_#labels', evaluated_reward, num_labels)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec892f1509623c435772f8336218a8bc59ddecf7 | 9,334 | ipynb | Jupyter Notebook | course/4_Python_III.ipynb | ebustosm6/python-basics | d29faf0578cb7fcb8fac1620b3798979d338f842 | [
"Apache-2.0"
] | null | null | null | course/4_Python_III.ipynb | ebustosm6/python-basics | d29faf0578cb7fcb8fac1620b3798979d338f842 | [
"Apache-2.0"
] | null | null | null | course/4_Python_III.ipynb | ebustosm6/python-basics | d29faf0578cb7fcb8fac1620b3798979d338f842 | [
"Apache-2.0"
] | null | null | null | 30.109677 | 288 | 0.58153 | [
[
[
"# Python nivel III: Bucles y condiciones\nAhora que ya sabemos lo que son las diferentes estructuras para guardar elementos, vamos a ver como iterar sobre ellas (recorrerlas), además de evaluar condiciones.\n\nA modo de introducción, diremos que hay una estructura fundamental de condiciones:\n- if / elif / else: Evalúa una expresión y se ejecuta una parte de código u otra.\nQue veremos en detalle.\n\nAdemás de los dos tipos de bucles:\n- Bucle while: Este bucle se ejecuta mientras (de ahí el while) una condición se cumpla.\n- Bucle for: Este bucle hace una iteración para cada elemento de la estructura.\n\nVamos a verlo más detalladamente.",
"_____no_output_____"
],
[
"## Antes de entrar en faena\nAhora que ya empezamos con cosas más complejas, es bueno mencionar una página que me ayudó mucho a entender cómo se ejecutan las cosas paso a paso y cómo está el estado de la memoria en cada momento. \nEs una herramienta muy útil al principio para ejecutar paso a paso cualquier programa y ver lo que está pasando.\n\nLa página es [Python tutor](http://www.pythontutor.com/). Para empezar, pinchar en [Start visualizing your code now](http://www.pythontutor.com/visualize.html#mode=edit)\n\nTambién sirve por si jupyter se quedá tostado, se puede copiar el código de aquí y ejecutarlo ahí.\n\n\n\n",
"_____no_output_____"
],
[
"## Condiciones: if / elif / else\nCuando estamos ejecutando un programa (recordamos que se ejecuta linea por línea de arriba a abajo) a veces queremos hacer diferentes cosas según una condición, para ello se utiliza el condicionante if.\n\nLa estructura del if / elif / else es:\n```python\nif condition_1:\n # ejecutar el código de estas líneas cuando cumple la condición 1\nelif condition_2:\n # ejecutar el código de estas líneas cuando no cumple la condición 1 pero sí cumple la condición 2\nelse:\n # ejecutar el código de estas líneas cuando no cumple ninguna condición\n```\n\nComo nota: Puede haber sólo un if, un if / else o un if / elif / elif / ... / else. Pero siempre se empieza por if, y nunca puede haber un else sin un if o elif antes. \n\nVamos a verlo con un ejemplo muy sencillo y ya de paso vamos a empezar a hacer ejemplos y programas un poco más complejos y que ya parezcan más útiles.\n\nCon los siguientes datos:\nQueremos hacer un programa que calcule el precio de la entrada del cine en función de la edad del comprador.\nLa entreda de adulto vale 8 euros, si es menor de edad, 6.5 euros y si es jubilado, 5.3, por ejemplo.\nEntonces lo que hay que hacer es definir dos variables (edad y precio) y con una estructura de condiciones, cambiar el valor de la variable del precio según lo que valga la de la edad. \n\nVamos a ver el programa:",
"_____no_output_____"
]
],
[
[
"age = 27\nprice = 8\n\nif age < 18:\n # is young, así que igualamos el precio a 6.5\n price = 6.5\n\nelif age >= 65:\n # is old, así que igualamos el precio a 5.3\n price = 5.3\n \nelse:\n # is adult (18 <= age < 65), así que igualamos el precio a 8\n # este else en realidad no hace falta porque \"price\" ya vale 8 al principio del programa, pero lo dejamos para el ejemplo\n price = 8\n \n# imprimimos el resultado con un poco de estilo (esto se llama string format) y las {} sirven para sustituir por los valores\n# Hay que poner tantas {} como cosas queramos imprimir\nprint('El precio de la entrada para la edad {} es {}'.format(age, price))\n",
"_____no_output_____"
]
],
[
[
"## Bucle while\nEl bucle while sirve para ejecutar un trozo de código indefinidamente mientras que se cumple una condición.\n\nUn ejemplo puede ser imprimir los primero 10 números:\nPara esto habría que:\n1. Definir una variable = 0 y un límite = 10\n2. Imprimir la variable\n3. Sumar 1 a la variable\n4. Repetir los pasos 2 y 3 hasta que la variable sea == al límite.\n\nVemos el ejemplo:",
"_____no_output_____"
]
],
[
[
"number = 0\nlimit = 10\n\nwhile number <= limit:\n # imprimir la variable y sumarle 1\n print(number)\n number += 1",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n"
],
[
"# Ejercicio 0: Con un bucle while, imprimir todos los nombres de la siguiente lista\n# Para saber la longitud de una estructura se utiliza la función len(<lista>)\nnames = ['Jhon', 'Scarlett', 'Vanessa', 'Isabel', 'Peter', 'Julius']\n\n# Escribir aquí:\n\n",
"_____no_output_____"
]
],
[
[
"## Bucle for\nEl bucle while sirve para ejecutar un trozo de código para cada elemento de una estructura iterable:\n\nUn ejemplo puede ser imprimir los valores de una lista:\nVeamos un ejemplo:",
"_____no_output_____"
]
],
[
[
"fruits = ['banana', 'orange', 'lemon']\n\nfor fruit in fruits:\n print(fruit)",
"banana\norange\nlemon\n"
],
[
"# Ejercicio 1: Crear una lista de elemntos que se quiera e imprimirla\n\n\n# Escribir aquí:\n\n",
"_____no_output_____"
]
],
[
[
"## Hasta aquí\nEn este punto ya hemos visto varias cosas interesantes:\n- tipos básicos de datos y sus operaciones\n- estructuras básicas\n- bucles\n\nCon esto ya podemos empezar a hacer programas un poco más complejo, combinar bucles con estructuras if / else, etc.\nEl siguiente jupyter notebook (.ipynb) vamos a plantear algunos ejercicios para coger un poco de práctica y para ver algunas cosas más concretas.\n\nLos ejercicios que se verán ya tendrán un poco más sentido para ver ejemplos prácticos de pequeños programas. Lo más difícil es pensar cómo resolver el problema, así que daremos unas pistas para enforcarlo. De todas formas, lo importante es equivocarse para aprender de los errores.",
"_____no_output_____"
],
[
"## Soluciones",
"_____no_output_____"
]
],
[
[
"# Ejercicio 0: Con un bucle while, imprimir todos los nombres de la siguiente lista\nnames = ['Jhon', 'Scarlett', 'Vanessa', 'Isabel', 'Peter', 'Julius']\n\nnames_index = 0\nnames_length = len(names)\n\nwhile names_index < names_length:\n print(names[names_index])\n names_index += 1",
"Jhon\nScarlett\nVanessa\nIsabel\nPeter\nJulius\n"
],
[
"# Ejercicio 1: Crear una lista de elemntos que se quiera e imprimirla\n\nexample_ids = ['asd324ada', 'oi234nl234', '2kl34naa24']\n\nfor example_id in example_ids:\n print(example_id)",
"asd324ada\noi234nl234\n2kl34naa24\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
ec8935008f54d06f193e41c44e0394a878627325 | 55,628 | ipynb | Jupyter Notebook | Data Science and Machine Learning Bootcamp - JP/04.Pandas-Exercises/01-SF Salaries Exercise-MySolutions.ipynb | ptyadana/probability-and-statistics-for-business-and-data-science | 6c4d09c70e4c8546461eb7ebc401bb95a0827ef2 | [
"MIT"
] | 10 | 2021-01-14T15:14:03.000Z | 2022-02-19T14:06:25.000Z | Data Science and Machine Learning Bootcamp - JP/04.Pandas-Exercises/01-SF Salaries Exercise-MySolutions.ipynb | ptyadana/probability-and-statistics-for-business-and-data-science | 6c4d09c70e4c8546461eb7ebc401bb95a0827ef2 | [
"MIT"
] | null | null | null | Data Science and Machine Learning Bootcamp - JP/04.Pandas-Exercises/01-SF Salaries Exercise-MySolutions.ipynb | ptyadana/probability-and-statistics-for-business-and-data-science | 6c4d09c70e4c8546461eb7ebc401bb95a0827ef2 | [
"MIT"
] | 8 | 2021-03-24T13:00:02.000Z | 2022-03-27T16:32:20.000Z | 40.515659 | 15,756 | 0.563961 | [
[
[
"___\n\n<a href='http://www.pieriandata.com'> <img src='../../Pierian_Data_Logo.png' /></a>\n___",
"_____no_output_____"
],
[
"# SF Salaries Exercise \n\nWelcome to a quick exercise for you to practice your pandas skills! We will be using the [SF Salaries Dataset](https://www.kaggle.com/kaggle/sf-salaries) from Kaggle! Just follow along and complete the tasks outlined in bold below. The tasks will get harder and harder as you go along.",
"_____no_output_____"
],
[
"** Import pandas as pd.**",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"** Read Salaries.csv as a dataframe called sal.**",
"_____no_output_____"
]
],
[
[
"sal = pd.read_csv(\"Salaries.csv\")",
"_____no_output_____"
]
],
[
[
"** Check the head of the DataFrame. **",
"_____no_output_____"
]
],
[
[
"sal.head()",
"_____no_output_____"
]
],
[
[
"** Use the .info() method to find out how many entries there are.**",
"_____no_output_____"
]
],
[
[
"sal.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 148654 entries, 0 to 148653\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Id 148654 non-null int64 \n 1 EmployeeName 148654 non-null object \n 2 JobTitle 148654 non-null object \n 3 BasePay 148045 non-null float64\n 4 OvertimePay 148650 non-null float64\n 5 OtherPay 148650 non-null float64\n 6 Benefits 112491 non-null float64\n 7 TotalPay 148654 non-null float64\n 8 TotalPayBenefits 148654 non-null float64\n 9 Year 148654 non-null int64 \n 10 Notes 0 non-null float64\n 11 Agency 148654 non-null object \n 12 Status 0 non-null float64\ndtypes: float64(8), int64(2), object(3)\nmemory usage: 14.7+ MB\n"
]
],
[
[
"**What is the average BasePay ?**",
"_____no_output_____"
]
],
[
[
"sal[\"BasePay\"].mean()",
"_____no_output_____"
]
],
[
[
"#### What is the highest amount of OvertimePay in the dataset ?",
"_____no_output_____"
]
],
[
[
"sal[\"OvertimePay\"].max()",
"_____no_output_____"
]
],
[
[
"#### What is the job title of JOSEPH DRISCOLL ? Note: Use all caps, otherwise you may get an answer that doesn't match up (there is also a lowercase Joseph Driscoll).",
"_____no_output_____"
]
],
[
[
"sal[sal[\"EmployeeName\"] == \"JOSEPH DRISCOLL\"][\"JobTitle\"]",
"_____no_output_____"
]
],
[
[
"#### How much does JOSEPH DRISCOLL make (including benefits)? ",
"_____no_output_____"
]
],
[
[
"sal[sal[\"EmployeeName\"] == \"JOSEPH DRISCOLL\"][\"TotalPayBenefits\"]",
"_____no_output_____"
]
],
[
[
"#### What is the name of highest paid person (including benefits)?",
"_____no_output_____"
]
],
[
[
"sal[sal[\"TotalPay\"] == sal[\"TotalPayBenefits\"].max()]",
"_____no_output_____"
],
[
"#alternative solution\nsal.iloc[sal['TotalPayBenefits'].idxmax()]",
"_____no_output_____"
]
],
[
[
"#### What is the name of lowest paid person (including benefits)? Do you notice something strange about how much he or she is paid?",
"_____no_output_____"
]
],
[
[
"sal[ sal[\"TotalPayBenefits\"] == sal[\"TotalPayBenefits\"].min()]",
"_____no_output_____"
],
[
"sal.iloc[sal[\"TotalPayBenefits\"].idxmin()]",
"_____no_output_____"
]
],
[
[
"#### What was the average (mean) BasePay of all employees per year? (2011-2014) ?",
"_____no_output_____"
]
],
[
[
"sal.head()",
"_____no_output_____"
],
[
"sal[\"Year\"].value_counts()",
"_____no_output_____"
],
[
"sal.groupby(\"Year\").mean()[\"BasePay\"]",
"_____no_output_____"
]
],
[
[
"#### How many unique job titles are there?",
"_____no_output_____"
]
],
[
[
"sal[\"JobTitle\"].nunique()",
"_____no_output_____"
]
],
[
[
"#### How many unique job titles are there?",
"_____no_output_____"
]
],
[
[
"sal[\"JobTitle\"].value_counts()",
"_____no_output_____"
],
[
"sal[\"JobTitle\"].value_counts().head(5)",
"_____no_output_____"
]
],
[
[
"#### How many Job Titles were represented by only one person in 2013? (e.g. Job Titles with only one occurence in 2013?) ",
"_____no_output_____"
]
],
[
[
"sal[sal[\"Year\"] == 2013][\"JobTitle\"].value_counts() == 1",
"_____no_output_____"
],
[
"sum(sal[sal[\"Year\"] == 2013][\"JobTitle\"].value_counts() == 1)",
"_____no_output_____"
]
],
[
[
"#### How many people have the word Chief in their job title? (This is pretty tricky)",
"_____no_output_____"
]
],
[
[
"sal.head()",
"_____no_output_____"
],
[
"def chief_finder(x):\n if \"chief\" in x.lower():\n return True\n else:\n return False\n \nsum(sal[\"JobTitle\"].apply(lambda x : chief_finder(x)))",
"_____no_output_____"
]
],
[
[
"#### Bonus: Is there a correlation between length of the Job Title string and Salary?",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"title_length = sal[\"JobTitle\"].apply(lambda x : len(x))\ntitle_length",
"_____no_output_____"
],
[
"salary = sal[\"TotalPayBenefits\"]\nsalary",
"_____no_output_____"
],
[
"plt.scatter(title_length,salary);",
"_____no_output_____"
],
[
"sal[\"title_len\"] = sal[\"JobTitle\"].apply(len)",
"_____no_output_____"
],
[
"sal[[\"title_len\", \"TotalPayBenefits\"]].corr()\n# seem like there is no correlation",
"_____no_output_____"
]
],
[
[
"# Great Job!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ec894444a6ef5c4dd1efa7db7310889223f0732b | 104,698 | ipynb | Jupyter Notebook | c5_polynomial_regression/04_why_tain_test_split.ipynb | Sea-Monster/MachineLearningClassicAlgorithm | 2aaad1965e7e4b8659b6296dfe938181825fa259 | [
"MIT"
] | 12 | 2018-04-01T13:28:46.000Z | 2021-03-26T10:53:25.000Z | c5_polynomial_regression/04_why_tain_test_split.ipynb | Sea-Monster/MachineLearningClassicAlgorithm | 2aaad1965e7e4b8659b6296dfe938181825fa259 | [
"MIT"
] | null | null | null | c5_polynomial_regression/04_why_tain_test_split.ipynb | Sea-Monster/MachineLearningClassicAlgorithm | 2aaad1965e7e4b8659b6296dfe938181825fa259 | [
"MIT"
] | 6 | 2018-09-03T23:08:02.000Z | 2020-07-10T00:37:06.000Z | 187.967684 | 22,592 | 0.91657 | [
[
[
"## (上一章节)过拟合与欠拟合",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.linear_model.base import LinearRegression\nfrom sklearn.metrics.scorer import mean_squared_error\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing.data import PolynomialFeatures\nfrom sklearn.preprocessing.data import StandardScaler\n",
"_____no_output_____"
],
[
"np.random.seed(666)\nx = np.random.uniform(-3.0, 3.0, size=100)\nX = x.reshape(-1,1)\ny = 0.5 * x**2 + x+2 + np.random.normal(0,1,size=100)\nplt.scatter(x,y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 使用线性回归",
"_____no_output_____"
]
],
[
[
"lin_reg = LinearRegression()\nlin_reg.fit(X,y)\nlin_reg.score(X, y)",
"/usr/local/seamonster/MachineLearningClassicAlgorithmEnv/lib/python3.6/site-packages/scipy/linalg/basic.py:1226: RuntimeWarning: internal gelsd driver lwork query error, required iwork dimension not returned. This is likely the result of LAPACK bug 0038, fixed in LAPACK 3.2.2 (released July 21, 2010). Falling back to 'gelss' driver.\n warnings.warn(mesg, RuntimeWarning)\n"
]
],
[
[
"可以看到R方值很低(欠拟合) \n画个图来看看效果",
"_____no_output_____"
]
],
[
[
"y_predict = lin_reg.predict(X)\nplt.scatter(x,y)\n# 直线就是拟合的线\nplt.plot(np.sort(x), y_predict[np.argsort(x)], color='r')\nplt.show()",
"_____no_output_____"
]
],
[
[
"对于多项式回归,使用R方值作为评估是没有问题的 \n但是不明白这老师到底出于什么原因,要使用MSE(均方误差)-- 姑且理解是为了方便吧",
"_____no_output_____"
]
],
[
[
"y_predict = lin_reg.predict(X)\nmean_squared_error(y, y_predict)",
"_____no_output_____"
]
],
[
[
"## 使用多项式回归",
"_____no_output_____"
]
],
[
[
"def PolynomialRegression(degree):\n return Pipeline(\n [\n ('poly', PolynomialFeatures(degree=degree)),\n ('std_scaler', StandardScaler()),\n ('lin_reg',LinearRegression())\n ]\n )",
"_____no_output_____"
],
[
"poly2_reg = PolynomialRegression(degree=2)\npoly2_reg.fit(X, y)",
"_____no_output_____"
],
[
"y2_predict = poly2_reg.predict(X)\nmean_squared_error(y, y2_predict)",
"_____no_output_____"
]
],
[
[
"可以看到使用多项式回归后,均方误差比使用线性回归低得多 \n数据可视化效果如下:",
"_____no_output_____"
]
],
[
[
"plt.scatter(x,y)\nplt.plot(np.sort(x), y2_predict[np.argsort(x)], color='r')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### degree为10的情况",
"_____no_output_____"
]
],
[
[
"poly10_reg = PolynomialRegression(degree=10)\npoly10_reg.fit(X, y)\ny10_predict = poly10_reg.predict(X)\nmean_squared_error(y, y10_predict)",
"_____no_output_____"
]
],
[
[
"均方误差更低了 \n拟合曲线是什么样子呢?",
"_____no_output_____"
]
],
[
[
"plt.scatter(x,y)\nplt.plot(np.sort(x), y10_predict[np.argsort(x)], color='r')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### degree为100的情况",
"_____no_output_____"
]
],
[
[
"poly100_reg = PolynomialRegression(degree=100)\npoly100_reg.fit(X, y)\ny100_predict = poly100_reg.predict(X)\nmean_squared_error(y, y100_predict)",
"_____no_output_____"
]
],
[
[
"均方误差的值更低了(注意,本章节没有使用训练集和测试集的分离,所以这个误差都是针对训练样本的,自然是degree越大误差越小)",
"_____no_output_____"
]
],
[
[
"plt.scatter(x,y)\nplt.plot(np.sort(x), y100_predict[np.argsort(x)], color='r')\nplt.show()",
"_____no_output_____"
]
],
[
[
"如果x在-3到3之间均匀分布,那么对应degree=100的拟合曲线,它的y值会是怎么样呢?",
"_____no_output_____"
]
],
[
[
"x_plot = np.linspace(-3,3,100).reshape(100,1)\ny_plot = poly100_reg.predict(x_plot)\n\nplt.scatter(x,y)\nplt.plot(x_plot[:,0], y_plot, color='r')\nplt.axis([-3,3,-1,10]) # 坐标轴,x轴取-3到3,y轴取-1到10\nplt.show()",
"_____no_output_____"
]
],
[
[
"# train test split的意义",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection._split import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X,y, random_state=666)",
"_____no_output_____"
],
[
"lin_reg = LinearRegression()\nlin_reg.fit(X_train, y_train)\ny_predict = lin_reg.predict(X_test)\nmean_squared_error(y_test, y_predict)",
"_____no_output_____"
],
[
"poly2_reg = PolynomialRegression(degree=2)\npoly2_reg.fit(X_train, y_train)\ny2_predict = poly2_reg.predict(X_test)\nmean_squared_error(y_test, y2_predict)",
"_____no_output_____"
]
],
[
[
"可以看到使用2阶多项式回归比线性回归的均方误差更小,泛化能力更强",
"_____no_output_____"
]
],
[
[
"poly10_reg = PolynomialRegression(degree=10)\npoly10_reg.fit(X_train, y_train)\ny10_predict = poly10_reg.predict(X_test)\nmean_squared_error(y_test, y10_predict)",
"_____no_output_____"
]
],
[
[
"改为10阶多项式回归时,均方误差比2阶多项式回归时反而更大了 \n换句话说,泛化能力变差了",
"_____no_output_____"
]
],
[
[
"poly100_reg = PolynomialRegression(degree=100)\npoly100_reg.fit(X_train, y_train)\ny100_predict = poly100_reg.predict(X_test)\nmean_squared_error(y_test, y100_predict)",
"_____no_output_____"
]
],
[
[
"改为100阶多项式回归时,均方误差高得吓人",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec89502baa1e1b967235804824b5ce0dbf9c094b | 171,618 | ipynb | Jupyter Notebook | Credit Risk Project/Credit Risk_Logistic Regression.ipynb | RevatiDineshDeshpande/machine-learning-projects | 5e5e11d7c299a029ddd4975e7f9d232c549004ca | [
"MIT"
] | null | null | null | Credit Risk Project/Credit Risk_Logistic Regression.ipynb | RevatiDineshDeshpande/machine-learning-projects | 5e5e11d7c299a029ddd4975e7f9d232c549004ca | [
"MIT"
] | null | null | null | Credit Risk Project/Credit Risk_Logistic Regression.ipynb | RevatiDineshDeshpande/machine-learning-projects | 5e5e11d7c299a029ddd4975e7f9d232c549004ca | [
"MIT"
] | null | null | null | 78.292883 | 37,928 | 0.70742 | [
[
[
"# Logistic regression",
"_____no_output_____"
],
[
"## Context\nThe original dataset contains 1000 entries with 20 categorial/symbolic attributes prepared by Prof. Hofmann. In this dataset, each entry represents a person who takes a credit by a bank. Each person is classified as good or bad credit risks according to the set of attributes. It is almost impossible to understand the original dataset due to its complicated system of categories and symbols. Thus, the attributes in this dataset is a subset of the original dataset. Several columns are simply ignored, and some of the important attritubes like age, account balance etc. are retained.",
"_____no_output_____"
],
[
"## Dataset: German Credit Risk\n\n- Age (Numeric : Age in years)\n- Sex (Categories : male, female)\n- Job (Categories : 0 - unskilled and non-resident, 1 - unskilled and resident, 2 - skilled, 3 - highly skilled)\n- Housing (Categories : own, rent, or free)\n- Saving accounts (Categories : little, moderate, quite rich, rich)\n- Checking account (Categories : little, moderate, rich)\n- Credit amount (Numeric : Amount of credit in DM - Deutsche Mark)\n- Duration (Numeric : Duration for which the credit is given in months)\n- Purpose (Categories: car, furniture/equipment, radio/TV, domestic appliances, repairs, education, business, vacation/others)\n- Risk (0 - Person is not at risk, 1 - Pesron is at risk(defaulter))",
"_____no_output_____"
],
[
"# Objective\n\nThe objective is to estimate probabilities whether a person would default or not using logistic regression. In this dataset, the target variable is 'Risk'.",
"_____no_output_____"
],
[
"# 1. Load Libraries and data",
"_____no_output_____"
]
],
[
[
"import numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\nimport matplotlib.pyplot as plt # matplotlib.pyplot plots data\n%matplotlib inline \nimport seaborn as sns\n\n# Remove scientific notations and display numbers with 2 decimal points instead\npd.options.display.float_format = '{:,.2f}'.format\n\npdata = pd.read_csv(\"German_Credit.csv\")\npdata.head()",
"_____no_output_____"
]
],
[
[
"# 2. Calculate the percentage of missing values for each column and fill those missing values.\n",
"_____no_output_____"
]
],
[
[
"totalvals = pd.DataFrame(pdata.count(), columns = ['Number of entries'])\nmisvals = pd.DataFrame(pdata.isnull().sum(), columns= ['Number of missing values'])\nnewdf = pd.merge(totalvals, misvals, how = 'outer', right_index = True, left_index=True)\nnewdf['percmis'] = (newdf['Number of missing values'] / newdf['Number of entries']) * 100\nprint(newdf)\n\n## This is wrong, teh number of entris for each column is 1000. df.count() counts without the nans\n#Correct code:\n# df.isna().sum()/len(df.isna())\n#the above code will be implemented on all columns even though it is not specified. ",
" Number of entries Number of missing values percmis\nAge 1000 0 0.00\nSex 1000 0 0.00\nJob 1000 0 0.00\nHousing 1000 0 0.00\nSaving accounts 817 183 22.40\nChecking account 606 394 65.02\nCredit amount 1000 0 0.00\nDuration 1000 0 0.00\nPurpose 1000 0 0.00\nRisk 1000 0 0.00\n"
],
[
"from sklearn.impute import SimpleImputer\nrep_0 = SimpleImputer(strategy=\"most_frequent\")\ncols=['Saving accounts','Checking account']\nimputer = rep_0.fit(pdata[cols])\npdata[cols] = imputer.transform(pdata[cols])\n\npdata.head()\n#pdata.head()\n\n\n#This is also wrong. Cannot impute unknown categorical data taht is dependant on other factors withy \"most frequent\"\n# Need to imput NaN with unknown as below: \n# df['Saving accounts'].fillna(value='unknown',inplace=True)\n# df['Checking account'].fillna(value='unknown',inplace=True)\n# df.head()",
"_____no_output_____"
]
],
[
[
"# 3. Plot histogram for columns 'Credit amount' and 'Age'",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(20,12))\ncreditplot = sns.barplot(x='Age', y='Credit amount', data=pdata);\ncreditplot.set_xticklabels(creditplot.get_xticklabels(), rotation=40, ha=\"right\")\nplt.tight_layout()\n\n#This is also wrong. Hiostogram to be plotted as below: \n#df['Age'].hist()",
"_____no_output_____"
]
],
[
[
"# 4. Create counplots for columns 'Duration', 'Purpose'. What durations are most common and for what purposes most credits are taken?",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(20,12))\ndurplot = sns.countplot(x='Duration', data=pdata)\ndurplot.set_xticklabels(rotation=40, ha=\"right\")\nplt.tight_layout()\n\n\n#Insight: most common duration - 12 months, 24 months\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,12))\npurplot = sns.countplot(x='Purpose', data=pdata)\npurplot.set_xticklabels(purplot.get_xticklabels(), rotation=40, ha=\"right\")\nplt.tight_layout()\n\n#Insight: most common purpose - car or radio/tv",
"_____no_output_____"
]
],
[
[
"# 5. Which 'Job', 'Savings account', 'Checking account' has highest number of defaulters?",
"_____no_output_____"
]
],
[
[
"jobdef = pdata.groupby(['Job', 'Risk'])[['Risk']].count()\nsavdef = pdata.groupby(['Saving accounts','Risk'])[['Risk']].count()\ncheckdef = pdata.groupby(['Checking account','Risk'])[['Risk']].count()\nprint(jobdef)\nprint(savdef)\nprint(checkdef)\n\n#A better way to do this: \n#pd.crosstab(df['Job'],df['Risk'],normalize=False)",
" Risk\nJob Risk \n0 0 15\n 1 7\n1 0 144\n 1 56\n2 0 444\n 1 186\n3 0 97\n 1 51\n Risk\nSaving accounts Risk \nlittle 0 537\n 1 249\nmoderate 0 69\n 1 34\nquite rich 0 52\n 1 11\nrich 0 42\n 1 6\n Risk\nChecking account Risk \nlittle 0 487\n 1 181\nmoderate 0 164\n 1 105\nrich 0 49\n 1 14\n"
],
[
"# Interpretation: \n# Job2 has the highest number of defaulters, \n# Those with 'little' money in their savings account are the highest defaulters\n# Those with 'little' money in their checking account are the highest defaulters",
"_____no_output_____"
]
],
[
[
"# 6. Convert 'Credit amount' and 'Age' into categorical features by grouping them into different intervals and drop the original columns.\nYou can decide the intervals using df.describe() method.",
"_____no_output_____"
]
],
[
[
"pdata.describe()",
"_____no_output_____"
],
[
"pdata['Credit amount cut'] = pd.qcut(pdata['Credit amount'], q=4)\npdata['Age cut'] = pd.qcut(pdata['Age'], q=4)\npdata.head()",
"_____no_output_____"
],
[
"pdata = pdata.drop(['Credit amount','Age'], axis=1)",
"_____no_output_____"
],
[
"# pdata['Job'] = pd.Categorical(pdata.Job)\n# pdata['Duration'] =pd.Categorical(pdata.Duration)\n# pdata.head()",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
],
[
[
"# 7. Find out total number of defaulters where for each category of 'Age' and 'Credit amount'.",
"_____no_output_____"
]
],
[
[
"creddef = pdata.groupby(['Credit amount cut', 'Risk'])[['Risk']].count()\nagedef = pdata.groupby(['Age cut', 'Risk'])[['Risk']].count()\nprint(creddef)\nprint(agedef)",
" Risk\nCredit amount cut Risk \n(249.999, 1365.5] 0 173\n 1 77\n(1365.5, 2319.5] 0 188\n 1 62\n(2319.5, 3972.25] 0 194\n 1 56\n(3972.25, 18424.0] 0 145\n 1 105\n Risk\nAge cut Risk \n(18.999, 27.0] 0 184\n 1 107\n(27.0, 33.0] 0 151\n 1 74\n(33.0, 42.0] 0 190\n 1 59\n(42.0, 75.0] 0 175\n 1 60\n"
]
],
[
[
"# 8. Prepare input data for the model",
"_____no_output_____"
],
[
"### Separate dependent and independent variables",
"_____no_output_____"
]
],
[
[
"X = pdata.drop('Risk',axis=1) # Predictor feature columns (9 X m)\nY = pdata['Risk'] # Predicted class (1=True, 0=False) (1 X m)",
"_____no_output_____"
]
],
[
[
"### One hot enocding for all the categorical variables",
"_____no_output_____"
]
],
[
[
"list(pdata.columns)",
"_____no_output_____"
],
[
"feature_cols = ['Sex',\n 'Job',\n 'Housing',\n 'Saving accounts',\n 'Checking account',\n 'Duration',\n 'Purpose',\n 'Credit amount cut',\n 'Age cut']\nfeatures = pdata[feature_cols]\nfeatures.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1000 entries, 0 to 999\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Sex 1000 non-null object \n 1 Job 1000 non-null int64 \n 2 Housing 1000 non-null object \n 3 Saving accounts 1000 non-null object \n 4 Checking account 1000 non-null object \n 5 Duration 1000 non-null int64 \n 6 Purpose 1000 non-null object \n 7 Credit amount cut 1000 non-null category\n 8 Age cut 1000 non-null category\ndtypes: category(2), int64(2), object(5)\nmemory usage: 57.2+ KB\n"
],
[
"features_dummies = pd.get_dummies(features)\nfeatures_dummies.head()",
"_____no_output_____"
]
],
[
[
"### Split data into train and test (maintain the ratio of both classes in train and test set same as the original dataset)",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nx_train, x_test, y_train, y_test = train_test_split(features_dummies, Y, test_size=0.3, random_state=1)\n# 1 is just any random seed number\nx_train.head()",
"_____no_output_____"
]
],
[
[
"# 9. Predict Target Variable for Test Data using Sklearn",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\n\nfrom sklearn.linear_model import LogisticRegression\n\n# Fit the model on train\nmodel = LogisticRegression(solver=\"liblinear\")\nmodel.fit(x_train, y_train)\n#predict on test\ny_predict = model.predict(x_test)\n\n\ncoef_df = pd.DataFrame(model.coef_)\ncoef_df['intercept'] = model.intercept_\nprint(coef_df)\n\nmodel_score = model.score(x_test, y_test)\nprint(model_score)\n",
" 0 1 2 3 4 5 6 \\\n0 -0.041797 0.043586 -0.157831 -0.422672 -0.034209 -0.563308 0.017014 \n\n 7 8 9 ... 21 22 23 24 \\\n0 0.346878 0.174819 -0.051326 ... -0.403625 0.294687 -0.187775 -0.502496 \n\n 25 26 27 28 29 intercept \n0 -0.184918 0.119631 0.249453 -0.603868 -0.345718 -0.580503 \n\n[1 rows x 31 columns]\n0.7333333333333333\n"
]
],
[
[
"# 10. Try calculating Probabilities of Target using Sklearn",
"_____no_output_____"
]
],
[
[
"pred = model.predict_proba(x_train)\nprint(pred)",
"[[0.64025335 0.35974665]\n [0.87817778 0.12182222]\n [0.56832555 0.43167445]\n ...\n [0.84020629 0.15979371]\n [0.75223322 0.24776678]\n [0.89498895 0.10501105]]\n"
],
[
"preddf = pd.DataFrame(data = y_predict, index = None, columns = ['Risk_pred'])\npreddf.head()\np_copy = pdata.copy()\ncomparedf = pd.merge(p_copy, preddf, how='outer', left_index=True, right_index=True)\ncomparedf.head(20)\n",
"_____no_output_____"
]
],
[
[
"# 11. Build a confusion matrix and display the same using heatmap plot",
"_____no_output_____"
]
],
[
[
"cm=metrics.confusion_matrix(y_test, y_predict, labels=[1, 0])\n\ndf_cm = pd.DataFrame(cm, index = [i for i in [\"1\",\"0\"]],\n columns = [i for i in [\"Predict 1\",\"Predict 0\"]])\nplt.figure(figsize = (10,8))\nsns.heatmap(df_cm, annot=True)",
"_____no_output_____"
]
],
[
[
"# 12. Make a predicted probability distribution plot for defaulters and non defaulters",
"_____no_output_____"
]
],
[
[
"preddf.head()",
"_____no_output_____"
]
],
[
[
"# 13. Give your conclusion for the problem. Which evaluation metric would you choose? ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec896e9c55c10d05c2f577613361e6b75f932972 | 12,274 | ipynb | Jupyter Notebook | 1. The AI behind Seeing AI.ipynb | AzureAdvocateBit/PracticalAI-1 | 80a5578ddda52c88cac90164389f3e0449818eca | [
"MIT"
] | 37 | 2018-12-07T13:36:30.000Z | 2022-01-01T06:31:05.000Z | 1. The AI behind Seeing AI.ipynb | AzureAdvocateBit/PracticalAI-1 | 80a5578ddda52c88cac90164389f3e0449818eca | [
"MIT"
] | 1 | 2021-02-25T07:55:48.000Z | 2021-02-25T07:55:56.000Z | 1. The AI behind Seeing AI.ipynb | AzureAdvocateBit/PracticalAI-1 | 80a5578ddda52c88cac90164389f3e0449818eca | [
"MIT"
] | 19 | 2018-12-07T14:01:59.000Z | 2021-12-30T09:19:29.000Z | 71.360465 | 519 | 0.708815 | [
[
[
"# The AI behind Seeing AI\n\nThe Seeing AI app (available on the iOS App Store) provides tools for blind and vision-impaired people to assist with everyday tasks. Many of the functions in the app are implemented with [Cognitive Services](https://azure.microsoft.com/en-us/services/cognitive-services/?WT.mc_id=ODSC-workshop-davidsmi) in Azure.\n\nIn this workshop, we'll experiment with various [Cognitive Service APIs](https://azure.microsoft.com/en-us/services/cognitive-services/directory/vision/?WT.mc_id=ODSC-workshop-davidsmi) used in the Seeing AI app to see how they behave and learn about the type of data they return. We will be using only the web demo interfaces, so you won't need an Azure subscription and you'll only need a web browser to try them out.\n\nNo programming will be required in this part of the workshop. If you'd like to learn how to use Cognitive Services from application via the API or language SDKs, try this Microsoft Learn module: [Process images with the Computer Vision service](https://docs.microsoft.com/en-us/learn/modules/create-computer-vision-service-to-classify-images/index?WT.mc_id=ODSC-workshop-davidsmi).",
"_____no_output_____"
],
[
"## Optional: Install Seeing AI\n\nIf you have an iPhone or iPad, search for \"Seeing AI\" in the App Store to install it. (Seeing AI is not available for Android.) The app is free.",
"_____no_output_____"
],
[
"## Finding data\n\nThe [Cognitive Services](https://azure.microsoft.com/en-us/services/cognitive-services/directory/vision/?WT.mc_id=AILive-workshop-davidsmi) demo pages typically include sample data (photos, etc) you can try, but we encourage you to find your own data and see what happens. You can use a file on your laptop or the URL of a file on the Web. Here are some things you can try:\n\n* [Random Wikimedia image page](https://commons.wikimedia.org/w/index.php?title=Special:Random&prop=images). Click the download button to get a direct URL (often with a choice of resolutions).\n* [ImageNet](http://www.image-net.org/). Type a word into the search box to find images by category.\n* [Google Images](https://images.google.com/). It can be tricky to get a link to an image: click on a result and right-click on a large image (_not_ a thumbnail) to get a URL.\n* Your own media files. Unless they are accessible by URL, you'll need to download them to your laptop first (in a suitable format/size) and use the \"Browse\" button on the demo page to upload them. Microsoft does not store media submitted on these pages.",
"_____no_output_____"
],
[
"# Computer Vision \n\nThe [Computer Vision services](https://docs.microsoft.com/en-us/azure/cognitive-services/computer-vision/home?WT.mc_id=AILive-workshop-davidsmi) provide APIs related to extracting information from images: people and faces, objects, text and more. \n\n## \"Person\" feature in Seeing AI\n\nIn the Seeing AI app, the \"Person\" feature detects people in the camera frame in real time, and describes them when pressing the camera button. Try it in the app now.\n\n* Click the \"Person\" button in the Seeing AI app\n* Point the camera at a face; an audio guide will alert you when a face is in frame. (There's a button to switch cameras for a selfie.)\n* Tap the screen to get detailed information about the faces on the screen: apparent age and gender, description, expression.\n\n## Face API\n\nThe [Face API](https://azure.microsoft.com/en-us/services/cognitive-services/face/?WT.mc_id=AILive-workshop-davidsmi#detection) will detect one or more human faces in an image, and return information about the location and attribute of those faces.\n\n* Visit the [Face Detection](https://azure.microsoft.com/en-us/services/cognitive-services/face/?WT.mc_id=AILive-workshop-davidsmi#detection) page.\n* Copy the URL of an image to the \"Image URL\" box, or click \"Browse\" to upload an image, then click Submit.\n* Your image will be shown on the page with a rectangle bounding all detected faces.\n* Look at the results in the right pane to see the detailed analysis. The JSON array will include one element for each detected face.\n* The `faceRectangle` property defines the bounding box of the face(s).\n* The `faceAttributes` propery includes details about the person's appearance: the person's apparent gender and age, and other visible attributes such as hair color/baldness, type of facial hair, presence of glasses and headwear, use of makeup, etc.\n* The `faceLandmarks` property locates specific parts of the face (pupils, tip of the nose, corner of the mouth, etc) within the image\n* Check the [FACE API Reference](https://docs.microsoft.com/en-us/azure/cognitive-services/face/apireference?WT.mc_id=AILive-workshop-davidsmi) for other attributes that may be of interest.\n\nThings to try with [Face Detection](https://azure.microsoft.com/en-us/services/cognitive-services/face/?WT.mc_id=AILive-workshop-davidsmi#detection): \n\n* [Sikh man](https://upload.wikimedia.org/wikipedia/commons/thumb/a/a4/Sikh_man%2C_Agra_10.jpg/648px-Sikh_man%2C_Agra_10.jpg)\n* [Woman in comic store](https://upload.wikimedia.org/wikipedia/commons/e/e7/7.9.10OliviaMunnByLuigiNovi53.jpg)\n* [Contemptuous man](https://upload.wikimedia.org/wikipedia/commons/thumb/4/40/Man_wearing_red_sunglasses.jpg/1024px-Man_wearing_red_sunglasses.jpg)\n\n## Emotion Recognition\n\nThe Face API returns information about the subject's apparent emotion, based on the facial expression. That information is included in the `faceAttributes` property also includes and intensity of emotion (anger, happiness, surprise, etc) on a scale from 0-1.\n\nYou can try this out in the [Emotion Recognition demo](https://azure.microsoft.com/en-us/services/cognitive-services/face/?WT.mc_id=AILive-workshop-davidsmi#recognition) app.\n\nThings to try with [Emotion Recognition](https://azure.microsoft.com/en-us/services/cognitive-services/face/?WT.mc_id=AILive-workshop-davidsmi#recognition):\n\n* [Happy man](https://upload.wikimedia.org/wikipedia/commons/4/4b/Sven.Littkowski.2011.JPG)\n* [Neutral boy](https://upload.wikimedia.org/wikipedia/commons/2/20/Bangalore%2C_India_%28844327901%29.jpg)\n* [Grimacing woman](https://upload.wikimedia.org/wikipedia/commons/e/e1/Grimace_2.jpg)\n* [Angry woman](https://upload.wikimedia.org/wikipedia/commons/0/08/Angry_woman.jpg)",
"_____no_output_____"
],
[
"# Documents\n\nThe Seeing AI app provides several tools to read written text aloud:\n\n* **Short Text** speaks aloud text as it becomes visible in the camera. It's intended for recognizing simple text, like signposts and labels.\n* **Document** is intended to read the text from a page. Audio cues help the user bring the entire page into frame, and then the entire document is scanned read aloud.\n* **Handwriting** (beta) analyzes a photo of a handwritten document and reads the text aloud. \"Document\" can also read some handwriting, but this works better with handwritten notes (and worse with printed documents).\n\n## Extract Printed Text\n\nThe [OCR Service](https://azure.microsoft.com/en-us/services/cognitive-services/computer-vision/?WT.mc_id=ODSC-workshop-davidsmi#text) detects text within images, and returns the extracted text along with the rectangle locating the text within the image. It can detect text in 25 languages, even if it's at an angle with respect to the page. [Detailed documentation](https://docs.microsoft.com/en-us/azure/cognitive-services/computer-vision/concept-extracting-text-ocr?WT.mc_id=AILive-workshop-davidsmi) here. \n\nThings to try with [OCR Service](https://azure.microsoft.com/en-us/services/cognitive-services/computer-vision/#text): \n\n* [Business Card](https://upload.wikimedia.org/wikipedia/commons/e/e0/Business_Card.jpg)\n* [letter of gratitude](https://upload.wikimedia.org/wikipedia/commons/b/bd/Ukranian_contemporary_architecture_and_desing_as_part_of_world_architecture_day_in_Rigga.jpg)\n* [White House menu](https://upload.wikimedia.org/wikipedia/commons/3/32/State_Dinner_Menu_King_Hussein_of_Jordan.jpg).\n\n## Handwriting\n\nThe \"Handwriting\" tool in Seeing AI can be used to read the text in a handwritten note.\n\nThe [Handwriting Recognition](https://azure.microsoft.com/en-us/services/cognitive-services/computer-vision/?WT.mc_id=AILive-workshop-davidsmi#handwriting) API performs the same task for a picture of handrwiting.\n\nThings to try with [Handwriting Recognition](https://azure.microsoft.com/en-us/services/cognitive-services/computer-vision/?WT.mc_id=ODSC-workshop-davidsmi#handwriting):\n\n* [Analytics notepad](https://upload.wikimedia.org/wikipedia/commons/b/bd/Handwritten_Text.jpg)\n* [Letter to Lt. Boomer](https://upload.wikimedia.org/wikipedia/commons/6/60/Letter_addressed_to_Lt._General_Boomer%2C_30_May_1991_%285840049586%29.jpg)",
"_____no_output_____"
],
[
"# Scene Description\n\nThe \"Scene\" feature in Seeing AI will provide a spoken description of an image. Point the camera at an interesting scene and tap the screen to generate a description (which will also be spoken aloud). \n\nYou can also use the \"hamburger\" menu to browse photos on your iPhone.\n\n## Image Analysis\n\nProvide an image to the [Analyze an image](https://azure.microsoft.com/en-us/services/cognitive-services/computer-vision/?WT.mc_id=AILive-workshop-davidsmi#analyze) tool, and the API returns\n\n* A natural-language description of the scene (with associated confidence score, from 0%-100%)\n* A list of objects detected in the scene (with associated confidence for each)\n* Attributes of the image (black and white? line drawing? clip art?)\n* Detection of \"racy\" and \"adult\" content (with associated scores)\n* Detected faces (location and apparent age/gender -- use Face API for more details)\n* Color analysis (dominant foreground, background and accent colors)\n\nThings to try with [Analyze an image](https://azure.microsoft.com/en-us/services/cognitive-services/computer-vision/?WT.mc_id=AILive-workshop-davidsmi#analyze):\n\n* [Scarborough Harbour](https://upload.wikimedia.org/wikipedia/commons/4/4c/Scarborough_MMB_24_Harbour.jpg): \"a harbor filled with lots of small boats in a body of water\"\n* [Husky](https://upload.wikimedia.org/wikipedia/commons/8/80/Siberian_husky1.JPG): \"a dog sitting in front of a fence\"\n* [Taj Mahal](https://upload.wikimedia.org/wikipedia/commons/thumb/a/a3/Taj_Mahal-11.jpg/1024px-Taj_Mahal-11.jpg): \"a group of people walking in front of Taj Mahal\"\n* [Pyramids](https://upload.wikimedia.org/wikipedia/commons/thumb/f/f1/Egypt-Archaeologists.jpg/2048px-Egypt-Archaeologists.jpg): \"a group of people on a beach\" (confidence 66%)\n\nWe'll look at generating scene descriptions via the Computer Vision API using R in the next section.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ec89756f6feecb99f083f5af1e9db56b47ea8bd7 | 10,522 | ipynb | Jupyter Notebook | 7.16-checkpoint.ipynb | hahaha789/Ahahaha | 752033e57d56464bf2852932f46b0198c510a972 | [
"Apache-2.0"
] | null | null | null | 7.16-checkpoint.ipynb | hahaha789/Ahahaha | 752033e57d56464bf2852932f46b0198c510a972 | [
"Apache-2.0"
] | null | null | null | 7.16-checkpoint.ipynb | hahaha789/Ahahaha | 752033e57d56464bf2852932f46b0198c510a972 | [
"Apache-2.0"
] | null | null | null | 19.306422 | 78 | 0.469492 | [
[
[
"# 基本程序设计\n- 一切代码输入,请使用英文输入法",
"_____no_output_____"
],
[
"## 编写一个简单的程序\n- 圆公式面积: area = radius \\* radius \\* 3.1415",
"_____no_output_____"
]
],
[
[
"radius = eval(input(\"请输入半径:\"))\narea = radius * radius *3.1415\nprint(area)",
"radius1.1\n3.801215000000001\n"
]
],
[
[
"### 在Python里面不需要定义数据的类型",
"_____no_output_____"
],
[
"## 控制台的读取与输入\n- input 输入进去的是字符串\n- eval",
"_____no_output_____"
],
[
"- 在jupyter用shift + tab 键可以跳出解释文档",
"_____no_output_____"
],
[
"## 变量命名的规范\n- 由字母、数字、下划线构成\n- 不能以数字开头 \\*\n- 标识符不能是关键词(实际上是可以强制改变的,但是对于代码规范而言是极其不适合)\n- 可以是任意长度\n- 驼峰式命名",
"_____no_output_____"
],
[
"## 变量、赋值语句和赋值表达式\n- 变量: 通俗理解为可以变化的量\n- x = 2 \\* x + 1 在数学中是一个方程,而在语言中它是一个表达式\n- test = test + 1 \\* 变量在赋值之前必须有值",
"_____no_output_____"
],
[
"## 同时赋值\nvar1, var2,var3... = exp1,exp2,exp3...",
"_____no_output_____"
],
[
"## 定义常量\n- 常量:表示一种定值标识符,适合于多次使用的场景。比如PI\n- 注意:在其他低级语言中如果定义了常量,那么,该常量是不可以被改变的,但是在Python中一切皆对象,常量也是可以被改变的",
"_____no_output_____"
],
[
"## 数值数据类型和运算符\n- 在Python中有两种数值类型(int 和 float)适用于加减乘除、模、幂次\n<img src = \"../Photo/01.jpg\"></img>",
"_____no_output_____"
],
[
"## 运算符 /、//、**",
"_____no_output_____"
],
[
"## 运算符 %",
"_____no_output_____"
],
[
"## EP:\n- 25/4 多少,如果要将其转变为整数该怎么改写\n- 输入一个数字判断是奇数还是偶数\n- 进阶: 输入一个秒数,写一个程序将其转换成分和秒:例如500秒等于8分20秒\n- 进阶: 如果今天是星期六,那么10天以后是星期几? 提示:每个星期的第0天是星期天",
"_____no_output_____"
]
],
[
[
"day=int(input(\"输入一个日期\"))\ndays=day/7\nif days=1:\n else ",
"_____no_output_____"
],
[
"time=int(input(\"请输入一个秒数:\"))\nmin=int(time/60)\nsecond=time%60\nprint(min,\"分\",second,\"秒\")",
"请输入一个秒数:50\n0 分 50 秒\n"
],
[
"num = 24/5\nprint (int(num))",
"4\n"
],
[
"number = int(input(\"输入一个数字\"))\nif(number%2==0):\n print(\"是偶数\")\nelse:\n print(\"是奇数\")",
"输入一个数字55\n是奇数\n"
]
],
[
[
"## 科学计数法\n- 1.234e+2\n- 1.234e-2",
"_____no_output_____"
],
[
"## 计算表达式和运算优先级\n<img src = \"../Photo/02.png\"></img>\n<img src = \"../Photo/03.png\"></img>",
"_____no_output_____"
]
],
[
[
"su=10*(y-5)(a+b+c)\nsu2=4/x\nsum=(3+4*x)/5-su/x+9*(su2+)",
"_____no_output_____"
]
],
[
[
"## 增强型赋值运算\n<img src = \"../Photo/04.png\"></img>",
"_____no_output_____"
],
[
"## 类型转换\n- float -> int\n- 四舍五入 round",
"_____no_output_____"
],
[
"## EP:\n- 如果一个年营业税为0.06%,那么对于197.55e+2的年收入,需要交税为多少?(结果保留2为小数)\n- 必须使用科学计数法",
"_____no_output_____"
],
[
"# Project\n- 用Python写一个贷款计算器程序:输入的是月供(monthlyPayment) 输出的是总还款数(totalpayment)\n",
"_____no_output_____"
],
[
"# Homework\n- 1\n<img src=\"../Photo/06.png\"></img>",
"_____no_output_____"
]
],
[
[
"ce=eval(input(\"Enter a degree in Celsius:\"))\nfa=(9/5)*ce+32\nprint(ce,\"Celsius is\",fa,\"Fahrenheit\")",
"Enter a degree in Celsius:43\n43 Celsius is 109.4 Fahrenheit\n"
]
],
[
[
"- 2\n<img src=\"../Photo/07.png\"></img>",
"_____no_output_____"
]
],
[
[
"import math\nr,le=eval(input(\"Enter the radius and length of a cylinder:\"))\narea=r*r*math.pi\nvolume = area * le\nprint(\"The area is\",round(area,4))\nprint(\"The volume is\",round(volume,1))",
"Enter the radius and length of a cylinder:5.5,12\nThe area is 95.0332\nThe volume is 1140.4\n"
]
],
[
[
"- 3\n<img src=\"../Photo/08.png\"></img>",
"_____no_output_____"
]
],
[
[
"feet=eval(input(\"Enter a value for feet:\"))\nprint(feet,\"feet is\",feet*0.305,\"meters\")",
"Enter a value for feet:16.5\n16.5 feet is 5.0325 meters\n"
]
],
[
[
"- 4\n<img src=\"../Photo/10.png\"></img>",
"_____no_output_____"
]
],
[
[
"M=eval(input(\"Enter the amount of water in kilograms:\"))\nini=eval(input(\"Enter ther initial temperature:\"))\nfinal=eval(input(\"Enter the final temperature:\"))\nQ=M*(final-ini)*4184\nprint(\"The energy needed is\",Q)",
"Enter the amount of water in kilograms:55.5\nEnter ther initial temperature:3.5\nEnter the final temperature:10.5\n<class 'float'>\nThe energy needed is 1625484.0\n"
]
],
[
[
"- 5\n<img src=\"../Photo/11.png\"></img>",
"_____no_output_____"
]
],
[
[
"c,n=eval(input(\"Enter balance and interest rate (e.g.,3 for 3%):\"))\nli=1000 * (3.5/1200)\nprint(\"The interest is\",round(li,5))",
"Enter balance and interest rate (e.g.,3 for 3%):1000,3.5\nThe interest is 2.91667\n"
]
],
[
[
"- 6\n<img src=\"../Photo/12.png\"></img>",
"_____no_output_____"
]
],
[
[
"v0,v1,t=eval(input(\"Enter v0,v1,and t :\"))\na=(v1-v0)/t\nprint(\"The average acceleration is\",round(a,4))",
"Enter v0,v1,and t :5.5,50.9,4.5\nThe average acceleration is 10.0889\n"
]
],
[
[
"- 7 进阶\n<img src=\"../Photo/13.png\"></img>",
"_____no_output_____"
]
],
[
[
"money=eval(input(\"Enter the monthly saving amount:\"))\nt1=money*(1+0.00417)\nt2=(money+t1)*(1+0.00417)\nt3=(money+t2)*(1+0.00417)\nt4=(money+t3)*(1+0.00417)\nt5=(money+t4)*(1+0.00417)\nt6=(money+t5)*(1+0.00417)\nprint(round(t6,2))",
"Enter the monthly saving amount:100\n608.82\n"
]
],
[
[
"- 8 进阶\n<img src=\"../Photo/14.png\"></img>",
"_____no_output_____"
]
],
[
[
"num=eval(input(\"Enter a number between 0 and 1000:\"))\nfirst_num=num//100\nthrid_num=num%10\nse_num=num//10%10\nif num>0 and num<1000:\n print(\"The sum of the digits is\",first_num+thrid_num+se_num)\nelse:\n print(\"输入有误\")",
"Enter a number between 0 and 1000:563\nThe sum of the digits is 14\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec899807e0cbb2fca8ff7d42a562f1bf85f6b3d5 | 31,928 | ipynb | Jupyter Notebook | 01-1. Python ch.01 ~ Q. ~21 (200512) .ipynb | Adrian123K/python_class | 11fb52d3ca3a84ee582ec9da68b463fac0f485fe | [
"MIT"
] | null | null | null | 01-1. Python ch.01 ~ Q. ~21 (200512) .ipynb | Adrian123K/python_class | 11fb52d3ca3a84ee582ec9da68b463fac0f485fe | [
"MIT"
] | null | null | null | 01-1. Python ch.01 ~ Q. ~21 (200512) .ipynb | Adrian123K/python_class | 11fb52d3ca3a84ee582ec9da68b463fac0f485fe | [
"MIT"
] | null | null | null | 19.696484 | 305 | 0.389439 | [
[
[
"#### ■ 파이썬 수업\n\n 목표 : 빅데이터를 활용한 데이터 분석가 또는 딥러닝 개발자, ML 엔지니어\n\n 갖추어야할 기본적 기술 \n\n 1. SQL\n 2. Python\n 3. R\n 4. ML\n 5. DL\n 6. 강화학습\n \n * 파이썬을 통해서 이루고자 하는 목표\n\n 1. ML 데이터 분석에 필요한 파이썬 활용능력\n 2. 데이터 분석을 위한 데이터 수집 (웹 크롤링)\n 3. DL을 통해 이미지 분류 (불량품 선별,)\n 4. 강화학습을 통해서 인공지능 게임 개발, 인공지능 주식 트레이딩\n\n#### ■ 파이썬 프로그램 실행하는 법\n\n 1. spider -> 한 라인씩 드래그해서 ctrl+enter, F9\n 2. jupyter notebook -> ctrl+enter\n 3. pycharm -> ctrl+shift+F10 / alt+shift+e (한 라인씩 실행)",
"_____no_output_____"
],
[
"### ■ 예제 1. 대화식 모드로 프로그래밍 하기\n\n* 파이썬을 실행하는 방법 2가지\n 1. 대화식 모드\n 2. 배치모드\n\n파이썬의 인터프리터가 위의 코드 문법의 오류가 없는지 체크하고 실행 \n* 아나콘다(Anaconda) : \n 기본 python이 내장되어있음 \n 각종 필요한 패키지들이 다 포함되어 있음",
"_____no_output_____"
],
[
"### ※ 문제1. 아래의 스크립트를 배치 모드로 실행하시오 \n2 \n3 \n4",
"_____no_output_____"
],
[
"### ■ 예제 2. 텍스트 에디터로 프로그래밍하기\n\n spyder 프로그램에서 ctrl+enter 로 코드를 실행 또는 F9를 누른다\n\n * #%% : 실행 영역 구분 시 사용",
"_____no_output_____"
],
[
"### ※ 문제2. 아래의 결과가 나오게 spyder에서 프로그래밍 하시오\n2 \n3 \n4",
"_____no_output_____"
]
],
[
[
"for i in[2,3,4]:\n print(i)",
"2\n3\n4\n"
]
],
[
[
"### ■ 예제 3. 변수명 만들기\n\n 어떤 값을 임시로 저장하는 변수의 이름을 만드는 방법과 규칙을 배운다\n\n#### ※ 예시\na=1 : a라는 변수에 1을 할당",
"_____no_output_____"
],
[
"#### ※ 변수 이름 생성 시 주의 사항\n 1. 변수 이름에는 다음 문자만 사용할 수 있다.\n - 소문자(a~z)\n - 대문자(A~Z)\n - 숫자 (0~9)\n - 언더스코어(_)\n 2. 변수 이름은 숫자로 시작할 수 없다.\n 3. 예약어를 사용할 수 없다. (파이썬에서 이미 사용되고 있는 단어들)",
"_____no_output_____"
],
[
"### ※ 문제3. 파이썬의 예약어가 무엇이 있는지 출력",
"_____no_output_____"
]
],
[
[
"import keyword\nprint(keyword.kwlist)",
"['False', 'None', 'True', 'and', 'as', 'assert', 'async', 'await', 'break', 'class', 'continue', 'def', 'del', 'elif', 'else', 'except', 'finally', 'for', 'from', 'global', 'if', 'import', 'in', 'is', 'lambda', 'nonlocal', 'not', 'or', 'pass', 'raise', 'return', 'try', 'while', 'with', 'yield']\n"
]
],
[
[
"#### ■ 예제4. 변수에 값 대입\n - 다양한 값을 변수에 대입하는 방법\n - 파이썬은 변수에 값을 대입할 때 = (assignment) 기호를 사용",
"_____no_output_____"
],
[
"### ※ 문제4. 위의 예약어를 변수로 사용하면 에러가 나는지 직접 테스트",
"_____no_output_____"
]
],
[
[
"and =1",
"_____no_output_____"
]
],
[
[
"### ※ 예시",
"_____no_output_____"
]
],
[
[
"a=1\nb='scott'\nprint(b)",
"scott\n"
]
],
[
[
"파이썬의 코드는 심플함을 철학으로 삼고 있다. \n파이썬의 기본 철학을 확인하는 방법",
"_____no_output_____"
]
],
[
[
"import this",
"The Zen of Python, by Tim Peters\n\nBeautiful is better than ugly.\nExplicit is better than implicit.\nSimple is better than complex.\nComplex is better than complicated.\nFlat is better than nested.\nSparse is better than dense.\nReadability counts.\nSpecial cases aren't special enough to break the rules.\nAlthough practicality beats purity.\nErrors should never pass silently.\nUnless explicitly silenced.\nIn the face of ambiguity, refuse the temptation to guess.\nThere should be one-- and preferably only one --obvious way to do it.\nAlthough that way may not be obvious at first unless you're Dutch.\nNow is better than never.\nAlthough never is often better than *right* now.\nIf the implementation is hard to explain, it's a bad idea.\nIf the implementation is easy to explain, it may be a good idea.\nNamespaces are one honking great idea -- let's do more of those!\n"
]
],
[
[
"#### 파이썬은 c/c++, java와는 달리 변수를 선언할 때 숫자형 자료인지 문자형 자료인지 \n#### 자료형을 명시하지 않아도 된다.",
"_____no_output_____"
],
[
" PL/SQL python \n a number(10) :=2; a=2",
"_____no_output_____"
],
[
"### ※ 문제5. 변수의 자료형을 확인하시오\nb='scott' \nprint(type(b))",
"_____no_output_____"
]
],
[
[
"b='scott'\nprint(type(b))",
"<class 'str'>\n"
]
],
[
[
"### ※ 문제6. 숫자형 변수의 자료형을 확인하시오",
"_____no_output_____"
]
],
[
[
"a=1\nprint(type(a))",
"<class 'int'>\n"
]
],
[
[
"### ■ 예제5. 주석처리하기(#) \n프로그램에서 주석부분은 인터프리터에 의해 무시되는 텍스트의 한 부분이다. \n코드를 설명하거나 나중에 어떤 문제를 고치기 위해 표시하는 등 다양항 목적으로 주석을 사용할 수 있다. \n코드를 작성할 때 주석을 잘 작성해두면 차후에 코드를 다시 보거나 타인이 코드를 검토할 때 매우 중요한 정보로 활용이 된다. \n그래서 주석을 항상 달아주는 습관을 가지고 있는 것이 좋다.",
"_____no_output_____"
]
],
[
[
"# 한줄 주석\na=1 #변수 a에 1을 할당합니다.\nprint(a) # a의 결과를 출력합니다",
"1\n"
],
[
"# 여러줄 주석\n\"\"\" 아래의 프로그램은 a 변수에 1을 할당해서 프린트하는 프로그램 입니다.\"\"\"",
"_____no_output_____"
]
],
[
[
"### ■ 예제6. 자료형 개념배우기 \n 자료형이란 프로그래밍을 할 때 쓰이는 숫자, 문자열 등의 자료 형태로 사용되는 모든 것을 뜻한다. \n#### - 파이썬에서 자주 다루게 되는 자료형이 5가지가 있다.\n #### 1. 숫자형 자료형 \n - 숫자를 표현하는 자료형. (예: a=1)\n #### 2. 문자형 자료형\n - 문자를 표현하는 자료형. (예: b='scott')\n #### 3. 리스트 자료형\n - []안에 임의 객체를 순서있게 나열한 자료형 \n d=[1,2,3] \n print(d) \n print(d[0])\n #### 4. 튜플 자료형\n - 리스트와 비슷하지만 요소값을 변경할 수 없다는 것이 리스트와 다른 점 \n c=(1,2,3) -> 안의 값을 변경할 수 없음 \n print(c)\n #### 5. 사전 자료형\n - {} 안에 키:값으로 된 쌍이 요소로 구성된 순서가 없는 자료형 \n m={ 'i':'나는', 'am':'입니다', 'boy':'소년'} \n print(m)",
"_____no_output_____"
]
],
[
[
"m={'i':'나는','am':'입니다','boy':'소년'}\nprint(m)",
"{'i': '나는', 'am': '입니다', 'boy': '소년'}\n"
]
],
[
[
"### ※ 문제7.(점심시간 문제) 아래의 리스트에서 문자 k를 출력하시오 \n['a','b','d','e','k','m','n','z']",
"_____no_output_____"
]
],
[
[
"t=['a','b','d','e','k','m','n','z']\nprint(t[4])",
"_____no_output_____"
]
],
[
[
"### ■ 예제7. 자료형 출력 개념 배우기(print) \nprint 함수를 이용하면 다양한 자료형을 화면에 출력할 수 있다.",
"_____no_output_____"
]
],
[
[
"# 예시\na=200\nb='i love python'\nc=['a','b','c']\nprint(a)\nprint(b)\nprint(c)",
"200\ni love python\n['a', 'b', 'c']\n"
]
],
[
[
"#### ※ 대화식 모드에서는 print 없이 출력이 가능",
"_____no_output_____"
],
[
"### ■ 예제8. 들여쓰기 개념 배우기 \n#### 파이썬에는 실행코드 부분을 묶어주는 () 괄호가 없다 \n-> 들여쓰기로 괄호를 대신 \n\n파이썬은 다른 프로그래밍 언어와 달리 if, for, while 등과 같은 제어문이나 루프문의 실행코드 부분을 구분해주는 괄호가 없음 \n들여쓰기로 괄호를 대신 \n\n파이썬에서 제어문이나 함수 이름, 클래스 이름 뒤에 : 콜론 으로 제어문, 함수이름, 클래스 이름의 끝을 표시하며 \n: 다음에 실행코드를 작성하는 데 이때 들여쓰기를 해야함",
"_____no_output_____"
],
[
"### 예제:\n 1. 실행코드를 다음 라인에 작성했을 경우",
"_____no_output_____"
]
],
[
[
"listdata=['a','b','c']\nif 'a' in listdata:\n print('a가 listdata에 있습니다.')",
"a가 listdata에 있습니다.\n"
]
],
[
[
"※ 콜론(:)을 쓴 다음 라인에서는 무조건 들여쓰기 4칸",
"_____no_output_____"
],
[
" 2. 실행코드를 한 라인에 작성한 경우",
"_____no_output_____"
]
],
[
[
"listdata=['a','b','c']\nif 'a' in listdata: print('a가 listdata에 있습니다.')",
"a가 listdata에 있습니다.\n"
]
],
[
[
"※ 콜론(:)으로 if문의 끝을 알린다.",
"_____no_output_____"
],
[
"### ■ 예제9. if문 개념 배우기 (if ~ else) \n#### 어떤 조건을 참과 거짓으로 판단할 때 if문을 사용 \n참과 거짓을 구분하여 코드를 실행하면 if~else를 사용 \n코드를 작성하다보면 조건에 따라 수행하는 일을 달리 해야하는 경우가 있는 데 \n조건이 참인지 거짓인지를 검사하고, 참인 경우와 거짓인 경우 실행해야 하는 일을 구분하여 처리할 수 있다.",
"_____no_output_____"
],
[
"#### ※ 예시\n```python \nif 조건: \n 실행코드1 \nelse: \n 실행코드2\n```",
"_____no_output_____"
],
[
"#### ※ 예제: \n```python\na=int(input('숫자를 입력하세요~'))\n```",
"_____no_output_____"
]
],
[
[
"a=int(input('숫자를 입력하세요~'))\nif a%2==0:\n print('짝수 입니다')\nelse:\n print('홀수 입니다')",
"숫자를 입력하세요~20\n짝수 입니다\n"
]
],
[
[
"### ※ 문제8. 숫자 두 개를 아래와 같이 각각 물어보게하고 아래처럼 메세지가 출력되게 하시오 \n첫번째 숫자를 입력하세요 ~ 2 \n두번째 숫자를 입력하세요 ~ 3 \n2는 3보다 작습니다.",
"_____no_output_____"
]
],
[
[
"a=int(input('첫번째 숫자를 입력하세요~ '))\nb=int(input('두번째 숫자를 입력하세요~ '))\nif b>a:\n print(str(a)+'는 '+str(b)+'보다 작습니다.')\nelse:\n print(str(a)+'는(은) '+str(b)+'보다 큽니다')",
"첫번째 숫자를 입력하세요~ 2\n두번째 숫자를 입력하세요~ 3\n2는 3보다 작습니다.\n"
]
],
[
[
"### ■ 예제10. if문 개념 배우기 (if ~elif) \n#### 여러 개의 조건을 순차적으로 체크하고 해당 조건이 참이면 특정 로직을 수행하고자 할 때 if~elif 문을 사용 ",
"_____no_output_____"
],
[
"#### ※ 예제:\n```python\nif 조건1:\n 실행코드1\nelif 조건2:\n 실행코드2\nelse:\n 실행코드3 # 위의 조건1과 조건2가 아니라면\n```",
"_____no_output_____"
],
[
"### ※ 문제9. 문제 8번을 다시 수행하는데 elif문을 이용해서 \n### 두 개의 숫자가 같을 때의 메세지도 아래와 같이 출력되게 하시오 \n첫번째 숫자를 입력하세요 ~ 5 \n두번째 숫자를 입력하세요 ~ 5 \n5와 5는 같습니다.",
"_____no_output_____"
]
],
[
[
"a=int(input('첫번째 숫자를 입력하세요~'))\nb=int(input('두번째 숫자를 입력하세요~'))\nif a>b:\n print(f'{a}는 {b}보다 작습니다')\nelif a<b:\n print(f'{a}는 {b}보다 큽니다')\nelse:\n print(f'{a}와 {b}는 같습니다')",
"첫번째 숫자를 입력하세요~5\n두번째 숫자를 입력하세요~5\n5와 5는 같습니다\n"
]
],
[
[
"### ■ 예제11. for문 개념 배우기 Ⅰ \n#### 특정 코드를 반복적으로 수행하기 위해서는 반복문을 사용해야 하는데 파이썬에서는 for문이 반복문을 수행하기 위해 가장 많이 사용되는 문법",
"_____no_output_____"
],
[
"### ※ 예제1)\n```python\nfor 변수 in 범위:\n 반복적으로 실행할 코드\n```\n#### 1. 리스트 범위인 경우\n```python\nfor i in [1,2,3]:\n print(i)\n```\n#### 2. 튜플 범위인 경우\n```python\nfor i in (1,2,3):\n print(i)\n```\n#### 3. range() 범위인 경우\n```python\nfor i in range(10):\n print(i)\n```\n#### 4. 사전형 범위인 경우\n```python\nm={'i':'나는','am':'입니다','boy':'소년'}\nfor i in m:\n print(i)\n```\n#### 5. 문자형이 범위인 경우\n```python\nfor i in 'I am a boy':\n print(i)\n```",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(i)",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n"
],
[
"m={'i':'나는','am':'입니다','boy':'소년'}\nfor i in m:\n print(i)",
"i\nam\nboy\n"
]
],
[
[
"### ■ for 루프문의 range 사용법 ",
"_____no_output_____"
]
],
[
[
"for i in range(6):\n print(i)",
"0\n1\n2\n3\n4\n5\n"
],
[
"for i in range(1,6):\n print(i)",
"1\n2\n3\n4\n5\n"
],
[
"for i in range(1,6,2):\n print(i)",
"1\n3\n5\n"
],
[
"for i in range(6,1,-1):\n print(i)",
"6\n5\n4\n3\n2\n"
]
],
[
[
"### ※ 문제10. ★ 별을 5개 출력하시오",
"_____no_output_____"
]
],
[
[
"print('★'*5)",
"★★★★★\n"
]
],
[
[
"### ※ 문제11.(파이썬 알고리즘 1번) 아래와 같이 결과가 출력되는 파이썬 코드를 작성하시오 \n숫자를 입력하세요 ~ 5 \n★ \n★★ \n★★★ \n★★★★ \n★★★★★",
"_____no_output_____"
]
],
[
[
"a=int(input('숫자를 입력하세요 ~ '))\nfor i in range(a+1):\n print('★'*i)",
"숫자를 입력하세요 ~ 10\n\n★\n★★\n★★★\n★★★★\n★★★★★\n★★★★★★\n★★★★★★★\n★★★★★★★★\n★★★★★★★★★\n★★★★★★★★★★\n"
]
],
[
[
"### ※ 문제12.(파이썬 알고리즘 2번) 아래와 같이 결과가 출력되는 파이썬 코드를 작성하시오 \n가로의 숫자를 입력하세요 ~ 5 \n세로의 숫자를 입력하세요 ~ 6 \n★★★★★ \n★★★★★ \n★★★★★ \n★★★★★ \n★★★★★ \n★★★★★",
"_____no_output_____"
]
],
[
[
"a=int(input('가로의 숫자를 입력하세요 ~ '))\nb=int(input('세로의 숫자를 입력하세요 ~ '))\nfor i in range(b):\n print('★'*a)",
"가로의 숫자를 입력하세요 ~ 5\n세로의 숫자를 입력하세요 ~ 6\n★★★★★\n★★★★★\n★★★★★\n★★★★★\n★★★★★\n★★★★★\n"
]
],
[
[
"### ※ 문제13. (파이썬 알고리즘 문제 3번)숫자를 물어보게 하고 숫자를 입력하면 ★가 아래와 같이 출력되게 하시오 \n숫자를 입력하세요 ~ 7 \n★★★★★★★ \n★★★★★★ \n★★★★★ \n★★★★ \n★★★ \n★★ \n★",
"_____no_output_____"
]
],
[
[
"a=int(input('숫자를 입력하세요 ~ '))\nfor i in range(a,0,-1):\n print('★'*i)",
"숫자를 입력하세요 ~ 7\n★★★★★★★\n★★★★★★\n★★★★★\n★★★★\n★★★\n★★\n★\n"
]
],
[
[
"### ※ 문제14.(파이썬 알고리즘 문제 4번) 아래와 같이 숫자를 물어보게 하고 ★를 출력하게 하시오 \n숫자를 입력하세요 ~ 5 \n ★ \n ★★ \n ★★★ \n ★★★★ \n★★★★★",
"_____no_output_____"
]
],
[
[
"a=int(input('숫자를 입력하세요 ~ '))\nfor i in range(a+1):\n print(' '*(a-i)+'★'*i)",
"숫자를 입력하세요 ~ 5\n \n ★\n ★★\n ★★★\n ★★★★\n★★★★★\n"
]
],
[
[
"### ※ 문제15.(파이썬 알고리즘 문제 5번) 구구단 2단을 출력하시오 \n2 x 1 = 2 \n2 x 2 = 4 \n...",
"_____no_output_____"
]
],
[
[
"for i in range(1,10):\n print(str(2)+' x '+str(i)+' = '+str(2*i))\nprint('----------------')\nfor i in range(1,10):\n print(2,'x',i,'=',2*i)",
"2 x 1 = 2\n2 x 2 = 4\n2 x 3 = 6\n2 x 4 = 8\n2 x 5 = 10\n2 x 6 = 12\n2 x 7 = 14\n2 x 8 = 16\n2 x 9 = 18\n----------------\n2 x 1 = 2\n2 x 2 = 4\n2 x 3 = 6\n2 x 4 = 8\n2 x 5 = 10\n2 x 6 = 12\n2 x 7 = 14\n2 x 8 = 16\n2 x 9 = 18\n"
]
],
[
[
"### ■ 중첩 for loop문 ",
"_____no_output_____"
],
[
"### ※ 문제16.(파이썬 알고리즘 문제 6번) 구구단 2단부터 9단까지 출력하시오 \n2 x 1 = 2 \n2 x 2 = 4 \n... \n9 x 9 = 81 ",
"_____no_output_____"
]
],
[
[
"for i in range(2,10):\n for j in range(1,10):\n print(i,'x',j,'=',i*j)\nprint('----------------------------')\nprint('\\n'.join([f'{i} x {j} = {i*j}' for i in range(2,10) for j in range(1,10)]))",
"2 x 1 = 2\n2 x 2 = 4\n2 x 3 = 6\n2 x 4 = 8\n2 x 5 = 10\n2 x 6 = 12\n2 x 7 = 14\n2 x 8 = 16\n2 x 9 = 18\n3 x 1 = 3\n3 x 2 = 6\n3 x 3 = 9\n3 x 4 = 12\n3 x 5 = 15\n3 x 6 = 18\n3 x 7 = 21\n3 x 8 = 24\n3 x 9 = 27\n4 x 1 = 4\n4 x 2 = 8\n4 x 3 = 12\n4 x 4 = 16\n4 x 5 = 20\n4 x 6 = 24\n4 x 7 = 28\n4 x 8 = 32\n4 x 9 = 36\n5 x 1 = 5\n5 x 2 = 10\n5 x 3 = 15\n5 x 4 = 20\n5 x 5 = 25\n5 x 6 = 30\n5 x 7 = 35\n5 x 8 = 40\n5 x 9 = 45\n6 x 1 = 6\n6 x 2 = 12\n6 x 3 = 18\n6 x 4 = 24\n6 x 5 = 30\n6 x 6 = 36\n6 x 7 = 42\n6 x 8 = 48\n6 x 9 = 54\n7 x 1 = 7\n7 x 2 = 14\n7 x 3 = 21\n7 x 4 = 28\n7 x 5 = 35\n7 x 6 = 42\n7 x 7 = 49\n7 x 8 = 56\n7 x 9 = 63\n8 x 1 = 8\n8 x 2 = 16\n8 x 3 = 24\n8 x 4 = 32\n8 x 5 = 40\n8 x 6 = 48\n8 x 7 = 56\n8 x 8 = 64\n8 x 9 = 72\n9 x 1 = 9\n9 x 2 = 18\n9 x 3 = 27\n9 x 4 = 36\n9 x 5 = 45\n9 x 6 = 54\n9 x 7 = 63\n9 x 8 = 72\n9 x 9 = 81\n---------\n2 x 1 = 2\n2 x 2 = 4\n2 x 3 = 6\n2 x 4 = 8\n2 x 5 = 10\n2 x 6 = 12\n2 x 7 = 14\n2 x 8 = 16\n2 x 9 = 18\n3 x 1 = 3\n3 x 2 = 6\n3 x 3 = 9\n3 x 4 = 12\n3 x 5 = 15\n3 x 6 = 18\n3 x 7 = 21\n3 x 8 = 24\n3 x 9 = 27\n4 x 1 = 4\n4 x 2 = 8\n4 x 3 = 12\n4 x 4 = 16\n4 x 5 = 20\n4 x 6 = 24\n4 x 7 = 28\n4 x 8 = 32\n4 x 9 = 36\n5 x 1 = 5\n5 x 2 = 10\n5 x 3 = 15\n5 x 4 = 20\n5 x 5 = 25\n5 x 6 = 30\n5 x 7 = 35\n5 x 8 = 40\n5 x 9 = 45\n6 x 1 = 6\n6 x 2 = 12\n6 x 3 = 18\n6 x 4 = 24\n6 x 5 = 30\n6 x 6 = 36\n6 x 7 = 42\n6 x 8 = 48\n6 x 9 = 54\n7 x 1 = 7\n7 x 2 = 14\n7 x 3 = 21\n7 x 4 = 28\n7 x 5 = 35\n7 x 6 = 42\n7 x 7 = 49\n7 x 8 = 56\n7 x 9 = 63\n8 x 1 = 8\n8 x 2 = 16\n8 x 3 = 24\n8 x 4 = 32\n8 x 5 = 40\n8 x 6 = 48\n8 x 7 = 56\n8 x 8 = 64\n8 x 9 = 72\n9 x 1 = 9\n9 x 2 = 18\n9 x 3 = 27\n9 x 4 = 36\n9 x 5 = 45\n9 x 6 = 54\n9 x 7 = 63\n9 x 8 = 72\n9 x 9 = 81\n"
]
],
[
[
"### ■ 예제12. for문 개념 배우기 Ⅱ (for ~ continue ~ break) \n#### for 반복문 내에서 continue를 만나면 그 다음 반복 실행으로 넘어가며 break를 만나면 for 반복문을 완전히 벗어난다. ",
"_____no_output_____"
],
[
"### 예제1: \n```python\nfor 변수 in 범위:\n ....\n continue # 다음 반복문 수행\n ....\n break # for 반복문을 탈출\n```",
"_____no_output_____"
],
[
"### 문제17. 숫자 1번부터 10번까지 출력하는데 중간에 숫자 5는 출력되지 않게 하시오",
"_____no_output_____"
]
],
[
[
"for i in range(1,11):\n if i==5:\n continue\n print(i)",
"1\n2\n3\n4\n6\n7\n8\n9\n10\n"
]
],
[
[
"### ※ 문제18. 숫자 0부터 9까지 출력하는데 홀수는 출력하지 마시오",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n if i%2==1: continue\n print(i)",
"0\n2\n4\n6\n8\n"
],
[
"print('\\n'.join([f'{i}' for i in range(10) if i%2==0]))",
"0\n2\n4\n6\n8\n"
]
],
[
[
"### ■ for 문에서 사용하는 break 문 \n#### loop문을 중단시키는 역할하는 키워드",
"_____no_output_____"
]
],
[
[
"# 예제\nscope=[1,2,3,4,5,6]\nfor i in scope:\n print(i)\n if i==3:\n break",
"1\n2\n3\n"
],
[
"# 예제\nscope=[1,2,3,4,5,6]\nfor i in scope:\n if i==3:\n continue\n elif i==5:\n break\n print(i)",
"1\n2\n4\n"
]
],
[
[
"### ※ 문제19. 숫자를 1번부터 100번까지 출력하는 for loop문을 작성하는데 \n### 숫자를 물어보게 하고 입력된 숫자까지 1번부터 출력되게 하시오 \n숫자를 입력하세요 ~ 7 \n1 \n2 \n3 \n4 \n5 \n6",
"_____no_output_____"
]
],
[
[
"a=int(input('숫자를 입력하세요 ~ '))\nfor i in range(1,a):\n if i==7:\n break\n print(i)",
"숫자를 입력하세요 ~ 7\n1\n2\n3\n4\n5\n6\n"
]
],
[
[
"### ※ 문제20.(파이썬 알고리즘 문제 7번) 아래와 같이 2개의 숫자를 물어보게 하고 \n### 두 개의 숫자의 최대공약수를 출력하시오 \n힌트 : 16%i==0 and 24%i==0 ",
"_____no_output_____"
]
],
[
[
"a=int(input('첫번째 숫자를 입력하세요 ~ '))\nb=int(input('두번째 숫자를 입력하세요 ~ '))\nfor i in range(max(a,b),1,-1):\n if a%i==0 and b%i==0:\n print('최대공약수는',i,'입니다')\n break\n else:\n continue",
"첫번째 숫자를 입력하세요 ~ 24\n두번째 숫자를 입력하세요 ~ 16\n최대공약수는 8 입니다\n"
]
],
[
[
"### ※ 문제21.(생각해야할 문제)(파이썬 알고리즘 문제 8번) \n두 숫자를 물어보게 하고 최소 공배수를 출력 \n첫번째 숫자를 입력하세요 ~ 16 \n두번째 숫자를 입력하세요 ~ 24 \n최소공배수는 48입니다.",
"_____no_output_____"
]
],
[
[
"a=int(input('첫번째 숫자를 입력하세요 ~ '))\nb=int(input('두번째 숫자를 입력하세요 ~ '))\nfor i in range(max(a,b),1,-1):\n if a%i==0 and b%i==0:\n print('최소공배수는',int(a*b/i),'입니다')\n break\n else:\n continue",
"첫번째 숫자를 입력하세요 ~ 16\n두번째 숫자를 입력하세요 ~ 24\n최소공배수는 48 입니다\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec89a1570a6433a022001273086aff3ffdd003a6 | 12,327 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Oneshot-checkpoint.ipynb | barnrang/omniglot | c93d333687b1d182e1c20aa7e6798c7a0bcc2474 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Oneshot-checkpoint.ipynb | barnrang/omniglot | c93d333687b1d182e1c20aa7e6798c7a0bcc2474 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Oneshot-checkpoint.ipynb | barnrang/omniglot | c93d333687b1d182e1c20aa7e6798c7a0bcc2474 | [
"MIT"
] | null | null | null | 86.202797 | 1,463 | 0.678186 | [
[
[
"# from tensorflow.keras.utils import np_utils\n# from tensorflow.keras import callbacks as cb\n# from tensorflow.keras.optimizers import Adam\n# from tensorflow.keras.models import load_model, Model\n# from tensorflow.keras.layers import *\n# from tensorflow.keras.models import Sequential\n# from tensorflow.keras import regularizers as rg\n# from tensorflow.keras.preprocessing.image import ImageDataGenerator\n# from tensorflow.keras import backend as K\n# from sklearn.preprocessing import LabelBinarizer\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib.image as img\nimport random\nimport tensorflow as tf\nfrom python.dataloader import loader\nfrom model import W_init, b_init, conv_net\n%matplotlib inline\n%load_ext autoreload\n%autoreload 2",
"Using TensorFlow backend.\n"
],
[
"model = conv_net()\nmodel.load_weights('model/omniglot_conv_950_1_20')",
"_____no_output_____"
],
[
"from oneshot import *\n\nall_acc = []\nfor i in range(1, 21):\n train, test = load_data(i)\n dist = retrieve_feature(model, train, test)\n label = load_label(i)\n all_acc.append(cal_acc(dist, label))\n\nnp.mean(all_acc)",
"_____no_output_____"
],
[
"0.984 ** 20",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
ec89b3b4489e92e60cf6489a315764ba0d99952a | 24,387 | ipynb | Jupyter Notebook | Avances de implementacion de algoritmos/DFS_segunda_version.ipynb | RodriCalle/cc41_tf_201915889_201910127_201917028_201718169_20141a449 | 2e69b7e95a73d5dce26ad95c5e4ef5559e773efd | [
"CC0-1.0"
] | null | null | null | Avances de implementacion de algoritmos/DFS_segunda_version.ipynb | RodriCalle/cc41_tf_201915889_201910127_201917028_201718169_20141a449 | 2e69b7e95a73d5dce26ad95c5e4ef5559e773efd | [
"CC0-1.0"
] | 41 | 2021-09-20T18:04:40.000Z | 2021-11-27T22:44:49.000Z | Avances de implementacion de algoritmos/DFS_segunda_version.ipynb | RodriCalle/cc41_tf_201915889_201910127_201917028_201718169 | 2e69b7e95a73d5dce26ad95c5e4ef5559e773efd | [
"CC0-1.0"
] | 7 | 2021-10-03T04:45:36.000Z | 2021-11-27T17:20:03.000Z | 75.036923 | 14,039 | 0.505064 | [
[
[
"import csv\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport random\nimport graphviz as gv\nimport math\nimport heapq as hq\nfrom google.colab import files",
"_____no_output_____"
],
[
"def adjlShow(L, labels=None, directed=False, weighted=False, path=[], layout=\"sfdp\"):\n g = gv.Digraph(\"G\") if directed else gv.Graph(\"G\")\n g.graph_attr[\"layout\"] = layout\n g.edge_attr[\"color\"] = \"gray\"\n g.node_attr[\"color\"] = \"orangered\"\n g.node_attr[\"width\"] = \"0.1\"\n g.node_attr[\"height\"] = \"0.1\"\n g.node_attr[\"fontsize\"] = \"8\"\n g.node_attr[\"fontcolor\"] = \"mediumslateblue\"\n g.node_attr[\"fontname\"] = \"monospace\"\n n = len(L)\n for u in range(n):\n g.node(str(u), labels[u] if labels else str(u))\n added = set()\n for v, u in enumerate(path):\n if u != None:\n g.edge(str(u), str(v), dir=\"forward\", penwidth=\"2\", color=\"orange\")\n added.add(f\"{u},{v}\")\n added.add(f\"{v},{u}\")\n if weighted:\n for u in range(n):\n for v, w in L[u]:\n if not directed and not f\"{u},{v}\" in added:\n added.add(f\"{u},{v}\")\n added.add(f\"{v},{u}\")\n g.edge(str(u), str(v), str(w))\n elif directed:\n g.edge(str(u), str(v), str(w))\n else:\n for u in range(n):\n for v in L[u]:\n if not directed and not f\"{u},{v}\" in added:\n added.add(f\"{u},{v}\")\n added.add(f\"{v},{u}\")\n g.edge(str(u), str(v))\n elif directed:\n g.edge(str(u), str(v))\n return g",
"_____no_output_____"
],
[
"def setEmptyGraph(size):\n n = size*size\n G = [ [] for _ in range(n) ]\n for x in range(size):\n for y in range(size):\n if(size > x + 1):\n G[ ((x * size) + y) ].append(((x+1) * size) + y)\n if(0 <= x - 1):\n G[ ((x * size) + y) ].append(((x-1) * size) + y)\n if(size > y + 1):\n G[ ((x * size) + y) ].append((x * size) + (y+1))\n if(0 <= y - 1):\n G[ ((x * size) + y) ].append((x * size) + (y-1))\n return G\n\ndef setKnownPoints(size, almacen, entrega):\n id = [\"empty\"]*(size*size)\n for a in almacen:\n x, y = a[0], a[1]\n pos = x*size + y\n id[pos] = \"A\"\n for a in entrega:\n x, y = a[0], a[1]\n pos = x*size + y\n id[pos] = \"E\"\n return id\n\ndef getGraphWeighted(G):\n adjlListWeighted = [[] for i in range(len(G))]\n for i in range(len(G)):\n for j in range(len(G[i])):\n adjlListWeighted[i].append( ( G[i][j], round( math.sqrt( (( (i // (len(G)/2) ) - (G[i][j] // (len(G)/2) ) )**2) + (( (i % (len(G)/2) ) - (G[i][j] % (len(G)/2) ) )**2) ) ) ) )\n return adjlListWeighted",
"_____no_output_____"
],
[
"alm = [[0,1],[1,2],[0,0],[3,0],[2,1]]\nent = [[1,0],[1,1],[2,0],[3,1]]\n\nsize = 4\n\nGp = setEmptyGraph(size)\nprint(Gp)\n\nlabels = setKnownPoints(size, alm, ent)\nprint(labels)\n\nGWp = getGraphWeighted(Gp)\nprint(GWp)\n\nadjlShow(GWp, labels=labels, weighted=True)",
"[[4, 1], [5, 2, 0], [6, 3, 1], [7, 2], [8, 0, 5], [9, 1, 6, 4], [10, 2, 7, 5], [11, 3, 6], [12, 4, 9], [13, 5, 10, 8], [14, 6, 11, 9], [15, 7, 10], [8, 13], [9, 14, 12], [10, 15, 13], [11, 14]]\n['A', 'A', 'empty', 'empty', 'E', 'E', 'A', 'empty', 'E', 'A', 'empty', 'empty', 'A', 'E', 'empty', 'empty']\n[[(4, 4), (1, 1)], [(5, 4), (2, 1), (0, 1)], [(6, 4), (3, 1), (1, 1)], [(7, 4), (2, 1)], [(8, 4), (0, 4), (5, 1)], [(9, 4), (1, 4), (6, 1), (4, 1)], [(10, 4), (2, 4), (7, 1), (5, 1)], [(11, 4), (3, 4), (6, 1)], [(12, 4), (4, 4), (9, 1)], [(13, 4), (5, 4), (10, 1), (8, 1)], [(14, 4), (6, 4), (11, 1), (9, 1)], [(15, 4), (7, 4), (10, 1)], [(8, 4), (13, 1)], [(9, 4), (14, 1), (12, 1)], [(10, 4), (15, 1), (13, 1)], [(11, 4), (14, 1)]]\n"
],
[
"graphFile = open('adjlistexample.txt', 'w')\nwith graphFile:\n writer = csv.writer(graphFile)\n writer.writerows(Gp)",
"_____no_output_____"
],
[
"def getAlmPoint(size, almacen):\n alm = []\n for a in almacen:\n x, y = a[0], a[1]\n pos = x*size + y\n alm.append(pos)\n return alm\n\ndef getCoord(pos, size):\n x = pos // size\n y = pos % size\n return [x, y]\n\ndef getEntPoint(size, entrega):\n ent = []\n for a in entrega:\n x, y = a[0], a[1]\n pos = x*size + y\n ent.append(pos)\n return ent",
"_____no_output_____"
],
[
"size = 4\n\narregloAlmP = getAlmPoint(size,alm)\nprint(arregloAlmP)\n\narregloEntP = getEntPoint(size,ent)\nprint(arregloEntP)\n",
"[1, 6, 0, 12, 9]\n[4, 5, 8, 13]\n"
],
[
"def dfs_stack(G, s):\n n = len(G)\n visited = [False]*n\n parent = [None]*n\n stack = [s]\n while stack:\n u = stack.pop()\n if not visited[u]:\n visited[u] = True\n for v in G[u]:\n if not visited[v]:\n parent[v] = u\n stack.append(v)\n\n return parent",
"_____no_output_____"
],
[
"def dfsEntregas(almacenes, entregas, G):\n caminos = []\n for i in almacenes:\n path = dfs_stack(G, i)\n caminos.append(path)\n return caminos\n",
"_____no_output_____"
],
[
"entregas = dfsEntregas(arregloAlmP, arregloEntP, Gp)\nfor i in range(len(arregloAlmP)):\n print(\"Almacen\", arregloAlmP[i],\":\",entregas[i])",
"Almacen 1 : [1, None, 3, 7, 0, 4, 5, 6, 9, 10, 11, 7, 8, 12, 13, 14]\nAlmacen 6 : [4, 0, 1, 2, 5, 6, None, 3, 9, 10, 11, 7, 8, 12, 13, 14]\nAlmacen 0 : [None, 0, 1, 2, 5, 6, 7, 3, 4, 8, 9, 10, 13, 14, 15, 11]\nAlmacen 12 : [1, 2, 3, 7, 8, 4, 5, 6, 9, 10, 11, 15, None, 12, 13, 14]\nAlmacen 9 : [1, 2, 3, 7, 8, 4, 5, 6, 9, None, 11, 7, 13, 14, 10, 14]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec89ba3c1ecbe2bedd17f5a343cc156582c5c5b8 | 73,023 | ipynb | Jupyter Notebook | code/figures/co2_number/co2_number.ipynb | ilopezgp/human_impacts | b2758245edac0946080a647f1dbfd1098c0f0b27 | [
"MIT"
] | 4 | 2020-08-25T00:52:01.000Z | 2020-11-16T16:57:46.000Z | code/figures/co2_number/co2_number.ipynb | ilopezgp/human_impacts | b2758245edac0946080a647f1dbfd1098c0f0b27 | [
"MIT"
] | 5 | 2020-10-30T21:22:55.000Z | 2021-12-30T02:07:02.000Z | code/figures/co2_number/co2_number.ipynb | ilopezgp/human_impacts | b2758245edac0946080a647f1dbfd1098c0f0b27 | [
"MIT"
] | 2 | 2020-08-28T10:11:28.000Z | 2020-11-11T07:58:46.000Z | 276.602273 | 33,404 | 0.920819 | [
[
[
"%matplotlib inline\nimport numpy as np \nimport pandas as pd \nimport matplotlib.pyplot as plt \nimport seaborn as sns\n\nfrom datetime import datetime",
"_____no_output_____"
],
[
"# Read in CO2 data from Mauna Loa - since 1958\natm_co2_contemporary_df = pd.read_csv('../../../data/atmosphere_biogeochemistry/mauna_loa/monthly_in_situ_co2_mlo_formatted.csv')\n\n# Calculate a timestamp for each row in order to plot all the data together. \n# Fits are normalized to the 15th at midnight.\nyr = atm_co2_contemporary_df.year\nmn = atm_co2_contemporary_df.month\ndatetimes = [datetime(year, month, 15, hour=23, minute=59, second=59) for year, month in zip(yr, mn)]\nts = [dt.timestamp() for dt in datetimes]\natm_co2_contemporary_df['timestamp'] = ts\n\n# Read in the historical ice core data that goes from 0 CE - 1960. \natm_co2_historical_df = pd.read_csv('../../../data/atmosphere_biogeochemistry/ice_cores/processed/law2006_by_year_clean.csv')\n\n# Calculate a timestamp for each row in order to plot all the data together. \n# Hack: python doesn't like the year 0, so I go forward 1 year and subtract off a year of seconds to get a timestamp.\nyr = atm_co2_historical_df.year_CE\ns_per_year = 60*60*24*365\ndatetimes = [datetime(year+1, month=1, day=1) for year in yr]\nts = [dt.timestamp() - s_per_year for dt in datetimes]\natm_co2_historical_df['timestamp'] = ts",
"_____no_output_____"
],
[
"plt.figure(figsize=(7.25, 5))\nsns.set_style('white')\n\npal = sns.color_palette('Blues')\nplt.plot(atm_co2_historical_df.timestamp, atm_co2_historical_df.CO2_spline_fit, c=pal[3])\n\nmask = atm_co2_contemporary_df.CO2_fit_ppm > 0\nmasked_co2_contemp = atm_co2_contemporary_df[mask]\nplt.plot(masked_co2_contemp.timestamp, masked_co2_contemp.CO2_fit_ppm, c=pal[4])\n\nxticks = np.arange(0, 2020, 100)\ns_per_year = 60*60*24*365\nyear_zero = atm_co2_historical_df.timestamp[0]\nxs = year_zero + (xticks*s_per_year)\nplt.xticks(xs, xticks, fontsize=8, rotation=45)\nplt.xlabel('Year', fontsize=9)\nplt.ylabel('Atmospheric CO$_2$ (parts per million)', fontsize=9)\nsns.despine()\n\nax = plt.gca()\naxin1 = ax.inset_axes([0.2, 0.4, 0.55, 0.55])\n\nmask = atm_co2_contemporary_df.CO2_fit_ppm > 0\nmasked_co2_contemp = atm_co2_contemporary_df[mask]\naxin1.plot(masked_co2_contemp.timestamp, masked_co2_contemp.CO2_fit_ppm, c=pal[4])\n\nxticks = np.arange(1960, 2021, 20)\ns_per_year = 60*60*24*365\nyear_zero = datetime(1960, 1, 1).timestamp()\nxs = year_zero + (xticks-1960)*s_per_year\naxin1.set_xticks(xs)\naxin1.set_xticklabels(xticks)\nsns.despine(ax=axin1)\n\nplt.savefig('keeling_curve.png', dpi=300)\nplt.show()",
"_____no_output_____"
],
[
"# Diagram of a line and exponential passing through the same endpoints. \ny0, yf = 2, 100\ndeltay = yf-y0\nx0, xf = 0, 220\ndeltax = xf-x0\n\n# Exponential \"fit\" \nxs = np.arange(x0, xf)\nexp_rate = np.log2(yf/y0)/deltax\nprint('%.2g' % exp_rate)\nexp_ys = y0*np.power(2, xs*exp_rate)\n\nx_intervals = np.linspace(0, xf, 5)\ny_intervals = y0*np.power(2, x_intervals*exp_rate)\n\nplt.figure(figsize=(7.25, 5))\nsns.set_style('white')\npal = sns.color_palette('muted')\n\nplt.plot(xs, exp_ys, color=pal[3], lw=4)\nplt.fill_between(xs, np.zeros(xs.size), exp_ys, color=pal[3], alpha=0.3)\n\nplt.scatter(x_intervals, y_intervals, zorder=10, color=pal[2], s=80, edgecolors='w')\n\nfor i in range(x_intervals.size):\n xv, yv = x_intervals[i], y_intervals[i]\n plt.plot([xv, xv], [0, yv], color=pal[2], lw=2)\n \n if i < x_intervals.size - 1:\n xv_next, yv_next = x_intervals[i+1], y_intervals[i+1]\n plt.plot([xv, xv_next], [yv, yv], c=pal[2], lw=2)\n plt.plot([xv, xv_next], [yv, yv_next], c=pal[2], lw=2)\n \n rect_len = xv_next - xv\n rect_height = yv\n rect_area = rect_len*rect_height \n\n rounded_rect_area = round(rect_area, -2)\n if i > 1:\n plt.text(xv + rect_len/2, rect_height/2, \n 'area ≈ %1.d' % rounded_rect_area, ha='center', va='center', fontsize=9)\n\n tri_len = rect_len\n tri_height = yv_next - yv\n tri_area = 0.5 * tri_len * tri_height \n\n rounded_tri_area = round(tri_area, -2)\n if i > 1:\n plt.text(xv + rect_len/1.3, yv + tri_height/3, \n 'area \\n≈ %1.d' % rounded_tri_area, ha='center', va='center', fontsize=9)\n \nplt.text(55, 10, \"area ≈ 700\", ha='center', fontsize=9)\n \nplt.xlim(-2, xf+3)\nplt.ylim(0, yf+2)\n\nxticks = np.arange(0, 221, 20)\nplt.xticks(xticks, xticks+1800, fontsize=8, rotation=45)\nplt.xlabel('Year', fontsize=9)\nplt.ylabel('Estimated Annual Energy Production ($10^{16}$ kJ)', fontsize=9)\n\nsns.despine()\nplt.savefig('emissions_est.png', dpi=300)\nplt.show()",
"0.026\n"
],
[
"'%.2g' % (60e21 * 2.77778e-16)",
"_____no_output_____"
],
[
"'%.2g' % (40/1.5e4)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec89e6db276edcaf091ee4e1245ba4ac3f420e49 | 86,977 | ipynb | Jupyter Notebook | mnist_classification02.ipynb | SolbiChoi/test_deeplearning | c1fb02bc0e14235c4696b4386c469603cc0c682c | [
"Apache-2.0"
] | null | null | null | mnist_classification02.ipynb | SolbiChoi/test_deeplearning | c1fb02bc0e14235c4696b4386c469603cc0c682c | [
"Apache-2.0"
] | null | null | null | mnist_classification02.ipynb | SolbiChoi/test_deeplearning | c1fb02bc0e14235c4696b4386c469603cc0c682c | [
"Apache-2.0"
] | null | null | null | 80.984171 | 22,710 | 0.680847 | [
[
[
"<a href=\"https://colab.research.google.com/github/SolbiChoi/test_deeplearning/blob/master/mnist_classification02.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.datasets.mnist import load_data",
"_____no_output_____"
],
[
"(x_train, y_train), (x_test, y_test) = load_data(path='mnist.npz')\nx_train.shape, y_train.shape, x_test.shape, y_test.shape # (행, 열, 밀어넣는..3차원)",
"_____no_output_____"
],
[
"# y_train[4], x_train[4]\ny_train[50000], x_train[50000]",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nplt.imshow(x_train[50000])\nprint(y_train[50000]), type(y_train[50000])",
"3\n"
],
[
"x_train = x_train.reshape(-1,28*28) / 255 # 255로 나누는 이유 : x_train max값, 값 scaling (min max scale)\nx_train.shape",
"_____no_output_____"
],
[
"x_test = x_test.reshape(-1,28*28) / 255\nx_test.shape",
"_____no_output_____"
],
[
"y_train.shape",
"_____no_output_____"
],
[
"y_train[2:10]",
"_____no_output_____"
],
[
"import numpy as np\nnp.unique(y_train)",
"_____no_output_____"
]
],
[
[
"## Apply model",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"model = tf.keras.models.Sequential()\n\nmodel.add(tf.keras.Input(shape=(784,))) # input layer - reshape후 열의 숫자 입력\nmodel.add(tf.keras.layers.Dense(64, activation='relu')) # hidden layer\nmodel.add(tf.keras.layers.Dense(64, activation='relu')) # hidden layer\n# 분류 3개 이상일 때 -> softmax, binary -> sigmoid (hidden에서만 사용)\nmodel.add(tf.keras.layers.Dense(10, activation='softmax')) # output layer\n\n# regression -> loss : mse\n# binary classification -> loss : binary crossentropy\n# over 3 classification -> loss : categorical cross entropy (sparse_categorical_crossentropy)\nmodel.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['acc'])",
"WARNING:tensorflow:Please add `keras.layers.InputLayer` instead of `keras.Input` to Sequential model. `keras.Input` is intended to be used by Functional model.\n"
],
[
"hist = model.fit(x_train, y_train, epochs=100, validation_split=0.3)",
"Epoch 1/100\n1313/1313 [==============================] - 4s 3ms/step - loss: 0.3202 - acc: 0.9065 - val_loss: 0.1785 - val_acc: 0.9476\nEpoch 2/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.1412 - acc: 0.9572 - val_loss: 0.1465 - val_acc: 0.9568\nEpoch 3/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.1025 - acc: 0.9693 - val_loss: 0.1240 - val_acc: 0.9647\nEpoch 4/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0797 - acc: 0.9753 - val_loss: 0.1296 - val_acc: 0.9636\nEpoch 5/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0625 - acc: 0.9811 - val_loss: 0.1130 - val_acc: 0.9665\nEpoch 6/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0524 - acc: 0.9837 - val_loss: 0.1142 - val_acc: 0.9688\nEpoch 7/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0427 - acc: 0.9860 - val_loss: 0.1212 - val_acc: 0.9678\nEpoch 8/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0370 - acc: 0.9874 - val_loss: 0.1248 - val_acc: 0.9679\nEpoch 9/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0293 - acc: 0.9903 - val_loss: 0.1205 - val_acc: 0.9695\nEpoch 10/100\n1313/1313 [==============================] - 4s 3ms/step - loss: 0.0275 - acc: 0.9907 - val_loss: 0.1341 - val_acc: 0.9689\nEpoch 11/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0236 - acc: 0.9921 - val_loss: 0.1288 - val_acc: 0.9702\nEpoch 12/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0207 - acc: 0.9928 - val_loss: 0.1433 - val_acc: 0.9666\nEpoch 13/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0184 - acc: 0.9941 - val_loss: 0.1402 - val_acc: 0.9704\nEpoch 14/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0191 - acc: 0.9940 - val_loss: 0.1544 - val_acc: 0.9667\nEpoch 15/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0160 - acc: 0.9944 - val_loss: 0.1505 - val_acc: 0.9691\nEpoch 16/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0150 - acc: 0.9951 - val_loss: 0.1530 - val_acc: 0.9689\nEpoch 17/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0106 - acc: 0.9965 - val_loss: 0.1526 - val_acc: 0.9716\nEpoch 18/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0136 - acc: 0.9953 - val_loss: 0.1680 - val_acc: 0.9706\nEpoch 19/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0134 - acc: 0.9955 - val_loss: 0.1696 - val_acc: 0.9677\nEpoch 20/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0133 - acc: 0.9955 - val_loss: 0.1798 - val_acc: 0.9667\nEpoch 21/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0085 - acc: 0.9973 - val_loss: 0.1688 - val_acc: 0.9712\nEpoch 22/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0126 - acc: 0.9959 - val_loss: 0.1606 - val_acc: 0.9727\nEpoch 23/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0081 - acc: 0.9972 - val_loss: 0.1706 - val_acc: 0.9693\nEpoch 24/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0111 - acc: 0.9964 - val_loss: 0.1862 - val_acc: 0.9710\nEpoch 25/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0093 - acc: 0.9967 - val_loss: 0.1977 - val_acc: 0.9695\nEpoch 26/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0115 - acc: 0.9964 - val_loss: 0.1964 - val_acc: 0.9667\nEpoch 27/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0069 - acc: 0.9977 - val_loss: 0.1929 - val_acc: 0.9707\nEpoch 28/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0112 - acc: 0.9962 - val_loss: 0.1914 - val_acc: 0.9701\nEpoch 29/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0085 - acc: 0.9975 - val_loss: 0.2062 - val_acc: 0.9699\nEpoch 30/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0098 - acc: 0.9972 - val_loss: 0.2045 - val_acc: 0.9701\nEpoch 31/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0088 - acc: 0.9970 - val_loss: 0.2100 - val_acc: 0.9708\nEpoch 32/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0076 - acc: 0.9977 - val_loss: 0.2156 - val_acc: 0.9699\nEpoch 33/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0055 - acc: 0.9980 - val_loss: 0.2134 - val_acc: 0.9717\nEpoch 34/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0088 - acc: 0.9973 - val_loss: 0.2056 - val_acc: 0.9714\nEpoch 35/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0062 - acc: 0.9980 - val_loss: 0.2540 - val_acc: 0.9671\nEpoch 36/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0097 - acc: 0.9966 - val_loss: 0.2078 - val_acc: 0.9721\nEpoch 37/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0097 - acc: 0.9970 - val_loss: 0.2489 - val_acc: 0.9694\nEpoch 38/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0049 - acc: 0.9985 - val_loss: 0.2078 - val_acc: 0.9736\nEpoch 39/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0048 - acc: 0.9987 - val_loss: 0.2404 - val_acc: 0.9689\nEpoch 40/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0118 - acc: 0.9962 - val_loss: 0.2025 - val_acc: 0.9721\nEpoch 41/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0048 - acc: 0.9986 - val_loss: 0.2186 - val_acc: 0.9722\nEpoch 42/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0068 - acc: 0.9977 - val_loss: 0.2417 - val_acc: 0.9704\nEpoch 43/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0075 - acc: 0.9979 - val_loss: 0.2493 - val_acc: 0.9706\nEpoch 44/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0064 - acc: 0.9979 - val_loss: 0.2422 - val_acc: 0.9717\nEpoch 45/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0087 - acc: 0.9975 - val_loss: 0.2440 - val_acc: 0.9704\nEpoch 46/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0048 - acc: 0.9985 - val_loss: 0.2634 - val_acc: 0.9694\nEpoch 47/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0054 - acc: 0.9983 - val_loss: 0.2845 - val_acc: 0.9677\nEpoch 48/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0076 - acc: 0.9978 - val_loss: 0.2637 - val_acc: 0.9698\nEpoch 49/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0060 - acc: 0.9982 - val_loss: 0.2650 - val_acc: 0.9693\nEpoch 50/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0049 - acc: 0.9984 - val_loss: 0.2618 - val_acc: 0.9714\nEpoch 51/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0076 - acc: 0.9977 - val_loss: 0.2767 - val_acc: 0.9698\nEpoch 52/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0077 - acc: 0.9977 - val_loss: 0.2881 - val_acc: 0.9666\nEpoch 53/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0073 - acc: 0.9978 - val_loss: 0.2699 - val_acc: 0.9714\nEpoch 54/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0054 - acc: 0.9985 - val_loss: 0.2735 - val_acc: 0.9713\nEpoch 55/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0068 - acc: 0.9980 - val_loss: 0.2734 - val_acc: 0.9718\nEpoch 56/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0064 - acc: 0.9978 - val_loss: 0.2621 - val_acc: 0.9708\nEpoch 57/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0046 - acc: 0.9987 - val_loss: 0.2741 - val_acc: 0.9713\nEpoch 58/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0067 - acc: 0.9983 - val_loss: 0.2922 - val_acc: 0.9697\nEpoch 59/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0049 - acc: 0.9985 - val_loss: 0.2875 - val_acc: 0.9704\nEpoch 60/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0097 - acc: 0.9974 - val_loss: 0.2767 - val_acc: 0.9706\nEpoch 61/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0059 - acc: 0.9982 - val_loss: 0.2800 - val_acc: 0.9732\nEpoch 62/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0038 - acc: 0.9989 - val_loss: 0.2818 - val_acc: 0.9730\nEpoch 63/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0055 - acc: 0.9986 - val_loss: 0.2922 - val_acc: 0.9709\nEpoch 64/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0085 - acc: 0.9978 - val_loss: 0.3070 - val_acc: 0.9706\nEpoch 65/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0058 - acc: 0.9982 - val_loss: 0.2832 - val_acc: 0.9708\nEpoch 66/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0057 - acc: 0.9983 - val_loss: 0.3176 - val_acc: 0.9712\nEpoch 67/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0071 - acc: 0.9980 - val_loss: 0.3206 - val_acc: 0.9714\nEpoch 68/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0028 - acc: 0.9991 - val_loss: 0.2850 - val_acc: 0.9721\nEpoch 69/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0029 - acc: 0.9992 - val_loss: 0.3309 - val_acc: 0.9677\nEpoch 70/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0084 - acc: 0.9978 - val_loss: 0.2981 - val_acc: 0.9708\nEpoch 71/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0043 - acc: 0.9987 - val_loss: 0.2909 - val_acc: 0.9719\nEpoch 72/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0080 - acc: 0.9978 - val_loss: 0.3256 - val_acc: 0.9698\nEpoch 73/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0043 - acc: 0.9987 - val_loss: 0.3181 - val_acc: 0.9723\nEpoch 74/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0061 - acc: 0.9982 - val_loss: 0.3246 - val_acc: 0.9714\nEpoch 75/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0058 - acc: 0.9987 - val_loss: 0.3061 - val_acc: 0.9726\nEpoch 76/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0040 - acc: 0.9989 - val_loss: 0.3439 - val_acc: 0.9688\nEpoch 77/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0088 - acc: 0.9980 - val_loss: 0.3359 - val_acc: 0.9697\nEpoch 78/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0039 - acc: 0.9987 - val_loss: 0.3075 - val_acc: 0.9725\nEpoch 79/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0022 - acc: 0.9994 - val_loss: 0.3648 - val_acc: 0.9692\nEpoch 80/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0082 - acc: 0.9978 - val_loss: 0.3423 - val_acc: 0.9706\nEpoch 81/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0071 - acc: 0.9986 - val_loss: 0.3139 - val_acc: 0.9711\nEpoch 82/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0054 - acc: 0.9985 - val_loss: 0.3261 - val_acc: 0.9714\nEpoch 83/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 7.1017e-04 - acc: 0.9999 - val_loss: 0.3046 - val_acc: 0.9733\nEpoch 84/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0107 - acc: 0.9974 - val_loss: 0.3572 - val_acc: 0.9697\nEpoch 85/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0036 - acc: 0.9992 - val_loss: 0.3830 - val_acc: 0.9688\nEpoch 86/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0067 - acc: 0.9983 - val_loss: 0.3375 - val_acc: 0.9695\nEpoch 87/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0044 - acc: 0.9987 - val_loss: 0.3391 - val_acc: 0.9719\nEpoch 88/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0058 - acc: 0.9989 - val_loss: 0.3692 - val_acc: 0.9706\nEpoch 89/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0063 - acc: 0.9980 - val_loss: 0.3703 - val_acc: 0.9700\nEpoch 90/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0047 - acc: 0.9986 - val_loss: 0.3528 - val_acc: 0.9709\nEpoch 91/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0022 - acc: 0.9993 - val_loss: 0.3317 - val_acc: 0.9725\nEpoch 92/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0067 - acc: 0.9985 - val_loss: 0.3480 - val_acc: 0.9708\nEpoch 93/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0064 - acc: 0.9984 - val_loss: 0.3829 - val_acc: 0.9700\nEpoch 94/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0050 - acc: 0.9985 - val_loss: 0.3544 - val_acc: 0.9713\nEpoch 95/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0034 - acc: 0.9990 - val_loss: 0.3849 - val_acc: 0.9694\nEpoch 96/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0075 - acc: 0.9982 - val_loss: 0.3605 - val_acc: 0.9718\nEpoch 97/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0027 - acc: 0.9992 - val_loss: 0.3734 - val_acc: 0.9721\nEpoch 98/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0043 - acc: 0.9987 - val_loss: 0.3877 - val_acc: 0.9720\nEpoch 99/100\n1313/1313 [==============================] - 3s 3ms/step - loss: 0.0064 - acc: 0.9986 - val_loss: 0.3528 - val_acc: 0.9729\nEpoch 100/100\n1313/1313 [==============================] - 3s 2ms/step - loss: 0.0051 - acc: 0.9988 - val_loss: 0.3844 - val_acc: 0.9702\n"
]
],
[
[
"## evaluation",
"_____no_output_____"
]
],
[
[
"hist.history.keys()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nplt.plot(hist.history['loss'])\nplt.plot(hist.history['val_loss'],'b-')\nplt.show()",
"_____no_output_____"
],
[
"plt.plot(hist.history['acc'])\nplt.plot(hist.history['val_acc'],'-r')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## service",
"_____no_output_____"
]
],
[
[
"# x_test[30]",
"_____no_output_____"
],
[
"# pred = model.predict([x_test[30:31]]) # 사이즈(영역)를 명확하게 지정해주어야 한다.",
"_____no_output_____"
],
[
"import numpy as np\n# np.set_printoptions(precision=8)\n\npred = model.predict([x_test[30:31]])\npred",
"_____no_output_____"
],
[
"np.argmax(pred, axis=1)",
"_____no_output_____"
],
[
"y_test[30]",
"_____no_output_____"
]
],
[
[
"### numpy.argmax 위치값을 가져옴 example",
"_____no_output_____"
]
],
[
[
"a = np.arange(6).reshape(2,3) + 10\nprint(a)\nnp.argmax(a, axis=0)",
"[[10 11 12]\n [13 14 15]]\n"
],
[
"np.argmax(np.array([[10,11,12]]), axis=1)",
"_____no_output_____"
]
],
[
[
"## save",
"_____no_output_____"
]
],
[
[
"model.save('./model_save')",
"INFO:tensorflow:Assets written to: ./model_save/assets\n"
],
[
"model.save('./model_save01.h5')",
"_____no_output_____"
],
[
"model_load = tf.keras.models.load_model('./model_save01.h5')\nmodel_load",
"_____no_output_____"
],
[
"model_load = tf.keras.models.load_model('./model_save01.h5')",
"_____no_output_____"
],
[
"load_pred = model_load.predict(x_test[30:31])\nload_pred",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec89e84cf3adfed0d88aeb85a94587c100ceba64 | 27,911 | ipynb | Jupyter Notebook | 13-Iterators-and-List-Comprehensions.ipynb | bagrow/WhirlwindTourOfPython | bd77cbd8b6a68f5b854d2b29ed266d36c8170d77 | [
"CC0-1.0"
] | 7 | 2017-01-16T19:36:35.000Z | 2021-11-08T08:54:35.000Z | 13-Iterators-and-List-Comprehensions.ipynb | bagrow/WhirlwindTourOfPython | bd77cbd8b6a68f5b854d2b29ed266d36c8170d77 | [
"CC0-1.0"
] | null | null | null | 13-Iterators-and-List-Comprehensions.ipynb | bagrow/WhirlwindTourOfPython | bd77cbd8b6a68f5b854d2b29ed266d36c8170d77 | [
"CC0-1.0"
] | null | null | null | 24.526362 | 372 | 0.534126 | [
[
[
"# Iterators and List Comprehensions\n\nOften an important piece of data analysis is repeating a similar calculation, over and over, in an automated fashion.\nFor example, you may have a table of a names that you'd like to split into first and last, or perhaps of dates that you'd like to convert to some standard format.\nOne of Python's answers to this is the *iterator* syntax.\nWe've seen this already with the ``range`` iterator:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(i, end=' ')",
"0 1 2 3 4 5 6 7 8 9 "
]
],
[
[
"Here we're going to dig a bit deeper.\nIt turns out that in Python 3, ``range`` is not a list, but is something called an *iterator*, and learning how it works is key to understanding a wide class of very useful Python functionality.",
"_____no_output_____"
],
[
"## Iterating over lists\nIterators are perhaps most easily understood in the concrete case of iterating through a list.\nConsider the following:",
"_____no_output_____"
]
],
[
[
"for value in [2, 4, 6, 8, 10]:\n # do some operation\n print(value + 1, end=' ')",
"3 5 7 9 11 "
]
],
[
[
"The familiar \"``for x in y``\" syntax allows us to repeat some operation for each value in the list.\nThe fact that the syntax of the code is so close to its English description (\"*for [each] value in [the] list*\") is just one of the syntactic choices that makes Python such an intuitive language to learn and use.\n\nBut the face-value behavior is not what's *really* happening.\nWhen you write something like \"``for val in L``\", the Python interpreter checks whether it has an *iterator* interface, which you can check yourself with the built-in ``iter`` function:",
"_____no_output_____"
]
],
[
[
"iter([2, 4, 6, 8, 10])",
"_____no_output_____"
]
],
[
[
"It is this iterator object that provides the functionality required by the ``for`` loop.\nThe ``iter`` object is a container that gives you access to the next object for as long as there **is** a next object, which can be seen with the built-in function ``next``:",
"_____no_output_____"
]
],
[
[
"I = iter([2, 4, 6, 8, 10])",
"_____no_output_____"
],
[
"print(next(I))",
"2\n"
],
[
"print(next(I))",
"4\n"
],
[
"print(next(I))",
"6\n"
]
],
[
[
"What is the purpose of this level of indirection?\nWell, it turns out this is incredibly useful, because it allows Python to treat things as lists that are *not actually lists*.",
"_____no_output_____"
],
[
"## ``range()``: A List Is Not Always a List\nPerhaps the most common example of this indirect iteration is the ``range()`` function in Python 3 (named ``xrange()`` in Python 2), which returns not a list, but a special ``range()`` object:",
"_____no_output_____"
]
],
[
[
"range(10)",
"_____no_output_____"
]
],
[
[
"``range``, like a list, exposes an iterator:",
"_____no_output_____"
]
],
[
[
"iter(range(10))",
"_____no_output_____"
]
],
[
[
"So Python knows to treat it *as if* it's a list:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(i, end=' ')",
"0 1 2 3 4 5 6 7 8 9 "
]
],
[
[
"The benefit of the iterator indirection is that **the full list is never explicitly created!**\nWe can see this by doing a range calculation that would overwhelm our system memory if we actually instantiated it (note that in Python 2, ``range`` creates a list, so running the following will not lead to good things!):",
"_____no_output_____"
]
],
[
[
"N = 10 ** 12\nfor i in range(N):\n if i >= 10: break\n print(i, end=', ')",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, "
]
],
[
[
"If ``range`` were to actually create that list of one trillion values, it would occupy tens of terabytes of machine memory: a waste, given the fact that we're ignoring all but the first 10 values!\n\nIn fact, there's no reason that iterators ever have to end at all!\nPython's ``itertools`` library contains a ``count`` function that acts as an infinite range:",
"_____no_output_____"
]
],
[
[
"from itertools import count\n\nfor i in count():\n if i >= 10:\n break\n print(i, end=', ')",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, "
]
],
[
[
"Had we not thrown in a loop break here, it would go on happily counting until the process is manually interrupted or killed (using, for example, ``ctrl-C``).",
"_____no_output_____"
],
[
"## Useful Iterators\nThis iterator syntax is used nearly universally in Python built-in types as well as the more data science-specific objects we'll explore in later sections.\nHere we'll cover some of the more useful iterators in the Python language:",
"_____no_output_____"
],
[
"### ``enumerate``\nOften you need to iterate not only the values in an array, but also keep track of the index.\nYou might be **tempted** to do things this way:",
"_____no_output_____"
]
],
[
[
"L = [2, 4, 6, 8, 10]\nfor i in range(len(L)):\n print(i, L[i])",
"0 2\n1 4\n2 6\n3 8\n4 10\n"
]
],
[
[
"Although this does work, Python provides a cleaner syntax using the ``enumerate`` iterator:",
"_____no_output_____"
]
],
[
[
"for i, val in enumerate(L):\n print(i, val)",
"0 2\n1 4\n2 6\n3 8\n4 10\n"
]
],
[
[
"This is the more \"Pythonic\" way to enumerate the indices and values in a list.",
"_____no_output_____"
],
[
"### ``zip``\nOther times, you may have multiple lists that you want to iterate over simultaneously.\nYou could certainly iterate over the index as in the non-Pythonic example we looked at previously, but it is better to use the ``zip`` iterator, which zips together iterables:",
"_____no_output_____"
]
],
[
[
"L = [2, 4, 6, 8, 10]\nR = [3, 6, 9, 12, 15]\nfor lval, rval in zip(L, R):\n print(lval, rval)",
"2 3\n4 6\n6 9\n8 12\n10 15\n"
]
],
[
[
"Any number of iterables can be zipped together, and if they are different lengths, the shortest will determine the length of the ``zip``.",
"_____no_output_____"
],
[
"### ``map`` and ``filter``\nThe ``map`` iterator takes a function and applies it to the values in an iterator:",
"_____no_output_____"
]
],
[
[
"# find the first 10 square numbers\nsquare = lambda x: x ** 2\nfor val in map(square, range(10)):\n print(val, end=' ')",
"0 1 4 9 16 25 36 49 64 81 "
]
],
[
[
"The ``filter`` iterator looks similar, except it only passes-through values for which the filter function evaluates to True:",
"_____no_output_____"
]
],
[
[
"# find values up to 10 for which x % 2 is zero\nis_even = lambda x: x % 2 == 0\nfor val in filter(is_even, range(10)):\n print(val, end=' ')",
"0 2 4 6 8 "
]
],
[
[
"The ``map`` and ``filter`` functions, along with the ``reduce`` function (which lives in Python's ``functools`` module) are fundamental components of the *functional programming* style, which, while not a dominant programming style in the Python world, has its outspoken proponents (see, for example, the [pytoolz](https://toolz.readthedocs.org/en/latest/) library).",
"_____no_output_____"
],
[
"### Iterators as function arguments\n\nWe saw in [``*args`` and ``**kwargs``: Flexible Arguments](#*args-and-**kwargs:-Flexible-Arguments). that ``*args`` and ``**kwargs`` can be used to pass sequences and dictionaries to functions.\nIt turns out that the ``*args`` syntax works not just with sequences, but with any iterator:",
"_____no_output_____"
]
],
[
[
"print(*range(10))",
"0 1 2 3 4 5 6 7 8 9\n"
]
],
[
[
"So, for example, we can get tricky and compress the ``map`` example from before into the following:",
"_____no_output_____"
]
],
[
[
"print(*map(lambda x: x ** 2, range(10)))",
"0 1 4 9 16 25 36 49 64 81\n"
]
],
[
[
"Using this trick lets us answer the age-old question that comes up in Python learners' forums: why is there no ``unzip()`` function which does the opposite of ``zip()``?\nIf you lock yourself in a dark closet and think about it for a while, you might realize that the opposite of ``zip()`` is... ``zip()``! The key is that ``zip()`` can zip-together any number of iterators or sequences. Observe:",
"_____no_output_____"
]
],
[
[
"L1 = (1, 2, 3, 4)\nL2 = ('a', 'b', 'c', 'd')",
"_____no_output_____"
],
[
"z = zip(L1, L2)\nprint(*z)",
"(1, 'a') (2, 'b') (3, 'c') (4, 'd')\n"
],
[
"z = zip(L1, L2)\nnew_L1, new_L2 = zip(*z)\nprint(new_L1, \" & \", new_L2)",
"(1, 2, 3, 4) & ('a', 'b', 'c', 'd')\n"
]
],
[
[
"Ponder this for a while. If you understand why it works, you'll have come a long way in understanding Python iterators!",
"_____no_output_____"
],
[
"## Specialized Iterators: ``itertools``\n\nWe briefly looked at the infinite ``range`` iterator, ``itertools.count``.\nThe ``itertools`` module contains a whole host of useful iterators; it's well worth your while to explore the module to see what's available.\nAs an example, consider the ``itertools.permutations`` function, which iterates over all permutations of a sequence:",
"_____no_output_____"
]
],
[
[
"from itertools import permutations\np = permutations(range(3))\nprint(*p)",
"(0, 1, 2) (0, 2, 1) (1, 0, 2) (1, 2, 0) (2, 0, 1) (2, 1, 0)\n"
]
],
[
[
"Similarly, the ``itertools.combinations`` function iterates over all unique combinations of ``N`` values within a list:",
"_____no_output_____"
]
],
[
[
"from itertools import combinations\nc = combinations(range(4), 2)\nprint(*c)",
"(0, 1) (0, 2) (0, 3) (1, 2) (1, 3) (2, 3)\n"
]
],
[
[
"Somewhat related is the ``product`` iterator, which iterates over all sets of pairs between two or more iterables:",
"_____no_output_____"
]
],
[
[
"from itertools import product\np = product('ab', range(3))\nprint(*p)",
"('a', 0) ('a', 1) ('a', 2) ('b', 0) ('b', 1) ('b', 2)\n"
]
],
[
[
"Many more useful iterators exist in ``itertools``: the full list can be found, along with some examples, in Python's [online documentation](https://docs.python.org/3.5/library/itertools.html).",
"_____no_output_____"
],
[
"## List Comprehensions\nIf you read enough Python code, you'll eventually come across the terse and efficient construction known as a *list comprehension*.\nThis is one feature of Python I expect **you will fall in love with** if you've not used it before; it looks something like this:",
"_____no_output_____"
]
],
[
[
"[i for i in range(20) if i % 3 > 0]",
"_____no_output_____"
]
],
[
[
"The result of this is a list of numbers which excludes multiples of 3.\nWhile this example may seem a bit dense and confusing at first, as familiarity with Python grows, reading and writing list comprehensions will become second nature.",
"_____no_output_____"
],
[
"### Basic List Comprehensions\n\nList comprehensions are simply a way to compress a list-building for-loop into a single short, readable line.\nFor example, here is a loop that constructs a list of the first 12 square integers:",
"_____no_output_____"
]
],
[
[
"L = []\nfor n in range(12):\n L.append(n ** 2)\nL",
"_____no_output_____"
]
],
[
[
"The list comprehension equivalent of this is the following:",
"_____no_output_____"
]
],
[
[
"[n ** 2 for n in range(12)]",
"_____no_output_____"
]
],
[
[
"As with many Python statements, you can almost read-off the meaning of this statement in plain English: \"construct a list consisting of the square of ``n`` for each ``n`` from zero to 12\".\n\nThis basic syntax, then, is ``[``*``expr``* ``for`` *``var``* ``in`` *``iterable``*``]``, where *``expr``* is any valid expression, *``var``* is a variable name, and *``iterable``* is any iterable Python object.",
"_____no_output_____"
],
[
"### Multiple Iteration\nSometimes you want to build a list not just from one value, but from two. To do this, simply add another ``for`` expression in the comprehension:",
"_____no_output_____"
]
],
[
[
"[(i, j) for i in range(2) for j in range(3)]",
"_____no_output_____"
]
],
[
[
"Notice that the second ``for`` expression acts as the interior index, varying the fastest in the resulting list.\nThis type of construction can be extended to three, four, or more iterators within the comprehension, though at some point code readibility will suffer!",
"_____no_output_____"
],
[
"### Conditionals on the Iterator\nYou can further control the iteration by adding a conditional to the end of the expression.\nIn the first example of the section, we iterated over all numbers from 1 to 20, but left-out multiples of 3.\nLook at this again, and notice the construction:",
"_____no_output_____"
]
],
[
[
"[val for val in range(20) if val % 3 > 0]",
"_____no_output_____"
]
],
[
[
"The expression ``(i % 3 > 0)`` evaluates to ``True`` unless ``val`` is divisible by 3.\nAgain, the English language meaning can be immediately read off: \"Construct a list of values for each value up to 20, but only if the value is not divisible by 3\".\nOnce you are comfortable with it, this is much easier to write – and to understand at a glance – than the equivalent loop syntax:",
"_____no_output_____"
]
],
[
[
"L = []\nfor val in range(20):\n if val % 3:\n L.append(val)\nL",
"_____no_output_____"
]
],
[
[
"### Conditionals on the Value\nIf you've programmed in C, you might be familiar with the single-line conditional enabled by the ``?`` operator:\n``` C\nint absval = (val < 0) ? -val : val\n```\nPython has something very similar to this, which is most often used within list comprehensions, ``lambda`` functions, and other places where a simple expression is desired:",
"_____no_output_____"
]
],
[
[
"val = -10\nval if val >= 0 else -val",
"_____no_output_____"
]
],
[
[
"We see that this simply duplicates the functionality of the built-in ``abs()`` function, but the construction lets you do some really interesting things within list comprehensions.\nThis is getting pretty complicated now, but you could do something like this:",
"_____no_output_____"
]
],
[
[
"[val if val % 2 else -val\n for val in range(20) if val % 3]",
"_____no_output_____"
]
],
[
[
"Note the line break within the list comprehension before the ``for`` expression: this is valid in Python, and is often a nice way to break-up long list comprehensions for greater readibility.\nLook this over: what we're doing is constructing a list, leaving out multiples of 3, and negating all mutliples of 2.",
"_____no_output_____"
],
[
"Once you understand the dynamics of list comprehensions, it's straightforward to move on to other types of comprehensions. The syntax is largely the same; the only difference is the type of bracket you use.\n\nFor example, with curly braces you can create a ``set`` with a *set comprehension*:",
"_____no_output_____"
]
],
[
[
"{n**2 for n in range(12)}",
"_____no_output_____"
]
],
[
[
"Recall that a ``set`` is a collection that contains no duplicates.\nThe set comprehension respects this rule, and eliminates any duplicate entries:",
"_____no_output_____"
]
],
[
[
"{a % 3 for a in range(1000)}",
"_____no_output_____"
]
],
[
[
"With a slight tweak, you can add a colon (``:``) to create a *dict comprehension*:",
"_____no_output_____"
]
],
[
[
"{n:n**2 for n in range(6)}",
"_____no_output_____"
]
],
[
[
"### Generators\n\nFinally, if you use parentheses rather than square brackets, you get what's called a **generator expression**:",
"_____no_output_____"
]
],
[
[
"(n**2 for n in range(12))",
"_____no_output_____"
]
],
[
[
"A generator expression is essentially a list comprehension in which **elements are generated as-needed rather than all at-once**, and the simplicity here belies the power of this language feature.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec89f734da1fc92f52ae882792e82c9089bbd544 | 273,512 | ipynb | Jupyter Notebook | Tutorials/DeepSets MnistSet/DeepSet Sum.ipynb | AhmedBegggaUA/SetXAI | b295982d75d24ddc7129f68e7d47b0be181c349f | [
"MIT"
] | null | null | null | Tutorials/DeepSets MnistSet/DeepSet Sum.ipynb | AhmedBegggaUA/SetXAI | b295982d75d24ddc7129f68e7d47b0be181c349f | [
"MIT"
] | null | null | null | Tutorials/DeepSets MnistSet/DeepSet Sum.ipynb | AhmedBegggaUA/SetXAI | b295982d75d24ddc7129f68e7d47b0be181c349f | [
"MIT"
] | null | null | null | 231.593565 | 59,164 | 0.906882 | [
[
[
"import sys\nsys.path.insert(0, '/Users/ahmedbegga/Desktop/TFG-Ahmed/SetXAI/')\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom src.utils import *\nfrom src.model import *\nfrom data.MnistSet import *\nfrom time import sleep\nfrom tqdm import tqdm",
"_____no_output_____"
]
],
[
[
"### Preparamos los datos de entreno y test",
"_____no_output_____"
]
],
[
[
"batch_size = 32",
"_____no_output_____"
],
[
"train_loader = get_loader(\n MNISTSet(train=True, full=True), batch_size=batch_size, num_workers=4)",
"_____no_output_____"
],
[
"test_loader = get_loader(\n MNISTSet(train=False, full=True), batch_size=batch_size, num_workers=4)",
"_____no_output_____"
],
[
"from time import sleep\nfrom tqdm import tqdm",
"_____no_output_____"
],
[
"set_channels = 2\nset_size = 342\n \nhidden_dim = 32\niters = 10\nlatent_dim = 16\nlr = 0.001\nn_epochs = 10\nnet = SumEncoder(set_channels,latent_dim,hidden_dim)",
"_____no_output_____"
],
[
"net",
"_____no_output_____"
],
[
"optimizer = torch.optim.Adam([p for p in net.parameters() if p.requires_grad], lr=0.001)",
"_____no_output_____"
],
[
"#net = net.cuda()",
"_____no_output_____"
],
[
"net.train()\nfor epoch in range(n_epochs):\n with tqdm(train_loader, unit=\"batch\") as tepoch:\n for i, sample in enumerate(tepoch):\n tepoch.set_description(f\"Epoch {epoch}\")\n input, target_set, target_mask = map(lambda x: x, sample)\n optimizer.zero_grad()\n output = net(target_set,target_mask)\n loss = F.cross_entropy(output, input)\n acc = (output.max(dim=1)[1] == input).float().mean()\n loss.backward()\n optimizer.step()\n tepoch.set_postfix(loss=loss.item(), acc=100. * acc.item())",
"Epoch 0: 100%|██████| 1875/1875 [01:19<00:00, 23.56batch/s, acc=15.6, loss=2.08]\nEpoch 1: 100%|██████| 1875/1875 [01:12<00:00, 25.75batch/s, acc=56.2, loss=1.33]\nEpoch 2: 100%|██████| 1875/1875 [01:11<00:00, 26.15batch/s, acc=28.1, loss=1.73]\nEpoch 3: 100%|████████| 1875/1875 [01:12<00:00, 26.01batch/s, acc=50, loss=1.64]\nEpoch 4: 100%|███████| 1875/1875 [01:45<00:00, 17.77batch/s, acc=53.1, loss=1.2]\nEpoch 5: 100%|█████| 1875/1875 [03:26<00:00, 9.09batch/s, acc=56.2, loss=0.998]\nEpoch 6: 100%|██████| 1875/1875 [01:21<00:00, 23.14batch/s, acc=59.4, loss=1.21]\nEpoch 7: 100%|█████| 1875/1875 [01:20<00:00, 23.37batch/s, acc=59.4, loss=0.986]\nEpoch 8: 100%|█████| 1875/1875 [01:19<00:00, 23.60batch/s, acc=65.6, loss=0.936]\nEpoch 9: 100%|█████| 1875/1875 [01:19<00:00, 23.51batch/s, acc=87.5, loss=0.466]\n"
],
[
"net.eval()\nwith tqdm(test_loader, unit=\"batch\") as tepoch:\n for i, sample in enumerate(tepoch):\n tepoch.set_description(f\"Epoch {1}\")\n input, target_set, target_mask = map(lambda x: x, sample)\n output = net(target_set,target_mask)\n loss = F.cross_entropy(output, input)\n acc = (output.max(dim=1)[1] == input).float().mean()\n tepoch.set_postfix(loss=loss.item(), acc=100. * acc.item())",
"Epoch 1: 100%|███████| 312/312 [00:08<00:00, 37.85batch/s, acc=78.1, loss=0.594]\n"
],
[
"#torch.save(net.state_dict(),'model_sum_encoder.pth')",
"_____no_output_____"
],
[
"model = torch.load('./SumEncoder_model_mnist.pth' ,map_location=torch.device('cpu'))\nmiNet = SumEncoder(set_channels,latent_dim,hidden_dim)",
"_____no_output_____"
],
[
"miNet.load_state_dict(torch.load('./SumEncoder_model_mnist.pth' ,map_location=torch.device('cpu')))",
"_____no_output_____"
],
[
"miNet",
"_____no_output_____"
],
[
"miNet = miNet\nmiNet.eval()\nlosses = []\naccs = []\nwith tqdm(test_loader, unit=\"batch\") as tepoch:\n for i, sample in enumerate(tepoch):\n tepoch.set_description(f\"Epoch {0}\")\n input, target_set, target_mask = map(lambda x: x, sample)\n output = miNet(target_set,target_mask)\n loss = F.cross_entropy(output, input)\n acc = (output.max(dim=1)[1] == input).float().mean()\n losses.append(loss.item())\n accs.append(acc.item())\n tepoch.set_postfix(loss=loss.item(), acc=100. * acc.item())\nprint('loss: {}, acc: {}'.format(round(sum(losses)/len(losses),2),round(sum(accs)/len(accs),2)))",
"Epoch 0: 100%|███████████████████████████████| 312/312 [00:08<00:00, 36.21batch/s, acc=87.5, loss=0.3]"
],
[
"for name, param in miNet.named_parameters():\n print(name, param.size())",
"conv.0.weight torch.Size([32, 2, 1])\nconv.0.bias torch.Size([32])\nconv.2.weight torch.Size([32, 32, 1])\nconv.2.bias torch.Size([32])\nlin.0.weight torch.Size([32, 32])\nlin.0.bias torch.Size([32])\nlin.2.weight torch.Size([16, 32])\nlin.2.bias torch.Size([16])\nclassifier.0.weight torch.Size([16, 16])\nclassifier.0.bias torch.Size([16])\nclassifier.2.weight torch.Size([10, 16])\nclassifier.2.bias torch.Size([10])\n"
],
[
"miNet",
"_____no_output_____"
],
[
"model1 = MaxEncoder(set_channels,latent_dim,hidden_dim)\nmodel1 = torch.nn.Sequential(*(list(miNet.children())[:-2]))\nprint(model1)",
"Sequential(\n (0): Sequential(\n (0): Conv1d(2, 32, kernel_size=(1,), stride=(1,))\n (1): ReLU(inplace=True)\n (2): Conv1d(32, 32, kernel_size=(1,), stride=(1,))\n )\n)\n"
],
[
"get_weight(miNet)",
"_____no_output_____"
],
[
"dataset_mnist_set = MNISTSet(full=True)\nclass_names = [ 0 , 1 , 2, 3 , 4 , 5 , 6 , 7 , 8 , 9]\nsampleSet = []\nsampleMask = []\nfor i in range(len(class_names)):\n for j in range(len(dataset_mnist_set)):\n if i == dataset_mnist_set[j][0]:\n sampleSet.append(dataset_mnist_set[j][1])\n sampleMask.append(dataset_mnist_set[j][2])\n break\nfig = plt.figure(figsize=(20, 20))\n\nfor i,set in enumerate(sampleSet):\n ax = fig.add_subplot(5, 5, i+1)\n x = set[1, :]\n y = set[0, :]\n plt.scatter(x, y,color = 'k')\n plt.axis([0, 1, 1, 0])\n plt.title('Label: {}'.format(i))",
"_____no_output_____"
],
[
"with torch.no_grad():\n for i in range(len(sampleSet)):\n pred = miNet(sampleSet[i].unsqueeze(0),sampleMask[i].unsqueeze(0))\n print(\"We obtained {}, and the expected is: {} with probaility\".format(np.argmax(pred).item(),class_names[i]))",
"We obtained 0, and the expected is: 0 with probaility\nWe obtained 1, and the expected is: 1 with probaility\nWe obtained 2, and the expected is: 2 with probaility\nWe obtained 3, and the expected is: 3 with probaility\nWe obtained 4, and the expected is: 4 with probaility\nWe obtained 5, and the expected is: 5 with probaility\nWe obtained 6, and the expected is: 6 with probaility\nWe obtained 7, and the expected is: 7 with probaility\nWe obtained 8, and the expected is: 8 with probaility\nWe obtained 9, and the expected is: 9 with probaility\n"
],
[
"with torch.no_grad():\n fig, ax = plt.subplots(1, 1, figsize=(15,5))\n plt.subplot(1,2,1) \n pred = model1(sampleSet[8].unsqueeze(0))\n feature_map = pred\n data = pred.squeeze(0).detach().numpy()\n sn.heatmap(data, annot=False, fmt='g')\n plt.title(\"Salida después de la primera capa\")\n plt.subplot(1,2,2) \n pred = pred.max(2)[0]\n max_map = pred\n print(max_map.shape)\n data = pred.detach().numpy()\n sn.heatmap(data, annot=False, fmt='g')\n plt.title(\"Salida después de la operación invariante al orden\")\n plt.show()",
"torch.Size([1, 32])\n"
],
[
"\ndef critical_points_MaxEncoder(model, sample):\n critical_points_y = []\n critical_points_x = []\n #Salida de la capa de convoluciones\n pred = model(sample.unsqueeze(0))\n features_map = abs(pred.squeeze(0))\n xsort,_ = torch.sort(features_map,descending=True)\n #print(xsort)\n xsortargs = torch.argsort(features_map,descending=True)\n #print(xsortargs[0][:][0])\n \n #xmax = features_map.max(axis=1)[0]\n #xmax,_ = torch.sort(xmax,descending=True)\n #print(xmax.shape)\n xmaxargs = features_map.argmax(axis=1)\n xmaxargs,_ = torch.sort(xmaxargs,descending=True)\n #No nos quedamos con las coordenadas repetidas\n newxmaxargs = np.unique(xmaxargs)\n xmin = features_map.min(axis=1)[0]\n #print(xmax.shape)\n xminargs = features_map.argmin(axis=1)\n #No nos quedamos con las coordenadas repetidas\n newxminargs = np.unique(xminargs)\n points = sample.numpy()\n '''\n critical = np.zeros(342)\n for f in range(32):\n for p in range(342):\n if features_map[f,p] == xmax[f]:\n critical[p] =critical[p]+1\n sorted_critical = np.sort(-critical)\n sorted_critical_args = -np.argsort(-critical)\n print(sorted_critical_args*-1)\n '''\n '''\n Vamos a probar cogiendo los 2 primeros maximos\n \n #print(features_map.shape)\n xsort,_ = torch.sort(features_map,descending=True)\n xsortargs= torch.argsort(features_map,descending=True)\n xsum = features_map.sum(1)\n print(xsortargs.shape)\n #print(xsortargs.shape)\n \n for i,tensor in enumerate(features_map):\n suma = 0\n z = 0\n #print(tensor.shape)\n while(suma<(xsum[i]*0.3) and z < 10):\n #print(xsortargs[z].shape)\n suma+= features_map[i][z]\n critical_points_y.append(points[0,xsortargs[i][z]])\n critical_points_x.append(points[1,xsortargs[i][z]])\n z += 1\n \n '''\n '''\n for k in range(len(newxmaxargs)):\n critical_points_y.append(points[0,newxmaxargs[k]])\n critical_points_x.append(points[1,newxmaxargs[k]])\n \n for k in range(len(newxminargs)):\n critical_points_y.append(points[0,newxminargs[k]])\n critical_points_x.append(points[1,newxminargs[k]])\n '''\n guardados = []\n for j in range(2):\n for i in range(32):\n if xsortargs[i][:][j] not in guardados:\n if j == 0 :\n critical_points_y.append(points[0,xsortargs[i][:][j+10]])\n critical_points_x.append(points[1,xsortargs[i][:][j+10]])\n guardados.append(xsortargs[i][:][j+10])\n \n else:\n \n critical_points_y.append(points[0,xsortargs[i][:][j]])\n critical_points_x.append(points[1,xsortargs[i][:][j]])\n #print(xsort[i][:][j])\n guardados.append(xsortargs[i][:][j])\n \n\n \n\n \n #print(len(critical_points_x))\n x = np.array(critical_points_x)\n y = np.array(critical_points_y)\n s = np.stack((y,x))\n return s ",
"_____no_output_____"
],
[
"def critical_points_MaxEncoder(model, sample):\n critical_points_y = []\n critical_points_x = []\n #Salida de la capa de convoluciones\n pred = model(sample.unsqueeze(0))\n features_map = pred.squeeze(0)\n #print(features_map.shape)\n #Aplicamos la operación invariante max\n xmax = features_map.max(axis=1)[0]\n #print(xmax.shape)\n xmaxargs = features_map.argmax(axis=1)\n #No nos quedamos con las coordenadas repetidas\n newxmaxargs = np.unique(xmaxargs)\n points = sample.numpy()\n '''\n critical = np.zeros(342)\n for f in range(32):\n for p in range(342):\n if features_map[f,p] == xmax[f]:\n critical[p] =critical[p]+1\n sorted_critical = np.sort(-critical)\n sorted_critical_args = -np.argsort(-critical)\n print(sorted_critical_args*-1)\n '''\n rango = len(newxmaxargs)\n for k in range(rango):\n #if not (points[0,newxmaxargs[k]]==0. and points[1,newxmaxargs[k]] ==0.):\n critical_points_y.append(points[0,newxmaxargs[k]])\n critical_points_x.append(points[1,newxmaxargs[k]])\n\n x = np.array(critical_points_x)\n y = np.array(critical_points_y)\n s = np.stack((y,x))\n return s ",
"_____no_output_____"
],
[
"crit = dataset_mnist_set[800][1]\ns = critical_points_MaxEncoder(model1,crit)\ns.shape",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(20, 20))\nfor i,set in enumerate(sampleSet):\n crit = critical_points_MaxEncoder(model1,set)\n ax = fig.add_subplot(5, 5, i+1)\n x = set[1, :]\n y = set[0, :]\n plt.scatter(x, y,color = 'c',s=40)\n x = crit[1, :]\n y = crit[0, :]\n plt.scatter(x, y,marker='o',color = 'r',s=70)\n plt.axis([0, 1, 1, 0])\n plt.title('Puntos Criticos')\nplt.show()",
"_____no_output_____"
],
[
"crit = dataset_mnist_set[88][1]\ns = critical_points_MaxEncoder(model1,crit)\nst = torch.Tensor(s).unsqueeze(0)\nprint(st.shape)\nmuestraSintetica = torch.zeros(1,2,0)\nmuestraSintetica = torch.cat((st,muestraSintetica,),2)\nmask_sintetica = torch.ones(1,st.size(2))\nprint(muestraSintetica.shape)\nprint(mask_sintetica.shape)",
"torch.Size([1, 2, 15])\ntorch.Size([1, 2, 15])\ntorch.Size([1, 15])\n"
],
[
"sam = muestraSintetica.squeeze(0)\n#print(sam)\nfig = plt.figure(figsize=(10, 5))\nax = fig.add_subplot(1, 2,1)\nx1 = s[1, :]\ny1 = s[0, :]\nplt.scatter(x1, y1,c ='r',s=40)\n#plt.scatter(x1, y1,marker='x',color ='r')\nplt.axis([0, 1, 1, 0])\nax = fig.add_subplot(1, 2,2)\nx = crit[1, :]\ny = crit[0, :]\nplt.scatter(x, y,marker='o',color = 'k',s=70)\nplt.axis([0, 1, 1, 0])\nplt.title('Puntos Criticos')\nplt.scatter(x1, y1,marker='o',color ='r')\nplt.show()",
"_____no_output_____"
],
[
"with torch.no_grad():\n pred = miNet(muestraSintetica,mask_sintetica)\n print(muestraSintetica.shape)\n print(mask_sintetica.shape)\n print(\"We obtained {}, and the expected is: {}\".format(np.argmax(pred).item(),0))\n predReal= miNet(crit.unsqueeze(0),dataset_mnist_set[98][1])\n print(pred)\n print(predReal)",
"torch.Size([1, 2, 15])\ntorch.Size([1, 15])\nWe obtained 0, and the expected is: 0\ntensor([[ 0.4071, -0.8904, 0.1836, -0.2046, -2.6372, -0.5906, -1.0604, -2.2175,\n -0.2494, -0.9318]])\ntensor([[ 5.5092, -9.4385, -2.3389, -3.3350, -10.9366, -2.3762, -7.7201,\n -3.4707, -0.8367, -4.8113]])\n"
],
[
"with torch.no_grad():\n for i in range(len(sampleSet)):\n #Preparacion de la muestra sintetica\n crit = sampleSet[i]\n s = critical_points_MaxEncoder(model1,crit)\n st = torch.Tensor(s).unsqueeze(0)\n muestraSintetica = torch.zeros(1,2,0)\n muestraSintetica = torch.cat((st,muestraSintetica,),2)\n mask_sintetica = torch.ones(1,st.size(2))\n #Prediccion de la red con la muestra\n pred = miNet(muestraSintetica,mask_sintetica)\n predReal= miNet(crit.unsqueeze(0),sampleMask[i].unsqueeze(0))\n print()\n print(\"We obtained {}, and the expected is: {}, with this trust {}, vs the real one: {}\".format(np.argmax(pred).item(), class_names[i],round(pred.max().item(),2),round(predReal.max().item(),2)))\n ",
"\nWe obtained 0, and the expected is: 0, with this trust 0.83, vs the real one: 5.07\n\nWe obtained 0, and the expected is: 1, with this trust 0.57, vs the real one: 2.8\n\nWe obtained 0, and the expected is: 2, with this trust 0.52, vs the real one: 1.34\n\nWe obtained 8, and the expected is: 3, with this trust -0.12, vs the real one: 3.91\n\nWe obtained 9, and the expected is: 4, with this trust -0.08, vs the real one: -0.91\n\nWe obtained 8, and the expected is: 5, with this trust 0.36, vs the real one: 4.59\n\nWe obtained 2, and the expected is: 6, with this trust 1.19, vs the real one: 5.08\n\nWe obtained 0, and the expected is: 7, with this trust 0.2, vs the real one: 3.87\n\nWe obtained 0, and the expected is: 8, with this trust 0.59, vs the real one: 0.97\n\nWe obtained 8, and the expected is: 9, with this trust 0.3, vs the real one: 3.32\n"
],
[
"muestraSintetica = torch.zeros(1,2,262)\na = torch.Tensor(x1).unsqueeze(0)\nk = torch.Tensor(y1).unsqueeze(0)\nprint(k.shape)\nf = torch.cat((k,a),0).unsqueeze(0)\nprint(f.shape)\nmuestraSintetica = torch.cat((f,muestraSintetica,),2)\nmuestraSintetica.shape\nmask_sintetica = torch.ones(1,342)\nprint(muestraSintetica.shape)\nprint(mask_sintetica.shape)",
"torch.Size([1, 32])\ntorch.Size([1, 2, 32])\ntorch.Size([1, 2, 294])\ntorch.Size([1, 342])\n"
],
[
"342-32",
"_____no_output_____"
],
[
"sam = muestraSintetica.squeeze(0)\nx = sam[1, :]\ny = sam[0, :]\npyplot.scatter(x, y,c ='b',s=40)\nx1 = critical_points_x\ny1 = critical_points_y\npyplot.scatter(x1, y1,marker='x',color ='r')\npyplot.axis([0, 1, 1, 0])\n#pyplot.title('Label: {}'.format(expected))\n#plt.savefig('test.png')\npyplot.show()",
"_____no_output_____"
],
[
"with torch.no_grad():\n pred = miNet(muestraSintetica,mask_sintetica)\n print(muestraSintetica.shape)\n print(mask_sintetica.shape)\n print(\"We obtained {}, and the expected is: {}\".format(np.argmax(pred).item(),expected.item()))",
"torch.Size([1, 2, 294])\ntorch.Size([1, 342])\nWe obtained 3, and the expected is: 9\n"
]
],
[
[
"## Comprobación de que funciona a la inversa, con los puntos menos informativos",
"_____no_output_____"
]
],
[
[
"sorted_noncritical = -np.sort(-critical)[::-1]\nsorted_noncritical_args = -np.argsort(-critical)[::-1]\nprint(sorted_noncritical)\n#print(sorted_critical_args)\nK = 100\nnoncritical_points_x = []\nnoncritical_points_y = []\nfor k in range(K):\n noncritical_points_x.append(points[0,sorted_noncritical_args[k]])\n noncritical_points_y.append(points[1,sorted_noncritical_args[k]])\n\n#criticos = critical_points.reshape(2,100)\n#print(critical_points_x)\nprint(len(noncritical_points_x))\n#print(critical_points_y)\nprint(np.sum(sorted_critical ==0))",
"[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.]\n100\n342\n"
],
[
"y1 = noncritical_points_x\nx1 = noncritical_points_y\npyplot.scatter(x1, y1,marker='o',color ='r',s=50)\nsam = sample.squeeze(0)\nx = sam[1, :]\ny = sam[0, :]\npyplot.scatter(x, y,c ='b',s=20)\npyplot.axis([0, 1, 1, 0])\npyplot.show()\n\ny = noncritical_points_x\nx = noncritical_points_y\npyplot.scatter(x, y,marker='x',color ='r')\npyplot.axis([0, 1, 1, 0])\npyplot.show()\n\nsam = sample.squeeze(0)\nx = sam[1, :]\ny = sam[0, :]\npyplot.scatter(x, y,c ='b',s=20)\npyplot.axis([0, 1, 1, 0])\n#pyplot.title('Label: {}'.format(expected))\n#plt.savefig('test.png')\npyplot.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec89f929b0820ee174a796b79cd2883099db5c13 | 13,023 | ipynb | Jupyter Notebook | notebook/flaml_pytorch_cifar10.ipynb | sugar-shay/FLAML-1 | db1fb9b47b52d82e24368a0527be88db7025c0f7 | [
"MIT"
] | 1 | 2021-12-03T06:48:31.000Z | 2021-12-03T06:48:31.000Z | notebook/flaml_pytorch_cifar10.ipynb | panda4698/FLAML | 3cf8cb0761e250fb01cfecbbf8ef403e487b8c89 | [
"MIT"
] | null | null | null | notebook/flaml_pytorch_cifar10.ipynb | panda4698/FLAML | 3cf8cb0761e250fb01cfecbbf8ef403e487b8c89 | [
"MIT"
] | 1 | 2021-11-20T01:59:46.000Z | 2021-11-20T01:59:46.000Z | 32.235149 | 175 | 0.518851 | [
[
[
"# Pytorch model tuning example on CIFAR10\nThis notebook uses flaml to tune a pytorch model on CIFAR10. It is modified based on [this example](https://docs.ray.io/en/master/tune/examples/cifar10_pytorch.html).\n\n**Requirements.** This notebook requires:",
"_____no_output_____"
]
],
[
[
"!pip install torchvision flaml[blendsearch,ray];",
"_____no_output_____"
]
],
[
[
"## Network Specification",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nfrom torch.utils.data import random_split\nimport torchvision\nimport torchvision.transforms as transforms\n\n\nclass Net(nn.Module):\n\n def __init__(self, l1=120, l2=84):\n super(Net, self).__init__()\n self.conv1 = nn.Conv2d(3, 6, 5)\n self.pool = nn.MaxPool2d(2, 2)\n self.conv2 = nn.Conv2d(6, 16, 5)\n self.fc1 = nn.Linear(16 * 5 * 5, l1)\n self.fc2 = nn.Linear(l1, l2)\n self.fc3 = nn.Linear(l2, 10)\n\n def forward(self, x):\n x = self.pool(F.relu(self.conv1(x)))\n x = self.pool(F.relu(self.conv2(x)))\n x = x.view(-1, 16 * 5 * 5)\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = self.fc3(x)\n return x",
"_____no_output_____"
]
],
[
[
"## Data",
"_____no_output_____"
]
],
[
[
"def load_data(data_dir=\"data\"):\n transform = transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))\n ])\n\n trainset = torchvision.datasets.CIFAR10(\n root=data_dir, train=True, download=True, transform=transform)\n\n testset = torchvision.datasets.CIFAR10(\n root=data_dir, train=False, download=True, transform=transform)\n\n return trainset, testset",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"from ray import tune\n\ndef train_cifar(config, checkpoint_dir=None, data_dir=None):\n if \"l1\" not in config:\n logger.warning(config)\n net = Net(2**config[\"l1\"], 2**config[\"l2\"])\n\n device = \"cpu\"\n if torch.cuda.is_available():\n device = \"cuda:0\"\n if torch.cuda.device_count() > 1:\n net = nn.DataParallel(net)\n net.to(device)\n\n criterion = nn.CrossEntropyLoss()\n optimizer = optim.SGD(net.parameters(), lr=config[\"lr\"], momentum=0.9)\n\n # The `checkpoint_dir` parameter gets passed by Ray Tune when a checkpoint\n # should be restored.\n if checkpoint_dir:\n checkpoint = os.path.join(checkpoint_dir, \"checkpoint\")\n model_state, optimizer_state = torch.load(checkpoint)\n net.load_state_dict(model_state)\n optimizer.load_state_dict(optimizer_state)\n\n trainset, testset = load_data(data_dir)\n\n test_abs = int(len(trainset) * 0.8)\n train_subset, val_subset = random_split(\n trainset, [test_abs, len(trainset) - test_abs])\n\n trainloader = torch.utils.data.DataLoader(\n train_subset,\n batch_size=int(2**config[\"batch_size\"]),\n shuffle=True,\n num_workers=4)\n valloader = torch.utils.data.DataLoader(\n val_subset,\n batch_size=int(2**config[\"batch_size\"]),\n shuffle=True,\n num_workers=4)\n\n for epoch in range(int(round(config[\"num_epochs\"]))): # loop over the dataset multiple times\n running_loss = 0.0\n epoch_steps = 0\n for i, data in enumerate(trainloader, 0):\n # get the inputs; data is a list of [inputs, labels]\n inputs, labels = data\n inputs, labels = inputs.to(device), labels.to(device)\n\n # zero the parameter gradients\n optimizer.zero_grad()\n\n # forward + backward + optimize\n outputs = net(inputs)\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n\n # print statistics\n running_loss += loss.item()\n epoch_steps += 1\n if i % 2000 == 1999: # print every 2000 mini-batches\n print(\"[%d, %5d] loss: %.3f\" % (epoch + 1, i + 1,\n running_loss / epoch_steps))\n running_loss = 0.0\n\n # Validation loss\n val_loss = 0.0\n val_steps = 0\n total = 0\n correct = 0\n for i, data in enumerate(valloader, 0):\n with torch.no_grad():\n inputs, labels = data\n inputs, labels = inputs.to(device), labels.to(device)\n\n outputs = net(inputs)\n _, predicted = torch.max(outputs.data, 1)\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n\n loss = criterion(outputs, labels)\n val_loss += loss.cpu().numpy()\n val_steps += 1\n\n # Here we save a checkpoint. It is automatically registered with\n # Ray Tune and will potentially be passed as the `checkpoint_dir`\n # parameter in future iterations.\n with tune.checkpoint_dir(step=epoch) as checkpoint_dir:\n path = os.path.join(checkpoint_dir, \"checkpoint\")\n torch.save(\n (net.state_dict(), optimizer.state_dict()), path)\n\n tune.report(loss=(val_loss / val_steps), accuracy=correct / total)\n print(\"Finished Training\")",
"_____no_output_____"
]
],
[
[
"## Test Accuracy",
"_____no_output_____"
]
],
[
[
"def _test_accuracy(net, device=\"cpu\"):\n trainset, testset = load_data()\n\n testloader = torch.utils.data.DataLoader(\n testset, batch_size=4, shuffle=False, num_workers=2)\n\n correct = 0\n total = 0\n with torch.no_grad():\n for data in testloader:\n images, labels = data\n images, labels = images.to(device), labels.to(device)\n outputs = net(images)\n _, predicted = torch.max(outputs.data, 1)\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n\n return correct / total",
"_____no_output_____"
]
],
[
[
"## Hyperparameter Optimization",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport flaml\nimport os\n\ndata_dir = os.path.abspath(\"data\")\nload_data(data_dir) # Download data for all trials before starting the run",
"_____no_output_____"
]
],
[
[
"### Search space",
"_____no_output_____"
]
],
[
[
"max_num_epoch = 100\nconfig = {\n \"l1\": tune.randint(2, 9), # log transformed with base 2\n \"l2\": tune.randint(2, 9), # log transformed with base 2\n \"lr\": tune.loguniform(1e-4, 1e-1),\n \"num_epochs\": tune.loguniform(1, max_num_epoch),\n \"batch_size\": tune.randint(1, 5) # log transformed with base 2\n}",
"_____no_output_____"
],
[
"time_budget_s = 600 # time budget in seconds\ngpus_per_trial = 0.5 # number of gpus for each trial; 0.5 means two training jobs can share one gpu\nnum_samples = 500 # maximal number of trials\nnp.random.seed(7654321)",
"_____no_output_____"
]
],
[
[
"### Launch the tuning",
"_____no_output_____"
]
],
[
[
"import time\nstart_time = time.time()\nresult = flaml.tune.run(\n tune.with_parameters(train_cifar, data_dir=data_dir),\n config=config,\n metric=\"loss\",\n mode=\"min\",\n low_cost_partial_config={\"num_epochs\": 1},\n max_resource=max_num_epoch,\n min_resource=1,\n report_intermediate_result=True, # only set to True when intermediate results are reported by tune.report\n resources_per_trial={\"cpu\": 1, \"gpu\": gpus_per_trial},\n local_dir='logs/',\n num_samples=num_samples,\n time_budget_s=time_budget_s,\n use_ray=True)\n",
"_____no_output_____"
],
[
"print(f\"#trials={len(result.trials)}\")\nprint(f\"time={time.time()-start_time}\")\nbest_trial = result.get_best_trial(\"loss\", \"min\", \"all\")\nprint(\"Best trial config: {}\".format(best_trial.config))\nprint(\"Best trial final validation loss: {}\".format(\n best_trial.metric_analysis[\"loss\"][\"min\"]))\nprint(\"Best trial final validation accuracy: {}\".format(\n best_trial.metric_analysis[\"accuracy\"][\"max\"]))\n\nbest_trained_model = Net(2**best_trial.config[\"l1\"],\n 2**best_trial.config[\"l2\"])\ndevice = \"cpu\"\nif torch.cuda.is_available():\n device = \"cuda:0\"\n if gpus_per_trial > 1:\n best_trained_model = nn.DataParallel(best_trained_model)\nbest_trained_model.to(device)\n\ncheckpoint_path = os.path.join(best_trial.checkpoint.value, \"checkpoint\")\n\nmodel_state, optimizer_state = torch.load(checkpoint_path)\nbest_trained_model.load_state_dict(model_state)\n\ntest_acc = _test_accuracy(best_trained_model, device)\nprint(\"Best trial test set accuracy: {}\".format(test_acc))",
"#trials=44\ntime=1193.913584947586\nBest trial config: {'l1': 8, 'l2': 8, 'lr': 0.0008818671030627281, 'num_epochs': 55.9513429004283, 'batch_size': 3}\nBest trial final validation loss: 1.0694482081472874\nBest trial final validation accuracy: 0.6389\nFiles already downloaded and verified\nFiles already downloaded and verified\nBest trial test set accuracy: 0.6294\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec8a0b27a976f27e1467b067e3556b2dc58da08b | 24,634 | ipynb | Jupyter Notebook | python_hw/Day_3_Conditionals_&_Loops_.ipynb | ibrahima1289/kura-labs-academy | 94e76962a0c6bdfd7657ce7c531d66238357608e | [
"MIT"
] | null | null | null | python_hw/Day_3_Conditionals_&_Loops_.ipynb | ibrahima1289/kura-labs-academy | 94e76962a0c6bdfd7657ce7c531d66238357608e | [
"MIT"
] | null | null | null | python_hw/Day_3_Conditionals_&_Loops_.ipynb | ibrahima1289/kura-labs-academy | 94e76962a0c6bdfd7657ce7c531d66238357608e | [
"MIT"
] | null | null | null | 27.432071 | 264 | 0.413615 | [
[
[
"<a href=\"https://colab.research.google.com/github/ibrahima1289/Python-Codes/blob/main/kura%20labs/Day_3_Conditionals_%26_Loops_.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Conditionals",
"_____no_output_____"
],
[
"## Overview\n- (Need to know) Indentation in Python\n- What are conditional statements? Why do we need them?\n- If statements in Python\n- Why, Why not Loops?\n- Loops in Python\n- Exercises",
"_____no_output_____"
],
[
"# [Tabs vs Spaces](https://www.youtube.com/watch?v=SsoOG6ZeyUI)\n(Worth Watching)",
"_____no_output_____"
],
[
"## Indentation in Python\n- Indentation has a very specific role in Python and is **important!**\n- It is used as alternative to parenthesis, and getting it wrong will cause errors. \n- Python 3 allows 4 spaces or tabs, but not both. [But in working in Jupyter seems to work fine.]\n- In Python, spaces are preferred according to the [PEP 8 style guide](https://www.python.org/dev/peps/pep-0008/).\n\n",
"_____no_output_____"
],
[
"# What are conditional statements? Why do we need them?",
"_____no_output_____"
],
[
"## `if` Statements\n- Enables logical branching and recoding of data.\n- BUT, `if statements` can result in long code branches, repeated code.\n- Best to keep if statements short.\n- Keep in mind the [Zen of Python](https://www.python.org/dev/peps/pep-0020/) when writing if statements. ",
"_____no_output_____"
],
[
"## Conditional Statements and Indentation\n- The syntax for control structures in Python use _colons_ and _indentation_.\n- Beware that indentation affects flow.\n- `if` statement starts the comparison. \n- `elif` give additional conditions.\n- `else` gives what to do if other conditions are not met.",
"_____no_output_____"
]
],
[
[
"y = 5\nx = 3\nif x > 0:\n print ('x is strictly positive')\n print (x) \nprint ('Finished.', x, y)\nx",
"x is strictly positive\n3\nFinished. 3 5\n"
],
[
"x = 1\ny = 0\nif x > 0:\n print('x is greater than 0')\n if y > 0:\n print('& y is also greater than 0')\n elif y < 0:\n print('& y is 0')\n else:\n print ('& y is equal 0')\nprint (\"x: \",x)\nprint ('Finished.')",
"x is greater than 0\n& y is equal 0\nx: 1\nFinished.\n"
],
[
"x > 0",
"_____no_output_____"
],
[
"x != 5 or ",
"_____no_output_____"
],
[
"x = 5\nx",
"_____no_output_____"
]
],
[
[
"## Python Logic and Conditions\n- Less than\t<\n- Greater than\t\t>\n- Less than or equal\t≤\t<=\n- Greater than or equal >=\n- Equals\t==\n- Not equal\t!=\n- `and` can be used to put multiple logical expressions together.\n- `or` can be used to put multiple logical expressions together.",
"_____no_output_____"
]
],
[
[
"x = -1\ny = 1\nif x >= 0 and y >= 0:\n print ('x and y are greater than 0 or 0')\nelif x >= 0 or y >= 0:\n if x > 0:\n print ('x is greater than 0')\n else:\n print ('y is greater than 0')\nelse:\n print ('neither x nor y greater than 0')\n",
"y is greater than 0\n"
]
],
[
[
"## Python Conditionals (Alt Syntax) \n- Clean syntax doesn't get complex branching \n- Python ternary conditional operator `(falseValue, trueValue)[<logicalTest>]`\n- Lambda Functions as if statement.",
"_____no_output_____"
]
],
[
[
"x=0\nz = 5 if x > 0 else 0\nprint(z)",
"0\n"
],
[
"# This is a form of if statement called ternary conditional operator \nx=1\n#The first value is the value if the conditional is false\nz=(0, 5)[x>0]\nprint(z)",
"5\n"
]
],
[
[
"# Why, Why Not Loops?",
"_____no_output_____"
],
[
"## Why, Why Not Loops?\n- Iterate over arrays or lists easily. `for` or `while` loops can be nested.\n- BUT, in many cases for loops don't scale well and are slower than alternate methods involving functions. \n- BUT, don't worry about prematurely optimizing code.\n- Often if you are doing a loop, there is a function that is faster. You might not care for small data applications.\n- Keep in mind the [Zen of Python](https://www.python.org/dev/peps/pep-0020/#id3) when writing `for` statements.",
"_____no_output_____"
]
],
[
[
"#Here we are iterating on lists.\nsum=0\nfor ad in [1, 2, 3]: \n sum+=ad #This is short hand for sum = sum+ad\n print(sum) \n \nfor country in ['England', 'Spain', 'India']:\n print(country)",
"1\n3\n6\nEngland\nSpain\nIndia\n"
],
[
"x=[0,1,2]\ny=['a','b','c']\n#Nested for loops\nfor a in x:\n for b in y:\n print(a,b)\n",
"0 a\n0 b\n0 c\n1 a\n1 b\n1 c\n2 a\n2 b\n2 c\n"
]
],
[
[
"## The `for` Loop\n- Can accept a `range(start, stop, step)` or `range(stop)` object\n- Can break out of it with a `break` command",
"_____no_output_____"
]
],
[
[
"z=range(5)\nz",
"_____no_output_____"
],
[
"#Range is a built in function that can be passed to a for loop\n#https://docs.python.org/3/library/functions.html#func-range\n#Range accepts a number and a (start/stop/step) like the arrange command.\nz=range(5)\nprint(z, type(z))\n#Range\nfor i in z:\n print('Printing ten')\nfor i in range(5):\n print('Print five more')\nfor i in range(5,20,2):\n print('i: %d is the value' % (i)) \n print(f'i:{i} is the value' ) #This is an alternate way of embedding values in text.",
"range(0, 5) <class 'range'>\nPrinting ten\nPrinting ten\nPrinting ten\nPrinting ten\nPrinting ten\nPrint five more\nPrint five more\nPrint five more\nPrint five more\nPrint five more\ni: 5 is the value\ni:5 is the value\ni: 7 is the value\ni:7 is the value\ni: 9 is the value\ni:9 is the value\ni: 11 is the value\ni:11 is the value\ni: 13 is the value\ni:13 is the value\ni: 15 is the value\ni:15 is the value\ni: 17 is the value\ni:17 is the value\ni: 19 is the value\ni:19 is the value\n"
],
[
"#Sometimes you need to break out of a loop\nfor x in range(3):\n print('x:',x)\n if x == 2:\n break\n",
"x: 0\nx: 1\nx: 2\n"
]
],
[
[
"### List, Set, and Dict Comprehension (Fancy for Loops)\n- Python has a special way of compressing list building to a single line. \n- Set Comprehension is very similar, but with the `{` bracket.\n- Can incorporate conditionals. \n- S\n",
"_____no_output_____"
]
],
[
[
"#This is the long way of building lists.\nL = []\nfor n in range(10):\n L.append(n ** 2)\nL",
"_____no_output_____"
],
[
"#With list comprehension. \nL=[n ** 2 for n in range(10)]\nL",
"_____no_output_____"
],
[
"#Any actions are on left side, any conditionals on right side \n[i for i in range(20) if i % 3 == 0]",
"_____no_output_____"
]
],
[
[
"### Multiple Interators\n- Iterating on multiple values",
"_____no_output_____"
]
],
[
[
"[(i, j) for i in range(2) for j in range(3)]",
"_____no_output_____"
]
],
[
[
"### Set Comprehension\n- Remember sets must have unique values.",
"_____no_output_____"
]
],
[
[
"#We can change it to a setby just changing the brackets.\n{n**2 for n in range(6)}",
"_____no_output_____"
]
],
[
[
"### Dict Comprehension\n- Remember sets must have unique values.",
"_____no_output_____"
]
],
[
[
"#We can change it to a dictionary by just changing the brackets and adding a colon.\n{n:n**2 for n in range(6)}",
"_____no_output_____"
]
],
[
[
"## While Loops\n- Performs a loop while a conditional is True.\n- Doesn't auto-increment.\n",
"_____no_output_____"
]
],
[
[
"# While loop is a very interesting \nx = 1\nsum=0\nwhile x<10:\n print (\"Printing x= %d sum= %d\" % (x, sum)) #Note this alternate way of specufiy\n x += 1\n sum+=10",
"Printing x= 1 sum= 0\nPrinting x= 2 sum= 10\nPrinting x= 3 sum= 20\nPrinting x= 4 sum= 30\nPrinting x= 5 sum= 40\nPrinting x= 6 sum= 50\nPrinting x= 7 sum= 60\nPrinting x= 8 sum= 70\nPrinting x= 9 sum= 80\n"
]
],
[
[
"## Recoding Variables/Creating Features with `for/if`\n- Often we want to recode data applying some type of conditional statement to each value of a series, list, or column of a data frame.\n- [Regular Expressions](https://docs.python.org/3/howto/regex.html) can be useful in recoding ",
"_____no_output_____"
]
],
[
[
"#Titanic Preview Women and Children first\n\ngender=['Female', 'Female','Female', 'Male', 'Male', 'Male' ]\nage=[75, 45, 15, 1, 45, 4 ]\nname = ['Ms. Sally White', 'Mrs. Susan King', 'Amanda Russ', 'Rev. John Smith' ]\nsurvived=[]\nfor i in range(len(gender)):\n#This is encoding a simple model that women survived. \n if gender[i]=='Female':\n survived.append('Survived')\n else:\n survived.append('Died') \nprint(survived)\n \n#BUT, we won't typically be using this type of recoding, so we aren't going to do a lot of exercises on it. ",
"['Survived', 'Survived', 'Survived', 'Died', 'Died', 'Died']\n"
]
],
[
[
"Copyright [AnalyticsDojo](http://rpi.analyticsdojo.com) 2016.\nThis work is licensed under the [Creative Commons Attribution 4.0 International](https://creativecommons.org/licenses/by/4.0/) license agreement.\nAdopted from [materials](https://github.com/phelps-sg/python-bigdata) Copyright [Steve Phelps](http://sphelps.net) 2014",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec8a169e3f5391dd4cbf371f246bbdfd7a14d095 | 21,629 | ipynb | Jupyter Notebook | Prediction/summary_pred.ipynb | choudhury722k/Text-summarizer | c0630ae57fb4e30e3c446eb9c95d45547751d1c8 | [
"Apache-2.0"
] | null | null | null | Prediction/summary_pred.ipynb | choudhury722k/Text-summarizer | c0630ae57fb4e30e3c446eb9c95d45547751d1c8 | [
"Apache-2.0"
] | null | null | null | Prediction/summary_pred.ipynb | choudhury722k/Text-summarizer | c0630ae57fb4e30e3c446eb9c95d45547751d1c8 | [
"Apache-2.0"
] | null | null | null | 81.31203 | 1,700 | 0.677655 | [
[
[
"from tensorflow.keras.preprocessing.text import Tokenizer\nfrom tensorflow.keras.preprocessing.sequence import pad_sequences\nfrom tensorflow.keras import models \nfrom tensorflow.keras.models import Model\nfrom tensorflow.keras.layers import Input,LSTM,Embedding,Dense,Concatenate,Attention\nimport numpy as np\nimport pickle\nimport nltk\nfrom nltk import word_tokenize\nfrom nltk.stem import LancasterStemmer\nfrom nltk.corpus import stopwords\nfrom bs4 import BeautifulSoup",
"_____no_output_____"
],
[
"# encoder inference\nlatent_dim=500\nmax_in_len = 74\nmax_tr_len = 17\n\n#load the model\nmodel = models.load_model(\"summarizer_model.h5\")\nin_tokenizer = pickle.load(open('in_tokenizer.pickle', 'rb'))\ntr_tokenizer = pickle.load(open('tr_tokenizer.pickle', 'rb'))\ncontractions=pickle.load(open(\"contractions.pkl\",\"rb\"))['contractions']\n#initialize stop words and LancasterStemmer\nstop_words=set(stopwords.words('english'))\nstemm=LancasterStemmer()\n \n \n#construct encoder model from the output of 6 layer i.e.last LSTM layer\nen_outputs,state_h_enc,state_c_enc = model.layers[6].output\nen_states=[state_h_enc,state_c_enc]\n#add input and state from the layer.\nen_model = Model(model.input[0],[en_outputs]+en_states)\n\n# decoder inference\n#create Input object for hidden and cell state for decoder\n#shape of layer with hidden or latent dimension\ndec_state_input_h = Input(shape=(latent_dim,))\ndec_state_input_c = Input(shape=(latent_dim,))\ndec_hidden_state_input = Input(shape=(max_in_len,latent_dim))\n\n# Get the embeddings and input layer from the model\ndec_inputs = model.input[1]\ndec_emb_layer = model.layers[5]\ndec_lstm = model.layers[7]\ndec_embedding= dec_emb_layer(dec_inputs)\n \n#add input and initialize LSTM layer with encoder LSTM states.\ndec_outputs2, state_h2, state_c2 = dec_lstm(dec_embedding, initial_state=[dec_state_input_h,dec_state_input_c])\n\n#Attention layer\nattention = model.layers[8]\nattn_out2 = attention([dec_outputs2,dec_hidden_state_input])\n \nmerge2 = Concatenate(axis=-1)([dec_outputs2, attn_out2])\n\n#Dense layer\ndec_dense = model.layers[10]\ndec_outputs2 = dec_dense(merge2)\n \n# Finally define the Model Class\ndec_model = Model([dec_inputs] + [dec_hidden_state_input,dec_state_input_h,dec_state_input_c],[dec_outputs2] + [state_h2, state_c2])\n\n#create a dictionary with a key as index and value as words.\nreverse_target_word_index = tr_tokenizer.index_word\nreverse_source_word_index = in_tokenizer.index_word\ntarget_word_index = tr_tokenizer.word_index\nreverse_target_word_index[0]=' '",
"_____no_output_____"
],
[
"def decode_sequence(input_seq):\n #get the encoder output and states by passing the input sequence\n en_out, en_h, en_c= en_model.predict(input_seq)\n\n #target sequence with inital word as 'sos'\n target_seq = np.zeros((1, 1))\n target_seq[0, 0] = target_word_index['sos']\n\n #if the iteration reaches the end of text than it will be stop the iteration\n stop_condition = False\n #append every predicted word in decoded sentence\n decoded_sentence = \"\"\n while not stop_condition: \n #get predicted output, hidden and cell state.\n output_words, dec_h, dec_c= dec_model.predict([target_seq] + [en_out,en_h, en_c])\n \n #get the index and from the dictionary get the word for that index.\n word_index = np.argmax(output_words[0, -1, :])\n text_word = reverse_target_word_index[word_index]\n decoded_sentence += text_word +\" \"\n\n # Exit condition: either hit max length\n # or find a stop word or last word.\n if text_word == \"eos\" or len(decoded_sentence) > max_tr_len:\n stop_condition = True\n \n #update target sequence to the current word index.\n target_seq = np.zeros((1, 1))\n target_seq[0, 0] = word_index\n en_h, en_c = dec_h, dec_c\n \n #return the deocded sentence\n return decoded_sentence",
"_____no_output_____"
],
[
"def clean(texts,src):\n #remove the html tags\n texts = BeautifulSoup(texts, \"html\").text\n #tokenize the text into words \n words=word_tokenize(texts.lower())\n words= list(filter(lambda w:(w.isalpha() and len(w)>=3),words))\n #filter words which contains \\ \n #integers or their length is less than or equal to 3\n #contraction file to expand shortened words\n words= [contractions[w] if w in contractions else w for w in words ]\n #stem the words to their root word and filter stop words\n if src==\"inputs\":\n words= [stemm.stem(w) for w in words if w not in stop_words]\n else:\n words= [w for w in words if w not in stop_words]\n return words",
"_____no_output_____"
],
[
"def summary_text(inp_review):\n print(\"Review :\",inp_review)\n inp_review = clean(inp_review,\"inputs\")\n inp_review = ' '.join(inp_review)\n inp_x= in_tokenizer.texts_to_sequences([inp_review]) \n inp_x= pad_sequences(inp_x, maxlen=max_in_len, padding='post')\n summary=decode_sequence(inp_x.reshape(1,max_in_len))\n print(inp_x)\n if 'eos' in summary :\n summary=summary.replace('eos','')\n print(\"\\nPredicted summary:\",summary);print(\"\\n\")\n return summary\n\n ",
"_____no_output_____"
],
[
"data = input(\"Enter : \")\n\nprint(summary_text(data))",
"Enter : tried several low carb breads search proven best one far regular bread texture works great sandwiches nice tender toasted piece good toasted also low carb bread husband low carb diet thought sandwich noon good bread think texture perfect course sandwiches\nReview : tried several low carb breads search proven best one far regular bread texture works great sandwiches nice tender toasted piece good toasted also low carb bread husband low carb diet thought sandwich noon good bread think texture perfect course sandwiches\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8a1cc290853ddf5a019899f086ae4745721825 | 1,925 | ipynb | Jupyter Notebook | 0.1_building_geometry_pre_processing.ipynb | GIScience/pybossa-mapswipe-2 | 920f3ed3707e450f023a30e6138976c790ffadd7 | [
"MIT"
] | 2 | 2020-05-02T10:47:55.000Z | 2021-07-10T06:38:34.000Z | 0.1_building_geometry_pre_processing.ipynb | GIScience/pybossa-mapswipe-2 | 920f3ed3707e450f023a30e6138976c790ffadd7 | [
"MIT"
] | null | null | null | 0.1_building_geometry_pre_processing.ipynb | GIScience/pybossa-mapswipe-2 | 920f3ed3707e450f023a30e6138976c790ffadd7 | [
"MIT"
] | null | null | null | 21.388889 | 410 | 0.579221 | [
[
[
"# Building Geometry Pre-Processing\nA pre-processing step might be needed if the building dataset was generated automatically from satellite imagery. This pre-processing might include simplifying the geometries and making building polygons rectangular. There is already a [great document](https://docs.google.com/document/d/1DEGJYlR-92idDsaP7qICxXUcpC0PZ7ldP48fEmZgxPY/edit?usp=sharing) with information to consider during pre-processing.\n",
"_____no_output_____"
],
[
"## Configuration",
"_____no_output_____"
],
[
"## Download raw Building Geometries from Geoserver",
"_____no_output_____"
],
[
"## Create oriented bounding box for building geometries",
"_____no_output_____"
],
[
"## Save processed geometries as geojson file",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ec8a2192b057514594f9a80184f518ee3c181fa1 | 120,367 | ipynb | Jupyter Notebook | heated_Solidsquare.ipynb | subhamkd/heat_equation | 8e7e299adaa5c355ffd6e82e1b5f7a2c569cb22b | [
"MIT"
] | null | null | null | heated_Solidsquare.ipynb | subhamkd/heat_equation | 8e7e299adaa5c355ffd6e82e1b5f7a2c569cb22b | [
"MIT"
] | null | null | null | heated_Solidsquare.ipynb | subhamkd/heat_equation | 8e7e299adaa5c355ffd6e82e1b5f7a2c569cb22b | [
"MIT"
] | null | null | null | 512.2 | 61,246 | 0.951349 | [
[
[
"## Constants and Geometry ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom matplotlib import pyplot as plt, cm\nfrom heat_equation import *\n\nlx = 5 #length in the x direction\nly = 5 #length in the y direction\nnx = 51 #grid points in x direction\nny = 51 #grid points in y direction\ntf = 3 #final time to calculate to\n\ndx = lx / (nx - 1)\ndy = ly / (ny - 1)\nx = np.linspace(0, lx, nx)\ny = np.linspace(0, ly, ny)\nX, Y = np.meshgrid(x, y)\n\nalpha = 1\n#nu = 0.5\ndt = 0.001 #time step size\nds = int(100) #number of steps after which the state of domain should be saved\n\nnt=int(tf/dt) # number of time steps",
"_____no_output_____"
]
],
[
[
"## Initial and boundary conditions",
"_____no_output_____"
]
],
[
[
"T = np.ones((nx,ny))*300 #initial condition\nnp.savetxt('T0.csv', T, delimiter=',')\n\n#initial condition\nfig = plt.figure(figsize=(11,7),dpi=100)\nlevels=np.linspace(0,300,50)\nplt.contourf(X, Y, T,levels, alpha=0.8, cmap=cm.rainbow)\nplt.colorbar()\n#plt.contour(X, Y, T,10, colors='black')\nax = plt.gca()\nax.set_aspect(1)\nplt.title('Initial condition of the domain')\nplt.show()\n\n#Boundary conditions\nTLeft=['D',300]\nTRight=['D',100]\nTTop=['D',100]\nTBottom=['D',300]\n\nTBCs=[TLeft,TRight,TTop,TBottom]\n\n#solving the equation for the given geometry\nT=heat_equation(T,dx,dy,alpha,dt,ds,nt,TBCs)\n\nclearResults()",
"_____no_output_____"
]
],
[
[
"## Plotting and visualization",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(11,7),dpi=100)\n# plotting the temperature field\nplt.contourf(X, Y, T, levels, alpha=0.8, cmap=cm.rainbow) \nplt.colorbar()\n#plt.contour(X, Y, T,10, colors='black')\n\nplt.xlabel('X')\nplt.ylabel('Y')\nax = plt.gca()\nax.set_aspect(1)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## saving the transient behaviour as an animation",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.animation as animation\n\nfig, ax = plt.subplots(dpi=100)\n\ndef animate(i):\n tmpName='T'+str(i*ds)+'.csv'\n Tfile=pd.read_csv(r\"Results/\"+str(tmpName),header=None)\n Tarr=Tfile.to_numpy()\n # plotting the temperature field\n re = plt.contourf(X, Y, Tarr, levels, alpha=0.8, cmap=cm.rainbow)\n plt.xlabel('X')\n plt.ylabel('Y')\n ax = plt.gca()\n ax.set_aspect(1)\n return re\n\nanim = animation.FuncAnimation(fig, animate, frames=nt//ds, repeat=False)\nanim.save(r'Gifs/SolidSquare.gif', writer='imagemagick', fps=10)\n",
"MovieWriter imagemagick unavailable; using Pillow instead.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec8a26a0e478b02be6979384150f8be3da6e9a43 | 33,093 | ipynb | Jupyter Notebook | notebooks/FD_SUP.data_exploration.ipynb | vCTO/FD_SUP | f05c4bbe9c8d2ecb463d8a68c2d7d20de2f08966 | [
"MIT"
] | null | null | null | notebooks/FD_SUP.data_exploration.ipynb | vCTO/FD_SUP | f05c4bbe9c8d2ecb463d8a68c2d7d20de2f08966 | [
"MIT"
] | null | null | null | notebooks/FD_SUP.data_exploration.ipynb | vCTO/FD_SUP | f05c4bbe9c8d2ecb463d8a68c2d7d20de2f08966 | [
"MIT"
] | null | null | null | 69.816456 | 18,732 | 0.738676 | [
[
[
"### Data Exploration for Foundations in Data Science Team Project\nStart by reading in the pickle file from the data load\n",
"_____no_output_____"
]
],
[
[
"sx ls ../data",
"_____no_output_____"
],
[
"import pickle\nimport os\nimport glob\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn\nfrom sklearn.decomposition import PCA\n\ndf = pd.read_pickle('../data/cleandf.pkl')",
"_____no_output_____"
],
[
"df.head(4)",
"_____no_output_____"
],
[
"new_df = df.filter(regex='zinc2_x|hud_med')\nnew_df.head(4)\nnew_df.describe()",
"_____no_output_____"
],
[
"corr_new_df = new_df.corr(method='pearson')\nprint('----------Correlations using Pearson Method----------')\nprint(corr_new_df)\nprint('----------Heatmap based on above correlations----------')\nmask = np.zeros_like(corr_new_df)\nmask[np.triu_indices_from(mask)] = True\nseaborn.heatmap(corr_new_df, cmap='RdYlGn_r', vmax=1.0, vmin=-1.0, mask = mask, linewidths=2.5)\n\nplt.yticks(rotation=0) \nplt.xticks(rotation=90) \nplt.show()",
"_____no_output_____"
],
[
"# investigate own_rent\ndf.own_rent_x.unique()",
"_____no_output_____"
],
[
"# Seperate out the features and set OWN_RENT_X as the target\ndf_y = df['own_rent_y']\n",
"_____no_output_____"
],
[
"features = ['assisted_y', 'structure_type_y', 'n_units_y', 'vacancy_y', 'cost12_rel_ami_cat_y', 'rooms_y', 'bedrms_y', 'value_y', 'cost08_rel_ami_cat_y', 'utility_y', 'cost12_rel_fmr_cat_y', 'abl_80_y', 'cost_12', 'fmr_y', 'cost12_rel_ami_pct_y', 'cost06_rel_ami_cat_y', 'cost12_rel_fmr_pct_y', 'cost12_rel_pov_pct_y', 'age1_y','cost_08_y', 'cost12_rel_pov_cat_y', 'inc_pel_ami_cat','abl_50_y','other_cost_y','cost08_rel_fmr_cat_y']\ndf_x = df.loc[:, features].values\ndf_x\n",
"_____no_output_____"
],
[
"df_features = pd.DataFrame(df_x)\ndf_next = df_features.replace([np.inf, -np.inf], np.nan)\n#df_features.head(1000)\n#df_cleaned = df_features.dropna(how='any') ",
"_____no_output_____"
],
[
"df_next.describe()\ndf_cleaned = df_next.dropna(how='any') \ndf_cleaned.isna().all()\ndf_cleaned = np.ndarray(df_cleaned)",
"_____no_output_____"
],
[
"np.nan_to_num(df_x)\n",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\ndf_cleaned = StandardScaler().fit_transform(df_x)",
"_____no_output_____"
],
[
"from sklearn.decomposition import PCA\npca = PCA(n_components=2)\nprincipalComponents = pca.fit_transform(df_cleaned)\n",
"_____no_output_____"
],
[
"principalDf = pd.DataFrame(data = principalComponents\n , columns = ['principal component 1', 'principal component 2'])\n\n",
"_____no_output_____"
],
[
"finalDf = pd.concat([principalDf, df[['own_rent_y']]], axis = 1)\n",
"_____no_output_____"
],
[
"fig = plt.figure(figsize = (8,8))\nax = fig.add_subplot(1,1,1) \nax.set_xlabel('Principal Component 1', fontsize = 15)\nax.set_ylabel('Principal Component 2', fontsize = 15)\n\nax.set_title('2 component PCA', fontsize = 20)\ntargets = ['Own', 'Rent']\ncolors = ['r', 'g']\nfor target, color in zip(targets,colors):\n indicesToKeep = finalDf['own_rent_y'] == target\n ax.scatter(finalDf.loc[indicesToKeep, 'principal component 1']\n , finalDf.loc[indicesToKeep, 'principal component 2']\n , c = color\n , s = 50)\nax.legend(targets)\nax.grid()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8a272e97a982e3bdf011a2a9dc36c3e9b5f0bc | 350,262 | ipynb | Jupyter Notebook | Sentiment_Classification_Projects.ipynb | Luke035/dlnd-sentiment-network | 0ef53072a6dfad7d11eb28688b297f65bd538032 | [
"MIT"
] | null | null | null | Sentiment_Classification_Projects.ipynb | Luke035/dlnd-sentiment-network | 0ef53072a6dfad7d11eb28688b297f65bd538032 | [
"MIT"
] | null | null | null | Sentiment_Classification_Projects.ipynb | Luke035/dlnd-sentiment-network | 0ef53072a6dfad7d11eb28688b297f65bd538032 | [
"MIT"
] | null | null | null | 55.85425 | 38,682 | 0.66929 | [
[
[
"# Sentiment Classification & How To \"Frame Problems\" for a Neural Network\n\nby Andrew Trask\n\n- **Twitter**: @iamtrask\n- **Blog**: http://iamtrask.github.io",
"_____no_output_____"
],
[
"### What You Should Already Know\n\n- neural networks, forward and back-propagation\n- stochastic gradient descent\n- mean squared error\n- and train/test splits\n\n### Where to Get Help if You Need it\n- Re-watch previous Udacity Lectures\n- Leverage the recommended Course Reading Material - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) (Check inside your classroom for a discount code)\n- Shoot me a tweet @iamtrask\n\n\n### Tutorial Outline:\n\n- Intro: The Importance of \"Framing a Problem\" (this lesson)\n\n- [Curate a Dataset](#lesson_1)\n- [Developing a \"Predictive Theory\"](#lesson_2)\n- [**PROJECT 1**: Quick Theory Validation](#project_1)\n\n\n- [Transforming Text to Numbers](#lesson_3)\n- [**PROJECT 2**: Creating the Input/Output Data](#project_2)\n\n\n- Putting it all together in a Neural Network (video only - nothing in notebook)\n- [**PROJECT 3**: Building our Neural Network](#project_3)\n\n\n- [Understanding Neural Noise](#lesson_4)\n- [**PROJECT 4**: Making Learning Faster by Reducing Noise](#project_4)\n\n\n- [Analyzing Inefficiencies in our Network](#lesson_5)\n- [**PROJECT 5**: Making our Network Train and Run Faster](#project_5)\n\n\n- [Further Noise Reduction](#lesson_6)\n- [**PROJECT 6**: Reducing Noise by Strategically Reducing the Vocabulary](#project_6)\n\n\n- [Analysis: What's going on in the weights?](#lesson_7)",
"_____no_output_____"
],
[
"# Lesson: Curate a Dataset<a id='lesson_1'></a>\nThe cells from here until Project 1 include code Andrew shows in the videos leading up to mini project 1. We've included them so you can run the code along with the videos without having to type in everything.",
"_____no_output_____"
]
],
[
[
"def pretty_print_review_and_label(i):\n print(labels[i] + \"\\t:\\t\" + reviews[i][:80] + \"...\")\n\ng = open('reviews.txt','r') # What we know!\nreviews = list(map(lambda x:x[:-1],g.readlines()))\ng.close()\n\ng = open('labels.txt','r') # What we WANT to know!\nlabels = list(map(lambda x:x[:-1].upper(),g.readlines()))\ng.close()",
"_____no_output_____"
]
],
[
[
"**Note:** The data in `reviews.txt` we're using has already been preprocessed a bit and contains only lower case characters. If we were working from raw data, where we didn't know it was all lower case, we would want to add a step here to convert it. That's so we treat different variations of the same word, like `The`, `the`, and `THE`, all the same way.",
"_____no_output_____"
]
],
[
[
"len(reviews)",
"_____no_output_____"
],
[
"reviews[1]",
"_____no_output_____"
],
[
"labels[1]",
"_____no_output_____"
]
],
[
[
"# Lesson: Develop a Predictive Theory<a id='lesson_2'></a>",
"_____no_output_____"
]
],
[
[
"print(\"labels.txt \\t : \\t reviews.txt\\n\")\npretty_print_review_and_label(2137)\npretty_print_review_and_label(12816)\npretty_print_review_and_label(6267)\npretty_print_review_and_label(21934)\npretty_print_review_and_label(5297)\npretty_print_review_and_label(4998)",
"labels.txt \t : \t reviews.txt\n\nNEGATIVE\t:\tthis movie is terrible but it has some good effects . ...\nPOSITIVE\t:\tadrian pasdar is excellent is this film . he makes a fascinating woman . ...\nNEGATIVE\t:\tcomment this movie is impossible . is terrible very improbable bad interpretat...\nPOSITIVE\t:\texcellent episode movie ala pulp fiction . days suicides . it doesnt get more...\nNEGATIVE\t:\tif you haven t seen this it s terrible . it is pure trash . i saw this about ...\nPOSITIVE\t:\tthis schiffer guy is a real genius the movie is of excellent quality and both e...\n"
]
],
[
[
"# Project 1: Quick Theory Validation<a id='project_1'></a>\n\nThere are multiple ways to implement these projects, but in order to get your code closer to what Andrew shows in his solutions, we've provided some hints and starter code throughout this notebook.\n\nYou'll find the [Counter](https://docs.python.org/2/library/collections.html#collections.Counter) class to be useful in this exercise, as well as the [numpy](https://docs.scipy.org/doc/numpy/reference/) library.",
"_____no_output_____"
]
],
[
[
"from collections import Counter\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"We'll create three `Counter` objects, one for words from postive reviews, one for words from negative reviews, and one for all the words.",
"_____no_output_____"
]
],
[
[
"test_counter = Counter()",
"_____no_output_____"
],
[
"test_counter['test'] += 1\ntest_counter['test']",
"_____no_output_____"
],
[
"# Create three Counter objects to store positive, negative and total counts\npositive_counts = Counter()\nnegative_counts = Counter()\ntotal_counts = Counter()",
"_____no_output_____"
]
],
[
[
"**TODO:** Examine all the reviews. For each word in a positive review, increase the count for that word in both your positive counter and the total words counter; likewise, for each word in a negative review, increase the count for that word in both your negative counter and the total words counter.\n\n**Note:** Throughout these projects, you should use `split(' ')` to divide a piece of text (such as a review) into individual words. If you use `split()` instead, you'll get slightly different results than what the videos and solutions show.",
"_____no_output_____"
]
],
[
[
"# TODO: Loop over all the words in all the reviews and increment the counts in the appropriate counter objects\nfor i in range(len(reviews)):\n splitted_review = reviews[i].split(' ')\n if labels[i] == 'POSITIVE':\n for gram in splitted_review:\n positive_counts[gram] += 1\n total_counts[gram] += 1\n if labels[i] == 'NEGATIVE':\n for gram in splitted_review:\n negative_counts[gram] += 1\n total_counts[gram] += 1",
"_____no_output_____"
]
],
[
[
"Run the following two cells to list the words used in positive reviews and negative reviews, respectively, ordered from most to least commonly used. ",
"_____no_output_____"
]
],
[
[
"# Examine the counts of the most common words in positive reviews\npositive_counts.most_common()",
"_____no_output_____"
],
[
"# Examine the counts of the most common words in negative reviews\nnegative_counts.most_common()",
"_____no_output_____"
]
],
[
[
"As you can see, common words like \"the\" appear very often in both positive and negative reviews. Instead of finding the most common words in positive or negative reviews, what you really want are the words found in positive reviews more often than in negative reviews, and vice versa. To accomplish this, you'll need to calculate the **ratios** of word usage between positive and negative reviews.\n\n**TODO:** Check all the words you've seen and calculate the ratio of postive to negative uses and store that ratio in `pos_neg_ratios`. \n>Hint: the positive-to-negative ratio for a given word can be calculated with `positive_counts[word] / float(negative_counts[word]+1)`. Notice the `+1` in the denominator – that ensures we don't divide by zero for words that are only seen in positive reviews.",
"_____no_output_____"
]
],
[
[
"# Create Counter object to store positive/negative ratios\npos_neg_ratios = Counter()\n\n# TODO: Calculate the ratios of positive and negative uses of the most common words\n# Consider words to be \"common\" if they've been used at least 100 times\nfor word in total_counts:\n if(total_counts[word] > 10):\n pos_neg_ratios[word] = positive_counts[word] / float(negative_counts[word] + 1)",
"_____no_output_____"
]
],
[
[
"Examine the ratios you've calculated for a few words:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 1.0607993145235326\nPos-to-neg ratio for 'amazing' = 4.022813688212928\nPos-to-neg ratio for 'terrible' = 0.17744252873563218\n"
]
],
[
[
"Looking closely at the values you just calculated, we see the following:\n\n* Words that you would expect to see more often in positive reviews – like \"amazing\" – have a ratio greater than 1. The more skewed a word is toward postive, the farther from 1 its positive-to-negative ratio will be.\n* Words that you would expect to see more often in negative reviews – like \"terrible\" – have positive values that are less than 1. The more skewed a word is toward negative, the closer to zero its positive-to-negative ratio will be.\n* Neutral words, which don't really convey any sentiment because you would expect to see them in all sorts of reviews – like \"the\" – have values very close to 1. A perfectly neutral word – one that was used in exactly the same number of positive reviews as negative reviews – would be almost exactly 1. The `+1` we suggested you add to the denominator slightly biases words toward negative, but it won't matter because it will be a tiny bias and later we'll be ignoring words that are too close to neutral anyway.\n\nOk, the ratios tell us which words are used more often in postive or negative reviews, but the specific values we've calculated are a bit difficult to work with. A very positive word like \"amazing\" has a value above 4, whereas a very negative word like \"terrible\" has a value around 0.18. Those values aren't easy to compare for a couple of reasons:\n\n* Right now, 1 is considered neutral, but the absolute value of the postive-to-negative rations of very postive words is larger than the absolute value of the ratios for the very negative words. So there is no way to directly compare two numbers and see if one word conveys the same magnitude of positive sentiment as another word conveys negative sentiment. So we should center all the values around netural so the absolute value fro neutral of the postive-to-negative ratio for a word would indicate how much sentiment (positive or negative) that word conveys.\n* When comparing absolute values it's easier to do that around zero than one. \n\nTo fix these issues, we'll convert all of our ratios to new values using logarithms.\n\n**TODO:** Go through all the ratios you calculated and convert them to logarithms. (i.e. use `np.log(ratio)`)\n\nIn the end, extremely positive and extremely negative words will have positive-to-negative ratios with similar magnitudes but opposite signs.",
"_____no_output_____"
]
],
[
[
"# TODO: Convert ratios to logs\nfor word in pos_neg_ratios:\n pos_neg_ratios[word] = np.log(pos_neg_ratios[word])",
"/Users/lucagrazioli/anaconda/envs/deepEnv/lib/python3.5/site-packages/ipykernel/__main__.py:3: RuntimeWarning: divide by zero encountered in log\n app.launch_new_instance()\n"
]
],
[
[
"Examine the new ratios you've calculated for the same words from before:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 0.05902269426102881\nPos-to-neg ratio for 'amazing' = 1.3919815802404802\nPos-to-neg ratio for 'terrible' = -1.7291085042663878\n"
]
],
[
[
"If everything worked, now you should see neutral words with values close to zero. In this case, \"the\" is near zero but slightly positive, so it was probably used in more positive reviews than negative reviews. But look at \"amazing\"'s ratio - it's above `1`, showing it is clearly a word with positive sentiment. And \"terrible\" has a similar score, but in the opposite direction, so it's below `-1`. It's now clear that both of these words are associated with specific, opposing sentiments.\n\nNow run the following cells to see more ratios. \n\nThe first cell displays all the words, ordered by how associated they are with postive reviews. (Your notebook will most likely truncate the output so you won't actually see *all* the words in the list.)\n\nThe second cell displays the 30 words most associated with negative reviews by reversing the order of the first list and then looking at the first 30 words. (If you want the second cell to display all the words, ordered by how associated they are with negative reviews, you could just write `reversed(pos_neg_ratios.most_common())`.)\n\nYou should continue to see values similar to the earlier ones we checked – neutral words will be close to `0`, words will get more positive as their ratios approach and go above `1`, and words will get more negative as their ratios approach and go below `-1`. That's why we decided to use the logs instead of the raw ratios.",
"_____no_output_____"
]
],
[
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]\n\n# Note: Above is the code Andrew uses in his solution video, \n# so we've included it here to avoid confusion.\n# If you explore the documentation for the Counter class, \n# you will see you could also find the 30 least common\n# words like this: pos_neg_ratios.most_common()[:-31:-1]",
"_____no_output_____"
]
],
[
[
"# End of Project 1. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n\n# Transforming Text into Numbers<a id='lesson_3'></a>\nThe cells here include code Andrew shows in the next video. We've included it so you can run the code along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\n\nreview = \"This was a horrible, terrible movie.\"\n\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"review = \"The movie was excellent\"\n\nImage(filename='sentiment_network_pos.png')",
"_____no_output_____"
]
],
[
[
"# Project 2: Creating the Input/Output Data<a id='project_2'></a>\n\n**TODO:** Create a [set](https://docs.python.org/3/tutorial/datastructures.html#sets) named `vocab` that contains every word in the vocabulary.",
"_____no_output_____"
]
],
[
[
"# TODO: Create set named \"vocab\" containing all of the words from all of the reviews\nvocab = total_counts.keys()",
"_____no_output_____"
]
],
[
[
"Run the following cell to check your vocabulary size. If everything worked correctly, it should print **74074**",
"_____no_output_____"
]
],
[
[
"vocab_size = len(vocab)\nprint(vocab_size)",
"74074\n"
]
],
[
[
"Take a look at the following image. It represents the layers of the neural network you'll be building throughout this notebook. `layer_0` is the input layer, `layer_1` is a hidden layer, and `layer_2` is the output layer.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network_2.png')",
"_____no_output_____"
]
],
[
[
"**TODO:** Create a numpy array called `layer_0` and initialize it to all zeros. You will find the [zeros](https://docs.scipy.org/doc/numpy/reference/generated/numpy.zeros.html) function particularly helpful here. Be sure you create `layer_0` as a 2-dimensional matrix with 1 row and `vocab_size` columns. ",
"_____no_output_____"
]
],
[
[
"# TODO: Create layer_0 matrix with dimensions 1 by vocab_size, initially filled with zeros\nlayer_0 = np.zeros(shape=(1, len(vocab)))",
"_____no_output_____"
]
],
[
[
"Run the following cell. It should display `(1, 74074)`",
"_____no_output_____"
]
],
[
[
"layer_0.shape",
"_____no_output_____"
],
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
]
],
[
[
"`layer_0` contains one entry for every word in the vocabulary, as shown in the above image. We need to make sure we know the index of each word, so run the following cell to create a lookup table that stores the index of every word.",
"_____no_output_____"
]
],
[
[
"# Create a dictionary of words in the vocabulary mapped to index positions\n# (to be used in layer_0)\nword2index = {}\nfor i,word in enumerate(vocab):\n word2index[word] = i\n \n# display the map of words to indices\nword2index",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `update_input_layer`. It should count \n how many times each word is used in the given review, and then store\n those counts at the appropriate indices inside `layer_0`.",
"_____no_output_____"
]
],
[
[
"def update_input_layer(review):\n \"\"\" Modify the global layer_0 to represent the vector form of review.\n The element at a given index of layer_0 should represent\n how many times the given word occurs in the review.\n Args:\n review(string) - the string of the review\n Returns:\n None\n \"\"\"\n global layer_0\n # clear out previous state by resetting the layer to be all 0s\n layer_0 *= 0\n \n # TODO: count how many times each word is used in the given review and store the results in layer_0 \n for word in review.split(' '):\n #print(word2index[word])\n layer_0[0][word2index[word]] += 1",
"_____no_output_____"
]
],
[
[
"Run the following cell to test updating the input layer with the first review. The indices assigned may not be the same as in the solution, but hopefully you'll see some non-zero values in `layer_0`. ",
"_____no_output_____"
]
],
[
[
"update_input_layer(reviews[0])\nlayer_0",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `get_target_for_labels`. It should return `0` or `1`, \n depending on whether the given label is `NEGATIVE` or `POSITIVE`, respectively.",
"_____no_output_____"
]
],
[
[
"def get_target_for_label(label):\n \"\"\"Convert a label to `0` or `1`.\n Args:\n label(string) - Either \"POSITIVE\" or \"NEGATIVE\".\n Returns:\n `0` or `1`.\n \"\"\"\n if label == 'POSITIVE':\n return 1\n if label == 'NEGATIVE':\n return 0\n # TODO: Your code here",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out`'POSITIVE'` and `1`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[0]",
"_____no_output_____"
],
[
"get_target_for_label(labels[0])",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out `'NEGATIVE'` and `0`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[1]",
"_____no_output_____"
],
[
"get_target_for_label(labels[1])",
"_____no_output_____"
]
],
[
[
"# End of Project 2. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Project 3: Building a Neural Network<a id='project_3'></a>",
"_____no_output_____"
],
[
"**TODO:** We've included the framework of a class called `SentimentNetork`. Implement all of the items marked `TODO` in the code. These include doing the following:\n- Create a basic neural network much like the networks you've seen in earlier lessons and in Project 1, with an input layer, a hidden layer, and an output layer. \n- Do **not** add a non-linearity in the hidden layer. That is, do not use an activation function when calculating the hidden layer outputs.\n- Re-use the code from earlier in this notebook to create the training data (see `TODO`s in the code)\n- Implement the `pre_process_data` function to create the vocabulary for our training data generating functions\n- Ensure `train` trains over the entire corpus",
"_____no_output_____"
],
[
"### Where to Get Help if You Need it\n- Re-watch earlier Udacity lectures\n- Chapters 3-5 - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) - (Check inside your classroom for a discount code)",
"_____no_output_____"
]
],
[
[
"import time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews,labels,hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n # populate review_vocab with all of the words in the given reviews\n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n review_vocab.add(word)\n\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n # populate label_vocab with all of the words in the given labels.\n label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Set number of nodes in input, hidden and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n\n # These are the weights between the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # The input layer, a two-dimensional matrix with shape 1 x input_nodes\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n\n # clear out previous state, reset the layer to be all 0s\n self.layer_0 *= 0\n \n for word in review.split(\" \"):\n # NOTE: This if-check was not in the version of this method created in Project 2,\n # and it appears in Andrew's Project 3 solution without explanation. \n # It simply ensures the word is actually a key in word2index before\n # accessing it, which is important because accessing an invalid key\n # with raise an exception in Python. This allows us to ignore unknown\n # words encountered in new reviews.\n if(word in self.word2index.keys()):\n ## New for Project 4: changed to set to 1 instead of add 1\n self.layer_0[0][self.word2index[word]] = 1\n \n def get_target_for_label(self,label):\n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n return output * (1 - output)\n \n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n\n # Remember when we started for printing time statistics\n start = time.time()\n \n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # Get the next review and its correct label\n review = training_reviews[i]\n label = training_labels[i]\n \n #### Implement the forward pass here ####\n ### Forward pass ###\n\n # Input Layer\n self.update_input_layer(review)\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n #### Implement the backward pass here ####\n ### Backward pass ###\n\n # Output error\n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n self.weights_1_2 -= layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n self.weights_0_1 -= self.layer_0.T.dot(layer_1_delta) * self.learning_rate # update input-to-hidden weights with gradient descent step\n\n # Keep track of correct predictions.\n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # Run a forward pass through the network, like in the \"train\" function.\n \n # Input Layer\n self.update_input_layer(review.lower())\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n # Return POSITIVE for values above greater-than-or-equal-to 0.5 in the output layer;\n # return NEGATIVE for other values\n if(layer_2[0] >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"\n ",
"_____no_output_____"
]
],
[
[
"Run the following cell to create a `SentimentNetwork` that will train on all but the last 1000 reviews (we're saving those for testing). Here we use a learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)",
"_____no_output_____"
]
],
[
[
"Run the following cell to test the network's performance against the last 1000 reviews (the ones we held out from our training set). \n\n**We have not trained the model yet, so the results should be about 50% as it will just be guessing and there are only two possible values to choose from.**",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"Progress:99.9% Speed(reviews/sec):653.8 #Correct:500 #Tested:1000 Testing Accuracy:50.0%"
]
],
[
[
"Run the following cell to actually train the network. During training, it will display the model's accuracy repeatedly as it trains so you can see how well it's doing.",
"_____no_output_____"
]
],
[
[
"mlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):121.5 #Correct:1788 #Trained:2501 Training Accuracy:71.4%\nProgress:20.8% Speed(reviews/sec):118.1 #Correct:3772 #Trained:5001 Training Accuracy:75.4%\nProgress:31.2% Speed(reviews/sec):112.2 #Correct:5875 #Trained:7501 Training Accuracy:78.3%\nProgress:41.6% Speed(reviews/sec):110.3 #Correct:8007 #Trained:10001 Training Accuracy:80.0%\nProgress:52.0% Speed(reviews/sec):112.1 #Correct:10135 #Trained:12501 Training Accuracy:81.0%\nProgress:62.5% Speed(reviews/sec):112.9 #Correct:12273 #Trained:15001 Training Accuracy:81.8%\nProgress:72.9% Speed(reviews/sec):113.5 #Correct:14392 #Trained:17501 Training Accuracy:82.2%\nProgress:83.3% Speed(reviews/sec):109.9 #Correct:16572 #Trained:20001 Training Accuracy:82.8%\nProgress:93.7% Speed(reviews/sec):106.7 #Correct:18758 #Trained:22501 Training Accuracy:83.3%\nProgress:99.9% Speed(reviews/sec):107.1 #Correct:20079 #Trained:24000 Training Accuracy:83.6%"
]
],
[
[
"That most likely didn't train very well. Part of the reason may be because the learning rate is too high. Run the following cell to recreate the network with a smaller learning rate, `0.01`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"That probably wasn't much different. Run the following cell to recreate the network one more time with an even smaller learning rate, `0.001`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.001)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"With a learning rate of `0.001`, the network should finall have started to improve during training. It's still not very good, but it shows that this solution has potential. We will improve it in the next lesson.",
"_____no_output_____"
],
[
"# End of Project 3. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Understanding Neural Noise<a id='lesson_4'></a>\n\nThe following cells include includes the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"def update_input_layer(review):\n \n global layer_0\n \n # clear out previous state, reset the layer to be all 0s\n layer_0 *= 0\n for word in review.split(\" \"):\n layer_0[0][word2index[word]] += 1\n\nupdate_input_layer(reviews[0])",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"review_counter = Counter()",
"_____no_output_____"
],
[
"for word in reviews[0].split(\" \"):\n review_counter[word] += 1",
"_____no_output_____"
],
[
"review_counter.most_common()",
"_____no_output_____"
]
],
[
[
"# Project 4: Reducing Noise in Our Input Data<a id='project_4'></a>\n\n**TODO:** Attempt to reduce the noise in the input data like Andrew did in the previous video. Specifically, do the following:\n* Copy the `SentimentNetwork` class you created earlier into the following cell.\n* Modify `update_input_layer` so it does not count how many times each word is used, but rather just stores whether or not a word was used. ",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Projet 3 lesson\n# -Modify it to reduce noise, like in the video ",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it. Notice we've gone back to the higher learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"That should have trained much better than the earlier attempts. It's still not wonderful, but it should have improved dramatically. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"# End of Project 4. \n## Andrew's solution was actually in the previous video, so rewatch that video if you had any problems with that project. Then continue on to the next lesson.\n# Analyzing Inefficiencies in our Network<a id='lesson_5'></a>\nThe following cells include the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"layer_0 = np.zeros(10)",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"layer_0[4] = 1\nlayer_0[9] = 1",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"weights_0_1 = np.random.randn(10,5)",
"_____no_output_____"
],
[
"layer_0.dot(weights_0_1)",
"_____no_output_____"
],
[
"indices = [4,9]",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (1 * weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
]
],
[
[
"# Project 5: Making our Network More Efficient<a id='project_5'></a>\n**TODO:** Make the `SentimentNetwork` class more efficient by eliminating unnecessary multiplications and additions that occur during forward and backward propagation. To do that, you can do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Remove the `update_input_layer` function - you will not need it in this version.\n* Modify `init_network`:\n>* You no longer need a separate input layer, so remove any mention of `self.layer_0`\n>* You will be dealing with the old hidden layer more directly, so create `self.layer_1`, a two-dimensional matrix with shape 1 x hidden_nodes, with all values initialized to zero\n* Modify `train`:\n>* Change the name of the input parameter `training_reviews` to `training_reviews_raw`. This will help with the next step.\n>* At the beginning of the function, you'll want to preprocess your reviews to convert them to a list of indices (from `word2index`) that are actually used in the review. This is equivalent to what you saw in the video when Andrew set specific indices to 1. Your code should create a local `list` variable named `training_reviews` that should contain a `list` for each review in `training_reviews_raw`. Those lists should contain the indices for words found in the review.\n>* Remove call to `update_input_layer`\n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* In the forward pass, replace the code that updates `layer_1` with new logic that only adds the weights for the indices used in the review.\n>* When updating `weights_0_1`, only update the individual weights that were used in the forward pass.\n* Modify `run`:\n>* Remove call to `update_input_layer` \n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* Much like you did in `train`, you will need to pre-process the `review` so you can work with word indices, then update `layer_1` by adding weights for the indices used in the review.",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Project 4 lesson\n# -Modify it according to the above instructions ",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it once again.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"That should have trained much better than the earlier attempts. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"# End of Project 5. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n# Further Noise Reduction<a id='lesson_6'></a>",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]",
"_____no_output_____"
],
[
"from bokeh.models import ColumnDataSource, LabelSet\nfrom bokeh.plotting import figure, show, output_file\nfrom bokeh.io import output_notebook\noutput_notebook()",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],pos_neg_ratios.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"Word Positive/Negative Affinity Distribution\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"_____no_output_____"
],
[
"frequency_frequency = Counter()\n\nfor word, cnt in total_counts.most_common():\n frequency_frequency[cnt] += 1",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],frequency_frequency.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"The frequency distribution of the words in our corpus\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"_____no_output_____"
]
],
[
[
"# Project 6: Reducing Noise by Strategically Reducing the Vocabulary<a id='project_6'></a>\n\n**TODO:** Improve `SentimentNetwork`'s performance by reducing more noise in the vocabulary. Specifically, do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Modify `pre_process_data`:\n>* Add two additional parameters: `min_count` and `polarity_cutoff`\n>* Calculate the positive-to-negative ratios of words used in the reviews. (You can use code you've written elsewhere in the notebook, but we are moving it into the class like we did with other helper code earlier.)\n>* Andrew's solution only calculates a postive-to-negative ratio for words that occur at least 50 times. This keeps the network from attributing too much sentiment to rarer words. You can choose to add this to your solution if you would like. \n>* Change so words are only added to the vocabulary if they occur in the vocabulary more than `min_count` times.\n>* Change so words are only added to the vocabulary if the absolute value of their postive-to-negative ratio is at least `polarity_cutoff`\n* Modify `__init__`:\n>* Add the same two parameters (`min_count` and `polarity_cutoff`) and use them when you call `pre_process_data`",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Project 5 lesson\n# -Modify it according to the above instructions ",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a small polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.05,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"And run the following cell to test it's performance. It should be ",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a much larger polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.8,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"And run the following cell to test it's performance.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"# End of Project 6. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Analysis: What's Going on in the Weights?<a id='lesson_7'></a>",
"_____no_output_____"
]
],
[
[
"mlp_full = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=0,polarity_cutoff=0,learning_rate=0.01)",
"_____no_output_____"
],
[
"mlp_full.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"def get_most_similar_words(focus = \"horrible\"):\n most_similar = Counter()\n\n for word in mlp_full.word2index.keys():\n most_similar[word] = np.dot(mlp_full.weights_0_1[mlp_full.word2index[word]],mlp_full.weights_0_1[mlp_full.word2index[focus]])\n \n return most_similar.most_common()",
"_____no_output_____"
],
[
"get_most_similar_words(\"excellent\")",
"_____no_output_____"
],
[
"get_most_similar_words(\"terrible\")",
"_____no_output_____"
],
[
"import matplotlib.colors as colors\n\nwords_to_visualize = list()\nfor word, ratio in pos_neg_ratios.most_common(500):\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)\n \nfor word, ratio in list(reversed(pos_neg_ratios.most_common()))[0:500]:\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)",
"_____no_output_____"
],
[
"pos = 0\nneg = 0\n\ncolors_list = list()\nvectors_list = list()\nfor word in words_to_visualize:\n if word in pos_neg_ratios.keys():\n vectors_list.append(mlp_full.weights_0_1[mlp_full.word2index[word]])\n if(pos_neg_ratios[word] > 0):\n pos+=1\n colors_list.append(\"#00ff00\")\n else:\n neg+=1\n colors_list.append(\"#000000\")",
"_____no_output_____"
],
[
"from sklearn.manifold import TSNE\ntsne = TSNE(n_components=2, random_state=0)\nwords_top_ted_tsne = tsne.fit_transform(vectors_list)",
"_____no_output_____"
],
[
"p = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"vector T-SNE for most polarized words\")\n\nsource = ColumnDataSource(data=dict(x1=words_top_ted_tsne[:,0],\n x2=words_top_ted_tsne[:,1],\n names=words_to_visualize,\n color=colors_list))\n\np.scatter(x=\"x1\", y=\"x2\", size=8, source=source, fill_color=\"color\")\n\nword_labels = LabelSet(x=\"x1\", y=\"x2\", text=\"names\", y_offset=6,\n text_font_size=\"8pt\", text_color=\"#555555\",\n source=source, text_align='center')\np.add_layout(word_labels)\n\nshow(p)\n\n# green indicates positive words, black indicates negative words",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8a41a87e547b13b7211b647ff8e2f02ffced3b | 382,539 | ipynb | Jupyter Notebook | ImageClassifierProject.ipynb | matehr/ImageClassifier | 2df84d98d73b0fce580d24f8ead9874b12d59d74 | [
"MIT"
] | null | null | null | ImageClassifierProject.ipynb | matehr/ImageClassifier | 2df84d98d73b0fce580d24f8ead9874b12d59d74 | [
"MIT"
] | null | null | null | ImageClassifierProject.ipynb | matehr/ImageClassifier | 2df84d98d73b0fce580d24f8ead9874b12d59d74 | [
"MIT"
] | null | null | null | 214.788883 | 134,688 | 0.853929 | [
[
[
"# Developing an AI application\n\nGoing forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications. \n\nIn this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below. \n\n<img src='assets/Flowers.png' width=500px>\n\nThe project is broken down into multiple steps:\n\n* Load and preprocess the image dataset\n* Train the image classifier on your dataset\n* Use the trained classifier to predict image content\n\nWe'll lead you through each part which you'll implement in Python.\n\nWhen you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.\n\nFirst up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.",
"_____no_output_____"
]
],
[
[
"# Imports here\n%config IPCompleter.greedy=True\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nimport torch\nfrom torch import nn\nfrom torch import optim\nfrom torchvision import datasets, transforms, models\n\nfrom PIL import Image\n\nfrom collections import OrderedDict\nfrom workspace_utils import active_session",
"_____no_output_____"
]
],
[
[
"## Load the data\n\nHere you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.\n\nThe validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.\n\nThe pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.\n ",
"_____no_output_____"
]
],
[
[
"data_dir = 'flowers'\ntrain_dir = data_dir + '/train'\nvalid_dir = data_dir + '/valid'\ntest_dir = data_dir + '/test'",
"_____no_output_____"
],
[
"# TODO: Define your transforms for the training, validation, and testing sets\ndata_transforms = {\"training\" : transforms.Compose([transforms.RandomRotation(30),\n transforms.RandomResizedCrop(224),\n transforms.RandomHorizontalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406],\n [0.229, 0.224, 0.225])]),\n \"testing\" : transforms.Compose([transforms.Resize(255),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406],\n [0.229, 0.224, 0.225])])}\n\n# TODO: Load the datasets with ImageFolder\nimage_datasets = {\"train_data\" : datasets.ImageFolder(train_dir, transform=data_transforms[\"training\"]),\n \"valid_data\" : datasets.ImageFolder(valid_dir, transform=data_transforms[\"testing\"]),\n \"test_data\" : datasets.ImageFolder(test_dir, transform=data_transforms[\"testing\"])}\n\n# TODO: Using the image datasets and the trainforms, define the dataloaders\ndataloaders = {\"trainloader\" : torch.utils.data.DataLoader(image_datasets[\"train_data\"], batch_size=64, shuffle=True),\n \"validloader\" : torch.utils.data.DataLoader(image_datasets[\"valid_data\"], batch_size=64),\n \"testloader\" : torch.utils.data.DataLoader(image_datasets[\"test_data\"], batch_size=64)} ",
"_____no_output_____"
]
],
[
[
"### Label mapping\n\nYou'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.",
"_____no_output_____"
]
],
[
[
"import json\n\nwith open('cat_to_name.json', 'r') as f:\n cat_to_name = json.load(f)",
"_____no_output_____"
]
],
[
[
"# Building and training the classifier\n\nNow that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.\n\nWe're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:\n\n* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)\n* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout\n* Train the classifier layers using backpropagation using the pre-trained network to get the features\n* Track the loss and accuracy on the validation set to determine the best hyperparameters\n\nWe've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!\n\nWhen training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.\n\nOne last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to\nGPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.\n\n**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.",
"_____no_output_____"
]
],
[
[
"# TODO: Build and train your network\n#Obtain the model and print to discover features\nmodel = models.densenet121(pretrained=True)\nmodel",
"/opt/conda/lib/python3.6/site-packages/torchvision-0.2.1-py3.6.egg/torchvision/models/densenet.py:212: UserWarning: nn.init.kaiming_normal is now deprecated in favor of nn.init.kaiming_normal_.\nDownloading: \"https://download.pytorch.org/models/densenet121-a639ec97.pth\" to /root/.torch/models/densenet121-a639ec97.pth\n100%|██████████| 32342954/32342954 [00:00<00:00, 72036804.98it/s]\n"
],
[
"#Freeze parameters\nfor param in model.parameters():\n param.requires_grad = False\n \nclassifier = nn.Sequential(OrderedDict([\n (\"fc1\", nn.Linear(1024, 512)),\n (\"relu\", nn.ReLU()),\n (\"drop\", nn.Dropout(p=0.2)),\n (\"fc2\", nn.Linear(512, 102)),\n (\"output\", nn.LogSoftmax(dim=1))\n]))\n\nmodel.classifier = classifier",
"_____no_output_____"
],
[
"#Move model to GPU/CPU\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nmodel = model.to(device)\n\n# Define loss function and optimizer\ncriterion = nn.NLLLoss()\nlearn_rate = 0.001\noptimizer = optim.Adam(model.classifier.parameters(), lr=learn_rate)",
"_____no_output_____"
],
[
"#Train the model and report stats as we go\nepochs = 5\nsteps = 0\nrunning_loss = 0\nprint_every = 10\n\nwith active_session():\n for epoch in range(epochs):\n for inputs, labels in dataloaders[\"trainloader\"]:\n steps += 1\n inputs, labels = inputs.to(device), labels.to(device)\n\n optimizer.zero_grad()\n\n logps = model.forward(inputs)\n loss = criterion(logps, labels)\n loss.backward()\n optimizer.step()\n\n running_loss += loss.item()\n\n if steps % print_every == 0:\n test_loss = 0\n accuracy = 0\n model.eval()\n\n with torch.no_grad():\n for inputs, labels in dataloaders[\"validloader\"]:\n inputs, labels = inputs.to(device), labels.to(device)\n\n logps = model.forward(inputs)\n batch_loss = criterion(logps, labels)\n\n test_loss += batch_loss.item()\n\n ps = torch.exp(logps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor)).item()\n\n print(f\"Epoch {epoch+1}/{epochs}.. \"\n f\"Train loss: {running_loss/print_every:.3f}.. \"\n f\"Test loss: {test_loss/len(dataloaders['validloader']):.3f} \"\n f\"Test accuracy: {accuracy/len(dataloaders['validloader']):.3f} \")\n running_loss = 0\n model.train()",
"Epoch 1/5.. Train loss: 4.503.. Test loss: 4.278 Test accuracy: 0.072 \nEpoch 1/5.. Train loss: 4.247.. Test loss: 3.967 Test accuracy: 0.196 \nEpoch 1/5.. Train loss: 3.935.. Test loss: 3.627 Test accuracy: 0.236 \nEpoch 1/5.. Train loss: 3.628.. Test loss: 3.270 Test accuracy: 0.343 \nEpoch 1/5.. Train loss: 3.304.. Test loss: 2.857 Test accuracy: 0.423 \nEpoch 1/5.. Train loss: 2.957.. Test loss: 2.510 Test accuracy: 0.483 \nEpoch 1/5.. Train loss: 2.697.. Test loss: 2.130 Test accuracy: 0.566 \nEpoch 1/5.. Train loss: 2.371.. Test loss: 1.902 Test accuracy: 0.617 \nEpoch 1/5.. Train loss: 2.272.. Test loss: 1.687 Test accuracy: 0.664 \nEpoch 1/5.. Train loss: 2.006.. Test loss: 1.538 Test accuracy: 0.674 \nEpoch 2/5.. Train loss: 1.853.. Test loss: 1.334 Test accuracy: 0.729 \nEpoch 2/5.. Train loss: 1.672.. Test loss: 1.188 Test accuracy: 0.764 \nEpoch 2/5.. Train loss: 1.495.. Test loss: 1.080 Test accuracy: 0.786 \nEpoch 2/5.. Train loss: 1.425.. Test loss: 0.999 Test accuracy: 0.792 \nEpoch 2/5.. Train loss: 1.376.. Test loss: 0.902 Test accuracy: 0.808 \nEpoch 2/5.. Train loss: 1.285.. Test loss: 0.896 Test accuracy: 0.796 \nEpoch 2/5.. Train loss: 1.182.. Test loss: 0.819 Test accuracy: 0.819 \nEpoch 2/5.. Train loss: 1.209.. Test loss: 0.774 Test accuracy: 0.833 \nEpoch 2/5.. Train loss: 1.110.. Test loss: 0.720 Test accuracy: 0.853 \nEpoch 2/5.. Train loss: 1.096.. Test loss: 0.705 Test accuracy: 0.853 \nEpoch 3/5.. Train loss: 1.017.. Test loss: 0.672 Test accuracy: 0.840 \nEpoch 3/5.. Train loss: 0.969.. Test loss: 0.628 Test accuracy: 0.857 \nEpoch 3/5.. Train loss: 0.950.. Test loss: 0.627 Test accuracy: 0.855 \nEpoch 3/5.. Train loss: 0.972.. Test loss: 0.596 Test accuracy: 0.862 \nEpoch 3/5.. Train loss: 0.894.. Test loss: 0.583 Test accuracy: 0.871 \nEpoch 3/5.. Train loss: 0.896.. Test loss: 0.526 Test accuracy: 0.882 \nEpoch 3/5.. Train loss: 0.915.. Test loss: 0.534 Test accuracy: 0.875 \nEpoch 3/5.. Train loss: 0.883.. Test loss: 0.534 Test accuracy: 0.868 \nEpoch 3/5.. Train loss: 0.837.. Test loss: 0.481 Test accuracy: 0.895 \nEpoch 3/5.. Train loss: 0.819.. Test loss: 0.473 Test accuracy: 0.894 \nEpoch 4/5.. Train loss: 0.787.. Test loss: 0.449 Test accuracy: 0.898 \nEpoch 4/5.. Train loss: 0.752.. Test loss: 0.468 Test accuracy: 0.879 \nEpoch 4/5.. Train loss: 0.723.. Test loss: 0.476 Test accuracy: 0.888 \nEpoch 4/5.. Train loss: 0.688.. Test loss: 0.434 Test accuracy: 0.900 \nEpoch 4/5.. Train loss: 0.738.. Test loss: 0.418 Test accuracy: 0.905 \nEpoch 4/5.. Train loss: 0.682.. Test loss: 0.412 Test accuracy: 0.903 \nEpoch 4/5.. Train loss: 0.777.. Test loss: 0.432 Test accuracy: 0.903 \nEpoch 4/5.. Train loss: 0.732.. Test loss: 0.414 Test accuracy: 0.902 \nEpoch 4/5.. Train loss: 0.646.. Test loss: 0.433 Test accuracy: 0.897 \nEpoch 4/5.. Train loss: 0.768.. Test loss: 0.406 Test accuracy: 0.900 \nEpoch 4/5.. Train loss: 0.690.. Test loss: 0.410 Test accuracy: 0.912 \nEpoch 5/5.. Train loss: 0.711.. Test loss: 0.415 Test accuracy: 0.894 \nEpoch 5/5.. Train loss: 0.652.. Test loss: 0.407 Test accuracy: 0.896 \nEpoch 5/5.. Train loss: 0.588.. Test loss: 0.382 Test accuracy: 0.915 \nEpoch 5/5.. Train loss: 0.622.. Test loss: 0.387 Test accuracy: 0.904 \nEpoch 5/5.. Train loss: 0.677.. Test loss: 0.353 Test accuracy: 0.912 \nEpoch 5/5.. Train loss: 0.580.. Test loss: 0.368 Test accuracy: 0.913 \nEpoch 5/5.. Train loss: 0.656.. Test loss: 0.325 Test accuracy: 0.923 \nEpoch 5/5.. Train loss: 0.579.. Test loss: 0.337 Test accuracy: 0.919 \nEpoch 5/5.. Train loss: 0.629.. Test loss: 0.370 Test accuracy: 0.911 \nEpoch 5/5.. Train loss: 0.636.. Test loss: 0.337 Test accuracy: 0.917 \n"
]
],
[
[
"## Testing your network\n\nIt's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.",
"_____no_output_____"
]
],
[
[
"# TODO: Do validation on the test set\ntest_loss = 0\naccuracy = 0\nmodel.eval()\n\nwith torch.no_grad():\n for inputs, labels in dataloaders[\"testloader\"]:\n inputs, labels = inputs.to(device), labels.to(device)\n\n logps = model.forward(inputs)\n batch_loss = criterion(logps, labels)\n\n test_loss += batch_loss.item()\n\n ps = torch.exp(logps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor)).item()\n\nprint(f\"Test loss: {test_loss/len(dataloaders['testloader']):.3f} \"\n f\"Test accuracy: {accuracy/len(dataloaders['testloader']):.3f} \")\nmodel.train()",
"Test loss: 0.432 Test accuracy: 0.879 \n"
]
],
[
[
"## Save the checkpoint\n\nNow that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.\n\n```model.class_to_idx = image_datasets['train'].class_to_idx```\n\nRemember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.",
"_____no_output_____"
]
],
[
[
"# TODO: Save the checkpoint \nmodel.class_to_idx = image_datasets['train_data'].class_to_idx\ncheckpoint = {'epochs' : epochs,\n 'learn_rate' : learn_rate,\n 'optimizer_state' : optimizer.state_dict(),\n 'classifier' : model.classifier,\n 'state_dict' : model.state_dict()}\ntorch.save(checkpoint, 'checkpoint.pth')",
"_____no_output_____"
]
],
[
[
"## Loading the checkpoint\n\nAt this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.",
"_____no_output_____"
]
],
[
[
"# TODO: Write a function that loads a checkpoint and rebuilds the model\ndef load_checkpoint(filepath):\n checkpoint = torch.load(filepath)\n model = models.densenet121(pretrained=True)\n model.classifier = checkpoint['classifier']\n model.load_state_dict(checkpoint['state_dict'])\n \n optimizer = optim.Adam(model.classifier.parameters(), lr=0.003)\n optimizer.load_state_dict(checkpoint['optimizer_state'])\n \n return model, optimizer",
"_____no_output_____"
],
[
"model, optimizer = load_checkpoint(\"checkpoint.pth\")",
"/opt/conda/lib/python3.6/site-packages/torchvision-0.2.1-py3.6.egg/torchvision/models/densenet.py:212: UserWarning: nn.init.kaiming_normal is now deprecated in favor of nn.init.kaiming_normal_.\nDownloading: \"https://download.pytorch.org/models/densenet121-a639ec97.pth\" to /root/.torch/models/densenet121-a639ec97.pth\n100%|██████████| 32342954/32342954 [00:00<00:00, 56286746.57it/s]\n"
]
],
[
[
"# Inference for classification\n\nNow you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like \n\n```python\nprobs, classes = predict(image_path, model)\nprint(probs)\nprint(classes)\n> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]\n> ['70', '3', '45', '62', '55']\n```\n\nFirst you'll need to handle processing the input image such that it can be used in your network. \n\n## Image Preprocessing\n\nYou'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training. \n\nFirst, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.\n\nColor channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.\n\nAs before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation. \n\nAnd finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.",
"_____no_output_____"
]
],
[
[
"def process_image(image):\n ''' Scales, crops, and normalizes a PIL image for a PyTorch model,\n returns an Numpy array\n '''\n \n # TODO: Process a PIL image for use in a PyTorch model\n # Open and resize\n im = Image.open(image)\n im.thumbnail((256, 256))\n \n # Center crop\n width, height = im.size\n desired_width, desired_height = 224, 224\n left = (width-desired_width) / 2\n upper = (height-desired_height) / 2\n right = (width+desired_width) / 2\n lower = (height+desired_height) / 2\n im = im.crop((left, upper, right, lower))\n \n # Normalize\n np_image = np.array(im)\n np_image = np_image / 255\n means = np.array([0.485, 0.456, 0.406])\n st_dev = np.array([0.229, 0.224, 0.225])\n np_image = np_image - means\n np_image = np_image / st_dev\n np_image = np_image.transpose(2, 0, 1)\n return np_image\n \n ",
"_____no_output_____"
]
],
[
[
"To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).",
"_____no_output_____"
]
],
[
[
"def imshow(image, ax=None, title=None):\n \"\"\"Imshow for Tensor.\"\"\"\n if ax is None:\n fig, ax = plt.subplots()\n \n # PyTorch tensors assume the color channel is the first dimension\n # but matplotlib assumes is the third dimension\n image = image.numpy().transpose((1, 2, 0))\n \n # Undo preprocessing\n mean = np.array([0.485, 0.456, 0.406])\n std = np.array([0.229, 0.224, 0.225])\n image = std * image + mean\n \n # Image needs to be clipped between 0 and 1 or it looks like noise when displayed\n image = np.clip(image, 0, 1)\n \n ax.imshow(image)\n \n return ax",
"_____no_output_____"
],
[
"train_image = process_image(\"flowers/train/1/image_06734.jpg\")\nprint(train_image.shape)\nimshow(torch.tensor(train_image))",
"(3, 224, 224)\n"
]
],
[
[
"## Class Prediction\n\nOnce you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.\n\nTo get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.\n\nAgain, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.\n\n```python\nprobs, classes = predict(image_path, model)\nprint(probs)\nprint(classes)\n> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]\n> ['70', '3', '45', '62', '55']\n```",
"_____no_output_____"
]
],
[
[
"def predict(image_path, model, topk=5):\n ''' Predict the class (or classes) of an image using a trained deep learning model.\n '''\n \n # TODO: Implement the code to predict the class from an image file\n device = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\n model = model.to(device)\n \n image = torch.tensor(process_image(image_path)).type(torch.FloatTensor)\n image.unsqueeze_(0)\n inputs = image.to(device)\n\n logps = model(inputs)\n ps = torch.exp(logps)\n top_ps, top_class = ps.topk(topk, dim=1)\n return image.squeeze(), top_ps, top_class\n ",
"_____no_output_____"
],
[
"image_tensor, top_ps, top_class = predict(\"flowers/train/1/image_06734.jpg\", model)\nprint(image_tensor.shape)",
"torch.Size([3, 224, 224])\n"
]
],
[
[
"## Sanity Checking\n\nNow that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:\n\n<img src='assets/inference_example.png' width=300px>\n\nYou can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.",
"_____no_output_____"
]
],
[
[
"# TODO: Display an image along with the top 5 classes\nimshow(image_tensor)",
"_____no_output_____"
],
[
"x = [cat_to_name[str(var)] for var in top_class.cpu().numpy()[0]]\ny = [var for var in top_ps.cpu().detach().numpy()[0]]\nplt.barh(x, y)\nplt.gca().invert_yaxis()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec8a56e304bce3d0830aab25378776007033d0de | 42,932 | ipynb | Jupyter Notebook | cups_mail_622/notes.ipynb | cut4cut/ml_comps | 39c6516f54ffe602abbf1d1992225f8c55ed2f47 | [
"MIT"
] | null | null | null | cups_mail_622/notes.ipynb | cut4cut/ml_comps | 39c6516f54ffe602abbf1d1992225f8c55ed2f47 | [
"MIT"
] | null | null | null | cups_mail_622/notes.ipynb | cut4cut/ml_comps | 39c6516f54ffe602abbf1d1992225f8c55ed2f47 | [
"MIT"
] | null | null | null | 81.1569 | 20,076 | 0.821019 | [
[
[
"import os\nimport lib\n\nimport numpy as np\nimport pandas as pd \n\nfrom PIL import Image\nfrom matplotlib import pyplot as plt\n\nfrom collections import OrderedDict\n\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\n\nfrom torchvision import datasets, transforms, models\nfrom torch.utils.data import DataLoader, random_split",
"_____no_output_____"
],
[
"SEED = 42\nVL_SIZE = 0.2",
"_____no_output_____"
],
[
"mean = [0.485, 0.456, 0.406]\nstd = [0.229, 0.224, 0.225]\n\nlearn_transform = transforms.Compose([\n transforms.Resize(255),\n transforms.RandomHorizontalFlip(),\n transforms.ColorJitter(),\n transforms.ToTensor(),\n transforms.Normalize(mean, std)])\n\ntest_transform = transforms.Compose([\n transforms.Resize(255),\n transforms.ColorJitter(),\n transforms.ToTensor(),\n transforms.Normalize(mean, std)])\n\n\nlearn_data = datasets.ImageFolder('./data/train', transform=learn_transform)\nsplits = [ int((1-VL_SIZE)*len(learn_data)), int((VL_SIZE)*len(learn_data)) ]\n\ntr_data, vl_data = random_split(learn_data, splits, generator=torch.Generator().manual_seed(SEED))\n\ntr_loader = DataLoader(tr_data, batch_size=64, shuffle=True)\nvl_loader = DataLoader(vl_data, batch_size=64, shuffle=True)",
"_____no_output_____"
],
[
"model = models.densenet161(pretrained=True)\nmodel.classifier",
"Downloading: \"https://download.pytorch.org/models/densenet161-8d451a50.pth\" to /tmp/xdg_cache/torch/hub/checkpoints/densenet161-8d451a50.pth\n"
],
[
"model = lib.freeze_parameters(model)",
"_____no_output_____"
],
[
"classifier = nn.Sequential(\n nn.Linear(in_features=2208, out_features=2208),\n nn.ReLU(),\n nn.Dropout(p=0.4),\n nn.Linear(in_features=2208, out_features=1024),\n nn.ReLU(),\n nn.Dropout(p=0.3),\n nn.Linear(in_features=1024, out_features=10),\n nn.LogSoftmax(dim=1) \n)\n \nmodel.classifier = classifier\nmodel.classifier",
"_____no_output_____"
],
[
"#!g1.1\nepoch = 5+5\n\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nmodel.to(device)\n\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.classifier.parameters(), lr=0.003)\n\nmodel, train_loss, test_loss = lib.train(model, tr_loader, vl_loader, epoch, optimizer, criterion)",
"Epoch: 1/10\n\t\tGoing for validation\n\tTrain loss:0.369909.. \tValid Loss:0.275122.. \tAccuracy: 88.8835\n\tValidation loss decreased (inf --> 0.275122). Saving model ...\n\tEpoch:1 completed in 3m 11s\nEpoch: 2/10\n\t\tGoing for validation\n\tTrain loss:0.294544.. \tValid Loss:0.266825.. \tAccuracy: 89.9775\n\tValidation loss decreased (0.275122 --> 0.266825). Saving model ...\n\tEpoch:2 completed in 2m 52s\nEpoch: 3/10\n\t\tGoing for validation\n\tTrain loss:0.271831.. \tValid Loss:0.228912.. \tAccuracy: 90.8623\n\tValidation loss decreased (0.266825 --> 0.228912). Saving model ...\n\tEpoch:3 completed in 2m 52s\nEpoch: 4/10\n\t\tGoing for validation\n\tTrain loss:0.285372.. \tValid Loss:0.233383.. \tAccuracy: 90.9266\n\tEpoch:4 completed in 2m 51s\nEpoch: 5/10\n\t\tGoing for validation\n\tTrain loss:0.255457.. \tValid Loss:0.252152.. \tAccuracy: 89.3501\n\tEpoch:5 completed in 2m 50s\nEpoch: 6/10\n\t\tGoing for validation\n\tTrain loss:0.270462.. \tValid Loss:0.246234.. \tAccuracy: 90.1544\n\tEpoch:6 completed in 2m 51s\nEpoch: 7/10\n\t\tGoing for validation\n\tTrain loss:0.250323.. \tValid Loss:0.229200.. \tAccuracy: 91.2323\n\tEpoch:7 completed in 2m 51s\nEpoch: 8/10\n\t\tGoing for validation\n\tTrain loss:0.262528.. \tValid Loss:0.254894.. \tAccuracy: 90.6210\n\tEpoch:8 completed in 2m 50s\nEpoch: 9/10\n\t\tGoing for validation\n\tTrain loss:0.273741.. \tValid Loss:0.252147.. \tAccuracy: 91.1036\n\tEpoch:9 completed in 2m 50s\nEpoch: 10/10\n\t\tGoing for validation\n\tTrain loss:0.266050.. \tValid Loss:0.218184.. \tAccuracy: 91.1840\n\tValidation loss decreased (0.228912 --> 0.218184). Saving model ...\n\tEpoch:10 completed in 2m 50s\nTraining completed in 28m 47s\n"
],
[
"#!g1.1\nmodel = lib.load_latest_model(model)",
"_____no_output_____"
],
[
"lib.check_overfitted(train_loss, test_loss)",
"_____no_output_____"
],
[
"def test(file):\n ids = {'airplane': 0, 'not_airplane': 1}\n with Image.open(file) as f:\n \n img = test_transform(f).unsqueeze(0)\n with torch.no_grad():\n out = model(img.to(device)).cpu().numpy()\n for key, value in ids.items():\n if value == np.argmax(out):\n #name = classes[int(key)]\n return value",
"_____no_output_____"
],
[
"name = '73391958-6b88-40a5-9191-5386b7925b40'\ndata_dir = './data/test/{}.png'\npath = data_dir.format(name)\n\nwith Image.open(path) as f:\n print(np.array(f).shape)\n plt.imshow(np.array(f))\n plt.show()",
"(20, 20, 4)\n"
],
[
"sumb = pd.read_csv('./data/sample_submission.csv')",
"_____no_output_____"
],
[
"def predict(s):\n name = s['filename']\n data_dir = './data/test/{}.png'\n path = data_dir.format(name)\n try:\n pred = test(path)\n except RuntimeError:\n print(name)\n pred = 0\n\n return pred",
"_____no_output_____"
],
[
"#!g1.1\npreds = sumb[:].apply(predict, axis=1)",
"73391958-6b88-40a5-9191-5386b7925b40\nb4f16723-149f-4179-a258-97b52442de74\n93a5da1c-3aea-44aa-9240-96cb8ed18994\n103c3b34-63b6-4a2f-a2f8-a6eb85c5be11\n71da2b41-4bbf-420b-b5ad-8f5352af0c50\n773d6063-5dd0-44f6-b5c1-4409b1e4b6c8\n3f895e00-8843-4eed-96ac-a22e8cad699e\n43af096e-2776-43f4-bc44-457fc3e33bba\n1e3baecb-49ef-456c-9c4c-cdf164b1faca\n176a953d-959f-416e-b6da-c47b3ff1e0f5\n9e0a7a8e-5412-4792-aeb1-cea4e12ea666\n81d83894-0e6f-431a-9971-583f99d94ab9\nb09197a8-64ad-436b-9dba-fe2d0d3c7077\ne33ca568-3517-4c8f-b96d-3eaaf025295d\n1bfd9c52-d8f9-496f-a0db-765eb8cd706e\n5e286847-3698-4048-b245-70aa4109afa6\n0c24093c-0d2c-4efd-9c29-c0a138946d70\n7ab51a9c-0fb9-40f5-bbe6-76370031faf4\n9674f377-915c-4059-8a89-2771a51b845d\n475adc32-815a-4f06-ae6a-61a992c42fbd\n4477ec27-4808-4a77-991e-5048a124095b\ndfd9000d-4100-448e-8c28-242d0ae24dcd\n01cf8479-f165-497d-9669-e7ad9fad33d5\n40ef19b7-f908-4a73-a84b-a6a9e5417b30\nfac2c36e-6395-44cc-ab73-d01f813eb605\n397b4845-0de8-4a83-9074-a945b85e39b9\n92c3951e-ab05-492f-90e8-ece27ffe3efa\ncba2e377-1120-45b3-9f9e-1a26f12a7b0b\n8184f255-2bf3-4d16-9cf5-e5296df8db9f\nde5f8bed-6a9b-4ece-9c57-345e923def22\n964624c7-4e79-4c9e-a86b-bd99678618b9\n61d5b980-b2f1-4314-ac9f-3fc19bcc4924\n83f5c499-358b-4c88-8065-40409fc28281\n073d35e0-f95e-491c-a190-7d5ab552c3ef\n0c2eb4b7-a85a-4807-a4e4-4491748a390d\nb1bbcdda-b279-4758-b61e-ea037a938745\n652419d4-fad5-47f0-bde8-66b376ed311b\n863ef7b1-8282-48e6-b777-c4f3da15b403\n0664e6a4-610a-4c97-b5e2-b05177de7163\n52ee5526-930d-4efc-b7bc-afe56d970278\ne8764332-4d6e-4d7b-ab72-6c6b8c107bb1\nb7cb6965-b63e-41e7-906f-2305176ab716\ne42c0702-e144-4ef9-af2b-0e0065f49378\n90952d65-f789-4d4c-abf6-71789a080b1b\ne6d95799-8c48-4a79-ba2a-09b985447158\n67d6890e-3de7-4366-b267-915f9b3ff02a\n2e82a554-98f6-456f-968a-706134810114\n618bc079-3baf-4f21-b92e-756d4ab38b83\nc79a5362-30cc-49c2-9faa-dc4577df8825\nebfaea85-562d-4b78-baa2-13f7622535f3\n92188a26-cfe7-41cc-b84a-729a89c3afc6\nfd08f9af-0bc8-4462-b4e9-3e77a783b49c\nf4c0f480-725d-4002-961c-703586636e55\n8d2f424c-d3ee-4641-a375-720694659f10\nd07b43d9-054a-4752-a3c8-ae8c780874a3\n3d231bb9-23d8-48f0-8545-44526608cd04\n71586e2b-f9c6-41c6-a26b-acb10eeaee32\nd70c0d57-6f62-41b1-9e1a-a6ac76118ea5\n1958ee34-cccd-4fe4-8038-4402736ae7fe\n2f423502-eb92-4123-b4b9-cbaaa6c01261\n"
],
[
"sumb['sign'] = -1 * (preds.values - 1)",
"_____no_output_____"
],
[
"sumb.to_csv('./data/submission.csv', index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8a8d85abe7a0df10be5410b3d2d8c7110e7ad6 | 80,723 | ipynb | Jupyter Notebook | Copy_of_DS_Unit_1_Sprint_Challenge_4_txt.ipynb | danhorsley/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments | 6828f11e5250c0832215b993c87227e71cfd781f | [
"MIT"
] | null | null | null | Copy_of_DS_Unit_1_Sprint_Challenge_4_txt.ipynb | danhorsley/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments | 6828f11e5250c0832215b993c87227e71cfd781f | [
"MIT"
] | null | null | null | Copy_of_DS_Unit_1_Sprint_Challenge_4_txt.ipynb | danhorsley/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments | 6828f11e5250c0832215b993c87227e71cfd781f | [
"MIT"
] | null | null | null | 50.045257 | 14,914 | 0.535808 | [
[
[
"<a href=\"https://colab.research.google.com/github/danhorsley/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments/blob/master/Copy_of_DS_Unit_1_Sprint_Challenge_4_txt.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Data Science Unit 1 Sprint Challenge 4\n\n## Exploring Data, Testing Hypotheses\n\nIn this sprint challenge you will look at a dataset of people being approved or rejected for credit.\n\nhttps://archive.ics.uci.edu/ml/datasets/Credit+Approval\n\nData Set Information: This file concerns credit card applications. All attribute names and values have been changed to meaningless symbols to protect confidentiality of the data. This dataset is interesting because there is a good mix of attributes -- continuous, nominal with small numbers of values, and nominal with larger numbers of values. There are also a few missing values.\n\nAttribute Information:\n- A1: b, a.\n- A2: continuous.\n- A3: continuous.\n- A4: u, y, l, t.\n- A5: g, p, gg.\n- A6: c, d, cc, i, j, k, m, r, q, w, x, e, aa, ff.\n- A7: v, h, bb, j, n, z, dd, ff, o.\n- A8: continuous.\n- A9: t, f.\n- A10: t, f.\n- A11: continuous.\n- A12: t, f.\n- A13: g, p, s.\n- A14: continuous.\n- A15: continuous.\n- A16: +,- (class attribute)\n\nYes, most of that doesn't mean anything. A16 (the class attribute) is the most interesting, as it separates the 307 approved cases from the 383 rejected cases. The remaining variables have been obfuscated for privacy - a challenge you may have to deal with in your data science career.\n\nSprint challenges are evaluated based on satisfactory completion of each part. It is suggested you work through it in order, getting each aspect reasonably working, before trying to deeply explore, iterate, or refine any given step. Once you get to the end, if you want to go back and improve things, go for it!",
"_____no_output_____"
],
[
"## Part 1 - Load and validate the data\n\n- Load the data as a `pandas` data frame.\n- Validate that it has the appropriate number of observations (you can check the raw file, and also read the dataset description from UCI).\n- UCI says there should be missing data - check, and if necessary change the data so pandas recognizes it as na\n- Make sure that the loaded features are of the types described above (continuous values should be treated as float), and correct as necessary\n\nThis is review, but skills that you'll use at the start of any data exploration. Further, you may have to do some investigation to figure out which file to load from - that is part of the puzzle.",
"_____no_output_____"
],
[
"# Part 1",
"_____no_output_____"
]
],
[
[
"from google.colab import files\nuploaded = files.upload()",
"_____no_output_____"
],
[
"import pandas as pd\ndf = pd.read_csv('crx.txt',header=None)",
"_____no_output_____"
],
[
"columns=[''.join(['A',str(i)]) for i in range(1,17)]\ndf.columns = columns\n",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"#number of instances should be 690 and number of attributes should be 15\ndf.shape",
"_____no_output_____"
],
[
"#checking for missing data\n# df.isna().sum() shows zero nans so will have to look at how they indicate data is missing\ndf['A1'].value_counts()",
"_____no_output_____"
],
[
"#ok so they are using question mark. let's reaplce it with np.nan\nimport numpy as np\ndf = df.replace('?',np.nan)",
"_____no_output_____"
],
[
"#now checking each column datatype\ndf.info()\n#mismatchbetween the following\n##A2: continuous.\n##A14: continuous.\n#changinf ints to floats too A11, A15\n",
"_____no_output_____"
],
[
"#changing dtypes of those columns\ndf['A2'] = df['A2'].astype('float')\ndf['A14'] = df['A14'].str.replace(r\"^0+(?!$)\", \"\").astype('float')\ndf['A11'] = df['A11'].astype('float')\ndf['A15'] = df['A15'].astype('float')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"# Part 2",
"_____no_output_____"
],
[
"## Part 2 - Exploring data, Testing hypotheses\n\nThe only thing we really know about this data is that A16 is the class label. Besides that, we have 6 continuous (float) features and 9 categorical features.\n\nExplore the data: you can use whatever approach (tables, utility functions, visualizations) to get an impression of the distributions and relationships of the variables. In general, your goal is to understand how the features are different when grouped by the two class labels (`+` and `-`).\n\nFor the 6 continuous features, how are they different when split between the two class labels? Choose two features to run t-tests (again split by class label) - specifically, select one feature that is *extremely* different between the classes, and another feature that is notably less different (though perhaps still \"statistically significantly\" different). You may have to explore more than two features to do this.\n\nFor the categorical features, explore by creating \"cross tabs\" (aka [contingency tables](https://en.wikipedia.org/wiki/Contingency_table)) between them and the class label, and apply the Chi-squared test to them. [pandas.crosstab](http://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.crosstab.html) can create contingency tables, and [scipy.stats.chi2_contingency](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.chi2_contingency.html) can calculate the Chi-squared statistic for them.\n\nThere are 9 categorical features - as with the t-test, try to find one where the Chi-squared test returns an extreme result (rejecting the null that the data are independent), and one where it is less extreme.\n\n**NOTE** - \"less extreme\" just means smaller test statistic/larger p-value. Even the least extreme differences may be strongly statistically significant.\n\nYour *main* goal is the hypothesis tests, so don't spend too much time on the exploration/visualization piece. That is just a means to an end - use simple visualizations, such as boxplots or a scatter matrix (both built in to pandas), to get a feel for the overall distribution of the variables.\n\nThis is challenging, so manage your time and aim for a baseline of at least running two t-tests and two Chi-squared tests before polishing. And don't forget to answer the questions in part 3, even if your results in this part aren't what you want them to be.",
"_____no_output_____"
]
],
[
[
"#encoding category data\ndf_coded=df.copy()\nobj_list = ['A1','A4','A5','A6','A7','A9','A10','A12','A13','A16']\nfor col in obj_list:\n df_coded[col] = pd.Categorical(df_coded[col])\n df_coded[col] = df_coded[col].cat.codes",
"_____no_output_____"
],
[
"df_coded.head()",
"_____no_output_____"
],
[
"#we can see A14 and A15 are teh most variable - i.e. have the highest standard deviation\ndf_coded.describe()",
"_____no_output_____"
],
[
"#now filter this into two dataframes: for + which is 0 in teh encoding and - which is 1 in the encoding and see what is looks like\ndf_plus = df_coded[df_coded['A16']==0].copy()\ndf_minus = df_coded[df_coded['A16']==1].copy()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nplus_means = df_plus.mean()\ndf_plot = pd.DataFrame(plus_means)\nminus_means = df_minus.mean()\ndf_plot[1] = minus_means\ndf_plot = df_plot[[0,1]].div(df_plot[0],axis=0)\n\ndf_plot.columns=[['plus','minus']]\ndf_plot=df_plot.drop(['A16'])\ndf_plot.plot(kind='bar');\n",
"_____no_output_____"
],
[
"#so we can see huge difference A5,A9,A10, A11 and A15 with A11 and A15 as teh biggest\n#so one feature that is different will choose A15 and one that is not so different will choose A2\nimport scipy\nfrom scipy import stats\nmean_A15 = df_coded['A15'].mean()\nscipy.stats.ttest_1samp(df_plus['A15'],mean_A15,nan_policy='omit')",
"_____no_output_____"
],
[
"scipy.stats.ttest_1samp(df_minus['A15'],mean_A15,nan_policy='omit')",
"_____no_output_____"
],
[
"mean_A1 = df_coded['A1'].mean()\nscipy.stats.ttest_1samp(df_plus['A1'],mean_A1,nan_policy='omit'),scipy.stats.ttest_1samp(df_minus['A1'],mean_A1,nan_policy='omit')",
"_____no_output_____"
],
[
"#so we can see clearly for A1 they are both drawn from teh same population with pscore 99, but for A15 teh populations\n#are very different for each group with p score much less than 0.01 for the minus group\nobj_list",
"_____no_output_____"
],
[
"#making crosstab to explore data\n\ndf_cat = df_coded[obj_list].copy()\nfor col in df_cat.columns:\n df_cat[col]=np.where(df_cat[col]==-1,2,df_cat[col])\n\nct = pd.crosstab(df_cat['A10'],df_cat['A16'])\nct",
"_____no_output_____"
],
[
"#running chi_squared test for each column\nchilist\nfor col in df_cat.columns:\n ",
"_____no_output_____"
],
[
"test_list = ['A1', 'A4', 'A5', 'A6', 'A7', 'A9', 'A10', 'A12', 'A13']\nchi_list = [] \nfor test in test_list:\n ct_temp = pd.crosstab(df_cat[test],df_cat['A16'])\n chi2statistic, pvalue, dof, observed = scipy.stats.chi2_contingency(ct_temp)\n aaa = (test, chi2statistic)\n chi_list.append(aaa)\nchi_list",
"_____no_output_____"
],
[
"#so we can see teh most extreme categories are A9 with 355 and A10 with 143 so must be dependent",
"_____no_output_____"
],
[
"#we can also check the chi squared tests to see if various inputs are dependent on each other\nchi_list2=[]\ncounter = 0\nfor i in test_list[0:7]:\n counter = counter +1\n for j in test_list[counter:]:\n ct_temp = pd.crosstab(df_cat[i],df_cat[j])\n chi2statistic, pvalue, dof, observed = scipy.stats.chi2_contingency(ct_temp)\n bbb = (i,j,chi2statistic)\n chi_list2.append(bbb)\n \nchi_list2",
"_____no_output_____"
],
[
"#so of the varaible we can see ('A4', 'A5', 1380.0), ('A6', 'A7', 1844.7402531958905),are the most extreme cases with noteable high numbers elsewhere",
"_____no_output_____"
]
],
[
[
"# part 3",
"_____no_output_____"
],
[
"## Part 3 - Analysis and Interpretation\n\nNow that you've looked at the data, answer the following questions:\n\n- Interpret and explain the two t-tests you ran - what do they tell you about the relationships between the continuous features you selected and the class labels?\n- Interpret and explain the two Chi-squared tests you ran - what do they tell you about the relationships between the categorical features you selected and the class labels?\n- What was the most challenging part of this sprint challenge?\n\nAnswer with text, but feel free to intersperse example code/results or refer to it from earlier.",
"_____no_output_____"
],
[
"first of all before we ran the ttests we could see some big differences between the attributes in the 2 classifier groups. the ttest showed that for thefirst feature (A15) tested they were not drawn form the same populations with such low pvalues, contrasting clearly with A1 which there was negligable difference. \n\n\n---\n\n\n\nFor the chi squared test we could see that some attributes clearly had a big relationship with the classifier, and when we tested them on each other, i.e. attribute to attribute (excluding classifier) we could also see they were heavily dependent on each other in some cases as you would expect with credit data.\n\n\n\n---\n\n\n\nI had two little hold ups on this, getting my bar chart to work and then getting crosstabs to work as i've used pivots where possible before, and also as i had categorized some attributes as -1 and i had to fix that before continuing as it was stopping the chiquared test from running\ni can also see how it's a little harder to work with data when you can't see what that attributes are, but mabye that gets rid of some unwanted biases.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ec8a8f9bb9c2911401e558ef63550ef80d949eff | 7,535 | ipynb | Jupyter Notebook | docs/installing_julia_and_ijulia.ipynb | larssp/winpython_afterdoc | c50e38ec0b68d74cba6b5da54a408a1f86aa9cd8 | [
"MIT"
] | null | null | null | docs/installing_julia_and_ijulia.ipynb | larssp/winpython_afterdoc | c50e38ec0b68d74cba6b5da54a408a1f86aa9cd8 | [
"MIT"
] | null | null | null | docs/installing_julia_and_ijulia.ipynb | larssp/winpython_afterdoc | c50e38ec0b68d74cba6b5da54a408a1f86aa9cd8 | [
"MIT"
] | null | null | null | 28.115672 | 132 | 0.542933 | [
[
[
"# Installating Julia/IJulia (version of 2019-08-25)\n\n### Warning: as of 2019-08-25, the Julia installation is not supposed to support a move of Winpython library\n\nread also https://pyjulia.readthedocs.io/en/latest/installation.html#step-1-install-julia\n\nother intesting note:\nor https://discourse.julialang.org/t/using-jupyterlab/20595/2\nor https://blog.jupyter.org/i-python-you-r-we-julia-baf064ca1fb6\n\n###0 - mandatory and only pre-requisites (prepared, in WinPython 2019-03+)\n\"pip install pyjulia\" pyjulia python package\n\"set PYTHON=%WINDPYDIR%\\python.exe\" in your %WINPYDIR%..\\scripts\\ENV.BAT\n\n###1 - Downloading and Installing the right Julia binary in the right place",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nimport io",
"_____no_output_____"
],
[
"# downloading julia (32Mo, may take 1 minute or 2)\ntry:\n import urllib.request as urllib2 # Python 3\nexcept:\n import urllib2 # Python 2\nif 'amd64' in sys.version.lower():\n julia_binary=\"julia-1.2.0-win64.exe\"\n julia_url=\"https://julialang-s3.julialang.org/bin/winnt/x64/1.2/julia-1.2.0-win64.exe\"\n hashes=(\"5ccb7d585f854f3a3d1c6d3db07b6c0f\", \"93A94ED6429463E585B7B7E2CAC079F92820F71D\" )\nelse:\n julia_binary=\"julia-1.2.0-win32.exe\"\n julia_url=\"https://julialang-s3.julialang.org/bin/winnt/x86/1.2/julia-1.2.0-win32.exe\"\n hashes=(\"a2a0ab81bd639339970248c808316f23\", \"b5e0e5b6f0e5a737f4de9baa758b3ab50e2d52cb\")\n \njulia_installer=os.environ[\"WINPYDIR\"]+\"\\\\..\\\\t\\\\\"+julia_binary\nos.environ[\"julia_installer\"]=julia_installer\ng = urllib2.urlopen(julia_url) \nwith io.open(julia_installer, 'wb') as f:\n f.write(g.read())\ng.close\ng = None",
"_____no_output_____"
],
[
"#checking it's there\n!dir %julia_installer%",
"_____no_output_____"
],
[
"# checking it's the official julia0.3.2\nimport hashlib\ndef give_hash(of_file, with_this):\n with io.open(julia_installer, 'rb') as f:\n return with_this(f.read()).hexdigest() \nprint (\" \"*12+\"MD5\"+\" \"*(32-12-3)+\" \"+\" \"*15+\"SHA-1\"+\" \"*(40-15-5)+\"\\n\"+\"-\"*32+\" \"+\"-\"*40)\n\nprint (\"%s %s %s\" % (give_hash(julia_installer, hashlib.md5) , give_hash(julia_installer, hashlib.sha1),julia_installer))\nassert give_hash(julia_installer, hashlib.md5) == hashes[0].lower() \nassert give_hash(julia_installer, hashlib.sha1) == hashes[1].lower()",
"_____no_output_____"
],
[
"# will be in env next time\nimport os\nos.environ[\"JULIA_HOME\"] = os.environ[\"WINPYDIR\"]+\"\\\\..\\\\t\\\\Julia\\\\bin\\\\\"\nos.environ[\"JULIA_EXE\"]=\"julia.exe\"\nos.environ[\"JULIA\"]=os.environ[\"JULIA_HOME\"]+os.environ[\"JULIA_EXE\"]\nos.environ[\"JULIA_PKGDIR\"]=os.environ[\"WINPYDIRBASE\"]+\"\\\\settings\\\\.julia\"\n# for installation we need this\nos.environ[\"JULIAROOT\"]=os.path.join(os.path.split(os.environ[\"WINPYDIR\"])[0] , 't','julia' ) ",
"_____no_output_____"
],
[
"# let's install it (add a /S before /D if you want silence mode installation)\n#nullsoft installers don't accept . or .. conventions\n\n# If you are \"USB life style\", or multi-winpython\n# ==> UN-CLICK the OPTION 'CREATE a StartMenuFolder and Shortcut' <== (when it will show up)\n!start cmd /C %julia_installer% /D=%JULIAROOT%\n",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/stonebig/winpython_afterdoc/master/examples/images/julia_setup_unclick_all.GIF\">",
"_____no_output_____"
],
[
"###2 - Initialize Julia , IJulia, and make them link to winpython",
"_____no_output_____"
]
],
[
[
"# connecting Julia to WinPython (only once, or everytime you move things)\nimport julia\njulia.install()",
"_____no_output_____"
],
[
"%load_ext julia.magic",
"_____no_output_____"
],
[
"%%julia \nimport Pkg;\nPkg.add(\"PyPlot\")\nPkg.add(\"Interact\")\nPkg.add(\"Compose\")\nPkg.add(\"SymPy\")\nPkg.add(\"JuMP\")\nPkg.add(\"Ipopt\")",
"_____no_output_____"
]
],
[
[
"###3 - Launching a Julia Notebook \n\nchoose a Julia Kernel from Notebook, or Julia from Jupyterlab Launcher\n\n",
"_____no_output_____"
],
[
"###4 - Julia Magic \nor use %load_ext julia.magic then %julia or %%julia",
"_____no_output_____"
]
],
[
[
"import julia\n%matplotlib inline\n%load_ext julia.magic",
"_____no_output_____"
],
[
"# since Julia 1.x ''@pyimport foo' is replaced per 'foo = pyimport(\"foo\")'' \n%julia plt = pyimport(\"matplotlib.pyplot\")\n%julia np = pyimport(\"numpy\")",
"_____no_output_____"
],
[
"%%julia \nt = np.linspace(0, 2*pi,1000); \ns = np.sin(3*t + 4*np.cos(2*t)); \nfig = plt.gcf() \nplt.plot(t, s, color=\"red\", linewidth=2.0, linestyle=\"--\", label=\"sin(3t+4.cos(2t))\")",
"_____no_output_____"
],
[
"#Alternative\nimport julia\nj=julia.Julia()\nj.eval(\"1 +31\")\nj.eval(\"sqrt(1 +31)\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ec8aa5ae7575609aa27d57fea06b7ed0b62aa31d | 1,312 | ipynb | Jupyter Notebook | gradient-descent/GradientDescentSolutions.ipynb | hrgupta/deep-learning | 3a7e9b96dea91d94b933610a6a0a1b263028de53 | [
"MIT"
] | 1 | 2020-05-22T14:32:49.000Z | 2020-05-22T14:32:49.000Z | gradient-descent/GradientDescentSolutions.ipynb | AlAlaa-Tashkandi/deep-learning | ea1316f1031ee2e7520c3e8f861406dee4e6438e | [
"MIT"
] | null | null | null | gradient-descent/GradientDescentSolutions.ipynb | AlAlaa-Tashkandi/deep-learning | ea1316f1031ee2e7520c3e8f861406dee4e6438e | [
"MIT"
] | null | null | null | 22.237288 | 67 | 0.521341 | [
[
[
"# Solutions",
"_____no_output_____"
]
],
[
[
"# Activation (sigmoid) function\ndef sigmoid(x):\n return 1 / (1 + np.exp(-x))\n\ndef output_formula(features, weights, bias):\n return sigmoid(np.dot(features, weights) + bias)\n\ndef error_formula(y, output):\n return - y*np.log(output) - (1 - y) * np.log(1-output)\n\ndef update_weights(x, y, weights, bias, learnrate):\n output = output_formula(x, weights, bias)\n d_error = -(y - output)\n weights -= learnrate * d_error * x\n bias -= learnrate * d_error\n return weights, bias",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
ec8ab4db807716b06e5453e1b8dd5ef046a0503b | 59,686 | ipynb | Jupyter Notebook | examples/PrimMST.ipynb | swyang50066/segmentation | 21aefb18bafaa8925b81fe9ef1fb0ab4d50b474e | [
"MIT"
] | 9 | 2021-09-06T12:54:14.000Z | 2022-02-25T03:33:24.000Z | examples/PrimMST.ipynb | swyang50066/medical-image-segmentation | bf53a19864bb31d8d4828b3bf42d9cf5f6bf361c | [
"MIT"
] | null | null | null | examples/PrimMST.ipynb | swyang50066/medical-image-segmentation | bf53a19864bb31d8d4828b3bf42d9cf5f6bf361c | [
"MIT"
] | 3 | 2021-06-18T15:49:16.000Z | 2022-02-24T13:13:41.000Z | 406.027211 | 55,884 | 0.933636 | [
[
[
"import sys\nsys.path.append(\"../utils\")\n\nimport cv2\nimport numpy as np\n\nimport matplotlib.pyplot as plt\n\nfrom minimum_spanning_tree_prim import GraphUndirectedWeighted\nfrom minimum_spanning_tree_prim import PrimMST",
"_____no_output_____"
],
[
"# Load sample image\n'''\nimsample = np.array([[0, 0, 1, 0, 0],\n [0, 1, 4, 1, 0],\n [1, 4, 6, 4, 1],\n [0, 1, 4, 1, 0],\n [0, 0, 1, 0, 0]])\n'''\n\nimsample = cv2.imread(\"../asset/cameraman.png\")[..., 0].astype(np.float32)\nimsample = (imsample - imsample.min())/(imsample.max() - imsample.min())\nimsample = imsample[251:271, 251:271]\n\n# Build undirected/weighted image graph\ngraph = GraphUndirectedWeighted(imsample)()\n\n# Print results\n#for k, v in graph.items():\n# print(\"Node => [%d]\"%k, v)\n#for node in nodes:\n# print(\"SortedWeighted Nodes\", node)",
"_____no_output_____"
],
[
"def drawMSF(x, leaves):\n # Get input shape\n height, width = x.shape\n \n # Gray-scale\n gray = 255*(x - x.min())/(x.max() - x.min())\n\n # Define plot components\n fig, axes = plt.subplots(nrows=1, ncols=1, figsize=(6, 6))\n\n # Plot MSF\n axes.imshow(gray, cmap=\"Greys\")\n for leaf in leaves:\n # Mark leaf node\n xa, ya = leaf.index//width, leaf.index%width\n axes.scatter(ya, xa, color=\"Blue\", s=20, marker=\"s\")\n\n while leaf.parent:\n # Mark branch node\n a, b = leaf.parent.index, leaf.index\n \n xa, ya = a//width, a%width\n xb, yb = b//width, b%width\n \n axes.scatter(ya, xa, color=\"Red\", s=20)\n axes.arrow(ya, xa, yb-ya, xb-xa, color=\"lime\", head_width=.05, head_length=.1)\n\n # Upstream\n leaf = leaf.parent\n\n # Mark root node\n if leaf.parent == None:\n axes.scatter(ya, xa, color=\"Green\", s=20, marker=\"D\")",
"_____no_output_____"
],
[
"# Find minimum spanning tree with Prim's algorithm\nleaves = PrimMST(graph)\n\n# Draw MSF\ndrawMSF(imsample, leaves)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
ec8ac5a3c2cc91536ba7ac418d1224ac392bc89a | 530,605 | ipynb | Jupyter Notebook | machine_learning/gan/create_tfrecords_cifar10.ipynb | ryangillard/artificial_intelligence | f7c21af221f366b075d351deeeb00a1b266ac3e3 | [
"Apache-2.0"
] | 4 | 2019-07-04T05:15:59.000Z | 2020-06-29T19:34:33.000Z | machine_learning/gan/create_tfrecords_cifar10.ipynb | ryangillard/artificial_intelligence | f7c21af221f366b075d351deeeb00a1b266ac3e3 | [
"Apache-2.0"
] | null | null | null | machine_learning/gan/create_tfrecords_cifar10.ipynb | ryangillard/artificial_intelligence | f7c21af221f366b075d351deeeb00a1b266ac3e3 | [
"Apache-2.0"
] | 1 | 2019-05-23T16:06:51.000Z | 2019-05-23T16:06:51.000Z | 720.930707 | 184,556 | 0.943998 | [
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport tensorflow as tf\nprint(tf.__version__)",
"2.3.0\n"
],
[
"(x_train, y_train), (x_test, y_test) = tf.keras.datasets.cifar10.load_data()",
"_____no_output_____"
],
[
"print(\"x_train.shape = {}\".format(x_train.shape))\nprint(\"y_train.shape = {}\".format(y_train.shape))\nprint(\"x_test.shape = {}\".format(x_test.shape))\nprint(\"y_test.shape = {}\".format(y_test.shape))",
"x_train.shape = (50000, 32, 32, 3)\ny_train.shape = (50000, 1)\nx_test.shape = (10000, 32, 32, 3)\ny_test.shape = (10000, 1)\n"
],
[
"def plot_images(images):\n num_images = len(images)\n\n plt.figure(figsize=(20, 20))\n for i in range(num_images):\n plt.subplot(1, num_images, i + 1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(images[i], cmap=plt.cm.binary)\n plt.show()",
"_____no_output_____"
],
[
"plot_images(images=x_train[:10])",
"_____no_output_____"
],
[
"plot_images(images=x_test[:10])",
"_____no_output_____"
]
],
[
[
"## Write",
"_____no_output_____"
]
],
[
[
"def write_tf_record(filepath, data):\n \"\"\"Writes TFRecord files of given data to given filepath.\n\n Args:\n filepath: str, filepath location.\n data: dict, np.arrays of feature and label data.\n \"\"\"\n with tf.io.TFRecordWriter(path=filepath) as ofp:\n for i in range(len(data[\"image_raw\"])):\n example = tf.train.Example(\n features=tf.train.Features(\n feature={\n \"image_raw\": tf.train.Feature(\n bytes_list=tf.train.BytesList(\n value=[data[\"image_raw\"][i].tobytes()]\n )\n ),\n \"label\": tf.train.Feature(\n int64_list=tf.train.Int64List(\n value=[data[\"label\"][i]]\n )\n )\n }\n )\n )\n ofp.write(example.SerializeToString())\n",
"_____no_output_____"
],
[
"def create_tf_record_shards(num_shards, filepath, data):\n \"\"\"Creates TFRecord shards of given data at given filepath.\n\n Args:\n num_shards: int, number of file shards to split data into.\n filepath: str, filepath location.\n data: dict, np.arrays of feature and label data.\n \"\"\"\n dataset_len = data[list(data.keys())[0]].shape[0]\n num_shards = min(num_shards, dataset_len)\n shard_size = dataset_len // num_shards\n remainder = dataset_len % num_shards\n loop_limit = num_shards if remainder == 0 else num_shards - 1\n for i in range(loop_limit):\n write_tf_record(\n filepath=\"{}_{}.tfrecord\".format(filepath, i),\n data={\n key: value[shard_size * i:shard_size * (i + 1)]\n for key, value in data.items()\n }\n )\n\n if remainder > 0:\n write_tf_record(\n filepath=\"{}_{}.tfrecord\".format(filepath, num_shards - 1),\n data={\n key: value[shard_size * (num_shards - 1):shard_size * num_shards + remainder]\n for key, value in data.items()\n }\n )\n",
"_____no_output_____"
],
[
"# Set number of shards to split dataset into.\nnum_shards = 10\n\n# Loop through each progressive resolution image size.\nfor i in range(4):\n img_sz = 4 * 2 ** i\n print(\"\\nWriting {0}_{0} TF Records!\".format(img_sz))\n\n create_tf_record_shards(\n num_shards=num_shards,\n filepath=\"data/cifar10/train_{0}x{0}\".format(img_sz),\n data={\n \"image_raw\": tf.image.resize(\n images=x_train, size=(img_sz, img_sz), method=\"nearest\"\n ).numpy(),\n \"label\": np.ones(shape=[x_train.shape[0]], dtype=np.int32)\n }\n )\n create_tf_record_shards(\n num_shards=num_shards,\n filepath=\"data/cifar10/test_{0}x{0}\".format(img_sz),\n data={\n \"image_raw\": tf.image.resize(\n images=x_test, size=(img_sz, img_sz), method=\"nearest\"\n ).numpy(),\n \"label\": np.ones(shape=[x_test.shape[0]], dtype=np.int32)\n }\n )\n",
"\nWriting 4_4 TF Records!\n\nWriting 8_8 TF Records!\n\nWriting 16_16 TF Records!\n\nWriting 32_32 TF Records!\n"
]
],
[
[
"## Read",
"_____no_output_____"
]
],
[
[
"def print_obj(function_name, object_name, object_value):\n \"\"\"Prints enclosing function, object name, and object value.\n\n Args:\n function_name: str, name of function.\n object_name: str, name of object.\n object_value: object, value of passed object.\n \"\"\"\n# pass\n print(\"{}: {} = {}\".format(function_name, object_name, object_value))",
"_____no_output_____"
],
[
"def decode_example(protos, params):\n \"\"\"Decodes TFRecord file into tensors.\n\n Given protobufs, decode into image and label tensors.\n\n Args:\n protos: protobufs from TFRecord file.\n params: dict, user passed parameters.\n\n Returns:\n Image and label tensors.\n \"\"\"\n # Create feature schema map for protos.\n features = {\n \"image_raw\": tf.io.FixedLenFeature(shape=[], dtype=tf.string),\n \"label\": tf.io.FixedLenFeature(shape=[], dtype=tf.int64)\n }\n\n # Parse features from tf.Example.\n parsed_features = tf.io.parse_single_example(\n serialized=protos, features=features\n )\n print_obj(\"\\ndecode_example\", \"features\", features)\n\n # Convert from a scalar string tensor (whose single string has\n # length height * width * depth) to a uint8 tensor with shape\n # [height * width * depth].\n image = tf.io.decode_raw(\n input_bytes=parsed_features[\"image_raw\"], out_type=tf.uint8\n )\n print_obj(\"decode_example\", \"image\", image)\n\n # Reshape flattened image back into normal dimensions.\n image = tf.reshape(\n tensor=image,\n shape=[params[\"height\"], params[\"width\"], params[\"depth\"]]\n )\n print_obj(\"decode_example\", \"image\", image)\n\n # Convert from [0, 255] -> [-1.0, 1.0] floats.\n image_scaled = tf.cast(x=image, dtype=tf.float32) * (2. / 255) - 1.0\n print_obj(\"decode_example\", \"image\", image)\n\n # Convert label from a scalar uint8 tensor to an int32 scalar.\n label = tf.cast(x=parsed_features[\"label\"], dtype=tf.int32)\n print_obj(\"decode_example\", \"label\", label)\n\n return {\"image\": image, \"image_scaled\": image_scaled}, label\n\ndef read_dataset(filename, mode, batch_size, params):\n \"\"\"Reads CSV time series data using tf.data, doing necessary preprocessing.\n\n Given filename, mode, batch size, and other parameters, read CSV dataset\n using Dataset API, apply necessary preprocessing, and return an input\n function to the Estimator API.\n\n Args:\n filename: str, file pattern that to read into our tf.data dataset.\n mode: The estimator ModeKeys. Can be TRAIN or EVAL.\n batch_size: int, number of examples per batch.\n params: dict, dictionary of user passed parameters.\n\n Returns:\n An input function.\n \"\"\"\n def _input_fn():\n \"\"\"Wrapper input function used by Estimator API to get data tensors.\n\n Returns:\n Batched dataset object of dictionary of feature tensors and label\n tensor.\n \"\"\"\n # Create list of files that match pattern.\n file_list = tf.io.gfile.glob(pattern=filename)\n\n # Create dataset from file list.\n dataset = tf.data.TFRecordDataset(\n filenames=file_list, num_parallel_reads=40\n )\n\n # Shuffle and repeat if training with fused op.\n if mode == tf.estimator.ModeKeys.TRAIN:\n dataset = dataset.apply(\n tf.contrib.data.shuffle_and_repeat(\n buffer_size=50 * batch_size,\n count=None # indefinitely\n )\n )\n\n # Decode example into a features dictionary of tensors, then batch.\n dataset = dataset.map(\n map_func=lambda x: decode_example(\n protos=x,\n params=params\n ),\n num_parallel_calls=4\n )\n\n dataset = dataset.batch(batch_size=batch_size)\n\n # Prefetch data to improve latency.\n dataset = dataset.prefetch(buffer_size=2)\n\n return dataset\n return _input_fn\n",
"_____no_output_____"
],
[
"def try_out_input_function(arguments, print_out=False):\n \"\"\"Trys out input function for testing purposes.\n\n Args:\n arguments: dict, user passed parameters.\n \"\"\"\n fn = read_dataset(\n filename=arguments[\"filename\"],\n mode=tf.estimator.ModeKeys.EVAL,\n batch_size=8,\n params=arguments\n )\n\n dataset = fn()\n\n batches = [batch for batch in dataset]\n features, labels = batches[0]\n print(\"features[image].shape = {}\".format(features[\"image\"].shape))\n print(\"labels.shape = {}\".format(labels.shape))\n# print(\"features = \\n{}\".format(features))\n print(\"labels = \\n{}\".format(labels))\n\n return features, labels\n",
"_____no_output_____"
],
[
"def plot_all_resolutions(filepath, starting_size, num_doublings, depth):\n \"\"\"Plots all resolutions of images.\n\n Args:\n filepath: str, filepath location.\n starting_size: int, smallest/initial size of image.\n num_doublings: int, number of times to double height/width of original\n image size.\n depth: int, number of image channels.\n \"\"\"\n for i in range(num_doublings):\n img_sz = starting_size * 2 ** i\n print(\"\\nImage size = {0}x{0}\".format(img_sz))\n\n features, labels = try_out_input_function(\n arguments={\n \"filename\": \"{0}_{1}x{1}_0.tfrecord\".format(\n filepath, img_sz\n ),\n \"height\": img_sz,\n \"width\": img_sz,\n \"depth\": depth\n },\n print_out=True\n )\n plot_images(features[\"image\"])\n",
"_____no_output_____"
],
[
"plot_all_resolutions(\n filepath=\"data/cifar10/train\",\n starting_size=4,\n num_doublings=4,\n depth=3\n)",
"\nImage size = 4x4\n\ndecode_example: features = {'image_raw': FixedLenFeature(shape=[], dtype=tf.string, default_value=None), 'label': FixedLenFeature(shape=[], dtype=tf.int64, default_value=None)}\ndecode_example: image = Tensor(\"DecodeRaw:0\", shape=(None,), dtype=uint8)\ndecode_example: image = Tensor(\"Reshape:0\", shape=(4, 4, 3), dtype=uint8)\ndecode_example: image = Tensor(\"Reshape:0\", shape=(4, 4, 3), dtype=uint8)\ndecode_example: label = Tensor(\"Cast_1:0\", shape=(), dtype=int32)\nfeatures[image].shape = (8, 4, 4, 3)\nlabels.shape = (8,)\nlabels = \n[1 1 1 1 1 1 1 1]\n"
],
[
"plot_all_resolutions(\n filepath=\"data/cifar10/test\",\n starting_size=4,\n num_doublings=4,\n depth=3\n)",
"\nImage size = 4x4\n\ndecode_example: features = {'image_raw': FixedLenFeature(shape=[], dtype=tf.string, default_value=None), 'label': FixedLenFeature(shape=[], dtype=tf.int64, default_value=None)}\ndecode_example: image = Tensor(\"DecodeRaw:0\", shape=(None,), dtype=uint8)\ndecode_example: image = Tensor(\"Reshape:0\", shape=(4, 4, 3), dtype=uint8)\ndecode_example: image = Tensor(\"Reshape:0\", shape=(4, 4, 3), dtype=uint8)\ndecode_example: label = Tensor(\"Cast_1:0\", shape=(), dtype=int32)\nfeatures[image].shape = (8, 4, 4, 3)\nlabels.shape = (8,)\nlabels = \n[1 1 1 1 1 1 1 1]\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8ac78e0704520e675093487ffc752e2fec17de | 32,553 | ipynb | Jupyter Notebook | notebook/_Agrupamento/kmeans_CartaoCredito_Clientes_2.ipynb | victorblois/py_datascience | bf5d05a0a4d9e26f55f9259d973f7f4f54432e24 | [
"MIT"
] | 1 | 2020-05-11T22:22:55.000Z | 2020-05-11T22:22:55.000Z | notebook/_Agrupamento/kmeans_CartaoCredito_Clientes_2.ipynb | victorblois/py_datascience | bf5d05a0a4d9e26f55f9259d973f7f4f54432e24 | [
"MIT"
] | null | null | null | notebook/_Agrupamento/kmeans_CartaoCredito_Clientes_2.ipynb | victorblois/py_datascience | bf5d05a0a4d9e26f55f9259d973f7f4f54432e24 | [
"MIT"
] | null | null | null | 64.846614 | 15,572 | 0.677418 | [
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\nfrom sklearn.cluster import KMeans\nfrom sklearn.preprocessing import StandardScaler\nimport numpy as np",
"_____no_output_____"
],
[
"import os.path\ndef path_base(base_name):\n current_dir = os.path.abspath(os.path.join(os.getcwd(), os.pardir))\n print(current_dir)\n data_dir = current_dir.replace('notebook','data')\n print(data_dir)\n data_base = data_dir + '\\\\' + base_name\n print(data_base)\n return data_base",
"_____no_output_____"
],
[
"base = pd.read_csv(path_base('db_cartaocredito_clientes.csv'), header = 1)",
"C:\\MyPhyton\\DataScience\\notebook\nC:\\MyPhyton\\DataScience\\data\nC:\\MyPhyton\\DataScience\\data\\db_cartaocredito_clientes.csv\n"
],
[
"base['BILL_TOTAL'] = base['BILL_AMT1'] + base['BILL_AMT2'] + base['BILL_AMT3'] + base['BILL_AMT4'] + base['BILL_AMT5'] + base['BILL_AMT6']",
"_____no_output_____"
],
[
"base.describe()",
"_____no_output_____"
],
[
"X = base.iloc[:,[1,2,3,4,5,25]].values\nscaler = StandardScaler()\nX = scaler.fit_transform(X)\n\n",
"_____no_output_____"
],
[
"wcss = []\nfor i in range(1, 11):\n kmeans = KMeans(n_clusters = i, random_state = 0)\n kmeans.fit(X)\n wcss.append(kmeans.inertia_)\nplt.plot(range(1, 11), wcss)\nplt.xlabel('Número de clusters')\nplt.ylabel('WCSS')\n\n",
"_____no_output_____"
],
[
"kmeans = KMeans(n_clusters = 4, random_state = 0)\nprevisoes = kmeans.fit_predict(X)",
"_____no_output_____"
],
[
"lista_clientes = np.column_stack((base, previsoes))\nlista_clientes = lista_clientes[lista_clientes[:,26].argsort()]",
"_____no_output_____"
],
[
"lista_clientes",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8accbe5243307a1b6754b66fc4a5f9c3d8ccdc | 4,127 | ipynb | Jupyter Notebook | cluster-mode-standalone/jupyterlab/starter-notebook.ipynb | cwilbar04/spark-kubernetes-jupyter | 6de0814b768080a38b0980cca98bdb3bc9203ba9 | [
"MIT"
] | null | null | null | cluster-mode-standalone/jupyterlab/starter-notebook.ipynb | cwilbar04/spark-kubernetes-jupyter | 6de0814b768080a38b0980cca98bdb3bc9203ba9 | [
"MIT"
] | null | null | null | cluster-mode-standalone/jupyterlab/starter-notebook.ipynb | cwilbar04/spark-kubernetes-jupyter | 6de0814b768080a38b0980cca98bdb3bc9203ba9 | [
"MIT"
] | null | null | null | 28.462069 | 132 | 0.548098 | [
[
[
"!gcloud auth activate-service-account --key-file=untitled.txt --project=spark-on-kubernetes-testing",
"_____no_output_____"
],
[
"!gcloud container clusters get-credentials spark-cluster --region=us-central1",
"_____no_output_____"
],
[
"!kubectl cluster-info",
"_____no_output_____"
],
[
"from pyspark.sql import SparkSession\nfrom pyspark import SparkFiles\nfrom pyspark.sql.types import StructType, StructField, StringType, DoubleType, BooleanType, ArrayType, LongType\nimport re\nimport requests\nfrom bs4 import BeautifulSoup",
"_____no_output_____"
],
[
"spark = SparkSession \\\n .builder \\\n .appName(\"kubernetes\") \\\n .getOrCreate()",
"_____no_output_____"
],
[
"spark.stop()",
"_____no_output_____"
],
[
"# Create Spark Session and Load BigQuery jar file\nspark = SparkSession \\\n .builder \\\n .appName(\"kubernetes\") \\\n .config(\"spark.master\",\"k8s://https://35.239.173.65:443\") \\\n .config(\"spark.submit.deployMode\",\"cluster\") \\\n .config(\"spark.executor.instances\",\"3\") \\\n .config(\"spark.kubernetes.container.image\",\"gcr.io/spark-on-kubernetes-testing/spark-cluster-spark-base:latest\") \\\n .config(\"spark.kubernetes.authenticate.driver.serviceAccountName\",\"spark\") \\\n .getOrCreate()",
"_____no_output_____"
],
[
"# Define schema of files to parse\nschema = StructType([ \n StructField(\"asin\",StringType(),True), \n StructField(\"image\",ArrayType(StringType()),True), \n StructField(\"overall\",DoubleType(),True),\n StructField(\"reviewText\",StringType(),True),\n StructField(\"reviewTime\",StringType(),True),\n StructField(\"reviewerID\",StringType(),True),\n StructField(\"reviewerName\",StringType(),True),\n StructField(\"summary\",StringType(),True),\n StructField(\"unixReviewTime\",LongType(),True),\n StructField(\"verified\",BooleanType(),True),\n StructField(\"vote\",StringType(),True),\n StructField(\"style\",ArrayType(StringType()),True)\n ])\n\n# URL to scrape to get files to download\nurl = \"https://nijianmo.github.io/amazon/index.html\"\nhtml = requests.get(url)\n\nif html.ok:\n soup = BeautifulSoup(html.content, 'html.parser') \n\noutput_final = []\nfiles = []\nlinks = soup.find_all('a',string='5-core')#.find('5-core')#.find_all('td', id='5-core')\nfor link in links[0:1]:\n url = link.get('href')\n file = url.split('/')[-1]\n print(url)\n print(url.split('/')[-1])\n spark.sparkContext.addFile(url)\n files.append(file)\n df = spark.read.json(\"file://\"+SparkFiles.get(file),schema)\n\ndf.show(5)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8ad41cc618d29a7a273279dc0131a0ae830cdd | 13,940 | ipynb | Jupyter Notebook | assignments/0205-Quiz1_Serial.ipynb | msu-cmse-courses/cmse401-S21-student | e7407d5f7860149606d9ea770eeafe61e93122c6 | [
"MIT"
] | null | null | null | assignments/0205-Quiz1_Serial.ipynb | msu-cmse-courses/cmse401-S21-student | e7407d5f7860149606d9ea770eeafe61e93122c6 | [
"MIT"
] | null | null | null | assignments/0205-Quiz1_Serial.ipynb | msu-cmse-courses/cmse401-S21-student | e7407d5f7860149606d9ea770eeafe61e93122c6 | [
"MIT"
] | 3 | 2021-01-23T18:15:41.000Z | 2022-03-08T21:18:44.000Z | 37.777778 | 442 | 0.611047 | [
[
[
"[Link to this document's Jupyter Notebook](./0205-Quiz1_Serial.ipynb)",
"_____no_output_____"
],
[
"----\n\n# CMSE401 Quiz Instructions\n\nThis quiz is designed to take approximately 20 minutes to complete (you will be given 50 Minutes). \n\nPlease read the following instructions before starting the quiz.\n",
"_____no_output_____"
],
[
"> This is an open Internet quiz. Feel free to use anything on the Internet with one important exception...\n> \n> - **DO NOT** communicate live with other people during the quiz (either verbally or on-line). The goal here is to find answers to problems as you would in the real world. \n> \n> You will be given **20 minutes** to complete this quiz. Use your time wisely. \n> \n> **HINTS:**\n> - Neatness and grammar is important. We will ignore all notes or code we can not read or understand.\n> - Read the entire quiz from beginning to end before starting. Not all questions are equal in **points vs. time** so plan your time accordingly. \n> - Some of the information provided my be a distraction. Do not assume you need to understand everything written to answer the questions. \n> - Spaces for answers are provided. Delete the prompting text such as \"Put your answer to the above question here\" and replace it with your answer. Do not leave the prompting text with your answer.\n> - Do not assume that the answer must be in the same format of the cell provided. Feel free to change the cell formatting (e.g., markdown to code, and vice versa) or add additional cells as needed to provide your answer.\n> - When a question asks for an answer \"**in your own words**\" it is still okay to search the Internet for the answer as a reminder. *However*, we would like you to do more than cut and paste. Make the answer your own. \n> - If you get stuck, try not to leave an answer blank. It is better to include some notes or stub functions so we have an idea about your thinking process so we can give you partial credit. \n> - Always provid links to any references you find helpful. \n> - Feel free to delete the provided check marks (✅) as a way to keep track of which questions you have successfully completed. ",
"_____no_output_____"
],
[
"> **Honor Code**\n> \n> I, agree to neither give nor receive any help on this quiz from other people. I also understand that providing answers to questions on this quiz to other students is also an academic misconduct violation as is live communication or receiving answers to questions on this quiz from other people. It is important to me to be a person of integrity and that means that ALL ANSWERS on this quiz are my answers.\n> \n> ✅ **<font color=red>DO THIS:</font>** Include your name in the line below to acknowledge the above statement:",
"_____no_output_____"
],
[
"Put your name here.",
"_____no_output_____"
],
[
"---\n\n# Scientific Image Analysis\n\n<img alt=\"ImageJ logo of a microscope. Motivating the reason behind ImageJ as a scientific tool\" src=\"https://miap.eu/fileadmin/primary/Public/user_uploads/Files/Workshops/2017-04_ImageJ_II/ImageJ.png\">\n\n[Logo from the ImageJ website](https://imagej.net/Welcome)\n\n\nImageJ is a software package developed with funding from the National Institute of Health (NIH). This is a well established tool written in java with decades of plugins and options to help researchers measure data inside of images (mostly medical images). For this quiz you will explore how to run large baches of ImageJ on the HPCC. ",
"_____no_output_____"
],
[
"✅ **<font color=red>Question 1</font>**: (10 points) What module command do you use to be able to load the ImageJ on the HPCC?",
"_____no_output_____"
],
[
"Put your answer here",
"_____no_output_____"
],
[
"✅ **<font color=red>Question 2</font>**: (10 points) What versions of ImageJ is installed on the HPCC. Type the command you used to figure this out? ",
"_____no_output_____"
],
[
"Put your answer here",
"_____no_output_____"
],
[
"----\n\nFor the following questions a researcher is trying to manually interact with the ImageJ Graphical User Interface (GUI) on the HPCC using the following command:\n\n```java -jar $EBROOTIMAGEJ/ij.jar```\n\nHowever, when they run the command on dev-intel18 through the ondemand server they get the following output:\n\n> ```bash\n$ java -jar $EBROOTIMAGEJ/ij.jar\nException in thread \"main\" java.awt.HeadlessException: \nNo X11 DISPLAY variable was set, but this program performed an operation which requires it.\n at java.awt.GraphicsEnvironment.checkHeadless(GraphicsEnvironment.java:204)\n at java.awt.Window.<init>(Window.java:536)\n at java.awt.Frame.<init>(Frame.java:420)\n at ij.ImageJ.<init>(ImageJ.java:143)\n at ij.ImageJ.main(ImageJ.java:703)\n```",
"_____no_output_____"
],
[
"✅ **<font color=red>Question 3</font>**: (20 points) Explain this error and describe one way you can fix or get around the problem and get the ImageJ GUI up and running on the HPCC. Make the instructions short but detailed enough for a new researcher to the HPCC. You are encouraged to provide links to websites as references to help answer your question. **_HINT_**: test your answers on the actual HPC to make sure they work.",
"_____no_output_____"
],
[
"Put your answer here",
"_____no_output_____"
],
[
"----\nWe can also run ImageJ with a \"macro\" that uses a custom language developed only for ImageJ. Here is an example:\n\n```\nprint(\"Inverting Image\");\nname = getArgument;\nif (name==\"\") exit (\"No argument!\");|\nsetBatchMode(true);\n\n//Set PATH\nvar patho=\"./\";\n\n//Open bot.tif file and make inverse\nprint(\"Opening File\");\nopen(patho+name);\n\nprint(\"Inverting Image\");\nrun(\"Invert\");\n\nprint(\"Saving Inverted File\");\nsaveAs(\"Png\", patho+\"Inv_\"+name);\n\n```\nThe above macro (named ```macro.ijm```) can be run on input file ```input1.png``` using the following command:\n\n java -jar $EBROOTIMAGEJ/ij.jar -batch ./macro.ijm input1.png\n\n\n\nAs we can tell from the error in the previous questions, one of the problems with ImageJ is that it needs a graphical user interface which requires a connected display. Unfortunately, if we try to run the macro using the scheduler in \"batch\" mode, we would get a similar error because none of the compute nodes have displays attached either. \n\nOne way to get around this problem is to use a \"fake\" display or \"virtual\" display. In Linux there is often a program called \"the X11 virtual frame buffer\" (aka ```Xvfb```). ",
"_____no_output_____"
],
[
"The following job script can be used with ImageJ and Xvfb to run a batch job on the hpcc:\n\n\n```bash\n#!/bin/bash\n#SBATCH --mem=4gb\n#SBATCH --time=00:10:00\n#SBATCH -n 1\n#SBATCH -c 1\n\nmodule load Java\nmodule load ImageJ\n\n#Remove left over xvfb lock files\nrm -rf /tmp/.X11-unix\nrm -rf /tmp/.X11-lock\n\n##### Specify the display, start the Xvfb server, and save the process ID.\nexport DISPLAY=\":1\"\nXvfb $DISPLAY -auth /dev/null &\nXVFB_PID=$!\n\n#Give system time to spin up X11 display (Probably not needed)\nsleep 5\n\n####\n#Run ImageJ script\njava -jar $EBROOTIMAGEJ/ij.jar -batch ./macro.ijm input1.png\n\n\n##### Stop the Xvfb server and remove the temporary lock files it created (if they don't remove themselves.\nkill -9 $XVFB_PID\nrm -rf /tmp/.X11-unix\nrm -rf /tmp/.X11-lock\n####\n\n```",
"_____no_output_____"
],
[
"✅ **<font color=red>Question 4</font>**: (20 points) If we remove the \"&\" at the end of the ```Xvfb``` line what will happen if we submit this script to the cluster? (explain why)",
"_____no_output_____"
],
[
"Put your answer to the above question here",
"_____no_output_____"
],
[
"-----\nNow let us assume we have a directory filled with image files we want to process using our ```macro.ijm``` in imagej. These files are located in your current directory with the following names:\n\n input1.png\n input2.png\n input3.png\n ...\n input300.png\n \nWe could use the following simple bash script to loop over all of the ```png``` files in the current directory and run ImageJ on each file:\n\n\n```bash\n\nfor file in *.png\ndo\n java -jar $EBROOTIMAGEJ/ij.jar -batch ./macro.ijm ${file}\ndone\n```\n\nHowever, lets predent that the time to process each file is 12 minutes and 31 seconds. \n\n✅ **<font color=red>Question 5</font>**: (10 points) How long (in seconds) will it take to run all 300 files using this loop?",
"_____no_output_____"
]
],
[
[
"# put your answer to the above question here.",
"_____no_output_____"
]
],
[
[
"----\n\nIf you think about it, the order of the loop does not matter and would be really easy to run in parallel. This type of problem is called pleasantly parallel. The idea is we can just process a different file on a different computer using a SLURM job array. \n\n\nThis type of workflow is also often called \"unrolling a loop\". Lets assume that we want to unroll the above loop and run it as a job array on the cluster. First, you would need to add the following resource request to the top of your script:\n\n```bash\n#SBATCH --array=1-300\n```\n\nThis ```--array``` request will tell SLURM to run 300 identical jobs. The only difference will be each job will run on a different node on the cluster and will be given a unique array task ID number (the numbers 1-300) inside a bash variable called ```SLURM_ARRAY_TASK_ID```.\n\n✅ **<font color=red>Question 6</font>**: (20 points) Modify the following java command (which would be inside your job script) so that it will use the ```SLURM_ARRAY_TASK_ID``` environment variable to select a different file name for each file in the array. (instead of all jobs trying to use the ```input1.png``` file)",
"_____no_output_____"
],
[
"#### Modify this code\n\n java -jar $EBROOTIMAGEJ/ij.jar -batch ./macro.ijm intput1.png\n",
"_____no_output_____"
],
[
"✅ **<font color=red>Question 7</font>**: (10 points) What is the fastest possible speed (in seconds) we could run the same job using our job array? ",
"_____no_output_____"
],
[
"put your answer to the above question here. ",
"_____no_output_____"
],
[
"# Congradulations\n\nYou are done with your quiz. Please save the file and upload the jupyter notebook to the D2L dropbox. ",
"_____no_output_____"
],
[
"Written by Dr. Dirk Colbry, Michigan State University\n<a rel=\"license\" href=\"http://creativecommons.org/licenses/by-nc/4.0/\"><img alt=\"Creative Commons License\" style=\"border-width:0\" src=\"https://i.creativecommons.org/l/by-nc/4.0/88x31.png\" /></a><br />This work is licensed under a <a rel=\"license\" href=\"http://creativecommons.org/licenses/by-nc/4.0/\">Creative Commons Attribution-NonCommercial 4.0 International License</a>.",
"_____no_output_____"
],
[
"----",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ec8adc935a587807222602aa5704b3331b3666a9 | 7,775 | ipynb | Jupyter Notebook | notebooks/benchmark/.ipynb_checkpoints/speed_benchmark_datacase_count-checkpoint.ipynb | jirifilip/cba_rules | 59168ef6fb4c9e319475f9a7498446ba5ff306e1 | [
"MIT"
] | 19 | 2019-08-05T12:02:58.000Z | 2022-01-20T08:26:56.000Z | notebooks/benchmark/.ipynb_checkpoints/speed_benchmark_datacase_count-checkpoint.ipynb | viaduct-ai/pyARC | 858e77a83b83da3910ee36f1c46540a6c45988a5 | [
"MIT"
] | 25 | 2019-08-05T14:08:32.000Z | 2021-10-08T07:58:34.000Z | notebooks/benchmark/.ipynb_checkpoints/speed_benchmark_datacase_count-checkpoint.ipynb | viaduct-ai/pyARC | 858e77a83b83da3910ee36f1c46540a6c45988a5 | [
"MIT"
] | 25 | 2018-08-09T06:41:04.000Z | 2021-10-17T07:37:11.000Z | 28.068592 | 121 | 0.517814 | [
[
[
"%run ../../main.py\n%matplotlib inline",
"_____no_output_____"
],
[
"import pandas as pd\n\nfrom pyarc.algorithms import M1Algorithm, M2Algorithm, top_rules, createCARs \nfrom pyarc.data_structures import TransactionDB",
"_____no_output_____"
],
[
"#\n#\n# =========================\n# Oveření běhu v závislosti na vložených pravidlech / instancích\n# =========================\n#\n#\n#\n\nimport time\n\nrule_count = 100\n\nbenchmark_data = {\n \"input rows\": [],\n \"input rules\": [],\n \"output rules M1 pyARC\": [],\n \"output rules M1 pyARC unique\": [],\n \"output rules M2 pyARC\": [],\n \"time M1 pyARC\": [],\n \"time M1 pyARC unique\": [],\n \"time M2 pyARC\": []\n}\n\nstop_m2 = True\n\nnumber_of_iterations = 30\n\ndirectory = \"c:/code/python/machine_learning/assoc_rules\"\n\ndataset_name_benchmark = \"lymph0\"\n\npd_ds = pd.read_csv(\"c:/code/python/machine_learning/assoc_rules/train/{}.csv\".format(dataset_name_benchmark))\n\nfor i in range(11):\n dataset_name_benchmark = \"lymph0\"\n \n pd_ds = pd.concat([pd_ds, pd_ds])\n \n txns = TransactionDB.from_DataFrame(pd_ds, unique_transactions=True)\n txns_unique = TransactionDB.from_DataFrame(pd_ds, unique_transactions=False) \n \n rules = top_rules(txns.string_representation, appearance=txns.appeardict, target_rule_count=rule_count)\n\n cars = createCARs(rules)\n \n if len(cars) > rule_count:\n cars = cars[:rule_count] \n\n \n m1t1 = time.time()\n m1clf_len = []\n for _ in range(number_of_iterations):\n m1 = M1Algorithm(cars, txns)\n clf = m1.build()\n m1clf_len.append(len(clf.rules) + 1)\n \n m1t2 = time.time()\n \n \n \n m1t1_unique = time.time()\n m1clf_len_unique = []\n for _ in range(number_of_iterations):\n m1 = M1Algorithm(cars, txns_unique)\n clf = m1.build()\n m1clf_len_unique.append(len(clf.rules) + 1)\n \n m1t2_unique = time.time()\n \n \n \n if not stop_m2:\n m2t1 = time.time()\n m2clf_len = []\n for _ in range(number_of_iterations):\n m2 = M2Algorithm(cars, txns)\n clf = m2.build()\n m2clf_len.append(len(clf.rules) + 1)\n\n m2t2 = time.time()\n \n \n m1duration = (m1t2 - m1t1) / number_of_iterations\n m1duration_unique = (m1t2_unique - m1t1_unique) / number_of_iterations\n outputrules_m1 = sum(m1clf_len) / len(m1clf_len)\n outputrules_m1_unique = sum(m1clf_len_unique) / len(m1clf_len_unique)\n \n if not stop_m2:\n m2duration = (m2t2 - m2t1) / number_of_iterations\n outputrules_m2 = sum(m2clf_len) / len(m2clf_len)\n if m2duration > 0.5:\n stop_m2 = True\n \n benchmark_data[\"input rows\"].append(len(txns))\n benchmark_data[\"input rules\"].append(rule_count)\n benchmark_data[\"output rules M1 pyARC\"].append(outputrules_m1)\n benchmark_data[\"output rules M1 pyARC unique\"].append(outputrules_m1_unique)\n benchmark_data[\"output rules M2 pyARC\"].append(None if stop_m2 else outputrules_m2)\n benchmark_data[\"time M1 pyARC\"].append(m1duration)\n benchmark_data[\"time M1 pyARC unique\"].append(m1duration_unique)\n benchmark_data[\"time M2 pyARC\"].append(None if stop_m2 else m2duration)\n\n print(\"data_count:\", len(txns))\n print(\"M1 duration:\", m1duration)\n print(\"M1 unique duration\", m1duration_unique)\n print(\"M1 output rules\", outputrules_m1)\n if not stop_m2:\n print(\"M2 duration:\", m2duration)\n print(\"M2 output rules\", outputrules_m2)\n print(\"\\n\\n\")",
"_____no_output_____"
],
[
"#benchmark_data.pop(\"M2_duration\")\n\nbenchmark_df = pd.DataFrame(benchmark_data)\n\nbenchmark_df.plot(x=[\"input rows\"], y=[\"time M1 pyARC\", \"time M2 pyARC\"])\n\n#benchmark_df.to_csv(\"../data/data_sensitivity.csv\")",
"_____no_output_____"
],
[
"benchmark_df",
"_____no_output_____"
],
[
"R_benchmark = pd.read_csv(\"../data/arc-data-size.csv\")\n\nR_benchmark[[\"input rows\"]] = R_benchmark[[\"input rows\"]].astype(str)\nR_benchmark.set_index(\"input rows\", inplace=True)",
"_____no_output_____"
],
[
"R_benchmark.head()",
"_____no_output_____"
],
[
"benchmark_df[[\"input rows\"]] = benchmark_df[[\"input rows\"]].astype(str)\nbenchmark_df = benchmark_df.set_index(\"input rows\")",
"_____no_output_____"
],
[
"benchmark_all = benchmark_df.join(R_benchmark, lsuffix=\"_py\", rsuffix=\"_R\")\nbenchmark_all",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nlabels = [\"pyARC - m1\", \"pyARC - m2\", \"arc\", \"rCBA\", \"arulesCBA\"]\n\nax = benchmark_all.plot(y=[\"time M1 pyARC\", \"time M2 pyARC\", \"time_arc\", \"time_acba\", \"time_rcba\"])\nax.legend(labels)\n\nplt.savefig(\"../data/data_size_sensitivity_plot.png\")",
"_____no_output_____"
],
[
"benchmark_all.plot(y=[\"time M1 pyARC\", \"time M2 pyARC\", \"time_arc\", \"time_acba\", \"time_rcba\"])",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8adce6a0b4cafcc22473320c0affcdd7f54d05 | 124,532 | ipynb | Jupyter Notebook | example_for_colocated_gauge_combination.ipynb | cchwala/cologauge | 4258ce3e86d362e547b0e92e3653321c3c9ea44a | [
"BSD-3-Clause"
] | null | null | null | example_for_colocated_gauge_combination.ipynb | cchwala/cologauge | 4258ce3e86d362e547b0e92e3653321c3c9ea44a | [
"BSD-3-Clause"
] | null | null | null | example_for_colocated_gauge_combination.ipynb | cchwala/cologauge | 4258ce3e86d362e547b0e92e3653321c3c9ea44a | [
"BSD-3-Clause"
] | null | null | null | 348.829132 | 81,256 | 0.921675 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"%matplotlib widget",
"_____no_output_____"
],
[
"import pandas as pd\nimport glob\nimport matplotlib.pyplot as plt\n\nimport gauge_proc_tools\nimport gauge_vis_tools",
"_____no_output_____"
]
],
[
[
"# Load data from three colocated gauges. \n\nThese David tiping bucket rain gauges have been installed approx. 1 meter apart from each other.",
"_____no_output_____"
]
],
[
[
"df_raw = pd.read_csv('example_data.csv.gz', parse_dates=True, index_col=0)\ndf_raw",
"_____no_output_____"
]
],
[
[
"# Plot accumulation of raw rainfall data",
"_____no_output_____"
]
],
[
[
"df_raw.cumsum().plot();",
"_____no_output_____"
]
],
[
[
"# Do combination of the three gauge time series\n\nThis combination excludes periods where individual gauges seem to produce erroneous results and averages the data of the remaining gauges.",
"_____no_output_____"
]
],
[
[
"df_combined = gauge_proc_tools.combine_gauges(\n df_three_gauges_R_1_min=df_raw*60,\n hours_to_average_for_diff=24,\n max_allowed_relative_diff=0.3,\n min_R=0.5,\n hours_to_average_for_zeros=1\n)/60",
"_____no_output_____"
],
[
"df_combined",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(2, 1, figsize=(10, 6), sharex=True)\ngauge_vis_tools.plot_three_gauges(df_three_gauges=df_raw.resample('H').sum(), df_gauge_combo=df_combined.resample('H').sum(), ax=axs[0])\ngauge_vis_tools.plot_three_gauges(df_three_gauges=df_raw.resample('H').sum().cumsum(), df_gauge_combo=df_combined.resample('H').sum().cumsum(), ax=axs[1]);",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ec8ae5fea29422bf6e7f39d10a4c9679de9ee429 | 15,719 | ipynb | Jupyter Notebook | tutorials/RandomNumberGeneration/RandomNumberGenerationTutorial.ipynb | cfhirsch/QuantumKatas | dd531c8c4b9034ef1dfbb303a6efe1d42fe7f2cb | [
"MIT"
] | null | null | null | tutorials/RandomNumberGeneration/RandomNumberGenerationTutorial.ipynb | cfhirsch/QuantumKatas | dd531c8c4b9034ef1dfbb303a6efe1d42fe7f2cb | [
"MIT"
] | null | null | null | tutorials/RandomNumberGeneration/RandomNumberGenerationTutorial.ipynb | cfhirsch/QuantumKatas | dd531c8c4b9034ef1dfbb303a6efe1d42fe7f2cb | [
"MIT"
] | null | null | null | 35.563348 | 382 | 0.575227 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ec8af5b76f205d2e40945ac57f55b0df5fa975f0 | 29,520 | ipynb | Jupyter Notebook | assets/all_html/2019_10_15_python-web-crawling-example.ipynb | dskw1/dskw1.github.io | ee85aaa7c99c4320cfac95e26063beaac3ae6fcb | [
"MIT"
] | null | null | null | assets/all_html/2019_10_15_python-web-crawling-example.ipynb | dskw1/dskw1.github.io | ee85aaa7c99c4320cfac95e26063beaac3ae6fcb | [
"MIT"
] | 1 | 2022-03-24T18:28:16.000Z | 2022-03-24T18:28:16.000Z | assets/all_html/2019_10_15_python-web-crawling-example.ipynb | dskw1/dskw1.github.io | ee85aaa7c99c4320cfac95e26063beaac3ae6fcb | [
"MIT"
] | 1 | 2021-09-01T16:54:38.000Z | 2021-09-01T16:54:38.000Z | 35.738499 | 3,444 | 0.428388 | [
[
[
"### Python web crawling example",
"_____no_output_____"
],
[
"This tutorial is written based on chapters 11-13 from the book \"Python for Everyone\" https://www.py4e.com\n",
"_____no_output_____"
],
[
"Step 1: import all necessary packages",
"_____no_output_____"
]
],
[
[
"import re\nimport urllib\nfrom bs4 import BeautifulSoup\nimport pprint\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"Step 2: download a sample webpage. \nYou can save the html page onto your computer and use text editor to view its content",
"_____no_output_____"
]
],
[
[
"url = \"http://www.metrolyrics.com/you-belong-with-me-lyrics-taylor-swift.html\"\nhtml = urllib.request.urlopen(url).read()",
"_____no_output_____"
]
],
[
[
"Step 3: use BeautifulSoup to parse the webpage and extract the lyrics content. The division that includes the lyrics starts from the html tag \"lyrics-body-text\"",
"_____no_output_____"
]
],
[
[
"soup = BeautifulSoup(html, 'html.parser')\nprint(soup.title.string)\n\ntext = soup.body.find_all(id='lyrics-body-text')\ntext = text[0].text\nprint(text)",
"Taylor Swift - You Belong With Me Lyrics | MetroLyrics\n\n\nYou're on the phone with your girlfriend—she's upset\nShe's going off about something that you said\n'Cause she doesn't get your humor like I do.I'm in the room, it's a typical Tuesday night.\nI'm listening to the kind of music she doesn't like.\nAnd she'll never know your story like I doBut she wears short skirts\nI wear t-shirt\nShe's cheer captain\nAnd I'm on the bleachersDreaming about the day when you wake up and find\nThat what you're looking for has been here the whole time.\n\n\n\nRelated\n\n\n\n\n \n\n\n\n\n\n\n11 Delicious Misheard Lyrics About Food\n\n\n\n\n\n\n\nNEW SONG: Taylor Swift - 'Lover' - LYRICS\n\n\n\n\n\n\n\nPrime Day concert by Amazon Music will be headlined by Taylor Swift\n\n\n\n\n\n\n \n \n\nIf you could see\nThat I'm the one\nWho understands you\nBeen here all along\nSo, why can't you see\nYou belong with me\nYou belong with me?Walk in the streets with you in your worn out jeans\nI can't help thinking this is how it ought to be.\nLaughing on a park bench thinking to myself\n\"Hey, isn't this easy?\"And you've got a smile\nThat can light up this whole town\nI haven't seen it in awhile\nSince she brought you down.You say you're fine—I know you better than that\nHey, what you doing with a girl like that?\n\n\n\n\n\n\n\n \n\n\n\n\nShe wears high heels\nI wear sneakers\nShe's cheer captain\nAnd I'm on the bleachersDreaming about the day when you wake up and find\nThat what you're looking for has been here the whole timeIf you could see\nThat I'm the one\nWho understands you\nBeen here all along\nSo, why can't you see\nYou belong with me?Standing by and waiting at your backdoor.\nAll this time how could you not know, baby?\nYou belong with me\nYou belong with meOh, I remember you driving to my house\nIn the middle of the night\nI'm the one who makes you laugh\nWhen you know you're 'bout to cryI know your favorite songs\nAnd you tell me about your dreams\nThink I know where you belong\nThink I know it's with meCan't you se that I'm the one\nWho understands you?\nBeen here all along\nSo, why can't you see\nYou belong with me?Standing by and waiting at your backdoor.\nAll this time how could you not know, baby?\nYou belong with me\nYou belong with me\nYou belong with me\nHave you ever thought just maybe\nYou belong with me?\nYou belong with me\n\n\n\n\n \n\n\n\n"
]
],
[
[
"Step 4: split text into individual words",
"_____no_output_____"
]
],
[
[
"words = text.split()\nprint(words)",
"[\"You're\", 'on', 'the', 'phone', 'with', 'your', \"girlfriend—she's\", 'upset', \"She's\", 'going', 'off', 'about', 'something', 'that', 'you', 'said', \"'Cause\", 'she', \"doesn't\", 'get', 'your', 'humor', 'like', 'I', \"do.I'm\", 'in', 'the', 'room,', \"it's\", 'a', 'typical', 'Tuesday', 'night.', \"I'm\", 'listening', 'to', 'the', 'kind', 'of', 'music', 'she', \"doesn't\", 'like.', 'And', \"she'll\", 'never', 'know', 'your', 'story', 'like', 'I', 'doBut', 'she', 'wears', 'short', 'skirts', 'I', 'wear', 't-shirt', \"She's\", 'cheer', 'captain', 'And', \"I'm\", 'on', 'the', 'bleachersDreaming', 'about', 'the', 'day', 'when', 'you', 'wake', 'up', 'and', 'find', 'That', 'what', \"you're\", 'looking', 'for', 'has', 'been', 'here', 'the', 'whole', 'time.', 'Related', '11', 'Delicious', 'Misheard', 'Lyrics', 'About', 'Food', 'NEW', 'SONG:', 'Taylor', 'Swift', '-', \"'Lover'\", '-', 'LYRICS', 'Prime', 'Day', 'concert', 'by', 'Amazon', 'Music', 'will', 'be', 'headlined', 'by', 'Taylor', 'Swift', 'If', 'you', 'could', 'see', 'That', \"I'm\", 'the', 'one', 'Who', 'understands', 'you', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me?Walk', 'in', 'the', 'streets', 'with', 'you', 'in', 'your', 'worn', 'out', 'jeans', 'I', \"can't\", 'help', 'thinking', 'this', 'is', 'how', 'it', 'ought', 'to', 'be.', 'Laughing', 'on', 'a', 'park', 'bench', 'thinking', 'to', 'myself', '\"Hey,', \"isn't\", 'this', 'easy?\"And', \"you've\", 'got', 'a', 'smile', 'That', 'can', 'light', 'up', 'this', 'whole', 'town', 'I', \"haven't\", 'seen', 'it', 'in', 'awhile', 'Since', 'she', 'brought', 'you', 'down.You', 'say', \"you're\", 'fine—I', 'know', 'you', 'better', 'than', 'that', 'Hey,', 'what', 'you', 'doing', 'with', 'a', 'girl', 'like', 'that?', 'She', 'wears', 'high', 'heels', 'I', 'wear', 'sneakers', \"She's\", 'cheer', 'captain', 'And', \"I'm\", 'on', 'the', 'bleachersDreaming', 'about', 'the', 'day', 'when', 'you', 'wake', 'up', 'and', 'find', 'That', 'what', \"you're\", 'looking', 'for', 'has', 'been', 'here', 'the', 'whole', 'timeIf', 'you', 'could', 'see', 'That', \"I'm\", 'the', 'one', 'Who', 'understands', 'you', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me?Standing', 'by', 'and', 'waiting', 'at', 'your', 'backdoor.', 'All', 'this', 'time', 'how', 'could', 'you', 'not', 'know,', 'baby?', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'meOh,', 'I', 'remember', 'you', 'driving', 'to', 'my', 'house', 'In', 'the', 'middle', 'of', 'the', 'night', \"I'm\", 'the', 'one', 'who', 'makes', 'you', 'laugh', 'When', 'you', 'know', \"you're\", \"'bout\", 'to', 'cryI', 'know', 'your', 'favorite', 'songs', 'And', 'you', 'tell', 'me', 'about', 'your', 'dreams', 'Think', 'I', 'know', 'where', 'you', 'belong', 'Think', 'I', 'know', \"it's\", 'with', \"meCan't\", 'you', 'se', 'that', \"I'm\", 'the', 'one', 'Who', 'understands', 'you?', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me?Standing', 'by', 'and', 'waiting', 'at', 'your', 'backdoor.', 'All', 'this', 'time', 'how', 'could', 'you', 'not', 'know,', 'baby?', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me', 'Have', 'you', 'ever', 'thought', 'just', 'maybe', 'You', 'belong', 'with', 'me?', 'You', 'belong', 'with', 'me']\n"
]
],
[
[
"Remove stopwords",
"_____no_output_____"
]
],
[
[
"stopwords = ['is', 'are', 'the', 'a', 'an']\ndef removeStopwords(wordlist, stopwords):\n return [w for w in wordlist if w not in stopwords]\nwords = removeStopwords(words, stopwords)\nprint(words)",
"[\"You're\", 'on', 'phone', 'with', 'your', \"girlfriend—she's\", 'upset', \"She's\", 'going', 'off', 'about', 'something', 'that', 'you', 'said', \"'Cause\", 'she', \"doesn't\", 'get', 'your', 'humor', 'like', 'I', \"do.I'm\", 'in', 'room,', \"it's\", 'typical', 'Tuesday', 'night.', \"I'm\", 'listening', 'to', 'kind', 'of', 'music', 'she', \"doesn't\", 'like.', 'And', \"she'll\", 'never', 'know', 'your', 'story', 'like', 'I', 'doBut', 'she', 'wears', 'short', 'skirts', 'I', 'wear', 't-shirt', \"She's\", 'cheer', 'captain', 'And', \"I'm\", 'on', 'bleachersDreaming', 'about', 'day', 'when', 'you', 'wake', 'up', 'and', 'find', 'That', 'what', \"you're\", 'looking', 'for', 'has', 'been', 'here', 'whole', 'time.', 'Related', '11', 'Delicious', 'Misheard', 'Lyrics', 'About', 'Food', 'NEW', 'SONG:', 'Taylor', 'Swift', '-', \"'Lover'\", '-', 'LYRICS', 'Prime', 'Day', 'concert', 'by', 'Amazon', 'Music', 'will', 'be', 'headlined', 'by', 'Taylor', 'Swift', 'If', 'you', 'could', 'see', 'That', \"I'm\", 'one', 'Who', 'understands', 'you', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me?Walk', 'in', 'streets', 'with', 'you', 'in', 'your', 'worn', 'out', 'jeans', 'I', \"can't\", 'help', 'thinking', 'this', 'how', 'it', 'ought', 'to', 'be.', 'Laughing', 'on', 'park', 'bench', 'thinking', 'to', 'myself', '\"Hey,', \"isn't\", 'this', 'easy?\"And', \"you've\", 'got', 'smile', 'That', 'can', 'light', 'up', 'this', 'whole', 'town', 'I', \"haven't\", 'seen', 'it', 'in', 'awhile', 'Since', 'she', 'brought', 'you', 'down.You', 'say', \"you're\", 'fine—I', 'know', 'you', 'better', 'than', 'that', 'Hey,', 'what', 'you', 'doing', 'with', 'girl', 'like', 'that?', 'She', 'wears', 'high', 'heels', 'I', 'wear', 'sneakers', \"She's\", 'cheer', 'captain', 'And', \"I'm\", 'on', 'bleachersDreaming', 'about', 'day', 'when', 'you', 'wake', 'up', 'and', 'find', 'That', 'what', \"you're\", 'looking', 'for', 'has', 'been', 'here', 'whole', 'timeIf', 'you', 'could', 'see', 'That', \"I'm\", 'one', 'Who', 'understands', 'you', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me?Standing', 'by', 'and', 'waiting', 'at', 'your', 'backdoor.', 'All', 'this', 'time', 'how', 'could', 'you', 'not', 'know,', 'baby?', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'meOh,', 'I', 'remember', 'you', 'driving', 'to', 'my', 'house', 'In', 'middle', 'of', 'night', \"I'm\", 'one', 'who', 'makes', 'you', 'laugh', 'When', 'you', 'know', \"you're\", \"'bout\", 'to', 'cryI', 'know', 'your', 'favorite', 'songs', 'And', 'you', 'tell', 'me', 'about', 'your', 'dreams', 'Think', 'I', 'know', 'where', 'you', 'belong', 'Think', 'I', 'know', \"it's\", 'with', \"meCan't\", 'you', 'se', 'that', \"I'm\", 'one', 'Who', 'understands', 'you?', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me?Standing', 'by', 'and', 'waiting', 'at', 'your', 'backdoor.', 'All', 'this', 'time', 'how', 'could', 'you', 'not', 'know,', 'baby?', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me', 'Have', 'you', 'ever', 'thought', 'just', 'maybe', 'You', 'belong', 'with', 'me?', 'You', 'belong', 'with', 'me']\n"
]
],
[
[
"count word frequency",
"_____no_output_____"
]
],
[
[
"counts = dict()\nfor word in words:\n counts[word] = counts.get(word,0) + 1\nsorted(counts, key=counts.__getitem__, reverse=True)\npprint.pprint(counts)\n",
"{'\"Hey,': 1,\n \"'Cause\": 1,\n \"'Lover'\": 1,\n \"'bout\": 1,\n '-': 2,\n '11': 1,\n 'About': 1,\n 'All': 2,\n 'Amazon': 1,\n 'And': 4,\n 'Been': 3,\n 'Day': 1,\n 'Delicious': 1,\n 'Food': 1,\n 'Have': 1,\n 'Hey,': 1,\n 'I': 9,\n \"I'm\": 7,\n 'If': 1,\n 'In': 1,\n 'LYRICS': 1,\n 'Laughing': 1,\n 'Lyrics': 1,\n 'Misheard': 1,\n 'Music': 1,\n 'NEW': 1,\n 'Prime': 1,\n 'Related': 1,\n 'SONG:': 1,\n 'She': 1,\n \"She's\": 3,\n 'Since': 1,\n 'So,': 3,\n 'Swift': 2,\n 'Taylor': 2,\n 'That': 5,\n 'Think': 2,\n 'Tuesday': 1,\n 'When': 1,\n 'Who': 3,\n 'You': 11,\n \"You're\": 1,\n 'about': 4,\n 'all': 3,\n 'along': 3,\n 'and': 4,\n 'at': 2,\n 'awhile': 1,\n 'baby?': 2,\n 'backdoor.': 2,\n 'be': 1,\n 'be.': 1,\n 'been': 2,\n 'belong': 12,\n 'bench': 1,\n 'better': 1,\n 'bleachersDreaming': 2,\n 'brought': 1,\n 'by': 4,\n 'can': 1,\n \"can't\": 4,\n 'captain': 2,\n 'cheer': 2,\n 'concert': 1,\n 'could': 4,\n 'cryI': 1,\n 'day': 2,\n \"do.I'm\": 1,\n 'doBut': 1,\n \"doesn't\": 2,\n 'doing': 1,\n 'down.You': 1,\n 'dreams': 1,\n 'driving': 1,\n 'easy?\"And': 1,\n 'ever': 1,\n 'favorite': 1,\n 'find': 2,\n 'fine—I': 1,\n 'for': 2,\n 'get': 1,\n 'girl': 1,\n \"girlfriend—she's\": 1,\n 'going': 1,\n 'got': 1,\n 'has': 2,\n \"haven't\": 1,\n 'headlined': 1,\n 'heels': 1,\n 'help': 1,\n 'here': 5,\n 'high': 1,\n 'house': 1,\n 'how': 3,\n 'humor': 1,\n 'in': 4,\n \"isn't\": 1,\n 'it': 2,\n \"it's\": 2,\n 'jeans': 1,\n 'just': 1,\n 'kind': 1,\n 'know': 6,\n 'know,': 2,\n 'laugh': 1,\n 'light': 1,\n 'like': 3,\n 'like.': 1,\n 'listening': 1,\n 'looking': 2,\n 'makes': 1,\n 'maybe': 1,\n 'me': 7,\n 'me?': 1,\n 'me?Standing': 2,\n 'me?Walk': 1,\n \"meCan't\": 1,\n 'meOh,': 1,\n 'middle': 1,\n 'music': 1,\n 'my': 1,\n 'myself': 1,\n 'never': 1,\n 'night': 1,\n 'night.': 1,\n 'not': 2,\n 'of': 2,\n 'off': 1,\n 'on': 4,\n 'one': 4,\n 'ought': 1,\n 'out': 1,\n 'park': 1,\n 'phone': 1,\n 'remember': 1,\n 'room,': 1,\n 'said': 1,\n 'say': 1,\n 'se': 1,\n 'see': 5,\n 'seen': 1,\n 'she': 4,\n \"she'll\": 1,\n 'short': 1,\n 'skirts': 1,\n 'smile': 1,\n 'sneakers': 1,\n 'something': 1,\n 'songs': 1,\n 'story': 1,\n 'streets': 1,\n 't-shirt': 1,\n 'tell': 1,\n 'than': 1,\n 'that': 3,\n 'that?': 1,\n 'thinking': 2,\n 'this': 5,\n 'thought': 1,\n 'time': 2,\n 'time.': 1,\n 'timeIf': 1,\n 'to': 5,\n 'town': 1,\n 'typical': 1,\n 'understands': 3,\n 'up': 3,\n 'upset': 1,\n 'waiting': 2,\n 'wake': 2,\n 'wear': 2,\n 'wears': 2,\n 'what': 3,\n 'when': 2,\n 'where': 1,\n 'who': 1,\n 'whole': 3,\n 'why': 3,\n 'will': 1,\n 'with': 15,\n 'worn': 1,\n 'you': 23,\n \"you're\": 4,\n \"you've\": 1,\n 'you?': 1,\n 'your': 8}\n"
]
],
[
[
"sort words by frequency",
"_____no_output_____"
],
[
"The above method uses loop, which needs quite a lot of programming, and is also slow. \nThe following method uses the dataframe data structure in the pandas package to quickly count and sort words by frequencies. \nPandas documentation includes more details on its powerful data structure\nhttps://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.html",
"_____no_output_____"
]
],
[
[
"df=pd.DataFrame(words, columns=['word'])\nx=df[\"word\"].value_counts()\npprint.pprint(x)",
"you 23\nwith 15\nbelong 12\nYou 11\nI 9\n ..\nlistening 1\nshort 1\ndo.I'm 1\ntime. 1\njust 1\nName: word, Length: 186, dtype: int64\n"
],
[
"import nltk\ntokens = nltk.word_tokenize(text)\ntags = nltk.pos_tag(tokens)\nprint(tags[0][0], tags[0][1])",
"You PRP\n"
]
],
[
[
"### Remove Stopwords",
"_____no_output_____"
]
],
[
[
"def removeStopwords(wordlist, stopwords):\n return [w for w in wordlist if w not in stopwords]\nwords = removeStopwords(words, stopwords)\nprint(words)",
"[\"You're\", 'on', 'phone', 'with', 'your', \"girlfriend—she's\", 'upset', \"She's\", 'going', 'off', 'about', 'something', 'that', 'you', 'said', \"'Cause\", 'she', \"doesn't\", 'get', 'your', 'humor', 'like', 'I', \"do.I'm\", 'in', 'room,', \"it's\", 'typical', 'Tuesday', 'night.', \"I'm\", 'listening', 'to', 'kind', 'of', 'music', 'she', \"doesn't\", 'like.', 'And', \"she'll\", 'never', 'know', 'your', 'story', 'like', 'I', 'doBut', 'she', 'wears', 'short', 'skirts', 'I', 'wear', 't-shirt', \"She's\", 'cheer', 'captain', 'And', \"I'm\", 'on', 'bleachersDreaming', 'about', 'day', 'when', 'you', 'wake', 'up', 'and', 'find', 'That', 'what', \"you're\", 'looking', 'for', 'has', 'been', 'here', 'whole', 'time.', 'Related', '11', 'Delicious', 'Misheard', 'Lyrics', 'About', 'Food', 'NEW', 'SONG:', 'Taylor', 'Swift', '-', \"'Lover'\", '-', 'LYRICS', 'Prime', 'Day', 'concert', 'by', 'Amazon', 'Music', 'will', 'be', 'headlined', 'by', 'Taylor', 'Swift', 'If', 'you', 'could', 'see', 'That', \"I'm\", 'one', 'Who', 'understands', 'you', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me?Walk', 'in', 'streets', 'with', 'you', 'in', 'your', 'worn', 'out', 'jeans', 'I', \"can't\", 'help', 'thinking', 'this', 'how', 'it', 'ought', 'to', 'be.', 'Laughing', 'on', 'park', 'bench', 'thinking', 'to', 'myself', '\"Hey,', \"isn't\", 'this', 'easy?\"And', \"you've\", 'got', 'smile', 'That', 'can', 'light', 'up', 'this', 'whole', 'town', 'I', \"haven't\", 'seen', 'it', 'in', 'awhile', 'Since', 'she', 'brought', 'you', 'down.You', 'say', \"you're\", 'fine—I', 'know', 'you', 'better', 'than', 'that', 'Hey,', 'what', 'you', 'doing', 'with', 'girl', 'like', 'that?', 'She', 'wears', 'high', 'heels', 'I', 'wear', 'sneakers', \"She's\", 'cheer', 'captain', 'And', \"I'm\", 'on', 'bleachersDreaming', 'about', 'day', 'when', 'you', 'wake', 'up', 'and', 'find', 'That', 'what', \"you're\", 'looking', 'for', 'has', 'been', 'here', 'whole', 'timeIf', 'you', 'could', 'see', 'That', \"I'm\", 'one', 'Who', 'understands', 'you', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me?Standing', 'by', 'and', 'waiting', 'at', 'your', 'backdoor.', 'All', 'this', 'time', 'how', 'could', 'you', 'not', 'know,', 'baby?', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'meOh,', 'I', 'remember', 'you', 'driving', 'to', 'my', 'house', 'In', 'middle', 'of', 'night', \"I'm\", 'one', 'who', 'makes', 'you', 'laugh', 'When', 'you', 'know', \"you're\", \"'bout\", 'to', 'cryI', 'know', 'your', 'favorite', 'songs', 'And', 'you', 'tell', 'me', 'about', 'your', 'dreams', 'Think', 'I', 'know', 'where', 'you', 'belong', 'Think', 'I', 'know', \"it's\", 'with', \"meCan't\", 'you', 'se', 'that', \"I'm\", 'one', 'Who', 'understands', 'you?', 'Been', 'here', 'all', 'along', 'So,', 'why', \"can't\", 'you', 'see', 'You', 'belong', 'with', 'me?Standing', 'by', 'and', 'waiting', 'at', 'your', 'backdoor.', 'All', 'this', 'time', 'how', 'could', 'you', 'not', 'know,', 'baby?', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me', 'You', 'belong', 'with', 'me', 'Have', 'you', 'ever', 'thought', 'just', 'maybe', 'You', 'belong', 'with', 'me?', 'You', 'belong', 'with', 'me']\n"
],
[
"counts = dict()\nfor word in words:\n counts[word] = counts.get(word, 0) + 1\nsorted(counts, key=counts.__getitem__, reverse=True)\npprint.pprint(counts)",
"{'\"Hey,': 1,\n \"'Cause\": 1,\n \"'Lover'\": 1,\n \"'bout\": 1,\n '-': 2,\n '11': 1,\n 'About': 1,\n 'All': 2,\n 'Amazon': 1,\n 'And': 4,\n 'Been': 3,\n 'Day': 1,\n 'Delicious': 1,\n 'Food': 1,\n 'Have': 1,\n 'Hey,': 1,\n 'I': 9,\n \"I'm\": 7,\n 'If': 1,\n 'In': 1,\n 'LYRICS': 1,\n 'Laughing': 1,\n 'Lyrics': 1,\n 'Misheard': 1,\n 'Music': 1,\n 'NEW': 1,\n 'Prime': 1,\n 'Related': 1,\n 'SONG:': 1,\n 'She': 1,\n \"She's\": 3,\n 'Since': 1,\n 'So,': 3,\n 'Swift': 2,\n 'Taylor': 2,\n 'That': 5,\n 'Think': 2,\n 'Tuesday': 1,\n 'When': 1,\n 'Who': 3,\n 'You': 11,\n \"You're\": 1,\n 'about': 4,\n 'all': 3,\n 'along': 3,\n 'and': 4,\n 'at': 2,\n 'awhile': 1,\n 'baby?': 2,\n 'backdoor.': 2,\n 'be': 1,\n 'be.': 1,\n 'been': 2,\n 'belong': 12,\n 'bench': 1,\n 'better': 1,\n 'bleachersDreaming': 2,\n 'brought': 1,\n 'by': 4,\n 'can': 1,\n \"can't\": 4,\n 'captain': 2,\n 'cheer': 2,\n 'concert': 1,\n 'could': 4,\n 'cryI': 1,\n 'day': 2,\n \"do.I'm\": 1,\n 'doBut': 1,\n \"doesn't\": 2,\n 'doing': 1,\n 'down.You': 1,\n 'dreams': 1,\n 'driving': 1,\n 'easy?\"And': 1,\n 'ever': 1,\n 'favorite': 1,\n 'find': 2,\n 'fine—I': 1,\n 'for': 2,\n 'get': 1,\n 'girl': 1,\n \"girlfriend—she's\": 1,\n 'going': 1,\n 'got': 1,\n 'has': 2,\n \"haven't\": 1,\n 'headlined': 1,\n 'heels': 1,\n 'help': 1,\n 'here': 5,\n 'high': 1,\n 'house': 1,\n 'how': 3,\n 'humor': 1,\n 'in': 4,\n \"isn't\": 1,\n 'it': 2,\n \"it's\": 2,\n 'jeans': 1,\n 'just': 1,\n 'kind': 1,\n 'know': 6,\n 'know,': 2,\n 'laugh': 1,\n 'light': 1,\n 'like': 3,\n 'like.': 1,\n 'listening': 1,\n 'looking': 2,\n 'makes': 1,\n 'maybe': 1,\n 'me': 7,\n 'me?': 1,\n 'me?Standing': 2,\n 'me?Walk': 1,\n \"meCan't\": 1,\n 'meOh,': 1,\n 'middle': 1,\n 'music': 1,\n 'my': 1,\n 'myself': 1,\n 'never': 1,\n 'night': 1,\n 'night.': 1,\n 'not': 2,\n 'of': 2,\n 'off': 1,\n 'on': 4,\n 'one': 4,\n 'ought': 1,\n 'out': 1,\n 'park': 1,\n 'phone': 1,\n 'remember': 1,\n 'room,': 1,\n 'said': 1,\n 'say': 1,\n 'se': 1,\n 'see': 5,\n 'seen': 1,\n 'she': 4,\n \"she'll\": 1,\n 'short': 1,\n 'skirts': 1,\n 'smile': 1,\n 'sneakers': 1,\n 'something': 1,\n 'songs': 1,\n 'story': 1,\n 'streets': 1,\n 't-shirt': 1,\n 'tell': 1,\n 'than': 1,\n 'that': 3,\n 'that?': 1,\n 'thinking': 2,\n 'this': 5,\n 'thought': 1,\n 'time': 2,\n 'time.': 1,\n 'timeIf': 1,\n 'to': 5,\n 'town': 1,\n 'typical': 1,\n 'understands': 3,\n 'up': 3,\n 'upset': 1,\n 'waiting': 2,\n 'wake': 2,\n 'wear': 2,\n 'wears': 2,\n 'what': 3,\n 'when': 2,\n 'where': 1,\n 'who': 1,\n 'whole': 3,\n 'why': 3,\n 'will': 1,\n 'with': 15,\n 'worn': 1,\n 'you': 23,\n \"you're\": 4,\n \"you've\": 1,\n 'you?': 1,\n 'your': 8}\n"
],
[
"df = pd.DataFrame(words, columns=['word'])\nx = df[\"word\"].value_counts()\npprint.pprint(x)",
"you 23\nwith 15\nbelong 12\nYou 11\nI 9\n ..\nlistening 1\nshort 1\ndo.I'm 1\ntime. 1\njust 1\nName: word, Length: 186, dtype: int64\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ec8afb43c1bcd751a783bddfdb14e1f1adf64f8d | 7,735 | ipynb | Jupyter Notebook | module5/s1_language_detection.ipynb | valerianebxl/tac | ac0dc50fc34ff5719325c922a5251b2d604542a5 | [
"MIT"
] | null | null | null | module5/s1_language_detection.ipynb | valerianebxl/tac | ac0dc50fc34ff5719325c922a5251b2d604542a5 | [
"MIT"
] | null | null | null | module5/s1_language_detection.ipynb | valerianebxl/tac | ac0dc50fc34ff5719325c922a5251b2d604542a5 | [
"MIT"
] | null | null | null | 27.823741 | 120 | 0.554363 | [
[
[
"# La détection de langue",
"_____no_output_____"
],
[
"Nous utilisons ici la librairie langid:\n \nhttps://pypi.org/project/langid/",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import os\nfrom collections import defaultdict\n\nimport langid\nimport pycountry",
"_____no_output_____"
]
],
[
[
"## Forcer l'algorithme à ne détecter que du Français et du Néerlandais",
"_____no_output_____"
]
],
[
[
"langid.set_languages(['fr', 'nl'])",
"_____no_output_____"
]
],
[
[
"## Lister tous les documents",
"_____no_output_____"
]
],
[
[
"root = \"../data/txt/\"\ntxts = os.listdir(root)\nprint(f\"{len(txts)} TXT files found\")",
"2828 TXT files found\n"
]
],
[
[
"## Détecter la langue pour tous les documents",
"_____no_output_____"
],
[
"Nous allons lire chaque fichier, détecter la langue, et incrémenter `lang_dict` lorsqu'une langue est détectée.\n\n**Important** : pour détecter les langues sur tous les documents, mettez `limit = None` ci-dessous.",
"_____no_output_____"
]
],
[
[
"# limit = 500\nlimit = None",
"_____no_output_____"
],
[
"lang_dict = defaultdict(int)\ntxts = txts[:limit] if limit else txts",
"_____no_output_____"
],
[
"for i, txt in enumerate(sorted(txts)):\n if txt.endswith(\"txt\"):\n if i % 50 == 0:\n print(f'{i} document(s) processed...')\n text = open(os.path.join(root, txt)).read()\n text_length = len(text)\n if text_length > 20:\n lang, conf = langid.classify(text)\n lang_dict[lang] += 1\n else:\n print(f\"{txt} contains only {text_length} characters, treating as unknown\")\n lang_dict['n/a'] += 1\nprint(\"Done\")",
"0 document(s) processed...\n50 document(s) processed...\n100 document(s) processed...\n150 document(s) processed...\n200 document(s) processed...\nBxl_1869_Tome_I1_Part_4.txt contains only 4 characters, treating as unknown\n250 document(s) processed...\n300 document(s) processed...\n350 document(s) processed...\n400 document(s) processed...\n450 document(s) processed...\n500 document(s) processed...\n550 document(s) processed...\n600 document(s) processed...\n650 document(s) processed...\n700 document(s) processed...\n750 document(s) processed...\nBxl_1903_Tome_I2_2_Part_12.txt contains only 19 characters, treating as unknown\n800 document(s) processed...\n850 document(s) processed...\n900 document(s) processed...\n950 document(s) processed...\n1000 document(s) processed...\n1050 document(s) processed...\n1100 document(s) processed...\n1150 document(s) processed...\nBxl_1925_Tome_II1_2_Part_8.txt contains only 9 characters, treating as unknown\n1200 document(s) processed...\n1250 document(s) processed...\nBxl_1929_Tome_I_Part_10.txt contains only 1 characters, treating as unknown\n1300 document(s) processed...\n1350 document(s) processed...\n1400 document(s) processed...\n1450 document(s) processed...\nBxl_1946_Tome_II_Part_14.txt contains only 1 characters, treating as unknown\n1500 document(s) processed...\n1550 document(s) processed...\n1600 document(s) processed...\nBxl_1952_Tome_I_Part_9.txt contains only 2 characters, treating as unknown\n1650 document(s) processed...\n1700 document(s) processed...\n1750 document(s) processed...\nBxl_1957_Tome_II2_Part_10.txt contains only 9 characters, treating as unknown\n1800 document(s) processed...\nBxl_1957_Tome_I_Part_12.txt contains only 2 characters, treating as unknown\nBxl_1958_Tome_RptAn_Part_10.txt contains only 9 characters, treating as unknown\n1850 document(s) processed...\n1900 document(s) processed...\n1950 document(s) processed...\n2000 document(s) processed...\n2050 document(s) processed...\n2100 document(s) processed...\n2150 document(s) processed...\nBxl_1967_Tome_I1_Part_11.txt contains only 1 characters, treating as unknown\n2200 document(s) processed...\n2250 document(s) processed...\nBxl_1969_Tome_II2_Part_14.txt contains only 8 characters, treating as unknown\n2300 document(s) processed...\nBxl_1970_Tome_II1_Part_11.txt contains only 3 characters, treating as unknown\n2350 document(s) processed...\n2400 document(s) processed...\n2450 document(s) processed...\n2500 document(s) processed...\n2550 document(s) processed...\nBxl_1976_Tome_I2_Part_7.txt contains only 11 characters, treating as unknown\n2600 document(s) processed...\n2650 document(s) processed...\n2700 document(s) processed...\n2750 document(s) processed...\n2800 document(s) processed...\nDone\n"
]
],
[
[
"## Afficher le nombre de documents par langue",
"_____no_output_____"
]
],
[
[
"for lang_code, nb_docs in lang_dict.items():\n language = pycountry.languages.get(alpha_2=lang_code)\n try:\n lang_name = language.name\n except AttributeError:\n lang_name = language\n print(f\"{lang_name}\\t{nb_docs}\")",
"French\t2739\nNone\t13\nDutch\t76\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec8afc02d6d44bd9b7cf9653d878b78f96b29108 | 6,216 | ipynb | Jupyter Notebook | ass_day5.ipynb | varunchenna1234/ass1-2-3-day5-b7 | 0028723e82ce7ec88e170b798a46f4396f00ce84 | [
"Apache-2.0"
] | null | null | null | ass_day5.ipynb | varunchenna1234/ass1-2-3-day5-b7 | 0028723e82ce7ec88e170b798a46f4396f00ce84 | [
"Apache-2.0"
] | null | null | null | ass_day5.ipynb | varunchenna1234/ass1-2-3-day5-b7 | 0028723e82ce7ec88e170b798a46f4396f00ce84 | [
"Apache-2.0"
] | null | null | null | 36.564706 | 2,051 | 0.498874 | [
[
[
"<a href=\"https://colab.research.google.com/github/varunchenna1234/ass1-2-3-day5-b7/blob/master/ass_day5.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"Assignment1 day5 batch 7\n",
"_____no_output_____"
]
],
[
[
"list1=input(print(\"Enter the actual list \"))\nlist2=input(print(\"ENter the list to be searched \"))\ncheck = any(item in list1 for item in list2)\n \nif check is True: \n print(\"it's a Match\")\nelse :\n print(\"It's Gone\")",
"Enter the actual list \n1,2,34,5\nENter the list to be searched \n2,5\nit's a Match\n"
]
],
[
[
"Assignment 2 day5",
"_____no_output_____"
]
],
[
[
"def isPrime(x):\n for n in range(2,x):\n if x%n==0:\n break\n else:\n return True\n\nfltrObj=filter(isPrime, range(2,2500))\nif fltrObj==True:\n fltrObj.append(x)\nprint ('Prime numbers between 1-2500:', list(fltrObj))",
"Prime numbers between 1-2500: [2, 3, 5, 7, 11, 13, 17, 19, 23, 29, 31, 37, 41, 43, 47, 53, 59, 61, 67, 71, 73, 79, 83, 89, 97, 101, 103, 107, 109, 113, 127, 131, 137, 139, 149, 151, 157, 163, 167, 173, 179, 181, 191, 193, 197, 199, 211, 223, 227, 229, 233, 239, 241, 251, 257, 263, 269, 271, 277, 281, 283, 293, 307, 311, 313, 317, 331, 337, 347, 349, 353, 359, 367, 373, 379, 383, 389, 397, 401, 409, 419, 421, 431, 433, 439, 443, 449, 457, 461, 463, 467, 479, 487, 491, 499, 503, 509, 521, 523, 541, 547, 557, 563, 569, 571, 577, 587, 593, 599, 601, 607, 613, 617, 619, 631, 641, 643, 647, 653, 659, 661, 673, 677, 683, 691, 701, 709, 719, 727, 733, 739, 743, 751, 757, 761, 769, 773, 787, 797, 809, 811, 821, 823, 827, 829, 839, 853, 857, 859, 863, 877, 881, 883, 887, 907, 911, 919, 929, 937, 941, 947, 953, 967, 971, 977, 983, 991, 997, 1009, 1013, 1019, 1021, 1031, 1033, 1039, 1049, 1051, 1061, 1063, 1069, 1087, 1091, 1093, 1097, 1103, 1109, 1117, 1123, 1129, 1151, 1153, 1163, 1171, 1181, 1187, 1193, 1201, 1213, 1217, 1223, 1229, 1231, 1237, 1249, 1259, 1277, 1279, 1283, 1289, 1291, 1297, 1301, 1303, 1307, 1319, 1321, 1327, 1361, 1367, 1373, 1381, 1399, 1409, 1423, 1427, 1429, 1433, 1439, 1447, 1451, 1453, 1459, 1471, 1481, 1483, 1487, 1489, 1493, 1499, 1511, 1523, 1531, 1543, 1549, 1553, 1559, 1567, 1571, 1579, 1583, 1597, 1601, 1607, 1609, 1613, 1619, 1621, 1627, 1637, 1657, 1663, 1667, 1669, 1693, 1697, 1699, 1709, 1721, 1723, 1733, 1741, 1747, 1753, 1759, 1777, 1783, 1787, 1789, 1801, 1811, 1823, 1831, 1847, 1861, 1867, 1871, 1873, 1877, 1879, 1889, 1901, 1907, 1913, 1931, 1933, 1949, 1951, 1973, 1979, 1987, 1993, 1997, 1999, 2003, 2011, 2017, 2027, 2029, 2039, 2053, 2063, 2069, 2081, 2083, 2087, 2089, 2099, 2111, 2113, 2129, 2131, 2137, 2141, 2143, 2153, 2161, 2179, 2203, 2207, 2213, 2221, 2237, 2239, 2243, 2251, 2267, 2269, 2273, 2281, 2287, 2293, 2297, 2309, 2311, 2333, 2339, 2341, 2347, 2351, 2357, 2371, 2377, 2381, 2383, 2389, 2393, 2399, 2411, 2417, 2423, 2437, 2441, 2447, 2459, 2467, 2473, 2477]\n"
]
],
[
[
"assignment-3day5,batch7",
"_____no_output_____"
]
],
[
[
"lst=[]\ns=input(print('Enter the words in list to capitlise'))\nlst.append(s)\ncaps=list(map(lambda x: x.title() , lst)) \nprint(caps)\n",
"Enter the words in list to capitlise\nhey i am varun,i am a student\n['Hey I Am Varun,I Am A Student']\n"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec8b17c4a2e36d56f361732105d5e2257613102c | 4,889 | ipynb | Jupyter Notebook | metatlas/interfaces/compounds/not_used/Troubleshoot SMARTS patterns.ipynb | biorack/metatlas | ce3f03817a0a7c5500452689ad587ce94af26a1a | [
"BSD-3-Clause"
] | 8 | 2016-08-10T22:29:39.000Z | 2021-04-06T22:49:46.000Z | metatlas/interfaces/compounds/not_used/Troubleshoot SMARTS patterns.ipynb | biorack/metatlas | ce3f03817a0a7c5500452689ad587ce94af26a1a | [
"BSD-3-Clause"
] | 111 | 2016-04-27T23:18:12.000Z | 2022-03-15T23:55:52.000Z | metatlas/interfaces/compounds/not_used/Troubleshoot SMARTS patterns.ipynb | biorack/metatlas | ce3f03817a0a7c5500452689ad587ce94af26a1a | [
"BSD-3-Clause"
] | 9 | 2016-08-21T16:23:02.000Z | 2021-04-06T22:49:50.000Z | 44.445455 | 1,179 | 0.635917 | [
[
[
"import sys,os\nsys.path.insert(0,'/global/project/projectdirs/metatlas/anaconda/lib/python2.7/site-packages' )\ncurr_ld_lib_path = ''\nos.environ['LD_LIBRARY_PATH'] = curr_ld_lib_path + ':/project/projectdirs/openmsi/jupyterhub_libs/boost_1_55_0/lib' + ':/project/projectdirs/openmsi/jupyterhub_libs/lib'\nsys.path.append('/global/project/projectdirs/openmsi/jupyterhub_libs/anaconda/lib/python2.7/site-packages')\nsys.path.insert(0,'/project/projectdirs/openmsi/projects/meta-iq/pactolus/pactolus' )\n\nfrom rdkit import Chem\nfrom rdkit.Chem import Descriptors\nfrom rdkit.Chem import rdMolDescriptors\nfrom rdkit.Chem import AllChem\nfrom rdkit.Chem.Fingerprints import FingerprintMols\nfrom rdkit.Chem import Draw\nfrom rdkit.Chem.Draw import MolDrawing\nfrom collections import defaultdict\nfrom matplotlib import pyplot as plt\n%matplotlib inline\n\nfrom rdkit import DataStructs\nimport numpy as np\n",
"_____no_output_____"
],
[
"molecule = Chem.MolFromSmiles('C1COC=[N+]1(CCC)',True)\n\nfragment = Chem.MolFromSmarts('[R;C]=[D3;N][R:C]')\n\nMolDrawing.elemDict=defaultdict(lambda : (0,0,0))\nmatching = molecule.GetSubstructMatches(fragment)\nprint matching\nprint len(matching)\nif len(matching)>0:\n plt.figure(figsize=(20,8*len(matching)))\n for i,f in enumerate(matching):\n plt.subplot(len(matching),2,i+1)\n i1 = Draw.MolToImage(molecule,highlightAtoms=f,highlightColor=[0.6,0.6,1],)\n plt.imshow(i1)\n plt.axis('off')\nelse:\n plt.figure(figsize=(20,8))\n i1 = Draw.MolToImage(molecule)\n plt.imshow(i1)\n plt.axis('off')\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ec8b1c86f71f0dca963b4c9cd8d1ba13810aa546 | 1,827 | ipynb | Jupyter Notebook | notebooks/Models/Tasks.ipynb | axxeny/cmf_rateslib | efefe45bfc349a18a4b318f0d524744e0140e155 | [
"MIT"
] | 3 | 2021-11-12T16:14:29.000Z | 2021-12-08T17:44:35.000Z | notebooks/Models/Tasks.ipynb | axxeny/cmf_rateslib | efefe45bfc349a18a4b318f0d524744e0140e155 | [
"MIT"
] | null | null | null | notebooks/Models/Tasks.ipynb | axxeny/cmf_rateslib | efefe45bfc349a18a4b318f0d524744e0140e155 | [
"MIT"
] | 13 | 2021-11-09T17:53:51.000Z | 2021-12-13T11:19:12.000Z | 22.280488 | 71 | 0.527641 | [
[
[
"cmf_rateslib_root = '../../'\nimport sys\nif cmf_rateslib_root not in sys.path:\n sys.path.append(cmf_rateslib_root)",
"_____no_output_____"
],
[
"from cmf_rateslib.rates.simple_pca import SimplePCAModel",
"_____no_output_____"
]
],
[
[
"# Tasks for Rates Group\nModels to implement:\n1. Simple PCA curve generator\n2. Mean Revetign PCA \n3. Vasicek Short Rate model (evolutio of theta + risk premium)\n $$dr = ((\\theta + \\eta) - r)dt + \\sigma dW$$\n $$d\\eta = ...$$\n4. Libor Market Model\n\nModels must do:\n 1. Evolve a single curve (or generate new)\n 2. Generate 2 (or more?) correlated curves!!!\n 3 factors for curve 1\n 3 factors for curve 2\n factors are correlated pairwise\n\n 3. Fit model parameters give existing zero curves\n ",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code"
],
[
"markdown"
]
] |
ec8b1e456a9668c97ea274041556756dbcb45a89 | 32,777 | ipynb | Jupyter Notebook | session_1/Intro_to_python_script_session_1.ipynb | idlaviV/intro-to-python | c3bd8e4f65ce6f1159f769a3da0fcc841b9d6d22 | [
"MIT"
] | null | null | null | session_1/Intro_to_python_script_session_1.ipynb | idlaviV/intro-to-python | c3bd8e4f65ce6f1159f769a3da0fcc841b9d6d22 | [
"MIT"
] | null | null | null | session_1/Intro_to_python_script_session_1.ipynb | idlaviV/intro-to-python | c3bd8e4f65ce6f1159f769a3da0fcc841b9d6d22 | [
"MIT"
] | 1 | 2021-07-05T07:12:20.000Z | 2021-07-05T07:12:20.000Z | 23.942294 | 188 | 0.443665 | [
[
[
"# Introduction to Python - Session 1\n",
"_____no_output_____"
],
[
"## Section 1: Printing, Calculator, Variables & Basic types",
"_____no_output_____"
],
[
"### Strings & Printing",
"_____no_output_____"
]
],
[
[
"# Basic printing\nprint(\"Hello World!\")\n\n# strings can be seperated/added via the plus sign\nprint(\"My name is \" + \"Tilman!\")",
"_____no_output_____"
]
],
[
[
"**Escaping quotes** <br>\nA String is a sequence of characters, identified/between quotes: '', \"\".\nThe second quote to a first one marks the end of the string. If we want to use quotes in our string we have to escape them or use other quotes as string delimiters",
"_____no_output_____"
]
],
[
[
"print('My name is O\\'Brian!')\n\nprint(\"My name is O'Brian!\")\n\nprint(\"\\\"Wow!\\\", he said.\")\n\nprint('\"Wow!\", he said.')",
"_____no_output_____"
]
],
[
[
"### Basic calculator\nWe can perform any basic operation via the respective symbols, e.g:",
"_____no_output_____"
]
],
[
[
"2+2\n1-1\n100*13\n123/98\n# modulo is supported as well\n30%2",
"_____no_output_____"
]
],
[
[
"The above works, however does not print anything (in normal python scripts) in order to display the result, we have to print it:",
"_____no_output_____"
]
],
[
[
"print(2+2)\nprint(123/98)\nprint(30%2)",
"_____no_output_____"
]
],
[
[
"**PEMDAS** - Python follows PEMDAS (Punkt-vor-Strich):",
"_____no_output_____"
]
],
[
[
"print(2+4*2)\nprint((2+4)*2)",
"_____no_output_____"
],
[
"# Potencies work as well easily:\nprint(2**3)\nprint(5**2)",
"_____no_output_____"
]
],
[
[
"### Variables\nIf we want to store the result of our calculation or simply our name we can assign a value to a variable.\n\n**Choose meaningfull names!** (yt py)\n\n**Good practice / standard:** in python is 'snake case': <br>\nmy_name, growth_per_year",
"_____no_output_____"
]
],
[
[
"name = \"Tilman\"\nage = 22\nheight = 1.78",
"_____no_output_____"
]
],
[
[
"We can use variables in calculations, suppose I grew every year of my life the same amount:",
"_____no_output_____"
]
],
[
[
"growth_per_year = height * 100 / age\n\nprint(growth_per_year)",
"_____no_output_____"
],
[
"# if we want to print our growth per year combined with a string, like this:\n# My growth per year is: 8.1, we might think the following will work:\nprint(\"My growth per year is: \" + growth_per_year) ",
"_____no_output_____"
]
],
[
[
"### Types\nThere are different types in Python. The ones we focus on for now are: *integers, floats and strings*:\n\n#### Integers/int",
"_____no_output_____"
]
],
[
[
"persons = 10\nnumber_of_apples = 1203\n\n# if we want to update a value we can do this in two ways:\npersons = persons + 1\nprint(persons)\npersons += 1\nprint(persons)\n\n# this works for all operations:\npersons *= 2\npersons /= 10\npersons **= 2\npersons -= 1000\nprint(persons)",
"_____no_output_____"
],
[
"# to check the type of a variable use the type function:\nprint(type(persons))",
"_____no_output_____"
]
],
[
[
"#### Floats/float",
"_____no_output_____"
]
],
[
[
"# floats or float\nheight = 2.02\nprint(type(growth_per_year))",
"_____no_output_____"
]
],
[
[
"For floats we might want to round - this ca be done via the round function: <br>\nsyntax: `round(number, round_position)` where: <br>\nround_position: 0 round 1er, -1 round 10er, -2 round 100er, \n1 round 0.x, 2 round 0.0x:",
"_____no_output_____"
]
],
[
[
"print(round(height, 1))\n\nval = 126.471\nprint(round(val, 0))\nprint(round(val, -1))\nprint(round(val, -2))\nprint(round(val, 1))\nprint(round(val, 2))",
"_____no_output_____"
]
],
[
[
"#### Strings/str",
"_____no_output_____"
]
],
[
[
"name = \"Leonhard Euler\"\nprint(type(name))\n\nfour = \"4\"\nfour_word = \"four\"\njob = 'Mathematician'\nbio = \"\"\"was a Swiss mathematician, physicist, astronomer, geographer, \nlogician and engineer who who made important and influential discoveries in \nmany branches of mathematics, such as infinitesimal calculus and graph theory, \nwhile also making pioneering contributions to several branches such as topology \nand analytic number theory.\n\"\"\"\nsource_url = \"https://en.wikipedia.org/wiki/Leonhard_Euler\"\n\nprint(four)\nprint(type(four))\nprint(4)\nprint(type(4))",
"_____no_output_____"
]
],
[
[
"#### Convert values/types",
"_____no_output_____"
]
],
[
[
"float_age = float(age)\nint_height = int(height)\nfour_int = int(four)\n# int + float = float\nstr_growth_per_year = str(growth_per_year)",
"_____no_output_____"
],
[
"four_word_int = int(four_word) # error",
"_____no_output_____"
],
[
"# so now we can print our growth:\nprint(\"My growth per year is: \" + str_growth_per_year) \n\n# If we don't want to handle conversions of types for printing we can use \n# formatted strings: f\"... {variable} {another_variable}\":\nprint(f\"My name is {name}\")\nprint(f\"My growth per year is: {growth_per_year}\") ",
"_____no_output_____"
]
],
[
[
"#### Boolean",
"_____no_output_____"
]
],
[
[
"# A further data type is bool. There are two possible values:\nTrue\nFalse\n\n# when talking about if- else statements, they will be more important\n# for now we just want to present them\n\nhas_pets = True",
"_____no_output_____"
]
],
[
[
"### input()\nWe can ask the user for input via the input function:",
"_____no_output_____"
]
],
[
[
"name = input()\nprint(f\"Hi {name}!\")",
"_____no_output_____"
],
[
"# we can add a string/info to the prompt:\n# For better formatting we have to add a space or a new line (\\n)\nname = input(\"What is your name? \")\nprint(f\"Hi {name}!\")",
"_____no_output_____"
],
[
"# everthing that the input function returns is a string\n# so we may have to convert to the respective type\nage = int(input(\"How old are you? \\n> \"))\nprint(f\"{age} years old, that's impressive!\")",
"_____no_output_____"
]
],
[
[
"## Section 2: More Types: Lists",
"_____no_output_____"
],
[
"### List init\nLists are a collection of elements of any type within two `[]` brackets:",
"_____no_output_____"
]
],
[
[
"friends = [\"Ben\", \"Maria\", \"Charly\"]\nprint(type(friends))\n\nmeassures = [13, 91, 84, 3.1, 83.4, 65]\nprint(meassures)",
"_____no_output_____"
],
[
"# initialize empty list\nmy_fruits = []",
"_____no_output_____"
]
],
[
[
"### List Methods",
"_____no_output_____"
],
[
"#### .append() \nAdd new entries (used e.g. in dynamic creation of lists in for-loop -> later).\n\n**The first element of a list has the index `0`!**",
"_____no_output_____"
]
],
[
[
"my_fruits.append(\"apple\")\nprint(my_fruits)",
"['apple']\n"
],
[
"my_fruits.append(\"apple\")\nmy_fruits.append(\"apple\")\nmy_fruits.append(\"melon\")\nmy_fruits.append(\"peach\")\nmy_fruits.append(\"strawberry\")\nprint(my_fruits)",
"['apple', 'apple', 'apple', 'melon', 'peach', 'strawberry']\n"
]
],
[
[
"#### .extend()\nConcat/merge two lists:",
"_____no_output_____"
]
],
[
[
"new_fruits = [\"pineapple\", \"banana\", \"kiwi\", \"lemon\", \"kiwi\"]\n\n# my_fruits.append(new_fruits)\nplus_list = my_fruits + new_fruits\nprint(plus_list)\nmy_fruits.extend(new_fruits)\nprint(my_fruits)",
"_____no_output_____"
]
],
[
[
"#### .count() \nCount the number of occources of an element in an list",
"_____no_output_____"
]
],
[
[
"print(my_fruits.count(\"apple\"))",
"_____no_output_____"
]
],
[
[
"#### .pop()\nRemove & return the last element - lists needs to be of min len 1. ",
"_____no_output_____"
]
],
[
[
"last_el = my_fruits.pop()\nprint(last_el)\nprint(my_fruits)\n\nmy_fruits.pop()\nprint(my_fruits)",
"_____no_output_____"
],
[
"# alternatively we can also pop a specific element via giving the pop method \n# the index of the to-be-poped item:\n\nmy_fruits.pop(0)",
"_____no_output_____"
]
],
[
[
"#### .remove() \nRemove a specific element from a list:",
"_____no_output_____"
]
],
[
[
"my_fruits.remove(\"apple\")\nprint(my_fruits)",
"_____no_output_____"
],
[
"# when there are multiple identical elements, only the first element gets removed\nmy_fruits.remove(\"kiwi\")\nprint(my_fruits)",
"_____no_output_____"
]
],
[
[
"#### .sort()\nSort a list alpha-numerical.",
"_____no_output_____"
]
],
[
[
"my_fruits.sort()\nprint(my_fruits)\n\nmeassures.sort()\nprint(meassures)",
"_____no_output_____"
],
[
"new_list = meassures + my_fruits\nprint(new_list)\nnew_list.sort()\n\n# TypeError: '<' not supported between instances of 'int' and 'str'\n# conversion of all elements in lists can be achieved using map or list comprehension (later)",
"_____no_output_____"
]
],
[
[
"#### .reverse()\n\nReverses a list",
"_____no_output_____"
]
],
[
[
"my_fruits.reverse()\nprint(my_fruits)",
"_____no_output_____"
]
],
[
[
"#### sum()\nSumms up all values in a list (*all values have to be ints or floats*)!",
"_____no_output_____"
]
],
[
[
"sum_of_meassures = sum(meassures)\nprint(sum_of_meassures)\n\nprint(sum([1,2,3,4,5,6,12]))",
"_____no_output_____"
]
],
[
[
"#### .index()\nSometimes you don't know the index of an element, in this case we can use \nthe index() method. The element has to be in the list (not like in JS), else: <br>\n> ValueError: 'x' is not in list",
"_____no_output_____"
]
],
[
[
"print(my_fruits.index(\"lemon\"))",
"_____no_output_____"
],
[
"# if a list contains one fruit/entry multiple times, the index for the first\n# occources gets returned (as it's the case for remove())\nmy_fruits.append(\"lemon\")\nprint(my_fruits.index(\"lemon\"))",
"_____no_output_____"
]
],
[
[
"#### .clear()\nIf we want to remove all elements from our list we can use the clear method:\n",
"_____no_output_____"
]
],
[
[
"my_fruits.clear()\nprint(my_fruits)",
"_____no_output_____"
]
],
[
[
"### Cutting & Accesing via the index",
"_____no_output_____"
]
],
[
[
"# access the first element\nfirst = my_fruits[0] # -> returns first element",
"_____no_output_____"
],
[
"# we can update entries in a list using the index like this:\nmy_fruits[0] = \"blue-berry\"\nprint(my_fruits)",
"_____no_output_____"
],
[
"# you can cut simply using the ':' char\n# cut of first element / return everything after the first element\n# the my_fruits list stays the same!\nsliced_list = my_fruits[1:]\nprint(sliced_list)",
"_____no_output_____"
],
[
"# cut of everything after the second element\nsliced_list = my_fruits[:2]",
"_____no_output_____"
]
],
[
[
"So the syntax is `[from:to]` with the defaults: <br>\nfrom = 0, to = len(list)-1 (last element)",
"_____no_output_____"
]
],
[
[
"# get the length of a list:\nnum_of_fruits = len(my_fruits)\n\n# last index is however:\nprint(len(my_fruits) - 1)",
"_____no_output_____"
]
],
[
[
"### Multiplying lists\nyou can mulitply lists, basically repeating all elements *x times:",
"_____no_output_____"
]
],
[
[
"print([1,2]*4)",
"_____no_output_____"
]
],
[
[
"### Complex lists \nYou can also combine types and create for example an info list for everybody:",
"_____no_output_____"
]
],
[
[
"age = 45\nname = \"Alex\"\ninfo = [age, name, friends]\n\nfirst_friend = info[2][0]\n\ninfo2 = [12, \"Ben\", [\"Alex\", \"Sena\", \"Leo\"]]\n\nall_infos = [info, info2]\n\nsecond_friend_from_ben = all_infos[1][2][1]",
"_____no_output_____"
]
],
[
[
"A little messy -> for structured data & informations we can use dicts.",
"_____no_output_____"
],
[
"## Section 3 - Tuples & Dictionaries",
"_____no_output_____"
],
[
"### Tuples\nTuples are similar to lists - \nthe difference between lists and tuples ist that tuples are not changeable and values ar within (), they are used to store multiple items in a single variable.",
"_____no_output_____"
]
],
[
[
"coords = (2, 4, 2)\nprint(type(coords))",
"_____no_output_____"
],
[
"# accesing and indicies work the identical to lists\nprint(coords[0])",
"_____no_output_____"
]
],
[
[
"The only methods tuples have are the index and count method we talked about for lists. They work identical:",
"_____no_output_____"
]
],
[
[
"print(coords.count(2))\n\n# again, .index() only shows the index of the first found element\nprint(coords.index(2))",
"_____no_output_____"
]
],
[
[
"Identical to lists we can also multiply tuples:",
"_____no_output_____"
]
],
[
[
"print(coords * 2)",
"_____no_output_____"
]
],
[
[
"### Dicts\nA dict is a associative list:<br>\nin a list we can access elements via the index ([1]), in a dict we have key:value pairs and access elemts/values using a defined key. <br>\n**One important thing: dicts do not allow duplicates!** <br>\nSyntax is as follows:",
"_____no_output_____"
]
],
[
[
"personal_info = {\"name\" : \"Tilman\"}\nprint(type(personal_info))\nprint(personal_info)",
"_____no_output_____"
]
],
[
[
"For dicts wich hold more than one key-value pair the following is cleaner and clearer. <br>\nOf course we can aso use variables in dicts:",
"_____no_output_____"
]
],
[
[
"my_job = \"student\"\n\npersonal_info = {\n \"name\": \"Tilman\",\n \"job\": my_job,\n \"height\": 1.78,\n}\n\nprint(personal_info)",
"_____no_output_____"
],
[
"# often lists of dictionaires are used, where every list entry represents one data point\n# e.g.: web scraping\npersons = [\n personal_info,\n {\n \"name\": \"Charly\",\n \"job\": \"Teacher\",\n \"height\": 1.68\n }\n]\n\nprint(persons)",
"_____no_output_____"
]
],
[
[
"> For naming the keys it is common to use snake_case with all lower chars (as for variables)",
"_____no_output_____"
],
[
"#### .update()\n\nUpdate a dict: <br>\nif we want to update (add new key:value paires / update existing) a dict, we can use the update method. if the key we specify already exists, the method updates, else it appends.",
"_____no_output_____"
]
],
[
[
"personal_info.update({\n \"height\": 1.69\n})\nprint(personal_info)",
"_____no_output_____"
],
[
"personal_info.update({\n \"has_pets\": True\n})\nprint(personal_info)",
"_____no_output_____"
],
[
"personal_info.update({\n \"has_pets\": False,\n \"job\": \"Taxi Driver\"\n})\n\nprint(personal_info)",
"_____no_output_____"
],
[
"# pay attention if you used a variable -> change the values here \n# -> updated everywhere\nprint(persons)",
"_____no_output_____"
]
],
[
[
"#### .keys()\nSometimes you may need all existing keys for a dict, you can get those via the keys method:",
"_____no_output_____"
]
],
[
[
"print(personal_info.keys())",
"_____no_output_____"
]
],
[
[
"#### Update & Access via index/key\nAnother way to update (similar to lists)",
"_____no_output_____"
]
],
[
[
"personal_info[\"height\"] = 1.70",
"_____no_output_____"
],
[
"# accessing values works similar:\nmy_height = personal_info[\"height\"]\nprint(f\"My height is {personal_info['height']}\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ec8b2264df8ed671f2324049481e5ed546b2252a | 107,325 | ipynb | Jupyter Notebook | doc/LectureNotes/_build/jupyter_execute/boltzmannmachines.ipynb | CompPhysics/ComputationalPhysics2 | 6caaa2a61a985b636f869ab11045f505fdaa95e9 | [
"CC0-1.0"
] | 87 | 2015-01-21T08:29:56.000Z | 2022-03-28T07:11:53.000Z | doc/LectureNotes/_build/jupyter_execute/boltzmannmachines.ipynb | hyc9527/ComputationalPhysics2 | 6caaa2a61a985b636f869ab11045f505fdaa95e9 | [
"CC0-1.0"
] | 3 | 2020-01-18T10:43:38.000Z | 2020-02-08T13:15:42.000Z | doc/LectureNotes/_build/jupyter_execute/boltzmannmachines.ipynb | hyc9527/ComputationalPhysics2 | 6caaa2a61a985b636f869ab11045f505fdaa95e9 | [
"CC0-1.0"
] | 54 | 2015-02-09T10:02:00.000Z | 2022-03-07T10:44:14.000Z | 31.640625 | 718 | 0.519394 | [
[
[
"# Boltzmann Machines\n\nWhy use a generative model rather than the more well known discriminative deep neural networks (DNN)? \n\n* Discriminitave methods have several limitations: They are mainly supervised learning methods, thus requiring labeled data. And there are tasks they cannot accomplish, like drawing new examples from an unknown probability distribution.\n\n* A generative model can learn to represent and sample from a probability distribution. The core idea is to learn a parametric model of the probability distribution from which the training data was drawn. As an example\n\na. A model for images could learn to draw new examples of cats and dogs, given a training dataset of images of cats and dogs.\n\nb. Generate a sample of an ordered or disordered Ising model phase, having been given samples of such phases.\n\nc. Model the trial function for Monte Carlo calculations\n\n\n4. Both use gradient-descent based learning procedures for minimizing cost functions\n\n5. Energy based models don't use backpropagation and automatic differentiation for computing gradients, instead turning to Markov Chain Monte Carlo methods.\n\n6. DNNs often have several hidden layers. A restricted Boltzmann machine has only one hidden layer, however several RBMs can be stacked to make up Deep Belief Networks, of which they constitute the building blocks.\n\nHistory: The RBM was developed by amongst others Geoffrey Hinton, called by some the \"Godfather of Deep Learning\", working with the University of Toronto and Google.\n\n\n\nA BM is what we would call an undirected probabilistic graphical model\nwith stochastic continuous or discrete units.\n\n\nIt is interpreted as a stochastic recurrent neural network where the\nstate of each unit(neurons/nodes) depends on the units it is connected\nto. The weights in the network represent thus the strength of the\ninteraction between various units/nodes.\n\n\nIt turns into a Hopfield network if we choose deterministic rather\nthan stochastic units. In contrast to a Hopfield network, a BM is a\nso-called generative model. It allows us to generate new samples from\nthe learned distribution.\n\n\n\nA standard BM network is divided into a set of observable and visible units $\\hat{x}$ and a set of unknown hidden units/nodes $\\hat{h}$.\n\n\n\nAdditionally there can be bias nodes for the hidden and visible layers. These biases are normally set to $1$.\n\n\n\nBMs are stackable, meaning they cwe can train a BM which serves as input to another BM. We can construct deep networks for learning complex PDFs. The layers can be trained one after another, a feature which makes them popular in deep learning\n\n\n\nHowever, they are often hard to train. This leads to the introduction of so-called restricted BMs, or RBMS.\nHere we take away all lateral connections between nodes in the visible layer as well as connections between nodes in the hidden layer. The network is illustrated in the figure below.\n\n<!-- dom:FIGURE: [figures/RBM.png, width=800 frac=1.0] -->\n<!-- begin figure -->\n<img src=\"figures/RBM.png\" width=800><p style=\"font-size: 0.9em\"><i>Figure 1: </i></p><!-- end figure -->\n\n\n\n\n\n## The network\n\n**The network layers**:\n1. A function $\\mathbf{x}$ that represents the visible layer, a vector of $M$ elements (nodes). This layer represents both what the RBM might be given as training input, and what we want it to be able to reconstruct. This might for example be the pixels of an image, the spin values of the Ising model, or coefficients representing speech.\n\n2. The function $\\mathbf{h}$ represents the hidden, or latent, layer. A vector of $N$ elements (nodes). Also called \"feature detectors\".\n\nThe goal of the hidden layer is to increase the model's expressive power. We encode complex interactions between visible variables by introducing additional, hidden variables that interact with visible degrees of freedom in a simple manner, yet still reproduce the complex correlations between visible degrees in the data once marginalized over (integrated out).\n\nExamples of this trick being employed in physics: \n1. The Hubbard-Stratonovich transformation\n\n2. The introduction of ghost fields in gauge theory\n\n3. Shadow wave functions in Quantum Monte Carlo simulations\n\n**The network parameters, to be optimized/learned**:\n1. $\\mathbf{a}$ represents the visible bias, a vector of same length as $\\mathbf{x}$.\n\n2. $\\mathbf{b}$ represents the hidden bias, a vector of same lenght as $\\mathbf{h}$.\n\n3. $W$ represents the interaction weights, a matrix of size $M\\times N$.\n\n### Joint distribution\n\nThe restricted Boltzmann machine is described by a Bolztmann distribution",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto1\"></div>\n\n$$\n\\begin{equation}\n\tP_{rbm}(\\mathbf{x},\\mathbf{h}) = \\frac{1}{Z} e^{-\\frac{1}{T_0}E(\\mathbf{x},\\mathbf{h})},\n\\label{_auto1} \\tag{1}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where $Z$ is the normalization constant or partition function, defined as",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto2\"></div>\n\n$$\n\\begin{equation}\n\tZ = \\int \\int e^{-\\frac{1}{T_0}E(\\mathbf{x},\\mathbf{h})} d\\mathbf{x} d\\mathbf{h}.\n\\label{_auto2} \\tag{2}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"It is common to ignore $T_0$ by setting it to one. \n\n\n### Network Elements, the energy function\n\nThe function $E(\\mathbf{x},\\mathbf{h})$ gives the **energy** of a\nconfiguration (pair of vectors) $(\\mathbf{x}, \\mathbf{h})$. The lower\nthe energy of a configuration, the higher the probability of it. This\nfunction also depends on the parameters $\\mathbf{a}$, $\\mathbf{b}$ and\n$W$. Thus, when we adjust them during the learning procedure, we are\nadjusting the energy function to best fit our problem.\n\n\n\n### Defining different types of RBMs\n\nThere are different variants of RBMs, and the differences lie in the types of visible and hidden units we choose as well as in the implementation of the energy function $E(\\mathbf{x},\\mathbf{h})$. The connection between the nodes in the two layers is given by the weights $w_{ij}$. \n\n**Binary-Binary RBM:**\n\n\nRBMs were first developed using binary units in both the visible and hidden layer. The corresponding energy function is defined as follows:",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto3\"></div>\n\n$$\n\\begin{equation}\n\tE(\\mathbf{x}, \\mathbf{h}) = - \\sum_i^M x_i a_i- \\sum_j^N b_j h_j - \\sum_{i,j}^{M,N} x_i w_{ij} h_j,\n\\label{_auto3} \\tag{3}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where the binary values taken on by the nodes are most commonly 0 and 1.\n\n\n**Gaussian-Binary RBM:**\n\n\nAnother varient is the RBM where the visible units are Gaussian while the hidden units remain binary:",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto4\"></div>\n\n$$\n\\begin{equation}\n\tE(\\mathbf{x}, \\mathbf{h}) = \\sum_i^M \\frac{(x_i - a_i)^2}{2\\sigma_i^2} - \\sum_j^N b_j h_j - \\sum_{i,j}^{M,N} \\frac{x_i w_{ij} h_j}{\\sigma_i^2}. \n\\label{_auto4} \\tag{4}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"1. RBMs are Useful when we model continuous data (i.e., we wish $\\mathbf{x}$ to be continuous)\n\n2. Requires a smaller learning rate, since there's no upper bound to the value a component might take in the reconstruction\n\nOther types of units include:\n1. Softmax and multinomial units\n\n2. Gaussian visible and hidden units\n\n3. Binomial units\n\n4. Rectified linear units\n\n### Cost function\n\nWhen working with a training dataset, the most common training approach is maximizing the log-likelihood of the training data. The log likelihood characterizes the log-probability of generating the observed data using our generative model. Using this method our cost function is chosen as the negative log-likelihood. The learning then consists of trying to find parameters that maximize the probability of the dataset, and is known as Maximum Likelihood Estimation (MLE).\nDenoting the parameters as $\\boldsymbol{\\theta} = a_1,...,a_M,b_1,...,b_N,w_{11},...,w_{MN}$, the log-likelihood is given by",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto5\"></div>\n\n$$\n\\begin{equation}\n\t\\mathcal{L}(\\{ \\theta_i \\}) = \\langle \\text{log} P_\\theta(\\boldsymbol{x}) \\rangle_{data} \n\\label{_auto5} \\tag{5}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto6\"></div>\n\n$$\n\\begin{equation} \n\t= - \\langle E(\\boldsymbol{x}; \\{ \\theta_i\\}) \\rangle_{data} - \\text{log} Z(\\{ \\theta_i\\}),\n\\label{_auto6} \\tag{6}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where we used that the normalization constant does not depend on the data, $\\langle \\text{log} Z(\\{ \\theta_i\\}) \\rangle = \\text{log} Z(\\{ \\theta_i\\})$\nOur cost function is the negative log-likelihood, $\\mathcal{C}(\\{ \\theta_i \\}) = - \\mathcal{L}(\\{ \\theta_i \\})$\n\n### Optimization / Training\n\nThe training procedure of choice often is Stochastic Gradient Descent (SGD). It consists of a series of iterations where we update the parameters according to the equation",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto7\"></div>\n\n$$\n\\begin{equation}\n\t\\boldsymbol{\\theta}_{k+1} = \\boldsymbol{\\theta}_k - \\eta \\nabla \\mathcal{C} (\\boldsymbol{\\theta}_k)\n\\label{_auto7} \\tag{7}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"at each $k$-th iteration. There are a range of variants of the algorithm which aim at making the learning rate $\\eta$ more adaptive so the method might be more efficient while remaining stable.\n\nWe now need the gradient of the cost function in order to minimize it. We find that",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto8\"></div>\n\n$$\n\\begin{equation}\n\t\\frac{\\partial \\mathcal{C}(\\{ \\theta_i\\})}{\\partial \\theta_i}\n\t= \\langle \\frac{\\partial E(\\boldsymbol{x}; \\theta_i)}{\\partial \\theta_i} \\rangle_{data}\n\t+ \\frac{\\partial \\text{log} Z(\\{ \\theta_i\\})}{\\partial \\theta_i} \n\\label{_auto8} \\tag{8}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto9\"></div>\n\n$$\n\\begin{equation} \n\t= \\langle O_i(\\boldsymbol{x}) \\rangle_{data} - \\langle O_i(\\boldsymbol{x}) \\rangle_{model},\n\\label{_auto9} \\tag{9}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where in order to simplify notation we defined the \"operator\"",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto10\"></div>\n\n$$\n\\begin{equation}\n\tO_i(\\boldsymbol{x}) = \\frac{\\partial E(\\boldsymbol{x}; \\theta_i)}{\\partial \\theta_i}, \n\\label{_auto10} \\tag{10}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"and used the statistical mechanics relationship between expectation values and the log-partition function:",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto11\"></div>\n\n$$\n\\begin{equation}\n\t\\langle O_i(\\boldsymbol{x}) \\rangle_{model} = \\text{Tr} P_\\theta(\\boldsymbol{x})O_i(\\boldsymbol{x}) = - \\frac{\\partial \\text{log} Z(\\{ \\theta_i\\})}{\\partial \\theta_i}.\n\\label{_auto11} \\tag{11}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The data-dependent term in the gradient is known as the positive phase\nof the gradient, while the model-dependent term is known as the\nnegative phase of the gradient. The aim of the training is to lower\nthe energy of configurations that are near observed data points\n(increasing their probability), and raising the energy of\nconfigurations that are far from observed data points (decreasing\ntheir probability).\n\nThe gradient of the negative log-likelihood cost function of a Binary-Binary RBM is then",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto12\"></div>\n\n$$\n\\begin{equation}\n\t\\frac{\\partial \\mathcal{C} (w_{ij}, a_i, b_j)}{\\partial w_{ij}} = \\langle x_i h_j \\rangle_{data} - \\langle x_i h_j \\rangle_{model} \n\\label{_auto12} \\tag{12}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto13\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial \\mathcal{C} (w_{ij}, a_i, b_j)}{\\partial a_{ij}} = \\langle x_i \\rangle_{data} - \\langle x_i \\rangle_{model} \n\\label{_auto13} \\tag{13}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto14\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial \\mathcal{C} (w_{ij}, a_i, b_j)}{\\partial b_{ij}} = \\langle h_i \\rangle_{data} - \\langle h_i \\rangle_{model}. \n\\label{_auto14} \\tag{14}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto15\"></div>\n\n$$\n\\begin{equation} \n\\label{_auto15} \\tag{15}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"To get the expectation values with respect to the *data*, we set the visible units to each of the observed samples in the training data, then update the hidden units according to the conditional probability found before. We then average over all samples in the training data to calculate expectation values with respect to the data. \n\n\n\n\n### Kullback-Leibler relative entropy\n\nWhen the goal of the training is to approximate a probability\ndistribution, as it is in generative modeling, another relevant\nmeasure is the **Kullback-Leibler divergence**, also known as the\nrelative entropy or Shannon entropy. It is a non-symmetric measure of the\ndissimilarity between two probability density functions $p$ and\n$q$. If $p$ is the unkown probability which we approximate with $q$,\nwe can measure the difference by",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto16\"></div>\n\n$$\n\\begin{equation}\n\t\\text{KL}(p||q) = \\int_{-\\infty}^{\\infty} p (\\boldsymbol{x}) \\log \\frac{p(\\boldsymbol{x})}{q(\\boldsymbol{x})} d\\boldsymbol{x}.\n\\label{_auto16} \\tag{16}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Thus, the Kullback-Leibler divergence between the distribution of the\ntraining data $f(\\boldsymbol{x})$ and the model distribution $p(\\boldsymbol{x}|\n\\boldsymbol{\\theta})$ is",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto17\"></div>\n\n$$\n\\begin{equation}\n\t\\text{KL} (f(\\boldsymbol{x})|| p(\\boldsymbol{x}| \\boldsymbol{\\theta})) = \\int_{-\\infty}^{\\infty}\n\tf (\\boldsymbol{x}) \\log \\frac{f(\\boldsymbol{x})}{p(\\boldsymbol{x}| \\boldsymbol{\\theta})} d\\boldsymbol{x} \n\\label{_auto17} \\tag{17}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto18\"></div>\n\n$$\n\\begin{equation} \n\t= \\int_{-\\infty}^{\\infty} f(\\boldsymbol{x}) \\log f(\\boldsymbol{x}) d\\boldsymbol{x} - \\int_{-\\infty}^{\\infty} f(\\boldsymbol{x}) \\log\n\tp(\\boldsymbol{x}| \\boldsymbol{\\theta}) d\\boldsymbol{x} \n\\label{_auto18} \\tag{18}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto19\"></div>\n\n$$\n\\begin{equation} \n\t%= \\mathbb{E}_{f(\\boldsymbol{x})} (\\log f(\\boldsymbol{x})) - \\mathbb{E}_{f(\\boldsymbol{x})} (\\log p(\\boldsymbol{x}| \\boldsymbol{\\theta}))\n\t= \\langle \\log f(\\boldsymbol{x}) \\rangle_{f(\\boldsymbol{x})} - \\langle \\log p(\\boldsymbol{x}| \\boldsymbol{\\theta}) \\rangle_{f(\\boldsymbol{x})} \n\\label{_auto19} \\tag{19}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto20\"></div>\n\n$$\n\\begin{equation} \n\t= \\langle \\log f(\\boldsymbol{x}) \\rangle_{data} + \\langle E(\\boldsymbol{x}) \\rangle_{data} + \\log Z \n\\label{_auto20} \\tag{20}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto21\"></div>\n\n$$\n\\begin{equation} \n\t= \\langle \\log f(\\boldsymbol{x}) \\rangle_{data} + \\mathcal{C}_{LL} .\n\\label{_auto21} \\tag{21}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The first term is constant with respect to $\\boldsymbol{\\theta}$ since $f(\\boldsymbol{x})$ is independent of $\\boldsymbol{\\theta}$. Thus the Kullback-Leibler Divergence is minimal when the second term is minimal. The second term is the log-likelihood cost function, hence minimizing the Kullback-Leibler divergence is equivalent to maximizing the log-likelihood.\n\n\nTo further understand generative models it is useful to study the\ngradient of the cost function which is needed in order to minimize it\nusing methods like stochastic gradient descent. \n\nThe partition function is the generating function of\nexpectation values, in particular there are mathematical relationships\nbetween expectation values and the log-partition function. In this\ncase we have",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto22\"></div>\n\n$$\n\\begin{equation}\n\t\\langle \\frac{ \\partial E(\\boldsymbol{x}; \\theta_i) } { \\partial \\theta_i} \\rangle_{model}\n\t= \\int p(\\boldsymbol{x}| \\boldsymbol{\\theta}) \\frac{ \\partial E(\\boldsymbol{x}; \\theta_i) } { \\partial \\theta_i} d\\boldsymbol{x} \n\t= -\\frac{\\partial \\log Z(\\theta_i)}{ \\partial \\theta_i} .\n\\label{_auto22} \\tag{22}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Here $\\langle \\cdot \\rangle_{model}$ is the expectation value over the model probability distribution $p(\\boldsymbol{x}| \\boldsymbol{\\theta})$.\n\n## Setting up for gradient descent calculations\n\nUsing the previous relationship we can express the gradient of the cost function as",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto23\"></div>\n\n$$\n\\begin{equation}\n\t\\frac{\\partial \\mathcal{C}_{LL}}{\\partial \\theta_i}\n\t= \\langle \\frac{ \\partial E(\\boldsymbol{x}; \\theta_i) } { \\partial \\theta_i} \\rangle_{data} + \\frac{\\partial \\log Z(\\theta_i)}{ \\partial \\theta_i} \n\\label{_auto23} \\tag{23}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto24\"></div>\n\n$$\n\\begin{equation} \n\t= \\langle \\frac{ \\partial E(\\boldsymbol{x}; \\theta_i) } { \\partial \\theta_i} \\rangle_{data} - \\langle \\frac{ \\partial E(\\boldsymbol{x}; \\theta_i) } { \\partial \\theta_i} \\rangle_{model} \n\\label{_auto24} \\tag{24}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto25\"></div>\n\n$$\n\\begin{equation} \n\t%= \\langle O_i(\\boldsymbol{x}) \\rangle_{data} - \\langle O_i(\\boldsymbol{x}) \\rangle_{model}\n\\label{_auto25} \\tag{25}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"This expression shows that the gradient of the log-likelihood cost\nfunction is a **difference of moments**, with one calculated from\nthe data and one calculated from the model. The data-dependent term is\ncalled the **positive phase** and the model-dependent term is\ncalled the **negative phase** of the gradient. We see now that\nminimizing the cost function results in lowering the energy of\nconfigurations $\\boldsymbol{x}$ near points in the training data and\nincreasing the energy of configurations not observed in the training\ndata. That means we increase the model's probability of configurations\nsimilar to those in the training data.\n\n\nThe gradient of the cost function also demonstrates why gradients of\nunsupervised, generative models must be computed differently from for\nthose of for example FNNs. While the data-dependent expectation value\nis easily calculated based on the samples $\\boldsymbol{x}_i$ in the training\ndata, we must sample from the model in order to generate samples from\nwhich to caclulate the model-dependent term. We sample from the model\nby using MCMC-based methods. We can not sample from the model directly\nbecause the partition function $Z$ is generally intractable.\n\nAs in supervised machine learning problems, the goal is also here to\nperform well on **unseen** data, that is to have good\ngeneralization from the training data. The distribution $f(x)$ we\napproximate is not the **true** distribution we wish to estimate,\nit is limited to the training data. Hence, in unsupervised training as\nwell it is important to prevent overfitting to the training data. Thus\nit is common to add regularizers to the cost function in the same\nmanner as we discussed for say linear regression.\n\n\n\n## RBMs for the quantum many body problem\n\nThe idea of applying RBMs to quantum many body problems was presented by G. Carleo and M. Troyer, working with ETH Zurich and Microsoft Research.\n\nSome of their motivation included\n\n* The wave function $\\Psi$ is a monolithic mathematical quantity that contains all the information on a quantum state, be it a single particle or a complex molecule. In principle, an exponential amount of information is needed to fully encode a generic many-body quantum state.\n\n* There are still interesting open problems, including fundamental questions ranging from the dynamical properties of high-dimensional systems to the exact ground-state properties of strongly interacting fermions.\n\n* The difficulty lies in finding a general strategy to reduce the exponential complexity of the full many-body wave function down to its most essential features. That is\n\na. Dimensional reduction\n\nb. Feature extraction\n\n\n* Among the most successful techniques to attack these challenges, artifical neural networks play a prominent role.\n\n* Want to understand whether an artifical neural network may adapt to describe a quantum system.\n\nCarleo and Troyer applied the RBM to the quantum mechanical spin lattice systems of the Ising model and Heisenberg model, with encouraging results. Our goal is to test the method on systems of moving particles. For the spin lattice systems it was natural to use a binary-binary RBM, with the nodes taking values of 1 and -1. For moving particles, on the other hand, we want the visible nodes to be continuous, representing position coordinates. Thus, we start by choosing a Gaussian-binary RBM, where the visible nodes are continuous and hidden nodes take on values of 0 or 1. If eventually we would like the hidden nodes to be continuous as well the rectified linear units seem like the most relevant choice.\n\n\n\n\n## Representing the wave function\n\nThe wavefunction should be a probability amplitude depending on\n $\\boldsymbol{x}$. The RBM model is given by the joint distribution of\n $\\boldsymbol{x}$ and $\\boldsymbol{h}$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto26\"></div>\n\n$$\n\\begin{equation}\n F_{rbm}(\\mathbf{x},\\mathbf{h}) = \\frac{1}{Z} e^{-\\frac{1}{T_0}E(\\mathbf{x},\\mathbf{h})}.\n\\label{_auto26} \\tag{26}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"To find the marginal distribution of $\\boldsymbol{x}$ we set:",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto27\"></div>\n\n$$\n\\begin{equation}\n F_{rbm}(\\mathbf{x}) = \\sum_\\mathbf{h} F_{rbm}(\\mathbf{x}, \\mathbf{h}) \n\\label{_auto27} \\tag{27}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto28\"></div>\n\n$$\n\\begin{equation} \n = \\frac{1}{Z}\\sum_\\mathbf{h} e^{-E(\\mathbf{x}, \\mathbf{h})}.\n\\label{_auto28} \\tag{28}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Now this is what we use to represent the wave function, calling it a neural-network quantum state (NQS)",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto29\"></div>\n\n$$\n\\begin{equation}\n \\Psi (\\mathbf{X}) = F_{rbm}(\\mathbf{x}) \n\\label{_auto29} \\tag{29}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto30\"></div>\n\n$$\n\\begin{equation} \n = \\frac{1}{Z}\\sum_{\\boldsymbol{h}} e^{-E(\\mathbf{x}, \\mathbf{h})} \n\\label{_auto30} \\tag{30}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto31\"></div>\n\n$$\n\\begin{equation} \n = \\frac{1}{Z} \\sum_{\\{h_j\\}} e^{-\\sum_i^M \\frac{(x_i - a_i)^2}{2\\sigma^2} + \\sum_j^N b_j h_j + \\sum_\\\n{i,j}^{M,N} \\frac{x_i w_{ij} h_j}{\\sigma^2}} \n\\label{_auto31} \\tag{31}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto32\"></div>\n\n$$\n\\begin{equation} \n = \\frac{1}{Z} e^{-\\sum_i^M \\frac{(x_i - a_i)^2}{2\\sigma^2}} \\prod_j^N (1 + e^{b_j + \\sum_i^M \\frac{x\\\n_i w_{ij}}{\\sigma^2}}). \n\\label{_auto32} \\tag{32}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto33\"></div>\n\n$$\n\\begin{equation} \n\\label{_auto33} \\tag{33}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"## Choose the cost function\n\nNow we don't necessarily have training data (unless we generate it by using some other method). However, what we do have is the variational principle which allows us to obtain the ground state wave function by minimizing the expectation value of the energy of a trial wavefunction (corresponding to the untrained NQS). Similarly to the traditional variational Monte Carlo method then, it is the local energy we wish to minimize. The gradient to use for the stochastic gradient descent procedure is",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto34\"></div>\n\n$$\n\\begin{equation}\n\tC_i = \\frac{\\partial \\langle E_L \\rangle}{\\partial \\theta_i}\n\t= 2(\\langle E_L \\frac{1}{\\Psi}\\frac{\\partial \\Psi}{\\partial \\theta_i} \\rangle - \\langle E_L \\rangle \\langle \\frac{1}{\\Psi}\\frac{\\partial \\Psi}{\\partial \\theta_i} \\rangle ),\n\\label{_auto34} \\tag{34}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where the local energy is given by",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto35\"></div>\n\n$$\n\\begin{equation}\n\tE_L = \\frac{1}{\\Psi} \\hat{\\mathbf{H}} \\Psi.\n\\label{_auto35} \\tag{35}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Mathematical details\n\nBecause we are restricted to potential functions which are positive it\nis convenient to express them as exponentials, so that",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto36\"></div>\n\n$$\n\\begin{equation}\n\t\\phi_C (\\boldsymbol{x}_C) = e^{-E_C(\\boldsymbol{x}_C)}\n\\label{_auto36} \\tag{36}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where $E(\\boldsymbol{x}_C)$ is called an *energy function*, and the\nexponential representation is the *Boltzmann distribution*. The\njoint distribution is defined as the product of potentials.\n\nThe joint distribution of the random variables is then",
"_____no_output_____"
],
[
"$$\np(\\boldsymbol{x}) = \\frac{1}{Z} \\prod_C \\phi_C (\\boldsymbol{x}_C) \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z} \\prod_C e^{-E_C(\\boldsymbol{x}_C)} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z} e^{-\\sum_C E_C(\\boldsymbol{x}_C)} \\nonumber\n$$",
"_____no_output_____"
],
[
"3\n9\n \n<\n<\n<\n!\n!\nM\nA\nT\nH\n_\nB\nL\nO\nC\nK",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto38\"></div>\n\n$$\n\\begin{equation}\n\tp_{BM}(\\boldsymbol{x}, \\boldsymbol{h}) = \\frac{1}{Z_{BM}} e^{-\\frac{1}{T}E_{BM}(\\boldsymbol{x}, \\boldsymbol{h})} ,\n\\label{_auto38} \\tag{38}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"with the partition function",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto39\"></div>\n\n$$\n\\begin{equation}\n\tZ_{BM} = \\int \\int e^{-\\frac{1}{T} E_{BM}(\\tilde{\\boldsymbol{x}}, \\tilde{\\boldsymbol{h}})} d\\tilde{\\boldsymbol{x}} d\\tilde{\\boldsymbol{h}} .\n\\label{_auto39} \\tag{39}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"$T$ is a physics-inspired parameter named temperature and will be assumed to be 1 unless otherwise stated. The energy function of the Boltzmann machine determines the interactions between the nodes and is defined",
"_____no_output_____"
],
[
"$$\nE_{BM}(\\boldsymbol{x}, \\boldsymbol{h}) = - \\sum_{i, k}^{M, K} a_i^k \\alpha_i^k (x_i)\n\t- \\sum_{j, l}^{N, L} b_j^l \\beta_j^l (h_j) \n\t- \\sum_{i,j,k,l}^{M,N,K,L} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (h_j) \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto40\"></div>\n\n$$\n\\begin{equation} \n\t- \\sum_{i, m=i+1, k}^{M, M, K} \\alpha_i^k (x_i) v_{im}^k \\alpha_m^k (x_m)\n\t- \\sum_{j,n=j+1,l}^{N,N,L} \\beta_j^l (h_j) u_{jn}^l \\beta_n^l (h_n).\n\\label{_auto40} \\tag{40}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Here $\\alpha_i^k (x_i)$ and $\\beta_j^l (h_j)$ are one-dimensional\ntransfer functions or mappings from the given input value to the\ndesired feature value. They can be arbitrary functions of the input\nvariables and are independent of the parameterization (parameters\nreferring to weight and biases), meaning they are not affected by\ntraining of the model. The indices $k$ and $l$ indicate that there can\nbe multiple transfer functions per variable. Furthermore, $a_i^k$ and\n$b_j^l$ are the visible and hidden bias. $w_{ij}^{kl}$ are weights of\nthe \\textbf{inter-layer} connection terms which connect visible and\nhidden units. $ v_{im}^k$ and $u_{jn}^l$ are weights of the\n\\textbf{intra-layer} connection terms which connect the visible units\nto each other and the hidden units to each other, respectively.\n\n\n\nWe remove the intra-layer connections by setting $v_{im}$ and $u_{jn}$\nto zero. The expression for the energy of the RBM is then",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto41\"></div>\n\n$$\n\\begin{equation}\n\tE_{RBM}(\\boldsymbol{x}, \\boldsymbol{h}) = - \\sum_{i, k}^{M, K} a_i^k \\alpha_i^k (x_i)\n\t- \\sum_{j, l}^{N, L} b_j^l \\beta_j^l (h_j) \n\t- \\sum_{i,j,k,l}^{M,N,K,L} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (h_j). \n\\label{_auto41} \\tag{41}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"resulting in",
"_____no_output_____"
],
[
"$$\nP_{RBM} (\\boldsymbol{x}) = \\int P_{RBM} (\\boldsymbol{x}, \\tilde{\\boldsymbol{h}}) d \\tilde{\\boldsymbol{h}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{RBM}} \\int e^{-E_{RBM} (\\boldsymbol{x}, \\tilde{\\boldsymbol{h}}) } d\\tilde{\\boldsymbol{h}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{RBM}} \\int e^{\\sum_{i, k} a_i^k \\alpha_i^k (x_i)\n\t+ \\sum_{j, l} b_j^l \\beta_j^l (\\tilde{h}_j) \n\t+ \\sum_{i,j,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (\\tilde{h}_j)} \n\td\\tilde{\\boldsymbol{h}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{RBM}} e^{\\sum_{i, k} a_i^k \\alpha_i^k (x_i)}\n\t\\int \\prod_j^N e^{\\sum_l b_j^l \\beta_j^l (\\tilde{h}_j) \n\t+ \\sum_{i,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (\\tilde{h}_j)} d\\tilde{\\boldsymbol{h}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{RBM}} e^{\\sum_{i, k} a_i^k \\alpha_i^k (x_i)}\n\t\\biggl( \\int e^{\\sum_l b_1^l \\beta_1^l (\\tilde{h}_1) + \\sum_{i,k,l} \\alpha_i^k (x_i) w_{i1}^{kl} \\beta_1^l (\\tilde{h}_1)} d \\tilde{h}_1 \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n\\times \\int e^{\\sum_l b_2^l \\beta_2^l (\\tilde{h}_2) + \\sum_{i,k,l} \\alpha_i^k (x_i) w_{i2}^{kl} \\beta_2^l (\\tilde{h}_2)} d \\tilde{h}_2 \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n\\times ... \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n\\times \\int e^{\\sum_l b_N^l \\beta_N^l (\\tilde{h}_N) + \\sum_{i,k,l} \\alpha_i^k (x_i) w_{iN}^{kl} \\beta_N^l (\\tilde{h}_N)} d \\tilde{h}_N \\biggr) \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto42\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z_{RBM}} e^{\\sum_{i, k} a_i^k \\alpha_i^k (x_i)}\n\t\\prod_j^N \\int e^{\\sum_l b_j^l \\beta_j^l (\\tilde{h}_j) + \\sum_{i,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (\\tilde{h}_j)} d\\tilde{h}_j\n\\label{_auto42} \\tag{42}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Similarly",
"_____no_output_____"
],
[
"$$\nP_{RBM} (\\boldsymbol{h}) = \\frac{1}{Z_{RBM}} \\int e^{-E_{RBM} (\\tilde{\\boldsymbol{x}}, \\boldsymbol{h})} d\\tilde{\\boldsymbol{x}} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto43\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z_{RBM}} e^{\\sum_{j, l} b_j^l \\beta_j^l (h_j)}\n\t\\prod_i^M \\int e^{\\sum_k a_i^k \\alpha_i^k (\\tilde{x}_i)\n\t+ \\sum_{j,k,l} \\alpha_i^k (\\tilde{x}_i) w_{ij}^{kl} \\beta_j^l (h_j)} d\\tilde{x}_i\n\\label{_auto43} \\tag{43}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Using Bayes theorem",
"_____no_output_____"
],
[
"$$\nP_{RBM} (\\boldsymbol{h}|\\boldsymbol{x}) = \\frac{P_{RBM} (\\boldsymbol{x}, \\boldsymbol{h})}{P_{RBM} (\\boldsymbol{x})} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{\\frac{1}{Z_{RBM}} e^{\\sum_{i, k} a_i^k \\alpha_i^k (x_i)\n\t+ \\sum_{j, l} b_j^l \\beta_j^l (h_j) \n\t+ \\sum_{i,j,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (h_j)}}\n\t{\\frac{1}{Z_{RBM}} e^{\\sum_{i, k} a_i^k \\alpha_i^k (x_i)}\n\t\\prod_j^N \\int e^{\\sum_l b_j^l \\beta_j^l (\\tilde{h}_j) + \\sum_{i,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (\\tilde{h}_j)} d\\tilde{h}_j} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto44\"></div>\n\n$$\n\\begin{equation} \n\t= \\prod_j^N \\frac{e^{\\sum_l b_j^l \\beta_j^l (h_j) + \\sum_{i,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (h_j)} }\n\t{\\int e^{\\sum_l b_j^l \\beta_j^l (\\tilde{h}_j) + \\sum_{i,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (\\tilde{h}_j)} d\\tilde{h}_j}\n\\label{_auto44} \\tag{44}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Similarly",
"_____no_output_____"
],
[
"$$\nP_{RBM} (\\boldsymbol{x}|\\boldsymbol{h}) = \\frac{P_{RBM} (\\boldsymbol{x}, \\boldsymbol{h})}{P_{RBM} (\\boldsymbol{h})} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto45\"></div>\n\n$$\n\\begin{equation} \n\t= \\prod_i^M \\frac{e^{\\sum_k a_i^k \\alpha_i^k (x_i)\n\t+ \\sum_{j,k,l} \\alpha_i^k (x_i) w_{ij}^{kl} \\beta_j^l (h_j)}}\n\t{\\int e^{\\sum_k a_i^k \\alpha_i^k (\\tilde{x}_i)\n\t+ \\sum_{j,k,l} \\alpha_i^k (\\tilde{x}_i) w_{ij}^{kl} \\beta_j^l (h_j)} d\\tilde{x}_i}\n\\label{_auto45} \\tag{45}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The original RBM had binary visible and hidden nodes. They were\nshowned to be universal approximators of discrete distributions.\nIt was also shown that adding hidden units yields\nstrictly improved modelling power. The common choice of binary values\nare 0 and 1. However, in some physics applications, -1 and 1 might be\na more natural choice. We will here use 0 and 1.",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto46\"></div>\n\n$$\n\\begin{equation}\n\tE_{BB}(\\boldsymbol{x}, \\mathbf{h}) = - \\sum_i^M x_i a_i- \\sum_j^N b_j h_j - \\sum_{i,j}^{M,N} x_i w_{ij} h_j.\n\\label{_auto46} \\tag{46}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto47\"></div>\n\n$$\n\\begin{equation}\n\tp_{BB}(\\boldsymbol{x}, \\boldsymbol{h}) = \\frac{1}{Z_{BB}} e^{\\sum_i^M a_i x_i + \\sum_j^N b_j h_j + \\sum_{ij}^{M,N} x_i w_{ij} h_j} \n\\label{_auto47} \\tag{47}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto48\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a} + \\boldsymbol{b}^T \\boldsymbol{h} + \\boldsymbol{x}^T \\boldsymbol{W} \\boldsymbol{h}}\n\\label{_auto48} \\tag{48}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"with the partition function",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto49\"></div>\n\n$$\n\\begin{equation}\n\tZ_{BB} = \\sum_{\\boldsymbol{x}, \\boldsymbol{h}} e^{\\boldsymbol{x}^T \\boldsymbol{a} + \\boldsymbol{b}^T \\boldsymbol{h} + \\boldsymbol{x}^T \\boldsymbol{W} \\boldsymbol{h}} .\n\\label{_auto49} \\tag{49}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Marginal Probability Density Functions\n\nIn order to find the probability of any configuration of the visible units we derive the marginal probability density function.",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto50\"></div>\n\n$$\n\\begin{equation}\n\tp_{BB} (\\boldsymbol{x}) = \\sum_{\\boldsymbol{h}} p_{BB} (\\boldsymbol{x}, \\boldsymbol{h}) \n\\label{_auto50} \\tag{50}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{BB}} \\sum_{\\boldsymbol{h}} e^{\\boldsymbol{x}^T \\boldsymbol{a} + \\boldsymbol{b}^T \\boldsymbol{h} + \\boldsymbol{x}^T \\boldsymbol{W} \\boldsymbol{h}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a}} \\sum_{\\boldsymbol{h}} e^{\\sum_j^N (b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j})h_j} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a}} \\sum_{\\boldsymbol{h}} \\prod_j^N e^{ (b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j})h_j} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a}} \\bigg ( \\sum_{h_1} e^{(b_1 + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast 1})h_1}\n\t\\times \\sum_{h_2} e^{(b_2 + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast 2})h_2} \\times \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n... \\times \\sum_{h_2} e^{(b_N + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast N})h_N} \\bigg ) \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a}} \\prod_j^N \\sum_{h_j} e^{(b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}) h_j} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto51\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a}} \\prod_j^N (1 + e^{b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}}) .\n\\label{_auto51} \\tag{51}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"A similar derivation yields the marginal probability of the hidden units",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto52\"></div>\n\n$$\n\\begin{equation}\n\tp_{BB} (\\boldsymbol{h}) = \\frac{1}{Z_{BB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M (1 + e^{a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}}) .\n\\label{_auto52} \\tag{52}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Conditional Probability Density Functions\n\nWe derive the probability of the hidden units given the visible units using Bayes' rule",
"_____no_output_____"
],
[
"$$\np_{BB} (\\boldsymbol{h}|\\boldsymbol{x}) = \\frac{p_{BB} (\\boldsymbol{x}, \\boldsymbol{h})}{p_{BB} (\\boldsymbol{x})} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{ \\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a} + \\boldsymbol{b}^T \\boldsymbol{h} + \\boldsymbol{x}^T \\boldsymbol{W} \\boldsymbol{h}} }\n\t {\\frac{1}{Z_{BB}} e^{\\boldsymbol{x}^T \\boldsymbol{a}} \\prod_j^N (1 + e^{b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}})} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{ e^{\\boldsymbol{x}^T \\boldsymbol{a}} e^{ \\sum_j^N (b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j} ) h_j} }\n\t { e^{\\boldsymbol{x}^T \\boldsymbol{a}} \\prod_j^N (1 + e^{b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}})} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\prod_j^N \\frac{ e^{(b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j} ) h_j} }\n\t{1 + e^{b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}}} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto53\"></div>\n\n$$\n\\begin{equation} \n\t= \\prod_j^N p_{BB} (h_j| \\boldsymbol{x}) .\n\\label{_auto53} \\tag{53}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"From this we find the probability of a hidden unit being \"on\" or \"off\":",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto54\"></div>\n\n$$\n\\begin{equation}\n\tp_{BB} (h_j=1 | \\boldsymbol{x}) = \\frac{ e^{(b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j} ) h_j} }\n\t{1 + e^{b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}}} \n\\label{_auto54} \\tag{54}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto55\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{ e^{(b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j} )} }\n\t{1 + e^{b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}}} \n\\label{_auto55} \\tag{55}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto56\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{ 1 }{1 + e^{-(b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j})} } ,\n\\label{_auto56} \\tag{56}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"and",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto57\"></div>\n\n$$\n\\begin{equation}\n\tp_{BB} (h_j=0 | \\boldsymbol{x}) =\\frac{ 1 }{1 + e^{b_j + \\boldsymbol{x}^T \\boldsymbol{w}_{\\ast j}} } .\n\\label{_auto57} \\tag{57}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Similarly we have that the conditional probability of the visible units given the hidden are",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto58\"></div>\n\n$$\n\\begin{equation}\n\tp_{BB} (\\boldsymbol{x}|\\boldsymbol{h}) = \\prod_i^M \\frac{ e^{ (a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}) x_i} }{ 1 + e^{a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}} } \n\\label{_auto58} \\tag{58}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto59\"></div>\n\n$$\n\\begin{equation} \n\t= \\prod_i^M p_{BB} (x_i | \\boldsymbol{h}) .\n\\label{_auto59} \\tag{59}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto60\"></div>\n\n$$\n\\begin{equation}\n\tp_{BB} (x_i=1 | \\boldsymbol{h}) = \\frac{1}{1 + e^{-(a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} )}} \n\\label{_auto60} \\tag{60}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto61\"></div>\n\n$$\n\\begin{equation} \n\tp_{BB} (x_i=0 | \\boldsymbol{h}) = \\frac{1}{1 + e^{a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} }} .\n\\label{_auto61} \\tag{61}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Gaussian-Binary Restricted Boltzmann Machines\n\nInserting into the expression for $E_{RBM}(\\boldsymbol{x},\\boldsymbol{h})$ in equation results in the energy",
"_____no_output_____"
],
[
"$$\nE_{GB}(\\boldsymbol{x}, \\boldsymbol{h}) = \\sum_i^M \\frac{(x_i - a_i)^2}{2\\sigma_i^2}\n\t- \\sum_j^N b_j h_j \n\t-\\sum_{ij}^{M,N} \\frac{x_i w_{ij} h_j}{\\sigma_i^2} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto62\"></div>\n\n$$\n\\begin{equation} \n\t= \\vert\\vert\\frac{\\boldsymbol{x} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2 - \\boldsymbol{b}^T \\boldsymbol{h} \n\t- (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{W}\\boldsymbol{h} . \n\\label{_auto62} \\tag{62}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Joint Probability Density Function",
"_____no_output_____"
],
[
"$$\np_{GB} (\\boldsymbol{x}, \\boldsymbol{h}) = \\frac{1}{Z_{GB}} e^{-\\vert\\vert\\frac{\\boldsymbol{x} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2 + \\boldsymbol{b}^T \\boldsymbol{h} \n\t+ (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{W}\\boldsymbol{h}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{- \\sum_i^M \\frac{(x_i - a_i)^2}{2\\sigma_i^2}\n\t+ \\sum_j^N b_j h_j \n\t+\\sum_{ij}^{M,N} \\frac{x_i w_{ij} h_j}{\\sigma_i^2}} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto63\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z_{GB}} \\prod_{ij}^{M,N} e^{-\\frac{(x_i - a_i)^2}{2\\sigma_i^2}\n\t+ b_j h_j \n\t+\\frac{x_i w_{ij} h_j}{\\sigma_i^2}} ,\n\\label{_auto63} \\tag{63}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"with the partition function given by",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto64\"></div>\n\n$$\n\\begin{equation}\n\tZ_{GB} = \\int \\sum_{\\tilde{\\boldsymbol{h}}}^{\\tilde{\\boldsymbol{H}}} e^{-\\vert\\vert\\frac{\\tilde{\\boldsymbol{x}} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2 + \\boldsymbol{b}^T \\tilde{\\boldsymbol{h}} \n\t+ (\\frac{\\tilde{\\boldsymbol{x}}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{W}\\tilde{\\boldsymbol{h}}} d\\tilde{\\boldsymbol{x}} .\n\\label{_auto64} \\tag{64}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Marginal Probability Density Functions\n\nWe proceed to find the marginal probability densitites of the\nGaussian-binary RBM. We first marginalize over the binary hidden units\nto find $p_{GB} (\\boldsymbol{x})$",
"_____no_output_____"
],
[
"$$\np_{GB} (\\boldsymbol{x}) = \\sum_{\\tilde{\\boldsymbol{h}}}^{\\tilde{\\boldsymbol{H}}} p_{GB} (\\boldsymbol{x}, \\tilde{\\boldsymbol{h}}) \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} \\sum_{\\tilde{\\boldsymbol{h}}}^{\\tilde{\\boldsymbol{H}}} \n\te^{-\\vert\\vert\\frac{\\boldsymbol{x} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2 + \\boldsymbol{b}^T \\tilde{\\boldsymbol{h}} \n\t+ (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{W}\\tilde{\\boldsymbol{h}}} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto65\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z_{GB}} e^{-\\vert\\vert\\frac{\\boldsymbol{x} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2}\n\t\\prod_j^N (1 + e^{b_j + (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j}} ) .\n\\label{_auto65} \\tag{65}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"We next marginalize over the visible units. This is the first time we\nmarginalize over continuous values. We rewrite the exponential factor\ndependent on $\\boldsymbol{x}$ as a Gaussian function before we integrate in\nthe last step.",
"_____no_output_____"
],
[
"$$\np_{GB} (\\boldsymbol{h}) = \\int p_{GB} (\\tilde{\\boldsymbol{x}}, \\boldsymbol{h}) d\\tilde{\\boldsymbol{x}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} \\int e^{-\\vert\\vert\\frac{\\tilde{\\boldsymbol{x}} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2 + \\boldsymbol{b}^T \\boldsymbol{h} \n\t+ (\\frac{\\tilde{\\boldsymbol{x}}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{W}\\boldsymbol{h}} d\\tilde{\\boldsymbol{x}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h} } \\int \\prod_i^M\n\te^{- \\frac{(\\tilde{x}_i - a_i)^2}{2\\sigma_i^2} + \\frac{\\tilde{x}_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}}{\\sigma_i^2} } d\\tilde{\\boldsymbol{x}} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h} }\n\t\\biggl( \\int e^{- \\frac{(\\tilde{x}_1 - a_1)^2}{2\\sigma_1^2} + \\frac{\\tilde{x}_1 \\boldsymbol{w}_{1\\ast}^T \\boldsymbol{h}}{\\sigma_1^2} } d\\tilde{x}_1 \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n\\times \\int e^{- \\frac{(\\tilde{x}_2 - a_2)^2}{2\\sigma_2^2} + \\frac{\\tilde{x}_2 \\boldsymbol{w}_{2\\ast}^T \\boldsymbol{h}}{\\sigma_2^2} } d\\tilde{x}_2 \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n\\times ... \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n\\times \\int e^{- \\frac{(\\tilde{x}_M - a_M)^2}{2\\sigma_M^2} + \\frac{\\tilde{x}_M \\boldsymbol{w}_{M\\ast}^T \\boldsymbol{h}}{\\sigma_M^2} } d\\tilde{x}_M \\biggr) \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M\n\t\\int e^{- \\frac{(\\tilde{x}_i - a_i)^2 - 2\\tilde{x}_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}}{2\\sigma_i^2} } d\\tilde{x}_i \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M\n\t\\int e^{- \\frac{\\tilde{x}_i^2 - 2\\tilde{x}_i(a_i + \\tilde{x}_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}) + a_i^2}{2\\sigma_i^2} } d\\tilde{x}_i \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M\n\t\\int e^{- \\frac{\\tilde{x}_i^2 - 2\\tilde{x}_i(a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}) + (a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 - (a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 + a_i^2}{2\\sigma_i^2} } d\\tilde{x}_i \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M\n\t\\int e^{- \\frac{(\\tilde{x}_i - (a_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}))^2 - a_i^2 -2a_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} - (\\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 + a_i^2}{2\\sigma_i^2} } d\\tilde{x}_i \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M\n\te^{\\frac{2a_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} +(\\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 }{2\\sigma_i^2}}\n\t\\int e^{- \\frac{(\\tilde{x}_i - a_i - \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2}{2\\sigma_i^2}}\n\td\\tilde{x}_i \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto66\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M\n\t\\sqrt{2\\pi \\sigma_i^2}\n\te^{\\frac{2a_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} +(\\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 }{2\\sigma_i^2}} .\n\\label{_auto66} \\tag{66}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Conditional Probability Density Functions\n\nWe finish by deriving the conditional probabilities.",
"_____no_output_____"
],
[
"$$\np_{GB} (\\boldsymbol{h}| \\boldsymbol{x}) = \\frac{p_{GB} (\\boldsymbol{x}, \\boldsymbol{h})}{p_{GB} (\\boldsymbol{x})} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{\\frac{1}{Z_{GB}} e^{-\\vert\\vert\\frac{\\boldsymbol{x} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2 + \\boldsymbol{b}^T \\boldsymbol{h} \n\t+ (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{W}\\boldsymbol{h}}}\n\t{\\frac{1}{Z_{GB}} e^{-\\vert\\vert\\frac{\\boldsymbol{x} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2}\n\t\\prod_j^N (1 + e^{b_j + (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j}} ) }\n\t\\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\prod_j^N \\frac{e^{(b_j + (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j})h_j } }\n\t{1 + e^{b_j + (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j}}} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto67\"></div>\n\n$$\n\\begin{equation} \n\t= \\prod_j^N p_{GB} (h_j|\\boldsymbol{x}).\n\\label{_auto67} \\tag{67}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The conditional probability of a binary hidden unit $h_j$ being on or off again takes the form of a sigmoid function",
"_____no_output_____"
],
[
"$$\np_{GB} (h_j =1 | \\boldsymbol{x}) = \\frac{e^{b_j + (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j} } }\n\t{1 + e^{b_j + (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j}}} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto68\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{1 + e^{-b_j - (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j}}} \n\\label{_auto68} \\tag{68}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto69\"></div>\n\n$$\n\\begin{equation} \n\tp_{GB} (h_j =0 | \\boldsymbol{x}) =\n\t\\frac{1}{1 + e^{b_j +(\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{w}_{\\ast j}}} .\n\\label{_auto69} \\tag{69}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The conditional probability of the continuous $\\boldsymbol{x}$ now has another form, however.",
"_____no_output_____"
],
[
"$$\np_{GB} (\\boldsymbol{x}|\\boldsymbol{h})\n\t= \\frac{p_{GB} (\\boldsymbol{x}, \\boldsymbol{h})}{p_{GB} (\\boldsymbol{h})} \\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\frac{\\frac{1}{Z_{GB}} e^{-\\vert\\vert\\frac{\\boldsymbol{x} -\\boldsymbol{a}}{2\\boldsymbol{\\sigma}}\\vert\\vert^2 + \\boldsymbol{b}^T \\boldsymbol{h} \n\t+ (\\frac{\\boldsymbol{x}}{\\boldsymbol{\\sigma}^2})^T \\boldsymbol{W}\\boldsymbol{h}}}\n\t{\\frac{1}{Z_{GB}} e^{\\boldsymbol{b}^T \\boldsymbol{h}} \\prod_i^M\n\t\\sqrt{2\\pi \\sigma_i^2}\n\te^{\\frac{2a_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} +(\\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 }{2\\sigma_i^2}}}\n\t\\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\prod_i^M \\frac{1}{\\sqrt{2\\pi \\sigma_i^2}}\n\t\\frac{e^{- \\frac{(x_i - a_i)^2}{2\\sigma_i^2} + \\frac{x_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}}{2\\sigma_i^2} }}\n\t{e^{\\frac{2a_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} +(\\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 }{2\\sigma_i^2}}}\n\t\\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\prod_i^M \\frac{1}{\\sqrt{2\\pi \\sigma_i^2}}\n\t\\frac{e^{-\\frac{x_i^2 - 2a_i x_i + a_i^2 - 2x_i \\boldsymbol{w}_{i\\ast}^T\\boldsymbol{h} }{2\\sigma_i^2} } }\n\t{e^{\\frac{2a_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} +(\\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2 }{2\\sigma_i^2}}}\n\t\\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\prod_i^M \\frac{1}{\\sqrt{2\\pi \\sigma_i^2}}\n\te^{- \\frac{x_i^2 - 2a_i x_i + a_i^2 - 2x_i \\boldsymbol{w}_{i\\ast}^T\\boldsymbol{h}\n\t+ 2a_i \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h} +(\\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2}\n\t{2\\sigma_i^2} }\n\t\\nonumber\n$$",
"_____no_output_____"
],
[
"$$\n= \\prod_i^M \\frac{1}{\\sqrt{2\\pi \\sigma_i^2}}\n\te^{ - \\frac{(x_i - b_i - \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h})^2}{2\\sigma_i^2}} \\nonumber\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto70\"></div>\n\n$$\n\\begin{equation} \n\t= \\prod_i^M \\mathcal{N}\n\t(x_i | b_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}, \\sigma_i^2) \n\\label{_auto70} \\tag{70}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto71\"></div>\n\n$$\n\\begin{equation} \n\t\\Rightarrow p_{GB} (x_i|\\boldsymbol{h}) = \\mathcal{N}\n\t(x_i | b_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}, \\sigma_i^2) .\n\\label{_auto71} \\tag{71}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The form of these conditional probabilities explains the name\n\"Gaussian\" and the form of the Gaussian-binary energy function. We see\nthat the conditional probability of $x_i$ given $\\boldsymbol{h}$ is a normal\ndistribution with mean $b_i + \\boldsymbol{w}_{i\\ast}^T \\boldsymbol{h}$ and variance\n$\\sigma_i^2$.\n\n\n## Neural Quantum States\n\n\nThe wavefunction should be a probability amplitude depending on $\\boldsymbol{x}$. The RBM model is given by the joint distribution of $\\boldsymbol{x}$ and $\\boldsymbol{h}$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto72\"></div>\n\n$$\n\\begin{equation}\n\tF_{rbm}(\\boldsymbol{x},\\mathbf{h}) = \\frac{1}{Z} e^{-\\frac{1}{T_0}E(\\boldsymbol{x},\\mathbf{h})}\n\\label{_auto72} \\tag{72}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"To find the marginal distribution of $\\boldsymbol{x}$ we set:",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto73\"></div>\n\n$$\n\\begin{equation}\n\tF_{rbm}(\\mathbf{x}) = \\sum_\\mathbf{h} F_{rbm}(\\mathbf{x}, \\mathbf{h}) \n\\label{_auto73} \\tag{73}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto74\"></div>\n\n$$\n\\begin{equation} \n\t\t\t\t= \\frac{1}{Z}\\sum_\\mathbf{h} e^{-E(\\mathbf{x}, \\mathbf{h})}\n\\label{_auto74} \\tag{74}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Now this is what we use to represent the wave function, calling it a neural-network quantum state (NQS)",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto75\"></div>\n\n$$\n\\begin{equation}\n\t\\Psi (\\mathbf{X}) = F_{rbm}(\\mathbf{x}) \n\\label{_auto75} \\tag{75}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto76\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z}\\sum_{\\boldsymbol{h}} e^{-E(\\mathbf{x}, \\mathbf{h})} \n\\label{_auto76} \\tag{76}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto77\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z} \\sum_{\\{h_j\\}} e^{-\\sum_i^M \\frac{(x_i - a_i)^2}{2\\sigma^2} + \\sum_j^N b_j h_j + \\sum_{i,j}^{M,N} \\frac{x_i w_{ij} h_j}{\\sigma^2}} \n\\label{_auto77} \\tag{77}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto78\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{Z} e^{-\\sum_i^M \\frac{(x_i - a_i)^2}{2\\sigma^2}} \\prod_j^N (1 + e^{b_j + \\sum_i^M \\frac{x_i w_{ij}}{\\sigma^2}}) \n\\label{_auto78} \\tag{78}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto79\"></div>\n\n$$\n\\begin{equation} \n\\label{_auto79} \\tag{79}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The above wavefunction is the most general one because it allows for\ncomplex valued wavefunctions. However it fundamentally changes the\nprobabilistic foundation of the RBM, because what is usually a\nprobability in the RBM framework is now a an amplitude. This means\nthat a lot of the theoretical framework usually used to interpret the\nmodel, i.e. graphical models, conditional probabilities, and Markov\nrandom fields, breaks down. If we assume the wavefunction to be\npostive definite, however, we can use the RBM to represent the squared\nwavefunction, and thereby a probability. This also makes it possible\nto sample from the model using Gibbs sampling, because we can obtain\nthe conditional probabilities.",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto80\"></div>\n\n$$\n\\begin{equation}\n\t|\\Psi (\\mathbf{X})|^2 = F_{rbm}(\\mathbf{X}) \n\\label{_auto80} \\tag{80}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto81\"></div>\n\n$$\n\\begin{equation} \n\t\\Rightarrow \\Psi (\\mathbf{X}) = \\sqrt{F_{rbm}(\\mathbf{X})} \n\\label{_auto81} \\tag{81}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto82\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{\\sqrt{Z}}\\sqrt{\\sum_{\\{h_j\\}} e^{-E(\\mathbf{X}, \\mathbf{h})}} \n\\label{_auto82} \\tag{82}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto83\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{\\sqrt{Z}} \\sqrt{\\sum_{\\{h_j\\}} e^{-\\sum_i^M \\frac{(X_i - a_i)^2}{2\\sigma^2} + \\sum_j^N b_j h_j + \\sum_{i,j}^{M,N} \\frac{X_i w_{ij} h_j}{\\sigma^2}} }\n\\label{_auto83} \\tag{83}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto84\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{\\sqrt{Z}} e^{-\\sum_i^M \\frac{(X_i - a_i)^2}{4\\sigma^2}} \\sqrt{\\sum_{\\{h_j\\}} \\prod_j^N e^{b_j h_j + \\sum_i^M \\frac{X_i w_{ij} h_j}{\\sigma^2}}} \n\\label{_auto84} \\tag{84}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto85\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{\\sqrt{Z}} e^{-\\sum_i^M \\frac{(X_i - a_i)^2}{4\\sigma^2}} \\sqrt{\\prod_j^N \\sum_{h_j} e^{b_j h_j + \\sum_i^M \\frac{X_i w_{ij} h_j}{\\sigma^2}}} \n\\label{_auto85} \\tag{85}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto86\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{\\sqrt{Z}} e^{-\\sum_i^M \\frac{(X_i - a_i)^2}{4\\sigma^2}} \\prod_j^N \\sqrt{e^0 + e^{b_j + \\sum_i^M \\frac{X_i w_{ij}}{\\sigma^2}}} \n\\label{_auto86} \\tag{86}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto87\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{\\sqrt{Z}} e^{-\\sum_i^M \\frac{(X_i - a_i)^2}{4\\sigma^2}} \\prod_j^N \\sqrt{1 + e^{b_j + \\sum_i^M \\frac{X_i w_{ij}}{\\sigma^2}}} \n\\label{_auto87} \\tag{87}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto88\"></div>\n\n$$\n\\begin{equation} \n\\label{_auto88} \\tag{88}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"### Cost function\n\nThis is where we deviate from what is common in machine\nlearning. Rather than defining a cost function based on some dataset,\nour cost function is the energy of the quantum mechanical system. From\nthe variational principle we know that minizing this energy should\nlead to the ground state wavefunction. As stated previously the local\nenergy is given by",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto89\"></div>\n\n$$\n\\begin{equation}\n\tE_L = \\frac{1}{\\Psi} \\hat{\\mathbf{H}} \\Psi,\n\\label{_auto89} \\tag{89}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"and the gradient is",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto90\"></div>\n\n$$\n\\begin{equation}\n\tG_i = \\frac{\\partial \\langle E_L \\rangle}{\\partial \\alpha_i}\n\t= 2(\\langle E_L \\frac{1}{\\Psi}\\frac{\\partial \\Psi}{\\partial \\alpha_i} \\rangle - \\langle E_L \\rangle \\langle \\frac{1}{\\Psi}\\frac{\\partial \\Psi}{\\partial \\alpha_i} \\rangle ),\n\\label{_auto90} \\tag{90}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where $\\alpha_i = a_1,...,a_M,b_1,...,b_N,w_{11},...,w_{MN}$.\n\n\nWe use that $\\frac{1}{\\Psi}\\frac{\\partial \\Psi}{\\partial \\alpha_i} \n\t= \\frac{\\partial \\ln{\\Psi}}{\\partial \\alpha_i}$,\nand find",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto91\"></div>\n\n$$\n\\begin{equation}\n\t\\ln{\\Psi({\\mathbf{X}})} = -\\ln{Z} - \\sum_m^M \\frac{(X_m - a_m)^2}{2\\sigma^2}\n\t+ \\sum_n^N \\ln({1 + e^{b_n + \\sum_i^M \\frac{X_i w_{in}}{\\sigma^2}})}.\n\\label{_auto91} \\tag{91}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"This gives",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto92\"></div>\n\n$$\n\\begin{equation}\n\t\\frac{\\partial }{\\partial a_m} \\ln\\Psi\n\t= \t\\frac{1}{\\sigma^2} (X_m - a_m) \n\\label{_auto92} \\tag{92}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto93\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial }{\\partial b_n} \\ln\\Psi\n\t=\n\t\\frac{1}{e^{-b_n-\\frac{1}{\\sigma^2}\\sum_i^M X_i w_{in}} + 1} \n\\label{_auto93} \\tag{93}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto94\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial }{\\partial w_{mn}} \\ln\\Psi\n\t= \\frac{X_m}{\\sigma^2(e^{-b_n-\\frac{1}{\\sigma^2}\\sum_i^M X_i w_{in}} + 1)}.\n\\label{_auto94} \\tag{94}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"If $\\Psi = \\sqrt{F_{rbm}}$ we have",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto95\"></div>\n\n$$\n\\begin{equation}\n\t\\ln{\\Psi({\\mathbf{X}})} = -\\frac{1}{2}\\ln{Z} - \\sum_m^M \\frac{(X_m - a_m)^2}{4\\sigma^2}\n\t+ \\frac{1}{2}\\sum_n^N \\ln({1 + e^{b_n + \\sum_i^M \\frac{X_i w_{in}}{\\sigma^2}})},\n\\label{_auto95} \\tag{95}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"which results in",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto96\"></div>\n\n$$\n\\begin{equation}\n\t\\frac{\\partial }{\\partial a_m} \\ln\\Psi\n\t= \t\\frac{1}{2\\sigma^2} (X_m - a_m) \n\\label{_auto96} \\tag{96}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto97\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial }{\\partial b_n} \\ln\\Psi\n\t=\n\t\\frac{1}{2(e^{-b_n-\\frac{1}{\\sigma^2}\\sum_i^M X_i w_{in}} + 1)} \n\\label{_auto97} \\tag{97}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto98\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial }{\\partial w_{mn}} \\ln\\Psi\n\t= \\frac{X_m}{2\\sigma^2(e^{-b_n-\\frac{1}{\\sigma^2}\\sum_i^M X_i w_{in}} + 1)}.\n\\label{_auto98} \\tag{98}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Let us assume again that our Hamiltonian is",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto99\"></div>\n\n$$\n\\begin{equation}\n\t\\hat{\\mathbf{H}} = \\sum_p^P (-\\frac{1}{2}\\nabla_p^2 + \\frac{1}{2}\\omega^2 r_p^2 ) + \\sum_{p<q} \\frac{1}{r_{pq}},\n\\label{_auto99} \\tag{99}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where the first summation term represents the standard harmonic\noscillator part and the latter the repulsive interaction between two\nelectrons. Natural units ($\\hbar=c=e=m_e=1$) are used, and $P$ is the\nnumber of particles. This gives us the following expression for the\nlocal energy ($D$ being the number of dimensions)",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto100\"></div>\n\n$$\n\\begin{equation}\n\tE_L = \\frac{1}{\\Psi} \\mathbf{H} \\Psi \n\\label{_auto100} \\tag{100}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto101\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{\\Psi} (\\sum_p^P (-\\frac{1}{2}\\nabla_p^2 + \\frac{1}{2}\\omega^2 r_p^2 ) + \\sum_{p<q} \\frac{1}{r_{pq}}) \\Psi \n\\label{_auto101} \\tag{101}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto102\"></div>\n\n$$\n\\begin{equation} \n\t= -\\frac{1}{2}\\frac{1}{\\Psi} \\sum_p^P \\nabla_p^2 \\Psi \n\t+ \\frac{1}{2}\\omega^2 \\sum_p^P r_p^2 + \\sum_{p<q} \\frac{1}{r_{pq}} \n\\label{_auto102} \\tag{102}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto103\"></div>\n\n$$\n\\begin{equation} \n\t= -\\frac{1}{2}\\frac{1}{\\Psi} \\sum_p^P \\sum_d^D \\frac{\\partial^2 \\Psi}{\\partial x_{pd}^2} + \\frac{1}{2}\\omega^2 \\sum_p^P r_p^2 + \\sum_{p<q} \\frac{1}{r_{pq}} \n\\label{_auto103} \\tag{103}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto104\"></div>\n\n$$\n\\begin{equation} \n\t= \\frac{1}{2} \\sum_p^P \\sum_d^D (-(\\frac{\\partial}{\\partial x_{pd}} \\ln\\Psi)^2 -\\frac{\\partial^2}{\\partial x_{pd}^2} \\ln\\Psi + \\omega^2 x_{pd}^2) + \\sum_{p<q} \\frac{1}{r_{pq}}. \n\\label{_auto104} \\tag{104}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto105\"></div>\n\n$$\n\\begin{equation} \n\\label{_auto105} \\tag{105}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"Letting each visible node in the Boltzmann machine \nrepresent one coordinate of one particle, we obtain",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto106\"></div>\n\n$$\n\\begin{equation}\n\tE_L =\n\t\\frac{1}{2} \\sum_m^M (-(\\frac{\\partial}{\\partial v_m} \\ln\\Psi)^2 -\\frac{\\partial^2}{\\partial v_m^2} \\ln\\Psi + \\omega^2 v_m^2) + \\sum_{p<q} \\frac{1}{r_{pq}},\n\\label{_auto106} \\tag{106}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"where we have that",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto107\"></div>\n\n$$\n\\begin{equation}\n\t\\frac{\\partial}{\\partial x_m} \\ln\\Psi\n\t= - \\frac{1}{\\sigma^2}(x_m - a_m) + \\frac{1}{\\sigma^2} \\sum_n^N \\frac{w_{mn}}{e^{-b_n - \\frac{1}{\\sigma^2}\\sum_i^M x_i w_{in}} + 1} \n\\label{_auto107} \\tag{107}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto108\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial^2}{\\partial x_m^2} \\ln\\Psi\n\t= - \\frac{1}{\\sigma^2} + \\frac{1}{\\sigma^4}\\sum_n^N \\omega_{mn}^2 \\frac{e^{b_n + \\frac{1}{\\sigma^2}\\sum_i^M x_i w_{in}}}{(e^{b_n + \\frac{1}{\\sigma^2}\\sum_i^M x_i w_{in}} + 1)^2}.\n\\label{_auto108} \\tag{108}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"We now have all the expressions neeeded to calculate the gradient of\nthe expected local energy with respect to the RBM parameters\n$\\frac{\\partial \\langle E_L \\rangle}{\\partial \\alpha_i}$.\n\nIf we use $\\Psi = \\sqrt{F_{rbm}}$ we obtain",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto109\"></div>\n\n$$\n\\begin{equation}\n\t\\frac{\\partial}{\\partial x_m} \\ln\\Psi\n\t= - \\frac{1}{2\\sigma^2}(x_m - a_m) + \\frac{1}{2\\sigma^2} \\sum_n^N\n \t\\frac{w_{mn}}{e^{-b_n-\\frac{1}{\\sigma^2}\\sum_i^M x_i w_{in}} + 1}\n\t\n\\label{_auto109} \\tag{109}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"<!-- Equation labels as ordinary links -->\n<div id=\"_auto110\"></div>\n\n$$\n\\begin{equation} \n\t\\frac{\\partial^2}{\\partial x_m^2} \\ln\\Psi\n\t= - \\frac{1}{2\\sigma^2} + \\frac{1}{2\\sigma^4}\\sum_n^N \\omega_{mn}^2 \\frac{e^{b_n + \\frac{1}{\\sigma^2}\\sum_i^M x_i w_{in}}}{(e^{b_n + \\frac{1}{\\sigma^2}\\sum_i^M x_i w_{in}} + 1)^2}.\n\\label{_auto110} \\tag{110}\n\\end{equation}\n$$",
"_____no_output_____"
],
[
"The difference between this equation and the previous one is that we multiply by a factor $1/2$.\n\n\n\n\n\n## Python version for the two non-interacting particles",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\n# 2-electron VMC code for 2dim quantum dot with importance sampling\n# Using gaussian rng for new positions and Metropolis- Hastings \n# Added restricted boltzmann machine method for dealing with the wavefunction\n# RBM code based heavily off of:\n# https://github.com/CompPhysics/ComputationalPhysics2/tree/gh-pages/doc/Programs/BoltzmannMachines/MLcpp/src/CppCode/ob\nfrom math import exp, sqrt\nfrom random import random, seed, normalvariate\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\nfrom matplotlib import cm\nfrom matplotlib.ticker import LinearLocator, FormatStrFormatter\nimport sys\n\n\n\n# Trial wave function for the 2-electron quantum dot in two dims\ndef WaveFunction(r,a,b,w):\n sigma=1.0\n sig2 = sigma**2\n Psi1 = 0.0\n Psi2 = 1.0\n Q = Qfac(r,b,w)\n \n for iq in range(NumberParticles):\n for ix in range(Dimension):\n Psi1 += (r[iq,ix]-a[iq,ix])**2\n \n for ih in range(NumberHidden):\n Psi2 *= (1.0 + np.exp(Q[ih]))\n \n Psi1 = np.exp(-Psi1/(2*sig2))\n\n return Psi1*Psi2\n\n# Local energy for the 2-electron quantum dot in two dims, using analytical local energy\ndef LocalEnergy(r,a,b,w):\n sigma=1.0\n sig2 = sigma**2\n locenergy = 0.0\n \n Q = Qfac(r,b,w)\n\n for iq in range(NumberParticles):\n for ix in range(Dimension):\n sum1 = 0.0\n sum2 = 0.0\n for ih in range(NumberHidden):\n sum1 += w[iq,ix,ih]/(1+np.exp(-Q[ih]))\n sum2 += w[iq,ix,ih]**2 * np.exp(Q[ih]) / (1.0 + np.exp(Q[ih]))**2\n \n dlnpsi1 = -(r[iq,ix] - a[iq,ix]) /sig2 + sum1/sig2\n dlnpsi2 = -1/sig2 + sum2/sig2**2\n locenergy += 0.5*(-dlnpsi1*dlnpsi1 - dlnpsi2 + r[iq,ix]**2)\n \n if(interaction==True):\n for iq1 in range(NumberParticles):\n for iq2 in range(iq1):\n distance = 0.0\n for ix in range(Dimension):\n distance += (r[iq1,ix] - r[iq2,ix])**2\n \n locenergy += 1/sqrt(distance)\n \n return locenergy\n\n# Derivate of wave function ansatz as function of variational parameters\ndef DerivativeWFansatz(r,a,b,w):\n \n sigma=1.0\n sig2 = sigma**2\n \n Q = Qfac(r,b,w)\n \n WfDer = np.empty((3,),dtype=object)\n WfDer = [np.copy(a),np.copy(b),np.copy(w)]\n \n WfDer[0] = (r-a)/sig2\n WfDer[1] = 1 / (1 + np.exp(-Q))\n \n for ih in range(NumberHidden):\n WfDer[2][:,:,ih] = w[:,:,ih] / (sig2*(1+np.exp(-Q[ih])))\n \n return WfDer\n\n# Setting up the quantum force for the two-electron quantum dot, recall that it is a vector\ndef QuantumForce(r,a,b,w):\n\n sigma=1.0\n sig2 = sigma**2\n \n qforce = np.zeros((NumberParticles,Dimension), np.double)\n sum1 = np.zeros((NumberParticles,Dimension), np.double)\n \n Q = Qfac(r,b,w)\n \n for ih in range(NumberHidden):\n sum1 += w[:,:,ih]/(1+np.exp(-Q[ih]))\n \n qforce = 2*(-(r-a)/sig2 + sum1/sig2)\n \n return qforce\n \ndef Qfac(r,b,w):\n Q = np.zeros((NumberHidden), np.double)\n temp = np.zeros((NumberHidden), np.double)\n \n for ih in range(NumberHidden):\n temp[ih] = (r*w[:,:,ih]).sum()\n \n Q = b + temp\n \n return Q\n \n# Computing the derivative of the energy and the energy \ndef EnergyMinimization(a,b,w):\n\n NumberMCcycles= 10000\n # Parameters in the Fokker-Planck simulation of the quantum force\n D = 0.5\n TimeStep = 0.05\n # positions\n PositionOld = np.zeros((NumberParticles,Dimension), np.double)\n PositionNew = np.zeros((NumberParticles,Dimension), np.double)\n # Quantum force\n QuantumForceOld = np.zeros((NumberParticles,Dimension), np.double)\n QuantumForceNew = np.zeros((NumberParticles,Dimension), np.double)\n\n # seed for rng generator \n seed()\n energy = 0.0\n DeltaE = 0.0\n\n EnergyDer = np.empty((3,),dtype=object)\n DeltaPsi = np.empty((3,),dtype=object)\n DerivativePsiE = np.empty((3,),dtype=object)\n EnergyDer = [np.copy(a),np.copy(b),np.copy(w)]\n DeltaPsi = [np.copy(a),np.copy(b),np.copy(w)]\n DerivativePsiE = [np.copy(a),np.copy(b),np.copy(w)]\n for i in range(3): EnergyDer[i].fill(0.0)\n for i in range(3): DeltaPsi[i].fill(0.0)\n for i in range(3): DerivativePsiE[i].fill(0.0)\n\n \n #Initial position\n for i in range(NumberParticles):\n for j in range(Dimension):\n PositionOld[i,j] = normalvariate(0.0,1.0)*sqrt(TimeStep)\n wfold = WaveFunction(PositionOld,a,b,w)\n QuantumForceOld = QuantumForce(PositionOld,a,b,w)\n\n #Loop over MC MCcycles\n for MCcycle in range(NumberMCcycles):\n #Trial position moving one particle at the time\n for i in range(NumberParticles):\n for j in range(Dimension):\n PositionNew[i,j] = PositionOld[i,j]+normalvariate(0.0,1.0)*sqrt(TimeStep)+\\\n QuantumForceOld[i,j]*TimeStep*D\n wfnew = WaveFunction(PositionNew,a,b,w)\n QuantumForceNew = QuantumForce(PositionNew,a,b,w)\n \n GreensFunction = 0.0\n for j in range(Dimension):\n GreensFunction += 0.5*(QuantumForceOld[i,j]+QuantumForceNew[i,j])*\\\n (D*TimeStep*0.5*(QuantumForceOld[i,j]-QuantumForceNew[i,j])-\\\n PositionNew[i,j]+PositionOld[i,j])\n \n GreensFunction = exp(GreensFunction)\n ProbabilityRatio = GreensFunction*wfnew**2/wfold**2\n #Metropolis-Hastings test to see whether we accept the move\n if random() <= ProbabilityRatio:\n for j in range(Dimension):\n PositionOld[i,j] = PositionNew[i,j]\n QuantumForceOld[i,j] = QuantumForceNew[i,j]\n wfold = wfnew\n #print(\"wf new: \", wfnew)\n #print(\"force on 1 new:\", QuantumForceNew[0,:])\n #print(\"pos of 1 new: \", PositionNew[0,:])\n #print(\"force on 2 new:\", QuantumForceNew[1,:])\n #print(\"pos of 2 new: \", PositionNew[1,:])\n DeltaE = LocalEnergy(PositionOld,a,b,w)\n DerPsi = DerivativeWFansatz(PositionOld,a,b,w)\n \n DeltaPsi[0] += DerPsi[0]\n DeltaPsi[1] += DerPsi[1]\n DeltaPsi[2] += DerPsi[2]\n \n energy += DeltaE\n\n DerivativePsiE[0] += DerPsi[0]*DeltaE\n DerivativePsiE[1] += DerPsi[1]*DeltaE\n DerivativePsiE[2] += DerPsi[2]*DeltaE\n \n # We calculate mean values\n energy /= NumberMCcycles\n DerivativePsiE[0] /= NumberMCcycles\n DerivativePsiE[1] /= NumberMCcycles\n DerivativePsiE[2] /= NumberMCcycles\n DeltaPsi[0] /= NumberMCcycles\n DeltaPsi[1] /= NumberMCcycles\n DeltaPsi[2] /= NumberMCcycles\n EnergyDer[0] = 2*(DerivativePsiE[0]-DeltaPsi[0]*energy)\n EnergyDer[1] = 2*(DerivativePsiE[1]-DeltaPsi[1]*energy)\n EnergyDer[2] = 2*(DerivativePsiE[2]-DeltaPsi[2]*energy)\n return energy, EnergyDer\n\n\n#Here starts the main program with variable declarations\nNumberParticles = 2\nDimension = 2\nNumberHidden = 2\n\ninteraction=False\n\n# guess for parameters\na=np.random.normal(loc=0.0, scale=0.001, size=(NumberParticles,Dimension))\nb=np.random.normal(loc=0.0, scale=0.001, size=(NumberHidden))\nw=np.random.normal(loc=0.0, scale=0.001, size=(NumberParticles,Dimension,NumberHidden))\n# Set up iteration using stochastic gradient method\nEnergy = 0\nEDerivative = np.empty((3,),dtype=object)\nEDerivative = [np.copy(a),np.copy(b),np.copy(w)]\n# Learning rate eta, max iterations, need to change to adaptive learning rate\neta = 0.001\nMaxIterations = 50\niter = 0\nnp.seterr(invalid='raise')\nEnergies = np.zeros(MaxIterations)\nEnergyDerivatives1 = np.zeros(MaxIterations)\nEnergyDerivatives2 = np.zeros(MaxIterations)\n\nwhile iter < MaxIterations:\n Energy, EDerivative = EnergyMinimization(a,b,w)\n agradient = EDerivative[0]\n bgradient = EDerivative[1]\n wgradient = EDerivative[2]\n a -= eta*agradient\n b -= eta*bgradient \n w -= eta*wgradient \n Energies[iter] = Energy\n print(\"Energy:\",Energy)\n #EnergyDerivatives1[iter] = EDerivative[0] \n #EnergyDerivatives2[iter] = EDerivative[1]\n #EnergyDerivatives3[iter] = EDerivative[2] \n\n\n iter += 1\n\n#nice printout with Pandas\nimport pandas as pd\nfrom pandas import DataFrame\npd.set_option('max_columns', 6)\ndata ={'Energy':Energies}#,'A Derivative':EnergyDerivatives1,'B Derivative':EnergyDerivatives2,'Weights Derivative':EnergyDerivatives3}\n\nframe = pd.DataFrame(data)\nprint(frame)",
"Energy: 1.9999776037459076\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
ec8b365105584cf44d5ad9012f99c7f380165263 | 464,876 | ipynb | Jupyter Notebook | demo/03-ap-threshold_vs_bias.ipynb | ehua7365/bn3d | b2ab7411c32c836fe5d0e48900461c1911408774 | [
"MIT"
] | null | null | null | demo/03-ap-threshold_vs_bias.ipynb | ehua7365/bn3d | b2ab7411c32c836fe5d0e48900461c1911408774 | [
"MIT"
] | null | null | null | demo/03-ap-threshold_vs_bias.ipynb | ehua7365/bn3d | b2ab7411c32c836fe5d0e48900461c1911408774 | [
"MIT"
] | null | null | null | 623.158177 | 64,308 | 0.940498 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom matplotlib.lines import Line2D\nfrom matplotlib.backends.backend_pdf import PdfPages\nimport os\nimport glob\nimport itertools\nimport pandas as pd\n\nfrom panqec.analysis import get_results_df, get_thresholds_df\nfrom panqec.plots._threshold import detailed_plot, update_plot, plot_crossing_collapse, plot_combined_threshold_vs_bias, xyz_sector_plot\nfrom panqec.plots._hashing_bound import get_hashing_bound",
"_____no_output_____"
]
],
[
[
"# Load results",
"_____no_output_____"
]
],
[
[
"dir_path = os.getcwd()\n\nexperiments1 = [\"det_rhombic_bposd_xzzx_xbias\", \"det_rhombic_bposd_undef_xbias\", \"det_unrot_bposd_xzzx_zbias\", \"det_unrot_bposd_undef_zbias\"]\n# experiments = [\"det_unrot_bposd_xzzx_zbias\", \"det_rot_bposd_undef_zbias\"]\nexperiments2 = ['det_xcube_bposd_undef_zbias', 'det_xcube_bposd_xzzx_zbias']\nexperiments = experiments1 + experiments2\n\nauthor1 = 'eric'\nauthor2 = 'arthur'\n\ninput_dir, output_dir, list_jobs = {}, {}, {}\nfor exp_name in experiments1:\n input_dir[exp_name] = os.path.join(dir_path, f\"../results/{author1}/threshold/{exp_name}/inputs\")\n output_dir[exp_name] = os.path.join(dir_path, f\"../results/{author1}/threshold/{exp_name}/results\")\n list_jobs[exp_name] = list(map(lambda x: os.path.basename(x).split('.json')[0], os.listdir(input_dir[exp_name])))\n \nfor exp_name in experiments2:\n input_dir[exp_name] = os.path.join(dir_path, f\"../results/{author2}/threshold/{exp_name}/inputs\")\n output_dir[exp_name] = os.path.join(dir_path, f\"../results/{author2}/threshold/{exp_name}/results\")\n list_jobs[exp_name] = list(map(lambda x: os.path.basename(x).split('.json')[0], os.listdir(input_dir[exp_name])))",
"_____no_output_____"
],
[
"results = {exp_name: get_results_df(list_jobs[exp_name], output_dir[exp_name], input_dir[exp_name]) for exp_name in experiments}\n\nfor exp_name in experiments1 + experiments2:\n print(exp_name)\n # Sort by probability\n results[exp_name].sort_values(\"probability\", inplace=True)",
"det_rhombic_bposd_xzzx_xbias\ndet_rhombic_bposd_undef_xbias\ndet_unrot_bposd_xzzx_zbias\ndet_unrot_bposd_undef_zbias\ndet_xcube_bposd_undef_zbias\ndet_xcube_bposd_xzzx_zbias\n"
]
],
[
[
"## Remove some sizes",
"_____no_output_____"
]
],
[
[
"for exp_name in experiments:\n results[exp_name].drop(results[exp_name][results[exp_name]['size'] == (9,9,9)].index, inplace=True)",
"_____no_output_____"
]
],
[
[
"# Analyze results",
"_____no_output_____"
],
[
"### Limit probability range manually",
"_____no_output_____"
]
],
[
[
"exp_name = \"det_xcube_bposd_undef_zbias\"\nproba_range = {'Pauli X0.0455Y0.0455Z0.9091': [0.085, 0.11],\n 'Pauli X0.0161Y0.0161Z0.9677': [0.085, 0.11],\n 'Pauli X0.0050Y0.0050Z0.9901': [0.085, 0.11],\n 'Pauli X0.0000Y0.0000Z1.0000': [0.085, 0.11]}\n\nif exp_name in results.keys():\n for error_model, (lower, upper) in proba_range.items():\n results[exp_name].drop(results[exp_name][(results[exp_name]['error_model'] == error_model) &\n ((results[exp_name]['probability'] > upper) |\n (results[exp_name]['probability'] < lower))].index,\n inplace=True)",
"_____no_output_____"
]
],
[
[
"### Get thresholds",
"_____no_output_____"
]
],
[
[
"thresholds_df, trunc_results_df, params_bs_list = {}, {}, {}\n\nfor exp_name in experiments:\n thresholds_df[exp_name], trunc_results_df[exp_name], params_bs_list[exp_name] = get_thresholds_df(results[exp_name], \n ftol_est=1e-5,\n ftol_std=1e-5,\n maxfev=10000)",
"_____no_output_____"
]
],
[
[
"## Sector plots",
"_____no_output_____"
]
],
[
[
"exp_index = 0\neta_key = 'eta_x'\nyscale = 'linear'\nsave = False\n\nexp_name = experiments[exp_index]\ntitle = exp_name.replace(\"_\", \"-\")\n\nif not os.path.exists(f\"../results/{author1}/images/{title}/\"):\n os.makedirs(f\"../results/{author1}/images/{title}/\")\n \nerror_models = np.sort(np.unique(results[exp_name][\"error_model\"]))\nif eta_key == 'eta_z':\n error_models = np.flip(error_models)\nprint(\"Error models\", error_models)\n \nfor i in range(len(error_models)):\n error_model = error_models[i]\n curr_results = results[exp_name][(results[exp_name]['error_model'] == error_model)]\n \n xlim = (np.min(curr_results[\"probability\"]), np.max(curr_results[\"probability\"]))\n# xlim = (0., 0.5)\n \n detailed_plot(plt, curr_results, error_model, thresholds_df=thresholds_df[exp_name], x_limits=[xlim, xlim, xlim],\n yscale=yscale, eta_key=eta_key, min_y_axis=1e-6)\n \n# detailed_plot(plt, curr_results, error_model, x_limits=[xlim, xlim, xlim],\n# yscale=yscale, eta_key=eta_key, min_y_axis=1e-6)\n\n# Saving\nif save:\n with PdfPages(f\"../results/{author}/images/{title}/sectors-{title}-{yscale}.pdf\") as pdf:\n for fig in range(1, plt.figure().number):\n pdf.savefig(fig)",
"Error models ['Deformed Rhombic Pauli X0.3333Y0.3333Z0.3333'\n 'Deformed Rhombic Pauli X0.7500Y0.1250Z0.1250'\n 'Deformed Rhombic Pauli X0.9091Y0.0455Z0.0455'\n 'Deformed Rhombic Pauli X0.9677Y0.0161Z0.0161'\n 'Deformed Rhombic Pauli X0.9901Y0.0050Z0.0050'\n 'Deformed Rhombic Pauli X0.9990Y0.0005Z0.0005'\n 'Deformed Rhombic Pauli X1.0000Y0.0000Z0.0000']\n"
]
],
[
[
"## XYZ sectors plot",
"_____no_output_____"
]
],
[
[
"# exp_index = 1\n# eta_key = 'eta_x'\n# yscale = 'linear'\n# save = False\n\n# exp_name = experiments[exp_index]\n# title = exp_name.replace(\"_\", \"-\")\n \n# if 'rot' or 'lay' in title:\n# if not os.path.exists(f\"../results/eric/images/{title}/\"):\n# os.makedirs(f\"../results/eric/images/{title}/\")\n\n# error_models = np.unique(results[exp_name]['error_model'])\n# print(\"Error models\", error_models)\n\n# for i in range(len(error_models)):\n# error_model = error_models[i]\n# curr_results = results[exp_name][(results[exp_name]['error_model'] == error_model)]\n\n# xlim = (np.min(curr_results[\"probability\"]), np.max(curr_results[\"probability\"]))\n# # xlim = (0, 0.5)\n\n# xyz_sector_plot(plt, curr_results, error_model, thresholds_df=thresholds_df[exp_name], \n# x_limits=[xlim, xlim, xlim, xlim], eta_key=eta_key,\n# yscale=yscale)\n\n# # Saving\n# if save:\n# with PdfPages(f\"../results/eric/images/{title}/xyz-sectors-{title}-{yscale}.pdf\") as pdf:\n# for fig in range(1, plt.figure().number):\n# pdf.savefig(fig)",
"_____no_output_____"
]
],
[
[
"## Unique plot",
"_____no_output_____"
]
],
[
[
"exp_index = 0\neta_key = 'eta_z'\nyscale = 'linear'\nsave = True\n\nexp_name = experiments[exp_index]\ntitle = exp_name.replace(\"_\", \"-\")\n \nif 'rot' or 'lay' in title:\n if not os.path.exists(f\"../results/eric/images/{title}/\"):\n os.makedirs(f\"../results/eric/images/{title}/\")\n\n error_models = np.unique(results[exp_name]['error_model'])\n print(\"Error models\", error_models)\n\n for i in range(len(error_models)):\n error_model = error_models[i]\n curr_results = results[exp_name][(results[exp_name]['error_model'] == error_model)]\n\n xlim = (np.min(curr_results[\"probability\"]), np.max(curr_results[\"probability\"]))\n xlim = (0., 0.5)\n ylim = (0., 0.55)\n\n update_plot(plt, curr_results, error_model,\n xlim=xlim, ylim=ylim, eta_key=eta_key,\n yscale=yscale)\n\n # Saving\n if save:\n with PdfPages(f\"../results/eric/images/{title}/simple-{title}-{yscale}.pdf\") as pdf:\n for fig in range(1, plt.figure().number):\n pdf.savefig(fig)",
"_____no_output_____"
]
],
[
[
"## Plot threshold vs bias",
"_____no_output_____"
],
[
"### Add infinite bias row for deformed models with 50% threshold",
"_____no_output_____"
]
],
[
[
"row_inf_bias = {'error_model': 'Deformed XZZX Pauli X0.0000Y0.0000Z1.0000',\n 'noise_direction': (0, 0, 1),\n 'r_x': 0, 'r_y': 0, 'r_z': 1,\n 'h': 0., 'v': 0.,\n 'eta_x': 0, 'eta_y': 0, 'eta_z': np.inf,\n 'p_th_sd': 0.,\n 'p_th_nearest': 0.5,\n 'p_left': 0.5,\n 'p_right': 0.5,\n 'p_th_fss': 0.5,\n 'p_th_fss_left': 0.5,\n 'p_th_fss_right': 0.5,\n 'p_th_fss_se': 0.,\n 'fss_params': (0.5,0,0,0,0)}\n# thresholds_df['det_rot_bposd_xzzx_zbias'] = thresholds_df['det_rot_bposd_xzzx_zbias'].append(row_inf_bias, ignore_index=True)\n# thresholds_df['det_unrot_bposd_xzzx_zbias'] = thresholds_df['det_unrot_bposd_xzzx_zbias'].append(row_inf_bias, ignore_index=True)\n\nfor exp_name in ['det_xcube_bposd_xzzx_zbias', 'det_unrot_bposd_xzzx_zbias']:\n thresholds_df[exp_name].drop(thresholds_df[exp_name][np.isclose(thresholds_df[exp_name]['r_z'], 1)].index, inplace=True)\n thresholds_df[exp_name] = thresholds_df[exp_name].append(row_inf_bias, ignore_index=True)",
"_____no_output_____"
]
],
[
[
"### Remove unwanted bias ratios",
"_____no_output_____"
]
],
[
[
"# Bias 1000\nfor exp_name in experiments:\n thresholds_df[exp_name].drop(thresholds_df[exp_name][np.isclose(thresholds_df[exp_name]['eta_x'], 1000)].index, inplace=True)\n thresholds_df[exp_name].drop(thresholds_df[exp_name][np.isclose(thresholds_df[exp_name]['eta_z'], 1000)].index, inplace=True)",
"_____no_output_____"
]
],
[
[
"### Get data from XZZX paper",
"_____no_output_____"
]
],
[
[
"csv_file_xzzx = \"../data/xzzx/xzzx_data.csv\"\ncsv_file_css = \"../data/xzzx/css_data.csv\"\n\nxzzx_data = pd.read_csv(csv_file_xzzx)\ncss_data = pd.read_csv(csv_file_css)",
"_____no_output_____"
],
[
"xzzx_data.iloc[-1, xzzx_data.columns.get_loc('bias')] = np.inf\ncss_data.iloc[-1, css_data.columns.get_loc('bias')] = np.inf",
"_____no_output_____"
],
[
"thresholds_df['2d_xzzx'] = pd.DataFrame({'eta_z': xzzx_data['bias'], \n 'p_th_fss': xzzx_data['threshold'],\n 'p_th_fss_left': xzzx_data['threshold'] - xzzx_data['threshold_error'],\n 'p_th_fss_right': xzzx_data['threshold'] + xzzx_data['threshold_error']})\n\n# thresholds_df['2d_css'] = pd.DataFrame({'eta_z': css_data['bias'], \n# 'p_th_fss': css_data['threshold'],\n# 'p_th_fss_left': css_data['threshold'] - css_data['threshold_error'],\n# 'p_th_fss_right': css_data['threshold'] + css_data['threshold_error']})",
"_____no_output_____"
]
],
[
[
"### Plot general",
"_____no_output_____"
]
],
[
[
"cmap = plt.cm.get_cmap('tab10')\nthresholds_df_list = [thresholds_df['det_unrot_bposd_xzzx_zbias'], \n thresholds_df['det_unrot_bposd_undef_zbias']\n thresholds_df['det_rhombic_bposd_xzzx_xbias'], \n thresholds_df['det_rhombic_bposd_undef_xbias'],\n thresholds_df['det_xcube_bposd_xzzx_zbias'], \n thresholds_df['det_xcube_bposd_undef_zbias']]\nplot_combined_threshold_vs_bias(plt, Line2D, thresholds_df_list,\n hashing=True,\n eta_keys=['eta_z', 'eta_z', 'eta_x', 'eta_x', 'eta_z', 'eta_z'], \n labels=['Toric deformed', 'Toric CSS', 'Rhombic deformed', 'Rhombic CSS', 'XCube deformed', 'XCube CSS'],\n colors=[cmap(0), cmap(0), cmap(0), cmap(0), cmap(1), cmap(1)],\n alphas=[1, 1, 1, 1, 1, 1],\n markers=['v', 's', 'o', 'x', '^', 's'],\n linestyles=['-', '--', '-', '--', '-', '--'],\n depolarizing_labels=[True, False, True, False, True, False],\n pdf='../figures/general-threshold-vs-bias.png')",
"_____no_output_____"
]
],
[
[
"## Plot toric",
"_____no_output_____"
]
],
[
[
"cmap = plt.cm.get_cmap('tab10')\nthresholds_df_list = [thresholds_df['det_unrot_bposd_xzzx_zbias'], \n thresholds_df['det_unrot_bposd_undef_zbias']]\nplot_combined_threshold_vs_bias(plt, Line2D, thresholds_df_list,\n hashing=True,\n eta_keys=['eta_z', 'eta_z'], \n labels=['Toric deformed', 'Toric CSS'],\n colors=[cmap(0), cmap(0)],\n alphas=[1, 1],\n markers=['o', 'x'],\n linestyles=['-', '--'],\n depolarizing_labels=[True, False],\n pdf='../figures/cubic-threshold-vs-bias.png')",
"_____no_output_____"
]
],
[
[
"## Plot rhombic",
"_____no_output_____"
]
],
[
[
"cmap = plt.cm.get_cmap('tab10')\nthresholds_df_list = [thresholds_df['det_rhombic_bposd_xzzx_xbias'], \n thresholds_df['det_rhombic_bposd_undef_xbias']]\nplot_combined_threshold_vs_bias(plt, Line2D, thresholds_df_list,\n hashing=True,\n eta_keys=['eta_x', 'eta_x'], \n labels=['Rhombic deformed', 'Rhombic CSS'],\n colors=[cmap(1), cmap(1)],\n alphas=[1, 1],\n markers=['o', 'x'],\n linestyles=['-', '--'],\n depolarizing_labels=[True, False],\n pdf='../figures/rhombic-threshold-vs-bias.png')",
"_____no_output_____"
]
],
[
[
"### Plot X-Cube",
"_____no_output_____"
]
],
[
[
"cmap = plt.cm.get_cmap('tab10')\nthresholds_df_list = [thresholds_df['det_xcube_bposd_xzzx_zbias'], \n thresholds_df['det_xcube_bposd_undef_zbias']]\nplot_combined_threshold_vs_bias(plt, Line2D, thresholds_df_list,\n hashing=True,\n eta_keys=['eta_z', 'eta_z'], \n labels=['XCube deformed', 'XCube CSS'],\n colors=[cmap(3), cmap(3)],\n alphas=[1, 1],\n markers=['o', 'x'],\n linestyles=['-', '--'],\n depolarizing_labels=[True, False],\n pdf='../figures/xcube-threshold-vs-bias.png')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec8b3a8d80497404460712bc406975951abcb3c5 | 98,295 | ipynb | Jupyter Notebook | examples/03_PlotSourceDistribution.ipynb | hluebbering/gravitational-waves | 0ef7d1d878690f4f309a9186dadb1a6e7d4e836a | [
"MIT"
] | null | null | null | examples/03_PlotSourceDistribution.ipynb | hluebbering/gravitational-waves | 0ef7d1d878690f4f309a9186dadb1a6e7d4e836a | [
"MIT"
] | null | null | null | examples/03_PlotSourceDistribution.ipynb | hluebbering/gravitational-waves | 0ef7d1d878690f4f309a9186dadb1a6e7d4e836a | [
"MIT"
] | null | null | null | 549.134078 | 55,394 | 0.943263 | [
[
[
"--------------------------------------------\n\n\n# Parameters Distributions\n\nThe first function is legwork.source.Source.plot_source_variables() which can create either 1D or 2D distributions of any subpopulation of sources.\n\nLet’s take it for a spin with a collection of stationary binaries.\n\n",
"_____no_output_____"
]
],
[
[
"from GravitationalWaves import source, utils, visualization\nimport numpy as np\nimport astropy.units as u\nimport matplotlib.pyplot as plt\nfrom astropy.coordinates import SkyCoord",
"_____no_output_____"
],
[
"# create a random collection of sources\nn_values = 15000\nm_1 = np.random.uniform(0, 10, n_values) * u.Msun\nm_2 = np.random.uniform(0, 10, n_values) * u.Msun\ndist = np.random.normal(8, 1.5, n_values) * u.kpc\nf_orb = 10**(np.random.normal(-5, 0.5, n_values)) * u.Hz\necc = 1 - np.random.power(3, n_values)\n\nsources = source.Source(m_1=m_1, m_2=m_2, ecc=ecc, dist=dist, f_orb=f_orb)",
"_____no_output_____"
]
],
[
[
"This function will let you plot any of several parameters (listed in the table below) and work out the units for the axes labels automatically based on the values in the source class.\n\n\nWe can start simple and just plot the orbital frequency distribution for all sources.\n",
"_____no_output_____"
]
],
[
[
"fig, ax = sources.plot_source_variables(xstr=\"f_orb\", disttype=\"kde\", log_scale=True, linewidth=3)",
"_____no_output_____"
]
],
[
[
"But we could also try to see how the detectable population is different from the entire population. Let’s create two frequency KDEs, one for the detectable binaries and another for all of them. For this we will use the which_sources parameter and pass a mask on the SNR.",
"_____no_output_____"
]
],
[
[
"# calculate the SNR\nsnr = sources.get_snr(verbose=True)\n\n# mask detectable binaries\ndetectable = snr > 7\n\n# plot all binaries\nfig, ax = sources.plot_source_variables(xstr=\"f_orb\", disttype=\"kde\", log_scale=True, linewidth=3,\n show=False, label=\"all binaries\")\n\n# plot all binaries\nfig, ax = sources.plot_source_variables(xstr=\"f_orb\", disttype=\"kde\", log_scale=True, linewidth=3, fig=fig,\n ax=ax, which_sources=detectable, label=\"detectable binaries\",\n show=False)\n\nax.legend()\n\nplt.show()",
"Calculating SNR for 15000 sources\n\t0 sources have already merged\n\t15000 sources are stationary\n\t\t2786 sources are stationary and circular\n\t\t12214 sources are stationary and eccentric\n"
]
],
[
[
"Here’s we can see that the distribution is shifted to higher frequencies for detectable binaries which makes sense since these are easier to detect.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ec8b444ce5367cf6bb198a6d0c0d3d68d97d60c3 | 2,286 | ipynb | Jupyter Notebook | gender-bias.ipynb | kandikits/gender-bias | e8e0a1fd7102d4777ba0e85768be368c0e72ed65 | [
"MIT"
] | null | null | null | gender-bias.ipynb | kandikits/gender-bias | e8e0a1fd7102d4777ba0e85768be368c0e72ed65 | [
"MIT"
] | null | null | null | gender-bias.ipynb | kandikits/gender-bias | e8e0a1fd7102d4777ba0e85768be368c0e72ed65 | [
"MIT"
] | null | null | null | 30.078947 | 197 | 0.609361 | [
[
[
"# To run this you need to have the 'genderbias' package installed\n\nfrom genderbias import ALL_SCANNED_DETECTORS, Document\nimport json, os",
"_____no_output_____"
],
[
"# Document can load text inline, as shown here\n#inline_example = Document(\"I thoroughly recommend PERSON due to their extreme effort in this endeavour.\")\ninput_file_location = \"data/input/letterofRecW\"\ninput_file = open(input_file_location,'r',encoding='utf-8')\ntext = input_file.read()\ninput_file.close()\nprint(\"input is read from location :\",os.path.abspath(input_file_location))\ninline_example = Document(text)\nreports_json = [detector().get_report(inline_example).to_dict() for key, detector in ALL_SCANNED_DETECTORS.items() if key in [\"FemaleDetector\",\"GenderedWordDetector\",\"MaleDetector\"]]\n#print(json.dumps(reports_json))\nout_file_location = \"data/output/gender-biased-words.txt\"\nwith open(out_file_location, 'w',encoding='utf-8') as f:\n f.write(json.dumps(reports_json))\n print(\"output is written to the location :\",os.path.abspath(out_file_location))",
"input is read from location : D:\\kandikits\\gender-bias-master\\data\\input\\letterofRecW\noutput is written to the location : D:\\kandikits\\gender-bias-master\\data\\output\\gender-biased-words.txt\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ec8b46d3fa61bc064aaad85f652b3ad4f67edbc0 | 21,493 | ipynb | Jupyter Notebook | exercises/florian/moutain_car/n-step.ipynb | NLeSC/reinforcement-learning-course | e800a7b7b9ada02f8d6b0438c4eb26a4eca8f31d | [
"Apache-2.0"
] | 2 | 2019-05-22T11:24:53.000Z | 2019-12-20T09:30:16.000Z | exercises/florian/moutain_car/n-step.ipynb | NLeSC/reinforcement-learning-course | e800a7b7b9ada02f8d6b0438c4eb26a4eca8f31d | [
"Apache-2.0"
] | null | null | null | exercises/florian/moutain_car/n-step.ipynb | NLeSC/reinforcement-learning-course | e800a7b7b9ada02f8d6b0438c4eb26a4eca8f31d | [
"Apache-2.0"
] | 1 | 2019-04-02T13:11:04.000Z | 2019-04-02T13:11:04.000Z | 48.298876 | 1,398 | 0.617643 | [
[
[
"import numpy as np\nimport gym\nfrom sklearn.linear_model import SGDRegressor\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.pipeline import FeatureUnion\nfrom gym import wrappers\nfrom datetime import datetime\n\n\nenv = gym.make('MountainCar-v0')",
"_____no_output_____"
],
[
"from mpl_toolkits.mplot3d import Axes3D\nfrom matplotlib import pyplot as plt\nimport matplotlib\n%matplotlib inline",
"_____no_output_____"
],
[
"# copied from lazyprogrammer\ndef plot_cost_to_go(env, estimator, num_tiles=20):\n x = np.linspace(env.observation_space.low[0], env.observation_space.high[0], num=num_tiles)\n y = np.linspace(env.observation_space.low[1], env.observation_space.high[1], num=num_tiles)\n X, Y = np.meshgrid(x, y)\n # both X and Y will be of shape (num_tiles, num_tiles)\n Z = np.apply_along_axis(lambda _: -np.max(estimator.predict(_)), 2, np.dstack([X, Y]))\n # Z will also be of shape (num_tiles, num_tiles)\n\n fig = plt.figure(figsize=(10, 5))\n ax = fig.add_subplot(111, projection='3d')\n surf = ax.plot_surface(X, Y, Z,\n rstride=1, cstride=1, cmap=matplotlib.cm.coolwarm, vmin=-1.0, vmax=1.0)\n ax.set_xlabel('Position')\n ax.set_ylabel('Velocity')\n ax.set_zlabel('Cost-To-Go == -V(s)')\n ax.set_title(\"Cost-To-Go Function\")\n fig.colorbar(surf)\n plt.show()\n\n\ndef plot_running_avg(totalrewards):\n N = len(totalrewards)\n running_avg = np.empty(N)\n for t in range(N):\n running_avg[t] = totalrewards[max(0, t-100):(t+1)].mean()\n plt.plot(running_avg)\n plt.title(\"Running Average\")\n plt.show()\n\n",
"_____no_output_____"
],
[
"#copied from lazyprogrammer\nfrom sklearn.kernel_approximation import RBFSampler\nclass FeatureTransformer:\n def __init__(self, env, n_components=500):\n observation_examples = np.array([env.observation_space.sample() for x in range(10000)])\n scaler = StandardScaler()\n scaler.fit(observation_examples)\n\n # Used to converte a state to a featurizes represenation.\n # We use RBF kernels with different variances to cover different parts of the space\n featurizer = FeatureUnion([\n (\"rbf1\", RBFSampler(gamma=5.0, n_components=n_components)),\n (\"rbf2\", RBFSampler(gamma=2.0, n_components=n_components)),\n (\"rbf3\", RBFSampler(gamma=1.0, n_components=n_components)),\n (\"rbf4\", RBFSampler(gamma=0.5, n_components=n_components))\n ])\n example_features = featurizer.fit_transform(scaler.transform(observation_examples))\n\n self.dimensions = example_features.shape[1]\n self.scaler = scaler\n self.featurizer = featurizer\n\n def transform(self, observation):\n observations = [observation]\n# print(\"observations:\", observations, observations.shape)\n scaled = self.scaler.transform(observations)\n # assert(len(scaled.shape) == 2)\n return self.featurizer.transform(scaled)\n\nfeature_transformer = FeatureTransformer(env)",
"_____no_output_____"
],
[
"np.array([env.observation_space.sample()]).shape\n# feature_transformer.transform(np.array([env.observation_space.sample()]))",
"_____no_output_____"
],
[
"feature_transformer.transform(env.observation_space.sample()).shape",
"_____no_output_____"
],
[
"10%5",
"_____no_output_____"
],
[
"# Holds one SGDRegressor for each action\nclass Model:\n def __init__(self, env, feature_transformer, learning_rate):\n self.env = env\n self.feature_transformer = feature_transformer\n self.learning_rate = learning_rate\n #self.regressors = [SGDRegressor(learning_rate=learning_rate) for _ in range(3)]\n self.regressors = [SGDRegressor() for _ in range(env.action_space.n)]\n# observation_examples = np.array([env.observation_space.sample() for x in range(10000)])\n# transformed_examples = [feature_transformer.transform(o) for o in observation_examples]\n for regressor in self.regressors:\n s = feature_transformer.transform(env.reset())\n regressor.partial_fit(s, [0])\n\n def estimate_state_value(self, state):\n return np.max(self.predict(state)) \n \n def predict(self, state):\n transformed_state = self.feature_transformer.transform(state)\n return [r.predict(transformed_state) for r in self.regressors]\n\n def update(self, state, a, G):\n transformed_state = self.feature_transformer.transform(state)\n self.regressors[a].partial_fit(transformed_state, [G])\n\n def sample_action(self, s, eps):\n # eps = 0\n # Technically, we don't need to do epsilon-greedy\n # because SGDRegressor predicts 0 for all states\n # until they are updated. This works as the\n # \"Optimistic Initial Values\" method, since all\n # the rewards for Mountain Car are -1.\n if np.random.random() < eps:\n return self.env.action_space.sample()\n else:\n return np.argmax(self.predict(s))\n\nlr = 'invscaling' #0.001\nmodel = Model(env, feature_transformer, lr)",
"C:\\Users\\FlorianHuber\\Anaconda3\\lib\\site-packages\\sklearn\\linear_model\\stochastic_gradient.py:128: FutureWarning: max_iter and tol parameters have been added in <class 'sklearn.linear_model.stochastic_gradient.SGDRegressor'> in 0.19. If both are left unset, they default to max_iter=5 and tol=None. If tol is not None, max_iter defaults to max_iter=1000. From 0.21, default max_iter will be 1000, and default tol will be 1e-3.\n \"and default tol will be 1e-3.\" % type(self), FutureWarning)\n"
],
[
"np.argmax(model.predict(env.reset()))",
"_____no_output_____"
],
[
"model.predict(env.reset())",
"_____no_output_____"
],
[
"test =[np.array([-92.96089602]), np.array([-91.99433263]), np.array([-93.03289979])]\nnp.argmax(test)",
"_____no_output_____"
],
[
"test",
"_____no_output_____"
],
[
"# returns a list of states_and_rewards, and the total reward\ndef play_all(model, eps, gamma, n=5, n_games=1):\n # Insert code here\n total_rewards = []\n \n for i in range(n_games):\n total_reward = play_one(eps, gamma, env, model) \n if i%100 == 0: #total_reward > -200:\n print(i, 'total_reward',total_reward)\n total_rewards += [total_reward]\n \n # after code plot some stuff:\n plt.plot(total_rewards)\n plt.title(\"Rewards\")\n plt.show()\n\n plot_running_avg(np.array(total_rewards))\n\n # plot the optimal state-value function\n plot_cost_to_go(env, model)\n\ndef play_one(eps, gamma, env, model):\n state = env.reset()\n history = [] #state, reward, is_done, info\n is_done = False\n \n total_reward = 0\n while not is_done: \n action = model.sample_action(state, eps)\n# print(action)\n history += [env.step(action)]\n state, reward, is_done, _info = history[-1]\n total_reward += reward\n \n model.update(state, action, reward + gamma*model.estimate_state_value(state))\n return total_reward\n",
"_____no_output_____"
],
[
"play_all(model, 0.1, 0.99, n_games=5000)",
"C:\\Users\\FlorianHuber\\Anaconda3\\lib\\site-packages\\sklearn\\linear_model\\stochastic_gradient.py:128: FutureWarning: max_iter and tol parameters have been added in <class 'sklearn.linear_model.stochastic_gradient.SGDRegressor'> in 0.19. If both are left unset, they default to max_iter=5 and tol=None. If tol is not None, max_iter defaults to max_iter=1000. From 0.21, default max_iter will be 1000, and default tol will be 1e-3.\n \"and default tol will be 1e-3.\" % type(self), FutureWarning)\n"
],
[
"env.step?",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8b4d89da3a31a315912ccc053b0a14d0992b55 | 25,775 | ipynb | Jupyter Notebook | 2019-04-16 BioIT World/2019-04-16_06 BioITWorld Classification and Prediction.ipynb | g2nb/workshop-notebooks | 1e22f9569438dd509f3148959ca5b87d78ea5157 | [
"BSD-3-Clause"
] | null | null | null | 2019-04-16 BioIT World/2019-04-16_06 BioITWorld Classification and Prediction.ipynb | g2nb/workshop-notebooks | 1e22f9569438dd509f3148959ca5b87d78ea5157 | [
"BSD-3-Clause"
] | 1 | 2019-05-03T18:53:07.000Z | 2019-05-03T18:54:59.000Z | Integrative_Genomics_Analysis_In_GenePattern/notebooks/2019-04-16_06 BioITWorld Classification and Prediction.ipynb | genepattern/tutorial-materials | 2ab7978fd46343274de999d96e6a65e568dd129f | [
"BSD-3-Clause"
] | 1 | 2022-01-12T20:17:50.000Z | 2022-01-12T20:17:50.000Z | 38.643178 | 751 | 0.585373 | [
[
[
"# Classification and Prediction in GenePattern Notebook\n\nThis notebook will show you how to use k-Nearest Neighbors (kNN) to build a predictor, use it to classify leukemia subtypes, and assess its accuracy in cross-validation.",
"_____no_output_____"
],
[
"### K-nearest-neighbors (KNN)\nKNN classifies an unknown sample by assigning it the phenotype label most frequently represented among the k nearest known samples. \n\nAdditionally, you can select a weighting factor for the 'votes' of the nearest neighbors. For example, one might weight the votes by the reciprocal of the distance between neighbors to give closer neighors a greater vote.",
"_____no_output_____"
],
[
"<h2>1. Log in to GenePattern</h2>",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<h3 style=\"margin-top: 0;\"> Instructions <i class=\"fa fa-info-circle\"></i></h3>\n<ul>\n\t<li>Enter your username and password <i>(if needed)</i> in the GenePattern login cell below.</li>\n\t<li>Click <em>Log into GenePattern</em>.</li>\n\t<li>Alternatively, if you are prompted to Login as your username, just click that button and give it a couple seconds to authenticate.</li>\n</ul>\n</div>",
"_____no_output_____"
]
],
[
[
"# Requires GenePattern Notebook: pip install genepattern-notebook\nimport gp\nimport genepattern\n\n# Username and password removed for security reasons.\ngenepattern.display(genepattern.session.register(\"https://cloud.genepattern.org/gp\", \"\", \"\"))",
"_____no_output_____"
]
],
[
[
"## 2. Run k-Nearest Neighbors Cross Validation",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<h3 style=\"margin-top: 0;\"> Instructions <i class=\"fa fa-info-circle\"></i></h3>\n<ol>\n<li>Select <a class=\"nbtools-markdown-file\" href=\"https://datasets.genepattern.org/data/ccmi_tutorial/2017-12-15/BRCA_HUGO_symbols.preprocessed.gct\">BRCA_HUGO_symbols.preprocessed.gct</a> in the <strong>data filename</strong> field below.\n</li><li>Select <a class=\"nbtools-markdown-file\" href=\"https://datasets.genepattern.org/data/ccmi_tutorial/2017-12-15/BRCA_HUGO_symbols.preprocessed.cls\">BRCA_HUGO_symbols.preprocessed.cls</a> in the <strong>class filename</strong> field.\n </li><li>Click <strong>Run</strong>.</li>\n</ol>\n</div>",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')\n\nknnxvalidation_task = gp.GPTask(genepattern.get_session(0), 'urn:lsid:broad.mit.edu:cancer.software.genepattern.module.analysis:00013')\nknnxvalidation_job_spec = knnxvalidation_task.make_job_spec()\nknnxvalidation_job_spec.set_parameter(\"data.filename\", \"\")\nknnxvalidation_job_spec.set_parameter(\"class.filename\", \"\")\nknnxvalidation_job_spec.set_parameter(\"num.features\", \"10\")\nknnxvalidation_job_spec.set_parameter(\"feature.selection.statistic\", \"0\")\nknnxvalidation_job_spec.set_parameter(\"min.std\", \"\")\nknnxvalidation_job_spec.set_parameter(\"num.neighbors\", \"3\")\nknnxvalidation_job_spec.set_parameter(\"weighting.type\", \"1\")\nknnxvalidation_job_spec.set_parameter(\"distance.measure\", \"1\")\nknnxvalidation_job_spec.set_parameter(\"pred.results.file\", \"<data.filename_basename>.pred.odf\")\nknnxvalidation_job_spec.set_parameter(\"feature.summary.file\", \"<data.filename_basename>.feat.odf\")\ngenepattern.display(knnxvalidation_task)",
"_____no_output_____"
]
],
[
[
"## 3. View a list of features used in the prediction model",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<h3 style=\"margin-top: 0;\"> Instructions <i class=\"fa fa-info-circle\"></i></h3>\n<ol>\n <li>Select the <em>KNNXvalidation</em> job result cell by clicking anywhere in it.\n </li><li>Click on the <i class=\"fa fa-info-circle\"></i> icon next to the <q><filename>.feat.odf</q> file.\n </li><li>Select <q>Send to DataFrame</q>.\n</li><li>You will see a new cell created below the job result cell.\n</li><li>You will see a table of features, descriptions, and the number of times each feature was included in a model in a cross-validation loop.\n</li></ol></div>",
"_____no_output_____"
],
[
"## 4. View prediction results",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<h3 style=\"margin-top: 0;\"> Instructions <i class=\"fa fa-info-circle\"></i></h3>\n<ol>\n <li>For the <q>prediction results file</q> parameter below, click the down arrow in the file input box.\n </li><li>Right click the <q>BRCA_HUGO_symbols.preprocessed.pred.odf</q> above and select <q>Copy link address</q>.</li>\n <li>Paste the link into the <q>Prediction Results File</q> parameter.</li>\n <li>Click <strong><em>Run</em></strong>.</li>\n <li>You will see the prediction results in an interactive viewer.</li>\n</ol>\n</div>\n",
"_____no_output_____"
]
],
[
[
"predictionresultsviewer_task = gp.GPTask(genepattern.session.get(0), 'urn:lsid:broad.mit.edu:cancer.software.genepattern.module.visualizer:00019')\npredictionresultsviewer_job_spec = predictionresultsviewer_task.make_job_spec()\npredictionresultsviewer_job_spec.set_parameter(\"prediction.results.file\", \"\")\ngenepattern.display(predictionresultsviewer_task)",
"_____no_output_____"
]
],
[
[
"<h2>Part 2, Compare classification of Preprocessed data to non-preprocessed data</h2>\n<p>\nBack in the Data Preparation section at the beginning of the workshop, we ran the RNA-Seq read counts data through the VoomNormalize module to preprocess RNA-Seq data into a form suitable for use downstream in other GenePattern analyses originally designed for microarray data. Many of these approaches assume that the data is distributed normally, yet this is not true of RNA-seq read count data. The PreprocessReadCounts module provides one approach to accommodate this. It uses a mean-variance modeling technique ('voom' from the limma Bioconductor package) to transform the dataset to fit an approximation of a normal distribution, with the goal of being able to apply statistical methods and workflows that assume a normal distribution.\n</p>\n<p>\nNow lets take a look at how well our KNNXValidation classifier works if we provide it with the RNA-Seq matrix from before this processing step.\n</p>\n\n<h2>5. Run KNNXValidation on the non-normalized data</h2>\nFor this exercise we want you to compare the results of the classifier we used above to using the same classifier and settings on the BRCA data that has not been run through VoomNormalize to normalize it first.",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<h3 style=\"margin-top: 0;\"> Instructions <i class=\"fa fa-info-circle\"></i></h3>\n<ol>\n <li>Select the blank cell below.</li> \n <li>Click the <q><i class=\"fa fa-th\"></i> Tools</q> button in the toolbar above and select the <em>KNNXValidation</em> module to insert the analysis.</li>\n\t<li>Select <a class=\"nbtools-markdown-file\" href=\"https://datasets.genepattern.org/data/ccmi_tutorial/2018-09-20/BRCA_Dataset.collapsed.gct\" target=\"_blank\">BRCA_Dataset.collapsed.gct</a> in the <strong>data filename</strong> field below. This is the version of the data that has been collapsed to HUGO symbols, but not run through VoomNormalize</li>\n\t<li>Select <a class=\"nbtools-markdown-file\" href=\"https://datasets.genepattern.org/data/ccmi_tutorial/2018-09-20/BRCA_Dataset.cls\" target=\"_blank\">BRCA_Dataset.cls</a> in the <strong>class filename</strong> field.</li>\n\t<li>Click <strong>Run</strong>.</li>\n <li>Examine the results using the PredictionResultsViewer module.</li>\n</ol>\n</div>",
"_____no_output_____"
],
[
"<h3>Discussion</h3>\n\n<p>As you can see, the KNNXvalidation classifier works just as well for the non-normalized data, though there is slightly less confidence in one of the calls. This is a reflection of the fact that from a gene-expression point of view, seperating tumor cells from normal cells is actually a fairly easy distinction to make which is why we us it in these exercises. </p>\n",
"_____no_output_____"
],
[
"## 6. Compare the selected features\n\nWe saw above that using the euclidean distance metric with the pre-normalized BRCA data, that KNNXValidation does not do as good a job classifying our tumor/normal samples. Here we will examine why that might be.\n\n<div class=\"alert alert-info\">\n<h3 style=\"margin-top: 0;\"> Instructions <i class=\"fa fa-info-circle\"></i></h3>\n<ol>\n <li>From Step 5 above, click on <strong>BRCA_Dataset.collapsed.feat.odf</strong> and choose <Strong>Send to Dataframe</Strong></li> \n <li>Move the resulting cell below this one using the down arrow (<i class=\"fa-arrow-down fa\"></i>) in the toolbar.</li>\n\t<li>Repeat these 2 steps for the <strong>BRCA_HUGO_symbols.preprocessed.feat.odf</strong> from the beginning of this notebook.</li>\n\t<li>Compare the lists of features. How many features are in both runs?</li>\n</ol>\n</div>\n\n",
"_____no_output_____"
],
[
"### The Answer (spoiler warning)",
"_____no_output_____"
],
[
"<p class=\"lead\">'MATN2',\n 'COMP',\n 'KLHL29',\n 'SPRY2',\n 'TMEM220',\n 'MME',\n 'INHBA',\n 'SLC50A1',\n 'TNFRSF10D',\n 'NTRK2'</p>",
"_____no_output_____"
],
[
"### Discussion\n\n<p>As you can see, the lists of features for the classifiers for the unprocessed and preprocessed data are not identical, but there is some overlap. Nonetheless each of the classifiers was able to perfectly classify the samples. As mentioned before, this is a reflection of the fact that this is a pretty easy classification problem and there are many genes that could be used to make the class distinction.</p>\n\n<p> </p>\n",
"_____no_output_____"
],
[
"<h3 id=\"Bonus-Content---comparing-the-processed-and-unprocessed-data\">Bonus Content - comparing the processed and unprocessed data</h3>\n\n<p><span style=\"color: inherit ; font-family: inherit\">To try to see the differences in the data sets, you can use the cell below to generate X-Y plots of genes from the two datasets to see how they differ.</span></p>\n\n<p>The cell below will use the pre-computed results from the spoilers above. In addition there is a third plot that shows the unprocessed dataset after it has been log transformed as a third refernce point.</p>\n\n<p> </p>\n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n\nimport numpy as np\nfrom gp.data import GCT\nimport matplotlib.pyplot as plt\n\[email protected]_ui(parameters={\n \"gene_1\": {\n \"name\": \"Gene 1\",\n \"description\": \"First gene for X-Y plots\",\n \"default\": \"MME\",\n \"type\": \"choice\",\n \"choices\":{\"ADAM33\":\"ADAM33\",\n\"ANXA1\":\"ANXA1\",\n\"CCNE2\":\"CCNE2\",\n\"CDCA8\":\"CDCA8\",\n\"CERS2\":\"CERS2\",\n\"COL10A1\":\"COL10A1\",\n\"COMP\":\"COMP\",\n\"CXCL2\":\"CXCL2\",\n\"DMD\":\"DMD\",\n\"EIF2AK1\":\"EIF2AK1\",\n\"FAXDC2\":\"FAXDC2\",\n\"FLVCR1\":\"FLVCR1\",\n\"GGTA1P\":\"GGTA1P\",\n\"HSD17B6\":\"HSD17B6\",\n\"IL11RA\":\"IL11RA\",\n\"INHBA\":\"INHBA\",\n\"KIF26B\":\"KIF26B\",\n\"KLHL29\":\"KLHL29\",\n\"LINC02408\":\"LINC02408\",\n\"LMOD1\":\"LMOD1\",\n\"MAMDC2\":\"MAMDC2\",\n\"MATN2\":\"MATN2\",\n\"MIR3153\":\"MIR3153\",\n\"MME\":\"MME\",\n\"MMP11\":\"MMP11\",\n\"NTRK2\":\"NTRK2\",\n \"RGN\":\"RGN\",\n\"ROBO3\":\"ROBO3\",\n\"SCN4B\":\"SCN4B\",\n\"SLC50A1\":\"SLC50A1\",\n\"SPRY2\":\"SPRY2\",\n\"TDRD10\":\"TDRD10\",\n\"TMEM220\":\"TMEM220\",\n\"TNFRSF10D\":\"TNFRSF10D\",\n\"TSLP\":\"TSLP\",\n\"UBE2T\":\"UBE2T\",\n\"UNC5B\":\"UNC5B\",\n\"VEGFD\":\"VEGFD\",\n\"WISP1\":\"WISP1\"}\n },\n \"gene_2\": {\n \"name\": \"Gene 2\",\n \"description\": \"Second gene for X-Y plots\",\n \"default\": \"SPRY2\",\n \"type\": \"choice\",\n \"choices\":{\"ADAM33\":\"ADAM33\",\n\"ANXA1\":\"ANXA1\",\n\"CCNE2\":\"CCNE2\",\n\"CDCA8\":\"CDCA8\",\n\"CERS2\":\"CERS2\",\n\"COL10A1\":\"COL10A1\",\n\"COMP\":\"COMP\",\n\"CXCL2\":\"CXCL2\",\n\"DMD\":\"DMD\",\n\"EIF2AK1\":\"EIF2AK1\",\n\"FAXDC2\":\"FAXDC2\",\n\"FLVCR1\":\"FLVCR1\",\n\"GGTA1P\":\"GGTA1P\",\n\"HSD17B6\":\"HSD17B6\",\n\"IL11RA\":\"IL11RA\",\n\"INHBA\":\"INHBA\",\n\"KIF26B\":\"KIF26B\",\n\"KLHL29\":\"KLHL29\",\n\"LINC02408\":\"LINC02408\",\n\"LMOD1\":\"LMOD1\",\n\"MAMDC2\":\"MAMDC2\",\n\"MATN2\":\"MATN2\",\n\"MIR3153\":\"MIR3153\",\n\"MME\":\"MME\",\n\"MMP11\":\"MMP11\",\n\"NTRK2\":\"NTRK2\",\n \"RGN\":\"RGN\",\n\"ROBO3\":\"ROBO3\",\n\"SCN4B\":\"SCN4B\",\n\"SLC50A1\":\"SLC50A1\",\n\"SPRY2\":\"SPRY2\",\n\"TDRD10\":\"TDRD10\",\n\"TMEM220\":\"TMEM220\",\n\"TNFRSF10D\":\"TNFRSF10D\",\n\"TSLP\":\"TSLP\",\n\"UBE2T\":\"UBE2T\",\n\"UNC5B\":\"UNC5B\",\n\"VEGFD\":\"VEGFD\",\n\"WISP1\":\"WISP1\"}\n },\n \"output_var\": {\n \"hide\": True,\n }\n})\ndef show_X_Y_plots(gene_1, gene_2):\n\n processedInput_GCT = GCT(gp.GPFile(genepattern.get_session(0), \"https://datasets.genepattern.org/data/ccmi_tutorial/2018-09-20/BRCA_HUGO_symbols.preprocessed.gct\"))\n UNprocessedInput_GCT = GCT(gp.GPFile(genepattern.get_session(0), \"https://datasets.genepattern.org/data/ccmi_tutorial/2018-09-20/BRCA_Dataset.collapsed.gct\"))\n ltgct = GCT(gp.GPFile(genepattern.get_session(0), \"https://cloud.genepattern.org/gp/jobResults/82746/BRCA_HUGO_symbols.preprocessed.gct \"))\n\n\n # drop the description from the index\n processedInput_GCT.reset_index(inplace=True)\n processedInput_GCT.set_index('Name', inplace=True)\n del processedInput_GCT['Description']\n\n UNprocessedInput_GCT.reset_index(inplace=True)\n UNprocessedInput_GCT.set_index('Name', inplace=True)\n del UNprocessedInput_GCT['Description']\n\n ltgct.reset_index(inplace=True)\n ltgct.set_index('Name', inplace=True)\n del ltgct['Description']\n ltgct = ltgct.astype(float)\n\n cls2 = gp.GPFile(genepattern.get_session(0), \"https://datasets.genepattern.org/data/ccmi_tutorial/2018-09-20/BRCA_Dataset.cls\")\n\n clsLines = cls2.read().splitlines()\n labels = np.asarray(clsLines[1].split(), dtype=str)[1:] \n classes = np.asarray(clsLines[2].strip('\\n').split(' '), dtype=int)\n\n\n dft = processedInput_GCT.transpose()\n dft['classes'] = [labels[i] for i in classes]\n\n UNdft = UNprocessedInput_GCT.transpose()\n UNdft['classes'] = [labels[i] for i in classes]\n\n lgdft = ltgct.transpose()\n lgdft['classes'] = [labels[i] for i in classes]\n\n\n # First 2 plots are for 2 of the top genes for the preprocessed data\n groups = dft.groupby('classes')\n fig = plt.figure()\n fig.set_size_inches(18,6)\n ax = fig.add_subplot(132)\n #plt.subplots()\n #fig.set_size_inches(12,8)\n ax.margins(0.05) # Optional, just adds 5% padding to the autoscaling\n for name, group in groups:\n ax.plot(group[gene_1], group[gene_2], marker='o', linestyle='', ms=5, label=name)\n ax.legend()\n plt.title(\"Preprocessed Data\")\n\n UNgroups = UNdft.groupby('classes')\n\n #fig2, ax2 = plt.subplots()\n #fig2.set_size_inches(12,8)\n ax2 = fig.add_subplot(131)\n ax2.margins(0.05) # Optional, just adds 5% padding to the autoscaling\n for name, group in UNgroups:\n ax2.plot(group[gene_1], group[gene_2], marker='o', linestyle='', ms=5, label=name)\n ax2.legend()\n plt.title(\"Un-preprocessed Data\")\n\n LGgroups = lgdft.groupby('classes')\n\n #fig3, ax3 = plt.subplots()\n #fig3.set_size_inches(12,8)\n ax3 = fig.add_subplot(133)\n ax3.margins(0.05) # Optional, just adds 5% padding to the autoscaling\n for name, group in LGgroups:\n ax3.plot(group[gene_1], group[gene_2], marker='o', linestyle='', ms=5, label=name)\n ax3.legend()\n \n\n ax.set_xlabel(gene_1)\n ax.set_ylabel(gene_2)\n ax2.set_xlabel(gene_1)\n ax2.set_ylabel(gene_2)\n ax3.set_xlabel(gene_1)\n ax3.set_ylabel(gene_2)\n\n ax.set_title(\"Preprocessed Data\")\n ax2.set_title(\"Unprocessed\")\n\n ax3.set_title(\"Unprocessed - log transformed\")\n \n plt.show()\n",
"_____no_output_____"
]
],
[
[
"<h3>Bonus - Discussion</h3>\n\n<p>As you can see from the plots, the processed (voom) data seperates the classes much more cleanly than the unprocessed data for most gene sets. Comparing it to the log-transformed unprocessed data we see that for many genes, the clusters on the X-Y plots appear a bit tighter, probably due to voom's variance correction.</p>\n",
"_____no_output_____"
],
[
"## References\n\nBreiman, L., Friedman, J. H., Olshen, R. A., & Stone, C. J. 1984. [Classification and regression trees](https://www.amazon.com/Classification-Regression-Wadsworth-Statistics-Probability/dp/0412048418?ie=UTF8&*Version*=1&*entries*=0). Wadsworth & Brooks/Cole Advanced Books & Software, Monterey, CA.\n\nGolub, T.R., Slonim, D.K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J.P., Coller, H., Loh, M., Downing, J.R., Caligiuri, M.A., Bloomfield, C.D., and Lander, E.S. 1999. Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression. [Science 286:531-537](http://science.sciencemag.org/content/286/5439/531.long).\n\nLu, J., Getz, G., Miska, E.A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B.L., Mak, R.H., Ferrando, A.A, Downing, J.R., Jacks, T., Horvitz, H.R., Golub, T.R. 2005. MicroRNA expression profiles classify human cancers. [Nature 435:834-838](http://www.nature.com/nature/journal/v435/n7043/full/nature03702.html).\n\nRifkin, R., Mukherjee, S., Tamayo, P., Ramaswamy, S., Yeang, C-H, Angelo, M., Reich, M., Poggio, T., Lander, E.S., Golub, T.R., Mesirov, J.P. 2003. An Analytical Method for Multiclass Molecular Cancer Classification. [SIAM Review 45(4):706-723](http://epubs.siam.org/doi/abs/10.1137/S0036144502411986).\n\nSlonim, D.K., Tamayo, P., Mesirov, J.P., Golub, T.R., Lander, E.S. 2000. Class prediction and discovery using gene expression data. In [Proceedings of the Fourth Annual International Conference on Computational Molecular Biology (RECOMB)](http://dl.acm.org/citation.cfm?id=332564). ACM Press, New York. pp. 263-272.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ec8b54b5ead9015ff1be1c89ca3dbc0d6ca0cf14 | 170,093 | ipynb | Jupyter Notebook | notebooks/ZernikePolynomials.ipynb | davidthomas5412/ForwardModelingLSSTDonuts | bd2d85bd1da998db42bdce734e19b4af2374552a | [
"MIT"
] | null | null | null | notebooks/ZernikePolynomials.ipynb | davidthomas5412/ForwardModelingLSSTDonuts | bd2d85bd1da998db42bdce734e19b4af2374552a | [
"MIT"
] | null | null | null | notebooks/ZernikePolynomials.ipynb | davidthomas5412/ForwardModelingLSSTDonuts | bd2d85bd1da998db42bdce734e19b4af2374552a | [
"MIT"
] | null | null | null | 193.72779 | 25,356 | 0.885598 | [
[
[
"# Zernike Polynomials\n*David Thomas and Emily Li 2019/06/27*",
"_____no_output_____"
],
[
"**Summary:** We usually decompose donut images and wavefronts into a special basis: the [zernike polynomials](https://en.wikipedia.org/wiki/Zernike_polynomials). This orthonormal set of polynomials is commonly used in optics, the low order terms correspond to common aberrations. For LSST we use the annular version of these polynomials which have a hole in the center. In this lesson we will play around with code to evaluate these polynomials and better understand their properties.",
"_____no_output_____"
],
[
"**Table of Contents:**\n- [Plotting](#Plotting)\n- [Orthogonality](#Orthogonality)\n- [Decomposition](#Decomposition)",
"_____no_output_____"
],
[
"### Plotting\nWe import the key packages. The `%matplotlib inline` command allows us to display figures in the notebook.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline \nimport matplotlib.pyplot as plt\nimport numpy as np\nimport galsim",
"_____no_output_____"
]
],
[
[
"We use the galsim.zernike.Zernike class to manipulate these polynomials. Let's start by plotting the 4th Zernike polynomial (which corresponds to astigmatism with our indexing). First we have to create a galsim.zernike.Zernike object.",
"_____no_output_____"
]
],
[
[
"zernikes = np.zeros(22)\nindex = 6\nzernikes[index] = 1\nzern = galsim.zernike.Zernike(zernikes)",
"_____no_output_____"
]
],
[
[
"Let's see what member variables and methods this object has.",
"_____no_output_____"
]
],
[
[
"help(zern)",
"Help on Zernike in module galsim.zernike object:\n\nclass Zernike(builtins.object)\n | A class to represent a Zernike polynomial series\n | (http://en.wikipedia.org/wiki/Zernike_polynomials#Zernike_polynomials).\n | \n | Zernike polynomials form an orthonormal basis over the unit circle. The convention used here is\n | for the normality constant to equal the area of integration, which is pi for the unit circle.\n | I.e.,\n | \n | \\int_{unit circle} Z_i Z_j dA = \\pi \\delta_{i, j}.\n | \n | Two generalizations of the unit circle Zernike polynomials are also available in this class:\n | annular Zernike polynomials, and polynomials defined over non-unit-radius circles.\n | \n | Annular Zernikes are orthonormal over an annulus instead of a circle (see Mahajan, J. Opt. Soc.\n | Am. 71, 1, (1981)). Similarly, the non-unit-radius polynomials are orthonormal over a region\n | with outer radius not equal to 1. Taken together, these generalizations yield the\n | orthonormality condition:\n | \n | \\int_{annulus} Z_i Z_j dA = \\pi (R_outer^2 - R_inner^2) \\delta_{i, j}\n | \n | where 0 <= R_inner < R_outer indicate the inner and outer radii of the annulus over which the\n | polynomials are orthonormal.\n | \n | The indexing convention for i and j above is that from Noll, J. Opt. Soc. Am. 66, 207-211(1976).\n | Note that the Noll indices begin at 1; there is no Z_0. Because of this, the series\n | coefficients argument `coef` effectively begins with `coef[1]` (`coef[0]` is ignored). This\n | convention is used consistently throughout GalSim, e.g., `OpticalPSF`, `OpticalScreen`,\n | `zernikeRotMatrix`, and `zernikeBasis`.\n | \n | As an example, the first few Zernike polynomials in terms of Cartesian coordinates x and y are\n | \n | Noll index | polynomial\n | --------------------------\n | 1 | 1\n | 2 | 2 x\n | 3 | 2 y\n | 4 | sqrt(3) (2 (x^2 + y^2) - 1)\n | \n | @param coef Zernike series coefficients. Note that coef[i] corresponds to Z_i under the\n | Noll index convention, and coef[0] is ignored. (I.e., coef[1] is 'piston',\n | coef[4] is 'defocus', ...)\n | @param R_outer Outer radius. [default: 1.0]\n | @param R_inner Inner radius. [default: 0.0]\n | \n | Methods defined here:\n | \n | __eq__(self, other)\n | Return self==value.\n | \n | __hash__(self)\n | Return hash(self).\n | \n | __init__(self, coef, R_outer=1.0, R_inner=0.0)\n | Initialize self. See help(type(self)) for accurate signature.\n | \n | __repr__(self)\n | Return repr(self).\n | \n | evalCartesian(self, x, y)\n | Evaluate this Zernike polynomial series at Cartesian coordinates x and y.\n | \n | @param x x-coordinate of evaluation points. Can be list-like.\n | @param y y-coordinate of evaluation points. Can be list-like.\n | @returns Series evaluations as numpy array.\n | \n | evalCartesianGrad(self, x, y)\n | \n | evalPolar(self, rho, theta)\n | Evaluate this Zernike polynomial series at polar coordinates rho and theta.\n | \n | @param rho radial coordinate of evaluation points. Can be list-like.\n | @param theta azimuthal coordinate in radians (or as Angle object) of evaluation points.\n | Can be list-like.\n | @returns Series evaluations as numpy array.\n | \n | gradX = <galsim.utilities.lazy_property object>\n | gradY = <galsim.utilities.lazy_property object>\n | rotate(self, theta)\n | Return new Zernike polynomial series rotated by angle theta. For example:\n | \n | >>> Z = Zernike(coefs)\n | >>> Zrot = Z.rotate(theta)\n | >>> Z.evalPolar(r, th) == Zrot.evalPolar(r, th + theta)\n | \n | @param theta Angle in radians.\n | @returns A new Zernike object.\n | \n | ----------------------------------------------------------------------\n | Data descriptors defined here:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n\n"
]
],
[
[
"We want to use the evalCartesian method. Let's start by iterating over all the values in a 2D grid with each dimension ranging from -1 to 1.",
"_____no_output_____"
]
],
[
[
"Z = np.zeros((100,100))\nfor i,x in enumerate(np.linspace(-1,1,100)):\n for j,y in enumerate(np.linspace(-1,1,100)):\n Z[j,i] = zern.evalCartesian(x,y)\n \nplt.figure()\nplt.title('First Attempt at Zernike Plot')\nplt.imshow(Z, origin='lower')\nplt.xticks([0,50,99],['-1','0','1'])\nplt.yticks([0,50,99],['-1','0','1'])\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"Wait ... this is a box! Aren't these polynomials supposed to be defined on the unit circle? Yes and, as we will see, they are only orthonormal over the unit circle. galsim.zernike.Zernike doesn't care, if we ask it to evaluate the polynomial at a location, it will evaluate the polynomial at that location. But we should create a mask so that only the relevant domain is defined. ",
"_____no_output_____"
]
],
[
[
"Z = np.zeros((100,100))\nfor i,x in enumerate(np.linspace(-1,1,100)):\n for j,y in enumerate(np.linspace(-1,1,100)):\n r = np.sqrt(x ** 2 + y ** 2)\n if abs(r) <= 1:\n Z[j,i] = zern.evalCartesian(x,y)\n else:\n Z[j,i] = np.nan\n \nplt.figure()\nplt.title('Second Attempt at Zernike Plot')\nplt.imshow(Z, origin='lower')\nplt.xticks([0,50,99],['-1','0','1'])\nplt.yticks([0,50,99],['-1','0','1'])\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"Woo! This looks exactly like $Z_2^{-2}$ from [wikipedia](https://en.wikipedia.org/wiki/Zernike_polynomials). However, we can make this a lot faster, and easier to read, if we use [vectorization](https://hackernoon.com/speeding-up-your-code-2-vectorizing-the-loops-with-numpy-e380e939bed3). Note that evalCartesian accepts list-like arguments, which is short for numpy.ndarrays. Here is the vectorized version of the code above.",
"_____no_output_____"
]
],
[
[
"X,Y = np.meshgrid(np.linspace(-1,1,100), np.linspace(-1,1,100))\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.abs(R) < 1\nZ = zern.evalCartesian(X,Y) \nZ[~mask] = np.nan\n\nplt.figure()\nplt.title('Second Attempt at Zernike Plot')\nplt.imshow(Z, origin='lower')\nplt.xticks([0,50,99],['-1','0','1'])\nplt.yticks([0,50,99],['-1','0','1'])\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"There is one more piece to add. LSST has an annular aperture. Therefore we want to use the *annular* zernike polynomials. Perhaps you noticed that galsim.zernike.Zernike takes R_outer and R_inner as keyword arguments. For LSST the inner radius is 0.61, so we just need to change this and update the mask accordingly.",
"_____no_output_____"
]
],
[
[
"obscuration = 0.61\nzern = galsim.zernike.Zernike(zernikes, R_inner=obscuration)\n\nX,Y = np.meshgrid(np.linspace(-1,1,100), np.linspace(-1,1,100))\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.logical_and(np.abs(R) < 1, np.abs(R) > obscuration)\nZ = zern.evalCartesian(X,Y) \nZ[~mask] = np.nan\n\nplt.figure()\nplt.title('Third Attempt at Zernike Plot')\nplt.imshow(Z, origin='lower')\nplt.xticks([0,50,99],['-1','0','1'])\nplt.yticks([0,50,99],['-1','0','1'])\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"**Trick question:** Is this the same as just applying the mask to our previous Z's? Hint: No. You can plot both of them and compare. Can you think of why they are different? Hint: look at the title of the next section (Orthogonality). ",
"_____no_output_____"
],
[
"**Problem 1:** Can you plot zernikes 1-10 in separate subplots and label them with the names provided below. You can look at the [matplotlib documentation](https://matplotlib.org/3.1.0/gallery/lines_bars_and_markers/markevery_demo.html#sphx-glr-gallery-lines-bars-and-markers-markevery-demo-py) to learn how to plot multiple plots within a single figure. Note: Usually in CS things are indexed by 0 but in this case it looks like the first two zernikes are piston.",
"_____no_output_____"
]
],
[
[
"indexToName = {\n 0: 'piston',\n 1: 'piston',\n 2: 'tilt',\n 3: 'tip',\n 4: 'defocus',\n 5: 'oblique astigmatism',\n 6: 'vertical astigmatism',\n 7: 'vertical coma',\n 8: 'horizontal coma',\n 9: 'vertical trefoil',\n 10: 'oblique trefoil',\n}\n\n# TODO: code here",
"_____no_output_____"
]
],
[
[
"### Orthogonality",
"_____no_output_____"
],
[
"A key property of this set of polynomials is that they are orthonormal, and the infinite set of them is complete. This means that we can decompose any continuous function into a set of zernike polynomials. Let's start by confirming that these polynomials are normalized - or that $\\iint Z_i(x,y)Z_i(x,y) d\\Omega = 1$, where $\\Omega$ is the annulus. To do this numerically we just multiply them element-wise and sum them, then divide by the total number of pixels in the annulus. ",
"_____no_output_____"
]
],
[
[
"obscuration = 0.61\nz5 = np.zeros(22)\nz5[5] = 1\nzern = galsim.zernike.Zernike(z5, R_inner=obscuration)\nX,Y = np.meshgrid(np.linspace(-1,1,100), np.linspace(-1,1,100))\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.logical_and(np.abs(R) < 1, np.abs(R) > obscuration)\nZ = zern.evalCartesian(X,Y) \nZ[~mask] = np.nan\n\nplt.figure()\nplt.title(r'$Z_5^2$')\nplt.imshow(Z*Z, origin='lower')\nplt.xticks([0,50,99],['-1','0','1'])\nplt.yticks([0,50,99],['-1','0','1'])\nplt.colorbar()\n\nintegral = np.sum(Z[mask] * Z[mask]) / np.sum(mask)\nprint('Integral = ', integral)",
"Integral = 0.993267951589\n"
]
],
[
[
"**Problem 2:** Can you repeat the calculation above for a few other zernikes?",
"_____no_output_____"
]
],
[
[
"# TODO: code here",
"_____no_output_____"
]
],
[
[
"Now what happens if we change the resolution, do you think this will impact the numerical result? It is dificult to tell without trying it out.",
"_____no_output_____"
]
],
[
[
"obscuration = 0.61\nz5 = np.zeros(22)\nz5[5] = 1\nzern = galsim.zernike.Zernike(z5, R_inner=obscuration)\n\nX,Y = np.meshgrid(np.linspace(-1,1,5), np.linspace(-1,1,5)) # Here is where we are changing the pixel resolution.\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.logical_and(np.abs(R) <=1, np.abs(R) > obscuration)\nZ = zern.evalCartesian(X,Y) \nZ[~mask] = np.nan\n\nplt.figure()\nplt.title(r'$Z_5^2$')\nplt.imshow(Z, origin='lower')\nplt.xticks([0,2,4],['-1','0','1'])\nplt.yticks([0,2,4],['-1','0','1'])\nplt.colorbar()\n\nintegral = np.sum(Z[mask] * Z[mask]) / np.sum(mask)\nprint('Integral = ', integral)",
"Integral = 0.496505130179\n"
]
],
[
[
"**Question:** Will this be a problem for LSST? Hint: LSST donuts will be 128 x 128 pixels.",
"_____no_output_____"
],
[
"If these polynomials are perfectly orthonormal then $\\iint Z_i(x,y) Z_j(x,y)d\\Omega = \\delta_{ij}$. Hence a matrix matrix $M$ where $M_{ij} = \\iint Z_i(x,y) Z_j(x,y)d\\Omega$ should be equal to the identity matrix.\n\n**Problem 3:** Can you fill in the code below to plot the matrix $M$ for $i,j <= 22$?",
"_____no_output_____"
]
],
[
[
"n = 22 \nM = np.zeros((n, n))\n\nX,Y = np.meshgrid(np.linspace(-1,1,100), np.linspace(-1,1,100))\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.logical_and(np.abs(R) <=1, np.abs(R) > obscuration) \n# note that X,Y,R,mask are the same for the different polynomials, so we only need to create them once.\n\nfor i in range(n):\n for j in range(n):\n #TODO: code here to create Zi, Zj\n M[i,j] = np.sum(Zi[mask] * Zj[mask]) / np.sum(mask)\n \n#TODO: code here to plot M. Hint: you will need plt.imshow().",
"_____no_output_____"
]
],
[
[
"### Decomposition",
"_____no_output_____"
],
[
"Our ultimate goal is to decompose functions like the wavefront, intensity, and surface maps into zernike polynomials. Let's start by creating a somewhat arbitrary annular surface $W$.",
"_____no_output_____"
]
],
[
[
"X,Y = np.meshgrid(np.linspace(-1,1,100), np.linspace(-1,1,100))\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.logical_and(np.abs(R) <=1, np.abs(R) > obscuration) \nW = X**2-Y+1\nW[~mask] = np.nan\n\nplt.figure()\nplt.title(r'$W$')\nplt.imshow(arbitrary, origin='lower')\nplt.xticks([0,50,99],['-1','0','1'])\nplt.yticks([0,50,99],['-1','0','1'])\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"We can take the inner product $W_i = \\iint W(x,y) Z_i(x,y) d\\Omega$ to see how much of $Z_i$ is in $W\n$. Let's start with $Z_5$.",
"_____no_output_____"
]
],
[
[
"X,Y = np.meshgrid(np.linspace(-1,1,100), np.linspace(-1,1,100))\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.logical_and(np.abs(R) <=1, np.abs(R) > obscuration) \n\nW = X**2-Y+1\nW[~mask] = np.nan\nz5 = np.zeros(22)\nz5[5] = 1\nzern = galsim.zernike.Zernike(z5, R_inner=obscuration)\nZ5 = zern.evalCartesian(X,Y)\n\nintegral = np.sum(W[mask] * Z5[mask]) / np.sum(mask)\nprint(\"Z5 Inner Product = \", integral)",
"Z5 Inner Product = -1.18768044495e-16\n"
]
],
[
[
"What about $Z_3$? \n**Problem 4:** Can you fill in the cell below.",
"_____no_output_____"
]
],
[
[
"X,Y = np.meshgrid(np.linspace(-1,1,100), np.linspace(-1,1,100))\nR = np.sqrt(X ** 2 + Y ** 2)\nmask = np.logical_and(np.abs(R) <=1, np.abs(R) > obscuration) \n\nW = X**2-Y+1\nW[~mask] = np.nan\n# TODO: code here to create Z3\n\nintegral = np.sum(W[mask] * Z3[mask]) / np.sum(mask)\nprint(\"Z3 Inner Product = \", integral)",
"_____no_output_____"
]
],
[
[
"**Problem 5:** Can you create a vector $\\vec{W}$ such that $\\vec{W}_i = \\iint W(x,y) Z_i(x,y) d\\Omega$?",
"_____no_output_____"
]
],
[
[
"# TODO: code here",
"_____no_output_____"
]
],
[
[
"**Problem 6:** Can you evaluate $\\vec{W}$ and see how close it is to our original $W$? The cell below provides a hint on how to evaluate $\\vec{W}$.",
"_____no_output_____"
]
],
[
[
"Wbar = np.random.poisson(5, size=10)\nzern = galsim.zernike.Zernike(Wbar, R_inner=obscuration)\nWbarEval = zern.evalCartesian(X,Y)\nWbarEval[~mask] = np.nan\n\nplt.figure()\nplt.title(r'$W$')\nplt.imshow(WbarEval, origin='lower')\nplt.xticks([0,50,99],['-1','0','1'])\nplt.yticks([0,50,99],['-1','0','1'])\nplt.colorbar()",
"_____no_output_____"
],
[
"# TODO: code here",
"_____no_output_____"
]
],
[
[
"**Problem 7:** How does the error in the reconstruction change if we add poisson noise to W? Hint: see np.random.poisson.",
"_____no_output_____"
]
],
[
[
"# TODO: code here",
"_____no_output_____"
]
],
[
[
"**Challenge Problem:** What do the X,Y gradients of zernike polynomials look like? Are they orthonormal? Hint: see help(zern) above to see if there is already a method that does this.",
"_____no_output_____"
]
],
[
[
"# TODO: code here",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ec8b54f26ccb8e31002f518571c2c0c1a2868271 | 404,211 | ipynb | Jupyter Notebook | habermans-survival-data-set eda/Habermans Dataset.ipynb | sagunesh/Machine-Learning-and-Data-Science | 2281688ee52825335a905be4289cc7b43189d294 | [
"MIT"
] | null | null | null | habermans-survival-data-set eda/Habermans Dataset.ipynb | sagunesh/Machine-Learning-and-Data-Science | 2281688ee52825335a905be4289cc7b43189d294 | [
"MIT"
] | null | null | null | habermans-survival-data-set eda/Habermans Dataset.ipynb | sagunesh/Machine-Learning-and-Data-Science | 2281688ee52825335a905be4289cc7b43189d294 | [
"MIT"
] | null | null | null | 488.177536 | 119,712 | 0.93466 | [
[
[
"# Plotting for Exploratory Data Analysis(EDA) for Cancer Patients",
"_____no_output_____"
],
[
"# Habermans Dataset",
"_____no_output_____"
],
[
"Sources: (a) Donor: Tjen-Sien Lim (b) Date: March 1999\n\nPast Usage:\n\nHaberman, S. J. (1976). Generalized Residuals for Log-Linear Models, Proceedings of the 9th International Biometrics Conference, Boston, pp. 104-122.\nLandwehr, J. M., Pregibon, D., and Shoemaker, A. C. (1984), Graphical Models for Assessing Logistic Regression Models (with discussion), Journal of the American Statistical Association 79: 61-83.\nLo, W.-D. (1993). Logistic Regression Trees, PhD thesis, Department of Statistics, University of Wisconsin, Madison, WI.\nRelevant Information: The dataset contains cases from a study that was conducted between 1958 and 1970 at the University of Chicago's Billings Hospital on the survival of patients who had undergone surgery for breast cancer.\n\n* Number of Instances: 306\n* Number of Attributes: 4 (including the class attribute)\n* Attribute Information:\n * Age of patient at time of operation (numerical)\n * Patients year of operation (year - 1900, numerical)\n * Number of positive axillary nodes detected (numerical)\n * Survival status (class attribute) 1 = the patient survived 5 years or longer 2 = the patient died within 5 year\n* Missing Attribute Values: None",
"_____no_output_____"
],
[
"# Objective",
"_____no_output_____"
],
[
"Classify a new patient according to one of the 2 classes that is whether it survived 5 years or longer or patient died within 5 years, given the 3 features",
"_____no_output_____"
]
],
[
[
"#importing all libraries \nimport pandas as pd\nimport seaborn as se\nimport numpy as np\nimport matplotlib.pyplot as plt\n",
"_____no_output_____"
],
[
"#reading the dataset\nhb = pd.read_csv(\"haberman.csv\")\n#hb",
"_____no_output_____"
],
[
"hb.shape\n#it shows we have 306 rows and 4 columns",
"_____no_output_____"
],
[
"hb.columns",
"_____no_output_____"
],
[
"hb['survival_status'].value_counts()",
"_____no_output_____"
]
],
[
[
"# Observations",
"_____no_output_____"
],
[
"This shows\n * Only 225 patients survived 5 years or longer\n * And 81 the patient died within 5 year",
"_____no_output_____"
],
[
"# Univariate Analysis",
"_____no_output_____"
],
[
"# Histogram",
"_____no_output_____"
]
],
[
[
"se.FacetGrid(hb,hue=\"survival_status\",size=5)\\\n .map(se.distplot,\"year\")\\\n .add_legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
" Observation : can't say much from the plot as points are overlapping",
"_____no_output_____"
]
],
[
[
"se.FacetGrid(hb,hue=\"survival_status\",size=5)\\\n .map(se.distplot,\"Age\")\\\n .add_legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
" Observation :\n * Patients with age less than 35 and greater than 30 have survived more than 5 years after operation\n * Patients with age less than 83 and greater than 78 have survived not more than 5 Years after operation\n * Patients from age 35 to 78 we can't say anything as point are almost overlapping.",
"_____no_output_____"
]
],
[
[
"se.FacetGrid(hb,hue=\"survival_status\",size=5)\\\n .map(se.distplot,\"positive_axillary_nodes\")\\\n .add_legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
" Observation : can't say much from the plot as points are overlapping but one thing we can infer is as the no. of positive auxillary nodes increases the survival status decreases less than 5 years .",
"_____no_output_____"
],
[
"# Box plot and Whiskers",
"_____no_output_____"
]
],
[
[
"se.boxplot(x = 'survival_status',y = 'year',data = hb)\nplt.show()",
"_____no_output_____"
],
[
"se.boxplot(x = 'survival_status',y = 'Age',data = hb)\nplt.show()",
"_____no_output_____"
],
[
"se.boxplot(x = 'survival_status',y = 'positive_axillary_nodes',data = hb)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Observations",
"_____no_output_____"
],
[
"* From the boxplot we can observe that most people who survived cancer have zero positive axillary nodes",
"_____no_output_____"
],
[
"# Violin plots",
"_____no_output_____"
]
],
[
[
"se.violinplot(x=\"survival_status\", y=\"year\", data=hb, size=8)\nplt.show()",
"_____no_output_____"
],
[
"se.violinplot(x=\"survival_status\", y=\"Age\", data=hb, size=8)\nplt.show()",
"_____no_output_____"
],
[
"se.violinplot(x=\"survival_status\", y=\"positive_axillary_nodes\", data=hb, size=8)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Observation",
"_____no_output_____"
],
[
"* From the violin plots we can observe that most people who survived cancer have zero positive axillary nodes",
"_____no_output_____"
],
[
"# PDF and CDF",
"_____no_output_____"
]
],
[
[
"#pdf cdf of year\n\ncounts,bin_edges = np.histogram(hb['year'],bins = 30, density = True)\npdf = counts/(sum(counts))\ncdf = np.cumsum(pdf)\nplt.plot(bin_edges[1:],pdf)\nplt.plot(bin_edges[1:],cdf)\nplt.legend()\n\ncounts,bin_edges = np.histogram(hb['year'],bins = 30, density = True)\npdf = counts/(sum(counts))\ncdf = np.cumsum(pdf)\nplt.plot(bin_edges[1:],pdf)\nplt.plot(bin_edges[1:],cdf)\n\n\nplt.xlabel('Year')\nplt.grid()\n\nplt.show()",
"C:\\Users\\sagun\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\matplotlib\\axes\\_axes.py:545: UserWarning: No labelled objects found. Use label='...' kwarg on individual plots.\n warnings.warn(\"No labelled objects found. \"\n"
],
[
"#pdf cdf of positive_axillary_nodes\n\ncounts,bin_edges = np.histogram(hb['positive_axillary_nodes'],bins = 30, density = True)\npdf = counts/(sum(counts))\ncdf = np.cumsum(pdf)\nplt.plot(bin_edges[1:],pdf)\nplt.plot(bin_edges[1:],cdf)\nplt.legend()\n\ncounts,bin_edges = np.histogram(hb['positive_axillary_nodes'],bins = 30, density = True)\npdf = counts/(sum(counts))\ncdf = np.cumsum(pdf)\nplt.plot(bin_edges[1:],pdf)\nplt.plot(bin_edges[1:],cdf)\n\nplt.xlabel('positive_axillary_nodes')\nplt.grid()\n\nplt.show()",
"C:\\Users\\sagun\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\matplotlib\\axes\\_axes.py:545: UserWarning: No labelled objects found. Use label='...' kwarg on individual plots.\n warnings.warn(\"No labelled objects found. \"\n"
],
[
"#pdf cdf of Age\n\ncounts,bin_edges = np.histogram(hb['Age'],bins = 30, density = True)\npdf = counts/(sum(counts))\ncdf = np.cumsum(pdf)\nplt.plot(bin_edges[1:],pdf)\nplt.plot(bin_edges[1:],cdf)\nplt.legend()\n\ncounts,bin_edges = np.histogram(hb['Age'],bins = 30, density = True)\npdf = counts/(sum(counts))\ncdf = np.cumsum(pdf)\nplt.plot(bin_edges[1:],pdf)\nplt.plot(bin_edges[1:],cdf)\n\nplt.xlabel('Age')\nplt.grid()\n\nplt.show()",
"C:\\Users\\sagun\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\matplotlib\\axes\\_axes.py:545: UserWarning: No labelled objects found. Use label='...' kwarg on individual plots.\n warnings.warn(\"No labelled objects found. \"\n"
]
],
[
[
"# Bivariate analysis",
"_____no_output_____"
],
[
"# 2-D Scatter Plot",
"_____no_output_____"
]
],
[
[
"se.set_style(\"darkgrid\");\nse.FacetGrid(hb,hue='survival_status',size=6)\\\n .map(plt.scatter,\"year\",\"Age\")\\\n .add_legend();\nplt.show()",
"_____no_output_____"
]
],
[
[
" Observation : can't say much from the plot as points overlapping",
"_____no_output_____"
]
],
[
[
"se.set_style(\"darkgrid\");\nse.FacetGrid(hb,hue='survival_status',size=6)\\\n .map(plt.scatter,\"positive_axillary_nodes\",\"Age\")\\\n .add_legend();\nplt.show()",
"_____no_output_____"
]
],
[
[
" Observation : can't say much from the plot as points overlapping",
"_____no_output_____"
],
[
"# Pair-Plot",
"_____no_output_____"
]
],
[
[
"plt.close();\nse.set_style(\"whitegrid\");\nse.pairplot(hb,hue=\"survival_status\",size=3)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"# Observations ",
"_____no_output_____"
],
[
"* Positive_axillary_nodes is a useful feature to identify the survival_status of cancer patients\n* Age and Year of operation have overlapping curves so we can't have a suitable observation that can classify survival_status\n",
"_____no_output_____"
],
[
"# Mean",
"_____no_output_____"
]
],
[
[
"#hb is the name of the data frame\nless_five = hb[hb['survival_status']==2]\nmore_five = hb[hb['survival_status']==1]",
"_____no_output_____"
],
[
"print(np.mean(more_five))",
"Age 52.017778\nyear 62.862222\npositive_axillary_nodes 2.791111\nsurvival_status 1.000000\ndtype: float64\n"
],
[
"print(np.mean(less_five))",
"Age 53.679012\nyear 62.827160\npositive_axillary_nodes 7.456790\nsurvival_status 2.000000\ndtype: float64\n"
]
],
[
[
"Observation\n* Mean age of patients who survived more than 5 years is 52 years and who didn't survive is 54 years\n* Those having more than 3 positive_axillary_nodes they have not survived more than 5 years\n* Those having less than 3 positive_axillary_nodes they have survived more than 5 years after the operation",
"_____no_output_____"
],
[
"# Final Conclusion",
"_____no_output_____"
],
[
"* Those having more than 3 positive_axillary_nodes they have not survived more than 5 years\n* Those having less than 3 positive_axillary_nodes they have survived more than 5 years after the operation\n* Positive_axillary_nodes is a useful feature to identify the survival_status of cancer patients\n* Age and Year of operation have overlapping curves so we can't classify patients for their survival_status using age ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ec8b57e2a343d0a9597835e1b0561a987babeec7 | 500,616 | ipynb | Jupyter Notebook | bayesian.ipynb | Guanxinyuan/Bayesian-classification | 1d148d6a5d3f654d3bd4c7a938dc4eaf36f8977d | [
"MIT"
] | null | null | null | bayesian.ipynb | Guanxinyuan/Bayesian-classification | 1d148d6a5d3f654d3bd4c7a938dc4eaf36f8977d | [
"MIT"
] | null | null | null | bayesian.ipynb | Guanxinyuan/Bayesian-classification | 1d148d6a5d3f654d3bd4c7a938dc4eaf36f8977d | [
"MIT"
] | null | null | null | 647.627426 | 435,224 | 0.944814 | [
[
[
"# Imports",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nimport gzip\nimport numpy as np\ntry:\n import cPickle as pickle\n kwargs = {}\nexcept:\n import _pickle as pickle\n kwargs = {'encoding':'bytes'}\n\nfeatures, labels, sample_ids, label_names = pickle.load( gzip.open( 'data.pgz', 'rb' ), **kwargs )\nprint('IDs and the corresponding national parks:')\nprint(label_names)",
"IDs and the corresponding national parks:\n{0: b'greatsanddunesnationalpark', 1: b'petrifiedforestnationalpark', 2: b'archesnationalpark', 3: b'congareenationalpark', 4: b'katmainationalpark'}\n"
],
[
"import numpy as np\nnp.random.seed(12345)\nN = features.shape[0]\narr = np.arange(N)\nnp.random.shuffle(arr)\ntrain_num = int(round(N*0.8))\ntest_num = features.shape[0]-train_num\n\ntrain_subset = arr[:train_num]\ntrain_features = features[train_subset,:]\ntrain_labels = labels[train_subset]\ntrain_sample_ids = [sample_ids[i] for i in train_subset]\n\ntest_subset = arr[train_num:]\ntest_features = features[test_subset,:]\ntest_labels = labels[test_subset]\ntest_sample_ids = [sample_ids[i] for i in test_subset]\nprint(\"Dataset split into {} training and {} test examples.\".format(train_num,test_num))",
"Dataset split into 403 training and 101 test examples.\n"
]
],
[
[
"# 1. Learning a tree structure\nChow-Liu structure learning algorithm uses mutual information to find the best tree. \n\n### Computing mutual information\n\nMutual information between variables distributed according to a categorical distribution. Assuming that the variables as are Gaussian distributed then mutual information is given \nby\n$$ \nI(X,Y) = -\\frac{1}{2} \\ln(1-Corr(X, Y)^2) \n$$\n",
"_____no_output_____"
]
],
[
[
"def corr(x,y):\n if x.shape[0] == 0:\n rowvar = 1\n else:\n rowvar = 0\n return np.corrcoef(np.asarray(x),np.asarray(y),rowvar = rowvar)[0,1]\n\ndef mutual_information(x,y):\n return -0.5*np.log(1-corr(x,y)**2)\n\ndef mutual_info_all(M):\n f_num = M.shape[1] #feature number\n mi_ary = np.zeros( (f_num, f_num) )\n for ix in np.arange(f_num):\n for jx in np.arange(ix+1,f_num):\n mi_ary[ix,jx] = mutual_information(M[:,ix], M[:,jx])\n \n return mi_ary\n\nnp.random.seed(1)\nx = np.random.randn(10)\ny = x + 0.01*np.random.randn(10)\nz = -x + 0.01*np.random.randn(10)\n\nprint((\"Mutual information {:1.3f} between variables \"+\n \"with correlation {:1.3f}.\").format(mutual_information(x,y),\n corr(x,y)))\nprint((\"Mutual information {:1.3f} between variables \"+\n \"with correlation {:1.3f}.\").format(mutual_information(x,z),\n corr(x,z)))\nfeatures = np.asmatrix([x,y,z]).transpose()\nMI = mutual_info_all(features)\nprint(MI)\nprint((\"MI(x,z) = {:1.3f} should be larger \" + \n \"than MI(y,z) = {:1.3f}.\").format(MI[0,2],MI[1,2]))\nassert(MI[0,2]>MI[1,2])",
"Mutual information 5.067 between variables with correlation 1.000.\nMutual information 5.037 between variables with correlation -1.000.\n[[0. 5.06715848 5.03662491]\n [0. 0. 4.79462788]\n [0. 0. 0. ]]\nMI(x,z) = 5.037 should be larger than MI(y,z) = 4.795.\n"
]
],
[
[
"### Chow-liu tree algorithm",
"_____no_output_____"
]
],
[
[
"from scipy.sparse.csgraph import minimum_spanning_tree\ndef chowliu(features):\n X = -mutual_info_all(features)\n adjacency_matrix = minimum_spanning_tree(X)\n return adjacency_matrix\n\ndef get_edge_list(mat):\n f_num = mat.shape[0]\n edges = []\n for k in range(f_num):\n lst = np.nonzero(mat[k,:])[1]\n # k is parent, j is child\n new_edges = [(k,j) for j in lst] \n edges.extend(new_edges)\n return edges\n \n\nnp.random.seed(1)\nx = np.random.randn(20)\ny = x + 0.1*np.random.randn(20)\nz = -x + 0.1*np.random.randn(20)\nq = z + 0.1*np.random.randn(20)\nw = y + 0.1*np.random.rand(20)\nfeatures = np.asmatrix([x,y,z,q,w]).transpose()\nadjacency = chowliu(features)\nnames = ['x','y','z','q','w']\nedges = get_edge_list(adjacency)\nprint('Edge list: ',[(names[i] + '->' + names[j]) for (i,j) in edges])\n\nedges = get_edge_list(adjacency)\n\nassert(set(edges)== set([(0, 1), (0, 2), (1, 4), (2, 3)]))",
"Edge list: ['x->y', 'x->z', 'y->w', 'z->q']\n"
]
],
[
[
"### Learning class-specific trees",
"_____no_output_____"
]
],
[
[
"def get_label_subsets(train_labels):\n label_set = np.unique(train_labels) #get 5 label numbers\n label_sample_map = {} #a label to sample index map \n \n for i in label_set:\n label_sample_map[i] = [index for index, value in enumerate(train_labels) if value == i]\n return label_sample_map\n \ndef get_edges_for_each_class(train_features, train_labels):\n label_set = np.unique(train_labels) #get 5 label numbers\n label_sample_map = get_label_subsets(train_labels) \n class_edges = {}\n for i in label_set:\n indices = label_sample_map[i]\n features = train_features[indices]\n adjacency = chowliu(features)\n edges = get_edge_list(adjacency)\n class_edges[i] = edges\n# print(class_edges[1])\n return class_edges\n \nclass_edges = get_edges_for_each_class(train_features, train_labels)",
"_____no_output_____"
]
],
[
[
"# 2. Learning parameters",
"_____no_output_____"
],
[
"### **Coordinate ascent**",
"_____no_output_____"
]
],
[
[
"#update for theta_{r,0}, r is the root_index (0 in our case)\ndef compute_theta_r( x ):\n return np.sum(x[:,0])/len(x)",
"_____no_output_____"
],
[
"#update for theta_{j, 0}, for link between j and k given theta_j_1. \n#k is parent, j is child\ndef compute_theta_j_k_0( j, k, x, theta_j_1 ):\n return np.sum(x[:,j] - theta_j_1*x[:,k])/len(x)",
"_____no_output_____"
],
[
"#update for theta_{j, 1}, for link between j and k given theta_j_0. \n#k is parent, j is child\ndef compute_theta_j_k_1( j, k, x, theta_j_0 ):\n return np.sum(x[:,j]*x[:,k]- theta_j_0*x[:,k])/np.sum(x[:,k]*x[:,k])",
"_____no_output_____"
],
[
"d = 1000\nx = np.zeros((d,2))\nx[:,0] = 1.0 + np.random.randn(d,)\nx[:,1] = 0.5 + 2.0*x[:,0] + np.random.randn(d,)\ntheta_j = [0.0,0.0]\n\ntheta_r = compute_theta_r( x )\nfor it in range(40):\n theta_j[0] = compute_theta_j_k_0( 1, 0, x, theta_j[1])\n theta_j[1] = compute_theta_j_k_1( 1, 0, x, theta_j[0])\nprint(\"theta_r_0:{} theta_j:{}\".format(theta_r,theta_j)) \nassert(abs(theta_r - 1.0)<0.1)\nassert(abs(theta_j[0] - 0.5)<0.1)\nassert(abs(theta_j[1] - 2.0)<0.1)",
"theta_r_0:1.0333542751510443 theta_j:[0.4950430649417859, 2.0243162317360426]\n"
]
],
[
[
"### Compute $\\theta^*_j$ and $\\theta^*_r$ for each class.",
"_____no_output_____"
]
],
[
[
"def get_thetas_for_each_class(train_features, train_labels, class_edges):\n label_sample_map = get_label_subsets(train_labels)\n label_set = np.unique(train_labels)\n class_thetas = {}\n f_num = train_features.shape[1]\n for lab in label_set:\n print( 'processing class label {}'.format(lab) )\n c_samples = train_features[label_sample_map[lab]]\n theta_r = compute_theta_r(c_samples)\n c_edge_list= class_edges[lab]\n thetas = np.zeros((f_num,2)) #the first column shoud be j_0, and the second column should be j_1\n #the first row (thetas[0,:]) is for theta_r\n\n for (k,j) in c_edge_list: \n theta_j_1 = 0\n #should do coordinate ascent using the function\n #compute_theta_j_k_0 and compute_theta_j_k_1 here\n for z in range(100):\n theta_j_0 = compute_theta_j_k_0(j, k, c_samples, theta_j_1)\n theta_j_1 = compute_theta_j_k_1(j, k, c_samples, theta_j_0)\n #set the optimal theta_j_0 and theta_j_1 for this the edge (k, j)\n thetas[j, 0] = theta_j_0\n thetas[j, 1] = theta_j_1\n thetas[0,0] = theta_r \n # root has no parents\n thetas[0,1] = np.nan\n\n class_thetas[lab] = thetas\n return class_thetas\n\nclass_thetas = get_thetas_for_each_class(train_features,train_labels,class_edges)\nclass_thetas",
"processing class label 0\nprocessing class label 1\nprocessing class label 2\nprocessing class label 3\nprocessing class label 4\n"
]
],
[
[
"# 3. Compute probability\n",
"_____no_output_____"
],
[
"### Compute probability of a feature vector x given a Bayes net's structure and parameters",
"_____no_output_____"
]
],
[
[
"# Compute probability of a feature vector x given a Bayes net's structure and parameters\n\n# compute log p(x_j|x_k,\\theta_j), for the edge between j and k, given \\theta_{j, 0} and \\theta_{j,1}\n# Let sigma^2 =1\ndef compute_lp_j_k( j, k, x, theta_j_0, theta_j_1 ):\n return -np.log(2*np.pi*1)-1/2*(x[j]-theta_j_0-theta_j_1*x[k])**2\n\n# compute log p(x_r|\\theta_r), for the root node, given theta_r. \ndef compute_lp_r( x, theta_r ):\n return -np.log(2*np.pi*1)-1/2*(x[0]-theta_r)**2",
"_____no_output_____"
],
[
"def compute_lp_x_given_Theta( x, thetas, edges):\n lp = compute_lp_r(x, thetas[0,0]) \n for (k,j) in edges: \n # k is parent, j is child\n lp = lp + compute_lp_j_k(j, k, x, thetas[j,0], thetas[j,1])\n return lp",
"_____no_output_____"
]
],
[
[
"### Compute probabilities that a feature vector x belongs to each class",
"_____no_output_____"
]
],
[
[
"from scipy.special import logsumexp\ndef get_log_p_h(train_labels):\n label_set = np.unique(train_labels)\n log_p_h = np.zeros(len(np.unique(labels)))\n for i in label_set:\n count = len(np.nonzero(train_labels==i)[0])\n log_p_h[i] = np.log( float(count) / float(len(train_labels) ) )\n return log_p_h\n\nlog_p_h = get_log_p_h(train_labels) \n\ndef logsumexp(vec):\n m = np.max(vec,axis=0) \n return np.log(np.sum(np.exp(vec-m),axis=0))+m\n\n\ndef p_h_given_x_theta( x, class_edges, class_thetas, log_p_h ):\n \n C = len(class_thetas)\n lognumerator = np.zeros(C)\n \n # implement Bayes rule here\n # compute log-numerators first and then normalize using logsumexp\n # there are more compact ways to do the normalization\n # feel free to rearrange the code, as long as you return correct\n # probabilities\n \n for i in range(C): \n edges = class_edges[i]\n thetas = class_thetas[i] \n lognumerator[i] = compute_lp_x_given_Theta(x, thetas, edges) + log_p_h[i]\n \n # use logsumexp to compute denominator\n logdenominator = logsumexp(lognumerator)\n \n probs = np.exp(lognumerator - logdenominator)\n \n assert(np.all(probs >= 0))\n assert(np.abs(np.sum(probs)-1.0)<1e-5)\n return probs",
"_____no_output_____"
]
],
[
[
"# 4. Make predictions based on probabilities",
"_____no_output_____"
]
],
[
[
"def evaluate_predictions(test_features,test_labels,class_edges,class_thetas,log_p_h):\n label_set = np.unique(train_labels)\n pred_lab = np.zeros((test_features.shape[0], 2)) \n #the first column shoud be the predicted label, and the second column should be the probability of that label.\n for i in range(test_num):\n x = test_features[i,:] \n res = p_h_given_x_theta(x, class_edges, class_thetas, log_p_h) \n # predicted label\n pred_lab[i, 0] = label_set[np.argmax(res)]\n # probability of that label\n pred_lab[i, 1] = res[np.argmax(res)]\n return pred_lab\n ",
"_____no_output_____"
],
[
"pred_lab = evaluate_predictions(test_features,test_labels,class_edges,class_thetas,log_p_h)\nprint(\"Prediction Accuracy: {}\".format(np.mean(pred_lab[:,0]==test_labels)))",
"Prediction Accuracy: 0.7326732673267327\n"
]
],
[
[
"# 5. Evaluate prediction performance",
"_____no_output_____"
],
[
"### Confusion matrix",
"_____no_output_____"
]
],
[
[
"import itertools\nimport matplotlib.pyplot as plt\n%matplotlib inline\ndef plot_confusion_matrix(cm, classes,\n normalize=False,\n title='Confusion matrix',\n cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45, rotation_mode='anchor', ha = 'right')\n plt.yticks(tick_marks, classes)\n\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n print(\"Normalized confusion matrix\")\n else:\n print('Confusion matrix, without normalization')\n\n print(cm)\n\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, cm[i, j],\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n #plt.tight_layout()\n plt.ylabel('True label')\n plt.xlabel('Predicted label')\n plt.show()",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\ncnf_matrix = confusion_matrix(test_labels, pred_lab[:, 0])\nlabel_set = np.unique(test_labels)\nplot_confusion_matrix(cnf_matrix, [label_names[i] for i in label_set],\n title='Confusion matrix, without normalization')",
"Confusion matrix, without normalization\n[[16 1 2 0 0]\n [ 4 15 3 0 1]\n [ 3 5 12 0 0]\n [ 1 1 0 21 0]\n [ 2 4 0 0 10]]\n"
],
[
"import matplotlib.image as mpimg\ndef gen_err_predicted_fig( test_labels, pred_lab, label_set, label_names, \n test_sample_ids, srcPath='images_5_classes', \n figsize=(10, 10)):\n errNameMap = {}\n maxErrNameMap = {}\n cnt = 0\n for lab in label_set:\n for plab in label_set:\n if lab == plab:\n continue\n else:\n cIdx = np.nonzero(test_labels==lab)[0]\n errIdx = cIdx[ np.nonzero(pred_lab[cIdx, 0]==plab)[0] ]\n errNameMap[(lab, plab)] = errIdx\n if len(errIdx) != 0:\n errProb = np.max( pred_lab[errIdx, 1] )\n errFIdx = errIdx[ np.argmax( pred_lab[errIdx, 1] ) ]\n maxErrNameMap[(lab, plab)] = errFIdx\n title_font = {'fontname':'Arial', 'size':'16', \n 'color':'black', 'weight':'bold'} \n fig = plt.figure(figsize=figsize)\n for i in range(25):\n rIdx = i // 5\n cIdx = i % 5\n #print(i, rIdx, cIdx)\n ax = plt.subplot(5, 5, i+1)\n if rIdx == 0:\n ax.xaxis.set_label_position('top') \n ax.set_xlabel( label_names[int(label_set[cIdx])].decode('utf-8'), \n rotation=45, rotation_mode='anchor', ha = 'left' )\n if cIdx == 0:\n ax.yaxis.set_label_position('left')\n ax.set_ylabel( label_names[int(label_set[rIdx])].decode('utf-8'), \n rotation=45, rotation_mode='anchor', ha = 'right' )\n ax.set_xticklabels([])\n ax.set_yticklabels([])\n ax.set_xticks([])\n ax.set_yticks([])\n plt.subplots_adjust(wspace=0, hspace=0)\n if (label_set[rIdx], label_set[cIdx]) in maxErrNameMap:\n fIdx = maxErrNameMap[(label_set[rIdx], label_set[cIdx])] \n folderName = label_names[test_labels[fIdx]] \n fID = test_sample_ids[fIdx]\n #int(fID.split('_')[0]) ]\n img=mpimg.imread( srcPath + '/' + folderName.decode('utf-8') + '/' + fID.decode('utf-8') + '.jpg') \n plt.imshow(img) # The AxesGrid object work as a list of axes.\n else:\n plt.text(0.5, 0.5, cnf_matrix[rIdx, cIdx],\n **title_font)\n \n plt.show()",
"_____no_output_____"
],
[
"label_set = np.unique(test_labels)\ngen_err_predicted_fig( test_labels, pred_lab, label_set, label_names, test_sample_ids)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ec8b5e515ce6b29b3f9cc2f03c6d7b359ca7b2e2 | 4,177 | ipynb | Jupyter Notebook | jupyter-notebooks/[01] [01] Data Cleaning.ipynb | AbnerBissolli/Malicious-DNS-Detection | 0764d719f9a361d2d4b615382fbed87db0f478a0 | [
"MIT"
] | null | null | null | jupyter-notebooks/[01] [01] Data Cleaning.ipynb | AbnerBissolli/Malicious-DNS-Detection | 0764d719f9a361d2d4b615382fbed87db0f478a0 | [
"MIT"
] | null | null | null | jupyter-notebooks/[01] [01] Data Cleaning.ipynb | AbnerBissolli/Malicious-DNS-Detection | 0764d719f9a361d2d4b615382fbed87db0f478a0 | [
"MIT"
] | null | null | null | 21.642487 | 98 | 0.52023 | [
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"# Reading datasets\n\ndf_be = pd.read_csv('../Dataset/Domains/Raw/benign_domains.csv',header=None)\ndf_ma = pd.read_csv('../Dataset/Domains/Raw/malware_domains.csv',header=None)\ndf_ph = pd.read_csv('../Dataset/Domains/Raw/phishing_domains.csv',header=None)\ndf_sp = pd.read_csv('../Dataset/Domains/Raw/spam_domains.csv',header=None)",
"_____no_output_____"
],
[
"# Cleaning phishing_domain dataset\n\ndef get_domain(string): \n string = string.replace('https://','')\n string = string.replace('http://','')\n string = string.split('/', 1)[0]\n \n return string\n\ndf_ph = df_ph[[1]]\ndf_ph[1] = df_ph[1].apply(get_domain)",
"_____no_output_____"
],
[
"# Dropping duplicates\n\ndf_be = df_be.drop_duplicates()\ndf_ma = df_ma.drop_duplicates()\ndf_ph = df_ph.drop_duplicates()\ndf_sp = df_sp.drop_duplicates()",
"_____no_output_____"
],
[
"# Setting classes\n\ndf_be[2] = 'benign'\ndf_ma[2] = 'malware'\ndf_ph[2] = 'phishing'\ndf_sp[2] = 'spam'",
"_____no_output_____"
],
[
"# Naming columns\n\ndf_be.columns = ['domain', 'class']\ndf_ma.columns = ['domain', 'class']\ndf_ph.columns = ['domain', 'class']\ndf_sp.columns = ['domain', 'class']",
"_____no_output_____"
],
[
"# Combaining datasets\n\ndf = pd.concat([df_be, df_ma, df_ph, df_sp])",
"_____no_output_____"
],
[
"# Dropping duplicates after combaining\n\ndf = df.drop_duplicates(subset=['domain'], keep='last')",
"_____no_output_____"
],
[
"# Reseting the index\n\ndf = df.reset_index(drop=True)",
"_____no_output_____"
],
[
"# Saving the dataset\n\ndf.to_csv('../Dataset/Domains/Clean/domains_raw.csv', index=False)",
"_____no_output_____"
],
[
"# Reducing The Benign Dataset\n\nnp.random.seed(42)\n\nremove_n = 914881\n\ndrop_indices = np.random.choice(df[df['class']=='benign'].index, remove_n, replace=False)\ndf_subset = df.drop(drop_indices)",
"_____no_output_____"
],
[
"# Reseting the index\n\ndf_subset = df_subset.reset_index(drop=True)",
"_____no_output_____"
],
[
"# Saving the dataset\n\ndf_subset.to_csv('../Dataset/Domains/Clean/domains.csv', index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ec8b65a653f2b6232560ad677370d004c0889ea2 | 1,079 | ipynb | Jupyter Notebook | examples/VTK_Example01.ipynb | radiasoft/jupyter-rs-vtk | b7c203863a664b9961fdcd272b4196946689eaaf | [
"Apache-2.0"
] | null | null | null | examples/VTK_Example01.ipynb | radiasoft/jupyter-rs-vtk | b7c203863a664b9961fdcd272b4196946689eaaf | [
"Apache-2.0"
] | 5 | 2020-01-10T04:31:21.000Z | 2021-05-10T21:33:18.000Z | examples/VTK_Example01.ipynb | radiasoft/jupyter-rs-vtk | b7c203863a664b9961fdcd272b4196946689eaaf | [
"Apache-2.0"
] | null | null | null | 18.288136 | 73 | 0.497683 | [
[
[
"# Comments\n\n# [cell]\n\n# imports\n\nfrom __future__ import absolute_import, division, print_function\n\nimport ipywidgets\n\nfrom jupyter_rs_vtk import vtk_viewer\n\n# [cell]\n\n# vars\n\n# radia viewer\nvtkv = vtk_viewer.Viewer()\nvtkv.test()\nvtkv.display()\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ec8b785b5342f1c09761fd593238547c3129d820 | 143,856 | ipynb | Jupyter Notebook | 1-DL course/9/o9_1 RNN v1.ipynb | v-mk-s/tinkoff-DL-course | f63045bed5a994bc038348c9b52ee7aa54ceb7af | [
"MIT"
] | null | null | null | 1-DL course/9/o9_1 RNN v1.ipynb | v-mk-s/tinkoff-DL-course | f63045bed5a994bc038348c9b52ee7aa54ceb7af | [
"MIT"
] | null | null | null | 1-DL course/9/o9_1 RNN v1.ipynb | v-mk-s/tinkoff-DL-course | f63045bed5a994bc038348c9b52ee7aa54ceb7af | [
"MIT"
] | null | null | null | 143,856 | 143,856 | 0.785668 | [
[
[
"# **Выводы в конце ноутбука:** ",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport os\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## Ссылки:\n* [Chris Olah's blog (LSTM/GRU)](http://colah.github.io/posts/2015-08-Understanding-LSTMs/)\n* [PyTorch tutorial - RNN for name classification](https://pytorch.org/tutorials/intermediate/char_rnn_classification_tutorial.html)\n* [MNIST classification with RNN tutorial](https://medium.com/dair-ai/building-rnns-is-fun-with-pytorch-and-google-colab-3903ea9a3a79)\n* [Good tutorials about Torch sentiment](https://github.com/bentrevett/pytorch-sentiment-analysis)",
"_____no_output_____"
],
[
"## Vanilla RNN",
"_____no_output_____"
],
[
"<img src=\"http://colah.github.io/posts/2015-08-Understanding-LSTMs/img/LSTM3-SimpleRNN.png\" width=\"600\">",
"_____no_output_____"
],
[
"$$\\Large h_{i+1} = tanh(W_x \\cdot X_{i+1} + W_y \\cdot h_{i})$$",
"_____no_output_____"
],
[
"Рекурретные нейросети нужны для работы с **последовательными данными** произвольной длины. Они представляют собой абстрактные ячейки, у которых есть какая-то **память** (hidden state), которая обновляется после обработки очередной порции данных.\n\nЕсли в самом простом виде, то в рекуррентных сетках для одного входного вектора $x_{(t)}$ и одного слоя рекуррентной сети справедливо такое соотношение:\n\n$$y_{(t)} = \\phi (x_{(t)}^T \\cdot w_x + y_{(t-1)}^T \\cdot w_y + b)$$\n\nгде \n* $x(t)$ — входной вектор на текущем шаге;\n* $y(t)$ — выходной вектор на текущем шаге;\n* $w_x$ — вектор весов нейронов для входа;\n* $w_y$ — вектор весов нейронов для выхода;\n* $y(t-1)$ — выходной вектор с прошлого шага (для первого шага этот вектор нулевой);\n* $b$ — bias;\n* $\\phi$ — какая-то функция активации (например, ReLU).\n\nЭту ячейку применяют по очереди ко всей последовательности, пробрасывая hidden state с предыдущего состояния. С точки зрения построения вычислительного графа это выглядит так:\n\n<img src=\"http://colah.github.io/posts/2015-08-Understanding-LSTMs/img/RNN-unrolled.png\" width=\"600\">\n\nТо есть если зафиксировать длину последовательности, то мы получим обычный фиксированный ациклический граф вычислений, в котором просто пошерены параметры всех ячеек.\n\n### Упрощение формулы\n\nСнова немножко математики чтобы привести формулу выше к более удобному виду.\n\nПредставим, что на вход подается не один вектор $x_{(t)}$, а целый мини-батч размера $m$ таких векторов $X_{(t)}$, соответственно все дальнейшие размышления мы уже производим в матричном виде:\n\n$$ Y_{(t)} = \\phi(X_{(t)}^T \\cdot W_x + Y_{(t-1)}^T \\cdot W_y + b) = \\phi([X_{(t)} Y_{(t-1)}] \\cdot W + b) $$\nгде\n$$ W = [W_x W_y]^T $$\n\n*Операция в квадратных скобках — конкатенация матриц\n\nПо размерностям:\n* $Y_{(t)}$ — матрица [$m$ x n_neurons]\n* $X_{(t)}$ — матрица [$m$ x n_features]\n* $b$ — вектор длины n_neurons\n* $W_x$ — веса между входами и нейронами размерностью [n_features x n_neurons]\n* $W_y$ — веса связей с прошлым выходом размерностью [n_neurons x n_neurons]",
"_____no_output_____"
],
[
"# RNN from scratch\n\n**Disclaimer:** не используйте самописные RNN-ки в реальной жизни.\n\nДавайте реализуем торчовый модуль, который это реализует.",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn\n\nclass RNN(nn.Module):\n def __init__(self, input_size, hidden_size):\n super().__init__()\n\n self.hidden_size = hidden_size\n # stdv = 1./np.sqrt(input_size)\n # self.Wx = np.random.uniform(-stdv, stdv, size=(input_size, hidden_size))\n # self.Wy = np.random.uniform(-stdv, stdv, size=(hidden_size, hidden_size))\n # self.b = np.random.uniform(-stdv, stdv, size=hidden_size)\n self.W = nn.Linear(input_size + hidden_size, hidden_size) # в качестве весов просто лин слой\n self.T = nn.Tanh()\n\n # self.hidden = init_hidden(self)\n\n def forward(self, input_data, hidden):\n # <использовать Wx, Wy для полученния нового hidden>\n A = self.W.forward(torch.cat([input_data, hidden], 1))\n hidden = self.T(A)\n\n #W = np.dot(self.Wx, self.Wy).T\n\n #self.hidden = np.tanh(np.dot(np.concatenate(input_data, hidden).T, W) + self.b)\n return hidden\n\n def init_hidden(self, batch_size):\n return torch.zeros(batch_size, self.hidden_size)",
"_____no_output_____"
],
[
"input_feature_size = 6\nhidden_size=5\nbatch_size=1",
"_____no_output_____"
],
[
"rnn = RNN(input_size=input_feature_size, hidden_size=hidden_size)\ninitial_hidden = rnn.init_hidden(batch_size)",
"_____no_output_____"
],
[
"input_example = torch.rand([batch_size, input_feature_size])\nnew_hidden = rnn(input_example, initial_hidden)",
"_____no_output_____"
],
[
"print(new_hidden.shape)",
"torch.Size([1, 5])\n"
],
[
"print(\"initial_hidden: \", initial_hidden.numpy())\nprint(\"new_hidden: \", new_hidden.detach().numpy())",
"initial_hidden: [[0. 0. 0. 0. 0.]]\nnew_hidden: [[ 0.13223282 -0.06284315 -0.33426526 0.1857464 -0.19251004]]\n"
],
[
"new_hidden = rnn(input_example, new_hidden)\nprint(\"new_hidden: \", new_hidden.detach().numpy())",
"new_hidden: [[ 0.27077615 -0.03183273 -0.3534962 0.28193203 -0.30477005]]\n"
]
],
[
[
"**Задание**. Модифицируйте код так, чтобы на вход можно было подавать батчи размером больше 1.",
"_____no_output_____"
],
[
"# **Классификация картинок с RNN**\n\nПредставьте, что у вас есть какая-то длинная картинка, в которой свёртки точно не зайдут. Например, снимки со спутника, спектрограмма или длиннокот.",
"_____no_output_____"
],
[
"Можно обработать их построчно с помощью рекуррентных сетей — просто подавать в качестве входа все пиксели очередной строки.\n\n<img src=\"https://cdn-images-1.medium.com/max/2000/1*wFYZpxTTiXVqncOLQd_CIQ.jpeg\" width=\"800\">",
"_____no_output_____"
]
],
[
[
"!mkdir data",
"_____no_output_____"
]
],
[
[
"Загружаем данные",
"_____no_output_____"
]
],
[
[
"import torchvision\nimport torchvision.transforms as transforms\n\nBATCH_SIZE = 64\n\n# переводим все в тензоры\ntransform = transforms.Compose(\n [transforms.ToTensor()])\n\ntrainset = torchvision.datasets.MNIST(root='./data', train=True,\n download=True, transform=transform)\n\ntestset = torchvision.datasets.MNIST(root='./data', train=False,\n download=True, transform=transform)\n\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=BATCH_SIZE,\n shuffle=True, num_workers=2)\n\ntestloader = torch.utils.data.DataLoader(testset, batch_size=BATCH_SIZE,\n shuffle=False, num_workers=2)",
"Downloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz\nDownloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz to ./data/MNIST/raw/train-images-idx3-ubyte.gz\n"
],
[
"!ls -lh data/",
"total 4.0K\ndrwxr-xr-x 3 root root 4.0K Feb 4 20:30 MNIST\n"
],
[
"%pylab inline\nimport numpy as np\n\ndef imshow(img):\n #img = img / 2 + 0.5 # unnormalize\n npimg = img.numpy()\n plt.imshow(np.transpose(npimg, (1, 2, 0)))\n\n# get some random training images\ndataiter = iter(trainloader)\nimages, labels = dataiter.next()\n\n# show images\nimshow(torchvision.utils.make_grid(images))",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"## Как выглядит классификация с RNN в общем виде ",
"_____no_output_____"
],
[
"<img src=\"https://cdn-images-1.medium.com/max/1600/1*vhAfRLlaeOXZ-bruv7Ostg.png\" width=\"400\">",
"_____no_output_____"
]
],
[
[
"class ImageRNN(nn.Module):\n def __init__(self, batch_size, n_steps, n_inputs, n_neurons, n_outputs):\n super().__init__()\n \n self.n_neurons = n_neurons\n self.batch_size = batch_size\n self.n_steps = n_steps\n self.n_inputs = n_inputs\n self.n_outputs = n_outputs\n \n self.basic_rnn = nn.RNN(self.n_inputs, self.n_neurons) \n \n self.FC = nn.Linear(self.n_neurons, self.n_outputs)\n \n def init_hidden(self,):\n # (num_layers, batch_size, n_neurons)\n return (torch.zeros(1, self.batch_size, self.n_neurons))\n \n def forward(self, X):\n # transforms X to dimensions: n_steps X batch_size X n_inputs\n X = X.permute(1, 0, 2) \n \n self.batch_size = X.size(1)\n self.hidden = self.init_hidden()\n \n lstm_out, self.hidden = self.basic_rnn(X, self.hidden) \n out = self.FC(self.hidden)\n \n return out.view(-1, self.n_outputs) # batch_size X n_output",
"_____no_output_____"
],
[
"N_STEPS = 28\nN_INPUTS = 28\nN_NEURONS = 150\nN_OUTPUTS = 10\nN_EPHOCS = 10",
"_____no_output_____"
],
[
"dataiter = iter(trainloader)\nimages, labels = dataiter.next()\nmodel = ImageRNN(BATCH_SIZE, N_STEPS, N_INPUTS, N_NEURONS, N_OUTPUTS)\nlogits = model(images.view(-1, 28,28))\nprint(logits[0:10])",
"tensor([[ 0.0653, 0.0367, 0.0443, 0.0432, -0.0721, 0.0012, 0.0674, -0.1200,\n -0.0966, -0.0399],\n [ 0.0464, 0.0159, 0.0466, 0.0295, -0.0752, -0.0066, 0.0663, -0.1007,\n -0.1376, -0.0278],\n [ 0.0478, 0.0183, 0.0476, 0.0310, -0.0897, -0.0032, 0.0704, -0.1145,\n -0.1291, -0.0384],\n [ 0.0422, 0.0136, 0.0452, 0.0347, -0.0862, 0.0051, 0.0753, -0.1082,\n -0.1272, -0.0388],\n [ 0.0495, 0.0093, 0.0541, 0.0372, -0.0700, 0.0019, 0.0626, -0.0980,\n -0.1407, -0.0256],\n [ 0.0522, 0.0100, 0.0539, 0.0339, -0.0754, 0.0017, 0.0610, -0.0961,\n -0.1441, -0.0271],\n [ 0.0416, 0.0067, 0.0521, 0.0337, -0.0761, -0.0013, 0.0603, -0.0984,\n -0.1372, -0.0256],\n [ 0.0857, 0.0294, 0.0855, 0.0180, -0.0717, -0.0492, 0.0711, -0.1305,\n -0.0891, -0.0816],\n [ 0.0440, 0.0086, 0.0524, 0.0329, -0.0751, -0.0054, 0.0621, -0.0975,\n -0.1364, -0.0260],\n [ 0.0462, 0.0194, 0.0449, 0.0309, -0.0785, -0.0012, 0.0709, -0.1109,\n -0.1327, -0.0402]], grad_fn=<SliceBackward0>)\n"
]
],
[
[
"## Обучаем",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\n\n# Device\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\n# Model instance\nmodel = ImageRNN(BATCH_SIZE, N_STEPS, N_INPUTS, N_NEURONS, N_OUTPUTS)\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.Adam(model.parameters(), lr=0.001)\n\ndef get_accuracy(logit, target, batch_size):\n ''' Obtain accuracy for training round '''\n corrects = (torch.max(logit, 1)[1].view(target.size()).data == target.data).sum()\n accuracy = 100.0 * corrects/batch_size\n return accuracy.item()",
"_____no_output_____"
],
[
"for epoch in range(N_EPHOCS):\n train_running_loss = 0.0\n train_acc = 0.0\n model.train()\n \n # TRAINING ROUND\n for i, data in enumerate(trainloader):\n # zero the parameter gradients\n optimizer.zero_grad()\n \n # reset hidden states\n model.hidden = model.init_hidden() \n \n # get the inputs\n inputs, labels = data\n inputs = inputs.view(-1, 28,28) \n\n # forward + backward + optimize\n outputs = model(inputs)\n\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n\n train_running_loss += loss.detach().item()\n train_acc += get_accuracy(outputs, labels, BATCH_SIZE)\n \n model.eval()\n print('Epoch: %d | Loss: %.4f | Train Accuracy: %.2f' \n %(epoch, train_running_loss / i, train_acc/i))",
"Epoch: 0 | Loss: 0.7357 | Train Accuracy: 75.37\nEpoch: 1 | Loss: 0.3086 | Train Accuracy: 91.03\nEpoch: 2 | Loss: 0.2221 | Train Accuracy: 93.71\nEpoch: 3 | Loss: 0.1837 | Train Accuracy: 94.76\nEpoch: 4 | Loss: 0.1615 | Train Accuracy: 95.46\nEpoch: 5 | Loss: 0.1460 | Train Accuracy: 95.86\nEpoch: 6 | Loss: 0.1369 | Train Accuracy: 96.19\nEpoch: 7 | Loss: 0.1283 | Train Accuracy: 96.43\nEpoch: 8 | Loss: 0.1197 | Train Accuracy: 96.66\nEpoch: 9 | Loss: 0.1190 | Train Accuracy: 96.70\n"
]
],
[
[
"### Смотрим что на тесте",
"_____no_output_____"
]
],
[
[
"test_acc = 0.0\nfor i, data in enumerate(testloader, 0):\n inputs, labels = data\n inputs = inputs.view(-1, 28, 28)\n\n outputs = model(inputs)\n\n test_acc += get_accuracy(outputs, labels, BATCH_SIZE)\n \nprint('Test Accuracy: %.2f'%( test_acc/i))",
"Test Accuracy: 96.39\n"
]
],
[
[
"# **Сентимент анализ**\n\nДомашка — классифицировать отзывы с IMDB на положительный / отрицательный только по тексту.\n\n<img src=\"https://github.com/bentrevett/pytorch-sentiment-analysis/raw/bf8cc46e4823ebf9af721b595501ad6231c73632/assets/sentiment1.png\">\n\nСуть такая же, только нужно предобработать тексты — каждому слову сопоставить обучаемый вектор (embedding), который пойдёт дальше в RNN.",
"_____no_output_____"
]
],
[
[
"# это уберет боль работы с текстами\n#!pip install --pre torchtext==0.11.0 -f https://download.pytorch.org/whl/nightly/cpu/torch_nightly.html\n#!python -m spacy download en",
"_____no_output_____"
],
[
"#!pip install --pre torch==1.09.0",
"_____no_output_____"
]
],
[
[
"**Примечание.** Torchtext уже не очень живой проект, а в spacy нет русского.\n",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchtext.legacy import data\n\nSEED = 1234\n\ntorch.manual_seed(SEED)\ntorch.backends.cudnn.deterministic = True\n\nTEXT = data.Field(tokenize = 'spacy',\n tokenizer_language = 'en_core_web_sm')\nLABEL = data.LabelField(dtype = torch.float)",
"_____no_output_____"
],
[
"from torchtext.legacy import datasets\n\ntrain_data0, test_data0 = datasets.IMDB.splits(TEXT, LABEL)",
"downloading aclImdb_v1.tar.gz\n"
]
],
[
[
"# start from here",
"_____no_output_____"
]
],
[
[
"import copy\n#train_data = copy.deepcopy(train_data0)\n#test_data = copy.deepcopy(test_data0)\n\ntrain_data = train_data0\ntest_data = test_data0\n\nprint(f'Number of training examples: {len(train_data)}')\nprint(f'Number of testing examples: {len(test_data)}')",
"Number of training examples: 25000\nNumber of testing examples: 25000\n"
],
[
"# ls -lh data/imdb/aclImdb/",
"_____no_output_____"
],
[
"print(vars(train_data.examples[0]))",
"{'text': ['This', 'film', 'reminds', 'me', 'of', '42nd', 'Street', 'starring', 'Bebe', 'Daniels', 'and', 'Ruby', 'Keeler', '.', 'When', 'I', 'watch', 'this', 'film', 'a', 'lot', 'of', 'it', 'reminded', 'me', 'of', '42nd', 'Street', ',', 'especially', 'the', 'character', 'Eloise', 'who', \"'s\", 'a', 'temperamental', 'star', 'and', 'she', 'ends', 'up', 'falling', 'and', 'breaks', 'her', 'ankle', ',', 'like', 'Bebe', 'Daniels', 'did', 'in', '42nd', 'Street', 'and', 'another', 'performer', 'gets', 'the', 'part', 'and', 'become', 'a', 'star', '.', 'This', 'film', ',', 'like', 'most', 'race', 'films', ',', 'keeps', 'people', 'watching', 'because', 'of', 'the', 'great', 'entertainment', '.', 'Race', 'films', 'always', 'showed', 'Black', 'Entertainment', 'as', 'it', 'truly', 'was', 'that', 'was', 'popular', 'in', 'that', 'time', 'era', '.', 'The', 'Dancing', 'Styles', ',', 'The', 'Music', ',', 'Dressing', 'Styles', ',', 'You', \"'ll\", 'Love', 'It', '.', 'This', 'movie', 'could', 'of', 'been', 'big', 'if', 'it', 'was', 'made', 'in', 'Hollywood', ',', 'it', 'would', 'of', 'had', 'better', 'scenery', ',', 'better', 'filming', ',', 'and', 'more', 'money', 'which', 'would', 'make', 'any', 'movie', 'better', '.', 'But', 'its', 'worth', 'watching', 'because', 'it', 'is', 'good', 'and', 'Micheaux', 'does', 'good', 'with', 'the', 'little', 'he', 'has', '.', 'I', 'have', 'to', 'say', 'out', 'of', 'all', 'Micheaux', \"'s\", 'films', ',', 'Swing', 'is', 'the', 'best', '!', 'The', 'movie', 'features', 'singers', ',', 'dancers', ',', 'actresses', ',', 'and', 'actors', 'who', 'were', 'popular', 'but', 'forgotten', 'today', '.', 'Doli', 'Armena', ',', 'a', 'awesome', 'female', 'trumpet', 'player', 'who', 'can', 'blow', 'the', 'horn', 'so', 'good', 'that', 'you', 'think', 'Gabriel', 'is', 'blowing', 'a', 'horn', 'in', 'the', 'sky', '.', 'The', 'sexy', ',', 'hot', 'female', 'dancer', 'Consuela', 'Harris', 'would', 'put', 'Ann', 'Miller', 'and', 'Gyspy', 'Rose', 'Lee', 'to', 'shame.<br', '/><br', '/>Adding', 'further', 'info', '...', 'Popular', 'blues', 'singer', 'of', 'the', '20', \"'s\", 'and', '30', \"'s\", 'Cora', 'Green', 'is', 'the', 'focus', 'of', 'the', 'film', ',', 'she', \"'s\", 'Mandy', ',', 'a', 'good', ',', 'hard', 'working', 'woman', 'with', 'a', 'no', 'good', 'man', 'who', 'takes', 'her', 'money', 'and', 'spend', 'it', 'on', 'other', 'women', '.', 'A', 'nosy', 'neighbor', 'played', 'by', 'Amanda', 'Randolph', 'tells', 'Mandy', 'what', 'she', 'seen', 'and', 'heard', 'and', 'Mandy', 'goes', 'down', 'to', 'the', 'club', 'and', 'catches', 'her', 'man', 'with', 'an', 'attractive', ',', 'curvy', 'woman', 'by', 'the', 'name', 'of', 'Eloise', '(', 'played', 'Hazel', 'Diaz', ',', 'a', 'Hot', '-', 'Cha', 'entertainer', 'in', 'the', '30', \"'s\", ')', 'and', 'a', 'fight', 'breaks', 'out', '.', 'Then', 'Mandy', 'goes', 'to', 'Harlem', 'where', 'she', 'reunites', 'with', 'a', 'somewhat', 'guardian', 'angel', 'Lena', 'played', 'by', 'one', 'of', 'the', 'most', 'beautiful', 'women', 'in', 'movies', 'Dorothy', 'Van', 'Engle', '.', 'Lena', 'provides', 'Mandy', 'with', 'a', 'home', ',', 'a', 'job', ',', 'and', 'helps', 'her', 'become', 'a', 'star', 'when', 'temperamental', 'Cora', 'Smith', '(', 'played', 'by', 'Hazel', ',', 'I', 'guess', 'she', \"'s\", 'playing', 'two', 'parts', 'or', 'maybe', 'she', 'changed', 'her', 'stage', 'name', ')', 'tries', 'to', 'ruin', 'the', 'show', 'with', 'her', 'bad', 'behavior', '.', 'When', 'Cora', 'gets', 'drunk', 'and', 'breaks', 'her', 'leg', ',', 'Lena', 'convinces', 'everyone', 'that', 'Mandy', 'is', 'right', 'for', 'the', 'job', 'and', 'Lena', 'is', 'right', 'and', 'a', 'star', 'is', 'born', 'in', 'Mandy', '.', 'Tall', ',', 'long', ',', 'lanky', ',', 'but', 'handsome', 'Carman', 'Newsome', 'is', 'the', 'cool', 'aspiring', 'producer', 'who', 'Lena', 'looks', 'out', 'for', 'as', 'well', '.', 'Pretty', 'boy', 'Larry', 'Seymour', 'plays', 'the', 'no', 'good', 'man', 'but', 'after', 'Lena', 'threatens', 'him', ',', 'he', 'might', 'shape', 'up', '.', 'There', 'are', 'a', 'few', 'highlights', 'but', 'the', 'one', 'that', 'sticks', 'out', 'to', 'me', 'is', 'the', 'part', 'where', 'Cora', 'Smith', '(', 'Hazel', 'Diaz', ')', 'struts', 'in', 'late', 'for', 'rehearsal', 'and', 'goes', 'off', 'on', 'everyone', 'and', 'then', 'her', 'man', 'comes', 'in', 'and', 'punches', 'her', 'in', 'the', 'jaw', 'but', 'that', \"'s\", 'not', 'enough', ',', 'she', 'almost', 'gets', 'into', 'a', 'fight', 'with', 'Mandy', 'again', '.', 'In', 'between', 'there', \"'s\", 'great', 'entertainment', 'by', 'chorus', 'girls', ',', 'tap', 'dancers', ',', 'shake', 'dancers', ',', 'swing', 'music', ',', 'and', 'blues', 'singing', '.', 'There', \"'s\", 'even', 'white', 'people', 'watching', 'the', 'entertainment', ',', 'I', 'wonder', 'where', 'Micheaux', 'found', 'them', ',', 'there', \"'s\", 'even', 'a', 'scene', 'where', 'there', \"'s\", 'blacks', 'and', 'whites', 'sitting', 'together', 'at', 'the', 'club', ',', 'Micheaux', 'frequently', 'integrated', 'blacks', 'and', 'whites', 'in', 'his', 'films', ',', 'he', 'should', 'be', 'commended', 'for', 'such', 'a', 'bold', 'move.<br', '/><br', '/>This', 'movie', 'was', 'the', 'first', 'race', 'film', 'I', 'really', 'enjoyed', 'and', 'it', 'helped', 'introduced', 'me', 'to', 'Oscar', 'Micheaux', '.', 'This', 'movie', 'is', 'one', 'of', 'the', 'best', 'of', 'the', 'race', 'film', 'genre', ',', 'its', 'a', 'behind', 'the', 'scenes', 'story', 'about', 'the', 'ups', 'and', 'downs', 'of', 'show', 'business.<br', '/><br', '/>No', 'these', 'early', 'race', 'films', 'may', 'not', 'be', 'the', 'best', ',', 'ca', \"n't\", 'be', 'compared', 'with', 'Hallelujah', ',', 'Green', 'Pastures', ',', 'Stormy', 'Weather', ',', 'Cabin', 'In', 'The', 'Sky', ',', 'Carmen', 'Jones', ',', 'or', 'any', 'other', 'Hollywood', 'films', 'but', 'their', 'great', 'to', 'watch', 'because', 'their', 'early', 'signs', 'of', 'black', 'film', '-', 'making', 'and', 'plus', 'these', 'films', 'provide', 'a', 'glimpse', 'into', 'black', 'life', 'and', 'black', 'entertainment', 'through', 'a', 'black', 'person', \"'s\", 'eyes', '.', 'These', 'films', 'gave', 'blacks', 'a', 'chance', 'to', 'play', 'people', 'from', 'all', 'walks', 'of', 'life', ',', 'be', 'beautiful', ',', 'classy', ',', 'and', 'elegant', ',', 'and', 'not', 'just', 'be', 'stereotypes', 'or', 'how', 'whites', 'felt', 'blacks', 'should', 'be', 'portrayed', 'like', 'in', 'Hollywood', '.', 'Most', 'of', 'the', 'actors', 'and', 'actresses', 'of', 'these', 'race', 'films', 'were', \"n't\", 'the', 'best', ',', 'but', 'they', 'were', 'the', 'only', 'ones', 'that', 'could', 'be', 'afforded', 'at', 'the', 'time', ',', 'Micheaux', 'and', 'Spencer', 'Williams', 'could', \"n't\", 'afford', 'Nina', 'Mae', 'McKinney', ',', 'Josephine', 'Baker', ',', 'Ethel', 'Waters', ',', 'Fredi', 'Washington', ',', 'Paul', 'Robeson', ',', 'Rex', 'Ingram', ',', 'and', 'more', 'of', 'the', 'bigger', 'stars', ',', 'so', 'Micheaux', 'and', 'other', 'black', 'and', 'white', 'race', 'film', '-', 'makers', 'would', 'use', 'nightclub', 'performers', 'in', 'their', 'movies', ',', 'some', 'were', 'good', ',', 'some', 'were', \"n't\", 'great', 'actors', 'and', 'actresses', ',', 'but', 'I', 'think', 'Micheaux', 'and', 'others', 'knew', 'most', 'were', \"n't\", 'good', 'actors', 'and', 'actresses', 'but', 'they', 'were', 'used', 'more', 'as', 'apart', 'of', 'an', 'experiment', 'than', 'for', 'true', 'talent', ',', 'they', 'just', 'wanted', 'their', 'stories', 'told', ',', 'and', 'in', 'return', 'many', 'black', 'performers', 'got', 'to', 'perform', 'their', 'true', 'talents', 'in', 'the', 'films', '.', 'For', 'some', 'true', 'actors', '/', 'actresses', 'race', 'films', 'were', 'the', 'only', 'type', 'of', 'films', 'they', 'could', 'get', 'work', ',', 'especially', 'if', 'they', 'did', \"n't\", 'want', 'to', 'play', 'Hollywood', 'stereotypes', ',', 'so', 'I', 'think', 'you', \"'ll\", 'be', 'able', 'to', 'spot', 'the', 'true', 'actors', '/', 'actresses', 'from', 'the', 'nightclub', 'performers', '.', 'These', 'race', 'films', 'are', 'very', 'historic', ',', 'they', 'could', 'have', 'been', 'lost', 'forever', ',', 'many', 'are', 'lost', ',', 'maybe', 'race', 'films', 'are', \"n't\", 'the', 'greatest', 'example', 'of', 'cinema', 'but', 'even', 'Hollywood', 'films', 'did', \"n't\", 'start', 'out', 'great', 'in', 'the', 'beginning', '.', 'I', 'think', 'if', 'the', 'race', 'film', 'genre', 'continued', ',', 'it', 'would', 'have', 'better', '.', 'If', 'your', 'looking', 'for', 'great', 'acting', ',', 'most', 'race', 'films', 'are', \"n't\", 'the', 'ones', ',', 'but', 'if', 'your', 'looking', 'for', 'a', 'real', 'example', 'of', 'black', 'entertainment', 'and', 'how', 'blacks', 'should', 'have', 'been', 'portrayed', 'in', 'films', ',', 'than', 'watch', 'race', 'films', '.', 'There', 'are', 'some', 'entertaining', 'race', 'films', 'with', 'a', 'good', 'acting', 'cast', ',', 'Moon', 'Over', 'Harlem', ',', 'Body', 'and', 'Soul', ',', 'Paradise', 'In', 'Harlem', ',', 'Keep', 'Punching', ',', 'Sunday', 'Sinners', ',', 'Dark', 'Manhattan', ',', 'Broken', 'Strings', ',', 'Boy', '!', 'What', 'A', 'Girl', ',', 'Mystery', 'In', 'Swing', ',', 'Miracle', 'In', 'Harlem', ',', 'and', 'Sepia', 'Cinderella', ',', 'that', 'not', 'only', 'has', 'good', 'entertainment', 'but', 'good', 'acting', '.'], 'label': 'pos'}\n"
],
[
"import random\n\ntrain_data, valid_data = train_data.split(random_state = random.seed(SEED))",
"_____no_output_____"
],
[
"print(f'Number of training examples: {len(train_data)}')\nprint(f'Number of validation examples: {len(valid_data)}')\nprint(f'Number of testing examples: {len(test_data)}')",
"Number of training examples: 17500\nNumber of validation examples: 7500\nNumber of testing examples: 25000\n"
],
[
"# Сделаем словарь\nMAX_VOCAB_SIZE = 25_000\n\nTEXT.build_vocab(train_data, max_size = MAX_VOCAB_SIZE)\nLABEL.build_vocab(train_data)",
"_____no_output_____"
],
[
"print(f\"Unique tokens in TEXT vocabulary: {len(TEXT.vocab)}\")\nprint(f\"Unique tokens in LABEL vocabulary: {len(LABEL.vocab)}\")",
"Unique tokens in TEXT vocabulary: 25002\nUnique tokens in LABEL vocabulary: 2\n"
],
[
"vars(LABEL.vocab)",
"_____no_output_____"
]
],
[
[
"Почему 25002, а не 25000?\nПотому что $<unk>$ и $<pad>$\n\n<img src=\"https://github.com/bentrevett/pytorch-sentiment-analysis/raw/bf8cc46e4823ebf9af721b595501ad6231c73632/assets/sentiment6.png\" width=\"160\">",
"_____no_output_____"
]
],
[
[
"print(TEXT.vocab.freqs.most_common(20))",
"[('the', 203371), (',', 192722), ('.', 166127), ('and', 109776), ('a', 109461), ('of', 100980), ('to', 94411), ('is', 76392), ('in', 61619), ('I', 54333), ('it', 53696), ('that', 49248), ('\"', 43554), (\"'s\", 43160), ('this', 42597), ('-', 36914), ('/><br', 35751), ('was', 35314), ('as', 30499), ('with', 29921)]\n"
]
],
[
[
"* stoi (string to int)\n* itos (int to string)",
"_____no_output_____"
]
],
[
[
"print(TEXT.vocab.itos[:10])",
"['<unk>', '<pad>', 'the', ',', '.', 'and', 'a', 'of', 'to', 'is']\n"
],
[
"print(LABEL.vocab.stoi)",
"defaultdict(None, {'neg': 0, 'pos': 1})\n"
]
],
[
[
"## Делаем модель",
"_____no_output_____"
],
[
"<img src=\"https://github.com/bentrevett/pytorch-sentiment-analysis/raw/bf8cc46e4823ebf9af721b595501ad6231c73632/assets/sentiment7.png\" width=\"450\">",
"_____no_output_____"
],
[
"* В эмбеддер (emb = [torch.nn.Embedding(num_embeddings, embedding_dim)](https://pytorch.org/docs/stable/nn.html?highlight=embedding#torch.nn.Embedding)) запихиваем тензор размерностью **[sentence length, batch size]**\n* Эмбеддер возвращает тензор размерностью **[sentence length, batch size, embedding dim]**\n* RNN (torch.nn.RNN(embedding_dim, hidden_dim)) возвращает 2 тензора, *output* размера [sentence length, batch size, hidden dim] и *hidden* размера [1, batch size, hidden dim]",
"_____no_output_____"
]
],
[
[
"# pretrained model\nMAX_VOCAB_SIZE = 25_000\n\nTEXT.build_vocab(train_data, \n max_size = MAX_VOCAB_SIZE, \n vectors = \"glove.6B.100d\", \n unk_init = torch.Tensor.normal_)\n\nLABEL.build_vocab(train_data)",
".vector_cache/glove.6B.zip: 862MB [02:42, 5.30MB/s] \n100%|█████████▉| 399999/400000 [00:14<00:00, 26701.26it/s]\n"
],
[
"BATCH_SIZE = 64\n\ndevice = torch.device('cuda' if torch.cuda.is_available() else 'cpu')\n\ntrain_iterator, valid_iterator, test_iterator = data.BucketIterator.splits(\n (train_data, valid_data, test_data), \n batch_size = BATCH_SIZE, \n device = device)",
"_____no_output_____"
],
[
"import torch.nn as nn\nimport torch.nn.functional as F\n\nclass BestText(nn.Module):\n def __init__(self, vocab_size, embedding_dim, output_dim, pad_idx):\n \n super().__init__()\n \n self.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx=pad_idx)\n \n self.fc = nn.Linear(embedding_dim, output_dim)\n \n def forward(self, text):\n \n #text = [sent len, batch size]\n \n embedded = self.embedding(text)\n \n #embedded = [sent len, batch size, emb dim]\n \n embedded = embedded.permute(1, 0, 2)\n \n #embedded = [batch size, sent len, emb dim]\n \n pooled = F.avg_pool2d(embedded, (embedded.shape[1], 1)).squeeze(1) \n \n #pooled = [batch size, embedding_dim]\n \n return self.fc(pooled)",
"_____no_output_____"
],
[
"import torch.nn as nn\n\nclass RNN(nn.Module):\n def __init__(self, input_dim, embedding_dim, hidden_dim, output_dim):\n super().__init__()\n \n ##self.embedding = nn.Embedding(input_dim, embedding_dim)\n self.embedding = nn.Embedding(input_dim, embedding_dim)\n # ## nn.Embedding.from_pretrained(weight)\n # nn_output_dim = hidden_dim\n # # nn.LSTM(5, 100, 1, bidirectional=True)\n # self.rnn = nn.LSTM(embedding_dim, nn_output_dim, bidirectional=True)\n # # линейный слой, который делает проекцию в 2 класса\n\n self.rnn = nn.RNN(embedding_dim, hidden_dim, bidirectional=True)\n self.fc = nn.Linear(hidden_dim, output_dim)\n \n def forward(self, text):\n\n #text, shape = [sent len, batch size]\n \n embedded = self.embedding(text)\n \n #embedded.shape = [sent len, batch size, emb dim]\n \n output, hidden = self.rnn(embedded)\n \n #output.shape = [sent len, batch size, hid dim]\n #hidden.shape = [1, batch size, hid dim]\n \n assert torch.equal(output[-1,:,:], hidden.squeeze(0))\n \n return self.fc(hidden.squeeze(0))",
"_____no_output_____"
],
[
"import torch.nn as nn\n\nclass RNN(nn.Module):\n def __init__(self, input_dim, embedding_dim, hidden_dim, output_dim):\n \n super().__init__()\n \n self.embedding = nn.Embedding(input_dim, embedding_dim)\n \n self.rnn1 = nn.RNN(embedding_dim, hidden_dim)\n self.rnn2 = nn.RNN(hidden_dim, hidden_dim)\n \n self.fc = nn.Linear(hidden_dim, output_dim)\n \n def forward(self, text):\n\n #text = [sent len, batch size]\n \n embedded = self.embedding(text)\n \n #embedded = [sent len, batch size, emb dim]\n \n output, hidden = self.rnn1(embedded)\n output, hidden = self.rnn2(output)\n \n #output = [sent len, batch size, hid dim]\n #hidden = [1, batch size, hid dim]\n \n assert torch.equal(output[-1,:,:], hidden.squeeze(0))\n \n return self.fc(hidden.squeeze(0))",
"_____no_output_____"
]
],
[
[
"https://cnvrg.io/pytorch-lstm/ чекнуть потом, реализация LSTM",
"_____no_output_____"
]
],
[
[
"class LSTM1(nn.Module):\n def __init__(self, num_classes, input_size, hidden_size, num_layers, seq_length):\n super(LSTM1, self).__init__()\n self.num_classes = num_classes #number of classes\n self.num_layers = num_layers #number of layers\n self.input_size = input_size #input size\n self.hidden_size = hidden_size #hidden state\n self.seq_length = seq_length #sequence length\n\n self.lstm = nn.LSTM(input_size=input_size, hidden_size=hidden_size,\n num_layers=num_layers, batch_first=True) #lstm\n self.fc_1 = nn.Linear(hidden_size, 128) #fully connected 1\n self.fc = nn.Linear(128, num_classes) #fully connected last layer\n\n self.relu = nn.ReLU()\n \n def forward(self,x):\n h_0 = Variable(torch.zeros(self.num_layers, x.size(0), self.hidden_size)) #hidden state\n c_0 = Variable(torch.zeros(self.num_layers, x.size(0), self.hidden_size)) #internal state\n # Propagate input through LSTM\n output, (hn, cn) = self.lstm(x, (h_0, c_0)) #lstm with input, hidden, and internal state\n hn = hn.view(-1, self.hidden_size) #reshaping the data for Dense layer next\n out = self.relu(hn)\n out = self.fc_1(out) #first Dense\n out = self.relu(out) #relu\n out = self.fc(out) #Final Output\n return out",
"_____no_output_____"
]
],
[
[
"## Тренировка модели",
"_____no_output_____"
]
],
[
[
"INPUT_DIM = len(TEXT.vocab)\nEMBEDDING_DIM = 100\nOUTPUT_DIM = 1\nPAD_IDX = TEXT.vocab.stoi[TEXT.pad_token]\n\nmodel = BestText(INPUT_DIM, EMBEDDING_DIM, OUTPUT_DIM, PAD_IDX)",
"_____no_output_____"
],
[
"# INPUT_DIM = len(TEXT.vocab)\n# EMBEDDING_DIM = 128\n# HIDDEN_DIM = 128\n# OUTPUT_DIM = 1\n\n# num_epochs = 10 #1000 epochs\n# learning_rate = 0.001 #0.001 lr\n\n# input_size = INPUT_DIM #number of features\n# hidden_size = HIDDEN_DIM #number of features in hidden state\n# num_layers = 2 #number of stacked lstm layers\n\n# num_classes = 2 #number of output classes \n\n# model = RNN(INPUT_DIM, EMBEDDING_DIM, HIDDEN_DIM, OUTPUT_DIM)\n# #model = LSTM1(num_classes, input_size, hidden_size, num_layers, X_train_tensors_final.shape[1])",
"_____no_output_____"
],
[
"def count_parameters(model):\n return sum(p.numel() for p in model.parameters() if p.requires_grad)\n\nprint(f'The model has {count_parameters(model):,} trainable parameters')",
"The model has 2,500,301 trainable parameters\n"
],
[
"# copy the pre-trained vectors to our embedding layer\npretrained_embeddings = TEXT.vocab.vectors\n\nmodel.embedding.weight.data.copy_(pretrained_embeddings)",
"_____no_output_____"
],
[
"# zero the initial weights of our unknown and padding tokens\nUNK_IDX = TEXT.vocab.stoi[TEXT.unk_token]\n\nmodel.embedding.weight.data[UNK_IDX] = torch.zeros(EMBEDDING_DIM)\nmodel.embedding.weight.data[PAD_IDX] = torch.zeros(EMBEDDING_DIM)",
"_____no_output_____"
],
[
"import torch.optim as optim\n\n# optimizer = optim.RMSprop(model.parameters(), lr=1e-3)\noptimizer = optim.Adam(model.parameters(), lr=1e-3)",
"_____no_output_____"
],
[
"criterion = nn.BCEWithLogitsLoss()",
"_____no_output_____"
],
[
"model = model.to(device)\ncriterion = criterion.to(device)",
"_____no_output_____"
],
[
"def binary_accuracy(preds, y):\n # округление к ближайшему целому round predictions to the closest integer\n rounded_preds = torch.round(torch.sigmoid(preds))\n correct = (rounded_preds == y).float() #convert into float for division \n acc = correct.sum() / len(correct)\n return acc",
"_____no_output_____"
],
[
"def train(model, iterator, optimizer, criterion):\n \n epoch_loss = 0\n epoch_acc = 0\n \n model.train()\n \n for batch in iterator:\n \n optimizer.zero_grad()\n \n predictions = model(batch.text).squeeze(1)\n \n loss = criterion(predictions, batch.label)\n \n acc = binary_accuracy(predictions, batch.label)\n \n loss.backward()\n \n optimizer.step()\n \n epoch_loss += loss.item()\n epoch_acc += acc.item()\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)",
"_____no_output_____"
],
[
"def evaluate(model, iterator, criterion):\n \n epoch_loss = 0\n epoch_acc = 0\n \n model.eval()\n \n with torch.no_grad():\n \n for batch in iterator:\n\n predictions = model(batch.text).squeeze(1)\n \n loss = criterion(predictions, batch.label)\n \n acc = binary_accuracy(predictions, batch.label)\n\n epoch_loss += loss.item()\n epoch_acc += acc.item()\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)",
"_____no_output_____"
],
[
"import time\n\ndef epoch_time(start_time, end_time):\n elapsed_time = end_time - start_time\n elapsed_mins = int(elapsed_time / 60)\n elapsed_secs = int(elapsed_time - (elapsed_mins * 60))\n return elapsed_mins, elapsed_secs",
"_____no_output_____"
],
[
"N_EPOCHS = 30\n\nbest_valid_loss = float('inf')\n\nfor epoch in range(N_EPOCHS):\n\n start_time = time.time()\n \n train_loss, train_acc = train(model, train_iterator, optimizer, criterion)\n valid_loss, valid_acc = evaluate(model, valid_iterator, criterion)\n \n end_time = time.time()\n\n epoch_mins, epoch_secs = epoch_time(start_time, end_time)\n \n if valid_loss < best_valid_loss:\n best_valid_loss = valid_loss\n torch.save(model.state_dict(), 'best_weights.pt')\n \n print(f'Epoch: {epoch+1:02} | Epoch Time: {epoch_mins}m {epoch_secs}s')\n print(f'\\tTrain Loss: {train_loss:.3f} | Train Acc: {train_acc*100:.2f}%')\n print(f'\\t Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:.2f}%')",
"Epoch: 01 | Epoch Time: 0m 4s\n\tTrain Loss: 0.384 | Train Acc: 87.84%\n\t Val. Loss: 0.430 | Val. Acc: 85.77%\nEpoch: 02 | Epoch Time: 0m 4s\n\tTrain Loss: 0.344 | Train Acc: 88.97%\n\t Val. Loss: 0.449 | Val. Acc: 86.50%\nEpoch: 03 | Epoch Time: 0m 4s\n\tTrain Loss: 0.314 | Train Acc: 89.98%\n\t Val. Loss: 0.455 | Val. Acc: 87.02%\nEpoch: 04 | Epoch Time: 0m 4s\n\tTrain Loss: 0.290 | Train Acc: 90.86%\n\t Val. Loss: 0.470 | Val. Acc: 87.54%\nEpoch: 05 | Epoch Time: 0m 4s\n\tTrain Loss: 0.270 | Train Acc: 91.50%\n\t Val. Loss: 0.483 | Val. Acc: 87.83%\nEpoch: 06 | Epoch Time: 0m 5s\n\tTrain Loss: 0.250 | Train Acc: 92.07%\n\t Val. Loss: 0.497 | Val. Acc: 87.93%\nEpoch: 07 | Epoch Time: 0m 4s\n\tTrain Loss: 0.236 | Train Acc: 92.59%\n\t Val. Loss: 0.509 | Val. Acc: 88.20%\nEpoch: 08 | Epoch Time: 0m 4s\n\tTrain Loss: 0.220 | Train Acc: 93.24%\n\t Val. Loss: 0.524 | Val. Acc: 88.50%\nEpoch: 09 | Epoch Time: 0m 4s\n\tTrain Loss: 0.208 | Train Acc: 93.63%\n\t Val. Loss: 0.533 | Val. Acc: 88.61%\nEpoch: 10 | Epoch Time: 0m 4s\n\tTrain Loss: 0.195 | Train Acc: 94.04%\n\t Val. Loss: 0.549 | Val. Acc: 88.85%\nEpoch: 11 | Epoch Time: 0m 4s\n\tTrain Loss: 0.185 | Train Acc: 94.39%\n\t Val. Loss: 0.564 | Val. Acc: 88.94%\nEpoch: 12 | Epoch Time: 0m 4s\n\tTrain Loss: 0.175 | Train Acc: 94.74%\n\t Val. Loss: 0.574 | Val. Acc: 89.09%\nEpoch: 13 | Epoch Time: 0m 4s\n\tTrain Loss: 0.166 | Train Acc: 95.05%\n\t Val. Loss: 0.592 | Val. Acc: 89.13%\nEpoch: 14 | Epoch Time: 0m 4s\n\tTrain Loss: 0.156 | Train Acc: 95.46%\n\t Val. Loss: 0.602 | Val. Acc: 89.16%\nEpoch: 15 | Epoch Time: 0m 4s\n\tTrain Loss: 0.148 | Train Acc: 95.71%\n\t Val. Loss: 0.618 | Val. Acc: 89.30%\nEpoch: 16 | Epoch Time: 0m 4s\n\tTrain Loss: 0.140 | Train Acc: 96.01%\n\t Val. Loss: 0.634 | Val. Acc: 89.30%\nEpoch: 17 | Epoch Time: 0m 4s\n\tTrain Loss: 0.132 | Train Acc: 96.36%\n\t Val. Loss: 0.650 | Val. Acc: 89.41%\nEpoch: 18 | Epoch Time: 0m 4s\n\tTrain Loss: 0.125 | Train Acc: 96.62%\n\t Val. Loss: 0.665 | Val. Acc: 89.38%\nEpoch: 19 | Epoch Time: 0m 4s\n\tTrain Loss: 0.118 | Train Acc: 96.86%\n\t Val. Loss: 0.683 | Val. Acc: 89.43%\nEpoch: 20 | Epoch Time: 0m 4s\n\tTrain Loss: 0.112 | Train Acc: 97.07%\n\t Val. Loss: 0.702 | Val. Acc: 89.46%\nEpoch: 21 | Epoch Time: 0m 4s\n\tTrain Loss: 0.105 | Train Acc: 97.29%\n\t Val. Loss: 0.719 | Val. Acc: 89.46%\nEpoch: 22 | Epoch Time: 0m 4s\n\tTrain Loss: 0.099 | Train Acc: 97.52%\n\t Val. Loss: 0.742 | Val. Acc: 89.39%\nEpoch: 23 | Epoch Time: 0m 4s\n\tTrain Loss: 0.094 | Train Acc: 97.66%\n\t Val. Loss: 0.760 | Val. Acc: 89.43%\nEpoch: 24 | Epoch Time: 0m 4s\n\tTrain Loss: 0.090 | Train Acc: 97.83%\n\t Val. Loss: 0.779 | Val. Acc: 89.54%\nEpoch: 25 | Epoch Time: 0m 4s\n\tTrain Loss: 0.085 | Train Acc: 97.97%\n\t Val. Loss: 0.797 | Val. Acc: 89.62%\nEpoch: 26 | Epoch Time: 0m 4s\n\tTrain Loss: 0.080 | Train Acc: 98.16%\n\t Val. Loss: 0.818 | Val. Acc: 89.53%\nEpoch: 27 | Epoch Time: 0m 4s\n\tTrain Loss: 0.075 | Train Acc: 98.29%\n\t Val. Loss: 0.839 | Val. Acc: 89.55%\nEpoch: 28 | Epoch Time: 0m 4s\n\tTrain Loss: 0.072 | Train Acc: 98.43%\n\t Val. Loss: 0.862 | Val. Acc: 89.51%\nEpoch: 29 | Epoch Time: 0m 4s\n\tTrain Loss: 0.068 | Train Acc: 98.55%\n\t Val. Loss: 0.884 | Val. Acc: 89.53%\nEpoch: 30 | Epoch Time: 0m 4s\n\tTrain Loss: 0.064 | Train Acc: 98.68%\n\t Val. Loss: 0.907 | Val. Acc: 89.42%\n"
],
[
"model.load_state_dict(torch.load('best_weights.pt'))\n\ntest_loss, test_acc = evaluate(model, test_iterator, criterion)\n\nprint(f'Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:.2f}%')",
"Test Loss: 0.448 | Test Acc: 85.27%\n"
]
],
[
[
"## Вывод:\n\nВзял предобученный embedding, оптимизатор Adam. RMSprop обычно лучше показывает себя в текстах, но по результатам эксперимента лучшим оказался Adam.\n\nДобился точности ~85.3% на тестовой выборке датасета IMBD.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ec8b886b8ce083284b63fad7d81ae4a3134a5626 | 264,337 | ipynb | Jupyter Notebook | Analysis/1_transcriptome_analysis.ipynb | BioinfoTongLI/NanoHack21-1 | 562deb4a8df200756fd47c942e2c25417f64c67a | [
"MIT"
] | null | null | null | Analysis/1_transcriptome_analysis.ipynb | BioinfoTongLI/NanoHack21-1 | 562deb4a8df200756fd47c942e2c25417f64c67a | [
"MIT"
] | null | null | null | Analysis/1_transcriptome_analysis.ipynb | BioinfoTongLI/NanoHack21-1 | 562deb4a8df200756fd47c942e2c25417f64c67a | [
"MIT"
] | 1 | 2021-05-02T15:07:14.000Z | 2021-05-02T15:07:14.000Z | 327.149752 | 109,616 | 0.916421 | [
[
[
"# Analysis of the kindey WTA dataset\n\nThis notebook provides analysis workflow of transcriptomic profiles of spatial regions of interest (ROIs) from the GeoMx® Digital Spatial Profiler data for human kidney which can be accessed through [here](http://nanostring-public-share.s3-website-us-west-2.amazonaws.com/GeoScriptHub/)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport scanpy as sc\nimport anndata\nimport matplotlib.pyplot as plt\nsc.settings.set_figure_params(dpi=80) # low dpi (dots per inch) yields small inline figures",
"_____no_output_____"
],
[
"path_data = './KidneyDataset/'",
"_____no_output_____"
]
],
[
[
"## Reading in tables with data that will come useful",
"_____no_output_____"
]
],
[
[
"# cell type expression profiles used for cell type deconvolution of the NanoString ROIs\nCell_Types_for_Spatial_Decon = pd.read_csv(path_data + 'Cell_Types_for_Spatial_Decon.txt',\n sep='\\t')\n\n# target QC’d, filtered, and quantile normalized (Q3) target level count data\nKidney_Q3Norm_TargetCountMatrix = pd.read_csv(path_data + 'Kidney_Q3Norm_TargetCountMatrix.txt',\n sep='\\t', index_col=0)\n\n# metadata for the ROIs\nKidney_Sample_Annotations = pd.read_csv(path_data + 'Kidney_Sample_Annotations.txt',\n sep='\\t')\nKidney_Sample_Annotations.set_index('SegmentDisplayName', inplace=True) \n\n# cell type deconvolution of the NanoString ROIs\n\nKidney_Spatial_Decon = pd.read_csv(path_data + 'Kidney_Spatial_Decon.txt',\n sep='\\t')",
"_____no_output_____"
]
],
[
[
"## Assigning exact region of ROI",
"_____no_output_____"
],
[
"#### For tubule ROIs, PanCK+ area was separately collected as distal tubules and PanCK- - as proximal tubules\n",
"_____no_output_____"
]
],
[
[
"# here in Kidney_Sample_Annotations column 'region' there is no indication of whether tubule is proximal or distal\n# need to add this depending on the SegmentLabel - neg means proximal tubules, PanCK - distal\n\ndef add_exact_region(idx, table):\n curr_SegmentLabel = table.loc[idx,'SegmentLabel']\n if curr_SegmentLabel == 'Geometric Segment':\n return('glomerulus')\n elif curr_SegmentLabel == 'neg':\n return('proximal tubule')\n elif curr_SegmentLabel == 'PanCK':\n return('distal tubule')\n else:\n return('error')\nKidney_Sample_Annotations['idx'] = Kidney_Sample_Annotations.index\nKidney_Sample_Annotations['region_exact'] = Kidney_Sample_Annotations['idx'].apply(lambda x: add_exact_region(x,Kidney_Sample_Annotations))",
"_____no_output_____"
],
[
"Kidney_Sample_Annotations['region_exact'].value_counts()",
"_____no_output_____"
]
],
[
[
"# Analysis workflow",
"_____no_output_____"
]
],
[
[
"# to get a cells x genes table\nKidney_Q3Norm_TargetCountMatrix = Kidney_Q3Norm_TargetCountMatrix.T\n\n# making a scanpy object for downstream analysis\nadata_ROI = anndata.AnnData(X = Kidney_Q3Norm_TargetCountMatrix.values,\n obs = pd.DataFrame(index = Kidney_Q3Norm_TargetCountMatrix.index),\n var = pd.DataFrame(index = Kidney_Q3Norm_TargetCountMatrix.columns))",
"_____no_output_____"
],
[
"adata_ROI",
"_____no_output_____"
],
[
"# quick scRNA-seq-like scanpy workflow for NanoString whole transcriptome data\n\n# this data is already normalised, but we need to log1p transform it\nsc.pp.log1p(adata_ROI)\n\n# calculating highly variable genes HVGs)\nsc.pp.highly_variable_genes(adata_ROI, min_mean=0.0125, max_mean=3, min_disp=0.5)\n# subsetting to HVGs\nadata_hvg = adata_ROI[:, adata_ROI.var['highly_variable']].copy()\n\n# scaling data and calculating PCs\nsc.pp.scale(adata_hvg, max_value=10)\nsc.tl.pca(adata_hvg, svd_solver='arpack', n_comps=50)\n# plot to decide how many PCs best explain the variance\nsc.pl.pca_variance_ratio(adata_hvg, n_pcs=50)",
"_____no_output_____"
],
[
"# claculating neighborhood graph and 2D UMAP embedding for visualisation\nn_PCs = 7\nsc.pp.neighbors(adata_hvg, n_neighbors = 7, n_pcs = n_PCs)\nsc.tl.umap(adata_hvg, random_state=0)",
"_____no_output_____"
],
[
"# make sure ROI indices match\nKidney_Sample_Annotations = Kidney_Sample_Annotations.loc[adata_hvg.obs_names,:]\n\n# add metadata\nfor col in Kidney_Sample_Annotations:\n #print(col)\n adata_hvg.obs.loc[Kidney_Sample_Annotations.index, col] = Kidney_Sample_Annotations.loc[:,col]",
"_____no_output_____"
],
[
"# visualise the 2D UMAP\nsc.pl.umap(adata_hvg, color = ['pathology','disease_status','region','region_exact','ScanName',\n ], ncols=3,\n size=50)\n\n# saving 2D UMAP coordinates for later visualisation\nUMAP_2d_coords = pd.DataFrame(adata_hvg.obsm['X_umap'], columns=['x_2d','y_2d'], index=adata_hvg.obs_names)",
"_____no_output_____"
],
[
"# calculating also a 3D UMAP\nsc.tl.umap(adata_hvg, random_state=0,\n n_components=3)\n\n# visualise the 3D UMAP\nsc.pl.umap(adata_hvg, color = ['pathology','disease_status','region','region_exact','ScanName',\n ], ncols=3,\n projection='3d',\n size=50)\n\n# saving 3D UMAP oordinates for later visualisation\nUMAP_3d_coords = pd.DataFrame(adata_hvg.obsm['X_umap'], columns=['x_3d','y_3d','z_3d'], index=adata_hvg.obs_names)",
"_____no_output_____"
]
],
[
[
"### Getting cell type deconvolution data for NanoString ROIs",
"_____no_output_____"
]
],
[
[
"# formatting columns\nKidney_Spatial_Decon.columns = ['Alias'] + [elem.replace('.','-') for elem in Kidney_Spatial_Decon.columns[1:]]\n\n# adding cell type names into Kidney_Spatial_Decon\nKidney_Spatial_Decon['cell_type_general'] = list(Cell_Types_for_Spatial_Decon.set_index('Alias').loc[Kidney_Spatial_Decon['Alias'],\n 'cell_type_general'])",
"_____no_output_____"
],
[
"# aggregating cell type proportions for cell types in 'cell_type_general'\naggregation_functions = {'Alias': 'first'}\nfor col in Kidney_Spatial_Decon[1:-1]:\n aggregation_functions[col] = 'sum'\n \nKidney_Spatial_Decon_merged = Kidney_Spatial_Decon.groupby(Kidney_Spatial_Decon['cell_type_general']).aggregate(aggregation_functions)",
"_____no_output_____"
],
[
"def add_ct_deconv(adata_obj, sample_ID, celltype):\n return(Kidney_Spatial_Decon_merged.loc[celltype,sample_ID])\n \nfor celltype in list(Kidney_Spatial_Decon_merged.index):\n #print(celltype)\n adata_hvg.obs['CT_deconv_' + celltype] = [Kidney_Spatial_Decon_merged.loc[celltype, sample_ID] for sample_ID in adata_hvg.obs['Sample_ID']]",
"_____no_output_____"
],
[
"# adding coordinates of the UMAP\nadata_hvg.obs['x_2d_UMAP'] = UMAP_2d_coords['x_2d']\nadata_hvg.obs['y_2d_UMAP'] = UMAP_2d_coords['y_2d']\n\nadata_hvg.obs['x_3d_UMAP'] = UMAP_3d_coords['x_3d']\nadata_hvg.obs['y_3d_UMAP'] = UMAP_3d_coords['y_3d']\nadata_hvg.obs['z_3d_UMAP'] = UMAP_3d_coords['z_3d']",
"_____no_output_____"
],
[
"# adding the corresponding image name to be displayed in ImJoy web browser\n# for all file names see: https://drive.google.com/drive/folders/1N3_ybrwg17SDsm0oCxvFOQUsfAsIDHM2\ndef add_img_names(adata_obj, barcode):\n \n curr_element_of_name = barcode.split(' | ')\n \n #print(curr_element_of_name)\n \n if curr_element_of_name[2] == 'Geometric Segment':\n addition = '' # don't add anything to the name in the end\n elif curr_element_of_name[2] == 'PanCK':\n addition = ' - PanCK'\n elif curr_element_of_name[2] == 'neg':\n addition = ' - neg'\n else:\n print('error')\n \n file_name = curr_element_of_name[0] + ' - ' + curr_element_of_name[1] + addition\n \n return(file_name)",
"_____no_output_____"
],
[
"adata_hvg.obs['barcode'] = adata_hvg.obs.index",
"_____no_output_____"
],
[
"adata_hvg.obs['Image_file_name'] = adata_hvg.obs['barcode'].apply(lambda x: add_img_names(adata_hvg, x))",
"_____no_output_____"
],
[
"adata_hvg.obs.head()",
"_____no_output_____"
],
[
"# convreting all column names to lowercase for java script\nadata_hvg.obs.columns = [elem.lower() for elem in adata_hvg.obs.columns]\n",
"_____no_output_____"
],
[
"adata_hvg.obs.to_csv('./table_for_visualisation_ROIs.csv')",
"_____no_output_____"
],
[
"adata_hvg.obs.columns",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.