hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
e71aa8a0c5b6763170dd14490fb601e27071c23c | 5,726 | ipynb | Jupyter Notebook | Bloque 1 - Ramp-Up/04_Python Basics/06_Ejercicios Python Basics II.ipynb | JuanDG5/bootcamp_thebridge_PTSep20 | 5116098ff3f3d15753585e3ee967a8e7ddfccf31 | [
"MIT"
] | 1 | 2020-10-16T16:13:02.000Z | 2020-10-16T16:13:02.000Z | Bloque 1 - Ramp-Up/04_Python Basics/06_Ejercicios Python Basics II.ipynb | JuanDG5/bootcamp_thebridge_PTSep20 | 5116098ff3f3d15753585e3ee967a8e7ddfccf31 | [
"MIT"
] | null | null | null | Bloque 1 - Ramp-Up/04_Python Basics/06_Ejercicios Python Basics II.ipynb | JuanDG5/bootcamp_thebridge_PTSep20 | 5116098ff3f3d15753585e3ee967a8e7ddfccf31 | [
"MIT"
] | 3 | 2020-10-15T18:53:54.000Z | 2020-10-16T17:25:28.000Z | 22.904 | 197 | 0.545407 | [
[
[
"",
"_____no_output_____"
],
[
"# Ejercicios Python Basics II",
"_____no_output_____"
],
[
"## Ejercicio 1\n* Crea dos variables numericas: un `int` y un `float`\n* Comprueba sus tipos\n* Súmalas en otra nueva\n* ¿De qué tipo es la nueva variable?\n* Elimina las dos primeras variables creadas",
"_____no_output_____"
],
[
"## Ejercicio 2\nEscribe un programa para pasar de grados a radianes. Hay que usar `input`. Recuerda que la conversión se realiza mediante\n\nradianes = grados*(pi/180)",
"_____no_output_____"
],
[
"## Ejercicio 3\nEscribe un programa que calcule el area de un paralelogramo (base x altura). Tambien con `input`",
"_____no_output_____"
],
[
"## Ejercicio 4\nTenemos las siguientes variables:\n\n```Python\nA = 4\nB = \"Text\"\nC = 4.1\n```\n\nComprueba:\n1. Si A y B son equivalentes\n2. Si A y C NO son equivalentes\n3. Si A es mayor que C \n4. Si C es menor o igual que A\n5. Si B NO es equivalente a C",
"_____no_output_____"
],
[
"## Ejercicio 5\nCrea un programa donde se recojan dos inputs del usuario, y el output del programa sea si esos inputs son iguales o no",
"_____no_output_____"
],
[
"## Ejercicio 6\nMismo programa que en 5, pero en esta ocasión tienen que ser tres inputs y dos salidas. Una de las salidas que nos indique si todos son iguales, y la otra si al menos dos inputs sí que lo son",
"_____no_output_____"
],
[
"## Ejercicio 7\nCrea un programa que recoja dos inputs. Tiene que comprobar si su suma es igual, superior o inferior a 10",
"_____no_output_____"
],
[
"## Ejercicio 8\nRazona sin ejecutar código el output que obtendremos de las siguientes sentencias\n1. True and True and False\n2. not(not (True or False)) or (True or False)\n3. (False or True or False or True) or False\n4. not True and True and True",
"_____no_output_____"
],
[
"1. False\n2. True\n3. True\n4. False",
"_____no_output_____"
],
[
"## Ejercicio 9\nPara este ejercicio vamos a poner en práctica [las funciones built in](https://docs.python.org/3/library/functions.html).\n\n1. Calcula el máximo de la lista: [4, 6, 8, -1]\n2. Suma todos los elementos de la lista anterior\n3. Redondea este float a 3 dígitos decimales: 63.451256965\n4. Valor absoluto de: -74",
"_____no_output_____"
],
[
"## Ejercicio 10\nPara el siguiente string se pide imprimir por pantalla los siguientes casos\n\n \"A quien madruga, dios le ayuda\"\n\n1. Pasarlo todo a mayusculas\n2. Pasarlo todo a minusculas\n3. Solo las iniciales de las palabras\n4. Crea una lista dividiendolo por sus espacios\n5. Sustituye las comas `,` por puntos y comas `;`\n6. Elimina las `a` minusculas",
"_____no_output_____"
],
[
"## Ejercicio 11\n1. Crea una lista con 3 elementos numéricos\n2. Añade un cuarto elemento\n3. Calcula la suma de todos\n4. Elimina el segundo elemento de la lista\n5. Añade otro elemento en la posicion 3 de la lista\n6. Crea otra lista con 4 elementos y concatenala a la que ya tenías.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e71ab5a8882fb9a4beff6be337148e403de256c7 | 212,345 | ipynb | Jupyter Notebook | Final_Model_Testing_Prediction_Matches_DT_Code_RPS_A01331212_.ipynb | ErickAxelMartinezRios/Statistics-and-Machine-Learning-Projects- | 23e159f789429090c99f40421a127242c4faf71b | [
"CC0-1.0"
] | null | null | null | Final_Model_Testing_Prediction_Matches_DT_Code_RPS_A01331212_.ipynb | ErickAxelMartinezRios/Statistics-and-Machine-Learning-Projects- | 23e159f789429090c99f40421a127242c4faf71b | [
"CC0-1.0"
] | null | null | null | Final_Model_Testing_Prediction_Matches_DT_Code_RPS_A01331212_.ipynb | ErickAxelMartinezRios/Statistics-and-Machine-Learning-Projects- | 23e159f789429090c99f40421a127242c4faf71b | [
"CC0-1.0"
] | null | null | null | 50.450226 | 68,426 | 0.610629 | [
[
[
"\n\n\n* Erick Axel Martinez Rios A01331212\n\n* November 20th, 2020 \n\n* Applied Computing \n\n* Final project \n\n* Testing of the model \n\n\n\n\n\n\n\n\n\n\n\n\n\n",
"_____no_output_____"
],
[
"Upload DATABASE CSV FILE and Actual_Results_FINAL csv FILE ",
"_____no_output_____"
]
],
[
[
"from google.colab import files\nuploaded = files.upload()",
"_____no_output_____"
]
],
[
[
"The CSV file imported here came from the real outcome of the fights according to the UFC stats website http://ufcstats.com/",
"_____no_output_____"
]
],
[
[
"import io\nimport numpy as np #Library to manage arrays \nimport pandas as pd #Lirary for mange dataframes \nimport matplotlib.pyplot as plt # Library to make plots. \n#Import csv into a dataframe \ndf = pd.read_csv(io.BytesIO(uploaded['data.csv'])) \nprint(\"Complete Data shape:\", df.shape) \nprint(\"Data shape withput NA values\",df.dropna().shape)",
"Complete Data shape: (5144, 145)\nData shape withput NA values (3202, 145)\n"
],
[
"actual_values = pd.read_csv(io.BytesIO(uploaded['Actual_Results_Final.csv'])) ",
"_____no_output_____"
],
[
"df_n = df.dropna(axis = 0, how ='any') \nprint(df_n.shape)",
"(3202, 145)\n"
],
[
"print('R_fighter numbers:',df_n['R_fighter'].value_counts().shape[0])\nprint('B_fighter numbers:',df_n['B_fighter'].value_counts().shape[0])\nprint('Referee numbers:',df_n['Referee'].value_counts().shape[0])\nprint('date:',df_n['date'].value_counts().shape[0])\nprint('location:',df_n['location'].value_counts().shape[0])\nprint('weight_class:',df_n['weight_class'].value_counts().shape[0])\nprint('R_Stance:',df_n['R_Stance'].value_counts().shape[0])\nprint('B_Stance:',df_n['B_Stance'].value_counts().shape[0])",
"R_fighter numbers: 875\nB_fighter numbers: 1048\nReferee numbers: 166\ndate: 445\nlocation: 146\nweight_class: 13\nR_Stance: 4\nB_Stance: 4\n"
],
[
"df_n = df_n.drop(['R_fighter', 'B_fighter', 'Referee', 'date', 'location'], axis=1)\n#df_n['R_fighter'] = df_n['R_fighter'].astype('category').cat.codes\n#df_n['B_fighter'] = df_n['B_fighter'].astype('category').cat.codes\n#df_n['location'] = df_n['location'].astype('category').cat.codes\n#f_n['Referee'] = df_n['Referee'].astype('category').cat.codes\n#df_n['date'] = df_n['date'].astype('category').cat.codes\ndf_n['title_bout'] = df_n[\"title_bout\"].astype('category').cat.codes\ndf_n = pd.get_dummies(df_n, columns=['R_Stance', 'B_Stance', 'weight_class'])\nprint(df_n.head())\nprint(df_n.shape)",
" Winner ... weight_class_Women's Strawweight\n0 Red ... 0\n1 Red ... 0\n2 Red ... 0\n3 Blue ... 0\n4 Blue ... 0\n\n[5 rows x 158 columns]\n(3202, 158)\n"
],
[
"df_filtered = df_n\n# Select input variables and separate target variable \nfeatures = df_filtered.loc[:, df_n.columns != 'Winner']\nprint(features.shape)",
"(3202, 157)\n"
]
],
[
[
"*Train* decision tree model with the inputs stats from the GINI importance criteria ",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier # Import Decision Tree Classifier\nfrom sklearn.model_selection import train_test_split # Import train_test_split function\nfrom sklearn.metrics import classification_report #Library for classification report \nfrom sklearn.metrics import confusion_matrix # Library to generate the confusion matrix of the model. \n#This are the final predictors that will be tested for the model \ninputs = ['R_age', 'R_avg_opp_SIG_STR_pct', 'R_avg_opp_SIG_STR_landed',\n 'B_avg_DISTANCE_landed', 'B_avg_SIG_STR_att', 'R_avg_opp_HEAD_landed',\n 'B_age', 'B_avg_opp_TOTAL_STR_landed', 'R_avg_opp_TOTAL_STR_landed',\n 'B_avg_HEAD_att', 'R_avg_GROUND_att', 'B_avg_opp_SIG_STR_pct',\n 'B_avg_BODY_att', 'R_avg_GROUND_landed', 'R_avg_opp_DISTANCE_landed']\n\nprint(inputs)\n\ndt_final =df_filtered[inputs]\n\nX = dt_final.values\ny = df_filtered.loc[:,['Winner']].values\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 1234) # 80% training and 20% test\n\nprint(X_train.shape, X_test.shape)\n# Create Decision Tree classifer object\ndt = DecisionTreeClassifier(criterion=\"gini\", max_depth=6)\n# Train Decision Tree Classifer\ndt = dt.fit(X_train,y_train)\n#Predict the response for test dataset\nprint('Accuracy of Decision Tree classifier on training set: {:.4f}'\n .format(dt.score(X_train, y_train)))\nprint('Accuracy of Decision Tree classifier on test set: {:.4f}'\n .format(dt.score(X_test, y_test)))\n\npred = dt.predict(X_test)\nprint(confusion_matrix(y_test, pred))\nprint(classification_report(y_test, pred))\n\nreport = classification_report(y_test, pred, output_dict=True)\nreport = pd.DataFrame(report).transpose()\nreport.to_csv(\"Report_DT_RFF_FINAL.csv\")\n\n\nf = dt.predict_proba(X_test) \n\nprob_DF = pd.DataFrame(f, columns=dt.classes_)\n\nprob_DF.head(10)",
"['R_age', 'R_avg_opp_SIG_STR_pct', 'R_avg_opp_SIG_STR_landed', 'B_avg_DISTANCE_landed', 'B_avg_SIG_STR_att', 'R_avg_opp_HEAD_landed', 'B_age', 'B_avg_opp_TOTAL_STR_landed', 'R_avg_opp_TOTAL_STR_landed', 'B_avg_HEAD_att', 'R_avg_GROUND_att', 'B_avg_opp_SIG_STR_pct', 'B_avg_BODY_att', 'R_avg_GROUND_landed', 'R_avg_opp_DISTANCE_landed']\n(2561, 15) (641, 15)\nAccuracy of Decision Tree classifier on training set: 0.6790\nAccuracy of Decision Tree classifier on test set: 0.6365\n[[ 62 1 158]\n [ 1 0 6]\n [ 67 0 346]]\n precision recall f1-score support\n\n Blue 0.48 0.28 0.35 221\n Draw 0.00 0.00 0.00 7\n Red 0.68 0.84 0.75 413\n\n accuracy 0.64 641\n macro avg 0.39 0.37 0.37 641\nweighted avg 0.60 0.64 0.60 641\n\n"
]
],
[
[
"Matches to test ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"Take input features from database to perform to compute the avarage of the stats for each of the fighters ",
"_____no_output_____"
]
],
[
[
"check_R = 'R_'\nres_R = [idx for idx in inputs if idx.lower().startswith(check_R.lower())] \nres_R.insert(0,'R_fighter')\nres_R.append('date')\nprint(res_R)\ncheck_B = 'B_'\nres_B = [idx for idx in inputs if idx.lower().startswith(check_B.lower())] \nres_B.insert(0,'B_fighter')\nres_B.append('date')\nprint(res_B)\ninput_dat = df\nR_stats = input_dat[res_R]\nB_stats = input_dat[res_B]\nactual_year = 2020 ",
"['R_fighter', 'R_age', 'R_avg_opp_SIG_STR_pct', 'R_avg_opp_SIG_STR_landed', 'R_avg_opp_HEAD_landed', 'R_avg_opp_TOTAL_STR_landed', 'R_avg_GROUND_att', 'R_avg_GROUND_landed', 'R_avg_opp_DISTANCE_landed', 'date']\n['B_fighter', 'B_avg_DISTANCE_landed', 'B_avg_SIG_STR_att', 'B_age', 'B_avg_opp_TOTAL_STR_landed', 'B_avg_HEAD_att', 'B_avg_opp_SIG_STR_pct', 'B_avg_BODY_att', 'date']\n"
]
],
[
[
"Match 1",
"_____no_output_____"
],
[
" Khabib Nurmagomedov\tvs\tJustin Gaethje",
"_____no_output_____"
]
],
[
[
"# Khabib Nurmagomedov\tvs\tJustin Gaethje (W:Khabib Nurmagomedov)\t\nmatch_1_R = R_stats.loc[R_stats['R_fighter'] == 'Khabib Nurmagomedov'].reset_index(drop=True)\nyears = actual_year - int(match_1_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_1_R.loc[0,'R_age'] + years\nmatch_1_R_mean = match_1_R.mean(skipna = True)\nmatch_1_R_mean.loc['R_age'] = n_years\nmatch_1_B = B_stats.loc[B_stats['B_fighter'] == 'Justin Gaethje'].reset_index(drop=True)\nyears = actual_year - int(match_1_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_1_B.loc[0,'B_age'] + years \nmatch_1_B_mean = match_1_B.mean(skipna = True)\nmatch_1_B_mean.loc['B_age'] = n_years\nmatch_1_input = pd.concat([match_1_R_mean,match_1_B_mean])\nmatch_1_input = pd.DataFrame(match_1_input).T\nmatch_1_input = match_1_input[inputs]\nf1 = dt.predict_proba(match_1_input.values) \nprob_DF_1 = pd.DataFrame(f1, columns=dt.classes_)\nprint(\"Probability of winning\")\nprob_DF_1.head(1)",
"Probability of winning\n"
],
[
"m1b = dt.predict(match_1_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 2",
"_____no_output_____"
]
],
[
[
"#Robert Whittaker\tvs\tJared Cannonier\t(W:)\nmatch_2_R = R_stats.loc[R_stats['R_fighter'] == 'Robert Whittaker'].reset_index(drop=True)\nyears = actual_year - int(match_2_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_2_R.loc[0,'R_age'] + years\nmatch_2_R_mean = match_2_R.mean(skipna = True)\nmatch_2_R_mean.loc['R_age'] = n_years\nmatch_2_B = B_stats.loc[B_stats['B_fighter'] == 'Jared Cannonier'].reset_index(drop=True)\nyears = actual_year - int(match_2_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_2_B.loc[0,'B_age'] + years \nmatch_2_B_mean = match_2_B.mean(skipna = True)\nmatch_2_B_mean.loc['B_age'] = n_years\nmatch_2_input = pd.concat([match_2_R_mean,match_2_B_mean])\nmatch_2_input = pd.DataFrame(match_2_input).T\nmatch_2_input = match_2_input[inputs]\nf2 = dt.predict_proba(match_2_input.values) \nprob_DF_2 = pd.DataFrame(f2, columns=dt.classes_)\nprob_DF_2.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_2_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 3 ",
"_____no_output_____"
]
],
[
[
"# Alexander Volkov\tvs\tWalt Harris\t\nmatch_3_R = R_stats.loc[R_stats['R_fighter'] == 'Alexander Volkov'].reset_index(drop=True)\nyears = actual_year - int(match_3_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_3_R.loc[0,'R_age'] + years\nmatch_3_R_mean = match_3_R.mean(skipna = True)\nmatch_3_R_mean.loc['R_age'] = n_years\nmatch_3_B = B_stats.loc[B_stats['B_fighter'] == 'Walt Harris'].reset_index(drop=True)\nyears = actual_year - int(match_3_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_3_B.loc[0,'B_age'] + years \nmatch_3_B_mean = match_3_B.mean(skipna = True)\nmatch_3_B_mean.loc['B_age'] = n_years\nmatch_3_input = pd.concat([match_3_R_mean,match_3_B_mean])\nmatch_3_input = pd.DataFrame(match_3_input).T\nmatch_3_input = match_3_input[inputs]\nf3 = dt.predict_proba(match_3_input.values) \nprob_DF_3 = pd.DataFrame(f3, columns=dt.classes_)\nprob_DF_3.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_3_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 4",
"_____no_output_____"
]
],
[
[
"# Magomed Ankalaev\tvs\tIon Cutelaba\t###Ok \nmatch_4_R = R_stats.loc[R_stats['R_fighter'] == 'Magomed Ankalaev'].reset_index(drop=True)\nyears = actual_year - int(match_4_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_4_R.loc[0,'R_age'] + years\nmatch_4_R_mean = match_4_R.mean(skipna = True)\nmatch_4_R_mean.loc['R_age'] = n_years\nmatch_4_B = B_stats.loc[B_stats['B_fighter'] == 'Ion Cutelaba'].reset_index(drop=True)\nyears = actual_year - int(match_4_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_4_B.loc[0,'B_age'] + years \nmatch_4_B_mean = match_4_B.mean(skipna = True)\nmatch_4_B_mean.loc['B_age'] = n_years\nmatch_4_input = pd.concat([match_4_R_mean,match_4_B_mean])\nmatch_4_input = pd.DataFrame(match_4_input).T\nmatch_4_input = match_4_input[inputs]\nf4 = dt.predict_proba(match_4_input.values) \nprob_DF_4 = pd.DataFrame(f4, columns=dt.classes_)\nprob_DF_4.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_4_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 5",
"_____no_output_____"
]
],
[
[
"# Tai Tuivasa\tvs\tStefan Struve\t\t\nmatch_5_R = R_stats.loc[R_stats['R_fighter'] == 'Tai Tuivasa'].reset_index(drop=True)\nyears = actual_year - int(match_5_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_5_R.loc[0,'R_age'] + years\nmatch_5_R_mean = match_5_R.mean(skipna = True)\nmatch_5_R_mean.loc['R_age'] = n_years\nmatch_5_B = B_stats.loc[B_stats['B_fighter'] == 'Stefan Struve'].reset_index(drop=True)\nyears = actual_year - int(match_5_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_5_B.loc[0,'B_age'] + years \nmatch_5_B_mean = match_5_B.mean(skipna = True)\nmatch_5_B_mean.loc['B_age'] = n_years\nmatch_5_input = pd.concat([match_5_R_mean,match_5_B_mean])\nmatch_5_input = pd.DataFrame(match_5_input).T\nmatch_5_input = match_5_input[inputs]\nf5 = dt.predict_proba(match_5_input.values) \nprob_DF_5 = pd.DataFrame(f5, columns=dt.classes_)\nprob_DF_5.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_5_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 6 ",
"_____no_output_____"
]
],
[
[
"# Joel Alvarez\tvs\tAlexander Yakovlev\t(Winner Joel Alvarez)\nmatch_6_R = R_stats.loc[R_stats['R_fighter'] == 'Joel Alvarez'].reset_index(drop=True)\nyears = actual_year - int(match_6_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_6_R.loc[0,'R_age'] + years\nmatch_6_R_mean = match_6_R.mean(skipna = True)\nmatch_6_R_mean.loc['R_age'] = n_years\nmatch_6_B = B_stats.loc[B_stats['B_fighter'] == 'Alexander Yakovlev'].reset_index(drop=True)\nyears = actual_year - int(match_6_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_6_B.loc[0,'B_age'] + years \nmatch_6_B_mean = match_6_B.mean(skipna = True)\nmatch_6_B_mean.loc['B_age'] = n_years\nmatch_6_input = pd.concat([match_6_R_mean,match_6_B_mean])\nmatch_6_input = pd.DataFrame(match_6_input).T\nmatch_6_input = match_6_input[inputs]\nf6 = dt.predict_proba(match_6_input.values) \nprob_DF_6 = pd.DataFrame(f6, columns=dt.classes_)\nprob_DF_6.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_6_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 7 ",
"_____no_output_____"
]
],
[
[
"# Israel Adesanya\tvs\tPaulo Costa\t (Winner Israel)\nmatch_7_R = R_stats.loc[R_stats['R_fighter'] == 'Israel Adesanya'].reset_index(drop=True)\nyears = actual_year - int(match_7_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_7_R.loc[0,'R_age'] + years\nmatch_7_R_mean = match_7_R.mean(skipna = True)\nmatch_7_R_mean.loc['R_age'] = n_years\nmatch_7_B = B_stats.loc[B_stats['B_fighter'] == 'Paulo Costa'].reset_index(drop=True)\nyears = actual_year - int(match_7_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_7_B.loc[0,'B_age'] + years \nmatch_7_B_mean = match_7_B.mean(skipna = True)\nmatch_7_B_mean.loc['B_age'] = n_years\nmatch_7_input = pd.concat([match_7_R_mean,match_7_B_mean])\nmatch_7_input = pd.DataFrame(match_7_input).T\nmatch_7_input = match_7_input[inputs]\nf7 = dt.predict_proba(match_7_input.values) \nprob_DF_7 = pd.DataFrame(f7, columns=dt.classes_)\nprob_DF_7.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_7_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 8 ",
"_____no_output_____"
]
],
[
[
"# Jake Matthews\tvs\tDiego Sanchez (Winner Jake Matthews)\nmatch_8_R = R_stats.loc[R_stats['R_fighter'] == 'Jake Matthews'].reset_index(drop=True)\nyears = actual_year - int(match_8_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_8_R.loc[0,'R_age'] + years\nmatch_8_R_mean = match_8_R.mean(skipna = True)\nmatch_8_R_mean.loc['R_age'] = n_years\nmatch_8_B = B_stats.loc[B_stats['B_fighter'] == 'Diego Sanchez'].reset_index(drop=True)\nyears = actual_year - int(match_8_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_8_B.loc[0,'B_age'] + years \nmatch_8_B_mean = match_8_B.mean(skipna = True)\nmatch_8_B_mean.loc['B_age'] = n_years\nmatch_8_input = pd.concat([match_8_R_mean,match_8_B_mean])\nmatch_8_input = pd.DataFrame(match_8_input).T\nmatch_8_input = match_8_input[inputs]\nf8 = dt.predict_proba(match_8_input.values) \nprob_DF_8 = pd.DataFrame(f8, columns=dt.classes_)\nprob_DF_8.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_8_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 9 ",
"_____no_output_____"
]
],
[
[
"# Islam Makhachev\tvs\tRafael Dos Anjos\t\nmatch_9_R = R_stats.loc[R_stats['R_fighter'] == 'Islam Makhachev'].reset_index(drop=True)\nyears = actual_year - int(match_9_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_9_R.loc[0,'R_age'] + years\nmatch_9_R_mean = match_9_R.mean(skipna = True)\nmatch_9_R_mean.loc['R_age'] = n_years\n\nmatch_9_B = B_stats.loc[B_stats['B_fighter'] == 'Rafael Dos Anjos'].reset_index(drop=True)\nyears = actual_year - int(match_9_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_9_B.loc[0,'B_age'] + years \nmatch_9_B_mean = match_9_B.mean(skipna = True)\nmatch_9_B_mean.loc['B_age'] = n_years\n\nmatch_9_input = pd.concat([match_9_R_mean,match_9_B_mean])\nmatch_9_input = pd.DataFrame(match_9_input).T\nmatch_9_input = match_9_input[inputs]\n\nf9 = dt.predict_proba(match_9_input.values) \nprob_DF_9 = pd.DataFrame(f9, columns=dt.classes_)\nprob_DF_9.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_9_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Mtach 10 ",
"_____no_output_____"
]
],
[
[
"# Jose Quinonez\tvs\tLouis Smolka\t\nmatch_10_R = R_stats.loc[R_stats['R_fighter'] == 'Jose Quinonez'].reset_index(drop=True)\nyears = actual_year - int(match_10_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_10_R.loc[0,'R_age'] + years\nmatch_10_R_mean = match_10_R.mean()\nmatch_10_R_mean.loc['R_age'] = n_years\nmatch_10_B = B_stats.loc[B_stats['B_fighter'] == 'Louis Smolka'].reset_index(drop=True)\nyears = actual_year - int(match_10_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_10_B.loc[0,'B_age'] + years \nmatch_10_B_mean = match_10_B.mean()\nmatch_10_B_mean.loc['B_age'] = n_years\nmatch_10_input = pd.concat([match_10_R_mean,match_10_B_mean])\nmatch_10_input = pd.DataFrame(match_10_input).T\nmatch_10_input = match_10_input[inputs]\nf10 = dt.predict_proba(match_10_input.values) \nprob_DF_10 = pd.DataFrame(f10, columns=dt.classes_)\nprob_DF_10.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_10_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 11",
"_____no_output_____"
]
],
[
[
"# Stipe Miocic\tvs\tDaniel Cormier (Winner: Miomic)\nmatch_11_R = R_stats.loc[R_stats['R_fighter'] == 'Stipe Miocic'].reset_index(drop=True)\nyears = actual_year - int(match_11_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_11_R.loc[0,'R_age'] + years\nmatch_11_R_mean = match_11_R.mean()\nmatch_11_R_mean.loc['R_age'] = n_years\nmatch_11_B = B_stats.loc[B_stats['B_fighter'] == 'Daniel Cormier'].reset_index(drop=True)\nyears = actual_year - int(match_11_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_11_B.loc[0,'B_age'] + years \nmatch_11_B_mean = match_11_B.mean()\nmatch_11_B_mean.loc['B_age'] = n_years\nmatch_11_input = pd.concat([match_11_R_mean,match_11_B_mean])\nmatch_11_input = pd.DataFrame(match_11_input).T\nmatch_11_input = match_11_input[inputs]\nf11 = dt.predict_proba(match_11_input.values) \nprob_DF_11 = pd.DataFrame(f11, columns=dt.classes_)\nprob_DF_11.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_11_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 12 Checar ",
"_____no_output_____"
]
],
[
[
"# Marlon Vera\tvs\tSean O'Malley\t\nmatch_12_R = R_stats.loc[R_stats['R_fighter'] == 'Marlon Vera'].reset_index(drop=True)\nyears = actual_year - int(match_12_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_12_R.loc[0,'R_age'] + years\nmatch_12_R_mean = match_12_R.mean()\nmatch_12_R_mean.loc['R_age'] = n_years\nk = []\nfor i in res_B:\n vr = i.replace(\"B_\", \"R_\")\n k.append(vr)\nk_stats = input_dat[k]\nmatch_12_B = k_stats.loc[k_stats['R_fighter'] == \"Sean O'Malley\"].reset_index(drop=True)\nyears = actual_year - int(match_12_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_12_B.loc[0,'R_age'] + years \nmatch_12_B_mean = match_12_B.mean()\nmatch_12_B_mean.loc['R_age'] = n_years\nmatch_12_B_mean = match_12_B_mean.rename(index={'R_avg_DISTANCE_landed':'B_avg_DISTANCE_landed', 'R_avg_SIG_STR_att':'B_avg_SIG_STR_att', 'R_age':'B_age',\n 'R_avg_opp_TOTAL_STR_landed':'B_avg_opp_TOTAL_STR_landed', 'R_avg_HEAD_att':'B_avg_HEAD_att', 'R_avg_opp_SIG_STR_pct':'B_avg_opp_SIG_STR_pct',\n 'R_avg_BODY_att':'B_avg_BODY_att'})\nmatch_12_input = pd.concat([match_12_R_mean,match_12_B_mean])\nmatch_12_input = pd.DataFrame(match_12_input).T\nmatch_12_input = match_12_input[inputs]\n\nf12 = dt.predict_proba(match_12_input.values) \nprob_DF_12 = pd.DataFrame(f12, columns=dt.classes_)\nprob_DF_12.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_12_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 13 ",
"_____no_output_____"
]
],
[
[
"# Merab Dvalishvili\tvs\tJohn Dodson\t(Winner: Merab)\nmatch_13_R = R_stats.loc[R_stats['R_fighter'] == 'Merab Dvalishvili'].reset_index(drop=True)\nyears = actual_year - int(match_13_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_13_R.loc[0,'R_age'] + years\nmatch_13_R_mean = match_13_R.mean()\nmatch_13_R_mean.loc['R_age'] = n_years\nmatch_13_B = B_stats.loc[B_stats['B_fighter'] == 'John Dodson'].reset_index(drop=True)\nyears = actual_year - int(match_13_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_13_B.loc[0,'B_age'] + years \nmatch_13_B_mean = match_13_B.mean()\nmatch_13_B_mean.loc['B_age'] = n_years\nmatch_13_input = pd.concat([match_13_R_mean,match_13_B_mean])\nmatch_13_input = pd.DataFrame(match_13_input).T\nmatch_13_input = match_13_input[inputs]\nf13 = dt.predict_proba(match_13_input.values) \nprob_DF_13 = pd.DataFrame(f13, columns=dt.classes_)\nprob_DF_13.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_13_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 14 ",
"_____no_output_____"
]
],
[
[
"# Vinc Pichel\tvs\tJim Miller\t(Winner: Vinc Pichel)\nmatch_14_R = R_stats.loc[R_stats['R_fighter'] == 'Vinc Pichel'].reset_index(drop=True)\nyears = actual_year - int(match_14_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_14_R.loc[0,'R_age'] + years\nmatch_14_R_mean = match_14_R.mean()\nmatch_14_R_mean.loc['R_age'] = n_years\nmatch_14_B = B_stats.loc[B_stats['B_fighter'] == 'Jim Miller'].reset_index(drop=True)\nyears = actual_year - int(match_14_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_14_B.loc[0,'B_age'] + years \nmatch_14_B_mean = match_14_B.mean()\nmatch_14_B_mean.loc['B_age'] = n_years\nmatch_14_input = pd.concat([match_14_R_mean,match_14_B_mean])\nmatch_14_input = pd.DataFrame(match_14_input).T\nmatch_14_input = match_14_input[inputs]\nf14 = dt.predict_proba(match_14_input.values) \nprob_DF_14 = pd.DataFrame(f14, columns=dt.classes_)\nprob_DF_14.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_14_input.values) \nprint(m1b)",
"['Blue']\n"
]
],
[
[
"Match 15 ",
"_____no_output_____"
]
],
[
[
"# Brian Ortega\tvs\tChan Sung Jung\t(Winner: Brian Ortega)\nmatch_15_R = R_stats.loc[R_stats['R_fighter'] == 'Brian Ortega'].reset_index(drop=True)\nyears = actual_year - int(match_15_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_15_R.loc[0,'R_age'] + years\nmatch_15_R_mean = match_15_R.mean()\nmatch_15_R_mean.loc['R_age'] = n_years\nmatch_15_B = B_stats.loc[B_stats['B_fighter'] == 'Chan Sung Jung'].reset_index(drop=True)\nyears = actual_year - int(match_15_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_15_B.loc[0,'B_age'] + years \nmatch_15_B_mean = match_15_B.mean()\nmatch_15_B_mean.loc['B_age'] = n_years\nmatch_15_input = pd.concat([match_15_R_mean,match_15_B_mean])\nmatch_15_input = pd.DataFrame(match_15_input).T\nmatch_15_input = match_15_input[inputs]\nf15 = dt.predict_proba(match_15_input.values) \nprob_DF_15 = pd.DataFrame(f15, columns=dt.classes_)\nprob_DF_15.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_15_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 16 ",
"_____no_output_____"
]
],
[
[
"# Jessica Andrade\tvs\tKatlyn Chookagian\t(Winner: Jessica Andrade)\nmatch_16_R = R_stats.loc[R_stats['R_fighter'] == 'Jessica Andrade'].reset_index(drop=True)\nyears = actual_year - int(match_16_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_16_R.loc[0,'R_age'] + years\nmatch_16_R_mean = match_16_R.mean()\nmatch_16_R_mean.loc['R_age'] = n_years\nmatch_16_B = B_stats.loc[B_stats['B_fighter'] == 'Katlyn Chookagian'].reset_index(drop=True)\nyears = actual_year - int(match_16_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_16_B.loc[0,'B_age'] + years \nmatch_16_B_mean = match_16_B.mean()\nmatch_16_B_mean.loc['B_age'] = n_years\nmatch_16_input = pd.concat([match_16_R_mean,match_16_B_mean])\nmatch_16_input = pd.DataFrame(match_16_input).T\nmatch_16_input = match_16_input[inputs]\nf16 = dt.predict_proba(match_16_input.values) \nprob_DF_16 = pd.DataFrame(f16, columns=dt.classes_)\nprob_DF_16.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_16_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 17 ",
"_____no_output_____"
]
],
[
[
"# James Krause\tvs\tClaudio Silva (Winner: Krause)\nmatch_17_R = R_stats.loc[R_stats['R_fighter'] == 'James Krause'].reset_index(drop=True)\nyears = actual_year - int(match_17_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_17_R.loc[0,'R_age'] + years\nmatch_17_R_mean = match_17_R.mean()\nmatch_17_R_mean.loc['R_age'] = n_years\nmatch_17_B = B_stats.loc[B_stats['B_fighter'] == 'Claudio Silva'].reset_index(drop=True)\nyears = actual_year - int(match_17_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_17_B.loc[0,'B_age'] + years \nmatch_17_B_mean = match_17_B.mean()\nmatch_17_B_mean.loc['B_age'] = n_years\nmatch_17_input = pd.concat([match_17_R_mean,match_17_B_mean])\nmatch_17_input = pd.DataFrame(match_17_input).T\nmatch_17_input = match_17_input[inputs]\nf17 = dt.predict_proba(match_17_input.values) \nprob_DF_17 = pd.DataFrame(f17, columns=dt.classes_)\nprob_DF_17.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_17_input.values) \nprint(m1b)",
"['Blue']\n"
]
],
[
[
"Match 18 ",
"_____no_output_____"
]
],
[
[
"# Gillian Robertson\tvs\tPoliana Botelho\nmatch_18_R = R_stats.loc[R_stats['R_fighter'] == 'Gillian Robertson'].reset_index(drop=True)\nyears = actual_year - int(match_18_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_18_R.loc[0,'R_age'] + years\nmatch_18_R_mean = match_18_R.mean()\nmatch_18_R_mean.loc['R_age'] = n_years\nmatch_18_B = B_stats.loc[B_stats['B_fighter'] == 'Poliana Botelho'].reset_index(drop=True)\nyears = actual_year - int(match_18_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_18_B.loc[0,'B_age'] + years \nmatch_18_B_mean = match_18_B.mean()\nmatch_18_B_mean.loc['B_age'] = n_years\nmatch_18_input = pd.concat([match_18_R_mean,match_18_B_mean])\nmatch_18_input = pd.DataFrame(match_18_input).T\nmatch_18_input = match_18_input[inputs]\nf18 = dt.predict_proba(match_18_input.values) \nprob_DF_18 = pd.DataFrame(f18, columns=dt.classes_)\nprob_DF_18.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_18_input.values) \nprint(m1b)",
"['Blue']\n"
]
],
[
[
"Match 19 ",
"_____no_output_____"
]
],
[
[
"# Neil Magny\tvs\tRobbie Lawler (W=Neil Magny)\nmatch_19_R = R_stats.loc[R_stats['R_fighter'] == 'Neil Magny'].reset_index(drop=True)\nyears = actual_year - int(match_19_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_19_R.loc[0,'R_age'] + years\nmatch_19_R_mean = match_19_R.mean()\nmatch_19_R_mean.loc['R_age'] = n_years\nmatch_19_B = B_stats.loc[B_stats['B_fighter'] == 'Robbie Lawler'].reset_index(drop=True)\nyears = actual_year - int(match_19_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_19_B.loc[0,'B_age'] + years \nmatch_19_B_mean = match_19_B.mean()\nmatch_19_B_mean.loc['B_age'] = n_years\nmatch_19_input = pd.concat([match_19_R_mean,match_19_B_mean])\nmatch_19_input = pd.DataFrame(match_19_input).T\nmatch_19_input = match_19_input[inputs]\nf19 = dt.predict_proba(match_19_input.values) \nprob_DF_19 = pd.DataFrame(f19, columns=dt.classes_)\nprob_DF_19.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_19_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 20 ",
"_____no_output_____"
]
],
[
[
"# Alexa Grasso\tvs\tJi Yeon Kim (#Alexa Grasso)\nmatch_20_R = R_stats.loc[R_stats['R_fighter'] == 'Alexa Grasso'].reset_index(drop=True)\nyears = actual_year - int(match_20_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_20_R.loc[0,'R_age'] + years\nmatch_20_R_mean = match_20_R.mean()\nmatch_20_R_mean.loc['R_age'] = n_years\nmatch_20_B = B_stats.loc[B_stats['B_fighter'] == 'Ji Yeon Kim'].reset_index(drop=True)\nyears = actual_year - int(match_20_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_20_B.loc[0,'B_age'] + years \nmatch_20_B_mean = match_20_B.mean()\nmatch_20_B_mean.loc['B_age'] = n_years\nmatch_20_input = pd.concat([match_20_R_mean,match_20_B_mean])\nmatch_20_input = pd.DataFrame(match_20_input).T\nmatch_20_input = match_20_input[inputs]\nf20 = dt.predict_proba(match_20_input.values) \nprob_DF_20 = pd.DataFrame(f20, columns=dt.classes_)\nprob_DF_20.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_20_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 21 ",
"_____no_output_____"
]
],
[
[
"# Zak Cummings\tvs\tAlessio Di Chirico (#Zak Cummings)\nmatch_21_R = R_stats.loc[R_stats['R_fighter'] == 'Zak Cummings'].reset_index(drop=True)\nyears = actual_year - int(match_21_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_21_R.loc[0,'R_age'] + years\nmatch_21_R_mean = match_21_R.mean()\nmatch_21_R_mean.loc['R_age'] = n_years\nmatch_21_B = B_stats.loc[B_stats['B_fighter'] == 'Alessio Di Chirico'].reset_index(drop=True)\nyears = actual_year - int(match_21_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_21_B.loc[0,'B_age'] + years \nmatch_21_B_mean = match_21_B.mean()\nmatch_21_B_mean.loc['B_age'] = n_years\nmatch_21_input = pd.concat([match_21_R_mean,match_21_B_mean])\nmatch_21_input = pd.DataFrame(match_21_input).T\nmatch_21_input = match_21_input[inputs]\nf21 = dt.predict_proba(match_21_input.values) \nprob_DF_21 = pd.DataFrame(f21, columns=dt.classes_)\nprob_DF_21.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_21_input.values) \nprint(m1b)",
"['Blue']\n"
]
],
[
[
"Match 22",
"_____no_output_____"
]
],
[
[
"# Colby Covington\tvs\tTyron Woodley (#Colby Covington)\nmatch_22_R = R_stats.loc[R_stats['R_fighter'] == 'Colby Covington'].reset_index(drop=True)\nyears = actual_year - int(match_22_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_22_R.loc[0,'R_age'] + years\nmatch_22_R_mean = match_22_R.mean()\nmatch_22_R_mean.loc['R_age'] = n_years\nmatch_22_B = B_stats.loc[B_stats['B_fighter'] == 'Tyron Woodley'].reset_index(drop=True)\nyears = actual_year - int(match_22_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_22_B.loc[0,'B_age'] + years \nmatch_22_B_mean = match_22_B.mean()\nmatch_22_B_mean.loc['B_age'] = n_years\nmatch_22_input = pd.concat([match_22_R_mean,match_22_B_mean])\nmatch_22_input = pd.DataFrame(match_22_input).T\nmatch_22_input = match_22_input[inputs]\nf22 = dt.predict_proba(match_22_input.values) \nprob_DF_22 = pd.DataFrame(f22, columns=dt.classes_)\nprob_DF_22.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_22_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 23 ",
"_____no_output_____"
]
],
[
[
"# Donald Cerrone\tvs\tNiko Price (#Draw)\nmatch_23_R = R_stats.loc[R_stats['R_fighter'] == 'Donald Cerrone'].reset_index(drop=True)\nyears = actual_year - int(match_23_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_23_R.loc[0,'R_age'] + years\nmatch_23_R_mean = match_23_R.mean()\nmatch_23_R_mean.loc['R_age'] = n_years\nmatch_23_B = B_stats.loc[B_stats['B_fighter'] == 'Niko Price'].reset_index(drop=True)\nyears = actual_year - int(match_23_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_23_B.loc[0,'B_age'] + years \nmatch_23_B_mean = match_23_B.mean()\nmatch_23_B_mean.loc['B_age'] = n_years\nmatch_23_input = pd.concat([match_23_R_mean,match_23_B_mean])\nmatch_23_input = pd.DataFrame(match_23_input).T\nmatch_23_input = match_23_input[inputs]\nf23 = dt.predict_proba(match_23_input.values) \nprob_DF_23 = pd.DataFrame(f23, columns=dt.classes_)\nprob_DF_23.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_23_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 24",
"_____no_output_____"
]
],
[
[
"# Mackenzie Dern\tvs\tRanda Markos (#Mackenzie Dern)\nmatch_24_R = R_stats.loc[R_stats['R_fighter'] == 'Mackenzie Dern'].reset_index(drop=True)\nyears = actual_year - int(match_24_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_24_R.loc[0,'R_age'] + years\nmatch_24_R_mean = match_24_R.mean()\nmatch_24_R_mean.loc['R_age'] = n_years\nmatch_24_B = B_stats.loc[B_stats['B_fighter'] == 'Randa Markos'].reset_index(drop=True)\nyears = actual_year - int(match_24_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_24_B.loc[0,'B_age'] + years \nmatch_24_B_mean = match_24_B.mean()\nmatch_24_B_mean.loc['B_age'] = n_years\nmatch_24_input = pd.concat([match_24_R_mean,match_24_B_mean])\nmatch_24_input = pd.DataFrame(match_24_input).T\nmatch_24_input = match_24_input[inputs]\nf24 = dt.predict_proba(match_24_input.values) \nprob_DF_24 = pd.DataFrame(f24, columns=dt.classes_)\nprob_DF_24.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_24_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 25 ",
"_____no_output_____"
]
],
[
[
"# Kevin Holland\tvs\tDarren Stewart (#Kevin Holland)\nmatch_25_R = R_stats.loc[R_stats['R_fighter'] == 'Kevin Holland'].reset_index(drop=True)\nmatch_25_R.head()\nyears = actual_year - int(match_25_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_25_R.loc[0,'R_age'] + years\nmatch_25_R_mean = match_25_R.mean()\nmatch_25_R_mean.loc['R_age'] = n_years\nmatch_25_B = B_stats.loc[B_stats['B_fighter'] == 'Darren Stewart'].reset_index(drop=True)\nyears = actual_year - int(match_25_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_25_B.loc[0,'B_age'] + years \nmatch_25_B_mean = match_25_B.mean()\nmatch_25_B_mean.loc['B_age'] = n_years\nmatch_25_input = pd.concat([match_25_R_mean,match_25_B_mean])\nmatch_25_input = pd.DataFrame(match_25_input).T\nmatch_25_input = match_25_input[inputs]\nf25 = dt.predict_proba(match_25_input.values) \nprob_DF_25 = pd.DataFrame(f25, columns=dt.classes_)\nprob_DF_25.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_25_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 26 ",
"_____no_output_____"
]
],
[
[
"# Damon Jackson\tvs\tMirsad Bektic (# Damon Jackson)\n\nmatch_26_R = R_stats.loc[R_stats['R_fighter'] == 'Damon Jackson'].reset_index(drop=True)\nyears = actual_year - int(match_26_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_26_R.loc[0,'R_age'] + years\nmatch_26_R_mean = match_26_R.mean()\nmatch_26_R_mean.loc['R_age'] = n_years\nmatch_26_B = B_stats.loc[B_stats['B_fighter'] == 'Mirsad Bektic'].reset_index(drop=True)\nyears = actual_year - int(match_26_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_26_B.loc[0,'B_age'] + years \nmatch_26_B_mean = match_26_B.mean()\nmatch_26_B_mean.loc['B_age'] = n_years\nmatch_26_input = pd.concat([match_26_R_mean,match_26_B_mean])\nmatch_26_input = pd.DataFrame(match_26_input).T\nmatch_26_input = match_26_input[inputs]\nf26 = dt.predict_proba(match_26_input.values) \nprob_DF_26 = pd.DataFrame(f26, columns=dt.classes_)\nprob_DF_26.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_26_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 27 ",
"_____no_output_____"
]
],
[
[
"# Mayra Bueno Silva\tvs\tMara Romero Borella\n\n# Mayra Bueno Silva only have one fight in the dataset, therefore another fighter\n# with a similar age (27), same gender, and similar height was chosen, in this case, this player was Livinha Souza \n# Her statistics were taken to prove the model. The age was chosen since it is important for the model\n\nmatch_27_R = R_stats.loc[R_stats['R_fighter'] == 'Livinha Souza'].reset_index(drop=True)\nyears = actual_year - int(match_27_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_27_R.loc[0,'R_age'] + years\nmatch_27_R_mean = match_27_R.mean()\nmatch_27_R_mean.loc['R_age'] = n_years\nmatch_27_B = B_stats.loc[B_stats['B_fighter'] == 'Mara Romero Borella'].reset_index(drop=True)\nyears = actual_year - int(match_27_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_27_B.loc[0,'B_age'] + years \nmatch_27_B_mean = match_27_B.mean()\nmatch_27_B_mean.loc['B_age'] = n_years\nmatch_27_input = pd.concat([match_27_R_mean,match_27_B_mean])\nmatch_27_input = pd.DataFrame(match_27_input).T\nmatch_27_input = match_27_input[inputs]\nf27 = dt.predict_proba(match_27_input.values) \nprob_DF_27 = pd.DataFrame(f27, columns=dt.classes_)\nprob_DF_27.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_27_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 28 ",
"_____no_output_____"
]
],
[
[
"# Michelle Waterson\tvs\tAngela Hill (Michelle Waterson)\n\nmatch_28_R = R_stats.loc[R_stats['R_fighter'] == 'Michelle Waterson'].reset_index(drop=True)\nyears = actual_year - int(match_28_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_28_R.loc[0,'R_age'] + years\nmatch_28_R_mean = match_28_R.mean()\nmatch_28_R_mean.loc['R_age'] = n_years\nmatch_28_B = B_stats.loc[B_stats['B_fighter'] == 'Angela Hill'].reset_index(drop=True)\nyears = actual_year - int(match_28_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_28_B.loc[0,'B_age'] + years \nmatch_28_B_mean = match_28_B.mean()\nmatch_28_B_mean.loc['B_age'] = n_years\nmatch_28_input = pd.concat([match_28_R_mean,match_28_B_mean])\nmatch_28_input = pd.DataFrame(match_28_input).T\nmatch_28_input = match_28_input[inputs]\nf28 = dt.predict_proba(match_28_input.values) \nprob_DF_28 = pd.DataFrame(f28, columns=dt.classes_)\nprob_DF_28.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_28_input.values) \nprint(m1b)",
"['Blue']\n"
]
],
[
[
"Match 29",
"_____no_output_____"
]
],
[
[
"# Bobby Green\tvs\tAlan Patrick (Winner: Bobby Green)\nmatch_29_R = R_stats.loc[R_stats['R_fighter'] == 'Bobby Green'].reset_index(drop=True)\nyears = actual_year - int(match_29_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_29_R.loc[0,'R_age'] + years\nmatch_29_R_mean = match_29_R.mean()\nmatch_29_R_mean.loc['R_age'] = n_years\nmatch_29_B = B_stats.loc[B_stats['B_fighter'] == 'Alan Patrick'].reset_index(drop=True)\nyears = actual_year - int(match_29_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_29_B.loc[0,'B_age'] + years \nmatch_29_B_mean = match_29_B.mean()\nmatch_29_B_mean.loc['B_age'] = n_years\nmatch_29_input = pd.concat([match_29_R_mean,match_29_B_mean])\nmatch_29_input = pd.DataFrame(match_29_input).T\nmatch_29_input = match_29_input[inputs]\nf29 = dt.predict_proba(match_29_input.values) \nprob_DF_29 = pd.DataFrame(f29, columns=dt.classes_)\nprob_DF_29.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_29_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 30",
"_____no_output_____"
]
],
[
[
"# Sijara Eubanks\tvs\tJulia Avila\n#\n# En este caso no existía registro de Julia Avila en la base de datos, por lo que se reemplazó con\n#Gina Mazany que tenía una categoría de peso similar, una edad similar y jugaba como la luchadora azul\n\nmatch_30_R = R_stats.loc[R_stats['R_fighter'] == 'Sijara Eubanks'].reset_index(drop=True)\nyears = actual_year - int(match_30_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_30_R.loc[0,'R_age'] + years\nmatch_30_R_mean = match_30_R.mean()\nmatch_30_R_mean.loc['R_age'] = n_years\nmatch_30_B = B_stats.loc[B_stats['B_fighter'] == 'Gina Mazany'].reset_index(drop=True)\nyears = actual_year - int(match_30_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_30_B.loc[0,'B_age'] + years\nmatch_30_B_mean = match_30_B.mean()\nmatch_30_B_mean.loc['B_age'] = n_years\nmatch_30_input = pd.concat([match_30_R_mean,match_30_B_mean])\nmatch_30_input = pd.DataFrame(match_30_input).T\nmatch_30_input = match_30_input[inputs]\nf30 = dt.predict_proba(match_30_input.values) \nprob_DF_30 = pd.DataFrame(f30, columns=dt.classes_)\nprob_DF_30.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_30_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Match 31",
"_____no_output_____"
]
],
[
[
"# Sabina Mazo\tvs\tJustine Kish #\n\n# Sabina Mazo only had one sample according to the database and in that sample, the stats needed were missing. \n\n# In this case a similar fighter was chosen to take the stats this was Montana De La Rosa the criteria was the weight class, the height, and the age similar to Sabina Mazo. \n\nmatch_31_R = R_stats.loc[R_stats['R_fighter'] == 'Montana De La Rosa'].reset_index(drop=True)\nyears = actual_year - int(match_31_R.loc[0,'date'].split(\"-\")[0]) \nn_years = match_31_R.loc[0,'R_age'] + years\nmatch_31_R_mean = match_31_R.mean()\nmatch_31_R_mean.loc['R_age'] = n_years\n\nmatch_31_B = B_stats.loc[B_stats['B_fighter'] == 'Justine Kish'].reset_index(drop=True)\nyears = actual_year - int(match_31_B.loc[0,'date'].split(\"-\")[0])\nn_years = match_31_B.loc[0,'B_age'] + years \nmatch_31_B_mean = match_31_B.mean()\nmatch_31_B_mean.loc['B_age'] = n_years\n\nmatch_31_input = pd.concat([match_31_R_mean,match_31_B_mean])\nmatch_31_input = pd.DataFrame(match_31_input).T\nmatch_31_input = match_31_input[inputs]\n\nf31 = dt.predict_proba(match_31_input.values) \nprob_DF_31 = pd.DataFrame(f31, columns=dt.classes_)\nprob_DF_31.head(1)",
"_____no_output_____"
],
[
"m1b = dt.predict(match_31_input.values) \nprint(m1b)",
"['Red']\n"
]
],
[
[
"Matches probability ",
"_____no_output_____"
]
],
[
[
"final_p_score = pd.concat([prob_DF_1,prob_DF_2, prob_DF_3,prob_DF_4,prob_DF_5,prob_DF_6,prob_DF_7,prob_DF_8,prob_DF_9,prob_DF_10,prob_DF_11,prob_DF_12,prob_DF_13,prob_DF_14,prob_DF_15,prob_DF_16,prob_DF_17,prob_DF_18,prob_DF_19,prob_DF_20,prob_DF_21,\n prob_DF_22, prob_DF_23,prob_DF_24,prob_DF_25,prob_DF_26,prob_DF_27,prob_DF_28,prob_DF_29,prob_DF_30,prob_DF_31]).reset_index(drop=True)\nfinal_p_score = final_p_score[['Red','Draw','Blue']]\n\nfinal_p_score.head(31)\n\nfinal_p_score.to_csv(\"Probability_Matches.csv\")",
"_____no_output_____"
]
],
[
[
"Compute Rank probability score RPS ",
"_____no_output_____"
]
],
[
[
"rps = ((final_p_score['Red']-actual_values['R_fighter'])**2+((final_p_score['Red']-actual_values['R_fighter'])+ final_p_score['Draw']-actual_values['Draw'])**2)/2\nprint(rps)\nprint('Avarage value:',rps.mean(axis=0))",
"0 0.025194\n1 0.162085\n2 0.162085\n3 0.162851\n4 0.017013\n5 0.162085\n6 0.243447\n7 0.042806\n8 0.048892\n9 0.243447\n10 0.115009\n11 0.243447\n12 0.042806\n13 0.658895\n14 0.243447\n15 0.243447\n16 0.259188\n17 1.000000\n18 0.115009\n19 0.243447\n20 0.259188\n21 0.062500\n22 0.252066\n23 0.042806\n24 0.003252\n25 0.243447\n26 0.082813\n27 0.259188\n28 0.115009\n29 0.003252\n30 0.243447\ndtype: float64\nAvarage value: 0.1935989378945617\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71adbb088bdb435a3bcc56578329f7051821d8c | 54,018 | ipynb | Jupyter Notebook | week4_approx_rl/homework_pytorch.ipynb | drewnoff/Practical_RL | 98f98f0e70e0aadcd86dd8991d606ffd36489c81 | [
"Unlicense"
] | null | null | null | week4_approx_rl/homework_pytorch.ipynb | drewnoff/Practical_RL | 98f98f0e70e0aadcd86dd8991d606ffd36489c81 | [
"Unlicense"
] | null | null | null | week4_approx_rl/homework_pytorch.ipynb | drewnoff/Practical_RL | 98f98f0e70e0aadcd86dd8991d606ffd36489c81 | [
"Unlicense"
] | 1 | 2019-11-12T01:38:43.000Z | 2019-11-12T01:38:43.000Z | 59.886918 | 8,738 | 0.735958 | [
[
[
"# Deep Q-Network implementation\n\nThis notebook shamelessly demands you to implement a DQN - an approximate q-learning algorithm with experience replay and target networks - and see if it works any better this way.",
"_____no_output_____"
]
],
[
[
"#XVFB will be launched if you run on a server\nimport os\nif os.environ.get(\"DISPLAY\") is not str or len(os.environ.get(\"DISPLAY\"))==0:\n !bash ../xvfb start\n %env DISPLAY=:1",
"Starting virtual X frame buffer: Xvfb../xvfb: line 8: start-stop-daemon: command not found\n.\nenv: DISPLAY=:1\n"
]
],
[
[
"__Frameworks__ - we'll accept this homework in any deep learning framework. This particular notebook was designed for tensorflow, but you will find it easy to adapt it to almost any python-based deep learning framework.",
"_____no_output_____"
]
],
[
[
"import gym\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Let's play some old videogames\n\n\nThis time we're gonna apply approximate q-learning to an atari game called Breakout. It's not the hardest thing out there, but it's definitely way more complex than anything we tried before.\n",
"_____no_output_____"
],
[
"### Processing game image \n\nRaw atari images are large, 210x160x3 by default. However, we don't need that level of detail in order to learn them.\n\nWe can thus save a lot of time by preprocessing game image, including\n* Resizing to a smaller shape, 64 x 64\n* Converting to grayscale\n* Cropping irrelevant image parts (top & bottom)",
"_____no_output_____"
]
],
[
[
"from gym.core import ObservationWrapper\nfrom gym.spaces import Box\n\nfrom scipy.misc import imresize\n\nclass PreprocessAtari(ObservationWrapper):\n def __init__(self, env):\n \"\"\"A gym wrapper that crops, scales image into the desired shapes and optionally grayscales it.\"\"\"\n ObservationWrapper.__init__(self,env)\n \n self.img_size = (1, 64, 64)\n self.observation_space = Box(0.0, 1.0, self.img_size)\n\n\n def _observation(self, img):\n \"\"\"what happens to each observation\"\"\"\n \n # Here's what you need to do:\n # * crop image, remove irrelevant parts\n # * resize image to self.img_size \n # (use imresize imported above or any library you want,\n # e.g. opencv, skimage, PIL, keras)\n # * cast image to grayscale\n # * convert image pixels to (0,1) range, float32 type\n \n <Your code here> \n return <...>",
"_____no_output_____"
],
[
"import gym\n#spawn game instance for tests\nenv = gym.make(\"BreakoutDeterministic-v0\") #create raw env\nenv = PreprocessAtari(env)\n\nobservation_shape = env.observation_space.shape\nn_actions = env.action_space.n\n\nenv.reset()\nobs, _, _, _ = env.step(env.action_space.sample())\n\n\n#test observation\nassert obs.ndim == 3, \"observation must be [batch, time, channels] even if there's just one channel\"\nassert obs.shape == observation_shape\nassert obs.dtype == 'float32'\nassert len(np.unique(obs))>2, \"your image must not be binary\"\nassert 0 <= np.min(obs) and np.max(obs) <=1, \"convert image pixels to (0,1) range\"\n\nprint(\"Formal tests seem fine. Here's an example of what you'll get.\")\n\nplt.title(\"what your network gonna see\")\nplt.imshow(obs[0, :, :],interpolation='none',cmap='gray');\n\n",
"Formal tests seem fine. Here's an example of what you'll get.\n"
]
],
[
[
"### Frame buffer\n\nOur agent can only process one observation at a time, so we gotta make sure it contains enough information to fing optimal actions. For instance, agent has to react to moving objects so he must be able to measure object's velocity.\n\nTo do so, we introduce a buffer that stores 4 last images. This time everything is pre-implemented for you.",
"_____no_output_____"
]
],
[
[
"from framebuffer import FrameBuffer\ndef make_env():\n env = gym.make(\"BreakoutDeterministic-v4\")\n env = PreprocessAtari(env)\n env = FrameBuffer(env, n_frames=4, dim_order='pytorch')\n return env\n\nenv = make_env()\nenv.reset()\nn_actions = env.action_space.n\nstate_dim = env.observation_space.shape",
"_____no_output_____"
],
[
"for _ in range(50):\n obs, _, _, _ = env.step(env.action_space.sample())\n\n\nplt.title(\"Game image\")\nplt.imshow(env.render(\"rgb_array\"))\nplt.show()\nplt.title(\"Agent observation (4 frames top to bottom)\")\nplt.imshow(obs.reshape([-1, state_dim[2]]));",
"_____no_output_____"
]
],
[
[
"### Building a network\n\nWe now need to build a neural network that can map images to state q-values. This network will be called on every agent's step so it better not be resnet-152 unless you have an array of GPUs. Instead, you can use strided convolutions with a small number of features to save time and memory.\n\nYou can build any architecture you want, but for reference, here's something that will more or less work:",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"import torch, torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.autograd import Variable",
"_____no_output_____"
],
[
"class DQNAgent(nn.Module):\n def __init__(self, state_shape, n_actions, epsilon=0):\n \"\"\"A simple DQN agent\"\"\"\n nn.Module.__init__(self)\n self.epsilon = epsilon\n self.n_actions = n_actions\n img_c, img_w, img_h = state_shape\n \n # Define your network body here. Please make sure agent is fully contained here\n \n <YOUR CODE>\n \n\n def forward(self, state_t):\n \"\"\"\n takes agent's observation (Variable), returns qvalues (Variable)\n :param state_t: a batch of 4-frame buffers, shape = [batch_size, 4, h, w]\n Hint: if you're running on GPU, use state_t.cuda() right here.\n \"\"\"\n \n # Use your network to compute qvalues for given state\n qvalues = <YOUR CODE>\n \n assert isinstance(qvalues, Variable) and qvalues.requires_grad, \"qvalues must be a torch variable with grad\"\n assert len(qvalues.shape) == 2 and qvalues.shape[0] == state_t.shape[0] and qvalues.shape[1] == n_actions\n \n return qvalues\n \n def get_qvalues(self, states):\n \"\"\"\n like forward, but works on numpy arrays, not Variables\n \"\"\"\n states = Variable(torch.FloatTensor(np.asarray(states)))\n qvalues = self.forward(states)\n return qvalues.data.cpu().numpy()\n \n def sample_actions(self, qvalues):\n \"\"\"pick actions given qvalues. Uses epsilon-greedy exploration strategy. \"\"\"\n epsilon = self.epsilon\n batch_size, n_actions = qvalues.shape\n \n random_actions = np.random.choice(n_actions, size=batch_size)\n best_actions = qvalues.argmax(axis=-1)\n \n should_explore = np.random.choice([0, 1], batch_size, p = [1-epsilon, epsilon])\n return np.where(should_explore, random_actions, best_actions)",
"_____no_output_____"
],
[
"agent = DQNAgent(state_dim, n_actions, epsilon=0.5)",
"_____no_output_____"
]
],
[
[
"Now let's try out our agent to see if it raises any errors.",
"_____no_output_____"
]
],
[
[
"def evaluate(env, agent, n_games=1, greedy=False, t_max=10000):\n \"\"\" Plays n_games full games. If greedy, picks actions as argmax(qvalues). Returns mean reward. \"\"\"\n rewards = []\n for _ in range(n_games):\n s = env.reset()\n reward = 0\n for _ in range(t_max):\n qvalues = agent.get_qvalues([s])\n action = qvalues.argmax(axis=-1)[0] if greedy else agent.sample_actions(qvalues)[0]\n s, r, done, _ = env.step(action)\n reward += r\n if done: break\n \n rewards.append(reward)\n return np.mean(rewards)",
"_____no_output_____"
],
[
"evaluate(env, agent, n_games=1)",
"_____no_output_____"
]
],
[
[
"### Experience replay\nFor this assignment, we provide you with experience replay buffer. If you implemented experience replay buffer in last week's assignment, you can copy-paste it here __to get 2 bonus points__.\n\n",
"_____no_output_____"
],
[
"#### The interface is fairly simple:\n* `exp_replay.add(obs, act, rw, next_obs, done)` - saves (s,a,r,s',done) tuple into the buffer\n* `exp_replay.sample(batch_size)` - returns observations, actions, rewards, next_observations and is_done for `batch_size` random samples.\n* `len(exp_replay)` - returns number of elements stored in replay buffer.",
"_____no_output_____"
]
],
[
[
"from replay_buffer import ReplayBuffer\nexp_replay = ReplayBuffer(10)\n\nfor _ in range(30):\n exp_replay.add(env.reset(), env.action_space.sample(), 1.0, env.reset(), done=False)\n\nobs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch = exp_replay.sample(5)\n\nassert len(exp_replay) == 10, \"experience replay size should be 10 because that's what maximum capacity is\"",
"_____no_output_____"
],
[
"def play_and_record(agent, env, exp_replay, n_steps=1):\n \"\"\"\n Play the game for exactly n steps, record every (s,a,r,s', done) to replay buffer. \n Whenever game ends, add record with done=True and reset the game.\n It is guaranteed that env has done=False when passed to this function.\n :returns: return sum of rewards over time\n \"\"\"\n\n # Play the game for n_steps as per instructions above\n <YOUR CODE>\n \n return <mean rewards>\n \n ",
"_____no_output_____"
],
[
"# testing your code. This may take a minute...\nexp_replay = ReplayBuffer(20000)\n\nplay_and_record(agent, env, exp_replay, n_steps=10000)\n\n# if you're using your own experience replay buffer, some of those tests may need correction. \n# just make sure you know what your code does\nassert len(exp_replay) == 10000, \"play_and_record should have added exactly 10000 steps, \"\\\n \"but instead added %i\" % len(exp_replay)\nis_dones = list(zip(*exp_replay._storage))[-1]\n\nassert 0 < np.mean(is_dones) < 0.1, \"Please make sure you restart the game whenever it is 'done' and record the is_done correctly into the buffer.\"\\\n \"Got %f is_done rate over %i steps. [If you think it's your tough luck, just re-run the test]\"%(np.mean(is_dones), len(exp_replay))\n \nfor _ in range(100):\n obs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch = exp_replay.sample(10)\n assert obs_batch.shape == next_obs_batch.shape == (10,) + state_dim\n assert act_batch.shape == (10,), \"actions batch should have shape (10,) but is instead %s\"%str(act_batch.shape)\n assert reward_batch.shape == (10,), \"rewards batch should have shape (10,) but is instead %s\"%str(reward_batch.shape)\n assert is_done_batch.shape == (10,), \"is_done batch should have shape (10,) but is instead %s\"%str(is_done_batch.shape)\n assert [int(i) in (0,1) for i in is_dones], \"is_done should be strictly True or False\"\n assert [0 <= a <= n_actions for a in act_batch], \"actions should be within [0, n_actions]\"\n \nprint(\"Well done!\")",
"_____no_output_____"
]
],
[
[
"### Target networks\n\nWe also employ the so called \"target network\" - a copy of neural network weights to be used for reference Q-values:\n\nThe network itself is an exact copy of agent network, but it's parameters are not trained. Instead, they are moved here from agent's actual network every so often.\n\n$$ Q_{reference}(s,a) = r + \\gamma \\cdot \\max _{a'} Q_{target}(s',a') $$\n\n\n\n",
"_____no_output_____"
]
],
[
[
"target_network = DQNAgent(state_dim, n_actions)",
"_____no_output_____"
],
[
"# This is how you can load weights from agent into target network\ntarget_network.load_state_dict(agent.state_dict())",
"_____no_output_____"
]
],
[
[
"### Learning with... Q-learning\nHere we write a function similar to `agent.update` from tabular q-learning.",
"_____no_output_____"
],
[
"Compute Q-learning TD error:\n\n$$ L = { 1 \\over N} \\sum_i [ Q_{\\theta}(s,a) - Q_{reference}(s,a) ] ^2 $$\n\nWith Q-reference defined as\n\n$$ Q_{reference}(s,a) = r(s,a) + \\gamma \\cdot max_{a'} Q_{target}(s', a') $$\n\nWhere\n* $Q_{target}(s',a')$ denotes q-value of next state and next action predicted by __target_network__\n* $s, a, r, s'$ are current state, action, reward and next state respectively\n* $\\gamma$ is a discount factor defined two cells above.\n\n\n__Note 1:__ there's an example input below. Feel free to experiment with it before you write the function.\n__Note 2:__ compute_td_loss is a source of 99% of bugs in this homework. If reward doesn't improve, it often helps to go through it line by line [with a rubber duck](https://rubberduckdebugging.com/).",
"_____no_output_____"
]
],
[
[
"def compute_td_loss(states, actions, rewards, next_states, is_done, gamma = 0.99, check_shapes = False):\n \"\"\" Compute td loss using torch operations only. Use the formula above. \"\"\"\n states = Variable(torch.FloatTensor(states)) # shape: [batch_size, c, h, w]\n actions = Variable(torch.LongTensor(actions)) # shape: [batch_size]\n rewards = Variable(torch.FloatTensor(rewards)) # shape: [batch_size]\n next_states = Variable(torch.FloatTensor(next_states)) # shape: [batch_size, c, h, w]\n is_done = Variable(torch.FloatTensor(is_done.astype('float32'))) # shape: [batch_size]\n is_not_done = 1 - is_done\n \n #get q-values for all actions in current states\n predicted_qvalues = agent(states)\n \n # compute q-values for all actions in next states\n predicted_next_qvalues = target_network(next_states)\n \n #select q-values for chosen actions\n predicted_qvalues_for_actions = predicted_qvalues[range(len(actions)), actions]\n \n\n # compute V*(next_states) using predicted next q-values\n next_state_values = < YOUR CODE >\n \n assert next_state_values.dim() == 1 and next_state_values.shape[0] == states.shape[0], \"must predict one value per state\"\n\n # compute \"target q-values\" for loss - it's what's inside square parentheses in the above formula.\n # at the last state use the simplified formula: Q(s,a) = r(s,a) since s' doesn't exist\n # you can multiply next state values by is_not_done to achieve this.\n target_qvalues_for_actions = <YOUR CODE>\n \n #mean squared error loss to minimize\n loss = torch.mean((predicted_qvalues_for_actions - target_qvalues_for_actions.detach()) ** 2 )\n \n if check_shapes:\n assert predicted_next_qvalues.data.dim() == 2, \"make sure you predicted q-values for all actions in next state\"\n assert next_state_values.data.dim() == 1, \"make sure you computed V(s') as maximum over just the actions axis and not all axes\"\n assert target_qvalues_for_actions.data.dim() == 1, \"there's something wrong with target q-values, they must be a vector\"\n \n return loss\n\n ",
"_____no_output_____"
],
[
"# sanity checks\nobs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch = exp_replay.sample(10)\n\nloss = compute_td_loss(obs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch, gamma=0.99,\n check_shapes=True)\nloss.backward()\n\nassert isinstance(loss, Variable) and tuple(loss.data.size()) == (1,), \"you must return scalar loss - mean over batch\"\nassert np.any(next(agent.parameters()).grad.data.numpy() != 0), \"loss must be differentiable w.r.t. network weights\"",
"_____no_output_____"
]
],
[
[
"### Main loop\n\nIt's time to put everything together and see if it learns anything.",
"_____no_output_____"
]
],
[
[
"from tqdm import trange\nfrom IPython.display import clear_output\nimport matplotlib.pyplot as plt\nfrom pandas import ewma\n%matplotlib inline\n\nmean_rw_history = []\ntd_loss_history = []",
"_____no_output_____"
],
[
"exp_replay = ReplayBuffer(10**5)\nplay_and_record(agent, env, exp_replay, n_steps=10000);",
"_____no_output_____"
],
[
"opt = < your favorite optimizer. Default to adam if you don't have one >",
"_____no_output_____"
],
[
"for i in trange(10**5):\n \n # play\n play_and_record(agent, env, exp_replay, 10)\n \n # train\n < sample data from experience replay>\n \n loss = < compute TD loss >\n \n < minimize loss by gradient descent >\n \n td_loss_history.append(loss.data.cpu().numpy()[0])\n \n # adjust agent parameters\n if i % 500 == 0:\n agent.epsilon = max(agent.epsilon * 0.99, 0.01)\n mean_rw_history.append(evaluate(make_env(), agent, n_games=3))\n \n #Load agent weights into target_network\n <YOUR CODE>\n \n \n if i % 100 == 0:\n clear_output(True)\n print(\"buffer size = %i, epsilon = %.5f\" % (len(exp_replay), agent.epsilon))\n plt.figure(figsize=[12, 4]) \n plt.subplot(1,2,1)\n plt.title(\"mean reward per game\")\n plt.plot(mean_rw_history)\n plt.grid()\n\n assert not np.isnan(td_loss_history[-1])\n plt.subplot(1,2,2)\n plt.title(\"TD loss history (moving average)\")\n plt.plot(pd.ewma(np.array(td_loss_history), span=100, min_periods=100))\n plt.grid()\n plt.show()\n \n ",
"_____no_output_____"
],
[
"assert np.mean(mean_rw_history[-10:]) > 10.\nprint(\"That's good enough for tutorial.\")",
"_____no_output_____"
]
],
[
[
"__ How to interpret plots: __\n\n\nThis aint no supervised learning so don't expect anything to improve monotonously. \n* __ TD loss __ is the MSE between agent's current Q-values and target Q-values. It may slowly increase or decrease, it's ok. The \"not ok\" behavior includes going NaN or stayng at exactly zero before agent has perfect performance.\n* __ mean reward__ is the expected sum of r(s,a) agent gets over the full game session. It will oscillate, but on average it should get higher over time (after a few thousand iterations...). \n * In basic q-learning implementation it takes 5-10k steps to \"warm up\" agent before it starts to get better.\n* __ buffer size__ - this one is simple. It should go up and cap at max size.\n* __ epsilon__ - agent's willingness to explore. If you see that agent's already at 0.01 epsilon before it's average reward is above 0 - __ it means you need to increase epsilon__. Set it back to some 0.2 - 0.5 and decrease the pace at which it goes down.\n* Also please ignore first 100-200 steps of each plot - they're just oscillations because of the way moving average works.\n\nAt first your agent will lose quickly. Then it will learn to suck less and at least hit the ball a few times before it loses. Finally it will learn to actually score points.\n\n__Training will take time.__ A lot of it actually. An optimistic estimate is to say it's gonna start winning (average reward > 10) after 20k steps. \n\nBut hey, look on the bright side of things:\n\n",
"_____no_output_____"
],
[
"### Video",
"_____no_output_____"
]
],
[
[
"agent.epsilon=0 # Don't forget to reset epsilon back to previous value if you want to go on training",
"_____no_output_____"
],
[
"#record sessions\nimport gym.wrappers\nenv_monitor = gym.wrappers.Monitor(make_env(),directory=\"videos\",force=True)\nsessions = [evaluate(env_monitor, agent, n_games=1) for _ in range(100)]\nenv_monitor.close()",
"_____no_output_____"
],
[
"#show video\nfrom IPython.display import HTML\nimport os\n\nvideo_names = list(filter(lambda s:s.endswith(\".mp4\"),os.listdir(\"./videos/\")))\n\nHTML(\"\"\"\n<video width=\"640\" height=\"480\" controls>\n <source src=\"{}\" type=\"video/mp4\">\n</video>\n\"\"\".format(\"./videos/\"+video_names[-1])) #this may or may not be _last_ video. Try other indices",
"_____no_output_____"
]
],
[
[
"## Assignment part I (5 pts)\n\nWe'll start by implementing target network to stabilize training.\n\nTo do that you should use TensorFlow functionality. \n\nWe recommend thoroughly debugging your code on simple tests before applying it in atari dqn.",
"_____no_output_____"
],
[
"## Bonus I (2+ pts)\n\nImplement and train double q-learning.\n\nThis task contains of\n* Implementing __double q-learning__ or __dueling q-learning__ or both (see tips below)\n* Training a network till convergence\n * Full points will be awarded if your network gets average score of >=10 (see \"evaluating results\")\n * Higher score = more points as usual\n * If you're running out of time, it's okay to submit a solution that hasn't converged yet and updating it when it converges. _Lateness penalty will not increase for second submission_, so submitting first one in time gets you no penalty.\n\n\n#### Tips:\n* Implementing __double q-learning__ shouldn't be a problem if you've already have target networks in place.\n * You will probably need `tf.argmax` to select best actions\n * Here's an original [article](https://arxiv.org/abs/1509.06461)\n\n* __Dueling__ architecture is also quite straightforward if you have standard DQN.\n * You will need to change network architecture, namely the q-values layer\n * It must now contain two heads: V(s) and A(s,a), both dense layers\n * You should then add them up via elemwise sum layer.\n * Here's an [article](https://arxiv.org/pdf/1511.06581.pdf)",
"_____no_output_____"
],
[
"## Bonus II (5+ pts): Prioritized experience replay\n\nIn this section, you're invited to implement prioritized experience replay\n\n* You will probably need to provide a custom data structure\n* Once pool.update is called, collect the pool.experience_replay.observations, actions, rewards and is_alive and store them in your data structure\n* You can now sample such transitions in proportion to the error (see [article](https://arxiv.org/abs/1511.05952)) for training.\n\nIt's probably more convenient to explicitly declare inputs for \"sample observations\", \"sample actions\" and so on to plug them into q-learning.\n\nPrioritized (and even normal) experience replay should greatly reduce amount of game sessions you need to play in order to achieve good performance. \n\nWhile it's effect on runtime is limited for atari, more complicated envs (further in the course) will certainly benefit for it.\n\nPrioritized experience replay only supports off-policy algorithms, so pls enforce `n_steps=1` in your q-learning reference computation (default is 10).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e71afde557a3ca38e9ef8a843a35f2b036897375 | 5,159 | ipynb | Jupyter Notebook | Part-2-Model_Building.ipynb | Shraeyas/Plagiarism-Detection | c47ba37a0f6ca5fdc3c012b228af05f5dba76962 | [
"MIT"
] | 1 | 2022-01-20T07:21:24.000Z | 2022-01-20T07:21:24.000Z | Part-2-Model_Building.ipynb | Shraeyas/Plagiarism-Detection | c47ba37a0f6ca5fdc3c012b228af05f5dba76962 | [
"MIT"
] | null | null | null | Part-2-Model_Building.ipynb | Shraeyas/Plagiarism-Detection | c47ba37a0f6ca5fdc3c012b228af05f5dba76962 | [
"MIT"
] | null | null | null | 26.869792 | 238 | 0.471215 | [
[
[
"import os\nimport numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"documents = pd.read_csv (os.path.join (\"dataset_processed\", \"documents_processed.csv\"))\ndocuments.head ()",
"_____no_output_____"
]
],
[
[
"### Converted all data to TaggedDocuments for Doc2Vec",
"_____no_output_____"
]
],
[
[
"from gensim.models.doc2vec import TaggedDocument\ntagged_documents = []\nfor index, row in documents.iterrows ():\n tagged_document = TaggedDocument (words = row['sentences'].split (\" \"), tags = [row['document_id']])\n tagged_documents.append (tagged_document)",
"_____no_output_____"
],
[
"tagged_documents[: 1]",
"_____no_output_____"
]
],
[
[
"### Train the final model",
"_____no_output_____"
]
],
[
[
"from gensim.models.doc2vec import Doc2Vec\nmodel = Doc2Vec (vector_size = 100, window = 3, min_count = 5, workers = 8, epochs = 10, alpha = 0.03, min_alpha = 0.002)\nmodel.build_vocab (tagged_documents)\nmodel.train (tagged_documents, total_examples = model.corpus_count, epochs = model.epochs)",
"_____no_output_____"
]
],
[
[
"### Save the model file on disk for future use",
"_____no_output_____"
]
],
[
[
"model_path = os.path.join (\"models\", \"doc2vec_20news.bin\")\nmodel.save (model_path)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71b07661a0d5301441e013f5071dadb36e10c4b | 21,070 | ipynb | Jupyter Notebook | tensorflow/lite/g3doc/models/style_transfer/overview.ipynb | bbbboom/tensorflow | 4d765c332e6c6046e7ab71f7aea9d5c0bd890913 | [
"Apache-2.0"
] | 50 | 2020-03-15T01:04:36.000Z | 2021-11-21T23:25:44.000Z | tensorflow/lite/g3doc/models/style_transfer/overview.ipynb | bbbboom/tensorflow | 4d765c332e6c6046e7ab71f7aea9d5c0bd890913 | [
"Apache-2.0"
] | 58 | 2021-11-22T05:41:28.000Z | 2022-01-19T01:33:40.000Z | tensorflow/lite/g3doc/models/style_transfer/overview.ipynb | Rawan19/tensorflow | 2452e74ca47ec398143ae47c4f14d542fa6ce71a | [
"Apache-2.0"
] | 66 | 2020-05-15T10:05:12.000Z | 2022-02-14T07:28:18.000Z | 39.679849 | 366 | 0.580731 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Artistic Style Transfer with TensorFlow Lite",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/lite/models/style_transfer/overview\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/tensorflow/blob/master/tensorflow/lite/g3doc/models/style_transfer/overview.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/tensorflow/blob/master/tensorflow/lite/g3doc/models/style_transfer/overview.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/tensorflow/tensorflow/lite/g3doc/models/style_transfer/overview.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"One of the most exciting developments in deep learning to come out recently is [artistic style transfer](https://arxiv.org/abs/1508.06576), or the ability to create a new image, known as a [pastiche](https://en.wikipedia.org/wiki/Pastiche), based on two input images: one representing the artistic style and one representing the content.\n\n\n\nUsing this technique, we can generate beautiful new artworks in a range of styles.\n\n\n\nIf you are new to TensorFlow Lite and are working with Android, we\nrecommend exploring the following example applications that can help you get\nstarted.\n\n<a class=\"button button-primary\" href=\"https://github.com/tensorflow/examples/tree/master/lite/examples/style_transfer/android\">Android\nexample</a>\n\nIf you are using a platform other than Android or iOS, or you are already\nfamiliar with the\n<a href=\"https://www.tensorflow.org/api_docs/python/tf/lite\">TensorFlow Lite\nAPIs</a>, you can follow this tutorial to learn how to apply style transfer on any pair of content and style image with a pre-trained TensorFlow Lite model. You can use the model to add style transfer to your own mobile applications.\n\nThe model is open-sourced on [GitHub](https://github.com/tensorflow/magenta/tree/master/magenta/models/arbitrary_image_stylization#train-a-model-on-a-large-dataset-with-data-augmentation-to-run-on-mobile). You can retrain the model with different parameters (e.g. increase content layers' weights to make the output image look more like the content image).",
"_____no_output_____"
],
[
"## Understand the model architecture",
"_____no_output_____"
],
[
"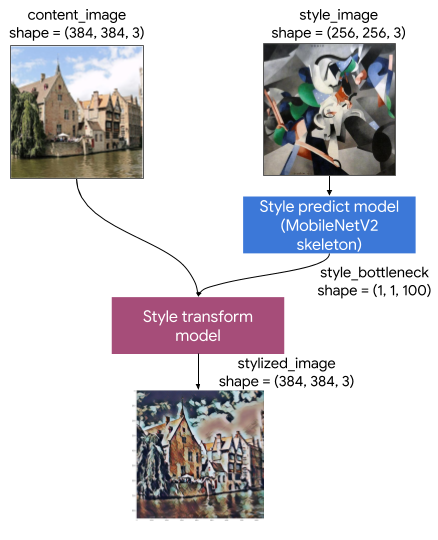\n\nThis Artistic Style Transfer model consists of two submodels:\n1. **Style Prediciton Model**: A MobilenetV2-based neural network that takes an input style image to a 100-dimension style bottleneck vector.\n1. **Style Transform Model**: A neural network that takes apply a style bottleneck vector to a content image and creates a stylized image.\n\nIf your app only needs to support a fixed set of style images, you can compute their style bottleneck vectors in advance, and exclude the Style Prediction Model from your app's binary.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
],
[
"Import dependencies.",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals",
"_____no_output_____"
],
[
"try:\n # %tensorflow_version only exists in Colab.\n import tensorflow.compat.v2 as tf\nexcept Exception:\n pass\ntf.enable_v2_behavior()",
"_____no_output_____"
],
[
"import IPython.display as display\n\nimport matplotlib.pyplot as plt\nimport matplotlib as mpl\nmpl.rcParams['figure.figsize'] = (12,12)\nmpl.rcParams['axes.grid'] = False\n\nimport numpy as np\nimport time\nimport functools",
"_____no_output_____"
]
],
[
[
"Download the content and style images, and the pre-trained TensorFlow Lite models.",
"_____no_output_____"
]
],
[
[
"content_path = tf.keras.utils.get_file('belfry.jpg','https://storage.googleapis.com/khanhlvg-public.appspot.com/arbitrary-style-transfer/belfry-2611573_1280.jpg')\nstyle_path = tf.keras.utils.get_file('style23.jpg','https://storage.googleapis.com/khanhlvg-public.appspot.com/arbitrary-style-transfer/style23.jpg')\n\nstyle_predict_path = tf.keras.utils.get_file('style_predict.tflite', 'https://storage.googleapis.com/download.tensorflow.org/models/tflite/arbitrary_style_transfer/style_predict_quantized_256.tflite')\nstyle_transform_path = tf.keras.utils.get_file('style_transform.tflite', 'https://storage.googleapis.com/download.tensorflow.org/models/tflite/arbitrary_style_transfer/style_transfer_quantized_dynamic.tflite')",
"_____no_output_____"
]
],
[
[
"## Pre-process the inputs\n\n* The content image and the style image must be RGB images with pixel values being float32 numbers between [0..1].\n* The style image size must be (1, 256, 256, 3). We central crop the image and resize it.\n* The content image can be any size. However, as we trained the model using square-cropped data, cropping the content image to a square results in better stylized image.",
"_____no_output_____"
]
],
[
[
"# Function to load an image from a file, and add a batch dimension.\ndef load_img(path_to_img):\n img = tf.io.read_file(path_to_img)\n img = tf.image.decode_image(img, channels=3)\n img = tf.image.convert_image_dtype(img, tf.float32)\n img = img[tf.newaxis, :]\n\n return img\n\n# Function to pre-process style image input.\ndef preprocess_style_image(style_image):\n # Resize the image so that the shorter dimension becomes 256px.\n target_dim = 256\n shape = tf.cast(tf.shape(style_image)[1:-1], tf.float32)\n short_dim = min(shape)\n scale = target_dim / short_dim\n new_shape = tf.cast(shape * scale, tf.int32)\n style_image = tf.image.resize(style_image, new_shape)\n\n # Central crop the image.\n style_image = tf.image.resize_with_crop_or_pad(style_image, target_dim, target_dim)\n\n return style_image\n\n# Function to pre-process content image input.\ndef preprocess_content_image(content_image):\n # Central crop the image.\n shape = tf.shape(content_image)[1:-1]\n short_dim = min(shape)\n content_image = tf.image.resize_with_crop_or_pad(content_image, short_dim, short_dim)\n\n return content_image\n\n# Load the input images.\ncontent_image = load_img(content_path)\nstyle_image = load_img(style_path)\n\n# Preprocess the input images.\npreprocessed_content_image = preprocess_content_image(content_image)\npreprocessed_style_image = preprocess_style_image(style_image)\n\nprint('Style Image Shape:', preprocessed_style_image.shape)\nprint('Content Image Shape:', preprocessed_content_image.shape)",
"_____no_output_____"
]
],
[
[
"## Visualize the inputs",
"_____no_output_____"
]
],
[
[
"def imshow(image, title=None):\n if len(image.shape) > 3:\n image = tf.squeeze(image, axis=0)\n\n plt.imshow(image)\n if title:\n plt.title(title)\n\nplt.subplot(1, 2, 1)\nimshow(preprocessed_content_image, 'Content Image')\n\nplt.subplot(1, 2, 2)\nimshow(preprocessed_style_image, 'Style Image')",
"_____no_output_____"
]
],
[
[
"## Run style transfer with TensorFlow Lite",
"_____no_output_____"
],
[
"### Style prediction",
"_____no_output_____"
]
],
[
[
"# Function to run style prediction on preprocessed style image.\ndef run_style_predict(preprocessed_style_image):\n # Load the model.\n interpreter = tf.lite.Interpreter(model_path=style_predict_path)\n\n # Set model input.\n interpreter.allocate_tensors()\n input_details = interpreter.get_input_details()\n interpreter.set_tensor(input_details[0][\"index\"], preprocessed_style_image)\n\n # Calculate style bottleneck.\n interpreter.invoke()\n style_bottleneck = interpreter.tensor(\n interpreter.get_output_details()[0][\"index\"]\n )()\n\n return style_bottleneck\n\n# Calculate style bottleneck for the preprocessed style image.\nstyle_bottleneck = run_style_predict(preprocessed_style_image)\nprint('Style Bottleneck Shape:', style_bottleneck.shape)",
"_____no_output_____"
]
],
[
[
"### Style transform",
"_____no_output_____"
]
],
[
[
"# Run style transform on preprocessed style image\ndef run_style_transform(style_bottleneck, preprocessed_content_image):\n # Load the model.\n interpreter = tf.lite.Interpreter(model_path=style_transform_path)\n\n # Set model input.\n input_details = interpreter.get_input_details()\n interpreter.resize_tensor_input(input_details[0][\"index\"],\n preprocessed_content_image.shape)\n interpreter.allocate_tensors()\n\n # Set model inputs.\n interpreter.set_tensor(input_details[0][\"index\"], preprocessed_content_image)\n interpreter.set_tensor(input_details[1][\"index\"], style_bottleneck)\n interpreter.invoke()\n\n # Transform content image.\n stylized_image = interpreter.tensor(\n interpreter.get_output_details()[0][\"index\"]\n )()\n\n return stylized_image\n\n# Stylize the content image using the style bottleneck.\nstylized_image = run_style_transform(style_bottleneck, preprocessed_content_image)\n\n# Visualize the output.\nimshow(stylized_image, 'Stylized Image')",
"_____no_output_____"
]
],
[
[
"### Style blending\n\nWe can blend the style of content image into the stylized output, which in turn making the output look more like the content image.",
"_____no_output_____"
]
],
[
[
"# Calculate style bottleneck of the content image.\nstyle_bottleneck_content = run_style_predict(\n preprocess_style_image(content_image)\n )",
"_____no_output_____"
],
[
"# Define content blending ratio between [0..1].\n# 0.0: 0% style extracts from content image.\n# 1.0: 100% style extracted from content image.\ncontent_blending_ratio = 0.5 #@param {type:\"slider\", min:0, max:1, step:0.01}\n\n# Blend the style bottleneck of style image and content image\nstyle_bottleneck_blended = content_blending_ratio * style_bottleneck_content \\\n + (1 - content_blending_ratio) * style_bottleneck\n\n# Stylize the content image using the style bottleneck.\nstylized_image_blended = run_style_transform(style_bottleneck_blended,\n preprocessed_content_image)\n\n# Visualize the output.\nimshow(stylized_image_blended, 'Blended Stylized Image')",
"_____no_output_____"
]
],
[
[
"## Performance Benchmarks\n\nPerformance benchmark numbers are generated with the tool [described here](https://www.tensorflow.org/lite/performance/benchmarks).\n<table ><thead><tr><th>Model name</th> <th>Model size</th> <th>Device </th> <th>NNAPI</th> <th>CPU</th></tr> </thead> \n<tr> <td rowspan = 3> <a href=\"https://storage.googleapis.com/download.tensorflow.org/models/tflite/arbitrary_style_transfer/style_predict_quantized_256.tflite\">Style prediction model</a> </td> \n<td rowspan = 3>2.8 Mb</td>\n<td>Pixel 3 (Android 10) </td> <td>142ms</td><td>14ms*</td></tr>\n<tr><td>Pixel 4 (Android 10) </td> <td>5.2ms</td><td>6.7ms*</td></tr>\n<tr><td>iPhone XS (iOS 12.4.1) </td> <td></td><td>10.7ms**</td></tr>\n<tr> <td rowspan = 3> <a href=\"https://storage.googleapis.com/download.tensorflow.org/models/tflite/arbitrary_style_transfer/style_transfer_quantized_dynamic.tflite\">Style transform model</a> </td> \n<td rowspan = 3>0.2s Mb</td>\n<td>Pixel 3 (Android 10) </td> <td></td><td>540ms*</td></tr>\n<tr><td>Pixel 4 (Android 10) </td> <td></td><td>405ms*</td></tr>\n<tr><td>iPhone XS (iOS 12.4.1) </td> <td></td><td>251ms**</td></tr></table>\n* 4 threads used.\n\n** 2 threads on iPhone for the best performance.\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e71b0cfd1a2bc2ec27ca2daa3e1f3a16ce92dcaf | 2,302 | ipynb | Jupyter Notebook | Filtro_Errores_Facturacion_Electronica.ipynb | RafaGrand/clinical_research_notebooks | d5ed32b2f4612aafd81f94d315e2f2252a3abb9a | [
"MIT"
] | null | null | null | Filtro_Errores_Facturacion_Electronica.ipynb | RafaGrand/clinical_research_notebooks | d5ed32b2f4612aafd81f94d315e2f2252a3abb9a | [
"MIT"
] | null | null | null | Filtro_Errores_Facturacion_Electronica.ipynb | RafaGrand/clinical_research_notebooks | d5ed32b2f4612aafd81f94d315e2f2252a3abb9a | [
"MIT"
] | null | null | null | 32.885714 | 186 | 0.593831 | [
[
[
"from IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = \"all\"\nfrom datetime import datetime\nimport pandas as pd\n#pd.options.display.max_seq_items = 2000\n## Usar linea de abajo para mostrar todas las filas de datos sin limitacion \npd.set_option('display.max_rows',None) \n\ndocs_pd = pd.read_csv(\"https://github.com/RafaGrand/clinical_research_notebooks/blob/master/Reporte_Fri%20Sep%2004%2003_06_55%20COT%202020.csv\", sep=';')\n\nfilter_df = docs_pd[(docs_pd.estado != 'Éxito en la Notificación del Adquiriente') \n & (docs_pd.estado !='Aceptación Tácita') \n & (docs_pd.estado !='Aceptación Adquiriente')\n & (docs_pd.estado !='Error en la Notificación del Adquiriente')]\n\ncb_df = filter_df[filter_df['Número Documento'].str.contains(\"CB\")] \ndf_trimmed = cb_df.apply(lambda x: x.str.strip() if x.dtype == \"object\" else x) # Trim whitespaces\n\n#df_trimmed.to_excel (r'C:\\Users\\SOPORTECNSDR\\jupyter\\Dataframes\\facturas_con_error_'+ str(datetime.now().strftime('%Y_%m_%d_%H_%M')) + '.xlsx', index = False, header=True)\n#df_trimmed.to_excel (r'C:\\Users\\SOPORTECNSDR\\jupyter\\Dataframes\\FE_septiembre'+ str(datetime.now().strftime('%Y_%m_%d_%H_%M')) + '.xlsx', index = False, header=True)\n\ndf_trimmed.head(2000)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
e71b2ba4727562ce533095296e23a5d15114c6f2 | 132,873 | ipynb | Jupyter Notebook | draw.ipynb | suyanzhou626/utility_on_segmentation | e8143c0cddb6a384cd4b23152700a02d39dd78d2 | [
"Apache-2.0"
] | null | null | null | draw.ipynb | suyanzhou626/utility_on_segmentation | e8143c0cddb6a384cd4b23152700a02d39dd78d2 | [
"Apache-2.0"
] | null | null | null | draw.ipynb | suyanzhou626/utility_on_segmentation | e8143c0cddb6a384cd4b23152700a02d39dd78d2 | [
"Apache-2.0"
] | null | null | null | 738.183333 | 128,440 | 0.952549 | [
[
[
"import os\nimport cv2\nfrom PIL import Image\nimport numpy as np\nimport skimage.io as io\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"%config Completer.use_jedi = False",
"_____no_output_____"
],
[
"# refer to: https://segmentfault.com/a/1190000015662096\ndef draw_counter(image_path, mask_path):\n \n image = io.imread(image_path)\n \n if os.path.splitext(mask_path)[-1] == \".tif\":\n mask = io.imread(mask_path, as_gray=True)\n else:\n mask = io.imread(mask_path, as_gray=True)*255\n mask = mask.astype(np.uint8)\n \n ret, thresh = cv2.threshold(mask, 127, 255, 0)\n contours, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)\n cnt = contours[0]\n \n # yellow: rgb [255, 255, 0]\n # red: rgb [255, 0, 0]\n # blue: rgb [0, 0, 255]\n # green: rgb [0, 255, 0]\n image_contour = cv2.drawContours(image, [cnt], 0, (0, 255, 0), 3)\n \n return image_contour\n\nif __name__ == \"__main__\":\n \n image_path = \"/media/hdd/PolySeg_old/data/oldKvasir-SEG/images/cju1f79yhsb5w0993txub59ol.jpg\"\n mask_path = \"/media/hdd/PolySeg_old/data/oldKvasir-SEG/masks/cju1f79yhsb5w0993txub59ol.jpg\"\n \n outimg = draw_counter(image_path, mask_path)\n outimg = Image.fromarray(outimg).resize((352, 352),Image.ANTIALIAS)\n \n #\n outimg.save('./img5.pdf')\n\n plt.imshow(outimg)\n plt.axis('off')\n ",
"_____no_output_____"
],
[
"# Kvasir-SEG\n# cju0qx73cjw570799j4n5cjze.jpg\n# cju76o55nymqd0871h31sph9w\n# cju1fyb1d69et0878muzdak9u\n# cju1f79yhsb5w0993txub59ol",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
e71b309dcfb5685cc220e817976fa57a53e01723 | 285,039 | ipynb | Jupyter Notebook | Requests.ipynb | Pytoddler/Web-scraping | c9ce8b84c2dc30917fc9a3ddc84eb8a8d6c583a3 | [
"MIT"
] | null | null | null | Requests.ipynb | Pytoddler/Web-scraping | c9ce8b84c2dc30917fc9a3ddc84eb8a8d6c583a3 | [
"MIT"
] | null | null | null | Requests.ipynb | Pytoddler/Web-scraping | c9ce8b84c2dc30917fc9a3ddc84eb8a8d6c583a3 | [
"MIT"
] | null | null | null | 93.241413 | 38,684 | 0.662815 | [
[
[
"# Requests 較urllib好用...",
"_____no_output_____"
],
[
"# 基本引入",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('http://www.baidu.com')\n\nprint(type(response))\nprint(response.status_code)\nprint(type(response.text))\nprint(response.text)\nprint(response.cookies)",
"<class 'requests.models.Response'>\n200\n<class 'str'>\n<!DOCTYPE html>\r\n<!--STATUS OK--><html> <head><meta http-equiv=content-type content=text/html;charset=utf-8><meta http-equiv=X-UA-Compatible content=IE=Edge><meta content=always name=referrer><link rel=stylesheet type=text/css href=http://s1.bdstatic.com/r/www/cache/bdorz/baidu.min.css><title>ç¾åº¦ä¸ä¸ï¼ä½ å°±ç¥é</title></head> <body link=#0000cc> <div id=wrapper> <div id=head> <div class=head_wrapper> <div class=s_form> <div class=s_form_wrapper> <div id=lg> <img hidefocus=true src=//www.baidu.com/img/bd_logo1.png width=270 height=129> </div> <form id=form name=f action=//www.baidu.com/s class=fm> <input type=hidden name=bdorz_come value=1> <input type=hidden name=ie value=utf-8> <input type=hidden name=f value=8> <input type=hidden name=rsv_bp value=1> <input type=hidden name=rsv_idx value=1> <input type=hidden name=tn value=baidu><span class=\"bg s_ipt_wr\"><input id=kw name=wd class=s_ipt value maxlength=255 autocomplete=off autofocus></span><span class=\"bg s_btn_wr\"><input type=submit id=su value=ç¾åº¦ä¸ä¸ class=\"bg s_btn\"></span> </form> </div> </div> <div id=u1> <a href=http://news.baidu.com name=tj_trnews class=mnav>æ°é»</a> <a href=http://www.hao123.com name=tj_trhao123 class=mnav>hao123</a> <a href=http://map.baidu.com name=tj_trmap class=mnav>å°å¾</a> <a href=http://v.baidu.com name=tj_trvideo class=mnav>è§é¢</a> <a href=http://tieba.baidu.com name=tj_trtieba class=mnav>è´´å§</a> <noscript> <a href=http://www.baidu.com/bdorz/login.gif?login&tpl=mn&u=http%3A%2F%2Fwww.baidu.com%2f%3fbdorz_come%3d1 name=tj_login class=lb>ç»å½</a> </noscript> <script>document.write('<a href=\"http://www.baidu.com/bdorz/login.gif?login&tpl=mn&u='+ encodeURIComponent(window.location.href+ (window.location.search === \"\" ? \"?\" : \"&\")+ \"bdorz_come=1\")+ '\" name=\"tj_login\" class=\"lb\">ç»å½</a>');</script> <a href=//www.baidu.com/more/ name=tj_briicon class=bri style=\"display: block;\">æ´å¤äº§å</a> </div> </div> </div> <div id=ftCon> <div id=ftConw> <p id=lh> <a href=http://home.baidu.com>å
³äºç¾åº¦</a> <a href=http://ir.baidu.com>About Baidu</a> </p> <p id=cp>©2017 Baidu <a href=http://www.baidu.com/duty/>使ç¨ç¾åº¦åå¿
读</a> <a href=http://jianyi.baidu.com/ class=cp-feedback>æè§åé¦</a> 京ICPè¯030173å· <img src=//www.baidu.com/img/gs.gif> </p> </div> </div> </div> </body> </html>\r\n\n<RequestsCookieJar[<Cookie BDORZ=27315 for .baidu.com/>]>\n"
]
],
[
[
"# 各種請求方式",
"_____no_output_____"
]
],
[
[
"import requests\nrequests.post('https://httpbin.org/post')\nrequests.put('https://httpbin.org/put')\nrequests.delete('https://httpbin.org/delete')\nrequests.head('https://httpbin.org/get')\nrequests.options('https://httpbin.org/get')",
"_____no_output_____"
]
],
[
[
"# requests",
"_____no_output_____"
],
[
"## 基本get請求",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('https://httpbin.org/get')\nprint(response.text)",
"{\n \"args\": {}, \n \"headers\": {\n \"Accept\": \"*/*\", \n \"Accept-Encoding\": \"gzip, deflate\", \n \"Connection\": \"close\", \n \"Host\": \"httpbin.org\", \n \"User-Agent\": \"python-requests/2.14.2\"\n }, \n \"origin\": \"140.112.73.184\", \n \"url\": \"https://httpbin.org/get\"\n}\n\n"
]
],
[
[
"## 帶參數請求",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('https://httpbin.org/get?name=germany&age=22')\nprint(response.text)",
"{\n \"args\": {\n \"age\": \"22\", \n \"name\": \"germany\"\n }, \n \"headers\": {\n \"Accept\": \"*/*\", \n \"Accept-Encoding\": \"gzip, deflate\", \n \"Connection\": \"close\", \n \"Host\": \"httpbin.org\", \n \"User-Agent\": \"python-requests/2.14.2\"\n }, \n \"origin\": \"140.112.73.184\", \n \"url\": \"https://httpbin.org/get?name=germany&age=22\"\n}\n\n"
],
[
"import requests\n\ndata = {\n 'name':'germany',\n 'age':22\n}\n\nresponse = requests.get('https://httpbin.org/get', params = data)\nprint(response.text)",
"{\n \"args\": {\n \"age\": \"22\", \n \"name\": \"germany\"\n }, \n \"headers\": {\n \"Accept\": \"*/*\", \n \"Accept-Encoding\": \"gzip, deflate\", \n \"Connection\": \"close\", \n \"Host\": \"httpbin.org\", \n \"User-Agent\": \"python-requests/2.14.2\"\n }, \n \"origin\": \"140.112.73.184\", \n \"url\": \"https://httpbin.org/get?name=germany&age=22\"\n}\n\n"
]
],
[
[
"## 解析 json",
"_____no_output_____"
]
],
[
[
"import requests\nimport json\n\nresponse = requests.get('https://httpbin.org/get')\nprint(type(response.text))\n\n#以下兩個是一樣的\nprint(response.json())\nprint(json.loads(response.text))\n\nprint(type(response.json))",
"<class 'str'>\n{'args': {}, 'headers': {'Accept': '*/*', 'Accept-Encoding': 'gzip, deflate', 'Connection': 'close', 'Host': 'httpbin.org', 'User-Agent': 'python-requests/2.14.2'}, 'origin': '140.112.73.184', 'url': 'https://httpbin.org/get'}\n{'args': {}, 'headers': {'Accept': '*/*', 'Accept-Encoding': 'gzip, deflate', 'Connection': 'close', 'Host': 'httpbin.org', 'User-Agent': 'python-requests/2.14.2'}, 'origin': '140.112.73.184', 'url': 'https://httpbin.org/get'}\n<class 'method'>\n"
]
],
[
[
"## 獲取二進位數據",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('http://github.com/favicon.ico')\nprint(type(response.text), type(response.content))\n\n#str\nprint(response.text)\n\n#bytes 二進制編碼\nprint(response.content) ",
"<class 'str'> <class 'bytes'>\n\u0000\u0000\u0001\u0000\u0002\u0000\u0010\u0010\u0000\u0000\u0001\u0000 \u0000(\u0005\u0000\u0000&\u0000\u0000\u0000 \u0000\u0000\u0001\u0000 \u0000(\u0014\u0000\u0000N\u0005\u0000\u0000(\u0000\u0000\u0000\u0010\u0000\u0000\u0000 \u0000\u0000\u0000\u0001\u0000 \u0000\u0000\u0000\u0000\u0000\u0000\u0005\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0011\u0011\u0013v\u0013\u0013\u0013�\u000e\u000e\u000e\u0012\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f\u0011\u0011\u0011\u0014�\u0013\u0013\u0013i\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0014\u0014\u0014�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0000\u0000\u0000\u0019\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0018\u0015\u0015\u0017�\u0015\u0015\u0017�\u0011\u0011\u0013�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0011\u0011\u0013�\u0013\u0013\u0014�\u0011\u0011\u0011\u001e\u000f\u000f\u000f\u0010\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\r\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0014\u0014\u0014�\u0015\u0015\u0017�\u0005\u0005\u0011,\r\r\r\\\u000e\u000e\u000f�\u000f\u000f\u000f\"\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000e\u000e\u000e4\u0010\u0010\u0010�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u0000\u0000\u0000\u0000\u000f\u000f\u000f0\u000f\r\u000f�\u0000\u0000\u0000�\u0001\u0001\u0001�\u0002\u0002\u0002�\u0002\u0002\u0002�\r\r\r8\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0007\u0007\u0007@\u0002\u0002\u0002�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0011\u0011\u0011-\u0013\u0013\u0016�\u0014\u0014\u0015�\u0001\u0001\u0001�\u000f\u000f\u0011�\f\f\u0011;\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\r\r\u0011:\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013O\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0010\u0010\u0010L\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0014\u0014\u0014�\u0000\u0000\u0000\u0002\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0002\u0013\u0013\u0014�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0014\u0014\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0014\u0014\u0014�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0013\u0013\u0014�\u000f\u000f\u000f!\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0010\u0010\u0010\u001f\u0013\u0013\u0014�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0011\u0011\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000e\u000e\u000e4\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f@\u0015\u0015\u0017�\u0015\u0015\u0017�\u0011\u0011\u0014�\r\r\r8\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0012�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\f\u0000\u0000\u0000\n\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\f\u0000\u0000\u0000\u0001\u0000\u0000\u0000\u0000\u0012\u0012\u0012�\u0015\u0015\u0017�\u0015\u0015\u0017�\r\r\r8\u0000\u0000\u0000\u0000\u0011\u0011\u0014�\u0015\u0015\u0017�\u0011\u0011\u0013�\u000e\u000e\u000e6\u0000\u0000\u0000�\r\r\r�\u0012\u0012\u0014�\u0012\u0012\u0014�\u0013\u0013\u0014�\u0000\u0000\u0000t\u0004\u0004\u00047\u0011\u0011\u0013�\u0015\u0015\u0017�\u0011\u0011\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0003\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u0000\u0000\u0000\u0003\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0003\u0011\u0011\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0011\u0011\u0014�\u0000\u0000\u0000\u0003\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0010\u0010\u0010>\u0012\u0012\u0012�\u0013\u0013\u0013�\u0012\u0012\u0014�\u0012\u0012\u0014�\u0013\u0013\u0013�\u0012\u0012\u0012�\u0010\u0010\u0010>\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000(\u0000\u0000\u0000 \u0000\u0000\u0000@\u0000\u0000\u0000\u0001\u0000 \u0000\u0000\u0000\u0000\u0000\u0000\u0014\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\f\u0011\u0011\u0013s\u0013\u0013\u0013�\u0013\u0013\u0014�\u0000\u0000\u0000\u0007\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0007\u0013\u0013\u0014�\u0013\u0013\u0013�\u0011\u0011\u0013t\u0000\u0000\u0000\f\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0001\u0011\u0011\u0014d\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000f\u000f\u000f\u0010\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f\u0010\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0011\u0011\u0014e\u0000\u0000\u0000\u0001\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000e\u000e\u000e\u0012\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000f\u000f\u000f\u0010\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f\u0010\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u000e\u000e\u000e\u0012\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0010\u0010\u0010\u001f\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0014\u0014\u0014�\u0011\u0011\u0013u\u000f\u000f\u000f`\u0013\u0013\u0013x\u0000\u0000\u0000\b\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f\u0010\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0010\u0010\u0010\u001f\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0018\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0012o\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f\u0010\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0018\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0004\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0000\u000f\u000f\u000f\u0010\u000f\u000f\u000fc\u0012\u0012\u0012z\u000e\u000e\u000eF\u0000\u0000\u0000\u0003\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\f\f\f\u0015\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u0000\u0000\u0000\u0004\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0012\u0012\u0012~\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\r\r\r8\u000f\u000f\u000f!\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0011\u0011\u0011<\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0010\u0010\u0010>\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0012~\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0011\u0011\u0011\u001e\u0013\u0013\u0014�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0011\u0011\u0013s\u0000\u0000\u0000\u0007\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0001\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0001\u0011\u0011\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0011\u0011\u0011\u001e\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0012\u0012\u0012�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0012�\u0010\u0010\u0010?\u0011\u0011\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0016�\u0011\u0011\u0014e\u000e\u000e\u000e4\u0000\u0000\u0000\u0003\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0004\r\r\u00119\u000e\u000e\u0013h\u0013\u0013\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0012�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\r\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0018\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0019\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\r\u000e\u000e\u0014Y\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000e\u000e\u0014Y\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0011\u0011\u0014�\u0000\u0000\u0000\u0004\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0004\u0011\u0011\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u0011\u0011\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0012^\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0012\u0012\u0012^\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0011\u0011\u0014�\u0012\u0012\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0000\u0000\u0000\u0016\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0016\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0012\u0012\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0014\u0014\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0014\u0014\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0011\u0011\u0011\u000f\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0011\u0011\u0011\u000f\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000f\u000f\u000fb\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000fb\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013�\u000e\u000e\u0013g\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0016�\u000e\u000e\u000e\u0012\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000e\u000e\u000e\u0012\u0013\u0013\u0013�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000e\u000e\u0013g\u0000\u0000\u0000\u0018\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0012\u0012\u0012_\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0012\u0012\u0012_\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0018\u0000\u0000\u0000\u0000\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000e\u000e\u000e5\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000e\u000e\u000e5\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000e\u000e\u000e4\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000f\u000f\u000f2\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0011\u0011\u0011\u000f\f\f\f(\u0000\u0000\u0000\u0002\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0002\f\f\f(\u0011\u0011\u0011\u000f\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f2\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u000e\u000e\u000e4\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0013\\\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0007\u000f\u000f\u000fc\u0013\u0013\u0014�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0014\u0014\u0014�\u0011\u0011\u0013�\u0011\u0011\u0013�\u0014\u0014\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0013\u0013\u0014�\u000f\u000f\u000fc\u0000\u0000\u0000\u0007\u0000\u0000\u0000\u0000\u0013\u0013\u0013\\\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f\u0011\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u000f\u000f\u000f\u0011\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f3\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u000f\u000f\u000f3\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000fB\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u000f\u000f\u000fB\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f1\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u000f\u000f\u000f1\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0011\u0011\u0011\u000f\u0013\u0013\u0013�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0013�\u0011\u0011\u0011\u000f\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u000f\u000f\u000f1\u0013\u0013\u0013�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0013\u0013\u0013�\u000f\u000f\u000f1\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\f\f\f\u0014\u000f\u000f\u000fc\u0011\u0011\u0014�\u0014\u0014\u0014�\u0013\u0013\u0014�\u0015\u0015\u0017�\u0015\u0015\u0017�\u0013\u0013\u0014�\u0014\u0014\u0014�\u0011\u0011\u0014�\u000f\u000f\u000fc\f\f\f\u0014\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\u0000\nb'\\x00\\x00\\x01\\x00\\x02\\x00\\x10\\x10\\x00\\x00\\x01\\x00 \\x00(\\x05\\x00\\x00&\\x00\\x00\\x00 \\x00\\x00\\x01\\x00 \\x00(\\x14\\x00\\x00N\\x05\\x00\\x00(\\x00\\x00\\x00\\x10\\x00\\x00\\x00 \\x00\\x00\\x00\\x01\\x00 \\x00\\x00\\x00\\x00\\x00\\x00\\x05\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x11\\x11\\x13v\\x13\\x13\\x13\\xc5\\x0e\\x0e\\x0e\\x12\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f\\x11\\x11\\x11\\x14\\xb1\\x13\\x13\\x13i\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x14\\x14\\x14\\x96\\x13\\x13\\x14\\xfc\\x13\\x13\\x14\\xed\\x00\\x00\\x00\\x19\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x18\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x11\\x11\\x13\\x85\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x11\\x11\\x13\\xc1\\x13\\x13\\x14\\xee\\x11\\x11\\x11\\x1e\\x0f\\x0f\\x0f\\x10\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\r\\x13\\x13\\x14\\xf5\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x14\\xaf\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x14\\x14\\x14\\x99\\x15\\x15\\x17\\xff\\x05\\x05\\x11,\\r\\r\\r\\\\\\x0e\\x0e\\x0f\\xc1\\x0f\\x0f\\x0f\"\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0e\\x0e\\x0e4\\x10\\x10\\x10\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\x8f\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f0\\x0f\\r\\x0f\\xff\\x00\\x00\\x00\\xf9\\x01\\x01\\x01\\xed\\x02\\x02\\x02\\xff\\x02\\x02\\x02\\xf6\\r\\r\\r8\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x07\\x07\\x07@\\x02\\x02\\x02\\xeb\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x11\\x11\\x11-\\x13\\x13\\x16\\x9c\\x14\\x14\\x15\\xff\\x01\\x01\\x01\\xfc\\x0f\\x0f\\x11\\xfb\\x0c\\x0c\\x11;\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\r\\r\\x11:\\x13\\x13\\x14\\xe7\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\x9a\\x13\\x13\\x13\\xd9\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13O\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x10\\x10\\x10L\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\xda\\x13\\x13\\x14\\xf6\\x15\\x15\\x17\\xff\\x14\\x14\\x14\\xf0\\x00\\x00\\x00\\x02\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x02\\x13\\x13\\x14\\xf1\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf6\\x13\\x13\\x14\\xf7\\x15\\x15\\x17\\xff\\x14\\x14\\x14\\xe1\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x14\\x14\\x14\\xe1\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf7\\x13\\x13\\x14\\xde\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf9\\x0f\\x0f\\x0f!\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x10\\x10\\x10\\x1f\\x13\\x13\\x14\\xf8\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xde\\x11\\x11\\x14\\xa2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0e\\x0e\\x0e4\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f@\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x11\\x11\\x14\\xa2\\r\\r\\r8\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x12\\x98\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0c\\x00\\x00\\x00\\n\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0c\\x00\\x00\\x00\\x01\\x00\\x00\\x00\\x00\\x12\\x12\\x12\\x98\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\r\\r\\r8\\x00\\x00\\x00\\x00\\x11\\x11\\x14\\xa4\\x15\\x15\\x17\\xff\\x11\\x11\\x13\\xc1\\x0e\\x0e\\x0e6\\x00\\x00\\x00\\x81\\r\\r\\r\\xdc\\x12\\x12\\x14\\xd8\\x12\\x12\\x14\\xd8\\x13\\x13\\x14\\xf7\\x00\\x00\\x00t\\x04\\x04\\x047\\x11\\x11\\x13\\xc1\\x15\\x15\\x17\\xff\\x11\\x11\\x14\\xa4\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x03\\x13\\x13\\x13\\xc6\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\xc6\\x00\\x00\\x00\\x03\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x03\\x11\\x11\\x14\\xa2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x11\\x11\\x14\\xa2\\x00\\x00\\x00\\x03\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x10\\x10\\x10>\\x12\\x12\\x12\\x97\\x13\\x13\\x13\\xd9\\x12\\x12\\x14\\xf2\\x12\\x12\\x14\\xf2\\x13\\x13\\x13\\xd9\\x12\\x12\\x12\\x97\\x10\\x10\\x10>\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00(\\x00\\x00\\x00 \\x00\\x00\\x00@\\x00\\x00\\x00\\x01\\x00 \\x00\\x00\\x00\\x00\\x00\\x00\\x14\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0c\\x11\\x11\\x13s\\x13\\x13\\x13\\xda\\x13\\x13\\x14\\xec\\x00\\x00\\x00\\x07\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x07\\x13\\x13\\x14\\xec\\x13\\x13\\x13\\xda\\x11\\x11\\x13t\\x00\\x00\\x00\\x0c\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x01\\x11\\x11\\x14d\\x13\\x13\\x14\\xea\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0f\\x0f\\x0f\\x10\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f\\x10\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xeb\\x11\\x11\\x14e\\x00\\x00\\x00\\x01\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0e\\x0e\\x0e\\x12\\x13\\x13\\x13\\xb7\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0f\\x0f\\x0f\\x10\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f\\x10\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\xb7\\x0e\\x0e\\x0e\\x12\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x10\\x10\\x10\\x1f\\x13\\x13\\x14\\xdb\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x14\\x14\\x14\\xc3\\x11\\x11\\x13u\\x0f\\x0f\\x0f`\\x13\\x13\\x13x\\x00\\x00\\x00\\x08\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f\\x10\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xdb\\x10\\x10\\x10\\x1f\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x18\\x13\\x13\\x14\\xdc\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x12o\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f\\x10\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xdc\\x00\\x00\\x00\\x18\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x04\\x13\\x13\\x13\\xc4\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xb9\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f\\x10\\x0f\\x0f\\x0fc\\x12\\x12\\x12z\\x0e\\x0e\\x0eF\\x00\\x00\\x00\\x03\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0c\\x0c\\x0c\\x15\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\xc4\\x00\\x00\\x00\\x04\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x12\\x12\\x12~\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\r\\r\\r8\\x0f\\x0f\\x0f!\\x13\\x13\\x13\\xe2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x11\\x11\\x11<\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x10\\x10\\x10>\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x12~\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x11\\x11\\x11\\x1e\\x13\\x13\\x14\\xf7\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xfe\\x11\\x11\\x13s\\x00\\x00\\x00\\x07\\x13\\x13\\x14\\xc9\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xae\\x00\\x00\\x00\\x01\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x01\\x11\\x11\\x14\\xb0\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf7\\x11\\x11\\x11\\x1e\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x12\\x12\\x12\\x97\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x12\\x8a\\x10\\x10\\x10?\\x11\\x11\\x13\\xc2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xfa\\x13\\x13\\x16\\xaa\\x11\\x11\\x14e\\x0e\\x0e\\x0e4\\x00\\x00\\x00\\x03\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x04\\r\\r\\x119\\x0e\\x0e\\x13h\\x13\\x13\\x14\\xae\\x13\\x13\\x14\\xfa\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x12\\x97\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\r\\x13\\x13\\x14\\xf5\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xab\\x00\\x00\\x00\\x18\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x19\\x13\\x13\\x14\\xab\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf5\\x00\\x00\\x00\\r\\x0e\\x0e\\x14Y\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\x8f\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x13\\x13\\x13\\x8f\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0e\\x0e\\x14Y\\x13\\x13\\x13\\x9d\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x11\\x11\\x14\\xd1\\x00\\x00\\x00\\x04\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x04\\x11\\x11\\x14\\xd1\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\x9d\\x11\\x11\\x14\\xd0\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x12^\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x12\\x12\\x12^\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x11\\x11\\x14\\xd0\\x12\\x12\\x14\\xf2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x00\\x00\\x00\\x16\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x16\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x14\\xf2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x14\\xf2\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x12\\x12\\x14\\xf2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xdb\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x13\\x13\\x14\\xdc\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf6\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x14\\x14\\x14\\xe1\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x14\\x14\\x14\\xe1\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf6\\x13\\x13\\x14\\xd6\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xfc\\x11\\x11\\x11\\x0f\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x11\\x11\\x11\\x0f\\x13\\x13\\x14\\xfc\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xd6\\x13\\x13\\x13\\xa9\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0f\\x0f\\x0fb\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0fb\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\xa9\\x0e\\x0e\\x13g\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x16\\xe3\\x0e\\x0e\\x0e\\x12\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0e\\x0e\\x0e\\x12\\x13\\x13\\x13\\xe2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0e\\x0e\\x13g\\x00\\x00\\x00\\x18\\x13\\x13\\x14\\xfc\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x12\\x12\\x12_\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x12\\x12\\x12_\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xfc\\x00\\x00\\x00\\x18\\x00\\x00\\x00\\x00\\x13\\x13\\x14\\xae\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0e\\x0e\\x0e5\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0e\\x0e\\x0e5\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xae\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0e\\x0e\\x0e4\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0f\\x0f\\x0f2\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x11\\x11\\x11\\x0f\\x0c\\x0c\\x0c(\\x00\\x00\\x00\\x02\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x02\\x0c\\x0c\\x0c(\\x11\\x11\\x11\\x0f\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f2\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x0e\\x0e\\x0e4\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x13\\x13\\x14\\x9e\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x13\\\\\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x07\\x0f\\x0f\\x0fc\\x13\\x13\\x14\\xe8\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf3\\x14\\x14\\x14\\xd2\\x11\\x11\\x13\\xc1\\x11\\x11\\x13\\xc1\\x14\\x14\\x14\\xd2\\x13\\x13\\x14\\xf3\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xe8\\x0f\\x0f\\x0fc\\x00\\x00\\x00\\x07\\x00\\x00\\x00\\x00\\x13\\x13\\x13\\\\\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\x9e\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f\\x11\\x13\\x13\\x14\\xdf\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xd6\\x13\\x13\\x14\\xad\\x13\\x13\\x14\\xf1\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf1\\x13\\x13\\x14\\xad\\x13\\x13\\x14\\xd6\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xdf\\x0f\\x0f\\x0f\\x11\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f3\\x13\\x13\\x14\\xf1\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf1\\x0f\\x0f\\x0f3\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0fB\\x13\\x13\\x14\\xf1\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf1\\x0f\\x0f\\x0fB\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f1\\x13\\x13\\x14\\xde\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xde\\x0f\\x0f\\x0f1\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x11\\x11\\x11\\x0f\\x13\\x13\\x13\\x9b\\x13\\x13\\x14\\xfe\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xfe\\x13\\x13\\x13\\x9b\\x11\\x11\\x11\\x0f\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0f\\x0f\\x0f1\\x13\\x13\\x13\\xa9\\x13\\x13\\x14\\xfb\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xfb\\x13\\x13\\x13\\xa9\\x0f\\x0f\\x0f1\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x0c\\x0c\\x0c\\x14\\x0f\\x0f\\x0fc\\x11\\x11\\x14\\xa4\\x14\\x14\\x14\\xd2\\x13\\x13\\x14\\xf3\\x15\\x15\\x17\\xff\\x15\\x15\\x17\\xff\\x13\\x13\\x14\\xf3\\x14\\x14\\x14\\xd2\\x11\\x11\\x14\\xa4\\x0f\\x0f\\x0fc\\x0c\\x0c\\x0c\\x14\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x00'\n"
]
],
[
[
"## 寫入二進位數據,儲存成檔案",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('http://github.com/favicon.ico')\n\n#寫入檔案,以二進為形式\nwith open('favicon.ico','wb') as f:\n f.write(response.content)\n f.close()",
"_____no_output_____"
]
],
[
[
"## 添加headers",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get(\"http://zhihu.com/explore\")\nprint(response.text)",
"<html><body><h1>500 Server Error</h1>\nAn internal server error occured.\n</body></html>\n\n"
],
[
"import requests\n\nheaders = {\n \"User-Agent\": \"Mozilla/5.0 (Windows NT 10.0; Win64; x64) \\\n AppleWebKit/537.36 (KHTML, like Gecko) Chrome/59.0.3071.115 Safari/537.36\"\n}\n\n#知乎這個網站不用headers偽裝過不去XD\nresponse = requests.get(\"http://zhihu.com/explore\", headers=headers)\nprint(response.text)",
"<!DOCTYPE html>\n<html lang=\"zh-CN\" dropEffect=\"none\" class=\"no-js no-auth \">\n<head>\n<meta charset=\"utf-8\" />\n\n<meta http-equiv=\"X-UA-Compatible\" content=\"IE=edge,chrome=1\" />\n<meta name=\"renderer\" content=\"webkit\" />\n<meta http-equiv=\"X-ZA-Response-Id\" content=\"12e6013417194862\">\n<meta http-equiv=\"X-ZA-Experiment\" content=\"default:None,ge2:ge2_1,nweb_sticky_sidebar:sticky,new_more:new,live_store:ls_a2_b2_c1_f2,topnavbar_qrcode:topnavbar_qrcode_hide,qa_live_recommendation:recommended_by_rule,ge120:ge120_2,zcm-lighting:zcm,fav_act:default,home_nweb:default,ge3:ge3_9,qrcode_login:qrcode,recommend_readings_on_share:wx_share_algo_recommend,qa_sticky_sidebar:sticky_sidebar\">\n\n<title>发现 - 知乎</title>\n\n<meta name=\"apple-itunes-app\" content=\"app-id=432274380, app-argument=zhihu://explore\">\n\n\n<meta name=\"viewport\" content=\"width=device-width, initial-scale=1, maximum-scale=1\"/>\n<meta http-equiv=\"mobile-agent\" content=\"format=html5;url=https://www.zhihu.com/explore\">\n<meta id=\"znonce\" name=\"znonce\" content=\"12ac55e2aba64796b10dba0c7c53d846\">\n\n\n\n<link rel=\"apple-touch-icon\" href=\"https://static.zhihu.com/static/revved/img/ios/touch-icon-152.87c020b9.png\" sizes=\"152x152\">\n<link rel=\"apple-touch-icon\" href=\"https://static.zhihu.com/static/revved/img/ios/touch-icon-120.496c913b.png\" sizes=\"120x120\">\n<link rel=\"apple-touch-icon\" href=\"https://static.zhihu.com/static/revved/img/ios/touch-icon-76.dcf79352.png\" sizes=\"76x76\">\n<link rel=\"apple-touch-icon\" href=\"https://static.zhihu.com/static/revved/img/ios/touch-icon-60.9911cffb.png\" sizes=\"60x60\">\n\n\n<link rel=\"shortcut icon\" href=\"https://static.zhihu.com/static/favicon.ico\" type=\"image/x-icon\">\n\n<link rel=\"search\" type=\"application/opensearchdescription+xml\" href=\"https://static.zhihu.com/static/search.xml\" title=\"知乎\" />\n<link rel=\"stylesheet\" href=\"https://static.zhihu.com/static/revved/-/css/z.7e3884f5.css\">\n\n\n\n<meta name=\"google-site-verification\" content=\"FTeR0c8arOPKh8c5DYh_9uu98_zJbaWw53J-Sch9MTg\" />\n\n\n\n<!--[if lt IE 9]>\n<script src=\"https://static.zhihu.com/static/components/respond/dest/respond.min.js\"></script>\n<link href=\"https://static.zhihu.com/static/components/respond/cross-domain/respond-proxy.html\" id=\"respond-proxy\" rel=\"respond-proxy\" />\n<link href=\"/static/components/respond/cross-domain/respond.proxy.gif\" id=\"respond-redirect\" rel=\"respond-redirect\" />\n<script src=\"/static/components/respond/cross-domain/respond.proxy.js\"></script>\n<![endif]-->\n<script src=\"https://static.zhihu.com/static/revved/-/js/instant.14757a4a.js\"></script>\n\n</head>\n\n<body class=\"zhi page-explore\">\n\n\n\n\n<div role=\"navigation\" class=\"zu-top\" data-za-module=\"TopNavBar\">\n<div class=\"zg-wrap modal-shifting clearfix\" id=\"zh-top-inner\">\n<a href=\"/\" class=\"zu-top-link-logo\" id=\"zh-top-link-logo\" data-za-c=\"view_home\" data-za-a=\"visit_home\" data-za-l=\"top_navigation_zhihu_logo\">知乎</a>\n\n\n\n<ul class=\"topnav-noauth clearfix\">\n<li>\n<a href=\"javascript:;\" class=\"js-signup-noauth\"><i class=\"zg-icon zg-icon-dd-home\"></i>注册知乎</a>\n</li>\n<li>\n<a href=\"javascript:;\" class=\"js-signin-noauth\">登录</a>\n</li>\n</ul>\n\n\n\n<button class=\"zu-top-add-question\" id=\"zu-top-add-question\">提问</button>\n\n\n<div role=\"search\" id=\"zh-top-search\" class=\"zu-top-search\">\n<form method=\"GET\" action=\"/search\" id=\"zh-top-search-form\" class=\"zu-top-search-form\">\n\n\n\n<input type=\"hidden\" name=\"type\" value=\"content\">\n<label for=\"q\" class=\"hide-text\">知乎搜索</label><input type=\"text\" class=\"zu-top-search-input\" id=\"q\" name=\"q\" autocomplete=\"off\" value=\"\" maxlength=\"100\" placeholder=\"搜索你感兴趣的内容...\">\n<button type=\"submit\" class=\"zu-top-search-button\"><span class=\"hide-text\">搜索</span><span class=\"sprite-global-icon-magnifier-dark\"></span></button>\n</form>\n</div>\n\n\n\n\n\n</div>\n</div>\n\n\n<div class=\"zu-global-notify\" id=\"zh-global-message\" style=\"display:none\">\n<div class=\"zg-wrap\">\n<div class=\"zu-global-nitify-inner\">\n<a class=\"zu-global-notify-close\" href=\"javascript:;\" title=\"关闭\" name=\"close\">x</a>\n<span class=\"zu-global-notify-icon\"></span>\n<span class=\"zu-global-notify-msg\"></span>\n</div>\n</div>\n</div>\n\n\n\n\n<div class=\"zg-wrap zu-main clearfix \" role=\"main\">\n<div class=\"zu-main-content\">\n<div class=\"zu-main-content-inner\">\n\n\n\n\n\n<div id=\"zh-recommend\">\n<div id=\"zh-recommend-title\" class=\"page-title\">\n<i class=\"zg-icon zg-icon-feedlist\"></i>\n<span>编辑推荐</span>\n<a href=\"/explore/recommendations\" class=\"zg-link-gray zg-right\" data-za-c=\"explore\" data-za-a=\"visit_explore_recommendations\" data-za-l=\"editor_recommendations_more\"><span>更多推荐 »</span></a>\n</div>\n<div id=\"zh-recommend-list\">\n\n\n<div class=\"top-recommend-feed feed-item\">\n<h2><a class=\"question_link\" href=\"/question/61945738/answer/193629131\" target=\"_blank\" data-id=\"17003001\" data-za-element-name=\"Title\">\n湖南政府为何给全省居民购买大灾保险?保险公司对于这种参保需求如何处理?\n</a></h2>\n<div class=\"avatar\">\n<a title=\"保险老金\"\ndata-hovercard=\"p$t$zhong-xin-jing-suan-shi\"\nclass=\"zm-item-link-avatar\"\ntarget=\"_blank\"\nhref=\"/people/zhong-xin-jing-suan-shi\">\n<img src=\"https://pic3.zhimg.com/v2-e66febe856dcae7f1c95cc9906922d9e_m.jpg\" class=\"zm-item-img-avatar\">\n</a>\n</div>\n\n<div class=\"feed-main\">\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$zhong-xin-jing-suan-shi\"\ntarget=\"_blank\" href=\"/people/zhong-xin-jing-suan-shi\"\n>保险老金</a></span><span title=\"保险精算师\" class=\"bio\">\n保险精算师\n</span>\n</span>\n</div>\n<div class=\"zm-item-rich-text\" data-resourceid=\"17003001\" data-action=\"/answer/content\">\n<div class=\"zh-summary summary clearfix\">\n<img src=\"https://pic1.zhimg.com/v2-333f8134f4da5e6d508e797c925e4fc8_200x112.png\" data-rawwidth=\"554\" data-rawheight=\"206\" class=\"origin_image inline-img zh-lightbox-thumb\" data-original=\"https://pic1.zhimg.com/v2-333f8134f4da5e6d508e797c925e4fc8_r.png\">\n谢谢邀请,中国的巨灾保障很值得一说。 <b>政府主导保险应对巨灾,以补偿市场保险覆盖率不足。</b>巨灾的显著特点是发生的频率很低,但一旦发生,其影响范围极广、损失程度巨大。因此,巨灾保险具有准公共产品的特点(群众通过保险分散风险的意识不够、依赖政府心…\n</div>\n</div>\n</div>\n</div>\n\n\n\n\n<div class=\"recommend-feed feed-item\">\n<span class=\"zg-right zg-gray-normal feed-meta\" >问答</span>\n<h2><a class=\"question_link\" href=\"/question/61741194/answer/195776203\" target=\"_blank\" data-id=\"16920940\" data-za-element-name=\"Title\">\n如何看待好莱坞不信任内地票房数据,聘请会计师事务所进行审计?\n</a></h2>\n</div>\n\n\n\n<div class=\"recommend-feed feed-item\">\n<span class=\"zg-right zg-gray-normal feed-meta\" >问答</span>\n<h2><a class=\"question_link\" href=\"/question/22698672/answer/195505484\" target=\"_blank\" data-id=\"1260229\" data-za-element-name=\"Title\">\n建筑学学生如何制作作品集?\n</a></h2>\n</div>\n\n\n\n<div class=\"recommend-feed feed-item\">\n<span class=\"zg-right zg-gray-normal feed-meta\" >问答</span>\n<h2><a class=\"question_link\" href=\"/question/51583052/answer/194923434\" target=\"_blank\" data-id=\"12851156\" data-za-element-name=\"Title\">\n互联网金融中需要关注的风控逾期指标有哪些?\n</a></h2>\n</div>\n\n\n\n<div class=\"recommend-feed feed-item\">\n\n<span class=\"zg-right zg-gray-normal feed-meta\" >老司机的生物学课堂</span>\n\n<h2><a class=\"post-link\" target=\"_blank\" href=\"https://zhuanlan.zhihu.com/p/27768900\" data-za-element-name=\"Title\">聊聊转录组测序——3.数据可视化及基因功能注释</a></h2>\n</div>\n\n\n</div>\n</div>\n<div class=\"explore-tab\" id=\"js-explore-tab\">\n<a class=\"zg-anchor-hidden\" name=\"daily-hot\"></a>\n<a class=\"zg-anchor-hidden\" name=\"monthly-hot\"></a>\n<ul class=\"tab-navs clearfix\">\n<li class=\"tab-nav\"><a class=\"anchor\" href=\"#daily-hot\" data-za-c=\"explore\" data-za-a=\"visit_explore_daily_trendings\" data-za-l=\"explore_daily_trendings\">今日最热</a></li>\n<li class=\"tab-nav\"><a class=\"anchor\" href=\"#monthly-hot\" data-za-c=\"explore\" data-za-a=\"visit_explore_monthly_trendings\" data-za-l=\"explore_monthly_trendings\">本月最热</a></li>\n</ul>\n<div class=\"tab-panel\">\n<div data-type=\"daily\">\n<div class=\"explore-feed feed-item\" data-offset=\"1\">\n<h2><a class=\"question_link\" href=\"/question/25524218/answer/196125448\" target=\"_blank\" data-id=\"2392308\" data-za-element-name=\"Title\">\n电影或电视剧中有哪些常识错误?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"65730051\"\ndata-atoken=\"196125448\"\ndata-collapsed=\"0\"\ndata-created=\"1499654984\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/25524218/answer/196125448\">\n<meta itemprop=\"answer-id\" content=\"65730051\">\n<meta itemprop=\"answer-url-token\" content=\"196125448\">\n<a class=\"zg-anchor-hidden\" name=\"answer-65730051\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>52</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">52</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$jian-dan-xin-li\"\ntarget=\"_blank\" href=\"/org/jian-dan-xin-li\"\n>简单心理</a><span class=\"OrgIcon sprite-global-icon-org-14\" data-tooltip=\"s$b$已认证的机构\"></span></span><span title=\"只提供高质量的心理服务\" class=\"bio\">\n只提供高质量的心理服务\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"52\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">52</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"2392308\" data-action=\"/answer/content\" data-author-name=\"简单心理\" data-entry-url=\"/question/25524218/answer/196125448\">\n\n<textarea hidden class=\"content\"><p>国内外的影视剧中,经常出现对于心理学词汇的滥用、心理疾病的误解以及咨询师这个行业的超夸张演绎。</p><br><p>为了总结出这篇文章,我们被集体气吐了血,想对这些编剧们说一句:别闹,好么。</p><br><p>下面,让我们一起来领略一下:</p><br><p>电视剧《亲爱的翻译官》中,杨幂饰演的乔菲,与心理咨询师高家明,进行了一次<b>匪夷所思</b>的治疗。</p><br><p>第一次咨询见面,这位大胆的咨询师开门见山地提出了第一个问题:</p><img src="https://pic2.zhimg.com/v2-dcd6b0996abb2a475204c82676ccf6b1_b.jpg" data-rawwidth="640" data-rawheight="337" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-dcd6b0996abb2a475204c82676ccf6b1_r.jpg"><br><p>从前几个画面的专业程度来看,我真的怕他下一句接:“说出来让我开心一下呗……”</p><img src="https://pic2.zhimg.com/v2-52e8f4133d52380543bf6614946d18cd_b.jpg" data-rawwidth="640" data-rawheight="316" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-52e8f4133d52380543bf6614946d18cd_r.jpg"><p>(换谁谁都不想说。)</p><br><p>结果,咨询师马上说是从别人口中得知来访者的情况,这简直就是在说:“全学校都知道你的事情了!你就别装了!” </p><img src="https://pic4.zhimg.com/v2-8bba9972e5106f0996d92e00173a2aeb_b.jpg" data-rawwidth="640" data-rawheight="459" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-8bba9972e5106f0996d92e00173a2aeb_r.jpg"><br><p>然后,又如此直白地表明自己的专业程度,和来访者建立信任关系:</p><img src="https://pic1.zhimg.com/v2-e7163e1ad12166ce0eeec9dcf3baf350_b.jpg" data-rawwidth="640" data-rawheight="344" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-e7163e1ad12166ce0eeec9dcf3baf350_r.jpg"><br><p>我很怀疑……(从始至终,乔菲连包都一直背在身上,可见她的防御和阻抗有多大。)\n</p><p>在一系列的装……之后(后面详述),第一次咨询就这样草率地结束了。我数了一下,乔菲一共说了5句话,点了两次头。咨询一共进行了3分6秒。</p><br><p>3分6秒,你逗我呢?</p><br><img src="https://pic3.zhimg.com/v2-4ac7fea5a5772e5996a43eb8a2140782_b.jpg" data-rawwidth="640" data-rawheight="350" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic3.zhimg.com/v2-4ac7fea5a5772e5996a43eb8a2140782_r.jpg"><br><p>(谁爱来谁来吧。)</p><br><p>一次标准的咨询正常来说大约是50-60分钟。一般情况下,以来访者表述为主,咨询师起到引导作用。而这位自我感觉良好的大哥直接为来访者进行了一次思想教育。</p><br><p>但高家明还是太年轻,<b>不够业余</b>,电视剧《好先生》中,徐丽可谓是把心理咨询师能犯的错误都犯了。好一个教课书级别的反例:</p><br><p>第一次与来访者见面,在一个环境嘈杂的咖啡厅,来访者是陆远(孙红雷),但在场居然还有另外二人,四人间关系更是错综复杂。</p><br><p>尤其是,当事人陆远完全没有意愿进行咨询,而咨询师才不管这些,总想强势分析别人。</p><br><img src="https://pic4.zhimg.com/v2-754a7db10850fb8c4bba4161edddae67_b.jpg" data-rawwidth="640" data-rawheight="356" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-754a7db10850fb8c4bba4161edddae67_r.jpg"><br><p>其次,徐丽把与来访者的工作内容告诉了来访者的朋友,“他不让我告诉你的,但我还是跟你说了吧。”</p><br><p>这种话就算平常朋友之间说出来都会产生隔阂,对于咨询师来说,更是严重违反了保密性原则。</p><br><img src="https://pic1.zhimg.com/v2-7c9d6e7910f92c1c16ec96ad5db84988_b.jpg" data-rawwidth="640" data-rawheight="717" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-7c9d6e7910f92c1c16ec96ad5db84988_r.jpg"><br><br><img src="https://pic4.zhimg.com/v2-9b50f7144a3b7064ff5c31c2a9a95d7f_b.jpg" data-rawwidth="647" data-rawheight="96" class="origin_image zh-lightbox-thumb" width="647" data-original="https://pic4.zhimg.com/v2-9b50f7144a3b7064ff5c31c2a9a95d7f_r.jpg"><br><br><h2><b>下诊断就是这么不羁,你咬我啊</b></h2><br><p>除了违反基本设置,<b>影视剧中的心理咨询师还酷爱为来访者下诊断,诊断下得越快越好,指定的病越严重越好。</b></p><br><p>依然是《亲爱的翻译官》,咨询师在首次咨询中,一上来就让来访者随便挑东西代表自己和家庭,我有点惶恐,这是要“显露功力”的节奏啊。</p><img src="https://pic2.zhimg.com/v2-2257fc9e096f3b2afbd1542157d9aa55_b.jpg" data-rawwidth="640" data-rawheight="697" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-2257fc9e096f3b2afbd1542157d9aa55_r.jpg"><br><p>话音还没落,就指派了花瓶给杨幂,不是说让来访者自己挑吗……</p><img src="https://pic1.zhimg.com/v2-e2e08dc3bed6e2daac69273579aa565c_b.jpg" data-rawwidth="640" data-rawheight="307" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-e2e08dc3bed6e2daac69273579aa565c_r.jpg"><img src="https://pic3.zhimg.com/v2-6b99be16af8b3dbf98d33ba8a3dbdb8e_b.jpg" data-rawwidth="640" data-rawheight="295" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic3.zhimg.com/v2-6b99be16af8b3dbf98d33ba8a3dbdb8e_r.jpg"><br><p>来访者表示不能接受这个草率的指派:</p><img src="https://pic1.zhimg.com/v2-e2e08dc3bed6e2daac69273579aa565c_b.jpg" data-rawwidth="640" data-rawheight="307" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-e2e08dc3bed6e2daac69273579aa565c_r.jpg"><br><p>最后杨幂挑了桌上摆在眼前的绿巨人代表自己,又挑了办公桌上的一棵绿植代表自己的家庭。</p><br><p>咨询师终于可以摆出一副大师的样子,胸有成竹地扯起淡来:</p><p>(前方高密度装逼,请大家保护好自己的双眼)</p><img src="https://pic1.zhimg.com/v2-5efb4e5aed1f094462bbe7535d7597d4_b.jpg" data-rawwidth="640" data-rawheight="1348" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-5efb4e5aed1f094462bbe7535d7597d4_r.jpg"><br><p>对对对……你说什么都对。(你看杨幂都不想说话了)</p><img src="https://pic4.zhimg.com/v2-e2495901b4d69395342248c0f75b3faf_b.jpg" data-rawwidth="640" data-rawheight="287" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-e2495901b4d69395342248c0f75b3faf_r.jpg"><br><p>然而,与电视剧《好先生》中的徐丽咨询师相比,高家明简直就是业界良心。</p><br><p>第一次咨询,是甘敬带彭佳禾前来,拜访自己的闺蜜:咨询师徐丽,这首先已经违背了避免双重关系的原则,但目的却是为一个不在场的人——陆远咨询,更是莫名其妙。</p><br><p>咨询师一句“说说吧”,结果彭佳禾念叨了一些有的没的,还把自己感动哭了,也许就是年轻女孩的中二病吧。</p><img src="https://pic4.zhimg.com/v2-d51012f42451315033e74c25fd33568b_b.jpg" data-rawwidth="640" data-rawheight="381" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-d51012f42451315033e74c25fd33568b_r.jpg"><br><p>但在情感还未平复的情况下, 徐丽咨询师直接请还在哭着的佳禾出去等着。 </p><img src="https://pic2.zhimg.com/v2-10791f95faf6cebff14ad40bc2b397c1_b.jpg" data-rawwidth="640" data-rawheight="382" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-10791f95faf6cebff14ad40bc2b397c1_r.jpg"><br><p>人家正抒发感情,你一句话不说,把人赶了出去?!</p><br><img src="https://pic2.zhimg.com/v2-5176dc4dd9868c1f3b8943ecb57e9f49_b.jpg" data-rawwidth="224" data-rawheight="217" class="content_image" width="224"><br><p>短暂接触了中二少女后,徐丽开始强行装逼:</p><img src="https://pic4.zhimg.com/v2-2cb9e5a11452719b8cfe3eebaa772273_b.jpg" data-rawwidth="550" data-rawheight="844" class="origin_image zh-lightbox-thumb" width="550" data-original="https://pic4.zhimg.com/v2-2cb9e5a11452719b8cfe3eebaa772273_r.jpg"><br><p>就说了五句话,就认定人家女孩是抑郁症……你开心就好……</p><br><p>同样是第一次见面,她还给孙红雷饰演的男主下了这样专业的诊断:</p><img src="https://pic2.zhimg.com/v2-94df5590f212d7fa1243d40266242ed9_b.jpg" data-rawwidth="640" data-rawheight="920" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-94df5590f212d7fa1243d40266242ed9_r.jpg"><br><p>这一席话,听得我忍不住想下跪。EXO ME?还负责任地说?</p><br><img src="https://pic1.zhimg.com/v2-c0f69522d7dacb8cd8451b7d280364ac_b.jpg" data-rawwidth="109" data-rawheight="120" class="content_image" width="109"><br><p>徐丽可谓是野蛮派分析的第一人,这一连串的吓人的专业词汇,要不是我上过学,差点就被唬住了呢。</p><br><p>除了一言不合就下诊断,影视剧里的咨询师们还有个毛病:包治百病。</p><p>徐峥在电影《催眠大师》中扮演的可能是个刑讯逼供的警察吧……</p><img src="https://pic2.zhimg.com/v2-96ba6c9f1d8560649de263d27866e8f9_b.jpg" data-rawwidth="640" data-rawheight="299" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-96ba6c9f1d8560649de263d27866e8f9_r.jpg"><br><img src="https://pic4.zhimg.com/v2-36065713e1adafc13036d2ff43c2d527_b.jpg" data-rawwidth="640" data-rawheight="315" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-36065713e1adafc13036d2ff43c2d527_r.jpg"><br><img src="https://pic4.zhimg.com/v2-6079ed11337a0a75296e4db2c1e0b37b_b.jpg" data-rawwidth="640" data-rawheight="311" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-6079ed11337a0a75296e4db2c1e0b37b_r.jpg"><br><p>病人:我在乎啊!</p><br><p>「谈天」不仅仅是普通意义上的闲聊,而是治疗性的谈话。大众对于心理咨询的误解也常常在于此,“我花这么多钱,结果咨询师一句话也不说,光看着我笑?”</p><br><p>咨询师应该抱着温暖和关怀的态度对待来访者,但徐峥一开口,我就觉得他在威胁……</p><br><img src="https://pic1.zhimg.com/v2-6e5084595607c9e9fe39dd47b35cae54_b.jpg" data-rawwidth="414" data-rawheight="527" class="content_image" width="414"><p>极端疗法,是师承杨主任吗,有点害怕呢:) </p><br><img src="https://pic4.zhimg.com/v2-360141804ed3728b742a558f841eb3af_b.jpg" data-rawwidth="645" data-rawheight="96" class="origin_image zh-lightbox-thumb" width="645" data-original="https://pic4.zhimg.com/v2-360141804ed3728b742a558f841eb3af_r.jpg"><br><h2><b>谁也别想拦住我跟来访者谈恋爱!</b></h2><br><p>不知道为什么,国内外影视剧中的咨询师,一定要和来访者发展双重关系,尤其是恋爱关系,拦都拦不住。</p><br><p>电视剧《好先生》中,女咨询师爱上孙红雷饰演的来访者,不仅在咨询室过程中喝红酒、进行暧昧的谈话,最后还留宿了来访者!没眼看……</p><br><img src="https://pic1.zhimg.com/v2-90a2b2403b12b98eaed5ad8b48d08b28_b.jpg" data-rawwidth="640" data-rawheight="724" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-90a2b2403b12b98eaed5ad8b48d08b28_r.jpg"><br><p>一脸骄傲是怎么回事……\n</p><p>而且还不顾世俗眼光,毅然决然地追求来访者:</p><br><img src="https://pic4.zhimg.com/v2-05b6f79a852c765ca705174772b7e72f_b.jpg" data-rawwidth="640" data-rawheight="426" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-05b6f79a852c765ca705174772b7e72f_r.jpg"><br><p>来访者在咨询过程中对于咨询师产生爱慕之情是可以理解的,毕竟生命中有一个人那么关心你、支持你、从来不批判你,产生好感是正常的。</p><br><p>但是作为专业的咨询师,把伦理道德抛在脑后,非要和来访者谈恋爱,那就是你的不对了!</p><br><img src="https://pic3.zhimg.com/v2-55380922880d6702f1e764a7a085622a_b.jpg" data-rawwidth="640" data-rawheight="427" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic3.zhimg.com/v2-55380922880d6702f1e764a7a085622a_r.jpg"><br><p>同样,《亲爱的翻译官》中,那位高家明咨询师都已经违反了那么多条职业规范,再多加一条又何妨,双重关系不吃亏,喜欢杨幂勇敢追。</p><br><p>在第二次咨询中,咨询师就为来访者准备了花束,动机不纯献殷勤,这种特殊的对待已经开始有触犯伦理道德的危险了:</p><br><img src="https://pic1.zhimg.com/v2-85bc21136b6f964a192530bbec8cc680_b.jpg" data-rawwidth="640" data-rawheight="347" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-85bc21136b6f964a192530bbec8cc680_r.jpg"><br><p>但终于有人看不下去了,作为精神科前辈的主任,提醒他不要忘了咨询师的伦理准则:</p><br><img src="https://pic1.zhimg.com/v2-c2a50f84bf8684559e349c9555296ef8_b.jpg" data-rawwidth="640" data-rawheight="300" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-c2a50f84bf8684559e349c9555296ef8_r.jpg"><br><img src="https://pic4.zhimg.com/v2-de98712d8178c4f5e40b1cb713e6f86f_b.jpg" data-rawwidth="640" data-rawheight="370" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-de98712d8178c4f5e40b1cb713e6f86f_r.jpg"><br><img src="https://pic2.zhimg.com/v2-0a612fb17ff76e069607791139196129_b.jpg" data-rawwidth="640" data-rawheight="337" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-0a612fb17ff76e069607791139196129_r.jpg"><br><p>亏他还记得……但他并没有听从前辈的话,居然还辩解:</p><br><img src="https://pic3.zhimg.com/v2-920763333079f81f6c6c9739922653e2_b.jpg" data-rawwidth="640" data-rawheight="274" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic3.zhimg.com/v2-920763333079f81f6c6c9739922653e2_r.jpg"><br><p>主任接着提出了恰当的解决办法,当咨询师与来访者的超出了一般治疗关系时,就需要转介,即把来访者转给别的咨询师继续咨询疗程。然而高家明再一次赤裸裸地拒绝:</p><br><img src="https://pic1.zhimg.com/v2-e109433dd774542a911ae3c8cc08b008_b.jpg" data-rawwidth="640" data-rawheight="367" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-e109433dd774542a911ae3c8cc08b008_r.jpg"><br><p>就这么顶撞主任……我佩服你的勇气,看来应该是真爱。</p><br><p>于是他终于没羞没臊地表白,并在人家试图拒绝时强行拥抱了来访者。</p><br><img src="https://pic4.zhimg.com/v2-f6c6c16de4943c621653d432cc839743_b.jpg" data-rawwidth="640" data-rawheight="302" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-f6c6c16de4943c621653d432cc839743_r.jpg"><br><img src="https://pic1.zhimg.com/v2-9fc116afd95773dd9191f3d30e4a07a8_b.jpg" data-rawwidth="640" data-rawheight="303" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic1.zhimg.com/v2-9fc116afd95773dd9191f3d30e4a07a8_r.jpg"><br><p>怎么说呢,祝福你们吧,如果这样还没被行业除名,那么心理咨询这个行业就没有什么存在的意义了。</p><br><p>当然了,国外也月亮也没有比较圆,该和来访者谈恋爱还是照样谈。疑犯追踪第四季里面,膝盖毁灭者李四就和他的治疗师谈起了恋爱,而且一谈就谈到了全剧终……</p><br><img src="https://pic1.zhimg.com/v2-a24b6e7d44055b8d752d1228f6e2b3a0_b.png" data-rawwidth="1024" data-rawheight="576" class="origin_image zh-lightbox-thumb" width="1024" data-original="https://pic1.zhimg.com/v2-a24b6e7d44055b8d752d1228f6e2b3a0_r.png"><br><p>双重/多重关系是指:咨询师与寻求专业心理服务的来访者之间,除了治疗关系,还存在其他利益或亲密情感等特点的人际关系状况。</p><br><p>避免双重关系是一名合格的咨询师最基本的职业伦理道德,更不用说和来访者发展恋爱关系。但影视剧中出现的咨询师,有一个算一个,都违反了这条基本原则,争先恐后地与来访者谈起恋爱。还有王法吗……还有法律吗?</p><br><br><img src="https://pic4.zhimg.com/v2-360141804ed3728b742a558f841eb3af_b.jpg" data-rawwidth="645" data-rawheight="96" class="origin_image zh-lightbox-thumb" width="645" data-original="https://pic4.zhimg.com/v2-360141804ed3728b742a558f841eb3af_r.jpg"><br><br><h2><b>不如催眠,谈恋爱不如催眠</b></h2><br><p>比起谈恋爱,还是和来访者睡觉,啊不,让来访者睡觉更吸引人。</p><br><p>催眠,这种充满神秘和奇幻感的技术,经常被编剧们用来大肆演绎,他们依据着自己的脑洞,把催眠想得和开灯一样简单:</p><br><p>电影《催眠大师》中,可以这样催眠:</p><br><img src="https://pic1.zhimg.com/v2-9aaf38469b00b3bfa26a871ae9e76fc8_b.jpg" data-rawwidth="600" data-rawheight="255" class="origin_image zh-lightbox-thumb" width="600" data-original="https://pic1.zhimg.com/v2-9aaf38469b00b3bfa26a871ae9e76fc8_r.jpg"><br><p>一个响指,立马睡着。 </p><br><p>还可以这样催眠:</p><br><img src="https://pic2.zhimg.com/v2-bc8d88bd9f98dcf6b127fafe9557b2e5_b.jpg" data-rawwidth="640" data-rawheight="402" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic2.zhimg.com/v2-bc8d88bd9f98dcf6b127fafe9557b2e5_r.jpg"><br><p>晃晃怀表,立刻晕倒</p><br><p>更可以这样催眠:</p><br><img src="https://pic3.zhimg.com/v2-2362e852d488a81ddd8cdd75f26484d2_b.jpg" data-rawwidth="640" data-rawheight="1356" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic3.zhimg.com/v2-2362e852d488a81ddd8cdd75f26484d2_r.jpg"><p>数三个数,你连亲妈都不认得。</p><br><p>电影《惊天魔盗团》中,一个词、一个响指,想催眠多少人,就催眠多少人:</p><br><img src="https://pic3.zhimg.com/v2-8eda921526be0b3f9016282dd4d4a59a_b.jpg" data-rawwidth="506" data-rawheight="314" class="origin_image zh-lightbox-thumb" width="506" data-original="https://pic3.zhimg.com/v2-8eda921526be0b3f9016282dd4d4a59a_r.jpg"><br><p>最牛的是,电视剧《爱情公寓》中,咨询师说自己没有带怀表,用悠悠球把子乔给催眠了……</p><br><img src="https://pic4.zhimg.com/v2-0f7ff8b644c334f153e82ef34827357f_b.jpg" data-rawwidth="640" data-rawheight="358" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic4.zhimg.com/v2-0f7ff8b644c334f153e82ef34827357f_r.jpg"><br><p>咋说呢,哄大人睡觉比哄小孩睡觉容易多了……</p><br><img src="https://pic3.zhimg.com/v2-a6d062b3dd544a5842edff878c784326_b.jpg" data-rawwidth="640" data-rawheight="76" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic3.zhimg.com/v2-a6d062b3dd544a5842edff878c784326_r.jpg"><br><p>虽然可以理解,出于戏剧冲突和角色的需要,是要对一个职业的内容进行一定改编,不可能完全照着真实生活的来。</p><br><p>但只要涉及心理咨询或者心理疾病的情节,但凡有一丁点良心的剧组,哪怕跟一个正经学过心理学的本科生问一下,也不会拍成这么夸张。</p><br><p>以上介绍的这些“咨询师”,谁更业余,不分伯仲,真是难以抉择。</p><br><p>也许有人会说,“哎呀只是影视作品嘛,大家就看一个乐,笑一笑完事,那么认真干什么?”</p><br><p>小编作为一个学过心理学的菜鸡,看到影视剧中这些随心所欲的误导,都常常会感到哭笑不得。可以想象那些从业的咨询师们,看到自己的职业被毫无尺度地演绎,会有多无奈和愤怒。</p><br><p>“艺术创作源于生活但高于生活”,但以上这些剧,既不知道源自的是哪里的生活,也没看出比真实生活高到哪里去。</p><br><img src="https://pic3.zhimg.com/v2-a6d062b3dd544a5842edff878c784326_b.jpg" data-rawwidth="640" data-rawheight="76" class="origin_image zh-lightbox-thumb" width="640" data-original="https://pic3.zhimg.com/v2-a6d062b3dd544a5842edff878c784326_r.jpg"><p>欢迎关注公众号:简单心理(janelee1231)</p><p>寻求专业的心理帮助戳:<a href="//link.zhihu.com/?target=https%3A//www.jiandanxinli.com/%3Futm_source%3Dzhihu%26utm_medium%3Djdxl%26utm_campaign%3Droutine%26utm_content%3Djdxl_homepage%26utm_term%3D25524218" class=" wrap external" target="_blank" rel="nofollow noreferrer">简单心理<i class="icon-external"></i></a></p></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n<img src=\"https://pic2.zhimg.com/v2-dcd6b0996abb2a475204c82676ccf6b1_200x112.jpg\" data-rawwidth=\"640\" data-rawheight=\"337\" class=\"origin_image inline-img zh-lightbox-thumb\" data-original=\"https://pic2.zhimg.com/v2-dcd6b0996abb2a475204c82676ccf6b1_r.jpg\">\n国内外的影视剧中,经常出现对于心理学词汇的滥用、心理疾病的误解以及咨询师这个行业的超夸张演绎。 为了总结出这篇文章,我们被集体气吐了血,想对这些编剧们说一句:别闹,好么。 下面,让我们一起来领略一下: 电视剧《亲爱的翻译官》中,杨幂饰演的乔…\n\n<a href=\"/question/25524218/answer/196125448\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 10:49\" target=\"_blank\" href=\"/question/25524218/answer/196125448\">编辑于 10:49</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-2392308\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>10 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"1\">\n<meta name=\"disableCopyAvatar\" content=\"\">\n\n<span class=\"zg-bull\">•</span>\n\n<a href=\"/terms#sec-licence-1\" target=\"_blank\" class=\"meta-item copyright\">\n作者保留权利\n</a>\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"2\">\n<h2><a class=\"question_link\" href=\"/question/59595588/answer/196014771\" target=\"_blank\" data-id=\"16060939\" data-za-element-name=\"Title\">\n网络小说里有哪些令人拍案叫绝的「智障桥段」?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"65710084\"\ndata-atoken=\"196014771\"\ndata-collapsed=\"0\"\ndata-created=\"1499619359\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/59595588/answer/196014771\">\n<meta itemprop=\"answer-id\" content=\"65710084\">\n<meta itemprop=\"answer-url-token\" content=\"196014771\">\n<a class=\"zg-anchor-hidden\" name=\"answer-65710084\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>711</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">711</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$jing-luo-32-10\"\ntarget=\"_blank\" href=\"/people/jing-luo-32-10\"\n>失迷沦彻</a></span><span title=\"沉迷二次元无法自拔的现实主义悲观人士\" class=\"bio\">\n沉迷二次元无法自拔的现实主义悲观人士\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"711\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">711</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"16060939\" data-action=\"/answer/content\" data-author-name=\"失迷沦彻\" data-entry-url=\"/question/59595588/answer/196014771\">\n\n<textarea hidden class=\"content\">男主的大屌长达【好几米】是的你没看错<br>和几个女人啪啪啪都把人家【从头插到尾】给【怼死了】。。。<br>男主感到万分悲伤因为没有女人能活着伺候完他那【一个月不倒】而且【长达几米直径五十厘米】的丁丁<br>于是归隐山林不闻世事<br>然后有次上山挖槽搞到一种蘑菇<br>和飞机杯差不多功效<br>男主超级开心用蘑菇自己动手解决生理问题<br>因为肾实在是太好撸了一个月才撸完。。。。神奇的是蘑菇没有烂。。。<br>于是男主超高兴啊心想撸蘑菇不好吗为什么要去找女朋友于是就【和蘑菇 拜天地结婚了。。。。】<br>最神奇的是后来<br>蘑菇因为吸收男主jingye和日月精华【成精了】。。。<br>于是男主角就有了一个会说话会做饭贤妻良母只重要的是【胸大屁股翘颜好腰细可以自动调节内部通道长度还不会被艹死】的女朋友。。。<br>快乐生活在一起之后生了个蘑菇宝。。成人之后也是女孩子。。于是母女两个就一起和男主玩起了3p。。。<br>再后来某个月黑风高的夜晚男主觉得这一想操人就操一个月的这个大jj太费时间了于是就切了。。。。切了。。。<br>切完之后修剪成了正常的尺寸。。结果不会操死人但不能满足蘑菇母女了。。。<br><br>故事的结局男主嫌弃蘑菇母女太能要就。。就。。就把母女宰了。。<br>母亲炖了汤女儿炒了菜。。。。</textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n\n男主的大屌长达【好几米】是的你没看错 和几个女人啪啪啪都把人家【从头插到尾】给【怼死了】。。。 男主感到万分悲伤因为没有女人能活着伺候完他那【一个月不倒】而且【长达几米直径五十厘米】的丁丁 于是归隐山林不闻世事 然后有次上山挖槽搞到一种蘑菇 …\n\n<a href=\"/question/59595588/answer/196014771\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" target=\"_blank\" href=\"/question/59595588/answer/196014771\">发布于 00:55</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-16060939\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>193 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"1\">\n<meta name=\"disableCopyAvatar\" content=\"\">\n\n<span class=\"zg-bull\">•</span>\n\n<a href=\"/terms#sec-licence-1\" target=\"_blank\" class=\"meta-item copyright\">\n作者保留权利\n</a>\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"3\">\n<h2><a class=\"question_link\" href=\"/question/62189690/answer/196054625\" target=\"_blank\" data-id=\"17100675\" data-za-element-name=\"Title\">\n小时候被姑姑和姨家的哥哥强奸,默默过了十几年,已经二十了,该不该告诉父母?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"65717257\"\ndata-atoken=\"196054625\"\ndata-collapsed=\"0\"\ndata-created=\"1499641057\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/62189690/answer/196054625\">\n<meta itemprop=\"answer-id\" content=\"65717257\">\n<meta itemprop=\"answer-url-token\" content=\"196054625\">\n<a class=\"zg-anchor-hidden\" name=\"answer-65717257\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>78</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">78</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$sparks\"\ntarget=\"_blank\" href=\"/people/sparks\"\n>花火</a></span><span title=\"休息一阵\" class=\"bio\">\n休息一阵\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"78\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">78</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"17100675\" data-action=\"/answer/content\" data-author-name=\"花火\" data-entry-url=\"/question/62189690/answer/196054625\">\n\n<textarea hidden class=\"content\"><p>从注册知乎以来一直在回应性侵类的求助,有过一些跟题主经历和年龄相仿的,童年遭到过猥亵、诱奸和强奸的求助者曾找我询问,该不该把这段经历告知他们的父母。在这儿做个泛答,希望能帮到题主和像题主这样的幸存者。</p><br><p><b>首先要强调的是,对于年满18岁、具有完全民事行为能力的成年人,该不该把这类创伤经历告知自己的亲属,这个主意只能靠当事人自己来拿。</b></p><p><b>没有人可以替代当事人来做这个决定,即使出面提供心理专业帮助的心理咨询师和社工都不可以这么做,决定权只能在当事人手里。</b>想提供当事人帮助,需要做到如「<a href="https://www.zhihu.com/question/21695988/answer/19107738" class="internal">如何劝导被强奸的女性?</a>」回答中提到的那样,<b>允许当事人替自己做决定。</b>同样,报警和发起法律诉讼也需经过当事人的同意,不可以由亲友和知情者来左右。<b>出于好意的替当事人拿主意,也会带来伤害。任何选择都有其利弊。当事人获得选择的自主权的同时,也需承担做出选择所带来的责任和风险。</b></p><br><p>童年遭到过猥亵、诱奸和强奸的当事人,事隔多年的经历让自己念念不忘,感到在心头压了很久,性情阴郁、暴饮暴食、回避与性侵害者性别相同的人群交往、不想或不敢与人婚恋等明显影响身心健康、人际交往和人生发展方向的情况,就有受到<b>早期心理创伤</b>影响的嫌疑。相关的问答见「<a href="https://www.zhihu.com/question/20553235" class="internal">儿童受到虐待会对以后的生活造成哪些影响?应当如何疏导?</a>」。</p><p>题主及其他有类似经历的求助者,<b>选择在网络公开求助,就要冒隐私泄露、被恶意攻击和得不到专业帮助的风险</b>,毕竟给回应的网友是什么人都有,也不全都具备性侵害救助相关的经验、受训经历和专业背景。<b>跟人的身体患病需要去正规的医疗机构看病,医生确诊按职业操守只能在工作场合进行一样,有困扰多年、疑似达到患病程度的心理问题也是有专门的求助渠道。</b>为了确保当事人的隐私保密性、操作的不受干扰和安全性等因素的考虑,此类求助和施助行为通常在非公开的场合进行。很重要的一点是,从性侵害的阴影中走出来通常有一段路要走,无论想获得专业或非专业的帮助,需要从固定的个人或机构处获得。</p><br><p>对于有早期心理创伤,未经任何治疗的当事人,无论是网络公开求助,告知亲友,还是接受精神医疗、心理治疗的专业帮助,<b>回忆和叙述早期创伤经历,「想起来」给自身所带来的负面影响可能不小,而且会持续一长段时间。</b></p><p>例如,「<a href="//link.zhihu.com/?target=https%3A//womany.net/read/article/10659" class=" wrap external" target="_blank" rel="nofollow noreferrer">妻子寫給被性侵「三歲老公」:提起隱藏的痛苦,才能放下悲傷 <i class="icon-external"></i></a>」中提到的性侵害幸存者恢复记忆后出现的情况:</p><blockquote>剛回復記憶的當下,三郎說不出一個字,整個身體顫抖著,哭了很久很久。後來,他用很小很小的聲音告訴我,他小時候是在奶媽家長大,不是在家裏成長。自那天起,他慢慢跟我述説在奶媽家的寂寞、奶媽一家人的暴力對待、哥哥們的排擠與感情疏離、爸爸媽媽回憶的匱乏……我靜靜聼著,重新理解他的童年。\n\n<b>三郎剛回復記憶的那段時間,他整個人變得很不一樣。他變得非常怕生,不敢獨自一人,也害怕出門。</b>那時,我像在照顧一個三歲的小孩一樣,要隨時把他帶在身邊,或要在他視線範圍内,他才安心。他會不停嚷著要吃冰淇淋,每次他想吃冰淇淋時,還會小心奕奕的問我可不可吃冰淇淋。他會在網路上找回小時候擁有的玩具,透過銀幕觀賞良久。他找回陪伴他成長的卡通,並希望我能一起觀看。他喜歡用被子包著自己,露出一雙眼睛,靜靜坐在家裏的角落。\n\n<b>因爲睡眠的節奏大亂,他吃的餐數變少,很難看到陽光,心情變得更不穩定,而且也越來越瘦。我嘗試了不少改善睡眠的方法,不過沒太大幫助。</b>後來我直接問他需要什麽協助。三郎雖然不是每次都能直接說出自己的需求,但他開始思考和表達自己的需要。他後來希望我能哄他入睡,所以我參照我哄姐姐小孩入睡的經驗,幫他建立一些睡前小儀式,然後為他說床邊故事。我也為他朗讀兒童性侵復原的書籍。雖然有時會朗讀到天亮,但當他開始累積安心入睡的身體感覺,他的睡眠素質就得到明顯改善。\n\n<b>努力了一年半,我知道三郎仍有困難,心靈的傷口依然沒完全癒合。</b>不過三郎現在會笑,會哭,可說出感受,重奪人生掌控權。復原期間的努力與投入,並沒有白費。他每天都比過往好一點點。</blockquote><p>作为当事人,建议优先考虑<b>找熟悉应激障碍、心理创伤治疗的正规医疗机构的精神科、临床心理科的医生,来判断自己的心理困扰是否达到患病的程度,接下去了解通过什么方式和途径可以获得专业帮助,再做出告知家属之类的举动会稳妥些。</b>比如,确诊患病后,接受药物治疗的同时接受心理治疗。接受个人的心理创伤治疗(eg. EMDR),同时与家属一起接受家庭治疗。次选是可以考虑通过12355等提供青少年心理辅导的热线获取帮助,但是热线通常只能提供单次的、较为简单的单次心理疏导,跟打补丁似的,无法替代治疗心理创伤所需的系统创伤治疗。有些创伤治疗的操作必须面对面地进行。</p><p><b>告知家属实情,来试图得到他们的理解、接纳和帮助,可以在心理治疗师、朋友或其他人的协助下进行。</b>告知前,类似 <a class="member_mention" href="//www.zhihu.com/people/null" data-hash="null" data-hovercard="p$b$null">@朵拉陈</a> 在「<a href="https://www.zhihu.com/question/35226712/answer/63654162" class="internal">成年后,孩子是否还有必要向父母表达对他们的「愤怒」? </a>」中描述的步骤,<b>需考虑相关的问题,必须要做的准备和明白的事情。</b>来避免告知父母后出现不理想的状况,可以得到及时的帮助和支持,避免造成严重的二次伤害,加重原本性侵害经历所带来的负面影响。</p><p>相关性侵害的医疗及公益方面的资源,我在「<a href="https://zhuanlan.zhihu.com/p/19838990" class="internal">医疗及公益方面的资源 - 知乎专栏</a>」中有提及。有疑问或求助上遇到困难,可给我发私信。</p><p><b>需要提醒的是,年轻女性幸存者无论通过哪个渠道求助,都有可能遭到别有用心的男性利用及侵害。</b>目前的心理咨询行业是乱象丛生,虚假宣传满天飞。例如「<a href="//link.zhihu.com/?target=http%3A//news.ifeng.com/society/1/detail_2014_04/02/35404173_0.shtml" class=" wrap external" target="_blank" rel="nofollow noreferrer">心理咨询师强奸女患者被抓 网络账号曾发帖称在外学习<i class="icon-external"></i></a>」中提到的这位,以性侵害作为治疗手段,被判刑了仍旧在网络上扯谎给自己辩解和继续做虚假宣传。为了避免被坑,从正规渠道寻找专业帮助之外,接受心理咨询的专业帮助时,可参考「<a href="https://zhuanlan.zhihu.com/p/27323778" class="internal">怀疑自己被心理咨询师剥削或虐待?这里有一篇心理咨询中的不恰当表现”速查表 - 知乎专栏</a>」来获悉自己是否遭到利用。部分条款也可以用来做其他求助与施助关系上出现利用情况的参照。</p><p>----</p><p><b>对于性侵害话题的公开讨论,每次都能见到些误解和迷思。</b></p><p>1、插入式的强奸行为,目前已知年龄最小的幸存者是仅六个月大的女婴。新闻见「<a href="//link.zhihu.com/?target=http%3A//news.sina.com.cn/c/2015-07-01/121532051977.shtml" class=" wrap external" target="_blank" rel="nofollow noreferrer">福建堂叔强奸女婴案宣判 嫌疑人获刑5年<i class="icon-external"></i></a>」。</p><p>2、如「<a href="//link.zhihu.com/?target=https%3A//womany.net/read/article/10659" class=" wrap external" target="_blank" rel="nofollow noreferrer">妻子寫給被性侵「三歲老公」:提起隱藏的痛苦,才能放下悲傷 <i class="icon-external"></i></a>」这篇文章的标题,<b>提起隐藏的痛苦,才能放下悲伤。</b></p><p>3、接触过为数不少,遭受过人为伤害的幸存者之后,我个人非常赞同陈三郎在「<a href="//link.zhihu.com/?target=https%3A//www.twreporter.org/a/sexual-assault-victims-01" class=" wrap external" target="_blank" rel="nofollow noreferrer">【不再讓你孤單】上篇:否定他人的痛苦,與殺人無異<i class="icon-external"></i></a>」这篇报道中所说的这段话:</p><blockquote>你不能否定一個人的痛苦。我覺得很少人可以理解到一件事情就是,如果你的整個人生都是痛苦的時候,人家否定你的痛苦,基本上是否定你所有的人生。這種事情是不能做的,這跟殺人我覺得沒什麼兩樣。</blockquote></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n\n从注册知乎以来一直在回应性侵类的求助,有过一些跟题主经历和年龄相仿的,童年遭到过猥亵、诱奸和强奸的求助者曾找我询问,该不该把这段经历告知他们的父母。在这儿做个泛答,希望能帮到题主和像题主这样的幸存者。 <b>首先要强调的是,对于年满18岁、具有完…</b>\n\n<a href=\"/question/62189690/answer/196054625\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 06:57\" target=\"_blank\" href=\"/question/62189690/answer/196054625\">编辑于 10:00</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-17100675\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>4 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"1\">\n<meta name=\"disableCopyAvatar\" content=\"\">\n\n<span class=\"zg-bull\">•</span>\n\n<a href=\"/terms#sec-licence-1\" target=\"_blank\" class=\"meta-item copyright\">\n作者保留权利\n</a>\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"4\">\n<h2><a class=\"question_link\" href=\"/question/62112909/answer/195752420\" target=\"_blank\" data-id=\"17070017\" data-za-element-name=\"Title\">\n有哪些看似不简单却人人都会的技能?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"65662475\"\ndata-atoken=\"195752420\"\ndata-collapsed=\"0\"\ndata-created=\"1499580049\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/62112909/answer/195752420\">\n<meta itemprop=\"answer-id\" content=\"65662475\">\n<meta itemprop=\"answer-url-token\" content=\"195752420\">\n<a class=\"zg-anchor-hidden\" name=\"answer-65662475\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>11K</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">11K</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$reenee\"\ntarget=\"_blank\" href=\"/people/reenee\"\n>Reenee</a></span><span title=\"Das Licht der Welt erblicken…\" class=\"bio\">\nDas Licht der Welt erblicken…\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"11044\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">11044</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"17070017\" data-action=\"/answer/content\" data-author-name=\"Reenee\" data-entry-url=\"/question/62112909/answer/195752420\">\n\n<textarea hidden class=\"content\">知乎小透明蟹蟹大家的赞(ฅωฅ*),强调下我是路人,并不是葛宇路本人,当时在毕业展上就觉得666好腻害呀脑洞真大…看到这个问题还挺贴切的就回答了。中间文字来源于链接视频,图片来源该视频及网络,侵删。<br><br><br>……………………………………………………………………………分割线下为原回答…………………………………………………………………………<br><p>拥有一条以自己名字命名的道路。</p><p>算得上666吧,如果还是在北京,还是免费呢?</p><br><p>下面说方法:</p><p>1.你要找到一条目前没有名字的道路</p><p>2.制作路牌,贴于该路段相关位置。</p><p>3.坐等网络地图收录。</p><p>4.加入“民政区划地名公共服务系统”正式转正。Over.</p><br><br><p>哈哈哈是不是简单的有点过分,别不相信,下面按时间轴来个案例分析。手动 <a class="member_mention" href="//www.zhihu.com/people/e20c150a8f86a5e16f9fd020eb12273a" data-hash="e20c150a8f86a5e16f9fd020eb12273a" data-hovercard="p$b$e20c150a8f86a5e16f9fd020eb12273a">@葛宇路</a> (虽然没有太多介绍,看头像应该是真身吧(๑・ิ◡・ิ๑)</p><p>2013年起</p><p>我开始寻找地图上空白的路段,并贴上我名字制成的路牌。</p><img src="https://pic3.zhimg.com/v2-8f4d8bf90863a5cfac3c231b974634ca_b.png" data-rawwidth="2560" data-rawheight="1600" class="origin_image zh-lightbox-thumb" width="2560" data-original="https://pic3.zhimg.com/v2-8f4d8bf90863a5cfac3c231b974634ca_r.png"><p>2014年</p><p>高德网络地图首先出现葛宇路,位于朝阳区双井附近(高德地图为中华人民共和国民政部,区划地名公共服务系统指定官方地图)。</p><img src="https://pic1.zhimg.com/v2-db7e68e66ae92aed24beffa4114f2dac_b.png" data-rawwidth="2560" data-rawheight="1600" class="origin_image zh-lightbox-thumb" width="2560" data-original="https://pic1.zhimg.com/v2-db7e68e66ae92aed24beffa4114f2dac_r.png"><br><p>2014年起</p><p>我开始去制作符合现场环境的正规路牌。</p><img src="https://pic2.zhimg.com/v2-bc66130799da088c0c09c34f5946f481_b.png" data-rawwidth="2560" data-rawheight="1600" class="origin_image zh-lightbox-thumb" width="2560" data-original="https://pic2.zhimg.com/v2-bc66130799da088c0c09c34f5946f481_r.png"><br><p>2015年</p><p>百度地图出现葛宇路</p><img src="https://pic3.zhimg.com/v2-9dbf3ab065f4ab1832b09532a4d9f7ea_b.png" data-rawwidth="2560" data-rawheight="1600" class="origin_image zh-lightbox-thumb" width="2560" data-original="https://pic3.zhimg.com/v2-9dbf3ab065f4ab1832b09532a4d9f7ea_r.png"><br><p>2015年底</p><p>路政工程对葛宇路上的路灯进行了统一编号</p><img src="https://pic4.zhimg.com/v2-57ebdcea9c5b52ccad167571be36af6b_b.png" data-rawwidth="2560" data-rawheight="1600" class="origin_image zh-lightbox-thumb" width="2560" data-original="https://pic4.zhimg.com/v2-57ebdcea9c5b52ccad167571be36af6b_r.png"><br><p>2016年</p><p>百度地图全景预览已经可以浏览该路段全貌,同年谷歌地图上出现葛宇路</p><img src="https://pic2.zhimg.com/v2-093067f1174480e2c1ed3013da1822f9_b.png" data-rawwidth="2560" data-rawheight="1600" class="origin_image zh-lightbox-thumb" width="2560" data-original="https://pic2.zhimg.com/v2-093067f1174480e2c1ed3013da1822f9_r.png"><br><img src="https://pic3.zhimg.com/v2-8354c78eac77a5865fcbf1b21248919a_b.jpg" data-rawwidth="1280" data-rawheight="720" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic3.zhimg.com/v2-8354c78eac77a5865fcbf1b21248919a_r.jpg"><br><p>截止目前</p><p>所有快递、外卖、导航、市政标示均可正常使用葛宇路进行定位</p><img src="https://pic1.zhimg.com/v2-e37c4cf89fa3b6cc8f19157e729e2c90_b.jpg" data-rawwidth="1280" data-rawheight="720" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic1.zhimg.com/v2-e37c4cf89fa3b6cc8f19157e729e2c90_r.jpg"><br><img src="https://pic1.zhimg.com/v2-023c415fb31d72a846c350b6b26b9a10_b.jpg" data-rawwidth="1280" data-rawheight="720" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic1.zhimg.com/v2-023c415fb31d72a846c350b6b26b9a10_r.jpg"><p>视频在这里</p><br><a class="video-box" href="//link.zhihu.com/?target=https%3A//v.qq.com/x/cover/f05067ku5vg/f05067ku5vg.html" target="_blank" data-video-id="867381541804847104" data-video-playable="true" data-name="葛宇路简介视频_腾讯视频" data-poster="https://pic1.zhimg.com/v2-a7149ab6fd992e1da01bb8af8ab69140.jpg" data-lens-id=""> <img class="thumbnail" src="https://pic1.zhimg.com/v2-a7149ab6fd992e1da01bb8af8ab69140.jpg"><span class="content"> <span class="title">葛宇路简介视频_腾讯视频<span class="z-ico-extern-gray"></span><span class="z-ico-extern-blue"></span></span> <span class="url"><span class="z-ico-video"></span>https://v.qq.com/x/cover/f05067ku5vg/f05067ku5vg.html</span> </span> </a><br><br><br><p>今年在央美研究生毕业展上看到这个葛宇路作品,感觉非常有趣。</p><img src="https://pic1.zhimg.com/v2-b7316a917c50b035970cf4f9029700d0_b.jpg" data-rawwidth="1280" data-rawheight="853" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic1.zhimg.com/v2-b7316a917c50b035970cf4f9029700d0_r.jpg"></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n<img src=\"https://pic3.zhimg.com/v2-8f4d8bf90863a5cfac3c231b974634ca_200x112.png\" data-rawwidth=\"2560\" data-rawheight=\"1600\" class=\"origin_image inline-img zh-lightbox-thumb\" data-original=\"https://pic3.zhimg.com/v2-8f4d8bf90863a5cfac3c231b974634ca_r.png\">\n知乎小透明蟹蟹大家的赞(ฅωฅ*),强调下我是路人,并不是葛宇路本人,当时在毕业展上就觉得666好腻害呀脑洞真大…看到这个问题还挺贴切的就回答了。中间文字来源于链接视频,图片来源该视频及网络,侵删。 ………………………………………………………\n\n<a href=\"/question/62112909/answer/195752420\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 昨天 14:00\" target=\"_blank\" href=\"/question/62112909/answer/195752420\">编辑于 08:33</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-17070017\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>632 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"1\">\n<meta name=\"disableCopyAvatar\" content=\"\">\n\n<span class=\"zg-bull\">•</span>\n\n<a href=\"/terms#sec-licence-1\" target=\"_blank\" class=\"meta-item copyright\">\n作者保留权利\n</a>\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"5\">\n<h2><a class=\"question_link\" href=\"/question/27427250/answer/195889163\" target=\"_blank\" data-id=\"3153750\" data-za-element-name=\"Title\">\n一个人旅行是什么体验?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"65687298\"\ndata-atoken=\"195889163\"\ndata-collapsed=\"0\"\ndata-created=\"1499602508\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/27427250/answer/195889163\">\n<meta itemprop=\"answer-id\" content=\"65687298\">\n<meta itemprop=\"answer-url-token\" content=\"195889163\">\n<a class=\"zg-anchor-hidden\" name=\"answer-65687298\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>146</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">146</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$yu-qing-74-5\"\ntarget=\"_blank\" href=\"/people/yu-qing-74-5\"\n>雨晴</a></span><span title=\"I'm Crystal. Designer, Traveller, Taster\" class=\"bio\">\nI'm Crystal. Designer, Traveller, Taster\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"146\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">146</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"3153750\" data-action=\"/answer/content\" data-author-name=\"雨晴\" data-entry-url=\"/question/27427250/answer/195889163\">\n\n<textarea hidden class=\"content\"><p>我很喜欢这段话:</p><p><b>你写PPT时,阿拉斯加的鳕鱼正跃出水面;你看报表时,梅里雪山的金丝猴刚好爬上树尖。 你挤进地铁时,西藏的山鹰一直盘旋云端;你在会议中吵架时,尼泊尔的背包客一起端起酒杯坐在火堆旁。 有一些穿高跟鞋走不到的路,有一些喷着香水闻不到的空气,有一些在写字楼里永远遇不见的人。看过阿拉斯加的鳕鱼跃出水面,在办公室埋头写PPT的时候好像也有海水打在脸上;感受过横断山脉的雾凇环绕,与客户绞尽脑汁的谈判也不过是山间刮过的一阵寒风;在尼泊尔的费瓦湖上荡起过双桨,会议室里闷坐再久也有一缕开阔萦绕心头。</b></p><p>--------------------</p><p>2017.6.2的那一天,开始了我为期一个月的孤独旅行。</p><p>以下所有照片均使用…我那已经摔烂的iPhone6s拍摄…</p><br><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-6de951b64c71c9d4a881ce2bfd45370a_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic3.zhimg.com/v2-6de951b64c71c9d4a881ce2bfd45370a_r.jpg"><br><p>我在美国的东部,呆了整整一个月。我去了纽约,新泽西,华盛顿,波士顿和费城。</p><p>拿着一个随身小包,一部手机,以及一个24寸的行李箱,我便出发了。</p><br><img data-rawheight="1052" src="https://pic2.zhimg.com/v2-8fa58c0cceb11455d94f0fdf477f5795_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic2.zhimg.com/v2-8fa58c0cceb11455d94f0fdf477f5795_r.png"><p><b>NEW YORK 纽约</b></p><p>梁启超当年到纽约后,曾感叹<b>“天下最繁盛者宜莫如纽约,天下最黑暗者殆亦莫如纽约”.</b></p><p><b>我只想说:深有同感 。</b></p><p>我这一个月里,在纽约经历了一次抢劫,一次打劫,以及纽约两次大游行。</p><p>并且与华盛顿的枪战擦肩而过。</p><p>所以说,只要你没有出国门,你真的不能道听途说别人口中的那句“美国是天堂”…</p><p>美国空气确实不错,但是!哪里甜了啊???</p><img data-rawheight="900" src="https://pic2.zhimg.com/v2-582a986d0af6697a21990a4da2d3e93d_b.png" data-rawwidth="1294" class="origin_image zh-lightbox-thumb" width="1294" data-original="https://pic2.zhimg.com/v2-582a986d0af6697a21990a4da2d3e93d_r.png"><br><p>不想说些负能量的话,本姑娘可是个正能量满满的耿直Girl啊~</p><p>现!在!开!始!上!图!</p><br><img data-rawheight="1208" src="https://pic3.zhimg.com/v2-dc183220ae9311b7435ae256a4c893d2_b.png" data-rawwidth="868" class="origin_image zh-lightbox-thumb" width="868" data-original="https://pic3.zhimg.com/v2-dc183220ae9311b7435ae256a4c893d2_r.png"><br><p>我是租住在与曼哈顿隔河相望的Newport,公寓叫Marbella。</p><p>和其他留学生住在一个三室一厅里,我租住的时候房东给我打了个折,一晚只要45美金。</p><p>整个房子两室两厅,$3000-$3500不等。</p><p>↓ 这是我在客厅拍摄到的外景</p><img data-rawheight="4724" src="https://pic3.zhimg.com/v2-98bc89ff05abcba66848ed07ea14c58a_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic3.zhimg.com/v2-98bc89ff05abcba66848ed07ea14c58a_r.jpg"><br><p>看着对面的曼哈顿时,我都不敢相信,一晚竟然只要45美金…</p><p>三百出头,在上海也就住个汉庭这类的吧?所以我每天睡觉时,都在偷笑… …</p><br><img data-rawheight="2362" src="https://pic1.zhimg.com/v2-ccc8f471459e4638d14974d96b63e64c_b.jpg" data-rawwidth="2362" class="origin_image zh-lightbox-thumb" width="2362" data-original="https://pic1.zhimg.com/v2-ccc8f471459e4638d14974d96b63e64c_r.jpg"><br><p>我每天都会去纽约,一般都是在33rd Street下车。</p><p>去纽约乘坐Path非常方便,开车的话要过隧道,15美金一次,性价比并不高~</p><p>所以,想要走遍纽约,买个MetroCard不要太方便喔~(单程统一价 $2.75)</p><br><img data-rawheight="1266" src="https://pic4.zhimg.com/v2-08050ad84f3153569eca9dfe5b2ba83f_b.png" data-rawwidth="1058" class="origin_image zh-lightbox-thumb" width="1058" data-original="https://pic4.zhimg.com/v2-08050ad84f3153569eca9dfe5b2ba83f_r.png"><p>到纽约之前,我建议大家买个CityPass,尤其适合大多数在纽约短期停留的游客~</p><p>里面包括<b>美国自然历史博物馆、大都会艺术博物馆、9/11纪念馆、帝国大厦、峭石之巅观景台(洛克菲勒大厦的顶层)、古根海姆博物馆、自由女神像等等。有效期9天。</b></p><p>我在某宝只花了450人民币,<b>性价比极高!(</b>当场购买不划算~)</p><img data-rawheight="771" src="https://pic4.zhimg.com/v2-595697c7eab8f7ab7a0ac30ce48dd493_b.png" data-rawwidth="600" class="origin_image zh-lightbox-thumb" width="600" data-original="https://pic4.zhimg.com/v2-595697c7eab8f7ab7a0ac30ce48dd493_r.png"><br><p>第一天,我去了世贸中心遗址,这个Path站点就叫“World Trade Center”。</p><p>刚出站的时候,我真的被震撼了一下…</p><p>(不过,在纽约…唯独这一站气势恢宏……)</p><br><img data-rawheight="4724" src="https://pic3.zhimg.com/v2-de401e8df3366559507c93e75f9464ba_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic3.zhimg.com/v2-de401e8df3366559507c93e75f9464ba_r.jpg"><p>世贸遗址</p><img data-rawheight="5315" src="https://pic1.zhimg.com/v2-445a93aadea086b3f2ed8b2781fa7e74_b.jpg" data-rawwidth="4134" class="origin_image zh-lightbox-thumb" width="4134" data-original="https://pic1.zhimg.com/v2-445a93aadea086b3f2ed8b2781fa7e74_r.jpg"><p>这里失去了3000多个生命…我站在这里,感到挺揪心的…</p><p>(有某个夕阳红旅行团集体比剪刀手,大笑拍照…我都无语了…<b>请注意素质</b>⚠️)</p><img data-rawheight="4724" src="https://pic3.zhimg.com/v2-1db5c9d23dc6bcd3da7becf5f395521e_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic3.zhimg.com/v2-1db5c9d23dc6bcd3da7becf5f395521e_r.jpg"><p>喜欢蓝天</p><img data-rawheight="4961" src="https://pic4.zhimg.com/v2-79bba37f1ec013818cd8f7ea423a378b_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic4.zhimg.com/v2-79bba37f1ec013818cd8f7ea423a378b_r.jpg"><br><p>因为本人很喜欢艺术,所以第二天就急不可耐地去了MET(我总共去了两趟)。</p><p>当天上午,恰好碰上游行~</p><p>(美国的游行形式多样……只有你想不到,没有他们做不到的~)</p><br><img data-rawheight="3543" src="https://pic1.zhimg.com/v2-d3f3deea4a2f7716ea6388d37f6e82c8_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic1.zhimg.com/v2-d3f3deea4a2f7716ea6388d37f6e82c8_r.jpg"><p>MET</p><p>如果你去MET可以不必买票,可以走Donation通道,非常划算!</p><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-991bf21cc4b2e0d239f7b67f08e05756_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic3.zhimg.com/v2-991bf21cc4b2e0d239f7b67f08e05756_r.jpg"><p>MET有太多让我惊叹的地方。</p><p>因为手机里的图片太多了,我就挑一些我喜欢的展品吧~</p><p>我这人喜欢直接点…</p><p>不废话,上图!</p><img data-rawheight="1012" src="https://pic1.zhimg.com/v2-82e3a061df54628d6e66390ae06848d8_b.png" data-rawwidth="1302" class="origin_image zh-lightbox-thumb" width="1302" data-original="https://pic1.zhimg.com/v2-82e3a061df54628d6e66390ae06848d8_r.png"><br><img data-rawheight="1050" src="https://pic1.zhimg.com/v2-93d3f594288a1028f9e6d6a63de6e258_b.png" data-rawwidth="1310" class="origin_image zh-lightbox-thumb" width="1310" data-original="https://pic1.zhimg.com/v2-93d3f594288a1028f9e6d6a63de6e258_r.png"><br><img data-rawheight="1044" src="https://pic1.zhimg.com/v2-e37b22e53e9ccc3f6852bf17884db3b8_b.png" data-rawwidth="1300" class="origin_image zh-lightbox-thumb" width="1300" data-original="https://pic1.zhimg.com/v2-e37b22e53e9ccc3f6852bf17884db3b8_r.png"><br><img data-rawheight="1046" src="https://pic3.zhimg.com/v2-6f85e58785cf67ae385ca289771df662_b.png" data-rawwidth="1310" class="origin_image zh-lightbox-thumb" width="1310" data-original="https://pic3.zhimg.com/v2-6f85e58785cf67ae385ca289771df662_r.png"><br><img data-rawheight="1206" src="https://pic1.zhimg.com/v2-1ab380e5820927c9f12880edeb8a9880_b.png" data-rawwidth="866" class="origin_image zh-lightbox-thumb" width="866" data-original="https://pic1.zhimg.com/v2-1ab380e5820927c9f12880edeb8a9880_r.png"><br><img data-rawheight="1042" src="https://pic1.zhimg.com/v2-81d9459eb136bff022958069fe9b5a7c_b.png" data-rawwidth="1310" class="origin_image zh-lightbox-thumb" width="1310" data-original="https://pic1.zhimg.com/v2-81d9459eb136bff022958069fe9b5a7c_r.png"><br><img data-rawheight="1046" src="https://pic2.zhimg.com/v2-f3b5937d94fdc92168c7925f77de1ce5_b.png" data-rawwidth="1312" class="origin_image zh-lightbox-thumb" width="1312" data-original="https://pic2.zhimg.com/v2-f3b5937d94fdc92168c7925f77de1ce5_r.png"><p>作为博物馆爱好者,我还去了MOMA。看到了梵高、毕加索等大师的真迹~哇咔咔</p><img data-rawheight="1176" src="https://pic2.zhimg.com/v2-bab885d1fe04a8255b7f38f981a6cb3d_b.png" data-rawwidth="858" class="origin_image zh-lightbox-thumb" width="858" data-original="https://pic2.zhimg.com/v2-bab885d1fe04a8255b7f38f981a6cb3d_r.png"><br><img data-rawheight="968" src="https://pic4.zhimg.com/v2-571a083c1979f6e9aaf561a72c454367_b.png" data-rawwidth="880" class="origin_image zh-lightbox-thumb" width="880" data-original="https://pic4.zhimg.com/v2-571a083c1979f6e9aaf561a72c454367_r.png"><br><img data-rawheight="972" src="https://pic2.zhimg.com/v2-cc9144dad42bca041012ccc88d4c16e9_b.png" data-rawwidth="772" class="origin_image zh-lightbox-thumb" width="772" data-original="https://pic2.zhimg.com/v2-cc9144dad42bca041012ccc88d4c16e9_r.png"><br><img data-rawheight="1280" src="https://pic3.zhimg.com/v2-ab2cb14eb262e146aafa9471d6fdcd62_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic3.zhimg.com/v2-ab2cb14eb262e146aafa9471d6fdcd62_r.png"><br><img data-rawheight="1280" src="https://pic4.zhimg.com/v2-7592f55ad315d78f5abf30380af8e663_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic4.zhimg.com/v2-7592f55ad315d78f5abf30380af8e663_r.png"><br><img data-rawheight="1280" src="https://pic2.zhimg.com/v2-63922e5dd2297b0ded96a817255153b9_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic2.zhimg.com/v2-63922e5dd2297b0ded96a817255153b9_r.png"><br><img data-rawheight="1132" src="https://pic2.zhimg.com/v2-1a6f5ee05bb1653679f287a0c8651799_b.png" data-rawwidth="886" class="origin_image zh-lightbox-thumb" width="886" data-original="https://pic2.zhimg.com/v2-1a6f5ee05bb1653679f287a0c8651799_r.png"><p>去了自然历史博物馆。</p><img data-rawheight="4724" src="https://pic2.zhimg.com/v2-4d0f0997f83310a354fab266a60b0609_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic2.zhimg.com/v2-4d0f0997f83310a354fab266a60b0609_r.jpg"><p>没有我签名的图片,您可随意复制使用~</p><img data-rawheight="960" src="https://pic3.zhimg.com/v2-4a009fb1a2607ca46809ada248ccf4ea_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic3.zhimg.com/v2-4a009fb1a2607ca46809ada248ccf4ea_r.png"><br><img data-rawheight="960" src="https://pic3.zhimg.com/v2-9bc4491bd9ae7b2dbdda1e1baa77a262_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic3.zhimg.com/v2-9bc4491bd9ae7b2dbdda1e1baa77a262_r.png"><br><img data-rawheight="960" src="https://pic4.zhimg.com/v2-d8419330a6f6ad6f8103c8d969d927f7_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic4.zhimg.com/v2-d8419330a6f6ad6f8103c8d969d927f7_r.png"><br><img data-rawheight="960" src="https://pic3.zhimg.com/v2-21a410e71785a1a36b26789dad050b62_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic3.zhimg.com/v2-21a410e71785a1a36b26789dad050b62_r.png"><p>在波士顿的时候去了MFA(Museum of Fine Arts)</p><p>由哈佛大学波士顿图书馆和马萨诸塞理工学院倡议,才筹建了这座博物馆。</p><p>里面有很多莫奈的真迹喔!</p><img data-rawheight="1038" src="https://pic1.zhimg.com/v2-1ab292a74914ffbc1d8eb9bf7df21bb8_b.png" data-rawwidth="1290" class="origin_image zh-lightbox-thumb" width="1290" data-original="https://pic1.zhimg.com/v2-1ab292a74914ffbc1d8eb9bf7df21bb8_r.png"><br><img data-rawheight="1048" src="https://pic3.zhimg.com/v2-7e6dbcec4464f92d044a21a606ff1d82_b.png" data-rawwidth="1298" class="origin_image zh-lightbox-thumb" width="1298" data-original="https://pic3.zhimg.com/v2-7e6dbcec4464f92d044a21a606ff1d82_r.png"><br><img data-rawheight="1280" src="https://pic4.zhimg.com/v2-16d37acc3c347c17b9b097addab19b87_b.png" data-rawwidth="904" class="origin_image zh-lightbox-thumb" width="904" data-original="https://pic4.zhimg.com/v2-16d37acc3c347c17b9b097addab19b87_r.png"><br><img data-rawheight="1134" src="https://pic2.zhimg.com/v2-d15de85a9b9d6dc798fa345a3d145cd5_b.png" data-rawwidth="1426" class="origin_image zh-lightbox-thumb" width="1426" data-original="https://pic2.zhimg.com/v2-d15de85a9b9d6dc798fa345a3d145cd5_r.png"><br><img data-rawheight="1116" src="https://pic1.zhimg.com/v2-87cbe3bdf1dfa1c549aba2eae9550c98_b.png" data-rawwidth="798" class="origin_image zh-lightbox-thumb" width="798" data-original="https://pic1.zhimg.com/v2-87cbe3bdf1dfa1c549aba2eae9550c98_r.png"><p>盛夏,迷醉在莫奈的睡莲里。</p><img data-rawheight="4961" src="https://pic3.zhimg.com/v2-311755be04f22132dcaec39c99b30276_b.jpg" data-rawwidth="3508" class="origin_image zh-lightbox-thumb" width="3508" data-original="https://pic3.zhimg.com/v2-311755be04f22132dcaec39c99b30276_r.jpg"><br><p><b>Harvard museum of Natural History 哈佛大学自然历史博物馆</b> (Adult $12)</p><p>博物馆的里面更像个小迷宫,你很容易一不小心就错过一个展区。</p><img data-rawheight="960" src="https://pic3.zhimg.com/v2-6f9ecae04ca1c655f2db21cb1cd46bea_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic3.zhimg.com/v2-6f9ecae04ca1c655f2db21cb1cd46bea_r.png"><br><img data-rawheight="960" src="https://pic2.zhimg.com/v2-d4d7c352b1d3b31e3b918e0e17ae6f29_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic2.zhimg.com/v2-d4d7c352b1d3b31e3b918e0e17ae6f29_r.png"><p>不得不提所有学霸梦寐以求的哈佛大学!</p><p>给你看看我的傻傻游客照~</p><img data-rawheight="1280" src="https://pic3.zhimg.com/v2-93049e2fcc790308905178790fb671ee_b.png" data-rawwidth="952" class="origin_image zh-lightbox-thumb" width="952" data-original="https://pic3.zhimg.com/v2-93049e2fcc790308905178790fb671ee_r.png"><p>哈佛大学周围真的很美啊!很像欧洲的小镇 有木有~</p><img data-rawheight="3543" src="https://pic1.zhimg.com/v2-4351fad37bf2e7bdf105dea61a3a9e2c_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic1.zhimg.com/v2-4351fad37bf2e7bdf105dea61a3a9e2c_r.jpg"><p>然后晚上去吃了波士顿大名鼎鼎的西班牙菜Toro</p><p>店面很小,通常需要排队。</p><p>Address:1704 Washington St, <i>Boston</i>, MA</p><img data-rawheight="958" src="https://pic3.zhimg.com/v2-eecc98ce3b41dac2f276e432b3038ada_b.png" data-rawwidth="1346" class="origin_image zh-lightbox-thumb" width="1346" data-original="https://pic3.zhimg.com/v2-eecc98ce3b41dac2f276e432b3038ada_r.png"><p>性价比极高,而且非常美味!(哼!馋死你~)</p><img data-rawheight="954" src="https://pic2.zhimg.com/v2-c71c936a9fd878682cdc3b01a3015e39_b.png" data-rawwidth="1352" class="origin_image zh-lightbox-thumb" width="1352" data-original="https://pic2.zhimg.com/v2-c71c936a9fd878682cdc3b01a3015e39_r.png"><br><p>Oh…我偏题了……</p><p>转到博物馆。</p><p>我去了华盛顿的美国财政部制版印刷局(Bureau of Engraving and Printing)</p><p>这里是了解美元印刷全过程的好地方~</p><p>我亲眼目睹了一大摞的空白纸张变成纸币的全过程(只可惜,不允许拍照)</p><img data-rawheight="1014" src="https://pic4.zhimg.com/v2-cf42be726f5f8b550efe79b7cd874d63_b.png" data-rawwidth="1324" class="origin_image zh-lightbox-thumb" width="1324" data-original="https://pic4.zhimg.com/v2-cf42be726f5f8b550efe79b7cd874d63_r.png"><p>里面也有个很小的展厅,类似博物馆。</p><p>2美金的纸币很稀有罕见哦~~</p><img data-rawheight="934" src="https://pic4.zhimg.com/v2-6e18907a155fdb9bf773949abfe416cb_b.png" data-rawwidth="1238" class="origin_image zh-lightbox-thumb" width="1238" data-original="https://pic4.zhimg.com/v2-6e18907a155fdb9bf773949abfe416cb_r.png"><p>旅行的时候,我最喜欢坐在公园里。</p><p>我可以在街边随便买个Gyros, 坐在中央公园的长椅上,发呆半个小时…</p><p>本人最喜欢Central Park 中央公园也算是纽约繁华中,最难得的一片静谧之地。</p><p>经常会有人在这里骑行,慢跑,也会有很多人在周末的闲暇之时,到这里野餐。</p><img data-rawheight="4961" src="https://pic3.zhimg.com/v2-780c588eab4efa21640f11eac6f4ae32_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic3.zhimg.com/v2-780c588eab4efa21640f11eac6f4ae32_r.jpg"><br><img data-rawheight="5315" src="https://pic4.zhimg.com/v2-736ea1a82c29ece804827931f4faa7a3_b.jpg" data-rawwidth="4134" class="origin_image zh-lightbox-thumb" width="4134" data-original="https://pic4.zhimg.com/v2-736ea1a82c29ece804827931f4faa7a3_r.jpg"><br><img data-rawheight="4724" src="https://pic2.zhimg.com/v2-d0379d7562c510c441414ecec851fc1d_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic2.zhimg.com/v2-d0379d7562c510c441414ecec851fc1d_r.jpg"><p>到处都有复古的马车。</p><img data-rawheight="4724" src="https://pic1.zhimg.com/v2-1201eb65cd25f2ad10dffe7bd542e5bc_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic1.zhimg.com/v2-1201eb65cd25f2ad10dffe7bd542e5bc_r.jpg"><br><p>很喜欢这对老夫妻坐在那里的感觉,所以偷拍了一张。</p><p>我真心希望我老了的那一天,也可以和我老公这样。</p><p>(我老公呢?我老公去哪了?咦呃,我哪有老公啊!!?)</p><br><img data-rawheight="1142" src="https://pic1.zhimg.com/v2-e9fff73b9faaccf9aaea97daecb98810_b.png" data-rawwidth="1430" class="origin_image zh-lightbox-thumb" width="1430" data-original="https://pic1.zhimg.com/v2-e9fff73b9faaccf9aaea97daecb98810_r.png"><p>现在穿越到波士顿</p><p><b>Boston Common Park 波士顿公园</b></p><p><b>波士顿公园是自由之路(Freedom Trail)的起始点。</b></p><br><img data-rawheight="776" src="https://pic4.zhimg.com/v2-cd896240a659cb028e9ce95130af8eff_b.png" data-rawwidth="776" class="origin_image zh-lightbox-thumb" width="776" data-original="https://pic4.zhimg.com/v2-cd896240a659cb028e9ce95130af8eff_r.png"><br><p><b>个人认为,自由之路经过的景点中,这个公园是我认为最美的景点。</b></p><p><b>(我照相不P图,只为还原最本真的样子)</b></p><img data-rawheight="3543" src="https://pic4.zhimg.com/v2-448d8ace725eaecdec348a4abe996bb3_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic4.zhimg.com/v2-448d8ace725eaecdec348a4abe996bb3_r.jpg"><br><img data-rawheight="1066" src="https://pic2.zhimg.com/v2-16d42f0f08598a610c9725ccfa794fe5_b.png" data-rawwidth="822" class="origin_image zh-lightbox-thumb" width="822" data-original="https://pic2.zhimg.com/v2-16d42f0f08598a610c9725ccfa794fe5_r.png"><p>这些鸭子…每天真是悠闲~~肥肥胖胖的~</p><img data-rawheight="972" src="https://pic3.zhimg.com/v2-fce53293d0614c39a69bc0f3a733d3ba_b.png" data-rawwidth="1122" class="origin_image zh-lightbox-thumb" width="1122" data-original="https://pic3.zhimg.com/v2-fce53293d0614c39a69bc0f3a733d3ba_r.png"><p>波士顿的鸭子很多。</p><p>但是你要知道,在波士顿,你还能出!海!看! 鲸!鱼!</p><p>我出海的那天,天气非常晴朗☀️</p><img data-rawheight="3543" src="https://pic4.zhimg.com/v2-2958ce0e389ff5de3946532ae85c8b6b_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic4.zhimg.com/v2-2958ce0e389ff5de3946532ae85c8b6b_r.jpg"><p>鲸鱼跃出水面的那一刻,整艘船都是</p><p>"Oh! My God!!!!!!" </p><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-3f2ff542adc3d5771e92e28ff7dd50d2_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic3.zhimg.com/v2-3f2ff542adc3d5771e92e28ff7dd50d2_r.jpg"><p>"WOW!!!!!!!!!"</p><p>抓拍到太不容易了,快给我点赞!</p><img data-rawheight="3543" src="https://pic1.zhimg.com/v2-d73713f45f5427956282464a2f228ebc_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic1.zhimg.com/v2-d73713f45f5427956282464a2f228ebc_r.jpg"><p>这条鱼…看到我比她漂亮,似乎比较嫉妒我。 表情萌萌哒</p><img data-rawheight="956" src="https://pic2.zhimg.com/v2-a44f4a4481e2815f67e9eebf98c79005_b.png" data-rawwidth="1350" class="origin_image zh-lightbox-thumb" width="1350" data-original="https://pic2.zhimg.com/v2-a44f4a4481e2815f67e9eebf98c79005_r.png"><p>你嫉妒我有什么用呢?有这功夫,应该去图书馆学习!</p><br><p><b>Boston Public Library 波士顿公共图书馆</b></p><img data-rawheight="3543" src="https://pic4.zhimg.com/v2-d316a2b078b23bd4e54df755a38de143_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic4.zhimg.com/v2-d316a2b078b23bd4e54df755a38de143_r.jpg"><br><img data-rawheight="4961" src="https://pic4.zhimg.com/v2-742a92cd5a8be2c4322561bf39321c4b_b.jpg" data-rawwidth="3508" class="origin_image zh-lightbox-thumb" width="3508" data-original="https://pic4.zhimg.com/v2-742a92cd5a8be2c4322561bf39321c4b_r.jpg"><p>~~~~~~~飞啊~~~~~~~ <b>穿越回到纽约。</b></p><p><b>Statue Of Liberty 自由女神</b></p><p>挑了个天气极好的周末,去看了自由女神。</p><p>很喜欢这艘船上的“MISS NEW JERSEY”</p><img data-rawheight="4724" src="https://pic4.zhimg.com/v2-232a0a5d0dfa5cafd8dc3bc0f2ead26f_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic4.zhimg.com/v2-232a0a5d0dfa5cafd8dc3bc0f2ead26f_r.jpg"><p>在去自由女神的路上,拍到的曼哈顿。</p><img data-rawheight="1340" src="https://pic4.zhimg.com/v2-be81995af4341c31431fea6fb24f8f63_b.png" data-rawwidth="1798" class="origin_image zh-lightbox-thumb" width="1798" data-original="https://pic4.zhimg.com/v2-be81995af4341c31431fea6fb24f8f63_r.png"><p>“照耀世界的自由女神”</p><img data-rawheight="4724" src="https://pic1.zhimg.com/v2-ca3fee35a2a1d18dbb6b0762ce8fd52c_b.jpg" data-rawwidth="5906" class="origin_image zh-lightbox-thumb" width="5906" data-original="https://pic1.zhimg.com/v2-ca3fee35a2a1d18dbb6b0762ce8fd52c_r.jpg"><p>我一定要推荐这个柠檬汁!这是我这辈子喝过的最棒的柠檬汁!没有之一!!!</p><p>不仅外表貌美,料还很足!里面有很多柠檬,看到了木有~~</p><img data-rawheight="5315" src="https://pic1.zhimg.com/v2-42a4f633fe60d1737e87e02883a0bc24_b.jpg" data-rawwidth="4134" class="origin_image zh-lightbox-thumb" width="4134" data-original="https://pic1.zhimg.com/v2-42a4f633fe60d1737e87e02883a0bc24_r.jpg"><p><b>TOP OF THE ROCK</b></p><p><b>洛克菲勒中心大厦</b></p><img data-rawheight="4961" src="https://pic3.zhimg.com/v2-6f439a79b0fc11c736d34707cf49dca2_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic3.zhimg.com/v2-6f439a79b0fc11c736d34707cf49dca2_r.jpg"><br><p>遥望着对面的帝国大厦,内心泛起波澜。</p><br><img data-rawheight="990" src="https://pic1.zhimg.com/v2-5683329599f8c3aa8e36d98161bdae88_b.png" data-rawwidth="1038" class="origin_image zh-lightbox-thumb" width="1038" data-original="https://pic1.zhimg.com/v2-5683329599f8c3aa8e36d98161bdae88_r.png"><p>只有黑夜,才能让帝国大厦更加闪耀。</p><p>只有黑夜,才能让你看到更加迷醉的纽约。</p><p>只有经历黑夜,才能让你看到光明。</p><img data-rawheight="1280" src="https://pic4.zhimg.com/v2-79ba79fb168c2e5c72fb8dcc03ebd423_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic4.zhimg.com/v2-79ba79fb168c2e5c72fb8dcc03ebd423_r.png"><br><img data-rawheight="1326" src="https://pic3.zhimg.com/v2-d9f4e36f7db6696fcba006abbd9d84b2_b.png" data-rawwidth="1794" class="origin_image zh-lightbox-thumb" width="1794" data-original="https://pic3.zhimg.com/v2-d9f4e36f7db6696fcba006abbd9d84b2_r.png"><p><b>Empire State Buiding</b></p><p>第二天,我就跑到帝国大厦去了。</p><p>在帝国大厦的天台上,没有心动的邂逅,也没有喜悦的重逢。</p><p>但是!我!可!以!自!拍!</p><p>(为了帝国大厦,还是上个素颜照吧~照片是一位白人老爷爷给我拍的)</p><img data-rawheight="4961" src="https://pic4.zhimg.com/v2-68e8177ed27bccd12d791c7fe2895c9f_b.jpg" data-rawwidth="3508" class="origin_image zh-lightbox-thumb" width="3508" data-original="https://pic4.zhimg.com/v2-68e8177ed27bccd12d791c7fe2895c9f_r.jpg"><p>哎,纽约确实要比上海更繁华(我真的不想承认这一点 )</p><img data-rawheight="3543" src="https://pic2.zhimg.com/v2-8280785c6b5ecd3da5a4dd07315b54b1_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic2.zhimg.com/v2-8280785c6b5ecd3da5a4dd07315b54b1_r.jpg"><p>久别的有情人在帝国大厦的天台上重逢,已经是经典的的电影桥段。</p><img data-rawheight="576" src="https://pic2.zhimg.com/v2-0d4f68f2ecffa2348b9b86d828901fbd_b.png" data-rawwidth="1024" class="origin_image zh-lightbox-thumb" width="1024" data-original="https://pic2.zhimg.com/v2-0d4f68f2ecffa2348b9b86d828901fbd_r.png"><p><b>Times Square</b></p><p>“世界的十字路口”——时代广场</p><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-bc90a539d55e1d5a5b30ef31b8d1cd7e_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic3.zhimg.com/v2-bc90a539d55e1d5a5b30ef31b8d1cd7e_r.jpg"><p>不得不吐槽,纽约的米妮穿得好土…哈哈哈</p><img data-rawheight="804" src="https://pic3.zhimg.com/v2-e39b4824efc144865b0c690a6b00d9b6_b.png" data-rawwidth="750" class="origin_image zh-lightbox-thumb" width="750" data-original="https://pic3.zhimg.com/v2-e39b4824efc144865b0c690a6b00d9b6_r.png"><p>纽约的时代广场和上海的南京路没法比~哦耶~</p><img data-rawheight="4961" src="https://pic3.zhimg.com/v2-ab7eea31d07626e180866a638e069daa_b.jpg" data-rawwidth="3508" class="origin_image zh-lightbox-thumb" width="3508" data-original="https://pic3.zhimg.com/v2-ab7eea31d07626e180866a638e069daa_r.jpg"><p><b>Washington, D.C. 华盛顿</b></p><p><b>带你去美国的心脏逛一逛。</b></p><p><b>United States Capitol 国会大厦</b></p><p>我在国会大厦的那一天,与枪击案擦肩而过。</p><img data-rawheight="956" src="https://pic3.zhimg.com/v2-30a269f87e4ca0cf1fc20d19b1b564ea_b.png" data-rawwidth="1280" class="origin_image zh-lightbox-thumb" width="1280" data-original="https://pic3.zhimg.com/v2-30a269f87e4ca0cf1fc20d19b1b564ea_r.png"><p>原来,自己可以和死神…如此接近。</p><img data-rawheight="3543" src="https://pic1.zhimg.com/v2-d4093ec97f4ea44eb64cf01194543e34_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic1.zhimg.com/v2-d4093ec97f4ea44eb64cf01194543e34_r.jpg"><p><b>所以那天人不多…估计都被吓跑了吧…</b></p><p><b>到处都是FBI…</b></p><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-55d6641896d497bae30ecec0aee8ee16_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic3.zhimg.com/v2-55d6641896d497bae30ecec0aee8ee16_r.jpg"><br><p><b>The White House 白宫</b></p><p>美国权力的象征。</p><br><img data-rawheight="756" src="https://pic4.zhimg.com/v2-be1552624cd58000a4825e55f449bc57_b.png" data-rawwidth="1186" class="origin_image zh-lightbox-thumb" width="1186" data-original="https://pic4.zhimg.com/v2-be1552624cd58000a4825e55f449bc57_r.png"><p>进不了白宫,只能在白宫前的栏杆边照相啦…</p><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-db28299ff7680bc20e78bf55c2cc07e2_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic3.zhimg.com/v2-db28299ff7680bc20e78bf55c2cc07e2_r.jpg"><p><b>Lincoln Memorial 林肯纪念堂</b></p><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-68075e7b8f9343db0443b490609b2dea_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic3.zhimg.com/v2-68075e7b8f9343db0443b490609b2dea_r.jpg"><p><b>Washington Monument 华盛顿纪念碑</b></p><p>位于华盛顿市中心,也是华府最老的纪念碑。在国会大厦、林肯纪念堂的轴线上。</p><p>顾名思义,是为纪念美国首任总统乔治·华盛顿而建造,高169.045米,重约9.1万吨。</p><p>(这是在坐快艇时速拍的,抱歉啦~像素有点渣)</p><img data-rawheight="3543" src="https://pic3.zhimg.com/v2-03d5e744bf45618b128e92a99bc66c4e_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic3.zhimg.com/v2-03d5e744bf45618b128e92a99bc66c4e_r.jpg"><p><b>费城 Philadelphia</b></p><p>美国最老、最具历史意义的城市,曾是国家首都。</p><p><b>Philadelphia Independence Hall 费城独立纪念堂</b></p><p>费城纪念堂被公认为美国建筑艺术最杰出的代表作之一,坐落在费城的Fairmount Park。</p><p><b>100美元上的建筑,就是费城独立纪念堂。</b></p><p>看!!我拍到了“爱心天空”~~</p><img data-rawheight="3543" src="https://pic4.zhimg.com/v2-1115fb90ea3524a726a01bd520799c27_b.jpg" data-rawwidth="3543" class="origin_image zh-lightbox-thumb" width="3543" data-original="https://pic4.zhimg.com/v2-1115fb90ea3524a726a01bd520799c27_r.jpg"><br><p>以上是美国的一些美景照片。</p><p>(艾玛…传图片传的我两眼昏花…)</p><p>估计有些人就会说,哇塞!美国好美啊!</p><p>PANG!我现在泼一盆冷水…让我跟你说些大实话:</p><p>在美国的街上走,你就知道什么叫做——没!有!安!全!感!</p><br><img data-rawheight="3543" src="https://pic4.zhimg.com/v2-2393733c4722dbe192b270724a1ddb5f_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic4.zhimg.com/v2-2393733c4722dbe192b270724a1ddb5f_r.jpg"><br><p><b>一个人旅行,才知道安全的重要性。大家应该多多关注自身的安全问题,无论男女。</b></p><p>刚到美国的时侯,我的朋友圈被“章莹颖绑架事件”刷屏了。爸妈每天叮嘱我一定要注意安全,奶奶每天祈祷我平安无事。所以,旅行不仅仅是自己一个人的勇气,更需要家人有足够的勇气。</p><p>1.我在这次旅行的中途,确实经历了一次抢劫和一次打劫(我到上海以后才敢告诉父母)。</p><p>2.我芝加哥的朋友告诉我,他的同学曾经被拿枪指着头,后来被劫的只剩下小裤衩。</p><p>3.我旅行时认识的一位女性朋友, 跟我坐在哈佛的长椅里,告诉了我她的一次不详遭遇…</p><p><b>听完以后,我的鸡皮疙瘩紧急集合:</b></p><p>在午夜凌晨时分,她抵达机场,当时她不太熟悉使用Uber, 只想找到等Taxi的地方,但是大晚上她根本找不到……于是便东张西望。有一位男士上前友善地告诉她,应该上电梯,3楼下去以后直走右拐,巴拉巴拉讲了一堆。朋友很感谢,便拖着沉重的箱子,准备按照他的指示前往。只是在朋友往电梯方向走的时候,发现这位男子一直跟着她,她上了电梯,这位男子也上了电梯,我朋友便马上警觉起来。等到电梯快要关上的一刹那,有一对母女走了上来,这对母女很复杂地看了她一眼,到了2楼她们便下了电梯,但是下电梯以后又回眸看了她一眼(事后,女性朋友说,这个眼神其实就是告诫她要小心)。等到三楼停止,门打开的一刹那,发现漆黑一片,然后那个男士就告诉他出去以后直走右拐,follow him. 我朋友有几秒的踌躇不定,不知道是否应该跟着他出电梯。但是,那位男子非常友善,她就将信将疑准备跟着他……等到快出去的一刹那,她突然想:哪有Taxi会在这么漆黑一片的地方??于是以迅雷不及掩耳的速度摁了关门键,到了一楼以后,像猎豹一样冲了出去。后来她跟我说:她想想很后怕,如果真的很愚蠢的相信他,跟着他出去了……</p><p>我可能现在就不能够坐在你身边聊天了。</p><p>………………………………………………</p><p>我用自己和身边人的亲身经历,想告诉大家:美国一点也不安全,不要把美国吹捧的太美好,出门在外,请不要抱有任何侥幸心理。在美国,一街相隔真的就是天堂与地狱的差别。不要贪图房租的便宜,把自己的性命都丢了。美国每年又上万的失踪案和杀人案没有曝光,倘若你在危险地带经常出没,却没摊上什么事儿,这真的就是你的运气好。</p><p>在国内外,请大家不要随便相信别人!!</p><p>有些看似友善的人,其实都是披着羊皮的狼。很多人就因为有些人的学历高或者是学识好,就误以为TA品德高尚。放下了顾虑<b>(例如<a href="//link.zhihu.com/?target=http%3A//www.baidu.com/link%3Furl%3Dwmv6U4DoUE8lPUEMrWPpRtKJX6sbEgpDSkaJNA559A69INaM5jj68m_8hxDkAU41q_j-YnCbPya1Xp3V0ExOvK7HxR1xVwatrWTVMrvScNzjy2T1LEG7s_OPDbwPQFno" class=" wrap external" target="_blank" rel="nofollow noreferrer">章莹颖<i class="icon-external"></i></a>事件,嫌疑犯是UIUC的博士生。)</b></p><p>以前,我被父母保护的很好,我很容易轻信别人,甚至从来不会怀疑别人。</p><p>旅行过后,我就更加坚信这一点:不要把人想太好,也不要把人想得太坏。</p><p><b>恶魔可以换上任意一副皮囊,利用你的需求,利用你的同情心,利用你的信任度...</b></p><p><b>把你拖入深渊。</b></p><br><img data-rawheight="3543" src="https://pic4.zhimg.com/v2-c5010ad3de6ffa5ae6a0bb05ccd5c8f7_b.jpg" data-rawwidth="4961" class="origin_image zh-lightbox-thumb" width="4961" data-original="https://pic4.zhimg.com/v2-c5010ad3de6ffa5ae6a0bb05ccd5c8f7_r.jpg"><p>(拍摄于2017.7.1早晨)</p><p><b>1. 只有你亲身经历的世界,才是最真实的世界。</b></p><p><b>2.一个人的旅行,能够让我在旅途上和陌生人的相识与相知,是旅行所能带给我的最大的快乐。</b></p><p><b>3.一个人的旅行,没有约束,没有顾虑,随性而为。</b></p><p>-----------------</p><p><b>我一直坚信,一个人的长途旅行,真的可以改变一个人的人生轨迹。</b></p><p><b>当我走过万里路,才会感受到自己在这个世界上的渺小;</b></p><p><b>才想去感受什么样的自己才是最真实的自己;</b></p><p><b>才能感受到以往的生活有多么弥足珍贵。</b></p><p><b>——雨晴Crystal</b></p><br><p>用一句我很喜欢的话,总结这个问题的回答吧。</p><p>You make millions of decisions that mean nothing and then one day your order takes out and it changes your life.<br>你每天都在做很多看起来毫无意义的决定,但某天你的某个决定就能改变你的一生.</p><p> ----《西雅图夜未眠》</p><br><p><b>小松鼠跟我说了个秘密:</b></p><p><b>“如果你给这个答主点赞,你下次去华盛顿,所有的……榛果……</b></p><p><b>都由我这个松鼠小队长</b></p><p><b>承包了!!!”</b></p><img data-rawheight="936" src="https://pic4.zhimg.com/v2-86bc546f0ebf40b91055e98b2083b267_b.png" data-rawwidth="938" class="origin_image zh-lightbox-thumb" width="938" data-original="https://pic4.zhimg.com/v2-86bc546f0ebf40b91055e98b2083b267_r.png"><p><b>打个广告:</b></p><p>我最大的梦想是做一位 <b>旅行博主。</b></p><p><b>本人擅长拍照修图与平面设计还有策划~</b></p><p><b>如果有这类工作,请速速私信我喔~比心 ❤️</b></p><br><p>Copyright Clarify:<br>Copyright ownership belongs to Yuqing (Crystal).</p><p>Please not be reproduced , copied, or used in other ways without permission.</p><p>版权声明:</p><p>此文归雨晴所有。</p><p>未经允许不得转载、复制或用作它途。本人有权追究法律责任的权利。</p><p><br></p><p>~~</p><p>补充:很多人好奇抢劫的经历</p><p>这经历很不好啊…点赞超过一千我就说。 </p></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n<img data-rawheight=\"3543\" src=\"https://pic3.zhimg.com/v2-6de951b64c71c9d4a881ce2bfd45370a_200x112.jpg\" data-rawwidth=\"3543\" class=\"origin_image inline-img zh-lightbox-thumb\" data-original=\"https://pic3.zhimg.com/v2-6de951b64c71c9d4a881ce2bfd45370a_r.jpg\">\n我很喜欢这段话:<b>你写PPT时,阿拉斯加的鳕鱼正跃出水面;你看报表时,梅里雪山的金丝猴刚好爬上树尖。 你挤进地铁时,西藏的山鹰一直盘旋云端;你在会议中吵架时,尼泊尔的背包客一起端起酒杯坐在火堆旁。 有一些穿高跟鞋走不到的路,有一些喷着香水闻不到…</b>\n\n<a href=\"/question/27427250/answer/195889163\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 昨天 20:15\" target=\"_blank\" href=\"/question/27427250/answer/195889163\">编辑于 09:24</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-3153750\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>39 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"1\">\n<meta name=\"disableCopyAvatar\" content=\"\">\n\n<span class=\"zg-bull\">•</span>\n\n<a href=\"/terms#sec-licence-1\" target=\"_blank\" class=\"meta-item copyright\">\n作者保留权利\n</a>\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div>\n</div>\n</div>\n<div class=\"tab-panel\">\n<div data-type=\"monthly\">\n<div class=\"explore-feed feed-item\" data-offset=\"1\">\n<h2><a class=\"question_link\" href=\"/question/21128507/answer/181695539\" target=\"_blank\" data-id=\"632044\" data-za-element-name=\"Title\">\n为什么富人越来越富,穷人越来越穷?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"63107073\"\ndata-atoken=\"181695539\"\ndata-collapsed=\"0\"\ndata-created=\"1497077097\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/21128507/answer/181695539\">\n<meta itemprop=\"answer-id\" content=\"63107073\">\n<meta itemprop=\"answer-url-token\" content=\"181695539\">\n<a class=\"zg-anchor-hidden\" name=\"answer-63107073\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>331</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">331</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$divinites\"\ntarget=\"_blank\" href=\"/people/divinites\"\n>司马懿</a><span class=\"icon icon-badge-id-an icon-badge\" data-tooltip=\"s$b$优秀回答者 · 已认证的个人\"></span></span><span class=\"badge-summary\"><a href=\"/people/divinites#hilightbadge\" target=\"_blank\">经济学、博弈论话题优秀回答者</a></span>\n\n\n\n<span title=\"《穿越历史聊经济》作者\" class=\"bio\">\n《穿越历史聊经济》作者\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"331\">\n\n\n<span class=\"answer-tag\">\n<span class=\"text\">收录于 </span>\n\n\n<a class=\"text\" data-tooltip=\"s$t$日常经济学 · 我为什么这么穷\" href=\"/roundtable/poor-econ\">知乎圆桌</a>\n\n\n<span class=\"text\">、</span>\n\n\n\n<span class=\"text\">编辑推荐</span>\n\n\n\n</span>\n\n\n<span class=\"zg-bull text\">•</span><span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">331</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"632044\" data-action=\"/answer/content\" data-author-name=\"司马懿\" data-entry-url=\"/question/21128507/answer/181695539\">\n\n<textarea hidden class=\"content\"><p>为什么富人越富,穷人越穷?这个声明里面我默认穷人富人是相对的,也就是说贫富差距在扩大。因为我们的绝对生活水平因为科技进步的缘故还是在提高。 这个问题光看劳动和资本的边际回报是会掩盖一些问题的。</p><p>首先,要承认人和人的天赋之间从经济上本来就是不平等的,生产率也是有差异的。有些天赋(比如判断股票市场涨跌的直觉)就是要比另外一些天赋(比如可以让自己的无名指直立起来)对市场来说更加的有价值。那么即便大家起初的资产一样,有些人通过天赋和运气,就会比另外一些人更加的富有,强行大锅饭只会减少人们创新和努力的动力,结果就是让大家都穷了。所以我们可以假定最初的一批人通过天赋差异,获得了一些资本,然后看资本和劳动的回报。</p><br><p>要明确的一点是,从微观上看,劳动和资本的边际回报率都是递减的,资本这方面并不例外。因为如果劳动力积压,资本不足,相当于100个工人需要轮流的用一个车床,这个时候再增加100个工人所带来的效率提升就远远不如增加一个车床来的快;同样的,如果10个工人有100个车床可以用,那么再增加1个车床,不如再增加100个工人。不同行业,不同企业都会有一个资本和劳动力的最优配比,资本太多了,资本回报率就会下降,劳动力太多了,劳动力的回报率就下降。根据市场工资和市场资产回报率的高低,在最优的配比下,完全有可能出现劳动力的边际回报率大于资本的边际回报率的情形。</p><br><p>但是问题在于,再多的资本,其产出的红利都可以由一个人或者少数几个人所占有,但是单个人所能提供的劳动力是有限的,因为每个人每天只有24个小时的时间。所以即便是在一个市场上,劳动力的边际回报很高,市场工资很高,但是劳动力边际回报的红利会因为员工能提供劳动力的限制依然可能出现贫富差距越来越大的情况。一旦如此,根据 <a class="member_mention" href="//www.zhihu.com/people/1d1c9784aab4f00964215f435e479924" data-hash="1d1c9784aab4f00964215f435e479924" data-hovercard="p$b$1d1c9784aab4f00964215f435e479924">@Manolo</a> 在本题下所提供的模拟,就无法阻止贫富差距进一步扩大。</p><br><p><b>所以要阻止少数人占有大多的资本本质上就意味着,能不能让每一代人都完全公平的起点进行公平的竞争?</b>这个其实很难。下面我从教育、社保和税收、立法等方面说明这些均贫富的努力最终的效果都是有限的。</p><br><p>教育对人有没有提高作用呢?之前 <a class="member_mention" href="//www.zhihu.com/people/b09ea18d0114f41330c2734814b3f0c0" data-hash="b09ea18d0114f41330c2734814b3f0c0" data-hovercard="p$b$b09ea18d0114f41330c2734814b3f0c0">@chenqin</a> 在本题的回答已经论证了教育的回报率是相当可观的。但是富有的人往往可以给自己的子女创造更好的受教育的机会,并且富有的人也可以给子女提供更多的实践的机会,让他们在更小的时候就可以接触到比如创业、资本的运作等等,这些条件是穷人很难享有的。所以教育对聪明的个体而言,确实是偶尔能够生产出逆天的存在,让自己从无到有,从穷到富的,但是就整体而言,教育并不能减少贫富差距。有的时候,我们观察到的教育让贫富差距的暂时性减少,是因为在特别贫困的基层,让教育从无到有,对人力资本的巨大提升,这种巨大的提升会短暂的拉升大批极端贫困的人的生活水平。</p><br><p>但是当所有人都可以享受到公立教育的时候,优质的教育资源无论何时总是稀缺的,我们永远不可能设计出一个制度,让富人比穷人在教育方面有劣势——因为富人总是可以模拟穷人的行为,而穷人无法模拟富人的有些行为。</p><br><p>社保和税收,其建立的目的并非是单一的劫富济贫,也有创造代际之间的资产转移的意思。古代的人们通过生孩子来完成代际转移——年轻的时候养孩子,年老的时候被孩子养,但是这个方面不稳定,风险也大,于是现代社会统一建立了保障体系,让年轻人交税养老年人。收税是基于收入的,而富人的收入更加的多元化和复杂,也更加有能力雇佣律师来避税,反而是穷人的收入来源比较单一,面对税法的时候很透明,该交的一点都不能少。就这方面而言,对社会保障贡献最大的拿着较高的固定薪水的劳动者,但是以资本作为自身财富主体的“富人”依然不是吃亏的一方。</p><br><p>最后还有立法的手段,比如可以征收100%的遗产税等等。但是法只要成文了,谁能够雇佣更好的律师来仔细研究法律的条文,在不违反法律的情况下把资产转移给后代呢?更何况国家和国家之间还存在着竞争富有移民的关系。富人的国际流动性也比穷人高。如果立法太严峻,把富人逼急了,富人可以移民到其他税率低的小国,结果就导致一点税也收不到了,法国、英国均存在不同程度的富人出逃的情况。所以从国家的角度上讲,收过高的税率,即便不考虑所带来的激励减少的负作用,结果就是导致富人大量出逃,过高的税率依然会压在中产和穷人身上。</p><br><p>最后的最后,说一点正面的希望:从更长期来看,打破代际优势传递的希望,还是落在养育社会化上。养老社会化我们已经做到了,并且效果也很好,现代父母对子女的控制力度和欲望已经远远的小很多了;因为养老的负担已经不由子女而由社会来共同承担了。</p><br><p>但是养育的负担依然落在了父母身上。如果把养育也社会化了,父母和子女之间的经济从小到大都是相对独立的,那么父母处心积虑的想给自己的子女创造竞争优势的心态就会得到一定的削弱。这方面目前做的比较好的是北欧模式——这种模式对于人类的发展好不好我有保留意见,但是就单纯的对于贫富差距和代际优势传递方面而言,北欧模式是有效的,见 <a class="member_mention" href="//www.zhihu.com/people/1d1c9784aab4f00964215f435e479924" data-hash="1d1c9784aab4f00964215f435e479924" data-hovercard="p$b$1d1c9784aab4f00964215f435e479924">@Manolo</a> 的专栏<a href="https://zhuanlan.zhihu.com/p/27169841" class="internal">北欧,真的更加平等吗 - 知乎专栏</a> 和Lindahl et al. (2015)</p><br><br><p>Lindahl M, Palme M, Massih S, et al. Long-Term Intergenerational Persistence of Human Capital: An Empirical Analysis of Four Generations[J]. <i>Journal of Human Resources</i>, 2015, 50(1): 1-33.</p></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n\n为什么富人越富,穷人越穷?这个声明里面我默认穷人富人是相对的,也就是说贫富差距在扩大。因为我们的绝对生活水平因为科技进步的缘故还是在提高。 这个问题光看劳动和资本的边际回报是会掩盖一些问题的。首先,要承认人和人的天赋之间从经济上本来就是不…\n\n<a href=\"/question/21128507/answer/181695539\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" target=\"_blank\" href=\"/question/21128507/answer/181695539\">发布于 2017-06-10</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-632044\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>99 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"2\">\n<meta name=\"disableCopyAvatar\" content=\"https://pic4.zhimg.com/v2-2136abf2d4081f2ee9493cd1f10109e3_s.jpg\">\n\n<span class=\"zg-bull\">•</span>\n\n\n<a href=\"/copyright/apply?answer=181695539\" target=\"_blank\" class=\"meta-item copyright\">申请转载</a>\n\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"2\">\n<h2><a class=\"question_link\" href=\"/question/49295364/answer/181661917\" target=\"_blank\" data-id=\"11934404\" data-za-element-name=\"Title\">\n亲身经历过的觉得最讽刺的事情是什么?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"63100936\"\ndata-atoken=\"181661917\"\ndata-collapsed=\"0\"\ndata-created=\"1497072376\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/49295364/answer/181661917\">\n<meta itemprop=\"answer-id\" content=\"63100936\">\n<meta itemprop=\"answer-url-token\" content=\"181661917\">\n<a class=\"zg-anchor-hidden\" name=\"answer-63100936\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>167</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">167</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$fan-xi-di\"\ntarget=\"_blank\" href=\"/people/fan-xi-di\"\n>范西迪</a></span><span title=\"伪文史政关注者,NBA段子手。\" class=\"bio\">\n伪文史政关注者,NBA段子手。\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"167\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">167</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"11934404\" data-action=\"/answer/content\" data-author-name=\"范西迪\" data-entry-url=\"/question/49295364/answer/181661917\">\n\n<textarea hidden class=\"content\"><p>谢邀<a href="https://www.zhihu.com/people/ca-ca-ca-31-28" class="internal">擦擦擦</a> </p><p><b>一、“我在马路边,捡到一分钱”</b></p><p>小学时候,老师总教育我们要拾金不昧,还教我们唱了一首歌。</p><blockquote>我在马路边 捡到一分钱 \n把它交到警察叔叔手里边 \n叔叔拿着钱 对我把头点 \n我高兴的说了声 叔叔再见 \n我在马路边 捡到一分钱 \n把它交到警察叔叔手里边 \n叔叔拿着钱 对我把头点 \n我高兴的说了声 叔叔再见 \n我在马路边 捡到一分钱 \n把它交到警察叔叔手里边 \n叔叔拿着钱 对我把头点 \n我高兴的说了声 叔叔再见</blockquote><p>这次我还真在操场上捡到一毛钱(九十年代初,一毛钱对于小孩子来说挺多的),我就思索起来:</p><p>“这一毛钱要交给谁呢?交给我们班主任老师吧。可班主任只管一个班级,在班里捡到的钱当然要给班主任。但是在操场上呢?交给班主任不合适,那怎么办?</p><p>“哦,对了,这钱应该交给教务处主任,因为他是管整个学校的。”</p><p>我就跑到教务处办公室找主任,说:“主任,您好!我捡到一毛钱,现在交给你!”</p><p>主任正在浏览文件,不耐烦的看了我一眼,说:“ 一毛钱就不用交了吧,你到校门口买根冰棒吃花掉算了。”</p><p>“可是……”</p><p>“可是什么,有什么可是的?你要捡五十块,一百块拿过来罢了。这一毛钱,我去哪儿找失主?”</p><p>“老师说要拾金不昧,捡到一毛钱也交给老师的啊!”</p><p>主任觉察过来,马上挤出一副笑容,说:“你真是好孩子,把一毛钱发在这里吧!到时我会找到失主给他的。你就回去吧!”</p><p>我走了后,还是不放心,心想:“看主任这样子,会不会不把钱交给失主?不对不对,我应该告诉他是我哪班级的。他找到失主后,也能通知我下。”</p><p>我快速折返办公室。却在门口听见主任爽朗的笑声,我躲在门口瞄了一眼。</p><p>主任正在和另一个老师聊天,主任说道:</p><p>“<b>有个傻逼学生,捡了一毛钱,居然也拿来交!</b>”</p><br><p><b>二、“没带就是没写”</b></p><p>小学时候我被班主任任命为组长,老师谆谆教导道:“要严格检查每个组员的作业,没完成的全部记录下来告诉我!”</p><p>我们当时作业繁重,每天田字格写生字要写近百个,还经常要抄试卷。虽然教育部年年喊“减负”,但用当时流行的话就是“减负就是加”。</p><p>我当组长后,像个酷吏一般。相对其他组组长敷衍了事,我一丝不苟地检查每个组员作业,没写的、漏写的全逮出来报告老师。所以,我们组的作业完成情况是最糟糕的,我在组里名声也臭得不得了。</p><p>那次又有一组员没交上作业,我质问他:“你作业呢?”</p><p>“我…………我没带,落家里了。”</p><p>“没带就是没写!我把你名字记上。”</p><p>“我写了的,不要记,不要记,我中午拿过来!”</p><p>我想了一下,说:“既然写了,那你回家拿,但你十二点之前就要把作业带过来(我们是十一点多放学),不然的话,就是在家里补写,我还是要记你名字。”</p><p>当天中午,那同学没来上学,反而是他家长过来找老师。</p><p>老师把我喊过去,责备道:“你怎么这么当组长,把同学吓得不敢来上学了?”</p><p>“老师,您不是说要严格检查每个组员作业吗?”</p><p><b>“我是让你严格检查作业,但没让你逼得人家不敢来上学啊!”</b></p><p>我流下了悔恨的泪水。从此以后,我再也没检查过组员作业,每次都敷衍了事。有同学没写作业,我也帮他遮掩过去,<b>我成为了最受欢迎的组长。</b></p></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n\n谢邀<a href=\"https://www.zhihu.com/people/ca-ca-ca-31-28\" class=\"internal\">擦擦擦</a> <b>一、“我在马路边,捡到一分钱”</b>小学时候,老师总教育我们要拾金不昧,还教我们唱了一首歌。我在马路边 捡到一分钱 \n把它交到警察叔叔手里边 \n叔叔拿着钱 对我把头点 \n我高兴的说了声 叔叔再见 \n我在马路边 捡到一分钱 \n把它交到警察叔叔手里边 …\n\n<a href=\"/question/49295364/answer/181661917\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 2017-06-10\" target=\"_blank\" href=\"/question/49295364/answer/181661917\">编辑于 2017-06-11</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-11934404\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>33 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"2\">\n<meta name=\"disableCopyAvatar\" content=\"https://pic4.zhimg.com/v2-4c479a184e94eb15a1653d677b269bc3_s.jpg\">\n\n<span class=\"zg-bull\">•</span>\n\n\n<a href=\"/copyright/apply?answer=181661917\" target=\"_blank\" class=\"meta-item copyright\">申请转载</a>\n\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"3\">\n<h2><a class=\"question_link\" href=\"/question/60774289/answer/181585578\" target=\"_blank\" data-id=\"16533311\" data-za-element-name=\"Title\">\n哪些行为看上去很帅,其实很傻?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"63087117\"\ndata-atoken=\"181585578\"\ndata-collapsed=\"0\"\ndata-created=\"1497062768\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/60774289/answer/181585578\">\n<meta itemprop=\"answer-id\" content=\"63087117\">\n<meta itemprop=\"answer-url-token\" content=\"181585578\">\n<a class=\"zg-anchor-hidden\" name=\"answer-63087117\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>1238</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">1238</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$hu-bu-gui-52\"\ntarget=\"_blank\" href=\"/people/hu-bu-gui-52\"\n>狐不归</a></span><span title=\"我不想谋生,我想生活\" class=\"bio\">\n我不想谋生,我想生活\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"1238\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">1238</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"16533311\" data-action=\"/answer/content\" data-author-name=\"狐不归\" data-entry-url=\"/question/60774289/answer/181585578\">\n\n<textarea hidden class=\"content\"><p>高中时,班上有一位号称“冷美人”的校花。全校大概有上百位男生暗恋她。</p><p>据我所知,其中有5位追她追得特别用功。</p><p>毕业后,语文老师曾经当着大家的面说:这五位同学要是学习也这么用功,现在已经上了清北了。</p><br><p>五位追她的男生里,两位体育特长生,一位小混混,还有一位文学青年。</p><p>我现在正在做高等数学题,抽空在知乎上回答这个题目。</p><p>下面我就简单地向大家说说当时的情况。</p><p>有一天,小混混对其他四位说:来来来,哥给你们几位看看最新的纹身。</p><p>说着他扒下衬衫,除了两只胳膊上原有的左青龙右白虎外,胸口又添了一处“我爱王露露”的字样。王露露就是校花。</p><br><p>两位体育生对视一眼,</p><p>不屑地说:</p><blockquote>这有什么?纹的时候还打麻醉吧?有种我们打个赌,谁有胆从二楼跳下去,其他人以后就不许再纠缠露露,怎么样?</blockquote><p>说着,他们俩先后从二楼跳到水泥地上。</p><p>后来的一个月,我们就没在学校见过他们了。</p><p>等他们来上学时,其中一位坐着轮椅,见到我们还哭了。</p><p>另一位还好,只是重重地崴了脚,不过后来再也当不成体育特长生了——高考当然也没考上。</p><br><p>五位同学最后那位,也就是前面说的文学青年,比较胆小腼腆。</p><p>他追王露露的方法比较含蓄,但更为执着。</p><p>他几乎一天一首情诗,趁人不注意,偷偷塞在王璐璐的抽屉里。</p><br><br><br><br><p>————————————————</p><p>上大学后,王露露终于和我们班上最有钱的同学在一起了。</p><p>当我多年后读着那位文学青年写给王璐璐的情诗时,一边赞叹,一边感慨不已。</p><p>有好几次还深深地感动了。</p><p>没错,我就是那位有钱的同学</p><br><br><br><br><br><br><br><br><br><br><br><br><p>最好的哥们。</p><p>我不是那位写诗的。打死我也不承认。</p><br><p>——————</p><br><p>相关链接</p><a href="https://www.zhihu.com/question/60815221/answer/186855899" class="internal">有哪些看似很傻,实则很帅的行为? - 知乎</a></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n\n高中时,班上有一位号称“冷美人”的校花。全校大概有上百位男生暗恋她。据我所知,其中有5位追她追得特别用功。毕业后,语文老师曾经当着大家的面说:这五位同学要是学习也这么用功,现在已经上了清北了。 五位追她的男生里,两位体育特长生,一位小混混,…\n\n<a href=\"/question/60774289/answer/181585578\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 2017-06-10\" target=\"_blank\" href=\"/question/60774289/answer/181585578\">编辑于 2017-06-22</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-16533311\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>180 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"2\">\n<meta name=\"disableCopyAvatar\" content=\"https://pic1.zhimg.com/5e48cf49ac163533c51f3da32716db3c_s.jpg\">\n\n<span class=\"zg-bull\">•</span>\n\n\n<a href=\"/copyright/apply?answer=181585578\" target=\"_blank\" class=\"meta-item copyright\">申请转载</a>\n\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"4\">\n<h2><a class=\"question_link\" href=\"/question/60823163/answer/181712433\" target=\"_blank\" data-id=\"16552946\" data-za-element-name=\"Title\">\n如何理解2017年浙江高考语文阅读理解《一种美味》最后那条鱼发出诡异的光?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"63110158\"\ndata-atoken=\"181712433\"\ndata-collapsed=\"0\"\ndata-created=\"1497079652\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/60823163/answer/181712433\">\n<meta itemprop=\"answer-id\" content=\"63110158\">\n<meta itemprop=\"answer-url-token\" content=\"181712433\">\n<a class=\"zg-anchor-hidden\" name=\"answer-63110158\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>50</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">50</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$ye-ge-3-51\"\ntarget=\"_blank\" href=\"/people/ye-ge-3-51\"\n>也鸽</a></span><span title=\"野鸽也歌\" class=\"bio\">\n野鸽也歌\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"50\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">50</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"16552946\" data-action=\"/answer/content\" data-author-name=\"也鸽\" data-entry-url=\"/question/60823163/answer/181712433\">\n\n<textarea hidden class=\"content\">作为一个已经一年没做阅读理解了的前浙江考生,我要来认(qiang)真(xing)从<b>浙江</b>这个角度来回答一下这个问题。<br><br>如果这篇文章的主题不是鱼,而是猪肉,或许能让大家觉得没有那么陌生吧。浙江是什么地方啊,得天独厚的鱼米之乡,缺什么肉最少缺的就是鱼了。不说沿海,哪个村子里没有条小河。那个生我养我的小村子更是以编织渔网为原始产业的。我爸还开玩笑的说过,我要是不会游泳,都不好意思说自己是xx村的。<br><br>但是如果是猪肉呢?<br><br>我曾经在另一个答案里提到我母亲的一个经历,这是她经常挂在嘴边来怀念以往生活不易的故事:那时候她七八岁,家里只有奶奶一个至亲。我妈说,那一天,家里真的是一分钱都没有了,但是她突然想吃肉。就像《一种美味》中豆腐和过节的联系。那时候,猪肉也是过节才能吃的宝贝。春节时杀头猪,熬出来的猪油是要吃一年多。但是我妈说,那一天,她非常非常想吃肉,想吃年糕。<br><br>于是她奶奶我太外婆想了个法子。她们一大早下农田,去拔一种长得像铜钱草的草药。她们拔了一个白天,两大篮子,拿到集市去换了五毛钱。两毛钱买了年糕,一毛钱买了一点点猪肉,一毛钱买了菜,就着一点点猪油,她们烧了一顿汤糕。<br><br>这个故事我妈从我小说道现在,每一次,她都会说,这是她这辈子吃过最好吃的汤糕了。<br><br>我母亲还说过另一个故事,以前骨头是可以卖钱的。她有时候馋坏了,就会去各种地方捡骨头,换来钱后花一毛钱可以买一大袋的瓜子。我妈说,那时候自己胆子真的大,都不怕是不是死人的骨头。<br><br><b>所以,在一个物质匮乏的时代,肉,和嘴里的一点点味道,有的时候真的是一个人唯一的希望和憧憬。</b><br><br>但是现在的浙江人,当我们听到这些老一辈的故事,会有多少感触?<br><br>浙江,这个几乎所有市县都超越了全国平均水平的地方早已不复当年的贫瘠。在这里,一个普通城市的房价都比西北省会高。在这里,我们有好山有好水还有好企业,在这里,我们的教育局可以很自信地说“浙江的素质教育不需要衡水中学”。这里也可能是全国农村发展的最好的地方,村村都有小洋房。去年有一个村卖了一块地,这个村每个人都能分到近二十万。<br><br><b>所以,当那些七十年代靠着划小渔船走私牛仔裤发家的父辈给自己的孩子讲曾经的物质匮乏,他们的孩子会不会觉得这种贫穷充满魔幻色彩,就像那条闪着诡异的光的鱼。</b><br><br>我不是说浙江人个个都有钱,也不是地图炮看不起别的省份。我是想表达:浙江就是这样一个天时地利人和的地方。前几天有一个“浙江给你最大的感受是什么”的问题一直出现在我的首页,靠前的答案没有一个黑浙江的,说到浙江,大家会想到的是“车让人”,是“好山好水”。是“家”。而我,作为一个浙江人,我是自豪的。<br><br>我的家乡有句方言,叫“白天当老板,晚上睡地板”。如果没有那一个个拼着命创业奋斗的老浙江人,就没有今天的浙江,就没有这个我作为年轻人为之自豪的浙江。<br><br>在开头我说道,浙江最不缺的肉就是鱼,就像现在的浙江,绝大多数人是不会像文中的那个“我”一样贫穷到不能上学没钱吃肉,这样看来,很多浙江普通人的起点,可能真的是一些贫困地区人的重点。<br><br>所以我不妨大胆揣测命题者选这篇文章的意图。标准答案已经放出,它并没有那么悲观,把靠读书来改变生活现状和阶级的困难撕裂开来,事实上,高考的答案是悲观不到哪里去的。我并不觉得出卷人是想给考生泼个冷水,我更愿意相信,<b>他们是希望这些即将去往五湖四海的浙江考生们珍惜你是一个浙江人。是希望他们理解,现在这个美丽浙江也曾贫瘠过,贫瘠到很多人都不知道肉是什么味道。而现在这个大家都吃得到鱼的浙江,是老一辈人用汗用泪用血拼出来的。</b>我们在为这片生我养我的土地自豪的同时,不能愧对十几年来她提供的物质精神条件。我们要更加奋发努力,不让身上这个“浙江人”的标签失色。因为它也代表着那些从贫穷走向温饱的传奇老浙江人。<br><br>原作者说,写出来的文章就像一个孩子,这篇文章他写了都七八年了,自己都忘了他长什么样。文学作品在创作完成后其实就脱离了作者的意愿,读者是什么,他就会从中看到什么。所以每个人的看法意见都可以不同,理解层次会有深浅高低,但觉不会分贵贱。<br><br><br>而这就是我的理解。<br><br><br><br>浙江人啊,杀克重。</textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n\n作为一个已经一年没做阅读理解了的前浙江考生,我要来认(qiang)真(xing)从<b>浙江</b>这个角度来回答一下这个问题。 如果这篇文章的主题不是鱼,而是猪肉,或许能让大家觉得没有那么陌生吧。浙江是什么地方啊,得天独厚的鱼米之乡,缺什么肉最少缺的就是鱼了。…\n\n<a href=\"/question/60823163/answer/181712433\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 2017-06-10\" target=\"_blank\" href=\"/question/60823163/answer/181712433\">编辑于 2017-06-10</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-16552946\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>28 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"1\">\n<meta name=\"disableCopyAvatar\" content=\"\">\n\n<span class=\"zg-bull\">•</span>\n\n<a href=\"/terms#sec-licence-1\" target=\"_blank\" class=\"meta-item copyright\">\n作者保留权利\n</a>\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div><div class=\"explore-feed feed-item\" data-offset=\"5\">\n<h2><a class=\"question_link\" href=\"/question/60810862/answer/181568571\" target=\"_blank\" data-id=\"16547953\" data-za-element-name=\"Title\">\n女生和她男朋友还没有分手就和我在一起了,该做的事都做了。她这是什么心态?\n</a></h2>\n<div tabindex=\"-1\" class=\"zm-item-answer \"\nitemscope itemtype=\"http://schema.org/Answer\"\ndata-aid=\"63084047\"\ndata-atoken=\"181568571\"\ndata-collapsed=\"0\"\ndata-created=\"1497060349\"\ndata-deleted=\"0\"\ndata-isowner=\"0\"\ndata-helpful=\"1\"\ndata-copyable=\"1\"\n>\n<link itemprop=\"url\" href=\"/question/60810862/answer/181568571\">\n<meta itemprop=\"answer-id\" content=\"63084047\">\n<meta itemprop=\"answer-url-token\" content=\"181568571\">\n<a class=\"zg-anchor-hidden\" name=\"answer-63084047\"></a>\n\n<div class=\"zm-item-vote\">\n<a class=\"zm-item-vote-count js-expand js-vote-count\" href=\"javascript:;\" data-bind-votecount>75</a>\n</div>\n\n<div class=\"zm-votebar\">\n<button class=\"up \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"count\">75</span>\n<span class=\"label sr-only\">赞同</span>\n</button>\n<button class=\"down \">\n<i class=\"icon vote-arrow\"></i>\n<span class=\"label sr-only\">反对</span>\n</button>\n</div>\n\n\n<div class=\"answer-head\">\n\n<div class=\"zm-item-answer-author-info\">\n\n<span class=\"summary-wrapper\">\n<span class=\"author-link-line\">\n\n<a class=\"author-link\"\ndata-hovercard=\"p$t$jin-zhi-75-54\"\ntarget=\"_blank\" href=\"/people/jin-zhi-75-54\"\n>金知</a></span><span title=\"研究控+八卦爱好者\" class=\"bio\">\n研究控+八卦爱好者\n</span>\n</span>\n</div>\n\n<div class=\"zm-item-vote-info\" data-votecount=\"75\">\n\n\n\n<span class=\"voters text\"><a href=\"#\" class=\"more text\"><span class=\"js-voteCount\">75</span> 人赞同</a></span>\n\n\n</div>\n</div>\n<div class=\"zm-item-rich-text expandable js-collapse-body\" data-resourceid=\"16547953\" data-action=\"/answer/content\" data-author-name=\"金知\" data-entry-url=\"/question/60810862/answer/181568571\">\n\n<textarea hidden class=\"content\"><p>谢邀。</p><br><p>喜欢是一定的。</p><p>但你要知道,人的喜欢是有层级的。</p><p>你喜欢自己的家,但你也喜欢出去玩的时候住的某一间精品酒店。</p><p>题主,你就是那个女生遇到的一间喜欢的酒店而已。就算再好,也不是家,没打算安身立命。</p><p>知道她喜欢过你,那么,谢谢她,也谢谢认真付出感情的自己。</p><br><p>假如你对自己的定位不是酒店,而希望成为一个家的话。那就把这位不够认真的房客赶出去,好好打扫干净,等待下一个人住进来。</p><br><br><br><p>我的婚姻情感类文章:</p><p>女性怎样越变越美?</p><a href="https://www.zhihu.com/question/60031007/answer/175838840" class="internal"><span class="invisible">https://www.</span><span class="visible">zhihu.com/question/6003</span><span class="invisible">1007/answer/175838840</span><span class="ellipsis"></span></a><p>对异性欣赏但并不想去认识是什么样的体验?</p><a href="https://www.zhihu.com/question/59111459/answer/161856905" class="internal"><span class="invisible">https://www.</span><span class="visible">zhihu.com/question/5911</span><span class="invisible">1459/answer/161856905</span><span class="ellipsis"></span></a><p>为什么其貌不扬的邓文迪梁洛施郭晶晶都能够被豪门青睐,女神林志玲却单身至今?</p><a href="https://www.zhihu.com/question/38114700/answer/155887241" class="internal"><span class="invisible">https://www.</span><span class="visible">zhihu.com/question/3811</span><span class="invisible">4700/answer/155887241</span><span class="ellipsis"></span></a><p>女友交往一年多感情很好,父母反对,该分手吗?</p><a href="https://www.zhihu.com/question/59015795/answer/161424685" class="internal"><span class="invisible">https://www.</span><span class="visible">zhihu.com/question/5901</span><span class="invisible">5795/answer/161424685</span><span class="ellipsis"></span></a></textarea>\n\n\n<div class=\"zh-summary summary clearfix\">\n\n谢邀。 喜欢是一定的。但你要知道,人的喜欢是有层级的。你喜欢自己的家,但你也喜欢出去玩的时候住的某一间精品酒店。题主,你就是那个女生遇到的一间喜欢的酒店而已。就算再好,也不是家,没打算安身立命。知道她喜欢过你,那么,谢谢她,也谢谢认真付出…\n\n<a href=\"/question/60810862/answer/181568571\" class=\"toggle-expand\">显示全部</a>\n\n</div>\n\n\n\n<p class=\"visible-expanded\"><a itemprop=\"url\" class=\"answer-date-link meta-item\" data-tooltip=\"s$t$发布于 2017-06-10\" target=\"_blank\" href=\"/question/60810862/answer/181568571\">编辑于 2017-06-13</a></p>\n\n</div>\n<div class=\"zm-item-meta answer-actions clearfix js-contentActions\">\n<div class=\"zm-meta-panel\">\n\n<a data-follow=\"q:link\" class=\"follow-link zg-follow meta-item\" href=\"javascript:;\" id=\"sfb-16547953\"><i class=\"z-icon-follow\"></i>关注问题</a>\n\n<a href=\"#\" name=\"addcomment\" class=\"meta-item toggle-comment js-toggleCommentBox\">\n<i class=\"z-icon-comment\"></i>23 条评论</a>\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-thank\" data-thanked=\"false\"><i class=\"z-icon-thank\"></i>感谢</a>\n\n\n\n<a href=\"#\" class=\"meta-item zu-autohide js-share\"><i class=\"z-icon-share\"></i>分享</a>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-collect\"><i class=\"z-icon-collect\"></i>收藏</a>\n\n\n\n\n\n<span class=\"zg-bull zu-autohide\">•</span>\n\n<a href=\"#\" class=\"meta-item zu-autohide js-noHelp\">没有帮助</a>\n\n<span class=\"zg-bull zu-autohide\">•</span>\n<a href=\"#\" class=\"meta-item zu-autohide js-report\">举报</a>\n\n\n\n\n<meta name=\"copyrightStatus\" content=\"1\">\n<meta name=\"disableCopyAvatar\" content=\"\">\n\n<span class=\"zg-bull\">•</span>\n\n<a href=\"/terms#sec-licence-1\" target=\"_blank\" class=\"meta-item copyright\">\n作者保留权利\n</a>\n\n\n<button class=\"meta-item item-collapse js-collapse\">\n<i class=\"z-icon-fold\"></i>收起\n</button>\n</div>\n</div>\n</div>\n</div>\n</div>\n</div>\n</div>\n\n\n</div>\n</div>\n\n\n<div class=\"zu-main-sidebar\" data-za-module=\"RightSideBar\">\n\n<div class=\"zm-side-section\">\n<div class=\"zm-side-section-inner\">\n<div class=\"SignFlow SignFlow--bordered\" id=\"SidebarSignFlow\">\n<div class=\"SignFlow-panel\">\n<div class=\"title register\">\n<h1>加入知乎</h1>\n<h2>与世界分享你的知识、经验和见解</h2>\n</div>\n<div class=\"view register SignupForm\" data-za-module=\"SignUpForm\">\n<form action=\"/register/email\" method=\"POST\">\n<div class=\"input-wrapper\">\n<input type=\"text\" name=\"fullname\" aria-label=\"姓名\" data-placeholder=\"姓名\" required>\n</div>\n<div class=\"input-wrapper\">\n\n<input type=\"text\" name=\"phone_num\" class=\"account\" aria-label=\"手机号\" data-placeholder=\"手机号\" required>\n\n</div>\n<div class=\"input-wrapper toggle-password\">\n<input type=\"password\" hidden> \n<input type=\"password\" name=\"password\" aria-label=\"密码\" data-placeholder=\"密码(不少于 6 位)\" required>\n<span class=\"z-ico-show-password\"></span>\n</div>\n<div class=\"input-wrapper captcha-module\" data-type=\"en\" >\n<input id=\"captcha\" name=\"captcha\" placeholder=\"验证码\" required data-rule-required=\"true\" data-msg-required=\"请填写验证码\">\n<div class=\"captcha-container\">\n\n<img class=\"js-refreshCaptcha captcha\" width=\"120\" height=\"30\" data-tooltip=\"s$t$看不清楚?换一张\" alt=\"验证码\">\n</div>\n</div>\n<div class=\"actions\">\n\n<input type=\"submit\" value=\"注册\" class=\"submit zg-btn-blue\">\n\n</div>\n<div class=\"sns clearfix\">\n<span>已有帐号?<a href=\"#\" class=\"switch-to-login\">登录</a></span>\n</div>\n</form>\n</div>\n</div>\n<div class=\"SignFlow-panel\" hidden>\n<div class=\"LoginForm\">\n<div class=\"title login\">\n<h1>登录知乎</h1>\n<h2>与世界分享你的知识、经验和见解</h2>\n</div>\n<div class=\"view login\" data-za-module=\"SignInForm\">\n<form method=\"POST\">\n<div class=\"input-wrapper\">\n<input type=\"text\" name=\"account\" class=\"account\" aria-label=\"手机号或邮箱\" placeholder=\"手机号或邮箱\" required>\n</div>\n<div class=\"input-wrapper toggle-password\">\n<input type=\"password\" name=\"password\" aria-label=\"密码\" placeholder=\"密码\" required>\n<span class=\"z-ico-show-password\"></span>\n</div>\n<div class=\"input-wrapper captcha-module\" data-type=\"en\" >\n<input id=\"captcha\" name=\"captcha\" placeholder=\"验证码\" required data-rule-required=\"true\" data-msg-required=\"请填写验证码\">\n<div class=\"captcha-container\">\n\n<img class=\"js-refreshCaptcha captcha\" width=\"120\" height=\"30\" data-tooltip=\"s$t$看不清楚?换一张\" alt=\"验证码\">\n</div>\n</div>\n<div class=\"actions\">\n\n<input type=\"submit\" value=\"登录\" class=\"submit zg-btn-blue\">\n\n</div>\n<div class=\"sns clearfix\">\n<label class=\"remember-me\"><input type=\"checkbox\" name=\"remember_me\" checked value=\"true\">记住我</label>\n<span class=\"middot\">·</span>\n<button class=\"unable-login\" type=\"button\">无法登录?</button>\n<button type=\"button\" class=\"js-show-sns-buttons is-visible\">社交帐号登录</button>\n<div class=\"sns-buttons\">\n<button type=\"button\" class=\"wechat\"><span class=\"ico sprite-global-icon-wechat-gray\"></span>微信</button>\n<button type=\"button\" class=\"weibo\"><span class=\"ico sprite-global-icon-weibo-gray\"></span>微博</button>\n<button type=\"button\" class=\"qq\"><span class=\"ico sprite-global-icon-qq-gray\"></span>QQ</button>\n</div>\n</div>\n</form>\n</div>\n</div>\n</div>\n</div>\n</div>\n</div>\n<div class=\"zm-side-section\">\n<div class=\"DownloadApp\">\n<button class=\"DownloadApp-button\">\n<span class=\"sprite-global-icon-qrcode\"></span>\n下载知乎 App\n</button>\n</div>\n</div>\n\n\n<div class=\"shameimaru-placeholder\" data-loc=\"explore_up\" data-params='{}'></div>\n\n<div class=\"zm-side-section explore-side-section explore-side-section-roundtable\">\n<div class=\"zm-side-section-inner\">\n<div class=\"section-title\">\n<a href=\"/roundtable\" class=\"zg-link-gray zg-right\" data-za-c=\"view_roundtable\" data-za-a=\"visit_roundtable_home\" data-za-l=\"explore_sidebar_trending_roundtable_more\">更多圆桌 »</a>\n<h3>热门圆桌</h3>\n</div>\n<ul class=\"list hot-roundtables\">\n\n<li class=\"clearfix\">\n<a target=\"_blank\" class=\"avatar-link\" href=\"/roundtable/caiwufenxi\"><img src=\"https://pic4.zhimg.com/v2-02147ca4010b8ac9a8f198b7a64c72ef_m.jpg\" alt=\"Path\" class=\"avatar 40\" /></a>\n<div class=\"content\">\n\n<span class=\"status zg-right zg-gray-normal\">\n还有 7 天结束\n</span>\n\n<a href=\"/roundtable/caiwufenxi\" target=\"_blank\" data-hovercard=\"r$t$caiwufenxi\">学一点财务分析</a>\n<div class=\"meta\">\n<span>2297 人关注</span>\n<span class=\"zg-bull\">•</span>\n<span>5 个问题</span>\n</div>\n</div>\n</li>\n\n<li class=\"clearfix\">\n<a target=\"_blank\" class=\"avatar-link\" href=\"/roundtable/jianzhu\"><img src=\"https://pic2.zhimg.com/v2-08091965921ed04731b2001c6e0bd411_m.jpg\" alt=\"Path\" class=\"avatar 40\" /></a>\n<div class=\"content\">\n\n<span class=\"status zg-right zg-gray-normal\">\n还有 4 天结束\n</span>\n\n<a href=\"/roundtable/jianzhu\" target=\"_blank\" data-hovercard=\"r$t$jianzhu\">看见建筑之美</a>\n<div class=\"meta\">\n<span>1430 人关注</span>\n<span class=\"zg-bull\">•</span>\n<span>5 个问题</span>\n</div>\n</div>\n</li>\n\n</ul>\n</div>\n</div>\n<div class=\"zm-side-section explore-side-section\">\n<div class=\"zm-side-section-inner\">\n<div class=\"section-title\">\n\n<a href=\"/topics\" class=\"zg-link-gray zg-right\">更多话题 »</a>\n\n<h3>热门话题</h3>\n</div>\n<ul class=\"list hot-topics\">\n\n<li class=\"clearfix\">\n<a target=\"_blank\" class=\"avatar-link\" href=\"/topic/19952767\" data-hovercard=\"t$b$19952767\"><img src=\"https://pic1.zhimg.com/693f18c3366811a20f1d1f3926361034_m.png\" alt=\"英雄联盟职业联赛(LPL)\" class=\"avatar 40\" /></a>\n<div class=\"content\">\n<a href=\"/topic/19952767\" target=\"_blank\" data-hovercard=\"t$b$19952767\">英雄联盟职业联赛(LPL)</a>\n<div class=\"meta\">\n<span>10119 人关注</span>\n\n</div>\n</div>\n<div class=\"bottom\">\n\n<a class=\"question_link\" href=\"/question/62215437\" target=\"_blank\" data-id=\"17111023\" data-za-element-name=\"Title\">\n如何评价 2017 年 LOL 亚洲对抗赛LPL中国拿下冠军?\n</a>\n\n</div>\n</li>\n\n<li class=\"clearfix\">\n<a target=\"_blank\" class=\"avatar-link\" href=\"/topic/19573413\" data-hovercard=\"t$b$19573413\"><img src=\"https://pic2.zhimg.com/v2-9036ee3d2e770be14882732dcf57ced9_m.jpg\" alt=\"鼓浪屿\" class=\"avatar 40\" /></a>\n<div class=\"content\">\n<a href=\"/topic/19573413\" target=\"_blank\" data-hovercard=\"t$b$19573413\">鼓浪屿</a>\n<div class=\"meta\">\n<span>961 人关注</span>\n\n</div>\n</div>\n<div class=\"bottom\">\n\n<a class=\"question_link\" href=\"/question/62185967\" target=\"_blank\" data-id=\"17099178\" data-za-element-name=\"Title\">\n如何评价厦门鼓浪屿申遗成功,列入「世界文化遗产」名录?\n</a>\n\n</div>\n</li>\n\n<li class=\"clearfix\">\n<a target=\"_blank\" class=\"avatar-link\" href=\"/topic/19553134\" data-hovercard=\"t$b$19553134\"><img src=\"https://pic3.zhimg.com/v2-b2803387bd34f4349a0640a20caf2372_m.jpg\" alt=\"雷军(人物)\" class=\"avatar 40\" /></a>\n<div class=\"content\">\n<a href=\"/topic/19553134\" target=\"_blank\" data-hovercard=\"t$b$19553134\">雷军(人物)</a>\n<div class=\"meta\">\n<span>22284 人关注</span>\n\n</div>\n</div>\n<div class=\"bottom\">\n\n<a class=\"question_link\" href=\"/question/62126661\" target=\"_blank\" data-id=\"17075547\" data-za-element-name=\"Title\">\n如何看待小米 2017 年第二季度出货量 2316 万台,环比增长 70%?\n</a>\n\n</div>\n</li>\n\n</ul>\n</div>\n</div>\n<div class=\"zm-side-section explore-side-section\">\n<div class=\"zm-side-section-inner\">\n<div class=\"section-title\">\n<a href=\"javascript:;\" id=\"js-hot-fav-switch\" class=\"zg-link-gray zg-right\" data-za-c=\"collection\" data-za-a=\"click_trending_collections_change\" data-za-l=\"explore_sidebar_trending_collections_collection_name\">换一换</a>\n<h3>热门收藏</h3>\n</div>\n<ul class=\"list hot-favlists\">\n<li>\n<div class=\"content\">\n<a href=\"/collection/131421063\" target=\"_blank\">健身知识</a>\n<div class=\"meta\">\n<span>30 人关注</span>\n<span class=\"zg-bull\">•</span>\n<span>154 条内容</span>\n</div>\n</div>\n</li>\n\n<li>\n<div class=\"content\">\n<a href=\"/collection/104823295\" target=\"_blank\">科幻和科学传播</a>\n<div class=\"meta\">\n<span>41 人关注</span>\n<span class=\"zg-bull\">•</span>\n<span>99 条内容</span>\n</div>\n</div>\n</li>\n\n<li>\n<div class=\"content\">\n<a href=\"/collection/19781236\" target=\"_blank\">日常实用技巧</a>\n<div class=\"meta\">\n<span>21585 人关注</span>\n<span class=\"zg-bull\">•</span>\n<span>581 条内容</span>\n</div>\n</div>\n</li>\n\n<li>\n<div class=\"content\">\n<a href=\"/collection/106360933\" target=\"_blank\">优秀的讽刺</a>\n<div class=\"meta\">\n<span>850 人关注</span>\n<span class=\"zg-bull\">•</span>\n<span>536 条内容</span>\n</div>\n</div>\n</li>\n\n<li>\n<div class=\"content\">\n<a href=\"/collection/20393860\" target=\"_blank\">爱学术欧耶</a>\n<div class=\"meta\">\n<span>29 人关注</span>\n<span class=\"zg-bull\">•</span>\n<span>83 条内容</span>\n</div>\n</div>\n</li>\n</ul>\n</div>\n</div>\n<div class=\"shameimaru-placeholder\" data-loc=\"explore_down\" data-params='{}'></div>\n<div id=\"zh-footer\" class=\"zh-footer\">\n<div class=\"content zg-wrap clearfix\">\n<ul>\n\n<li><a href=\"https://liukanshan.zhihu.com\" target=\"_blank\">刘看山</a></li>\n\n<li><a href=\"/app\" target=\"_blank\">移动应用</a></li>\n<li><a href=\"/careers\">加入知乎</a></li>\n<li><a href=\"/terms\" target=\"_blank\">知乎协议</a></li>\n<li><a href=\"/jubao\" target=\"_blank\">举报投诉</a></li>\n<li><a href=\"/contact\">联系我们</a></li>\n\n</ul>\n\n<span class=\"copy\">© 2017 知乎</span>\n\n</div>\n</div>\n\n\n</div>\n\n\n</div>\n\n\n\n<script type=\"text/json\" class=\"json-inline\" data-name=\"guiders2\">{}</script>\n<script type=\"text/json\" class=\"json-inline\" data-name=\"current_user\">[\"\",\"\",\"\",\"-1\",\"\",0,0]</script>\n<script type=\"text/json\" class=\"json-inline\" data-name=\"front_web_config\">{\"realname_win_config\":{\"timestamp\":1501344000,\"continue_time\":3600,\"continue\":1,\"tip\":\"\\u5e94\\u56fd\\u5bb6\\u6cd5\\u89c4\\u5bf9\\u4e8e\\u5e10\\u53f7\\u5b9e\\u540d\\u7684\\u8981\\u6c42\\uff0c\\u8fdb\\u884c\\u4e0b\\u4e00\\u6b65\\u64cd\\u4f5c\\u524d\\uff0c\\u9700\\u8981\\u5148\\u5b8c\\u6210\\u624b\\u673a\\u7ed1\\u5b9a\\u3002\",\"skip_ut_verification\":1}}</script>\n<script type=\"text/json\" class=\"json-inline\" data-name=\"user_status\">[null,null,false]</script>\n<script type=\"text/json\" class=\"json-inline\" data-name=\"env\">[\"zhihu.com\",\"comet.zhihu.com\",false,null,false,false]</script>\n<script type=\"text/json\" class=\"json-inline\" data-name=\"permissions\">[]</script>\n\n\n<script type=\"text/json\" class=\"json-inline\" data-name=\"ga_vars\">{\"user_created\":0,\"now\":1499661102000,\"abtest_mask\":\"------------------------------\",\"user_attr\":[0,0,0,\"-\",\"-\"],\"user_hash\":0}</script>\n\n<script type=\"text/json\" class=\"json-inline\" data-name=\"ra-urls\">{\"Copyright\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/Copyright.98ac6609.js\",\"PayUIApp\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/PayUIApp.ca78ba78.js\",\"CouponApp\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/CouponApp.8387f93f.js\",\"PaymentApp\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/PaymentApp.7f16d61c.js\",\"Community\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/Community.ea022099.js\",\"Report\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/Report.1a8d7f59.js\",\"OrgOpHelp\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/OrgOpHelp.f891eee7.js\",\"common\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/common.a4dd3236.js\",\"BalanceApp\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/BalanceApp.676e538e.js\",\"AnswerWarrant\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/AnswerWarrant.97f134aa.js\",\"CommentApp\":\"https:\\/\\/static.zhihu.com\\/static\\/revved\\/-\\/apps\\/CommentApp.966880f3.js\"}</script>\n\n<script src=\"https://static.zhihu.com/static/revved/-/js/vendor.cb14a042.js\"></script>\n<script src=\"https://static.zhihu.com/static/revved/-/js/closure/base.e0f055ab.js\"></script>\n\n<script src=\"https://static.zhihu.com/static/revved/-/js/closure/common.e4a2c5cd.js\"></script>\n\n\n<script src=\"https://static.zhihu.com/static/revved/-/js/closure/richtexteditor.5a4142fb.js\" async></script>\n<script src=\"https://static.zhihu.com/static/revved/-/js/closure/page-main.8a0625b5.js\"></script>\n<meta name=\"entry\" content=\"ZH.entryExplore\" data-module-id=\"page-main\">\n\n<script type=\"text/zscript\" znonce=\"12ac55e2aba64796b10dba0c7c53d846\"></script>\n\n<input type=\"hidden\" name=\"_xsrf\" value=\"605638cde53456f4a8ddcecd4b06ba35\"/>\n</body>\n</html>\n"
]
],
[
[
"## 基本POST請求",
"_____no_output_____"
]
],
[
[
"import requests\n\ndata = {\n 'name':'germany',\n 'age':22\n}\n\n#直接傳送一個字典\nresponse = requests.post(\"http://httpbin.org/post\", data=data)\nprint(response.text)",
"{\n \"args\": {}, \n \"data\": \"\", \n \"files\": {}, \n \"form\": {\n \"age\": \"22\", \n \"name\": \"germany\"\n }, \n \"headers\": {\n \"Accept\": \"*/*\", \n \"Accept-Encoding\": \"gzip, deflate\", \n \"Connection\": \"close\", \n \"Content-Length\": \"19\", \n \"Content-Type\": \"application/x-www-form-urlencoded\", \n \"Host\": \"httpbin.org\", \n \"User-Agent\": \"python-requests/2.14.2\"\n }, \n \"json\": null, \n \"origin\": \"140.112.73.184\", \n \"url\": \"http://httpbin.org/post\"\n}\n\n"
],
[
"import requests\n\ndata = {\n 'name':'germany',\n 'age':22\n}\n\nheaders = {\n \"User-Agent\": \"Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/59.0.3071.115 Safari/537.36\"\n}\n\n#直接傳送一個字典\nresponse = requests.post(\"http://httpbin.org/post\", data=data, headers=headers)\nprint(response.json())",
"{'args': {}, 'data': '', 'files': {}, 'form': {'age': '22', 'name': 'germany'}, 'headers': {'Accept': '*/*', 'Accept-Encoding': 'gzip, deflate', 'Connection': 'close', 'Content-Length': '19', 'Content-Type': 'application/x-www-form-urlencoded', 'Host': 'httpbin.org', 'User-Agent': 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/59.0.3071.115 Safari/537.36'}, 'json': None, 'origin': '140.112.73.184', 'url': 'http://httpbin.org/post'}\n"
]
],
[
[
"# response",
"_____no_output_____"
],
[
"## response屬性",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get(\"http://www.jianshu.com\")\nprint(type(response.status_code), response.status_code)\nprint(type(response.headers), response.headers)\nprint(type(response.cookies), response.cookies)\nprint(type(response.url), response.url)\nprint(type(response.history), response.history)",
"<class 'int'> 200\n<class 'requests.structures.CaseInsensitiveDict'> {'Server': 'nginx', 'Date': 'Mon, 10 Jul 2017 04:40:48 GMT', 'Content-Type': 'text/html; charset=utf-8', 'Transfer-Encoding': 'chunked', 'Connection': 'keep-alive', 'Vary': 'Accept-Encoding', 'X-Frame-Options': 'DENY', 'X-XSS-Protection': '1; mode=block', 'X-Content-Type-Options': 'nosniff', 'ETag': 'W/\"6af04d0add6ae79adc39a8b997c90c99\"', 'Cache-Control': 'max-age=0, private, must-revalidate', 'Set-Cookie': '_session_id=VkVheVg3bW05RUFSZllRTGNxQ0NEdVVRWG94UkZocG0rb2dwRkNtQWNDUVloTmh6T2FFZ0JnRkdJcXgvbWMyYlcxSGxwUVdzeW9mcDE2Zmh0ZHFjMFUwVmZNM2VrRW9sT05Bd2RMcmVkc0FodkNiYldEWDFpQmxyV2lZVC93V0l1T2xVNEg2K2I0QTZBRHRURWlkQ0l2WE1ZalMrUHhiaDB0NUw0ZEoxUXNwMzc2aHl1OUlHYmZyUjVyc2xkLzJsVnRFNmFlZmlYSVNPbDBveXJBTW1TVGV4aURPUGRLeHE1dnB2ZVlaeEJENTRMRllEOEN6NTNkb3VTZFMxditLMC0tcVRkNXZDM2I1WnFkTitUQy8ydnBSZz09--c75df867640c03a590153c591f32c528e32e8104; path=/; HttpOnly', 'X-Request-Id': '490f2cfb-94aa-43e8-81f5-2091cd44e3f7', 'X-Runtime': '0.006571', 'Content-Encoding': 'gzip'}\n<class 'requests.cookies.RequestsCookieJar'> <RequestsCookieJar[<Cookie _session_id=VkVheVg3bW05RUFSZllRTGNxQ0NEdVVRWG94UkZocG0rb2dwRkNtQWNDUVloTmh6T2FFZ0JnRkdJcXgvbWMyYlcxSGxwUVdzeW9mcDE2Zmh0ZHFjMFUwVmZNM2VrRW9sT05Bd2RMcmVkc0FodkNiYldEWDFpQmxyV2lZVC93V0l1T2xVNEg2K2I0QTZBRHRURWlkQ0l2WE1ZalMrUHhiaDB0NUw0ZEoxUXNwMzc2aHl1OUlHYmZyUjVyc2xkLzJsVnRFNmFlZmlYSVNPbDBveXJBTW1TVGV4aURPUGRLeHE1dnB2ZVlaeEJENTRMRllEOEN6NTNkb3VTZFMxditLMC0tcVRkNXZDM2I1WnFkTitUQy8ydnBSZz09--c75df867640c03a590153c591f32c528e32e8104 for www.jianshu.com/>]>\n<class 'str'> http://www.jianshu.com/\n<class 'list'> []\n"
]
],
[
[
"## status_code 判斷",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get(\"http://www.jianshu.com\")\n\nif not response.status_code == requests.codes.ok :\n exit() \nelse: \n print('Request successfully')",
"Request successfully\n"
],
[
"import requests\n\nresponse = requests.get(\"http://www.jianshu.com\")\n\nif not response.status_code == 200 :\n exit() \nelse: \n print('Request successfully')",
"Request successfully\n"
]
],
[
[
"status_code 的意義",
"_____no_output_____"
]
],
[
[
"1xx - 參考資訊 (Informational)\n這些狀態碼代表主機先暫時回應用戶端一個狀態,所以在接收一般的回應之前,用戶端應準備接收一個或多個 1xx 的回應。我以前在寫 ASP 的時候比較有看到 IIS 使用到這些狀態碼回應,在 Apache 的環境我還未曾遇到過。 \n100 - 繼續。 \n101 - 切換通訊協定。\n\n2xx - 成功 (OK)\n這類的狀態碼表示伺服器成功接收到用戶端要求、理解用戶端要求、以及接受用戶端要求。 \n200 - 確定。 用戶端要求成功。 \n201 - 已建立。 \n202 - 已接受。 \n203 - 非授權資訊。 \n204 - 無內容。 \n205 - 重設內容。 \n206 - 部分內容。 \n207 - 多重狀態 (WebDAV) -- 這好像只有在 IIS 中才有,HTTP/1.1 並沒有定義這個狀態。這狀態會出現在可以包含多個不同回應代碼 (視子要求數量而定) 的 XML 訊息之前。\n\n3xx - 重新導向 (Redirection)\n用戶端瀏覽器必須採取更多動作才能完成要求。例如:瀏覽器可能必須重新發出 HTTP Request 要求伺服器上的不同頁面。 \n301 - 要求的網頁已經永久改變網址。此狀態要求用戶端未來在連結此網址時應該導向至指定的 URI。 \n302 - 物件已移動,並告知移動過去的網址。針對表單架構驗證,這通常表示為「物件已移動」。 要求的資源暫時存於不同的 URI 底下。 由於重新導向可能偶而改變,用戶端應繼續使用要求 URI 來執行未來的要求。 除非以 Cache-Control 或 Expires 標頭欄位表示,此回應才能夠快取。 \nASP.NET 預設的 Response.Redirect 方法,就是以 302 Found 做回應。 \n303 - 通知 Client 連到另一個網址去查看上傳表單的結果(POST 變成 GET),當使用程式作網頁轉向時,會回應此訊息。\n在 ASP.NET 中要輸出 HTTP 303 轉向的程式碼如下: \nResponse.StatusCode = 303;\nResponse.RedirectLocation = \"/PageB.aspx\";\n304 - 未修改。用戶端要求該網頁時,其內容並沒有變更,應該回傳 304 告知網頁未修改。此時用戶端僅需要取得本地快取(Local Cache)的副本即可。 \n305 - 要求的網頁必須透過 Server 指定的 proxy 才能觀看 ( 透過 Location 標頭 ) \n306 - (未使用) 此代碼僅用來為了向前相容而已。 \n307 - 暫時重新導向。要求的網頁只是「暫時」改變網址而已。\n\n4xx - 用戶端錯誤 (Client Error)\n這代表錯誤發生,且這錯誤的發生的原因跟「用戶端」有關。例如:用戶端可能連結到不存在的頁面、用戶端的權限不足、或可能未提供有效的驗證資訊(輸入的帳號、密碼錯誤)。下次看到 4xx 的回應千萬不要傻傻的一直查程式哪裡寫錯誤了(不過也有可能是程式造成的)。 \n400 - 錯誤的要求。 \n401 - 拒絕存取。 IIS 定義數個不同的 401 錯誤,以表示更詳細的錯誤原因。 這些特定的錯誤碼會顯示在瀏覽器中,但不會顯示在 IIS 記錄檔中: \n401.1 - 登入失敗。 \n401.2 - 因為伺服器設定導致登入失敗。 \n401.3 - 因為資源上的 ACL 而沒有授權。 \n401.4 - 篩選授權失敗。 \n401.5 - ISAPI/CGI 應用程式授權失敗。 \n401.7 - Web 伺服器上的 URL 授權原則拒絕存取。 這是 IIS 6.0 專用的錯誤碼。\n403 - 禁止使用。 IIS 定義數個不同的 403 錯誤,以表示更詳細的錯誤原因: \n403.1 - 禁止執行存取。 \n403.2 - 禁止讀取存取。 \n403.3 - 禁止寫入存取。 \n403.4 - 需要 SSL。 \n403.5 - 需要 SSL 128。 \n403.6 - IP 位址遭拒。 \n403.7 - 需要用戶端憑證。 \n403.8 - 網站存取遭拒。 \n403.9 - 使用者過多。 \n403.10 - 設定無效。 \n403.11 - 密碼變更。 \n403.12 - 對應程式拒絕存取。 \n403.13 - 用戶端憑證已撤銷。 \n403.14 - 目錄清單遭拒。 \n403.15 - 超過用戶端存取授權數量。 \n403.16 - 用戶端憑證不受信任或無效。 \n403.17 - 用戶端憑證已經過期或尚未生效。 \n403.18 - 無法在目前的應用程式集區中執行要求的 URL。 這是 IIS 6.0 專用的代碼。 \n403.19 - 無法在這個應用程式集區中執行用戶端的 CGI。 這是 IIS 6.0 專用的代碼。 \n403.20 - Passport 登入失敗。 這是 IIS 6.0 專用的錯誤碼。\n404 - 找不到。 \n404.0 - (無) – 找不到檔案或目錄。 \n404.1 - 無法在要求的連接埠上存取網站。 \n404.2 - 網頁服務延伸鎖定原則阻止這個要求。 \n404.3 - MIME 對應原則阻止這個要求。\n405 - 用來存取這個頁面的 HTTP 動詞不受允許 (方法不受允許)。 \n406 - 用戶端瀏覽器不接受要求頁面的 MIME 類型。 \n407 - 需要 Proxy 驗證。 \n412 - 指定條件失敗。 \n413 - 要求的實體太大。 \n414 - 要求 URI 太長。 \n415 - 不支援的媒體類型。 \n416 - 無法滿足要求的範圍。 \n417 - 執行失敗。 \n423 - 鎖定錯誤。\n\n5xx - 伺服器錯誤 (Server Error)\n這代表錯誤發生,且這錯誤發生的原因跟「伺服器」有關。伺服器因為發生錯誤或例外狀況(Exception)而無法完成要求(Request)時,就會回應 5xx 的錯誤,且這肯定跟伺服器有關。 \n500 - 內部伺服器錯誤。 \n500.12 - 應用程式正忙於在 Web 伺服器上重新啟動。 \n500.13 - Web 伺服器過於忙碌。 \n500.15 - 不允許直接要求 Global.asa。 \n500.16 – UNC 授權認證不正確。 這是 IIS 6.0 專用的錯誤碼。 \n500.18 – 無法開啟 URL 授權存放區。 這是 IIS 6.0 專用的錯誤碼。 \n500.19 - 此檔案的資料在 Metabase 中設定不當。 \n500.100 - 內部的 ASP 錯誤。\n501 – 標頭值指定未實作的設定。 \n502 - Web 伺服器在作為閘道或 Proxy 時收到無效的回應。 \n502.1 - CGI 應用程式逾時。 \n502.2 - CGI 應用程式中發生錯誤。\n503 - 服務無法使用。 這是 IIS 6.0 專用的錯誤碼。 \n504 - 閘道逾時。 \n505 - 不支援的 HTTP 版本。",
"_____no_output_____"
]
],
[
[
"# 高級操作",
"_____no_output_____"
],
[
"## 文件上傳: files參數",
"_____no_output_____"
]
],
[
[
"import requests\n\nfiles = {'file': open('favicon.ico','rb')}\nresponse = requests.post('http://httpbin.org/post', files=files)\nprint(response.text)",
"{\n \"args\": {}, \n \"data\": \"\", \n \"files\": {\n \"file\": \"data:application/octet-stream;base64,AAABAAIAEBAAAAEAIAAoBQAAJgAAACAgAAABACAAKBQAAE4FAAAoAAAAEAAAACAAAAABACAAAAAAAAAFAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABERE3YTExPFDg4OEgAAAAAAAAAADw8PERERFLETExNpAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABQUFJYTExT8ExMU7QAAABkAAAAAAAAAAAAAABgVFRf/FRUX/xERE4UAAAAAAAAAAAAAAAAAAAAAAAAAABERE8ETExTuERERHg8PDxAAAAAAAAAAAAAAAAAAAAANExMU9RUVF/8VFRf/EhIUrwAAAAAAAAAAAAAAABQUFJkVFRf/BQURLA0NDVwODg/BDw8PIgAAAAAAAAAADg4ONBAQEP8VFRf/FRUX/xUVF/8TExOPAAAAAA8PDzAPDQ//AAAA+QEBAe0CAgL/AgIC9g0NDTgAAAAAAAAAAAcHB0ACAgLrFRUX/xUVF/8VFRf/FRUX/xERES0TExacFBQV/wEBAfwPDxH7DAwROwAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA0NEToTExTnFRUX/xUVF/8TExOaExMT2RUVF/8VFRf/ExMTTwAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAEBAQTBUVF/8VFRf/ExMT2hMTFPYVFRf/FBQU8AAAAAIAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAITExTxFRUX/xMTFPYTExT3FRUX/xQUFOEAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAFBQU4RUVF/8TExT3ExMU3hUVF/8TExT5Dw8PIQAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAEBAQHxMTFPgVFRf/ExMU3hERFKIVFRf/FRUX/w4ODjQAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA8PD0AVFRf/FRUX/xERFKINDQ04FRUX/xUVF/8SEhKYAAAAAAAAAAwAAAAKAAAAAAAAAAAAAAAMAAAAAQAAAAASEhKYFRUX/xUVF/8NDQ04AAAAABERFKQVFRf/ERETwQ4ODjYAAACBDQ0N3BISFNgSEhTYExMU9wAAAHQEBAQ3ERETwRUVF/8RERSkAAAAAAAAAAAAAAADExMTxhUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8TExPGAAAAAwAAAAAAAAAAAAAAAAAAAAMRERSiFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8RERSiAAAAAwAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABAQED4SEhKXExMT2RISFPISEhTyExMT2RISEpcQEBA+AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAoAAAAIAAAAEAAAAABACAAAAAAAAAUAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAwRERNzExMT2hMTFOwAAAAHAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABxMTFOwTExPaERETdAAAAAwAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAERERRkExMU6hUVF/8VFRf/FRUX/w8PDxAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAPDw8QFRUX/xUVF/8VFRf/ExMU6xERFGUAAAABAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAODg4SExMTtxUVF/8VFRf/FRUX/xUVF/8VFRf/Dw8PEAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA8PDxAVFRf/FRUX/xUVF/8VFRf/FRUX/xMTE7cODg4SAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAEBAQHxMTFNsVFRf/FRUX/xQUFMMRERN1Dw8PYBMTE3gAAAAIAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAADw8PEBUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xMTFNsQEBAfAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABgTExTcFRUX/xUVF/8SEhJvAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAPDw8QFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xMTFNwAAAAYAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAEExMTxBUVF/8VFRf/ExMUuQAAAAAPDw8QDw8PYxISEnoODg5GAAAAAwAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAwMDBUVFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xMTE8QAAAAEAAAAAAAAAAAAAAAAAAAAABISEn4VFRf/FRUX/xUVF/8NDQ04Dw8PIRMTE+IVFRf/FRUX/xUVF/8RERE8AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAEBAQPhUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xISEn4AAAAAAAAAAAAAAAAREREeExMU9xUVF/8TExT+ERETcwAAAAcTExTJFRUX/xUVF/8VFRf/FRUX/xMTFK4AAAABAAAAAAAAAAAAAAAAAAAAAAAAAAERERSwFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/ExMU9xERER4AAAAAAAAAABISEpcVFRf/FRUX/xISEooQEBA/ERETwhUVF/8VFRf/ExMU+hMTFqoRERRlDg4ONAAAAAMAAAAAAAAAAAAAAAAAAAAAAAAABA0NETkODhNoExMUrhMTFPoVFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/EhISlwAAAAAAAAANExMU9RUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xMTFKsAAAAYAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAGRMTFKsVFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8TExT1AAAADQ4OFFkVFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8TExOPAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABMTE48VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8ODhRZExMTnRUVF/8VFRf/FRUX/xUVF/8VFRf/EREU0QAAAAQAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABBERFNEVFRf/FRUX/xUVF/8VFRf/FRUX/xMTE50RERTQFRUX/xUVF/8VFRf/FRUX/xUVF/8SEhJeAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAEhISXhUVF/8VFRf/FRUX/xUVF/8VFRf/EREU0BISFPIVFRf/FRUX/xUVF/8VFRf/FRUX/wAAABYAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAWFRUX/xUVF/8VFRf/FRUX/xUVF/8SEhTyFRUX/xUVF/8VFRf/FRUX/xUVF/8SEhTyAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAASEhTyFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xMTFNsAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABMTFNwVFRf/FRUX/xUVF/8VFRf/FRUX/xMTFPYVFRf/FRUX/xUVF/8VFRf/FBQU4QAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAFBQU4RUVF/8VFRf/FRUX/xUVF/8TExT2ExMU1hUVF/8VFRf/FRUX/xUVF/8TExT8ERERDwAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABEREQ8TExT8FRUX/xUVF/8VFRf/FRUX/xMTFNYTExOpFRUX/xUVF/8VFRf/FRUX/xUVF/8PDw9iAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAADw8PYhUVF/8VFRf/FRUX/xUVF/8VFRf/ExMTqQ4OE2cVFRf/FRUX/xUVF/8VFRf/FRUX/xMTFuMODg4SAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA4ODhITExPiFRUX/xUVF/8VFRf/FRUX/xUVF/8ODhNnAAAAGBMTFPwVFRf/FRUX/xUVF/8VFRf/FRUX/xISEl8AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAEhISXxUVF/8VFRf/FRUX/xUVF/8VFRf/ExMU/AAAABgAAAAAExMUrhUVF/8VFRf/FRUX/xUVF/8VFRf/Dg4ONQAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAODg41FRUX/xUVF/8VFRf/FRUX/xUVF/8TExSuAAAAAAAAAAAODg40FRUX/xUVF/8VFRf/FRUX/xUVF/8PDw8yAAAAAAAAAAAAAAAAERERDwwMDCgAAAACAAAAAAAAAAAAAAAAAAAAAAAAAAIMDAwoERERDwAAAAAAAAAAAAAAAA8PDzIVFRf/FRUX/xUVF/8VFRf/FRUX/w4ODjQAAAAAAAAAAAAAAAATExSeFRUX/xUVF/8VFRf/FRUX/xMTE1wAAAAAAAAABw8PD2MTExToFRUX/xMTFPMUFBTSERETwRERE8EUFBTSExMU8xUVF/8TExToDw8PYwAAAAcAAAAAExMTXBUVF/8VFRf/FRUX/xUVF/8TExSeAAAAAAAAAAAAAAAAAAAAAA8PDxETExTfFRUX/xUVF/8VFRf/ExMU1hMTFK0TExTxFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/ExMU8RMTFK0TExTWFRUX/xUVF/8VFRf/ExMU3w8PDxEAAAAAAAAAAAAAAAAAAAAAAAAAAA8PDzMTExTxFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xMTFPEPDw8zAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA8PD0ITExTxFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8TExTxDw8PQgAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA8PDzETExTeFRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/ExMU3g8PDzEAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAABEREQ8TExObExMU/hUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/ExMU/hMTE5sREREPAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAPDw8xExMTqRMTFPsVFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/FRUX/xUVF/8VFRf/ExMU+xMTE6kPDw8xAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAADAwMFA8PD2MRERSkFBQU0hMTFPMVFRf/FRUX/xMTFPMUFBTSEREUpA8PD2MMDAwUAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA=\"\n }, \n \"form\": {}, \n \"headers\": {\n \"Accept\": \"*/*\", \n \"Accept-Encoding\": \"gzip, deflate\", \n \"Connection\": \"close\", \n \"Content-Length\": \"6665\", \n \"Content-Type\": \"multipart/form-data; boundary=549d4ce2ea40473ebae935e34c515925\", \n \"Host\": \"httpbin.org\", \n \"User-Agent\": \"python-requests/2.14.2\"\n }, \n \"json\": null, \n \"origin\": \"140.112.73.184\", \n \"url\": \"http://httpbin.org/post\"\n}\n\n"
]
],
[
[
"## 獲取cookies",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('http://www.baidu.com')\nprint(type(response.cookies))\nprint(response.cookies)\n\nfor key,value in response.cookies.items():\n print(key+\" = \"+value)",
"<class 'requests.cookies.RequestsCookieJar'>\n<RequestsCookieJar[<Cookie BDORZ=27315 for .baidu.com/>]>\nBDORZ = 27315\n"
]
],
[
[
"## 會話維持: requests.Session()",
"_____no_output_____"
],
[
"模擬登入",
"_____no_output_____"
]
],
[
[
"import requests\n\n#獨立瀏覽器 1\nrequests.get('https://httpbin.org/cookies/set/number/1234')\n\n#獨立瀏覽器 2\nresponse = requests.get('https://httpbin.org/cookies')\n\n#得不到我們要的cookies\nprint(response.text)",
"{\n \"cookies\": {}\n}\n\n"
],
[
"import requests\n\n#想像成把兩個事情在同一個瀏覽器完成\ns = requests.Session()\ns.get('https://httpbin.org/cookies/set/number/1234')\nresponse = s.get('https://httpbin.org/cookies')\n\nprint(response.text)",
"{\n \"cookies\": {\n \"number\": \"1234\"\n }\n}\n\n"
]
],
[
[
"## 證書驗證",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('https://www.12306.cn')\nprint(reponse.status_code)\n#網站證書不合法",
"_____no_output_____"
],
[
"import requests\n\nresponse = requests.get('https://www.12306.cn', verify=False)\nprint(response.status_code)\n#網站證書不合法",
"200\n"
],
[
"import requests\n\n#去除網站證書不合法警告\nfrom requests.packages import urllib3\nurllib3.disable_warnings()\n\nresponse = requests.get('https://www.12306.cn', verify=False)\nprint(response.status_code)",
"200\n"
]
],
[
[
"## 代理設置 VPN",
"_____no_output_____"
]
],
[
[
"import requests\n\nproxies = {\n 'http':'http://127.0.0.1:9743'\n 'https':'https://127.0.0.1:9743'\n}\n\nresponse = requests.get('http://www.baidu.com', proxies = proxies)\nprint(response.status_code)",
"_____no_output_____"
],
[
"import requests\n\n#proxy有密碼\nproxies = {\n 'http':'http://user:[email protected]:9743'\n}\n\nresponse = requests.get('http://www.baidu.com', proxies = proxies)\nprint(response.status_code)",
"_____no_output_____"
]
],
[
[
"## 設置超時",
"_____no_output_____"
]
],
[
[
"import requests\nfrom requests.exceptions import ReadTimeout\n\ntry:\n response = requests.get('https://www.baidu.com', timeout = 0.1)\n print(reponse.status_code)\n\nexcept ReadTimeout:\n print('Time out!!')",
"Time out!!\n"
]
],
[
[
"## 認證設置 ",
"_____no_output_____"
]
],
[
[
"import requests\nfrom requests.auth import HTTPBasicAuth\n\n#沒有帳號密碼會被禁止請求\nr = requests.get('http://120.27.34.24:9001')\nprint(r.status_code)",
"401\n"
],
[
"import requests\nfrom requests.auth import HTTPBasicAuth\n\nr = requests.get('http://120.27.34.24:9001', auth=HTTPBasicAuth('user','123'))\nprint(r.status_code)",
"200\n"
],
[
"import requests\nfrom requests.auth import HTTPBasicAuth\n\nr = requests.get('http://120.27.34.24:9001', auth=('user','123'))\nprint(r.status_code)",
"200\n"
]
],
[
[
"## 異常處理,可以保證程式不要被中斷的運行",
"_____no_output_____"
]
],
[
[
"import requests\nfrom requests.exceptions import ReadTimeout, HTTPError, RequestException\n\ntry:\n response = requests.get('https://www.baidu.com', timeout = 0.1)\n print(response.status_code)\n\nexcept ReadTimeout:\n print('Timeout')\nexcept HTTPError:\n print('Http error')\nexcept RequestException:\n print('Request error')",
"Timeout\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71b43e84c066739ac91453f2dc8c4e2e4fbbb1c | 58,487 | ipynb | Jupyter Notebook | action_recognition_pipeline.ipynb | Solvve/action_recognition | 2f5cef469b62ef17b3379637293924f2fddbc08c | [
"MIT"
] | 6 | 2021-01-11T16:47:16.000Z | 2021-07-26T11:46:58.000Z | action_recognition_pipeline.ipynb | Solvve/action_recognition | 2f5cef469b62ef17b3379637293924f2fddbc08c | [
"MIT"
] | 3 | 2020-11-04T06:51:54.000Z | 2021-11-28T12:45:03.000Z | action_recognition_pipeline.ipynb | Solvve/action_recognition | 2f5cef469b62ef17b3379637293924f2fddbc08c | [
"MIT"
] | 3 | 2021-06-25T01:02:20.000Z | 2022-03-02T16:29:39.000Z | 58,487 | 58,487 | 0.808419 | [
[
[
"## Description",
"_____no_output_____"
],
[
"This is an example of action recognition pipeline trained on [UCF-Crime dataset](https://www.crcv.ucf.edu/projects/real-world/) to detect anomaly behavior among next classes: 'Normal', 'Fighting', 'Robbery', 'Shoplifting', 'Stealing'.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade mxnet-cu100 gluoncv",
"_____no_output_____"
],
[
"!pip install decord",
"_____no_output_____"
],
[
"import mxnet as mx\nfrom mxnet import nd, gluon\nfrom mxnet import autograd as ag\n\nimport gluoncv\nfrom gluoncv.data import VideoClsCustom\nfrom gluoncv.data.transforms import video\nfrom gluoncv.model_zoo import get_model\nfrom gluoncv.utils import split_and_load, TrainingHistory\n\nimport decord\n\nfrom sklearn import metrics\n\nimport cv2\nimport numpy as np\nimport os\nimport shutil\nimport time\nimport tqdm\n\nimport matplotlib.pyplot as plt\nimport seaborn as sn",
"_____no_output_____"
],
[
"print(f'''Versions:\nmxnet: {mx.__version__}\ndecord: {decord.__version__}\ngluoncv: {gluoncv.__version__}\n''')",
"Versions:\nmxnet: 1.5.1\ndecord: 0.4.0\ngluoncv: 0.7.0\n\n"
],
[
"dataset_path = './dataset' # path to dataset\nmodels_path = './models' # path to model weights",
"_____no_output_____"
],
[
"num_gpus = 1\nctx = [mx.gpu(i) for i in range(num_gpus)]",
"_____no_output_____"
],
[
"classes = ['Normal', 'Fighting', 'Robbery', 'Shoplifting', 'Stealing']\n\nnum_segments = 4\nnum_frames = 48\n\nper_device_batch_size = 3\nnum_workers = 4\nbatch_size = num_gpus*per_device_batch_size",
"_____no_output_____"
]
],
[
[
"## Train",
"_____no_output_____"
]
],
[
[
"transform_train = video.VideoGroupTrainTransform(size=(224, 224), scale_ratios=[1.0, 0.85, 0.75], mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])\ntransform_valid = video.VideoGroupValTransform(size=224, mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])\n\ntrain_dataset = VideoClsCustom(root=dataset_path,\n setting=f'{dataset_path}/train_anomaly_detection.txt',\n train=True,\n new_length=num_frames,\n num_segments=num_segments,\n transform=transform_train,\n video_ext='mp4',\n video_loader=True,\n use_decord=True\n )\nprint('Load %d training samples.' % len(train_dataset))\ntrain_data = gluon.data.DataLoader(train_dataset, batch_size=batch_size,\n shuffle=True, num_workers=num_workers)\n\nvalid_dataset = VideoClsCustom(root=dataset_path,\n setting=f'{dataset_path}/valid_anomaly_detection.txt',\n train=False,\n new_length=num_frames,\n num_segments=num_segments,\n transform=transform_valid,\n video_ext='mp4',\n video_loader=True,\n use_decord=True)\nprint('Load %d valid samples.' % len(valid_dataset))\nvalid_data = gluon.data.DataLoader(valid_dataset, batch_size=batch_size,\n shuffle=True, num_workers=num_workers)",
"Load 451 training samples.\nLoad 219 valid samples.\n"
],
[
"net = get_model(name='slowfast_4x16_resnet50_custom', nclass=len(classes), num_segments=num_segments, ctx=ctx)\n\n# Learning rate decay factor\nlr_decay = 0.1\n# Epochs where learning rate decays\n\nlr_decay_epoch = [50, 80]\n\n# Stochastic gradient descent\noptimizer = 'sgd'\n# Set parameters\n\noptimizer_params = {'learning_rate': 0.0001, 'wd': 1e-5, 'momentum': 0.9}\n\n# Define our trainer for net\ntrainer = gluon.Trainer(net.collect_params(), optimizer, optimizer_params)\n\nloss_fn = gluon.loss.SoftmaxCrossEntropyLoss()\n\ntrain_metric = mx.metric.Accuracy()\nvalid_metric = mx.metric.Accuracy()\ntrain_history = TrainingHistory(['training-acc'])\nvalid_history = TrainingHistory(['valid-acc'])",
"_____no_output_____"
],
[
"epochs = 65\nlr_decay_count = 0\nvalid_loss_best = 1000\nvalid_acc_best = 10\nhist_prec_train = []\nhist_prec_test = []\nhist_recall_train = []\nhist_recall_test = []\nhist_loss = []\nhist_loss_valid = []\n\nfor epoch in range(epochs):\n tic = time.time()\n train_metric.reset()\n valid_metric.reset()\n train_loss = 0\n valid_loss = 0\n\n # Learning rate decay\n if epoch == lr_decay_epoch[lr_decay_count]:\n trainer.set_learning_rate(trainer.learning_rate*lr_decay)\n lr_decay_count += 1\n\n # Loop through each batch of training data\n y_true = np.array([], dtype='int')\n y_pred = np.array([], dtype='int')\n for i, batch in enumerate(train_data):\n # Extract data and label\n data = split_and_load(batch[0], ctx_list=ctx, batch_axis=0)\n label = split_and_load(batch[1], ctx_list=ctx, batch_axis=0)\n\n # AutoGrad\n with ag.record():\n output = []\n for _, X in enumerate(data):\n X = X.reshape((-1,) + X.shape[2:])\n pred = net(X)\n output.append(pred)\n\n loss = [loss_fn(yhat, y) for yhat, y in zip(output, label)]\n\n # Backpropagation\n for l in loss:\n l.backward()\n\n # Optimize\n trainer.step(batch_size)\n\n # Update metrics\n train_loss += sum([l.mean().asscalar() for l in loss])\n train_metric.update(label, output)\n\n y_true = np.concatenate((y_true, label[0].asnumpy()))\n y_pred = np.concatenate((y_pred, pred.argmax(axis=1).astype('int').asnumpy()))\n\n name, acc = train_metric.get()\n precisions = metrics.precision_score(y_true, y_pred, average=None, zero_division=False)\n recall = metrics.recall_score(y_true, y_pred, average=None, zero_division=False)\n\n # Update history and print metrics\n train_history.update([acc])\n print(f'[Epoch {epoch}] train={acc:.4f} loss={train_loss/(i+1):.4f} time: {time.time()-tic:.1f} sec')\n print('Train precision: ',{k:v for k,v in zip(classes, precisions)})\n print('Train recall: ',{k:v for k,v in zip(classes, recall)})\n hist_loss.append(train_loss/(i+1))\n hist_prec_train.append({k:v for k,v in zip(classes, precisions)})\n hist_recall_train.append({k:v for k,v in zip(classes, recall)})\n\n y_true_v = np.array([], dtype='int')\n y_pred_v = np.array([], dtype='int')\n for i, batch in enumerate(valid_data):\n # Extract data and label\n data = split_and_load(batch[0], ctx_list=ctx, batch_axis=0)\n label = split_and_load(batch[1], ctx_list=ctx, batch_axis=0)\n\n output = []\n for _, X in enumerate(data):\n X = X.reshape((-1,) + X.shape[2:])\n pred = net(X)\n output.append(pred)\n loss = [loss_fn(yhat, y) for yhat, y in zip(output, label)]\n\n # Update metrics\n valid_loss += sum([l.mean().asscalar() for l in loss])\n valid_metric.update(label, output)\n y_true_v = np.concatenate((y_true_v, label[0].asnumpy()))\n y_pred_v = np.concatenate((y_pred_v, pred.argmax(axis=1).astype('int').asnumpy()))\n \n name, acc = valid_metric.get()\n precisions_v = metrics.precision_score(y_true_v, y_pred_v, average=None, zero_division=False)\n recall_v = metrics.recall_score(y_true_v, y_pred_v, average=None, zero_division=False)\n\n # Update history and print metrics\n valid_history.update([acc])\n print(f'valid_acc: {acc}, valid_loss: {valid_loss/(i+1)}')\n hist_loss_valid.append(valid_loss/(i+1))\n print(f'valid precision:', {k:v for k,v in zip(classes, precisions_v)})\n print(f'valid recall:', {k:v for k,v in zip(classes, recall_v)})\n hist_prec_test.append({k:v for k,v in zip(classes, precisions_v)})\n hist_recall_test.append({k:v for k,v in zip(classes, recall_v)})\n \n if (valid_loss_best > valid_loss) or (valid_acc_best < acc) :\n valid_loss_best = valid_loss\n valid_acc_best = acc\n print(f'Best valid loss: {valid_loss_best}')\n file_name = f\"{models_path}/slowfast_ucf_{epoch}.params\"\n net.save_parameters(file_name)",
"_____no_output_____"
]
],
[
[
"## Validation",
"_____no_output_____"
]
],
[
[
"net = get_model(name='slowfast_4x16_resnet50_custom', nclass=len(classes), num_segments=num_segments, pretrainded=False, pretrained_base=False, ctx=ctx)\nnet.load_parameters(f'{models_path}/slowfast_ucf.params', ctx=ctx)",
"_____no_output_____"
],
[
"transform_valid = video.VideoGroupValTransform(size=224, mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])\n\n\nvalid_dataset = VideoClsCustom(root=dataset_path,\n setting=f'{dataset_path}/valid_anomaly_detection.txt',\n train=False,\n new_length=num_frames,\n num_segments=num_segments,\n transform=transform_valid,\n video_ext='mp4',\n video_loader=True,\n use_decord=True)\nprint('Load %d valid samples.' % len(valid_dataset))\nvalid_data = gluon.data.DataLoader(valid_dataset, batch_size=batch_size,\n shuffle=True, num_workers=num_workers)",
"Load 219 valid samples.\n"
],
[
"valid_loss = 0\nloss_fn = gluon.loss.SoftmaxCrossEntropyLoss()\ny_true = np.array([],dtype='int')\ny_pred = np.array([],dtype='int')\noutputs = []\nacc = mx.metric.Accuracy()\nfor i, batch in tqdm.tqdm(enumerate(valid_data)):\n # Extract data and label\n data = split_and_load(batch[0], ctx_list=ctx, batch_axis=0)\n label = split_and_load(batch[1], ctx_list=ctx, batch_axis=0)\n\n output = []\n for _, X in enumerate(data):\n X = X.reshape((-1,) + X.shape[2:])\n pred = net(X)\n output.append(pred)\n loss = [loss_fn(yhat, y) for yhat, y in zip(output, label)]\n acc.update(label, output)\n # Update metrics\n valid_loss += sum([l.mean().asscalar() for l in loss])\n y_true = np.concatenate((y_true, label[0].asnumpy()))\n y_pred = np.concatenate((y_pred, pred.argmax(axis=1).astype('int').asnumpy()))\n outputs.append((output,label))",
"_____no_output_____"
],
[
"y_true = np.ravel(np.array(y_true))\ny_pred = np.ravel(np.array(y_pred))",
"_____no_output_____"
]
],
[
[
"Metrics per class",
"_____no_output_____"
]
],
[
[
"precisions = metrics.precision_score(y_true, y_pred, average=None, zero_division=False)\nprint(f'Precision: ', {k:v for k,v in zip(classes, precisions)})\n\nrecalls = metrics.recall_score(y_true, y_pred, average=None, zero_division=False)\nprint(f'Recall: ', {k:v for k,v in zip(classes, recalls)})",
"Precision: {'Normal': 0.7522935779816514, 'Fighting': 0.75, 'Robbery': 0.03125, 'Shoplifting': 0.8636363636363636, 'Stealing': 0.5}\nRecall: {'Normal': 0.82, 'Fighting': 0.6, 'Robbery': 0.2, 'Shoplifting': 0.5480769230769231, 'Stealing': 0.8}\n"
],
[
"cm = metrics.confusion_matrix(y_true, y_pred)\nax = sn.heatmap(cm, annot=True, cmap=plt.cm.Blues, xticklabels=classes, yticklabels=classes)\nax.set_title('Valid confusion matrix')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Normal vs Anomalies",
"_____no_output_____"
]
],
[
[
"precisions = metrics.precision_score(y_true>0, y_pred>0)\nprint(f'Precision 2 classes: {precisions:.4f}')\n\nrecalls = metrics.recall_score(y_true>0, y_pred>0)\nprint(f'Recall 2 classes: {recalls:.4f}')",
"Precision 2 classes: 0.8364\nRecall 2 classes: 0.7731\n"
],
[
"cm = metrics.confusion_matrix(y_true>0, y_pred>0)\nax = sn.heatmap(cm, annot=True, cmap=plt.cm.Blues, xticklabels=['Normal', 'Anomaly'] , yticklabels=['Normal', 'Anomaly'] )\nax.set_title('Valid confusion matrix')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Inference",
"_____no_output_____"
]
],
[
[
"input_video_path = './test' # path to test video\noutput_video_path = './output' # path to the output results",
"_____no_output_____"
],
[
"transform_fn = video.VideoGroupValTransform(size=224, mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])\n\nnet = get_model(name='slowfast_4x16_resnet50_custom', nclass=len(classes), num_segments=num_segments, pretrainded=False, pretrained_base=False, ctx=ctx)\nnet.load_parameters(f'{models_path}/slowfast_ucf.params', ctx=ctx)",
"_____no_output_____"
],
[
"def process_video(video_name, input_path, output_path, net, transform_fn, classes, num_frames=48, verbose=True):\n '''\n Classify action on each num_frames*num_segments length clip from video and write output video with classification result.\n \n Args: \n video_name: video name\n input_path: path to input folder\n output_path: path to output folder\n net: the net to perform classification\n transform_fn: a function that transforms data\n classes: list of actions can be detected on video\n num_frames: the length of input video clip\n verbose: verbosity level\n\n Returns:\n dict: Classification result for each segment.\n '''\n if verbose:\n print(f'Video: {video_name}')\n vr = decord.VideoReader(f'{input_video_path}/{video_name}')\n segments = split_on_segments(vr, net.num_segments*num_frames)\n anomaly_classes = list(filter(lambda c: c != 'Normal', classes))\n \n temp_output_path = f'{output_path}/{video_name}'\n if not os.path.exists(temp_output_path):\n os.mkdir(temp_output_path)\n \n video_data = None\n results = {}\n\n for i, segment in enumerate(segments): \n video_data = vr.get_batch(segment).asnumpy()\n start_time = time.time()\n pred_class, pred_class_prob = predict(net, video_data, transform_fn, num_frames, classes)\n end_time = time.time()\n results[i] = {'predicted_class': pred_class, 'probability':pred_class_prob}\n\n add_result_on_clip(video_data, pred_class, pred_class_prob, anomaly_classes)\n write_video_output(f'{temp_output_path}/batch_{i:04d}.mp4', video_data)\n\n if verbose:\n print(f'[Segment: {i}] predicted_class: {pred_class}, probability: {pred_class_prob:.4f}, time: {end_time-start_time:0.1f} sec')\n\n if video_data is not None:\n height = video_data.shape[1]\n width = video_data.shape[2] \n merge_clips(temp_output_path, video_name, output_path, (width, height))\n shutil.rmtree(temp_output_path)\n\n return results\n\ndef split_on_segments(vr, segm_length=48, verbose=True):\n '''\n Split video on segments with *segm_length* length.\n Args: \n vr: decode.VideoReader\n segm_length: segment length\n verbose: verbosity level\n Returns:\n list: List of frame indexes, splitted on segments.\n '''\n n_frames = len(vr)\n fps = vr.get_avg_fps()\n duration = n_frames/fps\n all_idx = np.arange(n_frames)\n idx = range(0, len(all_idx), segm_length)\n\n segments = []\n for i in idx:\n segment = all_idx[i:i+segm_length]\n if len(segment) >= segm_length:\n segments.append(segment)\n\n if verbose:\n print(f'[Frames partitioning] total frames: {n_frames}, fps: {fps}, duration: {duration:.1f} sec, split on: {len(segments)} segments, segments length: {[i.shape[0] for i in segments]} frames')\n return segments\n\ndef predict(net, clip, transform_fn, num_frames, classes):\n '''\n Predict action on clip.\n \n Args:\n net: the net to perform classification\n clip: video clip for predicting action on it\n transform_fn: a function that transforms data\n num_frames: the length of input video clip\n classes: list of action that can be detected on video\n\n Returns:\n str, float: Class label with class probability.\n '''\n\n clip_input = transform_fn(clip)\n clip_input = np.stack(clip_input, axis=0)\n clip_input = clip_input.reshape((-1,) + (num_frames, 3, 224, 224))\n clip_input = np.transpose(clip_input, (0, 2, 1, 3, 4))\n \n pred = net(nd.array(clip_input).as_in_context(ctx[0]))\n ind = nd.topk(pred, k=1)[0].astype('int')\n pred_class = classes[ind[0].asscalar()]\n pred_class_prob = nd.softmax(pred)[0][ind[0]].asscalar()\n \n return pred_class, pred_class_prob\n\ndef add_result_on_clip(clip, pred_class, pred_class_prob, anomaly_classes):\n '''\n Add classification result on clip.\n \n Args:\n clip: video clip for adding classification results on it\n pred_class: predicted class\n pred_class_prob: probability of predicted action\n anomaly_classes: list of anomaly actions\n '''\n for frame in clip:\n draw_classification_result(frame, pred_class, pred_class_prob)\n if pred_class in anomaly_classes and pred_class_prob > 0.65:\n draw_alert_mark(frame)\n\ndef draw_alert_mark(frame):\n '''\n Add alert (triangle with \"!\" sign) to mark frame with anomaly.\n \n Args:\n frame: video frame \n '''\n\n pts = np.array([[165,15],[182,45],[148,45]])\n pts = pts.reshape((-1, 1, 2))\n cv2.fillPoly(frame,[pts],(255,0,0))\n cv2.putText(frame,\"!\",(160,42),cv2.FONT_HERSHEY_SIMPLEX,1,(255,255,255),2)\n\ndef draw_classification_result(frame, class_name, prob):\n '''\n Add classification result on frame.\n \n Args:\n frame: video frame\n class_name: predicted class\n prob: probability of predicted class\n '''\n cv2.rectangle(frame, (8, 5), (190, 55), (240, 240, 240), cv2.FILLED)\n\n text_color = (0,0,0)\n cv2.putText(frame,f'class: {class_name}',(10,25),cv2.FONT_HERSHEY_SIMPLEX,0.4,text_color,1)\n cv2.putText(frame,f'probability: {prob:0.4f}',(10,45),cv2.FONT_HERSHEY_SIMPLEX,0.4,text_color,1)\n\ndef write_video_output(output_path, video_data):\n '''\n Write classified video to file.\n \n Args:\n output_path: path to output video file\n video_data: video data that should be saved to file\n '''\n\n if video_data is None:\n print(f'{output_path} can\\'t write file.')\n return\n \n height = video_data.shape[1]\n width = video_data.shape[2]\n \n out = cv2.VideoWriter(f'{output_path}', cv2.VideoWriter_fourcc(*'MP4V'), 30.0, (width, height))\n \n for frame in video_data:\n out.write(cv2.cvtColor(frame, cv2.COLOR_BGR2RGB))\n \n out.release()\n\ndef merge_clips(clips_path, video_name, output_path, video_shape):\n '''\n Merge clips into one video.\n\n Args:\n clips_path: path to clips folder\n video_name: video name\n output_path: path to output folder\n video_shape: (width, height) shape of output video\n '''\n out = cv2.VideoWriter(f'{output_path}/output_{video_name}', cv2.VideoWriter_fourcc(*'MP4V'), 30.0, video_shape)\n clips_list = sorted(os.listdir(clips_path))\n for clip_name in clips_list:\n clip = cv2.VideoCapture(f'{clips_path}/{clip_name}')\n ret, frame = clip.read()\n while(ret):\n out.write(frame)\n ret, frame = clip.read() \n clip.release()\n\n out.release()\n",
"_____no_output_____"
],
[
"video_list = os.listdir(input_video_path)\n\nfor video_name in video_list:\n process_video(video_name, input_video_path, output_video_path, net, transform_fn, classes, num_frames=num_frames, verbose=1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e71b50f908706c8dc5018117ff1635c74e5257e1 | 205,360 | ipynb | Jupyter Notebook | Clustering_the_countries.ipynb | Asif1310/3005 | 7fffbae5821575f63e470b86fcc1dd4afc108cfc | [
"MIT"
] | null | null | null | Clustering_the_countries.ipynb | Asif1310/3005 | 7fffbae5821575f63e470b86fcc1dd4afc108cfc | [
"MIT"
] | null | null | null | Clustering_the_countries.ipynb | Asif1310/3005 | 7fffbae5821575f63e470b86fcc1dd4afc108cfc | [
"MIT"
] | null | null | null | 46.044843 | 14,810 | 0.375078 | [
[
[
"<a href=\"https://colab.research.google.com/github/Asif1310/3005/blob/main/Clustering_the_countries.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#importing necessary modules\nimport numpy as np #array processing\nimport pandas as pd #dataframes\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.cluster import KMeans\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"db1 = pd.read_html('https://www.numbeo.com/crime/rankings_by_country.jsp') #importing crime index statistics\ndb1",
"_____no_output_____"
],
[
"db1 = db1[1:][:] #slicing to remove the first row",
"_____no_output_____"
],
[
"db1",
"_____no_output_____"
],
[
"db1 = np.array(db1) #converting Python list to numpy array\ndb1.shape",
"_____no_output_____"
],
[
"db1 = pd.DataFrame(db1[0]) #converting numpy array to a dataframe",
"_____no_output_____"
],
[
"db1",
"_____no_output_____"
],
[
"db1.drop([0,3], axis =1, inplace = True) #dropping columns with 0 and 3 indices",
"_____no_output_____"
],
[
"db1",
"_____no_output_____"
],
[
"db2 = pd.read_html('https://worldpopulationreview.com/country-rankings/happiest-countries-in-the-world') #importing world happiness index report",
"_____no_output_____"
],
[
"db2 = np.array(db2) #converting python list to numpy array\ndb2.shape",
"_____no_output_____"
],
[
"db2 = pd.DataFrame(db2[0]) #converting numpy array to dataframe\ndb2",
"_____no_output_____"
],
[
"db2.drop([0,3,4], axis = 1, inplace = True) #dropping columns with indices 0,3 & 4\ndb2",
"_____no_output_____"
],
[
"db = pd.DataFrame.merge(db1 , db2 , on = 1 , how = 'inner') #merging two dataframes through an inner join\ndb",
"_____no_output_____"
],
[
"db.info() #check for null values",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 120 entries, 0 to 119\nData columns (total 3 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 1 120 non-null object\n 1 2_x 120 non-null object\n 2 2_y 120 non-null object\ndtypes: object(3)\nmemory usage: 3.8+ KB\n"
],
[
"db.rename(columns={1: 'Country', '2_x': 'crime index','2_y':'happiness index'}, inplace=True) #renaming the column headers of the dataframe\ndb",
"_____no_output_____"
],
[
"db3 = pd.read_html('https://worldpopulationreview.com/country-rankings/most-corrupt-countries') #importing corruption perception indices\n",
"_____no_output_____"
],
[
"db3 = np.array(db3) #converting python list to a numpy array\ndb3 = pd.DataFrame(db3[0]) #converting numpy array to a dataframe\ndb3",
"_____no_output_____"
],
[
"db3.drop(2, axis =1, inplace = True) #dropping column with index 2 from the dataframe\ndb3.rename(columns={0: 'Country', 1: 'corruption perception index'}, inplace=True) #renaming column headers",
"_____no_output_____"
],
[
"db = pd.DataFrame.merge(db , db3 , on = 'Country' , how = 'inner') #merging two dataframes through an inner join",
"_____no_output_____"
],
[
"db",
"_____no_output_____"
],
[
"db4 = pd.read_excel('https://github.com/Asif1310/3005/blob/main/FEMALE%20LITERACY%20FROM%20UNICEF.xlsx?raw=true') #importing the Excel file of youth literacy data from GitHub",
"_____no_output_____"
],
[
"db4.drop(0,axis = 0, inplace = True) #dropping the column with index 0 from dataframe",
"_____no_output_____"
],
[
"db4.dropna(inplace = True) #dropping all null values from dataframe\ndb4.drop('Time period', axis = 1, inplace = True) #dropping the Time period column from dataframe\ndb4.rename(columns={'Geographic area': 'Country', 'Unnamed: 2': 'Female literacy rate among the youth'}, inplace=True) #renaming the column headers of dataframe\ndb4",
"_____no_output_____"
],
[
"db = pd.DataFrame.merge(db , db4 , on = 'Country' , how = 'inner') #merging two dataframes through an inner join",
"_____no_output_____"
],
[
"db",
"_____no_output_____"
],
[
"data = db.iloc[:,1:] #separating the numeric columns of the dataframe from the country names\ndata",
"_____no_output_____"
],
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 63 entries, 0 to 62\nData columns (total 4 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 crime index 63 non-null object \n 1 happiness index 63 non-null object \n 2 corruption perception index 63 non-null object \n 3 Female literacy rate among the youth 63 non-null float64\ndtypes: float64(1), object(3)\nmemory usage: 2.5+ KB\n"
],
[
"data = data.astype('float64') #converting the data in all the columns of dataframe to float64 datatype\ndata.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 63 entries, 0 to 62\nData columns (total 4 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 crime index 63 non-null float64\n 1 happiness index 63 non-null float64\n 2 corruption perception index 63 non-null float64\n 3 Female literacy rate among the youth 63 non-null float64\ndtypes: float64(4)\nmemory usage: 2.5 KB\n"
],
[
"#feature scaling \nsc = StandardScaler() \ndata = sc.fit_transform(data)\ndata",
"_____no_output_____"
],
[
"metrics = [] #initializing a blank list\ntest_no_clusters = 6 \nfor num in range(1, test_no_clusters+1):\n kmeans = KMeans(n_clusters = num, init = 'k-means++', random_state = 32)\n kmeans.fit(data)\n metrics.append(kmeans.inertia_)",
"_____no_output_____"
],
[
"metrics",
"_____no_output_____"
],
[
"#elbow method to find the optimal number of clusters\nplt.plot(range(1, test_no_clusters+1), metrics)\nplt.xlabel('number of clusters')\nplt.ylabel('WCSS metric')",
"_____no_output_____"
],
[
"kmeans = KMeans(n_clusters = 4, init = 'k-means++', random_state = 42) #dividing the dataset into 4 clusters\nclusters = kmeans.fit_predict(data) #storing cluster numbers predicted",
"_____no_output_____"
],
[
"clusters = pd.Series(clusters)",
"_____no_output_____"
],
[
"db['Clusters'] = clusters\ndb",
"_____no_output_____"
],
[
"db[db['Clusters'] == 0] #cluster 1",
"_____no_output_____"
],
[
"db[db['Clusters'] == 1] #cluster 2",
"_____no_output_____"
],
[
"db[db['Clusters'] == 2] #cluster 3",
"_____no_output_____"
],
[
"db[db['Clusters'] == 3] #cluster 4",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71b6a3a790126b3c33db7185ea088d88c0cfe2a | 122,179 | ipynb | Jupyter Notebook | KAITMon.ipynb | kialio/FlareFinder | 503db208df35a5a1f0958b875d5f55c7105b5abf | [
"MIT"
] | null | null | null | KAITMon.ipynb | kialio/FlareFinder | 503db208df35a5a1f0958b875d5f55c7105b5abf | [
"MIT"
] | null | null | null | KAITMon.ipynb | kialio/FlareFinder | 503db208df35a5a1f0958b875d5f55c7105b5abf | [
"MIT"
] | null | null | null | 243.384462 | 55,163 | 0.866597 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e71b6f4eb1578a80061efed2c23c1510f6caf019 | 70,479 | ipynb | Jupyter Notebook | code/1_statistical_framework_graphs.ipynb | neurodata/MCC | 42c6b5fab09edf10bce2143c2366199531b2626b | [
"MIT"
] | 6 | 2021-01-15T02:11:57.000Z | 2021-12-31T04:06:58.000Z | code/1_statistical_framework_graphs.ipynb | neurodata/MCC | 42c6b5fab09edf10bce2143c2366199531b2626b | [
"MIT"
] | 3 | 2021-04-05T16:09:58.000Z | 2021-12-29T21:55:27.000Z | code/1_statistical_framework_graphs.ipynb | neurodata/MCC | 42c6b5fab09edf10bce2143c2366199531b2626b | [
"MIT"
] | 1 | 2021-04-28T14:41:22.000Z | 2021-04-28T14:41:22.000Z | 496.330986 | 41,623 | 0.949503 | [
[
[
"# Statistical Framework for Multiscale Comparative Connectomics\nThis notebook replicates the example graphs found in Figure 1 in _Multiscale Comparative Connectomics_.\n",
"_____no_output_____"
]
],
[
[
"%load_ext rpy2.ipython",
"_____no_output_____"
],
[
"%%R\nsuppressPackageStartupMessages(library(igraph))",
"_____no_output_____"
],
[
"%%R -w 3 -h 3 --units in -r 150\n\ng <- erdos.renyi.game(10, 0.5)\nV(g)$label <- NA\necol <- rep(\"black\", ecount(g))\nplot(g, edge.color=ecol)",
"_____no_output_____"
],
[
"%%R -w 3 -h 3 --units in -r 150\n\n# Read graph\nnodes <- read.csv(\"../data/nodes.csv\", header=T, as.is=T)\nlinks <- read.csv(\"../data/edges.csv\", header=T, as.is=T)\nnet <- graph_from_data_frame(d=links, vertices=nodes, directed=T) \n\n# Simplify graph\nnet <- simplify(net, remove.multiple = F, remove.loops = T) \nV(net)$size <- V(net)$audience.size*0.5\nV(net)$label <- NA\n\n# Plot graph\nceb <- cluster_edge_betweenness(as.undirected(net)) \nplot(ceb, as.undirected(net))",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e71ba177ca756d8cfd1e87db0a496aca6dd46fb1 | 29,056 | ipynb | Jupyter Notebook | metadata-translation/notebooks/test-pipeline.ipynb | elais/nmdc-runtime | 0c03684bc01fd1c0ce443aa271c234067d6d7c09 | [
"Apache-2.0"
] | 2 | 2021-09-14T16:24:58.000Z | 2022-02-22T21:45:08.000Z | metadata-translation/notebooks/test-pipeline.ipynb | elais/nmdc-runtime | 0c03684bc01fd1c0ce443aa271c234067d6d7c09 | [
"Apache-2.0"
] | 79 | 2021-05-04T22:00:22.000Z | 2022-03-31T16:35:46.000Z | metadata-translation/notebooks/test-pipeline.ipynb | elais/nmdc-runtime | 0c03684bc01fd1c0ce443aa271c234067d6d7c09 | [
"Apache-2.0"
] | 1 | 2021-04-21T16:07:21.000Z | 2021-04-21T16:07:21.000Z | 26.247516 | 559 | 0.547013 | [
[
[
"import os, sys\nsys.path.append(os.path.abspath('../src/bin/lib/')) # add path to data_opertations.py\nsys.path.append(os.path.abspath('../../schema/')) # add path nmdc.py",
"_____no_output_____"
],
[
"import yaml\nimport json\nfrom yaml import CLoader as Loader, CDumper as Dumper\nimport data_operations as dop\nfrom dotted_dict import DottedDict\nfrom collections import namedtuple\nimport nmdc\nimport data_operations as dop\nfrom pandasql import sqldf\nfrom pprint import pprint\nimport pandas as pds\nimport jsonasobj\n\nfrom pandasql import sqldf\ndef pysqldf(q):\n return sqldf(q, globals())",
"_____no_output_____"
]
],
[
[
"## Load yaml spec for data sources",
"_____no_output_____"
]
],
[
[
"spec_file = \"../src/bin/lib/nmdc_data_source.yaml\"\nwith open(spec_file, 'r') as input_file:\n spec = DottedDict(yaml.load(input_file, Loader=Loader))",
"_____no_output_____"
]
],
[
[
"## Create merged dataframe of all data sources",
"_____no_output_____"
]
],
[
[
"## build merged dataframe from data sources specified in the spec file\n# mdf = dop.make_dataframe_from_spec_file (spec_file)\n# mdf.to_csv('../src/data/nmdc_merged_data.tsv', sep='\\t', index=False) # save mergd data",
"_____no_output_____"
]
],
[
[
"## Load data from merged tsv (this can only be done after merged data tsv has been created)",
"_____no_output_____"
]
],
[
[
"# read data from saved file\nmdf = pds.read_csv('../src/data/nmdc_merged_data.tsv.zip', sep='\\t', dtype=str)\n# mdf = pds.read_csv('../src/data/nmdc_merged_data.tsv.zip', sep='\\t', dtype=str, nrows=100)",
"_____no_output_____"
],
[
"mdf.nmdc_data_source.unique() ## list of the data sources in merged",
"_____no_output_____"
]
],
[
[
"## Extract tables from merged dataset",
"_____no_output_____"
]
],
[
[
"study_table = dop.extract_table(mdf, 'study_table')\ncontact_table = dop.extract_table(mdf, 'contact_table')\nproposals_table = dop.extract_table(mdf, 'proposals_table')\nproject_table = dop.extract_table(mdf, 'project_table')\njgi_emsl_table = dop.extract_table(mdf, 'ficus_jgi_emsl')\nemsl_table = dop.extract_table(mdf, 'ficus_emsl')\nfaa_table = dop.extract_table(mdf, 'ficus_faa_table')\nfna_table = dop.extract_table(mdf, 'ficus_fna_table')\nfastq_table = dop.extract_table(mdf, 'ficus_fastq_table')\nproject_biosample_table = dop.extract_table(mdf, 'project_biosample_table')\nbiosample_table = dop.extract_table(mdf, 'biosample_table')\n# biosample_table.columns",
"_____no_output_____"
]
],
[
[
"## Test building study json",
"_____no_output_____"
]
],
[
[
"study = dop.make_study_dataframe(study_table, contact_table, proposals_table)\nstudy_dictdf = study.to_dict(orient=\"records\") # transorm dataframe to dictionary\n# study.gold_id",
"_____no_output_____"
],
[
"## specify attributes\nattributes = \\\n ['gold_study_name', 'principal_investigator_name', 'add_date', 'mod_date', 'doi',\n 'ecosystem', 'ecosystem_category', 'ecosystem_type', 'ecosystem_subtype', 'specific_ecosystem']\n\nconstructor = \\\n {\n 'id': 'gold_id',\n 'name': 'study_name',\n 'description': 'description'\n }\n\nstudy_json_list = dop.make_json_string_list\\\n (study_dictdf, nmdc.Study, constructor_map=constructor, attribute_fields=attributes)",
"_____no_output_____"
],
[
"# print(json.dumps(json.loads(study_json_list[0]), indent=4)) ## peek at data\n# print(nmdc.Study.class_class_curie)\n# len(study)",
"_____no_output_____"
]
],
[
[
"## Save study output",
"_____no_output_____"
]
],
[
[
"dop.save_json_string_list(\"output/test-pipeline/gold_study.json\", study_json_list) # save json string list to file",
"_____no_output_____"
]
],
[
[
"## Test building EMSL omics processing projects",
"_____no_output_____"
]
],
[
[
"emsl = dop.make_emsl_dataframe(emsl_table, jgi_emsl_table, study_table)\nemsl_dictdf = emsl.to_dict(orient=\"records\") # transorm dataframe to dictionary",
"_____no_output_____"
],
[
"# emsl.gold_study_id",
"_____no_output_____"
],
[
"# len(emsl)\n# emsl.head()\n# emsl.columns\n# len(emsl_table)",
"_____no_output_____"
],
[
"attributes = \\\n [\n #'file_size_bytes',\n # {'part_of': ({'id': 'gold_study_id'}, nmdc.Study)},\n # {'has_output': ({'id': 'data_object_id'}, nmdc.DataObject)}\n {'part_of': 'gold_study_id'},\n {'has_output': 'data_object_id'}\n ]\n\nconstructor = \\\n {\n 'id': 'dataset_id',\n 'name': 'dataset_name',\n 'description': 'dataset_type_description'\n }\n\nemsl_project_json_list = dop.make_json_string_list\\\n (emsl_dictdf, nmdc.OmicsProcessing, constructor_map=constructor, attribute_fields=attributes)",
"_____no_output_____"
],
[
"# print(json.dumps(json.loads(emsl_project_json_list[0]), indent=4)) ## peek at data",
"_____no_output_____"
]
],
[
[
"## Save EMSL omics processing projects",
"_____no_output_____"
]
],
[
[
"dop.save_json_string_list(\"output/test-pipeline/emsl_omics_processing.json\", emsl_project_json_list) # save json string list to file",
"_____no_output_____"
]
],
[
[
"## Test build EMSL data objects",
"_____no_output_____"
]
],
[
[
"attributes = \\\n [\n 'file_size_bytes'\n ]\n\nconstructor = \\\n {\n 'id': 'data_object_id',\n 'name': 'data_object_name',\n 'description': 'dataset_type_description'\n }\n\nemsl_data_object_json_list = dop.make_json_string_list\\\n (emsl_dictdf, nmdc.DataObject, constructor_map=constructor, attribute_fields=attributes)",
"_____no_output_____"
],
[
"# print(json.dumps(json.loads(emsl_data_object_json_list[0]), indent=4)) ## peek at data",
"_____no_output_____"
]
],
[
[
"## Save EMSL data objects",
"_____no_output_____"
]
],
[
[
"dop.save_json_string_list(\"output/test-pipeline/emsl_data_objects.json\", emsl_data_object_json_list) # save json string list to file",
"_____no_output_____"
]
],
[
[
"## Test building data obects (faa, fna, fastq)",
"_____no_output_____"
]
],
[
[
"# fastq_table.head() # peek at data",
"_____no_output_____"
],
[
"data_objects = dop.make_data_objects_dataframe(faa_table, fna_table, fastq_table, project_table)\ndata_objects_dictdf = data_objects.to_dict(orient=\"records\") # transorm dataframe to dictionary",
"_____no_output_____"
],
[
"len(data_objects)",
"_____no_output_____"
],
[
"attributes = \\\n [\n 'file_size_bytes'\n ]\n\nconstructor = \\\n {\n 'id': 'file_id',\n 'name': 'file_name',\n 'description': 'file_type_description'\n }\n\ndata_objects_json_list = dop.make_json_string_list\\\n (data_objects_dictdf, nmdc.DataObject, constructor_map=constructor, attribute_fields=attributes)",
"_____no_output_____"
],
[
"# print(json.dumps(json.loads(data_objects_json_list[0]), indent=4)) ## peek at data",
"_____no_output_____"
]
],
[
[
"## Save faa, fna, fastq data objects",
"_____no_output_____"
]
],
[
[
"dop.save_json_string_list(\"output/test-pipeline/faa_fna_fastq_data_objects.json\", data_objects_json_list) # save json string list to file",
"_____no_output_____"
]
],
[
[
"## Test building GOLD project json",
"_____no_output_____"
]
],
[
[
"# data_objects.head()",
"_____no_output_____"
],
[
"project = dop.make_project_dataframe(project_table, study_table, contact_table, data_objects)\n# project[pds.isnull(project.output_file_ids)]\n# project = project[project.nmdc_record_id == \"115128\"] # test if output_file_ids is null\n# project.output_file_ids.unique()\n# project.output_file_ids",
"_____no_output_____"
],
[
"project_dictdf = project.to_dict(orient=\"records\") # transorm dataframe to dictionary\n# project.columns",
"_____no_output_____"
],
[
"## specify characteristics\nattributes = \\\n [\n # {'part_of': ({'id': 'study_gold_id'}, nmdc.Study)},\n # {'has_output': ({'id': 'output_file_ids'}, nmdc.DataObject)},\n {'part_of': 'study_gold_id'},\n {'has_output': 'output_file_ids'},\n 'add_date', \n 'mod_date', \n 'completion_date', \n 'ncbi_project_name', \n 'omics_type', \n 'principal_investigator_name',\n 'processing_institution'\n ]\n\n\nconstructor = \\\n {\n 'id': 'gold_id',\n 'name': 'project_name',\n 'description': 'description'\n }\n\nproject_json_list = dop.make_json_string_list\\\n (project_dictdf, nmdc.OmicsProcessing, constructor_map=constructor, attribute_fields=attributes)",
"_____no_output_____"
],
[
"# print(json.dumps(json.loads(project_json_list[0]), indent=4)) ## peek at data",
"_____no_output_____"
]
],
[
[
"## Save output",
"_____no_output_____"
]
],
[
[
"dop.save_json_string_list(\"output/test-pipeline/gold_omics_processing.json\", project_json_list) # save json string list to file",
"_____no_output_____"
]
],
[
[
"## Test building biosample json",
"_____no_output_____"
]
],
[
[
"biosample = dop.make_biosample_dataframe(biosample_table, project_biosample_table, project_table)",
"_____no_output_____"
],
[
"biosample_dictdf = biosample.to_dict(orient=\"records\") # transorm dataframe to dictionary\n# biosample_dictdf[0] ## peek at dict data",
"_____no_output_____"
],
[
"## specify attributes\nattributes = \\\n [\n 'add_date',\n 'mod_date',\n 'collection_date',\n 'ecosystem',\n 'ecosystem_dcategory',\n 'ecosystem_type',\n 'ecosystem_subtype',\n 'specific_ecosystem',\n 'habitat',\n 'location',\n 'community',\n 'ncbi_taxonomy_name',\n 'geographic_location',\n 'sample_collection_site',\n 'identifier',\n 'host_name',\n 'depth',\n 'subsurface_depth',\n 'altitude',\n 'proport_woa_temperature',\n 'biogas_temperature',\n 'growth_temperature',\n 'water_samp_store_temp',\n 'biogas_retention_time',\n 'salinity',\n 'pressure',\n 'ph',\n 'chlorophyll_concentration',\n 'nitrate_concentration',\n 'oxygen_concentration',\n 'salinity_concentration',\n 'sample_volume',\n 'sample_weight_dna_ext',\n 'sampling_strategy',\n 'soil_link_climate_info',\n 'soil_misc_param',\n 'soil_misc_param ',\n 'soil_water_content',\n 'soluble_iron_micromol',\n 'subsurface_depth2',\n 'tot_nitrogen',\n 'tot_org_carbon',\n 'water_alkalinity',\n 'water_alkalinity_method',\n 'water_alkyl_diethers',\n 'water_aminopept_act',\n 'water_ammonium',\n 'water_bacterial_carbon_prod',\n 'water_bishomohopanol',\n 'water_bromide',\n 'water_calcium',\n 'water_carbon_nitrog_ratio',\n 'water_chem_administration',\n 'water_chloride',\n 'water_density',\n 'water_diether_lipids',\n 'water_diss_carbon_dioxide',\n 'water_diss_hydrogen',\n 'water_diss_inorg_carbon',\n 'water_diss_inorg_phosphorus',\n 'water_diss_org_carbon',\n 'water_diss_org_nitrogen',\n 'water_glucosidase_activity',\n 'water_magnesium',\n 'water_mean_frict_vel',\n 'water_mean_peak_frict_vel',\n 'water_misc_parameter',\n 'water_n_alkanes',\n 'water_nitrite',\n 'water_org_matter',\n 'water_org_nitrogen',\n 'water_organism_count',\n 'water_oxy_stat_sample',\n 'water_part_org_carbon',\n 'water_perturbation',\n 'water_petroleum_hydrocarbon',\n 'water_phaeopigments',\n 'water_phosplipid_fatt_acid',\n 'water_potassium',\n 'water_redox_potential',\n 'water_samp_store_dur',\n 'water_samp_store_loc',\n 'water_size_frac_low',\n 'water_size_frac_up',\n 'water_sodium',\n 'water_sulfate',\n 'water_sulfide',\n 'water_tidal_stage',\n 'water_tot_depth_water_col',\n 'water_tot_diss_nitro',\n 'water_tot_phosphorus',\n 'water_turbidity',\n {'part_of': 'project_gold_ids'}\n # {'part_of': ({'id': 'project_gold_ids'}, nmdc.OmicsProcessing)}\n ]\n\n# removed in version 5: 'temperature_range', 'soil_annual_season_temp'",
"_____no_output_____"
],
[
"# os.chdir('../..')\n# os.getcwd()",
"_____no_output_____"
],
[
"## create map betweeen gold fields and mixs terms\nmapping_df = dop.make_dataframe(\"../src/data/GOLD-to-mixs-map.tsv\")\nattr_map = dop.make_gold_to_mixs_map(attributes, mapping_df, 'biosample')",
"_____no_output_____"
],
[
"## create dict of constructor args\nconstructor = \\\n {\n 'id': 'gold_id',\n 'name': 'biosample_name',\n 'description': 'description',\n 'env_broad_scale': [{'has_raw_value':'env_broad_scale'}, nmdc.ControlledTermValue],\n 'env_local_scale': [{'has_raw_value':'env_local_scale'}, nmdc.ControlledTermValue],\n 'env_medium': [{'has_raw_value': 'env_medium'}, nmdc.ControlledTermValue],\n 'lat_lon': [{'latitude': 'latitude', 'longitude': 'longitude', 'has_raw_value': 'lat_lon'}, nmdc.GeolocationValue],\n }",
"_____no_output_____"
],
[
"## create list of json string objects\nbiosample_json_list = dop.make_json_string_list \\\n (biosample_dictdf, nmdc.Biosample, constructor_map=constructor, attribute_fields=attributes, attribute_map=attr_map)",
"_____no_output_____"
],
[
"# print(json.dumps(json.loads(biosample_json_list[0]), indent=4)) ## peek at data",
"_____no_output_____"
]
],
[
[
"## Save output",
"_____no_output_____"
]
],
[
[
"dop.save_json_string_list(\"output/test-pipeline/biosample.json\", biosample_json_list) # save json string list to file",
"_____no_output_____"
]
],
[
[
"## Test subset of output",
"_____no_output_____"
]
],
[
[
"## navigate to test output directory\nos.chdir('output/test-pipeline/')",
"_____no_output_____"
],
[
"os.getcwd()",
"_____no_output_____"
],
[
"## grab first five biosamples\n!jq '.[0:4]' biosample.json > '../test-five-biosamples/biosample.json'",
"_____no_output_____"
],
[
"biosample_set = None\nwith open('../test-five-biosamples/biosample.json', 'r') as f:\n biosample_set = json.load(f)",
"_____no_output_____"
],
[
"## find first 5 project ids of biosamples\n# !jq -c '.[] | {biosample:.id, project:.part_of[]}' biosample.json | head -n5\n!jq '.[] | .part_of[]' biosample.json | head -n5",
"\"gold:Gp0108335\"\n\"gold:Gp0108340\"\n\"gold:Gp0108341\"\n\"gold:Gp0108342\"\n\"gold:Gp0108344\"\n"
],
[
"## create project subset\n!jq '.[] | select(.id == (\"gold:Gp0108335\", \"gold:Gp0108340\", \"gold:Gp0108341\", \"gold:Gp0108342\", \"gold:Gp0108344\"))' \\\ngold_omics_processing.json \\\n| jq --slurp '.' \\\n> '../test-five-biosamples/project.json'",
"_____no_output_____"
],
[
"project_set = None\nwith open('../test-five-biosamples/project.json', 'r') as f:\n project_set = json.load(f)",
"_____no_output_____"
],
[
"## get the study ids\n!jq '.[] | .part_of[]' ../test-five-biosamples/project.json",
"\u001b[0;32m\"gold:Gs0112340\"\u001b[0m\n\u001b[0;32m\"gold:Gs0112340\"\u001b[0m\n\u001b[0;32m\"gold:Gs0112340\"\u001b[0m\n\u001b[0;32m\"gold:Gs0112340\"\u001b[0m\n\u001b[0;32m\"gold:Gs0112340\"\u001b[0m\n"
],
[
"## create study subset\n!jq '.[] | select(.id == \"gold:Gs0112340\")' \\\ngold_study.json \\\n| jq --slurp '.' \\\n> '../test-five-biosamples/study.json'",
"_____no_output_____"
],
[
"study_set = None\nwith open('../test-five-biosamples/study.json', 'r') as f:\n study_set = json.load(f)",
"_____no_output_____"
],
[
"## get outputs of projects\n!jq '.[] | .has_output[]' ../test-five-biosamples/project.json",
"\u001b[0;32m\"nmdc:5af44fd364d0b33747747ddb\"\u001b[0m\n\u001b[0;32m\"nmdc:5af44fd264d0b33747747dd9\"\u001b[0m\n\u001b[0;32m\"jgi:551a20d30d878525404e90d5\"\u001b[0m\n\u001b[0;32m\"nmdc:5af0d91764d0b3374773e07a\"\u001b[0m\n\u001b[0;32m\"nmdc:5af0d91764d0b3374773e078\"\u001b[0m\n\u001b[0;32m\"jgi:551a20d50d878525404e90d7\"\u001b[0m\n\u001b[0;32m\"nmdc:5af6f6bd64d0b3374774f9a7\"\u001b[0m\n\u001b[0;32m\"nmdc:5af6f6bc64d0b3374774f9a5\"\u001b[0m\n\u001b[0;32m\"jgi:551a20d90d878525404e90e1\"\u001b[0m\n\u001b[0;32m\"nmdc:5af0d80364d0b3374773e066\"\u001b[0m\n\u001b[0;32m\"nmdc:5af0d80264d0b3374773e064\"\u001b[0m\n\u001b[0;32m\"jgi:551a20d60d878525404e90d9\"\u001b[0m\n\u001b[0;32m\"nmdc:5af65c0864d0b3374774e587\"\u001b[0m\n\u001b[0;32m\"nmdc:5af65c0764d0b3374774e559\"\u001b[0m\n\u001b[0;32m\"jgi:551a20da0d878525404e90e4\"\u001b[0m\n"
],
[
"## create data objects subset\n!jq '.[] | select(.id == (\"nmdc:5af44fd364d0b33747747ddb\", \"nmdc:5af44fd264d0b33747747dd9\", \"jgi:551a20d30d878525404e90d5\", \"nmdc:5af0d91764d0b3374773e07a\", \"nmdc:5af0d91764d0b3374773e078\", \"jgi:551a20d50d878525404e90d7\", \"nmdc:5af6f6bd64d0b3374774f9a7\", \"nmdc:5af6f6bc64d0b3374774f9a5\", \"jgi:551a20d90d878525404e90e1\", \"nmdc:5af0d80364d0b3374773e066\", \"nmdc:5af0d80264d0b3374773e064\", \"jgi:551a20d60d878525404e90d9\", \"nmdc:5af65c0864d0b3374774e587\", \"nmdc:5af65c0764d0b3374774e559\", \"jgi:551a20da0d878525404e90e4\"))' \\\nfaa_fna_fastq_data_objects.json \\\n| jq --slurp '.' \\\n> '../test-five-biosamples/data_object.json'",
"_____no_output_____"
],
[
"data_object_set = None\nwith open('../test-five-biosamples/data_object.json', 'r') as f:\n data_object_set = json.load(f)",
"_____no_output_____"
],
[
"!ls",
"biosample.json gold_omics_processing.json\nemsl_data_objects.json gold_study.json\nemsl_omics_processing.json nmdc-02.json\nfaa_fna_fastq_data_objects.json\n"
],
[
"## emsl projects\n# !jq '.[0]' emsl_omics_processing.json ",
"_____no_output_____"
],
[
"# !jq '.[0]' emsl_data_objects.json",
"_____no_output_____"
],
[
"database = \\\n {\n \"study_set\": [*study_set], \n \"omics_processing_set\": [*project_set], \n \"biosample_set\": [*biosample_set], \n \"data_object_set\": [*data_object_set]\n }",
"_____no_output_____"
],
[
"with open('nmdc-02.json', 'w') as fp:\n json.dump(database, fp)",
"_____no_output_____"
],
[
"!ls",
"biosample.json gold_omics_processing.json\nemsl_data_objects.json gold_study.json\nemsl_omics_processing.json nmdc-02.json\nfaa_fna_fastq_data_objects.json\n"
],
[
"!pwd",
"/Users/wdduncan/repos/NMDC/nmdc-metadata/metadata-translation/notebooks/output/test-pipeline\n"
],
[
"# !jq '.' nmdc-02.json | head -n100",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71ba213a3af70b38653bab562636b831a255c5b | 12,076 | ipynb | Jupyter Notebook | dev/_downloads/5f84ce88b4773e5ca1f9b3502aef334a/plot_eeg_erp.ipynb | massich/mne-tools.github.io | 95650593ba0eca4ff8257ebcbdf05731038d8d4e | [
"BSD-3-Clause"
] | null | null | null | dev/_downloads/5f84ce88b4773e5ca1f9b3502aef334a/plot_eeg_erp.ipynb | massich/mne-tools.github.io | 95650593ba0eca4ff8257ebcbdf05731038d8d4e | [
"BSD-3-Clause"
] | null | null | null | dev/_downloads/5f84ce88b4773e5ca1f9b3502aef334a/plot_eeg_erp.ipynb | massich/mne-tools.github.io | 95650593ba0eca4ff8257ebcbdf05731038d8d4e | [
"BSD-3-Clause"
] | null | null | null | 33.544444 | 598 | 0.555565 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n\nEEG processing and Event Related Potentials (ERPs)\n==================================================\n\nFor a generic introduction to the computation of ERP and ERF\nsee `tut_epoching_and_averaging`.\n :depth: 1\n",
"_____no_output_____"
]
],
[
[
"import mne\nfrom mne.datasets import sample",
"_____no_output_____"
]
],
[
[
"Setup for reading the raw data\n\n",
"_____no_output_____"
]
],
[
[
"data_path = sample.data_path()\nraw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'\nevent_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'\n# these data already have an EEG average reference\nraw = mne.io.read_raw_fif(raw_fname, preload=True)",
"_____no_output_____"
]
],
[
[
"Let's restrict the data to the EEG channels\n\n",
"_____no_output_____"
]
],
[
[
"raw.pick_types(meg=False, eeg=True, eog=True)",
"_____no_output_____"
]
],
[
[
"By looking at the measurement info you will see that we have now\n59 EEG channels and 1 EOG channel\n\n",
"_____no_output_____"
]
],
[
[
"print(raw.info)",
"_____no_output_____"
]
],
[
[
"In practice it's quite common to have some EEG channels that are actually\nEOG channels. To change a channel type you can use the\n:func:`mne.io.Raw.set_channel_types` method. For example\nto treat an EOG channel as EEG you can change its type using\n\n",
"_____no_output_____"
]
],
[
[
"raw.set_channel_types(mapping={'EOG 061': 'eeg'})\nprint(raw.info)",
"_____no_output_____"
]
],
[
[
"And to change the name of the EOG channel\n\n",
"_____no_output_____"
]
],
[
[
"raw.rename_channels(mapping={'EOG 061': 'EOG'})",
"_____no_output_____"
]
],
[
[
"Let's reset the EOG channel back to EOG type.\n\n",
"_____no_output_____"
]
],
[
[
"raw.set_channel_types(mapping={'EOG': 'eog'})",
"_____no_output_____"
]
],
[
[
"The EEG channels in the sample dataset already have locations.\nThese locations are available in the 'loc' of each channel description.\nFor the first channel we get\n\n",
"_____no_output_____"
]
],
[
[
"print(raw.info['chs'][0]['loc'])",
"_____no_output_____"
]
],
[
[
"And it's actually possible to plot the channel locations using\n:func:`mne.io.Raw.plot_sensors`.\nIn the case where your data don't have locations you can use one of the\nstandard :class:`Montages <mne.channels.DigMontage>` shipped with MNE.\nSee `plot_montage` and `tut-eeg-fsaverage-source-modeling`.\n\n",
"_____no_output_____"
]
],
[
[
"raw.plot_sensors()\nraw.plot_sensors('3d') # in 3D",
"_____no_output_____"
]
],
[
[
"Setting EEG reference\n---------------------\n\nLet's first remove the reference from our Raw object.\n\nThis explicitly prevents MNE from adding a default EEG average reference\nrequired for source localization.\n\n",
"_____no_output_____"
]
],
[
[
"raw_no_ref, _ = mne.set_eeg_reference(raw, [])",
"_____no_output_____"
]
],
[
[
"We next define Epochs and compute an ERP for the left auditory condition.\n\n",
"_____no_output_____"
]
],
[
[
"reject = dict(eeg=180e-6, eog=150e-6)\nevent_id, tmin, tmax = {'left/auditory': 1}, -0.2, 0.5\nevents = mne.read_events(event_fname)\nepochs_params = dict(events=events, event_id=event_id, tmin=tmin, tmax=tmax,\n reject=reject)\n\nevoked_no_ref = mne.Epochs(raw_no_ref, **epochs_params).average()\ndel raw_no_ref # save memory\n\ntitle = 'EEG Original reference'\nevoked_no_ref.plot(titles=dict(eeg=title), time_unit='s')\nevoked_no_ref.plot_topomap(times=[0.1], size=3., title=title, time_unit='s')",
"_____no_output_____"
]
],
[
[
"**Average reference**: This is normally added by default, but can also\nbe added explicitly.\n\n",
"_____no_output_____"
]
],
[
[
"raw.del_proj()\nraw_car, _ = mne.set_eeg_reference(raw, 'average', projection=True)\nevoked_car = mne.Epochs(raw_car, **epochs_params).average()\ndel raw_car # save memory\n\ntitle = 'EEG Average reference'\nevoked_car.plot(titles=dict(eeg=title), time_unit='s')\nevoked_car.plot_topomap(times=[0.1], size=3., title=title, time_unit='s')",
"_____no_output_____"
]
],
[
[
"**Custom reference**: Use the mean of channels EEG 001 and EEG 002 as\na reference\n\n",
"_____no_output_____"
]
],
[
[
"raw_custom, _ = mne.set_eeg_reference(raw, ['EEG 001', 'EEG 002'])\nevoked_custom = mne.Epochs(raw_custom, **epochs_params).average()\ndel raw_custom # save memory\n\ntitle = 'EEG Custom reference'\nevoked_custom.plot(titles=dict(eeg=title), time_unit='s')\nevoked_custom.plot_topomap(times=[0.1], size=3., title=title, time_unit='s')",
"_____no_output_____"
]
],
[
[
"Evoked arithmetic (e.g. differences)\n------------------------------------\n\nTrial subsets from Epochs can be selected using 'tags' separated by '/'.\nEvoked objects support basic arithmetic.\nFirst, we create an Epochs object containing 4 conditions.\n\n",
"_____no_output_____"
]
],
[
[
"event_id = {'left/auditory': 1, 'right/auditory': 2,\n 'left/visual': 3, 'right/visual': 4}\nepochs_params = dict(events=events, event_id=event_id, tmin=tmin, tmax=tmax,\n reject=reject)\nepochs = mne.Epochs(raw, **epochs_params)\n\nprint(epochs)",
"_____no_output_____"
]
],
[
[
"Next, we create averages of stimulation-left vs stimulation-right trials.\nWe can use basic arithmetic to, for example, construct and plot\ndifference ERPs.\n\n",
"_____no_output_____"
]
],
[
[
"left, right = epochs[\"left\"].average(), epochs[\"right\"].average()\n\n# create and plot difference ERP\njoint_kwargs = dict(ts_args=dict(time_unit='s'),\n topomap_args=dict(time_unit='s'))\nmne.combine_evoked([left, -right], weights='equal').plot_joint(**joint_kwargs)",
"_____no_output_____"
]
],
[
[
"This is an equal-weighting difference. If you have imbalanced trial numbers,\nyou could also consider either equalizing the number of events per\ncondition (using\n:meth:`epochs.equalize_event_counts <mne.Epochs.equalize_event_counts>`).\nAs an example, first, we create individual ERPs for each condition.\n\n",
"_____no_output_____"
]
],
[
[
"aud_l = epochs[\"auditory\", \"left\"].average()\naud_r = epochs[\"auditory\", \"right\"].average()\nvis_l = epochs[\"visual\", \"left\"].average()\nvis_r = epochs[\"visual\", \"right\"].average()\n\nall_evokeds = [aud_l, aud_r, vis_l, vis_r]\nprint(all_evokeds)",
"_____no_output_____"
]
],
[
[
"This can be simplified with a Python list comprehension:\n\n",
"_____no_output_____"
]
],
[
[
"all_evokeds = [epochs[cond].average() for cond in sorted(event_id.keys())]\nprint(all_evokeds)\n\n# Then, we construct and plot an unweighted average of left vs. right trials\n# this way, too:\nmne.combine_evoked(\n [aud_l, -aud_r, vis_l, -vis_r], weights='equal').plot_joint(**joint_kwargs)",
"_____no_output_____"
]
],
[
[
"Often, it makes sense to store Evoked objects in a dictionary or a list -\neither different conditions, or different subjects.\n\n",
"_____no_output_____"
]
],
[
[
"# If they are stored in a list, they can be easily averaged, for example,\n# for a grand average across subjects (or conditions).\ngrand_average = mne.grand_average(all_evokeds)\nmne.write_evokeds('/tmp/tmp-ave.fif', all_evokeds)\n\n# If Evokeds objects are stored in a dictionary, they can be retrieved by name.\nall_evokeds = dict((cond, epochs[cond].average()) for cond in event_id)\nprint(all_evokeds['left/auditory'])\n\n# Besides for explicit access, this can be used for example to set titles.\nfor cond in all_evokeds:\n all_evokeds[cond].plot_joint(title=cond, **joint_kwargs)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71ba26bc2e982347c361ef90693dcff1f59fa20 | 7,636 | ipynb | Jupyter Notebook | MC/Blackjack Playground.ipynb | seanMahon/reinforcement-learning | 83d1340c0758f94b1dd2874b416e8c3a989753ce | [
"MIT"
] | null | null | null | MC/Blackjack Playground.ipynb | seanMahon/reinforcement-learning | 83d1340c0758f94b1dd2874b416e8c3a989753ce | [
"MIT"
] | null | null | null | MC/Blackjack Playground.ipynb | seanMahon/reinforcement-learning | 83d1340c0758f94b1dd2874b416e8c3a989753ce | [
"MIT"
] | null | null | null | 35.18894 | 83 | 0.544919 | [
[
[
"import numpy as np\nimport sys\nif \"../\" not in sys.path:\n sys.path.append(\"../\") \nfrom lib.envs.blackjack import BlackjackEnv",
"_____no_output_____"
],
[
"env = BlackjackEnv()",
"_____no_output_____"
],
[
"def print_observation(observation):\n score, dealer_score, usable_ace = observation\n print(\"Player Score: {} (Usable Ace: {}), Dealer Score: {}\".format(\n score, usable_ace, dealer_score))\n\ndef strategy(observation):\n score, dealer_score, usable_ace = observation\n # Stick (action 0) if the score is > 20, hit (action 1) otherwise\n return 0 if score >= 20 else 1\n\nfor i_episode in range(20):\n observation = env.reset()\n for t in range(100):\n print_observation(observation)\n action = strategy(observation)\n print(\"Taking action: {}\".format( [\"Stick\", \"Hit\"][action]))\n observation, reward, done, _ = env.step(action)\n if done:\n print_observation(observation)\n print(\"Game end. Reward: {}\\n\".format(float(reward)))\n break",
"Player Score: 20 (Usable Ace: False), Dealer Score: 9\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 9\nGame end. Reward: 1.0\n\nPlayer Score: 16 (Usable Ace: False), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 17 (Usable Ace: False), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 25 (Usable Ace: False), Dealer Score: 4\nGame end. Reward: -1.0\n\nPlayer Score: 19 (Usable Ace: True), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 17 (Usable Ace: False), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 19 (Usable Ace: False), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 29 (Usable Ace: False), Dealer Score: 4\nGame end. Reward: -1.0\n\nPlayer Score: 13 (Usable Ace: False), Dealer Score: 2\nTaking action: Hit 1\nPlayer Score: 16 (Usable Ace: False), Dealer Score: 2\nTaking action: Hit 1\nPlayer Score: 26 (Usable Ace: False), Dealer Score: 2\nGame end. Reward: -1.0\n\nPlayer Score: 17 (Usable Ace: False), Dealer Score: 7\nTaking action: Hit 1\nPlayer Score: 27 (Usable Ace: False), Dealer Score: 7\nGame end. Reward: -1.0\n\nPlayer Score: 19 (Usable Ace: True), Dealer Score: 1\nTaking action: Hit 1\nPlayer Score: 17 (Usable Ace: False), Dealer Score: 1\nTaking action: Hit 1\nPlayer Score: 27 (Usable Ace: False), Dealer Score: 1\nGame end. Reward: -1.0\n\nPlayer Score: 12 (Usable Ace: True), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 12 (Usable Ace: False), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 22 (Usable Ace: False), Dealer Score: 4\nGame end. Reward: -1.0\n\nPlayer Score: 12 (Usable Ace: False), Dealer Score: 10\nTaking action: Hit 1\nPlayer Score: 22 (Usable Ace: False), Dealer Score: 10\nGame end. Reward: -1.0\n\nPlayer Score: 17 (Usable Ace: False), Dealer Score: 10\nTaking action: Hit 1\nPlayer Score: 27 (Usable Ace: False), Dealer Score: 10\nGame end. Reward: -1.0\n\nPlayer Score: 17 (Usable Ace: False), Dealer Score: 7\nTaking action: Hit 1\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 7\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 7\nGame end. Reward: 1.0\n\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 10\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 10\nGame end. Reward: -1.0\n\nPlayer Score: 17 (Usable Ace: False), Dealer Score: 7\nTaking action: Hit 1\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 7\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 7\nGame end. Reward: 1.0\n\nPlayer Score: 12 (Usable Ace: True), Dealer Score: 4\nTaking action: Hit 1\nPlayer Score: 20 (Usable Ace: True), Dealer Score: 4\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: True), Dealer Score: 4\nGame end. Reward: 0.0\n\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 3\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 3\nGame end. Reward: 1.0\n\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 5\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 5\nGame end. Reward: 1.0\n\nPlayer Score: 17 (Usable Ace: True), Dealer Score: 10\nTaking action: Hit 1\nPlayer Score: 19 (Usable Ace: True), Dealer Score: 10\nTaking action: Hit 1\nPlayer Score: 12 (Usable Ace: False), Dealer Score: 10\nTaking action: Hit 1\nPlayer Score: 22 (Usable Ace: False), Dealer Score: 10\nGame end. Reward: -1.0\n\nPlayer Score: 16 (Usable Ace: True), Dealer Score: 1\nTaking action: Hit 1\nPlayer Score: 15 (Usable Ace: False), Dealer Score: 1\nTaking action: Hit 1\nPlayer Score: 21 (Usable Ace: False), Dealer Score: 1\nTaking action: Stick 0\nPlayer Score: 21 (Usable Ace: False), Dealer Score: 1\nGame end. Reward: 0.0\n\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 10\nTaking action: Stick 0\nPlayer Score: 20 (Usable Ace: False), Dealer Score: 10\nGame end. Reward: 0.0\n\nPlayer Score: 14 (Usable Ace: False), Dealer Score: 2\nTaking action: Hit 1\nPlayer Score: 24 (Usable Ace: False), Dealer Score: 2\nGame end. Reward: -1.0\n\nPlayer Score: 19 (Usable Ace: False), Dealer Score: 3\nTaking action: Hit 1\nPlayer Score: 28 (Usable Ace: False), Dealer Score: 3\nGame end. Reward: -1.0\n\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
e71baa25d7d35b815dd876fcfbec831d2d18c6bb | 464,380 | ipynb | Jupyter Notebook | look-into-the-data.ipynb | featherpeacock/pycon-ua-2018 | 420c73c74317085af7918318f6fc98ba4b14096b | [
"MIT"
] | 70 | 2018-04-29T21:27:05.000Z | 2022-03-29T10:24:29.000Z | look-into-the-data.ipynb | mayurmorin/pycon-ua-2018 | 420c73c74317085af7918318f6fc98ba4b14096b | [
"MIT"
] | 1 | 2020-05-04T09:21:04.000Z | 2020-05-05T16:13:36.000Z | look-into-the-data.ipynb | mayurmorin/pycon-ua-2018 | 420c73c74317085af7918318f6fc98ba4b14096b | [
"MIT"
] | 48 | 2018-05-08T23:30:19.000Z | 2022-02-23T09:17:16.000Z | 811.853147 | 101,524 | 0.955377 | [
[
[
"import IPython.core.display\nimport matplotlib\n\ndef apply_styles():\n matplotlib.rcParams['font.size'] = 12\n matplotlib.rcParams['figure.figsize'] = (18, 6)\n matplotlib.rcParams['lines.linewidth'] = 1\n\napply_styles()",
"_____no_output_____"
],
[
"%%html\n<style type=\"text/css\">\n@import url('https://fonts.googleapis.com/css?family=Playfair+Display');\n\n\ndiv.text_cell_render {font-family: 'Playfair Display', serif; color: #13213b; line-height: 145%; font-size:16px;}\n</style>",
"_____no_output_____"
]
],
[
[
"<span style=\"color:#13213b;\">[Andrii Gakhov](https://www.gakhov.com) / PyCon UA 2018</span>\n* * *\n## An Introduction to Time Series Forecasting with Python\nTime series is an important instrument to model, analyze and predict data collected over time. In this talk, we learn the basic theoretical concepts without going deep into mathematical aspects, study different models, and try them in practice using StatsModels, Prophet, scikit-learn, and keras.",
"_____no_output_____"
],
[
"# Part 1. Look into the data\n******",
"_____no_output_____"
],
[
"### OS visits to UK (All visits) \nThe dataset represents the monthly total number of visits to the UK by overseas residents (in thousands)<br>from January 1980 to October 2017. \n\n#### Source: [Office for National Statistics](https://www.ons.gov.uk/peoplepopulationandcommunity/leisureandtourism/timeseries/gmaa/ott)",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline\nsns.set(style=\"ticks\")",
"_____no_output_____"
],
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### Load the data into Pandas DataFrame",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.read_csv(\"data/GMAA-040218.csv\", header=None, skiprows=6, parse_dates=[0], names=['period', 'value'])\ndf.value.astype(int, copy=False);",
"_____no_output_____"
],
[
"df.head(5)",
"_____no_output_____"
],
[
"max_date = df.period.max()\nmin_date = df.period.min()\n\nnum_of_actual_points = df.index.shape[0]\nnum_of_expected_points = (max_date.year - min_date.year) * 12 + max_date.month - min_date.month + 1\n\nprint(\"Date range: {} - {}\".format(min_date.strftime(\"%d.%m.%Y\"), max_date.strftime(\"%d.%m.%Y\")))\nprint(\"Number of data points: {} of expected {}\".format(num_of_actual_points, num_of_expected_points))\n",
"Date range: 01.01.1980 - 01.10.2017\nNumber of data points: 454 of expected 454\n"
]
],
[
[
"### Visualize the data",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(18,6))\ndf.plot(x=\"period\", y=\"value\", ax=ax)\nplt.legend(loc='upper left')\n\nplt.savefig('images/intro-visualization.png');",
"_____no_output_____"
]
],
[
[
"In **2001** a combination of the outbreak of foot and mouth disease and the September 11 attacks in the US led to a dramatic slump.\n\nIn **2009** the global economic crisis, which started to bite in earnest in the autumn, was blamed as a factor for the fall. \n\nhttps://www.theguardian.com/business/2009/jul/16/tourism-uk-visitors-fall\n\nIn 2006 total visits to the UK by overseas residents were split fairly\nequally between three purposes: holiday, visiting friends or relatives,\nand business. This pattern is quite different compared with ten years\nago, when ‘holiday’ was the dominant reason\n\nhttps://www.ons.gov.uk/ons/rel/ott/travel-trends/2006/travel-trends---2006.pdf",
"_____no_output_____"
],
[
"The majority of visitors were from North America, followed by tourists from France and Germany.\nhttp://www.bbc.com/news/uk-england-london-27323755",
"_____no_output_____"
]
],
[
[
"zoom_range = df[(df.period >= '2010-01-01') & (df.period < '2012-01-01')].index\n\nfig, ax = plt.subplots(figsize=(18,6))\ndf.loc[zoom_range].plot(x=\"period\", y=\"value\", ax=ax, label=\"2010-2012 in zoom\")\nplt.legend(loc='upper left')\n\nplt.savefig('images/intro-zoom.png');",
"_____no_output_____"
]
],
[
[
"#### When is the best time to visit the UK?\n\nThe United Kingdom can be visited at any time of year ... Overall, **spring (late March to early June) and autumn (September to November) are the best times to visit**, when it’s usually warm and dry.\n\nhttps://www.audleytravel.com/us/uk/best-time-to-visit",
"_____no_output_____"
],
[
"## Trend and seasonality\n\nFrom the visualization it's already quite obvious that the OS visits have periodic fluctuations each year and overall tendency to grow up.\n\nThus, we can conclude that the time series has the **trend** and yearly **seasonality** components, and we can try to decompose them them using, for instance, **statsmodels** package.\n\nNote, from the data view we also can suspect that **additive** model better fits for data representation.",
"_____no_output_____"
]
],
[
[
"from statsmodels.tsa.seasonal import seasonal_decompose\n\ndecompfreq = 12 # 12 months seasonality\nmodel = 'additive'\n\ndecomposition = seasonal_decompose(\n df.set_index(\"period\").value.interpolate(\"linear\"),\n freq=decompfreq,\n model=model)",
"_____no_output_____"
],
[
"trend = decomposition.trend\nseasonal = decomposition.seasonal \nresidual = decomposition.resid ",
"_____no_output_____"
]
],
[
[
"### The Trend",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(18,6))\ndf.plot(x=\"period\", y=\"value\", ax=ax, label=\"observed\", c='lightgrey')\ntrend.plot(ax=ax, label=\"trend\")\nplt.legend(loc='upper left')\n\nplt.savefig('images/intro-trend.png');",
"_____no_output_____"
]
],
[
[
"### The Seasonality",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(18,4))\nseasonal.plot(ax=ax, label=\"seasonality\")\nplt.legend(loc='bottom left')\n\nplt.savefig('images/intro-seasonality.png');",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(18,6))\nseasonal[zoom_range].plot(x=\"period\", y=\"value\", ax=ax, label=\"2010-2012 in zoom\")\nplt.legend(loc='upper left')\n\nplt.savefig('images/intro-seasonality-zoom.png');",
"_____no_output_____"
]
],
[
[
"### The Residual",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(18,4))\nresidual.plot(ax=ax, legend=\"seasonality\")\nplt.legend(loc='upper left')\n\nplt.savefig('images/intro-residual.png');",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71bc129fcd430dea4628371b5f15de92bdd2f36 | 26,451 | ipynb | Jupyter Notebook | koho/sklearn/notebooks/user_guide.ipynb | AIWerkstatt/koho | 1136ac2de29a89052bf0f4f4747424eb0b6b0c2b | [
"BSD-3-Clause"
] | 2 | 2019-03-14T22:29:52.000Z | 2019-04-30T23:27:28.000Z | koho/sklearn/notebooks/user_guide.ipynb | AIWerkstatt/koho | 1136ac2de29a89052bf0f4f4747424eb0b6b0c2b | [
"BSD-3-Clause"
] | null | null | null | koho/sklearn/notebooks/user_guide.ipynb | AIWerkstatt/koho | 1136ac2de29a89052bf0f4f4747424eb0b6b0c2b | [
"BSD-3-Clause"
] | null | null | null | 43.504934 | 440 | 0.48501 | [
[
[
"from sklearn.datasets import load_iris\niris = load_iris()\nX, y = iris.data, iris.target",
"_____no_output_____"
],
[
"from koho.sklearn import DecisionTreeClassifier, DecisionForestClassifier\nclf = DecisionForestClassifier(random_state=0)",
"_____no_output_____"
],
[
"clf.fit(X, y)",
"_____no_output_____"
],
[
"feature_importances = clf.feature_importances_\nprint(feature_importances)",
"[0.09045256 0.00816573 0.38807981 0.5133019 ]\n"
],
[
"# $: conda install python-graphviz\nimport graphviz\ntree_idx = 0\ndot_data = clf.estimators_[tree_idx].export_graphviz(\n feature_names=iris.feature_names,\n class_names=iris.target_names,\n rotate=True)\ngraph = graphviz.Source(dot_data)\ngraph",
"_____no_output_____"
],
[
"graph.render(\"iris\", format='png')",
"_____no_output_____"
],
[
"t = clf.estimators_[tree_idx].export_text()\nprint(t)",
"0 X[3]<=0.8 [50, 50, 50]; 0->1; 0->2; 1 [50, 0, 0]; 2 X[3]<=1.75 [0, 50, 50]; 2->3; 2->6; 3 X[2]<=4.95 [0, 49, 5]; 3->4; 3->5; 4 [0, 47, 1]; 5 [0, 2, 4]; 6 X[3]<=1.85 [0, 1, 45]; 6->7; 6->8; 7 [0, 1, 11]; 8 [0, 0, 34]; \n"
],
[
"import pickle\nwith open(\"clf.pkl\", \"wb\") as f:\n pickle.dump(clf, f)\nwith open(\"clf.pkl\", \"rb\") as f:\n clf2 = pickle.load(f)",
"_____no_output_____"
],
[
"c = clf2.predict(X)\nprint(c)",
"[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1\n 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 1 2 2 2 2\n 2 2 2 2 2 2 2 2 1 2 2 2 2 2 2 2 2 2 2 2 2 2 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2\n 2 2]\n"
],
[
"cp = clf2.predict_proba(X)\nprint(cp)",
"[[1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [0.99833333 0.00166667 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [0.99833333 0.00166667 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [0.98322581 0.01677419 0. ]\n [0.98322581 0.01677419 0. ]\n [0.99833333 0.00166667 0. ]\n [1. 0. 0. ]\n [0.98322581 0.01677419 0. ]\n [1. 0. 0. ]\n [0.99833333 0.00166667 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [0.99833333 0.00166667 0. ]\n [1. 0. 0. ]\n [0.98322581 0.01677419 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [0.98322581 0.01677419 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [1. 0. 0. ]\n [0. 0.93816113 0.06183887]\n [0. 0.93816113 0.06183887]\n [0. 0.73006334 0.26993666]\n [0.00322581 0.96295065 0.03382354]\n [0. 0.93816113 0.06183887]\n [0.00322581 0.96602757 0.03074662]\n [0. 0.93816113 0.06183887]\n [0. 0.8998364 0.1001636 ]\n [0. 0.96123806 0.03876194]\n [0. 0.9548364 0.0451636 ]\n [0. 0.9748364 0.0251636 ]\n [0.00322581 0.93527208 0.06150211]\n [0.00322581 0.95527208 0.04150211]\n [0.00322581 0.93527208 0.06150211]\n [0.00322581 0.96602757 0.03074662]\n [0. 0.93816113 0.06183887]\n [0.00322581 0.94295065 0.05382354]\n [0.00322581 0.958349 0.03842519]\n [0. 0.93508421 0.06491579]\n [0.00322581 0.96295065 0.03382354]\n [0. 0.2040315 0.7959685 ]\n [0.00322581 0.958349 0.03842519]\n [0. 0.73006334 0.26993666]\n [0.00322581 0.958349 0.03842519]\n [0. 0.96123806 0.03876194]\n [0. 0.93816113 0.06183887]\n [0. 0.73006334 0.26993666]\n [0. 0.60735501 0.39264499]\n [0.00322581 0.93527208 0.06150211]\n [0.00322581 0.96602757 0.03074662]\n [0.00322581 0.96295065 0.03382354]\n [0.00322581 0.96295065 0.03382354]\n [0.00322581 0.958349 0.03842519]\n [0. 0.52643135 0.47356865]\n [0.00833333 0.9348364 0.05683026]\n [0.00322581 0.93527208 0.06150211]\n [0. 0.93816113 0.06183887]\n [0. 0.95816113 0.04183887]\n [0.00322581 0.96602757 0.03074662]\n [0.00322581 0.96295065 0.03382354]\n [0.00322581 0.96602757 0.03074662]\n [0.00322581 0.93527208 0.06150211]\n [0.00322581 0.958349 0.03842519]\n [0. 0.9748364 0.0251636 ]\n [0.00322581 0.96602757 0.03074662]\n [0.00322581 0.96602757 0.03074662]\n [0.00322581 0.96602757 0.03074662]\n [0. 0.96123806 0.03876194]\n [0. 0.9748364 0.0251636 ]\n [0.00322581 0.96602757 0.03074662]\n [0. 0.01935722 0.98064278]\n [0. 0.08072564 0.91927436]\n [0. 0.00540373 0.99459627]\n [0. 0.03019055 0.96980945]\n [0. 0.01935722 0.98064278]\n [0. 0.00540373 0.99459627]\n [0. 0.5880452 0.4119548 ]\n [0. 0.01623706 0.98376294]\n [0. 0.03019055 0.96980945]\n [0. 0.00540373 0.99459627]\n [0. 0.01935722 0.98064278]\n [0. 0.01935722 0.98064278]\n [0. 0.01935722 0.98064278]\n [0. 0.12087674 0.87912326]\n [0. 0.08072564 0.91927436]\n [0. 0.01935722 0.98064278]\n [0. 0.03019055 0.96980945]\n [0. 0.00540373 0.99459627]\n [0. 0.00540373 0.99459627]\n [0. 0.55890388 0.44109612]\n [0. 0.01935722 0.98064278]\n [0. 0.12087674 0.87912326]\n [0. 0.00540373 0.99459627]\n [0. 0.10476834 0.89523166]\n [0. 0.01935722 0.98064278]\n [0. 0.01623706 0.98376294]\n [0. 0.18476834 0.81523166]\n [0. 0.10260293 0.89739707]\n [0. 0.01935722 0.98064278]\n [0. 0.4263709 0.5736291 ]\n [0. 0.00540373 0.99459627]\n [0. 0.00540373 0.99459627]\n [0. 0.01935722 0.98064278]\n [0. 0.53277722 0.46722278]\n [0. 0.52643135 0.47356865]\n [0. 0.00540373 0.99459627]\n [0. 0.01935722 0.98064278]\n [0. 0.03019055 0.96980945]\n [0. 0.18260293 0.81739707]\n [0. 0.01935722 0.98064278]\n [0. 0.01935722 0.98064278]\n [0. 0.01935722 0.98064278]\n [0. 0.08072564 0.91927436]\n [0. 0.01935722 0.98064278]\n [0. 0.01935722 0.98064278]\n [0. 0.01935722 0.98064278]\n [0. 0.09393501 0.90606499]\n [0. 0.01935722 0.98064278]\n [0. 0.01935722 0.98064278]\n [0. 0.09155897 0.90844103]]\n"
],
[
"score = clf2.score(X, y)\nprint(\"Score: %f\" % score)",
"Score: 0.966667\n"
],
[
"from sklearn.pipeline import make_pipeline\npipe = make_pipeline(DecisionForestClassifier(random_state=0))\npipe.fit(X, y)\npipe.predict(X)\npipe.predict_proba(X)\nscore = pipe.score(X, y)\nprint(\"Score: %f\" % score)",
"Score: 0.966667\n"
],
[
"from sklearn.model_selection import GridSearchCV\nparameters = [{'n_estimators': [10, 20],\n 'bootstrap': [False, True],\n 'max_features': [None, 1],\n 'max_thresholds': [None, 1]}]\ngrid_search = GridSearchCV(DecisionForestClassifier(random_state=0), parameters, cv=3, iid=False)\ngrid_search.fit(X, y)\nprint(grid_search.best_params_)\nclf = DecisionForestClassifier(random_state=0)\nclf.set_params(**grid_search.best_params_)\nclf.fit(X, y)\nscore = clf.score(X, y)\nprint(\"Score: %f\" % score)",
"{'bootstrap': False, 'max_features': None, 'max_thresholds': 1, 'n_estimators': 10}\nScore: 0.966667\n"
],
[
"# $: conda install dask distributed\nfrom dask.distributed import Client\nclient = Client()\n# Firefox: http://localhost:8787/status",
"_____no_output_____"
],
[
"clf = DecisionForestClassifier(random_state=0)\nfrom sklearn.externals.joblib import parallel_backend\nwith parallel_backend('dask', n_jobs=-1): # 'loky' when not using dask\n clf.fit(X, y)\n score = clf.score(X, y)\nprint(\"Score: %f\" % score)",
"/home/drh/sw/anaconda3/envs/conda_test/lib/python3.7/site-packages/sklearn/externals/joblib/__init__.py:15: DeprecationWarning: sklearn.externals.joblib is deprecated in 0.21 and will be removed in 0.23. Please import this functionality directly from joblib, which can be installed with: pip install joblib. If this warning is raised when loading pickled models, you may need to re-serialize those models with scikit-learn 0.21+.\n warnings.warn(msg, category=DeprecationWarning)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71bc52d4e930c7e58befd4f282829ae3b89acf2 | 5,718 | ipynb | Jupyter Notebook | Preprocessing.ipynb | guillaumesimler/ML-Toolkit---Regression | 9eed0176625eecdf04805646fe73924b8a58af63 | [
"MIT"
] | null | null | null | Preprocessing.ipynb | guillaumesimler/ML-Toolkit---Regression | 9eed0176625eecdf04805646fe73924b8a58af63 | [
"MIT"
] | null | null | null | Preprocessing.ipynb | guillaumesimler/ML-Toolkit---Regression | 9eed0176625eecdf04805646fe73924b8a58af63 | [
"MIT"
] | null | null | null | 21.099631 | 190 | 0.526058 | [
[
[
"# Pre-processing template\n\nThis template allows a faster processing of the data. In the current setting, it is designed for \n- importing **csv** \n\n### Import the modules\n\nContrary to the usual data _scientist_ approach, the module are imported on top of the file and not in the specific cells.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.preprocessing import OneHotEncoder\nfrom sklearn.compose import ColumnTransformer",
"_____no_output_____"
]
],
[
[
"### Declaration of variables",
"_____no_output_____"
]
],
[
[
"# Main variables\n\nsrc_file = 'Data.csv'\ndepend_var_index = -1\ncateg_var_index = 3",
"_____no_output_____"
]
],
[
[
"### Importing the data sets",
"_____no_output_____"
]
],
[
[
"dataset = pd.read_csv(src_file)\nX = dataset.iloc[:, :depend_var_index].values\ny = dataset.iloc[:, depend_var_index].values",
"_____no_output_____"
]
],
[
[
"### Encoding categorical data (not always used)",
"_____no_output_____"
]
],
[
[
"transformer = ColumnTransformer([('one_hot_encoder', OneHotEncoder(), [categ_var_index])],remainder='passthrough')\n\nX = np.array(transformer.fit_transform(X), dtype=np.float)",
"_____no_output_____"
]
],
[
[
"### Avoiding the Dummy trap (not always used - used in combination with the encoded categorical data)",
"_____no_output_____"
]
],
[
[
"X = X[:, 1:]",
"_____no_output_____"
]
],
[
[
"### Splitting the dataset into the Training and Test set\n\nDon't use it with a small dataset. The parameter, random_state, is not used. \n\nIt ensured the same results for all members of the course.",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2)",
"_____no_output_____"
]
],
[
[
"### Feature Scaling\n\nActually, the regression data should be normalized or standardized. Most libraries do it natively, but not less frequent.\n\nThe library for SVM belongs to the latter. ",
"_____no_output_____"
]
],
[
[
"\"\"\"\nsc_X = StandardScaler()\nX_train = sc_X.fit_transform(X_train)\nX_test = sc_X.transform(X_test)\nsc_y = StandardScaler()\ny_train = sc_y.fit_transform(y_train)\"\"\"\n\n\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71bd5eab1f2d102f1d05f971f223aee341d0c2d | 113,531 | ipynb | Jupyter Notebook | analytics/workspace/.ipynb_checkpoints/【未使用だけど保存用】01_探索的データ解析-Copy1-checkpoint.ipynb | tomoya-innovator/NullSuck-AI | 1c2a697196245b45ad9a8a3c9e90e93073ece2d1 | [
"MIT"
] | 14 | 2019-02-11T07:56:18.000Z | 2021-11-15T12:34:39.000Z | analytics/workspace/.ipynb_checkpoints/【未使用だけど保存用】01_探索的データ解析-Copy1-checkpoint.ipynb | tomoya-innovator/NullSuck-AI | 1c2a697196245b45ad9a8a3c9e90e93073ece2d1 | [
"MIT"
] | 6 | 2019-02-19T16:03:22.000Z | 2021-11-18T01:34:31.000Z | analytics/workspace/.ipynb_checkpoints/【未使用だけど保存用】01_探索的データ解析-Copy1-checkpoint.ipynb | tomoya-innovator/NullSuck-AI | 1c2a697196245b45ad9a8a3c9e90e93073ece2d1 | [
"MIT"
] | 12 | 2019-04-25T13:44:02.000Z | 2021-11-17T05:45:22.000Z | 45.760177 | 5,964 | 0.479305 | [
[
[
"# 探索的データ解析\n---\n予測する前に、データセットの現状を把握する",
"_____no_output_____"
],
[
"### 分析環境構築",
"_____no_output_____"
]
],
[
[
"# 分析に必要なライブラリをインポート\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### データセットを読み込む",
"_____no_output_____"
]
],
[
[
"# データセットを読み込む\n# 読み込んだデータセットをdfという名前のデータフレームに代入する\ndf = pd.read_csv(\"../dataset/winequality-red.csv\")",
"_____no_output_____"
],
[
"# データの先頭行を表示する\ndf.head()",
"_____no_output_____"
]
],
[
[
"- 列ごとに表示されていないので、読み込み方を工夫する必要がある\n- ;(セミコロン)で列に分割することができそうだ\n- sep=';'を追記して、列に分割してみよう",
"_____no_output_____"
]
],
[
[
"# データセットを読み込む\ndf = pd.read_csv(\"../dataset/winequality-red.csv\", sep=\";\")\ndf.head()",
"_____no_output_____"
]
],
[
[
"- データテーブルとして綺麗に列を分割して表示されている",
"_____no_output_____"
],
[
"## 列名の意味を調べる\n---\n- 下記のリンクをクリックすると、公開用のスプレッドシートに飛びます\n- [スプレッドシートで翻訳関数を使ってみよう](https://docs.google.com/spreadsheets/d/1q2uugJ5_jtUcJeI8nHAX90Aa3TastjLwYW9NySmQ6ag/edit?usp=sharing)\n",
"_____no_output_____"
],
[
"### カラム名一覧",
"_____no_output_____"
],
[
"| 英語 | google翻訳 | 修正した日本語訳 |\n|:-----------|------------:|:------------:|\n| fixed acidity | 固定酸度 | This |\n| volatile acidity | 揮発性の酸味 | This |\n| citric acid | クエン酸 | This |\n| residual sugar | 残留糖 | This |\n| chlorides | 塩化物 | This |\n| free sulfur dioxide | 遊離二酸化硫黄 | This |\n| total sulfur dioxide | 総二酸化硫黄 | This |\n| density | 密度 | This |\n| pH | pH | This |\n| sulphates | 硫酸塩 | This |\n| alcohol | アルコール | This |\n| quality | 品質 | This |",
"_____no_output_____"
],
[
"## 集計",
"_____no_output_____"
],
[
"### データ量を把握する(shape)",
"_____no_output_____"
]
],
[
[
"df.shape",
"_____no_output_____"
]
],
[
[
"- およそ1600件のデータである\n- 1599行, 12列のデータテーブル",
"_____no_output_____"
],
[
"### データ個数を調べる(count)",
"_____no_output_____"
]
],
[
[
"df.count()",
"_____no_output_____"
]
],
[
[
"- 今回は欠損はなさそう\n- 異常値が入っていないかは注意して進める",
"_____no_output_____"
],
[
"## 記述統計量(describe)",
"_____no_output_____"
]
],
[
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"- 今回は、ワインの美味しさを「品質スコア」と定義する\n- 「品質スコア」はデータテーブルでは、「quality」に格納されている\n- 「quality」の最大値と最小値はそれぞれ<strong>8と3</strong>である\n - 一般的に最小値3, 最大値8の選択肢はなさそうだ\n - なので、0~10段階の選択肢があると推測する \n- 「quality」の平均値は<strong>約5.64</strong>である\n - 0〜10の10段階で、平均が5.6であるなら、「quality」の値は正しそうだ\n- 「quality」の標準偏差は<strong>約0.81</strong>である\n - 後述",
"_____no_output_____"
],
[
"## データ型",
"_____no_output_____"
]
],
[
[
"df.dtypes",
"_____no_output_____"
]
],
[
[
"- 64bitの浮動小数点数なので数値\n- 数値データで構成されているため、モデルは作りやすいと予想できる",
"_____no_output_____"
],
[
"## 可視化",
"_____no_output_____"
]
],
[
[
"# 可視化ライブラリのインポート\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"### 度数分布",
"_____no_output_____"
]
],
[
[
"plt.hist(df['quality'], bins=10 )",
"_____no_output_____"
]
],
[
[
"### 標準化(平均0分散1)",
"_____no_output_____"
]
],
[
[
"# StandardScalerクラスによる標準化\nfrom sklearn.preprocessing import StandardScaler\n\n#インスタンス化\nscaler = StandardScaler()\nscaler.fit(df)\nscaler.transform(df)\ndf_std = pd.DataFrame(scaler.transform(df), columns=df.columns)",
"_____no_output_____"
],
[
"df_std.describe()",
"_____no_output_____"
],
[
"plt.hist(df_std.quality)",
"_____no_output_____"
]
],
[
[
"### 正規化(最大1, 最小0)",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\nmmscaler = preprocessing.MinMaxScaler() # インスタンスの作成\n\nmmscaler.fit(df) # xの最大・最小を計算\ny = mmscaler.transform(df) # xを変換\ny.describe()",
"_____no_output_____"
],
[
"df0 = df.iloc[:,:-1] # 毎回スライスするのはくどいので別の変数に代入しておきます。\ndf3 = (df0 - df0.mean()) / (df0.max() - df0.min())\ndf3.head()",
"_____no_output_____"
],
[
"df5 = df.apply(lambda x: (x-x.mean())/x.std(), axis=0).fillna(0)\ndf5.describe()",
"_____no_output_____"
],
[
"\"\"\"\n1. z-score normalization\n以下の式で変換\nx_new = (x - x_mean) / x_std\n外れ値にもロバスト\n\"\"\"\nstandardized_sample_df = (df - df.mean()) / df.std()\nprint(standardized_sample_df)",
" fixed acidity volatile acidity citric acid residual sugar chlorides \\\n0 -0.528194 0.961576 -1.391037 -0.453077 -0.243630 \n1 -0.298454 1.966827 -1.391037 0.043403 0.223805 \n2 -0.298454 1.296660 -1.185699 -0.169374 0.096323 \n3 1.654339 -1.384011 1.483689 -0.453077 -0.264878 \n4 -0.528194 0.961576 -1.391037 -0.453077 -0.243630 \n5 -0.528194 0.738187 -1.391037 -0.524002 -0.264878 \n6 -0.241019 0.403103 -1.083031 -0.665853 -0.392360 \n7 -0.585629 0.682339 -1.391037 -0.949556 -0.477348 \n8 -0.298454 0.291408 -1.288368 -0.382151 -0.307372 \n9 -0.470759 -0.155370 0.457001 2.525799 -0.349866 \n10 -0.930240 0.291408 -0.980362 -0.524002 0.202558 \n11 -0.470759 -0.155370 0.457001 2.525799 -0.349866 \n12 -1.562026 0.486874 -1.391037 -0.665853 0.032582 \n13 -0.298454 0.458950 0.097661 -0.665853 0.563758 \n14 0.333332 0.514797 -0.467018 0.894510 1.881077 \n15 0.333332 0.514797 -0.415683 0.965435 1.753595 \n16 0.103591 -1.384011 1.483689 -0.524002 0.096323 \n17 -0.126149 0.179714 0.046326 -0.594928 5.960515 \n18 -0.528194 0.347256 -0.980362 1.320063 -0.031160 \n19 -0.241019 -1.160621 1.227017 -0.524002 5.386844 \n20 0.333332 -1.719094 1.073014 -0.524002 -0.222383 \n21 -0.413324 -0.769690 0.200329 -0.169374 -0.116148 \n22 -0.241019 -0.546301 -0.313015 -0.665853 0.393782 \n23 0.103591 -0.211217 -0.826359 -0.169374 -0.073654 \n24 -0.815370 -0.713843 -0.672355 -0.098449 -0.052407 \n25 -1.159980 -0.769690 -0.569687 -0.807705 -0.158642 \n26 -0.413324 -0.657996 -0.159011 -0.524002 -0.158642 \n27 -0.241019 -0.546301 -0.313015 -0.665853 0.393782 \n28 -0.700500 1.017423 -1.391037 -0.453077 -0.158642 \n29 -0.298454 0.654416 -1.391037 -0.382151 -0.116148 \n... ... ... ... ... ... \n1569 -1.217415 -0.099523 -0.672355 -0.453077 -0.668572 \n1570 -1.102545 -0.937232 1.329686 -0.240300 3.028419 \n1571 -1.102545 -0.825538 -0.672355 -0.240300 -1.051019 \n1572 -0.585629 0.905729 0.251664 -0.240300 -0.392360 \n1573 -1.332285 0.291408 -0.364349 -0.098449 -0.264878 \n1574 -1.562026 -1.216469 2.613046 8.057996 -0.286125 \n1575 -0.470759 -0.043675 0.662339 -0.240300 -0.583584 \n1576 -0.183584 -1.272316 1.843030 -0.665853 -0.137395 \n1577 -1.217415 0.961576 -0.621021 1.816543 -0.243630 \n1578 -0.872805 0.794034 -0.621021 -0.524002 0.648747 \n1579 -1.217415 0.179714 -0.929027 -0.594928 -0.732313 \n1580 -0.528194 -0.993079 0.302998 -0.098449 -0.413607 \n1581 -1.217415 0.179714 -0.929027 -0.594928 -0.732313 \n1582 -1.274850 1.045347 -0.877693 0.043403 -0.732313 \n1583 -1.217415 -0.378759 0.097661 -0.311225 -0.286125 \n1584 -0.930240 -1.160621 0.867677 -0.098449 -0.562337 \n1585 -0.643065 -0.769690 0.867677 0.043403 -0.456101 \n1586 -0.470759 -1.216469 0.713673 -0.098449 -0.477348 \n1587 -1.447156 0.458950 -0.826359 -0.524002 -0.456101 \n1588 -0.643065 0.738187 0.302998 -0.027523 -0.413607 \n1589 -0.987675 1.101194 -0.364349 3.731534 -0.307372 \n1590 -1.159980 0.123866 -0.621021 -0.524002 -0.222383 \n1591 -1.676896 1.184965 -0.929027 -0.594928 0.032582 \n1592 -1.159980 -0.099523 -0.723690 -0.169374 -0.243630 \n1593 -0.872805 0.514797 -0.980362 -0.453077 -0.413607 \n1594 -1.217415 0.403103 -0.980362 -0.382151 0.053829 \n1595 -1.389721 0.123866 -0.877693 -0.240300 -0.541090 \n1596 -1.159980 -0.099523 -0.723690 -0.169374 -0.243630 \n1597 -1.389721 0.654416 -0.775024 -0.382151 -0.264878 \n1598 -1.332285 -1.216469 1.021680 0.752659 -0.434854 \n\n free sulfur dioxide total sulfur dioxide density pH \\\n0 -0.466047 -0.379014 0.558100 1.288240 \n1 0.872365 0.624168 0.028252 -0.719708 \n2 -0.083643 0.228975 0.134222 -0.331073 \n3 0.107558 0.411372 0.664069 -0.978798 \n4 -0.466047 -0.379014 0.558100 1.288240 \n5 -0.274845 -0.196617 0.558100 1.288240 \n6 -0.083643 0.380972 -0.183687 -0.071983 \n7 -0.083643 -0.774207 -1.137414 0.510970 \n8 -0.657248 -0.865405 0.028252 0.316652 \n9 0.107558 1.688149 0.558100 0.251880 \n10 -0.083643 0.563369 -0.448611 -0.201528 \n11 0.107558 1.688149 0.558100 0.251880 \n12 0.011958 0.380972 -1.296368 1.741648 \n13 -0.657248 -0.531011 0.346161 -0.331073 \n14 3.453589 2.995326 0.981978 -0.978798 \n15 3.357988 3.086524 0.981978 -0.914026 \n16 1.828374 1.718548 0.081237 -0.071983 \n17 0.011958 0.289774 0.028252 -1.302661 \n18 -0.944051 -0.531011 0.346161 0.446197 \n19 0.107558 0.289774 0.081237 -1.756068 \n20 1.254769 0.411372 0.028252 0.510970 \n21 0.681164 0.745766 0.770039 1.353012 \n22 -0.561648 -0.287816 -0.077718 -0.914026 \n23 -0.657248 0.624168 0.028252 -0.914026 \n24 0.489962 -0.196617 0.028252 0.770060 \n25 -0.466047 -0.713408 -0.660550 0.187107 \n26 -1.135253 -1.078202 -0.289657 -0.201528 \n27 -0.561648 -0.287816 -0.077718 -0.914026 \n28 -0.179244 -0.348615 0.240191 1.029150 \n29 -0.752849 -0.926204 -0.183687 0.446197 \n... ... ... ... ... \n1569 -0.083643 -0.379014 -1.476516 1.093922 \n1570 0.298760 -0.348615 -1.773231 0.381425 \n1571 -0.083643 -0.652609 -0.851296 0.834832 \n1572 1.828374 1.748948 -0.226075 0.122335 \n1573 -0.083643 0.107377 -1.100324 1.741648 \n1574 0.681164 1.384154 0.012356 0.510970 \n1575 -0.370446 -0.804607 -1.063235 -0.331073 \n1576 0.011958 -0.531011 -0.459208 -0.071983 \n1577 -0.274845 -0.591810 -0.279060 1.482557 \n1578 -0.274845 -0.804607 -0.713535 0.705287 \n1579 0.776764 -0.439813 -1.444725 1.482557 \n1580 -0.657248 -0.622210 -1.084429 0.316652 \n1581 0.776764 -0.439813 -1.444725 1.482557 \n1582 -0.274845 -0.591810 -1.656665 1.676875 \n1583 1.541571 1.566551 -0.512193 0.122335 \n1584 0.776764 -0.379014 -1.010250 -0.136755 \n1585 0.585563 0.046578 -0.957265 -0.071983 \n1586 1.732773 0.411372 -0.967862 0.187107 \n1587 0.203159 -0.561411 -1.015549 1.547330 \n1588 1.732773 1.688149 -1.381144 -0.266301 \n1589 1.254769 0.988961 0.505115 -0.136755 \n1590 0.967966 -0.348615 -1.910992 0.057562 \n1591 0.011958 -0.622210 -1.444725 2.324600 \n1592 1.254769 -0.196617 -0.533387 0.705287 \n1593 1.159168 -0.257416 -0.125404 0.705287 \n1594 1.541571 -0.075020 -0.978459 0.899605 \n1595 2.210777 0.137777 -0.861893 1.353012 \n1596 1.254769 -0.196617 -0.533387 0.705287 \n1597 1.541571 -0.075020 -0.676446 1.676875 \n1598 0.203159 -0.135818 -0.665849 0.510970 \n\n sulphates alcohol quality \n0 -0.579025 -0.959946 -0.787576 \n1 0.128910 -0.584594 -0.787576 \n2 -0.048074 -0.584594 -0.787576 \n3 -0.461036 -0.584594 0.450707 \n4 -0.579025 -0.959946 -0.787576 \n5 -0.579025 -0.959946 -0.787576 \n6 -1.168972 -0.959946 -0.787576 \n7 -1.109977 -0.396918 1.688991 \n8 -0.520031 -0.866108 1.688991 \n9 0.836846 0.072271 -0.787576 \n10 -0.697015 -1.147622 -0.787576 \n11 0.836846 0.072271 -0.787576 \n12 -0.815004 -0.490756 -0.787576 \n13 5.320437 -1.241459 -0.787576 \n14 1.308802 -1.147622 -0.787576 \n15 1.603776 -1.147622 -0.787576 \n16 0.541872 0.072271 1.688991 \n17 3.668587 -1.053784 -0.787576 \n18 -0.932993 -1.335297 -2.025860 \n19 2.488695 -1.147622 0.450707 \n20 -0.756009 -0.959946 0.450707 \n21 -0.048074 -0.678432 -0.787576 \n22 1.485786 -0.866108 -0.787576 \n23 -0.756009 -0.959946 -0.787576 \n24 -0.166063 -0.678432 0.450707 \n25 -0.579025 -1.053784 -0.787576 \n26 -0.402042 -0.866108 -0.787576 \n27 1.485786 -0.866108 -0.787576 \n28 -0.638020 -0.959946 -0.787576 \n29 -0.402042 -0.584594 0.450707 \n... ... ... ... \n1569 -0.520031 1.010650 0.450707 \n1570 1.603776 1.855191 0.450707 \n1571 -0.048074 0.635298 0.450707 \n1572 -0.873998 -0.866108 -0.787576 \n1573 0.069915 1.949029 0.450707 \n1574 -1.050982 0.072271 0.450707 \n1575 -0.107068 1.292164 0.450707 \n1576 0.718856 0.353785 0.450707 \n1577 -0.343047 1.386002 0.450707 \n1578 0.069915 0.822974 0.450707 \n1579 -0.343047 0.822974 -0.787576 \n1580 -0.343047 1.386002 0.450707 \n1581 -0.343047 0.822974 -0.787576 \n1582 -0.932993 1.386002 -0.787576 \n1583 -0.225058 -0.584594 -0.787576 \n1584 0.836846 1.104488 1.688991 \n1585 1.072824 1.010650 0.450707 \n1586 1.131819 0.916812 0.450707 \n1587 0.010921 0.447623 0.450707 \n1588 0.718856 2.230543 0.450707 \n1589 -0.697015 -1.147622 -0.787576 \n1590 0.954835 1.104488 0.450707 \n1591 -0.579025 1.104488 0.450707 \n1592 0.541872 0.541460 0.450707 \n1593 0.954835 -0.866108 0.450707 \n1594 -0.461036 0.072271 -0.787576 \n1595 0.600867 0.729136 0.450707 \n1596 0.541872 0.541460 0.450707 \n1597 0.305894 -0.209243 -0.787576 \n1598 0.010921 0.541460 0.450707 \n\n[1599 rows x 12 columns]\n"
],
[
"standardized_sample_df.describe()",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"min_max_scaler = preprocessing.MinMaxScaler()\na_minmax = min_max_scaler.fit_transform(df)\n\nprint(a_minmax.max(axis=0)) # [1. 1. 1.]\nprint(a_minmax.min(axis=0)) # [0. 0. 0.]\nprint(a_minmax.mean(axis=0)) # [0.35185185 0.36206897 0.37096774]\nprint(a_minmax.var(axis=0)) # [0.10989941 0.10882767 0.1079566 ]\n",
"[1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n[0.32917144 0.27932912 0.27097561 0.11224695 0.12598755 0.20950594\n 0.14299573 0.49021139 0.44969543 0.19649631 0.31122817 0.5272045 ]\n[0.02372559 0.01503205 0.03792375 0.00932001 0.00616988 0.02169142\n 0.0135028 0.01918985 0.0147686 0.01029605 0.02686242 0.02607042]\n"
],
[
"df_a_minmax = pd.DataFrame(min_max_scaler.fit_transform(df))\ndf_a_minmax.head()",
"_____no_output_____"
],
[
"df_a_minmax.describe()",
"_____no_output_____"
],
[
"plt.hist(df_a_minmax[\"quality\"])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71bd62412683c816fd95bc2314fe28da38a373f | 2,002 | ipynb | Jupyter Notebook | extras/chapter_01_concepts/01_11_creating_a_face-on_ring.ipynb | samueltwum1/TiRiFiG | eafef39cff858851e051a38db6d1329388daebba | [
"MIT"
] | 1 | 2021-02-04T12:04:22.000Z | 2021-02-04T12:04:22.000Z | extras/chapter_01_concepts/01_11_creating_a_face-on_ring.ipynb | samueltwum1/TiRiFiG | eafef39cff858851e051a38db6d1329388daebba | [
"MIT"
] | 3 | 2018-11-27T13:29:31.000Z | 2019-01-17T14:07:28.000Z | extras/chapter_01_concepts/01_11_creating_a_face-on_ring.ipynb | samueltwum1/TiRiFiG | eafef39cff858851e051a38db6d1329388daebba | [
"MIT"
] | 1 | 2019-02-15T14:14:46.000Z | 2019-02-15T14:14:46.000Z | 19.821782 | 107 | 0.53996 | [
[
[
"***\n\n* [Outline](../0_Introduction/0_introduction.ipynb)\n* [Glossary](../0_Introduction/1_glossary.ipynb)\n* [1. Building the Concepts](01_00_introduction.ipynb) \n * Previous: [1.10 Convolution](01_10_convolution.ipynb)\n * Next: [1. Concepts: References and Further Reading](01_references_and_further_reading.ipynb)\n\n***",
"_____no_output_____"
],
[
"Import standard modules:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom IPython.display import HTML \nHTML('../style/course.css') #apply general CSS",
"_____no_output_____"
]
],
[
[
"Import section specific modules",
"_____no_output_____"
]
],
[
[
"pass",
"_____no_output_____"
]
],
[
[
"***\n\n* Next: [1. Concepts: References and Further Reading](01_references_and_further_reading.ipynb)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e71bd7e524dc87757d396e9c2906208995ff8e3b | 33,359 | ipynb | Jupyter Notebook | season1/01.MNIST.ipynb | minssoj/Study_MLDLCT | 5006f3b22b00d3aac752c11378fecff999569896 | [
"MIT"
] | 1 | 2021-12-16T06:55:25.000Z | 2021-12-16T06:55:25.000Z | season1/01.MNIST.ipynb | minssoj/Study_MLDLCT | 5006f3b22b00d3aac752c11378fecff999569896 | [
"MIT"
] | null | null | null | season1/01.MNIST.ipynb | minssoj/Study_MLDLCT | 5006f3b22b00d3aac752c11378fecff999569896 | [
"MIT"
] | 1 | 2021-12-29T13:48:31.000Z | 2021-12-29T13:48:31.000Z | 67.256048 | 20,171 | 0.782697 | [
[
[
"# [프로젝트] 손글씨 숫자 인식\n1. 모듈 불러오기\n2. 데이터셋 준비하기\n3. 모델 설계하기\n4. 모델 학습하기",
"_____no_output_____"
],
[
"## 1. 모듈 불러오기",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torchvision import transforms, datasets",
"_____no_output_____"
],
[
"if torch.cuda.is_available():\n DEVICE = torch.device('cuda')\nelse:\n DEVICE = torch.device('cpu')\nprint(f'PyTorch Vesion : {torch.__version__}')\nprint(f'Device : {DEVICE}')",
"PyTorch Vesion : 1.9.0+cu111\nDevice : cuda\n"
]
],
[
[
"## 2. 데이터셋 준비하기",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 32\ntrain_dataset = datasets.MNIST(root = \"../data/MNIST\",\n train = True,\n download = True,\n transform = transforms.ToTensor())\n\ntest_dataset = datasets.MNIST(root = \"../data/MNIST\",\n train = False,\n transform = transforms.ToTensor())\n\ntrain_loader = torch.utils.data.DataLoader(dataset = train_dataset,\n batch_size = BATCH_SIZE,\n shuffle = True)\n\ntest_loader = torch.utils.data.DataLoader(dataset = test_dataset,\n batch_size = BATCH_SIZE,\n shuffle = False)",
"Downloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz\nDownloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz to ../data/MNIST/MNIST/raw/train-images-idx3-ubyte.gz\n"
],
[
"for (X_train, y_train) in train_loader:\n print('X_train:', X_train.size(), 'type:', X_train.type())\n print('y_train:', y_train.size(), 'type:', y_train.type())\n break",
"X_train: torch.Size([32, 1, 28, 28]) type: torch.FloatTensor\ny_train: torch.Size([32]) type: torch.LongTensor\n"
],
[
"pltsize = 1\nplt.figure(figsize=(10 * pltsize, pltsize))\nfor i in range(10):\n plt.subplot(1, 10, i + 1)\n plt.axis('off')\n plt.imshow(X_train[i, :, :, :].numpy().reshape(28, 28), cmap = \"gray_r\")\n plt.title('Class: ' + str(y_train[i].item()))\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 3. 모델 설계하기",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 32\nEPOCHS = 10\nINPUT_SIZE = 784",
"_____no_output_____"
],
[
"class TwoLayerNet(nn.Module):\n def __init__(self, input_size, hidden_size, output_size):\n super(TwoLayerNet, self).__init__() # nn.Module 내에 있는 메서드 상속\n self.fc1 = nn.Linear(input_size, hidden_size)\n self.fc2 = nn.Linear(hidden_size, output_size)\n\n def forward(self, x, input_size=INPUT_SIZE):\n x = x.view(-1, input_size) # Flatten\n x = self.fc1(x)\n x = F.sigmoid(x)\n x = self.fc2(x)\n x = F.log_softmax(x, dim = 1)\n return x",
"_____no_output_____"
],
[
"model = TwoLayerNet(input_size=784, hidden_size=100, output_size=10).to(DEVICE)\noptimizer = torch.optim.SGD(model.parameters(), lr = 0.01, momentum = 0.5)\ncriterion = nn.CrossEntropyLoss()\n\nprint(model)",
"TwoLayerNet(\n (fc1): Linear(in_features=784, out_features=100, bias=True)\n (fc2): Linear(in_features=100, out_features=10, bias=True)\n)\n"
]
],
[
[
"## 4. 모델 학습하기",
"_____no_output_____"
]
],
[
[
"def train(model, train_loader, optimizer, log_interval):\n model.train()\n for batch_idx, (image, label) in enumerate(train_loader):\n image = image.to(DEVICE)\n label = label.to(DEVICE)\n optimizer.zero_grad()\n output = model(image)\n loss = criterion(output, label)\n loss.backward()\n optimizer.step()\n\n if batch_idx % log_interval == 0:\n print(\"Train Epoch: {} [{}/{} ({:.0f}%)]\\tTrain Loss: {:.6f}\".format(\n epoch, batch_idx * len(image),\n len(train_loader.dataset), 100. * batch_idx / len(train_loader),\n loss.item()))",
"_____no_output_____"
],
[
"def evaluate(model, test_loader):\n model.eval()\n test_loss = 0\n correct = 0\n\n with torch.no_grad():\n for image, label in test_loader:\n image = image.to(DEVICE)\n label = label.to(DEVICE)\n output = model(image)\n test_loss += criterion(output, label).item()\n prediction = output.max(1, keepdim = True)[1]\n print(prediction)\n print(prediction.eq(label.view_as(prediction)).sum())\n correct += prediction.eq(label.view_as(prediction)).sum().item()\n\n test_loss /= (len(test_loader.dataset) / BATCH_SIZE)\n test_accuracy = 100. * correct / len(test_loader.dataset)\n return test_loss, test_accuracy",
"_____no_output_____"
],
[
"for epoch in range(1, EPOCHS + 1):\n train(model, train_loader, optimizer, log_interval = 200)\n test_loss, test_accuracy = evaluate(model, test_loader)\n print(\"\\n[EPOCH: {}], \\tTest Loss: {:.4f}, \\tTest Accuracy: {:.2f} % \\n\".format(\n epoch, test_loss, test_accuracy))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e71be1e2b0c74f6d9ed088cfcb71f38effea24ff | 217,645 | ipynb | Jupyter Notebook | Lectures/Lecture03 -- Climate sensitivity and feedback.ipynb | pankajcivil/ClimateModeling_courseware | f5e277350cc0fb76301a3a7abd7cac4a5d58b1d7 | [
"MIT"
] | null | null | null | Lectures/Lecture03 -- Climate sensitivity and feedback.ipynb | pankajcivil/ClimateModeling_courseware | f5e277350cc0fb76301a3a7abd7cac4a5d58b1d7 | [
"MIT"
] | null | null | null | Lectures/Lecture03 -- Climate sensitivity and feedback.ipynb | pankajcivil/ClimateModeling_courseware | f5e277350cc0fb76301a3a7abd7cac4a5d58b1d7 | [
"MIT"
] | 1 | 2021-02-20T03:10:31.000Z | 2021-02-20T03:10:31.000Z | 176.373582 | 140,460 | 0.888791 | [
[
[
"# [ATM 623: Climate Modeling](../index.ipynb)\n\n[Brian E. J. Rose](http://www.atmos.albany.edu/facstaff/brose/index.html), University at Albany\n\n# Lecture 3: Climate sensitivity and feedback",
"_____no_output_____"
],
[
"### About these notes:\n\nThis document uses the interactive [`Jupyter notebook`](https://jupyter.org) format. The notes can be accessed in several different ways:\n\n- The interactive notebooks are hosted on `github` at https://github.com/brian-rose/ClimateModeling_courseware\n- The latest versions can be viewed as static web pages [rendered on nbviewer](http://nbviewer.ipython.org/github/brian-rose/ClimateModeling_courseware/blob/master/index.ipynb)\n- A complete snapshot of the notes as of May 2017 (end of spring semester) are [available on Brian's website](http://www.atmos.albany.edu/facstaff/brose/classes/ATM623_Spring2017/Notes/index.html).\n\n[Also here is a legacy version from 2015](http://www.atmos.albany.edu/facstaff/brose/classes/ATM623_Spring2015/Notes/index.html).\n\nMany of these notes make use of the `climlab` package, available at https://github.com/brian-rose/climlab",
"_____no_output_____"
]
],
[
[
"# Ensure compatibility with Python 2 and 3\nfrom __future__ import print_function, division",
"_____no_output_____"
]
],
[
[
"## Contents\n\n1. [Radiative forcing](#section1)\n2. [Climate sensitivity (without feedback)](#section2)\n3. [The feedback concept](#section3)\n4. [Climate feedback: some definitions](#section4)\n5. [Climate sensitivity with feedback](#section5)\n6. [Contribution of individual feedback processes to Equilibrium Climate Sensitivity](#section6)\n7. [Feedbacks diagnosed from complex climate models](#section7)\n8. [Feedback analysis of the zero-dimensional model with variable albedo](#section8)",
"_____no_output_____"
],
[
"____________\n<a id='section1'></a>\n\n## 1. Radiative forcing\n____________\n\nLet’s say we instantaneously double atmospheric CO$_2$. What happens?\n\n- The atmosphere is less efficient at radiating energy away to space.\n- OLR will decrease\n- The climate system will begin gaining energy.\n\n",
"_____no_output_____"
],
[
"We will call this abrupt decrease in OLR the **radiative forcing**, a positive number in W m$^{-2}$\n\n$$ \\Delta R = OLR_i - OLR_{2xCO2} $$\n\n$\\Delta R$ is a measure of the rate at which energy begins to accumulate in the climate system after an abrupt increase in greenhouse gases, but *before any change in climate* (i.e. temperature).\n\nWhat happens next?",
"_____no_output_____"
],
[
"____________\n<a id='section2'></a>\n\n## 2. Climate sensitivity (without feedback)\n____________\n\nLet’s use our simple zero-dimensional EBM to calculate the resulting change in **equilibrium temperature**. \n\nHow much warming will we get once the climate system has adjusted to the radiative forcing?",
"_____no_output_____"
],
[
"First note that at equilibrium we must have\n\n$$ASR = OLR$$\n\nand in our very simple model, there is no change in ASR, so \n$$ ASR_f = ASR_f $$\n(with standing for final.)\n\nFrom this we infer that\n$$ OLR_f = OLR_i $$\n\nThe new equilibrium will have **exactly the same OLR** as the old equilibrium, but a **different surface temperature**.\n\nThe climate system must warm up by a certain amount to get the OLR back up to its original value! The question is, **how much warming is necessary**? In other words, **what is the new equilibrium temperature**?\n",
"_____no_output_____"
],
[
"### Equilibrium Climate Sensitivity (ECS)\n\nWe now define the Equilibrium Climate Sensitivity (denoted ECS or $\\Delta T_{2xCO2}$):\n\n*The global mean surface warming necessary to balance the planetary energy budget after a doubling of atmospheric CO$_2$.*\n\nThe temperature must increase so that the increase in OLR is exactly equal to the radiative forcing:\n\n$$ OLR_f - OLR_{2xCO2} = \\Delta R $$",
"_____no_output_____"
],
[
"From last lecture, we have linearized our model for OLR with a slope $\\lambda_0 = 3.3$ W m$^{-2}$ K$^{-1}$. This means that a global warming of 1 degree causes a 3.3 W m$^{-2}$ increase in the OLR. So we can write:\n\n$$OLR_f \\approx OLR_{2xCO2} + \\lambda_0 \\Delta T_0 $$\n\nwhere we are writing the change in temperature as\n\n$$ \\Delta T_0 = T_f - T_i $$\n\n(and the subscript zero will remind us that this is the response in the simplest model, in the absence of any feedbacks)",
"_____no_output_____"
],
[
"To achieve energy balance, the planet must warm up by\n\n$$ \\Delta T_0 = \\frac{\\Delta R}{\\lambda_0} $$\n\nAs we will see later, the actual radiative forcing due CO$_2$ doubling is about 4 W m$^{-2}$. \n\nSo our model without feedback gives a prediction for climate sensitivity:",
"_____no_output_____"
]
],
[
[
"# Repeating code from Lecture 2\nsigma = 5.67E-8 # Stefan-Boltzmann constant in W/m2/K4\nQ = 341.3 # global mean insolation in W/m2\nalpha = 101.9 / Q # observed planetary albedo\nTe = ((1-alpha)*Q/sigma)**0.25 # Emission temperature (definition)\nTsbar = 288. # global mean surface temperature in K\nbeta = Te / Tsbar # Calculate value of beta from observations\nlambda_0 = 4 * sigma * beta**4 * Tsbar**3",
"_____no_output_____"
],
[
"DeltaR = 4. # Radiative forcing in W/m2\nDeltaT0 = DeltaR / lambda_0\nprint( 'The Equilibrium Climate Sensitivity in the absence of feedback is {:.1f} K.'.format(DeltaT0))",
"The Equilibrium Climate Sensitivity in the absence of feedback is 1.2 K.\n"
]
],
[
[
"Question: what are the current best estimates for the actual warming (including all feedbacks) in response to a doubling of CO$_2$?\n\nWe’ll now look at the feedback concept. Climate feedbacks tend to amplify the response to increased CO$_2$. But $\\Delta T_0$ is a meaningful climate sensitivity in the absence of feedback.\n\n$\\Delta T_0 = 1.2$ K is the ** warming that we would have if the Earth radiated the excess energy away to space as a blackbody**, and with no change in the planetary albedo.",
"_____no_output_____"
],
[
"____________\n<a id='section3'></a>\n\n## 3. The feedback concept\n____________\n\nA concept borrowed from electrical engineering. You have all heard or used the term before, but we’ll try take a more precise approach today.\n\nA feedback occurs when a portion of the output from the action of a system is added to the input and subsequently alters the output:",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='../images/feedback_sketch.png', width=500)",
"_____no_output_____"
]
],
[
[
"The result of a loop system can either be **amplification** or **dampening** of the process, depending on the sign of the gain in the loop, which we will denote $f$.\n\nWe will call amplifying feedbacks **positive** ($f>0$) and damping feedbacks **negative** ($f<0$).\n\nWe can think of the “process” here as the entire climate system, which contains many examples of both positive and negative feedback.",
"_____no_output_____"
],
[
"### Example: the water vapor feedback\n\nThe capacity of the atmosphere to hold water vapor (saturation specific humidity) increases exponentially with temperature. Warming is thus accompanied by moistening (more water vapor), which leads to more warming due to the enhanced water vapor greenhouse effect.\n\n**Positive or negative feedback?**",
"_____no_output_____"
],
[
"### Example: the ice-albedo feedback\n\nColder temperatures lead to expansion of the areas covered by ice and snow, which tend to be more reflective than water and vegetation. This causes a reduction in the absorbed solar radiation, which leads to more cooling. \n\n**Positive or negative feedback?**\n\n*Make sure it’s clear that the sign of the feedback is the same whether we are talking about warming or cooling.*",
"_____no_output_____"
],
[
"_____________\n<a id='section4'></a>\n## 4. Climate feedback: some definitions\n____________\n\nWe start with an initial radiative forcing , and get a response\n$$ \\Delta T_0 = \\frac{\\Delta R}{\\lambda_0} $$\n",
"_____no_output_____"
],
[
"Now consider what happens in the presence of a feedback process. For a concrete example, let’s take the **water vapor feedback**. For every degree of warming, there is an additional increase in the greenhouse effect, and thus additional energy added to the system.\n\nLet’s denote this extra energy as \n$$ f \\lambda_0 \\Delta T_0 $$\n\nwhere $f$ is the **feedback amount**, a number that represents what fraction of the output gets added back to the input. $f$ must be between $-\\infty$ and +1. \n\nFor the example of the water vapor feedback, $f$ is positive (between 0 and +1) – the process adds extra energy to the original radiative forcing.\n",
"_____no_output_____"
],
[
"The amount of energy in the full \"input\" is now\n$$ \\Delta R + f \\lambda_0 \\Delta T_0 $$\nor\n$$ (1+f) \\lambda_0 \\Delta T_0 $$",
"_____no_output_____"
],
[
"But now we need to consider the next loop. A fraction $f$ of the additional energy is also added to the input, giving us\n$$ (1+f+f^2) \\lambda_0 \\Delta T_0 $$",
"_____no_output_____"
],
[
"and we can go round and round, leading to the infinite series\n$$ (1+f+f^2+f^3+ ...) \\lambda_0 \\Delta T_0 = \\lambda_0 \\Delta T_0 \\sum_{n=0}^{\\infty} f^n $$\n\nQuestion: what happens if $f=1$?",
"_____no_output_____"
],
[
"It so happens that this infinite series has an exact solution\n\n$$ \\sum_{n=0}^{\\infty} f^n = \\frac{1}{1-f} $$",
"_____no_output_____"
],
[
"So the full response including all the effects of the feedback is actually\n\n$$ \\Delta T = \\frac{1}{1-f} \\Delta T_0 $$",
"_____no_output_____"
],
[
"This is also sometimes written as \n$$ \\Delta T = g \\Delta T_0 $$\n\nwhere \n\n$$ g = \\frac{1}{1-f} = \\frac{\\Delta T}{\\Delta T_0} $$\n\nis called the **system gain** -- the ratio of the actual warming (including all feedbacks) to the warming we would have in the absence of feedbacks.",
"_____no_output_____"
],
[
"So if the overall feedback is positive, then $f>0$ and $g>1$.\n\nAnd if the overall feedback is negative?",
"_____no_output_____"
],
[
"_____________\n<a id='section5'></a>\n## 5. Climate sensitivity with feedback\n____________\n\nECS is an important number. A major goal of climate modeling is to provide better estimates of ECS and its uncertainty.\n\nLatest IPCC report AR5 gives a likely range of 1.5 to 4.5 K. (There is lots of uncertainty in these numbers – we will definitely come back to this question)\n\nSo our simple estimate of the no-feedback change $\\Delta T_0$ is apparently underestimating climate sensitivity. \n\nSaying the same thing another way: the overall net climate feedback is positive, amplifying the response, and the system gain $g>1$.\n",
"_____no_output_____"
],
[
"Let’s assume that the true value is $\\Delta T_{2xCO2} = 3$ K (middle of the range). This implies that the gain is\n\n$$ g = \\frac{\\Delta T_{2xCO2}}{\\Delta T_0} = \\frac{3}{1.2} = 2.5 $$\n\nThe actual warming is substantially amplified!\n\nThere are lots of reasons for this, but the water vapor feedback is probably the most important.",
"_____no_output_____"
],
[
"Question: if $g=2.5$, what is the feedback amount $f$?\n\n$$ g = \\frac{1}{1-f} $$\n\nor rearranging,\n\n$$ f = 1 - 1/g = 0.6 $$\n\nThe overall feedback (due to water vapor, clouds, etc.) is **positive**.",
"_____no_output_____"
],
[
"_____________\n<a id='section6'></a>\n## 6. Contribution of individual feedback processes to Equilibrium Climate Sensitivity\n____________\n\n\nNow what if we have several individual feedback processes occurring simultaneously?\n\nWe can think of individual feedback amounts $f_1, f_2, f_3, ...$, with each representing a physically distinct mechanism, e.g. water vapor, surface snow and ice, cloud changes, etc.\n",
"_____no_output_____"
],
[
"Each individual process takes a fraction $f_i$ of the output and adds to the input. So the feedback amounts are additive,\n\n$$ f = f_1 + f_2 + f_3 + ... = \\sum_{i=0}^N f_i $$",
"_____no_output_____"
],
[
"This gives us a way to compare the importance of individual feedback processes!\n\nThe climate sensitivity is now\n\n$$ \\Delta T_{2xCO2} = \\frac{1}{1- \\sum_{i=0}^N f_i } \\Delta T_0 $$\n\nThe climate sensitivity is thus **increased by positive feedback processes**, and **decreased by negative feedback processes**.",
"_____no_output_____"
],
[
"### Climate feedback parameters\n\nWe can also write this in terms of the original radiative forcing as\t\n\n$$ \\Delta T_{2xCO2} = \\frac{\\Delta R}{\\lambda_0 - \\sum_{i=1}^{N} \\lambda_i} $$\n\nwhere\n\n$$ \\lambda_i = \\lambda_0 f_i $$\n\nknown as **climate feedback parameters**, in units of W m$^{-2}$ K$^{-1}$. \n\nWith this choice of sign conventions, $\\lambda_i > 0$ for a positive feedback process.",
"_____no_output_____"
],
[
"Individual feedback parameters $\\lambda_i$ are then additive, and can be compared to the no-feedback parameter $\\lambda_0$.\n\nBased on our earlier numbers, the net feedback necessary to get a climate sensitivity of 3 K is\n\n$$ \\sum_{i=1}^N \\lambda_i = \\lambda_0 \\sum_{i=1}^N f_i = (3.3 \\text{ W m}^{-2} \\text{ K}^{-1}) (0.6) = 2 \\text{ W m}^{-2} \\text{ K}^{-1} $$\n\nWe might decompose this net climate feedback into, for example\n\n- longwave and shortwave processes\n- cloud and non-cloud processes\n\nThese individual feedback processes may be positive or negative. This is very powerful, because we can **measure the relative importance of different feedback processes** simply by comparing their $\\lambda_i$ values.",
"_____no_output_____"
],
[
"### Every climate model has a Planck feedback\n\nThe \"Planck feedback\" represented by our reference parameter $\\lambda_0$ is not really a feedback at all.\n\nIt is the most basic and universal climate process, and is present in every climate model. It is simply an expression of the fact that a warm planet radiates more to space than a cold planet.\n\nAs we will see, our estimate of $\\lambda_0 = -3.3 ~\\text{W} ~\\text{m}^{-2} ~\\text{K}^{-1} $ is essentially the same as the Planck feedback diagnosed from complex GCMs. Unlike our simple zero-dimensional EBM, however, most other climate models (and the real climate system) have other radiative feedback processes, such that \n\n$$\\lambda = \\lambda_0 - \\sum_{i=1}^{N} \\lambda_i \\ne \\lambda_0 $$\n",
"_____no_output_____"
],
[
"____________\n<a id='section7'></a>\n## 7. Feedbacks diagnosed from complex climate models\n____________\n\n### Data from the IPCC AR5\n\nThis figure is reproduced from the recent IPCC AR5 report. It shows the feedbacks diagnosed from the various models that contributed to the assessment.\n\n(Later in the term we will discuss how the feedback diagnosis is actually done)\n\nSee below for complete citation information.",
"_____no_output_____"
]
],
[
[
"feedback_ar5 = 'http://www.climatechange2013.org/images/figures/WGI_AR5_Fig9-43.jpg'\nImage(url=feedback_ar5, width=800)",
"_____no_output_____"
]
],
[
[
"**Figure 9.43** | (a) Strengths of individual feedbacks for CMIP3 and CMIP5 models (left and right columns of symbols) for Planck (P), water vapour (WV), clouds (C), albedo (A), lapse rate (LR), combination of water vapour and lapse rate (WV+LR) and sum of all feedbacks except Planck (ALL), from Soden and Held (2006) and Vial et al. (2013), following Soden et al. (2008). CMIP5 feedbacks are derived from CMIP5 simulations for abrupt fourfold increases in CO2 concentrations (4 × CO2). (b) ECS obtained using regression techniques by Andrews et al. (2012) against ECS estimated from the ratio of CO2 ERF to the sum of all feedbacks. The CO2 ERF is one-half the 4 × CO2 forcings from Andrews et al. (2012), and the total feedback (ALL + Planck) is from Vial et al. (2013).\n\n*Figure caption reproduced from the AR5 WG1 report*",
"_____no_output_____"
],
[
"Legend:\n\n- P: Planck feedback\n- WV: Water vapor feedback\n- LR: Lapse rate feedback\n- WV+LR: combined water vapor plus lapse rate feedback\n- C: cloud feedback\n- A: surface albedo feedback\n- ALL: sum of all feedback except Plank, i.e. ALL = WV+LR+C+A",
"_____no_output_____"
],
[
"Things to note:\n\n- The models all agree strongly on the Planck feedback. \n- The Planck feedback is about $\\lambda_0 = -3.3 ~\\text{W} ~\\text{m}^{-2} ~\\text{K}^{-1} $ just like our above estimate (but with opposite sign convention -- watch carefully for that in the literature)\n- The water vapor feedback is strongly positive in every model.\n- The lapse rate feedback is something we will study later. It is slightly negative.\n- For reasons we will discuss later, the best way to measure the water vapor feedback is to combine it with lapse rate feedback.\n- Models agree strongly on the combined water vapor plus lapse rate feedback.",
"_____no_output_____"
],
[
"- The albedo feedback is slightly positive but rather small globally.\n- By far the largest spread across the models occurs in the cloud feedback.\n- Global cloud feedback ranges from slighly negative to strongly positive across the models.\n- Most of the spread in the total feedback is due to the spread in the cloud feedback.\n- Therefore, most of the spread in the ECS across the models is due to the spread in the cloud feedback.\n- Our estimate of $+2.0 ~\\text{W} ~\\text{m}^{-2} ~\\text{K}^{-1}$ for all the missing processes is consistent with the GCM ensemble.",
"_____no_output_____"
],
[
"### Citation\n\nThis is Figure 9.43 from Chapter 9 of the IPCC AR5 Working Group 1 report.\n\nThe report and images can be found online at\n<http://www.climatechange2013.org/report/full-report/>\n\nThe full citation is:\n\nFlato, G., J. Marotzke, B. Abiodun, P. Braconnot, S.C. Chou, W. Collins, P. Cox, F. Driouech, S. Emori, V. Eyring, C. Forest, P. Gleckler, E. Guilyardi, C. Jakob, V. Kattsov, C. Reason and M. Rummukainen, 2013: Evaluation of Climate Models. In: Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change [Stocker, T.F., D. Qin, G.-K. Plattner, M. Tignor, S.K. Allen, J. Boschung, A. Nauels, Y. Xia, V. Bex and P.M. Midgley (eds.)]. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA, pp. 741–866, doi:10.1017/CBO9781107415324.020",
"_____no_output_____"
],
[
"____________\n<a id='section8'></a>\n## 8. The zero-dimensional model with variable albedo\n____________\n",
"_____no_output_____"
],
[
"### The model\n\nIn homework you will be asked to include a new process in the zero-dimensional EBM: a temperature-dependent albedo.\n\nWe use the following formula:\n\n$$ \\alpha(T) = \\left\\{ \\begin{array}{ccc}\n\\alpha_i & & T \\le T_i \\\\\n\\alpha_o + (\\alpha_i-\\alpha_o) \\frac{(T-T_o)^2}{(T_i-T_o)^2} & & T_i < T < T_o \\\\\n\\alpha_o & & T \\ge T_o \\end{array} \\right\\}$$",
"_____no_output_____"
],
[
"with parameter values:\n\n- $\\alpha_o = 0.289$ is the albedo of a warm, ice-free planet\n- $\\alpha_i = 0.7$ is the albedo of a very cold, completely ice-covered planet\n- $T_o = 293$ K is the threshold temperature above which our model assumes the planet is ice-free\n- $T_i = 260$ K is the threshold temperature below which our model assumes the planet is completely ice covered. \n\nFor intermediate temperature, this formula gives a smooth variation in albedo with global mean temperature. It is tuned to reproduce the observed albedo $\\alpha = 0.299$ for $T = 288$ K. ",
"_____no_output_____"
],
[
"### Coding up the model in Python\n\nThis largely repeats what I asked you to do in your homework.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def albedo(T, alpha_o = 0.289, alpha_i = 0.7, To = 293., Ti = 260.):\n alb1 = alpha_o + (alpha_i-alpha_o)*(T-To)**2 / (Ti - To)**2\n alb2 = np.where(T>Ti, alb1, alpha_i)\n alb3 = np.where(T<To, alb2, alpha_o)\n return alb3",
"_____no_output_____"
],
[
"def ASR(T, Q=341.3):\n alpha = albedo(T)\n return Q * (1-alpha)\n\ndef OLR(T, sigma=5.67E-8, beta=0.885):\n return sigma * (beta*T)**4\n\ndef Ftoa(T):\n return ASR(T) - OLR(T)",
"_____no_output_____"
],
[
"T = np.linspace(220., 300., 100)\n\nplt.plot(T, albedo(T))\nplt.xlabel('Temperature (K)')\nplt.ylabel('albedo')\nplt.ylim(0,1)\nplt.title('Albedo as a function of global mean temperature')",
"_____no_output_____"
]
],
[
[
"### Graphical solution: TOA fluxes as functions of temperature",
"_____no_output_____"
]
],
[
[
"plt.plot(T, OLR(T), label='OLR')\nplt.plot(T, ASR(T), label='ASR')\nplt.plot(T, Ftoa(T), label='Ftoa')\nplt.xlabel('Surface temperature (K)')\nplt.ylabel('TOA flux (W m$^{-2}$)')\nplt.grid()\nplt.legend(loc='upper left')",
"_____no_output_____"
]
],
[
[
"The graphs meet at three different points! That means there are actually three different possible equilibrium temperatures in this model.",
"_____no_output_____"
],
[
"### Numerical solution to get the three equilibrium temperatures",
"_____no_output_____"
]
],
[
[
"# Use numerical root-finding to get the equilibria\nfrom scipy.optimize import brentq\n# brentq is a root-finding function\n# Need to give it a function and two end-points\n# It will look for a zero of the function between those end-points\nTeq1 = brentq(Ftoa, 280., 300.)\nTeq2 = brentq(Ftoa, 260., 280.)\nTeq3 = brentq(Ftoa, 200., 260.)\n\nprint( Teq1, Teq2, Teq3)",
"288.07486360356785 273.9423668460388 232.92995904643783\n"
]
],
[
[
"### Bonus exercise\n\nUsing numerical timestepping and different initial temperatures, can you get the model to converge on all three equilibria, or only some of them?\n\nWhat do you think this means?",
"_____no_output_____"
],
[
"<div class=\"alert alert-success\">\n[Back to ATM 623 notebook home](../index.ipynb)\n</div>",
"_____no_output_____"
],
[
"____________\n## Version information\n____________\n",
"_____no_output_____"
]
],
[
[
"%load_ext version_information\n%version_information numpy, scipy, matplotlib",
"Loading extensions from ~/.ipython/extensions is deprecated. We recommend managing extensions like any other Python packages, in site-packages.\n"
]
],
[
[
"____________\n\n## Credits\n\nThe author of this notebook is [Brian E. J. Rose](http://www.atmos.albany.edu/facstaff/brose/index.html), University at Albany.\n\nIt was developed in support of [ATM 623: Climate Modeling](http://www.atmos.albany.edu/facstaff/brose/classes/ATM623_Spring2015/), a graduate-level course in the [Department of Atmospheric and Envionmental Sciences](http://www.albany.edu/atmos/index.php)\n\nDevelopment of these notes and the [climlab software](https://github.com/brian-rose/climlab) is partially supported by the National Science Foundation under award AGS-1455071 to Brian Rose. Any opinions, findings, conclusions or recommendations expressed here are mine and do not necessarily reflect the views of the National Science Foundation.\n____________",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e71be4251ad400a5625d087bdd66c669cc8a1df3 | 3,649 | ipynb | Jupyter Notebook | Python-API/apl/notebooks/01_Check_APL_Version.ipynb | pbaumann76/hana-ml-samples | a2180c9d4cf4ec989234f79ec0d118ed658bd224 | [
"Apache-2.0"
] | 1 | 2022-03-24T07:51:05.000Z | 2022-03-24T07:51:05.000Z | Python-API/apl/notebooks/01_Check_APL_Version.ipynb | xinchen510/hana-ml-samples | c09ed92fcbe2af5c168bbd52923d431b0227113c | [
"Apache-2.0"
] | null | null | null | Python-API/apl/notebooks/01_Check_APL_Version.ipynb | xinchen510/hana-ml-samples | c09ed92fcbe2af5c168bbd52923d431b0227113c | [
"Apache-2.0"
] | null | null | null | 23.694805 | 226 | 0.5037 | [
[
[
"# Check Version for Python HANA ML APL",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n<b>Client side</b> <br>\n</div>",
"_____no_output_____"
],
[
"### Python version",
"_____no_output_____"
]
],
[
[
"from platform import python_version\nprint(python_version())",
"3.8.7\n"
]
],
[
[
"### HANA ML API version",
"_____no_output_____"
]
],
[
[
"import hana_ml\nprint(hana_ml.__version__)",
"2.11.21121103\n"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\n<b>Server side</b> <br>\n</div>",
"_____no_output_____"
],
[
"### Versions of HANA database and APL",
"_____no_output_____"
]
],
[
[
"from hana_ml import dataframe as hd\nconn = hd.ConnectionContext(userkey='MLMDA_KEY')\nimport hana_ml.algorithms.apl.apl_base as apl_base\ndf = apl_base.get_apl_version(conn)\ndf = df[df.name.isin(['HDB.Version','APL.Version.ServicePack'])]\ndf.columns = ['Item','Value']\ndf.style.hide_index()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e71be5704609564e310242d668dc472431960f48 | 133,730 | ipynb | Jupyter Notebook | Task 3.ipynb | HarshadaPatil317/The-Sparks-Foundation---GRIP---Task-3 | 3d693098fafb1d9233bb1c57f57418e126ce222e | [
"MIT"
] | null | null | null | Task 3.ipynb | HarshadaPatil317/The-Sparks-Foundation---GRIP---Task-3 | 3d693098fafb1d9233bb1c57f57418e126ce222e | [
"MIT"
] | null | null | null | Task 3.ipynb | HarshadaPatil317/The-Sparks-Foundation---GRIP---Task-3 | 3d693098fafb1d9233bb1c57f57418e126ce222e | [
"MIT"
] | null | null | null | 530.674603 | 52,684 | 0.70071 | [
[
[
"<h1> The Sparks Foundation</h1>\n<h2>Task 3 To Explore Unsupervised Machine Learning</h2>\n<h3> From the given ‘Iris’ dataset, predict the optimum number of\nclusters and represent it visually.\n\nDataset :\nhttps://drive.google.com/file/d/11Iq7YvbWZbt8VXjfm06brx6\n6b10YiwK-/view?usp=sharing\n </h3>",
"_____no_output_____"
],
[
"<h3>Importing Libraries</h3>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\ndata = open(\"D:\\Python tutorials\\Iris.csv\",\"r\")\ndf = pd.read_csv(data)\ndf",
"_____no_output_____"
],
[
"#Describe Data\ndf.describe()",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"<h3>Understanding the Data</h3>",
"_____no_output_____"
]
],
[
[
"Total_rows = len(df.axes[0])\nTotal_cols = len(df.axes[1])\nprint(\"Total no. of rows :\", Total_rows)\nprint(\"Total no. of Columns :\", Total_cols)",
"Total no. of rows : 150\nTotal no. of Columns : 6\n"
],
[
"x = df.iloc[:, [0, 1, 2, 3]].values\nfrom sklearn.cluster import KMeans\nwcss = []\nfor i in range(1, 11):\n kmeans = KMeans(n_clusters = i, init = 'k-means++', \n max_iter = 300, n_init = 10, random_state = 0)\n kmeans.fit(x)\n wcss.append(kmeans.inertia_)\n# Plotting the results onto a line graph, \nplt.plot(range(1, 11), wcss)\nplt.title('The Elbow method')\nplt.xlabel('Number of clusters')\nplt.ylabel('WCSS') \nplt.show()",
"_____no_output_____"
],
[
"# Apply kmeans to the Dataset\nkmeans = KMeans(n_clusters = 3, init = 'k-means++',\n\n max_iter = 300, n_init = 10, random_state = 0)\ny_kmeans = kmeans.fit_predict(x)",
"_____no_output_____"
],
[
"# Visualising the clusters on the first two columns\nplt.scatter(x[y_kmeans == 0, 0], x[y_kmeans == 0, 1], \n s = 50, c = 'black', label = 'Iris-setosa')\nplt.scatter(x[y_kmeans == 1, 0], x[y_kmeans == 1, 1], \n s = 50, c = 'green', label = 'Iris-versicolour')\nplt.scatter(x[y_kmeans == 2, 0], x[y_kmeans == 2, 1],\n s = 50, c = 'yellow', label = 'Iris-virginica')\n\n# Plotting the centroids of the clusters\nplt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:,1], \n s = 100, c = 'red', label = 'Centroids')\n\nplt.legend()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e71bf905855473aa026d72b90c45785ca57c8fe2 | 198,366 | ipynb | Jupyter Notebook | Regression/Linear_Regression_Methods.ipynb | thirupathi-chintu/Machine-Learning-with-Python | 0bb8753a5140c8e69a24f2ab95c7ef133ac025a6 | [
"BSD-2-Clause"
] | 1,803 | 2018-11-26T20:53:23.000Z | 2022-03-31T15:25:29.000Z | Regression/Linear_Regression_Methods.ipynb | thirupathi-chintu/Machine-Learning-with-Python | 0bb8753a5140c8e69a24f2ab95c7ef133ac025a6 | [
"BSD-2-Clause"
] | 8 | 2019-02-05T04:09:57.000Z | 2022-02-19T23:46:27.000Z | Regression/Linear_Regression_Methods.ipynb | thirupathi-chintu/Machine-Learning-with-Python | 0bb8753a5140c8e69a24f2ab95c7ef133ac025a6 | [
"BSD-2-Clause"
] | 1,237 | 2018-11-28T19:48:55.000Z | 2022-03-31T15:25:07.000Z | 179.516742 | 123,666 | 0.858564 | [
[
[
"# Linear regression with various methods\nThis is a very simple example of using two scipy tools for linear regression.\n* Scipy.Polyfit\n* Stats.linregress\n* Optimize.curve_fit\n* numpy.linalg.lstsq\n* statsmodels.OLS\n* Analytic solution using Moore-Penrose generalized inverse or simple multiplicative matrix inverse\n* sklearn.linear_model.LinearRegression",
"_____no_output_____"
],
[
"## Import libraries",
"_____no_output_____"
]
],
[
[
"from scipy import linspace, polyval, polyfit, sqrt, stats, randn, optimize\nimport statsmodels.api as sm\nimport matplotlib.pyplot as plt\nimport time\nimport numpy as np\nfrom sklearn.linear_model import LinearRegression\nimport pandas as pd\n%matplotlib inline",
"C:\\Users\\Tirtha\\Python\\Anaconda3\\lib\\site-packages\\statsmodels\\compat\\pandas.py:56: FutureWarning: The pandas.core.datetools module is deprecated and will be removed in a future version. Please use the pandas.tseries module instead.\n from pandas.core import datetools\n"
]
],
[
[
"## Generate random data of a sufficiently large size",
"_____no_output_____"
]
],
[
[
"#Sample data creation\n#number of points \nn=int(5e6)\nt=np.linspace(-10,10,n)\n#parameters\na=3.25; b=-6.5\nx=polyval([a,b],t)\n#add some noise\nxn=x+3*randn(n)",
"_____no_output_____"
]
],
[
[
"### Draw few random sample points and plot",
"_____no_output_____"
]
],
[
[
"xvar=np.random.choice(t,size=20)\nyvar=polyval([a,b],xvar)+3*randn(20)\nplt.scatter(xvar,yvar,c='green',edgecolors='k')\nplt.grid(True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Method: Scipy.Polyfit",
"_____no_output_____"
]
],
[
[
"#Linear regressison -polyfit - polyfit can be used other orders polynomials\nt1=time.time()\n(ar,br)=polyfit(t,xn,1)\nxr=polyval([ar,br],t)\n#compute the mean square error\nerr=sqrt(sum((xr-xn)**2)/n)\nt2=time.time()\nt_polyfit = float(t2-t1)\n\nprint('Linear regression using polyfit')\nprint('parameters: a=%.2f b=%.2f, ms error= %.3f' % (ar,br,err))\nprint(\"Time taken: {} seconds\".format(t_polyfit))",
"Linear regression using polyfit\nparameters: a=3.25 b=-6.50, ms error= 3.000\nTime taken: 1.7698638439178467 seconds\n"
]
],
[
[
"## Method: Stats.linregress",
"_____no_output_____"
]
],
[
[
"#Linear regression using stats.linregress\nt1=time.time()\n(a_s,b_s,r,tt,stderr)=stats.linregress(t,xn)\nt2=time.time()\nt_linregress = float(t2-t1)\n\nprint('Linear regression using stats.linregress')\nprint('a=%.2f b=%.2f, std error= %.3f, r^2 coefficient= %.3f' % (a_s,b_s,stderr,r))\nprint(\"Time taken: {} seconds\".format(t_linregress))",
"Linear regression using stats.linregress\na=3.25 b=-6.50, std error= 0.000, r^2 coefficient= 0.987\nTime taken: 0.15017366409301758 seconds\n"
]
],
[
[
"## Method: Optimize.curve_fit",
"_____no_output_____"
]
],
[
[
"def flin(t,a,b):\n result = a*t+b\n return(result)",
"_____no_output_____"
],
[
"t1=time.time()\np1,_=optimize.curve_fit(flin,xdata=t,ydata=xn,method='lm')\nt2=time.time()\nt_optimize_curve_fit = float(t2-t1)\n\nprint('Linear regression using optimize.curve_fit')\nprint('parameters: a=%.2f b=%.2f' % (p1[0],p1[1]))\nprint(\"Time taken: {} seconds\".format(t_optimize_curve_fit))",
"Linear regression using optimize.curve_fit\nparameters: a=3.25 b=-6.50\nTime taken: 1.2034447193145752 seconds\n"
]
],
[
[
"## Method: numpy.linalg.lstsq",
"_____no_output_____"
]
],
[
[
"t1=time.time()\nA = np.vstack([t, np.ones(len(t))]).T\nresult = np.linalg.lstsq(A, xn)\nar,br = result[0]\nerr = np.sqrt(result[1]/len(xn))\nt2=time.time()\nt_linalg_lstsq = float(t2-t1)\n\nprint('Linear regression using numpy.linalg.lstsq')\nprint('parameters: a=%.2f b=%.2f, ms error= %.3f' % (ar,br,err))\nprint(\"Time taken: {} seconds\".format(t_linalg_lstsq))",
"Linear regression using numpy.linalg.lstsq\nparameters: a=3.25 b=-6.50, ms error= 3.000\nTime taken: 0.3698573112487793 seconds\n"
]
],
[
[
"## Method: Statsmodels.OLS",
"_____no_output_____"
]
],
[
[
"t1=time.time()\nt=sm.add_constant(t)\nmodel = sm.OLS(x, t)\nresults = model.fit()\nar=results.params[1]\nbr=results.params[0]\nt2=time.time()\nt_OLS = float(t2-t1)\n\nprint('Linear regression using statsmodels.OLS')\nprint('parameters: a=%.2f b=%.2f'% (ar,br))\nprint(\"Time taken: {} seconds\".format(t_OLS)) ",
"Linear regression using statsmodels.OLS\nparameters: a=3.25 b=-6.50\nTime taken: 0.9167804718017578 seconds\n"
],
[
"print(results.summary())",
" OLS Regression Results \n==============================================================================\nDep. Variable: y R-squared: 1.000\nModel: OLS Adj. R-squared: 1.000\nMethod: Least Squares F-statistic: 4.287e+34\nDate: Fri, 08 Dec 2017 Prob (F-statistic): 0.00\nTime: 23:09:33 Log-Likelihood: 1.3904e+08\nNo. Observations: 5000000 AIC: -2.781e+08\nDf Residuals: 4999998 BIC: -2.781e+08\nDf Model: 1 \nCovariance Type: nonrobust \n==============================================================================\n coef std err t P>|t| [0.025 0.975]\n------------------------------------------------------------------------------\nconst -6.5000 9.06e-17 -7.17e+16 0.000 -6.500 -6.500\nx1 3.2500 1.57e-17 2.07e+17 0.000 3.250 3.250\n==============================================================================\nOmnibus: 4418788.703 Durbin-Watson: 0.000\nProb(Omnibus): 0.000 Jarque-Bera (JB): 299716.811\nSkew: -0.001 Prob(JB): 0.00\nKurtosis: 1.801 Cond. No. 5.77\n==============================================================================\n\nWarnings:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
]
],
[
[
"## Analytic solution using Moore-Penrose pseudoinverse",
"_____no_output_____"
]
],
[
[
"t1=time.time()\nmpinv = np.linalg.pinv(t)\nresult = mpinv.dot(x)\nar = result[1]\nbr = result[0]\nt2=time.time()\nt_inv_matrix = float(t2-t1)\n\nprint('Linear regression using Moore-Penrose inverse')\nprint('parameters: a=%.2f b=%.2f'% (ar,br))\nprint(\"Time taken: {} seconds\".format(t_inv_matrix)) ",
"Linear regression using Moore-Penrose inverse\nparameters: a=3.25 b=-6.50\nTime taken: 0.6019864082336426 seconds\n"
]
],
[
[
"## Analytic solution using simple multiplicative matrix inverse",
"_____no_output_____"
]
],
[
[
"t1=time.time()\nm = np.dot((np.dot(np.linalg.inv(np.dot(t.T,t)),t.T)),x)\nar = m[1]\nbr = m[0]\nt2=time.time()\nt_simple_inv = float(t2-t1)\n\nprint('Linear regression using simple inverse')\nprint('parameters: a=%.2f b=%.2f'% (ar,br))\nprint(\"Time taken: {} seconds\".format(t_simple_inv)) ",
"Linear regression using simple inverse\nparameters: a=3.25 b=-6.50\nTime taken: 0.13125276565551758 seconds\n"
]
],
[
[
"## Method: sklearn.linear_model.LinearRegression",
"_____no_output_____"
]
],
[
[
"t1=time.time()\nlm = LinearRegression()\nlm.fit(t,x)\nar=lm.coef_[1]\nbr=lm.intercept_\nt2=time.time()\nt_sklearn_linear = float(t2-t1)\n\nprint('Linear regression using sklearn.linear_model.LinearRegression')\nprint('parameters: a=%.2f b=%.2f'% (ar,br))\nprint(\"Time taken: {} seconds\".format(t_sklearn_linear)) ",
"Linear regression using sklearn.linear_model.LinearRegression\nparameters: a=3.25 b=-6.50\nTime taken: 0.5318112373352051 seconds\n"
]
],
[
[
"## Bucket all the execution times in a list and plot",
"_____no_output_____"
]
],
[
[
"times = [t_polyfit,t_linregress,t_optimize_curve_fit,t_linalg_lstsq,t_OLS,t_inv_matrix,t_simple_inv,t_sklearn_linear]",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,5))\nplt.grid(True)\nplt.bar(left=[l*0.8 for l in range(8)],height=times, width=0.4,\n tick_label=['Polyfit','Stats.linregress','Optimize.curve_fit',\n 'numpy.linalg.lstsq','statsmodels.OLS','Moore-Penrose matrix inverse',\n 'Simple matrix inverse','sklearn.linear_model'])\nplt.show()",
"_____no_output_____"
],
[
"n_min = 50000\nn_max = int(1e7)\nn_levels = 25\nr = np.log10(n_max/n_min)\nl = np.linspace(0,r,n_levels)\nn_data = list((n_min*np.power(10,l)))\nn_data = [int(n) for n in n_data]",
"_____no_output_____"
],
[
"#time_dict={'Polyfit':[],'Stats.lingress':[],'Optimize.curve_fit':[],'linalg.lstsq':[],'statsmodels.OLS':[],\n #'Moore-Penrose matrix inverse':[],'Simple matrix inverse':[], 'sklearn.linear_model':[]}\n\nl1=['Polyfit', 'Stats.lingress','Optimize.curve_fit', 'linalg.lstsq', \n 'statsmodels.OLS', 'Moore-Penrose matrix inverse', 'Simple matrix inverse', 'sklearn.linear_model']\ntime_dict = {key:[] for key in l1}\n\nfrom tqdm import tqdm\n\nfor i in tqdm(range(len(n_data))):\n t=np.linspace(-10,10,n_data[i])\n #parameters\n a=3.25; b=-6.5\n x=polyval([a,b],t)\n #add some noise\n xn=x+3*randn(n_data[i])\n \n #Linear regressison -polyfit - polyfit can be used other orders polynomials\n t1=time.time()\n (ar,br)=polyfit(t,xn,1)\n t2=time.time()\n t_polyfit = 1e3*float(t2-t1)\n time_dict['Polyfit'].append(t_polyfit)\n \n #Linear regression using stats.linregress\n t1=time.time()\n (a_s,b_s,r,tt,stderr)=stats.linregress(t,xn)\n t2=time.time()\n t_linregress = 1e3*float(t2-t1)\n time_dict['Stats.lingress'].append(t_linregress)\n \n #Linear regression using optimize.curve_fit\n t1=time.time()\n p1,_=optimize.curve_fit(flin,xdata=t,ydata=xn,method='lm')\n t2=time.time()\n t_optimize_curve_fit = 1e3*float(t2-t1)\n time_dict['Optimize.curve_fit'].append(t_optimize_curve_fit)\n \n # Linear regression using np.linalg.lstsq (solving Ax=B equation system)\n t1=time.time()\n A = np.vstack([t, np.ones(len(t))]).T\n result = np.linalg.lstsq(A, xn)\n ar,br = result[0]\n t2=time.time()\n t_linalg_lstsq = 1e3*float(t2-t1)\n time_dict['linalg.lstsq'].append(t_linalg_lstsq)\n \n # Linear regression using statsmodels.OLS\n t1=time.time()\n t=sm.add_constant(t)\n model = sm.OLS(x, t)\n results = model.fit()\n ar=results.params[1]\n br=results.params[0]\n t2=time.time()\n t_OLS = 1e3*float(t2-t1)\n time_dict['statsmodels.OLS'].append(t_OLS)\n \n # Linear regression using Moore-Penrose pseudoinverse matrix\n t1=time.time()\n mpinv = np.linalg.pinv(t)\n result = mpinv.dot(x)\n ar = result[1]\n br = result[0]\n t2=time.time()\n t_mpinverse = 1e3*float(t2-t1)\n time_dict['Moore-Penrose matrix inverse'].append(t_mpinverse)\n \n # Linear regression using simple multiplicative inverse matrix\n t1=time.time()\n m = np.dot((np.dot(np.linalg.inv(np.dot(t.T,t)),t.T)),x)\n ar = m[1]\n br = m[0]\n t2=time.time()\n t_simple_inv = 1e3*float(t2-t1)\n time_dict['Simple matrix inverse'].append(t_simple_inv)\n \n # Linear regression using scikit-learn's linear_model\n t1=time.time()\n lm = LinearRegression()\n lm.fit(t,x)\n ar=lm.coef_[1]\n br=lm.intercept_\n t2=time.time()\n t_sklearn_linear = 1e3*float(t2-t1)\n time_dict['sklearn.linear_model'].append(t_sklearn_linear)",
"_____no_output_____"
],
[
"df = pd.DataFrame(data=time_dict)\ndf",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,10))\nfor i in df.columns:\n plt.semilogx((n_data),df[i],lw=3)\nplt.xticks([1e5,2e5,5e5,1e6,2e6,5e6,1e7],fontsize=15)\nplt.xlabel(\"\\nSize of the data set (number of samples)\",fontsize=15)\nplt.yticks(fontsize=15)\nplt.ylabel(\"Milliseconds needed for simple linear regression model fit\\n\",fontsize=15)\nplt.grid(True)\nplt.legend([name for name in df.columns],fontsize=20)",
"_____no_output_____"
],
[
"a1=df.iloc[n_levels-1]",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,5))\nplt.grid(True)\nplt.bar(left=[l*0.8 for l in range(8)],height=a1, width=0.4,\n tick_label=list(a1.index))\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71c17677ea70e1d945d787c9c52657b672b8bcf | 17,343 | ipynb | Jupyter Notebook | notebooks/.ipynb_checkpoints/Tell Dor File Linking-checkpoint.ipynb | ekansa/open-context-jupyter | 5b5eebe7efea29bbc8eaf41bf4f10f251773f43a | [
"MIT"
] | 10 | 2017-04-08T01:25:28.000Z | 2021-09-06T13:46:24.000Z | notebooks/.ipynb_checkpoints/Tell Dor File Linking-checkpoint.ipynb | ekansa/open-context-jupyter | 5b5eebe7efea29bbc8eaf41bf4f10f251773f43a | [
"MIT"
] | 1 | 2021-08-23T20:50:21.000Z | 2021-08-23T20:50:21.000Z | notebooks/.ipynb_checkpoints/Tell Dor File Linking-checkpoint.ipynb | ekansa/open-context-jupyter | 5b5eebe7efea29bbc8eaf41bf4f10f251773f43a | [
"MIT"
] | 5 | 2018-05-22T23:27:58.000Z | 2021-09-10T14:11:33.000Z | 57.427152 | 1,999 | 0.615234 | [
[
[
"import itertools\nimport os\nimport numpy as np\nimport pandas as pd\n\n\n\n# Get the root_path for this jupyter notebook repo.\nrepo_path = os.path.dirname(os.path.abspath(os.getcwd()))\n\npath_files_locus_index = os.path.join(\n repo_path, 'files', 'tell-dor', 'tell-dor-area-g-locus-image-index.csv'\n)\n\n# Path to the Tell Dor file metadata CSV\npath_files = os.path.join(\n repo_path, 'files', 'tell-dor', 'tell-dor-files.csv'\n)\n# Path to the Tell Dor locus metadata CSV \npath_loci = os.path.join(\n repo_path, 'files', 'tell-dor', 'tell-dor-loci.csv'\n)\n# Output path for associations between the files and the loci.\npath_files_contexts = os.path.join(\n repo_path, 'files', 'tell-dor', 'tell-dor-files-contexts.csv'\n)\n\n\n# Read the file - locus index supplied by the Tell Dor team.\nfl_df = pd.read_csv(path_files_locus_index)\n\n# Read the file metadata CSV into dataframe f_df.\nf_df = pd.read_csv(path_files)\n\n# Read the locus (and wall) CSV into dataframe l_df.\nl_df = pd.read_csv(path_loci)\n\nfl_df['Locus_Wall'] = fl_df['Locus_Wall'].astype(str) \nfl_df['Locus ID'] = np.nan\nfor i, row in fl_df.iterrows():\n wall_id = 'Wall ' + row['Locus_Wall']\n locus_id = 'Locus ' + row['Locus_Wall']\n print('Look for {} or {}'.format(wall_id, locus_id))\n id_indx = ((l_df['Locus ID']==wall_id)|(l_df['Locus ID']==locus_id))\n if l_df[id_indx].empty:\n continue\n up_indx = (fl_df['Locus_Wall'] == row['Locus_Wall'])\n fl_df.loc[up_indx, 'Locus ID'] = l_df[id_indx]['Locus ID'].iloc[0]\n print('Update {} with {}'.format(row['Locus_Wall'], l_df[id_indx]['Locus ID'].iloc[0]))\n\nfl_df.to_csv(path_files_locus_index, index=False)\n\n\n\n# Set up a dict for File and Locus (and Wall) associations.\nfile_locus_data = {\n 'File ID':[], \n 'Locus ID': [],\n}\n\n# Set up a dict for File and Area associations.\n# NOTE: An \"Area\" is an aggregation of multiple squares in the locus/wall\n# datafile. Eric grouped these to make search / browsing easier. They\n# don't really have any purpose or value for interpretation.\nfile_square_data = {\n 'File ID':[], \n 'Area': [],\n}\n\n\ndef add_to_file_context_data(\n file_ids, \n context_ids, \n data,\n context_id_col='Locus ID'\n):\n \"\"\"Adds records of file and context associations to a data dict\"\"\"\n if not isinstance(context_ids, list):\n context_ids = [context_ids]\n # Get the cross product of all the file_ids and the\n # context_ids\n crossprod = list(itertools.product(file_ids, context_ids))\n data['File ID'] += [c[0] for c in crossprod]\n data[context_id_col] += [c[1] for c in crossprod]\n return data\n",
"Look for Wall 18839 or Locus 18839\nLook for Wall 18229 or Locus 18229\n"
],
[
"f_df.head(3)",
"_____no_output_____"
],
[
"l_df.head(3)",
"_____no_output_____"
],
[
"\n# Find matching Loci (including Wall Loci) by matching their IDs\n# with text in the file metadata 'Caption' column.\nfor locus_wall_id in l_df['Locus ID'].unique().tolist():\n l_w_id = locus_wall_id.replace('Locus ', 'L').replace('Wall ', 'W')\n \n # l_w_mum_id is for locus or wall IDs that are long unlikely to be\n # a false positive, and lack a \"L\" or \"W\" in the caption.\n l_w_num_id = l_w_id.replace('L', ' ').replace('W', ' ')\n if len(l_w_num_id) >= 6:\n # Catch cases where the Locus / Wall ID is long like \n # '18347'.\n l_w_indx = (\n f_df['Caption'].str.contains(l_w_id)\n | f_df['Caption'].str.contains(l_w_num_id)\n )\n else:\n # The locus / wall id is too short to trust without a \n # \"L\" or \"W\" prefix.\n l_w_indx = f_df['Caption'].str.contains(l_w_id)\n \n if f_df[l_w_indx].empty:\n # We didn't find a match, so continue.\n continue\n print('Found: {} for {} as {}'.format(\n len(f_df[l_w_indx]), \n locus_wall_id,\n l_w_id,\n )\n )\n file_ids = f_df[l_w_indx]['File ID'].unique().tolist()\n file_locus_data = add_to_file_context_data(\n file_ids, \n locus_wall_id, \n file_locus_data\n )\n\n# Now make a dataframe of the file - locus associations\nfile_locus_df = pd.DataFrame(data=file_locus_data)\nprint('File and Locus Associations (Found: {})'.format(\n len(file_locus_df.index))\n)",
"_____no_output_____"
],
[
"# Find matching Loci (including Wall Loci) by matching their Squares\n# with text in the file metadata 'Caption' column.\nl_df_sq = l_df[~l_df['Square'].isnull()]\nfor square in l_df_sq['Square'].astype(str).unique().tolist():\n sq_indx = f_df['Caption'].str.contains(square)\n if len(square) < 3 or f_df[sq_indx].empty:\n # Not enough characters for secure match.\n continue\n # Get all file_ids that have his square in their captions\n file_ids = f_df[sq_indx]['File ID'].unique().tolist()\n # Get all the locus ids that are associated with this square\n area_ids = l_df[\n l_df['Square']==square\n ]['Area'].unique().tolist()\n print('Found: {} files with square {} and {} areas'.format(\n len(f_df[sq_indx]), \n square,\n len(area_ids)\n )\n )\n # Now add to the file_locus_data.\n file_square_data = add_to_file_context_data(\n file_ids, \n area_ids, \n file_square_data,\n context_id_col='Area'\n )\n\n# Now make a dataframe of the file - area associations\nfile_area_df = pd.DataFrame(data=file_square_data)\nprint('File and Area Associations (Found: {})'.format(\n len(file_area_df.index))\n)",
"_____no_output_____"
],
[
"context_df = pd.merge(file_locus_df, file_area_df, on='File ID', how='outer')\ncontext_linked_files = context_df['File ID'].unique().tolist()\nprint('Found File and Context Associations for {} unique files (total rows: {})'.format(\n len(context_linked_files),\n len(context_df.index))\n)\n\n\n# Get a list of files that do NOT have context associations\nno_context_files = f_df[\n ~f_df['File ID'].isin(context_linked_files)\n]['File ID'].unique().tolist()\n\nfile_site_data = {\n 'File ID':[], \n 'Site Area': [],\n}\nfile_site_data = add_to_file_context_data(\n no_context_files, \n 'Area G', \n file_site_data,\n context_id_col='Site Area'\n)\nsite_df = pd.DataFrame(data=file_site_data)\ncontext_df = pd.concat([context_df, site_df], sort=False)\n\n# Set the column order for nice aesthetics\ncontext_df = context_df[['File ID', 'Site Area', 'Area', 'Locus ID']]\ncontext_df.sort_values(by=['File ID', 'Locus ID', 'Area'], inplace=True)\n\ncontext_df.to_csv(path_files_contexts, index=False)\ncontext_df.head(3)\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71c2fc8ce9b27d99fb84860b73151a839858285 | 337,855 | ipynb | Jupyter Notebook | Analisando_os_Dados_do_Airbnb(Bangkok).ipynb | rodrigowe1988/Portfolio-Data-Science | 9bf8afc788018ea04739082b02c70ab1dc10b54c | [
"MIT"
] | 1 | 2021-01-28T21:50:43.000Z | 2021-01-28T21:50:43.000Z | Analisando_os_Dados_do_Airbnb(Bangkok).ipynb | rodrigowe1988/Portfolio-Data-Science | 9bf8afc788018ea04739082b02c70ab1dc10b54c | [
"MIT"
] | null | null | null | Analisando_os_Dados_do_Airbnb(Bangkok).ipynb | rodrigowe1988/Portfolio-Data-Science | 9bf8afc788018ea04739082b02c70ab1dc10b54c | [
"MIT"
] | null | null | null | 239.613475 | 142,962 | 0.885113 | [
[
[
"<a href=\"https://colab.research.google.com/github/rodrigowe1988/Portfolio-Data-Science/blob/main/Analisando_os_Dados_do_Airbnb(Bangkok).ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"---\n\n# Análise dos Dados do Airbnb - (Bangkok)\n*\n\nO [Airbnb](https://www.airbnb.com.br/) já é considerado como sendo a **maior empresa hoteleira da atualidade**. Ah, o detalhe é que ele **não possui nenhum hotel**!\n\nConectando pessoas que querem viajar (e se hospedar) com anfitriões que querem alugar seus imóveis de maneira prática, o Airbnb fornece uma plataforma inovadora para tornar essa hospedagem alternativa.\n\nNo final de 2018, a Startup fundada 10 anos atrás, já havia **hospedado mais de 300 milhões** de pessoas ao redor de todo o mundo, desafiando as redes hoteleiras tradicionais.\n\nUma das iniciativas do Airbnb é disponibilizar dados do site, para algumas das principais cidades do mundo. Por meio do portal [Inside Airbnb](http://insideairbnb.com/get-the-data.html), é possível baixar uma grande quantidade de dados para desenvolver projetos e soluções de *Data Science*.\n\n<center><img alt=\"Analisando Airbnb\" width=\"100%\" src=\"https://quantocustaviajar.com/blog/wp-content/uploads/2016/12/como-funciona-o-airbnb.png\"></center>\n\n**Neste *notebook*, iremos analisar os dados referentes à cidade de Bangkok, e ver quais insights podem ser extraídos a partir de dados brutos.**",
"_____no_output_____"
],
[
"## Obtenção dos Dados\n\nTodos os dados usados aqui foram obtidos a partir do site [Inside Airbnb](http://insideairbnb.com/get-the-data.html).\n\nPara esta análise exploratória inicial, será baixado apenas o seguinte arquivo:\n\n* `listings.csv` - *Summary information and metrics for listings in Bangkok (good for visualisations).*\n",
"_____no_output_____"
]
],
[
[
"# importar os pacotes necessarios\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n%matplotlib inline ",
"_____no_output_____"
],
[
"# importar o arquivo listings.csv para um DataFrame\ndf = pd.read_csv(\"http://data.insideairbnb.com/thailand/central-thailand/bangkok/2020-12-23/visualisations/listings.csv\")\n",
"_____no_output_____"
]
],
[
[
"## Análise dos Dados\n",
"_____no_output_____"
],
[
"**Dicionário das variáveis**\n\n* id - número de id gerado para identificar o imóvel\n* name - nome da propriedade anunciada\n* host_id - número de id do proprietário (anfitrião) da propriedade\n* host_name - Nome do anfitrião\n* neighbourhood_group - esta coluna não contém nenhum valor válido\n* neighbourhood - nome do bairro\n* latitude - coordenada da latitude da propriedade\n* longitude - coordenada da longitude da propriedade\n* room_type - informa o tipo de quarto que é oferecido\n* price - preço para alugar o imóvel\n* minimum_nights - quantidade mínima de noites para reservar\n* number_of_reviews - número de reviews que a propriedade possui\n* last_review - data do último review\n* reviews_per_month - quantidade de reviews por mês\n* calculated_host_listings_count - quantidade de imóveis do mesmo anfitrião\n* availability_365 - número de dias de disponibilidade dentro de 365 dias\n\nAntes de iniciar qualquer análise, vamos verificar a cara do nosso *dataset*, analisando as 5 primeiras entradas.",
"_____no_output_____"
]
],
[
[
"# mostrar as 5 primeiras entradas\ndf.head()",
"_____no_output_____"
]
],
[
[
"### **Q1. Quantos atributos (variáveis) e quantas entradas o nosso conjunto de dados possui? Quais os tipos das variáveis?**",
"_____no_output_____"
]
],
[
[
"# identificar o volume de dados do DataFrame\nprint('Entradas:\\t {}'.format(df.shape[0]))\nprint('Variáveis:\\t {}'.format(df.shape[1]))\n\n# verificar as 5 primeiras entradas do dataset\ndisplay(df.dtypes)\n",
"Entradas:\t 19709\nVariáveis:\t 16\n"
]
],
[
[
"### **Q2. Qual a porcentagem de valores ausentes no *dataset*?**\n\nA qualidade de um *dataset* está diretamente relacionada à quantidade de valores ausentes. É importante entender logo no início se esses valores nulos são significativos comparados ao total de entradas.\n\n* É possível ver que a coluna `neighbourhood_group` possui 100% dos seus valores faltantes. \n* As variáveis `reviews_per_month` e `last_review` possuem valores nulos em quase metade das linhas.\n* As variáveis `name` e `host_name` têm aproximadamente 0,1% dos valores nulos.",
"_____no_output_____"
]
],
[
[
"# ordenar em ordem decrescente as variáveis por seus valores ausentes\n(df.isnull().sum() / df.shape[0]).sort_values(ascending=False)",
"_____no_output_____"
]
],
[
[
"### **Q3. Qual o tipo de distribuição das variáveis?** ",
"_____no_output_____"
]
],
[
[
"# plotar o histograma das variáveis numéricas\ndf.hist(bins=15, figsize=(15, 10));",
"_____no_output_____"
]
],
[
[
"### **Q4. Há outliers presentes?**\n\nPela distribuição do histograma, é possível verificar indícios da presença de *outliers*. Olhe por exemplo as variáveis `price`, `minimum_nights` e `calculated_host_listings_count`.\n\nOs valores não seguem uma distruição, e distorcem toda a representação gráfica. Para confirmar, há duas maneiras rápidas que auxiliam a detecção de *outliers*. São elas:\n\n* Resumo estatístico por meio do método `describe()`\n* Plotar `boxplots` para a variável.\n\n---\n\n",
"_____no_output_____"
]
],
[
[
"# ver o resumo estatístico das variáveis numéricas (somente das colunas que realmente importam para essa análise)\ndf[['price', 'minimum_nights','number_of_reviews', 'reviews_per_month', 'calculated_host_listings_count', 'availability_365']].describe()\n\n",
"_____no_output_____"
]
],
[
[
"Boxplot para minimum_nights\n\n* Separando em porcentagem as ofertas que requerem do hóspede **mais de 30 dias** como hospedagem mínima ",
"_____no_output_____"
]
],
[
[
"#minimum_nights\ndf.minimum_nights.plot(kind='box', vert=False, figsize=(15, 3))\nplt.show()\n\n#ver quantidade de valores acima de 30 dias para minimum_nights\nprint(\"minimum_nights: valores acima de 30: \")\nprint(\"{} entradas\".format(len(df[df.minimum_nights > 30])))\nprint(\"{:.4f}%\".format((len(df[df.minimum_nights > 30]) / df.shape[0])*100))",
"_____no_output_____"
]
],
[
[
"Boxplot para price\n\n* Separando em porcentagem as ofertas que tem seu preço **acima de 1900 baht** ",
"_____no_output_____"
]
],
[
[
"#price\ndf.price.plot(kind='box', vert=False, figsize=(15, 3))\nplt.show()\n\n#ver a quantidade de valores acima de 1900 na coluna price\nprint(\"\\nprice: valores acima de 1900 baht\")\nprint(\"{} entradas\".format(len(df[df.price > 1900])))\nprint(\"{:.4f}%\".format((len(df[df.price > 1900]) / df.shape[0])*100))",
"_____no_output_____"
]
],
[
[
"Histogramas sem outliers:\n\n* Já que foi identificada uma quantidade significativa de outliers nas variáveis price e minimum_nights, vamos agora 'limpar' o DataFrame delas e plotar novamente o histograma.",
"_____no_output_____"
]
],
[
[
"#remover os *outliers* em um novo DataFrame\ndf_clean = df.copy()\ndf_clean.drop(df_clean[df_clean.price > 1900].index, axis=0, inplace=True)\ndf_clean.drop(df_clean[df_clean.minimum_nights > 30].index, axis=0, inplace=True)\n\n#remover `neighbourhood_group`, pois está vazio\ndf_clean.drop('neighbourhood_group', axis=1, inplace=True)\n\n#plotar o histograma para as variáveis numéricas\ndf_clean.hist(bins=15, figsize=(15, 10));\n\n#média dos preços atualizada\nnew_media = df_clean.price.mean()\nprint(f'A nova média dos preços de diárias para a cidade a cidade de Bangkok (removendo os outliers) é de {new_media:.2f} baht.')\n",
"A nova média dos preços de diárias para a cidade a cidade de Bangkok (removendo os outliers) é de 963.77 baht.\n"
]
],
[
[
"\n### **Q4. Qual a correlação existente entre as variáveis**",
"_____no_output_____"
]
],
[
[
"# criar uma matriz de correlação\ncorr = df_clean[['price', 'minimum_nights', 'number_of_reviews', 'reviews_per_month', 'calculated_host_listings_count', 'availability_365']].corr()\n\n# mostrar a matriz de correlação\ndisplay(corr)\n",
"_____no_output_____"
],
[
"# plotar um heatmap a partir das correlações\nsns.heatmap(corr, cmap='RdBu', fmt='.2f', square=True, linecolor='white', annot=True);\n",
"_____no_output_____"
]
],
[
[
"### **Q5. Qual o tipo de imóvel mais alugado no Airbnb?**\n\nA coluna variável room_type indica o tipo de locação que está anunciada no Airbnb. Se você já alugou no site, sabe que existem opções de apartamentos/casas inteiras, apenas o aluguel de um quarto ou mesmo dividir o quarto com outras pessoas. \n\nUsando o método value_counts(), contaremos a quantidade de ocorrências de cada tipo de aluguel. ",
"_____no_output_____"
]
],
[
[
"df_clean.neighbourhood.value_counts().head()",
"_____no_output_____"
],
[
"# mostrar a quantidade de cada tipo de imóvel disponível\ndf_clean.room_type.value_counts()",
"_____no_output_____"
],
[
"# mostrar a porcentagem de cada tipo de imóvel disponível\ndf_clean.room_type.value_counts() / df_clean.shape[0]",
"_____no_output_____"
]
],
[
[
"### **Q6. Qual a localidade mais cara do dataset?**\n\nUma maneira de se verificar uma variável em função da outra é usando groupby(). No caso, queremos comparar os bairros(neighbourhoods) a partir do preço de locação. \n",
"_____no_output_____"
]
],
[
[
"# ver preços por bairros, na média\ndf_clean.groupby(['neighbourhood']).price.mean().sort_values(ascending=False)[:10]\n",
"_____no_output_____"
],
[
"#ver a quantidade de imóveis no bairro Parthum Wan\nqtd_imoveis = df_clean[df_clean.neighbourhood == 'Parthum Wan'].shape[0]\nprint(f'O bairro Parthum Wan possui {qtd_imoveis} imóveis cadastrados hoje no Airbnb.\\n\\n')\n\n#ver o .head() com as 5 primeiras respostas do bairro Parthum Wan\ndf_clean[df_clean.neighbourhood == 'Parthum Wan'].head()",
"O bairro Parthum Wan possui 302 imóveis cadastrados hoje no Airbnb.\n\n\n"
],
[
"# plotar os imóveis pela latitude-longitude\ndf_clean.plot(kind='scatter', x='longitude', y='latitude', alpha=0.4, c=df_clean['price'], s=8, cmap=plt.get_cmap('jet'), figsize=(12,8));\n",
"_____no_output_____"
]
],
[
[
"### **Q7. Qual é a média do mínimo de noites para aluguel (minimum_nights)?**",
"_____no_output_____"
]
],
[
[
"# ver a média da coluna `minimum_nights``\nmedia_noites = df_clean.minimum_nights.mean()\nprint('Usando os dados obtidos podemos ver que a média do número de noites mímimas que cada anúncio pede é de {:.2f} noites.'.format(media_noites))",
"Usando os dados obtidos podemos ver que a média do número de noites mímimas que cada anúncio pede é de 5.99 noites.\n"
]
],
[
[
"## Conclusões\n",
"_____no_output_____"
],
[
"Análise simples sobre a cidade de Bangkok e suas inúmeras ofertas de reserva no site Airbnb. Alguns detalhes importantes:\n\n* como o preço máximo de algumas dessas ofertas fugia muito da 'média real'(ocorrência de alguns outliers com valores que superam 20x o valor da mediana), optei por filtrar e exclui-los.\n\n* como pode ser facilmente notado, é um breve artigo de um aspirante a Cientista de Dados e com o tempo, garanto que meus artigos terão uma qualidade nitidamente maior.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e71c3891dcac0494e55fb5423506d909505c6953 | 1,416 | ipynb | Jupyter Notebook | 469-empty-chairs.ipynb | arkeros/projecteuler | c95db97583034af8fc61d5786692d82eabe50c12 | [
"MIT"
] | 2 | 2017-02-19T12:37:13.000Z | 2021-01-19T04:58:09.000Z | 469-empty-chairs.ipynb | arkeros/projecteuler | c95db97583034af8fc61d5786692d82eabe50c12 | [
"MIT"
] | null | null | null | 469-empty-chairs.ipynb | arkeros/projecteuler | c95db97583034af8fc61d5786692d82eabe50c12 | [
"MIT"
] | 4 | 2018-01-05T14:29:09.000Z | 2020-01-27T13:37:40.000Z | 24 | 115 | 0.53178 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e71c39ac9151dd76ca4f6fd8070040a22313d2a9 | 9,001 | ipynb | Jupyter Notebook | Major (1).ipynb | Blackwidow2981/Maskissue | 16347db197cd4ff2462b724a2c6c9228099c5d81 | [
"MIT"
] | null | null | null | Major (1).ipynb | Blackwidow2981/Maskissue | 16347db197cd4ff2462b724a2c6c9228099c5d81 | [
"MIT"
] | null | null | null | Major (1).ipynb | Blackwidow2981/Maskissue | 16347db197cd4ff2462b724a2c6c9228099c5d81 | [
"MIT"
] | null | null | null | 23.318653 | 99 | 0.467615 | [
[
[
"data_path=\"C:\\\\Users\\\\sneha\\\\Downloads\\\\dataset-20201208T142410Z-001\\\\dataset\"",
"_____no_output_____"
],
[
"import cv2,os\n",
"_____no_output_____"
],
[
"label_dict={\"with_mask\":0,\"without_mask\":1}",
"_____no_output_____"
],
[
"categories={\"with_mask\",\"without_mask\"}",
"_____no_output_____"
],
[
"data=[]\ntarget=[]\nfor category in categories:\n folder_path=os.path.join(data_path,category)\n img_names=os.listdir(folder_path)\n for img in img_names:\n img_path=os.path.join(folder_path,img)\n img=cv2.imread(img_path)\n try:\n gray=cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)\n resized=cv2.resize(gray,(100,100))\n data.append(resized)\n target.append(label_dict[category])\n except:\n pass",
"_____no_output_____"
],
[
"label_dict[\"without_mask\"]\n",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"data=np.array(data)",
"_____no_output_____"
],
[
"data=data/255",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"data=np.reshape(data,(1376,100,100,1))",
"_____no_output_____"
],
[
"target=np.array(target)",
"_____no_output_____"
],
[
"data.shape[1:]",
"_____no_output_____"
],
[
"target.shape",
"_____no_output_____"
],
[
"from keras.utils import np_utils",
"_____no_output_____"
],
[
"new_target=np_utils.to_categorical(target)",
"_____no_output_____"
],
[
"from keras.models import Sequential",
"_____no_output_____"
],
[
"from keras.layers import Dense,Activation,Flatten,Dropout",
"_____no_output_____"
],
[
"from keras.layers import Conv2D,MaxPooling2D",
"_____no_output_____"
],
[
"model=Sequential()",
"_____no_output_____"
],
[
"model.add(Conv2D(200,(3,3),activation=\"relu\",input_shape=data.shape[1:]))\nmodel.add(MaxPooling2D(pool_size=(2,2)))\nmodel.add(Conv2D(100,(3,3),activation=\"relu\"))\nmodel.add(MaxPooling2D(pool_size=(2,2)))\nmodel.add(Flatten())\nmodel.add(Dropout(0.5))\nmodel.add(Dense(50,activation=\"relu\"))\nmodel.add(Dense(2,activation=\"softmax\"))\n",
"_____no_output_____"
],
[
"model.compile(loss=\"categorical_crossentropy\",optimizer=\"adam\",metrics=[\"accuracy\"])",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"train_data,test_data,train_target,test_target=train_test_split(data,new_target,test_size=0.1)",
"_____no_output_____"
],
[
"model.fit(train_data,train_target,epochs=20)",
"Epoch 1/20\n39/39 [==============================] - 145s 4s/step - loss: 0.7018 - accuracy: 0.5392\nEpoch 2/20\n39/39 [==============================] - 120s 3s/step - loss: 0.5695 - accuracy: 0.7346\nEpoch 3/20\n39/39 [==============================] - 120s 3s/step - loss: 0.2758 - accuracy: 0.8814\nEpoch 4/20\n39/39 [==============================] - 119s 3s/step - loss: 0.1638 - accuracy: 0.9364\nEpoch 5/20\n39/39 [==============================] - 125s 3s/step - loss: 0.1182 - accuracy: 0.9549\nEpoch 6/20\n39/39 [==============================] - 96s 2s/step - loss: 0.1115 - accuracy: 0.9549\nEpoch 7/20\n39/39 [==============================] - 97s 2s/step - loss: 0.0966 - accuracy: 0.9641\nEpoch 8/20\n39/39 [==============================] - 123s 3s/step - loss: 0.0663 - accuracy: 0.9756\nEpoch 9/20\n39/39 [==============================] - 94s 2s/step - loss: 0.0629 - accuracy: 0.9751\nEpoch 10/20\n39/39 [==============================] - 92s 2s/step - loss: 0.0540 - accuracy: 0.9769\nEpoch 11/20\n39/39 [==============================] - 92s 2s/step - loss: 0.0467 - accuracy: 0.9854\nEpoch 12/20\n39/39 [==============================] - 91s 2s/step - loss: 0.0582 - accuracy: 0.9757\nEpoch 13/20\n39/39 [==============================] - 91s 2s/step - loss: 0.0462 - accuracy: 0.9795\nEpoch 14/20\n39/39 [==============================] - 92s 2s/step - loss: 0.0438 - accuracy: 0.9846\nEpoch 15/20\n39/39 [==============================] - 92s 2s/step - loss: 0.0336 - accuracy: 0.9901\nEpoch 16/20\n39/39 [==============================] - 229s 6s/step - loss: 0.0165 - accuracy: 0.9954\nEpoch 17/20\n39/39 [==============================] - 141s 4s/step - loss: 0.0203 - accuracy: 0.9932\nEpoch 18/20\n39/39 [==============================] - 113s 3s/step - loss: 0.0409 - accuracy: 0.9836\nEpoch 19/20\n39/39 [==============================] - 115s 3s/step - loss: 0.0266 - accuracy: 0.9930\nEpoch 20/20\n39/39 [==============================] - 119s 3s/step - loss: 0.0110 - accuracy: 0.9966\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71c539562cdbe80e0a8da4a1ff80ecc26bed881 | 22,215 | ipynb | Jupyter Notebook | data/data_aggregation.ipynb | minhrongcon2000/vn-aqi-viz | 52a170fb7ce4d5094570875b4c26ad3965384775 | [
"MIT"
] | 2 | 2021-04-17T10:06:20.000Z | 2021-04-19T10:33:18.000Z | data/data_aggregation.ipynb | minhrongcon2000/vn-aqi-viz | 52a170fb7ce4d5094570875b4c26ad3965384775 | [
"MIT"
] | null | null | null | data/data_aggregation.ipynb | minhrongcon2000/vn-aqi-viz | 52a170fb7ce4d5094570875b4c26ad3965384775 | [
"MIT"
] | 1 | 2021-04-25T14:40:33.000Z | 2021-04-25T14:40:33.000Z | 30.060893 | 158 | 0.314067 | [
[
[
"import pandas as pd\nimport glob\nimport numpy as np",
"_____no_output_____"
],
[
"filenames = glob.glob(\"*.csv\")",
"_____no_output_____"
],
[
"dfs = []\ncolumn_names = set()\nfor filename in filenames:\n if not filename.startswith(\"aqi\"):\n df = pd.read_csv(filename, na_values=[\" \"]).fillna(0)\n df.columns = [column.strip() for column in df.columns]\n df['province'] = filename[:-4]\n for column_name in df.columns:\n column_names.add(column_name)\n dfs.append(df)",
"_____no_output_____"
],
[
"for df in dfs:\n for column_name in column_names:\n if column_name not in df.columns:\n df[column_name] = 0",
"_____no_output_____"
],
[
"all_data = pd.concat(dfs, axis=0)",
"_____no_output_____"
],
[
"all_data",
"_____no_output_____"
],
[
"all_data.loc[all_data[\"aqi\"] == 0, \"aqi\"] = all_data.loc[all_data[\"aqi\"] == 0, [\"pm25\", \"pm10\", \"o3\", \"no2\", \"so2\", \"co\"]].max(axis=1)",
"_____no_output_____"
],
[
"all_data",
"_____no_output_____"
],
[
"all_data.to_csv(\"aqi.csv\", index=False)",
"_____no_output_____"
],
[
"all_data[\"province\"].unique()",
"_____no_output_____"
],
[
"all_data[all_data[\"province\"] == \"Thai Nguyen\"]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71c71db0f33693c94b84ac56c06991aaa1d9d5b | 217,890 | ipynb | Jupyter Notebook | tutorials/Certification_Trainings/Healthcare/11.Pretrained_Clinical_Pipelines.ipynb | AngelJavierSalazar/spark-nlp-workshop | 47e6c8f921b1fd2fcd99a7dce15e142d792c40a1 | [
"Apache-2.0"
] | 1 | 2022-01-25T17:24:13.000Z | 2022-01-25T17:24:13.000Z | tutorials/Certification_Trainings/Healthcare/11.Pretrained_Clinical_Pipelines.ipynb | AngelJavierSalazar/spark-nlp-workshop | 47e6c8f921b1fd2fcd99a7dce15e142d792c40a1 | [
"Apache-2.0"
] | null | null | null | tutorials/Certification_Trainings/Healthcare/11.Pretrained_Clinical_Pipelines.ipynb | AngelJavierSalazar/spark-nlp-workshop | 47e6c8f921b1fd2fcd99a7dce15e142d792c40a1 | [
"Apache-2.0"
] | null | null | null | 44.658742 | 2,565 | 0.385782 | [
[
[
"",
"_____no_output_____"
],
[
"[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/Certification_Trainings/Healthcare/11.Pretrained_Clinical_Pipelines.ipynb)",
"_____no_output_____"
],
[
"# 11. Pretrained_Clinical_Pipelines",
"_____no_output_____"
],
[
"## Colab Setup",
"_____no_output_____"
]
],
[
[
"import json\nimport os\n\nfrom google.colab import files\n\nlicense_keys = files.upload()\n\nwith open(list(license_keys.keys())[0]) as f:\n license_keys = json.load(f)\n\n# Defining license key-value pairs as local variables\nlocals().update(license_keys)\n\n# Adding license key-value pairs to environment variables\nos.environ.update(license_keys)",
"_____no_output_____"
],
[
"# Installing pyspark and spark-nlp\n! pip install --upgrade -q pyspark==3.1.2 spark-nlp==$PUBLIC_VERSION\n\n# Installing Spark NLP Healthcare\n! pip install --upgrade -q spark-nlp-jsl==$JSL_VERSION --extra-index-url https://pypi.johnsnowlabs.com/$SECRET",
"_____no_output_____"
],
[
"import json\nimport os\nfrom pyspark.ml import Pipeline\nfrom pyspark.sql import SparkSession\n\nfrom sparknlp.annotator import *\nfrom sparknlp_jsl.annotator import *\nfrom sparknlp.base import *\nimport sparknlp_jsl\nimport sparknlp\n\nparams = {\"spark.driver.memory\":\"16G\",\n\"spark.kryoserializer.buffer.max\":\"2000M\",\n\"spark.driver.maxResultSize\":\"2000M\"}\n\nspark = sparknlp_jsl.start(license_keys['SECRET'],params=params)\n\nprint (\"Spark NLP Version :\", sparknlp.version())\nprint (\"Spark NLP_JSL Version :\", sparknlp_jsl.version())",
"Spark NLP Version : 3.3.4\nSpark NLP_JSL Version : 3.3.4\n"
]
],
[
[
"\n<b> if you want to work with Spark 2.3 </b>\n```\nimport os\n\n# Install java\n! apt-get update -qq\n! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null\n\n!wget -q https://archive.apache.org/dist/spark/spark-2.3.0/spark-2.3.0-bin-hadoop2.7.tgz\n\n!tar xf spark-2.3.0-bin-hadoop2.7.tgz\n!pip install -q findspark\n\nos.environ[\"JAVA_HOME\"] = \"/usr/lib/jvm/java-8-openjdk-amd64\"\nos.environ[\"PATH\"] = os.environ[\"JAVA_HOME\"] + \"/bin:\" + os.environ[\"PATH\"]\nos.environ[\"SPARK_HOME\"] = \"/content/spark-2.3.0-bin-hadoop2.7\"\n! java -version\n\nimport findspark\nfindspark.init()\nfrom pyspark.sql import SparkSession\n\n! pip install --ignore-installed -q spark-nlp==2.7.5\nimport sparknlp\n\nspark = sparknlp.start(spark23=True)\n```",
"_____no_output_____"
],
[
"## Pretrained Pipelines",
"_____no_output_____"
],
[
"In order to save you from creating a pipeline from scratch, Spark NLP also has a pre-trained pipelines that are already fitted using certain annotators and transformers according to various use cases.\n\nHere is the list of clinical pre-trained pipelines: \n\n> These clinical pipelines are trained with `embeddings_healthcare_100d` and accuracies might be 1-2% lower than `embeddings_clinical` which is 200d.\n\n**1. explain_clinical_doc_carp** :\n\n> a pipeline with `ner_clinical`, `assertion_dl`, `re_clinical` and `ner_posology`. It will extract clinical and medication entities, assign assertion status and find relationships between clinical entities.\n\n**2. explain_clinical_doc_era** :\n\n> a pipeline with `ner_clinical_events`, `assertion_dl` and `re_temporal_events_clinical`. It will extract clinical entities, assign assertion status and find temporal relationships between clinical entities.\n\n**3. recognize_entities_posology** :\n\n> a pipeline with `ner_posology`. It will only extract medication entities.\n\n\n*Since 3rd pipeline is already a subset of 1st and 2nd pipeline, we will only cover the first two pipelines in this notebook.*\n\n**4. explain_clinical_doc_ade** :\n\n> a pipeline for `Adverse Drug Events (ADE)` with `ner_ade_biobert`, `assertiondl_biobert`, `classifierdl_ade_conversational_biobert` and `re_ade_biobert`. It will classify the document, extract `ADE` and `DRUG` entities, assign assertion status to `ADE` entities, and relate them with `DRUG` entities, then assign ADE status to a text (`True` means ADE, `False` means not related to ADE).\n\n**letter codes in the naming conventions:**\n\n> c : ner_clinical\n\n> e : ner_clinical_events\n\n> r : relation extraction\n\n> p : ner_posology\n\n> a : assertion\n\n> ade : adverse drug events\n\n**Relation Extraction types:**\n\n`re_clinical` >> TrIP (improved), TrWP (worsened), TrCP (caused problem), TrAP (administered), TrNAP (avoided), TeRP (revealed problem), TeCP (investigate problem), PIP (problems related)\n\n`re_temporal_events_clinical` >> `AFTER`, `BEFORE`, `OVERLAP`\n\n**5. ner_profiling_clinical and ner_profiling_biobert:**\n\n> pipelines for exploring all the available pretrained NER models at once.\n\n**6. ner_model_finder**\n\n> a pipeline trained with bert embeddings that can be used to find the most appropriate NER model given the entity name.\n\n**7. icd10cm_snomed_mapping**\n\n> a pipeline maps ICD10CM codes to SNOMED codes without using any text data. You’ll just feed a comma or white space delimited ICD10CM codes and it will return the corresponding SNOMED codes as a list. \n\n**8. snomed_icd10cm_mapping:**\n\n> a pipeline converts Snomed codes to ICD10CM codes. Just feed a comma or white space delimited SNOMED codes and it will return the corresponding ICD10CM codes as a list.\n",
"_____no_output_____"
],
[
"## 1.explain_clinical_doc_carp \n\na pipeline with ner_clinical, assertion_dl, re_clinical and ner_posology. It will extract clinical and medication entities, assign assertion status and find relationships between clinical entities.",
"_____no_output_____"
]
],
[
[
"from sparknlp.pretrained import PretrainedPipeline",
"_____no_output_____"
],
[
"pipeline = PretrainedPipeline('explain_clinical_doc_carp', 'en', 'clinical/models')",
"explain_clinical_doc_carp download started this may take some time.\nApprox size to download 1.6 GB\n[OK!]\n"
],
[
"pipeline.model.stages",
"_____no_output_____"
],
[
"# Load pretrained pipeline from local disk:\n\n# >> pipeline_local = PretrainedPipeline.from_disk('/root/cache_pretrained/explain_clinical_doc_carp_en_2.5.5_2.4_1597841630062')",
"_____no_output_____"
],
[
"text =\"\"\"A 28-year-old female with a history of gestational diabetes mellitus, used to take metformin 1000 mg two times a day, presented with a one-week history of polyuria , polydipsia , poor appetite , and vomiting .\nShe was seen by the endocrinology service and discharged on 40 units of insulin glargine at night, 12 units of insulin lispro with meals.\n\"\"\"\n\nannotations = pipeline.annotate(text)\n\nannotations.keys()\n",
"_____no_output_____"
],
[
"import pandas as pd\n\nrows = list(zip(annotations['tokens'], annotations['clinical_ner_tags'], annotations['posology_ner_tags'], annotations['pos_tags'], annotations['dependencies']))\n\ndf = pd.DataFrame(rows, columns = ['tokens','clinical_ner_tags','posology_ner_tags','POS_tags','dependencies'])\n\ndf.head(20)",
"_____no_output_____"
],
[
"text = 'Patient has a headache for the last 2 weeks and appears anxious when she walks fast. No alopecia noted. She denies pain'\n\nresult = pipeline.fullAnnotate(text)[0]\n\nchunks=[]\nentities=[]\nstatus=[]\n\nfor n,m in zip(result['clinical_ner_chunks'],result['assertion']):\n \n chunks.append(n.result)\n entities.append(n.metadata['entity']) \n status.append(m.result)\n \ndf = pd.DataFrame({'chunks':chunks, 'entities':entities, 'assertion':status})\n\ndf",
"_____no_output_____"
],
[
"text = \"\"\"\nThe patient was prescribed 1 unit of Advil for 5 days after meals. The patient was also \ngiven 1 unit of Metformin daily.\nHe was seen by the endocrinology service and she was discharged on 40 units of insulin glargine at night , \n12 units of insulin lispro with meals , and metformin 1000 mg two times a day.\n\"\"\"\n\nresult = pipeline.fullAnnotate(text)[0]\n\nchunks=[]\nentities=[]\nbegins=[]\nends=[]\n\nfor n in result['posology_ner_chunks']:\n \n chunks.append(n.result)\n begins.append(n.begin)\n ends.append(n.end)\n entities.append(n.metadata['entity']) \n \ndf = pd.DataFrame({'chunks':chunks, 'begin':begins, 'end':ends, 'entities':entities})\n\ndf",
"_____no_output_____"
]
],
[
[
"## **2. explain_clinical_doc_era** \n\n> a pipeline with `ner_clinical_events`, `assertion_dl` and `re_temporal_events_clinical`. It will extract clinical entities, assign assertion status and find temporal relationships between clinical entities.\n\n",
"_____no_output_____"
]
],
[
[
"era_pipeline = PretrainedPipeline('explain_clinical_doc_era', 'en', 'clinical/models')",
"explain_clinical_doc_era download started this may take some time.\nApprox size to download 1.6 GB\n[OK!]\n"
],
[
"era_pipeline.model.stages",
"_____no_output_____"
],
[
"text =\"\"\"She is admitted to The John Hopkins Hospital 2 days ago with a history of gestational diabetes mellitus diagnosed. She denied pain and any headache.\nShe was seen by the endocrinology service and she was discharged on 03/02/2018 on 40 units of insulin glargine, \n12 units of insulin lispro, and metformin 1000 mg two times a day. She had close follow-up with endocrinology post discharge. \n\"\"\"\n\nresult = era_pipeline.fullAnnotate(text)[0]\n",
"_____no_output_____"
],
[
"result.keys()",
"_____no_output_____"
],
[
"import pandas as pd\n\nchunks=[]\nentities=[]\nbegins=[]\nends=[]\n\nfor n in result['clinical_ner_chunks']:\n \n chunks.append(n.result)\n begins.append(n.begin)\n ends.append(n.end)\n entities.append(n.metadata['entity']) \n \ndf = pd.DataFrame({'chunks':chunks, 'begin':begins, 'end':ends, 'entities':entities})\n\ndf",
"_____no_output_____"
],
[
"chunks=[]\nentities=[]\nstatus=[]\n\nfor n,m in zip(result['clinical_ner_chunks'],result['assertion']):\n \n chunks.append(n.result)\n entities.append(n.metadata['entity']) \n status.append(m.result)\n \ndf = pd.DataFrame({'chunks':chunks, 'entities':entities, 'assertion':status})\n\ndf",
"_____no_output_____"
],
[
"import pandas as pd\n\ndef get_relations_df (results, col='relations'):\n rel_pairs=[]\n for rel in results[0][col]:\n rel_pairs.append((\n rel.result, \n rel.metadata['entity1'], \n rel.metadata['entity1_begin'],\n rel.metadata['entity1_end'],\n rel.metadata['chunk1'], \n rel.metadata['entity2'],\n rel.metadata['entity2_begin'],\n rel.metadata['entity2_end'],\n rel.metadata['chunk2'], \n rel.metadata['confidence']\n ))\n\n rel_df = pd.DataFrame(rel_pairs, columns=['relation','entity1','entity1_begin','entity1_end','chunk1','entity2','entity2_begin','entity2_end','chunk2', 'confidence'])\n\n rel_df.confidence = rel_df.confidence.astype(float)\n \n return rel_df",
"_____no_output_____"
],
[
"annotations = era_pipeline.fullAnnotate(text)\n\nrel_df = get_relations_df (annotations, 'clinical_relations')\n\nrel_df",
"_____no_output_____"
],
[
"annotations[0]['clinical_relations']",
"_____no_output_____"
]
],
[
[
"## 3.explain_clinical_doc_ade \n\nA pipeline for `Adverse Drug Events (ADE)` with `ner_ade_healthcare`, and `classifierdl_ade_biobert`. It will extract `ADE` and `DRUG` clinical entities, and then assign ADE status to a text(`True` means ADE, `False` means not related to ADE). Also extracts relations between `DRUG` and `ADE` entities (`1` means the adverse event and drug entities are related, `0` is not related).",
"_____no_output_____"
]
],
[
[
"ade_pipeline = PretrainedPipeline('explain_clinical_doc_ade', 'en', 'clinical/models')",
"explain_clinical_doc_ade download started this may take some time.\nApprox size to download 462.3 MB\n[OK!]\n"
],
[
"result = ade_pipeline.fullAnnotate(\"The main adverse effects of Leflunomide consist of diarrhea, nausea, liver enzyme elevation, hypertension, alopecia, and allergic skin reactions.\")\n\nresult[0].keys()",
"_____no_output_____"
],
[
"result[0]['class'][0].metadata",
"_____no_output_____"
],
[
"text = \"Jaw,neck, low back and hip pains. Numbness in legs and arms. Took about a month for the same symptoms to begin with Vytorin. The pravachol started the pains again in about 3 months. I stopped taking all statin drungs. Still hurting after 2 months of stopping. Be careful taking this drug.\"\n\nimport pandas as pd\n\nchunks = []\nentities = []\nbegin =[]\nend = []\n\nprint ('sentence:', text)\nprint()\n\nresult = ade_pipeline.fullAnnotate(text)\n\nprint ('ADE status:', result[0]['class'][0].result)\n\nprint ('prediction probability>> True : ', result[0]['class'][0].metadata['True'], \\\n 'False: ', result[0]['class'][0].metadata['False'])\n\nfor n in result[0]['ner_chunks_ade']:\n\n begin.append(n.begin)\n end.append(n.end)\n chunks.append(n.result)\n entities.append(n.metadata['entity']) \n\ndf = pd.DataFrame({'chunks':chunks, 'entities':entities,\n 'begin': begin, 'end': end})\n\ndf\n",
"sentence: Jaw,neck, low back and hip pains. Numbness in legs and arms. Took about a month for the same symptoms to begin with Vytorin. The pravachol started the pains again in about 3 months. I stopped taking all statin drungs. Still hurting after 2 months of stopping. Be careful taking this drug.\n\nADE status: True\nprediction probability>> True : 0.99863094 False: 0.0013689838\n"
]
],
[
[
"#### with AssertionDL",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ntext = \"\"\"I have an allergic reaction to vancomycin. \nMy skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums. \nI would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication.\"\"\"\n\nprint (text)\n\nlight_result = ade_pipeline.fullAnnotate(text)[0]\n\nchunks=[]\nentities=[]\nstatus=[]\n\nfor n,m in zip(light_result['ner_chunks_ade_assertion'],light_result['assertion_ade']):\n \n chunks.append(n.result)\n entities.append(n.metadata['entity']) \n status.append(m.result)\n \ndf = pd.DataFrame({'chunks':chunks, 'entities':entities, 'assertion':status})\n\ndf",
"I have an allergic reaction to vancomycin. \nMy skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums. \nI would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication.\n"
]
],
[
[
"#### with Relation Extraction",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ntext = \"\"\"I have Rhuematoid Arthritis for 35 yrs and have been on many arthritis meds. \nI currently am on Relefen for inflamation, Prednisone 5mg, every other day and Enbrel injections once a week. \nI have no problems from these drugs. Eight months ago, another doctor put me on Lipitor 10mg daily because my chol was 240. \nOver a period of 6 months, it went down to 159, which was great, BUT I started having terrible aching pain in my arms about that time which was radiating down my arms from my shoulder to my hands.\n\"\"\"\n \nprint (text)\n\nresults = ade_pipeline.fullAnnotate(text)\n\nrel_pairs=[]\n\nfor rel in results[0][\"relations_ade_drug\"]:\n rel_pairs.append((\n rel.result, \n rel.metadata['entity1'], \n rel.metadata['entity1_begin'],\n rel.metadata['entity1_end'],\n rel.metadata['chunk1'], \n rel.metadata['entity2'],\n rel.metadata['entity2_begin'],\n rel.metadata['entity2_end'],\n rel.metadata['chunk2'], \n rel.metadata['confidence']\n ))\n\nrel_df = pd.DataFrame(rel_pairs, columns=['relation','entity1','entity1_begin','entity1_end','chunk1','entity2','entity2_begin','entity2_end','chunk2', 'confidence'])\nrel_df",
"I have Rhuematoid Arthritis for 35 yrs and have been on many arthritis meds. \nI currently am on Relefen for inflamation, Prednisone 5mg, every other day and Enbrel injections once a week. \nI have no problems from these drugs. Eight months ago, another doctor put me on Lipitor 10mg daily because my chol was 240. \nOver a period of 6 months, it went down to 159, which was great, BUT I started having terrible aching pain in my arms about that time which was radiating down my arms from my shoulder to my hands.\n\n"
]
],
[
[
"## 4.Clinical Deidentification\n\nThis pipeline can be used to deidentify PHI information from medical texts. The PHI information will be masked and obfuscated in the resulting text. The pipeline can mask and obfuscate `AGE`, `CONTACT`, `DATE`, `ID`, `LOCATION`, `NAME`, `PROFESSION`, `CITY`, `COUNTRY`, `DOCTOR`, `HOSPITAL`, `IDNUM`, `MEDICALRECORD`, `ORGANIZATION`, `PATIENT`, `PHONE`, `PROFESSION`, `STREET`, `USERNAME`, `ZIP`, `ACCOUNT`, `LICENSE`, `VIN`, `SSN`, `DLN`, `PLATE`, `IPADDR` entities.",
"_____no_output_____"
]
],
[
[
"deid_pipeline = PretrainedPipeline(\"clinical_deidentification\", \"en\", \"clinical/models\")",
"clinical_deidentification download started this may take some time.\nApprox size to download 1.6 GB\n[OK!]\n"
],
[
"deid_res = deid_pipeline.annotate(\"Record date : 2093-01-13, David Hale, M.D. IP: 203.120.223.13. The driver's license no:A334455B. the SSN:324598674 and e-mail: [email protected]. Name : Hendrickson, Ora MR. 25 years-old # 719435 Date : 01/13/93. Signed by Oliveira Sander, . Record date : 2079-11-09, Patient's VIN : 1HGBH41JXMN109286.\")",
"_____no_output_____"
],
[
"deid_res.keys()",
"_____no_output_____"
],
[
"pd.set_option(\"display.max_colwidth\", 100)\n\ndf = pd.DataFrame(list(zip(deid_res['sentence'], deid_res['masked'], deid_res['obfuscated'])),\n columns = ['Sentence','Masked', 'Obfuscated'])\ndf",
"_____no_output_____"
]
],
[
[
"## 5.NER Profiling Pipelines\n\nWe can use pretrained NER profiling pipelines for exploring all the available pretrained NER models at once. In Spark NLP we have two different NER profiling pipelines;\n\n- `ner_profiling_clinical` : Returns results for clinical NER models trained with `embeddings_clinical`.\n- `ner_profiling_biobert` : Returns results for clinical NER models trained with `biobert_pubmed_base_cased`.\n\nFor more examples, please check [this notebook](https://github.com/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/Certification_Trainings/Healthcare/11.2.Pretrained_NER_Profiling_Pipelines.ipynb).\n\n\n\n",
"_____no_output_____"
],
[
"**`ner_profiling_clinical` Model List**\n\n| index | model | index | model | index | model | index | model |\n|--------:|:----------------------------------|--------:|:-----------------------------|--------:|:-------------------------------|--------:|:--------------------------------|\n| 1 | [ner_ade_clinical](https://nlp.johnsnowlabs.com/2021/04/01/ner_ade_clinical_en.html) | 13 | [nerdl_tumour_demo](https://nlp.johnsnowlabs.com/2021/04/01/nerdl_tumour_demo_en.html) | 25 | [ner_drugs](https://nlp.johnsnowlabs.com/2021/03/31/ner_drugs_en.html) | 37 | [ner_radiology_wip_clinical](https://nlp.johnsnowlabs.com/2021/04/01/ner_radiology_wip_clinical_en.html) |\n| 2 | [ner_posology_greedy](https://nlp.johnsnowlabs.com/2021/03/31/ner_posology_greedy_en.html) | 14 | [ner_deid_subentity_augmented](https://nlp.johnsnowlabs.com/2021/06/30/ner_deid_subentity_augmented_en.html) | 26 | [ner_deid_sd](https://nlp.johnsnowlabs.com/2021/04/01/ner_deid_sd_en.html) | 38 | [ner_clinical](https://nlp.johnsnowlabs.com/2020/01/30/ner_clinical_en.html) |\n| 3 | [ner_risk_factors](https://nlp.johnsnowlabs.com/2021/03/31/ner_risk_factors_en.html) | 15 | [ner_jsl_enriched](https://nlp.johnsnowlabs.com/2021/10/22/ner_jsl_enriched_en.html) | 27 | [ner_posology_large](https://nlp.johnsnowlabs.com/2021/03/31/ner_posology_large_en.html) | 39 | [ner_chemicals](https://nlp.johnsnowlabs.com/2021/04/01/ner_chemicals_en.html) |\n| 4 | [jsl_ner_wip_clinical](https://nlp.johnsnowlabs.com/2021/03/31/jsl_ner_wip_clinical_en.html) | 16 | [ner_genetic_variants](https://nlp.johnsnowlabs.com/2021/06/25/ner_genetic_variants_en.html) | 28 | [ner_deid_large](https://nlp.johnsnowlabs.com/2021/03/31/ner_deid_large_en.html) | 40 | [ner_deid_augmented](https://nlp.johnsnowlabs.com/2021/02/19/ner_deid_synthetic_en.html) |\n| 5 | [ner_human_phenotype_gene_clinical](https://nlp.johnsnowlabs.com/2021/03/31/ner_human_phenotype_gene_clinical_en.html) | 17 | [ner_bionlp](https://nlp.johnsnowlabs.com/2021/03/31/ner_bionlp_en.html) | 29 | [ner_posology](https://nlp.johnsnowlabs.com/2020/04/15/ner_posology_en.html) | 41 | [ner_events_clinical](https://nlp.johnsnowlabs.com/2021/03/31/ner_events_clinical_en.html) |\n| 6 | [jsl_ner_wip_greedy_clinical](https://nlp.johnsnowlabs.com/2021/03/31/jsl_ner_wip_greedy_clinical_en.html) | 18 | [ner_measurements_clinical](https://nlp.johnsnowlabs.com/2021/04/01/ner_measurements_clinical_en.html) | 30 | [ner_deidentify_dl](https://nlp.johnsnowlabs.com/2021/03/31/ner_deidentify_dl_en.html) | 42 | [ner_posology_small](https://nlp.johnsnowlabs.com/2021/03/31/ner_posology_small_en.html) |\n| 7 | [ner_cellular](https://nlp.johnsnowlabs.com/2021/03/31/ner_cellular_en.html) | 19 | [ner_diseases_large](https://nlp.johnsnowlabs.com/2021/04/01/ner_diseases_large_en.html) | 31 | [ner_deid_enriched](https://nlp.johnsnowlabs.com/2021/03/31/ner_deid_enriched_en.html) | 43 | [ner_anatomy_coarse](https://nlp.johnsnowlabs.com/2021/03/31/ner_anatomy_coarse_en.html) |\n| 8 | [ner_cancer_genetics](https://nlp.johnsnowlabs.com/2021/03/31/ner_cancer_genetics_en.html) | 20 | [ner_radiology](https://nlp.johnsnowlabs.com/2021/03/31/ner_radiology_en.html) | 32 | [ner_bacterial_species](https://nlp.johnsnowlabs.com/2021/04/01/ner_bacterial_species_en.html) | 44 | [ner_human_phenotype_go_clinical](https://nlp.johnsnowlabs.com/2020/09/21/ner_human_phenotype_go_clinical_en.html) |\n| 9 | [jsl_ner_wip_modifier_clinical](https://nlp.johnsnowlabs.com/2021/04/01/jsl_ner_wip_modifier_clinical_en.html) | 21 | [ner_deid_augmented](https://nlp.johnsnowlabs.com/2021/03/31/ner_deid_augmented_en.html) | 33 | [ner_drugs_large](https://nlp.johnsnowlabs.com/2021/03/31/ner_drugs_large_en.html) | 45 | [ner_jsl_slim](https://nlp.johnsnowlabs.com/2021/08/13/ner_jsl_slim_en.html) |\n| 10 | [ner_drugs_greedy](https://nlp.johnsnowlabs.com/2021/03/31/ner_drugs_greedy_en.html) | 22 | [ner_anatomy](https://nlp.johnsnowlabs.com/2021/03/31/ner_anatomy_en.html) | 34 | [ner_clinical_large](https://nlp.johnsnowlabs.com/2021/03/31/ner_clinical_large_en.html) | 46 | [ner_jsl](https://nlp.johnsnowlabs.com/2021/06/24/ner_jsl_en.html) |\n| 11 | [ner_deid_sd_large](https://nlp.johnsnowlabs.com/2021/04/01/ner_deid_sd_large_en.html) | 23 | [ner_chemprot_clinical](https://nlp.johnsnowlabs.com/2021/03/31/ner_chemprot_clinical_en.html) | 35 | [jsl_rd_ner_wip_greedy_clinical](https://nlp.johnsnowlabs.com/2021/04/01/jsl_rd_ner_wip_greedy_clinical_en.html) | 47 | [ner_jsl_greedy](https://nlp.johnsnowlabs.com/2021/06/24/ner_jsl_greedy_en.html) |\n| 12 | [ner_diseases](https://nlp.johnsnowlabs.com/2021/03/31/ner_diseases_en.html) | 24 | [ner_posology_experimental](https://nlp.johnsnowlabs.com/2021/09/01/ner_posology_experimental_en.html) | 36 | [ner_medmentions_coarse](https://nlp.johnsnowlabs.com/2021/04/01/ner_medmentions_coarse_en.html) | 48 | [ner_events_admission_clinical](https://nlp.johnsnowlabs.com/2021/03/31/ner_events_admission_clinical_en.html) |\n\n**`ner_profiling_BERT` Model List**\n\n\n\n| index | model | index | model | index | model |\n|--------:|:-----------------------|--------:|:---------------------------------|--------:|:------------------------------|\n| 1 | [ner_cellular_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_cellular_biobert_en.html) | 8 | [ner_jsl_enriched_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_jsl_enriched_biobert_en.html) | 15 | [ner_posology_large_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_posology_large_biobert_en.html) |\n| 2 | [ner_diseases_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_diseases_biobert_en.html) | 9 | [ner_human_phenotype_go_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_human_phenotype_go_biobert_en.html) | 16 | [jsl_rd_ner_wip_greedy_biobert](https://nlp.johnsnowlabs.com/2021/07/26/jsl_rd_ner_wip_greedy_biobert_en.html) |\n| 3 | [ner_events_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_events_biobert_en.html) | 10 | [ner_deid_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_deid_biobert_en.html) | 17 | [ner_posology_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_posology_biobert_en.html) |\n| 4 | [ner_bionlp_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_bionlp_biobert_en.html) | 11 | [ner_deid_enriched_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_deid_enriched_biobert_en.html) | 18 | [jsl_ner_wip_greedy_biobert](https://nlp.johnsnowlabs.com/2021/07/26/jsl_ner_wip_greedy_biobert_en.html) |\n| 5 | [ner_jsl_greedy_biobert](https://nlp.johnsnowlabs.com/2021/08/13/ner_jsl_greedy_biobert_en.html) | 12 | [ner_clinical_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_clinical_biobert_en.html) | 19 | [ner_chemprot_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_chemprot_biobert_en.html) |\n| 6 | [ner_jsl_biobert](https://nlp.johnsnowlabs.com/2021/09/05/ner_jsl_biobert_en.html) | 13 | [ner_anatomy_coarse_biobert](https://nlp.johnsnowlabs.com/2021/03/31/ner_anatomy_coarse_biobert_en.html) | 20 | [ner_ade_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_ade_biobert_en.html) |\n| 7 | [ner_anatomy_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_anatomy_biobert_en.html) | 14 | [ner_human_phenotype_gene_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_human_phenotype_gene_biobert_en.html) | 21 | [ner_risk_factors_biobert](https://nlp.johnsnowlabs.com/2021/04/01/ner_risk_factors_biobert_en.html) |",
"_____no_output_____"
]
],
[
[
"from sparknlp.pretrained import PretrainedPipeline\n\nclinical_profiling_pipeline = PretrainedPipeline(\"ner_profiling_clinical\", \"en\", \"clinical/models\")",
"ner_profiling_clinical download started this may take some time.\nApprox size to download 2.3 GB\n[OK!]\n"
],
[
"text = '''A 28-year-old female with a history of gestational diabetes mellitus diagnosed eight years prior to presentation and subsequent type two diabetes mellitus ( T2DM ), one prior episode of HTG-induced pancreatitis three years prior to presentation , associated with an acute hepatitis , and obesity with a body mass index ( BMI ) of 33.5 kg/m2 , presented with a one-week history of polyuria , polydipsia , poor appetite , and vomiting .'''",
"_____no_output_____"
],
[
"clinical_result = clinical_profiling_pipeline.fullAnnotate(text)[0]\nclinical_result.keys()",
"_____no_output_____"
],
[
"import pandas as pd\n\ndef get_token_results(light_result):\n\n tokens = [j.result for j in light_result[\"token\"]]\n sentences = [j.metadata[\"sentence\"] for j in light_result[\"token\"]]\n begins = [j.begin for j in light_result[\"token\"]]\n ends = [j.end for j in light_result[\"token\"]]\n model_list = [ a for a in light_result.keys() if (a not in [\"sentence\", \"token\"] and \"_chunks\" not in a)]\n\n df = pd.DataFrame({'sentence':sentences, 'begin': begins, 'end': ends, 'token':tokens})\n\n for model_name in model_list:\n \n temp_df = pd.DataFrame(light_result[model_name])\n temp_df[\"jsl_label\"] = temp_df.iloc[:,0].apply(lambda x : x.result)\n temp_df = temp_df[[\"jsl_label\"]]\n\n # temp_df = get_ner_result(model_name)\n temp_df.columns = [model_name]\n df = pd.concat([df, temp_df], axis=1)\n \n return df",
"_____no_output_____"
],
[
"get_token_results(clinical_result)",
"_____no_output_____"
]
],
[
[
"## 6.NER Model Finder Pretrained Pipeline\n`ner_model_finder` pretrained pipeline trained with bert embeddings that can be used to find the most appropriate NER model given the entity name.",
"_____no_output_____"
]
],
[
[
"from sparknlp.pretrained import PretrainedPipeline\nfinder_pipeline = PretrainedPipeline(\"ner_model_finder\", \"en\", \"clinical/models\")",
"ner_model_finder download started this may take some time.\nApprox size to download 148.6 MB\n[OK!]\n"
],
[
"result = finder_pipeline.fullAnnotate(\"oncology\")[0]\nresult.keys()",
"_____no_output_____"
]
],
[
[
"From the metadata in the 'model_names' column, we'll get to the top models to the given 'oncology' entity and oncology related categories.",
"_____no_output_____"
]
],
[
[
"df= pd.DataFrame(zip(result[\"model_names\"][0].metadata[\"all_k_resolutions\"].split(\":::\"), \n result[\"model_names\"][0].metadata[\"all_k_results\"].split(\":::\")), columns=[\"category\", \"top_models\"])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"## 7.ICD10CM to Snomed Code\n\nThis pretrained pipeline maps ICD10CM codes to SNOMED codes without using any text data. You’ll just feed a comma or white space delimited ICD10CM codes and it will return the corresponding SNOMED codes as a list. For the time being, it supports 132K Snomed codes and will be augmented & enriched in the next releases.",
"_____no_output_____"
]
],
[
[
"icd_snomed_pipeline = PretrainedPipeline(\"icd10cm_snomed_mapping\", \"en\", \"clinical/models\")",
"icd10cm_snomed_mapping download started this may take some time.\nApprox size to download 514.5 KB\n[OK!]\n"
],
[
"icd_snomed_pipeline.model.stages",
"_____no_output_____"
],
[
"icd_snomed_pipeline.annotate('M89.50 I288 H16269')",
"_____no_output_____"
]
],
[
[
"|**ICD10CM** | **Details** | \n| ---------- | -----------:|\n| M89.50 | Osteolysis, unspecified site |\n| I288 | Other diseases of pulmonary vessels |\n| H16269 | Vernal keratoconjunctivitis, with limbar and corneal involvement, unspecified eye |\n\n| **SNOMED** | **Details** |\n| ---------- | -----------:|\n| 733187009 | Osteolysis following surgical procedure on skeletal system |\n| 449433008 | Diffuse stenosis of left pulmonary artery |\n| 51264003 | Limbal AND/OR corneal involvement in vernal conjunctivitis |",
"_____no_output_____"
],
[
"## 8.Snomed to ICD10CM Code\nThis pretrained pipeline maps SNOMED codes to ICD10CM codes without using any text data. You'll just feed a comma or white space delimited SNOMED codes and it will return the corresponding candidate ICD10CM codes as a list (multiple ICD10 codes for each Snomed code). For the time being, it supports 132K Snomed codes and 30K ICD10 codes and will be augmented & enriched in the next releases.",
"_____no_output_____"
]
],
[
[
"snomed_icd_pipeline = PretrainedPipeline(\"snomed_icd10cm_mapping\",\"en\",\"clinical/models\")",
"snomed_icd10cm_mapping download started this may take some time.\nApprox size to download 1.8 MB\n[OK!]\n"
],
[
"snomed_icd_pipeline.model.stages",
"_____no_output_____"
],
[
"snomed_icd_pipeline.annotate('733187009 449433008 51264003')",
"_____no_output_____"
]
],
[
[
"| **SNOMED** | **Details** |\n| ------ | ------:|\n| 733187009| Osteolysis following surgical procedure on skeletal system |\n| 449433008 | Diffuse stenosis of left pulmonary artery |\n| 51264003 | Limbal AND/OR corneal involvement in vernal conjunctivitis|\n\n| **ICDM10CM** | **Details** | \n| ---------- | ---------:|\n| M89.59 | Osteolysis, multiple sites | \n| M89.50 | Osteolysis, unspecified site |\n| M96.89 | Other intraoperative and postprocedural complications and disorders of the musculoskeletal system | \n| Q25.6 | Stenosis of pulmonary artery | \n| I28.8 | Other diseases of pulmonary vessels |\n| H10.45 | Other chronic allergic conjunctivitis |\n| H10.1 | Acute atopic conjunctivitis | \n| H16.269 | Vernal keratoconjunctivitis, with limbar and corneal involvement, unspecified eye |",
"_____no_output_____"
],
[
"Also you can find these healthcare code mapping pretrained pipelines here: [Healthcare_Codes_Mapping](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/Certification_Trainings/Healthcare/11.1.Healthcare_Code_Mapping.ipynb)\n\n- ICD10CM to UMLS \n- Snomed to UMLS \n- RxNorm to UMLS\n- RxNorm to MeSH\n- MeSH to UMLS\n- ICD10 to ICD9",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e71c9cb978851dd9e6f544158452e773fef1c9e4 | 313,568 | ipynb | Jupyter Notebook | notebooks/demoHMMlearn.ipynb | davidbarber/Julia0p6ProbabilisticInferenceEngine | df932959bf88b53fc076633167df4e497367e7fc | [
"MIT"
] | 30 | 2017-09-07T22:15:49.000Z | 2022-03-18T20:30:14.000Z | notebooks/demoHMMlearn.ipynb | davidbarber/Julia0p6ProbabilisticInferenceEngine | df932959bf88b53fc076633167df4e497367e7fc | [
"MIT"
] | null | null | null | notebooks/demoHMMlearn.ipynb | davidbarber/Julia0p6ProbabilisticInferenceEngine | df932959bf88b53fc076633167df4e497367e7fc | [
"MIT"
] | 7 | 2017-10-22T20:10:59.000Z | 2020-06-01T22:52:22.000Z | 737.807059 | 128,450 | 0.947829 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e71c9e92e7b27be67a59d08ff3cf3d4488f4746b | 16,289 | ipynb | Jupyter Notebook | pml1/figure_notebooks/chapter23_graph_embeddings_figures.ipynb | hanchenresearch/pml-book | 2c1b327b983bb7d9c57dd99491e24bd77b870d5a | [
"MIT"
] | 1 | 2021-08-30T23:47:45.000Z | 2021-08-30T23:47:45.000Z | pml1/figure_notebooks/chapter23_graph_embeddings_figures.ipynb | luaburto/pml-book | d28d516434f5fef847d1402ee3c39660c60815e1 | [
"MIT"
] | null | null | null | pml1/figure_notebooks/chapter23_graph_embeddings_figures.ipynb | luaburto/pml-book | d28d516434f5fef847d1402ee3c39660c60815e1 | [
"MIT"
] | 1 | 2021-11-05T20:05:38.000Z | 2021-11-05T20:05:38.000Z | 31.085878 | 532 | 0.583768 | [
[
[
"# Copyright 2021 Google LLC\n# Use of this source code is governed by an MIT-style\n# license that can be found in the LICENSE file or at\n# https://opensource.org/licenses/MIT.\n# Notebook authors: Kevin P. Murphy ([email protected])\n# and Mahmoud Soliman ([email protected])\n\n# This notebook reproduces figures for chapter 23 from the book\n# \"Probabilistic Machine Learning: An Introduction\"\n# by Kevin Murphy (MIT Press, 2021).\n# Book pdf is available from http://probml.ai",
"_____no_output_____"
]
],
[
[
"<a href=\"https://opensource.org/licenses/MIT\" target=\"_parent\"><img src=\"https://img.shields.io/github/license/probml/pyprobml\"/></a>",
"_____no_output_____"
],
[
"<a href=\"https://colab.research.google.com/github/probml/pml-book/blob/main/pml1/figure_notebooks/chapter23_graph_embeddings_figures.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"## Figure 23.1:<a name='23.1'></a> <a name='non_euclidean_vs_euclidean'></a> ",
"_____no_output_____"
],
[
"\n An illustration of Euclidean vs. non-Euclidean graphs. Used with permission from \\cite chami2020machine .\\rela",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.1_A.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.1_B.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.2:<a name='23.2'></a> <a name='enc-dec'></a> ",
"_____no_output_____"
],
[
"\n Illustration of the \\textsc GraphEDM framework from \\citet chami2020machine . Based on the supervision available, methods will use some or all of the branches. In particular, unsupervised methods do not leverage label decoding for training and only optimize the similarity decoder (lower branch). On the other hand, semi-supervised and supervised methods leverage the additional supervision to learn models' parameters (upper branch). Reprinted with permission from \\cite chami2020machine ",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.2.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.3:<a name='23.3'></a> <a name='shallow'></a> ",
"_____no_output_____"
],
[
"\n Shallow embedding methods. The encoder is a simple embedding look-up and the graph structure is only used in the loss function. Reprinted with permission from \\cite chami2020machine ",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.3.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.4:<a name='23.4'></a> <a name='walk'></a> ",
"_____no_output_____"
],
[
"\n An overview of the pipeline for random-walk graph embedding methods. Reprinted with permission from <a href='#godec_2018'>[Pri18]</a> .\\rela",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.4.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.5:<a name='23.5'></a> <a name='graphSage'></a> ",
"_____no_output_____"
],
[
"\n Illustration of the GraphSAGE model. Reprinted with permission from <a href='#hamilton2017inductive'>[WZJ17]</a> .\\rela",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.5.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.6:<a name='23.6'></a> <a name='hgcn_viz'></a> ",
"_____no_output_____"
],
[
"\n Euclidean (left) and hyperbolic (right) embeddings of a tree graph. Hyperbolic embeddings learn natural hierarchies in the embedding space (depth indicated by color). Reprinted with permission from <a href='#chami2019hyperbolic'>[Ine+19]</a> .\\rela",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.6_A.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.6_B.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.7:<a name='23.7'></a> <a name='agg_unsup'></a> ",
"_____no_output_____"
],
[
"\n Unsupervised graph neural networks. Graph structure and input features are mapped to low-dimensional embeddings using a graph neural network encoder. Embeddings are then decoded to compute a graph regularization loss (unsupervised). Reprinted with permission from \\cite chami2020machine ",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.7.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.8:<a name='23.8'></a> <a name='fraudGraph'></a> ",
"_____no_output_____"
],
[
"\n A graph representation of some financial transactions. Adapted from http://pgql-lang.org/spec/1.2/ ",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.8.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## Figure 23.9:<a name='23.9'></a> <a name='smell'></a> ",
"_____no_output_____"
],
[
"\n Structurally similar molecules do not necessarily have similar odor descriptors. (A) Lyral, the reference molecule. (B) Molecules with similar structure can share similar odor descriptors. (C) However, a small structural change can render the molecule odorless. (D) Further, large structural changes can leave the odor of the molecule largely unchanged. From Figure 1 of <a href='#SanchezLengeling2019'>[Ben+19]</a> , originally from <a href='#Ohloff2012'>[GWP12]</a> . Used with kind permission of Benjamin Sanchez-Lengeling",
"_____no_output_____"
]
],
[
[
"#@title Click me to run setup { display-mode: \"form\" }\ntry:\n if PYPROBML_SETUP_ALREADY_RUN:\n print('skipping setup')\nexcept:\n PYPROBML_SETUP_ALREADY_RUN = True\n print('running setup...')\n !git clone --depth 1 https://github.com/probml/pyprobml /pyprobml &> /dev/null \n %cd -q /pyprobml/scripts\n %reload_ext autoreload \n %autoreload 2\n !pip install superimport deimport -qqq\n import superimport\n from deimport.deimport import deimport\n print('finished!')",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/probml/pml-book/main/pml1/figures/images/Figure_23.9.png\" width=\"256\"/>",
"_____no_output_____"
],
[
"## References:\n <a name='SanchezLengeling2019'>[Ben+19]</a> S. Benjamin, W. JenniferN, L. K, G. RichardC, A. Al'an and W. AlexanderB. \"Machine Learning for Scent: Learning GeneralizablePerceptual Representations of Small Molecules\". abs/1910.10685 (2019). arXiv: 1910.10685 \n\n<a name='Ohloff2012'>[GWP12]</a> O. Gunther, P. Wilhelm and K. Philip. \"Scent and Chemistry\". (2012). \n\n<a name='chami2019hyperbolic'>[Ine+19]</a> C. Ines, Y. Zhitao, R. Christopher and L. Jure. \"Hyperbolic graph convolutional neural networks\". (2019). \n\n<a name='godec_2018'>[Pri18]</a> G. Primož \"Graph Embeddings; The Summary\". (2018). \n\n<a name='hamilton2017inductive'>[WZJ17]</a> H. Will, Y. Zhitao and L. Jure. \"Inductive representation learning on large graphs\". (2017). \n\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e71ca57ca09a08b6a97cb848b090259e47273023 | 588,059 | ipynb | Jupyter Notebook | 06_NormalizationRegularization/06_NormalizationRegularization_CRUDE.ipynb | Septank766/TSAI-DeepVision-EVA4.0 | 02265d7e3e06789d0ee634a38399c6f0e01cfcbd | [
"MIT"
] | 22 | 2020-05-16T08:15:48.000Z | 2021-12-30T14:38:31.000Z | 06_NormalizationRegularization/06_NormalizationRegularization_CRUDE.ipynb | Septank766/TSAI-DeepVision-EVA4.0 | 02265d7e3e06789d0ee634a38399c6f0e01cfcbd | [
"MIT"
] | 1 | 2020-09-07T17:10:41.000Z | 2020-09-09T20:51:31.000Z | 06_NormalizationRegularization/06_NormalizationRegularization_CRUDE.ipynb | Septank766/TSAI-DeepVision-EVA4.0 | 02265d7e3e06789d0ee634a38399c6f0e01cfcbd | [
"MIT"
] | 43 | 2020-03-07T22:08:41.000Z | 2022-03-16T21:07:30.000Z | 52.234766 | 76,666 | 0.628667 | [
[
[
"<a href=\"https://colab.research.google.com/github/satyajitghana/TSAI-DeepVision-EVA4.0/blob/master/06_NormalizationRegularization/06_NormalizationRegularization_CRUDE.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Normalization & Regularization",
"_____no_output_____"
],
[
"## 1. Target\n\n- Fine Tune the Transforms, set rotation to `-10deg to 10deg`\n- Use the OneCycleLR Scheduler since the PyTorch Documentation says\n\n> Sets the learning rate of each parameter group according to the 1cycle learning rate policy. The 1cycle policy anneals the learning rate from an initial learning rate to some maximum learning rate and then from that maximum learning rate to some minimum learning rate much lower than the initial learning rate. This policy was initially described in the paper **Super-Convergence: Very Fast Training of Neural Networks Using Large Learning Rates.**\n\n> The 1cycle learning rate policy changes the learning rate after every batch. step should be called after a batch has been used for training.\n\n## 2. Result\n\n- Params: `9,962`\n- Best Train Accuracy: `98.85%`\n- Best Test Accuracy : `99.31%`\n\n## 3. Analysis\n\n- Now the learning is consistent, the accuracy increases",
"_____no_output_____"
],
[
"## Layer Added Model\n\n- Convolution Block 1\n - `Conv2D: 1, 14, 3; Out: 26x26x14`\n - `Conv2D: 14, 30, 3; Out: 24x24x30`\n\n- Transition Block\n - `Conv2D: 30, 10, 1; Out: 24x24x10`\n - `MaxPool2D: 2x2; Out: 12x12x10`\n\n- Convolution Block 2\n - `Conv2D: 10, 14, 3; Out: 10x10x14`\n - `Conv2D: 14, 15, 3; Out: 8x8x15`\n - `Conv2D: 15, 15, 3; Out: 6x6x15`\n\n- GAP\n - `AvgPool2D: 6x6; Out: 1x1x15`\n - `Conv2D: 15, 15, 1; Out: 1x1x15`\n - `Conv2D: 15, 10, 1; Out: 1x1x10`\n\n\nLegend\n- `Conv2D (input_channels), (output_channels), (kernel_size); Out: (output_dim)`\n- `MaxPool2D: (kernel_size); Out: (output_dim)`\n- `AvgPool2D: (kernel_size); Out: (output_dim)`",
"_____no_output_____"
],
[
"# Import Libraries",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nfrom torchvision import datasets, transforms",
"_____no_output_____"
]
],
[
[
"## Data Transformations\n\nWe first start with defining our data transformations. We need to think what our data is and how can we augment it to correct represent images which it might not see otherwise. \n",
"_____no_output_____"
]
],
[
[
"# Train Phase transformations\ntrain_transforms = transforms.Compose([\n transforms.RandomRotation((-10.0, 10.0), fill=(1,)),\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,)) # The mean and std have to be sequences (e.g., tuples), therefore you should add a comma after the values. \n # Note the difference between (0.1307) and (0.1307,)\n ])\n\n# Test Phase transformations\ntest_transforms = transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))\n ])\n",
"_____no_output_____"
]
],
[
[
"# Dataset and Creating Train/Test Split",
"_____no_output_____"
]
],
[
[
"train = datasets.MNIST('./data', train=True, download=True, transform=train_transforms)\ntest = datasets.MNIST('./data', train=False, download=True, transform=test_transforms)",
"Downloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz to ./data/MNIST/raw/train-images-idx3-ubyte.gz\n"
]
],
[
[
"# Dataloader Arguments & Test/Train Dataloaders\n",
"_____no_output_____"
]
],
[
[
"SEED = 1\n\n# CUDA?\ncuda = torch.cuda.is_available()\nprint(\"CUDA Available?\", cuda)\n\n# For reproducibility\ntorch.manual_seed(SEED)\n\nif cuda:\n torch.cuda.manual_seed(SEED)\n\n# note about pin_memory\n# If you load your samples in the Dataset on CPU and would like to push it\n# during training to the GPU, you can speed up the host to device transfer by\n# enabling pin_memory. This lets your DataLoader allocate the samples in\n# page-locked memory, which speeds-up the transfer.\n\n# dataloader arguments - something you'll fetch these from cmdprmt\ndataloader_args = dict(shuffle=True, batch_size=128, num_workers=4, pin_memory=True) if cuda else dict(shuffle=True, batch_size=64)\n\n# train dataloader\ntrain_loader = torch.utils.data.DataLoader(train, **dataloader_args)\n\n# test dataloader\ntest_loader = torch.utils.data.DataLoader(test, **dataloader_args)",
"CUDA Available? True\n"
]
],
[
[
"# The model\nLet's start with the model we first saw",
"_____no_output_____"
]
],
[
[
"import torch.nn.functional as F\n\ndropout_value = 0.08\n\nclass Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n # Input Block\n self.convblock1 = nn.Sequential(\n nn.Conv2d(in_channels=1, out_channels=14, kernel_size=(3, 3), padding=0, bias=False),\n nn.BatchNorm2d(14),\n nn.ReLU(),\n nn.Dropout(dropout_value)\n ) # output_size = 26\n\n # CONVOLUTION BLOCK 1\n self.convblock2 = nn.Sequential(\n nn.Conv2d(in_channels=14, out_channels=30, kernel_size=(3, 3), padding=0, bias=False),\n nn.BatchNorm2d(30),\n nn.ReLU(),\n nn.Dropout(dropout_value)\n ) # output_size = 24\n\n # TRANSITION BLOCK 1\n self.convblock3 = nn.Sequential(\n nn.Conv2d(in_channels=30, out_channels=10, kernel_size=(1, 1), padding=0, bias=False),\n ) # output_size = 24\n self.pool1 = nn.MaxPool2d(2, 2) # output_size = 12\n\n # CONVOLUTION BLOCK 2\n self.convblock4 = nn.Sequential(\n nn.Conv2d(in_channels=10, out_channels=14, kernel_size=(3, 3), padding=0, bias=False),\n nn.BatchNorm2d(14),\n nn.ReLU(), \n nn.Dropout(dropout_value)\n ) # output_size = 10\n self.convblock5 = nn.Sequential(\n nn.Conv2d(in_channels=14, out_channels=15, kernel_size=(3, 3), padding=0, bias=False),\n nn.BatchNorm2d(15),\n nn.ReLU(), \n nn.Dropout(dropout_value)\n ) # output_size = 8\n self.convblock6 = nn.Sequential(\n nn.Conv2d(in_channels=15, out_channels=15, kernel_size=(3, 3), padding=0, bias=False),\n nn.BatchNorm2d(15),\n nn.ReLU(), \n nn.Dropout(dropout_value)\n ) # output_size = 6\n \n # OUTPUT BLOCK\n self.gap = nn.Sequential(\n nn.AvgPool2d(kernel_size=6)\n ) # output_size = 1\n\n self.convblock7 = nn.Sequential(\n nn.Conv2d(in_channels=15, out_channels=15, kernel_size=(1, 1), padding=0, bias=False),\n nn.ReLU(),\n nn.BatchNorm2d(15),\n nn.Dropout(dropout_value)\n )\n\n self.convblock8 = nn.Sequential(\n nn.Conv2d(in_channels=15, out_channels=10, kernel_size=(1, 1), padding=0, bias=False),\n )\n\n self.dropout = nn.Dropout(dropout_value)\n\n def forward(self, x):\n x = self.convblock1(x)\n x = self.convblock2(x)\n x = self.convblock3(x)\n x = self.pool1(x)\n x = self.convblock4(x)\n x = self.convblock5(x)\n x = self.convblock6(x)\n x = self.gap(x) \n x = self.convblock7(x)\n x = self.convblock8(x)\n\n x = x.view(-1, 10)\n return F.log_softmax(x, dim=-1)",
"_____no_output_____"
]
],
[
[
"# Model Params\nCan't emphasize on how important viewing Model Summary is. \nUnfortunately, there is no in-built model visualizer, so we have to take external help",
"_____no_output_____"
]
],
[
[
"!pip install torchsummary\nfrom torchsummary import summary\nuse_cuda = torch.cuda.is_available()\ndevice = torch.device(\"cuda\" if use_cuda else \"cpu\")\nprint(device)\nmodel = Net().to(device)\nsummary(model, input_size=(1, 28, 28))",
"Requirement already satisfied: torchsummary in /usr/local/lib/python3.6/dist-packages (1.5.1)\ncuda\n----------------------------------------------------------------\n Layer (type) Output Shape Param #\n================================================================\n Conv2d-1 [-1, 14, 26, 26] 126\n BatchNorm2d-2 [-1, 14, 26, 26] 28\n ReLU-3 [-1, 14, 26, 26] 0\n Dropout-4 [-1, 14, 26, 26] 0\n Conv2d-5 [-1, 30, 24, 24] 3,780\n BatchNorm2d-6 [-1, 30, 24, 24] 60\n ReLU-7 [-1, 30, 24, 24] 0\n Dropout-8 [-1, 30, 24, 24] 0\n Conv2d-9 [-1, 10, 24, 24] 300\n MaxPool2d-10 [-1, 10, 12, 12] 0\n Conv2d-11 [-1, 14, 10, 10] 1,260\n BatchNorm2d-12 [-1, 14, 10, 10] 28\n ReLU-13 [-1, 14, 10, 10] 0\n Dropout-14 [-1, 14, 10, 10] 0\n Conv2d-15 [-1, 15, 8, 8] 1,890\n BatchNorm2d-16 [-1, 15, 8, 8] 30\n ReLU-17 [-1, 15, 8, 8] 0\n Dropout-18 [-1, 15, 8, 8] 0\n Conv2d-19 [-1, 15, 6, 6] 2,025\n BatchNorm2d-20 [-1, 15, 6, 6] 30\n ReLU-21 [-1, 15, 6, 6] 0\n Dropout-22 [-1, 15, 6, 6] 0\n AvgPool2d-23 [-1, 15, 1, 1] 0\n Conv2d-24 [-1, 15, 1, 1] 225\n ReLU-25 [-1, 15, 1, 1] 0\n BatchNorm2d-26 [-1, 15, 1, 1] 30\n Dropout-27 [-1, 15, 1, 1] 0\n Conv2d-28 [-1, 10, 1, 1] 150\n================================================================\nTotal params: 9,962\nTrainable params: 9,962\nNon-trainable params: 0\n----------------------------------------------------------------\nInput size (MB): 0.00\nForward/backward pass size (MB): 0.96\nParams size (MB): 0.04\nEstimated Total Size (MB): 1.00\n----------------------------------------------------------------\n"
]
],
[
[
"# Training and Testing\n\nLooking at logs can be boring, so we'll introduce **tqdm** progressbar to get cooler logs. \n\nLet's write train and test functions",
"_____no_output_____"
]
],
[
[
"# this automatically selects tqdm for colab_notebook\nfrom tqdm.auto import tqdm, trange\n\ntrain_losses = []\ntest_losses = []\ntrain_acc = []\ntest_acc = []\n\ndef train(model, device, train_loader, optimizer, epoch):\n model.train()\n pbar = tqdm(train_loader, ncols=\"80%\")\n correct = 0\n processed = 0\n for batch_idx, (data, target) in enumerate(pbar):\n # get samples\n data, target = data.to(device), target.to(device)\n\n # Init\n optimizer.zero_grad()\n # In PyTorch, we need to set the gradients to zero before starting to do \n # backpropragation because PyTorch accumulates the gradients on subsequent \n # backward passes. \n # Because of this, when you start your training loop, ideally you should \n # zero out the gradients so that you do the parameter update correctly.\n\n # Predict\n y_pred = model(data)\n\n # Calculate loss\n loss = F.nll_loss(y_pred, target)\n train_losses.append(loss)\n\n # Backpropagation\n loss.backward()\n optimizer.step()\n\n # Update the Learning Rate\n scheduler.step()\n \n # get the index of the max log-probability\n pred = y_pred.argmax(dim=1, keepdim=True)\n correct += pred.eq(target.view_as(pred)).sum().item()\n processed += len(data)\n\n # Update pbar-tqdm\n pbar.set_description(desc= f'Loss={loss.item()} Batch_id={batch_idx} Accuracy={100*correct/processed:0.2f}')\n train_acc.append(100*correct/processed)\n\ndef test(model, device, test_loader):\n model.eval()\n test_loss = 0\n correct = 0\n with torch.no_grad():\n for data, target in test_loader:\n data, target = data.to(device), target.to(device)\n output = model(data)\n # sum up batch loss\n test_loss += F.nll_loss(output, target, reduction='sum').item()\n # get the index of the max log-probability\n pred = output.argmax(dim=1, keepdim=True)\n correct += pred.eq(target.view_as(pred)).sum().item()\n\n test_loss /= len(test_loader.dataset)\n test_losses.append(test_loss)\n\n print('\\nTest set: Average loss: {:.4f}, Accuracy: {}/{} ({:.2f}%)\\n'.format(\n test_loss, correct, len(test_loader.dataset),\n 100. * correct / len(test_loader.dataset)))\n \n test_acc.append(100. * correct / len(test_loader.dataset))",
"_____no_output_____"
],
[
"from torch.optim.lr_scheduler import OneCycleLR\n\nmodel = Net().to(device)\n\nEPOCHS = 15\n\nlr_values = []\n\noptimizer = optim.SGD(model.parameters(), lr=0.01, momentum=0.95)\nscheduler = OneCycleLR(optimizer, max_lr=0.01, steps_per_epoch=len(train_loader), epochs=EPOCHS)\n\nfor epoch in range(EPOCHS):\n # store the current LR value\n lr_values.append(scheduler.get_lr())\n # Print Learning Rate\n print('EPOCH:', epoch+1, 'LR:', scheduler.get_lr())\n # Train the model\n train(model, device, train_loader, optimizer, epoch)\n # Validate the model\n test(model, device, test_loader)",
"EPOCH: 1 LR: [0.0003999999999999993]\n"
]
],
[
[
"# Plot the model's learning progress\n",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nimport matplotlib.pyplot as plt\nsns.set()\nplt.style.use(\"dark_background\")\n\nfig, axs = plt.subplots(3,2,figsize=(18,15))\naxs[0, 0].plot(train_losses)\naxs[0, 0].set_title(\"Training Loss\")\naxs[1, 0].plot(train_acc[4000:])\naxs[1, 0].set_title(\"Training Accuracy\")\naxs[0, 1].plot(test_losses)\naxs[0, 1].set_title(\"Test Loss\")\naxs[1, 1].plot(test_acc)\naxs[1, 1].set_title(\"Test Accuracy\")\naxs[2, 0].plot(lr_values)\naxs[2, 0].set_title(\"Learning Rate\")\nplt.show()",
"_____no_output_____"
],
[
"def get_misclassified(model, device, test_loader):\n misclassified = []\n misclassified_pred = []\n misclassified_target = []\n # put the model to evaluation mode\n model.eval()\n # turn off gradients\n with torch.no_grad():\n for data, target in test_loader:\n # move them to the respective device\n data, target = data.to(device), target.to(device)\n # do inferencing\n output = model(data)\n # get the predicted output\n pred = output.argmax(dim=1, keepdim=True)\n\n # get the current misclassified in this batch\n list_misclassified = (pred.eq(target.view_as(pred)) == False)\n batch_misclassified = data[list_misclassified]\n batch_mis_pred = pred[list_misclassified]\n batch_mis_target = target.view_as(pred)[list_misclassified]\n\n # batch_misclassified =\n\n misclassified.append(batch_misclassified)\n misclassified_pred.append(batch_mis_pred)\n misclassified_target.append(batch_mis_target)\n\n # group all the batched together\n misclassified = torch.cat(misclassified)\n misclassified_pred = torch.cat(misclassified_pred)\n misclassified_target = torch.cat(misclassified_target)\n\n return list(map(lambda x, y, z: (x, y, z), misclassified, misclassified_pred, misclassified_target))",
"_____no_output_____"
],
[
"misclassified = get_misclassified(model, device, test_loader)",
"_____no_output_____"
]
],
[
[
"## Misclassifications Plot",
"_____no_output_____"
],
[
"First 25 misclassifications",
"_____no_output_____"
]
],
[
[
"num_images = 25\nfig = plt.figure(figsize=(12, 12))\nfor idx, (image, pred, target) in enumerate(misclassified[:num_images]):\n image, pred, target = image.cpu().numpy(), pred.cpu(), target.cpu()\n ax = fig.add_subplot(5, 5, idx+1)\n ax.axis('off')\n ax.set_title('target {}\\npred {}'.format(target.item(), pred.item()), fontsize=12)\n ax.imshow(image.squeeze())\nplt.show()",
"_____no_output_____"
]
],
[
[
"Chosen Randomly 25 from the misclassifications",
"_____no_output_____"
]
],
[
[
"import random\n\nnum_images = 25\nfig = plt.figure(figsize=(12, 12))\nfor idx, (image, pred, target) in enumerate(random.choices(misclassified, k=num_images)):\n image, pred, target = image.cpu().numpy(), pred.cpu(), target.cpu()\n ax = fig.add_subplot(5, 5, idx+1)\n ax.axis('off')\n ax.set_title('target {}\\npred {}'.format(target.item(), pred.item()), fontsize=12)\n ax.imshow(image.squeeze())\nplt.show()",
"_____no_output_____"
]
],
[
[
"# L1 Loss",
"_____no_output_____"
]
],
[
[
"from tqdm.auto import tqdm, trange\n\ntrain_losses = []\ntest_losses = []\ntrain_acc = []\ntest_acc = []\n\nLAMBDA = 5e-4\n\ndef train(model, device, train_loader, optimizer, epoch):\n model.train()\n pbar = tqdm(train_loader, ncols=\"80%\")\n correct = 0\n processed = 0\n for batch_idx, (data, target) in enumerate(pbar):\n\n data, target = data.to(device), target.to(device) # get samples\n\n optimizer.zero_grad() # init\n\n # Predict\n y_pred = model(data) # predict\n\n # calculate losses\n nll_loss = F.nll_loss(y_pred, target)\n reg_loss = sum([torch.sum(abs(param)) for param in model.parameters()])\n\n # L1 Loss\n loss = nll_loss + LAMBDA * reg_loss\n \n train_losses.append(loss)\n print(type(loss))\n\n # Backpropagation\n loss.backward()\n optimizer.step()\n\n # Update the Learning Rate\n scheduler.step()\n \n # get the index of the max log-probability\n pred = y_pred.argmax(dim=1, keepdim=True)\n correct += pred.eq(target.view_as(pred)).sum().item()\n processed += len(data)\n\n # Update pbar-tqdm\n pbar.set_description(desc= f'Loss={loss.item()} Batch_id={batch_idx} Accuracy={100*correct/processed:0.2f}')\n train_acc.append(100*correct/processed)\n\ndef test(model, device, test_loader):\n model.eval()\n test_loss = 0\n correct = 0\n with torch.no_grad():\n for data, target in test_loader:\n data, target = data.to(device), target.to(device)\n output = model(data)\n # sum up batch loss\n test_loss += F.nll_loss(output, target, reduction='sum').item()\n # get the index of the max log-probability\n pred = output.argmax(dim=1, keepdim=True)\n correct += pred.eq(target.view_as(pred)).sum().item()\n\n test_loss /= len(test_loader.dataset)\n test_losses.append(test_loss)\n\n print('\\nTest set: Average loss: {:.4f}, Accuracy: {}/{} ({:.2f}%)\\n'.format(\n test_loss, correct, len(test_loader.dataset),\n 100. * correct / len(test_loader.dataset)))\n \n test_acc.append(100. * correct / len(test_loader.dataset))",
"_____no_output_____"
],
[
"from torch.optim.lr_scheduler import OneCycleLR\n\nmodel = Net().to(device)\n\nEPOCHS = 15\n\nlr_values = []\n\noptimizer = optim.SGD(model.parameters(), lr=0.01, momentum=0.95)\nscheduler = OneCycleLR(optimizer, max_lr=0.01, steps_per_epoch=len(train_loader), epochs=EPOCHS)\n\nfor epoch in range(EPOCHS):\n # store the current LR value\n lr_values.append(scheduler.get_lr())\n # Print Learning Rate\n print('EPOCH:', epoch+1, 'LR:', scheduler.get_lr())\n # Train the model\n train(model, device, train_loader, optimizer, epoch)\n # Validate the model\n test(model, device, test_loader)",
"EPOCH: 1 LR: [0.0003999999999999993]\n"
]
],
[
[
"# L2 Regularization",
"_____no_output_____"
]
],
[
[
"from torch.optim.lr_scheduler import OneCycleLR\n\nmodel = Net().to(device)\n\nEPOCHS = 15\n\nlr_values = []\n\noptimizer = optim.SGD(model.parameters(), lr=0.01, momentum=0.95, weight_decay=5e-3)\nscheduler = OneCycleLR(optimizer, max_lr=0.01, steps_per_epoch=len(train_loader), epochs=EPOCHS)\n\nfor epoch in range(EPOCHS):\n # store the current LR value\n lr_values.append(scheduler.get_lr())\n # Print Learning Rate\n print('EPOCH:', epoch+1, 'LR:', scheduler.get_lr())\n # Train the model\n train(model, device, train_loader, optimizer, epoch)\n # Validate the model\n test(model, device, test_loader)",
"EPOCH: 1 LR: [0.0003999999999999993]\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e71cb04a9572f5a8d0f2f4589ea505738f8ca7ee | 232,490 | ipynb | Jupyter Notebook | experiments/100_KNN/analysis/Analysis.ipynb | nathancfox/cfs-reu-2018 | dac4e7b795211c4f7c0b81777ef86a6ae8890cdd | [
"MIT"
] | 1 | 2020-03-26T02:39:48.000Z | 2020-03-26T02:39:48.000Z | experiments/100_KNN/analysis/Analysis.ipynb | nathancfox/cfs-reu-2018 | dac4e7b795211c4f7c0b81777ef86a6ae8890cdd | [
"MIT"
] | null | null | null | experiments/100_KNN/analysis/Analysis.ipynb | nathancfox/cfs-reu-2018 | dac4e7b795211c4f7c0b81777ef86a6ae8890cdd | [
"MIT"
] | null | null | null | 39.172704 | 37,592 | 0.491131 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy import stats",
"_____no_output_____"
],
[
"max = 0\nwith open('reported_kinases.csv', 'r') as f:\n for line in f:\n l = len(line.split(','))\n if l > max:\n max = l",
"_____no_output_____"
],
[
"max",
"_____no_output_____"
],
[
"kin_df = pd.read_csv('reported_kinases.csv', header=None, names=range(36))",
"_____no_output_____"
],
[
"kin_df",
"_____no_output_____"
],
[
"kin = []\nwith open('reported_kinases.csv', 'r') as f:\n for line in f:\n kin.append(line.strip().split(','))",
"_____no_output_____"
],
[
"kin",
"_____no_output_____"
],
[
"k_cnts = {}\nfor l in kin:\n for k in l:\n if k in k_cnts:\n k_cnts[k] += 1\n else:\n k_cnts[k] = 1",
"_____no_output_____"
],
[
"k_cnts",
"_____no_output_____"
],
[
"kin_cnt_ser = pd.Series(list(k_cnts.values()), index=k_cnts.keys())",
"_____no_output_____"
],
[
"kin_cnt_ser.sort_values(ascending=False, inplace=True)",
"_____no_output_____"
],
[
"kin_cnt_ser",
"_____no_output_____"
],
[
"groups = []\nwith open('linked_groups.csv', 'r') as f:\n for line in f:\n groups.append(line.strip().split(','))\nfor i in range(len(groups)):\n groups[i] = set(groups[i])",
"_____no_output_____"
],
[
"groups",
"_____no_output_____"
],
[
"sum = 0\nfor g in groups:\n sum += len(g)\nprint(sum)\nprint(190-sum)",
"46\n144\n"
],
[
"new_col = [-2 for i in range(len(kin_cnt_ser.index))]\nfor i in range(len(kin_cnt_ser.index)):\n for j in range(len(groups)):\n if kin_cnt_ser.index[i] in groups[j]:\n new_col[i] = j\n continue\n if new_col[i] == -2:\n new_col[i] = -1",
"_____no_output_____"
],
[
"-2 in new_col",
"_____no_output_____"
],
[
"kin_cnt_df = pd.DataFrame({'Count': kin_cnt_ser, 'Group': new_col})",
"_____no_output_____"
],
[
"kin_cnt_df",
"_____no_output_____"
],
[
"# Testing my code to make sure it does what I want on toy data\nresults = np.zeros(24).reshape((6, 4)).astype(bool)\na = [{1, 2, 3}, {4, 5}, {6, 7}]\nb = [[1, 2], [1, 4], [3, 4, 5, 6], [4, 6], [4, 23], [23, 56, 48]]\nfor i in range(len(b)):\n for j in range(len(b[i])):\n for k in range(len(a)):\n if b[i][j] in a[k]:\n results[i, k] = True\n break\n else:\n results[i, -1] = True\nresults",
"_____no_output_____"
],
[
"in_groups = np.zeros(1600).reshape((100, 16)).astype(bool)",
"_____no_output_____"
],
[
"for i in range(len(kin)):\n for j in range(len(kin[i])):\n for k in range(len(groups)):\n if kin[i][j] in groups[k]:\n in_groups[i, k] = True\n break\n else:\n in_groups[i, -1] = True",
"_____no_output_____"
],
[
"group_representation = np.zeros(16)\nfor i in range(in_groups.shape[0]):\n for j in range(in_groups.shape[1]):\n if in_groups[i, j]:\n group_representation[j] += 1",
"_____no_output_____"
],
[
"group_representation",
"_____no_output_____"
],
[
"group_frequencies = group_representation / np.array([len(g) for g in groups] + [144])",
"_____no_output_____"
],
[
"group_frequencies",
"_____no_output_____"
],
[
"plt.style.use('ggplot')\nlabels = [str(i+1) for i in range(15)] + ['N/A']\nfig, ax = plt.subplots()\nfig.set_size_inches((12, 8.25))\nax.bar([i+1 for i in range(16)], group_frequencies, tick_label=labels, align='center', color='#2f8cb5')\nax.set_xlabel('Group Number', size=14)\nax.set_xlim((0, 17))\nax.set_ylabel('Frequency of Inclusion (Normalized to Group Size)', size=14)\nax.set_title('Frequency of Inclusion of At Least 1 Member of Each Group',\n size='20')\nax.tick_params(axis='both', pad=8, labelsize=12)\nfig.set_facecolor('#ffffff')\nplt.savefig('figures/Frequency_of_Inclusion_Normalized.png', dpi=300, format='png',\n bbox_inches='tight')\nplt.show()",
"_____no_output_____"
],
[
"print(plt.style.available)",
"['classic', 'Solarize_Light2', 'grayscale', 'fast', 'fivethirtyeight', 'seaborn-dark-palette', 'seaborn-whitegrid', 'seaborn-dark', 'seaborn-colorblind', '_classic_test', 'ggplot', 'seaborn-talk', 'bmh', 'seaborn-darkgrid', 'seaborn-notebook', 'dark_background', 'seaborn-muted', 'seaborn', 'seaborn-pastel', 'seaborn-poster', 'seaborn-paper', 'seaborn-ticks', 'seaborn-bright', 'seaborn-deep', 'seaborn-white']\n"
],
[
"for s in plt.style.available:\n plt.style.use(s)\n labels = [str(i+1) for i in range(15)] + ['N/A']\n fig, ax = plt.subplots()\n fig.set_size_inches((12, 8.25))\n ax.bar([i for i in range(16)], group_frequencies, tick_label=labels)\n ax.set_xlabel('Group Number')\n ax.set_ylabel('Frequency of Inclusion (Normalized to Group Size)')\n ax.set_title(s)\n plt.savefig('test_styles/{}.png'.format(s))\n plt.close()",
"_____no_output_____"
],
[
"print(groups[1])\nprint(groups[11])\nprint(groups[13])",
"{'ROCK2', 'ROCK1'}\n{'MUSK'}\n{'ERBB2', 'EGFR', 'ERBB4'}\n"
],
[
"rock = 0\ndouble_rock = 0\nfor g in kin:\n if 'ROCK1' in g:\n rock += 1\n if 'ROCK2' in g:\n double_rock += 1\n continue\n elif 'ROCK2' in g:\n rock += 1\nprint(rock)\nprint(double_rock)",
"83\n0\n"
],
[
"{'ERBB2', 'ERBB4', 'EGFR'}\ne_count = [0, 0, 0]\nfor g in kin:\n flag = 0\n if 'ERBB2' in g:\n flag += 1\n if 'ERBB4' in g:\n flag += 1\n if 'EGFR' in g:\n flag += 1\n if flag > 0:\n e_count[flag - 1] += 1\ne_count",
"_____no_output_____"
],
[
"high_kin_cnt_df = kin_cnt_df[kin_cnt_df['Count'] >= 20]\nhigh_kin_cnt_df.shape",
"_____no_output_____"
],
[
"scores = pd.read_csv('numerical_data.csv')",
"_____no_output_____"
],
[
"scores.head()",
"_____no_output_____"
],
[
"scores.drop(['Unnamed: 0', 'g_fitness', 'g_accuracy', 'g_sensitivity', 'g_specificity'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"scores.head()",
"_____no_output_____"
],
[
"scores.describe()",
"_____no_output_____"
],
[
"acc = [75.1111, 79.3846, 85.564, 82.3111, 85.9777, 83.3386, 81.9413,79.7157, 82.0088, 82.2484]\nsen = [57, 71.9524, 86.6667, 75.8095, 84.2857, 78.6667, 72.381, 78.0952, 72.6667, 66.9524]\nspe = [80.7143, 82.8849, 84.4286, 84.3636, 86.619, 85.119, 86.9381, 81.7932, 84.7179, 86.2098]",
"_____no_output_____"
],
[
"t, p = stats.ttest_ind(acc, scores['a_accuracy'])\nprint('Accuracy\\n'+'-'*40)\nprint(' t: {}'.format(t))\nprint(' p: {}'.format(p))\nprint()\nt, p = stats.ttest_ind(sen, scores['a_sensitivity'])\nprint('Sensitivity\\n'+'-'*40)\nprint(' t: {}'.format(t))\nprint(' p: {}'.format(p))\nprint()\nt, p = stats.ttest_ind(spe, scores['a_specificity'])\nprint('Specificity\\n'+'-'*40)\nprint(' t: {}'.format(t))\nprint(' p: {}'.format(p))",
"Accuracy\n----------------------------------------\n t: 268.1387353105542\n p: 2.4625917909424514e-154\n\nSensitivity\n----------------------------------------\n t: 90.47780862558577\n p: 1.196687544670363e-103\n\nSpecificity\n----------------------------------------\n t: 424.2628928356859\n p: 7.772867889759253e-176\n"
],
[
"control_scores = pd.read_csv('control_scores.csv')\ncontrol_scores.head()",
"_____no_output_____"
],
[
"control_scores.describe()",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 10))\nax_acc = fig.add_subplot(311)\nax_acc.set_title('Scores')\nax_acc.errorbar([1, 2], [control_scores['accuracy'].mean(), scores['a_accuracy'].mean()], yerr = [control_scores['accuracy'].std(), scores['a_accuracy'].std()], linestyle='None', marker='o', markerfacecolor='r', ms=10, ecolor='k', capsize=10)\nax_acc.set_xlim((0.5, 2.5))\nax_acc.set_xticks([1, 2])\nax_acc.set_xticklabels(['', ''])\nax_acc.set_ylabel('Accuracy')\nax_sen = fig.add_subplot(312, sharex=ax_acc)\nax_sen.errorbar([1, 2], [control_scores['sensitivity'].mean(), scores['a_sensitivity'].mean()], yerr = [control_scores['sensitivity'].std(), scores['a_sensitivity'].std()], linestyle='None', marker='o', markerfacecolor='g', ms=10, ecolor='k', capsize=10)\nax_sen.set_xlim((0.5, 2.5))\nax_sen.set_xticks([1, 2])\nax_sen.set_xticklabels(['', ''])\nax_sen.set_ylabel('Sensitivity')\nax_spe = fig.add_subplot(313, sharex=ax_acc)\nax_spe.errorbar([1, 2], [control_scores['specificity'].mean(), scores['a_specificity'].mean()], yerr = [control_scores['specificity'].std(), scores['a_specificity'].std()], linestyle='None', marker='o', markerfacecolor='b', ms=10, ecolor='k', capsize=10)\nax_spe.set_xlim((0.5, 2.5))\nax_spe.set_xticks([1, 2])\nax_spe.set_ylabel('Specificity')\nax_spe.set_xticklabels(['Control', 'Experimental'])\nplt.savefig('figures/scores.png', dpi=300, format='png')\nplt.show()",
"_____no_output_____"
],
[
"t, p = stats.ttest_ind(control_scores['accuracy'], scores['a_accuracy'])\nprint('Accuracy\\n'+'-'*40)\nprint(' t: {}'.format(t))\nprint(' p: {}'.format(p))\nprint()\nt, p = stats.ttest_ind(control_scores['sensitivity'], scores['a_sensitivity'])\nprint('Sensitivity\\n'+'-'*40)\nprint(' t: {}'.format(t))\nprint(' p: {}'.format(p))\nprint()\nt, p = stats.ttest_ind(control_scores['specificity'], scores['a_specificity'])\nprint('Specificity\\n'+'-'*40)\nprint(' t: {}'.format(t))\nprint(' p: {}'.format(p))",
"Accuracy\n----------------------------------------\n t: -20.431367063427658\n p: 1.1932881481787495e-50\n\nSensitivity\n----------------------------------------\n t: -66.9322207524752\n p: 6.265591187632975e-138\n\nSpecificity\n----------------------------------------\n t: 12.025481361671337\n p: 2.3004228357067834e-25\n"
],
[
"best_runs = scores.nlargest(10, 'a_sensitivity')\nbest_runs",
"_____no_output_____"
],
[
"kin[76]",
"_____no_output_____"
],
[
"kin_cnt_df.loc[kin[76]]",
"_____no_output_____"
],
[
"kin_cnt_ser.head(10)",
"_____no_output_____"
],
[
"with open('Rational_Polypharm_Kinase_MAXIS_Frequencies.csv', 'r') as f:\n kinases = next(f).strip().split(',')\n freqs = next(f).strip().split(',')\nmaxis_kin_cnt = pd.Series(freqs, index=kinases, dtype=int)",
"_____no_output_____"
],
[
"maxis_kin_cnt.sort_values(ascending=False, inplace=True)",
"_____no_output_____"
],
[
"maxis_kin_cnt.head(10)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71cb3ce52b64859cbb67a93e1641724f84021c0 | 278,170 | ipynb | Jupyter Notebook | module4-makefeatures/LS_DS_124_Make-features.ipynb | JimKing100/DS-Unit-1-Sprint-1-Dealing-With-Data | f940453816fb895a52c5010833cf989ac8cffd01 | [
"MIT"
] | null | null | null | module4-makefeatures/LS_DS_124_Make-features.ipynb | JimKing100/DS-Unit-1-Sprint-1-Dealing-With-Data | f940453816fb895a52c5010833cf989ac8cffd01 | [
"MIT"
] | null | null | null | module4-makefeatures/LS_DS_124_Make-features.ipynb | JimKing100/DS-Unit-1-Sprint-1-Dealing-With-Data | f940453816fb895a52c5010833cf989ac8cffd01 | [
"MIT"
] | null | null | null | 47.380344 | 7,476 | 0.309056 | [
[
[
"<a href=\"https://colab.research.google.com/github/JimKing100/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/Copy_of_LS_DS_124_Make_features.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"_Lambda School Data Science_\n\n# Make features\n\nObjectives\n- understand the purpose of feature engineering\n- work with strings in pandas\n- work with dates and times in pandas\n\nLinks\n- [Feature Engineering](https://en.wikipedia.org/wiki/Feature_engineering)\n- Python Data Science Handbook\n - [Chapter 3.10](https://jakevdp.github.io/PythonDataScienceHandbook/03.10-working-with-strings.html), Vectorized String Operations\n - [Chapter 3.11](https://jakevdp.github.io/PythonDataScienceHandbook/03.11-working-with-time-series.html), Working with Time Series",
"_____no_output_____"
],
[
"## Get LendingClub data\n\n[Source](https://www.lendingclub.com/info/download-data.action)",
"_____no_output_____"
]
],
[
[
"!wget https://resources.lendingclub.com/LoanStats_2019Q1.csv.zip",
"--2019-07-19 02:40:14-- https://resources.lendingclub.com/LoanStats_2019Q1.csv.zip\nResolving resources.lendingclub.com (resources.lendingclub.com)... 64.48.1.20\nConnecting to resources.lendingclub.com (resources.lendingclub.com)|64.48.1.20|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: unspecified [application/zip]\nSaving to: ‘LoanStats_2019Q1.csv.zip.1’\n\nLoanStats_2019Q1.cs [ <=> ] 19.30M 910KB/s in 22s \n\n2019-07-19 02:40:36 (905 KB/s) - ‘LoanStats_2019Q1.csv.zip.1’ saved [20240936]\n\n"
],
[
"!unzip LoanStats_2019Q1.csv.zip",
"Archive: LoanStats_2019Q1.csv.zip\nreplace LoanStats_2019Q1.csv? [y]es, [n]o, [A]ll, [N]one, [r]ename: n\n"
],
[
"!head LoanStats_2019Q1.csv",
"Notes offered by Prospectus (https://www.lendingclub.com/info/prospectus.action)\n\"id\",\"member_id\",\"loan_amnt\",\"funded_amnt\",\"funded_amnt_inv\",\"term\",\"int_rate\",\"installment\",\"grade\",\"sub_grade\",\"emp_title\",\"emp_length\",\"home_ownership\",\"annual_inc\",\"verification_status\",\"issue_d\",\"loan_status\",\"pymnt_plan\",\"url\",\"desc\",\"purpose\",\"title\",\"zip_code\",\"addr_state\",\"dti\",\"delinq_2yrs\",\"earliest_cr_line\",\"inq_last_6mths\",\"mths_since_last_delinq\",\"mths_since_last_record\",\"open_acc\",\"pub_rec\",\"revol_bal\",\"revol_util\",\"total_acc\",\"initial_list_status\",\"out_prncp\",\"out_prncp_inv\",\"total_pymnt\",\"total_pymnt_inv\",\"total_rec_prncp\",\"total_rec_int\",\"total_rec_late_fee\",\"recoveries\",\"collection_recovery_fee\",\"last_pymnt_d\",\"last_pymnt_amnt\",\"next_pymnt_d\",\"last_credit_pull_d\",\"collections_12_mths_ex_med\",\"mths_since_last_major_derog\",\"policy_code\",\"application_type\",\"annual_inc_joint\",\"dti_joint\",\"verification_status_joint\",\"acc_now_delinq\",\"tot_coll_amt\",\"tot_cur_bal\",\"open_acc_6m\",\"open_act_il\",\"open_il_12m\",\"open_il_24m\",\"mths_since_rcnt_il\",\"total_bal_il\",\"il_util\",\"open_rv_12m\",\"open_rv_24m\",\"max_bal_bc\",\"all_util\",\"total_rev_hi_lim\",\"inq_fi\",\"total_cu_tl\",\"inq_last_12m\",\"acc_open_past_24mths\",\"avg_cur_bal\",\"bc_open_to_buy\",\"bc_util\",\"chargeoff_within_12_mths\",\"delinq_amnt\",\"mo_sin_old_il_acct\",\"mo_sin_old_rev_tl_op\",\"mo_sin_rcnt_rev_tl_op\",\"mo_sin_rcnt_tl\",\"mort_acc\",\"mths_since_recent_bc\",\"mths_since_recent_bc_dlq\",\"mths_since_recent_inq\",\"mths_since_recent_revol_delinq\",\"num_accts_ever_120_pd\",\"num_actv_bc_tl\",\"num_actv_rev_tl\",\"num_bc_sats\",\"num_bc_tl\",\"num_il_tl\",\"num_op_rev_tl\",\"num_rev_accts\",\"num_rev_tl_bal_gt_0\",\"num_sats\",\"num_tl_120dpd_2m\",\"num_tl_30dpd\",\"num_tl_90g_dpd_24m\",\"num_tl_op_past_12m\",\"pct_tl_nvr_dlq\",\"percent_bc_gt_75\",\"pub_rec_bankruptcies\",\"tax_liens\",\"tot_hi_cred_lim\",\"total_bal_ex_mort\",\"total_bc_limit\",\"total_il_high_credit_limit\",\"revol_bal_joint\",\"sec_app_earliest_cr_line\",\"sec_app_inq_last_6mths\",\"sec_app_mort_acc\",\"sec_app_open_acc\",\"sec_app_revol_util\",\"sec_app_open_act_il\",\"sec_app_num_rev_accts\",\"sec_app_chargeoff_within_12_mths\",\"sec_app_collections_12_mths_ex_med\",\"sec_app_mths_since_last_major_derog\",\"hardship_flag\",\"hardship_type\",\"hardship_reason\",\"hardship_status\",\"deferral_term\",\"hardship_amount\",\"hardship_start_date\",\"hardship_end_date\",\"payment_plan_start_date\",\"hardship_length\",\"hardship_dpd\",\"hardship_loan_status\",\"orig_projected_additional_accrued_interest\",\"hardship_payoff_balance_amount\",\"hardship_last_payment_amount\",\"debt_settlement_flag\",\"debt_settlement_flag_date\",\"settlement_status\",\"settlement_date\",\"settlement_amount\",\"settlement_percentage\",\"settlement_term\"\n\"\",\"\",\"20000\",\"20000\",\"20000\",\" 60 months\",\" 17.19%\",\"499.1\",\"C\",\"C5\",\"Front desk supervisor\",\"6 years\",\"RENT\",\"47000\",\"Source Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"debt_consolidation\",\"Debt consolidation\",\"958xx\",\"CA\",\"14.02\",\"0\",\"Sep-2006\",\"1\",\"50\",\"\",\"15\",\"0\",\"10687\",\"19.7%\",\"53\",\"w\",\"19254.76\",\"19254.76\",\"1459.1\",\"1459.10\",\"745.24\",\"713.86\",\"0.0\",\"0.0\",\"0.0\",\"Jun-2019\",\"499.1\",\"Jul-2019\",\"Jun-2019\",\"0\",\"50\",\"1\",\"Individual\",\"\",\"\",\"\",\"0\",\"0\",\"31546\",\"3\",\"2\",\"1\",\"2\",\"10\",\"20859\",\"97\",\"4\",\"9\",\"5909\",\"42\",\"54300\",\"6\",\"1\",\"3\",\"11\",\"2103\",\"23647\",\"30\",\"0\",\"0\",\"150\",\"100\",\"1\",\"1\",\"0\",\"5\",\"\",\"3\",\"50\",\"3\",\"3\",\"4\",\"8\",\"19\",\"19\",\"13\",\"33\",\"4\",\"15\",\"0\",\"0\",\"0\",\"5\",\"98\",\"12.5\",\"0\",\"0\",\"75824\",\"31546\",\"33800\",\"21524\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n\"\",\"\",\"12000\",\"12000\",\"12000\",\" 36 months\",\" 16.40%\",\"424.26\",\"C\",\"C4\",\"Executive Director\",\"4 years\",\"MORTGAGE\",\"95000\",\"Not Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"debt_consolidation\",\"Debt consolidation\",\"436xx\",\"OH\",\"29.5\",\"0\",\"Nov-1992\",\"0\",\"29\",\"\",\"19\",\"0\",\"16619\",\"64.9%\",\"36\",\"w\",\"11475.92\",\"11475.92\",\"826.65\",\"826.65\",\"524.08\",\"302.57\",\"0.0\",\"0.0\",\"0.0\",\"May-2019\",\"424.26\",\"Jul-2019\",\"Jun-2019\",\"0\",\"30\",\"1\",\"Individual\",\"\",\"\",\"\",\"0\",\"0\",\"176551\",\"1\",\"5\",\"2\",\"5\",\"7\",\"35115\",\"65\",\"3\",\"6\",\"4899\",\"65\",\"25600\",\"4\",\"0\",\"3\",\"12\",\"9292\",\"3231\",\"82.5\",\"0\",\"0\",\"316\",\"269\",\"5\",\"5\",\"2\",\"12\",\"58\",\"7\",\"29\",\"1\",\"6\",\"8\",\"6\",\"9\",\"10\",\"13\",\"23\",\"8\",\"19\",\"0\",\"0\",\"0\",\"6\",\"80.6\",\"66.7\",\"0\",\"0\",\"209488\",\"51734\",\"18500\",\"54263\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n\"\",\"\",\"3000\",\"3000\",\"3000\",\" 36 months\",\" 14.74%\",\"103.62\",\"C\",\"C2\",\"Office Manager\",\"4 years\",\"MORTGAGE\",\"58750\",\"Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"medical\",\"Medical expenses\",\"327xx\",\"FL\",\"30.91\",\"0\",\"Jun-2004\",\"0\",\"24\",\"\",\"16\",\"0\",\"20502\",\"60.1%\",\"25\",\"f\",\"2865.64\",\"2865.64\",\"202.33\",\"202.33\",\"134.36\",\"67.97\",\"0.0\",\"0.0\",\"0.0\",\"May-2019\",\"103.62\",\"Jul-2019\",\"Jun-2019\",\"0\",\"\",\"1\",\"Individual\",\"\",\"\",\"\",\"0\",\"0\",\"37816\",\"0\",\"3\",\"0\",\"0\",\"27\",\"17314\",\"48\",\"0\",\"0\",\"3962\",\"54\",\"34100\",\"0\",\"1\",\"0\",\"0\",\"2364\",\"1208\",\"89.4\",\"0\",\"0\",\"177\",\"164\",\"35\",\"27\",\"4\",\"40\",\"24\",\"\",\"24\",\"0\",\"7\",\"12\",\"7\",\"8\",\"6\",\"13\",\"15\",\"12\",\"16\",\"0\",\"0\",\"0\",\"0\",\"96\",\"71.4\",\"0\",\"0\",\"69911\",\"37816\",\"11400\",\"35811\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n\"\",\"\",\"35000\",\"35000\",\"35000\",\" 36 months\",\" 15.57%\",\"1223.08\",\"C\",\"C3\",\"Store Manager \",\"10+ years\",\"RENT\",\"122000\",\"Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"credit_card\",\"Credit card refinancing\",\"333xx\",\"FL\",\"22\",\"1\",\"Dec-2009\",\"0\",\"20\",\"\",\"5\",\"0\",\"1441\",\"24.4%\",\"18\",\"w\",\"33459.43\",\"33459.43\",\"2446.76\",\"2446.76\",\"1540.57\",\"845.04\",\"61.15\",\"0.0\",\"0.0\",\"Jun-2019\",\"1223.08\",\"Jul-2019\",\"Jun-2019\",\"0\",\"20\",\"1\",\"Individual\",\"\",\"\",\"\",\"0\",\"0\",\"24471\",\"0\",\"3\",\"0\",\"0\",\"26\",\"23030\",\"39\",\"0\",\"0\",\"1441\",\"37\",\"5900\",\"2\",\"4\",\"1\",\"1\",\"4894\",\"159\",\"90.1\",\"0\",\"0\",\"108\",\"111\",\"52\",\"24\",\"0\",\"85\",\"\",\"9\",\"\",\"1\",\"1\",\"1\",\"1\",\"3\",\"10\",\"2\",\"7\",\"1\",\"5\",\"0\",\"0\",\"1\",\"0\",\"94.4\",\"100\",\"0\",\"0\",\"65640\",\"24471\",\"1600\",\"59740\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n\"\",\"\",\"5000\",\"5000\",\"5000\",\" 36 months\",\" 15.57%\",\"174.73\",\"C\",\"C3\",\"Area Manager\",\"3 years\",\"OWN\",\"65000\",\"Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"house\",\"Home buying\",\"640xx\",\"MO\",\"16.28\",\"1\",\"Jul-2001\",\"0\",\"7\",\"\",\"9\",\"0\",\"5604\",\"64.4%\",\"25\",\"w\",\"4778.86\",\"4778.86\",\"340.81\",\"340.81\",\"221.14\",\"119.67\",\"0.0\",\"0.0\",\"0.0\",\"May-2019\",\"174.73\",\"Jul-2019\",\"Jun-2019\",\"0\",\"7\",\"1\",\"Individual\",\"\",\"\",\"\",\"0\",\"0\",\"29775\",\"0\",\"1\",\"0\",\"1\",\"17\",\"24171\",\"82\",\"1\",\"6\",\"1824\",\"78\",\"8700\",\"1\",\"0\",\"0\",\"7\",\"3722\",\"1070\",\"75.7\",\"0\",\"0\",\"111\",\"212\",\"10\",\"10\",\"4\",\"17\",\"69\",\"17\",\"69\",\"4\",\"3\",\"5\",\"5\",\"9\",\"7\",\"8\",\"14\",\"5\",\"9\",\"0\",\"0\",\"1\",\"1\",\"76\",\"50\",\"0\",\"0\",\"38190\",\"29775\",\"4400\",\"29490\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n\"\",\"\",\"40000\",\"40000\",\"40000\",\" 60 months\",\" 12.40%\",\"897.89\",\"B\",\"B4\",\"SVP\",\"10+ years\",\"MORTGAGE\",\"515000\",\"Source Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"debt_consolidation\",\"Debt consolidation\",\"117xx\",\"NY\",\"18.19\",\"0\",\"Feb-1993\",\"1\",\"\",\"\",\"24\",\"0\",\"130490\",\"86.4%\",\"49\",\"w\",\"39025.88\",\"39025.88\",\"1740.67\",\"1740.67\",\"974.12\",\"766.55\",\"0.0\",\"0.0\",\"0.0\",\"May-2019\",\"897.89\",\"Jul-2019\",\"Jun-2019\",\"0\",\"\",\"1\",\"Individual\",\"\",\"\",\"\",\"0\",\"205\",\"569565\",\"3\",\"6\",\"2\",\"3\",\"4\",\"72207\",\"39\",\"3\",\"3\",\"20896\",\"67\",\"151100\",\"1\",\"6\",\"2\",\"7\",\"23732\",\"4733\",\"94.8\",\"0\",\"0\",\"160\",\"304\",\"3\",\"3\",\"6\",\"10\",\"\",\"0\",\"\",\"0\",\"10\",\"14\",\"10\",\"17\",\"12\",\"15\",\"28\",\"14\",\"24\",\"0\",\"0\",\"0\",\"5\",\"100\",\"100\",\"0\",\"0\",\"752911\",\"204997\",\"90800\",\"144678\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n\"\",\"\",\"40000\",\"40000\",\"40000\",\" 60 months\",\" 11.02%\",\"870.1\",\"B\",\"B2\",\"VP of Sales\",\"3 years\",\"MORTGAGE\",\"288000\",\"Not Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"credit_card\",\"Credit card refinancing\",\"843xx\",\"UT\",\"19.74\",\"0\",\"May-2002\",\"0\",\"\",\"\",\"20\",\"0\",\"66240\",\"43.9%\",\"34\",\"w\",\"38989.85\",\"38989.85\",\"1691.22\",\"1691.22\",\"1010.15\",\"681.07\",\"0.0\",\"0.0\",\"0.0\",\"May-2019\",\"870.1\",\"Jul-2019\",\"Jun-2019\",\"0\",\"\",\"1\",\"Individual\",\"\",\"\",\"\",\"0\",\"0\",\"390598\",\"0\",\"5\",\"0\",\"2\",\"16\",\"140633\",\"58\",\"0\",\"2\",\"22577\",\"49\",\"150900\",\"0\",\"6\",\"1\",\"4\",\"19530\",\"81923\",\"43.1\",\"0\",\"0\",\"136\",\"202\",\"17\",\"16\",\"1\",\"17\",\"\",\"3\",\"\",\"0\",\"8\",\"9\",\"10\",\"15\",\"11\",\"13\",\"22\",\"10\",\"20\",\"0\",\"0\",\"0\",\"0\",\"100\",\"20\",\"0\",\"0\",\"543401\",\"206873\",\"143900\",\"197751\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n\"\",\"\",\"20000\",\"20000\",\"20000\",\" 60 months\",\" 12.40%\",\"448.95\",\"B\",\"B4\",\"Sales\",\"1 year\",\"MORTGAGE\",\"70000\",\"Source Verified\",\"Mar-2019\",\"Current\",\"n\",\"\",\"\",\"credit_card\",\"Credit card refinancing\",\"481xx\",\"MI\",\"32.33\",\"0\",\"May-1995\",\"0\",\"\",\"\",\"12\",\"0\",\"31036\",\"56.7%\",\"32\",\"w\",\"19512.93\",\"19512.93\",\"870.34\",\"870.34\",\"487.07\",\"383.27\",\"0.0\",\"0.0\",\"0.0\",\"May-2019\",\"448.95\",\"Jul-2019\",\"Jun-2019\",\"0\",\"\",\"1\",\"Joint App\",\"102400\",\"31.16\",\"Source Verified\",\"0\",\"0\",\"204292\",\"0\",\"3\",\"0\",\"1\",\"14\",\"16305\",\"33\",\"0\",\"0\",\"17389\",\"46\",\"54700\",\"1\",\"3\",\"0\",\"2\",\"17024\",\"268\",\"98.9\",\"0\",\"0\",\"133\",\"286\",\"41\",\"14\",\"4\",\"84\",\"\",\"15\",\"\",\"0\",\"3\",\"7\",\"3\",\"7\",\"9\",\"8\",\"19\",\"7\",\"12\",\"0\",\"0\",\"0\",\"0\",\"100\",\"100\",\"0\",\"0\",\"267991\",\"47341\",\"24800\",\"49291\",\"56647\",\"May-1995\",\"0\",\"4\",\"15\",\"61.3\",\"1\",\"31\",\"0\",\"0\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"\",\"N\",\"\",\"\",\"\",\"\",\"\",\"\"\n"
]
],
[
[
"## Load LendingClub data\n\npandas documentation\n- [`read_csv`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html)\n- [`options.display`](https://pandas.pydata.org/pandas-docs/stable/options.html#available-options)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.read_csv('LoanStats_2019Q1.csv', header=1, skipfooter=2, engine='python')\nprint(df.shape)\ndf.head()",
"(115675, 144)\n"
],
[
"df.isna().sum()",
"_____no_output_____"
],
[
"df[df.loan_amnt.isna()]",
"_____no_output_____"
],
[
"# default is !tail LoanStats_2019Q1.csv",
"_____no_output_____"
],
[
"# default is 30\npd.options.display.max_columns = 150\npd.options.display.max_rows = 150",
"_____no_output_____"
],
[
"df.head().T",
"_____no_output_____"
]
],
[
[
"## Work with strings",
"_____no_output_____"
],
[
"For machine learning, we usually want to replace strings with numbers.\n\nWe can get info about which columns have a datatype of \"object\" (strings)",
"_____no_output_____"
]
],
[
[
"df.describe(include='object')",
"_____no_output_____"
],
[
"df.emp_length.value_counts()",
"_____no_output_____"
]
],
[
[
"### Convert `int_rate`\n\nDefine a function to remove percent signs from strings and convert to floats",
"_____no_output_____"
]
],
[
[
"# vectorized version\n# df['int_rate'] = df['int_rate'].str.strip('%').astype(float)",
"_____no_output_____"
]
],
[
[
"Apply the function to the `int_rate` column",
"_____no_output_____"
]
],
[
[
"def remove_percent_sign(string):\n return float(string.strip('%'))\n\nx = '12.5%'\nremove_percent_sign(x)",
"_____no_output_____"
],
[
"df['int_rate'] = df['int_rate'].apply(remove_percent_sign)\ndf['int_rate'].head()",
"_____no_output_____"
],
[
"df.int_rate.hist()",
"_____no_output_____"
]
],
[
[
"### Clean `emp_title`\n\nLook at top 20 titles",
"_____no_output_____"
]
],
[
[
"df.emp_title.value_counts(dropna=False).head(20)",
"_____no_output_____"
]
],
[
[
"How often is `emp_title` null?",
"_____no_output_____"
]
],
[
[
"print(df.emp_title.isna().sum())\nprint(df.shape)\nprint(df.emp_title.isna().sum() / df.shape[0])",
"19518\n(115675, 144)\n0.1687313594121461\n"
]
],
[
[
"Clean the title and handle missing values\n- Capitalize\n- Strip spaces\n- Replace 'Nan' with missing",
"_____no_output_____"
]
],
[
[
"import numpy as np\nexample = ['owner','Supervisor ', ' Project Manager', 'missing', np.nan]\n\ndef clean_emp_title(x):\n if isinstance(x, str):\n return x.strip().title()\n else:\n return 'Missing'\n \n# for ex in example:\n# print(clean_emp_title(ex))\n \n[clean_emp_title(x) for x in example] ",
"_____no_output_____"
],
[
"df['emp_title'] = df['emp_title'].apply(clean_emp_title)\ndf['emp_title'].head(20)",
"_____no_output_____"
],
[
"df.emp_title.nunique()",
"_____no_output_____"
],
[
"df.emp_title.value_counts().head(10)",
"_____no_output_____"
]
],
[
[
"### Create `emp_title_manager`\n\npandas documentation: [`str.contains`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.Series.str.contains.html)",
"_____no_output_____"
]
],
[
[
"# Looks for an exact manager 'Manager' not 'manager' use case=False for all versions\ndf['emp_title'].str.contains('Manager').head(10)",
"_____no_output_____"
],
[
"df['emp_title'].iloc[0:10]",
"_____no_output_____"
],
[
"df['emp_title_manager'] = df.emp_title.str.contains('Manager')\ndf['emp_title_manager'].value_counts()",
"_____no_output_____"
],
[
"df.groupby('emp_title_manager').int_rate.mean()",
"_____no_output_____"
]
],
[
[
"## Work with dates",
"_____no_output_____"
],
[
"pandas documentation\n- [to_datetime](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.to_datetime.html)\n- [Time/Date Components](https://pandas.pydata.org/pandas-docs/stable/timeseries.html#time-date-components) \"You can access these properties via the `.dt` accessor\"",
"_____no_output_____"
]
],
[
[
"print(df['issue_d'].head().values)\ndf['issue_d'].describe()\n",
"['Mar-2019' 'Mar-2019' 'Mar-2019' 'Mar-2019' 'Mar-2019']\n"
],
[
"df['issue_d'] = pd.to_datetime(df['issue_d'], infer_datetime_format=True)\ndf['issue_d'].describe()",
"_____no_output_____"
],
[
"df['issue_d'].iloc[0:5].dt.month",
"_____no_output_____"
],
[
"df['earliest_cr_line'] = pd.to_datetime(df['earliest_cr_line'])",
"_____no_output_____"
],
[
"df['days_since_earliest_cr_line'] = (df['issue_d'] - df['earliest_cr_line']).dt.days\ndf['days_since_earliest_cr_line'].describe()",
"_____no_output_____"
],
[
"print(1124/365) # 3 years old!\nprint(27453/365) #95 year old! 75+18",
"3.0794520547945203\n75.21369863013699\n"
],
[
"[col for col in df if col.endswith('_d')]",
"_____no_output_____"
],
[
"for col in ['last_pymnt_d', 'next_pymnt_d', 'last_credit_pull_d']:\n df[col] = pd.to_datetime(df[col])\n \ndf.describe(include='datetime')\n",
"_____no_output_____"
]
],
[
[
"## Assignment",
"_____no_output_____"
]
],
[
[
"# Take a look at the term data\nprint(df.term.value_counts(dropna=False).head(20))\nprint(df['term'].head().values)\nprint(df['term'].describe())",
" 36 months 78429\n 60 months 37246\nName: term, dtype: int64\n[' 60 months' ' 36 months' ' 36 months' ' 36 months' ' 36 months']\ncount 115675\nunique 2\ntop 36 months\nfreq 78429\nName: term, dtype: object\n"
],
[
"# Convert the data from '36 Months' to '36' or '60 Months' to '60' \ndf['term'] = np.where(df.term.str.contains('36 months'), 36, 60)\nprint(df.term.value_counts(dropna=False).head(20))\nprint(df['term'].head().values)\nprint(df['term'].describe())",
"36 78429\n60 37246\nName: term, dtype: int64\n[60 36 36 36 36]\ncount 115675.000000\nmean 43.727720\nstd 11.213773\nmin 36.000000\n25% 36.000000\n50% 36.000000\n75% 60.000000\nmax 60.000000\nName: term, dtype: float64\n"
],
[
"# Convert the strings to integers\ndf['term'] = df['term'].astype(int)\nprint(df.term.value_counts(dropna=False).head(20))\nprint(df['term'].head().values)\n",
"36 78429\n60 37246\nName: term, dtype: int64\n[60 36 36 36 36]\n"
],
[
"# Check the loan_status data for NaN and number of values\ndf.loan_status.value_counts(dropna=False).head(20)",
"_____no_output_____"
],
[
"# Create a new column loan_status_is_great and check the counts to see if it works\ndf['loan_status_is_great'] = np.where((df.loan_status.str.contains('Current') | df.loan_status.str.contains('Fully Paid')), 1, 0)\ndf.loan_status_is_great.value_counts().head(20)",
"_____no_output_____"
],
[
"# Take a look at the last_pymnt_d data\nprint(df.last_pymnt_d.value_counts(dropna=False).head(20))\nprint(df['last_pymnt_d'].head().values)\ndf['last_pymnt_d'].describe()",
"2019-06-01 102101\n2019-05-01 9503\n2019-04-01 1581\n2019-03-01 1187\n2019-02-01 864\nNaT 230\n2019-01-01 208\n2019-07-01 1\nName: last_pymnt_d, dtype: int64\n['2019-06-01T00:00:00.000000000' '2019-05-01T00:00:00.000000000'\n '2019-05-01T00:00:00.000000000' '2019-06-01T00:00:00.000000000'\n '2019-05-01T00:00:00.000000000']\n"
],
[
"# Create the last_pymnt_d_month column and populate with month\ndf['last_pymnt_d_month'] = df['last_pymnt_d'].dt.month\ndf['last_pymnt_d_month'].head()",
"_____no_output_____"
],
[
"# Check the results\nprint(df.last_pymnt_d_month.value_counts(dropna=False).head(20))",
"6.0 102101\n5.0 9503\n4.0 1581\n3.0 1187\n2.0 864\nNaN 230\n1.0 208\n7.0 1\nName: last_pymnt_d_month, dtype: int64\n"
],
[
"# Create the last_pymnt_d_year column and populate with year\ndf['last_pymnt_d_year'] = df['last_pymnt_d'].dt.year\ndf['last_pymnt_d_year'].head()",
"_____no_output_____"
],
[
"# Check the results\nprint(df.last_pymnt_d_year.value_counts(dropna=False).head(20))",
"2019.0 115445\nNaN 230\nName: last_pymnt_d_year, dtype: int64\n"
]
],
[
[
"## Stretch Goals",
"_____no_output_____"
]
],
[
[
"# Take a look at the revol_util data\nprint(df.revol_util.value_counts(dropna=False).head(20))\ndf.revol_util.isna().sum()",
"0% 1054\n43% 210\n45% 210\n35% 207\n38% 207\n37% 202\n46% 199\n32% 196\n31% 196\n32.4% 195\n36% 195\n42% 194\n31.9% 193\n27% 193\n47% 193\n35.8% 192\n32.9% 190\n33.5% 190\n34% 190\n58% 190\nName: revol_util, dtype: int64\n"
],
[
"# Fill the 129 NaN/Missing with '0%' string - only .001% of the data\ndf['revol_util'].fillna('0%', inplace=True)\ndf.revol_util.isna().sum()",
"_____no_output_____"
],
[
"# Remove the percent sign\ndf['revol_util'] = df['revol_util'].apply(remove_percent_sign)\ndf['revol_util'].head()",
"_____no_output_____"
],
[
"df.emp_title.value_counts(dropna=False).head(20)",
"_____no_output_____"
],
[
"# Create a list of the top 20 names, exclude the top name 'Missing', so 21 rows needed\ntop20_list = df.emp_title.value_counts().head(21).index\nfor i in top20_list:\n print(i)",
"Missing\nTeacher\nManager\nRegistered Nurse\nDriver\nSupervisor\nOffice Manager\nSales\nTruck Driver\nGeneral Manager\nRn\nOwner\nProject Manager\nSales Manager\nDirector\nOperations Manager\nAdministrative Assistant\nPolice Officer\nTechnician\nEngineer\nStore Manager\n"
],
[
"# Create a dictionary mapping function for the top 20 names and test\ndef mapper(sval):\n namemap = {\n 'Teacher':'Teacher',\n 'Manager':'Manager',\n 'Registered Nurse':'Registered Nurse',\n 'Driver':'Driver',\n 'Supervisor':'Supervisor',\n 'Office Manager':'Office Manager',\n 'Sales':'Sales',\n 'Truck Driver':'Truck Driver',\n 'General Manager':'General Manager',\n 'Rn':'Rn',\n 'Owner':'Owner',\n 'Project Manager':'Project Manager',\n 'Sales Manager':'Sales Manager',\n 'Director':'Director',\n 'Operations Manager':'Operations Manager',\n 'Administrative Assistant':'Administrative Assistant',\n 'Police Officer':'Police Officer',\n 'Technician':'Technician',\n 'Engineer':'Engineer',\n 'Store Manager':'Store Manager'\n }\n return namemap.get(sval,'Other')\n \nprint(mapper('Teacher'))\nprint(mapper('Mechanic'))",
"Teacher\nOther\n"
],
[
"df['emp_title'] = df['emp_title'].apply(mapper)\ndf['emp_title'].value_counts()",
"_____no_output_____"
]
],
[
[
"# ASSIGNMENT\n\n- Replicate the lesson code.\n\n- Convert the `term` column from string to integer.\n\n- Make a column named `loan_status_is_great`. It should contain the integer 1 if `loan_status` is \"Current\" or \"Fully Paid.\" Else it should contain the integer 0.\n\n- Make `last_pymnt_d_month` and `last_pymnt_d_year` columns.",
"_____no_output_____"
],
[
"# STRETCH OPTIONS\n\nYou can do more with the LendingClub or Instacart datasets.\n\nLendingClub options:\n- There's one other column in the dataframe with percent signs. Remove them and convert to floats. You'll need to handle missing values.\n- Modify the `emp_title` column to replace titles with 'Other' if the title is not in the top 20. \n- Take initiatve and work on your own ideas!\n\nInstacart options:\n- Read [Instacart Market Basket Analysis, Winner's Interview: 2nd place, Kazuki Onodera](http://blog.kaggle.com/2017/09/21/instacart-market-basket-analysis-winners-interview-2nd-place-kazuki-onodera/), especially the **Feature Engineering** section. (Can you choose one feature from his bulleted lists, and try to engineer it with pandas code?)\n- Read and replicate parts of [Simple Exploration Notebook - Instacart](https://www.kaggle.com/sudalairajkumar/simple-exploration-notebook-instacart). (It's the Python Notebook with the most upvotes for this Kaggle competition.)\n- Take initiative and work on your own ideas!",
"_____no_output_____"
],
[
"You can uncomment and run the cells below to re-download and extract the Instacart data",
"_____no_output_____"
]
],
[
[
"# !wget https://s3.amazonaws.com/instacart-datasets/instacart_online_grocery_shopping_2017_05_01.tar.gz",
"_____no_output_____"
],
[
"# !tar --gunzip --extract --verbose --file=instacart_online_grocery_shopping_2017_05_01.tar.gz",
"_____no_output_____"
],
[
"# %cd instacart_2017_05_01",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
e71cb438724136d712f25577dd4a406f1d8a4b37 | 27,489 | ipynb | Jupyter Notebook | 2_csv_conversion.ipynb | iKhushPatel/anomaly-detection | 7dad17371dfca1f33967cb088bec463dfdc50d87 | [
"MIT"
] | 1 | 2021-01-15T11:20:16.000Z | 2021-01-15T11:20:16.000Z | 2_csv_conversion.ipynb | iKhushPatel/anomaly-detection | 7dad17371dfca1f33967cb088bec463dfdc50d87 | [
"MIT"
] | null | null | null | 2_csv_conversion.ipynb | iKhushPatel/anomaly-detection | 7dad17371dfca1f33967cb088bec463dfdc50d87 | [
"MIT"
] | null | null | null | 73.895161 | 1,806 | 0.720033 | [
[
[
"import pandas as pd\nimport shutil",
"_____no_output_____"
],
[
"#!ls Dataset2/\n!ls Dataset2/MURA-v1.1/elbow_negative",
"ls: cannot access 'Dataset2/MURA-v1.1/elbow_negative': No such file or directory\r\n"
],
[
"CSV_PATH = 'Dataset2/MURA-v1.1/valid_elbow_positive.csv'",
"_____no_output_____"
],
[
"df = pd.read_csv(CSV_PATH)\nprint(df.columns)\n#df = df[0:10]\nprint(len(df['image_name']))\nfor i in df['image_name']:\n print(i)",
"Index(['image_name'], dtype='object')\n230\nMURA-v1.1/valid/XR_ELBOW/patient11659/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11659/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11659/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11802/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11802/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11802/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11802/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11802/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11803/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11803/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11803/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11803/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11804/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11804/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11804/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11804/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11805/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11805/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11805/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11805/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11414/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11414/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11186/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11186/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11186/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11186/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11186/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11806/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11806/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11807/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11807/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11807/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11793/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11793/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11793/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11793/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11793/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11358/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11358/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11358/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11417/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11417/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11417/study2_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11417/study2_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11586/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11586/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11389/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11389/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11389/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11389/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11808/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11808/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11808/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11808/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11342/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11342/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11603/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11603/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11809/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11809/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11809/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11809/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11810/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11811/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11811/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11812/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11812/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11189/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11189/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11189/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11189/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11780/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11780/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11780/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11780/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11780/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11780/study1_positive/image6.png\nMURA-v1.1/valid/XR_ELBOW/patient11813/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11813/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11813/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11813/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11813/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11472/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11472/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11472/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11326/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11326/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11326/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11326/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11814/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11814/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11814/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11390/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11390/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11390/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11390/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11390/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11264/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11264/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11815/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11815/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11816/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11816/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11817/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11817/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11817/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11817/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11817/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11818/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11818/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11818/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11818/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11818/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11819/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11819/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11820/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11820/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11820/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11820/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11820/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11821/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11821/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11821/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11764/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11764/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11764/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11764/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11764/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11312/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11312/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11430/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11430/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11430/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11430/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11430/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11822/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11822/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11822/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11823/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11823/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11824/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11824/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11825/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11825/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11825/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11825/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11825/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11756/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11756/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11756/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11756/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11826/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11826/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11827/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11827/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11827/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11827/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11827/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11828/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11828/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11828/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11829/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11829/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11829/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image6.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image7.png\nMURA-v1.1/valid/XR_ELBOW/patient11830/study1_positive/image8.png\nMURA-v1.1/valid/XR_ELBOW/patient11236/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11236/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11236/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11831/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11831/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11831/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11831/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11831/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11831/study1_positive/image6.png\nMURA-v1.1/valid/XR_ELBOW/patient11831/study1_positive/image7.png\nMURA-v1.1/valid/XR_ELBOW/patient11832/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11832/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11832/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11832/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11833/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11834/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11834/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11834/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11835/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11835/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11836/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11836/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11836/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11836/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11836/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11837/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11837/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11837/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11838/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11838/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11838/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11421/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11421/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11421/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11421/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11839/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11839/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11839/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11839/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11839/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11840/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11840/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11840/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11840/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11840/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11841/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11841/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11841/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11842/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11842/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11842/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11842/study1_positive/image4.png\nMURA-v1.1/valid/XR_ELBOW/patient11842/study1_positive/image5.png\nMURA-v1.1/valid/XR_ELBOW/patient11843/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11843/study1_positive/image2.png\nMURA-v1.1/valid/XR_ELBOW/patient11843/study1_positive/image3.png\nMURA-v1.1/valid/XR_ELBOW/patient11844/study1_positive/image1.png\nMURA-v1.1/valid/XR_ELBOW/patient11844/study1_positive/image2.png\n"
],
[
"new_path = 'Dataset2/MURA-v1.1/valid/elbow_positive/'\ncount = 0\nfor i in df['image_name']:\n dest_path = new_path + 'image' + str(count) + '.png'\n image_name = 'Dataset2/' + i\n shutil.move(image_name, dest_path)\n count = count + 1",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimg=mpimg.imread('Dataset2/MURA-v1.1/train/XR_ELBOW/patient04938/study2_negative/image1.png')\nimgplot = plt.imshow(img)\nplt.show()",
"_____no_output_____"
],
[
"!",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71cd141ab35e14946c4bfa9da95d5a70faf31f6 | 261,242 | ipynb | Jupyter Notebook | nbs/01_tabular.pd.ipynb | hududed/fastinference | 172b175698517ddbfe2934c73cf5a55c1c08b6c7 | [
"Apache-2.0"
] | null | null | null | nbs/01_tabular.pd.ipynb | hududed/fastinference | 172b175698517ddbfe2934c73cf5a55c1c08b6c7 | [
"Apache-2.0"
] | null | null | null | nbs/01_tabular.pd.ipynb | hududed/fastinference | 172b175698517ddbfe2934c73cf5a55c1c08b6c7 | [
"Apache-2.0"
] | null | null | null | 285.822757 | 51,364 | 0.906298 | [
[
[
"# default_exp tabular.pd",
"_____no_output_____"
]
],
[
[
"# tabular.pd\n> This module calculates and plots partial dependence data.",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
],
[
"#export\nfrom fastai.tabular.all import *\nfrom fastinference.tabular.core import *",
"_____no_output_____"
],
[
"#export\nfrom plotnine import *",
"_____no_output_____"
],
[
"#export\nfrom IPython.display import clear_output",
"_____no_output_____"
]
],
[
[
"First let's train a model to analyze",
"_____no_output_____"
]
],
[
[
"path = untar_data(URLs.ADULT_SAMPLE)\ndf = pd.read_csv(path/'adult.csv')",
"_____no_output_____"
],
[
"dep_var = 'salary'\ncat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race', 'age']\ncont_names = ['fnlwgt', 'education-num']\nprocs = [Categorify, FillMissing, Normalize]",
"_____no_output_____"
],
[
"splits = IndexSplitter(list(range(800,1000)))(range_of(df))\nto = TabularPandas(df, procs, cat_names, cont_names, y_names=\"salary\", splits=splits)\ndls = to.dataloaders()",
"_____no_output_____"
],
[
"learn = tabular_learner(dls, layers=[200,100], metrics=accuracy)\nlearn.fit(1, 1e-2)",
"_____no_output_____"
],
[
"#export\nclass PartDep(Interpret):\n \"\"\"\n Calculate Partial Dependence. Countinious vars are divided into buckets and are analized as well\n Fields is a list of lists of what columns we want to test. The inner items are treated as connected fields.\n For ex. fields = [['Store','StoreType']] mean that Store and StoreType is treated as one entity\n (it's values are substitute as a pair, not as separate values)\n coef is useful when we don't want to deal with all the variants, but only with most common\n In short if coef for ex. is 0.9, then function outputs number of occurrences for all but least 10%\n of the least used\n If coef is more 1.0, then 'coef' itself is used as threshold (as min number of occurances)\n use_log=True is needed if we have transformed depended variable into log\n use_int=True is needed if we want to log-detransformed (exponented) var to me integer not float\n is_couninue=True helps with long calculation, it continues the last calculation from the saved file\n is_use_cache=True loads last fully calculated result. Can distinct caches that were mede with different\n fields and coef\n no_precalc=True -- don't calculate PartDep (usefull if you want to use `plot_raw` and `plot_model` only)\n \"\"\"\n\n def __init__(self, learn, df, model_name: str, fields: list = (), coef: float = 1.0,\n is_sorted: bool = True, use_log=False, use_int=False,\n cache_path=None, is_use_cache=True, is_continue=False, no_precalc=False):\n super().__init__(learn, df)\n self.use_log = use_log\n self.use_int = use_int\n self.coef = coef\n self.is_sorted = is_sorted\n\n if (fields is None) or (len(fields) == 0):\n self.fields = self._get_all_columns()\n else:\n self.fields = listify(fields)\n\n self.part_dep_df = None\n self.cache_path = ifnone(cache_path, learn.path / 'cache')\n self.save_name = f\"{model_name}_part_dep\"\n self.is_use_cache = is_use_cache\n self.is_continue = is_continue\n self.dep_var = self._get_dep_var()\n self.is_biclassification = True if (learn.dls.c == 2) else False\n\n if (no_precalc==False):\n self._load_or_calculate()\n\n @classmethod\n def what_cached(self, model_name: str, path=None, learn=None):\n \"\"\"\n Shows what keys are cached\n \"\"\"\n if isNone(path) and isNone(learn):\n print(\"path and learn cannot be None at the same time\")\n return\n elif isNone(path):\n path = learn.path\n\n name = f\"{model_name}_part_dep\"\n folder = 'cache'\n path = path / folder\n\n if not (Path(f\"{path / name}.pkl\").exists()):\n print(f\"No chache file\")\n else:\n f = open(path / f\"{name}.pkl\", \"rb\")\n var = load(f)\n f.close()\n for k in var.keys():\n print(k)\n\n @classmethod\n def empty_cache(self, model_name: str, path=None, learn=None):\n \"\"\"\n deletes the cache file\n \"\"\"\n if isNone(path) and isNone(learn):\n print(\"path and learn cannot be None at the same time\")\n return\n elif isNone(path):\n path = learn.path\n\n name = f\"{model_name}_part_dep\"\n folder = 'cache'\n path = path / folder\n\n files = (Path(f\"{path / name}.pkl\"), Path(path / 'pd_interm.pkl'))\n\n for file in files:\n if not (file.exists()):\n print(f\"No chache file {file}\")\n else:\n file.unlink()\n\n def _cont_into_buckets(self, df_init, CONT_COLS):\n \"\"\"\n Categorical values can be easily distiguished one from another\n But that doesn't work with continious values, we have to divede it's\n values into buckets and then use all values in a bucket as a single value\n that avarages the bucket. This way we convert cont feture into pseudo categorical\n and are able to apply partial dependense analysis to it\n \"\"\"\n fields = self.fields\n df = df_init.copy()\n if is_in_list(values=fields, in_list=CONT_COLS):\n for col in which_elms(values=fields, in_list=CONT_COLS):\n edges = np.histogram_bin_edges(a=df[col].dropna(), bins='auto')\n for x, y in zip(edges[::], edges[1::]):\n df.loc[(df[col] > x) & (df[col] < y), col] = (x + y) / 2\n return df\n\n def _get_field_uniq_x_coef(self, df: pd.DataFrame, fields: list, coef: float) -> list:\n '''\n This function outputs threshold to number of occurrences different variants of list of columns (fields)\n In short if coef for ex. is 0.9, then function outputs number of occurrences for all but least 10%\n of the least used\n If coef is more 1.0, then 'coef' itself is used as threshold\n '''\n if (coef > 1):\n return math.ceil(coef)\n coef = 0. if (coef < 0) else coef\n occs = df.groupby(fields).size().reset_index(name=\"Times\").sort_values(['Times'], ascending=False)\n num = math.ceil(coef * len(occs))\n if (num <= 0):\n # number of occurances is now = max_occs+1 (so it will be no items with this filter)\n return occs.iloc[0]['Times'] + 1\n else:\n return occs.iloc[num - 1]['Times']\n\n def _get_part_dep_one(self, fields: list, masterbar=None) -> pd.DataFrame:\n '''\n Function calculate partial dependency for column in fields.\n Fields is a list of lists of what columns we want to test. The inner items are treated as connected fields.\n For ex. fields = [['Store','StoreType']] mean that Store and StoreType is treated as one entity\n (it's values are substitute as a pair, not as separate values)\n coef is useful when we don't want to deal with all the variants, but only with most common\n '''\n NAN_SUBST = '###na###'\n cont_vars = self._get_cont_columns()\n fields = listify(fields)\n coef, is_sorted, use_log, use_int = self.coef, self.is_sorted, self.use_log, self.use_int\n dep_name = self._get_dep_var()\n\n df = self._cont_into_buckets(df_init=self.df, CONT_COLS=cont_vars)\n\n # here we prepare data to eliminate pairs that occure too little\n # and make NaN a separate value to appear in occures\n field_min_occ = self._get_field_uniq_x_coef(df=df, fields=fields, coef=coef)\n df[fields] = df[fields].fillna(NAN_SUBST) # to treat None as a separate field\n occs = df.groupby(fields).size().reset_index(name=\"Times\").sort_values(['Times'], ascending=False)\n occs[fields] = occs[fields].replace(to_replace=NAN_SUBST, value=np.nan) # get back Nones from NAN_SUBST\n df[fields] = df[fields].replace(to_replace=NAN_SUBST, value=np.nan) # get back Nones from NAN_SUBST\n occs = occs[occs['Times'] >= field_min_occ]\n df_copy = df.merge(occs[fields]).copy()\n\n # here for every pair of values of fields we substitute it's values in original df\n # with the current one and calculate predictions\n # So we predict mean dep_var for every pairs of value of fields on the whole dataset\n frame = []\n ln = len(occs)\n if (ln > 0):\n for _, row in progress_bar(occs.iterrows(), total=ln, parent=masterbar):\n # We don't need to do df_copy = df.merge(occs[field]).copy() every time\n # as every time we change the same column (set of columns)\n record = []\n for fld in fields:\n df_copy[fld] = row[fld]\n preds = self._predict_df(df=df_copy)\n preds = np.exp(np.mean(preds)) if (use_log == True) else np.mean(preds)\n preds = int(preds) if (use_int == True) else preds\n for fld in fields:\n record.append(row[fld])\n record.append(preds)\n record.append(row['Times'])\n frame.append(record)\n # Here for every pair of fields we calculate mean dep_var deviation\n # This devition is the score that shows how and where this partucular pair of fields\n # moves depend valiable\n # Added times to more easily understand the data (more times more sure we are)\n out = pd.DataFrame(frame, columns=fields + [dep_name, 'times'])\n median = out[dep_name].median()\n out[dep_name] /= median\n if (is_sorted == True):\n out = out.sort_values(by=dep_name, ascending=False)\n return out\n\n def _get_part_dep(self):\n '''\n Makes a datafreme with partial dependencies for every pair of columns in fields\n '''\n fields = self.fields\n learn = self.learn\n cache_path = self.cache_path\n dep_name = self._get_dep_var()\n is_continue = self.is_continue\n l2k = self._list_to_key\n result = []\n to_save = {}\n from_saved = {}\n\n # Load from cache\n if (is_continue == True):\n if Path(cache_path / 'pd_interm.pkl').exists():\n from_saved = ld_var(name='pd_interm', path=cache_path)\n else:\n is_continue = False\n\n elapsed = []\n left = []\n if (is_continue == True):\n for field in fields:\n if (l2k(field) in from_saved):\n elapsed.append(field)\n new_df = from_saved[l2k(field)]\n result.append(new_df)\n to_save[l2k(field)] = new_df\n\n for field in fields:\n if (l2k(field) not in from_saved):\n left.append(field)\n\n # Calculate\n pbar = master_bar(left)\n cache_path.mkdir(parents=True, exist_ok=True)\n sv_var(var=to_save, name='pd_interm', path=cache_path)\n for field in pbar:\n new_df = self._get_part_dep_one(fields=field, masterbar=pbar)\n new_df['feature'] = self._list_to_key(field)\n if is_listy(field):\n new_df['value'] = new_df[field].values.tolist()\n new_df.drop(columns=field, inplace=True)\n else:\n new_df = new_df.rename(index=str, columns={str(field): \"value\"})\n result.append(new_df)\n to_save[l2k(field)] = new_df\n sv_var(var=to_save, name='pd_interm', path=cache_path)\n clear_output()\n if Path(cache_path / 'pd_interm.pkl').exists():\n Path(cache_path / 'pd_interm.pkl').unlink() # delete intermediate file\n result = pd.concat(result, ignore_index=True, sort=True)\n result = result[['feature', 'value', dep_name, 'times']]\n clear_output()\n\n self.part_dep_df = result\n\n def _load_dict(self, name, path):\n if not (Path(f\"{path / name}.pkl\").exists()):\n return None\n return self._ld_var(name=name, path=path)\n\n def _save_cached(self):\n \"\"\"\n Saves calculated PartDep df into path.\n Can be saved more than one with as an dict with fields as key\n \"\"\"\n\n path = self.cache_path\n path.mkdir(parents=True, exist_ok=True)\n name = self.save_name\n\n sv_dict = self._load_dict(name=name, path=path)\n key = self._list_to_key(self.fields + [self.coef])\n if isNone(sv_dict):\n sv_dict = {key: self.part_dep_df}\n else:\n sv_dict[key] = self.part_dep_df\n\n self._sv_var(var=sv_dict, name=name, path=path)\n\n def _load_cached(self):\n \"\"\"\n Load calculated PartDep df if hash exist.\n \"\"\"\n name = self.save_name\n path = self.cache_path\n\n if not (Path(f\"{path / name}.pkl\").exists()):\n return None\n\n ld_dict = self._ld_var(name=name, path=path)\n key = self._list_to_key(self.fields + [self.coef])\n if (key not in ld_dict):\n return None\n\n return ld_dict[key]\n\n def _load_or_calculate(self):\n \"\"\"\n Calculates part dep or load it from cache if possible\n \"\"\"\n if (self.is_use_cache == False) or isNone(self._load_cached()):\n self._get_part_dep()\n return self._save_cached()\n else:\n self.part_dep_df = self._load_cached()\n \n def _general2partial(self, df):\n if (len(df) == 0):\n return None\n copy_df = df.copy()\n feature = copy_df['feature'].iloc[0]\n copy_df.drop(columns='feature', inplace=True)\n copy_df.rename(columns={\"value\": feature}, inplace=True)\n return copy_df\n \n\n def plot_raw(self, field, sample=1.0):\n \"\"\"\n Plot dependency graph from data itself\n field must be list of exactly one feature\n sample is a coef to len(df). Lower if kernel use to shut down on that\n \"\"\"\n df = self.df\n df = df.sample(int(len(df)*sample))\n field = field[0]\n dep_var = f\"{self._get_dep_var()}_orig\" if (self.use_log == True) else self._get_dep_var()\n return ggplot(df, aes(field, dep_var)) + stat_smooth(se=True, method='loess');\n\n def plot_model(self, field, strict_recalc=False, sample=1.0):\n '''\n Plot dependency graph from the model.\n It also take into account times, so plot becomes much more resilient, cause not every value treats as equal\n (more occurences means more power)\n field must be list of exactly one feature\n strict_recalc=True ignores precalculated `part_dep_df` and calculate it anyway\n sample is a coef to len(df). Lower if kernel use to shut down on that\n '''\n cached = self.get_pd(feature=self._list_to_key(field))\n \n if (strict_recalc == False) and isNotNone(cached):\n pd_table = cached\n else:\n pd_table = self._get_part_dep_one(fields=field)\n \n clear_output()\n field = field[0]\n dep_var = f\"{self._get_dep_var()}\"\n rearr = []\n for var, fee, times in zip(pd_table[field], pd_table[dep_var], pd_table['times']):\n for i in range(int(times)):\n rearr.append([var, fee])\n rearr = pd.DataFrame(rearr, columns=[field, dep_var])\n \n rearr = rearr.sample(int(len(rearr)*sample))\n return ggplot(rearr, aes(field, dep_var)) + stat_smooth(se=True, method='loess');\n\n def get_pd(self, feature, min_tm=1):\n \"\"\"\n Gets particular feature subtable from the whole one (min times is optional parameter)\n \"\"\"\n if isNone(self.part_dep_df):\n return None\n \n df = self.part_dep_df.query(f\"\"\"(feature == \"{feature}\") and (times > {min_tm})\"\"\")\n return self._general2partial(df=df)\n\n def get_pd_main_chained_feat(self, main_feat_idx=0, show_min=1):\n \"\"\"\n Transforms whole features table to get_part_dep_one output table format\n \"\"\"\n\n def get_xth_el(str_list: str, indexes: list):\n lst = str_list if is_listy(str_list) else ast.literal_eval(str_list)\n lst = listify(lst)\n if (len(lst) == 1):\n return lst[0]\n elif (len(lst) > 1):\n if (len(indexes) == 1):\n return lst[indexes[0]]\n else:\n return [lst[idx] for idx in indexes]\n else:\n return None\n\n feat_table = self.part_dep_df\n\n main_feat_idx = listify(main_feat_idx)\n feat_table_copy = feat_table.copy()\n func = functools.partial(get_xth_el, indexes=main_feat_idx)\n feat_table_copy['value'] = feat_table_copy['value'].apply(func)\n feat_table_copy.drop(columns='feature', inplace=True)\n return feat_table_copy.query(f'times > {show_min}')\n\n def plot_part_dep(self, fields, limit=20, asc=False):\n \"\"\"\n Plots partial dependency plot for sublist of connected `fields`\n `fields` must be sublist of `fields` given on initalization calculation\n \"\"\"\n\n def prepare_colors(df_pd: pd.DataFrame):\n heat_min = df_pd['times'].min()\n heat_max = df_pd['times'].max()\n dif = heat_max - heat_min\n colors = [((times - heat_min) / (dif), (times - heat_min) / (4 * dif), 0.75) for times in df_pd['times']]\n return colors\n\n df = self.part_dep_df.query(f\"feature == '{self._list_to_key(fields)}'\")\n dep_var = self.dep_var\n\n df_copy = df.copy()\n df_copy['feature'] = df_copy['feature'].str.slice(0, 45)\n df_copy = df_copy.sort_values(by=dep_var, ascending=asc)[:limit].sort_values(by=dep_var, ascending=not (asc))\n colors = prepare_colors(df_pd=df_copy)\n ax = df_copy.plot.barh(x=\"value\", y=dep_var, sort_columns=True, figsize=(10, 10), \n color=colors, title=self._list_to_key(fields))\n ax.set_ylabel(fields)\n \n if (self.is_biclassification):\n txt = f\"According to probability of {self._get_dep_var()} is '{learn.dls.vocab[0]}'\"\n ax.annotate(txt, (0,0), (0, -30), \n xycoords='axes fraction', textcoords='offset points', \n va='top')\n \n for (p, t) in zip(ax.patches, df_copy['times']):\n ax.annotate(f'{p.get_width():.4f}', ((p.get_width() * 1.005), p.get_y() * 1.005))\n ax.annotate(f'{int(t)}', ((p.get_width() * .45), p.get_y() + 0.1), color='white', weight='bold')",
"_____no_output_____"
]
],
[
[
"This module plots the relationship between the dependent variable and particulat values of a dependent variable. In other words what value variable should be to make dep_var bigger/smaller \nIt also supports caching as calculations on big data with huge variety can take more than a day, so saving intermediate result can help a lot here",
"_____no_output_____"
],
[
"- `model_name`\n what will be the name of saved cache-file. Calling init with the same `model_name` will result continuing calculations\n- `fields` \n fields is a list of lists of what columns we want to test. The inner items are treated as connected fields.\n For ex. fields = \\[\\['Store','StoreType'\\]\\] mean that Store and StoreType is treated as one entity (it's values are substitute as a pair, not as separate values)\n- `coef`\n coef is useful when we don't want to deal with all the variants, but only with most common\n In short if coef for ex. is 0.9, then function outputs number of occurrences for all but least 10% of the least used\n If coef is more 1.0, then 'coef' itself is used as threshold (as min number of occurances)\n- `is_sorted` \n if true sort values on dependence\n- `use_log=True `\n is needed if we have transformed depended variable into log\n- `use_int=True` \n is needed if we want to log-detransformed (exponented) var to me integer not float\n- `cache_path`\n sets different the paathe to cache intermediate calculations\n- `is_use_cache`\n if true loads last fully calculated result. Can distinct caches that were mede with different fields and coefs\n- `is_couninue`\n helps with long calculation, it continues the last calculation from the saved (cached) file\n- `no_precalc`\n if True don't calculate PartDep. Can be usefull if you want to use `plot_raw` and `plot_model` only without lond calculations that are needed for `plot_part_dep` ",
"_____no_output_____"
]
],
[
[
"fields = ['workclass', ['education', 'education-num'], 'marital-status', 'occupation', 'relationship', 'race', 'age', 'fnlwgt']",
"_____no_output_____"
],
[
"# PartDep.empty_cache(model_name='salary', learn=learn)",
"No chache file cache/pd_interm.pkl\n"
],
[
"#takes me 7 min to run\npart_dep = PartDep(learn=learn, df=df, model_name='salary', \n fields=fields, use_log=False, use_int=False, coef=0.8, \n is_continue=False)",
"_____no_output_____"
],
[
"part_dep.plot_part_dep(fields= ['marital-status'])",
"_____no_output_____"
],
[
"part_dep.plot_part_dep(fields= ['relationship'])",
"_____no_output_____"
],
[
"part_dep.plot_part_dep(fields= ['occupation'], asc=True)",
"_____no_output_____"
],
[
"part_dep.plot_part_dep(fields= ['age'])",
"_____no_output_____"
],
[
"part_dep.plot_part_dep(fields= ['education', 'education-num'], asc=True)",
"_____no_output_____"
],
[
"part_dep.get_pd(feature='marital-status')",
"_____no_output_____"
],
[
"# here we use short init version for a fast `plot_model` execution",
"_____no_output_____"
],
[
"part_dep = PartDep(learn=learn, df=df, model_name='salary', \n fields=fields, coef=0.8, no_precalc=True)",
"_____no_output_____"
],
[
"#takes me 3 min to run\npart_dep.plot_model(field=['age'])",
"_____no_output_____"
]
],
[
[
"Methods exposed: \n- `plot_part_dep` -- plots partial dependency plot for sublist of connected `fields` \n- `get_pd` -- outputs partial dependence of a single feature in dataframe-format\n- `plot_model` -- plot graph of partial dependence. Works only for number-like continious values (lake 'year made', 'age' and so on)\n- `plot_raw` -- plot dependency graph from data itself. Works only for regression data\n- `what_cached` -- shows what is cached\n- `empty_cache` -- deletes the cache file, if you want to rerun init from the scratch",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e71cd2193fe25ecd56dface944cd1bd8acc2c39b | 106,858 | ipynb | Jupyter Notebook | python/jupyter/py_tutorial.ipynb | sdsmt-robotics/tip-and-whatnot | 6c82d0664b68864a0e9ed50758d1a2e0cd64550c | [
"MIT"
] | null | null | null | python/jupyter/py_tutorial.ipynb | sdsmt-robotics/tip-and-whatnot | 6c82d0664b68864a0e9ed50758d1a2e0cd64550c | [
"MIT"
] | null | null | null | python/jupyter/py_tutorial.ipynb | sdsmt-robotics/tip-and-whatnot | 6c82d0664b68864a0e9ed50758d1a2e0cd64550c | [
"MIT"
] | null | null | null | 109.597949 | 31,186 | 0.820697 | [
[
[
"# This is a comment\n\nspam = 3\nprint(spam)\n\nspam = \"test\"\nprint(spam)\n\n# Python sees no difference between \" and '\nspam2 = 'another test'\nspam3 = 'contains \"'\nspam4 = \"contains '\"\nspam5 = 4*'n' # 'nnnn'\nprint(spam2, end='&\\n') # we can also customize the line endings\n\nspam = [1, 2, 3, 4]\nprint(spam)\n",
"3\ntest\nanother test&\n[1, 2, 3, 4]\n"
],
[
"# Python offers string formatting\nprint('spam: %s' % (spam))\nprint('spam: {} spam+[\"a\"]: {}'.format(spam, spam+['a']))",
"spam: [1, 2, 3, 4]\nspam: [1, 2, 3, 4] spam+[\"a\"]: [1, 2, 3, 4, 'a']\n"
],
[
"a = 1\na2 = 2\nb = 2.0\nc = 1 + 3j\nd = 3e-4\n\n# More on this later\nl = [a, b, c, d]\n[type(i) for i in l] # this is a 'list comprehension', it creates a new list from an old one in place",
"_____no_output_____"
],
[
"print(a + b)\nprint(a * b)\nprint(b + c)\nprint(a / a2)\nprint(b % a)\nprint(a // a2) # This is new in Python 3, it's the old style integer division truncation\n\nprint(b**c) # ** is the power operator\n\n# <<, >>, &, |, ~, and ^ are the C/C++ standard bitwise operators, but I'd recommend against using them\na += 1\na /= 2\na *= 3\na **= 4",
"3.0\n2.0\n(3+3j)\n0.5\n0.0\n0\n(-0.9739888359315625+1.746810163549743j)\n"
],
[
"# Types\nl = [type, None, True, 1, 0x7fffffff, 0x80000000, 0b1001, 0o17, 1.0, 1j, 'a', u'a', r'a', b'a', l]\n[type(i) for i in l]",
"_____no_output_____"
],
[
"# Conversions\na = 42\nl = [bin, hex, int, oct, int, chr, float]\n\n# We can also use complex(), list(), tuple(), set(), and dict() on things\n[i(a) for i in l]",
"_____no_output_____"
],
[
"# Control flow\na = 1\nb = 2\n\nif a < b:\n print('a < b')\nelif a > b:\n print('a > b')\nelse:\n print('else')\n \nwhile True:\n pass # does exactly nothing\n print('infinite loop?')\n break # also continue\n\nl = [1, 2, 3, 4]\nfor i in l: # a 'for each' loop similar to `for (auto i : seq)` in C++\n print(i)\n\nfor c in 'abcd':\n print(c)\n\n# multiple lists at once:\nfor i, j in zip('abc', 'xyz'): # will iterate over the shortest of the two lists\n print(i+j)\n \n# index and list at once:\nfor index, item in enumerate('abc'):\n print('index: {} item: {}'.format(index, item))",
"a < b\ninfinite loop?\n1\n2\n3\n4\na\nb\nc\nd\nax\nby\ncz\nindex: 0 item: a\nindex: 1 item: b\nindex: 2 item: c\n"
],
[
"# List comprehensions\nl = [i for i in range(10)]\nll = [i**2 for i in range(10)]\nlll = [i**2 for i in range(10) if i%2 != 0 ]\nllll = [i**2 if i%2 != 0 else i for i in range(10)]\n\na = 2 if False else 4 # Python ternary statement\nb = 2 if True else f\nprint(a)\nprint(b)\n\nc = [l, ll, lll, llll]\n[print(i) for i in c]",
"4\n2\n[0, 1, 2, 3, 4, 5, 6, 7, 8, 9]\n[0, 1, 4, 9, 16, 25, 36, 49, 64, 81]\n[1, 9, 25, 49, 81]\n[0, 1, 2, 9, 4, 25, 6, 49, 8, 81]\n"
],
[
"# functions\n\ndef func1(a=1, b=2):\n return a**b\n\nprint(func1(4, 2))\nprint(func1())\nprint(func1(a=4, b=2))\nprint(func1(4, b=2))\nprint(func1(b=2, a=4))\n\ndef func2(a=1, b=2):\n print(a**b)\n \nfunc2(4, 2)",
"16\n1\n16\n16\n16\n16\n"
],
[
"# Generators\n\ndef integers():\n \"\"\"Infinite sequence of integers starting at 10.\"\"\"\n i = 10\n while True:\n yield i\n i += 1\n\ngen = integers()\n \nfor i in gen:\n print(i)\n \n if i > 11:\n break\n \nprint('======')\n\nfor i in gen:\n print(i)\n \n if i > 14:\n break\n \nprint('======')\n\n# Generator expressions:\ngen1 = (i for i in range(6)) # redundant because how range() is implemented\n\nprint(next(gen1))\nprint(next(gen1))\nprint(next(gen1))\n\nprint('======')\n\nfor i in gen1:\n print(i)\n",
"10\n11\n12\n======\n13\n14\n15\n======\n0\n1\n2\n======\n3\n4\n5\n"
],
[
"# Strings\n\nmeat = \"\"\"Ground round fatback cow jerky porchetta hamburger shoulder short loin frankfurter salami short ribs corned beef pig shankle meatball. Chuck brisket pork loin shankle prosciutto swine. Tenderloin pastrami doner venison jowl jerky rump cow bacon leberkas capicola sausage pork ham hock. Ball tip salami ham brisket sausage. Rump ham hock shoulder sausage shank. Tri-tip ham venison, jerky landjaeger cow shank andouille alcatra shoulder. Flank salami spare ribs rump, ham hock t-bone pancetta strip steak.\nShankle sausage cow hamburger shoulder, fatback kevin chicken pork biltong porchetta jerky cupim jowl doner. Jerky pastrami alcatra rump pork belly tongue. Hamburger chuck bacon flank, pastrami bresaola alcatra turkey filet mignon pig brisket. Shank pork boudin tenderloin. Flank meatball ham, capicola hamburger landjaeger short loin beef ribs sausage. Chicken brisket turducken, bresaola pig swine hamburger shank rump ball tip venison fatback spare ribs bacon doner.\nKevin spare ribs pastrami tail capicola picanha meatball shankle short ribs drumstick jowl cow. Meatloaf capicola pastrami ball tip sausage short ribs cow turducken meatball. Andouille pork loin strip steak ribeye chicken turkey, cupim shank salami t-bone boudin tongue filet mignon. Sausage turkey tail chicken pig, spare ribs biltong fatback beef ribs. Shankle sirloin chuck meatloaf leberkas rump frankfurter, shoulder turducken brisket tongue porchetta filet mignon. Kevin fatback turkey meatloaf, pig pork chop andouille ball tip beef ribs beef chuck kielbasa short loin shoulder.\nCupim frankfurter biltong alcatra. Tail pork loin meatloaf brisket sirloin, tri-tip turkey drumstick alcatra kevin turducken chicken. Picanha bacon ground round strip steak. Fatback rump beef ribs frankfurter shoulder pork loin pig pastrami, ham hock cow. Fatback tongue short ribs bresaola salami chuck venison kevin strip steak sausage doner short loin drumstick.\nShank tri-tip boudin drumstick rump turducken jowl spare ribs. Pastrami doner picanha meatloaf beef ribs. Bresaola tongue leberkas turducken landjaeger pancetta swine cupim porchetta shank hamburger corned beef ball tip. Sirloin short ribs spare ribs, bresaola kielbasa rump jowl prosciutto boudin drumstick pork t-bone. Sausage landjaeger tenderloin jerky venison ribeye fatback chicken. Filet mignon leberkas shoulder andouille, biltong rump ribeye pancetta pork chop brisket spare ribs salami turducken.\nBoudin venison pancetta salami strip steak, cupim doner tongue ham hock turkey filet mignon rump biltong shank jerky. Kielbasa frankfurter cupim corned beef sirloin leberkas picanha beef ribs. Turducken beef ribs doner, ribeye venison drumstick beef jowl brisket filet mignon turkey. Ground round corned beef meatball capicola.\nFatback beef kevin meatball capicola porchetta. Short loin venison kielbasa shoulder chicken. Biltong pig corned beef fatback, capicola pancetta shankle pork belly tail shoulder ball tip swine cow tongue. Flank bacon rump pork loin tail brisket corned beef leberkas. Leberkas ground round beef doner corned beef pork loin kevin turkey picanha jowl filet mignon. Rump pastrami salami, chuck jowl biltong drumstick pork loin doner.\nPicanha pork chop chuck pork loin sirloin salami pig landjaeger venison ground round brisket jerky tri-tip swine. Prosciutto short loin porchetta shoulder brisket cupim corned beef jowl. Picanha chicken tri-tip pig tail, cupim short ribs ground round andouille pork belly sausage kevin ribeye. Sirloin picanha pastrami fatback tri-tip, meatloaf doner t-bone kevin bresaola.\nTurkey brisket filet mignon jerky biltong, prosciutto pork chop pork jowl picanha ball tip. Pastrami short loin sausage bresaola ham hock. Frankfurter filet mignon andouille shoulder. Prosciutto t-bone andouille ground round rump boudin capicola meatloaf spare ribs brisket. Pork loin turkey drumstick ham hock t-bone shankle. Bacon tail kielbasa pastrami, pork chop hamburger ground round turducken jowl strip steak.\nTri-tip filet mignon ham hock t-bone doner chuck biltong cupim bacon flank tail pastrami. Shoulder brisket pork chop, tail ham hock chuck doner filet mignon shank fatback rump prosciutto ribeye. Andouille jerky bacon chicken, tongue frankfurter ribeye spare ribs. Ham prosciutto meatball, drumstick biltong sausage strip steak andouille ground round fatback salami bacon jerky. Pig ham beef fatback prosciutto ground round tri-tip frankfurter landjaeger kielbasa.\nDrumstick biltong beef, shank pancetta pastrami cupim boudin. Tail sausage picanha tri-tip pork belly jerky. Drumstick chuck frankfurter prosciutto, picanha tri-tip filet mignon pig t-bone alcatra ground round bacon cow ham. Leberkas bresaola cupim shoulder tongue kevin. Chuck strip steak swine capicola short ribs, biltong bacon.\nStrip steak biltong drumstick pork loin tail. Cupim tongue pancetta biltong, prosciutto swine jowl brisket beef tri-tip ball tip tail venison doner. T-bone pig porchetta sirloin, ham hock venison tail salami pork chop pancetta shank boudin tenderloin fatback. Spare ribs shoulder hamburger meatloaf, beef turducken doner shankle pork chop. Beef ribs doner jowl, short loin short ribs tail drumstick porchetta ham hock flank ground round picanha turkey. Ball tip shoulder pastrami jowl turkey beef.\nPastrami bresaola biltong shank filet mignon spare ribs jerky jowl kevin picanha landjaeger frankfurter boudin sausage tenderloin. Kielbasa jowl short ribs, tri-tip doner picanha biltong alcatra pork loin. Short loin pork belly tail brisket. Capicola chuck biltong leberkas, doner porchetta cow jerky. Porchetta andouille ham shank ball tip capicola, jowl pancetta pork belly.\nShort loin alcatra swine chuck ham shoulder beef flank turkey landjaeger bacon pork belly. Pig pancetta short ribs capicola ground round beef, landjaeger pork belly porchetta swine meatloaf frankfurter bacon short loin. Chicken landjaeger fatback, ham strip steak pig kevin beef turducken pork loin meatball beef ribs pork chop pancetta doner. Leberkas porchetta spare ribs, alcatra doner filet mignon rump frankfurter beef. Alcatra ham salami sirloin tongue boudin jowl. Turkey short loin chuck, short ribs kevin ball tip shoulder. Picanha pig short ribs flank bacon rump strip steak pastrami short loin tongue pork pancetta kielbasa beef ribs bresaola.\nPork loin shoulder cow spare ribs. T-bone brisket flank, frankfurter ball tip ground round shankle porchetta tail tenderloin corned beef kevin short ribs. Cow shank beef ribs salami ball tip meatloaf porchetta cupim pastrami. Ham corned beef salami ham hock. Sausage bacon fatback beef ribs, drumstick andouille beef tail spare ribs venison kevin.\"\"\"",
"_____no_output_____"
],
[
"meat.replace('\\n', ' ')",
"_____no_output_____"
],
[
"meat.split('. ')",
"_____no_output_____"
],
[
"meat.splitlines()",
"_____no_output_____"
],
[
"meat = meat.replace('.', ' ')\nmeat = meat.replace(',', ' ')\nmeat = meat.replace('\\n', ' ')\nmeats = meat.split()\n\nmeats = [m.lower() for m in meats]",
"_____no_output_____"
],
[
"for m in set(meats): # a set is a data structure that contains no duplicates\n print('meat: {}\\n count: {}'.format(m, meats.count(m)))",
"meat: sausage\n count: 15\nmeat: ground\n count: 14\nmeat: tip\n count: 13\nmeat: tri-tip\n count: 12\nmeat: meatloaf\n count: 10\nmeat: loin\n count: 26\nmeat: hock\n count: 12\nmeat: brisket\n count: 17\nmeat: drumstick\n count: 14\nmeat: kielbasa\n count: 8\nmeat: belly\n count: 8\nmeat: kevin\n count: 15\nmeat: tongue\n count: 12\nmeat: bresaola\n count: 10\nmeat: filet\n count: 14\nmeat: hamburger\n count: 8\nmeat: porchetta\n count: 14\nmeat: shank\n count: 14\nmeat: steak\n count: 11\nmeat: ribeye\n count: 7\nmeat: cow\n count: 12\nmeat: sirloin\n count: 8\nmeat: pancetta\n count: 12\nmeat: ham\n count: 23\nmeat: capicola\n count: 12\nmeat: ribs\n count: 39\nmeat: jerky\n count: 14\nmeat: corned\n count: 10\nmeat: flank\n count: 9\nmeat: rump\n count: 17\nmeat: doner\n count: 19\nmeat: pastrami\n count: 17\nmeat: fatback\n count: 17\nmeat: chicken\n count: 10\nmeat: cupim\n count: 13\nmeat: chuck\n count: 14\nmeat: alcatra\n count: 10\nmeat: spare\n count: 14\nmeat: prosciutto\n count: 10\nmeat: swine\n count: 9\nmeat: leberkas\n count: 10\nmeat: turducken\n count: 11\nmeat: shankle\n count: 9\nmeat: frankfurter\n count: 13\nmeat: landjaeger\n count: 10\nmeat: round\n count: 14\nmeat: ball\n count: 13\nmeat: venison\n count: 12\nmeat: chop\n count: 9\nmeat: meatball\n count: 8\nmeat: t-bone\n count: 10\nmeat: turkey\n count: 14\nmeat: jowl\n count: 18\nmeat: short\n count: 26\nmeat: beef\n count: 36\nmeat: tail\n count: 17\nmeat: mignon\n count: 14\nmeat: shoulder\n count: 19\nmeat: bacon\n count: 15\nmeat: strip\n count: 11\nmeat: biltong\n count: 17\nmeat: pork\n count: 36\nmeat: boudin\n count: 10\nmeat: andouille\n count: 11\nmeat: picanha\n count: 15\nmeat: salami\n count: 14\nmeat: tenderloin\n count: 6\nmeat: pig\n count: 15\n"
],
[
"from collections import Counter\n\n# a dictionary is a hash table\ncounts = dict(Counter(meats)) # a better way\nprint(counts)",
"{'tri-tip': 12, 'ground': 14, 'meatloaf': 10, 'hock': 12, 'tip': 13, 'kielbasa': 8, 'filet': 14, 'drumstick': 14, 'hamburger': 8, 'porchetta': 14, 'shank': 14, 'pancetta': 12, 'jerky': 14, 'corned': 10, 'flank': 9, 'pastrami': 17, 'meatball': 8, 'rump': 17, 'alcatra': 10, 'spare': 14, 'swine': 9, 'leberkas': 10, 'shankle': 9, 'landjaeger': 10, 'round': 14, 'ball': 13, 'venison': 12, 'chicken': 10, 'jowl': 18, 'mignon': 14, 'shoulder': 19, 'strip': 11, 'pork': 36, 'picanha': 15, 'andouille': 11, 'tenderloin': 6, 'loin': 26, 'brisket': 17, 'beef': 36, 'belly': 8, 'kevin': 15, 'tongue': 12, 'sirloin': 8, 'ribeye': 7, 'cow': 12, 'ham': 23, 'capicola': 12, 'ribs': 39, 'bresaola': 10, 'doner': 19, 'fatback': 17, 'bacon': 15, 'cupim': 13, 'chuck': 14, 'prosciutto': 10, 'turducken': 11, 'frankfurter': 13, 'steak': 11, 'chop': 9, 't-bone': 10, 'turkey': 14, 'short': 26, 'tail': 17, 'sausage': 15, 'biltong': 17, 'boudin': 10, 'salami': 14, 'pig': 15}\n"
],
[
"# see https://docs.python.org/3/library/stdtypes.html#str for the whole list of string methods, they're super useful",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\n\n%matplotlib inline\n\ndistribution = list(counts.values())",
"_____no_output_____"
],
[
"sns.distplot(distribution) # meatsribution",
"_____no_output_____"
],
[
"sns.boxplot(distribution)",
"_____no_output_____"
],
[
"import numpy as np\nfrom time import time\n\nnp.random.seed(int(time()))\n\nx = np.linspace(start=0, stop=1.5*np.pi, num=150)\ny = 2*np.sin(np.pi*x) + 2*np.sin(x**2) + 3*np.random.rand(4, 1)\nsns.tsplot(y)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71cd6147ca9e75e7571eae64a197317014ed16f | 6,068 | ipynb | Jupyter Notebook | notes/03-conservation-laws/angular-momentum/example-barbell.ipynb | CapaFenLisesi/PHY3110 | 75375dda6aeb74e6ae40dfbbd2a6b44f6c76bab1 | [
"MIT"
] | null | null | null | notes/03-conservation-laws/angular-momentum/example-barbell.ipynb | CapaFenLisesi/PHY3110 | 75375dda6aeb74e6ae40dfbbd2a6b44f6c76bab1 | [
"MIT"
] | null | null | null | notes/03-conservation-laws/angular-momentum/example-barbell.ipynb | CapaFenLisesi/PHY3110 | 75375dda6aeb74e6ae40dfbbd2a6b44f6c76bab1 | [
"MIT"
] | 1 | 2018-07-17T16:39:44.000Z | 2018-07-17T16:39:44.000Z | 28.622642 | 169 | 0.493078 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e71ce4d3397ac701473316a5eda8ebc6157004a4 | 9,693 | ipynb | Jupyter Notebook | vanilla/RNN-Impl.ipynb | dbz10/rnn-impls | 1077671cb19d0dec4844b73671c53521d92f3ce7 | [
"MIT"
] | null | null | null | vanilla/RNN-Impl.ipynb | dbz10/rnn-impls | 1077671cb19d0dec4844b73671c53521d92f3ce7 | [
"MIT"
] | null | null | null | vanilla/RNN-Impl.ipynb | dbz10/rnn-impls | 1077671cb19d0dec4844b73671c53521d92f3ce7 | [
"MIT"
] | null | null | null | 30.009288 | 653 | 0.595481 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import preprocessing\nfrom vanilla_rnn import RNN\nfrom dio import load_data\nimport numpy as np\nimport joblib",
"_____no_output_____"
],
[
"snippet = load_data('../data/silmarillion.txt')",
"_____no_output_____"
]
],
[
[
"The data set consists of a list of lines which tells the story of the high elves struggle against Morgoth and his forces.",
"_____no_output_____"
]
],
[
[
"snippet[:3]",
"_____no_output_____"
],
[
"vocabulary = preprocessing.create_vocabulary(snippet)",
"_____no_output_____"
],
[
"vocabulary['vocab_size']",
"_____no_output_____"
]
],
[
[
"The vocab size here is relatively large for a couple of reasons\n\n1. There's a lot of unconvential characters like Ë and ú\n2. It has both uppercase and lower case letters.\n\nThe first is a feature, the second definitely makes life unecessarily difficult for the model since it has to learn upper case letter separate from lower case letters. It is unlikely that there's enough data here to facilitate that.",
"_____no_output_____"
],
[
"The character level RNN model is trained on (input, target) sequences that are constructed out of a sequence of text as follows:",
"_____no_output_____"
]
],
[
[
"# Split into train test and validation set while preserving individual lines intact.\ninput_lines, test_lines = preprocessing.train_test_split(snippet, test_size=0.1)\ntrain_lines, val_lines = preprocessing.train_test_split(input_lines, test_size=0.1)\n\nX_train, y_train = preprocessing.text_to_input_and_target(train_lines)\nX_val, y_val = preprocessing.text_to_input_and_target(val_lines)\nX_test, y_test = preprocessing.text_to_input_and_target(test_lines)\n",
"_____no_output_____"
],
[
"print(f\"'{X_test[:20]}'\", \"gets transformed into the following sequence of inputs and outputs: \\n\")\nprint([(input, target) for (input, target) in zip(X_test[:20],y_test[:20])])",
"'Then Turgon answered' gets transformed into the following sequence of inputs and outputs: \n\n[('T', 'h'), ('h', 'e'), ('e', 'n'), ('n', ' '), (' ', 'T'), ('T', 'u'), ('u', 'r'), ('r', 'g'), ('g', 'o'), ('o', 'n'), ('n', ' '), (' ', 'a'), ('a', 'n'), ('n', 's'), ('s', 'w'), ('w', 'e'), ('e', 'r'), ('r', 'e'), ('e', 'd'), ('d', ':')]\n"
]
],
[
[
"I already trained a model consisting of a single layer RNN with a hidden state size of 200 on the full text of the Silmarillion, various checkpoints are saved in `model_checkpoints`. For now we can just load one and have some fun.",
"_____no_output_____"
]
],
[
[
"network = joblib.load('model_checkpoints/model_40_1.393801855652784.joblib')",
"_____no_output_____"
]
],
[
[
"We can ask the RNN to start inventing some silmarillion like text. The \"temperature\" parameter used to sample text is quite important - at $\\beta = 1$ the text is more or less gibberish, although it must be said that it's very Tolkein-esque gibberish. ",
"_____no_output_____"
]
],
[
[
"''.join(network.sample(\" \", 1, 200))",
"_____no_output_____"
]
],
[
[
"At lower temperatures (higher $\\beta$) the model is more conservative and it produces a sequence consisting mostly of actual words. I like the part about how the house of Indwalt became the house of Indwald. ",
"_____no_output_____"
]
],
[
[
"''.join(network.sample(\" \", 3, 200))",
"_____no_output_____"
]
],
[
[
"We can also prime the RNN by feeding it a sequence of text to consume. This has the effect of setting up a specific hidden state prior to starting sampling. Again, the temperature is an important parameter in generating samples from the RNN.",
"_____no_output_____"
]
],
[
[
"prompt, _ = preprocessing.text_to_input_and_target(load_data('sample_input.txt'))",
"_____no_output_____"
],
[
"prompt",
"_____no_output_____"
],
[
"''.join(network.sample(prompt, 1, 200))",
"_____no_output_____"
],
[
"''.join(network.sample(prompt, 3, 200))",
"_____no_output_____"
]
],
[
[
"Hey that second generated line looks pretty reasonable! The RNN knows about the Noldor, love of the stars, and their fateful march to the west (with the many perils that followed). ",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e71cea73fd671176d4839279f99afe87e4e58e11 | 22,011 | ipynb | Jupyter Notebook | Chapter07/notebooks/user_churn_restricting.ipynb | balibaa/Practical-Data-Analysis-using-Jupyter-Notebook | 71f7892642a566444c1bff8a7bc80f717a269bf0 | [
"MIT"
] | 37 | 2020-06-18T14:55:41.000Z | 2022-03-11T19:14:44.000Z | Chapter07/notebooks/user_churn_restricting.ipynb | balibaa/Practical-Data-Analysis-using-Jupyter-Notebook | 71f7892642a566444c1bff8a7bc80f717a269bf0 | [
"MIT"
] | 1 | 2020-05-15T00:01:07.000Z | 2020-05-15T00:01:07.000Z | Chapter07/notebooks/user_churn_restricting.ipynb | balibaa/Practical-Data-Analysis-using-Jupyter-Notebook | 71f7892642a566444c1bff8a7bc80f717a269bf0 | [
"MIT"
] | 56 | 2020-06-26T09:33:29.000Z | 2022-03-16T05:16:11.000Z | 33.967593 | 1,427 | 0.459952 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df_user_churn = pd.read_csv('user_hits_export.csv', header=0, names=['userid','date'])",
"_____no_output_____"
],
[
"df_user_churn",
"_____no_output_____"
],
[
"df_user_restricted = df_user_churn[df_user_churn['userid']==1]",
"_____no_output_____"
],
[
"df_user_restricted.head()",
"_____no_output_____"
],
[
"df_user_restricted.sort_values(by='date')",
"_____no_output_____"
],
[
" df_user_restricted.sort_values(by='date', ascending=False)",
"_____no_output_____"
],
[
"df_usage_patterns = df_user_churn",
"_____no_output_____"
],
[
"import datetime\nimport numpy as np",
"_____no_output_____"
],
[
"now = pd.to_datetime('now')",
"_____no_output_____"
],
[
"df_usage_patterns['age'] = now - df_usage_patterns['date']",
"_____no_output_____"
],
[
"df_usage_patterns = pd.read_csv('user_hits_export.csv', header=0, names=['userid','date'], parse_dates=True)",
"_____no_output_____"
],
[
"df_usage_patterns.info()",
"<class 'pandas.core.frame.DataFrame'>\nIndex: 12 entries, 0 to 11\nData columns (total 2 columns):\nuserid 9 non-null float64\ndate 12 non-null object\ndtypes: float64(1), object(1)\nmemory usage: 288.0+ bytes\n"
],
[
"pd.isnull(df_usage_patterns)",
"_____no_output_____"
],
[
"df_user_churn_cleaned = df_user_churn.dropna()",
"_____no_output_____"
],
[
"df_user_churn_cleaned.head(10)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71cf1c6552fccb58816216e0ab8badc3a13d29c | 228,076 | ipynb | Jupyter Notebook | Stock_Analysis/Descriptive_Statistics.ipynb | Evans-colon/Time_Series_Analysis_Forcasting | cf7229d8558013ffcff0780b5ba7a2e2b95d7253 | [
"MIT"
] | null | null | null | Stock_Analysis/Descriptive_Statistics.ipynb | Evans-colon/Time_Series_Analysis_Forcasting | cf7229d8558013ffcff0780b5ba7a2e2b95d7253 | [
"MIT"
] | null | null | null | Stock_Analysis/Descriptive_Statistics.ipynb | Evans-colon/Time_Series_Analysis_Forcasting | cf7229d8558013ffcff0780b5ba7a2e2b95d7253 | [
"MIT"
] | null | null | null | 331.024673 | 156,342 | 0.916388 | [
[
[
"import pandas as pd\nimport numpy as np\nimport yfinance as yf\nimport seaborn as sns\nimport scipy.stats as scs\nimport datetime as dt\nimport seaborn as sns\nimport statsmodels.api as sm\nimport statsmodels.tsa.api as smt\nimport matplotlib.pyplot as plt\nfrom scipy import stats\nfrom scipy.stats import describe",
"_____no_output_____"
],
[
"#Download the prices\nsymbol = 'GOOGL'\nstart = dt.date.today() - dt.timedelta(days = 365*10)\nend = dt.date.today()\ndf_google = yf.download(symbol,start,end)\ndf_google.head()\ndf_google.head()",
"[*********************100%***********************] 1 of 1 completed\n"
],
[
"#Calculate the simple and log returns using the adjusted close prices\ndf_google.rename(columns={'Adj Close':'adj_close'}, inplace=True)\ndf_google['simple_return'] = df_google.adj_close.pct_change()\ndf_google['log_return'] = np.log(df_google.adj_close/df_google.adj_close.shift(1))\n\ndf_google.head()",
"_____no_output_____"
],
[
"df_google.describe()",
"_____no_output_____"
]
],
[
[
"Stylized facts are statistical properties that appear to be present in many empirical asset\nreturns (across time and markets).",
"_____no_output_____"
],
[
"Calculate the normal probability density function(PDF) using the mean and standard deviation of the observed returns",
"_____no_output_____"
]
],
[
[
"r_range =np.linspace(min(df_google.log_return), max(df_google.log_return))\nmu = df_google.log_return.mean()\nsigma = df_google.log_return.std()\nnorm_pdf = scs.norm.pdf(r_range, loc=mu, scale=sigma)\n",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 2, figsize=(16, 8))\n\n#Histogram\nsns.distplot(df_google.log_return, kde=False, norm_hist=True, ax=ax[0])\nax[0].set_title('Distribution of GOOGLE returns', fontsize = 16)\nax[0].plot(r_range, norm_pdf, 'g', lw=2,\n label=f'N({mu:.2f}, {sigma**2:.4f})')\nax[0].legend(loc='upper left');\n\n#Q-Q plot\nqq = sm.qqplot(df_google.log_return.values, line='s', ax=ax[1])\nax[1].set_title('Q-Q plot', fontsize=16)",
"c:\\Users\\USER\\projects_env\\time_series\\lib\\site-packages\\seaborn\\distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"#Volatility clustering\ndf_google.log_return.plot(title='Daily GOOGLE returns')",
"_____no_output_____"
]
],
[
[
"We can observe clear clusters of volatility—periods of higher positive and\nnegative returns",
"_____no_output_____"
],
[
"Leverage effect\n\nCalculate volatility measures as rolling standard deviations",
"_____no_output_____"
]
],
[
[
"df_google['moving_std_252'] = df_google[['log_return']].rolling(window=252).std()\ndf_google['moving_std_21'] = df_google[['log_return']].rolling(window=21).std()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(3, 1, figsize=(18, 15),\n sharex=True)\n\ndf_google.adj_close.plot(ax=ax[0])\nax[0].set(title='GOOGLE time series',\n ylabel='Stock price ($)')\n\ndf_google.log_return.plot(ax=ax[1])\nax[1].set(ylabel='Log returns (%)')\n\ndf_google.moving_std_252.plot(ax=ax[2], color='r',\n label='Moving Volatility 252d')\n\n\ndf_google.moving_std_21.plot(ax=ax[2], color='g',\nlabel='Moving Volatility 21d')\n\nax[2].set(ylabel='Moving Volatility',\n xlabel='Date')\nax[2].legend()",
"_____no_output_____"
]
],
[
[
"This fact states that most measures of an asset's volatility are negatively correlated\nwith its returns, and we can indeed observe a pattern of increased volatility when\nthe prices go down and decreased volatility when they are rising.\nIt also states that most measures of asset volatility are negatively correlated with their returns. To investigate it, we used the moving standard deviation (calculated using the rolling method of a pandas DataFrame) as a measure of historical volatility. We used\nwindows of 21 and 252 days, which correspond to one month and one year of trading data",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e71d156fa6bfad3bccf0f9041797dece70acfd62 | 11,167 | ipynb | Jupyter Notebook | Docs/Prashanth/.ipynb_checkpoints/Export-checkpoint.ipynb | infapy/infapy | 0cb11310130be70ce1b647aa5ede929c1eb9b2ce | [
"Apache-2.0"
] | null | null | null | Docs/Prashanth/.ipynb_checkpoints/Export-checkpoint.ipynb | infapy/infapy | 0cb11310130be70ce1b647aa5ede929c1eb9b2ce | [
"Apache-2.0"
] | null | null | null | Docs/Prashanth/.ipynb_checkpoints/Export-checkpoint.ipynb | infapy/infapy | 0cb11310130be70ce1b647aa5ede929c1eb9b2ce | [
"Apache-2.0"
] | 1 | 2021-09-23T10:31:56.000Z | 2021-09-23T10:31:56.000Z | 34.045732 | 1,146 | 0.582699 | [
[
[
"# Export",
"_____no_output_____"
],
[
"Use this resource with the import resource to migrate objects from one organization to another.",
"_____no_output_____"
],
[
"## Prerequisite - Identify the object ids",
"_____no_output_____"
],
[
"The first step in performing the migration is to identify the object id of the object you are trying to migrate. In this example, we will be using a test mapping task which we will be migrating.",
"_____no_output_____"
],
[
"### Example code to identify the object id of the mapping task we are trying to migrate",
"_____no_output_____"
]
],
[
[
"import infapy\n\ninfapy.setFileLogger(name=\"test\",level=\"DEBUG\")\ninfaHandler = infapy.connect(profile=\"DEV\")\n\nv3 = infaHandler.v3()\nmappingTaskDetails = v3.lookup(path=\"infapy/mt_infapy_test\",objectType=\"MTT\")\nprint(\"Mapping Task Details: \" + str(mappingTaskDetails))\nmappingTaskID = mappingTaskDetails[\"objects\"][0][\"id\"]\nprint(\"Mapping Task ID: \" + str(mappingTaskID))\n",
"Mapping Task Details: {'objects': [{'id': '1QcjcQ9hXwwcqW1P93354V', 'path': 'infapy/mt_infapy_test', 'type': 'MTT', 'description': '', 'updatedBy': 'prashanth-p', 'updateTime': '2021-09-26T08:41:04Z'}]}\nMapping Task ID: 1QcjcQ9hXwwcqW1P93354V\n"
]
],
[
[
"## Function: startExport()",
"_____no_output_____"
],
[
"> Used to export an object in IICS\n> This function initiates the export operation\n>\n> Args:\n> name (String): Name of your export job\n> id (String): This is the object id which you want to export\n> use the lookup function or object function to get the object id\n>\n> includeDependencies (bool, optional): If you want to include dependencies. Defaults to True.\n>\n> Returns:\n> json: confirmation or failure response from export operation",
"_____no_output_____"
],
[
"### Example Code to start the export operation",
"_____no_output_____"
]
],
[
[
"# name is just an identifier for your export operation\nexportObj = v3.exportObject()\nresponse = exportObj.startExport(name=\"MyTestJobFromPython\",ObjectId=\"1QcjcQ9hXwwcqW1P93354V\")\nprint(response)\nexportID = response[\"id\"]\nprint(\"Export Name: MyTestJobFromPython\")\nprint(\"Export ID \" + exportID)",
"{'id': '9SczgYW6zyufJkDB6Uaduz', 'createTime': '2021-09-26T09:06:38.898Z', 'updateTime': '2021-09-26T09:06:38.898Z', 'name': 'MyTestJobFromPython', 'startTime': '2021-09-26T09:06:38.803Z', 'endTime': None, 'status': {'state': 'IN_PROGRESS', 'message': 'In Progress'}, 'objects': None}\nExport Name: MyTestJobFromPython\nExport ID 9SczgYW6zyufJkDB6Uaduz\n"
]
],
[
[
"## Function: getStatusOfExportByExportID()",
"_____no_output_____"
],
[
"> use this method to get the status of the export\n> if it is a success or a failure\n>\n> Args:\n> exportID (exportID): provide the export id you recieved\n> from startExport Method used before this\n>\n> Returns:\n> json: Export operation status",
"_____no_output_____"
],
[
"### Example code to get the status of export",
"_____no_output_____"
]
],
[
[
"# exportObj variable is defined above\n# use the export id from the above code\n\nresponse = exportObj.getStatusOfExportByExportID(exportID=\"9SczgYW6zyufJkDB6Uaduz\")\nprint(response)",
"{'id': '9SczgYW6zyufJkDB6Uaduz', 'createTime': '2021-09-26T09:06:39.000Z', 'updateTime': '2021-09-26T09:06:40.000Z', 'name': 'MyTestJobFromPython', 'startTime': '2021-09-26T09:06:39.000Z', 'endTime': '2021-09-26T09:06:40.000Z', 'status': {'state': 'SUCCESSFUL', 'message': 'Export completed successfully'}, 'objects': [{'id': '1QcjcQ9hXwwcqW1P93354V', 'name': 'mt_infapy_test', 'path': '/infapy', 'type': 'MTT', 'description': '', 'status': {'state': 'SUCCESSFUL', 'message': None}}, {'id': '3vVj4xdOpKsgAqwRSyhQM3', 'name': 'm_infapy_test', 'path': '/infapy', 'type': 'DTEMPLATE', 'description': '', 'status': {'state': 'SUCCESSFUL', 'message': None}}, {'id': '848Au1yuOzAcdxJMgPkdqy', 'name': '__ff', 'path': None, 'type': 'Connection', 'description': None, 'status': {'state': 'SUCCESSFUL', 'message': None}}, {'id': '95OeUg6sjYVhH6zxQUB76k', 'name': 'prashanth-sbx', 'path': None, 'type': 'AgentGroup', 'description': None, 'status': {'state': 'SUCCESSFUL', 'message': None}}, {'id': 'aeOwF2U4Uauf5fdiFwaLCz', 'name': 'infapy', 'path': '/', 'type': 'Project', 'description': '', 'status': {'state': 'SUCCESSFUL', 'message': None}}]}\n"
]
],
[
[
"## Function: getExportLogsByExportID()",
"_____no_output_____"
],
[
"> use this method to get the export\n> logs\n>\n> Args:\n> exportID (exportID): provide the export id you recieved\n> from startExport Method used before this\n>\n> Returns:\n> string text: Export logs in text",
"_____no_output_____"
],
[
"### Example code to get exportLogsByExportID()",
"_____no_output_____"
]
],
[
[
"# exportObj variable is defined above\n# use the export id from the above code\n\nresponse = exportObj.getExportLogsByExportID(exportID=\"9SczgYW6zyufJkDB6Uaduz\")\nprint(response)",
"> OIE_002 INFO 2021-09-26T09:06:39.017Z Starting export operation.\nExecution Client: API\nJob Name: MyTestJobFromPython\nOrganization: GCS IICS\nRequestId: 9SczgYW6zyufJkDB6Uaduz\nUser: prashanth-p\n> OIE_004 INFO 2021-09-26T09:06:39.139Z Successfully exported object [/Explore/infapy] of type [Project] id [aeOwF2U4Uauf5fdiFwaLCz]\n> OIE_004 INFO 2021-09-26T09:06:39.139Z Successfully exported object [/SYS/_SYSTEM_PROJECT] of type [Project] id [hrRCl2TfTa1jvmKBfGC4Il]\n> OIE_004 INFO 2021-09-26T09:06:39.237Z Successfully exported object [/SYS/_SYSTEM_FOLDER] of type [Folder] id [cfvpgEOpQmhlh1qOwkFkkc]\n> OIE_004 INFO 2021-09-26T09:06:39.463Z Successfully exported object [/SYS/prashanth-sbx] of type [AgentGroup] id [95OeUg6sjYVhH6zxQUB76k]\n> OIE_004 INFO 2021-09-26T09:06:39.592Z Successfully exported object [/SYS/__ff] of type [Connection] id [848Au1yuOzAcdxJMgPkdqy]\n> OIE_004 INFO 2021-09-26T09:06:39.721Z Successfully exported object [/Explore/infapy/m_infapy_test] of type [DTEMPLATE] id [3vVj4xdOpKsgAqwRSyhQM3]\n> OIE_004 INFO 2021-09-26T09:06:39.943Z Successfully exported object [/Explore/infapy/mt_infapy_test] of type [MTT] id [1QcjcQ9hXwwcqW1P93354V]\n> OIE_003 INFO 2021-09-26T09:06:39.943Z Finished export operation.\nJob Name: MyTestJobFromPython\nStart Time: 9/26/21 9:06 AM\nEnd Time: 9/26/21 9:06 AM\nStarted by: prashanth-p\nStart Method: API\nSource Organization: GCS IICS\nStatus: SUCCESSFUL\n\n"
]
],
[
[
"## Function: getExportZipFileByExportID()",
"_____no_output_____"
],
[
"This is the final step in the export operation. After the above export was successful. We now need to download the package to our local computer / server, so the package can then be imported to QA or higher environments.\n\n> Use this method to download the export object as a zip file\n> 1. startExport()\n> 2. getStatusOfExportByExportID()\n> 3. getExportZipFileByExportID()\n> 4. getExportLogsByExportID()\n>\n> Args:\n> exportID (String): You recieve this id when you startExport()\n> filePath (String, optional): Path to download the object. Defaults to os.getcwd().\n> fileName (str, optional): exportObjectName.zip. Defaults to \"infapyExportDownloaded.zip\".\n>\n> Returns:\n> zipfile: downloaded to filepath/filename",
"_____no_output_____"
]
],
[
[
"# exportObj variable is defined above\n# use the export id from the above code\n# By default the zip file - \"infapyExportDownloaded.zip\" will get downloaded to your current working directory\n\nresponse = exportObj.getExportZipFileByExportID(exportID=\"9SczgYW6zyufJkDB6Uaduz\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e71d1cff53a3ffc133a0c15b5181024417acdca0 | 12,262 | ipynb | Jupyter Notebook | umaine_to_cf16.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
] | 32 | 2015-01-07T01:48:05.000Z | 2022-03-02T07:07:42.000Z | umaine_to_cf16.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
] | 1 | 2015-04-13T21:00:18.000Z | 2015-04-13T21:00:18.000Z | umaine_to_cf16.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
] | 30 | 2015-01-28T09:31:29.000Z | 2022-03-07T03:08:28.000Z | 26.483801 | 203 | 0.486544 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e71d1ec6715e504ec84c1adb3e71493dd2e0652c | 134,245 | ipynb | Jupyter Notebook | Taller 3 - Algebra Lineal/1_Lineal_Algebra_general_concepts.ipynb | deiry/DataScienceFEM | f579e54aad579e35f80656569781e39c2696e8ea | [
"MIT"
] | null | null | null | Taller 3 - Algebra Lineal/1_Lineal_Algebra_general_concepts.ipynb | deiry/DataScienceFEM | f579e54aad579e35f80656569781e39c2696e8ea | [
"MIT"
] | null | null | null | Taller 3 - Algebra Lineal/1_Lineal_Algebra_general_concepts.ipynb | deiry/DataScienceFEM | f579e54aad579e35f80656569781e39c2696e8ea | [
"MIT"
] | null | null | null | 193.436599 | 108,112 | 0.910432 | [
[
[
"<a href=\"https://colab.research.google.com/github/salvarezmeneses/DataScience2020/blob/master/1_Lineal_Algebra_general_concepts.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# LINEAL ALGEBRA.\n\n**Algebra Lineal:** It is the branch of mathematics that studies the operations between matrices and vectors.\n\n\n### Applications Requiered.\n\n- Jupyter Notebooks with Python 3.7.\n- Google Collaboraty.",
"_____no_output_____"
],
[
"# ARRAYS CLASSIFICATION\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# SCALARS, VECTORS, MATRIX AND TENSOR.\n\n* **Scalars:** A scalar in math is nothing more than a number.\nbut in python a scalar can be several things (integer, floating point, complex number, string, null type, boolean, etc)\n\n",
"_____no_output_____"
]
],
[
[
"# Scalares en python\na = 9\nb = 4.8\nc = 7.435\nprint(a) ",
"9\n"
]
],
[
[
"* **Vector**:(array) It is a set of numbers.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nvector = np.array([9,5,6,2,4,8,3]) # vector creation \nprint(vector) ",
"[9 5 6 2 4 8 3]\n"
]
],
[
[
" \n\n\n\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# Matriz\nmatriz = np.array([[9,5,7,2,4,8,3],[9,4,6,2,4,8,3],[9,1,6,2,4,8,3]])\nprint(matriz)",
"[[9 5 7 2 4 8 3]\n [9 4 6 2 4 8 3]\n [9 1 6 2 4 8 3]]\n"
]
],
[
[
"**Tensor**: It is the set of matrices. \n\n* What can be represented with a tensor?\n\n* If with a matrix it can represent an image, with a tensor I can represent a video.\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"tensor = np.array([[[255,255,255],[128,128,128],[80,80,80]],\n [[100,101,102],[103,104,105],[160,170,180]],\n [[190,200,210], [220,230,204], [255,227,238]]])\n\nprint(tensor)",
"[[[255 255 255]\n [128 128 128]\n [ 80 80 80]]\n\n [[100 101 102]\n [103 104 105]\n [160 170 180]]\n\n [[190 200 210]\n [220 230 204]\n [255 227 238]]]\n"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\nmethods = [None, 'none', 'nearest', 'bilinear', 'bicubic', 'spline16',\n 'spline36', 'hanning', 'hamming', 'hermite', 'kaiser', 'quadric',\n 'catrom', 'gaussian', 'bessel', 'mitchell', 'sinc', 'lanczos']\n\n# Fixing random state for reproducibility\nnp.random.seed(19680801)\n\n#grid = np.random.rand(4, 4)\n\nfig, axs = plt.subplots(nrows=3, ncols=6, figsize=(9, 6),\n subplot_kw={'xticks': [], 'yticks': []})\n\nfor ax, interp_method in zip(axs.flat, methods):\n ax.imshow(tensor, interpolation=interp_method, cmap='viridis')\n ax.set_title(str(interp_method))\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt # pyplot to make the graphics\n# imshow(to show the image like tensor)\nplt.imshow(tensor, interpolation='nearest') \n# show 0(black), 255(white).\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Practice 1.\n\n* Generate the scalar number 42.\n* Generate a vector containing 2, 3, 5, 7\n* Generate a 3 x 2 matrix\n* Generate a tensor that has in its first white row, second black and third gray.",
"_____no_output_____"
]
],
[
[
"# Practice 1\nscalar = 42\nprint(\"1. Generate scalar\",scalar)\n\nvector = np.array([2,3,5,7])\nprint(\"\\n2. Generate vector\",vector)\n\nmatrix = np.random.rand(3, 2)+1\nprint(\"\\n3. Generate matrix\")\nprint(matrix)\n\nwhite = np.ones((3,3), dtype=int)*255\nblack = np.zeros((3,3), dtype=int)\ngray = np.ones((3,3),dtype=int)*128\n\n\ntensor2 = np.array([white,black,gray])\n#tensor2 = tensor2.astype('uint8')\n\n\ntensor = np.array([[[255,255,255],\n [255,255,255],\n [255,255,255]],\n [[0,0,0],\n [0,0,0],\n [0,0,0]],\n [[128,128,128],\n [128,128,128],\n [128,128,128]]])\nprint(\"\\n4. Generate tensor\")\n\nprint(tensor2)\n\n\nplt.imshow(tensor2)\nplt.tight_layout()",
"1. Generate scalar 42\n\n2. Generate vector [2 3 5 7]\n\n3. Generate matrix\n[[1.88722812 1.11349096]\n [1.08942799 1.07520633]\n [1.58867013 1.90696054]]\n\n4. Generate tensor\n[[[255 255 255]\n [255 255 255]\n [255 255 255]]\n\n [[ 0 0 0]\n [ 0 0 0]\n [ 0 0 0]]\n\n [[128 128 128]\n [128 128 128]\n [128 128 128]]]\n"
]
],
[
[
"\nPractice 1: double click __Here__ for Solution.\n\n<!-- Your answer is below:\nescalar = 42\nprint(\"Escalar: \", escalar)\n\nvector = np.array([2,3,5,7])\nprint(\"\\nVector: \",vector)\n\nmatriz = np.array([[1,2,3], [4, 5, 6], [7, 8, 9]])\nprint(\"\\nMatriz: \",matriz)\n\ntensor = np.array([\n [[255, 255, 255], [255, 255, 255], [255, 255, 255]],\n [[0, 0, 0], [0, 0, 0], [0, 0 ,0]],\n [[128, 128, 128], [128, 128, 128], [128, 128, 128]],\n \n])\n\nprint(\"\\nTensor: \")\nplt.imshow(tensor, interpolation='nearest')\nplt.show()\n-->",
"_____no_output_____"
],
[
"# DIMENSION AND SIZE\n\n# Dimension of matrix , tensor \n\n**Matrix Dimension:** \n* the number of rows by the number of columns its called Matrix Dimension. if a Matrix have a rows and b columns, is a x b matrix .\n\n**Tensor Dimension:**\n* Tensor has 3 dimensions (depth, rows, columns.)\n\n\n",
"_____no_output_____"
],
[
"### Shape Funtion.\n\n* The shape function returns the dimensions (rows x columns) of the object.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nmatriz_A = np.array([[1,2,3], [4, 5, 6], [7, 8, 9]])\nmatriz_A.shape",
"_____no_output_____"
],
[
"v = np.array([8,7,5,9])\nv = np.reshape(v,(v.shape[0],1))\nv.shape",
"_____no_output_____"
]
],
[
[
"## size funcion\n\n* The size function returns the size (the number of elements).",
"_____no_output_____"
]
],
[
[
"import numpy as np\nmatriz_A = np.array([[1,2,3], [4, 5, 6], [7, 8, 9]])\nmatriz_A.size",
"_____no_output_____"
]
],
[
[
"## Shape and size function - Tensors\n\n* in tensor .shape will show us rows x columns x depth.\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ntensor2 = np.array([\n [[255, 255, 255], [255, 255, 255], [255, 255, 255]],\n [[0, 0, 0], [0, 0, 0], [0, 0 ,0]],\n [[128, 128, 128], [128, 128, 128], [128, 128, 128]],\n \n])",
"_____no_output_____"
],
[
"\nprint(tensor2.shape) ",
"(3, 3, 3)\n"
],
[
"print(tensor2.size)",
"27\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e71d265bd2d6a37635d91bb67f9f13d166a5a612 | 30,437 | ipynb | Jupyter Notebook | jupyter/topic04_linear_models/topic4_linear_models_part2_logit_likelihood_learning.ipynb | ivan-magda/mlcourse_open_homeworks | bc67fe6b872655e8e5628ec14b01fde407c5eb3c | [
"MIT"
] | 1 | 2018-10-24T08:35:29.000Z | 2018-10-24T08:35:29.000Z | jupyter/topic04_linear_models/topic4_linear_models_part2_logit_likelihood_learning.ipynb | ivan-magda/mlcourse_open_homeworks | bc67fe6b872655e8e5628ec14b01fde407c5eb3c | [
"MIT"
] | null | null | null | jupyter/topic04_linear_models/topic4_linear_models_part2_logit_likelihood_learning.ipynb | ivan-magda/mlcourse_open_homeworks | bc67fe6b872655e8e5628ec14b01fde407c5eb3c | [
"MIT"
] | 3 | 2019-10-03T22:32:24.000Z | 2021-01-13T10:09:22.000Z | 96.015773 | 13,784 | 0.779742 | [
[
[
"<center>\n<img src=\"../../img/ods_stickers.jpg\">\n## Открытый курс по машинному обучению\n<center>Автор материала: Юрий Кашницкий\n \nМатериал распространяется на условиях лицензии [Creative Commons CC BY-NC-SA 4.0](https://creativecommons.org/licenses/by-nc-sa/4.0/). Можно использовать в любых целях (редактировать, поправлять и брать за основу), кроме коммерческих, но с обязательным упоминанием автора материала.",
"_____no_output_____"
],
[
"# <center>Тема 4. Линейные модели классификации и регрессии\n## <center>Часть 2. Логистическая регрессия и метод максимального правдоподобия ",
"_____no_output_____"
],
[
"### Линейный классификатор",
"_____no_output_____"
],
[
"Основная идея линейного классификатора заключается в том, что признаковое пространство может быть разделено гиперплоскостью на две полуплоскости, в каждой из которых прогнозируется одно из двух значений целевого класса. \nЕсли это можно сделать без ошибок, то обучающая выборка называется *линейно разделимой*.\n\n<img src=\"../../img/logit.png\">\n\nМы уже знакомы с линейной регрессией и методом наименьших квадратов. Рассмотрим задачу бинарной классификации, причем метки целевого класса обозначим \"+1\" (положительные примеры) и \"-1\" (отрицательные примеры).\nОдин из самых простых линейных классификаторов получается на основе регрессии вот таким образом:\n\n$$\\Large a(\\textbf{x}) = \\text{sign}(\\textbf{w}^{\\text{T}}\\textbf x),$$\n\nгде\n - $\\textbf{x}$ – вектор признаков примера (вместе с единицей);\n - $\\textbf{w}$ – веса в линейной модели (вместе со смещением $w_0$);\n - $\\text{sign}(\\bullet)$ – функция \"сигнум\", возвращающая знак своего аргумента;\n - $a(\\textbf{x})$ – ответ классификатора на примере $\\textbf{x}$.\n\n",
"_____no_output_____"
],
[
"### Логистическая регрессия как линейный классификатор",
"_____no_output_____"
],
[
"Логистическая регрессия является частным случаем линейного классификатора, но она обладает хорошим \"умением\" – прогнозировать вероятность $p_+$ отнесения примера $\\textbf{x}_\\text{i}$ к классу \"+\":\n$$\\Large p_+ = \\text P\\left(y_i = 1 \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right) $$\n\nПрогнозирование не просто ответа (\"+1\" или \"-1\"), а именно *вероятности* отнесения к классу \"+1\" во многих задачах является очень важным бизнес-требованием. Например, в задаче кредитного скоринга, где традиционно применяется логистическая регрессия, часто прогнозируют вероятность невозврата кредита ($p_+$). Клиентов, обратившихся за кредитом, сортируют по этой предсказанной вероятности (по убыванию), и получается скоркарта — по сути, рейтинг клиентов от плохих к хорошим. Ниже приведен игрушечный пример такой скоркарты. \n <img src='../../img/toy_scorecard.png' width=60%>\n\nБанк выбирает для себя порог $p_*$ предсказанной вероятности невозврата кредита (на картинке – $0.15$) и начиная с этого значения уже не выдает кредит. Более того, можно умножить предсказнную вероятность на выданную сумму и получить матожидание потерь с клиента, что тоже будет хорошей бизнес-метрикой.",
"_____no_output_____"
],
[
"Итак, мы хотим прогнозировать вероятность $p_+ \\in [0,1]$, а пока умеем строить линейный прогноз с помощью МНК: $b(\\textbf{x}) = \\textbf{w}^\\text{T} \\textbf{x} \\in \\mathbb{R}$. Каким образом преобразовать полученное значение в вероятность, пределы которой – [0, 1]? Очевидно, для этого нужна некоторая функция $f: \\mathbb{R} \\rightarrow [0,1].$ В модели логистической регрессии для этого берется конкретная функция: $\\sigma(z) = \\frac{1}{1 + \\exp^{-z}}$. И сейчас разберемся, каковы для этого предпосылки. ",
"_____no_output_____"
]
],
[
[
"# отключим всякие предупреждения Anaconda\nimport warnings\nwarnings.filterwarnings('ignore')\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nimport seaborn as sns\nimport numpy as np",
"_____no_output_____"
],
[
"def sigma(z):\n return 1. / (1 + np.exp(-z))",
"_____no_output_____"
],
[
"xx = np.linspace(-10, 10, 1000)\nplt.plot(xx, [sigma(x) for x in xx]);\nplt.xlabel('z');\nplt.ylabel('sigmoid(z)')\nplt.title('Sigmoid function');",
"_____no_output_____"
]
],
[
[
"Обозначим $P(X)$ вероятностью происходящего события $X$. Тогда отношение вероятностей $OR(X)$ определяется из $\\frac{P(X)}{1-P(X)}$, а это — отношение вероятностей того, произойдет ли событие или не произойдет. Очевидно, что вероятность и отношение шансов содержат одинаковую информацию. Но в то время как $P(X)$ находится в пределах от 0 до 1, $OR(X)$ находится в пределах от 0 до $\\infty$.\n\nЕсли вычислить логарифм $OR(X)$ (то есть называется логарифм шансов, или логарифм отношения вероятностей), то легко заметить, что $\\log{OR(X)} \\in \\mathbb{R}$. Его то мы и будем прогнозировать с помощью МНК.\n\nПосмотрим, как логистическая регрессия будет делать прогноз $p_+ = \\text{P}\\left(y_i = 1 \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right)$ (пока считаем, что веса $\\textbf{w}$ мы как-то получили (т.е. обучили модель), далее разберемся, как именно). \n\n**Шаг 1.** Вычислить значение $w_{0}+w_{1}x_1 + w_{2}x_2 + ... = \\textbf{w}^\\text{T}\\textbf{x}$. (уравнение $\\textbf{w}^\\text{T}\\textbf{x} = 0$ задает гиперплоскость, разделяющую примеры на 2 класса);\n\n\n**Шаг 2.** Вычислить логарифм отношения шансов: $ \\log(OR_{+}) = \\textbf{w}^\\text{T}\\textbf{x}$.\n\n**Шаг 3.** Имея прогноз шансов на отнесение к классу \"+\" – $OR_{+}$, вычислить $p_{+}$ с помощью простой зависимости:\n\n$$\\Large p_{+} = \\frac{OR_{+}}{1 + OR_{+}} = \\frac{\\exp^{\\textbf{w}^\\text{T}\\textbf{x}}}{1 + \\exp^{\\textbf{w}^\\text{T}\\textbf{x}}} = \\frac{1}{1 + \\exp^{-\\textbf{w}^\\text{T}\\textbf{x}}} = \\sigma(\\textbf{w}^\\text{T}\\textbf{x})$$\n\n\nВ правой части мы получили как раз сигмоид-функцию.\n\nИтак, логистическая регрессия прогнозирует вероятность отнесения примера к классу \"+\" (при условии, что мы знаем его признаки и веса модели) как сигмоид-преобразование линейной комбинации вектора весов модели и вектора признаков примера:\n\n$$\\Large p_+(x_i) = \\text{P}\\left(y_i = 1 \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right) = \\sigma(\\textbf{w}^\\text{T}\\textbf{x}_\\text{i}). $$\n\nСледующий вопрос: как модель обучается. Тут мы опять обращаемся к принципу максимального правдоподобия.",
"_____no_output_____"
],
[
"### Принцип максимального правдоподобия и логистическая регрессия\nТеперь посмотрим, как из принципа максимального правдоподобия получается оптимизационная задача, которую решает логистическая регрессия, а именно, – минимизация *логистической* функции потерь. \nТолько что мы увидели, что логистическая регрессия моделирует вероятность отнесения примера к классу \"+\" как \n\n$$\\Large p_+(\\textbf{x}_\\text{i}) = \\text{P}\\left(y_i = 1 \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right) = \\sigma(\\textbf{w}^\\text{T}\\textbf{x}_\\text{i})$$\n\nТогда для класса \"-\" аналогичная вероятность:\n$$\\Large p_-(\\textbf{x}_\\text{i}) = \\text{P}\\left(y_i = -1 \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right) = 1 - \\sigma(\\textbf{w}^\\text{T}\\textbf{x}_\\text{i}) = \\sigma(-\\textbf{w}^\\text{T}\\textbf{x}_\\text{i}) $$\n\nОба этих выражения можно ловко объединить в одно (следите за моими руками – не обманывают ли вас):\n\n$$\\Large \\text{P}\\left(y = y_i \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right) = \\sigma(y_i\\textbf{w}^\\text{T}\\textbf{x}_\\text{i})$$\n\nВыражение $M(\\textbf{x}_\\text{i}) = y_i\\textbf{w}^\\text{T}\\textbf{x}_\\text{i}$ называется *отступом* (*margin*) классификации на объекте $\\textbf{x}_\\text{i}$ (не путать с зазором (тоже margin), про который чаще всего говорят в контексте SVM). Если он неотрицателен, модель не ошибается на объекте $\\textbf{x}_\\text{i}$, если же отрицателен – значит, класс для $\\textbf{x}_\\text{i}$ спрогнозирован неправильно. \nЗаметим, что отступ определен для объектов именно обучающей выборки, для которых известны реальные метки целевого класса $y_i$.\n\nЧтобы понять, почему это мы сделали такие выводы, обратимся к геометрической интерпретации линейного классификатора. Подробно про это можно почитать в материалах Евгения Соколова – [тут](https://github.com/esokolov/ml-course-msu/blob/master/ML16/lecture-notes/Sem09_linear.pdf). ",
"_____no_output_____"
],
[
"Рекомендую решить почти классическую задачу из начального курса линейной алгебры: найти расстояние от точки с радиус-вектором $\\textbf{x}_A$ до плоскости, которая задается уравнением $\\textbf{w}^\\text{T}\\textbf{x} = 0.$\n\n\nОтвет: \n$\\Large \\rho(\\textbf{x}_A, \\textbf{w}^\\text{T}\\textbf{x} = 0) = \\frac{\\textbf{w}^\\text{T}\\textbf{x}_A}{||\\textbf{w}||}$\n",
"_____no_output_____"
],
[
"<img src = '../../img/simple_linal_task.png' width=60%>",
"_____no_output_____"
],
[
"Когда получим (или посмотрим) ответ, то поймем, что чем больше по модулю выражение $\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}$, тем дальше точка $\\textbf{x}_\\text{i}$ находится от плоскости $\\textbf{w}^{\\text{T}}\\textbf{x} = 0.$\n\nЗначит, выражение $M(\\textbf{x}_\\text{i}) = y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}$ – это своего рода \"уверенность\" модели в классификации объекта $\\textbf{x}_\\text{i}$: \n\n- если отступ большой (по модулю) и положительный, это значит, что метка класса поставлена правильно, а объект находится далеко от разделяющей гиперплоскости (такой объект классифицируется уверенно). На рисунке – $x_3$.\n- если отступ большой (по модулю) и отрицательный, значит метка класса поставлена неправильно, а объект находится далеко от разделюящей гиперплоскости (скорее всего такой объект – аномалия, например, его метка в обучающей выборке поставлена неправильно). На рисунке – $x_1$.\n- если отступ малый (по модулю), то объект находится близко к разделюящей гиперплоскости, а знак отступа определяет, правильно ли объект классифицирован. На рисунке – $x_2$ и $x_4$.",
"_____no_output_____"
],
[
"<img src = '../../img/margin.png' width=60%>",
"_____no_output_____"
],
[
"Теперь распишем правдоподобие выборки, а именно, вероятность наблюдать данный вектор $\\textbf{y}$ у выборки $\\textbf X$. Делаем сильное предположение: объекты приходят независимо, из одного распределения (*i.i.d.*). Тогда\n\n$$\\Large \\text{P}\\left(\\textbf{y} \\mid \\textbf X, \\textbf{w}\\right) = \\prod_{i=1}^{\\ell} \\text{P}\\left(y = y_i \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right),$$\n\nгде $\\ell$ – длина выборки $\\textbf X$ (число строк).\n\nКак водится, возьмем логарифм данного выражения (сумму оптимизировать намного проще, чем произведение):\n\n$$\\Large \\log \\text{P}\\left(\\textbf{y} \\mid \\textbf X, \\textbf{w}\\right) = \\log \\sum_{i=1}^{\\ell} \\text{P}\\left(y = y_i \\mid \\textbf{x}_\\text{i}, \\textbf{w}\\right) = \\log \\prod_{i=1}^{\\ell} \\sigma(y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}) = $$\n\n$$\\Large = \\sum_{i=1}^{\\ell} \\log \\sigma(y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}) = \\sum_{i=1}^{\\ell} \\log \\frac{1}{1 + \\exp^{-y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}}} = - \\sum_{i=1}^{\\ell} \\log (1 + \\exp^{-y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}})$$",
"_____no_output_____"
],
[
"То есть в данном случае принцип максимизации правдоподобия приводит к минимизации выражения \n\n$$\\Large \\mathcal{L_{log}} (\\textbf X, \\textbf{y}, \\textbf{w}) = \\sum_{i=1}^{\\ell} \\log (1 + \\exp^{-y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}}).$$\n\nЭто *логистическая* функция потерь, просуммированная по всем объектам обучающей выборки.\n\nПосмотрим на новую фунцию как на функцию от отступа: $L(M) = \\log (1 + \\exp^{-M})$. Нарисуем ее график, а также график 1/0 функциий потерь (*zero-one loss*), которая просто штрафует модель на 1 за ошибку на каждом объекте (отступ отрицательный): $L_{1/0}(M) = [M < 0]$.",
"_____no_output_____"
],
[
"<img src = '../../img/logloss_margin.png' width=60%>",
"_____no_output_____"
],
[
"Картинка отражает общую идею, что в задаче классификации, не умея напрямую минимизировать число ошибок (по крайней мере, градиентными методами это не сделать – производная 1/0 функциий потерь в нуле обращается в бесконечность), мы минимизируем некоторую ее верхнюю оценку. В данном случае это логистическая функция потерь (где логарифм двоичный, но это не принципиально), и справедливо \n\n$$\\Large \\mathcal{L_{\\text{1/0}}} (\\textbf X, \\textbf{y}, \\textbf{w}) = \\sum_{i=1}^{\\ell} [M(\\textbf{x}_\\text{i}) < 0] \\leq \\sum_{i=1}^{\\ell} \\log (1 + \\exp^{-y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}}) = \\mathcal{L_{\\log}} (\\textbf X, \\textbf{y}, \\textbf{w}), $$\n\nгде $\\mathcal{L_{\\text{1/0}}} (\\textbf X, \\textbf{y}, \\textbf{w})$ – попросту число ошибок логистической регрессии с весами $\\textbf{w}$ на выборке $(\\textbf X, \\textbf{y})$.\n\nТо есть уменьшая верхнюю оценку $\\mathcal{L_{\\log}}$ на число ошибок классификации, мы таким образом надеемся уменьшить и само число ошибок.",
"_____no_output_____"
],
[
"### L2-регуляризация логистической функции потерь",
"_____no_output_____"
],
[
"$L2-регуляризация$ логистической регрессии устроена почти так же, как и в случае с гребневой (Ridge регрессией). Вместо функционала $\\mathcal{L_{\\log}} (X, \\textbf{y}, \\textbf{w})$ минимизируется следующий:\n\n$$\\Large J(\\textbf X, \\textbf{y}, \\textbf{w}) = \\mathcal{L_{\\log}} (\\textbf X, \\textbf{y}, \\textbf{w}) + \\lambda |\\textbf{w}|^2$$\n\nВ случае логистической регрессии принято введение обратного коэффициента регуляризации $C = \\frac{1}{\\lambda}$. И тогда решением задачи будет\n\n$$\\Large \\widehat{\\textbf{w}} = \\arg \\min_{\\textbf{w}} J(\\textbf X, \\textbf{y}, \\textbf{w}) = \\arg \\min_{\\textbf{w}}\\ (C\\sum_{i=1}^{\\ell} \\log (1 + \\exp^{-y_i\\textbf{w}^{\\text{T}}\\textbf{x}_\\text{i}})+ |\\textbf{w}|^2)$$ \n\nДалее рассмотрим пример, позволяющий интуитивно понять один из смыслов регуляризации. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e71d2bf0572da3d0e1b4e456102249de88173bea | 40,077 | ipynb | Jupyter Notebook | Chapter8/.ipynb_checkpoints/Chapter8-checkpoint.ipynb | raonetwo/AutomateTheBoringStuff | 537d7ac7234889a983530d0fc0f86e63753688c8 | [
"MIT"
] | null | null | null | Chapter8/.ipynb_checkpoints/Chapter8-checkpoint.ipynb | raonetwo/AutomateTheBoringStuff | 537d7ac7234889a983530d0fc0f86e63753688c8 | [
"MIT"
] | null | null | null | Chapter8/.ipynb_checkpoints/Chapter8-checkpoint.ipynb | raonetwo/AutomateTheBoringStuff | 537d7ac7234889a983530d0fc0f86e63753688c8 | [
"MIT"
] | null | null | null | 31.731591 | 1,132 | 0.54807 | [
[
[
"while True:\n print('Enter your age:')\n age = input()\n try:\n age = int(age)\n except:\n print('Please use numeric digits.')\n continue\n if age < 1:\n print('Please enter a positive number.')\n continue\n break\nprint(f'Your age is {age}.')",
"Enter your age:\n"
],
[
"# !pip install pyinputplus // uncomment this to install\nimport pyinputplus",
"Collecting pyinputplus\n Downloading PyInputPlus-0.2.12.tar.gz (20 kB)\n Installing build dependencies ... \u001b[?25ldone\n\u001b[?25h Getting requirements to build wheel ... \u001b[?25ldone\n\u001b[?25h Preparing wheel metadata ... \u001b[?25ldone\n\u001b[?25hCollecting pysimplevalidate>=0.2.7\n Downloading PySimpleValidate-0.2.12.tar.gz (22 kB)\n Installing build dependencies ... \u001b[?25ldone\n\u001b[?25h Getting requirements to build wheel ... \u001b[?25ldone\n\u001b[?25h Preparing wheel metadata ... \u001b[?25ldone\n\u001b[?25hCollecting stdiomask>=0.0.3\n Downloading stdiomask-0.0.6.tar.gz (3.6 kB)\n Installing build dependencies ... \u001b[?25ldone\n\u001b[?25h Getting requirements to build wheel ... \u001b[?25ldone\n\u001b[?25h Preparing wheel metadata ... \u001b[?25ldone\n\u001b[?25hBuilding wheels for collected packages: pyinputplus, pysimplevalidate, stdiomask\n Building wheel for pyinputplus (PEP 517) ... \u001b[?25ldone\n\u001b[?25h Created wheel for pyinputplus: filename=PyInputPlus-0.2.12-py3-none-any.whl size=11315 sha256=22a37f5b79f14987960d2679ecde6198faee39b88197860eb2e3015380f704c0\n Stored in directory: /home/yadav/.cache/pip/wheels/e7/33/73/719f8f1bd984d39face880fd22f6c5ef1b85f726e84c687094\n Building wheel for pysimplevalidate (PEP 517) ... \u001b[?25ldone\n\u001b[?25h Created wheel for pysimplevalidate: filename=PySimpleValidate-0.2.12-py3-none-any.whl size=16192 sha256=61a5b5ba46f5246249016350c75565559a337fbf0db3b258d532a3ce7d6b78bb\n Stored in directory: /home/yadav/.cache/pip/wheels/5a/8b/6c/2925d22b93860bdc9b8ce02c3d2cf79336606f7469cef66065\n Building wheel for stdiomask (PEP 517) ... \u001b[?25ldone\n\u001b[?25h Created wheel for stdiomask: filename=stdiomask-0.0.6-py3-none-any.whl size=3320 sha256=3c35d731d305b2af7e66bf344b6e4dc41a9b770868ce5fa77c413a07355ea484\n Stored in directory: /home/yadav/.cache/pip/wheels/cf/13/89/64d0d3b167759523f4c1e68f883bbc30e7bfa27050edeb418e\nSuccessfully built pyinputplus pysimplevalidate stdiomask\nInstalling collected packages: pysimplevalidate, stdiomask, pyinputplus\nSuccessfully installed pyinputplus-0.2.12 pysimplevalidate-0.2.12 stdiomask-0.0.6\n"
],
[
"import pyinputplus as pyip\nresponse = pyip.inputNum()",
" five\n"
],
[
"response",
"_____no_output_____"
],
[
"response = pyip.inputInt(prompt='Enter a number: ')",
"Enter a number: "
],
[
"response = pyip.inputNum('Enter num: ', min=4)",
"Enter num: "
],
[
"response = pyip.inputNum('Enter num: ', greaterThan=4)",
"Enter num: "
],
[
"response = pyip.inputNum('>', min=4, lessThan=6)",
">"
],
[
"response = pyip.inputNum('Enter num: ')",
"Enter num: "
],
[
"response = pyip.inputNum(blank=True)",
" \n"
],
[
"response",
"_____no_output_____"
],
[
"response = pyip.inputNum(limit=2)",
" test\n"
],
[
"response = pyip.inputNum(timeout=10)",
" 42\n"
],
[
"response = pyip.inputNum(allowRegexes=[r'(I|V|X|L|C|D|M)+', r'zero'])",
" xliii\n"
],
[
"response = pyip.inputNum(blockRegexes=[r'[02468]$'])",
" 42\n"
],
[
"def addsUpToTen(numbers):\n numberList = list(numbers)\n for i, digit in enumerate(numberList):\n numberList[i] = int(digit)\n if sum(numberList) != 10:\n raise Exception(\"The digit must add up to 10\")\n return int(numbers)",
"_____no_output_____"
],
[
"response = pyip.inputCustom(addsUpToTen)",
" 123\n"
],
[
"response",
"_____no_output_____"
],
[
"response = pyip.inputCustom(addsUpToTen)",
" hello\n"
],
[
"while True:\n prompt = 'Want to know how to keep an idiot busy for hours?\\n'\n response = pyip.inputYesNo(prompt)\n \n if response == 'no':\n break\nprint('Thank you. Have a nice day.')",
"Want to know how to keep an idiot busy for hours?\n"
],
[
"import random, time\nnumberOfQuestions = 10\ncorrectAnswers = 0\nfor questionNumber in range(numberOfQuestions):\n # Pick two random numbers:\n num1 = random.randint(0, 9)\n num2 = random.randint(0, 9)\n prompt = '#%s: %s x %s = ' % (questionNumber, num1, num2)\n try:\n # Right answers are handled by allowRegexes. # Wrong answers are handled by blockRegexes, with a custom message.\n pyip.inputStr(prompt, allowRegexes=['^%s$' % (num1 * num2)], blockRegexes=[('.*', 'Incorrect!')], timeout=8, limit=3)\n except pyip.TimeoutException:\n print('Out of time!')\n except pyip.RetryLimitException:\n print('Out of tries!')\n else : # try-except-else\n print('Correct!')\n correctAnswers+=1\n time.sleep(1)\nprint('Score: %s / %s' % (correctAnswers, numberOfQuestions))",
"#0: 9 x 6 = "
],
[
"pyip.inputMenu?",
"_____no_output_____"
],
[
"breadChoice = {'wheat' : 10, 'white' : 12, 'sourdough' : 15}\nproteinChoice = {'chicken' : 20, 'turkey' : 25, 'tofu' : 12}\ncheeseChoice = {'chedder' : 15, 'swiss' : 17, 'mozerella' : 20, '' : 0}\nbreadChosen = pyip.inputMenu(list(breadChoice.keys()), prompt=\"Choose your bread\\n\")\nproteinChosen = pyip.inputMenu(list(proteinChoice.keys()), prompt=\"Choose your protein\\n\")\ncheeseChosen = ''\nif pyip.inputYesNo(prompt = \"Do you want cheese in your sandwitch: \\n\")== 'yes':\n cheeseChosen = pyip.inputMenu(list(cheeseChoice.keys()), prompt=\"Choose your cheese\\n\")\nrepeat = pyip.inputInt(prompt=\"How many sandwitches do you want?\\n\")\nprint(f'''\nItem Type : Selected Item : Cost\n\nBread : {breadChosen.rjust(13)} : {breadChoice[breadChosen]}\nProtein : {proteinChosen.rjust(13)} : {proteinChoice[proteinChosen]}\nCheese : {cheeseChosen.rjust(13)} : {cheeseChoice[cheeseChosen]}\n\nNumber of sandwitches : {repeat}\nTotal Cost : {repeat*(breadChoice[breadChosen]+proteinChoice[proteinChosen]+cheeseChoice[cheeseChosen])}\n''')",
"Choose your bread\n* wheat\n* white\n* sourdough\n"
],
[
"pyip.inputYesNo?",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d2f0dc3e2d9e82578dbe35d93a6e5c7ffa5b8 | 133,233 | ipynb | Jupyter Notebook | Credict_Card_Fraud_Detection_.ipynb | Sachin-Kumar-Gupta/Credit-Card-Fraud-Detection | 05ff5c97577b5069c6ec673c39639d85dd053eec | [
"MIT"
] | null | null | null | Credict_Card_Fraud_Detection_.ipynb | Sachin-Kumar-Gupta/Credit-Card-Fraud-Detection | 05ff5c97577b5069c6ec673c39639d85dd053eec | [
"MIT"
] | null | null | null | Credict_Card_Fraud_Detection_.ipynb | Sachin-Kumar-Gupta/Credit-Card-Fraud-Detection | 05ff5c97577b5069c6ec673c39639d85dd053eec | [
"MIT"
] | 1 | 2022-02-26T13:07:10.000Z | 2022-02-26T13:07:10.000Z | 63.686902 | 18,782 | 0.623239 | [
[
[
"#Importing Libraries\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"data = pd.read_csv(\"creditcard.csv\")",
"_____no_output_____"
],
[
"data.head(5)",
"_____no_output_____"
],
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 284807 entries, 0 to 284806\nData columns (total 31 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Time 284807 non-null float64\n 1 V1 284807 non-null float64\n 2 V2 284807 non-null float64\n 3 V3 284807 non-null float64\n 4 V4 284807 non-null float64\n 5 V5 284807 non-null float64\n 6 V6 284807 non-null float64\n 7 V7 284807 non-null float64\n 8 V8 284807 non-null float64\n 9 V9 284807 non-null float64\n 10 V10 284807 non-null float64\n 11 V11 284807 non-null float64\n 12 V12 284807 non-null float64\n 13 V13 284807 non-null float64\n 14 V14 284807 non-null float64\n 15 V15 284807 non-null float64\n 16 V16 284807 non-null float64\n 17 V17 284807 non-null float64\n 18 V18 284807 non-null float64\n 19 V19 284807 non-null float64\n 20 V20 284807 non-null float64\n 21 V21 284807 non-null float64\n 22 V22 284807 non-null float64\n 23 V23 284807 non-null float64\n 24 V24 284807 non-null float64\n 25 V25 284807 non-null float64\n 26 V26 284807 non-null float64\n 27 V27 284807 non-null float64\n 28 V28 284807 non-null float64\n 29 Amount 284807 non-null float64\n 30 Class 284807 non-null int64 \ndtypes: float64(30), int64(1)\nmemory usage: 67.4 MB\n"
],
[
"data.describe()",
"_____no_output_____"
],
[
"data.isnull().sum()",
"_____no_output_____"
],
[
"data.tail(5)",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"print('Valid Transactions: ', round(data['Class'].value_counts()[0]/len(data) * 100,2), '% of the dataset')\n\nprint('Fraudulent Transactions: ', round(data['Class'].value_counts()[1]/len(data) * 100,2), '% of the dataset')",
"Valid Transactions: 99.83 % of the dataset\nFraudulent Transactions: 0.17 % of the dataset\n"
],
[
"data['Class'].value_counts()",
"_____no_output_____"
],
[
"sns.countplot(x = 'Class', data= data, palette = ['blue','red'])",
"_____no_output_____"
]
],
[
[
"# Splitting Dataset",
"_____no_output_____"
]
],
[
[
"X = data.loc[: , data.columns != 'Class']\ny = data.loc[:, data.columns == 'Class']",
"_____no_output_____"
],
[
"X.head(5)",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train,X_test,y_train,y_test = train_test_split(X,y, test_size = 0.3, random_state = 0)",
"_____no_output_____"
],
[
"print(\"Transactions in X_train dataset: \", X_train.shape)\nprint(\"Transaction classes in y_train dataset: \", y_train.shape)\n\nprint(\"Transactions in X_test dataset: \", X_test.shape)\nprint(\"Transaction classes in y_test dataset: \", y_test.shape)\n",
"Transactions in X_train dataset: (199364, 30)\nTransaction classes in y_train dataset: (199364, 1)\nTransactions in X_test dataset: (85443, 30)\nTransaction classes in y_test dataset: (85443, 1)\n"
]
],
[
[
"# Feature Scaling",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler\nscaler_amount = StandardScaler()\nscaler_time = StandardScaler()",
"_____no_output_____"
],
[
"X_train['normAmount']=scaler_amount.fit_transform(X_train['Amount'].values.reshape(-1,1))",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"X_test['normAmount'] = scaler_amount.transform(X_test['Amount'].values.reshape(-1,1))",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"X_train['normTime'] = scaler_time.fit_transform(X_train['Time'].values.reshape(-1,1))\nX_test['normTime'] = scaler_time.transform(X_test['Time'].values.reshape(-1,1))",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \n"
],
[
"X_train = X_train.drop(['Time', 'Amount'], axis=1)\nX_test = X_test.drop(['Time', 'Amount'], axis=1)",
"_____no_output_____"
]
],
[
[
"# Appying SMOTE(Synthetic Minority Over-sampling Technique) Technique",
"_____no_output_____"
]
],
[
[
"from imblearn.over_sampling import SMOTE",
"/usr/local/lib/python3.6/dist-packages/sklearn/externals/six.py:31: FutureWarning: The module is deprecated in version 0.21 and will be removed in version 0.23 since we've dropped support for Python 2.7. Please rely on the official version of six (https://pypi.org/project/six/).\n \"(https://pypi.org/project/six/).\", FutureWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/utils/deprecation.py:144: FutureWarning: The sklearn.neighbors.base module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.neighbors. Anything that cannot be imported from sklearn.neighbors is now part of the private API.\n warnings.warn(message, FutureWarning)\n"
],
[
"print(\"Before over-sampling:\\n\", y_train.value_counts())",
"Before over-sampling:\n Class\n0 199019\n1 345\ndtype: int64\n"
],
[
"sm = SMOTE()\nX_train_res, y_train_res = sm.fit_sample(X_train, y_train)",
"/usr/local/lib/python3.6/dist-packages/sklearn/utils/validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n/usr/local/lib/python3.6/dist-packages/sklearn/utils/deprecation.py:87: FutureWarning: Function safe_indexing is deprecated; safe_indexing is deprecated in version 0.22 and will be removed in version 0.24.\n warnings.warn(msg, category=FutureWarning)\n"
],
[
"from collections import Counter\nCounter(y_train_res)",
"_____no_output_____"
]
],
[
[
"# Model Building",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import confusion_matrix,auc, roc_curve",
"_____no_output_____"
],
[
"parameters = {\"penalty\": ['l1', 'l2'], 'C': [0.001, 0.01, 0.1, 1, 10, 100, 1000]}\nlr = LogisticRegression()\nclf = GridSearchCV(lr, parameters, cv=5, verbose=5, n_jobs=3)\nk = clf.fit(X_train_res, y_train_res)\nprint(k.best_params_)",
"Fitting 5 folds for each of 14 candidates, totalling 70 fits\n"
],
[
"lr_gridcv_best = clf.best_estimator_",
"_____no_output_____"
],
[
"y_test_pre = lr_gridcv_best.predict(X_test)",
"_____no_output_____"
],
[
"cnf_matrix_test = confusion_matrix(y_test, y_test_pre)",
"_____no_output_____"
],
[
"print(\"Recall metric in the test dataset:\", (cnf_matrix_test[1,1]/(cnf_matrix_test[1,0]+cnf_matrix_test[1,1] )))",
"Recall metric in the test dataset: 0.9183673469387755\n"
],
[
"y_train_pre = lr_gridcv_best.predict(X_train_res)",
"_____no_output_____"
],
[
"cnf_matrix_train = confusion_matrix(y_train_res, y_train_pre)",
"_____no_output_____"
],
[
"print(\"Recall metric in the train dataset:\", (cnf_matrix_train[1,1]/(cnf_matrix_train[1,0]+cnf_matrix_train[1,1] )))",
"Recall metric in the train dataset: 0.9187816238650581\n"
],
[
"from sklearn.metrics import plot_confusion_matrix\nclass_names = ['Not Fraud', 'Fraud']\nplot_confusion_matrix(k, X_test, y_test, values_format = '.5g', display_labels=class_names)\nplt.title(\"Test data Confusion Matrix\")\nplt.show()",
"_____no_output_____"
],
[
"plot_confusion_matrix(k, X_train_res, y_train_res, values_format = '.5g', display_labels=class_names) \nplt.title(\"Oversampled Train data Confusion Matrix\")\nplt.show()",
"_____no_output_____"
],
[
"y_k = k.decision_function(X_test)\nfpr, tpr, thresholds = roc_curve(y_test, y_k)\nroc_auc = auc(fpr, tpr)\nprint(\"ROC-AUC:\", roc_auc)",
"ROC-AUC: 0.9839061445249643\n"
],
[
"plt.title('Receiver Operating Characteristic')\nplt.plot(fpr, tpr, 'b',label='AUC = %0.3f'% roc_auc)\nplt.legend(loc='lower right')\nplt.plot([0,1],[0,1],'r--')\nplt.xlim([-0.1,1.0])\nplt.ylim([-0.1,1.01])\nplt.ylabel('True Positive Rate')\nplt.xlabel('False Positive Rate')\nplt.show()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d3722c34d4a4b731f5c94a6ed09caea96002b | 12,332 | ipynb | Jupyter Notebook | tutorial/20.mnist.ipynb | AndrewSerra/Knet.jl | cbbcb670667b1707ad1b1d7c637784e36a98b81e | [
"MIT"
] | null | null | null | tutorial/20.mnist.ipynb | AndrewSerra/Knet.jl | cbbcb670667b1707ad1b1d7c637784e36a98b81e | [
"MIT"
] | null | null | null | tutorial/20.mnist.ipynb | AndrewSerra/Knet.jl | cbbcb670667b1707ad1b1d7c637784e36a98b81e | [
"MIT"
] | null | null | null | 52.254237 | 5,005 | 0.71278 | [
[
[
"# Load and minibatch MNIST data\n(c) Deniz Yuret, 2019\n* Objective: Load the [MNIST](http://yann.lecun.com/exdb/mnist) dataset, convert into Julia arrays, split into minibatches using Knet's [minibatch](http://denizyuret.github.io/Knet.jl/latest/reference/#Knet.minibatch) function and [Data](https://github.com/denizyuret/Knet.jl/blob/master/src/data.jl) iterator type.\n* Prerequisites: [Julia arrays](https://docs.julialang.org/en/v1/manual/arrays)\n* New functions: [dir](http://denizyuret.github.io/Knet.jl/latest/reference/#Knet.dir), [minibatch, Data](http://denizyuret.github.io/Knet.jl/latest/reference/#Knet.minibatch)\n\nIn the next few notebooks, we build classification models for the MNIST handwritten digit recognition dataset. MNIST has 60000 training and 10000 test examples. Each input x consists of 784 pixels representing a 28x28 image. The corresponding output indicates the identity of the digit 0..9.\n\n\n\n[image source](http://yann.lecun.com/exdb/lenet)",
"_____no_output_____"
]
],
[
[
"# Load packages, import symbols\nusing Knet: minibatch\nusing MLDatasets: MNIST\nusing Images",
"_____no_output_____"
],
[
"# This loads the MNIST handwritten digit recognition dataset:\nxtrn,ytrn = MNIST.traindata(Float32)\nxtst,ytst = MNIST.testdata(Float32)\nprintln.(summary.((xtrn,ytrn,xtst,ytst)));",
"28×28×60000 Array{Float32,3}\n60000-element Array{Int64,1}\n28×28×10000 Array{Float32,3}\n10000-element Array{Int64,1}\n"
],
[
"# Here is the first five images from the test set:\n[MNIST.convert2image(xtst[:,:,i]) for i=1:5]",
"_____no_output_____"
],
[
"# Here are their labels\nprintln(ytst[1:5]);",
"[7, 2, 1, 0, 4]\n"
],
[
"# `minibatch` splits the data tensors to small chunks called minibatches.\n# It returns an iterator of (x,y) pairs.\ndtrn = minibatch(xtrn,ytrn,100)\ndtst = minibatch(xtst,ytst,100)",
"_____no_output_____"
],
[
"# Each minibatch is an (x,y) pair where x is 100 (28x28) images and y are the corresponding 100 labels.\n# Here is the first minibatch in the test set:\n(x,y) = first(dtst)\nprintln.(summary.((x,y)));",
"28×28×100 Array{Float32,3}\n100-element Array{Int64,1}\n"
],
[
"# Iterators can be used in for loops, e.g. `for (x,y) in dtrn`\n# dtrn generates 600 minibatches of 100 images (total 60000)\n# dtst generates 100 minibatches of 100 images (total 10000)\nn = 0\nfor (x,y) in dtrn\n global n += 1\nend\nn",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d3a470ecbc2a8563911075454f49035a11d0f | 1,602 | ipynb | Jupyter Notebook | DAY 201 ~ 300/DAY273_[leetCode] Sum of All odd Length Subarrays (Python).ipynb | SOMJANG/CODINGTEST_PRACTICE | 1a7304e9063579441b8a67765175c82b0ad93ac9 | [
"MIT"
] | 15 | 2020-03-17T01:18:33.000Z | 2021-12-24T06:31:06.000Z | DAY 201 ~ 300/DAY273_[leetCode] Sum of All odd Length Subarrays (Python).ipynb | SOMJANG/CODINGTEST_PRACTICE | 1a7304e9063579441b8a67765175c82b0ad93ac9 | [
"MIT"
] | null | null | null | DAY 201 ~ 300/DAY273_[leetCode] Sum of All odd Length Subarrays (Python).ipynb | SOMJANG/CODINGTEST_PRACTICE | 1a7304e9063579441b8a67765175c82b0ad93ac9 | [
"MIT"
] | 10 | 2020-03-17T01:18:34.000Z | 2022-03-30T10:53:07.000Z | 23.910448 | 102 | 0.474407 | [
[
[
"## 2020년 12월 7일 월요일\n### leetCode - Sum of All odd Length Subarrays (Python)\n### 문제 : https://leetcode.com/problems/sum-of-all-odd-length-subarrays/\n### 블로그 : https://somjang.tistory.com/entry/leetCode-1588-Sum-of-All-Odd-Length-Subarrays-Python",
"_____no_output_____"
],
[
"### 첫번째 시도",
"_____no_output_____"
]
],
[
[
"class Solution:\n def sumOddLengthSubarrays(self, arr: List[int]) -> int:\n max_len = len(arr)\n \n arr_len = len(arr)\n \n if max_len % 2 == 0:\n max_len = max_len - 1\n \n answer_list = []\n \n for i in range(1, max_len+1, 2):\n for j in range(arr_len - i + 1):\n answer_list.append(sum(arr[j:j+i])) \n \n return sum(answer_list)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
]
] |
e71d449be4c230101d31839f66bdbd3a38a7e85d | 100,298 | ipynb | Jupyter Notebook | nbs/01_basics.ipynb | michaelaye/fastcore | eeb59decfb3344cf83414795a5a8143a8957b420 | [
"Apache-2.0"
] | null | null | null | nbs/01_basics.ipynb | michaelaye/fastcore | eeb59decfb3344cf83414795a5a8143a8957b420 | [
"Apache-2.0"
] | null | null | null | nbs/01_basics.ipynb | michaelaye/fastcore | eeb59decfb3344cf83414795a5a8143a8957b420 | [
"Apache-2.0"
] | null | null | null | 24.906382 | 421 | 0.505892 | [
[
[
"#default_exp basics",
"_____no_output_____"
],
[
"#export\nfrom fastcore.imports import *",
"_____no_output_____"
],
[
"from fastcore.test import *\nfrom nbdev.showdoc import *\nfrom fastcore.nb_imports import *",
"_____no_output_____"
]
],
[
[
"# Basic functionality\n\n> Basic functionality used in the fastai library",
"_____no_output_____"
],
[
"## Basics",
"_____no_output_____"
]
],
[
[
"# export\ndefaults = SimpleNamespace()",
"_____no_output_____"
],
[
"# export\ndef ifnone(a, b):\n \"`b` if `a` is None else `a`\"\n return b if a is None else a",
"_____no_output_____"
]
],
[
[
"Since `b if a is None else a` is such a common pattern, we wrap it in a function. However, be careful, because python will evaluate *both* `a` and `b` when calling `ifnone` (which it doesn't do if using the `if` version directly).",
"_____no_output_____"
]
],
[
[
"test_eq(ifnone(None,1), 1)\ntest_eq(ifnone(2 ,1), 2)",
"_____no_output_____"
],
[
"#export\ndef maybe_attr(o, attr):\n \"`getattr(o,attr,o)`\"\n return getattr(o,attr,o)",
"_____no_output_____"
]
],
[
[
"Return the attribute `attr` for object `o`. If the attribute doesn't exist, then return the object `o` instead. ",
"_____no_output_____"
]
],
[
[
"class myobj: myattr='foo'\n\ntest_eq(maybe_attr(myobj, 'myattr'), 'foo')\ntest_eq(maybe_attr(myobj, 'another_attr'), myobj)",
"_____no_output_____"
],
[
"#export\ndef basic_repr(flds=None):\n if isinstance(flds, str): flds = re.split(', *', flds)\n flds = list(flds or [])\n def _f(self):\n sig = ', '.join(f'{o}={maybe_attr(getattr(self,o), \"__name__\")}' for o in flds)\n return f'{self.__class__.__name__}({sig})'\n return _f",
"_____no_output_____"
]
],
[
[
"Lookup a user-supplied list of attributes (`flds`) of an object and generate a string with the name of each attribute and its corresponding value. The format of this string is `key=value`, where `key` is the name of the attribute, and `value` is the value of the attribute. For each value, attempt to use the `__name__` attribute, otherwise fall back to using the value's `__repr__` when constructing the string. ",
"_____no_output_____"
]
],
[
[
"class SomeClass:\n a=1\n b='foo'\n __repr__=basic_repr('a,b')\n __name__='some-class'\n \nclass AnotherClass:\n c=SomeClass()\n d='bar'\n __repr__=basic_repr(['c', 'd'])\n \nsc = SomeClass() \nac = AnotherClass()\n\ntest_eq(repr(sc), 'SomeClass(a=1, b=foo)')\ntest_eq(repr(ac), 'AnotherClass(c=some-class, d=bar)')",
"_____no_output_____"
],
[
"#export\ndef is_array(x):\n \"`True` if `x` supports `__array__` or `iloc`\"\n return hasattr(x,'__array__') or hasattr(x,'iloc')",
"_____no_output_____"
],
[
"is_array(np.array(1)),is_array([1])",
"_____no_output_____"
],
[
"#export\ndef listify(o):\n \"Convert `o` to a `list`\"\n if o is None: return []\n if isinstance(o, list): return o\n if isinstance(o, str) or is_array(o): return [o]\n if is_iter(o): return list(o)\n return [o]",
"_____no_output_____"
]
],
[
[
"Conversion is designed to \"do what you mean\", e.g:",
"_____no_output_____"
]
],
[
[
"[(o,listify(o)) for o in\n ('hi',array(1),1, [1,2], range(3))]",
"_____no_output_____"
]
],
[
[
"Generators are turned into lists too:",
"_____no_output_____"
]
],
[
[
"gen = (o for o in range(3))\nlistify(gen)",
"_____no_output_____"
],
[
"#export\ndef true(x):\n \"Test whether `x` is truthy; collections with >0 elements are considered `True`\"\n try: return bool(len(x))\n except: return bool(x)",
"_____no_output_____"
],
[
"[(o,true(o)) for o in\n (array(0),array(1),array([0]),array([0,1]),1,0,'',None)]",
"_____no_output_____"
],
[
"#export\nclass NullType:\n \"An object that is `False` and can be called, chained, and indexed\"\n def __getattr__(self,*args):return null\n def __call__(self,*args, **kwargs):return null\n def __getitem__(self, *args):return null\n def __bool__(self): return False\n\nnull = NullType()",
"_____no_output_____"
],
[
"bool(null.hi().there[3])",
"_____no_output_____"
],
[
"#export\ndef tonull(x):\n \"Convert `None` to `null`\"\n return null if x is None else x",
"_____no_output_____"
],
[
"bool(tonull(None).hi().there[3])",
"_____no_output_____"
],
[
"#export\ndef get_class(nm, *fld_names, sup=None, doc=None, funcs=None, **flds):\n \"Dynamically create a class, optionally inheriting from `sup`, containing `fld_names`\"\n attrs = {}\n for f in fld_names: attrs[f] = None\n for f in listify(funcs): attrs[f.__name__] = f\n for k,v in flds.items(): attrs[k] = v\n sup = ifnone(sup, ())\n if not isinstance(sup, tuple): sup=(sup,)\n\n def _init(self, *args, **kwargs):\n for i,v in enumerate(args): setattr(self, list(attrs.keys())[i], v)\n for k,v in kwargs.items(): setattr(self,k,v)\n\n all_flds = [*fld_names,*flds.keys()]\n def _eq(self,b):\n return all([getattr(self,k)==getattr(b,k) for k in all_flds])\n\n if not sup: attrs['__repr__'] = basic_repr(all_flds)\n attrs['__init__'] = _init\n attrs['__eq__'] = _eq\n res = type(nm, sup, attrs)\n if doc is not None: res.__doc__ = doc\n return res",
"_____no_output_____"
],
[
"show_doc(get_class, title_level=4)",
"_____no_output_____"
],
[
"_t = get_class('_t', 'a', b=2)\nt = _t()\ntest_eq(t.a, None)\ntest_eq(t.b, 2)\nt = _t(1, b=3)\ntest_eq(t.a, 1)\ntest_eq(t.b, 3)\nt = _t(1, 3)\ntest_eq(t.a, 1)\ntest_eq(t.b, 3)\ntest_eq(repr(t), '_t(a=1, b=3)')\ntest_eq(t, pickle.loads(pickle.dumps(t)))",
"_____no_output_____"
]
],
[
[
"Most often you'll want to call `mk_class`, since it adds the class to your module. See `mk_class` for more details and examples of use (which also apply to `get_class`).",
"_____no_output_____"
]
],
[
[
"#export\ndef mk_class(nm, *fld_names, sup=None, doc=None, funcs=None, mod=None, **flds):\n \"Create a class using `get_class` and add to the caller's module\"\n if mod is None: mod = sys._getframe(1).f_locals\n res = get_class(nm, *fld_names, sup=sup, doc=doc, funcs=funcs, **flds)\n mod[nm] = res",
"_____no_output_____"
]
],
[
[
"Any `kwargs` will be added as class attributes, and `sup` is an optional (tuple of) base classes.",
"_____no_output_____"
]
],
[
[
"mk_class('_t', a=1, sup=dict)\nt = _t()\ntest_eq(t.a, 1)\nassert(isinstance(t,dict))",
"_____no_output_____"
]
],
[
[
"A `__init__` is provided that sets attrs for any `kwargs`, and for any `args` (matching by position to fields), along with a `__repr__` which prints all attrs. The docstring is set to `doc`. You can pass `funcs` which will be added as attrs with the function names.",
"_____no_output_____"
]
],
[
[
"def foo(self): return 1\nmk_class('_t', 'a', sup=dict, doc='test doc', funcs=foo)\n\nt = _t(3, b=2)\ntest_eq(t.a, 3)\ntest_eq(t.b, 2)\ntest_eq(t.foo(), 1)\ntest_eq(t.__doc__, 'test doc')\nt",
"_____no_output_____"
],
[
"#export\ndef wrap_class(nm, *fld_names, sup=None, doc=None, funcs=None, **flds):\n \"Decorator: makes function a method of a new class `nm` passing parameters to `mk_class`\"\n def _inner(f):\n mk_class(nm, *fld_names, sup=sup, doc=doc, funcs=listify(funcs)+[f], mod=f.__globals__, **flds)\n return f\n return _inner",
"_____no_output_____"
],
[
"@wrap_class('_t', a=2)\ndef bar(self,x): return x+1\n\nt = _t()\ntest_eq(t.a, 2)\ntest_eq(t.bar(3), 4)",
"_____no_output_____"
],
[
"#export\nclass ignore_exceptions:\n \"Context manager to ignore exceptions\"\n def __enter__(self): pass\n def __exit__(self, *args): return True",
"_____no_output_____"
],
[
"show_doc(ignore_exceptions, title_level=4)",
"_____no_output_____"
],
[
"with ignore_exceptions(): \n # Exception will be ignored\n raise Exception",
"_____no_output_____"
],
[
"#export\ndef exec_local(code, var_name):\n \"Call `exec` on `code` and return the var `var_name\"\n loc = {}\n exec(code, globals(), loc)\n return loc[var_name]",
"_____no_output_____"
],
[
"test_eq(exec_local(\"a=1\", \"a\"), 1)",
"_____no_output_____"
],
[
"#export\ndef risinstance(types, obj=None):\n \"Curried `isinstance` but with args reversed\"\n if obj is None: return partial(risinstance,types)\n return isinstance(obj, types)",
"_____no_output_____"
],
[
"assert risinstance(int, 1)\nassert not risinstance(str, 0)\nassert risinstance(int)(1)",
"_____no_output_____"
]
],
[
[
"## NoOp\n\nThese are used when you need a pass-through function.",
"_____no_output_____"
]
],
[
[
"show_doc(noop, title_level=4)",
"_____no_output_____"
],
[
"noop()\ntest_eq(noop(1),1)",
"_____no_output_____"
],
[
"show_doc(noops, title_level=4)",
"_____no_output_____"
],
[
"class _t: foo=noops\ntest_eq(_t().foo(1),1)",
"_____no_output_____"
]
],
[
[
"## Infinite Lists\n\nThese lists are useful for things like padding an array or adding index column(s) to arrays.",
"_____no_output_____"
]
],
[
[
"#export\n#hide\nclass _InfMeta(type):\n @property\n def count(self): return itertools.count()\n @property\n def zeros(self): return itertools.cycle([0])\n @property\n def ones(self): return itertools.cycle([1])\n @property\n def nones(self): return itertools.cycle([None])",
"_____no_output_____"
],
[
"#export\nclass Inf(metaclass=_InfMeta):\n \"Infinite lists\"\n pass",
"_____no_output_____"
],
[
"show_doc(Inf, title_level=4);",
"_____no_output_____"
]
],
[
[
"`Inf` defines the following properties:\n \n- `count: itertools.count()`\n- `zeros: itertools.cycle([0])`\n- `ones : itertools.cycle([1])`\n- `nones: itertools.cycle([None])`",
"_____no_output_____"
]
],
[
[
"test_eq([o for i,o in zip(range(5), Inf.count)],\n [0, 1, 2, 3, 4])\n\ntest_eq([o for i,o in zip(range(5), Inf.zeros)],\n [0]*5)\n\ntest_eq([o for i,o in zip(range(5), Inf.ones)],\n [1]*5)\n\ntest_eq([o for i,o in zip(range(5), Inf.nones)],\n [None]*5)",
"_____no_output_____"
]
],
[
[
"## Operator Functions",
"_____no_output_____"
]
],
[
[
"#export\n_dumobj = object()\ndef _oper(op,a,b=_dumobj): return (lambda o:op(o,a)) if b is _dumobj else op(a,b)\n\ndef _mk_op(nm, mod):\n \"Create an operator using `oper` and add to the caller's module\"\n op = getattr(operator,nm)\n def _inner(a, b=_dumobj): return _oper(op, a,b)\n _inner.__name__ = _inner.__qualname__ = nm\n _inner.__doc__ = f'Same as `operator.{nm}`, or returns partial if 1 arg'\n mod[nm] = _inner",
"_____no_output_____"
],
[
"#export\ndef in_(x, a):\n \"`True` if `x in a`\"\n return x in a\n\noperator.in_ = in_",
"_____no_output_____"
],
[
"#export\n_all_ = ['lt','gt','le','ge','eq','ne','add','sub','mul','truediv','is_','is_not','in_']",
"_____no_output_____"
],
[
"#export\nfor op in ['lt','gt','le','ge','eq','ne','add','sub','mul','truediv','is_','is_not','in_']: _mk_op(op, globals())",
"_____no_output_____"
],
[
"# test if element is in another\nassert in_('c', ('b', 'c', 'a'))\nassert in_(4, [2,3,4,5])\nassert in_('t', 'fastai')\ntest_fail(in_('h', 'fastai'))\n\n# use in_ as a partial\nassert in_('fastai')('t')\nassert in_([2,3,4,5])(4)\ntest_fail(in_('fastai')('h'))",
"_____no_output_____"
]
],
[
[
"In addition to `in_`, the following functions are provided matching the behavior of the equivalent versions in `operator`: *lt gt le ge eq ne add sub mul truediv is_ is_not*.",
"_____no_output_____"
]
],
[
[
"lt(3,5),gt(3,5),is_(None,None),in_(0,[1,2])",
"_____no_output_____"
]
],
[
[
"Similarly to `_in`, they also have additional functionality: if you only pass one param, they return a partial function that passes that param as the second positional parameter.",
"_____no_output_____"
]
],
[
[
"lt(5)(3),gt(5)(3),is_(None)(None),in_([1,2])(0)",
"_____no_output_____"
],
[
"#export\ndef true(*args, **kwargs):\n \"Predicate: always `True`\"\n return True",
"_____no_output_____"
],
[
"assert true(1,2,3)\nassert true(False)\nassert true(None)\nassert true([])",
"_____no_output_____"
],
[
"#export\ndef stop(e=StopIteration):\n \"Raises exception `e` (by default `StopException`)\"\n raise e",
"_____no_output_____"
],
[
"#export\ndef gen(func, seq, cond=true):\n \"Like `(func(o) for o in seq if cond(func(o)))` but handles `StopIteration`\"\n return itertools.takewhile(cond, map(func,seq))",
"_____no_output_____"
],
[
"test_eq(gen(noop, Inf.count, lt(5)),\n range(5))\ntest_eq(gen(operator.neg, Inf.count, gt(-5)),\n [0,-1,-2,-3,-4])\ntest_eq(gen(lambda o:o if o<5 else stop(), Inf.count),\n range(5))",
"_____no_output_____"
],
[
"#export\ndef chunked(it, chunk_sz=None, drop_last=False, n_chunks=None):\n \"Return batches from iterator `it` of size `chunk_sz` (or return `n_chunks` total)\"\n assert bool(chunk_sz) ^ bool(n_chunks)\n if n_chunks: chunk_sz = math.ceil(len(it)/n_chunks)\n if not isinstance(it, Iterator): it = iter(it)\n while True:\n res = list(itertools.islice(it, chunk_sz))\n if res and (len(res)==chunk_sz or not drop_last): yield res\n if len(res)<chunk_sz: return",
"_____no_output_____"
]
],
[
[
"Note that you must pass either `chunk_sz`, or `n_chunks`, but not both.",
"_____no_output_____"
]
],
[
[
"t = list(range(10))\ntest_eq(chunked(t,3), [[0,1,2], [3,4,5], [6,7,8], [9]])\ntest_eq(chunked(t,3,True), [[0,1,2], [3,4,5], [6,7,8], ])\n\nt = map(lambda o:stop() if o==6 else o, Inf.count)\ntest_eq(chunked(t,3), [[0, 1, 2], [3, 4, 5]])\nt = map(lambda o:stop() if o==7 else o, Inf.count)\ntest_eq(chunked(t,3), [[0, 1, 2], [3, 4, 5], [6]])\n\nt = np.arange(10)\ntest_eq(chunked(t,3), [[0,1,2], [3,4,5], [6,7,8], [9]])\ntest_eq(chunked(t,3,True), [[0,1,2], [3,4,5], [6,7,8], ])",
"_____no_output_____"
],
[
"#export\ndef otherwise(x, tst, y):\n \"`y if tst(x) else x`\"\n return y if tst(x) else x",
"_____no_output_____"
],
[
"test_eq(otherwise(2+1, gt(3), 4), 3)\ntest_eq(otherwise(2+1, gt(2), 4), 4)",
"_____no_output_____"
]
],
[
[
"## Attribute Helpers",
"_____no_output_____"
],
[
"These functions reduce boilerplate when setting or manipulating attributes or properties of objects.",
"_____no_output_____"
]
],
[
[
"#export\ndef custom_dir(c, add:list):\n \"Implement custom `__dir__`, adding `add` to `cls`\"\n return dir(type(c)) + list(c.__dict__.keys()) + add",
"_____no_output_____"
]
],
[
[
"`custom_dir` allows you extract the [`__dict__` property of a class](https://stackoverflow.com/questions/19907442/explain-dict-attribute) and appends the list `add` to it.",
"_____no_output_____"
]
],
[
[
"class _T: \n def f(): pass\n\ns = custom_dir(_T, add=['foo', 'bar']) # a list of everything in `__dict__` of `_T` with ['foo', 'bar'] appended.\nassert {'foo', 'bar', 'f'}.issubset(s)",
"_____no_output_____"
],
[
"#export\nclass AttrDict(dict):\n \"`dict` subclass that also provides access to keys as attrs\"\n def __getattr__(self,k): return self[k] if k in self else stop(AttributeError(k))\n def __setattr__(self, k, v): (self.__setitem__,super().__setattr__)[k[0]=='_'](k,v)\n def __dir__(self): return custom_dir(self, list(self.keys()))",
"_____no_output_____"
],
[
"show_doc(AttrDict, title_level=4)",
"_____no_output_____"
],
[
"d = AttrDict(a=1,b=\"two\")\ntest_eq(d.a, 1)\ntest_eq(d['b'], 'two')\ntest_eq(d.get('c','nope'), 'nope')\nd.b = 2\ntest_eq(d.b, 2)\ntest_eq(d['b'], 2)\nd['b'] = 3\ntest_eq(d['b'], 3)\ntest_eq(d.b, 3)\nassert 'a' in dir(d)",
"_____no_output_____"
],
[
"#exports\ndef type_hints(f):\n \"Same as `typing.get_type_hints` but returns `{}` if not allowed type\"\n return typing.get_type_hints(f) if isinstance(f, typing._allowed_types) else {}",
"_____no_output_____"
]
],
[
[
"Below is a list of allowed types for type hints in python:",
"_____no_output_____"
]
],
[
[
"list(typing._allowed_types)",
"_____no_output_____"
]
],
[
[
"For example, type `func` is allowed so `type_hints` returns the same value as `typing.get_hints`:",
"_____no_output_____"
]
],
[
[
"def f(a:int)->bool: ... # a function with type hints (allowed)\nexp = {'a':int,'return':bool}\ntest_eq(type_hints(f), typing.get_type_hints(f))\ntest_eq(type_hints(f), exp)",
"_____no_output_____"
]
],
[
[
"However, `class` is not an allowed type, so `type_hints` returns `{}`:",
"_____no_output_____"
]
],
[
[
"class _T:\n def __init__(self, a:int=0)->bool: ...\nassert not type_hints(_T)",
"_____no_output_____"
],
[
"#export\ndef annotations(o):\n \"Annotations for `o`, or `type(o)`\"\n res = {}\n if not o: return res\n res = type_hints(o)\n if not res: res = type_hints(getattr(o,'__init__',None))\n if not res: res = type_hints(type(o))\n return res",
"_____no_output_____"
]
],
[
[
"This supports a wider range of situations than `type_hints`, by checking `type()` and `__init__` for annotations too:",
"_____no_output_____"
]
],
[
[
"for o in _T,_T(),_T.__init__,f: test_eq(annotations(o), exp)\nassert not annotations(int)\nassert not annotations(print)",
"_____no_output_____"
],
[
"#export\ndef anno_ret(func):\n \"Get the return annotation of `func`\"\n return annotations(func).get('return', None) if func else None",
"_____no_output_____"
],
[
"def f(x) -> float: return x\ntest_eq(anno_ret(f), float)\n\ndef f(x) -> typing.Tuple[float,float]: return x\ntest_eq(anno_ret(f), typing.Tuple[float,float])",
"_____no_output_____"
]
],
[
[
"If your return annotation is `None`, `anno_ret` will return `NoneType` (and not `None`):",
"_____no_output_____"
]
],
[
[
"def f(x) -> None: return x\n\ntest_eq(anno_ret(f), NoneType)\nassert anno_ret(f) is not None # returns NoneType instead of None",
"_____no_output_____"
]
],
[
[
"If your function does not have a return type, or if you pass in `None` instead of a function, then `anno_ret` returns `None`:",
"_____no_output_____"
]
],
[
[
"def f(x): return x\n\ntest_eq(anno_ret(f), None)\ntest_eq(anno_ret(None), None) # instead of passing in a func, pass in None",
"_____no_output_____"
],
[
"#export\ndef argnames(f):\n \"Names of arguments to function `f`\"\n code = f.__code__\n return code.co_varnames[:code.co_argcount]",
"_____no_output_____"
],
[
"#export\ndef with_cast(f):\n \"Decorator which uses any parameter annotations as preprocessing functions\"\n anno,params = annotations(f),argnames(f)\n defaults = dict(zip(reversed(params), reversed(f.__defaults__ or {})))\n @functools.wraps(f)\n def _inner(*args, **kwargs):\n args = list(args)\n for i,v in enumerate(params):\n if v in anno:\n c = anno[v]\n if v in kwargs: kwargs[v] = c(kwargs[v])\n elif i<len(args): args[i] = c(args[i])\n elif v in defaults: kwargs[v] = c(defaults[v])\n return f(*args, **kwargs)\n return _inner",
"_____no_output_____"
],
[
"@with_cast\ndef _f(a, b:Path, c:str='', d=0)->bool: return (a,b,c,d)\n\ntest_eq(_f(1, '.', 3), (1,Path('.'),'3',0))\ntest_eq(_f(1, '.'), (1,Path('.'),'',0))",
"_____no_output_____"
],
[
"#export\ndef _store_attr(self, anno, **attrs):\n stored = self.__stored_args__\n for n,v in attrs.items():\n if n in anno: v = anno[n](v)\n setattr(self, n, v)\n stored[n] = v",
"_____no_output_____"
],
[
"#export\ndef store_attr(names=None, self=None, but='', cast=False, **attrs):\n \"Store params named in comma-separated `names` from calling context into attrs in `self`\"\n fr = sys._getframe(1)\n args = fr.f_code.co_varnames[:fr.f_code.co_argcount]\n if self: args = ('self', *args)\n else: self = fr.f_locals[args[0]]\n if not hasattr(self, '__stored_args__'): self.__stored_args__ = {}\n anno = annotations(self) if cast else {}\n if not attrs:\n ns = re.split(', *', names) if names else args[1:]\n attrs = {n:fr.f_locals[n] for n in ns}\n if isinstance(but,str): but = re.split(', *', but)\n attrs = {k:v for k,v in attrs.items() if k not in but}\n return _store_attr(self, anno, **attrs)",
"_____no_output_____"
]
],
[
[
"In it's most basic form, you can use `store_attr` to shorten code like this:",
"_____no_output_____"
]
],
[
[
"class T:\n def __init__(self, a,b,c): self.a,self.b,self.c = a,b,c",
"_____no_output_____"
]
],
[
[
"...to this:",
"_____no_output_____"
]
],
[
[
"class T:\n def __init__(self, a,b,c): store_attr('a,b,c', self)",
"_____no_output_____"
]
],
[
[
"This class behaves as if we'd used the first form:",
"_____no_output_____"
]
],
[
[
"t = T(1,c=2,b=3)\nassert t.a==1 and t.b==3 and t.c==2",
"_____no_output_____"
]
],
[
[
"In addition, it stores the attrs as a `dict` in `__stored_args__`, which you can use for display, logging, and so forth.",
"_____no_output_____"
]
],
[
[
"test_eq(t.__stored_args__, {'a':1, 'b':3, 'c':2})",
"_____no_output_____"
]
],
[
[
"Since you normally want to use the first argument (often called `self`) for storing attributes, it's optional:",
"_____no_output_____"
]
],
[
[
"class T:\n def __init__(self, a,b,c:str): store_attr('a,b,c')\n\nt = T(1,c=2,b=3)\nassert t.a==1 and t.b==3 and t.c==2",
"_____no_output_____"
],
[
"#hide\nclass _T:\n def __init__(self, a,b):\n c = 2\n store_attr('a,b,c')\n\nt = _T(1,b=3)\nassert t.a==1 and t.b==3 and t.c==2",
"_____no_output_____"
]
],
[
[
"With `cast=True` any parameter annotations will be used as preprocessing functions for the corresponding arguments:",
"_____no_output_____"
]
],
[
[
"class T:\n def __init__(self, a:listify, b, c:str): store_attr('a,b,c', cast=True)\n\nt = T(1,c=2,b=3)\nassert t.a==[1] and t.b==3 and t.c=='2'",
"_____no_output_____"
]
],
[
[
"You can inherit from a class using `store_attr`, and just call it again to add in any new attributes added in the derived class:",
"_____no_output_____"
]
],
[
[
"class T2(T):\n def __init__(self, d, **kwargs):\n super().__init__(**kwargs)\n store_attr('d')\n\nt = T2(d=1,a=2,b=3,c=4)\nassert t.a==2 and t.b==3 and t.c==4 and t.d==1",
"_____no_output_____"
]
],
[
[
"You can skip passing a list of attrs to store. In this case, all arguments passed to the method are stored:",
"_____no_output_____"
]
],
[
[
"class T:\n def __init__(self, a,b,c): store_attr()\n\nt = T(1,c=2,b=3)\nassert t.a==1 and t.b==3 and t.c==2",
"_____no_output_____"
],
[
"class T4(T):\n def __init__(self, d, **kwargs):\n super().__init__(**kwargs)\n store_attr()\n\nt = T4(4, a=1,c=2,b=3)\nassert t.a==1 and t.b==3 and t.c==2 and t.d==4",
"_____no_output_____"
],
[
"#hide\n# ensure that subclasses work with or without `store_attr`\nclass T4(T):\n def __init__(self, **kwargs):\n super().__init__(**kwargs)\n store_attr()\n\nt = T4(a=1,c=2,b=3)\nassert t.a==1 and t.b==3 and t.c==2\n\nclass T4(T): pass\n\nt = T4(a=1,c=2,b=3)\nassert t.a==1 and t.b==3 and t.c==2",
"_____no_output_____"
]
],
[
[
"You can skip some attrs by passing `but`:",
"_____no_output_____"
]
],
[
[
"class T:\n def __init__(self, a,b,c): store_attr(but='a')\n\nt = T(1,c=2,b=3)\nassert t.b==3 and t.c==2\nassert not hasattr(t,'a')",
"_____no_output_____"
]
],
[
[
"You can also pass keywords to `store_attr`, which is identical to setting the attrs directly, but also stores them in `__stored_args__`.",
"_____no_output_____"
]
],
[
[
"class T:\n def __init__(self): store_attr(a=1)\n\nt = T()\nassert t.a==1",
"_____no_output_____"
]
],
[
[
"You can also use store_attr inside functions.",
"_____no_output_____"
]
],
[
[
"def create_T(a, b):\n t = SimpleNamespace()\n store_attr(self=t)\n return t\n\nt = create_T(a=1, b=2)\nassert t.a==1 and t.b==2",
"_____no_output_____"
],
[
"#export\ndef attrdict(o, *ks, default=None):\n \"Dict from each `k` in `ks` to `getattr(o,k)`\"\n return {k:getattr(o, k, default) for k in ks}",
"_____no_output_____"
],
[
"class T:\n def __init__(self, a,b,c): store_attr()\n\nt = T(1,c=2,b=3)\ntest_eq(attrdict(t,'b','c'), {'b':3, 'c':2})",
"_____no_output_____"
],
[
"#export\ndef properties(cls, *ps):\n \"Change attrs in `cls` with names in `ps` to properties\"\n for p in ps: setattr(cls,p,property(getattr(cls,p)))",
"_____no_output_____"
],
[
"class T:\n def a(self): return 1\n def b(self): return 2\nproperties(T,'a')\n\ntest_eq(T().a,1)\ntest_eq(T().b(),2)",
"_____no_output_____"
],
[
"#export\n_camel_re1 = re.compile('(.)([A-Z][a-z]+)')\n_camel_re2 = re.compile('([a-z0-9])([A-Z])')",
"_____no_output_____"
],
[
"#export\ndef camel2snake(name):\n \"Convert CamelCase to snake_case\"\n s1 = re.sub(_camel_re1, r'\\1_\\2', name)\n return re.sub(_camel_re2, r'\\1_\\2', s1).lower()",
"_____no_output_____"
],
[
"test_eq(camel2snake('ClassAreCamel'), 'class_are_camel')\ntest_eq(camel2snake('Already_Snake'), 'already__snake')",
"_____no_output_____"
],
[
"#export\ndef snake2camel(s):\n \"Convert snake_case to CamelCase\"\n return ''.join(s.title().split('_'))",
"_____no_output_____"
],
[
"test_eq(snake2camel('a_b_cc'), 'ABCc')",
"_____no_output_____"
],
[
"#export\ndef class2attr(self, cls_name):\n \"Return the snake-cased name of the class; strip ending `cls_name` if it exists.\"\n return camel2snake(re.sub(rf'{cls_name}$', '', self.__class__.__name__) or cls_name.lower())",
"_____no_output_____"
],
[
"class Parent:\n @property\n def name(self): return class2attr(self, 'Parent')\n\nclass ChildOfParent(Parent): pass\nclass ParentChildOf(Parent): pass\n\np = Parent()\ncp = ChildOfParent()\ncp2 = ParentChildOf()\n\ntest_eq(p.name, 'parent')\ntest_eq(cp.name, 'child_of')\ntest_eq(cp2.name, 'parent_child_of')",
"_____no_output_____"
],
[
"#export\ndef getattrs(o, *attrs, default=None):\n \"List of all `attrs` in `o`\"\n return [getattr(o,attr,default) for attr in attrs]",
"_____no_output_____"
],
[
"from fractions import Fraction\ngetattrs(Fraction(1,2), 'numerator', 'denominator')",
"_____no_output_____"
],
[
"#export\ndef hasattrs(o,attrs):\n \"Test whether `o` contains all `attrs`\"\n return all(hasattr(o,attr) for attr in attrs)",
"_____no_output_____"
],
[
"assert hasattrs(1,('imag','real'))\nassert not hasattrs(1,('imag','foo'))",
"_____no_output_____"
],
[
"#export\ndef setattrs(dest, flds, src):\n f = dict.get if isinstance(src, dict) else getattr\n flds = re.split(r\",\\s*\", flds)\n for fld in flds: setattr(dest, fld, f(src, fld))",
"_____no_output_____"
],
[
"d = dict(a=1,bb=\"2\",ignore=3)\no = SimpleNamespace()\nsetattrs(o, \"a,bb\", d)\ntest_eq(o.a, 1)\ntest_eq(o.bb, \"2\")",
"_____no_output_____"
],
[
"d = SimpleNamespace(a=1,bb=\"2\",ignore=3)\no = SimpleNamespace()\nsetattrs(o, \"a,bb\", d)\ntest_eq(o.a, 1)\ntest_eq(o.bb, \"2\")",
"_____no_output_____"
],
[
"#export\ndef try_attrs(obj, *attrs):\n \"Return first attr that exists in `obj`\"\n for att in attrs:\n try: return getattr(obj, att)\n except: pass\n raise AttributeError(attrs)",
"_____no_output_____"
],
[
"test_eq(try_attrs(1, 'real'), 1)\ntest_eq(try_attrs(1, 'foobar', 'real'), 1)",
"_____no_output_____"
]
],
[
[
"## Extensible Types",
"_____no_output_____"
],
[
"`ShowPrint` is a base class that defines a `show` method, which is used primarily for callbacks in fastai that expect this method to be defined.",
"_____no_output_____"
]
],
[
[
"#export\n#hide\nclass ShowPrint:\n \"Base class that prints for `show`\"\n def show(self, *args, **kwargs): print(str(self))",
"_____no_output_____"
]
],
[
[
"`Int`, `Float`, and `Str` extend `int`, `float` and `str` respectively by adding an additional `show` method by inheriting from `ShowPrint`.\n\nThe code for `Int` is shown below:",
"_____no_output_____"
]
],
[
[
"#export\n#hide\nclass Int(int,ShowPrint):\n \"An extensible `int`\"\n pass",
"_____no_output_____"
],
[
"#export \n#hide\nclass Str(str,ShowPrint):\n \"An extensible `str`\"\n pass\nclass Float(float,ShowPrint):\n \"An extensible `float`\"\n pass",
"_____no_output_____"
]
],
[
[
"Examples:",
"_____no_output_____"
]
],
[
[
"Int(0).show()\nFloat(2.0).show()\nStr('Hello').show()",
"0\n2.0\nHello\n"
]
],
[
[
"## Collection functions\n\nFunctions that manipulate popular python collections.",
"_____no_output_____"
]
],
[
[
"#export\ndef detuplify(x):\n \"If `x` is a tuple with one thing, extract it\"\n return None if len(x)==0 else x[0] if len(x)==1 and getattr(x, 'ndim', 1)==1 else x",
"_____no_output_____"
],
[
"test_eq(detuplify(()),None)\ntest_eq(detuplify([1]),1)\ntest_eq(detuplify([1,2]), [1,2])\ntest_eq(detuplify(np.array([[1,2]])), np.array([[1,2]]))",
"_____no_output_____"
],
[
"#export\ndef replicate(item,match):\n \"Create tuple of `item` copied `len(match)` times\"\n return (item,)*len(match)",
"_____no_output_____"
],
[
"t = [1,1]\ntest_eq(replicate([1,2], t),([1,2],[1,2]))\ntest_eq(replicate(1, t),(1,1))",
"_____no_output_____"
],
[
"# export\ndef setify(o): \n \"Turn any list like-object into a set.\"\n return o if isinstance(o,set) else set(listify(o))",
"_____no_output_____"
],
[
"# test\ntest_eq(setify(None),set())\ntest_eq(setify('abc'),{'abc'})\ntest_eq(setify([1,2,2]),{1,2})\ntest_eq(setify(range(0,3)),{0,1,2})\ntest_eq(setify({1,2}),{1,2})",
"_____no_output_____"
],
[
"#export\ndef merge(*ds):\n \"Merge all dictionaries in `ds`\"\n return {k:v for d in ds if d is not None for k,v in d.items()}",
"_____no_output_____"
],
[
"test_eq(merge(), {})\ntest_eq(merge(dict(a=1,b=2)), dict(a=1,b=2))\ntest_eq(merge(dict(a=1,b=2), dict(b=3,c=4), None), dict(a=1, b=3, c=4))",
"_____no_output_____"
],
[
"#export\ndef range_of(x):\n \"All indices of collection `x` (i.e. `list(range(len(x)))`)\"\n return list(range(len(x)))",
"_____no_output_____"
],
[
"test_eq(range_of([1,1,1,1]), [0,1,2,3])",
"_____no_output_____"
],
[
"#export\ndef groupby(x, key, val=noop):\n \"Like `itertools.groupby` but doesn't need to be sorted, and isn't lazy, plus some extensions\"\n if isinstance(key,int): key = itemgetter(key)\n elif isinstance(key,str): key = attrgetter(key)\n if isinstance(val,int): val = itemgetter(val)\n elif isinstance(val,str): val = attrgetter(val)\n res = {}\n for o in x: res.setdefault(key(o), []).append(val(o))\n return res",
"_____no_output_____"
],
[
"test_eq(groupby('aa ab bb'.split(), itemgetter(0)), {'a':['aa','ab'], 'b':['bb']})",
"_____no_output_____"
]
],
[
[
"Here's an example of how to *invert* a grouping, using an `int` as `key` (which uses `itemgetter`; passing a `str` will use `attrgetter`), and using a `val` function:",
"_____no_output_____"
]
],
[
[
"d = {0: [1, 3, 7], 2: [3], 3: [5], 4: [8], 5: [4], 7: [5]}\ngroupby(((o,k) for k,v in d.items() for o in v), 0, 1)",
"_____no_output_____"
],
[
"#export\ndef last_index(x, o):\n \"Finds the last index of occurence of `x` in `o` (returns -1 if no occurence)\"\n try: return next(i for i in reversed(range(len(o))) if o[i] == x)\n except StopIteration: return -1",
"_____no_output_____"
],
[
"test_eq(last_index(9, [1, 2, 9, 3, 4, 9, 10]), 5)\ntest_eq(last_index(6, [1, 2, 9, 3, 4, 9, 10]), -1)",
"_____no_output_____"
],
[
"#export\ndef filter_dict(d, func):\n \"Filter a `dict` using `func`, applied to keys and values\"\n return {k:v for k,v in d.items() if func(k,v)}",
"_____no_output_____"
],
[
"letters = {o:chr(o) for o in range(65,73)}\nletters",
"_____no_output_____"
],
[
"filter_dict(letters, lambda k,v: k<67 or v in 'FG')",
"_____no_output_____"
],
[
"#export\ndef filter_keys(d, func):\n \"Filter a `dict` using `func`, applied to keys\"\n return {k:v for k,v in d.items() if func(k)}",
"_____no_output_____"
],
[
"filter_keys(letters, lt(67))",
"_____no_output_____"
],
[
"#export\ndef filter_values(d, func):\n \"Filter a `dict` using `func`, applied to values\"\n return {k:v for k,v in d.items() if func(v)}",
"_____no_output_____"
],
[
"filter_values(letters, in_('FG'))",
"_____no_output_____"
],
[
"#export\ndef cycle(o):\n \"Like `itertools.cycle` except creates list of `None`s if `o` is empty\"\n o = listify(o)\n return itertools.cycle(o) if o is not None and len(o) > 0 else itertools.cycle([None])",
"_____no_output_____"
],
[
"test_eq(itertools.islice(cycle([1,2,3]),5), [1,2,3,1,2])\ntest_eq(itertools.islice(cycle([]),3), [None]*3)\ntest_eq(itertools.islice(cycle(None),3), [None]*3)\ntest_eq(itertools.islice(cycle(1),3), [1,1,1])",
"_____no_output_____"
],
[
"#export\ndef zip_cycle(x, *args):\n \"Like `itertools.zip_longest` but `cycle`s through elements of all but first argument\"\n return zip(x, *map(cycle,args))",
"_____no_output_____"
],
[
"test_eq(zip_cycle([1,2,3,4],list('abc')), [(1, 'a'), (2, 'b'), (3, 'c'), (4, 'a')])",
"_____no_output_____"
],
[
"#export\ndef sorted_ex(iterable, key=None, reverse=False):\n \"Like `sorted`, but if key is str use `attrgetter`; if int use `itemgetter`\"\n if isinstance(key,str): k=lambda o:getattr(o,key,0)\n elif isinstance(key,int): k=itemgetter(key)\n else: k=key\n return sorted(iterable, key=k, reverse=reverse)",
"_____no_output_____"
],
[
"#export\ndef negate_func(f):\n \"Create new function that negates result of `f`\"\n def _f(*args, **kwargs): return not f(*args, **kwargs)\n return _f",
"_____no_output_____"
],
[
"def f(a): return a>0\ntest_eq(f(1),True)\ntest_eq(negate_func(f)(1),False)\ntest_eq(negate_func(f)(a=-1),True)",
"_____no_output_____"
],
[
"#export\ndef argwhere(iterable, f, negate=False, **kwargs):\n \"Like `filter_ex`, but return indices for matching items\"\n if kwargs: f = partial(f,**kwargs)\n if negate: f = negate_func(f)\n return [i for i,o in enumerate(iterable) if f(o)]",
"_____no_output_____"
],
[
"#export\ndef filter_ex(iterable, f=noop, negate=False, gen=False, **kwargs):\n \"Like `filter`, but passing `kwargs` to `f`, defaulting `f` to `noop`, and adding `negate` and `gen`\"\n if f is None: f = lambda _: True\n if kwargs: f = partial(f,**kwargs)\n if negate: f = negate_func(f)\n res = filter(f, iterable)\n if gen: return res\n return list(res)",
"_____no_output_____"
],
[
"#export\ndef range_of(a, b=None, step=None):\n \"All indices of collection `a`, if `a` is a collection, otherwise `range`\"\n if is_coll(a): a = len(a)\n return list(range(a,b,step) if step is not None else range(a,b) if b is not None else range(a))",
"_____no_output_____"
],
[
"test_eq(range_of([1,1,1,1]), [0,1,2,3])\ntest_eq(range_of(4), [0,1,2,3])",
"_____no_output_____"
],
[
"#export\ndef renumerate(iterable, start=0):\n \"Same as `enumerate`, but returns index as 2nd element instead of 1st\"\n return ((o,i) for i,o in enumerate(iterable, start=start))",
"_____no_output_____"
],
[
"test_eq(renumerate('abc'), (('a',0),('b',1),('c',2)))",
"_____no_output_____"
],
[
"#export\ndef first(x, f=None, negate=False, **kwargs):\n \"First element of `x`, optionally filtered by `f`, or None if missing\"\n x = iter(x)\n if f: x = filter_ex(x, f=f, negate=negate, gen=True, **kwargs)\n return next(x, None)",
"_____no_output_____"
],
[
"test_eq(first(['a', 'b', 'c', 'd', 'e']), 'a')\ntest_eq(first([False]), False)\ntest_eq(first([False], noop), None)",
"_____no_output_____"
],
[
"#export\ndef nested_attr(o, attr, default=None):\n \"Same as `getattr`, but if `attr` includes a `.`, then looks inside nested objects\"\n try:\n for a in attr.split(\".\"): o = getattr(o, a)\n except AttributeError: return default\n return o",
"_____no_output_____"
],
[
"a = SimpleNamespace(b=(SimpleNamespace(c=1)))\ntest_eq(nested_attr(a, 'b.c'), getattr(getattr(a, 'b'), 'c'))\ntest_eq(nested_attr(a, 'b.d'), None)",
"_____no_output_____"
],
[
"#export\ndef nested_idx(coll, *idxs):\n \"Index into nested collections, dicts, etc, with `idxs`\"\n if not coll or not idxs: return coll\n if isinstance(coll,str) or not isinstance(coll, typing.Collection): return None\n res = coll.get(idxs[0], None) if hasattr(coll, 'get') else coll[idxs[0]] if idxs[0]<len(coll) else None\n return nested_idx(res, *idxs[1:])",
"_____no_output_____"
],
[
"a = {'b':[1,{'c':2}]}\ntest_eq(nested_idx(a), a)\ntest_eq(nested_idx(a, 'b'), [1,{'c':2}])\ntest_eq(nested_idx(a, 'b', 1), {'c':2})\ntest_eq(nested_idx(a, 'b', 1, 'c'), 2)",
"_____no_output_____"
]
],
[
[
"## fastuple\n\nA tuple with extended functionality.",
"_____no_output_____"
]
],
[
[
"#export\nnum_methods = \"\"\"\n __add__ __sub__ __mul__ __matmul__ __truediv__ __floordiv__ __mod__ __divmod__ __pow__\n __lshift__ __rshift__ __and__ __xor__ __or__ __neg__ __pos__ __abs__\n\"\"\".split()\nrnum_methods = \"\"\"\n __radd__ __rsub__ __rmul__ __rmatmul__ __rtruediv__ __rfloordiv__ __rmod__ __rdivmod__\n __rpow__ __rlshift__ __rrshift__ __rand__ __rxor__ __ror__\n\"\"\".split()\ninum_methods = \"\"\"\n __iadd__ __isub__ __imul__ __imatmul__ __itruediv__\n __ifloordiv__ __imod__ __ipow__ __ilshift__ __irshift__ __iand__ __ixor__ __ior__\n\"\"\".split()",
"_____no_output_____"
],
[
"#export\nclass fastuple(tuple):\n \"A `tuple` with elementwise ops and more friendly __init__ behavior\"\n def __new__(cls, x=None, *rest):\n if x is None: x = ()\n if not isinstance(x,tuple):\n if len(rest): x = (x,)\n else:\n try: x = tuple(iter(x))\n except TypeError: x = (x,)\n return super().__new__(cls, x+rest if rest else x)\n\n def _op(self,op,*args):\n if not isinstance(self,fastuple): self = fastuple(self)\n return type(self)(map(op,self,*map(cycle, args)))\n\n def mul(self,*args):\n \"`*` is already defined in `tuple` for replicating, so use `mul` instead\"\n return fastuple._op(self, operator.mul,*args)\n\n def add(self,*args):\n \"`+` is already defined in `tuple` for concat, so use `add` instead\"\n return fastuple._op(self, operator.add,*args)\n\ndef _get_op(op):\n if isinstance(op,str): op = getattr(operator,op)\n def _f(self,*args): return self._op(op,*args)\n return _f\n\nfor n in num_methods:\n if not hasattr(fastuple, n) and hasattr(operator,n): setattr(fastuple,n,_get_op(n))\n\nfor n in 'eq ne lt le gt ge'.split(): setattr(fastuple,n,_get_op(n))\nsetattr(fastuple,'__invert__',_get_op('__not__'))\nsetattr(fastuple,'max',_get_op(max))\nsetattr(fastuple,'min',_get_op(min))",
"_____no_output_____"
],
[
"show_doc(fastuple, title_level=4)",
"_____no_output_____"
]
],
[
[
"#### Friendly init behavior",
"_____no_output_____"
],
[
"Common failure modes when trying to initialize a tuple in python:\n\n```py\ntuple(3)\n> TypeError: 'int' object is not iterable\n```\n\nor \n\n```py\ntuple(3, 4)\n> TypeError: tuple expected at most 1 arguments, got 2\n```\n\nHowever, `fastuple` allows you to define tuples like this and in the usual way:",
"_____no_output_____"
]
],
[
[
"test_eq(fastuple(3), (3,))\ntest_eq(fastuple(3,4), (3, 4))\ntest_eq(fastuple((3,4)), (3, 4))",
"_____no_output_____"
]
],
[
[
"#### Elementwise operations",
"_____no_output_____"
]
],
[
[
"show_doc(fastuple.add, title_level=5)",
"_____no_output_____"
],
[
"test_eq(fastuple.add((1,1),(2,2)), (3,3))\ntest_eq_type(fastuple(1,1).add(2), fastuple(3,3))\ntest_eq(fastuple('1','2').add('2'), fastuple('12','22'))",
"_____no_output_____"
],
[
"show_doc(fastuple.mul, title_level=5)",
"_____no_output_____"
],
[
"test_eq_type(fastuple(1,1).mul(2), fastuple(2,2))",
"_____no_output_____"
]
],
[
[
"#### Other Elementwise Operations\n\nAdditionally, the following elementwise operations are available:\n- `le`: less than\n- `eq`: equal\n- `gt`: greater than\n- `min`: minimum of",
"_____no_output_____"
]
],
[
[
"test_eq(fastuple(3,1).le(1), (False, True))\ntest_eq(fastuple(3,1).eq(1), (False, True))\ntest_eq(fastuple(3,1).gt(1), (True, False))\ntest_eq(fastuple(3,1).min(2), (2,1))",
"_____no_output_____"
]
],
[
[
"You can also do other elemntwise operations like negate a `fastuple`, or subtract two `fastuple`s:",
"_____no_output_____"
]
],
[
[
"test_eq(-fastuple(1,2), (-1,-2))\ntest_eq(~fastuple(1,0,1), (False,True,False))\n\ntest_eq(fastuple(1,1)-fastuple(2,2), (-1,-1))",
"_____no_output_____"
],
[
"test_eq(type(fastuple(1)), fastuple)\ntest_eq_type(fastuple(1,2), fastuple(1,2))\ntest_ne(fastuple(1,2), fastuple(1,3))\ntest_eq(fastuple(), ())",
"_____no_output_____"
]
],
[
[
"## Functions on Functions",
"_____no_output_____"
],
[
"Utilities for functional programming or for defining, modifying, or debugging functions. ",
"_____no_output_____"
]
],
[
[
"# export\nclass _Arg:\n def __init__(self,i): self.i = i\narg0 = _Arg(0)\narg1 = _Arg(1)\narg2 = _Arg(2)\narg3 = _Arg(3)\narg4 = _Arg(4)",
"_____no_output_____"
],
[
"#export\nclass bind:\n \"Same as `partial`, except you can use `arg0` `arg1` etc param placeholders\"\n def __init__(self, func, *pargs, **pkwargs):\n self.func,self.pargs,self.pkwargs = func,pargs,pkwargs\n self.maxi = max((x.i for x in pargs if isinstance(x, _Arg)), default=-1)\n\n def __call__(self, *args, **kwargs):\n args = list(args)\n kwargs = {**self.pkwargs,**kwargs}\n for k,v in kwargs.items():\n if isinstance(v,_Arg): kwargs[k] = args.pop(v.i)\n fargs = [args[x.i] if isinstance(x, _Arg) else x for x in self.pargs] + args[self.maxi+1:]\n return self.func(*fargs, **kwargs)",
"_____no_output_____"
],
[
"show_doc(bind, title_level=3)",
"_____no_output_____"
]
],
[
[
"`bind` is the same as `partial`, but also allows you to reorder positional arguments using variable name(s) `arg{i}` where i refers to the zero-indexed positional argument. `bind` as implemented currently only supports reordering of up to the first 5 positional arguments.\n\nConsider the function `myfunc` below, which has 3 positional arguments. These arguments can be referenced as `arg0`, `arg1`, and `arg1`, respectively. ",
"_____no_output_____"
]
],
[
[
"def myfn(a,b,c,d=1,e=2): return(a,b,c,d,e)",
"_____no_output_____"
]
],
[
[
"In the below example we bind the positional arguments of `myfn` as follows:\n\n- The second input `14`, referenced by `arg1`, is substituted for the first positional argument.\n- We supply a default value of `17` for the second positional argument.\n- The first input `19`, referenced by `arg0`, is subsituted for the third positional argument. ",
"_____no_output_____"
]
],
[
[
"test_eq(bind(myfn, arg1, 17, arg0, e=3)(19,14), (14,17,19,1,3))",
"_____no_output_____"
]
],
[
[
"In this next example:\n\n- We set the default value to `17` for the first positional argument.\n- The first input `19` refrenced by `arg0`, becomes the second positional argument.\n- The second input `14` becomes the third positional argument.\n- We override the default the value for named argument `e` to `3`.",
"_____no_output_____"
]
],
[
[
"test_eq(bind(myfn, 17, arg0, e=3)(19,14), (17,19,14,1,3))",
"_____no_output_____"
]
],
[
[
"This is an example of using `bind` like `partial` and do not reorder any arguments:",
"_____no_output_____"
]
],
[
[
"test_eq(bind(myfn)(17,19,14), (17,19,14,1,2))",
"_____no_output_____"
]
],
[
[
"`bind` can also be used to change default values. In the below example, we use the first input `3` to override the default value of the named argument `e`, and supply default values for the first three positional arguments:",
"_____no_output_____"
]
],
[
[
"test_eq(bind(myfn, 17,19,14,e=arg0)(3), (17,19,14,1,3))",
"_____no_output_____"
],
[
"#export\ndef map_ex(iterable, f, *args, gen=False, **kwargs):\n \"Like `map`, but use `bind`, and supports `str` and indexing\"\n g = (bind(f,*args,**kwargs) if callable(f)\n else f.format if isinstance(f,str)\n else f.__getitem__)\n res = map(g, iterable)\n if gen: return res\n return list(res)",
"_____no_output_____"
],
[
"t = [0,1,2,3]\ntest_eq(map_ex(t,operator.neg), [0,-1,-2,-3])",
"_____no_output_____"
]
],
[
[
"If `f` is a string then it is treated as a format string to create the mapping:",
"_____no_output_____"
]
],
[
[
"test_eq(map_ex(t, '#{}#'), ['#0#','#1#','#2#','#3#'])",
"_____no_output_____"
]
],
[
[
"If `f` is a dictionary (or anything supporting `__getitem__`) then it is indexed to create the mapping:",
"_____no_output_____"
]
],
[
[
"test_eq(map_ex(t, list('abcd')), list('abcd'))",
"_____no_output_____"
]
],
[
[
"You can also pass the same `arg` params that `bind` accepts:",
"_____no_output_____"
]
],
[
[
"def f(a=None,b=None): return b\ntest_eq(map_ex(t, f, b=arg0), range(4))",
"_____no_output_____"
],
[
"# export\ndef compose(*funcs, order=None):\n \"Create a function that composes all functions in `funcs`, passing along remaining `*args` and `**kwargs` to all\"\n funcs = listify(funcs)\n if len(funcs)==0: return noop\n if len(funcs)==1: return funcs[0]\n if order is not None: funcs = sorted_ex(funcs, key=order)\n def _inner(x, *args, **kwargs):\n for f in funcs: x = f(x, *args, **kwargs)\n return x\n return _inner",
"_____no_output_____"
],
[
"f1 = lambda o,p=0: (o*2)+p\nf2 = lambda o,p=1: (o+1)/p\ntest_eq(f2(f1(3)), compose(f1,f2)(3))\ntest_eq(f2(f1(3,p=3),p=3), compose(f1,f2)(3,p=3))\ntest_eq(f2(f1(3, 3), 3), compose(f1,f2)(3, 3))\n\nf1.order = 1\ntest_eq(f1(f2(3)), compose(f1,f2, order=\"order\")(3))",
"_____no_output_____"
],
[
"#export\ndef maps(*args, retain=noop):\n \"Like `map`, except funcs are composed first\"\n f = compose(*args[:-1])\n def _f(b): return retain(f(b), b)\n return map(_f, args[-1])",
"_____no_output_____"
],
[
"test_eq(maps([1]), [1])\ntest_eq(maps(operator.neg, [1,2]), [-1,-2])\ntest_eq(maps(operator.neg, operator.neg, [1,2]), [1,2])",
"_____no_output_____"
],
[
"#export\ndef partialler(f, *args, order=None, **kwargs):\n \"Like `functools.partial` but also copies over docstring\"\n fnew = partial(f,*args,**kwargs)\n fnew.__doc__ = f.__doc__\n if order is not None: fnew.order=order\n elif hasattr(f,'order'): fnew.order=f.order\n return fnew",
"_____no_output_____"
],
[
"def _f(x,a=1):\n \"test func\"\n return x-a\n_f.order=1\n\nf = partialler(_f, 2)\ntest_eq(f.order, 1)\ntest_eq(f(3), -1)\nf = partialler(_f, a=2, order=3)\ntest_eq(f.__doc__, \"test func\")\ntest_eq(f.order, 3)\ntest_eq(f(3), _f(3,2))",
"_____no_output_____"
],
[
"class partial0:\n \"Like `partialler`, but args passed to callable are inserted at started, instead of at end\"\n def __init__(self, f, *args, order=None, **kwargs):\n self.f,self.args,self.kwargs = f,args,kwargs\n self.order = ifnone(order, getattr(f,'order',None))\n self.__doc__ = f.__doc__\n\n def __call__(self, *args, **kwargs): return self.f(*args, *self.args, **kwargs, **self.kwargs)",
"_____no_output_____"
],
[
"f = partial0(_f, 2)\ntest_eq(f.order, 1)\ntest_eq(f(3), 1) # NB: different to `partialler` example",
"_____no_output_____"
],
[
"#export\ndef instantiate(t):\n \"Instantiate `t` if it's a type, otherwise do nothing\"\n return t() if isinstance(t, type) else t",
"_____no_output_____"
],
[
"test_eq_type(instantiate(int), 0)\ntest_eq_type(instantiate(1), 1)",
"_____no_output_____"
],
[
"#export\ndef _using_attr(f, attr, x): return f(getattr(x,attr))",
"_____no_output_____"
],
[
"#export\ndef using_attr(f, attr):\n \"Change function `f` to operate on `attr`\"\n return partial(_using_attr, f, attr)",
"_____no_output_____"
],
[
"t = Path('/a/b.txt')\nf = using_attr(str.upper, 'name')\ntest_eq(f(t), 'B.TXT')",
"_____no_output_____"
]
],
[
[
"### Self (with an _uppercase_ S)",
"_____no_output_____"
],
[
"A Concise Way To Create Lambdas",
"_____no_output_____"
]
],
[
[
"#export\nclass _Self:\n \"An alternative to `lambda` for calling methods on passed object.\"\n def __init__(self): self.nms,self.args,self.kwargs,self.ready = [],[],[],True\n def __repr__(self): return f'self: {self.nms}({self.args}, {self.kwargs})'\n\n def __call__(self, *args, **kwargs):\n if self.ready:\n x = args[0]\n for n,a,k in zip(self.nms,self.args,self.kwargs):\n x = getattr(x,n)\n if callable(x) and a is not None: x = x(*a, **k)\n return x\n else:\n self.args.append(args)\n self.kwargs.append(kwargs)\n self.ready = True\n return self\n\n def __getattr__(self,k):\n if not self.ready:\n self.args.append(None)\n self.kwargs.append(None)\n self.nms.append(k)\n self.ready = False\n return self\n \n def _call(self, *args, **kwargs):\n self.args,self.kwargs,self.nms = [args],[kwargs],['__call__']\n self.ready = True\n return self",
"_____no_output_____"
],
[
"#export\nclass _SelfCls:\n def __getattr__(self,k): return getattr(_Self(),k)\n def __getitem__(self,i): return self.__getattr__('__getitem__')(i)\n def __call__(self,*args,**kwargs): return self.__getattr__('_call')(*args,**kwargs)\n\nSelf = _SelfCls()",
"_____no_output_____"
],
[
"#export\n_all_ = ['Self']",
"_____no_output_____"
]
],
[
[
"This is a concise way to create lambdas that are calling methods on an object (note the capitalization!)\n\n`Self.sum()`, for instance, is a shortcut for `lambda o: o.sum()`.",
"_____no_output_____"
]
],
[
[
"f = Self.sum()\nx = np.array([3.,1])\ntest_eq(f(x), 4.)\n\n# This is equivalent to above\nf = lambda o: o.sum()\nx = np.array([3.,1])\ntest_eq(f(x), 4.)\n\nf = Self.argmin()\narr = np.array([1,2,3,4,5])\ntest_eq(f(arr), arr.argmin())\n\nf = Self.sum().is_integer()\nx = np.array([3.,1])\ntest_eq(f(x), True)\n\nf = Self.sum().real.is_integer()\nx = np.array([3.,1])\ntest_eq(f(x), True)\n\nf = Self.imag()\ntest_eq(f(3), 0)\n\nf = Self[1]\ntest_eq(f(x), 1)",
"_____no_output_____"
]
],
[
[
"`Self` is also callable, which creates a function which calls any function passed to it, using the arguments passed to `Self`:",
"_____no_output_____"
]
],
[
[
"def f(a, b=3): return a+b+2\ndef g(a, b=3): return a*b\nfg = Self(1,b=2)\nlist(map(fg, [f,g]))",
"_____no_output_____"
]
],
[
[
"## Other Helpers",
"_____no_output_____"
]
],
[
[
"#export\nclass PrettyString(str):\n \"Little hack to get strings to show properly in Jupyter.\"\n def __repr__(self): return self",
"_____no_output_____"
],
[
"show_doc(PrettyString, title_level=4)",
"_____no_output_____"
]
],
[
[
"Allow strings with special characters to render properly in Jupyter. Without calling `print()` strings with special characters are displayed like so:",
"_____no_output_____"
]
],
[
[
"with_special_chars='a string\\nwith\\nnew\\nlines and\\ttabs'\nwith_special_chars",
"_____no_output_____"
]
],
[
[
"We can correct this with `PrettyString`:",
"_____no_output_____"
]
],
[
[
"PrettyString(with_special_chars)",
"_____no_output_____"
],
[
"#export\ndef even_mults(start, stop, n):\n \"Build log-stepped array from `start` to `stop` in `n` steps.\"\n if n==1: return stop\n mult = stop/start\n step = mult**(1/(n-1))\n return [start*(step**i) for i in range(n)]",
"_____no_output_____"
],
[
"test_eq(even_mults(2,8,3), [2,4,8])\ntest_eq(even_mults(2,32,5), [2,4,8,16,32])\ntest_eq(even_mults(2,8,1), 8)",
"_____no_output_____"
],
[
"#export\ndef num_cpus():\n \"Get number of cpus\"\n try: return len(os.sched_getaffinity(0))\n except AttributeError: return os.cpu_count()\n\ndefaults.cpus = num_cpus()",
"_____no_output_____"
],
[
"num_cpus()",
"_____no_output_____"
],
[
"#export\ndef add_props(f, g=None, n=2):\n \"Create properties passing each of `range(n)` to f\"\n if g is None: return (property(partial(f,i)) for i in range(n))\n return (property(partial(f,i), partial(g,i)) for i in range(n))",
"_____no_output_____"
],
[
"class _T(): a,b = add_props(lambda i,x:i*2)\n\nt = _T()\ntest_eq(t.a,0)\ntest_eq(t.b,2)",
"_____no_output_____"
],
[
"class _T(): \n def __init__(self, v): self.v=v\n def _set(i, self, v): self.v[i] = v\n a,b = add_props(lambda i,x: x.v[i], _set)\n\nt = _T([0,2])\ntest_eq(t.a,0)\ntest_eq(t.b,2)\nt.a = t.a+1\nt.b = 3\ntest_eq(t.a,1)\ntest_eq(t.b,3)",
"_____no_output_____"
],
[
"#export\ndef _typeerr(arg, val, typ): return TypeError(f\"{arg}=={val} not {typ}\")",
"_____no_output_____"
],
[
"#export\ndef typed(f):\n \"Decorator to check param and return types at runtime\"\n names = f.__code__.co_varnames\n anno = annotations(f)\n ret = anno.pop('return',None)\n def _f(*args,**kwargs):\n kw = {**kwargs}\n if len(anno) > 0:\n for i,arg in enumerate(args): kw[names[i]] = arg\n for k,v in kw.items():\n if not isinstance(v,anno[k]): raise _typeerr(k, v, anno[k])\n res = f(*args,**kwargs)\n if ret is not None and not isinstance(res,ret): raise _typeerr(\"return\", res, ret)\n return res\n return functools.update_wrapper(_f, f)",
"_____no_output_____"
]
],
[
[
"`typed` validates argument types at **runtime**. This is in contrast to [MyPy](http://mypy-lang.org/) which only offers static type checking.\n\nFor example, a `TypeError` will be raised if we try to pass an integer into the first argument of the below function: ",
"_____no_output_____"
]
],
[
[
"@typed\ndef discount(price:int, pct:float): \n return (1-pct) * price\n\nwith ExceptionExpected(TypeError): discount(100.0, .1)",
"_____no_output_____"
]
],
[
[
"We can also optionally allow multiple types by enumarating the types in a tuple as illustrated below:",
"_____no_output_____"
]
],
[
[
"def discount(price:(int,float), pct:float): \n return (1-pct) * price\n\nassert 90.0 == discount(100.0, .1)",
"_____no_output_____"
],
[
"@typed\ndef foo(a:int, b:str='a'): return a\ntest_eq(foo(1, '2'), 1)\n\nwith ExceptionExpected(TypeError): foo(1,2)\n\n@typed\ndef foo()->str: return 1\nwith ExceptionExpected(TypeError): foo()\n\n@typed\ndef foo()->str: return '1'\nassert foo()",
"_____no_output_____"
]
],
[
[
"## Notebook functions",
"_____no_output_____"
]
],
[
[
"show_doc(ipython_shell)",
"_____no_output_____"
],
[
"show_doc(in_ipython)",
"_____no_output_____"
],
[
"show_doc(in_colab)",
"_____no_output_____"
],
[
"show_doc(in_jupyter)",
"_____no_output_____"
],
[
"show_doc(in_notebook)",
"_____no_output_____"
]
],
[
[
"These variables are availabe as booleans in `fastcore.basics` as `IN_IPYTHON`, `IN_JUPYTER`, `IN_COLAB` and `IN_NOTEBOOK`.",
"_____no_output_____"
]
],
[
[
"IN_IPYTHON, IN_JUPYTER, IN_COLAB, IN_NOTEBOOK",
"_____no_output_____"
]
],
[
[
"# Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.export import notebook2script\nnotebook2script()",
"Converted 00_test.ipynb.\nConverted 01_basics.ipynb.\nConverted 02_foundation.ipynb.\nConverted 03_xtras.ipynb.\nConverted 04_dispatch.ipynb.\nConverted 05_transform.ipynb.\nConverted 07_meta.ipynb.\nConverted 08_script.ipynb.\nConverted index.ipynb.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71d50a8f3c66d6962dffa710f8eccc68095d6d4 | 7,355 | ipynb | Jupyter Notebook | all_notebooks/17-Sorting and Searching/Implementation of a Hash Table.ipynb | robjpar/pdsa | 735a4cc2a18a0f7f42eec143d272a138925708fa | [
"MIT"
] | 1 | 2021-09-29T16:20:59.000Z | 2021-09-29T16:20:59.000Z | all_notebooks/17-Sorting and Searching/Implementation of a Hash Table.ipynb | robjpar/pdsa | 735a4cc2a18a0f7f42eec143d272a138925708fa | [
"MIT"
] | null | null | null | all_notebooks/17-Sorting and Searching/Implementation of a Hash Table.ipynb | robjpar/pdsa | 735a4cc2a18a0f7f42eec143d272a138925708fa | [
"MIT"
] | null | null | null | 27.240741 | 223 | 0.478178 | [
[
[
"# Implementation of a Hash Table\n\nIn this lecture we will be implementing our own Hash Table to complete our understanding of Hash Tables and Hash Functions! Make sure to review the video lecture before this to fully understand this implementation!\n\nKeep in mind that Python already has a built-in dictionary object that serves as a Hash Table, you would never actually need to implement your own hash table in Python.\n\n___\n## Map\nThe idea of a dictionary used as a hash table to get and retrieve items using **keys** is often referred to as a mapping. In our implementation we will have the following methods:\n\n\n* **HashTable()** Create a new, empty map. It returns an empty map collection.\n* **put(key,val)** Add a new key-value pair to the map. If the key is already in the map then replace the old value with the new value.\n* **get(key)** Given a key, return the value stored in the map or None otherwise.\n* **del** Delete the key-value pair from the map using a statement of the form del map[key].\n* **len()** Return the number of key-value pairs stored \n* **in** the map in Return True for a statement of the form **key in map**, if the given key is in the map, False otherwise.",
"_____no_output_____"
]
],
[
[
"class HashTable(object):\n \n def __init__(self,size):\n \n # Set up size and slots and data\n self.size = size\n self.slots = [None] * self.size\n self.data = [None] * self.size\n \n def put(self,key,data):\n #Note, we'll only use integer keys for ease of use with the Hash Function\n \n # Get the hash value\n hashvalue = self.hashfunction(key,len(self.slots))\n\n # If Slot is Empty\n if self.slots[hashvalue] == None:\n self.slots[hashvalue] = key\n self.data[hashvalue] = data\n \n else:\n \n # If key already exists, replace old value\n if self.slots[hashvalue] == key:\n self.data[hashvalue] = data \n \n # Otherwise, find the next available slot\n else:\n \n nextslot = self.rehash(hashvalue,len(self.slots))\n \n # Get to the next slot\n while self.slots[nextslot] != None and self.slots[nextslot] != key:\n nextslot = self.rehash(nextslot,len(self.slots))\n \n # Set new key, if NONE\n if self.slots[nextslot] == None:\n self.slots[nextslot]=key\n self.data[nextslot]=data\n \n # Otherwise replace old value\n else:\n self.data[nextslot] = data \n\n def hashfunction(self,key,size):\n # Remainder Method\n return key%size\n\n def rehash(self,oldhash,size):\n # For finding next possible positions\n return (oldhash+1)%size\n \n \n def get(self,key):\n \n # Getting items given a key\n \n # Set up variables for our search\n startslot = self.hashfunction(key,len(self.slots))\n data = None\n stop = False\n found = False\n position = startslot\n \n # Until we discern that its not empty or found (and haven't stopped yet)\n while self.slots[position] != None and not found and not stop:\n \n if self.slots[position] == key:\n found = True\n data = self.data[position]\n \n else:\n position=self.rehash(position,len(self.slots))\n if position == startslot:\n \n stop = True\n return data\n\n # Special Methods for use with Python indexing\n def __getitem__(self,key):\n return self.get(key)\n\n def __setitem__(self,key,data):\n self.put(key,data)",
"_____no_output_____"
]
],
[
[
"Let's see it in action!",
"_____no_output_____"
]
],
[
[
"h = HashTable(5)",
"_____no_output_____"
],
[
"# Put our first key in\nh[1] = 'one'",
"_____no_output_____"
],
[
"h[2] = 'two'",
"_____no_output_____"
],
[
"h[3] = 'three'",
"_____no_output_____"
],
[
"h[1]",
"_____no_output_____"
],
[
"h[1] = 'new_one'",
"_____no_output_____"
],
[
"h[1]",
"_____no_output_____"
],
[
"print h[4]",
"None\n"
]
],
[
[
"### Great Job!\n\nThat's it for this rudimentary implementation, try implementing a different hash function for practice!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e71d58fd7c382ff5231d41bf08117a87bed1c810 | 507,558 | ipynb | Jupyter Notebook | finalproject/Guething-006116832/Guething-006116832.ipynb | shionguha/cosc3570-introdatascience-fa19 | 8e8ca84baf3a3b1003e6531b43c7f84c9d0e6c7e | [
"MIT"
] | 4 | 2019-08-29T01:49:45.000Z | 2019-12-03T22:02:20.000Z | finalproject/Guething-006116832/Guething-006116832.ipynb | shionguha/cosc3570-introdatascience-fa19 | 8e8ca84baf3a3b1003e6531b43c7f84c9d0e6c7e | [
"MIT"
] | 2 | 2019-12-11T17:56:07.000Z | 2019-12-17T06:01:18.000Z | finalproject/Guething-006116832/Guething-006116832.ipynb | shionguha/cosc3570-introdatascience-fa19 | 8e8ca84baf3a3b1003e6531b43c7f84c9d0e6c7e | [
"MIT"
] | 25 | 2019-08-29T20:45:06.000Z | 2019-12-13T07:02:56.000Z | 190.524775 | 74,936 | 0.836976 | [
[
[
"import pandas as pd\nimport datetime\nfrom os import listdir\nfrom os.path import isfile, join\nimport glob\nimport re\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport seaborn as sns\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.metrics import r2_score\nimport statsmodels.api as sm\nsns.set(rc={'figure.figsize':(12.2,8.3)})",
"_____no_output_____"
],
[
"#loading datasets, deleting columns that are empty\ndf = pd.read_csv(\"Data/Guething_006116832A.csv\");\ndel df['Unnamed: 63']\ndel df['Additions']\ndel df['Applications 17/03/2016-09/06/2016']\ndel df['Duplicates (% of applications received) .2']\ndel df['Duplicate applications received 08/06/2016 - 09/06/2016']\ndel df['Applications received 08/06/2016-09/06/2016']\ndel df['Appliations received after the deadline']\ndf = df.fillna(0)\ndf.head(10)",
"_____no_output_____"
],
[
"#creating separate dataframe to count invalid vounts\n#Note: Two files were put together via mac terminal commands\ncol_names = ['Voting Area','Electorate','Votes Cast', 'Valid Votes']\nmy_df = pd.DataFrame(columns = col_names)\nmy_df['Electorate'] = df['Electorate'].str.replace(',', '').astype(int)\nmy_df['Voting Area'] = df['Voting Area']\nmy_df['Votes Cast'] = df['Votes cast'].str.replace(',', '').astype(int)\nmy_df['Valid Votes'] = df['Valid votes cast'].str.replace(',', '').astype(int)\nmy_df['Invalid Votes Count'] = my_df['Votes Cast'] - my_df['Valid Votes']",
"_____no_output_____"
],
[
"#creating sample\ncol_names2 = ['Electorate','Voting Area','No DOB', 'No Signature', 'Mismatched Signature','Both DOB and Signature mismatched']\nsample2 = pd.DataFrame(columns = col_names2)\nsample2['Electorate'] = df['Electorate'].str.replace(',', '').astype(int)\nsample2['Voting Area'] = df['Voting Area']\nsample2['No DOB'] = df['No DOB']\nsample2['No Signature'] = df['No signature']\nsample2['Mismatched DOB'] = df['Mismatched DOB']\nsample2['Mismatched Signature'] = df['Mismatched signature']\nsample2['Both DOB and Signature mismatched'] = df['Both mismatched']\nsample2['All Added'] = df['Both mismatched'] + sample2['Mismatched Signature'] +sample2['No DOB'] +sample2['No Signature'] + df['Mismatched DOB']\nsample2.sort_values(\"All Added\")\n",
"_____no_output_____"
],
[
"#comparing electorates and invalid votes\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"DarkBlue\", data=my_df,)\nplt.title('Invalid Vote Count versus Electorate Size')",
"_____no_output_____"
],
[
"#determining residuals\nsns.residplot(x='Invalid Votes Count', y=\"Electorate\",color=\"DarkBlue\",data=my_df);\nplt.xlim(0,200)\nplt.figure(figsize=(16, 8))",
"_____no_output_____"
],
[
"#determining linear regression equation from line 34\nX = my_df['Invalid Votes Count'].values.reshape(-1,1)\ny = my_df['Electorate'].values.reshape(-1,1)\nreg = LinearRegression()\nreg.fit(X, y)\nprint(\"Linear Line Equation -> y = {:.5} + {:.5}x\".format(reg.intercept_[0], reg.coef_[0][0]))",
"Linear Line Equation -> y = 3.2024e+04 + 1351.3x\n"
],
[
"#looking at R-Squared Values and P vales\nX = my_df['Invalid Votes Count']\ny = my_df['Electorate']\nX2 = sm.add_constant(X)\nest = sm.OLS(y, X2)\nest2 = est.fit()\nprint(est2.summary())",
" OLS Regression Results \n==============================================================================\nDep. Variable: Electorate R-squared: 0.685\nModel: OLS Adj. R-squared: 0.684\nMethod: Least Squares F-statistic: 826.5\nDate: Thu, 05 Dec 2019 Prob (F-statistic): 2.32e-97\nTime: 22:40:17 Log-Likelihood: -4707.4\nNo. Observations: 382 AIC: 9419.\nDf Residuals: 380 BIC: 9427.\nDf Model: 1 \nCovariance Type: nonrobust \n=======================================================================================\n coef std err t P>|t| [0.025 0.975]\n---------------------------------------------------------------------------------------\nconst 3.202e+04 4186.256 7.650 0.000 2.38e+04 4.03e+04\nInvalid Votes Count 1351.2670 47.004 28.748 0.000 1258.847 1443.687\n==============================================================================\nOmnibus: 492.554 Durbin-Watson: 1.964\nProb(Omnibus): 0.000 Jarque-Bera (JB): 110344.544\nSkew: 5.755 Prob(JB): 0.00\nKurtosis: 85.463 Cond. No. 134.\n==============================================================================\n\nWarnings:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"#looking at outliers\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"DarkBlue\", data=my_df)\nnew = my_df[my_df['Voting Area'].str.contains(r'^Northern')].copy()\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"Magenta\", data=new, fit_reg=False)\n\nnew = my_df[my_df['Voting Area'].str.contains(r'^Birmingham')].copy()\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"Orange\", data=new, fit_reg=False)\n\nnew = my_df[my_df['Voting Area'].str.contains(r'^Leicester')].copy()\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"Orange\", data=new, fit_reg=False)\n\nnew = my_df[my_df['Voting Area'].str.contains(r'^Brent')].copy()\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"turquoise\", data=new, fit_reg=False)\n\nnew = my_df[my_df['Voting Area'].str.contains(r'Bromsgrove')].copy()\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"turquoise\", data=new, fit_reg=False)\n\nnew = my_df[my_df['Voting Area'].str.contains(r'East Ayrshire')].copy()\nsns.regplot(x='Invalid Votes Count', y=\"Electorate\", color=\"Magenta\", data=new, fit_reg=False)\n\nplt.xlim(-10,625)\nplt.title('Major Outliers')",
"_____no_output_____"
],
[
"#More Outliers for Voting Reasons\nsns.regplot(x='Both DOB and Signature mismatched', y=\"Electorate\", color=\"DarkBlue\",fit_reg = False, data= sample2)\n\nnew = sample2[sample2['Voting Area'].str.contains(r'^Bromsgrove')].copy()\nsns.regplot(x='Both DOB and Signature mismatched', y=\"Electorate\", color=\"turquoise\",fit_reg = False, data=new)\n\nnew = sample2[sample2['Voting Area'].str.contains(r'^East Ayrshire')].copy()\nsns.regplot(x='Both DOB and Signature mismatched', y=\"Electorate\", color=\"Magenta\",fit_reg = False, data=new)\n\nnew = sample2[sample2['Voting Area'].str.contains(r'^Northern')].copy()\nsns.regplot(x='Both DOB and Signature mismatched', y=\"Electorate\", color=\"Magenta\",fit_reg = False, data=new)\n\nnew = sample2[sample2['Voting Area'].str.contains(r'^Birmingham')].copy()\nsns.regplot(x='Both DOB and Signature mismatched', y=\"Electorate\", color=\"Orange\",fit_reg = False, data=new)\n\nnew = sample2[sample2['Voting Area'].str.contains(r'^Leicester')].copy()\nsns.regplot(x='Both DOB and Signature mismatched', y=\"Electorate\", color=\"Orange\",fit_reg = False, data=new)\n\nnew = sample2[sample2['Voting Area'].str.contains(r'^Brent')].copy()\nsns.regplot(x='Both DOB and Signature mismatched', y=\"Electorate\", color=\"turquoise\",fit_reg = False, data=new)\n\nplt.title('Both Mismatched Signature & DOB versus Electorate Size')",
"_____no_output_____"
],
[
"#Dataset for Analysis of Rural versus Urban\ncol_names = ['Voting Area','Remain Votes', 'Leave Votes','Electorate']\ndf2 = pd.DataFrame(columns = col_names)\ndf2['Voting Area'] = df['Voting Area']\ndf2['Remain Votes'] = df['Remain']\ndf2['Leave Votes'] = df['Leave']\ndf2['Electorate'] = df['Electorate'].str.replace(',', '').astype(int)\ndf2[\"Decision Margin\"] = df2[\"Remain Votes\"] - df2[\"Leave Votes\"]\ndf2[\"Decided to Leave?\"] = df2[\"Remain Votes\"] < df2[\"Leave Votes\"]\ndf2.sort_values(\"Electorate\")",
"_____no_output_____"
],
[
"# Sorted by Electorate Size, purple shows stay while blue shows leave, analyzing rural versus major cities\nx = df2['Electorate']\ny = df2['Decision Margin']\nmy_color=np.where(y>=0, 'purple', 'Blue')\nplt.scatter(x, y, color=my_color, s=20, alpha=1)\nplt.xlim(0,400000)\nplt.vlines(x=x, ymin=0, ymax=y, color=my_color, alpha=0.4)\nplt.title(\"Comparison between Electorate Size and Decision Margin: Sorted by Electorate\")\nplt.xlabel(\"Number of Electorate\")\nplt.ylabel(\"Decision Margin\")",
"_____no_output_____"
],
[
"#Plotting on whether or not you can determine an error can be made \nax = sns.regplot(x='Electorate', y=\"Decision Margin\", color=\"DarkBlue\", data=df2,)\nax = ax.axhline(0, 0, color='red')\nplt.xlim(0,400000)\nplt.title(\"Electorate versus Decision Margin\")",
"_____no_output_____"
],
[
"#determining linear regression equation from line 34\nX = df2['Electorate'].values.reshape(-1,1)\ny = df2['Decision Margin'].values.reshape(-1,1)\nreg = LinearRegression()\nreg.fit(X, y)\nprint(\"Linear Line Equation -> y = {:.5} + {:.5}x\".format(reg.intercept_[0], reg.coef_[0][0]))",
"Linear Line Equation -> y = -9675.7 + 0.052185x\n"
],
[
"#looking at R-Squared Values and P vales\nX = df2['Electorate']\ny = df2['Decision Margin']\nX2 = sm.add_constant(X)\nest = sm.OLS(y, X2)\nest2 = est.fit()\nprint(est2.summary())",
" OLS Regression Results \n==============================================================================\nDep. Variable: Decision Margin R-squared: 0.056\nModel: OLS Adj. R-squared: 0.053\nMethod: Least Squares F-statistic: 22.49\nDate: Tue, 03 Dec 2019 Prob (F-statistic): 3.00e-06\nTime: 17:57:01 Log-Likelihood: -4340.1\nNo. Observations: 382 AIC: 8684.\nDf Residuals: 380 BIC: 8692.\nDf Model: 1 \nCovariance Type: nonrobust \n==============================================================================\n coef std err t P>|t| [0.025 0.975]\n------------------------------------------------------------------------------\nconst -9675.6843 1712.507 -5.650 0.000 -1.3e+04 -6308.508\nElectorate 0.0522 0.011 4.742 0.000 0.031 0.074\n==============================================================================\nOmnibus: 69.238 Durbin-Watson: 1.826\nProb(Omnibus): 0.000 Jarque-Bera (JB): 289.109\nSkew: 0.711 Prob(JB): 1.66e-63\nKurtosis: 7.018 Cond. No. 2.50e+05\n==============================================================================\n\nWarnings:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n[2] The condition number is large, 2.5e+05. This might indicate that there are\nstrong multicollinearity or other numerical problems.\n"
],
[
"sns.residplot(x='Electorate', y=\"Decision Margin\",color='DarkBlue', data=df2);\nplt.xlim(0,500000)\nplt.title(\"Residuals for Problem 1\")\nplt.figure(figsize=(16, 8))",
"_____no_output_____"
],
[
"#More Outliers for Voting Reasons\nsns.regplot(x='All Added', y=\"Electorate\", color=\"DarkBlue\",fit_reg = False, data= sample2)\nnew = sample2[sample2['Voting Area'].str.contains(r'^Bromsgrove')].copy()\nsns.regplot(x='All Added', y=\"Electorate\", color=\"turquoise\",fit_reg = False, data=new)\nnew = sample2[sample2['Voting Area'].str.contains(r'^East Ayrshire')].copy()\nsns.regplot(x='All Added', y=\"Electorate\", color=\"Magenta\",fit_reg = False, data=new)\nnew = sample2[sample2['Voting Area'].str.contains(r'^Northern')].copy()\nsns.regplot(x='All Added', y=\"Electorate\", color=\"Magenta\",fit_reg = False, data=new)\nnew = sample2[sample2['Voting Area'].str.contains(r'^Birmingham')].copy()\nsns.regplot(x='All Added', y=\"Electorate\", color=\"Orange\",fit_reg = False, data=new)\nnew = sample2[sample2['Voting Area'].str.contains(r'^Leicester')].copy()\nsns.regplot(x='All Added', y=\"Electorate\", color=\"Orange\",fit_reg = False, data=new)\nnew = sample2[sample2['Voting Area'].str.contains(r'^Brent')].copy()\nsns.regplot(x='All Added', y=\"Electorate\", color=\"turquoise\",fit_reg = False, data=new)\n\nplt.title('All Voting Issues Added versus Electorate Size')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d6aa22f60f3625c0b89205ebc9b386ee80d9a | 484,941 | ipynb | Jupyter Notebook | Matplotlib-lesson.ipynb | ljackson707/python-exercises | ae07485e0747457f9424202509c7248ae40e6617 | [
"MIT"
] | 1 | 2021-01-11T18:32:41.000Z | 2021-01-11T18:32:41.000Z | Matplotlib-lesson.ipynb | ljackson707/python-exercises | ae07485e0747457f9424202509c7248ae40e6617 | [
"MIT"
] | null | null | null | Matplotlib-lesson.ipynb | ljackson707/python-exercises | ae07485e0747457f9424202509c7248ae40e6617 | [
"MIT"
] | null | null | null | 656.212449 | 164,056 | 0.945014 | [
[
[
"### Matplotlib: - Default library/module in Python to create Visualizations\n- Most often used during exploration phase of data science pipeline",
"_____no_output_____"
],
[
"#### Why Visualize data?\n\nFor data science practioners, visualize to:\n1. Understand data/trends\n2. Communicate interesting findings to others ",
"_____no_output_____"
],
[
"#### Objectives of this lesson:\n1. How to plot using matplotlib - learn syntax \n2. Ability to understand other's code \n3. How to lookup documentation to customize your plots \n4. Think about what type of plot is appropriate for the data you have ",
"_____no_output_____"
],
[
"#### Different types of charts\nThe choice of the chart/plot type depends on what data type you are dealing with. here are some of the most common once we will focus today.\n\n\nLineplot \nScatter plots \nHistograms \nBar charts \n\n\nOther Examples: https://matplotlib.org/3.3.3/gallery/index.html\n\nSample plots: https://matplotlib.org/3.3.3/tutorials/introductory/sample_plots.html\n\nAnimations example: http://bit.ly/2G1NxuU",
"_____no_output_____"
],
[
"#### Elements of a plot",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"In Matplotlib, the figure (an instance of the class plt.Figure) can be thought of as a single container that contains all the objects representing axes, graphics, text, and labels. \n\nThe axes (an instance of the class plt.Axes) is what we see above: a bounding box with ticks and labels, which will eventually contain the plot elements that make up our visualization\n\n",
"_____no_output_____"
]
],
[
[
"#import libraries/modules\nimport numpy as np\nimport math\nfrom random import randint\nimport random\nimport pandas as pd\n",
"_____no_output_____"
]
],
[
[
"### Lineplot - Plot y versus x as lines.",
"_____no_output_____"
],
[
"##### In general, lineplot are ideal for showing trends in data at equal intervals or over time. With some datasets, you may want to understand changes in one variable as a function of time, or a similarly continuous variable. In this situation,using a line plot is a good choice.\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nx = [2,3,5,7,8]\n\nplt.plot(x)\nplt.show()",
"_____no_output_____"
],
[
"x = [1,2,3,4,5,6]\ny = [2,3,4,3,6,7]\nplt.style.use('seaborn-dark-palette')\nplt.plot(x, y)\nplt.show()",
"_____no_output_____"
],
[
"#Respondents age\nage = [25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35]\n# Overall Median Developer Salaries by Age\ndev_y = [38496, 42000, 46752, 49320, 53200,\n 56000, 62316, 64928, 67317, 68748, 73752]\n# Median Python Developer Salaries by Age\npy_dev_y = [45372, 48876, 53850, 57287, 63016,\n 65998, 70003, 70000, 71496, 75370, 83640]\n# Median JavaScript Developer Salaries by Age\njs_dev_y = [37810, 43515, 46823, 49293, 53437,\n 56373, 62375, 66674, 68745, 68746, 74583]",
"_____no_output_____"
],
[
"#Step 1: Define a figure (optional)\nplt.figure(figsize = (10,6))\n\n#Step 2: Plot data\n#All developers \nplt.plot(age, dev_y, '--bo', label = 'All Dev') # '--ko' this formats the line as a plack dash with dots'\n\n#Add python dev salaries\nplt.plot(age, py_dev_y,linestyle = '--', marker = 'o', color = 'g', label = 'Python Dev') # This way is superior when making legends\n\n#Add java devs\nplt.plot(age, js_dev_y,linestyle = '--', marker = 'o', color = 'k', label = 'Javascript')\n\n#Step 3: Customize plot \n#Add Title\nplt.title('Median Salary in USD by age')\n\n#Add x and y axis label\nplt.xlabel('Age')\nplt.ylabel('Meadian Salary (USD)')\n\n#Add legend\nplt.legend(['All dev', 'Python Dev'])\nplt.legend()\n\n#Add gridlines\nplt.grid(True, ls = '--')\n\n#Add annotation \nplt.annotate('This guy needs a salary bump', xy = (25, 38000), xytext = (28, 38000), arrowprops = dict(facecolor = 'red'))\n\n",
"_____no_output_____"
],
[
"# Line plot\n# Use for numeric and ordered data (typically x axis is time)\n# Use to show trends in data ",
"_____no_output_____"
]
],
[
[
"### Scatter plot: to explore relationship or correlation between two numeric variables",
"_____no_output_____"
]
],
[
[
"# Step 1: Define a figure\nplt.figure(figsize = (10, 8))\n\n#Step 2: plot the data \nplt.scatter(bmi_smoker, charges_smoker )\nplt.scatter(bmi_nonsmoker, charges_nonsmoker)\n\n#Step 3: Customize \nplt.title('BMI vs Insurance charges (USD)')\n\nplt.xlabel('BMI')\nplt.ylabel('Medical Charges (USD)')\n\nplt.legend()\n\nalpha = 0.7 # This allow you to change the transperancy of the dots\ns = 50 (Global) so do size_for_smokers = [charges for charges in charges_smoker] # Then set size = to size_for_smokers # Gradiant size change dpeanding of the growth of a variable ",
"_____no_output_____"
],
[
"# plot a mathematical equation y = x^2 \n\nx = range(-50, 50)\n\ny = [ n ** 2 for n in x]\nplt.scatter(x,y)\nplt.title('$y = x^2$')\n\nplt.xlabel('$x$', fontsize = 14)\nplt.ylabel('$x^2$', fontsize = 14)\n\nplt.xticks([-50,-40,-30,-20,-10,0,10,20,30,40,50], rotation = 45)\n\nplt.show()",
"_____no_output_____"
],
[
"y = sin(x)\n\nx = [x* 0.1 for x in range (-70, 70)]\ny = [math.sin(x) for x in x]\n\nplt.scatter(x,y)\n\nplt.grid(True, ls = '--')\n\nplt.text(0,0, '(0,0)', fontsize = 10, color = 'b')",
"_____no_output_____"
],
[
"# When you want to explore a relationship between numeric variables \n# Asking question: how are two variables related to each other. ",
"_____no_output_____"
]
],
[
[
"### Subplots",
"_____no_output_____"
],
[
"- multiple axes (plots) in a single figure",
"_____no_output_____"
]
],
[
[
"# Within a figure - create two subplots (one row and two columns)\n\nplt.subplot(1,2,1) #(number of rows, number of columns, number subplot)\nplt.text(0.5, 0.5, str((1,2,1)), ha = 'center', fontsize = 14)\n\n#plt.plot()\n\nplt.subplot(1,2,2) \nplt.text(0.5, 0.5, str((1,2,2)), ha = 'center', fontsize = 14)\n\n#plt.scatter()",
"_____no_output_____"
],
[
"x = [x * 0.25 for x in range(-25,26)]\ny1 = [math.sin(x) for x in x]\ny2 = [math.cos(x) for x in x]\ny3 = [math.sin(x) * -1 for x in x]\ny4 = [math.cos(x) * -1 for x in x]",
"_____no_output_____"
],
[
"# 2 columns and 2 rows\npi = math.pi\nplt.figure(figsize = (10, 8))\n\nplt.suptitle('Sin and Cos functions', fontsize = 16, color = 'w')\n\nplt.subplot(2,2,1)\nplt.plot( x, y1, ls = '-', marker = '*', color = 'red')\nplt.title('$sin(x)$', color = 'w')\nplt.grid(True, ls = '--')\n\n# X tick values to show up as gradiants multiples of five\nplt.xticks([-2 * pi , -1 * pi, 0, pi, 2 * pi]), ['$-2\\pi$', '$-\\pi$', '0', '$\\pi$', '$2\\pi$'] # Changes the scale for x axis\n\nplt.subplot(2,2,2)\nplt.plot( x, y2, ls = '-', marker = 'v', color = 'green')\nplt.title('$cos(x)$', color = 'w')\nplt.grid(True, ls = '--')\nplt.axhline(0, color = 'k', linewidth = 0.5) # Makes a horizontal line at y = 0 \n\nplt.subplot(2,2,3)\nplt.plot( x, y3, ls = '-', marker = '*', color = 'blue')\nplt.title('$-sin(x)$', color = 'w')\nplt.grid(True, ls = '--')\n\nplt.subplot(2,2,4)\nplt.plot( x, y4, ls = '-', marker = 'o', color = 'k')\nplt.title('$-cos(x)$', color = 'w')\nplt.grid(True, ls = '--')\nplt.axhline(0, color = 'k', linewidth = 0.5)\nplt.fill_between(x, y4, color = 'blue') # Fills the area in the graph with a color.\n\n#Add padding to avoid overlap of subplots \nplt.tight_layout()\n\nplt.show()\n\n\n",
"_____no_output_____"
],
[
"# one figure can have multpile subplots",
"_____no_output_____"
]
],
[
[
"### Histograms: explore distribution of a sample/population",
"_____no_output_____"
]
],
[
[
"%matplotlib inline # Takes you back to default figure\n%matplotlib notebook # Interactive figure\nplt.hist(data, bins = 50) # Start with default value of bins\nplt.show()",
"UsageError: unrecognized arguments: # Takes you back to default figure\n"
],
[
"plt.hist(data, bins = [0,10,20,30,40,50,60,70,80,90,100], edgecolor = 'k', color = '#30a2da', log = True) # Specifies which bins to show\nmedian = 29\nplt.axvline(median, color = 'r', ls = '--', label = 'median_age')\n\nplt.xlabel('ages')\nplt.ylabel('number of respondents')\n\nplt.legend()\nplt.show()\n# log tranformation makes the smaller numbers or outliers easier to see.",
"_____no_output_____"
],
[
"x1 = [random.gauss(0, 0.8) for _ in range(1000)] #(mean 0, sigma 0.8)\nx2 = [random.gauss(-2, 1) for _ in range(1000)]\nx3 = [random.gauss(3, 1.5) for _ in range(1000)]\nx4 = [random.gauss(7, 0.6) for _ in range(1000)]",
"_____no_output_____"
],
[
"plt.figure(figsize = (10, 6))\nx1 = [random.gauss(0, 0.8) for _ in range(1000)] #(mean 0, sigma 0.8)\nx2 = [random.gauss(-2, 1) for _ in range(1000)]\nx3 = [random.gauss(3, 1.5) for _ in range(1000)]\nx4 = [random.gauss(7, 0.6) for _ in range(1000)]\n\nkwargs = dict(alpha = 0.3, bins = 30, density = True)\n\nplt.hist(x1, **kwargs) # use ** to pass on kwargs to each histogram.\nplt.hist(x2, **kwargs)\nplt.hist(x3, **kwargs)\nplt.hist(x4, **kwargs)\n\nplt.show()",
"_____no_output_____"
],
[
"# use for loop to make subplots\nplt.figure(figsize = (10,6)) \n\n# Make list of list ti integrate\nx = [x1, x2, x3, x4]\n\nfor i in range(1, 5):\n plt.subplot(2,2,i)\n plt.hist(x[i-1], color = 'g', edgecolor = 'k') # Starts with 1 not 0 so use (i- 1)",
"_____no_output_____"
],
[
"# Explore single variable - look at the distributiomn outliers and range etc.",
"_____no_output_____"
],
[
"# BAR CHART \n\nlabels = ['Curie', 'Darden','Easley']\npython_score = [70, 80, 90]\nplt.bar(labels, python_score, color = '#30a2da') # plt.barh will make it hoizontal \nplt.xticks(rotation = 45)",
"_____no_output_____"
],
[
"# Marplotlib Styles \n\nprint(plt.style.available)\n\nplt.style.use('seaborn-dark-palette') # Changes global style\n\nplt.rcdefaults() # This will take you back to default\n",
"['Solarize_Light2', '_classic_test_patch', 'bmh', 'classic', 'dark_background', 'fast', 'fivethirtyeight', 'ggplot', 'grayscale', 'seaborn', 'seaborn-bright', 'seaborn-colorblind', 'seaborn-dark', 'seaborn-dark-palette', 'seaborn-darkgrid', 'seaborn-deep', 'seaborn-muted', 'seaborn-notebook', 'seaborn-paper', 'seaborn-pastel', 'seaborn-poster', 'seaborn-talk', 'seaborn-ticks', 'seaborn-white', 'seaborn-whitegrid', 'tableau-colorblind10']\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d6cfcc8cb97aa91d67c16905a816b8f236679 | 36,696 | ipynb | Jupyter Notebook | Wine_Quality.ipynb | heet2201/Projects | 346716935baa0171a81480a078d7f1aece6c5ba6 | [
"MIT"
] | null | null | null | Wine_Quality.ipynb | heet2201/Projects | 346716935baa0171a81480a078d7f1aece6c5ba6 | [
"MIT"
] | null | null | null | Wine_Quality.ipynb | heet2201/Projects | 346716935baa0171a81480a078d7f1aece6c5ba6 | [
"MIT"
] | null | null | null | 36,696 | 36,696 | 0.547635 | [
[
[
"import tensorflow as tf\nimport numpy as np",
"C:\\Users\\Admin\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"x=tf.Variable(tf.multiply(np.random.rand(10,10),np.sqrt(10)))",
"_____no_output_____"
],
[
"init=tf.global_variables_initializer()\nwith tf.Session() as sess:\n sess.run(init)\n print(sess.run(x))\n",
"[[1.14155281 2.14736078 0.94205657 0.08578865 0.80085464 0.66074418\n 0.65840868 0.8701922 0.5360241 1.50797304]\n [0.6656456 0.44341166 1.48976414 2.12074614 0.21279306 1.85736641\n 1.11191867 2.34613556 0.45985995 1.20952032]\n [3.14093983 3.05058856 0.37346527 0.03726475 0.4910576 2.2697432\n 2.49744048 2.45283321 0.35751836 3.0972507 ]\n [0.73792469 3.13740418 2.28607285 0.82499206 0.82119433 0.71009237\n 2.00154476 1.21430289 2.42757268 0.73194367]\n [0.12157121 0.83459414 0.21788662 1.11662563 0.17549444 0.19327188\n 1.49690773 2.66240334 2.53936647 1.66392261]\n [2.63462363 0.50229947 2.44121395 1.96906165 3.01696999 2.74758213\n 1.12426966 0.54580048 3.06739552 0.50107786]\n [3.10168991 2.12732737 1.68590435 0.76288957 3.07825753 1.47367169\n 1.38321756 2.70644669 3.15002524 2.80449599]\n [0.84270733 1.59484696 2.46256674 2.62742006 2.52530674 0.66649206\n 1.23446178 0.32757023 2.70787637 0.3924236 ]\n [0.78383599 0.54749664 1.68597983 0.91318847 3.01436815 2.89792298\n 2.5873932 0.55472669 1.55643363 0.47003767]\n [0.61313658 1.44617458 0.35465385 1.69145741 0.54208427 2.4548975\n 2.80460871 2.9509566 1.62091153 0.31132463]]\n"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"df=pd.read_csv(r\"C:\\Users\\Admin\\Desktop\\winequality-white (1).csv\")\ndf.head()",
"_____no_output_____"
],
[
"import keras\nimport tensorflow as tf\nimport numpy as np\nfrom keras.layers import Dense, Flatten , Activation ,Dropout, InputLayer ,LSTM, Conv1D, MaxPooling1D\nfrom keras.models import Sequential\nfrom sklearn.cross_validation import train_test_split\nfrom sklearn.preprocessing import normalize\nfrom sklearn.metrics import classification_report",
"_____no_output_____"
],
[
"data=np.array(df)\ndata.shape\n(x_t1,x_t2,y_t1,y_t2)=train_test_split(normalize(data[:,0:11]),data[:,11],test_size=0.15)\n#shuff=np.random.permutation(data.shape[0])\n\n#x_t1=data[shuff[:4500],0:data.shape[1]-1].reshape(4500,11)\n#y_t1=data[shuff[:4500],data.shape[1]-1].reshape(4500,1)\n#x_t2=data[shuff[4500:],0:data.shape[1]-1].reshape(398,11)\n#y_t2=data[shuff[4500:],data.shape[1]-1].reshape(398,1)\n#print(y_t1)\n\n#one_hot\n#y_t1=keras.utils.to_categorical(y_t1,num_classes=10)\n#y_t2=keras.utils.to_categorical(y_t2,num_classes=10)\ny_t2",
"_____no_output_____"
],
[
"model = Sequential()\n\nmodel.add(Dense(128,activation=\"relu\",input_dim=11))\nmodel.add(Dense(256,activation=\"relu\"))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(128,activation=\"relu\"))\nmodel.add(Dense(10,activation=\"softmax\"))",
"_____no_output_____"
],
[
"opt=keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999, epsilon=None, decay=0.0, amsgrad=False)\n\nmodel.compile(optimizer=opt, \n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.fit(x_t1, y_t1, epochs=100,batch_size=64,validation_data=(x_t2,y_t2),shuffle=True)",
"Train on 4163 samples, validate on 735 samples\nEpoch 1/100\n4163/4163 [==============================] - 0s 88us/step - loss: 1.0671 - acc: 0.5246 - val_loss: 1.1161 - val_acc: 0.5238\nEpoch 2/100\n4163/4163 [==============================] - 0s 81us/step - loss: 1.0669 - acc: 0.5205 - val_loss: 1.1262 - val_acc: 0.5238\nEpoch 3/100\n4163/4163 [==============================] - 0s 76us/step - loss: 1.0746 - acc: 0.5150 - val_loss: 1.1605 - val_acc: 0.4776\nEpoch 4/100\n4163/4163 [==============================] - 0s 78us/step - loss: 1.0724 - acc: 0.5241 - val_loss: 1.1470 - val_acc: 0.4925\nEpoch 5/100\n4163/4163 [==============================] - 0s 77us/step - loss: 1.0686 - acc: 0.5301 - val_loss: 1.1271 - val_acc: 0.5265\nEpoch 6/100\n4163/4163 [==============================] - 0s 78us/step - loss: 1.0767 - acc: 0.5263 - val_loss: 1.1601 - val_acc: 0.4680\nEpoch 7/100\n4163/4163 [==============================] - 0s 86us/step - loss: 1.0835 - acc: 0.5179 - val_loss: 1.1180 - val_acc: 0.5238\nEpoch 8/100\n4163/4163 [==============================] - 0s 84us/step - loss: 1.0691 - acc: 0.5239 - val_loss: 1.1588 - val_acc: 0.4884\nEpoch 9/100\n4163/4163 [==============================] - 0s 86us/step - loss: 1.0743 - acc: 0.5153 - val_loss: 1.1284 - val_acc: 0.5279\nEpoch 10/100\n4163/4163 [==============================] - 0s 79us/step - loss: 1.0724 - acc: 0.5198 - val_loss: 1.1178 - val_acc: 0.5320\nEpoch 11/100\n4163/4163 [==============================] - 0s 101us/step - loss: 1.0631 - acc: 0.5208 - val_loss: 1.1202 - val_acc: 0.5401\nEpoch 12/100\n4163/4163 [==============================] - 0s 88us/step - loss: 1.0638 - acc: 0.5210 - val_loss: 1.1187 - val_acc: 0.5306\nEpoch 13/100\n4163/4163 [==============================] - 0s 82us/step - loss: 1.0598 - acc: 0.5253 - val_loss: 1.1385 - val_acc: 0.5156\nEpoch 14/100\n4163/4163 [==============================] - 0s 80us/step - loss: 1.0667 - acc: 0.5167 - val_loss: 1.1311 - val_acc: 0.5415\nEpoch 15/100\n4163/4163 [==============================] - 0s 84us/step - loss: 1.0591 - acc: 0.5198 - val_loss: 1.1477 - val_acc: 0.4898\nEpoch 16/100\n4163/4163 [==============================] - 0s 84us/step - loss: 1.0680 - acc: 0.5169 - val_loss: 1.1121 - val_acc: 0.5293\nEpoch 17/100\n4163/4163 [==============================] - 0s 82us/step - loss: 1.0628 - acc: 0.5289 - val_loss: 1.1220 - val_acc: 0.5279\nEpoch 18/100\n4163/4163 [==============================] - 0s 80us/step - loss: 1.0644 - acc: 0.5309 - val_loss: 1.1121 - val_acc: 0.5361\nEpoch 19/100\n4163/4163 [==============================] - 0s 97us/step - loss: 1.0562 - acc: 0.5263 - val_loss: 1.1244 - val_acc: 0.5034\nEpoch 20/100\n4163/4163 [==============================] - 0s 85us/step - loss: 1.0568 - acc: 0.5359 - val_loss: 1.1267 - val_acc: 0.5361\nEpoch 21/100\n4163/4163 [==============================] - 0s 85us/step - loss: 1.0658 - acc: 0.5205 - val_loss: 1.1303 - val_acc: 0.5116\nEpoch 22/100\n4163/4163 [==============================] - 0s 84us/step - loss: 1.0593 - acc: 0.5179 - val_loss: 1.1205 - val_acc: 0.5347\nEpoch 23/100\n4163/4163 [==============================] - 0s 85us/step - loss: 1.0577 - acc: 0.5213 - val_loss: 1.1123 - val_acc: 0.5279\nEpoch 24/100\n4163/4163 [==============================] - 0s 90us/step - loss: 1.0640 - acc: 0.5246 - val_loss: 1.1060 - val_acc: 0.5333\nEpoch 25/100\n4163/4163 [==============================] - 0s 78us/step - loss: 1.0546 - acc: 0.5318 - val_loss: 1.1537 - val_acc: 0.4803\nEpoch 26/100\n4163/4163 [==============================] - 0s 82us/step - loss: 1.0602 - acc: 0.5301 - val_loss: 1.1055 - val_acc: 0.5265\nEpoch 27/100\n4163/4163 [==============================] - 0s 84us/step - loss: 1.0531 - acc: 0.5304 - val_loss: 1.1091 - val_acc: 0.5129\nEpoch 28/100\n4163/4163 [==============================] - 0s 111us/step - loss: 1.0536 - acc: 0.5261 - val_loss: 1.1221 - val_acc: 0.5293\nEpoch 29/100\n4163/4163 [==============================] - 0s 102us/step - loss: 1.0594 - acc: 0.5285 - val_loss: 1.1071 - val_acc: 0.5252\nEpoch 30/100\n4163/4163 [==============================] - 0s 83us/step - loss: 1.0505 - acc: 0.5347 - val_loss: 1.1270 - val_acc: 0.5156\nEpoch 31/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0594 - acc: 0.5306 - val_loss: 1.1082 - val_acc: 0.5238\nEpoch 32/100\n4163/4163 [==============================] - 0s 67us/step - loss: 1.0554 - acc: 0.5256 - val_loss: 1.1107 - val_acc: 0.5320\nEpoch 33/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0550 - acc: 0.5265 - val_loss: 1.1195 - val_acc: 0.5347\nEpoch 34/100\n4163/4163 [==============================] - 0s 82us/step - loss: 1.0503 - acc: 0.5306 - val_loss: 1.1232 - val_acc: 0.5374\nEpoch 35/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0524 - acc: 0.5323 - val_loss: 1.1072 - val_acc: 0.5184\nEpoch 36/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0483 - acc: 0.5299 - val_loss: 1.1103 - val_acc: 0.5075\nEpoch 37/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0497 - acc: 0.5301 - val_loss: 1.1151 - val_acc: 0.5252\nEpoch 38/100\n4163/4163 [==============================] - 0s 72us/step - loss: 1.0436 - acc: 0.5340 - val_loss: 1.1240 - val_acc: 0.5211\nEpoch 39/100\n4163/4163 [==============================] - 0s 67us/step - loss: 1.0439 - acc: 0.5273 - val_loss: 1.1161 - val_acc: 0.5320\nEpoch 40/100\n4163/4163 [==============================] - 0s 74us/step - loss: 1.0524 - acc: 0.5280 - val_loss: 1.1338 - val_acc: 0.5333\nEpoch 41/100\n4163/4163 [==============================] - 0s 71us/step - loss: 1.0502 - acc: 0.5345 - val_loss: 1.1267 - val_acc: 0.5061\nEpoch 42/100\n4163/4163 [==============================] - 0s 73us/step - loss: 1.0566 - acc: 0.5285 - val_loss: 1.1145 - val_acc: 0.5429\nEpoch 43/100\n4163/4163 [==============================] - 0s 70us/step - loss: 1.0468 - acc: 0.5330 - val_loss: 1.1234 - val_acc: 0.5333\nEpoch 44/100\n4163/4163 [==============================] - 0s 81us/step - loss: 1.0498 - acc: 0.5323 - val_loss: 1.1297 - val_acc: 0.5075\nEpoch 45/100\n4163/4163 [==============================] - 0s 75us/step - loss: 1.0538 - acc: 0.5299 - val_loss: 1.1598 - val_acc: 0.5143\nEpoch 46/100\n4163/4163 [==============================] - 0s 77us/step - loss: 1.0520 - acc: 0.5273 - val_loss: 1.1359 - val_acc: 0.5102\nEpoch 47/100\n4163/4163 [==============================] - 0s 76us/step - loss: 1.0430 - acc: 0.5287 - val_loss: 1.1114 - val_acc: 0.5211\nEpoch 48/100\n4163/4163 [==============================] - 0s 69us/step - loss: 1.0440 - acc: 0.5366 - val_loss: 1.1239 - val_acc: 0.5129\nEpoch 49/100\n4163/4163 [==============================] - 0s 71us/step - loss: 1.0464 - acc: 0.5359 - val_loss: 1.1339 - val_acc: 0.5252\nEpoch 50/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0410 - acc: 0.5371 - val_loss: 1.1028 - val_acc: 0.5524\nEpoch 51/100\n4163/4163 [==============================] - 0s 71us/step - loss: 1.0394 - acc: 0.5340 - val_loss: 1.1248 - val_acc: 0.5333\nEpoch 52/100\n4163/4163 [==============================] - 0s 74us/step - loss: 1.0565 - acc: 0.5311 - val_loss: 1.1312 - val_acc: 0.5143\nEpoch 53/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0528 - acc: 0.5265 - val_loss: 1.1260 - val_acc: 0.5306\nEpoch 54/100\n4163/4163 [==============================] - 0s 78us/step - loss: 1.0467 - acc: 0.5378 - val_loss: 1.1236 - val_acc: 0.5320\nEpoch 55/100\n4163/4163 [==============================] - 0s 85us/step - loss: 1.0525 - acc: 0.5301 - val_loss: 1.1108 - val_acc: 0.5510\nEpoch 56/100\n4163/4163 [==============================] - 0s 76us/step - loss: 1.0417 - acc: 0.5323 - val_loss: 1.1194 - val_acc: 0.5184\nEpoch 57/100\n4163/4163 [==============================] - 0s 74us/step - loss: 1.0378 - acc: 0.5386 - val_loss: 1.1319 - val_acc: 0.5129\nEpoch 58/100\n4163/4163 [==============================] - 0s 64us/step - loss: 1.0415 - acc: 0.5362 - val_loss: 1.1239 - val_acc: 0.5374\nEpoch 59/100\n4163/4163 [==============================] - 0s 68us/step - loss: 1.0360 - acc: 0.5328 - val_loss: 1.1317 - val_acc: 0.5211\nEpoch 60/100\n"
],
[
"model.evaluate(x_t2,y_t2)",
"398/398 [==============================] - 0s 63us/sample - loss: 1.0768 - accuracy: 0.5327\n"
],
[
"tf.keras.__version__",
"_____no_output_____"
],
[
"pre=model.predict(x_t2)\npre=np.argmax(pre,axis=1)\npre",
"_____no_output_____"
],
[
"print(classification_report(y_t2,pre))",
" precision recall f1-score support\n\n 3.0 0.00 0.00 0.00 5\n 4.0 0.50 0.08 0.14 24\n 5.0 0.59 0.40 0.48 221\n 6.0 0.51 0.83 0.63 329\n 7.0 0.48 0.17 0.25 121\n 8.0 1.00 0.03 0.06 34\n 9.0 0.00 0.00 0.00 1\n\navg / total 0.55 0.52 0.47 735\n\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d88df49e7b73509f54c5ea81451a7b2ae58e1 | 10,329 | ipynb | Jupyter Notebook | Course_1-PreLaunch_Preparatory_Content/Module_1-Introduction_to_Python_Programming/4-Object-Oriented_Programming_in_Python/Session.ipynb | Mathews-Tom/MSc_in_Machine_Learning_and_Artificial_Intelligence | 789bfeea203ccad5b5fd4bd10f4437a2752fe2e8 | [
"MIT"
] | null | null | null | Course_1-PreLaunch_Preparatory_Content/Module_1-Introduction_to_Python_Programming/4-Object-Oriented_Programming_in_Python/Session.ipynb | Mathews-Tom/MSc_in_Machine_Learning_and_Artificial_Intelligence | 789bfeea203ccad5b5fd4bd10f4437a2752fe2e8 | [
"MIT"
] | null | null | null | Course_1-PreLaunch_Preparatory_Content/Module_1-Introduction_to_Python_Programming/4-Object-Oriented_Programming_in_Python/Session.ipynb | Mathews-Tom/MSc_in_Machine_Learning_and_Artificial_Intelligence | 789bfeea203ccad5b5fd4bd10f4437a2752fe2e8 | [
"MIT"
] | 1 | 2022-03-23T03:54:55.000Z | 2022-03-23T03:54:55.000Z | 23.421769 | 139 | 0.486494 | [
[
[
"<h1 style = \"color : Blue\"> i. Creating Classes and Objects </h1>",
"_____no_output_____"
],
[
"<h2 style = \"color : Brown\">Defining a class</h2>\n\n- length and breadth as attributes\n- __init__() - constructor of class\n- self parameter - refers to the newly created instance of the class. \n- attributes length and breadth are associated with self-keyword to identify them as instance variables",
"_____no_output_____"
]
],
[
[
"class Rectangle :\n def __init__(self):\n self.length = 10\n self.breadth = 5",
"_____no_output_____"
]
],
[
[
"- create the object by calling name of the class followed by parenthesis. \n- print the values using dot operator",
"_____no_output_____"
]
],
[
[
"rect = Rectangle()\nprint(\"Length = \",rect.length, \"\\nBreadth = \" ,rect.breadth)",
"Length = 10 \nBreadth = 5\n"
]
],
[
[
"<h2 style = \"color : Brown\">Parametrised Constructor</h2>\n\n- parametrised constructor - dynamically assign the attribute values during object creation",
"_____no_output_____"
]
],
[
[
"class Rectangle :\n def __init__(self, length, breadth):\n self.length = length\n self.breadth = breadth\n \nrect = Rectangle(10, 5)\nprint(\"Length = \",rect.length, \"\\nBreadth = \" ,rect.breadth)",
"Length = 10 \nBreadth = 5\n"
]
],
[
[
"<h1 style = \"color : Blue\">ii. Class Variable and Instance variables ",
"_____no_output_____"
]
],
[
[
"class Circle :\n pi = 3.14\n def __init__(self, radius):\n self.radius = radius",
"_____no_output_____"
],
[
"circle_1 = Circle(5)\nprint(\"Radius = {} \\t pi = {}\".format(circle_1.radius,circle_1.pi))\n\ncircle_2 = Circle(2)\nprint(\"Radius = {} \\t pi = {}\".format(circle_2.radius,circle_2.pi))",
"Radius = 5 \t pi = 3.14\nRadius = 2 \t pi = 3.14\n"
],
[
"Circle.pi = 3.1436\n\ncircle_1 = Circle(5)\nprint(\"Radius = {} \\t pi = {}\".format(circle_1.radius,circle_1.pi))\n\ncircle_2 = Circle(2)\nprint(\"Radius = {} \\t pi = {}\".format(circle_2.radius,circle_2.pi))",
"Radius = 5 \t pi = 3.1436\nRadius = 2 \t pi = 3.1436\n"
]
],
[
[
"<h1 style = \"color : Blue\">iii. Adding a method to class</h1>\n\n- calculate_area() - retutns the product of attributes length and breadth \n- self - identifies its association with the instance",
"_____no_output_____"
]
],
[
[
"class Rectangle :\n def __init__(self, length, breadth):\n self.length = length\n self.breadth = breadth\n \n def calculate_area(self):\n return self.length * self.breadth\n \nrect = Rectangle(10, 5)\nprint(\"Length = \",rect.length, \"\\nBreadth = \" ,rect.breadth, \"\\nArea = \", rect.calculate_area())",
"Length = 10 \nBreadth = 5 \nArea = 50\n"
]
],
[
[
"<h2 style = \"color : Brown\"> Significance of self:</h2>\n\n- The attributes length and breadth are associated with an instance.\n- Self makes sure that each instance refers to its own copy of attributes",
"_____no_output_____"
]
],
[
[
"new_rect = Rectangle(15, 8)\nprint(\"Length = \",new_rect.length, \"\\nBreadth = \" ,new_rect.breadth, \"\\nArea = \", new_rect.calculate_area())",
"Length = 15 \nBreadth = 8 \nArea = 120\n"
],
[
"print(\"Length = \",rect.length, \"\\nBreadth = \" ,rect.breadth, \"\\nArea = \", rect.calculate_area())",
"Length = 10 \nBreadth = 5 \nArea = 50\n"
]
],
[
[
"<h1 style = \"color : Blue\">iv. Class Method and Static Method",
"_____no_output_____"
]
],
[
[
"class Circle :\n pi = 3.14\n def __init__(self, radius):\n self.radius = radius\n \n # Instance Method \n def calculate_area(self):\n return Circle.pi * self.radius\n \n # Class Method - I cannot access - radius\n @classmethod\n def access_pi(cls):\n pi = 3.1436\n return pi\n \n # Static Method - I cannot access - pi and radius\n @staticmethod\n def circle_static_method():\n print(\"This is circle's static method\")\n \ncir = Circle(5)\n\n# Calling methods \n\nprint(cir.calculate_area())\n\nprint(Circle.access_pi())\n\nCircle.circle_static_method()",
"15.700000000000001\n3.1436\nThis is circle's static method\n"
]
],
[
[
"<h1 style = \"color : Blue\">v. Inheritance and Overriding",
"_____no_output_____"
]
],
[
[
"class Shape :\n \n \n def set_color(self, color):\n self.color = color\n \n def calculate_area(self):\n pass\n \n def color_the_shape(self):\n color_price = {\"red\" : 10, \"blue\" : 15, \"green\" : 5}\n return self.calculate_area() * color_price[self.color]",
"_____no_output_____"
],
[
"class Circle(Shape) :\n pi = 3.14\n def __init__(self, radius):\n self.radius = radius\n \n # overriding\n def calculate_area(self):\n return Circle.pi * self.radius",
"_____no_output_____"
],
[
"c = Circle(5)\nc.set_color(\"red\")\nprint(\"Circle with radius =\",c.radius ,\"when colored\", c.color,\"costs $\",c.color_the_shape())",
"Circle with radius = 5 when colored red costs $ 157.0\n"
],
[
"class Rectangle(Shape) :\n def __init__(self, length, breadth):\n self.length = length\n self.breadth = breadth\n \n # Overriding user defined method \n def calculate_area(self):\n return self.length * self.breadth\n \n # Overriding python default method\n def __str__(self):\n return \"area of rectangle = \" + str(self.calculate_area())",
"_____no_output_____"
],
[
"r = Rectangle(5, 10)\nr.set_color(\"blue\")\nprint(\"Rectangle with length =\",r.length ,\" and breadth = \",r.breadth ,\"when colored\", r.color,\"costs $\",r.color_the_shape())",
"Rectangle with length = 5 and breadth = 10 when colored blue costs $ 750\n"
],
[
"print(r)",
"area of rectangle = 50\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d93a24bebdf4a455e902aec8fa65a445fcdd6 | 82,091 | ipynb | Jupyter Notebook | src/02_Pet_Stores_and_Services.ipynb | acdick/framing_data_with_dataframes | 8eccdb0db91fe2919235ead1f839668ae8968ca0 | [
"MIT"
] | 3 | 2019-04-14T00:05:59.000Z | 2020-12-02T15:15:38.000Z | src/02_Pet_Stores_and_Services.ipynb | acdick/framing_data_with_dataframes | 8eccdb0db91fe2919235ead1f839668ae8968ca0 | [
"MIT"
] | null | null | null | src/02_Pet_Stores_and_Services.ipynb | acdick/framing_data_with_dataframes | 8eccdb0db91fe2919235ead1f839668ae8968ca0 | [
"MIT"
] | null | null | null | 27.95063 | 105 | 0.330962 | [
[
[
"from config import client_id, api_key\n\nimport pandas as pd\nimport requests\nimport json\nimport time\nimport sqlite3",
"_____no_output_____"
]
],
[
[
"# Yelp Pet Stores and Pet Services in NYC (ETL): Extract",
"_____no_output_____"
]
],
[
[
"#function to parse Yelp Fusion API response for a list of Yelp businesses\ndef parse_response(response):\n response_json = response.json()\n businesses = []\n \n for business in response_json['businesses']:\n business_json = {}\n categories = []\n \n for category in business['categories']:\n categories.append(category['title'])\n \n business_json['categories'] = categories\n business_json['id'] = business['id']\n business_json['name'] = business['name']\n business_json['is_closed'] = business['is_closed']\n business_json['review_count'] = business['review_count']\n business_json['rating'] = business['rating']\n business_json['zip_code'] = business['location']['zip_code']\n \n businesses.append(business_json)\n \n return businesses\n \n#function to call Yelp Fusion API with given search term, location and max search results\ndef call_yelp(term, location, search_max):\n search_results = []\n search_limit = 50\n search_calls = int(search_max / search_limit)\n \n for i in range(search_calls):\n search_offset = i * search_limit\n url = 'https://api.yelp.com/v3/businesses/search'\n headers = {'Authorization': 'Bearer {}'.format(api_key),}\n url_params = {'term': term.replace(' ', '+'),\n 'location': location.replace(' ', '+'),\n 'limit': search_limit,\n 'offset': search_offset\n }\n \n response = requests.get(url, headers=headers, params=url_params)\n print('term: {}, offset: {}, response: {}'.format(term, search_offset, response))\n \n search_results.extend(parse_response(response))\n time.sleep(2)\n \n return search_results",
"_____no_output_____"
],
[
"#pull and save 1,000 (max) pet stores in NYC\npet_stores = 'Pet Stores'\nlocation = 'New York, NY'\nsearch_max = 1000\n\nyelp_pet_stores = call_yelp(pet_stores, location, search_max)",
"term: Pet Stores, offset: 0, response: <Response [200]>\nterm: Pet Stores, offset: 50, response: <Response [200]>\nterm: Pet Stores, offset: 100, response: <Response [200]>\nterm: Pet Stores, offset: 150, response: <Response [200]>\nterm: Pet Stores, offset: 200, response: <Response [200]>\nterm: Pet Stores, offset: 250, response: <Response [200]>\nterm: Pet Stores, offset: 300, response: <Response [200]>\nterm: Pet Stores, offset: 350, response: <Response [200]>\nterm: Pet Stores, offset: 400, response: <Response [200]>\nterm: Pet Stores, offset: 450, response: <Response [200]>\nterm: Pet Stores, offset: 500, response: <Response [200]>\nterm: Pet Stores, offset: 550, response: <Response [200]>\nterm: Pet Stores, offset: 600, response: <Response [200]>\nterm: Pet Stores, offset: 650, response: <Response [200]>\nterm: Pet Stores, offset: 700, response: <Response [200]>\nterm: Pet Stores, offset: 750, response: <Response [200]>\nterm: Pet Stores, offset: 800, response: <Response [200]>\nterm: Pet Stores, offset: 850, response: <Response [200]>\nterm: Pet Stores, offset: 900, response: <Response [200]>\nterm: Pet Stores, offset: 950, response: <Response [200]>\n"
],
[
"#pull and save 1,000 (max) pet services in NYC\npet_services = 'Pet Services'\nlocation = 'New York, NY'\nsearch_max = 1000\n\nyelp_pet_services = call_yelp(pet_services, location, search_max)",
"term: Pet Services, offset: 0, response: <Response [200]>\nterm: Pet Services, offset: 50, response: <Response [200]>\nterm: Pet Services, offset: 100, response: <Response [200]>\nterm: Pet Services, offset: 150, response: <Response [200]>\nterm: Pet Services, offset: 200, response: <Response [200]>\nterm: Pet Services, offset: 250, response: <Response [200]>\nterm: Pet Services, offset: 300, response: <Response [200]>\nterm: Pet Services, offset: 350, response: <Response [200]>\nterm: Pet Services, offset: 400, response: <Response [200]>\nterm: Pet Services, offset: 450, response: <Response [200]>\nterm: Pet Services, offset: 500, response: <Response [200]>\nterm: Pet Services, offset: 550, response: <Response [200]>\nterm: Pet Services, offset: 600, response: <Response [200]>\nterm: Pet Services, offset: 650, response: <Response [200]>\nterm: Pet Services, offset: 700, response: <Response [200]>\nterm: Pet Services, offset: 750, response: <Response [200]>\nterm: Pet Services, offset: 800, response: <Response [200]>\nterm: Pet Services, offset: 850, response: <Response [200]>\nterm: Pet Services, offset: 900, response: <Response [200]>\nterm: Pet Services, offset: 950, response: <Response [200]>\n"
]
],
[
[
"# Yelp Pet Stores and Pet Services in NYC (ETL): Transform",
"_____no_output_____"
]
],
[
[
"# create dataframe of pet stores\npet_stores = pd.DataFrame.from_dict(yelp_pet_stores)\npet_stores.head()",
"_____no_output_____"
],
[
"# create dataframe of pet services\npet_services = pd.DataFrame.from_dict(yelp_pet_services)\npet_services.head()",
"_____no_output_____"
],
[
"#create list of unique pet store and service categoories\npet_categories = []\n\nfor row in range(pet_stores.shape[0]):\n pet_categories.extend(pet_stores['categories'][row])\n \nfor row in range(pet_services.shape[0]):\n pet_categories.extend(pet_services['categories'][row])\n \npet_categories = sorted(list(set(pet_categories)))\npet_categories",
"_____no_output_____"
],
[
"#create junction table for businesses and categories\nbusiness_ids = []\ncategory_ids = []\n\nfor row in range(pet_stores.shape[0]):\n for category in pet_stores['categories'][row]:\n business_ids.append(pet_stores['id'][row])\n category_ids.append(pet_categories.index(category))\n \nfor row in range(pet_services.shape[0]):\n for category in pet_services['categories'][row]:\n business_ids.append(pet_services['id'][row])\n category_ids.append(pet_categories.index(category))\n \nprint(len(business_ids))\nprint(len(category_ids))",
"3274\n3274\n"
]
],
[
[
"# Yelp Pet Stores and Pet Services in NYC (ETL): Load",
"_____no_output_____"
]
],
[
[
"#creating SQL connection\nconn = sqlite3.connect('../Data/pet_care_industry.db')\nc = conn.cursor()\n\n#function to create table\ndef create_table(query):\n c.execute(query)\n\n#function to close connection\ndef close_c_conn():\n c.close()\n conn.close()",
"_____no_output_____"
],
[
"#create pet stores and services table\ncreate_query = \"\"\"CREATE TABLE stores_and_services\n (id TEXT PRIMARY KEY,\n Name TEXT,\n Rating REAL,\n Review_Count INTEGER,\n ZipCode INTEGER);\"\"\"\n\nc.execute('DROP TABLE IF EXISTS stores_and_services')\ncreate_table(create_query)",
"_____no_output_____"
],
[
"#function to insert businesses into table\ndef insert_businesses(businesses):\n for i in range(len(businesses.index)):\n if (not businesses.iloc[i]['is_closed']) & (businesses.iloc[i]['zip_code'].isnumeric()):\n c.execute(\"\"\"INSERT OR REPLACE INTO stores_and_services\n (id,\n Name,\n Rating,\n Review_Count,\n ZipCode)\n VALUES\n (?,?,?,?,?)\"\"\",\n (businesses.iloc[i]['id'],\n businesses.iloc[i]['name'],\n float(businesses.iloc[i]['rating']),\n int(businesses.iloc[i]['review_count']),\n int(businesses.iloc[i]['zip_code'])))\n \n conn.commit()\n \n#insert pet store and services into table\ninsert_businesses(pet_stores)\ninsert_businesses(pet_services)",
"_____no_output_____"
],
[
"#check SQL pet store and services table\nstores_and_services = pd.read_sql_query(\"\"\"SELECT Name, Rating, Review_Count, ZipCode\n FROM stores_and_services;\"\"\", conn)\nstores_and_services = stores_and_services.set_index('Name')\nstores_and_services",
"_____no_output_____"
],
[
"#create unique categories table\ncreate_query = \"\"\"CREATE TABLE categories\n (id TEXT PRIMARY KEY,\n Name TEXT);\"\"\"\n\nc.execute('DROP TABLE IF EXISTS categories')\ncreate_table(create_query)",
"_____no_output_____"
],
[
"#function to insert categories into table\ndef insert_categories(categories):\n for i in range(len(categories)):\n c.execute(\"\"\"INSERT INTO categories\n (id,\n Name)\n VALUES\n (?,?)\"\"\",\n (i,\n categories[i]))\n \n conn.commit()\n \n#insert categories into table\ninsert_categories(pet_categories)",
"_____no_output_____"
],
[
"#check SQL categories table\ncategories = pd.read_sql_query(\"\"\"SELECT * FROM categories;\"\"\", conn)\ncategories = categories.set_index('id')\ncategories",
"_____no_output_____"
],
[
"#defining SQL query to create junction table for businesses and categories\ncreate_query = \"\"\"CREATE TABLE IF NOT EXISTS businesses_categories\n( business_id TEXT NOT NULL,\n category_id INTEGER NOT NULL,\n PRIMARY KEY(business_id, category_id),\n FOREIGN KEY(business_id) REFERENCES stores_and_services(id),\n FOREIGN KEY(category_id) REFERENCES categories(id));\"\"\"\n\nc.execute('DROP TABLE IF EXISTS businesses_categories')\ncreate_table(create_query)",
"_____no_output_____"
],
[
"#function to insert businesses and categories into table\ndef insert_businesses_categories(business_ids, category_ids):\n for i in range(len(business_ids)):\n c.execute(\"\"\"INSERT OR REPLACE INTO businesses_categories\n (business_id,\n category_id)\n VALUES\n (?,?)\"\"\",\n (business_ids[i],\n category_ids[i]))\n \n conn.commit()\n \n#insert categories into table\ninsert_businesses_categories(business_ids, category_ids)",
"_____no_output_____"
],
[
"#querying SQL businesses and categories table\nbusinesses_categories = pd.read_sql_query(\"SELECT * FROM businesses_categories;\", conn)\nbusinesses_categories",
"_____no_output_____"
],
[
"#close connection\nclose_c_conn()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71d9439d69528958087b523047a934ed860f825 | 90,196 | ipynb | Jupyter Notebook | docs/source/tutorials/neurotransmitter_prediction.ipynb | SridharJagannathan/pyroglancer | 7b5e44289d24bc85269fe1305e22f2ae49948f36 | [
"BSD-3-Clause"
] | null | null | null | docs/source/tutorials/neurotransmitter_prediction.ipynb | SridharJagannathan/pyroglancer | 7b5e44289d24bc85269fe1305e22f2ae49948f36 | [
"BSD-3-Clause"
] | 12 | 2021-01-21T16:27:48.000Z | 2022-01-24T12:13:53.000Z | docs/source/tutorials/neurotransmitter_prediction.ipynb | SridharJagannathan/pyroglancer | 7b5e44289d24bc85269fe1305e22f2ae49948f36 | [
"BSD-3-Clause"
] | 2 | 2021-03-17T09:30:50.000Z | 2021-07-27T13:56:57.000Z | 99.007684 | 59,220 | 0.818185 | [
[
[
"# Co-plotting hemibrain skeleton with flywire dataset for predicting neurotransmitters of typed hemibrain neurons",
"_____no_output_____"
],
[
"### This section contains example of fetching skeleton from hemibrain dataset and co-plotting with a flywire instance",
"_____no_output_____"
],
[
"### Import neccesary library modules now",
"_____no_output_____"
]
],
[
[
"import navis\nimport fafbseg\nimport flybrains",
"_____no_output_____"
],
[
"from pyroglancer.localserver import startdataserver, closedataserver\nfrom pyroglancer.flywire import flywireurl2dict, add_flywirelayer, set_flywireviewerstate",
"_____no_output_____"
],
[
"import navis.interfaces.neuprint as neu\nfrom navis.interfaces.neuprint import NeuronCriteria as NC, SynapseCriteria as SC",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n#font = {'family': 'serif', 'serif': ['Palatino']}\n#font = {'family': 'serif', 'serif': ['Helvetica']}\n#plt.rc('font', **font)\n#plt.rc('text', usetex=False)\n\n# This makes it so the PDF export works\nimport matplotlib as mpl\nmpl.rcParams['pdf.use14corefonts']=True\nmpl.rcParams['font.sans-serif'] = ['Helvetica',\n 'DejaVu Sans',\n 'Bitstream Vera Sans',\n 'Computer Modern Sans Serif',\n 'Lucida Grande',\n 'Verdana',\n 'Geneva',\n 'Lucid',\n 'Arial', \n 'Avant Garde',\n 'sans-serif']\nmpl.rcParams['font.size'] = 11",
"_____no_output_____"
]
],
[
[
"### Set configurations to fetch from data from neuprint",
"_____no_output_____"
]
],
[
[
"client = neu.Client('https://neuprint.janelia.org/', dataset='hemibrain:v1.2.1')",
"_____no_output_____"
],
[
"client",
"_____no_output_____"
]
],
[
[
"### Get some typed neurons and neuropil meshes from neuprint",
"_____no_output_____"
]
],
[
[
"neurons_df, roi_counts_df = neu.fetch_neurons(NC(status='Traced',type='ER5',regex=True)) #get some ER5 neurons..",
"_____no_output_____"
],
[
"er5bodyidList = neurons_df[['bodyId']]",
"_____no_output_____"
],
[
"er5bodyidList",
"_____no_output_____"
],
[
"er5_skel = neu.fetch_skeletons(er5bodyidList, heal=1000, max_threads=30)",
"_____no_output_____"
],
[
"#fetch mesh for Right Antenna lobe..\nal_R = neu.fetch_roi('AL(R)')",
"_____no_output_____"
],
[
"hemibrain_mesh = navis.Volume(flybrains.JRCFIB2018Fraw.mesh)",
"_____no_output_____"
],
[
"figwidth_size = 5\nfigheigth_size = 5",
"_____no_output_____"
],
[
"#navis display options for frontal view..\nelev = -180\nazim = -90\ndist = 6",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(figwidth_size, figheigth_size))\nax = fig.add_subplot(1, 1, 1, projection = '3d')\nnavis.plot2d(er5_skel, method='3d', ax = ax, connectors=False)\nnavis.plot2d(hemibrain_mesh, method='3d', ax = ax, connectors=False)\n# Rotate to frontal view\nax.elev = elev\nax.azim = azim\nax.dist = dist",
"_____no_output_____"
]
],
[
[
"### Transform them to Flywire space..",
"_____no_output_____"
]
],
[
[
"flywire_neuron=navis.xform_brain(er5_skel,source='JRCFIB2018Fraw', target='FLYWIRE')",
"INFO : Pre-caching deformation field(s) for transforms... (navis)\nINFO - 2021-08-11 19:23:40,154 - base - Pre-caching deformation field(s) for transforms...\n"
],
[
"flywirevol = {}\nflywirevol['hemibrain']=navis.xform_brain(hemibrain_mesh, source='JRCFIB2018Fraw', target='FLYWIRE')\nflywirevol['AL_R']=navis.xform_brain(al_R, source='JRCFIB2018Fraw', target='FLYWIRE')",
"Transform path: JRCFIB2018Fraw -> JRCFIB2018F -> JRCFIB2018Fum -> JRC2018F -> FAFB14um -> FAFB14 -> FAFB14raw -> FLYWIREraw -> FLYWIRE\nTransform path: JRCFIB2018Fraw -> JRCFIB2018F -> JRCFIB2018Fum -> JRC2018F -> FAFB14um -> FAFB14 -> FAFB14raw -> FLYWIREraw -> FLYWIRE\n"
],
[
"flywirevol['hemibrain'].id = 200\nflywirevol['AL_R'].id = 300\nflywirevol",
"_____no_output_____"
]
],
[
[
"### Start the dataserver to host precomputed data..",
"_____no_output_____"
]
],
[
[
"startdataserver()",
"Serving data from: /var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0\nServing directory at http://127.0.0.1:8000\n"
]
],
[
[
"### Get the url from flywire to append data onto..",
"_____no_output_____"
]
],
[
[
"shorturl = 'https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4646812430368768'\n#This flywire segments has some stuff similar to the DA1 PNs, you will notice their closeness in the final plot",
"_____no_output_____"
]
],
[
[
"### Add bodyids to flywire layers..",
"_____no_output_____"
]
],
[
[
"tmpviewer = add_flywirelayer(flywireurl2dict(shorturl), layer_kws = {'type': 'skeletons',\n 'source': flywire_neuron,\n 'name': 'hemibrain_skels',\n 'color': 'red'}) \n#'alpha': 0.3, doesn't work yet in skeleton layers..",
"deleting.. /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1200057627\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230712894\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230738118\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230742552\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230738247\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230742517\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/5812979604\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230712956\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1231079482\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1231066741\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1231070792\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1200049187\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230738237\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1261751722\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1230742431\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1231070863\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1231066732\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/5813049931\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/5813020453\n/private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/1231066662\ncreating: /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_skels/skeletons/seg_props\n{'1200057627': ['#ff0000'], '1230712894': ['#ff0000'], '1230738118': ['#ff0000'], '1230742552': ['#ff0000'], '1230738247': ['#ff0000'], '1230742517': ['#ff0000'], '5812979604': ['#ff0000'], '1230712956': ['#ff0000'], '1231079482': ['#ff0000'], '1231066741': ['#ff0000'], '1231070792': ['#ff0000'], '1200049187': ['#ff0000'], '1230738237': ['#ff0000'], '1261751722': ['#ff0000'], '1230742431': ['#ff0000'], '1231070863': ['#ff0000'], '1231066732': ['#ff0000'], '5813049931': ['#ff0000'], '5813020453': ['#ff0000'], '1231066662': ['#ff0000']}\nflywire url at: https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6397512290140160\n"
]
],
[
[
"### Add neuropil meshes to flywire layers",
"_____no_output_____"
]
],
[
[
"shorturl = tmpviewer",
"_____no_output_____"
],
[
"tmpviewer = add_flywirelayer(flywireurl2dict(shorturl), layer_kws = {'type': 'volumes',\n 'source': [flywirevol['hemibrain'],\n flywirevol['AL_R']],\n 'name': 'hemibrain_neuropils',\n 'color': ['grey', 'magenta'], \n 'alpha': 0.3})",
"deleting.. /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_neuropils/mesh\nmesh/200\nSeg id is: 200\nFull filepath: /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_neuropils/mesh/200\nmesh/300\nSeg id is: 300\nFull filepath: /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_neuropils/mesh/300\ncreating: /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_neuropils/mesh/segment_properties\ncreating: /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0/precomputed/hemibrain_neuropils/mesh/segment_names\nflywire url at: https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4849669104467968\n"
],
[
"tmpviewer,shorturl = set_flywireviewerstate(tmpviewer, axis_lines=False, bounding_box=False)",
"flywire url at: https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5975569011310592\n"
],
[
"#some matches are.. 1231066732 --> 720575940630849069, 5812979604 --> 720575940624108862",
"_____no_output_____"
],
[
"#some of our matching er5 neurons are..\n#pre_syn = fafbseg.flywire.fetch_synapses([720575940620940045], transmitters=True, post=False)\n#fafbseg.flywire.predict_transmitter([720575940620940045], single_pred=True)",
"_____no_output_____"
],
[
"fafbseg.flywire.predict_transmitter([720575940630849069, 720575940624108862])",
"_____no_output_____"
]
],
[
[
"### Screenshot of the flywire instance",
"_____no_output_____"
],
[
"#### you can see the skeletons from hemibrain (red) plotted in the skeleton layer, and some similar neurons (meshes) of the flywire dataset plotted in green and cyan",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"### Close the dataserver",
"_____no_output_____"
]
],
[
[
"closedataserver()",
"Closing server at http://127.0.0.1:8000\nCleaning directory at /private/var/folders/4_/1td4h5lj50z735wckw6pjd5h0000gn/T/tmp_mub8so0\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
e71d959b4d278efe57ea2a308d9e81ecfa46e152 | 43,398 | ipynb | Jupyter Notebook | Notebooks/VQE.ipynb | cmleecm/QuantumCourse | 4e616a35cf502b28e55446780ca8f42414964073 | [
"Apache-2.0"
] | 16 | 2019-04-14T18:26:12.000Z | 2021-11-22T08:08:40.000Z | Notebooks/VQE.ipynb | cmleecm/QuantumCourse | 4e616a35cf502b28e55446780ca8f42414964073 | [
"Apache-2.0"
] | 1 | 2020-01-21T06:50:16.000Z | 2020-01-21T06:50:16.000Z | Notebooks/VQE.ipynb | gomeztato/QuantumCourse | 881d03635332ae4627975e713c30c1b833cabe21 | [
"Apache-2.0"
] | 3 | 2019-04-16T10:15:29.000Z | 2020-01-29T15:14:25.000Z | 78.053957 | 21,272 | 0.756648 | [
[
[
"# Implementation of a Variational Quantum Eigensolver (VQE).\n\nThis example is based on the exampled provided by [ProjectQ](https://raw.githubusercontent.com/ProjectQ-Framework/ProjectQ/develop/examples/variational_quantum_eigensolver.ipynb), however, in this notebook the simulation of a QPU will be used instead of geting the expected value of the full Hamiltonian.\n\nThe example shown here is from the paper \"Scalable Quantum Simulation of\nMolecular Energies\" by P.J.J. O'Malley et al. [arXiv:1512.06860v2](https://arxiv.org/abs/1512.06860v2)\n(Note that only the latest arXiv version contains the correct coefficients of\n the Hamiltonian)\n\nEq. 2 of the paper shows the functional which one needs to minimize and Eq. 3\nshows the coupled cluster ansatz for the trial wavefunction (using the unitary\ncoupled cluster approach). The Hamiltonian is given in Eq. 1. The coefficients\ncan be found in Table 1. Note that both the ansatz and the Hamiltonian can be\ncalculated using FermiLib which is a library for simulating quantum systems\non top of ProjectQ.",
"_____no_output_____"
],
[
"Remember that the Variational Quantum Eigensolver (VQE) is a hybrid algorithm. Part of the code is executed in the CPU (the optimisation) and part in the QPU (the calculus of the expected value of the Hamiltonian). \n",
"_____no_output_____"
]
],
[
[
"import projectq\nfrom projectq.ops import All, Measure, QubitOperator, TimeEvolution, X,H,S,Y,get_inverse,Rx,Ry\n\nimport matplotlib.pyplot as plt\nfrom scipy.optimize import minimize_scalar\n\n\n\n",
"_____no_output_____"
],
[
"HH=QubitOperator(\"X1\",0.2)+QubitOperator(\"Y1\")",
"_____no_output_____"
],
[
"import numpy as np\nHH.terms[list(HH.terms.keys())[0]]",
"_____no_output_____"
],
[
"ops=HH.terms\nfor i in HH.terms.keys(): \n for j in i:\n print(j[0])\n if j[1] =='X':\n print(\"X\")\n \n# if ops[0][1] == 'Y':\n# print(\"Y\")\n",
"1\nX\n1\n"
]
],
[
[
"Load the coffecientes for the Hamiltonian for each distance. This coefficientes must be calculated externally using other classical programs. \nthe spin\nHamiltonian for molecular hydrogen in the minimal\n(STO-6G) basis\nAs shown in the paper, the spin Hamiltonian for the Hydrogen Molecule in the minimal STO-6G basis, can be approximated as:\n\n$$H=g_o I+g_1 Z_0+g_2 Z_1+g_3 Z_0 Z_1+g_4 X_0 X_1+g_5 Y_0 Y_1$$\n\nwhere ${X,Y,Z}_{0,1}$ are the Pauli Matrices applied on spin 0 or 1. This table contents the calue of each $g_i$ for each bond lenght.",
"_____no_output_____"
]
],
[
[
"# Data from paper (arXiv:1512.06860v2) table 1: R, I, Z0, Z1, Z0Z1, X0X1, Y0Y1\nraw_data_table_1 = [\n [0.20, 2.8489, 0.5678, -1.4508, 0.6799, 0.0791, 0.0791],\n [0.25, 2.1868, 0.5449, -1.2870, 0.6719, 0.0798, 0.0798],\n [0.30, 1.7252, 0.5215, -1.1458, 0.6631, 0.0806, 0.0806],\n [0.35, 1.3827, 0.4982, -1.0226, 0.6537, 0.0815, 0.0815],\n [0.40, 1.1182, 0.4754, -0.9145, 0.6438, 0.0825, 0.0825],\n [0.45, 0.9083, 0.4534, -0.8194, 0.6336, 0.0835, 0.0835],\n [0.50, 0.7381, 0.4325, -0.7355, 0.6233, 0.0846, 0.0846],\n [0.55, 0.5979, 0.4125, -0.6612, 0.6129, 0.0858, 0.0858],\n [0.60, 0.4808, 0.3937, -0.5950, 0.6025, 0.0870, 0.0870],\n [0.65, 0.3819, 0.3760, -0.5358, 0.5921, 0.0883, 0.0883],\n [0.70, 0.2976, 0.3593, -0.4826, 0.5818, 0.0896, 0.0896],\n [0.75, 0.2252, 0.3435, -0.4347, 0.5716, 0.0910, 0.0910],\n [0.80, 0.1626, 0.3288, -0.3915, 0.5616, 0.0925, 0.0925],\n [0.85, 0.1083, 0.3149, -0.3523, 0.5518, 0.0939, 0.0939],\n [0.90, 0.0609, 0.3018, -0.3168, 0.5421, 0.0954, 0.0954],\n [0.95, 0.0193, 0.2895, -0.2845, 0.5327, 0.0970, 0.0970],\n [1.00, -0.0172, 0.2779, -0.2550, 0.5235, 0.0986, 0.0986],\n [1.05, -0.0493, 0.2669, -0.2282, 0.5146, 0.1002, 0.1002],\n [1.10, -0.0778, 0.2565, -0.2036, 0.5059, 0.1018, 0.1018],\n [1.15, -0.1029, 0.2467, -0.1810, 0.4974, 0.1034, 0.1034],\n [1.20, -0.1253, 0.2374, -0.1603, 0.4892, 0.1050, 0.1050],\n [1.25, -0.1452, 0.2286, -0.1413, 0.4812, 0.1067, 0.1067],\n [1.30, -0.1629, 0.2203, -0.1238, 0.4735, 0.1083, 0.1083],\n [1.35, -0.1786, 0.2123, -0.1077, 0.4660, 0.1100, 0.1100],\n [1.40, -0.1927, 0.2048, -0.0929, 0.4588, 0.1116, 0.1116],\n [1.45, -0.2053, 0.1976, -0.0792, 0.4518, 0.1133, 0.1133],\n [1.50, -0.2165, 0.1908, -0.0666, 0.4451, 0.1149, 0.1149],\n [1.55, -0.2265, 0.1843, -0.0549, 0.4386, 0.1165, 0.1165],\n [1.60, -0.2355, 0.1782, -0.0442, 0.4323, 0.1181, 0.1181],\n [1.65, -0.2436, 0.1723, -0.0342, 0.4262, 0.1196, 0.1196],\n [1.70, -0.2508, 0.1667, -0.0251, 0.4204, 0.1211, 0.1211],\n [1.75, -0.2573, 0.1615, -0.0166, 0.4148, 0.1226, 0.1226],\n [1.80, -0.2632, 0.1565, -0.0088, 0.4094, 0.1241, 0.1241],\n [1.85, -0.2684, 0.1517, -0.0015, 0.4042, 0.1256, 0.1256],\n [1.90, -0.2731, 0.1472, 0.0052, 0.3992, 0.1270, 0.1270],\n [1.95, -0.2774, 0.1430, 0.0114, 0.3944, 0.1284, 0.1284],\n [2.00, -0.2812, 0.1390, 0.0171, 0.3898, 0.1297, 0.1297],\n [2.05, -0.2847, 0.1352, 0.0223, 0.3853, 0.1310, 0.1310],\n [2.10, -0.2879, 0.1316, 0.0272, 0.3811, 0.1323, 0.1323],\n [2.15, -0.2908, 0.1282, 0.0317, 0.3769, 0.1335, 0.1335],\n [2.20, -0.2934, 0.1251, 0.0359, 0.3730, 0.1347, 0.1347],\n [2.25, -0.2958, 0.1221, 0.0397, 0.3692, 0.1359, 0.1359],\n [2.30, -0.2980, 0.1193, 0.0432, 0.3655, 0.1370, 0.1370],\n [2.35, -0.3000, 0.1167, 0.0465, 0.3620, 0.1381, 0.1381],\n [2.40, -0.3018, 0.1142, 0.0495, 0.3586, 0.1392, 0.1392],\n [2.45, -0.3035, 0.1119, 0.0523, 0.3553, 0.1402, 0.1402],\n [2.50, -0.3051, 0.1098, 0.0549, 0.3521, 0.1412, 0.1412],\n [2.55, -0.3066, 0.1078, 0.0572, 0.3491, 0.1422, 0.1422],\n [2.60, -0.3079, 0.1059, 0.0594, 0.3461, 0.1432, 0.1432],\n [2.65, -0.3092, 0.1042, 0.0614, 0.3433, 0.1441, 0.1441],\n [2.70, -0.3104, 0.1026, 0.0632, 0.3406, 0.1450, 0.1450],\n [2.75, -0.3115, 0.1011, 0.0649, 0.3379, 0.1458, 0.1458],\n [2.80, -0.3125, 0.0997, 0.0665, 0.3354, 0.1467, 0.1467],\n [2.85, -0.3135, 0.0984, 0.0679, 0.3329, 0.1475, 0.1475]]\n",
"_____no_output_____"
]
],
[
[
"Funtion to calculate the expectation value of one Hamiltonian H for a state defined by $\\theta$. This parameter controls the time of the evolution of the Hamiltonian $H=X_o Y_1$ applied to state $|01>$. So, the final state after this time evolution will be:\n\n$$|\\Psi\\rangle = e^{-i\\theta X_0 Y_1}|01\\rangle$$\n\nand the funtion will return\n\n$$\\langle H\\rangle = \\frac{\\langle \\Psi|H|\\Psi\\rangle}{\\langle \\Psi|\\Psi\\rangle}$$",
"_____no_output_____"
]
],
[
[
"def Expectation_H(theta, hamiltonian):\n \"\"\"\n Args:\n theta (float): variational parameter for ansatz wavefunction\n hamiltonian (QubitOperator): Hamiltonian of the system\n Returns:\n energy of the wavefunction for parameter theta\n \"\"\"\n # Create a ProjectQ compiler with a simulator as a backend\n eng = projectq.MainEngine()\n # Allocate 2 qubits in state |00>\n wavefunction = eng.allocate_qureg(2)\n # Initialize the Hartree Fock state |01>\n X | wavefunction[0]\n # build the operator for ansatz wavefunction\n ansatz_op = QubitOperator('X0 Y1')\n # Apply the unitary e^{-i * ansatz_op * t}\n TimeEvolution(theta, ansatz_op) | wavefunction\n # flush all gates\n eng.flush()\n # Calculate the energy.\n # The simulator can directly return expectation values, while on a\n # real quantum devices one would have to measure each term of the\n # Hamiltonian.\n energy = eng.backend.get_expectation_value(hamiltonian, wavefunction)\n # Measure in order to return to return to a classical state\n # (as otherwise the simulator will give an error)\n All(Measure) | wavefunction\n del eng\n return energy",
"_____no_output_____"
]
],
[
[
"To measure the results of applying Y to qubit *i*, a rotation around X by $\\pi/2$ is needed. \nSimilar, the result for $X_i$ needs first a rotation around Y by $-\\pi/2$.\n\nFinally, because \n\n<center>$\\langle 0|Z|0\\rangle = +1$ and $\\langle 1|Z|1\\rangle = -1$</center>\n\n<center>$\\langle 1|Z|0\\rangle = 0$ and $\\langle 0|Z|1\\rangle = 0$</center>\n\nThen, for $$|\\Psi>=\\sum_{i=0}^{2^N-1}a_i|i\\rangle $$\n\nmeasureing on Z:\n\n$$\\langle\\Psi|I^{\\otimes m}\\otimes Z^{\\otimes n} |\\Psi\\rangle = \\sum_{i=0}^{2^N-1} (-1)^{Parity\\space n\\space qubits\\space of\\space i} a_i^2$$ \n\nFor examples, for 2 qubits:\n\n$$\\langle\\Psi|Z\\otimes Z |\\Psi\\rangle = a_0^2-a_1^2-a_2^2+a_3^2$$\n$$\\langle\\Psi|I\\otimes Z |\\Psi\\rangle = a_0^2-a_1^2+a_2^2-a_3^2$$\n$$\\langle\\Psi|Z\\otimes I |\\Psi\\rangle = a_0^2+a_1^2-a_2^2-a_3^2$$\n",
"_____no_output_____"
]
],
[
[
"def Expectation_H_shots(theta, hamiltonian,shots=1024):\n \"\"\"\n Args:\n theta (float): variational parameter for ansatz wavefunction (the evolution time for the hamiltonian)\n hamiltonian (QubitOperator): Hamiltonian of the system\n Returns:\n energy of the wavefunction for parameter theta\n \"\"\"\n print(\"Hamiltonian:\",hamiltonian)\n # Create a ProjectQ compiler with a simulator as a backend\n import numpy as np\n results=np.zeros(4)\n energy1=0\n for i in range(shots):\n eng = projectq.MainEngine()\n # Allocate 2 qubits in state |00>\n wavefunction = eng.allocate_qureg(2)\n # Initialize the Hartree Fock state |01>\n X | wavefunction[0]\n # build the operator for ansatz wavefunction\n ansatz_op = QubitOperator('X0 Y1')\n # Apply the unitary e^{-i * ansatz_op * t}\n TimeEvolution(theta, ansatz_op) | wavefunction\n # flush all gates\n eng.flush()\n # Calculate the energy.\n # The simulator can directly return expectation values, while on a\n # real quantum devices one would have to measure each term of the\n # Hamiltonian.\n energy1 = eng.backend.get_expectation_value(hamiltonian, wavefunction)\n \n # However, using the results of previous cell, we can calculate using measuresements. \n\n for ops in hamiltonian.terms.keys():\n for paulis in ops:\n if paulis[1] =='X':\n #Applying Ry(-pi/2) is equivalent to apply H, but numerically introduce less error.\n #Ry(-np.pi/2)|wavefunction[paulis[0]]\n H | wavefunction[paulis[0]]\n if paulis[1] == 'Y':\n #Applying Rx(pi/2) is equivalent to apply A and H (because only introduce a phace to |1>), \n #but numerically introduces less error\n get_inverse(S) | wavefunction[paulis[0]]\n H | wavefunction[paulis[0]]\n #Rx(np.pi/2)|wavefunction[paulis[0]]\n\n All(Measure) | wavefunction\n eng.flush()\n results[int(wavefunction[1])*2+int(wavefunction[0])]+=1\n del eng\n print(results)\n coeff=hamiltonian.terms[list(hamiltonian.terms.keys())[0]]\n if (len(list(hamiltonian.terms.keys())[0])==0):\n print(\"I\")\n energy=results[0]/shots+results[3]/shots+results[1]/shots+results[2]/shots \n if (len(list(hamiltonian.terms.keys())[0])==1):\n ops=int(np.asarray(list(hamiltonian.terms.keys()))[0][0][0])\n print(\"Ops\",ops)\n if (ops>0):\n energy=results[0]/shots+results[1]/shots-results[2]/shots-results[3]/shots\n else:\n energy=results[0]/shots-results[1]/shots+results[2]/shots-results[3]/shots \n \n if (len(list(hamiltonian.terms.keys())[0])==2):\n energy=results[0]/shots-results[1]/shots-results[2]/shots+results[3]/shots \n print(\"Coeff:\",coeff, \" Measured energy: \",coeff*energy,\" Theoretical energy:\",energy1)\n return coeff*energy",
"_____no_output_____"
]
],
[
[
"Check the results when the expectation value of H is calcualted by rotations and measurements",
"_____no_output_____"
]
],
[
[
"expectedH=Expectation_H_shots(0.3,QubitOperator(\"X0 X1\"),1024)",
"Hamiltonian: 1.0 X0 X1\n[363. 117. 121. 423.]\nCoeff: 1.0 Measured energy: 0.53515625 Theoretical energy: 0.5646424733950346\n"
]
],
[
[
"Funtion to calculate the expectation value of the full Hamiltonian. This function uses the property:\n\n$$\\langle \\sum_{i=0}^{N}H_i\\rangle = \\sum_{i=0}^{N}\\frac{\\langle \\Psi|H_i|\\Psi\\rangle}{\\langle \\Psi|\\Psi\\rangle}$$\n\nUsing this property, the circuit can be shallow, allowing the parallelisation of the executions in the QPU. The expectation value of each term of the Hamiltonian can be calculated in parallel because of the linearlity of the expectation value operation",
"_____no_output_____"
]
],
[
[
"def variational_quantum_eigensolver(theta, hamiltonian,shots=0):\n #print(\"Theta:\",theta)\n vqe=0.\n if shots>0:\n for i in hamiltonian:\n vqe+=Expectation_H_shots(theta, i,shots)\n else:\n for i in hamiltonian:\n vqe+=Expectation_H(theta, i)\n \n return vqe",
"_____no_output_____"
]
],
[
[
"This is the main loop. For each bond length, the expectation value of the Hamiltonian is calculated, minimizing its value as function of the parameter $\\theta$",
"_____no_output_____"
]
],
[
[
"lowest_energies = []\nlowest_energies_shots = []\nbond_distances = []\nfor i in range(len(raw_data_table_1)):\n # Use data of paper to construct the Hamiltonian\n print(\"Calculating for Bond distance:\",i)\n bond_distances.append(raw_data_table_1[i][0])\n hamiltonian=[]\n hamiltonian.append(raw_data_table_1[i][1] * QubitOperator(())) # == identity\n hamiltonian.append(raw_data_table_1[i][2] * QubitOperator(\"Z0\"))\n hamiltonian.append(raw_data_table_1[i][3] * QubitOperator(\"Z1\"))\n hamiltonian.append(raw_data_table_1[i][4] * QubitOperator(\"Z0 Z1\"))\n hamiltonian.append(raw_data_table_1[i][5] * QubitOperator(\"X0 X1\"))\n hamiltonian.append(raw_data_table_1[i][6] * QubitOperator(\"Y0 Y1\"))\n\n # Use Scipy to perform the classical outerloop of the variational\n # eigensolver, i.e., the minimization of the parameter theta.\n # See documentation of Scipy for different optimizers.\n minimum = minimize_scalar(lambda theta: variational_quantum_eigensolver(theta, hamiltonian,shots=0))\n #minimumShots = minimize_scalar(lambda theta: variational_quantum_eigensolver(theta, hamiltonian,shots=1024))\n lowest_energies.append(minimum.fun)\n #lowest_energies_shots.append(minimumShots.fun)\n\n",
"Calculating for Bond distance: 0\nCalculating for Bond distance: 1\nCalculating for Bond distance: 2\nCalculating for Bond distance: 3\nCalculating for Bond distance: 4\nCalculating for Bond distance: 5\nCalculating for Bond distance: 6\nCalculating for Bond distance: 7\nCalculating for Bond distance: 8\nCalculating for Bond distance: 9\nCalculating for Bond distance: 10\nCalculating for Bond distance: 11\nCalculating for Bond distance: 12\nCalculating for Bond distance: 13\nCalculating for Bond distance: 14\nCalculating for Bond distance: 15\nCalculating for Bond distance: 16\nCalculating for Bond distance: 17\nCalculating for Bond distance: 18\nCalculating for Bond distance: 19\nCalculating for Bond distance: 20\nCalculating for Bond distance: 21\nCalculating for Bond distance: 22\nCalculating for Bond distance: 23\nCalculating for Bond distance: 24\nCalculating for Bond distance: 25\nCalculating for Bond distance: 26\nCalculating for Bond distance: 27\nCalculating for Bond distance: 28\nCalculating for Bond distance: 29\nCalculating for Bond distance: 30\nCalculating for Bond distance: 31\nCalculating for Bond distance: 32\nCalculating for Bond distance: 33\nCalculating for Bond distance: 34\nCalculating for Bond distance: 35\nCalculating for Bond distance: 36\nCalculating for Bond distance: 37\nCalculating for Bond distance: 38\nCalculating for Bond distance: 39\nCalculating for Bond distance: 40\nCalculating for Bond distance: 41\nCalculating for Bond distance: 42\nCalculating for Bond distance: 43\nCalculating for Bond distance: 44\nCalculating for Bond distance: 45\nCalculating for Bond distance: 46\nCalculating for Bond distance: 47\nCalculating for Bond distance: 48\nCalculating for Bond distance: 49\nCalculating for Bond distance: 50\nCalculating for Bond distance: 51\nCalculating for Bond distance: 52\nCalculating for Bond distance: 53\n"
]
],
[
[
"Ok. This is the end. Show the results",
"_____no_output_____"
]
],
[
[
"# print result\nplt.xlabel(\"Bond length [Angstrom]\")\nplt.ylabel(\"Total Energy [Hartree]\")\nplt.title(\"Variational Quantum Eigensolver\")\nplt.plot(bond_distances, lowest_energies, \"b.-\",\n label=\"Ground-state energy of molecular hydrogen\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71d974596c7f092b788f46ed07713507a45bf66 | 20,151 | ipynb | Jupyter Notebook | example.ipynb | defve1988/pyebas | 8db7e67a79788c2afaf5431808637a53e83ef7b1 | [
"MIT"
] | 1 | 2021-11-25T09:05:06.000Z | 2021-11-25T09:05:06.000Z | example.ipynb | defve1988/pyebas | 8db7e67a79788c2afaf5431808637a53e83ef7b1 | [
"MIT"
] | null | null | null | example.ipynb | defve1988/pyebas | 8db7e67a79788c2afaf5431808637a53e83ef7b1 | [
"MIT"
] | null | null | null | 29.119942 | 145 | 0.412287 | [
[
[
"### 1. Import pyebas",
"_____no_output_____"
]
],
[
[
"from pyebas import *",
"_____no_output_____"
]
],
[
[
"### 2. Download EBAS data (.nc files)",
"_____no_output_____"
]
],
[
[
"# set selection conditions\n# if you need the whole EBAS database, set conditions as None\nconditions = {\n \"start_year\": 1990,\n \"end_year\": 2021,\n \"site\": ['ES0010R', 'ES0011R'],\n \"matrix\": ['air'],\n \"components\": ['NOx'],\n}\n# set local stroage path\ndb_dir = r'ebas_db'\ndownloader = EbasDownloader(loc=db_dir)\n# download requires multiprocessing, error may occurs because of multiprocessing\n# use command line or Jupyter Notebook to prevent errors\ndownloader.get_raw_files(conditions=conditions, download=True)",
"Make data folder ebas_db\\raw_data...\n0 raw data (*.nc) files have been downloaded.\nRequesting data from ebas sever...\n13126 files found on ftp server.\n0 files need to be deleted...\n"
]
],
[
[
"### 3. Export to .csv file",
"_____no_output_____"
]
],
[
[
"# export all the downloaded .nc files in the output path to .csv \n# important: .csv file might be very large.\ncsv_exporter = csvExporter(loc=db_dir)\ncsv_exporter.export_csv('export.csv')",
"Processing files...: 100%|██████████| 5/5 [00:00<00:00, 19.52it/s]\n"
]
],
[
[
"### 4. Create local database",
"_____no_output_____"
]
],
[
[
"# set local stroage path, must be the same as previous path\ndb_dir = r'ebas_db'\n# local database object\ndb = EbasDB(dir=db_dir, dump='xz', detailed=True)\n# create/update database with new files\ndb.update_db()",
"Make data folder ebas_db\\dumps...\nGathering site information...\nUsing 5 threads...\n"
]
],
[
[
"### 5.Open local database",
"_____no_output_____"
]
],
[
[
"# set local stroage path\ndb_dir = r'ebas_db'\n# local database object\ndb = EbasDB(dir=db_dir, dump='xz', detailed=True)\n# open database if it is created\ndb.init_db()",
" 0%| | 0/2 [00:00<?, ?it/s]"
]
],
[
[
"### 6. Query data from local database as pandas.DataFrame",
"_____no_output_____"
]
],
[
[
"condition = {\n \"id\":[\"AM0001R\", \"EE0009R\", 'ES0010R', 'ES0011R'],\n \"component\":[\"NOx\", \"nitrate\", \"nitric_acid\"],\n \"matrix\":[\"air\", \"aerosol\"],\n \"stat\":['arithmetic mean',\"median\"],\n \"st\":np.datetime64(\"1970-01-01\"),\n \"ed\":np.datetime64(\"2021-10-01\"),\n # if you want to include all, just remove the condition\n #\"country\":[\"Denmark\",\"France\"],\n}\ndf = db.query(condition, use_number_indexing=False)\ndf.head(20)",
"seraching...: 100%|██████████| 2/2 [00:00<?, ?it/s]\n"
]
],
[
[
"### 7. Access detail information",
"_____no_output_____"
]
],
[
[
"# access information for one site\ndb.site_index[\"ES0011R\"]\ndb.site_index[\"ES0011R\"][\"components\"].keys()\ndb.site_index[\"ES0011R\"][\"files\"].keys()",
"_____no_output_____"
]
],
[
[
"### 8. Get summary",
"_____no_output_____"
]
],
[
[
"# get summary information\ndb.list_sites()\n# possible keys are: \"id\",\"name\",\"country\",\"station_setting\", \"lat\", \"lon\",\"alt\",\"land_use\", \"file_num\",\"components\"\ndb.list_sites(keys=[\"name\",\"lat\",\"lon\"])\n# if components are selected, set list_time=True to see the starting and ending time\ndb.list_sites(keys=[\"name\", \"components\"], list_time=True)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71db1af8df44674bc3a42a17fb6b38348d3dd75 | 3,725 | ipynb | Jupyter Notebook | ASSIGNMENT 1.ipynb | sahil8370/python | b7b2ebfd8d4e98f53478454bdd5ed98c8e0200bf | [
"Apache-2.0"
] | null | null | null | ASSIGNMENT 1.ipynb | sahil8370/python | b7b2ebfd8d4e98f53478454bdd5ed98c8e0200bf | [
"Apache-2.0"
] | null | null | null | ASSIGNMENT 1.ipynb | sahil8370/python | b7b2ebfd8d4e98f53478454bdd5ed98c8e0200bf | [
"Apache-2.0"
] | 1 | 2022-03-30T19:03:37.000Z | 2022-03-30T19:03:37.000Z | 20.694444 | 89 | 0.498523 | [
[
[
"1.Python program to check whether the given number is even or not.",
"_____no_output_____"
]
],
[
[
"x=int(input(\"Enter the number :\"))\nif x%2==0:\n print(\"The number is even\")\nelse:\n print(\"The number is not even\")",
"Enter the number :6\nThe number is even\n"
]
],
[
[
"2.Python program to convert the temperature in degree centigrade to fahrenheit.",
"_____no_output_____"
]
],
[
[
"C=float(input(\"Enter the celcius\"))\nf=(C*(9/5)+32)\nprint(\"The value of fahrenheit :\",f)",
"Enter the celcius0\nThe value of fahrenheit : 32.0\n"
]
],
[
[
"3.Python program to find the circumference and area of a circle with a given radius",
"_____no_output_____"
]
],
[
[
"R=float(input(\"Enter the radius of circle\"))\nArea=3.14*R**2\nCircum=2*3.14*R\nprint(\"The area of circle is : \",Area)\nprint(\"The circumference of circle is : \",Circum)",
"Enter the radius of circle3\nThe area of circle is : 28.26\nThe circumference of circle is : 18.84\n"
]
],
[
[
"4.Python program to check whether the given integer is multiple of 5.\n",
"_____no_output_____"
]
],
[
[
"X=int(input(\"Enter the number : \"))\nif(X%5==0):\n print(X,\"is multiple of 5\")\nelse:\n print(\"Not multiple of 5\")",
"Enter the number : 10\n10 is multiple of 5\n"
]
],
[
[
"5.Python program to check whether the number is multiple of 5 and 7",
"_____no_output_____"
]
],
[
[
"X=int(input(\"Enter the number\"))\nif(X%35)==0:\n print(X,\"is a multiple of both 5 and 7\")\nelse:\n print(x,\"is not a multiple of 5 and 7\")",
"Enter the number35\n35 is a multiple of both 5 and 7\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71db2d409c1cee1cdac9f4ff9cc8c488a0c1b00 | 5,227 | ipynb | Jupyter Notebook | create-new-models/make_ego_graph_dataset.ipynb | longland-m/wikigen | 459ba7bf9d3ca9584de65388cc9b9a15fa16a69f | [
"MIT"
] | null | null | null | create-new-models/make_ego_graph_dataset.ipynb | longland-m/wikigen | 459ba7bf9d3ca9584de65388cc9b9a15fa16a69f | [
"MIT"
] | 2 | 2021-08-25T16:04:29.000Z | 2022-02-10T01:50:44.000Z | create-new-models/make_ego_graph_dataset.ipynb | longland-m/wikigen | 459ba7bf9d3ca9584de65388cc9b9a15fa16a69f | [
"MIT"
] | null | null | null | 23.128319 | 171 | 0.540272 | [
[
[
"# Make a dataset of subgraphs sampled from a larger network",
"_____no_output_____"
],
[
"This notebook shows how to format the Wikipedia link dataset gathered from `get_wiki_data.ipynb` as a network using NetworkX and sample egocentric subgraphs from it.",
"_____no_output_____"
]
],
[
[
"import networkx as nx\nimport pickle as pkl\nimport random",
"_____no_output_____"
],
[
"def egoGraph(graph, node=None, returnNodeName=False):\n \"\"\" \n Get the ego graph centred at the specified node, or a random node if \n no node specified. Distance 1 from centre only.\n \n graph: the graph to sample from\n node: the name of the node at the centre of the graph, or None to choose a random node\n returnNodeName: whether to also return the name of the centre node. If randomly\n selecting a node, this will be necessary to know exactly which one was selected\n \"\"\"\n if node is None:\n node = random.choice(list(graph.nodes()))\n egoGraph = nx.generators.ego.ego_graph(graph, randnode)\n if returnNodeName:\n return egoGraph, node\n else:\n return egoGraph",
"_____no_output_____"
]
],
[
[
"## Load link dataset and format as edgelist",
"_____no_output_____"
]
],
[
[
"pageLinks = pkl.load(open('pageLinks.pkl', 'rb'))\npageTitles = pkl.load(open('pageTitles.pkl', 'rb'))",
"_____no_output_____"
]
],
[
[
"Create edgelist from all articles:",
"_____no_output_____"
]
],
[
[
"edgeList1 = []\nfor i in pageLinks:\n edgeList1.extend(list(zip([i[0]]*len(i[1]), i[1])))",
"_____no_output_____"
]
],
[
[
"Remove edges if it links to a page that wasn't in the original list (optional):",
"_____no_output_____"
]
],
[
[
"edgeList2 = [i for i in edgeList1 if i[0] in pageTitles and i[1] in pageTitles]",
"_____no_output_____"
]
],
[
[
"Remove self-edges:",
"_____no_output_____"
]
],
[
[
"edgeList3 = [i for i in edgeList2 if i[0] != i[1]]",
"_____no_output_____"
]
],
[
[
"Format as undirected NetworkX Graph:",
"_____no_output_____"
]
],
[
[
"graph = nx.Graph(edgeList3)",
"_____no_output_____"
]
],
[
[
"## Sample subgraphs",
"_____no_output_____"
],
[
"Single example:",
"_____no_output_____"
]
],
[
[
"egoNet, centrePage = egoGraph(graph = graph, node = None, returnNodeName = True)\nprint(centrePage)\nnx.draw(egoNet)",
"_____no_output_____"
]
],
[
[
"#### Make graph dataset\n\nNodes won't be sampled more than once.\n\n- numGraphs = number of graphs to sample\n- minNodes = minimum number of nodes the ego graphs should have\n- maxNodes = maximum number of nodes the ego graphs should have",
"_____no_output_____"
]
],
[
[
"numGraphs = 150\nminNodes = 15\nmaxNodes = 50\n\nnets = []\ncentrePages = []\nwhile len(nets) < numGraphs:\n tempNet, tempPage = egoGraph(graph = graph, node = None, returnNodeName = True)\n if len(tempNet) > minNodes and len(tempNet) < maxNodes and tempPage not in centrePages:\n nets.append(tempNet)\n centrePages.append(tempPage)",
"_____no_output_____"
],
[
"# save dataset\n#pkl.dump(nets, open('graphDataset.pkl', 'wb'))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e71dc212a95a9f27a72203701dec76b2339af675 | 4,747 | ipynb | Jupyter Notebook | clue/classification/tnews/train.ipynb | CLUEbenchmark/PyCLUE | c16af32dd7dc195e77f352b6b3d2d5b963e193ba | [
"MIT"
] | 122 | 2019-12-04T14:42:34.000Z | 2022-03-01T08:12:30.000Z | clue/classification/tnews/train.ipynb | CLUEbenchmark/PyCLUE | c16af32dd7dc195e77f352b6b3d2d5b963e193ba | [
"MIT"
] | 9 | 2020-06-05T00:42:13.000Z | 2022-02-09T23:39:31.000Z | clue/classification/tnews/train.ipynb | CLUEbenchmark/PyCLUE | c16af32dd7dc195e77f352b6b3d2d5b963e193ba | [
"MIT"
] | 12 | 2019-12-06T01:58:31.000Z | 2021-12-22T09:51:13.000Z | 28.42515 | 109 | 0.611123 | [
[
[
"# TNEWS",
"_____no_output_____"
],
[
"短文本分类\n\n该数据集来自今日头条的新闻版块,共提取了15个类别的新闻,包括旅游,教育,金融,军事等。",
"_____no_output_____"
],
[
"## 训练",
"_____no_output_____"
]
],
[
[
"from pyclue.tf1.tasks.classification.multi_class.train import Trainer",
"_____no_output_____"
],
[
"# 数据所在路径\ndata_dir = '/workspace/projects/PyCLUE_Corpus/classification/tnews'\n# 训练结果保存路径\noutput_dir = '/workspace/projects/PyCLUE_examples/tnews'\n\n# 是否重新创建tfrecord(会耗费一定时间,首次执行时需设置为True;当设为False时,可保证数据一致性)\nrecreate_tfrecord = True\n# 随机种子\nrandom_seed = 0\n\n# 使用内置的预训练语言模型时,需指定model_name,无需指定model_type, vocab_file, config_file和init_checkpoint_file\n# 使用自定义预训练语言模型时,需指定model_type, vocab_file, config_file和init_checkpoint_file,无需指定model_name\nmodel_name = 'albert_tiny_zh_brightmart'\nmodel_type = None\nvocab_file = None\nconfig_file = None\ninit_checkpoint_file = None\n\n# 任务类型,若为单句输入则为single,若为句子对输入则为pairs\ntask_type = 'pairs'\n\n# 训练时能接受的最长句长,注:该句长不能超过预训练时所指定的最大句长\nmax_seq_len = 128\n# 训练步数(num_train_steps) = 训练数据量(num_train_examples) * 训练轮次(num_train_epochs) / 每批次训练数据大小(batch_size)\n# 预热步数(num_warmup_steps) = 训练步数(num_train_steps) * 预热比例(warmup_proportion)\n# 训练轮次\nnum_train_epochs = 10\n# 预热比例\nwarmup_proportion = 0.1\n# 每批次训练数据大小\nbatch_size = 32\n\n# 初始学习率\nlearning_rate = 2e-5\n# 训练器名,可选adamw和lamb\noptimizer_name = 'adamw'\n\n# 验证指标,可选accuracy, premise, recall, f1\nmetric_name = 'accuracy'\n\n# 每若干训练步数保存一次checkpoint模型(最多保存最新的10个模型)\nsave_checkpoints_steps = 200\n# 每若干训练步数打印一次训练指标\nlog_steps = 50\n# 训练与验证执行策略,可选0(异步:先训练后验证,较快,CLUE默认采用此方式)或1(同步:同时执行训练与验证,较慢)\ntrain_and_evaluate_mode = 0\n# 是否使用最佳的checkpoint进行验证,当为True时,从已保存的checkpoint模型中选择最佳的模型进行预测;\n# 当为False时,使用最后一个checkpoint模型进行预测;仅在train_and_evaluate_mode=0时有效,CLUE默认采用False\napply_best_checkpoint = False\n# early stop参数,执行若干步时指标任未提升时停止训练模型,仅在train_and_evaluate_mode=1时有效\nmax_steps_without_increase = 500\n# early stop参数,最低训练若干步时才执行early stop策略,仅在train_and_evaluate_mode=1时有效\nmin_steps = 500",
"_____no_output_____"
],
[
"# 初始化训练器\ntrainer = Trainer(\n output_dir=output_dir, \n random_seed=random_seed)\n# 构建模型\ntrainer.build_model(\n model_name=model_name,\n model_type=model_type,\n vocab_file=vocab_file,\n config_file=config_file,\n init_checkpoint_file=init_checkpoint_file,\n max_seq_len=max_seq_len)\n# 加载数据\ntrainer.load_data(\n data_dir=data_dir,\n task_type=task_type,\n batch_size=batch_size,\n recreate_tfrecord=recreate_tfrecord)\n# 执行训练并输出保存好的模型路径(包含checkpoint模型和pb模型)\nmodel_file_dict = trainer.train_and_evaluate(\n num_train_epochs=num_train_epochs,\n warmup_proportion=warmup_proportion,\n learning_rate=learning_rate,\n optimizer_name=optimizer_name,\n log_steps=log_steps,\n metric_name=metric_name,\n save_checkpoints_steps=save_checkpoints_steps,\n max_steps_without_increase=max_steps_without_increase,\n min_steps=min_steps,\n mode=train_and_evaluate_mode,\n apply_best_checkpoint=apply_best_checkpoint)",
"_____no_output_____"
],
[
"print('model save path: \\n%s' % '\\n'.join(['%s: %s' % item for item in model_file_dict.items()]))",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e71dc25010572671417ce26fc2559b6f91e8d305 | 54,293 | ipynb | Jupyter Notebook | HMM Tagger.ipynb | oucler/HMM-Tagger | 0c8670692788078fbdcc652374c055f361fbee46 | [
"MIT"
] | 6 | 2019-06-29T04:26:08.000Z | 2022-03-13T08:47:32.000Z | HMM Tagger.ipynb | oucler/HMM-Tagger | 0c8670692788078fbdcc652374c055f361fbee46 | [
"MIT"
] | null | null | null | HMM Tagger.ipynb | oucler/HMM-Tagger | 0c8670692788078fbdcc652374c055f361fbee46 | [
"MIT"
] | 8 | 2019-05-09T06:50:12.000Z | 2021-12-04T13:45:20.000Z | 40.638473 | 660 | 0.581659 | [
[
[
"# Project: Part of Speech Tagging with Hidden Markov Models \n---\n### Introduction\n\nPart of speech tagging is the process of determining the syntactic category of a word from the words in its surrounding context. It is often used to help disambiguate natural language phrases because it can be done quickly with high accuracy. Tagging can be used for many NLP tasks like determining correct pronunciation during speech synthesis (for example, _dis_-count as a noun vs dis-_count_ as a verb), for information retrieval, and for word sense disambiguation.\n\nIn this notebook, you'll use the [Pomegranate](http://pomegranate.readthedocs.io/) library to build a hidden Markov model for part of speech tagging using a \"universal\" tagset. Hidden Markov models have been able to achieve [>96% tag accuracy with larger tagsets on realistic text corpora](http://www.coli.uni-saarland.de/~thorsten/publications/Brants-ANLP00.pdf). Hidden Markov models have also been used for speech recognition and speech generation, machine translation, gene recognition for bioinformatics, and human gesture recognition for computer vision, and more. \n\n\n\nThe notebook already contains some code to get you started. You only need to add some new functionality in the areas indicated to complete the project; you will not need to modify the included code beyond what is requested. Sections that begin with **'IMPLEMENTATION'** in the header indicate that you must provide code in the block that follows. Instructions will be provided for each section, and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n**Note:** Once you have completed all of the code implementations, you need to finalize your work by exporting the iPython Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You must then **export the notebook** by running the last cell in the notebook, or by using the menu above and navigating to **File -> Download as -> HTML (.html)** Your submissions should include both the `html` and `ipynb` files.\n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n**Note:** Code and Markdown cells can be executed using the `Shift + Enter` keyboard shortcut. Markdown cells can be edited by double-clicking the cell to enter edit mode.\n</div>",
"_____no_output_____"
],
[
"### The Road Ahead\nYou must complete Steps 1-3 below to pass the project. The section on Step 4 includes references & resources you can use to further explore HMM taggers.\n\n- [Step 1](#Step-1:-Read-and-preprocess-the-dataset): Review the provided interface to load and access the text corpus\n- [Step 2](#Step-2:-Build-a-Most-Frequent-Class-tagger): Build a Most Frequent Class tagger to use as a baseline\n- [Step 3](#Step-3:-Build-an-HMM-tagger): Build an HMM Part of Speech tagger and compare to the MFC baseline\n- [Step 4](#Step-4:-[Optional]-Improving-model-performance): (Optional) Improve the HMM tagger",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-warning\">\n**Note:** Make sure you have selected a **Python 3** kernel in Workspaces or the hmm-tagger conda environment if you are running the Jupyter server on your own machine.\n</div>",
"_____no_output_____"
]
],
[
[
"# Jupyter \"magic methods\" -- only need to be run once per kernel restart\n%load_ext autoreload\n%aimport helpers, tests\n%autoreload 1",
"_____no_output_____"
],
[
"# import python modules -- this cell needs to be run again if you make changes to any of the files\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom IPython.core.display import HTML\nfrom itertools import chain\nfrom collections import Counter, defaultdict\nfrom helpers import show_model, Dataset\nfrom pomegranate import State, HiddenMarkovModel, DiscreteDistribution",
"_____no_output_____"
]
],
[
[
"## Step 1: Read and preprocess the dataset\n---\nWe'll start by reading in a text corpus and splitting it into a training and testing dataset. The data set is a copy of the [Brown corpus](https://en.wikipedia.org/wiki/Brown_Corpus) (originally from the [NLTK](https://www.nltk.org/) library) that has already been pre-processed to only include the [universal tagset](https://arxiv.org/pdf/1104.2086.pdf). You should expect to get slightly higher accuracy using this simplified tagset than the same model would achieve on a larger tagset like the full [Penn treebank tagset](https://www.ling.upenn.edu/courses/Fall_2003/ling001/penn_treebank_pos.html), but the process you'll follow would be the same.\n\nThe `Dataset` class provided in helpers.py will read and parse the corpus. You can generate your own datasets compatible with the reader by writing them to the following format. The dataset is stored in plaintext as a collection of words and corresponding tags. Each sentence starts with a unique identifier on the first line, followed by one tab-separated word/tag pair on each following line. Sentences are separated by a single blank line.\n\nExample from the Brown corpus. \n```\nb100-38532\nPerhaps\tADV\nit\tPRON\nwas\tVERB\nright\tADJ\n;\t.\n;\t.\n\nb100-35577\n...\n```",
"_____no_output_____"
]
],
[
[
"data = Dataset(\"tags-universal.txt\", \"brown-universal.txt\", train_test_split=0.8)\n\nprint(\"There are {} sentences in the corpus.\".format(len(data)))\nprint(\"There are {} sentences in the training set.\".format(len(data.training_set)))\nprint(\"There are {} sentences in the testing set.\".format(len(data.testing_set)))\n\nassert len(data) == len(data.training_set) + len(data.testing_set), \\\n \"The number of sentences in the training set + testing set should sum to the number of sentences in the corpus\"",
"There are 57340 sentences in the corpus.\nThere are 45872 sentences in the training set.\nThere are 11468 sentences in the testing set.\n"
]
],
[
[
"### The Dataset Interface\n\nYou can access (mostly) immutable references to the dataset through a simple interface provided through the `Dataset` class, which represents an iterable collection of sentences along with easy access to partitions of the data for training & testing. Review the reference below, then run and review the next few cells to make sure you understand the interface before moving on to the next step.\n\n```\nDataset-only Attributes:\n training_set - reference to a Subset object containing the samples for training\n testing_set - reference to a Subset object containing the samples for testing\n\nDataset & Subset Attributes:\n sentences - a dictionary with an entry {sentence_key: Sentence()} for each sentence in the corpus\n keys - an immutable ordered (not sorted) collection of the sentence_keys for the corpus\n vocab - an immutable collection of the unique words in the corpus\n tagset - an immutable collection of the unique tags in the corpus\n X - returns an array of words grouped by sentences ((w11, w12, w13, ...), (w21, w22, w23, ...), ...)\n Y - returns an array of tags grouped by sentences ((t11, t12, t13, ...), (t21, t22, t23, ...), ...)\n N - returns the number of distinct samples (individual words or tags) in the dataset\n\nMethods:\n stream() - returns an flat iterable over all (word, tag) pairs across all sentences in the corpus\n __iter__() - returns an iterable over the data as (sentence_key, Sentence()) pairs\n __len__() - returns the nubmer of sentences in the dataset\n```\n\nFor example, consider a Subset, `subset`, of the sentences `{\"s0\": Sentence((\"See\", \"Spot\", \"run\"), (\"VERB\", \"NOUN\", \"VERB\")), \"s1\": Sentence((\"Spot\", \"ran\"), (\"NOUN\", \"VERB\"))}`. The subset will have these attributes:\n\n```\nsubset.keys == {\"s1\", \"s0\"} # unordered\nsubset.vocab == {\"See\", \"run\", \"ran\", \"Spot\"} # unordered\nsubset.tagset == {\"VERB\", \"NOUN\"} # unordered\nsubset.X == ((\"Spot\", \"ran\"), (\"See\", \"Spot\", \"run\")) # order matches .keys\nsubset.Y == ((\"NOUN\", \"VERB\"), (\"VERB\", \"NOUN\", \"VERB\")) # order matches .keys\nsubset.N == 7 # there are a total of seven observations over all sentences\nlen(subset) == 2 # because there are two sentences\n```\n\n<div class=\"alert alert-block alert-info\">\n**Note:** The `Dataset` class is _convenient_, but it is **not** efficient. It is not suitable for huge datasets because it stores multiple redundant copies of the same data.\n</div>",
"_____no_output_____"
],
[
"#### Sentences\n\n`Dataset.sentences` is a dictionary of all sentences in the training corpus, each keyed to a unique sentence identifier. Each `Sentence` is itself an object with two attributes: a tuple of the words in the sentence named `words` and a tuple of the tag corresponding to each word named `tags`.",
"_____no_output_____"
]
],
[
[
"key = 'b100-38532'\nprint(\"Sentence: {}\".format(key))\nprint(\"words:\\n\\t{!s}\".format(data.sentences[key].words))\nprint(\"tags:\\n\\t{!s}\".format(data.sentences[key].tags))",
"Sentence: b100-38532\nwords:\n\t('Perhaps', 'it', 'was', 'right', ';', ';')\ntags:\n\t('ADV', 'PRON', 'VERB', 'ADJ', '.', '.')\n"
],
[
"data.keys[:2]\n",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\n**Note:** The underlying iterable sequence is **unordered** over the sentences in the corpus; it is not guaranteed to return the sentences in a consistent order between calls. Use `Dataset.stream()`, `Dataset.keys`, `Dataset.X`, or `Dataset.Y` attributes if you need ordered access to the data.\n</div>\n\n#### Counting Unique Elements\n\nYou can access the list of unique words (the dataset vocabulary) via `Dataset.vocab` and the unique list of tags via `Dataset.tagset`.",
"_____no_output_____"
]
],
[
[
"print(\"There are a total of {} samples of {} unique words in the corpus.\"\n .format(data.N, len(data.vocab)))\nprint(\"There are {} samples of {} unique words in the training set.\"\n .format(data.training_set.N, len(data.training_set.vocab)))\nprint(\"There are {} samples of {} unique words in the testing set.\"\n .format(data.testing_set.N, len(data.testing_set.vocab)))\nprint(\"There are {} words in the test set that are missing in the training set.\"\n .format(len(data.testing_set.vocab - data.training_set.vocab)))\n\nassert data.N == data.training_set.N + data.testing_set.N, \\\n \"The number of training + test samples should sum to the total number of samples\"",
"There are a total of 1161192 samples of 56057 unique words in the corpus.\nThere are 928458 samples of 50536 unique words in the training set.\nThere are 232734 samples of 25112 unique words in the testing set.\nThere are 5521 words in the test set that are missing in the training set.\n"
]
],
[
[
"#### Accessing word and tag Sequences\nThe `Dataset.X` and `Dataset.Y` attributes provide access to ordered collections of matching word and tag sequences for each sentence in the dataset.",
"_____no_output_____"
]
],
[
[
"# accessing words with Dataset.X and tags with Dataset.Y \nfor i in range(2): \n print(\"Sentence {}:\".format(i + 1), data.X[i])\n print()\n print(\"Labels {}:\".format(i + 1), data.Y[i])\n print()",
"Sentence 1: ('Mr.', 'Podger', 'had', 'thanked', 'him', 'gravely', ',', 'and', 'now', 'he', 'made', 'use', 'of', 'the', 'advice', '.')\n\nLabels 1: ('NOUN', 'NOUN', 'VERB', 'VERB', 'PRON', 'ADV', '.', 'CONJ', 'ADV', 'PRON', 'VERB', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\nSentence 2: ('But', 'there', 'seemed', 'to', 'be', 'some', 'difference', 'of', 'opinion', 'as', 'to', 'how', 'far', 'the', 'board', 'should', 'go', ',', 'and', 'whose', 'advice', 'it', 'should', 'follow', '.')\n\nLabels 2: ('CONJ', 'PRT', 'VERB', 'PRT', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'ADP', 'ADV', 'ADV', 'DET', 'NOUN', 'VERB', 'VERB', '.', 'CONJ', 'DET', 'NOUN', 'PRON', 'VERB', 'VERB', '.')\n\n"
]
],
[
[
"#### Accessing (word, tag) Samples\nThe `Dataset.stream()` method returns an iterator that chains together every pair of (word, tag) entries across all sentences in the entire corpus.",
"_____no_output_____"
]
],
[
[
"# use Dataset.stream() (word, tag) samples for the entire corpus\nsqA = []\ndct = defaultdict(list)\ncount = Counter()\nprint(\"\\nStream (word, tag) pairs:\\n\")\nfor i, pair in enumerate(data.stream()):\n print(\"\\t\", pair)\n dct[pair].append(i)\n sqA.append(pair[0].lower())\n if i > 10: break",
"\nStream (word, tag) pairs:\n\n\t ('Mr.', 'NOUN')\n\t ('Podger', 'NOUN')\n\t ('had', 'VERB')\n\t ('thanked', 'VERB')\n\t ('him', 'PRON')\n\t ('gravely', 'ADV')\n\t (',', '.')\n\t ('and', 'CONJ')\n\t ('now', 'ADV')\n\t ('he', 'PRON')\n\t ('made', 'VERB')\n\t ('use', 'NOUN')\n"
],
[
"dictTrain = defaultdict(list)\nfor i, trainPair in enumerate(data.training_set.stream()):\n #print(\"\\t\", trainPair)\n dictTrain[trainPair].append(i)",
"_____no_output_____"
]
],
[
[
"\nFor both our baseline tagger and the HMM model we'll build, we need to estimate the frequency of tags & words from the frequency counts of observations in the training corpus. In the next several cells you will complete functions to compute the counts of several sets of counts. ",
"_____no_output_____"
],
[
"## Step 2: Build a Most Frequent Class tagger\n---\n\nPerhaps the simplest tagger (and a good baseline for tagger performance) is to simply choose the tag most frequently assigned to each word. This \"most frequent class\" tagger inspects each observed word in the sequence and assigns it the label that was most often assigned to that word in the corpus.",
"_____no_output_____"
],
[
"### IMPLEMENTATION: Pair Counts\n\nComplete the function below that computes the joint frequency counts for two input sequences.",
"_____no_output_____"
]
],
[
[
"tags = (tag for i, (word, tag) in enumerate(data.training_set.stream()))\nwords = (word for i, (word, tag) in enumerate(data.training_set.stream()))\ndict_tag_word = defaultdict(list)\ndict_word = defaultdict(list)",
"_____no_output_____"
],
[
"def pair_counts(sequences_A=None, sequences_B=None):\n \"\"\"Return a dictionary keyed to each unique value in the first sequence list\n that counts the number of occurrences of the corresponding value from the\n second sequences list.\n \n For example, if sequences_A is tags and sequences_B is the corresponding\n words, then if 1244 sequences contain the word \"time\" tagged as a NOUN, then\n you should return a dictionary such that pair_counts[NOUN][time] == 1244\n \"\"\"\n # TODO: Finish this function!\n for i, (tag,word) in enumerate(zip(sequences_A,sequences_B)):\n dict_tag_word[tag].append(word)\n for k in dict_tag_word.keys():\n dict_word[k] = Counter(dict_tag_word[k])\n \n return dict_word\n\n\n# Calculate C(t_i, w_i)\nemission_counts = pair_counts(tags, words)\n\nassert len(emission_counts) == 12, \\\n \"Uh oh. There should be 12 tags in your dictionary.\"\nassert max(emission_counts[\"NOUN\"], key=emission_counts[\"NOUN\"].get) == 'time', \\\n \"Hmmm...'time' is expected to be the most common NOUN.\"\nHTML('<div class=\"alert alert-block alert-success\">Your emission counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Most Frequent Class Tagger\n\nUse the `pair_counts()` function and the training dataset to find the most frequent class label for each word in the training data, and populate the `mfc_table` below. The table keys should be words, and the values should be the appropriate tag string.\n\nThe `MFCTagger` class is provided to mock the interface of Pomegranite HMM models so that they can be used interchangeably.",
"_____no_output_____"
]
],
[
[
"tags = (tag for i, (word, tag) in enumerate(data.training_set.stream()))\nwords = (word for i, (word, tag) in enumerate(data.training_set.stream()))\ndict_word_tag = defaultdict(list)\ndict_wt = defaultdict(list)",
"_____no_output_____"
],
[
"# Create a lookup table mfc_table where mfc_table[word] contains the tag label most frequently assigned to that word\nfrom collections import namedtuple\n\nFakeState = namedtuple(\"FakeState\", \"name\")\n\nclass MFCTagger:\n # NOTE: You should not need to modify this class or any of its methods\n missing = FakeState(name=\"<MISSING>\")\n \n def __init__(self, table):\n self.table = defaultdict(lambda: MFCTagger.missing)\n self.table.update({word: FakeState(name=tag) for word, tag in table.items()})\n \n def viterbi(self, seq):\n \"\"\"This method simplifies predictions by matching the Pomegranate viterbi() interface\"\"\"\n return 0., list(enumerate([\"<start>\"] + [self.table[w] for w in seq] + [\"<end>\"]))\n\n\n# TODO: calculate the frequency of each tag being assigned to each word (hint: similar, but not\n# the same as the emission probabilities) and use it to fill the mfc_table\nfor i, (tag,word) in enumerate(zip(tags,words)):\n dict_word_tag[word].append(tag)\nfor k in dict_word_tag.keys():\n dict_wt[k] = (Counter(dict_word_tag[k])).most_common(1)[0][0]\n\n#(Counter(dict_word_tag['time'])).most_common(1)[0][0]\nlen(dict_wt)\n#len(data.training_set.vocab)\n#len(dict_word_tag.keys())\nword_counts = pair_counts(tags,words)\n\nmfc_table = dict_wt\n\n# DO NOT MODIFY BELOW THIS LINE\nmfc_model = MFCTagger(mfc_table) # Create a Most Frequent Class tagger instance\n\nassert len(mfc_table) == len(data.training_set.vocab), \"\"\nassert all(k in data.training_set.vocab for k in mfc_table.keys()), \"\"\nassert sum(int(k not in mfc_table) for k in data.testing_set.vocab) == 5521, \"\"\nHTML('<div class=\"alert alert-block alert-success\">Your MFC tagger has all the correct words!</div>')",
"_____no_output_____"
]
],
[
[
"### Making Predictions with a Model\nThe helper functions provided below interface with Pomegranate network models & the mocked MFCTagger to take advantage of the [missing value](http://pomegranate.readthedocs.io/en/latest/nan.html) functionality in Pomegranate through a simple sequence decoding function. Run these functions, then run the next cell to see some of the predictions made by the MFC tagger.",
"_____no_output_____"
]
],
[
[
"def replace_unknown(sequence):\n \"\"\"Return a copy of the input sequence where each unknown word is replaced\n by the literal string value 'nan'. Pomegranate will ignore these values\n during computation.\n \"\"\"\n return [w if w in data.training_set.vocab else 'nan' for w in sequence]\n\ndef simplify_decoding(X, model):\n \"\"\"X should be a 1-D sequence of observations for the model to predict\"\"\"\n _, state_path = model.viterbi(replace_unknown(X))\n return [state[1].name for state in state_path[1:-1]] # do not show the start/end state predictions",
"_____no_output_____"
]
],
[
[
"### Example Decoding Sequences with MFC Tagger",
"_____no_output_____"
]
],
[
[
"for key in data.testing_set.keys[:3]:\n print(\"Sentence Key: {}\\n\".format(key))\n print(\"Predicted labels:\\n-----------------\")\n print(simplify_decoding(data.sentences[key].words, mfc_model))\n print()\n print(\"Actual labels:\\n--------------\")\n print(data.sentences[key].tags)\n print(\"\\n\")",
"Sentence Key: b100-28144\n\nPredicted labels:\n-----------------\n['CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.']\n\nActual labels:\n--------------\n('CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.')\n\n\nSentence Key: b100-23146\n\nPredicted labels:\n-----------------\n['PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.']\n\nActual labels:\n--------------\n('PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\n\nSentence Key: b100-35462\n\nPredicted labels:\n-----------------\n['DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', '<MISSING>', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADV', 'NOUN', '.']\n\nActual labels:\n--------------\n('DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADJ', 'NOUN', '.')\n\n\n"
]
],
[
[
"### Evaluating Model Accuracy\n\nThe function below will evaluate the accuracy of the MFC tagger on the collection of all sentences from a text corpus. ",
"_____no_output_____"
]
],
[
[
"def accuracy(X, Y, model):\n \"\"\"Calculate the prediction accuracy by using the model to decode each sequence\n in the input X and comparing the prediction with the true labels in Y.\n \n The X should be an array whose first dimension is the number of sentences to test,\n and each element of the array should be an iterable of the words in the sequence.\n The arrays X and Y should have the exact same shape.\n \n X = [(\"See\", \"Spot\", \"run\"), (\"Run\", \"Spot\", \"run\", \"fast\"), ...]\n Y = [(), (), ...]\n \"\"\"\n correct = total_predictions = 0\n for observations, actual_tags in zip(X, Y):\n \n # The model.viterbi call in simplify_decoding will return None if the HMM\n # raises an error (for example, if a test sentence contains a word that\n # is out of vocabulary for the training set). Any exception counts the\n # full sentence as an error (which makes this a conservative estimate).\n try:\n most_likely_tags = simplify_decoding(observations, model)\n correct += sum(p == t for p, t in zip(most_likely_tags, actual_tags))\n except:\n pass\n total_predictions += len(observations)\n return correct / total_predictions",
"_____no_output_____"
]
],
[
[
"#### Evaluate the accuracy of the MFC tagger\nRun the next cell to evaluate the accuracy of the tagger on the training and test corpus.",
"_____no_output_____"
]
],
[
[
"mfc_training_acc = accuracy(data.training_set.X, data.training_set.Y, mfc_model)\nprint(\"training accuracy mfc_model: {:.2f}%\".format(100 * mfc_training_acc))\n\nmfc_testing_acc = accuracy(data.testing_set.X, data.testing_set.Y, mfc_model)\nprint(\"testing accuracy mfc_model: {:.2f}%\".format(100 * mfc_testing_acc))\n\nassert mfc_training_acc >= 0.955, \"Uh oh. Your MFC accuracy on the training set doesn't look right.\"\nassert mfc_testing_acc >= 0.925, \"Uh oh. Your MFC accuracy on the testing set doesn't look right.\"\nHTML('<div class=\"alert alert-block alert-success\">Your MFC tagger accuracy looks correct!</div>')",
"training accuracy mfc_model: 95.72%\ntesting accuracy mfc_model: 93.00%\n"
]
],
[
[
"## Step 3: Build an HMM tagger\n---\nThe HMM tagger has one hidden state for each possible tag, and parameterized by two distributions: the emission probabilties giving the conditional probability of observing a given **word** from each hidden state, and the transition probabilities giving the conditional probability of moving between **tags** during the sequence.\n\nWe will also estimate the starting probability distribution (the probability of each **tag** being the first tag in a sequence), and the terminal probability distribution (the probability of each **tag** being the last tag in a sequence).\n\nThe maximum likelihood estimate of these distributions can be calculated from the frequency counts as described in the following sections where you'll implement functions to count the frequencies, and finally build the model. The HMM model will make predictions according to the formula:\n\n$$t_i^n = \\underset{t_i^n}{\\mathrm{argmin}} \\prod_{i=1}^n P(w_i|t_i) P(t_i|t_{i-1})$$\n\nRefer to Speech & Language Processing [Chapter 10](https://web.stanford.edu/~jurafsky/slp3/10.pdf) for more information.",
"_____no_output_____"
],
[
"### IMPLEMENTATION: Unigram Counts\n\nComplete the function below to estimate the co-occurrence frequency of each symbol over all of the input sequences. The unigram probabilities in our HMM model are estimated from the formula below, where N is the total number of samples in the input. (You only need to compute the counts for now.)\n\n$$P(tag_1) = \\frac{C(tag_1)}{N}$$",
"_____no_output_____"
]
],
[
[
"tags = [tag for i, (word, tag) in enumerate(data.training_set.stream())]\ndict_tag = {}",
"_____no_output_____"
],
[
"def unigram_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequence list that\n counts the number of occurrences of the value in the sequences list. The sequences\n collection should be a 2-dimensional array.\n \n For example, if the tag NOUN appears 275558 times over all the input sequences,\n then you should return a dictionary such that your_unigram_counts[NOUN] == 275558.\n \"\"\"\n # TODO: Finish this function!\n for tag in sequences:\n if tag in dict_tag.keys():\n dict_tag[tag] += 1\n else: dict_tag[tag] = 1\n #raise NotImplementedError\n return dict_tag\n\n# TODO: call unigram_counts with a list of tag sequences from the training set\n\ntag_unigrams = unigram_counts(tags)\n\nassert set(tag_unigrams.keys()) == data.training_set.tagset, \\\n \"Uh oh. It looks like your tag counts doesn't include all the tags!\"\nassert min(tag_unigrams, key=tag_unigrams.get) == 'X', \\\n \"Hmmm...'X' is expected to be the least common class\"\nassert max(tag_unigrams, key=tag_unigrams.get) == 'NOUN', \\\n \"Hmmm...'NOUN' is expected to be the most common class\"\nHTML('<div class=\"alert alert-block alert-success\">Your tag unigrams look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Bigram Counts\n\nComplete the function below to estimate the co-occurrence frequency of each pair of symbols in each of the input sequences. These counts are used in the HMM model to estimate the bigram probability of two tags from the frequency counts according to the formula: $$P(tag_2|tag_1) = \\frac{C(tag_2|tag_1)}{C(tag_2)}$$\n",
"_____no_output_____"
]
],
[
[
"tags = [tag for i, (word, tag) in enumerate(data.training_set.stream())]\nsq = list(zip(tags[:-1],tags[1:]))\ndict_sq = {}",
"_____no_output_____"
],
[
"def bigram_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique PAIR of values in the input sequences\n list that counts the number of occurrences of pair in the sequences list. The input\n should be a 2-dimensional array.\n \n For example, if the pair of tags (NOUN, VERB) appear 61582 times, then you should\n return a dictionary such that your_bigram_counts[(NOUN, VERB)] == 61582\n \"\"\"\n\n # TODO: Finish this function!\n for tag_pair in sequences:\n if tag_pair in dict_sq.keys():\n dict_sq[tag_pair] += 1\n else: dict_sq[tag_pair] = 1\n return dict_sq\n #raise NotImplementedError\n\n# TODO: call bigram_counts with a list of tag sequences from the training set\ntag_bigrams = bigram_counts(sq)\n\nassert len(tag_bigrams) == 144, \\\n \"Uh oh. There should be 144 pairs of bigrams (12 tags x 12 tags)\"\nassert min(tag_bigrams, key=tag_bigrams.get) in [('X', 'NUM'), ('PRON', 'X')], \\\n \"Hmmm...The least common bigram should be one of ('X', 'NUM') or ('PRON', 'X').\"\nassert max(tag_bigrams, key=tag_bigrams.get) in [('DET', 'NOUN')], \\\n \"Hmmm...('DET', 'NOUN') is expected to be the most common bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your tag bigrams look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Sequence Starting Counts\nComplete the code below to estimate the bigram probabilities of a sequence starting with each tag.",
"_____no_output_____"
]
],
[
[
"data.training_set.tagset\ndict_starting = defaultdict(list)",
"_____no_output_____"
],
[
"def starting_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequences list\n that counts the number of occurrences where that value is at the beginning of\n a sequence.\n \n For example, if 8093 sequences start with NOUN, then you should return a\n dictionary such that your_starting_counts[NOUN] == 8093\n \"\"\"\n # TODO: Finish this function!\n for tag in data.training_set.tagset:\n dict_starting[tag] = len([seq[0] for seq in sequences if seq[0]==tag])\n return dict_starting\n\n# TODO: Calculate the count of each tag starting a sequence\ntag_starts = starting_counts(data.training_set.Y)\n\n\nassert len(tag_starts) == 12, \"Uh oh. There should be 12 tags in your dictionary.\"\nassert min(tag_starts, key=tag_starts.get) == 'X', \"Hmmm...'X' is expected to be the least common starting bigram.\"\nassert max(tag_starts, key=tag_starts.get) == 'DET', \"Hmmm...'DET' is expected to be the most common starting bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your starting tag counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Sequence Ending Counts\nComplete the function below to estimate the bigram probabilities of a sequence ending with each tag.",
"_____no_output_____"
]
],
[
[
"dict_ending = defaultdict(list)",
"_____no_output_____"
],
[
"def ending_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequences list\n that counts the number of occurrences where that value is at the end of\n a sequence.\n \n For example, if 18 sequences end with DET, then you should return a\n dictionary such that your_starting_counts[DET] == 18\n \"\"\"\n # TODO: Finish this function!\n for tag in data.training_set.tagset:\n dict_ending[tag] = len([seq[-1] for seq in sequences if seq[-1]==tag])\n return dict_ending\n #raise NotImplementedError\n\n# TODO: Calculate the count of each tag ending a sequence\ntag_ends = ending_counts(data.training_set.Y)\n\nassert len(tag_ends) == 12, \"Uh oh. There should be 12 tags in your dictionary.\"\nassert min(tag_ends, key=tag_ends.get) in ['X', 'CONJ'], \"Hmmm...'X' or 'CONJ' should be the least common ending bigram.\"\nassert max(tag_ends, key=tag_ends.get) == '.', \"Hmmm...'.' is expected to be the most common ending bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your ending tag counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Basic HMM Tagger\nUse the tag unigrams and bigrams calculated above to construct a hidden Markov tagger.\n\n- Add one state per tag\n - The emission distribution at each state should be estimated with the formula: $P(w|t) = \\frac{C(t, w)}{C(t)}$\n- Add an edge from the starting state `basic_model.start` to each tag\n - The transition probability should be estimated with the formula: $P(t|start) = \\frac{C(start, t)}{C(start)}$\n- Add an edge from each tag to the end state `basic_model.end`\n - The transition probability should be estimated with the formula: $P(end|t) = \\frac{C(t, end)}{C(t)}$\n- Add an edge between _every_ pair of tags\n - The transition probability should be estimated with the formula: $P(t_2|t_1) = \\frac{C(t_1, t_2)}{C(t_1)}$",
"_____no_output_____"
]
],
[
[
"basic_model = HiddenMarkovModel(name=\"base-hmm-tagger\")\n\n# TODO: create states with emission probability distributions P(word | tag) and add to the model\n# (Hint: you may need to loop & create/add new states)\n\ncount_tag_and_word = pair_counts(tags, words)\nstates = {}\nfor tag, word_dict in count_tag_and_word.items(): #data.training_set.tagset\n p_words_given_tag_state = defaultdict(float)\n # for each tag/word, calculate P(word|tag)\n for word in word_dict.keys(): # data.training_set.vocab\n p_words_given_tag_state[word] = count_tag_and_word[tag][word] / tag_unigrams[tag] \n # create a new state for each tag from the dict of words that represents P(word|tag)\n emission = DiscreteDistribution(dict(p_words_given_tag_state))\n states[tag] = State(emission, name=tag)\n \nbasic_model.add_states(list(states.values()))\n\n# Adding start states\nfor tag in data.training_set.tagset:\n state = states[tag]\n basic_model.add_transition(basic_model.start, state, tag_starts[tag]/len(data.training_set))\n\n# Adding end states\nfor tag in data.training_set.tagset:\n state = states[tag]\n basic_model.add_transition(state, basic_model.end, tag_ends[tag]/tag_unigrams[tag])\n\n# Adding pairs\nfor tag1 in data.training_set.tagset:\n state1 = states[tag1]\n for tag2 in data.training_set.tagset:\n state2 = states[tag2]\n basic_model.add_transition(state1, state2, tag_bigrams[(tag1, tag2)]/tag_unigrams[tag1])\n\n# finalize the model\nbasic_model.bake()\n\nassert all(tag in set(s.name for s in basic_model.states) for tag in data.training_set.tagset), \\\n \"Every state in your network should use the name of the associated tag, which must be one of the training set tags.\"\nassert basic_model.edge_count() == 168, \\\n (\"Your network should have an edge from the start node to each state, one edge between every \" +\n \"pair of tags (states), and an edge from each state to the end node.\")\nHTML('<div class=\"alert alert-block alert-success\">Your HMM network topology looks good!</div>')",
"_____no_output_____"
],
[
"hmm_training_acc = accuracy(data.training_set.X, data.training_set.Y, basic_model)\nprint(\"training accuracy basic hmm model: {:.2f}%\".format(100 * hmm_training_acc))\n\nhmm_testing_acc = accuracy(data.testing_set.X, data.testing_set.Y, basic_model)\nprint(\"testing accuracy basic hmm model: {:.2f}%\".format(100 * hmm_testing_acc))\n\nassert hmm_training_acc > 0.97, \"Uh oh. Your HMM accuracy on the training set doesn't look right.\"\nassert hmm_testing_acc > 0.955, \"Uh oh. Your HMM accuracy on the testing set doesn't look right.\"\nHTML('<div class=\"alert alert-block alert-success\">Your HMM tagger accuracy looks correct! Congratulations, you\\'ve finished the project.</div>')",
"training accuracy basic hmm model: 97.52%\ntesting accuracy basic hmm model: 95.94%\n"
]
],
[
[
"### Example Decoding Sequences with the HMM Tagger",
"_____no_output_____"
]
],
[
[
"for key in data.testing_set.keys[:3]:\n print(\"Sentence Key: {}\\n\".format(key))\n print(\"Predicted labels:\\n-----------------\")\n print(simplify_decoding(data.sentences[key].words, basic_model))\n print()\n print(\"Actual labels:\\n--------------\")\n print(data.sentences[key].tags)\n print(\"\\n\")",
"Sentence Key: b100-28144\n\nPredicted labels:\n-----------------\n['CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.']\n\nActual labels:\n--------------\n('CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.')\n\n\nSentence Key: b100-23146\n\nPredicted labels:\n-----------------\n['PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.']\n\nActual labels:\n--------------\n('PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\n\nSentence Key: b100-35462\n\nPredicted labels:\n-----------------\n['DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADJ', 'NOUN', '.']\n\nActual labels:\n--------------\n('DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADJ', 'NOUN', '.')\n\n\n"
]
],
[
[
"\n## Finishing the project\n---\n\n<div class=\"alert alert-block alert-info\">\n**Note:** **SAVE YOUR NOTEBOOK**, then run the next cell to generate an HTML copy. You will zip & submit both this file and the HTML copy for review.\n</div>",
"_____no_output_____"
]
],
[
[
"!!jupyter nbconvert *.ipynb",
"_____no_output_____"
]
],
[
[
"## Step 4: [Optional] Improving model performance\n---\nThere are additional enhancements that can be incorporated into your tagger that improve performance on larger tagsets where the data sparsity problem is more significant. The data sparsity problem arises because the same amount of data split over more tags means there will be fewer samples in each tag, and there will be more missing data tags that have zero occurrences in the data. The techniques in this section are optional.\n\n- [Laplace Smoothing](https://en.wikipedia.org/wiki/Additive_smoothing) (pseudocounts)\n Laplace smoothing is a technique where you add a small, non-zero value to all observed counts to offset for unobserved values.\n\n- Backoff Smoothing\n Another smoothing technique is to interpolate between n-grams for missing data. This method is more effective than Laplace smoothing at combatting the data sparsity problem. Refer to chapters 4, 9, and 10 of the [Speech & Language Processing](https://web.stanford.edu/~jurafsky/slp3/) book for more information.\n\n- Extending to Trigrams\n HMM taggers have achieved better than 96% accuracy on this dataset with the full Penn treebank tagset using an architecture described in [this](http://www.coli.uni-saarland.de/~thorsten/publications/Brants-ANLP00.pdf) paper. Altering your HMM to achieve the same performance would require implementing deleted interpolation (described in the paper), incorporating trigram probabilities in your frequency tables, and re-implementing the Viterbi algorithm to consider three consecutive states instead of two.\n\n### Obtain the Brown Corpus with a Larger Tagset\nRun the code below to download a copy of the brown corpus with the full NLTK tagset. You will need to research the available tagset information in the NLTK docs and determine the best way to extract the subset of NLTK tags you want to explore. If you write the following the format specified in Step 1, then you can reload the data using all of the code above for comparison.\n\nRefer to [Chapter 5](http://www.nltk.org/book/ch05.html) of the NLTK book for more information on the available tagsets.",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk import pos_tag, word_tokenize\nfrom nltk.corpus import brown\n\nnltk.download('brown')\ntraining_corpus = nltk.corpus.brown\ntraining_corpus.tagged_sents()[0]",
"[nltk_data] Downloading package brown to\n[nltk_data] /home/ucleraiserver/nltk_data...\n[nltk_data] Package brown is already up-to-date!\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71dd824b2d88d5421d9b823a47a865c98220a2a | 24,162 | ipynb | Jupyter Notebook | Exercises/.ipynb_checkpoints/P3-MovieGenrePrediction-checkpoint.ipynb | zerodarkzone/AdvancedMethodsDataAnalysisClassGrupo4202119 | 99538d25a121a327cca3d18bc03a941aac82dca1 | [
"MIT"
] | 5 | 2021-07-07T17:19:39.000Z | 2022-03-13T23:30:51.000Z | Exercises/.ipynb_checkpoints/P3-MovieGenrePrediction-checkpoint.ipynb | davidzarruk/AdvancedMethodsDataAnalysisClass | 2c28ba737e422d7310abb2743a5f77b856ab3659 | [
"MIT"
] | null | null | null | Exercises/.ipynb_checkpoints/P3-MovieGenrePrediction-checkpoint.ipynb | davidzarruk/AdvancedMethodsDataAnalysisClass | 2c28ba737e422d7310abb2743a5f77b856ab3659 | [
"MIT"
] | 9 | 2021-07-09T01:06:37.000Z | 2021-10-17T08:16:18.000Z | 32.045093 | 487 | 0.424261 | [
[
[
"# Project 3\n\n\n# Movie Genre Classification\n\nClassify a movie genre based on its plot.\n\n<img src=\"moviegenre.png\"\n style=\"float: left; margin-right: 10px;\" />\n\n\n\n\nhttps://www.kaggle.com/c/miia4201-202019-p3-moviegenreclassification/overview\n\n### Data\n\nInput:\n- movie plot\n\nOutput:\n- Probability of the movie belonging to each genre\n\n\n### Evaluation\n\n- 50% Report with all the details of the solution, the analysis and the conclusions. The report cannot exceed 10 pages, must be send in PDF format and must be self-contained.\n- 50% Performance in the Kaggle competition (The grade for each group will be proportional to the ranking it occupies in the competition. The group in the first place will obtain 5 points, for each position below, 0.25 points will be subtracted, that is: first place: 5 points, second: 4.75 points, third place: 4.50 points ... eleventh place: 2.50 points, twelfth place: 2.25 points).\n\n\n### Deadline\n- The project must be carried out in the groups assigned.\n- Use clear and rigorous procedures.\n- The delivery of the project is on August 1st, 2021, 11:59 pm, through Bloque Neón.\n- No projects will be received after the delivery time or by any other means than the one established. \n\n\n\n\n### Acknowledgements\n\nWe thank Professor Fabio Gonzalez, Ph.D. and his student John Arevalo for providing this dataset.\n\nSee https://arxiv.org/abs/1702.01992",
"_____no_output_____"
],
[
"## Sample Submission",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport os\nimport numpy as np\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.preprocessing import MultiLabelBinarizer\nfrom sklearn.multiclass import OneVsRestClassifier\nfrom sklearn.ensemble import RandomForestRegressor, RandomForestClassifier\nfrom sklearn.metrics import r2_score, roc_auc_score\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"dataTraining = pd.read_csv('https://github.com/albahnsen/AdvancedMethodsDataAnalysisClass/raw/master/datasets/dataTraining.zip', encoding='UTF-8', index_col=0)\ndataTesting = pd.read_csv('https://github.com/albahnsen/AdvancedMethodsDataAnalysisClass/raw/master/datasets/dataTesting.zip', encoding='UTF-8', index_col=0)",
"_____no_output_____"
],
[
"dataTraining.head()",
"_____no_output_____"
],
[
"dataTesting.head()",
"_____no_output_____"
]
],
[
[
"### Create count vectorizer\n",
"_____no_output_____"
]
],
[
[
"vect = CountVectorizer(max_features=1000)\nX_dtm = vect.fit_transform(dataTraining['plot'])\nX_dtm.shape",
"_____no_output_____"
],
[
"print(vect.get_feature_names()[:50])",
"['able', 'about', 'accepts', 'accident', 'accidentally', 'across', 'act', 'action', 'actor', 'actress', 'actually', 'adam', 'adult', 'adventure', 'affair', 'after', 'again', 'against', 'age', 'agent', 'agents', 'ago', 'agrees', 'air', 'alan', 'alex', 'alice', 'alien', 'alive', 'all', 'almost', 'alone', 'along', 'already', 'also', 'although', 'always', 'america', 'american', 'among', 'an', 'and', 'angeles', 'ann', 'anna', 'another', 'any', 'anyone', 'anything', 'apartment']\n"
]
],
[
[
"### Create y",
"_____no_output_____"
]
],
[
[
"dataTraining['genres'] = dataTraining['genres'].map(lambda x: eval(x))\n\nle = MultiLabelBinarizer()\ny_genres = le.fit_transform(dataTraining['genres'])",
"_____no_output_____"
],
[
"y_genres",
"_____no_output_____"
],
[
"X_train, X_test, y_train_genres, y_test_genres = train_test_split(X_dtm, y_genres, test_size=0.33, random_state=42)",
"_____no_output_____"
]
],
[
[
"### Train multi-class multi-label model",
"_____no_output_____"
]
],
[
[
"clf = OneVsRestClassifier(RandomForestClassifier(n_jobs=-1, n_estimators=100, max_depth=10, random_state=42))",
"_____no_output_____"
],
[
"clf.fit(X_train, y_train_genres)",
"_____no_output_____"
],
[
"y_pred_genres = clf.predict_proba(X_test)",
"_____no_output_____"
],
[
"roc_auc_score(y_test_genres, y_pred_genres, average='macro')",
"_____no_output_____"
]
],
[
[
"### Predict the testing dataset",
"_____no_output_____"
]
],
[
[
"X_test_dtm = vect.transform(dataTesting['plot'])\n\ncols = ['p_Action', 'p_Adventure', 'p_Animation', 'p_Biography', 'p_Comedy', 'p_Crime', 'p_Documentary', 'p_Drama', 'p_Family',\n 'p_Fantasy', 'p_Film-Noir', 'p_History', 'p_Horror', 'p_Music', 'p_Musical', 'p_Mystery', 'p_News', 'p_Romance',\n 'p_Sci-Fi', 'p_Short', 'p_Sport', 'p_Thriller', 'p_War', 'p_Western']\n\ny_pred_test_genres = clf.predict_proba(X_test_dtm)\n",
"_____no_output_____"
],
[
"res = pd.DataFrame(y_pred_test_genres, index=dataTesting.index, columns=cols)",
"_____no_output_____"
],
[
"res.head()",
"_____no_output_____"
],
[
"res.to_csv('pred_genres_text_RF.csv', index_label='ID')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e71dddc1d14bba9b9b289e381a29530371d46fcf | 16,487 | ipynb | Jupyter Notebook | content/_build/jupyter_execute/notebooks/00-Intro.ipynb | zoeydy/CMEESamraat | 3c8e54d4878ff3f9b9272da73c3c8700902ddb21 | [
"MIT"
] | 1 | 2021-05-07T09:28:51.000Z | 2021-05-07T09:28:51.000Z | content/_build/jupyter_execute/notebooks/00-Intro.ipynb | zoeydy/CMEESamraat | 3c8e54d4878ff3f9b9272da73c3c8700902ddb21 | [
"MIT"
] | null | null | null | content/_build/jupyter_execute/notebooks/00-Intro.ipynb | zoeydy/CMEESamraat | 3c8e54d4878ff3f9b9272da73c3c8700902ddb21 | [
"MIT"
] | null | null | null | 63.411538 | 847 | 0.690969 | [
[
[
"# Introduction <a name=\"chap:Intro\"></a>\n\n> *It is hard for me to say confidently that, after fifty more years of explosive growth of computer science, there will still be a lot of fascinating unsolved problems at peoples' fingertips, that it won't be pretty much working on refinements of well-explored things. Maybe all of the simple stuff and the really great stuff has been discovered. It may not be true, but I can't predict an unending growth. I can't be as confident about computer science as I can about biology. Biology easily has 500 years of exciting problems to work on, it's at that level.*\n>\n> — *Donald Knuth*\n",
"_____no_output_____"
],
[
"## About\n \nThese online notes have emerged from the development of content for modules on Biological Computing taught in various past and present courses at the Department of Life Sciences, Imperial College London. These courses include Year 1 & 2 Computational Biostatistics modules at the South Kensington Campus, the Computational Methods in Ecology and Evolution [(CMEE) Masters program](http://www.imperial.ac.uk/life-sciences/postgraduate/masters-courses/computational-methods-in-ecology-and-evolution/) at the Silwood Park Campus, the Quantitative Methods in Ecology and Evolution Centre for Doctoral Training ([QMEE CDT](https://www.imperial.ac.uk/qmee-cdt/)), and the training workshops of the [VectorBiTE RCN](http://vectorbite.org).\n\nDifferent subsets of this document will be covered in different courses. Please look up your respective course guidebooks/handbooks to determine when the modules covered in these notes are scheduled in your course. You will be given instructions about which sections are covered in your course.\n\nAll the chapters of these notes are written as [jupyter](Appendix-JupyIntro.ipynb) notebooks. Each chapter/notebook is accompanied by data and code on which you can practice your skills in your own time and during practical sessions. These materials are available (and will be updated regularly) at a [git repository](https://github.com/mhasoba/TheMulQuaBio). We use git for hosting this course's materials because we want to version-control this course's content, which is constantly evolving to keep up with changing programming/computing technologies. That is, we are treating this course as any computing project that needs to be regularly updated and improved. Changes to the notes and content will also be made based upon student feedback. Blackboard is just not set up to handle dynamic updating and version control of this sort! \n\nIf you do not use git, you may download the code, data, these notes, and other course materials from the [repository](https://github.com/mhasoba/TheMulQuaBio) at one go look for the green \"Clone or Download\" button and then clicking on the \"Download repository\" link. You can then unzip the downloaded .zip and grab the files you need.\n\n---\n\n\n<small> <center>(Source: [http://xkcd.com/974](http://xkcd.com/974)) \n</center></small>\n\n---\n\nIt is important that you work through the exercises and problems in each chapter. This document does not tell you every single thing you need to know to perform the exercises in it. In programming and computing, you learn faster by trying to solve problems (including computer crashes!) on your own, often by liberally googling the problem!\n\n\n### Learning goals\n\nThe goal of these notes is to teach you to become (or at least show you the path towards becoming) a competent quantitative biologist. A large part of this involves learning computer programming. Why do biologists\nneed to write computer programs? Here are some (hopefully compelling!) reasons:\n\n* Short of fieldwork, programs can do anything (that can be specified). In fact, even fieldwork, if you could one day *program* a robot to do it for you <sup>[1](#intro:robot)</sup>!\n\n* As such, no software is typically available to perform exactly the analysis you are planning. You should be unhappy if you are trying to shoehorn your data into methods that don't quite seem right.\n\n* Biological problems and datasets are some of the most complicated imaginable. Programming permits success despite\ncomplexity through precise specification and modularization of complicated analyses.\n\n* Modularity – programming allows you to break up your complex analysis in smaller pieces, yet keep all the pieces in a single, functional analysis.\n\n* Reproducibility – you (or someone else) can just re-run the code to reproduce your analysis. This is also the key to maintaining scientific accountability, integrity, and accuracy.\n\n* Organised thinking – writing code requires you to do this!\n\n* Career prospects – good, scientific coders are in short supply in all fields, but most definitely in biology!\n\nThere are several hundred programming languages currently available – which ones should a biologist choose? Ideally, a quantitative biologist should be multilingual, knowing:\n\n1. A compiled (or semi-compiled) '[procedural](https://en.wikipedia.org/wiki/Procedural_programming)' language like `C`\n\n2. A modern, easy-to-write, interpreted (or semi-compiled) language that is still quite fast, like `python`\n\n3. Mathematical/statistical software with programming and graphing capabilities like `R`\n\nAnd all these because one language doesn't fit all purposes. Something like `C` is the first item in the list above because learning a procedural language forces you to deal with the real workings of your computer ubner the hood (memory management, pointers, etc.), which other languages hide (for ease of programming and running code). Without an understanding of these 'low-level' aspects of computer programming you will be limited if you try to develop an application that needs to run in a memory or performance constrained environment. Languages like Python and R obscure a lot of details of the underlying computer science. \n\nTherefore you will learn a few different languages in this course — hopefully, just the right number! Among the languages you will learn here — ` python`, `R`, and `C` are three of the [most popular currently](https://www.tiobe.com/tiobe-index) (also [see this page](https://spectrum.ieee.org/static/interactive-the-top-programming-languages-2018)), and with good reasons.\n\nOur goal is to teach you not just programming, but also good computing practices. In this course, you will write plenty of code, deal with different data files, and produce text and graphic outputs. You will learn to keep your project and coursework organized in logical, efficient, error-free and reproducible *workflows* (that's a mouthful, but an important mouthful).\n\n## Some guidelines, conventions and rules\n\n### Beware the dark forces\n\nYou will NOT be using spreadsheet software (e.g., Excel) on this course. There are times when you will feel the pull of the dark side (ahem!), and imagine a more \"comfortable\" world where you are mouse-clicking your way happily though Excel-based data manipulations and analyses. NO! You will be doing yourself a disservice. On the\nlong-ish run you will be much better off visualizing and manipulating data on your computer using a programming language like R. This is something you will learn, young [*padawan*](http://starwars.wikia.com/wiki/Padawan)!\n\n### Keep your workflow organized\n\nIn the following chapters, you will practice many examples where you are required to write large blocks of code. Please get into the habit of writing code into text files with an appropriate extension (e.g., `.R` for `R` code, `.py` for `python` code, etc.). Furthermore, please keep all your code files organized in one or more directories (e.g., named `Code`!). Similarly, some of these scripts will take data files as inputs, and output some results in the form of text or graphics. Please keep these inputs and outputs organized as well, in\nseparate directories (e.g., named `Data` and ` Results`) respectively. Your instructor(s) will help you get set up and abide by this \"workflow\".\n\n\n<small> \n <center>(Source: http://xkcd.com/1172) \n Logical workflows are important, but don't get married to yours!\n </center>\n</small>\n\n\n\n---\n\n### Conventions used in this document\n\nThroughout these sessions, directory paths will be specified in UNIX (Mac, Linux) style, using `/` instead of the `\\` used in Windows. Also, in general, we will be using [relative paths](https://en.wikipedia.org/wiki/Path_(computing)) throughout the exercises and practicals (more on this later, but google it!).\n\nYou will find all command line/console arguments, code snippets and output in boxes like this:",
"_____no_output_____"
]
],
[
[
"# Some code here!",
"_____no_output_____"
]
],
[
[
"You will type the commands/code that you see in such boxes into the relevant command line (don't copy-paste - you likely need all the command / syntax typing practice you can get !). \n\nNote that the commandline prompt you will see on your own terminal/console will vary with the programming language: `$` for UNIX, `>>>` for Python, `>` for R, etc. \n\nAlso note that:\n\n$\\star$ Lines marked with a star like this will be specific instructions for you to follow\n\n\nAnd there will be notes, tips and warnings that you should pay particular attention to, which will appear like this: \n\n\n```{note}\nThis is a note\n```\n\n```{tip}\nThis is a tip\n```\n\n```{warning}\nThis is a warning\n```\n\nSo here's your first (and perhaps most important) tip:\n\n```{tip}\n**Finding solutions online.** This document does not tell you every single thing you need to know to perform the exercises in it. In programming and computing, you learn faster by trying to solve problems on your own. The contemporary way to do this is to google the problem! Some suggestions: \n* Your typical approach should be to serach online the main keywords along the programming language name (e.g., \"unix mv vs cp\" or \"R create empty dataframe\"). \n* Look for [stackoverflow](https://stackoverflow.com/) or [stackexchange](https://stackexchange.com/) based results in particular. Look for number of endorsements for both the question that has been asked, and for its answer.\n* Be selective - there will often be multiple solutions to the same issue or problem - don't just blindly use the first one you find. \n* Also, every time a mysterious, geeky-sounding term like \"relative path\" or \"version control\" appears, please search (e.g., google) it online it as well!\n```",
"_____no_output_____"
],
[
"### To IDE or not to IDE?\n\nAs you embark on your journey to becoming a competent practitioner of biological computing, you will be faced with a Hamletian question: \"To IDE or not to IDE\" (anagram alert!). *OK, maybe not that dramatic or Hamletian...*\n\nAn interactive Development Environment (IDE) is a text editor with additional features that can make life easy by allowing auto-formatting of code, running code, providing a graphic view of the workspace (your active functions, variables, etc.), graphic debugging and profiling (you will see these delightful things later), and allowing integrated version control (e.g., using `git`). \n\nYou will benefit a lot if you use a code editor that can also offer an IDE. At the very least, your IDE should offer:\n\n* Auto-indentation\n\n* Automatic code wrapping (e.g., keeping lines <80 characters long)\n\n* [Syntax highlighting](https://en.wikipedia.org/wiki/Syntax_highlighting) (language elements such as variables, commands, and brackets are differently colored)\n\n* Code folding (fold large blocks of code, say an entire function or loop)\n\n* Keyboard control of commenting/uncommenting, code wrapping, etc.\n\n* Embedded terminal / shell / commandline console\n\n* Sending commands to terminal / shell\n\n* Debugging\n\n\nIf you end up using multiple programming languages, you will want an IDE that can handle them. Four commonly-used and very powerful multi-lingual code editors/IDEs are: Emacs, Vim, Visual Studio Code (AKA VS Code), and Atom. We will use [Visual Studio Code](https://code.visualstudio.com) in this course, because it is freely available, is relatively easy to master, and has many plugins that make multilingual code development easier. Atom, Vim and Emacs are also great alternatives. The last having a steeper learning curve, but are very powerful once you have mastered them. \n\n#### Gooey IDEs\n\nIDEs come with graphic user interfaces (GUI's, or \"gooeys\") of differing levels of sophistication and shiny-ness. Some go over and above the call of duty to offer further useful features like embedded data and plot views and package management. One such example is th freelty available [RStudio](http://rstudio.org), which is dedicated to R. However, there is a tradeoff – these are necessarily specialized to one language (e.g., RStudio). It is up to you if you want to use multiple, language specific IDEs, or one somewhat less shiny multi-lingual IDE. \n\n### Assessment\n\nYour computing coursework may be assessed. If you have been told that it will, please see [this Appendix](Appendix-Assessment.rst) if you are a *Masters student*. If you are an *Undergrad student*, you may have a computer based test, the format for which will be explained to you elsewhere.",
"_____no_output_____"
],
[
"---\n\n**Footnotes**\n\n<a name=\"intro:robot\">1</a>: That way you can traipse around the forest catching rare butterflies and frogs while the robot does the boring data collecting for you",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e71dee1234411f107f38e98cf4d304fd1959ff86 | 75,404 | ipynb | Jupyter Notebook | Question_2.ipynb | tianzixie/CAP5610_Exam | 5cd002383b7fd6a047a2f1a5571d1191e53438d4 | [
"MIT"
] | null | null | null | Question_2.ipynb | tianzixie/CAP5610_Exam | 5cd002383b7fd6a047a2f1a5571d1191e53438d4 | [
"MIT"
] | null | null | null | Question_2.ipynb | tianzixie/CAP5610_Exam | 5cd002383b7fd6a047a2f1a5571d1191e53438d4 | [
"MIT"
] | null | null | null | 139.896104 | 13,542 | 0.840128 | [
[
[
"Hard margin SVM with GridSearchCV(svm.SVC(), cv=5,param_grid={}) has best accuracy,it has highest R-Square value and lowest Mean squared error value. The performance measure actually tell me that I didn't get good result. I use a grid search to try out best parameter values. Attribute 'Class' is the target of prediction. I transformed \"classification_data.tsv\" to \"classification_data.csv\". I use small size of data to increase speed. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport seaborn as sns\nfrom sklearn.feature_selection import RFE, f_regression\nfrom sklearn.linear_model import (LinearRegression)\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.metrics import (r2_score, mean_squared_error)\n\nregression = pd.read_csv(\"./classification_data.csv\")\nregression.head()\n#regression = regression.drop(['Red','Green','Blue'],axis=1)\nY = regression.Class.values\nregression = regression.drop(['Class'], axis=1)\nX = regression.as_matrix()\nt = pd.DataFrame(X,index = Y )\nt.plot()\nplt.show()\n\nregression_X_train = X[:]\nregression_y_train = Y[:]\nlr = LinearRegression(normalize=True)\nlr.fit(regression_X_train, regression_y_train)\nprint('Coefficients: \\n', lr.coef_)\nprint('data size: 245058')",
"_____no_output_____"
],
[
"#KernelRidge with kernel='linear'\nfrom sklearn.kernel_ridge import KernelRidge\nfrom sklearn.model_selection import GridSearchCV\n#To reduce time, split the data set into a training set of 4000 (80%) and a testing set of 1000 (20%)\nregression_X_train = X[:-241058]\nregression_X_test = X[(245058-244058):245058]\n\nregression_y_train = Y[:-241058]\nregression_y_test = Y[(245058-244058):245058]\nlr = GridSearchCV(KernelRidge(kernel='linear', gamma=1), cv=5,\n param_grid={\"alpha\": [2,4,7],\n \"gamma\": [0.1, 0.5, 1, 2, 4]})\n#GridSearchCV is with 5-fold cross-validation and it includes grid search\nlr.fit = lr.fit(regression_X_train, regression_y_train)\nregression_y_train1 = lr.predict(regression_X_train)\ndataPred = lr.predict(regression_X_test)\nprint(\"R^2 on test dataset by Linear KRR: \",r2_score(regression_y_test, dataPred)) \nprint(\"Mean squared error: %.2f\" % mean_squared_error(regression_y_test, dataPred))\n#R^2 on test dataset by Linear KRR: -37.4341607128\n#Mean squared error: 6.25",
"R^2 on test dataset by Linear KRR: -37.4341607128\nMean squared error: 6.25\n"
],
[
"#KernelRidge with kernel='poly'\n#To reduce time, split the data set into a training set of 2000 (80%) and a testing set of 500 (20%)\nregression_X_train = X[:-243058]\nregression_X_test = X[(245058-244558):245058]\n\nregression_y_train = Y[:-243058]\nregression_y_test = Y[(245058-244558):245058]\nlr = GridSearchCV(KernelRidge(kernel='poly', gamma=1), cv=5,\n param_grid={\"alpha\": [2,4,7],\n \"gamma\": [0.1, 0.5, 1, 2, 4]})\n#GridSearchCV is with 5-fold cross-validation\nlr.fit = lr.fit(regression_X_train, regression_y_train)\nregression_y_train1 = lr.predict(regression_X_train)\ndataPred = lr.predict(regression_X_test)\nprint(\"R^2 on test dataset by poly KRR: \",r2_score(regression_y_test, dataPred)) \nprint(\"Mean squared error: %.2f\" % mean_squared_error(regression_y_test, dataPred))\n#R^2 on test dataset by poly KRR: -381.557505029\n#Mean squared error: 62.55",
"/Users/t0/anaconda3/lib/python3.6/site-packages/scipy/linalg/basic.py:40: RuntimeWarning: scipy.linalg.solve\nIll-conditioned matrix detected. Result is not guaranteed to be accurate.\nReciprocal condition number/precision: 8.878876873096602e-17 / 1.1102230246251565e-16\n RuntimeWarning)\n/Users/t0/anaconda3/lib/python3.6/site-packages/scipy/linalg/basic.py:40: RuntimeWarning: scipy.linalg.solve\nIll-conditioned matrix detected. Result is not guaranteed to be accurate.\nReciprocal condition number/precision: 8.32732582170335e-17 / 1.1102230246251565e-16\n RuntimeWarning)\n/Users/t0/anaconda3/lib/python3.6/site-packages/scipy/linalg/basic.py:40: RuntimeWarning: scipy.linalg.solve\nIll-conditioned matrix detected. Result is not guaranteed to be accurate.\nReciprocal condition number/precision: 9.037735687438652e-17 / 1.1102230246251565e-16\n RuntimeWarning)\n/Users/t0/anaconda3/lib/python3.6/site-packages/scipy/linalg/basic.py:40: RuntimeWarning: scipy.linalg.solve\nIll-conditioned matrix detected. Result is not guaranteed to be accurate.\nReciprocal condition number/precision: 8.691308353525568e-17 / 1.1102230246251565e-16\n RuntimeWarning)\n/Users/t0/anaconda3/lib/python3.6/site-packages/scipy/linalg/basic.py:40: RuntimeWarning: scipy.linalg.solve\nIll-conditioned matrix detected. Result is not guaranteed to be accurate.\nReciprocal condition number/precision: 8.000554187080201e-17 / 1.1102230246251565e-16\n RuntimeWarning)\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/sklearn/linear_model/ridge.py:154: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.\n warnings.warn(\"Singular matrix in solving dual problem. Using \"\n/Users/t0/anaconda3/lib/python3.6/site-packages/scipy/linalg/basic.py:40: RuntimeWarning: scipy.linalg.solve\nIll-conditioned matrix detected. Result is not guaranteed to be accurate.\nReciprocal condition number/precision: 6.390730617276815e-17 / 1.1102230246251565e-16\n RuntimeWarning)\n"
],
[
"#KernelRidge with kernel='rbf'\n#To reduce time, split the data set into a training set of 4000 (80%) and a testing set of 1000 (20%)\n\nregression_X_train = X[:-241058]\nregression_X_test = X[(245058-244058):245058]\n\nregression_y_train = Y[:-241058]\nregression_y_test = Y[(245058-244058):245058]\nlr = GridSearchCV(KernelRidge(kernel='rbf', gamma=1), cv=5,\n param_grid={\"alpha\": [2,4,7],\n \"gamma\": [0.1, 0.5, 1, 2, 4]})\n#GridSearchCV is with 5-fold cross-validation\nlr.fit = lr.fit(regression_X_train, regression_y_train)\nregression_y_train1 = lr.predict(regression_X_train)\ndataPred = lr.predict(regression_X_test)\nprint(\"R^2 on test dataset by rbf KRR: \",r2_score(regression_y_test, dataPred)) \nprint(\"Mean squared error: %.2f\" % mean_squared_error(regression_y_test, dataPred))\n#R^2 on test dataset by rbf KRR: -19.5871331947\n#Mean squared error: 3.35",
"R^2 on test dataset by rbf KRR: -19.5871331947\nMean squared error: 3.35\n"
],
[
"#Logistic Regression\nfrom sklearn.linear_model import LogisticRegression \n#Split the data set into a training set of 80% and a testing set of 20%\nregression_X_train = X[:-49012]\nregression_X_test = X[(245058-196046):245058]\n\nregression_y_train = Y[:-49012]\nregression_y_test = Y[(245058-196046):245058] \nlr = GridSearchCV(LogisticRegression(), cv=5,param_grid={}) \nlr.fit = lr.fit(regression_X_train, regression_y_train)\ndataPred = lr.predict(regression_X_test)\n\nfrom sklearn.metrics import confusion_matrix \nprint (dataPred)\nconfusion_matrix=confusion_matrix(regression_y_test,regression_y_train)\nprint (confusion_matrix)\nplt.matshow(confusion_matrix)\nplt.title(u'confusion_matrix')\nplt.colorbar()\nplt.ylabel(u'actural_Class')\nplt.xlabel(u'predict_Class')\nplt.show()\nfrom sklearn.cross_validation import cross_val_score\nprint(\"R^2 on test dataset by Logistic Regression: \",r2_score(regression_y_test, dataPred)) \nprint(\"Mean squared error: %.2f\" % mean_squared_error(regression_y_test, dataPred))\n#R^2 on test dataset by Logistic Regression: -6.00754704759\n#Mean squared error: 0.07",
"[1 1 1 ..., 2 2 2]\n[[ 1847 0]\n [ 49012 145186]]\n"
],
[
"#Hard margin SVM\nfrom sklearn import svm\n#Split the data set\nregression_X_train = X[40000:60000]\nregression_X_test = X[60000:80000]\n\nregression_y_train = Y[40000:60000]\nregression_y_test = Y[60000:80000] \n#lr = GridSearchCV(svm.SVC(), cv=5,param_grid={'C': [1e-3, 1e-2, 1e-1, 1, 10, 100, 1000], 'gamma': [0.001, 0.0001]})\nlr = GridSearchCV(svm.SVC(), cv=5,param_grid={})\n#lr.fit(regression_X_train, regression_y_train) \nlr.fit = lr.fit(regression_X_train, regression_y_train)\ndataPred = lr.predict(regression_X_test)\nfrom sklearn.metrics import confusion_matrix \nprint (dataPred)\nconfusion_matrix=confusion_matrix(regression_y_test,regression_y_train)\nprint (confusion_matrix)\nplt.matshow(confusion_matrix)\nplt.title(u'confusion_matrix')\nplt.colorbar()\nplt.ylabel(u'actural_Class')\nplt.xlabel(u'predict_Class')\nplt.show()\nfrom sklearn.cross_validation import cross_val_score\nprint(\"R^2 on test dataset by Hard margin SVM: \",r2_score(regression_y_test, dataPred)) \nprint(\"Mean squared error: %.2f\" % mean_squared_error(regression_y_test, dataPred))\n#R^2 on test dataset by Hard margin SVM: 0.0\n#Mean squared error: 0.36",
"[2 2 2 ..., 1 1 1]\n[[ 0 0]\n [10859 9141]]\n"
],
[
"#Soft margin SVM\nfrom sklearn import svm\n#Split the data set\nregression_X_train = X[40000:60000]\nregression_X_test = X[60000:80000]\n\nregression_y_train = Y[40000:60000]\nregression_y_test = Y[60000:80000] \nlr = GridSearchCV(svm.SVC(), cv=5,param_grid={'C': [0.1, 0.5, 1, 2, 5]})\n#lr = GridSearchCV(svm.SVC(), cv=5,param_grid={})\n#lr.fit(regression_X_train, regression_y_train) \nlr.fit = lr.fit(regression_X_train, regression_y_train)\ndataPred = lr.predict(regression_X_test)\n\nfrom sklearn.metrics import confusion_matrix \nprint (dataPred)\nconfusion_matrix=confusion_matrix(regression_y_test,regression_y_train)\nprint (confusion_matrix)\nplt.matshow(confusion_matrix)\nplt.title(u'confusion_matrix')\nplt.colorbar()\nplt.ylabel(u'actural_Class')\nplt.xlabel(u'predict_Class')\nplt.show()\nfrom sklearn.cross_validation import cross_val_score\n#scores=cross_val_score(regression_X_train,regression_y_train,cv=5)\n#print ('accuracy',np.mean(scores),scores)\nprint(\"R^2 on test dataset by Soft margin SVM: \",r2_score(regression_y_test, dataPred)) \nprint(\"Mean squared error: %.2f\" % mean_squared_error(regression_y_test, dataPred))\n#R^2 on test dataset by Soft margin SVM: 0.0\n#Mean squared error: 0.39",
"[2 2 2 ..., 1 1 1]\n[[ 0 0]\n [10859 9141]]\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71df727c6219391a2ae7ff5e6499a14f0270648 | 55,973 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive2/introduction_to_tensorflow/labs/sdk-custom-image-classification-online.ipynb | maureenm/training-data-analyst | 4ce1319bf0058ad83e929c500abbd2a47a14de5d | [
"Apache-2.0"
] | 1 | 2021-11-11T11:36:10.000Z | 2021-11-11T11:36:10.000Z | courses/machine_learning/deepdive2/introduction_to_tensorflow/labs/sdk-custom-image-classification-online.ipynb | maureenm/training-data-analyst | 4ce1319bf0058ad83e929c500abbd2a47a14de5d | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive2/introduction_to_tensorflow/labs/sdk-custom-image-classification-online.ipynb | maureenm/training-data-analyst | 4ce1319bf0058ad83e929c500abbd2a47a14de5d | [
"Apache-2.0"
] | null | null | null | 43.023059 | 416 | 0.636771 | [
[
[
"# Custom training and online prediction\n\n## Overview\n\nThis tutorial demonstrates how to use the Vertex SDK for Python to train and deploy a custom image classification model for online prediction.\n\n**Learning Objectives**\n\nIn this notebook, you create a custom-trained model from a Python script in a Docker container using the Vertex SDK for Python, and then do a prediction on the deployed model by sending data. Alternatively, you can create custom-trained models using `gcloud` command-line tool, or online using the Cloud Console.\n\n- Create a Vertex AI custom job for training a model.\n- Train a TensorFlow model.\n- Deploy the `Model` resource to a serving `Endpoint` resource.\n- Make a prediction.\n- Undeploy the `Model` resource.\n\n\n## Introduction \n\nIn this notebook, you create a custom-trained model from a Python script in a Docker container using the Vertex SDK for Python, and then do a prediction on the deployed model by sending data. Alternatively, you can create custom-trained models using `gcloud` command-line tool, or online using the Cloud Console.\n\nEach learning objective will correspond to a __#TODO__ in this student lab notebook -- try to complete this notebook first and then review the [solution notebook](../solutions/sdk-custom-image-classification-online.ipynb). \n\n### Dataset\n\nThe dataset used for this tutorial is the [cifar10 dataset](https://www.tensorflow.org/datasets/catalog/cifar10) from [TensorFlow Datasets](https://www.tensorflow.org/datasets/catalog/overview). The version of the dataset you will use is built into TensorFlow. The trained model predicts which type of class an image is from ten classes: airplane, automobile, bird, cat, deer, dog, frog, horse, ship, truck.\n\n**Make sure to enable the Vertex AI API and Compute Engine API.**",
"_____no_output_____"
],
[
"## Installation\n\nInstall the latest (preview) version of Vertex SDK for Python.",
"_____no_output_____"
]
],
[
[
"import os\n\n# The Google Cloud Notebook product has specific requirements\nIS_GOOGLE_CLOUD_NOTEBOOK = os.path.exists(\"/opt/deeplearning/metadata/env_version\")\n\n# Google Cloud Notebook requires dependencies to be installed with '--user'\nUSER_FLAG = \"\"\nif IS_GOOGLE_CLOUD_NOTEBOOK:\n USER_FLAG = \"--user\"",
"_____no_output_____"
],
[
"! pip install {USER_FLAG} --upgrade google-cloud-aiplatform",
"Requirement already satisfied: google-cloud-aiplatform in /opt/conda/lib/python3.7/site-packages (1.1.1)\nCollecting google-cloud-aiplatform\n Downloading google_cloud_aiplatform-1.3.0-py2.py3-none-any.whl (1.3 MB)\n\u001b[K |████████████████████████████████| 1.3 MB 7.7 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: google-cloud-storage<2.0.0dev,>=1.32.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-aiplatform) (1.41.1)\nRequirement already satisfied: google-api-core[grpc]<3.0.0dev,>=1.26.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-aiplatform) (1.31.1)\nRequirement already satisfied: proto-plus>=1.10.1 in /opt/conda/lib/python3.7/site-packages (from google-cloud-aiplatform) (1.19.0)\nRequirement already satisfied: packaging>=14.3 in /opt/conda/lib/python3.7/site-packages (from google-cloud-aiplatform) (21.0)\nRequirement already satisfied: google-cloud-bigquery<3.0.0dev,>=1.15.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-aiplatform) (2.23.2)\nRequirement already satisfied: six>=1.13.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (1.16.0)\nRequirement already satisfied: setuptools>=40.3.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (49.6.0.post20210108)\nRequirement already satisfied: requests<3.0.0dev,>=2.18.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (2.25.1)\nRequirement already satisfied: google-auth<2.0dev,>=1.25.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (1.34.0)\nRequirement already satisfied: pytz in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (2021.1)\nRequirement already satisfied: protobuf>=3.12.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (3.16.0)\nRequirement already satisfied: googleapis-common-protos<2.0dev,>=1.6.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (1.53.0)\nRequirement already satisfied: grpcio<2.0dev,>=1.29.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (1.38.1)\nRequirement already satisfied: pyasn1-modules>=0.2.1 in /opt/conda/lib/python3.7/site-packages (from google-auth<2.0dev,>=1.25.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (0.2.7)\nRequirement already satisfied: cachetools<5.0,>=2.0.0 in /opt/conda/lib/python3.7/site-packages (from google-auth<2.0dev,>=1.25.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (4.2.2)\nRequirement already satisfied: rsa<5,>=3.1.4 in /opt/conda/lib/python3.7/site-packages (from google-auth<2.0dev,>=1.25.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (4.7.2)\nRequirement already satisfied: google-resumable-media<3.0dev,>=0.6.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-bigquery<3.0.0dev,>=1.15.0->google-cloud-aiplatform) (1.3.2)\nRequirement already satisfied: google-cloud-core<3.0.0dev,>=1.4.1 in /opt/conda/lib/python3.7/site-packages (from google-cloud-bigquery<3.0.0dev,>=1.15.0->google-cloud-aiplatform) (1.7.2)\nRequirement already satisfied: google-crc32c<2.0dev,>=1.0 in /opt/conda/lib/python3.7/site-packages (from google-resumable-media<3.0dev,>=0.6.0->google-cloud-bigquery<3.0.0dev,>=1.15.0->google-cloud-aiplatform) (1.1.2)\nRequirement already satisfied: cffi>=1.0.0 in /opt/conda/lib/python3.7/site-packages (from google-crc32c<2.0dev,>=1.0->google-resumable-media<3.0dev,>=0.6.0->google-cloud-bigquery<3.0.0dev,>=1.15.0->google-cloud-aiplatform) (1.14.6)\nRequirement already satisfied: pycparser in /opt/conda/lib/python3.7/site-packages (from cffi>=1.0.0->google-crc32c<2.0dev,>=1.0->google-resumable-media<3.0dev,>=0.6.0->google-cloud-bigquery<3.0.0dev,>=1.15.0->google-cloud-aiplatform) (2.20)\nRequirement already satisfied: pyparsing>=2.0.2 in /opt/conda/lib/python3.7/site-packages (from packaging>=14.3->google-cloud-aiplatform) (2.4.7)\nRequirement already satisfied: pyasn1<0.5.0,>=0.4.6 in /opt/conda/lib/python3.7/site-packages (from pyasn1-modules>=0.2.1->google-auth<2.0dev,>=1.25.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (0.4.8)\nRequirement already satisfied: chardet<5,>=3.0.2 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (4.0.0)\nRequirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (1.26.6)\nRequirement already satisfied: certifi>=2017.4.17 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (2021.5.30)\nRequirement already satisfied: idna<3,>=2.5 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core[grpc]<3.0.0dev,>=1.26.0->google-cloud-aiplatform) (2.10)\nInstalling collected packages: google-cloud-aiplatform\n\u001b[33m WARNING: The script tb-gcp-uploader is installed in '/home/jupyter/.local/bin' which is not on PATH.\n Consider adding this directory to PATH or, if you prefer to suppress this warning, use --no-warn-script-location.\u001b[0m\nSuccessfully installed google-cloud-aiplatform-1.3.0\n"
]
],
[
[
"Install the latest GA version of *google-cloud-storage* library as well.",
"_____no_output_____"
]
],
[
[
"! pip install {USER_FLAG} --upgrade google-cloud-storage",
"Requirement already satisfied: google-cloud-storage in /opt/conda/lib/python3.7/site-packages (1.41.1)\nCollecting google-cloud-storage\n Downloading google_cloud_storage-1.42.0-py2.py3-none-any.whl (105 kB)\n\u001b[K |████████████████████████████████| 105 kB 8.0 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: google-auth<3.0dev,>=1.25.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-storage) (1.34.0)\nRequirement already satisfied: google-api-core<3.0dev,>=1.29.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-storage) (1.31.1)\nRequirement already satisfied: google-resumable-media<3.0dev,>=1.3.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-storage) (1.3.2)\nRequirement already satisfied: requests<3.0.0dev,>=2.18.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-storage) (2.25.1)\nRequirement already satisfied: google-cloud-core<3.0dev,>=1.6.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-storage) (1.7.2)\nRequirement already satisfied: setuptools>=40.3.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core<3.0dev,>=1.29.0->google-cloud-storage) (49.6.0.post20210108)\nRequirement already satisfied: protobuf>=3.12.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core<3.0dev,>=1.29.0->google-cloud-storage) (3.16.0)\nRequirement already satisfied: pytz in /opt/conda/lib/python3.7/site-packages (from google-api-core<3.0dev,>=1.29.0->google-cloud-storage) (2021.1)\nRequirement already satisfied: googleapis-common-protos<2.0dev,>=1.6.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core<3.0dev,>=1.29.0->google-cloud-storage) (1.53.0)\nRequirement already satisfied: packaging>=14.3 in /opt/conda/lib/python3.7/site-packages (from google-api-core<3.0dev,>=1.29.0->google-cloud-storage) (21.0)\nRequirement already satisfied: six>=1.13.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core<3.0dev,>=1.29.0->google-cloud-storage) (1.16.0)\nRequirement already satisfied: pyasn1-modules>=0.2.1 in /opt/conda/lib/python3.7/site-packages (from google-auth<3.0dev,>=1.25.0->google-cloud-storage) (0.2.7)\nRequirement already satisfied: rsa<5,>=3.1.4 in /opt/conda/lib/python3.7/site-packages (from google-auth<3.0dev,>=1.25.0->google-cloud-storage) (4.7.2)\nRequirement already satisfied: cachetools<5.0,>=2.0.0 in /opt/conda/lib/python3.7/site-packages (from google-auth<3.0dev,>=1.25.0->google-cloud-storage) (4.2.2)\nRequirement already satisfied: google-crc32c<2.0dev,>=1.0 in /opt/conda/lib/python3.7/site-packages (from google-resumable-media<3.0dev,>=1.3.0->google-cloud-storage) (1.1.2)\nRequirement already satisfied: cffi>=1.0.0 in /opt/conda/lib/python3.7/site-packages (from google-crc32c<2.0dev,>=1.0->google-resumable-media<3.0dev,>=1.3.0->google-cloud-storage) (1.14.6)\nRequirement already satisfied: pycparser in /opt/conda/lib/python3.7/site-packages (from cffi>=1.0.0->google-crc32c<2.0dev,>=1.0->google-resumable-media<3.0dev,>=1.3.0->google-cloud-storage) (2.20)\nRequirement already satisfied: pyparsing>=2.0.2 in /opt/conda/lib/python3.7/site-packages (from packaging>=14.3->google-api-core<3.0dev,>=1.29.0->google-cloud-storage) (2.4.7)\nRequirement already satisfied: pyasn1<0.5.0,>=0.4.6 in /opt/conda/lib/python3.7/site-packages (from pyasn1-modules>=0.2.1->google-auth<3.0dev,>=1.25.0->google-cloud-storage) (0.4.8)\nRequirement already satisfied: certifi>=2017.4.17 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-cloud-storage) (2021.5.30)\nRequirement already satisfied: idna<3,>=2.5 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-cloud-storage) (2.10)\nRequirement already satisfied: chardet<5,>=3.0.2 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-cloud-storage) (4.0.0)\nRequirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-cloud-storage) (1.26.6)\nInstalling collected packages: google-cloud-storage\nSuccessfully installed google-cloud-storage-1.42.0\n"
]
],
[
[
"Install the *pillow* library for loading images.",
"_____no_output_____"
]
],
[
[
"! pip install {USER_FLAG} --upgrade pillow",
"Requirement already satisfied: pillow in /opt/conda/lib/python3.7/site-packages (8.3.1)\n"
]
],
[
[
"Install the *numpy* library for manipulation of image data.",
"_____no_output_____"
]
],
[
[
"! pip install {USER_FLAG} --upgrade numpy",
"Requirement already satisfied: numpy in /opt/conda/lib/python3.7/site-packages (1.19.5)\nCollecting numpy\n Downloading numpy-1.21.2-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl (15.7 MB)\n\u001b[K |████████████████████████████████| 15.7 MB 6.9 MB/s eta 0:00:01\n\u001b[?25hInstalling collected packages: numpy\n\u001b[33m WARNING: The scripts f2py, f2py3 and f2py3.7 are installed in '/home/jupyter/.local/bin' which is not on PATH.\n Consider adding this directory to PATH or, if you prefer to suppress this warning, use --no-warn-script-location.\u001b[0m\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ntensorflow-io 0.18.0 requires tensorflow-io-gcs-filesystem==0.18.0, which is not installed.\ntfx-bsl 1.2.0 requires absl-py<0.13,>=0.9, but you have absl-py 0.13.0 which is incompatible.\ntfx-bsl 1.2.0 requires google-api-python-client<2,>=1.7.11, but you have google-api-python-client 2.15.0 which is incompatible.\ntfx-bsl 1.2.0 requires google-cloud-bigquery<2.21,>=1.28.0, but you have google-cloud-bigquery 2.23.2 which is incompatible.\ntfx-bsl 1.2.0 requires numpy<1.20,>=1.16, but you have numpy 1.21.2 which is incompatible.\ntfx-bsl 1.2.0 requires pyarrow<3,>=1, but you have pyarrow 5.0.0 which is incompatible.\ntensorflow 2.5.0 requires grpcio~=1.34.0, but you have grpcio 1.38.1 which is incompatible.\ntensorflow 2.5.0 requires numpy~=1.19.2, but you have numpy 1.21.2 which is incompatible.\ntensorflow 2.5.0 requires six~=1.15.0, but you have six 1.16.0 which is incompatible.\ntensorflow 2.5.0 requires typing-extensions~=3.7.4, but you have typing-extensions 3.10.0.0 which is incompatible.\ntensorflow-transform 1.2.0 requires absl-py<0.13,>=0.9, but you have absl-py 0.13.0 which is incompatible.\ntensorflow-transform 1.2.0 requires google-cloud-bigquery<2.21,>=1.28.0, but you have google-cloud-bigquery 2.23.2 which is incompatible.\ntensorflow-transform 1.2.0 requires numpy<1.20,>=1.16, but you have numpy 1.21.2 which is incompatible.\ntensorflow-transform 1.2.0 requires pyarrow<3,>=1, but you have pyarrow 5.0.0 which is incompatible.\napache-beam 2.31.0 requires dill<0.3.2,>=0.3.1.1, but you have dill 0.3.4 which is incompatible.\napache-beam 2.31.0 requires numpy<1.21.0,>=1.14.3, but you have numpy 1.21.2 which is incompatible.\napache-beam 2.31.0 requires pyarrow<5.0.0,>=0.15.1, but you have pyarrow 5.0.0 which is incompatible.\napache-beam 2.31.0 requires typing-extensions<3.8.0,>=3.7.0, but you have typing-extensions 3.10.0.0 which is incompatible.\u001b[0m\nSuccessfully installed numpy-1.21.2\n"
]
],
[
[
"**Note**: Please ignore any incompatibility warnings and errors.\n",
"_____no_output_____"
],
[
"### Restart the kernel\n\nOnce you've installed everything, you need to restart the notebook kernel so it can find the packages.",
"_____no_output_____"
]
],
[
[
"import os\n\nif not os.getenv(\"IS_TESTING\"):\n # Automatically restart kernel after installs\n import IPython\n\n app = IPython.Application.instance()\n app.kernel.do_shutdown(True)",
"_____no_output_____"
]
],
[
[
"#### Set your project ID\n\nUpdate `YOUR-PROJECT-ID` with your Project ID. **If you don't know your project ID**, you may be able to get your project ID using `gcloud`.",
"_____no_output_____"
]
],
[
[
"import os\n\nPROJECT_ID = \"YOUR-PROJECT-ID\"\n\nif not os.getenv(\"IS_TESTING\"):\n # Get your Google Cloud project ID from gcloud\n shell_output=!gcloud config list --format 'value(core.project)' 2>/dev/null\n PROJECT_ID = shell_output[0]\n print(\"Project ID: \", PROJECT_ID)",
"Project ID: qwiklabs-gcp-04-6c865137f72a\n"
]
],
[
[
"Otherwise, set your project ID here.",
"_____no_output_____"
]
],
[
[
"if PROJECT_ID == \"\" or PROJECT_ID is None:\n PROJECT_ID = \"YOUR-PROJECT-ID\" # @param {type:\"string\"}",
"_____no_output_____"
]
],
[
[
"#### Timestamp\n\nIf you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append it onto the name of resources you create in this tutorial.",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\n\nTIMESTAMP = datetime.now().strftime(\"%Y%m%d%H%M%S\")",
"_____no_output_____"
],
[
"import os\nimport sys\n\n# If you are running this notebook in Colab, run this cell and follow the\n# instructions to authenticate your GCP account. This provides access to your\n# Cloud Storage bucket and lets you submit training jobs and prediction\n# requests.\n\n# The Google Cloud Notebook product has specific requirements\nIS_GOOGLE_CLOUD_NOTEBOOK = os.path.exists(\"/opt/deeplearning/metadata/env_version\")\n\n# If on Google Cloud Notebooks, then don't execute this code\nif not IS_GOOGLE_CLOUD_NOTEBOOK:\n if \"google.colab\" in sys.modules:\n from google.colab import auth as google_auth\n\n google_auth.authenticate_user()\n\n # If you are running this notebook locally, replace the string below with the\n # path to your service account key and run this cell to authenticate your GCP\n # account.\n elif not os.getenv(\"IS_TESTING\"):\n %env GOOGLE_APPLICATION_CREDENTIALS ''",
"_____no_output_____"
]
],
[
[
"### Create a Cloud Storage bucket\n\n**The following steps are required, regardless of your notebook environment.**\n\nWhen you submit a training job using the Cloud SDK, you upload a Python package\ncontaining your training code to a Cloud Storage bucket. Vertex AI runs\nthe code from this package. In this tutorial, Vertex AI also saves the\ntrained model that results from your job in the same bucket. Using this model artifact, you can then\ncreate Vertex AI model and endpoint resources in order to serve\nonline predictions.\n\nSet the name of your Cloud Storage bucket below. It must be unique across all\nCloud Storage buckets.\n\nYou may also change the `REGION` variable, which is used for operations\nthroughout the rest of this notebook. Make sure to [choose a region where Vertex AI services are\navailable](https://cloud.google.com/vertex-ai/docs/general/locations#available_regions). You may\nnot use a Multi-Regional Storage bucket for training with Vertex AI.",
"_____no_output_____"
]
],
[
[
"BUCKET_NAME = \"gs://YOUR-BUCKET-NAME\" # @param {type:\"string\"}\nREGION = \"YOUR-REGION\" # @param {type:\"string\"}",
"_____no_output_____"
],
[
"if BUCKET_NAME == \"\" or BUCKET_NAME is None or BUCKET_NAME == \"gs://[your-bucket-name]\":\n BUCKET_NAME = \"gs://\" + PROJECT_ID + \"aip-\" + TIMESTAMP",
"_____no_output_____"
]
],
[
[
"**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.",
"_____no_output_____"
]
],
[
[
"! gsutil mb -l $REGION $BUCKET_NAME",
"Creating gs://qwiklabs-gcp-04-6c865137f72a/...\n"
]
],
[
[
"Finally, validate access to your Cloud Storage bucket by examining its contents:",
"_____no_output_____"
]
],
[
[
"! gsutil ls -al $BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"### Set up variables\n\nNext, set up some variables used throughout the tutorial.",
"_____no_output_____"
],
[
"#### Import Vertex SDK for Python\n\nImport the Vertex SDK for Python into your Python environment and initialize it.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\nfrom google.cloud import aiplatform\nfrom google.cloud.aiplatform import gapic as aip\n\naiplatform.init(project=PROJECT_ID, location=REGION, staging_bucket=BUCKET_NAME)",
"_____no_output_____"
]
],
[
[
"#### Set hardware accelerators\n\nYou can set hardware accelerators for both training and prediction.\n\nSet the variables `TRAIN_GPU/TRAIN_NGPU` and `DEPLOY_GPU/DEPLOY_NGPU` to use a container image supporting a GPU and the number of GPUs allocated to the virtual machine (VM) instance. For example, to use a GPU container image with 4 Nvidia Tesla K80 GPUs allocated to each VM, you would specify:\n\n (aip.AcceleratorType.NVIDIA_TESLA_K80, 4)\n\nSee the [locations where accelerators are available](https://cloud.google.com/vertex-ai/docs/general/locations#accelerators).\n\nOtherwise specify `(None, None)` to use a container image to run on a CPU.\n\n*Note*: TensorFlow releases earlier than 2.3 for GPU support fail to load the custom model in this tutorial. This issue is caused by static graph operations that are generated in the serving function. This is a known issue, which is fixed in TensorFlow 2.3. If you encounter this issue with your own custom models, use a container image for TensorFlow 2.3 or later with GPU support.",
"_____no_output_____"
]
],
[
[
"TRAIN_GPU, TRAIN_NGPU = (aip.AcceleratorType.NVIDIA_TESLA_K80, 1)\n\nDEPLOY_GPU, DEPLOY_NGPU = (aip.AcceleratorType.NVIDIA_TESLA_K80, 1)",
"_____no_output_____"
]
],
[
[
"#### Set pre-built containers\n\nVertex AI provides pre-built containers to run training and prediction.\n\nFor the latest list, see [Pre-built containers for training](https://cloud.google.com/vertex-ai/docs/training/pre-built-containers) and [Pre-built containers for prediction](https://cloud.google.com/vertex-ai/docs/predictions/pre-built-containers)",
"_____no_output_____"
]
],
[
[
"TRAIN_VERSION = \"tf-gpu.2-1\"\nDEPLOY_VERSION = \"tf2-gpu.2-1\"\n\nTRAIN_IMAGE = \"gcr.io/cloud-aiplatform/training/{}:latest\".format(TRAIN_VERSION)\nDEPLOY_IMAGE = \"gcr.io/cloud-aiplatform/prediction/{}:latest\".format(DEPLOY_VERSION)\n\nprint(\"Training:\", TRAIN_IMAGE, TRAIN_GPU, TRAIN_NGPU)\nprint(\"Deployment:\", DEPLOY_IMAGE, DEPLOY_GPU, DEPLOY_NGPU)",
"Training: gcr.io/cloud-aiplatform/training/tf-gpu.2-1:latest AcceleratorType.NVIDIA_TESLA_K80 1\nDeployment: gcr.io/cloud-aiplatform/prediction/tf2-gpu.2-1:latest AcceleratorType.NVIDIA_TESLA_K80 1\n"
]
],
[
[
"#### Set machine types\n\nNext, set the machine types to use for training and prediction.\n\n- Set the variables `TRAIN_COMPUTE` and `DEPLOY_COMPUTE` to configure your compute resources for training and prediction.\n - `machine type`\n - `n1-standard`: 3.75GB of memory per vCPU\n - `n1-highmem`: 6.5GB of memory per vCPU\n - `n1-highcpu`: 0.9 GB of memory per vCPU\n - `vCPUs`: number of \\[2, 4, 8, 16, 32, 64, 96 \\]\n\n*Note: The following is not supported for training:*\n\n - `standard`: 2 vCPUs\n - `highcpu`: 2, 4 and 8 vCPUs\n\n*Note: You may also use n2 and e2 machine types for training and deployment, but they do not support GPUs*.",
"_____no_output_____"
]
],
[
[
"MACHINE_TYPE = \"n1-standard\"\n\nVCPU = \"4\"\nTRAIN_COMPUTE = MACHINE_TYPE + \"-\" + VCPU\nprint(\"Train machine type\", TRAIN_COMPUTE)\n\nMACHINE_TYPE = \"n1-standard\"\n\nVCPU = \"4\"\nDEPLOY_COMPUTE = MACHINE_TYPE + \"-\" + VCPU\nprint(\"Deploy machine type\", DEPLOY_COMPUTE)",
"Train machine type n1-standard-4\nDeploy machine type n1-standard-4\n"
]
],
[
[
"# Tutorial\n\nNow you are ready to start creating your own custom-trained model with CIFAR10.",
"_____no_output_____"
],
[
"## Train a model\n\nThere are two ways you can train a custom model using a container image:\n\n- **Use a Google Cloud prebuilt container**. If you use a prebuilt container, you will additionally specify a Python package to install into the container image. This Python package contains your code for training a custom model.\n\n- **Use your own custom container image**. If you use your own container, the container needs to contain your code for training a custom model.",
"_____no_output_____"
],
[
"### Define the command args for the training script\n\nPrepare the command-line arguments to pass to your training script.\n- `args`: The command line arguments to pass to the corresponding Python module. In this example, they will be:\n - `\"--epochs=\" + EPOCHS`: The number of epochs for training.\n - `\"--steps=\" + STEPS`: The number of steps (batches) per epoch.\n - `\"--distribute=\" + TRAIN_STRATEGY\"` : The training distribution strategy to use for single or distributed training.\n - `\"single\"`: single device.\n - `\"mirror\"`: all GPU devices on a single compute instance.\n - `\"multi\"`: all GPU devices on all compute instances.",
"_____no_output_____"
]
],
[
[
"JOB_NAME = \"custom_job_\" + TIMESTAMP\nMODEL_DIR = \"{}/{}\".format(BUCKET_NAME, JOB_NAME)\n\nif not TRAIN_NGPU or TRAIN_NGPU < 2:\n TRAIN_STRATEGY = \"single\"\nelse:\n TRAIN_STRATEGY = \"mirror\"\n\nEPOCHS = 20\nSTEPS = 100\n\nCMDARGS = [\n \"--epochs=\" + str(EPOCHS),\n \"--steps=\" + str(STEPS),\n \"--distribute=\" + TRAIN_STRATEGY,\n]",
"_____no_output_____"
]
],
[
[
"#### Training script\n\nIn the next cell, you will write the contents of the training script, `task.py`. In summary:\n\n- Get the directory where to save the model artifacts from the environment variable `AIP_MODEL_DIR`. This variable is set by the training service.\n- Loads CIFAR10 dataset from TF Datasets (tfds).\n- Builds a model using TF.Keras model API.\n- Compiles the model (`compile()`).\n- Sets a training distribution strategy according to the argument `args.distribute`.\n- Trains the model (`fit()`) with epochs and steps according to the arguments `args.epochs` and `args.steps`\n- Saves the trained model (`save(MODEL_DIR)`) to the specified model directory.",
"_____no_output_____"
]
],
[
[
"%%writefile task.py\n# Single, Mirror and Multi-Machine Distributed Training for CIFAR-10\n\nimport tensorflow_datasets as tfds\nimport tensorflow as tf\nfrom tensorflow.python.client import device_lib\nimport argparse\nimport os\nimport sys\ntfds.disable_progress_bar()\n\nparser = argparse.ArgumentParser()\nparser.add_argument('--lr', dest='lr',\n default=0.01, type=float,\n help='Learning rate.')\nparser.add_argument('--epochs', dest='epochs',\n default=10, type=int,\n help='Number of epochs.')\nparser.add_argument('--steps', dest='steps',\n default=200, type=int,\n help='Number of steps per epoch.')\nparser.add_argument('--distribute', dest='distribute', type=str, default='single',\n help='distributed training strategy')\nargs = parser.parse_args()\n\nprint('Python Version = {}'.format(sys.version))\nprint('TensorFlow Version = {}'.format(tf.__version__))\nprint('TF_CONFIG = {}'.format(os.environ.get('TF_CONFIG', 'Not found')))\nprint('DEVICES', device_lib.list_local_devices())\n\n# Single Machine, single compute device\nif args.distribute == 'single':\n if tf.test.is_gpu_available():\n strategy = tf.distribute.OneDeviceStrategy(device=\"/gpu:0\")\n else:\n strategy = tf.distribute.OneDeviceStrategy(device=\"/cpu:0\")\n# Single Machine, multiple compute device\nelif args.distribute == 'mirror':\n strategy = tf.distribute.MirroredStrategy()\n# Multiple Machine, multiple compute device\nelif args.distribute == 'multi':\n strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy()\n\n# Multi-worker configuration\nprint('num_replicas_in_sync = {}'.format(strategy.num_replicas_in_sync))\n\n# Preparing dataset\nBUFFER_SIZE = 10000\nBATCH_SIZE = 64\n\ndef make_datasets_unbatched():\n # Scaling CIFAR10 data from (0, 255] to (0., 1.]\n def scale(image, label):\n image = tf.cast(image, tf.float32)\n image /= 255.0\n return image, label\n\n datasets, info = tfds.load(name='cifar10',\n with_info=True,\n as_supervised=True)\n return datasets['train'].map(scale).cache().shuffle(BUFFER_SIZE).repeat()\n\n\n# Build the Keras model\ndef build_and_compile_cnn_model():\n model = tf.keras.Sequential([\n tf.keras.layers.Conv2D(32, 3, activation='relu', input_shape=(32, 32, 3)),\n tf.keras.layers.MaxPooling2D(),\n tf.keras.layers.Conv2D(32, 3, activation='relu'),\n tf.keras.layers.MaxPooling2D(),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(10, activation='softmax')\n ])\n model.compile(\n loss=tf.keras.losses.sparse_categorical_crossentropy,\n optimizer=tf.keras.optimizers.SGD(learning_rate=args.lr),\n metrics=['accuracy'])\n return model\n\n# Train the model\nNUM_WORKERS = strategy.num_replicas_in_sync\n# Here the batch size scales up by number of workers since\n# `tf.data.Dataset.batch` expects the global batch size.\nGLOBAL_BATCH_SIZE = BATCH_SIZE * NUM_WORKERS\nMODEL_DIR = os.getenv(\"AIP_MODEL_DIR\")\n\ntrain_dataset = make_datasets_unbatched().batch(GLOBAL_BATCH_SIZE)\n\nwith strategy.scope():\n # Creation of dataset, and model building/compiling need to be within\n # `strategy.scope()`.\n model = build_and_compile_cnn_model()\n\nmodel.fit(x=train_dataset, epochs=args.epochs, steps_per_epoch=args.steps)\nmodel.save(MODEL_DIR)",
"Writing task.py\n"
]
],
[
[
"### Train the model\n\nDefine your custom training job on Vertex AI.\n\nUse the `CustomTrainingJob` class to define the job, which takes the following parameters:\n\n- `display_name`: The user-defined name of this training pipeline.\n- `script_path`: The local path to the training script.\n- `container_uri`: The URI of the training container image.\n- `requirements`: The list of Python package dependencies of the script.\n- `model_serving_container_image_uri`: The URI of a container that can serve predictions for your model — either a prebuilt container or a custom container.\n\nUse the `run` function to start training, which takes the following parameters:\n\n- `args`: The command line arguments to be passed to the Python script.\n- `replica_count`: The number of worker replicas.\n- `model_display_name`: The display name of the `Model` if the script produces a managed `Model`.\n- `machine_type`: The type of machine to use for training.\n- `accelerator_type`: The hardware accelerator type.\n- `accelerator_count`: The number of accelerators to attach to a worker replica.\n\nThe `run` function creates a training pipeline that trains and creates a `Model` object. After the training pipeline completes, the `run` function returns the `Model` object.",
"_____no_output_____"
]
],
[
[
"job = aiplatform.CustomTrainingJob(\n display_name=JOB_NAME,\n script_path=\"task.py\",\n container_uri=TRAIN_IMAGE,\n requirements=[\"tensorflow_datasets==1.3.0\"],\n model_serving_container_image_uri=DEPLOY_IMAGE,\n)\n\nMODEL_DISPLAY_NAME = \"cifar10-\" + TIMESTAMP\n\n# Start the training\nif TRAIN_GPU:\n model = job.run(\n model_display_name=MODEL_DISPLAY_NAME,\n args=CMDARGS,\n replica_count=1,\n machine_type=TRAIN_COMPUTE,\n accelerator_type=TRAIN_GPU.name,\n accelerator_count=TRAIN_NGPU,\n )\nelse:\n model = job.run(\n model_display_name=MODEL_DISPLAY_NAME,\n args=CMDARGS,\n replica_count=1,\n machine_type=TRAIN_COMPUTE,\n accelerator_count=0,\n )",
"INFO:google.cloud.aiplatform.utils.source_utils:Training script copied to:\ngs://qwiklabs-gcp-04-6c865137f72a/aiplatform-2021-08-26-14:26:46.974-aiplatform_custom_trainer_script-0.1.tar.gz.\nINFO:google.cloud.aiplatform.training_jobs:Training Output directory:\ngs://qwiklabs-gcp-04-6c865137f72a/aiplatform-custom-training-2021-08-26-14:26:47.099 \nINFO:google.cloud.aiplatform.training_jobs:View Training:\nhttps://console.cloud.google.com/ai/platform/locations/us-central1/training/7773250340236820480?project=21844552219\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480 current state:\nPipelineState.PIPELINE_STATE_PENDING\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480 current state:\nPipelineState.PIPELINE_STATE_PENDING\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480 current state:\nPipelineState.PIPELINE_STATE_PENDING\nINFO:google.cloud.aiplatform.training_jobs:View backing custom job:\nhttps://console.cloud.google.com/ai/platform/locations/us-central1/training/7937948385984643072?project=21844552219\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480 current state:\nPipelineState.PIPELINE_STATE_RUNNING\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480 current state:\nPipelineState.PIPELINE_STATE_RUNNING\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480 current state:\nPipelineState.PIPELINE_STATE_RUNNING\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480 current state:\nPipelineState.PIPELINE_STATE_RUNNING\nINFO:google.cloud.aiplatform.training_jobs:CustomTrainingJob run completed. Resource name: projects/21844552219/locations/us-central1/trainingPipelines/7773250340236820480\nINFO:google.cloud.aiplatform.training_jobs:Model available at projects/21844552219/locations/us-central1/models/6908020451084075008\n"
]
],
[
[
"### Deploy the model\n\nBefore you use your model to make predictions, you need to deploy it to an `Endpoint`. You can do this by calling the `deploy` function on the `Model` resource. This will do two things:\n\n1. Create an `Endpoint` resource for deploying the `Model` resource to.\n2. Deploy the `Model` resource to the `Endpoint` resource.\n\n\nThe function takes the following parameters:\n\n- `deployed_model_display_name`: A human readable name for the deployed model.\n- `traffic_split`: Percent of traffic at the endpoint that goes to this model, which is specified as a dictionary of one or more key/value pairs.\n - If only one model, then specify as **{ \"0\": 100 }**, where \"0\" refers to this model being uploaded and 100 means 100% of the traffic.\n - If there are existing models on the endpoint, for which the traffic will be split, then use `model_id` to specify as **{ \"0\": percent, model_id: percent, ... }**, where `model_id` is the model id of an existing model to the deployed endpoint. The percents must add up to 100.\n- `machine_type`: The type of machine to use for training.\n- `accelerator_type`: The hardware accelerator type.\n- `accelerator_count`: The number of accelerators to attach to a worker replica.\n- `starting_replica_count`: The number of compute instances to initially provision.\n- `max_replica_count`: The maximum number of compute instances to scale to. In this tutorial, only one instance is provisioned.\n\n### Traffic split\n\nThe `traffic_split` parameter is specified as a Python dictionary. You can deploy more than one instance of your model to an endpoint, and then set the percentage of traffic that goes to each instance.\n\nYou can use a traffic split to introduce a new model gradually into production. For example, if you had one existing model in production with 100% of the traffic, you could deploy a new model to the same endpoint, direct 10% of traffic to it, and reduce the original model's traffic to 90%. This allows you to monitor the new model's performance while minimizing the disruption to the majority of users.\n\n### Compute instance scaling\n\nYou can specify a single instance (or node) to serve your online prediction requests. This tutorial uses a single node, so the variables `MIN_NODES` and `MAX_NODES` are both set to `1`.\n\nIf you want to use multiple nodes to serve your online prediction requests, set `MAX_NODES` to the maximum number of nodes you want to use. Vertex AI autoscales the number of nodes used to serve your predictions, up to the maximum number you set. Refer to the [pricing page](https://cloud.google.com/vertex-ai/pricing#prediction-prices) to understand the costs of autoscaling with multiple nodes.\n\n### Endpoint\n\nThe method will block until the model is deployed and eventually return an `Endpoint` object. If this is the first time a model is deployed to the endpoint, it may take a few additional minutes to complete provisioning of resources.",
"_____no_output_____"
]
],
[
[
"DEPLOYED_NAME = \"cifar10_deployed-\" + TIMESTAMP\n\nTRAFFIC_SPLIT = {\"0\": 100}\n\nMIN_NODES = 1\nMAX_NODES = 1\n\nif DEPLOY_GPU:\n endpoint = model.deploy(\n deployed_model_display_name=DEPLOYED_NAME,\n traffic_split=TRAFFIC_SPLIT,\n machine_type=DEPLOY_COMPUTE,\n accelerator_type=DEPLOY_GPU.name,\n accelerator_count=DEPLOY_NGPU,\n min_replica_count=MIN_NODES,\n max_replica_count=MAX_NODES,\n )\nelse:\n endpoint = model.deploy(\n deployed_model_display_name=DEPLOYED_NAME,\n traffic_split=TRAFFIC_SPLIT,\n machine_type=DEPLOY_COMPUTE,\n accelerator_type=DEPLOY_COMPUTE.name,\n accelerator_count=0,\n min_replica_count=MIN_NODES,\n max_replica_count=MAX_NODES,\n )",
"INFO:google.cloud.aiplatform.models:Creating Endpoint\nINFO:google.cloud.aiplatform.models:Create Endpoint backing LRO: projects/21844552219/locations/us-central1/endpoints/3755562284575883264/operations/1226649531685273600\nINFO:google.cloud.aiplatform.models:Endpoint created. Resource name: projects/21844552219/locations/us-central1/endpoints/3755562284575883264\nINFO:google.cloud.aiplatform.models:To use this Endpoint in another session:\nINFO:google.cloud.aiplatform.models:endpoint = aiplatform.Endpoint('projects/21844552219/locations/us-central1/endpoints/3755562284575883264')\nINFO:google.cloud.aiplatform.models:Deploying model to Endpoint : projects/21844552219/locations/us-central1/endpoints/3755562284575883264\nINFO:google.cloud.aiplatform.models:Deploy Endpoint model backing LRO: projects/21844552219/locations/us-central1/endpoints/3755562284575883264/operations/7369559423418630144\nINFO:google.cloud.aiplatform.models:Endpoint model deployed. Resource name: projects/21844552219/locations/us-central1/endpoints/3755562284575883264\n"
]
],
[
[
"## Make an online prediction request\n\nSend an online prediction request to your deployed model.",
"_____no_output_____"
],
[
"### Get test data\n\nDownload images from the CIFAR dataset and preprocess them.\n\n#### Download the test images\n\nDownload the provided set of images from the CIFAR dataset:",
"_____no_output_____"
]
],
[
[
"# Download the images\n! gsutil -m cp -r gs://cloud-samples-data/ai-platform-unified/cifar_test_images .",
"_____no_output_____"
]
],
[
[
"#### Preprocess the images\nBefore you can run the data through the endpoint, you need to preprocess it to match the format that your custom model defined in `task.py` expects.\n\n`x_test`:\nNormalize (rescale) the pixel data by dividing each pixel by 255. This replaces each single byte integer pixel with a 32-bit floating point number between 0 and 1.\n\n`y_test`:\nYou can extract the labels from the image filenames. Each image's filename format is \"image_{LABEL}_{IMAGE_NUMBER}.jpg\"",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom PIL import Image\n\n# Load image data\nIMAGE_DIRECTORY = \"cifar_test_images\"\n\nimage_files = [file for file in os.listdir(IMAGE_DIRECTORY) if file.endswith(\".jpg\")]\n\n# Decode JPEG images into numpy arrays\nimage_data = [\n np.asarray(Image.open(os.path.join(IMAGE_DIRECTORY, file))) for file in image_files\n]\n\n# Scale and convert to expected format\nx_test = [(image / 255.0).astype(np.float32).tolist() for image in image_data]\n\n# Extract labels from image name\ny_test = [int(file.split(\"_\")[1]) for file in image_files]",
"_____no_output_____"
]
],
[
[
"### Send the prediction request\n\nNow that you have test images, you can use them to send a prediction request. Use the `Endpoint` object's `predict` function, which takes the following parameters:\n\n- `instances`: A list of image instances. According to your custom model, each image instance should be a 3-dimensional matrix of floats. This was prepared in the previous step.\n\nThe `predict` function returns a list, where each element in the list corresponds to the corresponding image in the request. You will see in the output for each prediction:\n\n- Confidence level for the prediction (`predictions`), between 0 and 1, for each of the ten classes.\n\nYou can then run a quick evaluation on the prediction results:\n1. `np.argmax`: Convert each list of confidence levels to a label\n2. Compare the predicted labels to the actual labels\n3. Calculate `accuracy` as `correct/total`",
"_____no_output_____"
]
],
[
[
"predictions = endpoint.predict(instances=x_test)\ny_predicted = np.argmax(predictions.predictions, axis=1)\n\ncorrect = sum(y_predicted == np.array(y_test))\naccuracy = len(y_predicted)\nprint(\n f\"Correct predictions = {correct}, Total predictions = {accuracy}, Accuracy = {correct/accuracy}\"\n)",
"_____no_output_____"
]
],
[
[
"## Undeploy the model\n\nTo undeploy your `Model` resource from the serving `Endpoint` resource, use the endpoint's `undeploy` method with the following parameter:\n\n- `deployed_model_id`: The model deployment identifier returned by the endpoint service when the `Model` resource was deployed. You can retrieve the deployed models using the endpoint's `deployed_models` property.\n\nSince this is the only deployed model on the `Endpoint` resource, you can omit `traffic_split`.",
"_____no_output_____"
]
],
[
[
"deployed_model_id = endpoint.list_models()[0].id\nendpoint.undeploy(deployed_model_id=deployed_model_id)",
"_____no_output_____"
]
],
[
[
"# Cleaning up\n\nTo clean up all Google Cloud resources used in this project, you can [delete the Google Cloud project](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.\n\nOtherwise, you can delete the individual resources you created in this tutorial:\n\n- Training Job\n- Model\n- Endpoint\n- Cloud Storage Bucket",
"_____no_output_____"
]
],
[
[
"delete_training_job = True\ndelete_model = True\ndelete_endpoint = True\n\n# Warning: Setting this to true will delete everything in your bucket\ndelete_bucket = False\n\n# Delete the training job\njob.delete()\n\n# Delete the model\nmodel.delete()\n\n# Delete the endpoint\nendpoint.delete()\n\nif delete_bucket and \"BUCKET_NAME\" in globals():\n ! gsutil -m rm -r $BUCKET_NAME",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71df99d9d206536305327a1e0690e922668a93b | 239,251 | ipynb | Jupyter Notebook | experiments/tl_1v2/cores-oracle.run1.framed/trials/7/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tl_1v2/cores-oracle.run1.framed/trials/7/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tl_1v2/cores-oracle.run1.framed/trials/7/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | 101.722364 | 73,256 | 0.747048 | [
[
[
"# Transfer Learning Template",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\n \nimport os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom torch.optim import Adam\nfrom easydict import EasyDict\nimport matplotlib.pyplot as plt\n\nfrom steves_models.steves_ptn import Steves_Prototypical_Network\n\nfrom steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper\nfrom steves_utils.iterable_aggregator import Iterable_Aggregator\nfrom steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader\nfrom steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)\nfrom steves_utils.PTN.utils import independent_accuracy_assesment\n\nfrom torch.utils.data import DataLoader\n\nfrom steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory\n\nfrom steves_utils.ptn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.transforms import get_chained_transform",
"_____no_output_____"
]
],
[
[
"# Allowed Parameters\nThese are allowed parameters, not defaults\nEach of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)\n\nPapermill uses the cell tag \"parameters\" to inject the real parameters below this cell.\nEnable tags to see what I mean",
"_____no_output_____"
]
],
[
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"seed\",\n \"dataset_seed\",\n \"n_shot\",\n \"n_query\",\n \"n_way\",\n \"train_k_factor\",\n \"val_k_factor\",\n \"test_k_factor\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"x_net\",\n \"datasets\",\n \"torch_default_dtype\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"x_shape\",\n}",
"_____no_output_____"
],
[
"from steves_utils.CORES.utils import (\n ALL_NODES,\n ALL_NODES_MINIMUM_1000_EXAMPLES,\n ALL_DAYS\n)\n\nfrom steves_utils.ORACLE.utils_v2 import (\n ALL_DISTANCES_FEET_NARROWED,\n ALL_RUNS,\n ALL_SERIAL_NUMBERS,\n)\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"STANDALONE PTN\"\nstandalone_parameters[\"lr\"] = 0.001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"dataset_seed\"] = 1337\n\nstandalone_parameters[\"n_way\"] = 8\nstandalone_parameters[\"n_shot\"] = 3\nstandalone_parameters[\"n_query\"] = 2\nstandalone_parameters[\"train_k_factor\"] = 1\nstandalone_parameters[\"val_k_factor\"] = 2\nstandalone_parameters[\"test_k_factor\"] = 2\n\n\nstandalone_parameters[\"n_epoch\"] = 50\n\nstandalone_parameters[\"patience\"] = 10\nstandalone_parameters[\"criteria_for_best\"] = \"source_loss\"\n\nstandalone_parameters[\"datasets\"] = [\n {\n \"labels\": ALL_SERIAL_NUMBERS,\n \"domains\": ALL_DISTANCES_FEET_NARROWED,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\", \"minus_two\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE_\"\n },\n {\n \"labels\": ALL_NODES,\n \"domains\": ALL_DAYS,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_power\", \"times_zero\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"CORES_\"\n } \n]\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\n\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n]\n\n# Parameters relevant to results\n# These parameters will basically never need to change\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"\n\n\n\n\n",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"tl_1v2:cores-oracle.run1.framed\",\n \"device\": \"cuda\",\n \"lr\": 0.0001,\n \"n_shot\": 3,\n \"n_query\": 2,\n \"train_k_factor\": 3,\n \"val_k_factor\": 2,\n \"test_k_factor\": 2,\n \"torch_default_dtype\": \"torch.float32\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_accuracy\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 256]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 20480, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"n_way\": 16,\n \"datasets\": [\n {\n \"labels\": [\n \"1-10.\",\n \"1-11.\",\n \"1-15.\",\n \"1-16.\",\n \"1-17.\",\n \"1-18.\",\n \"1-19.\",\n \"10-4.\",\n \"10-7.\",\n \"11-1.\",\n \"11-14.\",\n \"11-17.\",\n \"11-20.\",\n \"11-7.\",\n \"13-20.\",\n \"13-8.\",\n \"14-10.\",\n \"14-11.\",\n \"14-14.\",\n \"14-7.\",\n \"15-1.\",\n \"15-20.\",\n \"16-1.\",\n \"16-16.\",\n \"17-10.\",\n \"17-11.\",\n \"17-2.\",\n \"19-1.\",\n \"19-16.\",\n \"19-19.\",\n \"19-20.\",\n \"19-3.\",\n \"2-10.\",\n \"2-11.\",\n \"2-17.\",\n \"2-18.\",\n \"2-20.\",\n \"2-3.\",\n \"2-4.\",\n \"2-5.\",\n \"2-6.\",\n \"2-7.\",\n \"2-8.\",\n \"3-13.\",\n \"3-18.\",\n \"3-3.\",\n \"4-1.\",\n \"4-10.\",\n \"4-11.\",\n \"4-19.\",\n \"5-5.\",\n \"6-15.\",\n \"7-10.\",\n \"7-14.\",\n \"8-18.\",\n \"8-20.\",\n \"8-3.\",\n \"8-8.\",\n ],\n \"domains\": [1, 2, 3, 4, 5],\n \"num_examples_per_domain_per_label\": -1,\n \"pickle_path\": \"/root/csc500-main/datasets/cores.stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"CORES_\",\n },\n {\n \"labels\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"domains\": [32, 38, 8, 44, 14, 50, 20, 26],\n \"num_examples_per_domain_per_label\": 2000,\n \"pickle_path\": \"/root/csc500-main/datasets/oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_mag\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE.run1_\",\n },\n ],\n \"dataset_seed\": 420,\n \"seed\": 420,\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nif \"x_shape\" not in p:\n p.x_shape = [2,256] # Default to this if we dont supply x_shape\n\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"###########################################\n# The stratified datasets honor this\n###########################################\ntorch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"p.domains_source = []\np.domains_target = []\n\n\ntrain_original_source = []\nval_original_source = []\ntest_original_source = []\n\ntrain_original_target = []\nval_original_target = []\ntest_original_target = []",
"_____no_output_____"
],
[
"# global_x_transform_func = lambda x: normalize(x.to(torch.get_default_dtype()), \"unit_power\") # unit_power, unit_mag\n# global_x_transform_func = lambda x: normalize(x, \"unit_power\") # unit_power, unit_mag",
"_____no_output_____"
],
[
"def add_dataset(\n labels,\n domains,\n pickle_path,\n x_transforms,\n episode_transforms,\n domain_prefix,\n num_examples_per_domain_per_label,\n source_or_target_dataset:str,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n):\n \n if x_transforms == []: x_transform = None\n else: x_transform = get_chained_transform(x_transforms)\n \n if episode_transforms == []: episode_transform = None\n else: raise Exception(\"episode_transforms not implemented\")\n \n episode_transform = lambda tup, _prefix=domain_prefix: (_prefix + str(tup[0]), tup[1])\n\n\n eaf = Episodic_Accessor_Factory(\n labels=labels,\n domains=domains,\n num_examples_per_domain_per_label=num_examples_per_domain_per_label,\n iterator_seed=iterator_seed,\n dataset_seed=dataset_seed,\n n_shot=n_shot,\n n_way=n_way,\n n_query=n_query,\n train_val_test_k_factors=train_val_test_k_factors,\n pickle_path=pickle_path,\n x_transform_func=x_transform,\n )\n\n train, val, test = eaf.get_train(), eaf.get_val(), eaf.get_test()\n train = Lazy_Iterable_Wrapper(train, episode_transform)\n val = Lazy_Iterable_Wrapper(val, episode_transform)\n test = Lazy_Iterable_Wrapper(test, episode_transform)\n\n if source_or_target_dataset==\"source\":\n train_original_source.append(train)\n val_original_source.append(val)\n test_original_source.append(test)\n\n p.domains_source.extend(\n [domain_prefix + str(u) for u in domains]\n )\n elif source_or_target_dataset==\"target\":\n train_original_target.append(train)\n val_original_target.append(val)\n test_original_target.append(test)\n p.domains_target.extend(\n [domain_prefix + str(u) for u in domains]\n )\n else:\n raise Exception(f\"invalid source_or_target_dataset: {source_or_target_dataset}\")\n ",
"_____no_output_____"
],
[
"for ds in p.datasets:\n add_dataset(**ds)",
"_____no_output_____"
],
[
"# from steves_utils.CORES.utils import (\n# ALL_NODES,\n# ALL_NODES_MINIMUM_1000_EXAMPLES,\n# ALL_DAYS\n# )\n\n# add_dataset(\n# labels=ALL_NODES,\n# domains = ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"cores_{u}\"\n# )",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle1_{u}\"\n# )\n",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62,56}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle2_{u}\"\n# )",
"_____no_output_____"
],
[
"# add_dataset(\n# labels=list(range(19)),\n# domains = [0,1,2],\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"metehan.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"met_{u}\"\n# )",
"_____no_output_____"
],
[
"# # from steves_utils.wisig.utils import (\n# # ALL_NODES_MINIMUM_100_EXAMPLES,\n# # ALL_NODES_MINIMUM_500_EXAMPLES,\n# # ALL_NODES_MINIMUM_1000_EXAMPLES,\n# # ALL_DAYS\n# # )\n\n# import steves_utils.wisig.utils as wisig\n\n\n# add_dataset(\n# labels=wisig.ALL_NODES_MINIMUM_100_EXAMPLES,\n# domains = wisig.ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"wisig.node3-19.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"wisig_{u}\"\n# )",
"_____no_output_____"
],
[
"###################################\n# Build the dataset\n###################################\ntrain_original_source = Iterable_Aggregator(train_original_source, p.seed)\nval_original_source = Iterable_Aggregator(val_original_source, p.seed)\ntest_original_source = Iterable_Aggregator(test_original_source, p.seed)\n\n\ntrain_original_target = Iterable_Aggregator(train_original_target, p.seed)\nval_original_target = Iterable_Aggregator(val_original_target, p.seed)\ntest_original_target = Iterable_Aggregator(test_original_target, p.seed)\n\n# For CNN We only use X and Y. And we only train on the source.\n# Properly form the data using a transform lambda and Lazy_Iterable_Wrapper. Finally wrap them in a dataloader\n\ntransform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only\n\ntrain_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)\nval_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)\ntest_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)\n\ntrain_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)\nval_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)\ntest_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"from steves_utils.transforms import get_average_magnitude, get_average_power\n\nprint(set([u for u,_ in val_original_source]))\nprint(set([u for u,_ in val_original_target]))\n\ns_x, s_y, q_x, q_y, _ = next(iter(train_processed_source))\nprint(s_x)\n\n# for ds in [\n# train_processed_source,\n# val_processed_source,\n# test_processed_source,\n# train_processed_target,\n# val_processed_target,\n# test_processed_target\n# ]:\n# for s_x, s_y, q_x, q_y, _ in ds:\n# for X in (s_x, q_x):\n# for x in X:\n# assert np.isclose(get_average_magnitude(x.numpy()), 1.0)\n# assert np.isclose(get_average_power(x.numpy()), 1.0)\n ",
"{'CORES_5', 'CORES_3', 'CORES_1', 'CORES_4', 'CORES_2'}\n"
],
[
"###################################\n# Build the model\n###################################\n# easfsl only wants a tuple for the shape\nmodel = Steves_Prototypical_Network(x_net, device=p.device, x_shape=tuple(p.x_shape))\noptimizer = Adam(params=model.parameters(), lr=p.lr)",
"(2, 256)\n"
],
[
"###################################\n# train\n###################################\njig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n patience=p.patience,\n optimizer=optimizer,\n criteria_for_best=p.criteria_for_best,\n)",
"epoch: 1, [batch: 1 / 6315], examples_per_second: 35.1797, train_label_loss: 2.6447, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"###################################\n# Evaluate the model\n###################################\nsource_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))\n\nconfusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": dict(p),\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"ptn\"),\n}",
"_____no_output_____"
],
[
"ax = get_loss_curve(experiment)\nplt.show()",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.9990124153498872 Target Test Label Accuracy: 0.27177734375\nSource Val Label Accuracy: 0.9987142857142857 Target Val Label Accuracy: 0.2718098958333333\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71dfc648ca1c3b9c91d0e8e7f18e37a89994287 | 3,829 | ipynb | Jupyter Notebook | docs/source/simple_integration_tutorial.ipynb | Jedite/CommunicationWithoutMovement | 7de5da4cac5f4ecbd140b2eee32a2244e54981c8 | [
"MIT"
] | 44 | 2020-02-07T15:01:47.000Z | 2022-03-21T14:36:15.000Z | docs/source/simple_integration_tutorial.ipynb | Jedite/CommunicationWithoutMovement | 7de5da4cac5f4ecbd140b2eee32a2244e54981c8 | [
"MIT"
] | 17 | 2020-02-07T17:11:23.000Z | 2022-02-20T18:01:42.000Z | docs/source/simple_integration_tutorial.ipynb | Jedite/CommunicationWithoutMovement | 7de5da4cac5f4ecbd140b2eee32a2244e54981c8 | [
"MIT"
] | 19 | 2020-02-07T17:13:22.000Z | 2022-03-17T01:22:35.000Z | 26.776224 | 304 | 0.615043 | [
[
[
"# Running Custom Presentation\n\nThis tutorial shows how to integerate a custom presentation module (such as the custom presentation module made in the [Simple Presentation Tutorial](simple_presentation_tutorial.ipynb)), with the rest of the BCI run with the `online_bci.py` module. By the end of this tutorial you will know:\n * How to start the mindaffectBCI **without** automatically starting a presentation module\n * How to run your presentation module\n * How to cleanly shutdown the mindaffetBCI when you finish\n\nBefore running this tutorial you should have read [how an evoked bci works](https://mindaffect-bci.readthedocs.io/en/latest/how_an_evoked_bci_works.html) and run through the [Simple Presentation Tutorial](simple_presentation_tutorial.ipynb) to understand how to build a custom presentation module. ",
"_____no_output_____"
]
],
[
[
"# Import the mindaffectBCI decoder and other required modules. \n%load_ext autoreload\n%autoreload 2\n%gui qt\nimport mindaffectBCI.online_bci",
"_____no_output_____"
]
],
[
[
"## Start the EEG processing components\nThat is start the; 1) Hub, 2) Acquisation, 3) Decoder - by starting the `online_bci` script with presentation disabled. \n\nFor more information on these components and why we need them please consult the [Quickstart Tutorial](quickstart.ipynb)",
"_____no_output_____"
]
],
[
[
"# load the config-file to use. Here we use the default noisetag config file:\nconfig = mindaffectBCI.online_bci.load_config(\"noisetag_bci\")",
"_____no_output_____"
]
],
[
[
"Set the Presentation system to `None` and then start the online_bci.",
"_____no_output_____"
]
],
[
[
"config['presentation']=None\n#uncomment the following line to use to the fakedata acquisition\nconfig['acquisition']='fakedata'\nmindaffectBCI.online_bci.run(**config)",
"_____no_output_____"
]
],
[
[
"Check all is running correctly.",
"_____no_output_____"
]
],
[
[
"mindaffectBCI.online_bci.check_is_running()",
"_____no_output_____"
]
],
[
[
"## Run the Custom Presentation\n\nWe run our custom presentation module by first importing and then running it.",
"_____no_output_____"
]
],
[
[
"# import and run the presentation\nfrom mindaffectBCI.examples.presentation import minimal_presentation",
"_____no_output_____"
]
],
[
[
"## Shutdown the decoder",
"_____no_output_____"
]
],
[
[
"mindaffectBCI.online_bci.shutdown()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71e0a845c9dc82d84c595066565830260069314 | 17,052 | ipynb | Jupyter Notebook | Chapter03/Getting_Gene.ipynb | nalomran/Bioinformatics-with-Python-Cookbook-Second-Edition | b008b368e724d1963eadaf43f2f8657575998b47 | [
"MIT"
] | 244 | 2018-06-28T05:05:01.000Z | 2022-03-28T07:59:19.000Z | Chapter03/Getting_Gene.ipynb | nalomran/Bioinformatics-with-Python-Cookbook-Second-Edition | b008b368e724d1963eadaf43f2f8657575998b47 | [
"MIT"
] | 22 | 2019-01-19T08:30:18.000Z | 2021-11-02T20:15:48.000Z | Chapter03/Getting_Gene.ipynb | nalomran/Bioinformatics-with-Python-Cookbook-Second-Edition | b008b368e724d1963eadaf43f2f8657575998b47 | [
"MIT"
] | 178 | 2018-05-12T10:05:29.000Z | 2022-03-31T03:01:59.000Z | 69.317073 | 6,373 | 0.835796 | [
[
[
"import gffutils\nimport gzip\nfrom Bio import Alphabet, Seq, SeqIO",
"_____no_output_____"
]
],
[
[
"## Retrieving data",
"_____no_output_____"
]
],
[
[
"!rm -rf ag.db gambiae.fa.gz 2>/dev/null\n!wget ftp://ftp.vectorbase.org/public_data/organism_data/agambiae/Genome/agambiae.CHROMOSOMES-PEST.AgamP3.fa.gz -O gambiae.fa.gz",
"--2020-12-04 12:07:47-- ftp://ftp.vectorbase.org/public_data/organism_data/agambiae/Genome/agambiae.CHROMOSOMES-PEST.AgamP3.fa.gz\n => ‘gambiae.fa.gz’\nResolving ftp.vectorbase.org (ftp.vectorbase.org)... 129.74.255.228\nConnecting to ftp.vectorbase.org (ftp.vectorbase.org)|129.74.255.228|:21... connected.\nLogging in as anonymous ... Logged in!\n==> SYST ... done. ==> PWD ... done.\n==> TYPE I ... done. ==> CWD (1) /public_data/organism_data/agambiae/Genome ... done.\n==> SIZE agambiae.CHROMOSOMES-PEST.AgamP3.fa.gz ... 81591806\n==> PASV ... done. ==> RETR agambiae.CHROMOSOMES-PEST.AgamP3.fa.gz ... done.\nLength: 81591806 (78M) (unauthoritative)\n\nagambiae.CHROMOSOME 100%[===================>] 77.81M 2.42MB/s in 32s \n\n2020-12-04 12:08:20 (2.41 MB/s) - ‘gambiae.fa.gz’ saved [81591806]\n\n"
],
[
"!rm -f ag.db\n\ndb = gffutils.create_db('https://vectorbase.org/common/downloads/Pre-VEuPathDB%20VectorBase%20files/Anopheles-gambiae-PEST_BASEFEATURES_AgamP4.2.gff3.gz', 'ag.db')",
"_____no_output_____"
]
],
[
[
"# Getting a gene",
"_____no_output_____"
]
],
[
[
"gene_id = 'AGAP004707'",
"_____no_output_____"
],
[
"gene = db[gene_id]",
"_____no_output_____"
],
[
"print(gene)\nprint(gene.seqid, gene.strand)",
"2L\tVectorBase\tgene\t2358158\t2431617\t.\t+\t.\tID=AGAP004707;biotype=protein_coding\n2L +\n"
],
[
"recs = SeqIO.parse(gzip.open('gambiae.fa.gz', 'rt', encoding='utf-8'), 'fasta')\nfor rec in recs:\n print(rec.description)\n if rec.description.split(':')[2] == gene.seqid:\n my_seq = rec.seq\n break\nprint(my_seq.alphabet)",
"chromosome:AgamP3:2L:1:49364325:1 chromosome 2L\nSingleLetterAlphabet()\n"
],
[
"def get_sequence(chrom_seq, CDSs, strand):\n seq = Seq.Seq('', alphabet=Alphabet.IUPAC.unambiguous_dna)\n for CDS in CDSs:\n #FRAME???\n my_cds = Seq.Seq(str(chrom_seq[CDS.start - 1: CDS.end]), alphabet=Alphabet.IUPAC.unambiguous_dna)\n seq += my_cds\n return seq if strand == '+' else seq.reverse_complement()",
"_____no_output_____"
],
[
"mRNAs = db.children(gene, featuretype='mRNA')\nfor mRNA in mRNAs:\n print(mRNA.id)\n if mRNA.id.endswith('RA'):\n break\n\nCDSs = db.children(mRNA, featuretype='CDS', order_by='start')\ngene_seq = get_sequence(my_seq, CDSs, gene.strand)\n\nprint(len(gene_seq), gene_seq)\nprot = gene_seq.translate()\nprint(len(prot), prot)",
"AGAP004707-RA\n6357 ATGACCGAAGACTCCGATTCGATATCTGAGGAAGAACGTAGTTTGTTCCGTCCTTTCACTCGTGAATCATTACAAGCTATCGAAGCACGCATTGCAGATGAAGAAGCCAAACAGCGAGAATTGGAAAGAAAACGAGCTGAGGGGGAGGATGAGGATGAAGGTCCCCAACCGGACCCTACTCTTGAACAGGGTGTACCAGTCCCAGTTCGAATGCAAGGCAGCTTCCCCCCGGAGTTGGCCTCCACGCCTCTCGAGGATATTGACAGTTTCTATTCAAATCAAAGGACATTCGTAGTGATTAGTAAAGGAAAAGATATATTTCGTTTCTCCGCAACTAACGCATTATATGTACTTGATCCGTTTAACCCCATACGCCGCGTAGCTATTTATATTTTAGTACATCCACTGTTTTCACTTTTTATAATAACGACCATTCTTGTTAATTGTATATTGATGATTATGCCTACCACGCCGACAGTCGAATCTACCGAGGTGATATTCACCGGCATCTACACGTTCGAATCAGCTGTAAAAGTGATGGCGCGAGGTTTCATATTACAACCGTTTACTTATCTTAGAGATGCATGGAATTGGTTGGACTTCGTAGTAATAGCATTAGCATATGTAACTATGGGTATAGATTTGGGTAATCTTGCTGCGTTGAGAACATTCAGGGTATTACGAGCTCTTAAAACGGTAGCCATCGTTCCAGGCTTAAAAACCATCGTCGGAGCCGTTATAGAATCCGTAAAGAATCTCAGAGATGTGATAATTTTAACAATGTTTTCGTTATCCGTGTTTGCTTTGATGGGTCTACAAATCTACATGGGAGTACTAACACAAAAGTGCATAAAAGAGTTCCCATTGGATGGTTCCTGGGGTAATCTAACCGACGAAAGCTGGGAGCTGTTCAACAGCAATGACACAAATTGGTTCTATTCCGAGAGTGGCGACATTCCTCTTTGTGGAAACTCATCTGGAGCTGGACAATGTGATGAAGGCTACATTTGTTTACAAGGCTATGGCAAAAATCCAAATTACGGGTATACAAGTTTTGATACATTCGGATGGGCATTCTTGTCTGCCTTTCGTCTAATGACTCAAGATTATTGGGAGAATTTATATCAACTGGTGTTACGATCAGCTGGACCGTGGCACATGCTCTTCTTCATTGTGATTATCTTCTTGGGTTCGTTTTACCTTGTAAATTTAATCTTGGCCATTGTCGCCATGTCGTACGACGAACTCCAGAAGAAGGCCGAAGAGGAAGAGGCCGCCGAGGAAGAAGCACTTCGGGAAGCAGAAGAAGCTGCGGCAGCAAAGGCAGCTAAACTGGAAGCACAACAAGCGGCCGCAGCAGCAGCGGCGAACCCAGAAATCGCTAAAAGCCCTTCGGATTTCTCCTGTCACAGCTATGAGTTGTTTGTCGGACAGGAGAAAGGCAACGACGATAACAATAAGGAGAAAATGTCCATTAGAAGCGAAGGATTGGAGTCGGTGAGCGAAATCACAAGAACAACCGCACCAACAGCTACTGCAGCTGGCACTGCAAAAGCCCGTAAAGTGAGCGCGGCTTCACTTTCATTACCCGGTTCACCATTTAATCTTCGTAGAGGATCTAGAGGATCACACCAGTTCACGATACGTAACGGTAGAGGACGTTTCGTTGGTGTACCTGGTAGCGATAGAAAACCACTGGTACTATCAACATATCTCGATGCACAAGAACACCTGCCATACGCTGATGATTCCAACGCGGTAACGCCGATGTCGGAAGAAAATGGTGCAATCATCGTTCCAGTATACTATGCTAATTTAGGTTCAAGGCACTCGTCGTATACTTCGCATCAGTCGCGTATTTCGTACACATCTCACGGTGACCTGCTCGGGGGCATGACAAAAGAGAGCCGTCTGCGAAATCGATCAGCCCGTAACACTAACCATTCAATTGTACCACCTCCGAATGCTAACAATCTATCCTACGCTGAAACAAACCATAAAGGACAGCGAGATTTCGACTTGACACAGGACTGTACAGACGATGCCGGAAAAATAAAACATAACGACAATCCTTTCATAGAACCTGCTCAAACTCAAACTGTGGTAGATATGAAAGACGTGATGGTGTTAAATGACATTATTGAGCAAGCTGCTGGTCGGCACAGCAGGGCAAGTGATCACGGAGTCTCTGTTTATTACTTCCCCACAGAGGACGACGACGAAGATGGCCCAACGTTTAAGGACAAAGCCCTCGAGTTTCTGATGAAGATGATCGACATTTTCTGTGTGTGGGACTGTTGTTGGGTTTGGCTTAAATTTCAGGAATGGGTTGCTTTTATTGTGTTTGACCCATTTGTAGAGCTATTCATTACGCTCTGCATTGTGGTAAATACACTGTTTATGGCTTTAGATCATCACGATATGGATCCAGATATGGAAAAGGCACTGAAAAGTGGCAACTACTTCTTCACAGCTACATTTGCCATCGAAGCTACAATGAAGCTCATAGCAATGAGCCCCAAATATTACTTTCAAGAGGGTTGGAATATCTTCGATTTTATTATCGTAGCACTGTCTCTGCTAGAATTGGGACTTGAGGGTGTTCAAGGATTGTCAGTATTACGATCGTTCCGTTTGCTAAGAGTTTTCAAGCTGGCAAAATCATGGCCTACATTGAATCTTCTAATTTCCATCATGGGACGTACAGTTGGTGCCCTTGGTAATTTAACCTTCGTCTTATGCATCATCATTTTCATCTTCGCCGTGATGGGGATGCAACTTTTTGGCAAAAACTACACAGATAATGTGGATAGATTCCCCGACCATGATCTGCCAAGATGGAATTTTACAGATTTCATGCATTCCTTCATGATTGTGTTCCGTGTGCTATGCGGAGAATGGATTGAATCAATGTGGGATTGTATGCTTGTCGGTGATGTATCCTGCATACCATTTTTCTTGGCCACTGTAGTGATAGGAAATTTAGTCGTGCTTAACCTTTTCTTAGCCTTGCTTTTGTCAAATTTTGGTTCATCATCCTTGTCTGCACCAACGGCAGATAATGAGACCAACAAGATTGCAGAAGCGTTCAACAGAATATCACGCTTTTCTAACTGGATTAAAATGAATTTAGCAAACGCTCTCAAGTTTGTAAAAAATAAATTAACAAGCCAAATAGCATCCGTTCAACCGACAGGCAAAGGCGTATGTCCATGTATATCTTCAGAGCATGGTGAAAATGAACTGGAACTTACTCCAGACGATATTTTGGCGGATGGACTATTGAAGAAAGGAATCAAAGAGCACAACCAACTGGAAGTAGCGATTGGCGATGGCATGGAATTCACCATTCATGGTGATCTGAAGAACAAAGCAAAAAAGAATAAACAAATCATGAACAACTCTAAGGTGATAGGCAATTCTATTAGTAATCATCAAGATAATAAATTAGATCATGAACTGAATCATAGAGGCGTGTCCTTACAGGACGATGATACTGCTAGTATCAAATCTTATGGCAGTCACAAGAATCGCCCATTTAAGGATGAAAGCCACAAAGGCAGCGCCGAAACGATGGAGGGTGAAGAAAAACGTGATGCCAGCAAGGAGGATCTAGGAATTGACGAAGAACTCGACGACGAAGGCGAGGGAGATGAAGGTCCTCTGGATGGAGAGCTGATTATTCATGCAGAAGAAGACGAAGTGATTGAGGATTCACCGGCGGATTGCTGCCCGGACAACTGCTACAAAAAATTTCCTGTTCTTGCTGGGGATGATGACGCGCCGTTCTGGCAGGGTTGGGGAAATTTACGTCTCAAAACGTTTCAGCTAATAGAAAATAAGTATTTTGAGACAGCTGTAATTACAATGATTCTGCTTAGTAGCTTAGCTTTGGCCCTCGAAGATGTGCATCTTCCACAGCGCCCAATCCTTCAAGATATTCTTTATTACATGGATCGAATTTTCACAGTGATCttttttttAGAGATGTTAATCAAATGGTTAGCTTTAGGTTTTAAAGTATATTTTACAAATGCTTGGTGTTGGCTTGATTTCATTATCGTGATGGTATCTTTGATAAACTTCGTTGCTTCACTTTGTGGAGCTGGTGGTATTCAAGCATTCAAAACCATGCGAACTCTTAGAGCCCTGAGACCACTACGTGCCATGTCCCGTATGCAGGGAATGAGGGTCGTCGTGAATGCGTTGGTTCAAGCTATACCGTCCATCTTCAACGTGCTGCTGGTTTGTTTGATATTCTGGCTAATATTTGCAATCATGGGGGTGCAATTATTTGCTGGCAAATACTTCAAGTGTGTGGATAAAAATAAAACTACATTACCTCACGAAATTATACCGGATGTAAATGCTTGCAAAGCCGAAAACTATTCATGGGAAAATTCACCAATGAACTTCGATCATGTAGGTAAAGCATATTTGTGTCTGTTTCAAGTAGCCACATTCAAAGGATGGATACAAATAATGAACGATGCTATTGATTCTAGAGACGTAAGTTTTGTCGGTAAACAGCCTATACGGGAAACGAATATCTACATGTATCTGTACTTTGTGTTCTTTATTATCTTTGGGTCATTCTTCACGTTGAATCTATTCATTGGTGTTATAATTGACAACTTCAATGAACAGAAAAAGAAAGCTGGTGGATCGCTAGAAATGTTCATGACAGAGGATCAGAAAAAGTACTATAATGCAATGAAAAAAATGGGTTCGAAGAAACCTCTAAAGGCAATTCCTCGTCCAAGGTGGCGGCCTCAAGCAATAGTTTTTGAAATAGTGACGAACAAAAAGTTTGACATGATTATCATGTTGTTCATCGGATTCAATATGTTAACTATGACACTGGACCACTACAAACAATCAGAAACTTTTAGTGCTGTTTTGGATTACTTGAATATGATATTCATCTGCATATTCAGCAGCGAATGTTTAATGAAGATTTTTGCACTTCGTTATCATTACTTTATCGAGCCATGGAATTTGTTTGATTTTGTTGTCGTCATTCTTTCGATTTTGGGCCTTGTTCTAAGTGATATCATTGAAAAATATTTTGTATCTCCCACACTTCTACGAGTCGTGCGAGTGGCAAAAGTGGGCCGAGTATTGCGTTTGGTTAAAGGAGCCAAGGGTATCCGAACGTTGCTGTTTGCATTAGCAATGTCGCTACCTGCACTATTTAACATCTGCTTGTTACTCTTTTTGGTGATGTTTATATTTGCCATTTTTGGAATGTCATTTTTCATGCACGTCAAAGATAAGAGTGGCTTAGATGACGTGTACAATTTTAAAACGTTTGGCCAGAGCATGATTTTACTATTTCAGATGTCAACCTCGGCTGGGTGGGATGGTGTTTTAGATGGTATTATCAATGAAGAAGACTGTCTTCCACCAGACAATGATAAGGGCTATCCGGGAAATTGTGGTTCATCAACAATTGGCATAACGTACTTATTGGCGTATCTTGTAATAAGTTTCCTTATCGTTATTAACATGTACATTGCTGTTATCCTCGAAAACTACTCGCAAGCTACGGAAGATGTTCAAGAAGGCTTAACTGATGACGATTATGATATGTACTACGAAATATGGCAGCAATTCGATCCTGACGGTACACAATACGTTCGATATGATCAGCTATCAGACTTTTTGGATGTGCTGGAACCGCCTCTACAGATTCATAAACCAAATCGTTATAAGATTATTTCGATGGATATTCCGATATGCCGCGGAGATATGATGTTCTGTGTCGATATTCTAGATGCACTAACGAAAGATTTTTTTGCTAGAAAAGGAAATCCTATAGAAGAAACAGCCGAATTAGGTGAAGTTCAACAACGCCCAGACGAAGTTGGTTACGAACCAGTATCATCAACACTTTGGAGGCAGCGTGAAGAGTACTGTGCTCGACTGATACAGCATGCGTGGAAACGCTATAAACAGCGTCACGGAGGCGGAACAGACGCTTCAGGAGATGATCTTGAAATAGATGCCTGTGATAACGGTTGTGGTGGTGGTAATGGCAATGAAAATGATGATAGTGGAGATGGTGCAACAGGTAGTGGTGACAACGGAAGTCAGCATGGTGGTGGCAGCATAAGTGGCGGAGGAGGAACTCCTGGTGGTGGTAAAAGTAAAGGAATTATTGGCAGTACTCAGGCTAACATAGGCATAGTGGATAGTAATATATCACCAAAGGAATCACCGGATAGCATCGGCGATCCCCAAGGTCGTCAGACGGCCGTTCTTGTGGAGAGCGACGGATTTGTGACGAAAAACGGTCACCGTGTCGTCATACACTCTCGATCTCCCAGCATAACATCGCGAACGGCAGATGTCTGA\n2119 MTEDSDSISEEERSLFRPFTRESLQAIEARIADEEAKQRELERKRAEGEDEDEGPQPDPTLEQGVPVPVRMQGSFPPELASTPLEDIDSFYSNQRTFVVISKGKDIFRFSATNALYVLDPFNPIRRVAIYILVHPLFSLFIITTILVNCILMIMPTTPTVESTEVIFTGIYTFESAVKVMARGFILQPFTYLRDAWNWLDFVVIALAYVTMGIDLGNLAALRTFRVLRALKTVAIVPGLKTIVGAVIESVKNLRDVIILTMFSLSVFALMGLQIYMGVLTQKCIKEFPLDGSWGNLTDESWELFNSNDTNWFYSESGDIPLCGNSSGAGQCDEGYICLQGYGKNPNYGYTSFDTFGWAFLSAFRLMTQDYWENLYQLVLRSAGPWHMLFFIVIIFLGSFYLVNLILAIVAMSYDELQKKAEEEEAAEEEALREAEEAAAAKAAKLEAQQAAAAAAANPEIAKSPSDFSCHSYELFVGQEKGNDDNNKEKMSIRSEGLESVSEITRTTAPTATAAGTAKARKVSAASLSLPGSPFNLRRGSRGSHQFTIRNGRGRFVGVPGSDRKPLVLSTYLDAQEHLPYADDSNAVTPMSEENGAIIVPVYYANLGSRHSSYTSHQSRISYTSHGDLLGGMTKESRLRNRSARNTNHSIVPPPNANNLSYAETNHKGQRDFDLTQDCTDDAGKIKHNDNPFIEPAQTQTVVDMKDVMVLNDIIEQAAGRHSRASDHGVSVYYFPTEDDDEDGPTFKDKALEFLMKMIDIFCVWDCCWVWLKFQEWVAFIVFDPFVELFITLCIVVNTLFMALDHHDMDPDMEKALKSGNYFFTATFAIEATMKLIAMSPKYYFQEGWNIFDFIIVALSLLELGLEGVQGLSVLRSFRLLRVFKLAKSWPTLNLLISIMGRTVGALGNLTFVLCIIIFIFAVMGMQLFGKNYTDNVDRFPDHDLPRWNFTDFMHSFMIVFRVLCGEWIESMWDCMLVGDVSCIPFFLATVVIGNLVVLNLFLALLLSNFGSSSLSAPTADNETNKIAEAFNRISRFSNWIKMNLANALKFVKNKLTSQIASVQPTGKGVCPCISSEHGENELELTPDDILADGLLKKGIKEHNQLEVAIGDGMEFTIHGDLKNKAKKNKQIMNNSKVIGNSISNHQDNKLDHELNHRGVSLQDDDTASIKSYGSHKNRPFKDESHKGSAETMEGEEKRDASKEDLGIDEELDDEGEGDEGPLDGELIIHAEEDEVIEDSPADCCPDNCYKKFPVLAGDDDAPFWQGWGNLRLKTFQLIENKYFETAVITMILLSSLALALEDVHLPQRPILQDILYYMDRIFTVIFFLEMLIKWLALGFKVYFTNAWCWLDFIIVMVSLINFVASLCGAGGIQAFKTMRTLRALRPLRAMSRMQGMRVVVNALVQAIPSIFNVLLVCLIFWLIFAIMGVQLFAGKYFKCVDKNKTTLPHEIIPDVNACKAENYSWENSPMNFDHVGKAYLCLFQVATFKGWIQIMNDAIDSRDVSFVGKQPIRETNIYMYLYFVFFIIFGSFFTLNLFIGVIIDNFNEQKKKAGGSLEMFMTEDQKKYYNAMKKMGSKKPLKAIPRPRWRPQAIVFEIVTNKKFDMIIMLFIGFNMLTMTLDHYKQSETFSAVLDYLNMIFICIFSSECLMKIFALRYHYFIEPWNLFDFVVVILSILGLVLSDIIEKYFVSPTLLRVVRVAKVGRVLRLVKGAKGIRTLLFALAMSLPALFNICLLLFLVMFIFAIFGMSFFMHVKDKSGLDDVYNFKTFGQSMILLFQMSTSAGWDGVLDGIINEEDCLPPDNDKGYPGNCGSSTIGITYLLAYLVISFLIVINMYIAVILENYSQATEDVQEGLTDDDYDMYYEIWQQFDPDGTQYVRYDQLSDFLDVLEPPLQIHKPNRYKIISMDIPICRGDMMFCVDILDALTKDFFARKGNPIEETAELGEVQQRPDEVGYEPVSSTLWRQREEYCARLIQHAWKRYKQRHGGGTDASGDDLEIDACDNGCGGGNGNENDDSGDGATGSGDNGSQHGGGSISGGGGTPGGGKSKGIIGSTQANIGIVDSNISPKESPDSIGDPQGRQTAVLVESDGFVTKNGHRVVIHSRSPSITSRTADV*\n"
]
],
[
[
"# Reverse strand",
"_____no_output_____"
]
],
[
[
"reverse_transcript_id = 'AGAP004708-RA'",
"_____no_output_____"
],
[
"reverse_CDSs = db.children(reverse_transcript_id, featuretype='CDS', order_by='start')\nreverse_seq = get_sequence(my_seq, reverse_CDSs, '-')\n\nprint(len(reverse_seq), reverse_seq)\nreverse_prot = reverse_seq.translate()\nprint(len(reverse_prot), reverse_prot)",
"1992 ATGGCTGACTTCGATAGTGCCACTAAATGTATCAGAAACATTGAAAAAGAAATTCTTCTCTTGCAATCCGAAGTTTTGAAGACTCGTGAGGGGCTTGGGCTGGAAGATGATAACGTGGAACTTAAAAAGTTAATGGAGGAAAACACGAGATTAAAGCATCGTTTGGAGATAGTGCAATCGGCTATTGTACAGGAAGGCGGATCAATCGCATCCTCCGATTCTGGCAACCAATCCATTGTTGGCGAACTGCAGCAAGTATTTACCGAAGCCATTCAAAAAGCTTTTCCAAGTGTGTTGGTTGAGGCGGTTATTACTATTTCGTCATCCCCCAAGTTTGGCGATTATCAATGCAATAGTGCTATGCAGATTGCGCAGCATTTGAAGCAGTTATCTGTTAAATCGTCGCCACGTGAAGTGGCCCAAAAACTGGTAGCTGAATTGCAAAAACCAATACCTTGTGTCGATAGATTAGAAATCGCTGGAGCGGGATACGTTAATATTTTCCTGTCTAGATCTTATGGAGAACAACGCATTATGAGCATCTTGAGGCATGGGATTGTGGTACCATTAATAGAAAAGAAACGTGTGATAGTCGATTTTTCCTCGCCTAACGTAGCGAAAGAAATGCATGTCGGTCATTTACGTTCGACCATCATTGGTGATTCAATTTGTCGATTTTTGGAATATCTCGGACACGATGTGCTTCGTATTAACCATATCGGAGACTGGGGAACGCAATTTGGTATGTTAATTGCTCATTTGCAGGACCGTTTCCCTAATTTCCAAACCGAGTCCCCGCCTATCAGCGATTTGCAAGCATTTTACAAGGAGTCAAAGGTCCGATTTGACAGCGATGAAGTATTTAAAAAGCGTGCCTACGAATGTGTAGTCAAACTGCAAAGTGGAGAGCTGAGTTATTTGAAGGCCTGGAATCTAATTTGCGATGTTTCACGCAAAGAATTCCAAACCATCTACAACAGATTGGATGTGAAACTAGTTGAACGTGGTGAATCGTTTTATCAAAGCAGAATGGAAAAAATCGTAGAAGAACTTAAGCAGGATGGGTTCCTTGAAGAAGACGAAGGCCGTCTTATCATGTGGGGCGAAAATCGCGCTGGAATTCCTTTAACCATCGTAAAATCAGACGGAGGATTTACATATGATACTTCGGATATGGCCGCCATCAAACAACGCTTGCAAGAAGAAAAGGCTGATTGGTTGATATATGTAACTGACGCTGGGCAGGCGACTCATTTCCAAACAATTTTTTCTTGTGCAAAACGAGCCAAAATCCTACAAGAGAGCAAACATCGTGTGGATCACGTCGGATTTGGTGTGGTGCTAGGCGAAGATGGTAAAAAATTCAAGACTCGTTCTGGCGATACGGTGAAATTGACAGAACTTCTCAATGAAGGTTTGAGGAGGGCTATGGAAAAACTAGTTCAGAAGGAAAGGAACTTAGTGCTCACACAAGAGGAGCTAGTTGCAGCACAAGAATCAGTCGCCTACGGTTGTATTAAATATGCGGATCTGTCGCATAATCGTAACAACGAATATGTGTTCTCCTTCGATAAGATGCTGGAGGACAAAGGAAATACTGCCGTGTATCTGTTGTATGCCTATACCCGCATTCGTTCTATTGCAAGAAAATGTGGCGGAGATTTTGCAAATGACATGCAAAAGGTGATCGATTCCACAGTTATTAAATTAGATCATGAAAAGGAATGGAAACTCGCCAAGGTGTTGCTTCGTTTTACCGACGTTATGTTATTGATCATGAAAAATCTATCGTTACATCATCTTTGTGAGTTTGTGTACGAAATATGCACTGCTTTTAGTGAGTTTTATGACAGTTGTTATTGCATCGAAAAAAATAAGCAAGGTGAAATTATTACTGTTTATCCCTCTCGCGTCTTGCTATGCGAAGCAACATCAAAGGTGCTGGAAAAATGTTTCGATATTTTAGGACTGAAGCCTGTGCATAAAATATAA\n664 MADFDSATKCIRNIEKEILLLQSEVLKTREGLGLEDDNVELKKLMEENTRLKHRLEIVQSAIVQEGGSIASSDSGNQSIVGELQQVFTEAIQKAFPSVLVEAVITISSSPKFGDYQCNSAMQIAQHLKQLSVKSSPREVAQKLVAELQKPIPCVDRLEIAGAGYVNIFLSRSYGEQRIMSILRHGIVVPLIEKKRVIVDFSSPNVAKEMHVGHLRSTIIGDSICRFLEYLGHDVLRINHIGDWGTQFGMLIAHLQDRFPNFQTESPPISDLQAFYKESKVRFDSDEVFKKRAYECVVKLQSGELSYLKAWNLICDVSRKEFQTIYNRLDVKLVERGESFYQSRMEKIVEELKQDGFLEEDEGRLIMWGENRAGIPLTIVKSDGGFTYDTSDMAAIKQRLQEEKADWLIYVTDAGQATHFQTIFSCAKRAKILQESKHRVDHVGFGVVLGEDGKKFKTRSGDTVKLTELLNEGLRRAMEKLVQKERNLVLTQEELVAAQESVAYGCIKYADLSHNRNNEYVFSFDKMLEDKGNTAVYLLYAYTRIRSIARKCGGDFANDMQKVIDSTVIKLDHEKEWKLAKVLLRFTDVMLLIMKNLSLHHLCEFVYEICTAFSEFYDSCYCIEKNKQGEIITVYPSRVLLCEATSKVLEKCFDILGLKPVHKI*\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e71e1007e3ba5da82b006d68c577bcfe9d3b36b9 | 3,598 | ipynb | Jupyter Notebook | notebook/_Avaliar/Cross_NaiveBayes.ipynb | victorblois/py_datascience | bf5d05a0a4d9e26f55f9259d973f7f4f54432e24 | [
"MIT"
] | 1 | 2020-05-11T22:22:55.000Z | 2020-05-11T22:22:55.000Z | notebook/_Avaliar/Cross_NaiveBayes.ipynb | victorblois/py_datascience | bf5d05a0a4d9e26f55f9259d973f7f4f54432e24 | [
"MIT"
] | null | null | null | notebook/_Avaliar/Cross_NaiveBayes.ipynb | victorblois/py_datascience | bf5d05a0a4d9e26f55f9259d973f7f4f54432e24 | [
"MIT"
] | null | null | null | 20.918605 | 81 | 0.532518 | [
[
[
"import pandas as pd\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"import os.path\ndef path_base(base_name):\n current_dir = os.path.abspath(os.path.join(os.getcwd(), os.pardir))\n print(current_dir)\n data_dir = current_dir.replace('notebook','data')\n print(data_dir)\n data_base = data_dir + '\\\\' + base_name\n print(data_base)\n return data_base",
"_____no_output_____"
],
[
"base = pd.read_csv(path_base('db_dados_credito.csv'))",
"C:\\MyPhyton\\DataScience\\notebook\nC:\\MyPhyton\\DataScience\\data\nC:\\MyPhyton\\DataScience\\data\\db_dados_credito.csv\n"
],
[
"base.loc[base.age < 0, 'age'] = 40.92",
"_____no_output_____"
],
[
"previsores = base.iloc[:, 1:4]\nclasse = base.iloc[:, 4]",
"_____no_output_____"
],
[
"import numpy as np\nfrom sklearn.impute import SimpleImputer\nimputer = SimpleImputer(missing_values=np.nan, strategy='mean')\n",
"_____no_output_____"
],
[
"imputer = imputer.fit(previsores.iloc[:, 0:3])",
"_____no_output_____"
],
[
"previsores.iloc[:, 0:3] = imputer.transform(previsores.iloc[:,0:3])",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\nprevisores = scaler.fit_transform(previsores)",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\nfrom sklearn.naive_bayes import GaussianNB\nclassificador = GaussianNB()",
"_____no_output_____"
],
[
"resultados = cross_val_score(classificador, previsores, classe, cv = 10)\nprint(resultados.mean())\nprint(resultados.std())",
"0.924\n0.020223748416156664\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71e126494a37d5a784b2dacda5dd6b4ab5cc759 | 124,649 | ipynb | Jupyter Notebook | Pandas/5. Group By DataFrame.ipynb | Shaon2221/Learning-and-Experimenting_Data-Science | 817a402158c1cf5d77ce2ea92b3e91470851deec | [
"MIT"
] | 15 | 2020-08-20T17:45:12.000Z | 2022-03-08T20:06:49.000Z | Pandas/5. Group By DataFrame.ipynb | Shaon2221/Learning-and-Experiment_Data-Science | 7effb0b624ab478f33e1c688ca90319001555adb | [
"MIT"
] | null | null | null | Pandas/5. Group By DataFrame.ipynb | Shaon2221/Learning-and-Experiment_Data-Science | 7effb0b624ab478f33e1c688ca90319001555adb | [
"MIT"
] | 5 | 2020-08-20T18:41:49.000Z | 2020-09-03T09:10:24.000Z | 160.837419 | 63,132 | 0.858699 | [
[
[
"# groupby DataFrame",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.read_csv(\"weather_by_cities.csv\")\ndf",
"_____no_output_____"
],
[
"g = df.groupby(\"city\")\ng",
"_____no_output_____"
],
[
"from IPython.display import Image\nImage(filename='split_apply_combine.png') # DataFrameGroupBy object looks something like below",
"_____no_output_____"
],
[
"for city, data in g: #This is similar to SQL, SELECT * from weather_data GROUP BY city\n print(\"city:\",city)\n print(\"\\n\")\n print(\"data:\",data)",
"city: mumbai\n\n\ndata: day city temperature windspeed event\n4 1/1/2017 mumbai 90 5 Sunny\n5 1/2/2017 mumbai 85 12 Fog\n6 1/3/2017 mumbai 87 15 Fog\n7 1/4/2017 mumbai 92 5 Rain\ncity: new york\n\n\ndata: day city temperature windspeed event\n0 1/1/2017 new york 32 6 Rain\n1 1/2/2017 new york 36 7 Sunny\n2 1/3/2017 new york 28 12 Snow\n3 1/4/2017 new york 33 7 Sunny\ncity: paris\n\n\ndata: day city temperature windspeed event\n8 1/1/2017 paris 45 20 Sunny\n9 1/2/2017 paris 50 13 Cloudy\n10 1/3/2017 paris 54 8 Cloudy\n11 1/4/2017 paris 42 10 Cloudy\n"
],
[
"g.get_group('mumbai') # this function will return a dataframe cotaining mumbai;s cities information",
"_____no_output_____"
],
[
"g.describe()",
"_____no_output_____"
],
[
"g.size()",
"_____no_output_____"
],
[
"g.count()",
"_____no_output_____"
],
[
"%matplotlib inline\ng.plot()",
"_____no_output_____"
]
],
[
[
"### Custom grouping function and pass that to groupby",
"_____no_output_____"
]
],
[
[
"def grouper(df, idx, col):\n if 80 <= df[col].loc[idx] <= 90:\n return '80-90'\n elif 50 <= df[col].loc[idx] <= 60:\n return '50-60'\n else:\n return 'others'",
"_____no_output_____"
],
[
"g = df.groupby(lambda x: grouper(df, x, 'temperature'))\ng",
"_____no_output_____"
],
[
"for key, d in g:\n print(\"Group by Key: {}\\n\".format(key))\n print(d)",
"Group by Key: 50-60\n\n day city temperature windspeed event\n9 1/2/2017 paris 50 13 Cloudy\n10 1/3/2017 paris 54 8 Cloudy\nGroup by Key: 80-90\n\n day city temperature windspeed event\n4 1/1/2017 mumbai 90 5 Sunny\n5 1/2/2017 mumbai 85 12 Fog\n6 1/3/2017 mumbai 87 15 Fog\nGroup by Key: others\n\n day city temperature windspeed event\n0 1/1/2017 new york 32 6 Rain\n1 1/2/2017 new york 36 7 Sunny\n2 1/3/2017 new york 28 12 Snow\n3 1/4/2017 new york 33 7 Sunny\n7 1/4/2017 mumbai 92 5 Rain\n8 1/1/2017 paris 45 20 Sunny\n11 1/4/2017 paris 42 10 Cloudy\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e71e138db421361951b9eecab3895b3f66b5ffe8 | 51,716 | ipynb | Jupyter Notebook | notebooks/FluxPDF Example.ipynb | TGarfield17/FIRESONG | ad9e3688ed88563cfdb81b9f25aaa63850cc99f9 | [
"BSD-3-Clause"
] | null | null | null | notebooks/FluxPDF Example.ipynb | TGarfield17/FIRESONG | ad9e3688ed88563cfdb81b9f25aaa63850cc99f9 | [
"BSD-3-Clause"
] | null | null | null | notebooks/FluxPDF Example.ipynb | TGarfield17/FIRESONG | ad9e3688ed88563cfdb81b9f25aaa63850cc99f9 | [
"BSD-3-Clause"
] | null | null | null | 242.798122 | 26,664 | 0.911265 | [
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib as mpl\nimport matplotlib.pyplot as plt\nfrom matplotlib.gridspec import GridSpec\nimport numpy as np\n\nfrom firesong.FluxPDF import flux_pdf\nfrom firesong.Firesong import firesong_simulation\n\nmpl.style.use('./firesong_style.mplstyle')",
"_____no_output_____"
]
],
[
[
"## FluxPDF\nWhen simulating very large densities, it is cumbersome to look at the specifics of each source. However, it is still possible to extract some of the most important distributions from an observational perspective: for example, the $\\log(N) - \\log(S)$ distribution which describes the number of sources as a function of the flux on Earth.",
"_____no_output_____"
],
[
"First, we show how the two approaches – `Firesong.py` and `FluxPDF.py` – recover the same results for the same sets of physics parameters:",
"_____no_output_____"
]
],
[
[
"min_log_flux = -20.\nmax_log_flux = 5.\nnbins = 200\n\nfiresong_res = firesong_simulation(None, filename=None, \n density=1e-9,\n Evolution='MD2014SFR',\n verbose=False)\n\nflux_res = flux_pdf(None, filename=None,\n density=1e-9, \n logFMin=min_log_flux,\n logFMax=max_log_flux,\n nFluxBins=nbins,\n Evolution='MD2014SFR',\n verbose=False)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\n\nplt.plot(flux_res[0], flux_res[1], lw=4., ls='-', \n label=\"FluxPDF\")\nplt.hist(np.log10(firesong_res['sources']['flux']), \n bins=np.linspace(min_log_flux, max_log_flux, nbins + 1),\n label='Firesong', histtype='stepfilled', alpha=0.4)\n\nplt.xlabel(r'$\\log_{10}(\\phi)$ (a.u.)')\nplt.ylabel(r'$N$')\nplt.xlim(-14., -6.)\nplt.legend(loc=1, frameon=False)\nfig.set_facecolor('w')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Large density $\\log(N) - \\log(S)$\n\nWe now show the $\\log(N) - \\log(S)$ distributions that we get for a variety of densities, including large densities that would induce too large a memory requirement if using `Firesong`",
"_____no_output_____"
]
],
[
[
"min_log_flux = -20.\nmax_log_flux = 5.\nnbins = 150\n\nlow_density = 1e-10\nmed_density = 1e-7\nhigh_density = 1e-4\n\npdf_dict = dict()\n\nfor density in [low_density, med_density, high_density]:\n pdf_dict[density] = flux_pdf(None, filename=None,\n density=density, \n logFMin=min_log_flux,\n logFMax=max_log_flux,\n nFluxBins=nbins,\n Evolution='MD2014SFR',\n verbose=False)",
"_____no_output_____"
],
[
"for density, label in [(low_density, 'Low density'), \n (med_density, 'Medium density'), \n (high_density, 'High density')]:\n plt.plot(pdf_dict[density][0], np.cumsum(pdf_dict[density][1][::-1])[::-1],\n label = label)\n \nplt.xlabel(r'$\\log_{10}(\\phi)$')\nplt.ylabel(r'$N_{\\mathrm{sources}} > \\log_{10}(\\phi)$')\nplt.yscale('log')\nplt.legend(loc=1, frameon=False)\nplt.ylim(1e0, plt.ylim()[1])\nplt.xlim(-18, -5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"This plot shows the number of sources above a given flux threshold for three different densities, calculated by integrating the results from a `FluxPDF`.",
"_____no_output_____"
],
[
"Just as in `Firesong.py`, you can pass a variety of arguments to `FluxPDF`, including:\n* `Evolution`\n* `luminosity`\n* `density`\n* Transient vs. steady sources (transient sources have an associated `timescale`)\n\nThis list is not exhaustive. Please see the [API documentation](https://icecube.github.io/FIRESONG/) for the complete list of arguments.",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e71e1505b9c0006f104149b11053fca588ac4dce | 173,137 | ipynb | Jupyter Notebook | notebooks/Chebyshev.ipynb | EiffL/SSSOB | 45556c3baaf7384deb5cf8e51cdb7398d49579bc | [
"MIT"
] | null | null | null | notebooks/Chebyshev.ipynb | EiffL/SSSOB | 45556c3baaf7384deb5cf8e51cdb7398d49579bc | [
"MIT"
] | null | null | null | notebooks/Chebyshev.ipynb | EiffL/SSSOB | 45556c3baaf7384deb5cf8e51cdb7398d49579bc | [
"MIT"
] | null | null | null | 314.794545 | 34,144 | 0.931095 | [
[
[
"# Compute the log determinant and derivatives using Chebyshev expansion",
"_____no_output_____"
]
],
[
[
"%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"## Setting up the problem",
"_____no_output_____"
]
],
[
[
"import george\nfrom george import kernels\nimport numpy as np\nimport matplotlib.pyplot as pl\n\n\nnp.random.seed(1234)\nx = 10 * np.sort(np.random.rand(15))\nyerr = 0.2 * np.ones_like(x)\ny = np.sin(x) + yerr * np.random.randn(len(x))\n\npl.errorbar(x, y, yerr=yerr, fmt=\".k\", capsize=0)\npl.xlim(0, 10)\npl.ylim(-1.45, 1.45)\npl.xlabel(\"x\")\npl.ylabel(\"y\");",
"_____no_output_____"
]
],
[
[
"The fun part is here, choosing a particular model for the Kernel:",
"_____no_output_____"
]
],
[
[
"kernel = np.var(y)*kernels.ExpSquaredKernel(0.2)\n\ngp = george.GP(kernel, white_noise=0.2, fit_white_noise=True)\ngp.compute(x, yerr)",
"_____no_output_____"
],
[
"x_pred = np.linspace(0, 10, 500)\npred, pred_var = gp.predict(y, x_pred, return_var=True)\n\npl.fill_between(x_pred, pred - np.sqrt(pred_var), pred + np.sqrt(pred_var),\n color=\"k\", alpha=0.2)\npl.plot(x_pred, pred, \"k\", lw=1.5, alpha=0.5)\npl.errorbar(x, y, yerr=yerr, fmt=\".k\", capsize=0)\npl.plot(x_pred, np.sin(x_pred), \"--g\")\npl.xlim(0, 10)\npl.ylim(-1.45, 1.45)\npl.xlabel(\"x\")\npl.ylabel(\"y\");",
"_____no_output_____"
],
[
"from scipy.optimize import minimize\n\ndef neg_ln_like(p):\n gp.set_parameter_vector(p)\n return -gp.log_likelihood(y)\n\ndef grad_neg_ln_like(p):\n gp.set_parameter_vector(p)\n return -gp.grad_log_likelihood(y)\n\nresult = minimize(neg_ln_like, gp.get_parameter_vector(), jac=grad_neg_ln_like)\nprint(result)\n\ngp.set_parameter_vector(result.x)\nprint(\"\\nFinal ln-likelihood: {0:.2f}\".format(gp.log_likelihood(y)))",
" fun: 8.88350593800224\n hess_inv: array([[2.10275548, 0.03215612, 0.06337494],\n [0.03215612, 0.66360006, 0.42710708],\n [0.06337494, 0.42710708, 0.54832226]])\n jac: array([-1.32238738e-06, 4.50198803e-07, -1.52222154e-06])\n message: 'Optimization terminated successfully.'\n nfev: 14\n nit: 11\n njev: 14\n status: 0\n success: True\n x: array([-4.0135827 , -0.44568543, 0.66934464])\n\nFinal ln-likelihood: -8.88\n"
],
[
"gp.get_parameter_names()",
"_____no_output_____"
],
[
"x_pred = np.linspace(0, 10, 500)\npred, pred_var = gp.predict(y, x_pred, return_var=True)\n\npl.fill_between(x_pred, pred - np.sqrt(pred_var), pred + np.sqrt(pred_var),\n color=\"k\", alpha=0.2)\npl.plot(x_pred, pred, \"k\", lw=1.5, alpha=0.5)\npl.errorbar(x, y, yerr=yerr, fmt=\".k\", capsize=0)\npl.plot(x_pred, np.sin(x_pred), \"--g\")\npl.xlim(0, 10)\npl.ylim(-1.45, 1.45)\npl.xlabel(\"x\")\npl.ylabel(\"y\");",
"_____no_output_____"
]
],
[
[
"## Computing the gradients stochastically",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.insert(0,'..')",
"_____no_output_____"
],
[
"from sssob.chebyshev import chebyshev_logdet",
"_____no_output_____"
],
[
"# Step I: We extract the Kernel and it's derivatives from the kernel object\n# to define Matrix- vector-product operations\nM = kernel.get_value(atleast_2d(x).T) + np.exp(2*gp.get_parameter('white_noise:value')) * eye(len(x))\ndef mvp(x):\n return M.dot(x)\n\nMg = kernel.get_gradient(atleast_2d(x).T)\ndef gmvp(x):\n return np.einsum('ijk,jk->ik', Mg, x)",
"_____no_output_____"
],
[
"# Step II: Compute the log det and its derivatives using the Chebyshev expansion \nlog_det, glog_det = chebyshev_logdet(mvp, gmvp, shape=(15,1), m=10, deg=100, g=1.1, eps=1e-3)",
"_____no_output_____"
],
[
"hist(glog_det[:,0],64,alpha=0.75, range=[-50,50]);\naxvline(mean(glog_det[:,0]),color='cyan', label=\"est grad theta1\");\naxvline(trace(pinv(M).dot(Mg[:,:,0])),ls='--',color='blue', label=\"true grad theta1\");",
"_____no_output_____"
],
[
"hist(glog_det[:,1],64,alpha=0.75, range=[-50,50]);\naxvline(mean(glog_det[:,1]),color='cyan', label=\"est grad theta1\");\naxvline(trace(pinv(M).dot(Mg[:,:,1])),ls='--',color='blue', label=\"true grad theta1\");",
"_____no_output_____"
]
],
[
[
"## Evaluating the precision of the method as a function of number of vectors and polynomial order",
"_____no_output_____"
]
],
[
[
"n_tests = 20 # number of experiments at one point in parameter space\n\norders = linspace(10, 200, 32)\nsamples = linspace(1, 200, 32)\nxx,yy = meshgrid(orders, samples)\n\nres_mean = np.zeros((32,32))\nres_std = np.zeros((32,32))",
"_____no_output_____"
],
[
"for i in range(32):\n print(\"Line: \",i)\n for j in range(32):\n res = []\n for k in range(n_tests):\n log_det, glog_det = chebyshev_logdet(mvp, gmvp, shape=(15,1), m=int(yy[i,j]), deg=xx[i,j], g=1.1, eps=1e-3)\n res.append(glog_det[:,1].mean())\n \n res_mean[i,j] = array(res).mean()\n res_std[i,j] = array(res).std()",
"Line: 0\nLine: 1\nLine: 2\n"
],
[
"gtrue = trace(pinv(M).dot(Mg[:,:,1]))\n\nfigure(figsize=(12,4))\n\nsubplot(121)\n\nimshow(clip((res_mean[::-1,:] - gtrue) / gtrue,- 0.3,0.3),cmap='RdYlBu',vmin=-0.3,vmax=0.3,\n extent=[orders[0],orders[-1],samples[0],samples[-1]]) ; colorbar()\n\nxlabel('Order of Polynomial expansion')\nylabel('Number of random vectors')\ntitle('Residual error on the gradient estimate');\n\nsubplot(122)\n\nimshow(log10(res_std[::-1,:]),cmap='magma',\n extent=[orders[0],orders[-1],samples[0],samples[-1]]) ; colorbar()\n\nxlabel('Order of Polynomial expansion')\nylabel('Number of random vectors')\ntitle('Log10 of Standard Deviation of the gradient estimate');",
"_____no_output_____"
],
[
"# Just looking at the scaling with number of vectors\nloglog(samples, res_std[:,-1], label=\"Order %d\"%orders[-1]);\nloglog(samples, res_std[:,1], label=\"Order %d\"%orders[1]);\nxlabel('Number of samples')\nylabel('Standard deviation of estimate')\nlegend()",
"_____no_output_____"
],
[
"# Just looking at the scaling of the error in the mean\nplot(orders, (res_mean[-1,:] - gtrue) / gtrue);\nxlabel('Order of the polynomial approximation')",
"_____no_output_____"
],
[
"plot(orders, (res_mean[-1,:] - gtrue) / res_std[-1,:]);\naxhline(-3,ls='--') # 3 Sigma gap\naxhline(3,ls='--')\nxlabel('Order of the polynomial approximation')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71e307ecd3ab4d36769c88a8ec4527031e58414 | 8,773 | ipynb | Jupyter Notebook | jberkow713_assignment_4.ipynb | jberkow713/DS-Unit-3-Sprint-1-Software-Engineering | cfb9345121a86d667eb5a0a95bdbae33b699f4a6 | [
"MIT"
] | null | null | null | jberkow713_assignment_4.ipynb | jberkow713/DS-Unit-3-Sprint-1-Software-Engineering | cfb9345121a86d667eb5a0a95bdbae33b699f4a6 | [
"MIT"
] | null | null | null | jberkow713_assignment_4.ipynb | jberkow713/DS-Unit-3-Sprint-1-Software-Engineering | cfb9345121a86d667eb5a0a95bdbae33b699f4a6 | [
"MIT"
] | null | null | null | 35.808163 | 1,147 | 0.552605 | [
[
[
"<a href=\"https://colab.research.google.com/github/jberkow713/DS-Unit-3-Sprint-1-Software-Engineering/blob/master/jberkow713_assignment_4.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"pip install -i https://test.pypi.org/simple/ lambdata-jberkow713==1.3",
"Looking in indexes: https://test.pypi.org/simple/\nCollecting lambdata-jberkow713==1.3\n Downloading https://test-files.pythonhosted.org/packages/2c/d7/d01ae5bd1546a044957a2fc70b9e93ed16a0043c001b7057e916a4903496/lambdata_jberkow713-1.3-py3-none-any.whl\nInstalling collected packages: lambdata-jberkow713\nSuccessfully installed lambdata-jberkow713-1.3\n"
],
[
"from lambdata_jberkow713 import my_mod\nimport math\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"#This is the shrink function I created ",
"_____no_output_____"
],
[
"from lambdata_jberkow713.my_mod import shrink\nshrink(7)",
"_____no_output_____"
],
[
"#The shrink function takes a number(n) and multiplies it by 1/(n **3)\n#This next test function tests for this, and works in VS-Code through Anaconda Prompt",
"_____no_output_____"
],
[
"\n#import unittest\n\n#from lambdata_jberkow713.my_mod import shrink\n\n#class Testshrink(unittest.TestCase):\n\n # def Testshrink(self):\n # self.assertEqual(shrink(5), .04)\n\n \n#if __name__ == \"__main__\":\n # unittest.main()",
"_____no_output_____"
],
[
"'''\nMIT License\n\nCopyright (c) 2020 jberkow713.github.io\n\nPermission is hereby granted, free of charge, to any person obtaining a copy\nof this software and associated documentation files (the \"Software\"), to deal\nin the Software without restriction, including without limitation the rights\nto use, copy, modify, merge, publish, distribute, sublicense, and/or sell\ncopies of the Software, and to permit persons to whom the Software is\nfurnished to do so, subject to the following conditions:\n\nThe above copyright notice and this permission notice shall be included in all\ncopies or substantial portions of the Software.\n\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\nIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\nFITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\nAUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\nLIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\nOUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\nSOFTWARE.\n\n'''",
"_____no_output_____"
],
[
"# lambdata-jberkow713 Readme\n'''\nThis lambdata_jberkow713 file contains various invented mathematical functions in the my_mod.py file, \nand a Golf Class, in the golf.py folder, used to predict the distance golf balls travel based on various factors. \nThere are also various testing functions to be used on the Golf Class, as well as on the other math functions such as Shrink.\n'''\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71e3adc2a04d6cf0ab244a213b2e7bb2edb2038 | 6,936 | ipynb | Jupyter Notebook | scrape_craigslist.ipynb | nouyang/spatiotemporal-craigslist | b06fe5948ee2df6cb0c06b2a4c8e1091f4883eba | [
"MIT"
] | null | null | null | scrape_craigslist.ipynb | nouyang/spatiotemporal-craigslist | b06fe5948ee2df6cb0c06b2a4c8e1091f4883eba | [
"MIT"
] | null | null | null | scrape_craigslist.ipynb | nouyang/spatiotemporal-craigslist | b06fe5948ee2df6cb0c06b2a4c8e1091f4883eba | [
"MIT"
] | null | null | null | 33.346154 | 683 | 0.499856 | [
[
[
"keywords = ''\ncities = ''\nlatlonglookup = ''\ndaterange = ''\n#CraigslistCommunity\n#from craigslist import CraigslistHousing\n#cl_h = CraigslistHousing(site='sfbay', area='sfc', category='roo',\n# filters={'max_price': 1200, 'private_room': True})\n",
"_____no_output_____"
],
[
"from craigslist import CraigslistCommunity\nimport json",
"_____no_output_____"
],
[
"CraigslistCommunity.show_filters()",
"Base filters:\n* query = ...\n* search_titles = True/False\n* has_image = True/False\n* posted_today = True/False\n* bundle_duplicates = True/False\n* search_distance = ...\n* zip_code = ...\nSection specific filters:\n"
],
[
"#get_results(limit=2`)\n''' atl\nnat\neat\nsat\nwa\n'''\n'''\n boston\n gbs\n nwb\n bmw\n nos\n sob\n'''\nresults = []",
"_____no_output_____"
],
[
"atl_cl = CraigslistCommunity(site='atlanta', area='nat', category='mis')\nfor result in atl_cl.get_results(include_details=True, limit=2):\n print(result)\n results.append(result)",
"{'id': '7171431912', 'repost_of': None, 'name': '== 28Y/R Divorced white Mommy ==', 'url': 'https://atlanta.craigslist.org/nat/mis/d/smyrna-28y-divorced-white-mommy/7171431912.html', 'datetime': '2020-08-04 15:58', 'last_updated': '2020-08-04 15:58', 'price': None, 'where': None, 'has_image': False, 'geotag': None, 'deleted': False, 'body': \"28y Divorced MOmmy Board lets have fun Don't W Ste my time or yrs. If Ur serious and free now then hit me.\", 'created': '2020-08-04 14:25', 'images': [], 'attrs': []}\n{'id': '7171413090', 'repost_of': None, 'name': 'And Gus Makes Three', 'url': 'https://atlanta.craigslist.org/nat/mis/d/alpharetta-and-gus-makes-three/7171413090.html', 'datetime': '2020-08-04 14:50', 'last_updated': '2020-08-04 14:50', 'price': None, 'where': None, 'has_image': False, 'geotag': None, 'deleted': False, 'body': \"You're new to the neighborhood... I see you all the time with Gus. We passed each other today while I was jogging. You love basketball shorts and hats. HMU if you're ever in the mood to have a drink since we're neighbors. Black dude here...\", 'created': '2020-08-04 13:59', 'images': [], 'attrs': [], 'address': '13201 Deerfield Parkway'}\n"
],
[
"print(json.dumps(results, indent=2))",
"[\n {\n \"id\": \"7171431912\",\n \"repost_of\": null,\n \"name\": \"== 28Y/R Divorced white Mommy ==\",\n \"url\": \"https://atlanta.craigslist.org/nat/mis/d/smyrna-28y-divorced-white-mommy/7171431912.html\",\n \"datetime\": \"2020-08-04 15:58\",\n \"last_updated\": \"2020-08-04 15:58\",\n \"price\": null,\n \"where\": null,\n \"has_image\": false,\n \"geotag\": null,\n \"deleted\": false\n },\n {\n \"id\": \"7171413090\",\n \"repost_of\": null,\n \"name\": \"And Gus Makes Three\",\n \"url\": \"https://atlanta.craigslist.org/nat/mis/d/alpharetta-and-gus-makes-three/7171413090.html\",\n \"datetime\": \"2020-08-04 14:50\",\n \"last_updated\": \"2020-08-04 14:50\",\n \"price\": null,\n \"where\": null,\n \"has_image\": false,\n \"geotag\": null,\n \"deleted\": false\n },\n {\n \"id\": \"7171431912\",\n \"repost_of\": null,\n \"name\": \"== 28Y/R Divorced white Mommy ==\",\n \"url\": \"https://atlanta.craigslist.org/nat/mis/d/smyrna-28y-divorced-white-mommy/7171431912.html\",\n \"datetime\": \"2020-08-04 15:58\",\n \"last_updated\": \"2020-08-04 15:58\",\n \"price\": null,\n \"where\": null,\n \"has_image\": false,\n \"geotag\": null,\n \"deleted\": false,\n \"body\": \"28y Divorced MOmmy Board lets have fun Don't W Ste my time or yrs. If Ur serious and free now then hit me.\",\n \"created\": \"2020-08-04 14:25\",\n \"images\": [],\n \"attrs\": []\n },\n {\n \"id\": \"7171413090\",\n \"repost_of\": null,\n \"name\": \"And Gus Makes Three\",\n \"url\": \"https://atlanta.craigslist.org/nat/mis/d/alpharetta-and-gus-makes-three/7171413090.html\",\n \"datetime\": \"2020-08-04 14:50\",\n \"last_updated\": \"2020-08-04 14:50\",\n \"price\": null,\n \"where\": null,\n \"has_image\": false,\n \"geotag\": null,\n \"deleted\": false,\n \"body\": \"You're new to the neighborhood... I see you all the time with Gus. We passed each other today while I was jogging. You love basketball shorts and hats. HMU if you're ever in the mood to have a drink since we're neighbors. Black dude here...\",\n \"created\": \"2020-08-04 13:59\",\n \"images\": [],\n \"attrs\": [],\n \"address\": \"13201 Deerfield Parkway\"\n }\n]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71e5cb1a2e89320de7c032e9aaa9ff507649abf | 39,574 | ipynb | Jupyter Notebook | MessMLmodel.ipynb | batman004/Messlytix | 38e4518f6e0ddc1c873982d73df628901909ddee | [
"MIT"
] | 2 | 2020-07-28T08:01:32.000Z | 2020-10-21T11:10:49.000Z | MessMLmodel.ipynb | batman004/Messlytix | 38e4518f6e0ddc1c873982d73df628901909ddee | [
"MIT"
] | null | null | null | MessMLmodel.ipynb | batman004/Messlytix | 38e4518f6e0ddc1c873982d73df628901909ddee | [
"MIT"
] | 2 | 2020-06-21T10:13:30.000Z | 2020-06-21T15:12:12.000Z | 54.509642 | 3,877 | 0.395083 | [
[
[
"\n\nimport pandas as pd\nimport numpy as np\nimport sklearn\nfrom sklearn import linear_model\nfrom sklearn.model_selection import train_test_split\nimport matplotlib.pyplot as plt\nimport pickle\n",
"_____no_output_____"
],
[
"\ndata = pd.read_csv(r'DATASET.csv')\ndata.head()\n\n",
"_____no_output_____"
],
[
"data['Menu Rating'].fillna(data['Menu Rating'].mean(), inplace=True)\ndata['Amount Of Food Cooked'].fillna(data['Amount Of Food Cooked'].mean(), inplace=True)\ndata['Wastage'].fillna(data['Wastage'].mean(), inplace=True)\n",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"from sklearn.preprocessing import LabelEncoder as LE",
"_____no_output_____"
],
[
"#For days\n\ns=data['Day']\n \ndata.apply(lambda s: s.map({k:i for i,k in enumerate(s.unique())}))\n\n",
"_____no_output_____"
],
[
"predict = \"Amount Of Food Cooked\"\nX = np.array(data.drop([predict], 1))\ny = np.array(data[predict])\n\nX",
"_____no_output_____"
],
[
"# TRAIN MODEL MULTIPLE TIMES FOR BEST SCORE\nbest = 0\nlimit=97.0\nx=0.0\n\nfor _ in range(200):\n x_train, x_test, y_train, y_test = sklearn.model_selection.train_test_split(\n X, y, test_size=0.3)\n\n linear = linear_model.LinearRegression()\n\n linear.fit(x_train, y_train)\n acc = linear.score(x_test, y_test)\n print(\"Accuracy: \" + str(acc))\n\n if acc > best:\n best = acc\n with open(\"food.pickle\", \"wb\") as f:\n pickle.dump(linear, f)\n\n",
"Accuracy: 0.9568720424378561\nAccuracy: 0.9793796272521894\nAccuracy: 0.9675686884094415\nAccuracy: 0.955878381895554\nAccuracy: 0.9726107430832707\nAccuracy: 0.9466648390500924\nAccuracy: 0.9555785441316523\nAccuracy: 0.9723243023976227\nAccuracy: 0.9755495671482353\nAccuracy: 0.9790721377916785\nAccuracy: 0.9583982568019453\nAccuracy: 0.9598488958213097\nAccuracy: 0.943917884280722\nAccuracy: 0.9690135220746335\nAccuracy: 0.9659574104721697\nAccuracy: 0.9709638087626363\nAccuracy: 0.9766360007315125\nAccuracy: 0.959596274872894\nAccuracy: 0.9750001208214962\nAccuracy: 0.9717268760537726\nAccuracy: 0.94700519664461\nAccuracy: 0.9505832725884512\nAccuracy: 0.929023234927152\nAccuracy: 0.9502898000125046\nAccuracy: 0.9734654971246987\nAccuracy: 0.9728884789510521\nAccuracy: 0.9487886723112354\nAccuracy: 0.9663945413279206\nAccuracy: 0.9701348007205683\nAccuracy: 0.9660486182882815\nAccuracy: 0.9718098577982844\nAccuracy: 0.9729658632964032\nAccuracy: 0.9707728858375325\nAccuracy: 0.9573908824259841\nAccuracy: 0.972376460360265\nAccuracy: 0.9648123475086681\nAccuracy: 0.976833107990741\nAccuracy: 0.977969848136554\nAccuracy: 0.9749717979353951\nAccuracy: 0.9647992506277128\nAccuracy: 0.9736777919325788\nAccuracy: 0.9541713419797311\nAccuracy: 0.9661059522554722\nAccuracy: 0.9627654096853735\nAccuracy: 0.9482607825398309\nAccuracy: 0.9587443374611663\nAccuracy: 0.9587503652134071\nAccuracy: 0.9638335215612266\nAccuracy: 0.9294246198723066\nAccuracy: 0.9738721574051871\nAccuracy: 0.9633297709358488\nAccuracy: 0.9772888511543821\nAccuracy: 0.9593786847520512\nAccuracy: 0.979575207754223\nAccuracy: 0.9645005726744483\nAccuracy: 0.9598602446537386\nAccuracy: 0.9770157818288884\nAccuracy: 0.9763598309653958\nAccuracy: 0.9745436922272266\nAccuracy: 0.9680957585766234\nAccuracy: 0.9631788799175274\nAccuracy: 0.9678736771978927\nAccuracy: 0.981074682452279\nAccuracy: 0.979203914144145\nAccuracy: 0.9784178621346026\nAccuracy: 0.9787878820128523\nAccuracy: 0.9633297456680585\nAccuracy: 0.9756945085045299\nAccuracy: 0.9739762425316087\nAccuracy: 0.9692865361504334\nAccuracy: 0.9753413890085323\nAccuracy: 0.9708027374542773\nAccuracy: 0.9716335010634015\nAccuracy: 0.9488571617448062\nAccuracy: 0.9635392146200934\nAccuracy: 0.9636748414708817\nAccuracy: 0.9397980461154201\nAccuracy: 0.9737061866526989\nAccuracy: 0.9727427893538545\nAccuracy: 0.944006189040036\nAccuracy: 0.9748701396692694\nAccuracy: 0.9723719713528262\nAccuracy: 0.9630507814631171\nAccuracy: 0.9692948546280709\nAccuracy: 0.9692897717371961\nAccuracy: 0.9566773239789385\nAccuracy: 0.9700948936900761\nAccuracy: 0.9711828405027825\nAccuracy: 0.9705935827854683\nAccuracy: 0.9623692644130627\nAccuracy: 0.9706440991741774\nAccuracy: 0.9598633513459265\nAccuracy: 0.9723124060643112\nAccuracy: 0.9726328246364538\nAccuracy: 0.9709346145883218\nAccuracy: 0.9613945046091932\nAccuracy: 0.9715965180320367\nAccuracy: 0.9582171384265156\nAccuracy: 0.9771404097216706\nAccuracy: 0.9615487463649339\nAccuracy: 0.964679926173498\nAccuracy: 0.9662255583501222\nAccuracy: 0.9705917204135039\nAccuracy: 0.9736178888594655\nAccuracy: 0.943229223470682\nAccuracy: 0.9676193151663816\nAccuracy: 0.9758943815148828\nAccuracy: 0.9745811606141151\nAccuracy: 0.9630056772361308\nAccuracy: 0.9607301254558251\nAccuracy: 0.9389294728044073\nAccuracy: 0.9641946469561757\nAccuracy: 0.9673129382677157\nAccuracy: 0.9788765165871991\nAccuracy: 0.9736291283707562\nAccuracy: 0.9538910212907\nAccuracy: 0.9761586571611129\nAccuracy: 0.9405277443003598\nAccuracy: 0.9759844066329837\nAccuracy: 0.9669856725577056\nAccuracy: 0.9795573633859685\nAccuracy: 0.971562817347731\nAccuracy: 0.9760798106425691\nAccuracy: 0.9597144553969751\nAccuracy: 0.9399289575670347\nAccuracy: 0.9649503361580016\nAccuracy: 0.9584420452590742\nAccuracy: 0.9772902095013286\nAccuracy: 0.9430544145043482\nAccuracy: 0.9730897495839677\nAccuracy: 0.9740474088079992\nAccuracy: 0.9549387346837905\nAccuracy: 0.9647705064524124\nAccuracy: 0.9560802138472715\nAccuracy: 0.9182984252534164\nAccuracy: 0.9691999340210491\nAccuracy: 0.946426619016725\nAccuracy: 0.953348618889532\nAccuracy: 0.9660690530240655\nAccuracy: 0.9688605349962677\nAccuracy: 0.960320653982158\nAccuracy: 0.9818749385861355\nAccuracy: 0.9722148471521458\nAccuracy: 0.9728116482986605\nAccuracy: 0.9679554835543975\nAccuracy: 0.9568276973217438\nAccuracy: 0.9758267015105736\nAccuracy: 0.9759910799912223\nAccuracy: 0.9695428405309481\nAccuracy: 0.9706101084623017\nAccuracy: 0.9744704070515678\nAccuracy: 0.9679070631745705\nAccuracy: 0.9540910397673825\nAccuracy: 0.9781767038241225\nAccuracy: 0.9597896295644621\nAccuracy: 0.9650324402830036\nAccuracy: 0.9695209953482864\nAccuracy: 0.9796647380843623\nAccuracy: 0.9569133783565479\nAccuracy: 0.9636515442755937\nAccuracy: 0.9751309385768703\nAccuracy: 0.9711148431523379\nAccuracy: 0.9790821476210534\nAccuracy: 0.9461942002234465\nAccuracy: 0.9578000334160992\nAccuracy: 0.9760345065554573\nAccuracy: 0.9717883833513218\nAccuracy: 0.96648142093405\nAccuracy: 0.966163611380418\nAccuracy: 0.9597510745736932\nAccuracy: 0.9421086579132191\nAccuracy: 0.9740965874581724\nAccuracy: 0.9663733474522193\nAccuracy: 0.9674751385571907\nAccuracy: 0.9774421735495299\nAccuracy: 0.9626731360844347\nAccuracy: 0.9597299739204724\nAccuracy: 0.9740520071316723\nAccuracy: 0.9635909203005395\nAccuracy: 0.9665552958429088\nAccuracy: 0.9803711727696729\nAccuracy: 0.9682166302950534\nAccuracy: 0.9682334379035503\nAccuracy: 0.9515659237140127\nAccuracy: 0.9521970389923782\nAccuracy: 0.9719918471785065\nAccuracy: 0.9643134826298719\nAccuracy: 0.9783036387777843\nAccuracy: 0.9716115574572798\nAccuracy: 0.951925765239376\nAccuracy: 0.9606891219678608\nAccuracy: 0.9773324798236489\nAccuracy: 0.9498850367829614\nAccuracy: 0.9738727096104589\nAccuracy: 0.9688575334854572\nAccuracy: 0.9658906594328628\nAccuracy: 0.9688420037145729\nAccuracy: 0.970936049787412\nAccuracy: 0.9701398245750193\nAccuracy: 0.959459082938333\n"
],
[
"pickle_in = open(\"food.pickle\", \"rb\")\nlinear = pickle.load(pickle_in)\nacc\n",
"_____no_output_____"
],
[
"predicted= linear.predict(x_test)\nx_test1=np.array(x_test)\ny_test1=np.array(y_test)",
"_____no_output_____"
],
[
"#Comparing predicitons with actual results\nprint(\"Actual Result Input Params Predicted Result\")\nprint()\nfor w in range(len(y_test)):\n print(y_test1[w],'',x_test1[w],' ',\"{:.2f}\".format(predicted[w]))\n print()",
"Actual Result Input Params Predicted Result\n\n861.4285714 [ 7. 1. 8.14285714 180.5714286 ] 865.88\n\n800.0 [ 5. 0. 8.6 110. ] 804.47\n\n1000.0 [ 6. 1. 7. 350.] 1009.35\n\n861.4285714 [ 2. 0. 8.14285714 180.5714286 ] 856.52\n\n861.4285714 [ 7. 1. 8.14285714 180.5714286 ] 865.88\n\n861.4285714 [ 3. 0. 8.14285714 180.5714286 ] 859.73\n\n861.4285714 [ 1. 0. 8.14285714 180.5714286 ] 853.31\n\n861.4285714 [ 1. 0. 8.14285714 180.5714286 ] 853.31\n\n861.4285714 [ 7. 1. 8.14285714 180.5714286 ] 865.88\n\n800.0 [ 2. 0. 8.5 145. ] 829.38\n\n861.4285714 [ 2. 0. 8.14285714 180.5714286 ] 856.52\n\n861.4285714 [ 4. 0. 8.14285714 180.5714286 ] 862.94\n\n861.4285714 [ 2. 0. 8.14285714 180.5714286 ] 856.52\n\n820.0 [ 3. 0. 9.1 105. ] 808.25\n\n1000.0 [ 7. 1. 7.9 300. ] 986.81\n\n861.4285714 [ 3. 0. 8.14285714 180.5714286 ] 859.73\n\n861.4285714 [ 3. 0. 8.14285714 180.5714286 ] 859.73\n\n810.0 [ 4. 0. 8.9 100. ] 799.85\n\n820.0 [ 3. 0. 9.1 105. ] 808.25\n\n861.4285714 [ 5. 0. 8.14285714 180.5714286 ] 866.16\n\n861.4285714 [ 3. 0. 8.14285714 180.5714286 ] 859.73\n\n861.4285714 [ 6. 1. 8.14285714 180.5714286 ] 862.67\n\n820.0 [ 3. 0. 9.1 105. ] 808.25\n\n1000.0 [ 7. 1. 7.9 300. ] 986.81\n\n861.4285714 [ 4. 0. 8.14285714 180.5714286 ] 862.94\n\n861.4285714 [ 7. 1. 8.14285714 180.5714286 ] 865.88\n\n820.0 [ 3. 0. 9.1 105. ] 808.25\n\n861.4285714 [ 2. 0. 8.14285714 180.5714286 ] 856.52\n\n861.4285714 [ 4. 0. 8.14285714 180.5714286 ] 862.94\n\n861.4285714 [ 1. 0. 8.14285714 180.5714286 ] 853.31\n\n861.4285714 [ 1. 0. 8.14285714 180.5714286 ] 853.31\n\n861.4285714 [ 6. 1. 8.14285714 180.5714286 ] 862.67\n\n861.4285714 [ 7. 1. 8.14285714 180.5714286 ] 865.88\n\n800.0 [ 1. 0. 7. 154.] 789.10\n\n1000.0 [ 6. 1. 7. 350.] 1009.35\n\n861.4285714 [ 7. 1. 8.14285714 180.5714286 ] 865.88\n\n861.4285714 [ 5. 0. 8.14285714 180.5714286 ] 866.16\n\n1000.0 [ 6. 1. 7. 350.] 1009.35\n\n1000.0 [ 7. 1. 7.9 300. ] 986.81\n\n800.0 [ 5. 0. 8.6 110. ] 804.47\n\n800.0 [ 2. 0. 8.5 145. ] 829.38\n\n820.0 [ 3. 0. 9.1 105. ] 808.25\n\n800.0 [ 2. 0. 8.5 145. ] 829.38\n\n861.4285714 [ 7. 1. 8.14285714 180.5714286 ] 865.88\n\n1000.0 [ 7. 1. 7.9 300. ] 986.81\n\n800.0 [ 5. 0. 8.6 110. ] 804.47\n\n861.4285714 [ 5. 0. 8.14285714 180.5714286 ] 866.16\n\n800.0 [ 2. 0. 8.5 145. ] 829.38\n\n810.0 [ 4. 0. 8.9 100. ] 799.85\n\n861.4285714 [ 4. 0. 8.14285714 180.5714286 ] 862.94\n\n861.4285714 [ 2. 0. 8.14285714 180.5714286 ] 856.52\n\n800.0 [ 2. 0. 8.5 145. ] 829.38\n\n861.4285714 [ 1. 0. 8.14285714 180.5714286 ] 853.31\n\n861.4285714 [ 6. 1. 8.14285714 180.5714286 ] 862.67\n\n861.4285714 [ 5. 0. 8.14285714 180.5714286 ] 866.16\n\n"
],
[
"pred = linear.predict([[4, 0, 8.1, 0]])\npred",
"_____no_output_____"
],
[
"\ndef refactoring(day):\n word_dict = {'Monday':0, 'Tuesday':1, 'Wednesday':2, 'Thursday':3, 'Friday':4, 'Saturday': 5, 'Sunday':6}\n rating_dict = {0:7, 1:8.5, 2:9.1, 3:8.9, 4:8.6, 5: 7, 6:7.9}\n wastage_dict = {0:153.334, 1:143, 2:107.233, 3:102.223, 4:112.344, 5:349.456, 6:330.233}\n weekend = 1 if day in ['Saturday', 'Sunday'] else 0\n return list([word_dict[day],weekend,rating_dict[word_dict[day]],wastage_dict[word_dict[day]]])\n\nprint(refactoring('Monday'))\n\n# In[12]:\n\n\n# DAY 0 :MONDAY ; DAY 1:TUESDAY ; DAY 2 :WEDNESDAY ; DAY 3: THURSDAY ; DAY 4:FRIDAY ; DAY 5:SATURDAY ; DAY 6 =SUNDAY\n# IF IT IS A WEEKEND THEN TYPE 1, ELSE 0\n# CHECK THE AMOUNT THE OF FOOD YOU NEED TO COOK TO MINIMISE YOUR WASTAGE\n# By default wastage is 0 since we are trying to predict the ideal amounnt of food to be cooked\n\n\n# In[13]:\n\n\n# TESTING\n# ------------------\nWastage = 0\n# ------------------\ninput_list = refactoring(input('Enter the day of the week: '))\npred=linear.predict([input_list])\nprint('Menu rating for Today is :', input_list[2])\nprint('Average wastage on this day: ', input_list[3], 'Kgs')\nprint('To avoid this wastage,the predicted amount to be cooked :', pred, 'kgs')",
"[0, 0, 7, 153.334]\nMenu rating for Today is : 7\nAverage wastage on this day: 153.334 Kgs\nTo avoid this wastage,the predicted amount to be cooked : [785.17295269] kgs\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71e5d901c9e40d607589d2f4446910ee56fd851 | 36,275 | ipynb | Jupyter Notebook | HeatEquation_Solution_DeepXDE.ipynb | shadzzz90/Solution-Head-Conduction-using-Deep-Galerkin-Method | 59d7251a5526fd05477086f2187615e09ec9af0d | [
"MIT"
] | null | null | null | HeatEquation_Solution_DeepXDE.ipynb | shadzzz90/Solution-Head-Conduction-using-Deep-Galerkin-Method | 59d7251a5526fd05477086f2187615e09ec9af0d | [
"MIT"
] | null | null | null | HeatEquation_Solution_DeepXDE.ipynb | shadzzz90/Solution-Head-Conduction-using-Deep-Galerkin-Method | 59d7251a5526fd05477086f2187615e09ec9af0d | [
"MIT"
] | null | null | null | 112.306502 | 26,008 | 0.833604 | [
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nimport deepxde as dde\nfrom deepxde.backend import tf\nimport math\nfrom celluloid import Camera\n",
"_____no_output_____"
],
[
"L = 80\nT = 5\n\nrho = 8.92\nCp = 0.092\nk = 0.95\n\na = k/(rho*Cp) # Thermal diffusivity\n\nlambda_sqaure = (a*math.pi**2)/L**2\n\nn = 1 # Frequency of the sinusoidal initial conditions",
"_____no_output_____"
],
[
"u_x_t = lambda x, t: np.sin((x * np.pi) / L) * np.exp(-lambda_sqaure * t)",
"_____no_output_____"
],
[
"def pde(x, y):\n \"\"\"\n Expresses the PDE residual of the heat equation. \n \"\"\"\n dy_t = dde.grad.jacobian(y, x, i=0, j=1)\n dy_xx = dde.grad.hessian(y, x, i=0, j=0)\n return dy_t - a*dy_xx",
"_____no_output_____"
],
[
"# Computational geometry:\ngeom = dde.geometry.Interval(0, L)\ntimedomain = dde.geometry.TimeDomain(0, T)\ngeomtime = dde.geometry.GeometryXTime(geom, timedomain)\n\n# Initial and boundary conditions:\nbc = dde.DirichletBC(geomtime, lambda x: 0, lambda _, on_boundary: on_boundary)\nic = dde.IC(\n geomtime, lambda x: np.sin(n*np.pi*x[:, 0:1]/L), lambda _, on_initial: on_initial\n )\n\n# Define the PDE problem and configurations of the network:\ndata = dde.data.TimePDE(\n geomtime, pde, [bc, ic], num_domain=2540, num_boundary=80, num_initial=160, num_test=2540\n)\nnet = dde.nn.FNN([2] + [20] * 3 + [1], \"tanh\", \"Glorot normal\")\nmodel = dde.Model(data, net)\n\n# Build and train the model:\nmodel.compile(\"adam\", lr=1e-3)\nmodel.train(epochs=20000)",
"Warning: 2540 points required, but 2626 points sampled.\nCompiling model...\nBuilding feed-forward neural network...\n'build' took 0.048015 s\n\n"
],
[
"model.compile(\"L-BFGS\")",
"Compiling model...\n'compile' took 0.213653 s\n\n"
],
[
"losshistory, train_state = model.train()",
"Training model...\n\nStep Train loss Test loss Test metric\n20000 [5.16e-07, 5.65e-07, 3.38e-06] [3.90e-07, 5.65e-07, 3.38e-06] [] \nINFO:tensorflow:Optimization terminated with:\n Message: CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH\n Objective function value: 0.000004\n Number of iterations: 5\n Number of functions evaluations: 25\n20025 [5.16e-07, 5.91e-07, 3.35e-06] [3.90e-07, 5.91e-07, 3.35e-06] [] \n\nBest model at step 11000:\n train loss: 2.57e-06\n test loss: 2.46e-06\n test metric: []\n\n'train' took 0.515737 s\n\n"
],
[
"dde.saveplot(losshistory, train_state, issave=True, isplot=True)",
"_____no_output_____"
],
[
"x = np.linspace(0, L, 80)\nt = np.linspace(0, 15, 15)\n\n\nxx, tt = np.meshgrid(x, t)\n\n\nX = np.vstack((np.ravel(xx), np.ravel(tt))).T\n\nfinal_temp = model.predict(X)\n\nfinal_temps = np.split(final_temp,t.shape[0]) \n# plt.plot(x, final_temp[:x.shape[0]])\n \n# plt.show()\nfinal_temp_ana = []\n\nu_x_t = lambda x, t: np.sin((x * np.pi) / L) * np.exp(-lambda_sqaure * t)\n\nfig, ax = plt.subplots()\nfig.patch.set_facecolor('xkcd:white')\ncamera = Camera(fig)\n\nax.set_xlabel('Length (m)')\nax.set_ylabel('Tempreature ($^0$C)')\n\nfor i,_t in enumerate(t):\n\n\n final_temp_ana.append(u_x_t(x, _t))\n\n\n ax.plot(x, final_temps[i], '-b', x.flatten(), np.array(final_temp_ana)[i,:],'-r')\n\n ax.text(0.5, 1.01, \"Time = {} secs \".format(int(i)), transform=ax.transAxes)\n\n ax.legend(['DeepXDE', 'Analytical'])\n\n camera.snap()\n\n\nanim = camera.animate()\n\nanim.save('solution_DeepXDE_vs_Analytical_gen.gif', dpi=100)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71e65a4976ddbf8b7371f0294e6419d29999dae | 491,054 | ipynb | Jupyter Notebook | S2_3_Statistical_Forecasting_reno.ipynb | DuaneHsa/2022_ML_Earth_Env_Sci | 4627bb9cffbbc75718f60d2af69b93df770a962e | [
"MIT"
] | null | null | null | S2_3_Statistical_Forecasting_reno.ipynb | DuaneHsa/2022_ML_Earth_Env_Sci | 4627bb9cffbbc75718f60d2af69b93df770a962e | [
"MIT"
] | null | null | null | S2_3_Statistical_Forecasting_reno.ipynb | DuaneHsa/2022_ML_Earth_Env_Sci | 4627bb9cffbbc75718f60d2af69b93df770a962e | [
"MIT"
] | null | null | null | 632.801546 | 307,501 | 0.932954 | [
[
[
"<a href=\"https://colab.research.google.com/github/DuaneHsa/2022_ML_Earth_Env_Sci/blob/main/S2_3_Statistical_Forecasting_reno.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"This notebook will be used in the lab session for week 2 of the course and provides some hands-on experience applying the lessons to environmental science datasets.\n\nNeed a reminder of last week's labs? Click [_here_](https://github.com/tbeucler/2022_ML_Earth_Env_Sci/blob/main/Lab_Notebooks/Week_1_Basics_of_Python.ipynb) to go to notebook for week 1 of the course.",
"_____no_output_____"
],
[
"#Statistical Forecasting - Wilks\n\nWe will be using data from Wilks' book on Statistical Methods for the Atmospheric Sciences",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"# Python ≥3.5 is required\nimport sys\nassert sys.version_info >= (3, 5)\n\n# Is this notebook running on Colab or Kaggle?\nIS_COLAB = \"google.colab\" in sys.modules\n\n# Scikit-Learn ≥0.20 is required\nimport sklearn\nassert sklearn.__version__ >= \"0.20\"\n\n# Common imports\nimport numpy as np\nimport os\nimport pandas as pd\nimport pooch\n\n#Data Visalization Import\nfrom google.colab import data_table\n\n\n# to make this notebook's output stable across runs\nrnd_seed = 42\nrnd_gen = np.random.default_rng(rnd_seed)\n\n# To plot pretty figures\n%matplotlib inline\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nmpl.rc('axes', labelsize=14)\nmpl.rc('xtick', labelsize=12)\nmpl.rc('ytick', labelsize=12)\n\n# Where to save the figures\nPROJECT_ROOT_DIR = \".\"\nCHAPTER_ID = \"classification\"\nIMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, \"images\", CHAPTER_ID)\nos.makedirs(IMAGES_PATH, exist_ok=True)\n\ndef save_fig(fig_id, tight_layout=True, fig_extension=\"png\", resolution=300):\n path = os.path.join(IMAGES_PATH, fig_id + \".\" + fig_extension)\n print(\"Saving figure\", fig_id)\n if tight_layout:\n plt.tight_layout()\n plt.savefig(path, format=fig_extension, dpi=resolution)",
"_____no_output_____"
]
],
[
[
"Let's begin by loading relevant data from the cloud. ",
"_____no_output_____"
]
],
[
[
"#Loading Wilks' Table A-3 from the course datastore\ncsv_path = 'https://unils-my.sharepoint.com/:x:/g/personal/tom_beucler_unil_ch/EXG7Rht55mhPiwkUKEDSI8oBuXNe8OOLYJX3_5ACmK1w5A?download=1'\nhash = 'c158828a1bdf1aa521c61321842352cb1674e49187e21c504188ab976d3a41f2'\ncsv_file = pooch.retrieve(csv_path, known_hash=hash)\n\nA3_df = pd.read_csv(csv_file, index_col=0)\n\n#Display an interactive datatable for data visualization\ndata_table.DataTable(A3_df, num_rows_per_page=10)",
"Downloading data from 'https://unils-my.sharepoint.com/:x:/g/personal/tom_beucler_unil_ch/EXG7Rht55mhPiwkUKEDSI8oBuXNe8OOLYJX3_5ACmK1w5A?download=1' to file '/root/.cache/pooch/24ce34b8649699f6b858790e9bbb6a7c-EXG7Rht55mhPiwkUKEDSI8oBuXNe8OOLYJX3_5ACmK1w5A'.\n"
]
],
[
[
"##**Linear Regression**\n\nThe goal for this exercise is to train a linear regression model and a logistic regression model to forecast atmospheric temperature using atmospheric pressure. 🌡 \n\nFor the first case, we want to train linear regression to calculate June temperatures (the predictand) from June pressures (as the predictor) in Guayaquil, Ecuador.\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"**Caption** A beautiful day in Guayacil, Ecuador. Can you predict how hot it will be? 🌞",
"_____no_output_____"
],
[
"We can try addressing this question using a [linear regression model](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html) from scikit. \n\n**Q1) Import the LinearRegression model. Instantiate it and fit it using the A3 dataframes' pressure and temperature.**",
"_____no_output_____"
],
[
"Complete the code below",
"_____no_output_____"
]
],
[
[
"# Import the LinearRegression model\nfrom sklearn.linear_model import LinearRegression",
"_____no_output_____"
],
[
"# Instantiate the model\nlin_reg = LinearRegression()\n",
"_____no_output_____"
],
[
"# Load and reshape the input data\npressure = A3_df['Pressure (mb)'].values.reshape(-1,1)",
"_____no_output_____"
],
[
"# Load the truth data (i.e., the predictant)\ntemperature = A3_df['Temperature (°C)'].values.reshape(-1,1)",
"_____no_output_____"
],
[
"# Fit the model\nlin_reg.fit(pressure,temperature)",
"_____no_output_____"
]
],
[
[
"We now have a linear regression model for the temperature and pressure. Let's make some plots to visualize our data and get a qualitative sense of our model.\n\n**Q2) Generate a scatter plot with the linear regression plot for our data.**",
"_____no_output_____"
]
],
[
[
"#Complete the code below\n\n#Instantiate a figure having size 13,6\nfig, ax = plt.subplots()\n\n# Set figure title and axis labels\nfig.suptitle('June Temperature vs Pressure in Guayaquil, Ecuador')\nax.set_xlabel(\"Pressure (mb)\")\nax.set_ylabel(\"Temperature (°C)\")\n\n# The colors and styles suggested below are not compulsory, but please avoid \n# using the default settings.\n\n# Make a scatter plot for the pressure (x) and temperature (y). Use color=black,\n# marker size = 100, and set the marker style to '2'.\nax.scatter(pressure,temperature,c ='blue',marker = '2')\n\n# Make a 100 point numpy array between 1008 and 1014 and store it in reg_x. \n# Reshape it to (-1,1). Hint: numpy has a linear space generator\nreg_x = np.linspace(1008,1014,num = 100)\nreg_x = np.reshape(reg_x, (-1,1))\n\n# Let's produce a set of predictions from our linear space array.\nreg_y = lin_reg.predict(reg_x)\n# Let's plot the regression line using reg_x and reg_y. Set the color to red and\n# the linewidth to 1.5\nax.plot(reg_x,reg_y, c='red')\nax.autoscale(axis='x', tight=True)",
"_____no_output_____"
]
],
[
[
"We now have a qualitative verification of our model - but this is not enough. Let's do some quantitative analyses:\n\n**Q3) Print the slope of our model. Find the F-score, p-value, and $R^2$ statistics for our model.**",
"_____no_output_____"
]
],
[
[
"# Complete the code below\n\n# Fetch the slope directly from the linear model. Hint: check the attributes \n# section of the linear regression model documentation on scikit.\nslope = print((lin_reg.coef_))\n\n# Calculate the F-score and p-value for our dataset.\n# Hint: check the f_regression option in \nfrom sklearn.feature_selection import f_regression\n\nfscore, pvalue = (f_regression(pressure,temperature))\n# Fetch the R2 value from the lin_reg model. Hint: check built-in score methods\nR2 = lin_reg.score(pressure,temperature)",
"[[-0.92488103]]\n"
]
],
[
[
"##**Classification**\n\nLet's use the same dataset to train a classifier for El Niño years. We will use the June temperature and pressure in Guayaquil as the predictors for El Niño. Let's begin by setting up a training and testing dataset. Since the dataset is so small, we'll set aside one each of a random El Niño year and a non-El Niño year for our test dataset, and the remaining points as our training dataset.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"**Source:** [NOAA \"What is Enso?\"](https://www.climate.gov/enso)\n\n**Caption:** Can we predict whether we are in an El Niño phase based on June temperatures and pressures in Guayaquil, Ecuador?",
"_____no_output_____"
]
],
[
[
"# Let's make our train and test datasets\ndef test_train_split(df, rnd_gen):\n nino_years = df.loc[df['El Niño']==1].index.values\n not_nino_years = df.loc[df['El Niño']==0].index.values\n\n nino_idxs = rnd_gen.permutation(np.arange(0,nino_years.size))\n not_nino_idxs = rnd_gen.permutation(np.arange(0,not_nino_years.size))\n\n train = ( list(nino_years[nino_idxs[:-1]]) +\n list(not_nino_years[not_nino_idxs[:-1]]) )\n \n test = [nino_years[nino_idxs[-1]], not_nino_years[not_nino_idxs[-1]]]\n \n return (np.array(test), np.array(train))\n\n# Use test_train_split to make the testing and training datasets\ntest, train = test_train_split(A3_df, rnd_gen)\nprint(test, train)",
"[1951 1958] [1969 1957 1965 1953 1970 1963 1968 1961 1967 1964 1960 1962 1956 1952\n 1954 1959 1966 1955]\n"
]
],
[
[
"We're going to train a logistic regression classifier on the dataset, but in this exercise we'll rely on the scikit learn implementation!\n\n**Q4) Import and instantiate the logistic regression classifier from scikit. Fit it using the training dataset.**\n\nHint 1: Scikit-learn's `LogisticRegression` classifier is documented [at this link](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html).\n\nHint 2: Before training, use the dataframes' [`.loc` method](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.loc.html) with the test/train list, then convert the values to numpy (e.g., using [this method](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_numpy.html)) and [`.ravel()`](https://numpy.org/doc/stable/reference/generated/numpy.ravel.html) the truth if necessary.",
"_____no_output_____"
]
],
[
[
"# Build the training and test sets, using Hint 2 if necessary\n\nX_train = A3_df.iloc[:,[1,3]].loc[train].to_numpy()\ny_train = A3_df.iloc[:,0].loc[train].to_numpy()\nX_test = A3_df.iloc[:,[1,3]].loc[test].to_numpy()\n\n\ny_test = A3_df.iloc[:,0].loc[test].to_numpy()\n",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\n\nLR = LogisticRegression()\nLR.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"That should hopefully have felt much simpler than our previous exercise. Now that you have a trained model, let's see if our model is able to predict whether the test years are El Niño years!\n\n**Q5) Predict whether each of the two test years was an El Niño year using the logistic regression model, and print out the prediction alongside the truth.**\n\nHint: To find which method of your `LogisticRegression` classifier to use to make predictions, don't hesitate to consult its documentation [at this link](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html).",
"_____no_output_____"
]
],
[
[
"Prediction = LR.predict(X_test)\nprint('the prediction is ', Prediction)\nprint('the truth is ', y_test)",
"the prediction is [1 0]\nthe truth is [1 0]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e71e6a4c0b7f4a1003e20fe386922d22e323d4b0 | 31,881 | ipynb | Jupyter Notebook | Model backlog/Inference/91-tweet-inference-5fold-roberta-base-post6.ipynb | dimitreOliveira/Tweet-Sentiment-Extraction | 0a775abe9a92c4bc2db957519c523be7655df8d8 | [
"MIT"
] | 11 | 2020-06-17T07:30:20.000Z | 2022-03-25T16:56:01.000Z | Model backlog/Inference/91-tweet-inference-5fold-roberta-base-post6.ipynb | dimitreOliveira/Tweet-Sentiment-Extraction | 0a775abe9a92c4bc2db957519c523be7655df8d8 | [
"MIT"
] | null | null | null | Model backlog/Inference/91-tweet-inference-5fold-roberta-base-post6.ipynb | dimitreOliveira/Tweet-Sentiment-Extraction | 0a775abe9a92c4bc2db957519c523be7655df8d8 | [
"MIT"
] | null | null | null | 33.952077 | 149 | 0.412722 | [
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"import json, glob\nfrom tweet_utility_scripts import *\nfrom tweet_utility_preprocess_roberta_scripts import *\nfrom transformers import TFRobertaModel, RobertaConfig\nfrom tokenizers import ByteLevelBPETokenizer\nfrom tensorflow.keras import layers\nfrom tensorflow.keras.models import Model",
"_____no_output_____"
]
],
[
[
"# Load data",
"_____no_output_____"
]
],
[
[
"test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')\n\nprint('Test samples: %s' % len(test))\ndisplay(test.head())",
"Test samples: 3534\n"
]
],
[
[
"# Model parameters",
"_____no_output_____"
]
],
[
[
"input_base_path = '/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/'\nwith open(input_base_path + 'config.json') as json_file:\n config = json.load(json_file)\n\nconfig",
"_____no_output_____"
],
[
"vocab_path = input_base_path + 'vocab.json'\nmerges_path = input_base_path + 'merges.txt'\nbase_path = '/kaggle/input/qa-transformers/roberta/'\nmodel_path_list = glob.glob(input_base_path + '*.h5')\nmodel_path_list.sort()\nprint('Models to predict:')\nprint(*model_path_list, sep = \"\\n\")",
"Models to predict:\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_1.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_2.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_3.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_4.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_5.h5\n"
]
],
[
[
"# Tokenizer",
"_____no_output_____"
]
],
[
[
"tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path, \n lowercase=True, add_prefix_space=True)",
"_____no_output_____"
]
],
[
[
"# Pre process",
"_____no_output_____"
]
],
[
[
"test['text'].fillna('', inplace=True)\ntest[\"text\"] = test[\"text\"].apply(lambda x: x.lower())\ntest[\"text\"] = test[\"text\"].apply(lambda x: x.strip())\n\nx_test = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)",
"_____no_output_____"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)\n\ndef model_fn(MAX_LEN):\n input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')\n attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')\n \n base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name=\"base_model\")\n sequence_output = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})\n last_state = sequence_output[0]\n \n x_start = layers.Dropout(.1)(last_state) \n x_start = layers.Dense(1)(x_start)\n x_start = layers.Flatten()(x_start)\n y_start = layers.Activation('softmax', name='y_start')(x_start)\n\n x_end = layers.Dropout(.1)(last_state) \n x_end = layers.Dense(1)(x_end)\n x_end = layers.Flatten()(x_end)\n y_end = layers.Activation('softmax', name='y_end')(x_end)\n \n model = Model(inputs=[input_ids, attention_mask], outputs=[y_start, y_end])\n \n return model",
"_____no_output_____"
]
],
[
[
"# Make predictions",
"_____no_output_____"
]
],
[
[
"NUM_TEST_IMAGES = len(test)\ntest_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))\ntest_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))\n\nfor model_path in model_path_list:\n print(model_path)\n model = model_fn(config['MAX_LEN'])\n model.load_weights(model_path)\n \n test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE']))\n test_start_preds += test_preds[0] / len(model_path_list)\n test_end_preds += test_preds[1] / len(model_path_list)",
"/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_1.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_2.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_3.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_4.h5\n/kaggle/input/91-tweet-train-5fold-roberta-base-lbl-smooth02/model_fold_5.h5\n"
]
],
[
[
"# Post process",
"_____no_output_____"
]
],
[
[
"test['start'] = test_start_preds.argmax(axis=-1)\ntest['end'] = test_end_preds.argmax(axis=-1)\n\ntest['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)\n\n# Post-process\ntest[\"selected_text\"] = test.apply(lambda x: ' '.join([word for word in x['selected_text'].split() if word in x['text'].split()]), axis=1)\ntest['selected_text'] = test.apply(lambda x: x['text'] if (x['selected_text'] == '') else x['selected_text'], axis=1)\ntest['selected_text'].fillna(test['text'], inplace=True)",
"_____no_output_____"
]
],
[
[
"# Visualize predictions",
"_____no_output_____"
]
],
[
[
"test['text_len'] = test['text'].apply(lambda x : len(x))\ntest['label_len'] = test['selected_text'].apply(lambda x : len(x))\ntest['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))\ntest['label_wordCnt'] = test['selected_text'].apply(lambda x : len(x.split(' ')))\ntest['text_tokenCnt'] = test['text'].apply(lambda x : len(tokenizer.encode(x).ids))\ntest['label_tokenCnt'] = test['selected_text'].apply(lambda x : len(tokenizer.encode(x).ids))\ntest['jaccard'] = test.apply(lambda x: jaccard(x['text'], x['selected_text']), axis=1)\n\ndisplay(test.head(10))\ndisplay(test.describe())",
"_____no_output_____"
]
],
[
[
"# Test set predictions",
"_____no_output_____"
]
],
[
[
"submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')\nsubmission['selected_text'] = test[\"selected_text\"]\nsubmission.to_csv('submission.csv', index=False)\nsubmission.head(10)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71e6dd9c21f814aee255d847359ea4947841ca6 | 238,348 | ipynb | Jupyter Notebook | tabular-playground-fet2021/tabular-playground-feb2021-eda.ipynb | hayatochigi/Kaggle | ea882e25c4df9363142b51c41f651bce74522abd | [
"MIT"
] | 2 | 2021-03-01T07:43:26.000Z | 2021-03-31T22:29:53.000Z | tabular-playground-fet2021/tabular-playground-feb2021-eda.ipynb | hayatochigi/Kaggle | ea882e25c4df9363142b51c41f651bce74522abd | [
"MIT"
] | null | null | null | tabular-playground-fet2021/tabular-playground-feb2021-eda.ipynb | hayatochigi/Kaggle | ea882e25c4df9363142b51c41f651bce74522abd | [
"MIT"
] | null | null | null | 238,348 | 238,348 | 0.955078 | [
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"dataset = pd.read_csv('../input/tabular-playground-series-feb-2021/train.csv')\n\n# Check null data --> There is no null data.\n#print(dataset.isnull().sum())\n\n# category variable\ncategory = ['cat0', 'cat1', 'cat2', 'cat3', 'cat4', 'cat5', 'cat6', 'cat7', 'cat8', 'cat9']\n\n# continuous variable\ncontinuous = ['cont0', 'cont1', 'cont2', 'cont3', 'cont4', 'cont5', 'cont6', \n 'cont7', 'cont8', 'cont9', 'cont10', 'cont11', 'cont12', 'cont13']\n\n#outlier = [ 21248, 65710, 71166, 73335, 105049, 137574, 160168, 166042, 179361, 210708,\n# 240385, 252761, 304677, 306931, 342537, 343575, 373500, 376977, 394279, 438005,\n# 442278, 444922, 456587, 486120]\noutlier = [166042]\n\nfor x in outlier:\n dataset = dataset.loc[dataset['id'] != x, :]\n\n# One-Hot Encoding is better but for feature importance search,\n# use LabelEncoder\nfrom sklearn.preprocessing import LabelEncoder\nencoder = LabelEncoder()\nfor x in category:\n dataset[x] = encoder.fit_transform(dataset[x])\n\n# dataset['target'].plot()\nfig, axes = plt.subplots(nrows=1, ncols=2, figsize=(16,4))\ndataset['target'].hist(bins=50, ax=axes[0])\ndataset['target'].plot(ax=axes[1])\n\n# Calculate Mean and Std value\ntarget_mean = dataset.describe().loc['mean', 'target']\ntarget_std = dataset.describe().loc['std', 'target']\naxes[1].axhline(y=target_mean, color='g')\nup = target_mean+4*target_std\nlow=target_mean-4*target_std\n\n# Draw up and low lines\naxes[1].axhline(y=up, color='b')\naxes[1].axhline(y=low,color='r')\n\n# histogram shows bimodal\n\noutlier = dataset.loc[dataset['target'] < low, 'id'].values\nprint(outlier.shape)\n\n#for x in outlier:\n# dataset = dataset[dataset['id'] != x]\n",
"(23,)\n"
],
[
"X = dataset.drop(columns=['id', 'target'])\ny = dataset['target']\n\nimport xgboost as xgb\nimport shap\n\nxgb_params= {\n \"objective\": \"reg:squarederror\",\n \"max_depth\": 6,\n \"learning_rate\": 0.01,\n \"colsample_bytree\": 0.4,\n \"subsample\": 0.6,\n \"reg_alpha\" : 6,\n \"min_child_weight\": 100,\n \"n_jobs\": 2,\n \"seed\": 2001,\n 'tree_method': \"gpu_hist\"\n }\n\n\nmodel = xgb.XGBRegressor(objective=\"reg:squarederror\", max_depth=6, learning_rate=0.01,\n subsample=0.6, reg_alpha=6, min_child_weight=100, n_jobs=-1, random_state=0)\nmodel.fit(X,y)",
"_____no_output_____"
],
[
"shap.initjs()\nexplainer = shap.TreeExplainer(model=model, feature_perturbation='tree_path_dependent', model_output='raw')\nshap_values = explainer.shap_values(X=X)\nshap.summary_plot(shap_values, X, plot_type='bar')\nshap.summary_plot(shap_values, X)\n#shap.force_plot(base_value=explainer.expected_value, shap_values=shap_values, features=X)\n",
"_____no_output_____"
],
[
"import seaborn as sns\nfrom sklearn.preprocessing import LabelEncoder\nencoder = LabelEncoder()\nplace_holder = dataset\nfor x in category:\n place_holder[x] = encoder.fit_transform(dataset[x].values)\ncorrelation = dataset.corr()\nsns.heatmap(correlation)",
"_____no_output_____"
],
[
"dataset = dataset[dataset['id'] != 166042]\n\nfig, axes = plt.subplots(nrows=2, ncols=5, figsize=(22,8))\nfor i, x in enumerate(category):\n # 一列ごとに\n row = int(i/5)\n col = int(i%5)\n dataset[x].hist(ax=axes[row,col])\n axes[row, col].set_title(x)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=2, ncols=7, figsize=(22,10))\nskew_list = []\nfor i, x in enumerate(continuous):\n # 一列ごとに\n row = int(i/7)\n col = int(i%7)\n sns.distplot(dataset[x], ax=axes[row,col])\n #dataset[x].hist(ax=axes[row,col])\n axes[row,col].set_title(x)",
"/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n/opt/conda/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"fig, axes = plt.subplots(nrows=2, ncols=7, figsize=(20,8))\nfor i, x in enumerate(continuous):\n # 一列ごとに\n row = int(i/7)\n col = int(i%7)\n dataset.plot.scatter(x=x, y='target',ax=axes[row,col])\n axes[row,col].set_title(x)",
"_____no_output_____"
],
[
"features = ['cat1', 'cont11', 'cont13', 'cont0', 'cont2', 'cat8', 'cont8', 'cont9', 'cat3', 'cat6', 'cat5']\nfig, axes = plt.subplots(nrows=3, ncols=4, figsize=(20,8))\nfor i, x in enumerate(features):\n row = int(i/4)\n col = int(i%4)\n dataset[x].hist(ax=axes[row,col], bins=50)\n axes[row,col].set_title(x)",
"_____no_output_____"
],
[
"from sklearn.decomposition import PCA\npca = PCA(n_components=2)\npca.fit(dataset)\npca_result = pca.transform(dataset)\n\nfig = plt.figure(figsize=(6,4))\nplt.scatter(pca_result[:,0], pca_result[:,1], alpha=0.8)",
"_____no_output_____"
],
[
"cluster_feat = ['cat1', 'cont11', 'cont13', 'cont0']\nfeatures_val = dataset.loc[:,cluster_feat]\nn_components=4\n\ndef draw_cluster(features_val, pred):\n for i, x in enumerate(cluster_feat):\n fig = plt.figure(figsize=(5,4))\n dataA = features_val.loc[pred == 1, x]\n dataB = features_val.loc[pred == 0, x]\n plt.hist(dataA,bins=10)\n plt.hist(dataB, bins=10)\n\n# Gaussian Mixture\nfrom sklearn.mixture import GaussianMixture\ngm = GaussianMixture(n_components=n_components, random_state=0)\ngm.fit(features_val)\npred = (gm.predict(features_val) > 0.1)*1\n#draw_cluster(features_val, pred)\n\n \nfrom sklearn.cluster import KMeans\nkmeans = KMeans(n_clusters=n_components, random_state=0)\nkmeans.fit(features_val)\npred = (kmeans.predict(features_val) > 0.1)*1\nprint(pred)\ndraw_cluster(features_val, pred)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71e6f194c489802a9f45396db649129135e95ae | 297,937 | ipynb | Jupyter Notebook | examples/Toy_segmentation/simple_dltk_unet_multigpu.ipynb | mseitzer/DLTK | 3237aa6c7ed63aa177ca90eafcc076d144155a34 | [
"Apache-2.0"
] | null | null | null | examples/Toy_segmentation/simple_dltk_unet_multigpu.ipynb | mseitzer/DLTK | 3237aa6c7ed63aa177ca90eafcc076d144155a34 | [
"Apache-2.0"
] | null | null | null | examples/Toy_segmentation/simple_dltk_unet_multigpu.ipynb | mseitzer/DLTK | 3237aa6c7ed63aa177ca90eafcc076d144155a34 | [
"Apache-2.0"
] | null | null | null | 1,168.380392 | 146,602 | 0.941102 | [
[
[
"%matplotlib inline\nfrom __future__ import division, print_function\nimport sys,os\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nimport tensorflow as tf\n\nsys.path.append(os.path.join(os.getcwd(),'../..'))\n\nimport dltk.core.modules as modules\nfrom dltk.models.segmentation.unet import ResUNET\nfrom dltk.models.segmentation.fcn import ResNetFCN\nfrom dltk.core.modules import dice_loss\n\nplt.rcParams['image.cmap'] = 'gray'\n\nimwidth=256\nimheight=256\nbatchSize=5\ntrainSteps=200\ngpusets=(0,1) # GPU numbers to use ie. for devices /gpu:0 and /gpu:1, (0,) behaves like single GPU",
"_____no_output_____"
],
[
"def createTestImage(width,height,numObjs=12,radMax=30):\n '''Return a noisy 2D image with `numObj' circles and a 2D mask image.'''\n image=np.zeros((width,height))\n \n for i in range(numObjs):\n x=np.random.randint(radMax,width-radMax)\n y=np.random.randint(radMax,height-radMax)\n rad=np.random.randint(10,radMax)\n spy,spx = np.ogrid[-x:width-x, -y:height-y]\n circle=(spx*spx+spy*spy)<=rad*rad\n image[circle]=np.random.random()*0.5+0.5\n \n norm=np.random.uniform(0,0.25,size=image.shape)\n \n return np.maximum(image,norm),(image>0).astype(float)\n\n\ndef plotPair(im1,im2):\n '''Convenience function for plotting two images side-by-side.'''\n fig, ax = plt.subplots(1, 2, figsize=(10,5))\n ax[0].imshow(im1)\n ax[1].imshow(im2)\n \nim,mask=createTestImage(imwidth,imheight)\nplotPair(im,mask)",
"_____no_output_____"
],
[
"def averageGradients(tower_grads):\n '''This averages the tower gradients into one, copied directly from the Tensorflow MNIST example.'''\n average_grads = []\n for grad_and_vars in zip(*tower_grads):\n # Note that each grad_and_vars looks like the following:\n # ((grad0_gpu0, var0_gpu0), ... , (grad0_gpuN, var0_gpuN))\n grads = []\n for g, _ in grad_and_vars:\n if g is not None:\n # Add 0 dimension to the gradients to represent the tower.\n expanded_g = tf.expand_dims(g, 0)\n\n # Append on a 'tower' dimension which we will average over below.\n grads.append(expanded_g)\n\n # Average over the 'tower' dimension.\n grad = tf.concat(axis=0, values=grads)\n grad = tf.reduce_mean(grad, 0)\n\n # Keep in mind that the Variables are redundant because they are shared\n # across towers. So .. we will just return the first tower's pointer to\n # the Variable.\n v = grad_and_vars[0][1]\n grad_and_var = (grad, v)\n average_grads.append(grad_and_var)\n \n return average_grads\n\n\ndef fillTrainData(xt,yt):\n for i in range(batchSize):\n im,mask=createTestImage(imwidth,imheight)\n xt[i,:,:,0]=im\n yt[i,:,:]=mask\n \n \nwith tf.Graph().as_default(), tf.device('/cpu:0'):\n opt=tf.train.AdamOptimizer(1e-4)\n global_step = tf.get_variable('global_step', [], initializer=tf.constant_initializer(0), trainable=False)\n \n grads=[]\n xytrain=[]\n losses=[]\n feeddict={}\n \n xtrain=np.zeros((batchSize,imwidth,imheight,1))\n ytrain=np.zeros((batchSize,imwidth,imheight))\n\n # plug & play network definition:\n # net = ResNetFCN(num_classes=1,num_residual_units=3,\n # filters=(16, 32, 64),\n # strides=((1,1), (2,2), (2,2)))\n\n net = ResUNET(num_classes=1,num_residual_units=3,\n filters=(16, 32, 64),\n strides=((1,1), (2,2), (2,2)))\n \n with tf.variable_scope(tf.get_variable_scope()):\n for g in gpusets:\n with tf.device('/gpu:%i'%g),tf.name_scope('tower_%i'%g) as scope:\n xt=np.zeros(xtrain.shape)\n yt=np.zeros(ytrain.shape)\n x=tf.placeholder(tf.float32, xtrain.shape)\n y_=tf.placeholder(tf.int32, ytrain.shape)\n feeddict[x]=xt\n feeddict[y_]=yt\n xytrain.append((xt,yt)) \n\n y = net(x)['logits']\n loss = dice_loss(y, y_, 1)\n\n grad=opt.compute_gradients(loss)\n grads.append(grad)\n losses.append(loss)\n \n avggrads=averageGradients(grads)\n \n trainop = opt.apply_gradients(avggrads, global_step=global_step)\n\n soft_config = tf.ConfigProto(allow_soft_placement=True)\n soft_config.gpu_options.allow_growth = True\n soft_config.log_device_placement=True\n \n with tf.Session(config=soft_config) as sess:\n sess.run(tf.global_variables_initializer())\n \n # perform training steps\n for step in range(trainSteps):\n for xt,yt in xytrain:\n fillTrainData(xt,yt)\n \n trainop.run(feed_dict=feeddict)\n \n # evaluate result with new generated data\n fillTrainData(xtrain,ytrain)\n x=tf.placeholder(tf.float32, xtrain.shape)\n ypred=net(x, is_training=False)['y_']\n test=ypred.eval(feed_dict={x:xtrain})\n \n # plot prediction\n plotPair(np.squeeze(xtrain[0]),test[0]) ",
"INFO:tensorflow:(5, 256, 256, 16)\nINFO:tensorflow:feat_scale_1 shape (5, 128, 128, 32)\nINFO:tensorflow:feat_scale_2 shape (5, 64, 64, 64)\nINFO:tensorflow:Building upsampling for scale 1 with x ([5, 64, 64, 64]) x_up ([5, 128, 128, 32]) stride ((2, 2))\nINFO:tensorflow:up_1 shape (5, 128, 128, 32)\nINFO:tensorflow:Building upsampling for scale 0 with x ([5, 128, 128, 32]) x_up ([5, 256, 256, 16]) stride ((2, 2))\nINFO:tensorflow:up_0 shape (5, 256, 256, 16)\nINFO:tensorflow:last conv shape (5, 256, 256, 1)\nINFO:tensorflow:(5, 256, 256, 16)\nINFO:tensorflow:feat_scale_1 shape (5, 128, 128, 32)\nINFO:tensorflow:feat_scale_2 shape (5, 64, 64, 64)\nINFO:tensorflow:Building upsampling for scale 1 with x ([5, 64, 64, 64]) x_up ([5, 128, 128, 32]) stride ((2, 2))\nINFO:tensorflow:up_1 shape (5, 128, 128, 32)\nINFO:tensorflow:Building upsampling for scale 0 with x ([5, 128, 128, 32]) x_up ([5, 256, 256, 16]) stride ((2, 2))\nINFO:tensorflow:up_0 shape (5, 256, 256, 16)\nINFO:tensorflow:last conv shape (5, 256, 256, 1)\nINFO:tensorflow:(5, 256, 256, 16)\nINFO:tensorflow:feat_scale_1 shape (5, 128, 128, 32)\nINFO:tensorflow:feat_scale_2 shape (5, 64, 64, 64)\nINFO:tensorflow:Building upsampling for scale 1 with x ([5, 64, 64, 64]) x_up ([5, 128, 128, 32]) stride ((2, 2))\nINFO:tensorflow:up_1 shape (5, 128, 128, 32)\nINFO:tensorflow:Building upsampling for scale 0 with x ([5, 128, 128, 32]) x_up ([5, 256, 256, 16]) stride ((2, 2))\nINFO:tensorflow:up_0 shape (5, 256, 256, 16)\nINFO:tensorflow:last conv shape (5, 256, 256, 1)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
e71e84036cfaa21553770395f28f2fc753d88f7a | 25,437 | ipynb | Jupyter Notebook | docs/derivations.ipynb | davidreissmello/error_propagation | ce6600498dabfcf494243edd3a40557dbe8dc709 | [
"MIT"
] | 4 | 2021-09-14T02:21:06.000Z | 2021-11-11T15:33:08.000Z | docs/derivations.ipynb | davidreissmello/error_propagation | ce6600498dabfcf494243edd3a40557dbe8dc709 | [
"MIT"
] | null | null | null | docs/derivations.ipynb | davidreissmello/error_propagation | ce6600498dabfcf494243edd3a40557dbe8dc709 | [
"MIT"
] | null | null | null | 21.648511 | 180 | 0.462594 | [
[
[
"import numpy as np \nimport math \nfrom sympy import *",
"_____no_output_____"
]
],
[
[
"General formula for computing error: \nerror^2 = sum of [(partial derivative of f in respect to var)^2*(error of var)^2 for each var]",
"_____no_output_____"
]
],
[
[
"# Derivation of error for basic operators",
"_____no_output_____"
]
],
[
[
"a,b, a_error, b_error = symbols ('a b a_error b_error')",
"_____no_output_____"
],
[
"print(\"sum\")\nexpr_sum = a+b\nerror_sum = sqrt(diff(expr_sum, a)**2 * a_error**2 + diff(expr_sum, b)**2 * b_error**2)\nerror_sum",
"sum\n"
],
[
"print(\"subtraction\")\nexpr_subtraction = a-b\nerror_subtraction = sqrt(diff(expr_subtraction, a)**2 * a_error**2 + diff(expr_subtraction, b)**2 * b_error**2)\nerror_subtraction",
"subtraction\n"
],
[
"print(\"multiplication\")\nexpr_multiplication = a*b\nerror_multiplication = sqrt(diff(expr_multiplication, a)**2 * a_error**2 + diff(expr_multiplication, b)**2 * b_error**2)\nerror_multiplication",
"multiplication\n"
],
[
"print(\"division\")\nexpr_division = a/b\nerror_division = sqrt(diff(expr_division, a)**2 * a_error**2 + diff(expr_division, b)**2 * b_error**2)\nerror_division",
"division\n"
],
[
"print(\"power\")\nexpr_power = a**b\nerror_power = sqrt(diff(expr_power, a)**2 * a_error**2 + diff(expr_power, b)**2 * b_error**2)\nerror_power",
"power\n"
],
[
"print(\"log\")\nexpr_log = log(a,b)\nerror_log = sqrt(diff(expr_log, a)**2 * a_error**2 + diff(expr_log, b)**2 * b_error**2)\nerror_log",
"log\n"
],
[
"print(\"sin\")\nexpr_sin = sin(a)\nerror_sin = sqrt(diff(expr_sin, a)**2 * a_error**2)\nerror_sin",
"sin\n"
],
[
"print(\"cos\")\nexpr_cos = cos(a)\nerror_cos = sqrt(diff(expr_cos, a)**2 * a_error**2)\nerror_cos",
"cos\n"
],
[
"print(\"tan\")\nexpr_tan = tan(a)\nerror_tan = sqrt(diff(expr_tan, a)**2 * a_error**2)\nerror_tan",
"tan\n"
]
],
[
[
"# Derivation of more complex formulas",
"_____no_output_____"
]
],
[
[
"print(\"acos\")\nexp = acos(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"acos\n"
],
[
"print(\"acosh\")\nexp = acosh(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"acosh\n"
],
[
"print(\"acot\")\nexp = acot(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"acot\n"
],
[
"print(\"acoth\")\nexp = acoth(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"acoth\n"
],
[
"print(\"asin\")\nexp = asin(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"asin\n"
],
[
"print(\"asin\")\nexp = asinh(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"asin\n"
],
[
"print(\"atan\")\nexp = atan(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"atan\n"
],
[
"print(\"atanh\")\nexp = atanh(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"atanh\n"
],
[
"print(\"cosh\")\nexp = cosh(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"cosh\n"
],
[
"print(\"cot\")\nexp = cot(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"cot\n"
],
[
"print(\"coth\")\nexp = coth(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"coth\n"
],
[
"print(\"csc\")\nexp = csc(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"csc\n"
],
[
"print(\"csch\")\nexp = csch(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"csch\n"
],
[
"print(\"factorial\")\nexp = factorial(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"factorial\n"
],
[
"print(\"sec\")\nexp = sec(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"sec\n"
],
[
"print(\"sec\")\nexp = sech(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"sec\n"
],
[
"print(\"sinh\")\nexp = sinh(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"sinh\n"
],
[
"print(\"tanh\")\nexp = tanh(a)\nerror = sqrt(diff(exp, a)**2 * a_error**2)\nerror",
"tanh\n"
]
],
[
[
"# Creating functions",
"_____no_output_____"
]
],
[
[
"def sum_error(a, b, a_error, b_error):\n return np.sqrt(a_error**2 + b_error**2)\n\ndef subtraction_error(a, b, a_error, b_error):\n return sum_error(a, b, a_error, b_error)\n\ndef multiplication_error(a, b, a_error, b_error):\n return np.sqrt((a**2 * b_error**2) + (b**2 * a_error **2))\n\ndef division_error(a, b, a_error, b_error):\n return np.sqrt(((a**2 * b_error**2)/ (b**4)) + (a_error**2/b**2))\n\ndef power_error(a, b, a_error, b_error):\n return np.sqrt((a**(2*b) * b_error**2 * np.log(a)**2) + ((a**(2*b) * a_error**2 * b**2)/a**2))\n\ndef log_error(a, b, a_error, b_error):\n return np.sqrt((b_error**2 * np.log(a)**2)/(b**2 * np.log(b)**4) + a_error**2/(a**2 * np.log(b)**2))\n\ndef sin_error(a, b, a_error, b_error):\n return np.sqrt(a_error**2 * np.cos(a)**2)\n\ndef cos_error(a, b, a_error, b_error):\n return np.sqrt(a_error**2 * np.sin(a)**2)\n\ndef tan_error(a, b, a_error, b_error):\n return np.sqrt(a_error**2 * (np.tan(a)**2 + 1)**2)",
"_____no_output_____"
]
],
[
[
"# Testing of error formulas for basic operators",
"_____no_output_____"
]
],
[
[
"values = {a: 10, a_error:5, b:8, b_error:3}",
"_____no_output_____"
]
],
[
[
"Derived errors",
"_____no_output_____"
]
],
[
[
"print(\"sum =\", error_sum.evalf(subs=values))\nprint(\"subtraction =\", error_subtraction.evalf(subs=values))\nprint(\"multiplication =\", error_multiplication.evalf(subs=values))\nprint(\"division =\", error_division.evalf(subs=values))\nprint(\"power =\", error_power.evalf(subs=values))\nprint(\"log =\", error_log.evalf(subs=values))\nprint(\"sin =\", error_sin.evalf(subs=values))\nprint(\"cos =\", error_cos.evalf(subs=values))\nprint(\"tan =\", error_tan.evalf(subs=values))",
"sum = 5.83095189484530\nsubtraction = 5.83095189484530\nmultiplication = 50.0000000000000\ndivision = 0.781250000000000\npower = 798229810.232026\nlog = 0.312556216389312\nsin = 4.19535764538226\ncos = 2.72010555444685\ntan = 7.10185881291716\n"
]
],
[
[
"Errors from functions",
"_____no_output_____"
]
],
[
[
"a = 10\nb = 8\na_error = 5\nb_error = 3",
"_____no_output_____"
],
[
"print(\"sum =\", sum_error(a, b, a_error, b_error))\nprint(\"subtraction =\", subtraction_error(a, b, a_error, b_error))\nprint(\"multiplication =\", multiplication_error(a, b, a_error, b_error))\nprint(\"division =\", division_error(a, b, a_error, b_error))\nprint(\"power =\", power_error(a, b, a_error, b_error))\nprint(\"log =\", log_error(a, b, a_error, b_error))\nprint(\"sin =\", sin_error(a, b, a_error, b_error))\nprint(\"cos =\", cos_error(a, b, a_error, b_error))\nprint(\"tan =\", tan_error(a, b, a_error, b_error))",
"sum = 5.830951894845301\nsubtraction = 5.830951894845301\nmultiplication = 50.0\ndivision = 0.78125\npower = 798229810.2320257\nlog = 0.3125562163893125\nsin = 4.195357645382262\ncos = 2.7201055544468495\ntan = 7.101858812917159\n"
]
]
] | [
"code",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"raw"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e71e97e5b4c0e4e3b25f00e556b54e39de69ac6b | 4,255 | ipynb | Jupyter Notebook | introduction/javaIntroduction.ipynb | CCRCFISAT/Acharya-JavaProgramming | f0862938afdcac9aff94c5650581f58256c6a785 | [
"BSD-3-Clause"
] | null | null | null | introduction/javaIntroduction.ipynb | CCRCFISAT/Acharya-JavaProgramming | f0862938afdcac9aff94c5650581f58256c6a785 | [
"BSD-3-Clause"
] | null | null | null | introduction/javaIntroduction.ipynb | CCRCFISAT/Acharya-JavaProgramming | f0862938afdcac9aff94c5650581f58256c6a785 | [
"BSD-3-Clause"
] | 1 | 2020-10-26T16:50:16.000Z | 2020-10-26T16:50:16.000Z | 23.508287 | 278 | 0.580024 | [
[
[
"# Java",
"_____no_output_____"
],
[
"Java is a high-level programming language developed by Sun Microsystems. It was originally designed for developing programs for set-top boxes and handheld devices, but later became a popular choice for creating web applications.",
"_____no_output_____"
],
[
"The Java syntax is similar to C++, but is strictly an object-oriented programming language.",
"_____no_output_____"
],
[
"## Object oriented programming",
"_____no_output_____"
],
[
"<img src=\"img/1.png\" width=\"300px\" height=\"360px\">",
"_____no_output_____"
],
[
"It is a computer programming model that organizes software design around data, or objects, rather than functions and logic. That is we use class and its objects to do most of the things. We will learn more about class and objects in java in the coming days.",
"_____no_output_____"
],
[
"## Java features",
"_____no_output_____"
],
[
"Now let us checkout some of the features of `Java`",
"_____no_output_____"
],
[
"Java is :\n* Object Oriented\n* Simple\n* Secured\n* Platform independent\n* Robust\n* Portable\n* Architecture neutral\n* Dynamic\n* Interpreted\n* High performance\n* Multithreaded\n* Distributed",
"_____no_output_____"
],
[
"<img src=\"img/2.png\" width=\"400px\" height=\"400px\">",
"_____no_output_____"
],
[
"## Java Virtual Machine",
"_____no_output_____"
],
[
"`Java Virtual Machine (JVM)` is a engine that provides runtime environment to drive the Java Code or applications (run java programs). It converts Java bytecode into machines language the language that the computer understands. JVM is a part of Java Run Environment (JRE).",
"_____no_output_____"
],
[
"Now let us what happens in java virtual machine when we execute a program through an image.",
"_____no_output_____"
],
[
"<img src=\"img/3.jpg\" width=\"650\" height=\"100\">",
"_____no_output_____"
],
[
"That is when we execute our `java` program, the java compiler converts the program into a `.class` file and this file is interpreted by the interpreter and finally we gets our output.",
"_____no_output_____"
],
[
"## Main fields of application",
"_____no_output_____"
],
[
"### Mobile Applications",
"_____no_output_____"
],
[
"Java is considered as the official programming language for mobile app development.",
"_____no_output_____"
],
[
"### Desktop GUI Applications",
"_____no_output_____"
],
[
"All desktop applications can easily be developed in Java. Java also provides GUI development capability through various means mainly Abstract Windowing Toolkit (AWT), Swing and JavaFX.",
"_____no_output_____"
],
[
"***Java is a great programming language and its scope is not going to end in the near future.*",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e71ea8e33c1b4a3f1ed5b839ec92c1a650ca4852 | 34,351 | ipynb | Jupyter Notebook | comparisons.ipynb | erikfredner/hawthorne | 0315ffbe597aa021e2ffe5814ee81d39d095ad36 | [
"MIT"
] | 1 | 2020-11-26T12:23:33.000Z | 2020-11-26T12:23:33.000Z | comparisons.ipynb | erikfredner/hawthorne | 0315ffbe597aa021e2ffe5814ee81d39d095ad36 | [
"MIT"
] | null | null | null | comparisons.ipynb | erikfredner/hawthorne | 0315ffbe597aa021e2ffe5814ee81d39d095ad36 | [
"MIT"
] | 1 | 2020-11-24T23:05:27.000Z | 2020-11-24T23:05:27.000Z | 26.102584 | 149 | 0.513522 | [
[
[
"# Corpus comparisons\nEvaluating the difference Hawthorne's work makes between the corpora.",
"_____no_output_____"
],
[
"## Mean sentence length\nThis exemplifies how similar Hawthorne is to the rest of the corpus, as well as his small impact on corpus-wide measures.",
"_____no_output_____"
]
],
[
[
"import nltk\nimport random\nimport statistics\nimport re\nimport os\nimport scipy",
"_____no_output_____"
]
],
[
[
"### Hawthorne's sentences\nFor this test, I use the Library of America Hawthorne, as it is better transcribed than Gale.",
"_____no_output_____"
]
],
[
[
"nh_loa = '/Users/e/code/hawthorne/local/loa_nh/loa_hawthorne_all.txt'",
"_____no_output_____"
],
[
"def get_sent_lens(file):\n with open(file) as f:\n text = f.read()\n sents = nltk.tokenize.sent_tokenize(text)\n \n sent_lens = []\n\n for sent in sents:\n n_toks = len([x for x in re.split('\\s+', sent) if x])\n sent_lens.append(n_toks)\n \n return sent_lens",
"_____no_output_____"
],
[
"nh_sents = get_sent_lens(nh_loa)",
"_____no_output_____"
],
[
"statistics.mean(nh_sents)",
"_____no_output_____"
],
[
"statistics.quantiles(nh_sents)",
"_____no_output_____"
]
],
[
[
"# Hawthorne's peers' sentences\nComparing Hawthorne's sentence lengths against those of his contemporaries:",
"_____no_output_____"
]
],
[
[
"comparison = '/Users/e/code/hawthorne/local/corpus_no_nh'",
"_____no_output_____"
],
[
"files = [os.path.join(comparison, x) for x in os.listdir(comparison) if x.endswith('.txt')]",
"_____no_output_____"
],
[
"random.shuffle(files) # randomize sample\nsample_size = round(len(files) * 0.1) # get n for 10% sample\nsample = files[:sample_size]",
"_____no_output_____"
],
[
"sent_lens = []\n\nfor i, file in enumerate(sample):\n sent_lens.extend(get_sent_lens(file))\n \n if i % 25 == 0:\n print('\\r{} of {}'.format(i, len(sample)), end = '')",
"_____no_output_____"
],
[
"statistics.mean(sent_lens)",
"_____no_output_____"
],
[
"statistics.quantiles(sent_lens)",
"_____no_output_____"
]
],
[
[
"# Word Vectors\nHere, I use the [hyperhyper](https://github.com/jfilter/hyperhyper) implementation of the SVD PPMI method of creating word vectors.",
"_____no_output_____"
]
],
[
[
"import hyperhyper as hy",
"_____no_output_____"
]
],
[
[
"## Hawthorne",
"_____no_output_____"
]
],
[
[
"nh_corpus = '/Users/e/code/hawthorne/local/gale_nh'",
"_____no_output_____"
],
[
"# generate\n# corpus = hy.Corpus.from_text_files(nh_corpus, preproc_func = hy.tokenize_texts, keep_n = 5000)",
"build up vocab: 100%|██████████| 23/23 [00:03<00:00, 7.54it/s]\ntexts to ids: 100%|██████████| 23/23 [00:02<00:00, 10.55it/s]\n"
],
[
"# generate\n# bunch = hy.Bunch(\"results/hy_hawthorne\", corpus, force_overwrite = True)",
"_____no_output_____"
],
[
"# load\nbunch = hy.Bunch('results/hy_hawthorne/')",
"_____no_output_____"
],
[
"vectors_nh, results_nh = bunch.svd(keyed_vectors = True, subsample = None)",
"_____no_output_____"
],
[
"vectors_nh.most_similar('hester')",
"_____no_output_____"
],
[
"vectors_nh.most_similar('pearl')",
"_____no_output_____"
]
],
[
[
"## Gale 1828-1864 including NH",
"_____no_output_____"
]
],
[
[
"path = '/Users/e/code/hawthorne/local/corpus'",
"_____no_output_____"
],
[
"# generate\n# corpus = hy.Corpus.from_text_files(path, preproc_func = hy.tokenize_texts, keep_n = 10000)\n\n# load\ncorpus = hy.Corpus.load('results/corpus_subsample_none/corpus.pkl')",
"_____no_output_____"
],
[
"# generate\n# bunch = hy.Bunch(\"results/corpus_subsample_none\", corpus, force_overwrite = True)\n\n# load\nbunch = hy.Bunch('results/corpus_subsample_none//')",
"_____no_output_____"
],
[
"vectors, results = bunch.svd(keyed_vectors = True, subsample = None)",
"_____no_output_____"
],
[
"vectors.most_similar('puritan')",
"_____no_output_____"
],
[
"vectors.most_similar('hester')",
"_____no_output_____"
]
],
[
[
"## Gale 1828-1864 excluding NH",
"_____no_output_____"
]
],
[
[
"path = '/Users/e/code/hawthorne/local/corpus_no_nh'",
"_____no_output_____"
],
[
"# generate\n# corpus_no_nh = hy.Corpus.from_text_files(path, preproc_func = hy.tokenize_texts, keep_n = 10000)\n\n# load\ncorpus_no_nh = hy.Corpus.load('results/corpus_no_nh_subsample_none/corpus.pkl')",
"_____no_output_____"
],
[
"# generate\n# bunch = hy.Bunch(\"results/corpus_no_nh_subsample_none\", corpus, force_overwrite=True)\n\n# load\nbunch = hy.Bunch('results/corpus_no_nh_subsample_none//')",
"_____no_output_____"
],
[
"vectors_no_nh, results_no_nh = bunch.svd(keyed_vectors = True)",
"_____no_output_____"
],
[
"vectors_no_nh.most_similar('puritan')",
"_____no_output_____"
],
[
"vectors_no_nh.most_similar('hester')",
"_____no_output_____"
]
],
[
[
"# Comparing vectors across models\nIn order to compare across models, we take aggregate the differences of the cosine similarities between every vector in the model.\n\nBecause SVDs of PPMI are deterministic, differences between the two models are attributable to the presence or absence of Hawthorne's work.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom multiprocessing import Pool\nimport pickle\nimport time\nimport os",
"_____no_output_____"
],
[
"def get_vocab(c):\n '''\n Gets vocabulary from hyperhyper corpus object, c.\n '''\n return set([x for x in c.vocab.values()])",
"_____no_output_____"
],
[
"def get_shared_vocab(c1, c2):\n '''\n Returns shared vocab between hyperhyper corpus objects.\n '''\n return list(get_vocab(c1) & get_vocab(c2))",
"_____no_output_____"
],
[
"def diff_vector(word, v1, v2, vocab = None, topn = 10000, pkl = False):\n '''\n Takes hyperhyper vector objects and returns the difference between those vectors as a dict.\n '''\n # get cosine similarities\n sim1 = v1.most_similar(word, topn = topn)\n sim2 = v2.most_similar(word, topn = topn)\n\n # calculate differences\n sim1 = pd.Series(data = [x[1] for x in sim1], index = [x[0] for x in sim1])\n sim2 = pd.Series(data = [x[1] for x in sim2], index = [x[0] for x in sim2])\n diff = sim1 - sim2\n total = diff.abs().sum()\n \n # optionally pickle output\n if pkl:\n d = {'word' : word,\n 'abs_diff' : total,\n 'sim1' : sim1,\n 'sim2' : sim2}\n \n fn = str(round(time.time())) + '_' + word + '.pkl'\n path = os.path.join('results/vec_diffs', fn)\n\n with open(path, 'wb') as f:\n pickle.dump(d, f)\n f.close()\n print('\\r{} pickled at {}'.format(word, path))\n \n # return observation\n d = {'word' : word,\n 'abs_diff' : total}\n\n return d",
"_____no_output_____"
],
[
"# linear (takes 5-10 minutes)\n\ndef diff_vectors(v1, v2, vocab):\n '''\n Takes hyperhyper vector objects (v1, v2), and calculates the absolute difference between them.\n \n Corpus objects (c1, c2) used to extract vocabularies and calculate intersection.\n '''\n \n l = []\n \n for i, word in enumerate(vocab):\n l.append(diff_vector(word, v1, v2))\n if i % 100 == 0:\n pct = round((i / len(vocab)) * 100)\n print('\\r{}%'.format(pct), end = '')\n \n # pickle\n fn = str(round(time.time())) + '_diff_vectors.pkl'\n path = os.path.join('results/vec_diffs', fn)\n \n with open(path, 'wb') as f:\n pickle.dump(l, f)\n f.close()\n print('\\rpickle: {}'.format(path))\n \n return l",
"_____no_output_____"
],
[
"vocab = get_shared_vocab(corpus, corpus_no_nh)",
"_____no_output_____"
],
[
"# generate\ndiffs = diff_vectors(vectors, vectors_no_nh, vocab)",
"pickle: results/vec_diffs/1605914440_diff_vectors.pkl\n"
]
],
[
[
"# Munge results",
"_____no_output_____"
]
],
[
[
"# load\ndiffs = pickle.load(open('/Users/e/code/hawthorne/results/vec_diffs/1605914440_diff_vectors.pkl', 'rb'))",
"_____no_output_____"
],
[
"df = pd.DataFrame(diffs)",
"_____no_output_____"
],
[
"df['abs_diff'].describe()",
"_____no_output_____"
],
[
"# Add corpus raw frequency to output\nd = {v : corpus.counts[k] for k,v in corpus.vocab.items()}\ne = {v : corpus_no_nh.counts[k] for k,v in corpus_no_nh.vocab.items()}",
"_____no_output_____"
],
[
"# Make pandas objects\nds = pd.Series(d)\nds.name = '# Gale'\nes = pd.Series(e)\nes.name = '# Gale - NH'",
"_____no_output_____"
],
[
"counts = pd.merge(ds, es, left_on = ds.index, right_on = es.index)",
"_____no_output_____"
],
[
"counts.rename(columns = {'key_0':'word'}, inplace=True)",
"_____no_output_____"
],
[
"df = pd.merge(df, counts, on = 'word').set_index('word')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"Hawthorne accounts for about 1 million words (`1062724`) in the model:",
"_____no_output_____"
]
],
[
[
"nh_n = df['# Gale'].sum() - df['# Gale - NH'].sum()",
"_____no_output_____"
],
[
"nh_n",
"_____no_output_____"
]
],
[
[
"In percentage terms:",
"_____no_output_____"
]
],
[
[
"'{}%'.format(round((nh_n / df['# Gale'].sum()) * 100, 3))",
"_____no_output_____"
],
[
"# Per-word proportion\nobserved = 1 - (df['# Gale - NH'] / df['# Gale'])\nexpected = nh_n / df['# Gale'].sum()\ndf['obs / exp'] = observed / expected\n\n# Create status for obs/exp for plotting\ndf['o/e labels'] = ['more' if x > 1 else 'less' for x in df['obs / exp']]",
"_____no_output_____"
],
[
"# adapted from David Bamman's anlp19\nimport operator\n\ndef chi_square(one_counts, two_counts):\n\n one_sum=0.\n two_sum=0.\n vocab={}\n for word in one_counts.index:\n one_sum+=one_counts[word]\n vocab[word]=1\n for word in two_counts.index:\n vocab[word]=1\n two_sum+=two_counts[word]\n\n N=one_sum+two_sum\n vals={}\n \n for word in vocab:\n O11=one_counts[word]\n O12=two_counts[word]\n O21=one_sum-one_counts[word]\n O22=two_sum-two_counts[word]\n \n # We'll use the simpler form given in Manning and Schuetze (1999) \n # for 2x2 contingency tables: \n # https://nlp.stanford.edu/fsnlp/promo/colloc.pdf, equation 5.7\n \n vals[word]=(N*(O11*O22 - O12*O21)**2)/((O11 + O12)*(O11+O21)*(O12+O22)*(O21+O22))\n \n sorted_chi = sorted(vals.items(), key=operator.itemgetter(1), reverse=True)\n one=[]\n two=[]\n \n for k,v in sorted_chi:\n if one_counts[k]/one_sum > two_counts[k]/two_sum:\n one.append(k)\n else:\n two.append(k)\n \n return one, vals",
"_____no_output_____"
],
[
"dws, vals = chi_square(df['# Gale'], df['# Gale - NH'])",
"_____no_output_____"
],
[
"# per chi2\ndf['nh_distinctive'] = df.index.isin(dws)",
"_____no_output_____"
],
[
"df['chi2'] = pd.Series(vals)",
"_____no_output_____"
],
[
"df['n_diff'] = df['# Gale'] - df['# Gale - NH']",
"_____no_output_____"
],
[
"# add stopword metadata\nstopwords = nltk.corpus.stopwords.words('english')\ndf['stopword'] = df.index.isin(stopwords)",
"_____no_output_____"
],
[
"# dictionary validation\nwith open('/Users/e/Documents/Literary Lab/word lists/oed_wordlist.txt', 'r') as f:\n oed_words = [x for x in f.read().split('\\n') if x]",
"_____no_output_____"
],
[
"df['dict_word'] = df.index.isin(oed_words)",
"_____no_output_____"
]
],
[
[
"This is the dataset used to produce graphs in Tableau:",
"_____no_output_____"
]
],
[
[
"df.to_csv('/Users/e/code/hawthorne/hawthorne_diffs.csv')",
"_____no_output_____"
]
],
[
[
"# Evaluating dissimilarity by rank change\nHow does Hawthorne re-rank vector similarities?",
"_____no_output_____"
]
],
[
[
"# selected from Tableau\nkeywords = ['likewise', 'puritan', 'minister', 'artist', 'substance', 'methinks', 'whether', 'lifetime',\n 'world', 'mankind', 'pilgrims', 'fling', 'province', 'painter', 'brotherhood', 'clergyman',\n 'artist', 'fireside', 'might', 'hither', 'reverend', 'whatever', 'merely', 'fireside', 'antique',\n 'wrought', 'airy', 'discern', 'shadow', 'image', 'actual', 'flung', 'dusky']",
"_____no_output_____"
],
[
"def rank_similarities(word, vectors):\n c = 1\n d = {}\n \n for x in vectors.most_similar(word, topn=10000):\n d[x[0]] = c\n c += 1\n \n return d",
"_____no_output_____"
],
[
"def get_rank_change(word, v1, v2, topn = 20, spearman = True):\n d = rank_similarities(word, v1)\n e = rank_similarities(word, v2)\n data = pd.concat([pd.Series(d), pd.Series(e)], axis = 1)\n data.columns = ['v1_rank', 'v2_rank']\n data['rank_change'] = data['v1_rank'] - data['v2_rank']\n # filter\n data.dropna(axis=0, inplace=True)\n subset = data[data['v1_rank'] <= topn]\n subset.sort_values('rank_change', ascending = False, inplace = True)\n # write\n if spearman:\n sp = stats.spearmanr(subset['v1_rank'], subset['v2_rank'])[0]\n fp = '/Users/e/code/hawthorne/results/rank_changes'\n fp = os.path.join(fp, '{}_{}_rank_change.csv'.format(str(sp)[:4], word))\n subset.to_csv(fp)\n else:\n fp = '/Users/e/code/hawthorne/results/rank_changes'\n fp = os.path.join(fp, '{}_rank_change.csv'.format(word))\n # notify\n print(fp)\n return",
"_____no_output_____"
],
[
"for word in keywords:\n get_rank_change(word, vectors_no_nh, vectors, topn = 100)",
"<ipython-input-810-1be216334ace>:10: SettingWithCopyWarning:\n\n\nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n\n"
],
[
"# testing low volatility words\nfor word in ['five', 'six', 'twenty']:\n get_rank_change(word, vectors_no_nh, vectors, topn = 100)",
"/Users/e/code/hawthorne/results/rank_changes/0.95_five_rank_change.csv\n/Users/e/code/hawthorne/results/rank_changes/0.91_six_rank_change.csv\n/Users/e/code/hawthorne/results/rank_changes/0.88_twenty_rank_change.csv\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e71ebcf8e5287b1b14de92cafd43febf76b8aa53 | 10,734 | ipynb | Jupyter Notebook | content/lessons/03/Class-Coding-Lab/CCL-Variables-And-Types.ipynb | MahopacHS/spring2019-moorea0517 | a71f8f9c593881a0923f5811e655142e2037cb09 | [
"MIT"
] | null | null | null | content/lessons/03/Class-Coding-Lab/CCL-Variables-And-Types.ipynb | MahopacHS/spring2019-moorea0517 | a71f8f9c593881a0923f5811e655142e2037cb09 | [
"MIT"
] | null | null | null | content/lessons/03/Class-Coding-Lab/CCL-Variables-And-Types.ipynb | MahopacHS/spring2019-moorea0517 | a71f8f9c593881a0923f5811e655142e2037cb09 | [
"MIT"
] | null | null | null | 34.29393 | 702 | 0.510061 | [
[
[
"# Inter-Class Coding Lab: Variables And Types\n\nThe goals of this lab are to help you to understand:\n\n1. Python data types\n1. Getting input as different types\n1. Formatting output as different types\n1. Basic arithmetic operators\n1. How to create a program from an idea.",
"_____no_output_____"
],
[
"## Variable Types\n\nEvery Python variable has a **type**. The Type determines how the data is stored in the computer's memory: ",
"_____no_output_____"
]
],
[
[
"a = \"4\"\ntype(a) # should be str",
"_____no_output_____"
],
[
"a = 4\ntype(a) # should be int",
"_____no_output_____"
]
],
[
[
"### Types Matter \n\nPython's built in functions and operators work differently depending on the type of the variable.:",
"_____no_output_____"
]
],
[
[
"a = 4\nb = 5\na + b # this plus in this case means add so 9",
"_____no_output_____"
],
[
"a = \"4\"\nb = \"5\"\na + b # the plus + in this case means concatenation, so '45'",
"_____no_output_____"
]
],
[
[
"### Switching Types\n\nthere are built-in Python functions for switching types. For example:",
"_____no_output_____"
]
],
[
[
"x = \"45\" # x is a str\ny = int(x) # y is now an int\nz = float(x) # z is a float\nprint(x,y,z)",
"45 45 45.0\n"
]
],
[
[
"### Inputs type str\n\nWhen you use the `input()` function the result is of type `str`:\n",
"_____no_output_____"
]
],
[
[
"age = input(\"Enter your age: \")\ntype(age)",
"Enter your age: 45\n"
]
],
[
[
"We can use a built in Python function to convert the type from `str` to our desired type:",
"_____no_output_____"
]
],
[
[
"age = input(\"Enter your age: \")\nage = int(age)\ntype(age)",
"Enter your age: 45\n"
]
],
[
[
"We typically combine the first two lines into one expression like this:",
"_____no_output_____"
]
],
[
[
"age = int(input(\"Enter your age: \"))\ntype(age)",
"Enter your age: 45\n"
]
],
[
[
"## Now Try This:\n\nWrite a program to:\n\n- input your age, convert it to an int and store it in a variable\n- add one to your age, store it in another variable\n- print out your current age and your age next year.\n\nFor example:\n```\nEnter your age: 45\nToday you are 45 next year you will be 46\n```",
"_____no_output_____"
]
],
[
[
"# TODO: Write your code here\n",
"_____no_output_____"
]
],
[
[
"## Format Codes\n\nPython has some string format codes which allow us to control the output of our variables. \n\n- %s = format variable as str\n- %d = format variable as int\n- %f = format variable as float\n\nYou can also include the number of spaces to use for example `%5.2f` prints a float with 5 spaces 2 to the right of the decimal point.",
"_____no_output_____"
]
],
[
[
"name = \"Mike\"\nage = 45\ngpa = 3.4\nprint(\"%s is %d years old. His gpa is %.3f\" % (name, age,gpa))",
"Mike is 45 years old. His gpa is 3.400\n"
]
],
[
[
"## Now Try This:\n\nPrint the PI variable out 3 times. Once as a string, once as an int and once as a float to 4 decimal places:",
"_____no_output_____"
]
],
[
[
"PI = 3.1415927\n#TODO: Write Code Here\n",
"_____no_output_____"
]
],
[
[
"## Putting it all together: Fred's Fence Estimator\n\nFred's Fence has hired you to write a program to estimate the cost of their fencing projects. For a given length and width you will calculate the number of 6 foot fence sections, and total cost of the project. Each fence section costs $23.95. Assume the posts and labor are free.\n\nProgram Inputs:\n\n- Length of yard in feet\n- Width of yard in feet\n \nProgram Outputs:\n\n- Perimeter of yard ( 2 x (Length + Width))\n- Number of fence sections required (Permiemer divided by 6 )\n- Total cost for fence ( fence sections multiplied by $23.95 )\n \nNOTE: All outputs should be formatted to 2 decimal places: e.g. 123.05 ",
"_____no_output_____"
]
],
[
[
"#TODO:\n# 1. Input length of yard as float, assign to a variable\n# 2. Input Width of yard as float, assign to a variable\n# 3. Calculate perimeter of yar, assign to a variable\n# 4. calculate number of fence sections, assign to a variable \n# 5. calculate total cost, assign to variable\n# 6. print perimeter of yard\n# 7. print number of fence sections\n# 8. print total cost for fence. ",
"_____no_output_____"
]
],
[
[
"## Now Try This\n\nBased on the provided TODO, write the program in python in the cell below. Your solution should have 8 lines of code, one for each TODO.\n\n**HINT**: Don't try to write the program in one sitting. Instead write a line of code, run it, verify it works and fix any issues with it before writing the next line of code. ",
"_____no_output_____"
]
],
[
[
"# TODO: Write your code here\nlength = float(input(\"enter the length of your yard in feet\"))\nwidth = float(input(\"enter the width of your yard in feet\"))\nperimeter = (2 * (length + width))\nfencing = (perimeter / 6)\ncost = (fencing * 23.95)\nprint(\"the perimeter of your yard equals %s feet\" %(perimeter))\nprint(\"the total number of fence sections you will need is %.2f sections\" %(fencing))\nprint(\"the total cost for the fence will be $%.2f\" %(cost))",
"enter the length of your yard in feet17\nenter the width of your yard in feet18\nthe perimeter of your yard equals 70.0 feet\nthe total number of fence sections you will need is 11.67 sections\nthe total cost for the fence will be $279.42\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71ed4f03279a1a2bb5fa7cb691699fa55a8e466 | 18,589 | ipynb | Jupyter Notebook | MuRIL_on_original_dataset.ipynb | Aanisha/ACL_Abusive_Tamil_Comment_Classification | b330b82f7b3bcdf5791f64f043b830359162127c | [
"MIT"
] | null | null | null | MuRIL_on_original_dataset.ipynb | Aanisha/ACL_Abusive_Tamil_Comment_Classification | b330b82f7b3bcdf5791f64f043b830359162127c | [
"MIT"
] | null | null | null | MuRIL_on_original_dataset.ipynb | Aanisha/ACL_Abusive_Tamil_Comment_Classification | b330b82f7b3bcdf5791f64f043b830359162127c | [
"MIT"
] | null | null | null | 36.882937 | 272 | 0.500941 | [
[
[
"<a href=\"https://colab.research.google.com/github/Aanisha/ACL_Abusive_Tamil_Comment_Classification/blob/main/MuRIL_on_original_dataset.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Finetuned MuRIL on original dataset",
"_____no_output_____"
]
],
[
[
"!pip install transformers",
"Collecting transformers\n Downloading transformers-4.17.0-py3-none-any.whl (3.8 MB)\n\u001b[K |████████████████████████████████| 3.8 MB 5.2 MB/s \n\u001b[?25hRequirement already satisfied: numpy>=1.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (1.21.5)\nCollecting sacremoses\n Downloading sacremoses-0.0.47-py2.py3-none-any.whl (895 kB)\n\u001b[K |████████████████████████████████| 895 kB 33.8 MB/s \n\u001b[?25hCollecting pyyaml>=5.1\n Downloading PyYAML-6.0-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (596 kB)\n\u001b[K |████████████████████████████████| 596 kB 41.8 MB/s \n\u001b[?25hCollecting huggingface-hub<1.0,>=0.1.0\n Downloading huggingface_hub-0.4.0-py3-none-any.whl (67 kB)\n\u001b[K |████████████████████████████████| 67 kB 5.3 MB/s \n\u001b[?25hRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from transformers) (2.23.0)\nRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.7/dist-packages (from transformers) (4.63.0)\nRequirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.7/dist-packages (from transformers) (21.3)\nRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from transformers) (4.11.2)\nRequirement already satisfied: regex!=2019.12.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (2019.12.20)\nCollecting tokenizers!=0.11.3,>=0.11.1\n Downloading tokenizers-0.11.6-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl (6.5 MB)\n\u001b[K |████████████████████████████████| 6.5 MB 34.0 MB/s \n\u001b[?25hRequirement already satisfied: filelock in /usr/local/lib/python3.7/dist-packages (from transformers) (3.6.0)\nRequirement already satisfied: typing-extensions>=3.7.4.3 in /usr/local/lib/python3.7/dist-packages (from huggingface-hub<1.0,>=0.1.0->transformers) (3.10.0.2)\nRequirement already satisfied: pyparsing!=3.0.5,>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from packaging>=20.0->transformers) (3.0.7)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata->transformers) (3.7.0)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2.10)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2021.10.8)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (3.0.4)\nRequirement already satisfied: click in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (7.1.2)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (1.15.0)\nRequirement already satisfied: joblib in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (1.1.0)\nInstalling collected packages: pyyaml, tokenizers, sacremoses, huggingface-hub, transformers\n Attempting uninstall: pyyaml\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\nSuccessfully installed huggingface-hub-0.4.0 pyyaml-6.0 sacremoses-0.0.47 tokenizers-0.11.6 transformers-4.17.0\n"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport os\nimport os\nimport random\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom tqdm.auto import tqdm\n\nfrom transformers import AutoTokenizer\nfrom transformers import TFAutoModel\n\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\nimport tensorflow.keras.backend as K\nimport logging",
"_____no_output_____"
]
],
[
[
"#### Pre-processing the data",
"_____no_output_____"
]
],
[
[
"train = pd.read_csv('/content/Tamil_train_data.csv')\ntest = pd.read_csv('/content/Tamil_test_data.csv')\nvalid = pd.read_csv('/content/Tamil_valid_data.csv')",
"_____no_output_____"
],
[
"tags = {\"tag\": {'Hope-Speech':0, 'None-of-the-above':7, 'Homophobia':1, 'Misandry':2,\n 'Counter-speech':3, 'Misogyny':4, 'Xenophobia':5, 'Transphobic':6}}",
"_____no_output_____"
],
[
"test = test.replace(tags)",
"_____no_output_____"
]
],
[
[
"#### Loading the model",
"_____no_output_____"
]
],
[
[
"config = {\n \n 'seed' : 42,\n 'model': '/content/drive/MyDrive/Muril-base-cased',\n 'group': 'MURIL',\n \n 'batch_size': 16,\n 'max_length': 64,\n \n 'device' : 'GPU',\n 'epochs' : 2,\n\n 'test_size' : 0.1,\n 'lr': 5e-6,\n 'use_transfer_learning' : False,\n \n 'use_wandb': True,\n 'wandb_mode' : 'online',\n}",
"_____no_output_____"
],
[
"def get_keras_model():\n pretrained_model = TFAutoModel.from_pretrained(config['model'])\n \n input_ids = layers.Input(shape=(config['max_length']),\n name='input_ids', \n dtype=tf.int32)\n token_type_ids = layers.Input(shape=(config['max_length'],),\n name='token_type_ids', \n dtype=tf.int32)\n attention_mask = layers.Input(shape=(config['max_length'],),\n name='attention_mask', \n dtype=tf.int32)\n embedding = pretrained_model(input_ids, \n token_type_ids=token_type_ids, \n attention_mask=attention_mask)[0]\n\n \n\n x1 = tf.keras.layers.Dropout(0.2)(embedding) \n x1 = tf.keras.layers.Conv1D(1,1)(x1)\n x1 = tf.keras.layers.Flatten()(x1)\n x1 = tf.keras.layers.Dense(8, activation='softmax')(x1)\n \n #print(x1.shape)\n \n model = keras.Model(inputs=[input_ids, \n token_type_ids, \n attention_mask],\n outputs=x1)\n \n return model",
"_____no_output_____"
],
[
"model = get_keras_model()\n\nmodel.load_weights('/content/drive/MyDrive/Muril-base-cased/best_model_25.h5')",
"Some layers from the model checkpoint at /content/drive/MyDrive/Muril-base-cased were not used when initializing TFBertModel: ['mlm___cls']\n- This IS expected if you are initializing TFBertModel from the checkpoint of a model trained on another task or with another architecture (e.g. initializing a BertForSequenceClassification model from a BertForPreTraining model).\n- This IS NOT expected if you are initializing TFBertModel from the checkpoint of a model that you expect to be exactly identical (initializing a BertForSequenceClassification model from a BertForSequenceClassification model).\nSome layers of TFBertModel were not initialized from the model checkpoint at /content/drive/MyDrive/Muril-base-cased and are newly initialized: ['bert/pooler/dense/bias:0', 'bert/pooler/dense/kernel:0']\nYou should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.\n"
]
],
[
[
"#### Preparing test data",
"_____no_output_____"
]
],
[
[
"test_labels = pd.read_csv(\"/content/Tamil_test_labels_data.csv\")\n\ntest_labels = test_labels.replace(tags)\ntest_labels = pd.merge(test_labels, test, on=['comments'])\ntest_labels = test_labels.dropna()",
"_____no_output_____"
],
[
"test_labels.shape",
"_____no_output_____"
],
[
"tokenizer = AutoTokenizer.from_pretrained(config['model'])",
"_____no_output_____"
],
[
"x_test = tokenizer(\n text=test_labels.comments.tolist(),\n add_special_tokens=True,\n max_length = 64,\n padding='max_length',\n truncation=True, \n return_tensors='tf',\n return_token_type_ids = True,\n return_attention_mask = True,\n verbose = True)",
"_____no_output_____"
],
[
"len(x_test['input_ids'])",
"_____no_output_____"
],
[
"preds = model.predict(x = {'input_ids':x_test['input_ids'], \n 'token_type_ids':x_test['token_type_ids'], \n 'attention_mask': x_test['attention_mask']}, verbose = 1, workers=4)",
"80/80 [==============================] - 34s 288ms/step\n"
],
[
"pr = []\nfor p in preds:\n pr.append(np.argmax(p))",
"_____no_output_____"
]
],
[
[
"#### Testing the model on unseen test data",
"_____no_output_____"
]
],
[
[
"import sklearn\nimport pandas as pd, numpy as np\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.feature_extraction.text import TfidfVectorizer\n\nprint(sklearn.metrics.classification_report(test_labels['tag'], pr))",
" precision recall f1-score support\n\n 0 0.00 0.00 0.00 95\n 1 0.00 0.00 0.00 64\n 2 0.52 0.68 0.59 419\n 3 0.25 0.31 0.28 135\n 4 0.00 0.00 0.00 105\n 5 0.72 0.11 0.19 120\n 6 0.00 0.00 0.00 60\n 7 0.76 0.89 0.82 1557\n\n accuracy 0.68 2555\n macro avg 0.28 0.25 0.23 2555\nweighted avg 0.60 0.68 0.62 2555\n\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e71eed451753202bd704dd3e796cd7b89b010290 | 963,319 | ipynb | Jupyter Notebook | Small Projects/Auto Regression/Lecture_17_The_Regression_Problem_Example_(Part_1).ipynb | GleydsonHudson/FIS-Exercises-Unipd | 6af4d31d506a1fb69e512c47c8eddf050e5469b1 | [
"MIT"
] | null | null | null | Small Projects/Auto Regression/Lecture_17_The_Regression_Problem_Example_(Part_1).ipynb | GleydsonHudson/FIS-Exercises-Unipd | 6af4d31d506a1fb69e512c47c8eddf050e5469b1 | [
"MIT"
] | null | null | null | Small Projects/Auto Regression/Lecture_17_The_Regression_Problem_Example_(Part_1).ipynb | GleydsonHudson/FIS-Exercises-Unipd | 6af4d31d506a1fb69e512c47c8eddf050e5469b1 | [
"MIT"
] | null | null | null | 164.02503 | 177,876 | 0.845729 | [
[
[
"# Fundamentals of Information Systems\n\n## Python Programming (for Data Science)\n\n### Master's Degree in Data Science\n\n#### Giorgio Maria Di Nunzio\n#### (Courtesy of Gabriele Tolomei FIS 2018-2019)\n<a href=\"mailto:[email protected]\">[email protected]</a><br/>\nUniversity of Padua, Italy<br/>\n2019/2020<br/>",
"_____no_output_____"
],
[
"# Lecture 12: The Regression Problem - Example (Part 1)",
"_____no_output_____"
],
[
"## Instructions\n\n- We consider the dataset available at this [link](http://archive.ics.uci.edu/ml/machine-learning-databases/auto-mpg/auto-mpg.data), provided by the [UC Irvine Machine Learning Repository](http://archive.ics.uci.edu/ml/index.php)\n\n- The dataset comes with a [**README**](http://archive.ics.uci.edu/ml/machine-learning-databases/auto-mpg/auto-mpg.names) file, which contains information about the number of instances, the number and type of attributes, as well as the prediction goal.\n\n- To be able to work even without any network connection, I have stored both the dataset and README files locally on my machine.\n\n- \"The data concerns city-cycle fuel consumption in miles per gallon, to be predicted in terms of 3 multivalued discrete and 5 continuous attributes.\" (Quinlan, 1993)",
"_____no_output_____"
],
[
"## Additional Notes\n\n- The original file contains a mixture of whitespace and tab-separated fields. In order to transform it into a legitimate tab-separated file, I had to run the following shell commands:\n\n```bash\n> TAB=$(printf '\\t')\n> sed \"s/ \\{2,\\}/$TAB/g\" < ${ORIGINAL_DATASET_FILE} > ${NEW_DATASET_FILE}\n```\n\n- The commands above use <code>**sed**</code> to replace **2 or more** whitespaces with a tab character on **every line** of the original dataset file.\n\n- Linux and Mac OS X systems have <code>**sed**</code> natively installed. Windows users can install it from [here](http://gnuwin32.sourceforge.net/packages/sed.htm) ",
"_____no_output_____"
]
],
[
[
"import math\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n# Import stats module from scipy, which contains a large number \n# of probability distributions as well as an exhaustive library of statistical functions.\nimport scipy.stats as stats\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# 1. Data Collection",
"_____no_output_____"
]
],
[
[
"# Path to the local dataset file\nDATASET_PATH = \"./data/auto-mpg-regression/dataset.tsv\"",
"_____no_output_____"
],
[
"# Load the dataset with Pandas\ndata = pd.read_csv(DATASET_PATH, sep=\"\\t\")\nprint(\"Shape of the dataset: {}\".format(data.shape))\ndata.head()\n# NOTE: the first line of the file is considered as the header",
"Shape of the dataset: (397, 9)\n"
],
[
"# Load the dataset with Pandas, this time taking into account\n# the fact that there is no header line\ndata = pd.read_csv(DATASET_PATH, sep=\"\\t\", header=None)\nprint(\"Shape of the dataset: {}\".format(data.shape))\ndata.head()",
"Shape of the dataset: (398, 9)\n"
],
[
"# Column names (labels) are not so meaningful and we should use the attribute names\n# provided in the README file\n# Row index, instead, can be left as it is (i.e., default IndexRange)\ncolumns = ['mpg', 'cylinders', 'displacement', 'horsepower', 'weight', \n 'acceleration', 'model_year', 'origin', 'car_name']\ndata = pd.read_csv(DATASET_PATH, sep=\"\\t\", header=None, names=columns)\nprint(\"Shape of the dataset: {}\".format(data.shape))\ndata.head()",
"Shape of the dataset: (398, 9)\n"
]
],
[
[
"## Checking for any Missing Values",
"_____no_output_____"
]
],
[
[
"# Check if there is any missing value in the whole dataset\nprint(\"There are missing values in the dataset: {}\".\n format(data.isnull().any().any()))",
"There are missing values in the dataset: False\n"
],
[
"# Check if 'horsepower' has really 6 missing values\nprint(\"N. of missing values for attribute 'horsepower': {}\".\n format(data.horsepower.isnull().sum()))",
"N. of missing values for attribute 'horsepower': 0\n"
],
[
"# Weird... we know from the README file there must be 6 missing values\n# How come we are not able to spot those?\n# Possibly, because of the way in which NAs are 'encoded' in the file...\n# Let's see the set of values contained in the 'horsepower' column\nprint(sorted(data.horsepower.unique(), reverse=True))",
"['?', '98.00', '97.00', '96.00', '95.00', '94.00', '93.00', '92.00', '91.00', '90.00', '89.00', '88.00', '87.00', '86.00', '85.00', '84.00', '83.00', '82.00', '81.00', '80.00', '79.00', '78.00', '77.00', '76.00', '75.00', '74.00', '72.00', '71.00', '70.00', '69.00', '68.00', '67.00', '66.00', '65.00', '64.00', '63.00', '62.00', '61.00', '60.00', '58.00', '54.00', '53.00', '52.00', '49.00', '48.00', '46.00', '230.0', '225.0', '220.0', '215.0', '210.0', '208.0', '200.0', '198.0', '193.0', '190.0', '180.0', '175.0', '170.0', '167.0', '165.0', '160.0', '158.0', '155.0', '153.0', '152.0', '150.0', '149.0', '148.0', '145.0', '142.0', '140.0', '139.0', '138.0', '137.0', '135.0', '133.0', '132.0', '130.0', '129.0', '125.0', '122.0', '120.0', '116.0', '115.0', '113.0', '112.0', '110.0', '108.0', '107.0', '105.0', '103.0', '102.0', '100.0']\n"
],
[
"# Apparently, there are some question mark characters '?' in this column.\n# Let's see how many records have '?' in their 'horsepower' column.\n# Extract the sub-DataFrame using boolean indexing \n# on the 'horsepower' column and count the corresponding number of matching rows.\nprint(\"How many records have 'horsepower=?'?: {}\"\n .format(data[data.horsepower == '?'].shape[0]))",
"How many records have 'horsepower=?'?: 6\n"
],
[
"data.loc[data.horsepower == \"?\", :]",
"_____no_output_____"
],
[
"# We have therefore found that '?' is a sentinel value used to identify NAs\n# Let's reload the dataset using this value as a marker for NAs.\ndata = pd.read_csv(DATASET_PATH, sep=\"\\t\", header=None, \n names=columns,\n na_values={'horsepower':'?'})\n\n# Alternatively, we could simply replace '?' on the loaded dataset with np.nan\n# 1. Using 'loc':\n# data.loc[data.horsepower == '?', 'horsepower'] = np.nan\n# 2. Using 'replace':\n# data.horsepower.replace('?', np.nan, inplace=True)\nprint(\"Shape of the dataset: {}\".format(data.shape))\ndata.head()",
"Shape of the dataset: (398, 9)\n"
],
[
"# Let's repeat the same check as above on missing values.\n# Check if there is any missing value in the whole dataset\nprint(\"There are missing values in the dataset: {}\".\n format(data.isnull().any().any()))\n# Check if 'horsepower' has really 6 missing values\nprint(\"N. of missing values for attribute 'horsepower': {}\".\n format(data.horsepower.isnull().sum()))",
"There are missing values in the dataset: True\nN. of missing values for attribute 'horsepower': 6\n"
],
[
"# Let's have a look at the output of the 'describe()' function.\ndata.describe(include='all')",
"_____no_output_____"
],
[
"print(data[\"mpg\"])",
"0 18.0\n1 15.0\n2 18.0\n3 16.0\n4 17.0\n ... \n393 27.0\n394 44.0\n395 32.0\n396 28.0\n397 31.0\nName: mpg, Length: 398, dtype: float64\n"
]
],
[
[
"## Change the Column Layout",
"_____no_output_____"
]
],
[
[
"# Just as a convention, I prefer to place the column to be predicted\n# as the last one.\ncolumns = data.columns.tolist()\nprint(\"Orignal order of columns:\\n{}\".format(columns))\n# Popping out 'mpg' from the list and insert it back at the end.\ncolumns.insert(len(columns), columns.pop(columns.index('mpg')))\nprint(\"New order of columns:\\n{}\".format(columns))",
"Orignal order of columns:\n['mpg', 'cylinders', 'displacement', 'horsepower', 'weight', 'acceleration', 'model_year', 'origin', 'car_name']\nNew order of columns:\n['cylinders', 'displacement', 'horsepower', 'weight', 'acceleration', 'model_year', 'origin', 'car_name', 'mpg']\n"
],
[
"# Let's refactor the DataFrame using this new column index\ndata = data.loc[:, columns]\ndata.head()\n# Alternatively to 'loc' we can also use 'reindex()'\n# data = data.reindex(columns=columns)\n# data.head()",
"_____no_output_____"
]
],
[
[
"## Apply Some Simple Data Transformations",
"_____no_output_____"
]
],
[
[
"# Suppose we want to convert displacement unit from cubic inch to litre\n# There is a useful conversion table which tells us how to do that.\n# 1 cubic inch = 0.016387064 litre\nCI_TO_LITRE = 0.016387064\ndata.displacement *= CI_TO_LITRE\ndata.head()",
"_____no_output_____"
]
],
[
[
"# 2. Data Exploration",
"_____no_output_____"
],
[
"## 2.1 Analysis of Data Distributions: Continous Values\n\n- Let's start visualizing the distributions of the **5 continuous-valued** features:\n - <code>**displacement**</code>\n - <code>**horsepower**</code> (contains 6 <code>**NA**</code>s)\n - <code>**weight**</code>\n - <code>**acceleration**</code>",
"_____no_output_____"
]
],
[
[
"# Create a lambda function which will be applied to each entry\n# of the numpy 2-D array of AxesSubplot objects\n# x is a reference to an AxesSubplot object\ny_labeler = lambda x: x.set_ylabel('density')\n# np.vectorize() allows calling the function on each element\ny_labeler = np.vectorize(y_labeler)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(2, 2, figsize=(10,12))\ny_labeler(axes)",
"_____no_output_____"
],
[
"# Create a Figure containing 2x2 subplots\nfig, axes = plt.subplots(2, 2, figsize=(10,12))\n# Call the vectorized function for labeling all the y-axes\ny_labeler(axes)\n# Plot 'displacement' on the top-left subplot\nsns.distplot(data.displacement, color='#808080', ax=axes[0,0], \n hist_kws=dict(edgecolor=\"#404040\", linewidth=1))\n# Plot 'horsepower' (limited only to non-NA values) on the top-right subplot\nsns.distplot(data.loc[data.horsepower.notnull(), 'horsepower'], \n color='#df2020', ax=axes[0,1], \n hist_kws=dict(edgecolor=\"#404040\", linewidth=1))\n# Plot 'weight' on the bottom-left subplot\nsns.distplot(data.weight, color='#0033cc', ax=axes[1,0], \n hist_kws=dict(edgecolor=\"k\", linewidth=1))\n# Plot 'acceleration' on the bottom-right subplot\nsns.distplot(data.acceleration, color='#009933', ax=axes[1,1],\n hist_kws=dict(edgecolor=\"#006622\", linewidth=1))\n# Adjust space between plots\nplt.subplots_adjust(wspace=.3, hspace=.3)",
"_____no_output_____"
]
],
[
[
"## Additional Note on Binning\n\n- If we don't specify the number of **bins** as argument of <code>**sns.distplot**</code> function (i.e.,<code>**bins=None**</code>) the **Freedman-Diaconis** rule is used to devise the _best_ number of bins.\n\n- This rule starts from defining the **width** each bin should have on the basis of the range of values observed, as follows:\n\n$$\n\\texttt{bin}\\_\\texttt{width} = 2 * \\frac{\\texttt{IQR}}{n^{1/3}}\n$$\nwhere $\\texttt{IQR}$ stands for **interquartile range**, namely the length of the interval delimited by the 1st and the 3rd quartile, and $n$ is the **number of observations**.\n\n- Finally, the number of bins is computed as:\n\n$$\n\\texttt{bins} = (max−min)/\\texttt{bin}\\_\\texttt{width}\n$$\n\nwhere, $max$ ($min$) is the **maximum** (**minimum**) value observed.\n",
"_____no_output_____"
]
],
[
[
"# Let's produce the boxplots corresponding to the distribution plots above\n# Create a Figure containing 2x2 subplots\nfig, axes = plt.subplots(2, 2, figsize=(10,12))\n\nsns.boxplot(data.displacement, color='#808080', ax=axes[0,0])\nsns.boxplot(data.loc[data.horsepower.notnull(), 'horsepower'], \n color='#df2020', ax=axes[0,1])\nsns.boxplot(data.weight, color='#0033cc', ax=axes[1,0])\nsns.boxplot(data.acceleration, color='#009933', ax=axes[1,1])\nplt.subplots_adjust(wspace=.3, hspace=.3)",
"_____no_output_____"
]
],
[
[
"## Spotting Outliers\n\n- Generally speaking, an outlier is an observation that is numerically distant from the rest of the data. \n\n- Boxplots are useful to actually spot any possible **outlier**, as they show the distribution of values that are located within the $\\texttt{IQR}$ (i.e., any data point between the 1st and 3rd quartile).\n\n- A boxplot defines also 2 other values, called **fences** or **whiskers** which are used to define outliers (i.e., any data point that is located outside the fences).\n\n- Usually, fences are determined as follows: if $Q_1$ and $Q_3$ represents the 1st and 3rd quartile, respectively, we define $F_\\textrm{left}$ and $F_\\textrm{right}$ as the left and right fence point, respectively, so that:\n\n$$\nF_\\textrm{left} = Q_1 - 1.5 * \\texttt{IQR};~~F_\\textrm{right} = Q_3 + 1.5 * \\texttt{IQR}\n$$",
"_____no_output_____"
],
[
"## Few Observations from the Plots\n\n- <code>**displacement**</code> distribution is not uni-modal at all; in fact it is bi-, or possibly, tri-modal. Apparently, this is not affected by any outlier.\n\n- <code>**horsepower**</code> distribution is also not uni-modal, with a small bump around the value of 150Hp. However, here we can spot some possible outliers (i.e., large values over 200Hp).\n\n- <code>**weight**</code> distribution is uni-modal yet **right-skewed** (**positively skewed**), which means the mean is shifted to the right due to the presence of some large values towards the positive direction (not necessarily outliers).\n\n- <code>**acceleration**</code> essentially fits nicely to a **Normal** (**Gaussian**) **distribution** and contains some outliers.",
"_____no_output_____"
]
],
[
[
"# Check how many outliers we have\n# 1. 'horsepower'\nhp_q1, hp_q3 = data.loc[data.horsepower.notnull(), 'horsepower'].quantile([.25, .75])\nprint(\"1st Quartile of 'horsepower': {:.2f}\".format(hp_q1))\nprint(\"3rd Quartile of 'horsepower': {:.2f}\".format(hp_q3))\nhp_IQR = (hp_q3 - hp_q1)\nprint(\"IQR of 'horsepower': {:.2f}\".format(hp_IQR))\nhp_fence_left = hp_q1 - 1.5 * hp_IQR\nhp_fence_right = hp_q3 + 1.5 * hp_IQR\nprint(\"Fence range: [{:.2f}, {:.2f}]\".format(hp_fence_left, hp_fence_right))",
"1st Quartile of 'horsepower': 75.00\n3rd Quartile of 'horsepower': 126.00\nIQR of 'horsepower': 51.00\nFence range: [-1.50, 202.50]\n"
],
[
"print(\"N. of instances containing outlier of 'horsepower': {}\".\n format(data[data.horsepower > hp_fence_right].shape[0]))",
"N. of instances containing outlier of 'horsepower': 10\n"
],
[
"# Check how many outliers we have\n# 2. 'acceleration'\nacc_q1, acc_q3 = data['acceleration'].quantile([.25, .75])\nprint(\"1st Quartile of 'acceleration': {:.2f}\".format(acc_q1))\nprint(\"3rd Quartile of 'acceleration': {:.2f}\".format(acc_q3))\nacc_IQR = (acc_q3 - acc_q1)\nprint(\"IQR of 'acceleration': {:.2f}\".format(hp_IQR))\nacc_fence_left = acc_q1 - 1.5 * acc_IQR\nacc_fence_right = acc_q3 + 1.5 * acc_IQR\nprint(\"Fence range: [{:.2f}, {:.2f}]\".format(acc_fence_left, acc_fence_right))",
"1st Quartile of 'acceleration': 13.83\n3rd Quartile of 'acceleration': 17.17\nIQR of 'acceleration': 51.00\nFence range: [8.80, 22.20]\n"
],
[
"print(\"N. of instances containing outlier of 'acceleration': {}\".\n format(data[(data.acceleration < acc_fence_left) | \n (data.acceleration > acc_fence_right)].shape[0]))",
"N. of instances containing outlier of 'acceleration': 9\n"
],
[
"print(\"N. of instances containing outlier of 'horsepower' or 'acceleration': {}\".\n format(data[(data.horsepower > hp_fence_right) |\n (data.acceleration > acc_fence_right) |\n (data.acceleration < acc_fence_left)].shape[0]))",
"N. of instances containing outlier of 'horsepower' or 'acceleration': 18\n"
],
[
"# Let's see if we can spot where those outliers are located\n# w.r.t. other features (e.g., cylinders)\nfig, axes = plt.subplots(2, 1, figsize=(10,8))\n\nsns.boxplot(x=data.cylinders, y=data.loc[data.horsepower.notnull(), 'horsepower'], \n color='#df2020', ax=axes[0])\nsns.boxplot(x=data.cylinders, y=data.acceleration, color='#009933', ax=axes[1])\nplt.subplots_adjust(wspace=.3, hspace=.3)",
"_____no_output_____"
]
],
[
[
"## Other Few Observations from the Plots\n\n- <code>**horsepower**</code>: outliers seem to occur only on 6- and 8-cylinder vehicles, stronger in the former.\n\n- <code>**acceleration**</code>: outliers here are more evenly distributed over 3- 4- and 8-cylinder vehicles.",
"_____no_output_____"
]
],
[
[
"# Let's now plot the pairwise relationship between our continuous-valued features\nsns.pairplot(data.loc[data.horsepower.notnull(),\n ['displacement', 'horsepower', 'weight', \n 'acceleration']],\n kind=\"reg\",\n diag_kind='kde', \n diag_kws={'shade': True, 'color': '#ff6600'}, \n plot_kws={'color': '#ff6600'})",
"_____no_output_____"
]
],
[
[
"## How Continuous-valued Features Relate to Each Other\n\n- As <code>**displacement**</code> increases, so do <code>**weight**</code> and <code>**horsepower**</code>; also <code>**acceleration**</code> tends to decrease (pretty intuitive!).\n\n- As <code>**horsepower**</code> increases, <code>**acceleration**</code> decreases, whilst <code>**weight**</code> and <code>**displacement**</code> increase as well.\n\n- As <code>**weight**</code> increases, both <code>**horsepower**</code> and <code>**displacement**</code> increases but <code>**acceleration**</code> tends to decrease. Now, this might seem counterintuitive at first but the thing is that there might be a **latent factor** which affects this relationship (e.g., a heavy vehicle is also likely to have more horsepower and therefore this could be the actual reason why we observe such a phenomenon).",
"_____no_output_____"
]
],
[
[
"# Let's now plot the pairwise relationship between our continuous-valued features \n# this time also considering our target variable 'mpg'\nsns.pairplot(data.loc[data.horsepower.notnull(),\n ['displacement', 'horsepower', 'weight', 'acceleration', 'mpg']],\n kind=\"reg\",\n diag_kind='kde', \n diag_kws={'shade': True, 'color': '#33cccc'}, \n plot_kws={'color': '#33cccc'})",
"_____no_output_____"
]
],
[
[
"## 2.1 Analysis of Data Distributions: Categorical Values\n\n- Let's visualize the distributions of the **3 categorical** features:\n - <code>**cylinders**</code>\n - <code>**model_year**</code>\n - <code>**origin**</code>",
"_____no_output_____"
]
],
[
[
"# Let's see the frequency counts of each categorical variable\n# 'cylinders'\nprint(data.cylinders.value_counts())\nprint()\n# 'model_year'\nprint(data.model_year.value_counts())\nprint()\n# 'origin'\nprint(data.origin.value_counts())",
"4 204\n8 103\n6 84\n3 4\n5 3\nName: cylinders, dtype: int64\n\n73 40\n78 36\n76 34\n82 31\n75 30\n81 29\n80 29\n79 29\n70 29\n77 28\n72 28\n71 28\n74 27\nName: model_year, dtype: int64\n\n1 249\n3 79\n2 70\nName: origin, dtype: int64\n"
],
[
"# Let's produce the boxplots corresponding to the distribution plots above\n# Create a Figure containing 2x2 subplots\nfig, axes = plt.subplots(1, 3, figsize=(8,4))\ny_labeler(axes)\n\n# Plot 'cylinders'\nsns.distplot(data.cylinders, color='#006699', ax=axes[0], \n kde=False, norm_hist=True,\n hist_kws=dict(edgecolor=\"#404040\", linewidth=1))\n# Plot 'model_year'\nsns.distplot(data.model_year, color='#a6cc33', ax=axes[1], \n kde=False, norm_hist=True,\n hist_kws=dict(edgecolor=\"#85a329\", linewidth=1))\n# Plot 'origin'\nsns.distplot(data.origin, color='#cc3399', ax=axes[2], \n kde=False, norm_hist=True,\n hist_kws=dict(edgecolor=\"#8f246b\", linewidth=1))\n# Adjust space between plots\nplt.subplots_adjust(wspace=.5, hspace=.4)",
"_____no_output_____"
],
[
"# For categorical variables, 'countplot' is the way to go\n# Create a Figure containing 1x3 subplots\nfig, axes = plt.subplots(1, 3, figsize=(12,4))\n\n# Plot 'cylinders'\nsns.countplot(data.cylinders, ax=axes[0])\nsns.countplot(data.model_year, ax=axes[1])\nsns.countplot(data.origin, ax=axes[2])\nplt.subplots_adjust(wspace=.5, hspace=.4)",
"_____no_output_____"
],
[
"# stripplot is also another useful plot to relate categorical vs. target variable\n# Create a Figure containing 1x3 subplots\nfig, axes = plt.subplots(1, 3, figsize=(12,4))\n\nsns.stripplot(x=data.cylinders, y=data.mpg, color='#006699', ax=axes[0])\nsns.stripplot(x=data.model_year, y=data.mpg, color='#a6cc33', ax=axes[1])\nsns.stripplot(x=data.origin, y=data.mpg, color='#cc3399', ax=axes[2])\n\nplt.subplots_adjust(wspace=.5, hspace=.3)",
"_____no_output_____"
],
[
"# swarmplot is also another useful plot to relate categorical vs. target variable\n# Create a Figure containing 1x3 subplots\nfig, axes = plt.subplots(1, 3, figsize=(12,4))\n\nsns.swarmplot(x=data.cylinders, y=data.mpg, color='#006699', ax=axes[0])\nsns.swarmplot(x=data.model_year, y=data.mpg, color='#a6cc33', ax=axes[1])\nsns.swarmplot(x=data.origin, y=data.mpg, color='#cc3399', ax=axes[2])\n\nplt.subplots_adjust(wspace=.5, hspace=.3)",
"_____no_output_____"
],
[
"# boxplot is also another useful plot to relate categorical vs. target variable\n# Create a Figure containing 1x3 subplots\nfig, axes = plt.subplots(1, 3, figsize=(16,6))\n\nsns.boxplot(x=data.cylinders, y=data.mpg, color='#006699', ax=axes[0])\nsns.boxplot(x=data.model_year, y=data.mpg, color='#a6cc33', ax=axes[1])\nsns.boxplot(x=data.origin, y=data.mpg, color='#cc3399', ax=axes[2])\n\nplt.subplots_adjust(wspace=.5, hspace=.3)",
"_____no_output_____"
],
[
"# Let's overlay boxplot and swarmplot\n# Create a Figure containing 1x3 subplots\nfig, axes = plt.subplots(1, 3, figsize=(16,6))\n\nsns.boxplot(x=data.cylinders, y=data.mpg, color='#006699', ax=axes[0])\nsns.swarmplot(x=data.cylinders, y=data.mpg, color=\".5\", ax=axes[0])\nsns.boxplot(x=data.model_year, y=data.mpg, color='#a6cc33', ax=axes[1])\nsns.swarmplot(x=data.model_year, y=data.mpg, color=\".5\", ax=axes[1])\nsns.boxplot(x=data.origin, y=data.mpg, color='#cc3399', ax=axes[2])\nsns.swarmplot(x=data.origin, y=data.mpg, color=\".5\", ax=axes[2])\n\nplt.subplots_adjust(wspace=.5, hspace=.3)",
"_____no_output_____"
]
],
[
[
"# 3. Data Preprocessing (Munging)",
"_____no_output_____"
],
[
"## Summary of the Issues\n\n- From our exploratory data analysis above, **two** main issues are observed:\n 1. The presence of **6 missing values** for the attribute <code>**horsepower**</code>\n 2. The presence of a total of **18 outliers** on the attributes <code>**horsepower**</code> and <code>**acceleration**</code>\n- In addition to those, we should also consider how to properly handle different feature's scale as well as the fact that we are in presence of both continuous and categorical attributes.",
"_____no_output_____"
],
[
"## 3.1 Handling Missing Values (NA)\n\n- There are just **6 out of 398** (i.e., approximately **1.5%**) of records containing a missing value.\n\n- Since they do not represent a significant subset of the whole dataset, we can simply drop those records.\n\n- Otherwise, we could mark (i.e., replace) those missing value using one of the strategies discussed (e.g., replace them with the median as computed from observed values).",
"_____no_output_____"
]
],
[
[
"# Let's go for the second option, i.e., replacing missing values on 'horsepower'\n# using the median as computed from the other observations.\n# NOTE: here's a classical example where using the mean rather than the median\n# might affect the result, as the mean is more sensitive to outliers.\n# NOTE: by default, median() does not include NAs in the computation.\n# In other words, we don't need to explicitly tell pandas to work on non-NA values:\n# data.horsepower[data.horsepower.notnull()].median()\ndata.horsepower.fillna(data.horsepower.median(), inplace=True)\ndata.head()",
"_____no_output_____"
],
[
"data.describe(include='all')",
"_____no_output_____"
]
],
[
[
"## 3.2 Handling Outliers\n\n- There are **18 outliers** shared between <code>**horsepower**</code> and <code>**acceleration**</code>.\n\n- Like missing values, outliers can be simply discarded as well (i.e., a process which is also known as **trimming** or **truncation**).\n\n- Another approach is called **winsorizing** and consists of replacing outliers with a specified percentile of the data (e.g., a 90% winsorization would see all data below the 5th percentile set to the 5th percentile, and data above the 95th percentile set to the 95th percentile).",
"_____no_output_____"
]
],
[
[
"# Python can winsorize data using 'scipy.stats' module.\n# Example:\na = pd.Series([92, 19, 101, 58, 1053, 91, 26, 78, 10, 13, -40, 101, 86, 85, 15, 89, 89, 28, -5, 41])\nprint(\"Length of a: {}\".format(a.shape[0]))\nprint(\"Mean of a: {}\".format(a.mean()))\nprint(\"Median of a: {}\".format(a.median()))\nprint(\"Sorted a: {}\".format(np.sort(a)))\nq_005, q_95 = a.quantile([0.05, 0.95])\nprint(\"5th percentile of a: {:.2f}\".format(q_005))\nprint(\"95th percentile of a: {:.2f}\".format(q_95))\nstats.mstats.winsorize(a, limits=0.05, inplace=True)\nprint(\"Sorted a: {}\".format(np.sort(a)))",
"Length of a: 20\nMean of a: 101.5\nMedian of a: 68.0\nSorted a: [ -40 -5 10 13 15 19 26 28 41 58 78 85 86 89\n 89 91 92 101 101 1053]\n5th percentile of a: -6.75\n95th percentile of a: 148.60\nSorted a: [ -5 -5 10 13 15 19 26 28 41 58 78 85 86 89 89 91 92 101\n 101 101]\n"
],
[
"# Let's winsorize 'horsepower' and 'acceleration'\nstats.mstats.winsorize(data.horsepower, limits=0.0375, inplace=True)\nstats.mstats.winsorize(data.acceleration, limits=0.0375, inplace=True)",
"_____no_output_____"
],
[
"data.describe(include='all')",
"_____no_output_____"
],
[
"# Let's verify the outliers are actually gone\nfig, axes = plt.subplots(2, 2, figsize=(10,12))\ny_labeler(axes)\nsns.boxplot(data.displacement, color='#808080', ax=axes[0,0])\nsns.boxplot(data.horsepower, color='#df2020', ax=axes[0,1])\nsns.boxplot(data.weight, color='#0033cc', ax=axes[1,0])\nsns.boxplot(data.acceleration, color='#009933', ax=axes[1,1])\nplt.subplots_adjust(wspace=.3, hspace=.3)",
"_____no_output_____"
]
],
[
[
"## 3.3 Encoding Categorical Features\n\n- Categorical variables are typically stored as text values which represent various traits. \n\n- Some examples include <code>**color**</code> = {\"Red\", \"Yellow\", \"Blue\"), <code>**size**</code> = (\"Small\", \"Medium\", \"Large\"), etc.\n\n- Many ML algorithms can support categorical values without further manipulation but there are many others that do not. \n\n- Therefore, the analyst is faced with the challenge of figuring out how to turn these text attributes into **numerical values** for further processing.",
"_____no_output_____"
],
[
"## Approach 1: Label Encoding\n\n- Label encoding is simply converting each value in a column to a number. \n\n- For example, the <code>**model_year**</code> column contains 13 different values. We could choose to encode it like this:\n\n```python\n70 --> 0\n71 --> 1\n72 --> 2\n...\n```",
"_____no_output_____"
],
[
"## Approach 2: One-Hot Encoding\n\n- Label encoding is straightforward but it has the disadvantage that numeric values can be \"misinterpreted\" by the learning algorithms. \n\n- For example, the value of 0 is obviously less than the value of 4 but is that what we really aim for? For example, does a vehicle from 1974 have \"4x\" more weight than one from 1971?\n\n- A common alternative approach is called **one hot encoding**. Here, the basic strategy is to convert each category value into a new column and assigns a 1 or 0 (True/False) value to the column. \n\n- This has the benefit of not weighting a value improperly but does have the downside of adding more columns to the data set.",
"_____no_output_____"
]
],
[
[
"# In pandas we can achieve easily one-hot encoding using the 'get_dummies()' function\ncategorical_features = ['cylinders', 'model_year', 'origin']\ndata_with_dummies = pd.get_dummies(data, columns = categorical_features)\ndata_with_dummies.head()",
"_____no_output_____"
],
[
"# Just as a convention, I prefer to place the column to be predicted\n# as the last one.\ncolumns = data_with_dummies.columns.tolist()\n# Popping out 'mpg' from the list and insert it back at the end.\ncolumns.insert(len(columns), columns.pop(columns.index('mpg')))\n# Let's refactor the DataFrame using this new column index\ndata_with_dummies = data_with_dummies.loc[:, columns]\ndata_with_dummies.head()",
"_____no_output_____"
],
[
"# The categorical variable 'car_name' contains a lot of different values.\n# Using one-hot encoding might lead to a very sparse dataset, as we need\n# to map a single column to 305 columns!\ndata_with_dummies.car_name.value_counts()",
"_____no_output_____"
],
[
"\"\"\"\nThree solutions can be designed to tackle with this issue:\n1) Just drop the column 'car_name' (i.e., our model won't rely on that feature for prediction)\n2) Use one-hot encoding scheme and deal with sparsity data (i.e., possibly leading to overfitting)\n3) Trade-off: try to build another column which somehow reduces (i.e., cluster) similar values together\nand then apply one-hot encoding.\nLet's see how to perform 3)\n\"\"\"\n# Suppose we want to create another column called 'automaker_name', which simply contains\n# the name of the automaker, disregarding the model.\n# For example, automaker_name('ford gran torino') = automaker_name('ford f250') = 'ford'\ndata_with_dummies['automaker_name'] = data_with_dummies['car_name'].map(lambda x:\n x.split(' ')[0])\ndata_with_dummies.head()",
"_____no_output_____"
],
[
"data_with_dummies['car_name'].map(lambda x: x.split(' ')[0]).value_counts()",
"_____no_output_____"
],
[
"\"\"\"\nLet's see how many distinct values we have now for this nvalue_countsrical variable\n\"\"\"\ndata_with_dummies['automaker_name'].value_counts()",
"_____no_output_____"
],
[
"def sanitize_automaker_name(car_name):\n s = car_name.split(' ')[0]\n if s == 'vw' or s == 'vokswagen':\n return car_name.replace(s,'volkswagen')\n if s == 'chevroelt' or s == 'chevy':\n return car_name.replace(s,'chevrolet')\n if s == 'maxda':\n return car_name.replace(s,'mazda')\n if s == 'mercedes':\n return car_name.replace(s,'mercedes-benz')\n if s == 'toyouta':\n return car_name.replace(s,'toyota')\n return car_name",
"_____no_output_____"
],
[
"\"\"\"\nUse the 'sanitize_automaker_name' function to update 'car_name' values.\n\"\"\"\ndata_with_dummies['car_name'] = data_with_dummies['car_name'].map(lambda x:\n sanitize_automaker_name(x))\ndata_with_dummies.head()",
"_____no_output_____"
],
[
"\"\"\"\nRe-apply the function on the sanitized version of 'car_name'.\n\"\"\"\ndata_with_dummies['automaker_name'] = data_with_dummies['car_name'].map(lambda x: \n x.split(' ')[0])\ndata_with_dummies.head()",
"_____no_output_____"
],
[
"\"\"\"\nLet's see how many distinct values we have now for this new categorical variable.\n\"\"\"\ndata_with_dummies['automaker_name'].value_counts()",
"_____no_output_____"
],
[
"# Create the set of the top-10 automakers\ntop_10_automakers = set(data_with_dummies['automaker_name'].value_counts().index[:10])\n# Label with 'other' any automaker_name which is not in the list above\ndata_with_dummies['automaker_name'] = np.where(data_with_dummies.\n automaker_name.isin(top_10_automakers), \n data_with_dummies.automaker_name,\n 'other')",
"_____no_output_____"
],
[
"\"\"\"\nLet's verify we did it right!\n\"\"\"\ndata.loc[data_with_dummies.automaker_name == 'other', 'car_name'].map(lambda x: \n x.split(' ')[0] not in top_10_automakers).all()",
"_____no_output_____"
],
[
"\"\"\"\nWe now categorize (i.e., discretize) 'automaker_name' using the 10 + 1 discrete values above.\n\"\"\"\ncategorical_features = ['automaker_name']\ndata_with_dummies = pd.get_dummies(data_with_dummies, columns = categorical_features)\ndata_with_dummies.head()",
"_____no_output_____"
],
[
"# Just as a convention, I prefer to place the column to be predicted\n# as the last one.\ncolumns = data_with_dummies.columns.tolist()\n# Popping out 'mpg' from the list and insert it back at the end.\ncolumns.insert(len(columns), columns.pop(columns.index('mpg')))\n# Popping out 'automaker_name_other' from the list and insert it after 'automaker_name_volkswagen'.\ncolumns.insert(columns.index('automaker_name_volkswagen'), columns.pop(columns.index('automaker_name_other')))\n# Popping out 'car_name' from the list and insert it right before 'mpg'\ncolumns.insert(-1, columns.pop(columns.index('car_name')))\n# Let's refactor the DataFrame using this new column index\ndata_with_dummies = data_with_dummies.loc[:, columns]\ndata_with_dummies.head()",
"_____no_output_____"
]
],
[
[
"## 3.4 Standardize Feature Scale\n\n- Some learning models are sensitive to different feature scales appearing on the training dataset.\n\n- To overcome this issue, one typically standardize (i.e., normalize) the values of each continuous feature.\n\n- Two main strategies are usually enacted:\n - **min-max** normalization\n - **z-score** standardization",
"_____no_output_____"
]
],
[
[
"# Let's deep copy our DataFrame again\ndata_norm_0_1 = data_with_dummies.copy()",
"_____no_output_____"
],
[
"\"\"\"\nThe easiest way to normalize a (sub)set of features is as follows.\n\"\"\"\n# 1. Decide which list of features to standardize\nfeatures_to_standardize = ['displacement', 'horsepower', 'weight', 'acceleration']\n\n# 2. Select those features (i.e., DataFrame columns) and apply, for example, min-max normalization\ndata_norm_0_1[features_to_standardize] = ((data_norm_0_1[features_to_standardize] - \n data_norm_0_1[features_to_standardize].min()) \n / (data_norm_0_1[features_to_standardize].max() - \n data_norm_0_1[features_to_standardize].min()))\n# 3. Verify the result\ndata_norm_0_1.head()",
"_____no_output_____"
],
[
"# Let's make another deep copy of our DataFrame\ndata_std = data_with_dummies.copy()",
"_____no_output_____"
],
[
"\"\"\"\nThe following three functions are used to standardize/normalize features.\n\"\"\"\n# 1. z_score computes the standard z-score of a feature value x\ndef z_score(x, mu_X, sigma_X):\n return (x - mu_X)/sigma_X\n\n# 2. min_max computes the normalized value of a feature value x in the range [-1, 1]\ndef min_max(x, X_min, X_max):\n return (2*x - X_max - X_min)/(X_max - X_min)\n\n# 3. min_max_0_1 computes the normalized value of a feature value x in the range [0, 1]\ndef min_max_0_1(x, X_min, X_max):\n return (x - X_min)/(X_max - X_min)",
"_____no_output_____"
],
[
"\"\"\"\nWe use 'map' to call the z_score function above element-wise\non each Series object: displacement, horsepower, weight, and acceleration.\n\"\"\"\n# 1.a. Compute the mean and std deviation of 'displacement'\nmu_displacement = data_std.displacement.mean()\nsigma_displacement = data_std.displacement.std()\n# 1.b. Call the z_score function on the 'displacement' Series\ndata_std.displacement = data_std.displacement.map(lambda x: \n z_score(x, mu_displacement, \n sigma_displacement))\n\n# 2.a. Compute the mean and std deviation of 'horsepower'\nmu_horsepower = data_std.horsepower.mean()\nsigma_horsepower = data_std.horsepower.std()\n# 2.b. Call the z_score function on the 'horsepower' Series\ndata_std.horsepower = data_std.horsepower.map(lambda x: \n z_score(x, mu_horsepower, \n sigma_horsepower))\n\n# 3.a. Compute the mean and std deviation of 'weight'\nmu_weight = data_std.weight.mean()\nsigma_weight = data_std.weight.std()\n# 3.b. Call the z_score function on the 'weight' Series\ndata_std.weight = data_std.weight.map(lambda x: \n z_score(x, mu_weight, \n sigma_weight))\n\n# 4.a. Compute the mean and std deviation of 'acceleration'\nmu_acceleration = data_std.acceleration.mean()\nsigma_acceleration = data_std.acceleration.std()\n# 4.b. Call the z_score function on the 'acceleration' Series\ndata_std.acceleration = data_std.acceleration.map(lambda x: \n z_score(x, mu_acceleration, \n sigma_acceleration))\n\ndata_std.head()",
"_____no_output_____"
],
[
"# Let's create yet another deep copy of our DataFrame\ndata_std_z = data_with_dummies.copy()",
"_____no_output_____"
],
[
"\"\"\"\nThis is an even more general solution for standardizing multiple features\nin one shot, using customized \"normalizing\" functions.\n\"\"\"\ndef standardized_continuous_features(dataset, feature_names, func=z_score):\n \n if func != z_score and func != min_max and func != min_max_0_1:\n func = z_score\n \n for feature in feature_names:\n print(\"Standardized feature \\\"{}\\\" using [{}] function\".format(feature, func.__name__))\n if func == min_max or func == min_max_0_1:\n feature_min = dataset[feature].min()\n feature_max = dataset[feature].max()\n print(\"Min. = {}\".format(feature_min))\n print(\"Max. = {}\".format(feature_max))\n dataset[feature] = dataset[feature].map(lambda x: func(x, feature_min, feature_max))\n else:\n feature_mean = dataset[feature].mean()\n feature_std = dataset[feature].std()\n print(\"Mean = {}\".format(feature_mean))\n print(\"Std. Deviation = {}\".format(feature_std))\n dataset[feature] = dataset[feature].map(lambda x: func(x, feature_mean, feature_std))\n \n return dataset",
"_____no_output_____"
],
[
"\"\"\"\nWe call the function defined above on our deep copy, using the list of features\nthat needs to be standardized.\n\"\"\"\ndata_std_z = standardized_continuous_features(data_std_z, features_to_standardize)\ndata_std_z.head()",
"_____no_output_____"
],
[
"\"\"\"\nLet's verify the two different approaches above lead to the same result.\"\n\"\"\"\nprint((np.abs(data_std[features_to_standardize] - data_std_z[features_to_standardize]) < 0.0001).all())",
"_____no_output_____"
],
[
"\"\"\"\nStandardization and min-max scaling can be also performed using scikit-learn.\n\"\"\"\n# The following is the scikit-learn package which provides\n# various preprocessing capabilities\nfrom sklearn import preprocessing\n\n# Standardizing features using z-score\nstd_scale = preprocessing.StandardScaler().fit(data_with_dummies[features_to_standardize])\ndata_std = std_scale.transform(data_with_dummies[features_to_standardize])\n\n# Normalizing features using min-max\nminmax_scale = preprocessing.MinMaxScaler().fit(data_with_dummies[features_to_standardize])\ndata_minmax = minmax_scale.transform(data_with_dummies[features_to_standardize])\n\n# NOTE: 'data_std' and 'data_minmax' are numpy's ndarray (i.e., not pandas' DataFrame) objects",
"_____no_output_____"
]
],
[
[
"## 3.5 Feature Selection\n\n- This is a topic that would require an in-depth analysis.\n\n- A very simple approach to select highly discriminant features is given by measuring how each feature correlated with the target, and pick the ones with the highest correlation score.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nLet's compute feature correlation between each feature and our target 'mpg'.\nPandas 'corr()' function when applied to a DataFrame returns the whole correlation matrix.\nThis is a diagonal matrix having all 1's on its diagonal, whilst the entry (i,j) will contain\nthe correlation coefficient between feature i and feature j (i != j).\n\"\"\"\ncorr_matrix = data_with_dummies.corr()\nstrong_corr = corr_matrix[(corr_matrix['mpg'].abs() > .5) &\n (corr_matrix['mpg'] != 1)].loc[:, 'mpg']\n\nprint(\"Strongest correlated features:\\n{}\".format(strong_corr.sort_values()))\nprint()\nprint(\"Strongest correlated features (absolute values):\\n{}\".\n format(strong_corr.abs().sort_values(ascending=False)))",
"Strongest correlated features:\nweight -0.831741\ndisplacement -0.804203\nhorsepower -0.788402\ncylinders_8 -0.647308\norigin_1 -0.568192\ncylinders_4 0.758259\nName: mpg, dtype: float64\n\nStrongest correlated features (absolute values):\nweight 0.831741\ndisplacement 0.804203\nhorsepower 0.788402\ncylinders_4 0.758259\ncylinders_8 0.647308\norigin_1 0.568192\nName: mpg, dtype: float64\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71f19e501fb6beb94c5f386dfcf60b0cb55e63f | 97,814 | ipynb | Jupyter Notebook | Apple_Stock_Price_Prediction_with_fbprophet.ipynb | narc-kany/Apple-Stock-Prediction | a9e891e1a77ff5e6d35427fe43ce76b3f359718e | [
"Apache-2.0"
] | 1 | 2021-12-09T16:14:24.000Z | 2021-12-09T16:14:24.000Z | Apple_Stock_Price_Prediction_with_fbprophet.ipynb | narc-kany/Apple-Stock-Prediction | a9e891e1a77ff5e6d35427fe43ce76b3f359718e | [
"Apache-2.0"
] | null | null | null | Apple_Stock_Price_Prediction_with_fbprophet.ipynb | narc-kany/Apple-Stock-Prediction | a9e891e1a77ff5e6d35427fe43ce76b3f359718e | [
"Apache-2.0"
] | null | null | null | 107.84344 | 35,478 | 0.598381 | [
[
[
"<a href=\"https://colab.research.google.com/github/narc-kany/Apple-Stock-Prediction/blob/main/Apple_Stock_Price_Prediction_with_fbprophet.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# Description : Predict stock prices of Apple stock using facebook prophet package",
"_____no_output_____"
],
[
"# import pandas library\nimport pandas as pd",
"_____no_output_____"
],
[
"# Load Historical price data\nfrom google.colab import files\nfiles.upload()\n",
"_____no_output_____"
],
[
"# Read the uploaded data\n# Remove space from the header(column heading)\ndf = pd.read_csv('AAPL.csv', delimiter=',', header=None, skiprows=1, names=['Date','Open','High','Low','Close','Adj Close','Volume'])\n# print the read data\ndf",
"_____no_output_____"
],
[
"# Prophet is facebook time prediction library\n# Select the 2 columns \ndf = df[['Date','Close']]\n# Rename the columns to model requirements\ndf = df.rename(columns={'Date':'ds','Close':'y'})\n# Print the answer\ndf",
"_____no_output_____"
],
[
"# Get last 20 rows of data and store it in a new variable\nlast = df[len(df)-20:]\nlast",
"_____no_output_____"
],
[
"# Get all the rows expect for last 20\ndf = df[:-20]\ndf",
"_____no_output_____"
],
[
"# import fbprophet \nfrom fbprophet import Prophet\n# Create object for the Prophet library\nfpt = Prophet(daily_seasonality = True)\n# Fit and train the data model\nfpt.fit(df)\nfuture_pred = fpt.make_future_dataframe(periods=50)\nforecast_pred = fpt.predict(future_pred)\n",
"INFO:fbprophet:Disabling weekly seasonality. Run prophet with weekly_seasonality=True to override this.\n"
],
[
"# import fbptophet plotter library\nfrom fbprophet.plot import plot_plotly, plot_components_plotly\n# plot the data\nplot_plotly(fpt, forecast_pred)\n",
"_____no_output_____"
],
[
"# forecast for future predictions\nforecast_pred[forecast_pred.ds == '2018-01-01']['yhat']",
"_____no_output_____"
],
[
"# Show the actual value of AAP on 2017-12-15\nlast[last.ds == '2018-01-01']['y']",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71f2077d1b8f4ac4d45dcce6f2aece964bc18f6 | 139,199 | ipynb | Jupyter Notebook | python/2021-04-05.ipynb | risker93/king | b7cac65595960a81236cb0d9d004d4f3ffe1edf0 | [
"Apache-2.0"
] | 2 | 2021-05-02T12:23:27.000Z | 2021-05-02T12:56:25.000Z | python/2021-04-05.ipynb | risker93/king | b7cac65595960a81236cb0d9d004d4f3ffe1edf0 | [
"Apache-2.0"
] | null | null | null | python/2021-04-05.ipynb | risker93/king | b7cac65595960a81236cb0d9d004d4f3ffe1edf0 | [
"Apache-2.0"
] | null | null | null | 20.329926 | 913 | 0.381145 | [
[
[
"#패턴분석은 결국 차원축소.\n#함수에서 초기값을 정할떄 곱하기는 1 더하기는 \n#구구단, 팩토리얼 코딩데스트 많이나옴.\n\ndef fibonacci(n):\n if n == 1:\n return 1\n if n == 2:\n return 1\n else:\n return fibonacci(n-1) + fibonacci(n-2)\n \nprint(fibonacci(1))\nprint(fibonacci(2))\nprint(fibonacci(3))\nprint(fibonacci(4))\nprint(fibonacci(5))",
"1\n1\n2\n3\n5\n"
],
[
"#재귀 함수로 구현한 피보나치 수열2\ncounter = 0\n\n#gka\ndef fibonacci(n):\n #어떤 피보나치 수를 구하는지 출력 합니다.\n print(\"fibonacci({})를 구합니다.\".format(n))\n global counter\n counter += 1\n \n #피보나치 수를 구합니다\n if n == 1:\n return 1\n if n == 2:\n return 1\n else:\n return fibonacci(n-1) + fibonacci(n-2)\n \nfibonacci(10)\nprint(\"---\")\nprint(\"fiboonacci(10)계산에 활용된 덧셈 횟수는 {} 번 입니다.\".format(counter))",
"fibonacci(10)를 구합니다.\nfibonacci(9)를 구합니다.\nfibonacci(8)를 구합니다.\nfibonacci(7)를 구합니다.\nfibonacci(6)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(6)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(7)를 구합니다.\nfibonacci(6)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(8)를 구합니다.\nfibonacci(7)를 구합니다.\nfibonacci(6)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(6)를 구합니다.\nfibonacci(5)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(4)를 구합니다.\nfibonacci(3)를 구합니다.\nfibonacci(2)를 구합니다.\nfibonacci(1)를 구합니다.\nfibonacci(2)를 구합니다.\n---\nfiboonacci(10)계산에 활용된 덧셈 횟수는 109 번 입니다.\n"
],
[
"#재귀함수를 빠르게 실행하게 만들기\n#메모변수를 만듭니다.\n\ndictionary = {\n 1:1,\n 2:1\n}\n\n#함수를 선언합니다.\ndef fibonacci(n):\n if n in dictionary:\n #메모가 되어있으면 메모된 값을 리턴\n return dictionary[n]\n else:\n #메모가 되어 있지 않으면 값을구함\n output = fibonacci(n-1) + fibonacci(n-2)\n dictionary[n] = output\n return output\n \n#함수를 호출합니다.\nprint(fibonacci(10))\nprint(fibonacci(20))\n",
"55\n6765\n"
],
[
"#튜플 \n#함수와 함께 많이 사용되는 리스트와 비슷한 자료형으로\n#한번 결정된 요소를 바꿀 수 없다\n\n#람다\n#매개변수로 함수 구문 작성시 함수를 간단하고 쉽게 선언하는방법 \n\n#튜플 \na = (10,20,30)\nprint(a[0])\nprint(a[1])\nprint(a[2])\n\n#a[2] = 40 튜플은 한번 만들어진 요소는 변경이 불가능",
"10\n20\n30\n"
],
[
"#리스트와 튜플의 특이한 사용\n[a,b] = [10,20]\n(c,d) = (10,20)\n#출력합니다\nprint(a)\nprint(b)\nprint(c)\nprint(d)",
"10\n20\n10\n20\n"
],
[
"#괄호가 없는 튜플\ntuple_test = 10, 20, 30, 40\nprint(tuple_test)\nprint()\n\n#괄호가 없는 튜플 활용\na, b, c = 10, 20, 30\nprint(a)\nprint(b)\nprint(c)",
"(10, 20, 30, 40)\n\n10\n20\n30\n"
],
[
"#변수 값을 교환하는 튜플\na, b = 10, 20\n\nprint(a)\nprint(b)\n\n#값을 교환합니다\n\na, b = b, a\nprint(a)\nprint(b)",
"10\n20\n20\n10\n"
],
[
"def t ():\n return (10, 20)\na, b = t()\nprint(a)\nprint(b)\n\n",
"10\n20\n"
],
[
"#람다 ****\n#기능을 매개변수로 전달하는 코드를 더 효율적으로 작성\n####################################\n#함수의 매개변수로 함수 전달하기\n\ndef call_10(func):\n for i in range(10):\n func()\n \ndef print_hello():\n print(\"안녕하세요\")\n \ncall_10(print_hello)",
"안녕하세요\n안녕하세요\n안녕하세요\n안녕하세요\n안녕하세요\n안녕하세요\n안녕하세요\n안녕하세요\n안녕하세요\n안녕하세요\n"
],
[
"#filter() 함수와 map()함수 \n#함수를 매개변수로 전달하는 대표적인 표준함수\n#map(함수,리스트), filter(함수, 리스트)\n#함수를 선언합니다.\ndef power(item):\n return item *item\ndef under_3(item):\n return item < 3\n\n\n#람다함수로 표현\n# power = lambda x : x * x\n#under_3 = lambda x : x < 3\n\n#보통은 변수에 넣지않고 문장 안에 들어감\n\n\n\n#변수를 선언합니다\nlist_input_a = [1,2,3,4,5]\n\n#map 함수를 사용합니다\noutput_a = map(power, list_input_a)\nprint(output_a)\nprint(list(output_a))\n\n#filter() 함수를 사용합니다.\noutput_b = filter(under_3, list_input_a)\nprint(output_b)\nprint(list(output_b))\n\n#결과로 map object와 filter object가 나오는데 이를 제너레이터 라고부른다.\n#여기에 list() 함수를 적용해서 강제로 리스트 자료형으로 출력\n",
"<map object at 0x000002C1541071C8>\n[1, 4, 9, 16, 25]\n<filter object at 0x000002C154107248>\n[1, 2]\n"
],
[
"#람다의 개념\n#lambda 매개변수: 리턴값\n#인라인 람다\n\nlist_a = [1,2,3,4,5]\n\noutput_a = map(lambda x : x * x, list_a )\nprint(output_a)\nprint(list(output_a))\n\noutput_b = filter(lambda x: x < 3 , list_input_a)\nprint(output_b)\nprint(list(output_b))",
"<map object at 0x000002C154106088>\n[1, 4, 9, 16, 25]\n<filter object at 0x000002C154106CC8>\n[1, 2]\n"
],
[
"#파일처리 \n#파일 열고닫기\n#파일객체 = open(문자열 : 파일 경로, 문자열 : 읽기모드)\n#파일객체.close()\n#w : write 모드, a: append모드, r \"read모드\"\nf = open(\"basic.txt\", \"w\")\nf.write(\"hello stranger\")\nf.close()",
"_____no_output_____"
],
[
"#with 키워드\n#with open(문자열: 파일경로, 문자열: 모드) as 파일객체:\n# 문장\nwith open(\"apple.txt\", \"w\") as f:\n f.write(\"fine apple\")\n \n#위드문을 사용하면 with 구문이 종료될때 자동으로 파일이 닫힙니다.",
"_____no_output_____"
],
[
"#파일 객체.read() : 텍스트 읽기\nwith open(\"basic.txt\", \"r\") as f:\n contents = f.read()\nprint(contents)",
"hello stranger\n"
],
[
"#텍스트 한 줄씩 읽기\n#csv, xml, json 이 텍스트를 사용해 데이터를 구조적으로 표현함\n#csv 파일은 한줄에 하나의 데이터를 나타냄, 각각의 줄은 쉼표를 사용해 데이터를 구분\n# 이름, 키, 몸무게\n# 윤인성, 176, 62\n# 연하진, 169, 50\n#랜덤하게 1000명의 키와 몸무게 만들기\nimport random\n\nhanguls = list(\"가나다라마바사아자차카타파하\")\n\nwith open(\"info.txt\", \"w\") as f:\n for i in range(1000):\n name = random.choice(hanguls) + random.choice(hanguls) \n weight = random.randrange(40, 100)\n height = random.randrange(140, 200)\n \n f.write(\"{}, {}, {}\\n\".format(name, weight, height))",
"_____no_output_____"
],
[
"#반복문으로 파일 한 줄씩 읽기\nwith open(\"info.txt\", \"r\") as f:\n for line in f:\n #변수를 선언\n (name, weight, height) = line.strip().split(\",\")\n \n #데이터가 문제 없는지 확인 합니다.\n if (not name) or (not weight) or (not height):\n continue\n \n #결과를 계산합니다.\n bmi = int(weight)/ ((int(height) / 100) **2)\n result = \"\"\n if 25 <= bmi:\n result = \"과체중\"\n elif 18.5 <= bmi:\n result = \"정상체중\"\n else:\n result = \"저체중\"\n \n print('\\n'.join([\n \"이름: {}\",\n \"몸무게:{}\",\n \"키:{}\",\n \"BMI:{}\",\n \"결과:{}\"\n ]).format(name, weight, height, bmi, result))\n print()",
"이름: 가차\n몸무게: 65\n키: 152\nBMI:28.133656509695292\n결과:과체중\n\n이름: 마라\n몸무게: 82\n키: 190\nBMI:22.714681440443215\n결과:정상체중\n\n이름: 자자\n몸무게: 59\n키: 167\nBMI:21.15529420201513\n결과:정상체중\n\n이름: 자라\n몸무게: 88\n키: 194\nBMI:23.381868423849507\n결과:정상체중\n\n이름: 자차\n몸무게: 44\n키: 156\nBMI:18.080210387902692\n결과:저체중\n\n이름: 자아\n몸무게: 66\n키: 188\nBMI:18.673607967406067\n결과:정상체중\n\n이름: 아나\n몸무게: 63\n키: 170\nBMI:21.79930795847751\n결과:정상체중\n\n이름: 차나\n몸무게: 71\n키: 169\nBMI:24.859073561850078\n결과:정상체중\n\n이름: 다차\n몸무게: 86\n키: 189\nBMI:24.075473810923548\n결과:정상체중\n\n이름: 차아\n몸무게: 54\n키: 157\nBMI:21.907582457706194\n결과:정상체중\n\n이름: 바하\n몸무게: 52\n키: 141\nBMI:26.155625974548567\n결과:과체중\n\n이름: 하라\n몸무게: 98\n키: 169\nBMI:34.31252407128602\n결과:과체중\n\n이름: 마다\n몸무게: 97\n키: 150\nBMI:43.111111111111114\n결과:과체중\n\n이름: 자파\n몸무게: 69\n키: 169\nBMI:24.158817968558527\n결과:정상체중\n\n이름: 하파\n몸무게: 87\n키: 191\nBMI:23.84803048162057\n결과:정상체중\n\n이름: 카마\n몸무게: 68\n키: 163\nBMI:25.59373706198954\n결과:과체중\n\n이름: 타카\n몸무게: 82\n키: 141\nBMI:41.24541019063428\n결과:과체중\n\n이름: 카아\n몸무게: 88\n키: 174\nBMI:29.065926806711587\n결과:과체중\n\n이름: 라마\n몸무게: 49\n키: 150\nBMI:21.77777777777778\n결과:정상체중\n\n이름: 라라\n몸무게: 43\n키: 181\nBMI:13.125362473672965\n결과:저체중\n\n이름: 자아\n몸무게: 97\n키: 160\nBMI:37.89062499999999\n결과:과체중\n\n이름: 차다\n몸무게: 92\n키: 179\nBMI:28.71321119815237\n결과:과체중\n\n이름: 사다\n몸무게: 93\n키: 186\nBMI:26.881720430107524\n결과:과체중\n\n이름: 나가\n몸무게: 74\n키: 156\nBMI:30.407626561472714\n결과:과체중\n\n이름: 하하\n몸무게: 48\n키: 178\nBMI:15.149602322939023\n결과:저체중\n\n이름: 자카\n몸무게: 96\n키: 147\nBMI:44.42593363876163\n결과:과체중\n\n이름: 바아\n몸무게: 54\n키: 174\nBMI:17.8359096313912\n결과:저체중\n\n이름: 가가\n몸무게: 42\n키: 149\nBMI:18.91806675374983\n결과:정상체중\n\n이름: 아파\n몸무게: 55\n키: 153\nBMI:23.49523687470631\n결과:정상체중\n\n이름: 사다\n몸무게: 60\n키: 181\nBMI:18.314459265590184\n결과:저체중\n\n이름: 카자\n몸무게: 73\n키: 185\nBMI:21.32943754565376\n결과:정상체중\n\n이름: 하가\n몸무게: 80\n키: 174\nBMI:26.42356982428326\n결과:과체중\n\n이름: 라차\n몸무게: 78\n키: 181\nBMI:23.808797045267237\n결과:정상체중\n\n이름: 파하\n몸무게: 92\n키: 171\nBMI:31.462672275229988\n결과:과체중\n\n이름: 사다\n몸무게: 76\n키: 176\nBMI:24.535123966942148\n결과:정상체중\n\n이름: 다아\n몸무게: 54\n키: 165\nBMI:19.834710743801654\n결과:정상체중\n\n이름: 아마\n몸무게: 84\n키: 164\nBMI:31.231409875074366\n결과:과체중\n\n이름: 파자\n몸무게: 94\n키: 186\nBMI:27.170771187420506\n결과:과체중\n\n이름: 하하\n몸무게: 75\n키: 172\nBMI:25.351541373715524\n결과:과체중\n\n이름: 차마\n몸무게: 65\n키: 182\nBMI:19.623233908948194\n결과:정상체중\n\n이름: 마바\n몸무게: 86\n키: 194\nBMI:22.85046232330747\n결과:정상체중\n\n이름: 나카\n몸무게: 58\n키: 165\nBMI:21.30394857667585\n결과:정상체중\n\n이름: 타자\n몸무게: 44\n키: 181\nBMI:13.430603461432801\n결과:저체중\n\n이름: 라사\n몸무게: 84\n키: 191\nBMI:23.025684602944\n결과:정상체중\n\n이름: 가아\n몸무게: 87\n키: 165\nBMI:31.95592286501378\n결과:과체중\n\n이름: 차차\n몸무게: 48\n키: 173\nBMI:16.037956497043\n결과:저체중\n\n이름: 파사\n몸무게: 75\n키: 155\nBMI:31.217481789802285\n결과:과체중\n\n이름: 사하\n몸무게: 80\n키: 187\nBMI:22.877405702193368\n결과:정상체중\n\n이름: 사파\n몸무게: 93\n키: 150\nBMI:41.333333333333336\n결과:과체중\n\n이름: 아나\n몸무게: 86\n키: 159\nBMI:34.01764170721095\n결과:과체중\n\n이름: 카가\n몸무게: 88\n키: 187\nBMI:25.165146272412706\n결과:과체중\n\n이름: 마아\n몸무게: 97\n키: 172\nBMI:32.787993510005414\n결과:과체중\n\n이름: 파카\n몸무게: 72\n키: 160\nBMI:28.124999999999993\n결과:과체중\n\n이름: 나마\n몸무게: 51\n키: 176\nBMI:16.464359504132233\n결과:저체중\n\n이름: 가라\n몸무게: 69\n키: 143\nBMI:33.74248129492885\n결과:과체중\n\n이름: 사나\n몸무게: 62\n키: 172\nBMI:20.9572742022715\n결과:정상체중\n\n이름: 가가\n몸무게: 93\n키: 156\nBMI:38.21499013806706\n결과:과체중\n\n이름: 바타\n몸무게: 88\n키: 192\nBMI:23.87152777777778\n결과:정상체중\n\n이름: 카다\n몸무게: 44\n키: 165\nBMI:16.161616161616163\n결과:저체중\n\n이름: 차자\n몸무게: 88\n키: 152\nBMI:38.08864265927978\n결과:과체중\n\n이름: 파자\n몸무게: 67\n키: 198\nBMI:17.090092847668604\n결과:저체중\n\n이름: 사마\n몸무게: 67\n키: 184\nBMI:19.78969754253308\n결과:정상체중\n\n이름: 하사\n몸무게: 61\n키: 166\nBMI:22.136739730004358\n결과:정상체중\n\n이름: 나파\n몸무게: 76\n키: 180\nBMI:23.456790123456788\n결과:정상체중\n\n이름: 바나\n몸무게: 98\n키: 174\nBMI:32.368873034747\n결과:과체중\n\n이름: 바나\n몸무게: 99\n키: 153\nBMI:42.29142637447136\n결과:과체중\n\n이름: 가라\n몸무게: 55\n키: 189\nBMI:15.397105344195293\n결과:저체중\n\n이름: 가카\n몸무게: 81\n키: 165\nBMI:29.752066115702483\n결과:과체중\n\n이름: 차파\n몸무게: 51\n키: 180\nBMI:15.74074074074074\n결과:저체중\n\n이름: 자자\n몸무게: 42\n키: 164\nBMI:15.615704937537183\n결과:저체중\n\n이름: 가카\n몸무게: 92\n키: 166\nBMI:33.386558281318045\n결과:과체중\n\n이름: 사아\n몸무게: 66\n키: 145\nBMI:31.391200951248514\n결과:과체중\n\n이름: 사카\n몸무게: 63\n키: 188\nBMI:17.824807605251245\n결과:저체중\n\n이름: 가사\n몸무게: 84\n키: 196\nBMI:21.86588921282799\n결과:정상체중\n\n이름: 라카\n몸무게: 75\n키: 149\nBMI:33.782262060267556\n결과:과체중\n\n이름: 타하\n몸무게: 52\n키: 145\nBMI:24.732461355529132\n결과:정상체중\n\n이름: 하가\n몸무게: 71\n키: 174\nBMI:23.450918219051392\n결과:정상체중\n\n이름: 나파\n몸무게: 89\n키: 176\nBMI:28.731921487603305\n결과:과체중\n\n이름: 차사\n몸무게: 81\n키: 173\nBMI:27.064051588760066\n결과:과체중\n\n이름: 바카\n몸무게: 46\n키: 144\nBMI:22.183641975308642\n결과:정상체중\n\n이름: 라하\n몸무게: 59\n키: 162\nBMI:22.481329065691202\n결과:정상체중\n\n이름: 라카\n몸무게: 92\n키: 145\nBMI:43.75743162901308\n결과:과체중\n\n이름: 마카\n몸무게: 92\n키: 193\nBMI:24.69864962817794\n결과:정상체중\n\n이름: 차바\n몸무게: 71\n키: 148\nBMI:32.414170927684445\n결과:과체중\n\n이름: 차아\n몸무게: 71\n키: 183\nBMI:21.20099137030069\n결과:정상체중\n\n이름: 카다\n몸무게: 60\n키: 141\nBMI:30.179568432171422\n결과:과체중\n\n이름: 자아\n몸무게: 75\n키: 163\nBMI:28.228386465429637\n결과:과체중\n\n이름: 라라\n몸무게: 46\n키: 173\nBMI:15.36970830966621\n결과:저체중\n\n이름: 하카\n몸무게: 92\n키: 152\nBMI:39.81994459833795\n결과:과체중\n\n이름: 차아\n몸무게: 70\n키: 142\nBMI:34.71533425907558\n결과:과체중\n\n이름: 아파\n몸무게: 71\n키: 163\nBMI:26.722872520606725\n결과:과체중\n\n이름: 차하\n몸무게: 50\n키: 184\nBMI:14.768431001890358\n결과:저체중\n\n이름: 나카\n몸무게: 55\n키: 159\nBMI:21.75546853368142\n결과:정상체중\n\n이름: 아타\n몸무게: 72\n키: 150\nBMI:32.0\n결과:과체중\n\n이름: 마가\n몸무게: 42\n키: 154\nBMI:17.70956316410862\n결과:저체중\n\n이름: 아가\n몸무게: 94\n키: 168\nBMI:33.30498866213152\n결과:과체중\n\n이름: 라카\n몸무게: 44\n키: 158\nBMI:17.625380547989103\n결과:저체중\n\n이름: 차바\n몸무게: 74\n키: 169\nBMI:25.909456951787405\n결과:과체중\n\n이름: 사카\n몸무게: 86\n키: 192\nBMI:23.328993055555557\n결과:정상체중\n\n이름: 파카\n몸무게: 41\n키: 192\nBMI:11.121961805555555\n결과:저체중\n\n이름: 가아\n몸무게: 68\n키: 152\nBMI:29.43213296398892\n결과:과체중\n\n이름: 차파\n몸무게: 95\n키: 181\nBMI:28.997893837184456\n결과:과체중\n\n이름: 자라\n몸무게: 91\n키: 175\nBMI:29.714285714285715\n결과:과체중\n\n이름: 아카\n몸무게: 67\n키: 154\nBMI:28.25096980941137\n결과:과체중\n\n이름: 자라\n몸무게: 88\n키: 158\nBMI:35.250761095978206\n결과:과체중\n\n이름: 가바\n몸무게: 64\n키: 193\nBMI:17.181669306558565\n결과:저체중\n\n이름: 라다\n몸무게: 57\n키: 190\nBMI:15.789473684210527\n결과:저체중\n\n이름: 가가\n몸무게: 95\n키: 192\nBMI:25.770399305555557\n결과:과체중\n\n이름: 차차\n몸무게: 91\n키: 169\nBMI:31.861629494765594\n결과:과체중\n\n이름: 아차\n몸무게: 97\n키: 164\nBMI:36.06484235574064\n결과:과체중\n\n이름: 라사\n몸무게: 65\n키: 162\nBMI:24.76756591982929\n결과:정상체중\n\n이름: 다라\n몸무게: 96\n키: 197\nBMI:24.736530186296992\n결과:정상체중\n\n이름: 사사\n몸무게: 54\n키: 178\nBMI:17.043302613306402\n결과:저체중\n\n이름: 자카\n몸무게: 92\n키: 151\nBMI:40.349107495285295\n결과:과체중\n\n이름: 차라\n몸무게: 55\n키: 174\nBMI:18.166204254194742\n결과:저체중\n\n이름: 파자\n몸무게: 68\n키: 150\nBMI:30.22222222222222\n결과:과체중\n\n이름: 사사\n몸무게: 45\n키: 163\nBMI:16.937031879257784\n결과:저체중\n\n이름: 사라\n몸무게: 84\n키: 175\nBMI:27.428571428571427\n결과:과체중\n\n이름: 차나\n몸무게: 97\n키: 159\nBMI:38.36873541394723\n결과:과체중\n\n이름: 가카\n몸무게: 69\n키: 155\nBMI:28.720083246618103\n결과:과체중\n\n이름: 파차\n몸무게: 65\n키: 171\nBMI:22.229061933586404\n결과:정상체중\n\n이름: 자파\n몸무게: 91\n키: 158\nBMI:36.45249158788655\n결과:과체중\n\n이름: 하다\n몸무게: 61\n키: 171\nBMI:20.86111966075032\n결과:정상체중\n\n이름: 마다\n몸무게: 67\n키: 147\nBMI:31.005599518719055\n결과:과체중\n\n이름: 타바\n몸무게: 70\n키: 174\nBMI:23.120623596247853\n결과:정상체중\n\n이름: 마파\n몸무게: 82\n키: 199\nBMI:20.70654781444913\n결과:정상체중\n\n이름: 자카\n몸무게: 66\n키: 163\nBMI:24.840980089578082\n결과:정상체중\n\n이름: 하나\n몸무게: 58\n키: 192\nBMI:15.733506944444445\n결과:저체중\n\n이름: 라바\n몸무게: 51\n키: 172\nBMI:17.239048134126556\n결과:저체중\n\n이름: 카나\n몸무게: 49\n키: 162\nBMI:18.670934308794386\n결과:정상체중\n\n이름: 자가\n몸무게: 51\n키: 182\nBMI:15.396691220867044\n결과:저체중\n\n이름: 카가\n몸무게: 71\n키: 165\nBMI:26.07897153351699\n결과:과체중\n\n이름: 다가\n몸무게: 97\n키: 174\nBMI:32.038578411943455\n결과:과체중\n\n이름: 사마\n몸무게: 63\n키: 187\nBMI:18.015956990477278\n결과:저체중\n\n이름: 파하\n몸무게: 71\n키: 152\nBMI:30.730609418282548\n결과:과체중\n\n이름: 사나\n몸무게: 88\n키: 192\nBMI:23.87152777777778\n결과:정상체중\n\n이름: 카파\n몸무게: 62\n키: 197\nBMI:15.975675745316808\n결과:저체중\n\n이름: 자바\n몸무게: 78\n키: 169\nBMI:27.309968138370508\n결과:과체중\n\n이름: 가타\n몸무게: 63\n키: 194\nBMI:16.73929216707408\n결과:저체중\n\n이름: 다다\n몸무게: 65\n키: 154\nBMI:27.40765727778715\n결과:과체중\n\n이름: 사다\n몸무게: 48\n키: 193\nBMI:12.886251979918924\n결과:저체중\n\n이름: 가파\n몸무게: 84\n키: 172\nBMI:28.393726338561386\n결과:과체중\n\n이름: 나차\n몸무게: 47\n키: 183\nBMI:14.034459076114544\n결과:저체중\n\n이름: 차파\n몸무게: 43\n키: 176\nBMI:13.881714876033058\n결과:저체중\n\n이름: 타파\n몸무게: 79\n키: 178\nBMI:24.933720489837143\n결과:정상체중\n\n이름: 파파\n몸무게: 75\n키: 179\nBMI:23.40750912892856\n결과:정상체중\n\n이름: 나타\n몸무게: 54\n키: 189\nBMI:15.117157974300833\n결과:저체중\n\n이름: 마마\n몸무게: 80\n키: 180\nBMI:24.691358024691358\n결과:정상체중\n\n이름: 나하\n몸무게: 46\n키: 180\nBMI:14.19753086419753\n결과:저체중\n\n이름: 바아\n몸무게: 77\n키: 172\nBMI:26.027582477014604\n결과:과체중\n\n이름: 가하\n몸무게: 53\n키: 141\nBMI:26.658618781751425\n결과:과체중\n\n이름: 다파\n몸무게: 78\n키: 186\nBMI:22.54595907041276\n결과:정상체중\n\n이름: 타라\n몸무게: 93\n키: 154\nBMI:39.21403272052623\n결과:과체중\n\n이름: 자자\n몸무게: 65\n키: 176\nBMI:20.983987603305785\n결과:정상체중\n\n이름: 나마\n몸무게: 96\n키: 145\nBMI:45.65992865636147\n결과:과체중\n\n이름: 자마\n몸무게: 58\n키: 185\nBMI:16.946676406135865\n결과:저체중\n\n이름: 차타\n몸무게: 86\n키: 143\nBMI:42.05584625165045\n결과:과체중\n\n이름: 가가\n몸무게: 95\n키: 163\nBMI:35.75595618954421\n결과:과체중\n\n이름: 바아\n몸무게: 57\n키: 187\nBMI:16.300151562812776\n결과:저체중\n\n이름: 나나\n몸무게: 79\n키: 173\nBMI:26.395803401383272\n결과:과체중\n\n이름: 자타\n몸무게: 67\n키: 180\nBMI:20.679012345679013\n결과:정상체중\n\n이름: 하카\n몸무게: 83\n키: 169\nBMI:29.060607121599386\n결과:과체중\n\n이름: 카하\n몸무게: 53\n키: 160\nBMI:20.703124999999996\n결과:정상체중\n\n이름: 나바\n몸무게: 99\n키: 167\nBMI:35.49786654236437\n결과:과체중\n\n이름: 파라\n몸무게: 93\n키: 171\nBMI:31.80465784343901\n결과:과체중\n\n이름: 하카\n몸무게: 57\n키: 189\nBMI:15.957000083984212\n결과:저체중\n\n이름: 아아\n몸무게: 85\n키: 164\nBMI:31.603212373587155\n결과:과체중\n\n이름: 카다\n몸무게: 82\n키: 171\nBMI:28.042816593139772\n결과:과체중\n\n이름: 나타\n몸무게: 80\n키: 159\nBMI:31.644317867172973\n결과:과체중\n\n이름: 나자\n몸무게: 98\n키: 151\nBMI:42.98057102758651\n결과:과체중\n\n이름: 하다\n몸무게: 59\n키: 162\nBMI:22.481329065691202\n결과:정상체중\n\n이름: 바자\n몸무게: 51\n키: 177\nBMI:16.278847074595422\n결과:저체중\n\n이름: 바차\n몸무게: 72\n키: 164\nBMI:26.769779892920884\n결과:과체중\n\n이름: 하나\n몸무게: 63\n키: 159\nBMI:24.919900320398717\n결과:정상체중\n\n이름: 바다\n몸무게: 58\n키: 156\nBMI:23.83300460223537\n결과:정상체중\n\n이름: 아아\n몸무게: 44\n키: 147\nBMI:20.36188625109908\n결과:정상체중\n\n이름: 아타\n몸무게: 99\n키: 168\nBMI:35.0765306122449\n결과:과체중\n\n이름: 다아\n몸무게: 45\n키: 194\nBMI:11.95663726219577\n결과:저체중\n\n이름: 아자\n몸무게: 81\n키: 154\nBMI:34.15415753078091\n결과:과체중\n\n이름: 타아\n몸무게: 41\n키: 143\nBMI:20.049880189740332\n결과:정상체중\n\n이름: 카사\n몸무게: 81\n키: 187\nBMI:23.163373273470786\n결과:정상체중\n\n이름: 바라\n몸무게: 52\n키: 194\nBMI:13.81655861409289\n결과:저체중\n\n이름: 카사\n몸무게: 59\n키: 165\nBMI:21.6712580348944\n결과:정상체중\n\n이름: 타자\n몸무게: 68\n키: 152\nBMI:29.43213296398892\n결과:과체중\n\n이름: 자파\n몸무게: 91\n키: 143\nBMI:44.50095359186269\n결과:과체중\n\n이름: 가다\n몸무게: 75\n키: 180\nBMI:23.148148148148145\n결과:정상체중\n\n이름: 가라\n몸무게: 83\n키: 180\nBMI:25.61728395061728\n결과:과체중\n\n이름: 사카\n몸무게: 73\n키: 150\nBMI:32.44444444444444\n결과:과체중\n\n이름: 하다\n몸무게: 57\n키: 153\nBMI:24.34960912469563\n결과:정상체중\n\n이름: 파파\n몸무게: 43\n키: 161\nBMI:16.58886617028664\n결과:저체중\n\n이름: 사아\n몸무게: 51\n키: 185\nBMI:14.901387874360847\n결과:저체중\n\n이름: 카다\n몸무게: 95\n키: 193\nBMI:25.50404037692287\n결과:과체중\n\n이름: 바하\n몸무게: 45\n키: 142\nBMI:22.317000595120017\n결과:정상체중\n\n이름: 나파\n몸무게: 97\n키: 179\nBMI:30.273711806747606\n결과:과체중\n\n이름: 자마\n몸무게: 41\n키: 171\nBMI:14.021408296569886\n결과:저체중\n\n이름: 사차\n몸무게: 99\n키: 185\nBMI:28.926223520818112\n결과:과체중\n\n이름: 아나\n몸무게: 86\n키: 186\nBMI:24.858365128916635\n결과:정상체중\n\n이름: 카다\n몸무게: 51\n키: 198\nBMI:13.008876645240282\n결과:저체중\n\n이름: 타사\n몸무게: 83\n키: 171\nBMI:28.384802161348794\n결과:과체중\n\n이름: 아타\n몸무게: 66\n키: 155\nBMI:27.47138397502601\n결과:과체중\n\n이름: 아가\n몸무게: 68\n키: 197\nBMI:17.52170888196037\n결과:저체중\n\n이름: 차나\n몸무게: 70\n키: 190\nBMI:19.390581717451525\n결과:정상체중\n\n이름: 타바\n몸무게: 92\n키: 151\nBMI:40.349107495285295\n결과:과체중\n\n이름: 타다\n몸무게: 63\n키: 142\nBMI:31.243800833168024\n결과:과체중\n\n이름: 하바\n몸무게: 97\n키: 156\nBMI:39.85864562787639\n결과:과체중\n\n이름: 파나\n몸무게: 40\n키: 184\nBMI:11.814744801512287\n결과:저체중\n\n이름: 파차\n몸무게: 90\n키: 162\nBMI:34.293552812071326\n결과:과체중\n\n이름: 바사\n몸무게: 91\n키: 193\nBMI:24.43018604526296\n결과:정상체중\n\n이름: 바나\n몸무게: 41\n키: 156\nBMI:16.847468770545692\n결과:저체중\n\n이름: 카아\n몸무게: 77\n키: 192\nBMI:20.887586805555557\n결과:정상체중\n\n이름: 차바\n몸무게: 58\n키: 175\nBMI:18.93877551020408\n결과:정상체중\n\n이름: 사자\n몸무게: 70\n키: 166\nBMI:25.402816083611555\n결과:과체중\n\n이름: 하하\n몸무게: 89\n키: 154\nBMI:37.52740765727779\n결과:과체중\n\n이름: 타타\n몸무게: 89\n키: 172\nBMI:30.08382909680909\n결과:과체중\n\n이름: 사마\n몸무게: 47\n키: 161\nBMI:18.132016511708652\n결과:저체중\n\n이름: 차사\n몸무게: 90\n키: 193\nBMI:24.161722462347985\n결과:정상체중\n\n이름: 라사\n몸무게: 45\n키: 168\nBMI:15.94387755102041\n결과:저체중\n\n이름: 라바\n몸무게: 51\n키: 162\nBMI:19.43301326017375\n결과:정상체중\n\n이름: 타타\n몸무게: 57\n키: 187\nBMI:16.300151562812776\n결과:저체중\n\n이름: 다마\n몸무게: 88\n키: 167\nBMI:31.553659148768332\n결과:과체중\n\n이름: 다나\n몸무게: 63\n키: 198\nBMI:16.069788797061523\n결과:저체중\n\n이름: 아하\n몸무게: 76\n키: 146\nBMI:35.65396884969038\n결과:과체중\n\n이름: 아라\n몸무게: 79\n키: 194\nBMI:20.990540971410354\n결과:정상체중\n\n이름: 차카\n몸무게: 51\n키: 174\nBMI:16.84502576298058\n결과:저체중\n\n이름: 나마\n몸무게: 82\n키: 154\nBMI:34.57581379659302\n결과:과체중\n\n이름: 하카\n몸무게: 47\n키: 141\nBMI:23.640661938534283\n결과:정상체중\n\n이름: 타사\n몸무게: 67\n키: 145\nBMI:31.86682520808561\n결과:과체중\n\n이름: 바타\n몸무게: 41\n키: 164\nBMI:15.243902439024392\n결과:저체중\n\n이름: 바마\n몸무게: 82\n키: 162\nBMI:31.24523700655387\n결과:과체중\n\n이름: 파차\n몸무게: 64\n키: 147\nBMI:29.617289092507754\n결과:과체중\n\n이름: 타카\n몸무게: 92\n키: 189\nBMI:25.755158030290307\n결과:과체중\n\n이름: 아가\n몸무게: 55\n키: 196\nBMI:14.316951270304042\n결과:저체중\n\n이름: 자사\n몸무게: 94\n키: 145\nBMI:44.70868014268728\n결과:과체중\n\n이름: 다타\n몸무게: 78\n키: 199\nBMI:19.696472311305268\n결과:정상체중\n\n이름: 차자\n몸무게: 74\n키: 196\nBMI:19.2628071636818\n결과:정상체중\n\n이름: 파자\n몸무게: 69\n키: 176\nBMI:22.275309917355372\n결과:정상체중\n\n이름: 타차\n몸무게: 43\n키: 171\nBMI:14.705379432987929\n결과:저체중\n\n이름: 다라\n몸무게: 81\n키: 176\nBMI:26.149276859504134\n결과:과체중\n\n이름: 차다\n몸무게: 98\n키: 166\nBMI:35.56394251705618\n결과:과체중\n\n이름: 가다\n몸무게: 84\n키: 184\nBMI:24.8109640831758\n결과:정상체중\n\n이름: 파가\n몸무게: 78\n키: 180\nBMI:24.074074074074073\n결과:정상체중\n\n이름: 다타\n몸무게: 72\n키: 190\nBMI:19.94459833795014\n결과:정상체중\n\n이름: 파아\n몸무게: 45\n키: 168\nBMI:15.94387755102041\n결과:저체중\n\n이름: 마자\n몸무게: 85\n키: 146\nBMI:39.876149371364235\n결과:과체중\n\n이름: 차가\n몸무게: 59\n키: 157\nBMI:23.93606231490121\n결과:정상체중\n\n이름: 카사\n몸무게: 67\n키: 144\nBMI:32.310956790123456\n결과:과체중\n\n이름: 아마\n몸무게: 71\n키: 167\nBMI:25.458065904119906\n결과:과체중\n\n이름: 나사\n몸무게: 56\n키: 193\nBMI:15.033960643238744\n결과:저체중\n\n이름: 나하\n몸무게: 66\n키: 147\nBMI:30.542829376648623\n결과:과체중\n\n이름: 자사\n몸무게: 69\n키: 165\nBMI:25.344352617079892\n결과:과체중\n\n이름: 자라\n몸무게: 64\n키: 198\nBMI:16.324864809713294\n결과:저체중\n\n이름: 다아\n몸무게: 91\n키: 148\nBMI:41.54492330168006\n결과:과체중\n\n이름: 사나\n몸무게: 50\n키: 181\nBMI:15.26204938799182\n결과:저체중\n\n이름: 사아\n몸무게: 56\n키: 145\nBMI:26.634958382877528\n결과:과체중\n\n이름: 사마\n몸무게: 97\n키: 145\nBMI:46.13555291319857\n결과:과체중\n\n이름: 차파\n몸무게: 57\n키: 194\nBMI:15.145073865447976\n결과:저체중\n\n이름: 라마\n몸무게: 79\n키: 183\nBMI:23.589835468362743\n결과:정상체중\n\n이름: 가타\n몸무게: 87\n키: 169\nBMI:30.461118308182492\n결과:과체중\n\n이름: 나자\n몸무게: 58\n키: 196\nBMI:15.097875885047898\n결과:저체중\n\n이름: 파타\n몸무게: 92\n키: 147\nBMI:42.5748530704799\n결과:과체중\n\n이름: 파나\n몸무게: 89\n키: 140\nBMI:45.40816326530613\n결과:과체중\n\n이름: 하카\n몸무게: 94\n키: 181\nBMI:28.692652849424622\n결과:과체중\n\n이름: 바바\n몸무게: 43\n키: 175\nBMI:14.040816326530612\n결과:저체중\n\n이름: 차사\n몸무게: 68\n키: 184\nBMI:20.085066162570886\n결과:정상체중\n\n이름: 바아\n몸무게: 47\n키: 186\nBMI:13.585385593710253\n결과:저체중\n\n이름: 라라\n몸무게: 79\n키: 169\nBMI:27.660095935016283\n결과:과체중\n\n이름: 바마\n몸무게: 65\n키: 140\nBMI:33.163265306122454\n결과:과체중\n\n이름: 파아\n몸무게: 74\n키: 161\nBMI:28.54828131630724\n결과:과체중\n\n이름: 자라\n몸무게: 88\n키: 194\nBMI:23.381868423849507\n결과:정상체중\n\n이름: 파다\n몸무게: 77\n키: 171\nBMI:26.332888752094664\n결과:과체중\n\n이름: 라차\n몸무게: 82\n키: 155\nBMI:34.131113423517164\n결과:과체중\n\n이름: 타차\n몸무게: 68\n키: 196\nBMI:17.700957934194086\n결과:저체중\n\n이름: 다라\n몸무게: 52\n키: 168\nBMI:18.42403628117914\n결과:저체중\n\n이름: 아파\n몸무게: 65\n키: 194\nBMI:17.270698267616112\n결과:저체중\n\n이름: 자바\n몸무게: 61\n키: 194\nBMI:16.207886066532044\n결과:저체중\n\n이름: 차차\n몸무게: 50\n키: 161\nBMI:19.28937926777516\n결과:정상체중\n\n이름: 파파\n몸무게: 54\n키: 169\nBMI:18.906901018871892\n결과:정상체중\n\n이름: 하타\n몸무게: 95\n키: 141\nBMI:47.784316684271424\n결과:과체중\n\n이름: 다사\n몸무게: 78\n키: 180\nBMI:24.074074074074073\n결과:정상체중\n\n이름: 아마\n몸무게: 80\n키: 198\nBMI:20.40608101214162\n결과:정상체중\n\n이름: 파자\n몸무게: 94\n키: 157\nBMI:38.135421315266335\n결과:과체중\n\n이름: 사다\n몸무게: 41\n키: 181\nBMI:12.514880498153293\n결과:저체중\n\n이름: 하사\n몸무게: 76\n키: 195\nBMI:19.986850756081527\n결과:정상체중\n\n이름: 사자\n몸무게: 81\n키: 178\nBMI:25.5649539199596\n결과:과체중\n\n이름: 사마\n몸무게: 92\n키: 199\nBMI:23.23173657230878\n결과:정상체중\n\n이름: 마가\n몸무게: 63\n키: 161\nBMI:24.3046178773967\n결과:정상체중\n\n이름: 타가\n몸무게: 67\n키: 175\nBMI:21.877551020408163\n결과:정상체중\n\n이름: 바라\n몸무게: 65\n키: 153\nBMI:27.76709812465291\n결과:과체중\n\n이름: 다파\n몸무게: 46\n키: 177\nBMI:14.682881675125282\n결과:저체중\n\n이름: 아라\n몸무게: 64\n키: 145\nBMI:30.439952437574316\n결과:과체중\n\n이름: 바아\n몸무게: 82\n키: 188\nBMI:23.20054323223178\n결과:정상체중\n\n이름: 차사\n몸무게: 49\n키: 173\nBMI:16.372080590731397\n결과:저체중\n\n이름: 마자\n몸무게: 41\n키: 198\nBMI:10.45811651872258\n결과:저체중\n\n이름: 나다\n몸무게: 42\n키: 199\nBMI:10.60579278301053\n결과:저체중\n\n이름: 자나\n몸무게: 52\n키: 183\nBMI:15.527486637403324\n결과:저체중\n\n이름: 차마\n몸무게: 55\n키: 156\nBMI:22.600262984878366\n결과:정상체중\n\n이름: 파하\n몸무게: 40\n키: 160\nBMI:15.624999999999996\n결과:저체중\n\n이름: 하파\n몸무게: 85\n키: 193\nBMI:22.819404547773097\n결과:정상체중\n\n이름: 하아\n몸무게: 93\n키: 162\nBMI:35.43667123914037\n결과:과체중\n\n이름: 하바\n몸무게: 50\n키: 189\nBMI:13.997368494722993\n결과:저체중\n\n이름: 나카\n몸무게: 93\n키: 178\nBMI:29.352354500694357\n결과:과체중\n\n이름: 차하\n몸무게: 99\n키: 141\nBMI:49.79628791308285\n결과:과체중\n\n이름: 카자\n몸무게: 50\n키: 199\nBMI:12.62594378929825\n결과:저체중\n\n이름: 하아\n몸무게: 84\n키: 180\nBMI:25.925925925925924\n결과:과체중\n\n이름: 나다\n몸무게: 81\n키: 159\nBMI:32.03987184051263\n결과:과체중\n\n이름: 아나\n몸무게: 48\n키: 147\nBMI:22.212966819380814\n결과:정상체중\n\n이름: 바차\n몸무게: 48\n키: 153\nBMI:20.504933999743688\n결과:정상체중\n\n이름: 자아\n몸무게: 76\n키: 161\nBMI:29.319856487018246\n결과:과체중\n\n이름: 자차\n몸무게: 98\n키: 193\nBMI:26.309431125667803\n결과:과체중\n\n이름: 아라\n몸무게: 46\n키: 191\nBMI:12.609303473040761\n결과:저체중\n\n이름: 카나\n몸무게: 70\n키: 198\nBMI:17.855320885623918\n결과:저체중\n\n이름: 자차\n몸무게: 73\n키: 157\nBMI:29.61580591504726\n결과:과체중\n\n이름: 나나\n몸무게: 87\n키: 151\nBMI:38.15622121836761\n결과:과체중\n\n이름: 차사\n몸무게: 78\n키: 156\nBMI:32.05128205128205\n결과:과체중\n\n이름: 사하\n몸무게: 55\n키: 182\nBMI:16.604274846033086\n결과:저체중\n\n이름: 바바\n몸무게: 64\n키: 141\nBMI:32.191539660982855\n결과:과체중\n\n이름: 다바\n몸무게: 96\n키: 170\nBMI:33.21799307958478\n결과:과체중\n\n이름: 마라\n몸무게: 92\n키: 140\nBMI:46.93877551020409\n결과:과체중\n\n이름: 마아\n몸무게: 49\n키: 171\nBMI:16.75729284224206\n결과:저체중\n\n이름: 마카\n몸무게: 52\n키: 196\nBMI:13.536026655560184\n결과:저체중\n\n이름: 사타\n몸무게: 85\n키: 144\nBMI:40.99151234567901\n결과:과체중\n\n이름: 타파\n몸무게: 70\n키: 193\nBMI:18.792450804048432\n결과:정상체중\n\n이름: 마가\n몸무게: 57\n키: 173\nBMI:19.045073340238563\n결과:정상체중\n\n이름: 파바\n몸무게: 84\n키: 165\nBMI:30.85399449035813\n결과:과체중\n\n이름: 아카\n몸무게: 43\n키: 193\nBMI:11.543934065344036\n결과:저체중\n\n이름: 타가\n몸무게: 97\n키: 191\nBMI:26.589183410542475\n결과:과체중\n\n이름: 자카\n몸무게: 85\n키: 156\nBMI:34.927679158448385\n결과:과체중\n\n이름: 나다\n몸무게: 67\n키: 171\nBMI:22.91303307000445\n결과:정상체중\n\n이름: 자파\n몸무게: 68\n키: 153\nBMI:29.04865649963689\n결과:과체중\n\n이름: 타마\n몸무게: 93\n키: 167\nBMI:33.34648069131199\n결과:과체중\n\n이름: 아차\n몸무게: 94\n키: 167\nBMI:33.70504499982072\n결과:과체중\n\n이름: 하자\n몸무게: 63\n키: 179\nBMI:19.66230766829999\n결과:정상체중\n\n이름: 카나\n몸무게: 83\n키: 168\nBMI:29.40759637188209\n결과:과체중\n\n이름: 타라\n몸무게: 61\n키: 195\nBMI:16.04207758053912\n결과:저체중\n\n이름: 가라\n몸무게: 54\n키: 169\nBMI:18.906901018871892\n결과:정상체중\n\n이름: 자라\n몸무게: 59\n키: 155\nBMI:24.55775234131113\n결과:정상체중\n\n이름: 타카\n몸무게: 82\n키: 153\nBMI:35.02926224956214\n결과:과체중\n\n이름: 타카\n몸무게: 98\n키: 193\nBMI:26.309431125667803\n결과:과체중\n\n이름: 바다\n몸무게: 49\n키: 191\nBMI:13.431649351717333\n결과:저체중\n\n이름: 차바\n몸무게: 43\n키: 150\nBMI:19.11111111111111\n결과:정상체중\n\n이름: 바다\n몸무게: 82\n키: 164\nBMI:30.487804878048784\n결과:과체중\n\n이름: 마바\n몸무게: 60\n키: 149\nBMI:27.025809648214047\n결과:과체중\n\n이름: 카나\n몸무게: 66\n키: 168\nBMI:23.384353741496604\n결과:정상체중\n\n이름: 카아\n몸무게: 57\n키: 164\nBMI:21.192742415229034\n결과:정상체중\n\n이름: 마다\n몸무게: 72\n키: 170\nBMI:24.913494809688583\n결과:정상체중\n\n이름: 차라\n몸무게: 55\n키: 187\nBMI:15.72821642025794\n결과:저체중\n\n이름: 타가\n몸무게: 41\n키: 180\nBMI:12.65432098765432\n결과:저체중\n\n이름: 파가\n몸무게: 47\n키: 178\nBMI:14.833985607877793\n결과:저체중\n\n이름: 사차\n몸무게: 79\n키: 196\nBMI:20.564348188254897\n결과:정상체중\n\n이름: 라하\n몸무게: 91\n키: 159\nBMI:35.99541157390926\n결과:과체중\n\n이름: 마가\n몸무게: 93\n키: 180\nBMI:28.703703703703702\n결과:과체중\n\n이름: 다나\n몸무게: 62\n키: 169\nBMI:21.707923392038097\n결과:정상체중\n\n이름: 가아\n몸무게: 70\n키: 154\nBMI:29.515938606847698\n결과:과체중\n\n이름: 라아\n몸무게: 77\n키: 189\nBMI:21.55594748187341\n결과:정상체중\n\n이름: 마자\n몸무게: 64\n키: 177\nBMI:20.428357113217785\n결과:정상체중\n\n이름: 파사\n몸무게: 46\n키: 175\nBMI:15.020408163265307\n결과:저체중\n\n이름: 하마\n몸무게: 97\n키: 188\nBMI:27.44454504300589\n결과:과체중\n\n이름: 나차\n몸무게: 86\n키: 197\nBMI:22.159808291891057\n결과:정상체중\n\n이름: 마사\n몸무게: 96\n키: 148\nBMI:43.82761139517896\n결과:과체중\n\n이름: 나타\n몸무게: 81\n키: 156\nBMI:33.28402366863905\n결과:과체중\n\n이름: 마아\n몸무게: 59\n키: 159\nBMI:23.337684427040067\n결과:정상체중\n\n이름: 바파\n몸무게: 66\n키: 192\nBMI:17.903645833333332\n결과:저체중\n\n이름: 자타\n몸무게: 67\n키: 174\nBMI:22.12973972783723\n결과:정상체중\n\n이름: 카사\n몸무게: 87\n키: 178\nBMI:27.458654210326976\n결과:과체중\n\n이름: 나차\n몸무게: 77\n키: 151\nBMI:33.77044866453226\n결과:과체중\n\n이름: 카다\n몸무게: 56\n키: 153\nBMI:23.92242299970097\n결과:정상체중\n\n이름: 나타\n몸무게: 60\n키: 175\nBMI:19.591836734693878\n결과:정상체중\n\n이름: 하아\n몸무게: 72\n키: 169\nBMI:25.209201358495854\n결과:과체중\n\n이름: 다타\n몸무게: 93\n키: 194\nBMI:24.710383675204593\n결과:정상체중\n\n이름: 파아\n몸무게: 61\n키: 167\nBMI:21.872422819032593\n결과:정상체중\n\n이름: 라타\n몸무게: 70\n키: 162\nBMI:26.672763298277697\n결과:과체중\n\n이름: 차하\n몸무게: 78\n키: 186\nBMI:22.54595907041276\n결과:정상체중\n\n이름: 파카\n몸무게: 67\n키: 165\nBMI:24.609733700642796\n결과:정상체중\n\n이름: 파차\n몸무게: 53\n키: 142\nBMI:26.284467367585798\n결과:과체중\n\n이름: 사아\n몸무게: 98\n키: 172\nBMI:33.12601406165495\n결과:과체중\n\n이름: 자마\n몸무게: 75\n키: 147\nBMI:34.70776065528253\n결과:과체중\n\n이름: 바나\n몸무게: 60\n키: 160\nBMI:23.437499999999996\n결과:정상체중\n\n이름: 카나\n몸무게: 82\n키: 140\nBMI:41.83673469387756\n결과:과체중\n\n이름: 자카\n몸무게: 44\n키: 170\nBMI:15.224913494809691\n결과:저체중\n\n이름: 다바\n몸무게: 90\n키: 165\nBMI:33.057851239669425\n결과:과체중\n\n이름: 파아\n몸무게: 65\n키: 193\nBMI:17.450132889473544\n결과:저체중\n\n이름: 하파\n몸무게: 77\n키: 166\nBMI:27.94309769197271\n결과:과체중\n\n이름: 자차\n몸무게: 71\n키: 147\nBMI:32.85668008700079\n결과:과체중\n\n이름: 자다\n몸무게: 70\n키: 172\nBMI:23.661438615467823\n결과:정상체중\n\n이름: 차다\n몸무게: 51\n키: 173\nBMI:17.04032877810819\n결과:저체중\n\n이름: 라라\n몸무게: 46\n키: 173\nBMI:15.36970830966621\n결과:저체중\n\n이름: 다가\n몸무게: 40\n키: 140\nBMI:20.408163265306126\n결과:정상체중\n\n이름: 가바\n몸무게: 48\n키: 186\nBMI:13.874436351023238\n결과:저체중\n\n이름: 타사\n몸무게: 97\n키: 196\nBMI:25.249895876718035\n결과:과체중\n\n이름: 가차\n몸무게: 82\n키: 181\nBMI:25.029760996306585\n결과:과체중\n\n이름: 아아\n몸무게: 47\n키: 188\nBMI:13.297872340425533\n결과:저체중\n\n이름: 사아\n몸무게: 43\n키: 156\nBMI:17.66929651545036\n결과:저체중\n\n이름: 가바\n몸무게: 87\n키: 182\nBMI:26.26494384736143\n결과:과체중\n\n이름: 파사\n몸무게: 57\n키: 154\nBMI:24.034407151290267\n결과:정상체중\n\n이름: 하차\n몸무게: 51\n키: 170\nBMI:17.647058823529413\n결과:저체중\n\n이름: 하하\n몸무게: 57\n키: 188\nBMI:16.127206880941603\n결과:저체중\n\n이름: 하사\n몸무게: 75\n키: 170\nBMI:25.95155709342561\n결과:과체중\n\n이름: 라파\n몸무게: 69\n키: 155\nBMI:28.720083246618103\n결과:과체중\n\n이름: 타바\n몸무게: 64\n키: 167\nBMI:22.948115744558788\n결과:정상체중\n\n이름: 마나\n몸무게: 66\n키: 171\nBMI:22.571047501795427\n결과:정상체중\n\n이름: 하사\n몸무게: 64\n키: 152\nBMI:27.700831024930746\n결과:과체중\n\n이름: 다가\n몸무게: 56\n키: 185\nBMI:16.362308254200144\n결과:저체중\n\n이름: 아파\n몸무게: 88\n키: 177\nBMI:28.088991030674453\n결과:과체중\n\n이름: 나라\n몸무게: 89\n키: 148\nBMI:40.6318480642805\n결과:과체중\n\n이름: 자카\n몸무게: 91\n키: 150\nBMI:40.44444444444444\n결과:과체중\n\n이름: 타카\n몸무게: 86\n키: 162\nBMI:32.769394909312595\n결과:과체중\n\n이름: 나카\n몸무게: 84\n키: 189\nBMI:23.515579071134628\n결과:정상체중\n\n이름: 아자\n몸무게: 78\n키: 151\nBMI:34.209025919915796\n결과:과체중\n\n이름: 라가\n몸무게: 46\n키: 183\nBMI:13.735853563856788\n결과:저체중\n\n이름: 가자\n몸무게: 41\n키: 188\nBMI:11.60027161611589\n결과:저체중\n\n이름: 차다\n몸무게: 96\n키: 148\nBMI:43.82761139517896\n결과:과체중\n\n이름: 타하\n몸무게: 46\n키: 165\nBMI:16.896235078053262\n결과:저체중\n\n이름: 나마\n몸무게: 88\n키: 170\nBMI:30.449826989619382\n결과:과체중\n\n이름: 카다\n몸무게: 46\n키: 162\nBMI:17.527815881725342\n결과:저체중\n\n이름: 라다\n몸무게: 44\n키: 175\nBMI:14.36734693877551\n결과:저체중\n\n이름: 다마\n몸무게: 88\n키: 190\nBMI:24.37673130193906\n결과:정상체중\n\n이름: 가사\n몸무게: 62\n키: 172\nBMI:20.9572742022715\n결과:정상체중\n\n이름: 사카\n몸무게: 80\n키: 160\nBMI:31.249999999999993\n결과:과체중\n\n이름: 나자\n몸무게: 46\n키: 178\nBMI:14.518368892816563\n결과:저체중\n\n이름: 아하\n몸무게: 91\n키: 159\nBMI:35.99541157390926\n결과:과체중\n\n이름: 나하\n몸무게: 95\n키: 163\nBMI:35.75595618954421\n결과:과체중\n\n이름: 나카\n몸무게: 90\n키: 197\nBMI:23.19049704965343\n결과:정상체중\n\n이름: 카파\n몸무게: 98\n키: 199\nBMI:24.74684982702457\n결과:정상체중\n\n이름: 라라\n몸무게: 85\n키: 185\nBMI:24.835646457268076\n결과:정상체중\n\n이름: 하파\n몸무게: 67\n키: 169\nBMI:23.458562375266975\n결과:정상체중\n\n이름: 차마\n몸무게: 91\n키: 181\nBMI:27.776929886145112\n결과:과체중\n\n이름: 타타\n몸무게: 63\n키: 171\nBMI:21.545090797168363\n결과:정상체중\n\n이름: 카하\n몸무게: 88\n키: 145\nBMI:41.85493460166468\n결과:과체중\n\n이름: 카아\n몸무게: 81\n키: 177\nBMI:25.854639471416256\n결과:과체중\n\n이름: 사사\n몸무게: 73\n키: 160\nBMI:28.515624999999993\n결과:과체중\n\n이름: 카타\n몸무게: 95\n키: 196\nBMI:24.729279466888798\n결과:정상체중\n\n이름: 하마\n몸무게: 73\n키: 140\nBMI:37.24489795918368\n결과:과체중\n\n이름: 하카\n몸무게: 91\n키: 188\nBMI:25.746944318696244\n결과:과체중\n\n이름: 자타\n몸무게: 44\n키: 153\nBMI:18.79618949976505\n결과:정상체중\n\n이름: 사아\n몸무게: 83\n키: 197\nBMI:21.386791723569274\n결과:정상체중\n\n이름: 다나\n몸무게: 91\n키: 152\nBMI:39.387119113573405\n결과:과체중\n\n이름: 라아\n몸무게: 87\n키: 153\nBMI:37.165192874535435\n결과:과체중\n\n이름: 카가\n몸무게: 75\n키: 169\nBMI:26.25958474843318\n결과:과체중\n\n이름: 나카\n몸무게: 65\n키: 189\nBMI:18.19657904313989\n결과:저체중\n\n이름: 나타\n몸무게: 41\n키: 182\nBMI:12.377732157951938\n결과:저체중\n\n이름: 타라\n몸무게: 98\n키: 152\nBMI:42.41689750692521\n결과:과체중\n\n이름: 하다\n몸무게: 99\n키: 176\nBMI:31.960227272727273\n결과:과체중\n\n이름: 카다\n몸무게: 87\n키: 142\nBMI:43.14620115056537\n결과:과체중\n\n이름: 하마\n몸무게: 92\n키: 196\nBMI:23.94835485214494\n결과:정상체중\n\n이름: 자마\n몸무게: 46\n키: 180\nBMI:14.19753086419753\n결과:저체중\n\n이름: 사마\n몸무게: 73\n키: 163\nBMI:27.475629493018182\n결과:과체중\n\n이름: 바나\n몸무게: 59\n키: 198\nBMI:15.049484746454445\n결과:저체중\n\n이름: 아아\n몸무게: 79\n키: 162\nBMI:30.102118579484827\n결과:과체중\n\n이름: 바타\n몸무게: 46\n키: 149\nBMI:20.7197873969641\n결과:정상체중\n\n이름: 가자\n몸무게: 48\n키: 159\nBMI:18.986590720303784\n결과:정상체중\n\n이름: 사아\n몸무게: 63\n키: 159\nBMI:24.919900320398717\n결과:정상체중\n\n이름: 사하\n몸무게: 82\n키: 173\nBMI:27.39817568244846\n결과:과체중\n\n이름: 자차\n몸무게: 79\n키: 183\nBMI:23.589835468362743\n결과:정상체중\n\n이름: 카차\n몸무게: 76\n키: 186\nBMI:21.967857555786793\n결과:정상체중\n\n이름: 나가\n몸무게: 84\n키: 157\nBMI:34.0784616008763\n결과:과체중\n\n이름: 하다\n몸무게: 87\n키: 169\nBMI:30.461118308182492\n결과:과체중\n\n이름: 바다\n몸무게: 49\n키: 151\nBMI:21.490285513793253\n결과:정상체중\n\n이름: 다나\n몸무게: 44\n키: 187\nBMI:12.582573136206353\n결과:저체중\n\n이름: 다타\n몸무게: 82\n키: 153\nBMI:35.02926224956214\n결과:과체중\n\n이름: 나바\n몸무게: 90\n키: 188\nBMI:25.46401086464464\n결과:과체중\n\n이름: 자타\n몸무게: 66\n키: 192\nBMI:17.903645833333332\n결과:저체중\n\n이름: 나타\n몸무게: 82\n키: 148\nBMI:37.43608473338203\n결과:과체중\n\n이름: 사가\n몸무게: 47\n키: 154\nBMI:19.81784449316917\n결과:정상체중\n\n이름: 타자\n몸무게: 54\n키: 188\nBMI:15.278406518786783\n결과:저체중\n\n이름: 마아\n몸무게: 74\n키: 157\nBMI:30.021501886486266\n결과:과체중\n\n이름: 나가\n몸무게: 92\n키: 166\nBMI:33.386558281318045\n결과:과체중\n\n이름: 타바\n몸무게: 92\n키: 147\nBMI:42.5748530704799\n결과:과체중\n\n이름: 바하\n몸무게: 86\n키: 179\nBMI:26.840610467838083\n결과:과체중\n\n이름: 자하\n몸무게: 96\n키: 176\nBMI:30.991735537190085\n결과:과체중\n\n이름: 나나\n몸무게: 82\n키: 140\nBMI:41.83673469387756\n결과:과체중\n\n이름: 라차\n몸무게: 60\n키: 168\nBMI:21.258503401360546\n결과:정상체중\n\n이름: 마카\n몸무게: 69\n키: 186\nBMI:19.944502254595903\n결과:정상체중\n\n이름: 차카\n몸무게: 51\n키: 199\nBMI:12.878462665084214\n결과:저체중\n\n이름: 차다\n몸무게: 78\n키: 161\nBMI:30.091431657729252\n결과:과체중\n\n이름: 다파\n몸무게: 76\n키: 189\nBMI:21.27600011197895\n결과:정상체중\n\n이름: 타카\n몸무게: 81\n키: 147\nBMI:37.48438150770512\n결과:과체중\n\n이름: 카사\n몸무게: 93\n키: 174\nBMI:30.71739992072929\n결과:과체중\n\n이름: 나다\n몸무게: 44\n키: 144\nBMI:21.219135802469136\n결과:정상체중\n\n이름: 차하\n몸무게: 76\n키: 172\nBMI:25.689561925365066\n결과:과체중\n\n이름: 다자\n몸무게: 51\n키: 190\nBMI:14.127423822714682\n결과:저체중\n\n이름: 나하\n몸무게: 62\n키: 156\nBMI:25.476660092044707\n결과:과체중\n\n이름: 아마\n몸무게: 88\n키: 166\nBMI:31.934968790825955\n결과:과체중\n\n이름: 타바\n몸무게: 48\n키: 158\nBMI:19.227687870533565\n결과:정상체중\n\n이름: 차다\n몸무게: 89\n키: 148\nBMI:40.6318480642805\n결과:과체중\n\n이름: 타하\n몸무게: 51\n키: 162\nBMI:19.43301326017375\n결과:정상체중\n\n이름: 마마\n몸무게: 47\n키: 188\nBMI:13.297872340425533\n결과:저체중\n\n이름: 나다\n몸무게: 75\n키: 181\nBMI:22.89307408198773\n결과:정상체중\n\n이름: 자마\n몸무게: 49\n키: 185\nBMI:14.317019722425126\n결과:저체중\n\n이름: 하나\n몸무게: 40\n키: 166\nBMI:14.51589490492089\n결과:저체중\n\n이름: 바바\n몸무게: 53\n키: 193\nBMI:14.228569894493813\n결과:저체중\n\n이름: 파마\n몸무게: 46\n키: 178\nBMI:14.518368892816563\n결과:저체중\n\n이름: 파하\n몸무게: 98\n키: 165\nBMI:35.99632690541782\n결과:과체중\n\n이름: 타카\n몸무게: 90\n키: 195\nBMI:23.668639053254438\n결과:정상체중\n\n이름: 아바\n몸무게: 82\n키: 175\nBMI:26.775510204081634\n결과:과체중\n\n이름: 가바\n몸무게: 81\n키: 145\nBMI:38.525564803805\n결과:과체중\n\n이름: 하카\n몸무게: 76\n키: 199\nBMI:19.191434559733338\n결과:정상체중\n\n이름: 마차\n몸무게: 75\n키: 182\nBMI:22.6421929718633\n결과:정상체중\n\n이름: 파라\n몸무게: 66\n키: 178\nBMI:20.830703194041156\n결과:정상체중\n\n이름: 아나\n몸무게: 52\n키: 197\nBMI:13.398953850910871\n결과:저체중\n\n이름: 타하\n몸무게: 95\n키: 178\nBMI:29.983587930816814\n결과:과체중\n\n이름: 타카\n몸무게: 90\n키: 184\nBMI:26.583175803402646\n결과:과체중\n\n이름: 나자\n몸무게: 44\n키: 179\nBMI:13.73240535563809\n결과:저체중\n\n이름: 다가\n몸무게: 68\n키: 184\nBMI:20.085066162570886\n결과:정상체중\n\n이름: 가나\n몸무게: 78\n키: 196\nBMI:20.304039983340278\n결과:정상체중\n\n이름: 사가\n몸무게: 84\n키: 146\nBMI:39.407018202289365\n결과:과체중\n\n이름: 라바\n몸무게: 43\n키: 180\nBMI:13.271604938271604\n결과:저체중\n\n이름: 카라\n몸무게: 69\n키: 198\nBMI:17.600244872972148\n결과:저체중\n\n이름: 차사\n몸무게: 46\n키: 195\nBMI:12.097304404996713\n결과:저체중\n\n이름: 자아\n몸무게: 48\n키: 189\nBMI:13.437473754934073\n결과:저체중\n\n이름: 가나\n몸무게: 99\n키: 184\nBMI:29.24149338374291\n결과:과체중\n\n이름: 나라\n몸무게: 78\n키: 192\nBMI:21.158854166666668\n결과:정상체중\n\n이름: 나카\n몸무게: 78\n키: 178\nBMI:24.61810377477591\n결과:정상체중\n\n이름: 나카\n몸무게: 85\n키: 163\nBMI:31.992171327486922\n결과:과체중\n\n이름: 바차\n몸무게: 92\n키: 157\nBMI:37.32402937238833\n결과:과체중\n\n이름: 차하\n몸무게: 62\n키: 178\nBMI:19.568236333796236\n결과:정상체중\n\n이름: 자나\n몸무게: 63\n키: 159\nBMI:24.919900320398717\n결과:정상체중\n\n이름: 라바\n몸무게: 52\n키: 141\nBMI:26.155625974548567\n결과:과체중\n\n이름: 라자\n몸무게: 98\n키: 176\nBMI:31.63739669421488\n결과:과체중\n\n이름: 라하\n몸무게: 84\n키: 192\nBMI:22.786458333333336\n결과:정상체중\n\n이름: 하나\n몸무게: 98\n키: 198\nBMI:24.997449239873482\n결과:정상체중\n\n이름: 하타\n몸무게: 78\n키: 141\nBMI:39.23343896182285\n결과:과체중\n\n이름: 바바\n몸무게: 48\n키: 151\nBMI:21.051708258409718\n결과:정상체중\n\n이름: 파바\n몸무게: 51\n키: 155\nBMI:21.227887617065555\n결과:정상체중\n\n이름: 아라\n몸무게: 40\n키: 181\nBMI:12.209639510393455\n결과:저체중\n\n이름: 나사\n몸무게: 56\n키: 161\nBMI:21.60410477990818\n결과:정상체중\n\n이름: 나라\n몸무게: 49\n키: 185\nBMI:14.317019722425126\n결과:저체중\n\n이름: 차사\n몸무게: 63\n키: 149\nBMI:28.377100130624747\n결과:과체중\n\n이름: 마타\n몸무게: 96\n키: 160\nBMI:37.49999999999999\n결과:과체중\n\n이름: 마바\n몸무게: 64\n키: 166\nBMI:23.225431847873423\n결과:정상체중\n\n이름: 라차\n몸무게: 85\n키: 147\nBMI:39.33546207598686\n결과:과체중\n\n이름: 차파\n몸무게: 73\n키: 148\nBMI:33.327246165084006\n결과:과체중\n\n이름: 바사\n몸무게: 49\n키: 156\nBMI:20.134779750164363\n결과:정상체중\n\n이름: 사가\n몸무게: 86\n키: 197\nBMI:22.159808291891057\n결과:정상체중\n\n이름: 타파\n몸무게: 40\n키: 142\nBMI:19.837333862328904\n결과:정상체중\n\n이름: 다사\n몸무게: 78\n키: 184\nBMI:23.03875236294896\n결과:정상체중\n\n이름: 하파\n몸무게: 95\n키: 176\nBMI:30.668904958677686\n결과:과체중\n\n이름: 차마\n몸무게: 64\n키: 148\nBMI:29.218407596785976\n결과:과체중\n\n이름: 차자\n몸무게: 58\n키: 151\nBMI:25.437480812245077\n결과:과체중\n\n이름: 바마\n몸무게: 99\n키: 194\nBMI:26.304601976830696\n결과:과체중\n\n이름: 파아\n몸무게: 48\n키: 170\nBMI:16.60899653979239\n결과:저체중\n\n이름: 파자\n몸무게: 99\n키: 197\nBMI:25.509546754618775\n결과:과체중\n\n이름: 다나\n몸무게: 87\n키: 189\nBMI:24.355421180818006\n결과:정상체중\n\n이름: 마다\n몸무게: 77\n키: 148\nBMI:35.15339663988313\n결과:과체중\n\n이름: 파가\n몸무게: 45\n키: 176\nBMI:14.527376033057852\n결과:저체중\n\n이름: 카카\n몸무게: 99\n키: 151\nBMI:43.41914828297004\n결과:과체중\n\n이름: 사카\n몸무게: 92\n키: 197\nBMI:23.70584142853462\n결과:정상체중\n\n이름: 마하\n몸무게: 81\n키: 143\nBMI:39.61073891143822\n결과:과체중\n\n이름: 타파\n몸무게: 79\n키: 169\nBMI:27.660095935016283\n결과:과체중\n\n이름: 가사\n몸무게: 97\n키: 183\nBMI:28.964734689002356\n결과:과체중\n\n이름: 라라\n몸무게: 68\n키: 157\nBMI:27.587326057852245\n결과:과체중\n\n이름: 다마\n몸무게: 46\n키: 199\nBMI:11.61586828615439\n결과:저체중\n\n이름: 마가\n몸무게: 43\n키: 171\nBMI:14.705379432987929\n결과:저체중\n\n이름: 아카\n몸무게: 46\n키: 187\nBMI:13.154508278761186\n결과:저체중\n\n이름: 나자\n몸무게: 66\n키: 180\nBMI:20.37037037037037\n결과:정상체중\n\n이름: 차자\n몸무게: 69\n키: 141\nBMI:34.70650369699714\n결과:과체중\n\n이름: 타다\n몸무게: 87\n키: 176\nBMI:28.08626033057851\n결과:과체중\n\n이름: 파파\n몸무게: 58\n키: 194\nBMI:15.410776915718992\n결과:저체중\n\n이름: 다사\n몸무게: 75\n키: 178\nBMI:23.671253629592222\n결과:정상체중\n\n이름: 파아\n몸무게: 98\n키: 159\nBMI:38.764289387286894\n결과:과체중\n\n이름: 다타\n몸무게: 72\n키: 151\nBMI:31.577562387614577\n결과:과체중\n\n이름: 사카\n몸무게: 49\n키: 185\nBMI:14.317019722425126\n결과:저체중\n\n이름: 마타\n몸무게: 40\n키: 185\nBMI:11.68736303871439\n결과:저체중\n\n이름: 나가\n몸무게: 85\n키: 161\nBMI:32.79194475521777\n결과:과체중\n\n이름: 마자\n몸무게: 40\n키: 173\nBMI:13.364963747535834\n결과:저체중\n\n이름: 라바\n몸무게: 84\n키: 193\nBMI:22.550940964858118\n결과:정상체중\n\n이름: 파라\n몸무게: 60\n키: 142\nBMI:29.756000793493357\n결과:과체중\n\n이름: 자가\n몸무게: 43\n키: 174\nBMI:14.202668780552253\n결과:저체중\n\n이름: 라다\n몸무게: 73\n키: 173\nBMI:24.3910588392529\n결과:정상체중\n\n이름: 나다\n몸무게: 66\n키: 184\nBMI:19.494328922495274\n결과:정상체중\n\n이름: 파나\n몸무게: 89\n키: 163\nBMI:33.49768527230984\n결과:과체중\n\n이름: 가타\n몸무게: 51\n키: 180\nBMI:15.74074074074074\n결과:저체중\n\n이름: 타마\n몸무게: 71\n키: 196\nBMI:18.481882548937943\n결과:저체중\n\n이름: 자아\n몸무게: 96\n키: 171\nBMI:32.83061454806607\n결과:과체중\n\n이름: 바나\n몸무게: 76\n키: 163\nBMI:28.604764951635367\n결과:과체중\n\n이름: 마나\n몸무게: 99\n키: 192\nBMI:26.85546875\n결과:과체중\n\n이름: 가라\n몸무게: 46\n키: 196\nBMI:11.97417742607247\n결과:저체중\n\n이름: 파아\n몸무게: 79\n키: 183\nBMI:23.589835468362743\n결과:정상체중\n\n이름: 다나\n몸무게: 41\n키: 191\nBMI:11.23872700857981\n결과:저체중\n\n이름: 하자\n몸무게: 52\n키: 185\nBMI:15.193571950328705\n결과:저체중\n\n이름: 다바\n몸무게: 97\n키: 162\nBMI:36.96082914189909\n결과:과체중\n\n이름: 가자\n몸무게: 40\n키: 182\nBMI:12.075836251660427\n결과:저체중\n\n이름: 하자\n몸무게: 89\n키: 178\nBMI:28.089887640449437\n결과:과체중\n\n이름: 자가\n몸무게: 59\n키: 184\nBMI:17.426748582230623\n결과:저체중\n\n이름: 나라\n몸무게: 81\n키: 197\nBMI:20.87144734468809\n결과:정상체중\n\n이름: 하파\n몸무게: 54\n키: 181\nBMI:16.483013339031164\n결과:저체중\n\n이름: 라카\n몸무게: 66\n키: 183\nBMI:19.70796380901191\n결과:정상체중\n\n이름: 마사\n몸무게: 53\n키: 161\nBMI:20.446742023841672\n결과:정상체중\n\n이름: 카가\n몸무게: 54\n키: 173\nBMI:18.042701059173375\n결과:저체중\n\n이름: 파카\n몸무게: 40\n키: 164\nBMI:14.872099940511603\n결과:저체중\n\n이름: 파마\n몸무게: 96\n키: 164\nBMI:35.693039857227845\n결과:과체중\n\n이름: 바가\n몸무게: 75\n키: 156\nBMI:30.818540433925047\n결과:과체중\n\n이름: 차라\n몸무게: 53\n키: 198\nBMI:13.519028670543822\n결과:저체중\n\n이름: 사사\n몸무게: 49\n키: 183\nBMI:14.631670100630055\n결과:저체중\n\n이름: 카마\n몸무게: 46\n키: 185\nBMI:13.440467494521547\n결과:저체중\n\n이름: 가차\n몸무게: 41\n키: 195\nBMI:10.782380013149245\n결과:저체중\n\n이름: 하나\n몸무게: 99\n키: 163\nBMI:37.26147013436712\n결과:과체중\n\n이름: 자아\n몸무게: 47\n키: 149\nBMI:21.170217557767668\n결과:정상체중\n\n이름: 타파\n몸무게: 78\n키: 174\nBMI:25.762980578676178\n결과:과체중\n\n이름: 나차\n몸무게: 82\n키: 169\nBMI:28.71047932495361\n결과:과체중\n\n이름: 차마\n몸무게: 62\n키: 161\nBMI:23.9188302920412\n결과:정상체중\n\n이름: 다카\n몸무게: 67\n키: 164\nBMI:24.910767400356935\n결과:정상체중\n\n이름: 가나\n몸무게: 54\n키: 161\nBMI:20.832529609197174\n결과:정상체중\n\n이름: 다라\n몸무게: 68\n키: 199\nBMI:17.17128355344562\n결과:저체중\n\n이름: 아카\n몸무게: 86\n키: 173\nBMI:28.734672057202044\n결과:과체중\n\n이름: 자마\n몸무게: 62\n키: 145\nBMI:29.48870392390012\n결과:과체중\n\n이름: 바나\n몸무게: 57\n키: 195\nBMI:14.990138067061144\n결과:저체중\n\n이름: 차다\n몸무게: 80\n키: 153\nBMI:34.17488999957281\n결과:과체중\n\n이름: 하라\n몸무게: 99\n키: 186\nBMI:28.61602497398543\n결과:과체중\n\n이름: 라파\n몸무게: 53\n키: 185\nBMI:15.485756026296565\n결과:저체중\n\n이름: 카하\n몸무게: 47\n키: 179\nBMI:14.66870572079523\n결과:저체중\n\n이름: 타바\n몸무게: 65\n키: 165\nBMI:23.875114784205696\n결과:정상체중\n\n이름: 타다\n몸무게: 89\n키: 148\nBMI:40.6318480642805\n결과:과체중\n\n이름: 바카\n몸무게: 56\n키: 163\nBMI:21.077195227520797\n결과:정상체중\n\n이름: 카나\n몸무게: 48\n키: 140\nBMI:24.48979591836735\n결과:정상체중\n\n이름: 자바\n몸무게: 96\n키: 173\nBMI:32.075912994086\n결과:과체중\n\n이름: 하차\n몸무게: 55\n키: 167\nBMI:19.721036967980208\n결과:정상체중\n\n이름: 자마\n몸무게: 93\n키: 174\nBMI:30.71739992072929\n결과:과체중\n\n이름: 나카\n몸무게: 56\n키: 181\nBMI:17.09349531455084\n결과:저체중\n\n이름: 자타\n몸무게: 53\n키: 164\nBMI:19.705532421177875\n결과:정상체중\n\n이름: 다바\n몸무게: 99\n키: 159\nBMI:39.15984336062655\n결과:과체중\n\n이름: 가파\n몸무게: 65\n키: 154\nBMI:27.40765727778715\n결과:과체중\n\n이름: 가다\n몸무게: 95\n키: 153\nBMI:40.582681874492714\n결과:과체중\n\n이름: 바바\n몸무게: 54\n키: 186\nBMI:15.608740894901143\n결과:저체중\n\n이름: 파라\n몸무게: 94\n키: 167\nBMI:33.70504499982072\n결과:과체중\n\n이름: 마자\n몸무게: 85\n키: 160\nBMI:33.20312499999999\n결과:과체중\n\n이름: 카다\n몸무게: 52\n키: 146\nBMI:24.394820791893416\n결과:정상체중\n\n이름: 하하\n몸무게: 48\n키: 164\nBMI:17.846519928613922\n결과:저체중\n\n이름: 사다\n몸무게: 41\n키: 194\nBMI:10.893825061111702\n결과:저체중\n\n이름: 카카\n몸무게: 48\n키: 157\nBMI:19.473406629072173\n결과:정상체중\n\n이름: 파파\n몸무게: 87\n키: 194\nBMI:23.11616537357849\n결과:정상체중\n\n이름: 바나\n몸무게: 83\n키: 185\nBMI:24.251278305332356\n결과:정상체중\n\n이름: 파바\n몸무게: 51\n키: 194\nBMI:13.550855563821873\n결과:저체중\n\n이름: 나차\n몸무게: 53\n키: 148\nBMI:24.196493791088386\n결과:정상체중\n\n이름: 가아\n몸무게: 99\n키: 149\nBMI:44.59258591955317\n결과:과체중\n\n이름: 나라\n몸무게: 46\n키: 148\nBMI:21.00073046018992\n결과:정상체중\n\n이름: 나바\n몸무게: 41\n키: 196\nBMI:10.672636401499377\n결과:저체중\n\n이름: 라아\n몸무게: 99\n키: 175\nBMI:32.326530612244895\n결과:과체중\n\n이름: 마하\n몸무게: 84\n키: 149\nBMI:37.83613350749966\n결과:과체중\n\n이름: 나하\n몸무게: 50\n키: 172\nBMI:16.901027582477017\n결과:저체중\n\n이름: 나아\n몸무게: 90\n키: 182\nBMI:27.17063156623596\n결과:과체중\n\n이름: 가라\n몸무게: 58\n키: 171\nBMI:19.835162956123252\n결과:정상체중\n\n이름: 파카\n몸무게: 88\n키: 199\nBMI:22.22166106916492\n결과:정상체중\n\n이름: 차아\n몸무게: 81\n키: 177\nBMI:25.854639471416256\n결과:과체중\n\n이름: 바자\n몸무게: 76\n키: 152\nBMI:32.89473684210526\n결과:과체중\n\n이름: 나라\n몸무게: 79\n키: 172\nBMI:26.703623580313685\n결과:과체중\n\n이름: 카타\n몸무게: 64\n키: 164\nBMI:23.795359904818564\n결과:정상체중\n\n이름: 바사\n몸무게: 91\n키: 150\nBMI:40.44444444444444\n결과:과체중\n\n이름: 바나\n몸무게: 90\n키: 184\nBMI:26.583175803402646\n결과:과체중\n\n이름: 가파\n몸무게: 44\n키: 171\nBMI:15.047365001196951\n결과:저체중\n\n이름: 나자\n몸무게: 65\n키: 154\nBMI:27.40765727778715\n결과:과체중\n\n이름: 나나\n몸무게: 89\n키: 141\nBMI:44.76635984105428\n결과:과체중\n\n이름: 바아\n몸무게: 87\n키: 166\nBMI:31.572071418202935\n결과:과체중\n\n이름: 나파\n몸무게: 77\n키: 174\nBMI:25.43268595587264\n결과:과체중\n\n이름: 차나\n몸무게: 50\n키: 159\nBMI:19.77769866698311\n결과:정상체중\n\n이름: 카다\n몸무게: 42\n키: 172\nBMI:14.196863169280693\n결과:저체중\n\n이름: 나나\n몸무게: 70\n키: 157\nBMI:28.398718000730252\n결과:과체중\n\n이름: 사사\n몸무게: 65\n키: 165\nBMI:23.875114784205696\n결과:정상체중\n\n이름: 사파\n몸무게: 56\n키: 181\nBMI:17.09349531455084\n결과:저체중\n\n이름: 라라\n몸무게: 58\n키: 192\nBMI:15.733506944444445\n결과:저체중\n\n이름: 바하\n몸무게: 59\n키: 195\nBMI:15.516107823800132\n결과:저체중\n\n이름: 자라\n몸무게: 42\n키: 151\nBMI:18.420244726108503\n결과:저체중\n\n이름: 가가\n몸무게: 77\n키: 187\nBMI:22.019502988361115\n결과:정상체중\n\n이름: 바마\n몸무게: 80\n키: 177\nBMI:25.53544639152223\n결과:과체중\n\n이름: 자아\n몸무게: 41\n키: 176\nBMI:13.236053719008265\n결과:저체중\n\n이름: 카바\n몸무게: 41\n키: 174\nBMI:13.54207953494517\n결과:저체중\n\n이름: 라타\n몸무게: 89\n키: 197\nBMI:22.932824860212836\n결과:정상체중\n\n이름: 아마\n몸무게: 95\n키: 190\nBMI:26.315789473684212\n결과:과체중\n\n이름: 차타\n몸무게: 90\n키: 150\nBMI:40.0\n결과:과체중\n\n이름: 자다\n몸무게: 88\n키: 145\nBMI:41.85493460166468\n결과:과체중\n\n이름: 차자\n몸무게: 42\n키: 176\nBMI:13.558884297520661\n결과:저체중\n\n이름: 라가\n몸무게: 43\n키: 142\nBMI:21.32513390200357\n결과:정상체중\n\n이름: 카사\n몸무게: 70\n키: 174\nBMI:23.120623596247853\n결과:정상체중\n\n이름: 파가\n몸무게: 66\n키: 185\nBMI:19.284149013878743\n결과:정상체중\n\n이름: 마다\n몸무게: 44\n키: 173\nBMI:14.701460122289419\n결과:저체중\n\n이름: 카하\n몸무게: 46\n키: 165\nBMI:16.896235078053262\n결과:저체중\n\n이름: 사타\n몸무게: 66\n키: 187\nBMI:18.873859704309528\n결과:정상체중\n\n이름: 하라\n몸무게: 48\n키: 181\nBMI:14.651567412472147\n결과:저체중\n\n이름: 가가\n몸무게: 48\n키: 179\nBMI:14.980805842514279\n결과:저체중\n\n이름: 카다\n몸무게: 55\n키: 140\nBMI:28.061224489795922\n결과:과체중\n\n이름: 차사\n몸무게: 72\n키: 149\nBMI:32.43097157785685\n결과:과체중\n\n이름: 타타\n몸무게: 51\n키: 157\nBMI:20.690494543389182\n결과:정상체중\n\n이름: 카차\n몸무게: 91\n키: 166\nBMI:33.02366090869502\n결과:과체중\n\n이름: 사차\n몸무게: 45\n키: 199\nBMI:11.363349410368425\n결과:저체중\n\n이름: 차가\n몸무게: 91\n키: 151\nBMI:39.91053023990176\n결과:과체중\n\n이름: 차차\n몸무게: 89\n키: 166\nBMI:32.29786616344898\n결과:과체중\n\n이름: 가파\n몸무게: 78\n키: 172\nBMI:26.365603028664147\n결과:과체중\n\n이름: 마가\n몸무게: 94\n키: 182\nBMI:28.378215191402003\n결과:과체중\n\n이름: 가아\n몸무게: 96\n키: 173\nBMI:32.075912994086\n결과:과체중\n\n이름: 라자\n몸무게: 60\n키: 180\nBMI:18.51851851851852\n결과:정상체중\n\n이름: 다아\n몸무게: 99\n키: 177\nBMI:31.600114909508758\n결과:과체중\n\n이름: 나아\n몸무게: 65\n키: 186\nBMI:18.78829922534397\n결과:정상체중\n\n이름: 파하\n몸무게: 69\n키: 155\nBMI:28.720083246618103\n결과:과체중\n\n이름: 가바\n몸무게: 47\n키: 143\nBMI:22.984008997995016\n결과:정상체중\n\n이름: 나가\n몸무게: 97\n키: 194\nBMI:25.77319587628866\n결과:과체중\n\n이름: 바라\n몸무게: 43\n키: 152\nBMI:18.611495844875346\n결과:정상체중\n\n이름: 카나\n몸무게: 71\n키: 190\nBMI:19.667590027700832\n결과:정상체중\n\n이름: 라다\n몸무게: 79\n키: 175\nBMI:25.79591836734694\n결과:과체중\n\n이름: 아하\n몸무게: 74\n키: 154\nBMI:31.20256367009614\n결과:과체중\n\n이름: 다아\n몸무게: 66\n키: 175\nBMI:21.551020408163264\n결과:정상체중\n\n이름: 카마\n몸무게: 99\n키: 145\nBMI:47.08680142687277\n결과:과체중\n\n이름: 타카\n몸무게: 93\n키: 144\nBMI:44.84953703703704\n결과:과체중\n\n이름: 나차\n몸무게: 48\n키: 171\nBMI:16.415307274033037\n결과:저체중\n\n이름: 바하\n몸무게: 54\n키: 176\nBMI:17.43285123966942\n결과:저체중\n\n이름: 마나\n몸무게: 50\n키: 149\nBMI:22.52150804017837\n결과:정상체중\n\n이름: 차차\n몸무게: 61\n키: 150\nBMI:27.11111111111111\n결과:과체중\n\n이름: 마아\n몸무게: 40\n키: 159\nBMI:15.822158933586486\n결과:저체중\n\n이름: 라마\n몸무게: 90\n키: 158\nBMI:36.05191475725044\n결과:과체중\n\n이름: 차사\n몸무게: 43\n키: 143\nBMI:21.027923125825225\n결과:정상체중\n\n이름: 자다\n몸무게: 49\n키: 155\nBMI:20.39542143600416\n결과:정상체중\n\n이름: 타마\n몸무게: 69\n키: 187\nBMI:19.73176241814178\n결과:정상체중\n\n이름: 자아\n몸무게: 62\n키: 162\nBMI:23.624447492760247\n결과:정상체중\n\n이름: 차바\n몸무게: 61\n키: 178\nBMI:19.252619618735007\n결과:정상체중\n\n이름: 가바\n몸무게: 69\n키: 161\nBMI:26.61934338952972\n결과:과체중\n\n이름: 파라\n몸무게: 64\n키: 187\nBMI:18.301924561754696\n결과:저체중\n\n이름: 가나\n몸무게: 57\n키: 195\nBMI:14.990138067061144\n결과:저체중\n\n이름: 라하\n몸무게: 45\n키: 198\nBMI:11.47842056932966\n결과:저체중\n\n이름: 차마\n몸무게: 62\n키: 177\nBMI:19.789970953429727\n결과:정상체중\n\n이름: 하아\n몸무게: 41\n키: 141\nBMI:20.62270509531714\n결과:정상체중\n\n이름: 가하\n몸무게: 45\n키: 149\nBMI:20.269357236160534\n결과:정상체중\n\n이름: 마나\n몸무게: 90\n키: 173\nBMI:30.071168431955627\n결과:과체중\n\n이름: 나라\n몸무게: 56\n키: 146\nBMI:26.27134546819291\n결과:과체중\n\n이름: 하나\n몸무게: 40\n키: 181\nBMI:12.209639510393455\n결과:저체중\n\n이름: 카파\n몸무게: 41\n키: 151\nBMI:17.981667470724968\n결과:저체중\n\n이름: 하하\n몸무게: 63\n키: 150\nBMI:28.0\n결과:과체중\n\n이름: 라다\n몸무게: 70\n키: 161\nBMI:27.005130974885226\n결과:과체중\n\n이름: 타가\n몸무게: 77\n키: 168\nBMI:27.281746031746035\n결과:과체중\n\n이름: 다자\n몸무게: 78\n키: 150\nBMI:34.666666666666664\n결과:과체중\n\n이름: 바바\n몸무게: 78\n키: 162\nBMI:29.721079103795148\n결과:과체중\n\n이름: 사다\n몸무게: 45\n키: 160\nBMI:17.578124999999996\n결과:저체중\n\n이름: 가가\n몸무게: 63\n키: 175\nBMI:20.571428571428573\n결과:정상체중\n\n이름: 라아\n몸무게: 96\n키: 176\nBMI:30.991735537190085\n결과:과체중\n\n이름: 다타\n몸무게: 85\n키: 179\nBMI:26.528510346119035\n결과:과체중\n\n이름: 카카\n몸무게: 68\n키: 163\nBMI:25.59373706198954\n결과:과체중\n\n이름: 마차\n몸무게: 94\n키: 161\nBMI:36.264033023417305\n결과:과체중\n\n이름: 파마\n몸무게: 43\n키: 185\nBMI:12.563915266617968\n결과:저체중\n\n이름: 하카\n몸무게: 75\n키: 184\nBMI:22.152646502835537\n결과:정상체중\n\n이름: 타마\n몸무게: 89\n키: 175\nBMI:29.06122448979592\n결과:과체중\n\n이름: 다카\n몸무게: 77\n키: 184\nBMI:22.743383742911153\n결과:정상체중\n\n이름: 사가\n몸무게: 93\n키: 153\nBMI:39.728309624503396\n결과:과체중\n\n이름: 하아\n몸무게: 61\n키: 198\nBMI:15.559636771757985\n결과:저체중\n\n이름: 카라\n몸무게: 64\n키: 171\nBMI:21.887076365377382\n결과:정상체중\n\n이름: 아사\n몸무게: 67\n키: 177\nBMI:21.38593635289987\n결과:정상체중\n\n이름: 자나\n몸무게: 55\n키: 141\nBMI:27.664604396157138\n결과:과체중\n\n이름: 마사\n몸무게: 48\n키: 192\nBMI:13.020833333333334\n결과:저체중\n\n이름: 나차\n몸무게: 48\n키: 186\nBMI:13.874436351023238\n결과:저체중\n\n이름: 차가\n몸무게: 85\n키: 178\nBMI:26.82742078020452\n결과:과체중\n\n이름: 나라\n몸무게: 76\n키: 161\nBMI:29.319856487018246\n결과:과체중\n\n이름: 마가\n몸무게: 64\n키: 150\nBMI:28.444444444444443\n결과:과체중\n\n이름: 다차\n몸무게: 78\n키: 176\nBMI:25.180785123966942\n결과:과체중\n\n이름: 하아\n몸무게: 99\n키: 167\nBMI:35.49786654236437\n결과:과체중\n\n이름: 사라\n몸무게: 52\n키: 160\nBMI:20.312499999999996\n결과:정상체중\n\n이름: 라하\n몸무게: 61\n키: 175\nBMI:19.918367346938776\n결과:정상체중\n\n이름: 카마\n몸무게: 89\n키: 192\nBMI:24.14279513888889\n결과:정상체중\n\n이름: 나라\n몸무게: 60\n키: 181\nBMI:18.314459265590184\n결과:저체중\n\n이름: 다파\n몸무게: 99\n키: 176\nBMI:31.960227272727273\n결과:과체중\n\n이름: 나마\n몸무게: 62\n키: 161\nBMI:23.9188302920412\n결과:정상체중\n\n이름: 가아\n몸무게: 83\n키: 161\nBMI:32.02036958450677\n결과:과체중\n\n이름: 라하\n몸무게: 69\n키: 174\nBMI:22.79032897344431\n결과:정상체중\n\n이름: 파아\n몸무게: 48\n키: 141\nBMI:24.143654745737138\n결과:정상체중\n\n이름: 아마\n몸무게: 63\n키: 176\nBMI:20.33832644628099\n결과:정상체중\n\n이름: 아하\n몸무게: 53\n키: 148\nBMI:24.196493791088386\n결과:정상체중\n\n이름: 다아\n몸무게: 78\n키: 174\nBMI:25.762980578676178\n결과:과체중\n\n이름: 바사\n몸무게: 89\n키: 141\nBMI:44.76635984105428\n결과:과체중\n\n이름: 라자\n몸무게: 88\n키: 168\nBMI:31.17913832199547\n결과:과체중\n\n이름: 자가\n몸무게: 73\n키: 154\nBMI:30.78090740428403\n결과:과체중\n\n이름: 다타\n몸무게: 44\n키: 168\nBMI:15.589569160997735\n결과:저체중\n\n이름: 카가\n몸무게: 64\n키: 140\nBMI:32.653061224489804\n결과:과체중\n\n이름: 하다\n몸무게: 48\n키: 176\nBMI:15.495867768595042\n결과:저체중\n\n이름: 나자\n몸무게: 73\n키: 181\nBMI:22.282592106468055\n결과:정상체중\n\n이름: 마다\n몸무게: 96\n키: 163\nBMI:36.13233467574994\n결과:과체중\n\n이름: 사자\n몸무게: 88\n키: 195\nBMI:23.14266929651545\n결과:정상체중\n\n이름: 가파\n몸무게: 78\n키: 159\nBMI:30.853209920493647\n결과:과체중\n\n이름: 다카\n몸무게: 71\n키: 169\nBMI:24.859073561850078\n결과:정상체중\n\n이름: 파파\n몸무게: 64\n키: 140\nBMI:32.653061224489804\n결과:과체중\n\n이름: 타라\n몸무게: 64\n키: 159\nBMI:25.315454293738377\n결과:과체중\n\n이름: 타하\n몸무게: 61\n키: 144\nBMI:29.41743827160494\n결과:과체중\n\n이름: 가마\n몸무게: 75\n키: 177\nBMI:23.93948099205209\n결과:정상체중\n\n이름: 가자\n몸무게: 41\n키: 169\nBMI:14.355239662476805\n결과:저체중\n\n이름: 다하\n몸무게: 57\n키: 157\nBMI:23.124670372023203\n결과:정상체중\n\n이름: 카마\n몸무게: 79\n키: 176\nBMI:25.50361570247934\n결과:과체중\n\n이름: 바나\n몸무게: 55\n키: 161\nBMI:21.218317194552675\n결과:정상체중\n\n이름: 바바\n몸무게: 51\n키: 158\nBMI:20.429418362441915\n결과:정상체중\n\n이름: 나차\n몸무게: 89\n키: 161\nBMI:34.33509509663978\n결과:과체중\n\n이름: 가타\n몸무게: 62\n키: 197\nBMI:15.975675745316808\n결과:저체중\n\n이름: 사라\n몸무게: 65\n키: 197\nBMI:16.74869231363859\n결과:저체중\n\n이름: 라타\n몸무게: 99\n키: 168\nBMI:35.0765306122449\n결과:과체중\n\n이름: 바아\n몸무게: 75\n키: 157\nBMI:30.427197857925268\n결과:과체중\n\n이름: 카자\n몸무게: 50\n키: 152\nBMI:21.641274238227147\n결과:정상체중\n\n이름: 사파\n몸무게: 94\n키: 164\nBMI:34.94943486020227\n결과:과체중\n\n이름: 카카\n몸무게: 88\n키: 185\nBMI:25.712198685171657\n결과:과체중\n\n이름: 차파\n몸무게: 99\n키: 175\nBMI:32.326530612244895\n결과:과체중\n\n이름: 가카\n몸무게: 91\n키: 159\nBMI:35.99541157390926\n결과:과체중\n\n이름: 타마\n몸무게: 71\n키: 175\nBMI:23.183673469387756\n결과:정상체중\n\n이름: 가나\n몸무게: 78\n키: 178\nBMI:24.61810377477591\n결과:정상체중\n\n이름: 다타\n몸무게: 61\n키: 198\nBMI:15.559636771757985\n결과:저체중\n\n이름: 카파\n몸무게: 98\n키: 176\nBMI:31.63739669421488\n결과:과체중\n\n이름: 자카\n몸무게: 44\n키: 177\nBMI:14.044495515337227\n결과:저체중\n\n이름: 바라\n몸무게: 78\n키: 179\nBMI:24.343809494085704\n결과:정상체중\n\n이름: 차카\n몸무게: 40\n키: 189\nBMI:11.197894795778394\n결과:저체중\n\n이름: 마아\n몸무게: 50\n키: 168\nBMI:17.71541950113379\n결과:저체중\n\n이름: 다다\n몸무게: 61\n키: 152\nBMI:26.40235457063712\n결과:과체중\n\n이름: 사가\n몸무게: 97\n키: 166\nBMI:35.20104514443316\n결과:과체중\n\n이름: 카파\n몸무게: 63\n키: 175\nBMI:20.571428571428573\n결과:정상체중\n\n이름: 마나\n몸무게: 40\n키: 158\nBMI:16.023073225444637\n결과:저체중\n\n이름: 마아\n몸무게: 62\n키: 190\nBMI:17.174515235457065\n결과:저체중\n\n이름: 사사\n몸무게: 52\n키: 194\nBMI:13.81655861409289\n결과:저체중\n\n이름: 아파\n몸무게: 70\n키: 149\nBMI:31.530111256249718\n결과:과체중\n\n이름: 파나\n몸무게: 95\n키: 182\nBMI:28.680111097693512\n결과:과체중\n\n이름: 자가\n몸무게: 95\n키: 160\nBMI:37.10937499999999\n결과:과체중\n\n이름: 차사\n몸무게: 45\n키: 188\nBMI:12.73200543232232\n결과:저체중\n\n이름: 하마\n몸무게: 54\n키: 167\nBMI:19.362472659471475\n결과:정상체중\n\n이름: 카가\n몸무게: 58\n키: 179\nBMI:18.101807059704754\n결과:저체중\n\n이름: 사사\n몸무게: 54\n키: 149\nBMI:24.32322868339264\n결과:정상체중\n\n이름: 바자\n몸무게: 93\n키: 150\nBMI:41.333333333333336\n결과:과체중\n\n이름: 마자\n몸무게: 86\n키: 178\nBMI:27.143037495265748\n결과:과체중\n\n이름: 라라\n몸무게: 70\n키: 150\nBMI:31.11111111111111\n결과:과체중\n\n이름: 다바\n몸무게: 99\n키: 197\nBMI:25.509546754618775\n결과:과체중\n\n이름: 나자\n몸무게: 72\n키: 143\nBMI:35.20954569905619\n결과:과체중\n\n이름: 라차\n몸무게: 46\n키: 193\nBMI:12.34932481408897\n결과:저체중\n\n이름: 다타\n몸무게: 89\n키: 195\nBMI:23.405654174884944\n결과:정상체중\n\n이름: 사다\n몸무게: 50\n키: 187\nBMI:14.298378563870855\n결과:저체중\n\n이름: 다자\n몸무게: 79\n키: 147\nBMI:36.55884122356426\n결과:과체중\n\n이름: 카가\n몸무게: 85\n키: 163\nBMI:31.992171327486922\n결과:과체중\n\n이름: 다다\n몸무게: 93\n키: 145\nBMI:44.23305588585018\n결과:과체중\n\n이름: 나타\n몸무게: 71\n키: 167\nBMI:25.458065904119906\n결과:과체중\n\n이름: 바다\n몸무게: 85\n키: 156\nBMI:34.927679158448385\n결과:과체중\n\n이름: 카다\n몸무게: 84\n키: 181\nBMI:25.640242971826257\n결과:과체중\n\n이름: 다차\n몸무게: 95\n키: 194\nBMI:25.241789775746625\n결과:과체중\n\n이름: 나차\n몸무게: 71\n키: 170\nBMI:24.56747404844291\n결과:정상체중\n\n이름: 하하\n몸무게: 79\n키: 152\nBMI:34.193213296398895\n결과:과체중\n\n이름: 아차\n몸무게: 46\n키: 194\nBMI:12.222340312466788\n결과:저체중\n\n이름: 가아\n몸무게: 63\n키: 143\nBMI:30.80835248667417\n결과:과체중\n\n이름: 자자\n몸무게: 61\n키: 185\nBMI:17.82322863403944\n결과:저체중\n\n이름: 라바\n몸무게: 82\n키: 193\nBMI:22.014013799028163\n결과:정상체중\n\n이름: 다나\n몸무게: 99\n키: 158\nBMI:39.65710623297548\n결과:과체중\n\n이름: 다차\n몸무게: 94\n키: 181\nBMI:28.692652849424622\n결과:과체중\n\n이름: 가마\n몸무게: 75\n키: 146\nBMI:35.1848376806155\n결과:과체중\n\n이름: 타바\n몸무게: 61\n키: 169\nBMI:21.35779559539232\n결과:정상체중\n\n이름: 자타\n몸무게: 89\n키: 170\nBMI:30.795847750865054\n결과:과체중\n\n이름: 가카\n몸무게: 64\n키: 151\nBMI:28.068944344546292\n결과:과체중\n\n이름: 마타\n몸무게: 65\n키: 140\nBMI:33.163265306122454\n결과:과체중\n\n이름: 카카\n몸무게: 40\n키: 148\nBMI:18.261504747991236\n결과:저체중\n\n이름: 타차\n몸무게: 84\n키: 147\nBMI:38.87269193391643\n결과:과체중\n\n이름: 카파\n몸무게: 47\n키: 144\nBMI:22.665895061728396\n결과:정상체중\n\n이름: 바라\n몸무게: 72\n키: 156\nBMI:29.585798816568044\n결과:과체중\n\n이름: 사마\n몸무게: 86\n키: 173\nBMI:28.734672057202044\n결과:과체중\n\n이름: 사하\n몸무게: 40\n키: 165\nBMI:14.692378328741967\n결과:저체중\n\n이름: 하라\n몸무게: 62\n키: 182\nBMI:18.717546190073662\n결과:정상체중\n\n이름: 타자\n몸무게: 52\n키: 189\nBMI:14.557263234511913\n결과:저체중\n\n이름: 가다\n몸무게: 71\n키: 186\nBMI:20.522603769221874\n결과:정상체중\n\n이름: 바다\n몸무게: 75\n키: 157\nBMI:30.427197857925268\n결과:과체중\n\n이름: 아바\n몸무게: 57\n키: 188\nBMI:16.127206880941603\n결과:저체중\n\n이름: 아자\n몸무게: 42\n키: 149\nBMI:18.91806675374983\n결과:정상체중\n\n이름: 라사\n몸무게: 67\n키: 197\nBMI:17.264036692519777\n결과:저체중\n\n이름: 하라\n몸무게: 93\n키: 152\nBMI:40.25277008310249\n결과:과체중\n\n이름: 하카\n몸무게: 52\n키: 197\nBMI:13.398953850910871\n결과:저체중\n\n이름: 사마\n몸무게: 78\n키: 154\nBMI:32.889188733344575\n결과:과체중\n\n이름: 나타\n몸무게: 43\n키: 144\nBMI:20.736882716049383\n결과:정상체중\n\n이름: 자바\n몸무게: 56\n키: 157\nBMI:22.7189744005842\n결과:정상체중\n\n이름: 가차\n몸무게: 66\n키: 141\nBMI:33.197525275388564\n결과:과체중\n\n이름: 아하\n몸무게: 71\n키: 157\nBMI:28.804413972169254\n결과:과체중\n\n이름: 바자\n몸무게: 44\n키: 172\nBMI:14.872904272579774\n결과:저체중\n\n이름: 가타\n몸무게: 41\n키: 199\nBMI:10.353273907224565\n결과:저체중\n\n이름: 가차\n몸무게: 49\n키: 165\nBMI:17.99816345270891\n결과:저체중\n\n이름: 차아\n몸무게: 66\n키: 147\nBMI:30.542829376648623\n결과:과체중\n\n이름: 마타\n몸무게: 80\n키: 171\nBMI:27.358845456721728\n결과:과체중\n\n이름: 카카\n몸무게: 98\n키: 141\nBMI:49.29329510587999\n결과:과체중\n\n이름: 자마\n몸무게: 43\n키: 193\nBMI:11.543934065344036\n결과:저체중\n\n이름: 라차\n몸무게: 68\n키: 181\nBMI:20.756387167668876\n결과:정상체중\n\n이름: 다다\n몸무게: 79\n키: 193\nBMI:21.20862305028323\n결과:정상체중\n\n이름: 바다\n몸무게: 58\n키: 155\nBMI:24.141519250780433\n결과:정상체중\n\n이름: 차자\n몸무게: 79\n키: 140\nBMI:40.3061224489796\n결과:과체중\n\n이름: 아카\n몸무게: 89\n키: 151\nBMI:39.03337572913469\n결과:과체중\n\n이름: 사라\n몸무게: 81\n키: 190\nBMI:22.437673130193907\n결과:정상체중\n\n이름: 파마\n몸무게: 55\n키: 168\nBMI:19.48696145124717\n결과:정상체중\n\n이름: 타나\n몸무게: 68\n키: 140\nBMI:34.693877551020414\n결과:과체중\n\n이름: 타하\n몸무게: 46\n키: 152\nBMI:19.909972299168974\n결과:정상체중\n\n이름: 카자\n몸무게: 87\n키: 145\nBMI:41.37931034482759\n결과:과체중\n\n이름: 가라\n몸무게: 79\n키: 192\nBMI:21.43012152777778\n결과:정상체중\n\n이름: 다아\n몸무게: 58\n키: 174\nBMI:19.157088122605362\n결과:정상체중\n\n이름: 하라\n몸무게: 97\n키: 141\nBMI:48.79030229867713\n결과:과체중\n\n이름: 라마\n몸무게: 58\n키: 185\nBMI:16.946676406135865\n결과:저체중\n\n이름: 타카\n몸무게: 89\n키: 183\nBMI:26.575890590940304\n결과:과체중\n\n이름: 다타\n몸무게: 50\n키: 180\nBMI:15.432098765432098\n결과:저체중\n\n이름: 라차\n몸무게: 81\n키: 166\nBMI:29.3946871824648\n결과:과체중\n\n이름: 가타\n몸무게: 90\n키: 152\nBMI:38.95429362880886\n결과:과체중\n\n이름: 차마\n몸무게: 74\n키: 184\nBMI:21.85727788279773\n결과:정상체중\n\n이름: 자타\n몸무게: 52\n키: 158\nBMI:20.82999519307803\n결과:정상체중\n\n이름: 하하\n몸무게: 41\n키: 160\nBMI:16.015624999999996\n결과:저체중\n\n이름: 마다\n몸무게: 58\n키: 153\nBMI:24.77679524969029\n결과:정상체중\n\n이름: 마나\n몸무게: 69\n키: 177\nBMI:22.024322512687924\n결과:정상체중\n\n이름: 나타\n몸무게: 52\n키: 143\nBMI:25.42911633820725\n결과:과체중\n\n이름: 타타\n몸무게: 54\n키: 197\nBMI:13.914298229792058\n결과:저체중\n\n이름: 파다\n몸무게: 67\n키: 198\nBMI:17.090092847668604\n결과:저체중\n\n이름: 아가\n몸무게: 41\n키: 145\nBMI:19.500594530321045\n결과:정상체중\n\n이름: 하가\n몸무게: 78\n키: 187\nBMI:22.305470559638533\n결과:정상체중\n\n이름: 다카\n몸무게: 49\n키: 196\nBMI:12.755102040816327\n결과:저체중\n\n이름: 자바\n몸무게: 74\n키: 168\nBMI:26.218820861678008\n결과:과체중\n\n이름: 사파\n몸무게: 44\n키: 161\nBMI:16.97465375564214\n결과:저체중\n\n이름: 마파\n몸무게: 47\n키: 193\nBMI:12.617788397003947\n결과:저체중\n\n이름: 카카\n몸무게: 92\n키: 171\nBMI:31.462672275229988\n결과:과체중\n\n이름: 라바\n몸무게: 88\n키: 162\nBMI:33.53147386069196\n결과:과체중\n\n이름: 타라\n몸무게: 60\n키: 188\nBMI:16.976007243096426\n결과:저체중\n\n이름: 가사\n몸무게: 68\n키: 166\nBMI:24.677021338365513\n결과:정상체중\n\n이름: 다카\n몸무게: 45\n키: 158\nBMI:18.02595737862522\n결과:저체중\n\n이름: 다하\n몸무게: 75\n키: 172\nBMI:25.351541373715524\n결과:과체중\n\n이름: 바나\n몸무게: 91\n키: 152\nBMI:39.387119113573405\n결과:과체중\n\n이름: 카파\n몸무게: 97\n키: 193\nBMI:26.040967542752828\n결과:과체중\n\n이름: 라마\n몸무게: 49\n키: 154\nBMI:20.66115702479339\n결과:정상체중\n\n이름: 바자\n몸무게: 68\n키: 172\nBMI:22.985397512168742\n결과:정상체중\n\n이름: 하파\n몸무게: 49\n키: 168\nBMI:17.361111111111114\n결과:저체중\n\n이름: 타다\n몸무게: 70\n키: 166\nBMI:25.402816083611555\n결과:과체중\n\n이름: 카나\n몸무게: 51\n키: 195\nBMI:13.412228796844182\n결과:저체중\n\n이름: 바라\n몸무게: 43\n키: 194\nBMI:11.425231161653736\n결과:저체중\n\n이름: 나사\n몸무게: 42\n키: 157\nBMI:17.03923080043815\n결과:저체중\n\n이름: 라라\n몸무게: 73\n키: 169\nBMI:25.55932915514163\n결과:과체중\n\n이름: 사나\n몸무게: 57\n키: 166\nBMI:20.685150239512268\n결과:정상체중\n\n이름: 라아\n몸무게: 51\n키: 157\nBMI:20.690494543389182\n결과:정상체중\n\n이름: 가바\n몸무게: 79\n키: 199\nBMI:19.948991187091234\n결과:정상체중\n\n이름: 카하\n몸무게: 92\n키: 174\nBMI:30.38710529792575\n결과:과체중\n\n이름: 자하\n몸무게: 95\n키: 199\nBMI:23.989293199666673\n결과:정상체중\n\n이름: 아라\n몸무게: 72\n키: 140\nBMI:36.734693877551024\n결과:과체중\n\n이름: 하바\n몸무게: 97\n키: 162\nBMI:36.96082914189909\n결과:과체중\n\n이름: 다카\n몸무게: 57\n키: 198\nBMI:14.539332721150904\n결과:저체중\n\n이름: 파라\n몸무게: 54\n키: 183\nBMI:16.124697661918837\n결과:저체중\n\n이름: 라바\n몸무게: 91\n키: 144\nBMI:43.88503086419753\n결과:과체중\n\n이름: 하아\n몸무게: 80\n키: 197\nBMI:20.613775155247495\n결과:정상체중\n\n이름: 다사\n몸무게: 81\n키: 189\nBMI:22.67573696145125\n결과:정상체중\n\n이름: 차라\n몸무게: 96\n키: 174\nBMI:31.708283789139912\n결과:과체중\n\n이름: 카파\n몸무게: 94\n키: 178\nBMI:29.667971215755585\n결과:과체중\n\n이름: 카파\n몸무게: 95\n키: 165\nBMI:34.894398530762174\n결과:과체중\n\n이름: 아사\n몸무게: 94\n키: 183\nBMI:28.068918152229088\n결과:과체중\n\n이름: 가타\n몸무게: 40\n키: 176\nBMI:12.913223140495868\n결과:저체중\n\n이름: 아라\n몸무게: 82\n키: 149\nBMI:36.93527318589253\n결과:과체중\n\n이름: 라차\n몸무게: 88\n키: 164\nBMI:32.718619869125526\n결과:과체중\n\n이름: 아자\n몸무게: 52\n키: 182\nBMI:15.698587127158554\n결과:저체중\n\n이름: 차파\n몸무게: 96\n키: 183\nBMI:28.666129176744597\n결과:과체중\n\n이름: 바다\n몸무게: 54\n키: 196\nBMI:14.056643065389423\n결과:저체중\n\n이름: 자파\n몸무게: 56\n키: 193\nBMI:15.033960643238744\n결과:저체중\n\n이름: 파자\n몸무게: 58\n키: 192\nBMI:15.733506944444445\n결과:저체중\n\n이름: 자바\n몸무게: 55\n키: 141\nBMI:27.664604396157138\n결과:과체중\n\n이름: 차라\n몸무게: 81\n키: 145\nBMI:38.525564803805\n결과:과체중\n\n이름: 타카\n몸무게: 86\n키: 147\nBMI:39.798232218057294\n결과:과체중\n\n이름: 다가\n몸무게: 92\n키: 144\nBMI:44.367283950617285\n결과:과체중\n\n이름: 하마\n몸무게: 41\n키: 185\nBMI:11.979547114682248\n결과:저체중\n\n이름: 파마\n몸무게: 69\n키: 165\nBMI:25.344352617079892\n결과:과체중\n\n이름: 하차\n몸무게: 65\n키: 165\nBMI:23.875114784205696\n결과:정상체중\n\n이름: 다마\n몸무게: 48\n키: 173\nBMI:16.037956497043\n결과:저체중\n\n이름: 자파\n몸무게: 90\n키: 154\nBMI:37.949063923089895\n결과:과체중\n\n이름: 사카\n몸무게: 80\n키: 167\nBMI:28.685144680698485\n결과:과체중\n\n이름: 아차\n몸무게: 49\n키: 192\nBMI:13.292100694444445\n결과:저체중\n\n이름: 자다\n몸무게: 66\n키: 157\nBMI:26.775934114974238\n결과:과체중\n\n이름: 마가\n몸무게: 70\n키: 161\nBMI:27.005130974885226\n결과:과체중\n\n이름: 사마\n몸무게: 86\n키: 179\nBMI:26.840610467838083\n결과:과체중\n\n이름: 자가\n몸무게: 41\n키: 191\nBMI:11.23872700857981\n결과:저체중\n\n이름: 마하\n몸무게: 89\n키: 160\nBMI:34.76562499999999\n결과:과체중\n\n이름: 가가\n몸무게: 75\n키: 143\nBMI:36.676610103183535\n결과:과체중\n\n이름: 마아\n몸무게: 72\n키: 190\nBMI:19.94459833795014\n결과:정상체중\n\n이름: 하카\n몸무게: 46\n키: 186\nBMI:13.29633483639727\n결과:저체중\n\n이름: 아마\n몸무게: 75\n키: 140\nBMI:38.26530612244898\n결과:과체중\n\n이름: 자자\n몸무게: 59\n키: 194\nBMI:15.67647996599001\n결과:저체중\n\n이름: 차마\n몸무게: 94\n키: 140\nBMI:47.9591836734694\n결과:과체중\n\n이름: 하라\n몸무게: 52\n키: 185\nBMI:15.193571950328705\n결과:저체중\n\n이름: 아차\n몸무게: 54\n키: 155\nBMI:22.476586888657646\n결과:정상체중\n\n이름: 타차\n몸무게: 45\n키: 160\nBMI:17.578124999999996\n결과:저체중\n\n이름: 차가\n몸무게: 44\n키: 176\nBMI:14.204545454545455\n결과:저체중\n\n이름: 사나\n몸무게: 40\n키: 193\nBMI:10.738543316599104\n결과:저체중\n\n이름: 나카\n몸무게: 45\n키: 176\nBMI:14.527376033057852\n결과:저체중\n\n이름: 카가\n몸무게: 52\n키: 156\nBMI:21.367521367521366\n결과:정상체중\n\n이름: 라마\n몸무게: 48\n키: 140\nBMI:24.48979591836735\n결과:정상체중\n\n이름: 파다\n몸무게: 56\n키: 151\nBMI:24.560326301478007\n결과:정상체중\n\n이름: 파다\n몸무게: 70\n키: 196\nBMI:18.221574344023324\n결과:저체중\n\n이름: 가하\n몸무게: 85\n키: 170\nBMI:29.411764705882355\n결과:과체중\n\n이름: 아카\n몸무게: 75\n키: 154\nBMI:31.624219935908247\n결과:과체중\n\n이름: 다하\n몸무게: 86\n키: 188\nBMI:24.332277048438208\n결과:정상체중\n\n이름: 카카\n몸무게: 96\n키: 161\nBMI:37.03560819412831\n결과:과체중\n\n이름: 사카\n몸무게: 41\n키: 164\nBMI:15.243902439024392\n결과:저체중\n\n이름: 아나\n몸무게: 87\n키: 161\nBMI:33.56351992592878\n결과:과체중\n\n이름: 다파\n몸무게: 83\n키: 180\nBMI:25.61728395061728\n결과:과체중\n\n이름: 마마\n몸무게: 59\n키: 195\nBMI:15.516107823800132\n결과:저체중\n\n이름: 사아\n몸무게: 80\n키: 167\nBMI:28.685144680698485\n결과:과체중\n\n이름: 자가\n몸무게: 98\n키: 173\nBMI:32.74416118146279\n결과:과체중\n\n이름: 가하\n몸무게: 68\n키: 151\nBMI:29.823253366080436\n결과:과체중\n\n이름: 타라\n몸무게: 81\n키: 164\nBMI:30.116002379535995\n결과:과체중\n\n이름: 마가\n몸무게: 97\n키: 167\nBMI:34.78073792534691\n결과:과체중\n\n이름: 아하\n몸무게: 41\n키: 150\nBMI:18.22222222222222\n결과:저체중\n\n이름: 바파\n몸무게: 78\n키: 191\nBMI:21.380992845590857\n결과:정상체중\n\n이름: 카가\n몸무게: 99\n키: 189\nBMI:27.714789619551524\n결과:과체중\n\n이름: 차차\n몸무게: 94\n키: 170\nBMI:32.52595155709343\n결과:과체중\n\n이름: 나카\n몸무게: 64\n키: 177\nBMI:20.428357113217785\n결과:정상체중\n\n이름: 타마\n몸무게: 80\n키: 156\nBMI:32.87310979618672\n결과:과체중\n\n이름: 타자\n몸무게: 53\n키: 168\nBMI:18.77834467120182\n결과:정상체중\n\n이름: 다파\n몸무게: 58\n키: 177\nBMI:18.513198633853616\n결과:정상체중\n\n이름: 하다\n몸무게: 76\n키: 197\nBMI:19.58308639748512\n결과:정상체중\n\n이름: 라파\n몸무게: 57\n키: 156\nBMI:23.422090729783037\n결과:정상체중\n\n이름: 타마\n몸무게: 46\n키: 145\nBMI:21.87871581450654\n결과:정상체중\n\n이름: 마자\n몸무게: 76\n키: 169\nBMI:26.609712545078956\n결과:과체중\n\n이름: 다다\n몸무게: 69\n키: 173\nBMI:23.054562464499313\n결과:정상체중\n\n이름: 하카\n몸무게: 67\n키: 150\nBMI:29.77777777777778\n결과:과체중\n\n이름: 바아\n몸무게: 55\n키: 196\nBMI:14.316951270304042\n결과:저체중\n\n"
],
[
"#제너레이터\n#이터레이터를 직접 만들때 사용하는 코드\n#함수 내부에 yield 키워드를 사용하면(양보하다) 해당 함수는 제너레이터함수가돔\n#일반 함수와 달리 호출해도 함수 내부 코드가 실행되지 않음\n#제너레이터 객체는 next()함수를 사용해 함수 내부의 코드를 실행\n#이때 yield 키워드 부분 까지만 실행\n#next()함수의 리텅값으로 yield 키워드 뒤에 입력한 값이 출력됨.\n",
"_____no_output_____"
],
[
"#제너레이터 객체와 next()함수\n#함수를 선언합니다\ndef test():\n print(\"A 지점 통과\")\n yield 1 \n print(\"B 지점 통과\")\n yield 2\n print(\"C 지점 통과\")\n \n#함수를 호출합니다\noutput = test()\n\n#next()함수를 호출합니다\n\nprint(\"D 지점 통과\")\na = next(output)\nprint(a)\nprint(\"E 지점 통과\")\nb = next(output)\nprint(b)\nprint(\"F 지점 통과\")\nc = next(output)\nprint(c)\n\n#한번 더 실행하기\nnext(output)\n\n#마지막 next() 함수를 호출한 이후 yield 키워드를\n#만나지 못하고 함수가 끝나서 stoplteration 예외 발생",
"D 지점 통과\nA 지점 통과\n1\nE 지점 통과\nB 지점 통과\n2\nF 지점 통과\nC 지점 통과\n"
],
[
"#조건문으로 예외 처리하기\n#숫자를 입력 받습니다\nuser_input_a = input(\"정수입력>\")\npi = 3.14\n#사용자 입력이 숫자로만 구성되어 있을때\nif user_input_a.isdigit():\n #숫자로 변환합니다\n number_input_a = int(user_input_a)\n #출력합니다\n global pi\n print(\"원의 반지름: \", number_input_a)\n print(\"원의 둘레: \", 2 * pi * number_input_a)\n print(\"원의 넓이: \" ,pi*number_input_a *number_input_a)\nelse:\n print(\"정수를 입력하지 않았습니다.\")",
"_____no_output_____"
],
[
"input_a = input(\"정수를 입력해 주세요> \")\n\n\n\n\ntry:\n num_input_a = int(input_a) # 문자를 받아서 정수로 변경 \n \n global pi\n pi =3.14\n print(\"원의 반지름:\", num_input_a)\n print(\"원의 둘레:\", 2*pi*num_input_a)\n print(\"원의 넓이:\", pi*num_input_a*num_input_a)\n\nexcept:\n print(\"정수를 입력하라고요. 제발요\")\nfinally:\n print(\"임무 완수\")",
"정수를 입력해 주세요> 2\n원의 반지름: 2\n원의 둘레: 12.56\n원의 넓이: 12.56\n임무 완수\n"
],
[
"list_b = [\"52\", \"273\", \"32\", \"산업스파이\", \"210\"]\n\nlist_num =[]\n\nfor item in list_b:\n \n try:\n float(item)\n list_num.append(item)\n except:\n pass\nprint(list_b)\nprint(list_num)",
"['52', '273', '32', '산업스파이', '210']\n['52', '273', '32', '210']\n"
],
[
"#finally 키워드 활용\n#함수를 선언합니다.\ndef write_text_file(filename, text):\n #try except 구문을 사용합니다.\n \n try:\n #파일을 엽니다.\n file = open(filename, \"w\")\n \n #파일에 텍스트를 입력합니다.\n file.write(text)\n except Exception as e:\n print(e)\n finally:\n #파일을 닫습니다.\n file.close()\n \n#함수를 호출합니다.\nwrite_text_file(\"test.txt\", \"안녕하세요\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71f2534217874ee8f6d92da8c5f6bf762258a63 | 21,998 | ipynb | Jupyter Notebook | notebook/optimization_concepts/conic_programming.ipynb | frapac/JuMPTutorials.jl | 55374d470e4e1b16a3a16f2ec7088ecf317b48b2 | [
"MIT"
] | 2 | 2021-01-07T20:43:46.000Z | 2021-02-06T17:51:12.000Z | notebook/optimization_concepts/conic_programming.ipynb | frapac/JuMPTutorials.jl | 55374d470e4e1b16a3a16f2ec7088ecf317b48b2 | [
"MIT"
] | null | null | null | notebook/optimization_concepts/conic_programming.ipynb | frapac/JuMPTutorials.jl | 55374d470e4e1b16a3a16f2ec7088ecf317b48b2 | [
"MIT"
] | null | null | null | 29.213811 | 146 | 0.489454 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e71f2d1559f0e97048666b8be08c2b4ff32c0d1a | 30,929 | ipynb | Jupyter Notebook | notebooks/01_introduction.ipynb | LewisMcL/PYNQ_RFSOC_Workshop | 4ef17f842fb6109c3aac071412a6c0d4c7686f8c | [
"BSD-3-Clause"
] | 33 | 2019-07-16T21:53:18.000Z | 2022-03-23T20:45:39.000Z | notebooks/01_introduction.ipynb | LewisMcL/PYNQ_RFSOC_Workshop | 4ef17f842fb6109c3aac071412a6c0d4c7686f8c | [
"BSD-3-Clause"
] | 1 | 2021-03-26T02:26:10.000Z | 2021-09-09T01:44:28.000Z | notebooks/01_introduction.ipynb | LewisMcL/PYNQ_RFSOC_Workshop | 4ef17f842fb6109c3aac071412a6c0d4c7686f8c | [
"BSD-3-Clause"
] | 18 | 2019-07-11T20:18:18.000Z | 2022-02-10T11:12:20.000Z | 30.805777 | 507 | 0.558343 | [
[
[
"# Introduction to PYNQ RFSoC\n\n>Welcome to the PYNQ RFSoC Workshop! This collection of notebooks act as a guide on RFSoC development within the context of PYNQ, as well as exhibiting some of the RFSoC's unique features.\n> \n>We cover a lot of material but we strive to explain everything in as much detail as possible without being too overwhelming. To start off, we introduce the hardware and software that will be used over the course of the workshop. We then jump straight into the RFSoC hardware, interacting with both the RF data converters and the SD-FECs. Finally we spend some time showing what a Python/PYNQ development story looks like by doing some basic DSP in both software and FPGA hardware.\n\n<div style=\"background-color: #d9edf7; color: #31708f; padding: 15px;\">\n <b>Tip:</b> This workshop uses the JupyterLab interface.\n Please make sure you navigate to <b>http://<IP address>/lab</b>.\n</div>\n\n## RFSoC ZCU111 Overview\n\nThe RFSoC integrates multi-gigasample RF data converters and soft-decision forward error correction (SD-FEC) with the the Zynq Ultrascale+ SoC architecture. The RFSoC brings your digital data even closer to the antenna - allowing direct RF sampling!\n\nThe ZCU111 board includes:\n\n - ARM Cortex-A53\n - 8 Digital to Analog Converters\n - 8 Analog to Digital Converters\n - 8 SD-FECs\n\n",
"_____no_output_____"
],
[
"## Introduction to PYNQ\n\n### What is PYNQ?\n\nPYNQ is an open-source project from Xilinx Research Labs that makes it easy to design embedded systems with Zynq and Zynq Ultrascale+ SoCs. It \n\nUsing the Python language and libraries, designers can exploit the benefits of the PS and PL to build more capable and exciting embedded systems.\n\nThe PYNQ Python library provides support for low level control of an overlay (bitstream) including memory-mapped IO read and write, memory allocation, and control and management of these overlays.\n\n### Who is PYNQ for?\n\nPYNQ is intended to be used by a wide range of designers and developers including:\n\n- Software developers who want to take advantage of the capabilities of Zynq and Zynq Ultrascale+, and programmable hardware without having to use ASIC-style design tools to design hardware.\n- System architects who want an easy software interface and framework for rapid prototyping and development of their Zynq or Zynq Ultrascale+ design.\n- Hardware designers who want their designs to be used by the widest possible audience.\n\nYou can learn more about PYNQ by visiting the [website](http://pynq.io)",
"_____no_output_____"
],
[
"## Introduction to JupyterLab\n\nJupyterLab is an interactive environment that allows users to create and interact with notebooks that include features such as:\n- Live code\n- Interactive widgets\n- Plots\n- Narrative text\n- Equations\n- Images\n\nAlthough we will exclusively use Python in these notebooks, JupyterLab supports a range of different programming languages such as R, Haskell, Ruby and Scala. For each notebook that you open, the web application starts a kernel that runs the code for that notebook. It's interesting to note that the notebook server is running on the ARM processor of the board, as well as all the code you run in the cells. This means your laptop browser's job is just rendering the graphics, just like a web page!\n\nThe Jupyter environment is a feature-rich one and it would be impossible (and unnecessary) to learn everything about it in this workshop. This introduction aims to get you up to speed with enough knowledge to help you through the rest of the notebooks but, if you would like to learn more about Jupyter, then feel free to visit their [website](https://jupyter.org/) and read through the [documentation](https://jupyterlab.readthedocs.io/en/stable/).\n\n### Navigation\n\nThere are two different sets of keyboard shortcuts: one set that is active in edit mode and another in command mode.\n\nThe most important keyboard shortcuts are Enter, which enters edit mode, and Esc, which enters command mode.\n\nIn edit mode, most of the keyboard is dedicated to typing into the cell's editor. Thus, in edit mode there are relatively few shortcuts. In command mode, the entire keyboard is available for shortcuts, so there are many more:\n\n - Basic navigation: enter, shift-enter, up/k, down/j\n - Saving the notebook: s\n - Change Cell types: y, m, 1-6, t\n - Cell creation: a, b\n - Cell editing: x, c, v, d, z\n \nJupyterLab also has a toolbar on the left hand side of the screen which includes things like a file broswer and a list of running kernels. You can open this by clicking on one of the icons on the side, and close it by clicking again on the icons. It is good practice to shutdown kernels when you have finished with them, so keep an eye on what you have open from time to time.\n\nIf you need to stop or restart the kernel at any point; cut, copy or paste information to/from cells; or execute the current cell, these functions are available on the toolbar at the top of this notebook.\n\n### Running code\n\nFirst and foremost, the notebook is an interactive environment for writing and running code. Let's start off by running the code in the cells below. Navigate to the cell below and press Shift-Enter (or press the play button in the toolbar above) to run the cell and move on to the next one.",
"_____no_output_____"
]
],
[
[
"a = 10",
"_____no_output_____"
],
[
"print(a)",
"_____no_output_____"
]
],
[
[
"There are two other keyboard shortcuts for running code:\n\n- Alt-Enter runs the current cell and inserts a new one below.\n- Ctrl-Enter run the current cell and enters command mode.\n\n### Large outputs\n\nTo better handle large outputs, the output area can be collapsed. Run the following cell and then single or double click on the active area to the left of the output.",
"_____no_output_____"
]
],
[
[
"for i in range(50):\n print(i)",
"_____no_output_____"
]
],
[
[
"You can also clear the output of a cell by right clicking on it and selecting \"Clear Outputs\" from the drop down menu. You can also choose to clear the outputs of all cells in the notebook by selecting the \"Clear All Outputs\" from the same menu.",
"_____no_output_____"
],
[
"### Docstrings and **?**\n\nA docstring is a comment-like string that is the first statement in a function or class that has details about how the code operates. These comments can then be read by suffixing a `?` to the end of an object, giving the user a quick and simplistic way to read documentation without leaving the notebook.\n\nLet's look at a simple example by creating our own Hello World function and then using the `?` command to read the documentation.",
"_____no_output_____"
]
],
[
[
"def hello():\n \"\"\"Prints the phrase \"Hello, World\" to the console\"\"\" \n return \"Hello, World!\"",
"_____no_output_____"
]
],
[
[
"Before we check our docstring, it's a good time to mention the Jupyter console. If you right click anywhere in the notebook and select \"New Console for Notebook\" from the drop down menu, a new window will pop up at the bottom for you to type commands in. This is particularly useful if you want to run random snippets of code without disrupting the cells of your notebook. \n\nLet's try this out by opening a new console, entering `hello?` and running the command by pressing Shift-Enter. You can exit the console by pressing the *x* at the top of the window.\n\nTry testing this out on other Python functions and classes too.\n\n### IPython Widgets\n\nAnother great feature of JupyterLab worth mentioning are the widgets, which give users a graphical way of interacting with the notebook. A simple example of a slider can be run in the cell below.",
"_____no_output_____"
]
],
[
[
"import ipywidgets\n\[email protected]\ndef slider(val=(-10,10)):\n print(val)",
"_____no_output_____"
]
],
[
[
"### Introducing SciPy\n\nSciPy is a Python ecosystem of open-source software for mathematics, science and engineering. This includes libraries such as:\n\n- NumPy\n- SciPy library\n- Matplotlib\n- pandas\n\nWe'll be using these a lot in the following notebooks so it's best to introduce them first.\n\n#### NumPy\n\nNumPy is a fundamental package that give users, among other things, powerful N-dimensional arrays, similar in structure to a C array, allowing for really compact memory footprints as compared to Python's native lists. On top of this, NumPy has excellent functionality to manipulate these arrays in powerful ways, making it an indispensable tool when dealing with data sets. Let's look at a few quick examples...",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# create a random 4x4 matrix and print it\narr = np.random.rand(4,4)\nprint(arr)",
"_____no_output_____"
],
[
"# sum columns together\narr.sum(axis=1)",
"_____no_output_____"
],
[
"# remove even-indexed rows\narr[::2, :]",
"_____no_output_____"
]
],
[
[
"#### SciPy Library\n\nThe SciPy library contains a whole host of useful numerical functions such as optimisation, linear algebra, interpolation, decimation, and signal processing that is all built on top of NumPy. Due to the sheer amount of functionality available in SciPy it has become one of the most popular data processing libraries available, rivaling software such as R and MATLAB. \n\n#### Matplotlib\n\nMatplotlib is a plotting library which also takes a lot of inspiration from MATLAB. Although we won't be using it in the rest of the notebooks (we'll be using Plotly, but more on that later!), it is important to mention as it's the standard for visualising data in Python. It has a huge library of built-in plots such as line, histogram and scatter, to name a few, and enables users to plot data with only a few lines of code. Let's look at an example...",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# create two NumPy arrays with 10 elements each\nx = np.random.rand(10)\ny = np.arange(10)\n\n# plot a few different style\nplt.subplot(2,2,1)\nplt.plot(y,x)\nplt.subplot(2,2,2)\nplt.stem(y,x)\nplt.subplot(2,2,3)\nplt.scatter(y,x)\nplt.subplot(2,2,4)\nplt.hist(x)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Another neat feature of Jupyter is the ability to view the output of a cell in a new window. Try this out by right clicking the plot above and selecting \"Create New View for Output\". The window can then be dragged into another area of the notebook and JupyterLab will snap it into place for you. You can exit this new window (just like the console example above) by selecting the *x* at the top of it.",
"_____no_output_____"
],
[
"#### pandas\n\npandas is a data structure library written for Python used for data analysis and manipulation. Its main data structure is the \"data frame\" — a tabular, database-like collection. It offers features such as querying, reshaping, slicing, merging, indexing and a whole lot more. We can use a simple example to show off some of pandas features taken from [this website](https://pandas.pydata.org/pandas-docs/stable/getting_started/10min.html).",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n# create a date range between 1st-8th of june\nindex = pd.date_range('6/1/2019', periods=8)\n\n# create a data frame and populate it with random data over three columns\ndf = pd.DataFrame(np.random.randn(8, 3), index=index,columns=['A', 'B', 'C'])\n\n# display the data frame\ndf",
"_____no_output_____"
]
],
[
[
"You can see from the output that pandas automatically displays our data frame in a easy-to-read format. \n\nYou can also use pandas to select only a single column of data.",
"_____no_output_____"
]
],
[
[
"df.A",
"_____no_output_____"
]
],
[
[
"and even get a quick statistical summary of the data...",
"_____no_output_____"
]
],
[
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"# Summary\n\nWe've explored a lot of concepts in this first notebook so let's quickly recap what we've covered:\n\n - Introduced the workshop and what we'll be doing in the subsequent notebooks\n - Overview of RFSoC technology and the ZCU111 board\n - Introduced the concepts of PYNQ\n - Explored the features of JupyterLab and the IPython interpreter\n - Introduced the Python libraries that are featured throughout the workshop\n \nIn the next notebook we'll be diving straight into the RFSoC by sending and receiving real RF data through the data converters!\n\n⬅️ Previous 👩💻 [Next ➡️](02_pynq_and_data_converter.ipynb)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e71f6686c39fe8865b72ffcde36f04f5cc01e58a | 30,635 | ipynb | Jupyter Notebook | ANN_heart_disease_prediction.ipynb | rahul-lee/Heart-disease-prediction-ANN-artificial-neural-networks- | b57941e49955dfd8bd9628c62436c8967c3ba94d | [
"CC0-1.0"
] | null | null | null | ANN_heart_disease_prediction.ipynb | rahul-lee/Heart-disease-prediction-ANN-artificial-neural-networks- | b57941e49955dfd8bd9628c62436c8967c3ba94d | [
"CC0-1.0"
] | null | null | null | ANN_heart_disease_prediction.ipynb | rahul-lee/Heart-disease-prediction-ANN-artificial-neural-networks- | b57941e49955dfd8bd9628c62436c8967c3ba94d | [
"CC0-1.0"
] | null | null | null | 40.683931 | 6,420 | 0.707524 | [
[
[
"import numpy as np \nimport pandas as pd \nimport matplotlib.pyplot as plt\nimport seaborn as sns \nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import average_precision_score\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import auc\nfrom sklearn.svm import SVC\n%matplotlib inline",
"_____no_output_____"
],
[
"df = pd.read_csv(\"heart.csv\")",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df.info()",
"_____no_output_____"
],
[
"plt.figure(figsize=(14,10))\nsns.heatmap(df.corr(),annot=True,cmap='hsv',fmt='.3f',linewidths=2)\nplt.show()",
"_____no_output_____"
],
[
"df.groupby('cp',as_index=False)['target'].mean()",
"_____no_output_____"
],
[
"df.groupby('slope',as_index=False)['target'].mean()",
"_____no_output_____"
],
[
"df.groupby('thal',as_index=False)['target'].mean()",
"_____no_output_____"
],
[
"df.groupby('target').mean()",
"_____no_output_____"
],
[
"sns.distplot(df['target'],rug=True)\nplt.show()",
"_____no_output_____"
],
[
"pd.crosstab(df.age,df.target).plot(kind=\"bar\",figsize=(25,8),color=['gold','brown' ])\nplt.title('Heart Disease Frequency for Ages')\nplt.xlabel('Age')\nplt.ylabel('Frequency')\nplt.show()",
"_____no_output_____"
],
[
"pd.crosstab(df.sex,df.target).plot(kind=\"bar\",figsize=(10,5),color=['cyan','coral' ])\nplt.xlabel('Sex (0 = Female, 1 = Male)')\nplt.xticks(rotation=0)\nplt.legend([\"Haven't Disease\", \"Have Disease\"])\nplt.ylabel('Frequency')\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,8))\nsns.boxplot(df['target'], df['trestbps'], palette = 'rainbow')\nplt.title('Relation of tresbps with target', fontsize = 10)",
"_____no_output_____"
],
[
"# sns.pairplot(data=df)",
"_____no_output_____"
],
[
"pd.crosstab(df.cp,df.target).plot(kind=\"bar\",figsize=(10,5),color=['tomato','indigo' ])\nplt.xlabel('Chest Pain Type')\nplt.xticks(rotation = 0)\nplt.ylabel('Frequency of Disease or Not')\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(16,8))\nplt.subplot(2,2,1)\nplt.scatter(x=df.age[df.target==1],y=df.thalach[df.target==1],c='blue')\nplt.scatter(x=df.age[df.target==0],y=df.thalach[df.target==0],c='black')\nplt.xlabel('Age')\nplt.ylabel('Max Heart Rate')\nplt.legend(['Disease','No Disease'])\n\nplt.subplot(2,2,2)\nplt.scatter(x=df.age[df.target==1],y=df.chol[df.target==1],c='red')\nplt.scatter(x=df.age[df.target==0],y=df.chol[df.target==0],c='green')\nplt.xlabel('Age')\nplt.ylabel('Cholesterol')\nplt.legend(['Disease','No Disease'])\n\nplt.subplot(2,2,3)\nplt.scatter(x=df.age[df.target==1],y=df.trestbps[df.target==1],c='cyan')\nplt.scatter(x=df.age[df.target==0],y=df.trestbps[df.target==0],c='fuchsia')\nplt.xlabel('Age')\nplt.ylabel('Resting Blood Pressure')\nplt.legend(['Disease','No Disease'])\n\nplt.subplot(2,2,4)\nplt.scatter(x=df.age[df.target==1],y=df.oldpeak[df.target==1],c='grey')\nplt.scatter(x=df.age[df.target==0],y=df.oldpeak[df.target==0],c='navy')\nplt.xlabel('Age')\nplt.ylabel('ST depression')\nplt.legend(['Disease','No Disease'])\nplt.show()",
"_____no_output_____"
],
[
"chest_pain=pd.get_dummies(df['cp'],prefix='cp',drop_first=True)\ndf=pd.concat([df,chest_pain],axis=1)\ndf.drop(['cp'],axis=1,inplace=True)\nsp=pd.get_dummies(df['slope'],prefix='slope')\nth=pd.get_dummies(df['thal'],prefix='thal')\nrest_ecg=pd.get_dummies(df['restecg'],prefix='restecg')\nframes=[df,sp,th,rest_ecg]\ndf=pd.concat(frames,axis=1)\ndf.drop(['slope','thal','restecg'],axis=1,inplace=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"X = df.drop(['target'], axis = 1)\ny = df.target.values",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 0)",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\nsc = StandardScaler()\nX_train = sc.fit_transform(X_train)\nX_test = sc.transform(X_test)",
"_____no_output_____"
],
[
"from keras.models import Sequential\nfrom keras.layers import Conv2D, MaxPooling2D\nfrom keras.layers import Activation, Dropout, Flatten, Dense\nimport keras\nfrom keras.models import Sequential\nfrom keras.layers import Dense\nimport warnings",
"_____no_output_____"
],
[
"classifier = Sequential()\n\n# Adding the input layer and the first hidden layer\nclassifier.add(Dense(units = 11, kernel_initializer = 'uniform', activation = 'relu', input_dim = 22))\n\n# Adding the second hidden layer\nclassifier.add(Dense(units = 11, kernel_initializer = 'uniform', activation = 'relu'))\n\n# Adding the output layer\nclassifier.add(Dense(units = 1, kernel_initializer = 'uniform', activation = 'sigmoid'))\n\n# Compiling the ANN\nclassifier.compile(optimizer = 'adam', loss = 'binary_crossentropy', metrics = ['accuracy'])",
"_____no_output_____"
],
[
"# Fitting the model\nclassifier.fit(X_train, y_train, batch_size = 10, epochs = 100)",
"_____no_output_____"
],
[
"def get_model():\n inputs = keras.Input(shape=(22,))\n outputs = keras.layers.Dense(units = 1, kernel_initializer = 'uniform', activation = 'sigmoid')(inputs)\n model = keras.Model(inputs, outputs)\n model.compile(optimizer=\"adam\", loss=\"binary_crossentropy\", metrics = ['accuracy'])\n return model\n\n\nmodel = get_model()\n\ntest_input = X_train\ntest_target = y_train\nmodel.fit(test_input, test_target)",
"8/8 [==============================] - 1s 3ms/step - loss: 0.6881 - accuracy: 0.5408\n"
],
[
"from keras.models import model_from_json\nmodel.save(\"ANN_model.h5\")",
"_____no_output_____"
],
[
"load = keras.models.load_model(\"/content/ANN_model.h5\")\nnp.testing.assert_allclose(\n model.predict(test_input), load.predict(test_input)\n)\nload.fit(test_input, test_target)\n",
"8/8 [==============================] - 0s 2ms/step - loss: 0.6676 - accuracy: 0.6736\n<keras.callbacks.History object at 0x7f5c0dcd2450>\n"
],
[
"###########################\nfrom sklearn.preprocessing import MinMaxScaler\nimport numpy as np\nmodel = keras.models.load_model(\"/content/ANN_model.h5\")\nX = [[50.0, 90.0, 2.0, 1.02, 1.0, 1.0, 1.0, 0, 70.0,\n 107.0, 7.2, 3.7, 12100.0, 1.0, 1.0, 0.0, 0.0, 1.0,2.0,2.0,21.0, 5.0]]\nsc = MinMaxScaler(feature_range=(0, 1))\ndataset_scaled = sc.fit_transform(X)\n\nfloat_features = [float(x) for x in range(1,23)]\nfinal_features = [np.array(float_features)]\nprediction = model.predict(sc.transform(final_features))\n",
"_____no_output_____"
],
[
"# Prediction using artificial neural networks\n\nfrom sklearn.model_selection import train_test_split\nimport keras\nimport math\nfrom sklearn.preprocessing import MinMaxScaler\nimport numpy as np\nfrom flask import Flask, request, render_template\nfrom keras.models import load_model\nimport flask\nimport numpy as np\nimport tensorflow as tf\n\ndef init():\n global model\n model = load_model(\"/content/ANN_model.h5\")\n return model\n\n\nTest = [[50.0, 90.0, 2.0, 1.02, 1.0, 1.0, 1.0, 0, 70.0,\n 107.0, 7.2, 3.7, 12100.0, 1.0, 1.0, 0.0, 0.0, 1.0, 2.0, 2.0, 21.0, 5.0]]\nsc = MinMaxScaler(feature_range=(0, 1))\ndataset_scaled = sc.fit_transform(Test)\n\n\n\ndef predict():\n flask_user_inputs = 50.0, 90.0, 2.0, 1.02, 1.0, 1.0, 1.0, 0, 70.0,107.0, 7.2, 3.7, 12100.0, 1.0, 1.0, 0.0, 0.0, 1.0, 2.0, 2.0, 21.0, 5.0\n float_features = [float(x) for x in flask_user_inputs]\n final_features = [np.array(float_features)]\n prediction = init().predict(sc.transform(final_features))\n if (prediction > 0.5) or (prediction == 1.0):\n pred = \"You have a HeartDisease!\"\n elif (prediction < 0.5) or (prediction == 0):\n pred = \"You don't have a HeartDisease\"\n return pred\n \n\n\npredict()\n\n\n",
"_____no_output_____"
],
[
"# Predicting the Test set results\ny_pred = classifier.predict(X_test)\ny_pred = (y_pred > 0.5)",
"_____no_output_____"
],
[
"\nfrom sklearn.metrics import accuracy_score\nac=accuracy_score(y_test, y_pred.round())\nprint(\"ANN Accuracy: \", ac * 100)",
"ANN Accuracy: 86.88524590163934\n"
],
[
"cm = confusion_matrix(y_test, y_pred.round())\nsns.heatmap(cm,annot=True,cmap=\"Greens\",fmt=\"d\",cbar=False)\nplt.title(\"ANN Confusion Matrix\")",
"_____no_output_____"
],
[
"# Random forest model\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.ensemble import RandomForestClassifier\n\n\nrf = RandomForestClassifier(n_estimators=10,criterion='entropy',random_state=0)\nrf.fit(X_train, y_train)",
"_____no_output_____"
],
[
"rf_predict = rf.predict(X_test)",
"_____no_output_____"
],
[
"# rf accuracy\nfrom sklearn.metrics import accuracy_score\nac=accuracy_score(rf_predict, y_test)\nprint('Random Forest Accuracy: ',ac * 100)",
"Random Forest Accuracy: 80.32786885245902\n"
],
[
"# confusion matrix for random forest cancer\nfrom sklearn.metrics import confusion_matrix\ncm = confusion_matrix(y_test, rf_predict)\nsns.heatmap(cm,annot=True,cmap=\"Greens\",fmt=\"d\",cbar=False)\nplt.title(\"Random Forest Confusion Matrix\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71f71b46264dfd0ac8dff339c2ce003c0925a3e | 44,902 | ipynb | Jupyter Notebook | outputs/iris/128_sparse_pca.ipynb | cenkbircanoglu/scikit_learn_benchmarking | 09c5324148d2f52c2dc9e9dd803408bb8bd6c488 | [
"MIT"
] | null | null | null | outputs/iris/128_sparse_pca.ipynb | cenkbircanoglu/scikit_learn_benchmarking | 09c5324148d2f52c2dc9e9dd803408bb8bd6c488 | [
"MIT"
] | null | null | null | outputs/iris/128_sparse_pca.ipynb | cenkbircanoglu/scikit_learn_benchmarking | 09c5324148d2f52c2dc9e9dd803408bb8bd6c488 | [
"MIT"
] | null | null | null | 32.775182 | 125 | 0.347757 | [
[
[
"import os\nimport pandas as pd\nfrom sklearn.model_selection import StratifiedShuffleSplit\n\nfrom dimensionality_reduction import reduce_dimension\nimport load_database\nfrom algorithms import *",
"_____no_output_____"
],
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"database_name = os.environ['DATABASE']\nn_components = int(os.environ['N_COMPONENTS'])\ndimensionality_algorithm = os.environ['DIMENSIONALITY_ALGORITHM']",
"_____no_output_____"
],
[
"result_path = 'results/%s_%s_%s.csv' %(database_name, n_components, dimensionality_algorithm)",
"_____no_output_____"
],
[
"X, y = load_database.load(database_name)\nX = reduce_dimension(dimensionality_algorithm, X, n_components) if n_components else X",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"results = {}",
"_____no_output_____"
],
[
"sss = StratifiedShuffleSplit(n_splits=1, test_size=0.2)\nfor train_index, test_index in sss.split(X, y):\n X_train, X_test = X[train_index], X[test_index]\n y_train, y_test = y[train_index], y[test_index]",
"_____no_output_____"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'ada_boost')\nresults.update(result)",
"0.2503010000000001\n{'algorithm': 'SAMME', 'learning_rate': 0.3, 'n_estimators': 40}\n0.975\n0.9833333333333333 0.9333333333333333 0.9833333333333333 0.9326599326599326\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'bagging')\nresults.update(result)",
"0.03131799999999996\n{'bootstrap_features': 1, 'n_estimators': 10}\n0.9833333333333333\n1.0 0.9333333333333333 1.0 0.9333333333333333\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'extra_trees')\nresults.update(result)",
"0.05226200000000003\n{'criterion': 'gini', 'n_estimators': 20, 'warm_start': 0}\n0.9833333333333333\n1.0 0.9 1.0 0.8997493734335841\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'random_forest')\nresults.update(result)",
"0.08580200000000016\n{'criterion': 'gini', 'n_estimators': 5, 'oob_score': 0, 'warm_start': 0}\n0.975\n1.0 0.9 1.0 0.8997493734335841\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'logistic_regression')\nresults.update(result)",
"0.539447\n{'C': 1.1, 'solver': 'newton-cg', 'tol': 0.0001}\n0.9\n0.9166666666666666 0.8333333333333334 0.9158249158249159 0.8329156223893065\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'passive_aggressive')\nresults.update(result)",
"0.37297999999999965\n{'early_stopping': False, 'loss': 'hinge', 'tol': 2e-05, 'warm_start': 0}\n0.9\n0.9416666666666667 0.9 0.9418544818359575 0.8997493734335841\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'ridge')\nresults.update(result)",
"0.08810699999999994\n{'alpha': 0.8, 'tol': 0.0001}\n0.8416666666666667\n0.875 0.7666666666666667 0.874257137002065 0.7612958226768968\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'sgd')\nresults.update(result)",
"0.7585249999999997\n{'alpha': 0.0009, 'loss': 'squared_hinge', 'penalty': 'elasticnet', 'tol': 1.4285714285714285e-05}\n0.95\n0.9833333333333333 1.0 0.9833333333333333 1.0\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'bernoulli')\nresults.update(result)",
"0.01715800000000023\n{'alpha': 0.1}\n0.3333333333333333\n0.39166666666666666 0.36666666666666664 0.2775086233385778 0.23154623154623158\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'gaussian')\nresults.update(result)",
"0.01626900000000031\n{'var_smoothing': 1e-10}\n0.9583333333333334\n0.975 0.9 0.9749960931395532 0.8997493734335841\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'k_neighbors')\nresults.update(result)",
"0.0510420000000007\n{'algorithm': 'ball_tree', 'n_neighbors': 3, 'p': 2, 'weights': 'uniform'}\n0.975\n0.9666666666666667 0.9 0.9666458203043571 0.8997493734335841\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'nearest_centroid')\nresults.update(result)",
"0.007664999999999367\n{'metric': 'manhattan'}\n0.9333333333333333\n0.95 0.8666666666666667 0.9499687304565353 0.8666666666666667\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'mlp')\nresults.update(result)",
"2.816325999999999\n{'activation': 'logistic', 'alpha': 5e-06, 'early_stopping': True, 'learning_rate': 'constant', 'solver': 'lbfgs'}\n0.975\n1.0 0.9333333333333333 1.0 0.9326599326599326\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'linear_svc')\nresults.update(result)",
"0.492375\n{'C': 1.0, 'multi_class': 'crammer_singer', 'penalty': 'l2', 'tol': 0.0001}\n0.9166666666666666\n0.9583333333333334 0.9 0.9583268218992551 0.8997493734335841\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'decision_tree')\nresults.update(result)",
"0.010575999999999475\n{'criterion': 'gini', 'splitter': 'best'}\n0.975\n1.0 0.8666666666666667 1.0 0.8653198653198653\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'extra_tree')\nresults.update(result)",
"0.011077000000000226\n{'criterion': 'gini', 'splitter': 'best'}\n0.9333333333333333\n1.0 0.8666666666666667 1.0 0.8648287385129492\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'gradient_boosting')\nresults.update(result)",
"0.2568919999999997\n{'criterion': 'friedman_mse', 'learning_rate': 0.4, 'loss': 'deviance', 'tol': 3.3333333333333333e-06}\n0.975\n1.0 0.9 1.0 0.8997493734335841\n"
],
[
"result = train_test(X_train, y_train, X_test, y_test, 'hist_gradient_boosting')\nresults.update(result)",
"1.341927000000001\n{'l2_regularization': 0, 'tol': 1e-08}\n0.9666666666666667\n1.0 0.9333333333333333 1.0 0.9326599326599326\n"
],
[
"df = pd.DataFrame.from_records(results)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.to_csv(result_path)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e71f737f9a79909d91273d01464807975266b9a1 | 12,354 | ipynb | Jupyter Notebook | projeto/crimes_baltimore/testing_geopandas.ipynb | rmmariano/CAP386_intro_data_science | f7a720a22fd3df9aed30aaac44f2be79c3a3ceda | [
"MIT"
] | null | null | null | projeto/crimes_baltimore/testing_geopandas.ipynb | rmmariano/CAP386_intro_data_science | f7a720a22fd3df9aed30aaac44f2be79c3a3ceda | [
"MIT"
] | null | null | null | projeto/crimes_baltimore/testing_geopandas.ipynb | rmmariano/CAP386_intro_data_science | f7a720a22fd3df9aed30aaac44f2be79c3a3ceda | [
"MIT"
] | null | null | null | 30.655087 | 1,073 | 0.508418 | [
[
[
"# Testing GeoPandas",
"_____no_output_____"
],
[
"Crimes data: https://data.baltimorecity.gov/Public-Safety/BPD-Part-1-Victim-Based-Crime-Data/wsfq-mvij\n\nShapeFile: https://data.baltimorecity.gov/Geographic/Land-use-Shape/feax-3ycj",
"_____no_output_____"
],
[
"Trying to reproject the shapefile using geopandas:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport geopandas as gpd\nimport matplotlib.pyplot as plt\n\nshape_file_path = \"files/shp/landuse/landuse.shp\" \n\nshape_file_data = gpd.read_file(shape_file_path)\n\n# avoid missing values\nshape_file_data = shape_file_data[shape_file_data.geometry.notnull()]\n\n\n# import shapely\n# import geopandas\n# print (shapely.__version__)\n# print (geopandas.__version__)\n\n\n# print(shape_file_data.crs)\n\nprint(shape_file_data.head())\n\n# print(type(shape_file_data))\n\n\nshape_file_data.crs",
" LU_2008 Shape_Leng Shape_Area \\\n0 Low Density Residential 814.879889 39162.588802 \n1 Low Density Residential 1328.399489 108192.459482 \n2 Low Density Residential 1418.039915 75312.735463 \n3 Low Density Residential 2788.871558 455559.444337 \n4 Low Density Residential 898.177248 50353.730253 \n\n geometry \n0 POLYGON ((1416427.552987188 605118.4783347696,... \n1 POLYGON ((1425423.551399365 603273.2723795176,... \n2 POLYGON ((1427051.023538113 602637.8008814305,... \n3 POLYGON ((1410376.706821188 606914.4616194367,... \n4 POLYGON ((1443253.036081523 620145.7143563479,... \n"
],
[
"shape_file_data['geometry'] = shape_file_data['geometry'].to_crs(epsg=4326)\n\n# shape_file_data = shape_file_data.to_crs({'init': 'epsg:4326'})\n\n# shape_file_data = shape_file_data.to_crs(epsg=4326)\n\nprint(shape_file_data.head())\n\nshape_file_data.crs",
" LU_2008 Shape_Leng Shape_Area \\\n0 Low Density Residential 814.879889 39162.588802 \n1 Low Density Residential 1328.399489 108192.459482 \n2 Low Density Residential 1418.039915 75312.735463 \n3 Low Density Residential 2788.871558 455559.444337 \n4 Low Density Residential 898.177248 50353.730253 \n\n geometry \n0 POLYGON ((-76.63202678843902 39.32761753657134... \n1 POLYGON ((-76.60025487455859 39.32244777330502... \n2 POLYGON ((-76.59451222737225 39.32068341521045... \n3 POLYGON ((-76.6533922209007 39.33261332482494,... \n4 POLYGON ((-76.5369306671182 39.36853839332728,... \n"
],
[
"shape_file_data['geometry'] = shape_file_data['geometry'].to_crs({'init': 'epsg:4326'})\n\nshape_file_data.crs",
"_____no_output_____"
]
],
[
[
"## How it has not worked, I have reprojected the shapefile to EPSG:4326 using QGIS. ",
"_____no_output_____"
]
],
[
[
"import geopandas as gpd\nimport matplotlib.pyplot as plt\n\nshape_file_path = \"files/shp/landuse_4326/landuse_4326.shp\" \n\nshape_file_data = gpd.read_file(shape_file_path)\n\n# avoid missing values\n# shape_file_data = shape_file_data[shape_file_data.geometry.notnull()]\n\nshape_file_data.crs",
"_____no_output_____"
],
[
"shape_file_data.head()",
"_____no_output_____"
]
],
[
[
"Trying to plot the shapefile:",
"_____no_output_____"
]
],
[
[
"shape_file_data.plot(markersize=6, color=\"blue\", edgecolor=\"black\", figsize=(15,10));\n\nplt.title(\"WGS84 projection\");",
"_____no_output_____"
]
],
[
[
"Well, it has worked but has raised an exception...",
"_____no_output_____"
],
[
"Sources:\n\nhttp://geopandas.org/projections.html\n\nhttps://automating-gis-processes.github.io/2016/Lesson3-projections.html",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e71f755e86874a3945d52aa50acda515d96aa5d5 | 207,592 | ipynb | Jupyter Notebook | Lectures/15 - Runge-Kutta Methods.ipynb | josh-gree/NumericalMethods | 03cb91114b3f5eb1b56916920ad180d371fe5283 | [
"CC-BY-3.0"
] | 76 | 2015-02-12T19:51:52.000Z | 2022-03-26T15:34:11.000Z | Lectures/15 - Runge-Kutta Methods.ipynb | josh-gree/NumericalMethods | 03cb91114b3f5eb1b56916920ad180d371fe5283 | [
"CC-BY-3.0"
] | 2 | 2017-05-24T19:49:52.000Z | 2018-01-23T21:40:42.000Z | Lectures/15 - Runge-Kutta Methods.ipynb | josh-gree/NumericalMethods | 03cb91114b3f5eb1b56916920ad180d371fe5283 | [
"CC-BY-3.0"
] | 41 | 2015-01-05T13:30:47.000Z | 2022-02-15T09:59:39.000Z | 258.199005 | 50,961 | 0.890039 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e71f8bd89ce26803c94d19eb2d9c4df0aeb7085e | 462,851 | ipynb | Jupyter Notebook | AI_Class/023/Dnn-avocado.ipynb | easycastle/Cpprhtn-s_Deep_Learning | 98276236a83faa62533b35c7fb6a71a9fd51b3d2 | [
"MIT"
] | 12 | 2020-11-26T13:15:55.000Z | 2021-12-16T19:54:13.000Z | AI_Class/023/Dnn-avocado.ipynb | easycastle/Cpprhtn-s_Deep_Learning | 98276236a83faa62533b35c7fb6a71a9fd51b3d2 | [
"MIT"
] | 1 | 2021-01-05T16:26:23.000Z | 2021-01-15T12:44:08.000Z | AI_Class/023/Dnn-avocado.ipynb | easycastle/Cpprhtn-s_Deep_Learning | 98276236a83faa62533b35c7fb6a71a9fd51b3d2 | [
"MIT"
] | 3 | 2020-11-02T16:27:21.000Z | 2021-01-15T12:42:41.000Z | 408.518094 | 426,972 | 0.922217 | [
[
[
"import pandas as pd\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"df = pd.read_csv('avocado.csv')",
"_____no_output_____"
],
[
"df.head(100)",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 18249 entries, 0 to 18248\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 18249 non-null int64 \n 1 Date 18249 non-null object \n 2 AveragePrice 18249 non-null float64\n 3 Total Volume 18249 non-null float64\n 4 4046 18249 non-null float64\n 5 4225 18249 non-null float64\n 6 4770 18249 non-null float64\n 7 Total Bags 18249 non-null float64\n 8 Small Bags 18249 non-null float64\n 9 Large Bags 18249 non-null float64\n 10 XLarge Bags 18249 non-null float64\n 11 type 18249 non-null object \n 12 year 18249 non-null int64 \n 13 region 18249 non-null object \ndtypes: float64(9), int64(2), object(3)\nmemory usage: 1.9+ MB\n"
],
[
"df.isna().sum()",
"_____no_output_____"
],
[
"df[\"type\"].unique()",
"_____no_output_____"
],
[
"onehot = pd.get_dummies(df['type'])",
"_____no_output_____"
],
[
"df = pd.concat([df, onehot], axis=1)",
"_____no_output_____"
],
[
"df.drop([\"type\"], axis =1, inplace=True)",
"_____no_output_____"
],
[
"onehot_r = pd.get_dummies(df['region'])",
"_____no_output_____"
],
[
"df = pd.concat([df, onehot_r], axis=1)",
"_____no_output_____"
],
[
"df.drop([\"region\"], axis =1, inplace=True)",
"_____no_output_____"
],
[
"df[\"Date\"].unique()",
"_____no_output_____"
],
[
"df[\"region\"].unique()",
"_____no_output_____"
],
[
"df[\"XLarge Bags\"].unique()",
"_____no_output_____"
],
[
"df = df.drop([\"Unnamed: 0\", \"XLarge Bags\"], axis = 1)",
"_____no_output_____"
]
],
[
[
"## 날짜 전처리",
"_____no_output_____"
]
],
[
[
"date_split = df[\"Date\"].str.split(\"-\")\ndf[\"year\"] = date_split.str.get(0)\ndf[\"mon\"] = date_split.str.get(1)\ndf[\"dt\"] = date_split.str.get(2)",
"_____no_output_____"
],
[
"date_split",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df['year'] = df['year'].astype('int')",
"_____no_output_____"
],
[
"df['mon'] = df['mon'].astype('int')",
"_____no_output_____"
],
[
"df['dt'] = df['dt'].astype('int')",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 18249 entries, 0 to 18248\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Date 18249 non-null object \n 1 AveragePrice 18249 non-null float64\n 2 Total Volume 18249 non-null float64\n 3 4046 18249 non-null float64\n 4 4225 18249 non-null float64\n 5 4770 18249 non-null float64\n 6 Total Bags 18249 non-null float64\n 7 Small Bags 18249 non-null float64\n 8 Large Bags 18249 non-null float64\n 9 year 18249 non-null int64 \n 10 region 18249 non-null object \n 11 mon 18249 non-null int64 \n 12 dt 18249 non-null int64 \ndtypes: float64(8), int64(3), object(2)\nmemory usage: 1.8+ MB\n"
],
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nplt.figure(figsize=(15,15))\nsns.heatmap(data = df.corr(), annot=True, \nfmt = '.2f', linewidths=.5, cmap='Blues')",
"_____no_output_____"
],
[
"df.drop([\"Date\"], axis =1, inplace=True)",
"_____no_output_____"
],
[
"Y = df[\"AveragePrice\"]",
"_____no_output_____"
],
[
"X = df.drop([\"AveragePrice\", \"Small Bags\", \"Large Bags\", \"dt\",\"Total Volume\",\"4046\",\"4225\",\"4770\"], axis = 1)",
"_____no_output_____"
],
[
"X = df[[\"organic\"]]",
"_____no_output_____"
],
[
"from sklearn import preprocessing\nfrom sklearn.model_selection import train_test_split\n\nfrom keras.models import Sequential\nfrom keras.layers.core import Dense",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, \n Y, \n test_size=0.2, \n random_state=20)",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(Dense(256, input_shape=(1,), activation='relu'))\nmodel.add(Dense(256, activation='relu'))\nmodel.add(Dense(16, activation='relu'))\nmodel.add(Dense(16, activation='relu'))\nmodel.add(Dense((1), activation='linear'))\nmodel.compile(loss='mse', optimizer='Adam', metrics=['accuracy'])\nmodel.summary()",
"Model: \"sequential_26\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_126 (Dense) (None, 256) 512 \n_________________________________________________________________\ndense_127 (Dense) (None, 256) 65792 \n_________________________________________________________________\ndense_128 (Dense) (None, 16) 4112 \n_________________________________________________________________\ndense_129 (Dense) (None, 16) 272 \n_________________________________________________________________\ndense_130 (Dense) (None, 1) 17 \n=================================================================\nTotal params: 70,705\nTrainable params: 70,705\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"history = model.fit(X_train, y_train, validation_data=(X_test, y_test), epochs=5)",
"Train on 14599 samples, validate on 3650 samples\nEpoch 1/5\n14599/14599 [==============================] - 3s 175us/step - loss: 0.1499 - accuracy: 0.0107 - val_loss: 0.1013 - val_accuracy: 0.0112\nEpoch 2/5\n14599/14599 [==============================] - 1s 101us/step - loss: 0.1045 - accuracy: 0.0110 - val_loss: 0.1012 - val_accuracy: 0.0112\nEpoch 3/5\n14599/14599 [==============================] - 1s 91us/step - loss: 0.1034 - accuracy: 0.0110 - val_loss: 0.1003 - val_accuracy: 0.0112\nEpoch 4/5\n14599/14599 [==============================] - 1s 93us/step - loss: 0.1037 - accuracy: 0.0110 - val_loss: 0.1049 - val_accuracy: 0.0112\nEpoch 5/5\n14599/14599 [==============================] - 1s 93us/step - loss: 0.1041 - accuracy: 0.0110 - val_loss: 0.1041 - val_accuracy: 0.0112\n"
]
],
[
[
"### defining a way to find Mean Absolute Percentage Error",
"_____no_output_____"
]
],
[
[
"score = model.evaluate(X_test, y_test)\nscore[1] = 100 - 100*(score[1])\nprint(score[1])",
"3650/3650 [==============================] - 0s 32us/step\n98.8767122849822\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e71f92b3d0092dae1d7fb77690b79082b8df67ea | 70,797 | ipynb | Jupyter Notebook | 7 Factorization Machines/sdk1.7/movie_data_preparation.ipynb | jaypeeml/AWSSagemaker | 9ab931065e9f2af0b1c102476781a63c917e7c47 | [
"Apache-2.0"
] | 167 | 2019-04-07T16:33:56.000Z | 2022-03-24T12:13:13.000Z | 7 Factorization Machines/sdk1.7/movie_data_preparation.ipynb | jaypeeml/AWSSagemaker | 9ab931065e9f2af0b1c102476781a63c917e7c47 | [
"Apache-2.0"
] | 5 | 2019-04-13T06:39:43.000Z | 2019-11-09T06:09:56.000Z | 7 Factorization Machines/sdk1.7/movie_data_preparation.ipynb | jaypeeml/AWSSagemaker | 9ab931065e9f2af0b1c102476781a63c917e7c47 | [
"Apache-2.0"
] | 317 | 2019-04-07T16:34:00.000Z | 2022-03-31T11:20:32.000Z | 28.262275 | 210 | 0.347811 | [
[
[
"<h2>Movie Recommendations Dataset</h2>\n<h4>Hands-on: Classification with SageMaker</h4>\nInput Features: [userId, moveId] <br>\nTarget Feature: rating <br>\nObjective: Predict how a user would rate a particular movie<br>\n<h4>Movie Lens Overview: https://grouplens.org/datasets/movielens/</h4>\n<h4>Dataset: http://files.grouplens.org/datasets/movielens/ml-latest-small.zip</h4>\n<h4>F. Maxwell Harper and Joseph A. Konstan. 2015. The MovieLens Datasets: History and Context. ACM Transactions on Interactive Intelligent Systems (TiiS) 5, 4, Article 19 (December 2015), 19 pages. </h4>",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn import preprocessing\nfrom sklearn.datasets import dump_svmlight_file\n\n\nimport boto3\nimport sagemaker.amazon.common as smac",
"_____no_output_____"
]
],
[
[
"<h3>Load Movies and Parse Genre</h3>",
"_____no_output_____"
],
[
"#### Download Movie Dataset from grouplens",
"_____no_output_____"
]
],
[
[
"!wget http://files.grouplens.org/datasets/movielens/ml-latest-small.zip",
"--2020-11-20 12:22:05-- http://files.grouplens.org/datasets/movielens/ml-latest-small.zip\nResolving files.grouplens.org (files.grouplens.org)... 128.101.65.152\nConnecting to files.grouplens.org (files.grouplens.org)|128.101.65.152|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 978202 (955K) [application/zip]\nSaving to: ‘ml-latest-small.zip’\n\nml-latest-small.zip 100%[===================>] 955.28K 3.87MB/s in 0.2s \n\n2020-11-20 12:22:06 (3.87 MB/s) - ‘ml-latest-small.zip’ saved [978202/978202]\n\n"
],
[
"ls",
"fm_cloud_prediction_template.ipynb \u001b[0m\u001b[01;31mml-latest-small.zip\u001b[0m ReadMe.md\r\nfm_cloud_training_template.ipynb movie_data_preparation.ipynb\r\n"
],
[
"!unzip ml-latest-small.zip",
"Archive: ml-latest-small.zip\n creating: ml-latest-small/\n inflating: ml-latest-small/links.csv \n inflating: ml-latest-small/tags.csv \n inflating: ml-latest-small/ratings.csv \n inflating: ml-latest-small/README.txt \n inflating: ml-latest-small/movies.csv \n"
],
[
"df_movies = pd.read_csv(r'ml-latest-small/movies.csv')",
"_____no_output_____"
],
[
"df_movies.shape",
"_____no_output_____"
],
[
"df_movies.head()",
"_____no_output_____"
],
[
"genre_list = df_movies.genres.map(lambda value: value.split('|'))",
"_____no_output_____"
],
[
"genre_list[:10]",
"_____no_output_____"
],
[
"def get_unique_genres (genre_list):\n unique_list = set()\n \n for items in genre_list:\n for item in items:\n unique_list.add(item)\n \n return sorted(unique_list)",
"_____no_output_____"
],
[
"genre = get_unique_genres(genre_list)",
"_____no_output_____"
],
[
"genre, len(genre)",
"_____no_output_____"
],
[
"# Table of genre for each movie\ndf_genre = pd.DataFrame(index=range(df_movies.shape[0]),columns=genre)",
"_____no_output_____"
],
[
"df_genre = df_genre.fillna(0)",
"_____no_output_____"
],
[
"df_genre.shape",
"_____no_output_____"
],
[
"df_genre.head()",
"_____no_output_____"
],
[
"genre_list [:11]",
"_____no_output_____"
],
[
"# Fill genre for each movie\nfor row, movie_genre in enumerate(genre_list):\n df_genre.loc[row][movie_genre] = 1",
"_____no_output_____"
],
[
"df_genre.head()",
"_____no_output_____"
],
[
"# Some movies don't have genre listed\ndf_genre[df_genre['(no genres listed)'] > 0].head()",
"_____no_output_____"
],
[
"# Merge with movie description\ndf_movies = df_movies.join(df_genre)",
"_____no_output_____"
],
[
"df_movies.head()",
"_____no_output_____"
],
[
"df_movies.to_csv(r'ml-latest-small/movies_genre.csv', index=False)",
"_____no_output_____"
]
],
[
[
"<h3>Load Ratings given by each user for a movie</h3>",
"_____no_output_____"
]
],
[
[
"df_ratings = pd.read_csv(r'ml-latest-small/ratings.csv')",
"_____no_output_____"
],
[
"df_ratings.head()",
"_____no_output_____"
],
[
"df_ratings.userId.unique().shape",
"_____no_output_____"
],
[
"df_ratings.movieId.unique().shape",
"_____no_output_____"
],
[
"df_ratings.drop(axis=1,columns=['timestamp'],inplace=True)",
"_____no_output_____"
],
[
"# Merge rating and movie description\ndf_movie_ratings = pd.merge(df_ratings,df_movies,on='movieId')",
"_____no_output_____"
],
[
"df_movie_ratings.head(2)",
"_____no_output_____"
],
[
"df_movie_ratings.tail(2)",
"_____no_output_____"
]
],
[
[
"## Training and Validation Set\n### Target Variable as first column followed by input features:\n\n### Training, Validation files do not have a column header",
"_____no_output_____"
]
],
[
[
"# Training = 70% of the data\n# Validation = 30% of the data\n# Randomize the datset\nnp.random.seed(5)\nl = list(df_movie_ratings.index)\nnp.random.shuffle(l)\ndf = df_movie_ratings.iloc[l]",
"_____no_output_____"
],
[
"rows = df.shape[0]\ntrain = int(.7 * rows)\ntest = rows-train",
"_____no_output_____"
],
[
"rows,train,test",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.head(2)",
"_____no_output_____"
],
[
"# SageMaker Factorization Machine expects all columns to be of float32\n# Let's get the target variable as float32\ny = df['rating'].astype(np.float32).ravel()",
"_____no_output_____"
],
[
"len(y)",
"_____no_output_____"
],
[
"y.dtype",
"_____no_output_____"
],
[
"# We will create two different training datasets.\n# Training 1: rating, user id, movie id\n# Training 2: rating, user id, movie id, and movie genre attributes\ncolumns_user_movie = ['userId','movieId']\ncolumns_all = columns_user_movie + genre",
"_____no_output_____"
],
[
"columns_user_movie",
"_____no_output_____"
],
[
"columns_all",
"_____no_output_____"
],
[
"# Store a copy of user id, movie id and rating\n# Train and Test\ndf[['rating','userId','movieId']][:train].to_csv(r'ml-latest-small/user_movie_train.csv', index=False)\ndf[['rating','userId','movieId']][train:].to_csv(r'ml-latest-small/user_movie_test.csv',index=False)",
"_____no_output_____"
],
[
"# One Hot Encode\n# Training 1: user id, movie id\n# Training 2: user id, movie id, and movie genre attributes\nencoder = preprocessing.OneHotEncoder(dtype=np.float32)",
"_____no_output_____"
],
[
"X = encoder.fit_transform(df[columns_user_movie])",
"_____no_output_____"
],
[
"df.userId.unique().shape, df.movieId.unique().shape",
"_____no_output_____"
],
[
"# Write Dimensions - we need it for training and prediction\n# Number of unique users and movies\ndim_movie = df.userId.unique().shape[0] + df.movieId.unique().shape[0]\nwith open(r'ml-latest-small/movie_dimension.txt','w') as f:\n f.write(str(dim_movie))",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"X.shape[1]",
"_____no_output_____"
],
[
"# Create a spare matrix recordio file\ndef write_sparse_recordio_file (filename, x, y=None):\n with open(filename, 'wb') as f:\n smac.write_spmatrix_to_sparse_tensor (f, x, y)",
"_____no_output_____"
],
[
"# Training recordIO file\nwrite_sparse_recordio_file(r'ml-latest-small/user_movie_train.recordio',X[:train],y[:train])",
"_____no_output_____"
],
[
"# Test recordIO file\nwrite_sparse_recordio_file(r'ml-latest-small/user_movie_test.recordio',X[train:],y[train:])",
"_____no_output_____"
],
[
"# Create libSVM formatted file. Convenient text format\n# Output is stored as rating, user_index:value, movie_index:value\n# For example: 5.0 314:1 215:1 (user with index 314 and movie with index 215 in the one hot encoded table has a rating of 5 )\n\n# This file can be used for two purposes: \n# 1. directly traing with libFM binary in local mode\n# 2. It is easy to run inference with this format against sagemaker cloud as we need to\n# send only sparse input to sagemaker prediction service\n\n# \n# Store in libSVM format as well for directly testing with libFM\ndump_svmlight_file(X[:train],y[:train],r'ml-latest-small/user_movie_train.svm')\ndump_svmlight_file(X[train:],y[train:],r'ml-latest-small/user_movie_test.svm')",
"_____no_output_____"
],
[
"# Create two lookup files\n# File 1: Categorical Movie ID and corresponding Movie Index in One Hot Encoded Table\n# File 2: Categorical User ID and corresponding User Index in One Hot Encoded Table\n\n# This is useful for predicting how a particular user would rate all the movies\n# or all users rating one particular movie\n\nlist_of_movies = df.movieId.unique()\n# user 1 and all movies\ndf_user_movie = pd.DataFrame({'userId': np.full(len(list_of_movies),1), 'movieId' : list_of_movies})",
"_____no_output_____"
],
[
"df_user_movie[columns_user_movie].head()",
"_____no_output_____"
],
[
"list_of_movies",
"_____no_output_____"
],
[
"# Transform to one hot encoding (with existing encoder)\nX = encoder.transform(df_user_movie[columns_user_movie])",
"_____no_output_____"
],
[
"# Store movieId and corresponding one hot encoded entries\ndump_svmlight_file(X,list_of_movies,r'ml-latest-small/one_hot_enc_movies.svm')",
"_____no_output_____"
],
[
"# Now create \n# File 2: Categorical User ID and corresponding User Index in One Hot Encoded Table\nlist_of_users = df.userId.unique()",
"_____no_output_____"
],
[
"list_of_users.shape",
"_____no_output_____"
],
[
"list_of_users[:10]",
"_____no_output_____"
],
[
"# All users and movie 1\ndf_user_movie = pd.DataFrame({'userId': list_of_users, 'movieId' : np.full(len(list_of_users),1)})",
"_____no_output_____"
],
[
"df_user_movie.head()",
"_____no_output_____"
],
[
"# Transform to one hot encoding (with existing encoder)\nX = encoder.transform(df_user_movie[columns_user_movie])",
"_____no_output_____"
],
[
"# Store movieId and corresponding one hot encoded entries\ndump_svmlight_file(X,list_of_users,r'ml-latest-small/one_hot_enc_users.svm')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.