hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cb6d4f9f47465810569a2c3840cc605e08fce9a1 | 47,358 | ipynb | Jupyter Notebook | module3-cross-validation/LS_DS_223_assignment.ipynb | write2nk/DS-Unit-2-Kaggle-Challenge | 4b2c6d0ed8537815162047e41733dd7c627352b1 | [
"MIT"
]
| null | null | null | module3-cross-validation/LS_DS_223_assignment.ipynb | write2nk/DS-Unit-2-Kaggle-Challenge | 4b2c6d0ed8537815162047e41733dd7c627352b1 | [
"MIT"
]
| null | null | null | module3-cross-validation/LS_DS_223_assignment.ipynb | write2nk/DS-Unit-2-Kaggle-Challenge | 4b2c6d0ed8537815162047e41733dd7c627352b1 | [
"MIT"
]
| null | null | null | 36.683191 | 250 | 0.392669 | [
[
[
"Lambda School Data Science\n\n*Unit 2, Sprint 2, Module 3*\n\n---\n<p style=\"padding: 10px; border: 2px solid red;\">\n <b>Before you start:</b> Today is the day you should submit the dataset for your Unit 2 Build Week project. You can review the guidelines and make your submission in the Build Week course for your cohort on Canvas.</p>",
"_____no_output_____"
]
],
[
[
"%%capture\nimport sys\n\n# If you're on Colab:\nif 'google.colab' in sys.modules:\n DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge/main/data/'\n !pip install category_encoders==2.*\n !pip install pandas-profiling==2.*\n\n# If you're working locally:\nelse:\n DATA_PATH = '../data/'",
"_____no_output_____"
]
],
[
[
"# Module Project: Hyperparameter Tuning\n\nThis sprint, the module projects will focus on creating and improving a model for the Tanazania Water Pump dataset. Your goal is to create a model to predict whether a water pump is functional, non-functional, or needs repair.\n\nDataset source: [DrivenData.org](https://www.drivendata.org/competitions/7/pump-it-up-data-mining-the-water-table/).\n\n## Directions\n\nThe tasks for this project are as follows:\n\n- **Task 1:** Use `wrangle` function to import training and test data.\n- **Task 2:** Split training data into feature matrix `X` and target vector `y`.\n- **Task 3:** Establish the baseline accuracy score for your dataset.\n- **Task 4:** Build `clf_dt`.\n- **Task 5:** Build `clf_rf`.\n- **Task 6:** Evaluate classifiers using k-fold cross-validation.\n- **Task 7:** Tune hyperparameters for best performing classifier.\n- **Task 8:** Print out best score and params for model.\n- **Task 9:** Create `submission.csv` and upload to Kaggle.\n\nYou should limit yourself to the following libraries for this project:\n\n- `category_encoders`\n- `matplotlib`\n- `pandas`\n- `pandas-profiling`\n- `sklearn`\n\n# I. Wrangle Data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\n\n# Merge train_features.csv & train_labels.csv\ntrain = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'), \n pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv')).set_index('id')\n\n# Read test_features.csv & sample_submission.csv\ntest = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv').set_index('id')\nsample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')\n\n# Split train into train & val\n#train, val = train_test_split(train, train_size=0.80, test_size=0.20, \n #stratify=train['status_group'], random_state=42)",
"_____no_output_____"
],
[
"def wrangle(X):\n \"\"\"Wrangle train, validate, and test sets in the same way\"\"\"\n \n # Prevent SettingWithCopyWarning\n X = X.copy()\n \n # About 3% of the time, latitude has small values near zero,\n # outside Tanzania, so we'll treat these values like zero.\n X['latitude'] = X['latitude'].replace(-2e-08, 0)\n \n # When columns have zeros and shouldn't, they are like null values.\n # So we will replace the zeros with nulls, and impute missing values later.\n # Also create a \"missing indicator\" column, because the fact that\n # values are missing may be a predictive signal.\n cols_with_zeros = ['longitude', 'latitude', 'construction_year', \n 'gps_height', 'population']\n for col in cols_with_zeros:\n X[col] = X[col].replace(0, np.nan)\n X[col+'_MISSING'] = X[col].isnull()\n \n # Drop duplicate columns\n duplicates = ['quantity_group', 'payment_type']\n X = X.drop(columns=duplicates)\n \n # Drop recorded_by (never varies) and id (always varies, random)\n unusable_variance = ['recorded_by']\n X = X.drop(columns=unusable_variance)\n \n # Convert date_recorded to datetime\n X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)\n \n # Extract components from date_recorded, then drop the original column\n X['year_recorded'] = X['date_recorded'].dt.year\n X['month_recorded'] = X['date_recorded'].dt.month\n X['day_recorded'] = X['date_recorded'].dt.day\n X = X.drop(columns='date_recorded')\n \n # Engineer feature: how many years from construction_year to date_recorded\n X['years'] = X['year_recorded'] - X['construction_year']\n X['years_MISSING'] = X['years'].isnull()\n \n # return the wrangled dataframe\n return X",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Task 1:** Using the above `wrangle` function to read `train_features.csv` and `train_labels.csv` into the DataFrame `df`, and `test_features.csv` into the DataFrame `X_test`.",
"_____no_output_____"
]
],
[
[
"df = wrangle(train)\nX_test = wrangle(test)",
"_____no_output_____"
]
],
[
[
"# II. Split Data\n\n**Task 2:** Split your DataFrame `df` into a feature matrix `X` and the target vector `y`. You want to predict `'status_group'`.\n\n**Note:** You won't need to do a train-test split because you'll use cross-validation instead.",
"_____no_output_____"
]
],
[
[
"target = 'status_group'\ny = df[target]\nX = df.drop(columns=target)",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
],
[
[
"# III. Establish Baseline\n\n**Task 3:** Since this is a **classification** problem, you should establish a baseline accuracy score. Figure out what is the majority class in `y_train` and what percentage of your training observations it represents.",
"_____no_output_____"
]
],
[
[
"baseline_acc = y.value_counts(normalize = True)\nprint('Baseline Accuracy Score:', baseline_acc)",
"Baseline Accuracy Score: functional 0.543081\nnon functional 0.384242\nfunctional needs repair 0.072677\nName: status_group, dtype: float64\n"
]
],
[
[
"# IV. Build Models\n\n**Task 4:** Build a `Pipeline` named `clf_dt`. Your `Pipeline` should include:\n\n- an `OrdinalEncoder` transformer for categorical features.\n- a `SimpleImputer` transformer fot missing values.\n- a `DecisionTreeClassifier` Predictor.\n\n**Note:** Do not train `clf_dt`. You'll do that in a subsequent task. ",
"_____no_output_____"
]
],
[
[
"from sklearn.pipeline import make_pipeline\nimport category_encoders as ce\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.tree import DecisionTreeClassifier\nclf_dt = make_pipeline(\n ce.OrdinalEncoder(),\n SimpleImputer(strategy='median', verbose=0),\n DecisionTreeClassifier(random_state=42, max_depth=None,),\n)\nsorted(clf_dt.get_params().keys())",
"_____no_output_____"
]
],
[
[
"**Task 5:** Build a `Pipeline` named `clf_rf`. Your `Pipeline` should include:\n\n- an `OrdinalEncoder` transformer for categorical features.\n- a `SimpleImputer` transformer fot missing values.\n- a `RandomForestClassifier` predictor.\n\n**Note:** Do not train `clf_rf`. You'll do that in a subsequent task. ",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\nclf_rf = make_pipeline(\n ce.OrdinalEncoder(),\n SimpleImputer(),\n RandomForestClassifier()\n)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# V. Check Metrics\n\n**Task 6:** Evaluate the performance of both of your classifiers using k-fold cross-validation.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import KFold, cross_val_score\nk = 5\ncv_scores_dt = cross_val_score(clf_dt, X, y, cv = k)\ncv_scores_rf = cross_val_score(clf_rf, X, y, cv = k)",
"_____no_output_____"
],
[
"print('CV scores DecisionTreeClassifier')\nprint(cv_scores_dt)\nprint('Mean CV accuracy score:', cv_scores_dt.mean())\nprint('STD CV accuracy score:', cv_scores_dt.std())",
"CV scores DecisionTreeClassifier\n[0.69646465 0.71136364 0.71624579 0.70530303 0.71287879]\nMean CV accuracy score: 0.7084511784511784\nSTD CV accuracy score: 0.006963187643315079\n"
],
[
"print('CV score RandomForestClassifier')\nprint(cv_scores_rf)\nprint('Mean CV accuracy score:', cv_scores_rf.mean())\nprint('STD CV accuracy score:', cv_scores_rf.std())",
"CV score RandomForestClassifier\n[0.81228956 0.80698653 0.80934343 0.80833333 0.80841751]\nMean CV accuracy score: 0.8090740740740741\nSTD CV accuracy score: 0.0017747972668610615\n"
]
],
[
[
"# VI. Tune Model\n\n**Task 7:** Choose the best performing of your two models and tune its hyperparameters using a `RandomizedSearchCV` named `model`. Make sure that you include cross-validation and that `n_iter` is set to at least `25`.\n\n**Note:** If you're not sure which hyperparameters to tune, check the notes from today's guided project and the `sklearn` documentation. ",
"_____no_output_____"
]
],
[
[
"from sklearn.pipeline import Pipeline\nfrom category_encoders import OrdinalEncoder\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.tree import DecisionTreeClassifier",
"_____no_output_____"
],
[
"pipeline = Pipeline([\n ('encoder', OrdinalEncoder()),\n ('imputer', SimpleImputer(strategy='median')),\n ('classifier', DecisionTreeClassifier(random_state = 42))\n])",
"_____no_output_____"
],
[
"from scipy.stats import randint\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.model_selection import RandomizedSearchCV\n\nfeatures = df.columns.drop([target])\nX_train = df[features]\ny_train = df[target]\n\n# Setup the parameters and distributions to sample from: param_dist\nparam_dist = {\"classifier__max_depth\": [3, None],\n 'imputer__strategy': ['mean', 'median'],\n \"classifier__max_features\": randint(1, 9),\n \"classifier__min_samples_leaf\": randint(1, 9),\n \"classifier__criterion\": [\"gini\", \"entropy\"]}\n\n# Instantiate a Decision Tree classifier: tree\ntree = DecisionTreeClassifier()\n\n# Instantiate the RandomizedSearchCV object: tree_cv\n#tree_cv = RandomizedSearchCV(tree, param_dist, n_iter=25, cv=5)\ntree_cv = RandomizedSearchCV(\n pipeline, \n param_distributions=param_dist, \n n_iter=25, \n cv=3, \n)\n\ntree_cv.fit(X_train, y_train)\n #If you're on Colab, decrease n_iter & cv parameters\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Task 8:** Print out the best score and best params for `model`.",
"_____no_output_____"
]
],
[
[
"best_score = tree_cv.best_score_\nbest_params = tree_cv.best_params_\n\nprint('Best score for `model`:', best_score)\nprint('Best params for `model`:', best_params)",
"Best score for `model`: 0.7528114478114478\nBest params for `model`: {'classifier__criterion': 'entropy', 'classifier__max_depth': None, 'classifier__max_features': 7, 'classifier__min_samples_leaf': 6, 'imputer__strategy': 'mean'}\n"
]
],
[
[
"# Communicate Results",
"_____no_output_____"
],
[
"**Task 9:** Create a DataFrame `submission` whose index is the same as `X_test` and that has one column `'status_group'` with your predictions. Next, save this DataFrame as a CSV file and upload your submissions to our competition site. \n\n**Note:** Check the `sample_submission.csv` file on the competition website to make sure your submissions follows the same formatting. ",
"_____no_output_____"
]
],
[
[
"#since we have found the best parameters, we plug them in and then fit and make prediction\nbest_model = Pipeline([\n ('encoder', OrdinalEncoder()),\n ('imputer', SimpleImputer(strategy='mean')),\n ('classifier', DecisionTreeClassifier(criterion = 'gini', max_depth = None, max_features = 8, min_samples_leaf = 7,\n random_state = 42))\n])\n\nbest_model.fit(X_train, y_train)",
"_____no_output_____"
],
[
"print('X_test', X_test.shape)\nprint('X_train', X_train.shape)\nX_test.head()",
"X_test (14358, 45)\nX_train (59400, 45)\n"
],
[
"#make predictions first\npred = best_model.predict(X_test)",
"_____no_output_____"
],
[
"#Create a datafram submission\nsubmission = submission = pd.DataFrame(pred, columns = ['Status_group'], index = X_test.index)\nsubmission.head()",
"_____no_output_____"
],
[
"#Save into a csv file for upload\nsubmission.to_csv('Tanzania_water_pred_submission.csv')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6d516aab73e19bcc2fa9725153ffec30d6a00d | 12,662 | ipynb | Jupyter Notebook | Test_karl/material/session_16/exercise_16.ipynb | karlbindslev/sds_group29 | 6f5263b08b35f35374b7f01b31a0e90d1cf4d53e | [
"MIT",
"Unlicense"
]
| 24 | 2017-08-07T09:16:51.000Z | 2021-12-06T20:09:52.000Z | Test_karl/material/session_16/exercise_16.ipynb | karlbindslev/sds_group29 | 6f5263b08b35f35374b7f01b31a0e90d1cf4d53e | [
"MIT",
"Unlicense"
]
| 31 | 2018-08-11T09:34:52.000Z | 2018-09-17T09:40:02.000Z | Test_karl/material/session_16/exercise_16.ipynb | karlbindslev/sds_group29 | 6f5263b08b35f35374b7f01b31a0e90d1cf4d53e | [
"MIT",
"Unlicense"
]
| 39 | 2017-08-06T08:49:01.000Z | 2020-10-28T12:52:46.000Z | 45.221429 | 743 | 0.659296 | [
[
[
"> **Note:** In most sessions you will be solving exercises posed in a Jupyter notebook that looks like this one. Because you are cloning a Github repository that only we can push to, you should **NEVER EDIT** any of the files you pull from Github. Instead, what you should do, is either make a new notebook and write your solutions in there, or **make a copy of this notebook and save it somewhere else** on your computer, not inside the `sds` folder that you cloned, so you can write your answers in there. If you edit the notebook you pulled from Github, those edits (possible your solutions to the exercises) may be overwritten and lost the next time you pull from Github. This is important, so don't hesitate to ask if it is unclear.",
"_____no_output_____"
],
[
"# Exercise Set 16: Exploratory Data Analysis\n\n*Afternoon, August 22, 2018*\n\nIn this exercise set we will be practicing our skills wihtin Exploratory Data Analysis. Furthermore we will get a little deeper into the mechanics of the K-means clustering algorithm.",
"_____no_output_____"
],
[
"## Exercise Section 16.1: Exploratory data analysis with interactive plots\n\nIn the following exercise you will practice interactive plotting with the Plotly library.\n",
"_____no_output_____"
],
[
"** Preparation:** \nSetting up plotly:\n* Install plotly version (2.7.0) using conda: `conda install -c conda-forge plotly==2.7.0`\n* Intall cufflinks that bins the plotly to the dataframe. `conda install -c conda-forge cufflinks-py`\n* Create a user on \"https://plot.ly/\".\n* Login\n* Hover over your profile name, and click on settings.\n* Get the API key and copy it.\n* Run the following command in the notebook\n```python\n# First time you run it\nimport plotly \nusername = 'username' # your.username\napi_key = 'apikey' # find it under settings # your.apikey\nplotly.tools.set_credentials_file(username=username, api_key=api_key)\n```\n\n* Plotly is a sort of social media for Graphs, and automatically save all your figures. If you want to run it in offline mode, run the following in the notebook :\n\n```python\nimport plotly.offline as py # import plotly in offline mode\npy.init_notebook_mode(connected=True) # initialize the offline mode, with access to the internet or not.\nimport plotly.tools as tls \ntls.embed('https://plot.ly/~cufflinks/8') # embed cufflinks.\n# import cufflinks and make it offline\nimport cufflinks as cf\ncf.go_offline() # initialize cufflinks in offline mode\n```\n",
"_____no_output_____"
],
[
"> ** Ex. 16.1.1 ** Reproduce the plots made in the Lectures. This means doing a scatter plot where the colors (hue=) are the ratings, and the text comes when hovering over (text=).\n",
"_____no_output_____"
]
],
[
[
"#[Answer goes here]",
"_____no_output_____"
]
],
[
[
"## Exercise Section 16.2: Implementing the K-means Clustering algorithm\n\nIn the following exercise you will implement your own version of the K-means Clustering Algorithm. This will help you practice the basic matrix operations and syntax in python. ",
"_____no_output_____"
],
[
"> **Ex. 16.2.0:** First we need to load the dataset to practice on. For this task we will use the famous clustering dataset: of properties of 3 iris flower species. This is already build into many packages, including the plotting library seaborn, and can be loaded using the following command: ```df = sns.load_dataset('iris')```\nPlot the data as a scatter matrix to inspect that it indeed has some rather obvious clusters: search for seaborn and scatter matrix on google and figure out the command. Color the markers (the nodes in the graph) by setting the ```hue='species'```",
"_____no_output_____"
]
],
[
[
"# [Answer to Ex. 16.2.0]",
"_____no_output_____"
]
],
[
[
"If we weren't biologist and we had not already named the three flower species, we might want to find and define the natural groupings using a clustering method. Now you should implement the K-Means Clustering Algorithm.\n> **Ex. 16.2.1:** First define a matrix X, by extracting the four columns ('sepal_length','sepal_width','petal_length','petal_width') from the dataframe using the .values method.",
"_____no_output_____"
]
],
[
[
"# [Answer to Ex. 16.2.1]\n",
"_____no_output_____"
]
],
[
[
"Now we are ready to implement the algorithm. \n> **Ex. 16.2.2:** First we write the initialization, our first *Expectation*. This will initialize our first guess of the cluster centroids. This is done by picking K random points from the data. We do this by sampling a list of K numbers from the index. And then extracting datapoints using this index, same syntax as with a dataframe. \n***(hint: use the random.sample function and sample from a range(len(data)))***\n\nCheck that this works and wrap it in a function named `initialize_clusters`. The function should take the data and a value of K (number of clusters / intial samples) as input parameters. And return the initial cluster centroids.\n",
"_____no_output_____"
]
],
[
[
"#[Answer Ex. 16.2.2]",
"_____no_output_____"
]
],
[
[
"Now we will write the *Maximization* step.\n> **Ex.16.2.3.:** The maximization step is done by assigning each datapoint to the closests cluster centers/centroid. This means:\n* we need to calculate the distance from each point to each centroid (at first it is just our randomly initialized points). This can be done using the the sklearn.metrics.pairwise_distances() taking the two matrices as input. \n* Next run an argmin operation on the matrix to obtain the cluster_assignments, using the ```.argmin()``` method built into the matrix object. The argmin gives you the index of smallest value, and not the smallest value itself. Remember to choose the right axis to apply the argmin operation on - i.e. columns or rows to minimize. You do this setting the axis= argument. ```.argmin(axis=0)``` applies it on the columns and ```.argmin(axis=1)``` applies it on the rows. \n\nFinally wrap these operations into a function `maximize` that takes the cluster centers, and the data as input. And return the cluster assignments.\n",
"_____no_output_____"
]
],
[
[
"#[Answer Ex. 16.2.3]",
"_____no_output_____"
]
],
[
[
"> **Ex. 16.2.4:** Now we want to update our Expectation of the cluster centroids. \nWe calculate new cluster centroids, by applying the builtin ```.mean``` function on the subset of the data that is assigned to each cluster. \n\nFirst you define a container for the new centroids. Using the function: `np.zeros(shape)`. The `shape` parameter should be a tuple with the dimensions of matrix of cluster centroids i.e. (k, n_columns). \n>For each cluster you *(this can be done using a for loop from 0 to k number of clusters)*:\n* filter the data with a boolean vector that is True if the cluster assignment of the datapoint is equal to the cluster. The indexing is done in the same way as you would do with a dataframe. \n* calculate the mean on the subset of the data. Make sure you are doing it on the right axis. (axis=0) is on the columns, and axis=1 is on the rows.\n* store the it in a container\n\nEach cluster center should be a vector of 4 values [val,val2,val3,val4] so make sure you take the mean on the right axis. ```.mean(axis=?)```. \n\nFinally wrap these operations into a function `update_expectation` that takes the, `k`, the data `X`, and the `cluster_assignment` as input. And return the new cluster centers.\n",
"_____no_output_____"
]
],
[
[
"#[Answer 16.2.4]",
"_____no_output_____"
]
],
[
[
"Lastly we put it all together in the scikitlearn canonical \".fit()\" function. This function will use the other functions. The important new things here are setting the number of maximization steps, and checking if the solution has converged, i.e. it is stable with little to no change.\n\nPipeline is the following:\n* First we initialize our cluster centroids using the initialization function.\n* Then we run the maximazation function until convergence. Converged is checked by comparing if old_centroids from the previous step is equal to the new centroids.\n* Once convergence is reached we have our final cluster centroids, and the final cluster assignment.\n\n> **Ex. 16.2.5:** You should now implement it by doing the following:\n* Define a maximum number of iterations`max_iter` to 15.\n* Use the `initialize_clusters` function to define a variable `centroids`.\n* make a `for` loop from 0 to max_iter where you: \n * copy the current cluster centroids to a new variable: old_centroids. This will be used for checking convergence after the maximization step.\n * define the `cluster_assignment` by running the `maximize` function\n * define a new (i.e. overwrite) `centroids` variable by running the `update_expectation` function.\n * finally check if old_centroids is equal to new_centroids, using the np.array_equal() function. If they are: break.\n\nMake sure that it works and wrap it around a function `fit_transform()` that takes the data `X` as input, and the number of clusters `k` plus the maximum number of iterations `max_iter`. It should return the cluster assignments and the cluster centroids. \n",
"_____no_output_____"
]
],
[
[
"#[Answer exercise 16.2.5]",
"_____no_output_____"
]
],
[
[
"> **Ex. 16.2.6:** Run the algorithm and create a new variable `'cluster'` in your dataframe using the cluster_assignments. Count the overlap between the two variables, by using the `pd.pivot_table()` method. between the species and each cluster. Define the `aggfunc=` to the 'count' method.\n \n\nextra: To avoid a local minima (due to unlucky random initialization) you should run the algorithm more than once. Write a function that fits the algorithm N number of times, and evaluates the best solution by calculating ratio between the average distance between all points within the same cluster and the average distance to points outside ones cluster.",
"_____no_output_____"
]
],
[
[
"#[Answer to Ex. 16.2.6]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6d5ccc295d4380c7038e01e48cc8f66d1d93aa | 40,613 | ipynb | Jupyter Notebook | workflows/mashr_flashr_workflow.ipynb | stephens999/gtexresults_matrixash | f5d784e203be9cb7792e431d300896b0ac2300c7 | [
"MIT"
]
| 15 | 2018-05-08T14:07:39.000Z | 2022-01-28T11:32:15.000Z | workflows/mashr_flashr_workflow.ipynb | stephens999/gtexresults_matrixash | f5d784e203be9cb7792e431d300896b0ac2300c7 | [
"MIT"
]
| 16 | 2018-05-27T20:14:07.000Z | 2021-12-07T20:01:44.000Z | workflows/mashr_flashr_workflow.ipynb | stephens999/gtexresults_matrixash | f5d784e203be9cb7792e431d300896b0ac2300c7 | [
"MIT"
]
| 9 | 2018-07-02T16:02:49.000Z | 2022-03-03T01:29:01.000Z | 42.885956 | 454 | 0.580528 | [
[
[
"# MASH analysis pipeline with data-driven prior matrices\n\nThis notebook is a pipeline written in SoS to run `flashr + mashr` for multivariate analysis described in Urbut et al (2019). This pipeline was last applied to analyze GTEx V8 eQTL data, although it can be used as is to perform similar multivariate analysis for other association studies.",
"_____no_output_____"
],
[
"*Version: 2021.02.28 by Gao Wang and Yuxin Zou*",
"_____no_output_____"
]
],
[
[
"%revisions -s",
"_____no_output_____"
]
],
[
[
"## Data overview",
"_____no_output_____"
],
[
"`fastqtl` summary statistics data were obtained from dbGaP (data on CRI at UChicago Genetic Medicine). It has 49 tissues. [more description to come]",
"_____no_output_____"
],
[
"## Preparing MASH input",
"_____no_output_____"
],
[
"Using an established workflow (which takes 33hrs to run on a cluster system as configured by `midway2.yml`; see inside `fastqtl_to_mash.ipynb` for a note on computing environment),",
"_____no_output_____"
],
[
"```\nINPUT_DIR=/project/compbio/GTEx_dbGaP/GTEx_Analysis_2017-06-05_v8/eqtl/GTEx_Analysis_v8_eQTL_all_associations\nJOB_OPT=\"-c midway2.yml -q midway2\"\nsos run workflows/fastqtl_to_mash.ipynb --data-list $INPUT_DIR/FastQTLSumStats.list --common-suffix \".allpairs.txt\" $JOB_OPT\n```",
"_____no_output_____"
],
[
"As a result of command above I obtained the \"mashable\" data-set in the same format [as described here](https://stephenslab.github.io/gtexresults/gtexdata.html).",
"_____no_output_____"
],
[
"### Some data integrity check\n\n1. Check if I get the same number of groups (genes) at the end of HDF5 data conversion:\n\n```\n$ zcat Whole_Blood.allpairs.txt.gz | cut -f1 | sort -u | wc -l\n20316\n$ h5ls Whole_Blood.allpairs.txt.h5 | wc -l\n20315\n```\n\nThe results agreed on Whole Blood sample (the original data has a header thus one line more than the H5 version). We should be good (since the pipeline reported success for all other files).",
"_____no_output_____"
],
[
"### Data & job summary\n\nThe command above took 33 hours on UChicago RCC `midway2`. \n\n```\n[MW] cat FastQTLSumStats.log\n39832 out of 39832 groups merged!\n```\n\nSo we have a total of 39832 genes (union of 49 tissues).\n\n```\n[MW] cat FastQTLSumStats.portable.log\n15636 out of 39832 groups extracted!\n```\n\nWe have 15636 groups without missing data in any tissue. This will be used to train the MASH model.\n\nThe \"mashable\" data file is `FastQTLSumStats.mash.rds`, 124Mb serialized R file.",
"_____no_output_____"
],
[
"## Multivariate adaptive shrinkage (MASH) analysis of eQTL data\n\nBelow is a \"blackbox\" implementation of the `mashr` eQTL workflow -- blackbox in the sense that you can run this pipeline as an executable, without thinking too much about it, if you see your problem fits our GTEx analysis scheme. However when reading it as a notebook it is a good source of information to help developing your own `mashr` analysis procedures.",
"_____no_output_____"
],
[
"Since the submission to biorxiv of Urbut 2017 we have improved implementation of MASH algorithm and made a new R package, [`mashr`](https://github.com/stephenslab/mashr). Major improvements compared to Urbut 2019 are:\n\n1. Faster computation of likelihood and posterior quantities via matrix algebra tricks and a C++ implementation.\n2. Faster computation of MASH mixture via convex optimization.\n3. Replace `SFA` with `FLASH`, a new sparse factor analysis method to generate prior covariance candidates.\n4. Improve estimate of residual variance $\\hat{V}$.\n\nAt this point, the input data have already been converted from the original eQTL summary statistics to a format convenient for analysis in MASH, as a result of running the data conversion pipeline in `fastqtl_to_mash.ipynb`.",
"_____no_output_____"
],
[
"Example command:\n\n\n```bash\nJOB_OPT=\"-j 8\"\n#JOB_OPT=\"-c midway2.yml -q midway2\"\nsos run workflows/mashr_flashr_workflow.ipynb mash $JOB_OPT # --data ... --cwd ... --vhat ...\n```\n\n**FIXME: add comments on submitting jobs to HPC. Here we use the UChicago RCC cluster but other users can similarly configure their computating system to run the pipeline on HPC.**",
"_____no_output_____"
],
[
"### Global parameter settings",
"_____no_output_____"
]
],
[
[
"[global]\nparameter: cwd = path('./mashr_flashr_workflow_output')\n# Input summary statistics data\nparameter: data = path(\"fastqtl_to_mash_output/FastQTLSumStats.mash.rds\")\n# Prefix of output files. If not specified, it will derive it from data.\n# If it is specified, for example, `--output-prefix AnalysisResults`\n# It will save output files as `{cwd}/AnalysisResults*`.\nparameter: output_prefix = ''\n# Exchangable effect (EE) or exchangable z-scores (EZ)\nparameter: effect_model = 'EZ'\n# Identifier of $\\hat{V}$ estimate file\n# Options are \"identity\", \"simple\", \"mle\", \"vhat_corshrink_xcondition\", \"vhat_simple_specific\"\nparameter: vhat = 'mle'\nparameter: mixture_components = ['flash', 'flash_nonneg', 'pca']\n\ndata = data.absolute()\ncwd = cwd.absolute()\nif len(output_prefix) == 0:\n output_prefix = f\"{data:bn}\"\nprior_data = file_target(f\"{cwd:a}/{output_prefix}.{effect_model}.prior.rds\")\nvhat_data = file_target(f\"{cwd:a}/{output_prefix}.{effect_model}.V_{vhat}.rds\")\nmash_model = file_target(f\"{cwd:a}/{output_prefix}.{effect_model}.V_{vhat}.mash_model.rds\")\n\ndef sort_uniq(seq):\n seen = set()\n return [x for x in seq if not (x in seen or seen.add(x))]",
"_____no_output_____"
]
],
[
[
"### Command interface",
"_____no_output_____"
]
],
[
[
"sos run mashr_flashr_workflow.ipynb -h",
"usage: sos run mashr_flashr_workflow.ipynb\n [workflow_name | -t targets] [options] [workflow_options]\n workflow_name: Single or combined workflows defined in this script\n targets: One or more targets to generate\n options: Single-hyphen sos parameters (see \"sos run -h\" for details)\n workflow_options: Double-hyphen workflow-specific parameters\n\nWorkflows:\n flash\n flash_nonneg\n pca\n vhat_identity\n vhat_simple\n vhat_mle\n vhat_corshrink_xcondition\n vhat_simple_specific\n prior\n mash\n posterior\n\nGlobal Workflow Options:\n --cwd mashr_flashr_workflow_output (as path)\n --data fastqtl_to_mash_output/FastQTLSumStats.mash.rds (as path)\n Input summary statistics data\n --output-prefix ''\n Prefix of output files. If not specified, it will derive\n it from data. If it is specified, for example,\n `--output-prefix AnalysisResults` It will save output\n files as `{cwd}/AnalysisResults*`.\n --effect-model EZ\n Exchangable effect (EE) or exchangable z-scores (EZ)\n --vhat mle\n Identifier of $\\hat{V}$ estimate file Options are\n \"identity\", \"simple\", \"mle\",\n \"vhat_corshrink_xcondition\", \"vhat_simple_specific\"\n --mixture-components flash flash_nonneg pca (as list)\n\nSections\n flash: Perform FLASH analysis with non-negative factor\n constraint (time estimate: 20min)\n flash_nonneg: Perform FLASH analysis with non-negative factor\n constraint (time estimate: 20min)\n pca:\n Workflow Options:\n --npc 3 (as int)\n Number of components in PCA analysis for prior set to 3\n as in mash paper\n vhat_identity: V estimate: \"identity\" method\n vhat_simple: V estimate: \"simple\" method (using null z-scores)\n vhat_mle: V estimate: \"mle\" method\n Workflow Options:\n --n-subset 6000 (as int)\n number of samples to use\n --max-iter 6 (as int)\n maximum number of iterations\n vhat_corshrink_xcondition_1: Estimate each V separately via corshrink\n Workflow Options:\n --util-script /project/mstephens/gtex/scripts/SumstatQuery.R (as path)\n Utility script\n --gene-list . (as path)\n List of genes to analyze\n vhat_simple_specific_1: Estimate each V separately via \"simple\" method\n Workflow Options:\n --util-script /project/mstephens/gtex/scripts/SumstatQuery.R (as path)\n Utility script\n --gene-list . (as path)\n List of genes to analyze\n vhat_corshrink_xcondition_2, vhat_simple_specific_2: Consolidate Vhat into one\n file\n Workflow Options:\n --gene-list . (as path)\n List of genes to analyze\n prior: Compute data-driven / canonical prior matrices (time\n estimate: 2h ~ 12h for ~30 49 by 49 matrix mixture)\n mash_1: Fit MASH mixture model (time estimate: <15min for 70K by\n 49 matrix)\n mash_2: Compute posterior for the \"strong\" set of data as in\n Urbut et al 2017. This is optional because most of the\n time we want to apply the MASH model learned on much\n larger data-set.\n Workflow Options:\n --[no-]compute-posterior (default to True)\n default to True; use --no-compute-posterior to disable\n this\n posterior: Apply posterior calculations\n Workflow Options:\n --mash-model path(f\"{vhat_data:n}.mash_model.rds\")\n\n --posterior-input paths()\n\n --posterior-vhat-files paths()\n\n --data-table-name ''\n eg, if data is saved in R list as data$strong, then when\n you specify `--data-table-name strong` it will read the\n data as readRDS('{_input:r}')$strong\n --bhat-table-name Bhat\n --shat-table-name Shat\n"
]
],
[
[
"## Factor analyses",
"_____no_output_____"
]
],
[
[
"# Perform FLASH analysis with non-negative factor constraint (time estimate: 20min)\n[flash]\ninput: data\noutput: f\"{cwd}/{output_prefix}.flash.rds\"\ntask: trunk_workers = 1, walltime = '2h', trunk_size = 1, mem = '8G', cores = 2, tags = f'{_output:bn}'\nR: expand = \"${ }\", stderr = f'{_output:n}.stderr', stdout = f'{_output:n}.stdout'\n dat = readRDS(${_input:r})\n dat = mashr::mash_set_data(dat$strong.b, Shat=dat$strong.s, alpha=${1 if effect_model == 'EZ' else 0}, zero_Bhat_Shat_reset = 1E3)\n res = mashr::cov_flash(dat, factors=\"default\", remove_singleton=${\"TRUE\" if \"canonical\" in mixture_components else \"FALSE\"}, output_model=\"${_output:n}.model.rds\")\n saveRDS(res, ${_output:r})",
"_____no_output_____"
],
[
"# Perform FLASH analysis with non-negative factor constraint (time estimate: 20min)\n[flash_nonneg]\ninput: data\noutput: f\"{cwd}/{output_prefix}.flash_nonneg.rds\"\ntask: trunk_workers = 1, walltime = '2h', trunk_size = 1, mem = '8G', cores = 2, tags = f'{_output:bn}'\nR: expand = \"${ }\", stderr = f'{_output:n}.stderr', stdout = f'{_output:n}.stdout'\n dat = readRDS(${_input:r})\n dat = mashr::mash_set_data(dat$strong.b, Shat=dat$strong.s, alpha=${1 if effect_model == 'EZ' else 0}, zero_Bhat_Shat_reset = 1E3)\n res = mashr::cov_flash(dat, factors=\"nonneg\", remove_singleton=${\"TRUE\" if \"canonical\" in mixture_components else \"FALSE\"}, output_model=\"${_output:n}.model.rds\")\n saveRDS(res, ${_output:r})",
"_____no_output_____"
],
[
"[pca]\n# Number of components in PCA analysis for prior\n# set to 3 as in mash paper\nparameter: npc = 3\ninput: data\noutput: f\"{cwd}/{output_prefix}.pca.rds\"\ntask: trunk_workers = 1, walltime = '1h', trunk_size = 1, mem = '4G', cores = 2, tags = f'{_output:bn}'\nR: expand = \"${ }\", stderr = f'{_output:n}.stderr', stdout = f'{_output:n}.stdout'\n dat = readRDS(${_input:r})\n dat = mashr::mash_set_data(dat$strong.b, Shat=dat$strong.s, alpha=${1 if effect_model == 'EZ' else 0}, zero_Bhat_Shat_reset = 1E3)\n res = mashr::cov_pca(dat, ${npc})\n saveRDS(res, ${_output:r})",
"_____no_output_____"
]
],
[
[
"### Estimate residual variance\n\nFIXME: add some narratives here explaining what we do in each method.",
"_____no_output_____"
]
],
[
[
"# V estimate: \"identity\" method\n[vhat_identity]\ninput: data\noutput: f'{vhat_data:nn}.V_identity.rds'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n dat = readRDS(${_input:r})\n saveRDS(diag(ncol(dat$random.b)), ${_output:r})",
"_____no_output_____"
],
[
"# V estimate: \"simple\" method (using null z-scores)\n[vhat_simple]\ndepends: R_library(\"mashr\")\ninput: data\noutput: f'{vhat_data:nn}.V_simple.rds'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n library(mashr)\n dat = readRDS(${_input:r})\n vhat = estimate_null_correlation_simple(mash_set_data(dat$random.b, Shat=dat$random.s, alpha=${1 if effect_model == 'EZ' else 0}, zero_Bhat_Shat_reset = 1E3))\n saveRDS(vhat, ${_output:r})",
"_____no_output_____"
],
[
"# V estimate: \"mle\" method\n[vhat_mle]\n# number of samples to use\nparameter: n_subset = 6000\n# maximum number of iterations\nparameter: max_iter = 6\ndepends: R_library(\"mashr\")\ninput: data, prior_data\noutput: f'{vhat_data:nn}.V_mle.rds'\n\ntask: trunk_workers = 1, walltime = '36h', trunk_size = 1, mem = '4G', cores = 1, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n library(mashr)\n dat = readRDS(${_input[0]:r})\n # choose random subset\n set.seed(1)\n random.subset = sample(1:nrow(dat$random.b), min(${n_subset}, nrow(dat$random.b)))\n random.subset = mash_set_data(dat$random.b[random.subset,], dat$random.s[random.subset,], alpha=${1 if effect_model == 'EZ' else 0}, zero_Bhat_Shat_reset = 1E3)\n # estimate V mle\n vhat = estimate_null_correlation(random.subset, readRDS(${_input[1]:r}), max_iter = ${max_iter})\n saveRDS(vhat, ${_output:r})",
"_____no_output_____"
],
[
"# Estimate each V separately via corshrink\n[vhat_corshrink_xcondition_1]\n# Utility script\nparameter: util_script = path('/project/mstephens/gtex/scripts/SumstatQuery.R')\n# List of genes to analyze\nparameter: gene_list = path()\n\nfail_if(not gene_list.is_file(), msg = 'Please specify valid path for --gene-list')\nfail_if(not util_script.is_file() and len(str(util_script)), msg = 'Please specify valid path for --util-script')\ngenes = sort_uniq([x.strip().strip('\"') for x in open(f'{gene_list:a}').readlines() if not x.strip().startswith('#')])\n\n\ndepends: R_library(\"CorShrink\")\ninput: data, for_each = 'genes'\noutput: f'{vhat_data:nn}/{vhat_data:bnn}_V_corshrink_{_genes}.rds'\n\ntask: trunk_workers = 1, walltime = '3m', trunk_size = 500, mem = '3G', cores = 1, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n source(${util_script:r})\n CorShrink_sum = function(gene, database, z_thresh = 2){\n print(gene)\n dat <- GetSS(gene, database)\n z = dat$\"z-score\"\n max_absz = apply(abs(z), 1, max)\n nullish = which(max_absz < z_thresh)\n # if (length(nullish) < ncol(z)) {\n # stop(\"not enough null data to estimate null correlation\")\n # }\n if (length(nullish) <= 1){\n mat = diag(ncol(z))\n } else {\n nullish_z = z[nullish, ] \n mat = as.matrix(CorShrink::CorShrinkData(nullish_z, ash.control = list(mixcompdist = \"halfuniform\"))$cor)\n }\n return(mat)\n }\n V = Corshrink_sum(\"${_genes}\", ${data:r})\n saveRDS(V, ${_output:r})",
"_____no_output_____"
],
[
"# Estimate each V separately via \"simple\" method\n[vhat_simple_specific_1]\n# Utility script\nparameter: util_script = path('/project/mstephens/gtex/scripts/SumstatQuery.R')\n# List of genes to analyze\nparameter: gene_list = path()\n\nfail_if(not gene_list.is_file(), msg = 'Please specify valid path for --gene-list')\nfail_if(not util_script.is_file() and len(str(util_script)), msg = 'Please specify valid path for --util-script')\ngenes = sort_uniq([x.strip().strip('\"') for x in open(f'{gene_list:a}').readlines() if not x.strip().startswith('#')])\n\ndepends: R_library(\"Matrix\")\ninput: data, for_each = 'genes'\noutput: f'{vhat_data:nn}/{vhat_data:bnn}_V_simple_{_genes}.rds'\n\ntask: trunk_workers = 1, walltime = '1m', trunk_size = 500, mem = '3G', cores = 1, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n source(${util_script:r})\n simple_V = function(gene, database, z_thresh = 2){\n print(gene)\n dat <- GetSS(gene, database)\n z = dat$\"z-score\"\n max_absz = apply(abs(z), 1, max)\n nullish = which(max_absz < z_thresh)\n # if (length(nullish) < ncol(z)) {\n # stop(\"not enough null data to estimate null correlation\")\n # }\n if (length(nullish) <= 1){\n mat = diag(ncol(z))\n } else {\n nullish_z = z[nullish, ]\n mat = as.matrix(Matrix::nearPD(as.matrix(cov(nullish_z)), conv.tol=1e-06, doSym = TRUE, corr=TRUE)$mat)\n }\n return(mat)\n }\n V = simple_V(\"${_genes}\", ${data:r})\n saveRDS(V, ${_output:r})",
"_____no_output_____"
],
[
"# Consolidate Vhat into one file\n[vhat_corshrink_xcondition_2, vhat_simple_specific_2]\ndepends: R_library(\"parallel\")\n# List of genes to analyze\nparameter: gene_list = path()\n\nfail_if(not gene_list.is_file(), msg = 'Please specify valid path for --gene-list')\ngenes = paths([x.strip().strip('\"') for x in open(f'{gene_list:a}').readlines() if not x.strip().startswith('#')])\n\n\ninput: group_by = 'all'\noutput: f\"{vhat_data:nn}.V_{step_name.rsplit('_',1)[0]}.rds\"\n\ntask: trunk_workers = 1, walltime = '1h', trunk_size = 1, mem = '4G', cores = 1, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n library(parallel)\n files = sapply(c(${genes:r,}), function(g) paste0(c(${_input[0]:adr}), '/', g, '.rds'), USE.NAMES=FALSE)\n V = mclapply(files, function(i){ readRDS(i) }, mc.cores = 1)\n R = dim(V[[1]])[1]\n L = length(V)\n V.array = array(as.numeric(unlist(V)), dim=c(R, R, L))\n saveRDS(V.array, ${_output:ar})",
"_____no_output_____"
]
],
[
[
"### Compute MASH priors \n\nMain reference are our `mashr` vignettes [this for mashr eQTL outline](https://stephenslab.github.io/mashr/articles/eQTL_outline.html) and [this for using FLASH prior](https://github.com/stephenslab/mashr/blob/master/vignettes/flash_mash.Rmd). \n\nThe outcome of this workflow should be found under `./mashr_flashr_workflow_output` folder (can be configured). File names have pattern `*.mash_model_*.rds`. They can be used to computer posterior for input list of gene-SNP pairs (see next section).",
"_____no_output_____"
]
],
[
[
"# Compute data-driven / canonical prior matrices (time estimate: 2h ~ 12h for ~30 49 by 49 matrix mixture)\n[prior]\ndepends: R_library(\"mashr\")\n# if vhat method is `mle` it should use V_simple to analyze the data to provide a rough estimate, then later be refined via `mle`.\ninput: [data, vhat_data if vhat != \"mle\" else f'{vhat_data:nn}.V_simple.rds'] + [f\"{cwd}/{output_prefix}.{m}.rds\" for m in mixture_components]\noutput: prior_data\n\ntask: trunk_workers = 1, walltime = '36h', trunk_size = 1, mem = '4G', cores = 4, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n library(mashr)\n rds_files = c(${_input:r,})\n dat = readRDS(rds_files[1])\n vhat = readRDS(rds_files[2])\n mash_data = mash_set_data(dat$strong.b, Shat=dat$strong.s, V=vhat, alpha=${1 if effect_model == 'EZ' else 0}, zero_Bhat_Shat_reset = 1E3)\n # setup prior\n U = list(XtX = t(mash_data$Bhat) %*% mash_data$Bhat / nrow(mash_data$Bhat))\n for (f in rds_files[3:length(rds_files)]) U = c(U, readRDS(f))\n U.ed = cov_ed(mash_data, U, logfile=${_output:nr})\n # Canonical matrices\n U.can = cov_canonical(mash_data)\n saveRDS(c(U.ed, U.can), ${_output:r})",
"_____no_output_____"
]
],
[
[
"## `mashr` mixture model fitting",
"_____no_output_____"
]
],
[
[
"# Fit MASH mixture model (time estimate: <15min for 70K by 49 matrix)\n[mash_1]\ndepends: R_library(\"mashr\")\ninput: data, vhat_data, prior_data\noutput: mash_model\n\ntask: trunk_workers = 1, walltime = '36h', trunk_size = 1, mem = '4G', cores = 1, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n library(mashr)\n dat = readRDS(${_input[0]:r})\n vhat = readRDS(${_input[1]:r})\n U = readRDS(${_input[2]:r})\n mash_data = mash_set_data(dat$random.b, Shat=dat$random.s, alpha=${1 if effect_model == 'EZ' else 0}, V=vhat, zero_Bhat_Shat_reset = 1E3)\n saveRDS(mash(mash_data, Ulist = U, outputlevel = 1), ${_output:r})",
"_____no_output_____"
]
],
[
[
"### Optional posterior computations\n\nAdditionally provide posterior for the \"strong\" set in MASH input data.",
"_____no_output_____"
]
],
[
[
"# Compute posterior for the \"strong\" set of data as in Urbut et al 2017.\n# This is optional because most of the time we want to apply the \n# MASH model learned on much larger data-set.\n[mash_2]\n# default to True; use --no-compute-posterior to disable this\nparameter: compute_posterior = True\n# input Vhat file for the batch of posterior data\nskip_if(not compute_posterior)\ndepends: R_library(\"mashr\")\ninput: data, vhat_data, mash_model\noutput: f\"{cwd:a}/{output_prefix}.{effect_model}.posterior.rds\"\n\ntask: trunk_workers = 1, walltime = '36h', trunk_size = 1, mem = '4G', cores = 1, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n library(mashr)\n dat = readRDS(${_input[0]:r})\n vhat = readRDS(${_input[1]:r})\n mash_data = mash_set_data(dat$strong.b, Shat=dat$strong.s, alpha=${1 if effect_model == 'EZ' else 0}, V=vhat, zero_Bhat_Shat_reset = 1E3)\n mash_model = readRDS(${_input[2]:ar})\n saveRDS(mash_compute_posterior_matrices(mash_model, mash_data), ${_output:r})",
"_____no_output_____"
]
],
[
[
"## Compute MASH posteriors\n\nIn the GTEx V6 paper we assumed one eQTL per gene and applied the model learned above to those SNPs. Under that assumption, the input data for posterior calculation will be the `dat$strong.*` matrices.\nIt is a fairly straightforward procedure as shown in [this vignette](https://stephenslab.github.io/mashr/articles/eQTL_outline.html).\n\nBut it is often more interesting to apply MASH to given list of eQTLs, eg, from those from fine-mapping results. In GTEx V8 analysis we obtain such gene-SNP pairs from DAP-G fine-mapping analysis. See [this notebook](https://stephenslab.github.io/gtex-eqtls/analysis/Independent_eQTL_Results.html) for how the input data is prepared. The workflow below takes a number of input chunks (each chunk is a list of matrices `dat$Bhat` and `dat$Shat`) \nand computes posterior for each chunk. It is therefore suited for running in parallel posterior computation for all gene-SNP pairs, if input data chunks are provided.\n\n\n```\nJOB_OPT=\"-c midway2.yml -q midway2\"\nDATA_DIR=/project/compbio/GTEx_eQTL/independent_eQTL\nsos run workflows/mashr_flashr_workflow.ipynb posterior \\\n $JOB_OPT \\\n --posterior-input $DATA_DIR/DAPG_pip_gt_0.01-AllTissues/DAPG_pip_gt_0.01-AllTissues.*.rds \\\n $DATA_DIR/ConditionalAnalysis_AllTissues/ConditionalAnalysis_AllTissues.*.rds\n```",
"_____no_output_____"
]
],
[
[
"# Apply posterior calculations\n[posterior]\nparameter: mash_model = path(f\"{cwd:a}/{output_prefix}.{effect_model}.V_{vhat}.mash_model.rds\")\nparameter: posterior_input = paths()\nparameter: posterior_vhat_files = paths()\n# eg, if data is saved in R list as data$strong, then\n# when you specify `--data-table-name strong` it will read the data as\n# readRDS('{_input:r}')$strong\nparameter: data_table_name = ''\nparameter: bhat_table_name = 'Bhat'\nparameter: shat_table_name = 'Shat'\nmash_model = f\"{mash_model:a}\"\n\nskip_if(len(posterior_input) == 0, msg = \"No posterior input data to compute on. Please specify it using --posterior-input.\")\nfail_if(len(posterior_vhat_files) > 1 and len(posterior_vhat_files) != len(posterior_input), msg = \"length of --posterior-input and --posterior-vhat-files do not agree.\")\nfor p in posterior_input:\n fail_if(not p.is_file(), msg = f'Cannot find posterior input file ``{p}``')\n\ndepends: R_library(\"mashr\"), mash_model\ninput: posterior_input, group_by = 1\noutput: f\"{_input:n}.posterior.rds\"\n \ntask: trunk_workers = 1, walltime = '20h', trunk_size = 1, mem = '20G', cores = 1, tags = f'{_output:bn}'\nR: expand = \"${ }\", workdir = cwd, stderr = f\"{_output:n}.stderr\", stdout = f\"{_output:n}.stdout\"\n library(mashr)\n data = readRDS(${_input:r})${('$' + data_table_name) if data_table_name else ''}\n vhat = readRDS(\"${vhat_data if len(posterior_vhat_files) == 0 else posterior_vhat_files[_index]}\")\n mash_data = mash_set_data(data$${bhat_table_name}, Shat=data$${shat_table_name}, alpha=${1 if effect_model == 'EZ' else 0}, V=vhat, zero_Bhat_Shat_reset = 1E3)\n saveRDS(mash_compute_posterior_matrices(readRDS(${mash_model:r}), mash_data), ${_output:r})",
"_____no_output_____"
]
],
[
[
"### Posterior results",
"_____no_output_____"
],
[
"1. The outcome of the `[posterior]` step should produce a number of serialized R objects `*.batch_*.posterior.rds` (can be loaded to R via `readRDS()`) -- I chopped data to batches to take advantage of computing in multiple cluster nodes. It should be self-explanary but please let me know otherwise.\n2. Other posterior related files are:\n 1. `*.batch_*.yaml`: gene-SNP pairs of interest, identified elsewhere (eg. fine-mapping analysis). \n 2. The corresponding univariate analysis summary statistics for gene-SNPs from `*.batch_*.yaml` are extracted and saved to `*.batch_*.rds`, creating input to the `[posterior]` step.\n 3. Note the `*.batch_*.stdout` file documents some SNPs found in fine-mapping results but not found in the original `fastqtl` output.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
cb6d673c40aeb6d78a33eb24fca46fd6eac82ab8 | 74,210 | ipynb | Jupyter Notebook | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | yaoaaron/pandas_exercises | e98aacdf7e7d6a1c9d176ecb3b44f1d2899ddac5 | [
"BSD-3-Clause"
]
| null | null | null | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | yaoaaron/pandas_exercises | e98aacdf7e7d6a1c9d176ecb3b44f1d2899ddac5 | [
"BSD-3-Clause"
]
| null | null | null | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | yaoaaron/pandas_exercises | e98aacdf7e7d6a1c9d176ecb3b44f1d2899ddac5 | [
"BSD-3-Clause"
]
| null | null | null | 34.11954 | 185 | 0.256165 | [
[
[
"# Ex2 - Filtering and Sorting Data",
"_____no_output_____"
],
[
"This time we are going to pull data directly from the internet.\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/02_Filtering_%26_Sorting/Euro12/Euro_2012_stats_TEAM.csv). ",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable called euro12.",
"_____no_output_____"
]
],
[
[
"euro12 = pd.read_csv('https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/02_Filtering_%26_Sorting/Euro12/Euro_2012_stats_TEAM.csv', sep=',')\neuro12",
"_____no_output_____"
]
],
[
[
"### Step 4. Select only the Goal column.",
"_____no_output_____"
]
],
[
[
"euro12.Goals",
"_____no_output_____"
]
],
[
[
"### Step 5. How many team participated in the Euro2012?",
"_____no_output_____"
]
],
[
[
"euro12.shape[0]",
"_____no_output_____"
]
],
[
[
"### Step 6. What is the number of columns in the dataset?",
"_____no_output_____"
]
],
[
[
"euro12.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16 entries, 0 to 15\nData columns (total 35 columns):\nTeam 16 non-null object\nGoals 16 non-null int64\nShots on target 16 non-null int64\nShots off target 16 non-null int64\nShooting Accuracy 16 non-null object\n% Goals-to-shots 16 non-null object\nTotal shots (inc. Blocked) 16 non-null int64\nHit Woodwork 16 non-null int64\nPenalty goals 16 non-null int64\nPenalties not scored 16 non-null int64\nHeaded goals 16 non-null int64\nPasses 16 non-null int64\nPasses completed 16 non-null int64\nPassing Accuracy 16 non-null object\nTouches 16 non-null int64\nCrosses 16 non-null int64\nDribbles 16 non-null int64\nCorners Taken 16 non-null int64\nTackles 16 non-null int64\nClearances 16 non-null int64\nInterceptions 16 non-null int64\nClearances off line 15 non-null float64\nClean Sheets 16 non-null int64\nBlocks 16 non-null int64\nGoals conceded 16 non-null int64\nSaves made 16 non-null int64\nSaves-to-shots ratio 16 non-null object\nFouls Won 16 non-null int64\nFouls Conceded 16 non-null int64\nOffsides 16 non-null int64\nYellow Cards 16 non-null int64\nRed Cards 16 non-null int64\nSubs on 16 non-null int64\nSubs off 16 non-null int64\nPlayers Used 16 non-null int64\ndtypes: float64(1), int64(29), object(5)\nmemory usage: 4.4+ KB\n"
]
],
[
[
"### Step 7. View only the columns Team, Yellow Cards and Red Cards and assign them to a dataframe called discipline",
"_____no_output_____"
]
],
[
[
"# filter only giving the column names\n\ndiscipline = euro12[['Team', 'Yellow Cards', 'Red Cards']]\ndiscipline",
"_____no_output_____"
]
],
[
[
"### Step 8. Sort the teams by Red Cards, then to Yellow Cards",
"_____no_output_____"
]
],
[
[
"discipline.sort_values(['Red Cards', 'Yellow Cards'], ascending = False)",
"_____no_output_____"
]
],
[
[
"### Step 9. Calculate the mean Yellow Cards given per Team",
"_____no_output_____"
]
],
[
[
"round(discipline['Yellow Cards'].mean())",
"_____no_output_____"
]
],
[
[
"### Step 10. Filter teams that scored more than 6 goals",
"_____no_output_____"
]
],
[
[
"euro12[euro12.Goals > 6]",
"_____no_output_____"
]
],
[
[
"### Step 11. Select the teams that start with G",
"_____no_output_____"
]
],
[
[
"euro12[euro12.Team.str.startswith('G')]",
"_____no_output_____"
]
],
[
[
"### Step 12. Select the first 7 columns",
"_____no_output_____"
]
],
[
[
"# use .iloc to slices via the position of the passed integers\n# : means all, 0:7 means from 0 to 7\n\neuro12.iloc[: , 0:7]",
"_____no_output_____"
]
],
[
[
"### Step 13. Select all columns except the last 3.",
"_____no_output_____"
]
],
[
[
"# use negative to exclude the last 3 columns\n\neuro12.iloc[: , :-3]",
"_____no_output_____"
]
],
[
[
"### Step 14. Present only the Shooting Accuracy from England, Italy and Russia",
"_____no_output_____"
]
],
[
[
"# .loc is another way to slice, using the labels of the columns and indexes\n\neuro12.loc[euro12.Team.isin(['England', 'Italy', 'Russia']), ['Team','Shooting Accuracy']]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6d73377c35017e4d0a4a236b58722c50e5e9e2 | 285,588 | ipynb | Jupyter Notebook | Datascience_With_Python/Machine Learning/Cheatsheets/Matplotlib/Visualization_using_Matplotlib.ipynb | vishnupriya129/winter-of-contributing | 8632c74d0c2d55bb4fddee9d6faac30159f376e1 | [
"MIT"
]
| 1,078 | 2021-09-05T09:44:33.000Z | 2022-03-27T01:16:02.000Z | Datascience_With_Python/Machine Learning/Cheatsheets/Matplotlib/Visualization_using_Matplotlib.ipynb | vishnupriya129/winter-of-contributing | 8632c74d0c2d55bb4fddee9d6faac30159f376e1 | [
"MIT"
]
| 6,845 | 2021-09-05T12:49:50.000Z | 2022-03-12T16:41:13.000Z | Datascience_With_Python/Machine Learning/Cheatsheets/Matplotlib/Visualization_using_Matplotlib.ipynb | vishnupriya129/winter-of-contributing | 8632c74d0c2d55bb4fddee9d6faac30159f376e1 | [
"MIT"
]
| 2,629 | 2021-09-03T04:53:16.000Z | 2022-03-20T17:45:00.000Z | 327.509174 | 29,008 | 0.895734 | [
[
[
"# Visualisation in Python - Matplotlib",
"_____no_output_____"
],
[
"Here is the sales dataset for an online retailer. The data is collected over a period of three years: 2012 to 2015. It contains the information of sales made by the company.\n\nThe products captured belong to three categories:\n\nFurniture\n\nOffice Supplies\n\nTechnology\n\nAlso, the company caters to five different markets:\n\nUSCA\n\nLATAM\n\nASPAC\n\nEUR\n\nAFR\n\nWe will be using the 'pyplot' package of the Matplotlib library.",
"_____no_output_____"
]
],
[
[
"# importing numpy and the pyplot package of matplotlib\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Creating an array with product categories\nproduct_cat = np.array(['Furniture','Technology','Office Supplies'])",
"_____no_output_____"
],
[
"# Creating an array with the sales amount\n# Furniture: 4110451.90\n# Technology: 4744557.50\n# Office Supplies: 3787492.52\nsales_amt = np.array([4110451.90,4744557.50,3787492.52])\nprint(sales_amt)",
"[4110451.9 4744557.5 3787492.52]\n"
]
],
[
[
"## Bar Graph: Plotting sales across each product category",
"_____no_output_____"
]
],
[
[
"# plotting the bar graph with product categories on x-axis and sales amount of y-axis\nplt.bar(product_cat,sales_amt)\n\n# necessary command to display the created graph\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Adding title and labeling axes in the graph",
"_____no_output_____"
]
],
[
[
"# plotting the bar graph with product categories on x-axis and sales amount of y-axis\nplt.bar(product_cat, sales_amt)\n\n# adding title to the graph\nplt.title(\"Sales Across Product Categories\", fontdict={'fontsize': 20, 'fontweight' : 5, 'color' : 'Green'})\n\n# labeling axes\nplt.xlabel(\"Product Category\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\nplt.ylabel(\"Sales\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\n\n# necessary command to display the created graph\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Modifying the bars in the graph",
"_____no_output_____"
]
],
[
[
"# changing color of the bars in the bar graph\n# plotting the bar graph with product categories on x-axis and sales amount of y-axis\nplt.bar(product_cat, sales_amt, color='cyan', edgecolor='orange')\n\n# adding title to the graph\nplt.title(\"Sales Across Product Categories\", fontdict={'fontsize': 20, 'fontweight' : 5, 'color' : 'Green'})\n\n# labeling axes\nplt.xlabel(\"Product Category\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\nplt.ylabel(\"Sales\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\n\n# necessary command to display the created graph\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Adjusting tick values and the value labels",
"_____no_output_____"
]
],
[
[
"# plotting the bar graph with product categories on x-axis and sales amount of y-axis\nplt.bar(product_cat, sales_amt, color='cyan', edgecolor='orange')\n\n# adding title to the graph\nplt.title(\"Sales Across Product Categories\", fontdict={'fontsize': 20, 'fontweight' : 5, 'color' : 'Green'})\n\n# labeling axes\nplt.xlabel(\"Product Category\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\nplt.ylabel(\"Sales\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\n\n# Modifying the ticks to show information in (lakhs)\ntick_values = np.arange(0, 8000000, 1000000)\ntick_labels = [\"0L\", \"10L\", \"20L\", \"30L\", \"40L\", \"50L\", \"60L\", \"70L\"]\n\nplt.yticks(tick_values, tick_labels)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Scatter Chart: Plotting Sales vs Profits",
"_____no_output_____"
],
[
"Scatter plots are used when you want to show the relationship between two facts or measures.\n\nNow, you have the sales and profit data of different product categories across different countries. Let's try to build scatterplots to visualise the data at hand.",
"_____no_output_____"
]
],
[
[
"# Sales and Profit data for different product categories across different countries\nsales = np.array ([1013.14, 8298.48, 875.51, 22320.83, 9251.6, 4516.86, 585.16, 836154.03, 216748.48, 174.2, 27557.79, 563.25, 558.11, 37117.45, 357.36, 2206.96, 709.5, 35064.03, 7230.78, 235.33, 148.32, 3973.27, 11737.8, 7104.63, 83.67, 5569.83, 92.34, 107104.36, 1045.62, 9072.51, 42485.82, 5093.82, 14846.16, 943.92, 684.36, 15012.03, 38196.18, 2448.75, 28881.96, 13912.14, 4507.2, 4931.06, 12805.05, 67912.73, 4492.2, 1740.01, 458.04, 16904.32, 21744.53, 10417.26, 18665.33, 2808.42, 54195.57, 67332.5, 24390.95, 1790.43, 2234.19, 9917.5, 7408.14, 36051.99, 1352.22, 1907.7, 245722.14, 2154.66, 1078.21, 3391.65, 28262.73, 5177.04, 66.51, 2031.34, 1683.72, 1970.01, 6515.82, 1055.31, 1029.48, 5303.4, 1850.96, 1159.41, 39989.13, 1183.87, 96365.09, 8356.68, 7010.24, 23119.23, 46109.28, 146071.84, 242259.03, 9058.95, 1313.67, 31525.06, 2019.94, 703.04, 1868.79, 700.5, 55512.02, 243.5, 2113.18, 11781.81, 262189.49, 3487.29, 513.12, 312050.42, 5000.7, 121.02, 1302.78, 169.92, 124.29, 57366.05, 29445.93, 4614.3, 45009.98, 309.24, 3353.67, 41348.34, 2280.27, 61193.7, 1466.79, 12419.94, 445.12, 25188.65, 263514.92, 12351.23, 1152.3, 26298.81, 9900.78, 5355.57, 2325.66, 6282.81, 127707.92, 1283.1, 3560.15, 3723.84, 13715.01, 4887.9, 3396.89, 33348.42, 625.02, 1665.48, 32486.97, 340212.44, 20516.22, 8651.16, 13590.06, 2440.35, 6462.57, 1770.13, 7527.18, 1433.65, 423.3, 21601.72, 10035.72, 2378.49, 3062.38, 719469.32, 179366.79, 345.17, 30345.78, 300.71, 940.81, 36468.08, 1352.85, 1755.72, 2391.96, 19.98, 19792.8, 15633.88, 7.45, 521.67, 1118.24, 7231.68, 12399.32, 204.36, 23.64, 5916.48, 313.98, 108181.5, 9212.42, 27476.91, 1761.33, 289.5, 780.3, 15098.46, 813.27, 47.55, 8323.23, 22634.64, 1831.02, 28808.1, 10539.78, 588.99, 939.78, 7212.41, 15683.01, 41369.09, 5581.6, 403.36, 375.26, 12276.66, 15393.56, 76.65, 5884.38, 18005.49, 3094.71, 43642.78, 35554.83, 22977.11, 1026.33, 665.28, 9712.49, 6038.52, 30756.51, 3758.25, 4769.49, 2463.3, 160153.16, 967.11, 2311.74, 1414.83, 12764.91, 4191.24, 110.76, 637.34, 1195.12, 2271.63, 804.12, 196.17, 167.67, 131.77, 2842.05, 9969.12, 1784.35, 3098.49, 25005.54, 1300.1, 118697.39, 7920.54, 6471.78, 31707.57, 37636.47, 118777.77, 131170.76, 3980.88, 3339.39, 26563.9, 4038.73, 124.8, 196.65, 2797.77, 29832.76, 184.84, 79.08, 8047.83, 205313.25, 1726.98, 899.73, 224.06, 304763.54, 6101.31, 729.6, 896.07, 17.82, 26.22, 46429.78, 31167.27, 2455.94, 37714.3, 1506.93, 3812.78, 25223.34, 3795.96, 437.31, 41278.86, 2091.81, 6296.61, 468.82, 23629.64, 160435.53, 9725.46, 1317.03, 1225.26, 30034.08, 7893.45, 2036.07, 215.52, 3912.42, 82783.43, 253.14, 966.96, 3381.26, 164.07, 1984.23, 75.12, 25168.17, 3295.53, 991.12, 10772.1, 44.16, 1311.45, 35352.57, 245783.54, 20.49, 13471.06, 8171.16, 14075.67, 611.82, 3925.56, 981.84, 10209.84, 156.56, 243.06, 21287.52, 7300.51, 434.52, 6065.0, 741577.51, 132461.03, 224.75, 28953.6, 757.98, 528.15, 34922.41, 50.58, 2918.48, 1044.96, 22195.13, 3951.48, 6977.64, 219.12, 5908.38, 10987.46, 4852.26, 445.5, 71860.82, 14840.45, 24712.08, 1329.9, 1180.44, 85.02, 10341.63, 690.48, 1939.53, 20010.51, 914.31, 25223.82, 12804.66, 2124.24, 602.82, 2961.66, 15740.79, 74138.35, 7759.39, 447.0, 2094.84, 22358.95, 21734.53, 4223.73, 17679.53, 1019.85, 51848.72, 69133.3, 30146.9, 705.48, 14508.88, 7489.38, 20269.44, 246.12, 668.13, 768.93, 215677.35, 899.16, 2578.2, 4107.99, 20334.57, 366.84, 3249.27, 98.88, 3497.88, 3853.05, 786.75, 1573.68, 458.36, 1234.77, 1094.22, 2300.61, 970.14, 3068.25, 35792.85, 4277.82, 71080.28, 3016.86, 3157.49, 15888.0, 30000.36, 140037.89, 216056.25, 1214.22, 1493.94, 32036.69, 4979.66, 106.02, 46257.68, 1033.3, 937.32, 3442.62, 160633.45, 213.15, 338.88, 242117.13, 9602.34, 2280.99, 73759.08, 23526.12, 6272.74, 43416.3, 576.78, 1471.61, 20844.9, 3497.7, 56382.38, 902.58, 6235.26, 48.91, 32684.24, 276611.58, 13370.38, 10595.28, 4555.14, 10084.38, 267.72, 1012.95, 4630.5, 149433.51, 364.32, 349.2, 4647.56, 504.0, 10343.52, 5202.66, 2786.26, 34135.95, 2654.58, 24699.51, 339239.87, 136.26, 23524.51, 8731.68, 8425.86, 835.95, 11285.19])\nprofit = np.array([-1213.46, 1814.13, -1485.7, -2286.73, -2872.12, 946.8, 198.48, 145454.95, 49476.1, -245.56, 5980.77, -790.47, -895.72, -34572.08, 117.9, 561.96, 152.85, 1426.05, 1873.17, -251.03, 68.22, 635.11, 3722.4, -3168.63, 27.6, 952.11, 7.38, 20931.13, 186.36, -5395.38, 9738.45, 525.27, 3351.99, 120.78, 266.88, 3795.21, 8615.97, 609.54, 7710.57, 2930.43, 1047.96, -2733.32, 2873.73, -5957.89, -909.6, 163.41, -376.02, -6322.68, -10425.86, 2340.36, -28430.53, 756.12, 12633.33, 7382.54, -14327.69, 436.44, 683.85, -694.91, 1960.56, 10925.82, 334.08, 425.49, 53580.2, 1024.56, 110.93, 632.22, 8492.58, 1418.88, 19.26, -2567.57, 346.26, 601.86, 1318.68, 304.05, 428.37, 1416.24, -2878.18, 283.41, 12611.04, 261.95, -648.43, 1112.88, -2640.29, 6154.32, 11558.79, 15291.4, 56092.65, 1515.39, 342.03, -10865.66, -902.8, 351.52, 364.17, 87.72, 11565.66, 75.4, 289.33, 3129.63, 50795.72, 783.72, 215.46, 29196.89, 1147.26, 53.22, 286.56, 73.02, 42.24, 13914.85, 5754.54, 998.04, -1476.04, 86.58, -1636.35, 10511.91, 647.34, 13768.62, 338.67, 3095.67, 173.84, 5632.93, 64845.11, 3297.33, 338.61, 7246.62, 2255.52, 1326.36, 827.64, 1100.58, 9051.36, 412.23, 1063.91, 940.59, 3891.84, 1599.51, 1129.57, 8792.64, 6.24, 592.77, 8792.85, 47727.5, -4597.68, 2242.56, 3546.45, 321.87, 1536.72, -2463.29, 1906.08, -1916.99, 186.24, 3002.05, -3250.98, 554.7, 830.64, 122612.79, 33894.21, -559.03, 7528.05, -477.67, -1660.25, -33550.96, 481.68, 425.08, 450.3, 9.57, -3025.29, 2924.62, -11.84, 87.36, 26.51, 1727.19, -6131.18, 59.16, 3.06, 1693.47, 74.67, 24729.21, -4867.94, 6705.18, 410.79, 70.74, 101.7, 3264.3, 137.01, 6.18, 2100.21, 5295.24, 520.29, 7205.52, 2602.65, 116.67, 224.91, -5153.93, 3882.69, -6535.24, -1254.1, 84.56, -186.38, -3167.2, -7935.59, 37.02, 1908.06, -27087.84, 829.32, 8727.44, 2011.47, -11629.64, 234.96, 53.1, 1248.14, 1511.07, 7374.24, 1193.28, 1090.23, 553.86, 38483.86, 255.81, 528.54, 326.07, 3924.36, 1018.92, 36.48, 113.24, -1770.05, 527.64, 224.49, 79.53, 64.77, 38.08, 868.08, 2265.06, -2643.62, 833.73, 5100.03, 326.44, 18158.84, 1682.01, -3290.22, 8283.33, 7926.18, 1694.41, 30522.92, 1214.07, 900.6, -6860.8, -865.91, 26.16, 47.22, 863.52, 7061.26, 73.92, 33.12, 1801.23, 38815.44, 431.13, 216.81, 16.5, 53688.2, 1210.32, 236.94, 210.84, 3.18, 2.22, 10265.64, 7212.3, 343.56, 3898.28, 568.11, -1867.85, 5782.38, 697.29, -192.06, 10179.02, 616.32, 1090.47, 165.84, 6138.28, 39723.06, 2085.14, 90.0, 129.93, 7957.53, 2131.86, 562.44, 99.12, 1298.37, 7580.33, 113.73, 139.71, 456.0, 21.24, 292.68, 30.34, 5817.15, 1060.89, 252.9, 3060.61, 6.6, 219.09, 8735.82, 31481.09, 2.85, -3124.72, 2195.94, 3464.7, 141.12, 1125.69, -1752.03, 3281.52, -303.77, 114.18, -2412.63, -5099.61, 146.64, 660.22, 18329.28, 28529.84, -232.27, 7435.41, -1157.94, -746.73, -30324.2, 2.52, 1313.44, 213.72, -5708.95, 930.18, 1663.02, 31.59, 1787.88, -8219.56, 973.92, 4.32, 8729.78, -2529.52, 5361.06, 69.21, 519.3, 13.56, 2236.77, 213.96, 367.98, 5074.2, 206.61, 7620.36, 2093.19, 164.07, 230.01, -815.82, 4226.7, -3635.09, -3344.17, 167.26, 143.79, -8233.57, -4085.21, 919.35, -25232.35, 234.33, 12040.68, 7206.28, -15112.76, 206.04, -2662.49, 2346.81, 4461.36, 93.48, 82.11, 147.87, 10389.53, 395.58, 474.74, 1333.26, 3913.02, 117.36, 858.78, 6.9, -4628.49, 1170.6, 218.55, 539.58, -211.0, 438.87, 317.16, 310.8, -1578.09, 706.56, 6617.4, 803.84, 2475.26, 764.34, -1461.88, 3805.56, 7371.27, -1377.13, 42435.03, 472.47, 315.48, -11755.91, -2418.6, 6.36, 9317.76, 326.88, -287.31, 637.68, 17579.17, 70.83, 47.4, 26143.92, 1548.15, 612.78, 17842.76, 6735.39, 1206.5, -10035.74, 149.4, -777.85, 5566.29, 748.92, 14941.58, 348.93, 1944.06, -5.51, 7026.84, 46114.92, 2361.86, 2613.24, 1277.37, 2587.74, 103.08, 311.43, 1250.58, 13055.21, 18.21, 108.24, 709.44, 115.92, 1863.6, 1873.86, 817.32, 7577.64, 1019.19, 6813.03, 24698.84, 66.24, -10971.39, 2056.47, 2095.35, 246.33, 2797.89])\n",
"_____no_output_____"
]
],
[
[
"### Plotting a Scatterplot",
"_____no_output_____"
]
],
[
[
"# plotting scatterplot\nplt.scatter(sales,profit)\n\n# necessary command to display graph\nplt.show()\nplt.scatter(profit,sales)\nplt.show()",
"_____no_output_____"
],
[
"# Sales and Profit data for different product categories across different countries\nsales = np.array ([1013.14, 8298.48, 875.51, 22320.83, 9251.6, 4516.86, 585.16, 836154.03, 216748.48, 174.2, 27557.79, 563.25, 558.11, 37117.45, 357.36, 2206.96, 709.5, 35064.03, 7230.78, 235.33, 148.32, 3973.27, 11737.8, 7104.63, 83.67, 5569.83, 92.34, 107104.36, 1045.62, 9072.51, 42485.82, 5093.82, 14846.16, 943.92, 684.36, 15012.03, 38196.18, 2448.75, 28881.96, 13912.14, 4507.2, 4931.06, 12805.05, 67912.73, 4492.2, 1740.01, 458.04, 16904.32, 21744.53, 10417.26, 18665.33, 2808.42, 54195.57, 67332.5, 24390.95, 1790.43, 2234.19, 9917.5, 7408.14, 36051.99, 1352.22, 1907.7, 245722.14, 2154.66, 1078.21, 3391.65, 28262.73, 5177.04, 66.51, 2031.34, 1683.72, 1970.01, 6515.82, 1055.31, 1029.48, 5303.4, 1850.96, 1159.41, 39989.13, 1183.87, 96365.09, 8356.68, 7010.24, 23119.23, 46109.28, 146071.84, 242259.03, 9058.95, 1313.67, 31525.06, 2019.94, 703.04, 1868.79, 700.5, 55512.02, 243.5, 2113.18, 11781.81, 262189.49, 3487.29, 513.12, 312050.42, 5000.7, 121.02, 1302.78, 169.92, 124.29, 57366.05, 29445.93, 4614.3, 45009.98, 309.24, 3353.67, 41348.34, 2280.27, 61193.7, 1466.79, 12419.94, 445.12, 25188.65, 263514.92, 12351.23, 1152.3, 26298.81, 9900.78, 5355.57, 2325.66, 6282.81, 127707.92, 1283.1, 3560.15, 3723.84, 13715.01, 4887.9, 3396.89, 33348.42, 625.02, 1665.48, 32486.97, 340212.44, 20516.22, 8651.16, 13590.06, 2440.35, 6462.57, 1770.13, 7527.18, 1433.65, 423.3, 21601.72, 10035.72, 2378.49, 3062.38, 719469.32, 179366.79, 345.17, 30345.78, 300.71, 940.81, 36468.08, 1352.85, 1755.72, 2391.96, 19.98, 19792.8, 15633.88, 7.45, 521.67, 1118.24, 7231.68, 12399.32, 204.36, 23.64, 5916.48, 313.98, 108181.5, 9212.42, 27476.91, 1761.33, 289.5, 780.3, 15098.46, 813.27, 47.55, 8323.23, 22634.64, 1831.02, 28808.1, 10539.78, 588.99, 939.78, 7212.41, 15683.01, 41369.09, 5581.6, 403.36, 375.26, 12276.66, 15393.56, 76.65, 5884.38, 18005.49, 3094.71, 43642.78, 35554.83, 22977.11, 1026.33, 665.28, 9712.49, 6038.52, 30756.51, 3758.25, 4769.49, 2463.3, 160153.16, 967.11, 2311.74, 1414.83, 12764.91, 4191.24, 110.76, 637.34, 1195.12, 2271.63, 804.12, 196.17, 167.67, 131.77, 2842.05, 9969.12, 1784.35, 3098.49, 25005.54, 1300.1, 118697.39, 7920.54, 6471.78, 31707.57, 37636.47, 118777.77, 131170.76, 3980.88, 3339.39, 26563.9, 4038.73, 124.8, 196.65, 2797.77, 29832.76, 184.84, 79.08, 8047.83, 205313.25, 1726.98, 899.73, 224.06, 304763.54, 6101.31, 729.6, 896.07, 17.82, 26.22, 46429.78, 31167.27, 2455.94, 37714.3, 1506.93, 3812.78, 25223.34, 3795.96, 437.31, 41278.86, 2091.81, 6296.61, 468.82, 23629.64, 160435.53, 9725.46, 1317.03, 1225.26, 30034.08, 7893.45, 2036.07, 215.52, 3912.42, 82783.43, 253.14, 966.96, 3381.26, 164.07, 1984.23, 75.12, 25168.17, 3295.53, 991.12, 10772.1, 44.16, 1311.45, 35352.57, 245783.54, 20.49, 13471.06, 8171.16, 14075.67, 611.82, 3925.56, 981.84, 10209.84, 156.56, 243.06, 21287.52, 7300.51, 434.52, 6065.0, 741577.51, 132461.03, 224.75, 28953.6, 757.98, 528.15, 34922.41, 50.58, 2918.48, 1044.96, 22195.13, 3951.48, 6977.64, 219.12, 5908.38, 10987.46, 4852.26, 445.5, 71860.82, 14840.45, 24712.08, 1329.9, 1180.44, 85.02, 10341.63, 690.48, 1939.53, 20010.51, 914.31, 25223.82, 12804.66, 2124.24, 602.82, 2961.66, 15740.79, 74138.35, 7759.39, 447.0, 2094.84, 22358.95, 21734.53, 4223.73, 17679.53, 1019.85, 51848.72, 69133.3, 30146.9, 705.48, 14508.88, 7489.38, 20269.44, 246.12, 668.13, 768.93, 215677.35, 899.16, 2578.2, 4107.99, 20334.57, 366.84, 3249.27, 98.88, 3497.88, 3853.05, 786.75, 1573.68, 458.36, 1234.77, 1094.22, 2300.61, 970.14, 3068.25, 35792.85, 4277.82, 71080.28, 3016.86, 3157.49, 15888.0, 30000.36, 140037.89, 216056.25, 1214.22, 1493.94, 32036.69, 4979.66, 106.02, 46257.68, 1033.3, 937.32, 3442.62, 160633.45, 213.15, 338.88, 242117.13, 9602.34, 2280.99, 73759.08, 23526.12, 6272.74, 43416.3, 576.78, 1471.61, 20844.9, 3497.7, 56382.38, 902.58, 6235.26, 48.91, 32684.24, 276611.58, 13370.38, 10595.28, 4555.14, 10084.38, 267.72, 1012.95, 4630.5, 149433.51, 364.32, 349.2, 4647.56, 504.0, 10343.52, 5202.66, 2786.26, 34135.95, 2654.58, 24699.51, 339239.87, 136.26, 23524.51, 8731.68, 8425.86, 835.95, 11285.19])\nprofit = np.array([-1213.46, 1814.13, -1485.7, -2286.73, -2872.12, 946.8, 198.48, 145454.95, 49476.1, -245.56, 5980.77, -790.47, -895.72, -34572.08, 117.9, 561.96, 152.85, 1426.05, 1873.17, -251.03, 68.22, 635.11, 3722.4, -3168.63, 27.6, 952.11, 7.38, 20931.13, 186.36, -5395.38, 9738.45, 525.27, 3351.99, 120.78, 266.88, 3795.21, 8615.97, 609.54, 7710.57, 2930.43, 1047.96, -2733.32, 2873.73, -5957.89, -909.6, 163.41, -376.02, -6322.68, -10425.86, 2340.36, -28430.53, 756.12, 12633.33, 7382.54, -14327.69, 436.44, 683.85, -694.91, 1960.56, 10925.82, 334.08, 425.49, 53580.2, 1024.56, 110.93, 632.22, 8492.58, 1418.88, 19.26, -2567.57, 346.26, 601.86, 1318.68, 304.05, 428.37, 1416.24, -2878.18, 283.41, 12611.04, 261.95, -648.43, 1112.88, -2640.29, 6154.32, 11558.79, 15291.4, 56092.65, 1515.39, 342.03, -10865.66, -902.8, 351.52, 364.17, 87.72, 11565.66, 75.4, 289.33, 3129.63, 50795.72, 783.72, 215.46, 29196.89, 1147.26, 53.22, 286.56, 73.02, 42.24, 13914.85, 5754.54, 998.04, -1476.04, 86.58, -1636.35, 10511.91, 647.34, 13768.62, 338.67, 3095.67, 173.84, 5632.93, 64845.11, 3297.33, 338.61, 7246.62, 2255.52, 1326.36, 827.64, 1100.58, 9051.36, 412.23, 1063.91, 940.59, 3891.84, 1599.51, 1129.57, 8792.64, 6.24, 592.77, 8792.85, 47727.5, -4597.68, 2242.56, 3546.45, 321.87, 1536.72, -2463.29, 1906.08, -1916.99, 186.24, 3002.05, -3250.98, 554.7, 830.64, 122612.79, 33894.21, -559.03, 7528.05, -477.67, -1660.25, -33550.96, 481.68, 425.08, 450.3, 9.57, -3025.29, 2924.62, -11.84, 87.36, 26.51, 1727.19, -6131.18, 59.16, 3.06, 1693.47, 74.67, 24729.21, -4867.94, 6705.18, 410.79, 70.74, 101.7, 3264.3, 137.01, 6.18, 2100.21, 5295.24, 520.29, 7205.52, 2602.65, 116.67, 224.91, -5153.93, 3882.69, -6535.24, -1254.1, 84.56, -186.38, -3167.2, -7935.59, 37.02, 1908.06, -27087.84, 829.32, 8727.44, 2011.47, -11629.64, 234.96, 53.1, 1248.14, 1511.07, 7374.24, 1193.28, 1090.23, 553.86, 38483.86, 255.81, 528.54, 326.07, 3924.36, 1018.92, 36.48, 113.24, -1770.05, 527.64, 224.49, 79.53, 64.77, 38.08, 868.08, 2265.06, -2643.62, 833.73, 5100.03, 326.44, 18158.84, 1682.01, -3290.22, 8283.33, 7926.18, 1694.41, 30522.92, 1214.07, 900.6, -6860.8, -865.91, 26.16, 47.22, 863.52, 7061.26, 73.92, 33.12, 1801.23, 38815.44, 431.13, 216.81, 16.5, 53688.2, 1210.32, 236.94, 210.84, 3.18, 2.22, 10265.64, 7212.3, 343.56, 3898.28, 568.11, -1867.85, 5782.38, 697.29, -192.06, 10179.02, 616.32, 1090.47, 165.84, 6138.28, 39723.06, 2085.14, 90.0, 129.93, 7957.53, 2131.86, 562.44, 99.12, 1298.37, 7580.33, 113.73, 139.71, 456.0, 21.24, 292.68, 30.34, 5817.15, 1060.89, 252.9, 3060.61, 6.6, 219.09, 8735.82, 31481.09, 2.85, -3124.72, 2195.94, 3464.7, 141.12, 1125.69, -1752.03, 3281.52, -303.77, 114.18, -2412.63, -5099.61, 146.64, 660.22, 18329.28, 28529.84, -232.27, 7435.41, -1157.94, -746.73, -30324.2, 2.52, 1313.44, 213.72, -5708.95, 930.18, 1663.02, 31.59, 1787.88, -8219.56, 973.92, 4.32, 8729.78, -2529.52, 5361.06, 69.21, 519.3, 13.56, 2236.77, 213.96, 367.98, 5074.2, 206.61, 7620.36, 2093.19, 164.07, 230.01, -815.82, 4226.7, -3635.09, -3344.17, 167.26, 143.79, -8233.57, -4085.21, 919.35, -25232.35, 234.33, 12040.68, 7206.28, -15112.76, 206.04, -2662.49, 2346.81, 4461.36, 93.48, 82.11, 147.87, 10389.53, 395.58, 474.74, 1333.26, 3913.02, 117.36, 858.78, 6.9, -4628.49, 1170.6, 218.55, 539.58, -211.0, 438.87, 317.16, 310.8, -1578.09, 706.56, 6617.4, 803.84, 2475.26, 764.34, -1461.88, 3805.56, 7371.27, -1377.13, 42435.03, 472.47, 315.48, -11755.91, -2418.6, 6.36, 9317.76, 326.88, -287.31, 637.68, 17579.17, 70.83, 47.4, 26143.92, 1548.15, 612.78, 17842.76, 6735.39, 1206.5, -10035.74, 149.4, -777.85, 5566.29, 748.92, 14941.58, 348.93, 1944.06, -5.51, 7026.84, 46114.92, 2361.86, 2613.24, 1277.37, 2587.74, 103.08, 311.43, 1250.58, 13055.21, 18.21, 108.24, 709.44, 115.92, 1863.6, 1873.86, 817.32, 7577.64, 1019.19, 6813.03, 24698.84, 66.24, -10971.39, 2056.47, 2095.35, 246.33, 2797.89])\n\n# corresponding category and country value to the above arrays\nproduct_category = np.array(['Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Technology', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Office Supplies', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture', 'Furniture'])\ncountry = np.array(['Zimbabwe', 'Zambia', 'Yemen', 'Vietnam', 'Venezuela', 'Uzbekistan', 'Uruguay', 'United States', 'United Kingdom', 'United Arab Emirates', 'Ukraine', 'Uganda', 'Turkmenistan', 'Turkey', 'Tunisia', 'Trinidad and Tobago', 'Togo', 'Thailand', 'Tanzania', 'Tajikistan', 'Taiwan', 'Syria', 'Switzerland', 'Sweden', 'Swaziland', 'Sudan', 'Sri Lanka', 'Spain', 'South Sudan', 'South Korea', 'South Africa', 'Somalia', 'Singapore', 'Sierra Leone', 'Serbia', 'Senegal', 'Saudi Arabia', 'Rwanda', 'Russia', 'Romania', 'Qatar', 'Portugal', 'Poland', 'Philippines', 'Peru', 'Paraguay', 'Papua New Guinea', 'Panama', 'Pakistan', 'Norway', 'Nigeria', 'Niger', 'Nicaragua', 'New Zealand', 'Netherlands', 'Nepal', 'Namibia', 'Myanmar (Burma)', 'Mozambique', 'Morocco', 'Mongolia', 'Moldova', 'Mexico', 'Mauritania', 'Martinique', 'Mali', 'Malaysia', 'Madagascar', 'Luxembourg', 'Lithuania', 'Libya', 'Liberia', 'Lesotho', 'Lebanon', 'Kyrgyzstan', 'Kenya', 'Kazakhstan', 'Jordan', 'Japan', 'Jamaica', 'Italy', 'Israel', 'Ireland', 'Iraq', 'Iran', 'Indonesia', 'India', 'Hungary', 'Hong Kong', 'Honduras', 'Haiti', 'Guyana', 'Guinea-Bissau', 'Guinea', 'Guatemala', 'Guadeloupe', 'Greece', 'Ghana', 'Germany', 'Georgia', 'Gabon', 'France', 'Finland', 'Ethiopia', 'Estonia', 'Eritrea', 'Equatorial Guinea', 'El Salvador', 'Egypt', 'Ecuador', 'Dominican Republic', 'Djibouti', 'Denmark', 'Democratic Republic of the Congo', 'Czech Republic', 'Cuba', 'Croatia', \"Cote d'Ivoire\", 'Costa Rica', 'Colombia', 'China', 'Chile', 'Central African Republic', 'Canada', 'Cameroon', 'Cambodia', 'Burkina Faso', 'Bulgaria', 'Brazil', 'Bosnia and Herzegovina', 'Bolivia', 'Benin', 'Belgium', 'Belarus', 'Barbados', 'Bangladesh', 'Bahrain', 'Azerbaijan', 'Austria', 'Australia', 'Argentina', 'Angola', 'Algeria', 'Albania', 'Afghanistan', 'Zimbabwe', 'Zambia', 'Yemen', 'Western Sahara', 'Vietnam', 'Venezuela', 'Uzbekistan', 'Uruguay', 'United States', 'United Kingdom', 'United Arab Emirates', 'Ukraine', 'Uganda', 'Turkmenistan', 'Turkey', 'Tunisia', 'Trinidad and Tobago', 'Togo', 'The Gambia', 'Thailand', 'Tanzania', 'Tajikistan', 'Taiwan', 'Syria', 'Switzerland', 'Sweden', 'Swaziland', 'Suriname', 'Sudan', 'Sri Lanka', 'Spain', 'South Korea', 'South Africa', 'Somalia', 'Slovenia', 'Slovakia', 'Singapore', 'Sierra Leone', 'Serbia', 'Senegal', 'Saudi Arabia', 'Rwanda', 'Russia', 'Romania', 'Republic of the Congo', 'Qatar', 'Portugal', 'Poland', 'Philippines', 'Peru', 'Paraguay', 'Papua New Guinea', 'Panama', 'Pakistan', 'Oman', 'Norway', 'Nigeria', 'Niger', 'Nicaragua', 'New Zealand', 'Netherlands', 'Nepal', 'Namibia', 'Myanmar (Burma)', 'Mozambique', 'Morocco', 'Montenegro', 'Mongolia', 'Moldova', 'Mexico', 'Mauritania', 'Martinique', 'Mali', 'Malaysia', 'Madagascar', 'Macedonia', 'Luxembourg', 'Lithuania', 'Libya', 'Liberia', 'Lesotho', 'Lebanon', 'Laos', 'Kyrgyzstan', 'Kenya', 'Kazakhstan', 'Jordan', 'Japan', 'Jamaica', 'Italy', 'Israel', 'Ireland', 'Iraq', 'Iran', 'Indonesia', 'India', 'Hungary', 'Hong Kong', 'Honduras', 'Haiti', 'Guyana', 'Guinea-Bissau', 'Guinea', 'Guatemala', 'Guadeloupe', 'Greece', 'Ghana', 'Germany', 'Georgia', 'Gabon', 'French Guiana', 'France', 'Finland', 'Ethiopia', 'Estonia', 'Eritrea', 'Equatorial Guinea', 'El Salvador', 'Egypt', 'Ecuador', 'Dominican Republic', 'Djibouti', 'Denmark', 'Democratic Republic of the Congo', 'Czech Republic', 'Cyprus', 'Cuba', 'Croatia', \"Cote d'Ivoire\", 'Costa Rica', 'Colombia', 'China', 'Chile', 'Chad', 'Central African Republic', 'Canada', 'Cameroon', 'Cambodia', 'Burkina Faso', 'Bulgaria', 'Brazil', 'Botswana', 'Bosnia and Herzegovina', 'Bolivia', 'Bhutan', 'Benin', 'Belize', 'Belgium', 'Belarus', 'Barbados', 'Bangladesh', 'Bahrain', 'Azerbaijan', 'Austria', 'Australia', 'Armenia', 'Argentina', 'Angola', 'Algeria', 'Albania', 'Afghanistan', 'Zimbabwe', 'Zambia', 'Yemen', 'Western Sahara', 'Vietnam', 'Venezuela', 'Uzbekistan', 'Uruguay', 'United States', 'United Kingdom', 'United Arab Emirates', 'Ukraine', 'Uganda', 'Turkmenistan', 'Turkey', 'Tunisia', 'Trinidad and Tobago', 'Togo', 'Thailand', 'Tanzania', 'Taiwan', 'Syria', 'Switzerland', 'Sweden', 'Sudan', 'Sri Lanka', 'Spain', 'South Korea', 'South Africa', 'Somalia', 'Slovenia', 'Slovakia', 'Singapore', 'Sierra Leone', 'Senegal', 'Saudi Arabia', 'Rwanda', 'Russia', 'Romania', 'Republic of the Congo', 'Qatar', 'Portugal', 'Poland', 'Philippines', 'Peru', 'Paraguay', 'Papua New Guinea', 'Panama', 'Pakistan', 'Norway', 'Nigeria', 'Niger', 'Nicaragua', 'New Zealand', 'Netherlands', 'Nepal', 'Myanmar (Burma)', 'Mozambique', 'Morocco', 'Montenegro', 'Mongolia', 'Moldova', 'Mexico', 'Mauritania', 'Martinique', 'Mali', 'Malaysia', 'Malawi', 'Madagascar', 'Macedonia', 'Lithuania', 'Libya', 'Liberia', 'Lebanon', 'Laos', 'Kyrgyzstan', 'Kuwait', 'Kenya', 'Kazakhstan', 'Jordan', 'Japan', 'Jamaica', 'Italy', 'Israel', 'Ireland', 'Iraq', 'Iran', 'Indonesia', 'India', 'Hungary', 'Hong Kong', 'Honduras', 'Haiti', 'Guyana', 'Guatemala', 'Guadeloupe', 'Greece', 'Ghana', 'Germany', 'Georgia', 'Gabon', 'France', 'Finland', 'Estonia', 'El Salvador', 'Egypt', 'Ecuador', 'Dominican Republic', 'Djibouti', 'Denmark', 'Democratic Republic of the Congo', 'Czech Republic', 'Cuba', 'Croatia', \"Cote d'Ivoire\", 'Costa Rica', 'Colombia', 'China', 'Chile', 'Canada', 'Cameroon', 'Cambodia', 'Burundi', 'Burkina Faso', 'Bulgaria', 'Brazil', 'Botswana', 'Bosnia and Herzegovina', 'Bolivia', 'Benin', 'Belgium', 'Belarus', 'Barbados', 'Bangladesh', 'Azerbaijan', 'Austria', 'Australia', 'Armenia', 'Argentina', 'Angola', 'Algeria', 'Albania', 'Afghanistan'])",
"_____no_output_____"
]
],
[
[
"### Adding title and labeling axes",
"_____no_output_____"
]
],
[
[
"# plotting scatter chart\nplt.scatter(profit,sales)\n\n# Adding and formatting title\nplt.title(\"Sales Across Profit in various Counteries for different Product Categories\", fontdict={'fontsize': 20, 'fontweight' : 5, 'color' : 'Green'})\n\n# Labeling Axes\nplt.xlabel(\"Profit\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\nplt.ylabel(\"Sales\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Representing product categories using different colors",
"_____no_output_____"
]
],
[
[
"product_categories = np.array([\"Technology\", \"Furniture\", \"Office Supplies\"])\ncolors = np.array([\"cyan\", \"green\", \"yellow\"])\n\n# plotting the scatterplot with color coding the points belonging to different categories \nfor color,category in zip(colors,product_categories):\n sales_cat = sales[product_category==category]\n profit_cat = profit[product_category==category]\n plt.scatter(profit_cat,sales_cat,c=color,label=category)\n# Adding and formatting title\nplt.title(\"Sales Across Profit in various Counteries for different Product Categories\", fontdict={'fontsize': 20, 'fontweight' : 5, 'color' : 'Green'})\n\n# Labeling Axes\nplt.xlabel(\"Profit\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\nplt.ylabel(\"Sales\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\n\n\n# Adding legend for interpretation of points\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Adding labels to points belonging to specific country",
"_____no_output_____"
]
],
[
[
"# plotting the scatterplot with color coding the points belonging to different categories \nfor color,category in zip(colors,product_categories):\n sales_cat = sales[product_category==category]\n profit_cat = profit[product_category==category]\n plt.scatter(profit_cat,sales_cat,c=color,label=category)\n\n# labeling points that belong to country \"India\"\nfor xy in zip(profit[country == \"India\"],sales[country == \"India\"]):\n plt.annotate(text=\"India\",xy = xy)\n\n# Adding and formatting title\nplt.title(\"Sales Across Profit in various Counteries for different Product Categories\", fontdict={'fontsize': 20, 'fontweight' : 5, 'color' : 'Green'})\n\n# Labeling Axes\nplt.xlabel(\"Profit\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\nplt.ylabel(\"Sales\", fontdict={'fontsize': 10, 'fontweight' : 3, 'color' : 'Blue'})\n\n\n# Adding legend for interpretation of points\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Line Chart: Trend of sales over the 12 months",
"_____no_output_____"
],
[
"Can be used to present the trend with time variable on the x-axis\n\nIn some cases, can be used as an alternative to scatterplot to understand the relationship between 2 variables",
"_____no_output_____"
]
],
[
[
"# Sales data across months\nmonths = np.array(['January', 'February', 'March', 'April', 'May', 'June', 'July', 'August', 'September', 'October', 'November', 'December'])\nsales = np.array([241268.56, 184837.36, 263100.77, 242771.86, 288401.05, 401814.06, 258705.68, 456619.94, 481157.24, 422766.63, 555279.03, 503143.69])",
"_____no_output_____"
],
[
"# plotting a line chart\nplt.plot(months,sales,'bx')\n\n# adding title to the chart\nplt.title(\"Sales across months\")\n\n# labeling the axes\nplt.xlabel(\"Months\")\nplt.ylabel(\"Sales\")\n# rotating the tick values of x-axis\nplt.xticks(rotation = 90)\n\n# displating the created plot\nplt.show()",
"_____no_output_____"
],
[
"y = np.random.randint(1,100, 50)\nplt.plot(y, 'ro')\nplt.show()",
"_____no_output_____"
],
[
"# plotting a line chart\nplt.plot(months,sales,'b',marker='x')\n\n# adding title to the chart\nplt.title(\"Sales across months\")\n\n# labeling the axes\nplt.xlabel(\"Months\")\nplt.ylabel(\"Sales\")\n# rotating the tick values of x-axis\nplt.xticks(rotation = 90)\n\n# displating the created plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Histogram: Distibution of employees across different age groups",
"_____no_output_____"
],
[
"Useful in checking the distribution of data range\n\nBuilds a bar corresponding to each element in the data range showing its frequency",
"_____no_output_____"
]
],
[
[
"# data corresponding to age of the employees in the company\nage = np.array([23, 22, 24, 24, 23, 23, 22, 23, 24, 24, 24, 22, 24, 23, 24, 23, 22, 24, 23, 23, 22, 23, 23, 24, 23, 24, 23, 22, 24, 22, 23, 24, 23, 24, 22, 22, 24, 23, 22, 24, 24, 24, 23, 24, 24, 22, 23, 23, 24, 22, 22, 24, 22, 23, 22, 23, 22, 23, 23, 23, 23, 22, 22, 23, 23, 23, 23, 23, 23, 22, 29, 29, 27, 28, 28, 29, 28, 27, 26, 27, 28, 29, 26, 28, 26, 28, 27, 27, 28, 28, 26, 29, 28, 28, 26, 27, 26, 28, 27, 29, 29, 27, 27, 27, 28, 29, 29, 29, 27, 28, 28, 26, 28, 27, 26, 26, 27, 26, 29, 28, 28, 28, 29, 26, 26, 26, 29, 26, 28, 26, 28, 28, 27, 27, 27, 29, 27, 28, 27, 26, 29, 29, 27, 29, 26, 29, 26, 29, 29, 27, 28, 28, 27, 29, 26, 28, 26, 28, 27, 29, 29, 29, 27, 27, 29, 29, 26, 26, 26, 27, 28, 27, 28, 28, 29, 27, 26, 27, 29, 28, 29, 27, 27, 26, 26, 26, 26, 29, 28, 28, 33, 34, 33, 33, 34, 33, 31, 32, 33, 33, 32, 34, 32, 31, 33, 34, 31, 33, 34, 33, 34, 33, 32, 33, 31, 33, 32, 32, 31, 34, 33, 31, 34, 32, 32, 31, 32, 31, 32, 34, 33, 33, 31, 32, 32, 31, 32, 33, 34, 32, 34, 31, 32, 31, 33, 32, 34, 31, 32, 34, 31, 31, 34, 34, 34, 32, 34, 33, 33, 32, 32, 33, 31, 33, 31, 32, 34, 32, 32, 31, 34, 32, 32, 31, 32, 34, 32, 33, 31, 34, 31, 31, 32, 31, 33, 34, 34, 34, 31, 33, 34, 33, 34, 31, 34, 34, 33, 31, 32, 33, 31, 31, 33, 32, 34, 32, 34, 31, 31, 34, 32, 32, 31, 31, 32, 31, 31, 32, 33, 32, 31, 32, 32, 31, 31, 34, 31, 34, 33, 32, 31, 34, 34, 31, 34, 31, 32, 34, 33, 33, 34, 32, 33, 31, 31, 33, 32, 31, 31, 31, 37, 38, 37, 37, 36, 37, 36, 39, 37, 39, 37, 39, 38, 36, 37, 36, 38, 38, 36, 39, 39, 37, 39, 36, 37, 36, 36, 37, 38, 36, 38, 39, 39, 36, 38, 37, 39, 38, 39, 39, 36, 38, 37, 38, 39, 36, 37, 36, 36, 38, 38, 38, 39, 36, 37, 37, 39, 37, 37, 36, 36, 39, 37, 36, 36, 36, 39, 37, 37, 37, 37, 39, 36, 39, 37, 38, 37, 36, 36, 39, 39, 36, 36, 39, 39, 39, 37, 38, 36, 36, 37, 38, 37, 38, 37, 39, 39, 37, 39, 36, 36, 39, 39, 39, 36, 38, 39, 39, 39, 39, 38, 36, 37, 37, 38, 38, 39, 36, 37, 37, 39, 36, 37, 37, 36, 36, 36, 38, 39, 38, 36, 38, 36, 39, 38, 36, 36, 37, 39, 39, 37, 37, 37, 36, 37, 36, 36, 38, 38, 39, 36, 39, 36, 37, 37, 39, 39, 36, 38, 39, 39, 39, 37, 37, 37, 37, 39, 36, 37, 39, 38, 39, 36, 37, 38, 39, 38, 36, 37, 38, 42, 43, 44, 43, 41, 42, 41, 41, 42, 41, 43, 44, 43, 44, 44, 42, 43, 44, 43, 41, 44, 42, 43, 42, 42, 44, 43, 42, 41, 42, 41, 41, 41, 44, 44, 44, 41, 43, 42, 42, 43, 43, 44, 44, 44, 44, 44, 41, 42, 44, 43, 42, 42, 43, 44, 44, 44, 44, 41, 42, 43, 43, 43, 41, 43, 41, 42, 41, 42, 42, 41, 42, 44, 41, 43, 42, 41, 43, 41, 44, 44, 43, 43, 43, 41, 41, 41, 42, 43, 42, 48, 48, 48, 49, 47, 45, 46, 49, 46, 49, 49, 46, 47, 45, 47, 45, 47, 49, 47, 46, 46, 47, 45, 49, 49, 49, 45, 46, 47, 46, 45, 46, 45, 48, 48, 45, 49, 46, 48, 49, 47, 48, 45, 48, 46, 45, 48, 45, 46, 46, 48, 47, 46, 45, 48, 46, 49, 47, 46, 49, 48, 46, 47, 47, 46, 48, 47, 46, 46, 49, 50, 54, 53, 55, 51, 50, 51, 54, 54, 53, 53, 51, 51, 50, 54, 51, 51, 55, 50, 51, 50, 50, 53, 52, 54, 53, 55, 52, 52, 50, 52, 55, 54, 50, 50, 55, 52, 54, 52, 54])",
"_____no_output_____"
],
[
"# Checking the number of employees\nlen(age)",
"_____no_output_____"
],
[
"# plotting a histogram\nplt.hist(age)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Plotting a histogram with fixed number of bins",
"_____no_output_____"
]
],
[
[
"# plotting a histogram\nplt.hist(age,bins=5,color='green',edgecolor='black')\n\nplt.show()",
"_____no_output_____"
],
[
"list_1 = [48.49, 67.54, 57.47, 68.17, 51.18, 68.31, 50.33, 66.7, 45.62, 43.59, 53.64, 70.08, 47.69, 61.27, 44.14, 51.62, 48.72, 65.11]\nweights = np.array(list_1)",
"_____no_output_____"
],
[
"plt.hist(weights,bins = 4,range=[40,80],edgecolor='white')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Box plot: Understanding the spread of sales across different countries",
"_____no_output_____"
],
[
"Useful in understanding the spread of the data\n\nDivides the data based on the percentile values\n\nHelps identify the presence of outliers",
"_____no_output_____"
]
],
[
[
"# Creating arrays with sales in different countries across each category: 'Furniture', 'Technology' and 'Office Supplies'\nsales_technology = np.array ([1013.14, 8298.48, 875.51, 22320.83, 9251.6, 4516.86, 585.16, 174.2, 27557.79, 563.25, 558.11, 37117.45, 357.36, 2206.96, 709.5, 35064.03, 7230.78, 235.33, 148.32, 3973.27, 11737.8, 7104.63, 83.67, 5569.83, 92.34, 1045.62, 9072.51, 42485.82, 5093.82, 14846.16, 943.92, 684.36, 15012.03, 38196.18, 2448.75, 28881.96, 13912.14, 4507.2, 4931.06, 12805.05, 67912.73, 4492.2, 1740.01, 458.04, 16904.32, 21744.53, 10417.26, 18665.33, 2808.42, 54195.57, 67332.5, 24390.95, 1790.43, 2234.19, 9917.5, 7408.14, 36051.99, 1352.22, 1907.7, 2154.66, 1078.21, 3391.65, 28262.73, 5177.04, 66.51, 2031.34, 1683.72, 1970.01, 6515.82, 1055.31, 1029.48, 5303.4, 1850.96, 1159.41, 39989.13, 1183.87, 96365.09, 8356.68, 7010.24, 23119.23, 46109.28, 9058.95, 1313.67, 31525.06, 2019.94, 703.04, 1868.79, 700.5, 55512.02, 243.5, 2113.18, 11781.81, 3487.29, 513.12, 5000.7, 121.02, 1302.78, 169.92, 124.29, 57366.05, 29445.93, 4614.3, 45009.98, 309.24, 3353.67, 41348.34, 2280.27, 61193.7, 1466.79, 12419.94, 445.12, 25188.65, 12351.23, 1152.3, 26298.81, 9900.78, 5355.57, 2325.66, 6282.81, 1283.1, 3560.15, 3723.84, 13715.01, 4887.9, 3396.89, 33348.42, 625.02, 1665.48, 32486.97, 20516.22, 8651.16, 13590.06, 2440.35, 6462.57])\nsales_office_supplies = np.array ([1770.13, 7527.18, 1433.65, 423.3, 21601.72, 10035.72, 2378.49, 3062.38, 345.17, 30345.78, 300.71, 940.81, 36468.08, 1352.85, 1755.72, 2391.96, 19.98, 19792.8, 15633.88, 7.45, 521.67, 1118.24, 7231.68, 12399.32, 204.36, 23.64, 5916.48, 313.98, 9212.42, 27476.91, 1761.33, 289.5, 780.3, 15098.46, 813.27, 47.55, 8323.23, 22634.64, 1831.02, 28808.1, 10539.78, 588.99, 939.78, 7212.41, 15683.01, 41369.09, 5581.6, 403.36, 375.26, 12276.66, 15393.56, 76.65, 5884.38, 18005.49, 3094.71, 43642.78, 35554.83, 22977.11, 1026.33, 665.28, 9712.49, 6038.52, 30756.51, 3758.25, 4769.49, 2463.3, 967.11, 2311.74, 1414.83, 12764.91, 4191.24, 110.76, 637.34, 1195.12, 2271.63, 804.12, 196.17, 167.67, 131.77, 2842.05, 9969.12, 1784.35, 3098.49, 25005.54, 1300.1, 7920.54, 6471.78, 31707.57, 37636.47, 3980.88, 3339.39, 26563.9, 4038.73, 124.8, 196.65, 2797.77, 29832.76, 184.84, 79.08, 8047.83, 1726.98, 899.73, 224.06, 6101.31, 729.6, 896.07, 17.82, 26.22, 46429.78, 31167.27, 2455.94, 37714.3, 1506.93, 3812.78, 25223.34, 3795.96, 437.31, 41278.86, 2091.81, 6296.61, 468.82, 23629.64, 9725.46, 1317.03, 1225.26, 30034.08, 7893.45, 2036.07, 215.52, 3912.42, 82783.43, 253.14, 966.96, 3381.26, 164.07, 1984.23, 75.12, 25168.17, 3295.53, 991.12, 10772.1, 44.16, 1311.45, 35352.57, 20.49, 13471.06, 8171.16, 14075.67, 611.82, 3925.56])\nsales_furniture = np.array ([981.84, 10209.84, 156.56, 243.06, 21287.52, 7300.51, 434.52, 6065.0, 224.75, 28953.6, 757.98, 528.15, 34922.41, 50.58, 2918.48, 1044.96, 22195.13, 3951.48, 6977.64, 219.12, 5908.38, 10987.46, 4852.26, 445.5, 71860.82, 14840.45, 24712.08, 1329.9, 1180.44, 85.02, 10341.63, 690.48, 1939.53, 20010.51, 914.31, 25223.82, 12804.66, 2124.24, 602.82, 2961.66, 15740.79, 74138.35, 7759.39, 447.0, 2094.84, 22358.95, 21734.53, 4223.73, 17679.53, 1019.85, 51848.72, 69133.3, 30146.9, 705.48, 14508.88, 7489.38, 20269.44, 246.12, 668.13, 768.93, 899.16, 2578.2, 4107.99, 20334.57, 366.84, 3249.27, 98.88, 3497.88, 3853.05, 786.75, 1573.68, 458.36, 1234.77, 1094.22, 2300.61, 970.14, 3068.25, 35792.85, 4277.82, 71080.28, 3016.86, 3157.49, 15888.0, 30000.36, 1214.22, 1493.94, 32036.69, 4979.66, 106.02, 46257.68, 1033.3, 937.32, 3442.62, 213.15, 338.88, 9602.34, 2280.99, 73759.08, 23526.12, 6272.74, 43416.3, 576.78, 1471.61, 20844.9, 3497.7, 56382.38, 902.58, 6235.26, 48.91, 32684.24, 13370.38, 10595.28, 4555.14, 10084.38, 267.72, 1012.95, 4630.5, 364.32, 349.2, 4647.56, 504.0, 10343.52, 5202.66, 2786.26, 34135.95, 2654.58, 24699.51, 136.26, 23524.51, 8731.68, 8425.86, 835.95, 11285.19])",
"_____no_output_____"
],
[
"# plotting box plot for each category\nplt.boxplot([sales_technology,sales_office_supplies,sales_furniture])\n\n# adding title to the graph\nplt.title(\"Sales across country and product categories\")\n\n# labeling the axes\nplt.xlabel(\"Product Category\")\nplt.ylabel(\"Sales\")\n# Replacing the x ticks with respective category\nplt.xticks((1,2,3),[\"Technology\",\"Office_Supply\",\"Furniture\"])\n\nplt.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
cb6da0fba91297b3cdf01841cdadf947be01038a | 131,290 | ipynb | Jupyter Notebook | PythonCodes.ipynb | Shailyshaik2021/python | e4d59291288d9715306cf8a14fbd265cdba35828 | [
"Unlicense"
]
| null | null | null | PythonCodes.ipynb | Shailyshaik2021/python | e4d59291288d9715306cf8a14fbd265cdba35828 | [
"Unlicense"
]
| null | null | null | PythonCodes.ipynb | Shailyshaik2021/python | e4d59291288d9715306cf8a14fbd265cdba35828 | [
"Unlicense"
]
| null | null | null | 355.799458 | 86,165 | 0.922005 | [
[
[
"<a href=\"https://colab.research.google.com/github/Shailyshaik2021/python/blob/main/PythonCodes.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"<h1>Welcome to Colab!</h1>\n\nIf you're already familiar with Colab, check out this video to learn about interactive tables, the executed code history view, and the command palette.\n\n<center>\n <a href=\"https://www.youtube.com/watch?v=rNgswRZ2C1Y\" target=\"_blank\">\n <img alt='Thumbnail for a video showing 3 cool Google Colab features' src=\"data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAVIAAAC+CAYAAABnJIeiAAAMa2lDQ1BJQ0MgUHJvZmlsZQAASImVlwdYk1cXgO83kpCQsAJhyAh7CbKnjBBWBAGZgouQBBJGjAlBxI2WKli3iOBEqyKKVisgoiJqnUVxW0dRi0qlFrUoisp/M0Br//H853nud9+ce+45557cb1wAtAe4Ekk+qgNAgbhQmhgVxpyYnsEkdQNNYA7IgAQMuTyZhJWQEAugDPd/lzc3AaLor7kofP1z/L+KHl8g4wGATIacxZfxCiC3AYBv5EmkhQAQFXrrmYUSBS+ArC+FCUJeq+AcFe9WcJaKW5U2yYlsyFcA0KByudIcALTuQT2ziJcD/Wh9gOwm5ovEAGiPhhzME3L5kBW5jy4omK7gKsgO0F4CGeYD/LK+8JnzN/9ZI/653JwRVq1LKRrhIpkknzvr/yzN/5aCfPlwDDvYqEJpdKJi/bCGt/OmxyiYCrlXnBUXr6g15AERX1V3AFCKUB6dorJHTXkyNqwfYEB243PDYyCbQo4U58fFqvVZ2aJIDmS4W9BiUSEnGbIR5CUCWUSS2mardHqiOhbamC1ls9T6c1ypMq4i1gN5XgpL7f+VUMBR+8e0SoTJaZApkG2KRKlxkLUgu8rykmLUNmNLhOy4YRupPFGRvw3kRIE4KkzlHyvKlkYmqu3LC2TD68W2CkWcODUfLBQmR6vqg53mcZX5w7VgVwRiVsqwH4FsYuzwWviC8AjV2rFnAnFKktrPgKQwLFE1F6dI8hPU9riVID9KobeC7CUrSlLPxVML4eZU+cezJYUJyao88ZJc7rgEVT74ShAL2CAcMIEctiwwHeQCUUdvUy/8pRqJBFwgBTlAAFzUmuEZacoRMbwmgRLwByQBkI3MC1OOCkAR1H8c0aquLiBbOVqknJEHnkAuADEgH/6WK2eJR6Klgt+gRvSP6FzYeDDffNgU4/9eP6z9rGFBTaxaIx+OyNQetiRGEMOJ0cRIoiNuggfjgXgsvIbC5oH74f7D6/hsT3hC6CQ8ItwgdBHuTBOVSr/Kcjzogv4j1bXI+rIWuB306Y2H4UHQO/SMM3AT4IJ7wTgsPARG9oZatjpvRVWYX/n+2wq++DfUdmQ3Mko2JIeSHb6eqeWk5T3iRVHrL+ujyjVrpN7skZGv47O/qD4f9jFfW2JLsEPYWewkdh5rxZoAEzuBNWOXsGMKHtldvyl313C0RGU+edCP6B/xuOqYikrK3Ordetw+qMYKBcWFihuPPV0ySyrKERYyWfDtIGByxDzX0UwPNw93ABTvGtXj6zVD+Q5BGBc+60qPABDEGRoaav2sizkKwKFl8Pa/9VnnkKV6jp+r5smlRSodrrgQ4FNCG95pxvBdZg0c4Ho8gA8IBKEgAowD8SAZpIOpsMpCuM+lYCaYAxaCMlABVoJ1oBpsAdvBbrAPHARNoBWcBD+Bi+AKuAHuwt3TDZ6DPvAGDCIIQkJoCB0xRiwQW8QZ8UD8kGAkAolFEpF0JBPJQcSIHJmDLEIqkNVINbINqUN+QI4gJ5HzSCdyB3mI9CCvkPcohlJRfdQMtUPHoH4oC41Bk9EpaA46Ay1BF6PL0Sq0Ft2LNqIn0YvoDbQLfY72YwDTxBiYJeaC+WFsLB7LwLIxKTYPK8cqsVqsAWuB//M1rAvrxd7hRJyOM3EXuIOj8RSch8/A5+HL8Gp8N96In8av4Q/xPvwTgUYwJTgTAggcwkRCDmEmoYxQSdhJOEw4A++lbsIbIpHIINoTfeG9mE7MJc4mLiNuIu4nthE7iY+J/SQSyZjkTAoixZO4pEJSGWkDaS/pBOkqqZs0oKGpYaHhoRGpkaEh1ijVqNTYo3Fc46rGU41Bsg7ZlhxAjifzybPIK8g7yC3ky+Ru8iBFl2JPCaIkU3IpCylVlAbKGco9ymtNTU0rTX/NCZoizQWaVZoHNM9pPtR8R9WjOlHZ1MlUOXU5dRe1jXqH+ppGo9nRQmkZtELaclod7RTtAW1Ai67lqsXR4mvN16rRatS6qvVCm6xtq83Snqpdol2pfUj7snavDlnHToetw9WZp1Ojc0Tnlk6/Ll3XXTdet0B3me4e3fO6z/RIenZ6EXp8vcV62/VO6T2mY3RrOpvOoy+i76CfoXfrE/Xt9Tn6ufoV+vv0O/T7DPQMvAxSDYoNagyOGXQxMIYdg8PIZ6xgHGTcZLw3NDNkGQoMlxo2GF41fGs0yijUSGBUbrTf6IbRe2OmcYRxnvEq4ybj+ya4iZPJBJOZJptNzpj0jtIfFTiKN6p81MFRv5iipk6miaazTbebXjLtNzM3izKTmG0wO2XWa84wDzXPNV9rfty8x4JuEWwhslhrccLid6YBk8XMZ1YxTzP7LE0toy3lltssOywHreytUqxKrfZb3bemWPtZZ1uvtW637rOxsBlvM8em3uYXW7Ktn63Qdr3tWdu3dvZ2aXbf2jXZPbM3sufYl9jX299zoDmEOMxwqHW47kh09HPMc9zkeMUJdfJ2EjrVOF12Rp19nEXOm5w7RxNG+48Wj64dfcuF6sJyKXKpd3noynCNdS11bXJ9McZmTMaYVWPOjvnk5u2W77bD7a67nvs491L3FvdXHk4ePI8aj+ueNM9Iz/mezZ4vvZy9BF6bvW57073He3/r3e790cfXR+rT4NPja+Ob6bvR95afvl+C3zK/c/4E/zD/+f6t/u8CfAIKAw4G/BnoEpgXuCfw2Vj7sYKxO8Y+DrIK4gZtC+oKZgZnBm8N7gqxDOGG1IY8CrUO5YfuDH3KcmTlsvayXoS5hUnDDoe9ZQew57LbwrHwqPDy8I4IvYiUiOqIB5FWkTmR9ZF9Ud5Rs6PaognRMdGrom9xzDg8Th2nb5zvuLnjTsdQY5JiqmMexTrFSmNbxqPjx41fM/5enG2cOK4pHsRz4tfE30+wT5iRcHQCcULChJoJTxLdE+cknk2iJ01L2pP0JjkseUXy3RSHFHlKe6p26uTUutS3aeFpq9O6Jo6ZOHfixXSTdFF6cwYpIzVjZ0b/pIhJ6yZ1T/aeXDb55hT7KcVTzk81mZo/9dg07WncaYcyCZlpmXsyP3DjubXc/ixO1sasPh6bt573nB/KX8vvEQQJVgueZgdlr85+lhOUsyanRxgirBT2itiiatHL3OjcLblv8+LzduUN5afl7y/QKMgsOCLWE+eJT083n148vVPiLCmTdM0ImLFuRp80RrpThsimyJoL9eFH/SW5g/wb+cOi4KKaooGZqTMPFesWi4svzXKatXTW05LIku9n47N5s9vnWM5ZOOfhXNbcbfOQeVnz2udbz188v3tB1ILdCykL8xb+XOpWurr0r0Vpi1oWmy1esPjxN1Hf1JdplUnLbn0b+O2WJfgS0ZKOpZ5LNyz9VM4vv1DhVlFZ8WEZb9mF79y/q/puaHn28o4VPis2rySuFK+8uSpk1e7VuqtLVj9eM35N41rm2vK1f62btu58pVfllvWU9fL1XVWxVc0bbDas3PChWlh9oyasZv9G041LN77dxN90dXPo5oYtZlsqtrzfKtp6e1vUtsZau9rK7cTtRduf7EjdcfZ7v+/rdprsrNj5cZd4V9fuxN2n63zr6vaY7llRj9bL63v2Tt57ZV/4vuYGl4Zt+xn7Kw6AA/IDv/+Q+cPNgzEH2w/5HWr40fbHjYfph8sbkcZZjX1Nwqau5vTmziPjjrS3BLYcPup6dFerZWvNMYNjK45Tji8+PnSi5ER/m6St92TOycft09rvnpp46vrpCac7zsScOfdT5E+nzrLOnjgXdK71fMD5Ixf8LjRd9LnYeMn70uGfvX8+3OHT0XjZ93LzFf8rLZ1jO49fDbl68lr4tZ+uc65fvBF3o/Nmys3btybf6rrNv/3sTv6dl78U/TJ4d8E9wr3y+zr3Kx+YPqj91fHX/V0+Xccehj+89Cjp0d3HvMfPf5P99qF78RPak8qnFk/rnnk8a+2J7Lny+6Tfu59Lng/2lv2h+8fGFw4vfvwz9M9LfRP7ul9KXw69Wvba+PWuv7z+au9P6H/wpuDN4NvyAeOB3e/83p19n/b+6eDMD6QPVR8dP7Z8ivl0b6hgaEjClXKVnwIYbGh2NgCvdgFASweADs9tlEmqs6BSENX5VUngP7HqvKgUHwAaYKf4jGe3AXAANrs25VEFKD7hk0MB6uk50tQiy/b0UPmiwpMQYWBo6LUZAKQWAD5Kh4YGNw0NfdwBk70DQNsM1RlUIUR4ZtjqpaCrjOIF4CtRnU+/WOPXPVBkoJz+t/5fpM6PWp0rMUkAAAA4ZVhJZk1NACoAAAAIAAGHaQAEAAAAAQAAABoAAAAAAAKgAgAEAAAAAQAAAVKgAwAEAAAAAQAAAL4AAAAAvqIx7QAAQABJREFUeAHsvWeUXcd171m3c05oNBoZIAIBEswEJYpRFCmJFBUt07L8LMu2xmFsz3jeevNmzZq1Zr7Nh/kw857X8rOXbMt+tpIlipIoiZSYRZFizgkEkTPQOefu+f92nX3vubdvNxpgA6Sk3sDtc06dOnXq1Dn1r51qV2b7FTd1zWSmWzIzmZGwREstoBaYmpysblveEtavaQ8TE5OhvLws9PSPhP0HDoSQKf2V/054vtKyspFV7a3VlZXVYf/Bw/ZM45MT1R/csXJk7erl1UODI7/yz3muPubJycls0WVlZbafTsue/DXYmZienvcpSmZmqktLS0fUCjPV5JzJxO28Vy2d/I1ogakwExoaGg1EJyYnAr+66vJQWVkVBoaHq8vLy/lifoXaIhNKVNvxsTGr8/LWZaG5sba6f3AkHDx0lLTqyqoqDSAzoaSkJPaHkoxtS/SYmZKMXbeQPzMzuoCmSV0yM51rq5mkrEzSQTMlqhmXzNBhuSiXt9j9KD+TSRWeZLL7UoLO+X6x60+X5lUt9sicm5mZsmpmMqVW1JSeg7w8B+dI59jLSd/Pz7OlTbxducbKVWbfT58v3E+Xmd6fVpklujnlpts8nYd9zk9NRoAkf5oow2lK76Q0w5dTnDiv1gjT05kQh5Pi+ZZSf0NbwDr59Lg40WoDUZoBrhQaHR3TvjqL/k2fptPbBe/pnwigYxMTYVpcVEtzU1jR1hbGxsRdHzoWhkdGQ011lTpWaZgCoKxbxApPq5OUqBN5Z/fHmAukHNwMyArapaRUIKMOmgNLAZKAJ5PqtLF8OjEdO9eZ/b5s57q354l1mPt6zzfftgBX8rLauQRA01Wfmo4A6uDKRYXAyDkHSc6n29Xz+nUOgun8dg2AGybtWs9DupODYrFznoftQkCUfA6iXm4aZP08YAotAak1w9IfbwF1b9s9crwj1NXWCmiMMQtHT3SEpsY6pVWHUx3dYXJ6IlRUVgpMoeId3069R394jhl18JGx8VBTUxPa16wMpaVl4djxE6Gvvz9UVFSGuro6ASjDQXyKd1NVQM7B1MuJHVrppZF7iyDpZ+M21+nzOaP8XPGosPx0nhINBgA1v4wGgAja6RyLu++AC6AWAuhU6nNIg+SZ1MC4SrhfkYNuuiw7n0bzJF+uPe3Son8ARkCxEBiLZlbifPkAW76eJSCdq/V+U9MFCADk8PBIeH3X3tDYUB/GxdF19/SGqsoKcXStYcP6NaG3ry90dHaHEunIqioqjKN7fwBq5MhGpOKkvuvXrjLQpL7d3b32Vqs1OACfzk1EgEv1fuWCGy1GaTBLg2cxbtE5RD9nnT9VKOcd+PIZURfP8+uUulS7+ZxnvIfEWpUp/tBUGfn5T39k1ybg5EBZ7Ko0fjm4pcG0+DWqUyb3POn8XoZflwZJB9H0Od9/N1sHU8pwjpP9+UCT84XEN8SzLAFpYcssHSOXGZgiEp841RFKJYYCPlNT0zLMHDFwbV/RFurrag1M+/oHQlVVdSgrE1cknZlURuednJMGQKlve9vyUF9fpwFhWFzoEVNRVEsPqq/eQPRcV9BAUmAdwVIAojZxvjdPbTAtjiZpL8CLPAbhOcwpUtW5GjheZPzvu9SV+k0BzTSocgzwpYHQ8863zUgk12e1YEqD6UIvWgg3mi4rDaDp9LPZXwLSs2m1X+NrvFPDXZpRyQxLdHBE1BITh0dHx8M7e/ebznHNyuXaNoaTpzoNtOACSzRCk/98EeLVxHg0iqEHXdbSHMbHx8LhI0fDqER76lQmfW+s05nVy7lJniULism+HSfsWZpzIt2J6x1APc10rxpwTE8KSpFHnI1kWMti+Q29BPqki0pnYpledrpeliFp7ynVpzSNfPFkXt2TJNukgZIy05e67tPzcwyVSo0AGaiKFwMkz4QA4UIu1K/3cwbUJdEo5efe7RZusxh4prnQ3PevAeQ0n0pOolkS7d/tu/k1v77wS0IcnhEoldmvr68/8GtfsTysXbPagPTEyQ61yWQok3ogUmEZi91kGbsvelBUDtCJk6fCwOBQKC8rN/2occlnCOyliMh61jQBNA5knp4GUNLSAJfNW1CO5cPYpOKnsd4LveZSJWTTDVCjyJ8t1yuR2gKirh/1fNQpXa9UdgM02aazAJoGVsDMgRW9a01NlTw5ZLibioBqnKkKK5PuGZYbC74T3GFh2/i5+baUybUYlRZCZ8K5FgPRhdyjMA8AyuDtxiieeokjLWylpePTt0ACDNHYNB0OHz0eagVk+J5esHF96OruMZ0qIjZ5jHRNerQ//U1OnwNxHlHerfGdXZ2mauC+ACsAyu9sKA2i6RLcbJQu0/MCvsWouLa1WM5cmov/uZSo/4wgWfw+nhcAnQ88PR9bwHJ6SmI3AAyKQgJGvDQyUuWMyU2oulIDp3Thhw8fDmPDvaGsoipUlJVoW5dkL1V710n9I9WJCJ9SJIK0ddwBz7YF7k920Xv4B4B1rhQu9HTfKQDqYOp5l4D0PXyBv6q39o8H8R+NHtbvycmpcPDwMelPGwSoy2ThrzdQgzPEQl4qtUDMvzhPDYhOarIAelpcmpgsgBgfDUlycQHs58ebBVUkDaJc4KBZeHExEDUukCZK14NjiDR+fkzaHBR9TmMhcIZQBEv2ihfgYEqONHl6mvMEJMelGgFQmqSmgZA0IM1YCN1dXeHIvrdDX3dHaABg65vtnP8ZHJ0MdVUC3+raUN/YEOobmqVOqQpVCbDivzvOxI7Ee8HFdr8+vfVzDuoOwOk8vs+5c0FzifTO0Tro5vrBEkd6Lt7Db1iZUdwHKAGxwcFBcy9a3toikb9N3OJYOH6yyzjHSnUsAHCx9KfT0tmhn4UTBUSdC80DrgW/jdgpp2T8gWZzhAsrCKBK0yyROv909IlMX5DsZ0V6HRf6nObKLA6iufOzC/ZzYBA6z/LyCgPlEqFCmRDk4L7d4YR0y4dO9YeWhprQTPsePRB6+obC+tZGA1EAdWQ4Tm7wO3QKKKfH4rBTUlnChIfQunpDWLV2fVi2rDXLpXr+ubYu2vt5B1Q/9i0g6ty0p7GdD3jT+XzfQdGP59um88KVpgfZJY50vpZbOrfgFgAcGcld3MfVqKu3P6wUd7pJ4n5PT4841C4rr0LuUuoFQogIBOmRfcE3TDJqep64qUmz1J/ptQvJP9tpPnKDgHUhaBaW56CVTdfjprkoO680Nw6lud1CIDfRWwX5jCjtZYst3HGuk/TCOk4ixosdZSZXnTjIiYmpcOrkyXBSng3vvPlG6OqZCpMqukpANT1UEjqVDkACjgc7+8L00Z4wOS6vjvrSUCXudFQAWkhDw9NhaHggHDn+mpUJqF546eWhbeX6MK7ZY+iFMUimQbNE6oT0cWGZhcc8g+tR0+A5F/D69biGQbq7J+VtC7nOvJNFDmb08SIkLAFpkcZZSjq7FoiAyAcq7sZ0o9Ph6PGToRtAXdGa1Z92dnWHSon7GK0cTM/ujkH+rAOmSsDJHst9eYWmrxpAF+8op7tPaUmaz8jlLgSk3JnT7BXgXV5HLziXLinNkabT4U5d9IzjUPHnTIOpX49FH3Lx/e23Xg9HDh8PU8M9Ar0BO7e6rTJU11QaxwlIjg9MhYlSVVTgmCUdTwCWYTyblN6pliIZ4IUA4a5TA+HpXzwZtlzUG7Zuu9h0kGMa/DRBzsiAMHG+jynn5q+DKKWzD5g6cBbekfQ0B1p4nuOof48PUbp85fr/Q2UyeXqJllpgkVsgEyrkPoX/KeDJFpG/uakhjGiqKYYiZuTYfPMzvDMWb7jRAakS6uswdLWGvoFBTWEdFUCXmwqhOMTMdSOpKKTnXb2iRXre6oAPLWDEfWyLLAyxKQKAaaA1sOTmRfI5l2r5k7KAKK8rnCjnotdA3NfpPDLtga5V7ZTuV+Zl0VgS02lf8gCiGINqaqrNVe2hhx4Nzzy3K5RNDYbqqoqwpqU2VNaUS+8sjlS/4cHx0D+sMsoyAWAUIynxXHgKR6mtmir7oybCdzs/PiE+USNqVaV+Giin1IAZlQF1d3SEnt6+0CqddpN0qVZ12lgsnYFS8UeJXPwc56zg+c5ZhvhnWm2Sfk8R6lMZtAt4eh6aEEBNmjI/o47gibER6BEml4B0VvMsJSxmC/CpAZQA6rCMDr2aIVUhvVzrshYDQBzmxwSqnAcYFtgnFGRHH7K+YLjarq4eu37VynZxumWhX4aSKbnoYEShTMAwccOc99EA0rXtzQakk+Juuc4JYKIs/p+WyFMsn9KoMz3P6q9scWZMLjMA6kclyT7H6V/+/a2FC26oMrhWv0ndiDGgvr4mjCq2wOuvvBQeeuTJMDEwEFobIjfVvmp16B8atGIdREcElPV1vDe5l0mU79dvRAAJUzsjBJpSmYCrcETow6XUMAIPW8T7CVn8uR6uFDBle+JkXzgpnXZDbak433q9J3G9yme2SC4sQgZsqXdRJMuCkmhPquu/YhcBmmkgnQtE7Vo1rIoUZZaAtFhjLqUtfgvQD+AgmVLap+maWIXhjtpXtMsPscSmnMKVcN475cJqAZiWWnmDgwPidhvlkN+iDj9jaYjIJbrvQvphGkhRE7iFPFePBEyTBJvemQBIej+XP9mLGGMHhfXwTuvXZME7dY2fK7bl+sIyvP3w66xQ29Q1tYdDB94J9979w7DnwMmwvD4TGuA+dR795djIsBXd2LI8DEpF0i8UXdZYYeePdStegThQAKWqQtcJGCsl2vOrEjiWANQJt86rgzNVsZYGdyrG3kT5kZHo/sa5rt6hcOzgIaHxcKhvaQ01VYrZwIk5yJ6vsOHmyFssOc1lFjvvaYj7GT2ot+e8IKqLcoPzEpB6Gy5tz2MLwH3S+bpkkIIjxfLeumyZgd+gxHMs+4AusLUQojMzC4u+eKqj0yzEiPpwvUNSH8Rpo6VmkJqvPxYC6envna5fMUBLSkhlm+/+5IbDQWdYCJB+7J3c8ib54iiRLjk6xyPKV9ctD8//8oFw/89+KYNIRgBZatwicnnLsroAePaKo9/YtiJ09HQqOI3ULTWanTYzGQBRqEp1am8pDctqpQHUdRmxopUAarU4Vt0c3TgA6ioAB1O4V36oAXjfnl6h/QFxuCe6esPMxEho0ky0xqYmYEz59RLTj0IFCo9JOwNKtxmXpYGVfQCTHyAK+fF8Yr3ly77XJSC1hlv6c35bwD5XIUaVDFL4guLADzG1E/9TAqYMCWArNHcfjjJ+3nPXEc6APKgQKJOIT6gQEO3bV6wwcX9oaFgAO2FpcwF0GkjHVQaghghLuXgl2H2SY06SZvpdQz9mQqmTqh78hBU5Su2zy8/ETK4DadgmRLpb89MAkN73vPlb1S+pJ4BQ31BrvrsP/fRH4Zcv7A7NQsPlzRVhXOwlQLh8ZUNY07wsHDp61IxLYWoivH2wT6CnmWt6gq4hZu+E0CBd54plMgxKasD4VE1c2mr5jFYoUIfS0IEivk+gG9WjAKblcKX2SJFb5REdSNNgOqML+rr7w9jA8ZBBtSPLPfEa7Fl1zbkigHLavitUSfFroH7cl6HD37NOGc0FqJZPo7cklyXR/ly9rKVyT98CfMTMQoKbHBbQdQpQAT+4SeKEDirw8phmyGA8ivCTA5y5SrcyTYUQxf1+cbiNApVl4k4R9wd0TP+o0H3y+6o6kawpriPFQZ2ORWdh6z/K55+dS+1TH2XL/dIVLFZtbq6f38OzA6Skm042hcbkK0bRC0A1AkTFzVXLG6K6utK8JR555Odh954joVlgWFtTYqI6uspSWdQxMJ1EIpBRiaIPnZINXmAIwUGiPa1prBQXGvWopNfXV5shqksupAN94zLIyaJv4rtAVOK/G5oiiAqsNKIYMAuREfvJA5DC2JKH+wCynUPj4fiBw2Gk95i8PSpU/3qpYwTcGA65cJHIuU+K4x1aQ+svLevtyyAYVTq54RbgLUZ8G7CvS0BarHWW0s5rCxhQ6UPV0h+aGy/9qSJJDQwMhUbFPl0hbhKuZ1CGEeZ3lwkg+eDRI9o3zJ95CBUCgITHANMWAWi43hEZXVApEHwDII+dan4g9duQt5T789O13gH9vG8N96yHesrsLXWzoM+pU/ZI+lNYbuRyZz8w4rABgm5IOEPosYcfDg88+qym6Q5KB6opuuLyJsSJwjliSUc3Otw/Frrl2gSgjYxNaUKDHkiED6mwLixrFscpF6WJ8anQ0FRjU0ZxkTrRNymPi6lQKZAHFLluCIDU5WUCTICTY1i7CulUywTcEaBj3QFX7gmwA1CAacYAV1b9Xk3e0GSA1Wtadc9lGlyZFVeRJ4pTx7MluFA1bPK+U6XYN4BaIbZBGmA9FwOVX2ucaHyc5D0tifbeTkvb97AF7MPU/dnCfcpXO3QoePSIOBJcpTAeTajH9QtQ3XhEdck/H9EtALtKdUY4zA5NdQRc25c3h9raGgNsON4KBTfBmj4pUXNte6NZ7SNHqs6uewCe/o/7USZ6XMDSRXS2DvCkz0tWMfplzOjdl2sKL7WBQ/nIw4/7AKpJEdqL5eClUCP/T0D0xTcOmoN4vUBsWNM3pwV8jYmFHtEeKzxiOKAGPsApwhkCohCcKMBLnsYGGZ3EeR48OhwGlQ99aVut9KNiFAdHpQYQeNYkgElZPAGgSlm4SQGk/HCbKlPdq9VI0wJhOFmorE7qHQG506jq1C/pZIX8juvqGmTRlzpGg+likPOYqGFpejhPfwex/Fgn50jt3Stf/M6iFBJbvrA2S0Ba2CJLx++LFojg5/pT/E+Zvw+osjxIvv+pvvTTEN0D4xUqBAI892iCAB4Dy1UmU1vhgslDB3MgZdE/QCEN1lFfywwu71Rc452PHpdU5HRV0nkHSK7wy9jnUj+X7uSeByD1fb8NkxAA0RefezY8LX0osKOYIkIycYUCOjhDuFEIEIUb5BhDEfpSfEXhDLmE/IAt4IflvksiPFwoFvpl1fo1VxqH2t0/bnlwkSpTG1AM3CnNAbga2tsdI+dKuYBpXCpJg4HqZkCuaarci/v7jKqTsuqXzYyFzVu22CSLdDtQZNpYZLco8iedx0V6BkNr20Qfmr4sFzHLQdtbOS9X+iC1/z4HUkY2+7BU5fQHnXqC981uuq6/KnV+3zRekYrA3eG2BPgBnDhy43+KQ3+13GUwHhFhCF0n7X2674PyIMrDeIDHAOI+7lJ4DBBYo0cuWResbTWOdBSHfLFYdD4nOiHH3tXS6ZbNX7xOCCcsiaudi2SbJuvUKWDknBVRkA8VgNfCt+S1svUsWOYP7Hk1PPjwswZoFUkFa6QbhUvEADQjkRsQhXB9gsvDzQk1BRwid6ZdILboMTkPOArnQqNAtKYuqg4GNc9+YFD6WKXXKg3DU01VqQBb3Kws+Tjjs0VSzgKm6gGYIvZPqHHoLwC8ccMAvvbhYJkrxf2OneoLy1uqQvvK1QLT/BlUhcBqlS74w709H/sQ30guLb4hP46tmTtPejyXXByLmOPvOQZSxB9GcdU/+yHMUZNZyRbrTy0wrkZEbIHdNh3ZWZQ1q/AFJHD/hdbb8qrh4aDonLw4QMA5mAXc7rxnoc6Ru1nIh3Leq2cfPTXjh2+p+58ODo0I6KLxKMtNqrdG/9O560knsh9Z9Ozl6vX4irrHAPrTcukSV7TIWCOgntQ3l4OvWK4fUw5fR66zJfdVunfMdKv6vgFpcpA35z65nA3fnBH5/Efdk+T0BjAiRuiRQ/vC93/wkJ2qE1ghQvNDNwm3h5sSQAr32STZHJ3oiOR0QBRiC8Pqv2oBIfPpSWemJ79aAZ1f16sZT8018gKQ5Z9we/wQ/+FkuQ+WffSwcL1TOo4ifwLoajwgDMke7rRKeRxsHUx1OwNU3vWWTRt0vQxpZDoDog/6L3uZV8QSlCG79f3c+0tOZt9xGlRzKgG/7pwAqV4AAQnUXL29A2FQriyIY1jfZltKvbq5LR18WqJcV0+f9Dtj2VESl5a+PvkY8mLVCfwRclcuzh4dhA7T0aHgDBI76iUCpoNJpO9CXalHr0TFfhlIqBsvfEyA2t8/aJ2xUu44Md+5qnG6RqffTz8fsizuQryrVBc+fSHvUQ43HrF+FNGe2rSsMsajMQ22GI9y1v2FVdDFfSz5TBBo0NIkq1fUm7sU+lJTes5RFO80jxIQRdSHg1QPzJ4GA2xwVUoaQMlAB01/GXaVJ3CQFONJXANhoWchwl71k3/91r0mUjdKtzkqGXlC4AWQYnDH33NiRAONAHZ5nTh8bWuUVlelgZS+BJCJA+U8YMkx7kvmJ6p9tlUNuJRFPSb5qpUGdwp4dp8cDkMyXI2Jex2SbG4We9Whe2A6DIibnVQ9MrrUDE4Jp8vyxfiS8tmhE81oNlqJ2o12ghtGPUDdBgZGQp0Au619lXlUxCdf2F/aO02Uq6FE78Gn3RZkSDLb+8jmyZXgon9M4dr09ZnJxfMtsIKlexEnRgzKoZGxsPWCdWH5ssbA+uH75N7QIbBZvqzZPir+WKPn6mqA0yt91ZiWstixfVO47OKtYf2adn18pdZR3nx7X3jmhdc0V7grtCqIMKCwWCHZYjUiV8m0w499+IOhQz5uLKlRKXeS7Bed1JfnxN+xt38oXLBhdbjm8u1h3ZpVCaczHvYeOBKef+Utxeg8LmdjdU6JlPpOROkXkBR2HjfMIce6y/OdkEHn4KGjScSm97ZeC2kC89uUSEq4vlGBZ3a5k9WrLboU60sRqm+hRHlwdb4e1QHFU924qi60NNWGEakNIF/iI5aZaiMHuKTHCi4SrinmYZKAk2GGDrNz8DlBNtIKerx9I0qnXvF78VLilimklEPQl/blK8Jjj79gZSDOT8qVCeGbMUB8S6iHixSI1UsGb5W+s0FtV1EXV4UdV5/sF8NCGsT+sDjKYiT/fLVRzMf54XHl1b0gyuYecLAYkkYEpj0CUP23ZyyD9RT4ojc1zwBxu06mSxXoT4wRvQujlJ9RHtJ1ePidty0UX0N9o7lDlUr5CycbATHWqXDeGjrRNLbAQcZhQE2eDIBs88Ex3tvTfOs1Ot02W/XTZTz9+RhoARAd14j0u5/9mNjy9dnL8BO8+8cPhwMClgimtHSOGOEB0TI16Bd/986wft0aOwmngPW2Rbqsa6+5PFx12fbwk4eeNGCOAEe23MvJ7Xv5nCvc9/yk+3l0X3rZektVGq251zt7D4aXX9+VACn3iURdfbD4zO03hssvvchO8Ixd3d2hQYGOL9EgwO/lV98Mjz/1sgXBQD+XT14PT6XrFKb5ufTW65xO831/pnQ58fl5vkkBULmix/N8u97eExicCKmmryopwMv26xdaJ7//ud5G/pm2LNWA0NUzYNzk1k3rLL4m4fti5Cl/nvnrEzucuBQBSqWCES+UAEAX47km17G93fJLAhizXKr27XredfHsWSBwiRYODQJE4Wora5aFp597Prz+2stmoRcOCkDFiSagVS4govBlAsB2cY/1YmgqhVTlkkCMxMlvlOkdFYfTwMCwQE3cpdRTY+L4K+XT6cQx5ADs6b51EAZkRzSLAWAd0WACOFKXMoE69QF0SUM3CqFPBWBH9Vx1Oo8hjHn6cKq02cGOsdC++81wyVXXSn0jSVTLgKcbjXbXpcbJUh4gCjlgsu+gmE4jfTFpUYEULgEQ/YPP3xZWrV5rIPLqW3vD6vbWcOOHdoYv3fWJ8NV/vcespLVaM12PaM8CMCGaMZPly1/4pDlPP/fim+G5l1+XUQFH4SnpW0rD2tUrw+c+8eHwsZs/GP7x6yckTowZyHE9wR9GBGSTakjAuFruLTGdsci/VmZrlOolSok+OpLKq1kgKsOXpfBwY8Ua2sBWzzmgEf33Pn+7DRYA7qNPPGfP5XVt1pS3Oz96Xbj8km3iTN+WsaTXDB2xzKj+oL1Gpb6AyvWRVFVrap729Wnor9eZAV3OyXPmjW1IGVxD3iEZAyYm44dfJ9Bktgj6QHzhMiVMpYzjM0ElIDo1z8XR6Mi4XUsbwvVgEZ6yEGe5+thF7/mfGFC6VtbqMQ20Bw6fCGv0ffRJQoi83Lmvbz43mdwvfdv0q0naK4qO8YSDMYABF+lgmW7awjRAFIf7R+Tm9NwLL5n+MmEq4ypHgJbAdIWmgtZoBhIc58p1q0JLS5NcjuI3Ua04pJWSuhTrLn2r0NYW0XpcfQP1FEQ+9pkVxvUYfkbFQQ6qXxYSYOtA68DaJbWCupsRRi8IkDWLUjy0+sKBGseqr9AmD+jbHKIvK+srr+wLTcuXhws2b5fXxajUEDIw0l/V522AUjlmmaex1D4EfM57DSaqJyNRck82XkYqqSgA+/li+f3cIgKpnlhUK+6msqo2vPbG7vCtex6QVa8mvPTKm5rD2xm+9DufDJds3xwefPxZ+YjpUfWAEDpIgOm37rzFQPS+B34eHnzs6dCmRdUAgmr5rU0LTN/YtUcvUHNz5aw9oReLSMaHCSeLu8oq8ktvRJ6THV1q8JLQpHXZo44zgmi3dErQiuXLLC8zXzo05xdupKGuSi9g1M7P9Yf79Un3e8MHLzMQBfDvvveB0NBQZzo26orC/ZgWgfv2PT+zZTiIGs9yHIB9BCzpYDW/ubqyXIPMcg0S5XKe7tPMnl59/DX6kCqVVxZVfRBsOVelvOtWtZu7DgYS/CybZHSBm2TNHT4uwLJbz1NfXxtWtrUbh3b0BIvR6bvVB1aW4j7Sz4cFl5VBe3VtS4umDja1avCa0DvrUHDfYXM7iuDOxxjfc/r693Kfd4vTdjRKTpp+k1iXarp3R3qHegEFZXDM86fTpRNN0M48A07TPCbeU4K+I6MkP5bqwqaFW0YXar6znk/AVlLeFI4dO2SXE4AZbg4CdKANTZELZd9BlH0H0AotnT0XleshymoaQoVxfjGXlmgyAmAh4hcQTcqt6QArVCfmaEIMTKv24WoHuvr0PedUBnCrENypeKAs+ewntn3yTZWvlfxLdVoO+rhEjWmUeeb518OatReo7zeGIfUngDNpwahXTdqAQgkQvRByTjWdNxc4W+X4DZIMRfMboKvN0oW82330gPj8fe2bP9TDlYhDaDcLKOWyXC9Up4ZOE67NxJFcv3alicIHDx0JP//lSxYEGE4K8IHQW65VeXB2HQISABK9Cnq+ZdJBfkGqBKYBOnVpNsu3fviAARaqBPUK22+XbvV3PvNRzZxRPMSEWHXyu/c+YoBaIyCej+AMAUU4bNQOP3nwcfk4NtvaQeOy9BpXqA7IfYbEbVLfRtV1EtlLr56/3dK9XrR1Y/j07R82i6ffD1H7ez95zMQXrgEQCMKx88pLw+0f+VBeXlQG9z7wRKiV7Me6ReQlgDIA/5GbrvUiFWKuO3z7+z8Td3yD6tIffnD/4zZgeAYGG+rJbKJPffT6rJqC87gEfffeh8NerWVPG/q78Gvf+2380pmXv6KN7iv9nfTWZyLaL8YzONgBfETUz4LkAgp3rvR0WZEiwIq6mhjJ6eTJLuNGDYRSF2NN37IehkKgpvdngCdOEhCt1yBdLkvSTCZfxTSZAs0JoUe5bOayJ4k5iX3Pi6+Q1GSkLjKmSQwsUwKXGj0gIrdbhae+qFzgiiqhQgNx0DJQnVqqRLyoPqqkTDHDPbL+m57UrlAfV79BohyU8aoOw5fqAESzPdk5GJ578a1w9RXbktxRnDd9aAKiAN18XKOfzxbwLne4l9OiAinKe1xS4HzKJYpPjk3I2BIB9LO332z3fFtisE3LEycB8dHBTa5b3WbHT2nkgYslrmQEJktmmDGRPKoEYhqcJFzoV/7Dp1VQSXjq2ZeNi4LLu/aay8Jf/tEXwj/+2/cNeLlig8Aarpj7Pf7k8+GUuMKY9/Lwp1/6bPinb95rhqz6VSuSm+Zv4CaHxe1edOEFBmrPv/yGZWCtoHFxhXCQqCjgrgEonhMC7NknyhGc6NWXXhju+OhNBsRPPveG9FEjYfOGNTaQ/LGczr9x9/0GbnC+N193lQEjgIihbVicI0Y49LK0xbcEkiwuhw/kLddfHW687mpbjvjZl3ZZrMcbP3h5+Is//h1rn5GRPZZmlUr+0BZ4Sdz1yVvCtgs3m174FUkTrAp56w1Xht/77TvCN757n0TnY1b/9wuY8i4wnE2L8wFEW7Uu0AGFZnPuMP2MZ7U/ixv1UtSr56A8PegceUjOVwnkZywEVrhRCKxokATSeeJA+NZ3fmggWgIbK2MOXBvcKEC0tkWxXlsaQ01tXOGzTICHeA4XWia1Tux1uXsCoqQ7ZWYiIHIM6BYS4JoFVJ2kfMR+B9V0fkAVnSxqAC33FMZLo4ELsZ8AJUqRZT9e4Vwpz8HzRDE/PpeXuWfvgbBt6xqTRKkZIGpW/gRIyVeMa0ynLzaYUja0qECq3iqOixBb5cZlXnbx9nDdzottDnWNONHv/+SR8MrrssKtXGHiKEohdy3ClQVCLEfkjRycJdmfnBUuWlr9PHpIQPTv/vm76khHTcTmHi+99nb4HwUgn73jpvDVr//ARNvP3/kRtfR0+LuvfUeA1i2jUpVGuTfCG2/vD1/5/c+GOz5ybfhvX7tbaoMoquTuHvcAfWI8LhPYQSdO9Ui0lgLcBoWom1yjZ8NTQWNIHgGWu/YcMM4OEIUL/vt/ucfy4If39HOvhpu0rDHnbrx2p6kLLpbnAtwlXPo/f+vH2bzU+RO3XW+gefO1V4orfkIf2EY7Ju8/6nnhEgDIF6VWYfDA8Oc60XTFesTd3yadMyDKQPSdH/xM3HqjzUd/6dU3wn/+qy+Lm71eZX7fVAdICbl3kS7p/OyjA0aVgTscA1j7mpUavMvCgUOHTbzH3Yxv8N0S7jhpjoPyiHSUpSKiv4YkE8PJA5fqJGHcd1Nb6XgFGAClD07sx7on99FCfIzFGJx8qePHn3jSyjAxHou4yPa1xWreqviiTs2ydAtd7RCwBCSN41S+mUzkLqvE8Ewl+nQyEvMAIoDLfJQGU8/noMpxPrBKBaVyWZe0QUxGkO6WIKczqtqIdjEs+TNwbXqfY0gaM0lUPaaCYjE9ZqjZt5AC0fm40VjK7L8WA1ffz1wAzBWsy6QWEczofaQ+AUT/eJ3UZrOLfjcpubvQiYWH0bk50TY3yPXCHaER1fHXVFWK3jD3Gc4+zQMRxHfD2lW2UiXcJSC6Yf1qA7Wy0qawd/8R4zrh0FADVMkCCZijfwVEURMAxnCJGIvQde688iIrA65yPgL4IFZhhEyUE5iiC922+co88dgy6A9iMqCGmxR0/8NP2XaVDHEYt/BhfOr518IHrrokXK16oDLYvmWT5fnJg08ah4u6gLyoHx554vlw2Y6t4erLL9IA9XDYpGeHHvz5cwYsnhf96kPSSQOk5TZv0LJl/8DNXrwt3uexp140vXSDys9kms116/mXXjcLPzOKDh89ae3luu1sIedlR76Ouo8ZJVXn9Xr31J0O1dHZrWcT56WB0QyGfPfnmubgWAE9wG8+ovNZ/1cZacAtdo2Xh4THFNB9++ROJ04UvSj6UQgAhTa2R+MROsvlrepr4ijBWuc4DSQFkIAoAWKgyYnct+4gSjr7Dqa+Dwg7jafA19NimMIcd+vpvoX9SIPpsNRkZtFXHc1disEpRWlOm+RS6Ur37t5lcWZZ2hmO9HTE9wBeFCMbKHUqj0tNDYB+TUbo6clgqoMp18XyFeDFMy/OFusvo2qwDvfy67vDL55+xcRJF1FrZCDC2GR6S4FP5DCmTX9HHSok0o+IncMY5dxqrJs3GiKzxDqJuM3NcX3tw8dOGSdKPkZ0ALKpuUEGn6hWWL6sSVPY4gh8srNXoFVragIaBfsehqIjx0+EneEi1aspvJ0YpOJ9c3/hPBHRma4Iofjed+CohlmOZuQc3mQWejwV0BWj5sAd57PyNFizehWZzNeUcxh28C919QWcHoPM7r2HwrXS9cLVNom7QPRGB9gsnS7qAwiOHzp05ISpA5rlytLW2qQqSLEvHWG6XEAXKz7nSgU2hUR7u976P//ll63O2Tx8PeKSIPSwcOPvDdGB5U6jiR0AOmL88PBg2Lf/oNWpWgAq5LePerG5ZVbvTK8rP9fzu55Un9MsKibKg4HTAAdrKhH+SGSgyjoeBcT7ZrXPQ8c6BIoalMmiSxCH65MejJX+6ku3hqamNqmKFDXJDEoz0nXqHuJEnQMFFDOZeBGSV3m5vFsSQGaQnlHejICWrQMrx2XSh3LVZMaBN+caRXXdys8+BKhCcKZpcjCNGm3OSJWg6PkoQwFTdZk8jhQwRW8K1WkAYXnoETElFTAzBUAauUPLan8cRAFM3mH625iVl29dVOT1xfQEjB1ELTH1J3kNqZR3sYuOEFcUnNTpzIhecA1TatTHnnwhbN+8xnSXT7/0hnXumip0NnrZapQDR46HG3XvSyXOviEgAmijhZsH5GMmX7kZVboEQhAcr1Ouk0eRDI7YCXcnJ8CNFzNtVlM+HBYGA/RiU6Tz+jW+BaSxnu8/dMySLrvoAnGyrxooA4QzmlM3INAb7WYOuCKRyzSJZZO1hHpldBqUjy1uXNQBAGcSYiGVJQr96dToT32ZZVVudcxdM4vD5IPRC8+ovXnh8RlpvdzzF96PY2ad4QP72JPPmZEinWdwRB1ielz+sX1aZ6fa2it9/nzsgxuA6GqpTRoaGk2Mxy2uWvXBYSwrDs/VC866kvpO6H1Wbq7d5youb+nmeeriKoPI1Or7K6ImoN96p3Vu9NShE8aFTokbK9EAi06UzwW3onWKWgWIVthE+5x4T10jIOb6AyAJValv6oPU92KHBtJRjOU4crdpcCXVwdWQjwSjCJbmUpWksI91312uSHZDVKUAHFcpV0OMTI2HevUFuind1jwYknK81nwDffiIiTCkjYhhmIvSIj77OOxPqUFLnBEQaKa6V14xrmO3QS6x/uMR48R1gDL58M6Jy0jDQC4SAXpwPljD8a9skqhqLj6at8xa56w9PoQyRPkqGHYSIgWxFkd99HsYUdANIqobVydwhmulwhiX8P/ctnmDgS8uVdAF61fJJSKOlIA5xBRNZkVBXHfsZLft49Pa29NvdTBuWKlcu3FtzNspbhOwnE30DInVGhw65NaB1ZxJA9defUk4cvSEPTsiPrOGUCUw99v0jzd90OqKoQg6KH9HqEkDTbcMT8yQoh54AwC6WzetMS6UoBqdOs8g0yhuEMNTzCuDlnw9MWYxkwrPAe5zSpw2ZNMGNZAB7LQz1ni4SThLBrRC4r30qgzUHnhLoG998vlX7ffgz5+W3vQFUzmgtiBikpC0sIhzfkwMg8aGBgPR3e/ssZgG0egYdfLntAIJt1jsHm4IKnZuvjTnhkwlNEdG+jxG85mSKg0iY8aNmkFGfbpU3CnU2FBiUzW3rWs2ldTQUK/et74TgalxoqmyJ6fEQOgHlZUq4n21jFEC0ZIZcajqW9H4Ey8APCen5Hqkn4MuKgB+iPsu8pO70LIfS4h/ayV9pjlSd5NCX9qQeO8ApkwawCUKVYVDg1vzYRQTZlGAqPuNDImTnzIjU/pexfaNqVBDTicceLE86TQf4AxEkxOuFvA0QHRKaMqW6bJOOUTzlHexHRsbNZ0o+rhbrr/KSjp4+Gg4fuKU9I8Xm54OsIRjxfk8stpygVCnR2T+2aNPG4+GceS6D16pefrDBsZMCQWU69X4n7njFonKt4RLLtpq/qmUxywdjC2AL/nYAsakY9QxX1GBDaCD8WbTxjXS952wvIfFXV5x6TYTkdGVHj503MBydjPkAAT3pwd//oy5FmEcwvDDjAvu7T+s+3d95mNZA9BLr79j+sdX5eIEffrjNxl4UQ+e78TxDrPQMxBh9IED27XnoOX93J3RTcrzdsgl6rYbr7Hrn3zmJeP6d+87bHlvl8EMIi8/2vXOWz9gacWMTZx48bXddv4PNaMMUR91AqoTVA1/8cd3hb/+0y/aeXxNzz8JLMWN4zt88tQpuz369fcC0AuffT79JiDrv7SRClVBjLbEI+S+qcKyOS6TWF4Soll7ZXNNmJZPEtZsANWXO14mZnB526qwqn2tvDiaJKXluCfKcA4SQ1NZ6bSArU5+qNKBKx8gWoxsuqbA1kT6ZEs5/it2TToNMd9/GKBwvcKLAI4UlywnnzkFNw2Yout1QxPPmCYGlWoZqZatXCfPlVHxBTkQS+djn4EKUDTOVN8/y6g4EBbm9WPUAD7AkYZeNM2JApqUAYhCbDn2chdtOWYeGyv4IXGWPONlmtGzU9M50Udeu/OycIOsy3TQe37yqOk4WDkQsR5iywiFm9Ae6Qi3bV4fdly0OVx1yYVynG8Rx7lW1v8d4WMfuc70kDj7v/jKG9bRMYBcdvEW+ZddbL6bdXJm/9DOS5X3ersfrjt0RCLv7JEB6ioZe666/OLQoo5ZL6vmLdddEW6+4RoD2X+/9yHjgtFHXqm6A1hv7N5v7hbUEm6auqJvRWx/c9fesOWCtWH7hZvCNVdeEla1NYtLXB2u3LHZ/DY3rF9jQP6te34qUUJBI6TLPNXZYw72zHj6oAxLtYres6p9WfjYLdeGKy672Ljy+x75pfSAzeG4dLzDw0OWfrXAnqhEa1Ytt7w7NJDQDg89/pxE3uWWd0ofzKUXbwtXXLLVAHGDZrR85o4Pi/ttMXVCT09PeO2tPRY6jufj+J0DR1Snbj1Ttd7ZhfJv3WAc+eYNa+XnepMp9vGC2KPBicHPlhO2t3a+/vDBKkq79Ni8Aqbm4oAPkCbf9CJVRD6Mki09HumoGWUKiqZzpztwcgxg8l0glTmhmvQf9eTsjBL4B6XzGqBmy82VAZcDYPD877zxshanUxAfpcGtEQe0VNLEhtUrwroNGxVIRMoidX4J/aY7nBaAlJRI8ZHoB/DrhhO1WT/az0wjdcGtYXwS8HCdpkZn4FLlSZop1RRc06mL+zLlCWs0AaYYIqtMIuJ+GT0TRh9WG3DjKxM8MlOl0ulruqmYqyznrUczFZjugJ4T4pjobo2aDIP3AAGfUUnh1oQkz1gDl8r+cvlr77z6SrvnlGY+4e2QDk0YdczJRdaMam31/djmse15D/yP3CfnYXl1DcRW1+n2yXklJac9S8yY/1f5F3PNpmg4IpoQ87cBgC2bN0r8XWXgB2cIqJ1Up22TQQcdZrojMOODqOXMSHpO1m1m4qxZ1WauUgBFk8CNMn78wC9Ml8eaPi3NTTYbiI6+csWysHXLRnG9G2TJX26A9K///iMFTBk2dQMgBFC/KLeoVW0t4mA3GQi2yngBJ/qNu+8z1woWDePlrBVgHTpyMhw9qUAYJurbm7EWxAhGVCiiWj0ja/+EAlzgz7pSOjzqSpnogB5/6nlzgCc8G+DMbA9mauFRcEiuTljaN4t7v0CgxfPBid5z32Pm/sXzUee33tkvp/zucKEGl1zeenkkvBB++uhToUEDQpU+ftxjXhdIYqW/UqB7wYZ1Vi5W7a/+97vDDlnm+WBffHWXDW6rV7ba8+HBgAj26pu7dX5CFvyt4tjX2Xujb9/7s8fDk8++IiU/ZgJ9ge8B8dHDoa9sb7eOSbQmiwTFF79olA+k+KjOIvobjeKkfUDUOdO4lG/shJG74QI6cFQ3paNJ0aG9NQ1IY1Zd7KmxtQFSBrxDe96RlBCt/c1NfE+1QXFINAivCs2Kp1qSSeqrfmQksKxIKRsBUUR2A0Wli681ACUv3GmmTLP6JAKbISoxas4kW9AEIIlhAOH2/FigOq1nkwVsWoAKiMflW7AXEO1N2nndEz3pmFRyDAhQmfKXlOheKrRM9aRNHUwry1W+wG+IxxGI8Qi0CIdb1rdpmuhmqcFAt3wQ1ekEMNkTUWF+s0ggb68wOZfkMTVA3vcEyM66OJsAh8q74XJdNpnZfsWNwxop55/Ok7389Dvo+xjZcRCv1ZxgdFtYB+ngPmXTRgEVlbaiJQOnKhf9MYe11jbXo5NE1MBi3dcbjUyN8uMkCo5Po0Q3S36mN1aVK5TYxJjlRdfJ9X4/ymY6KSBOtHU64wizj9ARatYHU0SjZVzLxUq3yPx3n66p5ip4eAaOOG8ffSb+pD7biufH0m46Y+lC0ZtSV8qw51Tdmc0FcQ2uKBipTJ8pDt7zUz6g3acZSRipmO1UopEffWW/VAdMBSWviyUYLZjdRF3aNQV2QmCAeI+I/n/+pz8xDhZgpP0MjATw3j7UGU8CDEqoLuDiiRxP7IBce8/zZdnTnJs/+vTNfYx6YbgDWDo6u+xm6N8hdW3bnt0frpWeWrrnD12+TpNDmqVj10yclA5srnKzQAp3Sn5Z3c24kf5e0npWnU86Xy6f2hrCwEgfSRNh744c3h+ef+xJc17Xp2KR6xHp0S+2bdgWVjVWS6JQDFVmKOk9GundoidFHAdE0XdCxpXimK9yDEDFXUIzM+IcJ8SlzkRAdhBFZQUYumWfvBxDMxOxT0wKxCfH86dWj9O5p+M9fa7+8NCgzdO3i5M/Lup7oBSSmQXVqcj8p9xBQGmDYkl3bGwI19z0MQMwONJCcn/cmJ4MXoWZ5jG8uj40Z3CbdXFegov7jPPFlSR52c/sAMDI6OVjdceAwhRJxIrGpjoDHgOUPOSP5WdBVddXKTBsbU28HtCAYhkOoLD1vEzAdFogGAN+0BFGBDCel0/SGtcxUHkBLq4l6jqWX4J4tMptCYquSPFDZmodFF+OF2BJyZ94b2GdZtYss8EDH0fikXL/KonKdHzu5XXlQtcLx3pMiXOP1zAbLE7DJD8fSSyfa/B1BegANiiXd8rKHh1V3IHK0vDnf3RXOCY3LiYVYFBz+tTHb7FdZpU5AcLqCrpeHU/vo0RcSfqdkY+gJXV1MWiJ64b8+vO5BSSZ9sm3sP/AAZNSLti4Xpx6l9qk33xIqT/Pc07IQbJI4c6NOoiSZVKAyCfugJm9TCA6H5UI+DTK62OPIMD1eFRUKXZFiaIr12u4IA5otdRX6IkrqzWDSXrHaKmfXfLpQNSu0OJ2gOeMRHxuDzmIugFqZorYunwmGrQFnpOJmV/DmwbrHKABnlN5evTYb5iliO4WfWm5jEVEmYIAUWY+OZhaov6krflEi4LKdBsW39s+0C+GSb7Xkzlda8yR/jsXiOby5EAzV//IcJ3+Wi/F9aMcLzqQAgAQYIDFORc6LgcQlmHOP+nrmc+btqA7KOVf7NwIOjyZIpOT0rXkZ7OjCFICCYmzTn59PPbOGOvheebeOqBqbnCqTDq136vYtQ6W6Wvmyk86Yd6q5fbh5NdzzIc6rIFh7z5Nobtwc/hf/vyL0iG/ZVlRdWD8Yx7/2zJe4ZZG3SKlnzG2Le2da3N/hnS+5NLzvlFwEnU6LPjEtkWtwzpOyzQd8oSMdQxIhFWMXDxvXoOD/sZvwJ/3LCsNpzkPmBaW6nyCbzlPtPkEEwxkSRPLwWYWwdECxhArLiBRsPrnqMRxQNQJKcopPV+eNLjRtHUdThRy+9KM2hE8NBG/AAUAUHSnDqwYqJDytV6FuFgNauJc4UKzlHC/uBmMV+i+40nd4VTlKUJ+J6z49foEPeCJgyiWfA/VVyI9LFNKof0nxiwcHxb9LgU0GewnFnGb9S2k3+JES58eEF1SLSzDrfFpoCzMU3hc0ISFpxfj+N18xGd67ZnkP5O8C22HsylzodfMlS86/sOlflvLTdxyfa95Ctzx0bZspdG9MgkCsZ2Pby7AjhfMdZ9sce/ZDgMenCfcDR4Y/JZrajH+pQDpCemztYCwGUnS0x4B4PzB8iweATB1EmdZYqoaT4jbrFSVnywEUpsXpMFtQsa5ItYklFG5RLgvkToAUb+sTBybJLtRAR960QY9O9RQ1yin+zrNDFKYyXGpsFy01zlmNMGNOpA6iNqFihDmRBWQyNLWe+dGzT/SdaTKJ2wVJUyK9lm2BD3wTAJm8XFGbR7+uEDPxHrVC1YNMHV/UgdQSnO/UsAU0R4rPmA6PSmpUmCq/6GiXm9U8Ul12ri+5xRDorV9nVRdisWhNkmDqe8XSk8YxaAosse2VtVFuXbnyMmea86X6bnyt/Gt5KctHf1KtQDmA32UAlI579rU0Wdf3qXVHhUdS1wsM6twN2uRXvT9G1v0TBo8dgrTjQqNiIqPjnqVjHxbt2w21Qazu4g8xlIivX0x8PNZR4SCE3VKwBSOMZXqZ89o66K/A6pf7BMxfEs6wJIG0WzeSSIqyU9bfqO4Nk1KTPdAI4Coi/bkR0eKaJ7WdQKiOfFdxicAMikcQI1BlGOC60U5yon3kSN0/WhaL0q+UunqS6VPLZuJHKlHinLRnjwOpuyzDLNTeemUpuRXGieuocKSiQI1NDQaBqTOWdG2woDU87MFQEudjU6dyHGexYEzlZVS9Nz5KXMBbjrXeQJSfz3cmk/QP8N0erpav2r7PIc/03tR9yiCM5UQfS266U5Z4yE4VdKwDNsHlZY334uqLtY9EwTCWEYQk4OKTsU+U3zdIDUyOmjiPwNKl4xwUWVxhu8pzYlSdwErDjUwXacjqlisuQvBs7AcdKUz4lIdAMZlWBlhmm8iyddIP1jG5IhQa76jMfAI0z1zoi4gCrmRiX2ATejDrjnjMy3TOVCSEtUnu0YALZQG1LiIc46rtQz6w9x7RPrShGnFwFRm6olcQ7lzvov2OOgPaJBHT8rPZzxRJpxpdUV1qNa7tWj/Mt+jp13VUCpVlsL3JQNcvnSFcTZ3P6/b+dieMyB1KzwPEd0E0FnyETNqIF4WCjsLf1ysuPim8aEtnAPnvrgrqA76Lfw66f6t2vGec9XSA/Zy/kzKnqu8M0+PnCnPxpTCcjldY4kG4LMf23vzjZ35oyzgimwbwz7IuAeIIt4PJMtjHDtx0koh4DOR8/stojusxhk0QqFxCOu760tPU0e41tytEIP1zfEdJYYkLkeMhwDNNHEMAOOjiY4Ujg/daGPL8tDX3WFZu8V1s/CfifQqhqdKi/NkctG+srLWRHhuL5k46kZZMYFjEYBp3KiAFQI4HUQ5Zl8eSwmgavqx1K2ZaTnXI28nZNGghCbpYCZmzU9m07lo7/nZYtDFfJqOtu/ivecjyn+ZouwzmQv/2S5Z8zEU12uqcI5TdCzxrV/NlrbNb9/02dn7Z5I3d/U5A1JugWWMjxvndWbY4NqB6xFz23OR4Gfx0bnazdqLnYAAxRDWbIBDf+x4rj8AN1bvjsQli06nr2Ku7LPS/Tk8zuisDEpgSiyis3MRxfKcXVocALJgOG8h3g4OFn4870W/FifxjGBaLi5tvGsI7wn0XVNZXZ+3y+kf2cAslY3PrBh3mcoy564N3gVnHURJZt/BlG/NviFtvbZ8r6XyYnEinmeDxsna5hapXytDyVT05uC8c6J4sMChAqLQpDjGUo0+gKKL86Sb8Ul4WMiNOheaBlTyu4iPeC9Tcr7RSecBVMAUPTaEWD86Nay6RFYVFygIbpQfon1VZaP8wfuMCwVIfcaTgadmRVWXCjwTjGQZElbGWAhFDxpynh04LuQenuccAGn0r0T86ZQ1leUwrtt5ic0Lr1cYPVj2PfsPh9fe3GNTI+NqoFhXF9LpI/gxPZTOcVjRj0pMnPFPzh8rf4uoiw7x6ssvMR9EdGoLd5ch0lSMOrT9wlZVtMg0Sc0gwe0I3dxC6pNfu7mOYjty785fKx3n7OclhgDxFghIwtwHYrk2CymY3ZUmYg/09I9k8/T399n03+i2Fjlx3i0zyljfi6nBa1avlpqjU8YO9IeIqgv5ztJ3Pbt9t8gbZ5oqwsZ9agHXWYQjNbBOwNSBu7KmUcsny8CWcKNwaS0S7av1TWfkq0lkp3RAZjhRNzhx65x4r9mE9Hg1Q2ZCAWjKxV3yOQtn8kR8OFR8UkW+LQRUrPdGAmFXKeAKBYh6rKvVMCsAAEAASURBVFLnTqtKa7KA29DYLFBXIOgKBXWWG5SL9gQigcAHgHRCM6NcT5qRN061ApsQpqSS6Fbqz7XZwdEuK/qHgfR8gCg3X2QgFVegf3AEfNDXXLHdAhVnnxIuUNzhegX7uEFz6X/66DPh7Xf2WhSfmMcB0bek+ocfI+mTwtx2AOZvv/bvmtWm2RXZAWf2dYjlLDBHMOY7br3W4o7ef/CJxGeT0iC/Rzwq/IuxhjihzN2fiwhazaws/GX1kAXZ5is/nTedLwYyYcbT+nWrLeYmc91LJLa/O/L7pe9FiXOlF97N85FeWEZh3vmPmRZMKMJa+Q0TgIUZXi61NMjft9HEt1wZfQCnfGk9D25Qh44ck8QzYXFJCQouZY8G2KPSC7dayD3m52PdRwd3tvV18MP6bvtqgqxqIVc9NWFOyplRUGaAEEDNaS7TmVWb3IebPYHEBlfqz+gqo8HRnBhNZkBGU0/kQ6u1kxLDkhcCV4phCcoamaQfxQHfCRA1YtkR+ZGmuVTP41sHUz/GYAWwwuVCADmc50xFTnfqIFoqdyh0AZPT0f2qTLMAQR18SiF4aXeBcjAlneeDqmVIQ63hyz/XzDBRJvYBDEtzSWpMosLYuNg0rQ8gf1qqGB6941zLLsodCWkWbJ0hjz9KeLaHfvG8IiQdN4DlI2E65x2aN/+pj98Yvi3RnzXgmWnEtegx4SBZQgJRLe3T6FUERNP6GdeZ+nWoEbDSxg84dnYiSTlxHh0tq5CybwGB1UB0whygeG7UWrmu8PDPn7LI+DWaI58mggsTVIX1zNEJe138GSghV36sEz6Pno/XkVtFFG40o4XshsJVOy40d6a7733Q/EBRZ0T9slxclKf4h+Rgx31y/pTUgamPU3JdYdJD+hyioEqzNo8zq3LtBQAxQFL/MU3VjHXVvGvVv/j9VXQRYmFElpTZvHGtGYQu3LJZ02nF5SeECIzYRpR7ZmUhmqUJDoPvYUzuMujJrv3ANdnTnZ2dGpT3mGSAxLN73xEbbMnwbtyfDDizd5m94+cBzULyc6Qnk4EKs8x7bA76ypEXR1bfBLZt/C4LF7Fz8ATonAv1mUwZWFFwT8DpIMqceaCeNIUK074IB31RHsab1Z33IbWAztlrSV4NABq50+hf6h5SZsG3knJ/phRwZUQNVTaaAzjcnPgS0ZOaC5Sei22acPmK89hialVN5F4Lvz3/XvhOCkEUAExTIRimz53N/qICqXV+TQW9eNtmi7LE3Piv/tsPzA+uqTGGlptSoAXmxjNllHB4bAEgAh2wCBvggb6xSnohrLG90ocy1ZNQcKwkisogTTiqAw5Mr5zrunR+9gmx52XSYYnYtFCd7Zu7D1rYPAuikSqYZ6jSqM8idExhxGeTOlM+z8AUT5/phM4WAO3WLA1mGKEDAxzTU0EJTk14vN5kFkhv32Do1LTIOG222mY5EQC7RQ7pDCSoRtgyYPRofj35uD9cbJ8GM56XpbIBs3IFsKQtqW//4KiAadREa3wWGaRoj6zOVx8gc92ZWkv8A54BHd6QyvTpo3GQoDHy3w0p3I8gNKxzdcVll0p8bzCwpBz057SPf/TMtx7V/fmdjjA2pq9buXJl2LRpk81+AmT75SLz+htvBtafIorW0ALK9HuiBnAqBEg/TvdLE9HzkEdAZIiTlJMCWb+e8meVQZreoelMjc+Bo9Q70/eBaH+icyygHUS0r1SkJ6zazlA4iFKu6zEtTXp7sXBi+4ArkQBTLRf3k2M208QmFVm0KcAUcE0oUybH+iITibDg4+uJlI2fKU+r2PthTLOzLEKXjsen5QequfYj+t514zCpeAGTk6NaJVTB1UcxGHn4x9osmCa3nbUBX8fLmsyHtNgU0SjKc1nq+WaVsvAEwLYQgLnaudL0uUUFUkR6DEk3X3el1ZbliCFf1oNBDDVVQ127dWCWtrD58erQTH8kgPHtt1xvS37YhfpDQJGfPPC4Onev6VsBgDTBSZVJfi9cAZPwet9XpClzeyH6bUIdnR020+cPv/hpdcTIaRJF6YHHnjKwQAXg8+39mvQWf0xiojLv3T9YP++rcbKO0o1atM6+HP3lGe796WM2aLAMNPPqJ/VhFT4rdSYoyzHFWV2jBfj+9A9+y8RTyr/jlqvDbTft1JLUey0+KBw/C/c9/vTLNgDBqbGcCTFDf1crqiLyPvncawq+siLcfut14dEnnrNlWZACIJZcIf5ou/wv7/r4bdYmpAPETz79YuDdMBhQHm2/VlGGCP3nq6+yGN9PHnrS4sjGKbb5ILpu1cpw07WXG3iuWLHCwBNOEgI8+wT2TgDo2VD6unSZxCrlt2HDhvCRWz4cTp48qZCLr4afP/Wy4noeP6NbuY6T71rjiBEDi+b9xP1Ez8nUzrSoLl5dg5uAKbnGt6RRputQKSQaKGERYnc0H1KpwQBdJKa0AZMljfF8qq2OBWNsQk8KoBKzwbeAqPuMjgxJf6wuMMV0zmRKZ6lsC+UC1YmEAxX8aVpxr2ZEZQTcxCoVGCXn0vrTab27En1rpYoOlVHMOz/nKktAFHIA7RIjMChf3lL5sU4ls6sqpQKZVNxUlvzGOKiJeRrk5dZUwI2m9aRWqP5MKrAOIFoqNwJ7Hj9h2zMH0DQYpotyjnU+ME3nX0QglTg7Omwdt31FmwU+7hK3yUJ3hcCEcQAxlh9cLFwRIPqHX7jTOjuAgngGUDDl8S++8ru2+Jpzr/4AcGS1imP4+3d90pZiLrzuT770W1o07ocCgj5zUNcXGa77wBUCg3pFhzpq90DMvESrcrJ20r98+0cWpi1yiHzys4loSp0qj/pC6LQqK6vsWQi+/Ft3Klaqyjt29HB4a8+R7DP8+Zd/O3z1X79nelSW//j9uz6RfVaCPaPa4Lo/+8PfCX/zj98yzmple5txgNynqWmZQK3KAkMTaR+dIlM/n3j2NQNlfEhH5NIC0JFOEGcCVhNBiuO21maBf51FxUKZ369FyIgY9af/4bMW15TB5JQ4XtZvYp0r6vP9+x8zjwvWxmI1UcIgsj4WU39ZyZS4sQAysVajbrgkXLBurQH+zquuNO6TugNyBBzOhlMj8RwS94NwfWKu+po1a+x30w3Xh+deeNHWtdp36NCcNcB9TFOJ7DyAB/AZoKHjFwGqyL4AncnACXjYydQfB89Uku1yfRp0owivuyRjkQRzA9WYRy5QCUoxu8lv1ZTojytKNBMq+VRjEObIHJjPqO7WL3chA7VerXKQSDdVpZJohgfChBiCcluSRAyOARTLiyjmg1RvtZXSXy9viWAq8R11I9HBAFEjmyIa4QNQhYYnoiQxpAHz8IGD4qC7w0nNrR/uynkVmOGIaa6SICG46/r6Fg3aqBkoJ1+sNzAVo0WsgVGZ7onLaiBaJGiJc4qU61QsjXNzAWj6Ot8v3DrIpstYNCAFEBH1EOGhYyc6NOKx9gtfBy83n2PhGEMQqQSj+MztNxqw3PvTx235Dpb+wDKPAQLu8a5PfST8/X+/RyNuTqFNh4b2HTwh7uv18OjjT9kxf6647CLr6NdceZmtyGknxEkANOg5H5ChCyBhe9OHrjCj2EdvvjZ878ePaM42L5WaJeyEXRz/rF2z2oCTSPQWwV9cLSMrazdhXAMMWUjv7nsfsPIJ/0bg6C987uPh44o5+p0fPWIF7T94TCH99oYHH3k8WzpxTf/kDz4fbr1+p4Wv+7//v6+Fm67/gBnJ7v7xw8bZtsiS3appkU5wIC7SIHq7LtiDOPsxz41BjNVKUUsAigRyJjI+4Q1fFZAS1Jn2+PynPmpSAUFO4E7v+vRtdrv/8vf/Jh3kKYF7rcU1ZalnAJmBZLUGz88o4PaHPvCBLPeJiO3g6Vuv9/nYck+4Vn58h3CpH7vt1nDj9deFXz7zTPiB2oMwiYWUFu2dIyXPtIvvfBbpz7mIb6nlddTzG3gwkqQcOE1zdwKUnbO1ouN3TZrmJ2ngksAsNyAIQIHQj44rhB1ACsW59hHQLF1KgKGhgXBIuuNOxcDFF3VC6o2RlIzer28gDWhty1rtXjLrhgFFUSMC1joNohipmJtvgJmaYw83CsYDskRnIl4JU0BPnTweQTQJxD0m7nF8IJE6ZJc42RWNVBbIWT6ia0O36lVrngjm/zxdfBkR8SwBo5sb4OzBZ/2J95ku9AGele/sE/ieeG9pWjQgpVBG2srEqjwmUGUUtw6UjOTpG/s+nY2ZNywxgghMx127bqUkCwlH4vp2KbAyXBBc0iat/b7rnQN+qensCPrhoHXjdTvNOs969W/qOgxda1e3GkD4RXCtrB/FFEKCfaDTRMSFEwMEf/H0Sxb7kgXV0pZZgAdy0djLY0u9X1IMVYIlI17f+9NHQm1djek/cfl65vlXwlU6B3fd+sQLpkz/geKOAlxeZ1QQr2ixQERmuGN0mhMCdG2MiPPIwMTgYRxRTF7wX4KWsIopK60ycDEDCO8Jlkx5/qXXrD1YiZP4pA889ksD0p0Kgs376FVYM+gDCl794qtvqdNmLDwfS0QzSPxPX/ntcM3OnVLTtJgRyLnP9wI852oQ6gKn6lzqR26+WXW/NPzyqafDfQ8/qY7fk3epidN0lmQsTYviZIQbdQ4Sa77n80LsnANvNtF3Zm+N+yzST7iP1Z0lNsambX2msgRUKQXQhIP1lUJJy4irPHXquJYZ3xu6O44poM1kQCUA1cgKnt63xNQf9JnSsqmteiTBjIZ1m9ZGrtTzmOEpHkxIAoVGZMiaFIdYJs70RNcxTUs+JQkkzqzjXuMsbidiOZHJcTjseGx8qgB4Upb8+mrdt6JPWNBmKqWQAlOp9G2qqGA7DEiVRxyA6cQ9ywrO6kRlMDZpIR/W0pxjzP/u/jqIZlUuwqr8O76L8hnxAc7hJIwWukbAJ3KkxQqWWl3gMCrx/IINkcN6Z+8BAyCMMYj/cKysBuoL47EG0xu79uQVxlxyljH57Cc+opFR1wnIKqRghyMDdI6KM04TYjSARGRu7sEWzhRVAsDSJnHmHe3X0ImMS4g9yblft9rXyyAGxwzn3CWj0KqV+gBkQMHCyvIcvtwtouWIOAHOQehW4f4AoM9pdVEsyl7nOz96o9UZsC8kni3WIenZhRlOc8yaTjw3ejMkhzXSjUIb5XP5v/3Pf5itL2luwMD1Cu73sSefsQGJwYwfYP/Grn1Wnz//ypekdmgykPKQh+8nAOV50kTd4CioKxzqHbd/PHzo2g+Gf/iXb4cnnnvJsiLa01kAMRftOQE4kub7ce/M/mY7X3JZ4bFxqAKD2PlzrlPu/uSO6Vw+oQqh00RP6lSpPnWy42B46tXd4cSJHkVRqggNMkgSlhsi2AkEd+pgZwn6MyLQw93IZyERFKVXQcVpj5pyBX5Wf4LgThHzhxP1VhgftnFkeEwxbXtOBWZdAd6I7YjqrY1RfK9uiOEqR/qjgYmycE870MsUWM10Ess5LN/wjern6Oc155Qs5owfo15JF6x7Y4SGosO9+mmaFrg+U/qS0+3zjtLg6fvp6xYNSLEaI+Id09pD6CK3bl4nXdTTavAYnSjncB9n6lAJPmYaJs3FpisHR0hwZRdP0+fY75eeZf3a1RJFbzPu85sybhGfkjXsN27YYCIx1vLTEe5NwnMjLMHoPSMlvSY5YlNotXfucJkChgBUrBa6e88hWcFzIc64DlEewr923ZoVJurz7ETxx3WKOqM2gONFh+muHHZR8idXr3Rq3PcaOwgW5gCEGejg8q3OmkQAsZ5WjzwCqqWHShP1JTQfuk8cp//pG/eGK3Zskb51g+l9v/D5T1mQEOqJ9f3dEG3ubm5Y7NknzVUWXjb3AgThLFnWBmKftLMhruWHJ8H/+td/Fq6XuP/kL35hRRnApcB0oeUDtMaNLvSCgnwOrOjh0J2WlmrxOy2vDBGLtF8r1EKlZQq+rRs5mJJWXaOg3/K1fWvXbuXrMxDdtuWiUCMdJDpvPEucJmQYHhXTwSzBsZFBieiaK5/4hXoetqwq0Y6uNJGM0udYmK9U8QwG5fI3Kc+P7t7IhRJspNo4yypJZU2yH8gzRQzDZDK7qUxcr9OmjZvCmuNHwytvHzBuFS4VcK0WmPpsKL+1QyZeOrairoHm7D7qZZ/NthA0vQzS6TswIv6O/BzbRQNSCsN4dFIdk1BXO6+8SKLuRRLLnwsE4bW1WfTi0aVi3ScqPctzTE73ZFf4XKPo5+jwCDCMrMQqmCf7OiTSr6L40NHdbxygHegPK2BedvGFdgiIImIjsiMmH3riGfmqfkANHsUIv4b13z2UF+5CEEaZtSsjV8yyw4CvNVaRd9SomVqjctvAfccJVyYi1lMuQPb9nzwsXWyjPoTYvAAmVmqWpkacv+PW6+3Sb91zvy09TZ175ZK1RzreW2+4Oo87RNyGGEzS/qyWqD+0pVrV2qrP1Cmx0rlpkZ4zt6VeuGhB+w4eDQ8+9rQtzEc9AVvADIMWulAGqrGyMS39MBmefektA/lPf+JjoV3LfqCWYS74mXKg7rYEYOIzStu4FZ97n+zokOtXtzpmX7YtaNeWpsbQLPXBiuXLs0DKe6Ct8QgAFKEzBVaeA8nhumtZGHFj+MWTv5Q64x0riz/vBhizhSQ7HpDE04t1yuw5dKTj/aFZFWA991MHDpl4zHnEd3mye9ZQIUv7yPBg2PXmW+HQib6wheVi1l4QmlvVVuYzrHaRVQoARQzH+g0t1zIlIyNV+oZiWfh6jksPWibucnSeACCTssBnxgayIDqs9geIscK3Num9Khg1hitoVGqJoYEerVuvmAhJnwBcSwS4/NatXB3aJNUcPcFCkKfsGiJeiTPRO2Wqq7hqXceXzbuCWNMJiRXy912MU4w5FvbX3wXbYmW5F0Sx0hYVSBkxGhubwpPPvGDLCrPaJ9zm86++bVweHZjOCl22Y6utBoph5pt3/8isyYDvS6+9aWsaIdKj42TxvE/IfQcgeWfvflv6wwpI/vjsCgAKYuVRgPEzd9ysrw3fVOfV7LTpKbFiv/HWXlMbEEmeVUXRjyJSH5PxgfWVWIMGMC9GWOxRWTiXjcUcP1Z8Fpn9dNvNHzS9a62m9uFZwHP/9Z/9nj6CUXkR/DjbBnx0EHXGt/XOj91oxh/0lIWEQeuU6gaHDrneEmDnWcbG6qyMT6kMKGlm2/c/zj2zbtSBw8eyq6DuPXjcJlHw7vr6eg08f1vGvUeeeN70tl/4zK1Zw9kff/n3jVs8Gz0oAArwIYnQId5+++3wzAuvCLjHwn6pXFCRLNTfE/9UgnawjHaVPBM+cNVlYd26dWboAli5h3cwf/65ti7u80xMEPjUnZ8I9/30Z+HIobdngWgaVOE+oXRaej+ezf+bttbnnyl+BAtRXzkTXu2OKip0jHB8aTJQVULv4ICForviog2hbfVGDdq46I1a6LnOnu7Qp5l3ZYqiVFndZBwiZQCAAB0gCPDVKfxiBXpQqcfKBKjsY/VHlHffUET8Sn1g2OjhRPvU1uPiTAFKl//GRgWew6ey6gLAkO99TG5QfMsDA1G8b2puU/8oN65109pVAuF6fQsHg5wMwgqcZlNEc09qgGdBuxJ5HkzL8AznDug5CHr29DFrTEEzqp8DpJ/3Y7+ObbE0T/fr0vnZX7RVRClM8CIDTrmJ3Lv3HggXCrAuEce448KNAqd66dKalbZWq2B+MOy8YofpBp996XUTL0509ITLNYvnmisu0mjDKDNjjtx3ffqjauTqcM+PHwpHjp0SsNbbdE2+3qdkxBnV3NyrLr9IDt+bZIAcMnefO2+7znSQKLRwmQLgmsXNXKGVO9XiYcf2rWbMwVn96sulq7zzNrvn3fc+Yo7yLDzHswCk6GsHBczbtlxgC9tR1oCOEcWd19WrNMMX4LRpw2pb9bNJ4uKgdEWsnfS5T9wsTnml6n8i7BbXDHdFnXds3ywn5GEp6Utl0f+QuPhLbaQdHuyTVXyf3b9ORqvtWmq6Ts6DOGuv0EqKrPoJB8bzbN+ywe5DaDFWDN2qBQcZrTvE1b0ugMWQd6lWWT2itZvgPmtVDiIza1WxEOFlavNLtl2g9pbjtLgVViAFjNEfMnGCNZ+2b90Qrrri0vD5z37GuD7ufSZcKGI4K38Cbj//xRPhh+LYv/uD+8LDjz8ddksffVD3YA2rudQSfFuFRF6u4dq9B4/YwoG/eOq58M6e/dZJ28S11utbAbzhmhdCPBNcLddcJkPUwNBY6JSxZiEEqPJzypSIqxFnA7DyPorRXJ2SvIjtTfpWy/t6QnW9BiBxbIODveFoz2jYKAPqujWrxZFJbcZNBSyoKzuOHQulivq1ZsMme4aJCUlJw93h6KH9EvmHw/oLLpAxdpn5f9o6Z+L+eVa+K4B0Qpb9Eb2jWklOWORL1AfNT1RWd8CuAtEc/1JJivjRMrVzRIPP4FC/1EDDYUZlMFjzGx7qCaPyDW1e1hraxE1XVMX1xmiJaQ0E1QJjPHBOdXUJfCUJCcR5p5yXYBXKtTgexmDqBwD3i9no6FbUJwUlXbVWfslaAXg6cYFyNVjeN5k3ogG2qrfSyJNu9+xxOj/76WNeCFQkDXFfi/kt5iqi3EmV1D9WpeyRE+7Lr++yGTYXSMzAKZ+li1lV1NxnBEis3HlcgU3Wrmo3/8q3ZFnGMn+xgA6QQB9H9J5vfO9+rUy615zTaQTAeEhgs+fAMdPvoWMESAEmgGFZS1345vfkfpTo/RxId2jGlc2JVzDgD9/wAbOy4141Ku7o63ffL9HilPxNfQpm/PgZ8QCdNbKkY1DCso74CTBkgVQ76DWJcvX6m++YIecieQHwDNQJV6n7Hnxc1vBnwgr5hgLuBFzZJj3yju1bLN+KtpbwnR88YC+Z0fsNrcQKmB08clxL1VarTbYYIE6qo+/ac8AGqyF99Nv03Dw792J22Hc1lRSw7ekblgHtoK3+yT0OHDoaDh87aZwlU+t4R0e0lPV+pbPC6EXKg9cB7QG4o7sFpLZv2Rg+efuHw/Uf+pABIUCT98Hy2gsIbh1RGbGdvMfUwb9zz73hW9/7cfi5vABo52G1aVzLquDiszykLMqk7BfkWfDs8y+Ho0ePyn+2xUR/e1/KQ6ebr/6cg5MFfLdv2yYAqbb6T4srm490mfUztvx0I8tu+xqMrfPq/pRvaZzV8VzEGfFcITPeJ1CSZ8uWC0Of/Jf3HDkVVjZWhY0b1kdwSFyshgRkZfoG62TMZOFCfpMjPXJ9itLN5m07NMX4QPjFa6+EQU2weGnvXg3WB8Oew4fD2JC4XRmTYBjKMopWPyoH+ooqfU8usDJVm7WjpJfWNwaVCjQxFPcK6Ps0Qw8R3O/L1ubiC4h7JNIfkw70xd2HNGgeDwdPdcqftTcckdTVUFcf6irLQu/JYzKY0YDUG+u/JiSLi6QcgJQfQDqogDWb1rWECnxg9X3V1CkAit6VvU/aMmlfq2CqbZ1j9a2d9z+pfJ50Jlsul5S9+KuIeiXg5Cw+pPwMiQDFkhA4lBPdhSUhCASCaxDO73xk6FDRbSL6b1i7MrRKtMdHcb+4PIwsKMtpMDqp60m4FmJKI1MiAQGIYBaAGtM+mfUUo6Mjhqjz6l6Ew4NTW792jcThPpvrj/4xLkgXO4AVlP2jqWKqFyMonKiPbu4e5boanoF11ymfmUmrVrSYFwNBh0kj0hV6X0Yx6lwt3yas5uhO9wn00LNiraSOuF/x4ifkZM/qnrQJUz5Znwi9KFb8ThmuUEOsl0g0LhcSvA2gCtUDXScxBLBwwrWja40R8nm+OHee9gTUefaNKqNB76NTqo7DAlhP+49/8UcC660GorS9fbR2l7n/0OEA0gMHDoT7fvZQePblt0zknPuKc3eGwCjXyI3rjo/dGjZs2GAAyUB4OvJnRRWx6+3d4dFHfqZrT3+dl5vnF5qJAq9zQi46cgznyWBdSKQ3z0yGFQrPNDohYKtqDDi6f+cH95sR6ZMfv8P0pHCBGJxkbJDFQyCn7dCYjH8TWrAu5XP91Auvh8Pd4+GOT340HBMXOCI3oh69+2Oa8tzQUhmuufQyDXxlYffuXWGdnN5bNIGjLBNt/SUC1Qb14aYG9bckyAmc6kDPsMo6KXXYiawIX5vMg+/U5JRR9T+8DTqPHwlTlU1iWFSnwfGwSpNgljeUh92vSmJqLDWXJ7wFXPwnoAl1H9KS7p52WP6weCF8SNIZ1CuutnHThVI3iJOVrcQp3baedjZbNyyd7lryCfxHFlW0T98UDSIPWS/RdExTzZj/zfrsiGOsnd2k9dgRFwiEgU4bLqlGHRCDFbEJj5/UCpHKiy4M8RZ9ZPwHDOh9Cij4sY/YC0cCyAAuWM8b5Z5kKzzqIy038UX+bBME66BOtbZU8hFNF+QeXI/IG4OBzP6oMUpxH0Y8oi8hyjuIWrIu8Wdg5c1qlYW+7aimesKZUx9AmjJ4Tp4DPSWQhpiOTpQ6EvFoUp2L5ypJ6sxoXCVVA89F+xFbAFGHa6k3oMDkB+5DmaglWD+cZZsR4c1eoJeN750NRFQ4eR6XHhj0Tkk/SZujjqB9UEn8p7/6Y7PMs9qqXVWkw9sJ/XEulKAjGI6++rV/C9/+/n3hrb2HLBaC5zvfWyy8+w4fl+j/Ujh48GDYtnWTifxwPvNxpz5g4Hfa3r4itLW1h3379umaXKed71nANjU6H2o2m5XJCf/pjN/HM6HH4/3g+FQl56u6jFz0ZNgp1TUNy1vDmCSxk1KDXbhtqwZTcYxwbXovDKz1mntfJV/NaUWegsEr1bZ/sCc8/9puK/6OOz8bNly6M7z6/NNhj9Q2tVJ3rZSagBU5K2XEu+bGW0ONQL+nU+oMfTgtUk9VaKAuQ8TWc1RKvyp5WueYLz8ubnBYNoABAXJ3mFac0Uat7lkuUJvsGQhVUq1c/dHfEXfbH3a9sT+sXbtcagfFolC+1uVN4YZbPqLQeNKT75d0Ih3tsnoFcBZXzPfOt8u7oR0gGAsGkWnehVQcY2LCMnrOsmaVKdDN82ZReywGpQc3wJJXlk6L9eIl27e/2KK9lZv8iY0QAVUvQWIeRgFE4DLN86UKsRrkix8bHZvGIw+gWgH3h2ojSU8Kto+GBtdbtSTO8yFxHT/2PS3uk00NLxBkyzn2vT6AE/X0F0fuNFmt9GWWiNOL16fPpvdj2aTA/cGZpeuTzun18zpTtqfFZ4ttEdsI17JKi4YeAdZTeQ4tfKZnrtJ5mpEyiDjFktgxV6y3rVKQrkCyT36en3pQ32orqyL8xz//Utix42IZKiKIFrk0mwSIIjojDj/x1FPhb//h6+GNd/a/pwCarVyyA6CiqkDkr64qk5pGkz5UZ8T4QjArvBZ1Rpvceco12+fw4X2Fp7PHeeI7oel0xjmkXKb4VrLHqZ3InZIQ+8649KzVAq7qKvn+ygG9tLElLGtoDQcO7A4EaanSe7bc0i9ifspUEg+AQMkyFklMBlK65f1QJb3mmlVrBYjVoblWjIUMsIcO7pf0NxKmBIJSRoYrN28LJVIVDUsdICdT039WKuAyVv9yffsYnjThSWBIH0BzIdWBxHlUCqi+EPkR5wdH+5Smb6ZGa4QJxKdlhBrvPRGGNI+fmKk1VaVhvaTNUt2jX66CVSXyOZWHAH25UutO1ddKPytnIoCUfgCIQgAp8QuW1TWEKakUyuReWNLUakwZ31+O2NdvnkE/l3f2noMmRfrPc/mxb0nHkj89PXMugdRvb5+THyx4m2ua+a6PH9yCC50342KWNe+NzuJkrFuuTfKLgOuc61x+ztMfwQt95fc/F2684Xrjqk9/BUtG1Btn/F//2z+GH/7s58bVLuS69yIPHPcLL74hD41DYedVl1vd4TpPR+TZuHGjgmuEcPzYoVnZrd/qj4PyDC8FjrSAslxpQTqHvEPOs8SI9mxobywTx6l7jgv8SjVolq/ZGE4cPCBD0Lhmki0TGEr6UP4KxZyAZqRbhJvNzMDZzSg4dquMvC1hWoMA/u3dp7rN5WnlCs3sk96zXQFltq5ZL51oZeg6dQJfQOmVl2t24bowI79wIZkxJqgqplU2KiJil07jkC9RHV/UCRltGajK5GrYsmyFLO+tYlonNM+/K9RV14UN69bLDrIyrJfLHD9AbqRPUpiqXC0GAG60WRIqqjEGN7hM9j0Ikhub6lSHuhp5IuAJI+ktNDTrGTV9FWRzshehY9tS/eKqE8/ONg2e6fSF7HPrqqry8wGkC6nOUp73SwvcKVezuz7/uaxOdK56uSiPQQwd4v/zN/8QdslT4/1Ops9WJ2MiwrNyvbpAU2ZXqZPDmc4n6vNccNxbNm+Wr6PCG/ZEd6Q5nzdhWxxYyecca/oa8ysV9JlrjgAwSkYRgGekI0U8b6oCLDVpQcA8I/DIjPTKYHRcRkUB1uSwcUVlkoBYVhkQNYu7QKqsihl+AGyZDJa1MrrVi4uVH7fAhWA2iO7L4TgBLrUJ+ns8SKalShiX2xPjwAxLI4sbtfuLA0bsRrSfkliuTeRIhxWKUUBKG5aihqhmcorUDPIVRX2HM35ZbZ2VgbqvSgBekZwHoCsVIAUpARCFctH4ZSOQrnRULlYZxQ2o0oBQJeAESEul7psS14u1H3coB85YQI4p4jWkRfJiwJrGYbv+DP7AkWqa+RKQnkGb/dpnJYj0X/7ZV8yYB2ikQSD98A6iqBseeuTR8F8VzKRLetH3Pwmw1MemxMVFnJsJL8tvuULgAkDyXPOBKefcmr9HPs0YT+aiyFnGDg2AZo8Leq0UK0kRcQsQOhFsD1fKGsRofDrF3Y10HNW0zc4wKdBsbq6VRBz13wj5M+gwxUFmSUWVyCgEIsK5sgRJhXTY5dK7jksvCiZOykkfZ/ypMU29lPsTTvilUifA41UoSj1LPUOI1SWaS29AqmPASYoEcbiEw1PgGgE5YDouQ1eFuF93xqfNWBpENxdARlUEwIer1eS4VjRIRPdSjXDT0gpHsV7lC6CkF1A9eSdqB7k7rcDPeUzLBKk4gHS6rikBUj1zwoFaZZM/gCaUBtL43tVWOuW/JPtZbWIZ50W0P6v6LV10nlugWeL5//W//7VZ2xFjTweiGLIefuSR8M/f/OGsiObnueoLuh0GQ4wSM/oRY5U1otC/dctL4YWXXw9NdfL62LjRnnsuMKVNOIdHwqZNm8Mrrzy/oHtneyy9LkXoRHPtnAAporlEdWCFuKe4QFWVsKSIuEGl9nR0yLCj8JHyJkBfyfLDGYFQqUDKdPjCPRVhazMZh1oum4O4Np5VurwwI+5xRFxgiRzjR4ZxkRoUgAo4AVxRdYPWyhJAN9bL8IPRU5wwA40NBtOyb4iTjXpfhc2UG1KHOHOMm2VTmtYsHSdcqWqjVT6btLyL9Jh6lqjzV50EiAaiAlzfh3OtNS4YbjeCp3hm40rHNPkAbpRZTg0N8njRvacEvjOoOQSkJS1taqNkFqLuqoNs6zqIkuCAWdD82byn28lKEnnvK3eVBuDJOETk0pb2fgNbAIvsX/0PX7TgI8QKyHXu/MZwThRx7Uc/uS/82/d+qgz54JB/xfvhiM4lTlIcGQbABhkrILw14KAIyEHH/+o3fmiua5/99Kfs/FwcOW2D+x2BWm697dPhwQd+aPkNaAQ8Ttb5/GCOrXg/6+RznLbkIXSh4vBYc2pSXCPUKi+CMonpPQpOgp4Tj1MnCywiWxNPzZuRmci2NdKB2tIiAqYalTOmNZTKygWo45WKji+xWopYgp8Q9KREqlEANiPVAusx5RF6U0jTUX1dJg4zNQLssQgnYyN9oUMzPZfLE4AQ+oj2aWLW4GS97m9GpnimfKbfdibGywSe4o6ZIqr2ccIdamZcUZYE2EEzyKe1OioRrwBuoS4oHX9+gbVA9uBd7zDBIlLuHacLXQLSdGv8hu5ff81l4aqrrjK96FwgStO4k/199/9UIHr/+7y1IpRgcS8TODRp6iGhCIfk2M2MNrgqpvYi2DrkfPXrP1CA8OZw84032bNZJy3ylLQRs7Quvmh7ePHll0Pnif2WayHgWVgc4jsEqEJwqWmaLNXyxlOKWi//zRJZuBVySVZ2Apho0ovUtGPyb66QQcrXSuLauBZTupTUvoBQUKWI9UqTNV/TmJKTeMhotpFE80n5mKIGgMrEZUIsqMcAWkgspzwzHOtsIOeIojB4/b0nbUmUMCmPHbk2lUvXC5VKGqgTFz2G14Tm9qNSwF80TqOX3J6iCKh6JrlajQKaojGtHVUhg5a1FQAKkJ5D4lvhNpGyO/YN+Ts/tzXwey9t37ctgEj/R1/6XZtyChc2H+Ej+vCjj4Wv/fuP58v2np5DhDcxXgDDsjSsPcXyMMRqIPIWIIpFGLc42EHyMpVzRG5e+Po+/+xz4SlFgTrd2um0FVNlP3nH7eLsYjea5epUpCXIAwA4YAKgxpkKUB1U05cRxm8w0fV5es/wSdtlzXpzutfzzEesx+RrMs3M5CYVYFnHH9RXCZ2WrjNG2ReAlhOJPqIiy5bgyzqLCN8nkHSCK4VzZskQfuwDhDPTcsQXx+kTBFj9FKoUmErLkgVY0jIluWBAgCc0rsks/Zpt56uNliq8X4Vc9QzEnBu1nOf3j4Mod10C0vPb9u+7u31RS4i4+9Jc3Cgivc/w+aev36NnyOea3j8PhS5PhhQBHAv7LWuRMUKAyXpgzA4rEeCVSBRmMoVxouIsyUtcB2aYEex6XP6Nu15/xjwReGaevRjRVkyGoO2u2nlzsSxF01Q9A05OFgPO9EUz0mVCiPeTWhakRNNVW9Yt05RRLYR4TDFzpTNMk+lEbWXQdOrs/elMHDDdOm7uPwJVCMB00CwE0PmWbGbNJScAlB/cKj8mpMBxMmOJVUezpLWmpjSwFRIri0Jwnk74OUO9MpLNKFjNwmh+xqCwDIBxrl86L3pyN5J5+hKQekv8Bm5ZoI6lQXzKbbEmAEjgzpip9f/+7dcsGHaxfO9tGmI8c57lBC/xvUUAysqbiPEE/iZeQxRLo7iPTnhG4MlzswTL5k0bxS1N2XLXbNGbPvSgdKZ6Zp59LjDlnpRx6Y4dAos41Zm0+cjBsxgn6uI912NpZsYnxhSIReUwCJVrquimtdu0xIhiCwz3GzhpipHlmetPiaYP83MCcJ1w3+EHqPoPwCYNcqONg64+AFVGS3InwJuR3hlOtJCqShWDVC5P/NIg6uvcW344WlEeuOq4olRxYjXAeZT8CqkxfM17K6uCWYtqnEQvisGtOOWes9j5QtAslqdYGoZWfmnKP0qfWdr/NW+BTPjyFz5lQDGXLpAGQC8KQP3N3/1T6FR80HdLgBjz38+OohiOyM4PMCwmxhOnNivGi5OZ4cPXgGBivxhMYkBgTSY2A6H4jii4yeGjx+XPSIyA6CqEhf+H92opcXSsiYhbrM60HWB77bU3FDs9Ky0NloUnHWRJR/THFQgal350WIwb4DWt+KQV0uPWrdti6zYhLrOGPEuOwGmmRXe7WH/Soj360xkxd/8/e+8BZcd13nnezgndjZwTEZgDmEkxiVkiFRw0HllylMf2euz18Yz3eHzGnrOzZ7zn7J7ZPWfOzK7HM2N7ZcuWaUuWrERJlKhAEiIJggRJEAAJgCAAIqMbHdA57v/3VX3v3Vf93utuoBHZ3+nXVXXr1q2qW3X/9eXr3CWgyU+T3Xv1CctqWf1zpIgtiDnucY1qbpbuNuUgAT//Uae+SlOU6+ecaM248vGqLT4CTj5Jn2+PK7fE0GhNGNdxTHxH2/PmL7N2me8+EeubjHP0Y6r0LCEA1X++L7uMwTO7b7LtmAt1rtTLZoF0st67TPdfowxaNypVHEaTcoR4+8JPXgyvKjZ7JghO5w9/7zfDA3ferOZKcRLFziSxXeAGNStHLcYj0qwhmuPO5GI8U0f3KXoJQESMd0I3OSK9KVzqImWEumLtWkugQ45b6hNYkKVTijnHmEQflCP6kL6c25r/QExFX+ptxn6OXgbg4qTPPj4E7RIn4Q4BMDhCqGXhiqS6RORiVAxUqWfGKIFzBQBIEpIMmcEpbRIQBXStLhZ7fjqGifCwmsNN1smrAAL8EOkBvNNK39evOaYASnS5gKjRqI7XDzCNAXV8JPUGUCU3MLGEA21S/gjaTGYUFbcMcOatP0m7Rf8XivaA6FTJgdLrO2B6+SxH6j3zAV9+4olHTNwtJbZSTuw9eU3/9svfmrHeYp4s5oL67d/8XPgtzZi6VKA2NUr0V8yVhd6TZNCI72QVI6l3t7JuIcbDaXoCYo02NZ2ANVxolfIPbFi3RudvDfsPHAyHj5JWsMHmzfIhhmgf07ZXX7A+oC/K9RWqgxs35bnSeNBmDUwx58m54D6zYEodfm6Uol7vqBInp1Mfu6he3TDPLPfsh5hPyamY9T7HnWKxj0kO/5aIhGxSAi2mXcYPNAe2qh8fi5uVi+TV1cqML88Bfg0Nc0NLq5KrqI1+RSOhA60ViFYLUPnlKAXUyn4ZqBxkczv11FJDkyWDZiaAyoYwoulxRhTZVVqUjxpIV0txoP6hc4DMHulA6cCZ3Z89bpYjzfbQB2B73cqlpteDkyplYKIckfbzf/PUjKfA69Q0M4jMjz78UPiTP/79cO9tN0gvlhnYmeeA3QXOEwIwsb53ptFUXcoGRLpB8nHCvWFIwqDkYn+/LL5LFy8yLrRPDulwoQx0QNTq6n8pQsR/+tvfsr4o11f05VVXXVXAlXqbMah6WXaZZH3Klibb6EkR7y0ngADPnOfRVYqwfCPeV4grHVA+UeZsylFax0GXctOXqp5ztUJehYGS2BkQVrJm9QvO/9Wa7nlM0UTmIsWBasvbcQ8AwLahrtnS7SGCE5kEtWpmiKaKRrkr9RpXOlLEoGQV9W+sQfNOKQcqhIsUOlfagrt1gjOtq9TzVTo/DFexKspF+Sy4JgBaGt78mThg5s6VGpKyQOn7Sy1Ln6nUEbPll3wPfFjTViOulnN3ggPbv3+/5mnaeU7ul8GAMYeJ5/717/7L8K/+5S8HAL4U8eLDiWYnQhyVEYZMV6Q3zBNcaOI4D3d61UZS580xLvSYknozhTccZGn4zLfEGiI+fUGflCL6kj5dufb6UlUKyrNcKTvL6U/Z3yOfSwDP9JuprhLLPWJxMq+9XPMVKZSjLNeZ7shxrQ6iSkAypg8bBJdpOtH0WANNrXPMuGYMqBQI84tpQJ4OIwrlRARvbEpmCq0id6loTB+uUjQkAHWrPT6mtAPRltPQYOKo79uWC9g3Jizhxv03YWeuYLogmTuwzMoskJbpnMtxF4ae2269xXwgS3FY3DdA9/m//fI5tdJzfqzecHN33n57+A//7g/C4w/cZTlos32P61KPEl8j6mFpByAR0TEsYXgChvjBhcLtksQbLnTt6lUC7B7jQhHb4UITkZ/Bmh+w2ihLL2x+voATylbmXvArvfbqaxSTPnFYuegOgDpgxmDKerydbV/xoGFI3FjHgO5V3CkGI3SgtdXSSwqEhtJ56xHHzbLuDaRcqW+yrFCqulg36iAKhwlZtKj0sAPH3g8nX3srvP3mdsuW1SegM0AFVPXrUcLwU+KABxSvz1xQEM71UAyG3QrrzHKlbMdi/fBA/kPobVlDEulxocIValz5U42DLKEfHVeCF//ZsfpXDDTdP5Z92Z8fN91lXhs/3SNn61+SPXCNuDNyazLoSxEzD+zes9emNClVZ6bKHcwBU7jEX//VXwwP3Ht3+MJT/2hJofPnAZzGTJyHuyRxN3kw3bBk9aRrxJiE7pQ5spiH6l3NPMDsAXXilgBaRPmpUFZv2XHqWDh69KhNsFdq+mkAnL6dO2+pcbF+nljPSVkpwHS/Ueq42xPrEOBLlFO7fErHO9o1D5JmvRWnWF03YFwkGZsI8SSMVMnjJbonHKYDpovl1lgKroj0wwNEejG9dWJAIlc0me/7205LhdIVOjVbQoc8BY4ps/0mTW9y1c03hRqpOzimUyDar2Tj/eQ0FY0qi77zqsBqpbjSSs2KAQGmBOfGelJEevSjg3wVUid96kJwoviOzlF6wO5eBfHKCb9ViZz1fS9CAnaBaDHKiu7UcbG+WP3JynClcyIXglN+zUtml5d1D9xyE/MQpTHKZe6UKUIwDJ1PAogAqQ3r14U//oPfC5968sHIVYrkF8nr2qlpLE5pWhoDUXGmGA6w3vfLQX6VAHTtKk3JrYz/TDnDoPEpaabDgWbvm3bgSssRXDx9e8W6DQVGIj8GMIx/Xu5LwDMGULPYSzkMwAK+Y0raPCjAPDxeH44or2hHe5/ALIEukpBAQ/KjBSBzhAog5kodRMVRMjMogAjBjVZU6yMj0R5QJRS1X5x8ncDxiis2hrvXX2n+ou3t3Tp3m3Gi7d2d4WgkepO53n+06dZ31qEe3QWcaMydUuZcLK5P/CAs9Pxw9K+UuqButSb0k0O/WGHbf6b/Yg40BlkAMgZJ3+fLYueLj5nlSIv10GVaxrQtt968ycCq1C0CBCc0t/i2t/aUqnLOyxH3MXT94md+XgEDdwRUDG9q8kO4yUpxAebWKVFaXuMCylEZNIZCq3StizU9Clzo3vcOWx7LRIxPOZAIW870BuBK6Zt58+aZ+qBYO3wIrpHR6bWtL0zgfEpxotl2HExHo2lNAFMHWmFlOK6DxnTf9QJQpSMJLWkIZy2hU+kHB0A1cPQTRCDqnCigNmeu5qCX6kA5SsT6JkC1YPWy0KCJIBHdoZqlDaFVZ2L6ZXSZvZpaHCPXXLG/wwvl3iQVS688Jprky9k7joeEQnH7FD9fdTrURS5WaDwb6pIPIu5RzonS5hBpq1Lql6qidsWKULVwmQVZYIxjqiCpw2W114dFHGg+kYgflV+yjzoAZykwZJ9TzF1S5vtYxgDr9cf1bDwtIGWzHKn3zAdgefWGNZY811+SYrcM9/biy1umPL98sTbOtgxxH+6OuZ+Yq/6P/83vhV/82Y8q87qm4NVA4gfBhTJZIpP/LV+WcKF79x0wDhWDEsBrvxkAUc4HV7pj55sRh0tpIdG3JCieN3+pcaVZsb6wdvEt16dm9xpnKhCBmIOLFHSjmru+srrJ0uJRDiACnlj24TiNIq4U3SYg2tOJRf20idpwo9TlN9yfHIP+kwxTC5avCk0LFuuempUcWi5M8iPFut4oL4sWrTcqOKCuXg7y8qhgsslhuaJBGMYgLPeD8poY1DW44QkA5Qd4+s85UY5Bv1qjj37L3MVKDq25zvg46Hj8R0cVlRWDKOv8smR6XhUCojwT/2XrFdt2TtOXxerEIMr+iVdQ7KjZssuiB65Ys9xAAJAqRuSNhKPa+vqOYrsvSBlzRiHyk97u//j3fxDu3HSDXQflWMmv3LjBttHpdsmtCsd65qDSyDvr6wUE/eeNHT74jvWR59j0cl/St3yMFi5c7EUTlrEudMJOFXBOr+Nc6JiiuPhBvrSNzD/nQLHso+/0PKMmEguMANEOTfOBzhJnecR4wk/5YXQaTrlZwmQHBYYjmpWURNA1NeIGlcgEMGVW0Yam5rBi2Uqz0tfLEEgiZ35EMeHzCcGZ9vSPhW6lmupTvwCm+JYCnhCADDmI1trEfwBp+gHQ+zjIjKgRAablONGo6hmtFuM+yzUEZwrNivbleuky2oef5vp1V5QUc7hVxPo2TXt78PCJi+bOnTvFGIWr1O//7m+El195JXzvB5s1SIc1Z/oxJSVR2KQyHBHNxCytM03OIQJwXacHNGtmh4BSs2+W+CDBBTEX0tu78leSBcb8nsI1r+elvl2JTJsS6wmYKmRSxf0SievEpps/glgxD/9kCZASOkqekj6J9sOKjhpRX82XIadRyZEBULLnQ8AXXmR9JIAOmhJZHGo1AQAK+q9BlRIRYFol63+LONBExwl3mVjv8azgNyRghljaujhWo/7RHIgmBYX//TjaGGWa6ZRyvqLe73qnJ6Ny0pcfWw48syK8H+NL50xnOVLvkct8iSV7zapVZa31AMC77713QcX6co8B3SncKa5Sv/tbnwvLF7fqfgbMpQlf0kIQhXs7O67UATS+JsDzwPvvl/0g4RGxVJO8ERrp5Jylb5/NkrR/TjlwSQuifCRexUCUOH2c9qGW5oUmtvcIZIfhfsnkDFgKRdmGKjTNB1RXV6FZSJWARNyjkuIlZRVD0nsOmfV+jgB5fkurASOcKeGiiPxk30LUBwwhlnCazm06V8o+nPAhdKTOlVYsWKCPhPyGpcoYldtayoybekX6C6s/2b8YREvpSSdrw4EyWw+A9R/7ZoE020OX6TYuQ+WMJNw2HOnLr2y7aHvAuVMA1R35/93/8lth7col0TXjR4pvqfRi9oPbKuSmospTWs2K9+/ue8/6qtTBgD19bQmXi1SCy3RO03cXK/N95ZZcW1UqIgOC8pSdUJ3QT2L0cY8aEgBUabZPxPZhRTLJk1+i8pDsU4oqIqGIgLBeM3Xya5bPcYPADIAkJJQfgIrLkod1esgqJ3VRHWAEUBH1AVTnTi15iUR/wBTuFa7Wj3FDE0sxrIkKCunCuU+1X+C2VARMXQ8aA6h3RrGyKqksynGjfixLZWqwXwye8f5Z0T7ujct0HbF+2eKFZe/O9aNHTkQhhmWPuHA7AVTAlOWmTZvCNddcY6GsL7z8upzve0OzuO85TQmg4CZFspLCyKepXTsgVYxG+o/n9KSlxHuOq2tcEE73Hi7WREGZgypcq697hVgfytxMEFnjbF4nifNwzdJ2imPTLKOaa6ulSVn0K3Tv0keawSlR4Zm+NCiDFIawXqWSGlZdwi2HdVx9ms5uVJn4a6QeATiNPDJKKgGMVBij0KPGvqCnlREMy74bkvAIHUoYTGtnYJhIqeS6fUI82h4VuDvhAeBO+IT+QhZyCucdAeYE3ahAVnKIN6Pl1DhVP4A+k8bZNycsAc80T07BWagIpwqoOs0CqffEZbycLzeWdWtkRY5Ewuztun50cFAD8RIgQBQizBRwwJH/8YcfMFepztN9GvTJS7544TxNn9xmOkXE/6kSgAawFSMFWE2qJ6WvFy1eFk7L13JwsLdYMznQ9POUA1FBjwauN1N4XYjA0PDwkCa0U6o6qSKr0IuqDIOToleNmI9pWNZ4pnkm8mvotJK8iCscF4fZVFdn3GZFCqIkkh7U5HijCguFsPD3ylEfwsm+RR4DHR0C0VNyyE99WBtSFyyrlP4b074R6VER9+MQEAdVZi2FQ62S5wE0NNxrMfW1ivU3az2FEVfK5pkS3GfWzYntmCstB57xeWMQFafcMAukce9cpusYYhYsKJ9lCR0SyUROtiVRKpdKVwCoiNL8Vq5cGf7Nv/7t8NwLm8P3fvyKuMYeMTTi1KSysHmaQJUp6k0d3Ir1A8BIdvzJ9G7MO39cDKkwKweaxdrLAmixOvmyLIjmueZ+cZlNMhxVSWzGRgR5AuZkK/+f/C8KVlK2/c4w0hlCuzjY0DesOZZSg5CqYmWHYj9QtuE+O/XDrcnJHPFD4QeDiKhusXQt8gQYWrTUdKbUBzibmueFKulfh3vE4WLpTwmxvkJ+rfQteRRyIApnCqD60g+YwjIGysmqOwearRfrSgFRpoj2qamHBnr6Z4E022OX4TbcGXPQ8ysX3tglEe18RzPNZHe7Iz9Zpa6SW9RXvv50OHikzYCUKCjM0sTo41t6tjQgzg7wLkX0NQT3CsViu4O0A2g80MfdSTY5LPpfCKC+AwMb3O9wVaLK6GVeefnQVkikTvxIZT3XoMdqz3ekRgmg6QcE6LlypG9Uflj8SDt7OsLJvUcKwLEzDf0EEOfVtliUE4AJzZ03P8T7rbDIP0B0KU71DTqP4vH5oOF32txSbxPfCYTsKEGqAFZ6W6ZvaZxr9zSitIEmFACgzpX6ssi5ShWvycGKAABAAElEQVRluU7qeZ9nOdRSbcQcKHUcRL3+LJB6T1zGyxZN1dBkyTpK3ySDEd/MS5ngTtFZ8rGAO/293/mfwjPf+1741vc3m8GDOHwAlVDDs/U15aNTTlVCP9bLao2qFo50MsKQ4jkyJ6tbbj9caY18NcXyabQTLproHHFzqvN5lZjDRMVMwVyppCcA7MKFy8OC1sUBnSfRTsPSQc/XdMo1em9YhxDjG+bV5UR5wBRa0rSyKACzjxBTpiJh6hG+XyPq+/qGRIzvkkubE+5WXd0nQkOLpokhM5kBpq7zDICTNgHKGCQdTB1A/bwsfZ+tS/cJZYGTspgL9W2W0CyQJv1wWf9v1GBgUJcb+IhS7e2Xllhf7qE5d/rkE0+Ee++5J/z3v/xrJWE5aAlNOk4n+VBx9xHa6Dc1DtX1phh4OgQqJn5OMtAra0grV/iBck40e/2JVbo45xnXxQjGNZgxLFWcci1Y3sFsdKVDSoac9f3MtZH6ZmLFN22HdozbPPJJnH1PavABRFsWMYEgyUKGDVS9DTc4edz8sHSgvXJFO37okFcxrlWK19Ct8srqRMHrRqXT3QoJTblR9KUYnwgLnbtmkfXriHIAVFeKEy1DWWDLVo1B00E1BkgzGKX958fG+71sKstZIJ1KL13idRYqjK9e/n7lCF1iT6TzKlf3Utjn3ClhpuQR/e3f/LWw9bXXwle/8YyAodIyR2Hhx2qLI38sUZfK0+oiOQBG5in6rJR4z0eLPrfwyTSsM+63eJDH5aXXI1BBnC+C/b26CWKF4EpJEdLExHOmQ06OVRCo1BxKBCId3whJnAVUowTup1Rdp+k/mGhO2wNyWwIku092mpXeAdPr+rZzq4BoZ3eH7YYLNcOTQNR8Q5U53wnxvVIqEYBzUJFMdWl8/THp5uFGG1sWCmBlxU/sZ37YhOWIPTBZztUGbkxQxXjCfbPu811hPIL43lnQFkxuarQDNF33OVUABbxjQsQfU+OzQBr3yuz6ZdkDzp3iyH+L3KW++fS3w3d+8JLN8wQgEjWV1Xmdj44o8IucwgnLSRQAhTAzRybi1whYUlGVHSOyiA+NyaqvMjn2WF0T+VMwJQUffqE19Up/VyNFanNj6JMHhINmrvF0JQZRDE+AZ8OKifNbIdbLk9V0oBxqIDrQmzgrSdQ/Le8BqGnVFWWlJqukfwmYJdDFB2lEln8HTK+TA8YIkB1Avc50lwaaGSB1YJ0F0un25mz9y6IHmOsenWTLHHFPyjLPnE+9A9ItimzWUIHKuSAGPgal1GNpyqcoB6LmJhQ1OMh0yuK2u8WVtogrBTghcpZCcKRmKE/ZvhhMicX3BM/UJRafGKcsmAKi/WJmseDjDtWQ5h1NkpDk/LR0pOLlBfBkgvWoJl9irWe9u1cJqlesUISaptCWfneyj9p4BVmt8+fIgijXDSXc5uTPMQe6yWEpUKcbU1zMAukUO2q22qXbA6TTwwhFjP7ffumbATES9m1coYdM28xcUPOU6ciplGiPbtNF8lJ1vA1f+lTOsdVePFmMA141XRaKjuUAlANyvpY44Evc7xfINEjEbcRDo6dTYKqQUIEpBKACojERDUU0U09qUCLkE2B1IjbfmTpyhzph/IcAUXxRibfHJ5SIpWLkGfOJoyfSyV2eevt6LdPTPFn2+/vz7lTehnN8bDvA+jOwslR0Zx2O08R3rSfLvEM9+4vRTIAo7c4CabHevczK2to7Au465QigmRP5EJareynsY8ZPQIwMUZ2dneE//9e/CK/v2BPF46eJouX+gyWfX41CG9GXlpotNL7vZnGy5aKaqEuf9/YkkWKAcBKlVBxoqA9oYjRy8GQ93nYmLGI+dZTrOPPgR1v4aDboVP0Dco2qlSVf0xpDgGnfoHR6yubkhGXf4+xJDM28SJSRzMTUAUokrchyA8uhKoV16pSAJlRJOn6delD5SD1ayZ3tx0Z6dVTeRxQQxRDm8fd9Ak4zMG1cZZ4URFs5WPq1xUsHVQf2eJ+vn4n47u16G2eynAXSM+m1S+yYPnEbA3ppGZSliME7mdN+qWMvxnK4UO73a1//Rvj6d58LbXLryRL+pPQIvqVMqoeudGRkoZhVcatFDETx8fPkGuSAF5f7uvf1YF+7FwmglCCkNI5avbjNeJ2dhQAacaO5M6B/FKql2MpUzZoHRNFVUncyz0eKaYj1ABoEsOINm3gwqE0BaEyDUkMMS4fpSUaGBZhkk4LzBDgdNP0Y305ANTlh4iOagCiAHov0/SODoTnU5vpyKqDmYOncZ7zt6349M7XEoFRJMEAJmgXSEh1zORV39/TLMTz1DC9xYwx8cnleyuRcKJFMh+SG45n1y92TGX+lv8OvtKIyzz3F4qO7zsTttCrjEX1Wjivl48VUGXFbcRtTWXfgzYJoqWOrxFEryN52M1Vzvzjnng4SOBf/iBJr35hx6CCUFPEecJXLfuK8JfUxYIrLUp/0mi6iA5y52HxNo+zk3ClLrPOjafCCZ4FCpI8pp6KIC0usZwHUq00FRGOgLmY88rayy3IgSt1ZIM322GW4XSVnQVK7lZvwDvABHEhwcqlGN8GF4o70ne8+E778zWdlxJioc5vq48WiThb6mLKASJ+Vcn/yviYDVJ/aKkVwnc69Zus4iGbL2S63z+vDTTYoD0GY12xgytK2vYKWJGzWDB4FPqeuIwVQ6+qqgulN5zTrXnUvUgsw3QiE+gCqSKcgMT1pmrTZdugfxiRAFA7Y1QuAKCI91CDVgN441my71D9AshSAljrGywHPYqDpoFpsnx9bagmHGlPhmxLvmV2/bHqAWTQnc7ZnQM/VNMeLFs4Ph4+fvGTuHS6UpCWA2sGDB8P/89//Krx38Ijp+c7uJkpzkg0Nc62vsqJ39nzHjh8XkCTGHTwEpuvuFLeX59jQYbInP5AR56vVB3Xqgzr5zI72SpaXftIJ8BxRNBGcaQymOfFe+8cqpOvU9M4Yn9yRf2ikz2YoxYoPqKIaaCWIQVnyAVUMV7QBsDqIDioQwA1Lfn4/D9twsg6idZX6uMsdq1kRVUgGpqeGo85QzGnG65lqUg8kfVJMz+r7ssewXW5fXD8LnuyDU531I4176TJeP6UsPfsOHCsrisJZkUOzrq48Z3CxdBMAii5z7ty5ptv8q7/5oia9ez/0pVE5+I7WNeDuA+rkDStTuf7E2MSgzIvDgKCHcBL2OVluV7jMtrYTOZVKFkRjEI7X/frgOAHMiZxnYvF38MTpH06PYKXTnafC69v3irfrDvddd5U3ZWCHFT8oeqn/tGKfiB5VtiWMQcwX34xOuIr7Sw4hLym60lqFj5I2zzlUzwQ1ls7LRG3OPSppxy32o3K4d/cmA011o3NrJCQhlh5qaVIUVr9mfpWO9KVXtoebbrrWkmG3t53UxIUJB+kcqB0wjX8AYzEwLdXEZPrP+DgHTi9zcPV79PLZ5WXYA4jqR5VKrhyZ1V4WbrLO7ztYruaF3wfQIcaTGOQnL70UvvTVp8OuPfuVS7NGM4nOD6tWrhB30xeOnsTQoxhzcVRnm6jEQZS7J+yTeZmImipHpNArB6DZY7OgGW9jSU/iz3WUnicfEITq4yeOh4OH2pVA5Fjolm9s56mecPWaFnHpN4Qhi3BKwI7g+uoKOcbLP7ZPv1TzaWCq2VNCMyFRykeaUJIcYFjJbmrQHSuzPhGkDqBwohip6sVs96TWeyqgL0UnCoBCAGf1mDjWShmYtHQQbahNEJt56xvEir69852w793d4Y477wnXXb06nO45bdxpzmqWXFQuIindLFhkucrsdkHlzIaDY1YP6iAZV3fQze6bBdK4ly7j9dOao5y5hspxUnCld95+c3hh6/aLsicAUFyaEOO7NU/TF/7uy+HFrW+aThdgVeBkOHz0uIVmLlEi6/VrVoX3Dx222UarLRvT9DjTUp2w8cqrxeUlYFGsDtdHX4+OJtxXsTrZMgfNQi40P8WGGEYD0FqJ7nMEoodlTHtbU1SfOCn1gQCwWlnvIWY4rmpUdn75khba3223/Etxf0rBNF0HVAHTenHREAb9xib0vwJWC78UTKQGKUCU8FIio8Z1hlFl2h/SFM7D6RwnGJYA0FIEJ5qlOcoK1TU0Hn78/AvhwP5F4UP33GUeJKXUUTG3WQowp6v3dBB1oIyvMQZa9mdBlLqzQBr32GW8TuYj5hpiLqFSIICIuf6KK0KTBmtvKoJdTF0CWEIFjvVWQtwMVGmcKq5M7x04FJYvXWLc6b73DmgfQAPHdXZgigWfua+KieNq3AhO+dixY2FQUUJO5ep7nfwyQSKfp4hyRPlmGQOhnzz/XHhzxwG7k7lzUV8oM1OXcpHKwdOT2pOQRLBq+yrQfwr00JUSOgqYdihpiJNzqAMS95v17KG+3hSGdQyEjydEQmimZIZQC8jFX1ytpjGpHQk9XcwQmnf4hwOtFvcJN4peFBB1H9KhQUIFEhpF7ypdMjml979/Mhz/yjfDffc/GNavX27nG04NO6MyXHEdLvrHIIrBalRo78t4n58nu3RAdBD1/V6e3c6W+36Ws0Aa98ZlvI54z1xDt996a8m7dD3p6hWLw653Lw753rlQQJQ0bv/1z/86vPrWbt1DDIjRulZJSoIr15Fjx8MVdTVKBzc3tGvKETjFMyUX0VsVAVWOq6d9QPPE8aT/pgeg6ESLsHMmys8LPbJ2P/fcZsWm615qK+QEPx46OgftI7Jk4ZwwX2ntdu9JzusT3SGCy2IkFywMRHkwrROYDoozZQkBrHV1eRCkDA41D6CKXEJUVzITBw3296hsTHpOHPKdqhXp1C+1RkzMU58D0RLcvHBUYJ/c1/ee/WE4fPhKawI1SmNjU5jfyrNcYGXdmhkBYHMQjEE0Pi/rDoBeNy7LrrMNOReabE3+3/tk8pqzNS75HnjvwBHTHZK1qJj/o+tJb9t03UUDpAAiYPTVr309fPvZnxR1rI8fTOIXiqO9EllooA8OJiGJ8cybcf1i6w6avozrrF1/bVn9KH2Lfvbw8e4c18p1cA+I727QoU0X5+P2Yy7Uy1vl/H/yxMnw3PObrahZ3vWdnSc1dxOBA+MBEF2xZEE4cPiY2fLn1FdL1NfJhgWEzOWkjw6eDXCmAGOtjpHHkhFg6gRHCc1pSjjfAV2zE37+gCjg6eXE3wOip+ESNTWJc6MjChfFiIRrE/rRBjGcTZmP2PCorkvz3WO5r5K6Riyp6UAB00ZN3DEkYN6x6x0/vS1JDdDcMi/ceMN1YcMVSy2kFKOiAyRgOhVyYC1Xdyp14uO57ln6gPTA23sPmAGilN8i3QAI3H3nHSbeX6huARwApNbWVnNp+pP/8z+FL/zjtycF0fz1uqjPbJTijpThaVwAcLaEX+l1195ofVSqLfoWI09nO+qE0hSDqE05LCd6ljEx7VRzS3NoE4j+4Ic/UgSW5qMX19nWJv2rKtbI4X6BRHvKTnX3hS5xp0vFdG9ctiR0DiT3a36i6DqpL59RQkMRzd2f1DlS2695kgDT9lPtSqmYJHgGLD1hCUtAtF+p72x9tL8ARA1AxYki0gOicKGQc6K2kW5j7QdMoSWN+Xj+YXHZgGiV5teCO+VXp1sxqBWud3R2mC71h89tNeNWs96RqYIe9fg58I6nU6nYRehfse1smdfNLmc50myPXMbb6D1f3fZ6+Mhjj5a0OCPeL168ONx8/cYLZnRCjOc6vvDFvwvP/OjlaTnWw4miIx0Qh3TFmpX2NLs0F1XNDBibFi1ebX3Dx6YUIYaS93QEi01KLt7H3GiyCzic6DdZrY8IngZkij9y9ISBaH3DHMtUdUrTf/T1J/rEWrkhefIPxH3842+94YqwQOnvEOPdAd7BdHx80LjS6hqlDhysMV2pX2PMmTKhYEc7+s360NycT+ZidaNbz3KhbpWnHiDK8U3KeerX6G5Rfk5fLhAj2ddUEY71SgWhQmGdCC5a+UYHmAZZorYAtVLsOnlYcfXat3+/fKOPhcceeUB+qK3htDwoHCAnA9Z4P0BZUZGofHzpZb7N1UxGs0A6WQ9dZvtfe+Pt8PCDHzaOr5h477f7xOOPhC3bdp7XKCd0mPx279kbvvDUP05bvcBsmoBclbjCDevW6B6rA4amGsXSM0CnQ4BfIedepUz795VtwhM9v7dvb0G9mPsETONtwUauLtxntWBjWC5EPfKyOCC3pt3vbDcgqpFcC4h2a84k+MtGGW5GZaBxA1wQt7i8uSIsXb5MIn06hUfNfOOOmSFUE9ELXFN5XsfXVcrrIDI+cRFjStxyOjIWoeeEy4wJcMyWsd9B1LnQOWnGKVIe15nonm+lGKCuml8bTivJy2n5odalelLF12oSP0kXKlPSUbMVKseMcacoHbrkavDNbz0TPvnxx8Kc5jnWZ/mzJGvOUdamKQOdC2avA6XXiY8tVlaqPuWXLZDCmUB5LQ9DKSnjC1dY7sMs2c9xlyvt2iOXmRMnNEfPwqJ6Uu4bMLpi7Zpw9Ya14U252JxLQozHsZ4sTei7/ux//GV48dUd0/Qa4LklSUfmyTVo+bKlApwuuUK9L6yoM0f6hNcpfyfMuDkSAUlce9HC1rBs2bKyYj0fAfr25ImD8aEF63kQTQAUazx6YLjP9w6cDNu3v2WcVq9ydKKiXL1qkYElfTOQ6jMXSieKmN8VJWKpEbfWuKDFuNERJSqxhCU6sxmbtLRoJYEp7Jyk5xy3Snhn2+kO6U1lXVcI6bASOxunqVz7gGaWANFiYEp2e6heH62YPAN+XFZqvVk43yt1Bh8kHKyGpfCuNTcEieQAaUQ8cXj5ft3Md5/dIjB9UMayWkki+tCkHCbVWQcUh3TfcTn7IPYBsuyfjIqBqx9TeNdeekkupWeRczCO06ManMmLpzjfSGHutwXHUiuXiwaFqFRpmkIGM5R0ZeED82MulyXx51tffS188hMftz4CxIoRL/OvfPZT4Q//t//rnHGl9DscFb6hm1980aYB2XfoWLHLKVomFaE97wHpJDEmIco3NiIOH5MurdPmZ9LoYbT4N3RCO4kqQNE4AlHAvF7RUCNSThZyoyHc/+GP2wAv5TrGveD2tHPXThPr84Dpp8xzni7OA6LzpN/sVXKT7373WQUVFAIwRqQG6S37paawfqoZCzVzFS+vMrjTmIalU2xJQWxYGemzA9vzjo5XSOUg0ITMT1RuQwaiSmpCtFOrpgcZkCW/RqANoEJuRGLdwdWXlGXBs0bX58SYqo3eMU9mwn6MTXwbmGmUaZsxSqXh+2aUG0/nU+FdJNTWhqkaZITGoxS96fe+/+PwqMR8/Ef7+rpzAMp5nEoB4VRA1Nsotcz2d6l6F305gEjkB47QACWJeldI6b5oQWtoaW5V1IteHD00kseeUH7OEydPSTTo1cs0EuZKLKhnjho9zCQCpji4XPSdMMUL/NHmV8LDDz1oAFZKvGfe9rVr14Y7br72nOhKGRwA18mTJ8Pn/+apaasRTOLQY4J7bpA+buWKZRJjB00tgGGJtpmq2IZc0ccpnZtGJgMUw8bi+fPMKDUicKuSpRkrMoSIv2zFBusL0uyVIj4GRDq98/aOVHSPgTM5Km+RT/bNX7AgHD16NDz97WeNs6JWksJPBiFhnYvtDqaNDYlVekQT0ZUiy/hUZCfJm2NiojzCQselL21J/VMxJGGZV059We7VAxiXZFCKQTUGybi9Yuv1AtABQ79kr6fYKybaA6bt3J9CWOswvMlJf0TcJu+ncah6hu4ZNqZ2nVjjI3b4aLuB6QP33mmW/dPp/FHObWaXfvx0lmN8YDOJbPz4SxpIGUy8eHAkbZ2nTRd2wzUbwvVXrw/r1ipZrB5OSdIgO/D+kbDj7XfDjnf2hVPyM2TwNTUmL1wyCEsefUnvgOt78623wofuusuSHhfjSnk5AZlf+YVPhzd3vTstg89knUNyFMTgZ559Nvzj17+XZKyf7KCC/Rpk+gAOCjhxusev87hE6jbNglovboiUeOWfn9yG9NEkhrxZxqE5kkx6NAE9XOzIsiSbvBuIqmVdfuKjT6YcUaKGKLgUbdBXvDuvvvqqjB4TQ3HzAJoeKXAARHfvPRye+f4PrNCfQVXFuO6hWuJ+IlYDmtXMoxSJzGxnCdHeSOGcBCZALtbbRvqPOHrgWJegD6km6RjlfR8Shzco16Y6s+rjJjVQATT0FIAqTQCsTrGBysuy4Ml2McKSP2Tfg7xRbrw/v84x8ehVtxgXWqw17zvA9OvffCY8/tiHc2Dq3GZ2WeyaplPmoMqS6b2rFi1b80fCo4lPZjqtnue6AGglhgV1fFtbu4l3gOfPfuzBcIuSHyyQA3YlWulypAfMgN64fq0s1FepftBgbAsnNQ0FqgHEFc7BA7Sv3iTNlTvVxbjvVHt7eODeuw0gil0fLydAytzmjUqltvWNncWqTbsMg9AtN1yl+Phvhi8p1V2PDBxTJz13PY0B6QurJMJtWLfW9GIHDr6v+Oxe+R9KpORBFgh+SeumBtA98TzxpQTIF8xFUlGUjwAUDlxOV2GZxOnGRk0OB9KI7rrn0XDtNddoLiF0b8VfAsrhmv7pn76scFR8MRNQA9uSNWsq+ZeCKCGe3/rOD60M7nNeq7IgzanXPdTK7UuWcklYYybaKhuTBqqFPKb6AtaH9SHok5qGbE91MhqNSJxtluiPFFZDWGeVAFNzUeklNpUVhjenSoFkhVyYxqUK4z4rKyU2j2GQU9SSEpVAI3pOVfqI8BuT8atSWZn41UpvWjkmAB7XvE+6cP/V6Fqr0/6BlwdA6WP/IFmjtDvUH4alt6yE6xQHjOMTMQOjMix1yBvhtLCUKZtJ7UfnMY5Rs1A3tTfBtntzBUuew5ASTu9/7z3plpeHufMXarqVLt1nPuFMwQFT2AAo/Xhfchjr/KDKypqRSwpIjQPVQOKlZvqMBk3MdfNNN4SfffLDYdMNV2sA5HUzdodT/McUE3Cwm667UmqAJhuUJ091CaiJ9lC+RA2SCQNiim1frNXaOrrD1etWhtWrV5tlt9R1AqarFRJ5YP9+JT4hCcjZEfOxv7LtrbBHIZzTo5SLVMq2+fPm6ppWWtan/QcPaVxVmnsTDupFoMtOwyDDLWpMvxZZx5n0jufbKYMNCUEY9IQeAqStyuAxrPsmXv+RR56w/oHrLEVwo2+88UbYteOVgirZI3Brwsg3qBR03/rO8wbewH6tLNMkUU48BRJ9ICGOiJFVAhKAFBrLDVwlXhY7hx2gQe+8AelQT6jWva1duThU1iUO9caR5hgKwBhOVKCvFfoMMK0SivHN4B2vSifH41zVgl//jdfIjUmgWWlgJhck9VO1kmD7D1BVQwrLT+4YH9WaukaBcyKWJ20L/GWooqxaXzPulXLYlFE573eIKWrvU75TbdfoY0KaPr4AzF4A8ez8w1Tqg0Y99g2r4oH9B8T1LwtrlszXNraS7NOg9kRy4IyBkloOmtly9l0CQJrcvBmEdMHdcgnp7u6xgXTfXTeHjz12f7j26nUShRBPqFv8S6UdUyIAddWKpeHWm66WyLhIA2g4HBOX2i1dKjoWBlsyVM/+XFO6oHNciUinB+65w774pYCCF56Pya2bbggv/GSL0tQhOp45YT0HpKZDfEDJKDSuEb965TJxbnPC+4ePyuDSafpR/AdLW+WTd2JUAxPxeJ6kEB5ip3xLeb41GvRgDXo39KPOkVaIo/vMZ3/D7h0H+1KDFzDgY/O1rz0lq7oc/3Vj/vN7FFMjUX2OgKNGYbqHw+bNL4QTknwwSMNl1Zs/KK5bmrpD+lo4Tggw5Z0rBqTsA/iJWAJIq5R9qbtvJKxQ9qt5mld+VNxmLQCcAilcKEDKTzBmP85RLbCG46tQPHytpVBUFFYK2OyHAFS4eHSo7pyf7En+A7DY2R0wbR4n7YrDXTHSsc298eOp8G7BaQKkfYpAO654UzhRn2uej16Vxh2RTzjpg4WlnkN8PXkwfS+QMmDpkkXicmslLfSb9BrXZT0Gz+y+qWzr1am5KDlSF8MQrZkwrFPRFiR9XSFwe+T+O8JHHr5XHMkym6yMG+XFldA2lXsuU4dWILWjnkE9cL30rVeKa+NdbFdOz3ZxqSN6Gfji8vKVHrxJSxf7/y59mJYtnhuu3LjRuK5i12svpQYBjtkb160Oz//kFVN9FKs7k2X+DiCK9xMpIxF2/RVrxakMhQMHD5sPZb3KoCQs1FYL/gHAo9KlYv1tlqTR2KBkLDJOdfWI79G7hahaIU5zjJOorgPpHGU+evyjnworli8vK9JzMlyXtmzdGvbt2W7vCe+Kv0nCRJ1/KLS0zrUPwbefflq66Xekj9X0w8JKmFzAoVbZOmAGEHMHZOCJwRQRn23ADd2og2y/6lXquufM0Rz0AqYxRRINKHFzS2tTWLNigcJHldJOxxi4ybAEAaIxAa4QHOp4up6oxNQ3KeBx3nIgyvEAn4GkrsFBMC53T4cYWFmvxpdV5x0a7EuAtFtcKOoDOEg9HwdR/GUR2dWduR+3wnYihXC2QuK9pW9PSvW3X9zp6lVL9MHR/FR6lz0logMonD9lznUWtjT5VmVVXf9FAqS8enSLHraWzOSIzsqAS524fu3K8NiH7woPP3CnxK0FeshJXTtA/wq3vHS6S1qZ2NIciYAb168Nt9wo1YEGYqc4Yiz+hB1iqa3Vj6O4g0uR9u49GB66/y6zENtLphcwS7yUcGXL5eyN2Lv1jV3ZKjO+zTkTg9KQ6f2WKNoKt6ajx09YmrxqcWrwVZ4Os/ACUAPIWCYQa9Azmz+vVYNECT6kzhjU+2SShb1D8fF5IL3/wcfDLZs2meqA6yhGtIdVHTXTN//pi+KsBAIp0TQ/3JvmaIoOwPCfvvZMaDvVbSGPer0NRHlnGOxCIbWlOHa1B4cWgymc56i+FIj+DqKcBtEeAOADAwF4XEtLQ0VYv1pZkxRjDwGkDphZIIUz5YMC1crlCWYEvSw/jFwG0GqXSKAR3a/rSRHtISz8vt4gbl/Mo3HYtlP/iMOH4DwhB2fnTMd0X+wb0bM6qb5p6x0L9fqIAaajKoPId5DjRq0EjOC+1K3q4wRe0x2ZBc/O3l2B8JDyDlyrZNfoaAfl1gWIOqAWE9czTZXd1JTOFwtHmhgR3IB04mSb3r0qs75/4iP3hbtuv8k4RO6G7i3+ape917PeiXiB2H/7pmvC0kXzJMYN62vXYWI/OrY6qQW4roRLvRBXeGa3iMdDR3tbuE85IBFRS4n4tI4rysYNGzSHT2XYtmP3mZ1wSkclBiVCC9etXW0fTiKU4EwJwbTBr4FdCkT5IPAuLZRLU4M4vQ4kGlnl4ez4+CXPKHshCZB+9p9/Un1xr33Iy/UF7XAtf/8PTymp8vFcY4x/B1Gm/4Bj/c53npGKqN1A1Cvq8nNAwNuCkQmHcu4Z3SfYQ4gkBGDyZrmITxlASrSTlxnoaYqRKhmGNqxZKtcm1F15IK1K85VSlhibkKjyQJrz2RTa1gjIOL+7WQHGRCe5jtQBlQQmY9KN2nb6IYnB1DlQB1LODXk5S/YN9neHo6f6Q5ceKBwtgA+Q8r7xDCThm34UAKWvAFHj0lVeDkg5l1NiUByxPKdLUH3woVb7Ts6dOrh6eXYZA3C874JypIhviMgYINyANEfuR3feen345EcSAxIcYQKfXPbZC/DxzZ/Ruq51oeY1uvG6jTmxv+1Uh1n7x/TQG6RPTKz9cpXRCYoP9jM68zk76NCRExp8y8LatWsNQEpxYbz0vNhXXXllkIeOuUXN5EUhjTCC+6XGWbxoQVgp0Zo4efShxI2jDzQuNNOvrgbg2BEBTLPAC2MSelW4UHRtlRgt9Oz4EBenivCrn35SsduPGPfNR6VUP9AHLS0tYcfOXWHryz8wzhMgMjDS+4wul9G+bOliBRq8EXbKxY7kGxAAocPzlIJEtQAJ0K9X2r8RgWF//7CBTCU3x1WLq4U71S0Y90kZOl8AFMCDI62Vsz160lWLFoWFcq8akSUe0HH9qJ+UD9EYBiKRrwOqMbACpjAHjE8+GmpFIKc4KK07qHI8lnp+AMyInlGlrhEw5ed5RLNAyjU7V0qO0ROn+8LJrgEbL5XSeZCoGk7UP2R6FPbs6EH7qW2kUlWZEpD6czQx/733Qs9QlTx2GhVNJu8eeTjgfwwlHyy1GXGrtiP6FwOt1+O48wikuW6wy8INBuqQ/ybGHCyxGJCefPS+cNVGNyBZFf3zY3374li62O/W/t6+gXBcRoRCa7+PmovzHuhJOLQ3lePzw3JmRlwtJ+L7oECvuqClPmzfuWcGdKYyVOh9GJKxhmdtEUoNjZpC44j0412ae0m+oRo4pThJBgpqADgn3iNcmrrlDsXHGTG+vDFKnJ8Gwq995hMGonBBk4EofYRz/lNf/EsNeo3yGBm1zpTILUqk8fobezUVyhbThzLwXVylzxM9qO5Jr4cwWNYKMlXJ66RC96pXZUQfAa4FsIjBFAMTYr77SAOgEPpT15O2NlYqQGGF+kuW9dTnFC7PCcB0itddBcA+zpO4XyU1WXfRnHPGiT+oUak+d0Blm9ylrjs1Pa2ejb877AdIiXIaV16AdhkNT3XJmJT6cGOx594hQkP1+bB1/nMfuEdB9A0SgAOlFZb4Rx1+WPQB1L3KtwuAtrY0KunJQumJyTolY2HanzFgepMAp6sB2O91KDtPQMqN8/D0kvE1FIeA8QbAWblssRmQnnz0XmUzzxuQ/OIv5iUQyZ25tf8WWfuXLJhrYv9xzRfEB6JSogpf9uSF5Yjojb6Ibg4R/33Nwnm/RHw4AV76Yi8oZb4PznRuS0N4a+deM8Kdye3AhdI3+ES2istbs3qVRSuZW5P24TFQHEDpSTghcaHShSJCt7bMkcg7ZNZ8ngwg6lRKMqgXF/W5z/50ePThhw1AJwNRuDOA4Ut//+fhtOL5eaKGhn4iLWsU37/5+RfD1m3bLes7Az8GUT/E8RcgFbKYjlTSPRsJmCr+OwumLuY7kFIbAtwMTEd7wqCMiCtXLddHEWkuoRhIXbSHG41/1IRTRWfpNEJsvvrXwFPnGGTG0BTkvE68BFCNO02bcDClTgyogDJAOjDYK+ajJ3TCdeoDlOVG6RtEe947Rg734TYS40i1v9h7yvmy5Bwu5bTbJuPxu/sO5QC1qXmeLkFgq48Y4OhA6e04yLLt+5wrPS9AyguPuDskruGE3IkGlOVlwxWrwicev1dc0B1mQLI3x6/4Elkm38X4YvNi/3VXrrXB4WI/g8Z8UvVCaPWiJPxE++XAfJfykZbTlzqY8mICptcpGGLPuwcUcls6hJIb5j2AjRjT4BSDJi5AE0OICx3U+7BKIZ74WGJQOi4deb3yiKInLwmi6kQMYIjEC6QLdcd6vDuqhUa86BwLgJYC0dUynv3+73xO81TdZiqNciDK9QOi+Ix++StfDYcO7qUoeZb24ZFIK+DGIHdS7/iPN79m+xFxHTApSB0ECjhScItfS3OjGY+4Dn4GsDomBlMirfAt5WPmYGoAqoEPOM2p1UepZzAsFEe8ZPES1RPzEulHuQY+XABoTFXyHEA1YaK+n1gVspyog2GpJSALmFYyt1P6ortO1M9XpfmkquTDOjrSYz7bxzTJU79UF5D5j2ppH3M1MKyHZwCqMr0O1m/Ug0wY0HiaKvHexj+O41YB1H0CVAC0GIfqoBkDKeu+zf5zCqQOoB6BhN7lxuuvCZ/86H3hQ3dsssiipBPo8al3SHLMxf0fMW3j+rU5J3+s/ceOnVA467D8BmVUMJCAS7+47nuvXqi5sswTzTNoonbxfuaF5GV3a/69d90aDh44WNJpn3cB0AQA8N0cVj/gmYHBZuMVK8V1jIf39iNuaU4hlaVjsMjJdV6hDvq3uTjWN88xyQb/YgCLtksfm2/uVkWz/dvf/20DPsR07oV7KkfoRZ974YWw7dXnTA9qOlEdh1GJ5CMYL/a+ezS8tm2bAKLfRHoGatwsZ7BfJOqzn3q11fLZlZ6UaTvQhxI1RDnkYKp5kqWqUEhiBKYu3gOkg7LWVwBUOn758sVmOEpaKPwPYDp4ssfuJa2CO5SL9YkuMrkI2vdzOXj70lt3bjXLlVbVCr4dWeXMDzMVFM7adbIjHJM+eDhNpOKce4XmeLLoprRhA1P1EyoeQJznBWPMCIIme3ZJrcL/MajyIQNQd797WJFzPfJOaQhzWhYYhxqL/IUtJFvnAEilR+ELYtxA4kDf1dWj6AsZkG65Lnz88QfCTddfqa+6ixw8IH+1il3ipVyWvHwMbLf2L9d0EOieTigmnH5BJWDuU3opxp1VucC3zFUjqq+T391kUU9+qQAuhqDbb90UliuS5P33j+ZCP90QBIjCza1dszqsXCqdlLgWBvMyOUuT5+DQkWM5tyYQMctFAsRwmCSZIU57fupYf0o5FgBlImAqxZ1iMilHS2Uo/OVPfyL87Cc/ZqI/FvKpDEKufZsA8sc//KYAATlUwCfXI7L4Mwhf2/a2HO1flGFpTw5EdRv2dlM3B6bp684+J/bRBkEDtVJlQAApBJiiI0WMLQemABoE0BGCOTLWHxbJ7au5KZOY2WolXCkOuDGYprts4UDKRhZMAdSkPFk6uFIWA+lY1bCyOVWZ4WlUHKh6TJGrCaCSuAS3pnZ5v3T1DRqQuljPfdIfdJ73E91WrQgwKAFRvQ1WJymbyjO0g0v8c1Cl7VMdPWHPe3qHT3eJ6amxkF3URHCsxQjOdAY5UvSf+mLpTEQg9UrntVj6wg/dfqMs8A8Yd5ZEIMWXknRCXHL5rHNv0f1ptGDtx8l/4xUrTLQ9daozdfJPoofQHwMWF5oIOsD4dNWGNZp1dEnJF8ivk5eQlx/Rd8P69ZI2bgldysKFIRF1DoYgfIOvvVIzlOqjClc6V/HkNYrb3iO3pl4ZhurFhYrdsPsvBqK0AdfUKg4Uf178eDv1nuG07SqAcj3X0tQY7rrl+vCvfvtfhGuuusquoZRRze/Ll3DI7+zeE7799D8Y1wkjMEcBCj2KRHpz++7wwgublbH9kHGSONoLz41b4vHXSD/qYEB7pb6XlMOBjcsCT6ABulD/AXuAKfpGjFCAKro8LPlwpoj4MbihFqhXZvoGuUAtWrBY5+caYHD8jpIlIn6paIYYSKmd3Y5bijlVgJSYfWZN0dXlqrGuRyW3I1Q7xOUrhZ6kj24FSJzQtChDunkc7yHADE6c+4QYRYj1AHqOG2Wf6nBP1D1bIOU8UAyonZq+5b2Dx813vEk+s+RAKAaoM2K1d/GdrymcRa+iRjAgfeTDd4bHH/qQGZCSya2SC539r8nFNBA3rl8bbrx2o2UeIuHGEc3Hnoj9ysBjYv+F7SmMT29s3xWuXL/GxF9Ebl7wUi8s5ewHnDD8fOiu28Mmic84QB89cUrier0G9TzNVgkHyCiosHBT5i7HKm+jYsItM4TUpsRAOLWFijbjHJ3i5hMuFC4sqTPh0LQAAL3vjhvDr//yz2uKlYdzXGi5e+FQ3w8nCoh+59tfUfIW3JPqZPVt03Qiu8KLP9msLPZHcwBqxmRdji7RfnCaDPaYKOKX/Iv2qBBASKKcEid73+sGJsGQgSmuQXDmDqZZZ32AdFCuT/Ua+HMkKSxQwuZxkE2UNTT5OTAyxXpTB06MTb7udYstHUwTjlRgmpwuV7VKiUarqqX3FohWoT/VDyA9JPe2Tn2QxgB73RcfZMj7kHXwH24UEPX9w3KRopx+nEkgpUnIAZV11DTlABWOtOKam+/vk1g57WwfJrLpc8CA60pT2BGBdPdt18vyupLzz1LZHmCE2ZDSWzMW3t69L2x5fZfm9T5qlui5chqu1qC1l4V29GaVCoUse5qz3AkQ/dbnPm0p99AlMkhLgamfChCCO+XrzYtPzs3vPvtjc0citLFP4jQ5Yg8cOmypDB1IeaeMDIFwAUo4FMJ14bpwrB+UXtXSlk0ixiPC36QkNI8//IBltmcQAvLTuX5AlNR4L7/4fXX/uAxqR2zqj4Oae51PAc+GudizHDT3YECgZcI9JkaSnP5P9+nH4Abl5QQMwWEtkQoIB33AEhoWmI0MD+ra86Ll4GACOKTbwx2LnKWeco8ldom5cxeGq69cFW5eslAeARJNNVZjFydrPP3nfqXsR/3kBJBOhSrGBsRRk5gkSck3Opg8TACULPxOcKvs6+vtDAcVIXioo10/DI55lyfqMgmenxkOP56kD5UAl8iUJMMCZi5xsnfSz3+mS56/07y588It16+Sc38yKwA5cacNpHCgXDQHg9SIaldtXK8EwFcrOcBiP9fs8gx74JhCIF9TCOZu+bl1an4e+rdJoiUK7fI5Ns/whFM4DBehz/zM4+FjTz5hBii40+m8uCaq6vo7la7u9TffVKz8obD3vffDOwpPJb7dnOXFuhmQwtlKtQCIYLDj3nGsPy1XMsjrZi+7VkCJ4Wm9IqE23XCl9LW3CEjmCsgIuSycdyh7bLzNgEHfizj/vWefDXt2bbW8oTt2vGHJg6mLiIrIjojqgOjjjG+Agyh1AVJImg19DJIPpwNnvO2AOiSAsJlBlb4Qoh8cSK1A/wBU9ImIwkh79QrvJKMVelKPRuK4SmXKX33lTeGq+Q1hlXK1AKajCpdEL8oyS4Ap2aEgDMNZQEVfCwGuvu7bAOkQnKZmLAUsY/Bk5lIvp36/3p+O7oEwoqmVX9rymlRAp+1enNukfxzL+bDQ1zEBnuI99JwqxNXyDAr3x3Vner0YoLbMXdA/ZSB1B/rObriCIQvZvOnaDWY8akXxbwRqn7+bSk96mSz8i5f0H36VbygM8/Ude8JJpQzEkto6VxmEpBe7EIDK8ydhDIme4e6maqSJHw7HweXB2XL8QfmtbnvzLenUpYt6/5g533dqUAEqzQJF3KNwrDcxHiTSgHFjUpP0iLg9XbFqqSX8uPPWm8w4Bnfm5/CBGV9DuXUGCcdz3NPf+W544bkfGge6XxwoZPrPlIM0XafKgKPCJ0fNtEw7fR9lun27NwdSyqAYUAFSiGlGmGYZYMyCKUBKKClLMkah9kBdRDSXc6WDii0fVG7U+nkbpGK5MixUghDAtLlVKQLTk2TB1IEUdykH1ORqkv/OnU4A0UpZ4AWuDqTUBjyLEYC6/9DBsPbux8MRRax9/e+/GKrn1Np9eH2AstgHiP18vAbVr2C6utPWKZ/Oh536Z0sxoG5ct6w0R+rcARfIwEV854Eul8X15huuta9+nLWbbps+hCadLc2blPC8kjSiHorXk9IPxP+kDwt7kq298tF8VVzquzJo8Aya5zQkLkIa+A4s56uDcBv6tV/6tIxQSw0MpyIqZ68NQIUQ/fGtZYoOwBWCezx+8qTlCW0/dUrcCOGJiLdKMq1EzPPEqS1RCKRnfgI0sZzjOeBW1TMBUFQRgOixY8fCM9/5UvjiU18tyYECos6J2sXpX8yRUobezojXOn2lKcpypg6i1AVgaduBYuWKReojMhblRXzq4Z4DAaZMiIdHRGNjrT4+LSbis495nhYtXmmSY8vidcapjw2eCFdqTqZ5zYmoPZByp9R3UHUwpSxLMScar4+KG3Ug5Rgy7JNcGlGfKaChet3CgB7lEemUF950a1i87Mrw//6X/yhJo9tAFICsTDnPIaYY0TG8JXCdMbEPamio1vsnDlrr5xtE7QLSfymgOkcaCnSkzGWDAhVfr95+vaBixXHhuf+uTdK5rEvALm7tDNfpEiJTht76VqjZcGeo7jgcxpSgtmLdfdZiYRee4Ukuo8MQ+7fIzeadPe8qHdygif0NMuxUpR+7hP85972G3vTXf+lT4e4775y26FzscQCsgCoEkLJOGW50MSGmA5IApgMp69MFzrhN1l318OLLL4evfPkfwvPiRJmdEv2nhyM6yAGggCbfer75LlWWBNL0ZM4b+LmdO/Xt7JLzE5+/RPrNGEypBzfvXCnbrKNjRF/aNGdhqFOmfICU4+66+6GwZft7Yd2Gq6VTVJLysZ5wReNAWCXOFBY7y526rpR2nYpxp77PlwAr4j00rrR9vu77AdX9Rw6H5qXLwtW3PaR+/lJ465XN4kabQucp+fGqX/2jAliCDfFHh3Z4BjDTcwSiqDSYjvlCgqjfm8C0v6p53uI/QZwiKW2/Qjb5MX8NFuQmfaFXr1waHnvgDrPAL1y4wN4cbvJshyttjJ/Q9BVv/KdQvfudMH7N/fKbOhrCT76sh3sqhNalobLW/U39kj+gSxuFybxAuCQxNQrzDLUrIQcZqHAFatDsl0kCbOvZGXhCpft6UOC15dXtYb8c6K+/ZqM4ocRXEaA7E+KrDmfLD6Id1gHJ+EeZn8PrxyLWdM8NYAOicMT/5c8+H57+/vNh/943FPetCfUEYgaY6s4cSOoEvPf2E4gazKcDwQHVlzwFk/t9vwoAXgwntGeWZlXxcEeqx1QjFQKAQv5V3AYBRQxudv/4ZkpvPaSM82zDmdIekzdaFFNFvelND2pOsjtvvyGsW7M8vCk10QJcoZSntEvO7r19+hhpgr8G4vZ14nGdzyObsN4j67iFH7WSO+mjO/V1rpdtvDDGFASAT5/mUrXbqJC+tbZKHKPcuE6P1YaOwZrQLS711ns+EmAIvvfdr2l81ypRe699lKpSbhSw9L6pSf1GadBBFJG+Tq5ITFdtaOsdbme9MP/0iEeqP/Xxh9PEIX25qyBUDRcWEik3ivtIiFeDH1a46VFyVP7/aO+JMLbtG6Gm/c2kLcYhrhF6WYLWaw9vCRWntoShFR8JNdc8rAekkD9O6Z9167zpXsX0rvmiqs0IjIhncrciw267+Xqz9r8pB/r9GjSIgK2arpfIKQDmXIr9qHtefn17OPAnh8MTD98TPnT3XTa/U8ItTW7Zj27nvK7SLy7Gn5Lq4CcvvhSefnazTcCHUa2vX8CUuSLXh1LsnFNsaIqrx6Cbd7lQjegRwmjzKmNQIXF5TUZ89fYQX7FQY4yZp+fqRPo8DFCuJzXuVGho27LuQ3U10pnK8+OV13aGX/ilX5G/ckc4eHR/uO6qdaGte044rjptnT1hYfdwWNY4Jr9YuXaJBR/XOzSoi3XrvnOobAOaTvE6+/BnrUSMlxM+E+yNj1QpO31VODlYF4aaWsOWN34U7pZLHLRz+xthRLpvuFEA0gGFPuVbzMhOtT9Wn39pfIJCOBX5JXWGbv+i4Eb9AquvWLu6f83YWIFo7zsLl2cOXByJHnR0sDuMbv+GASWPRIlujMaZ/2wkb1mtSLwKQs3e74SxjudD5ZqPh/FVd+sA+QzwBvK2XgRfouTqL9x/uJQb5OLDz639u/bsD8c18Phqm8FG/XQujVPHlO3qL//+G+G7P3op/NSTD4UPSdyHQ0VnOV3r/rnsSQDUpuWQThawf/ZHPwr/9K0fhMPHE0MSb6gTfHElA1wUg6gV6F88+L3Ml95KqdHiQAuwVupV1p9ZnrNiLO0Bog3yv8U63ybJo1U6YneLwh0KAjwhwNTJXKVGa81Y9cbr2+QHvCI89MijoeMbX7MZc/Fs6BmU5b4yBVTppxeKQ22uVyJqAWKD5rh3y/64EosAjE4KpzCgZZt1p+qKkdCfcqOdip0/PVAX2saS2U9/8vwrobL/VLh+UwKkRyTi9+nQKrjKIsSHxsV8duMKxTNBpId6pBu92KjqySef/CPpQ/U0ErjLX2CpVyFfo+gaQGcgl3+lWBvZ+Wyo3vJnoVKdWCmgVBRbntQv4+vuV2d3hHD0rSDJxIhl5YBekPffCmNtz0uZtUy9uTj9EtEqokRaN11+UBfu5E9KP7JNdZ4+HdoUx8y8RHXitM51bP9peRm8sm2H3Jt2yRl7MCxauNCs53B/xh2L1Tjf+iznPjFoIcZj0HpOUUh//oWvhGd+9JPANcfE/EXdpxQYga9qykFOGAUqKPcdp74fk7yh8Rny61aHf6rEAnHWuS4X91nCfTUpazziPaK83NmN+3M1iLfokU3Vmr/es+o3NLYaZ/faq2+EG268Ptx62+1h947XQ58s+nX1ebUZ4v6poUrjHjtGKkKHooz6dCYCIRD88ZFHB8pvQFyjr0vuUFTSaOiW0epob3U4Ji70/b7qcKhbQRMatq1yydqz89Ww661t4TOf+UxYuGiJXe5LL/4kHDnepvsgYiuvQqEfEL6AD8pNzBec6JSmammWpNzTI7cpXc/5fpe8n0ssR1IgLTYdM7d1JsTrw9uBnkWdJD3o0A/+W6jZvy3ImGcgOqHVEkBq9fQRckCtOPZqGOpw/Wky/zh1Eos/5zzTa55wRZdsAfH7a1YvD3dsutYy+TPd8ZGjJ6VTkvFB+4jtJ5E2cxVlLc8zcdMd0je+KjB9VSnk+nu75GGgcEpZ1lEVAWwYkVzPORPnK9aGG6vwBQVwjh49Gn78/Avh//viV5SVaWs41d2tw+J3hXdWCYx1bVkgpRo16Sv4L2rGR2qzJJWqRxtOvLLxDz4E4AJU4coAlN7eEeU3bZYevF6O7KflH0qOVcT+REdKfzqQ5sNKpSKT3pGJ8U4oGUfnqXYB6R1hw8arwn4ll6mqUKz7qLhSnYwPiP8GANGR6tDZr58MWEcHKkLbiNoYqp7wOzZQJcCsDkf6G0LXaI0d19ur2UzrmsPCuQqrfWtLeOXVbVJLzAuf+MRPmYEIt77nnvu++aCjB4b4MHGvfEwg1gFRluxjltX5C+bb/XarL6YCohjMaT3+lfI3LVY3Ps4uqjy2OJCOp/zhhMOtjen8s4vVSdGDjr7wF6Hy9e+FKk1uVSlf/QIuNG60HJB6PdQAkjCqu8TRnng+DMk/e7R1tT0c3kTjTcvfrLf0wViqL4jtv0mGqSuvWKKXUgOhrUsZ47v05iqbugZY2mtpf/DsZ45OC7jfemdv+OFzLylF2Xuhu1NhojJeYonn3GRSAgD4uYUeoJ0OxccBnqgUAE6CRdrb28MPf/xc+PrT3w9PfeXb8lfdZR4oHi1FFqFCUqb3CEjN9qEqLtrbyGA78ytsoxBoi72O3KG1lTmQy3EPAdAa0HYwBVAwgFUpfn6+3L9wlEd8Bzz9h2jPOtxoElLqQCwXJFnT6Q+CKgnV3bn9tXBg395wk9wYh8RF9vd26HwJqHJZWPcBWOXpsl+l5roHYIfHZOUv8otvBU/fJjFMb23bLOnkLdtF1BXjc/MLz2u21S/bDAhwo+hE0XXaU9c/7hN1FYl+GuplxGLaEfULnCj31a7pSKz/inVsfBFap4+zVApIi9XNHmsPfkJhrmCk4k//9E/79DJPQUeaO6jkCjfpetCat7dYvQrpyF0XSgH60Hjby0Yf+aNQdepdpdF5KriO1BqI/8XqALnSjTY2hYrlHw0Va+9Up+f1OPEhH9R1e+EyNw83sPX1neHtvfvDMYXnYX0lGoiEIqMWHM0rxY+jz4yIfAMEsLYTAz5Pvo0QjvUQs7KSi3b5sqWKiNtgOUhth/4BpPhyIopzPCAbEyAMYKJ/Rc8ZcyZtin9/Z89ey2dK1NTBwydCr7wZaKdJYjET4AG8fZrFoF3JYogUq9DATYj71UyeavuQrPZdnR250E8HUurFHLxzTVnsj/ud8V5uf9zLWVznOG6fK3RjFFE8zUpxuFhzhmFYxAcT31EMTzEBsoj4OOhDHu20b99R87vE8s0h111zVbj5jg+bvnRQ2eGdxoYTlUdljRuafc/EpdcFyEk319g4R324LezY9U4ugAHAxDcW4pk1CSSHpbJwv1GeC+5MqKdMv6u20P3iI0vUFtQuFymuGe52bFTzO03i+gSXmSWm3C5GxerG9WipFAhbPbk/ZQ2U8fFntD72ypdCzX5Z4/NGRgNPGgNAsyA65ZPEIMpBkuyrhjU49345jGPtxxg1S7keNogEJwAALulJREFUKPbKYO2//57bwj133RL27M3H9nMQ0xXnrf25Zqa84gEco3LBYR4e5p7HRQvXrDi8c5dCX/lB+KPOlTN9k0IXV69cHtatXSVuSi4+aoOQx9aW1oLzt0s8JXGz19m3X4Cp6Uh6lc+SKUm6MzpP8pbWCRwXiTOH4LIwwDHjp0VQwcEV6yjVddBksJtxyVqY+C87NouMXzsoBtWJQzwB3Lgt1oX19klzMZdtAGRo8Jj5lxp3KkAFfPgAwc0BsDXVyTonrtR6rX5jmvfeQyoBbaK0ALvOzrZw131PhAXoH2WAMhJHWgxYk52l/+Plc/TgHmsXYzIfG4i+5Hx8lMg7C9XL57VaEVRuLLPC9B8+sV3dgwaiDfJWOHLkpIHo7bfeHB55/Inw9a9+OXTp2rNUDhA5a7n92bamuz2jQGrdhsxdhABQs85r37TBlI9lFkg5R1qGy0byyCicpcl6AH/Eq6/aYD+s/S9ufSvs3XcgHFEoaosAFX0mnMPUrf3qff0hNjN45uNvLDolNQKWe0R5RKOsOxbAx484+ZPtXUqpd0TAJyu2DBXzNIXJQmWLiqlN10fCE4g63Zrqg5SNQygVixDpAJlQDrUG3BI0qrTqhJ5avk+57SRQZbty/5y/s4GfWu/ZCUg62GXBlboAhr+HXo8B7GW5E0Qrvs/F+rhd2mA/58VBP+bGCFv1MFIAtFuh23SNAyqnsHBRLQFTyFQhZv+WxEAuUsH04aPt4QfPfDPccd/jYdWyBXKNSu4e8T5H8XqusHBlTt1YTidKr8JFx/eiR4HZxKQVzw/gLcB5Jin09JGQpwKEmxMgC4jihP+Ln/2sdLyb/JBpL72fp33gFA+YUSDlpQklnOjHTqgjxaUON68PtSMS4adIo0q8MFq9NNT2pMfkbUy5FvA/neyFzVWeXSnoARLN/LTcllzsf+vtd8NRZfLHUb11botFTZHerBTnhigPFzokLhSjUpYLJQ6coWncQIm3GSBs6+yyX8HFneVGrT4YzDEEiAIicKSAKh+SOt1fYSapwpPFIr3vKfeOOQdLXQdRW/eDM0vvCtrMivWZqrn2brzhuvAzn/pnYeuWLeG117Ya54dj+ipx88Tln5IhFnHfCSBKRHwJcMr2XlV9QjAqoBIwPfbIg+Hw4cPWRodUGc999+/DDXc8ptSOG0ykhjt1rrQAVL3xdEmdBc01ORD1fATsTjhQ3Z+4UX5Wpn/9Uq+MyqDF8yCunr6I57Vqlk8rKgv8ZwHRdWuVcvKmm/ShUCKbnF877RviWLvn8p89qzLn4ipmFEizNwPnCYBCI8vXh8p7fkXGokNh/MV3p8aVwkSgprnnN8NY17thfPffhCp0bRkwneVI6eGzIxf7cfQnpd+2t97RNMjHTVQs5eSPOA9IAZbz5yZcKI7fQwJWdJmQc6GlgPjsrrr80RUavXDJJ9s7wzxZvdEFc32oG0gQPSrDy2k4WulcjWtOm0PANe4yHadw2SPpgC93Rh9rXt/rxsDqZdmli+8cG3NybBuJM967d5d9tG67447A751d28Of/tmfywr/vmYeWCX3ohXKB3zK9MfoLAHZujplipLonyQzyQPPhg3rrFnEe6K40L9ueeG7oe3QO9Kb3iefzUaVZa+ycNtBlHSIWOezU06nioJCQFWh3xORWx5Lz4eusVERWwJ/rv243KM8rn6BJJNazcNVUyMxJCJ7TtH2uVxNn0LRU7DvnAIpIDrUqgQTNz8ZalbebF+eYSV+nvJJU+lC30ZZ/a8Nowv+JAy9/d1QJSf9qr48oFpEVNFbvMQLZV3HONIvv0bciACsc03lnPxJ6QfXiVEIgILTJPMX4jNhxfhpcjx6SQfQc329ZdvXdTIFCdmjAFOMS2NKQA5hcEIvTAYp0vR1KVSRxChQdtDEwAZYlgJGLzfQVSPuzoMdD7E22zawVuxc8TkcYOH0EMP37t4RrrrmBmuLJXpDQGyP9M7kMSUHaZMYjSEFvyDuA0r8bF0p8gA7AOqv/+rzUuHUm1GNc5BXgOe5b/9+WfiPhTvv/FBYvGJjzhCV5UoB0dYGpjY+GbY+/y2Z6gqJvnDY5qMkISAp0Drnn9NMkphg1+CivYNot/SjZPkzY6I6Y948VDzyMUkNyuh7IW/fNi7wv+z9z9jljJ8WF3qzQjyf+LdhPAVRGq/OfFXKnjBRa1kVOg2RrOa6j4bx2/8wDM6/Q3oCFaoOHGlCF1PXppd0FotjJ9pkDGi3Fo4qAgfx+3wSYv8Tjz0QfutX/ll46N7bzC3G0igKeLB6L1403+ZPgstLZu+s0cs/+bxJ5/MeUDCSwzSx0CfrNr+TkIOpSrh2aJGS9DaJQ4X8LYrFddsxjX+AoRMgyqb9QCsRQJMV6R2w2ZdVLTgX98orryYNpFd59dXX2jYWbYD20KEDZlhqVmJoxP36hjkmJmO8g0NF16rmpWsWp65pkB38aQQwAFAxaD3z/R+YuI7uExB1MZ96zol2SB2z+QdftzZJssz15+4hvX/uHQd6+tKt9HzKEO/r6hJ5H8s8elPi7js78yDq7nBLlizjtDkiReDFRlNmDqd64WMdR40LrX7sd0NNXUv6xaVXeXzTJDFgVaO9YVBhpeGGj4cqtQexrLrts1Ib3Gzi/qjSil1uzk9dmoIBWr5ilS3hSNvaTphujw/K+SQX+7H4v/7mzvDtH75kM3iS1q9WLk4Yp3Cuxjh1IUT4Un2Rv5Z0VNs7SA4CSM7q4pxJIs3cWXD7ixfOk6+rMvF3lmoxKXeO0cHNwaPUUX529gMsMXhlQTNWKRRr78gRzbSKKiKVTq5Yv8G4TACSwQwI9vYm3OmS+XNCg37sQ7RfvXqtNXno/f05DtLvhR12bi3hfgE/ON12GfgQ9VsVnjk83COn+xrTicKJPve9r1rbLtLTBh8A+pd7ZsRzf3gbSB1qkgzbwCfGLjhQyABewA7A8mbHbm3svxRoRoGUCKPqh34jVDUoBpREI9o2it+c6faK+rpOCUzCj/NJTIzFp22J+2Otfxgqc3H6ZwDW072e81Qfx3LPxckpaxWVVFktR2mJaVVV517Ez94mBqXX3nhbOVEPhk997GHpRJvDDze/Gt47cCickE9qYY7U9Lmfyccze+JzuS0UgYMGlLg/vAxIWsJAByQR0QtAMLoWM6SIA3Mw9V0FnKgKefUpi4HF69KGc70OoM6JernXZeni/YFDxzTn1xrbtUARP8uWrzGRHEs5OQIAQbjTU5oPi4gg4vNrxXwQ6dXT3a53qVo5TfV+qb4THwO/F/w+4TC5f0R9OM877nlQhkSpQmRY6u/vCy8993QBiPrHxMGYdmmPcjhvQNL7AGBF7wEHGk8VwtVQJ0vNmhq5GJWqX6zuuS6bUSDlc1vdpBAm6w51CZ/fsyXEd4xLWtYe/k4YlX50/Obfs/MQLVENl8oPYJ2J853t9c7Q8Q2yKiPWDw+3mN4RXSlTu1Du3MgMnWpiM7kPYPL83laE0gtbtsvnc054VCkVGbzQpz7xqFLQdYc3NOPojt3vheMn2u1aPZN/OWv/xJOe/5KYY0X09zBLvxL7/us19sHNksFr4JhWcgDxY7JL15Nmy+NtB01fxvvidQM6AdPRQ/tyQMr+VatWGuCxDhC7vhNOFEDFAb++7qQkxOYw0NeZgJu+Fg6eAGf8zXMRnKdPW1j1Ac677n+CU5i7FJyvc6LeTtwXfDzYZkl/yZaUrGvbP7OmZtBO62drufi/+qa8G9z42JA8MaQ3FKmp80Kcxx3y8RTgHcjSzAJprvVip8rtnP4KYIoUoB9W+7HeNmmpBdj+lGjxMgJRbgdxGuPHyRNHjROlDIfnbhl0enu6FBG0+NwBatqX+Jg++9xWM8Y8+sDtRSc1xNjkTv47396bs/aPymWq0Mmf4TPD7wWdMmM09WEZv3bx6SkvSbr9cYFS3AMAp3NtJY8rsuPggQMFpatXr9H2CwZcfgLOAwfLNWG4IWPSNSubw+Kl12kK6R0GjuyH/DpYdw6R9ZgAzi2bf2hFACuZmHBfgpyTtY3MP0Ay9zHRdQypAN0n4jvXOF0aJheApIaLjc4RkJ6j2wRQY6n2MgPPbK8BUohjzFCA3yO6UQAWoxPGpzq5hJBlaaZ1prT//ee3yq3mULj79k3h9lsSg4aGXHqJE4cA1xCn9MPJn1DUU6mTP9Z+KAlFZW1iG5ReCmS9oH/u+1gKDL237J4iEHXx3e+V412NQK9kOTTn+Lw+etJYAlu+IjHG4HMJOHr7ACRtwTnSxq49B8O7+w4asIGh4CD5UCH2A4uEjS5cuCicOHZQH/C8SN128nA43tZh3G0MohwHAdi2xgfDSvL/2Ab6XPfpy3yN8mtDqadF+VrJXvrPzx+vT+XYs6lz8QMpnGgyRU3ClQKmIjrLEpVcwgPSbmSSf7gT8YsJMF0jCzMuPIcOHwkrVyyfMTB9TtmRENOJif+tz/1cdO6kx+PrKLXuTv4u9r+xc+8EsX/U2Dd/5Uu1dD7LGXYTyUAtvUwW8RU7iPjSATAHKjSXMk84pFMOd8aZHIQLjo0b51hRltsDALuks+TZu4oF6YSoJzhFJ06Lny9gyjlop04XgP6U86On1F9OR2tcow7iw/3TP/NJZcw6LlexzrBy7fVS35wO//d//N+t6VE1iH6V++EY8gFA6SLZKPJ/OuDpoZwAe5bwDS5nECzShdkmprxdKja/WAOFI7RYjQtcNjqmRAbzr0sMTrhDpRxp8df+Al/s+Ty9uHEfSCeVsONsp8JGD/rcS69bco+f+8TDubbztzT9Hnex/67bbtDUywdM7N///lFr0sX+qYeh5q/k3Kzlh6DfKZwdQOQq45ypu8wF5MRYryO04ngPkYxdnmLO0b4rOgYw9vN6EywdDNF3Iqa3nTice0bozJcvW2xA6twx91Ap8ET/yT2YHlRlHO9EuZNf12uy1D/08MNh2bIlmhCPXAcV4e++8D8E1M1KhfezcpQ/aq5RsK/eFPrDqClvcsaWGFjPhPJPdOpH+31M91jvi6mf6XzWBDhF1Tf/fBi554/CUPX6MB67pvjbl1T7QP7HLcooN9on6wbQIc8/oAf92y89HV58dUd4/MG7zICUAPR0X6XS52WgI/b/0j//ePjln3si3HDNBg3sEeNSB5RwhKmek+m+Z+6cpa+m1B4fQuoeVQG4DICmeElZzjE+Cxoob8brOSfqYOb7Oc6NPXEbvu77Dh8+5kW2XLFihS0BTAdoB0/f9gM4p5/Xy1iiFoAL/MGzz8rkkDgU/vv/9d8rU9jp8MSTn7Sqt95xt4VsskGP5XvNdp/zf0yzUoxm6jp4DvGzKHauYmUXPUfKRY+PKFmsjEvjD/1uGN/3fLH7+MCWEe5oxGidEumVEweBHpTpQY4eOxluv1kRMjk9qDcyU6+mt5cs16xeaUYrzk9Kv9dl8T8iMZLYflL61Wk0M4nb+Y2MYujk+w8wQYTOcZf5XXYT9EyWc3SOcRwLeIaoaw1q4fUcqOEF7LsWncNBNm7GAdjLmK4jpmbld4UASAfOeD2uO9n6W9tfN1/VF55/2bjcB+67NzS3zFWI6j75NS8TJzzPvAToJ0R2RHFu0e/c12PRuEp1fH+p83MclK03zMwZRtqj97xO3geKTUvLLo7FJQGkdJV3LlM1W4dHXNXF0ZXn/yoAo065Hy1bsmjKJ8dfcvNLr4XXlOj4yvWrw7/4hZ86d9b/MlflTv7E9u/YtVti/55wIBX75ytZCtn8se4mgFpqiJU5wVnsijCtoBVeObuSIhVcpM69p9GRgCXEguNjMKXcMFxtZ6367HJQzQIpjvIxzZNrGsS5OMd0wNSv3RrQv/rGueGU9LBbt75oRVte3hyuu+7qcO/9D5rHyO7du6zc9Z5+z35/vu26Tip7mR1Y4l9cB76gSn0CWJMHwMhujkCKvETlTcXHetmZLum/qbbndS8ZIOWC7fbSzjSWwMuS19u2Pgj/iGwhyglapjDOxK/UH33SU8X6IfYH/dXPfNLi5JNXhmNLH1esrbMt8zNiSNt047X2O3DwUHht+25L6XdSzuRzbXoN5dLMmTJAML/Ps72C+PjCe2cLcDHROB2zubOmIGrgGB3mgOet+v35dnZJffSmVk9tAhzGuWYrRtsOwBQRD8+HlA8StGCRpi/XUt9JhcPql16wL61SiX9+7XZPqjNPk+x1nTou17uE68Pf8++e+sdA9qkD+9+x6Ck4dgyGnNO7wZd+mng713++s8jS63tdX+aqph0UexPk9s3gip83zi7lfqSlTnPJAGnuBqYswuaOuLRXjA3idYU7GZOTfqfSv/UqMUVrCoTsyT16NlLKl6EHfeZHL9vxE/1B/fX1487PsthZXex3a//2XXtCm6zTgG1+RlS/r3N3nQVnoOszDJDvB3jcSONX4/fldSh3gPR9qAx4rNShedrIcoUcBzkn6mCXlOpdkI6ZZDYOpExmRzSSObnHJ/cDSixp312v4ntBB4s7FdeHU363PAKY94ptU1Pq+gHTcjSNy0iaUeP0kQmbfBDSxvv6PcdxUoJnAUT7XscKZvgfbfPLPP6Cs/g9XmJAei67raB/Lp6N9MMBuOCIz4BZsnSFBhivtMiBVsvtO/doXqZ2m8oDUHI96GEB6Z2bboz0oOirzq2lNbm4M/vv1n4y+buTP9Z+MueXSul3ZmcqflQ8cGxQR9Wsu9lOuz/aVbBa6k11EPUmnC8AzCAHXdvQvwkAmvptJrk8PVkP7lTVJpn0a0K8LLlhKQbrnP5X7dk5HRF0cEND3lmb++B4IpMcOO0eVI4Llbs/WT2V+f1oNU/q0Kj5fHlmjb6lnrdhfaiytpMKwDGyvTa9tGZXTMsujsUlBqQXR6edz6swPWiaAapoNFP61j39vedDW0d32Kj5kH784rYwd+c+m35jxdKF4RMf+bk88KYXfyn44MZO/i724+TfpYS/OPgTQ46e7lyFotpATvtrSkAQVYpWNT11IZCwz/w40xM4qMXnK/eOAWhwnoQML12S1CSVYWUVDvSaslkNuUjvIEotQNnF9+y58Gt1Kz8cH/H0EAAakx0vcLN4ee2gHe4nd7/aZxR/aLRu50uBMq1RdkF9/6j0kTLzPJPdk1/AFM593oHUUt4pVHaq042Qd3SEpCTE03+AiLl32k+d0ss9qByTC3IiXLEu2L5jt0JH+829iP03ydXov/3NV8PHHrm3ICY7f2x2GOX3XKxrWbEfa38+k3+zuU/NpJN/jhMEEBwcSnSOc3o+7hysvHoOZNICQKjUE+BYb8ePZ+nnSNYBRV3WUD4bPomP3VHdQTQ+nna5jmLnpT4gaq5VAmhmYW3UxJIQIO8iP/WsjRgkrVb+n+3Pbxaucd+TgKldn+rQ5ROvNSlpSefyok4G5wvPdx63zjuQji25KlTJe6HYbKIF9y2vHuoML7gx1JBN6oNCkm+IWsnrQUkCU546OrvDmpVLc5XQm61XxnRS3F1KhA6Y5NBkviI5i+v/4ntwsR8n/53vvGvW/vePHLcqeSd/hnPxoRi3VW7dwTAr2rvYGR+bA13OyqmddAnxJsUcDzAZsGV2sungkQXU+BzefOFS1uwybkEGzpnzFQNs2uzvT13q2EjRykCUi4va4Hi75hRYs33F4RMIMFVhqbpR87lDT544lByQdv6c1iRpTq7CRbCSdsH5u5LqxoVh9CP/IQytuMOA0ifEC9VJUl2uBKd7ploOt3w61Nz7a3r5Lrdso8X7Gz3ogfePWLIS9KCAxlTouqvXhW3b3wkYlaAXt7wuMO4Ia1YtTw8v9nomg8DeaL3VVsPe7uJ104bO3ULnPn7ssIEo+j6s9qg1ShGeClj78T5wJ/8R5SQgA9VgOkEeTv6ESfKbLpU8BGDxX6ZRB1EDwzIgSj3a91/cjJXxT+Rgnl23nfo3pPmoipE77bOvGGDHx8DpIv7zKwbWfj3csoP7BADWTu7ZxX07Ju2jUoBpfRRdSDkuvev0iLLq56ObWubkdbhRExd09TxzpMkbQuo7EjOPXvNoGHv5qVB95F1lkpVuR9NVBIn9gzc8EqqufixUarBkO/yC9tY5OnmsB827M039ZEQi3XfXzeHbz76ovKCN0p31hU88fl/C/iQQWbQx05NqVLjhaTz94l+IPocLx5DmYa/o/HDxaiRrfXpdRW9ChS72048vbn0jvL1nfzh5MrH2z21pku5Qr7mhUopQCU9UqrmkHCAowmbELXgDsdgNyHKoU64vnROjgRJt+zEss2CV3abOybY4zI+S4sQpDQSL7y5aSkKcmGjDRfy4PLvO/VLXl+wHTK0f6E/vHPrD16lDxUyZ9b/qnO7uML9WD4M2j5XmenPD8nNx+IWk8wyk6a2qZzESTIhWmi/x9LHfDPVK2Jx7AS9k75zjc7sedFCzjC1aUF4POtmlEIK5fu1K4+TySUzKvWbJPv4PvPGNUNX+XhhnpknNRGA5Xic74TnYDyfq5Ims4UQSP1nfU2yZ3AuqgIcfuDvc/6Hbzcl/6xvvhEMS+yvl4eBif+Lk7yO49FvmPRcDQnxm9oMLgChkOK1mKXeyPYCDCgBC41hVh+8C23ZMus+PMcCzA71k4tLOKcORG4S8xphmkyhHfk9xnWJcKPt5J6H4Urj+eNv2614Aypz1Xtv6S/rB79W3o6W1Q8UsRWV+LqZRyRvWxhUBVy9jY5MBafbwC7Wdf3PPyxWoa3KfVh5rwgsRrVTJ07Ck0H4h/kp6d3r5ZbDUvcKB9YiDmqPBv3TxQvVL9Aad4S0CJGv0S8j7r3hj9Dw0tPVvQ5Wypldf/0gYPrgtjG3+fAgP/c/aw37aOPf9T4ABfVFTpUnRZI3HWs98VdDkIEqt+Bo1kVsJJ/8jigbCyR+rdAKoHFu6nxwcqJUlzsirHANR3JJdkR4pXJyJ8tppx2TU1rnhkJ4gu02xg6vrKeNzpodZgu02pbmDbL9dQHKs16GIa+SaaIuN2Krv9VhOsJRreEJEXzlxXQzbLFFG81Buf3qcv+ZebpcZtZkclfynDXZxit7e5CMxPsZ8YbU2yR9Jq9PbTA44z/9zIbDqiPMMpPGdZrrAezhXJbM/V35pr7gYz7Qhy5ctjdLUzfR9Td5/4yd2hgqBaO2Dv6ORpRhmSQJDL/xFGN75g1Bz7cMzfUET2sO4ROYqPBPgyCG2nci1On0qvG8X+9ulc31dU0zv3H0gSunn1n6ZwM9ySDowcL1cQZw2j7KsWGzcKTsccVhPKQZTwKoUxZPCHTtywLJC+YAudRzXZiBaqlGVw9n29aV6yBTkuB/IwDNZnfJ/biEW4+MDc/tKgCm6Uz5op5TtKoRrc4e6U36u4AKveL9f4Mu4/E8P10X88lTcmWa+N/KjMZEB/H9ypqxudHx8yMa3hcjFo3oGLwzDGnkC4MgXRBw5CazHNHKyOVjP+NQgnD4S6F5jsZ/Y/iNK2JLP5F/IKgI4+V4rf/YsaMETxIDjIGqcKexVEfJzcax/ChxsfZvDAEF9f4ziuYx27HjbyhzwkhqF//1Rci7nbl01QU04WXoBzrZDvrqObdaFhd1D9QKarL/8/goOSjfseortUJnfe1aN4e5ZJQ4778WzQHqOu9y5LnROc2WFb22d3J1ppi8pB5uMbo0m2z60LYzsfl7TtnSbjrTm6gfC2N7NgVlgK2//heQSfOTN8AXhXUB/FOPIEetzUVszcV6QKaKs2P/K67ssVypO/gvmT93NjsHvQOfGIwa9D3z2O4AyYyaEOiGLRw4iDqB0uYMcR7HfqZQYzv42zeoJEb8POUfMOu3QbmwUy7YFiPJ6wAGS77QyjIRqXazdo8psFlDapcHCLqUkRzFnniucwkrJ49JzHT6cZLuqqKyx1nx6FevntP24r6ZwyhmtMgukM9qdhY0hTs60HrTwDFPbAjoNPDWaRntPmKcEAFp758+E8QVXh5FtfxdGnvnPobKpJZkFVt4SUALAUzvHVGuh2oArX7NK+TP///auNTau4gof73ptL7YTHOKAQxOSlMRJQ4AmEBQDeRS1UhtSWt5CLeJHf1CkVuofWrWoqH8qtVJVqUUI9U9/lCJVtFWrFkF5BBJCCiSA84I4UmOSyE6ah+MYEyf7sPt9595zPXv32t6179op2kmu773zOHNmdua7Z87MnAmBXKk04opnw35Kx7s7DwBQj0JK9mbCufkyDHrhfAmUKnn6nZ2AZc5AVKVQRoJTa0n+s8Xj3cDAwI5+pgd1adKfIMvdwZyA4RI3c+5Q16qV9JQ4b0hnNA1QFSQBoHRaDj6ALsttO5pYDsbz8Vm/GQYaxhspWJ5hQHRYIPVJu9G1rV6utpaUwjnrI5zvpDMqIWFUmaxOSkhejVJqDbh6UJq4K23CpFTq5cbTZq771DP7X5Cao3sksWSN1K/8qhIiBiSwFM2OzuYw32uqXgdDFyk3w3Hj80A/6oeDnjdu7OkJ5HpdG/bverdNdrz5hp6+aeDBOgrXgr0TrAwbFbgQl+8EH5usYvqw9SB3zSVpWVoDO3ipM/qWh/6afub79+2XjlvXa7yWFu+kTQ3309rNaJMGny0PklEwtYi8g1nTZxK0LxKp4O7acqfMX7BYtr/+MlZEdKkRaPr7wR5e+y+kSz7UsfAWyfcq6+anDVu7ap07R8+i8k4iBUXkMxGYBjz5DJDPAl5LYIy/o7ULN3oVSN3aiOHZdH9TXc4UAys+CV0tKvltT6mkkfjK953lTV7TUvMlvkjhxbYmFh8XRimFJU6fQiLVVm9ijAXO8J3DfgITL5oc3P3ODvngA6hA/M5MTGDNqMOLSXZaW34AgYkd2jobpVA1fAwUs45sIEo6Jrl6RJEWkQh2Bpzm794N/Lq6uqS35xiMLS8Qd+KJ+dOUnuuMV/Mbkz7SGp82/L/n7m/I+vUbwFNOlrc/Js889Ss9SE8P2vMBzAUxS695+XVn+ZZ7N1o8xZTHk3u73UZUDTMHem/O3FudTUTb/ZiZiTzS509n+UxEY6xwto2qi6kGOKFEC01cxxm1vTGmbMomkz3woqZJ4YSBJDZDeI3GmpCPAAHV8HsQEMsD64UL77mDi/V1qbrl7dfKtx5+RH78xJNCC/E8OZOYQIBkzREAA8kOz9RNqh7RBw4CqA7l/fi4qf6RQ2YCpV0EOF50BATSNjul+uz7KbhqrFHw1fWVAxMvyidd49UnofmQfpHzEYFpaEaPh+otW75SDn70nvQc7cJHIiM3rb29KBm/iVHfxcg8ilJHe7BWSNO+Cb09xzUil0AxYP78hf67V2fRVCb2DfOo+U6crCBGVSItqI6pvXwy+KmCRKyTJVNjyUt9tldqFnXoM4fwHL5XQv9ZKqucQefiex4prbP2nOSJ6oWlEqxgPPJ69733yZ1bvi579+yRrVtfUimIIENnRwhRp2rASX+TePhMCTQsedLflRINROmvw2vc2ME9iAVQ+L2dN8Y1N5Lwjkw+evSIeZV0J12HTFEa4ikBjCeTUqfddnW7Cglcw8kD8AJnHw54kJ7xG6bNn9eVWoP0EQ/Gm9IDfaM5MHBOY+ezFzCqqhNXnREmYzTC/hO9B2tDJ4oYCmd9VV1MNUBDG9QBXnKuZb6M9O7x2EKL1nn7Ult1hQpDvSQldzqeP0SVyKXsqOe+ae1aefxHP5XHHv2Onv9OfikV2tA/zD87MydCXBCl3pEXHaXEQBr1vBQ0DDh8r6KbG55OezYqeA69OZJ3wdb8g7z8/M3fvbvNwj4Sz/7h92pEmiqEfZ275NVXt2qSqDyiZvTJr8uzm1/Uc5g9W9Jl51QlU16ZTZ3hf9MKSIVpFARW4KUqkcZYqRy2DsB6EZf3xLVbKQ72UivukOwbz+guprrV90H4wz5qbdlsbuU08Ti4GaVByZ0SX+Zio66xpVpkIpOBo6mn+mRdrfzyt69YJbz4O7/37r9l51vbdckQlzeZo4RCSVSH5L43n8MuMOgcDhjn3aVje+J50qc5ZqOASCbgGN+Vfuk3yinfPMc0mhavBk7Ug1IP+dtf/wJGtWdj7W2fYiX9neIaicg783IBOjLSOJ6Wj51TZUugWq8yozzRia0sDOWz6/juhrthk3n2q3oySatpompAARQBl46UhUE8gDO18VFl9+ILv5Ts4d3oSCqX+i08qltFla4yfpT2aJCCIMoJBQIU7RBUznnLwdzuNZkaIM+bt9wlT/7s5/LA/ffI1W3e7izyTXou4NkzJ3BsEsfi8T6WUxAKBRqw0JujIErzHIK7LqwpofTLdKYiKAAWAKjNrLv1wDiMT2nrPNaWElB5GqhJqlYmN99SngvyLiGBgfBZbGjhihj73bgum7rrKMcheh4X7/ZMdYtd5mfhdrdwVzUTRT/sF81FOFb1vfQaQAtmB+NECq0XUcqKtGxfOsWpx2RLBJjWY5nTMLaFZt75q4x8vFMStzwoSdg3KLdhT52haAqU6GntiXYIeo+fqJD+1ANRrqfNf/SKMpKEFTLWw2QdPwTubL8tEXL1qFF1TGAzfwMwex+LF4YbgM2G8Q6qSDjcpnOlIotDf6oSFMD5TI+Qs7wLvAmuJOjfqS/lRboKxqSJ9wJnaQo843lh3vxY0Lyi2ZOYDbsJlJIHh7w998aP3aNytjoyiTscR+sXnqQxHp1wuuTmzZt/giGJt10gHFp9n3QNJDGZ0gxL3slkSvr6TsunMJZbjw43/RNRkDzR+il/qkjSNE9S7etleGhQ8u//HdYLz0ntvMXoIGyq5qwJRXU7i1OhO3i97LI0zAE2weLPoAycw2QHUKABZvXicd6mhNzW38nInGtgqrER9fBPqfnccnxrPKvwU8lnLtY3rrnpZvni6jUAnhE5c6pXLmSHceImqOLicJ8Aao7AxIuunNommF2/apVcf8ON8srLr+iHx0CCdAokUni4oK2Z2R+CX4RTXpRQRCC8WASN45clKJNTtuiU5flqHkjCemOZFy5okwULr/GJ1Ej34W4tu8ULqFulBh6F9esuhXKiBL+BFiOChhvXec5VgdSpjUo88nz25uZm3RI5cK5PMtk8joTggvSin74S2StNBVE+aZ66alSSrUtEFt0oNd27JLf3XzKSukySmJQKXNDzpo/PIG88JDBDQ1Np9fUYuvb3ySefDOgzP1BTcewgeRhmqVlwndSv2oyPyFKsIEhJ7sDLklx8y1RIF6Ql7yu+sFJuv30jzL414hTOU3IOx8EEgIpq5c9B3aVWNZ4VCH1/8onHyAu4rO7Bh76t4X987jl9D4DUHrxokX85SAl+YieG5sn0YID86bCazPgX02imloYJGBdptHkxPMJZWcJB9A9/XIrikB/wy2LPnXOF1qvFOYbVCv853K11R1p0lpe9e4wxwBMqxgJRpmWYhmth6FOSy5VQ5SURqkYarwbQyjip0jqvTWf1qT/1dD3jJYozbLRpBY0LrZ92R+twAgG3iuYPvCSZrb+RHIa82hTZkLTXxMlH+bQ4bOZsMfWnVJVMVX9q5U/N8vbVs98PN89WmwNcIzkGDpTPuJ+C/K/fsEke/+ET8t3Hvqez/eyoBEMO/am7tFl8V3dqGVp/trv5L1rQqgatd2x/Xb3K7chWTrsbXb6zjlI42tl0kxYWdbf6VJQj0pXpmF+Yh0gSfgGPfNxVEHzttUv0vSQaBSnjfZna5z1eXj7z1LhzhvpT20LKiZWZ0p9yssm2hSZgPq9hC0zoYeE+h7zZhTdI+vrN6FFsvZ5OMegwM/Qrqf4UOlTaLwj0p/g4leYMHgCa0BHzRAYeXJG6eg3EoTrJ7X9VRlBmPludFI6NS8tl3FioSy7y58UybHv9NXlv99s62890VKxwEodgqjpIgJJy7WtcbJG+5bGsfZU+BsY8LGCcO8HYaDPaaK2MJqIfL34AMhFHO4/G9J4MbJlmPDdeuNEYL70CNSJ8fOyU8ERZ2kmgW7xkmW4aoP50JttodWivP8f0/tHhPvSn1P2dhrHhCxcu4CzxtA5np4sTr9Gheauo470lMcy14X5274s63E+0wLgInAcw7A7+FRaRNFbl/1B/2gjDzIMw9EujHXUY6rM+ix34hERN6Y//KGnnMKTPdXdKzdd+AFFwRLLv/Enyh7ZjvDhf6lff63fEwjoppjt1H5bBhv1zWmZhm+cJOY82wGE/L647tZ8Fr+rcwQH9OtZ1SFtbK3YcHdQPC/342eO3z9LbT8Qw/sK8G2iZH7yKHNe+trTMhgV+HP3j80JadrkJXL5c/1KfyddETtfegiduAOOHoLmxQZa1L0dZMlKbqpe+MwTXo1rGMD2+82Ld6JB9oswmF17VkU6u3uJJxQmUy2c1qf70HKTTeCdVSuDReqsTlQZFEgtXQ186F1ah/iEjPZ0imIxK1DX5oISWrJKqk2iaHzlhRx0kQZRSfbT+1NN15S8OSLbzzzK87zUtV91tj0gSk0oJ6Ih1gmnxGkktWqedTYsRUSeVKh71vZw42bhpoyzBMTHn8XE4dZrtwJcccSeo0bmABW9ZsWKZLFy4CPrdnBw+fEw/xhRew2CnxWF6XCbV8dEFHAUb5KOz+whLpWrk8pZ5MoDtpzS8YjQK0nEYr0TpO3nn8jEWFY2DvMgH6+b0qeOyes0t0pD2ToPgyoW339qpxfOrq4gUaVSBtKhaPkMeaB2UUDipwi2mnJDiTH+0lDUd5fbW2glm92sxuz/iz+5n/dl9mG7yOiFFG23h+mc6GCvKw5XsOSHFiTyujOBEFUWv7IcvSX7336SmuUVqOx6W2rYVAQ1K2DX4ONhM/cyVwmNpbuuVBbP9/WdPyoVMTiVUApYBBPkkhumky8qV0oIlQP04jpsSGSdtWHRzBF/89xwSBc/m59+D7yLTMg0uftSzHNrDzyEZPDMO/d2wENn4X5EZT4QdQr2kIJ6aVDoLxzMfOdKtHyGrJzdzJPHK7n4R3AhTf65KpFOvw3goUDqhlEUQpZQ1c8ulUB40OOskKrk5s/uC2f0EZ/cZR3vTtHalyMqmZM+VEVzEn8GRzMnefZLv/ItI9qIkOx6SOkiclLStTMqxU8aZL8FosWy2/1ZYoOKwv7/vJHbLDSl4EhAooVIqa2pMA3jXwsAJz7sakH37D6q/SbCkyG8df6IxERRBXl0wtv+M+KRPEGVSrTP8IQbxIk175p3gPR590o3LWb7krw9rjdeuXQtdrrdd9Fx/v3QdOhT8xpYny6CSKBNXzlWBtHJ1OznKJmXlYUqIw7w89u5P73IpNjj/Qi9UM3AAoSSG+4mI4f7kShl/KnaW2jzWxO56VoZ7DsjJK9dJ8+o7JdXQjMwMEuLPt1IUbdh/620bMKGyVIawseMM2wOLAke1cEfHOuFhn4MYgu/duzcAUk5MqZSJn1GB1EtS9NfiBAEASfxXy/hNs1qwv/5ioLc1/S3DCWR20Z9+xNOJljEhyqSdwqD+AQnkOQSdcvvSxUJJnm7OFa3S+UGn+ptUGkSvLIgy+1x11p7VcAk66n14oiYPg6PJOe+YklkepyoWWHOZDua9mXub3c9++Nro7P4NWwKBxBrudHDEPIgpzJOTDpn3n5eR491St7RDEsu/JFdBOj3T18dowkP0zGCHevyf/bHZfi792vnmNtn25g4ZhBpoCNs2aSQnA2tI5bpgOM+EREI41icdQTIh56WxIQkJPydp7DajpDwLV6I2jXw984f1qWEA1xC2Q5+SHMG7gg1AeQOf5Nuy6cEkHW0e8PfH/BMEjgR2P/llqCQzXhYFfz0gRcdMJGsxRVd1l1INJPCJ57lGON8oTVDgVxjv+J2mC0Sj86m/7suS+fzN6eFdz+vaU241rWsmX9Pvcif2p7nltaZtsdRvflyStQ3KB415sO7Onu1P//dEj67hhd+M8BhXrbA8997/gGzYdEf60MHOIbQHtaGXyWSH8j4YqlV+IA3vY0mjijGGmmQO8S0uQYrWrK5bdXN6y13fxITTgO7nb2puGrPusBwt/czTT3ML55C/WiuuIhfTAd/GPw7ES6OPsOzpk6exD39IK0H5dA3IGPAWE4vBp6aGv8HQ/wBHmhsC2gZn9AAAAABJRU5ErkJggg==\" height=\"188\" width=\"336\">\n </a>\n</center>",
"_____no_output_____"
],
[
"\n\n\n<h1>What is Colab?</h1>\n\nColab, or \"Colaboratory\", allows you to write and execute Python in your browser, with \n- Zero configuration required\n- Free access to GPUs\n- Easy sharing\n\nWhether you're a **student**, a **data scientist** or an **AI researcher**, Colab can make your work easier. Watch [Introduction to Colab](https://www.youtube.com/watch?v=inN8seMm7UI) to learn more, or just get started below!",
"_____no_output_____"
],
[
"## **Getting started**\n\nThe document you are reading is not a static web page, but an interactive environment called a **Colab notebook** that lets you write and execute code.\n\nFor example, here is a **code cell** with a short Python script that computes a value, stores it in a variable, and prints the result:",
"_____no_output_____"
]
],
[
[
"seconds_in_a_day = 24 * 60 * 60\nseconds_in_a_day",
"_____no_output_____"
]
],
[
[
"To execute the code in the above cell, select it with a click and then either press the play button to the left of the code, or use the keyboard shortcut \"Command/Ctrl+Enter\". To edit the code, just click the cell and start editing.\n\nVariables that you define in one cell can later be used in other cells:",
"_____no_output_____"
]
],
[
[
"seconds_in_a_week = 7 * seconds_in_a_day\nseconds_in_a_week",
"_____no_output_____"
]
],
[
[
"Colab notebooks allow you to combine **executable code** and **rich text** in a single document, along with **images**, **HTML**, **LaTeX** and more. When you create your own Colab notebooks, they are stored in your Google Drive account. You can easily share your Colab notebooks with co-workers or friends, allowing them to comment on your notebooks or even edit them. To learn more, see [Overview of Colab](/notebooks/basic_features_overview.ipynb). To create a new Colab notebook you can use the File menu above, or use the following link: [create a new Colab notebook](http://colab.research.google.com#create=true).\n\nColab notebooks are Jupyter notebooks that are hosted by Colab. To learn more about the Jupyter project, see [jupyter.org](https://www.jupyter.org).",
"_____no_output_____"
],
[
"## Data science\n\nWith Colab you can harness the full power of popular Python libraries to analyze and visualize data. The code cell below uses **numpy** to generate some random data, and uses **matplotlib** to visualize it. To edit the code, just click the cell and start editing.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom matplotlib import pyplot as plt\n\nys = 200 + np.random.randn(100)\nx = [x for x in range(len(ys))]\n\nplt.plot(x, ys, '-')\nplt.fill_between(x, ys, 195, where=(ys > 195), facecolor='g', alpha=0.6)\n\nplt.title(\"Sample Visualization\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"You can import your own data into Colab notebooks from your Google Drive account, including from spreadsheets, as well as from Github and many other sources. To learn more about importing data, and how Colab can be used for data science, see the links below under [Working with Data](#working-with-data).",
"_____no_output_____"
],
[
"## Machine learning\n\nWith Colab you can import an image dataset, train an image classifier on it, and evaluate the model, all in just [a few lines of code](https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/quickstart/beginner.ipynb). Colab notebooks execute code on Google's cloud servers, meaning you can leverage the power of Google hardware, including [GPUs and TPUs](#using-accelerated-hardware), regardless of the power of your machine. All you need is a browser.",
"_____no_output_____"
],
[
"Colab is used extensively in the machine learning community with applications including:\n- Getting started with TensorFlow\n- Developing and training neural networks\n- Experimenting with TPUs\n- Disseminating AI research\n- Creating tutorials\n\nTo see sample Colab notebooks that demonstrate machine learning applications, see the [machine learning examples](#machine-learning-examples) below.",
"_____no_output_____"
],
[
"## More Resources\n\n### Working with Notebooks in Colab\n- [Overview of Colaboratory](/notebooks/basic_features_overview.ipynb)\n- [Guide to Markdown](/notebooks/markdown_guide.ipynb)\n- [Importing libraries and installing dependencies](/notebooks/snippets/importing_libraries.ipynb)\n- [Saving and loading notebooks in GitHub](https://colab.research.google.com/github/googlecolab/colabtools/blob/main/notebooks/colab-github-demo.ipynb)\n- [Interactive forms](/notebooks/forms.ipynb)\n- [Interactive widgets](/notebooks/widgets.ipynb)\n- <img src=\"/img/new.png\" height=\"20px\" align=\"left\" hspace=\"4px\" alt=\"New\"></img>\n [TensorFlow 2 in Colab](/notebooks/tensorflow_version.ipynb)\n\n<a name=\"working-with-data\"></a>\n### Working with Data\n- [Loading data: Drive, Sheets, and Google Cloud Storage](/notebooks/io.ipynb) \n- [Charts: visualizing data](/notebooks/charts.ipynb)\n- [Getting started with BigQuery](/notebooks/bigquery.ipynb)\n\n### Machine Learning Crash Course\nThese are a few of the notebooks from Google's online Machine Learning course. See the [full course website](https://developers.google.com/machine-learning/crash-course/) for more.\n- [Intro to Pandas DataFrame](https://colab.research.google.com/github/google/eng-edu/blob/main/ml/cc/exercises/pandas_dataframe_ultraquick_tutorial.ipynb)\n- [Linear regression with tf.keras using synthetic data](https://colab.research.google.com/github/google/eng-edu/blob/main/ml/cc/exercises/linear_regression_with_synthetic_data.ipynb)\n\n\n<a name=\"using-accelerated-hardware\"></a>\n### Using Accelerated Hardware\n- [TensorFlow with GPUs](/notebooks/gpu.ipynb)\n- [TensorFlow with TPUs](/notebooks/tpu.ipynb)",
"_____no_output_____"
],
[
"<a name=\"machine-learning-examples\"></a>\n\n### Featured examples\n\n- [NeMo Voice Swap](https://colab.research.google.com/github/NVIDIA/NeMo/blob/stable/tutorials/VoiceSwapSample.ipynb): Use Nvidia's NeMo conversational AI Toolkit to swap a voice in an audio fragment with a computer generated one.\n\n- [Retraining an Image Classifier](https://tensorflow.org/hub/tutorials/tf2_image_retraining): Build a Keras model on top of a pre-trained image classifier to distinguish flowers.\n- [Text Classification](https://tensorflow.org/hub/tutorials/tf2_text_classification): Classify IMDB movie reviews as either *positive* or *negative*.\n- [Style Transfer](https://tensorflow.org/hub/tutorials/tf2_arbitrary_image_stylization): Use deep learning to transfer style between images.\n- [Multilingual Universal Sentence Encoder Q&A](https://tensorflow.org/hub/tutorials/retrieval_with_tf_hub_universal_encoder_qa): Use a machine learning model to answer questions from the SQuAD dataset.\n- [Video Interpolation](https://tensorflow.org/hub/tutorials/tweening_conv3d): Predict what happened in a video between the first and the last frame.\n",
"_____no_output_____"
],
[
"My first program",
"_____no_output_____"
]
],
[
[
"x=\"hello world\"\nprint(x)",
"hello world\n"
]
],
[
[
"Addition",
"_____no_output_____"
]
],
[
[
"x=1\ny=2\nz=x+y\nprint(z)",
"3\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6da44d6598d3c9f37937ece1004a4073a23b08 | 134,825 | ipynb | Jupyter Notebook | Tranformer_librispeech.ipynb | VRB01/capstone | 348c3810979ef62e552698190716f05b6f27cf16 | [
"Apache-2.0"
]
| null | null | null | Tranformer_librispeech.ipynb | VRB01/capstone | 348c3810979ef62e552698190716f05b6f27cf16 | [
"Apache-2.0"
]
| null | null | null | Tranformer_librispeech.ipynb | VRB01/capstone | 348c3810979ef62e552698190716f05b6f27cf16 | [
"Apache-2.0"
]
| 1 | 2021-04-21T07:25:32.000Z | 2021-04-21T07:25:32.000Z | 76.692264 | 56,682 | 0.65812 | [
[
[
"<a href=\"https://colab.research.google.com/github/VRB01/capstone/blob/main/Tranformer_librispeech.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import IPython.display as ipd\n# % pylab inline\nimport os\nimport pandas as pd\nimport librosa\nimport glob \nimport librosa.display\nimport random\n\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.model_selection import train_test_split\n\nfrom keras.utils.np_utils import to_categorical\n\nimport numpy as np\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Activation, Flatten\nfrom keras.layers import Convolution2D, MaxPooling2D\nfrom keras.optimizers import Adam\nfrom keras.utils import np_utils\nfrom sklearn import metrics \n\nfrom sklearn.datasets import make_regression\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn import metrics\nfrom sklearn.model_selection import train_test_split, GridSearchCV\n\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Dense, Dropout \nfrom tensorflow.keras.wrappers.scikit_learn import KerasRegressor\n\nfrom keras.callbacks import EarlyStopping\n\nfrom keras import regularizers\n\nfrom sklearn.preprocessing import LabelEncoder\n\nfrom datetime import datetime\n\nimport os\nimport numpy\nfrom keras.models import Sequential\nfrom keras.layers import LSTM\nfrom keras.datasets import imdb\nfrom keras.layers import Dense\nimport tensorflow as tf\nimport numpy as np\nimport pandas as pd \nimport os\nimport librosa\nimport matplotlib.pyplot as plt\nimport gc\n\nfrom tqdm import tqdm, tqdm_notebook\nfrom sklearn.metrics import label_ranking_average_precision_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import MultiLabelBinarizer\nfrom sklearn.model_selection import train_test_split\n\nimport zipfile\n\ntqdm.pandas()",
"/usr/local/lib/python3.7/dist-packages/tqdm/std.py:658: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version\n from pandas import Panel\n"
],
[
"from google.colab import drive\ndrive.mount('/content/gdrive')",
"Mounted at /content/gdrive\n"
],
[
"Directory = 'gdrive/MyDrive/Capstone Data/LibriSpeech/train-clean-100'\nDataset = os.listdir(Directory)",
"_____no_output_____"
],
[
"audio_list = []\nspeakers = []\nfor speaker in Dataset:\n chapters = os.listdir(Directory+'/'+speaker)\n for chapter in chapters:\n audios = os.listdir(Directory+'/'+speaker+'/'+chapter)\n for audio in audios:\n if(audio.endswith('.flac')):\n audio_list.append(Directory+'/'+speaker+'/'+chapter+'/'+audio)\n speakers.append(audio.split('-')[0])",
"_____no_output_____"
],
[
"audio_list = pd.DataFrame(audio_list)\naudio_list = audio_list.rename(columns={0:'file'})\n#len(audio_list)\nlen(speakers)",
"_____no_output_____"
],
[
"audio_list['speaker'] = speakers\ndf = audio_list.sample(frac=1, random_state=42).reset_index(drop=True)\ndf = df[:12000]\ndf_train = df[:8000] #19984:\ndf_validation = df[8000:11000] #19984:25694\ndf_test = df[11000:12000] #25694:\nlabels = df['speaker']\nCounter = 1\ndf",
"_____no_output_____"
],
[
"def scaled_dot_product_attention(query, key, value, mask):\n matmul_qk = tf.matmul(query, key, transpose_b=True)\n\n depth = tf.cast(tf.shape(key)[-1], tf.float32)\n logits = matmul_qk / tf.math.sqrt(depth)\n\n # add the mask zero out padding tokens.\n if mask is not None:\n logits += (mask * -1e9)\n\n attention_weights = tf.nn.softmax(logits, axis=-1)\n\n return tf.matmul(attention_weights, value)",
"_____no_output_____"
],
[
"class MultiHeadAttention(tf.keras.layers.Layer):\n\n def __init__(self, d_model, num_heads, name=\"multi_head_attention\"):\n super(MultiHeadAttention, self).__init__(name=name)\n self.num_heads = num_heads\n self.d_model = d_model\n\n assert d_model % self.num_heads == 0\n\n self.depth = d_model // self.num_heads\n\n self.query_dense = tf.keras.layers.Dense(units=d_model)\n self.key_dense = tf.keras.layers.Dense(units=d_model)\n self.value_dense = tf.keras.layers.Dense(units=d_model)\n\n self.dense = tf.keras.layers.Dense(units=d_model)\n\n def split_heads(self, inputs, batch_size):\n inputs = tf.reshape(\n inputs, shape=(batch_size, -1, self.num_heads, self.depth))\n return tf.transpose(inputs, perm=[0, 2, 1, 3])\n\n def call(self, inputs):\n query, key, value, mask = inputs['query'], inputs['key'], inputs[\n 'value'], inputs['mask']\n batch_size = tf.shape(query)[0]\n\n # linear layers\n query = self.query_dense(query)\n key = self.key_dense(key)\n value = self.value_dense(value)\n\n # split heads\n query = self.split_heads(query, batch_size)\n key = self.split_heads(key, batch_size)\n value = self.split_heads(value, batch_size)\n\n scaled_attention = scaled_dot_product_attention(query, key, value, mask)\n\n scaled_attention = tf.transpose(scaled_attention, perm=[0, 2, 1, 3])\n\n concat_attention = tf.reshape(scaled_attention,\n (batch_size, -1, self.d_model))\n\n outputs = self.dense(concat_attention)\n\n return outputs",
"_____no_output_____"
],
[
"class PositionalEncoding(tf.keras.layers.Layer):\n\n def __init__(self, position, d_model):\n super(PositionalEncoding, self).__init__()\n self.pos_encoding = self.positional_encoding(position, d_model)\n\n def get_angles(self, position, i, d_model):\n angles = 1 / tf.pow(10000, (2 * (i // 2)) / tf.cast(d_model, tf.float32))\n return position * angles\n\n def positional_encoding(self, position, d_model):\n angle_rads = self.get_angles(\n position=tf.range(position, dtype=tf.float32)[:, tf.newaxis],\n i=tf.range(d_model, dtype=tf.float32)[tf.newaxis, :],\n d_model=d_model)\n # apply sin to even index in the array\n sines = tf.math.sin(angle_rads[:, 0::2])\n # apply cos to odd index in the array\n cosines = tf.math.cos(angle_rads[:, 1::2])\n\n pos_encoding = tf.concat([sines, cosines], axis=-1)\n pos_encoding = pos_encoding[tf.newaxis, ...]\n return tf.cast(pos_encoding, tf.float32)\n\n def call(self, inputs):\n return inputs + self.pos_encoding[:, :tf.shape(inputs)[1], :]",
"_____no_output_____"
],
[
"# This allows to the transformer to know where there is real data and where it is padded\ndef create_padding_mask(seq):\n seq = tf.cast(tf.math.equal(seq, 0), tf.float32)\n \n # add extra dimensions to add the padding\n # to the attention logits.\n return seq[:, tf.newaxis, tf.newaxis, :] # (batch_size, 1, 1, seq_len)",
"_____no_output_____"
],
[
"def encoder_layer(units, d_model, num_heads, dropout,name=\"encoder_layer\"):\n inputs = tf.keras.Input(shape=(None,d_model ), name=\"inputs\")\n padding_mask = tf.keras.Input(shape=(1, 1, None), name=\"padding_mask\")\n\n attention = MultiHeadAttention(\n d_model, num_heads, name=\"attention\")({\n 'query': inputs,\n 'key': inputs,\n 'value': inputs,\n 'mask': padding_mask\n })\n attention = tf.keras.layers.Dropout(rate=dropout)(attention)\n attention = tf.keras.layers.LayerNormalization(\n epsilon=1e-6)(inputs + attention)\n\n outputs = tf.keras.layers.Dense(units=units, activation='relu')(attention)\n outputs = tf.keras.layers.Dense(units=d_model)(outputs)\n outputs = tf.keras.layers.Dropout(rate=dropout)(outputs)\n outputs = tf.keras.layers.LayerNormalization(\n epsilon=1e-6)(attention + outputs)\n\n return tf.keras.Model(\n inputs=[inputs, padding_mask], outputs=outputs, name=name)",
"_____no_output_____"
],
[
"def encoder(time_steps,\n num_layers,\n units,\n d_model,\n num_heads,\n dropout,\n projection,\n name=\"encoder\"):\n inputs = tf.keras.Input(shape=(None,d_model), name=\"inputs\")\n padding_mask = tf.keras.Input(shape=(1, 1, None), name=\"padding_mask\")\n \n if projection=='linear':\n ## We implement a linear projection based on Very Deep Self-Attention Networks for End-to-End Speech Recognition. Retrieved from https://arxiv.org/abs/1904.13377\n projection=tf.keras.layers.Dense( d_model,use_bias=True, activation='linear')(inputs)\n print('linear')\n \n else:\n projection=tf.identity(inputs)\n print('none')\n \n projection *= tf.math.sqrt(tf.cast(d_model, tf.float32))\n projection = PositionalEncoding(time_steps, d_model)(projection)\n\n outputs = tf.keras.layers.Dropout(rate=dropout)(projection)\n\n for i in range(num_layers):\n outputs = encoder_layer(\n units=units,\n d_model=d_model,\n num_heads=num_heads,\n dropout=dropout,\n name=\"encoder_layer_{}\".format(i),\n )([outputs, padding_mask])\n \n \n \n\n \n return tf.keras.Model(\n inputs=[inputs, padding_mask], outputs=outputs, name=name)",
"_____no_output_____"
],
[
"def transformer(time_steps,\n num_layers,\n units,\n d_model,\n num_heads,\n dropout,\n output_size,\n projection,\n name=\"transformer\"):\n inputs = tf.keras.Input(shape=(None,d_model), name=\"inputs\")\n \n \n enc_padding_mask = tf.keras.layers.Lambda(\n create_padding_mask, output_shape=(1, 1, None),\n name='enc_padding_mask')(tf.dtypes.cast(\n \n #Like our input has a dimension of length X d_model but the masking is applied to a vector\n # We get the sum for each row and result is a vector. So, if result is 0 it is because in that position was masked \n tf.math.reduce_sum(\n inputs,\n axis=2,\n keepdims=False,\n name=None\n), tf.int32))\n \n\n enc_outputs = encoder(\n time_steps=time_steps,\n num_layers=num_layers,\n units=units,\n d_model=d_model,\n num_heads=num_heads,\n dropout=dropout,\n projection=projection,\n name='encoder'\n )(inputs=[inputs, enc_padding_mask])\n\n #We reshape for feeding our FC in the next step\n outputs=tf.reshape(enc_outputs,(-1,time_steps*d_model))\n \n #We predict our class\n outputs = tf.keras.layers.Dense(units=output_size,use_bias=True,activation='softmax', name=\"outputs\")(outputs)\n\n return tf.keras.Model(inputs=[inputs], outputs=outputs, name='audio_class')",
"_____no_output_____"
],
[
"def extract_features(files):\n \n # Sets the name to be the path to where the file is in my computer\n file_name = os.path.join(str(files.file))\n global Counter\n if(Counter%10==0):\n print(Counter)\n Counter+=1\n\n # Loads the audio file as a floating point time series and assigns the default sample rate\n # Sample rate is set to 22050 by default\n X, sample_rate = librosa.load(file_name, res_type='kaiser_fast') \n\n # Generate Mel-frequency cepstral coefficients (MFCCs) from a time series \n #mfccs = np.mean(librosa.feature.mfcc(y=X, sr=sample_rate, n_mfcc=40).T,axis=0)\n\n # Generates a Short-time Fourier transform (STFT) to use in the chroma_stft\n #stft = np.abs(librosa.stft(X))\n\n # Computes a chromagram from a waveform or power spectrogram.\n #chroma = np.mean(librosa.feature.chroma_stft(S=stft, sr=sample_rate).T,axis=0)\n\n # Computes a mel-scaled spectrogram.\n mel = np.mean(librosa.feature.melspectrogram(X, sr=sample_rate).T,axis=0)\n\n # Computes spectral contrast\n #contrast = np.mean(librosa.feature.spectral_contrast(S=stft, sr=sample_rate).T,axis=0)\n\n # Computes the tonal centroid features (tonnetz)\n #tonnetz = np.mean(librosa.feature.tonnetz(y=librosa.effects.harmonic(X),\n #sr=sample_rate).T,axis=0)\n \n \n # We add also the classes of each file as a label at the end\n #label = files.label\n\n return mel",
"_____no_output_____"
],
[
"startTime = datetime.now()\n# Applying the function to the train data by accessing each row of the dataframe\nfeatures_label = df.apply(extract_features, axis=1)\nprint(datetime.now() - startTime)",
"10\n20\n30\n40\n50\n60\n70\n80\n90\n100\n110\n120\n130\n140\n150\n160\n170\n180\n190\n200\n210\n220\n230\n240\n250\n260\n270\n280\n290\n300\n310\n320\n330\n340\n350\n360\n370\n380\n390\n400\n410\n420\n430\n440\n450\n460\n470\n480\n490\n500\n510\n520\n530\n540\n550\n560\n570\n580\n590\n600\n610\n620\n630\n640\n650\n660\n670\n680\n690\n700\n710\n720\n730\n740\n750\n760\n770\n780\n790\n800\n810\n820\n830\n840\n850\n860\n870\n880\n890\n900\n910\n920\n930\n940\n950\n960\n970\n980\n990\n1000\n1010\n1020\n1030\n1040\n1050\n1060\n1070\n1080\n1090\n1100\n1110\n1120\n1130\n1140\n1150\n1160\n1170\n1180\n1190\n1200\n1210\n1220\n1230\n1240\n1250\n1260\n1270\n1280\n1290\n1300\n1310\n1320\n1330\n1340\n1350\n1360\n1370\n1380\n1390\n1400\n1410\n1420\n1430\n1440\n1450\n1460\n1470\n1480\n1490\n1500\n1510\n1520\n1530\n1540\n1550\n1560\n1570\n1580\n1590\n1600\n1610\n1620\n1630\n1640\n1650\n1660\n1670\n1680\n1690\n1700\n1710\n1720\n1730\n1740\n1750\n1760\n1770\n1780\n1790\n1800\n1810\n1820\n1830\n1840\n1850\n1860\n1870\n1880\n1890\n1900\n1910\n1920\n1930\n1940\n1950\n1960\n1970\n1980\n1990\n2000\n2010\n2020\n2030\n2040\n2050\n2060\n2070\n2080\n2090\n2100\n2110\n2120\n2130\n2140\n2150\n2160\n2170\n2180\n2190\n2200\n2210\n2220\n2230\n2240\n2250\n2260\n2270\n2280\n2290\n2300\n2310\n2320\n2330\n2340\n2350\n2360\n2370\n2380\n2390\n2400\n2410\n2420\n2430\n2440\n2450\n2460\n2470\n2480\n2490\n2500\n2510\n2520\n2530\n2540\n2550\n2560\n2570\n2580\n2590\n2600\n2610\n2620\n2630\n2640\n2650\n2660\n2670\n2680\n"
],
[
"# Saving the numpy array because it takes a long time to extract the features\nnp.save('features_label_libri', features_label)",
"_____no_output_____"
],
[
"# loading the features\nfeatures_label = np.load('features_label_libri.npy', allow_pickle=True)\nfeatures_label.shape",
"_____no_output_____"
],
[
"trial_features=[]\nfor i in range(0,len(features_label)):\n a=[]\n a.append(features_label[i])\n #a.append(features_label[i][1])\n trial_features.append(a)\nxxx = np.array(trial_features)\nxxx.shape",
"_____no_output_____"
],
[
"X = xxx\ny = np.array(labels)\nlb = LabelEncoder()\ny = to_categorical(lb.fit_transform(y))\nX.shape\ny.shape",
"_____no_output_____"
],
[
"limit_1 = int(X.shape[0]*0.7)\nlimit_2 = int(X.shape[0]*0.85)",
"_____no_output_____"
],
[
"X_train = X[:limit_1]\nY_train = y[:limit_1]\n\nX_val = X[limit_1:limit_2]\nY_val = y[limit_1:limit_2]\n\nX_test = X[limit_2:]\nY_test = y[limit_2:]",
"_____no_output_____"
],
[
"# #We get our train and test set\n# X_train,X_test, Y_train, Y_test =train_test_split(X,y, test_size=0.2, random_state=27)",
"_____no_output_____"
],
[
"projection=['linear','none']\naccuracy=[]\nproj_implemented=[]",
"_____no_output_____"
],
[
"for i in projection:\n NUM_LAYERS = 2\n D_MODEL = X.shape[2]\n NUM_HEADS = 4\n UNITS = 1024\n DROPOUT = 0.1\n TIME_STEPS= X.shape[1]\n OUTPUT_SIZE=251\n EPOCHS = 100\n EXPERIMENTS=1\n\n \n for j in range(EXPERIMENTS):\n \n \n model = transformer(time_steps=TIME_STEPS,\n num_layers=NUM_LAYERS,\n units=UNITS,\n d_model=D_MODEL,\n num_heads=NUM_HEADS,\n dropout=DROPOUT,\n output_size=OUTPUT_SIZE, \n projection=i)\n \n #model.compile(optimizer=tf.keras.optimizers.Adam(0.000001), loss='categorical_crossentropy', metrics=['accuracy'])\n model.compile(loss='categorical_crossentropy', metrics=['accuracy'], optimizer='adam')\n\n early_stop = EarlyStopping(monitor='val_loss', min_delta=0, patience=100, verbose=1, mode='auto')\n #history=model.fit(X_train,Y_train, epochs=EPOCHS, validation_data=(X_test, Y_test))\n history = model.fit(X_train, Y_train, batch_size=64, epochs=100, validation_data=(X_val, Y_val),callbacks=[early_stop])\n \n \n accuracy.append(sum(history.history['val_accuracy'])/len(history.history['val_accuracy']))\n \n proj_implemented.append(i)\n \n",
"linear\nEpoch 1/100\n132/132 [==============================] - 6s 22ms/step - loss: 4.8455 - accuracy: 0.0781 - val_loss: 2.9428 - val_accuracy: 0.3161\nEpoch 2/100\n132/132 [==============================] - 2s 18ms/step - loss: 2.7140 - accuracy: 0.3695 - val_loss: 1.8750 - val_accuracy: 0.5422\nEpoch 3/100\n132/132 [==============================] - 2s 18ms/step - loss: 1.8521 - accuracy: 0.5424 - val_loss: 1.3713 - val_accuracy: 0.6444\nEpoch 4/100\n132/132 [==============================] - 2s 18ms/step - loss: 1.4018 - accuracy: 0.6480 - val_loss: 1.2148 - val_accuracy: 0.6717\nEpoch 5/100\n132/132 [==============================] - 2s 18ms/step - loss: 1.1571 - accuracy: 0.6874 - val_loss: 1.0328 - val_accuracy: 0.7183\nEpoch 6/100\n132/132 [==============================] - 2s 18ms/step - loss: 1.0229 - accuracy: 0.7191 - val_loss: 0.9360 - val_accuracy: 0.7461\nEpoch 7/100\n132/132 [==============================] - 2s 17ms/step - loss: 0.8704 - accuracy: 0.7548 - val_loss: 0.8492 - val_accuracy: 0.7650\nEpoch 8/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.7440 - accuracy: 0.7914 - val_loss: 0.8110 - val_accuracy: 0.7739\nEpoch 9/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.6556 - accuracy: 0.8048 - val_loss: 0.7202 - val_accuracy: 0.7906\nEpoch 10/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.5798 - accuracy: 0.8313 - val_loss: 0.8110 - val_accuracy: 0.7750\nEpoch 11/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.5771 - accuracy: 0.8289 - val_loss: 0.7683 - val_accuracy: 0.7811\nEpoch 12/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.5570 - accuracy: 0.8245 - val_loss: 0.6585 - val_accuracy: 0.8150\nEpoch 13/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4777 - accuracy: 0.8562 - val_loss: 0.7419 - val_accuracy: 0.7933\nEpoch 14/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4538 - accuracy: 0.8570 - val_loss: 0.7103 - val_accuracy: 0.8044\nEpoch 15/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4520 - accuracy: 0.8614 - val_loss: 0.6962 - val_accuracy: 0.8033\nEpoch 16/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4184 - accuracy: 0.8631 - val_loss: 0.6866 - val_accuracy: 0.8122\nEpoch 17/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3831 - accuracy: 0.8786 - val_loss: 0.6559 - val_accuracy: 0.8311\nEpoch 18/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3252 - accuracy: 0.9008 - val_loss: 0.7489 - val_accuracy: 0.7961\nEpoch 19/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3541 - accuracy: 0.8939 - val_loss: 0.7950 - val_accuracy: 0.7950\nEpoch 20/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3313 - accuracy: 0.8970 - val_loss: 0.7187 - val_accuracy: 0.8028\nEpoch 21/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3543 - accuracy: 0.8872 - val_loss: 0.7109 - val_accuracy: 0.8217\nEpoch 22/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2956 - accuracy: 0.9108 - val_loss: 0.8206 - val_accuracy: 0.7856\nEpoch 23/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3106 - accuracy: 0.9000 - val_loss: 0.7584 - val_accuracy: 0.8061\nEpoch 24/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2423 - accuracy: 0.9228 - val_loss: 0.6907 - val_accuracy: 0.8244\nEpoch 25/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2651 - accuracy: 0.9183 - val_loss: 0.7692 - val_accuracy: 0.8094\nEpoch 26/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2765 - accuracy: 0.9111 - val_loss: 0.7401 - val_accuracy: 0.8133\nEpoch 27/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2514 - accuracy: 0.9184 - val_loss: 0.7297 - val_accuracy: 0.8222\nEpoch 28/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2437 - accuracy: 0.9222 - val_loss: 0.8924 - val_accuracy: 0.7822\nEpoch 29/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2770 - accuracy: 0.9105 - val_loss: 0.7733 - val_accuracy: 0.8211\nEpoch 30/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2237 - accuracy: 0.9303 - val_loss: 0.7396 - val_accuracy: 0.8211\nEpoch 31/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2424 - accuracy: 0.9220 - val_loss: 0.7768 - val_accuracy: 0.8111\nEpoch 32/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2455 - accuracy: 0.9224 - val_loss: 0.7093 - val_accuracy: 0.8172\nEpoch 33/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1998 - accuracy: 0.9442 - val_loss: 0.7491 - val_accuracy: 0.8256\nEpoch 34/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2134 - accuracy: 0.9303 - val_loss: 0.9013 - val_accuracy: 0.7872\nEpoch 35/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2443 - accuracy: 0.9167 - val_loss: 0.7683 - val_accuracy: 0.8189\nEpoch 36/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2452 - accuracy: 0.9195 - val_loss: 0.7610 - val_accuracy: 0.8300\nEpoch 37/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1866 - accuracy: 0.9397 - val_loss: 0.7727 - val_accuracy: 0.8139\nEpoch 38/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2023 - accuracy: 0.9333 - val_loss: 0.7860 - val_accuracy: 0.8256\nEpoch 39/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1962 - accuracy: 0.9377 - val_loss: 0.7429 - val_accuracy: 0.8256\nEpoch 40/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2007 - accuracy: 0.9337 - val_loss: 0.7880 - val_accuracy: 0.8167\nEpoch 41/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2328 - accuracy: 0.9260 - val_loss: 0.7359 - val_accuracy: 0.8261\nEpoch 42/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1843 - accuracy: 0.9387 - val_loss: 0.8955 - val_accuracy: 0.7900\nEpoch 43/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2403 - accuracy: 0.9236 - val_loss: 0.7488 - val_accuracy: 0.8228\nEpoch 44/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2007 - accuracy: 0.9389 - val_loss: 0.8617 - val_accuracy: 0.8117\nEpoch 45/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2050 - accuracy: 0.9343 - val_loss: 0.7854 - val_accuracy: 0.8250\nEpoch 46/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1827 - accuracy: 0.9417 - val_loss: 0.8236 - val_accuracy: 0.8183\nEpoch 47/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1517 - accuracy: 0.9517 - val_loss: 0.7856 - val_accuracy: 0.8206\nEpoch 48/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1800 - accuracy: 0.9441 - val_loss: 0.8231 - val_accuracy: 0.8228\nEpoch 49/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1623 - accuracy: 0.9467 - val_loss: 0.8813 - val_accuracy: 0.8144\nEpoch 50/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1683 - accuracy: 0.9482 - val_loss: 0.8488 - val_accuracy: 0.8150\nEpoch 51/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1691 - accuracy: 0.9460 - val_loss: 0.9480 - val_accuracy: 0.7956\nEpoch 52/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2381 - accuracy: 0.9237 - val_loss: 0.9664 - val_accuracy: 0.7972\nEpoch 53/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1854 - accuracy: 0.9382 - val_loss: 0.8772 - val_accuracy: 0.8056\nEpoch 54/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1850 - accuracy: 0.9408 - val_loss: 0.9049 - val_accuracy: 0.8100\nEpoch 55/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1854 - accuracy: 0.9430 - val_loss: 0.8121 - val_accuracy: 0.8261\nEpoch 56/100\n132/132 [==============================] - 2s 17ms/step - loss: 0.1605 - accuracy: 0.9469 - val_loss: 0.7862 - val_accuracy: 0.8167\nEpoch 57/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1519 - accuracy: 0.9508 - val_loss: 0.8173 - val_accuracy: 0.8194\nEpoch 58/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1401 - accuracy: 0.9553 - val_loss: 0.8291 - val_accuracy: 0.8194\nEpoch 59/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1694 - accuracy: 0.9424 - val_loss: 0.9036 - val_accuracy: 0.8072\nEpoch 60/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1533 - accuracy: 0.9485 - val_loss: 0.8355 - val_accuracy: 0.8167\nEpoch 61/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1750 - accuracy: 0.9449 - val_loss: 0.8567 - val_accuracy: 0.8133\nEpoch 62/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1808 - accuracy: 0.9403 - val_loss: 0.9194 - val_accuracy: 0.8111\nEpoch 63/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1443 - accuracy: 0.9572 - val_loss: 0.9375 - val_accuracy: 0.7989\nEpoch 64/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1720 - accuracy: 0.9427 - val_loss: 0.8276 - val_accuracy: 0.8289\nEpoch 65/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1802 - accuracy: 0.9430 - val_loss: 0.9028 - val_accuracy: 0.8222\nEpoch 66/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.1722 - accuracy: 0.9437 - val_loss: 0.8011 - val_accuracy: 0.8383\nEpoch 67/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1047 - accuracy: 0.9641 - val_loss: 0.8548 - val_accuracy: 0.8144\nEpoch 68/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1381 - accuracy: 0.9524 - val_loss: 0.8582 - val_accuracy: 0.8250\nEpoch 69/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1484 - accuracy: 0.9468 - val_loss: 0.8644 - val_accuracy: 0.8333\nEpoch 70/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1134 - accuracy: 0.9623 - val_loss: 0.7688 - val_accuracy: 0.8344\nEpoch 71/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1383 - accuracy: 0.9544 - val_loss: 0.8558 - val_accuracy: 0.8300\nEpoch 72/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1887 - accuracy: 0.9436 - val_loss: 0.8080 - val_accuracy: 0.8361\nEpoch 73/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1213 - accuracy: 0.9599 - val_loss: 0.9942 - val_accuracy: 0.8128\nEpoch 74/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1306 - accuracy: 0.9594 - val_loss: 0.8454 - val_accuracy: 0.8261\nEpoch 75/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1212 - accuracy: 0.9609 - val_loss: 0.8359 - val_accuracy: 0.8261\nEpoch 76/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.1463 - accuracy: 0.9483 - val_loss: 0.9228 - val_accuracy: 0.8189\nEpoch 77/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1102 - accuracy: 0.9640 - val_loss: 0.8861 - val_accuracy: 0.8289\nEpoch 78/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1241 - accuracy: 0.9619 - val_loss: 0.8519 - val_accuracy: 0.8317\nEpoch 79/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1329 - accuracy: 0.9559 - val_loss: 0.8393 - val_accuracy: 0.8256\nEpoch 80/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1313 - accuracy: 0.9560 - val_loss: 0.9059 - val_accuracy: 0.8172\nEpoch 81/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1308 - accuracy: 0.9565 - val_loss: 0.8844 - val_accuracy: 0.8250\nEpoch 82/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1307 - accuracy: 0.9558 - val_loss: 0.8035 - val_accuracy: 0.8322\nEpoch 83/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.1463 - accuracy: 0.9535 - val_loss: 0.8682 - val_accuracy: 0.8289\nEpoch 84/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1159 - accuracy: 0.9623 - val_loss: 0.8754 - val_accuracy: 0.8117\nEpoch 85/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1201 - accuracy: 0.9622 - val_loss: 0.8686 - val_accuracy: 0.8244\nEpoch 86/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.0959 - accuracy: 0.9689 - val_loss: 0.9308 - val_accuracy: 0.8228\nEpoch 87/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1237 - accuracy: 0.9602 - val_loss: 0.9020 - val_accuracy: 0.8206\nEpoch 88/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1034 - accuracy: 0.9675 - val_loss: 0.8965 - val_accuracy: 0.8239\nEpoch 89/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.0867 - accuracy: 0.9734 - val_loss: 0.8882 - val_accuracy: 0.8339\nEpoch 90/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1104 - accuracy: 0.9664 - val_loss: 0.8294 - val_accuracy: 0.8417\nEpoch 91/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.0967 - accuracy: 0.9660 - val_loss: 0.8227 - val_accuracy: 0.8339\nEpoch 92/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1288 - accuracy: 0.9604 - val_loss: 0.9676 - val_accuracy: 0.8200\nEpoch 93/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1479 - accuracy: 0.9540 - val_loss: 0.8641 - val_accuracy: 0.8294\nEpoch 94/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1354 - accuracy: 0.9559 - val_loss: 0.8306 - val_accuracy: 0.8417\nEpoch 95/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1140 - accuracy: 0.9650 - val_loss: 0.9555 - val_accuracy: 0.8150\nEpoch 96/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.0842 - accuracy: 0.9739 - val_loss: 0.8891 - val_accuracy: 0.8300\nEpoch 97/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1083 - accuracy: 0.9672 - val_loss: 0.8790 - val_accuracy: 0.8239\nEpoch 98/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1173 - accuracy: 0.9634 - val_loss: 0.9254 - val_accuracy: 0.8300\nEpoch 99/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.0952 - accuracy: 0.9706 - val_loss: 0.8215 - val_accuracy: 0.8328\nEpoch 100/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.1500 - accuracy: 0.9523 - val_loss: 0.8894 - val_accuracy: 0.8222\nnone\nEpoch 1/100\n132/132 [==============================] - 6s 25ms/step - loss: 4.8855 - accuracy: 0.0772 - val_loss: 2.9260 - val_accuracy: 0.3461\nEpoch 2/100\n132/132 [==============================] - 2s 18ms/step - loss: 2.9581 - accuracy: 0.3213 - val_loss: 1.9932 - val_accuracy: 0.5450\nEpoch 3/100\n132/132 [==============================] - 2s 18ms/step - loss: 2.1457 - accuracy: 0.4780 - val_loss: 1.4570 - val_accuracy: 0.6356\nEpoch 4/100\n132/132 [==============================] - 2s 18ms/step - loss: 1.7143 - accuracy: 0.5580 - val_loss: 1.2393 - val_accuracy: 0.6733\nEpoch 5/100\n132/132 [==============================] - 2s 19ms/step - loss: 1.4473 - accuracy: 0.6218 - val_loss: 1.0643 - val_accuracy: 0.7194\nEpoch 6/100\n132/132 [==============================] - 2s 19ms/step - loss: 1.2834 - accuracy: 0.6439 - val_loss: 0.9645 - val_accuracy: 0.7522\nEpoch 7/100\n132/132 [==============================] - 2s 18ms/step - loss: 1.1575 - accuracy: 0.6785 - val_loss: 0.8082 - val_accuracy: 0.7883\nEpoch 8/100\n132/132 [==============================] - 2s 18ms/step - loss: 1.0333 - accuracy: 0.7041 - val_loss: 0.8616 - val_accuracy: 0.7622\nEpoch 9/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.9169 - accuracy: 0.7473 - val_loss: 0.8123 - val_accuracy: 0.7750\nEpoch 10/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.9167 - accuracy: 0.7367 - val_loss: 0.7556 - val_accuracy: 0.7817\nEpoch 11/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.8644 - accuracy: 0.7524 - val_loss: 0.6612 - val_accuracy: 0.8139\nEpoch 12/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.7934 - accuracy: 0.7761 - val_loss: 0.6748 - val_accuracy: 0.8117\nEpoch 13/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.7336 - accuracy: 0.7913 - val_loss: 0.6668 - val_accuracy: 0.8172\nEpoch 14/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.7323 - accuracy: 0.7829 - val_loss: 0.6160 - val_accuracy: 0.8378\nEpoch 15/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.6565 - accuracy: 0.8059 - val_loss: 0.6324 - val_accuracy: 0.8144\nEpoch 16/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.6492 - accuracy: 0.8171 - val_loss: 0.6159 - val_accuracy: 0.8328\nEpoch 17/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.6312 - accuracy: 0.8136 - val_loss: 0.5908 - val_accuracy: 0.8322\nEpoch 18/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.5681 - accuracy: 0.8283 - val_loss: 0.6089 - val_accuracy: 0.8361\nEpoch 19/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.5522 - accuracy: 0.8334 - val_loss: 0.6645 - val_accuracy: 0.8156\nEpoch 20/100\n132/132 [==============================] - 2s 17ms/step - loss: 0.5701 - accuracy: 0.8289 - val_loss: 0.6319 - val_accuracy: 0.8233\nEpoch 21/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.5371 - accuracy: 0.8403 - val_loss: 0.6651 - val_accuracy: 0.8211\nEpoch 22/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.5098 - accuracy: 0.8397 - val_loss: 0.6127 - val_accuracy: 0.8317\nEpoch 23/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4856 - accuracy: 0.8554 - val_loss: 0.5891 - val_accuracy: 0.8439\nEpoch 24/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4601 - accuracy: 0.8615 - val_loss: 0.5547 - val_accuracy: 0.8544\nEpoch 25/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4718 - accuracy: 0.8590 - val_loss: 0.5637 - val_accuracy: 0.8400\nEpoch 26/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4085 - accuracy: 0.8727 - val_loss: 0.5811 - val_accuracy: 0.8394\nEpoch 27/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4304 - accuracy: 0.8735 - val_loss: 0.5479 - val_accuracy: 0.8494\nEpoch 28/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4395 - accuracy: 0.8663 - val_loss: 0.5440 - val_accuracy: 0.8467\nEpoch 29/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4307 - accuracy: 0.8725 - val_loss: 0.5596 - val_accuracy: 0.8439\nEpoch 30/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4311 - accuracy: 0.8651 - val_loss: 0.5281 - val_accuracy: 0.8528\nEpoch 31/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4386 - accuracy: 0.8723 - val_loss: 0.5479 - val_accuracy: 0.8528\nEpoch 32/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.4113 - accuracy: 0.8686 - val_loss: 0.5361 - val_accuracy: 0.8567\nEpoch 33/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3959 - accuracy: 0.8796 - val_loss: 0.5387 - val_accuracy: 0.8622\nEpoch 34/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3911 - accuracy: 0.8782 - val_loss: 0.4901 - val_accuracy: 0.8706\nEpoch 35/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3397 - accuracy: 0.8955 - val_loss: 0.5571 - val_accuracy: 0.8494\nEpoch 36/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3841 - accuracy: 0.8803 - val_loss: 0.5625 - val_accuracy: 0.8594\nEpoch 37/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3564 - accuracy: 0.8885 - val_loss: 0.5716 - val_accuracy: 0.8422\nEpoch 38/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.4113 - accuracy: 0.8759 - val_loss: 0.5696 - val_accuracy: 0.8461\nEpoch 39/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3628 - accuracy: 0.8857 - val_loss: 0.5211 - val_accuracy: 0.8589\nEpoch 40/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3327 - accuracy: 0.8969 - val_loss: 0.5424 - val_accuracy: 0.8622\nEpoch 41/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3340 - accuracy: 0.9019 - val_loss: 0.5492 - val_accuracy: 0.8572\nEpoch 42/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3199 - accuracy: 0.9037 - val_loss: 0.5510 - val_accuracy: 0.8633\nEpoch 43/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3687 - accuracy: 0.8919 - val_loss: 0.5033 - val_accuracy: 0.8694\nEpoch 44/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3310 - accuracy: 0.9023 - val_loss: 0.5614 - val_accuracy: 0.8606\nEpoch 45/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3399 - accuracy: 0.8946 - val_loss: 0.5677 - val_accuracy: 0.8506\nEpoch 46/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3140 - accuracy: 0.9013 - val_loss: 0.4967 - val_accuracy: 0.8744\nEpoch 47/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2778 - accuracy: 0.9121 - val_loss: 0.5538 - val_accuracy: 0.8572\nEpoch 48/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3036 - accuracy: 0.9089 - val_loss: 0.5605 - val_accuracy: 0.8522\nEpoch 49/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3257 - accuracy: 0.8993 - val_loss: 0.5159 - val_accuracy: 0.8706\nEpoch 50/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.3162 - accuracy: 0.9000 - val_loss: 0.5413 - val_accuracy: 0.8661\nEpoch 51/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.3066 - accuracy: 0.9043 - val_loss: 0.5910 - val_accuracy: 0.8478\nEpoch 52/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2923 - accuracy: 0.9041 - val_loss: 0.5634 - val_accuracy: 0.8478\nEpoch 53/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2821 - accuracy: 0.9140 - val_loss: 0.5207 - val_accuracy: 0.8672\nEpoch 54/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.3006 - accuracy: 0.9066 - val_loss: 0.6349 - val_accuracy: 0.8461\nEpoch 55/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2795 - accuracy: 0.9174 - val_loss: 0.5307 - val_accuracy: 0.8644\nEpoch 56/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2701 - accuracy: 0.9171 - val_loss: 0.5650 - val_accuracy: 0.8628\nEpoch 57/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2948 - accuracy: 0.9111 - val_loss: 0.5276 - val_accuracy: 0.8589\nEpoch 58/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2749 - accuracy: 0.9124 - val_loss: 0.5607 - val_accuracy: 0.8528\nEpoch 59/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2757 - accuracy: 0.9157 - val_loss: 0.5436 - val_accuracy: 0.8606\nEpoch 60/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2936 - accuracy: 0.9106 - val_loss: 0.5222 - val_accuracy: 0.8717\nEpoch 61/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2664 - accuracy: 0.9206 - val_loss: 0.6016 - val_accuracy: 0.8533\nEpoch 62/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.3295 - accuracy: 0.9005 - val_loss: 0.5363 - val_accuracy: 0.8633\nEpoch 63/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2512 - accuracy: 0.9250 - val_loss: 0.5101 - val_accuracy: 0.8644\nEpoch 64/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2679 - accuracy: 0.9149 - val_loss: 0.5263 - val_accuracy: 0.8650\nEpoch 65/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2412 - accuracy: 0.9262 - val_loss: 0.4766 - val_accuracy: 0.8678\nEpoch 66/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2538 - accuracy: 0.9259 - val_loss: 0.5107 - val_accuracy: 0.8722\nEpoch 67/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2565 - accuracy: 0.9242 - val_loss: 0.5320 - val_accuracy: 0.8644\nEpoch 68/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2356 - accuracy: 0.9276 - val_loss: 0.6057 - val_accuracy: 0.8489\nEpoch 69/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2312 - accuracy: 0.9278 - val_loss: 0.5606 - val_accuracy: 0.8600\nEpoch 70/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2213 - accuracy: 0.9320 - val_loss: 0.5961 - val_accuracy: 0.8600\nEpoch 71/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2479 - accuracy: 0.9255 - val_loss: 0.5579 - val_accuracy: 0.8628\nEpoch 72/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2462 - accuracy: 0.9216 - val_loss: 0.5648 - val_accuracy: 0.8656\nEpoch 73/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2305 - accuracy: 0.9324 - val_loss: 0.5288 - val_accuracy: 0.8661\nEpoch 74/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2559 - accuracy: 0.9227 - val_loss: 0.5753 - val_accuracy: 0.8611\nEpoch 75/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2329 - accuracy: 0.9259 - val_loss: 0.5182 - val_accuracy: 0.8767\nEpoch 76/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2384 - accuracy: 0.9261 - val_loss: 0.5836 - val_accuracy: 0.8661\nEpoch 77/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2563 - accuracy: 0.9241 - val_loss: 0.5325 - val_accuracy: 0.8667\nEpoch 78/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2614 - accuracy: 0.9255 - val_loss: 0.6201 - val_accuracy: 0.8417\nEpoch 79/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2444 - accuracy: 0.9214 - val_loss: 0.5769 - val_accuracy: 0.8656\nEpoch 80/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2184 - accuracy: 0.9326 - val_loss: 0.5065 - val_accuracy: 0.8711\nEpoch 81/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2384 - accuracy: 0.9264 - val_loss: 0.5523 - val_accuracy: 0.8628\nEpoch 82/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2346 - accuracy: 0.9299 - val_loss: 0.5678 - val_accuracy: 0.8639\nEpoch 83/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2694 - accuracy: 0.9220 - val_loss: 0.5469 - val_accuracy: 0.8578\nEpoch 84/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2098 - accuracy: 0.9305 - val_loss: 0.5520 - val_accuracy: 0.8628\nEpoch 85/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2459 - accuracy: 0.9271 - val_loss: 0.5672 - val_accuracy: 0.8583\nEpoch 86/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2483 - accuracy: 0.9262 - val_loss: 0.6077 - val_accuracy: 0.8494\nEpoch 87/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2232 - accuracy: 0.9294 - val_loss: 0.6494 - val_accuracy: 0.8467\nEpoch 88/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2190 - accuracy: 0.9351 - val_loss: 0.6562 - val_accuracy: 0.8400\nEpoch 89/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2395 - accuracy: 0.9302 - val_loss: 0.5531 - val_accuracy: 0.8689\nEpoch 90/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2183 - accuracy: 0.9322 - val_loss: 0.5629 - val_accuracy: 0.8650\nEpoch 91/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2309 - accuracy: 0.9250 - val_loss: 0.5682 - val_accuracy: 0.8561\nEpoch 92/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2286 - accuracy: 0.9297 - val_loss: 0.5734 - val_accuracy: 0.8728\nEpoch 93/100\n132/132 [==============================] - 3s 19ms/step - loss: 0.2003 - accuracy: 0.9422 - val_loss: 0.5538 - val_accuracy: 0.8628\nEpoch 94/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2036 - accuracy: 0.9354 - val_loss: 0.5850 - val_accuracy: 0.8567\nEpoch 95/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2016 - accuracy: 0.9372 - val_loss: 0.5641 - val_accuracy: 0.8672\nEpoch 96/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2174 - accuracy: 0.9330 - val_loss: 0.5581 - val_accuracy: 0.8694\nEpoch 97/100\n132/132 [==============================] - 2s 19ms/step - loss: 0.2200 - accuracy: 0.9346 - val_loss: 0.5727 - val_accuracy: 0.8661\nEpoch 98/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2033 - accuracy: 0.9392 - val_loss: 0.5561 - val_accuracy: 0.8661\nEpoch 99/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.1930 - accuracy: 0.9445 - val_loss: 0.5580 - val_accuracy: 0.8600\nEpoch 100/100\n132/132 [==============================] - 2s 18ms/step - loss: 0.2037 - accuracy: 0.9400 - val_loss: 0.5918 - val_accuracy: 0.8567\n"
],
[
"# Check out our train accuracy and validation accuracy over epochs.\ntrain_accuracy = history.history['accuracy']\nval_accuracy = history.history['val_accuracy']\nimport matplotlib.pyplot as plt\n# Set figure size.\nplt.figure(figsize=(12, 8))\n\n# Generate line plot of training, testing loss over epochs.\nplt.plot(train_accuracy, label='Training Accuracy', color='#185fad')\nplt.plot(val_accuracy, label='Validation Accuracy', color='orange')\n\n# Set title\nplt.title('Training and Validation Accuracy by Epoch', fontsize = 25)\nplt.xlabel('Epoch', fontsize = 18)\nplt.ylabel('Categorical Crossentropy', fontsize = 18)\nplt.xticks(range(0,100,5), range(0,100,5))\n\nplt.legend(fontsize = 18);",
"_____no_output_____"
],
[
"accuracy=pd.DataFrame(accuracy, columns=['accuracy'])\nproj_implemented=pd.DataFrame(proj_implemented, columns=['projection'])\nresults=pd.concat([accuracy,proj_implemented],axis=1)",
"_____no_output_____"
],
[
"results.groupby('projection').mean()",
"_____no_output_____"
],
[
"import keras\n\ny_prob = model.predict(X_test)\ny_classes = y_prob.argmax(axis=-1)\n\nres_list = y_classes.tolist()\nlabel_mapping = {0:'Aayush',1:'Kanishk',2:'Kayan',3:'Rohit'}#clarify\n\n\n# for i in range(len(res_list)):\n# print(\"prediction \",i,\" \",label_mapping[res_list[i]])\n\n\n",
"_____no_output_____"
],
[
"model.evaluate(X_test,Y_test)",
"57/57 [==============================] - 0s 4ms/step - loss: 0.6682 - accuracy: 0.8433\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6dbd356c9a37fdd28d8be0b716dd88ce3309ac | 50,243 | ipynb | Jupyter Notebook | HMM Tagger.ipynb | Laurans/hmm-tagger | 4ca1b33bbff8fa3ec1511018fa5e5953a0fa1f52 | [
"MIT"
]
| null | null | null | HMM Tagger.ipynb | Laurans/hmm-tagger | 4ca1b33bbff8fa3ec1511018fa5e5953a0fa1f52 | [
"MIT"
]
| null | null | null | HMM Tagger.ipynb | Laurans/hmm-tagger | 4ca1b33bbff8fa3ec1511018fa5e5953a0fa1f52 | [
"MIT"
]
| null | null | null | 42.434966 | 660 | 0.58842 | [
[
[
"# Project: Part of Speech Tagging with Hidden Markov Models \n---\n### Introduction\n\nPart of speech tagging is the process of determining the syntactic category of a word from the words in its surrounding context. It is often used to help disambiguate natural language phrases because it can be done quickly with high accuracy. Tagging can be used for many NLP tasks like determining correct pronunciation during speech synthesis (for example, _dis_-count as a noun vs dis-_count_ as a verb), for information retrieval, and for word sense disambiguation.\n\nIn this notebook, you'll use the [Pomegranate](http://pomegranate.readthedocs.io/) library to build a hidden Markov model for part of speech tagging using a \"universal\" tagset. Hidden Markov models have been able to achieve [>96% tag accuracy with larger tagsets on realistic text corpora](http://www.coli.uni-saarland.de/~thorsten/publications/Brants-ANLP00.pdf). Hidden Markov models have also been used for speech recognition and speech generation, machine translation, gene recognition for bioinformatics, and human gesture recognition for computer vision, and more. \n\n\n\nThe notebook already contains some code to get you started. You only need to add some new functionality in the areas indicated to complete the project; you will not need to modify the included code beyond what is requested. Sections that begin with **'IMPLEMENTATION'** in the header indicate that you must provide code in the block that follows. Instructions will be provided for each section, and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n**Note:** Once you have completed all of the code implementations, you need to finalize your work by exporting the iPython Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You must then **export the notebook** by running the last cell in the notebook, or by using the menu above and navigating to **File -> Download as -> HTML (.html)** Your submissions should include both the `html` and `ipynb` files.\n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n**Note:** Code and Markdown cells can be executed using the `Shift + Enter` keyboard shortcut. Markdown cells can be edited by double-clicking the cell to enter edit mode.\n</div>",
"_____no_output_____"
],
[
"### The Road Ahead\nYou must complete Steps 1-3 below to pass the project. The section on Step 4 includes references & resources you can use to further explore HMM taggers.\n\n- [Step 1](#Step-1:-Read-and-preprocess-the-dataset): Review the provided interface to load and access the text corpus\n- [Step 2](#Step-2:-Build-a-Most-Frequent-Class-tagger): Build a Most Frequent Class tagger to use as a baseline\n- [Step 3](#Step-3:-Build-an-HMM-tagger): Build an HMM Part of Speech tagger and compare to the MFC baseline\n- [Step 4](#Step-4:-[Optional]-Improving-model-performance): (Optional) Improve the HMM tagger",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-warning\">\n**Note:** Make sure you have selected a **Python 3** kernel in Workspaces or the hmm-tagger conda environment if you are running the Jupyter server on your own machine.\n</div>",
"_____no_output_____"
]
],
[
[
"# Jupyter \"magic methods\" -- only need to be run once per kernel restart\n%load_ext autoreload\n%aimport helpers, tests\n%autoreload 1",
"_____no_output_____"
],
[
"# import python modules -- this cell needs to be run again if you make changes to any of the files\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom IPython.core.display import HTML\nfrom itertools import chain\nfrom collections import Counter, defaultdict\nfrom helpers import show_model, Dataset\nfrom pomegranate import State, HiddenMarkovModel, DiscreteDistribution",
"_____no_output_____"
]
],
[
[
"## Step 1: Read and preprocess the dataset\n---\nWe'll start by reading in a text corpus and splitting it into a training and testing dataset. The data set is a copy of the [Brown corpus](https://en.wikipedia.org/wiki/Brown_Corpus) (originally from the [NLTK](https://www.nltk.org/) library) that has already been pre-processed to only include the [universal tagset](https://arxiv.org/pdf/1104.2086.pdf). You should expect to get slightly higher accuracy using this simplified tagset than the same model would achieve on a larger tagset like the full [Penn treebank tagset](https://www.ling.upenn.edu/courses/Fall_2003/ling001/penn_treebank_pos.html), but the process you'll follow would be the same.\n\nThe `Dataset` class provided in helpers.py will read and parse the corpus. You can generate your own datasets compatible with the reader by writing them to the following format. The dataset is stored in plaintext as a collection of words and corresponding tags. Each sentence starts with a unique identifier on the first line, followed by one tab-separated word/tag pair on each following line. Sentences are separated by a single blank line.\n\nExample from the Brown corpus. \n```\nb100-38532\nPerhaps\tADV\nit\tPRON\nwas\tVERB\nright\tADJ\n;\t.\n;\t.\n\nb100-35577\n...\n```",
"_____no_output_____"
]
],
[
[
"data = Dataset(\"tags-universal.txt\", \"brown-universal.txt\", train_test_split=0.8)\n\nprint(\"There are {} sentences in the corpus.\".format(len(data)))\nprint(\"There are {} sentences in the training set.\".format(len(data.training_set)))\nprint(\"There are {} sentences in the testing set.\".format(len(data.testing_set)))\n\nassert len(data) == len(data.training_set) + len(data.testing_set), \\\n \"The number of sentences in the training set + testing set should sum to the number of sentences in the corpus\"",
"There are 57340 sentences in the corpus.\nThere are 45872 sentences in the training set.\nThere are 11468 sentences in the testing set.\n"
]
],
[
[
"### The Dataset Interface\n\nYou can access (mostly) immutable references to the dataset through a simple interface provided through the `Dataset` class, which represents an iterable collection of sentences along with easy access to partitions of the data for training & testing. Review the reference below, then run and review the next few cells to make sure you understand the interface before moving on to the next step.\n\n```\nDataset-only Attributes:\n training_set - reference to a Subset object containing the samples for training\n testing_set - reference to a Subset object containing the samples for testing\n\nDataset & Subset Attributes:\n sentences - a dictionary with an entry {sentence_key: Sentence()} for each sentence in the corpus\n keys - an immutable ordered (not sorted) collection of the sentence_keys for the corpus\n vocab - an immutable collection of the unique words in the corpus\n tagset - an immutable collection of the unique tags in the corpus\n X - returns an array of words grouped by sentences ((w11, w12, w13, ...), (w21, w22, w23, ...), ...)\n Y - returns an array of tags grouped by sentences ((t11, t12, t13, ...), (t21, t22, t23, ...), ...)\n N - returns the number of distinct samples (individual words or tags) in the dataset\n\nMethods:\n stream() - returns an flat iterable over all (word, tag) pairs across all sentences in the corpus\n __iter__() - returns an iterable over the data as (sentence_key, Sentence()) pairs\n __len__() - returns the nubmer of sentences in the dataset\n```\n\nFor example, consider a Subset, `subset`, of the sentences `{\"s0\": Sentence((\"See\", \"Spot\", \"run\"), (\"VERB\", \"NOUN\", \"VERB\")), \"s1\": Sentence((\"Spot\", \"ran\"), (\"NOUN\", \"VERB\"))}`. The subset will have these attributes:\n\n```\nsubset.keys == {\"s1\", \"s0\"} # unordered\nsubset.vocab == {\"See\", \"run\", \"ran\", \"Spot\"} # unordered\nsubset.tagset == {\"VERB\", \"NOUN\"} # unordered\nsubset.X == ((\"Spot\", \"ran\"), (\"See\", \"Spot\", \"run\")) # order matches .keys\nsubset.Y == ((\"NOUN\", \"VERB\"), (\"VERB\", \"NOUN\", \"VERB\")) # order matches .keys\nsubset.N == 7 # there are a total of seven observations over all sentences\nlen(subset) == 2 # because there are two sentences\n```\n\n<div class=\"alert alert-block alert-info\">\n**Note:** The `Dataset` class is _convenient_, but it is **not** efficient. It is not suitable for huge datasets because it stores multiple redundant copies of the same data.\n</div>",
"_____no_output_____"
],
[
"#### Sentences\n\n`Dataset.sentences` is a dictionary of all sentences in the training corpus, each keyed to a unique sentence identifier. Each `Sentence` is itself an object with two attributes: a tuple of the words in the sentence named `words` and a tuple of the tag corresponding to each word named `tags`.",
"_____no_output_____"
]
],
[
[
"key = 'b100-38532'\nprint(\"Sentence: {}\".format(key))\nprint(\"words:\\n\\t{!s}\".format(data.sentences[key].words))\nprint(\"tags:\\n\\t{!s}\".format(data.sentences[key].tags))",
"Sentence: b100-38532\nwords:\n\t('Perhaps', 'it', 'was', 'right', ';', ';')\ntags:\n\t('ADV', 'PRON', 'VERB', 'ADJ', '.', '.')\n"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\n**Note:** The underlying iterable sequence is **unordered** over the sentences in the corpus; it is not guaranteed to return the sentences in a consistent order between calls. Use `Dataset.stream()`, `Dataset.keys`, `Dataset.X`, or `Dataset.Y` attributes if you need ordered access to the data.\n</div>\n\n#### Counting Unique Elements\n\nYou can access the list of unique words (the dataset vocabulary) via `Dataset.vocab` and the unique list of tags via `Dataset.tagset`.",
"_____no_output_____"
]
],
[
[
"print(\"There are a total of {} samples of {} unique words in the corpus.\"\n .format(data.N, len(data.vocab)))\nprint(\"There are {} samples of {} unique words in the training set.\"\n .format(data.training_set.N, len(data.training_set.vocab)))\nprint(\"There are {} samples of {} unique words in the testing set.\"\n .format(data.testing_set.N, len(data.testing_set.vocab)))\nprint(\"There are {} words in the test set that are missing in the training set.\"\n .format(len(data.testing_set.vocab - data.training_set.vocab)))\n\nassert data.N == data.training_set.N + data.testing_set.N, \\\n \"The number of training + test samples should sum to the total number of samples\"",
"There are a total of 1161192 samples of 56057 unique words in the corpus.\nThere are 928458 samples of 50536 unique words in the training set.\nThere are 232734 samples of 25112 unique words in the testing set.\nThere are 5521 words in the test set that are missing in the training set.\n"
]
],
[
[
"#### Accessing word and tag Sequences\nThe `Dataset.X` and `Dataset.Y` attributes provide access to ordered collections of matching word and tag sequences for each sentence in the dataset.",
"_____no_output_____"
]
],
[
[
"# accessing words with Dataset.X and tags with Dataset.Y \nfor i in range(2): \n print(\"Sentence {}:\".format(i + 1), data.X[i])\n print()\n print(\"Labels {}:\".format(i + 1), data.Y[i])\n print()",
"Sentence 1: ('Mr.', 'Podger', 'had', 'thanked', 'him', 'gravely', ',', 'and', 'now', 'he', 'made', 'use', 'of', 'the', 'advice', '.')\n\nLabels 1: ('NOUN', 'NOUN', 'VERB', 'VERB', 'PRON', 'ADV', '.', 'CONJ', 'ADV', 'PRON', 'VERB', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\nSentence 2: ('But', 'there', 'seemed', 'to', 'be', 'some', 'difference', 'of', 'opinion', 'as', 'to', 'how', 'far', 'the', 'board', 'should', 'go', ',', 'and', 'whose', 'advice', 'it', 'should', 'follow', '.')\n\nLabels 2: ('CONJ', 'PRT', 'VERB', 'PRT', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'ADP', 'ADV', 'ADV', 'DET', 'NOUN', 'VERB', 'VERB', '.', 'CONJ', 'DET', 'NOUN', 'PRON', 'VERB', 'VERB', '.')\n\n"
]
],
[
[
"#### Accessing (word, tag) Samples\nThe `Dataset.stream()` method returns an iterator that chains together every pair of (word, tag) entries across all sentences in the entire corpus.",
"_____no_output_____"
]
],
[
[
"# use Dataset.stream() (word, tag) samples for the entire corpus\nprint(\"\\nStream (word, tag) pairs:\\n\")\nfor i, pair in enumerate(data.stream()):\n print(\"\\t\", pair)\n if i > 5: break",
"\nStream (word, tag) pairs:\n\n\t ('Mr.', 'NOUN')\n\t ('Podger', 'NOUN')\n\t ('had', 'VERB')\n\t ('thanked', 'VERB')\n\t ('him', 'PRON')\n\t ('gravely', 'ADV')\n\t (',', '.')\n"
]
],
[
[
"\nFor both our baseline tagger and the HMM model we'll build, we need to estimate the frequency of tags & words from the frequency counts of observations in the training corpus. In the next several cells you will complete functions to compute the counts of several sets of counts. ",
"_____no_output_____"
],
[
"## Step 2: Build a Most Frequent Class tagger\n---\n\nPerhaps the simplest tagger (and a good baseline for tagger performance) is to simply choose the tag most frequently assigned to each word. This \"most frequent class\" tagger inspects each observed word in the sequence and assigns it the label that was most often assigned to that word in the corpus.",
"_____no_output_____"
],
[
"### IMPLEMENTATION: Pair Counts\n\nComplete the function below that computes the joint frequency counts for two input sequences.",
"_____no_output_____"
]
],
[
[
"from collections import defaultdict\n\ndef pair_counts(sequences_A, sequences_B):\n \"\"\"Return a dictionary keyed to each unique value in the first sequence list\n that counts the number of occurrences of the corresponding value from the\n second sequences list.\n \n For example, if sequences_A is tags and sequences_B is the corresponding\n words, then if 1244 sequences contain the word \"time\" tagged as a NOUN, then\n you should return a dictionary such that pair_counts[NOUN][time] == 1244\n \"\"\"\n # TODO: Finish this function!\n # Init dictionary\n tags_words_count = defaultdict(lambda : defaultdict(int))\n \n for i in range(len(sequences_B)):\n for itemA, itemB in zip(sequences_A[i], sequences_B[i]):\n tags_words_count[itemA][itemB] += 1\n \n return tags_words_count\n\n\n# Calculate C(t_i, w_i)\nemission_counts = pair_counts(data.Y, data.X)\n\nassert len(emission_counts) == 12, \\\n \"Uh oh. There should be 12 tags in your dictionary.\"\nassert max(emission_counts[\"NOUN\"], key=emission_counts[\"NOUN\"].get) == 'time', \\\n \"Hmmm...'time' is expected to be the most common NOUN.\"\nHTML('<div class=\"alert alert-block alert-success\">Your emission counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Most Frequent Class Tagger\n\nUse the `pair_counts()` function and the training dataset to find the most frequent class label for each word in the training data, and populate the `mfc_table` below. The table keys should be words, and the values should be the appropriate tag string.\n\nThe `MFCTagger` class is provided to mock the interface of Pomegranite HMM models so that they can be used interchangeably.",
"_____no_output_____"
]
],
[
[
"# Create a lookup table mfc_table where mfc_table[word] contains the tag label most frequently assigned to that word\nfrom collections import namedtuple\n\nFakeState = namedtuple(\"FakeState\", \"name\")\n\nclass MFCTagger:\n # NOTE: You should not need to modify this class or any of its methods\n missing = FakeState(name=\"<MISSING>\")\n \n def __init__(self, table):\n self.table = defaultdict(lambda: MFCTagger.missing)\n self.table.update({word: FakeState(name=tag) for word, tag in table.items()})\n \n def viterbi(self, seq):\n \"\"\"This method simplifies predictions by matching the Pomegranate viterbi() interface\"\"\"\n return 0., list(enumerate([\"<start>\"] + [self.table[w] for w in seq] + [\"<end>\"]))\n\n\n# TODO: calculate the frequency of each tag being assigned to each word (hint: similar, but not\n# the same as the emission probabilities) and use it to fill the mfc_table\n\nword_counts = pair_counts(data.training_set.X, data.training_set.Y)\n\nmfc_table = {word: max(subdict, key=subdict.get) for word, subdict in word_counts.items()} # TODO: YOUR CODE HERE\n\n# DO NOT MODIFY BELOW THIS LINE\nmfc_model = MFCTagger(mfc_table) # Create a Most Frequent Class tagger instance\n\nassert len(mfc_table) == len(data.training_set.vocab), \"\"\nassert all(k in data.training_set.vocab for k in mfc_table.keys()), \"\"\nassert sum(int(k not in mfc_table) for k in data.testing_set.vocab) == 5521, \"\"\nHTML('<div class=\"alert alert-block alert-success\">Your MFC tagger has all the correct words!</div>')",
"_____no_output_____"
]
],
[
[
"### Making Predictions with a Model\nThe helper functions provided below interface with Pomegranate network models & the mocked MFCTagger to take advantage of the [missing value](http://pomegranate.readthedocs.io/en/latest/nan.html) functionality in Pomegranate through a simple sequence decoding function. Run these functions, then run the next cell to see some of the predictions made by the MFC tagger.",
"_____no_output_____"
]
],
[
[
"def replace_unknown(sequence):\n \"\"\"Return a copy of the input sequence where each unknown word is replaced\n by the literal string value 'nan'. Pomegranate will ignore these values\n during computation.\n \"\"\"\n return [w if w in data.training_set.vocab else 'nan' for w in sequence]\n\ndef simplify_decoding(X, model):\n \"\"\"X should be a 1-D sequence of observations for the model to predict\"\"\"\n _, state_path = model.viterbi(replace_unknown(X))\n return [state[1].name for state in state_path[1:-1]] # do not show the start/end state predictions",
"_____no_output_____"
]
],
[
[
"### Example Decoding Sequences with MFC Tagger",
"_____no_output_____"
]
],
[
[
"for key in data.testing_set.keys[:3]:\n print(\"Sentence Key: {}\\n\".format(key))\n print(\"Predicted labels:\\n-----------------\")\n print(simplify_decoding(data.sentences[key].words, mfc_model))\n print()\n print(\"Actual labels:\\n--------------\")\n print(data.sentences[key].tags)\n print(\"\\n\")",
"Sentence Key: b100-28144\n\nPredicted labels:\n-----------------\n['CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.']\n\nActual labels:\n--------------\n('CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.')\n\n\nSentence Key: b100-23146\n\nPredicted labels:\n-----------------\n['PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.']\n\nActual labels:\n--------------\n('PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\n\nSentence Key: b100-35462\n\nPredicted labels:\n-----------------\n['DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', '<MISSING>', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADV', 'NOUN', '.']\n\nActual labels:\n--------------\n('DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADJ', 'NOUN', '.')\n\n\n"
]
],
[
[
"### Evaluating Model Accuracy\n\nThe function below will evaluate the accuracy of the MFC tagger on the collection of all sentences from a text corpus. ",
"_____no_output_____"
]
],
[
[
"def accuracy(X, Y, model):\n \"\"\"Calculate the prediction accuracy by using the model to decode each sequence\n in the input X and comparing the prediction with the true labels in Y.\n \n The X should be an array whose first dimension is the number of sentences to test,\n and each element of the array should be an iterable of the words in the sequence.\n The arrays X and Y should have the exact same shape.\n \n X = [(\"See\", \"Spot\", \"run\"), (\"Run\", \"Spot\", \"run\", \"fast\"), ...]\n Y = [(), (), ...]\n \"\"\"\n correct = total_predictions = 0\n for observations, actual_tags in zip(X, Y):\n \n # The model.viterbi call in simplify_decoding will return None if the HMM\n # raises an error (for example, if a test sentence contains a word that\n # is out of vocabulary for the training set). Any exception counts the\n # full sentence as an error (which makes this a conservative estimate).\n try:\n most_likely_tags = simplify_decoding(observations, model)\n correct += sum(p == t for p, t in zip(most_likely_tags, actual_tags))\n except:\n pass\n total_predictions += len(observations)\n return correct / total_predictions",
"_____no_output_____"
]
],
[
[
"#### Evaluate the accuracy of the MFC tagger\nRun the next cell to evaluate the accuracy of the tagger on the training and test corpus.",
"_____no_output_____"
]
],
[
[
"mfc_training_acc = accuracy(data.training_set.X, data.training_set.Y, mfc_model)\nprint(\"training accuracy mfc_model: {:.2f}%\".format(100 * mfc_training_acc))\n\nmfc_testing_acc = accuracy(data.testing_set.X, data.testing_set.Y, mfc_model)\nprint(\"testing accuracy mfc_model: {:.2f}%\".format(100 * mfc_testing_acc))\n\nassert mfc_training_acc >= 0.955, \"Uh oh. Your MFC accuracy on the training set doesn't look right.\"\nassert mfc_testing_acc >= 0.925, \"Uh oh. Your MFC accuracy on the testing set doesn't look right.\"\nHTML('<div class=\"alert alert-block alert-success\">Your MFC tagger accuracy looks correct!</div>')",
"training accuracy mfc_model: 95.72%\ntesting accuracy mfc_model: 93.01%\n"
]
],
[
[
"## Step 3: Build an HMM tagger\n---\nThe HMM tagger has one hidden state for each possible tag, and parameterized by two distributions: the emission probabilties giving the conditional probability of observing a given **word** from each hidden state, and the transition probabilities giving the conditional probability of moving between **tags** during the sequence.\n\nWe will also estimate the starting probability distribution (the probability of each **tag** being the first tag in a sequence), and the terminal probability distribution (the probability of each **tag** being the last tag in a sequence).\n\nThe maximum likelihood estimate of these distributions can be calculated from the frequency counts as described in the following sections where you'll implement functions to count the frequencies, and finally build the model. The HMM model will make predictions according to the formula:\n\n$$t_i^n = \\underset{t_i^n}{\\mathrm{argmin}} \\prod_{i=1}^n P(w_i|t_i) P(t_i|t_{i-1})$$\n\nRefer to Speech & Language Processing [Chapter 10](https://web.stanford.edu/~jurafsky/slp3/10.pdf) for more information.",
"_____no_output_____"
],
[
"### IMPLEMENTATION: Unigram Counts\n\nComplete the function below to estimate the co-occurrence frequency of each symbol over all of the input sequences. The unigram probabilities in our HMM model are estimated from the formula below, where N is the total number of samples in the input. (You only need to compute the counts for now.)\n\n$$P(tag_1) = \\frac{C(tag_1)}{N}$$",
"_____no_output_____"
]
],
[
[
"def unigram_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequence list that\n counts the number of occurrences of the value in the sequences list. The sequences\n collection should be a 2-dimensional array.\n \n For example, if the tag NOUN appears 275558 times over all the input sequences,\n then you should return a dictionary such that your_unigram_counts[NOUN] == 275558.\n \"\"\"\n counter = defaultdict(int)\n \n for i in range(len(sequences)):\n for element in sequences[i]:\n counter[element] += 1\n \n return counter\n\n# TODO: call unigram_counts with a list of tag sequences from the training set\ntag_unigrams = unigram_counts(data.training_set.Y)\n\nassert set(tag_unigrams.keys()) == data.training_set.tagset, \\\n \"Uh oh. It looks like your tag counts doesn't include all the tags!\"\nassert min(tag_unigrams, key=tag_unigrams.get) == 'X', \\\n \"Hmmm...'X' is expected to be the least common class\"\nassert max(tag_unigrams, key=tag_unigrams.get) == 'NOUN', \\\n \"Hmmm...'NOUN' is expected to be the most common class\"\nHTML('<div class=\"alert alert-block alert-success\">Your tag unigrams look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Bigram Counts\n\nComplete the function below to estimate the co-occurrence frequency of each pair of symbols in each of the input sequences. These counts are used in the HMM model to estimate the bigram probability of two tags from the frequency counts according to the formula: $$P(tag_2|tag_1) = \\frac{C(tag_2|tag_1)}{C(tag_2)}$$\n",
"_____no_output_____"
]
],
[
[
"def bigram_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique PAIR of values in the input sequences\n list that counts the number of occurrences of pair in the sequences list. The input\n should be a 2-dimensional array.\n \n For example, if the pair of tags (NOUN, VERB) appear 61582 times, then you should\n return a dictionary such that your_bigram_counts[(NOUN, VERB)] == 61582\n \"\"\"\n\n counter = defaultdict(int)\n \n for i in range(len(sequences)):\n seq = sequences[i]\n for element, next_element in zip(seq[:-1], seq[1:]):\n counter[(element, next_element)] += 1 \n \n return counter\n\n# TODO: call bigram_counts with a list of tag sequences from the training set\ntag_bigrams = bigram_counts(data.training_set.Y)\n\nassert len(tag_bigrams) == 144, \\\n \"Uh oh. There should be 144 pairs of bigrams (12 tags x 12 tags)\"\nassert min(tag_bigrams, key=tag_bigrams.get) in [('X', 'NUM'), ('PRON', 'X')], \\\n \"Hmmm...The least common bigram should be one of ('X', 'NUM') or ('PRON', 'X').\"\nassert max(tag_bigrams, key=tag_bigrams.get) in [('DET', 'NOUN')], \\\n \"Hmmm...('DET', 'NOUN') is expected to be the most common bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your tag bigrams look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Sequence Starting Counts\nComplete the code below to estimate the bigram probabilities of a sequence starting with each tag.",
"_____no_output_____"
]
],
[
[
"def starting_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequences list\n that counts the number of occurrences where that value is at the beginning of\n a sequence.\n \n For example, if 8093 sequences start with NOUN, then you should return a\n dictionary such that your_starting_counts[NOUN] == 8093\n \"\"\"\n counter = defaultdict(int)\n \n for i in range(len(sequences)):\n seq = sequences[i]\n counter[seq[0]] += 1 \n \n return counter\n \n\n# TODO: Calculate the count of each tag starting a sequence\ntag_starts = starting_counts(data.training_set.Y)\n\nassert len(tag_starts) == 12, \"Uh oh. There should be 12 tags in your dictionary.\"\nassert min(tag_starts, key=tag_starts.get) == 'X', \"Hmmm...'X' is expected to be the least common starting bigram.\"\nassert max(tag_starts, key=tag_starts.get) == 'DET', \"Hmmm...'DET' is expected to be the most common starting bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your starting tag counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Sequence Ending Counts\nComplete the function below to estimate the bigram probabilities of a sequence ending with each tag.",
"_____no_output_____"
]
],
[
[
"def ending_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequences list\n that counts the number of occurrences where that value is at the end of\n a sequence.\n \n For example, if 18 sequences end with DET, then you should return a\n dictionary such that your_starting_counts[DET] == 18\n \"\"\"\n counter = defaultdict(int)\n \n for i in range(len(sequences)):\n seq = sequences[i]\n counter[seq[-1]] += 1 \n \n return counter\n\n# TODO: Calculate the count of each tag ending a sequence\ntag_ends = ending_counts(data.training_set.Y)\n\nassert len(tag_ends) == 12, \"Uh oh. There should be 12 tags in your dictionary.\"\nassert min(tag_ends, key=tag_ends.get) in ['X', 'CONJ'], \"Hmmm...'X' or 'CONJ' should be the least common ending bigram.\"\nassert max(tag_ends, key=tag_ends.get) == '.', \"Hmmm...'.' is expected to be the most common ending bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your ending tag counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Basic HMM Tagger\nUse the tag unigrams and bigrams calculated above to construct a hidden Markov tagger.\n\n- Add one state per tag\n - The emission distribution at each state should be estimated with the formula: $P(w|t) = \\frac{C(t, w)}{C(t)}$\n- Add an edge from the starting state `basic_model.start` to each tag\n - The transition probability should be estimated with the formula: $P(t|start) = \\frac{C(start, t)}{C(start)}$\n- Add an edge from each tag to the end state `basic_model.end`\n - The transition probability should be estimated with the formula: $P(end|t) = \\frac{C(t, end)}{C(t)}$\n- Add an edge between _every_ pair of tags\n - The transition probability should be estimated with the formula: $P(t_2|t_1) = \\frac{C(t_1, t_2)}{C(t_1)}$",
"_____no_output_____"
]
],
[
[
"basic_model = HiddenMarkovModel(name=\"base-hmm-tagger\")\n\n# TODO: create states with emission probability distributions P(word | tag) and add to the model\n# (Hint: you may need to loop & create/add new states)\n\ntag_probabilities = defaultdict(dict)\n\nfor tag, subdict in emission_counts.items():\n for word, value in subdict.items():\n tag_probabilities[tag][word] = value / tag_unigrams[tag]\n\nstates = {}\nfor tag, prob in tag_probabilities.items():\n states[tag] = State(DiscreteDistribution(prob), name=tag)\n\nbasic_model.add_states(list(states.values()))\n\n# TODO: add edges between states for the observed transition frequencies P(tag_i | tag_i-1)\n# (Hint: you may need to loop & add transitions\ntotal = sum(tag_starts.values())\nfor tag, value in tag_starts.items():\n basic_model.add_transition(basic_model.start, states[tag], value / total)\n \ntotal = sum(tag_ends.values())\nfor tag, value in tag_ends.items():\n basic_model.add_transition(states[tag], basic_model.end, value / total) \n\nfor keys, value in tag_bigrams.items():\n tag1, tag2 = keys\n basic_model.add_transition(states[tag1], states[tag2], value / tag_unigrams[tag1])\n\n# NOTE: YOU SHOULD NOT NEED TO MODIFY ANYTHING BELOW THIS LINE\n# finalize the model\nbasic_model.bake()\n\nassert all(tag in set(s.name for s in basic_model.states) for tag in data.training_set.tagset), \\\n \"Every state in your network should use the name of the associated tag, which must be one of the training set tags.\"\nassert basic_model.edge_count() == 168, \\\n (\"Your network should have an edge from the start node to each state, one edge between every \" +\n \"pair of tags (states), and an edge from each state to the end node.\")\nHTML('<div class=\"alert alert-block alert-success\">Your HMM network topology looks good!</div>')",
"_____no_output_____"
],
[
"hmm_training_acc = accuracy(data.training_set.X, data.training_set.Y, basic_model)\nprint(\"training accuracy basic hmm model: {:.2f}%\".format(100 * hmm_training_acc))\n\nhmm_testing_acc = accuracy(data.testing_set.X, data.testing_set.Y, basic_model)\nprint(\"testing accuracy basic hmm model: {:.2f}%\".format(100 * hmm_testing_acc))\n\nassert hmm_training_acc > 0.97, \"Uh oh. Your HMM accuracy on the training set doesn't look right.\"\nassert hmm_training_acc > 0.955, \"Uh oh. Your HMM accuracy on the training set doesn't look right.\"\nHTML('<div class=\"alert alert-block alert-success\">Your HMM tagger accuracy looks correct! Congratulations, you\\'ve finished the project.</div>')",
"training accuracy basic hmm model: 97.54%\ntesting accuracy basic hmm model: 96.18%\n"
]
],
[
[
"### Example Decoding Sequences with the HMM Tagger",
"_____no_output_____"
]
],
[
[
"for key in data.testing_set.keys[:3]:\n print(\"Sentence Key: {}\\n\".format(key))\n print(\"Predicted labels:\\n-----------------\")\n print(simplify_decoding(data.sentences[key].words, basic_model))\n print()\n print(\"Actual labels:\\n--------------\")\n print(data.sentences[key].tags)\n print(\"\\n\")",
"Sentence Key: b100-28144\n\nPredicted labels:\n-----------------\n['CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.']\n\nActual labels:\n--------------\n('CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.')\n\n\nSentence Key: b100-23146\n\nPredicted labels:\n-----------------\n['PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.']\n\nActual labels:\n--------------\n('PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\n\nSentence Key: b100-35462\n\nPredicted labels:\n-----------------\n['DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADJ', 'NOUN', '.']\n\nActual labels:\n--------------\n('DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADJ', 'NOUN', '.')\n\n\n"
]
],
[
[
"\n## Finishing the project\n---\n\n<div class=\"alert alert-block alert-info\">\n**Note:** **SAVE YOUR NOTEBOOK**, then run the next cell to generate an HTML copy. You will zip & submit both this file and the HTML copy for review.\n</div>",
"_____no_output_____"
]
],
[
[
"!!jupyter nbconvert *.ipynb",
"_____no_output_____"
]
],
[
[
"## Step 4: [Optional] Improving model performance\n---\nThere are additional enhancements that can be incorporated into your tagger that improve performance on larger tagsets where the data sparsity problem is more significant. The data sparsity problem arises because the same amount of data split over more tags means there will be fewer samples in each tag, and there will be more missing data tags that have zero occurrences in the data. The techniques in this section are optional.\n\n- [Laplace Smoothing](https://en.wikipedia.org/wiki/Additive_smoothing) (pseudocounts)\n Laplace smoothing is a technique where you add a small, non-zero value to all observed counts to offset for unobserved values.\n\n- Backoff Smoothing\n Another smoothing technique is to interpolate between n-grams for missing data. This method is more effective than Laplace smoothing at combatting the data sparsity problem. Refer to chapters 4, 9, and 10 of the [Speech & Language Processing](https://web.stanford.edu/~jurafsky/slp3/) book for more information.\n\n- Extending to Trigrams\n HMM taggers have achieved better than 96% accuracy on this dataset with the full Penn treebank tagset using an architecture described in [this](http://www.coli.uni-saarland.de/~thorsten/publications/Brants-ANLP00.pdf) paper. Altering your HMM to achieve the same performance would require implementing deleted interpolation (described in the paper), incorporating trigram probabilities in your frequency tables, and re-implementing the Viterbi algorithm to consider three consecutive states instead of two.\n\n### Obtain the Brown Corpus with a Larger Tagset\nRun the code below to download a copy of the brown corpus with the full NLTK tagset. You will need to research the available tagset information in the NLTK docs and determine the best way to extract the subset of NLTK tags you want to explore. If you write the following the format specified in Step 1, then you can reload the data using all of the code above for comparison.\n\nRefer to [Chapter 5](http://www.nltk.org/book/ch05.html) of the NLTK book for more information on the available tagsets.",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk import pos_tag, word_tokenize\nfrom nltk.corpus import brown\n\nnltk.download('brown')\ntraining_corpus = nltk.corpus.brown\ntraining_corpus.tagged_sents()[0]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6dc3a4f64bce98032f9203e754f9ca4086192f | 6,272 | ipynb | Jupyter Notebook | Single_Object_Detection_CPU.ipynb | PM25/Object_Recognition_DL | e7e89643ce2b75e4f22724a156d91eb1a52d6b97 | [
"MIT"
]
| 1 | 2019-09-04T03:12:22.000Z | 2019-09-04T03:12:22.000Z | Single_Object_Detection_CPU.ipynb | PM25/Object_Recognition_DL | e7e89643ce2b75e4f22724a156d91eb1a52d6b97 | [
"MIT"
]
| null | null | null | Single_Object_Detection_CPU.ipynb | PM25/Object_Recognition_DL | e7e89643ce2b75e4f22724a156d91eb1a52d6b97 | [
"MIT"
]
| null | null | null | 23.757576 | 129 | 0.495536 | [
[
[
"### Set Data Path",
"_____no_output_____"
]
],
[
[
"from pathlib import Path\n\nbase_dir = Path(\"data\")\ntrain_dir = base_dir/Path(\"train\")\nvalidation_dir = base_dir/Path(\"validation\")\ntest_dir = base_dir/Path(\"test\")",
"_____no_output_____"
]
],
[
[
"### Image Transform Function",
"_____no_output_____"
]
],
[
[
"from torchvision import transforms\n\ntransform = transforms.Compose([ \n transforms.Resize((224, 224)),\n transforms.ToTensor(),\n transforms.Normalize(mean=(.5, .5, .5), std=(.5, .5, .5))\n])",
"_____no_output_____"
]
],
[
[
"### Load Training Data (x: features, y: labels)",
"_____no_output_____"
]
],
[
[
"import torch\nfrom PIL import Image\n\nx, y = [], []\nfor file_name in train_dir.glob(\"*.jpg\"):\n bounding_box_file = file_name.with_suffix('.txt')\n \n with open(bounding_box_file) as file:\n lines = file.readlines()\n if(len(lines) > 1):\n continue\n else:\n line = lines[0].strip('\\n')\n (classes, cen_x, cen_y, box_w, box_h) = list(map(float, line.split(' ')))\n torch_data = torch.FloatTensor([cen_x, cen_y, box_w, box_h])\n y.append(torch_data)\n \n img = Image.open(str(file_name)).convert('RGB')\n img = transform(img)\n x.append(img)",
"_____no_output_____"
]
],
[
[
"### Put Training Data into Torch Loader",
"_____no_output_____"
]
],
[
[
"import torch.utils.data as Data\n\ntensor_x = torch.stack(x)\ntensor_y = torch.stack(y)\ntorch_dataset = Data.TensorDataset(tensor_x, tensor_y)\nloader = Data.DataLoader(dataset=torch_dataset, batch_size=32, shuffle=True, num_workers=2)",
"_____no_output_____"
]
],
[
[
"### Load Pretrained RestNet18 Model",
"_____no_output_____"
]
],
[
[
"import torchvision\nfrom torch import nn\n\nmodel = torchvision.models.resnet18(pretrained=True)\nfc_in_size = model.fc.in_features\nmodel.fc = nn.Linear(fc_in_size, 4)",
"_____no_output_____"
]
],
[
[
"### Parameters",
"_____no_output_____"
]
],
[
[
"EPOCH = 10\nLR = 1e-3",
"_____no_output_____"
]
],
[
[
"### Loss Function & Optimizer",
"_____no_output_____"
]
],
[
[
"loss_func = nn.SmoothL1Loss()\nopt = torch.optim.Adam(model.parameters(), lr=LR)",
"_____no_output_____"
]
],
[
[
"### Training",
"_____no_output_____"
]
],
[
[
"for epoch in range(EPOCH):\n for step, (batch_x, batch_y) in enumerate(loader):\n batch_x = batch_x\n batch_y = batch_y\n output = model(batch_x)\n loss = loss_func(output, batch_y)\n opt.zero_grad()\n loss.backward()\n opt.step()\n \n if(step % 5 == 0):\n print(\"Epoch {} | Step {} | Loss {}\".format(epoch, step, loss))",
"Epoch 0 | Step 0 | Loss 0.3441098928451538\nEpoch 0 | Step 5 | Loss 0.17193390429019928\nEpoch 0 | Step 10 | Loss 0.10625818371772766\nEpoch 1 | Step 0 | Loss 0.0646333172917366\nEpoch 1 | Step 5 | Loss 0.03650979697704315\nEpoch 1 | Step 10 | Loss 0.017900140956044197\n"
]
],
[
[
"### Show some of the Prediction",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport cv2\nfrom matplotlib import pyplot as plt\nimport numpy as np\n\nmodel = model.cpu()\nfor batch_x, batch_y in loader:\n predict = model(batch_x)\n for x, pred, y in zip(batch_x, predict, batch_y):\n (pos_x, pos_y, box_w, box_h) = pred\n pos_x *= 224\n pos_y *= 224\n box_w *= 224\n box_h *= 224\n \n image = transforms.ToPILImage()(x)\n img = cv2.cvtColor(np.asarray(image), cv2.COLOR_RGB2BGR)\n img = cv2.rectangle(img, (pos_x - box_w/2, pos_y - box_h/2), (pos_x + box_w/2, pos_y + box_h/2), (255, 0, 0), 3)\n \n plt.imshow(img)\n plt.show()\n break",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6dc4a318bbc8cf8b0cf26053528cda3eb3156c | 65,160 | ipynb | Jupyter Notebook | tutorials/W1D2_LinearDeepLearning/student/W1D2_Tutorial2.ipynb | moekay/course-content-dl | 53e083d5d02d8fdc08a1bfb7740fd82116178fd0 | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| null | null | null | tutorials/W1D2_LinearDeepLearning/student/W1D2_Tutorial2.ipynb | moekay/course-content-dl | 53e083d5d02d8fdc08a1bfb7740fd82116178fd0 | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| null | null | null | tutorials/W1D2_LinearDeepLearning/student/W1D2_Tutorial2.ipynb | moekay/course-content-dl | 53e083d5d02d8fdc08a1bfb7740fd82116178fd0 | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| null | null | null | 34.84492 | 602 | 0.54647 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content-dl/blob/main/tutorials/W1D2_LinearDeepLearning/student/W1D2_Tutorial2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Tutorial 2: Learning Hyperparameters\n**Week 1, Day 2: Linear Deep Learning**\n\n**By Neuromatch Academy**\n\n__Content creators:__ Saeed Salehi, Andrew Saxe\n\n__Content reviewers:__ Polina Turishcheva, Antoine De Comite, Kelson Shilling-Scrivo\n\n__Content editors:__ Anoop Kulkarni\n\n__Production editors:__ Khalid Almubarak, Spiros Chavlis\n",
"_____no_output_____"
],
[
"**Our 2021 Sponsors, including Presenting Sponsor Facebook Reality Labs**\n\n<p align='center'><img src='https://github.com/NeuromatchAcademy/widgets/blob/master/sponsors.png?raw=True'/></p>",
"_____no_output_____"
],
[
"---\n# Tutorial Objectives\n\n* Training landscape\n* The effect of depth\n* Choosing a learning rate\n* Initialization matters\n",
"_____no_output_____"
]
],
[
[
"# @title Tutorial slides\n\n# @markdown These are the slides for the videos in the tutorial\nfrom IPython.display import IFrame\nIFrame(src=f\"https://mfr.ca-1.osf.io/render?url=https://osf.io/sne2m/?direct%26mode=render%26action=download%26mode=render\", width=854, height=480)",
"_____no_output_____"
]
],
[
[
"---\n# Setup\n\nThis a GPU-Free tutorial!",
"_____no_output_____"
]
],
[
[
"# @title Install dependencies\n!pip install git+https://github.com/NeuromatchAcademy/evaltools --quiet\n\nfrom evaltools.airtable import AirtableForm",
"_____no_output_____"
],
[
"# Imports\nimport time\nimport numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# @title Figure settings\n\nfrom ipywidgets import interact, IntSlider, FloatSlider, fixed\nfrom ipywidgets import HBox, interactive_output, ToggleButton, Layout\nfrom mpl_toolkits.axes_grid1 import make_axes_locatable\n\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/content-creation/main/nma.mplstyle\")",
"_____no_output_____"
],
[
"# @title Plotting functions\n\ndef plot_x_y_(x_t_, y_t_, x_ev_, y_ev_, loss_log_, weight_log_):\n \"\"\"\n \"\"\"\n plt.figure(figsize=(12, 4))\n plt.subplot(1, 3, 1)\n plt.scatter(x_t_, y_t_, c='r', label='training data')\n plt.plot(x_ev_, y_ev_, c='b', label='test results', linewidth=2)\n plt.xlabel('x')\n plt.ylabel('y')\n plt.legend()\n plt.subplot(1, 3, 2)\n plt.plot(loss_log_, c='r')\n plt.xlabel('epochs')\n plt.ylabel('mean squared error')\n plt.subplot(1, 3, 3)\n plt.plot(weight_log_)\n plt.xlabel('epochs')\n plt.ylabel('weights')\n plt.show()\n\n\ndef plot_vector_field(what, init_weights=None):\n \"\"\"\n \"\"\"\n n_epochs=40\n lr=0.15\n x_pos = np.linspace(2.0, 0.5, 100, endpoint=True)\n y_pos = 1. / x_pos\n xx, yy = np.mgrid[-1.9:2.0:0.2, -1.9:2.0:0.2]\n zz = np.empty_like(xx)\n x, y = xx[:, 0], yy[0]\n\n x_temp, y_temp = gen_samples(10, 1.0, 0.0)\n\n cmap = matplotlib.cm.plasma\n plt.figure(figsize=(8, 7))\n ax = plt.gca()\n\n if what == 'all' or what == 'vectors':\n for i, a in enumerate(x):\n for j, b in enumerate(y):\n temp_model = ShallowNarrowLNN([a, b])\n da, db = temp_model.dloss_dw(x_temp, y_temp)\n zz[i, j] = temp_model.loss(temp_model.forward(x_temp), y_temp)\n scale = min(40 * np.sqrt(da**2 + db**2), 50)\n ax.quiver(a, b, - da, - db, scale=scale, color=cmap(np.sqrt(da**2 + db**2)))\n\n if what == 'all' or what == 'trajectory':\n if init_weights is None:\n for init_weights in [[0.5, -0.5], [0.55, -0.45], [-1.8, 1.7]]:\n temp_model = ShallowNarrowLNN(init_weights)\n _, temp_records = temp_model.train(x_temp, y_temp, lr, n_epochs)\n ax.scatter(temp_records[:, 0], temp_records[:, 1],\n c=np.arange(len(temp_records)), cmap='Greys')\n ax.scatter(temp_records[0, 0], temp_records[0, 1], c='blue', zorder=9)\n ax.scatter(temp_records[-1, 0], temp_records[-1, 1], c='red', marker='X', s=100, zorder=9)\n else:\n temp_model = ShallowNarrowLNN(init_weights)\n _, temp_records = temp_model.train(x_temp, y_temp, lr, n_epochs)\n ax.scatter(temp_records[:, 0], temp_records[:, 1],\n c=np.arange(len(temp_records)), cmap='Greys')\n ax.scatter(temp_records[0, 0], temp_records[0, 1], c='blue', zorder=9)\n ax.scatter(temp_records[-1, 0], temp_records[-1, 1], c='red', marker='X', s=100, zorder=9)\n\n if what == 'all' or what == 'loss':\n contplt = ax.contourf(x, y, np.log(zz+0.001), zorder=-1, cmap='coolwarm', levels=100)\n divider = make_axes_locatable(ax)\n cax = divider.append_axes(\"right\", size=\"5%\", pad=0.05)\n cbar = plt.colorbar(contplt, cax=cax)\n cbar.set_label('log (Loss)')\n\n ax.set_xlabel(\"$w_1$\")\n ax.set_ylabel(\"$w_2$\")\n ax.set_xlim(-1.9, 1.9)\n ax.set_ylim(-1.9, 1.9)\n\n plt.show()\n\n\ndef plot_loss_landscape():\n \"\"\"\n \"\"\"\n x_temp, y_temp = gen_samples(10, 1.0, 0.0)\n\n xx, yy = np.mgrid[-1.9:2.0:0.2, -1.9:2.0:0.2]\n zz = np.empty_like(xx)\n x, y = xx[:, 0], yy[0]\n\n for i, a in enumerate(x):\n for j, b in enumerate(y):\n temp_model = ShallowNarrowLNN([a, b])\n zz[i, j] = temp_model.loss(temp_model.forward(x_temp), y_temp)\n\n temp_model = ShallowNarrowLNN([-1.8, 1.7])\n loss_rec_1, w_rec_1 = temp_model.train(x_temp, y_temp, 0.02, 240)\n\n temp_model = ShallowNarrowLNN([1.5, -1.5])\n loss_rec_2, w_rec_2 = temp_model.train(x_temp, y_temp, 0.02, 240)\n\n plt.figure(figsize=(12, 8))\n ax = plt.subplot(1, 1, 1, projection='3d')\n ax.plot_surface(xx, yy, np.log(zz+0.5), cmap='coolwarm', alpha=0.5)\n ax.scatter3D(w_rec_1[:, 0], w_rec_1[:, 1], np.log(loss_rec_1+0.5),\n c='k', s=50, zorder=9)\n ax.scatter3D(w_rec_2[:, 0], w_rec_2[:, 1], np.log(loss_rec_2+0.5),\n c='k', s=50, zorder=9)\n plt.axis(\"off\")\n ax.view_init(45, 260)\n\n plt.show()\n\n\ndef depth_widget(depth):\n if depth == 0:\n depth_lr_init_interplay(depth, 0.02, 0.9)\n else:\n depth_lr_init_interplay(depth, 0.01, 0.9)\n\n\ndef lr_widget(lr):\n depth_lr_init_interplay(50, lr, 0.9)\n\n\ndef depth_lr_interplay(depth, lr):\n depth_lr_init_interplay(depth, lr, 0.9)\n\n\ndef depth_lr_init_interplay(depth, lr, init_weights):\n n_epochs = 600\n\n x_train, y_train = gen_samples(100, 2.0, 0.1)\n model = DeepNarrowLNN(np.full((1, depth+1), init_weights))\n\n plt.figure(figsize=(10, 5))\n plt.plot(model.train(x_train, y_train, lr, n_epochs),\n linewidth=3.0, c='m')\n\n plt.title(\"Training a {}-layer LNN with\"\n \" $\\eta=${} initialized with $w_i=${}\".format(depth, lr, init_weights), pad=15)\n plt.yscale('log')\n plt.xlabel('epochs')\n plt.ylabel('Log mean squared error')\n plt.ylim(0.001, 1.0)\n plt.show()\n\n\ndef plot_init_effect():\n depth = 15\n n_epochs = 250\n lr = 0.02\n\n x_train, y_train = gen_samples(100, 2.0, 0.1)\n\n plt.figure(figsize=(12, 6))\n for init_w in np.arange(0.7, 1.09, 0.05):\n model = DeepNarrowLNN(np.full((1, depth), init_w))\n plt.plot(model.train(x_train, y_train, lr, n_epochs),\n linewidth=3.0, label=\"initial weights {:.2f}\".format(init_w))\n plt.title(\"Training a {}-layer narrow LNN with $\\eta=${}\".format(depth, lr), pad=15)\n plt.yscale('log')\n plt.xlabel('epochs')\n plt.ylabel('Log mean squared error')\n plt.legend(loc='lower left', ncol=4)\n plt.ylim(0.001, 1.0)\n plt.show()\n\n\nclass InterPlay:\n def __init__(self):\n self.lr = [None]\n self.depth = [None]\n self.success = [None]\n self.min_depth, self.max_depth = 5, 65\n self.depth_list = np.arange(10, 61, 10)\n self.i_depth = 0\n self.min_lr, self.max_lr = 0.001, 0.105\n self.n_epochs = 600\n self.x_train, self.y_train = gen_samples(100, 2.0, 0.1)\n self.converged = False\n self.button = None\n self.slider = None\n\n def train(self, lr, update=False, init_weights=0.9):\n if update and self.converged and self.i_depth < len(self.depth_list):\n depth = self.depth_list[self.i_depth]\n self.plot(depth, lr)\n self.i_depth += 1\n self.lr.append(None)\n self.depth.append(None)\n self.success.append(None)\n self.converged = False\n self.slider.value = 0.005\n if self.i_depth < len(self.depth_list):\n self.button.value = False\n self.button.description = 'Explore!'\n self.button.disabled = True\n self.button.button_style = 'danger'\n else:\n self.button.value = False\n self.button.button_style = ''\n self.button.disabled = True\n self.button.description = 'Done!'\n time.sleep(1.0)\n\n elif self.i_depth < len(self.depth_list):\n depth = self.depth_list[self.i_depth]\n # assert self.min_depth <= depth <= self.max_depth\n assert self.min_lr <= lr <= self.max_lr\n self.converged = False\n\n model = DeepNarrowLNN(np.full((1, depth), init_weights))\n self.losses = np.array(model.train(self.x_train, self.y_train, lr, self.n_epochs))\n if np.any(self.losses < 1e-2):\n success = np.argwhere(self.losses < 1e-2)[0][0]\n if np.all((self.losses[success:] < 1e-2)):\n self.converged = True\n self.success[-1] = success\n self.lr[-1] = lr\n self.depth[-1] = depth\n self.button.disabled = False\n self.button.button_style = 'success'\n self.button.description = 'Register!'\n else:\n self.button.disabled = True\n self.button.button_style = 'danger'\n self.button.description = 'Explore!'\n else:\n self.button.disabled = True\n self.button.button_style = 'danger'\n self.button.description = 'Explore!'\n self.plot(depth, lr)\n\n def plot(self, depth, lr):\n fig = plt.figure(constrained_layout=False, figsize=(10, 8))\n gs = fig.add_gridspec(2, 2)\n ax1 = fig.add_subplot(gs[0, :])\n ax2 = fig.add_subplot(gs[1, 0])\n ax3 = fig.add_subplot(gs[1, 1])\n\n ax1.plot(self.losses, linewidth=3.0, c='m')\n ax1.set_title(\"Training a {}-layer LNN with\"\n \" $\\eta=${}\".format(depth, lr), pad=15, fontsize=16)\n ax1.set_yscale('log')\n ax1.set_xlabel('epochs')\n ax1.set_ylabel('Log mean squared error')\n ax1.set_ylim(0.001, 1.0)\n\n ax2.set_xlim(self.min_depth, self.max_depth)\n ax2.set_ylim(-10, self.n_epochs)\n ax2.set_xlabel('Depth')\n ax2.set_ylabel('Learning time (Epochs)')\n ax2.set_title(\"Learning time vs depth\", fontsize=14)\n ax2.scatter(np.array(self.depth), np.array(self.success), c='r')\n\n # ax3.set_yscale('log')\n ax3.set_xlim(self.min_depth, self.max_depth)\n ax3.set_ylim(self.min_lr, self.max_lr)\n ax3.set_xlabel('Depth')\n ax3.set_ylabel('Optimial learning rate')\n ax3.set_title(\"Empirically optimal $\\eta$ vs depth\", fontsize=14)\n ax3.scatter(np.array(self.depth), np.array(self.lr), c='r')\n\n plt.show()",
"_____no_output_____"
],
[
"# @title Helper functions\n\natform = AirtableForm('appn7VdPRseSoMXEG','W1D2_T2','https://portal.neuromatchacademy.org/api/redirect/to/9c55f6cb-cdf9-4429-ac1c-ec44fe64c303')\n\n\ndef gen_samples(n, a, sigma):\n \"\"\"\n Generates `n` samples with `y = z * x + noise(sgma)` linear relation.\n\n Args:\n n : int\n a : float\n sigma : float\n Retutns:\n x : np.array\n y : np.array\n \"\"\"\n assert n > 0\n assert sigma >= 0\n\n if sigma > 0:\n x = np.random.rand(n)\n noise = np.random.normal(scale=sigma, size=(n))\n y = a * x + noise\n else:\n x = np.linspace(0.0, 1.0, n, endpoint=True)\n y = a * x\n return x, y\n\n\nclass ShallowNarrowLNN:\n \"\"\"\n Shallow and narrow (one neuron per layer) linear neural network\n \"\"\"\n def __init__(self, init_ws):\n \"\"\"\n init_ws: initial weights as a list\n \"\"\"\n assert isinstance(init_ws, list)\n assert len(init_ws) == 2\n self.w1 = init_ws[0]\n self.w2 = init_ws[1]\n\n def forward(self, x):\n \"\"\"\n The forward pass through netwrok y = x * w1 * w2\n \"\"\"\n y = x * self.w1 * self.w2\n return y\n\n def loss(self, y_p, y_t):\n \"\"\"\n Mean squared error (L2) with 1/2 for convenience\n \"\"\"\n assert y_p.shape == y_t.shape\n mse = ((y_t - y_p)**2).mean()\n return mse\n\n def dloss_dw(self, x, y_t):\n \"\"\"\n partial derivative of loss with respect to weights\n\n Args:\n x : np.array\n y_t : np.array\n \"\"\"\n assert x.shape == y_t.shape\n Error = y_t - self.w1 * self.w2 * x\n dloss_dw1 = - (2 * self.w2 * x * Error).mean()\n dloss_dw2 = - (2 * self.w1 * x * Error).mean()\n return dloss_dw1, dloss_dw2\n\n def train(self, x, y_t, eta, n_ep):\n \"\"\"\n Gradient descent algorithm\n\n Args:\n x : np.array\n y_t : np.array\n eta: float\n n_ep : int\n \"\"\"\n assert x.shape == y_t.shape\n\n loss_records = np.empty(n_ep) # pre allocation of loss records\n weight_records = np.empty((n_ep, 2)) # pre allocation of weight records\n\n for i in range(n_ep):\n y_p = self.forward(x)\n loss_records[i] = self.loss(y_p, y_t)\n dloss_dw1, dloss_dw2 = self.dloss_dw(x, y_t)\n self.w1 -= eta * dloss_dw1\n self.w2 -= eta * dloss_dw2\n weight_records[i] = [self.w1, self.w2]\n\n return loss_records, weight_records\n\n\nclass DeepNarrowLNN:\n \"\"\"\n Deep but thin (one neuron per layer) linear neural network\n \"\"\"\n def __init__(self, init_ws):\n \"\"\"\n init_ws: initial weights as a numpy array\n \"\"\"\n self.n = init_ws.size\n self.W = init_ws.reshape(1, -1)\n\n def forward(self, x):\n \"\"\"\n x : np.array\n input features\n \"\"\"\n y = np.prod(self.W) * x\n return y\n\n def loss(self, y_t, y_p):\n \"\"\"\n mean squared error (L2 loss)\n\n Args:\n y_t : np.array\n y_p : np.array\n \"\"\"\n assert y_p.shape == y_t.shape\n mse = ((y_t - y_p)**2 / 2).mean()\n return mse\n\n def dloss_dw(self, x, y_t, y_p):\n \"\"\"\n analytical gradient of weights\n\n Args:\n x : np.array\n y_t : np.array\n y_p : np.array\n \"\"\"\n E = y_t - y_p # = y_t - x * np.prod(self.W)\n Ex = np.multiply(x, E).mean()\n Wp = np.prod(self.W) / (self.W + 1e-9)\n dW = - Ex * Wp\n return dW\n\n def train(self, x, y_t, eta, n_epochs):\n \"\"\"\n training using gradient descent\n\n Args:\n x : np.array\n y_t : np.array\n eta: float\n n_epochs : int\n \"\"\"\n loss_records = np.empty(n_epochs)\n loss_records[:] = np.nan\n for i in range(n_epochs):\n y_p = self.forward(x)\n loss_records[i] = self.loss(y_t, y_p).mean()\n dloss_dw = self.dloss_dw(x, y_t, y_p)\n if np.isnan(dloss_dw).any() or np.isinf(dloss_dw).any():\n return loss_records\n self.W -= eta * dloss_dw\n return loss_records",
"_____no_output_____"
],
[
"#@title Set random seed\n\n#@markdown Executing `set_seed(seed=seed)` you are setting the seed\n\n# for DL its critical to set the random seed so that students can have a\n# baseline to compare their results to expected results.\n# Read more here: https://pytorch.org/docs/stable/notes/randomness.html\n\n# Call `set_seed` function in the exercises to ensure reproducibility.\nimport random\nimport torch\n\ndef set_seed(seed=None, seed_torch=True):\n if seed is None:\n seed = np.random.choice(2 ** 32)\n random.seed(seed)\n np.random.seed(seed)\n if seed_torch:\n torch.manual_seed(seed)\n torch.cuda.manual_seed_all(seed)\n torch.cuda.manual_seed(seed)\n torch.backends.cudnn.benchmark = False\n torch.backends.cudnn.deterministic = True\n\n print(f'Random seed {seed} has been set.')\n\n\n# In case that `DataLoader` is used\ndef seed_worker(worker_id):\n worker_seed = torch.initial_seed() % 2**32\n np.random.seed(worker_seed)\n random.seed(worker_seed)",
"_____no_output_____"
],
[
"#@title Set device (GPU or CPU). Execute `set_device()`\n# especially if torch modules used.\n\n# inform the user if the notebook uses GPU or CPU.\n\ndef set_device():\n device = \"cuda\" if torch.cuda.is_available() else \"cpu\"\n if device != \"cuda\":\n print(\"GPU is not enabled in this notebook. \\n\"\n \"If you want to enable it, in the menu under `Runtime` -> \\n\"\n \"`Hardware accelerator.` and select `GPU` from the dropdown menu\")\n else:\n print(\"GPU is enabled in this notebook. \\n\"\n \"If you want to disable it, in the menu under `Runtime` -> \\n\"\n \"`Hardware accelerator.` and select `None` from the dropdown menu\")\n\n return device",
"_____no_output_____"
],
[
"SEED = 2021\nset_seed(seed=SEED)\nDEVICE = set_device()",
"_____no_output_____"
]
],
[
[
"---\n# Section 1: A Shallow Narrow Linear Neural Network\n\n*Time estimate: ~30 mins*",
"_____no_output_____"
]
],
[
[
"# @title Video 1: Shallow Narrow Linear Net\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1F44y117ot\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"6e5JIYsqVvU\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('video 1: Shallow Narrow Linear Net')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"## Section 1.1: A Shallow Narrow Linear Net",
"_____no_output_____"
],
[
"To better understand the behavior of neural network training with gradient descent, we start with the incredibly simple case of a shallow narrow linear neural net, since state-of-the-art models are impossible to dissect and comprehend with our current mathematical tools.\n\nThe model we use has one hidden layer, with only one neuron, and two weights. We consider the squared error (or L2 loss) as the cost function. As you may have already guessed, we can visualize the model as a neural network:\n\n<center><img src=\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/W1D2_LinearDeepLearning/static/shallow_narrow_nn.png\" width=\"400\"/></center>\n\n<br/>\n\nor by its computation graph:\n\n<center><img src=\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/W1D2_LinearDeepLearning/static/shallow_narrow.png\" alt=\"Shallow Narrow Graph\" width=\"400\"/></center>\n\nor on a rare occasion, even as a reasonably compact mapping:\n\n$$ loss = (y - w_1 \\cdot w_2 \\cdot x)^2 $$\n\n<br/>\n\nImplementing a neural network from scratch without using any Automatic Differentiation tool is rarely necessary. The following two exercises are therefore **Bonus** (optional) exercises. Please ignore them if you have any time-limits or pressure and continue to Section 1.2.",
"_____no_output_____"
],
[
"### Analytical Exercise 1.1: Loss Gradients (Optional)\n\nOnce again, we ask you to calculate the network gradients analytically, since you will need them for the next exercise. We understand how annoying this is.\n\n$\\dfrac{\\partial{loss}}{\\partial{w_1}} = ?$\n\n$\\dfrac{\\partial{loss}}{\\partial{w_2}} = ?$\n\n<br/>\n\n---\n#### Solution\n\n$\\dfrac{\\partial{loss}}{\\partial{w_1}} = -2 \\cdot w_2 \\cdot x \\cdot (y - w_1 \\cdot w_2 \\cdot x)$\n\n$\\dfrac{\\partial{loss}}{\\partial{w_2}} = -2 \\cdot w_1 \\cdot x \\cdot (y - w_1 \\cdot w_2 \\cdot x)$\n\n---\n",
"_____no_output_____"
],
[
"### Coding Exercise 1.1: Implement simple narrow LNN (Optional)\n\nNext, we ask you to implement the `forward` pass for our model from scratch without using PyTorch.\n\nAlso, although our model gets a single input feature and outputs a single prediction, we could calculate the loss and perform training for multiple samples at once. This is the common practice for neural networks, since computers are incredibly fast doing matrix (or tensor) operations on batches of data, rather than processing samples one at a time through `for` loops. Therefore, for the `loss` function, please implement the **mean** squared error (MSE), and adjust your analytical gradients accordingly when implementing the `dloss_dw` function.\n\nFinally, complete the `train` function for the gradient descent algorithm:\n\n\\begin{equation}\n\\mathbf{w}^{(t+1)} = \\mathbf{w}^{(t)} - \\eta \\nabla loss (\\mathbf{w}^{(t)})\n\\end{equation}",
"_____no_output_____"
]
],
[
[
"class ShallowNarrowExercise:\n \"\"\"Shallow and narrow (one neuron per layer) linear neural network\n \"\"\"\n def __init__(self, init_weights):\n \"\"\"\n Args:\n init_weights (list): initial weights\n \"\"\"\n assert isinstance(init_weights, (list, np.ndarray, tuple))\n assert len(init_weights) == 2\n self.w1 = init_weights[0]\n self.w2 = init_weights[1]\n\n\n def forward(self, x):\n \"\"\"The forward pass through netwrok y = x * w1 * w2\n\n Args:\n x (np.ndarray): features (inputs) to neural net\n\n returns:\n (np.ndarray): neural network output (prediction)\n \"\"\"\n #################################################\n ## Implement the forward pass to calculate prediction\n ## Note that prediction is not the loss\n # Complete the function and remove or comment the line below\n raise NotImplementedError(\"Forward Pass `forward`\")\n #################################################\n y = ...\n return y\n\n\n def dloss_dw(self, x, y_true):\n \"\"\"Gradient of loss with respect to weights\n\n Args:\n x (np.ndarray): features (inputs) to neural net\n y_true (np.ndarray): true labels\n\n returns:\n (float): mean gradient of loss with respect to w1\n (float): mean gradient of loss with respect to w2\n \"\"\"\n assert x.shape == y_true.shape\n #################################################\n ## Implement the gradient computation function\n # Complete the function and remove or comment the line below\n raise NotImplementedError(\"Gradient of Loss `dloss_dw`\")\n #################################################\n dloss_dw1 = ...\n dloss_dw2 = ...\n return dloss_dw1, dloss_dw2\n\n\n def train(self, x, y_true, lr, n_ep):\n \"\"\"Training with Gradient descent algorithm\n\n Args:\n x (np.ndarray): features (inputs) to neural net\n y_true (np.ndarray): true labels\n lr (float): learning rate\n n_ep (int): number of epochs (training iterations)\n\n returns:\n (list): training loss records\n (list): training weight records (evolution of weights)\n \"\"\"\n assert x.shape == y_true.shape\n\n loss_records = np.empty(n_ep) # pre allocation of loss records\n weight_records = np.empty((n_ep, 2)) # pre allocation of weight records\n\n for i in range(n_ep):\n y_prediction = self.forward(x)\n loss_records[i] = loss(y_prediction, y_true)\n dloss_dw1, dloss_dw2 = self.dloss_dw(x, y_true)\n #################################################\n ## Implement the gradient descent step\n # Complete the function and remove or comment the line below\n raise NotImplementedError(\"Training loop `train`\")\n #################################################\n self.w1 -= ...\n self.w2 -= ...\n weight_records[i] = [self.w1, self.w2]\n\n return loss_records, weight_records\n\n\ndef loss(y_prediction, y_true):\n \"\"\"Mean squared error\n\n Args:\n y_prediction (np.ndarray): model output (prediction)\n y_true (np.ndarray): true label\n\n returns:\n (np.ndarray): mean squared error loss\n \"\"\"\n assert y_prediction.shape == y_true.shape\n #################################################\n ## Implement the MEAN squared error\n # Complete the function and remove or comment the line below\n raise NotImplementedError(\"Loss function `loss`\")\n #################################################\n mse = ...\n return mse\n\n\n#add event to airtable\natform.add_event('Coding Exercise 1.1: Implement simple narrow LNN')\n\nset_seed(seed=SEED)\nn_epochs = 211\nlearning_rate = 0.02\ninitial_weights = [1.4, -1.6]\nx_train, y_train = gen_samples(n=73, a=2.0, sigma=0.2)\nx_eval = np.linspace(0.0, 1.0, 37, endpoint=True)\n## Uncomment to run\n# sn_model = ShallowNarrowExercise(initial_weights)\n# loss_log, weight_log = sn_model.train(x_train, y_train, learning_rate, n_epochs)\n# y_eval = sn_model.forward(x_eval)\n# plot_x_y_(x_train, y_train, x_eval, y_eval, loss_log, weight_log)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D2_LinearDeepLearning/solutions/W1D2_Tutorial2_Solution_46492cd6.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=1696.0 height=544.0 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/W1D2_LinearDeepLearning/static/W1D2_Tutorial2_Solution_46492cd6_1.png>\n\n",
"_____no_output_____"
],
[
"## Section 1.2: Learning landscapes",
"_____no_output_____"
]
],
[
[
"# @title Video 2: Training Landscape\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1Nv411J71X\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"k28bnNAcOEg\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 2: Training Landscape')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"As you may have already asked yourself, we can analytically find $w_1$ and $w_2$ without using gradient descent:\n\n\\begin{equation}\nw_1 \\cdot w_2 = \\dfrac{y}{x}\n\\end{equation}\n\nIn fact, we can plot the gradients, the loss function and all the possible solutions in one figure. In this example, we use the $y = 1x$ mapping:\n\n**Blue ribbon**: shows all possible solutions: $~ w_1 w_2 = \\dfrac{y}{x} = \\dfrac{x}{x} = 1 \\Rightarrow w_1 = \\dfrac{1}{w_2}$\n\n**Contour background**: Shows the loss values, red being higher loss\n\n**Vector field (arrows)**: shows the gradient vector field. The larger yellow arrows show larger gradients, which correspond to bigger steps by gradient descent.\n\n**Scatter circles**: the trajectory (evolution) of weights during training for three different initializations, with blue dots marking the start of training and red crosses ( **x** ) marking the end of training. You can also try your own initializations (keep the initial values between `-2.0` and `2.0`) as shown here:\n```python\nplot_vector_field('all', [1.0, -1.0])\n```\n\nFinally, if the plot is too crowded, feel free to pass one of the following strings as argument:\n\n```python\nplot_vector_field('vectors') # for vector field\nplot_vector_field('trajectory') # for training trajectory\nplot_vector_field('loss') # for loss contour\n```\n\n**Think!**\n\nExplore the next two plots. Try different initial values. Can you find the saddle point? Why does training slow down near the minima?",
"_____no_output_____"
]
],
[
[
"plot_vector_field('all')",
"_____no_output_____"
]
],
[
[
"Here, we also visualize the loss landscape in a 3-D plot, with two training trajectories for different initial conditions.\nNote: the trajectories from the 3D plot and the previous plot are independent and different.",
"_____no_output_____"
]
],
[
[
"plot_loss_landscape()",
"_____no_output_____"
],
[
"# @title Student Response\nfrom ipywidgets import widgets\n\n\ntext=widgets.Textarea(\n value='Type your here and Push submit',\n placeholder='Type something',\n description='',\n disabled=False\n)\n\nbutton = widgets.Button(description=\"Submit!\")\n\ndisplay(text,button)\n\ndef on_button_clicked(b):\n atform.add_answer('q1', text.value)\n print(\"Submission successful!\")\n\nbutton.on_click(on_button_clicked)",
"_____no_output_____"
],
[
"# @title Video 3: Training Landscape - Discussion\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1py4y1j7cv\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"0EcUGgxOdkI\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 3: Training Landscape - Discussiond')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"---\n# Section 2: Depth, Learning rate, and initialization\n*Time estimate: ~45 mins*",
"_____no_output_____"
],
[
"Successful deep learning models are often developed by a team of very clever people, spending many many hours \"tuning\" learning hyperparameters, and finding effective initializations. In this section, we look at three basic (but often not simple) hyperparameters: depth, learning rate, and initialization.",
"_____no_output_____"
],
[
"## Section 2.1: The effect of depth",
"_____no_output_____"
]
],
[
[
"# @title Video 4: Effect of Depth\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1z341167di\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"Ii_As9cRR5Q\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 4: Effect of Depth')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"Why might depth be useful? What makes a network or learning system \"deep\"? The reality is that shallow neural nets are often incapable of learning complex functions due to data limitations. On the other hand, depth seems like magic. Depth can change the functions a network can represent, the way a network learns, and how a network generalizes to unseen data. \n\nSo let's look at the challenges that depth poses in training a neural network. Imagine a single input, single output linear network with 50 hidden layers and only one neuron per layer (i.e. a narrow deep neural network). The output of the network is easy to calculate:\n\n$$ prediction = x \\cdot w_1 \\cdot w_2 \\cdot \\cdot \\cdot w_{50} $$\n\nIf the initial value for all the weights is $w_i = 2$, the prediction for $x=1$ would be **exploding**: $y_p = 2^{50} \\approx 1.1256 \\times 10^{15}$. On the other hand, for weights initialized to $w_i = 0.5$, the output is **vanishing**: $y_p = 0.5^{50} \\approx 8.88 \\times 10^{-16}$. Similarly, if we recall the chain rule, as the graph gets deeper, the number of elements in the chain multiplication increases, which could lead to exploding or vanishing gradients. To avoid such numerical vulnerablities that could impair our training algorithm, we need to understand the effect of depth.\n",
"_____no_output_____"
],
[
"### Interactive Demo 2.1: Depth widget\n\nUse the widget to explore the impact of depth on the training curve (loss evolution) of a deep but narrow neural network.\n\n**Think!**\n\nWhich networks trained the fastest? Did all networks eventually \"work\" (converge)? What is the shape of their learning trajectory?",
"_____no_output_____"
]
],
[
[
"# @markdown Make sure you execute this cell to enable the widget!\n\n_ = interact(depth_widget,\n depth = IntSlider(min=0, max=51,\n step=5, value=0,\n continuous_update=False))",
"_____no_output_____"
],
[
"# @title Video 5: Effect of Depth - Discussion\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1Qq4y1H7uk\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"EqSDkwmSruk\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 5: Effect of Depth - Discussion')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"## Section 2.2: Choosing a learning rate",
"_____no_output_____"
],
[
"The learning rate is a common hyperparameter for most optimization algorithms. How should we set it? Sometimes the only option is to try all the possibilities, but sometimes knowing some key trade-offs will help guide our search for good hyperparameters.",
"_____no_output_____"
]
],
[
[
"# @title Video 6: Learning Rate\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV11f4y157MT\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"w_GrCVM-_Qo\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 6: Learning Rate')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"### Interactive Demo 2.2: Learning rate widget\n\nHere, we fix the network depth to 50 layers. Use the widget to explore the impact of learning rate $\\eta$ on the training curve (loss evolution) of a deep but narrow neural network.\n\n**Think!**\n\nCan we say that larger learning rates always lead to faster learning? Why not? ",
"_____no_output_____"
]
],
[
[
"# @markdown Make sure you execute this cell to enable the widget!\n\n_ = interact(lr_widget,\n lr = FloatSlider(min=0.005, max=0.045, step=0.005, value=0.005,\n continuous_update=False, readout_format='.3f',\n description='eta'))",
"_____no_output_____"
],
[
"# @title Video 7: Learning Rate - Discussion\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1Aq4y1p7bh\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"cmS0yqImz2E\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 7: Learning Rate - Discussion')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"## Section 2.3: Depth vs Learning Rate",
"_____no_output_____"
]
],
[
[
"# @title Video 8: Depth and Learning Rate\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1V44y1177e\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"J30phrux_3k\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 8: Depth and Learning Rate')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"### Interactive Demo 2.3: Depth and Learning-Rate\n",
"_____no_output_____"
],
[
"**Important instruction**\nThe exercise starts with 10 hidden layers. Your task is to find the learning rate that delivers fast but robust convergence (learning). When you are confident about the learning rate, you can **Register** the optimal learning rate for the given depth. Once you press register, a deeper model is instantiated, so you can find the next optimal learning rate. The Register button turns green only when the training converges, but does not imply the fastest convergence. Finally, be patient :) the widgets are slow.\n\n\n**Think!**\n\nCan you explain the relationship between the depth and optimal learning rate?",
"_____no_output_____"
]
],
[
[
"# @markdown Make sure you execute this cell to enable the widget!\nintpl_obj = InterPlay()\n\nintpl_obj.slider = FloatSlider(min=0.005, max=0.105, step=0.005, value=0.005,\n layout=Layout(width='500px'),\n continuous_update=False,\n readout_format='.3f',\n description='eta')\n\nintpl_obj.button = ToggleButton(value=intpl_obj.converged, description='Register')\n\nwidgets_ui = HBox([intpl_obj.slider, intpl_obj.button])\nwidgets_out = interactive_output(intpl_obj.train,\n {'lr': intpl_obj.slider,\n 'update': intpl_obj.button,\n 'init_weights': fixed(0.9)})\n\ndisplay(widgets_ui, widgets_out)",
"_____no_output_____"
],
[
"# @title Video 9: Depth and Learning Rate - Discussion\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV15q4y1p7Uq\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"7Fl8vH7cgco\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 9: Depth and Learning Rate - Discussion')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"## Section 2.4: Why initialization is important",
"_____no_output_____"
]
],
[
[
"# @title Video 10: Initialization Matters\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1UL411J7vu\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"KmqCz95AMzY\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 10: Initialization Matters')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"We’ve seen, even in the simplest of cases, that depth can slow learning. Why? From the chain rule, gradients are multiplied by the current weight at each layer, so the product can vanish or explode. Therefore, weight initialization is a fundamentally important hyperparameter.\n\nAlthough in practice initial values for learnable parameters are often sampled from different $\\mathcal{Uniform}$ or $\\mathcal{Normal}$ probability distribution, here we use a single value for all the parameters.\n\nThe figure below shows the effect of initialization on the speed of learning for the deep but narrow LNN. We have excluded initializations that lead to numerical errors such as `nan` or `inf`, which are the consequence of smaller or larger initializations.",
"_____no_output_____"
]
],
[
[
"# @markdown Make sure you execute this cell to see the figure!\n\nplot_init_effect()",
"_____no_output_____"
],
[
"# @title Video 11: Initialization Matters Explained\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1hM4y1T7gJ\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"vKktGdiQDsE\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 11: Initialization Matters Explained')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"---\n# Summary\n\nIn the second tutorial, we have learned what is the training landscape, and also we have see in depth the effect of the depth of the network and the learning rate, and their interplay. Finally, we have seen that initialization matters and why we need smart ways of initialization.",
"_____no_output_____"
]
],
[
[
"# @title Video 12: Tutorial 2 Wrap-up\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1P44y117Pd\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"r3K8gtak3wA\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n#add event to airtable\natform.add_event('Video 12: Tutorial 2 Wrap-up')\n\ndisplay(out)",
"_____no_output_____"
],
[
"\n# @title Airtable Submission Link\nfrom IPython import display as IPydisplay\nIPydisplay.HTML(\n f\"\"\"\n <div>\n <a href= \"{atform.url()}\" target=\"_blank\">\n <img src=\"https://github.com/NeuromatchAcademy/course-content-dl/blob/main/tutorials/static/AirtableSubmissionButton.png?raw=1\"\n alt=\"button link to Airtable\" style=\"width:410px\"></a>\n </div>\"\"\" )",
"_____no_output_____"
]
],
[
[
"---\n# Bonus",
"_____no_output_____"
],
[
"## Hyperparameter interaction\n\nFinally, let's put everything we learned together and find best initial weights and learning rate for a given depth. By now you should have learned the interactions and know how to find the optimal values quickly. If you get `numerical overflow` warnings, don't be discouraged! They are often caused by \"exploding\" or \"vanishing\" gradients.\n\n**Think!**\n\nDid you experience any surprising behaviour \nor difficulty finding the optimal parameters?",
"_____no_output_____"
]
],
[
[
"# @markdown Make sure you execute this cell to enable the widget!\n\n_ = interact(depth_lr_init_interplay,\n depth = IntSlider(min=10, max=51, step=5, value=25,\n continuous_update=False),\n lr = FloatSlider(min=0.001, max=0.1,\n step=0.005, value=0.005,\n continuous_update=False,\n readout_format='.3f',\n description='eta'),\n init_weights = FloatSlider(min=0.1, max=3.0,\n step=0.1, value=0.9,\n continuous_update=False,\n readout_format='.3f',\n description='initial weights'))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
cb6dc5a2b607587e8541e3b9b08e352e9d34bf29 | 8,234 | ipynb | Jupyter Notebook | NLP Projects/Requests Lib/requests_library_solution.ipynb | kcenan/AI_for_Trading | be28fa57fe932c17cd780ca9c571cebfe87bb5bd | [
"MIT"
]
| 1 | 2021-06-10T22:08:21.000Z | 2021-06-10T22:08:21.000Z | NLP Projects/Requests Lib/requests_library_solution.ipynb | kcenan/AI_for_Trading | be28fa57fe932c17cd780ca9c571cebfe87bb5bd | [
"MIT"
]
| null | null | null | NLP Projects/Requests Lib/requests_library_solution.ipynb | kcenan/AI_for_Trading | be28fa57fe932c17cd780ca9c571cebfe87bb5bd | [
"MIT"
]
| 1 | 2021-05-13T20:48:43.000Z | 2021-05-13T20:48:43.000Z | 32.936 | 82 | 0.355842 | [
[
[
"# Import libraries\nfrom bs4 import BeautifulSoup\nimport requests\nimport pandas as pd\nimport numpy as np\n\n# Create a Response object\nr = requests.get('https://en.wikipedia.org/wiki/Amazon_(company)')\n\n# Get HTML data\nhtml_data = r.text\n\n# Create a BeautifulSoup Object\npage_content = BeautifulSoup(html_data, 'html.parser')\n\n# Find financial table\nwikitable = page_content.find('table', {'class': 'wikitable float-left'})\n\n# Find all column titles\nwikicolumns = wikitable.tbody.findAll('tr')[0].findAll('th')\n\n# Loop through column titles and store into Python array\ndf_columns = []\nfor column in wikicolumns:\n text = column.get_text(strip=True, separator=\" \")\n df_columns.append(text)\n\n# Loop through the data rows and store into Python array\ndf_data = []\nfor row in wikitable.tbody.findAll('tr')[1:]:\n row_data = []\n for td in row.findAll('td'):\n text = td.get_text(strip=True, separator=\" \")\n row_data.append(text)\n df_data.append(np.array(row_data))\n\n# Print financial data in DataFrame format and set `Year` as index\ndataframe = pd.DataFrame(data=df_data, columns=df_columns)\ndataframe.set_index('Year', inplace=True)\ndataframe",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
cb6dd30108c5cb748b85710c8dbf500083a604c7 | 318,960 | ipynb | Jupyter Notebook | labs/trees/partitioning-feature-space.ipynb | Ming2010/msds621 | 4976b4c1547890b590383685ca7a7d665cc81ba5 | [
"MIT"
]
| 300 | 2019-07-23T17:30:45.000Z | 2022-03-28T18:45:16.000Z | labs/trees/partitioning-feature-space.ipynb | Ming2010/msds621 | 4976b4c1547890b590383685ca7a7d665cc81ba5 | [
"MIT"
]
| 1 | 2019-11-19T05:42:19.000Z | 2019-12-04T20:16:26.000Z | labs/trees/partitioning-feature-space.ipynb | Ming2010/msds621 | 4976b4c1547890b590383685ca7a7d665cc81ba5 | [
"MIT"
]
| 128 | 2019-08-02T20:11:35.000Z | 2022-03-27T19:12:24.000Z | 227.017794 | 112,076 | 0.908832 | [
[
[
"# Partitioning feature space",
"_____no_output_____"
],
[
"**Make sure to get latest dtreeviz**",
"_____no_output_____"
]
],
[
[
"! pip install -q -U dtreeviz\n! pip install -q graphviz==0.17 # 0.18 deletes the `run` func I need",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\n\nfrom sklearn.linear_model import LinearRegression, Ridge, Lasso, LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier, RandomForestRegressor\nfrom sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor\nfrom sklearn.datasets import load_boston, load_iris, load_wine, load_digits, \\\n load_breast_cancer, load_diabetes\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import mean_squared_error, accuracy_score\n\nimport matplotlib.pyplot as plt\n%config InlineBackend.figure_format = 'retina'\n\nfrom sklearn import tree\nfrom dtreeviz.trees import *\nfrom dtreeviz.models.shadow_decision_tree import ShadowDecTree",
"_____no_output_____"
],
[
"def show_mse_leaves(X,y,max_depth):\n t = DecisionTreeRegressor(max_depth=max_depth)\n t.fit(X,y)\n shadow = ShadowDecTree.get_shadow_tree(t, X, y, feature_names=['sqfeet'], target_name='rent')\n root, leaves, internal = shadow._get_tree_nodes()\n # node2samples = shadow._get_tree_nodes()_samples()\n # isleaf = shadow.get_node_type(t)\n n_node_samples = t.tree_.n_node_samples\n\n mse = 99.9#mean_squared_error(y, [np.mean(y)]*len(y))\n print(f\"Root {0:3d} has {n_node_samples[0]:3d} samples with MSE ={mse:6.2f}\")\n print(\"-----------------------------------------\")\n\n avg_mse_per_record = 0.0\n node2samples = shadow.get_node_samples()\n for node in leaves:\n leafy = y[node2samples[node.id]]\n n = len(leafy)\n mse = mean_squared_error(leafy, [np.mean(leafy)]*n)\n avg_mse_per_record += mse * n\n print(f\"Node {node.id:3d} has {n_node_samples[node.id]:3d} samples with MSE ={mse:6.2f}\")\n\n avg_mse_per_record /= len(y)\n print(f\"Average MSE per record is {avg_mse_per_record:.1f}\")",
"_____no_output_____"
]
],
[
[
"## Regression",
"_____no_output_____"
]
],
[
[
"df_cars = pd.read_csv(\"data/cars.csv\")\nX, y = df_cars[['ENG']], df_cars['MPG']\ndf_cars.head(3)",
"_____no_output_____"
],
[
"dt = DecisionTreeRegressor(max_depth=1)\ndt.fit(X, y)\n\nrtreeviz_univar(dt, X, y,\n feature_names='Horsepower',\n markersize=5,\n mean_linewidth=1,\n target_name='MPG',\n fontsize=9,\n show={})",
"_____no_output_____"
]
],
[
[
"**Q.** What is the MSE between y and predicted $\\hat{y} = \\overline{y}$?\n\nHints: You can use function `mean_squared_error(` $y$,$\\hat{y}$ `)`; create a vector of length $|y|$ with $\\overline{y}$ as elements.",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n<pre>\nmean_squared_error(y, [np.mean(y)]*len(y)) # about 60.76\n</pre>\n</details>",
"_____no_output_____"
],
[
"**Q.** Where would you split this if you could only split once? Set the `split` variable to a reasonable value.",
"_____no_output_____"
]
],
[
[
"split = ...",
"_____no_output_____"
]
],
[
[
"<details>\n<summary>Solution</summary>\nThe split location that gets most pure subregion might be about split = 200 HP because the region to the right has a relatively flat MPG average.\n</details>",
"_____no_output_____"
],
[
"**Alter the rtreeviz_univar() call to show the split with arg show={'splits'}**",
"_____no_output_____"
],
[
"<details>\n <summary>Solution</summary>\n<pre>\nrtreeviz_univar(dt, X, y,\n feature_names='Horsepower',\n markersize=5,\n mean_linewidth=1,\n target_name='MPG',\n fontsize=9,\n show={'splits'})\n</pre>\n</details>",
"_____no_output_____"
],
[
"**Q.** What are the MSE values for the left, right partitions?\n\nHints: Get the y values whose `X['ENG']` are less than `split` into `lefty` and those greater than or equal to `split` into `righty`. The split introduces two new children that are leaves until we (possibly) split them; the leaves predict the mean of their samples.",
"_____no_output_____"
]
],
[
[
"lefty = ...; mleft = ...\nrighty = ...; mright = ...\n\nmse_left = ...\nmse_right = ...\n\nmse_left, mse_right",
"_____no_output_____"
]
],
[
[
"<details>\n<summary>Solution</summary>\n Should be (35.68916307096633, 12.770261374699789)<p>\n<pre>\nlefty = y[X['ENG']<split]\nrighty = y[X['ENG']>=split]\nmleft = np.mean(lefty)\nmright = np.mean(righty)\n\nmse_left = mean_squared_error(lefty, [mleft]\\*len(lefty))\nmse_right = mean_squared_error(righty, [mright]\\*len(righty))\n</pre>\n</details>",
"_____no_output_____"
],
[
"**Q.** Compare the MSE values for overall y and the average of the left, right partition MSEs (which is about 24.2)?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nAfter the split the MSE of the children is much lower than before the split, therefore, it is a worthwhile split.\n</details>\n",
"_____no_output_____"
],
[
"**Q.** Set the split value to 100 and recompare MSE values for y, left, and right.",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nWith split=100, mse_left, mse_right become 33.6 and 41.0. These are still less than the y MSE of 60.7 so worthwhile but not nearly as splitting at 200.\n</details>\n",
"_____no_output_____"
],
[
"### Effect of deeper trees",
"_____no_output_____"
],
[
"Consider the sequence of tree depths 1..6 for horsepower vs MPG.",
"_____no_output_____"
]
],
[
[
"X = df_cars[['ENG']].values\ny = df_cars['MPG'].values\n\nfig, axes = plt.subplots(1,6, figsize=(14,3), sharey=True)\nfor i,ax in enumerate(axes.flatten()):\n dt = DecisionTreeRegressor(max_depth=i+1)\n dt.fit(X, y)\n t = rtreeviz_univar(dt,\n X, y,\n feature_names='Horsepower',\n markersize=5,\n mean_linewidth=1,\n target_name='MPG' if i==0 else None,\n fontsize=9,\n show={'splits'},\n ax=ax)\n ax.set_title(f\"Depth {i+1}\", fontsize=9)\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Q.** Focusing on the orange horizontal lines, what do you notice as more splits appear?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nWith depth 1, model is biased due to coarseness of the approximations (just 2 leaf means). Depth 2 gets much better approximation, so bias is lower. As we add more depth to tree, number of splits increases and these appear to be chasing details of the data, decreasing bias on training set but also hurting generality.\n</details>\n",
"_____no_output_____"
],
[
"**Q.** Consider the MSE for the 4 leaves of a depth 2 tree and 15 leaves of a depth 4 tree. What happens to the average MSE per leaf? What happens to the leaf sizes and how is it related to average MSE?",
"_____no_output_____"
]
],
[
[
"show_mse_leaves(df_cars[['ENG']], df_cars['MPG'], max_depth=2)",
"Root 0 has 392 samples with MSE = 99.90\n-----------------------------------------\nNode 2 has 120 samples with MSE = 30.45\nNode 3 has 102 samples with MSE = 20.07\nNode 5 has 72 samples with MSE = 9.23\nNode 6 has 98 samples with MSE = 6.76\nAverage MSE per record is 17.9\n"
],
[
"show_mse_leaves(df_cars[['ENG']], df_cars['MPG'], max_depth=4)",
"Root 0 has 392 samples with MSE = 99.90\n-----------------------------------------\nNode 4 has 1 samples with MSE = 0.00\nNode 5 has 3 samples with MSE = 6.18\nNode 7 has 51 samples with MSE = 29.27\nNode 8 has 65 samples with MSE = 20.59\nNode 11 has 68 samples with MSE = 20.26\nNode 12 has 16 samples with MSE = 9.32\nNode 14 has 13 samples with MSE = 23.93\nNode 15 has 5 samples with MSE = 3.21\nNode 19 has 44 samples with MSE = 2.91\nNode 20 has 25 samples with MSE = 4.35\nNode 22 has 2 samples with MSE = 81.00\nNode 23 has 1 samples with MSE = 0.00\nNode 26 has 22 samples with MSE = 6.03\nNode 27 has 47 samples with MSE = 8.26\nNode 29 has 20 samples with MSE = 3.81\nNode 30 has 9 samples with MSE = 1.51\nAverage MSE per record is 14.6\n"
]
],
[
[
"<details>\n<summary>Solution</summary>\nThe average MSE is much lower as we increase depth because that allows the tree to isolate pure/more-similar regions. This also shrinks leaf size since we are splitting more as the tree deepens.\n</details>\n",
"_____no_output_____"
],
[
"Consider the plot of the CYL feature (num cylinders) vs MPG:",
"_____no_output_____"
]
],
[
[
"X = df_cars[['CYL']].values\ny = df_cars['MPG'].values\n\nfig, axes = plt.subplots(1,3, figsize=(7,2.5), sharey=True)\ndepths = [1,2,10]\nfor i,ax in enumerate(axes.flatten()):\n dt = DecisionTreeRegressor(max_depth=depths[i])\n dt.fit(X, y)\n t = rtreeviz_univar(dt,\n X, y,\n feature_names='Horsepower',\n markersize=5,\n mean_linewidth=1,\n target_name='MPG' if i==0 else None,\n fontsize=9,\n show={'splits','title'},\n ax=ax)\n ax.set_title(f\"Depth {i+1}\", fontsize=9)\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Q.** Explain why the graph looks like a bunch of vertical bars.",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nThe x values are integers and will clump together. Since there are many MPG values at each int, you get vertical clumps of data.\n</details>",
"_____no_output_____"
],
[
"**Q.** Why don't we get many more splits for depth 10 vs depth 2?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nOnce each unique x value has a \"bin\", there are no more splits to do.\n</details>",
"_____no_output_____"
],
[
"**Q.** Why are the orange predictions bars at the levels they are in the plot?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nDecision tree leaves predict the average y for all samples in a leaf.\n</details>",
"_____no_output_____"
],
[
"## Classification",
"_____no_output_____"
]
],
[
[
"wine = load_wine()\ndf_wine = pd.DataFrame(data=wine.data, columns=wine.feature_names)\ndf_wine.head(3)",
"_____no_output_____"
],
[
"feature_names = list(wine.feature_names)\nclass_names = list(wine.target_names)",
"_____no_output_____"
]
],
[
[
"### 1 variable",
"_____no_output_____"
]
],
[
[
"X = df_wine[['flavanoids']].values\ny = wine.target\n\ndt = DecisionTreeClassifier(max_depth=1)\ndt.fit(X, y)\n\nfig, ax = plt.subplots(1,1, figsize=(4,1.8))\nct = ctreeviz_univar(dt, X, y,\n feature_names = 'flavanoids',\n class_names=class_names,\n target_name='Wine',\n nbins=40, gtype='strip',\n fontsize=9,\n show={},\n colors={'scatter_marker_alpha':1, 'scatter_marker_alpha':1},\n ax=ax)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Q.** Where would you split this (vertically) if you could only split once?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nThe split location that gets most pure subregion might be about 1.5 because it nicely carves off the left green samples.\n</details>",
"_____no_output_____"
],
[
"**Alter the code to show the split with arg show={'splits'}**",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n<pre>\nX = df_wine[['flavanoids']].values\ny = wine.target\n\ndt = DecisionTreeClassifier(max_depth=1)\ndt.fit(X, y)\n\nfig, ax = plt.subplots(1,1, figsize=(4,1.8))\nct = ctreeviz_univar(dt, X, y,\n feature_names = 'flavanoids',\n class_names=class_names,\n target_name='Wine',\n nbins=40, gtype='strip',\n fontsize=9,\n show={'splits'},\n colors={'scatter_marker_alpha':1, 'scatter_marker_alpha':1},\n ax=ax)\nplt.show()\n</pre>\n</details>",
"_____no_output_____"
],
[
"**Q.** For max_depth=2, how many splits will we get?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n3. We get one split for root and then with depth=2, we have 2 children that each get a split.\n</details>",
"_____no_output_____"
],
[
"**Q.** Where would you split this graph in that many places?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nOnce we carve off the leftmost green, we would want to isolate the blue in between 1.3 and 2.3. The other place to split is not obvious as there is no great choice. (sklearn will add a split point at 1.0)\n</details>\n",
"_____no_output_____"
],
[
"**Alter the code to show max_depth=2**",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n<pre>\nX = df_wine[['flavanoids']].values\ny = wine.target\n\ndt = DecisionTreeClassifier(max_depth=2)\ndt.fit(X, y)\n\nfig, ax = plt.subplots(1,1, figsize=(4,1.8))\nct = ctreeviz_univar(dt, X, y,\n feature_names = 'flavanoids',\n class_names=class_names,\n target_name='Wine',\n nbins=40, gtype='strip',\n fontsize=9,\n show={'splits'},\n colors={'scatter_marker_alpha':1, 'scatter_marker_alpha':1},\n ax=ax)\nplt.show()\n</pre>\n</details>",
"_____no_output_____"
],
[
"### Gini impurity",
"_____no_output_____"
],
[
"Let's compute the gini impurity for left and right sides for a depth=1 tree that splits flavanoids at 1.3. Here's a function that computes the value:\n\n$$\nGini({\\bf p}) = \\sum_{i=1}^{k} p_i \\left[ \\sum_{j \\ne i}^k p_j \\right] = \\sum_{i=1}^{k} p_i (1 - p_i) = 1 - \\sum_{i=1}^{k} p_i^2\n$$\n\nwhere $p_i = \\frac{|y[y==i]|}{|y|}$. Since $\\sum_{j \\ne i}^k p_j$ is the probability of \"not $p_i$\", we can summarize that as just $1-p_i$. The gini value is then computing $p_i$ times \"not $p_i$\" for $k$ classes. Value $p_i$ is the probability of seeing class $i$ in a list of target values, $y$. ",
"_____no_output_____"
]
],
[
[
"def gini(y):\n \"\"\"\n Compute gini impurity from y vector of class values (from k unique values).\n Result is in range 0..(k-1/k) inclusive; binary range is 0..1/2.\n See https://en.wikipedia.org/wiki/Decision_tree_learning#Gini_impurity\"\n \"\"\"\n _, counts = np.unique(y, return_counts=True)\n p = counts / len(y)\n return 1 - np.sum( p**2 )",
"_____no_output_____"
]
],
[
[
"**Q.** Using that function, what is the gini impurity for the overall y target",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\ngini(y) # about 0.66\n</details>\n",
"_____no_output_____"
],
[
"**Get all y values for rows where `df_wine['flavanoids']`<1.3 into variable `lefty` and `>=` into `righty`**",
"_____no_output_____"
]
],
[
[
"lefty = ...\nrighty = ...",
"_____no_output_____"
]
],
[
[
"<details>\n<summary>Solution</summary>\n<pre>\nlefty = y[df_wine['flavanoids']<1.3]\nrighty = y[df_wine['flavanoids']>=1.3]\n</pre>\n</details>\n",
"_____no_output_____"
],
[
"**Q.** What are the gini values for left and right partitions?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\ngini(lefty), gini(righty) # about 0.27, 0.53\n</details>",
"_____no_output_____"
],
[
"**Q.** What can we conclude about the purity of left and right? Also, compare to gini for all y values.",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nLeft partition is much more pure than right but right is still more pure than original gini(y). We can conclude that the split is worthwhile as the partition would let us give more accurate predictions.\n</details>",
"_____no_output_____"
],
[
"### 2 variables",
"_____no_output_____"
]
],
[
[
"X = df_wine[['alcohol','flavanoids']].values\ny = wine.target\n\ndt = DecisionTreeClassifier(max_depth=1)\ndt.fit(X, y)\n\nfig, ax = plt.subplots(1, 1, figsize=(4,3))\nct = ctreeviz_bivar(dt, X, y,\n feature_names = ['alcohol','flavanoid'], class_names=class_names,\n target_name='iris',\n show={},\n colors={'scatter_marker_alpha':1, 'scatter_marker_alpha':1},\n ax=ax\n )",
"_____no_output_____"
]
],
[
[
"**Q.** Which variable and split point would you choose if you could only split once?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nBecause the blue dots are spread vertically, a horizontal split won't be very good. Hence, we should choose variable proline. The best split will carve off the blue dots, leaving the yellow and green mixed up. A split at proline=12.7 seems pretty good.\n</details>\n",
"_____no_output_____"
],
[
"**Modify the code to view the splits and compare your answer**",
"_____no_output_____"
],
[
"**Q.** Which variable and split points would you choose next for depth=2?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nOnce we carve off most of the blue vertically, we should separate the yellow by choosing flavanoid=1.7 to split horizontally. NOTICE, however, that the 2nd split will not be across entire graph since we are splitting the region on the right. Splitting on the left can be at flavanoid=1 so we isolate the green from blue on left.\n</details>\n",
"_____no_output_____"
],
[
"**Modify the code to view the splits for depth=2 and compare your answer**",
"_____no_output_____"
],
[
"### Gini\n\nLet's examine gini impurity for a different pair of variables.",
"_____no_output_____"
]
],
[
[
"X = df_wine[['proline','flavanoids']].values\ny = wine.target\n\ndt = DecisionTreeClassifier(max_depth=1)\ndt.fit(X, y)\n\nfig, ax = plt.subplots(1, 1, figsize=(4,3))\nctreeviz_bivar(dt, X, y,\n feature_names = ['proline','flavanoid'],\n class_names=class_names,\n target_name='iris',\n show={'splits'},\n colors={'scatter_marker_alpha':1, 'scatter_marker_alpha':1},\n ax=ax)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Get all y values for rows where the split var is less than the split value into variable `lefty` and those `>=` into `righty`**",
"_____no_output_____"
]
],
[
[
"lefty = ...\nrighty = ...",
"_____no_output_____"
]
],
[
[
"<details>\n<summary>Solution</summary>\n<pre>\nlefty = y[df_wine['proline']<750]\nrighty = y[df_wine['proline']>=750]\n</pre>\n</details>\n",
"_____no_output_____"
],
[
"**Print out the gini for y, lefty, righty**",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n<pre>\ngini(y), gini(lefty), gini(righty)\n</pre>\n</details>\n",
"_____no_output_____"
],
[
"## Training a single tree and print out the training accuracy (num correct / total)",
"_____no_output_____"
]
],
[
[
"t = DecisionTreeClassifier()\nt.fit(df_wine, y)\naccuracy_score(y, t.predict(df_wine))",
"_____no_output_____"
]
],
[
[
"Take a look at the feature importance:",
"_____no_output_____"
]
],
[
[
"from rfpimp import *\nI = importances(t, df_wine, y)\nplot_importances(I)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6de366199b20af661ac21f1877e1e31ae65ea2 | 23,739 | ipynb | Jupyter Notebook | notebooks/string.ipynb | ManuelMBaumann/pymor | 9ad226a0a46c7ba30a18bdab27b8bbbfe8f83a31 | [
"Unlicense"
]
| null | null | null | notebooks/string.ipynb | ManuelMBaumann/pymor | 9ad226a0a46c7ba30a18bdab27b8bbbfe8f83a31 | [
"Unlicense"
]
| null | null | null | notebooks/string.ipynb | ManuelMBaumann/pymor | 9ad226a0a46c7ba30a18bdab27b8bbbfe8f83a31 | [
"Unlicense"
]
| null | null | null | 26.318182 | 172 | 0.532457 | [
[
[
"This file is part of the pyMOR project (http://www.pymor.org).\nCopyright 2013-2017 pyMOR developers and contributors. All rights reserved.\nLicense: BSD 2-Clause License (http://opensource.org/licenses/BSD-2-Clause)",
"_____no_output_____"
]
],
[
[
"# String equation example",
"_____no_output_____"
],
[
"## Analytic problem formulation\n\nWe consider a vibrating string on the segment $[0, 1]$, fixed on both sides, with input $u$ and output $\\tilde{y}$ in the middle:\n$$\n\\begin{align*}\n \\partial_{tt} \\xi(z, t)\n + d \\partial_t \\xi(z, t)\n - k \\partial_{zz} \\xi(z, t)\n & = \\delta(z - \\tfrac{1}{2}) u(t), & 0 < z < 1,\\ t > 0, \\\\\n \\partial_z \\xi(0, t) & = 0, & t > 0, \\\\\n \\partial_z \\xi(1, t) & = 0, & t > 0, \\\\\n \\tilde{y}(t) & = \\xi(1/2, t), & t > 0.\n\\end{align*}\n$$\n\n## Semidiscretized formulation\n\nUsing the finite volume method on the equidistant mesh $0 = z_1 < z_2 < \\ldots < z_{n + 1} = 1$, where $n = 2 n_2 - 1$, we obtain the semidiscretized formulation:\n$$\n\\begin{align*}\n \\ddot{x}_i(t)\n + d \\dot{x}_i(t)\n - k \\frac{x_{i - 1}(t) - 2 x_i(t) + x_{i + 1}(t)}{h^2}\n & = \\frac{1}{h} \\delta_{i, n_2} u(t), & i = 1, 2, 3, \\ldots, n - 1, n, \\\\\n x_0(t) & = 0, \\\\\n x_{n + 1}(t) & = 0, \\\\\n y(t) & = x_{n_2}(t),\n\\end{align*}\n$$\nwhere $h = \\frac{1}{n}$, $x_i(t) \\approx \\int_{z_i}^{z_{i + 1}} \\xi(z, t) \\, \\mathrm{d}z$, and $y(t) \\approx \\tilde{y}(t)$.\n\nSeparating cases $i = 1$ and $i = n$ in the first equation, we find:\n$$\n\\begin{alignat*}{6}\n \\ddot{x}_1(t)\n + d \\dot{x}_1(t)\n &\n && + 2 k n^2 x_1(t)\n && - k n^2 x_2(t)\n && = 0, \\\\\n \\ddot{x}_i(t)\n + d \\dot{x}_i(t)\n & - k n^2 x_{i - 1}(t)\n && + 2 k n^2 x_i(t)\n && - k n^2 x_{i + 1}(t)\n && = n \\delta_{i, n_2} u(t),\n & i = 2, 3, \\ldots, n - 1, \\\\\n \\ddot{x}_n(t)\n + d \\dot{x}_n(t)\n & - k n^2 x_{n - 1}(t)\n && + 2 k n^2 x_n(t)\n &&\n && = 0, \\\\\n &\n &&\n &&\n & y(t)\n & = x_{n_2}(t).\n\\end{alignat*}\n$$",
"_____no_output_____"
],
[
"## Import modules",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport scipy.sparse as sps\nimport matplotlib.pyplot as plt\n\nfrom pymor.core.config import config\nfrom pymor.models.iosys import SecondOrderModel\nfrom pymor.reductors.bt import BTReductor\nfrom pymor.reductors.h2 import IRKAReductor\nfrom pymor.reductors.sobt import (SOBTpReductor, SOBTvReductor, SOBTpvReductor, SOBTvpReductor,\n SOBTfvReductor, SOBTReductor)\nfrom pymor.reductors.sor_irka import SOR_IRKAReductor\n\nfrom pymor.core.logger import set_log_levels\nset_log_levels({'pymor.algorithms.gram_schmidt.gram_schmidt': 'WARNING'})",
"_____no_output_____"
]
],
[
[
"## Assemble $M$, $D$, $K$, $B$, $C_p$",
"_____no_output_____"
]
],
[
[
"n2 = 50\nn = 2 * n2 - 1 # dimension of the system\n\nd = 10 # damping\nk = 0.01 # stiffness\n\nM = sps.eye(n, format='csc')\n\nE = d * sps.eye(n, format='csc')\n\nK = sps.diags([n * [2 * k * n ** 2],\n (n - 1) * [-k * n ** 2],\n (n - 1) * [-k * n ** 2]],\n [0, -1, 1],\n format='csc')\n\nB = np.zeros((n, 1))\nB[n2 - 1, 0] = n\n\nCp = np.zeros((1, n))\nCp[0, n2 - 1] = 1",
"_____no_output_____"
]
],
[
[
"## Second-order system",
"_____no_output_____"
]
],
[
[
"so_sys = SecondOrderModel.from_matrices(M, E, K, B, Cp)\n\nprint(f'order of the model = {so_sys.order}')\nprint(f'number of inputs = {so_sys.input_dim}')\nprint(f'number of outputs = {so_sys.output_dim}')",
"_____no_output_____"
],
[
"poles = so_sys.poles()\nfig, ax = plt.subplots()\nax.plot(poles.real, poles.imag, '.')\nax.set_title('System poles')\nplt.show()",
"_____no_output_____"
],
[
"w = np.logspace(-4, 2, 200)\nfig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the full model')\nplt.show()",
"_____no_output_____"
],
[
"psv = so_sys.psv()\nvsv = so_sys.vsv()\npvsv = so_sys.pvsv()\nvpsv = so_sys.vpsv()\nfig, ax = plt.subplots(2, 2, figsize=(12, 8), sharey=True)\nax[0, 0].semilogy(range(1, len(psv) + 1), psv, '.-')\nax[0, 0].set_title('Position singular values')\nax[0, 1].semilogy(range(1, len(vsv) + 1), vsv, '.-')\nax[0, 1].set_title('Velocity singular values')\nax[1, 0].semilogy(range(1, len(pvsv) + 1), pvsv, '.-')\nax[1, 0].set_title('Position-velocity singular values')\nax[1, 1].semilogy(range(1, len(vpsv) + 1), vpsv, '.-')\nax[1, 1].set_title('Velocity-position singular values')\nplt.show()",
"_____no_output_____"
],
[
"print(f'H_2-norm of the full model: {so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'H_inf-norm of the full model: {so_sys.hinf_norm():e}')\nprint(f'Hankel-norm of the full model: {so_sys.hankel_norm():e}')",
"_____no_output_____"
]
],
[
[
"## Position Second-Order Balanced Truncation (SOBTp)",
"_____no_output_____"
]
],
[
[
"r = 5\nsobtp_reductor = SOBTpReductor(so_sys)\nrom_sobtp = sobtp_reductor.reduce(r)",
"_____no_output_____"
],
[
"poles_rom_sobtp = rom_sobtp.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_sobtp.real, poles_rom_sobtp.imag, '.')\nax.set_title(\"SOBTp reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_sobtp = so_sys - rom_sobtp\nprint(f'SOBTp relative H_2-error: {err_sobtp.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'SOBTp relative H_inf-error: {err_sobtp.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'SOBTp relative Hankel-error: {err_sobtp.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_sobtp.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and SOBTp reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_sobtp.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the SOBTp error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Velocity Second-Order Balanced Truncation (SOBTv)",
"_____no_output_____"
]
],
[
[
"r = 5\nsobtv_reductor = SOBTvReductor(so_sys)\nrom_sobtv = sobtv_reductor.reduce(r)",
"_____no_output_____"
],
[
"poles_rom_sobtv = rom_sobtv.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_sobtv.real, poles_rom_sobtv.imag, '.')\nax.set_title(\"SOBTv reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_sobtv = so_sys - rom_sobtv\nprint(f'SOBTv relative H_2-error: {err_sobtv.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'SOBTv relative H_inf-error: {err_sobtv.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'SOBTv relative Hankel-error: {err_sobtv.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_sobtv.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and SOBTv reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_sobtv.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the SOBTv error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Position-Velocity Second-Order Balanced Truncation (SOBTpv)",
"_____no_output_____"
]
],
[
[
"r = 5\nsobtpv_reductor = SOBTpvReductor(so_sys)\nrom_sobtpv = sobtpv_reductor.reduce(r)",
"_____no_output_____"
],
[
"poles_rom_sobtpv = rom_sobtpv.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_sobtpv.real, poles_rom_sobtpv.imag, '.')\nax.set_title(\"SOBTpv reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_sobtpv = so_sys - rom_sobtpv\nprint(f'SOBTpv relative H_2-error: {err_sobtpv.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'SOBTpv relative H_inf-error: {err_sobtpv.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'SOBTpv relative Hankel-error: {err_sobtpv.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_sobtpv.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and SOBTpv reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_sobtpv.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the SOBTpv error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Velocity-Position Second-Order Balanced Truncation (SOBTvp)",
"_____no_output_____"
]
],
[
[
"r = 5\nsobtvp_reductor = SOBTvpReductor(so_sys)\nrom_sobtvp = sobtvp_reductor.reduce(r)",
"_____no_output_____"
],
[
"poles_rom_sobtvp = rom_sobtvp.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_sobtvp.real, poles_rom_sobtvp.imag, '.')\nax.set_title(\"SOBTvp reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_sobtvp = so_sys - rom_sobtvp\nprint(f'SOBTvp relative H_2-error: {err_sobtvp.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'SOBTvp relative H_inf-error: {err_sobtvp.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'SOBTvp relative Hankel-error: {err_sobtvp.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_sobtvp.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and SOBTvp reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_sobtvp.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the SOBTvp error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Free-Velocity Second-Order Balanced Truncation (SOBTfv)",
"_____no_output_____"
]
],
[
[
"r = 5\nsobtfv_reductor = SOBTfvReductor(so_sys)\nrom_sobtfv = sobtfv_reductor.reduce(r)",
"_____no_output_____"
],
[
"poles_rom_sobtfv = rom_sobtfv.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_sobtfv.real, poles_rom_sobtfv.imag, '.')\nax.set_title(\"SOBTfv reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_sobtfv = so_sys - rom_sobtfv\nprint(f'SOBTfv relative H_2-error: {err_sobtfv.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'SOBTfv relative H_inf-error: {err_sobtfv.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'SOBTfv relative Hankel-error: {err_sobtfv.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_sobtfv.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and SOBTfv reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_sobtfv.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the SOBTfv error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Second-Order Balanced Truncation (SOBT)",
"_____no_output_____"
]
],
[
[
"r = 5\nsobt_reductor = SOBTReductor(so_sys)\nrom_sobt = sobt_reductor.reduce(r)",
"_____no_output_____"
],
[
"poles_rom_sobt = rom_sobt.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_sobt.real, poles_rom_sobt.imag, '.')\nax.set_title(\"SOBT reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_sobt = so_sys - rom_sobt\nprint(f'SOBT relative H_2-error: {err_sobt.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'SOBT relative H_inf-error: {err_sobt.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'SOBT relative Hankel-error: {err_sobt.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_sobt.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and SOBT reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_sobt.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the SOBT error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Balanced Truncation (BT)",
"_____no_output_____"
]
],
[
[
"r = 5\nbt_reductor = BTReductor(so_sys.to_lti())\nrom_bt = bt_reductor.reduce(r)",
"_____no_output_____"
],
[
"poles_rom_bt = rom_bt.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_bt.real, poles_rom_bt.imag, '.')\nax.set_title(\"BT reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_bt = so_sys - rom_bt\nprint(f'BT relative H_2-error: {err_bt.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'BT relative H_inf-error: {err_bt.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'BT relative Hankel-error: {err_bt.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_bt.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and BT reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_bt.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the BT error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Iterative Rational Krylov Algorithm (IRKA)",
"_____no_output_____"
]
],
[
[
"r = 5\nirka_reductor = IRKAReductor(so_sys.to_lti())\nrom_irka = irka_reductor.reduce(r)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.semilogy(irka_reductor.dist, '.-')\nax.set_title('IRKA convergence criterion')\nplt.show()",
"_____no_output_____"
],
[
"poles_rom_irka = rom_irka.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_irka.real, poles_rom_irka.imag, '.')\nax.set_title(\"IRKA reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_irka = so_sys - rom_irka\nprint(f'IRKA relative H_2-error: {err_irka.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'IRKA relative H_inf-error: {err_irka.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'IRKA relative Hankel-error: {err_irka.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_irka.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and IRKA reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_irka.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the IRKA error system')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Second-Order Iterative Rational Krylov Algorithm (SOR-IRKA)",
"_____no_output_____"
]
],
[
[
"r = 5\nsor_irka_reductor = SOR_IRKAReductor(so_sys)\nrom_sor_irka = sor_irka_reductor.reduce(r)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.semilogy(sor_irka_reductor.dist, '.-')\nax.set_title('SOR-IRKA convergence criterion')\nplt.show()",
"_____no_output_____"
],
[
"poles_rom_sor_irka = rom_sor_irka.poles()\nfig, ax = plt.subplots()\nax.plot(poles_rom_sor_irka.real, poles_rom_sor_irka.imag, '.')\nax.set_title(\"SOR-IRKA reduced model's poles\")\nplt.show()",
"_____no_output_____"
],
[
"err_sor_irka = so_sys - rom_sor_irka\nprint(f'SOR-IRKA relative H_2-error: {err_sor_irka.h2_norm() / so_sys.h2_norm():e}')\nif config.HAVE_SLYCOT:\n print(f'SOR-IRKA relative H_inf-error: {err_sor_irka.hinf_norm() / so_sys.hinf_norm():e}')\nprint(f'SOR-IRKA relative Hankel-error: {err_sor_irka.hankel_norm() / so_sys.hankel_norm():e}')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nso_sys.mag_plot(w, ax=ax)\nrom_sor_irka.mag_plot(w, ax=ax, linestyle='dashed')\nax.set_title('Bode plot of the full and SOR-IRKA reduced model')\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nerr_sor_irka.mag_plot(w, ax=ax)\nax.set_title('Bode plot of the SOR-IRKA error system')\nplt.show()",
"_____no_output_____"
]
]
]
| [
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"raw"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6df4e34d80a0254db97d42c8854737700d975d | 408,045 | ipynb | Jupyter Notebook | cifar10_cnn.ipynb | HemantJajoo/Ciphar-10 | db9fbb5011c42af1489e7fc921676a8097947dee | [
"Apache-2.0"
]
| 1 | 2020-08-08T11:46:47.000Z | 2020-08-08T11:46:47.000Z | cifar10_cnn.ipynb | HemantJajoo/Ciphar-10 | db9fbb5011c42af1489e7fc921676a8097947dee | [
"Apache-2.0"
]
| null | null | null | cifar10_cnn.ipynb | HemantJajoo/Ciphar-10 | db9fbb5011c42af1489e7fc921676a8097947dee | [
"Apache-2.0"
]
| null | null | null | 439.703664 | 187,012 | 0.921727 | [
[
[
"## Dataset\n\nThe CIFAR-10 dataset (Canadian Institute For Advanced Research) is a collection of images that are commonly used to train machine learning and computer vision algorithms. It is one of the most widely used datasets for machine learning research. The CIFAR-10 dataset contains 60,000 32x32 color images in 10 different classes. The 10 different classes represent airplanes, cars, birds, cats, deer, dogs, frogs, horses, ships, and trucks. There are 6,000 images of each class.\n\nComputer algorithms for recognizing objects in photos often learn by example. CIFAR-10 is a set of images that can be used to teach a computer how to recognize objects. Since the images in CIFAR-10 are low-resolution (32x32), this dataset can allow researchers to quickly try different algorithms to see what works. Various kinds of convolutional neural networks tend to be the best at recognizing the images in CIFAR-10.",
"_____no_output_____"
],
[
"<table>\n <tr>\n <td class=\"cifar-class-name\">airplane</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/airplane10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">automobile</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/automobile10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">bird</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/bird10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">cat</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/cat10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">deer</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/deer10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">dog</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/dog10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">frog</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/frog10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">horse</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/horse10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">ship</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/ship10.png\" class=\"cifar-sample\" /></td>\n </tr>\n <tr>\n <td class=\"cifar-class-name\">truck</td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck1.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck2.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck3.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck4.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck5.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck6.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck7.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck8.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck9.png\" class=\"cifar-sample\" /></td>\n <td><img src=\"https://www.cs.toronto.edu/~kriz/cifar-10-sample/truck10.png\" class=\"cifar-sample\" /></td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"[Dataset Download](https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz)",
"_____no_output_____"
],
[
"### 1. Load CIFAR-10 Database",
"_____no_output_____"
]
],
[
[
"import keras\nfrom keras.datasets import cifar10\n\n# load the pre-shuffled train and test data\n(x_train, y_train), (x_test, y_test) = cifar10.load_data()",
"C:\\Users\\jmo4cob\\AppData\\Local\\conda\\conda\\envs\\tf\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
]
],
[
[
"### 2. Visualize the First 24 Training Images",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nfig = plt.figure(figsize=(20,5))\nfor i in range(36):\n ax = fig.add_subplot(3, 12, i + 1, xticks=[], yticks=[])\n ax.imshow(np.squeeze(x_train[i]))",
"_____no_output_____"
]
],
[
[
"### 3. Rescale the Images by Dividing Every Pixel in Every Image by 255",
"_____no_output_____"
]
],
[
[
"# rescale [0,255] --> [0,1]\nx_train = x_train.astype('float32')/255\nx_test = x_test.astype('float32')/255",
"_____no_output_____"
]
],
[
[
"### 4. Break Dataset into Training, Testing, and Validation Sets",
"_____no_output_____"
]
],
[
[
"from keras.utils import np_utils\n\n# one-hot encode the labels\nnum_classes = len(np.unique(y_train))\ny_train = keras.utils.to_categorical(y_train, num_classes)\ny_test = keras.utils.to_categorical(y_test, num_classes)\n\n# break training set into training and validation sets\n(x_train, x_valid) = x_train[5000:], x_train[:5000]\n(y_train, y_valid) = y_train[5000:], y_train[:5000]\n\n# print shape of training set\nprint('x_train shape:', x_train.shape)\n\n# print number of training, validation, and test images\nprint(x_train.shape[0], 'train samples')\nprint(x_test.shape[0], 'test samples')\nprint(x_valid.shape[0], 'validation samples')",
"x_train shape: (45000, 32, 32, 3)\n45000 train samples\n10000 test samples\n5000 validation samples\n"
]
],
[
[
"### 5. Define the Model Architecture ",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers import Conv2D, MaxPooling2D, Flatten, Dense, Dropout\n\nmodel = Sequential()\nmodel.add(Conv2D(filters=16, kernel_size=2, padding='same', activation='relu', \n input_shape=(32, 32, 3)))\nmodel.add(MaxPooling2D(pool_size=2))\nmodel.add(Conv2D(filters=32, kernel_size=2, padding='same', activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\nmodel.add(Conv2D(filters=64, kernel_size=2, padding='same', activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\nmodel.add(Dropout(0.3))\nmodel.add(Flatten())\nmodel.add(Dense(500, activation='relu'))\nmodel.add(Dropout(0.4))\nmodel.add(Dense(10, activation='softmax'))\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_1 (Conv2D) (None, 32, 32, 16) 208 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 16, 16, 16) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 16, 16, 32) 2080 \n_________________________________________________________________\nmax_pooling2d_2 (MaxPooling2 (None, 8, 8, 32) 0 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 8, 8, 64) 8256 \n_________________________________________________________________\nmax_pooling2d_3 (MaxPooling2 (None, 4, 4, 64) 0 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 4, 4, 64) 0 \n_________________________________________________________________\nflatten_1 (Flatten) (None, 1024) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 500) 512500 \n_________________________________________________________________\ndropout_2 (Dropout) (None, 500) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 10) 5010 \n=================================================================\nTotal params: 528,054\nTrainable params: 528,054\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"### 6. Compile the Model ",
"_____no_output_____"
]
],
[
[
"# compile the model\nmodel.compile(loss='categorical_crossentropy', optimizer='rmsprop', \n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"### 7. Train the Model ",
"_____no_output_____"
]
],
[
[
"from keras.callbacks import ModelCheckpoint \n\n# train the model\ncheckpointer = ModelCheckpoint(filepath='model.weights.best.hdf5', verbose=1, \n save_best_only=True)\nhist = model.fit(x_train, y_train, batch_size=32, epochs=100,\n validation_data=(x_valid, y_valid), callbacks=[checkpointer], \n verbose=2, shuffle=True)",
"Train on 45000 samples, validate on 5000 samples\nEpoch 1/100\n - 12s - loss: 1.5927 - acc: 0.4211 - val_loss: 1.5076 - val_acc: 0.4614\n\nEpoch 00001: val_loss improved from inf to 1.50758, saving model to model.weights.best.hdf5\nEpoch 2/100\n - 9s - loss: 1.2664 - acc: 0.5472 - val_loss: 1.1566 - val_acc: 0.5934\n\nEpoch 00002: val_loss improved from 1.50758 to 1.15655, saving model to model.weights.best.hdf5\nEpoch 3/100\n - 8s - loss: 1.1441 - acc: 0.5953 - val_loss: 1.1665 - val_acc: 0.5756\n\nEpoch 00003: val_loss did not improve from 1.15655\nEpoch 4/100\n - 8s - loss: 1.0735 - acc: 0.6244 - val_loss: 0.9869 - val_acc: 0.6650\n\nEpoch 00004: val_loss improved from 1.15655 to 0.98695, saving model to model.weights.best.hdf5\nEpoch 5/100\n - 8s - loss: 1.0293 - acc: 0.6407 - val_loss: 1.0058 - val_acc: 0.6450\n\nEpoch 00005: val_loss did not improve from 0.98695\nEpoch 6/100\n - 8s - loss: 1.0118 - acc: 0.6507 - val_loss: 0.9553 - val_acc: 0.6758\n\nEpoch 00006: val_loss improved from 0.98695 to 0.95527, saving model to model.weights.best.hdf5\nEpoch 7/100\n - 8s - loss: 0.9968 - acc: 0.6551 - val_loss: 0.9545 - val_acc: 0.6658\n\nEpoch 00007: val_loss improved from 0.95527 to 0.95449, saving model to model.weights.best.hdf5\nEpoch 8/100\n - 8s - loss: 0.9842 - acc: 0.6612 - val_loss: 0.9239 - val_acc: 0.6760\n\nEpoch 00008: val_loss improved from 0.95449 to 0.92386, saving model to model.weights.best.hdf5\nEpoch 9/100\n - 8s - loss: 0.9804 - acc: 0.6660 - val_loss: 0.9957 - val_acc: 0.6764\n\nEpoch 00009: val_loss did not improve from 0.92386\nEpoch 10/100\n - 8s - loss: 0.9787 - acc: 0.6688 - val_loss: 1.0432 - val_acc: 0.6556\n\nEpoch 00010: val_loss did not improve from 0.92386\nEpoch 11/100\n - 9s - loss: 0.9878 - acc: 0.6650 - val_loss: 1.1084 - val_acc: 0.6382\n\nEpoch 00011: val_loss did not improve from 0.92386\nEpoch 12/100\n - 8s - loss: 0.9890 - acc: 0.6686 - val_loss: 0.9573 - val_acc: 0.6618\n\nEpoch 00012: val_loss did not improve from 0.92386\nEpoch 13/100\n - 8s - loss: 1.0116 - acc: 0.6605 - val_loss: 1.0875 - val_acc: 0.6714\n\nEpoch 00013: val_loss did not improve from 0.92386\nEpoch 14/100\n - 8s - loss: 1.0118 - acc: 0.6601 - val_loss: 1.0911 - val_acc: 0.6582\n\nEpoch 00014: val_loss did not improve from 0.92386\nEpoch 15/100\n - 8s - loss: 1.0280 - acc: 0.6580 - val_loss: 1.2182 - val_acc: 0.6108\n\nEpoch 00015: val_loss did not improve from 0.92386\nEpoch 16/100\n - 8s - loss: 1.0572 - acc: 0.6492 - val_loss: 1.0801 - val_acc: 0.6882\n\nEpoch 00016: val_loss did not improve from 0.92386\nEpoch 17/100\n - 8s - loss: 1.0789 - acc: 0.6417 - val_loss: 1.1487 - val_acc: 0.6406\n\nEpoch 00017: val_loss did not improve from 0.92386\nEpoch 18/100\n - 9s - loss: 1.0975 - acc: 0.6371 - val_loss: 1.0788 - val_acc: 0.6568\n\nEpoch 00018: val_loss did not improve from 0.92386\nEpoch 19/100\n - 8s - loss: 1.1293 - acc: 0.6277 - val_loss: 1.2891 - val_acc: 0.6280\n\nEpoch 00019: val_loss did not improve from 0.92386\nEpoch 20/100\n - 9s - loss: 1.1513 - acc: 0.6214 - val_loss: 1.2949 - val_acc: 0.6268\n\nEpoch 00020: val_loss did not improve from 0.92386\nEpoch 21/100\n - 8s - loss: 1.1849 - acc: 0.6112 - val_loss: 1.0050 - val_acc: 0.6766\n\nEpoch 00021: val_loss did not improve from 0.92386\nEpoch 22/100\n - 9s - loss: 1.2074 - acc: 0.6047 - val_loss: 1.0753 - val_acc: 0.6382\n\nEpoch 00022: val_loss did not improve from 0.92386\nEpoch 23/100\n - 8s - loss: 1.2222 - acc: 0.5981 - val_loss: 1.1969 - val_acc: 0.5846\n\nEpoch 00023: val_loss did not improve from 0.92386\nEpoch 24/100\n - 8s - loss: 1.2347 - acc: 0.5908 - val_loss: 1.2637 - val_acc: 0.5662\n\nEpoch 00024: val_loss did not improve from 0.92386\nEpoch 25/100\n - 8s - loss: 1.2653 - acc: 0.5817 - val_loss: 1.3072 - val_acc: 0.5814\n\nEpoch 00025: val_loss did not improve from 0.92386\nEpoch 26/100\n - 9s - loss: 1.2852 - acc: 0.5816 - val_loss: 1.3219 - val_acc: 0.6002\n\nEpoch 00026: val_loss did not improve from 0.92386\nEpoch 27/100\n - 8s - loss: 1.3083 - acc: 0.5735 - val_loss: 1.2315 - val_acc: 0.5870\n\nEpoch 00027: val_loss did not improve from 0.92386\nEpoch 28/100\n - 8s - loss: 1.3177 - acc: 0.5676 - val_loss: 1.1115 - val_acc: 0.6290\n\nEpoch 00028: val_loss did not improve from 0.92386\nEpoch 29/100\n - 8s - loss: 1.3381 - acc: 0.5576 - val_loss: 1.1374 - val_acc: 0.6234\n\nEpoch 00029: val_loss did not improve from 0.92386\nEpoch 30/100\n - 8s - loss: 1.3626 - acc: 0.5534 - val_loss: 1.1444 - val_acc: 0.6210\n\nEpoch 00030: val_loss did not improve from 0.92386\nEpoch 31/100\n - 8s - loss: 1.3703 - acc: 0.5470 - val_loss: 1.6556 - val_acc: 0.5432\n\nEpoch 00031: val_loss did not improve from 0.92386\nEpoch 32/100\n - 9s - loss: 1.3854 - acc: 0.5416 - val_loss: 1.1467 - val_acc: 0.6266\n\nEpoch 00032: val_loss did not improve from 0.92386\nEpoch 33/100\n - 8s - loss: 1.3979 - acc: 0.5355 - val_loss: 1.4332 - val_acc: 0.5186\n\nEpoch 00033: val_loss did not improve from 0.92386\nEpoch 34/100\n - 9s - loss: 1.4123 - acc: 0.5301 - val_loss: 1.2464 - val_acc: 0.5820\n\nEpoch 00034: val_loss did not improve from 0.92386\nEpoch 35/100\n - 9s - loss: 1.4291 - acc: 0.5260 - val_loss: 1.2079 - val_acc: 0.5958\n\nEpoch 00035: val_loss did not improve from 0.92386\nEpoch 36/100\n - 9s - loss: 1.4427 - acc: 0.5216 - val_loss: 1.5421 - val_acc: 0.5082\n\nEpoch 00036: val_loss did not improve from 0.92386\nEpoch 37/100\n - 8s - loss: 1.4523 - acc: 0.5184 - val_loss: 1.2119 - val_acc: 0.5906\n\nEpoch 00037: val_loss did not improve from 0.92386\nEpoch 38/100\n - 9s - loss: 1.4719 - acc: 0.5110 - val_loss: 1.3906 - val_acc: 0.5132\n\nEpoch 00038: val_loss did not improve from 0.92386\nEpoch 39/100\n - 8s - loss: 1.4741 - acc: 0.5103 - val_loss: 1.6496 - val_acc: 0.5638\n\nEpoch 00039: val_loss did not improve from 0.92386\nEpoch 40/100\n - 9s - loss: 1.4947 - acc: 0.5034 - val_loss: 1.9043 - val_acc: 0.4042\n\nEpoch 00040: val_loss did not improve from 0.92386\nEpoch 41/100\n - 9s - loss: 1.5053 - acc: 0.5010 - val_loss: 2.0755 - val_acc: 0.5002\n\nEpoch 00041: val_loss did not improve from 0.92386\nEpoch 42/100\n - 8s - loss: 1.5173 - acc: 0.4951 - val_loss: 1.4661 - val_acc: 0.4938\n\nEpoch 00042: val_loss did not improve from 0.92386\nEpoch 43/100\n - 8s - loss: 1.5253 - acc: 0.4960 - val_loss: 1.7256 - val_acc: 0.4402\n\nEpoch 00043: val_loss did not improve from 0.92386\nEpoch 44/100\n - 8s - loss: 1.5315 - acc: 0.4886 - val_loss: 1.3954 - val_acc: 0.5342\n\nEpoch 00044: val_loss did not improve from 0.92386\nEpoch 45/100\n - 8s - loss: 1.5362 - acc: 0.4867 - val_loss: 1.5772 - val_acc: 0.4526\n\nEpoch 00045: val_loss did not improve from 0.92386\nEpoch 46/100\n - 8s - loss: 1.5619 - acc: 0.4821 - val_loss: 1.4230 - val_acc: 0.5384\n\nEpoch 00046: val_loss did not improve from 0.92386\nEpoch 47/100\n - 9s - loss: 1.5679 - acc: 0.4742 - val_loss: 1.4077 - val_acc: 0.5062\n\nEpoch 00047: val_loss did not improve from 0.92386\nEpoch 48/100\n - 8s - loss: 1.5813 - acc: 0.4715 - val_loss: 1.2873 - val_acc: 0.5522\n\nEpoch 00048: val_loss did not improve from 0.92386\nEpoch 49/100\n - 9s - loss: 1.6023 - acc: 0.4651 - val_loss: 1.5283 - val_acc: 0.4866\n\nEpoch 00049: val_loss did not improve from 0.92386\nEpoch 50/100\n - 9s - loss: 1.6110 - acc: 0.4608 - val_loss: 1.7062 - val_acc: 0.4572\n\nEpoch 00050: val_loss did not improve from 0.92386\nEpoch 51/100\n - 9s - loss: 1.6281 - acc: 0.4502 - val_loss: 1.5190 - val_acc: 0.4766\n\nEpoch 00051: val_loss did not improve from 0.92386\nEpoch 52/100\n - 8s - loss: 1.6381 - acc: 0.4528 - val_loss: 2.0704 - val_acc: 0.3442\n\nEpoch 00052: val_loss did not improve from 0.92386\nEpoch 53/100\n - 8s - loss: 1.6637 - acc: 0.4432 - val_loss: 1.5559 - val_acc: 0.4486\n\nEpoch 00053: val_loss did not improve from 0.92386\nEpoch 54/100\n - 9s - loss: 1.6576 - acc: 0.4394 - val_loss: 1.4549 - val_acc: 0.5184\n\nEpoch 00054: val_loss did not improve from 0.92386\nEpoch 55/100\n - 9s - loss: 1.6777 - acc: 0.4331 - val_loss: 1.3620 - val_acc: 0.5168\n\nEpoch 00055: val_loss did not improve from 0.92386\nEpoch 56/100\n - 8s - loss: 1.6759 - acc: 0.4381 - val_loss: 1.4226 - val_acc: 0.5024\n\nEpoch 00056: val_loss did not improve from 0.92386\nEpoch 57/100\n - 9s - loss: 1.7010 - acc: 0.4268 - val_loss: 1.7691 - val_acc: 0.3948\n\nEpoch 00057: val_loss did not improve from 0.92386\nEpoch 58/100\n - 9s - loss: 1.7335 - acc: 0.4211 - val_loss: 1.7555 - val_acc: 0.3956\n"
]
],
[
[
"### 8. Load the Model with the Best Validation Accuracy",
"_____no_output_____"
]
],
[
[
"# load the weights that yielded the best validation accuracy\nmodel.load_weights('model.weights.best.hdf5')",
"_____no_output_____"
]
],
[
[
"### 9. Calculate Classification Accuracy on Test Set",
"_____no_output_____"
]
],
[
[
"# evaluate and print test accuracy\nscore = model.evaluate(x_test, y_test, verbose=0)\nprint('\\n', 'Test accuracy:', score[1])",
"\n Test accuracy: 0.6794\n"
]
],
[
[
"### 10. Visualize Some Predictions\n\nThis may give you some insight into why the network is misclassifying certain objects.",
"_____no_output_____"
]
],
[
[
"# get predictions on the test set\ny_hat = model.predict(x_test)\n\n# define text labels (source: https://www.cs.toronto.edu/~kriz/cifar.html)\ncifar10_labels = ['airplane', 'automobile', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']",
"_____no_output_____"
],
[
"# plot a random sample of test images, their predicted labels, and ground truth\nfig = plt.figure(figsize=(20, 8))\nfor i, idx in enumerate(np.random.choice(x_test.shape[0], size=32, replace=False)):\n ax = fig.add_subplot(4, 8, i + 1, xticks=[], yticks=[])\n ax.imshow(np.squeeze(x_test[idx]))\n pred_idx = np.argmax(y_hat[idx])\n true_idx = np.argmax(y_test[idx])\n ax.set_title(\"{} ({})\".format(cifar10_labels[pred_idx], cifar10_labels[true_idx]),\n color=(\"green\" if pred_idx == true_idx else \"red\"))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
cb6dfab944526aed3bfa9db68a6c667eb2bded57 | 2,069 | ipynb | Jupyter Notebook | metod kramera.ipynb | eztwokey/cross.laba3 | 3650f295222395cad67c7c62a5e596b05f4b223f | [
"MIT"
]
| null | null | null | metod kramera.ipynb | eztwokey/cross.laba3 | 3650f295222395cad67c7c62a5e596b05f4b223f | [
"MIT"
]
| null | null | null | metod kramera.ipynb | eztwokey/cross.laba3 | 3650f295222395cad67c7c62a5e596b05f4b223f | [
"MIT"
]
| null | null | null | 16.685484 | 54 | 0.462059 | [
[
[
"# Метод Крамера",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"M = np.array([[3., 2.], [1., -2.]])\nN = np.array([4., -6])",
"_____no_output_____"
]
],
[
[
"### Найдем определитель матрицы системы",
"_____no_output_____"
]
],
[
[
"O = round(np.linalg.det(M), 3)\nO",
"_____no_output_____"
]
],
[
[
"### Найдем вспомогательный определитель матрицы ",
"_____no_output_____"
]
],
[
[
"J = np.array([[12., -7], [4., 12.]])",
"_____no_output_____"
],
[
"O_vs = round(np.linalg.det(J), 3)\nO_vs",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
cb6dfde0c08b4a1a36bac08eaa43ff3e1bf1ae33 | 4,551 | ipynb | Jupyter Notebook | nbs/index.ipynb | adbmd/self_supervised | d87ebd9b4961c7da0efd6073c42782bbc61aaa2e | [
"Apache-2.0"
]
| 1 | 2020-09-22T14:29:07.000Z | 2020-09-22T14:29:07.000Z | nbs/index.ipynb | adbmd/self_supervised | d87ebd9b4961c7da0efd6073c42782bbc61aaa2e | [
"Apache-2.0"
]
| null | null | null | nbs/index.ipynb | adbmd/self_supervised | d87ebd9b4961c7da0efd6073c42782bbc61aaa2e | [
"Apache-2.0"
]
| null | null | null | 29.551948 | 367 | 0.601187 | [
[
[
"# Self Supervised Learning Fastai Extension\n\n> Implementation of popular SOTA self-supervised learning algorithms as Fastai Callbacks.",
"_____no_output_____"
],
[
"You may find documentation [here](https://keremturgutlu.github.io/self_supervised) and github repo [here](https://github.com/keremturgutlu/self_supervised/tree/master/)",
"_____no_output_____"
],
[
"## Install",
"_____no_output_____"
],
[
"`pip install self-supervised`",
"_____no_output_____"
],
[
"## Algorithms",
"_____no_output_____"
],
[
"Here are the list of implemented algorithms:\n\n- [SimCLR](https://arxiv.org/pdf/2002.05709.pdf)\n- [BYOL](https://arxiv.org/pdf/2006.07733.pdf)\n- [SwAV](https://arxiv.org/pdf/2006.09882.pdf)",
"_____no_output_____"
],
[
"## Simple Usage",
"_____no_output_____"
],
[
"```python\nfrom self_supervised.simclr import *\ndls = get_dls(resize, bs)\nmodel = create_simclr_model(arch=xresnet34, pretrained=False)\nlearn = Learner(dls, model, SimCLRLoss(temp=0.1), opt_func=opt_func, cbs=[SimCLR(size=size)])\nlearn.fit_flat_cos(100, 1e-2)\n```",
"_____no_output_____"
],
[
"```python\nfrom self_supervised.byol import *\ndls = get_dls(resize, bs)\nmodel = create_byol_model(arch=xresnet34, pretrained=False)\nlearn = Learner(dls, model, byol_loss, opt_func=opt_func, cbs=[BYOL(size=size, T=0.99)])\nlearn.fit_flat_cos(100, 1e-2)\n```",
"_____no_output_____"
],
[
"```python\nfrom self_supervised.swav import *\ndls = get_dls(resize, bs)\nmodel = create_swav_model(arch=xresnet34, pretrained=False)\nlearn = Learner(dls, model, SWAVLoss(), opt_func=opt_func, cbs=[SWAV(crop_sizes=[size,96], \n num_crops=[2,6],\n min_scales=[0.25,0.2],\n max_scales=[1.0,0.35])])\nlearn.fit_flat_cos(100, 1e-2)\n```",
"_____no_output_____"
],
[
"## ImageWang Benchmarks",
"_____no_output_____"
],
[
"All of the algorithms implemented in this library have been evaluated in [ImageWang Leaderboard](https://github.com/fastai/imagenette#image%E7%BD%91-leaderboard). \n\nIn overall superiority of the algorithms are as follows `SwAV > BYOL > SimCLR` in most of the benchmarks. For details you may inspect the history of [ImageWang Leaderboard](https://github.com/fastai/imagenette#image%E7%BD%91-leaderboard) through github. \n\nIt should be noted that during these experiments no hyperparameter selection/tuning was made beyond using `learn.lr_find()` or making sanity checks over data augmentations by visualizing batches. So, there is still space for improvement and overall rankings of the alogrithms may change based on your setup. Yet, the overall rankings are on par with the papers.",
"_____no_output_____"
],
[
"## Contributing",
"_____no_output_____"
],
[
"Contributions and or requests for new self-supervised algorithms are welcome. This repo will try to keep itself up-to-date with recent SOTA self-supervised algorithms.\n\nBefore raising a PR please create a new branch with name `<self-supervised-algorithm>`. You may refer to previous notebooks before implementing your Callback.\n\nPlease refer to sections `Developers Guide, Abbreviations Guide, and Style Guide` from https://docs.fast.ai/dev-setup and note that same rules apply for this library.",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
cb6e033fceb3e6d4df7ce31017ee055763dfcaf4 | 5,335 | ipynb | Jupyter Notebook | Mongo.ipynb | frburrue/tfm | 1e7d6943d436d45d0f5e95cd8c4e5826753e19ca | [
"MIT"
]
| null | null | null | Mongo.ipynb | frburrue/tfm | 1e7d6943d436d45d0f5e95cd8c4e5826753e19ca | [
"MIT"
]
| null | null | null | Mongo.ipynb | frburrue/tfm | 1e7d6943d436d45d0f5e95cd8c4e5826753e19ca | [
"MIT"
]
| null | null | null | 24.25 | 260 | 0.516214 | [
[
[
"# Dependencias",
"_____no_output_____"
]
],
[
[
"from pymongo import MongoClient\nimport os\nimport copy\n\nclass MongoWrapper:\n\n def __init__(self):\n\n self.client = MongoClient(\n 'mongodb://' + os.getenv('MONGO_USERNAME') + ':' + os.getenv('MONGO_PASSWORD') + \\\n '@' + os.getenv('MONGO', 'localhost:60222')\n )\n self.db = self.client[os.getenv('MONGO_BBDD', 'admin')]\n\n def set_one(self, collection, document):\n\n return self.db[collection].insert_one(document).acknowledged\n\n def get_one(self, collection, query):\n\n return self.db[collection].find_one(query)\n\n def get_many(self, collection, query):\n\n return self.db[collection].find(query)\n\n def __upsert(self, collection, query, document, options):\n\n return self.db[collection].update(query, document, **options).get('ok') == 1.0\n\n def upsert_one(self, collection, query, document):\n\n return self.__upsert(collection, query, document, {\"upsert\": True, \"multi\": False})\n\n def upsert_many(self, collection, query, document):\n\n return self.__upsert(collection, query, document, {\"upsert\": True, \"multi\": True})\n\n def delete_one(self, collection, query):\n\n self.db[collection].delete_one(query)\n\n def delete_many(self, collection, query):\n\n self.db[collection].delete_many(query)",
"_____no_output_____"
]
],
[
[
"# Recopilación de información de distintos canales",
"_____no_output_____"
]
],
[
[
"MONGO_CLIENT = MongoWrapper()\nUSERS_COLLECTION = 'users'\nDATA_COLLECTION = 'nodered'\nMQTT_COLLECTION = 'mqtt'\nTWTITTER_COLLECTION = 'twitter'",
"_____no_output_____"
],
[
"usernames = {user['user']: user['id'] for user in [copy.deepcopy(user) for user in MONGO_CLIENT.get_many(USERS_COLLECTION, {})]}",
"_____no_output_____"
],
[
"usernames",
"_____no_output_____"
],
[
"id_user ={'id': usernames['CALZADOS M&N'], 'user': 'CALZADOS M&N'}",
"_____no_output_____"
],
[
"id_user",
"_____no_output_____"
],
[
"for item in MONGO_CLIENT.get_many(MQTT_COLLECTION, {'mqttId': id_user['id']}):\n print(item)",
"{'_id': ObjectId('5fdb928f4ed3af001076a8b0'), 'mqttId': 265790648, 'content': {'ambiance': 22, 'ocupation': 3}, 'date': 1608225423}\n"
],
[
"for item in MONGO_CLIENT.get_many(TWTITTER_COLLECTION, {'tweetId': id_user['user'].replace(' ', '_')}):\n print(item)",
"{'_id': ObjectId('5fdb9d004ed3af001076a8b1'), 'content': '#fba_iot_tfm_2020_bot #CALZADOS_M&N Tax efficiency makes a big difference. Let’s talk before year-end.\\n https://t.co/LVVsG1lVmi', 'tweetId': 'CALZADOS_M&N', 'show': True, 'date': 1608228096}\n"
],
[
"id_user['user'].replace(' ', '_')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6e038a785e2b9f3e46793524fab55af8aebda4 | 44,132 | ipynb | Jupyter Notebook | Assignment10_BENITEZ_FERNANDEZ.ipynb | itsCiandrei/LinearAlgebra_2ndSem | a76ad0f3560de812967c5ff54410bbb0c7b8f24c | [
"Apache-2.0"
]
| null | null | null | Assignment10_BENITEZ_FERNANDEZ.ipynb | itsCiandrei/LinearAlgebra_2ndSem | a76ad0f3560de812967c5ff54410bbb0c7b8f24c | [
"Apache-2.0"
]
| null | null | null | Assignment10_BENITEZ_FERNANDEZ.ipynb | itsCiandrei/LinearAlgebra_2ndSem | a76ad0f3560de812967c5ff54410bbb0c7b8f24c | [
"Apache-2.0"
]
| null | null | null | 93.302326 | 14,997 | 0.821445 | [
[
[
"<a href=\"https://colab.research.google.com/github/itsCiandrei/LinearAlgebra_2ndSem/blob/main/Assignment10_BENITEZ_FERNANDEZ.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Linear Algebra for ECE\n## Laboratory 10 : Linear Combination and Vector Spaces",
"_____no_output_____"
],
[
"Now that you have a fundamental knowledge about linear combination, we'll try to visualize it using scientific programming.",
"_____no_output_____"
],
[
"### Objectives\nAt the end of this activity you will be able to:\n1. Be familiar with representing linear combinations in the 2-dimensional plane.\n2. Visualize spans using vector fields in Python.\n3. Perform vector fields operations using scientific programming.",
"_____no_output_____"
],
[
"## Discussion",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"##Linear Combination",
"_____no_output_____"
],
[
"Linear combination is the addition of two or more vectors. In order to properly understand the combination of vectors, it is necessary to to plot the involved vectors in the computation.\n\n$$R = \\begin{bmatrix} 1\\\\2 \\\\\\end{bmatrix} , P = \\begin{bmatrix} -2\\\\3 \\\\\\end{bmatrix} $$",
"_____no_output_____"
]
],
[
[
"vectR = np.array([1,2])\nvectP = np.array([-2,3])",
"_____no_output_____"
],
[
"vectR",
"_____no_output_____"
],
[
"vectP",
"_____no_output_____"
]
],
[
[
"## Span of single vectors",
"_____no_output_____"
],
[
"A vector of x and y is multiplied to three constants. The first one being the start of the point plot. While the second constant is where the plot will stop. Lastly, the last constant will be multiplied to the vector and serve increments or the point plots in the graph. The constants are multiplied to the vector using “np.arrange (c1,c2,c3)”. In addition, plt.scatter determines the plot as well as the constant multiplied to the vector. The [0] in the plt.scatter serves as the X value while [1] is for the Y value.",
"_____no_output_____"
],
[
"$$R = c\\cdot \\begin{bmatrix} 1\\\\2 \\\\\\end{bmatrix} $$",
"_____no_output_____"
]
],
[
[
"vectR = np.array([1,2])",
"_____no_output_____"
],
[
"c = np.arange(-5,5,.5)\n\nplt.scatter(c*vectR[0],c*vectR[1])\n\nplt.xlim(-10,10)\nplt.ylim(-10,10)\nplt.axhline(y=0, color='k')\nplt.axvline(x=0, color='k')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"$$P = c\\cdot \\begin{bmatrix} -2\\\\3 \\\\\\end{bmatrix} $$",
"_____no_output_____"
]
],
[
[
"vectP = np.array([-2,3])",
"_____no_output_____"
],
[
"c = np.arange(-10,10,.75)\n\nplt.scatter(c*vectP[0],c*vectP[1])\n\nplt.xlim(-31,31)\nplt.ylim(-31,31)\nplt.axhline(y=0, color='k')\nplt.axvline(x=0, color='k')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"##Span of a linear combination of vectors",
"_____no_output_____"
],
[
"Plotting the span of a linear combination of vectors will resulted to a two-dimensional plane. This span of a linear combination is the set of all possible combinations that can be formed from the elements of the given vectors multiplying to a set of scalars.",
"_____no_output_____"
],
[
"$$S = \\begin{Bmatrix} c_1 \\cdot\\begin{bmatrix} -3\\\\3 \\\\\\end{bmatrix}, \nc_2 \\cdot \\begin{bmatrix} 4\\\\2 \\\\\\end{bmatrix}\\end{Bmatrix} $$",
"_____no_output_____"
]
],
[
[
"vectRJ = np.array([-3,3])\nvectJP = np.array([4,2])\n\nR = np.arange(-5,5,1)\n\nc1, c2 = np.meshgrid(R,R)\n\nvectR = vectRJ + vectJP\nspanRx = c1*vectRJ[0] + c2*vectJP[0]\nspanRy = c1*vectRJ[1] + c2*vectJP[1]\nplt.scatter(R*vectRJ[0],R*vectRJ[1])\nplt.scatter(R*vectJP[0],R*vectJP[1])\nplt.scatter(spanRx,spanRy, s=10, alpha=1)\n\nplt.axhline(y=0, color='k')\nplt.axvline(x=0, color='k')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"$$S = \\begin{Bmatrix} c_1 \\cdot\\begin{bmatrix} -2\\\\3 \\\\\\end{bmatrix}, \nc_2 \\cdot \\begin{bmatrix} 1\\\\-4 \\\\\\end{bmatrix}\\end{Bmatrix} $$",
"_____no_output_____"
]
],
[
[
"vectA = np.array([-2,3])\nvectB = np.array([1,-4])\n\nR = np.arange(-5,5,1)\n\nc1, c2 = np.meshgrid(R,R)\n\nvectR = vectA + vectB\nspanRx = c1*vectA[0] + c2*vectB[0]\nspanRy = c1*vectA[1] + c2*vectB[1]\nplt.scatter(R*vectA[0],R*vectA[1])\nplt.scatter(R*vectB[0],R*vectB[1])\nplt.scatter(spanRx,spanRy, s=10, alpha=1)\n\nplt.axhline(y=0, color='k')\nplt.axvline(x=0, color='k')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Task 1",
"_____no_output_____"
]
],
[
[
"vectQ = np.array([4,2])\nvectW= np.array([8,6])\n\nR = np.arange(-5,5,0.75)\n\nc1, c2 = np.meshgrid(R,R)\n\nvectR = vectQ + vectW\nspanRx = c1*vectQ[0] + c2*vectW[0]\nspanRy = c1*vectQ[1] + c2*vectW[1]\nplt.scatter(R*vectQ[0],R*vectQ[1])\nplt.scatter(R*vectW[0],R*vectW[1])\nplt.scatter(spanRx,spanRy, s=10, alpha=.5)\n\nplt.axhline(y=0, color='k')\nplt.axvline(x=0, color='k')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"$$S = \\begin{Bmatrix} c_1 \\cdot\\begin{bmatrix} 4\\\\2 \\\\\\end{bmatrix}, \nc_2 \\cdot \\begin{bmatrix} 8\\\\6 \\\\\\end{bmatrix}\\end{Bmatrix} $$",
"_____no_output_____"
],
[
"$$ Q = 4\\hat{x} + 2\\hat{y} \\\\\nW = 8\\hat{x} + 6\\hat{y}$$",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
cb6e06d70a34571fb6ee1569a6d7036d0f3589e8 | 46,418 | ipynb | Jupyter Notebook | doc/ipython_notebooks_src/dev-computing-the-norm-from-dolfin.ipynb | davidcortesortuno/finmag | 9ac0268d2c0e45faf1284cee52a73525aa589e2b | [
"BSL-1.0"
]
| 10 | 2018-03-24T07:43:17.000Z | 2022-03-26T10:42:27.000Z | doc/ipython_notebooks_src/dev-computing-the-norm-from-dolfin.ipynb | davidcortesortuno/finmag | 9ac0268d2c0e45faf1284cee52a73525aa589e2b | [
"BSL-1.0"
]
| 21 | 2018-03-26T15:08:53.000Z | 2021-07-10T16:11:14.000Z | doc/ipython_notebooks_src/dev-computing-the-norm-from-dolfin.ipynb | davidcortesortuno/finmag | 9ac0268d2c0e45faf1284cee52a73525aa589e2b | [
"BSL-1.0"
]
| 7 | 2018-04-09T11:50:48.000Z | 2021-06-10T09:23:25.000Z | 40.646235 | 1,643 | 0.580486 | [
[
[
"# Compute norm from function space",
"_____no_output_____"
]
],
[
[
"from dolfin import * \nimport dolfin as df\nimport numpy as np\n\nimport logging\ndf.set_log_level(logging.INFO)\ndf.set_log_level(WARNING)\n\nmesh = RectangleMesh(0, 0, 1, 1, 10, 10) \n#mesh = Mesh(Rectangle(-10, -10, 10, 10) - Circle(0, 0, 0.1), 10) \nV = FunctionSpace(mesh, \"CG\", 1) \nW = VectorFunctionSpace(mesh, \"CG\", 1) \nw = interpolate(Expression([\"2\", \"1\"]), W)",
"DEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\n"
],
[
"%%timeit\nnorm_squared = 0\nfor i in range(2):\n norm_squared += w[i] ** 2\nnorm = norm_squared ** 0.5\n\nnorm = df.project(norm, V)\n#norm = df.interpolate(norm, V)\n",
"DEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\n"
]
],
[
[
"This next bit is fast, but doesn't compute the norm ;-|",
"_____no_output_____"
]
],
[
[
"%%timeit\nn = interpolate(Expression(\"sqrt(pow(x[0], 2) + pow(x[1], 2))\"), V)",
"100 loops, best of 3: 8.77 ms per loop\n"
]
],
[
[
"# Compute norm via dolfin vector norm function",
"_____no_output_____"
]
],
[
[
"vector = w.vector()",
"_____no_output_____"
],
[
"%%timeit\nnorm2 = vector.norm('l2')\n",
"1000000 loops, best of 3: 646 ns per loop\n"
],
[
"print(norm2)",
"35.3553390593\n"
]
],
[
[
"Okay, the method above is not suitable: it computes the norm of the whole vector, not the norm for the 2d vector at each node.",
"_____no_output_____"
],
[
"# Compute the norm using dolfin generic vector functions",
"_____no_output_____"
]
],
[
[
"mesh = RectangleMesh(0, 0, 1, 1, 10, 10) \nV = FunctionSpace(mesh, \"CG\", 1) \nW = VectorFunctionSpace(mesh, \"CG\", 1) \nw = interpolate(Expression([\"2\", \"1\"]), W)\nnorm = Function(V)\nnorm_vec = norm.vector()\nprint(\"Shape of w = {}\".format(w.vector().get_local().shape))\nprint(\"Shape of norm = {}\".format(norm.vector().get_local().shape))\n\n",
"DEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\n"
]
],
[
[
"Compute the norm-squared in dolfin vector:",
"_____no_output_____"
]
],
[
[
"%%timeit \nwx, wy = w.split(deepcopy=True)\nwnorm2 = (wx.vector() * wx.vector() + wy.vector() * wy.vector()) \n#At this point, I don't know how to compute the square root of wnorm2 (without numpy or other non-dolfin-generic-vector code).\nwnorm = np.sqrt(wnorm2.array())\nnorm_vec.set_local(wnorm)",
"100 loops, best of 3: 3 ms per loop\n"
]
],
[
[
"## plot some results",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport matplotlib.tri as tri\n",
"_____no_output_____"
],
[
"coords = mesh.coordinates()\nx = coords[:,0]\ny = coords[:,1]\ntriang = tri.Triangulation(x, y)\nz = norm2.vector().array()\nplt.tripcolor(triang, z, shading='flat', cmap=plt.cm.rainbow)\nplt.colorbar()",
"_____no_output_____"
],
[
"coords[:,0]",
"_____no_output_____"
]
],
[
[
"# Wacky stuff from webpage (works on the coordinate, not the field value)\nhttp://fenicsproject.org/qa/3693/parallel-vector-operations-something-akin-to-celliterator",
"_____no_output_____"
]
],
[
[
"from dolfin import *\nimport numpy as np\nimport math\n\nmesh = RectangleMesh(-1, -1, 1, 1, 10, 10)\nV = FunctionSpace(mesh, 'CG', 1)\n\nu = Function(V)\nuvec = u.vector()\n\ndofmap = V.dofmap()\ndof_x = dofmap.tabulate_all_coordinates(mesh).reshape((-1, 2))\nfirst_dof, last_dof = dofmap.ownership_range() # U.local_size()\n\n#rank = MPI.process_number()\nnew_values = np.zeros(last_dof - first_dof)\nfor i in range(len(new_values)):\n x, y = dof_x[i]\n new_values[i] = math.sqrt(x **2 + y **2)\nuvec.set_local(new_values)\nuvec.apply('insert')\n\n#plot(u, title=str(rank))\n#interactive()",
"DEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\n"
],
[
"dof_x[0]",
"_____no_output_____"
],
[
"mesh.coordinates()[0]",
"_____no_output_____"
]
],
[
[
"## Wacky stuff from http://fenicsproject.org/qa/3532/avoiding-assembly-vector-operations-scalar-vector-spaces",
"_____no_output_____"
]
],
[
[
"from dolfin import *\n\nmesh = RectangleMesh(0.0, 0.0, 1.0, 1.0, 10, 10)\nV = FunctionSpace(mesh, \"Lagrange\", 1)\nV_vec = VectorFunctionSpace(mesh, \"Lagrange\", 1)\nW = V_vec\n\nc = project(Expression('1.1'), V)\n\nv = as_vector((1,2))\n\nd = project(c*v,V_vec)",
"DEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nINFO:UFL:Adjusting missing element domain to <Domain built from <triangle cell in 2D> with label dolfin_mesh_with_id_69155>.\nINFO:UFL:Adjusting missing element degree to 1\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\n"
],
[
"d.vector().array()",
"_____no_output_____"
],
[
"W = VectorFunctionSpace(mesh, \"CG\", 1) \nw = interpolate(Expression([\"2\", \"1\"]), W)\n",
"DEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\n"
],
[
"%%timeit\n#dd = w #Function(V_vec)\ndofs0 = V_vec.sub(0).dofmap().dofs() # indices of x-components\ndofs1 = V_vec.sub(1).dofmap().dofs() # indices of y-components\nnorm = Function(V)\nnorm.vector()[:] = np.sqrt(w.vector()[dofs0] * w.vector()[dofs0] + w.vector()[dofs1] * w.vector()[dofs1])",
"100 loops, best of 3: 3.23 ms per loop\n"
],
[
"norm = Function(V)\n",
"_____no_output_____"
],
[
"\n%%timeit\nnorm.vector()[:] = np.sqrt(w.vector()[dofs0] * w.vector()[dofs0] + w.vector()[dofs1] * w.vector()[dofs1])",
"100 loops, best of 3: 2.72 ms per loop\n"
],
[
"norm.vector().array()",
"_____no_output_____"
]
],
[
[
"# Done a number of tests. Implement one or two versions as funcions",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef value_dim(w):\n if isinstance(w.function_space(), df.FunctionSpace):\n # Scalar field.\n return 1\n else:\n # value_shape() returns a tuple (N,) and int is required.\n return w.function_space().ufl_element().value_shape()[0]\n\n\ndef compute_pointwise_norm(w, target=None, method=1):\n \"\"\"Given a function vectior function w, compute the norm at each vertex, and store in scalar function target.\n If target is given (a scalar dolfin Function), then store the result in there, and return reference to it.\n If traget is not given, create the object and return reference to it.\n Method allows to choose which method we use.\n \"\"\"\n if not target:\n raise NotImplementeError(\"This is missing - could cerate a df.Function(V) here\")\n\n dim = value_dim(w)\n assert dim in [3], \"Only implemented for 2d and 3d vector field\" \n\n if method == 1:\n wx, wy, wz = w.split(deepcopy=True)\n wnorm = np.sqrt(wx.vector() * wx.vector() + wy.vector() * wy.vector() + wz.vector() * wz.vector()) \n target.vector().set_local(wnorm)\n elif method == 2:\n V_vec = w.function_space()\n dofs0 = V_vec.sub(0).dofmap().dofs() # indices of x-components\n dofs1 = V_vec.sub(1).dofmap().dofs() # indices of y-components\n dofs2 = V_vec.sub(2).dofmap().dofs() # indices of z-components\n\n target.vector()[:] = np.sqrt(w.vector()[dofs0] * w.vector()[dofs0] +\\\n w.vector()[dofs1] * w.vector()[dofs1] +\\\n w.vector()[dofs2] * w.vector()[dofs2])\n else:\n raise NotImplementedError(\"method {} unknown\".format(method))",
"_____no_output_____"
],
[
"import dolfin as df\n\ndef create_test_system(nx, ny=None):\n if not ny:\n ny = nx\n nz = ny\n mesh = df.BoxMesh(0, 0, 0, 1, 1, 1, nx, ny, nz) \n V = df.FunctionSpace(mesh, \"CG\", 1) \n W = df.VectorFunctionSpace(mesh, \"CG\", 1) \n w = df.interpolate(Expression([\"2\", \"1\", \"2\"]), W)\n target = df.Function(V)\n return w, mesh, V, W, target",
"_____no_output_____"
],
[
"w, mesh, V, W, norm = create_test_system(5)",
"DEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\nDEBUG:UFL:No integrals left after transformation, returning empty form.\nDEBUG:FFC:Reusing form from cache.\n"
],
[
"%timeit compute_pointwise_norm(w, norm, method=1)\nassert norm.vector().array()[0] == np.sqrt(2*2 + 1 + 2*2)",
"100 loops, best of 3: 10.2 ms per loop\n"
],
[
"assert norm.vector().array()[0] == np.sqrt(2*2 + 1 + 2*2)",
"_____no_output_____"
],
[
"%timeit compute_pointwise_norm(w, norm, method=2)\nassert norm.vector().array()[0] == np.sqrt(2*2 + 1 + 2*2)",
"10 loops, best of 3: 10.9 ms per loop\n"
],
[
"compute_pointwise_norm(w, norm, method=1)\nnorm.vector().array()[0]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6e072fa276d7666e90eaa08d68d71799c940c5 | 254,339 | ipynb | Jupyter Notebook | notebooks/Examples.ipynb | UriEdenLab/time_rescale | 32cbeb920185f1e5ac8ef28d9b26974005155fae | [
"MIT"
]
| 1 | 2017-12-11T19:28:15.000Z | 2017-12-11T19:28:15.000Z | notebooks/Examples.ipynb | UriEdenLab/time_rescale | 32cbeb920185f1e5ac8ef28d9b26974005155fae | [
"MIT"
]
| 2 | 2017-10-11T16:00:59.000Z | 2017-10-11T16:01:10.000Z | notebooks/Examples.ipynb | UriEdenLab/time_rescale | 32cbeb920185f1e5ac8ef28d9b26974005155fae | [
"MIT"
]
| 3 | 2018-05-22T16:48:00.000Z | 2021-09-09T16:14:41.000Z | 963.405303 | 79,668 | 0.953747 | [
[
[
"## Create Data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom patsy import dmatrix\nfrom statsmodels.api import GLM, families\n\ndef simulate_poisson_process(rate, sampling_frequency):\n return np.random.poisson(rate / sampling_frequency)\n\ndef plot_model_vs_true(time, spike_train, firing_rate, conditional_intensity, sampling_frequency):\n fig, axes = plt.subplots(2, 1, figsize=(12, 6), sharex=True, constrained_layout=True)\n\n s, t = np.nonzero(spike_train)\n axes[0].scatter(np.unique(time)[s], t, s=1, color='black')\n axes[0].set_ylabel('Trials')\n axes[0].set_title('Simulated Spikes')\n axes[0].set_xlim((0, 1))\n\n axes[1].plot(np.unique(time), firing_rate[:, 0],\n linestyle='--', color='black',\n linewidth=4, label='True Rate')\n axes[1].plot(time.ravel(), conditional_intensity * sampling_frequency,\n linewidth=4, label='model conditional intensity')\n axes[1].set_xlabel('Time')\n axes[1].set_ylabel('Firing Rate (Hz)')\n axes[1].set_title('True Rate vs. Model')\n axes[1].set_ylim((0, 15))\n plt.legend()\n\nn_time, n_trials = 1500, 1000\nsampling_frequency = 1500\n\n# Firing rate starts at 5 Hz and switches to 10 Hz\nfiring_rate = np.ones((n_time, n_trials)) * 10\nfiring_rate[:n_time // 2, :] = 5\nspike_train = simulate_poisson_process(\n firing_rate, sampling_frequency)\ntime = (np.arange(0, n_time)[:, np.newaxis] / sampling_frequency *\n np.ones((1, n_trials)))\ntrial_id = (np.arange(n_trials)[np.newaxis, :]\n * np.ones((n_time, 1)))",
"_____no_output_____"
]
],
[
[
"## Good Fit",
"_____no_output_____"
]
],
[
[
"# Fit a spline model to the firing rate\ndesign_matrix = dmatrix('bs(time, df=5)', dict(time=time.ravel()))\nfit = GLM(spike_train.ravel(), design_matrix,\n family=families.Poisson()).fit()\nconditional_intensity = fit.mu\n\nplot_model_vs_true(time, spike_train, firing_rate, conditional_intensity, sampling_frequency)\nplt.savefig('simulated_spikes_model.png')",
"_____no_output_____"
],
[
"from time_rescale import TimeRescaling\n\nconditional_intensity = fit.mu\nrescaled = TimeRescaling(conditional_intensity,\n spike_train.ravel(),\n trial_id.ravel())\n\nfig, axes = plt.subplots(1, 2, figsize=(12, 6))\nrescaled.plot_ks(ax=axes[0])\nrescaled.plot_rescaled_ISI_autocorrelation(ax=axes[1])\n\nplt.savefig('time_rescaling_ks_autocorrelation.png')",
"_____no_output_____"
]
],
[
[
"### Adjust for short trials",
"_____no_output_____"
]
],
[
[
"rescaled_adjusted = TimeRescaling(conditional_intensity,\n spike_train.ravel(),\n trial_id.ravel(),\n adjust_for_short_trials=True)\nfig, axes = plt.subplots(1, 2, figsize=(12, 6))\nrescaled_adjusted.plot_ks(ax=axes[0])\nrescaled_adjusted.plot_rescaled_ISI_autocorrelation(ax=axes[1])\n\nplt.savefig('time_rescaling_ks_autocorrelation_adjusted.png')",
"_____no_output_____"
]
],
[
[
"## Bad Fit",
"_____no_output_____"
]
],
[
[
"constant_fit = GLM(spike_train.ravel(),\n np.ones_like(spike_train.ravel()),\n family=families.Poisson()).fit()\n\nconditional_intensity = constant_fit.mu\n\nplot_model_vs_true(time, spike_train, firing_rate, conditional_intensity, sampling_frequency)\nplt.savefig('constant_model_fit.png')",
"_____no_output_____"
],
[
"bad_rescaled = TimeRescaling(constant_fit.mu,\n spike_train.ravel(),\n trial_id.ravel(),\n adjust_for_short_trials=True)\nfig, axes = plt.subplots(1, 2, figsize=(12, 6))\nbad_rescaled.plot_ks(ax=axes[0], scatter_kwargs=dict(s=10))\naxes[0].set_title('KS Plot')\nbad_rescaled.plot_rescaled_ISI_autocorrelation(ax=axes[1], scatter_kwargs=dict(s=10))\naxes[1].set_title('Autocorrelation');\nplt.savefig('time_rescaling_ks_autocorrelation_bad_fit.png')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
cb6e131b15b567bb8451006db6a07e2e5b647262 | 26,335 | ipynb | Jupyter Notebook | appendix/Preprocessing.ipynb | TobyTarrozaDXC/IRS_copy | 91be50f572a3cc293169f32364ecd6b04767a5de | [
"MIT"
]
| null | null | null | appendix/Preprocessing.ipynb | TobyTarrozaDXC/IRS_copy | 91be50f572a3cc293169f32364ecd6b04767a5de | [
"MIT"
]
| null | null | null | appendix/Preprocessing.ipynb | TobyTarrozaDXC/IRS_copy | 91be50f572a3cc293169f32364ecd6b04767a5de | [
"MIT"
]
| 1 | 2020-08-27T19:20:34.000Z | 2020-08-27T19:20:34.000Z | 43.964942 | 1,263 | 0.390089 | [
[
[
"import pandas as pd \nimport numpy as np \n\nmovies = pd.read_csv('IMDb movies.csv')",
"_____no_output_____"
],
[
"movies",
"_____no_output_____"
],
[
"movies_trimmed = movies[['imdb_title_id', 'original_title', 'year', 'genre', 'duration', 'country', \n 'language', 'director', 'writer', 'production_company', 'actors', 'description', 'avg_vote',]\n ].dropna(subset=['writer','year', 'duration', 'avg_vote'])",
"_____no_output_____"
],
[
"movies_trimmed.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 79780 entries, 0 to 81272\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 imdb_title_id 79780 non-null object \n 1 original_title 79780 non-null object \n 2 year 79780 non-null int64 \n 3 genre 79780 non-null object \n 4 duration 79780 non-null int64 \n 5 country 79750 non-null object \n 6 language 79060 non-null object \n 7 director 79754 non-null object \n 8 writer 79780 non-null object \n 9 production_company 75895 non-null object \n 10 actors 79750 non-null object \n 11 description 77452 non-null object \n 12 avg_vote 79780 non-null float64\ndtypes: float64(1), int64(2), object(10)\nmemory usage: 8.5+ MB\n"
],
[
"print(movies_trimmed[\"year\"].min(), movies_trimmed[\"year\"].max())\nprint(movies_trimmed[\"duration\"].min(), movies_trimmed[\"duration\"].max())\nprint(movies_trimmed[\"avg_vote\"].min(), movies_trimmed[\"avg_vote\"].max())\ngenre_list = movies_trimmed[\"genre\"].unique()\ngenre_unique = [ x for x in genre_list if \" \" not in x ]\nprint(genre_unique)",
"1906 2019\n40 3360\n1.0 10.0\n['Drama', 'Western', 'Comedy', 'Horror', 'Mystery', 'Fantasy', 'Adventure', 'Romance', 'Sci-Fi', 'Crime', 'Musical', 'Thriller', 'Music', 'Biography', 'Action', 'History', 'War', 'Family', 'Animation', 'Sport']\n"
],
[
"country_list = movies_trimmed[\"country\"].unique()\ncountry_unique = [ x for x in country_list if \" \" not in x ]\nprint(country_unique)",
"['USA', 'Italy', 'Germany', 'Denmark', 'France', 'Belgium', 'Hungary', 'Russia', 'Mexico', 'Sweden', 'Australia', 'Japan', 'UK', 'Austria', 'Spain', 'Czechoslovakia', 'India', 'Brazil', 'Portugal', 'Turkey', 'Netherlands', 'Finland', 'Norway', 'Poland', 'Switzerland', 'Argentina', 'Romania', 'Canada', 'China', 'Yugoslavia', 'Greece', 'Egypt', 'Israel', 'Ireland', 'Philippines', 'Cuba', 'Bulgaria', 'Lebanon', 'Bolivia', 'Chile', 'Iran', 'Croatia', 'Peru', 'Taiwan', 'Senegal', 'Syria', 'Jamaica', 'Algeria', 'Ethiopia', 'Venezuela', 'Mali', 'Indonesia', 'Vietnam', 'Iceland', 'Colombia', 'Tunisia', 'Gibraltar', 'Albania', 'Latvia', 'Ukraine', 'Kazakhstan', 'Estonia', 'Slovakia', 'Bangladesh', 'Georgia', 'Singapore', 'Slovenia', 'Thailand', 'Kuwait', 'Lithuania', 'Ecuador', 'Liechtenstein', 'Pakistan', 'Serbia', 'Uruguay', 'Moldova', 'Malta', 'Nepal', 'Malaysia', 'Armenia', 'Luxembourg', 'Bhutan', 'Iraq', 'Bahamas', 'Jordan', 'Morocco', 'Kosovo', 'Azerbaijan', 'Greenland', 'Palestine', 'Honduras', 'Uganda', 'Guatemala', 'Cyprus', 'Korea', 'Nigeria', 'Mongolia', 'Kyrgyzstan', 'Brunei', 'Panama', 'Yemen', 'Myanmar', 'Paraguay', 'Cambodia', 'Montenegro']\n"
],
[
"language_list = movies_trimmed[\"language\"].unique()\nlanguage_unique = [ x for x in language_list if \" \" not in x ]\nprint(language_unique)",
"['English', 'Italian', 'German', 'Danish', 'French', 'Hungarian', 'Russian', 'Spanish', 'Swedish', 'None', 'Japanese', 'Czech', 'Portuguese', 'Ukrainian', 'Turkish', 'Yiddish', 'Dutch', 'Finnish', 'Norwegian', 'Polish', 'Romanian', 'Hindi', 'Slovenian', 'Mandarin', 'Arabic', 'Serbo-Croatian', 'Bengali', 'Albanian', 'Greek', 'Croatian', 'Urdu', 'Hebrew', 'Sinhalese', 'Esperanto', 'Korean', 'Persian', 'Slovak', 'Estonian', 'Bulgarian', 'Georgian', 'Cantonese', 'Latin', 'Kannada', 'Armenian', 'Bambara', 'Indonesian', 'Vietnamese', 'Irish', 'Malayalam', 'Icelandic', 'Serbian', 'Tamil', 'Saami', 'Bosnian', 'More', 'Latvian', 'Kazakh', 'Haitian', 'Tajik', 'Khmer', 'Wolof', 'Catalan', 'Mongolian', 'Thai', 'Afrikaans', 'Telugu', 'Marathi', 'Kirghiz', 'Macedonian', 'Chinese', 'Tagalog', 'Tibetan', 'Lithuanian', 'Punjabi', 'Frisian', 'Maya', 'Basque', 'Malay', 'Inuktitut', 'Kabuverdianu', 'Kurdish', 'Nepali', 'Dari', 'Dzongkha', 'Zulu', 'Luxembourgish', 'Guarani', 'Tarahumara', 'Flemish', 'Maltese', 'Kinyarwanda', 'Gujarati', 'Tigrigna', 'Filipino', 'Pushto', 'Azerbaijani', 'Swahili', 'Samoan', 'Pular', 'Lao', 'Amharic', 'Aromanian', 'Maori', 'Burmese', 'Gallegan', 'Welsh', 'Aboriginal', 'Assamese', 'Rhaetian', 'Yakut', 'Aymara', 'Neapolitan']\n"
],
[
"movies_trimmed[:10000].to_csv('movies_trimmed1.csv', index=False)\nmovies_trimmed[10000:20000].to_csv('movies_trimmed2.csv', index=False)\nmovies_trimmed[20000:30000].to_csv('movies_trimmed3.csv', index=False)\nmovies_trimmed[30000:40000].to_csv('movies_trimmed4.csv', index=False)\nmovies_trimmed[40000:50000].to_csv('movies_trimmed5.csv', index=False)\nmovies_trimmed[60000:70000].to_csv('movies_trimmed6.csv', index=False)\nmovies_trimmed[70000:].to_csv('movies_trimmed7.csv', index=False)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6e27e42d4e41449037ad3111356049a88cff23 | 5,147 | ipynb | Jupyter Notebook | Lesson2-IntroAutoGrad.ipynb | HoustonDataScience/Intro-to-Image-Classification | 7417f273499b48bd07550bf61b4033bcc3a95e35 | [
"BSD-3-Clause"
]
| 11 | 2018-07-25T17:37:41.000Z | 2021-01-31T17:37:56.000Z | Lesson2-IntroAutoGrad.ipynb | RodeoBlues/Intro-to-Image-Classification | 7417f273499b48bd07550bf61b4033bcc3a95e35 | [
"BSD-3-Clause"
]
| null | null | null | Lesson2-IntroAutoGrad.ipynb | RodeoBlues/Intro-to-Image-Classification | 7417f273499b48bd07550bf61b4033bcc3a95e35 | [
"BSD-3-Clause"
]
| 8 | 2018-09-06T01:50:55.000Z | 2018-09-10T23:57:04.000Z | 23.828704 | 281 | 0.555858 | [
[
[
"# Automatic differentiation\n\nAutomatic differentiation is a core component of `torch`. `autograd` is essential to understand how neural networks can be trained. So, let us look at a few simple examples to understand how `autograd` works.\n\n`autograd` in `torch` provides automatic differentiation capabilities for all operations in tensor. It is an essential part to understand how backpropagation works in `torch`.\n\nLet us look at a few examples to carefully understand what this is all about.\n\nLet us start with an example from our previous notebook and define a tensor.",
"_____no_output_____"
]
],
[
[
"import torch",
"_____no_output_____"
]
],
[
[
"An important part of tensors in PyTorch is the ability to track operations as we go along. \n\nIf we set its attribute `.requires_grad` as `True`, it starts to track all operations on it. When we finish your computation you can call `.backward()` and have all the gradients computed automatically. The gradient for this tensor will be accumulated into `.grad` attribute.",
"_____no_output_____"
]
],
[
[
"x = torch.tensor([1., 2., 3.], requires_grad=True)\nprint(x)",
"tensor([ 1., 2., 3.])\n"
]
],
[
[
"Let us do a few operations on the tensor to see what it is all about.",
"_____no_output_____"
]
],
[
[
"y = x + 2\nz = y*y\nprint(z)",
"tensor([ 9., 16., 25.])\n"
],
[
"zmean = z.mean()\nprint(zmean)",
"tensor(16.6667)\n"
]
],
[
[
"## Computing gradients\n\nNow that we have computed a single value - `zmean` from a tensor `z`, let us run backpropagation and compute the gradients.",
"_____no_output_____"
]
],
[
[
"zmean.backward()",
"_____no_output_____"
]
],
[
[
"Let us look at the gradients now.",
"_____no_output_____"
]
],
[
[
"print(x.grad)",
"tensor([ 2.0000, 2.6667, 3.3333])\n"
]
],
[
[
"Of course, `torch` computed the gradients for us. Let us do this with hand and make sure we understand this well.\n\n\\begin{eqnarray}\ny &= x + 2 \\\\\nz &= y^2 \\\\\nz_{mean} &= \\sum_{i=1}^3 z_i\n\\end{eqnarray}",
"_____no_output_____"
],
[
"For simple functions like we defined above, we can compute it by hand. However, for complicated functions of several variables, it is a challenge to compute it by hand. That is where we see the true power of `autograd`.\n\nIn the next few notebooks, we will see the power of automatic differentiation.\n\nA final note: Although this tutorial uses `torch` to illustrate automatic differentiation, backward propagation and other concepts, these concepts are true for your favorite deep learning package.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
cb6e3b327cc0d17e687598d8427545924b2b55fb | 15,711 | ipynb | Jupyter Notebook | official_tutorial/lesson2a_autograd_tutorial.ipynb | zhennongchen/pytorch-tutorial | 7221bad5f85c6a937e3ad8d195bb6d7daf7803b6 | [
"MIT"
]
| null | null | null | official_tutorial/lesson2a_autograd_tutorial.ipynb | zhennongchen/pytorch-tutorial | 7221bad5f85c6a937e3ad8d195bb6d7daf7803b6 | [
"MIT"
]
| null | null | null | official_tutorial/lesson2a_autograd_tutorial.ipynb | zhennongchen/pytorch-tutorial | 7221bad5f85c6a937e3ad8d195bb6d7daf7803b6 | [
"MIT"
]
| null | null | null | 27.514886 | 164 | 0.5225 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"**Read Later:**\n\nDocument about ``autograd.Function`` is at\nhttps://pytorch.org/docs/stable/autograd.html#function",
"_____no_output_____"
],
[
"**Notes:**\n\nneeds to set requires_grad == True when define the tensor if you wanna autograd.\n\nassume z = f(x,y)\n\nz.backward() initiates the backward pass and computed all the gradient automatically.\n\nthe gradient will be accumulated into .grad attribute, that means x.grad will have dz/dx and y.grad will have dz/dy.\n\nif z is a scalar, then can use z.backward() and no argument needs to be passed.\n\nif z is a vector, then we have to pass a gradient argument into z.backward(), so if we wanna dz/dx and dz/dy\nit should be: z.backward(1). the argument can be any number and should have the same size as z\n\n\n",
"_____no_output_____"
],
[
"\nAutograd: Automatic Differentiation\n===================================\n\nCentral to all neural networks in PyTorch is the ``autograd`` package.\nLet’s first briefly visit this, and we will then go to training our\nfirst neural network.\n\n\nThe ``autograd`` package provides automatic differentiation for all operations\non Tensors. It is a define-by-run framework, which means that your backprop is\ndefined by how your code is run, and that every single iteration can be\ndifferent.\n\nLet us see this in more simple terms with some examples.\n\nTensor\n--------\n\n``torch.Tensor`` is the central class of the package. If you set its attribute\n``.requires_grad`` as ``True``, it starts to track all operations on it. When\nyou finish your computation you can call ``.backward()`` and have all the\ngradients computed automatically. The gradient for this tensor will be\naccumulated into ``.grad`` attribute.\n\nTo stop a tensor from tracking history, you can call ``.detach()`` to detach\nit from the computation history, and to prevent future computation from being\ntracked.\n\nTo prevent tracking history (and using memory), you can also wrap the code block\nin ``with torch.no_grad():``. This can be particularly helpful when evaluating a\nmodel because the model may have trainable parameters with\n``requires_grad=True``, but for which we don't need the gradients.\n\nThere’s one more class which is very important for autograd\nimplementation - a ``Function``.\n\n``Tensor`` and ``Function`` are interconnected and build up an acyclic\ngraph, that encodes a complete history of computation. Each tensor has\na ``.grad_fn`` attribute that references a ``Function`` that has created\nthe ``Tensor`` (except for Tensors created by the user - their\n``grad_fn is None``).\n\nIf you want to compute the derivatives, you can call ``.backward()`` on\na ``Tensor``. If ``Tensor`` is a scalar (i.e. it holds a one element\ndata), you don’t need to specify any arguments to ``backward()``,\nhowever if it has more elements, you need to specify a ``gradient``\nargument that is a tensor of matching shape.\n\n",
"_____no_output_____"
]
],
[
[
"import torch",
"_____no_output_____"
]
],
[
[
"Create a tensor and set ``requires_grad=True`` to track computation with it\n\n",
"_____no_output_____"
]
],
[
[
"x = torch.tensor([[1,3],[2,2]], requires_grad=True, dtype = torch.float32)\nprint(x)\nprint(x.size())",
"tensor([[1., 3.],\n [2., 2.]], requires_grad=True)\ntorch.Size([2, 2])\n"
]
],
[
[
"Do a tensor operation:\n\n",
"_____no_output_____"
]
],
[
[
"y = ( x + 2) \nz = 3 * (y ** 2)\nprint(y,z)",
"tensor([[3., 5.],\n [4., 4.]], grad_fn=<AddBackward0>) tensor([[27., 75.],\n [48., 48.]], grad_fn=<MulBackward0>)\n"
]
],
[
[
"``y`` was created as a result of an operation, so it has a ``grad_fn``.\n\n",
"_____no_output_____"
]
],
[
[
"print(y.grad_fn)\nprint(zz.grad_fn)",
"<AddBackward0 object at 0x7fef96385520>\n<MmBackward object at 0x7fef96385820>\n"
]
],
[
[
"Do more operations on ``y``\n\n",
"_____no_output_____"
]
],
[
[
"z = y * y # dot product\nout = z.mean()\nprint(z, out)",
"tensor([[ 9., 25.],\n [16., 16.]], grad_fn=<MulBackward0>) tensor(16.5000, grad_fn=<MeanBackward0>)\n"
]
],
[
[
"``.requires_grad_( ... )`` changes an existing Tensor's ``requires_grad``\nflag in-place. The input flag defaults to ``False`` if not given.\n\n",
"_____no_output_____"
]
],
[
[
"a = torch.tensor([2, 2],dtype = torch.float32)\na = ((a * 3))\nprint(a)\nprint(a.requires_grad)\na.requires_grad_(True)\nprint(a.requires_grad)\nb = (a * a).sum()\nprint(b)\nprint(b.grad_fn)",
"tensor([6., 6.])\nFalse\nTrue\ntensor(72., grad_fn=<SumBackward0>)\n<SumBackward0 object at 0x7fef94c668b0>\n"
]
],
[
[
" Get Gradients\n---------\nLet's backprop now.\nBecause ``out`` contains a single scalar, ``out.backward()`` is\nequivalent to ``out.backward(torch.tensor(1.))``.\n\nNote:\nout.backward() only works for scale\n\nout.backward(v) where v is a vector works for non-scale input, it gives Jacobian-vector product. here v is the gradient we have to put into the backwass pass.",
"_____no_output_____"
]
],
[
[
"# example: y = 2 * x + 3 * (x^2)\nx = torch.tensor([3],dtype = torch.float32,requires_grad = False)\nx.requires_grad_(True)\ny = 2 * x + x * x * 3\nprint(x,y)",
"tensor([3.], requires_grad=True) tensor([33.], grad_fn=<AddBackward0>)\n"
]
],
[
[
"Print gradients d(y)/dx, which should be y = 2 + 6 * x\n\n\n",
"_____no_output_____"
]
],
[
[
"y.backward() # first run y.backward(), then x.grad has the value\nprint(x.grad) # only works when x is scalar",
"tensor([20.])\n"
]
],
[
[
"Mathematically, if you have a vector valued function $\\vec{y}=f(\\vec{x})$,\nthen the gradient of $\\vec{y}$ with respect to $\\vec{x}$\nis a Jacobian matrix:\n\n\\begin{align}J=\\left(\\begin{array}{ccc}\n \\frac{\\partial y_{1}}{\\partial x_{1}} & \\cdots & \\frac{\\partial y_{1}}{\\partial x_{n}}\\\\\n \\vdots & \\ddots & \\vdots\\\\\n \\frac{\\partial y_{m}}{\\partial x_{1}} & \\cdots & \\frac{\\partial y_{m}}{\\partial x_{n}}\n \\end{array}\\right)\\end{align}\n\nGenerally speaking, ``torch.autograd`` is an engine for computing\nvector-Jacobian product. That is, given any vector\n$v=\\left(\\begin{array}{cccc} v_{1} & v_{2} & \\cdots & v_{m}\\end{array}\\right)^{T}$,\ncompute the product $v^{T}\\cdot J$. If $v$ happens to be\nthe gradient of a scalar function $l=g\\left(\\vec{y}\\right)$,\nthat is,\n$v=\\left(\\begin{array}{ccc}\\frac{\\partial l}{\\partial y_{1}} & \\cdots & \\frac{\\partial l}{\\partial y_{m}}\\end{array}\\right)^{T}$,\nthen by the chain rule, the vector-Jacobian product would be the\ngradient of $l$ with respect to $\\vec{x}$:\n\n\\begin{align}J^{T}\\cdot v=\\left(\\begin{array}{ccc}\n \\frac{\\partial y_{1}}{\\partial x_{1}} & \\cdots & \\frac{\\partial y_{m}}{\\partial x_{1}}\\\\\n \\vdots & \\ddots & \\vdots\\\\\n \\frac{\\partial y_{1}}{\\partial x_{n}} & \\cdots & \\frac{\\partial y_{m}}{\\partial x_{n}}\n \\end{array}\\right)\\left(\\begin{array}{c}\n \\frac{\\partial l}{\\partial y_{1}}\\\\\n \\vdots\\\\\n \\frac{\\partial l}{\\partial y_{m}}\n \\end{array}\\right)=\\left(\\begin{array}{c}\n \\frac{\\partial l}{\\partial x_{1}}\\\\\n \\vdots\\\\\n \\frac{\\partial l}{\\partial x_{n}}\n \\end{array}\\right)\\end{align}\n\n(Note that $v^{T}\\cdot J$ gives a row vector which can be\ntreated as a column vector by taking $J^{T}\\cdot v$.)\n\nThis characteristic of vector-Jacobian product makes it very\nconvenient to feed external gradients into a model that has\nnon-scalar output.\n\nNote: \n\n# vector-Jacobian product is matrix dot product!!!",
"_____no_output_____"
],
[
"Now let's take a look at an example of vector-Jacobian product:\n\n",
"_____no_output_____"
]
],
[
[
"# example 1:\nx = torch.tensor([3,4,6], dtype = torch.float32,requires_grad=True)\ny = x * x * 3\nv = torch.tensor([1,1,2], dtype=torch.float)\ny.backward(v) # here it does the derivative calulation dyi/dxi = 6 * xi * v\nprint(x.grad)",
"tensor([18., 24., 72.])\n"
],
[
"# example 2:\ng = torch.tensor([[1,1,1],[2,3,4]],dtype = torch.float32)\ng.add_(torch.ones_like(g))\ng.requires_grad_(True) # _ here means replace the variable with new value\nprint(g, g.requires_grad)\ngg = g*g*g\nggg = gg +3*g\nv = torch.tensor([[1,1,2],[2,2,2]], dtype=torch.float)\nggg.backward(v) # here we do the derivative calculation dggg/dg = 3g^2 + 3\nprint(g.grad)",
"tensor([[2., 2., 2.],\n [3., 4., 5.]], requires_grad=True) True\ntensor([[ 15., 15., 30.],\n [ 60., 102., 156.]])\n"
]
],
[
[
"in the above case if we do gg.backward(v), then the derivative is 3g^2 only",
"_____no_output_____"
]
],
[
[
"x = torch.randn(3, requires_grad=True)\nprint(x)\ny = x * 2\ncount = 0\nwhile y.data.norm() < 100: # L2 norm / Euclidean norm\n y = y * 2\n count += 1\n\nprint(y,count)\n\n# Now in this case ``y`` is no longer a scalar. ``torch.autograd``\n# could not compute the full Jacobian directly, but if we just\n# want the vector-Jacobian product, simply pass the vector to\n# ``backward`` as argument:\n \nv = torch.tensor([0.1, 1.0, 0.0001], dtype=torch.float)\ny.backward(v)\n\nprint(x.grad)",
"tensor([ 0.7491, 1.0516, -0.0123], requires_grad=True)\ntensor([ 95.8882, 134.6020, -1.5742], grad_fn=<MulBackward0>) 6\ntensor([1.2800e+01, 1.2800e+02, 1.2800e-02])\n"
]
],
[
[
"# Stop autograd\nYou can also stop autograd from tracking history on Tensors\nwith ``.requires_grad=True`` either by wrapping the code block in\n``with torch.no_grad():``\n\n",
"_____no_output_____"
]
],
[
[
"x = torch.randn(3, requires_grad=True)\nprint(x.requires_grad)\nprint((x ** 2).requires_grad)\n\nwith torch.no_grad():\n\tprint((x ** 2).requires_grad)\n \nx = torch.randn(3, requires_grad=True)\nx.requires_grad_(False)\nprint((x**2).requires_grad)",
"True\nTrue\nFalse\nFalse\n"
]
],
[
[
"Or by using ``.detach()`` to get a new Tensor with the same\ncontent but that does not require gradients:\n\n",
"_____no_output_____"
]
],
[
[
"x.requires_grad_(True)\nprint(x.requires_grad)\ny = x.detach()\nprint(y.requires_grad)\nprint(x.eq(y))",
"True\nFalse\ntensor([True, True, True])\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6e46c715a3f6ea7b89188a340685b5a6351c18 | 45,554 | ipynb | Jupyter Notebook | Additional Content/Homeworks/HW11_Data_Storage/Resources/climateAnalaysis.ipynb | kjordan18/kjordan18.github.io | d07014850000e254da84fdfb828e1a0587b4ffd1 | [
"MIT"
]
| 1 | 2019-04-15T21:33:14.000Z | 2019-04-15T21:33:14.000Z | Additional Content/Homeworks/HW11_Data_Storage/Resources/climateAnalaysis.ipynb | kjordan18/kjordan18.github.io | d07014850000e254da84fdfb828e1a0587b4ffd1 | [
"MIT"
]
| null | null | null | Additional Content/Homeworks/HW11_Data_Storage/Resources/climateAnalaysis.ipynb | kjordan18/kjordan18.github.io | d07014850000e254da84fdfb828e1a0587b4ffd1 | [
"MIT"
]
| null | null | null | 78.406196 | 12,056 | 0.736862 | [
[
[
"# Dependencies\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom datetime import timedelta\nimport time\nfrom datetime import date\n\n# Import SQL Alchemy\nfrom sqlalchemy import create_engine, ForeignKey, func\nfrom sqlalchemy.ext.automap import automap_base\nfrom sqlalchemy.orm import Session\n\n# Import PyMySQL (Not needed if mysqlclient is installed)\nimport pymysql\npymysql.install_as_MySQLdb()",
"_____no_output_____"
],
[
"firstDate = \"2017-07-17\"\nlastDate = \"2017-07-30\"",
"_____no_output_____"
],
[
"engine = create_engine(\"sqlite:///hawaii.sqlite\")\nconn = engine.connect()",
"_____no_output_____"
],
[
"Base = automap_base()\nBase.prepare(engine, reflect=True)",
"_____no_output_____"
],
[
"# mapped classes are now created with names by default\n# matching that of the table name.\nBase.classes.keys()",
"_____no_output_____"
],
[
"Measurement = Base.classes.Measurements\nStation = Base.classes.Stations",
"_____no_output_____"
],
[
"# To push the objects made and query the server we use a Session object\nsession = Session(bind=engine)",
"_____no_output_____"
],
[
"# Calculate the date 1 year ago from today\nprev_year = date.today() - timedelta(days=365)\n\n# Perform a query to retrieve the data and precipitation scores\nresults = session.query(Measurement.date, Measurement.prcp).filter(Measurement.date >= prev_year).all()\n\n# Save the query results as a Pandas DataFrame and set the index to the date column\ndf = pd.DataFrame(results, columns=['date', 'precipitation'])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"yAxis = df.precipitation\nxAxis = df.date\n\nplt.figure(figsize=(15,3))\nplt.bar(xAxis, yAxis, color='blue', alpha = 0.5, align='edge')\nplt.xticks(np.arange(12), df.date[1:13], rotation=90)\nplt.xlabel('Date')\nplt.ylabel('Precipitation (Inches)')\nplt.title('Precipitation')\n\nplt.show()\n\ndf.describe()",
"_____no_output_____"
],
[
"totalStations = session.query(Station.station).count()\ntotalStations",
"_____no_output_____"
],
[
"activeStations = session.query(Measurement.station, Measurement.tobs, func.count(Measurement.station)).group_by(Measurement.station).all()\ndfAS = pd.DataFrame(activeStations, columns = ['station', 'tobs', 'stationCount'])\nprint(dfAS)\n\nmaxObs = dfAS.loc[(dfAS['tobs'] == dfAS['tobs'].max())]\nmaxObs",
" station tobs stationCount\n0 USC00511918 64 1932\n1 USC00513117 77 2696\n2 USC00514830 82 1937\n3 USC00516128 76 2484\n4 USC00517948 80 683\n5 USC00518838 74 342\n6 USC00519281 79 2772\n7 USC00519397 81 2685\n8 USC00519523 82 2572\n"
],
[
"tobsData = session.query(Measurement.date, Measurement.tobs).filter(Measurement.date >= prev_year).all()\ndfTD = pd.DataFrame(tobsData, columns = ['Date', 'tobs']).sort_values('tobs', ascending = False)\ndfTD.head()",
"_____no_output_____"
],
[
"plt.hist(dfTD['tobs'], bins=12, color= \"blue\")\nplt.xlabel('Tobs (bins=12)')\nplt.ylabel('Frequency')\nplt.title('Tobs Frequency')\nplt.legend('Tobs')\nplt.show()",
"_____no_output_____"
],
[
"def calcTemps(x):\n session.query(x['Date'], x['tobs']).filter(lastDate >= x['Date'] >= firstDate).all()\n x['tobs'].max()\n x['tobs'].min()\n \ncalcTemps(dfTD)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6e5b8ab57287b3c3144509267557b0a2ecafb6 | 32,740 | ipynb | Jupyter Notebook | openaq-air-pollution-measure/air-quality-eda-using-sql-bigquery.ipynb | Rhilok/kaggle-datasets-and-kernels | d7d046d0072c6f202ebbfbc3cdd12684b1a7d576 | [
"MIT"
]
| 4 | 2021-01-29T13:39:23.000Z | 2021-12-30T02:56:16.000Z | openaq-air-pollution-measure/air-quality-eda-using-sql-bigquery.ipynb | Rhilok/kaggle-datasets-and-kernels | d7d046d0072c6f202ebbfbc3cdd12684b1a7d576 | [
"MIT"
]
| null | null | null | openaq-air-pollution-measure/air-quality-eda-using-sql-bigquery.ipynb | Rhilok/kaggle-datasets-and-kernels | d7d046d0072c6f202ebbfbc3cdd12684b1a7d576 | [
"MIT"
]
| 1 | 2021-02-27T07:27:54.000Z | 2021-02-27T07:27:54.000Z | 32,740 | 32,740 | 0.722999 | [
[
[
"## Global Air Pollution Measurements\n\n* [Air Quality Index - Wiki](https://en.wikipedia.org/wiki/Air_quality_index)\n* [BigQuery - Wiki](https://en.wikipedia.org/wiki/BigQuery)\n\nIn this notebook data is extracted from *BigQuery Public Data* assesible exclusively only in *Kaggle*. The BigQurey Helper Object will convert data in cloud storage into *Pandas DataFrame* object. The query syntax is same as *SQL*. As size of data is very high convert entire data to DataFrame is cumbersome. So query is written such that will be readly available for Visualization.\n***\n>**Baisc attributes of Air quality index** \n* Measurement units\n * $ug/m^3$: micro gram/cubic meter \n * $ppm$: Parts Per Million\n* Pollutant\n * $O3$: Ozone gas\n * $SO2$: Sulphur Dioxed\n * $NO2$: Nitrogen Dioxed\n * $PM 2.5$: Particles with an aerodynamic diameter less than $2.5 μm$\n * $PM 10$: Particles with an aerodynamic diameter less than $10 μm$\n * $CO$: Carbon monoxide\n \n**Steps**\n1. Load Packages\n2. Bigquery Object\n3. AQI range and Statistics \n4. Distribution of country listed in AQI\n5. Location\n6. Air Quality Index value distribution Map veiw\n7. Pollutant Statistics\n8. Distribution of pollutant and unit\n9. Distribution of Source name\n10. Sample AQI Averaged over in hours\n11. AQI variation with time\n12. Country Heatmap\n13. Animation",
"_____no_output_____"
],
[
"### Load packages",
"_____no_output_____"
]
],
[
[
"# Load packages\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom mpl_toolkits.basemap import Basemap\nimport folium\nimport folium.plugins as plugins\n\nimport warnings\nwarnings.filterwarnings('ignore')\npd.options.display.max_rows =10\n%matplotlib inline\n",
"_____no_output_____"
]
],
[
[
"### Bigquery\nBigQuery is a RESTful web service that enables interactive analysis of massively large datasets working in conjunction with Google Storage. It is an Infrastructure as a Service that may be used complementarily with MapReduce.",
"_____no_output_____"
]
],
[
[
"# Customized query helper function explosively in Kaggle\nimport bq_helper\n\n# Helper object\nopenAQ = bq_helper.BigQueryHelper(active_project='bigquery-public-data',\n dataset_name='openaq')\n# List of table\nopenAQ.list_tables()",
"_____no_output_____"
],
[
"#Schema \nopenAQ.table_schema('global_air_quality')",
"_____no_output_____"
]
],
[
[
"### Table display",
"_____no_output_____"
]
],
[
[
"openAQ.head('global_air_quality')",
"_____no_output_____"
],
[
"# Summary statics\nquery = \"\"\"SELECT value,averaged_over_in_hours\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³'\n \"\"\"\np1 = openAQ.query_to_pandas(query)\np1.describe()",
"_____no_output_____"
]
],
[
[
"# Air Quality Index Range\n* [AQI Range](http://aqicn.org/faq/2013-09-09/revised-pm25-aqi-breakpoints/)\n<center><img src = 'https://campuspress.yale.edu/datadriven/files/2012/03/AQI-1024x634-1ybtu6l.png '><center>\n\nThe range of AQI is 0 - 500, so lets limit data to that range, in previous kernel's these outlier data points are not removed",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT value,country \n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value < 0\n \"\"\"\np1 = openAQ.query_to_pandas(query)\np1.describe().T",
"_____no_output_____"
]
],
[
[
"There are more than 100 value having value less than 0. The lowest value is -999000, which is outlier data point. **Air Quality Meter** is digital a instruments, if meter is show error value then sensor is disconnected or faulty. ",
"_____no_output_____"
]
],
[
[
"query2 = \"\"\"SELECT value,country,pollutant\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value > 0\n \"\"\"\np2 = openAQ.query_to_pandas(query2)\nprint('0.99 Quantile',p2['value'].quantile(0.99))\np2.describe().T",
"_____no_output_____"
],
[
"p2[p2['value']>10000]",
"_____no_output_____"
]
],
[
[
"Country \n* MK is *Macedonia* [wiki](https://en.wikipedia.org/wiki/Republic_of_Macedonia)\n* CL is *Chile* [Wiki](https://en.wikipedia.org/wiki/Chile)\n>In both the countries some may some natural disaster happend so AQI is very high. \n We will disgrad value more than 10000, which are outlier data point ",
"_____no_output_____"
],
[
"### Distribution of country listed in AQI",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT country,COUNT(country) as `count`\n FROM `bigquery-public-data.openaq.global_air_quality`\n GROUP BY country\n HAVING COUNT(country) >10\n ORDER BY `count`\n \"\"\"\ncnt = openAQ.query_to_pandas_safe(query)\ncnt.tail()\n\nplt.style.use('bmh')\nplt.figure(figsize=(14,4))\nsns.barplot(cnt['country'], cnt['count'], palette='magma')\nplt.xticks(rotation=45)\nplt.title('Distribution of country listed in data');",
"_____no_output_____"
]
],
[
[
"## Location\nWe find find different location where air quality is taken. This location data consist of latitude and logitude, city.",
"_____no_output_____"
]
],
[
[
"#Average polution of air by countries\nquery = \"\"\"SELECT AVG(value) as `Average`,country\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY country\n ORDER BY Average DESC\n \"\"\"\ncnt = openAQ.query_to_pandas(query)",
"_____no_output_____"
],
[
"plt.figure(figsize=(14,4))\nsns.barplot(cnt['country'],cnt['Average'], palette= sns.color_palette('gist_heat',len(cnt)))\nplt.xticks(rotation=90)\nplt.title('Average polution of air by countries in unit $ug/m^3$')\nplt.ylabel('Average AQI in $ug/m^3$');",
"_____no_output_____"
]
],
[
[
"* Country PL ( Poland) and IN (India) are top pollutor of air\n***\n### AQI measurement center",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT city,latitude,longitude,\n AVG(value) as `Average`\n FROM `bigquery-public-data.openaq.global_air_quality`\n GROUP BY latitude,city,longitude \n \"\"\"\nlocation = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"#Location AQI measurement center\nm = folium.Map(location = [20,10],tiles='Mapbox Bright',zoom_start=2)\n\n# add marker one by on map\nfor i in range(0,500):\n folium.Marker(location = [location.iloc[i]['latitude'],location.iloc[i]['longitude']],\\\n popup=location.iloc[i]['city']).add_to(m)\n \nm # DRAW MAP",
"_____no_output_____"
]
],
[
[
"We find that thier are many air qulity index measurement unit across -US- and -Europe-. Thier are few measurement center in -African- continent. We are hardly find any measuring center in Mid East, Russia.",
"_____no_output_____"
],
[
"### Air Quality Index value distribution Map veiw",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT city,latitude,longitude,\n AVG(value) as `Average`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY latitude,city,longitude \n \"\"\"\nlocation = openAQ.query_to_pandas_safe(query)\nlocation.dropna(axis=0, inplace=True)",
"_____no_output_____"
],
[
"plt.style.use('ggplot')\nf,ax = plt.subplots(figsize=(14,10))\nm1 = Basemap(projection='cyl', llcrnrlon=-180, urcrnrlon=180, llcrnrlat=-90, urcrnrlat=90,\n resolution='c',lat_ts=True)\n\nm1.drawmapboundary(fill_color='#A6CAE0', linewidth=0)\nm1.fillcontinents(color='grey', alpha=0.3)\nm1.drawcoastlines(linewidth=0.1, color=\"white\")\nm1.shadedrelief()\nm1.bluemarble(alpha=0.4)\navg = location['Average']\nm1loc = m1(location['latitude'].tolist(),location['longitude'])\nm1.scatter(m1loc[1],m1loc[0],lw=3,alpha=0.5,zorder=3,cmap='coolwarm', c=avg)\nplt.title('Average Air qulity index in unit $ug/m^3$ value')\nm1.colorbar(label=' Average AQI value in unit $ug/m^3$');",
"_____no_output_____"
]
],
[
[
"### US",
"_____no_output_____"
]
],
[
[
"#USA location\nquery = \"\"\"SELECT \n MAX(latitude) as `max_lat`,\n MIN(latitude) as `min_lat`,\n MAX(longitude) as `max_lon`,\n MIN(longitude) as `min_lon`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE country = 'US' \"\"\"\nus_loc = openAQ.query_to_pandas_safe(query)\nus_loc",
"_____no_output_____"
],
[
"query = \"\"\" SELECT city,latitude,longitude,averaged_over_in_hours,\n AVG(value) as `Average`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE country = 'US' AND unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY latitude,city,longitude,averaged_over_in_hours,country \"\"\"\nus_aqi = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"# USA\nmin_lat = us_loc['min_lat']\nmax_lat = us_loc['max_lat']\nmin_lon = us_loc['min_lon']\nmax_lon = us_loc['max_lon']\n\nplt.figure(figsize=(14,8))\nm2 = Basemap(projection='cyl', llcrnrlon=min_lon, urcrnrlon=max_lon, llcrnrlat=min_lat, urcrnrlat=max_lat,\n resolution='c',lat_ts=True)\nm2.drawcounties()\nm2.drawmapboundary(fill_color='#A6CAE0', linewidth=0)\nm2.fillcontinents(color='grey', alpha=0.3)\nm2.drawcoastlines(linewidth=0.1, color=\"white\")\nm2.drawstates()\nm2.bluemarble(alpha=0.4)\navg = (us_aqi['Average'])\nm2loc = m2(us_aqi['latitude'].tolist(),us_aqi['longitude'])\nm2.scatter(m2loc[1],m2loc[0],c = avg,lw=3,alpha=0.5,zorder=3,cmap='rainbow')\nm1.colorbar(label = 'Average AQI value in unit $ug/m^3$')\nplt.title('Average Air qulity index in unit $ug/m^3$ of US');",
"_____no_output_____"
]
],
[
[
"AQI of US range 0 to 400, most of city data points are within 100\n### India",
"_____no_output_____"
]
],
[
[
"#INDIA location\nquery = \"\"\"SELECT \n MAX(latitude) as `max_lat`,\n MIN(latitude) as `min_lat`,\n MAX(longitude) as `max_lon`,\n MIN(longitude) as `min_lon`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE country = 'IN' \"\"\"\nin_loc = openAQ.query_to_pandas_safe(query)\nin_loc",
"_____no_output_____"
],
[
"query = \"\"\" SELECT city,latitude,longitude,\n AVG(value) as `Average`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE country = 'IN' AND unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY latitude,city,longitude,country \"\"\"\nin_aqi = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"# INDIA\nmin_lat = in_loc['min_lat']-5\nmax_lat = in_loc['max_lat']+5\nmin_lon = in_loc['min_lon']-5\nmax_lon = in_loc['max_lon']+5\n\nplt.figure(figsize=(14,8))\nm3 = Basemap(projection='cyl', llcrnrlon=min_lon, urcrnrlon=max_lon, llcrnrlat=min_lat, urcrnrlat=max_lat,\n resolution='c',lat_ts=True)\nm3.drawcounties()\nm3.drawmapboundary(fill_color='#A6CAE0', linewidth=0)\nm3.fillcontinents(color='grey', alpha=0.3)\nm3.drawcoastlines(linewidth=0.1, color=\"white\")\nm3.drawstates()\navg = in_aqi['Average']\nm3loc = m3(in_aqi['latitude'].tolist(),in_aqi['longitude'])\nm3.scatter(m3loc[1],m3loc[0],c = avg,alpha=0.5,zorder=5,cmap='rainbow')\nm1.colorbar(label = 'Average AQI value in unit $ug/m^3$')\nplt.title('Average Air qulity index in unit $ug/m^3$ of India');",
"_____no_output_____"
]
],
[
[
"### Distribution of pollutant and unit",
"_____no_output_____"
]
],
[
[
"# Unit query\nquery = \"\"\"SELECT unit,COUNT(unit) as `count`\n FROM `bigquery-public-data.openaq.global_air_quality`\n GROUP BY unit\n \"\"\"\nunit = openAQ.query_to_pandas(query)\n# Pollutant query\nquery = \"\"\"SELECT pollutant,COUNT(pollutant) as `count`\n FROM `bigquery-public-data.openaq.global_air_quality`\n GROUP BY pollutant\n \"\"\"\npoll_count = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"plt.style.use('fivethirtyeight')\nplt.style.use('bmh')\nf, ax = plt.subplots(1,2,figsize = (14,5))\nax1,ax2= ax.flatten()\nax1.pie(x=unit['count'],labels=unit['unit'],shadow=True,autopct='%1.1f%%',explode=[0,0.1],\\\n colors=sns.color_palette('hot',2),startangle=90,)\nax1.set_title('Distribution of measurement unit')\nexplode = np.arange(0,0.1)\nax2.pie(x=poll_count['count'],labels=poll_count['pollutant'], shadow=True, autopct='%1.1f%%',\\\n colors=sns.color_palette('Set2',5),startangle=60,)\nax2.set_title('Distribution of pollutants in air');",
"_____no_output_____"
]
],
[
[
"* The most polular unit of mesurement of air quality is $ug/m^3$\n* $O^3$ is share 23% pollution in air.\n***\n### Pollutant Statistics",
"_____no_output_____"
]
],
[
[
"query = \"\"\" SELECT pollutant,\n AVG(value) as `Average`,\n COUNT(value) as `Count`,\n MIN(value) as `Min`,\n MAX(value) as `Max`,\n SUM(value) as `Sum`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY pollutant\n \"\"\"\ncnt = openAQ.query_to_pandas_safe(query)\ncnt ",
"_____no_output_____"
]
],
[
[
" We find\n* The CO (carbon monoxide) having very wide range of value.\n* Look at sum of CO which is highest in list.\n* Except Average AQI of CO, all are below 54 $ug/m^3$",
"_____no_output_____"
],
[
"### Pollutants by Country",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT AVG(value) as`Average`,country, pollutant\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³'AND value BETWEEN 0 AND 10000\n GROUP BY country,pollutant\"\"\"\np1 = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"# By country\np1_pivot = p1.pivot(index = 'country',values='Average', columns= 'pollutant')\n\nplt.figure(figsize=(14,15))\nax = sns.heatmap(p1_pivot, lw=0.01, cmap=sns.color_palette('Reds',500))\nplt.yticks(rotation=30)\nplt.title('Heatmap average AQI by Pollutant');",
"_____no_output_____"
],
[
"f,ax = plt.subplots(figsize=(14,6))\nsns.barplot(p1[p1['pollutant']=='co']['country'],p1[p1['pollutant']=='co']['Average'],)\nplt.title('Co AQI in diffrent country')\nplt.xticks(rotation=90);",
"_____no_output_____"
],
[
"f,ax = plt.subplots(figsize=(14,6))\nsns.barplot(p1[p1['pollutant']=='pm25']['country'],p1[p1['pollutant']=='pm25']['Average'])\nplt.title('pm25 AQI in diffrent country')\nplt.xticks(rotation=90);",
"_____no_output_____"
]
],
[
[
"### Distribution of Source name\nThe institution where AQI is measure ",
"_____no_output_____"
]
],
[
[
"#source_name \nquery = \"\"\" SELECT source_name, COUNT(source_name) as `count`\n FROM `bigquery-public-data.openaq.global_air_quality`\n GROUP BY source_name\n ORDER BY count DESC\n \"\"\"\nsource_name = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"plt.figure(figsize=(14,10))\nsns.barplot(source_name['count'][:20], source_name['source_name'][:20],palette = sns.color_palette('YlOrBr'))\nplt.title('Distribution of Top 20 source_name')\n#plt.axvline(source_name['count'].median())\nplt.xticks(rotation=90);",
"_____no_output_____"
]
],
[
[
"We find \n* Airnow is top source unit in list\n* Europian country are top in the list, the instition name is starts with 'EEA country'.\n***\n\n### Sample AQI Averaged over in hours\nThe sample of AQI value taken in different hour",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT averaged_over_in_hours, COUNT(*) as `count`\n FROM `bigquery-public-data.openaq.global_air_quality`\n GROUP BY averaged_over_in_hours\n ORDER BY count DESC \"\"\"\ncnt = openAQ.query_to_pandas(query)",
"_____no_output_____"
],
[
"#cnt['averaged_over_in_hours'] = cnt['averaged_over_in_hours'].astype('category')\nplt.figure(figsize=(14,5))\nsns.barplot( cnt['averaged_over_in_hours'],cnt['count'], palette= sns.color_palette('brg'))\nplt.title('Distibution of quality measurement per hour ');",
"_____no_output_____"
]
],
[
[
"we find that air quality is measured every hour\n***\n### AQI in ppm",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT AVG(value) as`Average`,country,\n EXTRACT(YEAR FROM timestamp) as `Year`,\n pollutant\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'ppm' \n GROUP BY country,Year,pollutant\"\"\"\npol_aqi = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"# By month in year\nplt.figure(figsize=(14,8))\nsns.barplot(pol_aqi['country'], pol_aqi['Average'])\nplt.title('Distribution of average AQI by country $ppm$');",
"_____no_output_____"
]
],
[
[
" ### AQI variation with time",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT EXTRACT(YEAR FROM timestamp) as `Year`,\n AVG(value) as `Average`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY EXTRACT(YEAR FROM timestamp)\n \"\"\"\nquality = openAQ.query_to_pandas(query)\n\nquery = \"\"\"SELECT EXTRACT(MONTH FROM timestamp) as `Month`,\n AVG(value) as `Average`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY EXTRACT(MONTH FROM timestamp)\n \"\"\"\nquality1 = openAQ.query_to_pandas(query)",
"_____no_output_____"
],
[
"# plot\nf,ax = plt.subplots(1,2, figsize= (14,6),sharey=True)\nax1,ax2 = ax.flatten()\nsns.barplot(quality['Year'],quality['Average'],ax=ax1)\nax1.set_title('Distribution of average AQI by year')\nsns.barplot(quality1['Month'],quality['Average'], ax=ax2 )\nax2.set_title('Distribution of average AQI by month')\nax2.set_ylabel('');",
"_____no_output_____"
],
[
"# by year & month\nquery = \"\"\"SELECT EXTRACT(YEAR from timestamp) as `Year`,\n EXTRACT(MONTH FROM timestamp) as `Month`,\n AVG(value) as `Average`\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY year,Month\"\"\"\naqi_year = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"# By month in year\nplt.figure(figsize=(14,8))\nsns.pointplot(aqi_year['Month'],aqi_year['Average'],hue = aqi_year['Year'])\nplt.title('Distribution of average AQI by month');",
"_____no_output_____"
]
],
[
[
"We find \n* the data available for perticular year is incomplete\n* the year 2016, 2017 data is availabel completely",
"_____no_output_____"
],
[
"### Country Heatmap",
"_____no_output_____"
]
],
[
[
"# Heatmap by country \nquery = \"\"\"SELECT AVG(value) as `Average`,\n EXTRACT(YEAR FROM timestamp) as `Year`,\n country\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY country,Year\n \"\"\"\ncoun_aqi = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"coun_pivot = coun_aqi.pivot(index='country', columns='Year', values='Average').fillna(0)\n# By month in year\nplt.figure(figsize=(14,15))\nsns.heatmap(coun_pivot, lw=0.01, cmap=sns.color_palette('Reds',len(coun_pivot)))\nplt.yticks(rotation=30)\nplt.title('Heatmap average AQI by YEAR');",
"_____no_output_____"
]
],
[
[
"### Animation",
"_____no_output_____"
]
],
[
[
"query = \"\"\"SELECT EXTRACT(YEAR FROM timestamp) as `Year`,AVG(value) as `Average`,\n latitude,longitude\n FROM `bigquery-public-data.openaq.global_air_quality`\n WHERE unit = 'µg/m³' AND value BETWEEN 0 AND 10000\n GROUP BY Year, latitude,longitude\n \"\"\"\np1 = openAQ.query_to_pandas_safe(query)",
"_____no_output_____"
],
[
"from matplotlib import animation,rc\nimport io\nimport base64\nfrom IPython.display import HTML, display\nimport warnings\nwarnings.filterwarnings('ignore')\nfig = plt.figure(figsize=(14,10))\nplt.style.use('ggplot')\n\ndef animate(Year):\n ax = plt.axes()\n ax.clear()\n ax.set_title('Average AQI in Year: '+str(Year))\n m4 = Basemap(llcrnrlat=-90, urcrnrlat=90, llcrnrlon=-180,urcrnrlon=180,projection='cyl')\n m4.drawmapboundary(fill_color='#A6CAE0', linewidth=0)\n m4.fillcontinents(color='grey', alpha=0.3)\n m4.drawcoastlines(linewidth=0.1, color=\"white\")\n m4.shadedrelief()\n \n lat_y = list(p1[p1['Year'] == Year]['latitude'])\n lon_y = list(p1[p1['Year'] == Year]['longitude'])\n lat,lon = m4(lat_y,lon_y) \n avg = p1[p1['Year'] == Year]['Average']\n m4.scatter(lon,lat,c = avg,lw=2, alpha=0.3,cmap='hot_r')\n \n \nani = animation.FuncAnimation(fig,animate,list(p1['Year'].unique()), interval = 1500) \nani.save('animation.gif', writer='imagemagick', fps=1)\nplt.close(1)\nfilename = 'animation.gif'\nvideo = io.open(filename, 'r+b').read()\nencoded = base64.b64encode(video)\nHTML(data='''<img src=\"data:image/gif;base64,{0}\" type=\"gif\" />'''.format(encoded.decode('ascii')))",
"_____no_output_____"
],
[
"# Continued",
"_____no_output_____"
]
],
[
[
">>>>>> ### Thank you for visiting, please upvote if you like it. ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
]
|
cb6e9140c718f30d2c368372ebfa1786fcab69cb | 18,047 | ipynb | Jupyter Notebook | datasets_wcbc.ipynb | Chuyi1202/T81-558-Application-of-Deep-Neural-Networks | 55e2a272fb006237f776db4b63c980eff5a1d295 | [
"Apache-2.0"
]
| 1 | 2022-03-15T07:00:37.000Z | 2022-03-15T07:00:37.000Z | datasets_wcbc.ipynb | frankalcantara/t81_558_deep_learning | 0eeac399398a52a211a1ecdc0f65d6863aa8a9ae | [
"Apache-2.0"
]
| null | null | null | datasets_wcbc.ipynb | frankalcantara/t81_558_deep_learning | 0eeac399398a52a211a1ecdc0f65d6863aa8a9ae | [
"Apache-2.0"
]
| 1 | 2019-09-01T11:11:09.000Z | 2019-09-01T11:11:09.000Z | 39.663736 | 287 | 0.365989 | [
[
[
"# Breast Cancer Wisconsin (Diagnostic) Data Set\n* **[T81-558: Applications of Deep Learning](https://sites.wustl.edu/jeffheaton/t81-558/)**\n* Dataset provided by [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+%28Diagnostic%29)\n* [Download Here](https://raw.githubusercontent.com/jeffheaton/t81_558_deep_learning/master/data/wcbreast.csv)\n\nThis is a popular dataset that contains columns that might be useful to determine if a tumor is breast cancer or not. There are a total of 32 columns and 569 rows. This dataset is used in class to introduce binary (two class) classification. The following fields are present:\n\n* **id** - Identity column, not really useful to a neural network.\n* **diagnosis** - Diagnosis, B=Benign, M=Malignant.\n* **mean_radius** - Potentially predictive field.\n* **mean_texture** - Potentially predictive field.\n* **mean_perimeter** - Potentially predictive field.\n* **mean_area** - Potentially predictive field.\n* **mean_smoothness** - Potentially predictive field.\n* **mean_compactness** - Potentially predictive field.\n* **mean_concavity** - Potentially predictive field.\n* **mean_concave_points** - Potentially predictive field.\n* **mean_symmetry** - Potentially predictive field.\n* **mean_fractal_dimension** - Potentially predictive field.\n* **se_radius** - Potentially predictive field.\n* **se_texture** - Potentially predictive field.\n* **se_perimeter** - Potentially predictive field.\n* **se_area** - Potentially predictive field.\n* **se_smoothness** - Potentially predictive field.\n* **se_compactness** - Potentially predictive field.\n* **se_concavity** - Potentially predictive field.\n* **se_concave_points** - Potentially predictive field.\n* **se_symmetry** - Potentially predictive field.\n* **se_fractal_dimension** - Potentially predictive field.\n* **worst_radius** - Potentially predictive field.\n* **worst_texture** - Potentially predictive field.\n* **worst_perimeter** - Potentially predictive field.\n* **worst_area** - Potentially predictive field.\n* **worst_smoothness** - Potentially predictive field.\n* **worst_compactness** - Potentially predictive field.\n* **worst_concavity** - Potentially predictive field.\n* **worst_concave_points** - Potentially predictive field.\n* **worst_symmetry** - Potentially predictive field.\n* **worst_fractal_dimension** - Potentially predictive field.\n\n\nThe following code shows 10 sample rows.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\npath = \"./data/\"\n \nfilename = os.path.join(path,\"wcbreast_wdbc.csv\")\ndf = pd.read_csv(filename,na_values=['NA','?'])\n\n# Shuffle\nnp.random.seed(42)\ndf = df.reindex(np.random.permutation(df.index))\ndf.reset_index(inplace=True, drop=True)\n\ndf[0:10]\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
]
]
|
cb6e933de6959ebaa19951be3f4f397757e3f740 | 6,019 | ipynb | Jupyter Notebook | Lectures_Advanced-DSP/index.ipynb | lev1khachatryan/ASDS_DSP | 9059d737f6934b81a740c79b33756f7ec9ededb3 | [
"MIT"
]
| 1 | 2020-12-29T18:02:13.000Z | 2020-12-29T18:02:13.000Z | Lectures_Advanced-DSP/index.ipynb | lev1khachatryan/ASDS_DSP | 9059d737f6934b81a740c79b33756f7ec9ededb3 | [
"MIT"
]
| null | null | null | Lectures_Advanced-DSP/index.ipynb | lev1khachatryan/ASDS_DSP | 9059d737f6934b81a740c79b33756f7ec9ededb3 | [
"MIT"
]
| null | null | null | 49.336066 | 443 | 0.710915 | [
[
[
"# Digital Signal Processing\n\nThis collection of [jupyter](https://jupyter.org/) notebooks introduces various topics of [Digital Signal Processing](https://en.wikipedia.org/wiki/Digital_signal_processing). The theory is accompanied by computational examples written in [IPython 3](http://ipython.org/). The sources of the notebooks, as well as installation and usage instructions can be found on [GitHub](https://github.com/lev1khachatryan/Digital_Signal_Processing).",
"_____no_output_____"
],
[
"## Table of Contents\n\n#### 1. Introduction\n\n* [Introduction](introduction/introduction.ipynb)\n\n#### 2. Spectral Analysis of Deterministic Signals\n\n* [The Leakage-Effect](spectral_analysis_deterministic_signals/leakage_effect.ipynb)\n* [Window Functions](spectral_analysis_deterministic_signals/window_functions.ipynb)\n* [Zero-Padding](spectral_analysis_deterministic_signals/zero_padding.ipynb)\n* [Short-Time Fourier Transform](spectral_analysis_deterministic_signals/stft.ipynb)\n* [Summary](spectral_analysis_deterministic_signals/summary.ipynb)\n\n#### 3. Random Signals\n\n* [Introduction](random_signals/introduction.ipynb)\n* [Amplitude Distributions](random_signals/distributions.ipynb)\n* [Ensemble Averages](random_signals/ensemble_averages.ipynb)\n* [Stationary and Ergodic Processes](random_signals/stationary_ergodic.ipynb)\n* [Correlation Functions](random_signals/correlation_functions.ipynb)\n* [Power Spectral Densities](random_signals/power_spectral_densities.ipynb)\n* [Independent Processes](random_signals/independent.ipynb)\n* [Important Amplitude Distributions](random_signals/important_distributions.ipynb)\n* [White Noise](random_signals/white_noise.ipynb)\n* [Superposition of Random Signals](random_signals/superposition.ipynb)\n\n#### 4. Random Signals and LTI Systems\n\n* [Introduction](random_signals_LTI_systems/introduction.ipynb)\n* [Linear Mean](random_signals_LTI_systems/linear_mean.ipynb)\n* [Correlation Functions](random_signals_LTI_systems/correlation_functions.ipynb)\n* [Example: Measurement of Acoustic Impulse Responses](random_signals_LTI_systems/acoustic_impulse_response_measurement.ipynb)\n* [Power Spectral Densities](random_signals_LTI_systems/power_spectral_densities.ipynb)\n* [Wiener Filter](random_signals_LTI_systems/wiener_filter.ipynb)\n\n#### 5. Spectral Estimation of Random Signals\n\n* [Introduction](spectral_estimation_random_signals/introduction.ipynb)\n* [Periodogram](spectral_estimation_random_signals/periodogram.ipynb)\n* [Welch-Method](spectral_estimation_random_signals/welch_method.ipynb)\n* [Parametric Methods](spectral_estimation_random_signals/parametric_methods.ipynb)\n\n#### 6. Quantization\n\n* [Introduction](quantization/introduction.ipynb)\n* [Characteristic of Linear Uniform Quantization](quantization/linear_uniform_characteristic.ipynb)\n* [Quantization Error of Linear Uniform Quantization](quantization/linear_uniform_quantization_error.ipynb)\n* [Example: Requantization of a Speech Signal](quantization/requantization_speech_signal.ipynb)\n* [Noise Shaping](quantization/noise_shaping.ipynb)\n* [Oversampling](quantization/oversampling.ipynb)\n* [Example: Non-Linear Quantization of a Speech Signal](quantization/nonlinear_quantization_speech_signal.ipynb)\n\n#### 7. Realization of Non-Recursive Filters\n\n* [Introduction](nonrecursive_filters/introduction.ipynb)\n* [Fast Convolution](nonrecursive_filters/fast_convolution.ipynb)\n* [Segmented Convolution](nonrecursive_filters/segmented_convolution.ipynb)\n* [Quantization Effects](nonrecursive_filters/quantization_effects.ipynb)\n\n#### 8. Realization of Recursive Filters\n\n* [Introduction](recursive_filters/introduction.ipynb)\n* [Direct Form Structures](recursive_filters/direct_forms.ipynb)\n* [Cascaded Structures](recursive_filters/cascaded_structures.ipynb)\n* [Quantization of Filter Coefficients](recursive_filters/quantization_of_coefficients.ipynb)\n* [Quantization of Variables and Operations](recursive_filters/quantization_of_variables.ipynb)\n\n#### 9. Design of Digital Filters\n\n* [Design of Non-Recursive Filters by the Window Method](filter_design/window_method.ipynb)\n* [Design of Non-Recursive Filters by the Frequency Sampling Method](filter_design/frequency_sampling_method.ipynb)\n* [Design of Recursive Filters by the Bilinear Transform](filter_design/bilinear_transform.ipynb)\n* [Example: Non-Recursive versus Recursive Filter](filter_design/comparison_non_recursive.ipynb)\n* [Examples: Typical IIR-Filters in Audio](filter_design/audiofilter.ipynb)\n\n#### Reference Cards\n\n* [Reference Card Discrete Signals and Systems](reference_cards/RC_discrete_signals_and_systems.pdf)\n* [Reference Card Random Signals and LTI Systems](reference_cards/RC_random_signals_and_LTI_systems.pdf)",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown"
]
]
|
cb6e957ecfeed358d958a0ba4bd574222facd4d9 | 18,667 | ipynb | Jupyter Notebook | notebooks/raiderDownloadGNSS/.ipynb_checkpoints/raiderDownloadGNSS_tutorial-checkpoint.ipynb | sssangha/RAiDER-docs | 9f5e882a8abc3cb6a51672b8d8d257d5d9630b73 | [
"Apache-2.0"
]
| 1 | 2021-04-22T16:05:40.000Z | 2021-04-22T16:05:40.000Z | notebooks/raiderDownloadGNSS/.ipynb_checkpoints/raiderDownloadGNSS_tutorial-checkpoint.ipynb | sssangha/RAiDER-docs | 9f5e882a8abc3cb6a51672b8d8d257d5d9630b73 | [
"Apache-2.0"
]
| 4 | 2020-07-23T10:25:18.000Z | 2021-02-23T16:23:04.000Z | notebooks/raiderDownloadGNSS/.ipynb_checkpoints/raiderDownloadGNSS_tutorial-checkpoint.ipynb | sssangha/RAiDER-docs | 9f5e882a8abc3cb6a51672b8d8d257d5d9630b73 | [
"Apache-2.0"
]
| 6 | 2020-06-13T19:39:10.000Z | 2021-03-23T05:40:56.000Z | 30.40228 | 419 | 0.617935 | [
[
[
"# Downloading GNSS station locations and tropospheric zenith delays\n\n**Author**: Simran Sangha, David Bekaert - Jet Propulsion Laboratory\n\nThis notebook provides an overview of the functionality included in the **`raiderDownloadGNSS.py`** program. Specifically, we outline examples on how to access and store GNSS station location and tropospheric zenith delay information over a user defined area of interest and span of time. In this notebook, we query GNSS stations spanning northern California between 2016 and 2019. \n\nWe will outline the following downloading options to access station location and zenith delay information:\n- For a specified range of years\n- For a specified time of day\n- Confined to a specified geographic bounding box\n- Confined to an apriori defined list of GNSS stations\n\n<div class=\"alert alert-info\">\n <b>Terminology:</b>\n \n- *GNSS*: Stands for Global Navigation Satellite System. Describes any satellite constellation providing global or regional positioning, navigation, and timing services.\n- *tropospheric zenith delay*: The precise atmospheric delay satellite signals experience when propagating through the troposphere.\n \n </div>\n ",
"_____no_output_____"
],
[
"## Table of Contents:\n<a id='example_TOC'></a>",
"_____no_output_____"
],
[
"[**Overview of the raiderDownloadGNSS.py program**](#overview)\n- [1. Define spatial extent and/or apriori list of stations](#overview_1)\n- [2. Run parameters](#overview_2)\n\n[**Examples of the raiderDownloadGNSS.py program**](#examples)\n- [Example 1. Access data for specified year, time-step, and time of day, and across specified spatial subset](#example_1)\n- [Example 2. Access data for specified range of years and time of day, and across specified spatial subset, with the maximum allowed CPUs](#example_2)",
"_____no_output_____"
],
[
"## Prep: Initial setup of the notebook",
"_____no_output_____"
],
[
"Below we set up the directory structure for this notebook exercise. In addition, we load the required modules into our python environment using the **`import`** command.",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n## Defining the home and data directories\ntutorial_home_dir = os.path.abspath(os.getcwd())\nwork_dir = os.path.abspath(os.getcwd())\nprint(\"Tutorial directory: \", tutorial_home_dir)\nprint(\"Work directory: \", work_dir)\n\n# Verifying if RAiDER is installed correctly\ntry:\n from RAiDER import downloadGNSSDelays\nexcept:\n raise Exception('RAiDER is missing from your PYTHONPATH')\n\nos.chdir(work_dir)",
"_____no_output_____"
]
],
[
[
"# Supported GNSS provider\nCurrently **`raiderDownloadGNSS.py`** is able to access the UNR Geodetic Laboratory GNSS archive. The creation of a user account and/or special privileges are not necessary.\nData naming conventions are outlined here: http://geodesy.unr.edu/gps_timeseries/README_trop2.txt\nThis archive does not require a license agreement nor a setup of a user account.",
"_____no_output_____"
],
[
"## Overview of the raiderDownloadGNSS.py program\n<a id='overview'></a>",
"_____no_output_____"
],
[
"The **`raiderDownloadGNSS.py`** program allows for easy access of GNSS station locations and tropospheric zenith delays. Running **`raiderDownloadGNSS.py`** with the **`-h`** option will show the parameter options and outline several basic, practical examples. \n\nLet us explore these options:",
"_____no_output_____"
]
],
[
[
"!raiderDownloadGNSS.py -h",
"_____no_output_____"
]
],
[
[
"### 1. Define spatial extent and/or apriori list of stations\n<a id='overview_1'></a>",
"_____no_output_____"
],
[
"#### Geographic bounding box (**`--bounding_box BOUNDING_BOX`**)",
"_____no_output_____"
],
[
"An area of interest may be specified as `SNWE` coordinates using the **`--bounding_box`** option. Coordinates should be specified as a space delimited string surrounded by quotes. This example below would restrict the query to stations over northern California:\n**`--bounding_box '36 40 -124 -119'`**\n\nIf no area of interest is specified, the entire global archive will be queried.",
"_____no_output_____"
],
[
"#### Textfile with apriori list of station names (**`--station_file STATION_FILE`**)",
"_____no_output_____"
],
[
"The query may be restricted to an apropri list of stations. To pass this list to the program, a text file containing a list of 4-char station IDs separated by newlines must be passed as an argument for the **`--station_file`** option.\n\nIf used in conjunction with the **`--bounding_box`** option outlined above, then listed stations which fall outside of the specified geographic bounding box will be discarded.\n\nAs an example refer to the text-file below, which would be passed as so: **`--station_file support_docs/CA_subset.txt`**",
"_____no_output_____"
]
],
[
[
"!head support_docs/CA_subset.txt",
"_____no_output_____"
]
],
[
[
"### 2. Run parameters\n<a id='overview_2'></a>",
"_____no_output_____"
],
[
"#### Output directory (**`--out OUT`**)",
"_____no_output_____"
],
[
"Specify directory to deposit all outputs into with **`--out`**. Absolute and relative paths are both supported.\n\nBy default, outputs will be deposited into the current working directory where the program is launched.",
"_____no_output_____"
],
[
"#### GPS repository (**`--gpsrepo GPS_REPO`**)",
"_____no_output_____"
],
[
"Specify GPS repository you wish to query with **`--gpsrepo`**.\n\nNOTE that currently only the following archive is supported: UNR",
"_____no_output_____"
],
[
"#### Date(s) and step (**`----date DATELIST [DATELIST ...]`**)",
"_____no_output_____"
],
[
"**REQUIRED** argument. Specify valid year(s) and step in days **`--date DATE DATE STEP`** to access delays (format YYYYMMDD YYYYMMDD DD). Can be a single date (e.g. '20200101'), two dates between which data for every day between and inclusive is queried (e.g. '2017 2019'), or two dates and a step for which increment in days data is queried (e.g. '2019 2019 12').\n\nNote that this option mirrors a similar option as found in the script `raiderDelay.py`, is used to download weather model data for specified spatiotemporal constraints (i.e. the counterpart to the `raiderDownloadGNSS.py` which downloads GNSS data).",
"_____no_output_____"
],
[
"#### Time of day (**`--returntime RETURNTIME`**)",
"_____no_output_____"
],
[
"Return tropospheric zenith delays closest to 'HH:MM:SS' time specified with **`--returntime`**. \n\nNote that data is generally archived in 3 second increments. Thus if a time outside of this increment is specified (e.g. '00:00:02'), then the input is rounded to the closest 3 second increment (e.g. '00:00:03')\n\nIf not specified, the delays for all times of the day will be returned.",
"_____no_output_____"
],
[
"#### Physically download data (**`--download`**)",
"_____no_output_____"
],
[
"By default all data is virtually accessed from external zip and tarfiles. If **`--download`** is specified, these external files will be locally downloaded and stored. \n\nNote that this option is **not recommended** for most purposes as it is not neccesary to proceed with statistical analyses, as the code is designed to handle the data virtually.",
"_____no_output_____"
],
[
"#### Number of CPUs to be used (**`--cpus NUMCPUS`**)",
"_____no_output_____"
],
[
"Specify number of cpus to be used for multiprocessing with **`--cpus`**. For most cases, multiprocessing is essential in order to access data and perform statistical analyses within a reasonable amount of time.\n\nMay specify **`--cpus all`** at your own discretion in order to leverage all available CPUs on your system.\n\nBy default 8 CPUs will be used.",
"_____no_output_____"
],
[
"#### Verbose mode (**`--verbose`**)",
"_____no_output_____"
],
[
"Specify **`--verbose`** to print all statements through entire routine. For example, print each station and year within a loop as it is being accessed by the program.",
"_____no_output_____"
],
[
"## Examples of the **`raiderDownloadGNSS.py`** program\n<a id='examples'></a>",
"_____no_output_____"
],
[
"### Example 1. Access data for specified year, time-step, and time of day, and across specified spatial subset <a id='example_1'></a>",
"_____no_output_____"
],
[
"Virtually access GNSS station location and zenith delay information for the year '2016', for every 12 days, and at a UTC time of day 'HH:MM:SS' of '00:00:00', and across a geographic bounding box '36 40 -124 -119' spanning over Northern California.\n\nThe footprint of the specified geographic bounding box is depicted in **Fig. 1**.",
"_____no_output_____"
],
[
"<img src=\"support_docs/bbox_footprint.png\" alt=\"footprint\" width=\"700\">\n<center><b>Fig. 1</b> Footprint of geopraphic bounding box used in examples 1 and 2. </center>",
"_____no_output_____"
]
],
[
[
"!raiderDownloadGNSS.py --out products --date 20160101 20161231 12 --returntime '00:00:00' --bounding_box '36 40 -124 -119'",
"_____no_output_____"
]
],
[
[
"Now we can take a look at the generated products:",
"_____no_output_____"
]
],
[
[
"!ls products",
"_____no_output_____"
]
],
[
[
"A list of coordinates for all stations found within the specified geographic bounding box are recorded within **`gnssStationList_overbbox.csv`**:",
"_____no_output_____"
]
],
[
[
"!head products/gnssStationList_overbbox.csv",
"_____no_output_____"
]
],
[
[
"A list of all URL paths for zipfiles containing all tropospheric zenith delay information for a given station and year are recording within **`gnssStationList_overbbox_withpaths.csv`**:",
"_____no_output_____"
]
],
[
[
"!head products/gnssStationList_overbbox_withpaths.csv",
"_____no_output_____"
]
],
[
[
"The zipfiles listed within **`gnssStationList_overbbox_withpaths.csv`** are virtually accessed and queried for internal tarfiles that archive all tropospheric zenith delay information acquired for a given day of the year. \n\nSince we an explicit time of day '00:00:00' and time-step of 12 days was specified above, only data every 12 days from each tarfile corresponding to the time of day '00:00:00' is passed along. If no data is available at that time for a given day, empty strings are passed.\n\nThis information is then appended to a primary file allocated and named for a given GNSS station. **`GPS_delays`**:",
"_____no_output_____"
]
],
[
[
"!ls products/GPS_delays",
"_____no_output_____"
]
],
[
[
"Finally, all of the extracted tropospheric zenith delay information stored under **`GPS_delays`** is concatenated with the GNSS station location information stored under **`gnssStationList_overbbox.csv`** into a primary comprehensive file **`UNRcombinedGPS_ztd.csv`**. In this file, the prefix `UNR` denotes the GNSS repository that has been queried, which again may be toggled with the **`--gpsrepo`** option.\n\n**`UNRcombinedGPS_ztd.csv`** may in turn be directly used to perform basic statistical analyses using **`raiderStats.py`**. Please refer to the companion notebook **`raiderStats/raiderStats_tutorial.ipynb`** for a comprehensive outline of the program and examples.",
"_____no_output_____"
]
],
[
[
"!head products/UNRcombinedGPS_ztd.csv",
"_____no_output_____"
]
],
[
[
"### Example 2. Access data for specified range of years and time of day, and across specified spatial subset, with the maximum allowed CPUs <a id='example_2'></a>",
"_____no_output_____"
],
[
"Virtually access GNSS station location and zenith delay information for the years '2016-2019', for every day, at a UTC time of day 'HH:MM:SS' of '00:00:00', and across a geographic bounding box '36 40 -124 -119' spanning over Northern California.\n\nThe footprint of the specified geographic bounding box is again depicted in **Fig. 1**.\n\nIn addition to querying for multiple years, we will also experiment with using the maximum number of allowed CPUs to save some time! Recall again that the default number of CPUs used for parallelization is 8.",
"_____no_output_____"
]
],
[
[
"!rm -rf products\n!raiderDownloadGNSS.py --out products --date 20160101 20191231 --returntime '00:00:00' --bounding_box '36 40 -124 -119' --cpus all",
"_____no_output_____"
]
],
[
[
"Outputs are organized again in a fashion consistent with that outlined under **Ex. 1**.\n\nHowever now we have queried data spanning from the year 2016 up through 2019. Thus, **`UNRcombinedGPS_ztd.csv`** now contains GNSS station data recorded as late as in the year 2019:",
"_____no_output_____"
]
],
[
[
"!grep -m 10 '2019-' products/UNRcombinedGPS_ztd.csv",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6e96ee54d480c875b836343359836375e207a5 | 11,171 | ipynb | Jupyter Notebook | 4. Get Started! Handwritting Recognition, Simple Object Classification & OpenCV Demo/4.1 - Handwritten Digit Classification Demo (MNIST).ipynb | madhavjk/Computer-Vision-OpenCV | 9b88ae9c31008b5836cbfb8e90b952c5eec15318 | [
"Apache-2.0"
]
| null | null | null | 4. Get Started! Handwritting Recognition, Simple Object Classification & OpenCV Demo/4.1 - Handwritten Digit Classification Demo (MNIST).ipynb | madhavjk/Computer-Vision-OpenCV | 9b88ae9c31008b5836cbfb8e90b952c5eec15318 | [
"Apache-2.0"
]
| null | null | null | 4. Get Started! Handwritting Recognition, Simple Object Classification & OpenCV Demo/4.1 - Handwritten Digit Classification Demo (MNIST).ipynb | madhavjk/Computer-Vision-OpenCV | 9b88ae9c31008b5836cbfb8e90b952c5eec15318 | [
"Apache-2.0"
]
| null | null | null | 35.239748 | 137 | 0.539253 | [
[
[
"### Let's load a Handwritten Digit classifier we'll be building very soon!",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nfrom keras.datasets import mnist\nfrom keras.models import load_model\n\nclassifier = load_model('/home/deeplearningcv/DeepLearningCV/Trained Models/mnist_simple_cnn.h5')\n\n# loads the MNIST dataset\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\n\ndef draw_test(name, pred, input_im):\n BLACK = [0,0,0]\n expanded_image = cv2.copyMakeBorder(input_im, 0, 0, 0, imageL.shape[0] ,cv2.BORDER_CONSTANT,value=BLACK)\n expanded_image = cv2.cvtColor(expanded_image, cv2.COLOR_GRAY2BGR)\n cv2.putText(expanded_image, str(pred), (152, 70) , cv2.FONT_HERSHEY_COMPLEX_SMALL,4, (0,255,0), 2)\n cv2.imshow(name, expanded_image)\n\nfor i in range(0,10):\n rand = np.random.randint(0,len(x_test))\n input_im = x_test[rand]\n\n imageL = cv2.resize(input_im, None, fx=4, fy=4, interpolation = cv2.INTER_CUBIC) \n input_im = input_im.reshape(1,28,28,1) \n \n ## Get Prediction\n res = str(classifier.predict_classes(input_im, 1, verbose = 0)[0])\n draw_test(\"Prediction\", res, imageL) \n cv2.waitKey(0)\n\ncv2.destroyAllWindows()",
"Using TensorFlow backend.\n"
]
],
[
[
"### Testing our classifier on a real image",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport cv2\nfrom preprocessors import x_cord_contour, makeSquare, resize_to_pixel\n \nimage = cv2.imread('images/numbers.jpg')\ngray = cv2.cvtColor(image,cv2.COLOR_BGR2GRAY)\ncv2.imshow(\"image\", image)\ncv2.waitKey(0)\n\n# Blur image then find edges using Canny \nblurred = cv2.GaussianBlur(gray, (5, 5), 0)\n#cv2.imshow(\"blurred\", blurred)\n#cv2.waitKey(0)\n\nedged = cv2.Canny(blurred, 30, 150)\n#cv2.imshow(\"edged\", edged)\n#cv2.waitKey(0)\n\n# Find Contours\n_, contours, _ = cv2.findContours(edged.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)\n\n#Sort out contours left to right by using their x cordinates\ncontours = sorted(contours, key = x_cord_contour, reverse = False)\n\n# Create empty array to store entire number\nfull_number = []\n\n# loop over the contours\nfor c in contours:\n # compute the bounding box for the rectangle\n (x, y, w, h) = cv2.boundingRect(c) \n\n if w >= 5 and h >= 25:\n roi = blurred[y:y + h, x:x + w]\n ret, roi = cv2.threshold(roi, 127, 255,cv2.THRESH_BINARY_INV)\n roi = makeSquare(roi)\n roi = resize_to_pixel(28, roi)\n cv2.imshow(\"ROI\", roi)\n roi = roi / 255.0 \n roi = roi.reshape(1,28,28,1) \n\n ## Get Prediction\n res = str(classifier.predict_classes(roi, 1, verbose = 0)[0])\n full_number.append(res)\n cv2.rectangle(image, (x, y), (x + w, y + h), (0, 0, 255), 2)\n cv2.putText(image, res, (x , y + 155), cv2.FONT_HERSHEY_COMPLEX, 2, (255, 0, 0), 2)\n cv2.imshow(\"image\", image)\n cv2.waitKey(0) \n \ncv2.destroyAllWindows()\nprint (\"The number is: \" + ''.join(full_number))",
"The number is: 13540\n"
]
],
[
[
"### Training this Model",
"_____no_output_____"
]
],
[
[
"from keras.datasets import mnist\nfrom keras.utils import np_utils\nimport keras\nfrom keras.datasets import mnist\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Flatten\nfrom keras.layers import Conv2D, MaxPooling2D\nfrom keras import backend as K\n\n# Training Parameters\nbatch_size = 128\nepochs = 5\n\n# loads the MNIST dataset\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\n\n# Lets store the number of rows and columns\nimg_rows = x_train[0].shape[0]\nimg_cols = x_train[1].shape[0]\n\n# Getting our date in the right 'shape' needed for Keras\n# We need to add a 4th dimenion to our date thereby changing our\n# Our original image shape of (60000,28,28) to (60000,28,28,1)\nx_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)\nx_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)\n\n# store the shape of a single image \ninput_shape = (img_rows, img_cols, 1)\n\n# change our image type to float32 data type\nx_train = x_train.astype('float32')\nx_test = x_test.astype('float32')\n\n# Normalize our data by changing the range from (0 to 255) to (0 to 1)\nx_train /= 255\nx_test /= 255\n\nprint('x_train shape:', x_train.shape)\nprint(x_train.shape[0], 'train samples')\nprint(x_test.shape[0], 'test samples')\n\n# Now we one hot encode outputs\ny_train = np_utils.to_categorical(y_train)\ny_test = np_utils.to_categorical(y_test)\n\n# Let's count the number columns in our hot encoded matrix \nprint (\"Number of Classes: \" + str(y_test.shape[1]))\n\nnum_classes = y_test.shape[1]\nnum_pixels = x_train.shape[1] * x_train.shape[2]\n\n# create model\nmodel = Sequential()\n\nmodel.add(Conv2D(32, kernel_size=(3, 3),\n activation='relu',\n input_shape=input_shape))\nmodel.add(Conv2D(64, (3, 3), activation='relu'))\nmodel.add(MaxPooling2D(pool_size=(2, 2)))\nmodel.add(Dropout(0.25))\nmodel.add(Flatten())\nmodel.add(Dense(128, activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(num_classes, activation='softmax'))\n\nmodel.compile(loss = 'categorical_crossentropy',\n optimizer = keras.optimizers.Adadelta(),\n metrics = ['accuracy'])\n\nprint(model.summary())\n\nhistory = model.fit(x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n validation_data=(x_test, y_test))\n\nscore = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Using TensorFlow backend.\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6ea703449cb376deb384587ee92ce1464e845e | 10,369 | ipynb | Jupyter Notebook | 22-workout-solution_vectorized_string_operations.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
]
| 6 | 2020-04-13T19:22:18.000Z | 2021-04-20T18:20:13.000Z | 22-workout-solution_vectorized_string_operations.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
]
| null | null | null | 22-workout-solution_vectorized_string_operations.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
]
| 10 | 2020-05-12T01:02:32.000Z | 2022-02-28T17:04:37.000Z | 25.045894 | 82 | 0.417109 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"data = pd.read_csv('data/EC10.csv')",
"_____no_output_____"
],
[
"data.head()\n",
"_____no_output_____"
]
],
[
[
"1- Which is the most verbose sector (uses more words than others)",
"_____no_output_____"
]
],
[
[
"data['words'] = data['text'].str.split().str.len()",
"_____no_output_____"
],
[
"s = data.groupby('sector')['words'].sum()\ns",
"_____no_output_____"
],
[
"s.sort_values(ascending=False).index[0]",
"_____no_output_____"
]
],
[
[
"2- count the number of companies in each stock exchange",
"_____no_output_____"
]
],
[
[
"df=data['text'].str.extract(\"([A-Z]+):([A-Z]+)\")\ndf.columns = ['Stock market', 'Company']\ndf.groupby('Stock market').count()",
"_____no_output_____"
]
],
[
[
"3- What are the top 10 most frequent words?",
"_____no_output_____"
]
],
[
[
"from collections import Counter",
"_____no_output_____"
],
[
"Counter(['Hi', \"Hi\", \"There\"])",
"_____no_output_____"
],
[
"all_words = data['text'].str.split().sum()\ncount = Counter(all_words)",
"_____no_output_____"
],
[
"pd.Series(count).sort_values(ascending=False)[:10]",
"_____no_output_____"
]
],
[
[
"4- What are the top 10 most frequent words per sector?",
"_____no_output_____"
]
],
[
[
"def count_words(data_frame):\n all_words = data_frame['text'].str.split().sum()\n count = Counter(all_words)\n return pd.Series(count).sort_values(ascending=False)[:10].to_dict()",
"_____no_output_____"
],
[
"data.groupby('sector').apply(count_words)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
cb6ec754ea38d5a32874f84de3b2b7ebddeb615c | 35,248 | ipynb | Jupyter Notebook | notebooks/gizmo_read_tutorial.ipynb | caganze/popsims | 3fe2b62cd9c08b3b36c93d7c5a103f6f50965bf6 | [
"MIT"
]
| null | null | null | notebooks/gizmo_read_tutorial.ipynb | caganze/popsims | 3fe2b62cd9c08b3b36c93d7c5a103f6f50965bf6 | [
"MIT"
]
| null | null | null | notebooks/gizmo_read_tutorial.ipynb | caganze/popsims | 3fe2b62cd9c08b3b36c93d7c5a103f6f50965bf6 | [
"MIT"
]
| null | null | null | 29.845893 | 279 | 0.54369 | [
[
[
"# tutorial for reading a Gizmo snapshot\n\n@author: Andrew Wetzel <[email protected]>",
"_____no_output_____"
]
],
[
[
"# First, move within a simulation directory, or point 'directory' below to a simulation directory.\n# This directory should contain either a snapshot file\n# snapshot_???.hdf5\n# or a snapshot directory\n# snapdir_???\n\n# In general, the simulation directory also should contain a text file:\n# m12*_center.txt\n# that contains pre-computed galaxy center coordinates\n# and rotation vectors to align with the principal axes of the galaxy,\n# although that file is not required to read a snapshot.\n\n# The simulation directory also may contain text files:\n# m12*_LSR{0,1,2}.txt\n# that contains the local standard of rest (LSR) coordinates\n# used by Ananke in creating Gaia synthetic surveys.",
"_____no_output_____"
],
[
"# Ensure that your python path points to this python package, then:\n\nimport gizmo_read",
"_____no_output_____"
],
[
"directory = '.' # if running this notebook from within a simulation directory\n#directory = 'm12i/' # if running higher-level directory\n#directory = 'm12f/' # if running higher-level directory\n#directory = 'm12m/' # if running higher-level directory",
"_____no_output_____"
]
],
[
[
"# read particle data from a snapshot",
"_____no_output_____"
]
],
[
[
"# read star particles (all properties)\n\npart = gizmo_read.read.Read.read_snapshot(species='star', directory=directory)",
"reading header from:\n snapdir_600/snapshot_600.0.hdf5\n\nsnapshot contains the following number of particles:\n star (id = 4): 13976485 particles\n\nreading star properties:\n ['form.scalefactor', 'id', 'mass', 'massfraction', 'position', 'potential', 'velocity']\n\nreading particles from:\n snapshot_600.0.hdf5\n snapshot_600.1.hdf5\n snapshot_600.2.hdf5\n snapshot_600.3.hdf5\n\nreading galaxy center coordinates and principal axes from: m12i_res7100_center.txt\n center position [kpc] = 41792.145, 44131.235, 46267.676\n center velocity [km/s] = -52.5, 71.9, 95.2\n\nadjusting particle coordinates to be relative to galaxy center\n and aligned with the principal axes\n\n"
],
[
"# alternately, read all particle species (stars, gas, dark matter)\n\npart = gizmo_read.read.Read.read_snapshot(species='all', directory=directory)",
"reading header from:\n snapdir_600/snapshot_600.0.hdf5\n\nsnapshot contains the following number of particles:\n dark (id = 1): 70514272 particles\n dark.2 (id = 2): 5513331 particles\n gas (id = 0): 57060074 particles\n star (id = 4): 13976485 particles\n\nreading dark properties:\n ['id', 'mass', 'position', 'potential', 'velocity']\nreading dark.2 properties:\n ['id', 'mass', 'position', 'potential', 'velocity']\nreading gas properties:\n ['density', 'electron.fraction', 'hydrogen.neutral.fraction', 'id', 'mass', 'massfraction', 'position', 'potential', 'temperature', 'velocity']\nreading star properties:\n ['form.scalefactor', 'id', 'mass', 'massfraction', 'position', 'potential', 'velocity']\n\nreading particles from:\n snapshot_600.0.hdf5\n snapshot_600.1.hdf5\n snapshot_600.2.hdf5\n snapshot_600.3.hdf5\n\nreading galaxy center coordinates and principal axes from: m12i_res7100_center.txt\n center position [kpc] = 41792.145, 44131.235, 46267.676\n center velocity [km/s] = -52.5, 71.9, 95.2\n\nadjusting particle coordinates to be relative to galaxy center\n and aligned with the principal axes\n\n"
],
[
"# alternately, read just stars and dark matter (or any combination of species)\n\npart = gizmo_read.read.Read.read_snapshot(species=['star', 'dark'], directory=directory)",
"reading header from:\n snapdir_600/snapshot_600.0.hdf5\n\nsnapshot contains the following number of particles:\n star (id = 4): 13976485 particles\n dark (id = 1): 70514272 particles\n\nreading star properties:\n ['form.scalefactor', 'id', 'mass', 'massfraction', 'position', 'potential', 'velocity']\nreading dark properties:\n ['id', 'mass', 'position', 'potential', 'velocity']\n\nreading particles from:\n snapshot_600.0.hdf5\n snapshot_600.1.hdf5\n snapshot_600.2.hdf5\n snapshot_600.3.hdf5\n\nreading galaxy center coordinates and principal axes from: m12i_res7100_center.txt\n center position [kpc] = 41792.145, 44131.235, 46267.676\n center velocity [km/s] = -52.5, 71.9, 95.2\n\nadjusting particle coordinates to be relative to galaxy center\n and aligned with the principal axes\n\n"
],
[
"# alternately, read only a subset of particle properties (to save memory)\n\npart = gizmo_read.read.Read.read_snapshot(species='star', properties=['position', 'velocity', 'mass'], directory=directory)",
"reading header from:\n m12i/m12i_res7100/output/snapdir_600/snapshot_600.0.hdf5\n\nsnapshot contains the following number of particles:\n star (id = 4): 13976485 particles\n\nread star : ['mass', 'position', 'velocity']\nreading particles from:\n snapshot_600.0.hdf5\n snapshot_600.1.hdf5\n snapshot_600.2.hdf5\n snapshot_600.3.hdf5\n\nreading galaxy center coordinates and principal axes from:\n m12i/m12i_res7100/output/m12i_res7100_center.txt\n center position [kpc] = 41792.145, 44131.235, 46267.676\n center velocity [km/s] = -52.5, 71.9, 95.2\n\n"
],
[
"# also can use particle_subsample_factor to periodically sub-sample particles, to save memory\n\npart = gizmo_read.read.Read.read_snapshot(species='all', directory=directory, particle_subsample_factor=10)",
"reading header from:\n snapdir_600/snapshot_600.0.hdf5\n\nsnapshot contains the following number of particles:\n dark (id = 1): 70514272 particles\n dark.2 (id = 2): 5513331 particles\n gas (id = 0): 57060074 particles\n star (id = 4): 13976485 particles\n\nreading dark properties:\n ['id', 'mass', 'position', 'potential', 'velocity']\nreading dark.2 properties:\n ['id', 'mass', 'position', 'potential', 'velocity']\nreading gas properties:\n ['density', 'electron.fraction', 'hydrogen.neutral.fraction', 'id', 'mass', 'massfraction', 'position', 'potential', 'temperature', 'velocity']\nreading star properties:\n ['form.scalefactor', 'id', 'mass', 'massfraction', 'position', 'potential', 'velocity']\n\nreading particles from:\n snapshot_600.0.hdf5\n snapshot_600.1.hdf5\n snapshot_600.2.hdf5\n snapshot_600.3.hdf5\n\nperiodically subsampling all particles by factor = 10\n\nreading galaxy center coordinates and principal axes from: m12i_res7100_center.txt\n center position [kpc] = 41792.145, 44131.235, 46267.676\n center velocity [km/s] = -52.5, 71.9, 95.2\n\nadjusting particle coordinates to be relative to galaxy center\n and aligned with the principal axes\n\n"
]
],
[
[
"# species dictionary",
"_____no_output_____"
]
],
[
[
"# each particle species is stored as its own dictionary\n# 'star' = stars, 'gas' = gas, 'dark' = dark matter, 'dark.2' = low-resolution dark matter\n\npart.keys()",
"_____no_output_____"
],
[
"# properties of particles are stored as dictionary",
"_____no_output_____"
],
[
"# properties of star particles\n\nfor k in part['star'].keys():\n print(k)",
"position\nmass\nmassfraction\nid\npotential\nform.scalefactor\nvelocity\nage\nmetallicity.total\nmetallicity.he\nmetallicity.c\nmetallicity.n\nmetallicity.o\nmetallicity.ne\nmetallicity.mg\nmetallicity.si\nmetallicity.s\nmetallicity.ca\nmetallicity.fe\n"
],
[
"# properties of dark matter particles\n\nfor k in part['dark'].keys():\n print(k)",
"position\nmass\nid\npotential\nvelocity\n"
],
[
"# properties of gas particles\n\nfor k in part['gas'].keys():\n print(k)",
"position\ndensity\nelectron.fraction\ntemperature\nmass\nmassfraction\nhydrogen.neutral.fraction\nid\npotential\nvelocity\nmetallicity.total\nmetallicity.he\nmetallicity.c\nmetallicity.n\nmetallicity.o\nmetallicity.ne\nmetallicity.mg\nmetallicity.si\nmetallicity.s\nmetallicity.ca\nmetallicity.fe\n"
]
],
[
[
"# particle coordinates",
"_____no_output_____"
]
],
[
[
"# 3-D position of star particle (particle number x dimension number) in cartesian coordiantes [kpc physical]\n# if directory contains file m12*_center.txt, this reader automatically reads this file and \n# convert all positions to be in galactocentric coordinates, alined with principal axes of the galaxy\n\npart['star']['position']",
"_____no_output_____"
],
[
"# you can convert these to cylindrical coordiantes...\n\nstar_positions_cylindrical = gizmo_read.coordinate.get_positions_in_coordinate_system(\n part['star']['position'], system_to='cylindrical')\nprint(star_positions_cylindrical)",
"[[ 2.98728375e+04 9.79966007e+01 5.11315470e+00]\n [ 8.44916513e+00 -1.42074969e-01 4.81602573e+00]\n [ 8.56924321e+00 -9.42783421e-02 4.78167971e+00]\n ...\n [ 2.07095818e+03 3.64950773e+01 2.72158217e+00]\n [ 2.09849995e+03 4.84814836e+01 2.71389451e+00]\n [ 2.05142556e+03 1.05859972e+03 2.82709171e-01]]\n"
],
[
"# or spherical coordiantes\n\nstar_positions_spherical = gizmo_read.coordinate.get_positions_in_coordinate_system(\n part['star']['position'], system_to='spherical')\nprint(star_positions_spherical)",
"[[2.98729983e+04 1.56751588e+00 5.11315470e+00]\n [8.45035956e+00 1.58761001e+00 4.81602573e+00]\n [8.56976181e+00 1.58179783e+00 4.78167971e+00]\n ...\n [2.07127972e+03 1.55317584e+00 2.72158217e+00]\n [2.09905991e+03 1.54769751e+00 2.71389451e+00]\n [2.30845840e+03 1.09440612e+00 2.82709171e-01]]\n"
],
[
"# 3-D velocity of star particle (particle number x dimension number) in cartesian coordiantes [km/s]\n\npart['star']['velocity']",
"_____no_output_____"
],
[
"# you can convert these to cylindrical coordiantes...\n\nstar_velocities_cylindrical = gizmo_read.coordinate.get_velocities_in_coordinate_system(\n part['star']['velocity'], part['star']['position'], system_to='cylindrical')\nprint(star_velocities_cylindrical)",
"[[ 3.26827881e+03 7.08891220e+01 -2.79372520e+01]\n [-3.65740891e+01 1.09564304e+01 1.62347977e+02]\n [ 2.74234409e+01 -7.61478271e+01 2.27197754e+02]\n ...\n [ 1.33282959e+02 2.58180070e+00 2.25322895e+01]\n [ 1.26326935e+02 1.60031185e+01 1.44918041e+01]\n [ 1.65938049e+02 9.77062912e+01 -2.39563694e+01]]\n"
],
[
"# or spherical coordiantes\n\nstar_velocities_spherical = gizmo_read.coordinate.get_velocities_in_coordinate_system(\n part['star']['velocity'], part['star']['position'], system_to='spherical')\nprint(star_velocities_spherical)",
"[[ 3.2684939e+03 -6.0167347e+01 -2.7937252e+01]\n [-3.6753128e+01 -1.0339966e+01 1.6234798e+02]\n [ 2.8259504e+01 7.5841530e+01 2.2719775e+02]\n ...\n [ 1.3330775e+02 -2.3301035e-01 2.2532290e+01]\n [ 1.2666286e+02 -1.3081106e+01 1.4491804e+01]\n [ 1.9226746e+02 -1.0732359e+01 -2.3956369e+01]]\n"
],
[
"# the galaxy center position [kpc comoving] and velocity [km/s] are stored via\n\nprint(part.center_position)\nprint(part.center_velocity)",
"[41792.14534 44131.23473 46267.67629]\n[-52.45083 71.85282 95.19746]\n"
],
[
"# the rotation vectors to align with the principal axes are stored via\n\nprint(part.principal_axes_vectors)",
"[[ 0.11681398 -0.98166206 0.1506456 ]\n [-0.86026934 -0.02421714 0.50926436]\n [-0.49627729 -0.18908499 -0.84732267]]\n"
]
],
[
[
"# LSR coordinates for mock",
"_____no_output_____"
]
],
[
[
"# you can read the assumed local standard of rest (LSR) coordinates used in the Ananke mock catalogs\n# you need to input which LSR to use (currently 0, 1, or 2, because we use 3 per galaxy)\n\ngizmo_read.read.Read.read_lsr_coordinates(part, directory=directory, lsr_index=0)\ngizmo_read.read.Read.read_lsr_coordinates(part, directory=directory, lsr_index=1)\ngizmo_read.read.Read.read_lsr_coordinates(part, directory=directory, lsr_index=2)",
"reading LSR coordinates from:\n m12i_res7100_LSR0.txt\n LSR_0 position [kpc] = 0.000, 8.200, 0.000\n LSR_0 velocity [km/s] = -224.7, -20.4, 3.9\n\nreading LSR coordinates from:\n m12i_res7100_LSR1.txt\n LSR_1 position [kpc] = -7.101, -4.100, 0.000\n LSR_1 velocity [km/s] = 87.3, -186.9, -9.5\n\nreading LSR coordinates from:\n m12i_res7100_LSR2.txt\n LSR_2 position [kpc] = 7.101, -4.100, 0.000\n LSR_2 velocity [km/s] = 80.4, 191.7, 1.5\n\n"
],
[
"# the particle catalog can store one LSR at a time via\n\nprint(part.lsr_position)\nprint(part.lsr_velocity)",
"[ 7.1014 -4.1 0. ]\n[ 80.4269 191.724 1.5039]\n"
],
[
"# you can convert coordinates to be relative to LSR via\n\nstar_positions_wrt_lsr = part['star']['position'] - part.lsr_position\nstar_velocities_wrt_lsr = part['star']['velocity'] - part.lsr_velocity\nprint(star_positions_wrt_lsr)\nprint(star_velocities_wrt_lsr)",
"[[ 1.16469946e+04 -2.75016897e+04 9.79966007e+01]\n [-6.22732260e+00 -4.30383128e+00 -1.42074969e-01]\n [-6.50810594e+00 -4.44868009e+00 -9.42783421e-02]\n ...\n [-1.89806156e+03 8.48574680e+02 3.64950773e+01]\n [-1.91657460e+03 8.74510327e+02 4.84814836e+01]\n [ 1.96288917e+03 5.76362177e+02 1.05859972e+03]]\n[[ 1.1688820e+03 -3.2119316e+03 6.9385223e+01]\n [ 7.7266365e+01 -1.3855103e+02 9.4525299e+00]\n [ 1.4812433e+02 -2.0335153e+02 -7.7651726e+01]\n ...\n [-2.1131351e+02 -1.5794910e+02 1.0779006e+00]\n [-2.0138556e+02 -1.5251286e+02 1.4499218e+01]\n [ 8.5606773e+01 -1.6843958e+02 9.6202393e+01]]\n"
]
],
[
[
"# other particle properties",
"_____no_output_____"
]
],
[
[
"# mass of star particle [M_sun]\n# note that star particles are created with an initial mass of ~7070 Msun, \n# but because of stellar mass loss they can be less massive by z = 0\n# a few star particles form from slightly higher-mass gas particles\n# (because gas particles gain mass via stellar mass loss)\n# so some star particles are a little more massive than 7070 Msun\n\npart['star']['mass']",
"_____no_output_____"
],
[
"# formation scale-factor of star particle\n\npart['star']['form.scalefactor']",
"_____no_output_____"
],
[
"# or more usefully, the current age of star particle (the lookback time to when it formed) [Gyr]\n\npart['star']['age']",
"_____no_output_____"
],
[
"# gravitational potential at position of star particle [km^2 / s^2 physical]\n# note: normalization is arbitrary\n\npart['star']['potential']",
"_____no_output_____"
],
[
"# ID of star particle\n# NOTE: Ananke uses/references the *index* (within this array) of star particles, *not* their ID!\n# (because for technical reasons some star particles can end up with the same ID)\n# So you generally should never have to use this ID!\n\npart['star']['id']",
"_____no_output_____"
]
],
[
[
"# metallicities",
"_____no_output_____"
]
],
[
[
"# elemental abundance (metallicity) is stored natively as *linear mass fraction*\n# one value for each element, in a particle_number x element_number array\n# the first value is the mass fraction of all metals (everything not H, He)\n# 0 = all metals (everything not H, He), 1 = He, 2 = C, 3 = N, 4 = O, 5 = Ne, 6 = Mg, 7 = Si, 8 = S, 9 = Ca, 10 = Fe\n\npart['star']['massfraction']",
"_____no_output_____"
],
[
"# get individual elements by their index\n\n# total metal mass fraction (everything not H, He) is index 0\nprint(part['star']['massfraction'][:, 0])\n\n# iron is index 10\nprint(part['star']['massfraction'][:, 10])",
"[6.0437708e-03 3.2043904e-02 4.6177451e-02 ... 7.9349702e-04 5.3998221e-05\n 1.9502458e-03]\n[2.1929388e-04 1.3037791e-03 1.7796059e-03 ... 3.1033269e-05 8.7730750e-06\n 6.7164707e-05]\n"
],
[
"# for convenience, this reader also stores 'metallicity' := log10(mass_fraction / mass_fraction_solar)\n# where mass_fraction_solar is from Asplund et al 2009\n\nprint(part['star']['metallicity.total'])\nprint(part['star']['metallicity.fe'])\nprint(part['star']['metallicity.o'])",
"[-0.3457968 0.37864065 0.53732514 ... -1.2275594 -2.3947253\n -0.8370154 ]\n[-0.77062804 0.00354949 0.13866928 ... -1.619827 -2.1685026\n -1.2845135 ]\n[-0.23240621 0.4630599 0.62019897 ... -1.125268 -2.4857357\n -0.69058067]\n"
],
[
"# see gizmo_read.constant for assumed solar values (Asplund et al 2009) and other constants\n\ngizmo_read.constant.sun_composition",
"_____no_output_____"
]
],
[
[
"# additional information stored in sub-dictionaries",
"_____no_output_____"
]
],
[
[
"# dictionary of 'header' information about the simulation\n\npart.info",
"_____no_output_____"
],
[
"# dictionary of information about this snapshot's scale-factor, redshift, time, lookback-time\n\npart.snapshot",
"_____no_output_____"
],
[
"# dictionary class of cosmological parameters, with function for cosmological conversions\n\npart.Cosmology",
"_____no_output_____"
]
],
[
[
"See gizmo_read.constant for assumed (astro)physical constants used throughout.\n\nSee gizmo_read.coordinate for more coordiante transformation, zoom-in center ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
]
|
cb6ed3c9113ba6da1f1bebda131ee6ba655d048f | 2,808 | ipynb | Jupyter Notebook | lt787_cheapest_flights_dijkstra_algo.ipynb | devkosal/code_challenges | 0591c4839555376a231682db7cc12c8a70515b09 | [
"MIT"
]
| null | null | null | lt787_cheapest_flights_dijkstra_algo.ipynb | devkosal/code_challenges | 0591c4839555376a231682db7cc12c8a70515b09 | [
"MIT"
]
| null | null | null | lt787_cheapest_flights_dijkstra_algo.ipynb | devkosal/code_challenges | 0591c4839555376a231682db7cc12c8a70515b09 | [
"MIT"
]
| null | null | null | 20.8 | 114 | 0.466524 | [
[
[
"https://leetcode.com/problems/cheapest-flights-within-k-stops/",
"_____no_output_____"
],
[
"https://www.youtube.com/watch?v=UbP2Zek1c48&t=1442s",
"_____no_output_____"
],
[
"https://pastebin.com/t8uAaFud",
"_____no_output_____"
]
],
[
[
"import heapq",
"_____no_output_____"
],
[
"def findCheapestPrice(n: int, flights, src: int, dst: int, K: int) -> int:\n \"\"\"\n inspired by \n using djkistra here but we did not need to store the distances in an array. Need to investigate why..\n \"\"\"\n cons = [[] for _ in range(n)]\n for u,v,w in edges:\n cons[u].append((w,v))\n hp = [(0,0,src)]\n heapq.heapify(hp)\n while hp:\n cur_dist, stops, cur_node = heapq.heappop(hp)\n if cur_node == dst:\n return cur_dist\n if stops > K:\n continue\n for dist, n in cons[cur_node]:\n heapq.heappush(hp,(cur_dist+dist,stops+1,n))\n return -1\n \n ",
"_____no_output_____"
],
[
"n = 3\nedges = [[0,1,100],[1,2,100],[0,2,500]]\nsrc = 0\ndst = 2\nk = 1\nfindCheapestPrice(n,edges,src,dst,k)",
"_____no_output_____"
],
[
"n = 4\nedges = [[0,1,200],[1,2,100],[0,2,400],[2,3,100]]\nsrc = 0\ndst = 3\nk = 0\nfindCheapestPrice(n,edges,src,dst,k)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
cb6ed85d310df82d31a1b0b08ff17a247b6a42bb | 360,948 | ipynb | Jupyter Notebook | PRsimulation.ipynb | mostaphafakihi/Simulation | 2e8d1ea9eb7ce50604dc44175bcef5795f933ebf | [
"MIT"
]
| null | null | null | PRsimulation.ipynb | mostaphafakihi/Simulation | 2e8d1ea9eb7ce50604dc44175bcef5795f933ebf | [
"MIT"
]
| null | null | null | PRsimulation.ipynb | mostaphafakihi/Simulation | 2e8d1ea9eb7ce50604dc44175bcef5795f933ebf | [
"MIT"
]
| null | null | null | 145.837576 | 47,214 | 0.808313 | [
[
[
"<a href=\"https://colab.research.google.com/github/mostaphafakihi/Simulation/blob/main/PRsimulation.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# **Projet de simulation d'un super marché**\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom pandas import DataFrame\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport seaborn as sns\nimport statistics\nimport math \n#plot\nplt.rcParams['figure.figsize'] = (12, 9)\n",
"_____no_output_____"
],
[
"#la fonction alea\ndef aleas(IX,IY,IZ):\n IX[0] = 171 * ( IX[0] % 177 ) - 2 * (IX[0] // 177 ) \n IY[0] = 172 * ( IY[0] % 176 ) - 35 * ( IY[0] // 176 ) \n IZ[0] = 170 * ( IZ[0] % 178 ) - 63 * ( IZ[0] //178 ) \n if ( IX[0]<0 ):\n IX[0] = IX[0] + 30269 \n if ( IY[0]< 0 ) :\n IY[0] = IY[0] + 30307 \n if (IZ[0]< 0 ) :\n IZ[0] = IZ[0] + 30323 \n inter = ( ( IX[0] / 30269 ) + ( IY[0] / 30307 ) + ( IZ[0] / 30323 ) )\n alea = inter - int ( inter ) \n return alea\n ",
"_____no_output_____"
],
[
"def F1(alea):\n if alea >= 0 and alea < 0.3:\n return 1\n elif alea >= 0.3 and alea <= 0.8:\n return 2\n elif alea > 0.8 and alea <= 0.9:\n return 3\n elif alea > 0.9 and alea <= 0.95:\n return 4\n elif alea > 0.95 and alea <= 0.98:\n return 5\n else:\n return 6\n\n\ndef F2(alea):\n if alea >= 0 and alea < 0.1:\n return 2\n elif alea >= 0.1 and alea < 0.3:\n return 4\n elif alea >= 0.3 and alea <= 0.7:\n return 6\n elif alea > 0.7 and alea <= 0.9:\n return 8\n else:\n return 10\n\n\ndef F3(alea):\n if alea >= 0 and alea < 0.2:\n return 1\n elif alea >= 0.2 and alea <= 0.6:\n return 2\n elif alea > 0.6 and alea <= 0.85:\n return 3\n else:\n return 4",
"_____no_output_____"
],
[
"#Trier le calendrier\ndef Trier_cal(calendrier):\n l = len(calendrier)\n for i in range(0, l):\n for j in range(0, l-i-1):\n if (calendrier[j][2] > calendrier[j + 1][2]):\n tempo = calendrier[j]\n calendrier[j]= calendrier[j + 1]\n calendrier[j + 1]= tempo\n return calendrier",
"_____no_output_____"
],
[
"#pour planidier un événement\ndef planif_eve(evt=[]):\n cal_tri.append(evt)\n return cal_tri\n#selectionner un événement \ndef select_eve(cal_tri):\n evt_p=cal_tri[0]\n cal_tri.pop(0)\n return evt_p",
"_____no_output_____"
],
[
"def intervalle_confiance(NCP):\n moy_NCP=np.array(NCP)\n n=len(moy_NCP)\n m=np.mean(moy_NCP)\n s=statistics.stdev(moy_NCP,m)\n IC=[m-1.96*(s//math.sqrt(n)),m+1.96*(s//math.sqrt(n))]\n return IC",
"_____no_output_____"
]
],
[
[
"# **Scénario 1(simulation avec deux caisses)**",
"_____no_output_____"
]
],
[
[
" # Initialiser le calendrier par la 1ère arrivée du 1er client \n IX=[0]\n IY=[0]\n IZ=[0]\n IX[0]= int(input(\"Entrez la valeur du premier germe IX: \"))\n while (IX[0] <1 or IX[0] >30000):\n IX[0] = int(input(\"la valeur que vous avez saisie ne convient pas\"))\n \n IY[0]= int(input(\"Entrez la valeur du deuxieme germe IY: \"))\n while (IY[0] <1 or IY[0] >30000):\n IY[0] = int(input(\"la valeur que vous avez saisie ne convient pas\"))\n \n IZ[0]= int(input(\"Entrez1 la valeur du dernier germe IZ: \"))\n while (IZ[0] <1 or IZ[0] >30000):\n IZ[0] = int(input(\"la valeur que vous avez saisie ne convient pas\"))",
"Entrez la valeur du premier germe IX: 10\nEntrez la valeur du deuxieme germe IY: 100\nEntrez1 la valeur du dernier germe IZ: 1000\n"
],
[
"resultat=[]\nresultat_sanspi=[]\nfor k in range(40): \n H = 0 # Horloge de simulation \n i = 1 # numéro client arrivé à chaque fois \n LQ = 0 # Longueur Queue \n NCP = 0 # Nombre Clients Perdus\n NCE = 0 # Nombre Clients Entrés \n C1 = 0 # état caisse 1 libre \n C2 = 0 # état caisse 2 libre \n t1 = 0\n t2 = 0 \n s1 = 0\n s2 = 0\n DEQ = 0\n DSQ = 0\n tj = 0\n Q=[]\n tmp1=0\n tmp2=0\n TSmoy = 0\n TATmoy = 0 \n TauC1 =0 \n TauC2 = 0\n Qj=[]\n # Initialiser le calendrier par la 1ère arrivée du 1er client \n evt=[]\n a=aleas(IX,IY,IZ)\n evt1=[1,'A',F1(a)]\n cal_tri=[evt1]\n file=[]\n while (len(cal_tri)!=0):\n cal_tri=Trier_cal(cal_tri)\n evt_sel=select_eve(cal_tri)\n H=evt_sel[2]\n if (evt_sel[1] == 'A'):\n if (LQ <= 1):\n NCE = NCE+1\n planif_eve([evt_sel[0],'FM',H+F2(aleas(IX,IY,IZ))])\n else:\n NCP = NCP+1\n i=i+1\n DA=H+F1(aleas(IX,IY,IZ))\n if (DA<=720):\n planif_eve([i,'A',DA])\n \n if (evt_sel[1] == 'FM'):\n\n if (C1==0 or C2==0):\n if (C1==0):\n C1=evt_sel[0]\n t1=t1+(H-s1)\n \n else:\n C2=evt_sel[0] \n t2=t2+(H-s2)\n tmp1=H+F3(aleas(IX,IY,IZ))\n planif_eve([evt_sel[0],'FP',tmp1])\n DEQ=DEQ+tmp1\n else : \n LQ=LQ+1\n s1=H \n s2=H\n file.append(evt_sel[0])\n \n if (evt_sel[1] == 'FP'):\n\n if (LQ==0):\n if (C1==evt_sel[0]):\n C1=0\n \n else:\n C2=0\n else:\n j=file[0]\n file.pop(0)\n LQ=LQ-1\n tj=tj+(H-s1)\n Q.append(tj)\n if (C1==evt_sel[0]):\n C1=j\n else:\n C2=j\n tmp2=H+F3(aleas(IX,IY,IZ))\n planif_eve([j,'FP',tmp2])\n DSQ=DSQ+tmp2 \n DFS=H\n Qj=[element * (1/DFS) for element in Q]\n TauC1=t1/DFS\n TauC2=t2/DFS\n TATmoy=(DSQ-DEQ)/NCE\n TSmoy=(H-DA)/NCE\n resultat.extend([[DFS,NCE, NCP,TSmoy ,TATmoy ,TauC1 ,TauC2]])\n IX[0]=IX[0]+10+k*10\n IY[0]=IY[0]+30+k*30\n IZ[0]=IZ[0]+20+k*20\n",
"_____no_output_____"
],
[
"df1 = pd.DataFrame(resultat, columns =['DFS','NCE', 'NCP','TSmoy' ,'TATmoy' ,'TauC1' ,'TauC2'],index=['sim1','sim2','sim3','sim4','sim5','sim6','sim7','sim8','sim9','sim10','sim11','sim12','sim13','sim14','sim15','sim16','sim17','sim18','sim19','sim20','sim21','sim22','sim23','sim24','sim25','sim26','sim27','sim28','sim29','sim30','sim31','sim32','sim33','sim34','sim35','sim36','sim37','sim38','sim39','sim40'])\ndf1\n",
"_____no_output_____"
]
],
[
[
"# **Scénario 2(simulation avec 3 caisses)**",
"_____no_output_____"
]
],
[
[
" # Initialiser le calendrier par la 1ère arrivée du 1er client \n IX=[0]\n IY=[0]\n IZ=[0]\n IX[0]= int(input(\"Entrez la valeur du premier germe IX: \"))\n while (IX[0] <1 or IX[0] >30000):\n IX[0] = int(input(\"la valeur que vous avez saisie ne convient pas\"))\n \n IY[0]= int(input(\"Entrez la valeur du deuxieme germe IY: \"))\n while (IY[0] <1 or IY[0] >30000):\n IY[0] = int(input(\"la valeur que vous avez saisie ne convient pas\"))\n \n IZ[0]= int(input(\"Entrez la valeur du dernier germe IZ: \"))\n while (IZ[0] <1 or IZ[0] >30000):\n IZ[0] = int(input(\"la valeur que vous avez saisie ne convient pas\"))",
"Entrez la valeur du premier germe IX: 10\nEntrez la valeur du deuxieme germe IY: 100\nEntrez la valeur du dernier germe IZ: 1000\n"
],
[
"data=[]\ndata_sanspi=[]\nfor k in range(40): \n H = 0 #(* Horloge de simulation *)\n i = 1 #(* numéro client arrivé à chaque fois *)\n LQ = 0 #(* Longueur Queue *)\n NCP = 0 #(* Nombre Clients Perdus *)\n NCE = 0 #(* Nombre Clients Entrés *)\n C1 = 0 #(* état caisse 1 libre *)\n C2 = 0 #(* état caisse 2 libre *)\n C3 = 0\n t1 = 0\n t2 = 0 \n t3 = 0\n s1 = 0\n s2 = 0\n s3 = 0\n tmp1=0\n tmp2=0\n DEQ = 0\n DSQ = 0\n MTS=0\n TATmoy=0\n TauC1=0\n TauC2=0\n TauC3=0\n Qj=[]\n # Initialiser le calendrier par la 1ère arrivée du 1er client \n evt=[]\n a=aleas(IX,IY,IZ)\n evt1=[1,'A',F1(a)]\n cal_tri=[evt1]\n file=[]\n while (len(cal_tri)!=0):\n cal_tri=Trier_cal(cal_tri)\n evt_sel=select_eve(cal_tri)\n H=evt_sel[2]\n if (evt_sel[1] == 'A'):\n if (LQ <= 1):\n NCE = NCE+1\n planif_eve([evt_sel[0],'FM',H+F2(aleas(IX,IY,IZ))])\n else:\n NCP = NCP+1\n i=i+1\n DA=H+F1(aleas(IX,IY,IZ))\n if (DA<=720):\n planif_eve([i,'A',DA])\n \n if (evt_sel[1] == 'FM'):\n\n if (C1==0 or C2==0 or C3==0):\n if (C1==0):\n C1=evt_sel[0]\n t1=t1+(H-s1)\n \n if (C2==0):\n C2=evt_sel[0] \n t2=t2+(H-s2)\n else:\n C3=evt_sel[0] \n t3=t3+(H-s3)\n tmp1=H+F3(aleas(IX,IY,IZ))\n planif_eve([evt_sel[0],'FP',tmp1])\n DEQ=DEQ+tmp1\n else : \n LQ=LQ+1\n s1=H \n s2=H\n s3=H\n file.append(evt_sel[0])\n if (evt_sel[1] == 'FP'):\n\n if (LQ==0):\n if (C1==evt_sel[0]):\n C1=0\n \n if (C2==evt_sel[0]):\n C2=0\n else: \n C3=0\n else:\n j=file[0]\n file.pop(0)\n LQ=LQ-1\n tj=tj+(H-s1)\n Q.append(tj)\n if (C1==evt_sel[0]):\n C1=j\n if(C2==evt_sel[0]):\n C2=j\n else:\n C3=j\n tmp2=H+F3(aleas(IX,IY,IZ))\n planif_eve([j,'FP',tmp2])\n DSQ=DSQ+tmp2\n \n DFS=H\n Qj=[element * (1/DFS) for element in Q]\n pi=sum(Qj)\n p1=Qj[0]\n TauC1=t1/DFS\n TauC2=t2/DFS\n TauC3=t3/DFS\n TATmoy=(DSQ-DEQ)/NCE\n TSmoy=(H-DA)/NCE \n data.extend([[DFS,NCE, NCP,TATmoy,TSmoy,TauC1,TauC2,TauC3]])\n IX[0]=IX[0]+10+k*10\n IY[0]=IY[0]+30+k*30\n IZ[0]=IZ[0]+20+k*20\n",
"_____no_output_____"
],
[
"df2 = pd.DataFrame(data, columns =['DFS','NCE', 'NCP','TATmoy','TSmoy','TauC1','TauC2','TauC3',],index=['sim1','sim2','sim3','sim4','sim5','sim6','sim7','sim8','sim9','sim10','sim11','sim12','sim13','sim14','sim15','sim16','sim17','sim18','sim19','sim20','sim21','sim22','sim23','sim24','sim25','sim26','sim27','sim28','sim29','sim30','sim31','sim32','sim33','sim34','sim35','sim36','sim37','sim38','sim39','sim40'])\ndf2",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"description du scénario 1",
"_____no_output_____"
]
],
[
[
"df1.describe()",
"_____no_output_____"
]
],
[
[
"description des données du scénario 2\n\n\n\n",
"_____no_output_____"
]
],
[
[
"df2.describe()",
"_____no_output_____"
],
[
"IC1 = intervalle_confiance(df1['NCP'])\nIC1",
"_____no_output_____"
],
[
"IC2 = intervalle_confiance(df2['NCP'])\nIC2",
"_____no_output_____"
]
],
[
[
"# **Représentation graphique**",
"_____no_output_____"
],
[
"# *Scénario 1*",
"_____no_output_____"
]
],
[
[
"df1_sanspi.plot.bar(rot=0,figsize=(60,14))\nfrom google.colab import files\nplt.savefig(\"abc.png\")\nfiles.download(\"abc.png\") ",
"_____no_output_____"
],
[
"df1_sanspi.plot.bar(stacked=True,figsize=(24, 11))",
"_____no_output_____"
],
[
"df1['NCP'].plot.bar(rot=0)",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(df1['NCP'])\nplt.plot(df1['NCE'])",
"_____no_output_____"
],
[
"sns.displot(df1['NCP'])\nplt.title(\"Distribution de NCP\", fontsize=20)",
"_____no_output_____"
]
],
[
[
"# *Scénario 2*",
"_____no_output_____"
]
],
[
[
"df2_sanspi.plot.bar(rot=0,figsize=(44,24))",
"_____no_output_____"
],
[
"df2_sanspi.plot.bar(stacked=True,figsize=(24, 11))",
"_____no_output_____"
],
[
"df2['NCP'].plot.bar(rot=0)\n",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(df2['NCP'])\nplt.plot(df2['NCE'])\n",
"_____no_output_____"
],
[
"sns.displot(df2['NCP'])\nplt.title(\"Distribution de NCP\", fontsize=20)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6ee7b845adf6496ac670f08503d918146225aa | 423,138 | ipynb | Jupyter Notebook | 7th SEM/MACHINE LEARNING LABORATORY/4-BackPropogation Algorithm/LAB 4.ipynb | NisargaJN/VTU-CSE-LAB-SOLUTIONS | 5c977d3f31ec2225630b30a4c34f32d5700cbaf1 | [
"MIT"
]
| 80 | 2019-02-12T19:30:15.000Z | 2022-03-28T14:38:55.000Z | 4-BackPropogation Algorithm/LAB 4.ipynb | kshitij0209ag/Machine-Learning-Algorithms | 179556d23202f9b93bd129f0ca8d0eec93cbad0f | [
"MIT"
]
| 1 | 2020-03-29T09:17:14.000Z | 2020-03-29T09:17:15.000Z | 4-BackPropogation Algorithm/LAB 4.ipynb | kshitij0209ag/Machine-Learning-Algorithms | 179556d23202f9b93bd129f0ca8d0eec93cbad0f | [
"MIT"
]
| 115 | 2019-02-20T13:32:44.000Z | 2022-03-28T17:19:29.000Z | 23.301834 | 140 | 0.354402 | [
[
[
"# MACHINE LEARNING LAB - 4 ( Backpropagation Algorithm )",
"_____no_output_____"
],
[
"**4. Build an Artificial Neural Network by implementing the Backpropagation algorithm and test the same using appropriate data sets.**",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nX = np.array(([2, 9], [1, 5], [3, 6]), dtype=float) # X = (hours sleeping, hours studying)\ny = np.array(([92], [86], [89]), dtype=float) # y = score on test\n\n# scale units\nX = X/np.amax(X, axis=0) # maximum of X array\ny = y/100 # max test score is 100",
"_____no_output_____"
],
[
"class Neural_Network(object):\n def __init__(self):\n # Parameters\n self.inputSize = 2\n self.outputSize = 1\n self.hiddenSize = 3\n # Weights\n self.W1 = np.random.randn(self.inputSize, self.hiddenSize) # (3x2) weight matrix from input to hidden layer\n self.W2 = np.random.randn(self.hiddenSize, self.outputSize) # (3x1) weight matrix from hidden to output layer\n\n def forward(self, X):\n #forward propagation through our network\n self.z = np.dot(X, self.W1) # dot product of X (input) and first set of 3x2 weights\n self.z2 = self.sigmoid(self.z) # activation function\n self.z3 = np.dot(self.z2, self.W2) # dot product of hidden layer (z2) and second set of 3x1 weights\n o = self.sigmoid(self.z3) # final activation function\n return o \n\n def sigmoid(self, s):\n return 1/(1+np.exp(-s)) # activation function \n\n def sigmoidPrime(self, s):\n return s * (1 - s) # derivative of sigmoid\n \n def backward(self, X, y, o):\n # backward propgate through the network\n self.o_error = y - o # error in output\n self.o_delta = self.o_error*self.sigmoidPrime(o) # applying derivative of sigmoid to \n self.z2_error = self.o_delta.dot(self.W2.T) # z2 error: how much our hidden layer weights contributed to output error\n self.z2_delta = self.z2_error*self.sigmoidPrime(self.z2) # applying derivative of sigmoid to z2 error\n self.W1 += X.T.dot(self.z2_delta) # adjusting first set (input --> hidden) weights\n self.W2 += self.z2.T.dot(self.o_delta) # adjusting second set (hidden --> output) weights\n\n def train (self, X, y):\n o = self.forward(X)\n self.backward(X, y, o)",
"_____no_output_____"
],
[
"NN = Neural_Network()\nfor i in range(1000): # trains the NN 1,000 times\n print (\"\\nInput: \\n\" + str(X))\n print (\"\\nActual Output: \\n\" + str(y)) \n print (\"\\nPredicted Output: \\n\" + str(NN.forward(X)))\n print (\"\\nLoss: \\n\" + str(np.mean(np.square(y - NN.forward(X))))) # mean sum squared loss)\n NN.train(X, y)",
"\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.47212874]\n [0.42728946]\n [0.40891365]]\n\nLoss: \n0.20642371917499927\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.55398066]\n [0.49831918]\n [0.50254468]]\n\nLoss: \n0.13830159742519685\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.61884926]\n [0.55676904]\n [0.57857066]]\n\nLoss: \n0.09320967247558785\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.66796631]\n [0.60249504]\n [0.63636572]]\n\nLoss: \n0.06472004460391347\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.70497403]\n [0.63793606]\n [0.67970816]]\n\nLoss: \n0.04659040770460008\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.73326385]\n [0.66570437]\n [0.71259404]]\n\nLoss: \n0.0346980182977454\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.75534699]\n [0.68785267]\n [0.73806438]]\n\nLoss: \n0.026609916500395545\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.77295995]\n [0.70585464]\n [0.75822964]]\n\nLoss: \n0.020914999002099796\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.78728921]\n [0.72074616]\n [0.77452632]]\n\nLoss: \n0.0167793188604347\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.79915364]\n [0.73325905]\n [0.78793968]]\n\nLoss: \n0.013694472702822591\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.80912882]\n [0.74391816]\n [0.79915757]]\n\nLoss: \n0.01133992022831034\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.81762774]\n [0.75310683]\n [0.80867025]]\n\nLoss: \n0.009506918915787784\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.82495293]\n [0.76111037]\n [0.81683458]]\n\nLoss: \n0.008055427845679745\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.83133029]\n [0.76814492]\n [0.8239155 ]]\n\nLoss: \n0.006888944511310564\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.83693158]\n [0.77437703]\n [0.83011335]]\n\nLoss: \n0.0059393558595260245\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.84188954]\n [0.77993692]\n [0.83558217]]\n\nLoss: \n0.005157547368291981\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.84630826]\n [0.78492791]\n [0.84044224]]\n\nLoss: \n0.004507420891613049\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.85027052]\n [0.78943296]\n [0.84478878]]\n\nLoss: \n0.003961987439449809\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.85384288]\n [0.79351951]\n [0.84869808]]\n\nLoss: \n0.003500756236055738\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.85707954]\n [0.79724289]\n [0.85223203]]\n\nLoss: \n0.0031079532945512303\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.86002502]\n [0.80064897]\n [0.85544128]]\n\nLoss: \n0.0027712826775886886\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.86271625]\n [0.80377612]\n [0.85836776]]\n\nLoss: \n0.0024810500722398147\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.86518415]\n [0.80665667]\n [0.86104641]]\n\nLoss: \n0.002229532806726281\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.86745476]\n [0.80931805]\n [0.86350662]]\n\nLoss: \n0.0020105203895673285\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.8695502 ]\n [0.81178374]\n [0.86577328]]\n\nLoss: \n0.001818974914611737\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.87148937]\n [0.8140739 ]\n [0.86786761]]\n\nLoss: \n0.0016507769579183816\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.87328853]\n [0.81620601]\n [0.86980782]]\n\nLoss: \n0.0015025332748599648\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.87496174]\n [0.81819524]\n [0.87160961]]\n\nLoss: \n0.0013714297307684988\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.87652122]\n [0.82005488]\n [0.87328664]]\n\nLoss: \n0.0012551177241060967\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.87797766]\n [0.82179658]\n [0.87485078]]\n\nLoss: \n0.0011516256765708862\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.87934043]\n [0.82343063]\n [0.87631244]]\n\nLoss: \n0.0010592894729487847\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.8806178 ]\n [0.82496613]\n [0.87768081]]\n\nLoss: \n0.0009766973609146353\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88181707]\n [0.82641119]\n [0.87896397]]\n\nLoss: \n0.0009026459817299505\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88294475]\n [0.82777304]\n [0.88016912]]\n\nLoss: \n0.0008361050397846306\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88400663]\n [0.82905813]\n [0.88130264]]\n\nLoss: \n0.0007761887287067974\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88500788]\n [0.8302723 ]\n [0.88237025]]\n\nLoss: \n0.0007221324803102649\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88595317]\n [0.83142077]\n [0.88337707]]\n\nLoss: \n0.0006732739356062196\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88684668]\n [0.83250828]\n [0.8843277 ]]\n\nLoss: \n0.0006290372863893455\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88769219]\n [0.83353912]\n [0.8852263 ]]\n\nLoss: \n0.0005889203240633681\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88849312]\n [0.83451717]\n [0.88607662]]\n\nLoss: \n0.000552483675478915\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88925258]\n [0.83544598]\n [0.88688206]]\n\nLoss: \n0.0005193418151952424\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.88997338]\n [0.8363288 ]\n [0.8876457 ]]\n\nLoss: \n0.0004891555281542141\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89065811]\n [0.83716857]\n [0.88837036]]\n\nLoss: \n0.0004616255624265264\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89130911]\n [0.83796801]\n [0.88905862]]\n\nLoss: \n0.00043648726300042623\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89192855]\n [0.83872962]\n [0.88971282]]\n\nLoss: \n0.0004135060179099145\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89251839]\n [0.83945568]\n [0.89033512]]\n\nLoss: \n0.0003924733798735898\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89308046]\n [0.84014832]\n [0.89092749]]\n\nLoss: \n0.00037320375194367243\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89361643]\n [0.84080947]\n [0.89149176]]\n\nLoss: \n0.0003555315458958015\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89412784]\n [0.84144095]\n [0.89202961]]\n\nLoss: \n0.00033930873832851207\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89461613]\n [0.84204442]\n [0.89254258]]\n\nLoss: \n0.00032440276253704474\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89508261]\n [0.84262145]\n [0.89303211]]\n\nLoss: \n0.00031069468483416635\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89552851]\n [0.84317347]\n [0.89349952]]\n\nLoss: \n0.00029807762262127187\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89595496]\n [0.84370182]\n [0.89394606]]\n\nLoss: \n0.0002864553685636829\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89636302]\n [0.84420776]\n [0.89437286]]\n\nLoss: \n0.00027574119100667074\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89675368]\n [0.84469245]\n [0.89478099]]\n\nLoss: \n0.0002658567855299031\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89712785]\n [0.84515699]\n [0.89517144]]\n\nLoss: \n0.00025673135647190246\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89748639]\n [0.84560238]\n [0.89554514]]\n\nLoss: \n0.0002483008105187975\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89783011]\n [0.84602959]\n [0.89590295]]\n\nLoss: \n0.00024050704716584167\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89815974]\n [0.84643952]\n [0.89624568]]\n\nLoss: \n0.00023329733312642788\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.898476 ]\n [0.84683299]\n [0.89657408]]\n\nLoss: \n0.00022662374966078716\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89877953]\n [0.84721081]\n [0.89688887]]\n\nLoss: \n0.00022044270339060024\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89907097]\n [0.84757371]\n [0.89719072]]\n\nLoss: \n0.00021471449250844575\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89935088]\n [0.84792239]\n [0.89748023]]\n\nLoss: \n0.0002094029214254376\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89961981]\n [0.84825751]\n [0.89775801]]\n\nLoss: \n0.00020447495786141397\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.89987828]\n [0.8485797 ]\n [0.89802461]]\n\nLoss: \n0.00019990042719829142\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90012677]\n [0.84888952]\n [0.89828055]]\n\nLoss: \n0.00019565173961224\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90036575]\n [0.84918755]\n [0.89852631]]\n\nLoss: \n0.00019170364609375198\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90059562]\n [0.8494743 ]\n [0.89876236]]\n\nLoss: \n0.00018803301997234268\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90081682]\n [0.84975027]\n [0.89898914]]\n\nLoss: \n0.00018461866099827134\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90102972]\n [0.85001592]\n [0.89920706]]\n\nLoss: \n0.00018144111940787707\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90123468]\n [0.85027171]\n [0.89941652]]\n\nLoss: \n0.00017848253772174237\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90143206]\n [0.85051804]\n [0.89961787]]\n\nLoss: \n0.00017572650830334664\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90162217]\n [0.85075532]\n [0.89981148]]\n\nLoss: \n0.00017315794494673605\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90180533]\n [0.85098394]\n [0.89999767]]\n\nLoss: \n0.00017076296697055826\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90198183]\n [0.85120424]\n [0.90017676]]\n\nLoss: \n0.00016852879447720295\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90215195]\n [0.85141657]\n [0.90034905]]\n\nLoss: \n0.00016644365359351607\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90231596]\n [0.85162126]\n [0.90051482]]\n\nLoss: \n0.00016449669064728438\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.9024741 ]\n [0.85181862]\n [0.90067435]]\n\nLoss: \n0.00016267789435364343\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90262663]\n [0.85200894]\n [0.90082788]]\n\nLoss: \n0.00016097802519082598\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90277376]\n [0.8521925 ]\n [0.90097566]]\n\nLoss: \n0.0001593885512366694\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90291571]\n [0.85236957]\n [0.90111793]]\n\nLoss: \n0.00015790158981818478\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90305269]\n [0.85254041]\n [0.90125491]]\n\nLoss: \n0.0001565098543975822\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.9031849 ]\n [0.85270526]\n [0.9013868 ]]\n\nLoss: \n0.00015520660618062424\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90331253]\n [0.85286436]\n [0.90151381]]\n\nLoss: \n0.00015398560998850582\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90343575]\n [0.85301792]\n [0.90163613]]\n\nLoss: \n0.00015284109398307203\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90355474]\n [0.85316616]\n [0.90175394]]\n\nLoss: \n0.00015176771287831486\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90366966]\n [0.85330928]\n [0.90186742]]\n\nLoss: \n0.0001507605143093281\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90378066]\n [0.85344747]\n [0.90197674]]\n\nLoss: \n0.00014981490806362428\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90388791]\n [0.85358093]\n [0.90208204]]\n\nLoss: \n0.00014892663790993984\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90399153]\n [0.85370982]\n [0.9021835 ]]\n\nLoss: \n0.0001480917557861779\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90409166]\n [0.85383432]\n [0.90228125]]\n\nLoss: \n0.00014730659813218009\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90418845]\n [0.8539546 ]\n [0.90237543]]\n\nLoss: \n0.00014656776417402528\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.904282 ]\n [0.8540708 ]\n [0.90246617]]\n\nLoss: \n0.00014587209598558378\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90437245]\n [0.85418308]\n [0.90255361]]\n\nLoss: \n0.00014521666017000906\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90445991]\n [0.85429158]\n [0.90263787]]\n\nLoss: \n0.000144598731018849\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90454448]\n [0.85439644]\n [0.90271906]]\n\nLoss: \n0.00014401577502021188\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90462627]\n [0.8544978 ]\n [0.90279729]]\n\nLoss: \n0.00014346543659935423\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90470539]\n [0.85459576]\n [0.90287268]]\n\nLoss: \n0.00014294552498617322\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90478193]\n [0.85469047]\n [0.90294532]]\n\nLoss: \n0.00014245400211372123\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90485599]\n [0.85478203]\n [0.90301532]]\n\nLoss: \n0.00014198897146079238\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90492764]\n [0.85487057]\n [0.90308278]]\n\nLoss: \n0.00014154866775951163\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90499699]\n [0.85495617]\n [0.90314777]]\n\nLoss: \n0.00014113144749599842\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90506411]\n [0.85503896]\n [0.9032104 ]]\n\nLoss: \n0.00014073578013868744\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90512908]\n [0.85511902]\n [0.90327074]]\n\nLoss: \n0.00014036024003465206\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90519198]\n [0.85519646]\n [0.90332888]]\n\nLoss: \n0.00014000349891959613\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90525288]\n [0.85527136]\n [0.90338489]]\n\nLoss: \n0.00013966431899186193\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90531185]\n [0.85534382]\n [0.90343886]]\n\nLoss: \n0.00013934154650525947\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90536896]\n [0.85541392]\n [0.90349084]]\n\nLoss: \n0.00013903410583923918\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90542427]\n [0.85548174]\n [0.90354091]]\n\nLoss: \n0.00013874099400864923\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90547784]\n [0.85554736]\n [0.90358914]]\n\nLoss: \n0.0001384612755784036\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90552975]\n [0.85561085]\n [0.90363559]]\n\nLoss: \n0.00013819407795142728\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90558004]\n [0.8556723 ]\n [0.90368033]]\n\nLoss: \n0.00013793858700076936\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90562876]\n [0.85573176]\n [0.9037234 ]]\n\nLoss: \n0.0001376940430193154\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90567599]\n [0.85578931]\n [0.90376488]]\n\nLoss: \n0.00013745973696265964\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90572175]\n [0.85584501]\n [0.90380481]]\n\nLoss: \n0.00013723500696269172\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90576611]\n [0.85589893]\n [0.90384324]]\n\nLoss: \n0.000137019235091384\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90580912]\n [0.85595112]\n [0.90388024]]\n\nLoss: \n0.00013681184435576535\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90585081]\n [0.85600165]\n [0.90391584]]\n\nLoss: \n0.00013661229590674354\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90589124]\n [0.85605057]\n [0.9039501 ]]\n\nLoss: \n0.00013642008644577396\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90593045]\n [0.85609793]\n [0.90398305]]\n\nLoss: \n0.00013623474581460102\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90596847]\n [0.85614379]\n [0.90401475]]\n\nLoss: \n0.00013605583475454738\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90600535]\n [0.8561882 ]\n [0.90404524]]\n\nLoss: \n0.00013588294282281262\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90604113]\n [0.8562312 ]\n [0.90407455]]\n\nLoss: \n0.00013571568645429915\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90607584]\n [0.85627284]\n [0.90410273]]\n\nLoss: \n0.00013555370715826725\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90610952]\n [0.85631318]\n [0.90412982]]\n\nLoss: \n0.00013539666984010087\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.9061422 ]\n [0.85635224]\n [0.90415585]]\n\nLoss: \n0.00013524426123902867\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90617392]\n [0.85639008]\n [0.90418085]]\n\nLoss: \n0.00013509618847352836\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90620471]\n [0.85642674]\n [0.90420487]]\n\nLoss: \n0.00013495217768660816\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.9062346 ]\n [0.85646225]\n [0.90422793]]\n\nLoss: \n0.00013481197278387438\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90626362]\n [0.85649665]\n [0.90425006]]\n\nLoss: \n0.00013467533425775486\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90629179]\n [0.85652998]\n [0.90427131]]\n\nLoss: \n0.00013454203809176683\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90631916]\n [0.85656227]\n [0.90429168]]\n\nLoss: \n0.00013441187473916998\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90634574]\n [0.85659357]\n [0.90431123]]\n\nLoss: \n0.0001342846481707748\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90637155]\n [0.85662389]\n [0.90432996]]\n\nLoss: \n0.00013416017498705732\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90639663]\n [0.85665327]\n [0.90434792]]\n\nLoss: \n0.00013403828359005897\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.906421 ]\n [0.85668175]\n [0.90436512]]\n\nLoss: \n0.0001339188134109341\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90644469]\n [0.85670936]\n [0.90438158]]\n\nLoss: \n0.00013380161418927042\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.9064677 ]\n [0.85673611]\n [0.90439735]]\n\nLoss: \n0.00013368654530060618\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90649008]\n [0.85676205]\n [0.90441243]]\n\nLoss: \n0.00013357347512878487\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90651183]\n [0.85678719]\n [0.90442684]]\n\nLoss: \n0.00013346228048011493\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90653297]\n [0.85681156]\n [0.90444062]]\n\nLoss: \n0.00013335284603644128\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90655354]\n [0.85683519]\n [0.90445378]]\n\nLoss: \n0.00013324506384444187\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90657354]\n [0.8568581 ]\n [0.90446635]]\n\nLoss: \n0.00013313883283874758\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.906593 ]\n [0.85688032]\n [0.90447833]]\n\nLoss: \n0.00013303405839649825\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90661193]\n [0.85690187]\n [0.90448976]]\n\nLoss: \n0.0001329306519212959\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n\nPredicted Output: \n[[0.90663035]\n [0.85692276]\n [0.90450064]]\n\nLoss: \n0.00013282853045447942\n\nInput: \n[[0.66666667 1. ]\n [0.33333333 0.55555556]\n [1. 0.66666667]]\n\nActual Output: \n[[0.92]\n [0.86]\n [0.89]]\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
]
|
cb6eead41721ba73fe1cd804efde43bdbd441090 | 33,361 | ipynb | Jupyter Notebook | byoa-tutorial.ipynb | Aelvangunduz/amazon-sagemaker-byoa-prophet-tutorial | c889fa1da1db37b11e6edd46f2a51c7a30549ced | [
"MIT-0"
]
| null | null | null | byoa-tutorial.ipynb | Aelvangunduz/amazon-sagemaker-byoa-prophet-tutorial | c889fa1da1db37b11e6edd46f2a51c7a30549ced | [
"MIT-0"
]
| null | null | null | byoa-tutorial.ipynb | Aelvangunduz/amazon-sagemaker-byoa-prophet-tutorial | c889fa1da1db37b11e6edd46f2a51c7a30549ced | [
"MIT-0"
]
| null | null | null | 48.349275 | 2,619 | 0.587812 | [
[
[
"# BYOA Tutorial - Prophet Forecasting en Sagemaker\nThe following notebook shows how to integrate your own algorithms to Amazon Sagemaker.\nWe are going to go the way of putting together an inference pipeline on the Prophet algorithm for time series.\nThe algorithm is installed in a docker container and then it helps us to train the model and make inferences on an endpoint.\n",
"_____no_output_____"
],
[
"We are going to work with a public dataset that we must download from Kaggle.\nThis dataset is called:\n_Avocado Prices: Historical data on avocado prices and sales volume in multiple US markets_\nand can be downloaded from: https://www.kaggle.com/neuromusic/avocado-prices/download\nOnce downloaded, we must upload it to the same directory where we are running this notebook.\nThe following code prepares the dataset so that Prophet can understand it:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n# Nos quedamos solo con la fecha y las ventas\ndf = pd.read_csv('avocado.csv')\ndf = df[['Date', 'AveragePrice']].dropna()\n\ndf['Date'] = pd.to_datetime(df['Date'])\ndf = df.set_index('Date')\n\n# Dejamos 1 solo registro por día con el promedio de ventas\ndaily_df = df.resample('D').mean()\nd_df = daily_df.reset_index().dropna()\n\n# Formateamos los nombre de columnas como los espera Prophet\nd_df = d_df[['Date', 'AveragePrice']]\nd_df.columns = ['ds', 'y']\nd_df.head()\n\n# Guardamos el dataset resultante como avocado_daily.csv\nd_df.to_csv(\"avocado_daily.csv\",index = False , columns = ['ds', 'y'] )",
"_____no_output_____"
]
],
[
[
"# Step 2: Package and upload the algorithm for use with Amazon SageMaker\n\n### An overview of Docker\n\nDocker provides a simple way to package code into an _image_ that is completely self-contained. Once you have an image, you can use Docker to run a _container_ based on that image. Running a container is the same as running a program on the machine, except that the container creates a completely self-contained environment for the program to run. Containers are isolated from each other and from the host environment, so the way you configure the program is the way it runs, no matter where you run it.\n\nDocker is more powerful than environment managers like conda or virtualenv because (a) it is completely language independent and (b) it understands your entire operating environment, including startup commands, environment variables, etc.\n\nIn some ways, a Docker container is like a virtual machine, but it is much lighter. For example, a program that runs in a container can start in less than a second, and many containers can run on the same physical machine or virtual machine instance.\n\nDocker uses a simple file called `Dockerfile` to specify how the image is assembled.\nAmazon SagMaker uses Docker to allow users to train and implement algorithms.\n\nIn Amazon SageMaker, Docker containers are invoked in a certain way for training and in a slightly different way for hosting. The following sections describe how to create containers for the SageMaker environment.\n\n\n### How Amazon SageMaker runs the Docker container\n\nBecause it can run the same image in training or hosting, Amazon SageMaker runs the container with the `train` or` serve` argument. How your container processes this argument depends on the container:\n\n* In the example here, we did not define an ʻENTRYPOINT ʻin the Dockerfile for Docker to execute the `train` command at training time and` serve` at service time. In this example, we define them as executable Python scripts, but they could be any program that we want to start in that environment.\n* If you specify a program as \"ENTRYPOINT\" in the Dockerfile, that program will run at startup and its first argument will be either `train` or` serve`. The program can then examine that argument and decide what to do.\n* If you are building separate containers for training and hosting (or building just for one or the other), you can define a program as \"ENTRYPOINT\" in the Dockerfile and ignore (or check) the first argument passed.\n\n#### Run container during training\n\nWhen Amazon SageMaker runs the training, the `train` script runs like a regular Python program. A series of files are arranged for your use, under the `/ opt / ml` directory:\n\n /opt/ml\n ├── input\n │ ├── config\n │ │ ├── hyperparameters.json\n │ │ └── resourceConfig.json\n │ └── data\n │ └── <channel_name>\n │ └── <input data>\n ├── model\n │ └── <model files>\n └── output\n └── failure\n\n##### The entrance\n\n* `/ opt / ml / input / config` contains information to control how the program runs. `hyperparameters.json` is a JSON-formatted dictionary of hyperparameter names to values. These values will always be strings, so you may need to convert them. `ResourceConfig.json` is a JSON-formatted file that describes the network layout used for distributed training. Since scikit-learn does not support distributed training, we will ignore it here.\n* `/ opt / ml / input / data / <channel_name> /` (for File mode) contains the input data for that channel. Channels are created based on the call to CreateTrainingJob, but it is generally important that the channels match what the algorithm expects. The files for each channel will be copied from S3 to this directory, preserving the tree structure indicated by the S3 key structure.\n* `/ opt / ml / input / data / <channel_name> _ <epoch_number>` (for Pipe mode) is the pipe for a given epoch. The epochs start at zero and go up by one each time you read them. There is no limit to the number of epochs you can run, but you must close each pipe before reading the next epoch.\n \n##### The exit\n\n* `/ opt / ml / model /` is the directory where the model generated by your algorithm is written. Your model can be in any format you want. It can be a single file or an entire directory tree. SagMaker will package any files in this directory into a compressed tar file. This file will be available in the S3 location returned in the `DescribeTrainingJob` output.\n* `/ opt / ml / output` is a directory where the algorithm can write a` failure` file that describes why the job failed. The content of this file will be returned in the `FailureReason` field of the` DescribeTrainingJob` result. For successful jobs, there is no reason to write this file as it will be ignored.\n\n#### Running the container during hosting\n\nHosting has a very different model than training because it must respond to inference requests that arrive through HTTP. In this example, we use recommended Python code to provide a robust and scalable inference request service:\n\nAmazon SagMaker uses two URLs in the container:\n\n* `/ ping` will receive` GET` requests from the infrastructure. Returns 200 if the container is open and accepting requests.\n* `/ invocations` is the endpoint that receives inference` POST` requests from the client. The request and response format depends on the algorithm. If the client supplied the `ContentType` and ʻAccept` headers, these will also be passed.\n\nThe container will have the model files in the same place where they were written during training:\n\n / opt / ml\n └── model\n └── <model files>\n",
"_____no_output_____"
],
[
"### Container Parts\n\nIn the `container` directory are all the components you need to package the sample algorithm for Amazon SageManager:\n\n .\n ├── Dockerfile\n ├── build_and_push.sh\n └── decision_trees\n ├── nginx.conf\n ├── predictor.py\n ├── serve\n ├── train\n └── wsgi.py\n\n\nLet's see each one:\n\n* __`Dockerfile`__ describes how to build the Docker container image. More details below.\n* __`build_and_push.sh`__ is a script that uses Dockerfile to build its container images and then publishes (push) it to ECR. We will invoke the commands directly later in this notebook, but you can copy and run the script for other algorithms.\n* __`prophet`__ is the directory that contains the files to be installed in the container.\n* __`local_test`__ is a directory that shows how to test the new container on any machine that can run Docker, including an Amazon SageMaker Notebook Instance. With this method, you can quickly iterate using small data sets to eliminate any structural errors before using the container with Amazon SageMaker.\n\nThe files that we are going to put in the container are:\n\n* __`nginx.conf`__ is the configuration file for the nginx front-end. Generally, you should be able to take this file as is.\n* __`predictor.py`__ is the program that actually implements the Flask web server and Prophet predictions for this application.\n* __`serve`__ is the program started when the hosting container starts. It just launches the gunicorn server running multiple instances of the Flask application defined in `predictor.py`. You should be able to take this file as is.\n* __`train`__ is the program that is invoked when the container for training is executed.\n* __`wsgi.py`__ is a small wrapper used to invoke the Flask application. You should be able to take this file as is.\n\nIn summary, the two Prophet-specific code files are `train` and` predictor.py`.",
"_____no_output_____"
],
[
"### The Dockerfile file\n\nThe Dockerfile file describes the image we want to create. It is a description of the complete installation of the operating system of the system that you want to run. A running Docker container is significantly lighter than a full operating system, however, because it leverages Linux on the host machine for basic operations.\n\nFor this example, we'll start from a standard Ubuntu install and run the normal tools to install the things Prophet needs. Finally, we add the code that implements Prophet to the container and configure the correct environment to run correctly.\n\nThe following is the Dockerfile:",
"_____no_output_____"
]
],
[
[
"!cat container/Dockerfile",
"# Build an image that can do training and inference in SageMaker\n# This is a Python 3 image that uses the nginx, gunicorn, flask stack\n# for serving inferences in a stable way.\n\nFROM ubuntu:16.04\n\nMAINTAINER Amazon AI <[email protected]>\n\nRUN apt-get -y update && apt-get install -y --no-install-recommends \\\n wget \\\n curl \\\n python-dev \\\n build-essential libssl-dev libffi-dev \\\n libxml2-dev libxslt1-dev zlib1g-dev \\\n nginx \\\n ca-certificates \\\n && rm -rf /var/lib/apt/lists/*\n\nRUN curl -fSsL -O https://bootstrap.pypa.io/get-pip.py && \\\n python get-pip.py && \\\n rm get-pip.py\n \nRUN pip --no-cache-dir install \\\n numpy \\\n scipy \\\n sklearn \\\n pandas \\\n flask \\\n gevent \\\n gunicorn \\\n pystan \n\nRUN pip --no-cache-dir install \\\n fbprophet \n \nENV PYTHONUNBUFFERED=TRUE\nENV PYTHONDONTWRITEBYTECODE=TRUE\nENV PATH=\"/opt/program:${PATH}\"\n\n# Set up the program in the image\nCOPY prophet /opt/program\nWORKDIR /opt/program\n\n"
]
],
[
[
"### The train file\n\nThe train file describes the way we are going to do the training.\nThe Prophet-Docker / container / prophet / train file contains the specific training code for Prophet.\nWe must modify the train () function in the following way:\n\n def train():\n print('Starting the training.')\n try:\n # Read in any hyperparameters that the user passed with the training job\n with open(param_path, 'r') as tc:\n trainingParams = json.load(tc)\n # Take the set of files and read them all into a single pandas dataframe\n input_files = [ os.path.join(training_path, file) for file in os.listdir(training_path) ]\n if len(input_files) == 0:\n raise ValueError(('There are no files in {}.\\n' +\n 'This usually indicates that the channel ({}) was incorrectly specified,\\n' +\n 'the data specification in S3 was incorrectly specified or the role specified\\n' +\n 'does not have permission to access the data.').format(training_path, channel_name))\n raw_data = [ pd.read_csv(file, error_bad_lines=False ) for file in input_files ]\n train_data = pd.concat(raw_data)\n train_data.columns = ['ds', 'y']\n\n # Usamos Prophet para entrenar el modelo.\n clf = Prophet()\n clf = clf.fit(train_data)\n\n # save the model\n with open(os.path.join(model_path, 'prophet-model.pkl'), 'w') as out:\n pickle.dump(clf, out)\n print('Training complete.')\n",
"_____no_output_____"
],
[
"### The predictor.py file\n\nThe predictor.py file describes the way we are going to make predictions.\nThe file Prophet-Docker / container / prophet / predictor.py contains the specific prediction code for Prophet.\nWe must modify the predict () function in the following way:\n\n def predict(cls, input):\n \"\"\"For the input, do the predictions and return them.\n\n Args:\n input (a pandas dataframe): The data on which to do the predictions. There will be\n one prediction per row in the dataframe\"\"\"\n clf = cls.get_model()\n future = clf.make_future_dataframe(periods=int(input.iloc[0]))\n print(int(input.iloc[0]))\n print(input)\n forecast = clf.predict(future)\n \n return forecast.tail(int(input.iloc[0]))\n\n\nAnd then the transformation () function as follows:\n\n def transformation():\n \"\"\"Do an inference on a single batch of data. In this sample server, we take data as CSV, convert\n it to a pandas data frame for internal use and then convert the predictions back to CSV (which really\n just means one prediction per line, since there's a single column.\n \"\"\"\n data = None\n\n # Convert from CSV to pandas\n if flask.request.content_type == 'text/csv':\n data = flask.request.data.decode('utf-8')\n s = StringIO.StringIO(data)\n data = pd.read_csv(s, header=None)\n else:\n return flask.Response(response='This predictor only supports CSV data', status=415, mimetype='text/plain')\n\n print('Invoked with {} records'.format(data.shape[0]))\n\n # Do the prediction\n predictions = ScoringService.predict(data)\n\n # Convert from numpy back to CSV\n out = StringIO.StringIO()\n pd.DataFrame({'results':[predictions]}, index=[0]).to_csv(out, header=False, index=False)\n result = out.getvalue()\n\n return flask.Response(response=result, status=200, mimetype='text/csv')\n \n\nBasically we modify the line:\n\n pd.DataFrame({'results':predictions}).to_csv(out, header=False, index=False)\n \nBy the line:\n\n pd.DataFrame({'results':[predictions]}, index=[0]).to_csv(out, header=False, index=False)\n",
"_____no_output_____"
],
[
"# Part 3: Using Prophet in Amazon SageMaker\nNow that we have all the files created, we are going to use Prophet in Sagemaker\n\n## Container assembly\nWe start by building and registering the container",
"_____no_output_____"
]
],
[
[
"%%time\n%%sh\n\n# The name of our algorithm\nalgorithm_name=sagemaker-prophet\n\ncd container\n\nchmod +x prophet/train\nchmod +x prophet/serve\n\naccount=$(aws sts get-caller-identity --query Account --output text)\n\n# Get the region defined in the current configuration (default to us-west-2 if none defined)\nregion=$(aws configure get region)\nregion=${region:-us-west-2}\n\nfullname=\"${account}.dkr.ecr.${region}.amazonaws.com/${algorithm_name}:latest\"\n\n# If the repository doesn't exist in ECR, create it.\naws ecr describe-repositories --repository-names \"${algorithm_name}\" > /dev/null 2>&1\n\nif [ $? -ne 0 ]\nthen\n aws ecr create-repository --repository-name \"${algorithm_name}\" > /dev/null\nfi\n\n# Get the login command from ECR and execute it directly\n$(aws ecr get-login --region ${region} --no-include-email)\n\n# Build the docker image locally with the image name and then push it to ECR\n# with the full name.\n\ndocker build -t ${algorithm_name} .\ndocker tag ${algorithm_name} ${fullname}\n\ndocker push ${fullname}",
"Login Succeeded\nSending build context to Docker daemon 63.49kB\nStep 1/11 : FROM ubuntu:16.04\n ---> c6a43cd4801e\nStep 2/11 : MAINTAINER Amazon AI <[email protected]>\n ---> Using cache\n ---> c0ea7ed783e7\nStep 3/11 : RUN apt-get -y update && apt-get install -y --no-install-recommends wget curl python-dev build-essential libssl-dev libffi-dev libxml2-dev libxslt1-dev zlib1g-dev nginx ca-certificates && rm -rf /var/lib/apt/lists/*\n ---> Using cache\n ---> 17bd5ae1900b\nStep 4/11 : RUN curl -fSsL -O https://bootstrap.pypa.io/get-pip.py && python get-pip.py && rm get-pip.py\n ---> Using cache\n ---> e1f1939e31e1\nStep 5/11 : RUN pip --no-cache-dir install numpy scipy sklearn pandas flask gevent gunicorn pystan\n ---> Using cache\n ---> 8ff73a969fc2\nStep 6/11 : RUN pip --no-cache-dir install fbprophet\n ---> Using cache\n ---> 815dc3862860\nStep 7/11 : ENV PYTHONUNBUFFERED=TRUE\n ---> Using cache\n ---> 35c7a5aac761\nStep 8/11 : ENV PYTHONDONTWRITEBYTECODE=TRUE\n ---> Using cache\n ---> ef336e62f7f5\nStep 9/11 : ENV PATH=\"/opt/program:${PATH}\"\n ---> Using cache\n ---> 290851f5e67b\nStep 10/11 : COPY prophet /opt/program\n ---> Using cache\n ---> 4b6b9e299087\nStep 11/11 : WORKDIR /opt/program\n ---> Using cache\n ---> fe21109f123a\nSuccessfully built fe21109f123a\nSuccessfully tagged sagemaker-prophet:latest\nThe push refers to repository [563487891580.dkr.ecr.us-east-1.amazonaws.com/sagemaker-prophet]\n00236715c809: Preparing\n15d05e86afe8: Preparing\n168ec802cd02: Preparing\ne5e30443c428: Preparing\n022ecae92fcb: Preparing\n77008e118980: Preparing\n6cb741cb00b7: Preparing\nf36b28e4310d: Preparing\n91d23cf5425a: Preparing\nf36b28e4310d: Waiting\n91d23cf5425a: Waiting\n77008e118980: Waiting\n6cb741cb00b7: Waiting\ne5e30443c428: Layer already exists\n022ecae92fcb: Layer already exists\n168ec802cd02: Layer already exists\n15d05e86afe8: Layer already exists\n00236715c809: Layer already exists\n77008e118980: Layer already exists\n6cb741cb00b7: Layer already exists\n91d23cf5425a: Layer already exists\nf36b28e4310d: Layer already exists\nlatest: digest: sha256:92c9042b85d712100a6b73c3ab8257944a85f1e46a8d69f78b1dbbb72427f031 size: 2207\n"
]
],
[
[
"## Building the Training Environment\nWe initialize the session, execution role.",
"_____no_output_____"
]
],
[
[
"%%time\nimport boto3\nimport re\n\nimport os\nimport numpy as np\nimport pandas as pd\nfrom sagemaker import get_execution_role\n\nimport sagemaker as sage\nfrom time import gmtime, strftime\n\n\nprefix = 'DEMO-prophet-byo'\nrole = get_execution_role()\nsess = sage.Session()\n",
"CPU times: user 408 ms, sys: 40.3 ms, total: 448 ms\nWall time: 503 ms\n"
]
],
[
[
"# Upload the data to S3",
"_____no_output_____"
]
],
[
[
"WORK_DIRECTORY = 'data'\ndata_location = sess.upload_data(WORK_DIRECTORY, key_prefix=prefix)",
"_____no_output_____"
]
],
[
[
"## Train the model\nUsing the data uploaded to S3, we train the model by raising an ml.c4.2xlarge instance.\nSagemaker will leave the trained model in the / output directory",
"_____no_output_____"
]
],
[
[
"%%time\n\naccount = sess.boto_session.client('sts').get_caller_identity()['Account']\nregion = sess.boto_session.region_name\nimage = '{}.dkr.ecr.{}.amazonaws.com/sagemaker-prophet:latest'.format(account, region)\n\ntseries = sage.estimator.Estimator(image,\n role, \n 1, \n 'ml.c4.2xlarge',\n output_path=\"s3://{}/output\".format(sess.default_bucket()),\n sagemaker_session=sess)\n\ntseries.fit(data_location)",
"2019-12-27 16:00:08 Starting - Starting the training job...\n2019-12-27 16:00:09 Starting - Launching requested ML instances......\n2019-12-27 16:01:13 Starting - Preparing the instances for training...\n2019-12-27 16:01:58 Downloading - Downloading input data\n2019-12-27 16:01:58 Training - Downloading the training image...\n2019-12-27 16:02:34 Training - Training image download completed. Training in progress..\u001b[34mINFO:matplotlib.font_manager:font search path ['/usr/local/lib/python2.7/dist-packages/matplotlib/mpl-data/fonts/ttf', '/usr/local/lib/python2.7/dist-packages/matplotlib/mpl-data/fonts/afm', '/usr/local/lib/python2.7/dist-packages/matplotlib/mpl-data/fonts/pdfcorefonts']\u001b[0m\n\u001b[34mINFO:matplotlib.font_manager:generated new fontManager\u001b[0m\n\u001b[34mERROR:fbprophet:Importing matplotlib failed. Plotting will not work.\u001b[0m\n\u001b[34mERROR:fbprophet:Importing plotly failed. Interactive plots will not work.\u001b[0m\n\u001b[34mStarting the training.\u001b[0m\n\u001b[34mINFO:fbprophet:Disabling weekly seasonality. Run prophet with weekly_seasonality=True to override this.\u001b[0m\n\u001b[34mINFO:fbprophet:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.\u001b[0m\n\u001b[34mInitial log joint probability = -2.69053\n Iter log prob ||dx|| ||grad|| alpha alpha0 # evals Notes \n 99 450.532 0.00314344 85.9671 2.069 0.2069 135 \n Iter log prob ||dx|| ||grad|| alpha alpha0 # evals Notes \n 155 451.709 0.000185872 84.4313 3.9e-06 0.001 245 LS failed, Hessian reset \n 199 451.872 8.44141e-05 56.0574 0.5636 1 296 \n Iter log prob ||dx|| ||grad|| alpha alpha0 # evals Notes \n 299 452.598 0.0313561 102.286 0.9683 1 420 \n Iter log prob ||dx|| ||grad|| alpha alpha0 # evals Notes \n 300 452.617 0.000446882 108.925 4.369e-06 0.001 499 LS failed, Hessian reset \n 386 453.08 1.71918e-05 65.6637 2.482e-07 0.001 648 LS failed, Hessian reset \n 399 453.081 6.92601e-07 51.7009 1.002 0.2039 667 \n Iter log prob ||dx|| ||grad|| alpha alpha0 # evals Notes \n 408 453.081 3.47948e-09 59.4053 0.01718 1 682 \u001b[0m\n\u001b[34mOptimization terminated normally: \n Convergence detected: absolute parameter change was below tolerance\u001b[0m\n\u001b[34mTraining complete.\u001b[0m\n\n2019-12-27 16:02:45 Uploading - Uploading generated training model\n2019-12-27 16:02:45 Completed - Training job completed\nTraining seconds: 60\nBillable seconds: 60\nCPU times: user 362 ms, sys: 18 ms, total: 380 ms\nWall time: 3min 11s\n"
]
],
[
[
"## Endpoint assembly for inference\nUsing the newly trained model, we create an endpoint for inference hosted on an ml.c4.2xlarge instance",
"_____no_output_____"
]
],
[
[
"%%time\n\nfrom sagemaker.predictor import csv_serializer\npredictor = tseries.deploy(1, 'ml.m4.xlarge', serializer=csv_serializer)",
"---------------------------------------------------------------------------------------------------------------!CPU times: user 518 ms, sys: 39.5 ms, total: 557 ms\nWall time: 9min 20s\n"
]
],
[
[
"## Inference test\nFinally we ask the model to predict the sales for the next 30 days.",
"_____no_output_____"
]
],
[
[
"%%time\np = predictor.predict(\"30\")\nprint(p)",
"b'\" ds trend trend_lower ... yearly_lower yearly_upper yhat\\n169 2018-03-26 1.473312 1.473312 ... -0.076117 -0.076117 1.397195\\n170 2018-03-27 1.472971 1.472971 ... -0.072531 -0.072531 1.400440\\n171 2018-03-28 1.472631 1.472631 ... -0.068829 -0.068829 1.403802\\n172 2018-03-29 1.472291 1.472291 ... -0.065070 -0.065070 1.407221\\n173 2018-03-30 1.471950 1.471950 ... -0.061313 -0.061313 1.410637\\n174 2018-03-31 1.471610 1.471610 ... -0.057619 -0.057619 1.413991\\n175 2018-04-01 1.471270 1.471270 ... -0.054048 -0.054048 1.417222\\n176 2018-04-02 1.470929 1.470929 ... -0.050657 -0.050657 1.420273\\n177 2018-04-03 1.470589 1.470589 ... -0.047500 -0.047500 1.423089\\n178 2018-04-04 1.470248 1.470241 ... -0.044627 -0.044627 1.425622\\n179 2018-04-05 1.469908 1.469861 ... -0.042080 -0.042080 1.427828\\n180 2018-04-06 1.469568 1.469467 ... -0.039896 -0.039896 1.429672\\n181 2018-04-07 1.469227 1.469070 ... -0.038104 -0.038104 1.431123\\n182 2018-04-08 1.468887 1.468698 ... -0.036725 -0.036725 1.432162\\n183 2018-04-09 1.468547 1.468307 ... -0.035770 -0.035770 1.432776\\n184 2018-04-10 1.468206 1.467909 ... -0.035245 -0.035245 1.432961\\n185 2018-04-11 1.467866 1.467514 ... -0.035144 -0.035144 1.432722\\n186 2018-04-12 1.467526 1.467111 ... -0.035454 -0.035454 1.432072\\n187 2018-04-13 1.467185 1.466734 ... -0.036155 -0.036155 1.431030\\n188 2018-04-14 1.466845 1.466344 ... -0.037221 -0.037221 1.429624\\n189 2018-04-15 1.466505 1.465922 ... -0.038616 -0.038616 1.427889\\n190 2018-04-16 1.466164 1.465522 ... -0.040301 -0.040301 1.425863\\n191 2018-04-17 1.465824 1.465120 ... -0.042234 -0.042234 1.423590\\n192 2018-04-18 1.465483 1.464696 ... -0.044367 -0.044367 1.421117\\n193 2018-04-19 1.465143 1.464293 ... -0.046650 -0.046650 1.418493\\n194 2018-04-20 1.464803 1.463839 ... -0.049034 -0.049034 1.415769\\n195 2018-04-21 1.464462 1.463360 ... -0.051467 -0.051467 1.412995\\n196 2018-04-22 1.464122 1.462924 ... -0.053900 -0.053900 1.410222\\n197 2018-04-23 1.463782 1.462459 ... -0.056284 -0.056284 1.407497\\n198 2018-04-24 1.463441 1.461976 ... -0.058574 -0.058574 1.404867\\n\\n[30 rows x 16 columns]\"\\n'\nCPU times: user 11.5 ms, sys: 0 ns, total: 11.5 ms\nWall time: 3.03 s\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6ef11228f582529d051d7de4237c791f24981e | 2,008 | ipynb | Jupyter Notebook | lt1456_sliding_window_maximum_vowels.ipynb | devkosal/code_challenges | 0591c4839555376a231682db7cc12c8a70515b09 | [
"MIT"
]
| null | null | null | lt1456_sliding_window_maximum_vowels.ipynb | devkosal/code_challenges | 0591c4839555376a231682db7cc12c8a70515b09 | [
"MIT"
]
| null | null | null | lt1456_sliding_window_maximum_vowels.ipynb | devkosal/code_challenges | 0591c4839555376a231682db7cc12c8a70515b09 | [
"MIT"
]
| null | null | null | 20.916667 | 65 | 0.446215 | [
[
[
"def maxVowels(s: str, k: int) -> int:\n window_start = 0\n counts = {v:0 for v in \"aieou\".split()}\n highest = 0\n for window_end in range(len(s)):\n right_char = s[window_end]\n if right_char in counts:\n counts[right_char] += 1\n print(counts)\n if window_end >= k-1:\n highest = max(highest, sum(counts.values()))\n left_char = s[window_start]\n if left_char in counts:\n counts[left_char] -= 1\n window_start += 1\n return highest",
"_____no_output_____"
],
[
"maxVowels('oiwvb2ev', 2)",
"{'aieou': 0}\n{'aieou': 0}\n{'aieou': 0}\n{'aieou': 0}\n{'aieou': 0}\n{'aieou': 0}\n{'aieou': 0}\n{'aieou': 0}\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
cb6efa7e5bf687625ad33ebcfa32abc68ae3c49c | 135,424 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Updated_goodreads_script-checkpoint.ipynb | mmithil/Web-Mining-Repo | 62fe8f365c38be325dbce1e906a56b9545491150 | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Updated_goodreads_script-checkpoint.ipynb | mmithil/Web-Mining-Repo | 62fe8f365c38be325dbce1e906a56b9545491150 | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Updated_goodreads_script-checkpoint.ipynb | mmithil/Web-Mining-Repo | 62fe8f365c38be325dbce1e906a56b9545491150 | [
"MIT"
]
| null | null | null | 54.234682 | 259 | 0.570829 | [
[
[
"import urllib2\nfrom bs4 import BeautifulSoup\nimport csv\nimport time\nimport re\nimport urllib2\nimport csv\nimport time\nimport sys\nimport xml.etree.ElementTree as ET\nimport os\nimport random\nimport traceback\nfrom IPython.display import clear_output\n\n\ndef createUserDict(user_element):\n #userDict = []\n id = getval(user_element,'id')\n name = getval(user_element,'name')\n user_name = getval(user_element,'user_name')\n profile_url = getval(user_element,'link')\n image_url = getval(user_element,'image_url')\n about = getval(user_element,'about')\n age = getval(user_element,'age')\n gender = getval(user_element,'gender')\n location = getval(user_element,'location')\n joined = getval(user_element,'joined')\n last_active = getval(user_element,'last_active')\n userDict = dict ([('user_id', id), ('name', name) , ('user_name' , user_name),\n ('profile_url', profile_url), ('image_url', image_url),\n ('about', about), ('age', age), ('gender', gender), \n ('location', location) , ('joined', joined), ('last_active', last_active)])\n return userDict\n\ndef writeToCSV(writer, mydict):\n #writer = csv.DictWriter(csvfile, delimiter=',', lineterminator='\\n', fieldnames=insert_fieldnames)\n #for key, value in mydict.items():\n writer.writerow(mydict)\n\n\n\ndef getAmazonDetails(isbn):\n \n with open('csv_files/amazon_book_ratings.csv', 'a') as csvfile_ratings, open('csv_files/amazon_book_reviews.csv', 'a') as csvfile_reviews:\n ##Create file headers and writer\n ratings_fieldnames = ['book_isbn', 'avg_rating', 'five_rating', 'four_rating', 'three_rating', 'two_rating', 'one_rating' ]\n #writer = csv.DictWriter(csvfile_ratings, delimiter=',', lineterminator='\\n', fieldnames=ratings_fieldnames)\n ##writer.writeheader()\n \n reviews_fieldnames = ['book_isbn', 'review'] \n writer_book = csv.DictWriter(csvfile_reviews, delimiter=',', lineterminator='\\n', fieldnames=reviews_fieldnames)\n ##writer_book.writeheader()\n\n ##Get Overall details of the book \n req = urllib2.Request('http://www.amazon.com/product-reviews/' + isbn + '?ie=UTF8&showViewpoints=1&sortBy=helpful&pageNumber=1', headers={ 'User-Agent': 'Mozilla/5.0 (X11; U; Linux i686) Gecko/20071127 Firefox/2.0.0.11' })\n html = urllib2.urlopen(req).read()\n soup = BeautifulSoup(html, 'html.parser')\n \n avgRatingTemp = soup.find_all('div',{'class':\"a-row averageStarRatingNumerical\"})[0].get_text()\n avgRating = re.findall('\\d+\\.\\d+', avgRatingTemp)[0]\n \n try:\n fiveStarRatingTemp = soup.find_all('a',{'class':\"a-size-small a-link-normal 5star histogram-review-count\"})[0].get_text()\n fiveStarRating = fiveStarRatingTemp.strip('%')\n except:\n fiveStarRating = 0\n\n try:\n fourStarRatingTemp = soup.find_all('a',{'class':\"a-size-small a-link-normal 4star histogram-review-count\"})[0].get_text()\n fourStarRating = fourStarRatingTemp.strip('%')\n except:\n fourStarRating = 0\n\n try:\n threeStarRatingTemp = soup.find_all('a',{'class':\"a-size-small a-link-normal 3star histogram-review-count\"})[0].get_text()\n threeStarRating = threeStarRatingTemp.strip('%')\n except:\n threeStarRating = 0\n\n try:\n twoStarRatingTemp = soup.find_all('a',{'class':\"a-size-small a-link-normal 2star histogram-review-count\"})[0].get_text()\n twoStarRating = twoStarRatingTemp.strip('%')\n except:\n twoStarRating = 0\n\n try:\n oneStarRatingTemp = soup.find_all('a',{'class':\"a-size-small a-link-normal 1star histogram-review-count\"})[0].get_text()\n oneStarRating = oneStarRatingTemp.strip('%')\n except:\n oneStarRating = 0\n\n writer.writerow({'book_isbn': isbn, 'avg_rating': avgRating, 'five_rating': fiveStarRating, \n 'four_rating': fourStarRating, 'three_rating': threeStarRating, 'two_rating': twoStarRating,\n 'one_rating': oneStarRating})\n \n ##Get top 20 helpful review of book\n for pagenumber in range(1,3):\n req = urllib2.Request('http://www.amazon.com/product-reviews/' + isbn + '?ie=UTF8&showViewpoints=1&sortBy=helpful&pageNumber='+ str(pagenumber), headers={ 'User-Agent': 'Mozilla/5.0 (X11; U; Linux i686) Gecko/20071127 Firefox/2.0.0.11' })\n html = urllib2.urlopen(req).read()\n soup = BeautifulSoup(html, 'html.parser') \n for i in range(0,10):\n try:\n review = soup.find_all('div',{'class':\"a-section review\"})[i].contents[3].get_text().encode('UTF-8')\n writer_book.writerow({'book_isbn': isbn, 'review': review})\n except:\n print \"No Reviews ISBN - \" + isbn\n \ndef getval(root, element):\n try:\n ret = root.find(element).text\n if ret is None:\n return \"\"\n else:\n return ret.encode(\"utf8\")\n except:\n return \"\"\n \n\nwith open('csv_files/amazon_book_ratings.csv', 'w') as csvfile_ratings, open('csv_files/amazon_book_reviews.csv', 'w') as csvfile_reviews:\n ##Create file headers and writer\n ratings_fieldnames = ['book_isbn', 'avg_rating', 'five_rating', 'four_rating', 'three_rating', 'two_rating', 'one_rating' ]\n writer = csv.DictWriter(csvfile_ratings, delimiter=',', lineterminator='\\n', fieldnames=ratings_fieldnames)\n writer.writeheader()\n \n reviews_fieldnames = ['book_isbn', 'review'] \n writer_book = csv.DictWriter(csvfile_reviews, delimiter=',', lineterminator='\\n', fieldnames=reviews_fieldnames)\n writer_book.writeheader()\n\n\nwith open('csv_files/user_data.csv', 'w') as csvfile, open('csv_files/book_data.csv', 'w') as csvfile_book, open('csv_files/book_author.csv', 'w') as csvfile_author, open('csv_files/goodreads_user_reviews_ratings.csv', 'w') as gdrds_rr:\n fieldnames = ['user_id', 'name','user_name', 'profile_url','image_url', 'about', 'age', 'gender', \n 'location','joined','last_active' ]\n writer = csv.DictWriter(csvfile, delimiter = ',', lineterminator = '\\n', fieldnames=fieldnames)\n writer.writeheader()\n book_fieldnames = [\n 'user_id',\n 'b_id',\n 'shelf',\n 'isbn', \n 'isbn13',\n 'text_reviews_count',\n 'title',\n 'image_url',\n 'link',\n 'num_pages',\n 'b_format',\n 'publisher',\n 'publication_day', \n 'publication_year', \n 'publication_month',\n 'average_rating', \n 'ratings_count', \n 'description', \n 'published',\n\n 'fiction' ,\n 'fantasy' ,\n 'classics' ,\n 'young_adult' ,\n 'romance' ,\n 'non_fiction' ,\n 'historical_fiction' ,\n 'science_fiction' ,\n 'dystopian' ,\n 'horror' ,\n 'paranormal' ,\n 'contemporary' ,\n 'childrens' ,\n 'adult' ,\n 'adventure' ,\n 'novels' ,\n 'urban_fantasy' ,\n 'history' ,\n 'chick_lit' ,\n 'thriller' ,\n 'audiobook' ,\n 'drama' ,\n 'biography' ,\n 'vampires' ]\n \n writer_book = csv.DictWriter(csvfile_book, delimiter = ',', lineterminator = '\\n', fieldnames=book_fieldnames)\n writer_book.writeheader()\n \n goodreads_ratings_fieldnames = ['user_id', 'b_id', 'rating', 'review' ]\n rr_writer = csv.DictWriter(gdrds_rr, delimiter=',', lineterminator='\\n', fieldnames=goodreads_ratings_fieldnames)\n rr_writer.writeheader()\n\n author_fieldnames = [\n 'u_id',\n 'b_id',\n 'a_id',\n 'name',\n 'average_rating',\n 'ratings_count',\n 'text_reviews_count']\n writer_author = csv.DictWriter(csvfile_author, delimiter = ',', lineterminator = '\\n', fieldnames = author_fieldnames)\n writer_author.writeheader()\n\n\n lst = []\n i = 0\n \n while i < 50: \n try: \n \n #time.sleep(1)\n clear_output()\n c = random.randint(1, 2500000)\n #c = 23061285\n print \"random number: \" + str(c) \n\n if (c not in lst):\n print \"getting information for user id:\"+ str(c)\n lst.append(c)\n url = 'https://www.goodreads.com/user/show/'+ str(c) +'.xml?key=i3Zsl7r13oHEQCjv1vXw'\n response = urllib2.urlopen(url)\n user_data_xml = response.read()\n #write xml to file\n \n f = open(\"xml_docs/user\"+ str(c) +\".xml\", \"w\")\n try:\n f.write(user_data_xml)\n finally:\n f.close()\n \n #root = ET.fromstring()\n\n root = ET.parse(\"xml_docs/user\"+ str(c) +\".xml\").getroot()\n os.remove(\"xml_docs/user\"+ str(c) +\".xml\")\n user_element = root.find('user')\n user_shelf_to_count = user_element.find('user_shelves')\n b_count = 0\n for user_shelf in user_shelf_to_count.findall('user_shelf'):\n b_count = b_count + int(getval(user_shelf,'book_count'))\n \n print 'Book count is ' + str(b_count)\n if(b_count > 10):\n\n\n print 'Collecting data for user ' + str(c) \n '''id = getval(user_element,'id')\n name = getval(user_element,'name')\n user_name = getval(user_element,'user_name')\n profile_url = getval(user_element,'link')\n image_url = getval(user_element,'image_url')\n about = getval(user_element,'about')\n age = getval(user_element,'age')\n gender = getval(user_element,'gender')\n location = getval(user_element,'location')\n joined = getval(user_element,'joined')\n last_active = getval(user_element,'last_active')\n '''\n\n userDict = createUserDict(user_element) \n\n id = userDict['user_id']\n #writer.writerow({'id': id, 'name' : name,'user_name' : user_name,\n # 'profile_url' : profile_url,'image_url' : image_url,\n # 'about' : about, 'age': age, 'gender' : gender, \n # 'location' : location, 'joined' : joined, 'last_active': last_active})\n \n writeToCSV(writer,userDict)\n\n\n\n print \"Saved user data for user id:\" + str(c)\n \n \n # get list of user shelves\n \n user_shelves_root = user_element.find('user_shelves')\n \n user_shelf_list = []\n \n for user_shelf in user_shelves_root.findall(\"user_shelf\"):\n shelf = getval(user_shelf,\"name\")\n #Books on Shelf\n print \"Checking for books in shelf: \" + shelf + \" for user id:\" + str(c)\n \n shelf_url = \"https://www.goodreads.com/review/list/\"+ str(c) +\".xml?key=i3Zsl7r13oHEQCjv1vXw&v=2&per_page=200&shelf=\" + shelf\n #time.sleep(1)\n print shelf_url\n response = urllib2.urlopen(shelf_url)\n shelf_data_xml = response.read()\n # write xml to file\n f = open(\"xml_docs/user_shelf_\" + shelf + \"_\"+ str(c) + \".xml\", \"w\")\n try:\n f.write(shelf_data_xml)\n finally:\n f.close()\n \n shelf_root = ET.parse(\"xml_docs/user_shelf_\" + shelf + \"_\"+ str(c) + \".xml\").getroot()\n \n os.remove(\"xml_docs/user_shelf_\" + shelf + \"_\"+ str(c) + \".xml\")\n try:\n reviews = shelf_root.find(\"reviews\")\n \n \n for review in reviews.findall(\"review\"):\n\n for book in review.findall(\"book\"):\n b_id = getval(book,\"id\")\n isbn = getval(book,\"isbn\")\n print \"Fetching data for book with isbn:\" + str(isbn) + \" and id:\" + str(id)\n isbn13 = getval(book,\"isbn13\")\n text_reviews_count = getval(book,\"text_reviews_count\")\n title = getval(book,\"title\")\n image_url = getval(book,\"image_url\")\n link = getval(book,\"link\")\n num_pages = getval(book,\"num_pages\")\n b_format = getval(book,\"format\")\n publisher = getval(book,\"publisher\")\n publication_day = getval(book,\"publication_day\")\n publication_year = getval(book, \"publication_year\") \n publication_month = getval(book,\"publication_month\")\n average_rating = getval(book,\"average_rating\")\n ratings_count = getval(book,\"rating_count\")\n description = getval(book,\"description\")\n published = getval(book,\"published\")\n #getAmazonDetails(isbn)\n\n\n print \"Fetched review data from Amazon for book :\" + title\n\n #get number of books on each type of shelf\n book_url = 'https://www.goodreads.com/book/show/'+str(b_id)+'.xml?key=i3Zsl7r13oHEQCjv1vXw'\n response = urllib2.urlopen(book_url)\n book_data_xml = response.read()\n # write xml to file\n f = open(\"xml_docs/book_data_\" + str(b_id) + \".xml\", \"w\")\n try:\n f.write(book_data_xml)\n finally:\n f.close()\n \n book_root = ET.parse(\"xml_docs/book_data_\" + str(b_id) + \".xml\").getroot()\n os.remove(\"xml_docs/book_data_\" + str(b_id) + \".xml\")\n print \"checking count in shelf for book_id:\" + str(b_id) \n book_root = book_root.find(\"book\")\n book_shelves = book_root.find(\"popular_shelves\")\n \n fiction = 0\n fantasy = 0\n classics = 0\n young_adult = 0\n romance = 0\n non_fiction = 0\n historical_fiction = 0\n science_fiction = 0\n dystopian = 0\n horror = 0\n paranormal = 0\n contemporary = 0\n childrens = 0\n adult = 0\n adventure = 0\n novels = 0\n urban_fantasy = 0\n history = 0\n chick_lit = 0\n thriller = 0\n audiobook = 0\n drama = 0\n biography = 0\n vampires = 0\n cnt = 0\n \n for shelf_type in book_shelves.findall(\"shelf\"):\n attributes = shelf_type.attrib\n name = attributes['name']\n count = attributes['count']\n #print name + \":\" + count\n \n if ( name == 'fiction'):\n fiction = count\n cnt = cnt+count\n if ( name == 'fantasy'):\n fantasy = count\n cnt = cnt+count\n if ( name == 'classics' or name == 'classic'):\n classics = count\n cnt = cnt+count\n if ( name == 'young-adult'):\n young_adult = count\n cnt = cnt+count\n if ( name == 'romance'):\n romance = count\n cnt = cnt+count\n if ( name == 'non-fiction' or name == 'nonfiction'):\n non_fiction = count\n cnt = cnt+count\n if ( name == 'historical-fiction'):\n historical_fiction = count\n cnt = cnt+count\n if ( name == 'science-fiction' or name == 'sci-fi fantasy' or name == 'scifi' or name == 'fantasy-sci-fi' or name == 'sci-fi'):\n science_fiction = count\n cnt = cnt+count\n if ( name == 'dystopian' or name == 'dystopia'):\n dystopian = count\n cnt = cnt+count\n if ( name == 'horror'):\n horror = count\n cnt = cnt+count\n if ( name == 'paranormal'):\n paranormal = count\n cnt = cnt+count\n if ( name == 'contemporary' or name == 'contemporary-fiction'):\n contemporary = count\n cnt = cnt+count\n if ( name == 'childrens' or name == 'children' or name == 'kids' or name =='children-s-books'):\n childrens = count\n cnt = cnt+count\n if ( name == 'adult'):\n adult = count\n cnt = cnt+count\n if ( name == 'adventure'):\n adventure = count\n cnt = cnt+count\n if ( name == 'novels' or name == 'novel'):\n novels = count\n cnt = cnt+count\n if ( name == 'urban-fantasy'):\n urban_fantasy = count\n cnt = cnt+count\n if ( name == 'history' or name == 'historical'):\n history = count\n cnt = cnt+count\n if ( name == 'chick-lit'):\n chick_lit = count\n cnt = cnt+count\n if ( name == 'thriller'):\n thriller = count\n cnt = cnt+count\n if ( name == 'audiobook' or name == \"audio\"):\n audiobook = count\n cnt = cnt+count\n if ( name == 'drama'):\n drama = count\n cnt = cnt+count\n if ( name == 'biography' or name == 'memoirs'):\n biography = count\n cnt = cnt+count\n if ( name == 'vampires' or name == 'vampire'):\n vampires = count\n cnt = cnt+count\n\n fiction = fiction/cnt\n fantasy = fantasy/cnt\n classics = classics/cnt\n young_adult = young_adult/cnt\n romance = romance/cnt\n non_fiction = non_fiction/cnt\n historical_fiction = historical_fiction/cnt\n science_fiction = science_fiction/cnt\n dystopian = dystopian/cnt\n horror = horror/cnt\n paranormal = paranormal/cnt\n contemporary = contemporary/cnt\n childrens = childrens/cnt\n adult = adult/cnt\n adventure = adventures/cnt\n novels = novels/cnt\n urban_fantasy = urban_fantasy/cnt\n history = history/cnt\n chick_lit = chick_lit/cnt\n thriller = thriller/cnt\n audiobook = audiobook/cnt\n drama = drama/cnt\n biography = biography/cnt\n vampires = vampires/cnt\n writer_book.writerow({\n 'user_id': id,\n 'b_id' : b_id ,\n 'shelf' : shelf,\n 'isbn' : isbn, \n 'isbn13': isbn13,\n 'text_reviews_count' : text_reviews_count,\n 'title' : title,\n 'image_url' : image_url,\n 'link' : link,\n 'num_pages' : num_pages,\n 'b_format' : b_format,\n 'publisher' : publisher,\n 'publication_day' : publication_day, \n 'publication_year' : publication_year, \n 'publication_month' : publication_month,\n 'average_rating' : average_rating, \n 'ratings_count' : ratings_count, \n 'description' : description, \n \n 'fiction' : fiction , \n 'fantasy' : fantasy ,\n 'classics' : classics ,\n 'young_adult' : young_adult ,\n 'romance' : romance ,\n 'non_fiction' : non_fiction ,\n 'historical_fiction' : historical_fiction ,\n 'science_fiction' : science_fiction ,\n 'dystopian' : dystopian ,\n 'horror' : horror ,\n 'paranormal' : paranormal ,\n 'contemporary' : contemporary ,\n 'childrens' : childrens ,\n 'adult' : adult ,\n 'adventure' : adventure ,\n 'novels' : novels ,\n 'urban_fantasy' : urban_fantasy ,\n 'history' : history ,\n 'chick_lit' : chick_lit ,\n 'thriller' : thriller ,\n 'audiobook' : audiobook ,\n 'drama' : drama ,\n 'biography' : biography ,\n 'vampires' : vampires })\n\n\n #bookDict = createBookDict(book) \n\n print \"Data written on csv for book:\" + title\n\n print \"Getting reviews details from user: \" + str(id) + \" and book_id: \" + str(b_id)\n review_url = \"https://www.goodreads.com/review/show_by_user_and_book.xml?book_id=\" +str(b_id)+ \"&key=i3Zsl7r13oHEQCjv1vXw&user_id=\" + str(id)\n review_response = urllib2.urlopen(review_url)\n review_response_xml = review_response.read()\n review_root = ET.fromstring(review_response_xml)\n user_rr = review_root.find(\"review\")\n \n user_r_rating = getval(user_rr, \"rating\")\n print \"Got user review rating: \" + user_r_rating\n\n user_r_review = getval(user_rr, \"body\")\n print \"User review is: \" + user_r_review\n\n rr_writer.writerow({\n 'user_id': id,\n 'b_id' : b_id ,\n 'rating' : user_r_rating,\n 'review' : user_r_review })\n\n\n authors = book.find(\"authors\")\n for author in authors.findall(\"author\"):\n a_id = getval(author,\"id\")\n name = getval(author,\"name\")\n average_rating = getval(author,\"average_rating\")\n ratings_count = getval(author,\"ratings_count\")\n text_reviews_count = getval(author,\"text_reviews_count\")\n writer_author.writerow({'u_id': id,\n 'b_id' : b_id,\n 'a_id' : a_id,\n 'name' : name,\n 'average_rating' : average_rating,\n 'ratings_count' : ratings_count,\n 'text_reviews_count' : text_reviews_count})\n except Exception, e:\n traceback.print_exc()\n \n i = i + 1\n except:\n #time.sleep(1)\n print \"Exception!!\"\n traceback.print_exc()\n print \"End of Program\"",
"random number: 2373891\ngetting information for user id:2373891\nBook count is 257\nCollecting data for user 2373891\nSaved user data for user id:2373891\nChecking for books in shelf: read for user id:2373891\nhttps://www.goodreads.com/review/list/2373891.xml?key=i3Zsl7r13oHEQCjv1vXw&v=2&per_page=200&shelf=read\nFetching data for book with isbn:0778328791 and id:2373891\nFetched review data from Amazon for book :These Things Hidden\nchecking count in shelf for book_id:9166559\nData written on csv for book:These Things Hidden\nGetting reviews details from user: 2373891 and book_id: 9166559\nGot user review rating: 3\nUser review is: \n \nFetching data for book with isbn:1620610078 and id:2373891\nFetched review data from Amazon for book :Obsidian (Lux, #1)\nchecking count in shelf for book_id:12578077\nData written on csv for book:Obsidian (Lux, #1)\nGetting reviews details from user: 2373891 and book_id: 12578077\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743454154 and id:2373891\nFetched review data from Amazon for book :Blood Memory (Mississipi #5)\nchecking count in shelf for book_id:80631\nData written on csv for book:Blood Memory (Mississipi #5)\nGetting reviews details from user: 2373891 and book_id: 80631\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0345534352 and id:2373891\nFetched review data from Amazon for book :He's Gone\nchecking count in shelf for book_id:15841844\nData written on csv for book:He's Gone\nGetting reviews details from user: 2373891 and book_id: 15841844\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1595540547 and id:2373891\nFetched review data from Amazon for book :When Crickets Cry\nchecking count in shelf for book_id:241387\nData written on csv for book:When Crickets Cry\nGetting reviews details from user: 2373891 and book_id: 241387\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0007524277 and id:2373891\nFetched review data from Amazon for book :Allegiant (Divergent, #3)\nchecking count in shelf for book_id:18710190\nData written on csv for book:Allegiant (Divergent, #3)\nGetting reviews details from user: 2373891 and book_id: 18710190\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0007442912 and id:2373891\nFetched review data from Amazon for book :Insurgent (Divergent, #2)\nchecking count in shelf for book_id:11735983\nData written on csv for book:Insurgent (Divergent, #2)\nGetting reviews details from user: 2373891 and book_id: 11735983\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0062024035 and id:2373891\nFetched review data from Amazon for book :Divergent (Divergent, #1)\nchecking count in shelf for book_id:13335037\nData written on csv for book:Divergent (Divergent, #1)\nGetting reviews details from user: 2373891 and book_id: 13335037\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316099295 and id:2373891\nFetched review data from Amazon for book :Right As Rain: A Derek Strange Novel\nchecking count in shelf for book_id:10451507\nData written on csv for book:Right As Rain: A Derek Strange Novel\nGetting reviews details from user: 2373891 and book_id: 10451507\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476733953 and id:2373891\nFetched review data from Amazon for book :Wool (Silo, #1) (Wool, #1-5)\nchecking count in shelf for book_id:17164655\nData written on csv for book:Wool (Silo, #1) (Wool, #1-5)\nGetting reviews details from user: 2373891 and book_id: 17164655\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0061928178 and id:2373891\nFetched review data from Amazon for book :Beautiful Ruins\nchecking count in shelf for book_id:15818133\nData written on csv for book:Beautiful Ruins\nGetting reviews details from user: 2373891 and book_id: 15818133\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1439168857 and id:2373891\nFetched review data from Amazon for book :The Runaway Princess\nchecking count in shelf for book_id:13547080\nData written on csv for book:The Runaway Princess\nGetting reviews details from user: 2373891 and book_id: 13547080\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:006224454X and id:2373891\nFetched review data from Amazon for book :Before I Go To Sleep\nchecking count in shelf for book_id:15818923\nData written on csv for book:Before I Go To Sleep\nGetting reviews details from user: 2373891 and book_id: 15818923\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0679644199 and id:2373891\nFetched review data from Amazon for book :Tell the Wolves I'm Home\nchecking count in shelf for book_id:12875258\nData written on csv for book:Tell the Wolves I'm Home\nGetting reviews details from user: 2373891 and book_id: 12875258\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0316725366 and id:2373891\nFetched review data from Amazon for book :Derailed\nchecking count in shelf for book_id:314362\nData written on csv for book:Derailed\nGetting reviews details from user: 2373891 and book_id: 314362\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0385342063 and id:2373891\nFetched review data from Amazon for book :I've Got Your Number\nchecking count in shelf for book_id:12033455\nData written on csv for book:I've Got Your Number\nGetting reviews details from user: 2373891 and book_id: 12033455\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0739465511 and id:2373891\nFetched review data from Amazon for book :The Lincoln Lawyer (Mickey Haller, #1)\nchecking count in shelf for book_id:79885\nData written on csv for book:The Lincoln Lawyer (Mickey Haller, #1)\nGetting reviews details from user: 2373891 and book_id: 79885\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1416524797 and id:2373891\nFetched review data from Amazon for book :Angels & Demons (Robert Langdon, #1)\nchecking count in shelf for book_id:960\nData written on csv for book:Angels & Demons (Robert Langdon, #1)\nGetting reviews details from user: 2373891 and book_id: 960\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0449006530 and id:2373891\nFetched review data from Amazon for book :Midnight Voices\nchecking count in shelf for book_id:6552\nData written on csv for book:Midnight Voices\nGetting reviews details from user: 2373891 and book_id: 6552\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553288342 and id:2373891\nFetched review data from Amazon for book :Sleepwalk\nchecking count in shelf for book_id:760305\nData written on csv for book:Sleepwalk\nGetting reviews details from user: 2373891 and book_id: 760305\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:034548701X and id:2373891\nFetched review data from Amazon for book :In the Dark of the Night\nchecking count in shelf for book_id:6545\nData written on csv for book:In the Dark of the Night\nGetting reviews details from user: 2373891 and book_id: 6545\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0440170842 and id:2373891\nFetched review data from Amazon for book :Punish the Sinners\nchecking count in shelf for book_id:816858\nData written on csv for book:Punish the Sinners\nGetting reviews details from user: 2373891 and book_id: 816858\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0440114756 and id:2373891\nFetched review data from Amazon for book :Comes the Blind Fury\nchecking count in shelf for book_id:239887\nData written on csv for book:Comes the Blind Fury\nGetting reviews details from user: 2373891 and book_id: 239887\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553284118 and id:2373891\nFetched review data from Amazon for book :Creature\nchecking count in shelf for book_id:6562\nData written on csv for book:Creature\nGetting reviews details from user: 2373891 and book_id: 6562\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:044018293X and id:2373891\nFetched review data from Amazon for book :Suffer the Children\nchecking count in shelf for book_id:6572\nData written on csv for book:Suffer the Children\nGetting reviews details from user: 2373891 and book_id: 6572\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553560271 and id:2373891\nFetched review data from Amazon for book :Shadows\nchecking count in shelf for book_id:6556\nData written on csv for book:Shadows\nGetting reviews details from user: 2373891 and book_id: 6556\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425098605 and id:2373891\nFetched review data from Amazon for book :The Vision\nchecking count in shelf for book_id:481335\nData written on csv for book:The Vision\nGetting reviews details from user: 2373891 and book_id: 481335\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:042511984X and id:2373891\nFetched review data from Amazon for book :The Face of Fear\nchecking count in shelf for book_id:64960\nData written on csv for book:The Face of Fear\nGetting reviews details from user: 2373891 and book_id: 64960\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425127583 and id:2373891\nFetched review data from Amazon for book :The Mask\nchecking count in shelf for book_id:228221\nData written on csv for book:The Mask\nGetting reviews details from user: 2373891 and book_id: 228221\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425099334 and id:2373891\nFetched review data from Amazon for book :Shattered\nchecking count in shelf for book_id:32438\nData written on csv for book:Shattered\nGetting reviews details from user: 2373891 and book_id: 32438\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425142485 and id:2373891\nFetched review data from Amazon for book :The Funhouse\nchecking count in shelf for book_id:11166889\nData written on csv for book:The Funhouse\nGetting reviews details from user: 2373891 and book_id: 11166889\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553807145 and id:2373891\nFetched review data from Amazon for book :Relentless\nchecking count in shelf for book_id:4946005\nData written on csv for book:Relentless\nGetting reviews details from user: 2373891 and book_id: 4946005\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553584480 and id:2373891\nFetched review data from Amazon for book :The Face\nchecking count in shelf for book_id:32437\nData written on csv for book:The Face\nGetting reviews details from user: 2373891 and book_id: 32437\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425199584 and id:2373891\nFetched review data from Amazon for book :Cold Fire\nchecking count in shelf for book_id:32442\nData written on csv for book:Cold Fire\nGetting reviews details from user: 2373891 and book_id: 32442\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425208435 and id:2373891\nFetched review data from Amazon for book :Dragon Tears\nchecking count in shelf for book_id:32429\nData written on csv for book:Dragon Tears\nGetting reviews details from user: 2373891 and book_id: 32429\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425210758 and id:2373891\nFetched review data from Amazon for book :Mr. Murder\nchecking count in shelf for book_id:32434\nData written on csv for book:Mr. Murder\nGetting reviews details from user: 2373891 and book_id: 32434\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425100650 and id:2373891\nFetched review data from Amazon for book :Twilight Eyes\nchecking count in shelf for book_id:693172\nData written on csv for book:Twilight Eyes\nGetting reviews details from user: 2373891 and book_id: 693172\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553479016 and id:2373891\nFetched review data from Amazon for book :Seize the Night (Moonlight Bay, #2)\nchecking count in shelf for book_id:21362\nData written on csv for book:Seize the Night (Moonlight Bay, #2)\nGetting reviews details from user: 2373891 and book_id: 21362\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425203891 and id:2373891\nFetched review data from Amazon for book :Hideaway\nchecking count in shelf for book_id:32422\nData written on csv for book:Hideaway\nGetting reviews details from user: 2373891 and book_id: 32422\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:042520992X and id:2373891\nFetched review data from Amazon for book :Whispers\nchecking count in shelf for book_id:64948\nData written on csv for book:Whispers\nGetting reviews details from user: 2373891 and book_id: 64948\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0345405137 and id:2373891\nFetched review data from Amazon for book :Tick Tock\nchecking count in shelf for book_id:281433\nData written on csv for book:Tick Tock\nGetting reviews details from user: 2373891 and book_id: 281433\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553588249 and id:2373891\nFetched review data from Amazon for book :Life Expectancy\nchecking count in shelf for book_id:16435\nData written on csv for book:Life Expectancy\nGetting reviews details from user: 2373891 and book_id: 16435\nGot user review rating: 5\nUser review is: \n \nFetching data for book with isbn:0553804812 and id:2373891\nFetched review data from Amazon for book :The Good Guy\nchecking count in shelf for book_id:32441\nData written on csv for book:The Good Guy\nGetting reviews details from user: 2373891 and book_id: 32441\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553579754 and id:2373891\nFetched review data from Amazon for book :Fear Nothing (Moonlight Bay, #1)\nchecking count in shelf for book_id:32432\nData written on csv for book:Fear Nothing (Moonlight Bay, #1)\nGetting reviews details from user: 2373891 and book_id: 32432\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425181111 and id:2373891\nFetched review data from Amazon for book :Strangers\nchecking count in shelf for book_id:15676\nData written on csv for book:Strangers\nGetting reviews details from user: 2373891 and book_id: 15676\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553804790 and id:2373891\nFetched review data from Amazon for book :The Husband\nchecking count in shelf for book_id:16429\nData written on csv for book:The Husband\nGetting reviews details from user: 2373891 and book_id: 16429\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425192032 and id:2373891\nFetched review data from Amazon for book :Lightning\nchecking count in shelf for book_id:32424\nData written on csv for book:Lightning\nGetting reviews details from user: 2373891 and book_id: 32424\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553804804 and id:2373891\nFetched review data from Amazon for book :Brother Odd (Odd Thomas, #3)\nchecking count in shelf for book_id:14996\nData written on csv for book:Brother Odd (Odd Thomas, #3)\nGetting reviews details from user: 2373891 and book_id: 14996\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425181103 and id:2373891\nFetched review data from Amazon for book :Phantoms\nchecking count in shelf for book_id:32435\nData written on csv for book:Phantoms\nGetting reviews details from user: 2373891 and book_id: 32435\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0553588265 and id:2373891\nFetched review data from Amazon for book :Forever Odd (Odd Thomas, #2)\nchecking count in shelf for book_id:16433\nData written on csv for book:Forever Odd (Odd Thomas, #2)\nGetting reviews details from user: 2373891 and book_id: 16433\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :Watchers\nchecking count in shelf for book_id:32423\nData written on csv for book:Watchers\nGetting reviews details from user: 2373891 and book_id: 32423\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:067100042X and id:2373891\nFetched review data from Amazon for book :Silent Night\nchecking count in shelf for book_id:842355\nData written on csv for book:Silent Night\nGetting reviews details from user: 2373891 and book_id: 842355\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743484355 and id:2373891\nFetched review data from Amazon for book :A Cry In The Night\nchecking count in shelf for book_id:43345\nData written on csv for book:A Cry In The Night\nGetting reviews details from user: 2373891 and book_id: 43345\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743484304 and id:2373891\nFetched review data from Amazon for book :Moonlight Becomes You\nchecking count in shelf for book_id:237114\nData written on csv for book:Moonlight Becomes You\nGetting reviews details from user: 2373891 and book_id: 237114\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1416516743 and id:2373891\nFetched review data from Amazon for book :Let Me Call You Sweetheart\nchecking count in shelf for book_id:170632\nData written on csv for book:Let Me Call You Sweetheart\nGetting reviews details from user: 2373891 and book_id: 170632\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743484363 and id:2373891\nFetched review data from Amazon for book :Remember Me\nchecking count in shelf for book_id:35354\nData written on csv for book:Remember Me\nGetting reviews details from user: 2373891 and book_id: 35354\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0671004549 and id:2373891\nFetched review data from Amazon for book :You Belong To Me\nchecking count in shelf for book_id:43342\nData written on csv for book:You Belong To Me\nGetting reviews details from user: 2373891 and book_id: 43342\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1416516727 and id:2373891\nFetched review data from Amazon for book :All Around the Town\nchecking count in shelf for book_id:170619\nData written on csv for book:All Around the Town\nGetting reviews details from user: 2373891 and book_id: 170619\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0671673688 and id:2373891\nFetched review data from Amazon for book :While My Pretty One Sleeps\nchecking count in shelf for book_id:571555\nData written on csv for book:While My Pretty One Sleeps\nGetting reviews details from user: 2373891 and book_id: 571555\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0671758896 and id:2373891\nFetched review data from Amazon for book :Loves Music, Loves to Dance\nchecking count in shelf for book_id:170650\nData written on csv for book:Loves Music, Loves to Dance\nGetting reviews details from user: 2373891 and book_id: 170650\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0142000205 and id:2373891\nFetched review data from Amazon for book :Icy Sparks\nchecking count in shelf for book_id:3476\nData written on csv for book:Icy Sparks\nGetting reviews details from user: 2373891 and book_id: 3476\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0751529818 and id:2373891\nFetched review data from Amazon for book :Tuesdays with Morrie\nchecking count in shelf for book_id:6900\nData written on csv for book:Tuesdays with Morrie\nGetting reviews details from user: 2373891 and book_id: 6900\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :Taking Chances (Taking Chances, #1)\nchecking count in shelf for book_id:15739018\nData written on csv for book:Taking Chances (Taking Chances, #1)\nGetting reviews details from user: 2373891 and book_id: 15739018\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0749932775 and id:2373891\nFetched review data from Amazon for book :Dance Upon the Air (Three Sisters Island, #1)\nchecking count in shelf for book_id:59829\nData written on csv for book:Dance Upon the Air (Three Sisters Island, #1)\nGetting reviews details from user: 2373891 and book_id: 59829\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0749932988 and id:2373891\nFetched review data from Amazon for book :Face the Fire (Three Sisters Island, #3)\nchecking count in shelf for book_id:59822\nData written on csv for book:Face the Fire (Three Sisters Island, #3)\nGetting reviews details from user: 2373891 and book_id: 59822\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0749932821 and id:2373891\nFetched review data from Amazon for book :Heaven and Earth (Three Sisters Island, #2)\nchecking count in shelf for book_id:59830\nData written on csv for book:Heaven and Earth (Three Sisters Island, #2)\nGetting reviews details from user: 2373891 and book_id: 59830\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1416537775 and id:2373891\nFetched review data from Amazon for book :Hello, Darkness\nchecking count in shelf for book_id:30381\nData written on csv for book:Hello, Darkness\nGetting reviews details from user: 2373891 and book_id: 30381\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0340827688 and id:2373891\nFetched review data from Amazon for book :The Crush\nchecking count in shelf for book_id:96262\nData written on csv for book:The Crush\nGetting reviews details from user: 2373891 and book_id: 96262\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:044619154X and id:2373891\nFetched review data from Amazon for book :The Witness\nchecking count in shelf for book_id:323289\nData written on csv for book:The Witness\nGetting reviews details from user: 2373891 and book_id: 323289\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446353957 and id:2373891\nFetched review data from Amazon for book :Mirror Image\nchecking count in shelf for book_id:685788\nData written on csv for book:Mirror Image\nGetting reviews details from user: 2373891 and book_id: 685788\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743245539 and id:2373891\nFetched review data from Amazon for book :White Hot\nchecking count in shelf for book_id:710826\nData written on csv for book:White Hot\nGetting reviews details from user: 2373891 and book_id: 710826\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1416563067 and id:2373891\nFetched review data from Amazon for book :Smoke Screen\nchecking count in shelf for book_id:2306910\nData written on csv for book:Smoke Screen\nGetting reviews details from user: 2373891 and book_id: 2306910\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743466772 and id:2373891\nFetched review data from Amazon for book :Chill Factor\nchecking count in shelf for book_id:268275\nData written on csv for book:Chill Factor\nGetting reviews details from user: 2373891 and book_id: 268275\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0778325857 and id:2373891\nFetched review data from Amazon for book :Deadly Night (Flynn Brothers, #1)\nchecking count in shelf for book_id:2930659\nData written on csv for book:Deadly Night (Flynn Brothers, #1)\nGetting reviews details from user: 2373891 and book_id: 2930659\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0778325601 and id:2373891\nFetched review data from Amazon for book :Deadly Harvest (Flynn Brothers, #2)\nchecking count in shelf for book_id:3643502\nData written on csv for book:Deadly Harvest (Flynn Brothers, #2)\nGetting reviews details from user: 2373891 and book_id: 3643502\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0778325164 and id:2373891\nFetched review data from Amazon for book :The Widow (Boston Police/FBI, #1)\nchecking count in shelf for book_id:2071486\nData written on csv for book:The Widow (Boston Police/FBI, #1)\nGetting reviews details from user: 2373891 and book_id: 2071486\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0778328511 and id:2373891\nFetched review data from Amazon for book :The Whisper (Boston Police/FBI, #4)\nchecking count in shelf for book_id:7798635\nData written on csv for book:The Whisper (Boston Police/FBI, #4)\nGetting reviews details from user: 2373891 and book_id: 7798635\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0451933028 and id:2373891\nFetched review data from Amazon for book :The Green Mile\nchecking count in shelf for book_id:11566\nData written on csv for book:The Green Mile\nGetting reviews details from user: 2373891 and book_id: 11566\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425214435 and id:2373891\nFetched review data from Amazon for book :Eyes of Prey (Lucas Davenport, #3)\nchecking count in shelf for book_id:37297\nData written on csv for book:Eyes of Prey (Lucas Davenport, #3)\nGetting reviews details from user: 2373891 and book_id: 37297\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743484207 and id:2373891\nFetched review data from Amazon for book :Secret Prey (Lucas Davenport, #9)\nchecking count in shelf for book_id:216131\nData written on csv for book:Secret Prey (Lucas Davenport, #9)\nGetting reviews details from user: 2373891 and book_id: 216131\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425141233 and id:2373891\nFetched review data from Amazon for book :Winter Prey (Lucas Davenport, #5)\nchecking count in shelf for book_id:37304\nData written on csv for book:Winter Prey (Lucas Davenport, #5)\nGetting reviews details from user: 2373891 and book_id: 37304\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425205819 and id:2373891\nFetched review data from Amazon for book :Rules of Prey (Lucas Davenport, #1)\nchecking count in shelf for book_id:37301\nData written on csv for book:Rules of Prey (Lucas Davenport, #1)\nGetting reviews details from user: 2373891 and book_id: 37301\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425199606 and id:2373891\nFetched review data from Amazon for book :Hidden Prey (Lucas Davenport, #15)\nchecking count in shelf for book_id:888927\nData written on csv for book:Hidden Prey (Lucas Davenport, #15)\nGetting reviews details from user: 2373891 and book_id: 888927\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:031606792X and id:2373891\nFetched review data from Amazon for book :Breaking Dawn (Twilight, #4)\nchecking count in shelf for book_id:1162543\nData written on csv for book:Breaking Dawn (Twilight, #4)\nGetting reviews details from user: 2373891 and book_id: 1162543\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743453018 and id:2373891\nFetched review data from Amazon for book :Monday Mourning (Temperance Brennan, #7)\nchecking count in shelf for book_id:181116\nData written on csv for book:Monday Mourning (Temperance Brennan, #7)\nGetting reviews details from user: 2373891 and book_id: 181116\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0684859734 and id:2373891\nFetched review data from Amazon for book :Grave Secrets (Temperance Brennan, #5)\nchecking count in shelf for book_id:281350\nData written on csv for book:Grave Secrets (Temperance Brennan, #5)\nGetting reviews details from user: 2373891 and book_id: 281350\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0671028367 and id:2373891\nFetched review data from Amazon for book :Deadly Decisions (Temperance Brennan, #3)\nchecking count in shelf for book_id:128754\nData written on csv for book:Deadly Decisions (Temperance Brennan, #3)\nGetting reviews details from user: 2373891 and book_id: 128754\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0671011375 and id:2373891\nFetched review data from Amazon for book :Death du Jour (Temperance Brennan, #2)\nchecking count in shelf for book_id:128756\nData written on csv for book:Death du Jour (Temperance Brennan, #2)\nGetting reviews details from user: 2373891 and book_id: 128756\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1416510567 and id:2373891\nFetched review data from Amazon for book :Fatal Voyage (Temperance Brennan, #4)\nchecking count in shelf for book_id:128759\nData written on csv for book:Fatal Voyage (Temperance Brennan, #4)\nGetting reviews details from user: 2373891 and book_id: 128759\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:074345300X and id:2373891\nFetched review data from Amazon for book :Bare Bones (Temperance Brennan, #6)\nchecking count in shelf for book_id:128752\nData written on csv for book:Bare Bones (Temperance Brennan, #6)\nGetting reviews details from user: 2373891 and book_id: 128752\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425175405 and id:2373891\nFetched review data from Amazon for book :Black Notice (Kay Scarpetta, #10)\nchecking count in shelf for book_id:123598\nData written on csv for book:Black Notice (Kay Scarpetta, #10)\nGetting reviews details from user: 2373891 and book_id: 123598\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0751525359 and id:2373891\nFetched review data from Amazon for book :The Last Precinct (Kay Scarpetta, #11)\nchecking count in shelf for book_id:320167\nData written on csv for book:The Last Precinct (Kay Scarpetta, #11)\nGetting reviews details from user: 2373891 and book_id: 320167\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425213382 and id:2373891\nFetched review data from Amazon for book :Cause of Death (Kay Scarpetta, #7)\nchecking count in shelf for book_id:6541\nData written on csv for book:Cause of Death (Kay Scarpetta, #7)\nGetting reviews details from user: 2373891 and book_id: 6541\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0751530492 and id:2373891\nFetched review data from Amazon for book :Unnatural Exposure (Kay Scarpetta, #8)\nchecking count in shelf for book_id:232145\nData written on csv for book:Unnatural Exposure (Kay Scarpetta, #8)\nGetting reviews details from user: 2373891 and book_id: 232145\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425204693 and id:2373891\nFetched review data from Amazon for book :From Potter's Field (Kay Scarpetta, #6)\nchecking count in shelf for book_id:6537\nData written on csv for book:From Potter's Field (Kay Scarpetta, #6)\nGetting reviews details from user: 2373891 and book_id: 6537\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0751530484 and id:2373891\nFetched review data from Amazon for book :Point of Origin (Kay Scarpetta, #9)\nchecking count in shelf for book_id:6531\nData written on csv for book:Point of Origin (Kay Scarpetta, #9)\nGetting reviews details from user: 2373891 and book_id: 6531\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0684193957 and id:2373891\nFetched review data from Amazon for book :All That Remains (Kay Scarpetta, #3)\nchecking count in shelf for book_id:232123\nData written on csv for book:All That Remains (Kay Scarpetta, #3)\nGetting reviews details from user: 2373891 and book_id: 232123\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0425201449 and id:2373891\nFetched review data from Amazon for book :The Body Farm (Kay Scarpetta, #5)\nchecking count in shelf for book_id:6539\nData written on csv for book:The Body Farm (Kay Scarpetta, #5)\nGetting reviews details from user: 2373891 and book_id: 6539\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0380718340 and id:2373891\nFetched review data from Amazon for book :Cruel & Unusual (Kay Scarpetta, #4)\nchecking count in shelf for book_id:85379\nData written on csv for book:Cruel & Unusual (Kay Scarpetta, #4)\nGetting reviews details from user: 2373891 and book_id: 85379\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:006222543X and id:2373891\nFetched review data from Amazon for book :Reconstructing Amelia\nchecking count in shelf for book_id:15776309\nData written on csv for book:Reconstructing Amelia\nGetting reviews details from user: 2373891 and book_id: 15776309\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0061878251 and id:2373891\nFetched review data from Amazon for book :Into the Dark (Brenna Spector, #2)\nchecking count in shelf for book_id:15818129\nData written on csv for book:Into the Dark (Brenna Spector, #2)\nGetting reviews details from user: 2373891 and book_id: 15818129\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316176486 and id:2373891\nFetched review data from Amazon for book :Life After Life\nchecking count in shelf for book_id:15790842\nData written on csv for book:Life After Life\nGetting reviews details from user: 2373891 and book_id: 15790842\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1301949825 and id:2373891\nFetched review data from Amazon for book :Hopeless (Hopeless, #1)\nchecking count in shelf for book_id:15717943\nData written on csv for book:Hopeless (Hopeless, #1)\nGetting reviews details from user: 2373891 and book_id: 15717943\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0061726818 and id:2373891\nFetched review data from Amazon for book :Before I Fall\nchecking count in shelf for book_id:6482837\nData written on csv for book:Before I Fall\nGetting reviews details from user: 2373891 and book_id: 6482837\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0399159371 and id:2373891\nFetched review data from Amazon for book :The Witness\nchecking count in shelf for book_id:12716613\nData written on csv for book:The Witness\nGetting reviews details from user: 2373891 and book_id: 12716613\nGot user review rating: 3\nUser review is: \n \nFetching data for book with isbn:0446572993 and id:2373891\nFetched review data from Amazon for book :The Innocent (Will Robie, #1)\nchecking count in shelf for book_id:12849385\nData written on csv for book:The Innocent (Will Robie, #1)\nGetting reviews details from user: 2373891 and book_id: 12849385\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:1582341028 and id:2373891\nFetched review data from Amazon for book :Bone in the Throat\nchecking count in shelf for book_id:111129\nData written on csv for book:Bone in the Throat\nGetting reviews details from user: 2373891 and book_id: 111129\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0312315732 and id:2373891\nFetched review data from Amazon for book :Little Children\nchecking count in shelf for book_id:37426\nData written on csv for book:Little Children\nGetting reviews details from user: 2373891 and book_id: 37426\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :Flat-Out Love (Flat-Out Love, #1)\nchecking count in shelf for book_id:11096647\nData written on csv for book:Flat-Out Love (Flat-Out Love, #1)\nGetting reviews details from user: 2373891 and book_id: 11096647\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0061143316 and id:2373891\nFetched review data from Amazon for book :Promise Not to Tell\nchecking count in shelf for book_id:659546\nData written on csv for book:Promise Not to Tell\nGetting reviews details from user: 2373891 and book_id: 659546\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0374533571 and id:2373891\nFetched review data from Amazon for book :The Silver Linings Playbook\nchecking count in shelf for book_id:13539044\nData written on csv for book:The Silver Linings Playbook\nGetting reviews details from user: 2373891 and book_id: 13539044\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316160199 and id:2373891\nFetched review data from Amazon for book :New Moon (Twilight, #2)\nchecking count in shelf for book_id:49041\nData written on csv for book:New Moon (Twilight, #2)\nGetting reviews details from user: 2373891 and book_id: 49041\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316160202 and id:2373891\nFetched review data from Amazon for book :Eclipse (Twilight, #3)\nchecking count in shelf for book_id:428263\nData written on csv for book:Eclipse (Twilight, #3)\nGetting reviews details from user: 2373891 and book_id: 428263\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0061147931 and id:2373891\nFetched review data from Amazon for book :Heart-Shaped Box\nchecking count in shelf for book_id:153025\nData written on csv for book:Heart-Shaped Box\nGetting reviews details from user: 2373891 and book_id: 153025\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0446696110 and id:2373891\nFetched review data from Amazon for book :The Guardian\nchecking count in shelf for book_id:15925\nData written on csv for book:The Guardian\nGetting reviews details from user: 2373891 and book_id: 15925\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0446579939 and id:2373891\nFetched review data from Amazon for book :The Lucky One\nchecking count in shelf for book_id:3063499\nData written on csv for book:The Lucky One\nGetting reviews details from user: 2373891 and book_id: 3063499\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1846054729 and id:2373891\nFetched review data from Amazon for book :Don't Blink\nchecking count in shelf for book_id:6987558\nData written on csv for book:Don't Blink\nGetting reviews details from user: 2373891 and book_id: 6987558\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316014508 and id:2373891\nFetched review data from Amazon for book :You've Been Warned\nchecking count in shelf for book_id:13134\nData written on csv for book:You've Been Warned\nGetting reviews details from user: 2373891 and book_id: 13134\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316120553 and id:2373891\nFetched review data from Amazon for book :Now You See Her\nchecking count in shelf for book_id:7926569\nData written on csv for book:Now You See Her\nGetting reviews details from user: 2373891 and book_id: 7926569\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:044661761X and id:2373891\nFetched review data from Amazon for book :Lifeguard\nchecking count in shelf for book_id:86424\nData written on csv for book:Lifeguard\nGetting reviews details from user: 2373891 and book_id: 86424\nGot user review rating: 3\nUser review is: \n \nFetching data for book with isbn:0446609404 and id:2373891\nFetched review data from Amazon for book :Cradle and All\nchecking count in shelf for book_id:5575\nData written on csv for book:Cradle and All\nGetting reviews details from user: 2373891 and book_id: 5575\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316159786 and id:2373891\nFetched review data from Amazon for book :Beach Road\nchecking count in shelf for book_id:85733\nData written on csv for book:Beach Road\nGetting reviews details from user: 2373891 and book_id: 85733\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0446613355 and id:2373891\nFetched review data from Amazon for book :London Bridges (Alex Cross, #10)\nchecking count in shelf for book_id:13151\nData written on csv for book:London Bridges (Alex Cross, #10)\nGetting reviews details from user: 2373891 and book_id: 13151\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0747266921 and id:2373891\nFetched review data from Amazon for book :Four Blind Mice (Alex Cross, #8)\nchecking count in shelf for book_id:53625\nData written on csv for book:Four Blind Mice (Alex Cross, #8)\nGetting reviews details from user: 2373891 and book_id: 53625\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316013935 and id:2373891\nFetched review data from Amazon for book :Judge & Jury\nchecking count in shelf for book_id:13130\nData written on csv for book:Judge & Jury\nGetting reviews details from user: 2373891 and book_id: 13130\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446696587 and id:2373891\nFetched review data from Amazon for book :The Lake House (When the Wind Blows, #2)\nchecking count in shelf for book_id:110444\nData written on csv for book:The Lake House (When the Wind Blows, #2)\nGetting reviews details from user: 2373891 and book_id: 110444\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316117366 and id:2373891\nFetched review data from Amazon for book :The Quickie\nchecking count in shelf for book_id:13133\nData written on csv for book:The Quickie\nGetting reviews details from user: 2373891 and book_id: 13133\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446619035 and id:2373891\nFetched review data from Amazon for book :Mary, Mary (Alex Cross, #11)\nchecking count in shelf for book_id:84736\nData written on csv for book:Mary, Mary (Alex Cross, #11)\nGetting reviews details from user: 2373891 and book_id: 84736\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446610224 and id:2373891\nFetched review data from Amazon for book :The Big Bad Wolf (Alex Cross, #9)\nchecking count in shelf for book_id:6588\nData written on csv for book:The Big Bad Wolf (Alex Cross, #9)\nGetting reviews details from user: 2373891 and book_id: 6588\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316159794 and id:2373891\nFetched review data from Amazon for book :Cross (Alex Cross, #12)\nchecking count in shelf for book_id:13128\nData written on csv for book:Cross (Alex Cross, #12)\nGetting reviews details from user: 2373891 and book_id: 13128\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446676438 and id:2373891\nFetched review data from Amazon for book :When the Wind Blows (When the Wind Blows, #1)\nchecking count in shelf for book_id:13162\nData written on csv for book:When the Wind Blows (When the Wind Blows, #1)\nGetting reviews details from user: 2373891 and book_id: 13162\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446611212 and id:2373891\nFetched review data from Amazon for book :Violets Are Blue (Alex Cross, #7)\nchecking count in shelf for book_id:79379\nData written on csv for book:Violets Are Blue (Alex Cross, #7)\nGetting reviews details from user: 2373891 and book_id: 79379\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446605484 and id:2373891\nFetched review data from Amazon for book :Roses are Red (Alex Cross, #6)\nchecking count in shelf for book_id:79378\nData written on csv for book:Roses are Red (Alex Cross, #6)\nGetting reviews details from user: 2373891 and book_id: 79378\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1570425779 and id:2373891\nFetched review data from Amazon for book :Cat and Mouse (Alex Cross, #4)\nchecking count in shelf for book_id:21436\nData written on csv for book:Cat and Mouse (Alex Cross, #4)\nGetting reviews details from user: 2373891 and book_id: 21436\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446608815 and id:2373891\nFetched review data from Amazon for book :Pop Goes the Weasel (Alex Cross #5)\nchecking count in shelf for book_id:13143\nData written on csv for book:Pop Goes the Weasel (Alex Cross #5)\nGetting reviews details from user: 2373891 and book_id: 13143\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0446692654 and id:2373891\nFetched review data from Amazon for book :Jack & Jill (Alex Cross, #3)\nchecking count in shelf for book_id:13140\nData written on csv for book:Jack & Jill (Alex Cross, #3)\nGetting reviews details from user: 2373891 and book_id: 13140\nGot user review rating: 3\nUser review is: \n \nFetching data for book with isbn:0446612545 and id:2373891\nFetched review data from Amazon for book :The Beach House\nchecking count in shelf for book_id:7510\nData written on csv for book:The Beach House\nGetting reviews details from user: 2373891 and book_id: 7510\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:044654759X and id:2373891\nFetched review data from Amazon for book :Safe Haven\nchecking count in shelf for book_id:7812659\nData written on csv for book:Safe Haven\nGetting reviews details from user: 2373891 and book_id: 7812659\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0452286913 and id:2373891\nFetched review data from Amazon for book :The Doctor's Wife\nchecking count in shelf for book_id:227462\nData written on csv for book:The Doctor's Wife\nGetting reviews details from user: 2373891 and book_id: 227462\nGot user review rating: 3\nUser review is: \n \nFetching data for book with isbn:0345470990 and id:2373891\nFetched review data from Amazon for book :The Virgin of Small Plains\nchecking count in shelf for book_id:180648\nData written on csv for book:The Virgin of Small Plains\nGetting reviews details from user: 2373891 and book_id: 180648\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0345471016 and id:2373891\nFetched review data from Amazon for book :The Scent of Rain and Lightning\nchecking count in shelf for book_id:6606456\nData written on csv for book:The Scent of Rain and Lightning\nGetting reviews details from user: 2373891 and book_id: 6606456\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0385344082 and id:2373891\nFetched review data from Amazon for book :The Homecoming of Samuel Lake\nchecking count in shelf for book_id:7632310\nData written on csv for book:The Homecoming of Samuel Lake\nGetting reviews details from user: 2373891 and book_id: 7632310\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :Beautiful Disaster (Beautiful, #1)\nchecking count in shelf for book_id:11505797\nData written on csv for book:Beautiful Disaster (Beautiful, #1)\nGetting reviews details from user: 2373891 and book_id: 11505797\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743493915 and id:2373891\nFetched review data from Amazon for book :Body of Evidence (Kay Scarpetta, #2)\nchecking count in shelf for book_id:132336\nData written on csv for book:Body of Evidence (Kay Scarpetta, #2)\nGetting reviews details from user: 2373891 and book_id: 132336\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0312362080 and id:2373891\nFetched review data from Amazon for book :One for the Money (Stephanie Plum, #1)\nchecking count in shelf for book_id:6853\nData written on csv for book:One for the Money (Stephanie Plum, #1)\nGetting reviews details from user: 2373891 and book_id: 6853\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0743477154 and id:2373891\nFetched review data from Amazon for book :Postmortem (Kay Scarpetta, #1)\nchecking count in shelf for book_id:6534\nData written on csv for book:Postmortem (Kay Scarpetta, #1)\nGetting reviews details from user: 2373891 and book_id: 6534\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0671011367 and id:2373891\nFetched review data from Amazon for book :Déjà Dead (Temperance Brennan, #1)\nchecking count in shelf for book_id:231604\nData written on csv for book:Déjà Dead (Temperance Brennan, #1)\nGetting reviews details from user: 2373891 and book_id: 231604\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0297859382 and id:2373891\nFetched review data from Amazon for book :Gone Girl\nchecking count in shelf for book_id:8442457\nData written on csv for book:Gone Girl\nGetting reviews details from user: 2373891 and book_id: 8442457\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0743418174 and id:2373891\nFetched review data from Amazon for book :Good in Bed (Cannie Shapiro, #1)\nchecking count in shelf for book_id:14748\nData written on csv for book:Good in Bed (Cannie Shapiro, #1)\nGetting reviews details from user: 2373891 and book_id: 14748\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:006075995X and id:2373891\nFetched review data from Amazon for book :Divine Secrets of the Ya-Ya Sisterhood\nchecking count in shelf for book_id:137791\nData written on csv for book:Divine Secrets of the Ya-Ya Sisterhood\nGetting reviews details from user: 2373891 and book_id: 137791\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0316015849 and id:2373891\nFetched review data from Amazon for book :Twilight (Twilight, #1)\nchecking count in shelf for book_id:41865\nData written on csv for book:Twilight (Twilight, #1)\nGetting reviews details from user: 2373891 and book_id: 41865\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0439554934 and id:2373891\nFetched review data from Amazon for book :Harry Potter and the Sorcerer's Stone (Harry Potter, #1)\nchecking count in shelf for book_id:3\nData written on csv for book:Harry Potter and the Sorcerer's Stone (Harry Potter, #1)\nGetting reviews details from user: 2373891 and book_id: 3\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0553384287 and id:2373891\nFetched review data from Amazon for book :Odd Thomas (Odd Thomas, #1)\nchecking count in shelf for book_id:14995\nData written on csv for book:Odd Thomas (Odd Thomas, #1)\nGetting reviews details from user: 2373891 and book_id: 14995\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0446677388 and id:2373891\nFetched review data from Amazon for book :Kiss the Girls (Alex Cross, #2)\nchecking count in shelf for book_id:13148\nData written on csv for book:Kiss the Girls (Alex Cross, #2)\nGetting reviews details from user: 2373891 and book_id: 13148\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0385338600 and id:2373891\nFetched review data from Amazon for book :A Time to Kill (Jake Brigance, #1)\nchecking count in shelf for book_id:32542\nData written on csv for book:A Time to Kill (Jake Brigance, #1)\nGetting reviews details from user: 2373891 and book_id: 32542\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0450040186 and id:2373891\nFetched review data from Amazon for book :The Shining (The Shining, #1)\nchecking count in shelf for book_id:11588\nData written on csv for book:The Shining (The Shining, #1)\nGetting reviews details from user: 2373891 and book_id: 11588\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0307277674 and id:2373891\nFetched review data from Amazon for book :The Da Vinci Code (Robert Langdon, #2)\nchecking count in shelf for book_id:968\nData written on csv for book:The Da Vinci Code (Robert Langdon, #2)\nGetting reviews details from user: 2373891 and book_id: 968\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0143037145 and id:2373891\nFetched review data from Amazon for book :The Memory Keeper's Daughter\nchecking count in shelf for book_id:10441\nData written on csv for book:The Memory Keeper's Daughter\nGetting reviews details from user: 2373891 and book_id: 10441\nGot user review rating: 2\nUser review is: \n \nFetching data for book with isbn:0316166685 and id:2373891\nFetched review data from Amazon for book :The Lovely Bones\nchecking count in shelf for book_id:12232938\nData written on csv for book:The Lovely Bones\nGetting reviews details from user: 2373891 and book_id: 12232938\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0349113912 and id:2373891\nFetched review data from Amazon for book :Me Talk Pretty One Day\nchecking count in shelf for book_id:4137\nData written on csv for book:Me Talk Pretty One Day\nGetting reviews details from user: 2373891 and book_id: 4137\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0060513039 and id:2373891\nFetched review data from Amazon for book :Where the Sidewalk Ends: The Poems and Drawings of Shel Silverstein\nchecking count in shelf for book_id:30119\nData written on csv for book:Where the Sidewalk Ends: The Poems and Drawings of Shel Silverstein\nGetting reviews details from user: 2373891 and book_id: 30119\nGot user review rating: 4\nUser review is: \n \nFetching data for book with isbn:0316769177 and id:2373891\nFetched review data from Amazon for book :The Catcher in the Rye\nchecking count in shelf for book_id:5107\nData written on csv for book:The Catcher in the Rye\nGetting reviews details from user: 2373891 and book_id: 5107\nGot user review rating: 3\nUser review is: \n \nFetching data for book with isbn:0061120081 and id:2373891\nFetched review data from Amazon for book :To Kill a Mockingbird\nchecking count in shelf for book_id:2657\nData written on csv for book:To Kill a Mockingbird\nGetting reviews details from user: 2373891 and book_id: 2657\nGot user review rating: 4\nUser review is: \n \nChecking for books in shelf: currently-reading for user id:2373891\nhttps://www.goodreads.com/review/list/2373891.xml?key=i3Zsl7r13oHEQCjv1vXw&v=2&per_page=200&shelf=currently-reading\nFetching data for book with isbn:0385344228 and id:2373891\nFetched review data from Amazon for book :Defending Jacob\nchecking count in shelf for book_id:11367726\nData written on csv for book:Defending Jacob\nGetting reviews details from user: 2373891 and book_id: 11367726\nGot user review rating: 0\nUser review is: \n \nChecking for books in shelf: to-read for user id:2373891\nhttps://www.goodreads.com/review/list/2373891.xml?key=i3Zsl7r13oHEQCjv1vXw&v=2&per_page=200&shelf=to-read\nFetching data for book with isbn:1250077001 and id:2373891\nFetched review data from Amazon for book :Furiously Happy: A Funny Book About Horrible Things\nchecking count in shelf for book_id:23848559\nData written on csv for book:Furiously Happy: A Funny Book About Horrible Things\nGetting reviews details from user: 2373891 and book_id: 23848559\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1402298684 and id:2373891\nFetched review data from Amazon for book :The Magician's Lie\nchecking count in shelf for book_id:21897317\nData written on csv for book:The Magician's Lie\nGetting reviews details from user: 2373891 and book_id: 21897317\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476728747 and id:2373891\nFetched review data from Amazon for book :The Wright Brothers\nchecking count in shelf for book_id:22609391\nData written on csv for book:The Wright Brothers\nGetting reviews details from user: 2373891 and book_id: 22609391\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1501105779 and id:2373891\nFetched review data from Amazon for book :Before We Were Strangers\nchecking count in shelf for book_id:23309634\nData written on csv for book:Before We Were Strangers\nGetting reviews details from user: 2373891 and book_id: 23309634\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476755205 and id:2373891\nFetched review data from Amazon for book :All We Had\nchecking count in shelf for book_id:18775258\nData written on csv for book:All We Had\nGetting reviews details from user: 2373891 and book_id: 18775258\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316334529 and id:2373891\nFetched review data from Amazon for book :The Rumor\nchecking count in shelf for book_id:23341607\nData written on csv for book:The Rumor\nGetting reviews details from user: 2373891 and book_id: 23341607\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0804139024 and id:2373891\nFetched review data from Amazon for book :The Martian\nchecking count in shelf for book_id:18007564\nData written on csv for book:The Martian\nGetting reviews details from user: 2373891 and book_id: 18007564\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0778317706 and id:2373891\nFetched review data from Amazon for book :Pretty Baby\nchecking count in shelf for book_id:23638955\nData written on csv for book:Pretty Baby\nGetting reviews details from user: 2373891 and book_id: 23638955\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0062363239 and id:2373891\nFetched review data from Amazon for book :A Head Full of Ghosts\nchecking count in shelf for book_id:23019294\nData written on csv for book:A Head Full of Ghosts\nGetting reviews details from user: 2373891 and book_id: 23019294\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476789630 and id:2373891\nFetched review data from Amazon for book :Luckiest Girl Alive\nchecking count in shelf for book_id:22609317\nData written on csv for book:Luckiest Girl Alive\nGetting reviews details from user: 2373891 and book_id: 22609317\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0062225464 and id:2373891\nFetched review data from Amazon for book :Where They Found Her\nchecking count in shelf for book_id:22693182\nData written on csv for book:Where They Found Her\nGetting reviews details from user: 2373891 and book_id: 22693182\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1595549404 and id:2373891\nFetched review data from Amazon for book :Waking Hours (East Salem, #1)\nchecking count in shelf for book_id:10634346\nData written on csv for book:Waking Hours (East Salem, #1)\nGetting reviews details from user: 2373891 and book_id: 10634346\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :Never Never (Never Never, #1)\nchecking count in shelf for book_id:24378015\nData written on csv for book:Never Never (Never Never, #1)\nGetting reviews details from user: 2373891 and book_id: 24378015\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1602862729 and id:2373891\nFetched review data from Amazon for book :The Haunting of Sunshine Girl (The Haunting of Sunshine Girl, #1)\nchecking count in shelf for book_id:21413855\nData written on csv for book:The Haunting of Sunshine Girl (The Haunting of Sunshine Girl, #1)\nGetting reviews details from user: 2373891 and book_id: 21413855\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1594633665 and id:2373891\nFetched review data from Amazon for book :The Girl on the Train\nchecking count in shelf for book_id:22557272\nData written on csv for book:The Girl on the Train\nGetting reviews details from user: 2373891 and book_id: 22557272\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0812995201 and id:2373891\nFetched review data from Amazon for book :The Weight of Blood\nchecking count in shelf for book_id:18209468\nData written on csv for book:The Weight of Blood\nGetting reviews details from user: 2373891 and book_id: 18209468\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0345539931 and id:2373891\nFetched review data from Amazon for book :As Chimney Sweepers Come to Dust (Flavia de Luce, #7)\nchecking count in shelf for book_id:21874813\nData written on csv for book:As Chimney Sweepers Come to Dust (Flavia de Luce, #7)\nGetting reviews details from user: 2373891 and book_id: 21874813\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476790221 and id:2373891\nFetched review data from Amazon for book :The Accidental Empress\nchecking count in shelf for book_id:22609307\nData written on csv for book:The Accidental Empress\nGetting reviews details from user: 2373891 and book_id: 22609307\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0356500152 and id:2373891\nFetched review data from Amazon for book :The Girl with All the Gifts\nchecking count in shelf for book_id:17235026\nData written on csv for book:The Girl with All the Gifts\nGetting reviews details from user: 2373891 and book_id: 17235026\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476710791 and id:2373891\nFetched review data from Amazon for book :Wayfaring Stranger (Weldon Holland, #1)\nchecking count in shelf for book_id:18775356\nData written on csv for book:Wayfaring Stranger (Weldon Holland, #1)\nGetting reviews details from user: 2373891 and book_id: 18775356\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1414336241 and id:2373891\nFetched review data from Amazon for book :The Auschwitz Escape\nchecking count in shelf for book_id:18232495\nData written on csv for book:The Auschwitz Escape\nGetting reviews details from user: 2373891 and book_id: 18232495\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0062311077 and id:2373891\nFetched review data from Amazon for book :Natchez Burning (Penn Cage, #4)\nchecking count in shelf for book_id:18505832\nData written on csv for book:Natchez Burning (Penn Cage, #4)\nGetting reviews details from user: 2373891 and book_id: 18505832\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0525953493 and id:2373891\nFetched review data from Amazon for book :Missing You\nchecking count in shelf for book_id:18114060\nData written on csv for book:Missing You\nGetting reviews details from user: 2373891 and book_id: 18114060\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316206873 and id:2373891\nFetched review data from Amazon for book :The Silkworm (Cormoran Strike, #2)\nchecking count in shelf for book_id:18214414\nData written on csv for book:The Silkworm (Cormoran Strike, #2)\nGetting reviews details from user: 2373891 and book_id: 18214414\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0385539703 and id:2373891\nFetched review data from Amazon for book :The Children Act\nchecking count in shelf for book_id:21965107\nData written on csv for book:The Children Act\nGetting reviews details from user: 2373891 and book_id: 21965107\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1616203218 and id:2373891\nFetched review data from Amazon for book :The Storied Life of A.J. Fikry\nchecking count in shelf for book_id:18293427\nData written on csv for book:The Storied Life of A.J. Fikry\nGetting reviews details from user: 2373891 and book_id: 18293427\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :The Billionaire's Christmas (The Sinclairs, #0.5)\nchecking count in shelf for book_id:22883581\nData written on csv for book:The Billionaire's Christmas (The Sinclairs, #0.5)\nGetting reviews details from user: 2373891 and book_id: 22883581\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1592408605 and id:2373891\nFetched review data from Amazon for book :Graduates in Wonderland: The International Misadventures of Two (Almost) Adults\nchecking count in shelf for book_id:18668008\nData written on csv for book:Graduates in Wonderland: The International Misadventures of Two (Almost) Adults\nGetting reviews details from user: 2373891 and book_id: 18668008\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1471113876 and id:2373891\nFetched review data from Amazon for book :The Professional (The Game Maker, #1)\nchecking count in shelf for book_id:17558070\nData written on csv for book:The Professional (The Game Maker, #1)\nGetting reviews details from user: 2373891 and book_id: 17558070\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1444762443 and id:2373891\nFetched review data from Amazon for book :Remember Me This Way\nchecking count in shelf for book_id:22045253\nData written on csv for book:Remember Me This Way\nGetting reviews details from user: 2373891 and book_id: 22045253\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0345544927 and id:2373891\nFetched review data from Amazon for book :Leaving Time\nchecking count in shelf for book_id:18816603\nData written on csv for book:Leaving Time\nGetting reviews details from user: 2373891 and book_id: 18816603\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0141043768 and id:2373891\nFetched review data from Amazon for book :What Alice Forgot\nchecking count in shelf for book_id:6469165\nData written on csv for book:What Alice Forgot\nGetting reviews details from user: 2373891 and book_id: 6469165\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0399159347 and id:2373891\nFetched review data from Amazon for book :The Husband's Secret\nchecking count in shelf for book_id:17802724\nData written on csv for book:The Husband's Secret\nGetting reviews details from user: 2373891 and book_id: 17802724\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0399166238 and id:2373891\nFetched review data from Amazon for book :While Beauty Slept\nchecking count in shelf for book_id:18079665\nData written on csv for book:While Beauty Slept\nGetting reviews details from user: 2373891 and book_id: 18079665\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476746583 and id:2373891\nFetched review data from Amazon for book :All the Light We Cannot See\nchecking count in shelf for book_id:18143977\nData written on csv for book:All the Light We Cannot See\nGetting reviews details from user: 2373891 and book_id: 18143977\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0399167064 and id:2373891\nFetched review data from Amazon for book :Big Little Lies\nchecking count in shelf for book_id:19486412\nData written on csv for book:Big Little Lies\nGetting reviews details from user: 2373891 and book_id: 19486412\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0670026603 and id:2373891\nFetched review data from Amazon for book :Me Before You (Me Before You, #1)\nchecking count in shelf for book_id:15507958\nData written on csv for book:Me Before You (Me Before You, #1)\nGetting reviews details from user: 2373891 and book_id: 15507958\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0670014737 and id:2373891\nFetched review data from Amazon for book :Dollbaby\nchecking count in shelf for book_id:18693929\nData written on csv for book:Dollbaby\nGetting reviews details from user: 2373891 and book_id: 18693929\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476702993 and id:2373891\nFetched review data from Amazon for book :The House We Grew Up In\nchecking count in shelf for book_id:18764826\nData written on csv for book:The House We Grew Up In\nGetting reviews details from user: 2373891 and book_id: 18764826\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0060779632 and id:2373891\nFetched review data from Amazon for book :Help for the Haunted\nchecking count in shelf for book_id:17348985\nData written on csv for book:Help for the Haunted\nGetting reviews details from user: 2373891 and book_id: 17348985\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0062209841 and id:2373891\nFetched review data from Amazon for book :The Death of Bees\nchecking count in shelf for book_id:15818333\nData written on csv for book:The Death of Bees\nGetting reviews details from user: 2373891 and book_id: 15818333\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0545627044 and id:2373891\nFetched review data from Amazon for book :Catch a Falling Star\nchecking count in shelf for book_id:18527496\nData written on csv for book:Catch a Falling Star\nGetting reviews details from user: 2373891 and book_id: 18527496\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316204277 and id:2373891\nFetched review data from Amazon for book :Where'd You Go, Bernadette\nchecking count in shelf for book_id:13526165\nData written on csv for book:Where'd You Go, Bernadette\nGetting reviews details from user: 2373891 and book_id: 13526165\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0804138567 and id:2373891\nFetched review data from Amazon for book :Bittersweet\nchecking count in shelf for book_id:18339743\nData written on csv for book:Bittersweet\nGetting reviews details from user: 2373891 and book_id: 18339743\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0307589293 and id:2373891\nFetched review data from Amazon for book :A Triple Knot\nchecking count in shelf for book_id:18759930\nData written on csv for book:A Triple Knot\nGetting reviews details from user: 2373891 and book_id: 18759930\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :Guidebook to Murder (A Tourist Trap Mystery #1)\nchecking count in shelf for book_id:20817232\nData written on csv for book:Guidebook to Murder (A Tourist Trap Mystery #1)\nGetting reviews details from user: 2373891 and book_id: 20817232\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn: and id:2373891\nFetched review data from Amazon for book :Shame on You (Fool Me Once, #1)\nchecking count in shelf for book_id:18895851\nData written on csv for book:Shame on You (Fool Me Once, #1)\nGetting reviews details from user: 2373891 and book_id: 18895851\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1451667116 and id:2373891\nFetched review data from Amazon for book :The Sun and Other Stars\nchecking count in shelf for book_id:16130454\nData written on csv for book:The Sun and Other Stars\nGetting reviews details from user: 2373891 and book_id: 16130454\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1439164681 and id:2373891\nFetched review data from Amazon for book :Love Anthony\nchecking count in shelf for book_id:13547381\nData written on csv for book:Love Anthony\nGetting reviews details from user: 2373891 and book_id: 13547381\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1414375662 and id:2373891\nFetched review data from Amazon for book :A Little Salty to Cut the Sweet: Southern Stories of Faith, Family, and Fifteen Pounds of Bacon\nchecking count in shelf for book_id:17131044\nData written on csv for book:A Little Salty to Cut the Sweet: Southern Stories of Faith, Family, and Fifteen Pounds of Bacon\nGetting reviews details from user: 2373891 and book_id: 17131044\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0385537131 and id:2373891\nFetched review data from Amazon for book :Sycamore Row (Jake Brigance, #2)\nchecking count in shelf for book_id:17288661\nData written on csv for book:Sycamore Row (Jake Brigance, #2)\nGetting reviews details from user: 2373891 and book_id: 17288661\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1455501514 and id:2373891\nFetched review data from Amazon for book :Deadline\nchecking count in shelf for book_id:17333403\nData written on csv for book:Deadline\nGetting reviews details from user: 2373891 and book_id: 17333403\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0062217135 and id:2373891\nFetched review data from Amazon for book :The Beginning of Everything\nchecking count in shelf for book_id:13522285\nData written on csv for book:The Beginning of Everything\nGetting reviews details from user: 2373891 and book_id: 13522285\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0375842209 and id:2373891\nFetched review data from Amazon for book :The Book Thief\nchecking count in shelf for book_id:1118668\nData written on csv for book:The Book Thief\nGetting reviews details from user: 2373891 and book_id: 1118668\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0061827010 and id:2373891\nFetched review data from Amazon for book :The Bricklayer (Steve Vail, #1)\nchecking count in shelf for book_id:6497521\nData written on csv for book:The Bricklayer (Steve Vail, #1)\nGetting reviews details from user: 2373891 and book_id: 6497521\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0312577206 and id:2373891\nFetched review data from Amazon for book :Home Front\nchecking count in shelf for book_id:12022079\nData written on csv for book:Home Front\nGetting reviews details from user: 2373891 and book_id: 12022079\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1451651694 and id:2373891\nFetched review data from Amazon for book :A House in the Sky\nchecking count in shelf for book_id:18039963\nData written on csv for book:A House in the Sky\nGetting reviews details from user: 2373891 and book_id: 18039963\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:145162137X and id:2373891\nFetched review data from Amazon for book :Brain on Fire: My Month of Madness\nchecking count in shelf for book_id:13547180\nData written on csv for book:Brain on Fire: My Month of Madness\nGetting reviews details from user: 2373891 and book_id: 13547180\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1476707723 and id:2373891\nFetched review data from Amazon for book :Whistling Past the Graveyard\nchecking count in shelf for book_id:16058610\nData written on csv for book:Whistling Past the Graveyard\nGetting reviews details from user: 2373891 and book_id: 16058610\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0786031581 and id:2373891\nFetched review data from Amazon for book :Unspeakable\nchecking count in shelf for book_id:16281013\nData written on csv for book:Unspeakable\nGetting reviews details from user: 2373891 and book_id: 16281013\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0440241901 and id:2373891\nFetched review data from Amazon for book :Can You Keep a Secret?\nchecking count in shelf for book_id:33724\nData written on csv for book:Can You Keep a Secret?\nGetting reviews details from user: 2373891 and book_id: 33724\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0060899220 and id:2373891\nFetched review data from Amazon for book :Kitchen Confidential: Adventures in the Culinary Underbelly\nchecking count in shelf for book_id:33313\nData written on csv for book:Kitchen Confidential: Adventures in the Culinary Underbelly\nGetting reviews details from user: 2373891 and book_id: 33313\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316069353 and id:2373891\nFetched review data from Amazon for book :The Fifth Witness (Mickey Haller, #4)\nchecking count in shelf for book_id:9681098\nData written on csv for book:The Fifth Witness (Mickey Haller, #4)\nGetting reviews details from user: 2373891 and book_id: 9681098\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316069485 and id:2373891\nFetched review data from Amazon for book :The Reversal (Harry Bosch, #16; Mickey Haller, #3)\nchecking count in shelf for book_id:7936809\nData written on csv for book:The Reversal (Harry Bosch, #16; Mickey Haller, #3)\nGetting reviews details from user: 2373891 and book_id: 7936809\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0316166294 and id:2373891\nFetched review data from Amazon for book :The Brass Verdict (Harry Bosch, #14; Mickey Haller, #2)\nchecking count in shelf for book_id:2761626\nData written on csv for book:The Brass Verdict (Harry Bosch, #14; Mickey Haller, #2)\nGetting reviews details from user: 2373891 and book_id: 2761626\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1594480001 and id:2373891\nFetched review data from Amazon for book :The Kite Runner\nchecking count in shelf for book_id:77203\nData written on csv for book:The Kite Runner\nGetting reviews details from user: 2373891 and book_id: 77203\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0525953221 and id:2373891\nFetched review data from Amazon for book :The Shadow Tracer\nchecking count in shelf for book_id:16101033\nData written on csv for book:The Shadow Tracer\nGetting reviews details from user: 2373891 and book_id: 16101033\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:0399256601 and id:2373891\nFetched review data from Amazon for book :The Name of the Star (Shades of London, #1)\nchecking count in shelf for book_id:9802372\nData written on csv for book:The Name of the Star (Shades of London, #1)\nGetting reviews details from user: 2373891 and book_id: 9802372\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1451654967 and id:2373891\nFetched review data from Amazon for book :Who Owns the Future?\nchecking count in shelf for book_id:15802693\nData written on csv for book:Who Owns the Future?\nGetting reviews details from user: 2373891 and book_id: 15802693\nGot user review rating: 0\nUser review is: \n \nFetching data for book with isbn:1400032717 and id:2373891\nFetched review data from Amazon for book :The Curious Incident of the Dog in the Night-Time\nchecking count in shelf for book_id:1618\nData written on csv for book:The Curious Incident of the Dog in the Night-Time\nGetting reviews details from user: 2373891 and book_id: 1618\nGot user review rating: 0"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
cb6f0528d7220030e312b9bed20db43c17f6422e | 145,277 | ipynb | Jupyter Notebook | ROMS/Iris COAWST test 3d.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
]
| 32 | 2015-01-07T01:48:05.000Z | 2022-03-02T07:07:42.000Z | ROMS/Iris COAWST test 3d.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
]
| 1 | 2015-04-13T21:00:18.000Z | 2015-04-13T21:00:18.000Z | ROMS/Iris COAWST test 3d.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
]
| 30 | 2015-01-28T09:31:29.000Z | 2022-03-07T03:08:28.000Z | 408.081461 | 128,073 | 0.90704 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
cb6f0a5f124ecb643649916ba9494769a6d4956e | 8,744 | ipynb | Jupyter Notebook | sigEqExample.ipynb | 1ozturkbe/diSP | 78e1a484e569df1a0e32ab7a2ebc1b342ce756fb | [
"MIT"
]
| null | null | null | sigEqExample.ipynb | 1ozturkbe/diSP | 78e1a484e569df1a0e32ab7a2ebc1b342ce756fb | [
"MIT"
]
| null | null | null | sigEqExample.ipynb | 1ozturkbe/diSP | 78e1a484e569df1a0e32ab7a2ebc1b342ce756fb | [
"MIT"
]
| null | null | null | 98.247191 | 1,457 | 0.663312 | [
[
[
"# Playing with Max's signomial equality example\nfrom gpkit import Variable, Model, SignomialEquality, SignomialsEnabled, units\nfrom matplotlib import pyplot as plt\nfrom gpkit.interactive.sankey import Sankey",
"_____no_output_____"
],
[
"# Defining the problem\nx = Variable('x')\ny = Variable('y')\nwith SignomialsEnabled():\n objective = (x+1/4)**2 + (y-1/2)**2\n constraint = SignomialEquality(x**2 + x - y, 0)\nm = Model(objective, constraint)",
"_____no_output_____"
],
[
"# Solution\nsol = m.localsolve(verbosity = 2)",
"Beginning signomial solve.\nUsing solver 'mosek_cli'\nSolving for 2 variables.\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
cb6f1048585bf17ef148f03659451c208e72416e | 63,976 | ipynb | Jupyter Notebook | Guia_2/Problema_9.ipynb | MatiasNRamos/Astrometria | 28ef068d1bbb1e4d6dd543b2e3dfae56cb40ca5f | [
"MIT"
]
| null | null | null | Guia_2/Problema_9.ipynb | MatiasNRamos/Astrometria | 28ef068d1bbb1e4d6dd543b2e3dfae56cb40ca5f | [
"MIT"
]
| null | null | null | Guia_2/Problema_9.ipynb | MatiasNRamos/Astrometria | 28ef068d1bbb1e4d6dd543b2e3dfae56cb40ca5f | [
"MIT"
]
| null | null | null | 187.612903 | 29,472 | 0.895601 | [
[
[
"Este código crea una función que me permite estimar la varianza de una distribución y lo chequea con las distribuciones de Poisson y Gauss. Además, usa el método boostrap resampling que se basa en, a partir de una muestra, se crea una población y luego se toman muestras de la misma.\n\nÉsto permite medir un estadístico, por ej. la varianza, tantas veces como se quiera y luego tener un intervalo de confianza para el mismo.\n\n\n-----\nEl cógido genera una función que me premite calcular la varianza de una distribución y luego crea otra función que, usando el método de boostrap resampling, me devuelve el intervalo de confianza para cierto nivel de significancia y también me da la distribución de varianzas medidas",
"_____no_output_____"
]
],
[
[
"%matplotlib inline \nimport numpy as np \nimport matplotlib.pyplot as plt\n# Estilo de gráficos\nplt.style.use('dark_background')",
"_____no_output_____"
],
[
"# Estilo de gráficos\nplt.style.use('dark_background')",
"_____no_output_____"
],
[
"def Var(X):\n \"\"\" Devuelve la varianza un arreglo\n \n Parameters\n ----------\n X : np.darray()\n Arreglo de valores de una variable aleatoria\n \n Returns\n -------\n Var : .float\n Varianza del arreglo X\n \n \"\"\"\n import numpy as np \n # La varianza es el momento de orden 2 de la distribución discreta, la calculo:\n Var = np.sum((X-np.mean(np.array(X)))**2)/len(X)\n return Var",
"_____no_output_____"
]
],
[
[
"Para probarla uso dos distribuciones conocidas: gaussiana y Poisson. Para la de Poisson traigo mi propia función generadora",
"_____no_output_____"
]
],
[
[
"# Creo las dos distribuciones:\nPuntos = 10000\n# Poisson\nfrom Misfunciones import Poisson # Usar help(Poisson) para ver detalles\nLambd = 3 # Teóricamente es la media y la varianza\nXP = Poisson(lambd=Lambd, N=Puntos)\n# Gauss\ndev = 1 # desviación estándar\nXG = np.random.normal(0, dev, Puntos)\n\n# Grafico\nfig, ax = plt.subplots(1, 2, figsize = (14,6))\n\nax[0].hist(XP, color='cyan', bins=50);\nax[1].hist(XG, color='green', bins=50);\n\nax[0].set_title('Poisson', fontsize=20)\nax[0].set_ylabel('Frecuencia', fontsize=20)\nax[0].set_xlabel('k', fontsize=20)\nax[1].set_title('Gaussiana', fontsize=20)\nax[1].set_ylabel('Frecuencia', fontsize=20)\nax[1].set_xlabel('x', fontsize=20);",
"_____no_output_____"
],
[
"# Calculo varianzas:\nVarXP = Var(np.array(XP))\nprint('Varianza teórica Poisson =', Lambd)\nprint('Varianza obtenida Poisson =', VarXP)\nVarXG = Var(XG)\nprint('Varianza teórica Gauss =', dev**2)\nprint('Varianza obtenida Gauss =', VarXG)",
"Varianza teórica Poisson = 3\nVarianza obtenida Poisson = 2.803882714309116\nVarianza teórica Gauss = 1\nVarianza obtenida Gauss = 0.9891229637633558\n"
]
],
[
[
"No es extremadamente precisa pero funciona. Probé calcularla con np.var(XP) y da lo mismo",
"_____no_output_____"
],
[
"Ahora defino la función Boostrap_var() que requiere tener cargada la función Var(). Podría usar la función np.var() pero creo que eso no es lo que se pide.",
"_____no_output_____"
]
],
[
[
"def Boostrap_var(Sample, N, Mult, alpha):\n \"\"\" Usa el método boostrap resampling para tener la incerteza de la varianza de una muestra\n \n Parameters\n ----------\n \n Sample : np.ndarray()\n Muestra de la variable aleatoria\n N : int\n Cantidad de resamplings. Valor positivo mayor a cero\n Mult : int\n Multiplicador para crear la población de tamaño M*len(Sample)\n alpha : .float\n Nivel de significancia deseado, pertenece a (0,1). Si se quiere 95% => alpha=0.95. \n \n Returns\n -------\n .float\n Valor inferior del intervalo de confianza\n .float\n Valor superior del intervalo de confianza\n np.darray()\n Arreglo de las medias de las varianzas\n \"\"\"\n # Errores\n if N<1 or isinstance(N, int)==False:\n raise ValueError('Error: N debe ser una entero positivo')\n if alpha<0 or alpha>=1:\n raise ValueError('Error: Alpha debe pertenecer al intervalo (0,1)')\n # -------\n import numpy as np\n # Creo una población de tamaño M*len(Samples)\n # Básicamente copio y pego la muestra M veces\n Pop = []\n ij = 0\n while ij<Mult:\n ik = 0\n while ik<len(Sample):\n Pop.append(Sample[ik])\n ik = ik + 1\n ij = ij + 1\n # Tomo N samples DE ESA POBLACIÓN de tamaño len(Sample) y le calculo la varianza a c/u\n ij = 0\n Vars = []\n while ij<N:\n Resampling = np.random.choice(Pop, size=len(Sample))\n Vars.append( Var(Resampling) )\n ij = ij + 1\n # Transformo a array de Numpy\n Vars = np.array(Vars)\n \n # Calculos los intervalos de confianza ------------------------\n Dsort = np.sort(Vars) # Ordeno los valores de las varianzas\n # Encuentro el ij correspondiente a la media, lo llamo \"ijm\"\n ij = 0\n while ij<len(Dsort):\n EA = sum(Dsort<=Dsort[ij])/len(Dsort) # Estimación del área\n if EA>=0.5: # media --> 0.5 del área estimada\n ijm = ij\n break\n ij = ij + 1\n # Suponiendo intervalos de confianza simétricos busco el intervalo de confianza:\n ij = ijm\n while ij<len(Dsort):\n # Cuento los \"True\". Esto es la estimación de un área para una distrib. discreta\n EA = sum(Dsort<=Dsort[ij])/len(Dsort) # Empieza con el valor \"0.5\" para ij=ijm\n if EA>0.5+alpha/2:\n sup = Dsort[ij] # parte superior del intervalo \n inf = Dsort[ijm] - (sup - Dsort[ijm]) # Parte inferior del intervalo\n break \n ij = ij + 1\n return inf, sup, Dsort",
"_____no_output_____"
],
[
"# Ejemplo\nN1 = 1000\nM1 = 10\nalpha1 = 0.95\n\nD = Boostrap_var(XG, N=N1, Mult=M1, alpha=alpha1) # Distribución calculada",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize = (12,6))\n\nax.hist(D[2], color='green');\nax.axvline(D[0], ls='--', color='cyan', label='Límite inferior')\nax.axvline(D[1], ls='--', color='yellow', label='Límite superior')\nax.axvline(np.mean(D[2]), ls='--', color='white', label='Media')\n\nax.set_title('Distribución de medias de la varianza', fontsize=20)\nax.set_xlabel('Valor', fontsize=20)\nax.set_ylabel('Frecuencia', fontsize=20);\nax.legend();",
"_____no_output_____"
]
],
[
[
"Ahora veré si el nivel de significancia calculado contiene al valor correcto de la varianza",
"_____no_output_____"
]
],
[
[
"# Recordar que la varianza teórica de la gaussiana la definí como \"VarXG\"\nprint('El intervalo de confianza del %',100*alpha1, 'es: (', round(D[0],3),\n ',', round(D[1],3), ')')\nprint('La varianza teórica es:', round(VarXG,3))\n# Condición\nif VarXG>D[0] and VarXG<D[1]:\n print('Resultado: Si, los resultados con compatibles')\nelse: \n print('Resultado: No, los resultados no son compatibles')",
"El intervalo de confianza del % 95.0 es: ( 0.964 , 1.016 )\nLa varianza teórica es: 0.989\nResultado: Si, los resultados con compatibles\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6f215727c2235992ddf5ee31439f2654c40061 | 2,086 | ipynb | Jupyter Notebook | examples/notebooks/03_inspector_tool.ipynb | hugoledoux/geemap | 10c665d06976cf9ac2d732309334be1044df71b8 | [
"MIT"
]
| 3 | 2020-09-26T16:58:40.000Z | 2021-11-08T09:31:06.000Z | examples/notebooks/03_inspector_tool.ipynb | hugoledoux/geemap | 10c665d06976cf9ac2d732309334be1044df71b8 | [
"MIT"
]
| null | null | null | examples/notebooks/03_inspector_tool.ipynb | hugoledoux/geemap | 10c665d06976cf9ac2d732309334be1044df71b8 | [
"MIT"
]
| 1 | 2020-04-16T19:54:13.000Z | 2020-04-16T19:54:13.000Z | 21.957895 | 98 | 0.525887 | [
[
[
"import ee\nimport geemap",
"_____no_output_____"
],
[
"geemap.show_youtube('k477ksjkaXw')",
"_____no_output_____"
]
],
[
[
"## Create an interactive map",
"_____no_output_____"
]
],
[
[
"Map = geemap.Map(center=(40, -100), zoom=4)",
"_____no_output_____"
]
],
[
[
"## Add Earth Engine Python script",
"_____no_output_____"
]
],
[
[
"# Add Earth Engine dataset\ndem = ee.Image('USGS/SRTMGL1_003')\nlandcover = ee.Image(\"ESA/GLOBCOVER_L4_200901_200912_V2_3\").select('landcover')\nlandsat7 = ee.Image('LE7_TOA_5YEAR/1999_2003')\nstates = ee.FeatureCollection(\"TIGER/2018/States\")\n\n# Set visualization parameters.\nvis_params = {\n 'min': 0,\n 'max': 4000,\n 'palette': ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5']}\n\n# Add Earth Eninge layers to Map\nMap.addLayer(dem, vis_params, 'STRM DEM', True, 0.5)\nMap.addLayer(landcover, {}, 'Land cover')\nMap.addLayer(landsat7, {'bands': ['B4', 'B3', 'B2'], 'min': 20, 'max': 200}, 'Landsat 7')\nMap.addLayer(states, {}, \"US States\")\n\nMap",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6f258555056e291f3ce97abf9dc683dcc408eb | 9,782 | ipynb | Jupyter Notebook | pc4sbml.ipynb | rheiland/pc4sbml | 9b570ebd03fc7123e5caad2bccc3fc262b0a36f7 | [
"BSD-3-Clause"
]
| 2 | 2020-01-09T21:27:45.000Z | 2020-05-04T11:10:23.000Z | pc4sbml.ipynb | rheiland/pc4sbml | 9b570ebd03fc7123e5caad2bccc3fc262b0a36f7 | [
"BSD-3-Clause"
]
| null | null | null | pc4sbml.ipynb | rheiland/pc4sbml | 9b570ebd03fc7123e5caad2bccc3fc262b0a36f7 | [
"BSD-3-Clause"
]
| null | null | null | 32.606667 | 140 | 0.400736 | [
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import sys, os\nsys.path.insert(0, os.path.abspath('bin'))\nimport pc4sbml",
"_____no_output_____"
],
[
"pc4sbml.gui",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
cb6f26126b958662574cfc31252e80187cc33664 | 5,780 | ipynb | Jupyter Notebook | gensim/docs/notebooks/Varembed.ipynb | Abas-Khan/thesis | b733bd4382371203cc4992571890619a2e314047 | [
"MIT"
]
| 1 | 2021-04-04T13:40:10.000Z | 2021-04-04T13:40:10.000Z | gensim/docs/notebooks/Varembed.ipynb | YantianZha/Distr2Vec | 9316be830d450d453bcb70243a7829128fc17eaa | [
"MIT"
]
| null | null | null | gensim/docs/notebooks/Varembed.ipynb | YantianZha/Distr2Vec | 9316be830d450d453bcb70243a7829128fc17eaa | [
"MIT"
]
| null | null | null | 35.243902 | 531 | 0.646886 | [
[
[
"# VarEmbed Tutorial\n\nVarembed is a word embedding model incorporating morphological information, capturing shared sub-word features. Unlike previous work that constructs word embeddings directly from morphemes, varembed combines morphological and distributional information in a unified probabilistic framework. Varembed thus yields improvements on intrinsic word similarity evaluations. Check out the original paper, [arXiv:1608.01056](https://arxiv.org/abs/1608.01056) accepted in [EMNLP 2016](http://www.emnlp2016.net/accepted-papers.html).\n\nVarembed is now integrated into [Gensim](http://radimrehurek.com/gensim/) providing ability to load already trained varembed models into gensim with additional functionalities over word vectors already present in gensim.\n\n# This Tutorial\n\nIn this tutorial you will learn how to train, load and evaluate varembed model on your data.\n\n# Train Model\n\nThe authors provide their code to train a varembed model. Checkout the repository [MorphologicalPriorsForWordEmbeddings](https://github.com/rguthrie3/MorphologicalPriorsForWordEmbeddings) for to train a varembed model. You'll need to use that code if you want to train a model. \n\n# Load Varembed Model\n\nNow that you have an already trained varembed model, you can easily load the varembed word vectors directly into Gensim. <br>\nFor that, you need to provide the path to the word vectors pickle file generated after you train the model and run the script to [package varembed embeddings](https://github.com/rguthrie3/MorphologicalPriorsForWordEmbeddings/blob/master/package_embeddings.py) provided in the [varembed source code repository](https://github.com/rguthrie3/MorphologicalPriorsForWordEmbeddings).\n\nWe'll use a varembed model trained on [Lee Corpus](https://github.com/RaRe-Technologies/gensim/blob/develop/gensim/test/test_data/lee.cor) as the vocabulary, which is already available in gensim.\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"from gensim.models.wrappers import varembed\n\nvector_file = '../../gensim/test/test_data/varembed_leecorpus_vectors.pkl'\nmodel = varembed.VarEmbed.load_varembed_format(vectors=vector_file)",
"_____no_output_____"
]
],
[
[
"This loads a varembed model into Gensim. Also if you want to load with morphemes added into the varembed vectors, you just need to also provide the path to the trained morfessor model binary as an argument. This works as an optional parameter, if not provided, it would just load the varembed vectors without morphemes.",
"_____no_output_____"
]
],
[
[
"morfessor_file = '../../gensim/test/test_data/varembed_leecorpus_morfessor.bin'\nmodel_with_morphemes = varembed.VarEmbed.load_varembed_format(vectors=vector_file, morfessor_model=morfessor_file)",
"_____no_output_____"
]
],
[
[
"This helps load trained varembed models into Gensim. Now you can use this for any of the Keyed Vector functionalities, like 'most_similar', 'similarity' and so on, already provided in gensim. \n",
"_____no_output_____"
]
],
[
[
"model.most_similar('government')",
"_____no_output_____"
],
[
"model.similarity('peace', 'grim')",
"_____no_output_____"
]
],
[
[
"# Conclusion\nIn this tutorial, we learnt how to load already trained varembed models vectors into gensim and easily use and evaluate it. That's it!\n\n# Resources\n\n* [Varembed Source Code](https://github.com/rguthrie3/MorphologicalPriorsForWordEmbeddings)\n* [Gensim](http://radimrehurek.com/gensim/)\n* [Lee Corpus](https://github.com/RaRe-Technologies/gensim/blob/develop/gensim/test/test_data/lee.cor)\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
]
|
cb6f289b0b33e01f95faebebeb5914b6c20c9ea4 | 5,162 | ipynb | Jupyter Notebook | mini_book/exercises/fmskill_assignment.ipynb | DHI/book-learn-mikeio-fmskill | 595b95e341379f647e1dfd428a72516f699877c6 | [
"MIT"
]
| 1 | 2021-09-21T15:21:37.000Z | 2021-09-21T15:21:37.000Z | mini_book/exercises/fmskill_assignment.ipynb | DHI/book-learn-mikeio-fmskill | 595b95e341379f647e1dfd428a72516f699877c6 | [
"MIT"
]
| null | null | null | mini_book/exercises/fmskill_assignment.ipynb | DHI/book-learn-mikeio-fmskill | 595b95e341379f647e1dfd428a72516f699877c6 | [
"MIT"
]
| null | null | null | 31.096386 | 261 | 0.596087 | [
[
[
"# FMskill assignment\n\nYou are working on a project modelling waves in the Southern North Sea. You have done 6 different calibration runs and want to choose the \"best\". You would also like to see how your best model is performing compared to a third-party model in NetCDF. \n\nThe data: \n\n* SW model results: 6 dfs0 files ts_runX.dfs0 each with 4 items corresponding to 4 stations\n* observations: 4 dfs0 files with station data for (name, longitude, latitude):\n - F16: 4.0122, 54.1167\n - HKZA: 4.0090, 52.3066\n - K14: 3.6333, 53.2667\n - L9: 4.9667, 53.6167\n* A map observations_map.png showing the model domain and observation positions\n* Third party model: 1 NetCDF file\n\nThe tasks:\n\n1. Calibration - find the best run\n2. Validation - compare model to third-party model",
"_____no_output_____"
]
],
[
[
"fldr = \"../data/FMskill_assignment/\" # where have you put your data? ",
"_____no_output_____"
],
[
"import fmskill\nfrom fmskill import PointObservation, ModelResult, Connector",
"_____no_output_____"
]
],
[
[
"## 1. Calibration\n\n* 1.1 Start simple: compare F16 with SW1 (the first calibration run)\n* 1.2 Define all observations and all model results\n* 1.3 Create connector, plot temporal coverage\n* 1.4 Evaluate results\n* 1.5 Which model is best?",
"_____no_output_____"
],
[
"### 1.1 Simple compare\n\nUse fmskill.compare to do a quick comparison of F16 and SW1. \n\nWhat is the mean absolute error in cm? \nDo a time series plot.",
"_____no_output_____"
],
[
"### 1.2 Define all observations and all model results\n\n* Define 4 PointObservations o1, o2, o3, o4\n* Define 6 ModelResults mr1, mr2, ... (name them \"SW1\", \"SW2\", ...)\n* How many items do the ModelResults have? ",
"_____no_output_____"
],
[
"### 1.3 Create connector, plot temporal coverage\n\n* Create empty Connector con\n* The add the connections one observation at a time (start by matching o1 with the 6 models, then o2...)\n* Print con to screen - which observation has most observation points? \n* Plot the temporal coverage of observations and models\n* Save the Connector to an excel configuration file",
"_____no_output_____"
],
[
"### 1.4 Evaluate results\n\nDo relevant qualitative and quantitative analysis (e.g. time series plots, scatter plots, skill tables etc) to compare the models. ",
"_____no_output_____"
],
[
"### 1.5 Find the best\n\nWhich calibration run is best? \n\n* Which model performs best in terms of bias? \n* Which model has the smallest scatter index? \n* Which model has linear slope closest to 1.0 for the station HKZA? \n* Consider the last day only (Nov 19) - which model has the smallest bias for that day? \n* Weighted: Give observation F16 10-times more weight than the other observations - which has the smallest MAE? \n* Extremes: Which model has lowest rmse for Hs>4.0 (df = cc.all_df[cc.all_df.obs_val>4])?\n",
"_____no_output_____"
],
[
"## 2. Validation",
"_____no_output_____"
],
[
"We will now compare our best model against the UK MetOffice's North West Shelf model stored in NWS_HM0.nc. \n\n* 2.1 Create a ModelResult mr_NWS, evaluate mr_NWS.ds\n* 2.2 Plot the first time step (hint .isel(time=0)) of ds (hint: the item is called \"VHM0\")\n* 2.3 Create a Connector con_NWS with the 4 observations and mr_NWS \n* 2.4 Evaluate NWS - what is the mean rmse? \n* 2.5 Compare NWS to SW5 - which model is better? And is it so for all stations and all metrics? (hint: you can merge ComparisonCollections using the + operator)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
cb6f47bb9f233a1163cd289b5d113af320b18ba0 | 21,490 | ipynb | Jupyter Notebook | teoria/1.1_numpy.ipynb | aliciapj/intro_ml_python | 703e1050c2a8bce706e1d34076aaade032710ae3 | [
"CC0-1.0"
]
| 17 | 2019-03-29T15:18:36.000Z | 2020-04-03T11:00:02.000Z | teoria/1.1_numpy.ipynb | aliciapj/intro_ml_python | 703e1050c2a8bce706e1d34076aaade032710ae3 | [
"CC0-1.0"
]
| null | null | null | teoria/1.1_numpy.ipynb | aliciapj/intro_ml_python | 703e1050c2a8bce706e1d34076aaade032710ae3 | [
"CC0-1.0"
]
| 7 | 2019-03-29T15:18:28.000Z | 2020-11-03T19:22:48.000Z | 22.223371 | 156 | 0.475989 | [
[
[
"## Instalación de numpy",
"_____no_output_____"
]
],
[
[
"! pip install numpy",
"Requirement already satisfied: numpy in /home/alicia/workspace/intro_ml_python/venv/lib/python3.6/site-packages (1.16.2)\r\n"
],
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"### Array creation",
"_____no_output_____"
]
],
[
[
"my_int_list = [1, 2, 3, 4]\n\n#create numpy array from original python list\nmy_numpy_arr = np.array(my_int_list)\n\nprint(my_numpy_arr)",
"[1 2 3 4]\n"
],
[
"# Array of zeros\nprint(np.zeros(10))\n\n# Array of ones with type int\nprint(np.ones(10, dtype=int))\n\n# Range of numbers\nrangeArray = np.array(range(10), int)\nprint(rangeArray)",
"[0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n[1 1 1 1 1 1 1 1 1 1]\n[0 1 2 3 4 5 6 7 8 9]\n"
],
[
"# Random array\nprint(f\"Random array: {np.random.rand(5)}\\n\")\n\n# Random matrix\nprint(f\"Random matrix:\\n {np.random.rand(5,4)}\\n\")\n\n# Random array of integers in a range (say 0-9)\nrandomArray = np.floor(np.random.rand(10) * 10)\nprint(f\"Random integer array: {randomArray}\\n\")\n\n# Futher simplification\nprint(f\"Random matrix:\\n{np.random.randint(0, 10, (2,5))}\\n\")",
"Random array: [0.8843695 0.52825951 0.81974752 0.16345183 0.48115087]\n\nRandom matrix:\n [[0.08725599 0.71520207 0.02641297 0.60381681]\n [0.09491874 0.95021844 0.55669445 0.57031089]\n [0.06184176 0.77486812 0.62235827 0.27646621]\n [0.0926123 0.26080523 0.47212059 0.20246903]\n [0.42985942 0.95217091 0.68931885 0.86542322]]\n\nRandom integer array: [2. 0. 8. 5. 8. 2. 3. 5. 4. 2.]\n\nRandom matrix:\n[[6 8 6 5 0]\n [3 3 7 7 6]]\n\n"
],
[
"integerArray = np.array([1,2,3,4], int)\nintegerArray2 = np.array([5,6], int)\n\n# Concatenate two arrays\nprint(np.concatenate((integerArray, integerArray2)))\n",
"[1 2 3 4 5 6]\n"
],
[
"# Multidimensional array\nfloatArray = np.array([[1,2,3], [4,5,6]], float)\nprint(floatArray)\n",
"[[1. 2. 3.]\n [4. 5. 6.]]\n"
],
[
"# Convert one dimensional to multidimensional arrays\nrangeArray = rangeArray.reshape(5, 2)\nprint(rangeArray)",
"[[0 1]\n [2 3]\n [4 5]\n [6 7]\n [8 9]]\n"
],
[
"# Convert multidimensional to one dimensional array\nrangeArray = rangeArray.flatten()\nprint(rangeArray)",
"[0 1 2 3 4 5 6 7 8 9]\n"
],
[
"# Concatenation of multi-dimensional arrays\narr1 = np.array([[1,2], [3,4]], int)\narr2 = np.array([[5,6], [7,8]], int)\nprint(f'array1: \\n{arr1}\\n')\nprint(f'array2: \\n{arr2}')",
"array1: \n[[1 2]\n [3 4]]\n\narray2: \n[[5 6]\n [7 8]]\n"
],
[
"# Based on dimension 1\nprint(np.concatenate((arr1, arr2), axis=0))",
"[[1 2]\n [3 4]\n [5 6]\n [7 8]]\n"
],
[
"# Based on dimension 2\nprint(np.concatenate((arr1, arr2), axis=1))",
"[[1 2 5 6]\n [3 4 7 8]]\n"
]
],
[
[
"### Universal Functions\nThese functions are defined as functions that operate element-wise on the array elements whether it is a single or multidimensional array.",
"_____no_output_____"
]
],
[
[
"# we want to alter each element of the collection by multiplying each integer by 2\n\nmy_int_list = [1, 2, 3, 4]\n\n# python code\nfor i, val in enumerate(my_int_list):\n my_int_list[i] *= 2\n\nmy_int_list",
"_____no_output_____"
],
[
"#create numpy array from original python list\nmy_numpy_arr = np.array(my_int_list)\n\n#multiply each element by 2\nmy_numpy_arr * 2",
"_____no_output_____"
],
[
"# Addition\nprint(f\"Array 1 + Array 2\\n {arr1 + arr2}\\n\")\n\n# Multiplication\nprint(f\"Array 1 * Array 2\\n {arr1 * arr2}\\n\")\n\n# Square root\nprint(f\"Square root of Array 1\\n {np.sqrt(arr1)}\\n\")\n\n# Log\nprint(f\"Log of Array 1\\n {np.log(arr1)}\\n\")",
"Array 1 + Array 2\n [[ 6 8]\n [10 12]]\n\nArray 1 * Array 2\n [[ 5 12]\n [21 32]]\n\nSquare root of Array 1\n [[1. 1.41421356]\n [1.73205081 2. ]]\n\nLog of Array 1\n [[0. 0.69314718]\n [1.09861229 1.38629436]]\n\n"
]
],
[
[
"https://towardsdatascience.com/numpy-python-made-efficient-f82a2d84b6f7",
"_____no_output_____"
],
[
"### Aggregation Functions\nThese functions are useful when we wish to summarise the information contained in an array.",
"_____no_output_____"
]
],
[
[
"arr1 = np.arange(1,10).reshape(3,3)\nprint(f'Array 1: \\n{arr1}\\n')\n\nprint(f\"Sum of elements of Array 1: {arr1.sum()}\\n\")\n\nprint(f\"Sum by row elements of Array 1: {np.sum(arr1, axis=1)}\\n\")\n\nprint(f\"Sum by column elements of Array 1: {np.sum(arr1, axis=0)}\\n\")",
"Array 1: \n[[1 2 3]\n [4 5 6]\n [7 8 9]]\n\nSum of elements of Array 1: 45\n\nSum by row elements of Array 1: [ 6 15 24]\n\nSum by column elements of Array 1: [12 15 18]\n\n"
],
[
"print(f'Array 1: \\n{arr1}\\n')\n\n# Mean of array elements\nprint(f\"Mean of elements of Array 1: {arr1.mean()}\\n\")\n\n# Minimum of array elements\nprint(f\"Minimum of elements of Array 1: {arr1.min()}\\n\")\n# Minimum of elements of Array 1: 1\n\n# Index of maximum of array elements can be found using arg before the funciton name\nprint(f\"Index of minimum of elements of Array 1: {arr1.argmax()}\")",
"Array 1: \n[[1 2 3]\n [4 5 6]\n [7 8 9]]\n\nMean of elements of Array 1: 5.0\n\nMinimum of elements of Array 1: 1\n\nIndex of minimum of elements of Array 1: 8\n"
]
],
[
[
"### Broadcasting\nThese are a set of rules of how universal functions operate on numpy arrays.",
"_____no_output_____"
]
],
[
[
"sampleArray = np.array([[5,2,3], [3,4,5], [1,1,1]], int)\nprint(f\"Sample Array\\n {sampleArray}\\n\")\n\n# Get unqiue values\nprint(f\"Unique values: {np.unique(sampleArray)}\\n\")\n# Unique values: [1 2 3 4 5]\n\n# Get diagonal values\nprint(f\"Diagonal\\n {sampleArray.diagonal()}\\n\")\n# Diagonal\n# [5 4 1]\n\n# Sort values in the multidimensional array\nprint(f\"Sorted\\n {np.sort(sampleArray)}\\n\")",
"Sample Array\n [[5 2 3]\n [3 4 5]\n [1 1 1]]\n\nUnique values: [1 2 3 4 5]\n\nDiagonal\n [5 4 1]\n\nSorted\n [[2 3 5]\n [3 4 5]\n [1 1 1]]\n\n"
],
[
"sampleArray = np.array([[5,2,3], [3,4,5], [1,1,1]], int)\nprint(f\"Sample Array\\n {sampleArray}\\n\")\n\n# Get diagonal values\nprint(f\"Diagonal\\n {sampleArray.T.diagonal()}\\n\")",
"Sample Array\n [[5 2 3]\n [3 4 5]\n [1 1 1]]\n\nDiagonal\n [5 4 1]\n\n"
],
[
"vector = np.array([1,2,3,4], int)\nmatrix1 = np.array([[1,2,3], [4,5,6], [7,8,9]], int)\nmatrix2 = np.array([[1,1,1], [0,0,0], [1,1,1]], int)\n\n# Dot operator\nprint(f\"Dot of Matrix 1 and Matrix 2\\n {np.dot(matrix1, matrix2)}\\n\")\n\n# Cross operator\nprint(f\"Cross of Matrix 1 and Matrix 2\\n {np.cross(matrix1, matrix2)}\\n\")\n\n# Outer operator\nprint(f\"Outer of Matrix 1 and Matrix 2\\n {np.outer(matrix1, matrix2)}\\n\")\n\n# Inner operator\nprint(f\"Inner of Matrix 1 and Matrix 2\\n {np.inner(matrix1, matrix2)}\")",
"Dot of Matrix 1 and Matrix 2\n [[ 4 4 4]\n [10 10 10]\n [16 16 16]]\n\nCross of Matrix 1 and Matrix 2\n [[-1 2 -1]\n [ 0 0 0]\n [-1 2 -1]]\n\nOuter of Matrix 1 and Matrix 2\n [[1 1 1 0 0 0 1 1 1]\n [2 2 2 0 0 0 2 2 2]\n [3 3 3 0 0 0 3 3 3]\n [4 4 4 0 0 0 4 4 4]\n [5 5 5 0 0 0 5 5 5]\n [6 6 6 0 0 0 6 6 6]\n [7 7 7 0 0 0 7 7 7]\n [8 8 8 0 0 0 8 8 8]\n [9 9 9 0 0 0 9 9 9]]\n\nInner of Matrix 1 and Matrix 2\n [[ 6 0 6]\n [15 0 15]\n [24 0 24]]\n"
]
],
[
[
"### Slicing, masking and fancy indexing\nThe last strategy pools in a few tricks too",
"_____no_output_____"
]
],
[
[
"arr1 = np.array([[1,5], [7,8]], int)\narr2 = np.array([[6, 2], [7,8]], int)\n\nprint(f'Array 1: \\n{arr1}\\n')\nprint(f'Array 2: \\n{arr2}\\n\\n')\n\n# We can compare complete arrays of equal size element wise\nprint(f\"Array 1 > Array 2\\n{arr1 > arr2}\\n\")\n \n# We can compare elements of an array with a given value\nprint(f\"Array 1 == 2\\n {arr1 == arr2}\\n\")\n",
"Array 1: \n[[1 5]\n [7 8]]\n\nArray 2: \n[[6 2]\n [7 8]]\n\n\nArray 1 > Array 2\n[[False True]\n [False False]]\n\nArray 1 == 2\n [[False False]\n [ True True]]\n\n"
],
[
"bigArray = np.array(range(10))\nprint(\"Array: {}\".format(bigArray))\n\n# Slice array from index 0 to 4\nprint(\"Array value from index 0 to 4: {}\".format(bigArray[-5]))\n\n\n# Masking using boolean values and operators\nmask = (bigArray > 6) | (bigArray < 3)\nprint(mask)\nprint(\"Array values with mask as true: {}\".format(bigArray[mask]))\n\n# Fancy indexing\nind = [2,4,6]\nprint(\"Array values with index in list: {}\".format(bigArray[ind]))\n\n# Combine all three\nprint(\"Array values with index in list: {}\".format(bigArray[bigArray > 6][:1]))\n",
"Array: [0 1 2 3 4 5 6 7 8 9]\nArray value from index 0 to 4: 5\n[ True True True False False False False True True True]\nArray values with mask as true: [0 1 2 7 8 9]\nArray values with index in list: [2 4 6]\n"
]
],
[
[
"\n\n<img src=\"https://cdn-images-1.medium.com/max/800/1*cxbe7Omfj6Be0fbvD7gmGQ.png\">",
"_____no_output_____"
],
[
"<img src=\"https://cdn-images-1.medium.com/max/800/1*9FImAfjF6Z6Hyv9lm1WgjA.png\">",
"_____no_output_____"
],
[
"https://medium.com/@zachary.bedell/writing-beautiful-code-with-numpy-505f3b353174",
"_____no_output_____"
]
],
[
[
"# multiplying two matrices containing 60,000 and 80,000 integers\nimport time\nimport random as r\n\n\ntick = time.time()\n#create a 300x200 matrix of 60,000 random integers\nmy_list_1 = []\nfor row_index in range(300):\n new_row = []\n for col_index in range(200):\n new_row.append(r.randint(0, 20))\n my_list_1.append(new_row)\n#create a 200x400 matrix of 80,000 random integers\nmy_list_2 = []\nfor row_index in range(200):\n new_row = []\n for col_index in range(400):\n new_row.append(r.randint(0, 20))\n my_list_2.append(new_row)\n\n#create 2X3 array to hold results\nmy_result_arr = []\nfor row_index in range(300):\n new_row = []\n for col_index in range(400):\n new_row.append(0)\n my_result_arr.append(new_row)\n\n# iterate through rows of my_list_1\nfor i in range(len(my_list_1)):\n # iterate through columns of my_list_2\n for j in range(len(my_list_2[0])):\n # iterate through rows of my_list_2\n for k in range(len(my_list_2)):\n my_result_arr[i][j] += my_list_1[i][k] * my_list_2[k][j]\n\ntime_to_completion = time.time() - tick\n\nprint(\"execution time without NumPy: \", time_to_completion)",
"execution time without NumPy: 5.876939296722412\n"
]
],
[
[
"The code is difficult to read, and the solution requires double and triple nested loops, each of which have high time complexities of O(n²) and O(n³).",
"_____no_output_____"
]
],
[
[
"import time\ntick = time.time()\nnp_arr_1 = np.arange(0, 60000).reshape(300, 200)\nnp_arr_2 = np.arange(0, 80000).reshape(200, 400)\n\nmy_result_arr = np.matmul(np_arr_1, np_arr_2)\ntime_to_completion = time.time() - tick\n\nprint(\"execution time with NumPy: \", time_to_completion)",
"execution time with NumPy: 0.031472206115722656\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6f649e21bcdbd5b3a779aa79c6d53fa99e1da5 | 194,145 | ipynb | Jupyter Notebook | source/examples/demo/2020.ipynb | JetBrains/lets-plot-docs | 73583bce5308d34b341d9f8a7249ccb34a95f504 | [
"MIT"
]
| 2 | 2021-06-02T10:24:24.000Z | 2021-11-08T09:50:22.000Z | source/examples/demo/2020.ipynb | JetBrains/lets-plot-docs | 73583bce5308d34b341d9f8a7249ccb34a95f504 | [
"MIT"
]
| 13 | 2021-05-25T19:49:50.000Z | 2022-03-22T12:30:29.000Z | source/examples/demo/2020.ipynb | JetBrains/lets-plot-docs | 73583bce5308d34b341d9f8a7249ccb34a95f504 | [
"MIT"
]
| 4 | 2021-01-19T12:26:21.000Z | 2022-03-19T07:47:52.000Z | 94.751098 | 17,872 | 0.546061 | [
[
[
"# Lets-Plot in 2020",
"_____no_output_____"
],
[
"### Preparation",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport colorcet as cc\nfrom PIL import Image\n\nfrom lets_plot import *\nfrom lets_plot.bistro.corr import *\nLetsPlot.setup_html()",
"_____no_output_____"
],
[
"df = pd.read_csv(\"https://raw.githubusercontent.com/JetBrains/lets-plot-docs/master/data/lets_plot_git_history.csv\", sep=';')\n\ndf = df[['author_date', 'author_name', 'files_changed', 'insertions', 'deletions']]\ndf.author_date = pd.to_datetime(df.author_date, utc=True)\ndf.files_changed = df.files_changed.str.split(' ').str[0].astype(int)\ndf.insertions = df.insertions.str.split(' ').str[0].astype(int)\ndf.deletions = df.deletions.fillna('0').str.split(' ').str[0].astype(int)\n\ndf['diff'] = df.insertions - df.deletions\ndf['month'] = df.author_date.dt.month\ndf['day'] = df.author_date.dt.day\ndf['weekday'] = df.author_date.dt.weekday\ndf['hour'] = df.author_date.dt.hour\n\ndf = df[df.author_date.dt.year == 2020].sort_values(by='author_date').reset_index(drop=True)\n\ndf.head()",
"_____no_output_____"
]
],
[
[
"### General Analytics",
"_____no_output_____"
]
],
[
[
"agg_features = {'files_changed': ['sum', 'mean'], \\\n 'insertions': ['sum', 'mean'], \\\n 'deletions': ['sum', 'mean'], \\\n 'diff': ['sum']}\nagg_df = df.groupby('author_name').agg(agg_features).reset_index()\nagg_features['commits_number'] = ['sum']\nagg_df = pd.merge(agg_df, df.author_name.value_counts().to_frame(('commits_number', 'sum')).reset_index(), \\\n left_on='author_name', right_on='index')\nagg_df['color'] = cc.palette['glasbey_bw'][:agg_df.shape[0]]\n\nplots = []\nfor feature, agg in [(key, val) for key, vals in agg_features.items() for val in vals]:\n agg_df = agg_df.sort_values(by=(feature, agg), ascending=False)\n aes_name = ('total {0}' if agg == 'sum' else 'mean {0} per commit').format(feature.replace('_', ' '))\n plotted_df = agg_df[[('author_name', ''), (feature, agg), ('color', '')]]\n plotted_df.columns = plotted_df.columns.get_level_values(0)\n plots.append(ggplot(plotted_df) + \\\n geom_bar(aes(x='author_name', y=feature, color='color', fill='color'), \\\n stat='identity', alpha=.25, size=1, \\\n tooltips=layer_tooltips().line('^x')\n .line('{0}|^y'.format(aes_name))) + \\\n scale_color_identity() + scale_fill_identity() + \\\n xlab('') + ylab('') + \\\n ggtitle(aes_name.title()))\n\nw, h = 400, 300\nbunch = GGBunch()\nbunch.add_plot(plots[7], 0, 0, w, h)\nbunch.add_plot(plots[6], w, 0, w, h)\nbunch.add_plot(plots[0], 0, h, w, h)\nbunch.add_plot(plots[1], w, h, w, h)\nbunch.add_plot(plots[2], 0, 2 * h, w, h)\nbunch.add_plot(plots[3], w, 2 * h, w, h)\nbunch.add_plot(plots[4], 0, 3 * h, w, h)\nbunch.add_plot(plots[5], w, 3 * h, w, h)\nbunch.show()",
"_____no_output_____"
]
],
[
[
"Looking at the total values, we clearly see that Igor Alshannikov and Ivan Kupriyanov outcompete the rest. But there is a real intrigue as to who takes the third place.\n\nMeanwhile, we see more diversity in mean values of different contribution types.",
"_____no_output_____"
]
],
[
[
"ggplot(df.hour.value_counts().to_frame('count').reset_index().sort_values(by='index')) + \\\n geom_histogram(aes(x='index', y='count', color='index', fill='index'), \\\n stat='identity', show_legend=False, \\\n tooltips=layer_tooltips().line('^y')) + \\\n scale_x_discrete(breaks=list(range(24))) + \\\n scale_color_gradient(low='#e0ecf4', high='#8856a7') + \\\n scale_fill_gradient(low='#e0ecf4', high='#8856a7') + \\\n xlab('hour') + ylab('commits number') + \\\n ggtitle('Total Hourly Committing') + ggsize(600, 450)",
"_____no_output_____"
]
],
[
[
"The peak of commit activity is about 18 p.m. The evening seems to be a good time to save daily results.",
"_____no_output_____"
],
[
"### Higher Resolution",
"_____no_output_____"
]
],
[
[
"plotted_df = df[df.insertions > 0].reset_index(drop=True)\nplotted_df['insertions_unit'] = np.ones(plotted_df.shape[0])\nggplot(plotted_df) + \\\n geom_segment(aes(x='author_date', y='insertions_unit', xend='author_date', yend='insertions'), color='#8856a7') + \\\n geom_point(aes(x='author_date', y='insertions', fill='month'), shape=21, color='#8856a7', \\\n tooltips=layer_tooltips().line('@author_name').line('@|@insertions').line('@|@month')) + \\\n scale_x_datetime(name='date') + \\\n scale_y_log10(name='insertions (log)') + \\\n scale_fill_brewer(name='', type='qual', palette='Paired') + \\\n facet_grid(y='author_name') + \\\n ggtitle('Lollipop Plot of Commits by Authors') + ggsize(800, 1000)",
"_____no_output_____"
]
],
[
[
"Some of the team members started their work only a few months ago, so they still have time to catch up next year.",
"_____no_output_____"
]
],
[
[
"ggplot(df) + \\\n geom_point(aes(x='weekday', y='insertions', color='author_name', size='files_changed'), \\\n shape=8, alpha=.5, position='jitter', show_legend=False, \\\n tooltips=layer_tooltips().line('author|@author_name')\n .line('@|@insertions')\n .line('@|@deletions')\n .line('files changed|@files_changed')) + \\\n scale_x_discrete(labels=['Monday', 'Tuesday', 'Wednesday', 'Thursday', \\\n 'Friday', 'Saturday', 'Sunday']) + \\\n scale_y_log10(breaks=[2 ** n for n in range(16)]) + \\\n scale_size(range=[3, 7], trans='sqrt') + \\\n ggtitle('All Commits') + ggsize(800, 600) + \\\n theme(axis_tooltip='blank')",
"_____no_output_____"
]
],
[
[
"Usually no one works at the weekend. But if something needs to be done - it should be.",
"_____no_output_____"
],
[
"### And Finally...",
"_____no_output_____"
]
],
[
[
"r = df.groupby('day').insertions.median().values\nx = r * np.cos(np.linspace(0, 2 * np.pi, r.size))\ny = r * np.sin(np.linspace(0, 2 * np.pi, r.size))\ndaily_insertions_df = pd.DataFrame({'x': x, 'y': y})",
"_____no_output_____"
],
[
"MONTHS = ['Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun', 'Jul', 'Aug', 'Sep', 'Oct', 'Nov', 'Dec']\nmask_width, mask_height = 60, 80\n\nmask = np.array(Image.open(\"images/snowman_mask.bmp\").resize((mask_width, mask_height), Image.BILINEAR))\ngrid = [[(0 if color.mean() > 255 / 2 else 1) for color in row] for row in mask]\n\ngrid_df = pd.DataFrame(grid).stack().to_frame('month')\ngrid_df.index.set_names(['y', 'x'], inplace=True)\ngrid_df = grid_df.reset_index()\ngrid_df.y = grid_df.y.max() - grid_df.y\ngrid_df = grid_df[grid_df.month > 0].reset_index(drop=True)\n\nagg_df = np.round(df.month.value_counts() * grid_df.shape[0] / df.shape[0]).to_frame('commits_number')\nagg_df.iloc[0].commits_number += grid_df.shape[0] - agg_df.commits_number.sum()\nagg_df.commits_number = agg_df.commits_number.astype(int)\nagg_df.index.name = 'month'\nagg_df = agg_df.reset_index()\n\ngrid_df['commits_number'] = 0\nstart_idx = 0\nfor idx, (month, commits_number) in agg_df.iterrows():\n grid_df.loc[start_idx:(start_idx + commits_number), 'month'] = MONTHS[month - 1]\n grid_df.loc[start_idx:(start_idx + commits_number), 'commits_number'] = commits_number\n start_idx += commits_number",
"_____no_output_____"
],
[
"blank_theme = theme_classic() + theme(axis='blank', axis_ticks_x='blank', axis_ticks_y='blank', legend_position='none')\n\nps = ggplot(daily_insertions_df, aes(x='x', y='y')) + \\\n geom_polygon(color='#f03b20', fill='#fd8d3c', size=1) + coord_fixed() + blank_theme\np1l = corr_plot(data=df[['insertions', 'deletions']], flip=False).tiles(type='lower', diag=True)\\\n .palette_gradient(low='blue', mid='green', high='darkgreen').build() + blank_theme\np1r = corr_plot(data=df[['deletions', 'insertions']], flip=True).tiles(type='lower', diag=True)\\\n .palette_gradient(low='blue', mid='green', high='darkgreen').build() + blank_theme\np2l = corr_plot(data=df[['insertions', 'deletions', 'diff']], flip=False).tiles(type='lower', diag=True)\\\n .palette_gradient(low='blue', mid='green', high='darkgreen').build() + blank_theme\np2r = corr_plot(data=df[['diff', 'deletions', 'insertions']], flip=True).tiles(type='lower', diag=True)\\\n .palette_gradient(low='blue', mid='green', high='darkgreen').build() + blank_theme\np3l = corr_plot(data=df[['insertions', 'deletions', 'diff', 'files_changed']], flip=False)\\\n .tiles(type='lower', diag=True).palette_gradient(low='blue', mid='green', high='darkgreen').build() + blank_theme\np3r = corr_plot(data=df[['files_changed', 'diff', 'deletions', 'insertions']], flip=True)\\\n .tiles(type='lower', diag=True).palette_gradient(low='blue', mid='green', high='darkgreen').build() + blank_theme\npt = ggplot({'x': [0], 'y': [0], 'greetings': ['Happy New Year!']}, aes(x='x', y='y')) + \\\n geom_text(aes(label='greetings'), color='blue', size=20, family='Times New Roman', fontface='bold') + blank_theme\npm = ggplot(grid_df, aes(x='x', y='y')) + \\\n geom_tile(aes(fill='month'), width=.8, height=.8, \\\n tooltips=layer_tooltips().line('@|@month')\n .line('@|@commits_number')) + \\\n scale_fill_brewer(type='qual', palette='Set2') + \\\n blank_theme\n\nw, h = 50, 50\nbunch = GGBunch()\nbunch.add_plot(ps, 3 * w, 0, 2 * w, 2 * h)\nbunch.add_plot(p1l, 2 * w, 2 * h, 2 * w, 2 * h)\nbunch.add_plot(p1r, 4 * w, 2 * h, 2 * w, 2 * h)\nbunch.add_plot(p2l, w, 4 * h, 3 * w, 3 * h)\nbunch.add_plot(p2r, 4 * w, 4 * h, 3 * w, 3 * h)\nbunch.add_plot(p3l, 0, 7 * h, 4 * w, 4 * h)\nbunch.add_plot(p3r, 4 * w, 7 * h, 4 * w, 4 * h)\nbunch.add_plot(pt, 0, 11 * h, 16 * w, 2 * h)\nbunch.add_plot(pm, 8 * w, 3 * h, 8 * w, 8 * h)\nbunch.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
]
|
cb6f64e1aeb20edb907267e5684d45163250ce54 | 4,466 | ipynb | Jupyter Notebook | issa/cross_val.ipynb | phdinds-aim/resype | 322f272d600ed6831fb90c5d05794d0e5dd3eca2 | [
"MIT"
]
| null | null | null | issa/cross_val.ipynb | phdinds-aim/resype | 322f272d600ed6831fb90c5d05794d0e5dd3eca2 | [
"MIT"
]
| null | null | null | issa/cross_val.ipynb | phdinds-aim/resype | 322f272d600ed6831fb90c5d05794d0e5dd3eca2 | [
"MIT"
]
| null | null | null | 31.450704 | 141 | 0.500896 | [
[
[
"# Cross-validation \nThis notebook contains the function that performs cross validation tests. This is a dummy function that can be tested with the model/s.",
"_____no_output_____"
]
],
[
[
"def cross_val(df, k, model, split_method='random'):\n \"\"\"\n Performs cross-validation for different train and test sets.\n\n Parameters\n -----------\n df : the data to be split in the form of vanilla/transaction++ table (uid, iid, rating, timestamp)\n\n k : the number of times splitting and learning with the model is desired\n \n model : an unfitted sklearn model\n\n split_method : 'random' splitting or 'chronological' splitting of the data\n\n\n Returns\n --------\n mse and mae : error metrics using sklearn\n\n\n \"\"\"\n mse = []\n mae = []\n\n if split_method == 'random':\n\n for i in range(k):\n print(i)\n # 1. split\n print('Starting splitting')\n df_train, df_test, df_test_um, indx_train, indx_test = split_train_test(\n df, 0.7)\n print('Finished splitting')\n # 2. train with model\n model_clone = clone(model)\n print('Starting training')\n model_clone_fit = fit_ml_cb(df_train, model_clone)\n print('Finished training')\n print('Starting completing matrix')\n result = reco_ml_cb(user_df, list(df_test.index), item_df, model_clone_fit)\n print('Finished completing matrix')\n print('Starting computing MAE and MSE')\n # 3. evaluate results (result is in the form of utility matrix)\n mse_i, mae_i = evaluate(result, df_test_um)\n print('Finished computing MAE and MSE')\n\n mse.append(mse_i)\n mae.append(mae_i)\n\n elif split_method == 'chronological':\n\n # 1. split\n df_train, df_test, df_test_um, indx_train, indx_test = split_train_test_chronological(\n df, 0.7)\n\n print('Starting splitting')\n print('Finished splitting')\n # 2. train with model\n model_clone = clone(model)\n print('Starting training')\n model_clone_fit = fit_ml_cb(df_train, model_clone)\n print('Finished training')\n print('Starting completing matrix')\n result = reco_ml_cb(user_df, list(df_test.index), item_df, model_clone_fit)\n print('Finished completing matrix')\n print('Starting computing MAE and MSE')\n # 3. evaluate results (result is in the form of utility matrix)\n mse_i, mae_i = evaluate(result, df_test_um)\n print('Finished computing MAE and MSE')\n\n mse.append(mse_i)\n mae.append(mae_i)\n\n return mse, mae",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
]
]
|
cb6f726fb92e007287acc6d439f929262f3e7c0a | 17,018 | ipynb | Jupyter Notebook | notebooks/Gamma_Log_Facies_Type_Prediction/09_find_lag.ipynb | KirillVladimirov/kaggle_competitions | 2ae3144810729ec281fbc261d47c718308bc25cc | [
"MIT"
]
| null | null | null | notebooks/Gamma_Log_Facies_Type_Prediction/09_find_lag.ipynb | KirillVladimirov/kaggle_competitions | 2ae3144810729ec281fbc261d47c718308bc25cc | [
"MIT"
]
| null | null | null | notebooks/Gamma_Log_Facies_Type_Prediction/09_find_lag.ipynb | KirillVladimirov/kaggle_competitions | 2ae3144810729ec281fbc261d47c718308bc25cc | [
"MIT"
]
| null | null | null | 30.067138 | 271 | 0.494888 | [
[
[
"from tpot import TPOTClassifier\n\nimport os\nfrom tqdm import tqdm_notebook as tqdm\n\n# Ignore the warnings\nimport warnings\nwarnings.filterwarnings('always')\nwarnings.filterwarnings('ignore')\n\nimport numpy as np\nimport pandas as pd\n\nimport warnings\nimport matplotlib.pyplot as plt\nfrom matplotlib.pyplot import subplots\nimport matplotlib.patches as patches\nimport seaborn as sns\nfrom pylab import rcParams\n\n%matplotlib inline \nplt.style.use('seaborn')\nsns.set(style='whitegrid',color_codes=True)\n\n# classifiaction \nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier \nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.naive_bayes import GaussianNB\nimport xgboost as xgb\nimport catboost as ctb\n\n# for classification\nfrom sklearn.metrics import accuracy_score\n\n# model selection\nfrom sklearn.model_selection import train_test_split, cross_val_score, StratifiedKFold\nfrom sklearn.model_selection import GridSearchCV\n\n# Hp optimization imports\nfrom hyperopt import STATUS_OK, Trials, fmin, hp, tpe\nimport mlflow\n\nimport re\nimport eli5\nimport gc\nimport random \nimport math\nimport psutil\nimport pickle\nimport datetime\nfrom time import time\n\n# save/load models\nfrom joblib import dump\nfrom joblib import load\n\nimport timeit \nfrom sklearn.preprocessing import StandardScaler",
"/Users/kirillvladimirov/PycharmProjects/cv_projects/venv/lib/python3.7/site-packages/eli5/base_utils.py:28: DeprecationWarning: inspect.getargspec() is deprecated since Python 3.0, use inspect.signature() or inspect.getfullargspec()\n init_args = inspect.getargspec(class_.__init__)\n/Users/kirillvladimirov/PycharmProjects/cv_projects/venv/lib/python3.7/site-packages/eli5/base_utils.py:36: DeprecationWarning: The usage of `cmp` is deprecated and will be removed on or after 2021-06-01. Please use `eq` and `order` instead.\n return attr.s(class_, these=these, init=False, slots=True, **attrs_kwargs) # type: ignore\nUsing TensorFlow backend.\n/Users/kirillvladimirov/PycharmProjects/cv_projects/venv/lib/python3.7/site-packages/botocore/awsrequest.py:624: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated, and in 3.8 it will stop working\n class HeadersDict(collections.MutableMapping):\n"
],
[
"root = \"../../data/raw/Gamma_Log_Facies_Type_Prediction/\"\nmodels_root = \"../../models/Gamma_Log_Facies_Type_Prediction/\"\nRANDOM_STATE = 42\nnp.random.seed(RANDOM_STATE)\npd.set_option('max_columns', 150)\n# rcParams['figure.figsize'] = 16,8",
"_____no_output_____"
],
[
"%%time\nfull_train_df = pd.read_csv(root + \"Train_File.csv\")\nfull_test_df = pd.read_csv(root + \"Test_File.csv\")\nsubmit_df = pd.read_csv(root + \"Submission_File.csv\")\n",
"CPU times: user 2.82 s, sys: 400 ms, total: 3.22 s\nWall time: 3.25 s\n"
],
[
"def create_lags(df):\n for i in range(0, 25):\n df[\"lag_forward_{}\".format(i)] = df.GR.shift(i)\n df[\"lag_backward_{}\".format(i)] = df.GR.shift(-i)\n return df",
"_____no_output_____"
],
[
"train_df_ts = full_train_df[full_train_df[\"well_id\"] < 100]\nvalid_df_ts = full_train_df[full_train_df[\"well_id\"].isin(list(range(100,120)))]",
"_____no_output_____"
],
[
"train_df_ts.head()",
"_____no_output_____"
],
[
"width = 3\nshifted = train_df_ts.GR.shift(width - 1)\nwindow = shifted.rolling(window=width)\ndataframe = pd.concat([window.min(), window.mean(), window.max(), shifted], axis=1)\ndataframe.columns = ['min', 'mean', 'max', 't+1']\ndataframe = pd.concat([dataframe, train_df_ts])\nprint(dataframe.head(10))",
" GR label max mean min row_id t+1 well_id\n0 NaN NaN NaN NaN NaN NaN NaN NaN\n1 NaN NaN NaN NaN NaN NaN NaN NaN\n2 NaN NaN NaN NaN NaN NaN 143.510000 NaN\n3 NaN NaN NaN NaN NaN NaN 112.790928 NaN\n4 NaN NaN 143.510000 126.610928 112.790928 NaN 123.531856 NaN\n5 NaN NaN 123.531856 116.005190 111.692784 NaN 111.692784 NaN\n6 NaN NaN 123.613712 119.612784 111.692784 NaN 123.613712 NaN\n7 NaN NaN 123.613712 118.573712 111.692784 NaN 120.414641 NaN\n8 NaN NaN 123.613712 122.391307 120.414641 NaN 123.145569 NaN\n9 NaN NaN 123.145569 119.258902 114.216497 NaN 114.216497 NaN\n"
],
[
"train_df_ts.head()",
"_____no_output_____"
],
[
"window",
"_____no_output_____"
],
[
"window = train_df_ts.expanding()\ndataframe = pd.concat([window.min(), window.mean(), window.max(), train_df_ts.shift(-1)], axis=1)\n# dataframe.columns = ['min', 'mean', 'max', 't+1']\nprint(dataframe.head(5))",
" row_id well_id GR label row_id well_id GR label \\\n0 0.0 0.0 143.510000 0.0 0.0 0.0 143.510000 0.0 \n1 0.0 0.0 112.790928 0.0 0.5 0.0 128.150464 0.0 \n2 0.0 0.0 112.790928 0.0 1.0 0.0 126.610928 0.0 \n3 0.0 0.0 111.692784 0.0 1.5 0.0 122.881392 0.0 \n4 0.0 0.0 111.692784 0.0 2.0 0.0 123.027856 0.0 \n\n row_id well_id GR label row_id well_id GR label \n0 0.0 0.0 143.51 0.0 1.0 0.0 112.790928 0.0 \n1 1.0 0.0 143.51 0.0 2.0 0.0 123.531856 0.0 \n2 2.0 0.0 143.51 0.0 3.0 0.0 111.692784 0.0 \n3 3.0 0.0 143.51 0.0 4.0 0.0 123.613712 0.0 \n4 4.0 0.0 143.51 0.0 5.0 0.0 120.414641 0.0 \n"
],
[
"train_df_ts = train_df_ts.groupby(\"well_id\").apply(create_lags)\ntrain_df_ts = train_df_ts.fillna(0)\n\nvalid_df_ts = valid_df_ts.groupby(\"well_id\").apply(create_lags)\nvalid_df_ts = valid_df_ts.fillna(0)",
"_____no_output_____"
],
[
"X_train, y_train, X_test, y_test = train_df_ts.drop([\"label\"], axis=1), train_df_ts[\"label\"], \\\n valid_df_ts.drop([\"label\"], axis=1), valid_df_ts[\"label\"]",
"_____no_output_____"
],
[
"dataframe = concat([temps.shift(3), temps.shift(2), temps.shift(1), temps], axis=1)\ndataframe.columns = ['t-3', 't-2', 't-1', 't+1']",
"_____no_output_____"
],
[
"mlflow.set_experiment(\"xgboost_cls_feature_selecting\")\n\nclass HyperoptHPOptimizer:\n \n def __init__(self, hyperparameters_space, max_evals):\n self.trials = Trials()\n self.max_evals = max_evals\n self.hyperparameters_space = hyperparameters_space\n self.skf = StratifiedKFold(n_splits=3, shuffle=False, random_state=RANDOM_STATE)\n \n def get_loss(self, hyperparameters):\n # MLflow will track and save hyperparameters, loss, and scores. \n with mlflow.start_run(run_name='hyperopt_param'):\n params = {\n 'min_child_weight': 8,\n 'gamma': 3,\n 'subsample': 1,\n 'colsample_bytree': 0.6,\n 'eta': 0.3,\n 'max_depth': 4,\n 'random_state': RANDOM_STATE, \n 'verbosity': 1, \n 'n_jobs': -1, \n 'n_estimators': 10, \n 'learning_rate': 0.1, \n }\n cols = [col for col, is_use in hyperparameters.items() if is_use == 1]\n for k, v in hyperparameters.items():\n mlflow.log_param(k, v)\n model = xgb.XGBClassifier(**params)\n model.fit(X_train[cols], y_train)\n y_pred = model.predict(X_test[cols])\n loss = accuracy_score(y_test, y_pred)\n # Log the various losses and metrics (on train and validation)\n mlflow.log_metric(\"accuracy\", loss)\n # Use the last validation loss from the history object to optimize\n return {\n 'loss': -loss, \n 'status': STATUS_OK,\n 'eval_time': time()\n }\n\n def optimize(self):\n \"\"\"\n This is the optimization function that given a space of \n hyperparameters and a scoring function, finds the best hyperparameters.\n \"\"\"\n # Use the fmin function from Hyperopt to find the best hyperparameters\n # Here we use the tree-parzen estimator method. \n best = fmin(self.get_loss, self.hyperparameters_space, algo=tpe.suggest, \n trials=self.trials, max_evals=self.max_evals)\n return best\n\n\n\nMAX_EVALS = 200\n\nHYPERPARAMETERS_SPACE = {col: hp.choice(col, [0, 1]) for col in X_train.columns.values}\n\nhp_optimizer = HyperoptHPOptimizer(hyperparameters_space=HYPERPARAMETERS_SPACE, max_evals=MAX_EVALS)\noptimal_hyperparameters = hp_optimizer.optimize()\nprint(optimal_hyperparameters)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6f72ed85912a590f5298fa1b36be2437791c7a | 338,614 | ipynb | Jupyter Notebook | lsv_example.ipynb | kirkpsmith/voltammetry.py | 7ebeb132649dad5e5556720189ddd4bbe4628fe6 | [
"MIT"
]
| 3 | 2019-05-29T22:42:47.000Z | 2019-05-31T00:58:25.000Z | lsv_example.ipynb | kirkpsmith/voltammetry.py | 7ebeb132649dad5e5556720189ddd4bbe4628fe6 | [
"MIT"
]
| null | null | null | lsv_example.ipynb | kirkpsmith/voltammetry.py | 7ebeb132649dad5e5556720189ddd4bbe4628fe6 | [
"MIT"
]
| 1 | 2019-05-27T21:34:53.000Z | 2019-05-27T21:34:53.000Z | 27.106468 | 38,666 | 0.437356 | [
[
[
"# LSV Data Analysis and Parameter Estimation ",
"_____no_output_____"
],
[
"##### First, all relevent Python packages are imported",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom scipy.optimize import curve_fit\nfrom scipy.signal import savgol_filter, find_peaks, find_peaks_cwt\nimport pandas as pd\nimport math\nimport glob\nimport altair as alt\n\nfrom voltammetry import preprocessing, plotting, fitting",
"_____no_output_____"
]
],
[
[
"##### The user will be able to import experimental data for an LSV scan \n##### (Currently, we assume that the LSV sweep starts at equilibrium)",
"_____no_output_____"
]
],
[
[
"##Import Experimental Reversible Data:\n \nrev_exp_data = pd.read_csv(\"data/10mVs_Reversible.csv\")\ncurrent_exp=rev_exp_data['current(A)'].values\nvoltage_exp=rev_exp_data['voltage(mV)'].values\ntime_exp=rev_exp_data['time(s)'].values\n\n\n## all appropriate packages and the singular experimental data file is imported now\n",
"_____no_output_____"
]
],
[
[
"##### Next, the program will grab some simple quantitative information from the graph that may be hard to do by hand or over extensive datasets",
"_____no_output_____"
]
],
[
[
"t,i,v = preprocessing.readFile('data/10mM_F2CA_1M_KOH_pH_14_100mV.DTA',type='gamry',scan='first')\nlength = len(t)\nv1, v2 = v[0:int(length/2)], v[int(length/2):]\ni1, i2 = i[0:int(length/2)], i[int(length/2):]\nt1, t2 = t[0:int(length/2)], t[int(length/2):]\n\npeak_list = []\n\n_, v_peaks, i_peaks = fitting.peak_find(v1,i1,v2,i2)\nb1, b2 = fitting.baseline(v1,i1,v2,i2)\n\nfor n in range(len(v_peaks)):\n\n peak_list.append([i_peaks[n],v_peaks[n]])\n\n\n\nplotting.plot_voltammogram(t,i,v, peaks = peak_list).display()\n\nplt.plot(v1,b1)\nplt.plot(v1,i1)\nplt.plot(v2,b2)\nplt.plot(v2,i2)",
"_____no_output_____"
]
],
[
[
"##### This program can also return relevant parameters using a physics-based model.",
"_____no_output_____"
]
],
[
[
"# Import the dimensionless voltammagram (V I) for reversible reactions\n\nrev_dim_values = pd.read_csv(\"data/dimensionless_values_rev.csv\")\nrev_dim_current=rev_dim_values['dimensionless_current'].values\nrev_dim_voltage=rev_dim_values['dimensionless_Voltage'].values\n\n##We will now prompt the user to submit known parameters (THESE CAN BE CHANGED OR MADE MORE CONVENIENT)\nsweep_rate= float(input(\"What is the Voltage sweep rate in mV/s?(10)\"))\nelectrode_surface_area= float(input(\"What is the electrode surface area in cm^2?(.2)\"))\nconcentration_initial= float(input(\"What is the initial concentration in mol/cm^3?(.00001)\"))\nTemp= float(input(\"What is the temperature in K?(298)\"))\neq_pot= float(input(\"What is the equilibrium potential in V?(.10)\"))\n\n##we are inserting a diffusion coefficient to check math here, we will estimate this later:\nDiff_coeff=0.00001\n\n## Here we define constant variables, these can be made to user inputs if needed.\nn=1\nFaradays_const=96285\nR_const=8.314\nsigma=(n*Faradays_const*sweep_rate)/(R_const*Temp)\nPre=electrode_surface_area*concentration_initial*n*Faradays_const*math.sqrt(Diff_coeff*sigma)\n\n\n\noutput_voltage=(eq_pot+rev_dim_voltage/n)\noutput_current=Pre*rev_dim_current\n\nplt.plot(output_voltage,output_current)\n\n",
"What is the Voltage sweep rate in mV/s?(10) 10\nWhat is the electrode surface area in cm^2?(.2) .2\nWhat is the initial concentration in mol/cm^3?(.00001) .00001\nWhat is the temperature in K?(298) 298\nWhat is the equilibrium potential in V?(.10) .10\n"
]
],
[
[
"##### Then, we can back out a relevant parameter from the data:",
"_____no_output_____"
]
],
[
[
"# Fitting Diff_Coeff\n\ndef test_func(rev_dim_current, D):\n return electrode_surface_area*concentration_initial*n*Faradays_const*math.sqrt(D*sigma)*rev_dim_current\n\nparams, params_covariance = curve_fit(test_func, rev_dim_current, output_current,p0=None,bounds = (0,[1]))\n\nprint(\"Diffusion Coefficient (cm^2/s): {}\".format(params[0]))",
"Diffusion Coefficient (cm^2/s): 1.0000011458318317e-05\n"
]
],
[
[
"##### We can repeat this exercise on an LSV with an irreversible reaction to determine exchange current density.",
"_____no_output_____"
]
],
[
[
"##Import Experimental Irreversible Data:\n \nirrev_exp_data = pd.read_csv(\"data/10mVs_Irreversible.csv\")\ncurrent_exp=irrev_exp_data['current(A)'].values\nvoltage_exp=irrev_exp_data['voltage(mV)'].values\ntime_exp=irrev_exp_data['time(s)'].values\n\n## all appropriate packages and the singular experimental data file is imported now",
"_____no_output_____"
],
[
"# Import the dimensionless voltammagram (V I) for irreversible reactions\n\nirrev_dim_values = pd.read_csv(\"data/dimensionless_values_irrev.csv\")\nirrev_dim_current=irrev_dim_values['dimensionless_current'].values\nirrev_dim_voltage=irrev_dim_values['dimensionless_Voltage'].values",
"_____no_output_____"
],
[
"##We will now prompt the user to submit known parameters (THESE CAN BE CHANGED OR MADE MORE CONVENIENT)\nsweep_rate= float(input(\"What is the Voltage sweep rate in mV/s?(10)\"))\nelectrode_surface_area= float(input(\"What is the electrode surface area in cm^2?(.2)\"))\nconcentration_initial= float(input(\"What is the initial concentration in mol/cm^3?(.00001)\"))\nTemp= float(input(\"What is the temperature in K?(298)\"))\neq_pot= float(input(\"What is the equilibrium potential in mV?(100)\"))\n\n##we are inserting a diffusion coefficient to check math here, we will estimate this later:\nDiff_coeff=0.00001\n\n## Here we define constant variables, these can be made to user inputs if needed.\nn=1\nFaradays_const=96285\nR_const=8.314\nexchange_current_density=0.0002\nkinetic_coefficient=exchange_current_density/n/Faradays_const/electrode_surface_area/concentration_initial\ntransfer_coefficient=.6\neV_const=59.1\n\n\nbeta=transfer_coefficient*n*Faradays_const*sweep_rate/R_const/Temp/1000\nPre=(concentration_initial*n*Faradays_const*\n math.sqrt(Diff_coeff*sweep_rate*transfer_coefficient\n *Faradays_const/(R_const*Temp*1000)))\n\n\n\noutput_voltage=eq_pot+irrev_dim_voltage/transfer_coefficient-eV_const/transfer_coefficient*math.log(math.sqrt(math.pi*Diff_coeff*beta)/kinetic_coefficient)\noutput_current=Pre*irrev_dim_current\n\nplt.plot(output_voltage,output_current)\n\n\n",
"What is the Voltage sweep rate in mV/s?(10) 10\nWhat is the electrode surface area in cm^2?(.2) .2\nWhat is the initial concentration in mol/cm^3?(.00001) .00001\nWhat is the temperature in K?(298) 298\nWhat is the equilibrium potential in mV?(100) 100\n"
],
[
"# Fitting Diff_Coeff\nfrom scipy import optimize\n\n\ndef test_func(irrev_dim_voltage, exchange_current_density):\n return eq_pot+irrev_dim_voltage/transfer_coefficient-eV_const/transfer_coefficient*math.log(math.sqrt(math.pi*Diff_coeff*beta)/(exchange_current_density/n/Faradays_const/electrode_surface_area/concentration_initial))\n\nparams, params_covariance = optimize.curve_fit(test_func, irrev_dim_voltage, output_voltage,p0=None,bounds = (0,[1]))\n\nprint(\"Exchange current density (A/cm^2): {}\".format(params[0]))",
"Exchange current density (A/cm^2): 0.00020000000000000006\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
cb6f830b791efd8eae933ae45949d6f51ec91547 | 4,522 | ipynb | Jupyter Notebook | content/lessons/10/Now-You-Code/NYC1-Address.ipynb | MahopacHS/spring2019-Christian64Aguilar | 2e9ac4a5245d459f6d086c61ad3c0540db39b981 | [
"MIT"
]
| null | null | null | content/lessons/10/Now-You-Code/NYC1-Address.ipynb | MahopacHS/spring2019-Christian64Aguilar | 2e9ac4a5245d459f6d086c61ad3c0540db39b981 | [
"MIT"
]
| null | null | null | content/lessons/10/Now-You-Code/NYC1-Address.ipynb | MahopacHS/spring2019-Christian64Aguilar | 2e9ac4a5245d459f6d086c61ad3c0540db39b981 | [
"MIT"
]
| null | null | null | 47.6 | 667 | 0.613445 | [
[
[
"# Now You Code 1: Address\n\nWrite a Python program to input elements of your postal address and then output them as if they were an address label. The program should use a dictionary to store the address and complete two function defintions one for inputting the address and one for printing the address.\n\n**NOTE:** While you most certainly can write this program without using dictionaries or functions, the point of the exercise is to get used to using them!!!\n\nSample Run:\n\n```\nEnter Street: 314 Hinds Hall\nEnter City: Syracuse\nEnter State: NY\nEnter Postal Zip Code: 13244\nMailing Address:\n314 Hinds Hall\nSyracuse , NY 13244\n\n```",
"_____no_output_____"
],
[
"## Step 1: Problem Analysis `input_address` function\n\nThis function should get input from the user at run time and return the input address.\n\nInputs: None (gets input from user)\n\nOutputs: a Python dictionary of address info (street, city, state, postal_code)\n\nAlgorithm (Steps in Program):\n\n",
"_____no_output_____"
]
],
[
[
"## Step 2: Write input_address_ function\n#input: None (inputs from console)\n#output: dictionary of the address\ndef input_address():\n address= {}\n # todo: write code here to input the street, city, state and zip code and add to dictionary at runtime and store in a dictionary\n\n return address\n\n",
"_____no_output_____"
]
],
[
[
"## Step 3: Problem Analysis `print_address` function\n\nThis function should display a mailing address using the dictionary variable\n\nInputs: dictionary variable of address into (street, city, state, postal_code)\n\nOutputs: None (prints to screen)\n\nAlgorithm (Steps in Program):\n\n",
"_____no_output_____"
]
],
[
[
"## Step 4: write code\n# input: address dictionary\n# output: none (outputs to console)\ndef print_address(address):\n # todo: write code to print the address (leave empty return at the end\n \n return ",
"_____no_output_____"
]
],
[
[
"## Step 5: Problem Analysis main program\n\nShould be trivial at this point. \n\nInputs: \n\nOutputs: \n\nAlgorithm (Steps in Program):\n",
"_____no_output_____"
]
],
[
[
"## Step 6: write main program, use other 2 functions you made to solve this problem.\n \n# main program\n# todo: call input_address, then print_address\n\n\n",
"_____no_output_____"
]
],
[
[
"## Step 7: Questions\n\n1. Explain a strategy for a situation when an expected dictionary key, like 'state' for example does not exist?\n2. The program as it is written is not very useful. How can we make it more useful?\n",
"_____no_output_____"
],
[
"## Reminder of Evaluation Criteria\n\n1. What the problem attempted (analysis, code, and answered questions) ?\n2. What the problem analysis thought out? (does the program match the plan?)\n3. Does the code execute without syntax error?\n4. Does the code solve the intended problem?\n5. Is the code well written? (easy to understand, modular, and self-documenting, handles errors)\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
cb6f9fb7a460bfc51225c0c5b3ef0ffef4f4998f | 9,981 | ipynb | Jupyter Notebook | tutorial-ja/321_weakstrong_ja.ipynb | KeiichiroHiga/Blueqat-tutorials | 7d697453dc29e1f9f61e9a8be9c5c8ff900e751f | [
"Apache-2.0"
]
| null | null | null | tutorial-ja/321_weakstrong_ja.ipynb | KeiichiroHiga/Blueqat-tutorials | 7d697453dc29e1f9f61e9a8be9c5c8ff900e751f | [
"Apache-2.0"
]
| null | null | null | tutorial-ja/321_weakstrong_ja.ipynb | KeiichiroHiga/Blueqat-tutorials | 7d697453dc29e1f9f61e9a8be9c5c8ff900e751f | [
"Apache-2.0"
]
| null | null | null | 34.298969 | 377 | 0.551247 | [
[
[
"#Weak-Strong Cluster問題\n\n2015年にGoogleとNASAが共同でD-Waveマシンは既存マシンの1億倍高速という発表を行いました。その際に利用されたのが量子ビットのクラスタを作ってフリップさせるWeak-Strong Cluster問題です。今回は簡単なweak clusterとstrong clusterを作って見て計算を行います。\n\n論文は下記を参照します。\n\nWhat is the Computational Value of Finite Range Tunneling?\n\nhttps://arxiv.org/abs/1512.02206",
"_____no_output_____"
],
[
"##背景\n量子アニーリングは量子トンネル効果を利用した最適化マシンとして提案されていますが、ここでは、このトンネル効果がどのような計算上のメリットをもたらすかを検証しています。D-Wave2Xの量子アニーリングマシンは局所解同士を隔てるエネルギー障壁が高く、細い形状をしているような問題に対して有利で、Simulated Annealing(SA)にくらべても優位性があるといわれています。945量子ビットの場合で、SAにくらべて、およそ10^8倍高速(成功率99%)で、古典計算機でトンネル効果をシミュレートする量子モンテカルロ法(QMC)と比較しても同様に高速です。",
"_____no_output_____"
],
[
"##ハミルトニアンとSA、QA\n今回検証を行う際にシミュレーテッドアニーリング(以下SA)と量子アニーリング(今回は量子モンテカルロ法をつかっているので、以下QMC)が使用されています。\n\nときたい問題は一緒で、ハミルトニアンと呼ばれるコスト関数を最小にするようにアルゴリズムが働き、その最小基底状態に至る過程がSAとQMCでは原理が異なります。\n\nSAでは熱をシミュレートして、熱で基底状態の探索を行います。一方QMCでは熱の代わりに磁力を使って、量子トンネル効果を活用しながら探索を行います。\n\nSAではあるコスト関数がある場合、グラフの起伏をきちんとなぞるようにエネルギー障壁(以下エナジーバリア)を超えて探索を行うためエネルギー関数のコストをあげて探索を行わないといけませんが、QMCの場合にはトンネル効果によりエナジーバリアを越えるために熱のコストを上げる必要がなく、確率的にトンネル効果で起伏の向こう側に到達できると考えられます。\n\nこれらのエナジーバリアをトンネル効果で越える条件もありますので、できるだけエナジーバリアの高さが高くて、障壁の厚みが薄い方が確率的に超えやすいので、SAで行う場合には、かなり条件が厳しく、QMCや量子アニーリングに有利な条件となります。この条件を人為的に問題を作ることで、SAに対して速度優位性を持たせようという検証です。\n\nつまり、求めたいコスト関数に高くて薄いエナジーバリアがたくさんあるほどD-WaveマシンやQMCアルゴリズムが有利になると推測されます。",
"_____no_output_____"
],
[
"##Weak-Strong Cluster問題とは\nWeak-Strong Clusterという2つの量子ビットのクラスターを繋げる問題です。D-Waveはキメラグラフという接続を使用しており、8量子ビットで1ユニットセルという単位です。このユニットセルを2つ用意した、16量子ビットの2つのクラスターを構成する問題を用意しています。\n\n<img src='https://github.com/Blueqat/Wildqat/blob/master/examples_ja/img/023_1.png?raw=1'>\n\n\n全ての量子ビットはキメラグラフで接続されており、ferromagneticカップリングで結合されています。値が同じになるような結合です。一方局所磁場と呼ばれる量子ビットが-1か+1になりやすいように設定されたパラメータが工夫されています。右側のクラスターはすべてh2=−1という値が設定されている一方で、左側のクラスターの量子ビットにはh1=0.44というように設定されています。これにより、計算過程において、左側の量子ビットがまとめて8個同時にフリップして右側のクラスターと揃うという過程がおきます。局所磁場の値が、左がweak-clusterで右がstrong-clusterでweak-strong cluster問題です。\n\nこれをさらに巨大につなげることで大きなクラスタを作ることができています。クラスター16量子ビットの組みを複数用意し、strong-cluster同士を4量子ビットの結合でferro/anti-ferroをランダムで+1or-1でつなぐことで巨大なweak-strong clusterを作ったとのこと。\n\n<img src='https://github.com/Blueqat/Wildqat/blob/master/examples_ja/img/023_2.png?raw=1'>\n引用:https://arxiv.org/abs/1512.02206\n\nD-Waveにはところどころ不良量子ビットもあるので、それを避けるようにクラスターを配置し、上記の巨大クラスター構築では、黒丸が-1のstrong cluster。グレーが0.44のweak cluster。青い接続がferroで赤い接続がanti-ferroとなっています。",
"_____no_output_____"
],
[
"##実験の結果\n結果1億倍程度の速度差が生まれたとなっています。参考は下図。\n\n<img src='https://github.com/Blueqat/Wildqat/blob/master/examples_ja/img/023_4.png?raw=1'>\n引用:https://arxiv.org/abs/1512.02206\n",
"_____no_output_____"
],
[
"##部分的回路の実装\n少し実際のアルゴリズムでやって見ます。実用的に2クラスタを解いて見ます。とりあえず16量子ビットのクラスタを今回は検証して見たいと思います。\n\nまず面白いのは、量子ビット同士の結合がすべてferromagneticということです。設定する値は論文と符号が逆ですが、すべて-1を入れます。\n\n\n上記量子ビットで今回の実験の肝は量子ビットの局所磁場を設定するところで、上記オレンジの右側のクラスターの量子ビットの局所磁場の設定をすべて+1に。上記水色の左側のクラスターの量子ビットの局所磁場の設定を全て-0.44にします。また、便宜的に16量子ビットに通し番号をふりました。局所磁場はh0からh15まで、量子ビット間の相互作用強度はJijの表記をJ0,4のように量子ビットの番号で表現します。\n\n<img src='https://github.com/Blueqat/Wildqat/blob/master/examples_ja/img/023_1.png?raw=1'>\n\nまずは何も考えずにSAをかけて見たいと思います。",
"_____no_output_____"
],
[
"##キメラグラフの実装\nキメラグラフでの結合係数の決定をします。今回はせいぜい16量子ビットなので、そのまま16*16のmatrixを作って実現して見ます。wildqatに下記のQUBOmatrixをいれることで計算を行うことができます。\n\n<img src='https://github.com/Blueqat/Wildqat/blob/master/examples_ja/img/023_5.png?raw=1'>\n\nオレンジの-1がユニットセル内の16の結合。クラスタが2つあるので、合計32のユニットセル内部の結合があります。次に赤の-0.44はクラスタ1の局所磁場。青の+1はクラスタ2の局所磁場。紫はクラスタ間の-1の結合を表しています。論文とwildqatはプラスマイナスの符号が逆になっています。",
"_____no_output_____"
],
[
"##実行して見る\nこちらをwildqatに入力して実行してみます。",
"_____no_output_____"
]
],
[
[
"!pip install blueqat",
"Collecting blueqat\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/bb/86/1b72a7cbe500b861d63e84cc6383fbf3730f08ae69fcd85146ae8e3b8873/blueqat-0.3.10-py3-none-any.whl (46kB)\n\r\u001b[K |███████ | 10kB 19.5MB/s eta 0:00:01\r\u001b[K |██████████████ | 20kB 1.8MB/s eta 0:00:01\r\u001b[K |█████████████████████▏ | 30kB 2.6MB/s eta 0:00:01\r\u001b[K |████████████████████████████▏ | 40kB 1.7MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 51kB 1.7MB/s \n\u001b[?25hRequirement already satisfied: scipy>=1.1.0 in /usr/local/lib/python3.6/dist-packages (from blueqat) (1.3.3)\nRequirement already satisfied: numpy~=1.12 in /usr/local/lib/python3.6/dist-packages (from blueqat) (1.17.4)\nInstalling collected packages: blueqat\nSuccessfully installed blueqat-0.3.10\n"
],
[
"import blueqat.opt as wq\nimport numpy as np\na = wq.opt()\na.J = np.zeros((16,16))\n\nfor i in range(8):\n a.J[i][i] = -0.44\n\nfor i in range(8,16):\n a.J[i][i] = 1\n\nfor i in range(4,8):\n for j in range(0,4):\n a.J[j][i] = -1\n \nfor i in range(12,16):\n for j in range(8,12):\n a.J[j][i] = -1\n\na.J[4][12] = -1\na.J[5][13] = -1\na.J[6][14] = -1\na.J[7][15] = -1\n\na.sa()\n",
"_____no_output_____"
]
],
[
[
"すべて0になりました。時々実行すると左側だけすべて1となり、右側が0となる局所解にも落ちました。",
"_____no_output_____"
]
],
[
[
"a.sa()",
"_____no_output_____"
]
],
[
[
"##参考にD-Wave実機での実行結果\nまた、D-Wave本体でもやってみました。パラメータは論文と一緒です。\n\n<img src='https://github.com/Blueqat/Wildqat/blob/master/examples_ja/img/023_6.png?raw=1'>\n\n成功率98.6%で基底状態です。論文とほぼ同じ。shotは1000回にして見ました。\n\nこの問題はぜひ興味ある人は実装が難しくないので小さな問題からチャレンジして、大きな問題にチャレンジして欲しいです。実用というよりも研究の要素がとても大きかったと思います。正直SAでは局所解から最適解への相転移は容易ではないと思います。その辺りがD-WaveやQMCなどの量子アルゴリズムの利点なのかなと思いました。すべては左側のクラスターの量子ビットの局所磁場h0=−0.44という数字がポイントになるので、この値を調整して見ても勉強になるかと思います。",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
cb6f9ffe3f6fd47ec53641679d9f49dc4d4ab760 | 33,974 | ipynb | Jupyter Notebook | Codes/Bias_Mitigation_XGBM.ipynb | hshehjue/IntegrityM-Capstone_Project | 0859e2c3d95a6cdd81d743b4137c2bc24f7d4e18 | [
"MIT"
]
| null | null | null | Codes/Bias_Mitigation_XGBM.ipynb | hshehjue/IntegrityM-Capstone_Project | 0859e2c3d95a6cdd81d743b4137c2bc24f7d4e18 | [
"MIT"
]
| null | null | null | Codes/Bias_Mitigation_XGBM.ipynb | hshehjue/IntegrityM-Capstone_Project | 0859e2c3d95a6cdd81d743b4137c2bc24f7d4e18 | [
"MIT"
]
| null | null | null | 34.526423 | 315 | 0.462295 | [
[
[
"# Disparate Impact by Providers' Gender \n## the best model: XGBoost",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport time\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport glob\nimport copy\nfrom collections import Counter\nfrom numpy import where\nimport statsmodels.api as sm\nfrom sklearn.preprocessing import scale\nfrom sklearn.model_selection import train_test_split\nimport random\nimport itertools\nfrom interpret.glassbox import ExplainableBoostingClassifier \nimport xgboost as xgb\nfrom interpret.perf import ROC \nfrom imblearn import over_sampling\nfrom imblearn import under_sampling\nfrom imblearn.pipeline import Pipeline\nimport os # for directory and file manipulation\nimport numpy as np # for basic array manipulation\nimport pandas as pd # for dataframe manipulation\nimport datetime # for timestamp\n\n# for model eval\nfrom sklearn.metrics import accuracy_score, f1_score, log_loss, mean_squared_error, roc_auc_score\n\n# global constants \nROUND = 3 \n\n# set global random seed for better reproducibility\nSEED = 1234\nseed = 1234\nNTHREAD = 4\n\n#import sagemaker, boto3, os\n\nimport warnings\nwarnings.filterwarnings('ignore')\n",
"_____no_output_____"
],
[
"# import the cleaned dataset containing Gender feature\n\n#%cd /Users/alex/Desktop/Master/BA_Practicum_6217_10/Project/dataset\npartB = pd.read_csv(\"partB_new5.csv\")",
"_____no_output_____"
],
[
"partB.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 348887 entries, 0 to 348886\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 NPI 348887 non-null int64 \n 1 Gender 348887 non-null object \n 2 Type 348887 non-null object \n 3 Place_Of_Srvc 348887 non-null object \n 4 Tot_Benes 348887 non-null int64 \n 5 Tot_Srvcs 348887 non-null float64\n 6 Tot_Bene_Day_Srvcs 348887 non-null int64 \n 7 Avg_Sbmtd_Chrg 348887 non-null float64\n 8 Avg_Mdcr_Alowd_Amt 348887 non-null float64\n 9 Avg_Mdcr_Pymt_Amt 348887 non-null float64\n 10 Avg_Mdcr_Stdzd_Amt 348887 non-null float64\n 11 Fraud 348887 non-null int64 \ndtypes: float64(5), int64(4), object(3)\nmemory usage: 31.9+ MB\n"
],
[
"# One-Hot Encoding \n\n# Convert the Fraud variable to object datatype\npartB[\"Fraud\"] = partB[\"Fraud\"].astype(object)\n\n# Encoding\nencoded_partB = pd.get_dummies(partB, drop_first = True)\n\n# Rename some of the changed variable names\nencoded_partB.rename(columns = {\"Gender_M\":\"Gender\", \"Fraud_1\":\"Fraud\", \"Place_Of_Srvc_O\":\"Place_Of_Srvc\"}, inplace = True)",
"_____no_output_____"
]
],
[
[
"## Data Partitioning",
"_____no_output_____"
]
],
[
[
"# Assign X and y features\n\nX_var = list(encoded_partB.columns)\n\nfor var in [\"NPI\",\"Fraud\"]:\n X_var.remove(var)\n\ny_var = \"Fraud\"",
"_____no_output_____"
],
[
"# Split the whole dataset into train and test dataset\n# Using a stratified random sampling so that the Fraud-class (1) data are evenly split into train & test sets\nx_train, x_test, y_train, y_test = train_test_split(encoded_partB[X_var], \n encoded_partB[y_var], \n test_size=0.2, \n stratify=encoded_partB[\"Fraud\"])\n\n# Also concatenate the split x & y dataframes \ntr_df = pd.concat([x_train, y_train], axis = 1)\nte_df = pd.concat([x_test, y_test], axis = 1)",
"_____no_output_____"
]
],
[
[
"## Over-Sampling",
"_____no_output_____"
]
],
[
[
"# SMOTE the dataset\noversample = over_sampling.SMOTE()\ntr_X, tr_y = oversample.fit_resample(tr_df[X_var], tr_df[y_var])",
"_____no_output_____"
]
],
[
[
"## Modeling",
"_____no_output_____"
],
[
"### Data Partitioning (Train & Valid)",
"_____no_output_____"
]
],
[
[
"trans_tr_df = pd.concat([tr_X, tr_y], axis = 1)\n\n# Split train and validation sets \nnp.random.seed(SEED)\n\nratio = 0.7 # split train & validation sets with 7:3 ratio \n\nsplit = np.random.rand(len(trans_tr_df)) < ratio # define indices of 70% corresponding to the training set\n\ntrain = trans_tr_df[split]\nvalid = trans_tr_df[~split]\n\n# summarize split\nprint('Train data rows = %d, columns = %d' % (train.shape[0], train.shape[1]))\nprint('Validation data rows = %d, columns = %d' % (valid.shape[0], valid.shape[1]))",
"Train data rows = 355544, columns = 117\nValidation data rows = 151926, columns = 117\n"
],
[
"# reassign X_var\nX_var.remove(\"Gender\")",
"_____no_output_____"
]
],
[
[
"### XGBM",
"_____no_output_____"
]
],
[
[
"def xgb_grid(dtrain, dvalid, mono_constraints=None, gs_params=None, n_models=None,\n ntree=None, early_stopping_rounds=None, verbose=False, seed=None):\n \n \"\"\" Performs a random grid search over n_models and gs_params.\n\n :param dtrain: Training data in LightSVM format.\n :param dvalid: Validation data in LightSVM format.\n :param mono_constraints: User-supplied monotonicity constraints.\n :param gs_params: Dictionary of lists of potential XGBoost parameters over which to search.\n :param n_models: Number of random models to evaluate.\n :param ntree: Number of trees in XGBoost model.\n :param early_stopping_rounds: XGBoost early stopping rounds.\n :param verbose: Whether to display training iterations, default False.\n :param seed: Random seed for better interpretability.\n :return: Best candidate model from random grid search.\n\n \"\"\"\n\n # cartesian product of gs_params\n keys, values = zip(*gs_params.items())\n experiments = [dict(zip(keys, v)) for v in itertools.product(*values)]\n\n # preserve exact reproducibility for this function\n np.random.seed(SEED) \n \n # select randomly from cartesian product space\n selected_experiments = np.random.choice(len(experiments), n_models)\n\n # set global params for objective, etc.\n params = {'booster': 'gbtree',\n 'eval_metric': 'auc',\n 'nthread': NTHREAD,\n 'objective': 'binary:logistic',\n 'seed': SEED}\n\n # init grid search loop\n best_candidate = None\n best_score = 0\n\n # grid search loop\n for i, exp in enumerate(selected_experiments):\n\n params.update(experiments[exp]) # override global params with current grid run params\n\n print('Grid search run %d/%d:' % (int(i + 1), int(n_models)))\n print('Training with parameters:', params)\n\n # train on current params\n watchlist = [(dtrain, 'train'), (dvalid, 'eval')]\n \n if mono_constraints is not None:\n params['monotone_constraints'] = mono_constraints\n \n candidate = xgb.train(params,\n dtrain,\n ntree,\n early_stopping_rounds=early_stopping_rounds,\n evals=watchlist,\n verbose_eval=verbose) \n\n # determine if current model is better than previous best\n if candidate.best_score > best_score:\n best_candidate = candidate\n best_score = candidate.best_score\n print('Grid search new best score discovered at iteration %d/%d: %.4f.' %\n (int(i + 1), int(n_models), candidate.best_score))\n\n print('---------- ----------')\n \n return best_candidate",
"_____no_output_____"
],
[
"gs_params = {'colsample_bytree': [0.7],\n 'colsample_bylevel': [0.9],\n 'eta': [0.5],\n 'max_depth': [7], \n 'reg_alpha': [0.005],\n 'reg_lambda': [0.005],\n 'subsample': [0.9],\n 'min_child_weight': [1], \n 'gamma': [0.2]}\n\n# Convert data to SVMLight format\ndtrain = xgb.DMatrix(train[X_var], train[y_var])\ndvalid = xgb.DMatrix(valid[X_var], valid[y_var])\n\nbest_mxgb = xgb_grid(dtrain, dvalid, gs_params=gs_params, n_models=1, ntree=1000, early_stopping_rounds=100, seed=SEED)",
"Grid search run 1/1:\nTraining with parameters: {'booster': 'gbtree', 'eval_metric': 'auc', 'nthread': 4, 'objective': 'binary:logistic', 'seed': 1234, 'colsample_bytree': 0.7, 'colsample_bylevel': 0.9, 'eta': 0.5, 'max_depth': 7, 'reg_alpha': 0.005, 'reg_lambda': 0.005, 'subsample': 0.9, 'min_child_weight': 1, 'gamma': 0.2}\nGrid search new best score discovered at iteration 1/1: 0.9840.\n---------- ----------\n"
]
],
[
[
"### Combine valid set with the best prediction\n",
"_____no_output_____"
]
],
[
[
"dtest = xgb.DMatrix(te_df[X_var])\nbest_mxgb_phat = pd.DataFrame(best_mxgb.predict(dtest, iteration_range=(0, best_mxgb.best_ntree_limit)), columns=['phat'])\nbest_mxgb_phat = pd.concat([te_df.reset_index(drop=True), best_mxgb_phat], axis=1)\nbest_mxgb_phat.head()",
"_____no_output_____"
]
],
[
[
"## Mitigating Discrimination ",
"_____no_output_____"
],
[
"### Utility functions \n### Calculate confusion matrices by demographic group",
"_____no_output_____"
]
],
[
[
"def get_confusion_matrix(frame, y, yhat, by=None, level=None, cutoff=0.2, verbose=True):\n\n \"\"\" Creates confusion matrix from pandas dataframe of y and yhat values, can be sliced \n by a variable and level.\n \n :param frame: Pandas dataframe of actual (y) and predicted (yhat) values.\n :param y: Name of actual value column.\n :param yhat: Name of predicted value column.\n :param by: By variable to slice frame before creating confusion matrix, default None.\n :param level: Value of by variable to slice frame before creating confusion matrix, default None.\n :param cutoff: Cutoff threshold for confusion matrix, default 0.5. \n :param verbose: Whether to print confusion matrix titles, default True. \n :return: Confusion matrix as pandas dataframe. \n \n \"\"\"\n \n # determine levels of target (y) variable\n # sort for consistency\n level_list = list(frame[y].unique())\n level_list.sort(reverse=True) \n\n # init confusion matrix\n cm_frame = pd.DataFrame(columns=['actual: ' + str(i) for i in level_list], \n index=['predicted: ' + str(i) for i in level_list])\n \n # don't destroy original data\n frame_ = frame.copy(deep=True)\n \n # convert numeric predictions to binary decisions using cutoff\n dname = 'd_' + str(y)\n frame_[dname] = np.where(frame_[yhat] > cutoff , 1, 0)\n \n # slice frame\n if (by is not None) & (level is not None):\n frame_ = frame_[frame[by] == level]\n \n # calculate size of each confusion matrix value\n for i, lev_i in enumerate(level_list):\n for j, lev_j in enumerate(level_list):\n cm_frame.iat[j, i] = frame_[(frame_[y] == lev_i) & (frame_[dname] == lev_j)].shape[0]\n # i, j vs. j, i nasty little bug ... updated 8/30/19\n \n # output results\n if verbose:\n if by is None:\n print('Confusion matrix:')\n else:\n print('Confusion matrix by ' + by + '=' + str(level))\n \n return cm_frame",
"_____no_output_____"
]
],
[
[
"### Calculate Adverse Impact Ratio (AIR) ",
"_____no_output_____"
]
],
[
[
"def air(cm_dict, reference_key, protected_key, verbose=True):\n\n \"\"\" Calculates the adverse impact ratio as a quotient between protected and \n reference group acceptance rates: protected_prop/reference_prop. \n Optionally prints intermediate values. ASSUMES 0 IS \"POSITIVE\" OUTCOME!\n\n :param cm_dict: Dictionary of demographic group confusion matrices. \n :param reference_key: Name of reference group in cm_dict as a string.\n :param protected_key: Name of protected group in cm_dict as a string.\n :param verbose: Whether to print intermediate acceptance rates, default True. \n :return: AIR.\n \n \"\"\"\n\n eps = 1e-20 # numeric stability and divide by 0 protection\n \n # reference group summary\n reference_accepted = float(cm_dict[reference_key].iat[1,0] + cm_dict[reference_key].iat[1,1]) # predicted 0's\n reference_total = float(cm_dict[reference_key].sum().sum())\n reference_prop = reference_accepted/reference_total\n if verbose:\n print(reference_key.title() + ' proportion accepted: %.3f' % reference_prop)\n \n # protected group summary\n protected_accepted = float(cm_dict[protected_key].iat[1,0] + cm_dict[protected_key].iat[1,1]) # predicted 0's\n protected_total = float(cm_dict[protected_key].sum().sum())\n protected_prop = protected_accepted/protected_total\n if verbose:\n print(protected_key.title() + ' proportion accepted: %.3f' % protected_prop)\n\n # return adverse impact ratio\n return ((protected_prop + eps)/(reference_prop + eps))",
"_____no_output_____"
]
],
[
[
"### Select Probability Cutoff by F1-score",
"_____no_output_____"
]
],
[
[
"def get_max_f1_frame(frame, y, yhat, res=0.01, air_reference=None, air_protected=None): \n \n \"\"\" Utility function for finding max. F1. \n Coupled to get_confusion_matrix() and air(). \n Assumes 1 is the marker for class membership.\n \n :param frame: Pandas dataframe of actual (y) and predicted (yhat) values.\n :param y: Known y values.\n :param yhat: Model scores.\n :param res: Resolution over which to search for max. F1, default 0.01.\n :param air_reference: Reference group for AIR calculation, optional.\n :param air_protected: Protected group for AIR calculation, optional.\n :return: Pandas DataFrame of cutoffs to select from.\n \n \"\"\"\n \n do_air = all(v is not None for v in [air_reference, air_protected])\n \n # init frame to store f1 at different cutoffs\n if do_air:\n columns = ['cut', 'f1', 'acc', 'air']\n else:\n columns = ['cut', 'f1', 'acc']\n f1_frame = pd.DataFrame(columns=['cut', 'f1', 'acc'])\n \n # copy known y and score values into a temporary frame\n temp_df = frame[[y, yhat]].copy(deep=True)\n \n # find f1 at different cutoffs and store in acc_frame\n for cut in np.arange(0, 1 + res, res):\n temp_df['decision'] = np.where(temp_df.iloc[:, 1] > cut, 1, 0)\n f1 = f1_score(temp_df.iloc[:, 0], temp_df['decision'])\n acc = accuracy_score(temp_df.iloc[:, 0], temp_df['decision'])\n row_dict = {'cut': cut, 'f1': f1, 'acc': acc}\n if do_air:\n # conditionally calculate AIR \n cm_ref = get_confusion_matrix(frame, y, yhat, by=air_reference, level=1, cutoff=cut, verbose=False)\n cm_pro = get_confusion_matrix(frame, y, yhat, by=air_protected, level=1, cutoff=cut, verbose=False)\n air_ = air({air_reference: cm_ref, air_protected: cm_pro}, air_reference, air_protected, verbose=False)\n row_dict['air'] = air_\n \n f1_frame = f1_frame.append(row_dict, ignore_index=True)\n \n del temp_df\n \n return f1_frame",
"_____no_output_____"
]
],
[
[
"### Find optimal cutoff based on F1\n",
"_____no_output_____"
]
],
[
[
"f1_frame = get_max_f1_frame(best_mxgb_phat, y_var, 'phat')\n\nprint(f1_frame)\nprint()\n\nmax_f1 = f1_frame['f1'].max()\nbest_cut = f1_frame.loc[int(f1_frame['f1'].idxmax()), 'cut'] #idxmax() returns the index of the maximum value\nacc = f1_frame.loc[int(f1_frame['f1'].idxmax()), 'acc']\n\nprint('Best XGB F1: %.4f achieved at cutoff: %.2f with accuracy: %.4f.' % (max_f1, best_cut, acc))",
" cut f1 acc\n0 0.00 0.166656 0.090903\n1 0.01 0.283816 0.587520\n2 0.02 0.313475 0.661283\n3 0.03 0.332788 0.702041\n4 0.04 0.346562 0.728869\n.. ... ... ...\n96 0.96 0.113098 0.912566\n97 0.97 0.101147 0.912379\n98 0.98 0.084402 0.912007\n99 0.99 0.059897 0.911376\n100 1.00 0.000000 0.909097\n\n[101 rows x 3 columns]\n\nBest XGB F1: 0.4062 achieved at cutoff: 0.21 with accuracy: 0.8587.\n"
]
],
[
[
"### Specify Interesting Demographic Groups",
"_____no_output_____"
]
],
[
[
"best_mxgb_phat_copy = best_mxgb_phat.copy()\nbest_mxgb_phat_copy.rename(columns = {\"Gender\":\"male\"}, inplace = True)\nbest_mxgb_phat_copy[\"female\"] = np.where(best_mxgb_phat_copy[\"male\"] == 0, 1,0)",
"_____no_output_____"
]
],
[
[
"### Confusion Matrix by Groups",
"_____no_output_____"
]
],
[
[
"demographic_group_names = ['male', 'female']\ncm_dict = {}\n\nfor name in demographic_group_names:\n cm_dict[name] = get_confusion_matrix(best_mxgb_phat_copy, y_var, 'phat', by=name, level=1, cutoff=best_cut)\n print(cm_dict[name])\n print()",
"Confusion matrix by male=1\n actual: 1 actual: 0\npredicted: 1 3185 5133\npredicted: 0 2726 39566\n\nConfusion matrix by female=1\n actual: 1 actual: 0\npredicted: 1 187 1755\npredicted: 0 245 16981\n\n"
]
],
[
[
"### Find AIR for Female people\n* protect accepted: female providers\n* reference accepted: male providers",
"_____no_output_____"
]
],
[
[
"print('Adverse impact ratio(AIR) for Females vs. Males: %.3f' % air(cm_dict, 'male', 'female'))",
"Male proportion accepted: 0.836\nFemale proportion accepted: 0.899\nAdverse impact ratio(AIR) for Females vs. Males: 1.075\n"
]
],
[
[
"* Threshold: AIR >= 0.8 ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
cb6fa2f8193e08fd8a08e2d6f79b013231e6d15f | 1,622 | ipynb | Jupyter Notebook | I.Gentle_Overview_of_Big_Data_and_Spark/2.A_Gentle_Intro_to_Spark/example.ipynb | NikolayVaklinov10/Spark_The_Definitive_Guide | 36efd03133f169c227d715ce8921a0145962a228 | [
"Apache-2.0"
]
| null | null | null | I.Gentle_Overview_of_Big_Data_and_Spark/2.A_Gentle_Intro_to_Spark/example.ipynb | NikolayVaklinov10/Spark_The_Definitive_Guide | 36efd03133f169c227d715ce8921a0145962a228 | [
"Apache-2.0"
]
| null | null | null | I.Gentle_Overview_of_Big_Data_and_Spark/2.A_Gentle_Intro_to_Spark/example.ipynb | NikolayVaklinov10/Spark_The_Definitive_Guide | 36efd03133f169c227d715ce8921a0145962a228 | [
"Apache-2.0"
]
| null | null | null | 30.037037 | 506 | 0.557953 | [
[
[
"from pyspark ",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
cb6fa5f64db5097479bd5b20ef67386439af16a4 | 57,622 | ipynb | Jupyter Notebook | code/backtracking-algos.ipynb | vicb1/miscellaneous | 2c9762579abf75ef6cba75d1d1536a693d69e82a | [
"MIT"
]
| null | null | null | code/backtracking-algos.ipynb | vicb1/miscellaneous | 2c9762579abf75ef6cba75d1d1536a693d69e82a | [
"MIT"
]
| null | null | null | code/backtracking-algos.ipynb | vicb1/miscellaneous | 2c9762579abf75ef6cba75d1d1536a693d69e82a | [
"MIT"
]
| null | null | null | 110.811538 | 1,450 | 0.644216 | [
[
[
"# Python3 program to solve N Queen \n# Problem using backtracking \nglobal N \nN = 4\n \ndef printSolution(board): \n for i in range(N): \n for j in range(N): \n print (board[i][j], end = \" \") \n print() \n \n# A utility function to check if a queen can \n# be placed on board[row][col]. Note that this \n# function is called when \"col\" queens are \n# already placed in columns from 0 to col -1. \n# So we need to check only left side for \n# attacking queens \ndef isSafe(board, row, col): \n \n # Check this row on left side \n for i in range(col): \n if board[row][i] == 1: \n return False\n \n # Check upper diagonal on left side \n for i, j in zip(range(row, -1, -1), \n range(col, -1, -1)): \n if board[i][j] == 1: \n return False\n \n # Check lower diagonal on left side \n for i, j in zip(range(row, N, 1), \n range(col, -1, -1)): \n if board[i][j] == 1: \n return False\n \n return True\n \ndef solveNQUtil(board, col): \n \n # base case: If all queens are placed \n # then return true \n if col >= N: \n return True\n \n # Consider this column and try placing \n # this queen in all rows one by one \n for i in range(N): \n \n if isSafe(board, i, col): \n \n # Place this queen in board[i][col] \n board[i][col] = 1\n \n # recur to place rest of the queens \n if solveNQUtil(board, col + 1) == True: \n return True\n \n # If placing queen in board[i][col \n # doesn't lead to a solution, then \n # queen from board[i][col] \n board[i][col] = 0\n \n # if the queen can not be placed in any row in \n # this colum col then return false \n return False\n \n# This function solves the N Queen problem using \n# Backtracking. It mainly uses solveNQUtil() to \n# solve the problem. It returns false if queens \n# cannot be placed, otherwise return true and \n# placement of queens in the form of 1s. \n# note that there may be more than one \n# solutions, this function prints one of the \n# feasible solutions. \ndef solveNQ(): \n board = [ [0, 0, 0, 0], \n [0, 0, 0, 0], \n [0, 0, 0, 0], \n [0, 0, 0, 0] ] \n \n if solveNQUtil(board, 0) == False: \n print (\"Solution does not exist\") \n return False\n \n printSolution(board) \n return True\n \n# Driver Code \nsolveNQ() \n ",
"0 0 1 0 \n1 0 0 0 \n0 0 0 1 \n0 1 0 0 \n"
],
[
"def sudokutest(s,i,j,z):\n # z is the number\n isiValid = numpy.logical_or((i+1<1),(i+1>9));\n isjValid = numpy.logical_or((j+1<1),(j+1>9));\n iszValid = numpy.logical_or((z<1),(z>9));\n if s.shape!=(9,9):\n raise(Exception(\"Sudokumatrix not valid\"));\n if isiValid:\n raise(Exception(\"i not valid\"));\n if isjValid:\n raise(Exception(\"j not valid\"));\n if iszValid:\n raise(Exception(\"z not valid\"));\n\n if(s[i,j]!=0):\n return False;\n\n for ii in range(0,9):\n if(s[ii,j]==z):\n return False;\n\n for jj in range(0,9):\n if(s[i,jj]==z):\n return False;\n\n row = int(i/3) * 3;\n col = int(j/3) * 3;\n for ii in range(0,3):\n for jj in range(0,3):\n if(s[ii+row,jj+col]==z):\n return False;\n\n return True;\n\ndef possibleNums(s , i ,j):\n l = [];\n ind = 0;\n for k in range(1,10):\n if sudokutest(s,i,j,k):\n l.insert(ind,k);\n ind+=1;\n return l;\n\ndef sudokusolver(S):\n zeroFound = 0;\n for i in range(0,9):\n for j in range(0,9):\n if(S[i,j]==0):\n zeroFound=1;\n break;\n if(zeroFound==1):\n break;\n if(zeroFound==0):\n print(\"REALLY The end\")\n z = numpy.zeros(shape=(9,9))\n for x in range(0,9):\n for y in range(0,9):\n z[x,y] = S[x,y]\n print(z)\n return z\n\n\n x = possibleNums(S,i,j);\n\n for k in range(len(x)):\n S[i,j]=x[k];\n sudokusolver(S);\n S[i,j] = 0;\n\n\nif __name__ == \"__main__\":\n import numpy \n #s = numpy.zeros(shape=(9,9))\n\n k = numpy.matrix([0,0,0,0,0,9,0,7,8,5,1,0,0,0,0,0,6,9,9,0,8,0,2,5,0,0,0,0,3,2,0,0,0,0,0,0,0,0,9,3,0,0,0,1,0,0,0,0,4,0,0,0,8,0,8,0,0,0,9,0,7,0,0,6,0,1,0,0,0,0,0,0,0,0,0,0,7,0,8,0,1]).reshape(9,9)\n print(k)\n print('*'*80)\n %timeit sudokusolver(k)",
"[[0 0 0 0 0 9 0 7 8]\n [5 1 0 0 0 0 0 6 9]\n [9 0 8 0 2 5 0 0 0]\n [0 3 2 0 0 0 0 0 0]\n [0 0 9 3 0 0 0 1 0]\n [0 0 0 4 0 0 0 8 0]\n [8 0 0 0 9 0 7 0 0]\n [6 0 1 0 0 0 0 0 0]\n [0 0 0 0 7 0 8 0 1]]\n********************************************************************************\nREALLY The end\n[[3. 2. 4. 1. 6. 9. 5. 7. 8.]\n [5. 1. 7. 8. 3. 4. 2. 6. 9.]\n [9. 6. 8. 7. 2. 5. 1. 3. 4.]\n [1. 3. 2. 9. 8. 6. 4. 5. 7.]\n [4. 8. 9. 3. 5. 7. 6. 1. 2.]\n [7. 5. 6. 4. 1. 2. 9. 8. 3.]\n [8. 4. 3. 5. 9. 1. 7. 2. 6.]\n [6. 7. 1. 2. 4. 8. 3. 9. 5.]\n [2. 9. 5. 6. 7. 3. 8. 4. 1.]]\nREALLY The end\n[[3. 2. 4. 1. 6. 9. 5. 7. 8.]\n [5. 1. 7. 8. 3. 4. 2. 6. 9.]\n [9. 6. 8. 7. 2. 5. 1. 3. 4.]\n [1. 3. 2. 9. 8. 6. 4. 5. 7.]\n [4. 8. 9. 3. 5. 7. 6. 1. 2.]\n [7. 5. 6. 4. 1. 2. 9. 8. 3.]\n [8. 4. 3. 5. 9. 1. 7. 2. 6.]\n [6. 7. 1. 2. 4. 8. 3. 9. 5.]\n [2. 9. 5. 6. 7. 3. 8. 4. 1.]]\n"
],
[
"import numpy as np\nfrom functools import reduce\n\ndef solver_python(grid):\n numbers=np.arange(1,10)\n i,j = np.where(grid==0) \n if (i.size==0):\n return(True,grid)\n else:\n i,j=i[0],j[0] \n row = grid[i,:] \n col = grid[:,j]\n sqr = grid[(i//3)*3:(3+(i//3)*3),(j//3)*3:(3+(j//3)*3)].reshape(9)\n values = np.setdiff1d(numbers,reduce(np.union1d,(row,col,sqr)))\n\n grid_temp = np.copy(grid) \n\n for value in values:\n grid_temp[i,j] = value\n test = solver_python(grid_temp)\n if (test[0]):\n return(test)\n\n return(False,None)\n\nexample = np.array([[5,3,0,0,7,0,0,0,0],\n [6,0,0,1,9,5,0,0,0],\n [0,9,8,0,0,0,0,6,0],\n [8,0,0,0,6,0,0,0,3],\n [4,0,0,8,0,3,0,0,1],\n [7,0,0,0,2,0,0,0,6],\n [0,6,0,0,0,0,2,8,0],\n [0,0,0,4,1,9,0,0,5],\n [0,0,0,0,8,0,0,7,9]])\n\n%timeit solver_python(example)",
"491 ms ± 1.38 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"import sys\nimport numpy as np\nfrom functools import reduce\n\n# Instructions:\n# Linux>> python3 driver_3.py <soduku_str>\n# Windows py3\\> python driver_3.py <soduku_str>\n\n# Inputs\n# print(\"input was:\", sys.argv)\n\n\ndef BT(soduku, slices):\n \"Backtracking search to solve soduku\"\n # If soduku is complete return it.\n if isComplete(soduku):\n return soduku\n # Select the MRV variable to fill\n vars = [tuple(e) for e in np.transpose(np.where(soduku==0))]\n var, avail_d = selectMRVvar(vars, soduku, slices)\n # Fill in a value and solve further (recursively), \n # backtracking an assignment when stuck\n for value in avail_d:\n soduku[var] = value\n result = BT(soduku, slices)\n if np.any(result):\n return result\n else:\n soduku[var] = 0\n return False\n\n\ndef str2arr(soduku_str):\n \"Converts soduku_str to 2d array\"\n return np.array([int(s) for s in list(soduku_str)]).reshape((9,9))\n\n\ndef var2grid(var, slices):\n \"Returns the grid slice (3x3) to which the variable's coordinates belong \"\n row,col = var\n grid = ( slices[int(row/3)], slices[int(col/3)] )\n return grid\n\n\n# Constraints\ndef unique_rows(soduku):\n for row in soduku:\n if not np.array_equal(np.unique(row),np.array(range(1,10))) :\n return False\n return True\ndef unique_columns(soduku):\n for row in soduku.T: #transpose soduku to get columns\n if not np.array_equal(np.unique(row),np.array(range(1,10))) :\n return False\n return True\n\ndef unique_grids(soduku, slices):\n s1,s2,s3 = slices\n allgrids=[(si,sj) for si in [s1,s2,s3] for sj in [s1,s2,s3]] # Makes 2d slices for grids\n for grid in allgrids: \n if not np.array_equal(np.unique(soduku[grid]),np.array(range(1,10))) :\n return False\n return True\n\ndef isComplete(soduku):\n if 0 in soduku:\n return False\n else:\n return True\n\n\ndef checkCorrect(soduku, slices):\n if unique_columns(soduku):\n if unique_rows(soduku):\n if unique_grids(soduku, slices):\n return True\n return False\n\n\n# Search\ndef getDomain(var, soduku, slices):\n \"Gets the remaining legal values (available domain) for an unfilled box `var` in `soduku`\"\n row,col = var\n #ravail = np.setdiff1d(FULLDOMAIN, soduku[row,:])\n #cavail = np.setdiff1d(FULLDOMAIN, soduku[:,col])\n #gavail = np.setdiff1d(FULLDOMAIN, soduku[var2grid(var)])\n #avail_d = reduce(np.intersect1d, (ravail,cavail,gavail))\n used_d = reduce(np.union1d, (soduku[row,:], soduku[:,col], soduku[var2grid(var, slices)]))\n \n FULLDOMAIN = np.array(range(1,10)) #All possible values (1-9)\n avail_d = np.setdiff1d(FULLDOMAIN, used_d)\n #print(var, avail_d)\n return avail_d\n\ndef selectMRVvar(vars, soduku, slices):\n \"\"\"\n Returns the unfilled box `var` with minimum remaining [legal] values (MRV) \n and the corresponding values (available domain)\n \"\"\"\n #Could this be improved?\n avail_domains = [getDomain(var,soduku, slices) for var in vars]\n avail_sizes = [len(avail_d) for avail_d in avail_domains]\n index = np.argmin(avail_sizes)\n return vars[index], avail_domains[index]\n\n\n\n\n# Solve\n\ndef full_solution():\n soduku_str='000000000302540000050301070000000004409006005023054790000000050700810000080060009'\n\n soduku = str2arr(soduku_str)\n\n slices = [slice(0,3), slice(3,6), slice(6,9)]\n s1,s2,s3 = slices\n return BT(soduku, slices), soduku, slices\n\n%timeit sol, soduku, slices = full_solution()\nprint(\"solved:\\n\", sol)\nprint(\"correct:\", checkCorrect(soduku, slices))\n",
"351 ms ± 841 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
cb6fab6ebe3492f72e0630def9aab27466922c37 | 693,284 | ipynb | Jupyter Notebook | qnn/tests/test_minimum_error_discrimination.ipynb | LucianoPereiraValenzuela/QuantumNeuralNetworks_for_StateDiscrimination | 7e116e9631b9c83cc9b06de60d742c71031d9eed | [
"Apache-2.0"
]
| 6 | 2021-11-21T09:57:28.000Z | 2022-01-15T02:33:08.000Z | qnn/tests/test_minimum_error_discrimination.ipynb | LucianoPereiraValenzuela/QuantumNeuralNetworks_for_StateDiscrimination | 7e116e9631b9c83cc9b06de60d742c71031d9eed | [
"Apache-2.0"
]
| null | null | null | qnn/tests/test_minimum_error_discrimination.ipynb | LucianoPereiraValenzuela/QuantumNeuralNetworks_for_StateDiscrimination | 7e116e9631b9c83cc9b06de60d742c71031d9eed | [
"Apache-2.0"
]
| 1 | 2022-01-13T22:59:12.000Z | 2022-01-13T22:59:12.000Z | 1,716.049505 | 143,704 | 0.960642 | [
[
[
"# Test: Minimum error discrimination\n\nIn this notebook we are testing the evolution of the error probability with the number of evaluations.",
"_____no_output_____"
]
],
[
[
"\nimport sys \nsys.path.append('../../')\n\nimport itertools\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom numpy import pi\nfrom qiskit.algorithms.optimizers import SPSA\nfrom qnn.quantum_neural_networks import StateDiscriminativeQuantumNeuralNetworks as nnd\nfrom qnn.quantum_state import QuantumState \n\nplt.style.use('ggplot')",
"_____no_output_____"
],
[
"def callback(params, results, prob_error, prob_inc, prob):\n data.append(prob_error)",
"_____no_output_____"
],
[
"# Create random states\nψ = QuantumState.random(1)\nϕ = QuantumState.random(1)\n\n# Parameters\nth_u, fi_u, lam_u = [0], [0], [0]\nth1, th2 = [0], [pi]\nth_v1, th_v2 = [0], [0]\nfi_v1, fi_v2 = [0], [0]\nlam_v1, lam_v2 = [0], [0]\nparams = list(itertools.chain(th_u, fi_u, lam_u, th1, th2, th_v1, th_v2, fi_v1, fi_v2, lam_v1, lam_v2))\n\n# Initialize Discriminator\ndiscriminator = nnd([ψ, ϕ])\ndata = []\nresults = discriminator.discriminate(SPSA(100), params, callback=callback)\noptimal = nnd.helstrom_bound(ψ, ϕ)\n\nprint(f'Optimal results: {optimal}\\nActual results: {results}')",
"Optimal results: 0.1842391754983393\nActual results: (array([-1.00757705e+00, 3.32056507e+00, 1.16330850e+00, 1.98082431e-03,\n 3.23876708e+00, -1.86481792e+00, -2.30335836e+00, -9.06714945e-01,\n -1.46201088e+00, 1.96589739e-01, 4.87138113e-01]), 0.177734375, 200)\n"
],
[
"fig = plt.figure(figsize=(14, 6))\nplt.plot(data, '-')\nplt.xlabel('Number of evaluations')\nplt.ylabel('Probability')\nplt.legend(['Experimental'])\nplt.title('Evolution of error probability for 2 states')\nfig.savefig('twostates.png')\nplt.show()",
"_____no_output_____"
],
[
"th_u, fi_u, lam_u = results[0][:3]\nth1 = results[0][3]\nth2 = results[0][4]\nth_v1 = results[0][5]\nth_v2 = results[0][6]\nfi_v1 = results[0][7]\nfi_v2 = results[0][8]\nlam_v1 = results[0][9]\nlam_v2 = results[0][10]\n\nM = nnd.povm( 2,\n [th_u], [fi_u], [lam_u],\n [th1], [th2],\n [th_v1], [th_v2],\n [fi_v1], [fi_v2],\n [lam_v1], [lam_v2], output='povm' )\nplt.style.use('default')\nsphere = nnd.plot_bloch_sphere( M , [ψ, ϕ] )\nsphere.render()\nplt.savefig('sphere_2_states')\nplt.style.use('ggplot')",
"_____no_output_____"
],
[
"# Create random states\nψ = QuantumState.random(1)\nϕ = QuantumState.random(1)\nχ = QuantumState.random(1)\n\n# Parameters\nth_u, fi_u, lam_u = [0], [0], [0]\nth1, th2 = 2 * [0], 2 * [pi]\nth_v1, th_v2 = 2 * [0], 2 * [0]\nfi_v1, fi_v2 = 2 * [0], 2 * [0]\nlam_v1, lam_v2 = 2 * [0], 2 * [0]\nparams = list(itertools.chain(th_u, fi_u, lam_u, th1, th2, th_v1, th_v2, fi_v1, fi_v2, lam_v1, lam_v2))\n\n# Initialize Discriminator\ndiscriminator = nnd([ψ, ϕ, χ])\ndata = []\nresults = discriminator.discriminate(SPSA(100), params, callback=callback)\n\nprint(f'Results: {results}')",
"Results: (array([ 1.10538359, -1.70015769, -0.48348853, -2.11682256, -0.03103935,\n 3.19922651, 3.11729522, 0.31810485, -0.13160315, -0.18140742,\n -0.76371755, 0.27253825, 0.03474263, 0.01515616, -0.5763371 ,\n 0.10982771, 1.07646142, -0.01327666, -0.93122126]), 0.4046223958333333, 200)\n"
],
[
"fig = plt.figure(figsize=(14, 6))\nplt.plot(data, '-')\nplt.xlabel('Number of evaluations')\nplt.ylabel('Probability')\nplt.legend(['Experimental'])\nplt.title('Evolution of error probability for 3 states')\nfig.savefig('3states.png')\nplt.show()",
"_____no_output_____"
],
[
"th_u, fi_u, lam_u = results[0][:3]\nth1 = results[0][3:5]\nth2 = results[0][5:7]\nth_v1 = results[0][7:9]\nth_v2 = results[0][9:11]\nfi_v1 = results[0][11:13]\nfi_v2 = results[0][13:15]\nlam_v1 = results[0][15:17]\nlam_v2 = results[0][17:19]\n\nM = nnd.povm( 3,\n [th_u], [fi_u], [lam_u],\n th1, th2,\n th_v1, th_v2,\n fi_v1, fi_v2,\n lam_v1, lam_v2, output='povm' )\nplt.style.use('default')\nsphere = nnd.plot_bloch_sphere( M , [ψ, ϕ, χ] )\nsphere.render()\nplt.savefig('sphere_3_states.png')\nplt.style.use('ggplot')",
"_____no_output_____"
],
[
"# Create random states\nψ = QuantumState([ np.array([1,0]) ])\nϕ = QuantumState([ np.array([np.cos(np.pi/4), np.sin(np.pi/4)]), \n np.array([np.cos(0.1+np.pi/4),np.sin(0.1+np.pi/4)] ) ])\nχ = QuantumState([ np.array([np.cos(np.pi/4), 1j*np.sin(np.pi/4)]), \n np.array([np.cos(0.1+np.pi/4), 1j*np.sin(0.1+np.pi/4)] ),\n np.array([np.cos(-0.1+np.pi/4), 1j*np.sin(-0.1+np.pi/4)] )])\n\n# Parameters\nth_u, fi_u, lam_u = list(np.pi*np.random.randn(1)), list(np.pi*np.random.randn(1)), list(np.pi*np.random.randn(1))\nth1, th2 = list(np.pi*np.random.randn(2)), list(np.pi*np.random.randn(2))\nth_v1, th_v2 = list(np.pi*np.random.randn(2)), list(np.pi*np.random.randn(2))\nfi_v1, fi_v2 = list(np.pi*np.random.randn(2)), list(np.pi*np.random.randn(2))\nlam_v1, lam_v2 = list(np.pi*np.random.randn(2)), list(np.pi*np.random.randn(2))\nparams = list(itertools.chain(th_u, fi_u, lam_u, th1, th2, th_v1, th_v2, fi_v1, fi_v2, lam_v1, lam_v2))\n\n# Initialize Discriminator\ndiscriminator = nnd([ψ, ϕ, χ])\ndata = []\nresults = discriminator.discriminate(SPSA(100), params, callback=callback)\n\nprint(f'Results: {results}')",
"Results: (array([ 9.81810357, 0.59561506, -1.36035727, 3.18600335, -2.84531461,\n 0.99830421, -5.75338446, -0.4057666 , -3.25167801, -4.81894089,\n -6.46027888, -1.67360266, -1.92398161, 1.14189749, 5.38115967,\n -1.12673158, 2.53992157, 0.28855823, 1.09164108]), 0.4171549479166666, 200)\n"
],
[
"fig = plt.figure(figsize=(14, 6))\nplt.plot(data, '-')\nplt.xlabel('Number of evaluations')\nplt.ylabel('Probability')\nplt.legend(['Experimental'])\nplt.title('Evolution of error probability for 3 states with noise')\nfig.savefig('noisy.png')\nplt.show()",
"_____no_output_____"
],
[
"th_u, fi_u, lam_u = results[0][:3]\nth1 = results[0][3:5]\nth2 = results[0][5:7]\nth_v1 = results[0][7:9]\nth_v2 = results[0][9:11]\nfi_v1 = results[0][11:13]\nfi_v2 = results[0][13:15]\nlam_v1 = results[0][15:17]\nlam_v2 = results[0][17:19]\n\nM = nnd.povm( 3,\n [th_u], [fi_u], [lam_u],\n th1, th2,\n th_v1, th_v2,\n fi_v1, fi_v2,\n lam_v1, lam_v2, output='povm' )\nplt.style.use('default')\nsphere = nnd.plot_bloch_sphere( M , [ψ, ϕ, χ] )\nsphere.render()\nplt.savefig('sphere_3_states_noisy.png')\nplt.style.use('ggplot')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6fad61c65176246d1b281c29989d017dee1bdb | 3,377 | ipynb | Jupyter Notebook | week2/HoG.ipynb | Fabriceli/MachingLearning | d983f87c26f2ced2921030562a82dcd19c02171b | [
"MIT"
]
| null | null | null | week2/HoG.ipynb | Fabriceli/MachingLearning | d983f87c26f2ced2921030562a82dcd19c02171b | [
"MIT"
]
| null | null | null | week2/HoG.ipynb | Fabriceli/MachingLearning | d983f87c26f2ced2921030562a82dcd19c02171b | [
"MIT"
]
| null | null | null | 19.408046 | 83 | 0.498075 | [
[
[
"**Histogram of Oriented Gradients**",
"_____no_output_____"
]
],
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
],
[
"import cv2\nimport numpy as np",
"_____no_output_____"
],
[
"im = cv2.imread('lenna.jpeg')\nim = np.float32(im) / 255.0\n \n# Calculate gradient \ngx = cv2.Sobel(im, cv2.CV_32F, 1, 0, ksize=1)\ngy = cv2.Sobel(im, cv2.CV_32F, 0, 1, ksize=1)",
"_____no_output_____"
],
[
"mag, angle = cv2.cartToPolar(gx, gy, angleInDegrees=True)",
"_____no_output_____"
],
[
"# cv2.imshow('gx', gx)\n# cv2.imshow('gy', gy)\n# cv2.imshow('mag', mag)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
],
[
"np.random.seed(0)\nnp.random.randn(1, 5)",
"_____no_output_____"
],
[
"np.random.seed(1)\nnp.random.randn(1, 5)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt \nfrom sklearn.datasets import make_moons \nplt.subplot(122) \nx1,y1=make_moons(200,noise=0.1) \nplt.title('make_moons function example') \nplt.scatter(x1[:,0],x1[:,1],marker='o',c=y1) ",
"_____no_output_____"
],
[
"import torch\n ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb6fbfe851321feb2890772e32f924f5a84c1359 | 5,982 | ipynb | Jupyter Notebook | tutorial/2-Creating Your Own Initial Conditions.ipynb | dorisjlee/astroSim-tutorial | 0f0b07b4c4021c6e9f166b51e7dc8c3a1836920d | [
"BSD-3-Clause"
]
| 4 | 2016-07-24T17:28:34.000Z | 2022-03-03T14:08:18.000Z | tutorial/2-Creating Your Own Initial Conditions.ipynb | dorisjlee/astroSim-tutorial | 0f0b07b4c4021c6e9f166b51e7dc8c3a1836920d | [
"BSD-3-Clause"
]
| null | null | null | tutorial/2-Creating Your Own Initial Conditions.ipynb | dorisjlee/astroSim-tutorial | 0f0b07b4c4021c6e9f166b51e7dc8c3a1836920d | [
"BSD-3-Clause"
]
| null | null | null | 40.693878 | 393 | 0.625209 | [
[
[
"# Terminologies\n\n<img src=\"https://github.com/dorisjlee/remote/blob/master/astroSim-tutorial-img/terminology.jpg?raw=true\",width=20%>\n- __Domain__ (aka Grids): the whole simulation box.\n- __Block__(aka Zones): group of cells that make up a larger unit so that it is more easily handled. If the code is run in parallel, you could have one processor assigned to be in charge to work on several blocks (specified by iProcs,jProcs,kProcs in flash.par). In FLASH, the default block size in flash is $2^3$ = 8 cells. This means that level 0 in the AMR is 8 cells and so forth.\n\n<img src=\"https://github.com/dorisjlee/remote/blob/master/astroSim-tutorial-img/level_cells.jpg?raw=true\",width=20%>\n- __Cells__ : basic units that contain information about the fluid variables (often called primitives: $\\rho$, $P$, $v_{x,y,z}$,$B_{x,y,z}$)\n- __Ghost cells__ (abbrev as ``gc`` in FLASH): Could be thought of as an extra layer of padding outside the simulation domain. The alues of these gcs are mostly determined by what the boundary conditions you chose. Generally, you won't have to mess with these when specifying the initial conditions.\n\n\n",
"_____no_output_____"
],
[
"# Simulation_initBlock.F90",
"_____no_output_____"
],
[
"Simulation_initBlock is called by each block. First we compute the center based on the dimensions of the box (in cgs) from flash.par:\n~~~fortran\ncenter = abs(xmin-xmax)/2.\n~~~\n \n\nWe loop through all the coordinates of the cell within each block. ",
"_____no_output_____"
],
[
"~~~fortran\n do k = blkLimits(LOW,KAXIS),blkLimits(HIGH,KAXIS)\n ! get the coordinates of the cell center in the z-direction\n zz = zCoord(k)-center\n do j = blkLimits(LOW,JAXIS),blkLimits(HIGH,JAXIS)\n ! get the coordinates of the cell center in the y-direction\n yy = yCoord(j)-center\n do i = blkLimits(LOW,IAXIS),blkLimits(HIGH,IAXIS)\n ! get the cell center, left, and right positions in x\n xx = xCenter(i)-center\n~~~",
"_____no_output_____"
],
[
"``xCenter,yCoord,zCoord`` are functions that return the cell position (in cgs) given its cell index. These calculations are based on treating the bottom left corner of the box as the origin, so we minus the box center to get the origin to be at the center, as shown in Fig 3.",
"_____no_output_____"
],
[
"<img src=\"https://github.com/dorisjlee/remote/blob/master/astroSim-tutorial-img/user_coord.png?raw=true\",width=200,height=200>\n__Fig 3: The corrected ``xx,yy,zz`` are physical positions measured from the origin.__",
"_____no_output_____"
],
[
"Given the cell positions, you can specify values for initializing the fluid variables. \nThe fluid variables are stored inside the local variables (called rhoZone,presZone,velxZone, velyZone,velzZone in the example) which are then transferred into to the cell one at a time using the method Grid_putData:\n\n~~~fortran\n call Grid_putPointData(blockId, CENTER, DENS_VAR, EXTERIOR, axis, rhoZone)\n~~~ \n \nFor example, you may have an analytical radial density distribution ($\\rho= Ar^2$) that you would like to initialize the sphere with: \n~~~fortran\n rr = sqrt(xx**2 + yy**2 + zz**2)\n rhoZone = A*rr**2\n~~~\nOr maybe your initial conditions can not be expressed in closed form,then you could also read in precomputed-values for each cell. This optional tutorial will explain how to do linear interpolation to setup the numerical solution of the Lane-Emden Sphere. ",
"_____no_output_____"
],
[
"### Adding new RuntimeParameters to be read into Simulation_initBlock.F90",
"_____no_output_____"
],
[
"As we have already saw, to compute the center of the box, I need to read in the dimensions of the box (``xmin,xmax``) from flash.par. Some runtime parameters are used by other simulation modules and some are specific to the problem and defined by the users. \n\nTo add in a new runtime parameter: \n\n1) In ``Simulation_data.F90``, declare the variables to store these runtime parameters:\n~~~fortran\n real, save :: fattening_factor,beta_param,xmin,xmax\n~~~\n2) In ``Simulation_init.F90``, read in the values of the runtime parameter:\n~~~fortran \n call RuntimeParameters_get('xmin',xmin)\n call RuntimeParameters_get('xmax',xmax)\n~~~\n3) In ``Simulation_initBlock.F90``, use the data:\n~~~fortran\n use Simulation_data, ONLY: xmin,xmax\n~~~\nNote you should __NOT__ declare ``real::xmin,xmax`` again inside ``Simulation_initBlock.F90``, otherwise, the values that you read in will be overridden.",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
cb6fc042cd9bd318eea9ff897449d7c005244c84 | 597,192 | ipynb | Jupyter Notebook | corrections/single_layer_neural_network_cor.ipynb | heprom/cvml | aa5fb3f120ad5272cf8bd9c762b55c96c16e4356 | [
"CC0-1.0"
]
| 3 | 2021-02-22T09:06:32.000Z | 2022-02-20T23:00:09.000Z | corrections/single_layer_neural_network_cor.ipynb | heprom/cvml | aa5fb3f120ad5272cf8bd9c762b55c96c16e4356 | [
"CC0-1.0"
]
| null | null | null | corrections/single_layer_neural_network_cor.ipynb | heprom/cvml | aa5fb3f120ad5272cf8bd9c762b55c96c16e4356 | [
"CC0-1.0"
]
| 4 | 2021-02-22T09:10:56.000Z | 2022-02-17T14:41:13.000Z | 611.250768 | 178,674 | 0.940366 | [
[
[
"# Single layer Neural Network\n\nIn this notebook, we will code a single neuron and use it as a linear classifier with two inputs. The tuning of the neuron parameters is done by backpropagation using gradient descent.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_blobs\nimport numpy as np\n\n# matplotlib to display the data\nimport matplotlib\nmatplotlib.rc('font', size=16)\nmatplotlib.rc('xtick', labelsize=16) \nmatplotlib.rc('ytick', labelsize=16) \nfrom matplotlib import pyplot as plt, cm\nfrom matplotlib.colors import ListedColormap\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Dataset\n\nLet's create some labeled data in the form of (X, y) with an associated class which can be 0 or 1. For this we can use the function `make_blobs` in the `sklearn.datasets` module. Here we use 2 centers with coordinates (-0.5, -1.0) and (1.0, 1.0).",
"_____no_output_____"
]
],
[
[
"X, y = make_blobs(n_features=2, random_state=42, centers=[(-0.5, -1.0), (1.0, 1.0)])\ny = y.reshape((y.shape[0], 1))\nprint(X.shape)\nprint(y.shape)",
"(100, 2)\n(100, 1)\n"
]
],
[
[
"Plot our training data using `plt.scatter` to have a first visualization. Here we color the points with their labels stored in `y`.",
"_____no_output_____"
]
],
[
[
"plt.scatter(X[:, 0], X[:, 1], c=y.squeeze(), edgecolors='gray')\nplt.title('training data with labels')\nplt.axis('equal')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Activation functions\n\nHere we play with popular activation functions like tanh, ReLu or sigmoid.",
"_____no_output_____"
]
],
[
[
"def heaviside(x):\n return np.heaviside(x, np.zeros_like(x))\n\ndef sigmoid(x):\n return 1 / (1 + np.exp(-x))\n\ndef ReLU(x):\n return np.maximum(0, x)\n\ndef leaky_ReLU(x, alpha=0.1):\n return np.maximum(alpha * x, x)\n\ndef tanh(x):\n return np.tanh(x)",
"_____no_output_____"
],
[
"from math import pi\n\nplt.figure()\nx = np.arange(-pi, pi, 0.01)\nplt.axhline(y=0., color='gray', linestyle='dashed')\nplt.axhline(y=-1, color='gray', linestyle='dashed')\nplt.axhline(y=1., color='gray', linestyle='dashed')\nplt.axvline(x=0., color='gray', linestyle='dashed')\n\nplt.xlim(-pi, pi)\nplt.ylim(-1.2, 1.2)\nplt.title('activation functions', fontsize=16)\n\nplt.plot(x, heaviside(x), label='heavyside', linewidth=3)\nlegend = plt.legend(loc='lower right')\nplt.savefig('activation_functions_1.pdf')\nplt.plot(x, sigmoid(x), label='sigmoid', linewidth=3)\nplt.legend(loc='lower right')\nplt.savefig('activation_functions_2.pdf')\nplt.plot(x, tanh(x), label='tanh', linewidth=3)\nplt.legend(loc='lower right')\nplt.savefig('activation_functions_3.pdf')\nplt.plot(x, ReLU(x), label='ReLU', linewidth=3)\nplt.legend(loc='lower right')\nplt.savefig('activation_functions_4.pdf')\nplt.plot(x, leaky_ReLU(x), label='leaky ReLU', linewidth=3)\nplt.legend(loc='lower right')\nplt.savefig('activation_functions_5.pdf')\nplt.show()",
"_____no_output_____"
],
[
"# gradients of the activation functions\ndef sigmoid_grad(x):\n s = sigmoid(x)\n return s * (1 - s)\n\ndef relu_grad(x):\n return 1. * (x > 0)\n\ndef tanh_grad(x):\n return 1 - np.tanh(x) ** 2",
"_____no_output_____"
],
[
"plt.figure()\nx = np.arange(-pi, pi, 0.01)\nplt.plot(x, sigmoid_grad(x), label='sigmoid gradient', linewidth=3)\nplt.plot(x, relu_grad(x), label='ReLU gradient', linewidth=3)\nplt.plot(x, tanh_grad(x), label='tanh gradient', linewidth=3)\nplt.xlim(-pi, pi)\nplt.title('activation function derivatives', fontsize=16)\nlegend = plt.legend()\nlegend.get_frame().set_linewidth(2)\nplt.savefig('activation_functions_derivatives.pdf')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## ANN implementation\n\nA simple neuron with two inputs $(x_1, x_2)$ which applies an affine transform of weigths $(w_1, w_2)$ and bias $w_0$.\n\nThe neuron compute the quantity called activation $a=\\sum_i w_i x_i + w_0 = w_0 + w_1 x_1 + w_2 x_2$\n\nThis quantity is send to the activation function chosen to be a sigmoid function here: $f(a)=\\dfrac{1}{1+e^{-a}}$\n\n$f(a)$ is the output of the neuron bounded between 0 and 1.",
"_____no_output_____"
],
[
"### Quick implementation\n\nFirst let's implement our network in a concise fashion.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom numpy.random import randn\n\nX, y = make_blobs(n_samples= 100, n_features=2, random_state=42, centers=[[-0.5, -1], [1, 1]])\n# adjust the sizes of our arrays\nX = np.c_[np.ones(X.shape[0]), X]\nprint(X.shape)\ny = y.reshape((y.shape[0], 1))\n\nnp.random.seed(2)\nW = randn(3, 1)\nprint('* model params: {}'.format(W.tolist()))\neta = 1e-2 # learning rate\nn_epochs = 50\n\nfor t in range(n_epochs):\n # forward pass\n y_pred = sigmoid(X.dot(W))\n loss = np.sum((y_pred - y) ** 2)\n print(t, loss)\n\n # backprop\n grad_y_pred = 2 * (y_pred - y)\n grad_W = np.dot(X.T, grad_y_pred * y_pred * (1 - y_pred))\n\n # update rule\n W -= eta * grad_W\nprint('* new model params: {}'.format(W.tolist()))\n",
"(100, 3)\n* model params: [[-0.4167578474054706], [-0.056266827226329474], [-2.136196095668454]]\n0 69.888678007119\n1 68.10102729420157\n2 66.01234835808539\n3 63.52920474304009\n4 60.511931504891606\n5 56.781412908257316\n6 52.17672528838464\n7 46.67324119014275\n8 40.42792006624582\n9 33.65215682518604\n10 26.70717521140393\n11 20.544252734780706\n12 16.198710182740403\n13 13.580478268635446\n14 11.983692035385701\n15 10.922630988132667\n16 10.16098958742113\n17 9.58305898101223\n18 9.126953943573383\n19 8.756534709365818\n20 8.449122187454204\n21 8.189655164236605\n22 7.967673714087983\n23 7.77564748980427\n24 7.607993124138909\n25 7.460467877966521\n26 7.329780007443549\n27 7.213329709562506\n28 7.109031799422667\n29 7.0151912380112025\n30 6.93041381013332\n31 6.85354076229679\n32 6.783600131596645\n33 6.71976992958227\n34 6.661349894574377\n35 6.607739535904203\n36 6.558420865715362\n37 6.512944669754438\n38 6.470919482932122\n39 6.43200265563964\n40 6.395893053272442\n41 6.36232504406938\n42 6.331063512489355\n43 6.30189969588866\n44 6.274647687380679\n45 6.249141481725432\n46 6.225232466911465\n47 6.202787283890317\n48 6.1816859922367575\n49 6.161820491448388\n* new model params: [[-0.2559481613676354], [1.3472482576843752], [1.3640333350148732]]\n"
]
],
[
[
"### Modular implementation\n\nNow let's create a class to represent our neural network to have more flexibility and modularity. This will prove to be useful later when we add more layers.",
"_____no_output_____"
]
],
[
[
"class SingleLayerNeuralNetwork:\n \"\"\"A simple artificial neuron with a single layer and two inputs. \n \n This type of network is called a Single Layer Neural Network and belongs to \n the Feed-Forward Neural Networks. Here, the activation function is a sigmoid, \n the loss is computed using the squared error between the target and \n the prediction. Learning the parameters is achieved using back-propagation \n and gradient descent\n \"\"\"\n \n def __init__(self, eta=0.01, rand_seed=42):\n \"\"\"Initialisation routine.\"\"\"\n np.random.seed(rand_seed)\n self.W = np.random.randn(3, 1) # weigths\n self.eta = eta # learning rate\n self.loss_history = []\n \n def sigmoid(self, x):\n \"\"\"Our activation function.\"\"\"\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_grad(self, x):\n \"\"\"Gradient of the sigmoid function.\"\"\"\n return self.sigmoid(x) * (1 - self.sigmoid(x))\n \n def predict(self, X, bias_trick=True):\n X = np.atleast_2d(X)\n if bias_trick:\n # bias trick: add a column of 1 to X\n X = np.c_[np.ones((X.shape[0])), X]\n return self.sigmoid(np.dot(X, self.W))\n \n def loss(self, X, y, bias_trick=False):\n \"\"\"Compute the squared error loss for a given set of inputs.\"\"\"\n y_pred = self.predict(X, bias_trick=bias_trick)\n y_pred = y_pred.reshape((y_pred.shape[0], 1))\n loss = np.sum((y_pred - y) ** 2)\n return loss\n \n def back_propagation(self, X, y):\n \"\"\"Conduct backpropagation to update the weights.\"\"\"\n X = np.atleast_2d(X)\n y_pred = self.sigmoid(np.dot(X, self.W)).reshape((X.shape[0], 1))\n grad_y_pred = 2 * (y_pred - y)\n grad_W = np.dot(X.T, grad_y_pred * y_pred * (1 - y_pred))\n\n # update weights\n self.W -= eta * grad_W\n \n def fit(self, X, y, n_epochs=10, method='batch', save_fig=False):\n \"\"\"Perform gradient descent on a given number of epochs to update the weights.\"\"\"\n # bias trick: add a column of 1 to X\n X = np.c_[np.ones((X.shape[0])), X]\n self.loss_history.append(self.loss(X, y)) # initial loss\n for i_epoch in range(n_epochs):\n if method == 'batch':\n # perform backprop on the whole training set (batch)\n self.back_propagation(X, y)\n # weights were updated, compute the loss\n loss = self.loss(X, y)\n self.loss_history.append(loss)\n print(i_epoch, self.loss_history[-1])\n else:\n # here we update the weight for every data point (SGD)\n for (xi, yi) in zip(X, y):\n self.back_propagation(xi, yi)\n # weights were updated, compute the loss\n loss = self.loss(X, y)\n self.loss_history.append(loss)\n if save_fig:\n self.plot_model(i_epoch, save=True, display=False)\n\n def decision_boundary(self, x):\n \"\"\"Return the decision boundary in 2D.\"\"\"\n return -self.W[0] / self.W[2] - self.W[1] / self.W[2] * x\n \n def plot_model(self, i_epoch=-1, save=False, display=True):\n \"\"\"Build a figure to vizualise how the model perform.\"\"\"\n xx0, xx1 = np.arange(-3, 3.1, 0.1), np.arange(-3, 4.1, 0.1)\n XX0, XX1 = np.meshgrid(xx0, xx1)\n # apply the model to the grid\n y_an = np.empty(len(XX0.ravel()))\n i = 0\n for (x0, x1) in zip(XX0.ravel(), XX1.ravel()):\n y_an[i] = self.predict(np.array([x0, x1]))\n i += 1\n y_an = y_an.reshape((len(xx1), len(xx0)))\n figure = plt.figure(figsize=(12, 4))\n ax1 = plt.subplot(1, 3, 1)\n #ax1.set_title(r'$w_0=%.3f$, $w_1=%.3f$, $w_2=%.3f$' % (self.W[0], self.W[1], self.W[2]))\n ax1.set_title(\"current prediction\")\n ax1.contourf(XX0, XX1, y_an, alpha=.5)\n ax1.scatter(X[:, 0], X[:, 1], c=y.squeeze(), edgecolors='gray')\n ax1.set_xlim(-3, 3)\n ax1.set_ylim(-3, 4)\n print(ax1.get_xlim())\n x = np.array(ax1.get_xlim())\n ax1.plot(x, self.decision_boundary(x), 'k-', linewidth=2)\n ax2 = plt.subplot(1, 3, 2)\n x = np.arange(3) # the label locations\n rects1 = ax2.bar(x, [self.W[0, 0], self.W[1, 0], self.W[2, 0]])\n ax2.set_title('model parameters')\n ax2.set_xticks(x)\n ax2.set_xticklabels([r'$w_0$', r'$w_1$', r'$w_2$'])\n ax2.set_ylim(-1, 2)\n ax2.set_yticks([0, 2])\n ax2.axhline(xmin=0, xmax=2)\n ax3 = plt.subplot(1, 3, 3)\n ax3.plot(self.loss_history, c='lightgray', lw=2)\n if i_epoch < 0:\n i_epoch = len(self.loss_history) - 1\n ax3.plot(i_epoch, self.loss_history[i_epoch], 'o')\n ax3.set_title('loss evolution')\n ax3.set_yticks([])\n plt.subplots_adjust(left=0.05, right=0.98)\n if save:\n plt.savefig('an_%02d.png' % i_epoch)\n if display:\n plt.show()\n plt.close()\n",
"_____no_output_____"
]
],
[
[
"### Train our model on the data set\n\nCreate two blobs with $n=1000$ data points.\n\nInstantiate the model with $\\eta$=0.1 and a random seed of 2.\n\nTrain the model using the batch gradient descent on 20 epochs.",
"_____no_output_____"
]
],
[
[
"X, y = make_blobs(n_samples=10000, n_features=2, random_state=42, centers=[[-0.5, -1], [1, 1]])\ny = y.reshape((y.shape[0], 1))\n\nan1 = SingleLayerNeuralNetwork(eta=0.1, rand_seed=2)\nprint('* init model params: {}'.format(an1.W.tolist()))\nprint(an1.loss(X, y, bias_trick=True))\nan1.fit(X, y, n_epochs=100, method='batch', save_fig=False)\nprint('* new model params: {}'.format(an1.W.tolist()))",
"* init model params: [[-0.4167578474054706], [-0.056266827226329474], [-2.136196095668454]]\n6813.791619744032\n0 1065.5244005560853\n1 1004.2834940985097\n2 972.1020148641242\n3 952.1899581995993\n4 938.3234296045975\n5 927.9765410132449\n6 919.2260496173226\n7 911.8159986339842\n8 906.2581276170479\n9 902.2935497103937\n10 899.2937229391691\n11 896.835926605091\n12 894.6988964194702\n13 892.7681058525101\n14 890.9810531145381\n15 889.2998538790825\n16 887.6974929083249\n17 886.1513240313166\n18 884.6406287755832\n19 883.1462463333798\n20 881.6509308886809\n21 880.1396760889339\n22 878.5997412921436\n23 877.0204069660281\n24 875.3925836022929\n25 873.7083854938818\n26 871.9607354016401\n27 870.1430269399661\n28 868.2488478128737\n29 866.2717560113587\n30 864.2050976414034\n31 862.0418553064648\n32 859.7745176798607\n33 857.3949630082162\n34 854.8943514010089\n35 852.2630228507501\n36 849.4904001520829\n37 846.5648985409571\n38 843.4738474024027\n39 840.2034344600186\n40 836.7386904645787\n41 833.063544079041\n42 829.1609947726868\n43 825.0134796837673\n44 820.6035539374298\n45 815.9150703886328\n46 810.9351438457842\n47 805.6573257588102\n48 800.0865976476773\n49 794.2469781265381\n50 788.1925885333343\n51 782.0225479579994\n52 775.898199029485\n53 770.0564150296796\n54 764.8039653311268\n55 760.4688673389976\n56 757.2929187670451\n57 755.3023390375763\n58 754.2672366495963\n59 753.8256841536671\n60 753.6685438700401\n61 753.619993570175\n62 753.6063543359508\n63 753.6027346285179\n64 753.6018037351533\n65 753.601568267135\n66 753.6015092100265\n67 753.601494462039\n68 753.6014907887106\n69 753.6014898812546\n70 753.6014896826166\n71 753.6014897395092\n72 753.601490170193\n73 753.6014919095405\n74 753.6014987380806\n75 753.6015255006284\n76 753.6016303665517\n77 753.602041352367\n78 753.6036514124133\n79 753.6099638645105\n80 753.6346720066709\n81 753.7316650514454\n82 754.1097074452814\n83 755.5959994997058\n84 761.2170186342629\n85 782.3616728324131\n86 839.7242689213219\n87 947.4866771926982\n88 857.4699877558302\n89 802.3820107211028\n90 778.6472220261489\n91 771.2017368689822\n92 765.5866829418633\n93 761.1148555762665\n94 757.8825548351401\n95 756.0128641512408\n96 755.6007852090613\n97 757.447139475298\n98 764.9244410329911\n99 791.2484420481214\n* new model params: [[-0.014337638336448744], [1.8730398776699777], [1.9891406713721984]]\n"
]
],
[
[
"Now we have trained our model, plot the results",
"_____no_output_____"
]
],
[
[
"an1.plot_model()",
"(-3.0, 3.0)\n"
]
],
[
[
"Now try to train another network using SGD. Use only 1 epoch since with SGD, we are updating the weights with every training point (so $n$ times per epoch).",
"_____no_output_____"
]
],
[
[
"an2 = SingleLayerNeuralNetwork(eta=0.1, rand_seed=2)\nprint('* init model params: {}'.format(an2.W.tolist()))\nan2.fit(X, y, n_epochs=1, method='SGD', save_fig=False)\nprint('* new model params: {}'.format(an2.W.tolist()))",
"* init model params: [[-0.4167578474054706], [-0.056266827226329474], [-2.136196095668454]]\n* new model params: [[-0.3089337810149369], [1.2172382480136865], [1.6614794435241786]]\n"
]
],
[
[
"plot the difference in terms of loss evolution using batch or stochastic gradient descent",
"_____no_output_____"
]
],
[
[
"plt.plot(an1.loss_history[:], label='batch GD')\nplt.plot(an2.loss_history[::100], label='stochastic GD')\n#plt.ylim(0, 2000)\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"an2.plot_model()",
"(-3.0, 3.0)\n"
]
],
[
[
"## Logistic regression\n\nOur single layer network using the logistic function for activation is very similar to the logistic regression we saw in a previous tutorial. We can easily compare our result with the logistic regression using `sklearn` toolbox.",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\nX, y = make_blobs(n_samples=1000, n_features=2, random_state=42, centers=[[-0.5, -1], [1, 1]])\nlog_reg = LogisticRegression(solver='lbfgs')\nlog_reg.fit(X, y)\nprint(log_reg.coef_)\nprint(log_reg.intercept_)",
"[[1.5698506 1.81179711]]\n[-0.50179977]\n"
],
[
"x0, x1 = np.meshgrid(\n np.linspace(-3, 3.1, 62).reshape(-1, 1),\n np.linspace(-3, 4.1, 72).reshape(-1, 1),\n )\nX_new = np.c_[x0.ravel(), x1.ravel()]\n\ny_proba = log_reg.predict_proba(X_new)\nzz = y_proba[:, 1].reshape(x0.shape)\n\nplt.figure(figsize=(4, 4))\ncontour = plt.contourf(x0, x1, zz, alpha=0.5)\nplt.scatter(X[:, 0], X[:, 1], c=y, edgecolors='gray')\n\n# decision boundary\nx_bounds = np.array([-3, 3])\nboundary = -(log_reg.coef_[0][0] * x_bounds + log_reg.intercept_[0]) / log_reg.coef_[0][1]\nplt.plot(x_bounds, boundary, \"k-\", linewidth=3)\n\nplt.xlim(-3, 3)\nplt.ylim(-3, 4)\nplt.show()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
cb6fc19d5bf16f94c1c417ad84b3ed164bd83a53 | 23,957 | ipynb | Jupyter Notebook | scripts/workflow/notebooks/00_master_immob.ipynb | schwille-paint/SPT | e48c10402ae65b284acd9d049472a278462fe4cd | [
"MIT"
]
| 1 | 2020-07-29T02:56:41.000Z | 2020-07-29T02:56:41.000Z | scripts/workflow/notebooks/00_master_immob.ipynb | schwille-paint/SPT | e48c10402ae65b284acd9d049472a278462fe4cd | [
"MIT"
]
| 1 | 2021-05-25T16:46:57.000Z | 2021-08-08T16:37:48.000Z | scripts/workflow/notebooks/00_master_immob.ipynb | schwille-paint/SPT | e48c10402ae65b284acd9d049472a278462fe4cd | [
"MIT"
]
| 1 | 2020-05-29T16:56:06.000Z | 2020-05-29T16:56:06.000Z | 77.280645 | 1,659 | 0.644947 | [
[
[
"# From raw *.ome.tif file to kinetic properties for immobile particles\nThis notebook will run ... \n* picasso_addon.localize.main()\n* picasso_addon.autopick.main()\n* spt.immobile_props.main()\n\n... in a single run to get from the raw data to the fully evaluated data in a single stroke. We therefore: \n\n1. Define the full paths to the *ome.tif files\n2. Set the execution parameters\n3. Connect or start a local dask parallel computing cluster\n4. Run all sub-module main() functions for all defined datasets\n\nAs a result files with extension *_locs.hdf5, *_render.hdf5, *_autopick.yaml, *_tprops.hdf5 will be created in the same folder as the *.ome.tif file.",
"_____no_output_____"
]
],
[
[
"import os\nimport traceback\nimport importlib\nfrom dask.distributed import Client\nimport multiprocessing as mp\n\nimport picasso.io as io\nimport picasso_addon.localize as localize\nimport picasso_addon.autopick as autopick\nimport spt.immobile_props as improps\n\nimportlib.reload(localize)\nimportlib.reload(autopick)\nimportlib.reload(improps)",
"_____no_output_____"
]
],
[
[
"### 1. Define the full paths to the *ome.tif files",
"_____no_output_____"
]
],
[
[
"dir_names=[]\ndir_names.extend([r'C:\\Data\\p06.SP-tracking\\20-03-11_pseries_fix_B21_rep\\id140_B_exp200_p114uW_T21_1\\test'])\n\nfile_names=[]\nfile_names.extend(['id140_B_exp200_p114uW_T21_1_MMStack_Pos0.ome.tif'])",
"_____no_output_____"
]
],
[
[
"### 2. Set the execution parameters",
"_____no_output_____"
]
],
[
[
"### Valid for all evaluations\nparams_all={'undrift':False,\n 'min_n_locs':5,\n 'filter':'fix',\n }\n\n### Exceptions\nparams_special={}",
"_____no_output_____"
]
],
[
[
"All possible parameters for ...\n* picasso_addon.localize.main()\n* picasso_addon.autopick.main()\n* spt.immobile_props.main() \n\n... can be given. Please run `help(localize.main)` or `help(autopick.main)` or `help(improps.main)` or readthedocs. If not stated otherwise standard values are used (indicated in brackets).",
"_____no_output_____"
]
],
[
[
"help(localize.main)",
"Help on function main in module picasso_addon.localize:\n\nmain(file, info, path, **params)\n Localize movie (least squares, GPU fitting if available) and undrift resulting localizations using rcc.\n \n Args:\n file(picasso.io): Either raw movie loaded with picasso.io.load_movie() or_locs.hdf5 loaded with picasso.io.load_locs()\n info(list(dicts)): Info to raw movie/_locs.hdf5 loaded with picasso.io.load_movie() or picasso.io.load_locs()\n \n Keyword Arguments:\n localize(bool=True) Localize raw movie (see picasso.localize)\n baseline(int=70): Camera spec. baseline (see picasso.localize).\n gain(float=1): Camera spec. EM gain (see picasso.localize)\n sensitivity(float=0.56): Camera spec. sensitivity (see picasso.localize)\n qe(float=0.82): Camera spec. sensitivity (see picasso.localize)\n box(int=5): Box length (uneven!) of fitted spots (see picasso.localize)\n mng(int or str='auto'): Minimal net-gradient spot detection threshold(see picasso.localize. If set to 'auto' minimal net_gradient is determined by autodetect_mng().\n undrift(bool=True) Apply RCC drift correction (see picasso.render)\n segments(int=1000): Segment length (frames) for undrifting by RCC (see picasso.render)\n \n Returns: \n list:\n - [0][0] Dict of **kwargs passed to localize\n - | [0][1] Localizations (numpy.array) as created by picasso.localize\n | Will be saved with extension '_locs.hdf5' for usage in picasso.render\n - [1][0] Dict of **kwargs passed to undriftrcc_locs \n - | [1][1] Undrifted(RCC) localizations as created by picasso.render. \n | If undrifing was not succesfull just corresponds to original loclizations.\n | Will be saved with extension '_locs_render.hdf5' for usage in picasso.render \n | If undrifting was not succesfull only _locs will be saved.\n\n"
]
],
[
[
"### 3. Connect or start a local dask parallel computing cluster\nThis is only necessary if you want to use parallel computing for the spt.immobile.props.main() execution (standard). If not set `params_all={'parallel':False}`",
"_____no_output_____"
]
],
[
[
"try:\n client = Client('localhost:8787')\n print('Connecting to existing cluster...')\nexcept OSError:\n improps.cluster_setup_howto()",
"Connecting to existing cluster...\n"
]
],
[
[
"If we execute the prompt (see below) a local cluster is started, and we only have to execute the cell above to reconnect to it the next time. If you try to create a new cluster under the same address this will throw an error!",
"_____no_output_____"
]
],
[
[
"Client(n_workers=max(1,int(0.8 * mp.cpu_count())),\n processes=True,\n threads_per_worker=1,\n scheduler_port=8787,\n dashboard_address=\":1234\")",
"_____no_output_____"
]
],
[
[
"### 4. Run all sub-module main() functions for all defined datasets",
"_____no_output_____"
]
],
[
[
"failed_path=[]\nfor i in range(0,len(file_names)):\n ### Create path\n path=os.path.join(dir_names[i],file_names[i])\n \n ### Set paramters for each run\n params=params_all.copy()\n for key, value in params_special.items():\n params[key]=value[i]\n \n ### Run main function\n try:\n ### Load movie\n movie,info=io.load_movie(path)\n \n ### Localize and undrift\n out=localize.main(movie,info,path,**params)\n info=info+[out[0][0]]+[out[0][1]] # Update info to used params\n path=out[-1] # Update path\n \n ### Autopick\n print()\n locs=out[1]\n out=autopick.main(locs,info,path,**params)\n info=info+[out[0]] # Update info to used params\n path=out[-1] # Update path\n \n ### Immobile kinetics analysis\n print()\n locs=out[1]\n out=improps.main(locs,info,path,**params)\n \n except Exception:\n traceback.print_exc()\n failed_path.extend([path])\n\nprint() \nprint('Failed attempts: %i'%(len(failed_path)))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6fe81c53256902adbaa9a7d9f5d5356ef9fb6d | 138,243 | ipynb | Jupyter Notebook | pytorch/Pytorch Examples Numpy, Tensors and Autograd.ipynb | HiteshDhola/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials | a0b839d412d2e7e4d8f3b3d885e318650399b857 | [
"Apache-2.0"
]
| 3,266 | 2017-08-06T16:51:46.000Z | 2022-03-30T07:34:24.000Z | pytorch/Pytorch Examples Numpy, Tensors and Autograd.ipynb | HiteshDhola/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials | a0b839d412d2e7e4d8f3b3d885e318650399b857 | [
"Apache-2.0"
]
| 150 | 2017-08-28T14:59:36.000Z | 2022-03-11T23:21:35.000Z | pytorch/Pytorch Examples Numpy, Tensors and Autograd.ipynb | HiteshDhola/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials | a0b839d412d2e7e4d8f3b3d885e318650399b857 | [
"Apache-2.0"
]
| 1,449 | 2017-08-06T17:40:59.000Z | 2022-03-31T12:03:24.000Z | 33.808511 | 419 | 0.601817 | [
[
[
"# What are Tensors?",
"_____no_output_____"
]
],
[
[
"# -*- coding: utf-8 -*-\nimport numpy as np\n\n# N is batch size; D_in is input dimension;\n# H is hidden dimension; D_out is output dimension.\nN, D_in, H, D_out = 64, 1000, 100, 10\n\n# Create random input and output data\nx = np.random.randn(N, D_in)\ny = np.random.randn(N, D_out)\n\n# Randomly initialize weights\nw1 = np.random.randn(D_in, H)\nw2 = np.random.randn(H, D_out)\n\nlearning_rate = 1e-6\nfor t in range(500):\n # Forward pass: compute predicted y\n h = x.dot(w1)\n h_relu = np.maximum(h, 0)\n y_pred = h_relu.dot(w2)\n\n # Compute and print loss\n loss = np.square(y_pred - y).sum()\n print(t, loss)\n\n # Backprop to compute gradients of w1 and w2 with respect to loss\n grad_y_pred = 2.0 * (y_pred - y)\n grad_w2 = h_relu.T.dot(grad_y_pred)\n grad_h_relu = grad_y_pred.dot(w2.T)\n grad_h = grad_h_relu.copy()\n grad_h[h < 0] = 0\n grad_w1 = x.T.dot(grad_h)\n\n # Update weights\n w1 -= learning_rate * grad_w1\n w2 -= learning_rate * grad_w2",
"0 29261998.9383\n1 23584624.4749\n2 21318274.0133\n3 19389745.5408\n4 16479856.1687\n5 12805039.2482\n6 9059166.91546\n7 6042659.8759\n8 3908408.60775\n9 2553920.39789\n10 1723204.06721\n11 1219705.10145\n12 906659.056268\n13 704582.301008\n14 567415.897123\n15 469502.722688\n16 396243.703489\n17 339183.787367\n18 293384.908371\n19 255753.24473\n20 224375.289442\n21 197817.587324\n22 175121.073496\n23 155577.723508\n24 138727.89154\n25 124054.575745\n26 111219.330545\n27 99943.0384346\n28 90002.3975585\n29 81206.7719005\n30 73409.0380627\n31 66473.3112012\n32 60296.3106408\n33 54785.7768329\n34 49859.0677676\n35 45441.2604793\n36 41474.757966\n37 37900.6254289\n38 34674.8838041\n39 31756.4912462\n40 29118.0035071\n41 26731.4581525\n42 24563.2300185\n43 22591.0640449\n44 20795.1155897\n45 19157.9008332\n46 17663.3262804\n47 16297.9786927\n48 15048.7541864\n49 13904.2761665\n50 12855.5370557\n51 11893.5831871\n52 11009.8840228\n53 10198.5198944\n54 9452.97741562\n55 8766.63119037\n56 8134.73416199\n57 7552.22098812\n58 7015.3292248\n59 6519.75327917\n60 6061.87657874\n61 5638.66314253\n62 5247.43856354\n63 4885.45681905\n64 4550.32597631\n65 4239.90233531\n66 3952.19792216\n67 3685.42375585\n68 3437.99282102\n69 3208.32364349\n70 2995.0123591\n71 2796.90365889\n72 2612.77458434\n73 2441.61153781\n74 2282.34987655\n75 2134.12359301\n76 1996.15517682\n77 1867.71870124\n78 1748.02217447\n79 1636.43626907\n80 1532.41129335\n81 1435.3645166\n82 1344.82770496\n83 1260.38274162\n84 1181.52556187\n85 1107.86359066\n86 1039.03117769\n87 974.722224799\n88 914.826976987\n89 858.928900525\n90 806.610252238\n91 757.653403984\n92 711.841667209\n93 668.955674145\n94 628.786703973\n95 591.166198315\n96 555.91219558\n97 522.862764788\n98 491.872428375\n99 462.809720147\n100 435.556280818\n101 409.988779832\n102 385.991214644\n103 363.455455472\n104 342.303748325\n105 322.439528753\n106 303.77815213\n107 286.25152638\n108 269.777995546\n109 254.289212316\n110 239.728038759\n111 226.036798421\n112 213.162389858\n113 201.050099682\n114 189.657042403\n115 178.930752507\n116 168.839022075\n117 159.335671601\n118 150.386617916\n119 141.960890819\n120 134.028933026\n121 126.552752973\n122 119.508618317\n123 112.871599105\n124 106.615401142\n125 100.718185857\n126 95.1601396318\n127 89.9172743676\n128 84.9725273635\n129 80.3094850327\n130 75.9095180326\n131 71.7595557249\n132 67.8449877975\n133 64.1482616471\n134 60.6589306595\n135 57.3666798893\n136 54.257828578\n137 51.3223084883\n138 48.5512904041\n139 45.9332963261\n140 43.4597612423\n141 41.1234738552\n142 38.9164620437\n143 36.8310989551\n144 34.8614349703\n145 32.9989944077\n146 31.2389494061\n147 29.5751657677\n148 28.0018533674\n149 26.5143723378\n150 25.1090290368\n151 23.7789949676\n152 22.5209671013\n153 21.3316066277\n154 20.2065400681\n155 19.1417900359\n156 18.1352582169\n157 17.1825260105\n158 16.2808499372\n159 15.4275713396\n160 14.6199024945\n161 13.8558667482\n162 13.1326730867\n163 12.4476228187\n164 11.7992270904\n165 11.1852772281\n166 10.6039207665\n167 10.053368564\n168 9.53252947626\n169 9.0388943525\n170 8.57125552508\n171 8.12835146959\n172 7.70876529188\n173 7.31119727339\n174 6.93478135637\n175 6.57803922866\n176 6.23990443082\n177 5.91946669864\n178 5.61584117512\n179 5.32809852758\n180 5.05546438442\n181 4.79691466999\n182 4.55190170806\n183 4.31959471325\n184 4.0993672564\n185 3.89053306571\n186 3.69272848442\n187 3.5049605073\n188 3.32690498544\n189 3.15811754743\n190 2.99800272266\n191 2.84612083648\n192 2.70218120603\n193 2.56559847877\n194 2.43598627756\n195 2.31303903422\n196 2.19641792427\n197 2.08576686945\n198 1.98084058378\n199 1.88122059939\n200 1.78671463098\n201 1.69700035\n202 1.61185950487\n203 1.53108818397\n204 1.45446845079\n205 1.38168499608\n206 1.31259415864\n207 1.24704228715\n208 1.18479076767\n209 1.12569966367\n210 1.06963559835\n211 1.01638970173\n212 0.965825057948\n213 0.91780839967\n214 0.872233916761\n215 0.828936944529\n216 0.787844706919\n217 0.748810185424\n218 0.711734247058\n219 0.676516900494\n220 0.643068334746\n221 0.611308116249\n222 0.581137457877\n223 0.552687236904\n224 0.525699566311\n225 0.500057688773\n226 0.475694935078\n227 0.452550412934\n228 0.430568156872\n229 0.409673907048\n230 0.389806573715\n231 0.370932381921\n232 0.352983443987\n233 0.335920213981\n234 0.319708565298\n235 0.304296629709\n236 0.289633110114\n237 0.275692202732\n238 0.262442493992\n239 0.249835379143\n240 0.23784557329\n241 0.226452367874\n242 0.215608440517\n243 0.205291982434\n244 0.195480341079\n245 0.186148690515\n246 0.177270139497\n247 0.168825227757\n248 0.160792294175\n249 0.153144502297\n250 0.145867509368\n251 0.138945619351\n252 0.132357061522\n253 0.126087562687\n254 0.120122854459\n255 0.114444458516\n256 0.109038268771\n257 0.103892027299\n258 0.098995553132\n259 0.0943323574882\n260 0.0898952639829\n261 0.0856698048795\n262 0.0816465504529\n263 0.0778154553274\n264 0.0741678325738\n265 0.0706947627847\n266 0.0673880124412\n267 0.064238458385\n268 0.0612391624809\n269 0.0583816564353\n270 0.0556601823579\n271 0.053069427739\n272 0.0506003196627\n273 0.0482491518176\n274 0.0460090812469\n275 0.0438747302262\n276 0.0418408493085\n277 0.0399035163839\n278 0.0380575463064\n279 0.036299097922\n280 0.0346233229447\n281 0.0330262284609\n282 0.031503877848\n283 0.0300531029465\n284 0.0286707587626\n285 0.0273528067848\n286 0.0260970026726\n287 0.0249004123643\n288 0.0237589699625\n289 0.0226708018116\n290 0.021633820431\n291 0.0206448922209\n292 0.0197019772237\n293 0.0188035993027\n294 0.0179464410754\n295 0.0171291505117\n296 0.0163498514344\n297 0.0156066053911\n298 0.0148977251492\n299 0.014221758725\n300 0.0135773068144\n301 0.012962249886\n302 0.0123757145187\n303 0.0118163369769\n304 0.0112825023947\n305 0.0107732945335\n306 0.0102876221896\n307 0.00982427812751\n308 0.00938206972078\n309 0.00896026628877\n310 0.00855771803633\n311 0.00817356616853\n312 0.00780707377096\n313 0.00745720068811\n314 0.00712345076459\n315 0.00680492145638\n316 0.0065007764508\n317 0.00621051064797\n318 0.0059335532637\n319 0.00566905975894\n320 0.0054165748066\n321 0.00517568064705\n322 0.00494562595128\n323 0.00472595134395\n324 0.004516286571\n325 0.00431606748656\n326 0.00412485701368\n327 0.00394233640381\n328 0.00376798636873\n329 0.00360153498254\n330 0.0034425879938\n331 0.00329072705371\n332 0.00314572164753\n333 0.00300724330687\n334 0.00287492825219\n335 0.00274855064807\n336 0.0026278818479\n337 0.0025125794175\n338 0.00240242046594\n339 0.00229721881855\n340 0.00219666068919\n341 0.00210058824046\n342 0.00200881735521\n343 0.00192110469496\n344 0.00183730831055\n345 0.00175725174098\n346 0.00168073646648\n347 0.0016076003348\n348 0.00153771359982\n349 0.00147090447885\n350 0.0014070576511\n351 0.00134603797493\n352 0.00128770467549\n353 0.00123197628558\n354 0.00117868085568\n355 0.00112772121847\n356 0.00107901471112\n357 0.00103244508717\n358 0.000987915862711\n359 0.000945353945088\n360 0.000904654397546\n361 0.000865735304185\n362 0.000828532206376\n363 0.000792939748134\n364 0.000758904541284\n365 0.000726362131352\n366 0.000695237114628\n367 0.000665469366721\n368 0.000636998610063\n369 0.000609763414406\n370 0.000583720319318\n371 0.000558805930044\n372 0.000534968044314\n373 0.000512173467121\n374 0.000490358710071\n375 0.000469488609342\n376 0.000449526687963\n377 0.000430422479868\n378 0.000412146230894\n379 0.00039466322235\n380 0.000377932326339\n381 0.000361920938159\n382 0.00034659873668\n383 0.000331934151076\n384 0.000317903619901\n385 0.000304474691838\n386 0.000291623739083\n387 0.000279323742851\n388 0.000267548222855\n389 0.000256278541801\n390 0.000245493571213\n391 0.000235166229351\n392 0.000225284297347\n393 0.00021582135579\n394 0.000206761919814\n395 0.00019809028382\n396 0.000189786130103\n397 0.000181836438281\n398 0.00017422642703\n399 0.000166938234581\n400 0.000159960525744\n401 0.000153278944243\n402 0.000146880325055\n403 0.000140753610841\n404 0.000134885748538\n405 0.000129266522935\n406 0.000123884484981\n407 0.000118729332324\n408 0.000113793045321\n409 0.000109064256962\n410 0.000104535895706\n411 0.000100198028952\n412 9.60419382144e-05\n413 9.20618879821e-05\n414 8.82483093451e-05\n415 8.45950482713e-05\n416 8.10960474992e-05\n417 7.77430445771e-05\n418 7.45311414851e-05\n419 7.14534254255e-05\n420 6.85046590699e-05\n421 6.56795112e-05\n422 6.29724733817e-05\n423 6.03785890755e-05\n424 5.7892722874e-05\n425 5.55106718637e-05\n426 5.32282361494e-05\n427 5.10406825078e-05\n428 4.89448352152e-05\n429 4.6935677075e-05\n430 4.50100354238e-05\n431 4.31646099667e-05\n432 4.13956138296e-05\n433 3.97004647487e-05\n434 3.80755850664e-05\n435 3.65178860272e-05\n436 3.50247961582e-05\n437 3.35933714026e-05\n438 3.22215532328e-05\n439 3.09063452968e-05\n440 2.9645585819e-05\n441 2.84367405606e-05\n442 2.72777668962e-05\n443 2.61667329407e-05\n444 2.51013501797e-05\n445 2.40802574239e-05\n446 2.31009022636e-05\n447 2.21618921174e-05\n448 2.12615223444e-05\n449 2.03981352425e-05\n450 1.95703954971e-05\n451 1.877653602e-05\n452 1.8015266419e-05\n453 1.72851702352e-05\n454 1.65850141998e-05\n455 1.59137366858e-05\n456 1.52708808101e-05\n457 1.46533379366e-05\n458 1.40608956291e-05\n459 1.34927646819e-05\n460 1.29478110483e-05\n461 1.24251453509e-05\n"
]
],
[
[
"# PyTorch Tensors\n\nClearly modern deep neural networks are in need of more than what our beloved numpy can offer.\n\nHere we introduce the most fundamental PyTorch concept: the *Tensor*. A PyTorch Tensor is conceptually identical to a numpy array: a Tensor is an n-dimensional array, and PyTorch provides many functions for operating on these Tensors. Like numpy arrays, PyTorch Tensors do not know anything about deep learning or computational graphs or gradients; they are a generic tool for scientific computing.\n\nHowever unlike numpy, PyTorch Tensors can utilize GPUs to accelerate their numeric computations. To run a PyTorch Tensor on GPU, you simply need to cast it to a new datatype.\n\nHere we use PyTorch Tensors to fit a two-layer network to random data. Like the numpy example above we need to manually implement the forward and backward passes through the network:",
"_____no_output_____"
]
],
[
[
"import torch\ndtype = torch.FloatTensor\n# dtype = torch.cuda.FloatTensor # Uncomment this to run on GPU\n\n# N is batch size; D_in is input dimension;\n# H is hidden dimension; D_out is output dimension.\nN, D_in, H, D_out = 64, 1000, 100, 10\n\n# Create random input and output data\nx = torch.randn(N, D_in).type(dtype)\ny = torch.randn(N, D_out).type(dtype)\n\n# Randomly initialize weights\nw1 = torch.randn(D_in, H).type(dtype)\nw2 = torch.randn(H, D_out).type(dtype)\n\nlearning_rate = 1e-6\nfor t in range(500):\n # Forward pass: compute predicted y\n h = x.mm(w1)\n h_relu = h.clamp(min=0)\n y_pred = h_relu.mm(w2)\n\n # Compute and print loss\n loss = (y_pred - y).pow(2).sum()\n print(t, loss)\n\n # Backprop to compute gradients of w1 and w2 with respect to loss\n grad_y_pred = 2.0 * (y_pred - y)\n grad_w2 = h_relu.t().mm(grad_y_pred)\n grad_h_relu = grad_y_pred.mm(w2.t())\n grad_h = grad_h_relu.clone()\n grad_h[h < 0] = 0\n grad_w1 = x.t().mm(grad_h)\n\n # Update weights using gradient descent\n w1 -= learning_rate * grad_w1\n w2 -= learning_rate * grad_w2",
"0 28214897.691271067\n1 25380405.792548403\n2 26288556.067442656\n3 27187362.93774879\n4 25326431.49736169\n5 20070726.423171997\n6 13438367.445337629\n7 7935834.941528201\n8 4453037.240495725\n9 2567232.1655493514\n10 1604364.933374187\n11 1106295.9881061036\n12 831370.3628886025\n13 664479.3320915042\n14 552383.0191260207\n15 470307.21917449264\n16 406323.70261433884\n17 354377.92758273566\n18 311124.330613622\n19 274515.3858363455\n20 243215.3152763464\n21 216254.64485477417\n22 192876.48988408546\n23 172511.55349881982\n24 154696.59197369026\n25 139077.64419030334\n26 125326.40331724554\n27 113168.27359832195\n28 102388.44114990594\n29 92802.5217316554\n30 84252.873688431\n31 76614.83165994265\n32 69777.57502200827\n33 63643.55059441269\n34 58122.45377116208\n35 53149.5297017009\n36 48661.48595352931\n37 44605.11924878636\n38 40936.86403570355\n39 37612.1624785422\n40 34589.84976270138\n41 31842.023658028404\n42 29339.426460701798\n43 27055.76113430076\n44 24971.357019224655\n45 23066.443739543673\n46 21322.47401335786\n47 19723.635119302293\n48 18257.593847584038\n49 16911.857851812914\n50 15676.901621120574\n51 14541.234468931158\n52 13495.409479309936\n53 12531.688689953091\n54 11642.912229433834\n55 10823.396586809435\n56 10066.756259321584\n57 9368.032180714887\n58 8722.69606901206\n59 8125.648766315389\n60 7573.201240118957\n61 7061.110367171321\n62 6586.37116674181\n63 6146.2882079665205\n64 5738.038465707713\n65 5359.140022478372\n66 5007.218182836571\n67 4679.821472426938\n68 4375.545563822141\n69 4092.5672241640546\n70 3829.2538478331044\n71 3583.9982694811915\n72 3355.5044128053214\n73 3142.522992099788\n74 2944.0477814121896\n75 2758.9096522632453\n76 2586.098822437947\n77 2424.783757172412\n78 2274.162424382146\n79 2133.472267201043\n80 2001.943758391455\n81 1879.0327707577635\n82 1764.1420179859847\n83 1656.6765891071607\n84 1556.0836619963645\n85 1461.960692876407\n86 1373.8333980444747\n87 1291.3251499255507\n88 1214.077519632569\n89 1141.6407961478803\n90 1073.7122116708274\n91 1010.047731572995\n92 950.3514467849104\n93 894.3273352336082\n94 841.7842243861196\n95 792.4734904819334\n96 746.1964596283701\n97 702.7443149700078\n98 661.9300986860596\n99 623.5698773736967\n100 587.5490628035759\n101 553.7059624342619\n102 521.87459074208\n103 491.95096067483627\n104 463.81437045894427\n105 437.3619707183019\n106 412.4634959739533\n107 389.03197571304185\n108 366.98684185984854\n109 346.2511910920458\n110 326.7168373228138\n111 308.3204062757866\n112 291.0116837719783\n113 274.708204616996\n114 259.3530469133465\n115 244.89019768539188\n116 231.2530311334451\n117 218.40625489775357\n118 206.29494907575645\n119 194.87408803031087\n120 184.10918123054637\n121 173.95670181258504\n122 164.3796226045149\n123 155.34510760042053\n124 146.82159396161046\n125 138.78259947243896\n126 131.19334880439965\n127 124.03206751540091\n128 117.27145023435516\n129 110.8922230492454\n130 104.86954096430226\n131 99.17821632714708\n132 93.80321977845797\n133 88.73180206294792\n134 83.93784170194142\n135 79.41110559695994\n136 75.13158900665832\n137 71.09239467909009\n138 67.27303860367512\n139 63.661083760649944\n140 60.24877063365615\n141 57.02609438798197\n142 53.97754052526591\n143 51.095064315871184\n144 48.37058476978203\n145 45.794669434952766\n146 43.357149485835066\n147 41.053389754456276\n148 38.87377426878407\n149 36.81266361362863\n150 34.86269390504242\n151 33.019418138638315\n152 31.27372891445308\n153 29.623019411905716\n154 28.06002445682043\n155 26.58085827334935\n156 25.182066968294635\n157 23.856794249429644\n158 22.603709343965306\n159 21.416727958537756\n160 20.294056803979785\n161 19.230226081371868\n162 18.22286712818012\n163 17.26986351281531\n164 16.36696748965665\n165 15.512043060681435\n166 14.70277965469339\n167 13.936395793035047\n168 13.21058501636503\n169 12.522788125846773\n170 11.871329149475358\n171 11.254089594353673\n172 10.669772994995135\n173 10.115961444046548\n174 9.591341026183215\n175 9.094685662630582\n176 8.623675345308872\n177 8.177212815510206\n178 7.754271122965591\n179 7.354052512118528\n180 6.974105103205304\n181 6.613862763094033\n182 6.273167739637028\n183 5.949956651557034\n184 5.643680276344654\n185 5.353149802081873\n186 5.077408776123896\n187 4.8164800713806315\n188 4.568759942421966\n189 4.334537105201893\n190 4.112015826773195\n191 3.9009179414881707\n192 3.7012154731272986\n193 3.511612145634661\n194 3.331681329765537\n195 3.1611259816769106\n196 2.9996718148188464\n197 2.8461790457236766\n198 2.7007109757500025\n199 2.562890156220522\n200 2.4321240546360414\n201 2.308078948186587\n202 2.1904870139545665\n203 2.0787757790351513\n204 1.972721255352237\n205 1.8724816279031096\n206 1.776974327720918\n207 1.6867990743287722\n208 1.601016306899063\n209 1.5197483114327683\n210 1.442438605099003\n211 1.369157533522884\n212 1.2998218626227995\n213 1.2339273899186163\n214 1.17146151531626\n215 1.1119642766772915\n216 1.0557099815853666\n217 1.0022163466049716\n218 0.9514815204819733\n219 0.9033794087224507\n220 0.8576643044382202\n221 0.8143504655566967\n222 0.7732258198716373\n223 0.7342158760394923\n224 0.6971644104382229\n225 0.6619967066271535\n226 0.6285948940725881\n227 0.5968096996362284\n228 0.5667985106167974\n229 0.5382560311909526\n230 0.5111128765857158\n231 0.48532747688128897\n232 0.4609265227778163\n233 0.4378205148075356\n234 0.4157447156268157\n235 0.39488582392669613\n236 0.3749829100757234\n237 0.35613537196222556\n238 0.3382650005067456\n239 0.32128029946794356\n240 0.30518656196033334\n241 0.2898877071115251\n242 0.2753985487457893\n243 0.26155083612243324\n244 0.2484203549989168\n245 0.23601150551252115\n246 0.22414258684202437\n247 0.21293119006192796\n248 0.20228290632133167\n249 0.19217315565631465\n250 0.18254562652399353\n251 0.17339564711978817\n252 0.16472807684149715\n253 0.15650172744047652\n254 0.14871153441717966\n255 0.141252334180054\n256 0.13420798837495873\n257 0.1275148040973093\n258 0.12115617519511047\n259 0.11513308130563794\n260 0.10940513697614086\n261 0.10392053471471474\n262 0.09873369084591621\n263 0.0938313782580984\n264 0.08916601624925224\n265 0.08473324384660685\n266 0.08052892574508519\n267 0.0765146490751043\n268 0.07271481811263403\n269 0.06908563553494673\n270 0.06569151383855609\n271 0.06243397875781054\n272 0.059288574380887527\n273 0.056329070592443964\n274 0.053548219642252315\n275 0.050900103240740124\n276 0.04838548394463982\n277 0.0459671132623376\n278 0.0437052950628003\n279 0.04152813333603267\n280 0.03946723512516925\n281 0.037533394479267956\n282 0.03568910868574027\n283 0.03390143972920545\n284 0.03224024862347741\n285 0.030643039031359953\n286 0.029140048215710534\n287 0.027684004166977916\n288 0.026314751884853438\n289 0.025033289714943285\n290 0.023797433226710796\n291 0.02262336258232206\n292 0.021504991155510966\n293 0.02045539104525429\n294 0.01946322911578169\n295 0.018508058725132337\n296 0.017591603927018307\n297 0.0167319201745022\n298 0.01589690324248605\n299 0.015139217233493873\n300 0.014406465492699028\n301 0.013696118249792555\n302 0.013027044818570366\n303 0.012396137516559824\n304 0.011789959562407637\n305 0.011218404833911066\n306 0.010683310567349003\n307 0.01017237445438407\n308 0.009680823881585532\n309 0.009207409667161714\n310 0.008765478168209662\n311 0.008343950057827287\n312 0.007942077827435279\n313 0.007565309388805952\n314 0.007207875892412341\n315 0.006865805795088331\n316 0.006540033828446479\n317 0.006230358566105432\n318 0.005935793194971506\n319 0.0056569668626278435\n320 0.005392627609972722\n321 0.005142164502647928\n322 0.004903280543345323\n323 0.0046710665883187286\n324 0.0044596153714386855\n325 0.004254983110094868\n326 0.004053112385123736\n327 0.0038716554215878496\n328 0.003693314754828869\n329 0.0035276847824143864\n330 0.0033722945089047496\n331 0.003224334560254949\n332 0.0030797497759889048\n333 0.0029394659957044933\n334 0.002811574650088522\n335 0.002687472415199954\n336 0.0025703362047908573\n337 0.002459747191192241\n338 0.002356102818997119\n339 0.002255833670969709\n340 0.002162025463318118\n341 0.0020706938958603427\n342 0.0019804429030327864\n343 0.001900624922958949\n344 0.0018219768719105467\n345 0.0017470823932357327\n346 0.00167403269153521\n347 0.0016074784101616224\n348 0.0015434483287186662\n349 0.0014800083688997212\n350 0.0014237042363174357\n351 0.0013673234525823919\n352 0.001313357088474909\n353 0.001260321802043718\n354 0.0012110123492108382\n355 0.0011649015229149295\n356 0.0011204107047679823\n357 0.0010796027719008894\n358 0.0010387537130391866\n359 0.0010000977633572994\n360 0.0009621068961130352\n361 0.0009280829840783711\n362 0.0008925718210586187\n363 0.000861557630299048\n364 0.0008311890107142728\n365 0.0008009163000355368\n366 0.0007749579994773548\n367 0.0007484465107412408\n368 0.0007222093569896337\n369 0.0006973693140788217\n370 0.0006740144106807122\n371 0.0006497241727248526\n372 0.0006289278786107411\n373 0.0006074793578099702\n374 0.0005876308963297938\n375 0.0005683213433220757\n376 0.0005498372268836205\n377 0.0005331871459254289\n378 0.000514447040892041\n379 0.0004995928681320039\n380 0.0004830170838928116\n381 0.00046774268823562837\n382 0.0004539691694929737\n383 0.000439916381955785\n384 0.0004264477815108525\n385 0.00041461180957180765\n386 0.0004007517174830638\n387 0.0003885748281060031\n388 0.00037587470802746825\n389 0.00036685178849288347\n390 0.0003563211267975097\n391 0.00034580960975350017\n392 0.0003373833671450055\n393 0.00032734962350788877\n394 0.0003172536302761264\n395 0.0003087773417211892\n396 0.0003003043221507795\n397 0.00029157922913469747\n398 0.0002838640100005785\n399 0.0002773871556948082\n400 0.0002693207433572403\n401 0.00026274296119056795\n402 0.00025629517436694116\n403 0.00024865622459442627\n404 0.00024191564002837285\n405 0.00023580017504160056\n406 0.00023021821139053433\n407 0.00022492894727565993\n408 0.0002187235492969869\n409 0.0002130760149380989\n410 0.00020846465683301008\n411 0.00020339997672215449\n412 0.00019872765675885856\n413 0.0001944256389800475\n414 0.00019000826152877626\n415 0.00018525978015550282\n416 0.00018077204148654602\n417 0.00017703581449693417\n418 0.00017288089631457837\n419 0.00016859326936614905\n420 0.00016520088735737237\n421 0.00016153575890172356\n422 0.00015850395108210624\n423 0.00015459131808931437\n424 0.00015143964321755188\n425 0.00014752541333357128\n426 0.00014445116156402982\n427 0.000141356974335205\n428 0.00013808374274071355\n429 0.0001353787969475273\n430 0.00013243990439421038\n431 0.00012966755098536842\n432 0.00012675479128350375\n433 0.0001242446317876872\n434 0.00012147723341672523\n435 0.00011897251544801257\n436 0.00011670839108181286\n437 0.00011433415206264785\n438 0.00011219214203966876\n439 0.00011027887981625295\n440 0.00010749669199967837\n441 0.00010553591091314041\n442 0.00010367609742491235\n443 0.00010109258945595334\n444 9.932886336866398e-05\n445 9.749088251564952e-05\n446 9.520718197066069e-05\n447 9.345653584887093e-05\n448 9.182967794658936e-05\n449 8.988625574320175e-05\n450 8.847123125822753e-05\n451 8.713054826821331e-05\n452 8.549155933869346e-05\n"
]
],
[
[
"# Autograd\nPyTorch variables and autograd. Autograd package provides cool functionality as the forward pass of your network defines the computational graph; nodes in the graph will be Tensors and edges will be functions that produce output Tensors from input Tensors. Backprop through this graph then allows us to easily compue gradients.\n\nHere we wrap the PyTorch Tensor in a Variable object; where Vaiabel represents a node in the computational graph. if x is a variable then x.data is a Tensor and x.grad is another Varialble holding the gradient of x w.r.t to some scalar value.\n\nPyTorch Variables have samer API as PyTorch Tensots: any operation that you can do with Tensor, also works fine with Variables, difference only being that the Variable defines a computational graph, allowing us to automatically compute gradients.",
"_____no_output_____"
]
],
[
[
"# Use of Vaiables and Autograd in a 2-layer network with no need to manually implement backprop!\nimport torch\nfrom torch.autograd import Variable\ndtype = torch.FloatTensor\n\n# N is batch size; D_in is input dimension;\n# H is hidden dimension; D_out is output dimension.\nN, D_in, H, D_out = 64, 1000, 100, 10\n\n# Create random Tensors to hold input and outputs and wrap them in Variables.\n\nx = Variable(torch.randn(N, D_in).type(dtype), requires_grad=False) # requires_grad=False means no need to compute gradients\ny = Variable(torch.randn(N, D_out).type(dtype), requires_grad=False)\n\n# Create random Tensors to hold weights and wrap them in Variables.\n# requires_grad=True here to compute gradients w.r.t Variables during a backprop pass.\n\nw1 = Variable(torch.randn(D_in, H).type(dtype), requires_grad=True) # requires_grad=False means no need to compute gradients\nw2 = Variable(torch.randn(H, D_out).type(dtype), requires_grad=True)\n\nlearning_rate = 1e-6\nfor t in range(500):\n # Forward pass: compute predicted y using operations on Variables; these\n # are exactly the same operations we used to compute the forward pass using\n # Tensors, but we do not need to keep references to intermediate values since\n # we are not implementing the backward pass by hand.\n y_pred = x.mm(w1).clamp(min=0).mm(w2)\n\n # Compute and print loss using operations on Variables.\n # Now loss is a Variable of shape (1,) and loss.data is a Tensor of shape\n # (1,); loss.data[0] is a scalar value holding the loss.\n loss = (y_pred - y).pow(2).sum()\n print(t, loss.data[0])\n\n # Use autograd to compute the backward pass. This call will compute the\n # gradient of loss with respect to all Variables with requires_grad=True.\n # After this call w1.grad and w2.grad will be Variables holding the gradient\n # of the loss with respect to w1 and w2 respectively.\n loss.backward()\n\n # Update weights using gradient descent; w1.data and w2.data are Tensors,\n # w1.grad and w2.grad are Variables and w1.grad.data and w2.grad.data are\n # Tensors.\n w1.data -= learning_rate * w1.grad.data\n w2.data -= learning_rate * w2.grad.data\n\n # Manually zero the gradients after updating weights\n w1.grad.data.zero_()\n w2.grad.data.zero_()",
"0 35878500.0\n1 33502642.0\n2 31638146.0\n3 26216880.0\n4 18097450.0\n5 10643111.0\n6 5868223.0\n7 3356485.0\n8 2129793.5\n9 1508282.875\n10 1160753.375\n11 940967.3125\n12 785975.375\n13 668166.125\n14 574389.8125\n15 497736.9375\n16 433985.8125\n17 380330.0\n18 334801.71875\n19 295919.6875\n20 262469.40625\n21 233624.078125\n22 208602.84375\n23 186785.21875\n24 167705.6875\n25 150947.25\n26 136179.03125\n27 123118.4375\n28 111543.015625\n29 101252.5\n30 92084.828125\n31 83890.2109375\n32 76550.203125\n33 69970.4609375\n34 64056.62109375\n35 58728.921875\n36 53917.6015625\n37 49565.42578125\n38 45631.03515625\n39 42059.48046875\n40 38813.0390625\n41 35858.09765625\n42 33163.74609375\n43 30702.73828125\n44 28452.41796875\n45 26393.0234375\n46 24505.55078125\n47 22772.90234375\n48 21181.724609375\n49 19717.416015625\n50 18369.517578125\n51 17127.080078125\n52 15980.390625\n53 14921.2587890625\n54 13942.4697265625\n55 13036.6015625\n56 12197.0810546875\n57 11419.048828125\n58 10696.9755859375\n59 10026.7861328125\n60 9403.92578125\n61 8824.4482421875\n62 8285.025390625\n63 7782.73583984375\n64 7314.56591796875\n65 6878.09619140625\n66 6470.642578125\n67 6090.18603515625\n68 5734.34912109375\n69 5401.63623046875\n70 5090.5068359375\n71 4799.25390625\n72 4526.57177734375\n73 4271.24853515625\n74 4031.948974609375\n75 3807.61865234375\n76 3597.07275390625\n77 3399.272216796875\n78 3213.479248046875\n79 3038.89013671875\n80 2874.7353515625\n81 2720.358642578125\n82 2575.041015625\n83 2438.2685546875\n84 2309.4228515625\n85 2188.03125\n86 2073.676513671875\n87 1965.752685546875\n88 1863.9710693359375\n89 1767.9381103515625\n90 1677.3052978515625\n91 1591.765869140625\n92 1510.904541015625\n93 1434.5537109375\n94 1362.3419189453125\n95 1294.068115234375\n96 1229.5435791015625\n97 1168.46240234375\n98 1110.672607421875\n99 1055.9576416015625\n100 1004.15771484375\n101 955.1239013671875\n102 908.6265869140625\n103 864.5731811523438\n104 822.8087158203125\n105 783.23779296875\n106 745.7089233398438\n107 710.0859985351562\n108 676.2913818359375\n109 644.2283935546875\n110 613.8003540039062\n111 584.9096069335938\n112 557.466796875\n113 531.3948364257812\n114 506.61962890625\n115 483.0841369628906\n116 460.7171936035156\n117 439.4493408203125\n118 419.2239074707031\n119 399.9928283691406\n120 381.705322265625\n121 364.3070373535156\n122 347.7393798828125\n123 331.97283935546875\n124 316.9671630859375\n125 302.68853759765625\n126 289.0873718261719\n127 276.12823486328125\n128 263.7820129394531\n129 252.02195739746094\n130 240.82232666015625\n131 230.1421661376953\n132 219.9631805419922\n133 210.26112365722656\n134 201.00880432128906\n135 192.19129943847656\n136 183.77542114257812\n137 175.74905395507812\n138 168.08860778808594\n139 160.7852325439453\n140 153.8109893798828\n141 147.15760803222656\n142 140.80355834960938\n143 134.73838806152344\n144 128.94873046875\n145 123.42112731933594\n146 118.14532470703125\n147 113.09927368164062\n148 108.27923583984375\n149 103.67829132080078\n150 99.27864837646484\n151 95.07437133789062\n152 91.05731964111328\n153 87.2179183959961\n154 83.54911804199219\n155 80.0405044555664\n156 76.684814453125\n157 73.47515106201172\n158 70.40677642822266\n159 67.47286224365234\n160 64.66546630859375\n161 61.97762680053711\n162 59.40721893310547\n163 56.94810485839844\n164 54.59606170654297\n165 52.34416961669922\n166 50.18893814086914\n167 48.12546157836914\n168 46.15074920654297\n169 44.2588996887207\n170 42.44817352294922\n171 40.71442794799805\n172 39.05426788330078\n173 37.46466064453125\n174 35.94228744506836\n175 34.483211517333984\n176 33.08485794067383\n177 31.74663734436035\n178 30.463415145874023\n179 29.23353385925293\n180 28.055572509765625\n181 26.927295684814453\n182 25.845623016357422\n183 24.808975219726562\n184 23.814783096313477\n185 22.86162757873535\n186 21.947311401367188\n187 21.071706771850586\n188 20.23161506652832\n189 19.426319122314453\n190 18.65383529663086\n191 17.913501739501953\n192 17.202938079833984\n193 16.521442413330078\n194 15.867642402648926\n195 15.240697860717773\n196 14.638861656188965\n197 14.062265396118164\n198 13.50815200805664\n199 12.976459503173828\n200 12.466854095458984\n201 11.977287292480469\n202 11.508007049560547\n203 11.057541847229004\n204 10.624938011169434\n205 10.209487915039062\n206 9.811256408691406\n207 9.42843246459961\n208 9.060935020446777\n209 8.708433151245117\n210 8.369855880737305\n211 8.044754028320312\n212 7.732644081115723\n213 7.432569980621338\n214 7.144754886627197\n215 6.868185997009277\n216 6.602710723876953\n217 6.347681522369385\n218 6.103111267089844\n219 5.8678483963012695\n220 5.64181661605835\n221 5.424355506896973\n222 5.215670108795166\n223 5.015231132507324\n224 4.822762966156006\n225 4.637856483459473\n226 4.460170269012451\n227 4.28932523727417\n228 4.125042915344238\n229 3.967371940612793\n230 3.8158133029937744\n231 3.670203924179077\n232 3.530052661895752\n233 3.3955368995666504\n234 3.2662713527679443\n235 3.141659736633301\n236 3.022263526916504\n237 2.9074623584747314\n238 2.7971487045288086\n239 2.6909444332122803\n240 2.5889720916748047\n241 2.4906997680664062\n242 2.3964099884033203\n243 2.305689573287964\n244 2.2184183597564697\n245 2.134580612182617\n246 2.053964376449585\n247 1.9763656854629517\n248 1.9018102884292603\n249 1.830053687095642\n250 1.7611491680145264\n251 1.6948093175888062\n252 1.6310014724731445\n253 1.5695786476135254\n254 1.5105568170547485\n255 1.453961730003357\n256 1.3993768692016602\n257 1.3468921184539795\n258 1.296314001083374\n259 1.2476303577423096\n260 1.2009528875350952\n261 1.1559319496154785\n262 1.1127043962478638\n263 1.071056604385376\n264 1.0309712886810303\n265 0.9924764037132263\n266 0.9553502202033997\n267 0.9197673797607422\n268 0.8854739665985107\n269 0.8523190021514893\n270 0.8206137418746948\n271 0.7899705767631531\n272 0.7606277465820312\n273 0.73226398229599\n274 0.7050141096115112\n275 0.678828239440918\n276 0.6536497473716736\n277 0.6293545365333557\n278 0.6060177683830261\n279 0.5834728479385376\n280 0.561832070350647\n281 0.5409616231918335\n282 0.5209071636199951\n283 0.5015677213668823\n284 0.48301637172698975\n285 0.46511176228523254\n286 0.4478737413883209\n287 0.43125540018081665\n288 0.4153783321380615\n289 0.3999677896499634\n290 0.3852301836013794\n291 0.3709765374660492\n292 0.3573264181613922\n293 0.34412410855293274\n294 0.3314245641231537\n295 0.3191765248775482\n296 0.3074215054512024\n297 0.296056866645813\n298 0.28519463539123535\n299 0.2746979594230652\n300 0.26459330320358276\n301 0.25485098361968994\n302 0.24548396468162537\n303 0.23641419410705566\n304 0.22769007086753845\n305 0.21935638785362244\n306 0.21129179000854492\n307 0.20352837443351746\n308 0.19601596891880035\n309 0.18884600698947906\n310 0.1819111704826355\n311 0.17525847256183624\n312 0.16882126033306122\n313 0.16263249516487122\n314 0.15666751563549042\n315 0.15092256665229797\n316 0.14540348947048187\n317 0.14006322622299194\n318 0.13494110107421875\n319 0.13001160323619843\n320 0.12526486814022064\n321 0.1206771731376648\n322 0.11626103520393372\n323 0.11202862858772278\n324 0.10792234539985657\n325 0.10398980975151062\n326 0.1001921221613884\n327 0.09651487320661545\n328 0.09299999475479126\n329 0.08962738513946533\n330 0.08636265993118286\n331 0.08319984376430511\n332 0.08016426116228104\n333 0.07726199924945831\n334 0.07444174587726593\n335 0.07173093408346176\n336 0.0690966546535492\n337 0.06658675521612167\n338 0.06416875869035721\n339 0.06185092404484749\n340 0.05959108844399452\n341 0.057433173060417175\n342 0.0553474947810173\n343 0.053334783762693405\n344 0.051402702927589417\n345 0.04955539479851723\n346 0.04775090888142586\n347 0.04602515324950218\n348 0.044351741671562195\n349 0.04274814575910568\n350 0.041199490427970886\n351 0.03970283269882202\n352 0.03825933113694191\n353 0.036878347396850586\n354 0.0355573333799839\n355 0.03427095338702202\n356 0.03304218873381615\n357 0.03185059875249863\n358 0.03069448284804821\n359 0.029578279703855515\n360 0.028519731014966965\n361 0.02748997136950493\n362 0.026511413976550102\n363 0.025562353432178497\n364 0.02463531121611595\n365 0.023760242387652397\n366 0.02290988340973854\n367 0.022078199312090874\n368 0.021282397210597992\n369 0.020530449226498604\n370 0.019799597561359406\n371 0.019093159586191177\n372 0.018408171832561493\n373 0.017752142623066902\n374 0.017124634236097336\n375 0.0165147352963686\n376 0.0159267857670784\n377 0.015361725352704525\n378 0.014817671850323677\n379 0.014293329790234566\n380 0.013794535771012306\n381 0.013302133418619633\n382 0.012831700965762138\n383 0.012388093397021294\n384 0.011951521039009094\n385 0.011539716273546219\n386 0.011130605824291706\n387 0.010747026652097702\n388 0.010368922725319862\n389 0.010003476403653622\n390 0.009659701958298683\n391 0.009321259334683418\n392 0.009002749808132648\n393 0.008687980473041534\n394 0.008389437571167946\n395 0.008094895631074905\n396 0.00781853124499321\n397 0.00755091430619359\n398 0.007287600077688694\n399 0.007037787232547998\n400 0.006801604758948088\n"
]
],
[
[
"# PyTorch: Defining new autograd functions\nUnder the hood, each primitive autograd operator is really two functions that operate on Tensors. The forward function computes output Tensors from input Tensors. The backward function receives the gradient of the output Tensors with respect to some scalar value, and computes the gradient of the input Tensors with respect to that same scalar value.\n\nIn PyTorch we can easily define our own autograd operator by defining a subclass of torch.autograd.Function and implementing the forward and backward functions. We can then use our new autograd operator by constructing an instance and calling it like a function, passing Variables containing input data.\n\nIn this example we define our own custom autograd function for performing the ReLU nonlinearity, and use it to implement our two-layer network:\n\n",
"_____no_output_____"
]
],
[
[
"# -*- coding: utf-8 -*-\nimport torch\nfrom torch.autograd import Variable\n\n\nclass MyReLU(torch.autograd.Function):\n \"\"\"\n We can implement our own custom autograd Functions by subclassing\n torch.autograd.Function and implementing the forward and backward passes\n which operate on Tensors.\n \"\"\"\n\n def forward(self, input):\n \"\"\"\n In the forward pass we receive a Tensor containing the input and return a\n Tensor containing the output. You can cache arbitrary Tensors for use in the\n backward pass using the save_for_backward method.\n \"\"\"\n self.save_for_backward(input)\n return input.clamp(min=0)\n\n def backward(self, grad_output):\n \"\"\"\n In the backward pass we receive a Tensor containing the gradient of the loss\n with respect to the output, and we need to compute the gradient of the loss\n with respect to the input.\n \"\"\"\n input, = self.saved_tensors\n grad_input = grad_output.clone()\n grad_input[input < 0] = 0\n return grad_input\n\n\ndtype = torch.FloatTensor\n# dtype = torch.cuda.FloatTensor # Uncomment this to run on GPU\n\n# N is batch size; D_in is input dimension;\n# H is hidden dimension; D_out is output dimension.\nN, D_in, H, D_out = 64, 1000, 100, 10\n\n# Create random Tensors to hold input and outputs, and wrap them in Variables.\nx = Variable(torch.randn(N, D_in).type(dtype), requires_grad=False)\ny = Variable(torch.randn(N, D_out).type(dtype), requires_grad=False)\n\n# Create random Tensors for weights, and wrap them in Variables.\nw1 = Variable(torch.randn(D_in, H).type(dtype), requires_grad=True)\nw2 = Variable(torch.randn(H, D_out).type(dtype), requires_grad=True)\n\nlearning_rate = 1e-6\nfor t in range(500):\n # Construct an instance of our MyReLU class to use in our network\n relu = MyReLU()\n\n # Forward pass: compute predicted y using operations on Variables; we compute\n # ReLU using our custom autograd operation.\n y_pred = relu(x.mm(w1)).mm(w2)\n\n # Compute and print loss\n loss = (y_pred - y).pow(2).sum()\n print(t, loss.data[0])\n\n # Use autograd to compute the backward pass.\n loss.backward()\n\n # Update weights using gradient descent\n w1.data -= learning_rate * w1.grad.data\n w2.data -= learning_rate * w2.grad.data\n\n # Manually zero the gradients after updating weights\n w1.grad.data.zero_()\n w2.grad.data.zero_()",
"0 37267740.0\n1 35764716.0\n2 35199480.0\n3 30134798.0\n4 20876230.0\n5 11940865.0\n6 6248357.5\n7 3411474.25\n8 2109929.75\n9 1486262.5\n10 1147416.0\n11 933659.25\n12 781843.3125\n13 665453.875\n14 572186.3125\n15 495587.15625\n16 431790.71875\n17 378165.53125\n18 332707.5\n19 293927.75\n20 260602.078125\n21 231805.1875\n22 206837.75\n23 185119.71875\n24 166225.28125\n25 149666.796875\n26 135110.765625\n27 122273.4140625\n28 110904.5703125\n29 100806.375\n30 91813.03125\n31 83787.9921875\n32 76605.484375\n33 70168.1953125\n34 64381.640625\n35 59169.4453125\n36 54462.21875\n37 50205.0546875\n38 46347.6328125\n39 42845.19921875\n40 39659.359375\n41 36757.4609375\n42 34109.640625\n43 31690.05078125\n44 29476.259765625\n45 27446.322265625\n46 25583.3671875\n47 23871.591796875\n48 22297.443359375\n49 20848.37109375\n50 19513.296875\n51 18281.951171875\n52 17144.08203125\n53 16093.103515625\n54 15118.5703125\n55 14213.7685546875\n56 13372.318359375\n57 12589.6640625\n58 11861.248046875\n59 11182.630859375\n60 10549.470703125\n61 9958.626953125\n62 9406.4560546875\n63 8890.7177734375\n64 8409.7861328125\n65 7959.3427734375\n66 7536.99755859375\n67 7140.72119140625\n68 6768.61767578125\n69 6418.8779296875\n70 6090.21875\n71 5781.171875\n72 5489.9794921875\n73 5215.68603515625\n74 4957.0380859375\n75 4713.0322265625\n76 4482.77490234375\n77 4265.33984375\n78 4059.960205078125\n79 3865.8935546875\n80 3682.364990234375\n81 3508.66455078125\n82 3344.263427734375\n83 3188.444580078125\n84 3040.791015625\n85 2900.77099609375\n86 2767.92041015625\n87 2641.862060546875\n88 2522.1953125\n89 2408.60107421875\n90 2300.690185546875\n91 2198.137451171875\n92 2100.65966796875\n93 2007.908447265625\n94 1919.6483154296875\n95 1835.62548828125\n96 1755.661865234375\n97 1679.4940185546875\n98 1606.9317626953125\n99 1537.7978515625\n100 1471.900146484375\n101 1409.1009521484375\n102 1349.2012939453125\n103 1292.034423828125\n104 1237.503662109375\n105 1185.4390869140625\n106 1135.744384765625\n107 1088.301513671875\n108 1042.9739990234375\n109 999.6783447265625\n110 958.3234252929688\n111 918.8175659179688\n112 881.0344848632812\n113 844.9067993164062\n114 810.3673095703125\n115 777.3306274414062\n116 745.7218627929688\n117 715.4826049804688\n118 686.55517578125\n119 658.874755859375\n120 632.377197265625\n121 606.9979858398438\n122 582.705322265625\n123 559.4349975585938\n124 537.1435546875\n125 515.7855834960938\n126 495.33056640625\n127 475.72637939453125\n128 456.9494323730469\n129 438.9481506347656\n130 421.68475341796875\n131 405.1681823730469\n132 389.3519287109375\n133 374.1879577636719\n134 359.64312744140625\n135 345.6932678222656\n136 332.3123779296875\n137 319.4747009277344\n138 307.150634765625\n139 295.32281494140625\n140 283.9774169921875\n141 273.079833984375\n142 262.6209411621094\n143 252.57992553710938\n144 242.93710327148438\n145 233.6918487548828\n146 224.80181884765625\n147 216.26210021972656\n148 208.05758666992188\n149 200.1812744140625\n150 192.6105499267578\n151 185.34080505371094\n152 178.35305786132812\n153 171.64593505859375\n154 165.1945343017578\n155 158.99392700195312\n156 153.0364532470703\n157 147.30772399902344\n158 141.80587768554688\n159 136.51861572265625\n160 131.43280029296875\n161 126.54752349853516\n162 121.8475112915039\n163 117.32698822021484\n164 112.97860717773438\n165 108.79804992675781\n166 104.7764892578125\n167 100.90899658203125\n168 97.18679809570312\n169 93.61103820800781\n170 90.16815948486328\n171 86.85511779785156\n172 83.66851043701172\n173 80.60234832763672\n174 77.6517562866211\n175 74.81199645996094\n176 72.0802230834961\n177 69.45210266113281\n178 66.92032623291016\n179 64.4840087890625\n180 62.139137268066406\n181 59.881126403808594\n182 57.707550048828125\n183 55.61520004272461\n184 53.60150146484375\n185 51.66324996948242\n186 49.795257568359375\n187 47.99642562866211\n188 46.26518249511719\n189 44.59707260131836\n190 42.99076843261719\n191 41.4441032409668\n192 39.95560073852539\n193 38.5213623046875\n194 37.1389274597168\n195 35.80757522583008\n196 34.524593353271484\n197 33.28947067260742\n198 32.09878158569336\n199 30.953550338745117\n200 29.848520278930664\n201 28.78449249267578\n202 27.759124755859375\n203 26.77107810974121\n204 25.818552017211914\n205 24.90070152282715\n206 24.016618728637695\n207 23.165571212768555\n208 22.34392738342285\n209 21.552274703979492\n210 20.789934158325195\n211 20.05422019958496\n212 19.345535278320312\n213 18.662813186645508\n214 18.00432777404785\n215 17.369583129882812\n216 16.757579803466797\n217 16.167564392089844\n218 15.598984718322754\n219 15.05059814453125\n220 14.521815299987793\n221 14.012593269348145\n222 13.521288871765137\n223 13.04751205444336\n224 12.590230941772461\n225 12.14965534210205\n226 11.725013732910156\n227 11.314926147460938\n228 10.919958114624023\n229 10.539161682128906\n230 10.171355247497559\n231 9.817110061645508\n232 9.474847793579102\n233 9.145466804504395\n234 8.827065467834473\n235 8.520536422729492\n236 8.224677085876465\n237 7.939168453216553\n238 7.663815021514893\n239 7.398035049438477\n240 7.141916751861572\n241 6.894689083099365\n242 6.656137943267822\n243 6.42626428604126\n244 6.204166889190674\n245 5.989865303039551\n246 5.7830491065979\n247 5.583761692047119\n248 5.391234397888184\n249 5.205582618713379\n250 5.026453018188477\n251 4.853508472442627\n252 4.686666011810303\n253 4.525530815124512\n254 4.370087146759033\n255 4.220409393310547\n256 4.075485706329346\n257 3.935882568359375\n258 3.800877332687378\n259 3.6708292961120605\n260 3.5451159477233887\n261 3.423879623413086\n262 3.306863307952881\n263 3.194089651107788\n264 3.084909439086914\n265 2.979776382446289\n266 2.8781092166900635\n267 2.7799901962280273\n268 2.6851348876953125\n269 2.593947172164917\n270 2.50581693649292\n271 2.4204869270324707\n272 2.3381664752960205\n273 2.2585439682006836\n274 2.18206787109375\n275 2.1080169677734375\n276 2.036513566970825\n277 1.9676604270935059\n278 1.9008911848068237\n279 1.8365049362182617\n280 1.7742794752120972\n281 1.714285135269165\n282 1.6563860177993774\n283 1.6004620790481567\n284 1.5464372634887695\n285 1.494178295135498\n286 1.4437446594238281\n287 1.3950902223587036\n288 1.3480584621429443\n289 1.3026072978973389\n290 1.258941411972046\n291 1.2163642644882202\n292 1.1755170822143555\n293 1.1359773874282837\n294 1.0978280305862427\n295 1.06088387966156\n296 1.0252677202224731\n297 0.990941047668457\n298 0.9577256441116333\n299 0.9256399273872375\n300 0.8946157097816467\n301 0.8645932674407959\n302 0.8356277346611023\n303 0.8076440095901489\n304 0.7806621789932251\n305 0.7545725703239441\n306 0.7293268442153931\n307 0.7049664855003357\n308 0.6814430952072144\n309 0.658761739730835\n310 0.6367996335029602\n311 0.6155775785446167\n312 0.5950506925582886\n313 0.5752754211425781\n314 0.5561575889587402\n315 0.5376402139663696\n316 0.5197628736495972\n317 0.5025719404220581\n318 0.48587459325790405\n319 0.46972161531448364\n320 0.4541400969028473\n321 0.43905285000801086\n322 0.4244878590106964\n323 0.4104618728160858\n324 0.39690709114074707\n325 0.38367366790771484\n326 0.37096306681632996\n327 0.35865893959999084\n328 0.34682440757751465\n329 0.3353811800479889\n330 0.3242852985858917\n331 0.3135945498943329\n332 0.3032459616661072\n333 0.29322201013565063\n334 0.2835618853569031\n335 0.2741992473602295\n336 0.26517972350120544\n337 0.25643640756607056\n338 0.2479628622531891\n339 0.23979204893112183\n340 0.23193234205245972\n341 0.2242671400308609\n342 0.2168930619955063\n343 0.2097444087266922\n344 0.2028605192899704\n345 0.1962338536977768\n346 0.18978500366210938\n347 0.1835404634475708\n348 0.17750103771686554\n349 0.17172792553901672\n350 0.16605545580387115\n351 0.16060921549797058\n352 0.15534861385822296\n353 0.15024448931217194\n354 0.14536623656749725\n355 0.14057379961013794\n356 0.13597580790519714\n357 0.13154900074005127\n358 0.1272878497838974\n359 0.12308774888515472\n360 0.1190754845738411\n361 0.11518353223800659\n362 0.11140834540128708\n363 0.1077791303396225\n364 0.10426264256238937\n365 0.10088305175304413\n366 0.09758631885051727\n367 0.09441451728343964\n368 0.09134842455387115\n369 0.08836644142866135\n370 0.08551565557718277\n371 0.0827186331152916\n372 0.0800226628780365\n373 0.07741819322109222\n374 0.0749141052365303\n375 0.0724942609667778\n376 0.07012748718261719\n377 0.06782606244087219\n378 0.06563150137662888\n379 0.06352625042200089\n380 0.06144631654024124\n381 0.05948108434677124\n382 0.05756570026278496\n383 0.055704500526189804\n384 0.05389977991580963\n385 0.052152279764413834\n386 0.050462037324905396\n387 0.0488210991024971\n388 0.04726396128535271\n389 0.045748334378004074\n390 0.04426315799355507\n391 0.04283710569143295\n392 0.041462723165750504\n393 0.04012970253825188\n394 0.03883979097008705\n395 0.0375945121049881\n396 0.036392029374837875\n397 0.035216353833675385\n398 0.0340835303068161\n399 0.0330033153295517\n400 0.0319441556930542\n"
]
],
[
[
"## What is a nn module\nWhen building neural networks we frequently think of arranging the computation into layers, some of which have learnable parameters which will be optimized during learning.\n\nIn TensorFlow, packages like Keras, TensorFlow-Slim, and TFLearn provide higher-level abstractions over raw computational graphs that are useful for building neural networks.\n\nIn PyTorch, the nn package serves this same purpose. The nn package defines a set of Modules, which are roughly equivalent to neural network layers. A Module receives input Variables and computes output Variables, but may also hold internal state such as Variables containing learnable parameters. The nn package also defines a set of useful loss functions that are commonly used when training neural networks.\n\nIn this example we use the nn package to implement our two-layer network:",
"_____no_output_____"
]
],
[
[
"# -*- coding: utf-8 -*-\nimport torch\nfrom torch.autograd import Variable\n\n# N is batch size; D_in is input dimension;\n# H is hidden dimension; D_out is output dimension.\nN, D_in, H, D_out = 64, 1000, 100, 10\n\n# Create random Tensors to hold inputs and outputs, and wrap them in Variables.\nx = Variable(torch.randn(N, D_in))\ny = Variable(torch.randn(N, D_out), requires_grad=False)\n\n# Use the nn package to define our model as a sequence of layers. nn.Sequential\n# is a Module which contains other Modules, and applies them in sequence to\n# produce its output. Each Linear Module computes output from input using a\n# linear function, and holds internal Variables for its weight and bias.\nmodel = torch.nn.Sequential(\n torch.nn.Linear(D_in, H),\n torch.nn.ReLU(),\n torch.nn.Linear(H, D_out),\n)\n\n# The nn package also contains definitions of popular loss functions; in this\n# case we will use Mean Squared Error (MSE) as our loss function.\nloss_fn = torch.nn.MSELoss(size_average=False)\n\nlearning_rate = 1e-4\nfor t in range(500):\n # Forward pass: compute predicted y by passing x to the model. Module objects\n # override the __call__ operator so you can call them like functions. When\n # doing so you pass a Variable of input data to the Module and it produces\n # a Variable of output data.\n y_pred = model(x)\n\n # Compute and print loss. We pass Variables containing the predicted and true\n # values of y, and the loss function returns a Variable containing the\n # loss.\n loss = loss_fn(y_pred, y)\n print(t, loss.data[0])\n\n # Zero the gradients before running the backward pass.\n model.zero_grad()\n\n # Backward pass: compute gradient of the loss with respect to all the learnable\n # parameters of the model. Internally, the parameters of each Module are stored\n # in Variables with requires_grad=True, so this call will compute gradients for\n # all learnable parameters in the model.\n loss.backward()\n\n # Update the weights using gradient descent. Each parameter is a Variable, so\n # we can access its data and gradients like we did before.\n for param in model.parameters():\n param.data -= learning_rate * param.grad.data",
"0 680.08154296875\n1 628.8499755859375\n2 584.1482543945312\n3 544.8362426757812\n4 509.60052490234375\n5 477.94586181640625\n6 449.4169616699219\n7 423.177734375\n8 398.7761535644531\n9 376.30096435546875\n10 355.3580322265625\n11 335.78729248046875\n12 317.5242614746094\n13 300.2922058105469\n14 283.9698181152344\n15 268.5042419433594\n16 253.79812622070312\n17 239.88864135742188\n18 226.66624450683594\n19 214.0961151123047\n20 202.15972900390625\n21 190.82431030273438\n22 180.07672119140625\n23 169.84349060058594\n24 160.177734375\n25 151.01641845703125\n26 142.33706665039062\n27 134.12139892578125\n28 126.35624694824219\n29 119.03170776367188\n30 112.15123748779297\n31 105.64317321777344\n32 99.51466369628906\n33 93.72786712646484\n34 88.27943420410156\n35 83.15460205078125\n36 78.34161376953125\n37 73.80974578857422\n38 69.55596923828125\n39 65.56876373291016\n40 61.8112907409668\n41 58.278228759765625\n42 54.950050354003906\n43 51.81352615356445\n44 48.865779876708984\n45 46.08952713012695\n46 43.48237609863281\n47 41.032230377197266\n48 38.72626495361328\n49 36.560401916503906\n50 34.52272415161133\n51 32.603416442871094\n52 30.797603607177734\n53 29.0950984954834\n54 27.492034912109375\n55 25.980615615844727\n56 24.55516242980957\n57 23.21516990661621\n58 21.952560424804688\n59 20.762468338012695\n60 19.642051696777344\n61 18.584585189819336\n62 17.585891723632812\n63 16.644123077392578\n64 15.755452156066895\n65 14.916620254516602\n66 14.125481605529785\n67 13.380037307739258\n68 12.675837516784668\n69 12.011648178100586\n70 11.383655548095703\n71 10.7904634475708\n72 10.233356475830078\n73 9.707324981689453\n74 9.2102689743042\n75 8.740628242492676\n76 8.298053741455078\n77 7.879985809326172\n78 7.484074592590332\n79 7.109870910644531\n80 6.756218433380127\n81 6.421789646148682\n82 6.105353355407715\n83 5.8055315017700195\n84 5.52149772644043\n85 5.252496242523193\n86 4.997690677642822\n87 4.756503582000732\n88 4.527294158935547\n89 4.309894561767578\n90 4.103759765625\n91 3.9084742069244385\n92 3.7226548194885254\n93 3.5464107990264893\n94 3.37937068939209\n95 3.2209417819976807\n96 3.070256471633911\n97 2.9272148609161377\n98 2.791459083557129\n99 2.6623592376708984\n100 2.5399575233459473\n101 2.4237000942230225\n102 2.313190460205078\n103 2.2082247734069824\n104 2.1084320545196533\n105 2.0137369632720947\n106 1.9235584735870361\n107 1.8378454446792603\n108 1.7563918828964233\n109 1.6788384914398193\n110 1.6050490140914917\n111 1.5347329378128052\n112 1.4678232669830322\n113 1.4041197299957275\n114 1.3435639142990112\n115 1.2858960628509521\n116 1.2308998107910156\n117 1.1784309148788452\n118 1.1284685134887695\n119 1.0808899402618408\n120 1.0353796482086182\n121 0.9919747710227966\n122 0.9504337310791016\n123 0.9108781814575195\n124 0.8731096386909485\n125 0.8370406627655029\n126 0.8026205897331238\n127 0.7697471380233765\n128 0.7383762001991272\n129 0.7084004282951355\n130 0.679768979549408\n131 0.6523947715759277\n132 0.626214861869812\n133 0.6011669635772705\n134 0.5772068500518799\n135 0.5543199777603149\n136 0.5323988795280457\n137 0.5114548206329346\n138 0.49139833450317383\n139 0.4721781611442566\n140 0.45376724004745483\n141 0.43613412976264954\n142 0.41924476623535156\n143 0.40306606888771057\n144 0.38756275177001953\n145 0.3727158010005951\n146 0.35854285955429077\n147 0.34496134519577026\n148 0.33194538950920105\n149 0.31945526599884033\n150 0.3074859082698822\n151 0.29600128531455994\n152 0.2849758565425873\n153 0.2743982970714569\n154 0.2642490863800049\n155 0.2545064389705658\n156 0.2451404333114624\n157 0.23615539073944092\n158 0.22752535343170166\n159 0.21923471987247467\n160 0.2112797051668167\n161 0.20362289249897003\n162 0.19626261293888092\n163 0.1891934722661972\n164 0.18240153789520264\n165 0.17586886882781982\n166 0.16958405077457428\n167 0.16354581713676453\n168 0.15773969888687134\n169 0.15215399861335754\n170 0.14677844941616058\n171 0.14159975945949554\n172 0.13661399483680725\n173 0.13181616365909576\n174 0.1271965205669403\n175 0.12274857610464096\n176 0.11846866458654404\n177 0.11434680968523026\n178 0.11037838459014893\n179 0.10655605047941208\n180 0.10287366062402725\n181 0.09932510554790497\n182 0.0959087535738945\n183 0.09261389821767807\n184 0.08943838626146317\n185 0.08637817949056625\n186 0.08342777192592621\n187 0.0805828794836998\n188 0.07784154266119003\n189 0.07519736886024475\n190 0.0726480707526207\n191 0.07019388675689697\n192 0.06782343983650208\n193 0.06553709506988525\n194 0.06333591789007187\n195 0.06121001020073891\n196 0.059156980365514755\n197 0.05717690289020538\n198 0.055265795439481735\n199 0.053421154618263245\n200 0.05164136365056038\n201 0.04992407187819481\n202 0.04826623946428299\n203 0.046666938811540604\n204 0.04512207210063934\n205 0.04363016411662102\n206 0.04219016805291176\n207 0.04079950973391533\n208 0.03945689648389816\n209 0.038160402327775955\n210 0.036908261477947235\n211 0.03569883480668068\n212 0.034530479460954666\n213 0.03340240567922592\n214 0.032312240451574326\n215 0.031259190291166306\n216 0.030241671949625015\n217 0.02925860695540905\n218 0.028309127315878868\n219 0.02739332616329193\n220 0.026506047695875168\n221 0.025648759678006172\n222 0.02482016757130623\n223 0.024018865078687668\n224 0.023244598880410194\n225 0.022496270015835762\n226 0.02177284099161625\n227 0.021073248237371445\n228 0.020396927371621132\n229 0.01974322460591793\n230 0.019111426547169685\n231 0.0185005571693182\n232 0.017909277230501175\n233 0.017337419092655182\n234 0.016784587875008583\n235 0.016250004991889\n236 0.01573282666504383\n237 0.015232610516250134\n238 0.014748821035027504\n239 0.014280819334089756\n240 0.013828235678374767\n241 0.013390136882662773\n242 0.012966613285243511\n243 0.012556690722703934\n244 0.012161037884652615\n245 0.0117774223908782\n246 0.011406159959733486\n247 0.011046788655221462\n248 0.010699193924665451\n249 0.010362686589360237\n250 0.010037034749984741\n251 0.00972204003483057\n252 0.009417165070772171\n253 0.009121924638748169\n254 0.008836277760565281\n255 0.00855974666774273\n256 0.008291949518024921\n257 0.00803307630121708\n258 0.007782185450196266\n259 0.007539329584687948\n260 0.007304261904209852\n261 0.007076835259795189\n262 0.00685644056648016\n263 0.006643133703619242\n264 0.006436587776988745\n265 0.006236482877284288\n266 0.006042997818440199\n267 0.005855487193912268\n268 0.005673850420862436\n269 0.005498659797012806\n270 0.005328337661921978\n271 0.005163470283150673\n272 0.0050038304179906845\n273 0.004849148914217949\n274 0.004699397832155228\n275 0.00455437321215868\n276 0.004413901828229427\n277 0.0042778486385941505\n278 0.004146031104028225\n279 0.004018353298306465\n280 0.0038947267457842827\n281 0.0037749370094388723\n282 0.003658916801214218\n283 0.0035464726388454437\n284 0.003437580307945609\n285 0.0033320633228868246\n286 0.0032298287842422724\n287 0.003130849450826645\n288 0.003034892724826932\n289 0.0029419672209769487\n290 0.0028519199695438147\n291 0.002764653880149126\n292 0.0026801559142768383\n293 0.002598251448944211\n294 0.002518962835893035\n295 0.002442245604470372\n296 0.0023677151184529066\n297 0.0022955138701945543\n298 0.0022255151998251677\n299 0.002157705370336771\n300 0.002091981703415513\n301 0.002028322545811534\n302 0.00196659192442894\n303 0.0019068144029006362\n304 0.0018488289788365364\n305 0.001792663475498557\n306 0.0017382020596414804\n307 0.0016854503192007542\n308 0.0016342989401891828\n309 0.0015847444301471114\n310 0.0015366816660389304\n311 0.0014901245012879372\n312 0.0014449851587414742\n313 0.001401250483468175\n314 0.0013588427100330591\n315 0.0013177691726014018\n316 0.001277906121686101\n317 0.0012393008219078183\n318 0.0012018403504043818\n319 0.001165554509498179\n320 0.0011303661158308387\n321 0.001096343039534986\n322 0.0010632890043780208\n323 0.001031213440001011\n324 0.0010001431219279766\n325 0.0009700573864392936\n326 0.0009408452315256\n327 0.000912512477952987\n328 0.0008850548765622079\n329 0.0008584235911257565\n330 0.0008326029637828469\n331 0.0008075683144852519\n332 0.0007833010167814791\n333 0.0007597761577926576\n334 0.0007369595696218312\n335 0.0007148331496864557\n336 0.0006933821714483202\n337 0.0006725748535245657\n338 0.0006523994379676878\n339 0.0006328612216748297\n340 0.0006138771423138678\n341 0.0005954877706244588\n342 0.0005776527686975896\n343 0.0005603585159406066\n344 0.0005436040228232741\n345 0.0005273337010294199\n346 0.000511560239829123\n347 0.0004962603561580181\n348 0.00048146690824069083\n349 0.0004670854832511395\n350 0.0004531279264483601\n351 0.0004396018339321017\n352 0.0004264691669959575\n353 0.00041373888961970806\n354 0.0004013977595604956\n355 0.0003894170222338289\n356 0.00037781387800350785\n357 0.0003665469994302839\n358 0.0003556182491593063\n359 0.0003450293734204024\n360 0.00033474891097284853\n361 0.0003247867280151695\n362 0.00031511206179857254\n363 0.0003057269495911896\n364 0.00029663191526196897\n365 0.00028781587025150657\n366 0.00027925128233619034\n367 0.0002709476975724101\n368 0.00026289623929187655\n369 0.00025508779799565673\n370 0.0002475187066011131\n371 0.00024016370298340917\n372 0.00023303109628614038\n373 0.00022611598251387477\n374 0.00021940314036328346\n375 0.00021291013399604708\n376 0.0002065994485747069\n377 0.00020047678845003247\n"
]
],
[
[
"## PyTorch - optim\nWith learning rate of $1e-4$\n",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch.autograd import Variable\n\nN, D_in, H, D_out = 64, 1000, 100, 10\n\nx = Variable(torch.randn(N, D_in))\ny = Variable(torch.randn(N, D_out), requires_grad=False)\n\nmodel = torch.nn.Sequential( torch.nn.Linear(D_in, H),\n torch.nn.ReLU(),\n torch.nn.Linear(H, D_out)\n )\n\nloss_fxn = torch.nn.MSELoss(size_average=False)\n\nlearning_rate = 1e-4\noptimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)",
"_____no_output_____"
],
[
"# We loop\n\nfor i in range(500):\n y_pred = model(x)\n loss = loss_fxn(y_pred, y)\n print(t, loss.data[0])\n \n optimizer.zero_grad()\n loss.backward()\n optimizer.step()",
"499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n499 5.173239514988381e-06\n"
]
],
[
[
"## Custom nn module\n\nFor more complex computation, you can define your own module by subclassing nn.Module",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch.autograd import Variable\n\nclass DoubleLayerNet(torch.nn.Module):\n def __init__(self, D_in, H, D_out):\n # initialize 2 instances of nn.Linear mods\n super(DoubleLayerNet, self).__init__()\n self.linear1 = torch.nn.Linear(D_in, H)\n self.linear2 = torch.nn.Linear(H, D_out)\n \n def forward(self, x):\n # in this fxn we accept a Var of input data and\n # return a Var of output data.\n h_relu = self.linear1(x).clamp(min=0)\n y_pred = self.linear2(h_relu)\n return y_pred\n\n# Next, again as usual, define batch size, input dimensions, hidden dimension and output dimension\n\nN, D_in, H, D_out = 64, 1000, 100, 10\n\n# Create some random tensors to hold both input and output\n\nx = Variable(torch.randn(N, D_in))\ny = Variable(torch.randn(N, D_out), requires_grad=False)\n\n# Build model by instantiating class defined above\nmy_model = DoubleLayerNet(D_in, H, D_out)\n\n# Build loss fxn and optimizer\n\ncriterion = torch.nn.MSELoss(size_average=False)\noptimizer = torch.optim.SGD(model.parameters(), lr=1e-4)\n\n# and then we loop\n\nfor i in range(500):\n # fwd pass, calculate predicted y by passing x to the model\n y_pred = my_model(x)\n \n #calculate and print loss\n loss = criteria(y_pred, y)\n print(t, loss.data[0])\n \n # Zero gradients, performs a backprop pass and update the weights as it goe along\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()",
"0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n0 656.797607421875\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6ff5e3daee3025347279cf5d20a94552487b35 | 13,772 | ipynb | Jupyter Notebook | _posts/python-v3/statistical/error-bar/error-bars.ipynb | bmb804/documentation | 57826d25e0afea7fff6a8da9abab8be2f7a4b48c | [
"CC-BY-3.0"
]
| 2 | 2019-06-24T23:55:53.000Z | 2019-07-08T12:22:56.000Z | _posts/python-v3/statistical/error-bar/error-bars.ipynb | bmb804/documentation | 57826d25e0afea7fff6a8da9abab8be2f7a4b48c | [
"CC-BY-3.0"
]
| 15 | 2020-06-30T21:21:30.000Z | 2021-08-02T21:16:33.000Z | _posts/python-v3/statistical/error-bar/error-bars.ipynb | bmb804/documentation | 57826d25e0afea7fff6a8da9abab8be2f7a4b48c | [
"CC-BY-3.0"
]
| 1 | 2019-11-10T04:01:48.000Z | 2019-11-10T04:01:48.000Z | 28.395876 | 316 | 0.509076 | [
[
[
"#### New to Plotly?\nPlotly's Python library is free and open source! [Get started](https://plot.ly/python/getting-started/) by downloading the client and [reading the primer](https://plot.ly/python/getting-started/).\n<br>You can set up Plotly to work in [online](https://plot.ly/python/getting-started/#initialization-for-online-plotting) or [offline](https://plot.ly/python/getting-started/#initialization-for-offline-plotting) mode, or in [jupyter notebooks](https://plot.ly/python/getting-started/#start-plotting-online).\n<br>We also have a quick-reference [cheatsheet](https://images.plot.ly/plotly-documentation/images/python_cheat_sheet.pdf) (new!) to help you get started!",
"_____no_output_____"
],
[
"#### Basic Symmetric Error Bars",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ndata = [\n go.Scatter(\n x=[0, 1, 2],\n y=[6, 10, 2],\n error_y=dict(\n type='data',\n array=[1, 2, 3],\n visible=True\n )\n )\n]\n\npy.iplot(data, filename='basic-error-bar')",
"_____no_output_____"
]
],
[
[
"#### Asymmetric Error Bars",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ndata = [\n go.Scatter(\n x=[1, 2, 3, 4],\n y=[2, 1, 3, 4],\n error_y=dict(\n type='data',\n symmetric=False,\n array=[0.1, 0.2, 0.1, 0.1],\n arrayminus=[0.2, 0.4, 1, 0.2]\n )\n )\n]\npy.iplot(data, filename='error-bar-asymmetric-array')",
"_____no_output_____"
]
],
[
[
"#### Error Bars as a Percentage of the y Value",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ndata = [\n go.Scatter(\n x=[0, 1, 2],\n y=[6, 10, 2],\n error_y=dict(\n type='percent',\n value=50,\n visible=True\n )\n )\n]\npy.iplot(data, filename='percent-error-bar')",
"_____no_output_____"
]
],
[
[
"#### Asymmetric Error Bars with a Constant Offset",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ndata = [\n go.Scatter(\n x=[1, 2, 3, 4],\n y=[2, 1, 3, 4],\n error_y=dict(\n type='percent',\n symmetric=False,\n value=15,\n valueminus=25\n )\n )\n]\npy.iplot(data, filename='error-bar-asymmetric-constant')",
"_____no_output_____"
]
],
[
[
"#### Horizontal Error Bars",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ndata = [\n go.Scatter(\n x=[1, 2, 3, 4],\n y=[2, 1, 3, 4],\n error_x=dict(\n type='percent',\n value=10\n )\n )\n]\npy.iplot(data, filename='error-bar-horizontal')",
"_____no_output_____"
]
],
[
[
"#### Bar Chart with Error Bars",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Bar(\n x=['Trial 1', 'Trial 2', 'Trial 3'],\n y=[3, 6, 4],\n name='Control',\n error_y=dict(\n type='data',\n array=[1, 0.5, 1.5],\n visible=True\n )\n)\ntrace2 = go.Bar(\n x=['Trial 1', 'Trial 2', 'Trial 3'],\n y=[4, 7, 3],\n name='Experimental',\n error_y=dict(\n type='data',\n array=[0.5, 1, 2],\n visible=True\n )\n)\ndata = [trace1, trace2]\nlayout = go.Layout(\n barmode='group'\n)\nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig, filename='error-bar-bar')",
"_____no_output_____"
]
],
[
[
"#### Colored and Styled Error Bars",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\nimport numpy as np\n\nx_theo = np.linspace(-4, 4, 100)\nsincx = np.sinc(x_theo)\nx = [-3.8, -3.03, -1.91, -1.46, -0.89, -0.24, -0.0, 0.41, 0.89, 1.01, 1.91, 2.28, 2.79, 3.56]\ny = [-0.02, 0.04, -0.01, -0.27, 0.36, 0.75, 1.03, 0.65, 0.28, 0.02, -0.11, 0.16, 0.04, -0.15]\n\ntrace1 = go.Scatter(\n x=x_theo,\n y=sincx,\n name='sinc(x)'\n)\ntrace2 = go.Scatter(\n x=x,\n y=y,\n mode='markers',\n name='measured',\n error_y=dict(\n type='constant',\n value=0.1,\n color='#85144B',\n thickness=1.5,\n width=3,\n ),\n error_x=dict(\n type='constant',\n value=0.2,\n color='#85144B',\n thickness=1.5,\n width=3,\n ),\n marker=dict(\n color='#85144B',\n size=8\n )\n)\ndata = [trace1, trace2]\npy.iplot(data, filename='error-bar-style')",
"_____no_output_____"
]
],
[
[
"#### Reference\nSee https://plot.ly/python/reference/#scatter for more information and chart attribute options!",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, HTML\n\ndisplay(HTML('<link href=\"//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700\" rel=\"stylesheet\" type=\"text/css\" />'))\ndisplay(HTML('<link rel=\"stylesheet\" type=\"text/css\" href=\"http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css\">'))\n\n! pip install git+https://github.com/plotly/publisher.git --upgrade\nimport publisher\npublisher.publish(\n 'error-bars.ipynb', 'python/error-bars/', 'Error Bars | plotly',\n 'How to add error-bars to charts in Python with Plotly.',\n title = 'Error Bars | plotly',\n name = 'Error Bars',\n thumbnail='thumbnail/error-bar.jpg', language='python',\n page_type='example_index', has_thumbnail='true', display_as='statistical', order=1,\n ipynb='~notebook_demo/18')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb6ff98c84de39ce3e19b6820d230009b1c63cdf | 26,878 | ipynb | Jupyter Notebook | module3-permutation-boosting/LS_DS_233_assignment.ipynb | bendevera/DS-Unit-2-Applied-Modeling | 4aa93a8466b950ff45da977c2022a7d843d18c48 | [
"MIT"
]
| null | null | null | module3-permutation-boosting/LS_DS_233_assignment.ipynb | bendevera/DS-Unit-2-Applied-Modeling | 4aa93a8466b950ff45da977c2022a7d843d18c48 | [
"MIT"
]
| null | null | null | module3-permutation-boosting/LS_DS_233_assignment.ipynb | bendevera/DS-Unit-2-Applied-Modeling | 4aa93a8466b950ff45da977c2022a7d843d18c48 | [
"MIT"
]
| null | null | null | 35.319317 | 823 | 0.409889 | [
[
[
"Lambda School Data Science\n\n*Unit 2, Sprint 3, Module 3*\n\n---\n\n\n# Permutation & Boosting\n\nYou will use your portfolio project dataset for all assignments this sprint.\n\n## Assignment\n\nComplete these tasks for your project, and document your work.\n\n- [ ] If you haven't completed assignment #1, please do so first.\n- [ ] Continue to clean and explore your data. Make exploratory visualizations.\n- [ ] Fit a model. Does it beat your baseline? \n- [ ] Try xgboost.\n- [ ] Get your model's permutation importances.\n\nYou should try to complete an initial model today, because the rest of the week, we're making model interpretation visualizations.\n\nBut, if you aren't ready to try xgboost and permutation importances with your dataset today, that's okay. You can practice with another dataset instead. You may choose any dataset you've worked with previously.\n\nThe data subdirectory includes the Titanic dataset for classification and the NYC apartments dataset for regression. You may want to choose one of these datasets, because example solutions will be available for each.\n\n\n## Reading\n\nTop recommendations in _**bold italic:**_\n\n#### Permutation Importances\n- _**[Kaggle / Dan Becker: Machine Learning Explainability](https://www.kaggle.com/dansbecker/permutation-importance)**_\n- [Christoph Molnar: Interpretable Machine Learning](https://christophm.github.io/interpretable-ml-book/feature-importance.html)\n\n#### (Default) Feature Importances\n - [Ando Saabas: Selecting good features, Part 3, Random Forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/)\n - [Terence Parr, et al: Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)\n\n#### Gradient Boosting\n - [A Gentle Introduction to the Gradient Boosting Algorithm for Machine Learning](https://machinelearningmastery.com/gentle-introduction-gradient-boosting-algorithm-machine-learning/)\n - _**[A Kaggle Master Explains Gradient Boosting](http://blog.kaggle.com/2017/01/23/a-kaggle-master-explains-gradient-boosting/)**_\n - [_An Introduction to Statistical Learning_](http://www-bcf.usc.edu/~gareth/ISL/ISLR%20Seventh%20Printing.pdf) Chapter 8\n - [Gradient Boosting Explained](http://arogozhnikov.github.io/2016/06/24/gradient_boosting_explained.html)\n - _**[Boosting](https://www.youtube.com/watch?v=GM3CDQfQ4sw) (2.5 minute video)**_",
"_____no_output_____"
]
],
[
[
"%%capture\n!pip install category_encoders",
"_____no_output_____"
],
[
"import pandas as pd\nimport seaborn as sns\nfrom sklearn.model_selection import train_test_split\n\ntitanic = sns.load_dataset('titanic')\n\ntrain, test = train_test_split(titanic, test_size=.2)\n\nfeatures = ['age', 'class', 'deck', 'embarked', 'fare', 'sex']\ntarget = 'survived'\n\nX_train = train[features]\ny_train = train[target]\nX_test = test[features]\ny_test = test[target]\n\nX_train.shape, X_test.shape, y_train.shape, y_test.shape",
"_____no_output_____"
],
[
"X_train.isnull().sum()\n# we're dealing with some null values",
"_____no_output_____"
],
[
"# what is our baseline\nmax(1-y_train.mean(), y_train.mean())",
"_____no_output_____"
],
[
"from sklearn.pipeline import Pipeline\nimport category_encoders as ce\nfrom xgboost import XGBClassifier\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.impute import SimpleImputer",
"_____no_output_____"
],
[
"# create base pipeline\npipeline = Pipeline([\n ('encoder', ce.OrdinalEncoder()),\n ('model', XGBClassifier())\n])",
"_____no_output_____"
],
[
"# fit base pipeline \ntrain_size = .8\ncutoff = int(train_size*X_train.shape[0])\nsmall_X_train = X_train[:cutoff]\nX_val = X_train[cutoff:]\nsmall_y_train = y_train[:cutoff]\ny_val = y_train[cutoff:]\n\npipeline.fit(small_X_train, small_y_train)\npipeline.score(X_val, y_val)",
"569\n"
]
],
[
[
"## Baseline model beat the baseline by about 16%!",
"_____no_output_____"
]
],
[
[
"# now lets tune some hyperparameters! \nparams = {\n 'model__n_estimators': [50, 70, 90],\n 'model__max_depth': [3, 5]\n}\n\nsearch = GridSearchCV(pipeline, params, n_jobs=-1)\nsearch.fit(X_train, y_train)\nprint(f\"Best params: \\n{search.best_params_}\")\nprint(f\"Best score: \\n{search.best_score_}\")",
"Best params: \n{'model__max_depth': 3, 'model__n_estimators': 90}\nBest score: \n0.8215699793164581\n"
],
[
"pipeline = Pipeline([\n ('encoder', ce.OrdinalEncoder()),\n ('model', XGBClassifier(n_estimators=90,\n max_depth=3))\n])",
"_____no_output_____"
],
[
"pipeline.fit(X_train, y_train)\npipeline.score(X_test, y_test)",
"_____no_output_____"
],
[
"# get model and encoded data seperate for permutation importance eval\nmodel = XGBClassifier(n_estimators=90, max_depth=3)\ntransformer = Pipeline([\n ('encoder', ce.OrdinalEncoder()),\n ('imputer', SimpleImputer())\n])\n\nX_train_transformed = transformer.fit_transform(X_train)\nX_test_transformed = transformer.transform(X_test)\nmodel.fit(X_train_transformed, y_train)",
"_____no_output_____"
],
[
"!pip install eli5",
"Collecting eli5\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/97/2f/c85c7d8f8548e460829971785347e14e45fa5c6617da374711dec8cb38cc/eli5-0.10.1-py2.py3-none-any.whl (105kB)\n\r\u001b[K |███ | 10kB 18.5MB/s eta 0:00:01\r\u001b[K |██████▏ | 20kB 1.7MB/s eta 0:00:01\r\u001b[K |█████████▎ | 30kB 2.5MB/s eta 0:00:01\r\u001b[K |████████████▍ | 40kB 1.7MB/s eta 0:00:01\r\u001b[K |███████████████▌ | 51kB 2.1MB/s eta 0:00:01\r\u001b[K |██████████████████▋ | 61kB 2.5MB/s eta 0:00:01\r\u001b[K |█████████████████████▊ | 71kB 2.9MB/s eta 0:00:01\r\u001b[K |████████████████████████▊ | 81kB 3.2MB/s eta 0:00:01\r\u001b[K |███████████████████████████▉ | 92kB 3.6MB/s eta 0:00:01\r\u001b[K |███████████████████████████████ | 102kB 2.8MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 112kB 2.8MB/s \n\u001b[?25hRequirement already satisfied: jinja2 in /usr/local/lib/python3.6/dist-packages (from eli5) (2.10.3)\nRequirement already satisfied: numpy>=1.9.0 in /usr/local/lib/python3.6/dist-packages (from eli5) (1.17.5)\nRequirement already satisfied: graphviz in /usr/local/lib/python3.6/dist-packages (from eli5) (0.10.1)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from eli5) (1.12.0)\nRequirement already satisfied: tabulate>=0.7.7 in /usr/local/lib/python3.6/dist-packages (from eli5) (0.8.6)\nRequirement already satisfied: scikit-learn>=0.18 in /usr/local/lib/python3.6/dist-packages (from eli5) (0.22.1)\nRequirement already satisfied: attrs>16.0.0 in /usr/local/lib/python3.6/dist-packages (from eli5) (19.3.0)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from eli5) (1.4.1)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.6/dist-packages (from jinja2->eli5) (1.1.1)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.18->eli5) (0.14.1)\nInstalling collected packages: eli5\nSuccessfully installed eli5-0.10.1\n"
],
[
"import eli5 \nfrom eli5.sklearn import PermutationImportance\n\npermuter = PermutationImportance(\n model,\n scoring=\"accuracy\",\n n_iter=10,\n random_state=42\n)\n\npermuter.fit(X_test_transformed, y_test)",
"_____no_output_____"
],
[
"feature_names = X_train.columns.tolist()\npd.Series(permuter.feature_importances_, feature_names).sort_values()",
"_____no_output_____"
],
[
"eli5.show_weights(\n permuter,\n top=None, # includes all features\n feature_names=feature_names\n)",
"_____no_output_____"
],
[
"# I may be cautious about embarked given the standard error is larger than \n# the permutation importance value",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb7006afb77d899eded4aff7c83bbcc80d9baa51 | 170,529 | ipynb | Jupyter Notebook | model_training.ipynb | jeongwhanchoi/CarND-Vehicle-Detection | 3c8af313b2dee8cabaa1039c225b6a44db8f1081 | [
"MIT"
]
| null | null | null | model_training.ipynb | jeongwhanchoi/CarND-Vehicle-Detection | 3c8af313b2dee8cabaa1039c225b6a44db8f1081 | [
"MIT"
]
| null | null | null | model_training.ipynb | jeongwhanchoi/CarND-Vehicle-Detection | 3c8af313b2dee8cabaa1039c225b6a44db8f1081 | [
"MIT"
]
| null | null | null | 606.864769 | 162,288 | 0.946736 | [
[
[
"import glob\nimport time\n\n# Divide up into cars and notcars\nimages = glob.glob('dataset/**/*.png', recursive=True)\ncars = []\nnotcars = []\nfor image in images:\n if 'non-vehicles' in image:\n notcars.append(image)\n else:\n cars.append(image)",
"_____no_output_____"
],
[
"from hog import *\n\ncolor_space='YCrCb'\nspatial_size=(32, 32)\nhist_bins=32\norient=9\npix_per_cell=8\ncell_per_block=2\nhog_channel='ALL'\nspatial_feat=True\nhist_feat=True\nhog_feat=True\n\nt=time.time()\ncar_features = extract_features(cars, color_space=color_space, spatial_size=spatial_size,\n hist_bins=hist_bins, orient=orient, pix_per_cell=pix_per_cell,\n cell_per_block=cell_per_block, hog_channel=hog_channel,\n spatial_feat=spatial_feat, hist_feat=hist_feat, hog_feat=hog_feat)\n\nnotcar_features = extract_features(notcars, color_space=color_space, spatial_size=spatial_size,\n hist_bins=hist_bins, orient=orient, pix_per_cell=pix_per_cell,\n cell_per_block=cell_per_block, hog_channel=hog_channel,\n spatial_feat=spatial_feat, hist_feat=hist_feat, hog_feat=hog_feat)\nt2 = time.time()\nprint(round(t2-t, 2), 'Seconds to extract features...')\n\n# Create an array stack of feature vectors\nX = np.vstack((car_features, notcar_features)).astype(np.float64) \n\n# Define the labels vector\ny = np.hstack((np.ones(len(car_features)), np.zeros(len(notcar_features))))",
"/Users/jeongwhanchoi/anaconda3/envs/carnd-term1/lib/python3.5/site-packages/skimage/feature/_hog.py:150: skimage_deprecation: Default value of `block_norm`==`L1` is deprecated and will be changed to `L2-Hys` in v0.15. To supress this message specify explicitly the normalization method.\n skimage_deprecation)\n/Users/jeongwhanchoi/anaconda3/envs/carnd-term1/lib/python3.5/site-packages/skimage/feature/_hog.py:248: skimage_deprecation: Argument `visualise` is deprecated and will be changed to `visualize` in v0.16\n 'be changed to `visualize` in v0.16', skimage_deprecation)\n"
],
[
"from sklearn.preprocessing import StandardScaler\n\n# Fit a per-column scaler\nX_scaler = StandardScaler().fit(X)\n# Apply the scaler to X\nscaled_X = X_scaler.transform(X)",
"_____no_output_____"
],
[
"# Split up data into randomized training and test set\nfrom sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(scaled_X, y, test_size=0.2, stratify =y)\n\nprint('Feature vector length:', len(X_train[0]))\n",
"Feature vector length: 8460\n"
],
[
"from sklearn.svm import LinearSVC\n\n# Use a linear SVC \nsvc = LinearSVC()\n\n# Check the training time for the SVC\nt=time.time()\nsvc.fit(X_train, y_train)\nt2 = time.time()\n\nprint(round(t2-t, 2), 'Seconds to train SVC...')",
"14.53 Seconds to train SVC...\n"
],
[
"# Check the score of the SVC\nprint('Test Accuracy of SVC = ', round(svc.score(X_test, y_test), 4))\n\n# Check the prediction time for a single sample\nt=time.time()\nn_predict = 10\nprint('My SVC predicts: ', svc.predict(X_test[0:n_predict]))\nprint('For these',n_predict, 'labels: ', y_test[0:n_predict])\nt2 = time.time()\nprint(round(t2-t, 5), 'Seconds to predict', n_predict,'labels with SVC')",
"Test Accuracy of SVC = 0.9921\nMy SVC predicts: [ 1. 1. 1. 1. 0. 1. 1. 0. 1. 1.]\nFor these 10 labels: [ 1. 1. 1. 1. 0. 1. 1. 0. 1. 1.]\n0.00766 Seconds to predict 10 labels with SVC\n"
],
[
"from hog import *\nimport matplotlib.image as mpimg\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pickle\nimport cv2\n%matplotlib inline\n\nimg = mpimg.imread('test_images/test4.jpg')\n\n\nystart = 400\nystop = 656\nscale = 1.5\n \nout_img = find_cars(img, ystart, ystop, scale, svc, X_scaler, color_space, orient, pix_per_cell, cell_per_block, hog_channel, spatial_size, hist_bins)\nplt.imshow(out_img)",
"/Users/jeongwhanchoi/anaconda3/envs/carnd-term1/lib/python3.5/site-packages/skimage/feature/_hog.py:150: skimage_deprecation: Default value of `block_norm`==`L1` is deprecated and will be changed to `L2-Hys` in v0.15. To supress this message specify explicitly the normalization method.\n skimage_deprecation)\n/Users/jeongwhanchoi/anaconda3/envs/carnd-term1/lib/python3.5/site-packages/skimage/feature/_hog.py:248: skimage_deprecation: Argument `visualise` is deprecated and will be changed to `visualize` in v0.16\n 'be changed to `visualize` in v0.16', skimage_deprecation)\n"
],
[
"import pickle\n\ndata={\n 'svc': svc,\n 'X_scaler': X_scaler,\n 'color_space': color_space,\n 'orient': orient,\n 'pix_per_cell': pix_per_cell,\n 'cell_per_block': cell_per_block,\n 'spatial_size' : spatial_size,\n 'hist_bins': hist_bins,\n 'hog_channel': hog_channel\n }\n\nwith open('svc_model.p', 'wb') as pFile:\n pickle.dump(data, pFile)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb702226b0324ce7821367b29095521650b4ad5c | 651,613 | ipynb | Jupyter Notebook | 017_Seaborn_FacetGrid_Plot.ipynb | milaan9/12_Python_Seaborn_Module | e2edc3b9d48997162568c3196fd32538c6e859c9 | [
"MIT"
]
| 53 | 2021-07-16T06:47:22.000Z | 2022-03-16T18:58:03.000Z | 017_Seaborn_FacetGrid_Plot.ipynb | EEonn/12_Python_Seaborn_Module | e2edc3b9d48997162568c3196fd32538c6e859c9 | [
"MIT"
]
| null | null | null | 017_Seaborn_FacetGrid_Plot.ipynb | EEonn/12_Python_Seaborn_Module | e2edc3b9d48997162568c3196fd32538c6e859c9 | [
"MIT"
]
| 50 | 2021-10-02T04:50:43.000Z | 2022-03-16T18:58:06.000Z | 469.12383 | 163,072 | 0.936444 | [
[
[
"<small><small><i>\nAll the IPython Notebooks in **[Python Seaborn Module](https://github.com/milaan9/12_Python_Seaborn_Module)** lecture series by **[Dr. Milaan Parmar](https://www.linkedin.com/in/milaanparmar/)** are available @ **[GitHub](https://github.com/milaan9)**\n</i></small></small>",
"_____no_output_____"
],
[
"<a href=\"https://colab.research.google.com/github/milaan9/12_Python_Seaborn_Module/blob/main/017_Seaborn_FacetGrid_Plot.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# FacetGrid",
"_____no_output_____"
],
[
"Welcome to another lecture on *Seaborn*! Our journey began with assigning *style* and *color* to our plots as per our requirement. Then we moved on to *visualize distribution of a dataset*, and *Linear relationships*, and further we dived into topics covering *plots for Categorical data*. Every now and then, we've also roughly touched customization aspects using underlying Matplotlib code. That indeed is the end of the types of plots offered by Seaborn, and only leaves us with widening the scope of usage of all the plots that we have learnt till now.\n\nOur discussion in upcoming lectures is majorly going to focus on using the core of Seaborn, based on which, *Seaborn* allows us to plot these amazing figures, that we had been detailing previously. This ofcourse isn't going to be a brand new topic because every now & then I have used these in previous lectures but hereon we're going to specifically deal with each one of those.\n\nTo introduce our new topic, i.e. **<span style=\"color:red\">Grids</span>**, we shall at first list the options available. Majorly, there are just two aspects to our discussion on *Grids* that includes:\n- **<span style=\"color:red\">FacetGrid</span>**\n- **<span style=\"color:red\">PairGrid</span>**\nAdditionally, we also have a companion function for *PairGrid* to enhance execution speed of *PairGrid*, i.e.\n- **<span style=\"color:red\">Pairplot</span>**\n\nOur discourse shall detail each one of these topics in-length for better understanding. As we have already covered the statistical inference of each type of plot, our emphasis shall mostly be on scaling and parameter variety of known plots on these grids. So let us commence our journey with **[FacetGrid](http://seaborn.pydata.org/generated/seaborn.FacetGrid.html?highlight=facetgrid#seaborn.FacetGrid)** in this lecture.",
"_____no_output_____"
],
[
"## FacetGrid",
"_____no_output_____"
],
[
"The term **Facet** here refers to *a dimension* or say, an *aspect* or a feature of a *multi-dimensional dataset*. This analysis is extremely useful when working with a multi-variate dataset which has a varied blend of datatypes, specially in *Data Science* & *Machine Learning* domain, where generally you would be dealing with huge datasets. If you're a *working pofessional*, you know what I am talking about. And if you're a *fresher* or a *student*, just to give you an idea, in this era of *Big Data*, an average *CSV file* (which is generally the most common form), or even a RDBMS size would vary from Gigabytes to Terabytes of data. If you are dealing with *Image/Video/Audio datasets*, then you may easily expect those to be in *hundreds of gigabyte*.",
"_____no_output_____"
],
[
"On the other hand, the term **Grid** refers to any *framework with spaced bars that are parallel to or cross each other, to form a series of squares or rectangles*. Statistically, these *Grids* are also used to represent and understand an entire *population* or just a *sample space* out of it. In general, these are pretty powerful tool for presentation, to describe our dataset and to study the *interrelationship*, or *correlation* between *each facet* of any *environment*.",
"_____no_output_____"
],
[
"Subplot grid for plotting conditional relationships.\n\nThe FacetGrid is an object that links a Pandas DataFrame to a matplotlib figure with a particular structure.\n\nIn particular, FacetGrid is used to draw plots with multiple Axes where each Axes shows the same relationship conditioned on different levels of some variable. It’s possible to condition on up to three variables by assigning variables to the rows and columns of the grid and using different colors for the plot elements.\n\nThe general approach to plotting here is called “small multiples”, where the same kind of plot is repeated multiple times, and the specific use of small multiples to display the same relationship conditioned on one ore more other variables is often called a “trellis plot”.\n\nThe basic workflow is to initialize the FacetGrid object with the dataset and the variables that are used to structure the grid. Then one or more plotting functions can be applied to each subset by calling **`FacetGrid.map()`** or **`FacetGrid.map_dataframe()`**. Finally, the plot can be tweaked with other methods to do things like change the axis labels, use different ticks, or add a legend. See the detailed code examples below for more information.",
"_____no_output_____"
],
[
"To kill our curiousity, let us plot a simple **<span style=\"color:red\">FacetGrid</span>** before continuing on with our discussion. And to do that, we shall once again quickly import our package dependencies and set the aesthetics for future use with built-in datasets.",
"_____no_output_____"
]
],
[
[
"# Importing intrinsic libraries:\nimport numpy as np\nimport pandas as pd\nnp.random.seed(101)\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline\nsns.set(style=\"whitegrid\", palette=\"rocket\")\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\n# Let us also get tableau colors we defined earlier:\ntableau_20 = [(31, 119, 180), (174, 199, 232), (255, 127, 14), (255, 187, 120),\n (44, 160, 44), (152, 223, 138), (214, 39, 40), (255, 152, 150),\n (148, 103, 189), (197, 176, 213), (140, 86, 75), (196, 156, 148),\n (227, 119, 194), (247, 182, 210), (127, 127, 127), (199, 199, 199),\n (188, 189, 34), (219, 219, 141), (23, 190, 207), (158, 218, 229)]\n\n# Scaling above RGB values to [0, 1] range, which is Matplotlib acceptable format:\nfor i in range(len(tableau_20)):\n r, g, b = tableau_20[i]\n tableau_20[i] = (r / 255., g / 255., b / 255.)",
"_____no_output_____"
],
[
"# Loading built-in Tips dataset:\ntips = sns.load_dataset(\"tips\")\ntips.head()",
"_____no_output_____"
],
[
"# Initialize a 2x2 grid of facets using the tips dataset:\nsns.set(style=\"ticks\", color_codes=True)\nsns.FacetGrid(tips, row='time', col='smoker')",
"_____no_output_____"
],
[
"# Draw a univariate plot on each facet:\nx = sns.FacetGrid(tips, col='time',row='smoker')\nx = x.map(plt.hist,\"total_bill\")",
"_____no_output_____"
],
[
"bins = np.arange(0,65,5)\nx = sns.FacetGrid(tips, col=\"time\", row=\"smoker\")\nx =x.map(plt.hist, \"total_bill\", bins=bins, color=\"g\")",
"_____no_output_____"
],
[
"# Plot a bivariate function on each facet:\n\nx = sns.FacetGrid(tips, col=\"time\", row=\"smoker\")\nx = x.map(plt.scatter, \"total_bill\", \"tip\", edgecolor=\"w\")",
"_____no_output_____"
],
[
"# Assign one of the variables to the color of the plot elements:\n\nx = sns.FacetGrid(tips, col=\"time\", hue=\"smoker\")\nx = x.map(plt.scatter,\"total_bill\",\"tip\",edgecolor = \"w\")\nx =x.add_legend()",
"_____no_output_____"
],
[
"# Plotting a basic FacetGrid with Scatterplot representation:\nax = sns.FacetGrid(tips, col=\"sex\", hue=\"smoker\", size=5)\nax.map(plt.scatter, \"total_bill\", \"tip\", alpha=.6)\nax.add_legend()",
"_____no_output_____"
]
],
[
[
"This is a combined scatter representation of Tips dataset that we have seen earlier as well, where Total tip generated against Total Bill amount is drawn in accordance with their Gender and Smoking practice. With this we can conclude how **FacetGrid** helps us visualize distribution of a variable or the relationship between multiple variables separately within subsets of our dataset. Important to note here is that Seaborn FacetGrid can only support upto **3-Dimensional figures**, using `row`, `column` and `hue` dimensions of the grid for *Categorical* and *Discrete* variables within our dataset.",
"_____no_output_____"
],
[
"Let us now have a look at the *parameters* offered or supported by Seaborn for a **FacetGrid**:\n**`seaborn.FacetGrid(data, row=None, col=None, hue=None, col_wrap=None, sharex=True, sharey=True, size=3, aspect=1, palette=None, row_order=None, col_order=None, hue_order=None, hue_kws=None, dropna=True, legend_out=True, despine=True, margin_titles=False, xlim=None, ylim=None, subplot_kws=None, gridspec_kws=None`**",
"_____no_output_____"
],
[
"There seems to be few new parameters out here for us, so let us one-by-one understand their scope before we start experimenting with those on our plots:\n- We are well acquainted with mandatory **`data`**, **`row`**, **`col`** and **`hue`** parameters.\n- Next is **`col_wrap`** that defines the **width of our variable** selected as **`col`** dimension, so that the *column facets* can span multiple rows.\n- **`sharex`** helps us **draft dedicated Y-axis** for each sub-plot, if declared **`False`**. Same concept holds good for **`sharey`** as well.\n- **`size`** helps us determine the size of our grid-frame.\n- We may also declare **`hue_kws`** parameter that lets us **control other aesthetics** of our plot.\n- **`dropna`** drops all the **NULL variables** from the selected features; and **`legend_out`** places the Legend either inside or outside our plot, as we've already seen.\n- **`margin_titles`** fetch the **feature names** from our dataset; and **`xlim`** & **`ylim`** additionally offers Matplotlib style limitation to each of our axes on the grid.\n\nThat pretty much seems to cover *intrinsic parameters* so let us now try to use them one-by-one with slight modifications:",
"_____no_output_____"
],
[
"Let us begin by pulling the *Legend inside* our FacetGrid and *creating a Header* for our grid:",
"_____no_output_____"
]
],
[
[
"ax = sns.FacetGrid(tips, col=\"sex\", hue=\"smoker\", size=5, legend_out=False)\nax.map(plt.scatter, \"total_bill\", \"tip\", alpha=.6)\nax.add_legend()\n\nplt.suptitle('Tip Collection based on Gender and Smoking', fontsize=11)",
"_____no_output_____"
]
],
[
[
"So declaring **`legend_out`** as **`False`** and creating a **Superhead title** using *Matplotlib* seems to be working great on our Grid. Customization on *Header size* gives us an add-on capability as well. Right now, we are going by default **`palette`** for **marker colors** which can be customized by setting to a different one. Let us try other parameters as well: ",
"_____no_output_____"
],
[
"Actually, before we jump further into utilization of other parameters, let me quickly take you behind the curtain of this plot. As visible, we assigned **`ax`** as a variable to our **FacetGrid** for creating a visualizaion figure, and then plotted a **Scatterplot** on top of it, before decorating further with a *Legend* and a *Super Title*. So when we initialized the assignment of **`ax`**, the grid actually gets created using backend *Matplotlib figure and axes*, though doesn't plot anything on top of it. This is when we call Scatterplot on our sample data, that in turn at the backend calls **`FacetGrid.map()`** function to map this grid to our Scatterplot. We intended to draw a linear relation plot, and thus entered multiple variable names, i.e. **`Total Bill`** and associated **`Tip`** to form *facets*, or dimensions of our grid.",
"_____no_output_____"
]
],
[
[
"# Change the size and aspect ratio of each facet:\n\nx = sns.FacetGrid(tips, col=\"day\", size=5, aspect=.5)\nx =x.map(plt.hist, \"total_bill\", bins=bins)",
"_____no_output_____"
],
[
"# Specify the order for plot elements:\n\ng = sns.FacetGrid(tips, col=\"smoker\", col_order=[\"Yes\", \"No\"])\ng = g.map(plt.hist, \"total_bill\", bins=bins, color=\"m\")",
"_____no_output_____"
],
[
"# Use a different color palette:\n\nkws = dict(s=50, linewidth=.5, edgecolor=\"w\")\ng =sns.FacetGrid(tips, col=\"sex\", hue=\"time\", palette=\"Set1\",\\\n hue_order=[\"Dinner\", \"Lunch\"]) \n\ng = g.map(plt.scatter, \"total_bill\", \"tip\", **kws)\ng.add_legend()",
"_____no_output_____"
],
[
"# Use a dictionary mapping hue levels to colors:\n\npal = dict(Lunch=\"seagreen\", Dinner=\"gray\")\ng = sns.FacetGrid(tips, col=\"sex\", hue=\"time\", palette=pal,\\\n hue_order=[\"Dinner\", \"Lunch\"])\n\ng = g.map(plt.scatter, \"total_bill\", \"tip\", **kws)\ng.add_legend()",
"_____no_output_____"
],
[
"# FacetGrid with boxplot\nx = sns.FacetGrid(tips,col= 'day')\nx = x.map(sns.boxplot,\"total_bill\",\"time\")",
"_____no_output_____"
]
],
[
[
"Also important to note is the use the **[matplotlib.pyplot.gca()](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.gca.html)** function, if required to *set the current axes* on our Grid. This shall fetch the current Axes instance on our current figure matching the given keyword arguments or params, & if unavailable, it shall even create one.",
"_____no_output_____"
]
],
[
[
"# Let us create a dummy DataFrame:\nfootball = pd.DataFrame({\n \"Wins\": [76, 64, 38, 78, 63, 45, 32, 46, 13, 40, 59, 80],\n \"Loss\": [55, 67, 70, 56, 59, 69, 72, 24, 45, 21, 58, 22],\n \"Team\": [\"Arsenal\"] * 4 + [\"Liverpool\"] * 4 + [\"Chelsea\"] * 4,\n \"Year\": [2015, 2016, 2017, 2018] * 3})",
"_____no_output_____"
]
],
[
[
"Before I begin illustration using this DataFrame, on a lighter note, I would add a disclosure that this is a dummy dataset and holds no resemblance whatsoever to actual records of respective Soccer clubs. So if you're one among those die-hard fans of any of these clubs, kindly excuse me if the numbers don't tally, as they are all fabricated.\n\nHere, **football** is kind of a *Time-series Pandas DataFrame* that in entirety reflects 4 features, where **`Wins`** and **`Loss`** variables represent the quarterly Scorecard of three soccer **`Teams`** for last four **`Years`**, from 2015 to 2018. Let us check how this DataFrame looks like:",
"_____no_output_____"
]
],
[
[
"football",
"_____no_output_____"
]
],
[
[
"This looks pretty good for our purpose so now let us initialize our FacetGrid on top of it and try to obtain a time-indexed with further plotting. In production environment, to keep our solution scalable, this is generally done by defining a function for data manipulation so we shall try that in this example:",
"_____no_output_____"
]
],
[
[
"# Defining a customizable function to be precise with our requirements & shall discuss it a little later:\n# We shall be using a new type of plot here that I shall discuss in detail later on.\ndef football_plot(data, color):\n sns.heatmap(data[[\"Wins\", \"Loss\"]])\n\n# 'margin_titles' won't necessarily guarantee desired results so better to be cautious:\nax = sns.FacetGrid(football, col=\"Team\", size=5, margin_titles=True)\nax.map_dataframe(football_plot)",
"_____no_output_____"
],
[
"ax = sns.FacetGrid(football, col=\"Team\", size=5)\nax.map(sns.kdeplot, \"Wins\", \"Year\", hist=True, lw=2)",
"_____no_output_____"
]
],
[
[
"As visible, **Heatmap** plots rectangular boxes for data points as a color-encoded matrix, and this is a topic we shall be discussing in detail in another Lecture but for now, I just wanted you to have a preview of it, and hence used it on top of our **FacetGrid**. Another good thing to know with *FacetGrid* is **gridspec** module which allows Matplotlib params to be passed for drawing attention to a particular facet by increasing its size. To better understand, let us try to use this module now:",
"_____no_output_____"
]
],
[
[
"# Loading built-in Titanic Dataset:\ntitanic = sns.load_dataset(\"titanic\")\n\n# Assigning reformed `deck` column:\ntitanic = titanic.assign(deck=titanic.deck.astype(object)).sort_values(\"deck\")\n\n# Creating Grid and Plot:\nax = sns.FacetGrid(titanic, col=\"class\", sharex=False, size=7,\n gridspec_kws={\"width_ratios\": [3.5, 2, 2]})\nax.map(sns.boxplot, \"deck\", \"age\")\n\nax.set_titles(fontweight='bold', size=17)",
"_____no_output_____"
]
],
[
[
"Breaking it down, at first we import our built-in Titanic dataset, and then assign a new column, i.e. **`deck`** using Pandas **`.assign()`** function. Here we declare this new column as a component of pre-existing **`deck`** column from Titanic dataset, but as a sorted object. Then we create our *FacetGrid* mentioning the DataFrame, the column on which Grids get segregated but with shared across *Y-axis*; for **`chosen deck`** against **`Age`** of passengers. Next in action is our **grid keyword specifications**, where we decide the *width ratio* of the plot that shall be passed on to these grids. Finally, we have our **Box Plot** representing values of **`Age`** feature across respective decks.",
"_____no_output_____"
],
[
"Now let us try to use different axes with same size for multivariate plotting on Tips dataset:",
"_____no_output_____"
]
],
[
[
"# Loading built-in Tips dataset:\ntips = sns.load_dataset(\"tips\")\n\n# Mapping a Scatterplot to our FacetGrid:\nax = sns.FacetGrid(tips, col=\"smoker\", row=\"sex\", size=3.5)\nax = (ax.map(plt.scatter, \"total_bill\", \"tip\", color=tableau_20[6]).set_axis_labels(\"Total Bill Generated (USD)\", \"Tip Amount\"))\n\n# Increasing size for subplot Titles & making it appear Bolder:\nax.set_titles(fontweight='bold', size=11)",
"_____no_output_____"
]
],
[
[
"**Scatterplot** dealing with data that has multiple variables is no new science for us so instead let me highlight what **`.map()`** does for us. This function actually allows us to project our figure axes, in accordance to which our Scatterplot spreads the feature datapoints across the grids, depending upon the segregators. Here we have **`sex`** and **`smoker`** as our segregators (When I use the general term \"segregator\", it just refers to the columns on which we decide to determine the layout). This comes in really handy as we can pass *Matplotlib parrameters* for further customization of our plot. At the end, when we add **`.set_axis_labels()`** it gets easy for us to label our axes but please note that this method shall work for you only when you're dealing with grids, hence you didn't observe me adapting to this function, while detailing various other plots.",
"_____no_output_____"
],
[
"- Let us now talk about the **`football_plot`** function we defined earlier with **football** DataFrame. The only reason I didn't speak of it then was because I wanted you to go through a few more parameter implementation before getting into this. There are **3 important rules for defining such functions** that are supported by **[FacetGrid.map](http://xarray.pydata.org/en/stable/generated/xarray.plot.FacetGrid.map.html)**:\n\n -They must take array-like inputs as positional arguments, with the first argument corresponding to the **`X-Axis`**, and the second argument corresponding to **`y-Axis`**.\n -They must also accept two keyword arguments: **`color`**, and **`label`**. If you want to use a **`hue`** variable, than these should get passed to the underlying plotting function (As a side note: You may just catch **`**kwargs`** and not do anything with them, if it's not relevant to the specific plot you're making.\n -Lastly, when called, they must draw a plot on the \"currently active\" matplotlib Axes.\n\n- Important to note is that there may be cases where your function draws a plot that looks correct without taking `x`, `y`, positional inputs and then it is better to just call the plot, like: **`ax.set_axis_labels(\"Column_1\", \"Column_2\")`** after you use **`.map()`**, which should rename your axes properly. Alternatively, you may also want to do something like `ax.set(xticklabels=)` to get more meaningful ticks.\n\n- Well I am also quite stoked to mention another important function (though not that comonly used), that is **[`FacetGrid.map_dataframe()`](http://nullege.com/codes/search/axisgrid.FacetGrid.map_dataframe)**. The rules here are similar to **`FacetGrid.map`** but the function you pass must accept a DataFrame input in a parameter called `data`, and instead of taking *array-like positional* inputs it takes *strings* that correspond to variables in that dataframe. Then on each iteration through the *facets*, the function will be called with the *Input dataframe*, masked to just the values for that combination of **`row`**, **`col`**, and **`hue`** levels.\n\nAnother important to note with both the above-mentioned functions is that the **`return`** value is ignored so you don't really have to worry about it. Just for illustration purpose, let us consider drafting a function that just *draws a horizontal line* in each **`facet`** at **`y=2`** and ignores all the Input data*:",
"_____no_output_____"
]
],
[
[
"# That is all you require in your function:\ndef plot_func(x, y, color=None, label=None):\n ax.map(plt.axhline, y=2)",
"_____no_output_____"
]
],
[
[
"I know this function concept might look little hazy at the moment but once you have covered more on dates and maptplotlib syntax in particular, the picture shall get much more clearer for you.",
"_____no_output_____"
],
[
"Let us look at one more example of **`FacetGrid()`** and this time let us again create a synthetic DataFrame for this demonstration:",
"_____no_output_____"
]
],
[
[
"# Creating synthetic Data (Don't focus on how it's getting created):\nunits = np.linspace(0, 50)\nA = [1., 18., 40., 100.]\n\ndf = []\nfor i in A:\n V1 = np.sin(i * units)\n V2 = np.cos(i * units)\n df.append(pd.DataFrame({\"units\": units, \"V_1\": V1, \"V_2\": V2, \"A\": i}))\n\nsample = pd.concat(df, axis=0)",
"_____no_output_____"
],
[
"# Previewing DataFrame:\nsample.head(10)\nsample.describe()",
"_____no_output_____"
],
[
"# Melting our sample DataFrame: \nsample_melt = sample.melt(id_vars=['A', 'units'], value_vars=['V_1', 'V_2'])\n\n# Creating plot:\nax = sns.FacetGrid(sample_melt, col='A', hue='A', palette=\"icefire\", row='variable', sharey='row', margin_titles=True)\nax.map(plt.plot, 'units', 'value')\nax.add_legend()",
"_____no_output_____"
]
],
[
[
"This process shall come in handy if you ever wish to vertically stack rows of subplots on top of one another. You do not really have to focus on the process of creating dataset, as generally you will have your dataset provided with a problem statement. For our plot, you may just consider these visual variations as **[Sinusoidal waves](https://en.wikipedia.org/wiki/Sine_wave)**. I shall attach a link in our notebook, if you wish to dig deeper into what these are and how are they actually computed. ",
"_____no_output_____"
],
[
"Our next lecture would be pretty much a small follow up to this lecture, where we would try to bring more of *Categorical data* to our **`FacetGrid()`**. Meanwhile, I would again suggest you to play around with analyzing and plotting datasets, as much as you can because visualization is a very important facet of *Data Science & Research*. And, I shall see you in our next lecture with **[Heat Map](https://github.com/milaan9/12_Python_Seaborn_Module/blob/main/018_Seaborn_Heat_Map.ipynb)**.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
]
|
cb702f5bf96ea54c5779df336c312264e4dcbfb7 | 341 | ipynb | Jupyter Notebook | docs/html/examples/rawdata/bolometer.ipynb | nifs-software/nifs-retrieve | 4ff9d70a1d2301d7b5762162586388ae67046ad2 | [
"MIT"
]
| null | null | null | docs/html/examples/rawdata/bolometer.ipynb | nifs-software/nifs-retrieve | 4ff9d70a1d2301d7b5762162586388ae67046ad2 | [
"MIT"
]
| 2 | 2021-12-16T04:50:00.000Z | 2021-12-22T11:55:01.000Z | docs/html/examples/rawdata/bolometer.ipynb | nifs-software/nifs-retrieve | 4ff9d70a1d2301d7b5762162586388ae67046ad2 | [
"MIT"
]
| null | null | null | 12.62963 | 36 | 0.463343 | [
[
[
"Retrieve Bolometer Raw Data\n===",
"_____no_output_____"
],
[
"comming soon...",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown"
]
]
|
cb70603d6367bd816aa4123ea0ca99c8def4cf2f | 55,488 | ipynb | Jupyter Notebook | analysis/.ipynb_checkpoints/analyzeFrequencyPeaksTemplates292021-checkpoint.ipynb | zieglerad/romulus | e65c5a10f51f55301dc07a8c2b786f733d293b8e | [
"MIT"
]
| null | null | null | analysis/.ipynb_checkpoints/analyzeFrequencyPeaksTemplates292021-checkpoint.ipynb | zieglerad/romulus | e65c5a10f51f55301dc07a8c2b786f733d293b8e | [
"MIT"
]
| null | null | null | analysis/.ipynb_checkpoints/analyzeFrequencyPeaksTemplates292021-checkpoint.ipynb | zieglerad/romulus | e65c5a10f51f55301dc07a8c2b786f733d293b8e | [
"MIT"
]
| null | null | null | 107.743689 | 10,648 | 0.789702 | [
[
[
"## imports ##\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport pickle as pkl\n####\n\n## global ##\ndataPath='/Users/ziegler/repos/mayfly/output/templatePeaks1252021.pkl'\ntemplatePitchAngles=np.linspace(85,90,51)\ntemplatePos=np.linspace(0,5e-2,21)\nradius=0.0\nnPeaks=5\nkeysAmp=[]\nkeysInd=[]\nkeysR=[]\nkeysI=[]\nfor i in range(nPeaks):\n keysAmp.append('pAmp'+str(i))\n keysInd.append('pInd'+str(i))\n keysR.append('pR'+str(i))\n keysI.append('pI'+str(i))\n\ncolors=['r','b','g','c','m','k']\nfrequencyConversion=200e6/8192\n####\n\n## definitions ##\n\n####\n\nsimulationPeaks=pd.read_pickle(dataPath)\npeaksAtRadius=simulationPeaks[simulationPeaks[\"r\"]==radius].sort_values('pa')\nnEntries=peaksAtRadius['pa'].size\n\nfig,axs=plt.subplots()\nfor i,key in enumerate(keysInd):\n axi=axs.plot(peaksAtRadius['pa'],peaksAtRadius[key],c=peaksAtRadius[keysAmp[i]],cmap='inferno',s=25)\nplt.colorbar(axi)\n\n#fig,axs=plt.subplots()\n#for i,key in enumerate(keysInd):\n# axs.scatter(peaksAtRadius['pa'],np.arctan2(peaksAtRadius[keysI[i]],peaksAtRadius[keysR[i]]))\n \n#fig,axs=plt.subplots()\n#for i,key in enumerate(keysInd):\n# axs.scatter(peaksAtRadius['pa'],np.sqrt(peaksAtRadius[keysR[i]]**2+peaksAtRadius[keysI[i]]**2))\n#plt.colorbar(axi)\n\n\n#lines={}\n#for entry in range(nEntries):\n# peaks=peaksAtRadius[['pa','pInd0','pInd1','pInd2','pInd3','pInd4']].iloc[entry]\n# nextPeaks=peaksAtRadius[['pa','pInd0','pInd1','pInd2','pInd3','pInd4']].iloc[entry+1]\n# if entry==0:\n# for i,key in enumerate(keysInd):\n# lines.update({i:[peaks[key]]})\n# else:\n# for i,keyLines in enumerate(lines): #loop over the lines that exist\n# iLine=lines[keyLine][-1] # get the last point in that line\n# distToNextPeaks=[]\n# for i,key2 in enumerate(keysInd):\n# distToNextPeaks.append((nextPeaks[key2]-iPeak)**2) # calculate the distance from the next \n \n ",
"_____no_output_____"
],
[
"# get entries with r == radius sort by pitch angle\npeaksAtRadius=simulationPeaks[simulationPeaks[\"r\"]==radius].sort_values(by=['pa'])\npeaksAtRadius.reset_index(inplace=True)\n#print(len(np.where(peaksAtRadius['pAmp0']>10.)[0]))\n\nlines={} # holds the (pitch angle, frequency) lines, permanent\nlinePeakIndexes=[] # holds the amplitude index of the peak (0-4) in the line, temporary\nlineFreqIndexes=[] # holds the frequency index of the peak (0-8191) in the line, y coordinant, temporary, saved in lines\npitchAngles=[] # holds the pitch angle of the point (85-90) in the line, x coordinant, temporary, saved in lines\nrowInds=[] # holds the row index that contains the pitch angle, used for setting the used elements to zero\n\n# iterate through all the rows\nfor irow in range(len(peaksAtRadius['pa'])): # iterate through all the rows/pitch angles\n \n # check if the row contains any peaks with amplitude above zero\n # if not skip the row\n hasPeak=False\n for i in range(5):\n if peaksAtRadius.iloc[irow]['pAmp'+str(i)]>0: # check if any peak amplitudes is not zero in the row. This means that this row contains a point in a line\n hasPeak=True # we found a non-zero element in the row\n \n if not hasPeak:\n continue # otherwise skip the row\n \n rowInds.append(irow) # add row index to the list of rows we are using.\n\n # find highest frequency index in the row\n maxPeakInd=0\n maxFreqInd=peaksAtRadius.iloc[irow]['pInd'+str(0)]\n for i in range(5):\n if peaksAtRadius.iloc[irow]['pInd'+str(i)] > maxFreqInd:\n maxPeakInd=i\n maxFreqInd=peaksAtRadius.iloc[irow]['pInd'+str(i)]\n \n linePeakIndexes.append(maxPeakInd) # the number 0-4 of the peak that is in the line\n lineFreqIndexes.append(maxFreqInd) # the y coordinate of the line\n pitchAngles.append(peaksAtRadius.iloc[irow]['pa']) # the x coordinate of the line point\n \n# find the start of the rightmost line\nlineStart=np.max(np.where(np.diff(indexes,prepend=0)>100)) # find the rightmost disconsinuity that marks the start of the line\n\n# create a numpy array of the line\nline=np.array([pitchAngles[lineStart:],lineFreqIndexes[lineStart:]]) # create 2D array with the line x and y coordinates\n# add the line to the dict of lines\nlines.update({1:line}) # put the line in some sort of dictionary\n\n\nprint(rowInds[lineStart:])\n# set the amplitude of the peaks in our line to zero\nfor irow in rowInds[lineStart:]:\n print(peaksAtRadius.iloc[irow]['pa'])\n peaksAtRadius.at[irow,'pAmp'+str(maxPeakInd)]=0.\n \n# check if all amplitudes are zeros\n#print(peaksAtRadius[keysAmp])\n\nprint\n\n\nfig,axs=plt.subplots()\naxs.plot(peaksAtRadius['pa'],indexes,'.')\naxs.plot(peaksAtRadius['pa'],np.diff(indexes,prepend=0),'.')\naxs.plot(peaksAtRadius['pa'][lineStart:],indexes[lineStart:],'.')\naxs.set_ylim(0,8192)\n\n\n# plot the highest peak index vs pitch angle\nfor i in range(1):\n fig,axs=plt.subplots()\n axs.plot(peaksAtRadius['pa'],peaksAtRadius['pInd'+str(i)],'r.')\naxs.set_ylim(0,8192)\n\n\n# scatter plot\nfig,axs=plt.subplots()\nfor i in range(5):\n axi=axs.scatter(peaksAtRadius['pa'],peaksAtRadius['pInd'+str(i)],c=peaksAtRadius['pAmp'+str(i)],\n cmap='inferno',s=50)\n \nplt.colorbar(axi)\naxs.set_ylim(0,8192)",
"_____no_output_____"
],
[
"\n#print(peaks90deg)\npa1cm=peaks1cm['pa']\npeak0Ind1cm=peaks1cm['pInd0']\npeak1Ind1cm=peaks1cm['pInd1']\n\npeak0Amp1cm=peaks1cm['pAmp0']\npeak1Amp1cm=peaks1cm['pAmp1']\n\npeak01IndDiff=abs(peak0Ind1cm-peak1Ind1cm)\npeak01AmpDiv=peak0Amp1cm/peak1Amp1cm\n#peak0Ind1cm=peaks1cm[['pInd0','pInd1','pInd2','pInd3','pInd4']]\n#peak0Amp1cm=peaks1cm[['pAmp0','pAmp1','pAmp2','pAmp3','pAmp4']]\npeak0Amp1cm=peaks1cm[['pAmp0','pAmp1']]\n\n\n\npeaks0cm=simulationPeaks[simulationPeaks[\"r\"]==0.0].sort_values(by=['pa'])\n#print(peaks90deg)\npa0cm=peaks0cm['pa']\npeak0Ind0cm=peaks0cm['pInd0']\n\n\n\n",
"_____no_output_____"
],
[
"fig,axs=plt.subplots()\n#axs.plot(pa0cm,peak0Ind0cm,'.')\naxs.plot(pa1cm,peak0Ind1cm,'.')\n\n#axs.set_yscale('log')\n\nfig,axs=plt.subplots()\n#axs.plot(pa0cm,peak0Ind0cm,'.')\naxs.plot(pa1cm,peak0Amp1cm,'.')\n\nfig,axs=plt.subplots()\n#axs.plot(pa0cm,peak0Ind0cm,'.')\naxs.plot(pa1cm,peak01AmpDiv,'.')",
"_____no_output_____"
],
[
"testDF=pd.DataFrame({'a':[1,2,3],'b':[4,5,6],'c':[7,8,9]})\nprint(testDF.take(,axis=0))\nprint(testDF)",
"_____no_output_____"
],
[
"def getFrequencyPitchAngleBehavior(peaksAtRadius,lineDict,lineNumber=0):\n \n allAmplitudesAreZero=False\n while not allAmplitudesAreZero:\n linePeakIndexes=[] # holds the amplitude index of the peak (0-4) in the line, temporary\n lineFreqIndexes=[] # holds the frequency index of the peak (0-8191) in the line, y coordinant, temporary, saved in lines\n pitchAngles=[] # holds the pitch angle of the point (85-90) in the line, x coordinant, temporary, saved in lines\n rowInds=[] # holds the row index that contains the pitch angle, used for setting the used elements to zero\n\n # iterate through all the rows\n for irow in range(len(peaksAtRadius['pa'])): # iterate through all the rows/pitch angles\n\n # check if the row contains any peaks with amplitude above zero\n # if not skip the row\n hasPeak=False\n for i in range(5):\n if peaksAtRadius.iloc[irow]['pAmp'+str(i)]>0:# check if any peak amplitudes is not zero in the row. This means that this row contains a point in a line\n print(peaksAtRadius.iloc[irow]['pAmp'+str(i)])\n hasPeak=True # we found a non-zero element in the row\n if not hasPeak:\n continue # otherwise skip the row\n if hasPeak:\n rowInds.append(irow) # add row index to the list of rows we are using.\n\n # find highest frequency index in the row\n maxPeakInd=0\n maxFreqInd=peaksAtRadius.iloc[irow]['pInd'+str(0)]\n for i in range(5):\n if peaksAtRadius.iloc[irow]['pInd'+str(i)] > maxFreqInd:\n maxPeakInd=i\n maxFreqInd=peaksAtRadius.iloc[irow]['pInd'+str(i)]\n\n linePeakIndexes.append(maxPeakInd) # the number 0-4 of the peak that is in the line\n lineFreqIndexes.append(maxFreqInd) # the y coordinate of the line\n pitchAngles.append(peaksAtRadius.iloc[irow]['pa']) # the x coordinate of the line point\n\n # find the start of the rightmost line\n lineStart=np.max(np.where(np.diff(lineFreqIndexes,prepend=0)>100)) # find the rightmost disconsinuity that marks the start of the line\n\n # create a numpy array of the line\n line=np.array([pitchAngles[lineStart:],lineFreqIndexes[lineStart:]]) # create 2D array with the line x and y coordinates\n # add the line to the dict of lines\n lineDict.update({lineNumber:line}) # put the line in some sort of dictionary\n\n #print(peaksAtRadius[keysAmp])\n #print(rowInds[lineStart:])\n # set the amplitude of the peaks in our line to zero\n for irow in rowInds[lineStart:]:\n #print(peaksAtRadius.iloc[irow]['pa'])\n peaksAtRadius.at[irow,'pAmp'+str(maxPeakInd)]=\n #print(peaksAtRadius[keysAmp])\n\n # check if all amplitudes are zeros\n allAmplitudesAreZero=True\n for key in keysAmp:\n if len(np.where(peaksAtRadius[key]>0)[0])>0:\n allAmplitudesAreZero=False\n\n if not allAmplitudesAreZero:\n lineNumber+=1\n \n\n return True\n \n# get entries with r == radius sort by pitch angle\npeaksAtRadius=simulationPeaks[simulationPeaks[\"r\"]==radius].sort_values(by=['pa'])\npeaksAtRadius.reset_index(inplace=True)\nlines={}\ngetFrequencyPitchAngleBehavior(peaksAtRadius,lines)\n\nprint(lines)",
"0 1\n1 2\n2 3\nName: a, dtype: int64\n"
],
[
"peaksAtRadius=simulationPeaks[simulationPeaks[\"r\"]==radius].sort_values(by=['pa'])\npeaksAtRadius.reset_index(inplace=True)\ndataDict=peaksAtRadius.to_dict()\n\nrowInds=np.array(list(dataDict['pa'].keys()))\n\n\ndef findLines(lines,dataDict,nLine=0):\n potentialRowsInLine=[]\n linePeakIndexes=[] # holds the amplitude index of the peak (0-4) in the line, temporary\n lineFreqIndexes=[] # holds the frequency index of the peak (0-8191) in the line, y coordinant, temporary, saved in lines\n pitchAngles=[]\n for irow in rowInds:\n\n\n hasPeak=False\n\n for key in keysAmp:\n if dataDict[key][irow]>0: # check if any peak amplitudes is not zero in the row. \n # This means that this row contains a point in a line\n hasPeak=True # we found a non-zero element in the row\n if hasPeak:\n potentialRowsInLine.append(irow)\n else:\n continue # otherwise skip the row\n\n # find highest frequency index in the row\n maxPeakInd=0\n maxFreqInd=dataDict['pInd'+str(0)][irow]\n for i,key in enumerate(keysInd):\n if dataDict[key][irow] > maxFreqInd:\n maxPeakInd=i\n maxFreqInd=dataDict[key][irow]\n\n #print(maxPeakInd,maxFreqInd)\n\n linePeakIndexes.append(maxPeakInd) # the number 0-4 of the peak that is in the line\n lineFreqIndexes.append(maxFreqInd) # the y coordinate of the line\n pitchAngles.append(dataDict['pa'][irow]) # the x coordinate of the line point\n\n # find the start of the rightmost line\n #print(lineFreqIndexes)\n if len(lineFreqIndexes)==0:\n return True\n #print(lineFreqIndexes)\n lineStart=np.max(np.where(np.diff(lineFreqIndexes,prepend=0)>150)) # find the rightmost disconsinuity that marks the start of the line\n #print(lineStart)\n # create a numpy array of the line\n line=np.array([pitchAngles[lineStart:],lineFreqIndexes[lineStart:]]) # create 2D array with the line x and y coordinates\n # add the line to the dict of lines\n lines.update({nLine:line}) # put the line in a dictionary\n\n\n # remove the points in the line from the data dictionary\n for i,irow in enumerate(potentialRowsInLine):\n if i>=lineStart:\n dataDict['pAmp'+str(linePeakIndexes[i])][irow]=0\n dataDict['pInd'+str(linePeakIndexes[i])][irow]=-1\n\n allPeaksDone=True\n for key in keysAmp:\n if len(list(dataDict[key].keys()))>0:\n allPeaksDone=False\n\n if not allPeaksDone:\n nLine+=1\n #print(line)\n findLines(lines,dataDict,nLine=nLine)\n else:\n return True\n\n \nlines={}\nfindLines(lines,dataDict)\n\nfig,axs=plt.subplots()\nfor i,key in enumerate(lines):\n axs.plot(lines[key][0,:],lines[key][1,:],'.')\n ",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb706519f0a32ac46e8137c3a1526f0cd1f057d4 | 36,810 | ipynb | Jupyter Notebook | textcnn.ipynb | zhiming-xu/weibo-emoji-predict | 5d4636a6f79ea88ec5da8d36592d605857ca37b4 | [
"Apache-2.0"
]
| 2 | 2019-10-21T05:52:07.000Z | 2019-12-04T04:27:28.000Z | textcnn.ipynb | StevenBirdBrown/weibo_emoji_predict | 5d4636a6f79ea88ec5da8d36592d605857ca37b4 | [
"Apache-2.0"
]
| 4 | 2020-03-31T03:42:59.000Z | 2022-01-13T01:18:38.000Z | textcnn.ipynb | StevenBirdBrown/weibo-emoji-predict | 5d4636a6f79ea88ec5da8d36592d605857ca37b4 | [
"Apache-2.0"
]
| null | null | null | 36.590457 | 143 | 0.540234 | [
[
[
"import pandas as pd\nimport numpy as np\nimport mxnet as mx\nfrom mxnet import nd, autograd, gluon, init\nfrom mxnet.gluon import nn, rnn\nimport gluonnlp as nlp\nimport pkuseg\nimport multiprocessing as mp\nimport time\nfrom d2l import try_gpu\nimport itertools\nimport jieba\nfrom sklearn.metrics import accuracy_score, f1_score\nimport d2l\nimport re\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n# fixed random number seed\nnp.random.seed(2333)\nmx.random.seed(2333)",
"_____no_output_____"
],
[
"DATA_FOLDER = 'data/'\nTRAIN_DATA = 'train.csv'\nWORD_EMBED = 'sgns.weibo.bigram-char'\nLABEL_FILE = 'train.label'\nN_ROWS=1000\nctx = mx.gpu(0)\nseg = pkuseg.pkuseg(model_name='web')",
"_____no_output_____"
],
[
"train_df = pd.read_csv(DATA_FOLDER+TRAIN_DATA, sep='|')\ntrain_df = train_df.sample(frac=1)\ntrain_df.head()",
"_____no_output_____"
],
[
"dataset =[ [row[0], row[1]] for _, row in train_df.iterrows()]\ntrain_dataset, valid_dataset = nlp.data.train_valid_split(dataset)\nlen(train_dataset), len(valid_dataset)",
"_____no_output_____"
],
[
"def tokenizer(x):\n tweet, label = x\n if type(tweet) != str:\n print(tweet)\n tweet = str(tweet)\n word_list = jieba.lcut(tweet)\n if len(word_list)==0:\n word_list=['<unk>']\n return word_list, label\n\ndef get_length(x):\n return float(len(x[0]))\n\ndef to_word_list(dataset):\n start = time.time()\n with mp.Pool() as pool:\n # Each sample is processed in an asynchronous manner.\n dataset = gluon.data.ArrayDataset(pool.map(tokenizer, dataset))\n lengths = gluon.data.ArrayDataset(pool.map(get_length, dataset))\n end = time.time()\n\n print('Done! Tokenizing Time={:.2f}s, #Sentences={}'.format(end - start, len(dataset)))\n return dataset, lengths\n\ntrain_word_list, train_word_lengths = to_word_list(train_dataset)\nvalid_word_list, valid_word_lengths = to_word_list(valid_dataset)",
"Building prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nBuilding prefix dict from the default dictionary ...\nDumping model to file cache /tmp/jieba.cache\nDumping model to file cache /tmp/jieba.cache\nLoading model cost 1.420 seconds.\nPrefix dict has been built succesfully.\nDumping model to file cache /tmp/jieba.cache\nDumping model to file cache /tmp/jieba.cache\nLoading model cost 1.514 seconds.\nPrefix dict has been built succesfully.\nDumping model to file cache /tmp/jieba.cache\nDumping model to file cache /tmp/jieba.cache\nLoading model cost 1.551 seconds.\nLoading model cost 1.507 seconds.\nDumping model to file cache /tmp/jieba.cache\nPrefix dict has been built succesfully.\nPrefix dict has been built succesfully.\nDumping model to file cache /tmp/jieba.cache\nLoading model cost 1.617 seconds.\nPrefix dict has been built succesfully.\nLoading model cost 1.625 seconds.\nPrefix dict has been built succesfully.\nDumping model to file cache /tmp/jieba.cache\nLoading model cost 1.550 seconds.\nDumping model to file cache /tmp/jieba.cache\nPrefix dict has been built succesfully.\nDumping model to file cache /tmp/jieba.cache\nLoading model cost 1.673 seconds.\nPrefix dict has been built succesfully.\nLoading model cost 1.647 seconds.\nPrefix dict has been built succesfully.\nLoading model cost 1.508 seconds.\nDumping model to file cache /tmp/jieba.cache\nLoading model cost 1.573 seconds.\nPrefix dict has been built succesfully.\nPrefix dict has been built succesfully.\nLoading model cost 1.640 seconds.\nPrefix dict has been built succesfully.\n"
],
[
"train_seqs = [sample[0] for sample in train_word_list]\ncounter = nlp.data.count_tokens(list(itertools.chain.from_iterable(train_seqs)))\n\nvocab = nlp.Vocab(counter, max_size=200000)\n\n# load customed pre-trained embedding\nembedding_weights = nlp.embedding.TokenEmbedding.from_file(file_path=DATA_FOLDER+WORD_EMBED)\nvocab.set_embedding(embedding_weights)\nprint(vocab)",
"Vocab(size=200004, unk=\"<unk>\", reserved=\"['<pad>', '<bos>', '<eos>']\")\n"
],
[
"def token_to_idx(x):\n return vocab[x[0]], x[1]\n\n# A token index or a list of token indices is returned according to the vocabulary.\nwith mp.Pool() as pool:\n train_dataset = pool.map(token_to_idx, train_word_list)\n valid_dataset = pool.map(token_to_idx, valid_word_list)",
"_____no_output_____"
],
[
"batch_size = 1024\nbucket_num = 20\nbucket_ratio = 0.1\n\n\ndef get_dataloader():\n # Construct the DataLoader Pad data, stack label and lengths\n batchify_fn = nlp.data.batchify.Tuple(nlp.data.batchify.Pad(axis=0), \\\n nlp.data.batchify.Stack())\n\n # in this example, we use a FixedBucketSampler,\n # which assigns each data sample to a fixed bucket based on its length.\n batch_sampler = nlp.data.sampler.FixedBucketSampler(\n train_word_lengths,\n batch_size=batch_size,\n num_buckets=bucket_num,\n ratio=bucket_ratio,\n shuffle=True)\n print(batch_sampler.stats())\n\n # train_dataloader\n train_dataloader = gluon.data.DataLoader(\n dataset=train_dataset,\n batch_sampler=batch_sampler,\n batchify_fn=batchify_fn)\n # valid_dataloader\n valid_dataloader = gluon.data.DataLoader(\n dataset=valid_dataset,\n batch_size=batch_size,\n shuffle=False,\n batchify_fn=batchify_fn)\n return train_dataloader, valid_dataloader\n\ntrain_dataloader, valid_dataloader = get_dataloader()",
"FixedBucketSampler:\n sample_num=820005, batch_num=632\n key=[15, 26, 37, 48, 59, 70, 81, 92, 103, 114, 125, 136, 147, 158, 169, 180, 191, 202, 213, 224]\n cnt=[573412, 124268, 51415, 27570, 17000, 11311, 8113, 5220, 1308, 205, 83, 48, 15, 11, 7, 7, 6, 2, 2, 2]\n batch_size=[1529, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024, 1024]\n"
],
[
"for tweet, label in train_dataloader:\n print(tweet, label)\n break",
"\n[[2.8000e+01 4.7000e+01 8.0000e+00 ... 0.0000e+00 0.0000e+00 0.0000e+00]\n [7.0900e+02 4.2430e+03 1.9200e+03 ... 3.3310e+03 4.5660e+03 3.6580e+03]\n [7.4600e+02 2.5000e+01 1.4900e+02 ... 0.0000e+00 0.0000e+00 0.0000e+00]\n ...\n [1.2120e+03 5.3500e+02 9.0000e+00 ... 0.0000e+00 0.0000e+00 0.0000e+00]\n [1.4000e+01 8.5540e+03 9.0000e+01 ... 0.0000e+00 0.0000e+00 0.0000e+00]\n [3.3770e+03 5.0000e+00 1.0984e+04 ... 1.2000e+02 2.3700e+02 1.1000e+01]]\n<NDArray 1529x15 @cpu_shared(0)> \n[ 3 31 3 ... 35 31 10]\n<NDArray 1529 @cpu_shared(0)>\n"
]
],
[
[
"## Model contruction\nSelf attention layer, weighted cross entropy, and whole model",
"_____no_output_____"
]
],
[
[
"class TextCNN(nn.Block):\n def __init__(self, vocab_len, embed_size, kernel_sizes, num_channels, \\\n dropout, nclass, **kwargs):\n super(TextCNN, self).__init__(**kwargs)\n self.embedding = nn.Embedding(vocab_len, embed_size)\n \n self.constant_embedding = nn.Embedding(vocab_len, embed_size)\n self.dropout = nn.Dropout(dropout)\n self.decoder = nn.Dense(nclass)\n self.pool = nn.GlobalMaxPool1D()\n self.convs = nn.Sequential() \n for c, k in zip(num_channels, kernel_sizes):\n self.convs.add(nn.Conv1D(c, k, activation='relu'))\n \n def forward(self, inputs):\n embeddings = nd.concat(\n self.embedding(inputs), self.constant_embedding(inputs), dim=2)\n embeddings = embeddings.transpose((0, 2, 1))\n \n encoding = nd.concat(*[nd.flatten(\n self.pool(conv(embeddings))) for conv in self.convs], dim=1)\n outputs = self.decoder(self.dropout(encoding))\n return outputs",
"_____no_output_____"
],
[
"vocab_len = len(vocab)\nemsize = 300 # word embedding size\nnhidden = 400 # lstm hidden_dim\nnlayers = 4 # lstm layers\nnatt_unit = 400 # the hidden_units of attention layer\nnatt_hops = 20 # the channels of attention\nnfc = 256 # last dense layer size\nnclass = 72 # we have 72 emoji in total\n\ndrop_prob = 0.2\npool_way = 'flatten' # # The way to handle M\nprune_p = None\nprune_q = None\n\nctx = try_gpu()\n\nkernel_sizes, nums_channels = [2, 3, 4, 5], [100, 100, 100, 100]\nmodel = TextCNN(vocab_len, emsize, kernel_sizes, nums_channels, drop_prob, nclass)\nmodel.initialize(init.Xavier(), ctx=ctx)\n\nprint(model)\nmodel.embedding.weight.set_data(vocab.embedding.idx_to_vec)\nmodel.constant_embedding.weight.set_data(vocab.embedding.idx_to_vec)\nmodel.constant_embedding.collect_params().setattr('grad_req', 'null')",
"TextCNN(\n (embedding): Embedding(200004 -> 300, float32)\n (constant_embedding): Embedding(200004 -> 300, float32)\n (dropout): Dropout(p = 0.2, axes=())\n (decoder): Dense(None -> 72, linear)\n (pool): GlobalMaxPool1D(size=(1,), stride=(1,), padding=(0,), ceil_mode=True)\n (convs): Sequential(\n (0): Conv1D(None -> 100, kernel_size=(2,), stride=(1,), Activation(relu))\n (1): Conv1D(None -> 100, kernel_size=(3,), stride=(1,), Activation(relu))\n (2): Conv1D(None -> 100, kernel_size=(4,), stride=(1,), Activation(relu))\n (3): Conv1D(None -> 100, kernel_size=(5,), stride=(1,), Activation(relu))\n )\n)\n"
],
[
"tmp = nd.array([10, 20, 30, 40, 50, 60], ctx=ctx).reshape(1, -1)\nmodel(tmp)",
"_____no_output_____"
],
[
"class WeightedSoftmaxCE(nn.HybridBlock):\n def __init__(self, sparse_label=True, from_logits=False, **kwargs):\n super(WeightedSoftmaxCE, self).__init__(**kwargs)\n with self.name_scope():\n self.sparse_label = sparse_label\n self.from_logits = from_logits\n\n def hybrid_forward(self, F, pred, label, class_weight, depth=None):\n if self.sparse_label:\n label = F.reshape(label, shape=(-1, ))\n label = F.one_hot(label, depth)\n if not self.from_logits:\n pred = F.log_softmax(pred, -1)\n\n weight_label = F.broadcast_mul(label, class_weight)\n loss = -F.sum(pred * weight_label, axis=-1)\n\n # return F.mean(loss, axis=0, exclude=True)\n return loss",
"_____no_output_____"
],
[
"def calculate_loss(x, y, model, loss, class_weight):\n pred = model(x)\n y = nd.array(y.asnumpy().astype('int32')).as_in_context(ctx)\n if loss_name == 'sce':\n l = loss(pred, y)\n elif loss_name == 'wsce':\n l = loss(pred, y, class_weight, class_weight.shape[0])\n else:\n raise NotImplemented\n return pred, l",
"_____no_output_____"
],
[
"def one_epoch(data_iter, model, loss, trainer, ctx, is_train, epoch,\n clip=None, class_weight=None, loss_name='sce'):\n\n loss_val = 0.\n total_pred = []\n total_true = []\n n_batch = 0\n\n for batch_x, batch_y in data_iter:\n batch_x = batch_x.as_in_context(ctx)\n batch_y = batch_y.as_in_context(ctx)\n\n if is_train:\n with autograd.record():\n batch_pred, l = calculate_loss(batch_x, batch_y, model, \\\n loss, class_weight)\n\n # backward calculate\n l.backward()\n\n # clip gradient\n clip_params = [p.data() for p in model.collect_params().values()]\n if clip is not None:\n norm = nd.array([0.0], ctx)\n for param in clip_params:\n if param.grad is not None:\n norm += (param.grad ** 2).sum()\n norm = norm.sqrt().asscalar()\n if norm > clip:\n for param in clip_params:\n if param.grad is not None:\n param.grad[:] *= clip / norm\n\n # update parmas\n trainer.step(batch_x.shape[0])\n\n else:\n batch_pred, l = calculate_loss(batch_x, batch_y, model, \\\n loss, class_weight)\n\n # keep result for metric\n batch_pred = nd.argmax(nd.softmax(batch_pred, axis=1), axis=1).asnumpy()\n batch_true = np.reshape(batch_y.asnumpy(), (-1, ))\n total_pred.extend(batch_pred.tolist())\n total_true.extend(batch_true.tolist())\n \n batch_loss = l.mean().asscalar()\n\n n_batch += 1\n loss_val += batch_loss\n\n # check the result of traing phase\n if is_train and n_batch % 400 == 0:\n print('epoch %d, batch %d, batch_train_loss %.4f, batch_train_acc %.3f' %\n (epoch, n_batch, batch_loss, accuracy_score(batch_true, batch_pred)))\n\n # metric\n F1 = f1_score(np.array(total_true), np.array(total_pred), average='weighted')\n acc = accuracy_score(np.array(total_true), np.array(total_pred))\n loss_val /= n_batch\n\n if is_train:\n print('epoch %d, learning_rate %.5f \\n\\t train_loss %.4f, acc_train %.3f, F1_train %.3f, ' %\n (epoch, trainer.learning_rate, loss_val, acc, F1))\n # declay lr\n if epoch % 3 == 0:\n trainer.set_learning_rate(trainer.learning_rate * 0.9)\n else:\n print('\\t valid_loss %.4f, acc_valid %.3f, F1_valid %.3f, ' % (loss_val, acc, F1))",
"_____no_output_____"
],
[
"def train_valid(data_iter_train, data_iter_valid, model, loss, trainer, ctx, nepochs,\n clip=None, class_weight=None, loss_name='sce'):\n\n for epoch in range(1, nepochs+1):\n start = time.time()\n # train\n is_train = True\n one_epoch(data_iter_train, model, loss, trainer, ctx, is_train,\n epoch, clip, class_weight, loss_name)\n\n # valid\n is_train = False\n one_epoch(data_iter_valid, model, loss, trainer, ctx, is_train,\n epoch, clip, class_weight, loss_name)\n end = time.time()\n print('time %.2f sec' % (end-start))\n print(\"*\"*100)",
"_____no_output_____"
],
[
"from util import get_weight\nweight_list = get_weight(DATA_FOLDER, LABEL_FILE)\n\nclass_weight = None\nloss_name = 'sce'\noptim = 'adam'\nlr, wd = .001, .999\nclip = None\nnepochs = 5\n\ntrainer = gluon.Trainer(model.collect_params(), optim, {'learning_rate': lr})\n\nif loss_name == 'sce':\n loss = gluon.loss.SoftmaxCrossEntropyLoss()\nelif loss_name == 'wsce':\n loss = WeightedSoftmaxCE()\n # the value of class_weight is obtained by counting data in advance. It can be seen as a hyperparameter.\n class_weight = nd.array(weight_list, ctx=ctx)",
"_____no_output_____"
],
[
"# train and valid\nprint(ctx)\ntrain_valid(train_dataloader, valid_dataloader, model, loss, \\\n trainer, ctx, nepochs, clip=clip, class_weight=class_weight, \\\n loss_name=loss_name)",
"gpu(0)\nepoch 1, batch 400, batch_train_loss 3.4517, batch_train_acc 0.155\nepoch 1, learning_rate 0.00100 \n\t train_loss 3.5593, acc_train 0.152, F1_train 0.101, \n\t valid_loss 3.4231, acc_valid 0.170, F1_valid 0.106, \ntime 105.61 sec\n****************************************************************************************************\nepoch 2, batch 400, batch_train_loss 3.3264, batch_train_acc 0.178\nepoch 2, learning_rate 0.00100 \n\t train_loss 3.3224, acc_train 0.181, F1_train 0.128, \n\t valid_loss 3.3578, acc_valid 0.179, F1_valid 0.125, \ntime 106.17 sec\n****************************************************************************************************\nepoch 3, batch 400, batch_train_loss 3.2211, batch_train_acc 0.206\nepoch 3, learning_rate 0.00100 \n\t train_loss 3.1981, acc_train 0.200, F1_train 0.148, \n\t valid_loss 3.3231, acc_valid 0.186, F1_valid 0.135, \ntime 106.75 sec\n****************************************************************************************************\nepoch 4, batch 400, batch_train_loss 3.1442, batch_train_acc 0.219\nepoch 4, learning_rate 0.00090 \n\t train_loss 3.0581, acc_train 0.224, F1_train 0.174, \n\t valid_loss 3.3449, acc_valid 0.182, F1_valid 0.133, \ntime 106.78 sec\n****************************************************************************************************\nepoch 5, batch 400, batch_train_loss 3.0234, batch_train_acc 0.243\n"
],
[
"model.save_parameters(\"model/textcnn.params\")",
"_____no_output_____"
],
[
"kernel_sizes, nums_channels = [2, 3, 4, 5], [100, 100, 100, 100]\nmodel = TextCNN(vocab_len, emsize, kernel_sizes, nums_channels, 0, nclass)\nmodel.load_parameters('model/textcnn.params', ctx=ctx)",
"_____no_output_____"
],
[
"TEST_DATA = 'test.csv'\npredictions = []\ntest_df = pd.read_csv(DATA_FOLDER+TEST_DATA, header=None, sep='\\t')\nlen(test_df)",
"_____no_output_____"
],
[
"start = time.time()\nfor _, tweet in test_df.iterrows():\n token = vocab[jieba.lcut(tweet[1])]\n if len(token)<5:\n token += [0.]*(5-len(token))\n inp = nd.array(token, ctx=ctx).reshape(1,-1)\n pred = model(inp)\n pred = nd.argmax(pred, axis=1).asscalar()\n predictions.append(int(pred))\n if len(predictions)%2000==0:\n ckpt = time.time()\n print('current pred len %d, time %.2fs' % (len(predictions), ckpt-start))\n start = ckpt\nsubmit = pd.DataFrame({'Expected': predictions})\nsubmit.to_csv('submission.csv', sep=',', index_label='ID')",
"Building prefix dict from the default dictionary ...\nDEBUG:jieba:Building prefix dict from the default dictionary ...\nLoading model from cache /tmp/jieba.cache\nDEBUG:jieba:Loading model from cache /tmp/jieba.cache\nDumping model to file cache /tmp/jieba.cache\nDEBUG:jieba:Dumping model to file cache /tmp/jieba.cache\nDump cache file failed.\nTraceback (most recent call last):\n File \"/home/user_data/anaconda3/lib/python3.6/site-packages/jieba/__init__.py\", line 152, in initialize\n _replace_file(fpath, cache_file)\nPermissionError: [Errno 1] Operation not permitted: '/tmp/tmp2d02kovo' -> '/tmp/jieba.cache'\nERROR:jieba:Dump cache file failed.\nTraceback (most recent call last):\n File \"/home/user_data/anaconda3/lib/python3.6/site-packages/jieba/__init__.py\", line 152, in initialize\n _replace_file(fpath, cache_file)\nPermissionError: [Errno 1] Operation not permitted: '/tmp/tmp2d02kovo' -> '/tmp/jieba.cache'\nLoading model cost 0.884 seconds.\nDEBUG:jieba:Loading model cost 0.884 seconds.\nPrefix dict has been built succesfully.\nDEBUG:jieba:Prefix dict has been built succesfully.\n"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb70747f6ab776752b757e389317f1c13fc29b93 | 6,991 | ipynb | Jupyter Notebook | Big-Data-Clusters/CU14/public/content/log-analyzers/tsg088-get-datanode-logs.ipynb | glienard/tigertoolbox | 766f28d23139a25c884f60ba50cbd39438e52aca | [
"MIT"
]
| 1 | 2022-01-19T20:05:24.000Z | 2022-01-19T20:05:24.000Z | Big-Data-Clusters/CU14/public/content/log-analyzers/tsg088-get-datanode-logs.ipynb | glienard/tigertoolbox | 766f28d23139a25c884f60ba50cbd39438e52aca | [
"MIT"
]
| null | null | null | Big-Data-Clusters/CU14/public/content/log-analyzers/tsg088-get-datanode-logs.ipynb | glienard/tigertoolbox | 766f28d23139a25c884f60ba50cbd39438e52aca | [
"MIT"
]
| null | null | null | 6,991 | 6,991 | 0.631526 | [
[
[
"TSG088 - Hadoop datanode logs\n=============================\n\nSteps\n-----\n\n### Parameters",
"_____no_output_____"
]
],
[
[
"import re\n\ntail_lines = 500\n\npod = None # All\ncontainer = \"hadoop\"\nlog_files = [ \"/var/log/supervisor/log/datanode*.log\" ]\n\nexpressions_to_analyze = [\n re.compile(\".{23} WARN \"),\n re.compile(\".{23} ERROR \")\n]\n\nlog_analyzer_rules = []",
"_____no_output_____"
]
],
[
[
"### Instantiate Kubernetes client",
"_____no_output_____"
]
],
[
[
"# Instantiate the Python Kubernetes client into 'api' variable\n\nimport os\nfrom IPython.display import Markdown\n\ntry:\n from kubernetes import client, config\n from kubernetes.stream import stream\nexcept ImportError: \n\n # Install the Kubernetes module\n import sys\n !{sys.executable} -m pip install kubernetes \n \n try:\n from kubernetes import client, config\n from kubernetes.stream import stream\n except ImportError:\n display(Markdown(f'HINT: Use [SOP059 - Install Kubernetes Python module](../install/sop059-install-kubernetes-module.ipynb) to resolve this issue.'))\n raise\n\nif \"KUBERNETES_SERVICE_PORT\" in os.environ and \"KUBERNETES_SERVICE_HOST\" in os.environ:\n config.load_incluster_config()\nelse:\n try:\n config.load_kube_config()\n except:\n display(Markdown(f'HINT: Use [TSG118 - Configure Kubernetes config](../repair/tsg118-configure-kube-config.ipynb) to resolve this issue.'))\n raise\n\napi = client.CoreV1Api()\n\nprint('Kubernetes client instantiated')",
"_____no_output_____"
]
],
[
[
"### Get the namespace for the big data cluster\n\nGet the namespace of the Big Data Cluster from the Kuberenetes API.\n\n**NOTE:**\n\nIf there is more than one Big Data Cluster in the target Kubernetes\ncluster, then either:\n\n- set \\[0\\] to the correct value for the big data cluster.\n- set the environment variable AZDATA\\_NAMESPACE, before starting\n Azure Data Studio.",
"_____no_output_____"
]
],
[
[
"# Place Kubernetes namespace name for BDC into 'namespace' variable\n\nif \"AZDATA_NAMESPACE\" in os.environ:\n namespace = os.environ[\"AZDATA_NAMESPACE\"]\nelse:\n try:\n namespace = api.list_namespace(label_selector='MSSQL_CLUSTER').items[0].metadata.name\n except IndexError:\n from IPython.display import Markdown\n display(Markdown(f'HINT: Use [TSG081 - Get namespaces (Kubernetes)](../monitor-k8s/tsg081-get-kubernetes-namespaces.ipynb) to resolve this issue.'))\n display(Markdown(f'HINT: Use [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb) to resolve this issue.'))\n display(Markdown(f'HINT: Use [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) to resolve this issue.'))\n raise\n\nprint('The kubernetes namespace for your big data cluster is: ' + namespace)",
"_____no_output_____"
]
],
[
[
"### Get the Hadoop datanode logs from the hadoop container\n\n### Get tail for log",
"_____no_output_____"
]
],
[
[
"# Display the last 'tail_lines' of files in 'log_files' list\n\npods = api.list_namespaced_pod(namespace)\n\nentries_for_analysis = []\n\nfor p in pods.items:\n if pod is None or p.metadata.name == pod:\n for c in p.spec.containers:\n if container is None or c.name == container:\n for log_file in log_files:\n print (f\"- LOGS: '{log_file}' for CONTAINER: '{c.name}' in POD: '{p.metadata.name}'\")\n try:\n output = stream(api.connect_get_namespaced_pod_exec, p.metadata.name, namespace, command=['/bin/sh', '-c', f'tail -n {tail_lines} {log_file}'], container=c.name, stderr=True, stdout=True)\n except Exception:\n print (f\"FAILED to get LOGS for CONTAINER: {c.name} in POD: {p.metadata.name}\")\n else:\n for line in output.split('\\n'):\n for expression in expressions_to_analyze:\n if expression.match(line):\n entries_for_analysis.append(line)\n print(line)\nprint(\"\")\nprint(f\"{len(entries_for_analysis)} log entries found for further analysis.\")",
"_____no_output_____"
]
],
[
[
"### Analyze log entries and suggest relevant Troubleshooting Guides",
"_____no_output_____"
]
],
[
[
"# Analyze log entries and suggest further relevant troubleshooting guides\nfrom IPython.display import Markdown\n\nprint(f\"Applying the following {len(log_analyzer_rules)} rules to {len(entries_for_analysis)} log entries for analysis, looking for HINTs to further troubleshooting.\")\nprint(log_analyzer_rules)\nhints = 0\nif len(log_analyzer_rules) > 0:\n for entry in entries_for_analysis:\n for rule in log_analyzer_rules:\n if entry.find(rule[0]) != -1:\n print (entry)\n\n display(Markdown(f'HINT: Use [{rule[2]}]({rule[3]}) to resolve this issue.'))\n hints = hints + 1\n\nprint(\"\")\nprint(f\"{len(entries_for_analysis)} log entries analyzed (using {len(log_analyzer_rules)} rules). {hints} further troubleshooting hints made inline.\")",
"_____no_output_____"
],
[
"print(\"Notebook execution is complete.\")",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
cb707a83fcf87c6710437128ee08fd829fbe40b4 | 10,982 | ipynb | Jupyter Notebook | completed/02. Working with data files.ipynb | cjwinchester/2018-03-09-nicar-class | 7d05fe55e5181aa2c44d8989d2851d22196633e9 | [
"MIT"
]
| null | null | null | completed/02. Working with data files.ipynb | cjwinchester/2018-03-09-nicar-class | 7d05fe55e5181aa2c44d8989d2851d22196633e9 | [
"MIT"
]
| 5 | 2020-03-24T15:37:22.000Z | 2021-06-01T21:58:15.000Z | completed/02. Working with data files.ipynb | cjwinchester/2018-03-09-nicar-class | 7d05fe55e5181aa2c44d8989d2851d22196633e9 | [
"MIT"
]
| null | null | null | 32.587537 | 351 | 0.549353 | [
[
[
"# Working with data files\n\nReading and writing data files is a common task, and Python offers native support for working with many kinds of data files. Today, we're going to be working mainly with CSVs.",
"_____no_output_____"
],
[
"### Import the csv module\n\nWe're going to be working with delimited text files, so the first thing we need to do is import this functionality from the standard library.",
"_____no_output_____"
]
],
[
[
"import csv",
"_____no_output_____"
]
],
[
[
"### Opening a file to read the contents\n\nWe're going to use something called a [`with`](https://docs.python.org/3/reference/compound_stmts.html#with) statement to open a file and read the contents. The `open()` function takes at least two arguments: The path to the file you're opening and what [\"mode\"](https://docs.python.org/3/library/functions.html#open) you're opening it in.\n\nTo start with, we're going to use the `'r'` mode to read the data. We'll use the default arguments for delimiter -- comma -- and we don't need to specify a quote character.\n\n**Important:** If you open a data file in `w` (write) mode, anything that's already in the file will be erased.\n\nThe file we're using -- MLB roster data from 2017 -- lives at `data/mlb.csv`.\n\nOnce we have the file open, we're going to use some functionality from the `csv` module to iterate over the lines of data and print each one.\n\nSpecifically, we're going to use the `csv.reader` method, which returns a list of lines in the data file. Each line, in turn, is a list of the \"cells\" of data in that line.\n\nThen we're going to loop over the lines of data and print each line. We can also use bracket notation to retrieve elements from inside each line of data.",
"_____no_output_____"
]
],
[
[
"# open the MLB data file `as` mlb\nwith open('data/mlb.csv', 'r') as mlb:\n \n # create a reader object\n reader = csv.reader(mlb)\n \n # loop over the rows in the file\n for row in reader:\n \n # assign variables to each element in the row (shortcut!)\n name, team, position, salary, start_year, end_year, years = row\n \n # print the row, which is a list\n print(row)",
"_____no_output_____"
]
],
[
[
"### Simple filtering\n\nIf you wanted to filter your data, you could use an `if` statement inside your `with` block.",
"_____no_output_____"
]
],
[
[
"# open the MLB data file `as` mlb\nwith open('data/mlb.csv', 'r') as mlb:\n \n # create a reader object\n reader = csv.reader(mlb)\n\n # move past the header row\n next(reader)\n \n # loop over the rows in the file\n for row in reader:\n\n # assign variables to each element in the row (shortcut!)\n name, team, position, salary, start_year, end_year, years = row\n \n # print the line of data ~only~ if the player is on the Twins\n if team == 'MIN':\n \n # print the row, which is a list\n print(row)",
"_____no_output_____"
]
],
[
[
"### _Exercise_\n\nRead in the MLB data, print only the names and salaries of players who make at least $1 million. (Hint: Use type coercion!)",
"_____no_output_____"
]
],
[
[
"# open the MLB data file `as` mlb\nwith open('data/mlb.csv', 'r') as mlb:\n \n # create a reader object\n reader = csv.reader(mlb)\n \n # move past the header row\n next(reader)\n \n # loop over the rows in the file\n for row in reader:\n\n # assign variables to each element in the row (shortcut!)\n name, team, position, salary, start_year, end_year, years = row\n \n # print the line of data ~only~ if the player is on the Twins\n if int(salary) >= 1000000:\n \n # print the row, which is a list\n print(name, salary)",
"_____no_output_____"
]
],
[
[
"### DictReader: Another way to read CSV files\n\nSometimes it's more convenient to work with data files as a list of dictionaries instead of a list of lists. That way, you don't have to remember the position of each \"column\" of data -- you can just reference the column name. To do it, we'll use a `csv.DictReader` object instead of a `csv.reader` object. Otherwise the code is much the same.",
"_____no_output_____"
]
],
[
[
"# open the MLB data file `as` mlb\nwith open('data/mlb.csv', 'r') as mlb:\n \n # create a reader object\n reader = csv.DictReader(mlb)\n \n # loop over the rows in the file\n for row in reader:\n\n # print just the player's name (the column header is \"NAME\")\n print(row['NAME'])",
"_____no_output_____"
]
],
[
[
"### Writing to CSV files\n\nYou can also use the `csv` module to _create_ csv files -- same idea, you just need to change the mode to `'w'`. As with reading, there's a list-based writing method and a dictionary-based method.",
"_____no_output_____"
]
],
[
[
"# define the column names\nCOLNAMES = ['name', 'org', 'position']\n\n# let's make a few rows of data to write\nDATA_TO_WRITE = [\n ['Cody', 'IRE', 'Training Director'],\n ['Maggie', 'The New York Times', 'Reporter'],\n ['Donald', 'The White House', 'President']\n]\n\n# open an output file in write mode\nwith open('people-list.csv', 'w') as outfile:\n \n # create a writer object\n writer = csv.writer(outfile)\n \n # write the header row\n writer.writerow(COLNAMES)\n \n # loop over the data and write to file\n for human in DATA_TO_WRITE:\n writer.writerow(human)",
"_____no_output_____"
]
],
[
[
"### Using DictWriter to write data\n\nSimilar to using the list-based method, except that you need to ensure that the keys in your dictionaries of data match exactly a list of fieldnames.",
"_____no_output_____"
]
],
[
[
"# define the column names\nCOLNAMES = ['name', 'org', 'position']\n\n# let's make a few rows of data to write\nDATA_TO_WRITE = [\n {'name': 'Cody', 'org': 'IRE', 'position': 'Training Director'},\n {'name': 'Maggie', 'org': 'The New York Times', 'position': 'Reporter'},\n {'name': 'Donald', 'org': 'The White House', 'position': 'President'}\n]\n\n# open an output file in write mode\nwith open('people-dict.csv', 'w') as outfile:\n \n # create a writer object -- pass the list of column names to the `fieldnames` keyword argument\n writer = csv.DictWriter(outfile, fieldnames=COLNAMES)\n \n # use the writeheader method to write the header row\n writer.writeheader()\n \n # loop over the data and write to file\n for human in DATA_TO_WRITE:\n writer.writerow(human)",
"_____no_output_____"
]
],
[
[
"### You can open multiple files for reading/writing\n\nSometimes you want to open multiple files at the same time. One thing you might want to do: Opening a file of raw data in read mode, clean each row in a loop and write out the clean data to a new file.\n\nYou can open multiple files in the same `with` block -- just separate your `open()` functions with a comma.\n\nFor this example, we're not going to do any cleaning -- we're just going to copy the contents of one file to another.",
"_____no_output_____"
]
],
[
[
"# open the MLB data file `as` mlb\n# also, open `mlb-copy.csv` to write to\nwith open('data/mlb.csv', 'r') as mlb, open('mlb-copy.csv', 'w') as mlb_copy:\n \n # create a reader object\n reader = csv.DictReader(mlb)\n \n # create a writer object\n # we're going to use the `fieldnames` attribute of the DictReader object\n # as our output headers, as well\n # b/c we're basically just making a copy\n writer = csv.DictWriter(mlb_copy, fieldnames=reader.fieldnames)\n \n # write header row\n writer.writeheader()\n \n # loop over the rows in the file\n for row in reader:\n \n # what type of object is `row`?\n # how would we find out?\n \n # write row to output file\n writer.writerow(row)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb708aeab1ee152baf37a36301e59c9dab40375f | 33,759 | ipynb | Jupyter Notebook | Lesson2-Dataloaders.ipynb | dxgp/Learning-PyTorch | 97aa48c19b36a4ea5f6d2abdc49613acedd6ffd0 | [
"MIT"
]
| null | null | null | Lesson2-Dataloaders.ipynb | dxgp/Learning-PyTorch | 97aa48c19b36a4ea5f6d2abdc49613acedd6ffd0 | [
"MIT"
]
| null | null | null | Lesson2-Dataloaders.ipynb | dxgp/Learning-PyTorch | 97aa48c19b36a4ea5f6d2abdc49613acedd6ffd0 | [
"MIT"
]
| null | null | null | 113.285235 | 26,008 | 0.861133 | [
[
[
"# Datasets and Dataloaders",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch.utils.data import Dataset\nfrom torchvision import datasets\nfrom torchvision.transforms import ToTensor\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"training_data = datasets.FashionMNIST(\n root=\"data\",\n train=True,\n download=True,\n transform=ToTensor()\n)",
"Downloading http://fashion-mnist.s3-website.eu-central-1.amazonaws.com/train-images-idx3-ubyte.gz to data/FashionMNIST/raw/train-images-idx3-ubyte.gz\n"
],
[
"test_data = datasets.FashionMNIST(\n root=\"data\",\n train=False,\n download=True,\n transform=ToTensor()\n)",
"_____no_output_____"
],
[
"training_data.train_data[0].shape",
"/Users/gunjanpayal/miniforge3/envs/torch/lib/python3.8/site-packages/torchvision/datasets/mnist.py:58: UserWarning: train_data has been renamed data\n warnings.warn(\"train_data has been renamed data\")\n"
],
[
"labels_map = {\n 0: \"T-Shirt\",\n 1: \"Trouser\",\n 2: \"Pullover\",\n 3: \"Dress\",\n 4: \"Coat\",\n 5: \"Sandal\",\n 6: \"Shirt\",\n 7: \"Sneaker\",\n 8: \"Bag\",\n 9: \"Ankle Boot\",\n}\n\nfigure = plt.figure(figsize=(8, 8))\n\ncols, rows = 3, 3\n\nfor i in range(1, cols * rows + 1):\n sample_idx = torch.randint(len(training_data), size=(1,)).item()\n img, label = training_data[sample_idx]\n figure.add_subplot(rows, cols, i)\n plt.title(labels_map[label])\n plt.axis(\"off\")\n plt.imshow(img.squeeze(), cmap=\"gray\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## How to Create a Custom Dataset",
"_____no_output_____"
],
[
"A custom dataset class must implement __init__,__len__ and __getitem__.",
"_____no_output_____"
]
],
[
[
"import os\nimport pandas as pd\nfrom torchvision.io import read_image",
"_____no_output_____"
],
[
"class CustomImageDataset(Dataset):\n def __init__(self,annotations_file,img_dir,transform=None,target_transform=None):\n self.img_labels = pd.read_csv(annotations_file)\n self.img_dir = img_dir\n self.transform = transform\n self.target_transform = target_transform\n def __len__(self):\n return len(self.img_labels)\n def __getitem__(self,idx):\n img_path = os.path.join(self.img_dir,self.img_labels.iloc[idx,0])\n image = read_image(img_path)\n label = self.img_labels.iloc[idx,1]\n if self.transform:\n image = self.transform(image)\n if self.target_transform:\n label = self.target_transform(label)\n return image,label",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
cb708fb9de69bb3df5fe529f9bad95bc3aae6215 | 2,548 | ipynb | Jupyter Notebook | 2022/Udacity_Cloud-Architect-using-Microsoft-Azure-Scholarship-Program/cu02le01.ipynb | IsFilimonov/Learning | c8e8ca8b0006746423eb1f6daa5e46f6bced5167 | [
"MIT"
]
| null | null | null | 2022/Udacity_Cloud-Architect-using-Microsoft-Azure-Scholarship-Program/cu02le01.ipynb | IsFilimonov/Learning | c8e8ca8b0006746423eb1f6daa5e46f6bced5167 | [
"MIT"
]
| null | null | null | 2022/Udacity_Cloud-Architect-using-Microsoft-Azure-Scholarship-Program/cu02le01.ipynb | IsFilimonov/Learning | c8e8ca8b0006746423eb1f6daa5e46f6bced5167 | [
"MIT"
]
| null | null | null | 24.980392 | 158 | 0.582418 | [
[
[
"# Curriculum 2: Designing Infrastructure and Managing Migration",
"_____no_output_____"
],
[
"## Lesson 1: Introduction to Designing Infrastructure and Managing Migration",
"_____no_output_____"
],
[
"### Concept 7: Business Stakeholders",
"_____no_output_____"
],
[
"CLO (Chief Legal Officer) –– директор по юридическим вопросам. Должен быть вовлечен в процесс определения политик внедрения Azure Politics/Blueprint.\nCFO (Chief Finance Officer) –– директор по финансам. Должен быть обеспокоин стоимостью и затратами.\n\n<img src=\"imgs/cu02le01co07.png\" title=\"Стейкхолдеры\" width=\"400\" height=\"400\" />",
"_____no_output_____"
],
[
"### Concept 13: Tools and Environments",
"_____no_output_____"
],
[
"<img src=\"imgs/cu02le01co13.png\" title=\"Инструменты и среда\" width=\"400\" height=\"400\" />\n\nКонкретные ресурсы:\n- Azure Migrate\n- Azure AD\n- Azure Backup\n- Azure Virtual Machine\n- Virtual Network Gateway\n- Virtual network\n- local network gateway\n- Azure PowerShell\n- Azure CLI\n- App Service\n- DSC\n- Azure SQL Database\n- Azure SQL Managed instance\n- Azure Blueprint/Azure Policy\n- RBAC\n- Kubernetes\n- Azure Container Instance",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
cb708fc479bee8e96f975664088859c2a1a1c04a | 48,692 | ipynb | Jupyter Notebook | Chinenye Vivian James WT-21-049/Project 2 Ugandan Milk ImportExport.ipynb | ruthwaiharo/Week-5-Assessment | f320a9e553c9b723fff996128fcdca45bbe0f2b0 | [
"MIT"
]
| 1 | 2021-06-18T22:08:40.000Z | 2021-06-18T22:08:40.000Z | Chinenye Vivian James WT-21-049/Project 2 Ugandan Milk ImportExport.ipynb | ruthwaiharo/Week-5-Assessment | f320a9e553c9b723fff996128fcdca45bbe0f2b0 | [
"MIT"
]
| 4 | 2021-06-19T00:36:02.000Z | 2021-07-05T08:48:08.000Z | Chinenye Vivian James WT-21-049/Project 2 Ugandan Milk ImportExport.ipynb | ruthwaiharo/Week-5-Assessment | f320a9e553c9b723fff996128fcdca45bbe0f2b0 | [
"MIT"
]
| 68 | 2021-06-12T09:24:30.000Z | 2021-08-31T12:14:36.000Z | 44.960295 | 13,528 | 0.588577 | [
[
[
"## Project 2: Exploring the Uganda's milk imports and exports\nA country's economy depends, sometimes heavily, on its exports and imports. The United Nations Comtrade database provides data on global trade. It will be used to analyse the Uganda's imports and exports of milk in 2015:\n\n* How much does the Uganda export and import and is the balance positive (more exports than imports)?\n* Which are the main trading partners, i.e. from/to which countries does the Uganda import/export the most?\n* Which are the regular customers, i.e. which countries buy milk from the Uganda every month?\n* Which countries does the Uganda both import from and export to?",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.simplefilter('ignore', FutureWarning)\n\nfrom pandas import *\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Getting and preparing the data\n\nThe data is obtained from the [United Nations Comtrade](http://comtrade.un.org/data/) website, by selecting the following configuration:\n\n- Type of Product: goods\n- Frequency: monthly \n- Periods: Jan - May 2018\n- Reporter: Uganda\n- Partners: all\n- Flows: imports and exports\n- HS (as reported) commodity codes: 401 (Milk and cream, neither concentrated nor sweetened) and 402 (Milk and cream, concentrated or sweetened)",
"_____no_output_____"
]
],
[
[
"LOCATION = 'comrade_milk_ug_jan_dec_2015.csv'",
"_____no_output_____"
]
],
[
[
"On reading in the data, the commodity code has to be read as a string, to not lose the leading zero.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nmilk = pd.read_csv(LOCATION, dtype={'Commodity Code':str})\nmilk.tail(2)",
"_____no_output_____"
]
],
[
[
"The data only covers the first five months of 2015. Most columns are irrelevant for this analysis, or contain always the same value, like the year and reporter columns. The commodity code is transformed into a short but descriptive text and only the relevant columns are selected.",
"_____no_output_____"
]
],
[
[
"def milkType(code):\n if code == '401': # neither concentrated nor sweetened\n return 'unprocessed'\n if code == '402': # concentrated or sweetened\n return 'processed' \n return 'unknown'\n\nCOMMODITY = 'Milk and cream'\nmilk[COMMODITY] = milk['Commodity Code'].apply(milkType)\nMONTH = 'Period'\nPARTNER = 'Partner'\nFLOW = 'Trade Flow'\nVALUE = 'Trade Value (US$)'\nheadings = [MONTH, PARTNER, FLOW, COMMODITY, VALUE]\nmilk = milk[headings]\nmilk.head()",
"_____no_output_____"
]
],
[
[
"The data contains the total imports and exports per month, under the 'World' partner. Those rows are removed to keep only the per-country data.",
"_____no_output_____"
]
],
[
[
"milk = milk[milk[PARTNER] != 'World']\nmilk.head()",
"_____no_output_____"
],
[
"milk.tail()",
"_____no_output_____"
]
],
[
[
"## Total trade flow\nTo answer the first question, 'how much does the Uganda export and import and is the balance positive (more exports than imports)?', the dataframe is split into two groups: exports from the Uganda and imports into the Uganda. The trade values within each group are summed up to get the total trading.",
"_____no_output_____"
]
],
[
[
"grouped = milk.groupby([FLOW])\ngrouped[VALUE].aggregate(sum)",
"_____no_output_____"
]
],
[
[
"This shows a trade surplus of over 30 million dollars.",
"_____no_output_____"
],
[
"## Main trade partners\n\nTo address the second question, 'Which are the main trading partners, i.e. from/to which countries does the Uganda import/export the most?', the dataframe is split by country instead, and then each group aggregated for the total trade value. This is done separately for imports and exports. The result is sorted in descending order so that the main partners are at the top.",
"_____no_output_____"
]
],
[
[
"imports = milk[milk[FLOW] == 'Imports']\ngrouped = imports.groupby([PARTNER])\nprint('The Uganda imports from', len(grouped), 'countries.')\nprint('The 5 biggest exporters to the Uganda are:')\ntotalImports = grouped[VALUE].aggregate(sum).sort_values(inplace=False,ascending=False)\ntotalImports.head()",
"The Uganda imports from 24 countries.\nThe 5 biggest exporters to the Uganda are:\n"
]
],
[
[
"The export values can be plotted as a bar chart, making differences between countries easier to see.",
"_____no_output_____"
]
],
[
[
"totalImports.head(10).plot(kind='barh')",
"_____no_output_____"
]
],
[
[
"We can deduce that Switzerland is the lowest partnering company of milk to Uganda for imports.",
"_____no_output_____"
]
],
[
[
"exports = milk[milk[FLOW] == 'Exports']\ngrouped = exports.groupby([PARTNER])\nprint('The Uganda exports to', len(grouped), 'countries.')\nprint('The 5 biggest importers from the Uganda are:')\ngrouped[VALUE].aggregate(sum).sort_values(ascending=False,inplace=False).head()",
"The Uganda exports to 10 countries.\nThe 5 biggest importers from the Uganda are:\n"
]
],
[
[
"## Regular importers\n\nGiven that there are two commodities, the third question, 'Which are the regular customers, i.e. which countries buy milk from the Uganda every month?', is meant in the sense that a regular customer imports both commodities every month. This means that if the exports dataframe is grouped by country, each group has exactly ten rows (two commodities bought each of the five months). To see the countries, only the first month of one commodity has to be listed, as by definition it's the same countries every month and for the other commodity.",
"_____no_output_____"
]
],
[
[
"def buysEveryMonth(group):\n reply = len(group) == 20\n return reply\n\ngrouped = exports.groupby([PARTNER])\nregular = grouped.filter(buysEveryMonth)\nprint(regular)\nregular[(regular[MONTH] == 201501) & (regular[COMMODITY] == 'processed')]\n\n",
" Period Partner Trade Flow Milk and cream Trade Value (US$)\n5 201501 Rwanda Exports unprocessed 89479\n15 201502 Rwanda Exports unprocessed 40316\n28 201501 Rwanda Exports processed 28170\n30 201501 Kenya Exports processed 164116\n46 201502 Rwanda Exports processed 9870\n53 201503 Kenya Exports unprocessed 427005\n54 201503 Rwanda Exports unprocessed 60944\n62 201504 Kenya Exports unprocessed 164518\n65 201504 Rwanda Exports unprocessed 55548\n81 201503 Kenya Exports processed 2889812\n93 201504 Rwanda Exports processed 8794\n96 201504 Kenya Exports processed 491255\n102 201505 Kenya Exports unprocessed 823719\n104 201505 Rwanda Exports unprocessed 51333\n115 201506 Kenya Exports unprocessed 1329559\n117 201506 Rwanda Exports unprocessed 58566\n133 201505 Kenya Exports processed 4121189\n134 201505 Rwanda Exports processed 105002\n145 201506 Rwanda Exports processed 1898\n146 201506 Kenya Exports processed 4838396\n153 201507 Kenya Exports unprocessed 1588589\n154 201507 Rwanda Exports unprocessed 12670\n163 201508 Kenya Exports unprocessed 1347178\n164 201508 Rwanda Exports unprocessed 77258\n180 201507 Kenya Exports processed 4975538\n181 201507 Rwanda Exports processed 32000\n193 201508 Kenya Exports processed 1569400\n199 201509 Kenya Exports unprocessed 708175\n200 201509 Rwanda Exports unprocessed 46005\n212 201509 Kenya Exports processed 1748002\n223 201510 Kenya Exports unprocessed 748874\n224 201510 Rwanda Exports unprocessed 39260\n240 201510 Kenya Exports processed 313792\n247 201511 Kenya Exports unprocessed 691882\n248 201511 Rwanda Exports unprocessed 43399\n260 201511 Rwanda Exports processed 2364\n261 201511 Kenya Exports processed 624220\n270 201512 Kenya Exports unprocessed 671814\n271 201512 Rwanda Exports unprocessed 42565\n287 201512 Rwanda Exports processed 64025\n"
]
],
[
[
"Just over 5% of the total Uganda exports are due to these regular customers.",
"_____no_output_____"
]
],
[
[
"regular[VALUE].sum() / exports[VALUE].sum()",
"_____no_output_____"
]
],
[
[
"## Bi-directional trade\n\nTo address the fourth question, \n'Which countries does the Uganda both import from and export to?', a pivot table is used to list the total export and import value for each country. ",
"_____no_output_____"
]
],
[
[
"countries = pivot_table(milk, index=[PARTNER], columns=[FLOW], \n values=VALUE, aggfunc=sum)\ncountries.head()",
"_____no_output_____"
]
],
[
[
"Removing the rows with a missing value will result in only those countries with bi-directional trade flow with the Uganda.",
"_____no_output_____"
]
],
[
[
"countries.dropna()",
"_____no_output_____"
]
],
[
[
"## Conclusions\n\nThe milk and cream trade of the Uganda from January to December 2015 was analysed in terms of which countries the Uganda mostly depends on for income (exports) and goods (imports). Over the period, the Uganda had a trade surplus of over 1 million US dollars.\n\nKenya is the main partner, but it exported from the Uganda almost the triple in value than it imported to the Uganda. \n\nThe Uganda exported to over 100 countries during the period, but only imported from 24 countries, the main ones (top five by trade value) being not so geographically close (Kenya, Netherlands, United Arab Emirates, Oman, and South Africa). Kenya and Netherlands are the main importers that are not also main exporters except Kenya. \n\nThe Uganda is heavily dependent on its regular customers, the 10 countries that buy all types of milk and cream every month. They contribute three quarters of the total export value.\n\nAlthough for some, the trade value (in US dollars) is suspiciously low, which raises questions about the data's accuracy.\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
cb7090ce06e0f7e1099d52bb6ff0894468f4636f | 444,160 | ipynb | Jupyter Notebook | MLCourse/LinearRegression.ipynb | jashburn8020/ml-course | c360f84ea43fcd550120e98189076f30e85084cb | [
"Apache-2.0"
]
| null | null | null | MLCourse/LinearRegression.ipynb | jashburn8020/ml-course | c360f84ea43fcd550120e98189076f30e85084cb | [
"Apache-2.0"
]
| null | null | null | MLCourse/LinearRegression.ipynb | jashburn8020/ml-course | c360f84ea43fcd550120e98189076f30e85084cb | [
"Apache-2.0"
]
| null | null | null | 2,075.514019 | 131,511 | 0.656385 | [
[
[
"# Linear Regression",
"_____no_output_____"
],
[
"Let's fabricate some data that shows a roughly linear relationship between page speed and amount purchased:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nfrom pylab import *\n\npageSpeeds = np.random.normal(3.0, 1.0, 1000)\npurchaseAmount = 100 - (pageSpeeds + np.random.normal(0, 0.1, 1000)) * 3\n\nscatter(pageSpeeds, purchaseAmount)",
"_____no_output_____"
]
],
[
[
"As we only have two features, we can keep it simple and just use scipy.state.linregress:",
"_____no_output_____"
]
],
[
[
"from scipy import stats\n\nslope, intercept, r_value, p_value, std_err = stats.linregress(pageSpeeds, purchaseAmount)\n",
"_____no_output_____"
]
],
[
[
"Not surprisngly, our R-squared value shows a really good fit:",
"_____no_output_____"
]
],
[
[
"r_value ** 2",
"_____no_output_____"
]
],
[
[
"Let's use the slope and intercept we got from the regression to plot predicted values vs. observed:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\ndef predict(x):\n return slope * x + intercept\n\nfitLine = predict(pageSpeeds)\n\nplt.scatter(pageSpeeds, purchaseAmount)\nplt.plot(pageSpeeds, fitLine, c='r')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Activity",
"_____no_output_____"
],
[
"Try increasing the random variation in the test data, and see what effect it has on the r-squared error value.",
"_____no_output_____"
]
],
[
[
"pageSpeeds = np.random.normal(3.0, 1.0, 1000)\npurchaseAmount = 100 - (pageSpeeds + np.random.normal(0, 1, 1000)) * 3\n\nscatter(pageSpeeds, purchaseAmount)\n\nslope, intercept, r_value, p_value, std_err = stats.linregress(pageSpeeds, purchaseAmount)\nprint(f\"r-squared: {r_value ** 2}\")",
"r-squared: 0.4814458616713199\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
cb70a48eaa0179243b452a2e2701a892174eb6de | 331,831 | ipynb | Jupyter Notebook | Projects_Jupyter/advertising_in_e-learning_markets.ipynb | FlorentCLMichel/learning_data_science | d9ccc0a85609406b2c77a91db96dba8c97fc9ac4 | [
"MIT"
]
| null | null | null | Projects_Jupyter/advertising_in_e-learning_markets.ipynb | FlorentCLMichel/learning_data_science | d9ccc0a85609406b2c77a91db96dba8c97fc9ac4 | [
"MIT"
]
| null | null | null | Projects_Jupyter/advertising_in_e-learning_markets.ipynb | FlorentCLMichel/learning_data_science | d9ccc0a85609406b2c77a91db96dba8c97fc9ac4 | [
"MIT"
]
| null | null | null | 77.349883 | 24,224 | 0.594601 | [
[
[
"# Finding the best market to adverts in e-learning\n\nThe aim of this project is to give examples of how to use basic concepts in Statistics, such as mean values, medians, ranges, and standard deviations, to answer questions using real-world data. \nTo be concrete, we will focus on Programming courses markets. \nUsing a real-world dataset, we will determine what are the best markets to advertise in and estimate the extent to which these results should be trusted.\n\n## The dataset\n\nThe dataset we will use, `2017-fCC-New-Coders-Survey-Data.csv`, was downloaded from [this Github repository](https://github.com/freeCodeCamp/2017-new-coder-survey). \nIt was used by Quincy Larson, founder of the e-learning platform [freeCodeCamp](https://www.freecodecamp.org/), to write [this article on Medium](https://www.freecodecamp.org/news/we-asked-20-000-people-who-they-are-and-how-theyre-learning-to-code-fff5d668969/) about new coders (defined as people who had been coding for less than 5 years) in 2017. \nThe questions asked in the survey can be found in `2017-fCC-New-Coders-Survey-Data_questions.csv`.\n\nLet us first import the modules we will need:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"We then import the dataset, find its shape, and print the five first rows:",
"_____no_output_____"
]
],
[
[
"path_to_csv = '../Data/2017-fCC-New-Coders-Survey-Data.csv'\ndf = pd.read_csv(path_to_csv)\nprint(df.shape)\ndf.head()",
"(18175, 136)\n"
]
],
[
[
"This dataframe has 18175 rows and 136 columns. \nLet us list them and their main properties: ",
"_____no_output_____"
]
],
[
[
"pd.set_option('display.max_columns', 200) # to print all columns\ndf.describe(include='all')",
"_____no_output_____"
]
],
[
[
"## Job role interests\n\nWe first want to determine which jobs new coders are interested in, which will in turn determine the topics they should be interested in learning. \nTo this end, we generate a relative frequency table for the `JobInterest` columns and represent it graphically as a bar plot.",
"_____no_output_____"
]
],
[
[
"# number of bars in the plot\nnbars = 11\n\n# job interests with their own column\nlist_jobs = ['BackEnd', 'DataEngr', 'DataSci', 'DevOps', 'FrontEnd', \n 'FullStack', 'GameDev', 'InfoSec', 'Mobile', 'ProjMngr',\n 'QAEngr', 'UX']\nfreqs = {}\nnrows = df.shape[0]\nfor job in list_jobs: \n name_col = 'JobInterest' + job\n freqs[name_col] = df[name_col].sum() / nrows\n \n# job interests in the 'other' column\nfor job in df['JobInterestOther']:\n if pd.isna(job):\n pass\n elif job in freqs.keys(): \n freqs[job] += 1 / nrows\n else:\n freqs[job] = 1 / nrows\n\n# turn the dictionary to two lists, ordered by the y value in descending order\nx, y = zip(*sorted(freqs.items(), key=lambda item: -item[1])) \nplt.bar(x, y)\nplt.xlim(-0.6, nbars-0.4)\nplt.xticks(rotation=90)\nplt.grid()\nplt.title('Frequency of job interests')\nplt.show()",
"_____no_output_____"
]
],
[
[
"We find that: \n* a plurality (more than 20%) are interested in Full Stack roles,\n* the next two most popular careers are Front End Developer and Back End Developer.\n\nLet us now sum the values:",
"_____no_output_____"
]
],
[
[
"sum(y)",
"_____no_output_____"
]
],
[
[
"Values add up to more than 1.4, meaning that, on average, a new coder contemplates approximately 1.4 different careers. \n*It may thus be valuable to put forward learning opportunities which may lead to different careers.* \nTo help selecting them, let us plot the correlation map between expressions of interests in different career opportunities. \nFor simplicity, we focus on those having their own columns, which from the graph above covers at least the 11 most popular career opportunities. ",
"_____no_output_____"
]
],
[
[
"df[['JobInterest' + job for job in list_jobs]].fillna(0).corr().style.background_gradient(cmap='coolwarm')",
"_____no_output_____"
]
],
[
[
"There is a significant (larger than 0.4) positive correlation between interests in Full Stack, Front End, Back End, and Mobile development. \nSince these are also the four most popular career opportunities, *it would make sense to advertise e-learning content which can be useful to these four roles*.\n\nThere is also a significant correlation between interest for Data Scientist and Data Engineer careers, which are both among the ten most popular career opportunities. \n*It would thus also make sense to advertise content relevent to these two roles.*\n\n## Best countries to advertise in\n\nWe now focus on the geographic location of new coders to determine which countries an advertising campaign should focus on. \nWe focus on coders who have expressed interest for at least one of the 10 most popular careers and plot a frequency table of the `CountryLive` column, indicating in which country each new coder lives.",
"_____no_output_____"
]
],
[
[
"# list of the 10 most popular careers\nmost_popular_careers = list(x[:10])\n\n# dataframe containing only the rows for coders having expressed interest in at \n# least one of the 10 most popular careers\ndf_interested_10 = df[df[most_popular_careers].sum(axis=1) > 0.]\n\n# frequency table\nfreq_table_country = df_interested_10['CountryLive'].value_counts()\nfreq_table_country_per = df_interested_10['CountryLive'].value_counts(normalize=True) * 100\n\n# bar plot of the frequency table, showing only the 5 most represented \n# countries\n\nnbars = 5\n\nfreq_table_country.plot.bar()\nplt.xlim(-0.5,nbars-0.5)\nplt.grid()\nplt.title('Number of new coders living in each country')\nplt.show()\n\nfreq_table_country_per.plot.bar()\nplt.xlim(-0.5,nbars-0.5)\nplt.ylabel('%')\nplt.grid()\nplt.title('Percentage of new coders living in each country')\nplt.show()",
"_____no_output_____"
]
],
[
[
"A plurality (more than 40%, corresponding to more than 3000 coders in the survey) of new coders live in the United States of America (USA).\nThe next two most represented countries are India and the United Kingdom (UK), with less than 10% each, followed by Canada and Poland.\nMoreover, the first four are all English-speaking countries, which could make it easy to extent a campaign from one to another.\n*An advertising campagign could thus start in the USA, before being potentially extended to India, the UK, and Canada.* \n\nLet us check that the previous results about job interests remain true when restricting to coders living in the USA, which could be the primary target of a campaign.",
"_____no_output_____"
]
],
[
[
"# number of bars in the plot\nnbars = 11\n\n# country where \ncountry = 'United States of America'\n\n# number of rows\nnrows_USA = df[df['CountryLive'] == country].shape[0]\n\n# job interests with their own column\nfreqs_USA = {}\nfor job in list_jobs: \n name_col = 'JobInterest' + job\n freqs_USA[name_col] = df[df['CountryLive'] == country][name_col].sum() / nrows_USA\n \n# job interests in the 'other' column\nfor job in df[df['CountryLive'] == country]['JobInterestOther']:\n if pd.isna(job):\n pass\n elif job in freqs_USA.keys(): \n freqs_USA[job] += 1 / nrows_USA\n else:\n freqs_USA[job] = 1 / nrows_USA\n\n# turn the dictionary to two lists, ordered by the y value in descending order\nx, y = zip(*sorted(freqs_USA.items(), key=lambda item: -item[1])) \nplt.bar(x, y)\nplt.xlim(-0.6, nbars-0.4)\nplt.xticks(rotation=90)\nplt.grid()\nplt.title('Frequency of job interests in the USA')\nplt.show()",
"_____no_output_____"
]
],
[
[
"This frequency table looks very similar to the one obtaned above from the full dataset. \nAll frequencies tend to be a bit larger, which does not affect our conclusions. \nLet us check that the correlations we found above are also still present: ",
"_____no_output_____"
]
],
[
[
"df[df['CountryLive'] == country][['JobInterest' + job for job in list_jobs]].fillna(0).corr().style.background_gradient(cmap='coolwarm')",
"_____no_output_____"
]
],
[
[
"There are still strong positive correlations between interest for the Full Stack, Front End, and Back End careers, as well as the data Scientist and Data Engineer ones.\n\nAnother important information for deciding which countries to invest in is how much coders are willign to pay for learning. \nTo estimate it, we first add a new column `MoneyPerMonth` showing, in US Dollars, how much each coder has spent per month since they started programming. \nIt is obtained by dividng the `MoneyForLearning` column by the `MonthsProgramming` one, after replacing 0s in the later by 1s. \n(Since most subscriptions are monthly, one can expect the total amount spent by someone who has just started coding to be a fair estimation of what they pay for the first month.)",
"_____no_output_____"
]
],
[
[
"df['MoneyPerMonth'] = df['MoneyForLearning'] / df['MonthsProgramming'].replace(0, 1)",
"_____no_output_____"
]
],
[
[
"We then group the results by country and show summary statistics for the four ones where the number of new coders is highest.",
"_____no_output_____"
]
],
[
[
"# column to consider\ncolumn = 'MoneyPerMonth'\n\n# countries to be considered\nlist_countries = ['United States of America', 'India', 'United Kingdom', 'Canada']\n\n# dataframe grouped by country\ndf_country = df.groupby(['CountryLive'])[column]\n\n# mean values\nmean_money_per_month = df_country.mean()\n\n# standard deviations\nstd_money_per_month = df_country.std()\n\n# stanard errors\nste_money_per_month = std_money_per_month / np.sqrt(df_country.size())\n\nphrase = '{}: mean = ${:.0f}±{:.0f}, std = ${:.0f}'\nfor country in list_countries: \n print(phrase.format(country, mean_money_per_month[country], ste_money_per_month[country], std_money_per_month[country]))",
"United States of America: mean = $206±24, std = $1808\nIndia: mean = $67±12, std = $442\nUnited Kingdom: mean = $56±9, std = $239\nCanada: mean = $118±15, std = $366\n"
]
],
[
[
"It seems that coders in the USA spend more for learning than coders in the other three countries on average. \nHowever, the large standard deviation may indicate that the result is biased by a few high-payers. \nLet us show box plots of the distributions of monthly spending in each of these four countries:",
"_____no_output_____"
]
],
[
[
"# dataframe grouped by country\ndf[df['CountryLive'].isin(list_countries)].boxplot(column, 'CountryLive')\nplt.xticks(rotation=90)\nplt.ylabel('$')\nplt.title('Money spent per month')\nplt.suptitle('')\nplt.show()",
"_____no_output_____"
]
],
[
[
"There seems to be a significant number of outliers, which may well bias the analysis. \nTo get a better sense of the data, let us focus on coders spending less than a given threshold, which we choose as $1000, as it is unlikely that many coders will regularly spend more than that on learning each month. \nHigher values may point to misreportings, other errors, or a lot of money spent on attending bootcamps, which are not covered by our advertising campaign.",
"_____no_output_____"
]
],
[
[
"threshold = 1000\n\n# dataframe grouped by country\ndf[(df['CountryLive'].isin(list_countries)) & (df[column] < threshold)].boxplot(column, 'CountryLive')\nplt.xticks(rotation=90)\nplt.ylabel('$')\nplt.title('Money spent per month')\nplt.suptitle('')\nplt.show()",
"_____no_output_____"
]
],
[
[
"These box plots seem more sensible. \nWe can alread see that more than half of new coders in each of these countries do not spend money on learning (the median is zero in all cases). \n\nLet us re-compute the means and standard deviations with this threshold:",
"_____no_output_____"
]
],
[
[
"# dataframe grouped by country, keepong only the rows where MoneyPerMonth is\n# below the threshold\ndf_country = df[df[column] < threshold].groupby(['CountryLive'])[column]\n\n# mean values\nmean_money_per_month = df_country.mean()\n\n# standard deviations\nstd_money_per_month = df_country.std()\n\n# stanard errors\nste_money_per_month = std_money_per_month / np.sqrt(df_country.size())\n\nphrase = '{}: mean = ${:.1f}±{:.1f}, std = ${:.1f}'\nfor country in list_countries: \n print(phrase.format(country, mean_money_per_month[country], ste_money_per_month[country], std_money_per_month[country]))",
"United States of America: mean = $59.1±2.0, std = $146.8\nIndia: mean = $21.9±2.1, std = $74.3\nUnited Kingdom: mean = $29.4±3.6, std = $94.0\nCanada: mean = $56.9±6.6, std = $154.9\n"
]
],
[
[
"The average amounts of money spent per month by new coders is largest in the USA and smallest in India.\n\n## Conclusions\n\nFrom this brief study, it seems clear that the best market to advertise in is the USA, and that the campaign should focus on skills relevent to Full Stack, Front End, and Back End development. \nPossible future extensions of the campaign, if successful, could include the UK, Canada, and India, as well as skills relevant to Data Science and Data Engineering. ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
cb70d65d8599fcd489a8ccc8d883fa07dcb53ff2 | 35,730 | ipynb | Jupyter Notebook | TwitterSentimentAnalysis.ipynb | AliKarasneh/create-react-app | a09b387b44dad9f97b5cfbb5d03ffcfb10c4832c | [
"MIT"
]
| null | null | null | TwitterSentimentAnalysis.ipynb | AliKarasneh/create-react-app | a09b387b44dad9f97b5cfbb5d03ffcfb10c4832c | [
"MIT"
]
| null | null | null | TwitterSentimentAnalysis.ipynb | AliKarasneh/create-react-app | a09b387b44dad9f97b5cfbb5d03ffcfb10c4832c | [
"MIT"
]
| null | null | null | 112.00627 | 15,714 | 0.670221 | [
[
[
"<a href=\"https://colab.research.google.com/github/AliKarasneh/create-react-app/blob/master/TwitterSentimentAnalysis.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip install langdetect\n",
"Collecting langdetect\n Downloading langdetect-1.0.9.tar.gz (981 kB)\n\u001b[?25l\r\u001b[K |▍ | 10 kB 23.3 MB/s eta 0:00:01\r\u001b[K |▊ | 20 kB 28.2 MB/s eta 0:00:01\r\u001b[K |█ | 30 kB 21.4 MB/s eta 0:00:01\r\u001b[K |█▍ | 40 kB 17.1 MB/s eta 0:00:01\r\u001b[K |█▊ | 51 kB 8.5 MB/s eta 0:00:01\r\u001b[K |██ | 61 kB 8.0 MB/s eta 0:00:01\r\u001b[K |██▍ | 71 kB 8.3 MB/s eta 0:00:01\r\u001b[K |██▊ | 81 kB 9.3 MB/s eta 0:00:01\r\u001b[K |███ | 92 kB 7.1 MB/s eta 0:00:01\r\u001b[K |███▍ | 102 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███▊ | 112 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████ | 122 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████▍ | 133 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████▊ | 143 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████ | 153 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████▍ | 163 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████▊ | 174 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████ | 184 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████▍ | 194 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████▊ | 204 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████ | 215 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████▍ | 225 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████▊ | 235 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████ | 245 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████▍ | 256 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████▊ | 266 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████ | 276 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████▍ | 286 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████▊ | 296 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████ | 307 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████▍ | 317 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████▊ | 327 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████ | 337 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████▍ | 348 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████▊ | 358 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████ | 368 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████▍ | 378 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████▊ | 389 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████ | 399 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████▍ | 409 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████▊ | 419 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████ | 430 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████▍ | 440 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████▊ | 450 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████ | 460 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████▍ | 471 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████▊ | 481 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████ | 491 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████▍ | 501 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████▊ | 512 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████ | 522 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████▍ | 532 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████▊ | 542 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████ | 552 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████▍ | 563 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████▊ | 573 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████ | 583 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████▍ | 593 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████▊ | 604 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████ | 614 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████▍ | 624 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████▊ | 634 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████ | 645 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████▍ | 655 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████▊ | 665 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████ | 675 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████▍ | 686 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████▊ | 696 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████ | 706 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████▍ | 716 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████▊ | 727 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████████ | 737 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████████▍ | 747 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████████▊ | 757 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████████ | 768 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████████▍ | 778 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████████▊ | 788 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████████ | 798 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████████▍ | 808 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████████▊ | 819 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████████ | 829 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████████▍ | 839 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████████▊ | 849 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████████████ | 860 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████████████▍ | 870 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████████████▊ | 880 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████████████ | 890 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████████████▍ | 901 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████████████████████████████▊ | 911 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████ | 921 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▍ | 931 kB 7.7 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▊ | 942 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████████████ | 952 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████████████▍| 962 kB 7.7 MB/s eta 0:00:01\r\u001b[K |███████████████████████████████▊| 972 kB 7.7 MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 981 kB 7.7 MB/s \n\u001b[?25hRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from langdetect) (1.15.0)\nBuilding wheels for collected packages: langdetect\n Building wheel for langdetect (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for langdetect: filename=langdetect-1.0.9-py3-none-any.whl size=993241 sha256=1b6a5f48a643fb0574494a0696bca180fa4035e6c0c53a883b58fcfc8e69be54\n Stored in directory: /root/.cache/pip/wheels/c5/96/8a/f90c59ed25d75e50a8c10a1b1c2d4c402e4dacfa87f3aff36a\nSuccessfully built langdetect\nInstalling collected packages: langdetect\nSuccessfully installed langdetect-1.0.9\n"
],
[
"!pip install tweepy",
"Requirement already satisfied: tweepy in /usr/local/lib/python3.7/dist-packages (3.10.0)\nRequirement already satisfied: six>=1.10.0 in /usr/local/lib/python3.7/dist-packages (from tweepy) (1.15.0)\nRequirement already satisfied: requests[socks]>=2.11.1 in /usr/local/lib/python3.7/dist-packages (from tweepy) (2.23.0)\nRequirement already satisfied: requests-oauthlib>=0.7.0 in /usr/local/lib/python3.7/dist-packages (from tweepy) (1.3.0)\nRequirement already satisfied: oauthlib>=3.0.0 in /usr/local/lib/python3.7/dist-packages (from requests-oauthlib>=0.7.0->tweepy) (3.1.1)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests[socks]>=2.11.1->tweepy) (2.10)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests[socks]>=2.11.1->tweepy) (2021.5.30)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests[socks]>=2.11.1->tweepy) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests[socks]>=2.11.1->tweepy) (3.0.4)\nRequirement already satisfied: PySocks!=1.5.7,>=1.5.6 in /usr/local/lib/python3.7/dist-packages (from requests[socks]>=2.11.1->tweepy) (1.7.1)\n"
],
[
"from PIL import Image\nfrom nltk.sentiment.vader import SentimentIntensityAnalyzer\nfrom langdetect import detect\nfrom nltk.stem import SnowballStemmer\nfrom nltk.sentiment.vader import SentimentIntensityAnalyzer\nfrom sklearn.feature_extraction.text import CountVectorizer\nimport tweepy\nfrom textblob import TextBlob\nimport nltk\nimport pandas as pd\nfrom matplotlib import pyplot as plt\n\n\n# Authentication\nconsumerKey = 'rZms1g0HlFHDxlUmTbMV20vFk'\nconsumerSecret = '8ZygdjdRd0YdKXTvIvh07fD2SwCp1hFhnLNcM5qgD6GZ1dl02Q'\naccessToken = '176112318-wVeHbbHx6A8wxgrJBHoLgVFitlUjzsqfe5OfSOcL'\naccessTokenSecret = 'UMPsVRWULXSVEWKouANKjf1cmiIjLatwdCUUNQs5tXUm9'\nauth = tweepy.OAuthHandler(consumerKey, consumerSecret)\nauth.set_access_token(accessToken, accessTokenSecret)\napi = tweepy.API(auth)",
"_____no_output_____"
],
[
"#Sentiment Analysis\ndef percentage(part,whole):\n return 100 * float(part)/float(whole)\nkeyword = input('Please enter keyword or hashtag to search: ')\nnoOfTweet = int(input ('Please enter how many tweets to analyze: '))\ntweets = tweepy.Cursor(api.search, q=keyword).items(noOfTweet)\npositive = 0\nnegative = 0\nneutral = 0\npolarity = 0\ntweet_list = []\nneutral_list = []\nnegative_list = []\npositive_list = []\nfor tweet in tweets:\n nltk.download('vader_lexicon')\n #nltk.download()\n #print(tweet.text)\n tweet_list.append(tweet.text)\n analysis = TextBlob(tweet.text)\n score = SentimentIntensityAnalyzer().polarity_scores(tweet.text)\n neg = score['neg']\n neu = score['neu']\n pos = score['pos']\n comp = score['compound']\n polarity += analysis.sentiment.polarity\n \nif neg > pos:\n negative_list.append(tweet.text)\n negative += 1\n\nelif pos > neg:\n positive_list.append(tweet.text)\n positive += 1\n\nelif pos == neg:\n neutral_list.append(tweet.text)\n neutral += 1\npositive = percentage(positive, noOfTweet)\nnegative = percentage(negative, noOfTweet)\nneutral = percentage(neutral, noOfTweet)\npolarity = percentage(polarity, noOfTweet)\npositive = format(positive, '.1f')\nnegative = format(negative, '.1f')\nneutral = format(neutral, '.1f')\n\n#Number of Tweets (Total, Positive, Negative, Neutral)\ntweet_list = pd.DataFrame(tweet_list)\nneutral_list = pd.DataFrame(neutral_list)\nnegative_list = pd.DataFrame(negative_list)\npositive_list = pd.DataFrame(positive_list)\nprint('total number: ',len(tweet_list))\nprint('positive number :',len(positive_list))\nprint('negative number: ', len(negative_list))\nprint('neutral number: ',len(neutral_list))\n\n#Creating PieCart\nlabels = ['Positive ['+str(positive)+'%]' , 'Neutral ['+str(neutral)+'%]','Negative ['+str(negative)+'%]']\nsizes = [positive, neutral, negative]\ncolors = ['yellowgreen', 'blue','red']\npatches, texts = plt.pie(sizes,colors=colors, startangle=90)\nplt.style.use('default')\nplt.legend(labels)\nplt.title('Sentiment Analysis Result for keyword= '+keyword+'' )\nplt.axis('equal')\nplt.show()\ntweet_list",
"Please enter keyword or hashtag to search: tesla\nPlease enter how many tweets to analyze: 5\n[nltk_data] Downloading package vader_lexicon to /root/nltk_data...\n[nltk_data] Package vader_lexicon is already up-to-date!\n[nltk_data] Downloading package vader_lexicon to /root/nltk_data...\n[nltk_data] Package vader_lexicon is already up-to-date!\n[nltk_data] Downloading package vader_lexicon to /root/nltk_data...\n[nltk_data] Package vader_lexicon is already up-to-date!\n[nltk_data] Downloading package vader_lexicon to /root/nltk_data...\n[nltk_data] Package vader_lexicon is already up-to-date!\n[nltk_data] Downloading package vader_lexicon to /root/nltk_data...\n[nltk_data] Package vader_lexicon is already up-to-date!\ntotal number: 5\npositive number : 0\nnegative number: 0\nneutral number: 1\n"
],
[
"\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
cb70ed28d0594ba376c8b717c71e33c733d38cc1 | 23,992 | ipynb | Jupyter Notebook | notes_on_feature_extraction/Local_nuclear_density.ipynb | GVS-Lab/genomic-scoring-breast-cancer-progression | 5b8b8d9945a1bec5f6d5b5fc03a23f6835cbc036 | [
"MIT"
]
| null | null | null | notes_on_feature_extraction/Local_nuclear_density.ipynb | GVS-Lab/genomic-scoring-breast-cancer-progression | 5b8b8d9945a1bec5f6d5b5fc03a23f6835cbc036 | [
"MIT"
]
| null | null | null | notes_on_feature_extraction/Local_nuclear_density.ipynb | GVS-Lab/genomic-scoring-breast-cancer-progression | 5b8b8d9945a1bec5f6d5b5fc03a23f6835cbc036 | [
"MIT"
]
| null | null | null | 34.973761 | 213 | 0.363079 | [
[
[
"## Local nuclear density\n\nHere we aim to characterise the crowding experienced by each nuclei and use distribution descriptors of these features at the tissue level to describe crowding at the tissue scale. ",
"_____no_output_____"
]
],
[
[
"# import libraries\n%load_ext autoreload\nimport sys\nsys.path.append(\"..\")\n\nfrom tifffile import imread\nimport pandas as pd \nimport numpy as np \nfrom skimage import measure\nimport src.spatial_features.Nuclear_neighbourhood_density as NND",
"_____no_output_____"
]
],
[
[
"#### Approach:\n\nWe have used two approaches to achieve this goal. \n\n1. We obtain the number of neighbours of each nucleus within a fixed radius R, were R is 20, 40, 50, 100, 200 pixels. \n2. The other approach we used is to obtain the distance of between a nuclei at its Kth nearest neighbour, where K is 1, 3, 5, 10, 20. \n\nNote that here we have used centroid to centroid distances. ",
"_____no_output_____"
]
],
[
[
"image_path = '/home/pathy_s/Documents/TMA/Stardist_seg_results/Instance_segmentation/BR2082B_H15.tif'\n# Read in the image\nimg = imread(image_path)\n\n#Get features for the individual nuclei in the image\nfeat = measure.regionprops_table(img,properties = ('label','centroid'))\n\nknn_dist = NND.distance_to_k_nneigh(feat,[1,3,5,10,20])\nnum_neigh_rad = NND.num_neigbours_in_Radius(feat, [20,50,100,150,200])\npd.merge(num_neigh_rad,knn_dist, on=\"label\", how=\"outer\")",
"_____no_output_____"
]
],
[
[
"For a quick extraction of all features given a segmented image use the following code:",
"_____no_output_____"
]
],
[
[
"from src.utlis.Run_nuclear_local_density_estimation import local_nuclear_density\nlocal_nuclear_density(image_path)",
"_____no_output_____"
]
],
[
[
"#### Tissue level summary:\n\nIn order to characterise the nuclear density/crowding in a given tissue, we compute the distribution characteristics of each of the above features. \n\nThe measures available are: Median, Min, Max, Standard Deviation (SD) Coefficient of Variation (CV) and Coefficient of Dispersion (CD), Inter_Quartile_Range(IQR) and Quartile Coeeffient of Dispersrion (QCD).",
"_____no_output_____"
]
],
[
[
"from src.utlis.summarising_features import summarise_feature_table\nsummarise_feature_table(local_nuclear_density(image_path))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb70effff4822f7e2699618bc36e3f6d8caf755f | 69,306 | ipynb | Jupyter Notebook | Assignment 1 I310D .ipynb | pablo-carbajo1/Assignment1-I310D | 8a2c9ccd2f3bca1d07e6334767714651fbccd0c1 | [
"MIT"
]
| null | null | null | Assignment 1 I310D .ipynb | pablo-carbajo1/Assignment1-I310D | 8a2c9ccd2f3bca1d07e6334767714651fbccd0c1 | [
"MIT"
]
| null | null | null | Assignment 1 I310D .ipynb | pablo-carbajo1/Assignment1-I310D | 8a2c9ccd2f3bca1d07e6334767714651fbccd0c1 | [
"MIT"
]
| 1 | 2022-02-13T23:42:02.000Z | 2022-02-13T23:42:02.000Z | 291.201681 | 26,060 | 0.926615 | [
[
[
" ## Visualizing and Comparing hours enrolled by UT students during the Fall 2020 semester \n \n This project aims to see what are the different hours that UT students were enrolled in during the Fall 2020 semester. It will try to see if there any trends or differences based on the gender\n \n ### Libraries used for the visualization of the data ",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nimport pandas as pd \nimport numpy as np\n",
"_____no_output_____"
]
],
[
[
"## Data from UT's statistical handbook for the Fall 2020 semester \n\n###### Data about women enrollment (Fall 2020):\n\nPart-time (< 12 hours) |Full-time (> 12 hours) \n:-----:|:-----:| \n1,318|21,014|\n5,9 %|94,1 %|",
"_____no_output_____"
],
[
"###### Data about men enrollment (Fall 2020):\n\nPart-time (< 12 hours) |Full-time (> 12 hours) \n:-----:|:-----:| \n1,326|16,390|\n7,5 %|92,5 %|",
"_____no_output_____"
],
[
"## Visualization for this data ",
"_____no_output_____"
]
],
[
[
"fig, (ax1,ax2) = plt.subplots(1,2,figsize=(10,10)) #ax1,ax2 refer to your two pies\n# 1,2 denotes 1 row, 2 columns\nfig.subplots_adjust(wspace=1)\n\n\nlabels = 'Full-time', 'Part-time'\nsizes = [94.1, 5.9]\nexplode = (0.3, 0)\nax1.pie (sizes,labels = labels, autopct = '%.1f%%', shadow = True, explode = explode) #plot first pie\nax1.set_title ('Women enrolled in Full-time hours vs. Part-time hours')\nax1.axis = ('equal')\n\n\nlabels = 'Full-time', 'Part-time'\nsizes = [92.5, 7.5]\nexplode = (0.3, 0)\nax2.pie(sizes,labels = labels, autopct = '%.1f%%', shadow = True, explode = explode) #plot second pie\nax2.set_title('Women enrolled in Full-time hours vs. Part-time hours')\nax2.axis = (\"equal\")\n\n",
"_____no_output_____"
]
],
[
[
"As we can see, there were more women enrolled Full-time than men enrolled Full-time and the number of Part-time students was also higher for men.\n\n---\n## Data collected from the form to know how many hours students were enrolled in and they were Full-time students (> 12 hours) \n\nI led a form asking for the number of hours current seniors were enrolled during the Fall 2020 semester. I collected 20 responses and these were the results.",
"_____no_output_____"
]
],
[
[
"labels = '12 hours', '15 hours', '18 hours'\nsizes = [30, 65, 5]\nexplode = (0, 0, 0.3)\nplt.pie (sizes,labels = labels, autopct = '%.1f%%', shadow = True, explode = explode) \nplt.title ('Enrolled hours by Full-time UT students during the Fall 2020 semester')\nplt.axis = ('equal')\n",
"_____no_output_____"
]
],
[
[
"---\n\n## Representing and visualizing this data \n\nI decided to represent these values with binary numbers (0 and 1). 0 means \"Yes\" and 1 means \"No\". I represented this numbers on a Google Sheets spreadsheet and these were the results that I found.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(8,5))\nx=['Part-time', '12 hours', '15 hours', '18 hours']\ny= [7,28,60,5]\nplt.bar(x,y, color=\"#bdeb13\")\nplt.xlabel(\"Hours enrolled by students (Fall 2020)\")\nplt.ylabel(\"Number of students out of 100\")\nplt.title('Number of hours enrolled by UT students (Fall 2020)')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Analyzing number of enrollment hours through each Fall semester from 2011-2020\n\n I collected this data from UT's statistical handbook ([UT Statistical Handbook](https://utexas.app.box.com/s/gflzag1a3f88jelotrdv9uendvmdf1kt)) and the data can be found on page 12 of this handbook. This data has been verified by the Admissions Office at the University of Texas and the aim of this section is to see if there are have been any trends for the number of enrolled students over the years. Furthermore, I will also compare enrollment over the years between genders.",
"_____no_output_____"
],
[
"## Reproducibility of this project\n\nIn order to reproduce this project, you would have to use UT's statistical handbook. A good idea to further this research would be observe if there are any trends in enrollment of there the years and compare the difference in enrollment between men and women.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
cb70f77a49f4d09c1bb7d1e8c7afca3fddde0bca | 29,104 | ipynb | Jupyter Notebook | coursera_nlp/reviews/week5-project2.ipynb | o3c9/playgrounds | 1363dca0ece0283b5cb1f5c4e26b7a12cf5badab | [
"MIT"
]
| null | null | null | coursera_nlp/reviews/week5-project2.ipynb | o3c9/playgrounds | 1363dca0ece0283b5cb1f5c4e26b7a12cf5badab | [
"MIT"
]
| 2 | 2021-12-09T01:31:34.000Z | 2022-02-17T20:48:11.000Z | coursera_nlp/reviews/week5-project2.ipynb | o3c9/playgrounds | 1363dca0ece0283b5cb1f5c4e26b7a12cf5badab | [
"MIT"
]
| null | null | null | 31.429806 | 534 | 0.540476 | [
[
[
"# Final project: StackOverflow assistant bot\n\nCongratulations on coming this far and solving the programming assignments! In this final project, we will combine everything we have learned about Natural Language Processing to construct a *dialogue chat bot*, which will be able to:\n* answer programming-related questions (using StackOverflow dataset);\n* chit-chat and simulate dialogue on all non programming-related questions.\n\nFor a chit-chat mode we will use a pre-trained neural network engine available from [ChatterBot](https://github.com/gunthercox/ChatterBot).\nThose who aim at honor certificates for our course or are just curious, will train their own models for chit-chat.\n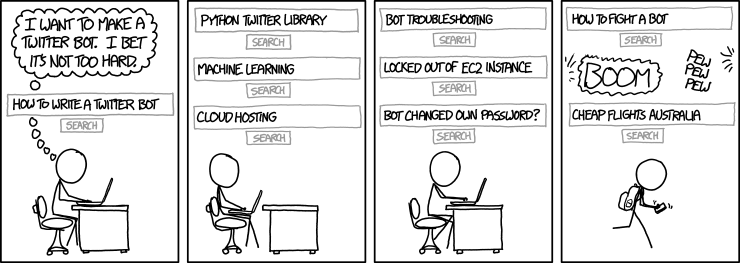\n©[xkcd](https://xkcd.com)",
"_____no_output_____"
],
[
"### Data description\n\nTo detect *intent* of users questions we will need two text collections:\n- `tagged_posts.tsv` — StackOverflow posts, tagged with one programming language (*positive samples*).\n- `dialogues.tsv` — dialogue phrases from movie subtitles (*negative samples*).\n",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import sys\nsys.path.append(\"..\")\nfrom common.download_utils import download_project_resources\n\ndownload_project_resources()",
"File data/dialogues.tsv is already downloaded.\nFile data/tagged_posts.tsv is already downloaded.\n"
]
],
[
[
"For those questions, that have programming-related intent, we will proceed as follow predict programming language (only one tag per question allowed here) and rank candidates within the tag using embeddings.\nFor the ranking part, you will need:\n- `word_embeddings.tsv` — word embeddings, that you trained with StarSpace in the 3rd assignment. It's not a problem if you didn't do it, because we can offer an alternative solution for you.",
"_____no_output_____"
],
[
"As a result of this notebook, you should obtain the following new objects that you will then use in the running bot:\n\n- `intent_recognizer.pkl` — intent recognition model;\n- `tag_classifier.pkl` — programming language classification model;\n- `tfidf_vectorizer.pkl` — vectorizer used during training;\n- `thread_embeddings_by_tags` — folder with thread embeddings, arranged by tags.\n ",
"_____no_output_____"
],
[
"Some functions will be reused by this notebook and the scripts, so we put them into *utils.py* file. Don't forget to open it and fill in the gaps!",
"_____no_output_____"
]
],
[
[
"from utils import *",
"[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
]
],
[
[
"## Part I. Intent and language recognition",
"_____no_output_____"
],
[
"We want to write a bot, which will not only **answer programming-related questions**, but also will be able to **maintain a dialogue**. We would also like to detect the *intent* of the user from the question (we could have had a 'Question answering mode' check-box in the bot, but it wouldn't fun at all, would it?). So the first thing we need to do is to **distinguish programming-related questions from general ones**.\n\nIt would also be good to predict which programming language a particular question referees to. By doing so, we will speed up question search by a factor of the number of languages (10 here), and exercise our *text classification* skill a bit. :)",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport pickle\nimport re\n\nfrom sklearn.feature_extraction.text import TfidfVectorizer",
"_____no_output_____"
]
],
[
[
"### Data preparation",
"_____no_output_____"
],
[
"In the first assignment (Predict tags on StackOverflow with linear models), you have already learnt how to preprocess texts and do TF-IDF tranformations. Reuse your code here. In addition, you will also need to [dump](https://docs.python.org/3/library/pickle.html#pickle.dump) the TF-IDF vectorizer with pickle to use it later in the running bot.",
"_____no_output_____"
]
],
[
[
"def tfidf_features(X_train, X_test, vectorizer_path):\n \"\"\"Performs TF-IDF transformation and dumps the model.\"\"\"\n \n # Train a vectorizer on X_train data.\n # Transform X_train and X_test data.\n \n # Pickle the trained vectorizer to 'vectorizer_path'\n # Don't forget to open the file in writing bytes mode.\n \n ######################################\n tfidf_vectorizer = TfidfVectorizer(\n analyzer='word',\n token_pattern='(\\S+)',\n min_df=5,\n max_df=0.9,\n ngram_range=(1, 2)\n )\n \n X_train = tfidf_vectorizer.fit_transform(X_train)\n X_test = tfidf_vectorizer.transform(X_test)\n pickle.dump(tfidf_vectorizer, open(vectorizer_path, \"wb\"))\n ######################################\n \n return X_train, X_test",
"_____no_output_____"
]
],
[
[
"Now, load examples of two classes. Use a subsample of stackoverflow data to balance the classes. You will need the full data later.",
"_____no_output_____"
]
],
[
[
"sample_size = 200000\n\ndialogue_df = pd.read_csv('data/dialogues.tsv', sep='\\t').sample(sample_size, random_state=0)\nstackoverflow_df = pd.read_csv('data/tagged_posts.tsv', sep='\\t').sample(sample_size, random_state=0)",
"_____no_output_____"
]
],
[
[
"Check how the data look like:",
"_____no_output_____"
]
],
[
[
"dialogue_df.head()",
"_____no_output_____"
],
[
"stackoverflow_df.head()",
"_____no_output_____"
]
],
[
[
"Apply *text_prepare* function to preprocess the data:",
"_____no_output_____"
]
],
[
[
"from utils import text_prepare",
"_____no_output_____"
],
[
"dialogue_df['text'] = list(map(text_prepare, dialogue_df.text.values))\nstackoverflow_df['title'] = list(map(text_prepare, stackoverflow_df.title.values))",
"_____no_output_____"
]
],
[
[
"### Intent recognition",
"_____no_output_____"
],
[
"We will do a binary classification on TF-IDF representations of texts. Labels will be either `dialogue` for general questions or `stackoverflow` for programming-related questions. First, prepare the data for this task:\n- concatenate `dialogue` and `stackoverflow` examples into one sample\n- split it into train and test in proportion 9:1, use *random_state=0* for reproducibility\n- transform it into TF-IDF features",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X = np.concatenate([dialogue_df['text'].values, stackoverflow_df['title'].values])\ny = ['dialogue'] * dialogue_df.shape[0] + ['stackoverflow'] * stackoverflow_df.shape[0]\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.1, random_state = 0)\nprint('Train size = {}, test size = {}'.format(len(X_train), len(X_test)))\n\nX_train_tfidf, X_test_tfidf = tfidf_features(X_train, X_test, RESOURCE_PATH['TFIDF_VECTORIZER'])",
"Train size = 360000, test size = 40000\n"
]
],
[
[
"Train the **intent recognizer** using LogisticRegression on the train set with the following parameters: *penalty='l2'*, *C=10*, *random_state=0*. Print out the accuracy on the test set to check whether everything looks good.",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"######################################\nintent_recognizer = LogisticRegression(penalty='l2', C=10, random_state=0)\nintent_recognizer.fit(X_train_tfidf, y_train)\n######################################",
"_____no_output_____"
],
[
"# Check test accuracy.\ny_test_pred = intent_recognizer.predict(X_test_tfidf)\ntest_accuracy = accuracy_score(y_test, y_test_pred)\nprint('Test accuracy = {}'.format(test_accuracy))",
"Test accuracy = 0.991575\n"
]
],
[
[
"Dump the classifier to use it in the running bot.",
"_____no_output_____"
]
],
[
[
"pickle.dump(intent_recognizer, open(RESOURCE_PATH['INTENT_RECOGNIZER'], 'wb'))",
"_____no_output_____"
]
],
[
[
"### Programming language classification ",
"_____no_output_____"
],
[
"We will train one more classifier for the programming-related questions. It will predict exactly one tag (=programming language) and will be also based on Logistic Regression with TF-IDF features. \n\nFirst, let us prepare the data for this task.",
"_____no_output_____"
]
],
[
[
"X = stackoverflow_df['title'].values\ny = stackoverflow_df['tag'].values",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)\nprint('Train size = {}, test size = {}'.format(len(X_train), len(X_test)))",
"Train size = 160000, test size = 40000\n"
]
],
[
[
"Let us reuse the TF-IDF vectorizer that we have already created above. It should not make a huge difference which data was used to train it.",
"_____no_output_____"
]
],
[
[
"vectorizer = pickle.load(open(RESOURCE_PATH['TFIDF_VECTORIZER'], 'rb'))\n\nX_train_tfidf, X_test_tfidf = vectorizer.transform(X_train), vectorizer.transform(X_test)",
"_____no_output_____"
]
],
[
[
"Train the **tag classifier** using OneVsRestClassifier wrapper over LogisticRegression. Use the following parameters: *penalty='l2'*, *C=5*, *random_state=0*.",
"_____no_output_____"
]
],
[
[
"from sklearn.multiclass import OneVsRestClassifier",
"_____no_output_____"
],
[
"######################################\ntag_classifier = OneVsRestClassifier(LogisticRegression(penalty='l2', C=5, random_state=0))\ntag_classifier.fit(X_train_tfidf, y_train)\n######################################",
"_____no_output_____"
],
[
"# Check test accuracy.\ny_test_pred = tag_classifier.predict(X_test_tfidf)\ntest_accuracy = accuracy_score(y_test, y_test_pred)\nprint('Test accuracy = {}'.format(test_accuracy))",
"Test accuracy = 0.800725\n"
]
],
[
[
"Dump the classifier to use it in the running bot.",
"_____no_output_____"
]
],
[
[
"pickle.dump(tag_classifier, open(RESOURCE_PATH['TAG_CLASSIFIER'], 'wb'))",
"_____no_output_____"
]
],
[
[
"## Part II. Ranking questions with embeddings",
"_____no_output_____"
],
[
"To find a relevant answer (a thread from StackOverflow) on a question you will use vector representations to calculate similarity between the question and existing threads. We already had `question_to_vec` function from the assignment 3, which can create such a representation based on word vectors. \n\nHowever, it would be costly to compute such a representation for all possible answers in *online mode* of the bot (e.g. when bot is running and answering questions from many users). This is the reason why you will create a *database* with pre-computed representations. These representations will be arranged by non-overlaping tags (programming languages), so that the search of the answer can be performed only within one tag each time. This will make our bot even more efficient and allow not to store all the database in RAM. ",
"_____no_output_____"
],
[
"Load StarSpace embeddings which were trained on Stack Overflow posts. These embeddings were trained in *supervised mode* for duplicates detection on the same corpus that is used in search. We can account on that these representations will allow us to find closely related answers for a question. \n\nIf for some reasons you didn't train StarSpace embeddings in the assignment 3, you can use [pre-trained word vectors](https://code.google.com/archive/p/word2vec/) from Google. All instructions about how to work with these vectors were provided in the same assignment. However, we highly recommend to use StartSpace's embeddings, because it contains more appropriate embeddings. If you chose to use Google's embeddings, delete the words, which is not in Stackoverflow data.",
"_____no_output_____"
]
],
[
[
"starspace_embeddings, embeddings_dim = load_embeddings(RESOURCE_PATH['WORD_EMBEDDINGS'])",
"_____no_output_____"
]
],
[
[
"Since we want to precompute representations for all possible answers, we need to load the whole posts dataset, unlike we did for the intent classifier:",
"_____no_output_____"
]
],
[
[
"posts_df = pd.read_csv('data/tagged_posts.tsv', sep='\\t')",
"_____no_output_____"
]
],
[
[
"Look at the distribution of posts for programming languages (tags) and find the most common ones. \nYou might want to use pandas [groupby](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.groupby.html) and [count](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.count.html) methods:",
"_____no_output_____"
]
],
[
[
"posts_df.head()",
"_____no_output_____"
],
[
"# Because of diskspace issues on the 1GB EC2 instance\n# we need to reduce the size of the pickle files\n#import math\nposts_df = posts_df.sample(200000)\ncounts_by_tag = posts_df.tag.value_counts()",
"_____no_output_____"
]
],
[
[
"Now for each `tag` you need to create two data structures, which will serve as online search index:\n* `tag_post_ids` — a list of post_ids with shape `(counts_by_tag[tag],)`. It will be needed to show the title and link to the thread;\n* `tag_vectors` — a matrix with shape `(counts_by_tag[tag], embeddings_dim)` where embeddings for each answer are stored.\n\nImplement the code which will calculate the mentioned structures and dump it to files. It should take several minutes to compute it.",
"_____no_output_____"
]
],
[
[
"import os\nos.makedirs(RESOURCE_PATH['THREAD_EMBEDDINGS_FOLDER'], exist_ok=True)\n\nfor tag, count in counts_by_tag.items():\n print(tag, count)\n tag_posts = posts_df[posts_df['tag'] == tag]\n \n tag_post_ids = tag_posts.post_id.tolist()\n \n tag_vectors = np.zeros((count, embeddings_dim), dtype=np.float32)\n for i, title in enumerate(tag_posts['title']):\n tag_vectors[i, :] = question_to_vec(title, starspace_embeddings, embeddings_dim)\n\n # Dump post ids and vectors to a file.\n filename = os.path.join(RESOURCE_PATH['THREAD_EMBEDDINGS_FOLDER'], os.path.normpath('%s.pkl' % tag))\n pickle.dump((tag_post_ids, tag_vectors), open(filename, 'wb'))",
"c# 36521\njava 35333\njavascript 34320\nphp 29504\nc_cpp 26197\npython 19339\nruby 9150\nr 3277\nswift 3211\nvb 3148\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb70f98f1c488bfce87420efc669fcc697828ee8 | 118,387 | ipynb | Jupyter Notebook | module3-autoencoders/LS_DS_433_Autoencoders_Lecture.ipynb | lucguittard/DS-Unit-4-Sprint-3-Deep-Learning | ab5a72e39a4738b09dbebdfba35276599bebf61c | [
"MIT"
]
| null | null | null | module3-autoencoders/LS_DS_433_Autoencoders_Lecture.ipynb | lucguittard/DS-Unit-4-Sprint-3-Deep-Learning | ab5a72e39a4738b09dbebdfba35276599bebf61c | [
"MIT"
]
| null | null | null | module3-autoencoders/LS_DS_433_Autoencoders_Lecture.ipynb | lucguittard/DS-Unit-4-Sprint-3-Deep-Learning | ab5a72e39a4738b09dbebdfba35276599bebf61c | [
"MIT"
]
| null | null | null | 125.277249 | 44,076 | 0.800662 | [
[
[
"Lambda School Data Science\n\n*Unit 4, Sprint 3, Module 3*\n\n---",
"_____no_output_____"
],
[
"# Autoencoders\n\n> An autoencoder is a type of artificial neural network used to learn efficient data codings in an unsupervised manner.[1][2] The aim of an autoencoder is to learn a representation (encoding) for a set of data, typically for dimensionality reduction, by training the network to ignore signal “noise”. Along with the reduction side, a reconstructing side is learnt, where the autoencoder tries to generate from the reduced encoding a representation as close as possible to its original input, hence its name. ",
"_____no_output_____"
],
[
"## Learning Objectives\n*At the end of the lecture you should be to*:\n* <a href=\"#p1\">Part 1</a>: Describe the componenets of an autoencoder\n* <a href=\"#p2\">Part 2</a>: Train an autoencoder\n* <a href=\"#p3\">Part 3</a>: Apply an autoenocder to a basic information retrieval problem\n\n__Problem:__ Is it possible to automatically represent an image as a fixed-sized vector even if it isn’t labeled?\n\n__Solution:__ Use an autoencoder\n\nWhy do we need to represent an image as a fixed-sized vector do you ask? \n\n* __Information Retrieval__\n - [Reverse Image Search](https://en.wikipedia.org/wiki/Reverse_image_search)\n - [Recommendation Systems - Content Based Filtering](https://en.wikipedia.org/wiki/Recommender_system#Content-based_filtering)\n* __Dimensionality Reduction__\n - [Feature Extraction](https://www.kaggle.com/c/vsb-power-line-fault-detection/discussion/78285)\n - [Manifold Learning](https://en.wikipedia.org/wiki/Nonlinear_dimensionality_reduction)\n\nWe've already seen *representation learning* when we talked about word embedding modelings during our NLP week. Today we're going to achieve a similiar goal on images using *autoencoders*. An autoencoder is a neural network that is trained to attempt to copy its input to its output. Usually they are restricted in ways that allow them to copy only approximately. The model often learns useful properties of the data, because it is forced to prioritize which aspecs of the input should be copied. The properties of autoencoders have made them an important part of modern generative modeling approaches. Consider autoencoders a special case of feed-forward networks (the kind we've been studying); backpropagation and gradient descent still work. ",
"_____no_output_____"
],
[
"# Autoencoder Architecture (Learn)\n<a id=\"p1\"></a>",
"_____no_output_____"
],
[
"## Overview\n\nThe *encoder* compresses the input data and the *decoder* does the reverse to produce the uncompressed version of the data to create a reconstruction of the input as accurately as possible:\n\n<img src='https://miro.medium.com/max/1400/1*[email protected]' width=800/>\n\nThe learning process gis described simply as minimizing a loss function: \n$ L(x, g(f(x))) $\n\n- $L$ is a loss function penalizing $g(f(x))$ for being dissimiliar from $x$ (such as mean squared error)\n- $f$ is the encoder function\n- $g$ is the decoder function",
"_____no_output_____"
],
[
"## Follow Along\n### Extremely Simple Autoencoder",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.layers import Input, Dense\nfrom tensorflow.keras.models import Model\n# import wandb\n# from wandb.keras import WandbCallback\n\n# this is the size of our encoded representations\nencoding_dim = 32 # 32 floats -> compression of factor 24.5, assuming the input is 784 floats\n\n# this is our input placeholder\ninput_img = Input(shape=(784,))\n\n# \"encoded\" is the encoded representation of the input\nencoded = Dense(encoding_dim, activation='sigmoid')(input_img)\n\n# \"decoded\" is the lossy reconstruction of the input\ndecoded = Dense(784, activation = 'sigmoid')(encoded)\n\n# this model maps an input to its reconstruction\nautoencoder = Model(input_img, decoded)",
"_____no_output_____"
],
[
"# this model maps an input to its encoded representation\nencoder = Model(input_img, encoded)",
"_____no_output_____"
],
[
"# create a placeholder for an encoded (32-dimensional) input\n\n\n# retrieve the last layer of the autoencoder model\n\n\n# create the decoder model\n",
"_____no_output_____"
],
[
"autoencoder.compile(optimizer='adadelta', loss='binary_crossentropy')",
"_____no_output_____"
],
[
"from tensorflow.keras.datasets import mnist\nimport numpy as np\n(x_train, _), (x_test, _) = mnist.load_data()",
"_____no_output_____"
],
[
"x_train = x_train.astype('float32') / 255.\nx_test = x_test.astype('float32') / 255.\nx_train = x_train.reshape((len(x_train), np.prod(x_train.shape[1:])))\nx_test = x_test.reshape((len(x_test), np.prod(x_test.shape[1:])))\nprint(x_train.shape)\nprint(x_test.shape)",
"(60000, 784)\n(10000, 784)\n"
],
[
"#wandb.init(project=\"mnist_autoencoder\", entity=\"ds5\")\n\nautoencoder.fit(x_train, x_train,\n epochs=10,\n batch_size=256,\n shuffle=True,\n validation_data=(x_test, x_test),\n verbose = True)\n\n# can stop running/training of model at any point and training will be preserved",
"Train on 60000 samples, validate on 10000 samples\nEpoch 1/10\n60000/60000 [==============================] - 5s 88us/sample - loss: 0.6945 - val_loss: 0.6943\nEpoch 2/10\n60000/60000 [==============================] - 4s 70us/sample - loss: 0.6940 - val_loss: 0.6938\nEpoch 3/10\n60000/60000 [==============================] - 5s 91us/sample - loss: 0.6935 - val_loss: 0.6934\nEpoch 4/10\n60000/60000 [==============================] - 6s 97us/sample - loss: 0.6931 - val_loss: 0.6929\nEpoch 5/10\n60000/60000 [==============================] - 5s 91us/sample - loss: 0.6927 - val_loss: 0.6925\nEpoch 6/10\n60000/60000 [==============================] - 6s 107us/sample - loss: 0.6922 - val_loss: 0.6920\nEpoch 7/10\n60000/60000 [==============================] - 5s 87us/sample - loss: 0.6918 - val_loss: 0.6916\nEpoch 8/10\n60000/60000 [==============================] - 3s 54us/sample - loss: 0.6913 - val_loss: 0.6911\nEpoch 9/10\n60000/60000 [==============================] - 4s 61us/sample - loss: 0.6909 - val_loss: 0.6907\nEpoch 10/10\n60000/60000 [==============================] - 3s 56us/sample - loss: 0.6904 - val_loss: 0.6903\n"
],
[
"# encode and decode some digits\n# note that we take them from the *test* set\n\n# visualize the results\n\n#encoded_images = encoder.predict(x_test)\ndecoded_imgs = autoencoder.predict(x_test)",
"_____no_output_____"
],
[
"# use Matplotlib (don't ask)\nimport matplotlib.pyplot as plt\n\nn = 10 # how many digits we will display\nplt.figure(figsize=(20, 4))\nfor i in range(n):\n # display original\n ax = plt.subplot(2, n, i + 1)\n plt.imshow(x_test[i].reshape(28, 28))\n plt.gray()\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\n # display reconstruction\n ax = plt.subplot(2, n, i + 1 + n)\n plt.imshow(decoded_imgs[i].reshape(28, 28))\n plt.gray()\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\nplt.show()\n\n# poor results; highly dependent on training time",
"_____no_output_____"
]
],
[
[
"## Challenge\n\nExpected to talk about the components of autoencoder and their purpose. ",
"_____no_output_____"
],
[
"# Train an Autoencoder (Learn)\n<a id=\"p2\"></a>",
"_____no_output_____"
],
[
"## Overview\n\nAs long as our architecture maintains an hourglass shape, we can continue to add layers and create a deeper network. ",
"_____no_output_____"
],
[
"## Follow Along",
"_____no_output_____"
],
[
"### Deep Autoencoder",
"_____no_output_____"
]
],
[
[
"input_img = Input(shape=(784,)) # first layer of the neural network\nencoded = Dense(128, activation= 'relu')(input_img) # input_img - the data getting pushed to the next layer\nencoded = Dense(64, activation= 'relu')(encoded)\nencoded = Dense(32, activation= 'relu')(encoded) # fully dehydrated layer\n\ndecoded = Dense(64, activation= 'relu')(encoded)\ndecoded = Dense(128, activation= 'relu')(decoded)\ndecoded = Dense(784, activation= 'sigmoid')(decoded)\n",
"_____no_output_____"
],
[
"# compile & fit model\n\nautoencoder = Model(input_img, decoded)\n\nautoencoder.compile(optimizer='adam', loss='binary_crossentropy')\n\nautoencoder.fit(x_train, x_train,\n epochs=20,\n batch_size=784,\n shuffle=True,\n validation_data=(x_test,x_test),\n verbose= True)",
"Train on 60000 samples, validate on 10000 samples\nEpoch 1/20\n60000/60000 [==============================] - 5s 85us/sample - loss: 0.1046 - val_loss: 0.0974\nEpoch 2/20\n60000/60000 [==============================] - 4s 60us/sample - loss: 0.0983 - val_loss: 0.0971\nEpoch 3/20\n60000/60000 [==============================] - 4s 63us/sample - loss: 0.0980 - val_loss: 0.0969\nEpoch 4/20\n60000/60000 [==============================] - 4s 66us/sample - loss: 0.0977 - val_loss: 0.0965\nEpoch 5/20\n60000/60000 [==============================] - 5s 79us/sample - loss: 0.0974 - val_loss: 0.0965\nEpoch 6/20\n60000/60000 [==============================] - 5s 84us/sample - loss: 0.0971 - val_loss: 0.0959\nEpoch 7/20\n60000/60000 [==============================] - 4s 63us/sample - loss: 0.0968 - val_loss: 0.0957\nEpoch 8/20\n60000/60000 [==============================] - 4s 59us/sample - loss: 0.0965 - val_loss: 0.0953\nEpoch 9/20\n60000/60000 [==============================] - 4s 64us/sample - loss: 0.0962 - val_loss: 0.0950\nEpoch 10/20\n60000/60000 [==============================] - 5s 85us/sample - loss: 0.0959 - val_loss: 0.0947\nEpoch 11/20\n60000/60000 [==============================] - 5s 82us/sample - loss: 0.0956 - val_loss: 0.0946\nEpoch 12/20\n60000/60000 [==============================] - 4s 71us/sample - loss: 0.0953 - val_loss: 0.0942\nEpoch 13/20\n60000/60000 [==============================] - 4s 65us/sample - loss: 0.0950 - val_loss: 0.0939\nEpoch 14/20\n60000/60000 [==============================] - 4s 58us/sample - loss: 0.0947 - val_loss: 0.0937\nEpoch 15/20\n60000/60000 [==============================] - 5s 78us/sample - loss: 0.0945 - val_loss: 0.0935\nEpoch 16/20\n60000/60000 [==============================] - 5s 76us/sample - loss: 0.0941 - val_loss: 0.0930\nEpoch 17/20\n60000/60000 [==============================] - 4s 70us/sample - loss: 0.0939 - val_loss: 0.0930\nEpoch 18/20\n60000/60000 [==============================] - 4s 72us/sample - loss: 0.0936 - val_loss: 0.0927\nEpoch 19/20\n60000/60000 [==============================] - 5s 86us/sample - loss: 0.0935 - val_loss: 0.0925\nEpoch 20/20\n60000/60000 [==============================] - 4s 69us/sample - loss: 0.0932 - val_loss: 0.0921\n"
],
[
"decoded_imgs = autoencoder.predict(x_test)",
"_____no_output_____"
],
[
"# use Matplotlib (don't ask)\nimport matplotlib.pyplot as plt\n\nn = 10 # how many digits we will display\nplt.figure(figsize=(20, 4))\nfor i in range(n):\n # display original\n ax = plt.subplot(2, n, i + 1)\n plt.imshow(x_test[i].reshape(28, 28))\n plt.gray()\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\n # display reconstruction\n ax = plt.subplot(2, n, i + 1 + n)\n plt.imshow(decoded_imgs[i].reshape(28, 28))\n plt.gray()\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Convolutional autoencoder\n\n> Since our inputs are images, it makes sense to use convolutional neural networks (convnets) as encoders and decoders. In practical settings, autoencoders applied to images are always convolutional autoencoders --they simply perform much better.\n\n> Let's implement one. The encoder will consist in a stack of Conv2D and MaxPooling2D layers (max pooling being used for spatial down-sampling), while the decoder will consist in a stack of Conv2D and UpSampling2D layers.",
"_____no_output_____"
]
],
[
[
"# Working with upsampling example\n\nfrom keras.layers import Input, Dense, Conv2D, MaxPooling2D, UpSampling2D\nfrom keras.models import Model\nfrom keras import backend as K\n\n# Create Model \n# Create Model \ninput_img = Input(shape=(28,28,1))\n\nx = Conv2D(16,(3,3), activation='relu', padding='same')(input_img)\nx = MaxPooling2D((2,2), padding='same')(x)\nx = Conv2D(8, (3, 3), activation='relu', padding='same')(x)\nx = MaxPooling2D((2, 2), padding='same')(x)\nx = Conv2D(8, (3, 3), activation='relu', padding='same')(x)\nencoded = MaxPooling2D((2, 2), padding='same')(x)\n\n# at this point the representation is (4, 4, 8) i.e. 128-dimensional representation\n\nx = Conv2D(8, (3, 3), activation='relu', padding='same')(encoded)\nx = UpSampling2D((2, 2))(x)\nx = Conv2D(8, (3, 3), activation='relu', padding='same')(x)\nx = UpSampling2D((2, 2))(x)\nx = Conv2D(16, (3, 3), activation='relu')(x)\nx = UpSampling2D((2, 2))(x)\ndecoded = Conv2D(1, (3, 3), activation='sigmoid', padding='same')(x)\n\nautoencoder = Model(input_img, decoded)\nautoencoder.compile(optimizer='adam', loss='binary_crossentropy')",
"_____no_output_____"
],
[
"autoencoder.summary()",
"Model: \"model_2\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_8 (InputLayer) (None, 28, 28, 1) 0 \n_________________________________________________________________\nconv2d_8 (Conv2D) (None, 28, 28, 16) 160 \n_________________________________________________________________\nmax_pooling2d_4 (MaxPooling2 (None, 14, 14, 16) 0 \n_________________________________________________________________\nconv2d_9 (Conv2D) (None, 14, 14, 8) 1160 \n_________________________________________________________________\nmax_pooling2d_5 (MaxPooling2 (None, 7, 7, 8) 0 \n_________________________________________________________________\nconv2d_10 (Conv2D) (None, 7, 7, 8) 584 \n_________________________________________________________________\nmax_pooling2d_6 (MaxPooling2 (None, 4, 4, 8) 0 \n_________________________________________________________________\nconv2d_11 (Conv2D) (None, 4, 4, 8) 584 \n_________________________________________________________________\nup_sampling2d_4 (UpSampling2 (None, 8, 8, 8) 0 \n_________________________________________________________________\nconv2d_12 (Conv2D) (None, 8, 8, 8) 584 \n_________________________________________________________________\nup_sampling2d_5 (UpSampling2 (None, 16, 16, 8) 0 \n_________________________________________________________________\nconv2d_13 (Conv2D) (None, 14, 14, 16) 1168 \n_________________________________________________________________\nup_sampling2d_6 (UpSampling2 (None, 28, 28, 16) 0 \n_________________________________________________________________\nconv2d_14 (Conv2D) (None, 28, 28, 1) 145 \n=================================================================\nTotal params: 4,385\nTrainable params: 4,385\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"from keras.datasets import mnist\nimport numpy as np\n\n(x_train, _), (x_test, _) = mnist.load_data()\n\nx_train = x_train.astype('float32') / 255.\nx_test = x_test.astype('float32') / 255.\nx_train = np.reshape(x_train, (len(x_train), 28, 28, 1)) # adapt this if using `channels_first` image data format\nx_test = np.reshape(x_test, (len(x_test), 28, 28, 1)) # adapt this if using `channels_first` image data format",
"_____no_output_____"
],
[
"#wandb.init(project=\"mnist_autoencoder\", entity=\"ds5\")\n\nautoencoder.fit(x_train, x_train,\n epochs=10,\n batch_size=784,\n shuffle=True,\n validation_data=(x_test, x_test),\n verbose=True)",
"Train on 60000 samples, validate on 10000 samples\nEpoch 1/20\n60000/60000 [==============================] - 66s 1ms/step - loss: 0.2405 - val_loss: 0.2059\nEpoch 2/20\n60000/60000 [==============================] - 59s 984us/step - loss: 0.1931 - val_loss: 0.1794\nEpoch 3/20\n47040/60000 [======================>.......] - ETA: 12s - loss: 0.1736"
],
[
"decoded_imgs = autoencoder.predict(x_test)\n\nn = 10\nplt.figure(figsize=(20, 4))\nfor i in range(n):\n # display original\n ax = plt.subplot(2, n, i)\n plt.imshow(x_test[i].reshape(28, 28))\n plt.gray()\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\n # display reconstruction\n ax = plt.subplot(2, n, i + n)\n plt.imshow(decoded_imgs[i].reshape(28, 28))\n plt.gray()\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Visualization of the Representations",
"_____no_output_____"
]
],
[
[
"encoder = Model(input_img, encoded)\nencoder.predict(x_train)\n\nn = 10\nplt.figure(figsize=(20, 8))\nfor i in range(n):\n ax = plt.subplot(1, n, i)\n plt.imshow(encoded_imgs[i].reshape(4, 4 * 8).T)\n plt.gray()\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Challenge\n\nYou will train an autoencoder at some point in the near future. ",
"_____no_output_____"
],
[
"# Information Retrieval with Autoencoders (Learn)\n<a id=\"p3\"></a>",
"_____no_output_____"
],
[
"## Overview\n\nA common usecase for autoencoders is for reverse image search. Let's try to draw an image and see what's most similiar in our dataset. \n\nTo accomplish this we will need to slice our autoendoer in half to extract our reduced features. :) ",
"_____no_output_____"
],
[
"## Follow Along",
"_____no_output_____"
]
],
[
[
"encoder = Model(input_img, encoded)\nencoded_imgs = encoder.predict(x_train)",
"_____no_output_____"
],
[
"encoded_imgs[0].reshape((128,)) #shape before reshape: 4,4,8",
"_____no_output_____"
],
[
"from sklearn.neighbors import NearestNeighbors\n\nnn = NearestNeighbors(n_neighbors=10, algorithm='ball_tree')\nnn.fit(encoded_imgs)",
"_____no_output_____"
],
[
"nn.kneighbors(...)",
"_____no_output_____"
]
],
[
[
"## Challenge\n\nYou should already be familiar with KNN and similarity queries, so the key component of this section is know what to 'slice' from your autoencoder (the encoder) to extract features from your data. ",
"_____no_output_____"
],
[
"# Review\n\n* <a href=\"#p1\">Part 1</a>: Describe the componenets of an autoencoder\n - Enocder\n - Decoder\n* <a href=\"#p2\">Part 2</a>: Train an autoencoder\n - Can do in Keras Easily\n - Can use a variety of architectures\n - Architectures must follow hourglass shape\n* <a href=\"#p3\">Part 3</a>: Apply an autoenocder to a basic information retrieval problem\n - Extract just the encoder to use for various tasks\n - AE ares good for dimensionality reduction, reverse image search, and may more things. \n",
"_____no_output_____"
],
[
"# Sources\n\n__References__\n- [Building Autoencoders in Keras](https://blog.keras.io/building-autoencoders-in-keras.html)\n- [Deep Learning Cookbook](http://shop.oreilly.com/product/0636920097471.do)\n\n__Additional Material__",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
cb7100cccc5c9e6f1aadaf91a2b4f9efaf6ab8bc | 23,991 | ipynb | Jupyter Notebook | Q1.ipynb | RaeChen07/hcde-410-final | ce92b7890621b9af261db5bbec3b31aea5a623bc | [
"MIT"
]
| null | null | null | Q1.ipynb | RaeChen07/hcde-410-final | ce92b7890621b9af261db5bbec3b31aea5a623bc | [
"MIT"
]
| null | null | null | Q1.ipynb | RaeChen07/hcde-410-final | ce92b7890621b9af261db5bbec3b31aea5a623bc | [
"MIT"
]
| null | null | null | 110.050459 | 16,756 | 0.836272 | [
[
[
"## Q1: What is the impact of days in a week on the number of Burglaries?",
"_____no_output_____"
],
[
"Use pandas to cleaning up the data to contain only 'Report Number','Offense Start DateTime','Offense Parent Group',and 'Offense'. Sort all the 'Offense' being 'Burglary/Breaking & Entering' out.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport calendar\n\n\nall_crime_data = pd.read_csv('SPD_Crime_Data__2008-Present.csv')\nall_crime_data_clean = all_crime_data[['Report Number','Offense Start DateTime','Offense Parent Group','Offense']]\nburglary_crime = all_crime_data_clean[all_crime_data_clean['Offense'] == 'Burglary/Breaking & Entering']\n",
"_____no_output_____"
]
],
[
[
"Drop all row without a 'Offense Start DateTime'. Use calendaer to add column 'day_of_week' to the dataframe and save it in a new csv file called Burglary_Crime_Q1.csv.",
"_____no_output_____"
]
],
[
[
"\nburglary_crime['Offense Start DateTime'] = pd.to_datetime(burglary_crime['Offense Start DateTime'])\n\nburglary_crime['day_of_week'] = burglary_crime['Offense Start DateTime'].apply(lambda x: x.weekday()) # get the weekday index, between 0 and 6\nnan_value = float(\"NaN\")\nburglary_crime.replace(\"\", nan_value, inplace=True)\nburglary_crime = burglary_crime.dropna(subset=['day_of_week'])\nburglary_crime['day_of_week'] = burglary_crime['day_of_week'].apply(lambda x: calendar.day_name[int(x)])\nburglary_crime.to_csv('Burglary_Crime_Q1.csv')\nburglary_crime.head()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\n\nsns.catplot(x='day_of_week', kind='count', data=burglary_crime, order=['Monday','Tuesday','Wednesday','Thursday','Friday','Saturday','Sunday'])\nplt.title('Day in a Week v.s. Number of Burglaries')\nplt.xlabel('Day of Week')\nplt.ylabel('Number of Burglaries')\n# Save the plot to a file\nplt.savefig('day_of_week_burglary.png', bbox_inches='tight')",
"_____no_output_____"
]
],
[
[
"Use matplotlib and seaborn to plot the graph.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
]
|
cb7102e3ede6bea9c3aa91cc3f586945c02fbe7a | 4,967 | ipynb | Jupyter Notebook | 6. Question.ipynb | ramyuva/WEB2 | 17d7e513038199de282f37a074e234f05814faef | [
"Apache-2.0"
]
| null | null | null | 6. Question.ipynb | ramyuva/WEB2 | 17d7e513038199de282f37a074e234f05814faef | [
"Apache-2.0"
]
| null | null | null | 6. Question.ipynb | ramyuva/WEB2 | 17d7e513038199de282f37a074e234f05814faef | [
"Apache-2.0"
]
| null | null | null | 34.493056 | 172 | 0.481981 | [
[
[
"# Importing Libraries\nimport pandas as pd\nfrom bs4 import BeautifulSoup\nimport requests\nimport selenium\nfrom selenium import webdriver\nimport time",
"_____no_output_____"
],
[
"# Function Definition\ndef flipkart_sunglasses(url):\n driver=webdriver.Chrome('chromedriver.exe')\n start_page=0\n end_page=2\n urls = []\n brand=[]\n product_desc=[]\n price=[]\n discount=[]\n #loop to fetch urls of each mobile till page 3\n for page in range(start_page,end_page+1):\n driver.get(url)\n soup= BeautifulSoup(driver.page_source, 'html.parser')\n prod_urls = soup.find_all('a', attrs ={'class':'IRpwTa'})\n for prod in prod_urls:\n urls.append('https://www.flipkart.com'+prod.get('href'))\n \n #loop to scrap required details from each mobile page\n for url in urls:\n driver.get(url)\n soup = BeautifulSoup(driver.page_source, 'html.parser')\n n = soup.find('h1',attrs={'class':'yhB1nd'})\n if n is not None:\n brand.append(n.find('span').text.replace('\\n',''))\n else:\n brand.append('-')\n desc = soup.find('div', attrs = {'class':'_2yIA0Y'})\n if desc is not None:\n product_desc.append(desc.text)\n else:\n product_desc.append('-')\n p = soup.find('div', attrs = {'class':'_30jeq3 _16Jk6d'})\n if p is not None:\n price.append(p.text)\n else:\n price.append('-')\n dis = soup.find('div', attrs = {'class':'_3Ay6Sb _31Dcoz pZkvcx'})\n if dis is not None:\n discount.append(dis.find('span').text)\n else:\n discount.append('-')\n sun_df =pd.DataFrame({'Brand':brand,\n 'product_desc':product_desc,\n 'price':price,\n 'Discount':discount})\n print(sun_df)\n sun_df.to_csv('flipkart.csv', index = False)\n \n \n# Calling Function\nflipkart_sunglasses('https://www.flipkart.com/search?q=sunglasses&otracker=search&otracker1=search&marketplace=FLIPKART&as-show=on&as=off&as-pos=1&as-type=HISTORY')\n",
" Brand product_desc price \\\n0 ROYAL SON Product DetailsSizeThis product is sold as Med... ₹854 \n1 Royal Son Product DetailsSizeThis product is sold as Med... ₹379 \n2 ROZZETTA CRAFT Product DetailsSizeThis product is sold as Fre... ₹499 \n3 Fastrack Product DetailsSizeThis product is sold as Fre... ₹499 \n4 Fastrack Product DetailsSizeThis product is sold as Fre... ₹499 \n.. ... ... ... \n115 LOPO Product DetailsSizeThis product is sold as Fre... ₹179 \n116 Fastrack Product DetailsSizeThis product is sold as Fre... ₹695 \n117 Ayezent Product DetailsSizeThis product is sold as Sma... ₹282 \n118 Royal Son Product DetailsSizeThis product is sold as Lar... ₹279 \n119 Elegante Product DetailsSizeThis product is sold as Fre... ₹449 \n\n Discount \n0 57% off \n1 74% off \n2 77% off \n3 37% off \n4 37% off \n.. ... \n115 74% off \n116 13% off \n117 85% off \n118 78% off \n119 70% off \n\n[120 rows x 4 columns]\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
cb710b7e771a240e75b7dfe1537bb4864ec2a733 | 4,457 | ipynb | Jupyter Notebook | explore.ipynb | j-berg/explore_colon | 515cec58abec3fb916ca36c86bdf296af3c2a4b8 | [
"MIT"
]
| null | null | null | explore.ipynb | j-berg/explore_colon | 515cec58abec3fb916ca36c86bdf296af3c2a4b8 | [
"MIT"
]
| null | null | null | explore.ipynb | j-berg/explore_colon | 515cec58abec3fb916ca36c86bdf296af3c2a4b8 | [
"MIT"
]
| null | null | null | 32.532847 | 163 | 0.583128 | [
[
[
"# Analyzing colon tumor gene expression data\nData source: \n- https://dx.doi.org/10.1038%2Fsdata.2018.61\n- https://www.ncbi.nlm.nih.gov/gds?term=GSE8671\n- https://www.ncbi.nlm.nih.gov/gds?term=GSE20916",
"_____no_output_____"
],
[
"### 1. Initialize the environment and variables\nUpon launching this page, run the below code to initialize the analysis environment by selecting the cell and pressing `Shift + Enter`",
"_____no_output_____"
]
],
[
[
"#Set path to this directory for accessing and saving files\nimport os\nimport warnings\nwarnings.filterwarnings('ignore')\n\n__path__ = os.getcwd() + os.path.sep\nprint('Current path: ' + __path__)\n\nfrom local_utils import init_tcga, init_GSE8671, init_GSE20916, sort_data\nfrom local_utils import eval_gene, make_heatmap\n\n%matplotlib inline\n\n# Read data \nprint(\"Loading data. Please wait...\")\ntcga_scaled, tcga_data, tcga_info, tcga_palette = init_tcga()\nGSE8671_scaled, GSE8671_data, GSE8671_info, GSE8671_palette = init_GSE8671()\nGSE20916_scaled, GSE20916_data, GSE20916_info, GSE20916_palette = init_GSE20916()\nprint(\"Data import complete. Continue below...\")",
"_____no_output_____"
]
],
[
[
"### 2a. Explore a gene of interest in the Unified TCGA data or GSE8671 and GSE20916\n- In the first line, edit the gene name (human) within the quotes\n- Press `Shift + Enter`",
"_____no_output_____"
]
],
[
[
"gene = \"FABP1\" # <-- edit between the quotation marks here\n\n# Do not edit below this line\n# ------------------------------------------------------------------------\nprint(\"Running analysis. Please wait...\\n\\n\")\neval_gene(gene, tcga_data, tcga_info, tcga_palette, 'TCGA (unified)')\neval_gene(gene, GSE8671_data, GSE8671_info, GSE8671_palette, 'GSE8671')\neval_gene(gene, GSE20916_data, GSE20916_info, GSE20916_palette, 'GSE20916')",
"_____no_output_____"
]
],
[
[
"### 2a. Explore a set of genes in the Unified TCGA data or GSE8671 and GSE20916\n- Between the brackets, edit the gene names (human) within the quotes\n- If you want to have less than the provided number of genes, remove the necessary number of lines \n- If you want to have more than the provided number of genes, add lines with the gene name in quotes, followed by a comma outside of the quotes\n- Press `Shift + Enter`",
"_____no_output_____"
]
],
[
[
"gene_list = [\n \"FABP1\", # <-- edit between the quote marks here\n \"ME1\",\n \"ME2\",\n \"PC\", # <-- add more genes by adding a line, the gene name between quotes, and a comma after that quote\n \n]\n\n# Do not edit below this line\n# ------------------------------------------------------------------------\nprint(\"Running analysis. Please wait...\\n\\n\")\nmake_heatmap(gene_list, tcga_scaled, tcga_info, tcga_palette, 'TCGA (unified)')\nmake_heatmap(gene_list, GSE8671_scaled, GSE8671_info, GSE8671_palette, 'GSE8671')\nmake_heatmap(gene_list, sort_data(GSE20916_scaled, GSE20916_info, ['adenoma', 'adenocarcinoma','normal_colon']), GSE20916_info, GSE20916_palette, 'GSE20916')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb7110f4b6ce9f259c0c6ee94163fe6df0a4f9b9 | 918,576 | ipynb | Jupyter Notebook | test/norm_develop.ipynb | VU-Cog-Sci/prfpy_tools | 6729ec21ae6f4fc6494a34807efa8350893dbdee | [
"MIT"
]
| null | null | null | test/norm_develop.ipynb | VU-Cog-Sci/prfpy_tools | 6729ec21ae6f4fc6494a34807efa8350893dbdee | [
"MIT"
]
| null | null | null | test/norm_develop.ipynb | VU-Cog-Sci/prfpy_tools | 6729ec21ae6f4fc6494a34807efa8350893dbdee | [
"MIT"
]
| null | null | null | 385.14717 | 696,297 | 0.914563 | [
[
[
"%load_ext autoreload\n%autoreload 2\n\nimport numpy as np\nimport nibabel as nb\nimport scipy as sp\nimport matplotlib.pyplot as pl\nimport os\nopj = os.path.join\n%matplotlib notebook\npl.ion()\n\nimport sys\nsys.path.append(\"..\")",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"from prfpy.stimulus import PRFStimulus2D\nfrom prfpy.grid import Iso2DGaussianGridder, Norm_Iso2DGaussianGridder, DoG_Iso2DGaussianGridder\nfrom prfpy.fit import Iso2DGaussianFitter, Norm_Iso2DGaussianFitter, DoG_Iso2DGaussianFitter\nfrom prfpy.timecourse import sgfilter_predictions\nfrom utils.utils import *\nfrom prfpy.fit import iterative_search",
"Using TensorFlow backend.\n"
],
[
"#some params needed (stimulus size, initial volumes to discard, savgol window length)\nn_pix=40\ndiscard_volumes = 5\nwindow_length=121",
"_____no_output_____"
],
[
"#create stimulus\ndm_1R = create_dm_from_screenshots(screenshot_path='/Users/marcoaqil/PRFMapping/PRFMapping-Raw/sub-001/ses-1/rawdata/sub-001_ses-1_Logs/sub-001_ses-1_task-1R_run-1_Logs/Screenshots', n_pix=n_pix)\ndm_1S = create_dm_from_screenshots(screenshot_path='/Users/marcoaqil/PRFMapping/PRFMapping-Raw/sub-001/ses-1/rawdata/sub-001_ses-1_Logs/sub-001_ses-1_task-1S_run-1_Logs/Screenshots', n_pix=n_pix)\ndm_2R = create_dm_from_screenshots(screenshot_path='/Users/marcoaqil/PRFMapping/PRFMapping-Raw/sub-001/ses-1/rawdata/sub-001_ses-1_Logs/sub-001_ses-1_task-2R_run-1_Logs/Screenshots', n_pix=n_pix)\ndm_4F = create_dm_from_screenshots(screenshot_path='/Users/marcoaqil/PRFMapping/PRFMapping-Raw/sub-001/ses-1/rawdata/sub-001_ses-1_Logs/sub-001_ses-1_task-4F_run-1_Logs/Screenshots', n_pix=n_pix)\ndm_4R = create_dm_from_screenshots(screenshot_path='/Users/marcoaqil/PRFMapping/PRFMapping-Raw/sub-001/ses-1/rawdata/sub-001_ses-1_Logs/sub-001_ses-1_task-4R_run-1_Logs/Screenshots', n_pix=n_pix)\n\ntask_lengths=[dm_1R.shape[2]-discard_volumes, \n dm_1S.shape[2]-discard_volumes, \n dm_2R.shape[2]-discard_volumes, \n dm_4F.shape[2]-discard_volumes, \n dm_4R.shape[2]-discard_volumes]\n\ndm_full = np.concatenate((dm_1R[:,:,discard_volumes:],\n dm_1S[:,:,discard_volumes:],\n dm_2R[:,:,discard_volumes:],\n dm_4F[:,:,discard_volumes:],\n dm_4R[:,:,discard_volumes:]), axis=-1)\n\nprf_stim = PRFStimulus2D(screen_size_cm=70, \n screen_distance_cm=210, \n design_matrix=dm_full,\n TR=1.5)",
"_____no_output_____"
],
[
"#calculating the average BOLD baseline so that it is the same throughout the timecourse (BOLD has arbtirary units)\niso_periods = np.where(np.sum(dm_full, axis=(0,1))==0)[0]\nshifted_dm = np.zeros_like(dm_full)\n#number of TRs in which activity may linger (hrf)\nshifted_dm[:,:,7:] = dm_full[:,:,:-7]\n\nlate_iso_periods = np.where((np.sum(dm_full, axis=(0,1))==0) & (np.sum(shifted_dm, axis=(0,1))==0))[0]\n\nlate_iso_dict={}\nlate_iso_dict['1R'] = np.split(late_iso_periods,5)[0]\nlate_iso_dict['1S'] = np.split(late_iso_periods,5)[1]\nlate_iso_dict['2R'] = np.split(late_iso_periods,5)[2]\nlate_iso_dict['4F'] = np.split(late_iso_periods,5)[3]\nlate_iso_dict['4R'] = np.split(late_iso_periods,5)[4]",
"_____no_output_____"
],
[
"################preparing the data (SURFACE FITTING)\ndata_path = '/Users/marcoaqil/PRFMapping/PRFMapping-Deriv-noflairtse-manual-hires'\nsubj = 'sub-001'\ntc_dict = {}\ntc_full_iso_nonzerovar_dict = {}\nfor hemi in ['L', 'R']:\n tc_dict[hemi] = {}\n for task_name in ['1R', '1S', '2R', '4F', '4R']:\n data_ses1 = nb.load(opj(data_path, 'fmriprep/'+subj+'/ses-1/func/'+subj+'_ses-1_task-'+task_name+'_run-1_space-fsaverage_hemi-'+hemi+'.func.gii'))\n data_ses2 = nb.load(opj(data_path, 'fmriprep/'+subj+'/ses-2/func/'+subj+'_ses-2_task-'+task_name+'_run-1_space-fsaverage_hemi-'+hemi+'.func.gii'))\n \n tc_ses_1 = sgfilter_predictions(np.array([arr.data for arr in data_ses1.darrays]).T[...,discard_volumes:],\n window_length=window_length)\n\n tc_ses_2 = sgfilter_predictions(np.array([arr.data for arr in data_ses2.darrays]).T[...,discard_volumes:],\n window_length=window_length)\n\n tc_dict[hemi][task_name] = (tc_ses_1+tc_ses_2)/2.0\n \n print('Finished filtering hemi '+hemi)\n #when scanning sub-001 i mistakenly set the length of the 4F scan to 147, while it should have been 145\n #therefore, there are two extra images at the end to discard in that time series.\n #from sub-002 onwards, this was corrected.\n if subj == 'sub-001':\n tc_dict[hemi]['4F'] = tc_dict[hemi]['4F'][...,:-2]\n \n tc_full=np.concatenate((tc_dict[hemi]['1R'],\n tc_dict[hemi]['1S'],\n tc_dict[hemi]['2R'],\n tc_dict[hemi]['4F'],\n tc_dict[hemi]['4R']), axis=-1)\n \n \n #shift timeseries so they have the same average value in proper baseline periods across conditions\n iso_full = np.mean(tc_full[...,late_iso_periods], axis=-1)\n iso_1R_diff = iso_full - np.mean(tc_full[...,late_iso_dict['1R']], axis=-1)\n iso_1S_diff = iso_full - np.mean(tc_full[...,late_iso_dict['1S']], axis=-1)\n iso_2R_diff = iso_full - np.mean(tc_full[...,late_iso_dict['2R']], axis=-1)\n iso_4F_diff = iso_full - np.mean(tc_full[...,late_iso_dict['4F']], axis=-1)\n iso_4R_diff = iso_full - np.mean(tc_full[...,late_iso_dict['4R']], axis=-1)\n \n tc_full_iso=np.concatenate((tc_dict[hemi]['1R'] + iso_1R_diff[...,np.newaxis],\n tc_dict[hemi]['1S'] + iso_1S_diff[...,np.newaxis],\n tc_dict[hemi]['2R'] + iso_2R_diff[...,np.newaxis],\n tc_dict[hemi]['4F'] + iso_4F_diff[...,np.newaxis],\n tc_dict[hemi]['4R'] + iso_4R_diff[...,np.newaxis]), axis=-1)\n \n \n tc_full_iso_nonzerovar_dict['indices_'+hemi] = np.where(np.var(tc_full_iso, axis=-1)>0)\n tc_full_iso_nonzerovar_dict['tc_'+hemi] = tc_full_iso[np.where(np.var(tc_full_iso, axis=-1)>0)]\n \n \n \n \n ",
"Finished filtering hemi L\nFinished filtering hemi R\n"
],
[
"#############preparing the data (VOLUME FITTING)\n############VOLUME MASK\ndata_path = '/Users/marcoaqil/PRFMapping/PRFMapping-Deriv-noflairtse-manual-hires'\nsubj = 'sub-001'\n\n#create a single brain mask in epi space \nmask_dict = {}\nfor task_name in ['1R', '1S', '2R', '4F', '4R']:\n mask_ses_1 = nb.load(opj(data_path,'fmriprep/'+subj+'/ses-1/func/'+subj+'_ses-1_task-'+task_name+'_run-1_space-T1w_desc-brain_mask.nii.gz')).get_data().astype(bool)\n mask_ses_2 = nb.load(opj(data_path, 'fmriprep/'+subj+'/ses-2/func/'+subj+'_ses-2_task-'+task_name+'_run-1_space-T1w_desc-brain_mask.nii.gz')).get_data().astype(bool)\n \n mask_dict[task_name] = mask_ses_1 & mask_ses_2\n \nfinal_mask = mask_dict['1R'] & mask_dict['1S'] & mask_dict['2R'] & mask_dict['4F'] & mask_dict['4R']\n",
"_____no_output_____"
],
[
"#############preparing the data (VOLUME FITTING)\ntc_dict = {}\nfor task_name in ['1R', '1S', '2R', '4F', '4R']:\n timecoursefile_ses_1 = nb.load(opj(data_path, 'fmriprep/'+subj+'/ses-1/func/'+subj+'_ses-1_task-'+task_name+'_run-1_space-T1w_desc-preproc_bold.nii.gz'))\n timecoursefile_ses_2 = nb.load(opj(data_path, 'fmriprep/'+subj+'/ses-2/func/'+subj+'_ses-2_task-'+task_name+'_run-1_space-T1w_desc-preproc_bold.nii.gz'))\n \n tc_ses_1 = sgfilter_predictions(timecoursefile_ses_1.get_data()[...,discard_volumes:],\n window_length=window_length)\n tc_ses_2 = sgfilter_predictions(timecoursefile_ses_2.get_data()[...,discard_volumes:],\n window_length=window_length)\n \n tc_dict[task_name] = (tc_ses_1+tc_ses_2)/2.0\n \n \n \n \n\n#when scanning sub-001 i mistakenly set the length of the 4F-task scan to 147, while it should have been 145\n#therefore, there are two extra images at the end to discard in that time series.\n#from sub-002 onwards, this was corrected.\nif subj == 'sub-001':\n tc_dict['4F'] = tc_dict['4F'][...,:-2]\n \n\ntimecourse_full=np.concatenate((tc_dict['1R'],\n tc_dict['1S'],\n tc_dict['2R'],\n tc_dict['4F'],\n tc_dict['4R']), axis=-1)\n\n\n#shift timeseries so they have the same average value in baseline periods across conditions\niso_full = np.mean(timecourse_full[...,late_iso_periods], axis=-1)\niso_1R_diff = iso_full - np.mean(timecourse_full[...,late_iso_dict['1R']], axis=-1)\niso_1S_diff = iso_full - np.mean(timecourse_full[...,late_iso_dict['1S']], axis=-1)\niso_2R_diff = iso_full - np.mean(timecourse_full[...,late_iso_dict['2R']], axis=-1)\niso_4F_diff = iso_full - np.mean(timecourse_full[...,late_iso_dict['4F']], axis=-1)\niso_4R_diff = iso_full - np.mean(timecourse_full[...,late_iso_dict['4R']], axis=-1)\n\n\ntimecourse_full_iso=np.concatenate((tc_dict['1R'] + iso_1R_diff[...,np.newaxis],\n tc_dict['1S'] + iso_1S_diff[...,np.newaxis],\n tc_dict['2R'] + iso_2R_diff[...,np.newaxis],\n tc_dict['4F'] + iso_4F_diff[...,np.newaxis],\n tc_dict['4R'] + iso_4R_diff[...,np.newaxis]), axis=-1)\n\n \n#brain mask \ntimecourse_brain = timecourse_full_iso[final_mask]\n#exclude timecourses with zero variance\ntimecourse_brain_nonzerovar = timecourse_brain[np.where(np.var(timecourse_brain, axis=-1)>0)]",
"_____no_output_____"
],
[
"#np.save('/Users/marcoaqil/PRFMapping/timecourse_brain_nonzerovar_sub-001.npy', timecourse_brain_nonzerovar)\ntimecourse_brain_nonzerovar = np.load('/Users/marcoaqil/PRFMapping/timecourse_brain_nonzerovar_sub-001.npy')",
"_____no_output_____"
],
[
"#create gaussian grid\ngrid_nr = 20\nmax_ecc_size = 16\nsizes, eccs, polars = max_ecc_size * np.linspace(0.25,1,grid_nr)**2, \\\n max_ecc_size * np.linspace(0.1,1,grid_nr)**2, \\\n np.linspace(0, 2*np.pi, grid_nr)\n\ngg = Iso2DGaussianGridder(stimulus=prf_stim,\n filter_predictions=True,\n window_length=window_length,\n task_lengths=task_lengths)",
"_____no_output_____"
],
[
"%%time\ngf = Iso2DGaussianFitter(data=timecourse_brain_nonzerovar, gridder=gg, n_jobs=10)\n\ngf.grid_fit(ecc_grid=eccs,\n polar_grid=polars,\n size_grid=sizes)",
"100%|██████████| 8000/8000 [8:00:42<00:00, 3.56s/it] "
],
[
"np.save('/Users/marcoaqil/PRFMapping/gauss_grid_sub-001.npy', gf.gridsearch_params)",
"_____no_output_____"
],
[
"%%time\n#refine Gaussian fits\ngf.iterative_fit(rsq_threshold=0.1, verbose=True)",
"[Parallel(n_jobs=10)]: Using backend LokyBackend with 10 concurrent workers.\n[Parallel(n_jobs=10)]: Done 30 tasks | elapsed: 1.3min\n[Parallel(n_jobs=10)]: Done 180 tasks | elapsed: 9.7min\n[Parallel(n_jobs=10)]: Done 430 tasks | elapsed: 18.6min\n[Parallel(n_jobs=10)]: Done 780 tasks | elapsed: 29.3min\n[Parallel(n_jobs=10)]: Done 1230 tasks | elapsed: 42.8min\n[Parallel(n_jobs=10)]: Done 1780 tasks | elapsed: 60.7min\n[Parallel(n_jobs=10)]: Done 2430 tasks | elapsed: 79.5min\n[Parallel(n_jobs=10)]: Done 3180 tasks | elapsed: 105.7min\n[Parallel(n_jobs=10)]: Done 4030 tasks | elapsed: 136.5min\n[Parallel(n_jobs=10)]: Done 4980 tasks | elapsed: 162.9min\n"
],
[
"np.save('/Users/marcoaqil/PRFMapping/gauss_iter_sub-001.npy', gf.iterative_search_params)",
"_____no_output_____"
],
[
"gridsearch_params =np.load('/Users/marcoaqil/PRFMapping/gauss_grid_sub-001.npy')",
"_____no_output_____"
],
[
"%%time\n#now refit normalization model, starting from results of iterated Gaussian fitting\ngg_norm = Norm_Iso2DGaussianGridder(stimulus=prf_stim,\n hrf=[1,1,0],\n filter_predictions=True,\n window_length=window_length,\n task_lengths=task_lengths)\ninf=np.inf\neps=1e-4 #to avoid dividing by zero\n\ngf_norm = Norm_Iso2DGaussianFitter(data=timecourse_brain_nonzerovar,\n gridder=gg_norm,\n n_jobs=10,\n bounds=[(-10*n_pix,10*n_pix), #x\n (-10*n_pix,10*n_pix), #y\n (eps,10*n_pix), #prf size\n (-inf,+inf), #prf amplitude\n (0,+inf), #bold baseline\n (0,+inf), #neural baseline\n (0,+inf), #surround amplitude \n (eps,10*n_pix), #surround size\n (eps,+inf)], #surround baseline \n gradient_method='numerical') \n \n\n#have to add a column since in current code syntax\n#gridsearch_params always contains the CSS exponent parameter, even if it is not fit.\n#whereas iterative_search_params does not contain it if it is not fit)\n#starting_params = np.insert(gf.iterative_search_params, -1, 1.0, axis=-1)\n\n#starting_params = np.insert(current_result_numerical, -1, 1.0, axis=-1)#gridsearch_params\n\nstarting_params = gridsearch_params\n\ngf_norm.iterative_fit(rsq_threshold=0.0, gridsearch_params=starting_params, verbose=True)\n\ncurrent_result_numerical=np.copy(gf_norm.iterative_search_params)",
"[Parallel(n_jobs=10)]: Using backend LokyBackend with 10 concurrent workers.\n[Parallel(n_jobs=10)]: Done 30 tasks | elapsed: 12.3min\n[Parallel(n_jobs=10)]: Done 180 tasks | elapsed: 59.9min\n[Parallel(n_jobs=10)]: Done 430 tasks | elapsed: 135.3min\n[Parallel(n_jobs=10)]: Done 780 tasks | elapsed: 240.3min\n[Parallel(n_jobs=10)]: Done 1230 tasks | elapsed: 376.0min\n[Parallel(n_jobs=10)]: Done 1780 tasks | elapsed: 540.2min\n[Parallel(n_jobs=10)]: Done 2430 tasks | elapsed: 738.3min\n[Parallel(n_jobs=10)]: Done 3180 tasks | elapsed: 958.5min\n[Parallel(n_jobs=10)]: Done 4030 tasks | elapsed: 1205.8min\n[Parallel(n_jobs=10)]: Done 4980 tasks | elapsed: 1493.1min\n[Parallel(n_jobs=10)]: Done 6030 tasks | elapsed: 1807.0min\n[Parallel(n_jobs=10)]: Done 7180 tasks | elapsed: 2145.0min\n"
],
[
"#analytic LBFGSB in 1.1minutes, with tol 1e-80, maxls=300 (best rsq=0.758464 with spm hrf derivative)\nprint(gridsearch_params[np.where(gridsearch_params[:,-1]>0.1),-1].mean())\nprint(current_result[np.where(gridsearch_params[:,-1]>0.1),-1].mean())",
"0.6817100110484308\n0.7584493981057998\n"
],
[
"#numerical L BFGS B in 3.7min, tol 1e-30 maxls=200 (best rsq 0.7801825 with spm hrf derivative)\nprint(gridsearch_params[np.where(gridsearch_params[:,-1]>0.66),-1].mean())\nprint(current_result_numerical[np.where(gridsearch_params[:,-1]>0.66),-1].mean())",
"_____no_output_____"
],
[
"#trust constr standard settings, in 7.4minutes (rsq 0.72019)\nprint(gridsearch_params[np.where(gridsearch_params[:,-1]>0.66),-1].mean())\nprint(current_result_numerical[np.where(gridsearch_params[:,-1]>0.66),-1].mean())",
"0.6817100110484308\n0.7201935822421683\n"
],
[
"print(current_result[np.where(gridsearch_params[:,-1]>0.66),:] - current_result_numerical[np.where(gridsearch_params[:,-1]>0.66),:])\n\n",
"[[[-1.58215457e-02 2.94665645e-02 1.44762422e-01 -2.46626912e+00\n -6.59789638e-01 0.00000000e+00 -1.94087511e-04 1.11924969e+00\n -7.39021304e-03 4.07435824e-04]\n [-1.33285664e+00 -8.14345968e-01 -3.34551595e-02 1.05584280e+03\n 1.41176340e+02 3.07487969e+00 6.17810303e-01 -1.27487557e-01\n 1.52991622e+00 -6.39042293e-02]\n [-5.73344851e-01 -2.48086891e-01 -1.75844267e-01 5.31424911e+02\n 8.82159669e+01 9.99631162e-01 6.20582040e-01 -7.37582050e-01\n 1.48076989e+00 -2.34156135e-02]\n [-1.00378477e-01 3.02112017e-01 7.88803326e-02 8.72723548e+02\n 4.02929175e+02 9.99369668e-01 1.31540694e-01 2.74884942e-01\n 1.09900734e+00 -1.59084632e-02]\n [-2.18296688e-01 -1.03045885e-01 -5.37273931e-03 6.11873125e+01\n 5.32015501e+01 -1.10601350e+01 -6.49870494e+02 6.33744522e-01\n 8.64191175e-03 -1.93111923e-03]\n [-1.27228778e-01 2.00012523e-01 7.69821207e-02 7.54245343e+02\n 3.54478046e+02 9.99301094e-01 -1.83296374e+00 1.91348018e-01\n 9.52495924e-01 -1.19047342e-02]\n [ 9.03320340e-02 -5.97520027e-02 2.04367311e-01 2.95970026e+02\n 2.62245665e+02 -9.32121433e-01 -1.68410441e+02 -6.64412872e-02\n 9.68830083e-01 -6.75230370e-02]\n [-2.59192605e-02 3.01191552e-01 1.70515569e-01 7.57482134e+02\n 3.58216911e+02 9.99007556e-01 4.92711105e-01 -5.43270303e-01\n 8.54545951e-01 -2.15699787e-02]\n [ 1.60990488e-01 -2.13473369e-01 4.48814463e-04 9.65320842e+02\n 1.38709430e+02 9.99818963e-01 -1.18118024e+00 3.19081130e-01\n 1.44572358e+00 -8.92618159e-03]\n [ 2.83551928e-01 -2.19066362e-01 -1.93936879e-01 8.98781541e+02\n 2.13012609e+02 9.99213443e-01 5.28012455e-01 -1.34330509e+00\n 1.21233549e+00 -2.20185975e-02]]]\n"
],
[
"#current_result[np.where(gridsearch_params[:,-1]>0.66),:][0,0,:-1]\nvox=6\nfig=pl.figure()\npl.plot(timecourse_brain_nonzerovar[np.where(gridsearch_params[:,-1]>0.66),:][0,vox])\n#pl.plot(gg_norm.return_single_prediction(*list(current_result[np.where(gridsearch_params[:,-1]>0.66),:][0,vox,:-1])))\npl.plot(gg_norm.return_single_prediction(*list(current_result_numerical[np.where(gridsearch_params[:,-1]>0.66),:][0,vox,:-1])))\n",
"_____no_output_____"
],
[
"from nistats.hemodynamic_models import spm_hrf, spm_time_derivative, spm_dispersion_derivative\n\nfig=pl.figure()\n\npl.plot(spm_hrf(tr=1.5, oversampling=1, time_length=40)+\n 5*spm_time_derivative(tr=1.5, oversampling=1, time_length=40))",
"_____no_output_____"
],
[
"%%time\n#now refit dog model, starting from results of iterated Gaussian fitting\ngg_dog = DoG_Iso2DGaussianGridder(stimulus=prf_stim,\n filter_predictions=True,\n window_length=window_length,\n cond_lengths=cond_lengths)\ninf=np.inf\neps=1e-6 #to avoid dividing by zero\n\ngf_dog = DoG_Iso2DGaussianFitter(data=timecourse_brain_nonzerovar,\n gridder=gg_dog,\n n_jobs=10,\n bounds=[(-10*n_pix,10*n_pix), #x\n (-10*n_pix,10*n_pix), #y\n (eps,10*n_pix), #prf size\n (0,+inf), #prf amplitude\n (0,+inf), #bold baseline\n (0,+inf), #surround amplitude \n (eps,10*n_pix)]) #surround size\n \n\nstarting_params = gf.gridsearch_params\n\ngf_dog.iterative_fit(rsq_threshold=0.0, gridsearch_params=starting_params, verbose=True)",
"_____no_output_____"
],
[
"#compare rsq between models (ideally should be crossvalidated AIC or BIC)\nrsq_mask=np.where(gf_norm.iterative_search_params[:,-1]>0.1)\nprint(np.mean(gf.gridsearch_params[gf.rsq_mask,-1]))\nprint(np.mean(gf.iterative_search_params[gf.rsq_mask,-1]))\nprint(np.mean(gf_norm.iterative_search_params[gf.rsq_mask,-1]))",
"0.24096577922649914\n0.24490732239713542\n0.2686311162490467\n"
],
[
"np.save('/Users/marcoaqil/PRFMapping/norm_bounded_iterparams_sub-001.npy', current_result)",
"_____no_output_____"
],
[
"#convert to ecc/polar and save results for plotting\necc = np.sqrt(gf_norm.iterative_search_params[:,1]**2 + gf_norm.iterative_search_params[:,0]**2)\npolar = np.arctan2(gf_norm.iterative_search_params[:,1], gf_norm.iterative_search_params[:,0])\n\npolar[gf.rsq_mask]+=np.pi\n\nattempt = np.zeros((final_mask.shape[0],final_mask.shape[1],final_mask.shape[2],10))\nha = attempt.reshape((-1,10))\n\ncombined_mask = np.ravel(np.var(timecourse_full_iso, axis=-1)>0) & np.ravel(final_mask)\n\nha[combined_mask,2:]=gf_norm.iterative_search_params[:,2:]\nha[combined_mask,0] = ecc\nha[combined_mask,1] = polar\n\nhaha = ha.reshape((final_mask.shape[0],final_mask.shape[1],final_mask.shape[2],10))\n\n\nfor i in range(0,10):\n nb.Nifti1Image(haha[:,:,:,i], timecoursefile_ses_1.affine).to_filename('norm_bounded{}.nii.gz'.format(i))\n",
"_____no_output_____"
],
[
"###old plotting cells\n\n#print(timecourse_brain_nonzerovar.shape)\n#a nice voxel for testing should be 185537\nfig2=pl.figure()\npl.plot(timecourse_brain_nonzerovar[185537,:])\n#pl.plot(sgfilter_predictions(timecourse_brain_nonzerovar[185537,:],\n# window_length=121))\n#pl.plot(np.load('/Users/marcoaqil/PRFMapping/timecourse_brain_nonzerovar_sub-001.npy')[185537,:])\n\n#print(np.min(gf.gridsearch_params[gf.rsq_mask,-1]))\n#print(np.min(gf.iterative_search_params[gf.rsq_mask,-1]))\nprint(np.argmax(gf.gridsearch_params[np.where((gf.gridsearch_params[gf.rsq_mask,-1]<gf.iterative_search_params[gf.rsq_mask,-1])),-1]))\n#combined_params = np.copy(gf.iterative_search_params)\n#combined_params[np.where(gf.gridsearch_params[gf.rsq_mask,-1]>gf.iterative_search_params[gf.rsq_mask,-1])] = \nfig=pl.figure()\n\nvoxel_nr = 1716\n\nprint(gf.gridsearch_params[voxel_nr,-1])\nprint(gf.iterative_search_params[voxel_nr,-1])\n#print(gf_norm.gridsearch_params[voxel_nr,-1])\n\npl.plot(np.load('/Users/marcoaqil/PRFMapping/timecourse_brain_nonzerovar_sub-001.npy')[voxel_nr,:])\npl.plot(gg.return_single_prediction(*list(gf.gridsearch_params[voxel_nr,:-1])))\npl.plot(gg.return_single_prediction(*list(gf.iterative_search_params[voxel_nr,:-1])))\n#pl.plot(gg_norm.return_single_prediction(*list(gf_norm.iterative_search_params[voxel_nr,:-1])))\nfig = pl.figure()\ngg_norm.add_mean=True\npl.plot(gg_norm.return_single_prediction(*list(gf_norm.iterative_search_params[185537,:-1])))\npl.plot(sgfilter_predictions(timecourse_brain_nonzerovar[185537,:],\n window_length=121,add_mean=False)\n )\n\n#print(np.argmax(gf.iterative_search_params[:,-1]))",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb7114eccf1c3d03611f55d55889dbfe131057fe | 55,005 | ipynb | Jupyter Notebook | site/_build/jupyter_execute/notebooks/graphical-max.ipynb | rpi-techfundamentals/ms-website-fall-2020 | 517b24801286140af5f1e10ee9099cf5d0a28b7c | [
"MIT"
]
| null | null | null | site/_build/jupyter_execute/notebooks/graphical-max.ipynb | rpi-techfundamentals/ms-website-fall-2020 | 517b24801286140af5f1e10ee9099cf5d0a28b7c | [
"MIT"
]
| null | null | null | site/_build/jupyter_execute/notebooks/graphical-max.ipynb | rpi-techfundamentals/ms-website-fall-2020 | 517b24801286140af5f1e10ee9099cf5d0a28b7c | [
"MIT"
]
| null | null | null | 105.982659 | 17,020 | 0.845796 | [
[
[
"# Graphical Solutions \n## Introduction to Linear Programming",
"_____no_output_____"
]
],
[
[
"#Import some required packages. \nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Graphical solution is limited to linear programming models containing only two decision variables (can be used with three variables but only with great difficulty).\n\nGraphical methods provide a picture of how a solution for a linear programming problem is obtained.\n",
"_____no_output_____"
],
[
"## Product mix problem - Beaver Creek Pottery Company\nHow many bowls and mugs should be produced to maximize profits given labor and materials constraints?\n\nProduct resource requirements and unit profit:\n\nDecision Variables:\n\n$x_{1}$ = number of bowls to produce per day\n\n$x_{2}$ = number of mugs to produce per day\n\n\nProfit (Z) Mazimization\n\nZ = 40$x_{1}$ + 50$x_{2}$\n\nLabor Constraint Check\n\n1$x_{1}$ + 2$x_{2}$ <= 40\n\nClay (Physicial Resource) Constraint Check\n\n4$x_{1}$ + 3$x_{2}$ <= 120\n\nNegative Production Constaint Check\n\n$x_{1}$ > 0\n\n$x_{2}$ > 0\n\n",
"_____no_output_____"
]
],
[
[
"#Create an Array X2 from 0 to 60, and it should have a length of 61.\nx2 = np.linspace(0, 60, 61) ",
"_____no_output_____"
],
[
"#This is the same as starting your Excel Spreadsheet with incrementing X2\nx2",
"_____no_output_____"
],
[
"#Labor Constraint Check\n# 1x1 + 2x2 <= 40\n#x1 = 40 - 2*x2\nc1 = 40 - 2*x2\nc1",
"_____no_output_____"
],
[
"#Clay (Physicial Resource) Constraint Check\n#4x1 + 3x2 <= 120\n#x1 = (120 - 3*x2)/4\nc2 = (120 - 3*x2)/4\nc2",
"_____no_output_____"
],
[
"#Calculate the minimum of X1 you can make per the 2 different constraints.\nct = np.minimum(c1,c2)\nct",
"_____no_output_____"
],
[
"#remove those valuese that don't follow non-negativity constraint.\nct= ct[0:21]\nx2= x2[0:21] #Shape of array must be the same.\nct",
"_____no_output_____"
],
[
"#Calculate the profit from the constrained \nprofit = 40*ct+50*x2 \nprofit",
"_____no_output_____"
],
[
"# Make plot for the labor constraint\nplt.plot(c1, x2, label=r'1$x_{1}$ + 2$x_{2}$ <= 40')\nplt.xlim((0, 60))\nplt.ylim((0, 60))\nplt.xlabel(r'$x_{1}$') #Latex way of writing X subscript 1 (See Markdown)\nplt.ylabel(r'$x_{2}$') #Latex way of writing X subscript 1 (See Markdown)\nplt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\nplt.fill_between(c1, x2, color='grey', alpha=0.5)\n",
"_____no_output_____"
],
[
"#Graph Resource Constraint\nplt.plot(c2, x2, label=r'4$x_{1}$ + 3$x_{2}$ <= 120')\nplt.xlim((0, 60))\nplt.ylim((0, 60))\nplt.xlabel(r'$x_{1}$') #Latex way of writing X subscript 1 (See Markdown)\nplt.ylabel(r'$x_{2}$') #Latex way of writing X subscript 1 (See Markdown)\nplt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\nplt.fill_between(c2, x2, color='grey', alpha=0.5)",
"_____no_output_____"
],
[
"# Make plot for the combined constraints.\nplt.plot(c1, x2, label=r'1$x_{1}$ + 2$x_{2}$ <= 40')\nplt.plot(c2, x2, label=r'4$x_{1}$ + 3$x_{2}$ <= 120')\n#plt.plot(ct, x2, label=r'min(x$x_{1}$)')\nplt.xlim((0, 60))\nplt.ylim((0, 60))\nplt.xlabel(r'$x_{1}$') #Latex way of writing X subscript 1 (See Markdown)\nplt.ylabel(r'$x_{2}$') #Latex way of writing X subscript 1 (See Markdown)\nplt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\nplt.fill_between(ct, x2, color='grey', alpha=0.5)",
"_____no_output_____"
]
],
[
[
"Our solution must be in in lies somewhere in the grey feasible region in the graph above. However, according to the fundamental theorum of Linear programming we know it is at a vertex. \n\n\"In mathematical optimization, the fundamental theorem of linear programming states, in a weak formulation, that the maxima and minima of a linear function over a convex polygonal region occur at the region's corners. Further, if an extreme value occurs at two corners, then it must also occur everywhere on the line segment between them.\"\n\n- [Wikipedia](https://en.wikipedia.org/wiki/Fundamental_theorem_of_linear_programming)",
"_____no_output_____"
]
],
[
[
"#This returns the index position of the maximum value\nmax_value = np.argmax(profit)\nmax_value",
"_____no_output_____"
],
[
"#Calculate The max Profit that is made. \nprofit_answer=profit[max_value]\nprofit_answer",
"_____no_output_____"
],
[
"# Verify all constraints are integers\nx2_answer = x2[max_value]\nx2_answer",
"_____no_output_____"
],
[
"# Verify all constraints are integers\nct_answer = ct[max_value]\nct_answer",
"_____no_output_____"
]
],
[
[
"## Q1 Challenge\n\nWhat if the profit function is:\n\nZ = 70$x_{1}$ + 20$x_{2}$ \n\nFind the optimal solution using Python. Assign the answers to: \n\nq1_profit_answer\nq1_x1_answer\nq1_x2_answer\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
cb7118f05643856ad38ccb7ae40dc02254a76f70 | 15,298 | ipynb | Jupyter Notebook | experiment-preprocessing.ipynb | achimkoh/midi-classification | bed1fdc31006092a32dac84d8c4ab34ca243e92c | [
"MIT"
]
| 7 | 2017-05-31T07:09:34.000Z | 2020-09-20T18:59:52.000Z | experiment-preprocessing.ipynb | achimkoh/midi-classification | bed1fdc31006092a32dac84d8c4ab34ca243e92c | [
"MIT"
]
| 1 | 2019-01-10T09:56:46.000Z | 2019-01-10T09:56:46.000Z | experiment-preprocessing.ipynb | achimkoh/midi-classification | bed1fdc31006092a32dac84d8c4ab34ca243e92c | [
"MIT"
]
| null | null | null | 38.340852 | 406 | 0.544058 | [
[
[
"#################\n# Preprocessing #\n#################\n# Scores by other composers from the Bach family have been removed beforehand. \n# Miscellaneous scores like mass pieces have also been removed; the assumption here is that\n# since different interpretations of the same piece (e.g. Ave Maria, etc) exist, including\n# theses pieces might hurt the prediction accuracy, here mostly based on chord progression. \n# (more exactly, a reduced version of the chord progression.)\n\n# In shell, find and copy midi files to target data directory and convert to mxl:\n'''\ncd {TARGETDIR}\nfind {MIDIFILEDIR} \\( -name \"bach*.mid\" -o -name \"beethoven*.mid\" -o -name \"scarlatti*.mid\" \\) -type f -exec cp {} . \\;\nfind . -type f -name \"*.mid\" -exec /Applications/MuseScore\\ 2.app/Contents/MacOS/mscore {} --export-to {}.mxl \\;\nfor f in *.mxl; do mv \"$f\" \"${f%.mid.mxl}.mxl\"; done\nls *.mxl > mxl_list.txt\n'''",
"_____no_output_____"
],
[
"from music21 import *\nfrom os import listdir\nfrom os.path import isfile, getsize",
"_____no_output_____"
],
[
"# timeout function that lets move on beyond too big files.\n# by Thomas Ahle: http://stackoverflow.com/a/22348885\nimport signal\n\nclass timeout:\n def __init__(self, seconds=1, error_message='Timeout'):\n self.seconds = seconds\n self.error_message = error_message\n def handle_timeout(self, signum, frame):\n raise TimeoutError(self.error_message)\n def __enter__(self):\n signal.signal(signal.SIGALRM, self.handle_timeout)\n signal.alarm(self.seconds)\n def __exit__(self, type, value, traceback):\n signal.alarm(0)",
"_____no_output_____"
],
[
"def parse(mxllist, composer):\n composer_list = [f for f in mxllist if f.replace('-', '_').split('_')[0] == composer]\n for file in composer_list:\n if (getsize(file)>10000): # remove too short scores that may contain no notes\n with timeout(seconds=6000):\n try:\n s = converter.parse(mxldir+file)\n try:\n k = s.flat.keySignature.sharps\n except AttributeError:\n k = s.analyze('key').sharps\n except:\n with open('{}-parsed.txt'.format(composer), 'a') as output_file: \n output_file.write('key could not by analyzed\\n')\n with open('{}-transposed.txt'.format(composer), 'a') as output_file: \n output_file.write('key could not by analyzed\\n')\n continue\n t = s.transpose((k*5)%12)\n except:\n with open('{}-parsed.txt'.format(composer), 'a') as output_file:\n output_file.write('timeout\\n')\n with open('{}-transposed.txt'.format(composer), 'a') as output_file: \n output_file.write('timeout\\n')\n continue\n\n fp_s = converter.freeze(s, fmt='pickle')\n fp_t = converter.freeze(t, fmt='pickle')\n\n with open('{}-parsed.txt'.format(composer), 'a') as output_file:\n output_file.write(fp_s+'\\n')\n with open('{}-transposed.txt'.format(composer), 'a') as output_file: \n output_file.write(fp_t+'\\n')",
"_____no_output_____"
],
[
"with open('mxl_list.txt', 'r') as f:\n mxllist = [line.strip() for line in f.readlines()]\n\nparse(mxllist, 'bach')\nparse(mxllist, 'beethoven')\nparse(mxllist, 'debussy')\nparse(mxllist, 'scarlatti')\nparse(mxllist, 'victoria')",
"_____no_output_____"
],
[
"######################\n# Feature Extraction #\n######################",
"_____no_output_____"
],
[
"import itertools\nfrom collections import Counter\n\nflatten = lambda l: [item for sublist in l for item in sublist] # by Alex Martinelli & Guillaume Jacquenot: http://stackoverflow.com/a/952952\nuniqify = lambda seq: list(set(seq))",
"_____no_output_____"
],
[
"# Define known chords\nmajor, minor, suspended, augmented, diminished, major_sixth, minor_sixth, dominant_seventh, major_seventh, minor_seventh, half_diminished_seventh, diminished_seventh, major_ninth, dominant_ninth, dominant_minor_ninth, minor_ninth = [0,4,7],[0,3,7],[0,5,7],[0,4,8],[0,3,6],[0,4,7,9],[0,3,7,9],[0,4,7,10],[0,4,7,11],[0,3,7,10],[0,3,6,10],[0,3,6,9],[0,2,4,7,11],[0,2,4,7,10],[0,1,4,7,10],[0,2,3,7,10]\nchord_types_list = [major, minor, suspended, augmented, diminished, major_sixth, minor_sixth, dominant_seventh, major_seventh, minor_seventh, half_diminished_seventh, diminished_seventh, major_ninth, dominant_ninth, dominant_minor_ninth, minor_ninth]\nchord_types_string = ['major', 'minor', 'suspended', 'augmented', 'diminished', 'major_sixth', 'minor_sixth', 'dominant_seventh', 'major_seventh', 'minor_seventh', 'half_diminished_seventh', 'diminished_seventh', 'major_ninth', 'dominant_ninth', 'dominant_minor_ninth', 'minor_ninth']\n\nroots = list(range(12))\nchord_orders = flatten([[{(n+r)%12 for n in v} for v in chord_types_list] for r in roots])\nunique_orders = []\nfor i in range(192):\n if chord_orders[i] not in unique_orders:\n unique_orders.append(chord_orders[i])",
"_____no_output_____"
],
[
"def merge_chords(s):\n sf = s.flat\n chords_by_offset = []\n for i in range(int(sf.highestTime)):\n chords_by_offset.append(chord.Chord(sf.getElementsByOffset(i,i+1, includeEndBoundary=False, mustFinishInSpan=False, mustBeginInSpan=False).notes))\n return chords_by_offset\n\ndef find_neighbor_note(n, k):\n # find notes k steps away from n\n return (roots[n-6:]+roots[:(n+6)%12])[6+k], (roots[n-6:]+roots[:(n+6)%12])[6-k]\n\ndef find_note_distance(n1, n2):\n return abs(6 - (roots[n1-6:]+roots[:(n1+6)%12]).index(n2))\n\ndef find_chord_distance(set1, set2):\n d1, d2 = set1.difference(set2), set2.difference(set1)\n if len(d1) < len(d2):\n longer, shorter = d2, list(d1)\n else:\n longer, shorter = d1, list(d2)\n distances = []\n for combination in itertools.combinations(longer, len(shorter)):\n for permutation in itertools.permutations(combination):\n dist_p = abs(len(d1)-len(d2))*3 # length difference means notes need to be added/deleted. weighted by 3\n for i in range(len(shorter)):\n dist_p += find_note_distance(shorter[i], permutation[i])\n distances.append(dist_p)\n return min(distances)",
"_____no_output_____"
],
[
"CACHE = dict()\n\ndef find_closest_chord(c, cache=CACHE):\n if len(c) == 0:\n return -1 # use -1 for rest (chords are 0 to 191)\n \n # retrieve from existing knowledge\n o_str, o, p = str(c.normalOrder), set(c.normalOrder), c.pitchClasses\n if o in chord_orders:\n return chord_orders.index(o)\n # the above root sometimes differs from c.findRoot(), which might be more reliable.\n # however, the errors are rare and it should be good enough for now.\n if o_str in cache.keys():\n return cache[o_str]\n \n # find closest chord from scratch\n chord_distances = dict()\n most_common_note = Counter(c.pitchClasses).most_common(1)[0][0]\n\n for i in range(192):\n d = find_chord_distance(o, chord_orders[i])\n # prioritize found chord's root note if most common note of the chord.\n if int(i/16) == most_common_note:\n d += -1\n if chord_distances.get(d) == None:\n chord_distances[d] = []\n chord_distances[d].append(i)\n\n # if multiple chords are tied, use first one (could be better)\n closest_chord = chord_distances[min(chord_distances.keys())][0]\n \n cache[o_str] = closest_chord\n return closest_chord",
"_____no_output_____"
],
[
"def extract_features(parsed_list, idx):\n s = converter.thaw(parsed_list[idx])\n chords_by_offset = merge_chords(s)\n\n chord_sequence = []\n for i in range(len(chords_by_offset)):\n chord_sequence.append(find_closest_chord(chords_by_offset[i], CACHE))\n return chord_sequence",
"_____no_output_____"
],
[
"with open('bach-parsed.txt', 'r') as f:\n FILES_BACH = [line.strip() for line in f.readlines()]\nwith open('beethoven-parsed.txt', 'r') as f:\n FILES_BEETHOVEN = [line.strip() for line in f.readlines()]\nwith open('debussy-parsed.txt', 'r') as f:\n FILES_DEBUSSY = [line.strip() for line in f.readlines()]\nwith open('scarlatti-parsed.txt', 'r') as f:\n FILES_SCARLATTI = [line.strip() for line in f.readlines()]\nwith open('victoria-parsed.txt', 'r') as f:\n FILES_VICTORIA = [line.strip() for line in f.readlines()]\n \nfor i in range(len(FILES_BACH)):\n with open('bach-chordsequence.txt', 'a') as f:\n f.write(str(extract_features(FILES_BACH, i))+'\\n')\nfor i in range(len(FILES_BEETHOVEN)):\n with open('beethoven-chordsequence.txt', 'a') as f:\n f.write(str(extract_features(FILES_BEETHOVEN, i))+'\\n')\nfor i in range(len(FILES_DEBUSSY)):\n with open('debussy-chordsequence.txt', 'a') as f:\n f.write(str(extract_features(FILES_DEBUSSY, i))+'\\n')\nfor i in range(len(FILES_SCARLATTI)):\n with open('scarlatti-chordsequence.txt', 'a') as f:\n f.write(str(extract_features(FILES_SCARLATTI, i))+'\\n')\nfor i in range(len(FILES_VICTORIA)):\n with open('victoria-chordsequence.txt', 'a') as f:\n f.write(str(extract_features(FILES_VICTORIA, i))+'\\n')",
"_____no_output_____"
],
[
"# Additional feature set: extract durations of notes, chords, and rests\ndef find_length_add_to_list(cnr, out_list):\n try:\n out_list.append(cnr.duration.fullName)\n except:\n out_list.append(str(cnr.duration.quarterLength))\n\ndef extract_cnr_duration(piece):\n s = converter.thaw(piece).flat\n chords, notes, rests = [], [], []\n for c in s.getElementsByClass(chord.Chord):\n find_length_add_to_list(c, chords)\n for n in s.getElementsByClass(note.Note):\n find_length_add_to_list(n, notes)\n for r in s.getElementsByClass(note.Rest):\n find_length_add_to_list(r, rests)\n elements = ['chord|'+d for d in chords] + ['note|'+d for d in notes] + ['rest|'+d for d in rests]\n return ';'.join(elements)",
"_____no_output_____"
],
[
"for piece in FILES_BACH:\n with open('bach-durations.txt', 'a') as f:\n f.write(extract_cnr_duration(piece)+'\\n')\nfor piece in FILES_BEETHOVEN:\n with open('beethoven-durations.txt', 'a') as f:\n f.write(extract_cnr_duration(piece)+'\\n')\nfor piece in FILES_DEBUSSY:\n with open('debussy-durations.txt', 'a') as f:\n f.write(extract_cnr_duration(piece)+'\\n')\nfor piece in FILES_SCARLATTI:\n with open('scarlatti-durations.txt', 'a') as f:\n f.write(extract_cnr_duration(piece)+'\\n')\nfor piece in FILES_VICTORIA:\n with open('victoria-durations.txt', 'a') as f:\n f.write(extract_cnr_duration(piece)+'\\n')",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb713b957e68cc36ecfec3469df2f99c0e5936fb | 32,559 | ipynb | Jupyter Notebook | src/user_guide/sos_ruby.ipynb | andrewrocks/sos-docs | fb742052f761e6c3532d8e849a8d07167bc2b4ef | [
"MIT"
]
| null | null | null | src/user_guide/sos_ruby.ipynb | andrewrocks/sos-docs | fb742052f761e6c3532d8e849a8d07167bc2b4ef | [
"MIT"
]
| null | null | null | src/user_guide/sos_ruby.ipynb | andrewrocks/sos-docs | fb742052f761e6c3532d8e849a8d07167bc2b4ef | [
"MIT"
]
| null | null | null | 26.406326 | 240 | 0.238029 | [
[
[
"# How to work with Ruby",
"_____no_output_____"
],
[
"* **Difficulty level**: easy\n* **Time need to lean**: 10 minutes or less\n ",
"_____no_output_____"
],
[
"## Ruby ",
"_____no_output_____"
],
[
"Basic data types recognised in Ruby are similar with Python's data types and there is a one-to-one correspondence for these types.\n\nThe convertion of datatype from SoS to Ruby (e.g. `%get` from Ruby) is as followings:\n\n \n | Python | condition | Ruby |\n | --- | --- |---|\n | `None` | | `nil` |\n | `boolean` | | `TrueClass or FalseClass` |\n | `integer` | | `Integer` |\n | `float` | | `Float` |\n | `complex` | | `Complex` |\n | `str` | | `String` |\n | Sequence (`list`, `tuple`, ...) | | `Array` |\n | `set` | | `Set` |\n | `dict` | | `Hash` |\n | `range` | | `Range` |\n | `numpy.ndarray` | | `Array` |\n | `numpy.matrix` | | `NMatrix` |\n | `pandas.Series` | | `Hash` |\n | `pandas.DataFrame` | | `Daru::DataFrame` |\n\n Python objects in other datatypes are transferred as string `\"Unsupported datatype\"`. Please [let us know](https://github.com/vatlab/sos-ruby/issues) if there is a natural corresponding data type in Ruby to convert this data type.",
"_____no_output_____"
],
[
"Conversion of datatypes from Ruby to SoS (`%get var --from Ruby` from SoS) follows the following rules:\n\n | Ruby | condition | Python |\n | --- | ---| ---|\n | `nil` | | `None` |\n | `Float::NAN` | | `numpy.nan` |\n | `TrueClass or FalseClass` | | `boolean` |\n | `Integer` | | `integer` |\n | `String` | | `str` |\n | `Complex` | | `complex` |\n | `Float` | | `float` |\n | `Array` | | `numpy.ndarray` |\n | `Range` | | `range` |\n | `Set` | | `set` |\n | `Hash` | | `dict` |\n | `NMatrix` | | `numpy.matrix` |\n | `Array` | | `numpy.ndarray` |\n | `Daru::DataFrame` | | `pandas.DataFrame` |\n Ruby objects in other datatypes are transferred as string `\"Unsupported datatype\"`. ",
"_____no_output_____"
],
[
"For example, the scalar data is converted from SoS to Ruby as follows:",
"_____no_output_____"
]
],
[
[
"null_var = None\nnum_var = 123\nlogic_var = True\nchar_var = '1\"23'\ncomp_var = 1+2j",
"_____no_output_____"
],
[
"%get null_var num_var logic_var char_var comp_var\nputs [null_var, num_var, logic_var, char_var, comp_var]",
"[nil, 123, true, \"1\\\"23\", (1.0+2.0i)]\n"
]
],
[
[
"Ruby supports DataFrame from its daru (Data Analysis in RUby) library so you will need to install this library before using the Ruby kernel. For example, a R dataframe is transfered as Daru::DataFrame to Ruby.",
"_____no_output_____"
]
],
[
[
"%get mtcars --from R\nmtcars",
"Loading required package: feather\n"
]
],
[
[
"Also, we choose NMatrix library in Ruby becuase its fast performance. Same as daru (Data Analysis in RUby), you will need to install nmatrix library before using the Ruby kernel.",
"_____no_output_____"
]
],
[
[
"mat_var = N[ [2, 3, 4], [7, 8, 9] ]",
"_____no_output_____"
],
[
"%put mat_var",
"'numpy.matrix([[2, 3, 4], [7, 8, 9]])'\n"
],
[
"mat_var",
"_____no_output_____"
]
],
[
[
"## Further reading\n\n* ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
]
|
cb715c90106560a1f27f03151eb94b4dee87fb91 | 77,446 | ipynb | Jupyter Notebook | explore/Multilayer.ipynb | ai-maker/ntk | 5a32b5bb9f545f140223f98d09ce42141d717015 | [
"MIT"
]
| 2 | 2017-01-28T21:16:25.000Z | 2020-03-18T13:12:41.000Z | explore/Multilayer.ipynb | ai-maker/ntk | 5a32b5bb9f545f140223f98d09ce42141d717015 | [
"MIT"
]
| null | null | null | explore/Multilayer.ipynb | ai-maker/ntk | 5a32b5bb9f545f140223f98d09ce42141d717015 | [
"MIT"
]
| 1 | 2020-03-19T09:49:05.000Z | 2020-03-19T09:49:05.000Z | 98.531807 | 23,444 | 0.80234 | [
[
[
"# Multilayer Perceptron\n\nSome say that 9 out of 10 people who use neural networks apply a Multilayer Perceptron (MLP). A MLP is basically a feed-forward network with 3 layers (at least): an input layer, an output layer, and a hidden layer in between. Thus, the MLP has no structural loops: information always flows from left (input)to right (output). The lack of inherent feedback saves a lot of headaches. Its analysis is totally straightforward given that the output of the network is always a function of the input, it does not depend on any former state of the model or previous input.\n\n\n\nRegarding the topology of a MLP it is normally assumed to be a densely-meshed one-to-many link model between the layers. This is mathematically represented by two matrices of parameters named “the thetas”. In any case, if a certain connection is of little relevance with respect to the observable training data, the network will automatically pay little attention to its contribution and assign it a low weight close to zero.\n\n## Prediction\n\nThe evaluation of the output of a MLP, i.e., its prediction, given an input vector of data is a matter of matrix multiplication. To that end, the following variables are described for convenience:\n* $N$ is the dimension of the input layer.\n* $H$ is the dimension of the hidden layer.\n* $K$ is the dimension of the output layer.\n* $M$ is the dimension of the corpus (number of examples).\n\nGiven the variables above, the parameters of the network, i.e., the thetas matrices, are defined as follows:\n* $\\theta^{(IN)} \\rightarrow H \\times (N+1)$\n* $\\theta^{(OUT)} \\rightarrow K \\times (H+1)$",
"_____no_output_____"
]
],
[
[
"import NeuralNetwork\n\n# 2 input neurons, 3 hidden neurons, 1 output neuron\nnn = NeuralNetwork.MLP([2,3,1])\n\n# nn[0] -> ThetaIN, nn[1] -> ThetaOUT\nprint(nn)",
"[array([[ 0.46134229, 0.42459821, 0.31052926],\n [ 0.04318592, 0.51459231, 0.18476152],\n [-0.98272842, 0.0665426 , 0.87261338]]), array([[ 0.86890272, -0.21121202, 0.70903383, -1.05713378]])]\n"
]
],
[
[
"What follows are the ordered steps that need to be followed in order to evaluate the network prediction.\n\n### Input Feature Expansion\n\nThe first step to attain a successful operation of the neural network is to add a bias term to the input feature space (mapped to the input layer):\n\n$$a^{(IN)} = [1;\\ x]$$\n\nThe feature expansion of the input space with the bias term increases the learning effectiveness of the model because it adds a degree of freedom to the adaptation process. Note that $a^{(IN)}$ directly represents the activation values of the input layer. Thus, the input layer is linear with the input vector $x$ (it is defined by a linear activation function).\n\n### Transit to the Hidden Layer\n\nOnce the activations (outputs) of the input layer are determined, their values flow into the hidden layer through the weights defined in $\\theta^{(IN)}$:\n\n$$z^{(HID)} = \\theta^{(IN)}\\;a^{(IN)}$$\n\nSimilarly, the dimensionality of the hidden layer is expanded with a bias term to increase its learning effectiveness:\n\n$$a^{(HID)} = [1;\\ g(z^{(HID)})]$$\n\nHere, a new function $g()$ is introduced. This is the generic activation function of a neuron, and generally it is non-linear. Its application yields the output values of the hidden layer $a^{(HID)}$ and provides the true learning power to the neural model.\n\n### Output\n\nThen, the activation values of the output layer, i.e., the network prediction, are calculated as follows:\n\n$$z^{(OUT)} = \\theta^{(OUT)}\\;a^{(HID)}$$\n\nand finally\n\n$$a^{(OUT)} = g(z^{(OUT)}) = y$$\n\n### Activation Function\n\nThe activation function of the neuron is (usually) a non-linear function that provides the expressive power to the neural network. It is recommended this function to be smooth, differentiable and monotonically non-decreasing (for learning purposes). Typically, the logistic sigmoid function is used.\n\n$$g(z) = \\frac{1}{(1 + \\exp^{-z})}$$\n\nNote that the range of this function varies from 0 to 1. Therefore, the output values of the neurons will always be bounded by the upper and the lower limits of this range. This entails considering a scaling process if a broader range of predicted values is needed. Other activation functions can be used with the \"af\" parameter. For example, the range of the hyperbolic tangent (\"HyperTan\" function) goes from -1 to 1.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Random instance with 2 values\nx = np.array([1.0, 2.0])\ny = NeuralNetwork.MLP_Predict(nn, x)\n\n# intermediate results are available\n# y[0] -> input result, y[1] -> hidden result, y[2] -> output result\nprint(y)",
"[array([ 1., 2.]), array([ 0.81861604, 0.71652745, 0.6961521 ]), array([ 0.61493809])]\n"
],
[
"z = np.arange(-8, 8, 0.1)\ng = NeuralNetwork.Logistic(z)\n\n%matplotlib inline\nimport matplotlib.pyplot as plt\n\nplt.figure()\nplt.plot(z, g, 'b-', label='g(z)')\nplt.legend(loc='upper left')\nplt.xlabel('Input [z]')\nplt.ylabel('Output [g]')\nplt.title('Logistic sigmoid activation function')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Training\n\nTraining a neural network essentially means fitting its parameters to a set of example data considering an objective function, aka cost function. This process is also known as supervised learning. It is usually implemented as an iterative procedure.\n\n### Cost Function\n\nThe cost function somehow encodes the objective or goal that should be attained with the network. It is usually defined as a classification or a regression evaluation function. However, the actual form of the cost function is effectively the same, which is an error or fitting function. A cost function measures the discrepancy between the desired output for a pattern and the output produced by the network.\n\nThe cost function $J$ quantifies the amount of squared error (or misfitting) that the network displays with respect to a set of data. Thus, in order to achieve a successfully working model, this cost function must be minimised with an adequate set of parameter values. To do so, several solutions are valid as long as this cost function be a convex function (i.e., a bowl-like shape). A well known example of such is the quadratic function, which trains the neural network considering a minimum squared error criterion over the whole dataset of training examples:\n\n$$J(\\theta, x) = \\frac{1}{M} \\sum_{m=1}^M \\sum_{k=1}^K \\left(Error_k^{(m)}\\right)^2 = \\frac{1}{M} \\sum_{m=1}^M \\sum_{k=1}^K \\left(t_k^{(m)}-y_k^{(m)}\\right)^2$$\n\nNote that the term $t$ in the cost function represents the target value of the network (i.e., the ideal/desired network output) for a given input data value $x$. Now that the cost function can be expressed, a convex optimisation procedure (e.g., a gradient-based method) must be conducted in order to minimise its value. Note that this is essentially a least-squares regression.\n\n### Regularisation\n\nThe mean squared-error cost function described above does not incorporate any knowledge or constraint about the characteristics of the parameters being adjusted through the optimisation training strategy. This may develop into a generalisation problem because the space of solutions is large and some of these solutions may turn the model unstable with new unseen data. Therefore, there is the need to smooth the performance of the model over a wide range of input data.\n\nNeural networks usually generalise well as long as the weights are kept small. Thus, the Tikhonov regularisation function, aka ridge regression, is introduced as a means to control complexity of the model in favour of its increased general performance. This regularisation approach, which is used in conjunction with the aforementioned cost function, favours small weight values (it is a cost over large weight values):\n\n$$R(\\theta) = \\frac{\\lambda}{2 M} \\sum_{\\forall \\theta \\notin bias} \\theta^2$$\n\nThere is a typical trade-off in Machine Learning, known as the bias-variance trade-off, which has a direct relationship with the complexity of the model, the nature of the data and the amount of available training data to adjust it. This ability of the model to learn more or less complex scenarios raises an issue with respect to its fitting (memorisation v. generalisation): if the data is simple to explain, a complex model is said to overfit the data, causing its overall performance to drop (high variance model). Similarly, if complex data is tackled with a simple model, such model is said to underfit the data, also causing its overall performance to drop (high bias model). As it is usual in engineering, a compromise must be reached with an adequate $\\lambda$ value.\n\n### Parameter Initialisation\n\nThe initial weights of the thetas assigned by the training process are critical with respect to the success of the learning strategy. They determine the starting point of the optimisation procedure, and depending on their value, the adjusted parameter values may end up in different places if the cost function has multiple (local) minima.\n\nThe parameter initialisation process is based on a uniform distribution between two small numbers that take into account the amount of input and output units of the adjacent layers:\n\n$$\\theta_{init} = U[-\\sigma, +\\sigma]\\ \\ where\\ \\ \\sigma = \\frac{\\sqrt{6}}{\\sqrt{in + out}}$$\n\nIn order to ensure a proper learning procedure, the weights of the parameters need to be randomly assigned in order to prevent any symmetry in the topology of the network model (that would be likely to end in convergence problems).\n\n### Gradient Descent\n\nGiven the convex shape of the cost function (which usually also includes the regularisation), the minimisation objective boils down to finding the extremum of this function using its derivative in the continuos space of the weights. To this end you may use the analytic form of the derivative of the cost function (a nightmare), a numerical finite difference, or automatic differentiation.\n\nGradient descent is a first-order optimisation algorithm, complete but non-optimal. It first starts with some arbitrarily chosen parameters and computes the derivative of the cost function with respect to each of them $\\frac{\\partial J(\\theta,x)}{\\partial \\theta}$. The model parameters are then updated by moving them some distance (determined by the so called learning rate $\\eta$) from the former initial point in the direction of the steepest descent, i.e., along the negative of the gradient. If $\\eta$ is set too small, though, convergence is needlessly slow, whereas if it is too large, the update correction process may overshoot and even diverge.\n\n$$\\theta^{t+1} \\leftarrow \\theta^t - \\eta \\frac{\\partial^t J(\\theta,x)}{\\partial \\theta} $$\n\nThese steps are iterated in a loop until some stopping criterion is met, e.g., a determined number of epochs (i.e., the processing of all patterns in the training example set) is reached, or when no significant improvement is observed.\n\n#### Stochastic versus Batch Learning\n\nOne last remark should be made about the amount of examples $M$ used in the cost function for learning. If the training procedure considers several instances at once per cost gradient computation and parameter update, i.e., $M \\gg 1$, the approach is called batch learning. Batch learning is usually slow because each cost computation accounts for all the available training instances, and especially if the data redundancy is high (similar patterns). However, the conditions of convergence are well understood.\n\nAlternatively, it is usual to consider only one single training instance at a time, i.e., $M=1$, to estimate the gradient in order to speed up the iterative learning process. This procedure is called stochastic (online) learning. Online learning steps are faster to compute, but this noisy single-instance approximation of the cost gradient function makes it a little inaccurate around the optimum. However, stochastic learning often results in better solutions because of the noise in the updates, and thus it is very convenient in most cases.",
"_____no_output_____"
]
],
[
[
"# Load Iris dataset\nfrom sklearn import datasets as dset\nimport copy\n\niris = dset.load_iris()\n\n# build network with 4 input, 1 output\nnn = NeuralNetwork.MLP([4,4,1])\n\n# keep original for further experiments\norig = copy.deepcopy(nn)\n\n# Target needs to be divided by 2 because of the sigmoid, values 0, 0.5, 1\nidat, itar = iris.data, iris.target/2.0\n\n# regularisation parameter of 0.2\ntcost = NeuralNetwork.MLP_Cost(nn, idat, itar, 0.2)\n\n# Cost value for an untrained network\nprint(\"J(ini) = \" + str(tcost))\n\n# Train with numerical gradient, 20 rounds, batch\n# learning rate is 0.1\nNeuralNetwork.MLP_NumGradDesc(nn, idat, itar, 0.2, 20, 0.1)",
"J(ini) = 0.190225295146\nJ(0) = 0.176597276551\nJ(1) = 0.166449794007\nJ(2) = 0.15978678443\nJ(3) = 0.155663219807\nJ(4) = 0.153082110438\nJ(5) = 0.151361935359\nJ(6) = 0.150096475413\nJ(7) = 0.149046019548\nJ(8) = 0.148059359187\nJ(9) = 0.147027418784\nJ(10) = 0.145855924682\nJ(11) = 0.14444895802\nJ(12) = 0.142701554528\nJ(13) = 0.140507269294\nJ(14) = 0.137795918163\nJ(15) = 0.134614743415\nJ(16) = 0.131214474997\nJ(17) = 0.128005994046\nJ(18) = 0.125326101813\nJ(19) = 0.123255428432\nElapsed time = 3.6347489357 seconds\n"
]
],
[
[
"### Backpropagation\n\nThe backpropagation algorithm estimates the error for each neuron unit so as to effectively deploy the gradient descent optimisation procedure. It is a popular algorithm, conceptually simple, computationally efficient, and it often works. In order to conduct the estimation of the neuron-wise errors, it first propagates the training data through the network, then it computes the error with the predictions and the target values, and afterwards it backpropagates the error from the output to the input, generally speaking, from a given layer $(n)$ to the immediately former one $(n-1)$:\n\n$$Error^{(n-1)} = Error^{(n)} \\; \\theta^{(n)}$$\n\nNote that the bias neurons don't backpropagate, they are not connected to the former layer.\n\nFinally, the gradient is computed so that the weights may be updated. Each weight links an input unit $I$ to an output unit $O$, which also provides the error feedback. The general formula that is derived for a logistic sigmoid activation function is shown as folows:\n\n$$\\theta^{(t+1)} \\leftarrow \\theta^{(t)} + \\eta \\; I \\; Error \\; O \\; (1 - O)$$\n\nFrom a computational complexity perspective, Backpropagation is much more effective than the numerical gradient applied above because it computes the errors for all the weights in 2 network traversals, whereas numerical gradient needs to compute 2 traversals per parameter. In addition, stochastic learning is generally the preferred method for Backprop.",
"_____no_output_____"
]
],
[
[
"# Iris example with Backprop\n\n# load original network\nnn = copy.deepcopy(orig)\n\n# Cost value for an untrained network\ntcost = NeuralNetwork.MLP_Cost(nn, idat, itar, 0.2)\nprint(\"J(ini) = \" + str(tcost))\n\n# Train with numerical gradient, 20 rounds\n# learning rate is 0.1\nNeuralNetwork.MLP_Backprop(nn, idat, itar, 0.2, 20, 0.1)",
"J(ini) = 0.190225295146\nJ(0) = 0.210753967513\nJ(1) = 0.171440585973\nJ(2) = 0.123691544254\nJ(3) = 0.0864647094107\nJ(4) = 0.0671969287148\nJ(5) = 0.058489862934\nJ(6) = 0.0545650791155\nJ(7) = 0.0527572869884\nJ(8) = 0.0519049332513\nJ(9) = 0.0514797932695\nJ(10) = 0.0512677581317\nJ(11) = 0.0511888931484\nJ(12) = 0.0512013148995\nJ(13) = 0.0512708124973\nJ(14) = 0.051366940638\nJ(15) = 0.0514662247609\nJ(16) = 0.0515536760846\nJ(17) = 0.051621011596\nJ(18) = 0.0516639229285\nJ(19) = 0.0516801295206\nElapsed time = 0.912276029587 seconds\n"
]
],
[
[
"### Practical Techniques\n\nBackpropagation learning can be tricky particularly for multilayered networks where the cost surface is non-quadratic, non-convex, and high dimensional with many local minima and/or flat regions. Its successful convergence is not guarateed. Designing and training a MLP using Backprop requires making choices such as the number and type of nodes, layers, learning rates, training and test sets, etc, and many undesirable behaviours can be avoided with practical techniques.\n\n#### Instance Shuffling\n\nIn stochastic learning neural networks learn the most from the unexpected instances. Therefore, it is advisable to iterate over instances that are the most unfamiliar to the system (i.e., have the maximum information content). As a means to progress towards getting more chances for learning better, it is recommended to shuffle the training set so that successive training instances rarely belong to the same class.",
"_____no_output_____"
]
],
[
[
"from sklearn.utils import shuffle\n\n# load original network\nnn = copy.deepcopy(orig)\n\n# shuffle instances\nidat, itar = shuffle(idat, itar)\n\nNeuralNetwork.MLP_Backprop(nn, idat, itar, 0.2, 20, 0.1)",
"J(0) = 0.0682368810819\nJ(1) = 0.0417199133578\nJ(2) = 0.0315061299042\nJ(3) = 0.0264972274942\nJ(4) = 0.0234335034004\nJ(5) = 0.0213585773284\nJ(6) = 0.0198743228208\nJ(7) = 0.0187738718684\nJ(8) = 0.0179348376091\nJ(9) = 0.017278820927\nJ(10) = 0.016752915272\nJ(11) = 0.0163201147669\nJ(12) = 0.015953898303\nJ(13) = 0.0156350062017\nJ(14) = 0.0153494683224\nJ(15) = 0.0150873667407\nJ(16) = 0.0148419918866\nJ(17) = 0.0146091530341\nJ(18) = 0.0143865166372\nJ(19) = 0.0141729656303\nElapsed time = 0.88114118576 seconds\n"
]
],
[
[
"#### Feature Standardisation\n\nConvergence is usually faster if the average of each input feature over the training set is close to zero, otherwise the updates will be biased in a particular direction and thus will slow learning.\n\nAdditionally, scaling the features so that all have about the same covariance speeds learning because it helps to balance out the rate at which the weights connected to the input nodes learn.",
"_____no_output_____"
]
],
[
[
"# feature stats\nmu_idat = np.mean(idat, axis=0)\nstd_idat = np.std(idat, axis=0)\n\n# standardise\ns_idat = (idat - mu_idat) / std_idat\n\n# eval\ntest = copy.deepcopy(orig)\nNeuralNetwork.MLP_Backprop(test, s_idat, itar, 0.2, 20, 0.1)",
"J(0) = 0.0523370538733\nJ(1) = 0.0337756553682\nJ(2) = 0.0270653270919\nJ(3) = 0.0237540248914\nJ(4) = 0.0217543183134\nJ(5) = 0.0203610671667\nJ(6) = 0.0192934022444\nJ(7) = 0.0184263828903\nJ(8) = 0.01769901178\nJ(9) = 0.0170782698247\nJ(10) = 0.0165439066423\nJ(11) = 0.0160817888099\nJ(12) = 0.0156809844911\nJ(13) = 0.0153324934183\nJ(14) = 0.0150286798281\nJ(15) = 0.0147629899769\nJ(16) = 0.0145297789721\nJ(17) = 0.0143241801037\nJ(18) = 0.0141419944603\nJ(19) = 0.0139795948063\nElapsed time = 0.779333114624 seconds\n"
]
],
[
[
"#### Feature Decorrelation\n\nIf inputs are uncorrelated then it is possible to solve for the weight values independently. With correlated inputs, the solution must be searched simultaneously, which is a much harder problem. Principal Component Analysis (aka the Karhunen-Loeve expansion) can be used to remove linear correlations in inputs.",
"_____no_output_____"
]
],
[
[
"# construct orthogonal basis with principal vectors\ncovmat = np.cov(s_idat.T)\nl,v = np.linalg.eig(covmat)\n\n# reproject\nd_s_idat = s_idat.dot(v)\n\n# eval\ntest = copy.deepcopy(orig)\nNeuralNetwork.MLP_Backprop(test, d_s_idat, itar, 0.2, 20, 0.1)",
"J(0) = 0.06554149571\nJ(1) = 0.037324025947\nJ(2) = 0.028728921096\nJ(3) = 0.0249406884732\nJ(4) = 0.0228213629049\nJ(5) = 0.021415706904\nJ(6) = 0.0203630759712\nJ(7) = 0.0195081056515\nJ(8) = 0.0187778935497\nJ(9) = 0.0181360491059\nJ(10) = 0.0175632977734\nJ(11) = 0.0170485881247\nJ(12) = 0.0165848154958\nJ(13) = 0.0161667397594\nJ(14) = 0.0157899953346\nJ(15) = 0.0154506502998\nJ(16) = 0.0151450337491\nJ(17) = 0.0148696850601\nJ(18) = 0.0146213512749\nJ(19) = 0.0143969979774\nElapsed time = 0.779126882553 seconds\n"
]
],
[
[
"#### Target Values\n\nTarget values at the sigmoid asymptotes need to be driven by large weights, which can result in instabilities. Instead, target values at the points of the extrema of the second derivative of the sigmoid activation function avoid saturating the output units. The second derivative of the logistic sigmoid is $g''(z) = g(z)(1 - g(z))(1 - 2g(z))$, shown below.",
"_____no_output_____"
]
],
[
[
"g = NeuralNetwork.Logistic\nddg = g(z)*(1 - g(z))*(1 - 2*g(z))\n\nplt.figure()\nplt.plot(z, ddg, 'b-', label='g\\'\\'(z)')\nplt.legend(loc='upper left')\nplt.xlabel('Input [z]')\nplt.ylabel('Output [g\\'\\']')\nplt.title('Second derivative of the logistic sigmoid activation function')\nplt.show()\n\n# max min target values\nmx = max(ddg)\nmi = min(ddg)\nc = 0\nfor i in ddg:\n if i == mx:\n print(\"Max target \" + str(z[c]) + \" -> \" + str(g(z[c])))\n if i == mi:\n print(\"Min target \" + str(z[c]) + \" -> \" + str(g(z[c])))\n c += 1",
"_____no_output_____"
]
],
[
[
"Therefore, optimum target values must be at 0.21 and 0.79.",
"_____no_output_____"
]
],
[
[
"for i in xrange(len(itar)):\n if itar[i] == 0:\n itar[i] = 0.21\n if itar[i] == 1:\n itar[i] = 0.79\ntest = copy.deepcopy(orig)\nNeuralNetwork.MLP_Backprop(test, d_s_idat, itar, 0.2, 20, 0.1)",
"J(0) = 0.023827172164\nJ(1) = 0.0105960582951\nJ(2) = 0.00689965410664\nJ(3) = 0.00564065279975\nJ(4) = 0.00511918611814\nJ(5) = 0.00485322132217\nJ(6) = 0.00468529456735\nJ(7) = 0.00455867238725\nJ(8) = 0.00445156683676\nJ(9) = 0.00435525075903\nJ(10) = 0.00426611403831\nJ(11) = 0.00418260562598\nJ(12) = 0.00410401130004\nJ(13) = 0.00402995467467\nJ(14) = 0.00396019200834\nJ(15) = 0.00389452846363\nJ(16) = 0.00383278490424\nJ(17) = 0.00377478563797\nJ(18) = 0.00372035469683\nJ(19) = 0.00366931546655\nElapsed time = 0.78206205368 seconds\n"
]
],
[
[
"#### Target Vectors\n\nWhen designing a learning system, it is suitable to take into account the nature of the problem at hand (e.g., whether if it is a classification problem or a regression problem) to determine the number of output units $K$.\n\nIn the case of classification, $K$ should be the amount of different classes, and the target output should be a binary vector. Given an instance, only the output unit that corresponds to the instance class should be set. This approach is usually referred to as \"one-hot\" encoding. The decision rule for classification is then driven by the maximum output unit.\n\nIn the case of a regression problem, $K$ should be equal to the number of dependent variables.",
"_____no_output_____"
]
],
[
[
"# Iris is a classification problem, K=3\n\n# build network with 4 input, 3 outputs\ntest3 = NeuralNetwork.MLP([4,4,3])\n\n# modify targets\nt = []\nfor i in itar:\n if i == 0.21:\n t.append([0.79,0.21,0.21])\n elif i == 0.5:\n t.append([0.21,0.79,0.21])\n else:\n t.append([0.21,0.21,0.79])\n\nt = np.array(t)\n\nNeuralNetwork.MLP_Backprop(test3, d_s_idat, t, 0.2, 20, 0.1)",
"J(0) = 0.146039962848\nJ(1) = 0.123696062492\nJ(2) = 0.112324148354\nJ(3) = 0.105530905347\nJ(4) = 0.100608446283\nJ(5) = 0.0964054172643\nJ(6) = 0.092489051251\nJ(7) = 0.0887326759989\nJ(8) = 0.0851165841373\nJ(9) = 0.0816346452278\nJ(10) = 0.078265934502\nJ(11) = 0.074979629429\nJ(12) = 0.0717474249261\nJ(13) = 0.068551693191\nJ(14) = 0.0653878281377\nJ(15) = 0.062262700165\nJ(16) = 0.0591912813568\nJ(17) = 0.0561928474591\nJ(18) = 0.0532875724887\nJ(19) = 0.0504939199725\nElapsed time = 0.821996927261 seconds\n"
]
],
[
[
"Finally, the effectiveness/performance of each approach should be scored with an appropriate metric: squared-error residuals like the cost function for regression problems, and competitive selection for classification.",
"_____no_output_____"
]
],
[
[
"# compare accuracies between single K and multiple K\nsingle = 0\nmultiple = 0\nfor x,y in zip(d_s_idat, itar):\n ps = NeuralNetwork.MLP_Predict(test, x)\n ps = ps[-1][0]\n pm = NeuralNetwork.MLP_Predict(test3, x)\n pm = [pm[-1][0], pm[-1][1], pm[-1][2]]\n if y == 0.21: # class 0\n if np.abs(ps - 0.21) < np.abs(ps - 0.5):\n if np.abs(ps - 0.21) < np.abs(ps - 0.79):\n single += 1\n if pm[0] > pm[1]:\n if pm[0] > pm[2]:\n multiple += 1\n elif y == 0.5: # class 1\n if np.abs(ps - 0.5) < np.abs(ps - 0.21):\n if np.abs(ps - 0.5) < np.abs(ps - 0.79):\n single += 1\n if pm[1] > pm[0]:\n if pm[1] > pm[2]:\n multiple += 1\n else: # class 2\n if np.abs(ps - 0.79) < np.abs(ps - 0.21):\n if np.abs(ps - 0.79) < np.abs(ps - 0.5):\n single += 1\n if pm[2] > pm[0]:\n if pm[2] > pm[1]:\n multiple += 1\nprint(\"Accuracy single: \" + str(single))\nprint(\"Accuracy multiple: \" + str(multiple))",
"Accuracy single: 145\nAccuracy multiple: 142\n"
]
],
[
[
"#### Hidden Units\n\nThe number of hidden units determines the expressive power of the network, and thus, the complexity of its transfer function. The more complex a model is, the more complicated data structures it can learn. Nevertheless, this argument cannot be extended ad infinitum because a shortage of training data with respect to the amount of parameters to be learnt may lead the model to overfit the data. That’s why the aforementioned regularisation function is also used to avoid this situation.\n\nThus, it is common to have a skew toward suggesting a slightly more complex model than strictly necessary (regularisation will compensate for the extra complexity if necessary). Some heuristic guidelines to guess this optimum number of hidden units indicate an amount somewhat related to the number of input and output units. This is an experimental issue, though. There is no rule of thumb for this. Apply a configuration that works for your problem and you’re done.",
"_____no_output_____"
],
[
"#### Final Remarks\n\n* Tweak the network: different activation function, adaptive learning rate, momentum, annealing, noise, etc.\n* Focus on model generalisation: keep a separate self-validation set of data (not used to train the model) to test and estimate the actual performance of the model. See [test_iris.py](test_iris.py)\n* Incorporate as much knowledge as possible. Expertise is a key indicator of success. Data driven models don’t do magic, the more information that is available, the greater the performance of the model.\n* Feature Engineering is of utmost importance. This relates to the former point: the more useful information that can be extracted from the input data, the better performance can be expected. Salient indicators are keys to success. This may lead to selecting only the most informative features (mutual information, chi-square...), or to change the feature space that is used to represent the instance data (Principal Component Analysis for feature extraction and dimensionality reduction). And always standardise your data and exclude outliers.\n* Get more data if the model is not good enough. Related to “the curse of dimensionality” principle: if good data is lacking, no successful model can be obtained. There must be a coherent relation between the parameters of the model (i.e., its complexity) and the amount of available data to train them.\n* Ensemble models, integrate criteria. Bearing in mind that the optimum model structure is not known in advance, one of the most reasonable approaches to obtain a fairly good guess is to apply different models (with different learning features) to the same problem and combine/weight their outputs. Related techniques to this are also known as “boosting”.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
cb71660e7c2a8ffc6beed6072824122fc42fcd2e | 2,983 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Untitled-checkpoint.ipynb | ageva2022/thesisspring22 | 4c9d40a8f01aad87f205403bdc124ae818cf7f6d | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Untitled-checkpoint.ipynb | ageva2022/thesisspring22 | 4c9d40a8f01aad87f205403bdc124ae818cf7f6d | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Untitled-checkpoint.ipynb | ageva2022/thesisspring22 | 4c9d40a8f01aad87f205403bdc124ae818cf7f6d | [
"MIT"
]
| null | null | null | 25.067227 | 218 | 0.478377 | [
[
[
"import numpy as np \nimport matplotlib.pyplot as plt\nimport random",
"_____no_output_____"
],
[
"# UVMRec inputs\n\ntrue_mean = 190 #normalize data? mean needs to be 0\ntrue_sd = 30\ntrue_var = true_sd**2\n\n\n\nt = 10\nrho = 0.5\nbeta = 0.1\nbounds = [180, 200]",
"_____no_output_____"
],
[
"def UVM(data, lower, upper, var, rho, beta):\n\n n = len(data)\n \n data_sq = data**2\n \n clamped_data = np.copy(data_sq)\n \n T_upper = upper * (1 + 2*np.sqrt(np.log(1/beta)) + 2*np.log(1/beta))\n T_lower = 0\n \n clamped_data[clamped_data>T_upper] = T_upper\n clamped_data[clamped_data<T_lower] = T_lower\n \n delta = (1/len) * upper * (1 + 2*np.sqrt(np.log(1/beta)) + 2*np.log(1/beta))\n (upper-lower+2*sd*np.sqrt(2*np.log(2*n/beta)))/n\n \n Y = np.random.normal(0, (delta/np.sqrt(2*rho))**2)\n \n Z = np.mean(data_sq) + Y\n \n new_lower = \n ",
"_____no_output_____"
],
[
"def UVVRec(data, bounds, true_var, t, rho, beta):\n lower = bounds[0]\n upper = bounds[1]\n \n for i in range(t-1):\n \n lower, upper = UVV(data, lower, upper, true_var, rho/(4*(t-1)), (beta/(4*(t-1))))\n# print(\"after it \", lower, \" \", upper)\n# print(\"diff \", upper-lower)\n lower, upper = UVV(data, lower, upper, true_var, 3*rho/4, beta/4)\n \n \n \n return ((lower+upper)/2)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
cb716bd4f5770a280e40ad7277774d4f9d1a4d46 | 362,336 | ipynb | Jupyter Notebook | steps/step58.ipynb | choiking10/mytorch | 67140b608b14e2ec6ecca1638705af91d2d71b6b | [
"MIT"
]
| null | null | null | steps/step58.ipynb | choiking10/mytorch | 67140b608b14e2ec6ecca1638705af91d2d71b6b | [
"MIT"
]
| null | null | null | steps/step58.ipynb | choiking10/mytorch | 67140b608b14e2ec6ecca1638705af91d2d71b6b | [
"MIT"
]
| null | null | null | 1,172.608414 | 233,004 | 0.95399 | [
[
[
"import sys\nsys.path.append(\"..\")",
"_____no_output_____"
],
[
"import numpy as np\nfrom mytorch.models import VGG16\n\nmodel = VGG16(pretrained=True)\n\nx = np.random.randn(1, 3, 224, 224).astype(np.float32)\nmodel.plot(x)",
"_____no_output_____"
],
[
"!apt-get update -y\n!apt-get install -y graphviz\n!dot /root/.mytorch/tmp_graph.dot -T png -o /root/.mytorch/graph.png",
"Ign:1 https://developer.download.nvidia.com/compute/cuda/repos/ubuntu1804/x86_64 InRelease\nIgn:2 https://developer.download.nvidia.com/compute/machine-learning/repos/ubuntu1804/x86_64 InRelease\nHit:3 https://developer.download.nvidia.com/compute/cuda/repos/ubuntu1804/x86_64 Release\nHit:4 https://developer.download.nvidia.com/compute/machine-learning/repos/ubuntu1804/x86_64 Release\nHit:7 http://security.ubuntu.com/ubuntu bionic-security InRelease \nHit:8 http://archive.ubuntu.com/ubuntu bionic InRelease\nHit:9 http://archive.ubuntu.com/ubuntu bionic-updates InRelease\nHit:10 http://archive.ubuntu.com/ubuntu bionic-backports InRelease\nReading package lists... Done \nReading package lists... Done\nBuilding dependency tree \nReading state information... Done\ngraphviz is already the newest version (2.40.1-2).\n0 upgraded, 0 newly installed, 0 to remove and 23 not upgraded.\n"
],
[
"\nimport matplotlib.pyplot as plt\nimg = plt.imread('/root/.mytorch/graph.png')\nplt.figure(figsize=(20, 20))\nplt.imshow(img)",
"_____no_output_____"
],
[
"!pip install ipywidgets",
"Requirement already satisfied: ipywidgets in /usr/local/lib/python3.6/dist-packages (7.6.3)\nRequirement already satisfied: nbformat>=4.2.0 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (5.1.3)\nRequirement already satisfied: ipykernel>=4.5.1 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (5.5.5)\nRequirement already satisfied: widgetsnbextension~=3.5.0 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (3.5.1)\nRequirement already satisfied: jupyterlab-widgets>=1.0.0 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (1.0.0)\nRequirement already satisfied: ipython>=4.0.0 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (7.16.1)\nRequirement already satisfied: traitlets>=4.3.1 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (4.3.3)\nRequirement already satisfied: jupyter-client in /usr/local/lib/python3.6/dist-packages (from ipykernel>=4.5.1->ipywidgets) (6.1.12)\nRequirement already satisfied: tornado>=4.2 in /usr/local/lib/python3.6/dist-packages (from ipykernel>=4.5.1->ipywidgets) (6.1)\nRequirement already satisfied: decorator in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (5.0.9)\nRequirement already satisfied: jedi>=0.10 in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (0.18.0)\nRequirement already satisfied: setuptools>=18.5 in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (54.2.0)\nRequirement already satisfied: pickleshare in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (0.7.5)\nRequirement already satisfied: pygments in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (2.9.0)\nRequirement already satisfied: pexpect in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (4.8.0)\nRequirement already satisfied: prompt-toolkit!=3.0.0,!=3.0.1,<3.1.0,>=2.0.0 in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (3.0.18)\nRequirement already satisfied: backcall in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0->ipywidgets) (0.2.0)\nRequirement already satisfied: parso<0.9.0,>=0.8.0 in /usr/local/lib/python3.6/dist-packages (from jedi>=0.10->ipython>=4.0.0->ipywidgets) (0.8.2)\nRequirement already satisfied: jupyter-core in /usr/local/lib/python3.6/dist-packages (from nbformat>=4.2.0->ipywidgets) (4.7.1)\nRequirement already satisfied: ipython-genutils in /usr/local/lib/python3.6/dist-packages (from nbformat>=4.2.0->ipywidgets) (0.2.0)\nRequirement already satisfied: jsonschema!=2.5.0,>=2.4 in /usr/local/lib/python3.6/dist-packages (from nbformat>=4.2.0->ipywidgets) (3.2.0)\nRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.6/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets) (3.7.3)\nRequirement already satisfied: attrs>=17.4.0 in /usr/local/lib/python3.6/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets) (20.3.0)\nRequirement already satisfied: six>=1.11.0 in /usr/local/lib/python3.6/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets) (1.15.0)\nRequirement already satisfied: pyrsistent>=0.14.0 in /usr/local/lib/python3.6/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets) (0.17.3)\nRequirement already satisfied: wcwidth in /usr/local/lib/python3.6/dist-packages (from prompt-toolkit!=3.0.0,!=3.0.1,<3.1.0,>=2.0.0->ipython>=4.0.0->ipywidgets) (0.2.5)\nRequirement already satisfied: notebook>=4.4.1 in /usr/local/lib/python3.6/dist-packages (from widgetsnbextension~=3.5.0->ipywidgets) (6.4.0)\nRequirement already satisfied: argon2-cffi in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (20.1.0)\nRequirement already satisfied: terminado>=0.8.3 in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.10.0)\nRequirement already satisfied: Send2Trash>=1.5.0 in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (1.5.0)\nRequirement already satisfied: prometheus-client in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.11.0)\nRequirement already satisfied: nbconvert in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (6.0.7)\nRequirement already satisfied: pyzmq>=17 in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (22.1.0)\nRequirement already satisfied: jinja2 in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (3.0.1)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from jupyter-client->ipykernel>=4.5.1->ipywidgets) (2.8.1)\nRequirement already satisfied: ptyprocess in /usr/local/lib/python3.6/dist-packages (from terminado>=0.8.3->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.7.0)\nRequirement already satisfied: cffi>=1.0.0 in /usr/local/lib/python3.6/dist-packages (from argon2-cffi->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (1.14.5)\nRequirement already satisfied: pycparser in /usr/local/lib/python3.6/dist-packages (from cffi>=1.0.0->argon2-cffi->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (2.20)\nRequirement already satisfied: typing-extensions>=3.6.4 in /usr/local/lib/python3.6/dist-packages (from importlib-metadata->jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets) (3.7.4.3)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.6/dist-packages (from importlib-metadata->jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets) (3.4.1)\nRequirement already satisfied: MarkupSafe>=2.0 in /usr/local/lib/python3.6/dist-packages (from jinja2->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (2.0.1)\nRequirement already satisfied: defusedxml in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.7.1)\nRequirement already satisfied: nbclient<0.6.0,>=0.5.0 in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.5.3)\nRequirement already satisfied: mistune<2,>=0.8.1 in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.8.4)\nRequirement already satisfied: jupyterlab-pygments in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.1.2)\nRequirement already satisfied: bleach in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (3.3.0)\nRequirement already satisfied: pandocfilters>=1.4.1 in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (1.4.3)\nRequirement already satisfied: entrypoints>=0.2.2 in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.3)\nRequirement already satisfied: testpath in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.5.0)\nRequirement already satisfied: nest-asyncio in /usr/local/lib/python3.6/dist-packages (from nbclient<0.6.0,>=0.5.0->nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (1.5.1)\nRequirement already satisfied: async-generator in /usr/local/lib/python3.6/dist-packages (from nbclient<0.6.0,>=0.5.0->nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (1.10)\nRequirement already satisfied: webencodings in /usr/local/lib/python3.6/dist-packages (from bleach->nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (0.5.1)\nRequirement already satisfied: packaging in /usr/local/lib/python3.6/dist-packages (from bleach->nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (20.9)\nRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from packaging->bleach->nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets) (2.4.7)\n\u001b[33mWARNING: You are using pip version 21.0.1; however, version 21.1.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\n"
],
[
"from ipywidgets import FileUpload\nfrom IPython.display import display\nupload = FileUpload(accept='.jpg')\ndisplay(upload)",
"_____no_output_____"
],
[
"import io\nimport matplotlib.image as mpimg\nfrom PIL import Image\nfor key, value in upload.value.items():\n print(f\"image : {key}\")\n fp = io.BytesIO(value['content'])\n with fp:\n img = Image.open(io.BytesIO(value['content']))\n plt.imshow(np.array(img))\n \n",
"image : zebra.jpg\n"
],
[
"import mytorch\nmodel = VGG16(pretrained=True)",
"_____no_output_____"
],
[
"for key, value in upload.value.items():\n print(f\"image : {key}\")\n fp = io.BytesIO(value['content'])\n with fp:\n img = Image.open(io.BytesIO(value['content']))\n x = VGG16.preprocess(img)\n x = x[np.newaxis]\n with mytorch.test_mode():\n y = model(x)\n predict_id = np.argmax(y.data)\n \n labels = mytorch.datasets.ImageNet.labels()\n print(labels[predict_id])",
"image : zebra.jpg\nzebra\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb716cc7f2d786a552ec7d7deabe8a287f8d10b0 | 107,047 | ipynb | Jupyter Notebook | Week 3+4/Starter_Code/.ipynb_checkpoints/whale_analysis-Copy13-checkpoint.ipynb | maricheklin/python-stock-analysis-base | a5f0fc3ab1de93291fc23cc4268ecf4094ec3bb9 | [
"MIT"
]
| null | null | null | Week 3+4/Starter_Code/.ipynb_checkpoints/whale_analysis-Copy13-checkpoint.ipynb | maricheklin/python-stock-analysis-base | a5f0fc3ab1de93291fc23cc4268ecf4094ec3bb9 | [
"MIT"
]
| null | null | null | Week 3+4/Starter_Code/.ipynb_checkpoints/whale_analysis-Copy13-checkpoint.ipynb | maricheklin/python-stock-analysis-base | a5f0fc3ab1de93291fc23cc4268ecf4094ec3bb9 | [
"MIT"
]
| null | null | null | 27.079939 | 1,566 | 0.591815 | [
[
[
" # A Whale off the Port(folio)\n ---\n\n In this assignment, you'll get to use what you've learned this week to evaluate the performance among various algorithmic, hedge, and mutual fund portfolios and compare them against the S&P TSX 60 Index.",
"_____no_output_____"
],
[
"## Assumptions and limitations\n\n1. Limitation: Only dates that overlap between portfolios will be compared\n2. Assumption: There are no significant anomalous price impacting events during the time window such as share split, trading halt\n3. Assumption: S&P TSX 60 is representative of the market as a whole, acting as an index\n4. Assumption: Each portfolio (new shares, Whale, and Algos) will have an even spread of weights across all sub-portfolios",
"_____no_output_____"
],
[
"## 0. Import Required Libraries",
"_____no_output_____"
]
],
[
[
"# Initial imports\nimport pandas as pd # daataframe manipulation\nimport numpy as np # calc and numeric manipulatino\nimport datetime as dt # date and time \nfrom pathlib import Path # setting the path for file manipulation\nimport datetime\nimport seaborn as sns # advanced plotting/charting library\nimport matplotlib as plt\npd.options.display.float_format = '{:.6f}'.format # float format to 6 decimal places",
"_____no_output_____"
]
],
[
[
"# I. Data Cleaning\n\nIn this section, you will need to read the CSV files into DataFrames and perform any necessary data cleaning steps. After cleaning, combine all DataFrames into a single DataFrame.\n\nFiles:\n\n* `whale_returns.csv`: Contains returns of some famous \"whale\" investors' portfolios.\n\n* `algo_returns.csv`: Contains returns from the in-house trading algorithms from Harold's company.\n\n* `sp_tsx_history.csv`: Contains historical closing prices of the S&P TSX 60 Index.",
"_____no_output_____"
],
[
"## A. Whale Returns\n\nRead the Whale Portfolio daily returns and clean the data.",
"_____no_output_____"
],
[
"### 1. import whale csv and set index to date",
"_____no_output_____"
]
],
[
[
"df_wr = pd.read_csv('Resources/whale_returns.csv', index_col=\"Date\")",
"_____no_output_____"
]
],
[
[
"### 2. Inspect imported data",
"_____no_output_____"
]
],
[
[
"# look at colums and value head\ndf_wr.head(3)",
"_____no_output_____"
],
[
"# look at last few values\ndf_wr.tail(3)",
"_____no_output_____"
],
[
"# check dimensions of df\ndf_wr.shape",
"_____no_output_____"
],
[
"# get index datatype - for later merging\ndf_wr.index.dtype",
"_____no_output_____"
],
[
"# get datatypes of all values\ndf_wr.dtypes",
"_____no_output_____"
]
],
[
[
"### 3. Count and drop any null values",
"_____no_output_____"
]
],
[
[
"# Count nulls\ndf_wr.isna().sum()",
"_____no_output_____"
],
[
"# Drop nulls \ndf_wr.dropna(inplace=True)",
"_____no_output_____"
],
[
"# Count nulls -again to ensure they're removed\ndf_wr.isna().sum()",
"_____no_output_____"
],
[
"df_wr.count() #double check all values are equal in length",
"_____no_output_____"
]
],
[
[
"### 4. Sort the index to ensure the correct date order for calculations",
"_____no_output_____"
]
],
[
[
"df_wr.sort_index(inplace=True)",
"_____no_output_____"
]
],
[
[
"### 5. Rename columns - shorten and make consistent with other tables",
"_____no_output_____"
]
],
[
[
"# change columns to be consistent and informative\ndf_wr.columns",
"_____no_output_____"
],
[
"df_wr.columns = ['Whale_Soros_Fund_Daily_Returns', 'Whale_Paulson_Daily_Returns',\n 'Whale_Tiger_Daily_Returns', 'Whale_Berekshire_Daily_Returns']",
"_____no_output_____"
]
],
[
[
"### 6. Create copy dataframe with new column for cumulative returns",
"_____no_output_____"
]
],
[
[
"# copy the dataframe to store cumprod in a new view\ndf_wr_cumulative = df_wr.copy()",
"_____no_output_____"
],
[
"# create a new column in new df for each cumulative daily return using the cumprod function\ndf_wr_cumulative['Whale_Soros_Fund_Daily_CumReturns'] = (1 + df_wr_cumulative['Whale_Soros_Fund_Daily_Returns']).cumprod()",
"_____no_output_____"
],
[
"df_wr_cumulative['Whale_Paulson_Daily_CumReturns'] = (1 + df_wr_cumulative['Whale_Paulson_Daily_Returns']).cumprod()",
"_____no_output_____"
],
[
"df_wr_cumulative['Whale_Tiger_Daily_CumReturns'] = (1 + df_wr_cumulative['Whale_Tiger_Daily_Returns']).cumprod()",
"_____no_output_____"
],
[
"df_wr_cumulative['Whale_Berekshire_Daily_CumReturns'] = (1 + df_wr_cumulative['Whale_Berekshire_Daily_Returns']).cumprod()",
"_____no_output_____"
],
[
"df_wr_cumulative.head() # check result is consistent against original column ie adds up",
"_____no_output_____"
],
[
"# drop returns columns from cumulative df",
"_____no_output_____"
],
[
"df_wr_cumulative.columns",
"_____no_output_____"
],
[
"df_wr_cumulative = df_wr_cumulative[['Whale_Soros_Fund_Daily_CumReturns', 'Whale_Paulson_Daily_CumReturns','Whale_Tiger_Daily_CumReturns', 'Whale_Berekshire_Daily_CumReturns']]",
"_____no_output_____"
],
[
"df_wr_cumulative.head()",
"_____no_output_____"
]
],
[
[
"### 7. Look at high level stats & plot for whale portfolios",
"_____no_output_____"
]
],
[
[
"df_wr.describe(include='all') # basic stats for daily whale returns",
"_____no_output_____"
],
[
"df_wr_cumulative.describe(include='all') # basic stats for daily cumulative whale returns",
"_____no_output_____"
],
[
"# plot daily returns - whales\ndf_wr.plot(figsize=(10,5))",
"_____no_output_____"
],
[
"# Plot cumulative returns - individual subportfolios\ndf_wr_cumulative.plot(figsize=(10,5), title='Cumulative Returns - Whale Sub-Portfolios')",
"_____no_output_____"
]
],
[
[
"### 8. Calculate the overall portfolio returns, given equal weight to sub-portfolios",
"_____no_output_____"
]
],
[
[
"# Set weights\nweights_wr = [0.25, 0.25, 0.25, 0.25] # equal weights across all 4 portfolios",
"_____no_output_____"
],
[
"# use the dot function to cross multiple the daily rturns of individual stocks against the weights\nportfolio_df_wr = df_wr.dot(weights_wr)",
"_____no_output_____"
],
[
"portfolio_df_wr.plot(figsize=(10,5), title='Daily Returns for Overall Whale Portfolio (Equal Weighting)')",
"_____no_output_____"
]
],
[
[
"### 9. Calculate the overall portfolio cumultative returns, given equal weight to sub-portfolios",
"_____no_output_____"
]
],
[
[
"# Use the `cumprod` function to cumulatively multiply each element in the Series by it's preceding element until the end\nwr_cumulative_returns = (1 + portfolio_df_wr).cumprod() - 1\n",
"_____no_output_____"
],
[
"wr_cumulative_returns.head()",
"_____no_output_____"
],
[
"wr_cumulative_returns.plot(figsize=(10,5), title='Cumulative Daily Returns for Overall Whale Portfolio (Equal Weighting)')",
"_____no_output_____"
]
],
[
[
"### 10. Initial data overview for Whales ",
"_____no_output_____"
],
[
"<li> Lack of gaps in chart indicate there are no data gaps, and the lack of extreme fluctuations indicates the data is consistent. The data looks consistent and there are no obvious data errors identified. \n<li> Initial high level observations of standalone daily returns data for whale portfolio: At initial glance, the mean daily return indicates that Berkshire portfolio performed best (mean daily returns of 0.000501, mean cumulative daily returns 1.159732), while Paulson worst (-0.000203). The standard deviation indicates highest risk for Berkshire (0.012831 STD), while lowest risk/volatility is Paulson (std 0.006977)\n<li> By looking at the cumulative chart, it is evident that all portfolios were vulnerable to a loss at the same time around 2019-02-16, but that Berkshir was able to increas the most over time and climb the steepest after the downturn.\n<li> A more thorough portfolio comparison analysis will be done in the following analysis section, so no conclusions are drawn yet. ",
"_____no_output_____"
],
[
"## B. Algorithmic Daily Returns\n\nRead the algorithmic daily returns and clean the data.",
"_____no_output_____"
],
[
"### 1. import algo csv and set index to date",
"_____no_output_____"
]
],
[
[
"# Reading algorithmic returns\ndf_ar = pd.read_csv('Resources/algo_returns.csv', index_col='Date')",
"_____no_output_____"
]
],
[
[
"### 2. Inspect resulting dataframe and contained data",
"_____no_output_____"
]
],
[
[
"# look at colums and value first 3 rows\ndf_ar.head(3)",
"_____no_output_____"
],
[
"# look at colums and value last 3 rows\ndf_ar.tail(3)",
"_____no_output_____"
],
[
"# get dimensions of df\ndf_ar.shape",
"_____no_output_____"
],
[
"# get index datatype - for later merging\ndf_ar.index.dtype",
"_____no_output_____"
],
[
"# get datatypes\ndf_ar.dtypes",
"_____no_output_____"
]
],
[
[
"### 3. Count and remove null values",
"_____no_output_____"
]
],
[
[
"# Count nulls\ndf_ar.isna().sum()",
"_____no_output_____"
],
[
"# Drop nulls\ndf_ar.dropna(inplace=True)",
"_____no_output_____"
],
[
"# Count nulls -again to ensure that nulls actually are removed\ndf_ar.isna().sum()",
"_____no_output_____"
],
[
"df_ar.count()",
"_____no_output_____"
]
],
[
[
"### 4. Sort index to ensure correct date order for calculations",
"_____no_output_____"
]
],
[
[
"df_ar.sort_index(inplace=True)",
"_____no_output_____"
]
],
[
[
"### 5. Rename columns to be consistent with future merge",
"_____no_output_____"
]
],
[
[
"df_ar.columns",
"_____no_output_____"
],
[
"df_ar.columns = ['Algo1_Daily_Returns', 'Algo2_Daily_Returns']",
"_____no_output_____"
]
],
[
[
"### 6. Create new column in a copy df for cumulative returns per Algo daily return",
"_____no_output_____"
]
],
[
[
"# create a df copy to store cumulative data\ndf_ar_cumulative = df_ar.copy() ",
"_____no_output_____"
],
[
"# use cumprod to get the daily cumulative returns for each of the algos 1 and 2\ndf_ar_cumulative['Algo1_Daily_CumReturns'] = (1 + df_ar_cumulative['Algo1_Daily_Returns']).cumprod()",
"_____no_output_____"
],
[
"df_ar_cumulative['Algo2_Daily_CumReturns'] = (1 + df_ar_cumulative['Algo2_Daily_Returns']).cumprod()",
"_____no_output_____"
],
[
"# check the result is consistent with the daily returns for first few columns\ndf_ar_cumulative.head(10)",
"_____no_output_____"
],
[
"# drop columns that are not required",
"_____no_output_____"
],
[
"df_ar_cumulative.columns # get the columns",
"_____no_output_____"
],
[
"df_ar_cumulative = df_ar_cumulative[['Algo1_Daily_CumReturns','Algo2_Daily_CumReturns']]",
"_____no_output_____"
],
[
"# check result - first few lines\ndf_ar_cumulative.head(10)",
"_____no_output_____"
]
],
[
[
"### 7. Look at high level stats & plot for algo portfolios",
"_____no_output_____"
]
],
[
[
"df_ar.describe(include='all') # stats for daily returns",
"_____no_output_____"
],
[
"df_ar_cumulative.describe(include='all') # stats for daily cumulative returns",
"_____no_output_____"
],
[
"# plot daily returns - algos\ndf_ar.plot(figsize=(10,5))",
"_____no_output_____"
],
[
"# plot daily cumulative returns - algos\ndf_ar_cumulative.plot(figsize=(10,5))",
"_____no_output_____"
]
],
[
[
"### 8. Calculate the overall portfolio returns, given equal weight to sub-portfolios",
"_____no_output_____"
]
],
[
[
"# Set weights\nweights_ar = [0.5, 0.5] # equal weights across 2 algo sub-portfolios\n",
"_____no_output_____"
],
[
"# use the dot function to cross multiple the daily rturns of individual stocks against the weights\nportfolio_df_ar = df_ar.dot(weights_wr)",
"_____no_output_____"
],
[
"portfolio_df_ar.plot(figsize=(10,5), title='Daily Returns for Overall Algos Portfolio (Equal Weighting)')",
"_____no_output_____"
]
],
[
[
"### 9. Calculate the overall portfolio cumultative returns, given equal weight to sub-portfolios",
"_____no_output_____"
]
],
[
[
"# Use the `cumprod` function to cumulatively multiply each element in the Series by it's preceding element until the end\nar_cumulative_returns = (1 + portfolio_df_ar).cumprod() - 1\n",
"_____no_output_____"
],
[
"ar_cumulative_returns.head()",
"_____no_output_____"
],
[
"ar_cumulative_returns.plot(figsize=(10,5), title='Cumulative Daily Returns for Overall Algos Portfolio (Equal Weighting)')",
"_____no_output_____"
]
],
[
[
"### 10. Quick data overview - Algos",
"_____no_output_____"
],
[
"Initial observations of standalone daily returns data for Algo 1 vs Algo 2: \n\n<li> mean daily return indicates that Algo 1 (mean daily return 0.000654) performs slightly better than Algo 2 (mean daily return 0.000341), which is alo evident in the cumulative daily returns plot. \n \n<li> When looking at just daily returns, Algo 2 is more risky, but when looking at cumulative returns, Algo 1 is more risky (ie higher standard deviation). \n \n<li> Lack of gaps in chart indicate there are no data gaps, and the lack of extreme fluctuations indicates the data is consistent\n\n<li> Cumulative portfio level returns appear steeper compared with Whales at initial glance",
"_____no_output_____"
],
[
"## C. S&P TSX 60 Returns\n\nRead the S&P TSX 60 historic closing prices and create a new daily returns DataFrame from the data. \nNote: this contains daily closing and not returns - needs to be converted",
"_____no_output_____"
],
[
"### 1. Import S&P csv daily closing price (not returns)",
"_____no_output_____"
]
],
[
[
"# Reading S&P TSX 60 Closing Prices\n\ndf_sr = pd.read_csv('Resources/sp_tsx_history.csv')",
"_____no_output_____"
]
],
[
[
"### 2. Inspect columns of dataframe",
"_____no_output_____"
]
],
[
[
"# look at colums and value head\ndf_sr.head(3)",
"_____no_output_____"
],
[
"# look at tail values\ndf_sr.tail(3)",
"_____no_output_____"
]
],
[
[
"#### Note from dataframe inspection: \n#### 1. date column was not immediated converted because it is in\n#### a different format to the other csv files and \n#### needs to bee converted to consistent format first\n#### 2. Close cannot be explicitly converted to float as it has\n#### dollar and commas. \n#### 3. A new column for returns will need to be created from \n#### return calculations. ",
"_____no_output_____"
]
],
[
[
"# check dimension of df\ndf_sr.shape",
"_____no_output_____"
],
[
"# Check Data Types\ndf_sr.dtypes",
"_____no_output_____"
]
],
[
[
"### 3. Convert the date into a consistent format with other tables",
"_____no_output_____"
]
],
[
[
"df_sr['Date']= pd.to_datetime(df_sr['Date']).dt.strftime('%Y-%m-%d')\n",
"_____no_output_____"
]
],
[
[
"### 4. Convert the date data to index and check format and data type",
"_____no_output_____"
]
],
[
[
"# set date as index\ndf_sr.set_index('Date', inplace=True)",
"_____no_output_____"
],
[
"df_sr.head(2)",
"_____no_output_____"
],
[
"df_sr.index.dtype",
"_____no_output_____"
]
],
[
[
"### 5. Check for null values",
"_____no_output_____"
]
],
[
[
"# Count nulls - none observed\ndf_ar.isna().sum()",
"_____no_output_____"
]
],
[
[
"### 6. Convert daily closing price to float (from string)",
"_____no_output_____"
]
],
[
[
"# Change the Closing column to b float type\ndf_sr['Close']= df_sr['Close'].str.replace('$','')\ndf_sr['Close']= df_sr['Close'].str.replace(',','')\ndf_sr['Close']= df_sr['Close'].astype(float)",
"_____no_output_____"
],
[
"# Check Data Types\ndf_sr.dtypes",
"_____no_output_____"
],
[
"# test \ndf_sr.iloc[0]",
"_____no_output_____"
],
[
"# check null values\ndf_sr.isna().sum()",
"_____no_output_____"
],
[
"df_sr.count()",
"_____no_output_____"
]
],
[
[
"### 7. Sort the index for calculations of returns",
"_____no_output_____"
]
],
[
[
"# sort_index \ndf_sr.sort_index(inplace=True)",
"_____no_output_____"
],
[
"df_sr.head(2)",
"_____no_output_____"
]
],
[
[
"df_sr.tail(2)",
"_____no_output_____"
]
],
[
[
"### 8. Calculate daily returns and store in new column",
"_____no_output_____"
],
[
"Equation: $r=\\frac{{p_{t}} - {p_{t-1}}}{p_{t-1}}$\n\nThe daily return is the (current closing price minus the previous day closing price) all divided by the previous day closing price. The initial value has no daily return as there is no prior period to compare it with. \n\nHere the calculation uses the python shift function ",
"_____no_output_____"
]
],
[
[
"\ndf_sr['SnP_TSX_60_Returns'] = (df_sr['Close'] - df_sr['Close'].shift(1))/ df_sr['Close'].shift(1)",
"_____no_output_____"
],
[
"df_sr.head(10)",
"_____no_output_____"
]
],
[
[
"### 9. Cross check conversion to daily returns against alternative method - pct_change function",
"_____no_output_____"
]
],
[
[
"df_sr['SnP_TSX_60_Returns'] = df_sr['Close'].pct_change()\ndf_sr.head(10)",
"_____no_output_____"
]
],
[
[
"#### Methods cross check - looks good - continue",
"_____no_output_____"
]
],
[
[
"# check for null - first row would have null\ndf_sr.isna().sum()",
"_____no_output_____"
],
[
"# Drop nulls - first row\ndf_sr.dropna(inplace=True)",
"_____no_output_____"
],
[
"# Rename `Close` Column to be specific to this portfolio.\ndf_sr.columns",
"_____no_output_____"
],
[
"df_sr.head()",
"_____no_output_____"
]
],
[
[
"### 10. Drop original Closing column - not needed for comparison",
"_____no_output_____"
]
],
[
[
"df_sr = df_sr[['SnP_TSX_60_Returns']] ",
"_____no_output_____"
],
[
"df_sr.columns",
"_____no_output_____"
]
],
[
[
"### 11. Create new column in a copy df for cumulative returns per daily return S&P TSX 60",
"_____no_output_____"
]
],
[
[
"df_sr_cumulative = df_sr.copy()",
"_____no_output_____"
],
[
"# use cumprod to get the daily cumulative returns for each of the algos 1 and 2\ndf_sr_cumulative['SnP_TSX_60_CumReturns'] = (1+df_sr_cumulative['SnP_TSX_60_Returns']).cumprod()",
"_____no_output_____"
],
[
"# visually check first 10 rows to ensure that results make sense\ndf_sr_cumulative.head(10)",
"_____no_output_____"
],
[
"# drop daily returns column from cumulative df\ndf_sr_cumulative = df_sr_cumulative[['SnP_TSX_60_CumReturns']]",
"_____no_output_____"
],
[
"df_sr_cumulative.head()",
"_____no_output_____"
]
],
[
[
"### 12. Look at high level stats & plot for algo portfolios",
"_____no_output_____"
]
],
[
[
"df_sr.describe()",
"_____no_output_____"
],
[
"df_sr_cumulative.describe()",
"_____no_output_____"
],
[
"# plot daily returns - S&P TSX 60\ndf_sr.plot(figsize=(10,5))",
"_____no_output_____"
],
[
"# plot daily returns - S&P TSX 60\ndf_sr_cumulative.plot(figsize=(10,5))",
"_____no_output_____"
]
],
[
[
"### 13. Initial Data Overview - S&P (Market Representation)",
"_____no_output_____"
],
[
"<li> The standard deviation, as expected is the lowst of all portfolios, as this represents the market index and so should not fluctuate as much as other portfolios. It is the leeast risky. The closest other sub-portfolio with lowest risk is Whale_Paulson_Daily_Returns.\n \n<li> Lack of gaps in chart indicate there are no data gaps, and the lack of extreme fluctuations indicates the data is consistent\n\n<li>The returns of individual portfolios would be expected to be higher than the SnP (given higher risk) but this is not always the case, as shall be explored in the analysis sectiond",
"_____no_output_____"
],
[
"## D. Combine Whale, Algorithmic, and S&P TSX 60 Returns",
"_____no_output_____"
],
[
"### 1. Merge daily returns dataframes from all portfolios",
"_____no_output_____"
]
],
[
[
"# Use the `concat` function to combine the two DataFrames by matching indexes (or in this case `Date`)\nmerged_analysis_df_tmp = pd.concat([df_wr, df_ar ], axis=\"columns\", join=\"inner\")",
"_____no_output_____"
],
[
"merged_analysis_df_tmp.head(3)",
"_____no_output_____"
],
[
"# Use the `concat` function to combine the two DataFrames by matching indexes\nmerged_daily_returns_df = pd.concat([merged_analysis_df_tmp, df_sr ], axis=\"columns\", join=\"inner\")",
"_____no_output_____"
],
[
"merged_daily_returns_df.head(3)",
"_____no_output_____"
],
[
"merged_daily_returns_df.tail(3)",
"_____no_output_____"
],
[
"merged_daily_returns_df.shape",
"_____no_output_____"
]
],
[
[
"# II Conduct Quantitative Analysis\n\nIn this section, you will calculate and visualize performance and risk metrics for the portfolios.\n\n<li> First a daily returns comparison is reviewed for individual sub-portfolios\n<li> Second a daily returns comparison is reviewed for portfolio level - comparing Whales and Algos",
"_____no_output_____"
],
[
"## A. Performance Anlysis\n\n#### Calculate and Plot the daily returns",
"_____no_output_____"
],
[
"### 1. Compare daily returns of individual sub-portfolios",
"_____no_output_____"
]
],
[
[
"# Plot daily returns of all portfolios\ndrp = merged_daily_returns_df.plot(figsize=(20,10), rot=45, title='Comparison of Daily Returns on Stock Portfolios')\ndrp.set_xlabel(\"Daily Returns\")\ndrp.set_ylabel(\"Date\")",
"_____no_output_____"
]
],
[
[
"#### Calculate and Plot cumulative returns.",
"_____no_output_____"
],
[
"### 2. Compare Cumulative Daily Returns\n\nCalculations were already done in the first section",
"_____no_output_____"
]
],
[
[
"# Use the `concat` function to combine the two DataFrames by matching indexes\nmerged_cumulative__df_tmp = pd.concat([df_wr_cumulative, df_ar_cumulative ], axis=\"columns\", join=\"inner\")",
"_____no_output_____"
],
[
"merged_daily_cumreturns_df = pd.concat([merged_cumulative__df_tmp, df_sr_cumulative ], axis=\"columns\", join=\"inner\")",
"_____no_output_____"
],
[
"merged_daily_cumreturns_df.head()",
"_____no_output_____"
],
[
"# Plot cumulative returns\n\ndcrp = merged_daily_cumreturns_df.plot(figsize=(20,10), rot=45, title='Comparison of Daily Cumulative Returns on Stock Portfolios')\ndcrp.set_xlabel(\"Daily Cumulative Returns\")\ndcrp.set_ylabel(\"Date\")\n",
"_____no_output_____"
]
],
[
[
"### 3. Compare portfolio level daily returns",
"_____no_output_____"
]
],
[
[
"# create a copy of the daily returns and add the portoflio level columns\nportfolio_daily_return = merged_daily_returns_df.copy()",
"_____no_output_____"
],
[
"\nportfolio_daily_return['Whale_Portfolio_Daily_Returns'] = portfolio_df_wr",
"_____no_output_____"
],
[
"portfolio_daily_return['Algo_Portfolio_Daily_Returns'] = portfolio_df_ar ",
"_____no_output_____"
],
[
"portfolio_daily_return.head(3)",
"_____no_output_____"
],
[
"portfolio_daily_return.tail(3)",
"_____no_output_____"
],
[
"portfolio_daily_return.describe()",
"_____no_output_____"
],
[
"# Plot portfolio vs individual daily returns\n\ndcrp = portfolio_daily_return.plot(figsize=(20,10), rot=45, title='Comparison of Daily Returns on Stock Portfolios - Individual Sub Portfolios vs Portfolio')\ndcrp.set_xlabel(\"Daily Returns\")\ndcrp.set_ylabel(\"Date\")\n\n",
"_____no_output_____"
],
[
"# Plot portfolio only (remove individual sub-portfolios)\nportfolio_daily_return.columns",
"_____no_output_____"
],
[
"portfolio_daily_return_only = portfolio_daily_return[['SnP_TSX_60_Returns','Whale_Portfolio_Daily_Returns', 'Algo_Portfolio_Daily_Returns']]",
"_____no_output_____"
],
[
"dcrp = portfolio_daily_return_only.plot(figsize=(20,10), rot=45, title='Comparison of Daily Returns on Stock Portfolios - Whale vs Algos')\ndcrp.set_xlabel(\"Daily Returns\")\ndcrp.set_ylabel(\"Date\")",
"_____no_output_____"
],
[
"dcrp = portfolio_daily_return.plot(figsize=(20,10), rot=45, title='Comparison of Daily Returns on Stock Portfolios - Individual Sub Portfolios vs Portfolio')\ndcrp.set_xlabel(\"Daily Returns\")\ndcrp.set_ylabel(\"Date\")",
"_____no_output_____"
]
],
[
[
"### 4. Compare portfolio level cumulative daily returns",
"_____no_output_____"
]
],
[
[
"# Copy cumulative daily retrurns df to include portfolio\nportfolio_daily_cumreturns = merged_daily_cumreturns_df.copy()",
"_____no_output_____"
],
[
"# add porrtfolio level cumulative daily returned for whales and algos",
"_____no_output_____"
],
[
"portfolio_daily_cumreturns['Whale_Portfolio_CumRet'] = wr_cumulative_returns",
"_____no_output_____"
],
[
"portfolio_daily_cumreturns['Algos_Portfolio_CumRet'] = ar_cumulative_returns",
"_____no_output_____"
],
[
"dcrp = portfolio_daily_cumreturns.plot(figsize=(20,10), rot=45, title='Comparison of Cumulative Daily Returns on Stock Portfolios - Individual Sub Portfolios vs Portfolio')\ndcrp.set_xlabel(\"Daily Cumulative Returns\")\ndcrp.set_ylabel(\"Date\")",
"_____no_output_____"
],
[
"portfolio_daily_cumreturns.tail(1)",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"## B. Risk Analysis\n\nDetermine the _risk_ of each portfolio:\n\n1. Create a box plot for each portfolio. \n2. Calculate the standard deviation for all portfolios.\n4. Determine which portfolios are riskier than the S&P TSX 60.\n5. Calculate the Annualized Standard Deviation.",
"_____no_output_____"
],
[
"### 1. Create a box plot for each portfolio\n",
"_____no_output_____"
]
],
[
[
"# Box plot to visually show risk\nmcrb = merged_daily_returns_df.plot.box(figsize=(20,10), rot=45, title='Boxplot Comparison of Daily Returns on Stock Portfolios')\ndcrp.set_xlabel(\"Daily Returns\")\ndcrp.set_ylabel(\"Date\")",
"_____no_output_____"
]
],
[
[
"### 2. Calculate Standard Deviations",
"_____no_output_____"
]
],
[
[
"# Daily standard deviation of daily returns sorted in ascending ordeer\ndaily_std = merged_daily_returns_df.std()\ndaily_std.sort_values()\n\n",
"_____no_output_____"
],
[
"mcrb = daily_std.plot.hist(figsize=(20,10), rot=45, title='Comparison of Standard Deviation of Daily Returns on Stock Portfolios')\nmcrb.set_xlabel(\"Returns Standard Deviation\")\nmcrb.set_ylabel(\"Portfolio\")",
"_____no_output_____"
]
],
[
[
"### 3. Standard deviation for S&P TSX 60",
"_____no_output_____"
]
],
[
[
"# Calculate the daily standard deviation of S&P TSX 60\n\ndaily_SnP60_std = merged_daily_returns_df['SnP_TSX_60_Returns'].std()\ndaily_SnP60_std.sort_values()\n\n",
"_____no_output_____"
]
],
[
[
"### 4. Determine which portfolios are riskier than the S&P TSX 60",
"_____no_output_____"
],
[
"The S&P TSX 60 is a market indicator, acting as a benchmark to represent m",
"_____no_output_____"
],
[
"By sorting in ordere of srd deviation on daily return above, the riskier portfolios than S&P TSX 60 are all eexcept Whale Paulson portfolio, as all others have higher std deviation than S&P TSX 60",
"_____no_output_____"
],
[
"### 5. Calculate the Annualized Standard Deviation",
"_____no_output_____"
]
],
[
[
"# Calculate the annualized standard deviation (252 trading days)\nannualized_std = daily_std * np.sqrt(252)\nannualized_std\n\n\n",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"## D. Rolling Statistics\n\nRisk changes over time. Analyze the rolling statistics for Risk and Beta. \n\n1. Calculate and plot the rolling standard deviation for the S&P TSX 60 using a 21-day window.\n2. Calculate the correlation between each stock to determine which portfolios may mimick the S&P TSX 60.\n3. Choose one portfolio, then calculate and plot the 60-day rolling beta for it and the S&P TSX 60.",
"_____no_output_____"
],
[
"### 1. Calculate and plot rolling `std` for all portfolios with 21-day window",
"_____no_output_____"
]
],
[
[
"# Calculate the rolling standard deviation for all portfolios using a 21-day window\n\nroll21_std = merged_daily_returns_df.rolling(window=21).std()\n\nroll21_std",
"_____no_output_____"
],
[
"# Plot the rolling standard deviation on all daily return (not closing price)\nrollsp = roll21_std.rolling(window=21).std().plot(figsize=(20,10), rot=45, title='21 Day Rolling Standard Deviation on Daily Returns on Stock Portfolios')\nrollsp.set_xlabel(\"21 Day Rolling Dates\")\nrollsp.set_ylabel(\"Standard Deviation\")",
"_____no_output_____"
]
],
[
[
"### 2. Calculate and plot the correlation",
"_____no_output_____"
]
],
[
[
"# Calculate the correlation between each column\ncorrelation = merged_daily_returns_df.corr()\ncorrelation.sort_values(ascending=False)\n",
"_____no_output_____"
],
[
"# Display correlation matrix\n\nimport matplotlib.pyplot as plt\n\nfig = plt.gcf()\n\n# Set the title\nplt.title('Inter-Portfolio Correlations')\n\n# Change seaborn plot size\nfig.set_size_inches(12, 8)\n\n\nsns.heatmap(correlation, vmin=-1, vmax=1)",
"_____no_output_____"
]
],
[
[
"### 3. Calculate and Plot Beta for a chosen portfolio and the S&P 60 TSX",
"_____no_output_____"
]
],
[
[
"\n# Covariance of Whales against SnP TSX 60 Returns\nWhale_Soros_Covariance = df_wr[\"Whale_Soros_Fund_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\nWhale_Paulson_Covariance = df_wr[\"Whale_Paulson_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\nWhale_Tiger_Covariance = df_wr[\"Whale_Tiger_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\nWhale_Berekshire_Covariance = df_wr[\"Whale_Berekshire_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# Display the covariance of each whale sub-portfolio\nprint(\"Soros Covariance: \", \"%.16f\" % Whale_Soros_Covariance)\nprint(\"Paulson Covariance: \", \"%.16f\" % Whale_Paulson_Covariance)\nprint(\"Tiger Covariance: \", \"%.16f\" % Whale_Tiger_Covariance)\nprint(\"Berekshire Covariance: \", \"%.16f\" % Whale_Berekshire_Covariance)\n",
"_____no_output_____"
],
[
"# Covariance of Whales against SnP TSX 60 Returns\nAlgo1_Covariance = df_ar[\"Algo1_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\nAlgo2_Covariance = df_ar[\"Algo2_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# Display the covariance of each whale sub-portfolio\nprint(\"Algo1 Covariance: \", \"%.16f\" % Algo1_Covariance)\nprint(\"Algo2 Covariance: \", \"%.16f\" % Algo2_Covariance)\n\n",
"_____no_output_____"
],
[
"# covariance of algos portfolio (within the portfolio)\ncovariance_algo = df_ar.cov()\ncovariance_algo",
"_____no_output_____"
],
[
"# covariance of s&p 60 TSR portfolio\ncovariance_snp = df_sr.cov()\ncovariance_snp",
"_____no_output_____"
],
[
"# Calculate covariance of a single sub-portfolio streams in portfolios\n# how each individual sub-portfolios covary with other sub-portfolios\n# similar evaluation to correlation heat map\ncovariance_a = merged_daily_returns_df.cov()\ncovariance_a",
"_____no_output_____"
],
[
"# Calculate variance of S&P TSX\nvariance_snp = df_sr.var()\nvariance_snp",
"_____no_output_____"
],
[
"# Beta Values for Whales Sub-Portfolios\n# Calculate beta of all daily returns of whale portfolio\nSoros_beta = Whale_Soros_Covariance / variance_snp\nPaulson_beta = Whale_Paulson_Covariance / variance_snp\nTiger_beta = Whale_Tiger_Covariance / variance_snp\nBerekshire_beta = Whale_Berekshire_Covariance / variance_snp\n\n\n# Display the covariance of each Whale sub-portfolio\nprint(\"Soros Beta: \", \"%.16f\" % Soros_beta)\nprint(\"Paulson Beta: \", \"%.16f\" % Paulson_beta)\nprint(\"Tiger Beta: \", \"%.16f\" % Tiger_beta)\nprint(\"Berekshire Beta: \", \"%.16f\" % Berekshire_beta)\nprint(\"--------------------\")\n\nAverage_Whale_beta = (Soros_beta + Paulson_beta + Tiger_beta + Berekshire_beta)/4\nprint(\"Average Whale Beta: \", \"%.16f\" % Average_Whale_beta)",
"_____no_output_____"
],
[
"# Beta Values for Algos Sub-Portfolios\n# Calculate beta of all daily returns of Algos portfolio\nAlgo1_beta = Algo1_Covariance / variance_snp\nAlgo2_beta = Algo2_Covariance / variance_snp\n\n\n# Display the covariance of each Algos sub-portfolio\nprint(\"Algo1 Beta: \", \"%.16f\" % Algo1_beta)\nprint(\"Algo2 Beta: \", \"%.16f\" % Algo2_beta)\n\nprint(\"--------------------\")\n\nAverage_Algo_beta = (Algo1_beta + Algo2_beta)/2\nprint(\"Average Algo Beta: \", \"%.16f\" % Average_Algo_beta)",
"_____no_output_____"
],
[
"# 21 day rolling covariance of algo portfolio stocks vs. S&P TSX 60\nrolling_algo1_covariance = merged_daily_returns_df[\"Algo1_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_algo2_covariance = merged_daily_returns_df[\"Algo2_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# 21 day rolling covariance of whale portfolio stocks vs. S&P TSX 60\nrolling_Soros_covariance = merged_daily_returns_df[\"Whale_Soros_Fund_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Paulson_covariance = merged_daily_returns_df[\"Whale_Paulson_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Tiger_covariance = merged_daily_returns_df[\"Whale_Tiger_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Berkshire_covariance = merged_daily_returns_df[\"Whale_Berekshire_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# 21 day rolling S&P TSX 60 covariance\nrolling_SnP_covariance = merged_daily_returns_df[\"SnP_TSX_60_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n\n# 21 day rolling variance of S&P TSX 60\nrolling_variance = merged_daily_returns_df[\"SnP_TSX_60_Returns\"].rolling(window=21).var()\n\n# 21 day rolling beta of algo portfolio stocks vs. S&P TSX 60\nrolling_algo1_beta = rolling_algo1_covariance / rolling_variance\nrolling_algo2_beta = rolling_algo2_covariance / rolling_variance\n\n# 21 day average beta for algo portfolio\nrolling_average_algo_beta = (rolling_algo1_beta + rolling_algo1_beta)/2\n\n# 21 day rolling beta of whale portfolio stocks vs. S&P TSX 60\nrolling_Soros_beta = rolling_Soros_covariance / rolling_variance\nrolling_Paulson_beta = rolling_Paulson_covariance / rolling_variance\nrolling_Tiger_beta = rolling_Tiger_covariance / rolling_variance\nrolling_Berkshire_beta = rolling_Berkshire_covariance / rolling_variance\nrolling_SnP_Beta = rolling_SnP_covariance/ rolling_variance\n\n# 21 day average beta for whale portfolio\nrolling_average_whale_beta = (rolling_Soros_beta + rolling_Paulson_beta + rolling_Tiger_beta + rolling_Berkshire_beta)/4\n",
"_____no_output_____"
],
[
"# Set the figure and plot the different social media beta values as multiple trends on the same figure\nax = rolling_algo1_covariance.plot(figsize=(20, 10), title=\"Rolling 21 Day Covariance of Sub-Portfolio Returns vs. S&P TSX 60 Returns\")\nrolling_algo2_covariance.plot(ax=ax)\nrolling_Soros_covariance.plot(ax=ax)\nrolling_Paulson_covariance.plot(ax=ax)\nrolling_Tiger_covariance.plot(ax=ax)\nrolling_Berkshire_covariance.plot(ax=ax)\n\n# Set the legend of the figure\nax.legend([\"Algo1 Covariance\", \"Algo2 Covariance\", \"Whale Soros Covariance\", \"Whale Paulson Covariance\", \"Whale Tiger Covariance\",\"Whale Berkshire Covariance\"])",
"_____no_output_____"
],
[
"\nrolling_algo1_covariance.plot(figsize=(20, 10), title='Rolling 21 Day Covariance of Sub-Portfolio Returns vs. S&P TSX 60 Returns')",
"_____no_output_____"
],
[
"rolling_algo1_beta.describe()",
"_____no_output_____"
],
[
"\n\n# Plot beta trend\n# Set the figure and plot the different social media beta values as multiple trends on the same figure\nax = rolling_algo1_beta.plot(figsize=(20, 10), title=\"Rolling 21 Day Beta of Sub-Portfolio Returns vs. S&P TSX 60 Returns\")\nrolling_algo2_beta.plot(ax=ax)\nrolling_Soros_beta.plot(ax=ax)\nrolling_Paulson_beta.plot(ax=ax)\nrolling_Tiger_beta.plot(ax=ax)\nrolling_Berkshire_beta.plot(ax=ax)\nrolling_SnP_Beta.plot(ax=ax)\n\n# Set the legend of the figure\nax.legend([\"Algo1 Beta\", \"Algo2 Beta\", \"Whale Soros Beta\", \"Whale Paulson Beta\", \"Whale Tiger Beta\",\"Whale Berkshire Beta\"])",
"_____no_output_____"
]
],
[
[
"## E. Rolling Statistics Challenge: Exponentially Weighted Average \n\nAn alternative way to calculate a rolling window is to take the exponentially weighted moving average. This is like a moving window average, but it assigns greater importance to more recent observations. Try calculating the [`ewm`](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.ewm.html) with a 21-day half-life.",
"_____no_output_____"
]
],
[
[
"### 1. Show mean and stanadrd deviation exponentially weighted average",
"_____no_output_____"
]
],
[
[
"\newm_21_mean = merged_daily_returns_df.ewm(halflife=21).mean()",
"_____no_output_____"
]
],
[
[
"ewm_21_mean.head()",
"_____no_output_____"
],
[
"ewm_21_mean.plot(figsize=(20, 10), title=\"Mean EWM\")",
"_____no_output_____"
],
[
"ewm_21_std = merged_daily_returns_df.ewm(halflife=21).std()",
"_____no_output_____"
],
[
"ewm_21_std.head()",
"_____no_output_____"
],
[
"ewm_21_std.plot(figsize=(20, 10), title=\"Standard Deviation EWM\")",
"_____no_output_____"
]
],
[
[
"### 2. EWM end of section discussion",
"_____no_output_____"
],
[
"Exponentially Weighted Mean analysis provides an emphasis on more recent events by providing an exponential \"decay\" of weights values going back in time, growing smaller further back in times. In this instance a half life of 21 days is used, meaning that at that point the weights would have \"decayed\" exponentially to 50% of the most recent value. The theory behind this is that more recent events are more relevant and accurate to the current market conditions and should therefore have more weight. ",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"# III Sharpe Ratios\nIn reality, investment managers and thier institutional investors look at the ratio of return-to-risk, and not just returns alone. After all, if you could invest in one of two portfolios, and each offered the same 10% return, yet one offered lower risk, you'd take that one, right?\n\n### Using the daily returns, calculate and visualise the Sharpe ratios using a bar plot",
"_____no_output_____"
]
],
[
[
"# Annualised Sharpe Ratios\nsharpe_ratios = (merged_daily_returns_df.mean() * 252) / (merged_daily_returns_df.std() * np.sqrt(252))\n\n",
"_____no_output_____"
],
[
"sharpe_ratios.sort_values(ascending = False)",
"_____no_output_____"
],
[
"# Visualize the sharpe ratios as a bar plot\n# Plot sharpe ratios\nsharpe_ratios.plot(kind=\"bar\", title=\"Sharpe Ratios\")",
"_____no_output_____"
]
],
[
[
"### [[TODO]] Get individual portfolio average sharp ratios to compare overall portfolio types",
"_____no_output_____"
]
],
[
[
"# Calculate standar deviaton for all investments for each portfolio\nharold_std_annual = harold_returns.std() * np.sqrt(252)\nmy_std_annual = my_returns.std() * np.sqrt(252)\n\n\n\n\nalgo1_sharp = merged_daily_returns_df[\"Algo1_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_algo2_covariance = merged_daily_returns_df[\"Algo2_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# 21 day rolling covariance of whale portfolio stocks vs. S&P TSX 60\nrolling_Soros_covariance = merged_daily_returns_df[\"Whale_Soros_Fund_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Paulson_covariance = merged_daily_returns_df[\"Whale_Paulson_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Tiger_covariance = merged_daily_returns_df[\"Whale_Tiger_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Berkshire_covariance = merged_daily_returns_df[\"Whale_Berekshire_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n",
"_____no_output_____"
],
[
"# Calculate standar deviaton for all investments for each portfolio\nharold_std_annual = harold_returns.std() * np.sqrt(252)\nmy_std_annual = my_returns.std() * np.sqrt(252)",
"_____no_output_____"
],
[
"# Calculate sharpe ratio\nharold_sharpe_ratios = (harold_returns.mean() * 252) / (harold_std_annual)\nmy_sharpe_ratios = (my_returns.mean() * 252) / (my_std_annual)",
"_____no_output_____"
]
],
[
[
"### Determine whether the algorithmic strategies outperform both the market (S&P TSX 60) and the whales portfolios.\n\nWrite your answer here!",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"# Create Custom Portfolio\n\nIn this section, you will build your own portfolio of stocks, calculate the returns, and compare the results to the Whale Portfolios and the S&P TSX 60. \n\n1. Choose 3-5 custom stocks with at last 1 year's worth of historic prices and create a DataFrame of the closing prices and dates for each stock.\n2. Calculate the weighted returns for the portfolio assuming an equal number of shares for each stock.\n3. Join your portfolio returns to the DataFrame that contains all of the portfolio returns.\n4. Re-run the performance and risk analysis with your portfolio to see how it compares to the others.\n5. Include correlation analysis to determine which stocks (if any) are correlated.",
"_____no_output_____"
],
[
"## Choose 3-5 custom stocks with at last 1 year's worth of historic prices and create a DataFrame of the closing prices and dates for each stock.\n\nFor this demo solution, we fetch data from three companies listes in the S&P TSX 60 index.\n\n* `SHOP` - [Shopify Inc](https://en.wikipedia.org/wiki/Shopify)\n\n* `OTEX` - [Open Text Corporation](https://en.wikipedia.org/wiki/OpenText)\n\n* `L` - [Loblaw Companies Limited](https://en.wikipedia.org/wiki/Loblaw_Companies)",
"_____no_output_____"
]
],
[
[
"merged_daily_returns_df.head(1)",
"_____no_output_____"
]
],
[
[
"## A. Get Daily Returns for Shopify Stocks ",
"_____no_output_____"
],
[
"### 1. Read in csv shopify data",
"_____no_output_____"
]
],
[
[
"# Reading data from 1st stock - shopify\ndf_shop = pd.read_csv('Resources/Shopify.csv')",
"_____no_output_____"
]
],
[
[
"### 2. Inspect data",
"_____no_output_____"
]
],
[
[
"df_shop.shape",
"_____no_output_____"
],
[
"df_shop.head(3)",
"_____no_output_____"
],
[
"df_shop.dtypes",
"_____no_output_____"
],
[
"df_shop.count()",
"_____no_output_____"
]
],
[
[
"### 3. Convert date to index",
"_____no_output_____"
]
],
[
[
"df_shop['Date']= pd.to_datetime(df_shop['Date']).dt.strftime('%Y-%m-%d')",
"_____no_output_____"
],
[
"df_shop.head(3)",
"_____no_output_____"
],
[
"# set date as index\ndf_shop.set_index('Date', inplace=True)",
"_____no_output_____"
],
[
"df_shop.head(3)",
"_____no_output_____"
]
],
[
[
"### 4. Remove unwanted columns",
"_____no_output_____"
]
],
[
[
"df_shop.columns",
"_____no_output_____"
],
[
"df_shop = df_shop[['Close']]",
"_____no_output_____"
]
],
[
[
"### 5. Sort date index ascending just in case",
"_____no_output_____"
]
],
[
[
"df_shop.sort_index(inplace=True) # probably not necssary but just in case",
"_____no_output_____"
]
],
[
[
"### 6. Get daily returns & remove closing cost",
"_____no_output_____"
]
],
[
[
"df_shop['Shop_Daily_Returns'] = df_shop['Close'].pct_change()",
"_____no_output_____"
],
[
"df_shop = df_shop[['Shop_Daily_Returns']]",
"_____no_output_____"
]
],
[
[
"### 7. Review and drop nulls",
"_____no_output_____"
]
],
[
[
"df_shop.isna().sum() # first row would be null",
"_____no_output_____"
],
[
"df_shop.dropna(inplace=True)",
"_____no_output_____"
],
[
"df_shop.isna().sum() #null should be gone",
"_____no_output_____"
]
],
[
[
"## B. Get Daily Returns For Open Text Stocks ",
"_____no_output_____"
],
[
"### 1. Read in csv for Open Text",
"_____no_output_____"
]
],
[
[
"# Reading data from 2nd stock - Otex\ndf_otex = pd.read_csv('Resources/Otex.csv')",
"_____no_output_____"
]
],
[
[
"### 2. Inspect dataframe",
"_____no_output_____"
]
],
[
[
"df_otex.heaad(3)",
"_____no_output_____"
],
[
"df_otex.tail(3)",
"_____no_output_____"
],
[
"df_otex.shape",
"_____no_output_____"
],
[
"df_otex.count()",
"_____no_output_____"
]
],
[
[
"### 3. Convert date to index",
"_____no_output_____"
]
],
[
[
"df_otex['Date']= pd.to_datetime(df_otex['Date']).dt.strftime('%Y-%m-%d')",
"_____no_output_____"
],
[
"# set date as index\ndf_otex.set_index('Date', inplace=True)",
"_____no_output_____"
],
[
"### 4. Remove unwanted columns -declutter",
"_____no_output_____"
],
[
"df_otex = df_otex[['Close']]",
"_____no_output_____"
],
[
"### 5. Sort date index ascending just in case",
"_____no_output_____"
],
[
"df_otex.sort_index(inplace=True) # probably not necssary but just in case",
"_____no_output_____"
],
[
"### 6. Get daily returns & remove closing cost",
"_____no_output_____"
],
[
"df_otex['Otex_Daily_Returns'] = df_otex['Close'].pct_change()",
"_____no_output_____"
],
[
"df_otex = df_otex[['Otex_Daily_Returns']]",
"_____no_output_____"
],
[
"### 7. Review and drop nulls",
"_____no_output_____"
],
[
"df_otex.isna().sum() # first row would be null",
"_____no_output_____"
],
[
"df_otex.dropna(inplace=True)",
"_____no_output_____"
]
],
[
[
"## C. Get Returns for Loblaw Stocks",
"_____no_output_____"
],
[
"### 1. Read in csv for Loblaw",
"_____no_output_____"
]
],
[
[
"# Reading data from 3rd stock - Loblaw\ndf_lob = pd.read_csv('Resources/TSE_L.csv')",
"_____no_output_____"
]
],
[
[
"### 2. Inspect dataframe",
"_____no_output_____"
]
],
[
[
"df_lob.head(3)",
"_____no_output_____"
],
[
"df_lob.tail(3)",
"_____no_output_____"
],
[
"df_lob.shape",
"_____no_output_____"
],
[
"df_lob.dtypes",
"_____no_output_____"
]
],
[
[
"### 3. Convert date to index",
"_____no_output_____"
]
],
[
[
"df_lob['Date']= pd.to_datetime(df_lob['Date']).dt.strftime('%Y-%m-%d')",
"_____no_output_____"
],
[
"# set date as index\ndf_lob.set_index('Date', inplace=True)",
"_____no_output_____"
],
[
"### 4. Remove unwanted columns -declutter",
"_____no_output_____"
],
[
"df_lob = df_lob[['Close']]",
"_____no_output_____"
],
[
"### 5. Sort date index ascending just in case",
"_____no_output_____"
],
[
"df_lob.sort_index(inplace=True) # probably not necssary but just in case",
"_____no_output_____"
],
[
"### 6. Get daily returns & remove closing cost",
"_____no_output_____"
],
[
"df_lob['Loblaw_Daily_Returns'] = df_lob['Close'].pct_change()",
"_____no_output_____"
],
[
"df_lob = df_lob[['Loblaw_Daily_Returns']]",
"_____no_output_____"
]
],
[
[
"### 7. Review and drop nulls",
"_____no_output_____"
]
],
[
[
"df_lob.isna().sum() # first row would be null",
"_____no_output_____"
],
[
"df_lob.dropna(inplace = True)",
"_____no_output_____"
]
],
[
[
"### 8. Have a final look at the data - plot and describe",
"_____no_output_____"
]
],
[
[
"df_lob.describe()",
"_____no_output_____"
],
[
"df_lob.plot() # have a quick look that it is centred around zero",
"_____no_output_____"
]
],
[
[
"## D. Concat New Stock Daily Returns into Single Dataframe",
"_____no_output_____"
],
[
"### 1. Perform inner concat to ensure dates line up",
"_____no_output_____"
]
],
[
[
"# Use the `concat` function to combine the two DataFrames by matching indexes (or in this case `Date`)\nmerged_analysis_newstock_df_tmp = pd.concat([df_lob, df_otex], axis=\"columns\", join=\"inner\")",
"_____no_output_____"
],
[
"merged_newstock_daily_returns_df = pd.concat([merged_analysis_newstock_df_tmp, df_shop], axis=\"columns\", join=\"inner\")",
"_____no_output_____"
]
],
[
[
"### 2. Inspect data in newly merged ",
"_____no_output_____"
]
],
[
[
"merged_newstock_daily_returns_df.head(5)",
"_____no_output_____"
],
[
"merged_newstock_daily_returns_df.tail(5)",
"_____no_output_____"
],
[
"merged_newstock_daily_returns_df.shape",
"_____no_output_____"
],
[
"merged_newstock_daily_returns_df.dtypes",
"_____no_output_____"
],
[
"merged_newstock_daily_returns_df.index.dtype # check the data type of the index",
"_____no_output_____"
],
[
"merged_newstock_daily_returns_df.isna().sum() # no nulls found, already removed in last step",
"_____no_output_____"
],
[
"### 3. Plot Merged Daily Returns Data",
"_____no_output_____"
],
[
"drp = merged_newstock_daily_returns_df.plot(figsize=(20,10), rot=45, title='Comparison of Daily Returns on Stocks in New Portfolio')\ndrp.set_xlabel(\"Daily Returns\")\ndrp.set_ylabel(\"Date\")\n",
"_____no_output_____"
]
],
[
[
"## E. Calculate the weighted returns for the portfolio assuming an equal number of shares for each stock",
"_____no_output_____"
],
[
"### 1. Portfolio returns. - Set even weights across the 3 new portfolios",
"_____no_output_____"
]
],
[
[
"# Set weights\nweights = [1/3, 1/3, 1/3]\n\n",
"_____no_output_____"
]
],
[
[
"### 2. Calculate portfolio return",
"_____no_output_____"
]
],
[
[
"# use the dot function to cross multiple the daily rturns of individual stocks against the weights\nportfolio_newstock_daily_returns_df = merged_newstock_daily_returns_df.dot(weights)\n",
"_____no_output_____"
],
[
"portfolio_newstock_daily_returns_df.shape",
"_____no_output_____"
],
[
"portfolio_newstock_daily_returns_df.head()",
"_____no_output_____"
]
],
[
[
"### 3. Plot Portfolio Returns",
"_____no_output_____"
]
],
[
[
"drp = portfolio_newstock_daily_returns_df.plot(figsize=(20,10), rot=45, title='Comparison of Daily Portfolio Returns on Stocks in New Portfolio')\ndrp.set_xlabel(\"Portfolio Daily Returns\")\ndrp.set_ylabel(\"Date\")\n\n",
"_____no_output_____"
]
],
[
[
"## F. Join your portfolio returns to the DataFrame that contains all of the portfolio returns",
"_____no_output_____"
]
],
[
[
"# Join your returns DataFrame to the original returns DataFrame\n\nmerged_orig_vs_new_returns = pd.concat([merged_daily_returns_df, portfolio_newstock_daily_returns_df], axis=\"columns\", join=\"inner\")",
"_____no_output_____"
],
[
"len(merged_orig_vs_new_returns) # there are 966 overlappting dates after inner join",
"_____no_output_____"
],
[
"merged_orig_vs_new_returns.head()",
"_____no_output_____"
],
[
"# Only compare dates where return data exists for all the stocks (drop NaNs)\n",
"_____no_output_____"
],
[
"drp = merged_orig_vs_new_returns.plot(figsize=(20,10), rot=45, title='Comparison of Daily Portfolio Returns on Stocks in New Portfolio')\ndrp.set_xlabel(\"Portfolio Daily Returns\")\ndrp.set_ylabel(\"Date\")",
"_____no_output_____"
]
],
[
[
"## G. Re-run the risk analysis with your portfolio to see how it compares to the others",
"_____no_output_____"
],
[
"### 1. Calculate the Annualized Standard Deviation",
"_____no_output_____"
]
],
[
[
"# Daily standard deviation of new and old daily returns sorted in ascending ordeer\ndaily_std_new_old = merged_orig_vs_new_returns.std()\ndaily_std_new_old.sort_values()",
"_____no_output_____"
],
[
"nomcrb = daily_std_new_old.plot.hist(figsize=(20,10), rot=45, title='Comparison of Standard Deviation of Daily Returns on New vs Original Stock Portfolios')\nnomcrb.set_xlabel(\"Daily Returns Standard Deviation\")\nnomcrb.set_ylabel(\"Portfolio\")",
"_____no_output_____"
],
[
"# Calculate the annualised `std`\n# Calculate the annualised standard deviation (252 trading days)\nannualized_new_old_std = daily_std_new_old * np.sqrt(252)\nannualized_new_old_std.sort_values()",
"_____no_output_____"
],
[
"# plot annualised standard deviation old vs new\n\n\nnomcrb = annualized_new_old_std.hist(figsize=(20,10), rot=45, title='Comparison of Annualised Standard Deviation of Daily Returns on New vs Original Stock Portfolios')\n\n",
"_____no_output_____"
]
],
[
[
"### 2. Calculate and plot rolling `std` with 21-day window",
"_____no_output_____"
]
],
[
[
"# Calculate rolling standard deviation\n\n# Calculate the rolling standard deviation for all portfolios using a 21-day window\n\nroll21_old_new_std = merged_daily_returns_df.rolling(window=21).std()\n\nroll21_old_new_std\n\n\n\n",
"_____no_output_____"
],
[
"\n# Plot the rolling standard deviation on all daily return (not closing price)\nrollsp = roll21_old_new_std.rolling(window=21).std().plot(figsize=(20,10), rot=45, title='21 Day Rolling Standard Deviation on Old vs New Daily Returns on Stock Portfolios')\nrollsp.set_xlabel(\"21 Day Rolling Dates\")\nrollsp.set_ylabel(\"Standard Deviation\")",
"_____no_output_____"
]
],
[
[
"### 3. Calculate and plot the correlation",
"_____no_output_____"
]
],
[
[
"# Calculate and plot the correlation\n# Calculate the correlation between each column\ncorrelation_oldnew = merged_orig_vs_new_returns.corr()\ncorrelation_oldnew.sort_values(ascending=False)\n\n\n",
"_____no_output_____"
],
[
"# Display correlation matrix\n\nimport matplotlib.pyplot as plt\n\nfig = plt.gcf()\n\n# Set the title\nplt.title('Inter-Portfolio Correlations')\n\n# Change seaborn plot size\nfig.set_size_inches(12, 8)\n\n\nsns.heatmap(correlation_oldnewcorrelation, vmin=-1, vmax=1)",
"_____no_output_____"
]
],
[
[
"### 4. Calculate and Plot the 60-day Rolling Beta for Your Portfolio compared to the S&P 60 TSX",
"_____no_output_____"
]
],
[
[
"# Calculate and plot Beta\n\n# Covariance of Whales against SnP TSX 60 Return\nNew_Stocks_Covariance = df_wr[\"Whale_Berekshire_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# Display the covariance of each whale sub-portfolio\nprint(\"Soros Covariance: \", \"%.16f\" % Whale_Soros_Covariance)\nprint(\"Paulson Covariance: \", \"%.16f\" % Whale_Paulson_Covariance)\nprint(\"Tiger Covariance: \", \"%.16f\" % Whale_Tiger_Covariance)\nprint(\"Berekshire Covariance: \", \"%.16f\" % Whale_Berekshire_Covariance)\n\n# Covariance of Whales against SnP TSX 60 Returns\nAlgo1_Covariance = df_ar[\"Algo1_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\nAlgo2_Covariance = df_ar[\"Algo2_Daily_Returns\"].cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# Display the covariance of each whale sub-portfolio\nprint(\"Algo1 Covariance: \", \"%.16f\" % Algo1_Covariance)\nprint(\"Algo2 Covariance: \", \"%.16f\" % Algo2_Covariance\n \n \n# covariance of algos portfolio (within the portfolio)\ncovariance_algo = df_ar.cov()\ncovariance_algo\n \n# covariance of s&p 60 TSR portfolio\ncovariance_snp = df_sr.cov()\ncovariance_snp\n \n \n # Calculate covariance of a single sub-portfolio streams in portfolios\n# how each individual sub-portfolios covary with other sub-portfolios\n# similar evaluation to correlation heat map\ncovariance_a = merged_daily_returns_df.cov()\ncovariance_a\n \n \n# Calculate variance of S&P TSX\nvariance_snp = df_sr.var()\nvariance_snp\n \n# Beta Values for Whales Sub-Portfolios\n# Calculate beta of all daily returns of whale portfolio\nSoros_beta = Whale_Soros_Covariance / variance_snp\nPaulson_beta = Whale_Paulson_Covariance / variance_snp\nTiger_beta = Whale_Tiger_Covariance / variance_snp\nBerekshire_beta = Whale_Berekshire_Covariance / variance_snp\n\n\n# Display the covariance of each Whale sub-portfolio\nprint(\"Soros Beta: \", \"%.16f\" % Soros_beta)\nprint(\"Paulson Beta: \", \"%.16f\" % Paulson_beta)\nprint(\"Tiger Beta: \", \"%.16f\" % Tiger_beta)\nprint(\"Berekshire Beta: \", \"%.16f\" % Berekshire_beta)\nprint(\"--------------------\")\n\nAverage_Whale_beta = (Soros_beta + Paulson_beta + Tiger_beta + Berekshire_beta)/4\nprint(\"Average Whale Beta: \", \"%.16f\" % Average_Whale_beta)\n \n# Beta Values for Algos Sub-Portfolios\n# Calculate beta of all daily returns of Algos portfolio\nAlgo1_beta = Algo1_Covariance / variance_snp\nAlgo2_beta = Algo2_Covariance / variance_snp\n\n\n# Display the covariance of each Algos sub-portfolio\nprint(\"Algo1 Beta: \", \"%.16f\" % Algo1_beta)\nprint(\"Algo2 Beta: \", \"%.16f\" % Algo2_beta)\n\nprint(\"--------------------\")\n\nAverage_Algo_beta = (Algo1_beta + Algo2_beta)/2\nprint(\"Average Algo Beta: \", \"%.16f\" % Average_Algo_beta)\n \n \n \n \n \n# 21 day rolling covariance of algo portfolio stocks vs. S&P TSX 60\nrolling_algo1_covariance = merged_daily_returns_df[\"Algo1_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_algo2_covariance = merged_daily_returns_df[\"Algo2_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# 21 day rolling covariance of whale portfolio stocks vs. S&P TSX 60\nrolling_Soros_covariance = merged_daily_returns_df[\"Whale_Soros_Fund_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Paulson_covariance = merged_daily_returns_df[\"Whale_Paulson_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Tiger_covariance = merged_daily_returns_df[\"Whale_Tiger_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\nrolling_Berkshire_covariance = merged_daily_returns_df[\"Whale_Berekshire_Daily_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n# 21 day rolling S&P TSX 60 covariance\nrolling_SnP_covariance = merged_daily_returns_df[\"SnP_TSX_60_Returns\"].rolling(window=21, min_periods=1).cov(df_sr[\"SnP_TSX_60_Returns\"])\n\n\n# 21 day rolling variance of S&P TSX 60\nrolling_variance = merged_daily_returns_df[\"SnP_TSX_60_Returns\"].rolling(window=21).var()\n\n# 21 day rolling beta of algo portfolio stocks vs. S&P TSX 60\nrolling_algo1_beta = rolling_algo1_covariance / rolling_variance\nrolling_algo2_beta = rolling_algo2_covariance / rolling_variance\n\n# 21 day average beta for algo portfolio\nrolling_average_algo_beta = (rolling_algo1_beta + rolling_algo1_beta)/2\n\n# 21 day rolling beta of whale portfolio stocks vs. S&P TSX 60\nrolling_Soros_beta = rolling_Soros_covariance / rolling_variance\nrolling_Paulson_beta = rolling_Paulson_covariance / rolling_variance\nrolling_Tiger_beta = rolling_Tiger_covariance / rolling_variance\nrolling_Berkshire_beta = rolling_Berkshire_covariance / rolling_variance\nrolling_SnP_Beta = rolling_SnP_covariance/ rolling_variance\n\n# 21 day average beta for whale portfolio\nrolling_average_whale_beta = (rolling_Soros_beta + rolling_Paulson_beta + rolling_Tiger_beta + rolling_Berkshire_beta)/4\n\n\n# Set the figure and plot the different social media beta values as multiple trends on the same figure\nax = rolling_algo1_covariance.plot(figsize=(20, 10), title=\"Rolling 21 Day Covariance of Sub-Portfolio Returns vs. S&P TSX 60 Returns\")\nrolling_algo2_covariance.plot(ax=ax)\nrolling_Soros_covariance.plot(ax=ax)\nrolling_Paulson_covariance.plot(ax=ax)\nrolling_Tiger_covariance.plot(ax=ax)\nrolling_Berkshire_covariance.plot(ax=ax)\n\n# Set the legend of the figure\nax.legend([\"Algo1 Covariance\", \"Algo2 Covariance\", \"Whale Soros Covariance\", \"Whale Paulson Covariance\", \"Whale Tiger Covariance\",\"Whale Berkshire Covariance\"])",
"_____no_output_____"
]
],
[
[
"### 5. Using the daily returns, calculate and visualize the Sharpe ratios using a bar plot",
"_____no_output_____"
]
],
[
[
"# Calculate Annualised Sharpe Ratios\n# Annualised Sharpe Ratios\nsharpe_ratios_old_new = (merged_daily_returns_df.mean() * 252) / (merged_daily_returns_df.std() * np.sqrt(252))\n\n\n",
"_____no_output_____"
],
[
"sharpe_ratios_old_new.sort_values(ascending=False)",
"_____no_output_____"
],
[
"# Visualise the sharpe ratios as a bar plot\n\n# Plot sharpe ratios\nsharpe_ratios_old_new.plot(kind=\"bar\", title=\"Sharpe Ratios\")",
"_____no_output_____"
]
],
[
[
"### 6. How does your portfolio do?\n\n\n\nHowever, the individual risk appetite of the individual needs to be taken into consideration. For relatively low risk and reasonable returns, a market index such as the S&P is a good options. \nFor others who have more liquid cash to gamble, might simple ignore risk and go for short term highest rewards. \n\nHowever, overall it is possible to look at the best stock as one that offer the best return for the lowest risk. This is what the sharpe ratio uncovers. For that reason, of all the portfolios, the best investment would be \n\nMore data would be needed to see to what extent the intrenal portfolio stock eg Algo 1, correlate with one another. Highly correlated stocks move togeether, and the net effect is an amplification of gains and losses. From a risk mitigation perspective, highly correlated stock portfolios can be diversified with those that have none or negative correlation. ",
"_____no_output_____"
],
[
"## References\n\nShift function in pandas - \nhttps://stackoverflow.com/questions/20000726/calculate-daily-returns-with-pandas-dataframe\n\nConditional line color - \nhttps://stackoverflow.com/questions/31590184/plot-multicolored-line-based-on-conditional-in-python\n\nhttps://stackoverflow.com/questions/40803570/python-matplotlib-scatter-plot-specify-color-points-depending-on-conditions/40804861\n\nhttps://stackoverflow.com/questions/42453649/conditional-color-with-matplotlib-scatter\n\nhttps://stackoverflow.com/questions/3832809/how-to-change-the-color-of-a-single-bar-if-condition-is-true-matplotlib\n\nhttps://stackoverflow.com/questions/56779975/conditional-coloring-in-matplotlib-using-numpys-where\n\nGoogle finance - https://support.google.com/docs/answer/3093281?hl=en\n\nBoxplots - https://towardsdatascience.com/understanding-boxplots-5e2df7bcbd51\n\nEWM - https://www.youtube.com/watch?v=lAq96T8FkTw\n\nPEP 8 - Standards - https://www.python.org/dev/peps/pep-0008/\n\n",
"_____no_output_____"
],
[
"# Instructions: Unit 4 Homework Assignment: A Whale Off the Port(folio)\n\n\n\n## Background\n\nHarold's company has been investing in algorithmic trading strategies. Some of the investment managers love them, some hate them, but they all think their way is best.\n\nYou just learned these quantitative analysis techniques with Python and Pandas, so Harold has come to you with a challenge—to help him determine which portfolio is performing the best across multiple areas: volatility, returns, risk, and Sharpe ratios.\n\nYou need to create a tool (an analysis notebook) that analyzes and visualizes the major metrics of the portfolios across all of these areas, and determine which portfolio outperformed the others. You will be given the historical daily returns of several portfolios: some from the firm's algorithmic portfolios, some that represent the portfolios of famous \"whale\" investors like Warren Buffett, and some from the big hedge and mutual funds. You will then use this analysis to create a custom portfolio of stocks and compare its performance to that of the other portfolios, as well as the larger market ([S&P TSX 60 Index](https://en.wikipedia.org/wiki/S%26P/TSX_60)).\n\nFor this homework assignment, you have three main tasks:\n\n1. [Read in and Wrangle Returns Data](#Prepare-the-Data)\n\n2. [Determine Success of Each Portfolio](#Conduct-Quantitative-Analysis)\n\n3. [Choose and Evaluate a Custom Portfolio](#Create-a-Custom-Portfolio)\n\n---\n\n## Instructions\n\n**Files:**\n\n* [Whale Analysis Starter Code](Starter_Code/whale_analysis.ipynb)\n\n* [algo_returns.csv](Starter_Code/Resources/algo_returns.csv)\n\n* [otex_historical.csv](Starter_Code/Resources/otex_historical.csv)\n\n* [sp_tsx_history.csv](Starter_Code/Resources/sp_tsx_history.csv)\n\n* [l_historical.csv](Starter_Code/Resources/l_historical.csv)\n\n* [shop_historical.csv](Starter_Code/Resources/shop_historical.csv)\n\n* [whale_returns.csv](Starter_Code/Resources/whale_returns.csv)\n\n### Prepare the Data\n\nFirst, read and clean several CSV files for analysis. The CSV files include whale portfolio returns, algorithmic trading portfolio returns, and S&P TSX 60 Index historical prices. Use the starter code to complete the following steps:\n\n1. Use Pandas to read the following CSV files as a DataFrame. Be sure to convert the dates to a `DateTimeIndex`.\n\n * `whale_returns.csv`: Contains returns of some famous \"whale\" investors' portfolios.\n\n * `algo_returns.csv`: Contains returns from the in-house trading algorithms from Harold's company.\n\n * `sp_tsx_history.csv`: Contains historical closing prices of the S&P TSX 60 Index.\n\n2. Detect and remove null values.\n\n3. If any columns have dollar signs or characters other than numeric values, remove those characters and convert the data types as needed.\n\n4. The whale portfolios and algorithmic portfolio CSV files contain daily returns, but the S&P TSX 60 CSV file contains closing prices. Convert the S&P TSX 60 closing prices to daily returns.\n\n5. Join `Whale Returns`, `Algorithmic Returns`, and the `S&P TSX 60 Returns` into a single DataFrame with columns for each portfolio's returns.\n\n \n\n### Conduct Quantitative Analysis\n\nAnalyze the data to see if any of the portfolios outperform the stock market (i.e., the S&P TSX 60).\n\n#### Performance Analysis\n\n1. Calculate and plot daily returns of all portfolios.\n\n2. Calculate and plot cumulative returns for all portfolios. Does any portfolio outperform the S&P TSX 60?\n\n#### Risk Analysis\n\n1. Create a box plot for each of the returns. \n\n2. Calculate the standard deviation for each portfolio. \n\n3. Determine which portfolios are riskier than the S&P TSX 60\n\n4. Calculate the Annualized Standard Deviation.\n\n#### Rolling Statistics\n\n1. Calculate and plot the rolling standard deviation for all portfolios using a 21-day window.\n\n2. Calculate and plot the correlation between each stock to determine which portfolios may mimick the S&P TSX 60.\n\n3. Choose one portfolio, then calculate and plot the 60-day rolling beta for it and the S&P TSX 60.\n\n#### Rolling Statistics Challenge: Exponentially Weighted Average\n\nAn alternative method to calculate a rolling window is to take the exponentially weighted moving average. This is like a moving window average, but it assigns greater importance to more recent observations. Try calculating the [`ewm`](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.ewm.html) with a 21-day half-life.\n\n### Sharpe Ratios\n\nInvestment managers and their institutional investors look at the return-to-risk ratio, not just the returns. After all, if you have two portfolios that each offer a 10% return, yet one is lower risk, you would invest in the lower-risk portfolio, right?\n\n1. Using the daily returns, calculate and visualize the Sharpe ratios using a bar plot.\n\n2. Determine whether the algorithmic strategies outperform both the market (S&P TSX 60) and the whales portfolios.\n\n### Create a Custom Portfolio\n\nHarold is ecstatic that you were able to help him prove that the algorithmic trading portfolios are doing so well compared to the market and whales portfolios. However, now you are wondering whether you can choose your own portfolio that performs just as well as the algorithmic portfolios. Investigate by doing the following:\n\n1. Visit [Google Sheets](https://docs.google.com/spreadsheets/) and use the built-in Google Finance function to choose 3-5 stocks for your portfolio.\n\n2. Download the data as CSV files and calculate the portfolio returns.\n\n3. Calculate the weighted returns for your portfolio, assuming equal number of shares per stock.\n\n4. Add your portfolio returns to the DataFrame with the other portfolios.\n\n5. Run the following analyses:\n\n * Calculate the Annualized Standard Deviation.\n * Calculate and plot rolling `std` with a 21-day window.\n * Calculate and plot the correlation.\n * Calculate and plot the 60-day rolling beta for your portfolio compared to the S&P 60 TSX.\n * Calculate the Sharpe ratios and generate a bar plot.\n\n4. How does your portfolio do?\n\n---\n\n## Resources\n\n* [Pandas API Docs](https://pandas.pydata.org/pandas-docs/stable/reference/index.html)\n\n* [Exponential weighted function in Pandas](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.ewm.html)\n\n* [`GOOGLEFINANCE` function help](https://support.google.com/docs/answer/3093281)\n\n* [Supplemental Guide: Fetching Stock Data Using Google Sheets](../../../01-Lesson-Plans/04-Pandas/Supplemental/googlefinance_guide.md)\n\n---\n\n## Hints\n\n* After reading each CSV file, don't forget to sort each DataFrame in ascending order by the Date using `sort_index`. This is especially important when working with time series data, as we want to make sure Date indexes go from earliest to latest.\n\n* The Pandas functions used in class this week will be useful for this assignment.\n\n* Be sure to use `head()` or `tail()` when you want to look at your data, but don't want to print to a large DataFrame.\n\n---\n\n## Submission\n\n1. Use the provided starter Jupyter Notebook to house the code for your data preparation, analysis, and visualizations. Put any analysis or answers to assignment questions in raw text (markdown) cells in the report.\n\n2. Submit your notebook to a new GitHub repository.\n\n3. Add the URL of your GitHub repository to your assignment when submitting via Bootcamp Spot.\n\n---\n\n© 2020 Trilogy Education Services, a 2U, Inc. brand. All Rights Reserved.\n\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"raw"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
cb718edfed91d1b9e5e6305b02d415aaf03e35d7 | 43,904 | ipynb | Jupyter Notebook | New Jupyter Notebooks/.ipynb_checkpoints/DecisionTreesWithIris-checkpoint.ipynb | FlyN-Nick/introToML | a61c330f97755fdc446171c4a39db3d3ff5c879a | [
"MIT"
]
| 3 | 2020-10-21T17:45:20.000Z | 2021-06-05T10:38:45.000Z | New Jupyter Notebooks/Classification/Iris Dataset/Decision Trees/.ipynb_checkpoints/DecisionTreesWithIris-checkpoint.ipynb | FlyN-Nick/introToML | a61c330f97755fdc446171c4a39db3d3ff5c879a | [
"MIT"
]
| null | null | null | New Jupyter Notebooks/Classification/Iris Dataset/Decision Trees/.ipynb_checkpoints/DecisionTreesWithIris-checkpoint.ipynb | FlyN-Nick/introToML | a61c330f97755fdc446171c4a39db3d3ff5c879a | [
"MIT"
]
| 1 | 2020-11-10T03:33:23.000Z | 2020-11-10T03:33:23.000Z | 59.010753 | 19,052 | 0.662035 | [
[
[
"from sklearn.datasets import load_iris # iris dataset\nfrom sklearn import tree # for fitting model\n\n# for the particular visualization used\nfrom six import StringIO\nimport pydot\nimport os.path\n\n# to display graphs \n%matplotlib inline\nimport matplotlib.pyplot",
"_____no_output_____"
],
[
"# get dataset \niris = load_iris()\niris.keys()",
"_____no_output_____"
],
[
"import pandas\niris_df = pandas.DataFrame(iris.data)\niris_df.columns = iris.feature_names\niris_df['target'] = [iris.target_names[target] for target in iris.target]\niris_df.head()",
"_____no_output_____"
],
[
"iris_df.describe()",
"_____no_output_____"
],
[
"print(iris_df)",
" sepal length (cm) sepal width (cm) petal length (cm) petal width (cm) \\\n0 5.1 3.5 1.4 0.2 \n1 4.9 3.0 1.4 0.2 \n2 4.7 3.2 1.3 0.2 \n3 4.6 3.1 1.5 0.2 \n4 5.0 3.6 1.4 0.2 \n.. ... ... ... ... \n145 6.7 3.0 5.2 2.3 \n146 6.3 2.5 5.0 1.9 \n147 6.5 3.0 5.2 2.0 \n148 6.2 3.4 5.4 2.3 \n149 5.9 3.0 5.1 1.8 \n\n target \n0 setosa \n1 setosa \n2 setosa \n3 setosa \n4 setosa \n.. ... \n145 virginica \n146 virginica \n147 virginica \n148 virginica \n149 virginica \n\n[150 rows x 5 columns]\n"
],
[
"# choose two features to plot\nx_feature = 0\ny_feature = 3\n\n#x = list(list(zip(*iris.data))[x_feature])\n#y = list(list(zip(*iris.data))[y_feature])\nx = iris.data[:, x_feature]\ny = iris.data[:, y_feature]\n\n# The data are in order by type (types of irises). Find out the border indexes of the types. \nend_type_one = list(iris.target).index(1)\nend_type_two = list(iris.target).index(2)\n\nfig = matplotlib.pyplot.figure() # create graph\nfig.suptitle('Two Features of the Iris Data Set') # set title \n# set axis labels \nmatplotlib.pyplot.xlabel(iris.feature_names[x_feature])\nmatplotlib.pyplot.ylabel(iris.feature_names[y_feature])\n\n# put the input data on the graph, with different colors and shapes for each type\nscatter_0 = matplotlib.pyplot.scatter(x[:end_type_one], y[:end_type_one],\n c=\"red\", marker=\"o\", label=iris.target_names[0])\nscatter_1 = matplotlib.pyplot.scatter(x[end_type_one:end_type_two], y[end_type_one:end_type_two],\n c=\"blue\", marker=\"^\", label=iris.target_names[1])\nscatter_2 = matplotlib.pyplot.scatter(x[end_type_two:], y[end_type_two:],\n c=\"green\", marker=\"*\", label=iris.target_names[2])\n\nmatplotlib.pyplot.legend(handles=[scatter_0, scatter_1, scatter_2]) # make legend\n\nmatplotlib.pyplot.show() # show the graph ",
"_____no_output_____"
],
[
"print(iris.data)\nprint(x)",
"[[5.1 3.5 1.4 0.2]\n [4.9 3. 1.4 0.2]\n [4.7 3.2 1.3 0.2]\n [4.6 3.1 1.5 0.2]\n [5. 3.6 1.4 0.2]\n [5.4 3.9 1.7 0.4]\n [4.6 3.4 1.4 0.3]\n [5. 3.4 1.5 0.2]\n [4.4 2.9 1.4 0.2]\n [4.9 3.1 1.5 0.1]\n [5.4 3.7 1.5 0.2]\n [4.8 3.4 1.6 0.2]\n [4.8 3. 1.4 0.1]\n [4.3 3. 1.1 0.1]\n [5.8 4. 1.2 0.2]\n [5.7 4.4 1.5 0.4]\n [5.4 3.9 1.3 0.4]\n [5.1 3.5 1.4 0.3]\n [5.7 3.8 1.7 0.3]\n [5.1 3.8 1.5 0.3]\n [5.4 3.4 1.7 0.2]\n [5.1 3.7 1.5 0.4]\n [4.6 3.6 1. 0.2]\n [5.1 3.3 1.7 0.5]\n [4.8 3.4 1.9 0.2]\n [5. 3. 1.6 0.2]\n [5. 3.4 1.6 0.4]\n [5.2 3.5 1.5 0.2]\n [5.2 3.4 1.4 0.2]\n [4.7 3.2 1.6 0.2]\n [4.8 3.1 1.6 0.2]\n [5.4 3.4 1.5 0.4]\n [5.2 4.1 1.5 0.1]\n [5.5 4.2 1.4 0.2]\n [4.9 3.1 1.5 0.2]\n [5. 3.2 1.2 0.2]\n [5.5 3.5 1.3 0.2]\n [4.9 3.6 1.4 0.1]\n [4.4 3. 1.3 0.2]\n [5.1 3.4 1.5 0.2]\n [5. 3.5 1.3 0.3]\n [4.5 2.3 1.3 0.3]\n [4.4 3.2 1.3 0.2]\n [5. 3.5 1.6 0.6]\n [5.1 3.8 1.9 0.4]\n [4.8 3. 1.4 0.3]\n [5.1 3.8 1.6 0.2]\n [4.6 3.2 1.4 0.2]\n [5.3 3.7 1.5 0.2]\n [5. 3.3 1.4 0.2]\n [7. 3.2 4.7 1.4]\n [6.4 3.2 4.5 1.5]\n [6.9 3.1 4.9 1.5]\n [5.5 2.3 4. 1.3]\n [6.5 2.8 4.6 1.5]\n [5.7 2.8 4.5 1.3]\n [6.3 3.3 4.7 1.6]\n [4.9 2.4 3.3 1. ]\n [6.6 2.9 4.6 1.3]\n [5.2 2.7 3.9 1.4]\n [5. 2. 3.5 1. ]\n [5.9 3. 4.2 1.5]\n [6. 2.2 4. 1. ]\n [6.1 2.9 4.7 1.4]\n [5.6 2.9 3.6 1.3]\n [6.7 3.1 4.4 1.4]\n [5.6 3. 4.5 1.5]\n [5.8 2.7 4.1 1. ]\n [6.2 2.2 4.5 1.5]\n [5.6 2.5 3.9 1.1]\n [5.9 3.2 4.8 1.8]\n [6.1 2.8 4. 1.3]\n [6.3 2.5 4.9 1.5]\n [6.1 2.8 4.7 1.2]\n [6.4 2.9 4.3 1.3]\n [6.6 3. 4.4 1.4]\n [6.8 2.8 4.8 1.4]\n [6.7 3. 5. 1.7]\n [6. 2.9 4.5 1.5]\n [5.7 2.6 3.5 1. ]\n [5.5 2.4 3.8 1.1]\n [5.5 2.4 3.7 1. ]\n [5.8 2.7 3.9 1.2]\n [6. 2.7 5.1 1.6]\n [5.4 3. 4.5 1.5]\n [6. 3.4 4.5 1.6]\n [6.7 3.1 4.7 1.5]\n [6.3 2.3 4.4 1.3]\n [5.6 3. 4.1 1.3]\n [5.5 2.5 4. 1.3]\n [5.5 2.6 4.4 1.2]\n [6.1 3. 4.6 1.4]\n [5.8 2.6 4. 1.2]\n [5. 2.3 3.3 1. ]\n [5.6 2.7 4.2 1.3]\n [5.7 3. 4.2 1.2]\n [5.7 2.9 4.2 1.3]\n [6.2 2.9 4.3 1.3]\n [5.1 2.5 3. 1.1]\n [5.7 2.8 4.1 1.3]\n [6.3 3.3 6. 2.5]\n [5.8 2.7 5.1 1.9]\n [7.1 3. 5.9 2.1]\n [6.3 2.9 5.6 1.8]\n [6.5 3. 5.8 2.2]\n [7.6 3. 6.6 2.1]\n [4.9 2.5 4.5 1.7]\n [7.3 2.9 6.3 1.8]\n [6.7 2.5 5.8 1.8]\n [7.2 3.6 6.1 2.5]\n [6.5 3.2 5.1 2. ]\n [6.4 2.7 5.3 1.9]\n [6.8 3. 5.5 2.1]\n [5.7 2.5 5. 2. ]\n [5.8 2.8 5.1 2.4]\n [6.4 3.2 5.3 2.3]\n [6.5 3. 5.5 1.8]\n [7.7 3.8 6.7 2.2]\n [7.7 2.6 6.9 2.3]\n [6. 2.2 5. 1.5]\n [6.9 3.2 5.7 2.3]\n [5.6 2.8 4.9 2. ]\n [7.7 2.8 6.7 2. ]\n [6.3 2.7 4.9 1.8]\n [6.7 3.3 5.7 2.1]\n [7.2 3.2 6. 1.8]\n [6.2 2.8 4.8 1.8]\n [6.1 3. 4.9 1.8]\n [6.4 2.8 5.6 2.1]\n [7.2 3. 5.8 1.6]\n [7.4 2.8 6.1 1.9]\n [7.9 3.8 6.4 2. ]\n [6.4 2.8 5.6 2.2]\n [6.3 2.8 5.1 1.5]\n [6.1 2.6 5.6 1.4]\n [7.7 3. 6.1 2.3]\n [6.3 3.4 5.6 2.4]\n [6.4 3.1 5.5 1.8]\n [6. 3. 4.8 1.8]\n [6.9 3.1 5.4 2.1]\n [6.7 3.1 5.6 2.4]\n [6.9 3.1 5.1 2.3]\n [5.8 2.7 5.1 1.9]\n [6.8 3.2 5.9 2.3]\n [6.7 3.3 5.7 2.5]\n [6.7 3. 5.2 2.3]\n [6.3 2.5 5. 1.9]\n [6.5 3. 5.2 2. ]\n [6.2 3.4 5.4 2.3]\n [5.9 3. 5.1 1.8]]\n[5.1 4.9 4.7 4.6 5. 5.4 4.6 5. 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1\n 5.7 5.1 5.4 5.1 4.6 5.1 4.8 5. 5. 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.\n 5.5 4.9 4.4 5.1 5. 4.5 4.4 5. 5.1 4.8 5.1 4.6 5.3 5. 7. 6.4 6.9 5.5\n 6.5 5.7 6.3 4.9 6.6 5.2 5. 5.9 6. 6.1 5.6 6.7 5.6 5.8 6.2 5.6 5.9 6.1\n 6.3 6.1 6.4 6.6 6.8 6.7 6. 5.7 5.5 5.5 5.8 6. 5.4 6. 6.7 6.3 5.6 5.5\n 5.5 6.1 5.8 5. 5.6 5.7 5.7 6.2 5.1 5.7 6.3 5.8 7.1 6.3 6.5 7.6 4.9 7.3\n 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7 7.7 6. 6.9 5.6 7.7 6.3 6.7 7.2\n 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4 6. 6.9 6.7 6.9 5.8 6.8\n 6.7 6.7 6.3 6.5 6.2 5.9]\n"
],
[
"decision_tree = tree.DecisionTreeClassifier() # make model \ndecision_tree.fit(iris.data, iris.target) # fit model to data ",
"_____no_output_____"
],
[
"# make pdf diagram of decision tree\ndot_data = StringIO()\ntree.export_graphviz(decision_tree, out_file=dot_data, feature_names=iris.feature_names, class_names=iris.target_names,\n filled=True, rounded=True, special_characters=True)\ngraph = pydot.graph_from_dot_data(dot_data.getvalue())[0]\ngraph.write_pdf(os.path.expanduser(\"~/Desktop/introToML/ML/New Jupyter Notebooks/iris_decision_tree_regular.pdf\"))",
"_____no_output_____"
],
[
"inputs = [iris.data[0], iris.data[end_type_one], iris.data[end_type_two]] # use the first input of each class \nprint('Class predictions: {0}'.format(list(iris.target_names[prediction] for prediction in decision_tree.predict(inputs)))) # print predictions\nprint('Probabilities:\\n{0}'.format(decision_tree.predict_proba(inputs))) # print prediction probabilities \n",
"Class predictions: ['setosa', 'versicolor', 'virginica']\nProbabilities:\n[[1. 0. 0.]\n [0. 1. 0.]\n [0. 0. 1.]]\n"
]
],
[
[
"# Exercise Option #1 - Standard Difficulty\n\n0. Submit the PDF you generated as a separate file in Canvas.\n1. According to the PDF, a petal width <= 0.8 cm would tell you with high (100%) probability that you are looking at a setosa iris. \n2. According to the PDF, you're supposed to look at the petal length, petal width, and sepal length to tell a virginica from a versicolor.\n3. The array value at each node in the pdf shows how many data values of each class passed through the node. \n4. The predictions are always have a 100% probability because any data value you give will end up at one end node. Each end node has one class prediction. \n5. Below I use a subset of the features (3/4). The new decision tree was completely different than the original: it had more nodes and a different overall shape. When looking at the original decision tree, most of the nodes separated data based on petal length or petal width. The one feature that the new tree does not use is petal width, which is the most likely cause for why the second tree had to use more nodes (it lacked a feature that would make it easy to distinguish the classes). ",
"_____no_output_____"
]
],
[
[
"# Use 3/4 columns (the first, second, & third)\nfirst_feature = 0\nsecond_feature = 1\nthird_feature = 2\niris_inputs = iris.data[:,[first_feature, second_feature, third_feature]] # use only two collumns of the data \n\ndecision_tree_with_portion = tree.DecisionTreeClassifier() # make model \ndecision_tree_with_portion.fit(iris_inputs, iris.target) # fit model to data \n\n# make pdf diagram of decision tree\ndot_data = StringIO()\ntree.export_graphviz(decision_tree_with_portion, out_file=dot_data, feature_names=iris.feature_names[:3], class_names=iris.target_names,\n filled=True, rounded=True, special_characters=True)\ngraph = pydot.graph_from_dot_data(dot_data.getvalue())[0]\ngraph.write_pdf(os.path.expanduser(\"~/Desktop/introToML/ML/New Jupyter Notebooks/iris_decision_tree_with_portion.pdf\"))\n\nnew_inputs = [iris_inputs[0], iris_inputs[end_type_one], iris_inputs[end_type_two]] # make new inputs with iris_inputs, which only has two features per input\nprint('Class predictions: {0}'.format(list(iris.target_names[prediction] for prediction in decision_tree_with_portion.predict(new_inputs)))) # print predictions\nprint('Probabilities:\\n{0}'.format(decision_tree_with_portion.predict_proba(new_inputs))) # print prediction probabilities ",
"Class predictions: ['setosa', 'versicolor', 'virginica']\nProbabilities:\n[[1. 0. 0.]\n [0. 1. 0.]\n [0. 0. 1.]]\n"
]
],
[
[
"# Exercise Option #2 - Advanced Difficulty\nTry fitting a Random Forest model to the iris data. See [this example](http://scikit-learn.org/stable/modules/ensemble.html#forest).\n\nAs seen below, the random forest & decision tree had the same F1 score (a perfect 1.0), meaning that they performed the same. ",
"_____no_output_____"
]
],
[
[
"# https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html?highlight=random%20forest#sklearn.ensemble.RandomForestClassifier\nfrom sklearn.ensemble import RandomForestClassifier \nrand_forst = RandomForestClassifier() # make model \nrand_forst = rand_forst.fit(iris.data, iris.target) # fit model \nprint('Class predictions: {0}'.format(list(iris.target_names[prediction] for prediction in rand_forst.predict(inputs)))) # print class predictions\n",
"Class predictions: ['setosa', 'versicolor', 'virginica']\n"
],
[
"# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html?highlight=f1#sklearn.metrics.f1_score\nfrom sklearn.metrics import f1_score\n\n# get predictions for whole dataset\ndecision_tree_predictions = decision_tree.predict(iris.data) \nrand_forst_predictions = rand_forst.predict(iris.data)\n\n# print F1 scores\nprint ('Decision tree F1 score: {}'.format(f1_score(iris.target, decision_tree_predictions, average='weighted')))\nprint ('Random forest F1 score: {}'.format(f1_score(iris.target, rand_forst_predictions, average='weighted')))",
"Decision tree F1 score: 1.0\nRandom forest F1 score: 1.0\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
cb7196bc85a4995bf5bb1a30fcd726f200a14a61 | 62,970 | ipynb | Jupyter Notebook | DesafioDSA/Missao5/missao5.ipynb | DaniloGR91/Python_Fundamentos_DataScienceAcademy | 56ef0f3ca7feca901ad17d8c450db4d4b7fafe15 | [
"MIT"
]
| null | null | null | DesafioDSA/Missao5/missao5.ipynb | DaniloGR91/Python_Fundamentos_DataScienceAcademy | 56ef0f3ca7feca901ad17d8c450db4d4b7fafe15 | [
"MIT"
]
| 1 | 2020-06-12T22:22:06.000Z | 2020-06-12T22:22:06.000Z | DesafioDSA/Missao5/missao5.ipynb | DaniloGR91/Python_Fundamentos_DataScienceAcademy | 56ef0f3ca7feca901ad17d8c450db4d4b7fafe15 | [
"MIT"
]
| null | null | null | 66.918172 | 3,818 | 0.489963 | [
[
[
"# <font color='blue'>Data Science Academy - Python Fundamentos - Capítulo 7</font>\n\n## Download: http://github.com/dsacademybr",
"_____no_output_____"
]
],
[
[
"# Versão da Linguagem Python\nfrom platform import python_version\nprint('Versão da Linguagem Python Usada Neste Jupyter Notebook:', python_version())",
"Versão da Linguagem Python Usada Neste Jupyter Notebook: 3.7.6\n"
]
],
[
[
"## Missão: Analisar o Comportamento de Compra de Consumidores.",
"_____no_output_____"
],
[
"## Nível de Dificuldade: Alto",
"_____no_output_____"
],
[
"Você recebeu a tarefa de analisar os dados de compras de um web site! Os dados estão no formato JSON e disponíveis junto com este notebook.\n\nNo site, cada usuário efetua login usando sua conta pessoal e pode adquirir produtos à medida que navega pela lista de produtos oferecidos. Cada produto possui um valor de venda. Dados de idade e sexo de cada usuário foram coletados e estão fornecidos no arquivo JSON.\n\nSeu trabalho é entregar uma análise de comportamento de compra dos consumidores. Esse é um tipo de atividade comum realizado por Cientistas de Dados e o resultado deste trabalho pode ser usado, por exemplo, para alimentar um modelo de Machine Learning e fazer previsões sobre comportamentos futuros.\n\nMas nesta missão você vai analisar o comportamento de compra dos consumidores usando o pacote Pandas da linguagem Python e seu relatório final deve incluir cada um dos seguintes itens:\n\n** Contagem de Consumidores **\n\n* Número total de consumidores\n\n\n** Análise Geral de Compras **\n\n* Número de itens exclusivos\n* Preço médio de compra\n* Número total de compras\n* Rendimento total\n\n\n** Informações Demográficas Por Gênero **\n\n* Porcentagem e contagem de compradores masculinos\n* Porcentagem e contagem de compradores do sexo feminino\n* Porcentagem e contagem de outros / não divulgados\n\n\n** Análise de Compras Por Gênero **\n\n* Número de compras\n* Preço médio de compra\n* Valor Total de Compra\n* Compras for faixa etária\n\n\n** Identifique os 5 principais compradores pelo valor total de compra e, em seguida, liste (em uma tabela): **\n\n* Login\n* Número de compras\n* Preço médio de compra\n* Valor Total de Compra\n* Itens mais populares\n\n\n** Identifique os 5 itens mais populares por contagem de compras e, em seguida, liste (em uma tabela): **\n\n* ID do item\n* Nome do item\n* Número de compras\n* Preço do item\n* Valor Total de Compra\n* Itens mais lucrativos\n\n\n** Identifique os 5 itens mais lucrativos pelo valor total de compra e, em seguida, liste (em uma tabela): **\n\n* ID do item\n* Nome do item\n* Número de compras\n* Preço do item\n* Valor Total de Compra\n\n\n** Como considerações finais: **\n\n* Seu script deve funcionar para o conjunto de dados fornecido.\n* Você deve usar a Biblioteca Pandas e o Jupyter Notebook.\n",
"_____no_output_____"
]
],
[
[
"# Imports\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Carrega o arquivo\nload_file = \"dados_compras.json\"\ndf = pd.read_json(load_file, orient = \"records\")\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Informações Sobre os Consumidores",
"_____no_output_____"
]
],
[
[
"df",
"_____no_output_____"
],
[
"len(df['Login'].unique())",
"_____no_output_____"
]
],
[
[
"### O WebSite teve um total de 573 logins diferentes realizando compras",
"_____no_output_____"
],
[
"## Análise Geral de Compras\n- Número de itens exclusivos\n- Preço médio de compra\n- Número total de compras\n- Rendimento total",
"_____no_output_____"
],
[
"### Os seguintes itens foram vendidos",
"_____no_output_____"
]
],
[
[
"for prod in df['Nome do Item'].unique():\n print(prod)",
"Bone Crushing Silver Skewer\nStormbringer, Dark Blade of Ending Misery\nPrimitive Blade\nFinal Critic\nStormfury Mace\nSleepwalker\nMercenary Sabre\nInterrogator, Blood Blade of the Queen\nGhost Reaver, Longsword of Magic\nExpiration, Warscythe Of Lost Worlds\nDespair, Favor of Due Diligence\nAlpha, Reach of Ending Hope\nDreamkiss\nPiety, Guardian of Riddles\nBonecarvin Battle Axe\nBlood-Forged Skeletal Spine\nTwilight's Carver\nLightning, Etcher of the King\nCeleste\nWinterthorn, Defender of Shifting Worlds\nGlimmer, Ender of the Moon\nPhantomlight\nBrimstone\nDragon's Greatsword\nConqueror Adamantite Mace\nPersuasion\nCrying Steel Sickle\nThe Oculus, Token of Lost Worlds\nGlinting Glass Edge\nWar-Forged Gold Deflector\nShadow Strike, Glory of Ending Hope\nRage, Legacy of the Lone Victor\nBetrayer\nSerenity\nScalpel\nHero Cane\nThorn, Satchel of Dark Souls\nVenom Claymore\nStorm-Weaver, Slayer of Inception\nMalificent Bag\nAgatha\nLazarus, Terror of the Earth\nRitual Mace\nSeverance\nOrenmir\nEndbringer\nStriker\nTrickster\nRetribution Axe\nVerdict\nStormfury Lantern\nBetrayal, Whisper of Grieving Widows\nFlux, Destroyer of Due Diligence\nAzurewrath\nUndead Crusader\nLifebender\nThe Void, Vengeance of Dark Magic\nDarkheart, Butcher of the Champion\nSuspension\nUnending Tyranny\nEternal Cleaver\nHailstorm Shadowsteel Scythe\nDeluge, Edge of the West\nCrucifer\nBlood Infused Guardian\nBlazeguard, Reach of Eternity\nWoeful Adamantite Claymore\nHopeless Ebon Dualblade\nDeathraze\nDarkheart\nExtraction, Quickblade Of Trembling Hands\nBlade of the Grave\nMalice, Legacy of the Queen\nReaper's Toll\nNetherbane\nAlpha\nMisery's End\nSinged Scalpel\nStormcaller\nMercy, Katana of Dismay\nStormfury Longsword\nVindictive Glass Edge\nSpectral Diamond Doomblade\nHeartless Bone Dualblade\nThe Decapitator\nThorn, Conqueror of the Corrupted\nArcane Gem\nAvenger\nVengeance Cleaver\nTorchlight, Bond of Storms\nYearning Crusher\nExiled Mithril Longsword\nDawn\nPiece Maker\nWhistling Mithril Warblade\nMassacre\nFate, Vengeance of Eternal Justice\nTranquility, Razor of Black Magic\nDawne\nDemise\nPossessed Core\nFreak's Bite, Favor of Holy Might\nRiddle, Tribute of Ended Dreams\nHatred\nVictor Iron Spikes\nBlindscythe\nWorldbreaker\nWarped Fetish\nMourning Blade\nSplitter, Foe Of Subtlety\nSplinter\nToothpick\nSwan Song, Gouger Of Terror\nHope's End\nBlazefury, Protector of Delusions\nShadowsteel\nApocalyptic Battlescythe\nWolf, Promise of the Moonwalker\nSpectral Bone Axe\nBrutality Ivory Warmace\nFrenzied Scimitar\nAetherius, Boon of the Blessed\nRelentless Iron Skewer\nWarmonger, Gift of Suffering's End\nHeartseeker, Reaver of Souls\nHellreaver, Heirloom of Inception\nDevine\nFeral Katana\nSpada, Etcher of Hatred\nRenewed Skeletal Katana\nRagnarok\nFury\nOrbit\nFiery Glass Crusader\nWolf\nAbyssal Shard\nWarped Diamond Crusader\nPurgatory, Gem of Regret\nSecond Chance\nSoul Infused Crystal\nChaos, Ender of the End\nWinter's Bite\nYearning Mageblade\nHeartstriker, Legacy of the Light\nAlpha, Oath of Zeal\nEmberling, Defender of Delusions\nGladiator's Glaive\nSinged Onyx Warscythe\nFusion Pummel\nDeadline, Voice Of Subtlety\nCurved Axe\nHaunted Bronzed Bludgeon\nWarped Iron Scimitar\nThirsty Iron Reaver\nFoul Titanium Battle Axe\nAmnesia\nSouleater\nRestored Bauble\nCeleste, Incarnation of the Corrupted\nFaith's Scimitar\nFrenzy, Defender of the Harvest\nOathbreaker, Spellblade of Trials\nNirvana\nSolitude's Reaver\nExiled Doomblade\nRusty Skull\nThunderfury Scimitar\nPutrid Fan\nPursuit, Cudgel of Necromancy\nSun Strike, Jaws of Twisted Visions\nGhastly Adamantite Protector\nDreamsong\nUnholy Wand\nRighteous Might\nOathbreaker, Last Hope of the Breaking Storm\nSoul-Forged Steel Shortsword\nDownfall, Scalpel Of The Emperor\nFoul Edge\nBloodlord's Fetish\n"
]
],
[
[
"### O número total de compras foi 780, cada uma tendo valor médio de 2.93",
"_____no_output_____"
]
],
[
[
"df.describe()",
"_____no_output_____"
],
[
"df['Valor'].sum()",
"_____no_output_____"
]
],
[
[
"### Totalizando uma renda total de 2286,33",
"_____no_output_____"
],
[
"## Análise Demográfica",
"_____no_output_____"
],
[
"## Informações Demográficas Por Gênero\n- Porcentagem e contagem de compradores masculinos\n- Porcentagem e contagem de compradores do sexo feminino\n- Porcentagem e contagem de outros / não divulgados",
"_____no_output_____"
]
],
[
[
"dfLoginSex = df[['Login', 'Sexo']].drop_duplicates()\ndfLoginSex['Sexo'].value_counts()\ntotal = 465 + 100 + 8\nMasc = 465\npercMasc = 100*Masc / total\nFem = 100\npercFem = 100 * Fem / total\noutro = 8\npercOutro = 100 * outro / total",
"_____no_output_____"
],
[
"print(f'Foram um total de {total} consumidores, sendo: \\n \\\n- {Masc} do sexo Masculino ({percMasc:.2f}%) \\n \\\n- {Fem} do sexo Feminino ({percFem:.2f}%) \\n \\\n- {outro} do sexo outro/não divulgado ({percOutro:.2f}%)')",
"Foram um total de 573 consumidores, sendo: \n - 465 do sexo Masculino (81.15%) \n - 100 do sexo Feminino (17.45%) \n - 8 do sexo outro/não divulgado (1.40%)\n"
]
],
[
[
"## Análise de Compras Por Gênero\n- Número de compras\n- Preço médio de compra\n- Valor Total de Compra\n- Compras for faixa etária",
"_____no_output_____"
]
],
[
[
"df['FaixaEtaria'] = ''\nfor i, v in enumerate(df['Idade']):\n if v < 18:\n df['FaixaEtaria'][i] = 'Até 18 anos'\n elif 18 <= v <= 25:\n df['FaixaEtaria'][i] = '18 - 25 anos'\n elif 25 < v <= 40:\n df['FaixaEtaria'][i] = '26 - 40 anos'\n elif 40 < v <= 60:\n df['FaixaEtaria'][i] = '41 - 60 anos'\n elif v > 60:\n df['FaixaEtaria'][i] = 'Mais de 60 anos'",
"_____no_output_____"
],
[
"df_sexGroup = df[['Idade', 'Valor', 'FaixaEtaria']].groupby(by = df['Sexo'])\ndf_sexGroup.describe()",
"_____no_output_____"
],
[
"df_sexGroup['Valor'].sum()",
"_____no_output_____"
],
[
"df_sexGroup['FaixaEtaria'].value_counts()",
"_____no_output_____"
],
[
"df_FaixaEtariaGroup = df[['Idade', 'Valor', 'Sexo']].groupby(by=df['FaixaEtaria'])",
"_____no_output_____"
],
[
"sexo = df['Sexo'].unique()\ndf_FaixaMasc = df[['Idade', 'Valor', 'Sexo']].query(f\"Sexo == '{sexo[0]}'\").groupby(by=df['FaixaEtaria'])\ndf_FaixaFem = df[['Idade', 'Valor', 'Sexo']].query(f\"Sexo == '{sexo[1]}'\").groupby(by=df['FaixaEtaria'])\ndf_FaixaOutro = df[['Idade', 'Valor', 'Sexo']].query(f\"Sexo == '{sexo[2]}'\").groupby(by=df['FaixaEtaria'])",
"_____no_output_____"
],
[
"df_FaixaMasc.describe().round(2)",
"_____no_output_____"
],
[
"df_FaixaFem.describe().round(2)",
"_____no_output_____"
],
[
"df_FaixaOutro.describe().round(2)",
"_____no_output_____"
]
],
[
[
"### Total de compras:\n- Feminno: 136\n- Masculino: 633\n- Outro / Não Divulgado: 11\n\n### Preço Médio de compras\n- Feminino: 2.81\n- Masculino: 2.95\n- Outro / Não Divulgado: 3.25\n\n### Valor Total\n- Feminino: 382.91\n- Masculino: 1867.68\n- Outro / Não Divulgado: 35.74\n\n### Perfil de comprar por faixa etária (Média do valor)\nFeminino\n- Até 18 anos: 2.83\n- 18 - 25 anos: 2.80\n- 26 - 40 anos: 2.83\n- 41 - 60 anos: Nenhum\n- Mais de 60 anos: Nenhum\n\nMasculino\n- Até 18 anos: 2.89\n- 18 - 25 anos: 2.96\n- 26 - 40 anos: 2.97\n- 41 - 60 anos: 2.88\n- Mais de 60 anos: Nenhum\n\nOutro / Não Divulgado\n- Até 18 anos: 4.00\n- 18 - 25 anos: 3.30\n- 26 - 40 anos: 3.15\n- 41 - 60 anos: Nenhum\n- Mais de 60 anos: Nenhum",
"_____no_output_____"
],
[
"## Consumidores Mais Populares (Top 5)\n- Login\n- Número de compras\n- Preço médio de compra\n- Valor Total de Compra\n- Itens mais populares",
"_____no_output_____"
],
[
"### Top 5 - Login com número de compras",
"_____no_output_____"
]
],
[
[
"df['Login'].value_counts().sort_values(ascending=False).head(5)",
"_____no_output_____"
],
[
"top5Consumers = []\nfor k, v in df['Login'].value_counts().sort_values(ascending=False).head().items():\n top5Consumers.append(k)\n\nprint('Valor médio de compra dos 5 principais consumidores (com base no número de compras)')\ndf[['Login','Valor']].query(f'Login in {top5Consumers}').sort_values(by='Login').groupby(by = 'Login').mean()",
"Valor médio de compra dos 5 principais consumidores (com base no número de compras)\n"
],
[
"print('Valor total de compra dos 5 principais consumidores (com base no número de compras)')\n\ndf[['Login', 'Valor']].query(f'Login in {top5Consumers}').sort_values(by='Login').groupby(by='Login').sum()",
"Valor total de compra dos 5 principais consumidores (com base no número de compras)\n"
],
[
"print('Os 5 produtos mais comprados')\ndf['Nome do Item'].value_counts().sort_values(ascending=False).head()",
"Os 5 produtos mais comprados\n"
]
],
[
[
"## Itens Mais Populares\n- ID do item\n- Nome do item\n- Número de compras\n- Preço do item\n- Valor Total de Compra\n- Itens mais lucrativos",
"_____no_output_____"
]
],
[
[
"df = df[['Login', 'Idade', 'Sexo', 'Item ID', 'Item', 'Valor', 'FaixaEtaria']]\n",
"_____no_output_____"
],
[
"itensMaisPopulares = []\nfor k, v in df['Item'].value_counts().sort_values(ascending=False).head().items():\n itensMaisPopulares.append(k)\n\ndfTopItens = df.query(f'Item in {itensMaisPopulares}').sort_values(by = 'Item')\ndfTopItens[['Item ID', 'Item']].groupby(by = 'Item ID').describe()",
"_____no_output_____"
]
],
[
[
"- O DataFrame acima mostra os protudos mais comprados, com ItemID e a quantidade de vezes em que foram comprados.\n- Inclusive pode-se perceber um erro em que um produto (Final Critic) possui dois IDs diferentes.",
"_____no_output_____"
]
],
[
[
"dfTopItens[['Item', 'Valor']].groupby(by='Valor').describe()",
"_____no_output_____"
]
],
[
[
"- O DataFrame acima mostra os principais produtos com os valores.\n- Pode-se observar que também existem diferenças nos valores, os produtos StormCaller e Final Critic possuem dois valores diferentes",
"_____no_output_____"
]
],
[
[
"dfTopItens[['Item', 'Valor']].groupby(by='Item').sum()",
"_____no_output_____"
]
],
[
[
"- Com o DataFrame acima pode-se observar o valor total vendido dos 5 produtos mais vendidos",
"_____no_output_____"
]
],
[
[
"df[['Item', 'Valor']].groupby(by='Item').sum().sort_values(by = 'Valor', ascending=False).head()",
"_____no_output_____"
]
],
[
[
"## Itens Mais Lucrativos\n- ID do item\n- Nome do item\n- Número de compras\n- Preço do item\n- Valor Total de Compra",
"_____no_output_____"
]
],
[
[
"prodMaisLucrativos = []\nfor k,v in df[['Item', 'Valor']].groupby(by='Item').sum().sort_values(by = 'Valor', ascending=False).head().items():\n for prod in v.items():\n prodMaisLucrativos.append(prod[0])\n\ndf_prodMaisLucrativos = df.query(f'Item in {prodMaisLucrativos}')\ndf_prodMaisLucrativos[['Item ID', 'Item']].groupby(by='Item ID').describe()",
"_____no_output_____"
]
],
[
[
"- O DataFrame acima mostra os produtos mais lucrativos, com o número de vendas e o Item ID.\n- Observa-se mais uma vez os dois IDs do produto Final Critic",
"_____no_output_____"
]
],
[
[
"df_prodMaisLucrativos[['Item', 'Valor']].groupby(by='Valor').describe()",
"_____no_output_____"
]
],
[
[
"- O DataFrame acima mostra o valor dos produtos mais lucrativos\n- Observa-se mais uma vez os dois valores dos produtos Final Critic e Stormcaller",
"_____no_output_____"
]
],
[
[
"df_prodMaisLucrativos[['Item', 'Valor']].groupby(by='Item').sum().sort_values(by='Valor', ascending=False)",
"_____no_output_____"
]
],
[
[
"- A tabela acima mostra o valor total vendido dos produtos mais lucrativos.",
"_____no_output_____"
],
[
"## Fim",
"_____no_output_____"
],
[
"### Obrigado - Data Science Academy - <a href=\"http://facebook.com/dsacademybr\">facebook.com/dsacademybr</a>",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
cb71a4786ce4cc7e119dc951796315aa0759257b | 847,929 | ipynb | Jupyter Notebook | Modeling the Volatility of US Bond Yields/notebook.ipynb | veeralakrishna/DataCamp-Portofolio-Projects-R | e1d76d66f55c65129c02d7c13fee462229674299 | [
"MIT"
]
| 13 | 2020-08-09T07:38:56.000Z | 2022-02-19T22:58:10.000Z | Modeling the Volatility of US Bond Yields/notebook.ipynb | Anandlytics/Datacamp-Project-Solutions-R | 15ad9190440c67c98c001e2b7f3e8c6363140c6f | [
"MIT"
]
| null | null | null | Modeling the Volatility of US Bond Yields/notebook.ipynb | Anandlytics/Datacamp-Project-Solutions-R | 15ad9190440c67c98c001e2b7f3e8c6363140c6f | [
"MIT"
]
| 27 | 2020-06-05T15:49:06.000Z | 2022-02-25T20:31:31.000Z | 847,929 | 847,929 | 0.949748 | [
[
[
"## 1. Volatility changes over time\n<p>What is financial risk? </p>\n<p>Financial risk has many faces, and we measure it in many ways, but for now, let's agree that it is a measure of the possible loss on an investment. In financial markets, where we measure prices frequently, volatility (which is analogous to <em>standard deviation</em>) is an obvious choice to measure risk. But in real markets, volatility changes with the market itself. </p>\n<p><img src=\"https://assets.datacamp.com/production/project_738/img/VolaClusteringAssetClasses.png\" alt=\"\"></p>\n<p>In the picture above, we see the returns of four very different assets. All of them exhibit alternating regimes of low and high volatilities. The highest volatility is observed around the end of 2008 - the most severe period of the recent financial crisis.</p>\n<p>In this notebook, we will build a model to study the nature of volatility in the case of US government bond yields.</p>",
"_____no_output_____"
]
],
[
[
"# Load the packages\nlibrary(xts)\nlibrary(readr)\n\n# Load the data\nyc_raw <- read_csv(\"datasets/FED-SVENY.csv\")\n\n# Convert the data into xts format\nyc_all <- as.xts(x = yc_raw[, -1], order.by = yc_raw$Date)\n\n# Show only the tail of the 1st, 5th, 10th, 20th and 30th columns\nyc_all_tail <- tail(yc_all[,c(1,5,10, 20, 30)])\nyc_all_tail",
"Loading required package: zoo\n\nAttaching package: 'zoo'\n\nThe following objects are masked from 'package:base':\n\n as.Date, as.Date.numeric\n\nParsed with column specification:\ncols(\n .default = col_double(),\n Date = \u001b[34mcol_date(format = \"\")\u001b[39m\n)\nSee spec(...) for full column specifications.\n"
]
],
[
[
"## 2. Plotting the evolution of bond yields\n<p>In the output table of the previous task, we see the yields for some maturities.</p>\n<p>These data include the whole yield curve. The yield of a bond is the price of the money lent. The higher the yield, the more money you receive on your investment. The yield curve has many maturities; in this case, it ranges from 1 year to 30 years. Different maturities have different yields, but yields of neighboring maturities are relatively close to each other and also move together.</p>\n<p>Let's visualize the yields over time. We will see that the long yields (e.g. SVENY30) tend to be more stable in the long term, while the short yields (e.g. SVENY01) vary a lot. These movements are related to the monetary policy of the FED and economic cycles.</p>",
"_____no_output_____"
]
],
[
[
"library(viridis)\n\n# Define plot arguments\nyields <- yc_all\nplot.type <- \"single\"\nplot.palette <- viridis(n = 30)\nasset.names <- colnames(yc_all)\n\n# Plot the time series\nplot.zoo(x = yc_all, plot.type = plot.type, col = plot.palette)\n\n# Add the legend\nlegend(x = \"topleft\", legend = asset.names,\n col = plot.palette, cex = 0.45, lwd = 3)",
"Loading required package: viridisLite\n"
]
],
[
[
"## 3. Make the difference\n<p>In the output of the previous task, we see the level of bond yields for some maturities, but to understand how volatility evolves we have to examine the changes in the time series. Currently, we have yield levels; we need to calculate the changes in the yield levels. This is called \"differentiation\" in time series analysis. Differentiation has the added benefit of making a time series independent of time.</p>",
"_____no_output_____"
]
],
[
[
"# Differentiate the time series \nycc_all <- diff.xts(yc_all)\n\n# Show the tail of the 1st, 5th, 10th, 20th and 30th columns\nycc_all_tail <- tail(ycc_all[, c(1, 5, 10, 20, 30)])\nycc_all_tail",
"_____no_output_____"
]
],
[
[
"## 4. The US yields are no exceptions, but maturity matters\n<p>Now that we have a time series of the changes in US government yields let's examine it visually.</p>\n<p>By taking a look at the time series from the previous plots, we see hints that the returns following each other have some unique properties:</p>\n<ul>\n<li>The direction (positive or negative) of a return is mostly independent of the previous day's return. In other words, you don't know if the next day's return will be positive or negative just by looking at the time series.</li>\n<li>The magnitude of the return is similar to the previous day's return. That means, if markets are calm today, we expect the same tomorrow. However, in a volatile market (crisis), you should expect a similarly turbulent tomorrow.</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"# Define the plot parameters\nyield.changes <- ycc_all\nplot.type <- \"multiple\"\n\n\n# Plot the differentiated time series\nplot.zoo(x = yield.changes, plot.type = plot.type, \n ylim = c(-0.5, 0.5), cex.axis = 0.7, \n ylab = 1:30, col = plot.palette)",
"_____no_output_____"
]
],
[
[
"## 5. Let's dive into some statistics\n<p>The statistical properties visualized earlier can be measured by analytical tools. The simplest method is to test for autocorrelation. Autocorrelation measures how a datapoint's past determines the future of a time series. </p>\n<ul>\n<li>If the autocorrelation is close to 1, the next day's value will be very close to today's value. </li>\n<li>If the autocorrelation is close to 0, the next day's value will be unaffected by today's value.</li>\n</ul>\n<p>Because we are interested in the recent evolution of bond yields, we will filter the time series for data from 2000 onward.</p>",
"_____no_output_____"
]
],
[
[
"# Filter for changes in and after 2000\nycc <- ycc_all[\"2000/\",]\n\n# Save the 1-year and 20-year maturity yield changes into separate variables\nx_1 <- ycc[,\"SVENY01\"]\nx_20 <- ycc[, \"SVENY20\"]\n\n# Plot the autocorrelations of the yield changes\npar(mfrow=c(2,2))\nacf_1 <- acf(x_1)\nacf_20 <- acf(x_20)\n\n# Plot the autocorrelations of the absolute changes of yields\nacf_abs_1 <- acf(abs(x_1))\nacf_abs_20 <- acf(abs(x_20))",
"_____no_output_____"
]
],
[
[
"## 6. GARCH in action\n<p>A Generalized AutoRegressive Conditional Heteroskedasticity (<a href=\"https://en.wikipedia.org/wiki/Autoregressive_conditional_heteroskedasticity\">GARCH</a>) model is the most well known econometric tool to handle changing volatility in financial time series data. It assumes a hidden volatility variable that has a long-run average it tries to return to while the short-run behavior is affected by the past returns.</p>\n<p>The most popular form of the GARCH model assumes that the volatility follows this process:\n</p><p></p>\n<math>\n σ<sup>2</sup><sub>t</sub> = ω + α ⋅ ε<sup>2</sup><sub>t-1</sub> + β ⋅ σ<sup>2</sup><sub>t-1</sub>\n</math>\n<p></p><p></p>\n<math> \nwhere σ is the current volatility, σ<sub>t-1</sub> the last day's volatility and ε<sub>t-1</sub> is the last day's return. The estimated parameters are ω, α, and β.\n</math>\n<p>For GARCH modeling we will use <a href=\"https://cran.r-project.org/web/packages/rugarch/index.html\"><code>rugarch</code></a> package developed by Alexios Ghalanos.</p>",
"_____no_output_____"
]
],
[
[
"library(rugarch)\n\n# Specify the GARCH model with the skewed t-distribution\nspec <- ugarchspec(distribution.model = \"sstd\")\n\n# Fit the model\nfit_1 <- ugarchfit(x_1, spec = spec)\n\n# Save the volatilities and the rescaled residuals\nvol_1 <- sigma(fit_1)\nres_1 <- scale(residuals(fit_1, standardize = TRUE)) * sd(x_1) + mean(x_1)\n\n# Plot the yield changes with the estimated volatilities and residuals\nmerge_1 <- merge.xts(x_1, vol_1, res_1)\nplot.zoo(merge_1)",
"Loading required package: parallel\n\nAttaching package: 'rugarch'\n\nThe following object is masked from 'package:stats':\n\n sigma\n\n"
]
],
[
[
"## 7. Fitting the 20-year maturity\n<p>Let's do the same for the 20-year maturity. As we can see in the plot from Task 6, the bond yields of various maturities show similar but slightly different characteristics. These different characteristics can be the result of multiple factors such as the monetary policy of the FED or the fact that the investors might be different.</p>\n<p>Are there differences between the 1-year maturity and 20-year maturity plots?</p>",
"_____no_output_____"
]
],
[
[
"# Fit the model\nfit_20 <- ugarchfit(x_20, spec = spec)\n\n# Save the volatilities and the rescaled residuals\nvol_20 <- sigma(fit_20)\nres_20 <- scale(residuals(fit_20, standardize = TRUE)) * sd(x_20) + mean(x_20)\n\n# Plot the yield changes with the estimated volatilities and residuals\nmerge_20 <- merge.xts(x_20, vol_20, res_20)\nplot.zoo(merge_20)",
"_____no_output_____"
]
],
[
[
"## 8. What about the distributions? (Part 1)\n<p>From the plots in Task 6 and Task 7, we can see that the 1-year GARCH model shows a similar but more erratic behavior compared to the 20-year GARCH model. Not only does the 1-year model have greater volatility, but the volatility of its volatility is larger than the 20-year model. That brings us to two statistical facts of financial markets not mentioned yet. </p>\n<ul>\n<li>The unconditional (before GARCH) distribution of the yield differences has heavier tails than the normal distribution.</li>\n<li>The distribution of the yield differences adjusted by the GARCH model has lighter tails than the unconditional distribution, but they are still heavier than the normal distribution.</li>\n</ul>\n<p>Let's find out what the fitted GARCH model did with the distribution we examined.</p>",
"_____no_output_____"
]
],
[
[
"# Calculate the kernel density for the 1-year maturity and residuals\ndensity_x_1 <- density(x_1)\ndensity_res_1 <- density(res_1)\n\n# Plot the density diagram for the 1-year maturity and residuals\nplot(density_x_1)\nlines(density_res_1, col = \"red\")\n\n# Add the normal distribution to the plot\nnorm_dist <- dnorm(seq(-0.4, 0.4, by = .01), mean = mean(x_1), sd = sd(x_1))\nlines(seq(-0.4, 0.4, by = .01), \n norm_dist, \n col = \"darkgreen\"\n )\n\n# Add legend\nlegend <- c(\"Before GARCH\", \"After GARCH\", \"Normal distribution\")\nlegend(\"topleft\", legend = legend, \n col = c(\"black\", \"red\", \"darkgreen\"), lty=c(1,1))",
"_____no_output_____"
]
],
[
[
"## 9. What about the distributions? (Part 2)\n<p>In the previous plot, we see that the two distributions from the GARCH models are different from the normal distribution of the data, but the tails, where the differences are the most profound, are hard to see. Using a Q-Q plot will help us focus in on the tails.</p>\n<p>You can read an excellent summary of Q-Q plots <a href=\"https://stats.stackexchange.com/questions/101274/how-to-interpret-a-qq-plot\">here</a>.</p>",
"_____no_output_____"
]
],
[
[
"# Define the data to plot: the 1-year maturity yield changes and residuals \ndata_orig <- x_1\ndata_res <- res_1\n\n# Define the benchmark distribution\ndistribution <- qnorm\n\n# Make the Q-Q plot of original data with the line of normal distribution\nqqnorm(data_orig, ylim = c(-0.5, 0.5))\nqqline(data_orig, distribution = distribution, col = \"darkgreen\")\n\n# Make the Q-Q plot of GARCH residuals with the line of normal distribution\npar(new=TRUE)\nqqnorm(data_res * 0.614256270265139, col = \"red\", ylim = c(-0.5, 0.5))\nqqline(data_res * 0.614256270265139, distribution = distribution, col = \"darkgreen\")\nlegend(\"topleft\", c(\"Before GARCH\", \"After GARCH\"), col = c(\"black\", \"red\"), pch=c(1,1))",
"_____no_output_____"
]
],
[
[
"## 10. A final quiz\n<p>In this project, we fitted a GARCH model to develop a better understanding of how bond volatility evolves and how it affects the probability distribution. In the final task, we will evaluate our model. Did the model succeed, or did it fail?</p>",
"_____no_output_____"
]
],
[
[
"# Q1: Did GARCH revealed how volatility changed over time? # Yes or No?\n(Q1 <- \"Yes\")\n\n# Q2: Did GARCH bring the residuals closer to normal distribution? Yes or No?\n(Q2 <- \"Yes\")\n\n# Q3: Which time series of yield changes deviates more \n# from a normally distributed white noise process? Choose 1 or 20.\n(Q3 <- 1)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb71af8a6fe4714f46917c1faefca72f920eafee | 20,052 | ipynb | Jupyter Notebook | samples/core/kubeflow_tf_serving/kubeflow_tf_serving.ipynb | rui5i/pipelines | f53a1e59b589fa88971c56a582275a5818d0782c | [
"Apache-2.0"
]
| 1 | 2021-09-05T17:31:13.000Z | 2021-09-05T17:31:13.000Z | samples/core/kubeflow_tf_serving/kubeflow_tf_serving.ipynb | rui5i/pipelines | f53a1e59b589fa88971c56a582275a5818d0782c | [
"Apache-2.0"
]
| null | null | null | samples/core/kubeflow_tf_serving/kubeflow_tf_serving.ipynb | rui5i/pipelines | f53a1e59b589fa88971c56a582275a5818d0782c | [
"Apache-2.0"
]
| 1 | 2022-03-04T14:26:55.000Z | 2022-03-04T14:26:55.000Z | 57.128205 | 259 | 0.644674 | [
[
[
"# Copyright 2019 The Kubeflow Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"# Install Pipeline SDK - This only needs to be ran once in the enviroment. \n!python3 -m pip install 'kfp>=0.1.31' --quiet\n!pip3 install tensorflow==1.14 --upgrade",
"Requirement already up-to-date: kfp in /opt/conda/lib/python3.6/site-packages (0.1.30)\nRequirement already satisfied, skipping upgrade: kfp-server-api<=0.1.25,>=0.1.18 in /opt/conda/lib/python3.6/site-packages (from kfp) (0.1.18.3)\nRequirement already satisfied, skipping upgrade: tabulate==0.8.3 in /opt/conda/lib/python3.6/site-packages (from kfp) (0.8.3)\nRequirement already satisfied, skipping upgrade: cloudpickle in /opt/conda/lib/python3.6/site-packages (from kfp) (0.8.1)\nRequirement already satisfied, skipping upgrade: six>=1.10 in /opt/conda/lib/python3.6/site-packages (from kfp) (1.12.0)\nRequirement already satisfied, skipping upgrade: cryptography>=2.4.2 in /opt/conda/lib/python3.6/site-packages (from kfp) (2.6.1)\nRequirement already satisfied, skipping upgrade: PyYAML in /opt/conda/lib/python3.6/site-packages (from kfp) (3.13)\nRequirement already satisfied, skipping upgrade: python-dateutil in /opt/conda/lib/python3.6/site-packages (from kfp) (2.8.0)\nRequirement already satisfied, skipping upgrade: google-auth>=1.6.1 in /opt/conda/lib/python3.6/site-packages (from kfp) (1.6.3)\nRequirement already satisfied, skipping upgrade: PyJWT>=1.6.4 in /opt/conda/lib/python3.6/site-packages (from kfp) (1.7.1)\nRequirement already satisfied, skipping upgrade: requests-toolbelt>=0.8.0 in /opt/conda/lib/python3.6/site-packages (from kfp) (0.9.1)\nRequirement already satisfied, skipping upgrade: argo-models==2.2.1a in /opt/conda/lib/python3.6/site-packages (from kfp) (2.2.1a0)\nRequirement already satisfied, skipping upgrade: Deprecated in /opt/conda/lib/python3.6/site-packages (from kfp) (1.2.6)\nRequirement already satisfied, skipping upgrade: kubernetes<=9.0.0,>=8.0.0 in /opt/conda/lib/python3.6/site-packages (from kfp) (9.0.0)\nRequirement already satisfied, skipping upgrade: urllib3<1.25,>=1.15 in /opt/conda/lib/python3.6/site-packages (from kfp) (1.24.1)\nRequirement already satisfied, skipping upgrade: jsonschema>=3.0.1 in /opt/conda/lib/python3.6/site-packages (from kfp) (3.0.1)\nRequirement already satisfied, skipping upgrade: click==7.0 in /opt/conda/lib/python3.6/site-packages (from kfp) (7.0)\nRequirement already satisfied, skipping upgrade: certifi in /opt/conda/lib/python3.6/site-packages (from kfp) (2019.3.9)\nRequirement already satisfied, skipping upgrade: google-cloud-storage>=1.13.0 in /opt/conda/lib/python3.6/site-packages (from kfp) (1.18.0)\nRequirement already satisfied, skipping upgrade: asn1crypto>=0.21.0 in /opt/conda/lib/python3.6/site-packages (from cryptography>=2.4.2->kfp) (0.24.0)\nRequirement already satisfied, skipping upgrade: cffi!=1.11.3,>=1.8 in /opt/conda/lib/python3.6/site-packages (from cryptography>=2.4.2->kfp) (1.12.2)\nRequirement already satisfied, skipping upgrade: cachetools>=2.0.0 in /opt/conda/lib/python3.6/site-packages (from google-auth>=1.6.1->kfp) (3.1.0)\nRequirement already satisfied, skipping upgrade: rsa>=3.1.4 in /opt/conda/lib/python3.6/site-packages (from google-auth>=1.6.1->kfp) (4.0)\nRequirement already satisfied, skipping upgrade: pyasn1-modules>=0.2.1 in /opt/conda/lib/python3.6/site-packages (from google-auth>=1.6.1->kfp) (0.2.4)\nRequirement already satisfied, skipping upgrade: requests<3.0.0,>=2.0.1 in /opt/conda/lib/python3.6/site-packages (from requests-toolbelt>=0.8.0->kfp) (2.21.0)\nRequirement already satisfied, skipping upgrade: wrapt<2,>=1.10 in /opt/conda/lib/python3.6/site-packages (from Deprecated->kfp) (1.11.2)\nRequirement already satisfied, skipping upgrade: setuptools>=21.0.0 in /opt/conda/lib/python3.6/site-packages (from kubernetes<=9.0.0,>=8.0.0->kfp) (40.9.0)\nRequirement already satisfied, skipping upgrade: requests-oauthlib in /opt/conda/lib/python3.6/site-packages (from kubernetes<=9.0.0,>=8.0.0->kfp) (1.2.0)\nRequirement already satisfied, skipping upgrade: websocket-client!=0.40.0,!=0.41.*,!=0.42.*,>=0.32.0 in /opt/conda/lib/python3.6/site-packages (from kubernetes<=9.0.0,>=8.0.0->kfp) (0.56.0)\nRequirement already satisfied, skipping upgrade: attrs>=17.4.0 in /opt/conda/lib/python3.6/site-packages (from jsonschema>=3.0.1->kfp) (19.1.0)\nRequirement already satisfied, skipping upgrade: pyrsistent>=0.14.0 in /opt/conda/lib/python3.6/site-packages (from jsonschema>=3.0.1->kfp) (0.14.11)\nRequirement already satisfied, skipping upgrade: google-resumable-media>=0.3.1 in /opt/conda/lib/python3.6/site-packages (from google-cloud-storage>=1.13.0->kfp) (0.3.2)\nRequirement already satisfied, skipping upgrade: google-cloud-core<2.0dev,>=1.0.0 in /opt/conda/lib/python3.6/site-packages (from google-cloud-storage>=1.13.0->kfp) (1.0.3)\nRequirement already satisfied, skipping upgrade: pycparser in /opt/conda/lib/python3.6/site-packages (from cffi!=1.11.3,>=1.8->cryptography>=2.4.2->kfp) (2.19)\nRequirement already satisfied, skipping upgrade: pyasn1>=0.1.3 in /opt/conda/lib/python3.6/site-packages (from rsa>=3.1.4->google-auth>=1.6.1->kfp) (0.4.5)\nRequirement already satisfied, skipping upgrade: chardet<3.1.0,>=3.0.2 in /opt/conda/lib/python3.6/site-packages (from requests<3.0.0,>=2.0.1->requests-toolbelt>=0.8.0->kfp) (3.0.4)\nRequirement already satisfied, skipping upgrade: idna<2.9,>=2.5 in /opt/conda/lib/python3.6/site-packages (from requests<3.0.0,>=2.0.1->requests-toolbelt>=0.8.0->kfp) (2.8)\nRequirement already satisfied, skipping upgrade: oauthlib>=3.0.0 in /opt/conda/lib/python3.6/site-packages (from requests-oauthlib->kubernetes<=9.0.0,>=8.0.0->kfp) (3.0.1)\nRequirement already satisfied, skipping upgrade: google-api-core<2.0.0dev,>=1.14.0 in /opt/conda/lib/python3.6/site-packages (from google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage>=1.13.0->kfp) (1.14.2)\nRequirement already satisfied, skipping upgrade: pytz in /opt/conda/lib/python3.6/site-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage>=1.13.0->kfp) (2018.9)\nRequirement already satisfied, skipping upgrade: protobuf>=3.4.0 in /opt/conda/lib/python3.6/site-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage>=1.13.0->kfp) (3.7.1)\nRequirement already satisfied, skipping upgrade: googleapis-common-protos<2.0dev,>=1.6.0 in /opt/conda/lib/python3.6/site-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage>=1.13.0->kfp) (1.6.0)\n\u001b[33mYou are using pip version 19.0.1, however version 19.2.3 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\nCollecting tensorflow==1.14\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/de/f0/96fb2e0412ae9692dbf400e5b04432885f677ad6241c088ccc5fe7724d69/tensorflow-1.14.0-cp36-cp36m-manylinux1_x86_64.whl (109.2MB)\n\u001b[K 100% |████████████████████████████████| 109.2MB 279kB/s eta 0:00:01\n\u001b[?25hCollecting tensorboard<1.15.0,>=1.14.0 (from tensorflow==1.14)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/91/2d/2ed263449a078cd9c8a9ba50ebd50123adf1f8cfbea1492f9084169b89d9/tensorboard-1.14.0-py3-none-any.whl (3.1MB)\n\u001b[K 100% |████████████████████████████████| 3.2MB 7.2MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied, skipping upgrade: keras-applications>=1.0.6 in /opt/conda/lib/python3.6/site-packages (from tensorflow==1.14) (1.0.7)\nRequirement already satisfied, skipping upgrade: numpy<2.0,>=1.14.5 in /opt/conda/lib/python3.6/site-packages (from tensorflow==1.14) (1.16.2)\nRequirement already satisfied, skipping upgrade: absl-py>=0.7.0 in /opt/conda/lib/python3.6/site-packages (from tensorflow==1.14) (0.7.1)\nRequirement already satisfied, skipping upgrade: grpcio>=1.8.6 in /opt/conda/lib/python3.6/site-packages (from tensorflow==1.14) (1.19.0)\nCollecting google-pasta>=0.1.6 (from tensorflow==1.14)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/d0/33/376510eb8d6246f3c30545f416b2263eee461e40940c2a4413c711bdf62d/google_pasta-0.1.7-py3-none-any.whl (52kB)\n\u001b[K 100% |████████████████████████████████| 61kB 25.5MB/s ta 0:00:01\n\u001b[?25hCollecting tensorflow-estimator<1.15.0rc0,>=1.14.0rc0 (from tensorflow==1.14)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/3c/d5/21860a5b11caf0678fbc8319341b0ae21a07156911132e0e71bffed0510d/tensorflow_estimator-1.14.0-py2.py3-none-any.whl (488kB)\n\u001b[K 100% |████████████████████████████████| 491kB 25.5MB/s ta 0:00:01\n"
]
],
[
[
"## KubeFlow Pipelines Serving Component\nIn this notebook, we will demo:\n\n* Saving a Keras model in a format compatible with TF Serving\n* Creating a pipeline to serve a trained model within a KubeFlow cluster\n\nReference documentation:\n* https://www.tensorflow.org/tfx/serving/architecture\n* https://www.tensorflow.org/beta/guide/keras/saving_and_serializing\n* https://www.kubeflow.org/docs/components/serving/tfserving_new/",
"_____no_output_____"
],
[
"### Setup\n",
"_____no_output_____"
]
],
[
[
"# Set your output and project. !!!Must Do before you can proceed!!!\nproject = 'Your-Gcp-Project-ID' #'Your-GCP-Project-ID'\nmodel_name = 'model-name' # Model name matching TF_serve naming requirements \nimport time\nts = int(time.time())\nmodel_version = str(ts) # Here we use timestamp as version to avoid conflict \noutput = 'Your-Gcs-Path' # A GCS bucket for asset outputs\nKUBEFLOW_DEPLOYER_IMAGE = 'gcr.io/ml-pipeline/ml-pipeline-kubeflow-deployer:1.7.0-rc.3'",
"_____no_output_____"
],
[
"model_path = '%s/%s' % (output,model_name) \nmodel_version_path = '%s/%s/%s' % (output,model_name,model_version)",
"_____no_output_____"
]
],
[
[
"### Load a Keras Model \nLoading a pretrained Keras model to use as an example. ",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"model = tf.keras.applications.NASNetMobile(input_shape=None,\n include_top=True,\n weights='imagenet',\n input_tensor=None,\n pooling=None,\n classes=1000)",
"Exception ignored in: <bound method _CheckpointRestoreCoordinator.__del__ of <tensorflow.python.training.tracking.util._CheckpointRestoreCoordinator object at 0x7f31dc197f98>>\nTraceback (most recent call last):\n File \"/opt/conda/lib/python3.6/site-packages/tensorflow/python/training/tracking/util.py\", line 244, in __del__\n .format(pretty_printer.node_names[node_id]))\n File \"/opt/conda/lib/python3.6/site-packages/tensorflow/python/training/tracking/util.py\", line 93, in node_names\n path_to_root[node_id] + (child.local_name,))\n File \"/opt/conda/lib/python3.6/site-packages/tensorflow/python/training/tracking/object_identity.py\", line 76, in __getitem__\n return self._storage[self._wrap_key(key)]\nKeyError: (<tensorflow.python.training.tracking.object_identity._ObjectIdentityWrapper object at 0x7f32aca065f8>,)\n"
]
],
[
[
"### Saved the Model for TF-Serve\nSave the model using keras export_saved_model function. Note that specifically for TF-Serve the output directory should be structure as model_name/model_version/saved_model.",
"_____no_output_____"
]
],
[
[
"tf.keras.experimental.export_saved_model(model, model_version_path)",
"_____no_output_____"
]
],
[
[
"### Create a pipeline using KFP TF-Serve component\n",
"_____no_output_____"
]
],
[
[
"def kubeflow_deploy_op():\n return dsl.ContainerOp(\n name = 'deploy',\n image = KUBEFLOW_DEPLOYER_IMAGE,\n arguments = [\n '--model-export-path', model_path,\n '--server-name', model_name,\n ]\n )",
"_____no_output_____"
],
[
"import kfp\nimport kfp.dsl as dsl\n\n# The pipeline definition\[email protected](\n name='sample-model-deployer',\n description='Sample for deploying models using KFP model serving component'\n)\ndef model_server():\n deploy = kubeflow_deploy_op()",
"_____no_output_____"
]
],
[
[
"Submit pipeline for execution on Kubeflow Pipelines cluster",
"_____no_output_____"
]
],
[
[
"kfp.Client().create_run_from_pipeline_func(model_server, arguments={})\n\n#vvvvvvvvv This link leads to the run information page. (Note: There is a bug in JupyterLab that modifies the URL and makes the link stop working)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb71b5c5d094cea495d890cb4e3d63ea89893802 | 599,834 | ipynb | Jupyter Notebook | 4_mol_similarity/T4_mol_similarity_JP.ipynb | magattaca/TeachOpenCADD_JP | b1a2879eba59bc1d218853ea58cf9965e8f8d0b4 | [
"CC-BY-4.0"
]
| 2 | 2021-07-29T05:53:34.000Z | 2021-07-29T08:14:54.000Z | 4_mol_similarity/T4_mol_similarity_JP.ipynb | magattaca/TeachOpenCADD_JP | b1a2879eba59bc1d218853ea58cf9965e8f8d0b4 | [
"CC-BY-4.0"
]
| null | null | null | 4_mol_similarity/T4_mol_similarity_JP.ipynb | magattaca/TeachOpenCADD_JP | b1a2879eba59bc1d218853ea58cf9965e8f8d0b4 | [
"CC-BY-4.0"
]
| null | null | null | 228.595274 | 90,824 | 0.900536 | [
[
[
"# トークトリアル 4\n\n# リガンドベーススクリーニング:化合物類似性\n\n#### Developed in the CADD seminars 2017 and 2018, AG Volkamer, Charité/FU Berlin \n\nAndrea Morger and Franziska Fritz",
"_____no_output_____"
],
[
"## このトークトリアルの目的\n\nこのトークトリアルでは、化合物をエンコード(記述子、フィンガープリント)し、比較(類似性評価)する様々なアプローチを取り扱います。さらに、バーチャルスクリーニングを実施します。バーチャルスクリーニングは、ChEMBLデータベースから取得し、リピンスキーのルールオブファイブでフィルタリングをかけた、EGFRに対して評価済みの化合物データセット(**トークトリアル 2** 参照) に対して、EGFR阻害剤ゲフィチニブ(Gefitinib)との類似性検索を実施するという形で実施します。\n\n## 学習の目標\n\n### 理論\n\n* 化合物類似性(Molecular similarity)\n* 化合物記述子(Molecular descriptors)\n* 化合物フィンガープリント(Molecular fingerprints)\n * 部分構造ベースのフィンガープリント(Substructure-based fingerprints)\n * MACCSフィンガープリント(MACCS fingerprints)\n * Morganフィンガープリント、サーキュラーフィンガープリント(Morgan fingerprints, circular fingerprints)\n* 化合物類似性評価(Molecular similarity measures)\n * タニモト係数(Tanimoto coefficient)\n * Dice係数(Dice coefficient)\n* バーチャルスクリーニング(Virtual screening)\n * 類似性検索(similarity search)によるバーチャルスクリーニング\n \n### 実践\n\n* 分子の読み込みと描画\n* 化合物記述子の計算\n * 1D 化合物記述子:分子量\n * 2D 化合物記述子:MACCS ファインガープリント\n * 2D 化合物記述子:Morgan フィンガープリント\n* 化合物類似性の計算\n * MACCS フィンガープリント:タニモト類似性とDice類似性\n * Morgan フィンガープリント:タニモト類似性とDice類似性\n* 類似性検索によるバーチャルスクリーニング\n * データセットの全化合物に対する化合物クエリの比較\n * 類似度の分布\n * 最も類似した分子の描画\n * エンリッチメントプロットの作成\n\n## レファレンス\n\n* レビュー\"Molecular similarity in medicinal chemistry\" ([<i>J. Med. Chem.</i> (2014), <b>57</b>, 3186-3204](http://pubs.acs.org/doi/abs/10.1021/jm401411z))\n* RDKitのMorganフィンガープリント ([RDKit tutorial on Morgan fingerprints](http://www.rdkit.org/docs/GettingStartedInPython.html#morgan-fingerprints-circular-fingerprints))\n* ECFP - extended-connectivity fingerprints ([<i>J. Chem. Inf. Model.</i> (2010), <b>50</b>,742-754](https://pubs.acs.org/doi/abs/10.1021/ci100050t))\n* ケミカルスペース\n([<i>ACS Chem. Neurosci.</i> (2012), <b>19</b>, 649-57](https://www.ncbi.nlm.nih.gov/pubmed/23019491))\n* RDKitの化合物記述子リスト ([RDKit documentation: Descriptors](https://www.rdkit.org/docs/GettingStartedInPython.html#list-of-available-descriptors))\n* RDKitのフィンガープリントのリスト ([RDKit documentation: Fingerprints](https://www.rdkit.org/docs/GettingStartedInPython.html#list-of-available-fingerprints))\n* エンリッチメントプロット([Applied Chemoinformatics, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, (2018), **1**, 313-31](https://onlinelibrary.wiley.com/doi/10.1002/9783527806539.ch6h))",
"_____no_output_____"
],
[
"_____________________________________________________________________________________________________________________\n\n\n## 理論\n\n### 化合物類似性\n\n化合物類似性は化学情報学(ケモインフォマティクス、chemical informatics)の中でよく知られており、頻繁に用いられる考え方です。化合物とその特性の比較はいろいろな用途で使用でき、望みの特性と生理活性をもつ新しい化合物を見つけるのに役立つかもしれません。\n\n構造的に類似の化合物は類似の特性、そして類似の生理活性を示すという考え方は、類似性質原則(similar property principle、SPP)や構造活性相関(structure activity relationship、SAR)に表れています。この文脈において、バーチャルスクリーニングは、結合親和性のわかっている化合物セットがあれば、そのような化合物をさらに探すことができる、というアイデアに基づいています。\n\n### 化合物記述子\n\n類似度は適用範囲に応じて様々な方法で評価することができます(参考 <a href=\"http://pubs.acs.org/doi/abs/10.1021/jm401411z\"><i>J. Med. Chem.</i> (2014), <b>57</b>, 3186-3204</a>):\n\n* **1D 化合物記述子**: 溶解度、logP、分子量、融点 etc.\n * グローバル記述子(Global descriptor):分子全体を一つの値だけで表現する <br>\n * 通常、機械学習(machine learning、ML)を適用するには分子を特定するのに十分な特性とはならない\n * 機械学習のための化合物エンコーディングを改良するために2Dフィンガープリントに付け加えることができる\n* **2D 化合物記述子**: 分子グラフ(Molecular graph)、経路(path)、フラグメント、原子環境(atom environment)\n * 分子の個々の部位の詳細な表現\n * 一つの分子に対して多数のフィンガープリントと呼ばれる特徴/ビット\n * 類似性検索と機械学習で非常によく使われる\n* **3D 化合物記述子**: 形状(Shape), 立体化学\n * 化学者は通常2次元表現で訓練されている <br>\n * 化合物の自由度(flexibility、化合物の「正しい」配座はどれか?)のため、2次元表現と比べて頑健性が低い\n* **生物学的類似性**\n * 生物学的フィンガープリント(例、個々のビットが異なるターゲット分子に対して評価された生理活性を表す)\n * 化合物構造からは独立\n * 実験データ(あるいは予測値)が必要\n\nすでに **トークトリアル 2** で、分子量やlogPといった1D 物理化学パラメーターを計算する方法を学びました。RDKitに実装されているそのような記述子は [RDKit documentation: Descriptors](https://www.rdkit.org/docs/GettingStartedInPython.html#list-of-available-descriptors) で見つけることができます。\n\n以降では、2D(あるいは3D)化合物記述子の定義に焦点を当てます。多くの場合、分子ごとに固有の(ユニークな)ものとなるので、これらの記述子はフィンガープリント(指紋)ともよばれます。",
"_____no_output_____"
],
[
"### 化合物フィンガープリント(Molecular fingerprints)\n\n#### 部分構造に基づくフィンガープリント(Substructure-based fingerprints)\n\n化合物フィンガープリントは化学的な特徴と分子の特徴をビット文字列(bitstring)やビットベクトル(bitvector)、配列(array)の形でエンコードします。各ビットは、事前に定義された分子の特徴あるいは環境に相当し、「1」はその特徴が存在していることを、「0」は存在していないことを示します。実装の仕方によっては、数え上げベース(count-based)となっていて、ある特定の特徴がいくつ存在しているかを数えるようになっていることに注意してください。\n\nフィンガープリントのデザインには複数の方法があります。ここではよく使われる2Dフィンガープリントのとして、MACCSキーとMorganフィンガープリントの2種類を導入します。\n [RDKit documentation: Fingerprints](https://www.rdkit.org/docs/GettingStartedInPython.html#list-of-available-fingerprints)に記載されているように、RDKitではこの2つ以外にも多数のフィンガープリントを提供しています。\n\n#### MACCS フィンガープリント(MACCS fingerprints)\n\nMolecular ACCess System (MACCS) フィンガープリント、あるいはMACCS構造キーとも名付けられている手法は、あらかじめ定義された166個の構造フラグメントから構成されています。各位置は、ある特定の構造フラグメントあるいはキーが存在しているかいないかを問い合わせた(クエリ)結果を格納しています。それぞれのキーは創薬化学者によって経験的に定義されたもので、利用、解釈が容易です。([RDKit documentation: MACCS keys](http://rdkit.org/Python_Docs/rdkit.Chem.MACCSkeys-module.html)).\n\n<img src=\"images/maccs_fp.png\" align=\"above\" alt=\"Image cannot be shown\" width=\"250\">\n<div align=\"center\"> Figure 2: MACCSフィンガープリントの図例(Andrea Morgerによる図)</div>\n\n#### Morganフィンガープリントとサーキュラーフィンガープリント(Morgan fingerprints and circular fingerprints) \n\nこの一連のフィンガープリントはMorganアルゴリズムに基づいています。ビットは分子の各原子の円形状の環境(circular environment)に相当しています。半径(radius)によって、環境として考慮にいれる近接の結合と原子の数を設定します。ビット文字列の長さを定義することもでき、より長いビット文字列を希望する長さに縮められます。従って、Morganフィンガープリントはある特定の数のビットには制限されません。Morganフィンガープリントに関してもっと知りたい場合は[RDKit documentation: Morgan fingerprints](http://www.rdkit.org/docs/GettingStartedInPython.html#morgan-fingerprints-circular-fingerprints) を参照してください。Extended connectivity fingerprints (ECFP)もよく使われるフィンガープリントで、Morganアルゴリズムのバリエーションから導かれています。さらなる情報は([<i>J. Chem. Inf. Model.</i> (2010), <b>50</b>,742-754](https://pubs.acs.org/doi/abs/10.1021/ci100050t))を参照してください。\n\n<img src=\"images/morgan_fp.png\" align=\"above\" alt=\"Image cannot be shown\" width=\"270\">\n<div align=\"center\">Figure 3: Morganサーキュラーフィンガープリントの図例(Andrea Morgerによる図)</div>",
"_____no_output_____"
],
[
"### 化合物類似性評価\n\n記述子/フィンガープリントの計算ができれば、それらを比較することで、二つの分子の間の類似度を評価することができます。化合物類似度は様々な類似度係数で定量化することができますが、よく使われる2つの指標はタニモト係数とDice係数です(Tanimoto and Dice index) ([<i>J. Med. Chem.</i> (2014), <b>57</b>, 3186-3204](http://pubs.acs.org/doi/abs/10.1021/jm401411z))。\n\n#### タニモト係数(Tanimoto coefficient)\n\n$$T _{c}(A,B) = \\frac{c}{a+b-c}$$\n\na: 化合物Aに存在する特徴の数 <br>\nb: 化合物Bに存在する特徴の数 <br>\nc: 化合物AとBで共有されている特徴の数\n\n#### Dice係数(Dice coefficient)\n\n$$D_{c}(A,B) = \\frac{c}{\\frac{1}{2}(a+b)}$$\n\na: 化合物Aに存在する特徴の数 <br>\nb: 化合物Bに存在する特徴の数 <br>\nc: 化合物AとBで共有されている特徴の数\n\n類似度評価は通常、それぞれのフィンガープリントの正の(1の)ビットの数と、両者が共通してもつ正のビットの数を考慮します。Dice類似度は通常タニモト類似度よりも大きな値を返し、それはそれぞれの分母の違いに由来します。:\n\n$$\\frac{c}{a+b-c} \\leq \\frac{c}{\\frac{1}{2}(a+b)}$$\n\n\n### バーチャルスクリーニング(Virtual screening) \n\n医薬品探索の初期段階における課題は、低分子(化合物)のセットを、有りうる巨大なケミカルスペースから、研究対象のターゲット分子に結合するポテンシャルのあるものに範囲を狭めることです。このケミカルスペースは非常に大きく、低分子化合物群は部分構造(chemical moiety)の10<sup>20</sup> の組み合わせにまでいたります ([<i>ACS Chem. Neurosci.</i> (2012), <b>19</b>, 649-57](https://www.ncbi.nlm.nih.gov/pubmed/23019491)) 。\n\n目的のターゲット分子に対するこれら低分子の活性を評価するハイスループットスクリーニング(HTS)実験は費用と時間が非常にかかるので、計算機に支援された(computer-aided)手法により、試験にかける低分子のリストをより絞り込む(focused list)ことが期待されています。このプロセスはバーチャル(ハイスループット)スクリーニングと呼ばれていて、研究対象のターゲット分子に結合する可能性の最も高い低分子を見つけるために、巨大な低分子ライブラリーをルールとパターンのどちらか、あるいは両方によってフィルタリングします。\n\n#### 類似度検索を用いたバーチャルスクリーニング\n\nバーチャルスクリーニングの簡単な方法として、既知の活性化合物(群)と新規な化合物セットを比較して、最も類似しているものを探すことが行われます。類似性質原則(similar property principle、SPP)に基づき、(例えば既知の阻害剤に)最も類似した化合物は類似の効果を有すると推測されます。類似性検索に必要となるものは次の通りです(上でも詳細に議論しています)。\n\n* 化学/分子の特徴をエンコードした表現\n* 特徴のポテンシャルの重み付け(オプション)\n* 類似度評価\n\n類似性検索はある特定のデータベースの全ての化合物と一つの化合物との間の類似度を計算することで実行することができます。データベースの化合物の類似度係数によるランク付けにより、最も類似度の高い分子が得られます。\n\n#### エンリッチメントプロット(Enrichment plots)\n\nエンリッチメントプロットはバーチャルスクリーニングの結果の妥当性を評価するために使われ、ランク付けされたリストの上位x%の中に含まれる活性化合物の比率を表します。すなわち、 \n\n* データセット全体のうち、トップにランクした化合物の比率(x-axis) vs. \n* データセット全体のうち活性化合物(y-axis)の比率\n\n<img src=\"images/enrichment_plot.png\" align=\"above\" alt=\"Image cannot be shown\" width=\"270\">\n<div align=\"center\">Figure 4: バーチャルスクリーニングの結果のエンリッチメントプロットの例</div>",
"_____no_output_____"
],
[
"## 実践\n\n実践編の最初のパートでは、RDKitを使って化合物のエンコード(化合物フィンガープリント)をしたのち、上の理論編で議論したように、類似度(化合物類似性評価)を計算するため、それらの比較を実行します。\n\n2番目のパートではこれらのエンコーディングと比較手法を使って類似度検索(バーチャルスクリーニング)を実施します。既知のEGFR阻害剤ゲフィチニブ(Gefitinib)をクエリとして使用し、EGFRに対して試験済みの化合物データセットの中から類似した化合物を検索します。このデータセットは **トークトリアル1**でChEMBLから抽出し、**トークトリアル2**でリピンスキーのルールオブファイブによりフィルタリングをおこなったものです。\n\n### 化合物の読み込みと描画\n\nまず、8個の化合物例を定義し描画します。後ほど、これらの分子をエンコードし比較します。SMILES形式の分子をRDKitのmolオブジェクトに変換し、RDKitの`Draw`関数で可視化します。",
"_____no_output_____"
]
],
[
[
"# 関連するPythonパッケージのimport\n# 基本的な分子を取り扱う機能はモジュール rdkti.Chem にあります\nfrom rdkit import Chem\n# 描画関係\nfrom rdkit.Chem.Draw import IPythonConsole\nfrom rdkit.Chem import Draw\nfrom rdkit.Chem import Descriptors\nfrom rdkit.Chem import AllChem\nfrom rdkit.Chem import MACCSkeys\nfrom rdkit.Chem import rdFingerprintGenerator\nfrom rdkit import DataStructs\n\nimport math\nimport numpy as np\nimport pandas as pd\nfrom rdkit.Chem import PandasTools\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# SMILES形式の化合物\nsmiles1 = 'CC1C2C(C3C(C(=O)C(=C(C3(C(=O)C2=C(C4=C1C=CC=C4O)O)O)O)C(=O)N)N(C)C)O' # Doxycycline\nsmiles2 = 'CC1(C(N2C(S1)C(C2=O)NC(=O)C(C3=CC=C(C=C3)O)N)C(=O)O)C' # Amoxicilline\nsmiles3 = 'C1=COC(=C1)CNC2=CC(=C(C=C2C(=O)O)S(=O)(=O)N)Cl' # Furosemide\nsmiles4 = 'CCCCCCCCCCCC(=O)OCCOC(=O)CCCCCCCCCCC' # Glycol dilaurate\nsmiles5 = 'C1NC2=CC(=C(C=C2S(=O)(=O)N1)S(=O)(=O)N)Cl' # Hydrochlorothiazide\nsmiles6 = 'CC1=C(C(CCC1)(C)C)C=CC(=CC=CC(=CC(=O)O)C)C' # Isotretinoine\nsmiles7 = 'CC1(C2CC3C(C(=O)C(=C(C3(C(=O)C2=C(C4=C1C=CC=C4O)O)O)O)C(=O)N)N(C)C)O' # Tetracycline\nsmiles8 = 'CC1C(CC(=O)C2=C1C=CC=C2O)C(=O)O' # Hemi-cycline D\n\n# 化合物SMILESのリストを作成\nsmiles = [smiles1, smiles2, smiles3, smiles4, smiles5, smiles6, smiles7, smiles8]\n\n# ROMolオブジェクトのリストを作成\nmols = [Chem.MolFromSmiles(i) for i in smiles]\n\n# 化合物名称のリストを作成\nmol_names = ['Doxycycline', 'Amoxicilline', 'Furosemide', 'Glycol dilaurate',\n 'Hydrochlorothiazide', 'Isotretinoine', 'Tetracycline', 'Hemi-cycline D']\n\n# 化合物の描画\nDraw.MolsToGridImage(mols, molsPerRow=2, subImgSize=(450,150), legends=mol_names)",
"_____no_output_____"
]
],
[
[
"### 化合物記述子の計算\n\n化合物の比較を行うために1Dと2Dの化合物記述子を抽出、生成します。2D記述子については、あとで化合物の類似度の計算に使用するので、異なるタイプのフィンガープリントを生成します。",
"_____no_output_____"
],
[
"#### 1D 化合物記述子:分子量\n\n例示構造の分子量を計算します。",
"_____no_output_____"
]
],
[
[
"# 化合物の分子量を計算\nmol_weights = [Descriptors.MolWt(mol) for mol in mols]",
"_____no_output_____"
]
],
[
[
"視覚的に比較するために、類似の分子量の化合物構造を描画します。分子量は化合物の類似度にとって有用な記述子となるでしょうか?",
"_____no_output_____"
]
],
[
[
"# 結果を格納するデータフレームの生成\nsim_mw_df = pd.DataFrame({'smiles': smiles, 'name': mol_names, 'mw': mol_weights, \"Mol\": mols})\n\n# 分子量でソート\nsim_mw_df.sort_values(['mw'], ascending=False, inplace=True)\nsim_mw_df[[\"smiles\", \"name\", \"mw\"]]",
"_____no_output_____"
],
[
"# 分子量とともに化合物を描画\nDraw.MolsToGridImage(sim_mw_df[\"Mol\"], \n legends=[i+': '+str(round(j, 2))+\" Da\" for i,j in zip(sim_mw_df[\"name\"], sim_mw_df[\"mw\"])],\n molsPerRow=2, subImgSize=(450, 150))",
"_____no_output_____"
]
],
[
[
"見てわかるように類似の分子量を持つ化合物は類似の構造をもつことがあります(例 Doxycycline/Tetracycline)。一方で、類似の数の原子を持ちながらも全く異なる原子の配置となっているものもあります(例 Doxycycline/Glycol dilaurate あるいはHydrochlorothiazide/Isotretinoine)。\n\n次に、より詳細な分子の特徴を説明するために、2D化合物記述子を見て見ましょう。",
"_____no_output_____"
],
[
"#### 2D 化合物記述子:MACCSフィンガープリント\n\nMACCSフィンガープリントはRDKitを使って簡単に生成することができます。明示的なビットベクトル(explicit bitvector)は我々人間が読めるものではないので、さらにビット文字配列(bitstring)へと変換します。",
"_____no_output_____"
]
],
[
[
"# MACCSフィンガープリントの生成\nmaccs_fp1 = MACCSkeys.GenMACCSKeys(mols[0]) # Doxycycline\nmaccs_fp2 = MACCSkeys.GenMACCSKeys(mols[1]) # Amoxicilline\nmaccs_fp1",
"_____no_output_____"
],
[
"# フィンガープリントをビット文字配列としてプリント\nmaccs_fp1.ToBitString()",
"_____no_output_____"
],
[
"# 全化合物のMACCS fingerprintsを生成\nmaccs_fp_list = []\nfor i in range(len(mols)):\n maccs_fp_list.append(MACCSkeys.GenMACCSKeys(mols[i]))",
"_____no_output_____"
]
],
[
[
"#### 2D 化合物記述子:Morganフィンガープリント\n\nRDKitを使ってMorganサーキュラーフィンガープリントも計算します。2つの異なる関数により、Morganフィンガープリントは整数(int)あるいはビット(bit)ベクトルとして計算することができます。",
"_____no_output_____"
]
],
[
[
"# Morganフィンガープリントを生成(int vector)、デフォルトでは半径2でベクトルの長さは2048\ncirc_fp1 = rdFingerprintGenerator.GetCountFPs(mols[:1])[0]\ncirc_fp1",
"_____no_output_____"
],
[
"# セットされた値をみてみます:\ncirc_fp1.GetNonzeroElements()",
"_____no_output_____"
],
[
"# Morganフィンガープリントを(bit vectorとして)生成、デフォルトでは半径2でフィンガープリントの長さは2048\ncirc_b_fp1 = rdFingerprintGenerator.GetFPs(mols[:1])[0]\ncirc_b_fp1",
"_____no_output_____"
],
[
"# フィンガープリントをビット文字列としてプリント\ncirc_b_fp1.ToBitString()",
"_____no_output_____"
],
[
"# 全化合物のMorganフィンガープリントを生成\ncirc_fp_list = rdFingerprintGenerator.GetFPs(mols)",
"_____no_output_____"
]
],
[
[
"### 化合物類似度の計算\n\n次では、2つの類似度評価、すなわち**Tanimoto**と**Dice**を、2つのタイプのフィンガープリント、すなわち**MACCS**と**Morgan**フィンガープリントに適用します。\n\n例:2つのMACCSフィンガープリントをタニモト類似度で比較",
"_____no_output_____"
]
],
[
[
"# 2つの化合物のタニモト係数を計算\nDataStructs.TanimotoSimilarity(maccs_fp1, maccs_fp2)",
"_____no_output_____"
],
[
"# 同じ化合物のタニモト係数を計算\nDataStructs.TanimotoSimilarity(maccs_fp1, maccs_fp1)",
"_____no_output_____"
]
],
[
[
"次に、クエリ化合物を我々の化合物リストと比較したいと思います。\nそこで、RDKitの ```BulkTanimotoSimilarity```関数と```BulkDiceSimilarity```関数を使って、タニモトあるいはDice類似度の類似度評価に基づいて、クエリのフィンガープリントと、リストに格納されたフィンガープリントとの類似度を計算します。",
"_____no_output_____"
],
[
"類似度を計算したあとで、次の関数を使ってランク付けした化合物を描画したいと思います。:",
"_____no_output_____"
]
],
[
[
"def draw_ranked_molecules(sim_df_sorted, sorted_column):\n \"\"\"\n (ソートした)データフレームの分子を描画する関数\n \"\"\"\n # ラベルを定義:最初の分子はクエリ(Query)で、次の分子はランク1から始まる\n rank = [\"#\"+str(i)+\": \" for i in range(0, len(sim_df_sorted))]\n rank[0] = \"Query: \"\n\n # Doxycyclineと最も類似した化合物(Tanimoto と MACCS フィンガープリント)\n top_smiles = sim_df_sorted[\"smiles\"].tolist()\n top_mols = [Chem.MolFromSmiles(i) for i in top_smiles]\n top_names = [i+j+\" (\"+str(round(k, 2))+\")\" for i, j, k in zip(rank, sim_df_sorted[\"name\"].tolist(), \n sim_df_sorted[sorted_column])]\n\n return Draw.MolsToGridImage(top_mols, legends=top_names, molsPerRow=2, subImgSize=(450, 150))",
"_____no_output_____"
]
],
[
[
"次に、タニモト/Dice類似度評価に基づいて、MACCS/Morganフィンガープリントの比較の全ての組み合わせを調べます。そこで、結果を要約するデータフレームを作成します。",
"_____no_output_____"
]
],
[
[
"# 結果を格納するデータフレームの生成\nsim_df = pd.DataFrame({'smiles': smiles, 'name': mol_names})",
"_____no_output_____"
]
],
[
[
"#### MACCSフィンガープリント:タニモト類似度",
"_____no_output_____"
]
],
[
[
"# 類似度評価スコアをデータフレームに追加\nsim_df['tanimoto_MACCS'] = DataStructs.BulkTanimotoSimilarity(maccs_fp1,maccs_fp_list)",
"_____no_output_____"
],
[
"# MACCSフィンガープリントのタニモト類似度で並べ替えたデータフレーム\nsim_df_sorted_t_ma = sim_df.copy()\nsim_df_sorted_t_ma.sort_values(['tanimoto_MACCS'], ascending=False, inplace=True)\nsim_df_sorted_t_ma",
"_____no_output_____"
],
[
"# MACCSフィンガープリントのタニモト類似度でランクした分子の描画\ndraw_ranked_molecules(sim_df_sorted_t_ma, \"tanimoto_MACCS\")",
"_____no_output_____"
]
],
[
[
"MACCSフィンガープリントを使用した場合、Tetracyclineは最も類似した分子(スコアが高い)で、ついでAmoxicillineでした。1D 化合物記述子の分子量とは対照的に、線形分子のGlucol dilaurateは類似していない(ランクが最も低い)と認識されました。",
"_____no_output_____"
],
[
"#### MACCSフィンガープリント:Dice類似度",
"_____no_output_____"
]
],
[
[
"# データフレームへの類似度スコアの追加\nsim_df['dice_MACCS'] = DataStructs.BulkDiceSimilarity(maccs_fp1, maccs_fp_list)",
"_____no_output_____"
],
[
"# MACCSフィンガープリントのDice類似度でソートしたデータフレーム\nsim_df_sorted_d_ma = sim_df.copy()\nsim_df_sorted_d_ma.sort_values(['dice_MACCS'], ascending=False, inplace=True)\nsim_df_sorted_d_ma",
"_____no_output_____"
]
],
[
[
"定義より、タニモトとDice類似度評価は同じランキング結果になりますが、Dice類似度の値の方が大きくなります(タニモトとDiceを求める式はこのトークトリアルの理論編を参照してください)。",
"_____no_output_____"
],
[
"#### Morganフィンガープリント:タニモト類似度",
"_____no_output_____"
]
],
[
[
"# データフレームへの類似度スコアの追加\nsim_df['tanimoto_morgan'] = DataStructs.BulkTanimotoSimilarity(circ_b_fp1, circ_fp_list)\nsim_df['dice_morgan'] = DataStructs.BulkDiceSimilarity(circ_b_fp1, circ_fp_list)",
"_____no_output_____"
],
[
"# Morganフィンガープリントのタニモト類似度で並べ替えたデータフレーム\nsim_df_sorted_t_mo = sim_df.copy()\nsim_df_sorted_t_mo.sort_values(['tanimoto_morgan'], ascending=False, inplace=True)\nsim_df_sorted_t_mo",
"_____no_output_____"
],
[
"# Morganフィンガープリントのタニモト類似度による化合物ランキングの描画\ndraw_ranked_molecules(sim_df_sorted_t_mo, \"tanimoto_morgan\")",
"_____no_output_____"
]
],
[
[
"MACCSとMorganの類似度をタニモト(Morgan) vs タニモト(MACCS)でプロットし比較します。",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(figsize=(6,6), nrows=1, ncols=1)\nsim_df_sorted_t_mo.plot('tanimoto_MACCS','tanimoto_morgan',kind='scatter',ax=axes)\nplt.plot([0,1],[0,1],'k--')\naxes.set_xlabel(\"MACCS\")\naxes.set_ylabel(\"Morgan\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"異なるフィンガープリント(ここでは、MACCSフィンガープリントとMorganフィンガープリント)を用いると、異なる類似度の値(ここでは、タニモト係数)となり、ここで示したように、潜在的には化合物類似度のランキングが異なるものとなります。\n\nMorganフィンガープリントはDoxycyclineに対してTetracyclineを(スコアはより低かったでしたが)最も類似した化合物として認識し、Glycol dilaurateを最も似ていない化合物として認識しました。一方で、2番目にランク付されたのはHemi-cycline Dでした。この化合物はサイクリン系化合物の部分構造で、Morganフィンガープリントのアルゴリズムが原子の環境に基づくものであることがその理由であるかもしれません(一方で、MACCSフィンガープリントは特定の特徴の出現頻度を求めるものとなっています)。",
"_____no_output_____"
],
[
"### 類似度検索を使ったバーチャルスクリーニング\n\nフィンガープリントと類似度の計算方法を学んだので、この知識を化合物セット全体からのクエリ化合物の類似度検索に応用することができます。\n\n既知のEGFR阻害剤ゲフィチニブ(Gefitinib)をクエリとして使用し、EGFRに対して試験済みの化合物データセットの中から類似した化合物を検索します。このデータセットは **トークトリアル1**でChEMBLから抽出し、**トークトリアル2**でリピンスキーのルールオブファイブによりフィルタリングをおこなったものです。\n\n#### クエリ化合物をデータセットの全化合物と比較\n\n**トークトリアル2**で取得したChEMBLデータベースから取り出したEGFRに対して評価済みのフィルタリングされた化合物を含むcsvファイルから化合物を読み込みます。1つのクエリ化合物(ここではゲフィチニブ)を使って、類似の化合物をデータセットの中から探し出します。",
"_____no_output_____"
]
],
[
[
"# SMILES形式の化合物を含むcsvファイルからデータを読み込む\nfiltered_df = pd.read_csv('../data/T2/EGFR_compounds_lipinski.csv', delimiter=';', usecols=['molecule_chembl_id', 'smiles', 'pIC50'])\nfiltered_df.head() ",
"_____no_output_____"
],
[
"# クエリ化合物のSMILESからMolオブジェクトを生成\nquery = Chem.MolFromSmiles('COC1=C(OCCCN2CCOCC2)C=C2C(NC3=CC(Cl)=C(F)C=C3)=NC=NC2=C1'); # Gefitinib, Iressa\nquery",
"_____no_output_____"
],
[
"# クエリ化合物のMACCSフィンガープリントとMorganフィンガープリントを生成\nmaccs_fp_query = MACCSkeys.GenMACCSKeys(query)\ncirc_fp_query = rdFingerprintGenerator.GetCountFPs([query])[0]",
"_____no_output_____"
],
[
"# ファイルの全化合物のMACCSフィンガープリントとMorganフィンガープリントを生成\nms = [Chem.MolFromSmiles(i) for i in filtered_df.smiles]\ncirc_fp_list = rdFingerprintGenerator.GetCountFPs(ms)\nmaccs_fp_list = [MACCSkeys.GenMACCSKeys(m) for m in ms]",
"_____no_output_____"
],
[
"# クエリ化合物(Gefitinib)とファイルの全化合物のタニモト類似性を計算(MACCS、Morgan)\ntanimoto_maccs = DataStructs.BulkTanimotoSimilarity(maccs_fp_query,maccs_fp_list)\ntanimoto_circ = DataStructs.BulkTanimotoSimilarity(circ_fp_query,circ_fp_list)",
"_____no_output_____"
],
[
"# クエリ化合物(Gefitinib)とファイルの全化合物のDice類似性を計算(MACCS、Morgan)\ndice_maccs = DataStructs.BulkDiceSimilarity(maccs_fp_query,maccs_fp_list)\ndice_circ = DataStructs.BulkDiceSimilarity(circ_fp_query,circ_fp_list)",
"_____no_output_____"
],
[
"# ChEMBL IDとSMILES、Gefitinibに対する化合物のタニモト類似性のテーブルを作成\nsimilarity_df = pd.DataFrame({'ChEMBL_ID':filtered_df.molecule_chembl_id,\n 'bioactivity':filtered_df.pIC50,\n 'tanimoto_MACCS': tanimoto_maccs, \n 'tanimoto_morgan': tanimoto_circ, \n 'dice_MACCS': dice_maccs,\n 'dice_morgan': dice_circ,\n 'smiles': filtered_df.smiles,})",
"_____no_output_____"
],
[
"# データフレームを表示\nsimilarity_df.head()",
"_____no_output_____"
]
],
[
[
"#### 類似性評価の値の分布\n\n理論編で述べたように、同じフィンガープリント(例 MACCSフィンガープリント)について比較すれば、タニモト類似度の値はDIce類似度の値よりも小さくなります。また、2つの異なるフィンガープリント(例 MACCSフィンガープリントとMorganフィンガープリント)を比較すると、類似性評価の値(例 タニモト類似度)は変化します。\n\nヒストグラムをプロットすることで分布を見ることができます。",
"_____no_output_____"
]
],
[
[
"# MACCSフィンガープリントのタニモト類似度の分布をプロット\n%matplotlib inline\nfig, axes = plt.subplots(figsize=(10,6), nrows=2, ncols=2)\nsimilarity_df.hist([\"tanimoto_MACCS\"], ax=axes[0,0])\nsimilarity_df.hist([\"tanimoto_morgan\"], ax=axes[0,1])\nsimilarity_df.hist([\"dice_MACCS\"], ax=axes[1,0])\nsimilarity_df.hist([\"dice_morgan\"], ax=axes[1,1])\naxes[1,0].set_xlabel(\"similarity value\")\naxes[1,0].set_ylabel(\"# molecules\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"ここでも類似度を比較します。今回は直接、2つのフィンガープリントに関するタニモト類似度とDice類似度を比較します。",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(figsize=(12,6), nrows=1, ncols=2)\n\nsimilarity_df.plot('tanimoto_MACCS','dice_MACCS',kind='scatter',ax=axes[0])\naxes[0].plot([0,1],[0,1],'k--')\naxes[0].set_xlabel(\"Tanimoto(MACCS)\")\naxes[0].set_ylabel(\"Dice(MACCS)\")\n\nsimilarity_df.plot('tanimoto_morgan','dice_morgan',kind='scatter',ax=axes[1])\naxes[1].plot([0,1],[0,1],'k--')\naxes[1].set_xlabel(\"Tanimoto(Morgan)\")\naxes[1].set_ylabel(\"Dice(Morgan)\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"類似度分布は類似度を解釈するのに重要です(例 MACCSフィンガープリントとMorganフィンガープリント、タニモト類似度とDice類似度について値0.6は異なる評価を与えられる必要があります)\n\n次では、Morganフィンガープリントに基づき、タニモト類似度で最もよく似た化合物を描画します。",
"_____no_output_____"
],
[
"#### 最も類似の化合物を描画",
"_____no_output_____"
],
[
"私たちの作成したランキングにおいて最も類似した化合物との比較として、ゲフィチニブ(Gefitinib)の構造を視覚的に調べます。生理活性の情報(**トークトリアル1**でChEMBLから抽出したpIC50)も含めます。",
"_____no_output_____"
]
],
[
[
"# tanimoto_morganでソートしたデータフレーム\nsimilarity_df.sort_values(['tanimoto_morgan'], ascending=False, inplace=True)\nsimilarity_df.head()",
"_____no_output_____"
],
[
"# データフレームにSMILES文字列の構造表現(ROMol - RDKit オブジェクト Mol)を追加\nPandasTools.AddMoleculeColumnToFrame(similarity_df, 'smiles')",
"_____no_output_____"
],
[
"# クエリ構造とトップランクの化合物群(+生理活性)の描画\nsim_mols = [Chem.MolFromSmiles(i) for i in similarity_df.smiles][:11]\n\nlegend = ['#' + str(a) + ' ' + b + ' ('+str(round(c,2))+')' for a, b, c in zip(range(1,len(sim_mols)+1),\n similarity_df.ChEMBL_ID, \n similarity_df.bioactivity)]\nChem.Draw.MolsToGridImage(mols = [query] + sim_mols[:11], \n legends = (['Gefitinib'] + legend), \n molsPerRow = 4)",
"_____no_output_____"
]
],
[
[
"データセットにおいてゲフィチニブと比較してトップにランクした化合物群は、最初は我々のデータセットに含まれるゲフィチニブのエントリー(rank1 と rank2)で、続いてゲフィチニブの変換体(例 benzole置換基パターンが異なるもの)です。\n\n注:ChEMBLにはゲフィチニブ(よく研究された化合物なので)の完全な構造活性相関分析がふくまれていて、したがって私たちが取得したデータセットにゲフィチニブ様化合物が多く含まれていることは驚くべきことではありません。\n\nそれでは、類似度検索がどの程度、データセット上の活性化合物と不活性化合物を区別することができるか、その性能をチェックしたいと思います。そこで、**トークトリアル1** でChEMBLから取得した化合物の(EGFRに対する)生理活性の値を使用します。",
"_____no_output_____"
],
[
"#### エンリッチメントプロットの生成\n\nバーチャルスクリーニングの妥当性を評価し、見つかった活性化合物の比率を見るためにエンリッチメントプロットを作成します。\n\nエンリッチメントプロットが示すのは;\n* データセット全体のうち、トップにランクした化合物の比率(x-axis) vs. \n* データセット全体のうち活性化合物(y-axis)の比率\n\nMACCSフィンガープリントとMorganフィンガープリントのタニモト類似度を比較します。\n\n化合物を活性化合物あるいは不活性化合物のいずれとして取り扱うかを決めるために、一般に使用されるpIC50のカットオフ値6.3を適用します。文献中にはpIC50カットオフ値として5〜7にわたる範囲でいくつか提案がなされていて、データポイントをとらない排除範囲を定義しているものもありますが、私たちはこのカットオフ(6.3)は合理的と考えています。\n同じカットオフを**トークトリアル10**の機械学習でも用います。",
"_____no_output_____"
]
],
[
[
"# 活性化合物と不活性化合物を区別するpIC50 カットオフ値\nthreshold = 6.3",
"_____no_output_____"
],
[
"similarity_df.head()",
"_____no_output_____"
],
[
"def get_enrichment_data(similarity_df, similarity_measure, threshold):\n \"\"\"\n エンリッチメントプロットのxとyの値を計算する関数:\n x - データセットで何%にランクしているか\n y - 何%の本当に活性な化合物が見つかったか\n \"\"\"\n \n # データセットの化合物の数を取得\n mols_all = len(similarity_df)\n \n # データセットの活性化合物の数を取得\n actives_all = sum(similarity_df.bioactivity >= threshold)\n\n # データセット全体を処理している間、活性化合物のカウンターを保持するリストを初期化\n actives_counter_list = []\n \n # 活性化合物のカウンターを初期化\n actives_counter = 0\n \n # 注: エンリッチメントプロットのためデータをランク付けしなければなりません。\n # 選択した類似度評価によって化合物を並べ替えます。\n similarity_df.sort_values([similarity_measure], ascending=False, inplace=True)\n\n # ランク付けされたデータセットを一つずつ処理し、(生理活性をチェックすることで)各化合物が活性化合物どうか確認します\n for value in similarity_df.bioactivity:\n if value >= threshold:\n actives_counter += 1\n actives_counter_list.append(actives_counter)\n\n # 化合物の数をデータセットのランク何%になるかに変換\n mols_perc_list = [i/mols_all for i in list(range(1, mols_all+1))]\n\n # 活性化合物の数を本当の活性化合物の何%が見つかったかに変換\n actives_perc_list = [i/actives_all for i in actives_counter_list]\n\n # xとyの値とラベルをもつデータフレームを生成\n enrich_df = pd.DataFrame({'% ranked dataset':mols_perc_list, \n '% true actives identified':actives_perc_list,\n 'similarity_measure': similarity_measure})\n \n return enrich_df",
"_____no_output_____"
],
[
"# プロットする類似度評価を定義\nsim_measures = ['tanimoto_MACCS', 'tanimoto_morgan']\n\n# 全類似度評価についてエンリッチメントプロットのデータを持つデータフレームのリストを作成\nenrich_data = [get_enrichment_data(similarity_df, i, threshold) for i in sim_measures]",
"_____no_output_____"
],
[
"# プロットのためのデータセットを準備:\n# 類似度評価毎のデータフレームを一つのデータフレームに連結\n# …異なる類似度評価は「similarity_measure」列によって区別可能です\nenrich_df = pd.concat(enrich_data)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(6, 6))\n\nfontsize = 20\n\nfor key, grp in enrich_df.groupby(['similarity_measure']):\n ax = grp.plot(ax = ax,\n x = '% ranked dataset',\n y = '% true actives identified',\n label=key,\n alpha=0.5, linewidth=4)\nax.set_ylabel('% True actives identified', size=fontsize)\nax.set_xlabel('% Ranked dataset', size=fontsize)\n\n# データセットの活性化合物比率\nratio = sum(similarity_df.bioactivity >= threshold) / len(similarity_df)\n\n# 理想的な場合のカーブをプロット\nax.plot([0,ratio,1], [0,1,1], label=\"Optimal curve\", color=\"black\", linestyle=\"--\")\n\n# ランダムな場合のカーブをプロット\nax.plot([0,1], [0,1], label=\"Random curve\", color=\"grey\", linestyle=\"--\")\n\nplt.tick_params(labelsize=16)\nplt.legend(labels=['MACCS', 'Morgan', \"Optimal\", \"Random\"], loc=(.5, 0.08), \n fontsize=fontsize, labelspacing=0.3)\n\n# プロットを保存ーテキストボックスを含めるためにbbox_inchesを使用:\n# https://stackoverflow.com/questions/44642082/text-or-legend-cut-from-matplotlib-figure-on-savefig?rq=1\nplt.savefig(\"../data/T4/enrichment_plot.png\", dpi=300, bbox_inches=\"tight\", transparent=True)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"エンリッチメントプロットによるとMACCSフィンガープリントよりもMorganフィンガープリント基づく比較の方が少し良いパフォーマンスを示しています。",
"_____no_output_____"
]
],
[
[
"# ランク付されたデータセットのx%についてEFを取得\ndef print_data_ef(perc_ranked_dataset, enrich_df):\n data_ef = enrich_df[enrich_df['% ranked dataset'] <= perc_ranked_dataset].tail(1)\n data_ef = round(float(data_ef['% true actives identified']), 1)\n print(\"Experimental EF for \", perc_ranked_dataset, \"% of ranked dataset: \", data_ef, \"%\", sep=\"\")\n\n# ランク付されたデータセットのx%についてランダムEFを取得\ndef print_random_ef(perc_ranked_dataset):\n random_ef = round(float(perc_ranked_dataset), 1)\n print(\"Random EF for \", perc_ranked_dataset, \"% of ranked dataset: \", random_ef, \"%\", sep=\"\")\n\n# ランク付されたデータセットのx%について理想的な場合のEFを取得\ndef print_optimal_ef(perc_ranked_dataset, similarity_df, threshold):\n ratio = sum(similarity_df.bioactivity >= threshold) / len(similarity_df) * 100\n if perc_ranked_dataset <= ratio:\n optimal_ef = round(100/ratio * perc_ranked_dataset, 1)\n else:\n optimal_ef = round(float(100), 2)\n print(\"Optimal EF for \", perc_ranked_dataset, \"% of ranked dataset: \", optimal_ef, \"%\", sep=\"\")",
"_____no_output_____"
],
[
"# パーセンテージを選択\nperc_ranked_list = 5\n\n# EFデータを取得\nprint_data_ef(perc_ranked_list, enrich_df)\nprint_random_ef(perc_ranked_list)\nprint_optimal_ef(perc_ranked_list, similarity_df, threshold)",
"Experimental EF for 5% of ranked dataset: 1.0%\nRandom EF for 5% of ranked dataset: 5.0%\nOptimal EF for 5% of ranked dataset: 8.9%\n"
]
],
[
[
"**訳注(04/2020)**\n\nオリジナルの実践編はここまでですが、このEFの結果は少しおかしい気がします。エンリッチメントプロットを見ると、エンリッチメントファクターは「**optimal** > **Experimental** > **Random**」となると思われます。**Random**よりも**Experimental**が低いということは、むしろ不活性化合物へと選択のバイアスがかかっていることになってしまいます。\n\nどこかおかしいところがないか?順番に見ていきます。 \n\nまずEFの算出に使われているDataFrame **enrich_df**は2つの類似度評価基準のデータを繋げたものなので、それぞれ別々にしてみます。",
"_____no_output_____"
]
],
[
[
"# tanimoto_MACCS\nenrich_df_taMA = enrich_df[enrich_df['similarity_measure'] == 'tanimoto_MACCS']\n\n# tanimoto_morgan\nenrich_df_tamo = enrich_df[enrich_df['similarity_measure'] == 'tanimoto_morgan']\n\nprint(\"Size of enrich_df: \", len(enrich_df))\nprint(\"Size of tanimoto_MACCS DataFrame: \", len(enrich_df_taMA))\nprint(\"Size of tanimoto_morgan DataFrame: \", len(enrich_df_tamo))",
"Size of enrich_df: 9046\nSize of tanimoto_MACCS DataFrame: 4523\nSize of tanimoto_morgan DataFrame: 4523\n"
]
],
[
[
"DataFrameの5% ranked Datasetに相当する箇所を見てみます。",
"_____no_output_____"
]
],
[
[
"# 5% に相当する数\nindex_5perc = round(len(enrich_df_taMA)*0.05)\n\n# DataFrameのindexは0から始まるので-1した行を表示\nenrich_df_taMA[index_5perc-1:index_5perc]",
"_____no_output_____"
]
],
[
[
"見やすさのためsliceでDataFrameとして取り出しています。 \nランク上位5%(0.049)に相当する数のなかに、実際の活性評価でactiveだったものは7.3%(% true actives identified, 0.07319)となっています。この値は先の**Random**、**Optimal**と比較して妥当な値に思います。\n\nDataFrameのデータ自体には問題なさそうなので、値の取り出し方(関数`print_data_ef`)に問題があったのでしょう。\n関数の中身を順番に実行してみます。 ",
"_____no_output_____"
]
],
[
[
"# 5%に設定\nperc_ranked_dataset = 5\n\n# 5%内のDataFrameを取り出し、その一番最後の行(tail)を取り出す\nenrich_df[enrich_df['% ranked dataset'] <= perc_ranked_dataset].tail(1)",
"_____no_output_____"
]
],
[
[
"取り出されたのは「index:4522」で4523番目の行です。この`% true actives identifed`列がEFとして取り出されていた値(1.0)です。 \n閾値5以下で取り出されたのは`similarity_measure`:**tanimoto_morgan**の全化合物でした。単純にDataFrameのデータは%に換算していないのに、取り出す際に%換算の値を使ってしまったのが原因のようです。 \n\nそれではそれぞれの類似度について正しい値を確認してみます。",
"_____no_output_____"
]
],
[
[
"# 関数の再定義\ndef print_data_ef2(perc_ranked_dataset, enrich_df):\n perc_ranked_dataset_100 = perc_ranked_dataset / 100\n data_ef = enrich_df[enrich_df['% ranked dataset'] <= perc_ranked_dataset_100].tail(1)\n data_ef = round(float(data_ef['% true actives identified'] * 100), 1)\n print(\"Experimental EF for \", perc_ranked_dataset, \"% of ranked dataset: \", data_ef, \"%\", sep=\"\")",
"_____no_output_____"
],
[
"# MACCS keyの場合\n# パーセンテージを選択\nperc_ranked_list = 5\n\n# EFデータを取得\nprint_data_ef2(perc_ranked_list, enrich_df_taMA)\nprint_random_ef(perc_ranked_list)\nprint_optimal_ef(perc_ranked_list, similarity_df, threshold)",
"Experimental EF for 5% of ranked dataset: 7.3%\nRandom EF for 5% of ranked dataset: 5.0%\nOptimal EF for 5% of ranked dataset: 8.9%\n"
],
[
"# Morganフィンガープリントの場合\n# パーセンテージを選択\nperc_ranked_list = 5\n\n# EFデータを取得\nprint_data_ef2(perc_ranked_list, enrich_df_tamo)\nprint_random_ef(perc_ranked_list)\nprint_optimal_ef(perc_ranked_list, similarity_df, threshold)",
"Experimental EF for 5% of ranked dataset: 7.9%\nRandom EF for 5% of ranked dataset: 5.0%\nOptimal EF for 5% of ranked dataset: 8.9%\n"
]
],
[
[
"いずれも「**optimal** > **Experimental** > **Random**」となっており、**Morgan**の方が**MACCS**よりも若干良い値となっています。無事エンリッチメントプロットと比較してもおかしくない値が得られました。\n\n**訳注ここまで**",
"_____no_output_____"
],
[
"## ディスカッション\n\nここではタニモト類似度を使ってバーチャルスクリーニングを実施しました。もちろん、Dice類似度や他の類似度評価を使っても行うことができます。\n\n化合物フィンガープリントを使用した類似度検索の欠点は、化合物類似度に基づくものなので新規な構造を生み出さないことです。化合物類似度を扱う上でのその他の課題としては、いわゆるアクティビティクリフ(activity cllif)があります。分子の官能基におけるちょっとした変化が生理活性の大きな変化を起こすことがあります。",
"_____no_output_____"
],
[
"## クイズ\n\n* アクティビティクリフを回避するにはどこから始めれば良いでしょうか?\n* MACCSフィンガープリントとMorganフィンガープリントを互いに比較した場合の利点と欠点は何でしょう?\n* 使用したフィンガープリントによっておこる、類似度データフレームにおける順序の違いをどう説明できるでしょうか?",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
cb71bae7f476d1f1db41189a7b061f2138434e36 | 51,629 | ipynb | Jupyter Notebook | seminar13/gp.ipynb | EAkeweje/ML2021_seminars | 1ec37da8057841de7dbfbd73bc03a2bb485bbb57 | [
"MIT"
]
| 11 | 2021-01-30T18:47:43.000Z | 2022-01-23T22:52:24.000Z | seminar13/gp.ipynb | EAkeweje/ML2021_seminars | 1ec37da8057841de7dbfbd73bc03a2bb485bbb57 | [
"MIT"
]
| null | null | null | seminar13/gp.ipynb | EAkeweje/ML2021_seminars | 1ec37da8057841de7dbfbd73bc03a2bb485bbb57 | [
"MIT"
]
| 14 | 2021-02-02T11:11:29.000Z | 2021-09-01T19:23:17.000Z | 27.579594 | 326 | 0.522342 | [
[
[
"<a href=\"https://colab.research.google.com/github/adasegroup/ML2021_seminars/blob/master/seminar13/gp.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"## Gaussian Processes (GP) with GPy\n\nIn this notebook we are going to use GPy library for GP modeling [SheffieldML github page](https://github.com/SheffieldML/GPy).\n\nWhy **GPy**?\n\n* Specialized library of GP models (regression, classification, GPLVM)\n* Variety of covariance functions is implemented\n* There are GP models for large-scale problems\n* Easy to use",
"_____no_output_____"
],
[
"Run the following line to install GPy library",
"_____no_output_____"
]
],
[
[
"!pip install GPy",
"_____no_output_____"
],
[
"from __future__ import print_function\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport GPy\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Current documentation of GPy library can be found [here](http://gpy.readthedocs.org/en/latest/).",
"_____no_output_____"
],
[
"## Gaussian Process Regression\n\nA data set $\\left (X, \\mathbf{y} \\right ) = \\left \\{ (x_i, y_i), x_i \\in \\mathbb{R}^d, y_i \\in \\mathbb{R} \\right \\}_{i = 1}^N$ is given. \n\nAssumption:\n$$\ny = f(x) + \\varepsilon,\n$$\nwhere $f(x)$ is a Gaussian Processes and $\\varepsilon \\sim \\mathcal{N}(0, \\sigma_n^2)$ is a Gaussian noise .",
"_____no_output_____"
],
[
"Posterior distribution of function value $y^*$ at point $x^*$\n$$\ny_* | X, \\mathbf{y}, x_* \\sim \\mathcal{N}(m(x_*), \\sigma(x_*)),\n$$\nwith predictive mean and variance given by\n$$\nm(x_*) = \\mathbf{k}^T \\mathbf{K}_y^{-1} \\mathbf{y} = \\sum_{i = 1}^N \\alpha_i k(x_*, x_i),\n$$\n$$\n\\sigma^2(x_*) = k(x_*, x_*) - \\mathbf{k}^T\\mathbf{K}_y^{-1}\\mathbf{k},\n$$\nwhere\n$$\n\\mathbf{k} = \\left ( k(x_*, x_1), \\ldots, k(x_*, x_N) \\right )^T\n$$\n$$\n\\mathbf{K}_y = \\|k(x_i, x_j)\\|_{i, j = 1}^N + \\sigma_n^2 \\mathbf{I}\n$$",
"_____no_output_____"
],
[
"### Exercises\n\n1. What the posterior variance at the points from the training set is equal to? What if the noise variance is equal to 0?\n\n2. Suppose that we want to minimize some unknown function $f(\\mathbf{x})$.\n We are given a set of observations $y_i = f(\\mathbf{x}_i) + \\varepsilon_i$, where $\\varepsilon_i \\sim \\mathcal{N}(0, \\sigma^2)$.\n Using the observations we built a GP model $\\hat{f}(\\mathbf{x})$.\n Now, let us consider the value called *improvement*: \n$$\nI(\\mathbf{x}) = \\max(0, y^* - f(\\mathbf{x})),\n$$\n where $y^*$ is currently found minimum value of $f(\\mathbf{x})$. \n To choose the next candidate for the minimum we would like to maximize the *Expected Improvement*\n$$\n EI(x) = \\mathbb{E}_f I(\\mathbf{x})\n$$\n\n 1. Express the $EI(\\mathbf{x})$ in terms $\\Phi(\\cdot)$ and $\\phi(\\cdot)$ - the pdf and cdf of the standard normal distribution $\\mathcal{N}(0, 1)$.\n 2. Assuming $\\sigma = 0$ what is the value of $EI(\\mathbf{x})$ for any value $y_i$ from the dataset?",
"_____no_output_____"
],
[
"## Building GPR model\n\nLets fit GPR model for function $f(x) = − \\cos(\\pi x) + \\sin(4\\pi x)$ in $[0, 1]$,\nwith noise $y(x) = f(x) + \\epsilon$, $\\epsilon \\sim \\mathcal{N}(0, 0.1)$. ",
"_____no_output_____"
]
],
[
[
"N = 10\nX = np.linspace(0.05, 0.95, N).reshape(-1, 1)\nY = -np.cos(np.pi * X) + np.sin(4 * np.pi * X) + \\\n np.random.normal(loc=0.0, scale=0.1, size=(N, 1))\nplt.figure(figsize=(5, 3))\nplt.plot(X, Y, '.')",
"_____no_output_____"
]
],
[
[
"#### 1. Define covariance function\n\nThe most popular kernel - RBF kernel - has 2 parameters: `variance` and `lengthscale`, $k(x, y) = \\sigma^2 \\exp\\left ( -\\dfrac{\\|x - y\\|^2}{2l^2}\\right )$,\nwhere `variance` is $\\sigma^2$, and `lengthscale` - $l$.",
"_____no_output_____"
]
],
[
[
"input_dim = 1\nvariance = 1\nlengthscale = 0.2\nkernel = GPy.kern.RBF(input_dim, variance=variance,\n lengthscale=lengthscale)",
"_____no_output_____"
]
],
[
[
"#### 2. Create GPR model",
"_____no_output_____"
]
],
[
[
"model = GPy.models.GPRegression(X, Y, kernel)\nprint(model)\nmodel.plot(figsize=(5, 3))",
"_____no_output_____"
]
],
[
[
"### Parameters of the covariance function\n\nValues of parameters of covariance function can be set like: `k.lengthscale = 0.1`.\n\nLet's change the value of `lengthscale` parameter and see how it changes the covariance function.",
"_____no_output_____"
]
],
[
[
"k = GPy.kern.RBF(1)\ntheta = np.asarray([0.2, 0.5, 1, 2, 4, 10])\nfigure, axes = plt.subplots(2, 3, figsize=(8, 4))\nfor t, ax in zip(theta, axes.ravel()):\n k.lengthscale = t\n k.plot(ax=ax)\n ax.set_ylim([0, 1])\n ax.set_xlim([-4, 4])\n ax.legend([t])",
"_____no_output_____"
]
],
[
[
"### Task\nTry to change parameters to obtain more accurate model.",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nkernel = \nmodel = ",
"_____no_output_____"
],
[
"model.Gaussian_noise.variance.fix(0.01)\nprint(model)\nmodel.plot()",
"_____no_output_____"
]
],
[
[
"### Tuning parameters of the covariance function\n\nThe parameters are tuned by maximizing likelihood. To do it just use `optimize()` method of the model.",
"_____no_output_____"
]
],
[
[
"model = GPy.models.GPRegression(X, Y, kernel)\nmodel.optimize()\nprint(model)\nmodel.plot(figsize=(5, 3))",
"_____no_output_____"
]
],
[
[
"### Noise variance\n\nNoise variance acts like a regularization in GP models. Larger values of noise variance lead to more smooth model. \nLet's check it: try to change noise variance to some large value, to some small value and see the results.\n\nNoise variance accessed like this: `model.Gaussian_noise.variance = 1`",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nmodel.Gaussian_noise.variance = \nmodel.plot(figsize=(5, 3))",
"_____no_output_____"
]
],
[
[
"Now, let's generate more noisy data and try to fit model.",
"_____no_output_____"
]
],
[
[
"N = 40\nX = np.linspace(0.05, 0.95, N).reshape(-1, 1)\nY = -np.cos(np.pi * X) + np.sin(4 * np.pi * X) + \\\n np.random.normal(loc=0.0, scale=0.5, size=(N, 1))\n\nkernel = GPy.kern.RBF(1)\nmodel = GPy.models.GPRegression(X, Y, kernel)\nmodel.optimize()\nprint(model)\nmodel.plot(figsize=(5, 3))",
"_____no_output_____"
]
],
[
[
"Now, let's fix noise variance to some small value and fit the model",
"_____no_output_____"
]
],
[
[
"kernel = GPy.kern.RBF(1)\nmodel = GPy.models.GPRegression(X, Y, kernel)\nmodel.Gaussian_noise.variance.fix(0.01)\nmodel.optimize()\nmodel.plot(figsize=(5, 3))",
"_____no_output_____"
]
],
[
[
"## Approximate multi-dimensional function",
"_____no_output_____"
]
],
[
[
"def rosenbrock(x):\n x = 0.5 * (4 * x - 2)\n y = np.sum((1 - x[:, :-1])**2 +\n 100 * (x[:, 1:] - x[:, :-1]**2)**2, axis=1)\n return y",
"_____no_output_____"
],
[
"from mpl_toolkits.mplot3d import Axes3D\nfrom matplotlib import cm\nfrom sklearn.metrics import mean_squared_error\n\ndef plot_2d_func(func, n_rows=1, n_cols=1, title=None):\n grid_size = 100\n x_grid = np.meshgrid(np.linspace(0, 1, grid_size), np.linspace(0, 1, grid_size))\n x_grid = np.hstack((x_grid[0].reshape(-1, 1), x_grid[1].reshape(-1, 1)))\n y = func(x_grid)\n fig = plt.figure(figsize=(n_cols * 6, n_rows * 6))\n ax = fig.add_subplot(n_rows, n_cols, 1, projection='3d')\n ax.plot_surface(x_grid[:, 0].reshape(grid_size, grid_size), x_grid[:, 1].reshape(grid_size, grid_size),\n y.reshape(grid_size, grid_size),\n cmap=cm.jet, rstride=1, cstride=1)\n if title is not None:\n ax.set_title(title)\n return fig",
"_____no_output_____"
]
],
[
[
"#### Here how the function looks like in 2D",
"_____no_output_____"
]
],
[
[
"fig = plot_2d_func(rosenbrock)",
"_____no_output_____"
]
],
[
[
"### Training set\nNote that it is 3-dimensional now",
"_____no_output_____"
]
],
[
[
"dim = 3\nx_train = np.random.rand(300, dim)\ny_train = rosenbrock(x_train).reshape(-1, 1)",
"_____no_output_____"
]
],
[
[
"### Task\n\nTry to approximate Rosenbrock function using RBF kernel. MSE (mean squared error) should be $<10^{-2}$.\n**Hint**: if results are not good maybe it is due to bad local minimum. You can do one of the following things:\n1. Try to use multi-start by calling `model.optimize_restarts(n_restarts)` method of the model.\n2. Constrain model parameters to some reasonable bounds. You can do it for example as follows:\n`model.Gaussian_noise.variance.constrain_bounded(0, 1)`\n",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nmodel = ",
"_____no_output_____"
],
[
"x_test = np.random.rand(3000, dim)\ny_test = rosenbrock(x_test)\ny_pr = model.predict(x_test)[0]\n\nmse = mean_squared_error(y_test.ravel(), y_pr.ravel())\nprint('\\nMSE: {}'.format(mse))",
"_____no_output_____"
]
],
[
[
"### Covariance functions\n\nShort info about covariance function can be printed using `print(k)`. ",
"_____no_output_____"
]
],
[
[
"k = GPy.kern.RBF(1)\nprint(k)",
"_____no_output_____"
]
],
[
[
"You can plot the covariance function using `plot()` method.",
"_____no_output_____"
]
],
[
[
"k.plot(figsize=(5, 3))",
"_____no_output_____"
]
],
[
[
"## More \"complex\" functions\nThe most popular covariance function is RBF. However, not all the functions can be modelled using RBF covariance function. For example, approximations of discontinuous functions will suffer from oscillations, approximation of curvy function may suffer from oversmoothing.",
"_____no_output_____"
]
],
[
[
"def heaviside(x):\n return np.asfarray(x > 0)\n\n\ndef rastrigin(x):\n \"\"\"\n Parameters\n ==========\n x : ndarray - 2D array in [0, 1]\n \n Returns\n =======\n 1D array of values of Rastrigin function\n \"\"\"\n scale = 8 # 10.24\n x = scale * x - scale / 2\n y = 10 * x.shape[1] + (x**2).sum(axis=1) - 10 * np.cos(2 * np.pi * x).sum(axis=1)\n return y",
"_____no_output_____"
],
[
"fig = plot_2d_func(rastrigin, 1, 2, title='Rastrigin function')\n\nx = np.linspace(-1, 1, 100)\ny = heaviside(x)\n\nax = fig.add_subplot(1, 2, 2)\nax.plot(x, y)\nax.set_title('Heaviside function')\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Example of oscillations\nAs you can see there are oscillations in viscinity of discontinuity because we are trying to approximate\ndiscontinuous function using infinitily smooth function.",
"_____no_output_____"
]
],
[
[
"np.random.seed(42)\nX = np.random.rand(30, 1) * 2 - 1\ny = heaviside(X)\n\nk = GPy.kern.RBF(input_dim=1, variance=1., lengthscale=1.)\n\nm = GPy.models.GPRegression(X, y, k)\nm.optimize()\nm.plot(figsize=(5, 3))\nplt.ylim([-0.2, 1.2])",
"_____no_output_____"
]
],
[
[
"#### Example of oversmoothing\nActually, the GP model only approximates trend of the function.\nAll the curves are treated as noise.\nThe knowledge about this (in fact there is some repeated structure) should be incorporated into the model via kernel function.",
"_____no_output_____"
]
],
[
[
"np.random.seed(42)\n\nX = np.random.rand(300, 2)\ny = rastrigin(X)\n\nk = GPy.kern.RBF(input_dim=2)\n\nm = GPy.models.GPRegression(X, y.reshape(-1, 1), k)\nm.optimize()\nfig = plot_2d_func(lambda x: m.predict(x)[0])",
"_____no_output_____"
]
],
[
[
"### Covariance functions in GPy\n\nPopular covariance functions: `Exponential`, `Matern32`, `Matern52`, `RatQuad`, `Linear`, `StdPeriodic`. \n\n* Exponential:\n$$\nk(x, x') = \\sigma^2 \\exp \\left (-\\frac{r}{l} \\right), \\quad r = \\|x - x'\\|\n$$\n\n* Matern32\n$$\nk(x, x') = \\sigma^2 \\left (1 + \\sqrt{3}\\frac{r}{l} \\right )\\exp \\left (-\\sqrt{3}\\frac{r}{l} \\right )\n$$\n\n* Matern52\n$$\nk(x, x') = \\sigma^2 \\left (1 + \\sqrt{5}\\frac{r}{l} + \\frac{5}{3}\\frac{r^2}{l^2} \\right ) \\exp \\left (-\\sqrt{5}\\frac{r}{l} \\right )\n$$\n\n* RatQuad\n$$\nk(x, x') = \\left ( 1 + \\frac{r^2}{2\\alpha l^2}\\right )^{-\\alpha}\n$$\n\n* Linear\n$$\nk(x, x') = \\sum_i \\sigma_i^2 x_i x_i'\n$$\n\n* Poly\n$$\nk(x, x') = \\sigma^2 (x^T x' + c)^d\n$$\n\n* StdPeriodic\n$$\nk(x, x') = \\sigma^2 \\exp\\left ( -2 \\frac{\\sin^2(\\pi r)}{l^2}\\right )\n$$",
"_____no_output_____"
]
],
[
[
"covariance_functions = [GPy.kern.Exponential(1), GPy.kern.Matern32(1),\n GPy.kern.RatQuad(1), GPy.kern.Linear(1),\n GPy.kern.Poly(1), GPy.kern.StdPeriodic(1)]\nfigure, axes = plt.subplots(2, 3, figsize=(9, 6))\naxes = axes.ravel()\nfor i, k in enumerate(covariance_functions):\n k.plot(ax=axes[i])\n axes[i].set_title(k.name)\nfigure.tight_layout()",
"_____no_output_____"
]
],
[
[
"## Combination of covariance functions\n\n* Sum of covariance function is a valid covariance function:\n\n$$\nk(x, x') = k_1(x, x') + k_2(x, x')\n$$\n\n* Product of covariance functions is a valid covariance funciton:\n$$\nk(x, x') = k_1(x, x') k_2(x, x')\n$$\n\n### Combinations of covariance functions in GPy\n\nIn GPy to combine covariance functions you can just use operators `+` and `*`.",
"_____no_output_____"
],
[
"Let's plot some of the combinations",
"_____no_output_____"
]
],
[
[
"covariance_functions = [GPy.kern.Linear(input_dim=1), GPy.kern.StdPeriodic(input_dim=1), GPy.kern.RBF(input_dim=1, lengthscale=1)]\noperations = {'+': lambda x, y: x + y, '*': lambda x, y: x * y}\n\nfigure, axes = plt.subplots(len(operations), len(covariance_functions), figsize=(9, 6))\n\nimport itertools\naxes = axes.ravel()\ncount = 0\nfor j, base_kernels in enumerate(itertools.combinations(covariance_functions, 2)):\n for k, (op_name, op) in enumerate(operations.items()):\n kernel = op(base_kernels[0], base_kernels[1])\n kernel.plot(ax=axes[count])\n axes[count].set_title('{} {} {}'.format(base_kernels[0].name, op_name, base_kernels[1].name),\n fontsize=14)\n count += 1\nfigure.tight_layout()",
"_____no_output_____"
]
],
[
[
"### Additive kernels\n\nOne of the popular approach to model the function of interest is\n$$\nf(x) = \\sum_{i=1}^d f_i(x_i) + \\sum_{i < j} f_{ij}(x_i, x_j) + \\ldots\n$$\n\n**Example**: $\\quad f(x_1, x_2) = f_1(x_1) + f_2(x_2)$ \nTo model it using GP use additive kernel $\\quad k(x, y) = k_1(x_1, y_1) + k_2(x_2, y_2)$.\n\nMore general - add kernels each depending on subset of inputs\n$$\nk(x, y) = k_1(x, y) + \\ldots + k_D(x, y),\n$$\nwhere, for example, $k_1(x, x') = k_1(x_1, x_1'), \\; k_2(x, x') = k_2((x_1, x_3), (x_1', x_3'))$, etc.\n\nHere is an example of ${\\rm RBF}(x_1) + {\\rm RBF}(x_2)$",
"_____no_output_____"
]
],
[
[
"k1 = GPy.kern.RBF(1, active_dims=[0])\nk2 = GPy.kern.RBF(1, active_dims=[1])\n\nkernel = k1 + k2\n\nx = np.meshgrid(np.linspace(-3, 3, 50), np.linspace(-3, 3, 50))\nx = np.hstack((x[0].reshape(-1, 1), x[1].reshape(-1, 1)))\nz = kernel.K(x, np.array([[0, 0]]))\n\nfrom mpl_toolkits.mplot3d import Axes3D\nfrom matplotlib import cm\n\nfigure = plt.figure()\nax = figure.add_subplot(111, projection='3d')\nax.plot_surface(x[:, 0].reshape(50, 50), x[:, 1].reshape(50, 50), z.reshape(50, 50), cmap=cm.jet)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Kernels on arbitrary types of objects\n\nKernels can be defined over all types of data structures: text, images, matrices, graphs, etc. You just need to define similarity between objects.\n\n#### Kernels on categorical data\n\n* Represent your categorical variable as a by a one-of-k encoding: $\\quad x = (x_1, \\ldots, x_k)$.\n* Use RBF kernel with `ARD=True`: $\\quad k(x , x') = \\sigma^2 \\prod_{i = 1}^k\\exp{\\left ( -\\dfrac{(x_i - x_i')^2}{\\sigma_i^2} \\right )}$. The lengthscale will now encode whether the rest of the function changes.\n* Short lengthscales for categorical variables means your model is not sharing any information between data of different categories. ",
"_____no_output_____"
],
[
"## 2 Sampling from GP\n\nSo, you have defined some complex kernel.\nYou can plot it to see how it looks and guess what kind of functions it can approximate.\nAnother way to do it is to actually generate random functions using this kernel.\n\nGP defines distribution over functions, which is defined by its *mean function* $m(x)$ and *covariance function* $k(x, y)$: for any set $\\mathbf{x}_1, \\ldots, \\mathbf{x}_N \\in \\mathbb{R}^d \\rightarrow$ $\\left (f(\\mathbf{x}_1), \\ldots, f(\\mathbf{x}_N) \\right ) \\sim \\mathcal{N}(\\mathbf{m}, \\mathbf{K})$,\nwhere $\\mathcal{m} = (m(\\mathbf{x}_1, \\ldots, \\mathbf{x}_N)$, $\\mathbf{K} = \\|k(\\mathbf{x}_i, \\mathbf{x}_j)\\|_{i,j=1}^N$.\n\nSampling procedure:\n\n1. Generate set of points $\\mathbf{x}_1, \\ldots, \\mathbf{x}_N$.\n2. Calculate mean and covariance matrix $\\mathcal{m} = (m(\\mathbf{x}_1, \\ldots, \\mathbf{x}_N)$, $\\mathbf{K} = \\|k(\\mathbf{x}_i, \\mathbf{x}_j)\\|_{i,j=1}^N$.\n3. Generate vector from multivariate normal distribution $\\mathcal{N}(\\mathbf{m}, \\mathbf{K})$.\n\nBelow try to change RBF kernel to some other kernel and see the results.",
"_____no_output_____"
]
],
[
[
"k = GPy.kern.RBF(input_dim=1, lengthscale=0.3)\n\nX = np.linspace(0, 5, 500).reshape(-1, 1)\n\nmu = np.zeros(500)\nC = k.K(X, X)\n\nZ = np.random.multivariate_normal(mu, C, 3)\n\nplt.figure()\nfor i in range(3):\n plt.plot(X, Z[i, :])",
"_____no_output_____"
]
],
[
[
"### Task\n\nBuild a GP model that predicts airline passenger counts on international flights.",
"_____no_output_____"
]
],
[
[
"!wget https://github.com/adasegroup/ML2020_seminars/raw/master/seminar11/data/airline.npz",
"_____no_output_____"
],
[
"data = np.load('airline.npz')\n\nX = data['X']\ny = data['y']\n\ntrain_indices = list(range(70)) + list(range(90, 129))\ntest_indices = range(70, 90)\nX_train = X[train_indices]\ny_train = y[train_indices]\n\nX_test = X[test_indices]\ny_test = y[test_indices]\n\nplt.figure(figsize=(5, 3))\nplt.plot(X_train, y_train, '.')",
"_____no_output_____"
]
],
[
[
"You need to obtain something like this\n\n<img src=https://github.com/adasegroup/ML2020_seminars/raw/master/seminar11/imgs/airline_result.png>",
"_____no_output_____"
]
],
[
[
"def plot_model(X, y, model):\n x = np.linspace(1948, 1964, 400).reshape(-1, 1)\n prediction_mean, prediction_var = model.predict(x)\n prediction_std = np.sqrt(prediction_var).ravel()\n prediction_mean = prediction_mean.ravel()\n \n plt.figure(figsize=(5, 3))\n plt.plot(X, y, '.', label='Train data')\n plt.plot(x, prediction_mean, label='Prediction')\n plt.fill_between(x.ravel(), prediction_mean - prediction_std, prediction_mean + prediction_std, alpha=0.3)",
"_____no_output_____"
]
],
[
[
"#### Let's try RBF kernel",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nk_rbf = ",
"_____no_output_____"
]
],
[
[
"As you can see below it doesn't work ;(",
"_____no_output_____"
]
],
[
[
"model = GPy.models.GPRegression(X, y, k_rbf)\nmodel.optimize()\nprint(model)\nplot_model(X_train, y_train, model)",
"_____no_output_____"
]
],
[
[
"We will try to model this data set using 3 additive components: trend, seasonality and noise. \nSo, the kernel should be a sum of 3 kernels: \n`kernel = kernel_trend + kernel_seasonality + kernel_noise`\n\n#### Let's first try to model trend\n\nTrend is almost linear with some small nonlinearity, so you can use sum of linear kernel with some other which gives this small nonlinearity.",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nk_trend = ",
"_____no_output_____"
],
[
"model = GPy.models.GPRegression(X, y, k_trend)\nmodel.optimize()\nprint(model)\nplot_model(X_train, y_train, model)",
"_____no_output_____"
]
],
[
[
"#### Let's model periodicity\nJust periodic kernel will not work (why?).\nTry to use product of periodic kernel with some other kernel (or maybe 2 other kernels).\nNote that the amplitude increases with x.",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nk_trend = \nk_seasonal = ",
"_____no_output_____"
],
[
"kernel = k_trend + k_seasonal\nmodel = GPy.models.GPRegression(X, y, kernel)\nmodel.optimize()\nprint(model)\nplot_model(X_train, y_train, model)",
"_____no_output_____"
]
],
[
[
"#### Let's add noise model\nThe dataset is heteroscedastic, i.e. noise variance depends on x: it increases linearly with x.\nNoise can be modeled using `GPy.kern.White(1)`, but it assumes that noise variance is the same at every x.\nBy what kernel it should be multiplied?",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nk_trend = \nk_periodicity = \nk_noise = ",
"_____no_output_____"
],
[
"kernel = k_trend + k_periodicity + k_noise\nmodel = GPy.models.GPRegression(X, y, kernel)\nmodel.optimize()\nprint(model)\nplot_model(X_train, y_train, model)",
"_____no_output_____"
]
],
[
[
"# Automatic covariance structure search\nWe can construct kernel is automatic way.\nHere is our data set (almost the same)",
"_____no_output_____"
]
],
[
[
"idx_test = np.where((X[:,0] > 1957))[0]\nidx_train = np.where((X[:,0] <= 1957))[0]\nX_train = X[idx_train]\ny_train = y[idx_train]\n\nX_test = X[idx_test]\ny_test = y[idx_test]\n\nplt.figure(figsize=(7, 5))\nplt.plot(X_train, y_train, '.', color='red');\nplt.plot(X_test, y_test, '.', color='green');",
"_____no_output_____"
],
[
"def plot_model_learned(X, y, train_idx, test_idx, model):\n prediction_mean, prediction_var = model.predict(X)\n prediction_std = np.sqrt(prediction_var).ravel()\n prediction_mean = prediction_mean.ravel()\n \n plt.figure(figsize=(7, 5))\n plt.plot(X, y, '.')\n plt.plot(X[train_idx], y[train_idx], '.', color='green')\n plt.plot(X, prediction_mean, color='red')\n plt.fill_between(X.ravel(), prediction_mean - prediction_std, prediction_mean + prediction_std, alpha=0.3)",
"_____no_output_____"
]
],
[
[
"## Expressing Sturcture Through Kernels",
"_____no_output_____"
],
[
"For example:\n\n$$\n\\underbrace{\\text{RBF}\\times\\text{Lin}}_\\text{increasing trend} + \\underbrace{\\text{RBF}\\times\\text{Per}}_\\text{varying-amplitude periodic} + \\underbrace{\\text{RBF}}_\\text{residual}\n$$",
"_____no_output_____"
],
[
"## Greedy Searching for the Optimum Kernel Combination",
"_____no_output_____"
],
[
"One can wonder: how to automatically search the kernel structure? We can optimize some criteria, which balance between a loss function value and the complexity of the model.\nReasinobale candidate for this is BIC-criteria:\n\n$$\nBIC = - 2. \\text{Log-Liklihood} + m \\cdot\\log{n}\n$$\n\nwhere $n$ sample size and $m$ number of the parameters.\n\nHowever, the procedure of fitting Gaussian Process is quite expensive $O(n^3)$. Hence, instead of the combinatorial search through all possible combinations, we grow the kernel structure greedy.\n\nYou can find more details at the https://github.com/jamesrobertlloyd/gp-structure-search. For now, we present toy-example algorithm.",
"_____no_output_____"
],
[
"Consider the set of operations:\n\n$$\n\\text{Algebra: } +,\\times\n$$\n\nand the set of basic kernels:\n\n$$\n\\text{Kernels: } \\text{Poly}, \\text{RBF}, \\text{Periodic}\n$$",
"_____no_output_____"
],
[
"For each level we select extenstion of our current kernel with the lowest BIC. This is an example of the possible kernel grow process (mark notes the lowest BIC at the level):",
"_____no_output_____"
],
[
"<img src='https://github.com/adasegroup/ML2020_seminars/raw/master/seminar11/imgs/gp.png'>",
"_____no_output_____"
],
[
"### Task*\nImplement function that trains a model with given kernel and dataset, calculates and returns BIC\nThe log-lilkelihood of the model can be calculated using `model.log_likelihood()` method,\nnumber of parameters of the model you can get via `len(model.param_array)`.",
"_____no_output_____"
]
],
[
[
"def train_model_get_bic(X_train, y_train, kernel, num_restarts=1):\n '''\n Input:\n X_train: numpy array of train features, n*d (d>=1)\n y_train: numpy array n*1\n kernel: GPy object kern\n num_restars: number of the restarts of the optimization routine\n Output:\n bic value\n '''\n kernel = kernel.copy()\n \n ######## Your code here ########\n\n return bic ",
"_____no_output_____"
]
],
[
[
"Here is a utility function which take list of kernels and operations between them, calculates all product kernels\nand returns a list of them.\nAfter that we need only take sum of the kernels from this list.",
"_____no_output_____"
]
],
[
[
"def _get_all_product_kernels(op_list, kernel_list):\n '''\n Find product pairs and calculate them.\n For example, if we are given expression:\n K = k1 * k2 + k3 * k4 * k5\n the function will calculate all the product kernels\n k_mul_1 = k1 * k2\n k_mul_2 = k3 * k4 * k5\n and return list [k_mul_1, k_mul_2].\n '''\n product_index = np.where(np.array(op_list) == '*')[0]\n if len(product_index) == 0:\n return kernel_list\n\n product_index = product_index[0]\n product_kernel = kernel_list[product_index] * kernel_list[product_index + 1]\n \n if len(op_list) == product_index + 1:\n kernel_list_copy = kernel_list[:product_index] + [product_kernel]\n op_list_copy = op_list[:product_index]\n else:\n kernel_list_copy = kernel_list[:product_index] + [product_kernel] + kernel_list[product_index + 2:]\n op_list_copy = op_list[:product_index] + op_list[product_index + 1:]\n \n return _get_all_product_kernels(op_list_copy, kernel_list_copy)",
"_____no_output_____"
]
],
[
[
"### Task*\n\nThis is the main class, you need to implement several methods inside\n1. method `init_kernel()` - this function constructs initial model, i.e. the model with one kernel. You need just iterate through the list of base kernels and choose the best one according to BIC\n2. method `grow_level()` - this function adds new level. You need to iterate through all base kernels and all operations,\napply each operation to the previously constructed kernel and each base kernel (use method `_make_kernel()` for this) and then choose the best one according to BIC.",
"_____no_output_____"
]
],
[
[
"class GreedyKernel:\n '''\n Class for greedy growing kernel structure\n '''\n def __init__(self, algebra, base_kernels):\n self.algebra = algebra\n self.base_kernels = base_kernels\n self.kernel = None\n self.kernel_list = []\n self.op_list = []\n self.str_kernel = None\n \n def _make_kernel(self, op_list, kernel_list):\n '''\n Sumation in kernel experssion\n '''\n kernels_to_sum = _get_all_product_kernels(op_list, kernel_list)\n new_kernel = kernels_to_sum[0]\n for k in kernels_to_sum[1:]:\n new_kernel = new_kernel + k\n return new_kernel\n \n def init_kernel(self, X_train, y_train):\n '''\n Initialization of first kernel\n '''\n best_kernel = None\n \n ###### Your code here ######\n\n # You need just iterate through the list of base kernels and choose the best one according to BIC\n # save the kernel in `best_kernel` variable\n \n # base kernels are given by self.base_kernels --- list of kernel objects\n \n \n \n \n ############################\n \n assert best_kernel is not None\n \n self.kernel_list.append(best_kernel)\n self.str_kernel = str(best_kernel.name)\n \n def grow_level(self, X_train, y_train):\n '''\n Select optimal extension of current kernel \n '''\n \n best_kernel = None # should be kernel object\n best_op = None # should be operation name, i.e. \"+\" or \"*\"\n \n ###### Your code here ######\n \n # You need to iterate through all base kernels and all operations,\n # apply each operation to the previously constructed kernel and each base kernel\n # (use method `_make_kernel()` for this) and then choose the best one according to BIC.\n \n # base kernels are given by self.base_kernels --- list of kernel objects\n # operations are given by self.algebra --- dictionary:\n # {\"+\": lambda x, y: x + y\n # \"*\": lambda x, y: x * y}\n\n # best_kernel - kernel object, store in this variable the best found kernel\n # best_op - '+' or '*', store in this variable the best found operation\n \n \n \n \n ############################\n\n assert best_kernel is not None\n assert best_op is not None\n \n self.kernel_list.append(best_kernel)\n self.op_list.append(best_op)\n \n new_kernel = self._make_kernel(self.op_list, self.kernel_list)\n str_new_kernel = '{} {} {}'.format(self.str_kernel, best_op, best_kernel.name)\n \n return new_kernel, str_new_kernel\n \n def grow_tree(self, X_train, y_train, max_depth):\n '''\n Greedy kernel growing\n '''\n if self.kernel == None:\n self.init_kernel(X_train, y_train)\n \n for i in range(max_depth):\n self.kernel, self.str_kernel = self.grow_level(X_train, y_train)\n print(self.str_kernel)\n \n def fit_model(self, X_train, y_train, kernel, num_restarts=1):\n model = GPy.models.GPRegression(X_train, y_train, kernel)\n model.optimize_restarts(num_restarts, verbose=False)\n return model",
"_____no_output_____"
]
],
[
[
"Now let us define the algebra and list of base kernels.\nTo make learning process more robust we constrain some parameters of the kernels to lie within\nsome reasonable intervals",
"_____no_output_____"
]
],
[
[
"# operations under kernels:\n\nalgebra = {'+': lambda x, y: x + y,\n '*': lambda x, y: x * y\n }\n\n# basic kernels list:\npoly_kern = GPy.kern.Poly(input_dim=1, order=1)\n\nperiodic_kern = GPy.kern.StdPeriodic(input_dim=1)\nperiodic_kern.period.constrain_bounded(1e-2, 1e1)\nperiodic_kern.lengthscale.constrain_bounded(1e-2, 1e1)\n\nrbf_kern = GPy.kern.RBF(input_dim=1)\nrbf_kern.lengthscale.constrain_bounded(1e-2, 1e1)\n\nbias_kern = GPy.kern.Bias(1)\n\nkernels_list = [poly_kern, periodic_kern, rbf_kern]",
"_____no_output_____"
]
],
[
[
"Let's train the model.\nYou should obtain something which is more accurate than the trend model ;)",
"_____no_output_____"
]
],
[
[
"GK = GreedyKernel(algebra, kernels_list)\nGK.grow_tree(X_train, y_train, 4)\nmodel = GK.fit_model(X_train, y_train, GK.kernel)\nplot_model_learned(X, y, idx_train, idx_test, model)",
"_____no_output_____"
]
],
[
[
"## Bonus Task\nTry to approximate rastrigin function",
"_____no_output_____"
]
],
[
[
"fig = plot_2d_func(rastrigin)",
"_____no_output_____"
]
],
[
[
"### Training set",
"_____no_output_____"
]
],
[
[
"np.random.rand(42)\nx_train = np.random.rand(200, 2)\ny_train = rastrigin(x_train)",
"_____no_output_____"
]
],
[
[
"#### Hint: you can constrain parameters of the covariance functions, for example\n`model.std_periodic.period.constrain_bounded(0, 0.2)`.",
"_____no_output_____"
]
],
[
[
"######## Your code here ########\nmodel = ",
"_____no_output_____"
],
[
"print(model)\n\nx_test = np.random.rand(1000, 2)\ny_test = rastrigin(x_test)\ny_pr = model.predict(x_test)[0]\n\nmse = mean_squared_error(y_test.ravel(), y_pr.ravel())\nprint('MSE: {}'.format(mse))\n\nfig = plot_2d_func(lambda x: model.predict(x)[0])",
"_____no_output_____"
]
],
[
[
"# Appendix: Gaussian Process Classification",
"_____no_output_____"
],
[
"### Classification\n\nA data set $\\left (X, \\mathbf{y} \\right ) = \\left \\{ (x_i, y_i), x_i \\in \\mathbb{R}^d, y_i \\in \\{+1, -1\\} \\right \\}_{i = 1}^N$ is given. \n\nAssumption:\n$$\np(y = +1 \\; | \\; x) = \\sigma(f(x)) = \\pi(x),\n$$\nwhere latent function $f(x)$ is a Gaussian Processes.",
"_____no_output_____"
],
[
"We need to produce a probabilistic prediction\n$$\n\\pi_* = p(y_* \\; | \\; X, \\mathbf{y}, x_*) = \\int \\sigma(f_*) p(f_* \\; | \\; X, \\mathbf{y}, x_*) df_*,\n$$\n$$\np(f_* \\; | \\; X, \\mathbf{y}, x_*) = \\int p(f_* \\; | \\; X, x_*, \\mathbf{f}) p(\\mathbf{f} \\; | \\; X, \\mathbf{y}) d\\mathbf{f},\n$$\nwhere $p(\\mathbf{f} \\; |\\; X, \\mathbf{y}) = \\dfrac{p(\\mathbf{y} | X, \\mathbf{f}) p(\\mathbf{f} | X)}{p(\\mathbf{y} | X)}$ is the posterior over the latent variables.\n\nBoth integrals are intractable.\n\nUse approximation technique like Laplace approximation or Expectation Propagation.",
"_____no_output_____"
]
],
[
[
"from matplotlib import cm\n\ndef cylinder(x):\n y = (1 / 7.0 - (x[:, 0] - 0.5)**2 - (x[:, 1] - 0.5)**2) > 0\n return y\n\nnp.random.seed(42)\nX = np.random.rand(40, 2)\ny = cylinder(X)\n\nx_grid = np.meshgrid(np.linspace(0, 1, 100), np.linspace(0, 1, 100))\ny_grid = cylinder(np.hstack((x_grid[0].reshape(-1, 1), x_grid[1].reshape(-1, 1)))).reshape(x_grid[0].shape)\n\npositive_idx = y == 1\nplt.figure(figsize=(5, 3))\nplt.plot(X[positive_idx, 0], X[positive_idx, 1], '.', markersize=10, label='Positive')\nplt.plot(X[~positive_idx, 0], X[~positive_idx, 1], '.', markersize=10, label='Negative')\nim = plt.contour(x_grid[0], x_grid[1], y_grid, 10, cmap=cm.hot)\nplt.colorbar(im)\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"kernel = GPy.kern.RBF(2, variance=1., lengthscale=0.2, ARD=True)\n\nmodel = GPy.models.GPClassification(X, y.reshape(-1, 1), kernel=kernel)\nmodel.optimize()\nprint(model)\n\n\ndef plot_model_2d(model):\n\n model.plot(levels=40, resolution=80, plot_data=False, figsize=(5, 3))\n plt.plot(X[positive_idx, 0], X[positive_idx, 1], '.', markersize=10, label='Positive')\n plt.plot(X[~positive_idx, 0], X[~positive_idx, 1], '.', markersize=10, label='Negative')\n plt.legend()\n plt.show()\n \nplot_model_2d(model)",
"_____no_output_____"
]
],
[
[
"Let's change lengthscale to some small value",
"_____no_output_____"
]
],
[
[
"model.rbf.lengthscale = [0.05, 0.05]\nplot_model_2d(model)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
cb71c929de4d4acd2e1979b387c547a8ba23f13c | 598,021 | ipynb | Jupyter Notebook | DebayeringTests.ipynb | newsch/wonky-bobbin | 325c47914bc2f1348af58101dc1c0592c400fc63 | [
"MIT"
]
| null | null | null | DebayeringTests.ipynb | newsch/wonky-bobbin | 325c47914bc2f1348af58101dc1c0592c400fc63 | [
"MIT"
]
| null | null | null | DebayeringTests.ipynb | newsch/wonky-bobbin | 325c47914bc2f1348af58101dc1c0592c400fc63 | [
"MIT"
]
| null | null | null | 1,377.928571 | 163,314 | 0.945841 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom PIL import Image\nimport scipy\nfrom scipy.signal import convolve\nfrom scipy import ndimage\nimport getBayer\n% matplotlib inline\nimport io\nimport time\nimport copy\nfrom numpy.lib.stride_tricks import as_strided",
"_____no_output_____"
],
[
"Im = getBayer.getBayer('pic2.jpeg')\nbayer = getBayer.bayerGrid \ntestIm = copy.deepcopy(Im)\n\nplt.imshow(Im/255)",
"_____no_output_____"
],
[
"# trying out a method where you interpolate along a direction based on which seems most constant\n(m,n) = testIm[:,:,0].shape\nR = testIm[:,:,0].astype(np.int32)\nG = testIm[:,:,1].astype(np.int32)\nB = testIm[:,:,2].astype(np.int32)\nG[0:3,0:3]",
"_____no_output_____"
],
[
"G_n = copy.deepcopy(G)\nR_n = copy.deepcopy(R)\nB_n = copy.deepcopy(B)",
"_____no_output_____"
]
],
[
[
"I'm basing the below pixel patterning algorithm on the one detailed in Chuan-kai Lin's website https://sites.google.com/site/chklin/demosaic",
"_____no_output_____"
]
],
[
[
"\nfor i in range(2,m -3):\n for j in range(2,n-3):\n if G[i,j] == 0:\n if B[i,j] == 0: # if this isn't a blue or green pixel, its a red one\n M = R\n elif R[i,j] == 0:\n M = B\n north = 2*abs(M[i,j] -M[i-2,j]) + abs(G[i-1,j] - G[i+1,j])\n east = 2*abs(M[i,j] -M[i,j+2]) + abs(G[i,j+1] - G[i,j-1])\n south = 2*abs(M[i,j] -M[i+2,j]) + abs(G[i-1,j] - G[i+1,j])\n west = 2*abs(M[i,j] -M[i,j-2]) + abs(G[i,j+1] - G[i,j-1])\n# print(north)\n grads = [north, east, south, west]\n if min(grads) == north:\n G_n[i,j] = (3*G[i-1,j] + G[i+1,j] + M[i,j] -M[i-2,j])/4\n elif min(grads) == east:\n G_n[i,j] = (3*G[i,j+1] + G[i,j-1] + M[i,j] -M[i,j+2])/4\n elif min(grads) == south:\n G_n[i,j] = (3*G[i+1,j] + G[i-1,j] + M[i,j] -M[i+2,j])/4\n elif min(grads) == west:\n G_n[i,j] = (3*G[i,j-1] + G[i,j+1] + M[i,j] -M[i,j-2])/4\n \n ",
"_____no_output_____"
],
[
"temp = testIm.copy()\ntemp[:,:,1] = G_n[:,:]\n\nplt.imshow(temp/255)",
"_____no_output_____"
],
[
"plt.imshow(G_n[100:200,100:200])",
"_____no_output_____"
],
[
"#make a hue gradient function\ndef hueGrad(c1, c2, c3, p1, p3):\n if c1< c2 and c2 < c3 or c3< c2 and c2 < c1: \n return p1 + (p3-p1)*(c2 - c1)/(c3-c1)\n else:\n return (p1+p3)/2 + (2*c2 + c1 + c3)/4",
"_____no_output_____"
]
],
[
[
"I'm not following the algorithm I was before here, I'm mostly just doing bilinear interpolation",
"_____no_output_____"
]
],
[
[
"\nfor i in range(1,m-2):\n for j in range(1,n-2):\n if R[i,j] == 0 and B[i,j] ==0: #Green sencel location\n if R[i+1,j] == 0: # this means that the next pixel in the bayer grid directly below is a blue one\n R_n[i,j] = abs(R[i,j-1] + R[i,j+1])//2\n B_n[i,j] = abs(B[i-1,j] + B[i+1,j])//2\n# R_n[i,j] = hueGrad(G_n[i,j-1], G_n[i,j], G_n[i,j+1], R[i,j-1], R[i,j+1]) \n# B_n[i,j] = hueGrad(G_n[i-1,j], G_n[i,j], G_n[i+1,j], B[i-1,j], B[i+1,j])\n elif B[i+1,j] == 0:\n R_n[i,j] = abs(R[i-1,j] + R[i+1,j])//2 \n B_n[i,j] = abs(B[i,j-1] + B[i,j+1])//2\n# B_n[i,j] = hueGrad(G_n[i,j-1], G_n[i,j], G_n[i,j+1], B[i,j-1], B[i,j+1]) \n# R_n[i,j] = hueGrad(G_n[i-1,j], G_n[i,j], G_n[i+1,j], R[i-1,j], R[i+1,j])\n elif B[i,j] == 0 and G[i,j] ==0: # the sencel location is a Blue one\n NE = abs(B[i-1,j+1] - B[i+1, j-1])\n NW = abs(B[i-1, j-1] - B[i+1,j+1])\n if NW > 2*NE:\n B_n[i,j] = abs(B[i-1,j+1] + B[i+1, j-1])//2\n elif NE > 2*NW: \n B_n[i,j] = abs(B[i-1, j-1] + B[i+1,j+1])//2\n else: \n B_n[i,j] = abs(B[i-1, j-1] + B[i+1,j+1] + B[i-1,j+1] + B[i+1, j-1])//4\n \n elif R[i,j] == 0 and G[i,j]==0: #Sencel in this location is Red \n NE = abs(R[i-1,j+1] - R[i+1, j-1])\n NW = abs(R[i-1, j-1] - R[i+1,j+1])\n if NW > 2*NE:\n R_n[i,j] = abs(R[i-1,j+1] + R[i+1, j-1])//2\n elif NE > 2*NW: \n R_n[i,j] = abs(R[i-1, j-1] + R[i+1,j+1])//2\n else: \n R_n[i,j] = abs(R[i-1, j-1] + R[i+1,j+1] + R[i-1,j+1] + R[i+1, j-1])//4",
"_____no_output_____"
],
[
"temp[:,:,0] = R_n[:,:]\ntemp[:,:,2] = B_n[:,:]\n\nplt.imshow(temp/255)",
"_____no_output_____"
],
[
"# plt.imshow(testIm[100:120,100:120,0]/255, cmap = 'gray')\nB_n[50:60,50:60]",
"_____no_output_____"
],
[
"# B[50:60,50:60]\nB_n.shape",
"_____no_output_____"
],
[
"plt.imshow(temp[850:880,870:900]/255)",
"_____no_output_____"
],
[
"rgbIm = getBayer.get_rgb_array('pic2.jpeg')\nplt.imshow(rgbIm/255)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb71ce42fd12a0adaf137fe9df8f906747e75f77 | 1,514 | ipynb | Jupyter Notebook | numpy/np.random.ipynb | ybdesire/machinelearning | 0224746332e1085336e0b02e0ca3b11d74bd9a91 | [
"MIT"
]
| 30 | 2017-02-28T13:52:58.000Z | 2022-03-24T10:28:43.000Z | numpy/np.random.ipynb | ybdesire/machinelearning | 0224746332e1085336e0b02e0ca3b11d74bd9a91 | [
"MIT"
]
| null | null | null | numpy/np.random.ipynb | ybdesire/machinelearning | 0224746332e1085336e0b02e0ca3b11d74bd9a91 | [
"MIT"
]
| 17 | 2017-03-03T12:38:04.000Z | 2022-03-11T01:53:20.000Z | 17.811765 | 71 | 0.484808 | [
[
[
"import numpy as np\n\n# random float [0,-1)\nrandom01 = np.random.random_sample()\nprint(random01)",
"0.4249824904344124\n"
],
[
"# 5 number list random float [0,-1)\nrandom01 = np.random.random_sample(5)\nprint(random01)",
"[ 0.6056018 0.35899653 0.87123477 0.60738734 0.03525969]\n"
],
[
"# get random int<5, (0,1,2,3,4)\nr = np.random.randint(5)\nprint(r)",
"3\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
cb71d0b8799c190d4db10ab077ff7644b5338b02 | 26,795 | ipynb | Jupyter Notebook | Models/m5-validation-detrend-d2d-predict(1).ipynb | esmeluo/kaggle_m5_accuracy | edbee6ddb83bd35871f9ea06e6c23b8c9131f9a7 | [
"MIT"
]
| null | null | null | Models/m5-validation-detrend-d2d-predict(1).ipynb | esmeluo/kaggle_m5_accuracy | edbee6ddb83bd35871f9ea06e6c23b8c9131f9a7 | [
"MIT"
]
| null | null | null | Models/m5-validation-detrend-d2d-predict(1).ipynb | esmeluo/kaggle_m5_accuracy | edbee6ddb83bd35871f9ea06e6c23b8c9131f9a7 | [
"MIT"
]
| null | null | null | 37.267038 | 1,266 | 0.534727 | [
[
[
"# General imports\nimport numpy as np\nimport pandas as pd\nimport os, sys, gc, time, warnings, pickle, psutil, random\n\n# custom imports\nfrom multiprocessing import Pool # Multiprocess Runs\n\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"########################### Helpers\n#################################################################################\n## Seeder\n# :seed to make all processes deterministic # type: int\ndef seed_everything(seed=0):\n random.seed(seed)\n np.random.seed(seed)\n\n \n## Multiprocess Runs\ndef df_parallelize_run(func, t_split):\n num_cores = np.min([N_CORES,len(t_split)])\n pool = Pool(num_cores)\n df = pd.concat(pool.map(func, t_split), axis=1)\n pool.close()\n pool.join()\n return df",
"_____no_output_____"
],
[
"########################### Helper to load data by store ID\n#################################################################################\n# Read data\ndef get_data_by_store(store):\n \n # Read and contact basic feature\n df = pd.concat([pd.read_pickle(BASE),\n pd.read_pickle(PRICE).iloc[:,2:],\n pd.read_pickle(CALENDAR).iloc[:,2:]],\n axis=1)\n \n # Leave only relevant store\n df = df[df['store_id']==store]\n\n # With memory limits we have to read \n # lags and mean encoding features\n # separately and drop items that we don't need.\n # As our Features Grids are aligned \n # we can use index to keep only necessary rows\n # Alignment is good for us as concat uses less memory than merge.\n df2 = pd.read_pickle(MEAN_ENC)[mean_features]\n df2 = df2[df2.index.isin(df.index)]\n \n df3 = pd.read_pickle(LAGS).iloc[:,3:]\n df3 = df3[df3.index.isin(df.index)]\n \n df = pd.concat([df, df2], axis=1)\n del df2 # to not reach memory limit \n \n df = pd.concat([df, df3], axis=1)\n del df3 # to not reach memory limit \n \n # Create features list\n features = [col for col in list(df) if col not in remove_features]\n df = df[['id','d',TARGET]+features]\n \n # Skipping first n rows\n df = df[df['d']>=START_TRAIN].reset_index(drop=True)\n \n return df, features\n\n# Recombine Test set after training\ndef get_base_test():\n base_test = pd.DataFrame()\n\n for store_id in STORES_IDS:\n temp_df = pd.read_pickle('test_'+store_id+'.pkl')\n temp_df['store_id'] = store_id\n base_test = pd.concat([base_test, temp_df]).reset_index(drop=True)\n \n return base_test\n# -------------------------------------\n# def get_base_valid():\n# base_test = pd.DataFrame()\n\n# for store_id in STORES_IDS:\n# temp_df = pd.read_pickle('valid_'+store_id+'.pkl')\n# temp_df['store_id'] = store_id\n# base_test = pd.concat([base_test, temp_df]).reset_index(drop=True)\n \n# return base_test\n# -------------------------------------\n\n########################### Helper to make dynamic rolling lags\n#################################################################################\ndef make_lag(LAG_DAY):\n lag_df = base_test[['id','d',TARGET]]\n col_name = 'sales_lag_'+str(LAG_DAY)\n lag_df[col_name] = lag_df.groupby(['id'])[TARGET].transform(lambda x: x.shift(LAG_DAY)).astype(np.float16)\n return lag_df[[col_name]]\n\n\ndef make_lag_roll(LAG_DAY):\n shift_day = LAG_DAY[0]\n roll_wind = LAG_DAY[1]\n lag_df = base_test[['id','d',TARGET]]\n col_name = 'rolling_mean_tmp_'+str(shift_day)+'_'+str(roll_wind)\n lag_df[col_name] = lag_df.groupby(['id'])[TARGET].transform(lambda x: x.shift(shift_day).rolling(roll_wind).mean())\n return lag_df[[col_name]]",
"_____no_output_____"
],
[
"########################### Model params\n#################################################################################\nimport lightgbm as lgb\nlgb_params = {\n 'boosting_type': 'gbdt',\n 'objective': 'tweedie',\n 'tweedie_variance_power': 1.1,\n 'metric': 'rmse',\n 'subsample': 0.5,\n 'subsample_freq': 1,\n 'learning_rate': 0.05,\n 'num_leaves': 2**11-1,\n 'min_data_in_leaf': 2**12-1,\n 'feature_fraction': 0.5,\n 'max_bin': 100,\n 'n_estimators': 1400,\n 'boost_from_average': False,\n 'verbose': -1,\n 'n_jobs':40\n } \n\n# Let's look closer on params\n\n## 'boosting_type': 'gbdt'\n# we have 'goss' option for faster training\n# but it normally leads to underfit.\n# Also there is good 'dart' mode\n# but it takes forever to train\n# and model performance depends \n# a lot on random factor \n# https://www.kaggle.com/c/home-credit-default-risk/discussion/60921\n\n## 'objective': 'tweedie'\n# Tweedie Gradient Boosting for Extremely\n# Unbalanced Zero-inflated Data\n# https://arxiv.org/pdf/1811.10192.pdf\n# and many more articles about tweediie\n#\n# Strange (for me) but Tweedie is close in results\n# to my own ugly loss.\n# My advice here - make OWN LOSS function\n# https://www.kaggle.com/c/m5-forecasting-accuracy/discussion/140564\n# https://www.kaggle.com/c/m5-forecasting-accuracy/discussion/143070\n# I think many of you already using it (after poisson kernel appeared) \n# (kagglers are very good with \"params\" testing and tuning).\n# Try to figure out why Tweedie works.\n# probably it will show you new features options\n# or data transformation (Target transformation?).\n\n## 'tweedie_variance_power': 1.1\n# default = 1.5\n# set this closer to 2 to shift towards a Gamma distribution\n# set this closer to 1 to shift towards a Poisson distribution\n# my CV shows 1.1 is optimal \n# but you can make your own choice\n\n## 'metric': 'rmse'\n# Doesn't mean anything to us\n# as competition metric is different\n# and we don't use early stoppings here.\n# So rmse serves just for general \n# model performance overview.\n# Also we use \"fake\" validation set\n# (as it makes part of the training set)\n# so even general rmse score doesn't mean anything))\n# https://www.kaggle.com/c/m5-forecasting-accuracy/discussion/133834\n\n## 'subsample': 0.5\n# Serves to fight with overfit\n# this will randomly select part of data without resampling\n# Chosen by CV (my CV can be wrong!)\n# Next kernel will be about CV\n\n##'subsample_freq': 1\n# frequency for bagging\n# default value - seems ok\n\n## 'learning_rate': 0.03\n# Chosen by CV\n# Smaller - longer training\n# but there is an option to stop \n# in \"local minimum\"\n# Bigger - faster training\n# but there is a chance to\n# not find \"global minimum\" minimum\n\n## 'num_leaves': 2**11-1\n## 'min_data_in_leaf': 2**12-1\n# Force model to use more features\n# We need it to reduce \"recursive\"\n# error impact.\n# Also it leads to overfit\n# that's why we use small \n# 'max_bin': 100\n\n## l1, l2 regularizations\n# https://towardsdatascience.com/l1-and-l2-regularization-methods-ce25e7fc831c\n# Good tiny explanation\n# l2 can work with bigger num_leaves\n# but my CV doesn't show boost\n \n## 'n_estimators': 1400\n# CV shows that there should be\n# different values for each state/store.\n# Current value was chosen \n# for general purpose.\n# As we don't use any early stopings\n# careful to not overfit Public LB.\n\n##'feature_fraction': 0.5\n# LightGBM will randomly select \n# part of features on each iteration (tree).\n# We have maaaany features\n# and many of them are \"duplicates\"\n# and many just \"noise\"\n# good values here - 0.5-0.7 (by CV)\n\n## 'boost_from_average': False\n# There is some \"problem\"\n# to code boost_from_average for \n# custom loss\n# 'True' makes training faster\n# BUT carefull use it\n# https://github.com/microsoft/LightGBM/issues/1514\n# not our case but good to know cons",
"_____no_output_____"
],
[
"########################### Vars\n#################################################################################\nVER = 1 # Our model version\nSEED = 42 # We want all things\nseed_everything(SEED) # to be as deterministic \nlgb_params['seed'] = SEED # as possible\nN_CORES = psutil.cpu_count() # Available CPU cores\n\n\n#LIMITS and const\nTARGET = 'sales' # Our target\nSTART_TRAIN = 0 # We can skip some rows (Nans/faster training)\nEND_TRAIN = 1941 # End day of our train set\nSTART_VALID = 1913\nP_HORIZON = 28 # Prediction horizon\nUSE_AUX = False # Use or not pretrained models\n\n#FEATURES to remove\n## These features lead to overfit\n## or values not present in test set\nremove_features = ['id','state_id','store_id',\n 'date','wm_yr_wk','d',TARGET]\nmean_features = ['enc_cat_id_mean','enc_cat_id_std',\n 'enc_dept_id_mean','enc_dept_id_std',\n 'enc_item_id_mean','enc_item_id_std'] \n\n#PATHS for Features\nORIGINAL = 'data/m5-detrend/'\nBASE = 'data/m5-detrend/grid_part_1.pkl'\nPRICE = 'data/m5-detrend/grid_part_2.pkl'\nCALENDAR = 'data/m5-detrend/grid_part_3.pkl'\nLAGS = 'data/m5-detrend/lags_df_28.pkl'\nMEAN_ENC = 'data/m5-custom-features/mean_encoding_df.pkl'\n\n\n# AUX(pretrained) Models paths\nAUX_MODELS = 'data/m5-aux-models/'\n\n\n#STORES ids\nSTORES_IDS = pd.read_pickle(ORIGINAL+'final_sales_train_evaluation.pkl')['store_id']\nSTORES_IDS = list(STORES_IDS.unique())\n\n\n#SPLITS for lags creation\nSHIFT_DAY = 28\nN_LAGS = 15\nLAGS_SPLIT = [col for col in range(SHIFT_DAY,SHIFT_DAY+N_LAGS)]\nROLS_SPLIT = []\nfor i in [1,7,14]:\n for j in [7,14,30,60]:\n ROLS_SPLIT.append([i,j])",
"_____no_output_____"
]
],
[
[
"### Train and Valid",
"_____no_output_____"
]
],
[
[
"MODEL_PATH = 'models/1914_1941_valid_detrend_d2d/'\nfor day in range(1,28):\n\n for store_id in STORES_IDS:\n print('Train', store_id)\n\n # Get grid for current store\n grid_df, features_columns = get_data_by_store(store_id)\n grid_df['sales'] = grid_df.groupby('item_id')['sales'].shift(-day)\n grid_df = grid_df[grid_df.groupby('item_id').cumcount(ascending=False) > day-1]\n\n grid_df['sales'] = grid_df['sales'].values * grid_df['sell_price'].values\n # break\n # Masks for \n # Train (All data less than 1913)\n # \"Validation\" (Last 28 days - not real validatio set)\n # Test (All data greater than 1913 day, \n # with some gap for recursive features)\n train_mask = grid_df['d']<=END_TRAIN-P_HORIZON\n # valid_mask = grid_df['d']>(END_TRAIN-100)\n preds_mask = (grid_df['d']<=END_TRAIN)&(grid_df['d']>(END_TRAIN-P_HORIZON-100))\n\n # Apply masks and save lgb dataset as bin\n # to reduce memory spikes during dtype convertations\n # https://github.com/Microsoft/LightGBM/issues/1032\n # \"To avoid any conversions, you should always use np.float32\"\n # or save to bin before start training\n # https://www.kaggle.com/c/talkingdata-adtracking-fraud-detection/discussion/53773\n train_data = lgb.Dataset(grid_df[train_mask][features_columns], \n label=grid_df[train_mask][TARGET])\n # train_data.save_binary('train_data.bin')\n # train_data = lgb.Dataset('train_data.bin')\n\n ## valid_data = lgb.Dataset(grid_df[valid_mask][features_columns], \n ## label=grid_df[valid_mask][TARGET])\n # break\n # Saving part of the dataset for later predictions\n # Removing features that we need to calculate recursively \n grid_df = grid_df[preds_mask].reset_index(drop=True)\n keep_cols = [col for col in list(grid_df) if '_tmp_' not in col]\n grid_df = grid_df[keep_cols]\n if day==1:\n grid_df.to_pickle(MODEL_PATH+'valid_'+store_id+'.pkl')\n\n del grid_df\n\n # Launch seeder again to make lgb training 100% deterministic\n # with each \"code line\" np.random \"evolves\" \n # so we need (may want) to \"reset\" it\n seed_everything(SEED)\n estimator = lgb.train(lgb_params,\n train_data,\n valid_sets = [train_data],\n verbose_eval = 100,\n )\n\n # Save model - it's not real '.bin' but a pickle file\n # estimator = lgb.Booster(model_file='model.txt')\n # can only predict with the best iteration (or the saving iteration)\n # pickle.dump gives us more flexibility\n # like estimator.predict(TEST, num_iteration=100)\n # num_iteration - number of iteration want to predict with, \n # NULL or <= 0 means use best iteration\n model_name = MODEL_PATH+'lgb_model_'+store_id+'_v'+str(VER)+ '_valid' +'_d_'+ str(day+1) +'.bin'\n pickle.dump(estimator, open(model_name, 'wb'))\n\n # Remove temporary files and objects \n # to free some hdd space and ram memory\n # !rm train_data.bin\n del train_data, estimator\n gc.collect()\n\n # \"Keep\" models features for predictions\n MODEL_FEATURES = features_columns",
"_____no_output_____"
],
[
"features_columns = ['item_id', 'dept_id', 'cat_id', 'release', 'sell_price', 'price_max', 'price_min', 'price_std', 'price_mean', 'price_norm', 'price_nunique', 'item_nunique', 'price_momentum', 'price_momentum_m', 'price_momentum_y', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2', 'snap_CA', 'snap_TX', 'snap_WI', 'tm_d', 'tm_w', 'tm_m', 'tm_y', 'tm_wm', 'tm_dw', 'tm_w_end', 'enc_cat_id_mean', 'enc_cat_id_std', 'enc_dept_id_mean', 'enc_dept_id_std', 'enc_item_id_mean', 'enc_item_id_std', 'sales_lag_28', 'sales_lag_29', 'sales_lag_30', 'sales_lag_31', 'sales_lag_32', 'sales_lag_33', 'sales_lag_34', 'sales_lag_35', 'sales_lag_36', 'sales_lag_37', 'sales_lag_38', 'sales_lag_39', 'sales_lag_40', 'sales_lag_41', 'sales_lag_42', 'sales_lag_43', 'sales_lag_44', 'sales_lag_45', 'sales_lag_46', 'sales_lag_47', 'sales_lag_48', 'sales_lag_49', 'sales_lag_50', 'sales_lag_51', 'sales_lag_52', 'sales_lag_53', 'sales_lag_54', 'sales_lag_55', 'rolling_mean_tmp_1_7', 'rolling_mean_tmp_1_14', 'rolling_mean_tmp_1_30', 'rolling_mean_tmp_1_60', 'rolling_mean_tmp_7_7', 'rolling_mean_tmp_7_14', 'rolling_mean_tmp_7_30', 'rolling_mean_tmp_7_60', 'rolling_mean_tmp_14_7', 'rolling_mean_tmp_14_14', 'rolling_mean_tmp_14_30', 'rolling_mean_tmp_14_60']\nMODEL_FEATURES = features_columns",
"_____no_output_____"
],
[
"MODEL_PATH = 'models/1914_1941_valid_detrend_d2d/'",
"_____no_output_____"
],
[
"def get_base_valid():\n base_test = pd.DataFrame()\n\n for store_id in STORES_IDS:\n temp_df = pd.read_pickle(MODEL_PATH+'valid_'+store_id+'.pkl')\n temp_df['store_id'] = store_id\n base_test = pd.concat([base_test, temp_df]).reset_index(drop=True)\n \n return base_test",
"_____no_output_____"
],
[
"all_preds = pd.DataFrame()\n\n# Join back the Test dataset with \n# a small part of the training data \n# to make recursive features\nbase_test = get_base_valid()\nbase_test = base_test[base_test['d']<=END_TRAIN]\nindex = base_test[base_test['d']>END_TRAIN-P_HORIZON].index\nbase_test.loc[index,'sales']=np.NaN\n\n# Timer to measure predictions time \nmain_time = time.time()\n\nPREDICT_DAY = 2",
"_____no_output_____"
],
[
"base_test[base_test['d']==1840]['sales'].sum()",
"_____no_output_____"
],
[
"END_TRAIN-P_HORIZON+PREDICT_DAY",
"_____no_output_____"
],
[
"start_time = time.time()\ngrid_df = base_test.copy()\ngrid_df = pd.concat([grid_df, df_parallelize_run(make_lag_roll, ROLS_SPLIT)], axis=1)\n\nfor store_id in STORES_IDS:\n\n # Read all our models and make predictions\n # for each day/store pairs\n model_path = MODEL_PATH + 'lgb_model_'+store_id+'_v'+str(VER)+'_valid'+'_d_'+str(PREDICT_DAY)+'.bin' \n if USE_AUX:\n model_path = AUX_MODELS + model_path\n\n estimator = pickle.load(open(model_path, 'rb'))\n\n day_mask = base_test['d']==(END_TRAIN-P_HORIZON+PREDICT_DAY)\n store_mask = base_test['store_id']==store_id\n\n mask = (day_mask)&(store_mask)\n base_test[TARGET][mask] = estimator.predict(grid_df[mask][MODEL_FEATURES])\n\n# Make good column naming and add \n# to all_preds DataFrame\ntemp_df = base_test[day_mask][['id',TARGET]]\ntemp_df.columns = ['id','F'+str(PREDICT_DAY)]\nif 'id' in list(all_preds):\n all_preds = all_preds.merge(temp_df, on=['id'], how='left')\nelse:\n all_preds = temp_df.copy()\n\nprint('#'*10, ' %0.2f min round |' % ((time.time() - start_time) / 60),\n ' %0.2f min total |' % ((time.time() - main_time) / 60),\n ' %0.2f day sales |' % (temp_df['F'+str(PREDICT_DAY)].sum()))\ndel temp_df",
"########## 0.55 min round | 0.55 min total | 175328.04 day sales |\n"
],
[
"all_preds",
"_____no_output_____"
],
[
"all_preds.to_pickle('revenue_1914_1941_valid_detrend_d2.pkl')",
"_____no_output_____"
],
[
"END_TRAIN-P_HORIZON+PREDICT_DAY",
"_____no_output_____"
],
[
"########################### Export\n#################################################################################\n# Reading competition sample submission and\n# merging our predictions\n# As we have predictions only for \"_validation\" data\n# we need to do fillna() for \"_evaluation\" items\nsubmission = pd.read_csv(ORIGINAL+'sample_submission.csv')[['id']]\nsubmission = submission.merge(all_preds, on=['id'], how='left').fillna(0)\nsubmission.to_csv('submission_v'+str(VER)+'.csv', index=False)",
"_____no_output_____"
],
[
"# Summary\n\n# Of course here is no magic at all.\n# No \"Novel\" features and no brilliant ideas.\n# We just carefully joined all\n# our previous fe work and created a model.\n\n# Also!\n# In my opinion this strategy is a \"dead end\".\n# Overfits a lot LB and with 1 final submission \n# you have no option to risk.\n\n\n# Improvement should come from:\n# Loss function\n# Data representation\n# Stable CV\n# Good features reduction strategy\n# Predictions stabilization with NN\n# Trend prediction\n# Real zero sales detection/classification\n\n\n# Good kernels references \n## (the order is random and the list is not complete):\n# https://www.kaggle.com/ragnar123/simple-lgbm-groupkfold-cv\n# https://www.kaggle.com/jpmiller/grouping-items-by-stockout-pattern\n# https://www.kaggle.com/headsortails/back-to-predict-the-future-interactive-m5-eda\n# https://www.kaggle.com/sibmike/m5-out-of-stock-feature\n# https://www.kaggle.com/mayer79/m5-forecast-attack-of-the-data-table\n# https://www.kaggle.com/yassinealouini/seq2seq\n# https://www.kaggle.com/kailex/m5-forecaster-v2\n# https://www.kaggle.com/aerdem4/m5-lofo-importance-on-gpu-via-rapids-xgboost\n\n\n# Features were created in these kernels:\n## \n# Mean encodings and PCA options\n# https://www.kaggle.com/kyakovlev/m5-custom-features\n##\n# Lags and rolling lags\n# https://www.kaggle.com/kyakovlev/m5-lags-features\n##\n# Base Grid and base features (calendar/price/etc)\n# https://www.kaggle.com/kyakovlev/m5-simple-fe\n\n\n# Personal request\n# Please don't upvote any ensemble and copypaste kernels\n## The worst case is ensemble without any analyse.\n## The best choice - just ignore it.\n## I would like to see more kernels with interesting and original approaches.\n## Don't feed copypasters with upvotes.\n\n## It doesn't mean that you should not fork and improve others kernels\n## but I would like to see params and code tuning based on some CV and analyse\n## and not only on LB probing.\n## Small changes could be shared in comments and authors can improve their kernel.\n\n## Feel free to criticize this kernel as my knowlege is very limited\n## and I can be wrong in code and descriptions. \n## Thank you.",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
cb71fddf4ef746bd9b8e0662dda9cc75e2b4819a | 59,458 | ipynb | Jupyter Notebook | sklearn_basics/imbalanced.ipynb | KellyHwong/deeplearning-basics | 0c0f45daaab42a25d2cd047cbecdca4f4bc7df59 | [
"MIT"
]
| 1 | 2020-07-02T06:29:11.000Z | 2020-07-02T06:29:11.000Z | sklearn_basics/imbalanced.ipynb | KellyHwong/deeplearning-basics | 0c0f45daaab42a25d2cd047cbecdca4f4bc7df59 | [
"MIT"
]
| null | null | null | sklearn_basics/imbalanced.ipynb | KellyHwong/deeplearning-basics | 0c0f45daaab42a25d2cd047cbecdca4f4bc7df59 | [
"MIT"
]
| null | null | null | 396.386667 | 32,751 | 0.763985 | [
[
[
"#!/usr/bin/env python3\n# -*- coding: utf-8 -*-\n# @Date : Dec-02-20 00:19\n# @Author : Kan HUANG ([email protected])",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nnum_samples = 30000\nnum_pos = 3000\nnum_neg = 27000\nnum_classes = 2",
"_____no_output_____"
],
[
"pos_weight = num_samples / (num_classes * num_pos)\nneg_weight = num_samples / (num_classes * num_neg)\nprint(pos_weight, neg_weight)",
"5.0 0.5555555555555556\n"
],
[
"def get_class_weights(num_samples=30000, num_pos=3000):\n pos_weight = num_samples / (num_classes * num_pos)\n num_neg = num_samples - num_pos\n neg_weight = num_samples / (num_classes * num_neg)\n return (pos_weight, neg_weight)",
"_____no_output_____"
],
[
"# test_space = np.arange(1, 30000)\ntest_space = np.arange(3000, 27000)\nweights = np.zeros((len(test_space), 2))\nfor i, num_pos in enumerate(test_space):\n weights[i, 0], weights[i, 1] = get_class_weights(num_pos=num_pos)",
"_____no_output_____"
],
[
"print(test_space[2999])\nprint(weights[2999])",
"5999\n[2.50041674 0.62497396]\n"
],
[
"plt.plot(test_space, weights[:, 0], label=\"pos weight\")\nplt.plot(test_space, weights[:, 1], label=\"neg weight\")\nplt.plot(test_space, weights[:, 0]/weights[:, 1], label=\"pos/neg weight ratio\")\nplt.legend()\nplt.grid()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.