prompt
stringlengths 19
1.03M
| completion
stringlengths 4
2.12k
| api
stringlengths 8
90
|
|---|---|---|
"""{{ cookiecutter.project }} PSII analysis."""
# %% Setup
# Export .png to outdir from LemnaBase using LT-db_extractor.py
from plantcv import plantcv as pcv
import cppcpyutils as cppc
import importlib
import os
import cv2 as cv2
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import warnings
import importlib
from skimage import filters
from skimage import morphology
from skimage import segmentation
warnings.filterwarnings("ignore", module="matplotlib")
warnings.filterwarnings("ignore", module='plotnine')
# %% io directories
indir = os.path.join('data', 'psII')
# snapshotdir = indir
outdir = os.path.join('output', 'psII')
debugdir = os.path.join('debug', 'psII')
maskdir = os.path.join(outdir, 'masks')
fluordir = os.path.join(outdir, 'fluorescence')
os.makedirs(outdir, exist_ok=True)
os.makedirs(debugdir, exist_ok=True)
os.makedirs(maskdir, exist_ok=True)
outfile = os.path.join(outdir,'output_psII_level0.csv')
# %% pixel pixel_resolution
# mm (this is approx and should only be used for scalebar)
cppc.pixelresolution = 0.3
# %% Import tif file information based on the filenames. If extract_frames=True it will save each frame form the multiframe TIF to a separate file in data/pimframes/ with a numeric suffix
fdf = cppc.io.import_snapshots(indir, camera='psii')
# %% Define the frames from the PSII measurements and merge this information with the filename information
pimframes = pd.read_csv(os.path.join('data', 'pimframes_map.csv'),
skipinitialspace=True)
# this eliminate weird whitespace around any of the character fields
fdf_dark = (pd.merge(fdf.reset_index(), pimframes, on=['frameid'],
how='right'))
# %% remove absorptivity measurements which are blank images
# also remove Ft_FRon measurements. THere is no Far Red light.
df = (fdf_dark.query(
'~parameter.str.contains("Abs") and ~parameter.str.contains("FRon")',
engine='python'))
# %% remove the duplicate Fm and Fo frames where frame = Fmp and Fp from frameid 5,6
df = (df.query(
'(parameter!="FvFm") or (parameter=="FvFm" and (frame=="Fo" or frame=="Fm") )'
))
# %% Arrange dataframe of metadata so Fv/Fm comes first
param_order = pimframes.parameter.unique()
df['parameter'] = pd.Categorical(df.parameter,
categories=param_order,
ordered=True)
# %% Check for existing output file and only analyze new files
newheader = True
defaultcols = df.columns.values.tolist()
if os.path.exists(outfile):
# reading existing results file
existingdf = pd.read_csv(outfile)
# format dates consistently and NOT in data format becuase pandas doesn't handle datetimes well in merges(!?)...
existingdf.jobdate = pd.to_datetime(existingdf.jobdate).dt.strftime('%Y-%m-%d')
df.loc[:,'jobdate'] = df.jobdate.dt.strftime('%Y-%m-%d')
# set common index
mergecols = ['plantbarcode','jobdate','frame','frameid','parameter']
df = df.set_index(mergecols)
existingdf = existingdf.set_index(mergecols)
# filter and sort df
df = df[~df.index.isin(existingdf.index)].reset_index()
df.jobdate =
|
pd.to_datetime(df.jobdate)
|
pandas.to_datetime
|
# -*- coding: utf-8 -*-
"""
This is a parameter search module. With this module, a certain firing pattern
can be searched randomly with AN model, SAN model and X model. In order to
lessen dependence of models on parameters, it's important to make various
parameter sets (models) and then extract common features among them.\
In this script, parameter sets that recapitulate slow wave sleep (SWS) firing
pattern can be searched with algorithms as described in Tatsuki et al., 2016
and Yoshida et al., 2018.
"""
__author__ = '<NAME>, <NAME>, <NAME>, \
<NAME>, <NAME>, <NAME>'
__status__ = 'Published'
__version__ = '1.0.0'
__date__ = '15 May 2020'
import os
import sys
"""
LIMIT THE NUMBER OF THREADS!
change local env variables BEFORE importing numpy
"""
os.environ["OMP_NUM_THREADS"] = "1" # 2nd likely
os.environ["NUMEXPR_NUM_THREADS"] = "1"
os.environ["MKL_NUM_THREADS"] = "1" # most likely
from datetime import datetime
from multiprocessing import Pool
import numpy as np
import pandas as pd
from pathlib import Path
import pickle
from time import time
from typing import Dict, List, Optional
import warnings
warnings.filterwarnings('ignore')
import models
import analysis
class RandomParamSearch():
""" Random parameter search.
Generate parameter sets randomly, and pick up those which recapitulate a cirtain
firing pattern.
Parameters
----------
model : str
model in which parameter search is conducted
pattern : str
searched firing pattern
ncore : int
number of cores you are going to use
time : int or str
how long to run parameter search (hour), default 48 (2 days)
channel_bool : list (bool) or None
WHEN YOU USE X MODEL, YOU MUST DESIGNATE THIS LIST.\
Channel lists that X model contains. True means channels incorporated
in the model and False means not. The order of the list is the same
as other lists or dictionaries that contain channel information in
AN model. Example: \
channel_bool = [
1, # leak channel
0, # voltage-gated sodium channel
1, # HH-type delayed rectifier potassium channel
0, # fast A-type potassium channel
0, # slowly inactivating potassium channel
1, # voltage-gated calcium channel
1, # calcium-dependent potassium channel
1, # persistent sodium channel
0, # inwardly rectifier potassium channel
0, # AMPA receptor
0, # NMDA receptor
0, # GABA receptor
1, # calcium pump
]\
This is SAN model, default None
model_name : str or None
name of the X model, default None
ion : bool
whether you make equiribrium potential variable or not,
default False
concentration : dictionary or str or None
dictionary of ion concentration, or 'sleep'/'awake' that
designate typical ion concentrations, default None
Attributes
----------
wave_check : object
Keep attributes and helper functions needed for parameter search.
pattern : str
searched firing pattern
time : int
how long to run parameter search (hour)
model_name : str
model name
model : object
Simulation model object. See anmodel.models.py
"""
def __init__(self, model: str, pattern: str='SWS', ncore: int=1,
hr: int=48, samp_freq: int=1000, samp_len: int=10,
channel_bool: Optional[List[bool]]=None,
model_name: Optional[str]=None,
ion: bool=False, concentration: Optional[Dict]=None) -> None:
self.wave_check = analysis.WaveCheck(samp_freq=samp_freq)
self.pattern = pattern
self.ncore = ncore
self.hr = int(hr)
self.samp_freq = samp_freq
self.samp_len = samp_len
if model == 'AN':
self.model_name = 'AN'
self.model = models.ANmodel(ion, concentration)
if model == 'SAN':
self.model_name = 'SAN'
self.model = models.SANmodel(ion, concentration)
if model == "X":
if channel_bool is None:
raise TypeError('Designate channel in argument of X model.')
self.model_name = model_name
self.model = models.Xmodel(channel_bool, ion, concentration)
def singleprocess(self, args: List) -> None:
""" Random parameter search using single core.
Search parameter sets which recapitulate a cirtain firing pattern randomly,
and save them every 1 hour. After self.time hours, this process terminates.
Parameters
----------
args : list
core : int
n th core of designated number of cores
now : datetime.datetime
datetime.datetime.now() when simulation starts
time_start : float
time() when simulation starts
rand_seed : int
random seed for generating random parameters. 0 ~ 2**32-1.
"""
core, now, time_start, rand_seed = args
date: str = f'{now.year}_{now.month}_{now.day}'
p: Path = Path.cwd()
res_p: Path = p / 'results' / f'{self.pattern}_params' / f'{date}_{self.model_name}'
res_p.mkdir(parents=True, exist_ok=True)
save_p: Path = res_p / f'{self.pattern}_{date}_{core}.pickle'
param_df: pd.DataFrame =
|
pd.DataFrame([])
|
pandas.DataFrame
|
import numpy as np
import pandas as pd
from mpl_toolkits.axes_grid1 import make_axes_locatable
import os
import platform
import matplotlib.pyplot as plt
import matplotlib as mpl
import matplotlib.ticker as mticker
import matplotlib.gridspec as gridspec
from tqdm import trange
from matplotlib.ticker import ScalarFormatter
import pidsim.parameter_span as pspan
from scipy import interpolate
import pnptransport.utils as utils
import re
import json
base_path = r'G:\My Drive\Research\PVRD1\Manuscripts\Device_Simulations_draft\simulations\inputs_20201028'
span_database = r'G:\My Drive\Research\PVRD1\Manuscripts\Device_Simulations_draft\simulations\inputs_20201028\one_factor_at_a_time_lower_20201028_h=1E-12.csv'
parameter = 'h'
output_folder = 'ofat_comparison_20201121'
batch_analysis = 'batch_analysis_rfr_20201121'
t_max_h = 96.
parameter_units = {
'sigma_s': 'cm^{-2}',
'zeta': 's^{-1}',
'DSF': 'cm^2/s',
'E': 'V/cm',
'm': '',
'h': 'cm/s',
'recovery time': 's',
'recovery electric field': 'V/cm'
}
map_parameter_names = {
'sigma_s': 'S_0',
'zeta': 'k',
'DSF': r'D_{{\mathrm{{SF}}}}',
'E': 'E',
'm': 'm',
'h': 'h',
}
def slugify(value):
"""
Normalizes string, converts to lowercase, removes non-alpha characters,
and converts spaces to hyphens.
Parameters
----------
value: str
The string
Returns
-------
str
Normalized string
"""
value = re.sub('[^\w\s\-]', '', value).strip().lower()
value = re.sub('[-\s]+', '-', value)
return value
if __name__ == '__main__':
output_path = os.path.join(base_path, output_folder)
if platform.system() == 'Windows':
base_path = r'\\?\\' + base_path
output_path = os.path.join(base_path, output_folder)
if not os.path.exists(output_path):
os.makedirs(output_path)
span_df = pd.read_csv(span_database, index_col=0)
ofat_df = pd.read_csv(os.path.join(base_path, 'ofat_db.csv'), index_col=None)
# Get the row corresponding to the values to compare
parameter_info = span_df.loc[parameter]
parameter_span = pspan.string_list_to_float(parameter_info['span'])
units = parameter_units[parameter]
# Get the values of every parameter from the ofat_db file
ofat_constant_parameters = ofat_df.iloc[0]
time_s = float(ofat_constant_parameters['time (s)'])
temp_c = float(ofat_constant_parameters['temp (C)'])
bias = float(ofat_constant_parameters['bias (V)'])
thickness_sin = float(ofat_constant_parameters['thickness sin (um)'])
thickness_si = float(ofat_constant_parameters['thickness si (um)'])
er = float(ofat_constant_parameters['er'])
thickness_si = float(ofat_constant_parameters['thickness si (um)'])
data_files = []
for p in parameter_span:
converged = False
if parameter == 'sigma_s':
file_tag = pspan.create_filetag(
time_s=time_s, temp_c=temp_c, sigma_s=p, zeta=span_df.loc['zeta']['base'],
d_sf=span_df.loc['DSF']['base'], ef=span_df.loc['E']['base'], m=span_df.loc['m']['base'],
h=span_df.loc['h']['base'], recovery_time=span_df.loc['recovery time']['base'],
recovery_e_field=span_df.loc['recovery electric field']['base']
)
# Determine whether the simulation converged
simulation_parameters = ofat_df[ofat_df['config file'] == file_tag + '.ini'].reset_index(drop=True)
converged = bool(simulation_parameters['converged'][0])
if parameter == 'zeta':
file_tag = pspan.create_filetag(
time_s=time_s, temp_c=temp_c, sigma_s=span_df.loc['sigma_s']['base'], zeta=p,
d_sf=span_df.loc['DSF']['base'], ef=span_df.loc['E']['base'], m=span_df.loc['m']['base'],
h=span_df.loc['h']['base'], recovery_time=span_df.loc['recovery time']['base'],
recovery_e_field=span_df.loc['recovery electric field']['base']
)
# Determine whether the simulation converged
simulation_parameters = ofat_df[ofat_df['config file'] == file_tag + '.ini'].reset_index(drop=True)
converged = bool(simulation_parameters['converged'][0])
if parameter == 'DSF':
file_tag = pspan.create_filetag(
time_s=time_s, temp_c=temp_c, sigma_s=span_df.loc['sigma_s']['base'], zeta=span_df.loc['zeta']['base'],
d_sf=p, ef=span_df.loc['E']['base'], m=span_df.loc['m']['base'],
h=span_df.loc['h']['base'], recovery_time=span_df.loc['recovery time']['base'],
recovery_e_field=span_df.loc['recovery electric field']['base']
)
# Determine whether the simulation converged
simulation_parameters = ofat_df[ofat_df['config file'] == file_tag + '.ini'].reset_index(drop=True)
converged = bool(simulation_parameters['converged'][0])
if parameter == 'E':
file_tag = pspan.create_filetag(
time_s=time_s, temp_c=temp_c, sigma_s=span_df.loc['sigma_s']['base'], zeta=span_df.loc['zeta']['base'],
d_sf=span_df.loc['DSF']['base'], ef=p, m=span_df.loc['m']['base'],
h=span_df.loc['h']['base'], recovery_time=span_df.loc['recovery time']['base'],
recovery_e_field=-p
)
# Determine whether the simulation converged
simulation_parameters = ofat_df[ofat_df['config file'] == file_tag + '.ini'].reset_index(drop=True)
if len(simulation_parameters) > 0:
converged = bool(simulation_parameters['converged'][0])
else:
converged = False
if parameter == 'm':
file_tag = pspan.create_filetag(
time_s=time_s, temp_c=temp_c, sigma_s=span_df.loc['sigma_s']['base'], zeta=span_df.loc['zeta']['base'],
d_sf=span_df.loc['DSF']['base'], ef=span_df.loc['E']['base'], m=p,
h=span_df.loc['h']['base'], recovery_time=span_df.loc['recovery time']['base'],
recovery_e_field=span_df.loc['recovery electric field']['base']
)
# Determine whether the simulation converged
simulation_parameters = ofat_df[ofat_df['config file'] == file_tag + '.ini'].reset_index(drop=True)
converged = bool(simulation_parameters['converged'][0])
if parameter == 'h':
file_tag = pspan.create_filetag(
time_s=time_s, temp_c=temp_c, sigma_s=span_df.loc['sigma_s']['base'], zeta=span_df.loc['zeta']['base'],
d_sf=span_df.loc['DSF']['base'], ef=span_df.loc['E']['base'], m=span_df.loc['m']['base'],
h=p, recovery_time=span_df.loc['recovery time']['base'],
recovery_e_field=span_df.loc['recovery electric field']['base']
)
# Determine whether the simulation converged
simulation_parameters = ofat_df[ofat_df['config file'] == file_tag + '.ini'].reset_index(drop=True)
converged = bool(simulation_parameters['converged'][0])
if converged:
data_files.append({
'parameter': parameter, 'value': p, 'pid_file': file_tag + '_simulated_pid.csv',
'units': parameter_units[parameter]
})
# for f in data_files:
# print(f)
n_files = len(data_files)
# c_map1 = mpl.cm.get_cmap('RdYlGn_r')
c_map1 = mpl.cm.get_cmap('rainbow')
if parameter == 'DSF':
c_map1 = mpl.cm.get_cmap('rainbow_r')
normalize = mpl.colors.Normalize(vmin=0, vmax=(n_files-1))
plot_colors = [c_map1(normalize(i)) for i in range(n_files)]
t_max = t_max_h * 3600.
failure_times = np.empty(
n_files, dtype=np.dtype([
(r'{0} ({1})'.format(parameter, parameter_units[parameter]), 'd'),
('t 1000 (s)', 'd'), ('Rsh 96h (Ohm cm2)', 'd')
])
)
with open('plotstyle.json', 'r') as style_file:
mpl.rcParams.update(json.load(style_file)['defaultPlotStyle'])
# mpl.rcParams.update(defaultPlotStyle)
xfmt = ScalarFormatter(useMathText=True)
xfmt.set_powerlimits((-3, 3))
fig_p = plt.figure(1)
fig_p.set_size_inches(4.75, 2.5, forward=True)
# fig_p.subplots_adjust(hspace=0.0, wspace=0.0)
# gs0_p = gridspec.GridSpec(ncols=1, nrows=1, figure=fig_p, width_ratios=[1])
# gs00_p = gridspec.GridSpecFromSubplotSpec(nrows=1, ncols=1, subplot_spec=gs0_p[0])
# ax1_p = fig_p.add_subplot(gs00_p[0, 0])
ax1_p = fig_p.add_subplot(1, 1, 1)
fig_r = plt.figure(2)
fig_r.set_size_inches(4.75, 2.5, forward=True)
# fig_r.subplots_adjust(hspace=0.0, wspace=0.0)
gs0_r = gridspec.GridSpec(ncols=1, nrows=1, figure=fig_r, width_ratios=[1])
gs00_r = gridspec.GridSpecFromSubplotSpec(nrows=1, ncols=1, subplot_spec=gs0_r[0])
ax1_r = fig_r.add_subplot(1, 1, 1)
pbar = trange(n_files, desc='Analyzing file', leave=True)
for i, file_info in enumerate(data_files):
# Read the simulated data from the csv file
csv_file = os.path.join(base_path, batch_analysis, file_info['pid_file'])
pid_df = pd.read_csv(csv_file)
# print('Analysing file \'{0}\':'.format(csv_file))
# print(pid_df.head())
time_s = pid_df['time (s)'].to_numpy(dtype=float)
power = pid_df['Pmpp (mW/cm^2)'].to_numpy(dtype=float)
rsh = pid_df['Rsh (Ohm cm^2)'].to_numpy(dtype=float)
time_h = time_s / 3600.
t_interp = np.linspace(np.amin(time_s), np.amax(time_s), num=1000)
f_r_interp = interpolate.interp1d(time_s, rsh, kind='linear')
rsh_interp = f_r_interp(t_interp)
if rsh_interp.min() <= 1000:
idx_1000 = (np.abs(rsh_interp - 1000)).argmin()
failure_times[i] = (file_info['value'], t_interp[idx_1000].copy(), f_r_interp(96.*3600.))
else:
failure_times[i] = (file_info['value'], np.inf, f_r_interp(96.*3600.))
sv_txt = r'${0}$ = ${1}$ $\mathregular{{{2}}}$'.format(
map_parameter_names[file_info['parameter']], utils.latex_order_of_magnitude(
file_info['value'], dollar=False
),
file_info['units']
)
ax1_p.plot(
time_h, power / power[0], color=plot_colors[i], ls='-', label=sv_txt, zorder=(i+1)
)
ax1_r.plot(
time_h, rsh, color=plot_colors[i], ls='-', label=sv_txt, zorder=(i+1)
)
ptx = t_interp[idx_1000]/3600.
ax1_r.scatter(
ptx, 1000, marker='o', color='k', zorder=(1+n_files), lw=1,
s=10
)
# ax1_r.plot(
# [ptx, ptx], [0, 1000],
# lw=1.5, ls='-', color=(0.95, 0.95, 0.95), zorder=0,
# )
# print('1000 Ohms cm2 failure: ({0:.3f}) h'.format(t_interp[idx_1000]/3600.))
pbar.set_description('Analyzing parameter {0}: {1}'.format(
parameter, file_info['value'], file_info['units']
))
pbar.update()
pbar.refresh()
ax1_p.set_ylabel('Normalized Power')
ax1_r.set_ylabel(r'$R_{\mathrm{sh}}$ ($\Omega\cdot$ cm$^{2})$' )
ax1_r.set_title(r'${0}$'.format(map_parameter_names[parameter]))
ax1_p.set_xlim(0, t_max_h)
ax1_r.set_xlim(0, t_max_h)
# ax1_p.set_ylim(top=1.1, bottom=0.2)
ax1_p.set_xlabel('Time (hr)')
ax1_r.set_xlabel('Time (hr)')
# ax1.tick_params(labelbottom=True, top=False, right=True, which='both', labeltop=False)
# ax2.tick_params(labelbottom=True, top=False, right=True, which='both')
ax1_r.set_yscale('log')
ax1_r.yaxis.set_major_locator(mpl.ticker.LogLocator(base=10.0, numticks=5))
ax1_r.yaxis.set_minor_locator(mpl.ticker.LogLocator(base=10.0, numticks=50, subs=np.arange(2, 10) * .1))
ax1_r.axhline(y=1000, lw=1.5, ls='--', color=(0.9, 0.9, 0.9), zorder=0)
ax1_p.xaxis.set_major_formatter(xfmt)
ax1_p.xaxis.set_major_locator(mticker.MaxNLocator(12, prune=None))
ax1_p.xaxis.set_minor_locator(mticker.AutoMinorLocator(2))
ax1_p.yaxis.set_major_formatter(xfmt)
ax1_p.yaxis.set_major_locator(mticker.MaxNLocator(5, prune=None))
ax1_p.yaxis.set_minor_locator(mticker.AutoMinorLocator(2))
ax1_r.xaxis.set_major_formatter(xfmt)
ax1_r.xaxis.set_major_locator(mticker.MaxNLocator(12, prune=None))
ax1_r.xaxis.set_minor_locator(mticker.AutoMinorLocator(2))
# leg1 = ax1_p.legend(bbox_to_anchor=(1.05, 1.), loc='upper left', borderaxespad=0., ncol=1, frameon=False)
# leg2 = ax1_r.legend(bbox_to_anchor=(1.05, 1.), loc='upper left', borderaxespad=0., ncol=1, frameon=False)
if parameter == 'DSF':
leg_cols = 2
else:
leg_cols = 1
leg1 = ax1_p.legend(loc='upper right', frameon=False, ncol=leg_cols, fontsize=8)
leg2 = ax1_r.legend(loc='upper right', frameon=False, ncol=leg_cols, fontsize=8)
fig_p.tight_layout()
fig_r.tight_layout()
plt.show()
output_file_tag = 'ofat_parameter_{}'.format(slugify(value=parameter))
fig_p.savefig(os.path.join(output_path, output_file_tag + '_power.png'), dpi=600)
fig_p.savefig(os.path.join(output_path, output_file_tag + '_power.svg'), dpi=600)
fig_r.savefig(os.path.join(output_path, output_file_tag + '_rsh.png'), dpi=600)
fig_r.savefig(os.path.join(output_path, output_file_tag + '_rsh.svg'), dpi=600)
df_degradation =
|
pd.DataFrame(failure_times)
|
pandas.DataFrame
|
'''
get a list of unique SNP/TAG/Gene
'''
import pandas as pd
from tqdm import tqdm
df =
|
pd.read_csv('gwas_RS_IDs_tagged.csv', low_memory=False)
|
pandas.read_csv
|
import pandas as pd
import numpy as np
from datetime import date
"""
dataset split:
(date_received)
dateset3: 20160701~20160731 (113640),features3 from 20160315~20160630 (off_test)
dateset2: 20160515~20160615 (258446),features2 from 20160201~20160514
dateset1: 20160414~20160514 (138303),features1 from 20160101~20160413
1.merchant related:
sales_use_coupon. total_coupon
transfer_rate = sales_use_coupon/total_coupon.
merchant_avg_distance,merchant_min_distance,merchant_max_distance of those use coupon
total_sales. coupon_rate = sales_use_coupon/total_sales.
2.coupon related:
discount_rate. discount_man. discount_jian. is_man_jian
day_of_week,day_of_month. (date_received)
3.user related:
distance.
user_avg_distance, user_min_distance,user_max_distance.
buy_use_coupon. buy_total. coupon_received.
buy_use_coupon/coupon_received.
avg_diff_date_datereceived. min_diff_date_datereceived. max_diff_date_datereceived.
count_merchant.
4.user_merchant:
times_user_buy_merchant_before.
5. other feature:
this_month_user_receive_all_coupon_count
this_month_user_receive_same_coupon_count
this_month_user_receive_same_coupon_lastone
this_month_user_receive_same_coupon_firstone
this_day_user_receive_all_coupon_count
this_day_user_receive_same_coupon_count
day_gap_before, day_gap_after (receive the same coupon)
"""
#1754884 record,1053282 with coupon_id,9738 coupon. date_received:20160101~20160615,date:20160101~20160630, 539438 users, 8415 merchants
off_train = pd.read_csv('data/ccf_offline_stage1_train.csv',header=None)
off_train.columns = ['user_id','merchant_id','coupon_id','discount_rate','distance','date_received','date']
#2050 coupon_id. date_received:20160701~20160731, 76309 users(76307 in trainset, 35965 in online_trainset), 1559 merchants(1558 in trainset)
off_test = pd.read_csv('data/ccf_offline_stage1_test_revised.csv',header=None)
off_test.columns = ['user_id','merchant_id','coupon_id','discount_rate','distance','date_received']
#11429826 record(872357 with coupon_id),762858 user(267448 in off_train)
on_train = pd.read_csv('data/ccf_online_stage1_train.csv',header=None)
on_train.columns = ['user_id','merchant_id','action','coupon_id','discount_rate','date_received','date']
dataset3 = off_test
feature3 = off_train[((off_train.date>='20160315')&(off_train.date<='20160630'))|((off_train.date=='null')&(off_train.date_received>='20160315')&(off_train.date_received<='20160630'))]
dataset2 = off_train[(off_train.date_received>='20160515')&(off_train.date_received<='20160615')]
feature2 = off_train[(off_train.date>='20160201')&(off_train.date<='20160514')|((off_train.date=='null')&(off_train.date_received>='20160201')&(off_train.date_received<='20160514'))]
dataset1 = off_train[(off_train.date_received>='20160414')&(off_train.date_received<='20160514')]
feature1 = off_train[(off_train.date>='20160101')&(off_train.date<='20160413')|((off_train.date=='null')&(off_train.date_received>='20160101')&(off_train.date_received<='20160413'))]
############# other feature ##################3
"""
5. other feature:
this_month_user_receive_all_coupon_count
this_month_user_receive_same_coupon_count
this_month_user_receive_same_coupon_lastone
this_month_user_receive_same_coupon_firstone
this_day_user_receive_all_coupon_count
this_day_user_receive_same_coupon_count
day_gap_before, day_gap_after (receive the same coupon)
"""
#for dataset3
t = dataset3[['user_id']]
t['this_month_user_receive_all_coupon_count'] = 1
t = t.groupby('user_id').agg('sum').reset_index()
t1 = dataset3[['user_id','coupon_id']]
t1['this_month_user_receive_same_coupon_count'] = 1
t1 = t1.groupby(['user_id','coupon_id']).agg('sum').reset_index()
t2 = dataset3[['user_id','coupon_id','date_received']]
t2.date_received = t2.date_received.astype('str')
t2 = t2.groupby(['user_id','coupon_id'])['date_received'].agg(lambda x:':'.join(x)).reset_index()
t2['receive_number'] = t2.date_received.apply(lambda s:len(s.split(':')))
t2 = t2[t2.receive_number>1]
t2['max_date_received'] = t2.date_received.apply(lambda s:max([int(d) for d in s.split(':')]))
t2['min_date_received'] = t2.date_received.apply(lambda s:min([int(d) for d in s.split(':')]))
t2 = t2[['user_id','coupon_id','max_date_received','min_date_received']]
t3 = dataset3[['user_id','coupon_id','date_received']]
t3 = pd.merge(t3,t2,on=['user_id','coupon_id'],how='left')
t3['this_month_user_receive_same_coupon_lastone'] = t3.max_date_received - t3.date_received
t3['this_month_user_receive_same_coupon_firstone'] = t3.date_received - t3.min_date_received
def is_firstlastone(x):
if x==0:
return 1
elif x>0:
return 0
else:
return -1 #those only receive once
t3.this_month_user_receive_same_coupon_lastone = t3.this_month_user_receive_same_coupon_lastone.apply(is_firstlastone)
t3.this_month_user_receive_same_coupon_firstone = t3.this_month_user_receive_same_coupon_firstone.apply(is_firstlastone)
t3 = t3[['user_id','coupon_id','date_received','this_month_user_receive_same_coupon_lastone','this_month_user_receive_same_coupon_firstone']]
t4 = dataset3[['user_id','date_received']]
t4['this_day_user_receive_all_coupon_count'] = 1
t4 = t4.groupby(['user_id','date_received']).agg('sum').reset_index()
t5 = dataset3[['user_id','coupon_id','date_received']]
t5['this_day_user_receive_same_coupon_count'] = 1
t5 = t5.groupby(['user_id','coupon_id','date_received']).agg('sum').reset_index()
t6 = dataset3[['user_id','coupon_id','date_received']]
t6.date_received = t6.date_received.astype('str')
t6 = t6.groupby(['user_id','coupon_id'])['date_received'].agg(lambda x:':'.join(x)).reset_index()
t6.rename(columns={'date_received':'dates'},inplace=True)
def get_day_gap_before(s):
date_received,dates = s.split('-')
dates = dates.split(':')
gaps = []
for d in dates:
this_gap = (date(int(date_received[0:4]),int(date_received[4:6]),int(date_received[6:8]))-date(int(d[0:4]),int(d[4:6]),int(d[6:8]))).days
if this_gap>0:
gaps.append(this_gap)
if len(gaps)==0:
return -1
else:
return min(gaps)
def get_day_gap_after(s):
date_received,dates = s.split('-')
dates = dates.split(':')
gaps = []
for d in dates:
this_gap = (date(int(d[0:4]),int(d[4:6]),int(d[6:8]))-date(int(date_received[0:4]),int(date_received[4:6]),int(date_received[6:8]))).days
if this_gap>0:
gaps.append(this_gap)
if len(gaps)==0:
return -1
else:
return min(gaps)
t7 = dataset3[['user_id','coupon_id','date_received']]
t7 = pd.merge(t7,t6,on=['user_id','coupon_id'],how='left')
t7['date_received_date'] = t7.date_received.astype('str') + '-' + t7.dates
t7['day_gap_before'] = t7.date_received_date.apply(get_day_gap_before)
t7['day_gap_after'] = t7.date_received_date.apply(get_day_gap_after)
t7 = t7[['user_id','coupon_id','date_received','day_gap_before','day_gap_after']]
other_feature3 = pd.merge(t1,t,on='user_id')
other_feature3 = pd.merge(other_feature3,t3,on=['user_id','coupon_id'])
other_feature3 = pd.merge(other_feature3,t4,on=['user_id','date_received'])
other_feature3 = pd.merge(other_feature3,t5,on=['user_id','coupon_id','date_received'])
other_feature3 = pd.merge(other_feature3,t7,on=['user_id','coupon_id','date_received'])
other_feature3.to_csv('data/other_feature3.csv',index=None)
print(other_feature3.shape)
#for dataset2
t = dataset2[['user_id']]
t['this_month_user_receive_all_coupon_count'] = 1
t = t.groupby('user_id').agg('sum').reset_index()
t1 = dataset2[['user_id','coupon_id']]
t1['this_month_user_receive_same_coupon_count'] = 1
t1 = t1.groupby(['user_id','coupon_id']).agg('sum').reset_index()
t2 = dataset2[['user_id','coupon_id','date_received']]
t2.date_received = t2.date_received.astype('str')
t2 = t2.groupby(['user_id','coupon_id'])['date_received'].agg(lambda x:':'.join(x)).reset_index()
t2['receive_number'] = t2.date_received.apply(lambda s:len(s.split(':')))
t2 = t2[t2.receive_number>1]
t2['max_date_received'] = t2.date_received.apply(lambda s:max([int(d) for d in s.split(':')]))
t2['min_date_received'] = t2.date_received.apply(lambda s:min([int(d) for d in s.split(':')]))
t2 = t2[['user_id','coupon_id','max_date_received','min_date_received']]
t3 = dataset2[['user_id','coupon_id','date_received']]
t3 = pd.merge(t3,t2,on=['user_id','coupon_id'],how='left')
t3['this_month_user_receive_same_coupon_lastone'] = t3.max_date_received - t3.date_received.astype('int')
t3['this_month_user_receive_same_coupon_firstone'] = t3.date_received.astype('int') - t3.min_date_received
def is_firstlastone(x):
if x==0:
return 1
elif x>0:
return 0
else:
return -1 #those only receive once
t3.this_month_user_receive_same_coupon_lastone = t3.this_month_user_receive_same_coupon_lastone.apply(is_firstlastone)
t3.this_month_user_receive_same_coupon_firstone = t3.this_month_user_receive_same_coupon_firstone.apply(is_firstlastone)
t3 = t3[['user_id','coupon_id','date_received','this_month_user_receive_same_coupon_lastone','this_month_user_receive_same_coupon_firstone']]
t4 = dataset2[['user_id','date_received']]
t4['this_day_user_receive_all_coupon_count'] = 1
t4 = t4.groupby(['user_id','date_received']).agg('sum').reset_index()
t5 = dataset2[['user_id','coupon_id','date_received']]
t5['this_day_user_receive_same_coupon_count'] = 1
t5 = t5.groupby(['user_id','coupon_id','date_received']).agg('sum').reset_index()
t6 = dataset2[['user_id','coupon_id','date_received']]
t6.date_received = t6.date_received.astype('str')
t6 = t6.groupby(['user_id','coupon_id'])['date_received'].agg(lambda x:':'.join(x)).reset_index()
t6.rename(columns={'date_received':'dates'},inplace=True)
def get_day_gap_before(s):
date_received,dates = s.split('-')
dates = dates.split(':')
gaps = []
for d in dates:
this_gap = (date(int(date_received[0:4]),int(date_received[4:6]),int(date_received[6:8]))-date(int(d[0:4]),int(d[4:6]),int(d[6:8]))).days
if this_gap>0:
gaps.append(this_gap)
if len(gaps)==0:
return -1
else:
return min(gaps)
def get_day_gap_after(s):
date_received,dates = s.split('-')
dates = dates.split(':')
gaps = []
for d in dates:
this_gap = (date(int(d[0:4]),int(d[4:6]),int(d[6:8]))-date(int(date_received[0:4]),int(date_received[4:6]),int(date_received[6:8]))).days
if this_gap>0:
gaps.append(this_gap)
if len(gaps)==0:
return -1
else:
return min(gaps)
t7 = dataset2[['user_id','coupon_id','date_received']]
t7 = pd.merge(t7,t6,on=['user_id','coupon_id'],how='left')
t7['date_received_date'] = t7.date_received.astype('str') + '-' + t7.dates
t7['day_gap_before'] = t7.date_received_date.apply(get_day_gap_before)
t7['day_gap_after'] = t7.date_received_date.apply(get_day_gap_after)
t7 = t7[['user_id','coupon_id','date_received','day_gap_before','day_gap_after']]
other_feature2 = pd.merge(t1,t,on='user_id')
other_feature2 = pd.merge(other_feature2,t3,on=['user_id','coupon_id'])
other_feature2 = pd.merge(other_feature2,t4,on=['user_id','date_received'])
other_feature2 = pd.merge(other_feature2,t5,on=['user_id','coupon_id','date_received'])
other_feature2 = pd.merge(other_feature2,t7,on=['user_id','coupon_id','date_received'])
other_feature2.to_csv('data/other_feature2.csv',index=None)
print(other_feature2.shape)
#for dataset1
t = dataset1[['user_id']]
t['this_month_user_receive_all_coupon_count'] = 1
t = t.groupby('user_id').agg('sum').reset_index()
t1 = dataset1[['user_id','coupon_id']]
t1['this_month_user_receive_same_coupon_count'] = 1
t1 = t1.groupby(['user_id','coupon_id']).agg('sum').reset_index()
t2 = dataset1[['user_id','coupon_id','date_received']]
t2.date_received = t2.date_received.astype('str')
t2 = t2.groupby(['user_id','coupon_id'])['date_received'].agg(lambda x:':'.join(x)).reset_index()
t2['receive_number'] = t2.date_received.apply(lambda s:len(s.split(':')))
t2 = t2[t2.receive_number>1]
t2['max_date_received'] = t2.date_received.apply(lambda s:max([int(d) for d in s.split(':')]))
t2['min_date_received'] = t2.date_received.apply(lambda s:min([int(d) for d in s.split(':')]))
t2 = t2[['user_id','coupon_id','max_date_received','min_date_received']]
t3 = dataset1[['user_id','coupon_id','date_received']]
t3 = pd.merge(t3,t2,on=['user_id','coupon_id'],how='left')
t3['this_month_user_receive_same_coupon_lastone'] = t3.max_date_received - t3.date_received.astype('int')
t3['this_month_user_receive_same_coupon_firstone'] = t3.date_received.astype('int') - t3.min_date_received
def is_firstlastone(x):
if x==0:
return 1
elif x>0:
return 0
else:
return -1 #those only receive once
t3.this_month_user_receive_same_coupon_lastone = t3.this_month_user_receive_same_coupon_lastone.apply(is_firstlastone)
t3.this_month_user_receive_same_coupon_firstone = t3.this_month_user_receive_same_coupon_firstone.apply(is_firstlastone)
t3 = t3[['user_id','coupon_id','date_received','this_month_user_receive_same_coupon_lastone','this_month_user_receive_same_coupon_firstone']]
t4 = dataset1[['user_id','date_received']]
t4['this_day_user_receive_all_coupon_count'] = 1
t4 = t4.groupby(['user_id','date_received']).agg('sum').reset_index()
t5 = dataset1[['user_id','coupon_id','date_received']]
t5['this_day_user_receive_same_coupon_count'] = 1
t5 = t5.groupby(['user_id','coupon_id','date_received']).agg('sum').reset_index()
t6 = dataset1[['user_id','coupon_id','date_received']]
t6.date_received = t6.date_received.astype('str')
t6 = t6.groupby(['user_id','coupon_id'])['date_received'].agg(lambda x:':'.join(x)).reset_index()
t6.rename(columns={'date_received':'dates'},inplace=True)
def get_day_gap_before(s):
date_received,dates = s.split('-')
dates = dates.split(':')
gaps = []
for d in dates:
this_gap = (date(int(date_received[0:4]),int(date_received[4:6]),int(date_received[6:8]))-date(int(d[0:4]),int(d[4:6]),int(d[6:8]))).days
if this_gap>0:
gaps.append(this_gap)
if len(gaps)==0:
return -1
else:
return min(gaps)
def get_day_gap_after(s):
date_received,dates = s.split('-')
dates = dates.split(':')
gaps = []
for d in dates:
this_gap = (date(int(d[0:4]),int(d[4:6]),int(d[6:8]))-date(int(date_received[0:4]),int(date_received[4:6]),int(date_received[6:8]))).days
if this_gap>0:
gaps.append(this_gap)
if len(gaps)==0:
return -1
else:
return min(gaps)
t7 = dataset1[['user_id','coupon_id','date_received']]
t7 = pd.merge(t7,t6,on=['user_id','coupon_id'],how='left')
t7['date_received_date'] = t7.date_received.astype('str') + '-' + t7.dates
t7['day_gap_before'] = t7.date_received_date.apply(get_day_gap_before)
t7['day_gap_after'] = t7.date_received_date.apply(get_day_gap_after)
t7 = t7[['user_id','coupon_id','date_received','day_gap_before','day_gap_after']]
other_feature1 = pd.merge(t1,t,on='user_id')
other_feature1 = pd.merge(other_feature1,t3,on=['user_id','coupon_id'])
other_feature1 = pd.merge(other_feature1,t4,on=['user_id','date_received'])
other_feature1 = pd.merge(other_feature1,t5,on=['user_id','coupon_id','date_received'])
other_feature1 = pd.merge(other_feature1,t7,on=['user_id','coupon_id','date_received'])
other_feature1.to_csv('data/other_feature1.csv',index=None)
print(other_feature1.shape)
############# coupon related feature #############
"""
2.coupon related:
discount_rate. discount_man. discount_jian. is_man_jian
day_of_week,day_of_month. (date_received)
"""
def calc_discount_rate(s):
s =str(s)
s = s.split(':')
if len(s)==1:
return float(s[0])
else:
return 1.0-float(s[1])/float(s[0])
def get_discount_man(s):
s =str(s)
s = s.split(':')
if len(s)==1:
return 'null'
else:
return int(s[0])
def get_discount_jian(s):
s =str(s)
s = s.split(':')
if len(s)==1:
return 'null'
else:
return int(s[1])
def is_man_jian(s):
s =str(s)
s = s.split(':')
if len(s)==1:
return 0
else:
return 1
#dataset3
dataset3['day_of_week'] = dataset3.date_received.astype('str').apply(lambda x:date(int(x[0:4]),int(x[4:6]),int(x[6:8])).weekday()+1)
dataset3['day_of_month'] = dataset3.date_received.astype('str').apply(lambda x:int(x[6:8]))
dataset3['days_distance'] = dataset3.date_received.astype('str').apply(lambda x:(date(int(x[0:4]),int(x[4:6]),int(x[6:8]))-date(2016,6,30)).days)
dataset3['discount_man'] = dataset3.discount_rate.apply(get_discount_man)
dataset3['discount_jian'] = dataset3.discount_rate.apply(get_discount_jian)
dataset3['is_man_jian'] = dataset3.discount_rate.apply(is_man_jian)
dataset3['discount_rate'] = dataset3.discount_rate.apply(calc_discount_rate)
d = dataset3[['coupon_id']]
d['coupon_count'] = 1
d = d.groupby('coupon_id').agg('sum').reset_index()
dataset3 = pd.merge(dataset3,d,on='coupon_id',how='left')
dataset3.to_csv('data/coupon3_feature.csv',index=None)
#dataset2
dataset2['day_of_week'] = dataset2.date_received.astype('str').apply(lambda x:date(int(x[0:4]),int(x[4:6]),int(x[6:8])).weekday()+1)
dataset2['day_of_month'] = dataset2.date_received.astype('str').apply(lambda x:int(x[6:8]))
dataset2['days_distance'] = dataset2.date_received.astype('str').apply(lambda x:(date(int(x[0:4]),int(x[4:6]),int(x[6:8]))-date(2016,5,14)).days)
dataset2['discount_man'] = dataset2.discount_rate.apply(get_discount_man)
dataset2['discount_jian'] = dataset2.discount_rate.apply(get_discount_jian)
dataset2['is_man_jian'] = dataset2.discount_rate.apply(is_man_jian)
dataset2['discount_rate'] = dataset2.discount_rate.apply(calc_discount_rate)
d = dataset2[['coupon_id']]
d['coupon_count'] = 1
d = d.groupby('coupon_id').agg('sum').reset_index()
dataset2 = pd.merge(dataset2,d,on='coupon_id',how='left')
dataset2.to_csv('data/coupon2_feature.csv',index=None)
#dataset1
dataset1['day_of_week'] = dataset1.date_received.astype('str').apply(lambda x:date(int(x[0:4]),int(x[4:6]),int(x[6:8])).weekday()+1)
dataset1['day_of_month'] = dataset1.date_received.astype('str').apply(lambda x:int(x[6:8]))
dataset1['days_distance'] = dataset1.date_received.astype('str').apply(lambda x:(date(int(x[0:4]),int(x[4:6]),int(x[6:8]))-date(2016,4,13)).days)
dataset1['discount_man'] = dataset1.discount_rate.apply(get_discount_man)
dataset1['discount_jian'] = dataset1.discount_rate.apply(get_discount_jian)
dataset1['is_man_jian'] = dataset1.discount_rate.apply(is_man_jian)
dataset1['discount_rate'] = dataset1.discount_rate.apply(calc_discount_rate)
d = dataset1[['coupon_id']]
d['coupon_count'] = 1
d = d.groupby('coupon_id').agg('sum').reset_index()
dataset1 = pd.merge(dataset1,d,on='coupon_id',how='left')
dataset1.to_csv('data/coupon1_feature.csv',index=None)
############# merchant related feature #############
"""
1.merchant related:
total_sales. sales_use_coupon. total_coupon
coupon_rate = sales_use_coupon/total_sales.
transfer_rate = sales_use_coupon/total_coupon.
merchant_avg_distance,merchant_min_distance,merchant_max_distance of those use coupon
"""
#for dataset3
merchant3 = feature3[['merchant_id','coupon_id','distance','date_received','date']]
t = merchant3[['merchant_id']]
t.drop_duplicates(inplace=True)
t1 = merchant3[merchant3.date!='null'][['merchant_id']]
t1['total_sales'] = 1
t1 = t1.groupby('merchant_id').agg('sum').reset_index()
t2 = merchant3[(merchant3.date!='null')&(merchant3.coupon_id!='null')][['merchant_id']]
t2['sales_use_coupon'] = 1
t2 = t2.groupby('merchant_id').agg('sum').reset_index()
t3 = merchant3[merchant3.coupon_id!='null'][['merchant_id']]
t3['total_coupon'] = 1
t3 = t3.groupby('merchant_id').agg('sum').reset_index()
t4 = merchant3[(merchant3.date!='null')&(merchant3.coupon_id!='null')][['merchant_id','distance']]
t4.replace('null',-1,inplace=True)
t4.distance = t4.distance.astype('int')
t4.replace(-1,np.nan,inplace=True)
t5 = t4.groupby('merchant_id').agg('min').reset_index()
t5.rename(columns={'distance':'merchant_min_distance'},inplace=True)
t6 = t4.groupby('merchant_id').agg('max').reset_index()
t6.rename(columns={'distance':'merchant_max_distance'},inplace=True)
t7 = t4.groupby('merchant_id').agg('mean').reset_index()
t7.rename(columns={'distance':'merchant_mean_distance'},inplace=True)
t8 = t4.groupby('merchant_id').agg('median').reset_index()
t8.rename(columns={'distance':'merchant_median_distance'},inplace=True)
merchant3_feature = pd.merge(t,t1,on='merchant_id',how='left')
merchant3_feature = pd.merge(merchant3_feature,t2,on='merchant_id',how='left')
merchant3_feature = pd.merge(merchant3_feature,t3,on='merchant_id',how='left')
merchant3_feature = pd.merge(merchant3_feature,t5,on='merchant_id',how='left')
merchant3_feature = pd.merge(merchant3_feature,t6,on='merchant_id',how='left')
merchant3_feature = pd.merge(merchant3_feature,t7,on='merchant_id',how='left')
merchant3_feature = pd.merge(merchant3_feature,t8,on='merchant_id',how='left')
merchant3_feature.sales_use_coupon = merchant3_feature.sales_use_coupon.replace(np.nan,0) #fillna with 0
merchant3_feature['merchant_coupon_transfer_rate'] = merchant3_feature.sales_use_coupon.astype('float') / merchant3_feature.total_coupon
merchant3_feature['coupon_rate'] = merchant3_feature.sales_use_coupon.astype('float') / merchant3_feature.total_sales
merchant3_feature.total_coupon = merchant3_feature.total_coupon.replace(np.nan,0) #fillna with 0
merchant3_feature.to_csv('data/merchant3_feature.csv',index=None)
#for dataset2
merchant2 = feature2[['merchant_id','coupon_id','distance','date_received','date']]
t = merchant2[['merchant_id']]
t.drop_duplicates(inplace=True)
t1 = merchant2[merchant2.date!='null'][['merchant_id']]
t1['total_sales'] = 1
t1 = t1.groupby('merchant_id').agg('sum').reset_index()
t2 = merchant2[(merchant2.date!='null')&(merchant2.coupon_id!='null')][['merchant_id']]
t2['sales_use_coupon'] = 1
t2 = t2.groupby('merchant_id').agg('sum').reset_index()
t3 = merchant2[merchant2.coupon_id!='null'][['merchant_id']]
t3['total_coupon'] = 1
t3 = t3.groupby('merchant_id').agg('sum').reset_index()
t4 = merchant2[(merchant2.date!='null')&(merchant2.coupon_id!='null')][['merchant_id','distance']]
t4.replace('null',-1,inplace=True)
t4.distance = t4.distance.astype('int')
t4.replace(-1,np.nan,inplace=True)
t5 = t4.groupby('merchant_id').agg('min').reset_index()
t5.rename(columns={'distance':'merchant_min_distance'},inplace=True)
t6 = t4.groupby('merchant_id').agg('max').reset_index()
t6.rename(columns={'distance':'merchant_max_distance'},inplace=True)
t7 = t4.groupby('merchant_id').agg('mean').reset_index()
t7.rename(columns={'distance':'merchant_mean_distance'},inplace=True)
t8 = t4.groupby('merchant_id').agg('median').reset_index()
t8.rename(columns={'distance':'merchant_median_distance'},inplace=True)
merchant2_feature = pd.merge(t,t1,on='merchant_id',how='left')
merchant2_feature = pd.merge(merchant2_feature,t2,on='merchant_id',how='left')
merchant2_feature = pd.merge(merchant2_feature,t3,on='merchant_id',how='left')
merchant2_feature = pd.merge(merchant2_feature,t5,on='merchant_id',how='left')
merchant2_feature = pd.merge(merchant2_feature,t6,on='merchant_id',how='left')
merchant2_feature = pd.merge(merchant2_feature,t7,on='merchant_id',how='left')
merchant2_feature = pd.merge(merchant2_feature,t8,on='merchant_id',how='left')
merchant2_feature.sales_use_coupon = merchant2_feature.sales_use_coupon.replace(np.nan,0) #fillna with 0
merchant2_feature['merchant_coupon_transfer_rate'] = merchant2_feature.sales_use_coupon.astype('float') / merchant2_feature.total_coupon
merchant2_feature['coupon_rate'] = merchant2_feature.sales_use_coupon.astype('float') / merchant2_feature.total_sales
merchant2_feature.total_coupon = merchant2_feature.total_coupon.replace(np.nan,0) #fillna with 0
merchant2_feature.to_csv('data/merchant2_feature.csv',index=None)
#for dataset1
merchant1 = feature1[['merchant_id','coupon_id','distance','date_received','date']]
t = merchant1[['merchant_id']]
t.drop_duplicates(inplace=True)
t1 = merchant1[merchant1.date!='null'][['merchant_id']]
t1['total_sales'] = 1
t1 = t1.groupby('merchant_id').agg('sum').reset_index()
t2 = merchant1[(merchant1.date!='null')&(merchant1.coupon_id!='null')][['merchant_id']]
t2['sales_use_coupon'] = 1
t2 = t2.groupby('merchant_id').agg('sum').reset_index()
t3 = merchant1[merchant1.coupon_id!='null'][['merchant_id']]
t3['total_coupon'] = 1
t3 = t3.groupby('merchant_id').agg('sum').reset_index()
t4 = merchant1[(merchant1.date!='null')&(merchant1.coupon_id!='null')][['merchant_id','distance']]
t4.replace('null',-1,inplace=True)
t4.distance = t4.distance.astype('int')
t4.replace(-1,np.nan,inplace=True)
t5 = t4.groupby('merchant_id').agg('min').reset_index()
t5.rename(columns={'distance':'merchant_min_distance'},inplace=True)
t6 = t4.groupby('merchant_id').agg('max').reset_index()
t6.rename(columns={'distance':'merchant_max_distance'},inplace=True)
t7 = t4.groupby('merchant_id').agg('mean').reset_index()
t7.rename(columns={'distance':'merchant_mean_distance'},inplace=True)
t8 = t4.groupby('merchant_id').agg('median').reset_index()
t8.rename(columns={'distance':'merchant_median_distance'},inplace=True)
merchant1_feature = pd.merge(t,t1,on='merchant_id',how='left')
merchant1_feature = pd.merge(merchant1_feature,t2,on='merchant_id',how='left')
merchant1_feature = pd.merge(merchant1_feature,t3,on='merchant_id',how='left')
merchant1_feature = pd.merge(merchant1_feature,t5,on='merchant_id',how='left')
merchant1_feature =
|
pd.merge(merchant1_feature,t6,on='merchant_id',how='left')
|
pandas.merge
|
""" Sixth version, make the code easier and more modifiable """
# Define the main programme
from funcs import store_namespace
from funcs import load_namespace
from funcs import emulate_jmod
import os
import datetime
import time
import pandas as pd
#from multiprocessing import Pool
from mpcpy import units
from mpcpy import variables
from mpcpy import models_mod as models
from Simulator_HP_mod3 import SimHandler
if __name__ == "__main__":
# Naming conventions for the simulation
community = 'ResidentialCommunityUK_rad_2elements'
sim_id = 'MinEne'
model_id = 'R2CW_HP'
bldg_list = load_namespace(os.path.join('path_to_models', 'teaser_bldgs_residentialUK_10bldgs_fallback'))
folder = 'results'
bldg_index_start = 0
bldg_index_end = 10
# Overall options
date = '1/7/2017 '
start = date + '16:30:00'
end = date + '19:00:00'
meas_sampl = '300'
horizon = 2*3600/float(meas_sampl) #time horizon for optimization in multiples of the sample
mon = 'jan'
DRstart = datetime.datetime.strptime(date + '17:30:00', '%m/%d/%Y %H:%M:%S') # hour to start DR - ramp down 30 mins before
DRend = datetime.datetime.strptime(date + '18:30:00', '%m/%d/%Y %H:%M:%S') # hour to end DR - ramp 30 mins later
DR_call_start = datetime.datetime.strptime(date + '17:00:00', '%m/%d/%Y %H:%M:%S') # Round of loop to implement the call
DR_ramp_start = datetime.datetime.strptime(date + '17:30:00', '%m/%d/%Y %H:%M:%S')
DR_ramp_end = datetime.datetime.strptime(date + '18:30:00', '%m/%d/%Y %H:%M:%S') # Round of loop to stop implementing the call
flex_cost = 150 # Cost for flexibility
compr_capacity=float(3000)
ramp_modifier = float(2000/compr_capacity) # to further modify the load profile
max_modifier = float(2000/compr_capacity)
dyn_price = 1
stat_cost = 50
#set_change = 1
sim_range = pd.date_range(start, end, freq = meas_sampl+'S')
opt_start_str = start
opt_end = datetime.datetime.strptime(end, '%m/%d/%Y %H:%M:%S') + datetime.timedelta(seconds = horizon*int(meas_sampl))
opt_end_str = opt_end.strftime('%m/%d/%Y %H:%M:%S')
init_start = sim_range[0] - datetime.timedelta(seconds = 0.5*3600)
init_start_str = init_start.strftime('%m/%d/%Y %H:%M:%S')
print(init_start_str)
# Instantiate Simulator for aggregated optimisation
SimAggr = SimHandler(sim_start = start,
sim_end = end,
meas_sampl = meas_sampl
)
SimAggr.moinfo_mpc = (os.path.join(SimAggr.simu_path, 'AggrMPC_ResUK_10bldgs_heatpump_rad_fallback.mo'),
'AggrMPC_ResUK_10bldgs_heatpump_rad_fallback.Residential',
{}
)
SimAggr.building = 'AggrMPC_ResUK_10bldgs_heatpump_rad_fallback'
SimAggr.target_variable = 'TotalHeatPower'
# Not really used in this case ...
SimAggr.fmupath_emu = os.path.join(SimAggr.simu_path, 'fmus', community, 'AggrMPC_ResUK_10bldgs_heatpump_rad_fallback_Residential.fmu')
SimAggr.fmupath_mpc = os.path.join(SimAggr.simu_path, 'fmus', community, 'AggrMPC_ResUK_10bldgs_heatpump_rad_fallback_Residential.fmu')
SimAggr.moinfo_emu = (os.path.join(SimAggr.simu_path, 'AggrMPC_ResUK_10bldgs_heatpump_rad_fallback.mo'),
'AggrMPC_ResUK_10bldgs_heatpump_rad_fallback.Residential',
{}
)
# Initialise aggregated model
# Control
bldg_control = load_namespace(os.path.join(SimAggr.simu_path, 'ibpsa_paper', '10bldgs_decentr_nodyn_jan', 'control_SemiDetached_2_R2CW_HP'))
# Constraints
bldg_constraints = load_namespace(os.path.join(SimAggr.simu_path, 'ibpsa_paper', '10bldgs_decentr_nodyn_jan', 'constraints_SemiDetached_2_R2CW_HP'))
# Optimisation constraints variable map
SimAggr.optcon_varmap = {}
SimAggr.contr_varmap = {}
SimAggr.addobj_varmap = {}
SimAggr.slack_var = []
for bldg in bldg_list:
model_name = bldg+'_'+model_id
for key in bldg_control.data:
SimAggr.contr_varmap[key+'_'+bldg] = (key+'_'+bldg, bldg_control.data[key].get_display_unit())
for key in bldg_constraints.data:
for key1 in bldg_constraints.data[key]:
if key1 != 'Slack_GTE' and key1 != 'Slack_LTE':
SimAggr.optcon_varmap[model_name+'_'+key1] = (model_name+'.'+key, key1, bldg_constraints.data[key][key1].get_display_unit())
else:
SimAggr.optcon_varmap[model_name+'_'+key1] = (model_name + '_TAir' + '_Slack', key1[-3:], units.degC)
SimAggr.slack_var.append(model_name +'_TAir'+ '_Slack')
SimAggr.addobj_varmap[model_name + '_TAir' + '_Slack'] = (model_name + '_TAir' + '_Slack', units.unit1)
#exit()
index = pd.date_range(init_start, opt_end_str, freq = meas_sampl+'S')
SimAggr.constraint_csv = os.path.join(SimAggr.simu_path,'csvs','Constraints_AggrRes.csv')
SimAggr.control_file = os.path.join(SimAggr.simu_path,'csvs','ControlSignal_AggrRes.csv')
SimAggr.price_file = os.path.join(SimAggr.simu_path,'csvs','PriceSignal.csv')
SimAggr.param_file = os.path.join(SimAggr.simu_path,'csvs','Parameters.csv')
# Initialise exogenous data sources
SimAggr.update_weather(init_start_str,opt_end_str)
SimAggr.get_DRinfo(init_start_str,opt_end_str)
SimAggr.get_control()
SimAggr.get_params()
SimAggr.get_other_input(init_start_str,opt_end_str)
SimAggr.get_constraints(init_start_str,opt_end_str,upd_control = 1)
# Empty old data
SimAggr.parameters.data = {}
SimAggr.control.data = {}
SimAggr.constraints.data = {}
SimAggr.meas_varmap_mpc = {}
SimAggr.meas_vars_mpc = {}
SimAggr.other_input.data = {}
index = pd.date_range(init_start_str, opt_end_str, freq = meas_sampl+'S', tz=SimAggr.weather.tz_name)
for bldg in bldg_list:
#Parameters from system id
bldg_params = load_namespace(os.path.join(SimAggr.simu_path, 'sysid', 'sysid_HPrad_2element_'+mon+'_600S','est_params_'+bldg+'_'+model_id))
bldg_other_input = load_namespace(os.path.join(SimAggr.simu_path, 'ibpsa_paper', 'decentr_enemin_'+mon, 'other_input_'+bldg+'_'+model_id))
bldg_constraints = load_namespace(os.path.join(SimAggr.simu_path, 'ibpsa_paper', 'decentr_enemin_'+mon, 'constraints_'+bldg+'_'+model_id))
model_name = bldg+'_'+model_id
pheat_min = pd.Series(0,index=index)
pheat_max = pd.Series(1,index=index)
bldg_constraints.data['HPPower']= {'GTE': variables.Timeseries('HPPower_GTE', pheat_min, units.W, tz_name=SimAggr.weather.tz_name),
'LTE': variables.Timeseries('HPPower_LTE', pheat_max, units.W,tz_name=SimAggr.weather.tz_name)}
#print(SimAggr.start_temp)
for key in bldg_params:
SimAggr.parameters.data[model_name+'.'+key] = {'Free': variables.Static('FreeOrNot', bldg_params[key]['Free'].data, units.boolean),
'Minimum': variables.Static('Min', bldg_params[key]['Minimum'].data, bldg_params[key]['Minimum'].get_display_unit()),
'Covariance': variables.Static('Covar', bldg_params[key]['Covariance'].data, bldg_params[key]['Covariance'].get_display_unit()),
'Value': variables.Static(model_name+'.'+key, bldg_params[key]['Value'].data, bldg_params[key]['Value'].get_display_unit()),
'Maximum': variables.Static('Max', bldg_params[key]['Maximum'].data, bldg_params[key]['Maximum'].get_display_unit())
}
SimAggr.update_params(model_name+'.heatCapacitor.T.start',SimAggr.start_temp,unit=units.degC)
SimAggr.update_params(model_name+'.heatCapacitor1.T.start',SimAggr.start_temp, unit=units.degC)
if dyn_price == 0:
bldg_control = load_namespace(os.path.join(SimAggr.simu_path, 'ibpsa_paper', '10bldgs_decentr_'+'nodyn_'+mon, 'control_'+bldg+'_'+model_id))
else:
bldg_control = load_namespace(os.path.join(SimAggr.simu_path, 'ibpsa_paper', '10bldgs_decentr_'+'dyn_'+mon, 'control_'+bldg+'_'+model_id))
for key in bldg_control.data:
SimAggr.control.data[key+'_'+bldg] = variables.Timeseries(
name = key+'_'+bldg,
timeseries = bldg_control.data[key].display_data(), # use the same initial guess as with decentralised
display_unit = bldg_control.data[key].get_display_unit(),
tz_name = SimAggr.weather.tz_name
)
for key in bldg_constraints.data:
if key == 'HPPower':
SimAggr.constraints.data[key+'_'+bldg] = {}
else:
SimAggr.constraints.data[model_name+'.'+key] = {}
for key1 in bldg_constraints.data[key]:
if key == 'HPPower':
SimAggr.constraints.data[key+'_'+bldg][key1] = variables.Timeseries(
name = key+'_'+bldg+'_'+key1,
timeseries = bldg_constraints.data[key][key1].display_data().loc[index],
display_unit = bldg_constraints.data[key][key1].get_display_unit(),
tz_name = SimAggr.weather.tz_name
)
else:
if key1 == 'Slack_GTE' or key1 == 'Slack_LTE':
SimAggr.constraints.data[model_name+'.'+key][key1] = variables.Timeseries(
name = model_name+'_'+key+'_'+key1,
timeseries = bldg_constraints.data[key][key1].display_data().loc[index],
display_unit = bldg_constraints.data[key][key1].get_display_unit(),
tz_name = SimAggr.weather.tz_name
)
else:
SimAggr.constraints.data[model_name+'.'+key][key1] = variables.Timeseries(
name = model_name+'_'+key+'_'+key1,
timeseries = bldg_constraints.data[key][key1].display_data().loc[index],
display_unit = bldg_constraints.data[key][key1].get_display_unit(),
tz_name = SimAggr.weather.tz_name
)
SimAggr.meas_varmap_mpc[model_name+'.'+'TAir'] = (model_name+'.'+'TAir', units.K)
SimAggr.meas_vars_mpc[model_name+'.'+'TAir'] = {}
SimAggr.meas_vars_mpc[model_name+'.'+'TAir']['Sample'] = variables.Static('sample_rate_TAir', int(meas_sampl), units.s)
SimAggr.meas_varmap_emu = SimAggr.meas_varmap_mpc
SimAggr.meas_vars_emu = SimAggr.meas_vars_mpc
index = pd.date_range(init_start_str, opt_end_str, freq = meas_sampl+'S')
SimAggr.price = load_namespace(os.path.join('prices','sim_price_'+mon))
if dyn_price == 0:
index = pd.date_range(init_start_str, opt_end_str, freq = '1800S')
price_signal = pd.Series(50,index=index)
SimAggr.price.data = {"pi_e": variables.Timeseries('pi_e', price_signal,units.cents_kWh,tz_name=SimAggr.weather.tz_name)
}
store_namespace(os.path.join(folder, 'sim_price'), SimAggr.price)
store_namespace(os.path.join(folder, 'params_'+SimAggr.building), SimAggr.parameters)
store_namespace(os.path.join(folder, 'control_'+SimAggr.building), SimAggr.control)
store_namespace(os.path.join(folder, 'constraints_'+SimAggr.building), SimAggr.constraints)
SimAggr.init_models(use_ukf=0, use_fmu_emu=0, use_fmu_mpc=0) # Use for initialising models
#SimAggr.init_refmodel(use_fmu=1)
# Instantiate Simulator
Emu_list = []
i = 0
for bldg in bldg_list[bldg_index_start:bldg_index_end]:
i = i+1
print('Instantiating emulation models, loop: ' + str(i))
Sim = SimHandler(sim_start = start,
sim_end = end,
meas_sampl = meas_sampl
)
Sim.moinfo_mpc = (os.path.join(Sim.simu_path, 'Tutorial_'+model_id+'.mo'),
'Tutorial_'+model_id+'.'+model_id,
{}
)
Sim.building = bldg+'_'+model_id
Sim.fmupath_mpc = os.path.join(Sim.simu_path, 'fmus',community, 'Tutorial_'+model_id+'_'+model_id+'.fmu')
Sim.fmupath_emu = os.path.join(Sim.simu_path, 'fmus', community, community+'_'+bldg+'_'+bldg+'_Models_'+bldg+'_House_mpc.fmu')
Sim.fmupath_ref = os.path.join(Sim.simu_path, 'fmus', community, community+'_'+bldg+'_'+bldg+'_Models_'+bldg+'_House_PI.fmu')
Sim.moinfo_emu = (os.path.join(Sim.mod_path, community, bldg,bldg+'_Models',bldg+'_House_mpc.mo'), community+'.'+bldg+'.'+bldg+'_Models.'+bldg+'_House_mpc',
{}
)
Sim.moinfo_emu_ref = (os.path.join(Sim.mod_path, community, bldg,bldg+'_Models',bldg+'_House_PI.mo'), community+'.'+bldg+'.'+bldg+'_Models.'+bldg+'_House_PI',
{}
)
# Initialise exogenous data sources
if i == 1:
Sim.weather = SimAggr.weather
Sim.get_DRinfo(init_start_str,opt_end_str)
Sim.flex_cost = SimAggr.flex_cost
Sim.price = SimAggr.price
Sim.rho = SimAggr.rho
#Sim.addobj = SimAggr.addobj
else:
Sim.weather = Emu_list[i-2].weather
Sim.flex_cost = Emu_list[i-2].flex_cost
Sim.price = Emu_list[i-2].price
Sim.rho = Emu_list[i-2].rho
Sim.addobj = Emu_list[i-2].addobj
#Sim.sim_start= '1/1/2017 00:00'
Sim.get_control()
#Sim.sim_start= start
Sim.get_other_input(init_start_str,opt_end_str)
Sim.get_constraints(init_start_str,opt_end_str,upd_control=1)
Sim.param_file = os.path.join(Sim.simu_path,'csvs','Parameters_R2CW.csv')
Sim.get_params()
Sim.parameters.data = load_namespace(os.path.join(Sim.simu_path, 'sysid', 'sysid_HPrad_2element_'+mon+'_600S','est_params_'+Sim.building))
Sim.other_input = load_namespace(os.path.join(Sim.simu_path, 'ibpsa_paper', 'decentr_enemin_'+mon, 'other_input_'+Sim.building))
Sim.constraints = load_namespace(os.path.join(Sim.simu_path, 'ibpsa_paper', 'decentr_enemin_'+mon, 'constraints_'+Sim.building))
# Add to list of simulations
Emu_list.append(Sim)
# Start the hourly loop
i = 0
emutemps = {}
mpctemps = {}
controlseq = {}
power = {}
opt_stats = {}
refheat = []
reftemps = []
index = pd.date_range(start, opt_end_str, freq = meas_sampl+'S')
flex_cost_signal = pd.Series(0,index=index)
start_temps = []
for Sim in Emu_list:
while True:
try:
# Initialise models
Sim.init_models(use_ukf=1, use_fmu_mpc=1, use_fmu_emu=1) # Use for initialising
emulate_jmod(Sim.emu, Sim.meas_vars_emu, Sim.meas_sampl, init_start_str, start)
Sim.start_temp = Sim.emu.display_measurements('Measured').values[-1][-1]-273.15
print(Sim.emu.display_measurements('Measured'))
out_temp=Sim.weather.display_data()['weaTDryBul'].resample(meas_sampl+'S').ffill()[start]
start_temp = Sim.start_temp
print(out_temp)
print(Sim.start_temp)
Sim.mpc.measurements = {}
Sim.mpc.measurements['TAir'] = Sim.emu.measurements['TAir']
SimAggr.update_params(Sim.building+'.heatCapacitor.T.start',Sim.start_temp+273.15, units.K)
SimAggr.update_params(Sim.building+'.heatCapacitor1.T.start',(7*Sim.start_temp+out_temp)/8+273.15, units.K)
Sim.update_params('C1start', start_temp+273.15, units.K)
Sim.update_params('C2start', (7*start_temp+out_temp)/8+273.15, units.K)
break
except:
print('%%%%%%%%%%%%%%%%%% Failed, trying again! %%%%%%%%%%%%%%%%%%%%%%')
continue
print(sim_range)
for simtime in sim_range:
i = i + 1
print('%%%%%%%%% IN LOOP: ' + str(i) + ' %%%%%%%%%%%%%%%%%')
if i == 1:
simtime_str = 'continue'
else:
simtime_str = 'continue'
opt_start_str = simtime.strftime('%m/%d/%Y %H:%M:%S')
opt_end = simtime + datetime.timedelta(seconds = horizon*int(SimAggr.meas_sampl))
emu_end = simtime + datetime.timedelta(seconds = int(SimAggr.meas_sampl))
opt_end_str = opt_end.strftime('%m/%d/%Y %H:%M:%S')
emu_end_str = emu_end.strftime('%m/%d/%Y %H:%M:%S')
simtime_naive = simtime.replace(tzinfo=None)
print('---- Simulation time: ' + str(simtime) + ' -------')
print('---- Next time step: ' + str(emu_end) + ' -------')
print('---- Optimisation horizon end: ' + str(opt_end) + ' -------')
emutemps = {}
mpctemps = {}
controlseq = {}
power = {}
opt_stats = {}
while True:
try:
# Optimise for next time step
print("%%%%%% --- Optimising --- %%%%%%")
SimAggr.opt_start = opt_start_str
SimAggr.opt_end = opt_end_str
if simtime.hour == DR_call_start.hour and simtime.minute == DR_call_start.minute:
print('%%%%%%%%%%%%%%% DR event called - flexibility profile defined %%%%%%%%%%%%%%%%%%%%%')
flex_cost_signal = pd.Series(0,index=index)
j = 0
for bldg in bldg_list:
if j == 0:
load_profiles = pd.Series(SimAggr.opt_controlseq['HPPower_' + bldg].display_data().values, index = SimAggr.opt_controlseq['HPPower_' + bldg].display_data().index)
else:
load_profile = pd.Series(SimAggr.opt_controlseq['HPPower_' + bldg].display_data().values, index = SimAggr.opt_controlseq['HPPower_' + bldg].display_data().index)
load_profiles =
|
pd.concat([load_profiles, load_profile], axis=1)
|
pandas.concat
|
# -*- coding: utf-8 -*-
import re
import warnings
from datetime import timedelta
from itertools import product
import pytest
import numpy as np
import pandas as pd
from pandas import (CategoricalIndex, DataFrame, Index, MultiIndex,
compat, date_range, period_range)
from pandas.compat import PY3, long, lrange, lzip, range, u, PYPY
from pandas.errors import PerformanceWarning, UnsortedIndexError
from pandas.core.dtypes.dtypes import CategoricalDtype
from pandas.core.indexes.base import InvalidIndexError
from pandas.core.dtypes.cast import construct_1d_object_array_from_listlike
from pandas._libs.tslib import Timestamp
import pandas.util.testing as tm
from pandas.util.testing import assert_almost_equal, assert_copy
from .common import Base
class TestMultiIndex(Base):
_holder = MultiIndex
_compat_props = ['shape', 'ndim', 'size']
def setup_method(self, method):
major_axis = Index(['foo', 'bar', 'baz', 'qux'])
minor_axis = Index(['one', 'two'])
major_labels = np.array([0, 0, 1, 2, 3, 3])
minor_labels = np.array([0, 1, 0, 1, 0, 1])
self.index_names = ['first', 'second']
self.indices = dict(index=MultiIndex(levels=[major_axis, minor_axis],
labels=[major_labels, minor_labels
], names=self.index_names,
verify_integrity=False))
self.setup_indices()
def create_index(self):
return self.index
def test_can_hold_identifiers(self):
idx = self.create_index()
key = idx[0]
assert idx._can_hold_identifiers_and_holds_name(key) is True
def test_boolean_context_compat2(self):
# boolean context compat
# GH7897
i1 = MultiIndex.from_tuples([('A', 1), ('A', 2)])
i2 = MultiIndex.from_tuples([('A', 1), ('A', 3)])
common = i1.intersection(i2)
def f():
if common:
pass
tm.assert_raises_regex(ValueError, 'The truth value of a', f)
def test_labels_dtypes(self):
# GH 8456
i = MultiIndex.from_tuples([('A', 1), ('A', 2)])
assert i.labels[0].dtype == 'int8'
assert i.labels[1].dtype == 'int8'
i = MultiIndex.from_product([['a'], range(40)])
assert i.labels[1].dtype == 'int8'
i = MultiIndex.from_product([['a'], range(400)])
assert i.labels[1].dtype == 'int16'
i = MultiIndex.from_product([['a'], range(40000)])
assert i.labels[1].dtype == 'int32'
i = pd.MultiIndex.from_product([['a'], range(1000)])
assert (i.labels[0] >= 0).all()
assert (i.labels[1] >= 0).all()
def test_where(self):
i = MultiIndex.from_tuples([('A', 1), ('A', 2)])
def f():
i.where(True)
pytest.raises(NotImplementedError, f)
def test_where_array_like(self):
i = MultiIndex.from_tuples([('A', 1), ('A', 2)])
klasses = [list, tuple, np.array, pd.Series]
cond = [False, True]
for klass in klasses:
def f():
return i.where(klass(cond))
pytest.raises(NotImplementedError, f)
def test_repeat(self):
reps = 2
numbers = [1, 2, 3]
names = np.array(['foo', 'bar'])
m = MultiIndex.from_product([
numbers, names], names=names)
expected = MultiIndex.from_product([
numbers, names.repeat(reps)], names=names)
tm.assert_index_equal(m.repeat(reps), expected)
with tm.assert_produces_warning(FutureWarning):
result = m.repeat(n=reps)
tm.assert_index_equal(result, expected)
def test_numpy_repeat(self):
reps = 2
numbers = [1, 2, 3]
names = np.array(['foo', 'bar'])
m = MultiIndex.from_product([
numbers, names], names=names)
expected = MultiIndex.from_product([
numbers, names.repeat(reps)], names=names)
tm.assert_index_equal(np.repeat(m, reps), expected)
msg = "the 'axis' parameter is not supported"
tm.assert_raises_regex(
ValueError, msg, np.repeat, m, reps, axis=1)
def test_set_name_methods(self):
# so long as these are synonyms, we don't need to test set_names
assert self.index.rename == self.index.set_names
new_names = [name + "SUFFIX" for name in self.index_names]
ind = self.index.set_names(new_names)
assert self.index.names == self.index_names
assert ind.names == new_names
with tm.assert_raises_regex(ValueError, "^Length"):
ind.set_names(new_names + new_names)
new_names2 = [name + "SUFFIX2" for name in new_names]
res = ind.set_names(new_names2, inplace=True)
assert res is None
assert ind.names == new_names2
# set names for specific level (# GH7792)
ind = self.index.set_names(new_names[0], level=0)
assert self.index.names == self.index_names
assert ind.names == [new_names[0], self.index_names[1]]
res = ind.set_names(new_names2[0], level=0, inplace=True)
assert res is None
assert ind.names == [new_names2[0], self.index_names[1]]
# set names for multiple levels
ind = self.index.set_names(new_names, level=[0, 1])
assert self.index.names == self.index_names
assert ind.names == new_names
res = ind.set_names(new_names2, level=[0, 1], inplace=True)
assert res is None
assert ind.names == new_names2
@pytest.mark.parametrize('inplace', [True, False])
def test_set_names_with_nlevel_1(self, inplace):
# GH 21149
# Ensure that .set_names for MultiIndex with
# nlevels == 1 does not raise any errors
expected = pd.MultiIndex(levels=[[0, 1]],
labels=[[0, 1]],
names=['first'])
m = pd.MultiIndex.from_product([[0, 1]])
result = m.set_names('first', level=0, inplace=inplace)
if inplace:
result = m
tm.assert_index_equal(result, expected)
def test_set_levels_labels_directly(self):
# setting levels/labels directly raises AttributeError
levels = self.index.levels
new_levels = [[lev + 'a' for lev in level] for level in levels]
labels = self.index.labels
major_labels, minor_labels = labels
major_labels = [(x + 1) % 3 for x in major_labels]
minor_labels = [(x + 1) % 1 for x in minor_labels]
new_labels = [major_labels, minor_labels]
with pytest.raises(AttributeError):
self.index.levels = new_levels
with pytest.raises(AttributeError):
self.index.labels = new_labels
def test_set_levels(self):
# side note - you probably wouldn't want to use levels and labels
# directly like this - but it is possible.
levels = self.index.levels
new_levels = [[lev + 'a' for lev in level] for level in levels]
def assert_matching(actual, expected, check_dtype=False):
# avoid specifying internal representation
# as much as possible
assert len(actual) == len(expected)
for act, exp in zip(actual, expected):
act = np.asarray(act)
exp = np.asarray(exp)
tm.assert_numpy_array_equal(act, exp, check_dtype=check_dtype)
# level changing [w/o mutation]
ind2 = self.index.set_levels(new_levels)
assert_matching(ind2.levels, new_levels)
assert_matching(self.index.levels, levels)
# level changing [w/ mutation]
ind2 = self.index.copy()
inplace_return = ind2.set_levels(new_levels, inplace=True)
assert inplace_return is None
assert_matching(ind2.levels, new_levels)
# level changing specific level [w/o mutation]
ind2 = self.index.set_levels(new_levels[0], level=0)
assert_matching(ind2.levels, [new_levels[0], levels[1]])
assert_matching(self.index.levels, levels)
ind2 = self.index.set_levels(new_levels[1], level=1)
assert_matching(ind2.levels, [levels[0], new_levels[1]])
assert_matching(self.index.levels, levels)
# level changing multiple levels [w/o mutation]
ind2 = self.index.set_levels(new_levels, level=[0, 1])
assert_matching(ind2.levels, new_levels)
assert_matching(self.index.levels, levels)
# level changing specific level [w/ mutation]
ind2 = self.index.copy()
inplace_return = ind2.set_levels(new_levels[0], level=0, inplace=True)
assert inplace_return is None
assert_matching(ind2.levels, [new_levels[0], levels[1]])
assert_matching(self.index.levels, levels)
ind2 = self.index.copy()
inplace_return = ind2.set_levels(new_levels[1], level=1, inplace=True)
assert inplace_return is None
assert_matching(ind2.levels, [levels[0], new_levels[1]])
assert_matching(self.index.levels, levels)
# level changing multiple levels [w/ mutation]
ind2 = self.index.copy()
inplace_return = ind2.set_levels(new_levels, level=[0, 1],
inplace=True)
assert inplace_return is None
assert_matching(ind2.levels, new_levels)
assert_matching(self.index.levels, levels)
# illegal level changing should not change levels
# GH 13754
original_index = self.index.copy()
for inplace in [True, False]:
with tm.assert_raises_regex(ValueError, "^On"):
self.index.set_levels(['c'], level=0, inplace=inplace)
assert_matching(self.index.levels, original_index.levels,
check_dtype=True)
with tm.assert_raises_regex(ValueError, "^On"):
self.index.set_labels([0, 1, 2, 3, 4, 5], level=0,
inplace=inplace)
assert_matching(self.index.labels, original_index.labels,
check_dtype=True)
with tm.assert_raises_regex(TypeError, "^Levels"):
self.index.set_levels('c', level=0, inplace=inplace)
assert_matching(self.index.levels, original_index.levels,
check_dtype=True)
with tm.assert_raises_regex(TypeError, "^Labels"):
self.index.set_labels(1, level=0, inplace=inplace)
assert_matching(self.index.labels, original_index.labels,
check_dtype=True)
def test_set_labels(self):
# side note - you probably wouldn't want to use levels and labels
# directly like this - but it is possible.
labels = self.index.labels
major_labels, minor_labels = labels
major_labels = [(x + 1) % 3 for x in major_labels]
minor_labels = [(x + 1) % 1 for x in minor_labels]
new_labels = [major_labels, minor_labels]
def assert_matching(actual, expected):
# avoid specifying internal representation
# as much as possible
assert len(actual) == len(expected)
for act, exp in zip(actual, expected):
act = np.asarray(act)
exp = np.asarray(exp, dtype=np.int8)
tm.assert_numpy_array_equal(act, exp)
# label changing [w/o mutation]
ind2 = self.index.set_labels(new_labels)
assert_matching(ind2.labels, new_labels)
assert_matching(self.index.labels, labels)
# label changing [w/ mutation]
ind2 = self.index.copy()
inplace_return = ind2.set_labels(new_labels, inplace=True)
assert inplace_return is None
assert_matching(ind2.labels, new_labels)
# label changing specific level [w/o mutation]
ind2 = self.index.set_labels(new_labels[0], level=0)
assert_matching(ind2.labels, [new_labels[0], labels[1]])
assert_matching(self.index.labels, labels)
ind2 = self.index.set_labels(new_labels[1], level=1)
assert_matching(ind2.labels, [labels[0], new_labels[1]])
assert_matching(self.index.labels, labels)
# label changing multiple levels [w/o mutation]
ind2 = self.index.set_labels(new_labels, level=[0, 1])
assert_matching(ind2.labels, new_labels)
assert_matching(self.index.labels, labels)
# label changing specific level [w/ mutation]
ind2 = self.index.copy()
inplace_return = ind2.set_labels(new_labels[0], level=0, inplace=True)
assert inplace_return is None
assert_matching(ind2.labels, [new_labels[0], labels[1]])
assert_matching(self.index.labels, labels)
ind2 = self.index.copy()
inplace_return = ind2.set_labels(new_labels[1], level=1, inplace=True)
assert inplace_return is None
assert_matching(ind2.labels, [labels[0], new_labels[1]])
assert_matching(self.index.labels, labels)
# label changing multiple levels [w/ mutation]
ind2 = self.index.copy()
inplace_return = ind2.set_labels(new_labels, level=[0, 1],
inplace=True)
assert inplace_return is None
assert_matching(ind2.labels, new_labels)
assert_matching(self.index.labels, labels)
# label changing for levels of different magnitude of categories
ind = pd.MultiIndex.from_tuples([(0, i) for i in range(130)])
new_labels = range(129, -1, -1)
expected = pd.MultiIndex.from_tuples(
[(0, i) for i in new_labels])
# [w/o mutation]
result = ind.set_labels(labels=new_labels, level=1)
assert result.equals(expected)
# [w/ mutation]
result = ind.copy()
result.set_labels(labels=new_labels, level=1, inplace=True)
assert result.equals(expected)
def test_set_levels_labels_names_bad_input(self):
levels, labels = self.index.levels, self.index.labels
names = self.index.names
with tm.assert_raises_regex(ValueError, 'Length of levels'):
self.index.set_levels([levels[0]])
with tm.assert_raises_regex(ValueError, 'Length of labels'):
self.index.set_labels([labels[0]])
with tm.assert_raises_regex(ValueError, 'Length of names'):
self.index.set_names([names[0]])
# shouldn't scalar data error, instead should demand list-like
with tm.assert_raises_regex(TypeError, 'list of lists-like'):
self.index.set_levels(levels[0])
# shouldn't scalar data error, instead should demand list-like
with tm.assert_raises_regex(TypeError, 'list of lists-like'):
self.index.set_labels(labels[0])
# shouldn't scalar data error, instead should demand list-like
with tm.assert_raises_regex(TypeError, 'list-like'):
self.index.set_names(names[0])
# should have equal lengths
with tm.assert_raises_regex(TypeError, 'list of lists-like'):
self.index.set_levels(levels[0], level=[0, 1])
with tm.assert_raises_regex(TypeError, 'list-like'):
self.index.set_levels(levels, level=0)
# should have equal lengths
with tm.assert_raises_regex(TypeError, 'list of lists-like'):
self.index.set_labels(labels[0], level=[0, 1])
with tm.assert_raises_regex(TypeError, 'list-like'):
self.index.set_labels(labels, level=0)
# should have equal lengths
with tm.assert_raises_regex(ValueError, 'Length of names'):
self.index.set_names(names[0], level=[0, 1])
with tm.assert_raises_regex(TypeError, 'string'):
self.index.set_names(names, level=0)
def test_set_levels_categorical(self):
# GH13854
index = MultiIndex.from_arrays([list("xyzx"), [0, 1, 2, 3]])
for ordered in [False, True]:
cidx = CategoricalIndex(list("bac"), ordered=ordered)
result = index.set_levels(cidx, 0)
expected = MultiIndex(levels=[cidx, [0, 1, 2, 3]],
labels=index.labels)
tm.assert_index_equal(result, expected)
result_lvl = result.get_level_values(0)
expected_lvl = CategoricalIndex(list("bacb"),
categories=cidx.categories,
ordered=cidx.ordered)
tm.assert_index_equal(result_lvl, expected_lvl)
def test_metadata_immutable(self):
levels, labels = self.index.levels, self.index.labels
# shouldn't be able to set at either the top level or base level
mutable_regex = re.compile('does not support mutable operations')
with tm.assert_raises_regex(TypeError, mutable_regex):
levels[0] = levels[0]
with tm.assert_raises_regex(TypeError, mutable_regex):
levels[0][0] = levels[0][0]
# ditto for labels
with tm.assert_raises_regex(TypeError, mutable_regex):
labels[0] = labels[0]
with tm.assert_raises_regex(TypeError, mutable_regex):
labels[0][0] = labels[0][0]
# and for names
names = self.index.names
with tm.assert_raises_regex(TypeError, mutable_regex):
names[0] = names[0]
def test_inplace_mutation_resets_values(self):
levels = [['a', 'b', 'c'], [4]]
levels2 = [[1, 2, 3], ['a']]
labels = [[0, 1, 0, 2, 2, 0], [0, 0, 0, 0, 0, 0]]
mi1 = MultiIndex(levels=levels, labels=labels)
mi2 = MultiIndex(levels=levels2, labels=labels)
vals = mi1.values.copy()
vals2 = mi2.values.copy()
assert mi1._tuples is not None
# Make sure level setting works
new_vals = mi1.set_levels(levels2).values
tm.assert_almost_equal(vals2, new_vals)
# Non-inplace doesn't kill _tuples [implementation detail]
tm.assert_almost_equal(mi1._tuples, vals)
# ...and values is still same too
tm.assert_almost_equal(mi1.values, vals)
# Inplace should kill _tuples
mi1.set_levels(levels2, inplace=True)
tm.assert_almost_equal(mi1.values, vals2)
# Make sure label setting works too
labels2 = [[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0]]
exp_values = np.empty((6,), dtype=object)
exp_values[:] = [(long(1), 'a')] * 6
# Must be 1d array of tuples
assert exp_values.shape == (6,)
new_values = mi2.set_labels(labels2).values
# Not inplace shouldn't change
tm.assert_almost_equal(mi2._tuples, vals2)
# Should have correct values
tm.assert_almost_equal(exp_values, new_values)
# ...and again setting inplace should kill _tuples, etc
mi2.set_labels(labels2, inplace=True)
tm.assert_almost_equal(mi2.values, new_values)
def test_copy_in_constructor(self):
levels = np.array(["a", "b", "c"])
labels = np.array([1, 1, 2, 0, 0, 1, 1])
val = labels[0]
mi = MultiIndex(levels=[levels, levels], labels=[labels, labels],
copy=True)
assert mi.labels[0][0] == val
labels[0] = 15
assert mi.labels[0][0] == val
val = levels[0]
levels[0] = "PANDA"
assert mi.levels[0][0] == val
def test_set_value_keeps_names(self):
# motivating example from #3742
lev1 = ['hans', 'hans', 'hans', 'grethe', 'grethe', 'grethe']
lev2 = ['1', '2', '3'] * 2
idx = pd.MultiIndex.from_arrays([lev1, lev2], names=['Name', 'Number'])
df = pd.DataFrame(
np.random.randn(6, 4),
columns=['one', 'two', 'three', 'four'],
index=idx)
df = df.sort_index()
assert df._is_copy is None
assert df.index.names == ('Name', 'Number')
df.at[('grethe', '4'), 'one'] = 99.34
assert df._is_copy is None
assert df.index.names == ('Name', 'Number')
def test_copy_names(self):
# Check that adding a "names" parameter to the copy is honored
# GH14302
multi_idx = pd.Index([(1, 2), (3, 4)], names=['MyName1', 'MyName2'])
multi_idx1 = multi_idx.copy()
assert multi_idx.equals(multi_idx1)
assert multi_idx.names == ['MyName1', 'MyName2']
assert multi_idx1.names == ['MyName1', 'MyName2']
multi_idx2 = multi_idx.copy(names=['NewName1', 'NewName2'])
assert multi_idx.equals(multi_idx2)
assert multi_idx.names == ['MyName1', 'MyName2']
assert multi_idx2.names == ['NewName1', 'NewName2']
multi_idx3 = multi_idx.copy(name=['NewName1', 'NewName2'])
assert multi_idx.equals(multi_idx3)
assert multi_idx.names == ['MyName1', 'MyName2']
assert multi_idx3.names == ['NewName1', 'NewName2']
def test_names(self):
# names are assigned in setup
names = self.index_names
level_names = [level.name for level in self.index.levels]
assert names == level_names
# setting bad names on existing
index = self.index
tm.assert_raises_regex(ValueError, "^Length of names",
setattr, index, "names",
list(index.names) + ["third"])
tm.assert_raises_regex(ValueError, "^Length of names",
setattr, index, "names", [])
# initializing with bad names (should always be equivalent)
major_axis, minor_axis = self.index.levels
major_labels, minor_labels = self.index.labels
tm.assert_raises_regex(ValueError, "^Length of names", MultiIndex,
levels=[major_axis, minor_axis],
labels=[major_labels, minor_labels],
names=['first'])
tm.assert_raises_regex(ValueError, "^Length of names", MultiIndex,
levels=[major_axis, minor_axis],
labels=[major_labels, minor_labels],
names=['first', 'second', 'third'])
# names are assigned
index.names = ["a", "b"]
ind_names = list(index.names)
level_names = [level.name for level in index.levels]
assert ind_names == level_names
def test_astype(self):
expected = self.index.copy()
actual = self.index.astype('O')
assert_copy(actual.levels, expected.levels)
assert_copy(actual.labels, expected.labels)
self.check_level_names(actual, expected.names)
with tm.assert_raises_regex(TypeError, "^Setting.*dtype.*object"):
self.index.astype(np.dtype(int))
@pytest.mark.parametrize('ordered', [True, False])
def test_astype_category(self, ordered):
# GH 18630
msg = '> 1 ndim Categorical are not supported at this time'
with tm.assert_raises_regex(NotImplementedError, msg):
self.index.astype(CategoricalDtype(ordered=ordered))
if ordered is False:
# dtype='category' defaults to ordered=False, so only test once
with tm.assert_raises_regex(NotImplementedError, msg):
self.index.astype('category')
def test_constructor_single_level(self):
result = MultiIndex(levels=[['foo', 'bar', 'baz', 'qux']],
labels=[[0, 1, 2, 3]], names=['first'])
assert isinstance(result, MultiIndex)
expected = Index(['foo', 'bar', 'baz', 'qux'], name='first')
tm.assert_index_equal(result.levels[0], expected)
assert result.names == ['first']
def test_constructor_no_levels(self):
tm.assert_raises_regex(ValueError, "non-zero number "
"of levels/labels",
MultiIndex, levels=[], labels=[])
both_re = re.compile('Must pass both levels and labels')
with tm.assert_raises_regex(TypeError, both_re):
MultiIndex(levels=[])
with tm.assert_raises_regex(TypeError, both_re):
MultiIndex(labels=[])
def test_constructor_mismatched_label_levels(self):
labels = [np.array([1]), np.array([2]), np.array([3])]
levels = ["a"]
tm.assert_raises_regex(ValueError, "Length of levels and labels "
"must be the same", MultiIndex,
levels=levels, labels=labels)
length_error = re.compile('>= length of level')
label_error = re.compile(r'Unequal label lengths: \[4, 2\]')
# important to check that it's looking at the right thing.
with tm.assert_raises_regex(ValueError, length_error):
MultiIndex(levels=[['a'], ['b']],
labels=[[0, 1, 2, 3], [0, 3, 4, 1]])
with tm.assert_raises_regex(ValueError, label_error):
MultiIndex(levels=[['a'], ['b']], labels=[[0, 0, 0, 0], [0, 0]])
# external API
with tm.assert_raises_regex(ValueError, length_error):
self.index.copy().set_levels([['a'], ['b']])
with tm.assert_raises_regex(ValueError, label_error):
self.index.copy().set_labels([[0, 0, 0, 0], [0, 0]])
def test_constructor_nonhashable_names(self):
# GH 20527
levels = [[1, 2], [u'one', u'two']]
labels = [[0, 0, 1, 1], [0, 1, 0, 1]]
names = ((['foo'], ['bar']))
message = "MultiIndex.name must be a hashable type"
tm.assert_raises_regex(TypeError, message,
MultiIndex, levels=levels,
labels=labels, names=names)
# With .rename()
mi = MultiIndex(levels=[[1, 2], [u'one', u'two']],
labels=[[0, 0, 1, 1], [0, 1, 0, 1]],
names=('foo', 'bar'))
renamed = [['foor'], ['barr']]
tm.assert_raises_regex(TypeError, message, mi.rename, names=renamed)
# With .set_names()
tm.assert_raises_regex(TypeError, message, mi.set_names, names=renamed)
@pytest.mark.parametrize('names', [['a', 'b', 'a'], ['1', '1', '2'],
['1', 'a', '1']])
def test_duplicate_level_names(self, names):
# GH18872
pytest.raises(ValueError, pd.MultiIndex.from_product,
[[0, 1]] * 3, names=names)
# With .rename()
mi = pd.MultiIndex.from_product([[0, 1]] * 3)
tm.assert_raises_regex(ValueError, "Duplicated level name:",
mi.rename, names)
# With .rename(., level=)
mi.rename(names[0], level=1, inplace=True)
tm.assert_raises_regex(ValueError, "Duplicated level name:",
mi.rename, names[:2], level=[0, 2])
def assert_multiindex_copied(self, copy, original):
# Levels should be (at least, shallow copied)
tm.assert_copy(copy.levels, original.levels)
tm.assert_almost_equal(copy.labels, original.labels)
# Labels doesn't matter which way copied
tm.assert_almost_equal(copy.labels, original.labels)
assert copy.labels is not original.labels
# Names doesn't matter which way copied
assert copy.names == original.names
assert copy.names is not original.names
# Sort order should be copied
assert copy.sortorder == original.sortorder
def test_copy(self):
i_copy = self.index.copy()
self.assert_multiindex_copied(i_copy, self.index)
def test_shallow_copy(self):
i_copy = self.index._shallow_copy()
self.assert_multiindex_copied(i_copy, self.index)
def test_view(self):
i_view = self.index.view()
self.assert_multiindex_copied(i_view, self.index)
def check_level_names(self, index, names):
assert [level.name for level in index.levels] == list(names)
def test_changing_names(self):
# names should be applied to levels
level_names = [level.name for level in self.index.levels]
self.check_level_names(self.index, self.index.names)
view = self.index.view()
copy = self.index.copy()
shallow_copy = self.index._shallow_copy()
# changing names should change level names on object
new_names = [name + "a" for name in self.index.names]
self.index.names = new_names
self.check_level_names(self.index, new_names)
# but not on copies
self.check_level_names(view, level_names)
self.check_level_names(copy, level_names)
self.check_level_names(shallow_copy, level_names)
# and copies shouldn't change original
shallow_copy.names = [name + "c" for name in shallow_copy.names]
self.check_level_names(self.index, new_names)
def test_get_level_number_integer(self):
self.index.names = [1, 0]
assert self.index._get_level_number(1) == 0
assert self.index._get_level_number(0) == 1
pytest.raises(IndexError, self.index._get_level_number, 2)
tm.assert_raises_regex(KeyError, 'Level fourth not found',
self.index._get_level_number, 'fourth')
def test_from_arrays(self):
arrays = []
for lev, lab in zip(self.index.levels, self.index.labels):
arrays.append(np.asarray(lev).take(lab))
# list of arrays as input
result = MultiIndex.from_arrays(arrays, names=self.index.names)
tm.assert_index_equal(result, self.index)
# infer correctly
result = MultiIndex.from_arrays([[pd.NaT, Timestamp('20130101')],
['a', 'b']])
assert result.levels[0].equals(Index([Timestamp('20130101')]))
assert result.levels[1].equals(Index(['a', 'b']))
def test_from_arrays_iterator(self):
# GH 18434
arrays = []
for lev, lab in zip(self.index.levels, self.index.labels):
arrays.append(np.asarray(lev).take(lab))
# iterator as input
result = MultiIndex.from_arrays(iter(arrays), names=self.index.names)
tm.assert_index_equal(result, self.index)
# invalid iterator input
with tm.assert_raises_regex(
TypeError, "Input must be a list / sequence of array-likes."):
MultiIndex.from_arrays(0)
def test_from_arrays_index_series_datetimetz(self):
idx1 = pd.date_range('2015-01-01 10:00', freq='D', periods=3,
tz='US/Eastern')
idx2 = pd.date_range('2015-01-01 10:00', freq='H', periods=3,
tz='Asia/Tokyo')
result = pd.MultiIndex.from_arrays([idx1, idx2])
tm.assert_index_equal(result.get_level_values(0), idx1)
tm.assert_index_equal(result.get_level_values(1), idx2)
result2 = pd.MultiIndex.from_arrays([pd.Series(idx1), pd.Series(idx2)])
tm.assert_index_equal(result2.get_level_values(0), idx1)
tm.assert_index_equal(result2.get_level_values(1), idx2)
tm.assert_index_equal(result, result2)
def test_from_arrays_index_series_timedelta(self):
idx1 = pd.timedelta_range('1 days', freq='D', periods=3)
idx2 = pd.timedelta_range('2 hours', freq='H', periods=3)
result = pd.MultiIndex.from_arrays([idx1, idx2])
tm.assert_index_equal(result.get_level_values(0), idx1)
tm.assert_index_equal(result.get_level_values(1), idx2)
result2 = pd.MultiIndex.from_arrays([pd.Series(idx1), pd.Series(idx2)])
tm.assert_index_equal(result2.get_level_values(0), idx1)
tm.assert_index_equal(result2.get_level_values(1), idx2)
tm.assert_index_equal(result, result2)
def test_from_arrays_index_series_period(self):
idx1 = pd.period_range('2011-01-01', freq='D', periods=3)
idx2 = pd.period_range('2015-01-01', freq='H', periods=3)
result = pd.MultiIndex.from_arrays([idx1, idx2])
tm.assert_index_equal(result.get_level_values(0), idx1)
tm.assert_index_equal(result.get_level_values(1), idx2)
result2 = pd.MultiIndex.from_arrays([pd.Series(idx1), pd.Series(idx2)])
tm.assert_index_equal(result2.get_level_values(0), idx1)
tm.assert_index_equal(result2.get_level_values(1), idx2)
tm.assert_index_equal(result, result2)
def test_from_arrays_index_datetimelike_mixed(self):
idx1 = pd.date_range('2015-01-01 10:00', freq='D', periods=3,
tz='US/Eastern')
idx2 = pd.date_range('2015-01-01 10:00', freq='H', periods=3)
idx3 = pd.timedelta_range('1 days', freq='D', periods=3)
idx4 = pd.period_range('2011-01-01', freq='D', periods=3)
result = pd.MultiIndex.from_arrays([idx1, idx2, idx3, idx4])
tm.assert_index_equal(result.get_level_values(0), idx1)
tm.assert_index_equal(result.get_level_values(1), idx2)
tm.assert_index_equal(result.get_level_values(2), idx3)
tm.assert_index_equal(result.get_level_values(3), idx4)
result2 = pd.MultiIndex.from_arrays([pd.Series(idx1),
pd.Series(idx2),
pd.Series(idx3),
pd.Series(idx4)])
tm.assert_index_equal(result2.get_level_values(0), idx1)
tm.assert_index_equal(result2.get_level_values(1), idx2)
tm.assert_index_equal(result2.get_level_values(2), idx3)
tm.assert_index_equal(result2.get_level_values(3), idx4)
tm.assert_index_equal(result, result2)
def test_from_arrays_index_series_categorical(self):
# GH13743
idx1 = pd.CategoricalIndex(list("abcaab"), categories=list("bac"),
ordered=False)
idx2 = pd.CategoricalIndex(list("abcaab"), categories=list("bac"),
ordered=True)
result = pd.MultiIndex.from_arrays([idx1, idx2])
tm.assert_index_equal(result.get_level_values(0), idx1)
tm.assert_index_equal(result.get_level_values(1), idx2)
result2 = pd.MultiIndex.from_arrays([pd.Series(idx1), pd.Series(idx2)])
tm.assert_index_equal(result2.get_level_values(0), idx1)
tm.assert_index_equal(result2.get_level_values(1), idx2)
result3 = pd.MultiIndex.from_arrays([idx1.values, idx2.values])
tm.assert_index_equal(result3.get_level_values(0), idx1)
tm.assert_index_equal(result3.get_level_values(1), idx2)
def test_from_arrays_empty(self):
# 0 levels
with tm.assert_raises_regex(
ValueError, "Must pass non-zero number of levels/labels"):
MultiIndex.from_arrays(arrays=[])
# 1 level
result = MultiIndex.from_arrays(arrays=[[]], names=['A'])
assert isinstance(result, MultiIndex)
expected = Index([], name='A')
tm.assert_index_equal(result.levels[0], expected)
# N levels
for N in [2, 3]:
arrays = [[]] * N
names = list('ABC')[:N]
result = MultiIndex.from_arrays(arrays=arrays, names=names)
expected = MultiIndex(levels=[[]] * N, labels=[[]] * N,
names=names)
tm.assert_index_equal(result, expected)
def test_from_arrays_invalid_input(self):
invalid_inputs = [1, [1], [1, 2], [[1], 2],
'a', ['a'], ['a', 'b'], [['a'], 'b']]
for i in invalid_inputs:
pytest.raises(TypeError, MultiIndex.from_arrays, arrays=i)
def test_from_arrays_different_lengths(self):
# see gh-13599
idx1 = [1, 2, 3]
idx2 = ['a', 'b']
tm.assert_raises_regex(ValueError, '^all arrays must '
'be same length$',
MultiIndex.from_arrays, [idx1, idx2])
idx1 = []
idx2 = ['a', 'b']
tm.assert_raises_regex(ValueError, '^all arrays must '
'be same length$',
MultiIndex.from_arrays, [idx1, idx2])
idx1 = [1, 2, 3]
idx2 = []
tm.assert_raises_regex(ValueError, '^all arrays must '
'be same length$',
MultiIndex.from_arrays, [idx1, idx2])
def test_from_product(self):
first = ['foo', 'bar', 'buz']
second = ['a', 'b', 'c']
names = ['first', 'second']
result = MultiIndex.from_product([first, second], names=names)
tuples = [('foo', 'a'), ('foo', 'b'), ('foo', 'c'), ('bar', 'a'),
('bar', 'b'), ('bar', 'c'), ('buz', 'a'), ('buz', 'b'),
('buz', 'c')]
expected = MultiIndex.from_tuples(tuples, names=names)
tm.assert_index_equal(result, expected)
def test_from_product_iterator(self):
# GH 18434
first = ['foo', 'bar', 'buz']
second = ['a', 'b', 'c']
names = ['first', 'second']
tuples = [('foo', 'a'), ('foo', 'b'), ('foo', 'c'), ('bar', 'a'),
('bar', 'b'), ('bar', 'c'), ('buz', 'a'), ('buz', 'b'),
('buz', 'c')]
expected = MultiIndex.from_tuples(tuples, names=names)
# iterator as input
result = MultiIndex.from_product(iter([first, second]), names=names)
tm.assert_index_equal(result, expected)
# Invalid non-iterable input
with tm.assert_raises_regex(
TypeError, "Input must be a list / sequence of iterables."):
MultiIndex.from_product(0)
def test_from_product_empty(self):
# 0 levels
with tm.assert_raises_regex(
ValueError, "Must pass non-zero number of levels/labels"):
MultiIndex.from_product([])
# 1 level
result = MultiIndex.from_product([[]], names=['A'])
expected = pd.Index([], name='A')
tm.assert_index_equal(result.levels[0], expected)
# 2 levels
l1 = [[], ['foo', 'bar', 'baz'], []]
l2 = [[], [], ['a', 'b', 'c']]
names = ['A', 'B']
for first, second in zip(l1, l2):
result = MultiIndex.from_product([first, second], names=names)
expected = MultiIndex(levels=[first, second],
labels=[[], []], names=names)
tm.assert_index_equal(result, expected)
# GH12258
names = ['A', 'B', 'C']
for N in range(4):
lvl2 = lrange(N)
result = MultiIndex.from_product([[], lvl2, []], names=names)
expected = MultiIndex(levels=[[], lvl2, []],
labels=[[], [], []], names=names)
tm.assert_index_equal(result, expected)
def test_from_product_invalid_input(self):
invalid_inputs = [1, [1], [1, 2], [[1], 2],
'a', ['a'], ['a', 'b'], [['a'], 'b']]
for i in invalid_inputs:
pytest.raises(TypeError, MultiIndex.from_product, iterables=i)
def test_from_product_datetimeindex(self):
dt_index = date_range('2000-01-01', periods=2)
mi = pd.MultiIndex.from_product([[1, 2], dt_index])
etalon = construct_1d_object_array_from_listlike([(1, pd.Timestamp(
'2000-01-01')), (1, pd.Timestamp('2000-01-02')), (2, pd.Timestamp(
'2000-01-01')), (2, pd.Timestamp('2000-01-02'))])
tm.assert_numpy_array_equal(mi.values, etalon)
def test_from_product_index_series_categorical(self):
# GH13743
first = ['foo', 'bar']
for ordered in [False, True]:
idx = pd.CategoricalIndex(list("abcaab"), categories=list("bac"),
ordered=ordered)
expected = pd.CategoricalIndex(list("abcaab") + list("abcaab"),
categories=list("bac"),
ordered=ordered)
for arr in [idx, pd.Series(idx), idx.values]:
result = pd.MultiIndex.from_product([first, arr])
tm.assert_index_equal(result.get_level_values(1), expected)
def test_values_boxed(self):
tuples = [(1, pd.Timestamp('2000-01-01')), (2, pd.NaT),
(3, pd.Timestamp('2000-01-03')),
(1, pd.Timestamp('2000-01-04')),
(2, pd.Timestamp('2000-01-02')),
(3, pd.Timestamp('2000-01-03'))]
result = pd.MultiIndex.from_tuples(tuples)
expected = construct_1d_object_array_from_listlike(tuples)
tm.assert_numpy_array_equal(result.values, expected)
# Check that code branches for boxed values produce identical results
tm.assert_numpy_array_equal(result.values[:4], result[:4].values)
def test_values_multiindex_datetimeindex(self):
# Test to ensure we hit the boxing / nobox part of MI.values
ints = np.arange(10 ** 18, 10 ** 18 + 5)
naive = pd.DatetimeIndex(ints)
aware = pd.DatetimeIndex(ints, tz='US/Central')
idx = pd.MultiIndex.from_arrays([naive, aware])
result = idx.values
outer = pd.DatetimeIndex([x[0] for x in result])
tm.assert_index_equal(outer, naive)
inner = pd.DatetimeIndex([x[1] for x in result])
tm.assert_index_equal(inner, aware)
# n_lev > n_lab
result = idx[:2].values
outer = pd.DatetimeIndex([x[0] for x in result])
tm.assert_index_equal(outer, naive[:2])
inner = pd.DatetimeIndex([x[1] for x in result])
tm.assert_index_equal(inner, aware[:2])
def test_values_multiindex_periodindex(self):
# Test to ensure we hit the boxing / nobox part of MI.values
ints = np.arange(2007, 2012)
pidx = pd.PeriodIndex(ints, freq='D')
idx = pd.MultiIndex.from_arrays([ints, pidx])
result = idx.values
outer = pd.Int64Index([x[0] for x in result])
tm.assert_index_equal(outer, pd.Int64Index(ints))
inner = pd.PeriodIndex([x[1] for x in result])
tm.assert_index_equal(inner, pidx)
# n_lev > n_lab
result = idx[:2].values
outer = pd.Int64Index([x[0] for x in result])
tm.assert_index_equal(outer, pd.Int64Index(ints[:2]))
inner = pd.PeriodIndex([x[1] for x in result])
tm.assert_index_equal(inner, pidx[:2])
def test_append(self):
result = self.index[:3].append(self.index[3:])
assert result.equals(self.index)
foos = [self.index[:1], self.index[1:3], self.index[3:]]
result = foos[0].append(foos[1:])
assert result.equals(self.index)
# empty
result = self.index.append([])
assert result.equals(self.index)
def test_append_mixed_dtypes(self):
# GH 13660
dti = date_range('2011-01-01', freq='M', periods=3, )
dti_tz = date_range('2011-01-01', freq='M', periods=3, tz='US/Eastern')
pi = period_range('2011-01', freq='M', periods=3)
mi = MultiIndex.from_arrays([[1, 2, 3],
[1.1, np.nan, 3.3],
['a', 'b', 'c'],
dti, dti_tz, pi])
assert mi.nlevels == 6
res = mi.append(mi)
exp = MultiIndex.from_arrays([[1, 2, 3, 1, 2, 3],
[1.1, np.nan, 3.3, 1.1, np.nan, 3.3],
['a', 'b', 'c', 'a', 'b', 'c'],
dti.append(dti),
dti_tz.append(dti_tz),
pi.append(pi)])
tm.assert_index_equal(res, exp)
other = MultiIndex.from_arrays([['x', 'y', 'z'], ['x', 'y', 'z'],
['x', 'y', 'z'], ['x', 'y', 'z'],
['x', 'y', 'z'], ['x', 'y', 'z']])
res = mi.append(other)
exp = MultiIndex.from_arrays([[1, 2, 3, 'x', 'y', 'z'],
[1.1, np.nan, 3.3, 'x', 'y', 'z'],
['a', 'b', 'c', 'x', 'y', 'z'],
dti.append(pd.Index(['x', 'y', 'z'])),
dti_tz.append(pd.Index(['x', 'y', 'z'])),
pi.append(pd.Index(['x', 'y', 'z']))])
tm.assert_index_equal(res, exp)
def test_get_level_values(self):
result = self.index.get_level_values(0)
expected = Index(['foo', 'foo', 'bar', 'baz', 'qux', 'qux'],
name='first')
tm.assert_index_equal(result, expected)
assert result.name == 'first'
result = self.index.get_level_values('first')
expected = self.index.get_level_values(0)
tm.assert_index_equal(result, expected)
# GH 10460
index = MultiIndex(
levels=[CategoricalIndex(['A', 'B']),
CategoricalIndex([1, 2, 3])],
labels=[np.array([0, 0, 0, 1, 1, 1]),
np.array([0, 1, 2, 0, 1, 2])])
exp = CategoricalIndex(['A', 'A', 'A', 'B', 'B', 'B'])
tm.assert_index_equal(index.get_level_values(0), exp)
exp = CategoricalIndex([1, 2, 3, 1, 2, 3])
tm.assert_index_equal(index.get_level_values(1), exp)
def test_get_level_values_int_with_na(self):
# GH 17924
arrays = [['a', 'b', 'b'], [1, np.nan, 2]]
index = pd.MultiIndex.from_arrays(arrays)
result = index.get_level_values(1)
expected = Index([1, np.nan, 2])
tm.assert_index_equal(result, expected)
arrays = [['a', 'b', 'b'], [np.nan, np.nan, 2]]
index = pd.MultiIndex.from_arrays(arrays)
result = index.get_level_values(1)
expected = Index([np.nan, np.nan, 2])
tm.assert_index_equal(result, expected)
def test_get_level_values_na(self):
arrays = [[np.nan, np.nan, np.nan], ['a', np.nan, 1]]
index = pd.MultiIndex.from_arrays(arrays)
result = index.get_level_values(0)
expected = pd.Index([np.nan, np.nan, np.nan])
tm.assert_index_equal(result, expected)
result = index.get_level_values(1)
expected = pd.Index(['a', np.nan, 1])
tm.assert_index_equal(result, expected)
arrays = [['a', 'b', 'b'], pd.DatetimeIndex([0, 1, pd.NaT])]
index = pd.MultiIndex.from_arrays(arrays)
result = index.get_level_values(1)
expected = pd.DatetimeIndex([0, 1, pd.NaT])
tm.assert_index_equal(result, expected)
arrays = [[], []]
index = pd.MultiIndex.from_arrays(arrays)
result = index.get_level_values(0)
expected = pd.Index([], dtype=object)
tm.assert_index_equal(result, expected)
def test_get_level_values_all_na(self):
# GH 17924 when level entirely consists of nan
arrays = [[np.nan, np.nan, np.nan], ['a', np.nan, 1]]
index = pd.MultiIndex.from_arrays(arrays)
result = index.get_level_values(0)
expected = pd.Index([np.nan, np.nan, np.nan], dtype=np.float64)
tm.assert_index_equal(result, expected)
result = index.get_level_values(1)
expected = pd.Index(['a', np.nan, 1], dtype=object)
tm.assert_index_equal(result, expected)
def test_reorder_levels(self):
# this blows up
tm.assert_raises_regex(IndexError, '^Too many levels',
self.index.reorder_levels, [2, 1, 0])
def test_nlevels(self):
assert self.index.nlevels == 2
def test_iter(self):
result = list(self.index)
expected = [('foo', 'one'), ('foo', 'two'), ('bar', 'one'),
('baz', 'two'), ('qux', 'one'), ('qux', 'two')]
assert result == expected
def test_legacy_pickle(self):
if PY3:
pytest.skip("testing for legacy pickles not "
"support on py3")
path = tm.get_data_path('multiindex_v1.pickle')
obj = pd.read_pickle(path)
obj2 = MultiIndex.from_tuples(obj.values)
assert obj.equals(obj2)
res = obj.get_indexer(obj)
exp = np.arange(len(obj), dtype=np.intp)
assert_almost_equal(res, exp)
res = obj.get_indexer(obj2[::-1])
exp = obj.get_indexer(obj[::-1])
exp2 = obj2.get_indexer(obj2[::-1])
assert_almost_equal(res, exp)
assert_almost_equal(exp, exp2)
def test_legacy_v2_unpickle(self):
# 0.7.3 -> 0.8.0 format manage
path = tm.get_data_path('mindex_073.pickle')
obj = pd.read_pickle(path)
obj2 = MultiIndex.from_tuples(obj.values)
assert obj.equals(obj2)
res = obj.get_indexer(obj)
exp = np.arange(len(obj), dtype=np.intp)
assert_almost_equal(res, exp)
res = obj.get_indexer(obj2[::-1])
exp = obj.get_indexer(obj[::-1])
exp2 = obj2.get_indexer(obj2[::-1])
assert_almost_equal(res, exp)
assert_almost_equal(exp, exp2)
def test_roundtrip_pickle_with_tz(self):
# GH 8367
# round-trip of timezone
index = MultiIndex.from_product(
[[1, 2], ['a', 'b'], date_range('20130101', periods=3,
tz='US/Eastern')
], names=['one', 'two', 'three'])
unpickled = tm.round_trip_pickle(index)
assert index.equal_levels(unpickled)
def test_from_tuples_index_values(self):
result = MultiIndex.from_tuples(self.index)
assert (result.values == self.index.values).all()
def test_contains(self):
assert ('foo', 'two') in self.index
assert ('bar', 'two') not in self.index
assert None not in self.index
def test_contains_top_level(self):
midx = MultiIndex.from_product([['A', 'B'], [1, 2]])
assert 'A' in midx
assert 'A' not in midx._engine
def test_contains_with_nat(self):
# MI with a NaT
mi = MultiIndex(levels=[['C'],
pd.date_range('2012-01-01', periods=5)],
labels=[[0, 0, 0, 0, 0, 0], [-1, 0, 1, 2, 3, 4]],
names=[None, 'B'])
assert ('C', pd.Timestamp('2012-01-01')) in mi
for val in mi.values:
assert val in mi
def test_is_all_dates(self):
assert not self.index.is_all_dates
def test_is_numeric(self):
# MultiIndex is never numeric
assert not self.index.is_numeric()
def test_getitem(self):
# scalar
assert self.index[2] == ('bar', 'one')
# slice
result = self.index[2:5]
expected = self.index[[2, 3, 4]]
assert result.equals(expected)
# boolean
result = self.index[[True, False, True, False, True, True]]
result2 = self.index[np.array([True, False, True, False, True, True])]
expected = self.index[[0, 2, 4, 5]]
assert result.equals(expected)
assert result2.equals(expected)
def test_getitem_group_select(self):
sorted_idx, _ = self.index.sortlevel(0)
assert sorted_idx.get_loc('baz') == slice(3, 4)
assert sorted_idx.get_loc('foo') == slice(0, 2)
def test_get_loc(self):
assert self.index.get_loc(('foo', 'two')) == 1
assert self.index.get_loc(('baz', 'two')) == 3
pytest.raises(KeyError, self.index.get_loc, ('bar', 'two'))
pytest.raises(KeyError, self.index.get_loc, 'quux')
pytest.raises(NotImplementedError, self.index.get_loc, 'foo',
method='nearest')
# 3 levels
index = MultiIndex(levels=[Index(lrange(4)), Index(lrange(4)), Index(
lrange(4))], labels=[np.array([0, 0, 1, 2, 2, 2, 3, 3]), np.array(
[0, 1, 0, 0, 0, 1, 0, 1]), np.array([1, 0, 1, 1, 0, 0, 1, 0])])
pytest.raises(KeyError, index.get_loc, (1, 1))
assert index.get_loc((2, 0)) == slice(3, 5)
def test_get_loc_duplicates(self):
index = Index([2, 2, 2, 2])
result = index.get_loc(2)
expected = slice(0, 4)
assert result == expected
# pytest.raises(Exception, index.get_loc, 2)
index = Index(['c', 'a', 'a', 'b', 'b'])
rs = index.get_loc('c')
xp = 0
assert rs == xp
def test_get_value_duplicates(self):
index = MultiIndex(levels=[['D', 'B', 'C'],
[0, 26, 27, 37, 57, 67, 75, 82]],
labels=[[0, 0, 0, 1, 2, 2, 2, 2, 2, 2],
[1, 3, 4, 6, 0, 2, 2, 3, 5, 7]],
names=['tag', 'day'])
assert index.get_loc('D') == slice(0, 3)
with pytest.raises(KeyError):
index._engine.get_value(np.array([]), 'D')
def test_get_loc_level(self):
index = MultiIndex(levels=[Index(lrange(4)), Index(lrange(4)), Index(
lrange(4))], labels=[np.array([0, 0, 1, 2, 2, 2, 3, 3]), np.array(
[0, 1, 0, 0, 0, 1, 0, 1]), np.array([1, 0, 1, 1, 0, 0, 1, 0])])
loc, new_index = index.get_loc_level((0, 1))
expected = slice(1, 2)
exp_index = index[expected].droplevel(0).droplevel(0)
assert loc == expected
assert new_index.equals(exp_index)
loc, new_index = index.get_loc_level((0, 1, 0))
expected = 1
assert loc == expected
assert new_index is None
pytest.raises(KeyError, index.get_loc_level, (2, 2))
index = MultiIndex(levels=[[2000], lrange(4)], labels=[np.array(
[0, 0, 0, 0]), np.array([0, 1, 2, 3])])
result, new_index = index.get_loc_level((2000, slice(None, None)))
expected = slice(None, None)
assert result == expected
assert new_index.equals(index.droplevel(0))
@pytest.mark.parametrize('level', [0, 1])
@pytest.mark.parametrize('null_val', [np.nan, pd.NaT, None])
def test_get_loc_nan(self, level, null_val):
# GH 18485 : NaN in MultiIndex
levels = [['a', 'b'], ['c', 'd']]
key = ['b', 'd']
levels[level] = np.array([0, null_val], dtype=type(null_val))
key[level] = null_val
idx = MultiIndex.from_product(levels)
assert idx.get_loc(tuple(key)) == 3
def test_get_loc_missing_nan(self):
# GH 8569
idx = MultiIndex.from_arrays([[1.0, 2.0], [3.0, 4.0]])
assert isinstance(idx.get_loc(1), slice)
pytest.raises(KeyError, idx.get_loc, 3)
pytest.raises(KeyError, idx.get_loc, np.nan)
pytest.raises(KeyError, idx.get_loc, [np.nan])
@pytest.mark.parametrize('dtype1', [int, float, bool, str])
@pytest.mark.parametrize('dtype2', [int, float, bool, str])
def test_get_loc_multiple_dtypes(self, dtype1, dtype2):
# GH 18520
levels = [np.array([0, 1]).astype(dtype1),
np.array([0, 1]).astype(dtype2)]
idx = pd.MultiIndex.from_product(levels)
assert idx.get_loc(idx[2]) == 2
@pytest.mark.parametrize('level', [0, 1])
@pytest.mark.parametrize('dtypes', [[int, float], [float, int]])
def test_get_loc_implicit_cast(self, level, dtypes):
# GH 18818, GH 15994 : as flat index, cast int to float and vice-versa
levels = [['a', 'b'], ['c', 'd']]
key = ['b', 'd']
lev_dtype, key_dtype = dtypes
levels[level] = np.array([0, 1], dtype=lev_dtype)
key[level] = key_dtype(1)
idx = MultiIndex.from_product(levels)
assert idx.get_loc(tuple(key)) == 3
def test_get_loc_cast_bool(self):
# GH 19086 : int is casted to bool, but not vice-versa
levels = [[False, True], np.arange(2, dtype='int64')]
idx = MultiIndex.from_product(levels)
assert idx.get_loc((0, 1)) == 1
assert idx.get_loc((1, 0)) == 2
pytest.raises(KeyError, idx.get_loc, (False, True))
pytest.raises(KeyError, idx.get_loc, (True, False))
def test_slice_locs(self):
df = tm.makeTimeDataFrame()
stacked = df.stack()
idx = stacked.index
slob = slice(*idx.slice_locs(df.index[5], df.index[15]))
sliced = stacked[slob]
expected = df[5:16].stack()
tm.assert_almost_equal(sliced.values, expected.values)
slob = slice(*idx.slice_locs(df.index[5] + timedelta(seconds=30),
df.index[15] - timedelta(seconds=30)))
sliced = stacked[slob]
expected = df[6:15].stack()
tm.assert_almost_equal(sliced.values, expected.values)
def test_slice_locs_with_type_mismatch(self):
df = tm.makeTimeDataFrame()
stacked = df.stack()
idx = stacked.index
tm.assert_raises_regex(TypeError, '^Level type mismatch',
idx.slice_locs, (1, 3))
tm.assert_raises_regex(TypeError, '^Level type mismatch',
idx.slice_locs,
df.index[5] + timedelta(
seconds=30), (5, 2))
df = tm.makeCustomDataframe(5, 5)
stacked = df.stack()
idx = stacked.index
with tm.assert_raises_regex(TypeError, '^Level type mismatch'):
idx.slice_locs(timedelta(seconds=30))
# TODO: Try creating a UnicodeDecodeError in exception message
with tm.assert_raises_regex(TypeError, '^Level type mismatch'):
idx.slice_locs(df.index[1], (16, "a"))
def test_slice_locs_not_sorted(self):
index = MultiIndex(levels=[Index(lrange(4)), Index(lrange(4)), Index(
lrange(4))], labels=[np.array([0, 0, 1, 2, 2, 2, 3, 3]), np.array(
[0, 1, 0, 0, 0, 1, 0, 1]), np.array([1, 0, 1, 1, 0, 0, 1, 0])])
tm.assert_raises_regex(KeyError, "[Kk]ey length.*greater than "
"MultiIndex lexsort depth",
index.slice_locs, (1, 0, 1), (2, 1, 0))
# works
sorted_index, _ = index.sortlevel(0)
# should there be a test case here???
sorted_index.slice_locs((1, 0, 1), (2, 1, 0))
def test_slice_locs_partial(self):
sorted_idx, _ = self.index.sortlevel(0)
result = sorted_idx.slice_locs(('foo', 'two'), ('qux', 'one'))
assert result == (1, 5)
result = sorted_idx.slice_locs(None, ('qux', 'one'))
assert result == (0, 5)
result = sorted_idx.slice_locs(('foo', 'two'), None)
assert result == (1, len(sorted_idx))
result = sorted_idx.slice_locs('bar', 'baz')
assert result == (2, 4)
def test_slice_locs_not_contained(self):
# some searchsorted action
index = MultiIndex(levels=[[0, 2, 4, 6], [0, 2, 4]],
labels=[[0, 0, 0, 1, 1, 2, 3, 3, 3],
[0, 1, 2, 1, 2, 2, 0, 1, 2]], sortorder=0)
result = index.slice_locs((1, 0), (5, 2))
assert result == (3, 6)
result = index.slice_locs(1, 5)
assert result == (3, 6)
result = index.slice_locs((2, 2), (5, 2))
assert result == (3, 6)
result = index.slice_locs(2, 5)
assert result == (3, 6)
result = index.slice_locs((1, 0), (6, 3))
assert result == (3, 8)
result = index.slice_locs(-1, 10)
assert result == (0, len(index))
def test_consistency(self):
# need to construct an overflow
major_axis = lrange(70000)
minor_axis = lrange(10)
major_labels = np.arange(70000)
minor_labels = np.repeat(lrange(10), 7000)
# the fact that is works means it's consistent
index = MultiIndex(levels=[major_axis, minor_axis],
labels=[major_labels, minor_labels])
# inconsistent
major_labels = np.array([0, 0, 1, 1, 1, 2, 2, 3, 3])
minor_labels = np.array([0, 1, 0, 1, 1, 0, 1, 0, 1])
index = MultiIndex(levels=[major_axis, minor_axis],
labels=[major_labels, minor_labels])
assert not index.is_unique
def test_truncate(self):
major_axis = Index(
|
lrange(4)
|
pandas.compat.lrange
|
import pandas as pd
import numpy as np
class logistic:
def __init__(self,x,y):
self.data = x
self.target = y
self.theta = np.array([0,0,0,0])
self.cost = 0
self.thresh = 0.5
def hypo(self,theta,X):
z = (np.matrix(theta)*np.matrix(X).T)
print("hypo : ",1/1+np.exp(-z))
return 1/1+np.exp(-z)
def gradient(self,m,alpha):
n = 0
while n<=1500:
n=n+1
self.theta = self.theta - (alpha/m)*(self.hypo(self.theta,self.data)-self.target)*np.matrix(self.data)
print("Iteration :: ",n)
print(self.theta)
break
print("Values of Theta :: ",self.theta)
def predict(self,x):
prediction = self.hypo(self.theta,x)
print("The prediction probability is :: ",prediction)
if __name__ == "__main__":
iris_data = pd.read_csv('/home/proton/Desktop/My/Might/MachineLearning/Python Implementation/Dataset/Iris.csv')
iris_data=iris_data[:100]
df =
|
pd.DataFrame(iris_data)
|
pandas.DataFrame
|
import numpy as np
import pandas as pd
from sapextractor.algo.o2c import o2c_common
from sapextractor.utils import constants
from sapextractor.utils.change_tables import extract_change
from sapextractor.utils.filters import case_filter
from sapextractor.utils.graph_building import build_graph
from sapextractor.algo.o2c import payment_part
def extract_changes_vbfa(con, dataframe, mandt="800"):
if len(dataframe) > 0:
case_vbeln = dataframe[["case:concept:name", "VBELN"]].to_dict("records")
else:
case_vbeln = []
case_vbeln_dict = {}
for x in case_vbeln:
caseid = x["case:concept:name"]
vbeln = x["VBELN"]
if vbeln not in case_vbeln_dict:
case_vbeln_dict[vbeln] = set()
case_vbeln_dict[vbeln].add(caseid)
ret = []
for tup in [("VERKBELEG", "VBAK"), ("VERKBELEG", "VBAP"), ("VERKBELEG", "VBUK"), ("LIEFERUNG", "LIKP"),
("LIEFERUNG", "LIPS"), ("LIEFERUNG", "VBUK")]:
changes = extract_change.apply(con, objectclas=tup[0], tabname=tup[1], mandt=mandt)
changes = {x: y for x, y in changes.items() if x in case_vbeln_dict}
for x, y in changes.items():
y = y[[xx for xx in y.columns if xx.startswith("event_")]]
cols = {x: x.split("event_")[-1] for x in y.columns}
cols["event_timestamp"] = "time:timestamp"
y = y.rename(columns=cols)
y["VBELN"] = y["AWKEY"]
y["concept:name"] = y["CHANGEDESC"]
for cc in case_vbeln_dict[x]:
z = y.copy()
z["case:concept:name"] = cc
ret.append(z)
if ret:
ret = pd.concat(ret)
else:
ret = pd.DataFrame()
return ret
def extract_bkpf_bsak(con, dataframe, gjahr="2020", mandt="800"):
if len(dataframe) > 0:
case_vbeln = dataframe[["case:concept:name", "VBELN"]].to_dict("records")
else:
case_vbeln = []
case_vbeln_dict = {}
for x in case_vbeln:
caseid = x["case:concept:name"]
vbeln = x["VBELN"]
if vbeln not in case_vbeln_dict:
case_vbeln_dict[vbeln] = set()
case_vbeln_dict[vbeln].add(caseid)
dict_awkey, clearance_docs_dates, blart_vals = payment_part.apply(con, gjahr=gjahr, mandt=mandt)
intersect = set(case_vbeln_dict.keys()).intersection(dict_awkey.keys())
ret = []
for k in intersect:
for belnr in dict_awkey[k]:
if belnr in clearance_docs_dates:
for clearingdoc in clearance_docs_dates[belnr]:
for cas in case_vbeln_dict[k]:
ret.append(
{"case:concept:name": cas, "concept:name": "Clearance (" + blart_vals[clearingdoc[2]] + ")",
"AUGBL": clearingdoc[0], "time:timestamp": clearingdoc[1]})
ret = pd.DataFrame(ret)
if len(ret) > 0:
if "time:timestamp" in ret.columns:
ret["time:timestamp"] = ret["time:timestamp"] + pd.Timedelta(np.timedelta64(86399, 's'))
ret = ret.groupby(["case:concept:name", "AUGBL"]).first().reset_index()
return ret
def apply(con, ref_type="Order", keep_first=True, min_extr_date="2020-01-01 00:00:00", gjahr="2020", enable_changes=True, enable_payments=True, allowed_act_doc_types=None, allowed_act_changes=None, mandt="800"):
dataframe = o2c_common.apply(con, keep_first=keep_first, min_extr_date=min_extr_date, mandt=mandt)
dataframe = dataframe[[x for x in dataframe.columns if x.startswith("event_")]]
cols = {x: x.split("event_")[-1] for x in dataframe.columns}
cols["event_activity"] = "concept:name"
cols["event_timestamp"] = "time:timestamp"
dataframe = dataframe.rename(columns=cols)
if len(dataframe) > 0:
all_docs = set(dataframe[dataframe["VBTYP_N"] == ref_type]["VBELN"].unique())
ancest_succ = build_graph.get_ancestors_successors(dataframe, "VBELV", "VBELN", "VBTYP_V", "VBTYP_N",
ref_type=ref_type, all_docs=all_docs)
# ancest_succ = build_graph.get_conn_comp(dataframe, "VBELV", "VBELN", "VBTYP_V", "VBTYP_N", ref_type=ref_type)
dataframe = dataframe.merge(ancest_succ, left_on="VBELN", right_on="node", suffixes=('', '_r'), how="right")
dataframe = dataframe.reset_index()
if keep_first:
dataframe = dataframe.groupby(["case:concept:name", "VBELN"]).first().reset_index()
if allowed_act_doc_types is not None:
allowed_act_doc_types = set(allowed_act_doc_types)
dataframe = dataframe[dataframe["concept:name"].isin(allowed_act_doc_types)]
if enable_changes:
changes = extract_changes_vbfa(con, dataframe, mandt=mandt)
else:
changes = pd.DataFrame()
if enable_payments:
payments = extract_bkpf_bsak(con, dataframe, gjahr=gjahr, mandt=mandt)
else:
payments = pd.DataFrame()
if allowed_act_changes is not None:
allowed_act_changes = set(allowed_act_changes)
changes = changes[changes["concept:name"].isin(allowed_act_changes)]
dataframe =
|
pd.concat([dataframe, changes, payments])
|
pandas.concat
|
import os
import json
import requests
import thingspeak
import datetime
import pandas as pd
from functools import reduce
from itertools import tee
def pairwise(iterable):
"s -> (s0,s1), (s1,s2), (s2, s3), ..."
a, b = tee(iterable)
next(b, None)
return zip(a, b)
def get_block(start, end):
params = { 'start': str(start),
'end': str(end),
'average': '60'
}
try:
r = channel.get(params)
except:
raise
print('error')
data = [list(feed.values()) for feed in json.loads(r)['feeds']]
block_df = pd.DataFrame(data, columns = cols)
return block_df
sensor_list = pd.read_csv('pa_sensor_list.csv')
#sensor_list = sensor_list.drop(columns=['Unnamed: 0', 'DEVICE_LOCATIONTYPE'])
#group the channel ID and key into pairs
pairs = list(zip(sensor_list['THINGSPEAK_PRIMARY_ID'],
sensor_list['THINGSPEAK_PRIMARY_ID_READ_KEY']))
#split data into 7.2 days, ~ 8000 data points @ 80 second resolution. 8000 is the Thingspeak limit
dates = pd.date_range(start='1/1/2018', end='1/4/2019', freq='7.2D')
def download_pa():
#TODO desperately needs to be async
for cid, key in pairs:
channel_id = cid
read_key = key
channel = thingspeak.Channel(id=channel_id,api_key=read_key)
#prep columns for dataframe
cols = json.loads(channel.get({'results': '1'}))['channel'] # get the json for channel columns
cols = [(key, value) for key, value in cols.items() if key.startswith('field')] # extract only field values
cols = [unit for field, unit in cols]
cols = [col + f'_{channel_id}' for col in cols]
cols.insert(0,'datetime')
df = pd.DataFrame(columns = cols)
for start, end in pairwise(dates):
df = df.append(get_block(start,end))
# ['datetime', 'PM1.0 (ATM)_{channel_id}', 'PM2.5 (ATM)_{channel_id}',
# 'PM10.0 (ATM)_248887', 'Mem_248887',
# 'Unused_248887', 'PM2.5 (CF=1)_248887']
# # df = df.drop(columns = [f'Uptime_{channel_id}',
# # f'RSSI_{channel_id}',
# # f'Temperature_{channel_id}',
# # f'Humidity_{channel_id}'
# # ])
df.to_csv(f'data/purple_air/{channel_id}.csv')
print(f'Channel: {channel_id} processed')
def clean_pa(data, path):
thingspeak_id = int(data[:6])
df = pd.read_csv(path + data)
df = df.drop(df[df['datetime'].duplicated() == True].index)
df = df.drop(df[df['datetime'] > '2018-12-31T23:00:00Z'].index)
if df.shape[0] >= 6000:
try:
lat, lon = zip(*sensor_list[
sensor_list['THINGSPEAK_PRIMARY_ID'] == thingspeak_id][['lat', 'lon']].values)
df['lat'] = lat[0]
df['lon'] = lon[0]
df['id'] = thingspeak_id
except:
print('lat / lon not found, skipping')
return pd.DataFrame()
#filter for only the columns we want
df = df[df.columns[df.columns.str.startswith((
'datetime','lat','lon','PM','id'))]]
return df
else:
return pd.DataFrame()
def pa_concat(path):
files = os.listdir(path)
cols = ['datetime', 'PM1.0 (ATM)',
'PM2.5 (ATM)', 'PM10.0 (ATM)',
'PM2.5 (CF=1)', 'lat',
'lon', 'id'
]
df = pd.concat([
pd.read_csv(path+file, names = cols, skiprows=1, parse_dates=['datetime']) for file in files])
df['datetime'] =
|
pd.to_datetime(df['datetime'])
|
pandas.to_datetime
|
import pandas as pd
import numpy as np
import os
import requests
import logging
import argparse
import re
import pathlib
API_KEY = '<KEY>'
MAX_VARS = 50
STATE_CODES = {'Alabama': ('AL', '01'), 'Alaska': ('AK', '02'),
'Arizona': ('AZ', '04'), 'Arkansas': ('AR', '05'),
'California': ('CA', '06'), 'Colorado': ('CO', '08'),
'Connecticut': ('CT', '09'), 'Delaware': ('DE', '10'),
'District of Columbia': ('DC', '11'), 'Florida': ('FL', '12'),
'Georgia': ('GA', '13'), 'Hawaii': ('HI', '15'),
'Idaho': ('ID', '16'), 'Illinois': ('IL', '17'),
'Indiana': ('IN', '18'), 'Iowa': ('IA', '19'),
'Kansas': ('KS', '20'), 'Kentucky': ('KY', '21'),
'Louisiana': ('LA', '22'), 'Maine': ('ME', '23'),
'Maryland': ('MD', '24'), 'Massachusetts': ('MA', '25'),
'Michigan': ('MI', '26'), 'Minnesota': ('MN', '27'),
'Mississippi': ('MS', '28'), 'Missouri': ('MO', '29'),
'Montana': ('MT', '30'), 'Nebraska': ('NE', '31'),
'Nevada': ('NV', '32'), 'New Hampshire': ('NH', '33'),
'New Jersey': ('NJ', '34'), 'New Mexico': ('NM', '35'),
'New York': ('NY', '36'), 'North Carolina': ('NC', '37'),
'North Dakota': ('ND', '38'), 'Ohio': ('OH', '39'),
'Oklahoma': ('OK', '40'), 'Oregon': ('OR', '41'),
'Pennsylvania': ('PA', '42'), 'Rhode Island': ('RI', '44'),
'South Carolina': ('SC', '45'), 'South Dakota': ('SD', '46'),
'Tennessee': ('TN', '47'), 'Texas': ('TX', '48'),
'Utah': ('UT', '49'), 'Vermont': ('VT', '50'),
'Virginia': ('VA', '51'), 'Washington': ('WA', '53'),
'West Virginia': ('WV', '54'), 'Wisconsin': ('WI', '55'),
'Wyoming': ('WY', '56')}
def extract_vars(vars_file):
""" extract vars to be downloaded from munging file
:param vars_file: string path and filename to list of variables
should have column header: variable_name
if doing transformations, headers should be (tab delimited):
variable_name, operator, argument1, argument2
:returns: list of sorted variable names
"""
# get vars/transform file
do_transforms = False
try:
transforms =
|
pd.read_csv(vars_file, sep='\t')
|
pandas.read_csv
|
# https://colab.research.google.com/notebooks/mlcc/first_steps_with_tensor_flow.ipynb
from __future__ import print_function
import math
import os
from IPython import display
from matplotlib import cm
from matplotlib import gridspec
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
from sklearn import metrics
import tensorflow as tf
from tensorflow.python.data import Dataset
from utils.input_fn import my_input_fn
dirname = os.path.dirname(__file__)
tf.logging.set_verbosity(tf.logging.ERROR)
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:.1f}'.format
csv = os.path.join(dirname, '../datasets/california_housing_train.csv')
california_housing_dataframe = pd.read_csv(csv, sep=",")
# Randomizing the DataFrame
california_housing_dataframe = california_housing_dataframe.reindex(
np.random.permutation(california_housing_dataframe.index)
)
california_housing_dataframe["median_house_value"] /= 1000.0
# print(california_housing_dataframe.describe())
# Define the input feature: total_rooms.
my_feature = california_housing_dataframe[["total_rooms"]]
# Configure a numeric feature column for total_rooms.
feature_columns = [tf.feature_column.numeric_column("total_rooms")]
# Define the label.
targets = california_housing_dataframe["median_house_value"]
# Use gradient descent as the optimizer for training the model.
my_optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.0000001)
my_optimizer = tf.contrib.estimator.clip_gradients_by_norm(my_optimizer, 5.0)
# Configure the linear regression model with our feature columns and optimizer.
# Set a learning rate of 0.0000001 for Gradient Descent.
linear_regressor = tf.estimator.LinearRegressor(
feature_columns=feature_columns,
optimizer=my_optimizer
)
linear_regressor.train(
input_fn = lambda:my_input_fn(my_feature, targets),
steps=100
)
# Evaluating the model
# Create an input function for predictions.
# Note: Since we're making just one prediction for each example, we don't
# need to repeat or shuffle the data here.
prediction_input_fn = lambda: my_input_fn(my_feature, targets, num_epochs=1, shuffle=False)
# Call predict() on the linear_regressor to make predictions.
predictions = linear_regressor.predict(input_fn=prediction_input_fn)
# Format predictions as a NumPy array, so we can calculate error metrics.
predictions = np.array([item['predictions'][0] for item in predictions])
# Print Mean Squared Error and Root Mean Squared Error.
mean_squared_error = metrics.mean_squared_error(predictions, targets)
root_mean_squared_error = math.sqrt(mean_squared_error)
print("Mean Squared Error (on training data): %0.3f" % mean_squared_error)
print("Root Mean Squared Error (on training data): %0.3f" % root_mean_squared_error)
min_house_value = california_housing_dataframe["median_house_value"].min()
max_house_value = california_housing_dataframe["median_house_value"].max()
min_max_difference = max_house_value - min_house_value
print("Min. Median House Value: %0.3f" % min_house_value)
print("Max. Median House Value: %0.3f" % max_house_value)
print("Difference between Min. and Max.: %0.3f" % min_max_difference)
print("Root Mean Squared Error: %0.3f" % root_mean_squared_error)
# Reducing model error
calibration_data = pd.DataFrame()
calibration_data["predictions"] = pd.Series(predictions)
calibration_data["targets"] =
|
pd.Series(targets)
|
pandas.Series
|
from flask import Flask, render_template, request, redirect, url_for, session
import pandas as pd
import pymysql
import os
import io
#from werkzeug.utils import secure_filename
from pulp import *
import numpy as np
import pymysql
import pymysql.cursors
from pandas.io import sql
#from sqlalchemy import create_engine
import pandas as pd
import numpy as np
#import io
import statsmodels.formula.api as smf
import statsmodels.api as sm
import scipy.optimize as optimize
import matplotlib.mlab as mlab
import matplotlib.pyplot as plt
#from flask import Flask, render_template, request, redirect, url_for, session, g
from sklearn.linear_model import LogisticRegression
from math import sin, cos, sqrt, atan2, radians
from statsmodels.tsa.arima_model import ARIMA
#from sqlalchemy import create_engine
from collections import defaultdict
from sklearn import linear_model
import statsmodels.api as sm
import scipy.stats as st
import pandas as pd
import numpy as np
from pulp import *
import pymysql
import math
app = Flask(__name__)
app.secret_key = os.urandom(24)
localaddress="D:\\home\\site\\wwwroot"
localpath=localaddress
os.chdir(localaddress)
@app.route('/')
def index():
return redirect(url_for('home'))
@app.route('/home')
def home():
return render_template('home.html')
@app.route('/demandplanning')
def demandplanning():
return render_template("Demand_Planning.html")
@app.route("/elasticopt",methods = ['GET','POST'])
def elasticopt():
if request.method== 'POST':
start_date =request.form['from']
end_date=request.form['to']
prdct_name=request.form['typedf']
# connection = pymysql.connect(host='localhost',
# user='user',
# password='',
# db='test',
# charset='utf8mb4',
# cursorclass=pymysql.cursors.DictCursor)
#
# x=connection.cursor()
# x.execute("select * from `transcdata`")
# connection.commit()
# datass=pd.DataFrame(x.fetchall())
datass = pd.read_csv("C:\\Users\\1026819\\Downloads\\optimizdata.csv")
# datas = datass[(datass['Week']>=start_date) & (datass['Week']<=end_date )]
datas=datass
df = datas[datas['Product'] == prdct_name]
df=datass
changeData=pd.concat([df['Product_Price'],df['Product_Qty']],axis=1)
changep=[]
changed=[]
for i in range(0,len(changeData)-1):
changep.append(changeData['Product_Price'].iloc[i]-changeData['Product_Price'].iloc[i+1])
changed.append(changeData['Product_Qty'].iloc[1]-changeData['Product_Qty'].iloc[i+1])
cpd=pd.concat([pd.DataFrame(changep),pd.DataFrame(changed)],axis=1)
cpd.columns=['Product_Price','Product_Qty']
sortedpricedata=df.sort_values(['Product_Price'], ascending=[True])
spq=pd.concat([sortedpricedata['Product_Price'],sortedpricedata['Product_Qty']],axis=1).reset_index(drop=True)
pint=[]
dint=[]
x = spq['Product_Price']
num_bins = 5
# n, pint, patches = plt.hist(x, num_bins, facecolor='blue', alpha=0.5)
y = spq['Product_Qty']
num_bins = 5
# n, dint, patches = plt.hist(y, num_bins, facecolor='blue', alpha=0.5)
arr= np.zeros(shape=(len(pint),len(dint)))
count=0
for i in range(0, len(pint)):
lbp=pint[i]
if i==len(pint)-1:
ubp=pint[i]+1
else:
ubp=pint[i+1]
for j in range(0, len(dint)):
lbd=dint[j]
if j==len(dint)-1:
ubd=dint[j]+1
else:
ubd=dint[j+1]
print(lbd,ubd)
for k in range(0, len(spq)):
if (spq['Product_Price'].iloc[k]>=lbp\
and spq['Product_Price'].iloc[k]<ubp):
if(spq['Product_Qty'].iloc[k]>=lbd\
and spq['Product_Qty'].iloc[k]<ubd):
count+=1
arr[i][j]+=1
price_range=np.zeros(shape=(len(pint),2))
for j in range(0,len(pint)):
lbp=pint[j]
price_range[j][0]=lbp
if j==len(pint)-1:
ubp=pint[j]+1
price_range[j][1]=ubp
else:
ubp=pint[j+1]
price_range[j][1]=ubp
demand_range=np.zeros(shape=(len(dint),2))
for j in range(0,len(dint)):
lbd=dint[j]
demand_range[j][0]=lbd
if j==len(dint)-1:
ubd=dint[j]+1
demand_range[j][1]=ubd
else:
ubd=dint[j+1]
demand_range[j][1]=ubd
pr=pd.DataFrame(price_range)
pr.columns=['Price','Demand']
dr=pd.DataFrame(demand_range)
dr.columns=['Price','Demand']
priceranges=pr.Price.astype(str).str.cat(pr.Demand.astype(str), sep='-')
demandranges=dr.Price.astype(str).str.cat(dr.Demand.astype(str), sep='-')
price=pd.DataFrame(arr)
price.columns=demandranges
price.index=priceranges
pp=price.reset_index()
global data
data=
|
pd.concat([df['Week'],df['Product_Qty'],df['Product_Price'],df['Comp_Prod_Price'],df['Promo1'],df['Promo2'],df['overallsale']],axis=1)
|
pandas.concat
|
"""Module containing implementations for various psychological questionnaires.
Each function at least expects a dataframe containing the required columns in a specified order
(see function documentations for specifics) to be passed to the ``data`` argument.
If ``data`` is a dataframe that contains more than the required two columns, e.g., if the complete questionnaire
dataframe is passed, the required columns can be sliced by specifying them in the ``columns`` parameter.
Also, if the columns in the dataframe are not in the correct order, the order can be specified
using the ``columns`` parameter.
Some questionnaire functions also allow the possibility to only compute certain subscales. To do this, a dictionary
with subscale names as keys and the corresponding column names (as list of str) or column indices
(as list of ints) can be passed to the ``subscales`` parameter.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
"""
from typing import Dict, Optional, Sequence, Union
import numpy as np
import pandas as pd
from typing_extensions import Literal
from biopsykit.questionnaires.utils import (
_compute_questionnaire_subscales,
_invert_subscales,
bin_scale,
invert,
to_idx,
)
from biopsykit.utils._datatype_validation_helper import _assert_has_columns, _assert_num_columns, _assert_value_range
from biopsykit.utils.exceptions import ValueRangeError
from biopsykit.utils.time import time_to_datetime
def psqi(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Pittsburgh Sleep Quality Index (PSQI)**.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
..warning::
The PSQI has a slightly different score name format than other questionnaires since it has several
subquestions (denoted "a", "b", ..., "j") for question 5, as well as one free-text question. When using this
function to compute the PSQI the make sure your column names adhere to the following naming convention for
the function to work properly:
* Questions 1 - 10 (except Question 5): suffix "01", "02", ..., "10"
* Subquestions of Question 5: suffix "05a", "05b", ..., "05j"
* Free-text subquestion of Question 5: suffix "05j_text"
Returns
-------
:class:`~pandas.DataFrame`
PSQI score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
"""
score_name = "PSQI"
score_range = [0, 3]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
data = data.loc[:, columns]
# Bedtime Start: Question 1
bed_time_start = data.filter(regex="01").iloc[:, 0]
bed_time_start = time_to_datetime(bed_time_start)
# Bedtime End: Question 3
bed_time_end = data.filter(regex="03").iloc[:, 0]
bed_time_end = time_to_datetime(bed_time_end)
# Compute Hours in Bed (needed for habitual sleep efficiency)
bed_time_diff = bed_time_end - bed_time_start
hours_bed = ((bed_time_diff.view(np.int64) / 1e9) / 3600) % 24
# Sleep Duration: Question 4
sd = data.filter(regex="04").iloc[:, 0]
# Sleep Latency: Question 2
sl = data.filter(regex="02").iloc[:, 0]
# Bin scale: 0-15 = 0, 16-30 = 1, 31-60 = 2, >=61 = 3
bin_scale(sl, bins=[0, 15, 30, 60], last_max=True, inplace=True)
data = data.drop(columns=data.filter(regex="0[1234]"))
data = data.drop(columns=data.filter(regex="05j_text"))
_assert_value_range(data, score_range)
# Subjective Sleep Quality
ssq = data.filter(regex="06").iloc[:, 0]
# Sleep Disturbances: Use all questions from 5, except 05a and 05j_text
sdist = data.filter(regex="05").iloc[:, :]
# 05j_text does not need to be dropped since it was already excluded previously
sdist = sdist.drop(columns=sdist.filter(regex="05a")).sum(axis=1)
# Bin scale: 0 = 0, 1-9 = 1, 10-18 = 2, 19-27 = 3
sdist = bin_scale(sdist, bins=[-1, 0, 9, 18, 27])
# Use of Sleep Medication: Use question 7
sm = data.filter(regex="07").iloc[:, 0]
# Daytime Dysfunction: Sum questions 8 and 9
dd = data.filter(regex="0[89]").sum(axis=1)
# Bin scale: 0 = 0, 1-2 = 1, 3-4 = 2, 5-6 = 3
dd = bin_scale(dd, bins=[-1, 0, 2, 4], inplace=False, last_max=True)
# Sleep Latency: Question 2 and 5a, sum them
sl = sl + data.filter(regex="05a").iloc[:, 0]
# Bin scale: 0 = 0, 1-2 = 1, 3-4 = 2, 5-6 = 3
sl = bin_scale(sl, bins=[-1, 0, 2, 4, 6])
# Habitual Sleep Efficiency
hse = ((sd / hours_bed) * 100.0).round().astype(int)
# Bin scale: >= 85% = 0, 75%-84% = 1, 65%-74% = 2, < 65% = 3
hse = invert(bin_scale(hse, bins=[0, 64, 74, 84], last_max=True), score_range=score_range)
# Sleep Duration: Bin scale: > 7 = 0, 6-7 = 1, 5-6 = 2, < 5 = 3
sd = invert(bin_scale(sd, bins=[0, 4.9, 6, 7], last_max=True), score_range=score_range)
psqi_data = {
score_name + "_SubjectiveSleepQuality": ssq,
score_name + "_SleepLatency": sl,
score_name + "_SleepDuration": sd,
score_name + "_HabitualSleepEfficiency": hse,
score_name + "_SleepDisturbances": sdist,
score_name + "_UseSleepMedication": sm,
score_name + "_DaytimeDysfunction": dd,
}
data = pd.DataFrame(psqi_data, index=data.index)
data[score_name + "_TotalIndex"] = data.sum(axis=1)
return data
def mves(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Maastricht Vital Exhaustion Scale (MVES)**.
The MVES uses 23 items to assess the concept of Vital Exhaustion (VE), which is characterized by feelings of
excessive fatigue, lack of energy, irritability, and feelings of demoralization. Higher scores indicate greater
vital exhaustion.
.. note::
This implementation assumes a score range of [0, 2].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
MVES score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., & <NAME>. (1987). A questionnaire to assess premonitory symptoms of myocardial
infarction. *International Journal of Cardiology*, 17(1), 15–24. https://doi.org/10.1016/0167-5273(87)90029-5
"""
score_name = "MVES"
score_range = [0, 2]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 23)
_assert_value_range(data, score_range)
# Reverse scores 9, 14
data = invert(data, cols=to_idx([9, 14]), score_range=score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name])
def tics_l(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[str, int]]]] = None,
) -> pd.DataFrame:
"""Compute the **Trier Inventory for Chronic Stress (Long Version) (TICS_L)**.
The TICS assesses frequency of various types of stressful experiences in the past 3 months.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Work Overload``: [50, 38, 44, 54, 17, 4, 27, 1]
* ``Social Overload``: [39, 28, 49, 19, 7, 57]
* ``Excessive Demands at Work``: [55, 24, 20, 35, 47, 3]
* ``Lack of Social Recognition``: [31, 18, 46, 2]
* ``Work Discontent``: [21, 53, 10, 48, 41, 13, 37, 5]
* ``Social Tension``: [26, 15, 45, 52, 6, 33]
* ``Performance Pressure at Work``: [23, 43, 32, 22, 12, 14, 8, 40, 30]
* ``Performance Pressure in Social Interactions``: [6, 15, 22]
* ``Social Isolation``: [42, 51, 34, 56, 11, 29]
* ``Worry Propensity``: [36, 25, 16, 9]
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
TICS_L score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
Examples
--------
>>> from biopsykit.questionnaires import tics_s
>>> # compute only a subset of subscales; questionnaire items additionally have custom indices
>>> subscales = {
>>> 'WorkOverload': [1, 2, 3],
>>> 'SocialOverload': [4, 5, 6],
>>> }
>>> tics_s_result = tics_s(data, subscales=subscales)
References
----------
<NAME>., <NAME>., & <NAME>. (2004). Trierer Inventar zum chronischen Stress: TICS. *Hogrefe*.
"""
score_name = "TICS_L"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 57)
subscales = {
"WorkOverload": [1, 4, 17, 27, 38, 44, 50, 54], # Arbeitsüberlastung
"SocialOverload": [7, 19, 28, 39, 49, 57], # Soziale Überlastung
"PressureToPerform": [8, 12, 14, 22, 23, 30, 32, 40, 43], # Erfolgsdruck
"WorkDiscontent": [5, 10, 13, 21, 37, 41, 48, 53], # Unzufriedenheit mit der Arbeit
"DemandsWork": [3, 20, 24, 35, 47, 55], # Überforderung bei der Arbeit
"LackSocialRec": [2, 18, 31, 46], # Mangel an sozialer Anerkennung
"SocialTension": [6, 15, 26, 33, 45, 52], # Soziale Spannungen
"SocialIsolation": [11, 29, 34, 42, 51, 56], # Soziale Isolation
"ChronicWorry": [9, 16, 25, 36], # Chronische Besorgnis
}
_assert_value_range(data, score_range)
tics_data = _compute_questionnaire_subscales(data, score_name, subscales)
if len(data.columns) == 57:
# compute total score if all columns are present
tics_data[score_name] = data.sum(axis=1)
return pd.DataFrame(tics_data, index=data.index)
def tics_s(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[str, int]]]] = None,
) -> pd.DataFrame:
"""Compute the **Trier Inventory for Chronic Stress (Short Version) (TICS_S)**.
The TICS assesses frequency of various types of stressful experiences in the past 3 months.
It consists of the subscales (the name in the brackets indicate the name in the returned dataframe),
with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Work Overload``: [1, 3, 21]
* ``Social Overload``: [11, 18, 28]
* ``Excessive Demands at Work``: [12, 16, 27]
* ``Lack of Social Recognition``: [2, 20, 23]
* ``Work Discontent``: [8, 13, 24]
* ``Social Tension``: [4, 9, 26]
* ``Performance Pressure at Work``: [5, 14, 29]
* ``Performance Pressure in Social Interactions``: [6, 15, 22]
* ``Social Isolation``: [19, 25, 30]
* ``Worry Propensity``: [7, 10, 17]
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
TICS_S score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
Examples
--------
>>> from biopsykit.questionnaires import tics_s
>>> # compute only a subset of subscales; questionnaire items additionally have custom indices
>>> subscales = {
>>> 'WorkOverload': [1, 2, 3],
>>> 'SocialOverload': [4, 5, 6],
>>> }
>>> tics_s_result = tics_s(data, subscales=subscales)
References
----------
<NAME>., <NAME>., & <NAME>. (2004). Trierer Inventar zum chronischen Stress: TICS. *Hogrefe*.
"""
score_name = "TICS_S"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 30)
subscales = {
"WorkOverload": [1, 3, 21],
"SocialOverload": [11, 18, 28],
"PressureToPerform": [5, 14, 29],
"WorkDiscontent": [8, 13, 24],
"DemandsWork": [12, 16, 27],
"PressureSocial": [6, 15, 22],
"LackSocialRec": [2, 20, 23],
"SocialTension": [4, 9, 26],
"SocialIsolation": [19, 25, 30],
"ChronicWorry": [7, 10, 17],
}
_assert_value_range(data, score_range)
tics_data = _compute_questionnaire_subscales(data, score_name, subscales)
if len(data.columns) == 30:
# compute total score if all columns are present
tics_data[score_name] = data.sum(axis=1)
return pd.DataFrame(tics_data, index=data.index)
def pss(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[str, int]]]] = None,
) -> pd.DataFrame:
"""Compute the **Perceived Stress Scale (PSS)**.
The PSS is a widely used self-report questionnaire with adequate reliability and validity asking
about how stressful a person has found his/her life during the previous month.
The PSS consists of the subscales with the item indices
(count-by-one, i.e., the first question has the index 1!):
* Perceived Helplessness (Hilflosigkeit - ``Helpness``): [1, 2, 3, 6, 9, 10]
* Perceived Self-Efficacy (Selbstwirksamkeit - ``SelfEff``): [4, 5, 7, 8]
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
PSS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., & <NAME>. (1983). A Global Measure of Perceived Stress.
*Journal of Health and Social Behavior*, 24(4), 385. https://doi.org/10.2307/2136404
"""
score_name = "PSS"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 10)
subscales = {"Helpless": [1, 2, 3, 6, 9, 10], "SelfEff": [4, 5, 7, 8]}
_assert_value_range(data, score_range)
# Reverse scores 4, 5, 7, 8
data = invert(data, cols=to_idx([4, 5, 7, 8]), score_range=score_range)
pss_data = _compute_questionnaire_subscales(data, score_name, subscales)
pss_data["{}_Total".format(score_name)] = data.sum(axis=1)
return pd.DataFrame(pss_data, index=data.index)
def cesd(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Center for Epidemiological Studies Depression Scale (CES-D)**.
The CES-D asks about depressive symptoms experienced over the past week.
Higher scores indicate greater depressive symptoms.
.. note::
This implementation assumes a score range of [0, 3].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
CES-D score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (1977). The CES-D Scale: A Self-Report Depression Scale for Research in the General Population.
Applied Psychological Measurement, 1(3), 385–401. https://doi.org/10.1177/014662167700100306
"""
score_name = "CESD"
score_range = [0, 3]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 20)
_assert_value_range(data, score_range)
# Reverse scores 4, 8, 12, 16
data = invert(data, cols=to_idx([4, 8, 12, 16]), score_range=score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name])
def ads_l(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Allgemeine Depressionsskala - Langform (ADS-L)** (General Depression Scale – Long Version).
The General Depression Scale (ADS) is a self-report instrument that can be used to assess the impairment caused by
depressive symptoms within the last week. Emotional, motivational, cognitive, somatic as well as motor symptoms are
assessed, motivational, cognitive, somatic, and motor/interactional complaints.
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
ADS-L score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (2001). Allgemeine Depressions-Skala (ADS). Normierung an Minderjährigen und
Erweiterung zur Erfassung manischer Symptome (ADMS). Diagnostica.
"""
score_name = "ADS_L"
score_range = [0, 3]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 20)
_assert_value_range(data, score_range)
# Reverse scores 4, 8, 12, 16
data = invert(data, cols=to_idx([4, 8, 12, 16]), score_range=score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name])
def ghq(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **General Health Questionnaire (GHQ)**.
The GHQ-12 is a widely used tool for detecting psychological and mental health and as a screening tool for
excluding psychological and psychiatric morbidity. Higher scores indicate *lower* health.
A summed score above 4 is considered an indicator of psychological morbidity.
.. note::
This implementation assumes a score range of [0, 3].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
GHQ score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (1972). The detection of psychiatric illness by questionnaire. *Maudsley monograph*, 21.
"""
score_name = "GHQ"
score_range = [0, 3]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 12)
_assert_value_range(data, score_range)
# Reverse scores 1, 3, 4, 7, 8, 12
data = invert(data, cols=to_idx([1, 3, 4, 7, 8, 12]), score_range=score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name])
def hads(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[str, int]]]] = None,
) -> pd.DataFrame:
"""Compute the **Hospital Anxiety and Depression Scale (HADS)**.
The HADS is a brief and widely used instrument to measure psychological distress in patients
and in the general population. It has two subscales: anxiety and depression.
Higher scores indicate greater distress.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Anxiety``: [1, 3, 5, 7, 9, 11, 13]
* ``Depression``: [2, 4, 6, 8, 10, 12, 14]
.. note::
This implementation assumes a score range of [0, 3].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
HADS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns do not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (1983). The hospital anxiety and depression scale.
*Acta psychiatrica scandinavica*, 67(6), 361-370.
"""
score_name = "HADS"
score_range = [0, 3]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 14)
subscales = {
"Anxiety": [1, 3, 5, 7, 9, 11, 13],
"Depression": [2, 4, 6, 8, 10, 12, 14],
}
_assert_value_range(data, score_range)
# Reverse scores 2, 4, 6, 7, 12, 14
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data, subscales=subscales, idx_dict={"Anxiety": [3], "Depression": [0, 1, 2, 5, 6]}, score_range=score_range
)
hads_data = _compute_questionnaire_subscales(data, score_name, subscales)
if len(data.columns) == 14:
# compute total score if all columns are present
hads_data[score_name] = data.sum(axis=1)
return pd.DataFrame(hads_data, index=data.index)
def type_d(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Type D Personality Scale**.
Type D personality is a personality trait characterized by negative affectivity (NA) and social
inhibition (SI). Individuals who are high in both NA and SI have a *distressed* or Type D personality.
It consists of the subscales, with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Negative Affect``: [2, 4, 5, 7, 9, 12, 13]
* ``Social Inhibition``: [1, 3, 6, 8, 10, 11, 14]
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
TypeD score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (2005). DS14: standard assessment of negative affectivity, social inhibition, and Type D personality.
*Psychosomatic medicine*, 67(1), 89-97.
"""
score_name = "Type_D"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 14)
subscales = {
"NegativeAffect": [2, 4, 5, 7, 9, 12, 13],
"SocialInhibition": [1, 3, 6, 8, 10, 11, 14],
}
_assert_value_range(data, score_range)
# Reverse scores 1, 3
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(data, subscales=subscales, idx_dict={"SocialInhibition": [0, 1]}, score_range=score_range)
ds_data = _compute_questionnaire_subscales(data, score_name, subscales)
if len(data.columns) == 14:
# compute total score if all columns are present
ds_data[score_name] = data.sum(axis=1)
return pd.DataFrame(ds_data, index=data.index)
def rse(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Rosenberg Self-Esteem Inventory**.
The RSE is the most frequently used measure of global self-esteem. Higher scores indicate greater self-esteem.
.. note::
This implementation assumes a score range of [0, 3].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
RSE score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (1965). Society and the Adolescent Self-Image. *Princeton University Press*, Princeton, NJ.
"""
score_name = "RSE"
score_range = [0, 3]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 10)
_assert_value_range(data, score_range)
# Reverse scores 2, 5, 6, 8, 9
data = invert(data, cols=to_idx([2, 5, 6, 8, 9]), score_range=score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name])
def scs(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Self-Compassion Scale (SCS)**.
The Self-Compassion Scale measures the tendency to be compassionate rather than critical
toward the self in difficult times. It is typically assessed as a composite but can be broken down
into subscales. Higher scores indicate greater self-compassion.
It consists of the subscales, with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``SelfKindness``: [5, 12, 19, 23, 26]
* ``SelfJudgment``: [1, 8, 11, 16, 21]
* ``CommonHumanity``: [3, 7, 10, 15]
* ``Isolation``: [4, 13, 18, 25]
* ``Mindfulness``: [9, 14, 17, 22]
* ``OverIdentified`` [2, 6, 20, 24]
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
SCS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (2003). The development and validation of a scale to measure self-compassion.
*Self and identity*, 2(3), 223-250.
https://www.academia.edu/2040459
"""
score_name = "SCS"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 26)
subscales = {
"SelfKindness": [5, 12, 19, 23, 26],
"SelfJudgment": [1, 8, 11, 16, 21],
"CommonHumanity": [3, 7, 10, 15],
"Isolation": [4, 13, 18, 25],
"Mindfulness": [9, 14, 17, 22],
"OverIdentified": [2, 6, 20, 24],
}
_assert_value_range(data, score_range)
# Reverse scores 1, 2, 4, 6, 8, 11, 13, 16, 18, 20, 21, 24, 25
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data,
subscales=subscales,
idx_dict={"SelfJudgment": [0, 1, 2, 3, 4], "Isolation": [0, 1, 2, 3], "OverIdentified": [0, 1, 2, 3]},
score_range=score_range,
)
# SCS is a mean, not a sum score!
scs_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
if len(data.columns) == 26:
# compute total score if all columns are present
scs_data[score_name] = data.mean(axis=1)
return pd.DataFrame(scs_data, index=data.index)
def midi(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Midlife Development Inventory (MIDI) Sense of Control Scale**.
The Midlife Development Inventory (MIDI) sense of control scale assesses perceived control,
that is, how much an individual perceives to be in control of his or her environment. Higher scores indicate
greater sense of control.
.. note::
This implementation assumes a score range of [1, 7].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
MIDI score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (1998). The sense of control as a moderator of social class differences in
health and well-being. *Journal of personality and social psychology*, 74(3), 763.
"""
score_name = "MIDI"
score_range = [1, 7]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 12)
_assert_value_range(data, score_range)
# Reverse scores 1, 2, 4, 5, 7, 9, 10, 11
data = invert(data, cols=to_idx([1, 2, 4, 5, 7, 9, 10, 11]), score_range=score_range)
# MIDI is a mean, not a sum score!
return pd.DataFrame(data.mean(axis=1), columns=[score_name])
def tsgs(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[str, int]]]] = None,
) -> pd.DataFrame:
"""Compute the **Trait Shame and Guilt Scale**.
The TSGS assesses the experience of shame, guilt, and pride over the past few months with three separate subscales.
Shame and guilt are considered distinct emotions, with shame being a global negative feeling about the self,
and guilt being a negative feeling about a specific event rather than the self. Higher scores on each subscale
indicate higher shame, guilt, or pride.
It consists of the subscales,
with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Shame``: [2, 5, 8, 11, 14]
* ``Guilt``: [3, 6, 9, 12, 15]
* ``Pride``: [1, 4, 7, 10, 13]
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
TSGS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., & <NAME>. (2008). The psychobiology of trait shame in young women:
Extending the social self preservation theory. *Health Psychology*, 27(5), 523.
"""
score_name = "TSGS"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 15)
subscales = {
"Pride": [1, 4, 7, 10, 13],
"Shame": [2, 5, 8, 11, 14],
"Guilt": [3, 6, 9, 12, 15],
}
_assert_value_range(data, score_range)
tsgs_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(tsgs_data, index=data.index)
def rmidi(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[str, int]]]] = None,
) -> pd.DataFrame:
"""Compute the **Revised Midlife Development Inventory (MIDI) Personality Scale**.
The Midlife Development Inventory (MIDI) includes 6 personality trait scales: Neuroticism,
Extraversion, Openness to Experience, Conscientiousness, Agreeableness, and Agency. Higher scores
indicate higher endorsement of each personality trait.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Neuroticism``: [3, 8, 13, 19]
* ``Extraversion``: [1, 6, 11, 23, 27]
* ``Openness``: [14, 17, 21, 22, 25, 28, 29]
* ``Conscientiousness``: [4, 9, 16, 24, 31]
* ``Agreeableness``: [2, 7, 12, 18, 26]
* ``Agency``: [5, 10, 15, 20, 30]
.. note::
This implementation assumes a score range of [1, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
RMIDI score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (2001). Planning for the future: a life management strategy for increasing control
and life satisfaction in adulthood. *Psychology and aging*, 16(2), 206.
"""
score_name = "RMIDI"
score_range = [1, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 31)
subscales = {
"Neuroticism": [3, 8, 13, 19],
"Extraversion": [1, 6, 11, 23, 27],
"Openness": [14, 17, 21, 22, 25, 28, 29],
"Conscientiousness": [4, 9, 16, 24, 31],
"Agreeableness": [2, 7, 12, 18, 26],
"Agency": [5, 10, 15, 20, 30],
}
_assert_value_range(data, score_range)
# "most items need to be reverse scored before subscales are computed => reverse all"
data = invert(data, score_range=score_range)
# Re-reverse scores 19, 24
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data, subscales=subscales, idx_dict={"Neuroticism": [3], "Conscientiousness": [3]}, score_range=score_range
)
# RMIDI is a mean, not a sum score!
rmidi_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
return pd.DataFrame(rmidi_data, index=data.index)
def lsq(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Sequence[str]] = None,
) -> pd.DataFrame:
"""Compute the **Life Stress Questionnaire**.
The LSQ asks participants about stressful life events that they and their close relatives have experienced
throughout their entire life, what age they were when the event occurred, and how much it impacted them.
Higher scores indicate more stress.
It consists of the subscales:
* ``PartnerStress``: columns with suffix ``_Partner``
* ``ParentStress``: columns with suffix ``_Parent``
* ``ChildStress``: columns with suffix ``_Child``
.. note::
This implementation assumes a score range of [0, 1].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : list of str, optional
List of subscales (``Partner``, ``Parent``, ``Child``) to compute or ``None`` to compute all subscales.
Default: ``None``
Returns
-------
:class:`~pandas.DataFrame`
LSQ score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (2001). Planning for the future: a life management strategy for increasing control
and life satisfaction in adulthood. *Psychology and aging*, 16(2), 206.
"""
score_name = "LSQ"
score_range = [0, 1]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 30)
subscales = ["Partner", "Parent", "Child"]
if isinstance(subscales, str):
subscales = [subscales]
_assert_value_range(data, score_range)
lsq_data = {"{}_{}".format(score_name, subscale): data.filter(like=subscale).sum(axis=1) for subscale in subscales}
return pd.DataFrame(lsq_data, index=data.index)
def ctq(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Childhood Trauma Questionnaire (CTQ)**.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``PhysicalAbuse``: [9, 11, 12, 15, 17]
* ``SexualAbuse``: [20, 21, 23, 24, 27]
* ``EmotionalNeglect``: [5, 7, 13, 19, 28]
* ``PhysicalNeglect``: [1, 2, 4, 6, 26]
* ``EmotionalAbuse``: [3, 8, 14, 18, 25]
Additionally, three items assess the validity of the responses (high scores on these items could be grounds for
exclusion of a given participants’ responses):
* ``Validity``: [10, 16, 22]
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
CTQ score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., <NAME>., <NAME>., <NAME>., ... & <NAME>. (1994).
Initial reliability and validity of a new retrospective measure of child abuse and neglect.
*The American journal of psychiatry*.
"""
score_name = "CTQ"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
_assert_has_columns(data, [columns])
# if columns parameter is supplied: slice columns from dataframe
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 28)
subscales = {
"PhysicalAbuse": [9, 11, 12, 15, 17],
"SexualAbuse": [20, 21, 23, 24, 27],
"EmotionalNeglect": [5, 7, 13, 19, 28],
"PhysicalNeglect": [1, 2, 4, 6, 26],
"EmotionalAbuse": [3, 8, 14, 18, 25],
"Validity": [10, 16, 22],
}
_assert_value_range(data, score_range)
# reverse scores 2, 5, 7, 13, 19, 26, 28
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data,
subscales=subscales,
idx_dict={
"PhysicalNeglect": [1, 4],
"EmotionalNeglect": [0, 1, 2, 3, 4],
},
score_range=score_range,
)
ctq_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(ctq_data, index=data.index)
def peat(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Pittsburgh Enjoyable Activities Test (PEAT)**.
The PEAT is a self-report measure of engagement in leisure activities. It asks participants to report how often
over the last month they have engaged in each of the activities. Higher scores indicate more time spent in
leisure activities.
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
PEAT score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., <NAME>., <NAME>., <NAME>., & <NAME>. (2009).
Association of enjoyable leisure activities with psychological and physical well-being.
*Psychosomatic medicine*, 71(7), 725.
"""
score_name = "PEAT"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 10)
_assert_value_range(data, score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name])
def purpose_life(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Purpose in Life** questionnaire.
Purpose in life refers to the psychological tendency to derive meaning from life’s experiences
and to possess a sense of intentionality and goal directedness that guides behavior.
Higher scores indicate greater purpose in life.
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
TICS score
References
----------
<NAME>., <NAME>., <NAME>., & <NAME>. (2009). Purpose in life is associated with
mortality among community-dwelling older persons. *Psychosomatic medicine*, 71(5), 574.
"""
score_name = "PurposeLife"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 10)
_assert_value_range(data, score_range)
# reverse scores 2, 3, 5, 6, 10
data = invert(data, cols=to_idx([2, 3, 5, 6, 10]), score_range=score_range)
# Purpose in Life is a mean, not a sum score!
return pd.DataFrame(data.mean(axis=1), columns=[score_name])
def trait_rumination(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Trait Rumination**.
Higher scores indicate greater rumination.
.. note::
This implementation assumes a score range of [0, 1], where 0 = no rumination, 1 = rumination.
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
TraitRumination score
References
----------
<NAME>., <NAME>., & <NAME>. (1993). Response styles and the duration of episodes of
depressed mood. *Journal of abnormal psychology*, 102(1), 20.
"""
score_name = "TraitRumination"
score_range = [0, 1]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 14)
_assert_value_range(data, score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name])
def besaa(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[int, str]]]] = None,
) -> pd.DataFrame:
"""Compute the **Body-Esteem Scale for Adolescents and Adults (BESAA)**.
Body Esteem refers to self-evaluations of one’s body or appearance. The BESAA is based on
the idea that feelings about one’s weight can be differentiated from feelings about one’s general appearance,
and that one’s own opinions may be differentiated from the opinions attributed to others.
Higher scores indicate higher body esteem.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Appearance``: [1, 6, 9, 7, 11, 13, 15, 17, 21, 23]
* ``Weight``: [3, 4, 8, 10, 16, 18, 19, 22]
* ``Attribution``: [2, 5, 12, 14, 20]
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
BESAA score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., & <NAME>. (2001). Body-esteem scale for adolescents and adults.
*Journal of personality assessment*, 76(1), 90-106.
"""
score_name = "BESAA"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 23)
subscales = {
"Appearance": [1, 6, 7, 9, 11, 13, 15, 17, 21, 23],
"Weight": [3, 4, 8, 10, 16, 18, 19, 22],
"Attribution": [2, 5, 12, 14, 20],
}
_assert_value_range(data, score_range)
# reverse scores 4, 7, 9, 11, 13, 17, 18, 19, 21
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data,
subscales=subscales,
idx_dict={"Appearance": [2, 3, 4, 5, 7, 8], "Weight": [1, 5, 6]},
score_range=score_range,
)
# BESAA is a mean, not a sum score!
besaa_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
return pd.DataFrame(besaa_data, index=data.index)
def fscrs(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Forms of Self-Criticizing/Attacking and Self-Reassuring Scale (FSCRS)**.
Self-criticism describes the internal relationship with the self in which part of the self shames
and puts down, while the other part of the self responds and submits to such attacks.
Self-reassurance refers to the opposing idea that many individuals focus on positive aspects of self and defend
against self-criticism. The FSCRS exemplifies some of the self-statements made by either those who are
self-critical or by those who self-reassure.
The scale measures these two traits on a continuum with self-criticism at one end and
self-reassurance at the other. Higher scores on each subscale indicate higher self-criticizing ("Inadequate Self"),
self-attacking ("Hated Self"), and self-reassuring "Reassuring Self", respectively.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``InadequateSelf``: [1, 2, 4, 6, 7, 14, 17, 18, 20]
* ``HatedSelf``: [9, 10, 12, 15, 22]
* ``ReassuringSelf``: [3, 5, 8, 11, 13, 16, 19, 21]
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
FSCRS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., <NAME>., & <NAME>. (2004). Criticizing and reassuring oneself:
An exploration of forms, styles and reasons in female students. *British Journal of Clinical Psychology*,
43(1), 31-50.
"""
score_name = "FSCRS"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 22)
subscales = {
"InadequateSelf": [1, 2, 4, 6, 7, 14, 17, 18, 20],
"HatedSelf": [9, 10, 12, 15, 22],
"ReassuringSelf": [3, 5, 8, 11, 13, 16, 19, 21],
}
_assert_value_range(data, score_range)
fscrs_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(fscrs_data, index=data.index)
def pasa(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Primary Appraisal Secondary Appraisal Scale (PASA)**.
The PASA assesses each of the four cognitive appraisal processes relevant for acute stress protocols,
such as the TSST: primary stress appraisal (threat and challenge) and secondary stress appraisal
(self-concept of own abilities and control expectancy). Higher scores indicate greater appraisals for each sub-type.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Threat``: [1, 9, 5, 13]
* ``Challenge``: [6, 10, 2, 14]
* ``SelfConcept``: [7, 3, 11, 15]
* ``ControlExp``: [4, 8, 12, 16]
.. note::
This implementation assumes a score range of [1, 6].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
PASA score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., & <NAME>. (2005). Psychological determinants of the cortisol stress
response: the role of anticipatory cognitive appraisal. *Psychoneuroendocrinology*, 30(6), 599-610.
"""
score_name = "PASA"
score_range = [1, 6]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 16)
subscales = {
"Threat": [1, 5, 9, 13],
"Challenge": [2, 6, 10, 14],
"SelfConcept": [3, 7, 11, 15],
"ControlExp": [4, 8, 12, 16],
}
_assert_value_range(data, score_range)
# reverse scores 1, 6, 7, 9, 10
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data,
subscales=subscales,
idx_dict={"Threat": [0, 2], "Challenge": [1, 2], "SelfConcept": [1]},
score_range=score_range,
)
pasa_data = _compute_questionnaire_subscales(data, score_name, subscales)
if all(s in subscales for s in ["Threat", "Challenge"]):
pasa_data[score_name + "_Primary"] = (
pasa_data[score_name + "_Threat"] + pasa_data[score_name + "_Challenge"]
) / 2
if all(s in subscales for s in ["SelfConcept", "ControlExp"]):
pasa_data[score_name + "_Secondary"] = (
pasa_data[score_name + "_SelfConcept"] + pasa_data[score_name + "_ControlExp"]
) / 2
if all("{}_{}".format(score_name, s) in pasa_data for s in ["Primary", "Secondary"]):
pasa_data[score_name + "_StressComposite"] = (
pasa_data[score_name + "_Primary"] - pasa_data[score_name + "_Secondary"]
)
return pd.DataFrame(pasa_data, index=data.index)
def ssgs(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **State Shame and Guilt Scale (SSGS)**.
The SSGS assesses the experience of shame, guilt, and pride experienced during an acute stress protocol with three
separate subscales. Shame and guilt are considered distinct emotions, with shame being a global negative feeling
about the self, and guilt being a negative feeling about a specific event rather than the self.
This scale is a modified version from the State Shame and Guilt scale by Marschall et al. (1994).
Higher scores on each subscale indicate higher shame, guilt, or pride.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``Pride``: [1, 4, 7, 10, 13]
* ``Shame``: [2, 5, 8, 11, 14]
* ``Guilt``: [3, 6, 9, 12, 15]
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
SSGS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., & <NAME>. (2008). The psychobiology of trait shame in young women:
Extending the social self preservation theory. *Health Psychology*, 27(5), 523.
<NAME>., <NAME>., & <NAME>. (1994). The state shame and guilt scale.
*Fairfax, VA: George Mason University*.
"""
score_name = "SSGS"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 15)
subscales = {
"Pride": [1, 4, 7, 10, 13],
"Shame": [2, 5, 8, 11, 14],
"Guilt": [3, 6, 9, 12, 15],
}
_assert_value_range(data, score_range)
ssgs_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(ssgs_data, index=data.index)
def panas(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
language: Optional[Literal["english", "german"]] = None,
) -> pd.DataFrame:
"""Compute the **Positive and Negative Affect Schedule (PANAS)**.
The PANAS assesses *positive affect* (interested, excited, strong, enthusiastic, proud, alert, inspired,
determined, attentive, and active) and *negative affect* (distressed, upset, guilty, scared, hostile, irritable,
ashamed, nervous, jittery, and afraid).
Higher scores on each subscale indicate greater positive or negative affect.
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
language : "english" or "german", optional
Language of the questionnaire used since index items differ between the german and the english version.
Default: ``english``
Returns
-------
:class:`~pandas.DataFrame`
PANAS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., & <NAME>. (1988). Development and validation of brief measures of positive and
negative affect: the PANAS scales. *Journal of personality and social psychology*, 54(6), 1063.
"""
score_name = "PANAS"
score_range = [1, 5]
supported_versions = ["english", "german"]
# create copy of data
data = data.copy()
if language is None:
language = "english"
if language not in supported_versions:
raise ValueError("questionnaire_version must be one of {}, not {}.".format(supported_versions, language))
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 20)
_assert_value_range(data, score_range)
if language == "german":
# German Version has other item indices
subscales = {
"NegativeAffect": [2, 5, 7, 8, 9, 12, 14, 16, 19, 20],
"PositiveAffect": [1, 3, 4, 6, 10, 11, 13, 15, 17, 18],
}
else:
subscales = {
"NegativeAffect": [2, 4, 6, 7, 8, 11, 13, 15, 18, 20],
"PositiveAffect": [1, 3, 5, 9, 10, 12, 14, 16, 17, 19],
}
# PANAS is a mean, not a sum score!
panas_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
data = _invert_subscales(
data, subscales, {"NegativeAffect": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]}, score_range=score_range
)
panas_data[score_name + "_Total"] = data.mean(axis=1)
return pd.DataFrame(panas_data, index=data.index)
def state_rumination(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **State Rumination** scale.
Rumination is the tendency to dwell on negative thoughts and emotions.
Higher scores indicate greater rumination.
.. note::
This implementation assumes a score range of [0, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
State Rumination score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (1998). The relationship between emotional rumination and cortisol secretion
under stress. *Personality and Individual Differences*, 24(4), 531-538.
"""
score_name = "StateRumination"
score_range = [0, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 27)
_assert_value_range(data, score_range)
# reverse scores 1, 6, 9, 12, 15, 17, 18, 20, 27
data = invert(data, cols=to_idx([1, 6, 9, 12, 15, 17, 18, 20, 27]), score_range=score_range)
return pd.DataFrame(data.sum(axis=1), columns=[score_name], index=data.index)
# HABIT DATASET
def abi(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
"""Compute the **Angstbewältigungsinventar (ABI)** (Anxiety Management Inventory).
The ABI measures two key personality constructs in the area of stress or anxiety management:
*Vigilance (VIG)* and *Cognitive Avoidance (KOV)*. *VIG* is defined as a class of coping strategies whose
use aims to, to reduce uncertainty in threatening situations.
In contrast, *KOV* refers to strategies aimed at shielding the organism from arousal-inducing stimuli.
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
Returns
-------
:class:`~pandas.DataFrame`
ABI score
Raises
------
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., Angstbewältigung, <NAME>., & VIG, V. (1999).
Das Angstbewältigungs-Inventar (ABI). *Frankfurt am Main*.
"""
score_name = "ABI"
score_range = [1, 2]
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
_assert_num_columns(data, 80)
_assert_value_range(data, score_range)
# split into 8 subitems, consisting of 10 questions each
items = np.split(data, 8, axis=1)
abi_raw = pd.concat(items, keys=[str(i) for i in range(1, len(items) + 1)], axis=1)
idx_kov = {
# ABI-P
"2": [2, 3, 7, 8, 9],
"4": [1, 4, 5, 8, 10],
"6": [2, 3, 5, 6, 7],
"8": [2, 4, 6, 8, 10],
# ABI-E
"1": [2, 3, 6, 8, 10],
"3": [2, 4, 5, 7, 9],
"5": [3, 4, 5, 9, 10],
"7": [1, 5, 6, 7, 9],
}
idx_kov = {key: np.array(val) for key, val in idx_kov.items()}
idx_vig = {key: np.setdiff1d(np.arange(1, 11), np.array(val), assume_unique=True) for key, val in idx_kov.items()}
abi_kov, abi_vig = [
pd.concat(
[abi_raw.loc[:, key].iloc[:, idx[key] - 1] for key in idx],
axis=1,
keys=idx_kov.keys(),
)
for idx in [idx_kov, idx_vig]
]
abi_data = {
score_name + "_KOV_T": abi_kov.sum(axis=1),
score_name + "_VIG_T": abi_vig.sum(axis=1),
score_name + "_KOV_P": abi_kov.loc[:, ["2", "4", "6", "8"]].sum(axis=1),
score_name + "_VIG_P": abi_vig.loc[:, ["2", "4", "6", "8"]].sum(axis=1),
score_name + "_KOV_E": abi_kov.loc[:, ["1", "3", "5", "7"]].sum(axis=1),
score_name + "_VIG_E": abi_vig.loc[:, ["1", "3", "5", "7"]].sum(axis=1),
}
return pd.DataFrame(abi_data, index=data.index)
def stadi(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
stadi_type: Optional[Literal["state", "trait", "state_trait"]] = None,
) -> pd.DataFrame:
"""Compute the **State-Trait Anxiety-Depression Inventory (STADI)**.
With the STADI, anxiety and depression can be recorded, both as state and as trait.
Two self-report questionnaires with 20 items each are available for this purpose.
The state part measures the degree of anxiety and depression currently experienced by a person, which varies
depending on internal or external influences. It can be used in a variety of situations of different types.
This includes not only the whole spectrum of highly heterogeneous stressful situations, but also situations of
neutral or positive ("euthymic") character. The trait part is used to record trait expressions, i.e. the
enduring tendency to experience anxiety and depression.
The STADI can either be computed only for state, only for trait, or for state and trait.
The state and trait scales both consist of the subscales with the item indices
(count-by-one, i.e., the first question has the index 1!):
* Emotionality (Aufgeregtheit - affektive Komponente – ``AU``): [1, 5, 9, 13, 17]
* Worry (Besorgnis - kognitive Komponente - ``BE``): [2, 6, 10, 14, 18]
* Anhedonia (Euthymie - positive Stimmung - ``EU``): [3, 7, 11, 15, 19]
* Dysthymia (Dysthymie - depressive Stimmung - ``DY``): [4, 8, 12, 16, 20]
.. note::
This implementation assumes a score range of [1, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. note::
If both state and trait score are present it is assumed that all *state* items are first,
followed by all *trait* items. If all subscales are present this adds up to 20 state items and 20 trait items.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
stadi_type : any of ``state``, ``trait``, or ``state_trait``
which type of STADI subscale should be computed. Default: ``state_trait``
Returns
-------
:class:`~pandas.DataFrame`
STADI score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
if invalid parameter was passed to ``stadi_type``
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., <NAME>., & <NAME>. (2013).
Das State-Trait-Angst-Depressions-Inventar: STADI; Manual.
<NAME>., <NAME>., <NAME>., & <NAME>. (2018). Differentiating anxiety and depression:
the state-trait anxiety-depression inventory. *Cognition and Emotion*, 32(7), 1409-1423.
"""
score_name = "STADI"
score_range = [1, 4]
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
stadi_type = _get_stadi_type(stadi_type)
if subscales is None:
_assert_num_columns(data, 20 * len(stadi_type))
subscales = {
"AU": [1, 5, 9, 13, 17],
"BE": [2, 6, 10, 14, 18],
"EU": [3, 7, 11, 15, 19],
"DY": [4, 8, 12, 16, 20],
}
_assert_value_range(data, score_range)
# split into n subitems (either "State", "Trait" or "State and Trait")
items = np.split(data, len(stadi_type), axis=1)
data = pd.concat(items, keys=stadi_type, axis=1)
stadi_data = {}
for st in stadi_type:
stadi_data.update(_compute_questionnaire_subscales(data[st], "{}_{}".format(score_name, st), subscales))
if all("{}_{}_{}".format(score_name, st, subtype) in stadi_data for subtype in ["AU", "BE"]):
stadi_data.update(
{
"{}_{}_Anxiety".format(score_name, st): stadi_data["{}_{}_AU".format(score_name, st)]
+ stadi_data["{}_{}_BE".format(score_name, st)]
}
)
if all("{}_{}_{}".format(score_name, st, subtype) in stadi_data for subtype in ["EU", "DY"]):
stadi_data.update(
{
"{}_{}_Depression".format(score_name, st): stadi_data["{}_{}_EU".format(score_name, st)]
+ stadi_data["{}_{}_DY".format(score_name, st)]
}
)
if all("{}_{}_{}".format(score_name, st, subtype) in stadi_data for subtype in ["Anxiety", "Depression"]):
stadi_data.update(
{
"{}_{}_Total".format(score_name, st): stadi_data["{}_{}_Anxiety".format(score_name, st)]
+ stadi_data["{}_{}_Depression".format(score_name, st)]
}
)
df_stadi = pd.DataFrame(stadi_data, index=data.index)
return df_stadi
def _get_stadi_type(stadi_type: str) -> Sequence[str]:
if stadi_type is None:
stadi_type = ["State", "Trait"]
elif stadi_type == "state_trait":
stadi_type = ["State", "Trait"]
elif stadi_type == "state":
stadi_type = ["State"]
elif stadi_type == "trait":
stadi_type = ["Trait"]
else:
raise ValueError(
"Invalid 'stadi_type'! Must be one of 'state_trait', 'state', or 'trait', not {}.".format(stadi_type)
)
return stadi_type
def svf_120(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Stressverarbeitungsfragebogen - 120 item version (SVF120)**.
The stress processing questionnaire enables the assessment of coping or processing measures in stressful
situations. The SVF is not a singular test instrument, but rather an inventory of methods that relate to various
aspects of stress processing and coping and from which individual procedures can be selected depending on
the study objective/question.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* Trivialization/Minimalization (Bagatellisierung – ``Bag``): [10, 31, 50, 67, 88, 106]
* De-Emphasis by Comparison with Others (Herunterspielen – ``Her``): [17, 38, 52, 77, 97, 113]
* Rejection of Guilt (Schuldabwehr – ``Schab``): [5, 30, 43, 65, 104, 119]
* Distraction/Deflection from a Situation (Ablenkung – ``Abl``): [1, 20, 45, 86, 101, 111]
* Vicarious Satisfaction (Ersatzbefriedigung –``Ers``): [22, 36, 64, 74, 80, 103]
* Search for Self-Affirmation (Selbstbestätigung – ``Sebest``): [34, 47, 59, 78, 95, 115]
* Relaxation (Entspannung –``Entsp``): [12, 28, 58, 81, 99, 114]
* Attempt to Control Situation (Situationskontrolle – ``Sitkon``): [11, 18, 39, 66, 91, 116]
* Response Control (Reaktionskontrolle – ``Rekon``): [2, 26, 54, 68, 85, 109]
* Positive Self-Instruction (Positive Selbstinstruktion – ``Posi``): [15, 37, 56, 71, 83, 96]
* Need for Social Support (Soziales Unterstützungsbedürfnis – ``Sozube``): [3, 21, 42, 63, 84, 102]
* Avoidance Tendencies (Vermeidung – ``Verm``): [8, 29, 48, 69, 98, 118]
* Escapist Tendencies (Flucht – ``Flu``): [14, 24, 40, 62, 73, 120]
* Social Isolation (Soziale Abkapselung – ``Soza``): [6, 27, 49, 76, 92, 107]
* Mental Perseveration (Gedankliche Weiterbeschäftigung – ``Gedw``): [16, 23, 55, 72, 100, 110]
* Resignation (Resignation – ``Res``): [4, 32, 46, 60, 89, 105]
* Self-Pity (Selbstbemitleidung – ``Selmit``): [13, 41, 51, 79, 94, 117]
* Self-Incrimination (Selbstbeschuldigung – ``Sesch``): [9, 25, 35, 57, 75, 87]
* Aggression (Aggression – ``Agg``): [33, 44, 61, 82, 93, 112]
* Medicine-Taking (Pharmakaeinnahme – ``Pha``): [7, 19, 53, 70, 90, 108]
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
SFV120 score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
"""
score_name = "SVF120"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 120)
subscales = {
"Bag": [10, 31, 50, 67, 88, 106], # Bagatellisierung
"Her": [17, 38, 52, 77, 97, 113], # Herunterspielen
"Schab": [5, 30, 43, 65, 104, 119], # Schuldabwehr
"Abl": [1, 20, 45, 86, 101, 111], # Ablenkung
"Ers": [22, 36, 64, 74, 80, 103], # Ersatzbefriedigung
"Sebest": [34, 47, 59, 78, 95, 115], # Selbstbestätigung
"Entsp": [12, 28, 58, 81, 99, 114], # Entspannung
"Sitkon": [11, 18, 39, 66, 91, 116], # Situationskontrolle
"Rekon": [2, 26, 54, 68, 85, 109], # Reaktionskontrolle
"Posi": [15, 37, 56, 71, 83, 96], # Positive Selbstinstruktion
"Sozube": [3, 21, 42, 63, 84, 102], # Soziales Unterstützungsbedürfnis
"Verm": [8, 29, 48, 69, 98, 118], # Vermeidung
"Flu": [14, 24, 40, 62, 73, 120], # Flucht
"Soza": [6, 27, 49, 76, 92, 107], # Soziale Abkapselung
"Gedw": [16, 23, 55, 72, 100, 110], # Gedankliche Weiterbeschäftigung
"Res": [4, 32, 46, 60, 89, 105], # Resignation
"Selmit": [13, 41, 51, 79, 94, 117], # Selbstbemitleidung
"Sesch": [9, 25, 35, 57, 75, 87], # Selbstbeschuldigung
"Agg": [33, 44, 61, 82, 93, 112], # Aggression
"Pha": [7, 19, 53, 70, 90, 108], # Pharmakaeinnahme
}
_assert_value_range(data, score_range)
svf_data = _compute_questionnaire_subscales(data, score_name, subscales)
svf_data = pd.DataFrame(svf_data, index=data.index)
meta_scales = {
"Pos1": ("Bag", "Her", "Schab"),
"Pos2": ("Abl", "Ers", "Sebest", "Entsp"),
"Pos3": ("Sitkon", "Rekon", "Posi"),
"PosGesamt": (
"Bag",
"Her",
"Schab",
"Abl",
"Ers",
"Sebest",
"Entsp",
"Sitkon",
"Rekon",
"Posi",
),
"NegGesamt": ("Flu", "Soza", "Gedw", "Res", "Selmit", "Sesch"),
}
for name, scale_items in meta_scales.items():
if all(scale in subscales for scale in scale_items):
svf_data["{}_{}".format(score_name, name)] = svf_data[
["{}_{}".format(score_name, s) for s in scale_items]
].mean(axis=1)
return svf_data
def svf_42(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Stressverarbeitungsfragebogen - 42 item version (SVF42)**.
The stress processing questionnaire enables the assessment of coping or processing measures in stressful
situations. The SVF is not a singular test instrument, but rather an inventory of methods that relate to various
aspects of stress processing and coping and from which individual procedures can be selected depending on
the study objective/question.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* Trivialization/Minimalization (Bagatellisierung – ``Bag``): [7, 22]
* De-Emphasis by Comparison with Others (Herunterspielen – ``Her``): [11, 35]
* Rejection of Guilt (Schuldabwehr – ``Schab``): [2, 34]
* Distraction/Deflection from a Situation (Ablenkung – ``Abl``): [1, 32]
* Vicarious Satisfaction (Ersatzbefriedigung –``Ers``): [12, 42]
* Search for Self-Affirmation (Selbstbestätigung – ``Sebest``): [19, 37]
* Relaxation (Entspannung –``Entsp``): [13, 26]
* Attempt to Control Situation (Situationskontrolle – ``Sitkon``): [4, 23]
* Response Control (Reaktionskontrolle – ``Rekon``): [17, 33]
* Positive Self-Instruction (Positive Selbstinstruktion – ``Posi``): [9, 24]
* Need for Social Support (Soziales Unterstützungsbedürfnis – ``Sozube``): [14, 27]
* Avoidance Tendencies (Vermeidung – ``Verm``): [6, 30]
* Escapist Tendencies (Flucht – ``Flu``): [16, 40]
* Social Isolation (Soziale Abkapselung – ``Soza``): [20, 29]
* Mental Perseveration (Gedankliche Weiterbeschäftigung – ``Gedw``): [10, 25]
* Resignation (Resignation – ``Res``): [38, 15]
* Self-Pity (Selbstbemitleidung – ``Selmit``): [18, 28]
* Self-Incrimination (Selbstbeschuldigung – ``Sesch``): [8, 31]
* Aggression (Aggression – ``Agg``): [21, 36]
* Medicine-Taking (Pharmakaeinnahme – ``Pha``): [3, 39]
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
SFV42 score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
"""
score_name = "SVF42"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 42)
subscales = {
"Bag": [7, 22], # Bagatellisierung
"Her": [11, 35], # Herunterspielen
"Schab": [2, 34], # Schuldabwehr
"Abl": [1, 32], # Ablenkung
"Ers": [12, 42], # Ersatzbefriedigung
"Sebest": [19, 37], # Selbstbestätigung
"Entsp": [13, 26], # Entspannung
"Sitkon": [4, 23], # Situationskontrolle
"Rekon": [17, 33], # Reaktionskontrolle
"Posi": [9, 24], # Positive Selbstinstruktion
"Sozube": [14, 27], # Soziales Unterstützungsbedürfnis
"Verm": [6, 30], # Vermeidung
"Flu": [16, 40], # Flucht
"Soza": [20, 29], # Soziale Abkapselung
"Gedw": [10, 25], # Gedankliche Weiterbeschäftigung
"Res": [15, 38], # Resignation
"Hilf": [18, 28], # Hilflosigkeit
"Selmit": [8, 31], # Selbstbemitleidung
"Sesch": [21, 36], # Selbstbeschuldigung
"Agg": [3, 39], # Aggression
"Pha": [5, 41], # Pharmakaeinnahme
}
_assert_value_range(data, score_range)
svf_data = _compute_questionnaire_subscales(data, score_name, subscales)
svf_data = pd.DataFrame(svf_data, index=data.index)
meta_scales = {
"Denial": ["Verm", "Flu", "Soza"],
"Distraction": ["Ers", "Entsp", "Sozube"],
"Stressordevaluation": ["Bag", "Her", "Posi"],
}
for name, scale_items in meta_scales.items():
if all(scale in subscales.keys() for scale in scale_items):
svf_data["{}_{}".format(score_name, name)] = svf_data[
["{}_{}".format(score_name, s) for s in scale_items]
].mean(axis=1)
return svf_data
def brief_cope(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Brief-COPE (28 items) Questionnaire (Brief_COPE)**.
The Brief-COPE is a 28 item self-report questionnaire designed to measure effective and ineffective ways to cope
with a stressful life event. "Coping" is defined broadly as an effort used to minimize distress associated with
negative life experiences. The scale is often used in health-care settings to ascertain how patients are
responding to a serious diagnosis. It can be used to measure how someone is coping with a wide range of
adversity, including cancer diagnosis, heart failure, injuries, assaults, natural disasters and financial stress.
The scale can determine someone’s primary coping styles as either Approach Coping, or Avoidant Coping.
Higher scores indicate better coping capabilities.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``SelfDistraction``: [1, 19]
* ``ActiveCoping``: [2, 7]
* ``Denial``: [3, 8]
* ``SubstanceUse``: [4, 11]
* ``EmotionalSupport``: [5, 15]
* ``InstrumentalSupport``: [10, 23]
* ``BehavioralDisengagement``: [6, 16]
* ``Venting``: [9, 21]
* ``PosReframing``: [12, 17]
* ``Planning``: [14, 25]
* ``Humor``: [18, 28]
* ``Acceptance``: [20, 24]
* ``Religion``: [22, 27]
* ``SelfBlame``: [13, 26]
.. note::
This implementation assumes a score range of [1, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
Brief_COPE score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (1997). You want to measure coping but your protocol’too long: Consider the brief cope.
*International journal of behavioral medicine*, 4(1), 92-100.
"""
score_name = "Brief_COPE"
score_range = [1, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 28)
subscales = {
"SelfDistraction": [1, 19], # Ablenkung
"ActiveCoping": [2, 7], # Aktive Bewältigung
"Denial": [3, 8], # Verleugnung
"SubstanceUse": [4, 11], # Alkohol/Drogen
"EmotionalSupport": [5, 15], # Emotionale Unterstützung
"InstrumentalSupport": [10, 23], # Instrumentelle Unterstützung
"BehavioralDisengagement": [6, 16], # Verhaltensrückzug
"Venting": [9, 21], # Ausleben von Emotionen
"PosReframing": [12, 17], # Positive Umdeutung
"Planning": [14, 25], # Planung
"Humor": [18, 28], # Humor
"Acceptance": [20, 24], # Akzeptanz
"Religion": [22, 27], # Religion
"SelfBlame": [13, 26], # Selbstbeschuldigung
}
_assert_value_range(data, score_range)
cope_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(cope_data, index=data.index)
def bfi_k(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Big Five Inventory (short version) (BFI-K)**.
The BFI measures an individual on the Big Five Factors (dimensions) of personality (Goldberg, 1993).
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* Extraversion (``E``): [1, 6, 11, 16]
* Agreeableness (``A``): [2, 7, 12, 17]
* Conscientiousness (``C``): [3, 8, 13, 18]
* Neuroticism (``N``): [4, 9, 14, 19]
* Openness (``O``): [5, 10, 15, 20, 21]
.. note::
This implementation assumes a score range of [1, 5].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
BFI_K score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (2005). Kurzversion des big five inventory (BFI-K). *Diagnostica*, 51(4), 195-206.
"""
score_name = "BFI_K"
score_range = [1, 5]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 21)
subscales = {
"E": [1, 6, 11, 16], # Extraversion (Extraversion vs. Introversion)
"A": [2, 7, 12, 17], # Verträglichkeit (Agreeableness vs. Antagonism)
"C": [3, 8, 13, 18], # Gewissenhaftigkeit (Conscientiousness vs. lack of direction)
"N": [4, 9, 14, 19], # Neurotizismus (Neuroticism vs. Emotional stability)
"O": [5, 10, 15, 20, 21], # Offenheit für neue Erfahrungen (Openness vs. Closedness to experience)
}
_assert_value_range(data, score_range)
# Reverse scores 1, 2, 8, 9, 11, 12, 17, 21
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data,
subscales=subscales,
idx_dict={"E": [0, 2], "A": [0, 2, 3], "C": [1], "N": [1], "O": [4]},
score_range=score_range,
)
# BFI is a mean score, not a sum score!
bfi_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
return pd.DataFrame(bfi_data, index=data.index)
def rsq(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Response Styles Questionnaire (RSQ)**.
The RSQ is a questionnaire that measures cognitive and behavioral coping styles in dealing with depressed or
dysphoric mood and was developed based on <NAME>'s Response Styles Theory.
The theory postulates that rumination about symptoms and negative aspects of self (rumination) prolongs or
exacerbates depressed moods, whereas cognitive and behavioral distraction (distraction) shortens or
attenuates them.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``SymptomRumination``: [2, 3, 4, 8, 11, 12, 13, 25]
* ``SelfRumination``: [1, 19, 26, 28, 30, 31, 32]
* ``Distraction``: [5, 6, 7, 9, 14, 16, 18, 20]
.. note::
This implementation assumes a score range of [1, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
RSQ score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., & <NAME>. (1993). Response styles and the duration of episodes of
depressed mood. *Journal of abnormal psychology*, 102(1), 20.
"""
score_name = "RSQ"
score_range = [1, 4]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 32)
subscales = {
"SymptomRumination": [2, 3, 4, 8, 11, 12, 13, 25], # Symptombezogene Rumination
"SelfRumination": [1, 10, 19, 26, 28, 30, 31, 32], # Selbstfokussierte Rumination
"Distraction": [5, 6, 7, 9, 14, 16, 18, 20], # Distraktion
}
_assert_value_range(data, score_range)
# RSQ is a mean score, not a sum score!
rsq_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
rsq_data = pd.DataFrame(rsq_data, index=data.index)
if all(s in subscales for s in ["SymptomRumination", "SelfRumination", "Distraction"]):
# compute total score if all subscales are present
# invert "Distraction" subscale and then add it to total score
rsq_data["{}_{}".format(score_name, "Total")] = pd.concat(
[
(score_range[1] - rsq_data["{}_{}".format(score_name, "Distraction")] + score_range[0]),
rsq_data["{}_{}".format(score_name, "SymptomRumination")],
rsq_data["{}_{}".format(score_name, "SelfRumination")],
],
axis=1,
).mean(axis=1)
return rsq_data
def sss(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[Union[str, int]]]] = None,
) -> pd.DataFrame:
"""Compute the **Subjective Social Status (SSS)**.
The MacArthur Scale of Subjective Social Status (MacArthur SSS Scale) is a single-item measure that assesses a
person's perceived rank relative to others in their group.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* Socioeconomic Status Ladder (``SocioeconomicStatus``): [1]
* Community Ladder (``Community``): [2]
.. note::
This implementation assumes a score range of [0, 10].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
SSS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
"""
score_name = "SSS"
score_range = [1, 10]
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 2)
subscales = {
"SocioeconomicStatus": [1],
"Community": [2],
}
_assert_value_range(data, score_range)
sss_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(sss_data, index=data.index)
def fkk(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Fragebogen zur Kompetenz- und Kontrollüberzeugungen (FKK)** (Competence and Control Beliefs).
The questionnaire on competence and control beliefs can be used to assess
(1) the generalized self-concept of own abilities,
(2) internality in generalized control beliefs,
(3) socially conditioned externality, and
(4) fatalistic externality in adolescents and adults.
In addition to profile evaluations according to these four primary scales, evaluations according to secondary and
tertiary scales are possible (generalized self-efficacy; generalized externality;
internality versus externality in control beliefs).
It consists of the primary subscales with the item indices (count-by-one, i.e.,
the first question has the index 1!):
* Self-concept of Own Abilities (Selbstkonzept eigener Fähigkeiten – ``SK``): [4, 8, 12, 24, 16, 20, 28, 32]
* Internality (Internalität – ``I``): [1, 5, 6, 11, 23, 25, 27, 30]
* Socially Induced Externality (Sozial bedingte Externalität – ``P``) (P = powerful others control orientation):
[3, 10, 14, 17, 19, 22, 26, 29]
* Fatalistic Externality (Fatalistische Externalität – ``C``) (C = chance control orientation):
[2, 7, 9, 13, 15, 18, 21, 31]
Further, the following secondary subscales can be computed:
* Self-Efficacy / Generalized self-efficacy Beliefs (Selbstwirksamkeit / generalisierte
Selbstwirksamkeitsüberzeugung – ``SKI``): ``SK`` + ``I``
* Generalized Externality in Control Beliefs (Generalisierte Externalität in Kontrollüberzeugungen – ``PC``):
``P`` + ``C``
Further, the following tertiary subscale can be computed:
* Generalized Internality (Generalisierte Internalität) vs. Externality in control beliefs
(Externalität in Kontrollüberzeugungen – ``SKI_PC``): ``SKI`` - ``PC``
.. note::
This implementation assumes a score range of [1, 6].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
FKK score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (1991). Fragebogen zu Kompetenz-und Kontrollüberzeugungen: (FKK). *Hogrefe, Verlag für Psychologie*.
"""
score_name = "FKK"
score_range = [1, 6]
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 32)
# Primärskalenwerte
subscales = {
"SK": [4, 8, 12, 16, 20, 24, 28, 32],
"I": [1, 5, 6, 11, 23, 25, 27, 30],
"P": [3, 10, 14, 17, 19, 22, 26, 29],
"C": [2, 7, 9, 13, 15, 18, 21, 31],
}
_assert_value_range(data, score_range)
# Reverse scores 4, 8, 12, 24
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(data, subscales=subscales, idx_dict={"SK": [0, 1, 2, 5]}, score_range=score_range)
fkk_data = _compute_questionnaire_subscales(data, score_name, subscales)
fkk_data = pd.DataFrame(fkk_data, index=data.index)
# Sekundärskalenwerte
if all("{}_{}".format(score_name, s) in fkk_data.columns for s in ["SK", "I"]):
fkk_data["{}_{}".format(score_name, "SKI")] = (
fkk_data["{}_{}".format(score_name, "SK")] + fkk_data["{}_{}".format(score_name, "I")]
)
if all("{}_{}".format(score_name, s) in fkk_data.columns for s in ["P", "C"]):
fkk_data["{}_{}".format(score_name, "PC")] = (
fkk_data["{}_{}".format(score_name, "P")] + fkk_data["{}_{}".format(score_name, "C")]
)
# Tertiärskalenwerte
if all("{}_{}".format(score_name, s) in fkk_data.columns for s in ["SKI", "PC"]):
fkk_data["{}_{}".format(score_name, "SKI_PC")] = (
fkk_data["{}_{}".format(score_name, "SKI")] - fkk_data["{}_{}".format(score_name, "PC")]
)
return fkk_data
def bidr(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Balanced Inventory of Desirable Responding (BIDR)**.
The BIDR is a 40-item instrument that is used to measure 2 constructs:
* Self-deceptive positivity – described as the tendency to give self-reports that are believed but have a
positivety bias
* Impression management – deliberate self-presentation to an audience.
The BIDR emphasizes exaggerated claims of positive cognitive attributes (overconfidence in one’s judgments and
rationality). It is viewed as a measure of defense, i.e., people who score high on self-deceptive positivity
tend to defend against negative self-evaluations and seek out inflated positive self-evaluations.
.. note::
This implementation assumes a score range of [1, 7].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
BIDR score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>. (1988). Balanced inventory of desirable responding (BIDR).
*Acceptance and Commitment Therapy. Measures Package*, 41, 79586-7.
"""
score_name = "BIDR"
score_range = [1, 7]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 20)
subscales = {
"ST": list(range(1, 11)), # Selbsttäuschung
"FT": list(range(11, 21)), # Fremdtäuschung
}
_assert_value_range(data, score_range)
# invert items 2, 4, 5, 7, 9, 10, 11, 12, 14, 15, 17, 18, 20
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(
data,
subscales=subscales,
idx_dict={"ST": [1, 3, 4, 6, 8, 9], "FT": [0, 1, 3, 4, 6, 7, 9]},
score_range=score_range,
)
bidr_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(bidr_data, index=data.index)
def kkg(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Kontrollüberzeugungen zu Krankheit und Gesundheit Questionnaire (KKG)**.
The KKG is a health attitude test and assesses the locus of control about disease and health.
3 health- or illness-related locus of control are evaluated:
(1) internality: attitudes that health and illness are controllable by oneself,
(2) social externality: attitudes that they are controllable by other outside persons, and
(3) fatalistic externality: attitudes that they are not controllable (chance or fate dependence of one's
health status).
.. note::
This implementation assumes a score range of [1, 6].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
KKG score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., & <NAME>. (1989). *Kontrollüberzeugungen zu Krankheit und Gesundheit (KKG): Testverfahren und
Testmanual*. Göttingen: Hogrefe.
"""
score_name = "KKG"
score_range = [1, 6]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 21)
subscales = {
"I": [1, 5, 8, 16, 17, 18, 21],
"P": [2, 4, 6, 10, 12, 14, 20],
"C": [3, 7, 9, 11, 13, 15, 19],
}
_assert_value_range(data, score_range)
kkg_data = _compute_questionnaire_subscales(data, score_name, subscales)
return pd.DataFrame(kkg_data, index=data.index)
def fee(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
language: Optional[Literal["german", "english"]] = None,
) -> pd.DataFrame:
"""Compute the **Fragebogen zum erinnerten elterlichen Erziehungsverhalten (FEE)**.
The FEE allows for the recording of memories of the parenting behavior (separately for father and mother) with
regard to the factor-analytically dimensions "rejection and punishment", "emotional warmth" and "control and
overprotection", as well as "control and overprotection".
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``RejectionPunishment``: [1, 3, 6, 8, 16, 18, 20, 22]
* ``EmotionalWarmth``: [2, 7, 9, 12, 14, 15, 17, 24]
* ``ControlOverprotection``: [4, 5, 10, 11, 13, 19, 21, 23]
.. note::
This implementation assumes a score range of [1, 4].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. note::
All columns corresponding to the parenting behavior of the *Father* are expected to have ``Father``
(or ``Vater`` if ``language`` is ``german``) included in the column names, all *Mother* columns
are expected to have ``Mother`` (or ``Mutter`` if ``language`` is ``german``) included in the column names.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
language : "english" or "german", optional
Language of the questionnaire used to extract ``Mother`` and ``Father`` columns. Default: ``english``
Returns
-------
:class:`~pandas.DataFrame`
TICS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints or
if ``language`` is not supported
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., & <NAME>. (1999). Rückblick auf die Eltern: Der Fragebogen zum erinnerten
elterlichen Erziehungsverhalten (FEE). *Diagnostica*, 45(4), 194-204.
"""
score_name = "FEE"
score_range = [1, 4]
if language is None:
language = "english"
supported_versions = ["english", "german"]
if language not in supported_versions:
raise ValueError("questionnaire_version must be one of {}, not {}.".format(supported_versions, language))
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if language == "german":
mother = "Mutter"
father = "Vater"
else:
mother = "Mother"
father = "Father"
df_mother = data.filter(like=mother).copy()
df_father = data.filter(like=father).copy()
if subscales is None:
_assert_num_columns(df_father, 24)
_assert_num_columns(df_mother, 24)
subscales = {
"RejectionPunishment": [1, 3, 6, 8, 16, 18, 20, 22],
"EmotionalWarmth": [2, 7, 9, 12, 14, 15, 17, 24],
"ControlOverprotection": [4, 5, 10, 11, 13, 19, 21, 23],
}
_assert_value_range(data, score_range)
# FEE is a mean score, not a sum score!
fee_mother = _compute_questionnaire_subscales(df_mother, score_name, subscales, agg_type="mean")
fee_mother = {"{}_{}".format(key, mother): val for key, val in fee_mother.items()}
fee_father = _compute_questionnaire_subscales(df_father, score_name, subscales, agg_type="mean")
fee_father = {"{}_{}".format(key, father): val for key, val in fee_father.items()}
fee_mother.update(fee_father)
return pd.DataFrame(fee_mother, index=data.index)
def mbi_gs(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Maslach Burnout Inventory – General Survey (MBI-GS)**.
The MBI measures burnout as defined by the World Health Organization (WHO) and in the ICD-11.
The MBI-GS is a psychological assessment instrument comprising 16 symptom items pertaining to occupational burnout.
It is designed for use with occupational groups other than human services and education, including those working
in jobs such as customer service, maintenance, manufacturing, management, and most other professions.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* Emotional Exhaustion (``EE``): [1, 2, 3, 4, 5]
* Personal Accomplishment (``PA``): [6, 7, 8, 11, 12, 16]
* Depersonalization / Cynicism (``DC``): [9, 10, 13, 14, 15]
.. note::
This implementation assumes a score range of [0, 6].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
MBI-GS score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
"""
score_name = "MBI_GS"
score_range = [0, 6]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 16)
subscales = {
"EE": [1, 2, 3, 4, 5], # Emotional Exhaustion
"PA": [6, 7, 8, 11, 12, 16], # Personal Accomplishment
"DC": [9, 10, 13, 14, 15], # Depersonalization / Cynicism
}
_assert_value_range(data, score_range)
# MBI is a mean score, not a sum score!
mbi_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
data = pd.DataFrame(mbi_data, index=data.index)
return data
def mbi_gss(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Maslach Burnout Inventory – General Survey for Students (MBI-GS (S))**.
The MBI measures burnout as defined by the World Health Organization (WHO) and in the ICD-11.
The MBI-GS (S) is an adaptation of the MBI-GS designed to assess burnout in college and university students.
It is available for use but its psychometric properties are not yet documented.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* Emotional Exhaustion (``EE``): [1, 2, 3, 4, 5]
* Personal Accomplishment (``PA``): [6, 7, 8, 11, 12, 16]
* Depersonalization / Cynicism (``DC``): [9, 10, 13, 14, 15]
.. note::
This implementation assumes a score range of [0, 6].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
MBI-GS (S) score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
"""
score_name = "MBI_GSS"
score_range = [0, 6]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 16)
subscales = {
"EE": [1, 2, 3, 4, 5],
"PA": [6, 7, 8, 11, 12, 16],
"DC": [9, 10, 13, 14, 15],
}
_assert_value_range(data, score_range)
# MBI is a mean score, not a sum score!
mbi_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
data = pd.DataFrame(mbi_data, index=data.index)
return data
def mlq(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Meaning in Life Questionnaire (MLQ)**.
The MLQ is a 10-item measure of the Presence of Meaning in Life, and the Search for Meaning in Life.
The MLQ has been used to help people understand and track their perceptions about their lives.
It has been included in numerous studies around the world, and in several internet-based resources concerning
happiness and fulfillment.
It consists of the subscales with the item indices (count-by-one, i.e., the first question has the index 1!):
* ``PresenceMeaning``: [1, 4, 5, 6, 9]
* ``SearchMeaning``: [2, 3, 7, 8, 10]
.. note::
This implementation assumes a score range of [1, 7].
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
MLQ score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., <NAME>., & <NAME>. (2006). The meaning in life questionnaire: Assessing the
presence of and search for meaning in life. *Journal of counseling psychology*, 53(1), 80.
"""
score_name = "MLQ"
score_range = [1, 7]
# create copy of data
data = data.copy()
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 10)
subscales = {
"PresenceMeaning": [1, 4, 5, 6, 9],
"SearchMeaning": [2, 3, 7, 8, 10],
}
_assert_value_range(data, score_range)
# Reverse scores 9
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(data, subscales=subscales, idx_dict={"PresenceMeaning": [4]}, score_range=score_range)
# MLQ is a mean score, not a sum score!
mlq_data = _compute_questionnaire_subscales(data, score_name, subscales, agg_type="mean")
return pd.DataFrame(mlq_data, index=data.index)
# def ceca(data: pd.DataFrame, columns: Optional[Union[Sequence[str], pd.Index]] = None) -> pd.DataFrame:
# """Compute the **Childhood Experiences of Care and Abuse Questionnaire (CECA)**.
#
# The CECA is a measure of childhood and adolescent experience of neglect and abuse. Its original use was to
# investigate lifetime risk factors for psychological disorder.
#
# .. note::
# This implementation assumes a score range of [0, 4].
# Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
# beforehand.
#
# .. warning::
# Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
# questionnaire item columns, which typically also start with index 1!
#
#
# Parameters
# ----------
# data : :class:`~pandas.DataFrame`
# dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
# a complete dataframe if ``columns`` parameter is supplied
# columns : list of str or :class:`pandas.Index`, optional
# list with column names in correct order.
# This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
# passed as ``data``.
#
#
# Returns
# -------
# :class:`~pandas.DataFrame`
# CECA score
#
#
# Raises
# ------
# `biopsykit.exceptions.ValidationError`
# if number of columns does not match
# `biopsykit.exceptions.ValueRangeError`
# if values are not within the required score range
#
# References
# ----------
# <NAME>., <NAME>., & <NAME>. (1994). Childhood Experience of Care and Abuse (CECA): a retrospective
# interview measure. *Journal of Child Psychology and Psychiatry*, 35(8), 1419-1435.
#
# """
# score_name = "CECA"
#
# # create copy of data
# data = data.copy()
#
# if columns is not None:
# # if columns parameter is supplied: slice columns from dataframe
# _assert_has_columns(data, [columns])
# data = data.loc[:, columns]
#
# ceca_data = [
# data.filter(like="Q3_05"),
# data.filter(like="Q3_07"),
# data.filter(like="Q3_09"),
# data.filter(like="Q3_12").iloc[:, to_idx([5, 6])],
# data.filter(like="Q3_13"),
# data.filter(like="Q3_16").iloc[:, to_idx([5, 6])],
# ]
#
# ceca_data = pd.concat(ceca_data, axis=1).sum(axis=1)
# return pd.DataFrame(ceca_data, index=data.index, columns=[score_name])
def pfb(
data: pd.DataFrame,
columns: Optional[Union[Sequence[str], pd.Index]] = None,
subscales: Optional[Dict[str, Sequence[int]]] = None,
) -> pd.DataFrame:
"""Compute the **Partnerschaftsfragebogen (PFB)**.
.. note::
This implementation assumes a score range of [1, 4], except for the ``Glueck`` column, which has a score range
of [1, 6]
Use :func:`~biopsykit.questionnaires.utils.convert_scale()` to convert the items into the correct range
beforehand.
.. warning::
Column indices in ``subscales`` are assumed to start at 1 (instead of 0) to avoid confusion with
questionnaire item columns, which typically also start with index 1!
Parameters
----------
data : :class:`~pandas.DataFrame`
dataframe containing questionnaire data. Can either be only the relevant columns for computing this score or
a complete dataframe if ``columns`` parameter is supplied.
columns : list of str or :class:`pandas.Index`, optional
list with column names in correct order.
This can be used if columns in the dataframe are not in the correct order or if a complete dataframe is
passed as ``data``.
subscales : dict, optional
A dictionary with subscale names (keys) and column names or column indices (count-by-1) (values)
if only specific subscales should be computed.
Returns
-------
:class:`~pandas.DataFrame`
PFB score
Raises
------
ValueError
if ``subscales`` is supplied and dict values are something else than a list of strings or a list of ints
:exc:`~biopsykit.utils.exceptions.ValidationError`
if number of columns does not match
:exc:`~biopsykit.utils.exceptions.ValueRangeError`
if values are not within the required score range
References
----------
<NAME>., <NAME>., & <NAME>. (2001). Der Partnerschaftsfragebogen (PFB).
*Diagnostica*, 47(3), 132–141.
"""
score_name = "PFB"
score_range = [1, 4]
if columns is not None:
# if columns parameter is supplied: slice columns from dataframe
_assert_has_columns(data, [columns])
data = data.loc[:, columns]
if subscales is None:
_assert_num_columns(data, 31)
subscales = {
"Zaertlichkeit": [2, 3, 5, 9, 13, 14, 16, 23, 27, 28],
"Streitverhalten": [1, 6, 8, 17, 18, 19, 21, 22, 24, 26],
"Gemeinsamkeit": [4, 7, 10, 11, 12, 15, 20, 25, 29, 30],
"Glueck": [31],
}
_pfb_assert_value_range(data, subscales, score_range)
# invert item 19
# (numbers in the dictionary correspond to the *positions* of the items to be reversed in the item list specified
# by the subscale dict)
data = _invert_subscales(data, subscales=subscales, idx_dict={"Streitverhalten": [5]}, score_range=score_range)
pfb_data = _compute_questionnaire_subscales(data, score_name, subscales)
if len(data.columns) == 31:
pfb_data[score_name] = data.iloc[:, 0:30].sum(axis=1)
return
|
pd.DataFrame(pfb_data, index=data.index)
|
pandas.DataFrame
|
from datetime import date, datetime
from pathlib import Path
import matplotlib.pylab as plt
import numpy as np
import pandas as pd
import seaborn as sns
def load_data(data_dir: Path) -> pd.DataFrame:
body_frames = [
pd.read_csv(
fname,
converters={
0: lambda x: datetime.strptime(x, "%Y-%m-%d").date().toordinal()
},
skiprows=1,
)
for fname in data_dir.glob("*.csv")
]
body_frame =
|
pd.concat(body_frames)
|
pandas.concat
|
from io import StringIO
import streamlit as st
from PIL import Image
import pickle
import base64
import pandas as pd
import numpy as np
# File download
def filedownload(df):
csv = df.to_csv()
b64 = base64.b64encode(csv.encode()).decode() # strings <-> bytes conversions
href = f'<a href="data:file/csv;base64,{b64}" download="prediction.csv">Download Predictions</a>'
return href
def taxa_extractor(input_seq,level=3):
input_df=pd.read_csv("Profile_sequences_5.csv")
input_taxa=pd.read_csv("Taxa_info.csv")
from scipy.spatial import distance
distance_holder=[]
for i in range(input_df.shape[0]):
distance_holder.append(distance.euclidean(input_seq, input_df.iloc[i,:].values[:-1]))
return(input_taxa.iloc[np.argmin(distance_holder),level])
def significance(input_value):
input_df=pd.read_csv("baseline_PKJK.csv")
return(int(len(np.where(input_value > input_df.Value)[0])/len(input_df.Value)*100))
def processor(file_input, label="labels"):
labels=[]
sequences=[]
for i in file_input:
if ">" in i:
labels.append(i.replace(">", "").replace('\n', ''))
else:
sequences.append(i.replace('\n', ''))
if label=="labels":
return labels
elif label=="sequence":
return sequences
# Model building
def build_model(file_input, rank):
# Reads in saved regression model
unique_sequences=pd.read_csv("unique_sequences_5.csv")
unique_sequences=unique_sequences.iloc[:,1].values
input_sequence_list=processor(file_input,label="sequence")
label_list=processor(file_input, label="labels")
prediction_output=[]
significance_output=[]
taxa_output=[]
for j in input_sequence_list:
unique_sequences_freq=[]
unique_sequences_freq.append([j.count(i) for i in unique_sequences])
pickle_file=open('finalized_model_5_PKJK.pkl','rb')
model=pickle.load(pickle_file)
prediction = model.predict(
|
pd.DataFrame(unique_sequences_freq)
|
pandas.DataFrame
|
import pandas as pd
import config
import dataset
import sys
def join_csv_files(flist, ofname):
pd.concat([pd.read_csv(f) for f in flist])\
.to_csv(ofname, index=None, compression='gzip')
def create_app_flags_file():
dlist = []
for k, v in config.source_files.items():
d = pd.read_csv(v, index_col='appId')
if k == 'offstore':
d['relevant'] = 'y'
elif (('relevant' not in d.columns) or (d.relevant.count() < len(d) * 0.5)) \
and ('ml_score' in d.columns):
## TODO: Remove this or set 0.5 to 0.2 or something
print("----> Relevant column is missing or unpopulated... recreating", k, v)
d['relevant'] = ((d['ml_score'] > 0.4) | (d.get('relevant',
|
pd.Series([])
|
pandas.Series
|
from __future__ import division, print_function, absolute_import
import os
import traceback
import scipy.misc as misc
import matplotlib.pyplot as plt
import numpy as np
import glob
import pandas as pd
import random
from PIL import Image, ImageOps
def get_data_A1A4(data_path, split_load):
# Getting images (x data)
imgname_train_A1 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A1'+str(h)+'/*.png') for h in split_load[0]])
imgname_train_A4 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A4'+str(h)+'/*.png') for h in split_load[0]])
imgname_val_A1 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A1'+str(split_load[1])+'/*.png')])
imgname_val_A4 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A4'+str(split_load[1])+'/*.png')])
imgname_test_A1 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A1'+str(split_load[2])+'/*.png')])
imgname_test_A4 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A4'+str(split_load[2])+'/*.png')])
filelist_train_A1 = list(np.sort(imgname_train_A1.flat)[1::2])
filelist_train_A4 = list(np.sort(imgname_train_A4.flat)[1::2])
filelist_train_A1_fg = list(np.sort(imgname_train_A1.flat)[0::2])
filelist_train_A4_fg = list(np.sort(imgname_train_A4.flat)[0::2])
filelist_train_A1_img = np.array([np.array(filelist_train_A1[h][-16:]) for h in range(0,len(filelist_train_A1))])
filelist_train_A4_img = np.array([np.array(filelist_train_A4[h][-17:]) for h in range(0,len(filelist_train_A4))])
filelist_train_A1_set = np.array([np.array(filelist_train_A1[h][-20:-18]) for h in range(0,len(filelist_train_A1))])
filelist_train_A4_set = np.array([np.array(filelist_train_A4[h][-20:-18]) for h in range(0,len(filelist_train_A4))])
filelist_val_A1 = list(np.sort(imgname_val_A1.flat)[1::2])
filelist_val_A4 = list(np.sort(imgname_val_A4.flat)[1::2])
filelist_val_A1_fg = list(np.sort(imgname_val_A1.flat)[0::2])
filelist_val_A4_fg = list(np.sort(imgname_val_A4.flat)[0::2])
filelist_val_A1_img = np.array([np.array(filelist_val_A1[h][-16:]) for h in range(0,len(filelist_val_A1))])
filelist_val_A4_img = np.array([np.array(filelist_val_A4[h][-17:]) for h in range(0,len(filelist_val_A4))])
filelist_val_A1_set = np.array([np.array(filelist_val_A1[h][-20:-18]) for h in range(0,len(filelist_val_A1))])
filelist_val_A4_set = np.array([np.array(filelist_val_A4[h][-20:-18]) for h in range(0,len(filelist_val_A4))])
filelist_test_A1 = list(np.sort(imgname_test_A1.flat)[1::2])
filelist_test_A4 = list(np.sort(imgname_test_A4.flat)[1::2])
filelist_test_A1_fg = list(np.sort(imgname_test_A1.flat)[0::2])
filelist_test_A4_fg = list(np.sort(imgname_test_A4.flat)[0::2])
filelist_test_A1_img = np.array([np.array(filelist_test_A1[h][-16:]) for h in range(0,len(filelist_test_A1))])
filelist_test_A4_img = np.array([np.array(filelist_test_A4[h][-17:]) for h in range(0,len(filelist_test_A4))])
filelist_test_A1_set = np.array([np.array(filelist_test_A1[h][-20:-18]) for h in range(0,len(filelist_test_A1))])
filelist_test_A4_set = np.array([np.array(filelist_test_A4[h][-20:-18]) for h in range(0,len(filelist_test_A4))])
x_train_A1 = np.array([np.array(Image.open(fname)) for fname in filelist_train_A1])
x_train_A1 = np.delete(x_train_A1,3,3)
x_train_A4 = np.array([np.array(Image.open(fname)) for fname in filelist_train_A4])
x_train_A1_fg = np.array([np.array(Image.open(fname)) for fname in filelist_train_A1_fg])
x_train_A4_fg = np.array([np.array(Image.open(fname)) for fname in filelist_train_A4_fg])
x_val_A1 = np.array([np.array(Image.open(fname)) for fname in filelist_val_A1])
x_val_A1 = np.delete(x_val_A1,3,3)
x_val_A4 = np.array([np.array(Image.open(fname)) for fname in filelist_val_A4])
x_val_A1_fg = np.array([np.array(Image.open(fname)) for fname in filelist_val_A1_fg])
x_val_A4_fg = np.array([np.array(Image.open(fname)) for fname in filelist_val_A4_fg])
x_test_A1 = np.array([np.array(Image.open(fname)) for fname in filelist_test_A1])
x_test_A1 = np.delete(x_test_A1,3,3)
x_test_A4 = np.array([np.array(Image.open(fname)) for fname in filelist_test_A4])
x_test_A1_fg = np.array([np.array(Image.open(fname)) for fname in filelist_test_A1_fg])
x_test_A4_fg = np.array([np.array(Image.open(fname)) for fname in filelist_test_A4_fg])
x_train_res_A1 = np.array([misc.imresize(x_train_A1[i],[317,309,3]) for i in range(0,len(x_train_A1))])
x_train_res_A4 = np.array([misc.imresize(x_train_A4[i],[317,309,3]) for i in range(0,len(x_train_A4))])
x_val_res_A1 = np.array([misc.imresize(x_val_A1[i],[317,309,3]) for i in range(0,len(x_val_A1))])
x_val_res_A4 = np.array([misc.imresize(x_val_A4[i],[317,309,3]) for i in range(0,len(x_val_A4))])
x_test_res_A1 = np.array([misc.imresize(x_test_A1[i],[317,309,3]) for i in range(0,len(x_test_A1))])
x_test_res_A4 = np.array([misc.imresize(x_test_A4[i],[317,309,3]) for i in range(0,len(x_test_A4))])
x_train_res_A1_fg = np.array([misc.imresize(x_train_A1_fg[i],[317,309,3]) for i in range(0,len(x_train_A1_fg))])
x_train_res_A4_fg = np.array([misc.imresize(x_train_A4_fg[i],[317,309,3]) for i in range(0,len(x_train_A4_fg))])
x_val_res_A1_fg = np.array([misc.imresize(x_val_A1_fg[i],[317,309,3]) for i in range(0,len(x_val_A1))])
x_val_res_A4_fg = np.array([misc.imresize(x_val_A4_fg[i],[317,309,3]) for i in range(0,len(x_val_A4))])
x_test_res_A1_fg = np.array([misc.imresize(x_test_A1_fg[i],[317,309,3]) for i in range(0,len(x_test_A1_fg))])
x_test_res_A4_fg = np.array([misc.imresize(x_test_A4_fg[i],[317,309,3]) for i in range(0,len(x_test_A4_fg))])
x_train_all = np.concatenate((x_train_res_A1, x_train_res_A4), axis=0)
x_val_all = np.concatenate((x_val_res_A1, x_val_res_A4), axis=0)
x_test_all = np.concatenate((x_test_res_A1, x_test_res_A4), axis=0)
for h in range(0,len(x_train_all)):
x_img = x_train_all[h]
x_img_pil = Image.fromarray(x_img)
x_img_pil = ImageOps.autocontrast(x_img_pil)
x_img_ar = np.array(x_img_pil)
x_train_all[h] = x_img_ar
for h in range(0,len(x_val_all)):
x_img = x_val_all[h]
x_img_pil = Image.fromarray(x_img)
x_img_pil = ImageOps.autocontrast(x_img_pil)
x_img_ar = np.array(x_img_pil)
x_val_all[h] = x_img_ar
for h in range(0,len(x_test_all)):
x_img = x_test_all[h]
x_img_pil = Image.fromarray(x_img)
x_img_pil = ImageOps.autocontrast(x_img_pil)
x_img_ar = np.array(x_img_pil)
x_test_all[h] = x_img_ar
x_train_all_fg = np.concatenate((x_train_res_A1_fg, x_train_res_A4_fg), axis=0)
x_val_all_fg = np.concatenate((x_val_res_A1_fg, x_val_res_A4_fg), axis=0)
x_test_all_fg = np.concatenate((x_test_res_A1_fg, x_test_res_A4_fg), axis=0)
sum_train_all = np.zeros((len(x_train_all_fg),1))
sum_val_all = np.zeros((len(x_val_all_fg),1))
sum_test_all = np.zeros((len(x_test_all_fg),1))
for i in range(0, len(x_train_all_fg)):
x_train_all_fg[i][x_train_all_fg[i] > 0] = 1
sum_train_all[i] = np.sum(x_train_all_fg[i])
for i in range(0, len(x_val_all_fg)):
x_val_all_fg[i][x_val_all_fg[i] > 0] = 1
sum_val_all[i] = np.sum(x_val_all_fg[i])
for i in range(0, len(x_test_all_fg)):
x_test_all_fg[i][x_test_all_fg[i] > 0] = 1
sum_test_all[i] = np.sum(x_test_all_fg[i])
x_train_img = np.concatenate((filelist_train_A1_img, filelist_train_A4_img), axis=0)
x_val_img = np.concatenate((filelist_val_A1_img, filelist_val_A4_img), axis=0)
x_test_img = np.concatenate((filelist_test_A1_img, filelist_test_A4_img), axis=0)
x_train_set = np.concatenate((filelist_train_A1_set, filelist_train_A4_set), axis=0)
x_val_set = np.concatenate((filelist_val_A1_set, filelist_val_A4_set), axis=0)
x_test_set = np.concatenate((filelist_test_A1_set, filelist_test_A4_set), axis=0)
# Getting targets (y data) #
counts_A1 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A1.xlsx')])
counts_A4 = np.array([glob.glob(data_path+'/CVPPP2017_LCC_training/TrainingSplits/A4.xlsx')])
counts_train_flat_A1 = list(counts_A1.flat)
train_labels_A1 = pd.DataFrame()
y_train_A1_list = []
y_val_A1_list = []
y_test_A1_list = []
for f in counts_train_flat_A1:
frame =
|
pd.read_excel(f, header=None)
|
pandas.read_excel
|
# -*- coding: utf-8 -*-
""" This module is designed for the use with the coastdat2 weather data set
of the Helmholtz-Zentrum Geesthacht.
A description of the coastdat2 data set can be found here:
https://www.earth-syst-sci-data.net/6/147/2014/
SPDX-FileCopyrightText: 2016-2019 <NAME> <<EMAIL>>
SPDX-License-Identifier: MIT
"""
__copyright__ = "<NAME> <<EMAIL>>"
__license__ = "MIT"
import os
import pandas as pd
import pvlib
from nose.tools import eq_
from windpowerlib.wind_turbine import WindTurbine
from reegis import coastdat, feedin, config as cfg
import warnings
warnings.filterwarnings("ignore", category=RuntimeWarning)
def feedin_wind_sets_tests():
fn = os.path.join(
os.path.dirname(__file__),
os.pardir,
"tests",
"data",
"test_coastdat_weather.csv",
)
wind_sets = feedin.create_windpowerlib_sets()
weather = pd.read_csv(fn, header=[0, 1])["1126088"]
data_height = cfg.get_dict("coastdat_data_height")
wind_weather = coastdat.adapt_coastdat_weather_to_windpowerlib(
weather, data_height
)
df = pd.DataFrame()
for wind_key, wind_set in wind_sets.items():
df[str(wind_key).replace(" ", "_")] = (
feedin.feedin_wind_sets(wind_weather, wind_set).sum().sort_index()
)
s1 = df.transpose()["1"]
s2 = pd.Series(
{
"ENERCON_127_hub135_7500": 1277.28988,
"ENERCON_82_hub138_2300": 1681.47858,
"ENERCON_82_hub78_3000": 1057.03957,
"ENERCON_82_hub98_2300": 1496.55769,
}
)
pd.testing.assert_series_equal(
s1.sort_index(), s2.sort_index(), check_names=False
)
def feedin_windpowerlib_test():
fn = os.path.join(
os.path.dirname(__file__),
os.pardir,
"tests",
"data",
"test_coastdat_weather.csv",
)
weather = pd.read_csv(fn, header=[0, 1])["1126088"]
turbine = {"hub_height": 135, "turbine_type": "E-141/4200"}
data_height = cfg.get_dict("coastdat_data_height")
wind_weather = coastdat.adapt_coastdat_weather_to_windpowerlib(
weather, data_height
) # doctest: +SKIP
eq_(int(feedin.feedin_windpowerlib(wind_weather, turbine).sum()), 2164)
turbine = WindTurbine(**turbine)
eq_(int(feedin.feedin_windpowerlib(wind_weather, turbine).sum()), 2164)
def feedin_pvlib_test():
fn = os.path.join(
os.path.dirname(__file__),
os.pardir,
"tests",
"data",
"test_coastdat_weather.csv",
)
coastdat_id = "1126088"
weather = pd.read_csv(
fn,
header=[0, 1],
index_col=[0],
date_parser=lambda idx: pd.to_datetime(idx, utc=True),
)[coastdat_id]
c = coastdat.fetch_data_coordinates_by_id(coastdat_id)
location = pvlib.location.Location(**getattr(c, "_asdict")())
pv_weather = coastdat.adapt_coastdat_weather_to_pvlib(weather, location)
sandia_modules = pvlib.pvsystem.retrieve_sam("sandiamod")
sapm_inverters = pvlib.pvsystem.retrieve_sam("sandiainverter")
pv = {
"pv_set_name": "M_LG290G3__I_ABB_MICRO_025_US208",
"module_name": "LG_LG290N1C_G3__2013_",
"module_key": "LG290G3",
"inverter_name": "ABB__MICRO_0_25_I_OUTD_US_208_208V__CEC_2014_",
"surface_azimuth": 180,
"surface_tilt": 35,
"albedo": 0.2,
}
pv["module_parameters"] = sandia_modules[pv["module_name"]]
pv["inverter_parameters"] = sapm_inverters[pv["inverter_name"]]
pv["p_peak"] = pv["module_parameters"].Impo * pv["module_parameters"].Vmpo
eq_(int(feedin.feedin_pvlib(location, pv, pv_weather).sum()), 1025)
eq_(int(feedin.feedin_pvlib(location, pv, pv_weather).sum()), 1025)
def feedin_pv_sets_tests():
fn = os.path.join(
os.path.dirname(__file__),
os.pardir,
"tests",
"data",
"test_coastdat_weather.csv",
)
pv_sets = feedin.create_pvlib_sets()
coastdat_id = "1126088"
weather = pd.read_csv(
fn,
header=[0, 1],
index_col=[0],
date_parser=lambda idx: pd.to_datetime(idx, utc=True),
)[coastdat_id]
c = coastdat.fetch_data_coordinates_by_id(coastdat_id)
location = pvlib.location.Location(**getattr(c, "_asdict")())
pv_weather = coastdat.adapt_coastdat_weather_to_pvlib(weather, location)
s1 =
|
pd.Series()
|
pandas.Series
|
import wandb
from wandb import data_types
import numpy as np
import pytest
import PIL
import os
import six
import sys
import glob
import platform
from click.testing import CliRunner
from . import utils
from .utils import dummy_data
import matplotlib
import rdkit.Chem
from wandb import Api
import time
matplotlib.use("Agg")
import matplotlib.pyplot as plt # noqa: E402
data = np.random.randint(255, size=(1000))
@pytest.fixture
def api(runner):
return Api()
def test_wb_value(live_mock_server, test_settings):
run = wandb.init(settings=test_settings)
local_art = wandb.Artifact("N", "T")
public_art = run.use_artifact("N:latest")
wbvalue = data_types.WBValue()
with pytest.raises(NotImplementedError):
wbvalue.to_json(local_art)
with pytest.raises(NotImplementedError):
data_types.WBValue.from_json({}, public_art)
assert data_types.WBValue.with_suffix("item") == "item.json"
table = data_types.WBValue.init_from_json(
{
"_type": "table",
"data": [[]],
"columns": [],
"column_types": wandb.data_types._dtypes.TypedDictType({}).to_json(),
},
public_art,
)
assert isinstance(table, data_types.WBValue) and isinstance(
table, wandb.data_types.Table
)
type_mapping = data_types.WBValue.type_mapping()
assert all(
[issubclass(type_mapping[key], data_types.WBValue) for key in type_mapping]
)
assert wbvalue == wbvalue
assert wbvalue != data_types.WBValue()
run.finish()
@pytest.mark.skipif(sys.version_info >= (3, 10), reason="no pandas py3.10 wheel")
def test_log_dataframe(live_mock_server, test_settings):
import pandas as pd
run = wandb.init(settings=test_settings)
cv_results = pd.DataFrame(data={"test_col": [1, 2, 3], "test_col2": [4, 5, 6]})
run.log({"results_df": cv_results})
run.finish()
ctx = live_mock_server.get_ctx()
assert len(ctx["artifacts"]) == 1
def test_raw_data():
wbhist = wandb.Histogram(data)
assert len(wbhist.histogram) == 64
def test_np_histogram():
wbhist = wandb.Histogram(np_histogram=np.histogram(data))
assert len(wbhist.histogram) == 10
def test_manual_histogram():
wbhist = wandb.Histogram(np_histogram=([1, 2, 4], [3, 10, 20, 0]))
assert len(wbhist.histogram) == 3
def test_invalid_histogram():
with pytest.raises(ValueError):
wandb.Histogram(np_histogram=([1, 2, 3], [1]))
image = np.zeros((28, 28))
def test_captions():
wbone = wandb.Image(image, caption="Cool")
wbtwo = wandb.Image(image, caption="Nice")
assert wandb.Image.all_captions([wbone, wbtwo]) == ["Cool", "Nice"]
def test_bind_image(mocked_run):
wb_image = wandb.Image(image)
wb_image.bind_to_run(mocked_run, "stuff", 10)
assert wb_image.is_bound()
full_box = {
"position": {"middle": (0.5, 0.5), "width": 0.1, "height": 0.2},
"class_id": 2,
"box_caption": "This is a big car",
"scores": {"acc": 0.3},
}
# Helper function return a new dictionary with the key removed
def dissoc(d, key):
new_d = d.copy()
new_d.pop(key)
return new_d
optional_keys = ["box_caption", "scores"]
boxes_with_removed_optional_args = [dissoc(full_box, k) for k in optional_keys]
def test_image_accepts_other_images(mocked_run):
image_a = wandb.Image(np.random.random((300, 300, 3)))
image_b = wandb.Image(image_a)
assert image_a == image_b
def test_image_accepts_bounding_boxes(mocked_run):
img = wandb.Image(image, boxes={"predictions": {"box_data": [full_box]}})
img.bind_to_run(mocked_run, "images", 0)
img_json = img.to_json(mocked_run)
path = img_json["boxes"]["predictions"]["path"]
assert os.path.exists(os.path.join(mocked_run.dir, path))
def test_image_accepts_bounding_boxes_optional_args(mocked_run):
img = data_types.Image(
image, boxes={"predictions": {"box_data": boxes_with_removed_optional_args}}
)
img.bind_to_run(mocked_run, "images", 0)
img_json = img.to_json(mocked_run)
path = img_json["boxes"]["predictions"]["path"]
assert os.path.exists(os.path.join(mocked_run.dir, path))
standard_mask = {
"mask_data": np.array([[1, 2, 2, 2], [2, 3, 3, 4], [4, 4, 4, 4], [4, 4, 4, 2]]),
"class_labels": {1: "car", 2: "pedestrian", 3: "tractor", 4: "cthululu"},
}
def test_image_accepts_masks(mocked_run):
img = wandb.Image(image, masks={"overlay": standard_mask})
img.bind_to_run(mocked_run, "images", 0)
img_json = img.to_json(mocked_run)
path = img_json["masks"]["overlay"]["path"]
assert os.path.exists(os.path.join(mocked_run.dir, path))
def test_image_accepts_masks_without_class_labels(mocked_run):
img = wandb.Image(image, masks={"overlay": dissoc(standard_mask, "class_labels")})
img.bind_to_run(mocked_run, "images", 0)
img_json = img.to_json(mocked_run)
path = img_json["masks"]["overlay"]["path"]
assert os.path.exists(os.path.join(mocked_run.dir, path))
def test_cant_serialize_to_other_run(mocked_run, test_settings):
"""This isn't implemented yet. Should work eventually."""
other_run = wandb.wandb_sdk.wandb_run.Run(settings=test_settings)
other_run._set_backend(mocked_run._backend)
wb_image = wandb.Image(image)
wb_image.bind_to_run(mocked_run, "stuff", 10)
with pytest.raises(AssertionError):
wb_image.to_json(other_run)
def test_image_seq_to_json(mocked_run):
wb_image = wandb.Image(image)
wb_image.bind_to_run(mocked_run, "test", 0, 0)
meta = wandb.Image.seq_to_json([wb_image], mocked_run, "test", 0)
assert os.path.exists(
os.path.join(mocked_run.dir, "media", "images", "test_0_0.png")
)
meta_expected = {
"_type": "images/separated",
"count": 1,
"height": 28,
"width": 28,
}
assert utils.subdict(meta, meta_expected) == meta_expected
def test_max_images(caplog, mocked_run):
large_image = np.random.randint(255, size=(10, 10))
large_list = [wandb.Image(large_image)] * 200
large_list[0].bind_to_run(mocked_run, "test2", 0, 0)
meta = wandb.Image.seq_to_json(
wandb.wandb_sdk.data_types._prune_max_seq(large_list), mocked_run, "test2", 0
)
expected = {
"_type": "images/separated",
"count": data_types.Image.MAX_ITEMS,
"height": 10,
"width": 10,
}
path = os.path.join(mocked_run.dir, "media/images/test2_0_0.png")
assert utils.subdict(meta, expected) == expected
assert os.path.exists(os.path.join(mocked_run.dir, "media/images/test2_0_0.png"))
def test_audio_sample_rates():
audio1 = np.random.uniform(-1, 1, 44100)
audio2 = np.random.uniform(-1, 1, 88200)
wbaudio1 = wandb.Audio(audio1, sample_rate=44100)
wbaudio2 = wandb.Audio(audio2, sample_rate=88200)
assert wandb.Audio.sample_rates([wbaudio1, wbaudio2]) == [44100, 88200]
# test with missing sample rate
with pytest.raises(ValueError):
wandb.Audio(audio1)
def test_audio_durations():
audio1 = np.random.uniform(-1, 1, 44100)
audio2 = np.random.uniform(-1, 1, 88200)
wbaudio1 = wandb.Audio(audio1, sample_rate=44100)
wbaudio2 = wandb.Audio(audio2, sample_rate=44100)
assert wandb.Audio.durations([wbaudio1, wbaudio2]) == [1.0, 2.0]
def test_audio_captions():
audio = np.random.uniform(-1, 1, 44100)
sample_rate = 44100
caption1 = "This is what a dog sounds like"
caption2 = "This is what a chicken sounds like"
# test with all captions
wbaudio1 = wandb.Audio(audio, sample_rate=sample_rate, caption=caption1)
wbaudio2 = wandb.Audio(audio, sample_rate=sample_rate, caption=caption2)
assert wandb.Audio.captions([wbaudio1, wbaudio2]) == [caption1, caption2]
# test with no captions
wbaudio3 = wandb.Audio(audio, sample_rate=sample_rate)
wbaudio4 = wandb.Audio(audio, sample_rate=sample_rate)
assert wandb.Audio.captions([wbaudio3, wbaudio4]) is False
# test with some captions
wbaudio5 = wandb.Audio(audio, sample_rate=sample_rate)
wbaudio6 = wandb.Audio(audio, sample_rate=sample_rate, caption=caption2)
assert wandb.Audio.captions([wbaudio5, wbaudio6]) == ["", caption2]
def test_audio_to_json(mocked_run):
audio = np.zeros(44100)
audioObj = wandb.Audio(audio, sample_rate=44100)
audioObj.bind_to_run(mocked_run, "test", 0)
meta = wandb.Audio.seq_to_json([audioObj], mocked_run, "test", 0)
assert os.path.exists(os.path.join(mocked_run.dir, meta["audio"][0]["path"]))
meta_expected = {
"_type": "audio",
"count": 1,
"sampleRates": [44100],
"durations": [1.0],
}
assert utils.subdict(meta, meta_expected) == meta_expected
audio_expected = {
"_type": "audio-file",
"caption": None,
"size": 88244,
}
assert utils.subdict(meta["audio"][0], audio_expected) == audio_expected
wandb.finish()
def test_audio_refs():
audioObj = wandb.Audio(
"https://wandb-artifacts-refs-public-test.s3-us-west-2.amazonaws.com/StarWars3.wav"
)
art = wandb.Artifact("audio_ref_test", "dataset")
art.add(audioObj, "audio_ref")
audio_expected = {
"_type": "audio-file",
"caption": None,
}
assert utils.subdict(audioObj.to_json(art), audio_expected) == audio_expected
def test_guess_mode():
image = np.random.randint(255, size=(28, 28, 3))
wbimg = wandb.Image(image)
assert wbimg.image.mode == "RGB"
def test_pil():
pil = PIL.Image.new("L", (28, 28))
img = wandb.Image(pil)
assert list(img.image.getdata()) == list(pil.getdata())
def test_matplotlib_image():
plt.plot([1, 2, 2, 4])
img = wandb.Image(plt)
assert img.image.width == 640
def test_matplotlib_image_with_multiple_axes():
"""Ensures that wandb.Image constructor can accept a pyplot or figure
reference in which the figure has multiple axes. Importantly, there is
no requirement that any of the axes have plotted data.
"""
for fig in utils.matplotlib_multiple_axes_figures():
wandb.Image(fig) # this should not error.
for fig in utils.matplotlib_multiple_axes_figures():
wandb.Image(plt) # this should not error.
@pytest.mark.skipif(
sys.version_info >= (3, 9), reason="plotly doesn't support py3.9 yet"
)
def test_matplotlib_plotly_with_multiple_axes():
"""Ensures that wandb.Plotly constructor can accept a plotly figure
reference in which the figure has multiple axes. Importantly, there is
no requirement that any of the axes have plotted data.
"""
for fig in utils.matplotlib_multiple_axes_figures():
wandb.Plotly(fig) # this should not error.
for fig in utils.matplotlib_multiple_axes_figures():
wandb.Plotly(plt) # this should not error.
def test_plotly_from_matplotlib_with_image():
"""Ensures that wandb.Plotly constructor properly errors when
a pyplot with image is passed
"""
# try the figure version
fig = utils.matplotlib_with_image()
with pytest.raises(ValueError):
wandb.Plotly(fig)
plt.close()
# try the plt version
fig = utils.matplotlib_with_image()
with pytest.raises(ValueError):
wandb.Plotly(plt)
plt.close()
def test_image_from_matplotlib_with_image():
"""Ensures that wandb.Image constructor supports a pyplot with image is passed"""
# try the figure version
fig = utils.matplotlib_with_image()
wandb.Image(fig) # this should not error.
plt.close()
# try the plt version
fig = utils.matplotlib_with_image()
wandb.Image(plt) # this should not error.
plt.close()
@pytest.mark.skipif(
sys.version_info >= (3, 9), reason="plotly doesn't support py3.9 yet"
)
def test_make_plot_media_from_matplotlib_without_image():
"""Ensures that wand.Plotly.make_plot_media() returns a Plotly object when
there is no image
"""
fig = utils.matplotlib_without_image()
assert type(wandb.Plotly.make_plot_media(fig)) == wandb.Plotly
plt.close()
fig = utils.matplotlib_without_image()
assert type(wandb.Plotly.make_plot_media(plt)) == wandb.Plotly
plt.close()
def test_make_plot_media_from_matplotlib_with_image():
"""Ensures that wand.Plotly.make_plot_media() returns an Image object when
there is an image in the matplotlib figure
"""
fig = utils.matplotlib_with_image()
assert type(wandb.Plotly.make_plot_media(fig)) == wandb.Image
plt.close()
fig = utils.matplotlib_with_image()
assert type(wandb.Plotly.make_plot_media(plt)) == wandb.Image
plt.close()
def test_create_bokeh_plot(mocked_run):
"""Ensures that wandb.Bokeh constructor accepts a bokeh plot"""
bp = dummy_data.bokeh_plot()
bp = wandb.data_types.Bokeh(bp)
bp.bind_to_run(mocked_run, "bokeh", 0)
@pytest.mark.skipif(sys.version_info < (3, 6), reason="No moviepy.editor in py2")
def test_video_numpy_gif(mocked_run):
video = np.random.randint(255, size=(10, 3, 28, 28))
vid = wandb.Video(video, format="gif")
vid.bind_to_run(mocked_run, "videos", 0)
assert vid.to_json(mocked_run)["path"].endswith(".gif")
@pytest.mark.skipif(sys.version_info < (3, 6), reason="No moviepy.editor in py2")
def test_video_numpy_mp4(mocked_run):
video = np.random.randint(255, size=(10, 3, 28, 28))
vid = wandb.Video(video, format="mp4")
vid.bind_to_run(mocked_run, "videos", 0)
assert vid.to_json(mocked_run)["path"].endswith(".mp4")
@pytest.mark.skipif(sys.version_info < (3, 6), reason="No moviepy.editor in py2")
def test_video_numpy_multi(mocked_run):
video = np.random.random(size=(2, 10, 3, 28, 28))
vid = wandb.Video(video)
vid.bind_to_run(mocked_run, "videos", 0)
assert vid.to_json(mocked_run)["path"].endswith(".gif")
@pytest.mark.skipif(sys.version_info < (3, 6), reason="No moviepy.editor in py2")
def test_video_numpy_invalid():
video = np.random.random(size=(3, 28, 28))
with pytest.raises(ValueError):
wandb.Video(video)
def test_video_path(mocked_run):
with open("video.mp4", "w") as f:
f.write("00000")
vid = wandb.Video("video.mp4")
vid.bind_to_run(mocked_run, "videos", 0)
assert vid.to_json(mocked_run)["path"].endswith(".mp4")
def test_video_path_invalid(runner):
with runner.isolated_filesystem():
with open("video.avi", "w") as f:
f.write("00000")
with pytest.raises(ValueError):
wandb.Video("video.avi")
def test_molecule(mocked_run):
with open("test.pdb", "w") as f:
f.write("00000")
mol = wandb.Molecule("test.pdb")
mol.bind_to_run(mocked_run, "rad", "summary")
wandb.Molecule.seq_to_json([mol], mocked_run, "rad", "summary")
assert os.path.exists(mol._path)
wandb.finish()
def test_molecule_file(mocked_run):
with open("test.pdb", "w") as f:
f.write("00000")
mol = wandb.Molecule(open("test.pdb", "r"))
mol.bind_to_run(mocked_run, "rad", "summary")
wandb.Molecule.seq_to_json([mol], mocked_run, "rad", "summary")
assert os.path.exists(mol._path)
wandb.finish()
def test_molecule_from_smiles(mocked_run):
"""Ensures that wandb.Molecule.from_smiles supports valid SMILES molecule string representations"""
mol = wandb.Molecule.from_smiles("CC(=O)Nc1ccc(O)cc1")
mol.bind_to_run(mocked_run, "rad", "summary")
wandb.Molecule.seq_to_json([mol], mocked_run, "rad", "summary")
assert os.path.exists(mol._path)
wandb.finish()
def test_molecule_from_invalid_smiles(mocked_run):
"""Ensures that wandb.Molecule.from_smiles errs if passed an invalid SMILES string"""
with pytest.raises(ValueError):
wandb.Molecule.from_smiles("TEST")
wandb.finish()
def test_molecule_from_rdkit_mol_object(mocked_run):
"""Ensures that wandb.Molecule.from_rdkit supports rdkit.Chem.rdchem.Mol objects"""
mol = wandb.Molecule.from_rdkit(rdkit.Chem.MolFromSmiles("CC(=O)Nc1ccc(O)cc1"))
mol.bind_to_run(mocked_run, "rad", "summary")
wandb.Molecule.seq_to_json([mol], mocked_run, "rad", "summary")
assert os.path.exists(mol._path)
wandb.finish()
def test_molecule_from_rdkit_mol_file(mocked_run):
"""Ensures that wandb.Molecule.from_rdkit supports .mol files"""
substance = rdkit.Chem.MolFromSmiles("CC(=O)Nc1ccc(O)cc1")
mol_file_name = "test.mol"
rdkit.Chem.rdmolfiles.MolToMolFile(substance, mol_file_name)
mol = wandb.Molecule.from_rdkit(mol_file_name)
mol.bind_to_run(mocked_run, "rad", "summary")
wandb.Molecule.seq_to_json([mol], mocked_run, "rad", "summary")
assert os.path.exists(mol._path)
wandb.finish()
def test_molecule_from_rdkit_invalid_input(mocked_run):
"""Ensures that wandb.Molecule.from_rdkit errs on invalid input"""
mol_file_name = "test"
with pytest.raises(ValueError):
wandb.Molecule.from_rdkit(mol_file_name)
wandb.finish()
def test_html_str(mocked_run):
html = wandb.Html("<html><body><h1>Hello</h1></body></html>")
html.bind_to_run(mocked_run, "rad", "summary")
wandb.Html.seq_to_json([html], mocked_run, "rad", "summary")
assert os.path.exists(html._path)
wandb.finish()
def test_html_styles():
with CliRunner().isolated_filesystem():
pre = (
'<base target="_blank"><link rel="stylesheet" type="text/css" '
'href="https://app.wandb.ai/normalize.css" />'
)
html = wandb.Html("<html><body><h1>Hello</h1></body></html>")
assert (
html.html
== "<html><head>" + pre + "</head><body><h1>Hello</h1></body></html>"
)
html = wandb.Html("<html><head></head><body><h1>Hello</h1></body></html>")
assert (
html.html
== "<html><head>" + pre + "</head><body><h1>Hello</h1></body></html>"
)
html = wandb.Html("<h1>Hello</h1>")
assert html.html == pre + "<h1>Hello</h1>"
html = wandb.Html("<h1>Hello</h1>", inject=False)
assert html.html == "<h1>Hello</h1>"
def test_html_file(mocked_run):
with open("test.html", "w") as f:
f.write("<html><body><h1>Hello</h1></body></html>")
html = wandb.Html(open("test.html"))
html.bind_to_run(mocked_run, "rad", "summary")
wandb.Html.seq_to_json([html, html], mocked_run, "rad", "summary")
assert os.path.exists(html._path)
def test_html_file_path(mocked_run):
with open("test.html", "w") as f:
f.write("<html><body><h1>Hello</h1></body></html>")
html = wandb.Html("test.html")
html.bind_to_run(mocked_run, "rad", "summary")
wandb.Html.seq_to_json([html, html], mocked_run, "rad", "summary")
assert os.path.exists(html._path)
def test_table_default():
table = wandb.Table()
table.add_data("Some awesome text", "Positive", "Negative")
assert table._to_table_json() == {
"data": [["Some awesome text", "Positive", "Negative"]],
"columns": ["Input", "Output", "Expected"],
}
def test_table_eq_debug():
# Invalid Type
a = wandb.Table(data=[[1, 2, 3], [4, 5, 6]])
b = {}
with pytest.raises(AssertionError):
a._eq_debug(b, True)
assert a != b
# Mismatch Rows
a = wandb.Table(data=[[1, 2, 3], [4, 5, 6]])
b = wandb.Table(data=[[1, 2, 3]])
with pytest.raises(AssertionError):
a._eq_debug(b, True)
assert a != b
# Mismatch Columns
a = wandb.Table(data=[[1, 2, 3], [4, 5, 6]])
b = wandb.Table(data=[[1, 2, 3], [4, 5, 6]], columns=["a", "b", "c"])
with pytest.raises(AssertionError):
a._eq_debug(b, True)
assert a != b
# Mismatch Types
a = wandb.Table(data=[[1, 2, 3]])
b = wandb.Table(data=[["1", "2", "3"]])
with pytest.raises(AssertionError):
a._eq_debug(b, True)
assert a != b
# Mismatch Data
a = wandb.Table(data=[[1, 2, 3], [4, 5, 6]])
b = wandb.Table(data=[[1, 2, 3], [4, 5, 100]])
with pytest.raises(AssertionError):
a._eq_debug(b, True)
assert a != b
a = wandb.Table(data=[[1, 2, 3], [4, 5, 6]])
b = wandb.Table(data=[[1, 2, 3], [4, 5, 6]])
a._eq_debug(b, True)
assert a == b
@pytest.mark.skipif(sys.version_info >= (3, 10), reason="no pandas py3.10 wheel")
def test_table_custom():
import pandas as pd
table = wandb.Table(["Foo", "Bar"])
table.add_data("So", "Cool")
table.add_row("&", "Rad")
assert table._to_table_json() == {
"data": [["So", "Cool"], ["&", "Rad"]],
"columns": ["Foo", "Bar"],
}
df =
|
pd.DataFrame(columns=["Foo", "Bar"], data=[["So", "Cool"], ["&", "Rad"]])
|
pandas.DataFrame
|
from tqdm import tqdm
from model.QACGBERT import *
from util.tokenization import *
import numpy as np
import pandas as pd
import random
from torch.utils.data import DataLoader, TensorDataset
label_list = ['Yes', 'No']
context_id_map_fiqa = {"legal": 0, "m&a": 1, "regulatory": 2, "risks": 3, "rumors": 4, "company communication": 5,
"trade": 6, "central banks": 7, "market": 8, "volatility": 9, "financial": 10,
"fundamentals": 11, "price action": 12, "insider activity": 13, "ipo": 14, "others": 15}
class InputExample(object):
def __init__(self, guid, text_a, text_b=None, label=None):
self.guid = guid
self.text_a = text_a
self.text_b = text_b
self.label = label
class InputFeatures(object):
def __init__(self, input_ids, input_mask, segment_ids, label_id, seq_len,
context_ids):
self.input_ids = input_ids
self.input_mask = input_mask
self.segment_ids = segment_ids
self.label_id = label_id
self.seq_len = seq_len
self.context_ids = context_ids
def truncate_seq_pair(tokens_a, tokens_b, max_length):
while True:
total_length = len(tokens_a) + len(tokens_b)
if total_length <= max_length:
break
if len(tokens_a) > len(tokens_b):
tokens_a.pop()
else:
tokens_b.pop()
def convert_to_unicode(text):
if six.PY3:
if isinstance(text, str):
return text
elif isinstance(text, bytes):
return text.decode("utf-8", "ignore")
else:
raise ValueError("Unsupported string type: %s" % (type(text)))
else:
raise ValueError("Not running on Python 3")
def get_test_examples(path):
test_data =
|
pd.read_csv(path, header=None)
|
pandas.read_csv
|
# -*- coding: utf-8 -*-
from __future__ import division
from functools import wraps
import numpy as np
from pandas import DataFrame, Series
#from pandas.stats import moments
import pandas as pd
def simple_moving_average(prices, period=26):
"""
:param df: pandas dataframe object
:param period: periods for calculating SMA
:return: a pandas series
"""
weights = np.repeat(1.0, period) / period
sma = np.convolve(prices, weights, 'valid')
return sma
def stochastic_oscillator_k(df):
"""Calculate stochastic oscillator %K for given data.
:param df: pandas.DataFrame
:return: pandas.DataFrame
"""
SOk = pd.Series((df['close'] - df['low']) / (df['high'] - df['low']), name='SO%k')
df = df.join(SOk)
return df
def stochastic_oscillator_d(df, n):
"""Calculate stochastic oscillator %D for given data.
:param df: pandas.DataFrame
:param n:
:return: pandas.DataFrame
"""
SOk = pd.Series((df['close'] - df['low']) / (df['high'] - df['low']), name='SO%k')
SOd = pd.Series(SOk.ewm(span=n, min_periods=n).mean(), name='SO%d')
df = df.join(SOd)
return df
def bollinger_bands(df, n, std, add_ave=True):
"""
:param df: pandas.DataFrame
:param n:
:return: pandas.DataFrame
"""
ave = df['close'].rolling(window=n, center=False).mean()
sd = df['close'].rolling(window=n, center=False).std()
upband = pd.Series(ave + (sd * std), name='bband_upper_' + str(n))
dnband = pd.Series(ave - (sd * std), name='bband_lower_' + str(n))
if add_ave:
ave = pd.Series(ave, name='bband_ave_' + str(n))
df = df.join(pd.concat([upband, dnband, ave], axis=1))
else:
df = df.join(pd.concat([upband, dnband], axis=1))
return df
def money_flow_index(df, n):
"""Calculate Money Flow Index and Ratio for given data.
:param df: pandas.DataFrame
:param n:
:return: pandas.DataFrame
"""
PP = (df['high'] + df['low'] + df['close']) / 3
i = 0
PosMF = [0]
while i < df.index[-1]:
if PP[i + 1] > PP[i]:
PosMF.append(PP[i + 1] * df.loc[i + 1, 'volume'])
else:
PosMF.append(0)
i = i + 1
PosMF = pd.Series(PosMF)
TotMF = PP * df['volume']
MFR = pd.Series(PosMF / TotMF)
MFI = pd.Series(MFR.rolling(n, min_periods=n).mean())
# df = df.join(MFI)
return MFI
def series_indicator(col):
def inner_series_indicator(f):
@wraps(f)
def wrapper(s, *args, **kwargs):
if isinstance(s, DataFrame):
s = s[col]
return f(s, *args, **kwargs)
return wrapper
return inner_series_indicator
def _wilder_sum(s, n):
s = s.dropna()
nf = (n - 1) / n
ws = [np.nan] * (n - 1) + [s[n - 1] + nf * sum(s[:n - 1])]
for v in s[n:]:
ws.append(v + ws[-1] * nf)
return Series(ws, index=s.index)
@series_indicator('high')
def hhv(s, n):
return pd.rolling_max(s, n)
@series_indicator('low')
def llv(s, n):
return pd.rolling_min(s, n)
@series_indicator('close')
def ema(s, n, wilder=False):
span = n if not wilder else 2 * n - 1
return pd.ewma(s, span=span)
@series_indicator('close')
def macd(s, nfast=12, nslow=26, nsig=9, percent=True):
fast, slow = ema(s, nfast), ema(s, nslow)
if percent:
macd = 100 * (fast / slow - 1)
else:
macd = fast - slow
sig = ema(macd, nsig)
hist = macd - sig
return DataFrame(dict(macd=macd, signal=sig, hist=hist,
fast=fast, slow=slow))
def aroon(s, n=25):
up = 100 * pd.rolling_apply(s.high, n + 1, lambda x: x.argmax()) / n
dn = 100 * pd.rolling_apply(s.low, n + 1, lambda x: x.argmin()) / n
return DataFrame(dict(up=up, down=dn))
@series_indicator('close')
def rsi(s, n=14):
diff = s.diff()
which_dn = diff < 0
up, dn = diff, diff * 0
up[which_dn], dn[which_dn] = 0, -up[which_dn]
emaup = ema(up, n, wilder=True)
emadn = ema(dn, n, wilder=True)
return 100 * emaup / (emaup + emadn)
def stoch(s, nfastk=14, nfullk=3, nfulld=3):
if not isinstance(s, DataFrame):
s = DataFrame(dict(high=s, low=s, close=s))
hmax, lmin = hhv(s, nfastk), llv(s, nfastk)
fastk = 100 * (s.close - lmin) / (hmax - lmin)
fullk = pd.rolling_mean(fastk, nfullk)
fulld = pd.rolling_mean(fullk, nfulld)
return DataFrame(dict(fastk=fastk, fullk=fullk, fulld=fulld))
@series_indicator('close')
def dtosc(s, nrsi=13, nfastk=8, nfullk=5, nfulld=3):
srsi = stoch(rsi(s, nrsi), nfastk, nfullk, nfulld)
return DataFrame(dict(fast=srsi.fullk, slow=srsi.fulld))
def atr(s, n=14):
cs = s.close.shift(1)
tr = s.high.combine(cs, max) - s.low.combine(cs, min)
return ema(tr, n, wilder=True)
def cci(s, n=20, c=0.015):
if isinstance(s, DataFrame):
s = s[['high', 'low', 'close']].mean(axis=1)
mavg = pd.rolling_mean(s, n)
mdev = pd.rolling_apply(s, n, lambda x: np.fabs(x - x.mean()).mean())
return (s - mavg) / (c * mdev)
def cmf(s, n=20):
clv = (2 * s.close - s.high - s.low) / (s.high - s.low)
vol = s.volume
return pd.rolling_sum(clv * vol, n) / pd.rolling_sum(vol, n)
def force(s, n=2):
return ema(s.close.diff() * s.volume, n)
@series_indicator('close')
def kst(s, r1=10, r2=15, r3=20, r4=30, n1=10, n2=10, n3=10, n4=15, nsig=9):
rocma1 = pd.rolling_mean(s / s.shift(r1) - 1, n1)
rocma2 = pd.rolling_mean(s / s.shift(r2) - 1, n2)
rocma3 = pd.rolling_mean(s / s.shift(r3) - 1, n3)
rocma4 = pd.rolling_mean(s / s.shift(r4) - 1, n4)
kst = 100 * (rocma1 + 2 * rocma2 + 3 * rocma3 + 4 * rocma4)
sig = pd.rolling_mean(kst, nsig)
return DataFrame(dict(kst=kst, signal=sig))
def ichimoku(s, n1=9, n2=26, n3=52):
conv = (hhv(s, n1) + llv(s, n1)) / 2
base = (hhv(s, n2) + llv(s, n2)) / 2
spana = (conv + base) / 2
spanb = (hhv(s, n3) + llv(s, n3)) / 2
return DataFrame(dict(conv=conv, base=base, spana=spana.shift(n2),
spanb=spanb.shift(n2), lspan=s.close.shift(-n2)))
def ultimate(s, n1=7, n2=14, n3=28):
cs = s.close.shift(1)
bp = s.close - s.low.combine(cs, min)
tr = s.high.combine(cs, max) - s.low.combine(cs, min)
avg1 = pd.rolling_sum(bp, n1) / pd.rolling_sum(tr, n1)
avg2 = pd.rolling_sum(bp, n2) / pd.rolling_sum(tr, n2)
avg3 =
|
pd.rolling_sum(bp, n3)
|
pandas.rolling_sum
|
import re
import numpy as np
import pandas as pd
import pytest
from woodwork.datacolumn import DataColumn
from woodwork.exceptions import ColumnNameMismatchWarning, DuplicateTagsWarning
from woodwork.logical_types import (
Categorical,
CountryCode,
Datetime,
Double,
Integer,
NaturalLanguage,
Ordinal,
SubRegionCode,
ZIPCode
)
from woodwork.tests.testing_utils import to_pandas
from woodwork.utils import import_or_none
dd = import_or_none('dask.dataframe')
ks = import_or_none('databricks.koalas')
def test_datacolumn_init(sample_series):
data_col = DataColumn(sample_series, use_standard_tags=False)
# Koalas doesn't support category dtype
if not (ks and isinstance(sample_series, ks.Series)):
sample_series = sample_series.astype('category')
pd.testing.assert_series_equal(to_pandas(data_col.to_series()), to_pandas(sample_series))
assert data_col.name == sample_series.name
assert data_col.logical_type == Categorical
assert data_col.semantic_tags == set()
def test_datacolumn_init_with_logical_type(sample_series):
data_col = DataColumn(sample_series, NaturalLanguage)
assert data_col.logical_type == NaturalLanguage
assert data_col.semantic_tags == set()
data_col = DataColumn(sample_series, "natural_language")
assert data_col.logical_type == NaturalLanguage
assert data_col.semantic_tags == set()
data_col = DataColumn(sample_series, "NaturalLanguage")
assert data_col.logical_type == NaturalLanguage
assert data_col.semantic_tags == set()
def test_datacolumn_init_with_semantic_tags(sample_series):
semantic_tags = ['tag1', 'tag2']
data_col = DataColumn(sample_series, semantic_tags=semantic_tags, use_standard_tags=False)
assert data_col.semantic_tags == set(semantic_tags)
def test_datacolumn_init_wrong_series():
error = 'Series must be one of: pandas.Series, dask.Series, koalas.Series, numpy.ndarray, or pandas.ExtensionArray'
with pytest.raises(TypeError, match=error):
DataColumn([1, 2, 3, 4])
with pytest.raises(TypeError, match=error):
DataColumn({1, 2, 3, 4})
def test_datacolumn_init_with_name(sample_series, sample_datetime_series):
name = 'sample_series'
changed_name = 'changed_name'
dc_use_series_name = DataColumn(sample_series)
assert dc_use_series_name.name == name
assert dc_use_series_name.to_series().name == name
warning = 'Name mismatch between sample_series and changed_name. DataColumn and underlying series name are now changed_name'
with pytest.warns(ColumnNameMismatchWarning, match=warning):
dc_use_input_name = DataColumn(sample_series, name=changed_name)
assert dc_use_input_name.name == changed_name
assert dc_use_input_name.to_series().name == changed_name
warning = 'Name mismatch between sample_datetime_series and changed_name. DataColumn and underlying series name are now changed_name'
with pytest.warns(ColumnNameMismatchWarning, match=warning):
dc_with_ltype_change = DataColumn(sample_datetime_series, name=changed_name)
assert dc_with_ltype_change.name == changed_name
assert dc_with_ltype_change.to_series().name == changed_name
def test_datacolumn_inity_with_falsy_name(sample_series):
falsy_name = 0
warning = 'Name mismatch between sample_series and 0. DataColumn and underlying series name are now 0'
with pytest.warns(ColumnNameMismatchWarning, match=warning):
dc_falsy_name = DataColumn(sample_series.copy(), name=falsy_name)
assert dc_falsy_name.name == falsy_name
assert dc_falsy_name.to_series().name == falsy_name
def test_datacolumn_init_with_extension_array():
series_categories = pd.Series([1, 2, 3], dtype='category')
extension_categories = pd.Categorical([1, 2, 3])
data_col = DataColumn(extension_categories)
series = data_col.to_series()
assert series.equals(series_categories)
assert series.name is None
assert data_col.name is None
assert data_col.dtype == 'category'
assert data_col.logical_type == Categorical
series_ints =
|
pd.Series([1, 2, None, 4], dtype='Int64')
|
pandas.Series
|
import importlib
import sys
import numpy as np
import pandas as pd
import pytest
from sweat import utils
from sweat.metrics import power
@pytest.fixture()
def reload_power_module():
yield
key_values = [(key, value) for key, value in sys.modules.items()]
for key, value in key_values:
if (
key.startswith("sweat.hrm")
or key.startswith("sweat.pdm")
or key.startswith("sweat.metrics")
):
importlib.reload(value)
def test_enable_type_casting_module(reload_power_module):
pwr = [1, 2, 3]
wap = [1, 2, 3]
weight = 80
threshold_power = 80
assert isinstance(power.wpk(np.asarray(pwr), weight), np.ndarray)
assert isinstance(
power.relative_intensity(np.asarray(wap), threshold_power), np.ndarray
)
with pytest.raises(TypeError):
power.wpk(pwr, weight)
with pytest.raises(TypeError):
power.relative_intensity(wap, threshold_power)
doc_string = power.wpk.__doc__
utils.enable_type_casting(power)
assert isinstance(power.wpk(pwr, weight), list)
assert isinstance(power.wpk(pd.Series(pwr), weight), pd.Series)
assert isinstance(power.wpk(np.asarray(pwr), weight), np.ndarray)
assert isinstance(power.relative_intensity(wap, threshold_power), list)
assert isinstance(
power.relative_intensity(pd.Series(wap), threshold_power), pd.Series
)
assert isinstance(
power.relative_intensity(np.asarray(wap), threshold_power), np.ndarray
)
assert power.wpk.__doc__ == doc_string
def test_enable_type_casting_func(reload_power_module):
pwr = [1, 2, 3]
wap = [1, 2, 3]
weight = 80
threshold_power = 80
assert isinstance(power.wpk(np.asarray(pwr), weight), np.ndarray)
assert isinstance(
power.relative_intensity(np.asarray(wap), threshold_power), np.ndarray
)
with pytest.raises(TypeError):
power.wpk(pwr, weight)
with pytest.raises(TypeError):
power.relative_intensity(wap, threshold_power)
doc_string = power.wpk.__doc__
wpk = utils.enable_type_casting(power.wpk)
assert isinstance(wpk(pwr, weight), list)
assert isinstance(wpk(pd.Series(pwr), weight), pd.Series)
assert isinstance(wpk(np.asarray(pwr), weight), np.ndarray)
with pytest.raises(TypeError):
power.relative_intensity(wap, threshold_power)
assert power.wpk.__doc__ == doc_string
def test_enable_type_casting_all(reload_power_module):
pwr = [1, 2, 3]
wap = [1, 2, 3]
weight = 80
threshold_power = 80
assert isinstance(power.wpk(np.asarray(pwr), weight), np.ndarray)
assert isinstance(
power.relative_intensity(np.asarray(wap), threshold_power), np.ndarray
)
with pytest.raises(TypeError):
power.wpk(pwr, weight)
with pytest.raises(TypeError):
power.relative_intensity(wap, threshold_power)
doc_string = power.wpk.__doc__
utils.enable_type_casting()
assert isinstance(power.wpk(pwr, weight), list)
assert isinstance(power.wpk(pd.Series(pwr), weight), pd.Series)
assert isinstance(power.wpk(np.asarray(pwr), weight), np.ndarray)
assert isinstance(power.relative_intensity(wap, threshold_power), list)
assert isinstance(
power.relative_intensity(
|
pd.Series(wap)
|
pandas.Series
|
# coding=utf-8
import pathlib
import numpy as np
import pandas as pd
import plotly.express as px
def main():
'''
生成树状图
'''
df = px.data.gapminder().query("year == 2007")
fig = px.treemap(df, path=['continent', 'country'], values='pop',
color='lifeExp', hover_data=['iso_alpha'],
color_continuous_scale='RdBu',
color_continuous_midpoint=np.average(df['lifeExp'], weights=df['pop']))
fig.show()
def gen_report(file_path):
comps = ['腾讯', '阿里', '百度', '京东', '美团', '小米', '字节跳动', '滴滴']
df_dict = pd.read_excel(file_path, comps)
for k, v in df_dict.items():
v['投资主体'] = k
# domain_set = set().union(* [set(df['行业']) for df in df_dict.values()])
# df_all = pd.concat(df_dict.values())
df_all =
|
pd.concat(df_dict)
|
pandas.concat
|
# -*- coding: utf-8 -*-
# script
import wikilanguages_utils
# app
import flask
from flask import send_from_directory
from dash import Dash
import dash
import dash_html_components as html
import dash_core_components as dcc
import dash_table_experiments as dt
from dash.dependencies import Input, Output, State
# viz
import plotly
import plotly.plotly as py
import plotly.figure_factory as ff
# data
from urllib.parse import urlparse, parse_qs
import pandas as pd
import sqlite3
# other
import logging
from logging.handlers import RotatingFileHandler
import datetime
import time
print ('\n\n\n*** START WCDO APP:'+str(datetime.datetime.now()))
databases_path = '/srv/wcdo/databases/'
territories = wikilanguages_utils.load_languageterritories_mapping()
languages = wikilanguages_utils.load_wiki_projects_information(territories);
wikilanguagecodes = languages.index.tolist()
wikipedialanguage_numberarticles = wikilanguages_utils.load_wikipedia_language_editions_numberofarticles(wikilanguagecodes)
for languagecode in wikilanguagecodes:
if languagecode not in wikipedialanguage_numberarticles: wikilanguagecodes.remove(languagecode)
languageswithoutterritory=['eo','got','ia','ie','io','jbo','lfn','nov','vo']
# Only those with a geographical context
for languagecode in languageswithoutterritory: wikilanguagecodes.remove(languagecode)
closest_langs = wikilanguages_utils.obtain_closest_for_all_languages(wikipedialanguage_numberarticles, wikilanguagecodes, 4)
country_names, regions, subregions = wikilanguages_utils.load_iso_3166_to_geographical_regions()
# LET THE SHOW START
app = flask.Flask(__name__)
if __name__ == '__main__':
handler = RotatingFileHandler('wcdo_app.log', maxBytes=10000, backupCount=1)
handler.setLevel(logging.INFO)
app.logger.addHandler(handler)
app.run(host='0.0.0.0')
@app.route('/')
def main():
return flask.redirect('https://meta.wikimedia.org/wiki/Wikipedia_Cultural_Diversity_Observatory')
### DASH APP ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ###
dash_app1 = Dash(__name__, server = app, url_base_pathname='/language_territories_mapping/')
df = pd.read_csv(databases_path + 'language_territories_mapping.csv',sep='\t',na_filter = False)
#df = df[['territoryname','territorynameNative','QitemTerritory','WikimediaLanguagecode','demonym','demonymNative','ISO3166','ISO31662']]
df.WikimediaLanguagecode = df['WikimediaLanguagecode'].str.replace('-','_')
df.WikimediaLanguagecode = df['WikimediaLanguagecode'].str.replace('be_tarask', 'be_x_old')
df.WikimediaLanguagecode = df['WikimediaLanguagecode'].str.replace('nan', 'zh_min_nan')
df = df.set_index('WikimediaLanguagecode')
df['Language Name'] = pd.Series(languages[['languagename']].to_dict('dict')['languagename'])
df = df.reset_index()
columns_dict = {'Language Name':'Language','WikimediaLanguagecode':'Wiki','QitemTerritory':'WD Qitem','territoryname':'Territory','territorynameNative':'Territory (Local)','demonymNative':'Demonyms (Local)','ISO3166':'ISO 3166', 'ISO3662':'ISO 3166-2','country':'Country'}
df=df.rename(columns=columns_dict)
title = 'Language Territories Mapping'
dash_app1.title = title
dash_app1.layout = html.Div([
html.H3(title, style={'textAlign':'center'}),
dt.DataTable(
columns=['Wiki','Language','WD Qitem','Territory (Local)','Demonyms (Local)','ISO3166','ISO31662'],
rows=df.to_dict('records'),
filterable=True,
sortable=True,
id='datatable-languageterritories'
),
html.A(html.H5('Home - Wikipedia Cultural Diverstiy Observatory'), href='https://meta.wikimedia.org/wiki/Wikipedia_Cultural_Diversity_Observatory', target="_blank", style={'textAlign': 'right', 'text-decoration':'none'})
], className="container")
### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ###
### DASH APP ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ###
dash_app2 = Dash(__name__, server = app, url_base_pathname='/list_of_wikipedias_by_cultural_context_content/')
conn = sqlite3.connect(databases_path + 'wcdo_stats.db'); cursor = conn.cursor()
df = pd.DataFrame(wikilanguagecodes)
df = df.set_index(0)
df['wp_number_articles']= pd.Series(wikipedialanguage_numberarticles)
# CCC %
query = 'SELECT set1, abs_value, rel_value FROM wcdo_intersections WHERE set1descriptor = "wp" AND set2descriptor = "ccc" AND content = "articles" AND set1=set2 AND measurement_date IN (SELECT MAX(measurement_date) FROM wcdo_intersections) ORDER BY abs_value DESC;'
rank_dict = {}; i=1
lang_dict = {}
abs_rel_value_dict = {}
abs_value_dict = {}
for row in cursor.execute(query):
lang_dict[row[0]]=languages.loc[row[0]]['languagename']
abs_rel_value_dict[row[0]]=round(row[2],2)
abs_value_dict[row[0]]=int(row[1])
rank_dict[row[0]]=i
i=i+1
df['Language'] = pd.Series(lang_dict)
df['Nº'] = pd.Series(rank_dict)
df['ccc_percent'] = pd.Series(abs_rel_value_dict)
df['ccc_number_articles'] = pd.Series(abs_value_dict)
df['Region']=languages.region
for x in df.index.values.tolist():
if ';' in df.loc[x]['Region']: df.at[x, 'Region'] = df.loc[x]['Region'].split(';')[0]
df['Subregion']=languages.subregion
for x in df.index.values.tolist():
if ';' in df.loc[x]['Subregion']: df.at[x, 'Subregion'] = df.loc[x]['Subregion'].split(';')[0]
# Renaming the columns
columns_dict = {'Language':'Language','wp_number_articles':'Articles','ccc_number_articles':'CCC art.','ccc_percent':'CCC %'}
df=df.rename(columns=columns_dict)
df = df.reset_index()
df = df.rename(columns={0: 'Wiki'})
df = df.fillna('')
title = 'Lists of Wikipedias by Cultural Context Content'
dash_app2.title = title
dash_app2.layout = html.Div([
html.H3(title, style={'textAlign':'center'}),
dt.DataTable(
rows=df.to_dict('records'),
columns = ['Nº','Language','Wiki','Articles','CCC art.','CCC %','Subregion','Region'],
row_selectable=True,
filterable=True,
sortable=True,
selected_row_indices=[],
id='datatable-cccextent'
),
html.Div(id='selected-indexes'),
dcc.Graph(
id='graph-cccextent'
),
html.A(html.H5('Home - Wikipedia Cultural Diverstiy Observatory'), href='https://meta.wikimedia.org/wiki/Wikipedia_Cultural_Diversity_Observatory', target="_blank", style={'textAlign': 'right', 'text-decoration':'none'})
], className="container")
@dash_app2.callback(
Output('datatable-cccextent', 'selected_row_indices'),
[Input('graph-cccextent', 'clickData')],
[State('datatable-cccextent', 'selected_row_indices')])
def app2_update_selected_row_indices(clickData, selected_row_indices):
if clickData:
for point in clickData['points']:
if point['pointNumber'] in selected_row_indices:
selected_row_indices.remove(point['pointNumber'])
else:
selected_row_indices.append(point['pointNumber'])
return selected_row_indices
@dash_app2.callback(
Output('graph-cccextent', 'figure'),
[Input('datatable-cccextent', 'rows'),
Input('datatable-cccextent', 'selected_row_indices')])
def app2_update_figure(rows, selected_row_indices):
dff = pd.DataFrame(rows)
fig = plotly.tools.make_subplots(
rows=3, cols=1,
subplot_titles=('CCC Articles', 'CCC %', 'Wikipedia articles',),
shared_xaxes=True)
marker = {'color': ['#0074D9']*len(dff)}
for i in (selected_row_indices or []):
marker['color'][i] = '#FF851B'
fig.append_trace({
'name':'',
'hovertext':'articles',
'x': dff['Language'],
'y': dff['CCC art.'],
'type': 'bar',
'marker': marker
}, 1, 1)
fig.append_trace({
'name':'',
'hovertext':'percent',
'x': dff['Language'],
'y': dff['CCC %'],
'type': 'bar',
'marker': marker
}, 2, 1)
fig.append_trace({
'name':'',
'hovertext':'articles',
'x': dff['Language'],
'y': dff['Articles'],
'type': 'bar',
'marker': marker
}, 3, 1)
fig['layout']['showlegend'] = False
fig['layout']['height'] = 800
fig['layout']['margin'] = {
'l': 40,
'r': 10,
't': 60,
'b': 200
}
fig['layout']['yaxis3']['type'] = 'log'
return fig
### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ###
### DASH APP ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ### ###
dash_app3 = Dash(__name__, server = app, url_base_pathname='/list_of_language_territories_by_cultural_context_content/')
# QUESTION: What is the extent of Cultural Context Content in each language edition broken down to territories? # OBTAIN THE DATA.
conn = sqlite3.connect(databases_path + 'wcdo_stats.db'); cursor = conn.cursor()
# CCC
query = 'SELECT set1 as languagecode, set2descriptor as Qitem, abs_value as CCC_articles, ROUND(rel_value,2) CCC_percent FROM wcdo_intersections WHERE set1descriptor = "ccc" AND set2 = "ccc" AND content = "articles" AND measurement_date IN (SELECT MAX(measurement_date) FROM wcdo_intersections) ORDER BY set1, set2descriptor DESC;'
df1 = pd.read_sql_query(query, conn)
# GL
query = 'SELECT set1 as languagecode2, set2descriptor as Qitem2, abs_value as CCC_articles_GL, ROUND(rel_value,2) CCC_percent_GL FROM wcdo_intersections WHERE set1descriptor = "ccc" AND set2 = "ccc_geolocated" AND content = "articles" AND measurement_date IN (SELECT MAX(measurement_date) FROM wcdo_intersections) ORDER BY set1, set2descriptor DESC;'
df2 = pd.read_sql_query(query, conn)
# KW
query = 'SELECT set1 as languagecode3, set2descriptor as Qitem3, abs_value as CCC_articles_KW, ROUND(rel_value,2) CCC_percent_KW FROM wcdo_intersections WHERE set1descriptor = "ccc" AND set2 = "ccc_keywords" AND content = "articles" AND measurement_date IN (SELECT MAX(measurement_date) FROM wcdo_intersections) ORDER BY set1, set2descriptor DESC;'
df3 = pd.read_sql_query(query, conn)
dfx = pd.concat([df1, df2, df3], axis=1)
dfx = dfx[['languagecode','Qitem','CCC_articles','CCC_percent','CCC_articles_GL','CCC_percent_GL','CCC_articles_KW','CCC_percent_KW']]
columns = ['territoryname','territorynameNative','country','ISO3166','ISO31662','subregion','region']
territoriesx = list()
for index in dfx.index.values:
qitem = dfx.loc[index]['Qitem']
languagecode = dfx.loc[index]['languagecode']
languagename = languages.loc[languagecode]['languagename']
current = []
try:
current = territories.loc[territories['QitemTerritory'] == qitem].loc[languagecode][columns].values.tolist()
current.append(languagename)
except:
current = [None,None,None,None,None,None,None,languagename]
pass
territoriesx.append(current)
columns.append('languagename')
all_territories = pd.DataFrame.from_records(territoriesx, columns=columns)
df =
|
pd.concat([dfx, all_territories], axis=1)
|
pandas.concat
|
from dash import dcc, html, Input, Output, callback
from mplcursors import HoverMode
import numpy as np
import pandas as pd
from plotly.subplots import make_subplots
import plotly.graph_objects as go
import dash_bootstrap_components as dbc
layout = html.Div(
[
html.H2('Bitcoin ROI As Measured From The Market Cycle Bottom'),
html.Div(dbc.Spinner(size='lg', color='black', type='border'), id='btc_roi_cycle_bottom'),
html.Hr(),
html.Br(),
html.Div(
[
]
)
]
)
@callback(
Output('btc_roi_cycle_bottom', 'children'),
Input('df-data', 'data'))
def monthly_roi(data):
df = pd.DataFrame(data)
df['Date'] =
|
pd.to_datetime(df['Date'])
|
pandas.to_datetime
|
# Copyright (c) 2020 Civic Knowledge. This file is licensed under the terms of the
# MIT license included in this distribution as LICENSE
import logging
import re
from collections import defaultdict, deque
from pathlib import Path
from time import time
import pandas as pd
from synpums.util import *
''
_logger = logging.getLogger(__name__)
def sample_to_sum(N, df, col, weights):
"""Sample a number of records from a dataset, then return the smallest set of
rows at the front of the dataset where the weight sums to more than N"""
t = df.sample(n=N, weights=weights, replace=True)
# Get the number of records that sum to N.
arg = t[col].cumsum().sub(N).abs().astype(int).argmin()
return t.iloc[:arg + 1]
def rms(s):
"""Root mean square"""
return np.sqrt(np.sum(np.square(s)))
def vector_walk_callback(puma_task, tract_task, data, memo):
pass
def make_acs_target_df(acs, columns, geoid):
t = acs.loc[geoid]
target_map = {c + '_m90': c for c in columns if "WGTP" not in columns}
target_df = pd.DataFrame({
'est': t[target_map.values()],
'm90': t[target_map.keys()].rename(target_map)
})
target_df['est_min'] = target_df.est - target_df.m90
target_df['est_max'] = target_df.est + target_df.m90
target_df.loc[target_df.est_min < 0, 'est_min'] = 0
return target_df.astype('Int64')
def geoid_path(geoid):
from pathlib import Path
from geoid.acs import AcsGeoid
go = AcsGeoid.parse(geoid)
try:
return Path(f"{go.level}/{go.stusab}/{go.county:03d}/{str(go)}.csv")
except AttributeError:
return Path(f"{go.level}/{go.stusab}/{str(go)}.csv")
class AllocationTask(object):
"""Represents the allocation process to one tract"""
def __init__(self, region_geoid, puma_geoid, acs_ref, hh_ref, cache_dir):
self.region_geoid = region_geoid
self.puma_geoid = puma_geoid
self.acs_ref = acs_ref
self.hh_ref = hh_ref
self.cache_dir = cache_dir
self.sample_pop = None
self.sample_weights = None
self.unallocated_weights = None # Initialized to the puma weights, gets decremented
self.target_marginals = None
self.allocated_weights = None
self.household_count = None
self.population_count = None
self.gq_count = None
self.gq_cols = None
self.sex_age_cols = None
self.hh_size_cols = None
self.hh_race_type_cols = None
self.hh_eth_type_cols = None
self.hh_income_cols = None
self._init = False
self.running_allocated_marginals = None
# A version of the sample_pop constructed by map_cp, added as an instance var so
# the probabilities can be manipulated during the vector walk.
self.cp_df = None
self.cp_prob = None
@property
def row(self):
from geoid.acs import AcsGeoid
tract = AcsGeoid.parse(self.region_geoid)
return [tract.state, tract.stusab, tract.county, self.region_geoid, self.puma_geoid, str(self.acs_ref),
str(self.hh_ref)]
def init(self, use_sample_weights=False, puma_weights=None):
"""Load all of the data, just before running the allocation"""
if isinstance(self.hh_ref, pd.DataFrame):
hh_source = self.hh_ref
else:
hh_source = pd.read_csv(self.hh_ref, index_col='SERIALNO', low_memory=False) \
.drop(columns=['geoid'], errors='ignore').astype('Int64')
if isinstance(self.acs_ref, pd.DataFrame):
acs = self.acs_ref
else:
acs = pd.read_csv(self.acs_ref, index_col='geoid', low_memory=False)
# These are only for debugging.
#self.hh_source = hh_source
#self.tract_acs = acs
return self._do_init(hh_source, acs, puma_weights=puma_weights)
def _do_init(self, hh_source, acs, puma_weights=None):
self.serialno = hh_source.index
# Col 0 is the WGTP column
w_cols = [c for c in hh_source.columns if "WGTP" in c]
not_w_cols = [c for c in hh_source.columns if "WGTP" not in c]
# Not actually a sample pop --- populations are supposed to be unweighted
self.sample_pop = hh_source[['WGTP'] + not_w_cols].iloc[:, 1:].reset_index(drop=True).astype(int)
# Shouldn't this be:
# self.sample_pop = hh_source[not_w_cols].reset_index(drop=True).astype(int)
self.sample_weights = hh_source.iloc[:, 0].reset_index(drop=True).astype(int)
assert self.sample_pop.shape[0] == self.sample_weights.shape[0]
not_w_cols = [c for c in hh_source.columns if "WGTP" not in c]
self.target_marginals = make_acs_target_df(acs, not_w_cols, self.region_geoid)
self.household_count = acs.loc[self.region_geoid].b11016_001
self.population_count = acs.loc[self.region_geoid].b01003_001
self.gq_count = acs.loc[self.region_geoid].b26001_001
self.total_count = self.household_count + self.gq_count
self.allocated_weights = np.zeros(len(self.sample_pop))
self.unallocated_weights = puma_weights if puma_weights is not None else self.sample_weights.copy()
self.running_allocated_marginals = pd.Series(0, index=self.target_marginals.index)
# Sample pop, normalized to unit length to speed up cosine similarity
self.sample_pop_norm = vectors_normalize(self.sample_pop.values)
# Column sets
self.gq_cols = ['b26001_001']
self.sex_age_cols = [c for c in hh_source.columns if c.startswith('b01001')]
self.hh_size_cols = [c for c in hh_source.columns if c.startswith('b11016')]
p = re.compile(r'b11001[^hi]_')
self.hh_race_type_cols = [c for c in hh_source.columns if p.match(c)]
p = re.compile(r'b11001[hi]_')
self.hh_eth_type_cols = [c for c in hh_source.columns if p.match(c)]
p = re.compile(r'b19025')
self.hh_income_cols = [c for c in hh_source.columns if p.match(c)]
# We will use this identity in the numpy version of step_scjhedule
# assert all((self.cp.index / 2).astype(int) == self['index'])
self.rng = np.random.default_rng()
self.make_cp(self.sample_pop)
self._init = True
return acs
def make_cp(self, sp):
"""Make a version of the sample population with two records for each
row, one the negative of the one before it. This is used to generate
rows that can be used in the vector walk."""
self.cp = pd.concat([sp, sp]).sort_index().reset_index()
self.cp.insert(1, 'sign', 1)
self.cp.insert(2, 'select_weight', 0)
self.cp.iloc[0::2, 1:] = self.cp.iloc[0::2, 1:] * -1 # flip sign on the marginal counts
self.update_cp()
return self.cp
def update_cp(self):
self.cp.loc[0::2, 'select_weight'] = self.allocated_weights.tolist()
self.cp.loc[1::2, 'select_weight'] = self.unallocated_weights.tolist()
def set_cp_prob(self, cp_prob):
pass
@property
def path(self):
return Path(self.cache_dir).joinpath(geoid_path(str(self.region_geoid))).resolve()
@property
def pums(self):
"""Return the PUMS household and personal records for this PUMA"""
from .pums import build_pums_dfp_dfh
from geoid.acs import Puma
puma = Puma.parse(self.puma_geoid)
dfp, dfh = build_pums_dfp_dfh(puma.stusab, year=2018, release=5)
return dfp, dfh
def get_saved_frame(self):
if self.path.exists():
return pd.read_csv(self.path.resolve(), low_memory=False)
else:
return None
@property
def results_frame(self):
return pd.DataFrame({
'geoid': self.region_geoid,
'serialno': self.serialno,
'weight': self.allocated_weights
})
def save_frame(self):
self.path.parent.mkdir(parents=True, exist_ok=True)
df = pd.DataFrame({
'serialno': self.serialno,
'weight': self.allocated_weights
})
df = df[df.weight > 0]
df.to_csv(self.path, index=False)
def load_frame(self):
df = pd.read_csv(self.path, low_memory=False)
self.init()
aw, _ = df.align(self.sample_weights, axis=0)
self.allocated_weights = df.set_index('serialno').reindex(self.serialno).fillna(0).values[:, 0]
def inc(self, rown, n=1):
if self.allocated_weights[rown] > 0 or n > 0:
self.allocated_weights[rown] += n # Increment the count column
self.running_allocated_marginals += n * self.sample_pop.iloc[rown]
@property
def allocated_pop(self):
return self.sample_pop.mul(self.allocated_weights, axis=0)
@property
def allocated_marginals(self):
t = self.allocated_pop.sum()
t.name = 'allocated_marginals'
return t
def calc_region_sum(self):
return self.allocated_weights.sum()
def column_diff(self, column):
return (self.target_marginals.est[column] - self.allocated_marginals[column])
@property
def target_diff(self):
return self.target_marginals.est - self.allocated_marginals
@property
def rel_target_diff(self):
return ((self.target_marginals.est - self.allocated_marginals) / self.target_marginals.est) \
.replace({np.inf: 0, -np.inf: 0})
@property
def running_target_diff(self):
return self.target_marginals.est - self.running_allocated_marginals
@property
def error_frame(self):
return self.target_marginals \
.join(self.allocated_marginals.to_frame('allocated')) \
.join(self.m90_error.to_frame('m_90')) \
.join(self.target_diff.to_frame('diff')) \
.join(self.rel_target_diff.to_frame('rel'))
@property
def total_error(self):
"""Magnitude of the error vector"""
return np.sqrt(np.sum(np.square(self.target_diff)))
@property
def running_total_error(self):
"""Magnitude of the error vector"""
return np.sqrt(np.sum(np.square(self.running_target_diff)))
@property
def m90_error(self):
"""Error that is relative to the m90 limits. Any value within the m90 limits is an error of 0"""
# There the allocated marginal is withing the m90 range, return the target marginal estimate
# otherwise, return amount of the allocated marginals that is outside of the m90 range
t = self.allocated_marginals - self.target_marginals.est
t[self.allocated_marginals.between(self.target_marginals.est_min, self.target_marginals.est_max)] = 0
t[t > self.target_marginals.m90] = t - self.target_marginals.m90
t[t < -1 * self.target_marginals.m90] = t + self.target_marginals.m90
return t
@property
def m90_total_error(self):
return np.sqrt(np.sum(np.square(self.m90_error)))
@property
def m90_rms_error(self):
"""RMS error of the m90 differences. Like m90 total error, but divides
by the number of marginal value variables"""
return np.sqrt(np.sum(np.square(self.m90_total_error)) / len(self.target_marginals))
# Equivalent to cosine similarity when the vectors are both normalized
def cosine_similarities(self):
'''Calculate the cosine similaries for all of the sample population records
to the normalized error vector'''
return self.sample_pop_norm.dot(vector_normalize(self.target_diff.values).T)
def sample_multicol(self, columns):
targets = self.target_marginals.est
frames = []
for col in columns:
target = targets.loc[col]
if target > 0:
t = self.sample_pop[self.sample_pop[col] > 0]
w = self.sample_weights[self.sample_pop[col] > 0]
if len(t) > 0 and w.sum() > 0:
frames.append(sample_to_sum(target, t, col, w))
if frames:
return pd.concat(frames)
else:
return None
def _pop_to_weights(self, pop):
'''Return weights by counting the records in a population'''
t = pop.copy()
t.insert(0, 'dummy', 1)
t = t.groupby(t.index).dummy.count()
t = t.align(self.sample_weights)[0].fillna(0).values
return t
def initialize_weights_set_sample(self, f=0.85):
"""Sample from the sample population one column at a time, in groups of
columns that describe exclusive measures ( a household can contribute to
only one marginal column) Then, resample the population to match the correct number of
households"""
assert self._init
if f == 0:
return
frames = [
self.sample_multicol(self.hh_race_type_cols + self.gq_cols),
self.sample_multicol(self.hh_eth_type_cols),
self.sample_multicol(self.sex_age_cols),
]
frames = [f for f in frames if f is not None]
if len(frames) == 0:
return
# An initial population, which is of the wrong size, so just
# convert it to weights
t = pd.concat(frames)
initial_weights = self._pop_to_weights(t)
# These use those weights to re-sample the population.
target_count = self.household_count + self.gq_count
# Sample some fraction less than the target count, so we can vector walk to the final value
target_count = int(target_count * f)
t = self.sample_pop.sample(target_count, weights=initial_weights, replace=True)
self.allocated_weights = self._pop_to_weights(t)
self.unallocated_weights -= self.allocated_weights
self.running_allocated_marginals = self.allocated_marginals
def _rake(self, f=1):
# Sort the columns by the error
cols = list(self.error_frame.sort_values('diff', ascending=False).index)
# cols = random.sample(list(self.sample_pop.columns), len(self.sample_pop.columns)):
for col in cols:
b = self.sample_pop[col].mul(self.allocated_weights, axis=0).sum()
if b != 0:
a = self.target_marginals.loc[col].replace({pd.NA: 0}).est
r = a / b * f
self.allocated_weights[self.sample_pop[col] > 0] *= r
self.allocated_weights = np.round(self.allocated_weights, 0)
def initialize_weights_raking(self, n_iter=5, initial_weights='sample'):
"""Set the allocated weights to an initial value by 1-D raking, adjusting the
weights to fit the target marginal value for each column. """
if initial_weights == 'sample':
assert self.allocated_weights.shape == self.sample_weights.shape
self.allocated_weights = self.sample_weights
else:
self.allocated_weights = np.ones(self.allocated_weights.shape)
for i in range(n_iter):
# Sort the columns by the error
cols = list(self.error_frame.sort_values('diff', ascending=False).index)
# cols = random.sample(list(self.sample_pop.columns), len(self.sample_pop.columns)):
for col in cols:
b = self.sample_pop[col].mul(self.allocated_weights, axis=0).sum()
if b != 0:
a = self.target_marginals.loc[col].replace({pd.NA: 0}).est
r = a / b
self.allocated_weights[self.sample_pop[col] > 0] *= r
self.allocated_weights = np.round(self.allocated_weights, 0)
try:
self.allocated_weights = self.allocated_weights.values
except AttributeError:
pass
def initialize_weights_sample(self):
"""Initialize the allocated weights proportional to the sample population weights,
adjusted to the total population. """
self.allocated_weights = (self.sample_weights / (self.sample_weights.sum())).multiply(
self.household_count).values.round(0).astype(float)
self.unallocated_weights -= self.allocated_weights
def step_schedule_np(self, i, N, te, td, step_size_max, step_size_min, reversal_rate):
""" Return the next set of samples to add or remove
:param i: Loop index
:param N: Max number of iterations
:param cp: Sample population, transformed by make_cp
:param te: Total error
:param td: Marginals difference vector
:param step_size_max: Maximum step size
:param step_size_min: Minimum step size
:param reversal_rate: Probability to allow an increase in error
:param p: Probability to select each sample row. If None, use column 2 of cp
:return: Records to add or remove from the allocated population
"""
# Compute change in each column of the error vector for adding or subtracting in
# each of the sample population records
# idx 0 is the index of the row in self.sample_pop
# idx 1 is the sign, 1 or -1
# idx 2 is the selection weight
# idx 3 and up are the census count columns
expanded_pop = self.cp.values.astype(int)
p = expanded_pop[:, 2]
# For each new error vector, compute total error ( via vector length). By
# removing the current total error, we get the change in total error for
# adding or removing each row. ( positive values are better )
total_errors = (np.sqrt(np.square(expanded_pop[:, 3:] + td).sum(axis=1))) - te
# For error reducing records, sort them and then mutliply
# the weights by a linear ramp, so the larger values of
# reduction get a relative preference over the lower reduction values.
gt0 = np.argwhere(total_errors > 0).flatten() # Error reducing records
srt = np.argsort(total_errors) # Sorted by error
reducing_error = srt[np.in1d(srt, gt0)][::-1] # get the intersection. These are index values into self.cp
# Selection probabilities, multiply by linear ramp to preference higher values.
reducing_p = ((p[reducing_error]) * np.linspace(1, 0, len(reducing_error)))
rps = np.sum(reducing_p)
if rps > 0:
reducing_p = np.nan_to_num(reducing_p / rps)
else:
reducing_p = []
increasing_error = np.argwhere(total_errors < 0).flatten() # Error increasing indexes
increasing_p = p[increasing_error].flatten().clip(min=0)
ips = np.sum(increasing_p)
if ips != 0:
increasing_p = np.nan_to_num(increasing_p / ips) # normalize to 1
else:
increasing_p =[]
# Min number of record to return in this step. The error-increasing records are in
# addition to this number
step_size = int((step_size_max - step_size_min) * ((N - i) / N) + step_size_min)
# Randomly select from each group of increasing or reducing indexes.
cc = []
if len(increasing_error) > 0 and ips > 0:
cc.append(self.rng.choice(increasing_error, int(step_size * reversal_rate), p=increasing_p))
if len(reducing_error) > 0 and rps > 0:
cc.append(self.rng.choice(reducing_error, int(step_size), p=reducing_p))
idx = np.concatenate(cc)
# Columns are : 'index', 'sign', 'delta_err'
delta_err = total_errors[idx].reshape(-1, 1).round(0).astype(int)
return np.hstack([expanded_pop[idx][:, 0:2], delta_err]) # Return the index and sign columns of cp
def _loop_asignment(self, ss):
for j, (idx, sgn, *_) in enumerate(ss):
idx = int(idx)
if (self.allocated_weights[idx] > 0 and sgn < 0) or \
(self.unallocated_weights[idx]>0 and sgn > 0) :
self.running_allocated_marginals += (sgn * self.sample_pop.iloc[idx])
self.allocated_weights[idx] += sgn # Increment the count column
self.unallocated_weights[idx] -= sgn
def _numpy_assignment(self, ss):
# The following code is the numpy equivalent of the loop version of
# assignment to the allocated marginals. It is about 20% faster than the loop
# This selection on ss is the equivalent to this if statement in the loop version:
# if self.allocated_weights[idx] > 0 or sgn > 0:
#
ss = ss[np.logical_or(
np.isin(ss[:, 0], np.nonzero(self.allocated_weights > 0)), # self.allocated_weights[idx] > 0
ss[:, 1] > 0) # sgn > 0
]
# Assign the steps from the step schedule into the allocated weights
if len(ss):
idx = ss[:, 0].astype(int)
sgn = ss[:, 1]
# Update all weights by the array of signs
self.allocated_weights[idx] += sgn
# Don't allow negative weights
self.allocated_weights[self.allocated_weights < 0] = 0
# Add in the signed sampled to the running marginal, to save the cost
# of re-calculating the marginals.
self.running_allocated_marginals += \
np.multiply(self.sample_pop.iloc[idx], sgn.reshape(ss.shape[0], -1)).sum()
def _vector_walk(self, N=2000, min_iter=750, target_error=0.03,
step_size_min=3, step_size_max=15, reversal_rate=.3,
max_ssm=250, cb=None, memo=None):
"""Allocate PUMS records to this object's region.
Args:
N:
min_iter:
target_error:
step_size_min:
step_size_max:
reversal_rate:
max_ssm:
"""
assert self._init
if target_error < 1:
target_error = self.household_count * target_error
min_allocation = None # allocated weights at last minimum
steps_since_min = 0
min_error = self.total_error
self.running_allocated_marginals = self.allocated_marginals
if cb:
# vector_walk_callback(puma_task, tract_task, data, memo):
cb(memo.get('puma_task'), self, None, memo)
for i in range(N):
td = self.running_target_diff.values.astype(int)
te = vector_length(td)
# The unallocated weights can be updated both internally and externally --
# the array can be shared among all tracts in the puma
self.update_cp()
if te < min_error:
min_error = te
min_allocation = self.allocated_weights
steps_since_min = 0
else:
steps_since_min += 1
min_error = min(te, min_error)
if (i > min_iter and te < target_error) or steps_since_min > max_ssm:
break
try:
ss = self.step_schedule_np(i, N, te, td,
step_size_max, step_size_min, reversal_rate)
self._loop_asignment(ss)
yield (i, te, min_error, steps_since_min, len(ss))
except ValueError as e:
# Usually b/c numpy choice() got an empty array
pass
print(e)
raise
if min_allocation is not None:
self.allocated_weights = min_allocation
def vector_walk(self, N=2000, min_iter=750, target_error=0.03, step_size_min=3, step_size_max=10,
reversal_rate=.3, max_ssm=250, callback=None, memo=None,
stats = True):
"""Consider the target state and each household to be a vector. For each iteration
select a household vector with the best cosine similarity to the vector to the
target and add that household to the population. """
assert self._init
rows = []
ts = time()
errors = deque(maxlen=20)
errors.extend([self.total_error] * 20)
g = self._vector_walk(
N=N, min_iter=min_iter, target_error=target_error,
step_size_min=step_size_min, step_size_max=step_size_max,
reversal_rate=reversal_rate, max_ssm=max_ssm,
cb=callback, memo=memo)
if stats is not True:
list(g)
return []
else:
for i, te, min_error, steps_since_min, n_iter in g :
d = {'i': i, 'time': time() - ts, 'step_size': n_iter, 'error': te,
'target_error': target_error,
'total_error': te,
'size': np.sum(self.allocated_weights),
'ssm': steps_since_min,
'min_error': min_error,
'mean_error': np.mean(errors),
'std_error': np.std(errors),
'uw_sum': np.sum(self.unallocated_weights),
'total_count': self.total_count
}
rows.append(d)
errors.append(te)
if callback and i % 10 == 0:
# vector_walk_callback(puma_task, tract_task, data, memo):
callback(None, self, None, memo)
return rows
@classmethod
def get_us_tasks(cls, cache_dir, sl='tract', year=2018, release=5, limit=None, ignore_completed=True):
"""Return all of the tasks for all US states"""
from geoid.censusnames import stusab
tasks = []
for state in stusab.values():
state_tasks = cls.get_state_tasks(cache_dir, state, sl, year, release, limit, ignore_completed)
tasks.extend(state_tasks)
return tasks
@classmethod
def get_tasks(cls, cache_dir, state, sl='tract', year=2018, release=5,
limit=None, use_tqdm=False, ignore_completed=True):
if state.upper() == 'US':
return cls.get_us_tasks(cache_dir, sl, year, release, limit, use_tqdm, ignore_completed)
else:
return cls.get_state_tasks(cache_dir, state, sl, year, release, limit, ignore_completed)
@classmethod
def get_state_tasks(cls, cache_dir, state, sl='tract', year=2018, release=5,
limit=None, ignore_completed=True):
"""Fetch ( possibly download) the source data to generate allocation tasks,
and cache the data if a cache_dir is provided"""
from .acs import puma_tract_map
from synpums import build_acs, build_pums_households
from functools import partial
import pickle
_logger.info(f'Loading tasks for {state} from cache {cache_dir}')
cp = Path(cache_dir).joinpath('tasks', 'source', f"{state}-{year}-{release}/")
cp.mkdir(parents=True, exist_ok=True)
asc_p = cp.joinpath("acs.csv")
hh_p = cp.joinpath("households.csv")
tasks_p = cp.joinpath("tasks.pkl")
if limit:
from itertools import islice
limiter = partial(islice, limit)
else:
def limiter(g, *args, **kwargs):
yield from g
if tasks_p and tasks_p.exists():
with tasks_p.open('rb') as f:
_logger.debug(f"Returning cached tasks from {str(tasks_p)}")
return pickle.load(f)
# Cached ACS files
if asc_p and asc_p.exists():
tract_acs = pd.read_csv(asc_p, index_col='geoid', low_memory=False)
else:
tract_acs = build_acs(state, sl, year, release)
if asc_p:
tract_acs.to_csv(asc_p, index=True)
# Cached Households
if hh_p and hh_p.exists():
households = pd.read_csv(hh_p, index_col='SERIALNO', low_memory=False)
else:
households = build_pums_households(state, year=year, release=release)
if hh_p:
households.to_csv(hh_p, index=True)
hh = households.groupby('geoid')
hh_file_map = {}
for key, g in hh:
puma_p = cp.joinpath(f"pumas/{key}.csv")
puma_p.parent.mkdir(parents=True, exist_ok=True)
_logger.debug(f"Write puma file {str(puma_p)}")
g.to_csv(puma_p)
hh_file_map[key] = puma_p
pt_map = puma_tract_map()
tasks = []
for tract_geoid, targets in limiter(tract_acs.iterrows(), desc='Generate Tasks'):
try:
puma_geoid = pt_map[tract_geoid]
t = AllocationTask(tract_geoid, puma_geoid, asc_p, hh_file_map[puma_geoid], cache_dir)
if not t.path.exists() or ignore_completed is False:
tasks.append(t)
except Exception as e:
print("Error", tract_geoid, type(e), e)
if tasks_p:
with tasks_p.open('wb') as f:
_logger.debug(f"Write tasks file {str(tasks_p)}")
pickle.dump(tasks, f, pickle.HIGHEST_PROTOCOL)
return tasks
def run(self, *args, callback=None, memo=None, **kwargs):
self.init()
self.initialize_weights_sample()
rows = self.vector_walk(*args, callback=callback, memo=memo, **kwargs)
self.save_frame()
return rows
class PumaAllocator(object):
"""Simultaneously allocate all of the tracts in a pums, attempting to reduce the
error between the sum of the allocated weights and the PUMS weights"""
def __init__(self, puma_geoid, tasks, cache_dir, state, year=2018, release=5):
self.cache_dir = cache_dir
self.puma_geoid = puma_geoid
self.tasks = tasks
self.year = year
self.release = release
self.state = state
pums_files = [task.hh_ref for task in self.tasks]
assert all([e == pums_files[0] for e in pums_files])
self.pums_file = pums_files[0]
self._puma_target_marginals = None
self._puma_allocated_marginals = None
self._puma_max_weights = None
self._puma_allocated_weights = None
self._puma_unallocated_weights = None
self.pums = pd.read_csv(pums_files[0], low_memory=False)
self.weights = pd.DataFrame({
'allocated': 0,
'pums': self.pums.WGTP, # Original PUMS weights
'remaining': self.pums.WGTP # Remaining
})
self.prob = None
self.gq_cols = None
self.sex_age_cols = None
self.hh_size_cols = None
self.hh_race_type_cols = None
self.hh_eth_type_cols = None
self.hh_income_cols = None
self.replicate = 0
def init(self, init_method='sample'):
"""Initialize the weights of all of the tasks"""
from tqdm import tqdm
self.hh_ref = hh_source = pd.read_csv(self.tasks[0].hh_ref, index_col='SERIALNO', low_memory=False) \
.drop(columns=['geoid'], errors='ignore').astype('Int64')
self._puma_max_weights = hh_source.iloc[:, 0].reset_index(drop=True).astype(int)
self._puma_unallocated_weights = self._puma_max_weights.copy()
for task in tqdm(self.tasks):
task.init(puma_weights=self._puma_unallocated_weights)
if init_method == 'sample':
self.initialize_weights_sample(task)
if init_method == 'set':
task.initialize_weights_set_sample()
t0 = self.tasks[0] # Just to copy out some internal info.
self.gq_cols = t0.gq_cols
self.sex_age_cols = t0.sex_age_cols
self.hh_size_cols = t0.hh_size_cols
self.hh_race_type_cols = t0.hh_race_type_cols
self.hh_eth_type_cols = t0.hh_eth_type_cols
p = re.compile(r'b19025')
self.hh_income_cols = [c for c in t0.hh_source.columns if p.match(c)]
@classmethod
def get_tasks(cls, cache_dir, state, year=2018, release=5):
tasks = AllocationTask.get_state_tasks(cache_dir, state, sl='tract', year=2018, release=5)
puma_tasks = defaultdict(list)
for task in tasks:
puma_tasks[task.puma_geoid].append(task)
return puma_tasks
@classmethod
def get_allocators(cls, cache_dir, state, year=2018, release=5):
tasks = AllocationTask.get_state_tasks(cache_dir, state, sl='tract', year=2018, release=5)
puma_tasks = defaultdict(list)
for task in tasks:
puma_tasks[task.puma_geoid].append(task)
return [PumaAllocator(puma_geoid, tasks, cache_dir, state, year, release) for puma_geoid, tasks in
puma_tasks.items()]
def initialize_weights_sample(self, task, frac=.7):
"""Initialize the allocated weights proportional to the sample population weights,
adjusted to the total population. """
wf = self.weights_frame
assert wf.remaining.sum() != 0
wn1 = wf.remaining / wf.remaining.sum() # weights normalized to 1
task.allocated_weights = rand_round(wn1.multiply(task.household_count).values.astype(float))
task.unallocated_weights = np.clip(task.unallocated_weights-task.allocated_weights, a_min=0, a_max=None)
assert not any(task.unallocated_weights<0)
def vector_walk(self, N=1200, min_iter=5000, target_error=0.03, step_size_min=1,
step_size_max=10, reversal_rate=.3, max_ssm=150,
callback=None, memo=None):
"""Run a vector walk on all of the tracts tasks in this puma """
from itertools import cycle
rows = []
ts = time()
memo['puma_task'] = self
def make_vw(task):
return iter(task._vector_walk(
N=N, min_iter=min_iter, target_error=target_error,
step_size_min=step_size_min, step_size_max=step_size_max,
reversal_rate=reversal_rate, max_ssm=max_ssm,
cb=callback, memo=memo))
task_iters = [(task, make_vw(task)) for task in self.tasks]
stopped = set()
running = set([e[0] for e in task_iters])
memo['n_stopped'] = len(stopped)
memo['n_running'] = len(running)
memo['n_calls'] = 0
while True:
for task, task_iter in task_iters:
if task in running:
try:
i, te, min_error, steps_since_min, n_iter = next(task_iter)
memo['n_calls'] += 1
d = {'i': i, 'time': time() - ts, 'step_size': n_iter, 'error': te,
'target_error': target_error,
'size': np.sum(task.allocated_weights),
'ssm': steps_since_min,
'min_error': min_error,
'task': task
}
rows.append(d)
if callback and i % 10 == 0:
callback(self, task, d, memo)
except StopIteration:
stopped.add(task)
running.remove(task)
memo['n_stopped'] = len(stopped)
memo['n_running'] = len(running)
if len(running) == 0:
return rows
if callback:
# vector_walk_callback(puma_task, tract_task, data, memo):
callback(self, None, None, memo)
assert False # Should never get here.
def run(self, *args, callback=None, memo=None, **kwargs):
self.init(init_method='sample')
rows = self.vector_walk(*args, callback=callback, memo=memo, **kwargs)
self.save_frame()
return rows
def get_task(self, geoid):
for task in self.tasks:
if geoid == task.region_geoid:
return task
return None
def tune_puma_allocation(self):
"""Re-run all of the tasks in the puma, trying to reduce the discrepancy
between the """
task_iters = [(task, iter(task._vector_walk())) for task in self.tasks]
for task, task_iter in task_iters:
try:
task.cp_prob = self._update_probabilities()
row = next(task_iter)
print(task.region_geoid, self.rms_error, self.rms_weight_error, np.sum(task.cp_prob))
except StopIteration:
print(task.region_geoid, 'stopped')
@property
def weights_frame(self):
self.weights[
'allocated'] = self.puma_allocated_weights # np.sum(np.array([task.allocated_weights for task in self.tasks]), axis=0)
self.weights['remaining'] = self.weights.pums - self.weights.allocated
self.weights['dff'] = self.weights.allocated - self.weights.pums
self.weights['rdff'] = (self.weights.dff / self.weights.pums).fillna(0)
self.weights['p'] = self.weights.rdff
return self.weights
def _update_probabilities(self):
"""Update the running cp_probs, the probabilities for selecting each PUMS
household from the sample_pop, based on the error in weights for
the households at the Puma level"""
w = self.weights_frame
w['p_pos'] = - w.p.where(w.p < 0, 0)
w['p_neg'] = w.p.where(w.p > 0, 0)
self.prob = np.array(w[['p_neg', 'p_pos']].values.flat)
return self.prob
@property
def puma_target_marginals(self):
from .acs import build_acs
if self._puma_target_marginals is None:
_puma_marginals = build_acs(state=self.state, sl='puma', year=self.year, release=self.release)
cols = self.tasks[
0].target_marginals.index # [c for c in _puma_marginals.columns if c.startswith('b') and not c.endswith('_m90')]
self._puma_target_marginals = _puma_marginals.loc[self.puma_geoid][cols]
return self._puma_target_marginals
@property
def puma_allocated_marginals(self):
return self.allocated_marginals.sum()
@property
def allocated_marginals(self):
series = {task.region_geoid: task.allocated_marginals for task in self.tasks}
return pd.DataFrame(series).T
@property
def allocated_weights(self):
series = {task.region_geoid: task.allocated_weights for task in self.tasks}
return
|
pd.DataFrame(series)
|
pandas.DataFrame
|
#!/usr/bin/env python
# coding: utf-8
# # Manipulação de dados - II
# ## *DataFrames*
#
# O *DataFrame* é a segunda estrutura basilar do *pandas*. Um *DataFrame*:
# - é uma tabela, ou seja, é bidimensional;
# - tem cada coluna formada como uma *Series* do *pandas*;
# - pode ter *Series* contendo tipos de dado diferentes.
# In[1]:
import numpy as np
import pandas as pd
# ## Criação de um *DataFrame*
# O método padrão para criarmos um *DataFrame* é através de uma função com mesmo nome.
#
# ```python
# df_exemplo = pd.DataFrame(dados_de_interesse, index = indice_de_interesse,
# columns = colunas_de_interesse)
# ```
# Ao criar um *DataFrame*, podemos informar
# - `index`: rótulos para as linhas (atributos *index* das *Series*).
# - `columns`: rótulos para as colunas (atributos *name* das *Series*).
# No _template_, `dados_de_interesse` pode ser
#
# * um dicionário de:
# * *arrays* unidimensionais do *numpy*;
# * listas;
# * dicionários;
# * *Series* do *pandas*.
# * um *array* bidimensional do *numpy*;
# * uma *Series* do *Pandas*;
# * outro *DataFrame*.
# ### *DataFrame* a partir de dicionários de *Series*
#
# Neste método de criação, as *Series* do dicionário não precisam possuir o mesmo número de elementos. O *index* do *DataFrame* será dado pela **união** dos *index* de todas as *Series* contidas no dicionário.
# Exemplo:
# In[2]:
serie_Idade = pd.Series({'Ana':20, 'João': 19, 'Maria': 21, 'Pedro': 22}, name="Idade")
# In[3]:
serie_Peso = pd.Series({'Ana':55, 'João': 80, 'Maria': 62, 'Pedro': 67, 'Túlio': 73}, name="Peso")
# In[4]:
serie_Altura = pd.Series({'Ana':162, 'João': 178, 'Maria': 162, 'Pedro': 165, 'Túlio': 171}, name="Altura")
# In[5]:
dicionario_series_exemplo = {'Idade': serie_Idade, 'Peso': serie_Peso, 'Altura': serie_Altura}
# In[6]:
df_dict_series = pd.DataFrame(dicionario_series_exemplo)
# In[7]:
df_dict_series
# > Compare este resultado com a criação de uma planilha pelos métodos usuais. Veja que há muita flexibilidade para criarmos ou modificarmos uma tabela.
#
# Vejamos exemplos sobre como acessar intervalos de dados na tabela.
# In[8]:
pd.DataFrame(dicionario_series_exemplo, index=['João','Ana','Maria'])
# In[9]:
|
pd.DataFrame(dicionario_series_exemplo, index=['Ana','Maria'], columns=['Altura','Peso'])
|
pandas.DataFrame
|
# coding=utf-8
# pylint: disable-msg=E1101,W0612
from datetime import datetime, timedelta
from numpy import nan
import numpy as np
import pandas as pd
from pandas.types.common import is_integer, is_scalar
from pandas import Index, Series, DataFrame, isnull, date_range
from pandas.core.index import MultiIndex
from pandas.core.indexing import IndexingError
from pandas.tseries.index import Timestamp
from pandas.tseries.offsets import BDay
from pandas.tseries.tdi import Timedelta
from pandas.compat import lrange, range
from pandas import compat
from pandas.util.testing import assert_series_equal, assert_almost_equal
import pandas.util.testing as tm
from pandas.tests.series.common import TestData
JOIN_TYPES = ['inner', 'outer', 'left', 'right']
class TestSeriesIndexing(TestData, tm.TestCase):
_multiprocess_can_split_ = True
def test_get(self):
# GH 6383
s = Series(np.array([43, 48, 60, 48, 50, 51, 50, 45, 57, 48, 56, 45,
51, 39, 55, 43, 54, 52, 51, 54]))
result = s.get(25, 0)
expected = 0
self.assertEqual(result, expected)
s = Series(np.array([43, 48, 60, 48, 50, 51, 50, 45, 57, 48, 56,
45, 51, 39, 55, 43, 54, 52, 51, 54]),
index=pd.Float64Index(
[25.0, 36.0, 49.0, 64.0, 81.0, 100.0,
121.0, 144.0, 169.0, 196.0, 1225.0,
1296.0, 1369.0, 1444.0, 1521.0, 1600.0,
1681.0, 1764.0, 1849.0, 1936.0],
dtype='object'))
result = s.get(25, 0)
expected = 43
self.assertEqual(result, expected)
# GH 7407
# with a boolean accessor
df = pd.DataFrame({'i': [0] * 3, 'b': [False] * 3})
vc = df.i.value_counts()
result = vc.get(99, default='Missing')
self.assertEqual(result, 'Missing')
vc = df.b.value_counts()
result = vc.get(False, default='Missing')
self.assertEqual(result, 3)
result = vc.get(True, default='Missing')
self.assertEqual(result, 'Missing')
def test_delitem(self):
# GH 5542
# should delete the item inplace
s = Series(lrange(5))
del s[0]
expected = Series(lrange(1, 5), index=lrange(1, 5))
assert_series_equal(s, expected)
del s[1]
expected = Series(lrange(2, 5), index=lrange(2, 5))
assert_series_equal(s, expected)
# empty
s = Series()
def f():
del s[0]
self.assertRaises(KeyError, f)
# only 1 left, del, add, del
s = Series(1)
del s[0]
assert_series_equal(s, Series(dtype='int64', index=Index(
[], dtype='int64')))
s[0] = 1
assert_series_equal(s, Series(1))
del s[0]
assert_series_equal(s, Series(dtype='int64', index=Index(
[], dtype='int64')))
# Index(dtype=object)
s = Series(1, index=['a'])
del s['a']
assert_series_equal(s, Series(dtype='int64', index=Index(
[], dtype='object')))
s['a'] = 1
assert_series_equal(s, Series(1, index=['a']))
del s['a']
assert_series_equal(s, Series(dtype='int64', index=Index(
[], dtype='object')))
def test_getitem_setitem_ellipsis(self):
s = Series(np.random.randn(10))
np.fix(s)
result = s[...]
assert_series_equal(result, s)
s[...] = 5
self.assertTrue((result == 5).all())
def test_getitem_negative_out_of_bounds(self):
s = Series(tm.rands_array(5, 10), index=tm.rands_array(10, 10))
self.assertRaises(IndexError, s.__getitem__, -11)
self.assertRaises(IndexError, s.__setitem__, -11, 'foo')
def test_pop(self):
# GH 6600
df = DataFrame({'A': 0, 'B': np.arange(5, dtype='int64'), 'C': 0, })
k = df.iloc[4]
result = k.pop('B')
self.assertEqual(result, 4)
expected = Series([0, 0], index=['A', 'C'], name=4)
assert_series_equal(k, expected)
def test_getitem_get(self):
idx1 = self.series.index[5]
idx2 = self.objSeries.index[5]
self.assertEqual(self.series[idx1], self.series.get(idx1))
self.assertEqual(self.objSeries[idx2], self.objSeries.get(idx2))
self.assertEqual(self.series[idx1], self.series[5])
self.assertEqual(self.objSeries[idx2], self.objSeries[5])
self.assertEqual(
self.series.get(-1), self.series.get(self.series.index[-1]))
self.assertEqual(self.series[5], self.series.get(self.series.index[5]))
# missing
d = self.ts.index[0] - BDay()
self.assertRaises(KeyError, self.ts.__getitem__, d)
# None
# GH 5652
for s in [Series(), Series(index=list('abc'))]:
result = s.get(None)
self.assertIsNone(result)
def test_iget(self):
s = Series(np.random.randn(10), index=lrange(0, 20, 2))
# 10711, deprecated
with tm.assert_produces_warning(FutureWarning):
s.iget(1)
# 10711, deprecated
with tm.assert_produces_warning(FutureWarning):
s.irow(1)
# 10711, deprecated
with tm.assert_produces_warning(FutureWarning):
s.iget_value(1)
for i in range(len(s)):
result = s.iloc[i]
exp = s[s.index[i]]
assert_almost_equal(result, exp)
# pass a slice
result = s.iloc[slice(1, 3)]
expected = s.ix[2:4]
assert_series_equal(result, expected)
# test slice is a view
result[:] = 0
self.assertTrue((s[1:3] == 0).all())
# list of integers
result = s.iloc[[0, 2, 3, 4, 5]]
expected = s.reindex(s.index[[0, 2, 3, 4, 5]])
assert_series_equal(result, expected)
def test_iget_nonunique(self):
s = Series([0, 1, 2], index=[0, 1, 0])
self.assertEqual(s.iloc[2], 2)
def test_getitem_regression(self):
s = Series(lrange(5), index=lrange(5))
result = s[lrange(5)]
assert_series_equal(result, s)
def test_getitem_setitem_slice_bug(self):
s = Series(lrange(10), lrange(10))
result = s[-12:]
assert_series_equal(result, s)
result = s[-7:]
assert_series_equal(result, s[3:])
result = s[:-12]
assert_series_equal(result, s[:0])
s = Series(lrange(10), lrange(10))
s[-12:] = 0
self.assertTrue((s == 0).all())
s[:-12] = 5
self.assertTrue((s == 0).all())
def test_getitem_int64(self):
idx = np.int64(5)
self.assertEqual(self.ts[idx], self.ts[5])
def test_getitem_fancy(self):
slice1 = self.series[[1, 2, 3]]
slice2 = self.objSeries[[1, 2, 3]]
self.assertEqual(self.series.index[2], slice1.index[1])
self.assertEqual(self.objSeries.index[2], slice2.index[1])
self.assertEqual(self.series[2], slice1[1])
self.assertEqual(self.objSeries[2], slice2[1])
def test_getitem_boolean(self):
s = self.series
mask = s > s.median()
# passing list is OK
result = s[list(mask)]
expected = s[mask]
assert_series_equal(result, expected)
self.assert_index_equal(result.index, s.index[mask])
def test_getitem_boolean_empty(self):
s = Series([], dtype=np.int64)
s.index.name = 'index_name'
s = s[s.isnull()]
self.assertEqual(s.index.name, 'index_name')
self.assertEqual(s.dtype, np.int64)
# GH5877
# indexing with empty series
s = Series(['A', 'B'])
expected = Series(np.nan, index=['C'], dtype=object)
result = s[Series(['C'], dtype=object)]
assert_series_equal(result, expected)
s = Series(['A', 'B'])
expected = Series(dtype=object, index=Index([], dtype='int64'))
result = s[Series([], dtype=object)]
assert_series_equal(result, expected)
# invalid because of the boolean indexer
# that's empty or not-aligned
def f():
s[Series([], dtype=bool)]
self.assertRaises(IndexingError, f)
def f():
s[Series([True], dtype=bool)]
self.assertRaises(IndexingError, f)
def test_getitem_generator(self):
gen = (x > 0 for x in self.series)
result = self.series[gen]
result2 = self.series[iter(self.series > 0)]
expected = self.series[self.series > 0]
assert_series_equal(result, expected)
assert_series_equal(result2, expected)
def test_type_promotion(self):
# GH12599
s = pd.Series()
s["a"] = pd.Timestamp("2016-01-01")
s["b"] = 3.0
s["c"] = "foo"
expected = Series([pd.Timestamp("2016-01-01"), 3.0, "foo"],
index=["a", "b", "c"])
assert_series_equal(s, expected)
def test_getitem_boolean_object(self):
# using column from DataFrame
s = self.series
mask = s > s.median()
omask = mask.astype(object)
# getitem
result = s[omask]
expected = s[mask]
assert_series_equal(result, expected)
# setitem
s2 = s.copy()
cop = s.copy()
cop[omask] = 5
s2[mask] = 5
assert_series_equal(cop, s2)
# nans raise exception
omask[5:10] = np.nan
self.assertRaises(Exception, s.__getitem__, omask)
self.assertRaises(Exception, s.__setitem__, omask, 5)
def test_getitem_setitem_boolean_corner(self):
ts = self.ts
mask_shifted = ts.shift(1, freq=BDay()) > ts.median()
# these used to raise...??
self.assertRaises(Exception, ts.__getitem__, mask_shifted)
self.assertRaises(Exception, ts.__setitem__, mask_shifted, 1)
# ts[mask_shifted]
# ts[mask_shifted] = 1
self.assertRaises(Exception, ts.ix.__getitem__, mask_shifted)
self.assertRaises(Exception, ts.ix.__setitem__, mask_shifted, 1)
# ts.ix[mask_shifted]
# ts.ix[mask_shifted] = 2
def test_getitem_setitem_slice_integers(self):
s = Series(np.random.randn(8), index=[2, 4, 6, 8, 10, 12, 14, 16])
result = s[:4]
expected = s.reindex([2, 4, 6, 8])
assert_series_equal(result, expected)
s[:4] = 0
self.assertTrue((s[:4] == 0).all())
self.assertTrue(not (s[4:] == 0).any())
def test_getitem_out_of_bounds(self):
# don't segfault, GH #495
self.assertRaises(IndexError, self.ts.__getitem__, len(self.ts))
# GH #917
s = Series([])
self.assertRaises(IndexError, s.__getitem__, -1)
def test_getitem_setitem_integers(self):
# caused bug without test
s = Series([1, 2, 3], ['a', 'b', 'c'])
self.assertEqual(s.ix[0], s['a'])
s.ix[0] = 5
self.assertAlmostEqual(s['a'], 5)
def test_getitem_box_float64(self):
value = self.ts[5]
tm.assertIsInstance(value, np.float64)
def test_getitem_ambiguous_keyerror(self):
s = Series(lrange(10), index=lrange(0, 20, 2))
self.assertRaises(KeyError, s.__getitem__, 1)
self.assertRaises(KeyError, s.ix.__getitem__, 1)
def test_getitem_unordered_dup(self):
obj = Series(lrange(5), index=['c', 'a', 'a', 'b', 'b'])
self.assertTrue(is_scalar(obj['c']))
self.assertEqual(obj['c'], 0)
def test_getitem_dups_with_missing(self):
# breaks reindex, so need to use .ix internally
# GH 4246
s = Series([1, 2, 3, 4], ['foo', 'bar', 'foo', 'bah'])
expected = s.ix[['foo', 'bar', 'bah', 'bam']]
result = s[['foo', 'bar', 'bah', 'bam']]
assert_series_equal(result, expected)
def test_getitem_dups(self):
s = Series(range(5), index=['A', 'A', 'B', 'C', 'C'], dtype=np.int64)
expected = Series([3, 4], index=['C', 'C'], dtype=np.int64)
result = s['C']
assert_series_equal(result, expected)
def test_getitem_dataframe(self):
rng = list(range(10))
s = pd.Series(10, index=rng)
df = pd.DataFrame(rng, index=rng)
self.assertRaises(TypeError, s.__getitem__, df > 5)
def test_getitem_callable(self):
# GH 12533
s = pd.Series(4, index=list('ABCD'))
result = s[lambda x: 'A']
self.assertEqual(result, s.loc['A'])
result = s[lambda x: ['A', 'B']]
tm.assert_series_equal(result, s.loc[['A', 'B']])
result = s[lambda x: [True, False, True, True]]
tm.assert_series_equal(result, s.iloc[[0, 2, 3]])
def test_setitem_ambiguous_keyerror(self):
s = Series(lrange(10), index=lrange(0, 20, 2))
# equivalent of an append
s2 = s.copy()
s2[1] = 5
expected = s.append(Series([5], index=[1]))
assert_series_equal(s2, expected)
s2 = s.copy()
s2.ix[1] = 5
expected = s.append(Series([5], index=[1]))
assert_series_equal(s2, expected)
def test_setitem_float_labels(self):
# note labels are floats
s = Series(['a', 'b', 'c'], index=[0, 0.5, 1])
tmp = s.copy()
s.ix[1] = 'zoo'
tmp.iloc[2] = 'zoo'
assert_series_equal(s, tmp)
def test_setitem_callable(self):
# GH 12533
s = pd.Series([1, 2, 3, 4], index=list('ABCD'))
s[lambda x: 'A'] = -1
tm.assert_series_equal(s, pd.Series([-1, 2, 3, 4], index=list('ABCD')))
def test_setitem_other_callable(self):
# GH 13299
inc = lambda x: x + 1
s = pd.Series([1, 2, -1, 4])
s[s < 0] = inc
expected = pd.Series([1, 2, inc, 4])
tm.assert_series_equal(s, expected)
def test_slice(self):
numSlice = self.series[10:20]
numSliceEnd = self.series[-10:]
objSlice = self.objSeries[10:20]
self.assertNotIn(self.series.index[9], numSlice.index)
self.assertNotIn(self.objSeries.index[9], objSlice.index)
self.assertEqual(len(numSlice), len(numSlice.index))
self.assertEqual(self.series[numSlice.index[0]],
numSlice[numSlice.index[0]])
self.assertEqual(numSlice.index[1], self.series.index[11])
self.assertTrue(tm.equalContents(numSliceEnd, np.array(self.series)[
-10:]))
# test return view
sl = self.series[10:20]
sl[:] = 0
self.assertTrue((self.series[10:20] == 0).all())
def test_slice_can_reorder_not_uniquely_indexed(self):
s = Series(1, index=['a', 'a', 'b', 'b', 'c'])
s[::-1] # it works!
def test_slice_float_get_set(self):
self.assertRaises(TypeError, lambda: self.ts[4.0:10.0])
def f():
self.ts[4.0:10.0] = 0
self.assertRaises(TypeError, f)
self.assertRaises(TypeError, self.ts.__getitem__, slice(4.5, 10.0))
self.assertRaises(TypeError, self.ts.__setitem__, slice(4.5, 10.0), 0)
def test_slice_floats2(self):
s = Series(np.random.rand(10), index=np.arange(10, 20, dtype=float))
self.assertEqual(len(s.ix[12.0:]), 8)
self.assertEqual(len(s.ix[12.5:]), 7)
i = np.arange(10, 20, dtype=float)
i[2] = 12.2
s.index = i
self.assertEqual(len(s.ix[12.0:]), 8)
self.assertEqual(len(s.ix[12.5:]), 7)
def test_slice_float64(self):
values = np.arange(10., 50., 2)
index = Index(values)
start, end = values[[5, 15]]
s = Series(np.random.randn(20), index=index)
result = s[start:end]
expected = s.iloc[5:16]
assert_series_equal(result, expected)
result = s.loc[start:end]
assert_series_equal(result, expected)
df = DataFrame(np.random.randn(20, 3), index=index)
result = df[start:end]
expected = df.iloc[5:16]
tm.assert_frame_equal(result, expected)
result = df.loc[start:end]
tm.assert_frame_equal(result, expected)
def test_setitem(self):
self.ts[self.ts.index[5]] = np.NaN
self.ts[[1, 2, 17]] = np.NaN
self.ts[6] = np.NaN
self.assertTrue(np.isnan(self.ts[6]))
self.assertTrue(np.isnan(self.ts[2]))
self.ts[np.isnan(self.ts)] = 5
self.assertFalse(np.isnan(self.ts[2]))
# caught this bug when writing tests
series = Series(tm.makeIntIndex(20).astype(float),
index=tm.makeIntIndex(20))
series[::2] = 0
self.assertTrue((series[::2] == 0).all())
# set item that's not contained
s = self.series.copy()
s['foobar'] = 1
app = Series([1], index=['foobar'], name='series')
expected = self.series.append(app)
assert_series_equal(s, expected)
# Test for issue #10193
key = pd.Timestamp('2012-01-01')
series = pd.Series()
series[key] = 47
expected = pd.Series(47, [key])
assert_series_equal(series, expected)
series = pd.Series([], pd.DatetimeIndex([], freq='D'))
series[key] = 47
expected = pd.Series(47, pd.DatetimeIndex([key], freq='D'))
assert_series_equal(series, expected)
def test_setitem_dtypes(self):
# change dtypes
# GH 4463
expected = Series([np.nan, 2, 3])
s = Series([1, 2, 3])
s.iloc[0] = np.nan
assert_series_equal(s, expected)
s = Series([1, 2, 3])
s.loc[0] = np.nan
assert_series_equal(s, expected)
s = Series([1, 2, 3])
s[0] = np.nan
assert_series_equal(s, expected)
s = Series([False])
s.loc[0] = np.nan
assert_series_equal(s, Series([np.nan]))
s = Series([False, True])
s.loc[0] = np.nan
assert_series_equal(s, Series([np.nan, 1.0]))
def test_set_value(self):
idx = self.ts.index[10]
res = self.ts.set_value(idx, 0)
self.assertIs(res, self.ts)
self.assertEqual(self.ts[idx], 0)
# equiv
s = self.series.copy()
res = s.set_value('foobar', 0)
self.assertIs(res, s)
self.assertEqual(res.index[-1], 'foobar')
self.assertEqual(res['foobar'], 0)
s = self.series.copy()
s.loc['foobar'] = 0
self.assertEqual(s.index[-1], 'foobar')
self.assertEqual(s['foobar'], 0)
def test_setslice(self):
sl = self.ts[5:20]
self.assertEqual(len(sl), len(sl.index))
self.assertTrue(sl.index.is_unique)
def test_basic_getitem_setitem_corner(self):
# invalid tuples, e.g. self.ts[:, None] vs. self.ts[:, 2]
with tm.assertRaisesRegexp(ValueError, 'tuple-index'):
self.ts[:, 2]
with tm.assertRaisesRegexp(ValueError, 'tuple-index'):
self.ts[:, 2] = 2
# weird lists. [slice(0, 5)] will work but not two slices
result = self.ts[[slice(None, 5)]]
expected = self.ts[:5]
assert_series_equal(result, expected)
# OK
self.assertRaises(Exception, self.ts.__getitem__,
[5, slice(None, None)])
self.assertRaises(Exception, self.ts.__setitem__,
[5, slice(None, None)], 2)
def test_basic_getitem_with_labels(self):
indices = self.ts.index[[5, 10, 15]]
result = self.ts[indices]
expected = self.ts.reindex(indices)
assert_series_equal(result, expected)
result = self.ts[indices[0]:indices[2]]
expected = self.ts.ix[indices[0]:indices[2]]
assert_series_equal(result, expected)
# integer indexes, be careful
s = Series(np.random.randn(10), index=lrange(0, 20, 2))
inds = [0, 2, 5, 7, 8]
arr_inds = np.array([0, 2, 5, 7, 8])
result = s[inds]
expected = s.reindex(inds)
assert_series_equal(result, expected)
result = s[arr_inds]
expected = s.reindex(arr_inds)
assert_series_equal(result, expected)
# GH12089
# with tz for values
s = Series(pd.date_range("2011-01-01", periods=3, tz="US/Eastern"),
index=['a', 'b', 'c'])
expected = Timestamp('2011-01-01', tz='US/Eastern')
result = s.loc['a']
self.assertEqual(result, expected)
result = s.iloc[0]
self.assertEqual(result, expected)
result = s['a']
self.assertEqual(result, expected)
def test_basic_setitem_with_labels(self):
indices = self.ts.index[[5, 10, 15]]
cp = self.ts.copy()
exp = self.ts.copy()
cp[indices] = 0
exp.ix[indices] = 0
assert_series_equal(cp, exp)
cp = self.ts.copy()
exp = self.ts.copy()
cp[indices[0]:indices[2]] = 0
exp.ix[indices[0]:indices[2]] = 0
assert_series_equal(cp, exp)
# integer indexes, be careful
s = Series(np.random.randn(10), index=lrange(0, 20, 2))
inds = [0, 4, 6]
arr_inds = np.array([0, 4, 6])
cp = s.copy()
exp = s.copy()
s[inds] = 0
s.ix[inds] = 0
assert_series_equal(cp, exp)
cp = s.copy()
exp = s.copy()
s[arr_inds] = 0
s.ix[arr_inds] = 0
assert_series_equal(cp, exp)
inds_notfound = [0, 4, 5, 6]
arr_inds_notfound = np.array([0, 4, 5, 6])
self.assertRaises(Exception, s.__setitem__, inds_notfound, 0)
self.assertRaises(Exception, s.__setitem__, arr_inds_notfound, 0)
# GH12089
# with tz for values
s = Series(pd.date_range("2011-01-01", periods=3, tz="US/Eastern"),
index=['a', 'b', 'c'])
s2 = s.copy()
expected = Timestamp('2011-01-03', tz='US/Eastern')
s2.loc['a'] = expected
result = s2.loc['a']
self.assertEqual(result, expected)
s2 = s.copy()
s2.iloc[0] = expected
result = s2.iloc[0]
self.assertEqual(result, expected)
s2 = s.copy()
s2['a'] = expected
result = s2['a']
self.assertEqual(result, expected)
def test_ix_getitem(self):
inds = self.series.index[[3, 4, 7]]
assert_series_equal(self.series.ix[inds], self.series.reindex(inds))
assert_series_equal(self.series.ix[5::2], self.series[5::2])
# slice with indices
d1, d2 = self.ts.index[[5, 15]]
result = self.ts.ix[d1:d2]
expected = self.ts.truncate(d1, d2)
assert_series_equal(result, expected)
# boolean
mask = self.series > self.series.median()
assert_series_equal(self.series.ix[mask], self.series[mask])
# ask for index value
self.assertEqual(self.ts.ix[d1], self.ts[d1])
self.assertEqual(self.ts.ix[d2], self.ts[d2])
def test_ix_getitem_not_monotonic(self):
d1, d2 = self.ts.index[[5, 15]]
ts2 = self.ts[::2][[1, 2, 0]]
self.assertRaises(KeyError, ts2.ix.__getitem__, slice(d1, d2))
self.assertRaises(KeyError, ts2.ix.__setitem__, slice(d1, d2), 0)
def test_ix_getitem_setitem_integer_slice_keyerrors(self):
s = Series(np.random.randn(10), index=lrange(0, 20, 2))
# this is OK
cp = s.copy()
cp.ix[4:10] = 0
self.assertTrue((cp.ix[4:10] == 0).all())
# so is this
cp = s.copy()
cp.ix[3:11] = 0
self.assertTrue((cp.ix[3:11] == 0).values.all())
result = s.ix[4:10]
result2 = s.ix[3:11]
expected = s.reindex([4, 6, 8, 10])
assert_series_equal(result, expected)
assert_series_equal(result2, expected)
# non-monotonic, raise KeyError
s2 = s.iloc[lrange(5) + lrange(5, 10)[::-1]]
self.assertRaises(KeyError, s2.ix.__getitem__, slice(3, 11))
self.assertRaises(KeyError, s2.ix.__setitem__, slice(3, 11), 0)
def test_ix_getitem_iterator(self):
idx = iter(self.series.index[:10])
result = self.series.ix[idx]
assert_series_equal(result, self.series[:10])
def test_setitem_with_tz(self):
for tz in ['US/Eastern', 'UTC', 'Asia/Tokyo']:
orig = pd.Series(pd.date_range('2016-01-01', freq='H', periods=3,
tz=tz))
self.assertEqual(orig.dtype, 'datetime64[ns, {0}]'.format(tz))
# scalar
s = orig.copy()
s[1] = pd.Timestamp('2011-01-01', tz=tz)
exp = pd.Series([pd.Timestamp('2016-01-01 00:00', tz=tz),
pd.Timestamp('2011-01-01 00:00', tz=tz),
pd.Timestamp('2016-01-01 02:00', tz=tz)])
tm.assert_series_equal(s, exp)
s = orig.copy()
s.loc[1] = pd.Timestamp('2011-01-01', tz=tz)
tm.assert_series_equal(s, exp)
s = orig.copy()
s.iloc[1] = pd.Timestamp('2011-01-01', tz=tz)
tm.assert_series_equal(s, exp)
# vector
vals = pd.Series([pd.Timestamp('2011-01-01', tz=tz),
pd.Timestamp('2012-01-01', tz=tz)], index=[1, 2])
self.assertEqual(vals.dtype, 'datetime64[ns, {0}]'.format(tz))
s[[1, 2]] = vals
exp = pd.Series([pd.Timestamp('2016-01-01 00:00', tz=tz),
pd.Timestamp('2011-01-01 00:00', tz=tz),
pd.Timestamp('2012-01-01 00:00', tz=tz)])
tm.assert_series_equal(s, exp)
s = orig.copy()
s.loc[[1, 2]] = vals
tm.assert_series_equal(s, exp)
s = orig.copy()
s.iloc[[1, 2]] = vals
tm.assert_series_equal(s, exp)
def test_setitem_with_tz_dst(self):
# GH XXX
tz = 'US/Eastern'
orig = pd.Series(pd.date_range('2016-11-06', freq='H', periods=3,
tz=tz))
self.assertEqual(orig.dtype, 'datetime64[ns, {0}]'.format(tz))
# scalar
s = orig.copy()
s[1] = pd.Timestamp('2011-01-01', tz=tz)
exp = pd.Series([pd.Timestamp('2016-11-06 00:00', tz=tz),
pd.Timestamp('2011-01-01 00:00', tz=tz),
pd.Timestamp('2016-11-06 02:00', tz=tz)])
tm.assert_series_equal(s, exp)
s = orig.copy()
s.loc[1] = pd.Timestamp('2011-01-01', tz=tz)
tm.assert_series_equal(s, exp)
s = orig.copy()
s.iloc[1] = pd.Timestamp('2011-01-01', tz=tz)
tm.assert_series_equal(s, exp)
# vector
vals = pd.Series([pd.Timestamp('2011-01-01', tz=tz),
pd.Timestamp('2012-01-01', tz=tz)], index=[1, 2])
self.assertEqual(vals.dtype, 'datetime64[ns, {0}]'.format(tz))
s[[1, 2]] = vals
exp = pd.Series([pd.Timestamp('2016-11-06 00:00', tz=tz),
pd.Timestamp('2011-01-01 00:00', tz=tz),
pd.Timestamp('2012-01-01 00:00', tz=tz)])
tm.assert_series_equal(s, exp)
s = orig.copy()
s.loc[[1, 2]] = vals
tm.assert_series_equal(s, exp)
s = orig.copy()
s.iloc[[1, 2]] = vals
tm.assert_series_equal(s, exp)
def test_where(self):
s = Series(np.random.randn(5))
cond = s > 0
rs = s.where(cond).dropna()
rs2 = s[cond]
assert_series_equal(rs, rs2)
rs = s.where(cond, -s)
assert_series_equal(rs, s.abs())
rs = s.where(cond)
assert (s.shape == rs.shape)
assert (rs is not s)
# test alignment
cond = Series([True, False, False, True, False], index=s.index)
s2 = -(s.abs())
expected = s2[cond].reindex(s2.index[:3]).reindex(s2.index)
rs = s2.where(cond[:3])
assert_series_equal(rs, expected)
expected = s2.abs()
expected.ix[0] = s2[0]
rs = s2.where(cond[:3], -s2)
assert_series_equal(rs, expected)
self.assertRaises(ValueError, s.where, 1)
self.assertRaises(ValueError, s.where, cond[:3].values, -s)
# GH 2745
s = Series([1, 2])
s[[True, False]] = [0, 1]
expected = Series([0, 2])
assert_series_equal(s, expected)
# failures
self.assertRaises(ValueError, s.__setitem__, tuple([[[True, False]]]),
[0, 2, 3])
self.assertRaises(ValueError, s.__setitem__, tuple([[[True, False]]]),
[])
# unsafe dtype changes
for dtype in [np.int8, np.int16, np.int32, np.int64, np.float16,
np.float32, np.float64]:
s = Series(np.arange(10), dtype=dtype)
mask = s < 5
s[mask] = lrange(2, 7)
expected = Series(lrange(2, 7) + lrange(5, 10), dtype=dtype)
assert_series_equal(s, expected)
self.assertEqual(s.dtype, expected.dtype)
# these are allowed operations, but are upcasted
for dtype in [np.int64, np.float64]:
s = Series(np.arange(10), dtype=dtype)
mask = s < 5
values = [2.5, 3.5, 4.5, 5.5, 6.5]
s[mask] = values
expected = Series(values + lrange(5, 10), dtype='float64')
assert_series_equal(s, expected)
self.assertEqual(s.dtype, expected.dtype)
# GH 9731
s = Series(np.arange(10), dtype='int64')
mask = s > 5
values = [2.5, 3.5, 4.5, 5.5]
s[mask] = values
expected = Series(lrange(6) + values, dtype='float64')
assert_series_equal(s, expected)
# can't do these as we are forced to change the itemsize of the input
# to something we cannot
for dtype in [np.int8, np.int16, np.int32, np.float16, np.float32]:
s = Series(np.arange(10), dtype=dtype)
mask = s < 5
values = [2.5, 3.5, 4.5, 5.5, 6.5]
self.assertRaises(Exception, s.__setitem__, tuple(mask), values)
# GH3235
s = Series(np.arange(10), dtype='int64')
mask = s < 5
s[mask] = lrange(2, 7)
expected = Series(lrange(2, 7) + lrange(5, 10), dtype='int64')
assert_series_equal(s, expected)
self.assertEqual(s.dtype, expected.dtype)
s = Series(np.arange(10), dtype='int64')
mask = s > 5
s[mask] = [0] * 4
expected = Series([0, 1, 2, 3, 4, 5] + [0] * 4, dtype='int64')
assert_series_equal(s, expected)
s = Series(np.arange(10))
mask = s > 5
def f():
s[mask] = [5, 4, 3, 2, 1]
self.assertRaises(ValueError, f)
def f():
s[mask] = [0] * 5
self.assertRaises(ValueError, f)
# dtype changes
s = Series([1, 2, 3, 4])
result = s.where(s > 2, np.nan)
expected = Series([np.nan, np.nan, 3, 4])
assert_series_equal(result, expected)
# GH 4667
# setting with None changes dtype
s = Series(range(10)).astype(float)
s[8] = None
result = s[8]
self.assertTrue(isnull(result))
s = Series(range(10)).astype(float)
s[s > 8] = None
result = s[isnull(s)]
expected = Series(np.nan, index=[9])
assert_series_equal(result, expected)
def test_where_setitem_invalid(self):
# GH 2702
# make sure correct exceptions are raised on invalid list assignment
# slice
s = Series(list('abc'))
def f():
s[0:3] = list(range(27))
self.assertRaises(ValueError, f)
s[0:3] = list(range(3))
expected = Series([0, 1, 2])
assert_series_equal(s.astype(np.int64), expected, )
# slice with step
s = Series(list('abcdef'))
def f():
s[0:4:2] = list(range(27))
self.assertRaises(ValueError, f)
s = Series(list('abcdef'))
s[0:4:2] = list(range(2))
expected = Series([0, 'b', 1, 'd', 'e', 'f'])
assert_series_equal(s, expected)
# neg slices
s = Series(list('abcdef'))
def f():
s[:-1] = list(range(27))
self.assertRaises(ValueError, f)
s[-3:-1] = list(range(2))
expected = Series(['a', 'b', 'c', 0, 1, 'f'])
assert_series_equal(s, expected)
# list
s = Series(list('abc'))
def f():
s[[0, 1, 2]] = list(range(27))
self.assertRaises(ValueError, f)
s = Series(list('abc'))
def f():
s[[0, 1, 2]] = list(range(2))
self.assertRaises(ValueError, f)
# scalar
s = Series(list('abc'))
s[0] = list(range(10))
expected = Series([list(range(10)), 'b', 'c'])
assert_series_equal(s, expected)
def test_where_broadcast(self):
# Test a variety of differently sized series
for size in range(2, 6):
# Test a variety of boolean indices
for selection in [
# First element should be set
np.resize([True, False, False, False, False], size),
# Set alternating elements]
np.resize([True, False], size),
# No element should be set
np.resize([False], size)]:
# Test a variety of different numbers as content
for item in [2.0, np.nan, np.finfo(np.float).max,
np.finfo(np.float).min]:
# Test numpy arrays, lists and tuples as the input to be
# broadcast
for arr in [np.array([item]), [item], (item, )]:
data = np.arange(size, dtype=float)
s = Series(data)
s[selection] = arr
# Construct the expected series by taking the source
# data or item based on the selection
expected = Series([item if use_item else data[
i] for i, use_item in enumerate(selection)])
assert_series_equal(s, expected)
s = Series(data)
result = s.where(~selection, arr)
assert_series_equal(result, expected)
def test_where_inplace(self):
s = Series(np.random.randn(5))
cond = s > 0
rs = s.copy()
rs.where(cond, inplace=True)
assert_series_equal(rs.dropna(), s[cond])
assert_series_equal(rs, s.where(cond))
rs = s.copy()
rs.where(cond, -s, inplace=True)
assert_series_equal(rs, s.where(cond, -s))
def test_where_dups(self):
# GH 4550
# where crashes with dups in index
s1 = Series(list(range(3)))
s2 = Series(list(range(3)))
comb = pd.concat([s1, s2])
result = comb.where(comb < 2)
expected = Series([0, 1, np.nan, 0, 1, np.nan],
index=[0, 1, 2, 0, 1, 2])
assert_series_equal(result, expected)
# GH 4548
# inplace updating not working with dups
comb[comb < 1] = 5
expected = Series([5, 1, 2, 5, 1, 2], index=[0, 1, 2, 0, 1, 2])
assert_series_equal(comb, expected)
comb[comb < 2] += 10
expected = Series([5, 11, 2, 5, 11, 2], index=[0, 1, 2, 0, 1, 2])
assert_series_equal(comb, expected)
def test_where_datetime(self):
s = Series(date_range('20130102', periods=2))
expected = Series([10, 10], dtype='datetime64[ns]')
mask = np.array([False, False])
rs = s.where(mask, [10, 10])
assert_series_equal(rs, expected)
rs = s.where(mask, 10)
assert_series_equal(rs, expected)
rs = s.where(mask, 10.0)
assert_series_equal(rs, expected)
rs = s.where(mask, [10.0, 10.0])
assert_series_equal(rs, expected)
rs = s.where(mask, [10.0, np.nan])
expected = Series([10, None], dtype='datetime64[ns]')
assert_series_equal(rs, expected)
def test_where_timedelta(self):
s = Series([1, 2], dtype='timedelta64[ns]')
expected = Series([10, 10], dtype='timedelta64[ns]')
mask = np.array([False, False])
rs = s.where(mask, [10, 10])
assert_series_equal(rs, expected)
rs = s.where(mask, 10)
assert_series_equal(rs, expected)
rs = s.where(mask, 10.0)
assert_series_equal(rs, expected)
rs = s.where(mask, [10.0, 10.0])
assert_series_equal(rs, expected)
rs = s.where(mask, [10.0, np.nan])
expected = Series([10, None], dtype='timedelta64[ns]')
assert_series_equal(rs, expected)
def test_mask(self):
# compare with tested results in test_where
s = Series(np.random.randn(5))
cond = s > 0
rs = s.where(~cond, np.nan)
assert_series_equal(rs, s.mask(cond))
rs = s.where(~cond)
rs2 = s.mask(cond)
assert_series_equal(rs, rs2)
rs = s.where(~cond, -s)
rs2 = s.mask(cond, -s)
assert_series_equal(rs, rs2)
cond = Series([True, False, False, True, False], index=s.index)
s2 = -(s.abs())
rs = s2.where(~cond[:3])
rs2 = s2.mask(cond[:3])
assert_series_equal(rs, rs2)
rs = s2.where(~cond[:3], -s2)
rs2 = s2.mask(cond[:3], -s2)
assert_series_equal(rs, rs2)
self.assertRaises(ValueError, s.mask, 1)
self.assertRaises(ValueError, s.mask, cond[:3].values, -s)
# dtype changes
s = Series([1, 2, 3, 4])
result = s.mask(s > 2, np.nan)
expected = Series([1, 2, np.nan, np.nan])
assert_series_equal(result, expected)
def test_mask_broadcast(self):
# GH 8801
# copied from test_where_broadcast
for size in range(2, 6):
for selection in [
# First element should be set
np.resize([True, False, False, False, False], size),
# Set alternating elements]
np.resize([True, False], size),
# No element should be set
np.resize([False], size)]:
for item in [2.0, np.nan, np.finfo(np.float).max,
np.finfo(np.float).min]:
for arr in [np.array([item]), [item], (item, )]:
data = np.arange(size, dtype=float)
s = Series(data)
result = s.mask(selection, arr)
expected = Series([item if use_item else data[
i] for i, use_item in enumerate(selection)])
assert_series_equal(result, expected)
def test_mask_inplace(self):
s = Series(np.random.randn(5))
cond = s > 0
rs = s.copy()
rs.mask(cond, inplace=True)
assert_series_equal(rs.dropna(), s[~cond])
assert_series_equal(rs, s.mask(cond))
rs = s.copy()
rs.mask(cond, -s, inplace=True)
assert_series_equal(rs, s.mask(cond, -s))
def test_ix_setitem(self):
inds = self.series.index[[3, 4, 7]]
result = self.series.copy()
result.ix[inds] = 5
expected = self.series.copy()
expected[[3, 4, 7]] = 5
assert_series_equal(result, expected)
result.ix[5:10] = 10
expected[5:10] = 10
assert_series_equal(result, expected)
# set slice with indices
d1, d2 = self.series.index[[5, 15]]
result.ix[d1:d2] = 6
expected[5:16] = 6 # because it's inclusive
assert_series_equal(result, expected)
# set index value
self.series.ix[d1] = 4
self.series.ix[d2] = 6
self.assertEqual(self.series[d1], 4)
self.assertEqual(self.series[d2], 6)
def test_where_numeric_with_string(self):
# GH 9280
s = pd.Series([1, 2, 3])
w = s.where(s > 1, 'X')
self.assertFalse(is_integer(w[0]))
self.assertTrue(is_integer(w[1]))
self.assertTrue(is_integer(w[2]))
self.assertTrue(isinstance(w[0], str))
self.assertTrue(w.dtype == 'object')
w = s.where(s > 1, ['X', 'Y', 'Z'])
self.assertFalse(is_integer(w[0]))
self.assertTrue(is_integer(w[1]))
self.assertTrue(is_integer(w[2]))
self.assertTrue(isinstance(w[0], str))
self.assertTrue(w.dtype == 'object')
w = s.where(s > 1, np.array(['X', 'Y', 'Z']))
self.assertFalse(is_integer(w[0]))
self.assertTrue(is_integer(w[1]))
self.assertTrue(is_integer(w[2]))
self.assertTrue(isinstance(w[0], str))
self.assertTrue(w.dtype == 'object')
def test_setitem_boolean(self):
mask = self.series > self.series.median()
# similiar indexed series
result = self.series.copy()
result[mask] = self.series * 2
expected = self.series * 2
assert_series_equal(result[mask], expected[mask])
# needs alignment
result = self.series.copy()
result[mask] = (self.series * 2)[0:5]
expected = (self.series * 2)[0:5].reindex_like(self.series)
expected[-mask] = self.series[mask]
assert_series_equal(result[mask], expected[mask])
def test_ix_setitem_boolean(self):
mask = self.series > self.series.median()
result = self.series.copy()
result.ix[mask] = 0
expected = self.series
expected[mask] = 0
assert_series_equal(result, expected)
def test_ix_setitem_corner(self):
inds = list(self.series.index[[5, 8, 12]])
self.series.ix[inds] = 5
self.assertRaises(Exception, self.series.ix.__setitem__,
inds + ['foo'], 5)
def test_get_set_boolean_different_order(self):
ordered = self.series.sort_values()
# setting
copy = self.series.copy()
copy[ordered > 0] = 0
expected = self.series.copy()
expected[expected > 0] = 0
assert_series_equal(copy, expected)
# getting
sel = self.series[ordered > 0]
exp = self.series[self.series > 0]
assert_series_equal(sel, exp)
def test_setitem_na(self):
# these induce dtype changes
expected = Series([np.nan, 3, np.nan, 5, np.nan, 7, np.nan, 9, np.nan])
s = Series([2, 3, 4, 5, 6, 7, 8, 9, 10])
s[::2] = np.nan
assert_series_equal(s, expected)
# get's coerced to float, right?
expected = Series([np.nan, 1, np.nan, 0])
s = Series([True, True, False, False])
s[::2] = np.nan
assert_series_equal(s, expected)
expected = Series([np.nan, np.nan, np.nan, np.nan, np.nan, 5, 6, 7, 8,
9])
s = Series(np.arange(10))
s[:5] = np.nan
assert_series_equal(s, expected)
def test_basic_indexing(self):
s = Series(np.random.randn(5), index=['a', 'b', 'a', 'a', 'b'])
self.assertRaises(IndexError, s.__getitem__, 5)
self.assertRaises(IndexError, s.__setitem__, 5, 0)
self.assertRaises(KeyError, s.__getitem__, 'c')
s = s.sort_index()
self.assertRaises(IndexError, s.__getitem__, 5)
self.assertRaises(IndexError, s.__setitem__, 5, 0)
def test_int_indexing(self):
s = Series(np.random.randn(6), index=[0, 0, 1, 1, 2, 2])
self.assertRaises(KeyError, s.__getitem__, 5)
self.assertRaises(KeyError, s.__getitem__, 'c')
# not monotonic
s = Series(np.random.randn(6), index=[2, 2, 0, 0, 1, 1])
self.assertRaises(KeyError, s.__getitem__, 5)
self.assertRaises(KeyError, s.__getitem__, 'c')
def test_datetime_indexing(self):
from pandas import date_range
index = date_range('1/1/2000', '1/7/2000')
index = index.repeat(3)
s = Series(len(index), index=index)
stamp = Timestamp('1/8/2000')
self.assertRaises(KeyError, s.__getitem__, stamp)
s[stamp] = 0
self.assertEqual(s[stamp], 0)
# not monotonic
s = Series(len(index), index=index)
s = s[::-1]
self.assertRaises(KeyError, s.__getitem__, stamp)
s[stamp] = 0
self.assertEqual(s[stamp], 0)
def test_timedelta_assignment(self):
# GH 8209
s = Series([])
s.loc['B'] = timedelta(1)
tm.assert_series_equal(s, Series(Timedelta('1 days'), index=['B']))
s = s.reindex(s.index.insert(0, 'A'))
tm.assert_series_equal(s, Series(
[np.nan, Timedelta('1 days')], index=['A', 'B']))
result = s.fillna(timedelta(1))
expected = Series(Timedelta('1 days'), index=['A', 'B'])
tm.assert_series_equal(result, expected)
s.loc['A'] = timedelta(1)
tm.assert_series_equal(s, expected)
# GH 14155
s = Series(10 * [np.timedelta64(10, 'm')])
s.loc[[1, 2, 3]] = np.timedelta64(20, 'm')
expected = pd.Series(10 * [np.timedelta64(10, 'm')])
expected.loc[[1, 2, 3]] = pd.Timedelta(np.timedelta64(20, 'm'))
tm.assert_series_equal(s, expected)
def test_underlying_data_conversion(self):
# GH 4080
df = DataFrame(dict((c, [1, 2, 3]) for c in ['a', 'b', 'c']))
df.set_index(['a', 'b', 'c'], inplace=True)
s = Series([1], index=[(2, 2, 2)])
df['val'] = 0
df
df['val'].update(s)
expected = DataFrame(
dict(a=[1, 2, 3], b=[1, 2, 3], c=[1, 2, 3], val=[0, 1, 0]))
expected.set_index(['a', 'b', 'c'], inplace=True)
tm.assert_frame_equal(df, expected)
# GH 3970
# these are chained assignments as well
pd.set_option('chained_assignment', None)
df = DataFrame({"aa": range(5), "bb": [2.2] * 5})
df["cc"] = 0.0
ck = [True] * len(df)
df["bb"].iloc[0] = .13
# TODO: unused
df_tmp = df.iloc[ck] # noqa
df["bb"].iloc[0] = .15
self.assertEqual(df['bb'].iloc[0], 0.15)
pd.set_option('chained_assignment', 'raise')
# GH 3217
df = DataFrame(dict(a=[1, 3], b=[np.nan, 2]))
df['c'] = np.nan
df['c'].update(pd.Series(['foo'], index=[0]))
expected = DataFrame(dict(a=[1, 3], b=[np.nan, 2], c=['foo', np.nan]))
tm.assert_frame_equal(df, expected)
def test_preserveRefs(self):
seq = self.ts[[5, 10, 15]]
seq[1] = np.NaN
self.assertFalse(np.isnan(self.ts[10]))
def test_drop(self):
# unique
s = Series([1, 2], index=['one', 'two'])
expected = Series([1], index=['one'])
result = s.drop(['two'])
assert_series_equal(result, expected)
result = s.drop('two', axis='rows')
assert_series_equal(result, expected)
# non-unique
# GH 5248
s = Series([1, 1, 2], index=['one', 'two', 'one'])
expected = Series([1, 2], index=['one', 'one'])
result = s.drop(['two'], axis=0)
assert_series_equal(result, expected)
result = s.drop('two')
assert_series_equal(result, expected)
expected = Series([1], index=['two'])
result = s.drop(['one'])
assert_series_equal(result, expected)
result = s.drop('one')
assert_series_equal(result, expected)
# single string/tuple-like
s = Series(range(3), index=list('abc'))
self.assertRaises(ValueError, s.drop, 'bc')
self.assertRaises(ValueError, s.drop, ('a', ))
# errors='ignore'
s = Series(range(3), index=list('abc'))
result = s.drop('bc', errors='ignore')
assert_series_equal(result, s)
result = s.drop(['a', 'd'], errors='ignore')
expected = s.ix[1:]
assert_series_equal(result, expected)
# bad axis
self.assertRaises(ValueError, s.drop, 'one', axis='columns')
# GH 8522
s = Series([2, 3], index=[True, False])
self.assertTrue(s.index.is_object())
result = s.drop(True)
expected = Series([3], index=[False])
assert_series_equal(result, expected)
def test_align(self):
def _check_align(a, b, how='left', fill=None):
aa, ab = a.align(b, join=how, fill_value=fill)
join_index = a.index.join(b.index, how=how)
if fill is not None:
diff_a = aa.index.difference(join_index)
diff_b = ab.index.difference(join_index)
if len(diff_a) > 0:
self.assertTrue((aa.reindex(diff_a) == fill).all())
if len(diff_b) > 0:
self.assertTrue((ab.reindex(diff_b) == fill).all())
ea = a.reindex(join_index)
eb = b.reindex(join_index)
if fill is not None:
ea = ea.fillna(fill)
eb = eb.fillna(fill)
assert_series_equal(aa, ea)
assert_series_equal(ab, eb)
self.assertEqual(aa.name, 'ts')
self.assertEqual(ea.name, 'ts')
self.assertEqual(ab.name, 'ts')
self.assertEqual(eb.name, 'ts')
for kind in JOIN_TYPES:
_check_align(self.ts[2:], self.ts[:-5], how=kind)
_check_align(self.ts[2:], self.ts[:-5], how=kind, fill=-1)
# empty left
_check_align(self.ts[:0], self.ts[:-5], how=kind)
_check_align(self.ts[:0], self.ts[:-5], how=kind, fill=-1)
# empty right
_check_align(self.ts[:-5], self.ts[:0], how=kind)
_check_align(self.ts[:-5], self.ts[:0], how=kind, fill=-1)
# both empty
_check_align(self.ts[:0], self.ts[:0], how=kind)
_check_align(self.ts[:0], self.ts[:0], how=kind, fill=-1)
def test_align_fill_method(self):
def _check_align(a, b, how='left', method='pad', limit=None):
aa, ab = a.align(b, join=how, method=method, limit=limit)
join_index = a.index.join(b.index, how=how)
ea = a.reindex(join_index)
eb = b.reindex(join_index)
ea = ea.fillna(method=method, limit=limit)
eb = eb.fillna(method=method, limit=limit)
assert_series_equal(aa, ea)
assert_series_equal(ab, eb)
for kind in JOIN_TYPES:
for meth in ['pad', 'bfill']:
_check_align(self.ts[2:], self.ts[:-5], how=kind, method=meth)
_check_align(self.ts[2:], self.ts[:-5], how=kind, method=meth,
limit=1)
# empty left
_check_align(self.ts[:0], self.ts[:-5], how=kind, method=meth)
_check_align(self.ts[:0], self.ts[:-5], how=kind, method=meth,
limit=1)
# empty right
_check_align(self.ts[:-5], self.ts[:0], how=kind, method=meth)
_check_align(self.ts[:-5], self.ts[:0], how=kind, method=meth,
limit=1)
# both empty
_check_align(self.ts[:0], self.ts[:0], how=kind, method=meth)
_check_align(self.ts[:0], self.ts[:0], how=kind, method=meth,
limit=1)
def test_align_nocopy(self):
b = self.ts[:5].copy()
# do copy
a = self.ts.copy()
ra, _ = a.align(b, join='left')
ra[:5] = 5
self.assertFalse((a[:5] == 5).any())
# do not copy
a = self.ts.copy()
ra, _ = a.align(b, join='left', copy=False)
ra[:5] = 5
self.assertTrue((a[:5] == 5).all())
# do copy
a = self.ts.copy()
b = self.ts[:5].copy()
_, rb = a.align(b, join='right')
rb[:3] = 5
self.assertFalse((b[:3] == 5).any())
# do not copy
a = self.ts.copy()
b = self.ts[:5].copy()
_, rb = a.align(b, join='right', copy=False)
rb[:2] = 5
self.assertTrue((b[:2] == 5).all())
def test_align_sameindex(self):
a, b = self.ts.align(self.ts, copy=False)
self.assertIs(a.index, self.ts.index)
self.assertIs(b.index, self.ts.index)
# a, b = self.ts.align(self.ts, copy=True)
# self.assertIsNot(a.index, self.ts.index)
# self.assertIsNot(b.index, self.ts.index)
def test_align_multiindex(self):
# GH 10665
midx = pd.MultiIndex.from_product([range(2), range(3), range(2)],
names=('a', 'b', 'c'))
idx = pd.Index(range(2), name='b')
s1 = pd.Series(np.arange(12, dtype='int64'), index=midx)
s2 = pd.Series(np.arange(2, dtype='int64'), index=idx)
# these must be the same results (but flipped)
res1l, res1r = s1.align(s2, join='left')
res2l, res2r = s2.align(s1, join='right')
expl = s1
tm.assert_series_equal(expl, res1l)
tm.assert_series_equal(expl, res2r)
expr = pd.Series([0, 0, 1, 1, np.nan, np.nan] * 2, index=midx)
tm.assert_series_equal(expr, res1r)
tm.assert_series_equal(expr, res2l)
res1l, res1r = s1.align(s2, join='right')
res2l, res2r = s2.align(s1, join='left')
exp_idx = pd.MultiIndex.from_product([range(2), range(2), range(2)],
names=('a', 'b', 'c'))
expl = pd.Series([0, 1, 2, 3, 6, 7, 8, 9], index=exp_idx)
tm.assert_series_equal(expl, res1l)
tm.assert_series_equal(expl, res2r)
expr = pd.Series([0, 0, 1, 1] * 2, index=exp_idx)
tm.assert_series_equal(expr, res1r)
tm.assert_series_equal(expr, res2l)
def test_reindex(self):
identity = self.series.reindex(self.series.index)
# __array_interface__ is not defined for older numpies
# and on some pythons
try:
self.assertTrue(np.may_share_memory(self.series.index,
identity.index))
except (AttributeError):
pass
self.assertTrue(identity.index.is_(self.series.index))
self.assertTrue(identity.index.identical(self.series.index))
subIndex = self.series.index[10:20]
subSeries = self.series.reindex(subIndex)
for idx, val in compat.iteritems(subSeries):
self.assertEqual(val, self.series[idx])
subIndex2 = self.ts.index[10:20]
subTS = self.ts.reindex(subIndex2)
for idx, val in compat.iteritems(subTS):
self.assertEqual(val, self.ts[idx])
stuffSeries = self.ts.reindex(subIndex)
self.assertTrue(np.isnan(stuffSeries).all())
# This is extremely important for the Cython code to not screw up
nonContigIndex = self.ts.index[::2]
subNonContig = self.ts.reindex(nonContigIndex)
for idx, val in compat.iteritems(subNonContig):
self.assertEqual(val, self.ts[idx])
# return a copy the same index here
result = self.ts.reindex()
self.assertFalse((result is self.ts))
def test_reindex_nan(self):
ts = Series([2, 3, 5, 7], index=[1, 4, nan, 8])
i, j = [nan, 1, nan, 8, 4, nan], [2, 0, 2, 3, 1, 2]
assert_series_equal(ts.reindex(i), ts.iloc[j])
ts.index = ts.index.astype('object')
# reindex coerces index.dtype to float, loc/iloc doesn't
assert_series_equal(ts.reindex(i), ts.iloc[j], check_index_type=False)
def test_reindex_corner(self):
# (don't forget to fix this) I think it's fixed
self.empty.reindex(self.ts.index, method='pad') # it works
# corner case: pad empty series
reindexed = self.empty.reindex(self.ts.index, method='pad')
# pass non-Index
reindexed = self.ts.reindex(list(self.ts.index))
assert_series_equal(self.ts, reindexed)
# bad fill method
ts = self.ts[::2]
self.assertRaises(Exception, ts.reindex, self.ts.index, method='foo')
def test_reindex_pad(self):
s = Series(np.arange(10), dtype='int64')
s2 = s[::2]
reindexed = s2.reindex(s.index, method='pad')
reindexed2 = s2.reindex(s.index, method='ffill')
assert_series_equal(reindexed, reindexed2)
expected = Series([0, 0, 2, 2, 4, 4, 6, 6, 8, 8], index=np.arange(10))
assert_series_equal(reindexed, expected)
# GH4604
s = Series([1, 2, 3, 4, 5], index=['a', 'b', 'c', 'd', 'e'])
new_index = ['a', 'g', 'c', 'f']
expected = Series([1, 1, 3, 3], index=new_index)
# this changes dtype because the ffill happens after
result = s.reindex(new_index).ffill()
assert_series_equal(result, expected.astype('float64'))
result = s.reindex(new_index).ffill(downcast='infer')
assert_series_equal(result, expected)
expected = Series([1, 5, 3, 5], index=new_index)
result = s.reindex(new_index, method='ffill')
assert_series_equal(result, expected)
# inferrence of new dtype
s = Series([True, False, False, True], index=list('abcd'))
new_index = 'agc'
result = s.reindex(list(new_index)).ffill()
expected = Series([True, True, False], index=list(new_index))
assert_series_equal(result, expected)
# GH4618 shifted series downcasting
s = Series(False, index=lrange(0, 5))
result = s.shift(1).fillna(method='bfill')
expected = Series(False, index=lrange(0, 5))
assert_series_equal(result, expected)
def test_reindex_nearest(self):
s = Series(np.arange(10, dtype='int64'))
target = [0.1, 0.9, 1.5, 2.0]
actual = s.reindex(target, method='nearest')
expected = Series(np.around(target).astype('int64'), target)
assert_series_equal(expected, actual)
actual = s.reindex_like(actual, method='nearest')
assert_series_equal(expected, actual)
actual = s.reindex_like(actual, method='nearest', tolerance=1)
assert_series_equal(expected, actual)
actual = s.reindex(target, method='nearest', tolerance=0.2)
expected = Series([0, 1, np.nan, 2], target)
assert_series_equal(expected, actual)
def test_reindex_backfill(self):
pass
def test_reindex_int(self):
ts = self.ts[::2]
int_ts = Series(np.zeros(len(ts), dtype=int), index=ts.index)
# this should work fine
reindexed_int = int_ts.reindex(self.ts.index)
# if NaNs introduced
self.assertEqual(reindexed_int.dtype, np.float_)
# NO NaNs introduced
reindexed_int = int_ts.reindex(int_ts.index[::2])
self.assertEqual(reindexed_int.dtype, np.int_)
def test_reindex_bool(self):
# A series other than float, int, string, or object
ts = self.ts[::2]
bool_ts = Series(np.zeros(len(ts), dtype=bool), index=ts.index)
# this should work fine
reindexed_bool = bool_ts.reindex(self.ts.index)
# if NaNs introduced
self.assertEqual(reindexed_bool.dtype, np.object_)
# NO NaNs introduced
reindexed_bool = bool_ts.reindex(bool_ts.index[::2])
self.assertEqual(reindexed_bool.dtype, np.bool_)
def test_reindex_bool_pad(self):
# fail
ts = self.ts[5:]
bool_ts = Series(np.zeros(len(ts), dtype=bool), index=ts.index)
filled_bool = bool_ts.reindex(self.ts.index, method='pad')
self.assertTrue(isnull(filled_bool[:5]).all())
def test_reindex_like(self):
other = self.ts[::2]
assert_series_equal(self.ts.reindex(other.index),
self.ts.reindex_like(other))
# GH 7179
day1 = datetime(2013, 3, 5)
day2 = datetime(2013, 5, 5)
day3 = datetime(2014, 3, 5)
series1 = Series([5, None, None], [day1, day2, day3])
series2 = Series([None, None], [day1, day3])
result = series1.reindex_like(series2, method='pad')
expected =
|
Series([5, np.nan], index=[day1, day3])
|
pandas.Series
|
# Author: <NAME>
#
# License: BSD 3 clause
import logging
import numpy as np
import pandas as pd
from gensim.models.doc2vec import Doc2Vec, TaggedDocument
from gensim.utils import simple_preprocess
from gensim.parsing.preprocessing import strip_tags
import umap
import hdbscan
from sklearn.metrics.pairwise import cosine_similarity
from wordcloud import WordCloud
import matplotlib.pyplot as plt
from joblib import dump, load
from sklearn.cluster import dbscan
import tempfile
logger = logging.getLogger('top2vec')
logger.setLevel(logging.WARNING)
sh = logging.StreamHandler()
sh.setFormatter(logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s'))
logger.addHandler(sh)
def default_tokenizer(doc):
"""Tokenize documents for training and remove too long/short words"""
return simple_preprocess(strip_tags(doc), deacc=True)
class Top2Vec:
"""
Top2Vec
Creates jointly embedded topic, document and word vectors.
Parameters
----------
documents: List of str
Input corpus, should be a list of strings.
min_count: int (Optional, default 50)
Ignores all words with total frequency lower than this. For smaller
corpora a smaller min_count will be necessary.
speed: string (Optional, default 'learn')
This parameter will determine how fast the model takes to train. The
fast-learn option is the fastest and will generate the lowest quality
vectors. The learn option will learn better quality vectors but take
a longer time to train. The deep-learn option will learn the best quality
vectors but will take significant time to train. The valid string speed
options are:
* fast-learn
* learn
* deep-learn
use_corpus_file: bool (Optional, default False)
Setting use_corpus_file to True can sometimes provide speedup for large
datasets when multiple worker threads are available. Documents are still
passed to the model as a list of str, the model will create a temporary
corpus file for training.
document_ids: List of str, int (Optional)
A unique value per document that will be used for referring to documents
in search results. If ids are not given to the model, the index of each
document in the original corpus will become the id.
keep_documents: bool (Optional, default True)
If set to False documents will only be used for training and not saved
as part of the model. This will reduce model size. When using search
functions only document ids will be returned, not the actual documents.
workers: int (Optional)
The amount of worker threads to be used in training the model. Larger
amount will lead to faster training.
tokenizer: callable (Optional, default None)
Override the default tokenization method. If None then gensim.utils.simple_preprocess
will be used.
verbose: bool (Optional, default False)
Whether to print status data during training.
"""
def __init__(self, documents, min_count=50, speed="learn", use_corpus_file=False, document_ids=None,
keep_documents=True, workers=None, tokenizer=None, verbose=False):
if verbose:
logger.setLevel(logging.DEBUG)
else:
logger.setLevel(logging.WARNING)
# validate training inputs
if speed == "fast-learn":
hs = 0
negative = 5
epochs = 40
elif speed == "learn":
hs = 1
negative = 0
epochs = 40
elif speed == "deep-learn":
hs = 1
negative = 0
epochs = 400
elif speed == "test-learn":
hs = 0
negative = 5
epochs = 1
else:
raise ValueError("speed parameter needs to be one of: fast-learn, learn or deep-learn")
if workers is None:
pass
elif isinstance(workers, int):
pass
else:
raise ValueError("workers needs to be an int")
if tokenizer is not None:
self._tokenizer = tokenizer
else:
self._tokenizer = default_tokenizer
# validate documents
if not all((isinstance(doc, str) or isinstance(doc, np.str_)) for doc in documents):
raise ValueError("Documents need to be a list of strings")
if keep_documents:
self.documents = np.array(documents, dtype="object")
else:
self.documents = None
# validate document ids
if document_ids is not None:
if len(documents) != len(document_ids):
raise ValueError("Document ids need to match number of documents")
elif len(document_ids) != len(set(document_ids)):
raise ValueError("Document ids need to be unique")
if all((isinstance(doc_id, str) or isinstance(doc_id, np.str_)) for doc_id in document_ids):
self.doc_id_type = np.str_
elif all((isinstance(doc_id, int) or isinstance(doc_id, np.int_)) for doc_id in document_ids):
self.doc_id_type = np.int_
else:
raise ValueError("Document ids need to be str or int")
self.document_ids = np.array(document_ids)
self.doc_id2index = dict(zip(document_ids, list(range(0, len(document_ids)))))
else:
self.document_ids = None
self.doc_id2index = None
self.doc_id_type = np.int_
if use_corpus_file:
logger.info('Pre-processing documents for training')
processed = [' '.join(self._tokenizer(doc)) for doc in documents]
lines = "\n".join(processed)
temp = tempfile.NamedTemporaryFile(mode='w+t')
temp.write(lines)
logger.info('Creating joint document/word embedding')
if workers is None:
self.model = Doc2Vec(corpus_file=temp.name,
vector_size=300,
min_count=min_count,
window=15,
sample=1e-5,
negative=negative,
hs=hs,
epochs=epochs,
dm=0,
dbow_words=1)
else:
self.model = Doc2Vec(corpus_file=temp.name,
vector_size=300,
min_count=min_count,
window=15,
sample=1e-5,
negative=negative,
hs=hs,
workers=workers,
epochs=epochs,
dm=0,
dbow_words=1)
temp.close()
else:
logger.info('Pre-processing documents for training')
train_corpus = [TaggedDocument(self._tokenizer(doc), [i])
for i, doc in enumerate(documents)]
logger.info('Creating joint document/word embedding')
if workers is None:
self.model = Doc2Vec(documents=train_corpus,
vector_size=300,
min_count=min_count,
window=15,
sample=1e-5,
negative=negative,
hs=hs,
epochs=epochs,
dm=0,
dbow_words=1)
else:
self.model = Doc2Vec(documents=train_corpus,
vector_size=300,
min_count=min_count,
window=15,
sample=1e-5,
negative=negative,
hs=hs,
workers=workers,
epochs=epochs,
dm=0,
dbow_words=1)
# create 5D embeddings of documents
logger.info('Creating lower dimension embedding of documents')
umap_model = umap.UMAP(n_neighbors=15,
n_components=5,
metric='cosine').fit(self.model.docvecs.vectors_docs)
# find dense areas of document vectors
logger.info('Finding dense areas of documents')
cluster = hdbscan.HDBSCAN(min_cluster_size=15,
metric='euclidean',
cluster_selection_method='eom').fit(umap_model.embedding_)
# calculate topic vectors from dense areas of documents
logger.info('Finding topics')
self._create_topic_vectors(cluster.labels_)
# deduplicate topics
self._deduplicate_topics()
# calculate topic sizes and index nearest topic for each document
self.topic_vectors, self.doc_top, self.doc_dist, self.topic_sizes = self._calculate_topic_sizes(
self.topic_vectors)
# find topic words and scores
self.topic_words, self.topic_word_scores = self._find_topic_words_scores(topic_vectors=self.topic_vectors)
# initialize variables for hierarchical topic reduction
self.topic_vectors_reduced = None
self.doc_top_reduced = None
self.doc_dist_reduced = None
self.topic_sizes_reduced = None
self.topic_words_reduced = None
self.topic_word_scores_reduced = None
self.hierarchy = None
def _create_topic_vectors(self, cluster_labels):
unique_labels = set(cluster_labels)
if -1 in unique_labels:
unique_labels.remove(-1)
self.topic_vectors = np.vstack([self.model.docvecs.vectors_docs[np.where(cluster_labels == label)[0]]
.mean(axis=0) for label in unique_labels])
def _deduplicate_topics(self):
core_samples, labels = dbscan(X=self.topic_vectors,
eps=0.1,
min_samples=2,
metric="cosine")
duplicate_clusters = set(labels)
if len(duplicate_clusters) > 1 or -1 not in duplicate_clusters:
# unique topics
unique_topics = self.topic_vectors[np.where(labels == -1)[0]]
if -1 in duplicate_clusters:
duplicate_clusters.remove(-1)
# merge duplicate topics
for unique_label in duplicate_clusters:
unique_topics = np.vstack(
[unique_topics, self.topic_vectors[np.where(labels == unique_label)[0]]
.mean(axis=0)])
self.topic_vectors = unique_topics
def _calculate_topic_sizes(self, topic_vectors, hierarchy=None):
# find nearest topic of each document
doc_top, doc_dist = self._calculate_documents_topic(topic_vectors=topic_vectors,
document_vectors=self.model.docvecs.vectors_docs)
topic_sizes = pd.Series(doc_top).value_counts()
return self._reorder_topics(topic_vectors, topic_sizes, doc_top, doc_dist, hierarchy)
@staticmethod
def _reorder_topics(topic_vectors, topic_sizes, doc_top, doc_dist, hierarchy=None):
topic_vectors = topic_vectors[topic_sizes.index]
old2new = dict(zip(topic_sizes.index, range(topic_sizes.shape[0])))
doc_top = np.array([old2new[i] for i in doc_top])
if hierarchy is None:
topic_sizes.reset_index(drop=True, inplace=True)
return topic_vectors, doc_top, doc_dist, topic_sizes
else:
hierarchy = [hierarchy[i] for i in topic_sizes.index]
topic_sizes.reset_index(drop=True, inplace=True)
return topic_vectors, doc_top, doc_dist, topic_sizes, hierarchy
@staticmethod
def _calculate_documents_topic(topic_vectors, document_vectors, dist=True):
batch_size = 10000
doc_top = []
if dist:
doc_dist = []
if document_vectors.shape[0] > batch_size:
current = 0
batches = int(document_vectors.shape[0] / batch_size)
extra = document_vectors.shape[0] % batch_size
for ind in range(0, batches):
res = cosine_similarity(document_vectors[current:current + batch_size], topic_vectors)
doc_top.extend(np.argmax(res, axis=1))
if dist:
doc_dist.extend(np.max(res, axis=1))
current += batch_size
if extra > 0:
res = cosine_similarity(document_vectors[current:current + extra], topic_vectors)
doc_top.extend(np.argmax(res, axis=1))
if dist:
doc_dist.extend(np.max(res, axis=1))
if dist:
doc_dist = np.array(doc_dist)
else:
res = cosine_similarity(document_vectors, topic_vectors)
doc_top = np.argmax(res, axis=1)
if dist:
doc_dist = np.max(res, axis=1)
if dist:
return doc_top, doc_dist
else:
return doc_top
def _find_topic_words_scores(self, topic_vectors):
topic_words = []
topic_word_scores = []
for topic_vector in topic_vectors:
sim_words = self.model.wv.most_similar(positive=[topic_vector], topn=50)
topic_words.append([word[0] for word in sim_words])
topic_word_scores.append([round(word[1], 4) for word in sim_words])
topic_words = np.array(topic_words)
topic_word_scores = np.array(topic_word_scores)
return topic_words, topic_word_scores
def save(self, file):
"""
Saves the current model to the specified file.
Parameters
----------
file: str
File where model will be saved.
"""
dump(self, file)
@classmethod
def load(cls, file):
"""
Load a pre-trained model from the specified file.
Parameters
----------
file: str
File where model will be loaded from.
"""
return load(file)
@staticmethod
def _less_than_zero(num, var_name):
if num < 0:
raise ValueError(f"{var_name} cannot be less than 0.")
def _validate_hierarchical_reduction(self):
if self.hierarchy is None:
raise ValueError("Hierarchical topic reduction has not been performed.")
def _validate_hierarchical_reduction_num_topics(self, num_topics):
current_num_topics = len(self.topic_vectors)
if num_topics >= current_num_topics:
raise ValueError(f"Number of topics must be less than {current_num_topics}.")
def _validate_num_docs(self, num_docs):
self._less_than_zero(num_docs, "num_docs")
document_count = self.model.docvecs.count
if num_docs > self.model.docvecs.count:
raise ValueError(f"num_docs cannot exceed the number of documents: {document_count}.")
def _validate_num_topics(self, num_topics, reduced):
self._less_than_zero(num_topics, "num_topics")
if reduced:
topic_count = len(self.topic_vectors_reduced)
if num_topics > topic_count:
raise ValueError(f"num_topics cannot exceed the number of reduced topics: {topic_count}.")
else:
topic_count = len(self.topic_vectors)
if num_topics > topic_count:
raise ValueError(f"num_topics cannot exceed the number of topics: {topic_count}.")
def _validate_topic_num(self, topic_num, reduced):
self._less_than_zero(topic_num, "topic_num")
if reduced:
topic_count = len(self.topic_vectors_reduced) - 1
if topic_num > topic_count:
raise ValueError(f"Invalid topic number: valid reduced topics numbers are 0 to {topic_count}.")
else:
topic_count = len(self.topic_vectors) - 1
if topic_num > topic_count:
raise ValueError(f"Invalid topic number: valid original topics numbers are 0 to {topic_count}.")
def _validate_topic_search(self, topic_num, num_docs, reduced):
self._less_than_zero(num_docs, "num_docs")
if reduced:
if num_docs > self.topic_sizes_reduced[topic_num]:
raise ValueError(f"Invalid number of documents: reduced topic {topic_num}"
f" only has {self.topic_sizes_reduced[topic_num]} documents.")
else:
if num_docs > self.topic_sizes[topic_num]:
raise ValueError(f"Invalid number of documents: original topic {topic_num}"
f" only has {self.topic_sizes[topic_num]} documents.")
def _validate_doc_ids(self, doc_ids, doc_ids_neg):
if not isinstance(doc_ids, list):
raise ValueError("doc_ids must be a list of string or int.")
if not isinstance(doc_ids_neg, list):
raise ValueError("doc_ids_neg must be a list of string or int.")
doc_ids_all = doc_ids + doc_ids_neg
for doc_id in doc_ids_all:
if self.document_ids is not None:
if doc_id not in self.document_ids:
raise ValueError(f"{doc_id} is not a valid document id.")
elif doc_id < 0 or doc_id > self.model.docvecs.count - 1:
raise ValueError(f"{doc_id} is not a valid document id.")
def _validate_keywords(self, keywords, keywords_neg):
if not (isinstance(keywords, list) or isinstance(keywords, np.ndarray)):
raise ValueError("keywords must be a list of strings.")
if not (isinstance(keywords_neg, list) or isinstance(keywords_neg, np.ndarray)):
raise ValueError("keywords_neg must be a list of strings.")
keywords_lower = [keyword.lower() for keyword in keywords]
keywords_neg_lower = [keyword.lower() for keyword in keywords_neg]
for word in keywords_lower + keywords_neg_lower:
if word not in self.model.wv.vocab:
raise ValueError(f"'{word}' has not been learned by the model so it cannot be searched.")
return keywords_lower, keywords_neg_lower
def _get_document_ids(self, doc_index):
if self.document_ids is None:
return doc_index
else:
return self.document_ids[doc_index]
def _get_document_indexes(self, doc_ids):
if self.document_ids is None:
return doc_ids
else:
return [self.doc_id2index[doc_id] for doc_id in doc_ids]
def _get_word_vectors(self, keywords):
return [self.model[word] for word in keywords]
def _validate_document_ids_add_doc(self, documents, document_ids):
if document_ids is None:
raise ValueError("Document ids need to be provided.")
if len(documents) != len(document_ids):
raise ValueError("Document ids need to match number of documents.")
elif len(document_ids) != len(set(document_ids)):
raise ValueError("Document ids need to be unique.")
if all((isinstance(doc_id, str) or isinstance(doc_id, np.str_)) for doc_id in document_ids):
if self.doc_id_type == np.int_:
raise ValueError("Document ids need to be of type int.")
elif all((isinstance(doc_id, int) or isinstance(doc_id, np.int_)) for doc_id in document_ids):
if self.doc_id_type == np.str_:
raise ValueError("Document ids need to be of type str.")
if len(set(document_ids).intersection(self.document_ids)) > 0:
raise ValueError("Some document ids already exist in model.")
@staticmethod
def _validate_documents(documents):
if not all((isinstance(doc, str) or isinstance(doc, np.str_)) for doc in documents):
raise ValueError("Documents need to be a list of strings.")
def _assign_documents_to_topic(self, document_vectors, topic_vectors, topic_sizes, doc_top, doc_dist,
hierarchy=None):
doc_top_new, doc_dist_new = self._calculate_documents_topic(topic_vectors, document_vectors, dist=True)
doc_top = np.append(doc_top, doc_top_new)
doc_dist = np.append(doc_dist, doc_dist_new)
topic_sizes_new =
|
pd.Series(doc_top_new)
|
pandas.Series
|
# $Id$
# $HeadURL$
################################################################
# The contents of this file are subject to the BSD 3Clause (New)
# you may not use this file except in
# compliance with the License. You may obtain a copy of the License at
# http://directory.fsf.org/wiki/License:BSD_3Clause
# Software distributed under the License is distributed on an "AS IS"
# basis, WITHOUT WARRANTY OF ANY KIND, either express or implied. See the
# License for the specific language governing rights and limitations
# under the License.
# The Original Code is part of the PyRadi toolkit.
# The Initial Developer of the Original Code is <NAME>,
# Portions created by <NAME> are Copyright (C) 2006-2012
# All Rights Reserved.
# Contributor(s): <NAME>, <NAME>, <NAME>, <NAME>
################################################################
"""
This module provides functions for plotting cartesian and polar plots.
This class provides a basic plotting capability, with a minimum
number of lines. These are all wrapper functions,
based on existing functions in other Python classes.
Provision is made for combinations of linear and log scales, as well
as polar plots for two-dimensional graphs.
The Plotter class can save files to disk in a number of formats.
For more examples of use see:
https://github.com/NelisW/ComputationalRadiometry
See the __main__ function for examples of use.
This package was partly developed to provide additional material in support of students
and readers of the book Electro-Optical System Analysis and Design: A Radiometry
Perspective, <NAME>, ISBN 9780819495693, SPIE Monograph Volume
PM236, SPIE Press, 2013. http://spie.org/x648.html?product_id=2021423&origin_id=x646
"""
__version__ = "$Revision$"
__author__ = 'pyradi team'
__all__ = ['Plotter','cubehelixcmap', 'FilledMarker', 'Markers','ProcessImage',
'savePlot']
import numpy as np
import pandas as pd
import math
import sys
import itertools
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from matplotlib.font_manager import FontProperties
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
from matplotlib.backends.backend_pdf import PdfPages
import matplotlib.dates as mdates
from mpl_toolkits.axes_grid1 import make_axes_locatable
# following for the pie plots
from matplotlib.transforms import Affine2D
import mpl_toolkits.axisartist.floating_axes as floating_axes
import mpl_toolkits.axisartist.angle_helper as angle_helper
from matplotlib.projections import PolarAxes
from mpl_toolkits.axisartist.grid_finder import MaxNLocator
from matplotlib.ticker import FormatStrFormatter
from matplotlib.colors import LinearSegmentedColormap as LSC
# see if plotly is available
try:
__import__('plotly.tools')
imported_plotly = True
from plotly import tools
from plotly.offline import download_plotlyjs, offline
from plotly.graph_objs import Scatter, Layout, Figure,Scatter3d,Mesh3d,ColorBar,Contour
except ImportError:
imported_plotly = False
from datetime import datetime
####################################################################
##
class FilledMarker:
"""Filled marker user-settable values.
This class encapsulates a few variables describing a Filled marker.
Default values are provided that can be overridden in user plots.
Values relevant to filled makers are as follows:
| marker = ['o', 'v', '^', '<', '>', '8', 's', 'p', '*', 'h', 'H', 'D', 'd']
| fillstyle = ['full', 'left', 'right', 'bottom', 'top', 'none']
| colour names = http://www.w3schools.com/html/html_colornames.asp
"""
def __init__(self, markerfacecolor=None, markerfacecoloralt=None,
markeredgecolor=None, marker=None, markersize=None,
fillstyle=None):
"""Define marker default values.
Args:
| markerfacecolor (colour): main colour for marker (optional)
| markerfacecoloralt (colour): alterive colour for marker (optional)
| markeredgecolor (colour): edge colour for marker (optional)
| marker (string): string to specify the marker (optional)
| markersize (int)): size of the marker (optional)
| fillstyle (string): string to define fill style (optional)
Returns:
| Nothing. Creates the figure for subequent use.
Raises:
| No exception is raised.
"""
__all__ = ['__init__']
if markerfacecolor is None:
self.markerfacecolor = 'r'
else:
self.markerfacecolor = markerfacecolor
if markerfacecoloralt is None:
self.markerfacecoloralt = 'b'
else:
self.markerfacecoloralt = markerfacecoloralt
if markeredgecolor is None:
self.markeredgecolor = 'k'
else:
self.markeredgecolor = markeredgecolor
if marker is None:
self.marker = 'o'
else:
self.marker = marker
if markersize is None:
self.markersize = 20
else:
self.markersize = markersize
if fillstyle is None:
self.fillstyle = 'full'
else:
self.fillstyle = fillstyle
###################################################################################
###################################################################################
class Markers:
"""Collect marker location and types and mark subplot.
Build a list of markers at plot locations with the specified marker.
"""
####################################################################
##
def __init__(self, markerfacecolor = None, markerfacecoloralt = None,
markeredgecolor = None, marker = None, markersize = None,
fillstyle = None):
"""Set default marker values for this collection
Specify default marker properties to be used for all markers
in this instance. If no marker properties are specified here,
the default FilledMarker marker properties will be used.
Args:
| markerfacecolor (colour): main colour for marker (optional)
| markerfacecoloralt (colour): alternative colour for marker (optional)
| markeredgecolor (colour): edge colour for marker (optional)
| marker (string): string to specify the marker (optional)
| markersize (int)): size of the marker (optional)
| fillstyle (string): string to define fill style (optional)
Returns:
| Nothing. Creates the figure for subequent use.
Raises:
| No exception is raised.
"""
__all__ = ['__init__', 'add', 'plot']
if markerfacecolor is None:
self.markerfacecolor = None
else:
self.markerfacecolor = markerfacecolor
if markerfacecoloralt is None:
self.markerfacecoloralt = None
else:
self.markerfacecoloralt = markerfacecoloralt
if markeredgecolor is None:
self.markeredgecolor = None
else:
self.markeredgecolor = markeredgecolor
if marker is None:
self.marker = None
else:
self.marker = marker
if markersize is None:
self.markersize = markersize
else:
self.markersize = markersize
if fillstyle is None:
self.fillstyle = None
else:
self.fillstyle = fillstyle
#list if markers to be drawn
self.markers = []
####################################################################
##
def add(self,x,y,markerfacecolor = None, markerfacecoloralt = None,
markeredgecolor = None, marker = None, markersize = None,
fillstyle = None):
"""Add a marker to the list, overridding properties if necessary.
Specify location and any specific marker properties to be used.
The location can be (xy,y) for cartesian plots or (theta,rad) for polars.
If no marker properties are specified, the current marker class
properties will be used. If the current maker instance does not
specify properties, the default marker properties will be used.
Args:
| x (float): the x/theta location for the marker
| y (float): the y/radial location for the marker
| markerfacecolor (colour): main colour for marker (optional)
| markerfacecoloralt (colour): alterive colour for marker (optional)
| markeredgecolor (colour): edge colour for marker (optional)
| marker (string): string to specify the marker (optional)
| markersize (int)): size of the marker (optional)
| fillstyle (string): string to define fill style (optional)
Returns:
| Nothing. Creates the figure for subequent use.
Raises:
| No exception is raised.
"""
if markerfacecolor is None:
if self.markerfacecolor is not None:
markerfacecolor = self.markerfacecolor
if markerfacecoloralt is None:
if self.markerfacecoloralt is not None:
markerfacecoloralt = self.markerfacecoloralt
if markeredgecolor is None:
if self.markeredgecolor is not None:
markeredgecolor = self.markeredgecolor
if marker is None:
if self.marker is not None:
marker = self.marker
if markersize is None:
if self.markersize is not None:
markersize = self.markersize
if fillstyle is None:
if self.fillstyle is not None:
fillstyle = self.fillstyle
marker = FilledMarker(markerfacecolor, markerfacecoloralt ,
markeredgecolor , marker, markersize , fillstyle)
self.markers.append((x,y,marker))
####################################################################
##
def plot(self,ax):
"""Plot the current list of markers on the given axes.
All the markers currently stored in the class will be
drawn.
Args:
| ax (axes): an axes handle for the plot
Returns:
| Nothing. Creates the figure for subsequent use.
Raises:
| No exception is raised.
"""
usetex = plt.rcParams['text.usetex']
plt.rcParams['text.usetex'] = False # otherwise, '^' will cause trouble
for marker in self.markers:
ax.plot(marker[0], marker[1],
color = marker[2].markerfacecolor,
markerfacecoloralt = marker[2].markerfacecoloralt,
markeredgecolor = marker[2].markeredgecolor,
marker = marker[2].marker,
markersize = marker[2].markersize,
fillstyle = marker[2].fillstyle,
linewidth=0)
plt.rcParams['text.usetex'] = usetex
###################################################################################
###################################################################################
class ProcessImage:
"""This class provides a functions to assist in the optimal display of images.
"""
#define the compression rule to be used in the equalisation function
compressSet = [
[lambda x : x , lambda x : x, 'Linear'],
[np.log, np.exp, 'Natural Log'],
[np.sqrt, np.square, 'Square Root']]
############################################################
def __init__(self):
"""Class constructor
Sets up some variables for use in this class
Args:
| None
Returns:
| Nothing
Raises:
| No exception is raised.
"""
__all__ = ['__init__', 'compressEqualizeImage', 'reprojectImageIntoPolar']
############################################################
def compressEqualizeImage(self, image, selectCompressSet=2, numCbarlevels=20,
cbarformat='.3f'):
"""Compress an image (and then inversely expand the color bar values),
prior to histogram equalisation to ensure that the two keep in step,
we store the compression function names as pairs, and invoke the
compression function as follows: linear, log. sqrt. Note that the
image is histogram equalised in all cases.
Args:
| image (np.ndarray): the image to be processed
| selectCompressSet (int): compression selection [0,1,2] (optional)
| numCbarlevels (int): number of labels in the colourbar (optional)
| cbarformat (string): colourbar label format, e.g., '10.3f', '.5e' (optional)
Returns:
| imgHEQ (np.ndarray): the equalised image array
| customticksz (zip(float, string)): colourbar levels and associated levels
Raises:
| No exception is raised.
"""
#compress the input image - rescale color bar tick to match below
#also collapse into single dimension
imgFlat = self.compressSet[selectCompressSet][0](image.flatten())
imgFlatSort = np.sort(imgFlat)
#cumulative distribution
cdf = imgFlatSort.cumsum()/imgFlatSort[-1]
#remap image values to achieve histogram equalisation
y=np.interp(imgFlat,imgFlatSort, cdf )
#and reshape to original image shape
imgHEQ = y.reshape(image.shape)
# #plot the histogram mapping
# minData = np.min(imgFlat)
# maxData = np.max(imgFlat)
# print('Image irradiance range minimum={0} maximum={1}'.format(minData, maxData))
# irradRange=np.linspace(minData, maxData, 100)
# normalRange = np.interp(irradRange,imgFlatSort, cdf )
# H = ryplot.Plotter(1, 1, 1,'Mapping Input Irradiance to Equalised Value',
# figsize=(10, 10))
# H.plot(1, "","Irradiance [W/(m$^2$)]", "Equalised value",irradRange,
# normalRange, powerLimits = [-4, 2, -10, 2])
# #H.getPlot().show()
# H.saveFig('cumhist{0}.png'.format(entry), dpi=300)
#prepare the color bar tick labels from image values (as plotted)
imgLevels = np.linspace(np.min(imgHEQ), np.max(imgHEQ), numCbarlevels)
#map back from image values to original values as read it (inverse to above)
irrLevels=np.interp(imgLevels,cdf, imgFlatSort)
#uncompress the tick labels - match with compression above
fstr = '{0:' + cbarformat + '}'
customticksz = list(zip(imgLevels, [fstr.format(self.compressSet[selectCompressSet][1](x)) for x in irrLevels]))
return imgHEQ, customticksz
##############################################################################
##
def reprojectImageIntoPolar(self, data, origin=None, framesFirst=True,cval=0.0):
"""Reprojects a 3D numpy array into a polar coordinate system, relative to some origin.
This function reprojects an image from cartesian to polar coordinates.
The origin of the new coordinate system defaults to the center of the image,
unless the user supplies a new origin.
The data format can be data.shape = (rows, cols, frames) or
data.shape = (frames, rows, cols), the format of which is indicated by the
framesFirst parameter.
The reprojectImageIntoPolar function maps radial to cartesian coords.
The radial image is however presented in a cartesian grid, the corners have no meaning.
The radial coordinates are mapped to the radius, not the corners.
This means that in order to map corners, the frequency is scaled with sqrt(2),
The corners are filled with the value specified in cval.
Args:
| data (np.array): 3-D array to which transformation must be applied.
| origin ( (x-orig, y-orig) ): data-coordinates of where origin should be placed
| framesFirst (bool): True if data.shape is (frames, rows, cols), False if
data.shape is (rows, cols, frames)
| cval (float): the fill value to be used in coords outside the mapped range(optional)
Returns:
| output (float np.array): transformed images/array data in the same sequence as input sequence.
| r_i (np.array[N,]): radial values for returned image.
| theta_i (np.array[M,]): angular values for returned image.
Raises:
| No exception is raised.
original code by <NAME>
https://stackoverflow.com/questions/3798333/image-information-along-a-polar-coordinate-system
"""
import pyradi.ryutils as ryutils
# import scipy as sp
import scipy.ndimage as spndi
if framesFirst:
data = ryutils.framesLast(data)
ny, nx = data.shape[:2]
if origin is None:
origin = (nx//2, ny//2)
# Determine what the min and max r and theta coords will be
x, y = ryutils.index_coords(data, origin=origin, framesFirst=framesFirst )
r, theta = ryutils.cart2polar(x, y)
# Make a regular (in polar space) grid based on the min and max r & theta
r_i = np.linspace(r.min(), r.max(), nx)
theta_i = np.linspace(theta.min(), theta.max(), ny)
theta_grid, r_grid = np.meshgrid(theta_i, r_i)
# Project the r and theta grid back into pixel coordinates
xi, yi = ryutils.polar2cart(r_grid, theta_grid)
xi += origin[0] # We need to shift the origin back to
yi += origin[1] # back to the lower-left corner...
xi, yi = xi.flatten(), yi.flatten()
coords = np.vstack((xi, yi)) # (map_coordinates requires a 2xn array)
# Reproject each band individually and the restack
# (uses less memory than reprojection the 3-dimensional array in one step)
bands = []
for band in data.T:
zi = spndi.map_coordinates(band, coords, order=1,cval=cval)
bands.append(zi.reshape((nx, ny)))
output = np.dstack(bands)
if framesFirst:
output = ryutils.framesFirst(output)
return output, r_i, theta_i
###################################################################################
###################################################################################
class Plotter:
""" Encapsulates a plotting environment, optimized for compact code.
This class provides a wrapper around Matplotlib to provide a plotting
environment specialised towards typical pyradi visualisation.
These functions were developed to provide sophisticated plots by entering
the various plot options on a few lines, instead of typing many commands.
Provision is made for plots containing subplots (i.e., multiple plots on
the same figure), linear scale and log scale plots, images, and cartesian,
3-D and polar plots.
"""
############################################################
##
def __init__(self,fignumber=0,subpltnrow=1,subpltncol=1,\
figuretitle=None, figsize=(9,9), titlefontsize=14,
useplotly = False,doWarning=True):
"""Class constructor
The constructor defines the number for this figure, allowing future reference
to this figure. The number of subplot rows and columns allow the user to define
the subplot configuration. The user can also provide a title to be
used for the figure (centred on top) and finally, the size of the figure in inches
can be specified to scale the text relative to the figure.
Args:
| fignumber (int): the plt figure number, must be supplied
| subpltnrow (int): subplot number of rows
| subpltncol (int): subplot number of columns
| figuretitle (string): the overall heading for the figure
| figsize ((w,h)): the figure size in inches
| titlefontsize (int): the figure title size in points
| useplotly (bool): Plotly activation parameter
| doWarning (bool): print warning messages to the screen
Returns:
| Nothing. Creates the figure for subequent use.
Raises:
| No exception is raised.
"""
__all__ = ['__init__', 'saveFig', 'getPlot', 'plot', 'logLog', 'semilogX',
'semilogY', 'polar', 'showImage', 'plot3d', 'buildPlotCol',
'getSubPlot', 'meshContour', 'nextPlotCol', 'plotArray',
'polarMesh', 'resetPlotCol', 'mesh3D', 'polar3d', 'labelSubplot',
'emptyPlot','setup_pie_axes','pie']
version=mpl.__version__.split('.')
vnum=float(version[0]+'.'+version[1])
if vnum<1.1:
print('Install Matplotlib 1.1 or later')
print('current version is {0}'.format(vnum))
sys.exit(-1)
self.figurenumber = fignumber
self.fig = plt.figure(self.figurenumber)
self.fig.set_size_inches(figsize[0], figsize[1])
self.fig.clear()
self.figuretitle = figuretitle
self.doWarning = doWarning
#Plotly variables initialization
self.useplotly = useplotly
if self.useplotly:
self.Plotlyfig = []
self.Plotlydata = []
self.Plotlylayout = []
self.PlotlyXaxisTitles = []
self.PlotlyYaxisTitles = []
self.PlotlySubPlotTitles = []
self.PlotlySubPlotLabels = []
self.PlotlySubPlotNumbers = []
self.PlotlyPlotCalls = 0
self.PLcolor=''
self.PLwidth=0
self.PLdash=''
self.PLmultiAxisTitle=''
self.PLmultipleYAxis=False
self.PLyAxisSide=''
self.PLyAxisOverlaying=''
self.PLmultipleXAxis=False
self.PLxAxisSide=''
self.PLxAxisOverlaying=''
self.PLIs3D=False
self.PLType=''
self.nrow=subpltnrow
self.ncol=subpltncol
# width reserved for space between subplots
self.fig.subplots_adjust(wspace=0.25)
#height reserved for space between subplots
self.fig.subplots_adjust(hspace=0.4)
#height reserved for top of the subplots of the figure
self.fig.subplots_adjust(top=0.88)
# define the default line colour and style
self.buildPlotCol(plotCol=None, n=None)
self.bbox_extra_artists = []
self.subplots = {}
self.gridSpecsOuter = {}
self.arrayRows = {}
self.gridSpecsInner = {}
if figuretitle:
self.figtitle=plt.gcf().text(.5,.95,figuretitle,\
horizontalalignment='center',\
fontproperties=FontProperties(size=titlefontsize))
self.bbox_extra_artists.append(self.figtitle)
############################################################
##
def buildPlotCol(self, plotCol=None, n=None):
"""Set a sequence of default colour styles of
appropriate length.
The constructor provides a sequence with length
14 pre-defined plot styles.
The user can define a new sequence if required.
This function modulus-folds either sequence, in
case longer sequences are required.
Colours can be one of the basic colours:
['b', 'g', 'r', 'c', 'm', 'y', 'k']
or it can be a gray shade float value between 0 and 1,
such as '0.75', or it can be in hex format '#eeefff'
or it can be one of the legal html colours.
See http://html-color-codes.info/ and
http://www.computerhope.com/htmcolor.htm.
http://latexcolor.com/
Args:
| plotCol ([strings]): User-supplied list
| of plotting styles(can be empty []).
| n (int): Length of required sequence.
Returns:
| A list with sequence of plot styles, of required length.
Raises:
| No exception is raised.
"""
# assemble the list as requested, use default if not specified
if plotCol is None:
plotCol = ['b', 'g', 'r', 'c', 'm', 'y', 'k',
'#5D8AA8','#E52B50','#FF7E00','#9966CC','#CD9575','#915C83',
'#008000','#4B5320','#B2BEB5','#A1CAF1','#FE6F5E','#333399',
'#DE5D83','#800020','#1E4D2B','#00BFFF','#007BA7','#FFBCD9']
if n is None:
n = len(plotCol)
self.plotCol = [plotCol[i % len(plotCol)] for i in range(n)]
# copy this to circular list as well
self.plotColCirc = itertools.cycle(self.plotCol)
return self.plotCol
############################################################
##
def nextPlotCol(self):
"""Returns the next entry in a sequence of default
plot line colour styles in circular list.
One day I want to do this with a generator....
Args:
| None
Returns:
| The next plot colour in the sequence.
Raises:
| No exception is raised.
"""
col = next(self.plotColCirc)
return col
############################################################
##
def resetPlotCol(self):
"""Resets the plot colours to start at the beginning of
the cycle.
Args:
| None
Returns:
| None.
Raises:
| No exception is raised.
"""
self.plotColCirc = itertools.cycle(self.plotCol)
############################################################
##
def saveFig(self, filename='mpl.png',dpi=300,bbox_inches='tight',\
pad_inches=0.1, useTrueType = True):
"""Save the plot to a disk file, using filename, dpi specification and bounding box limits.
One of matplotlib's design choices is a bounding box strategy which may result in a bounding box
that is smaller than the size of all the objects on the page. It took a while to figure this out,
but the current default values for bbox_inches and pad_inches seem to create meaningful
bounding boxes. These are however larger than the true bounding box. You still need a
tool such as epstools or Adobe Acrobat to trim eps files to the true bounding box.
The type of file written is picked up in the filename.
Most backends support png, pdf, ps, eps and svg.
Args:
| filename (string): output filename to write plot, file ext
| dpi (int): the resolution of the graph in dots per inch
| bbox_inches: see matplotlib docs for more detail.
| pad_inches: see matplotlib docs for more detail.
| useTrueType: if True, truetype fonts are used in eps/pdf files, otherwise Type3
Returns:
| Nothing. Saves a file to disk.
Raises:
| No exception is raised.
"""
# http://matplotlib.1069221.n5.nabble.com/TrueType-font-embedding-in-eps-problem-td12691.html
# http://stackoverflow.com/questions/5956182/cannot-edit-text-in-chart-exported-by-matplotlib-and-opened-in-illustrator
# http://newsgroups.derkeiler.com/Archive/Comp/comp.soft-sys.matlab/2008-07/msg02038.html
if useTrueType:
mpl.rcParams['pdf.fonttype'] = 42
mpl.rcParams['ps.fonttype'] = 42
#http://stackoverflow.com/questions/15341757/how-to-check-that-pylab-backend-of-matplotlib-runs-inline/17826459#17826459
# print(mpl.get_backend())
if 'inline' in mpl.get_backend() and self.doWarning:
print('**** If saveFig does not work inside the notebook please comment out the line "%matplotlib inline" ')
print('To disable ryplot warnings, set doWarning=False')
# return
if len(filename)>0:
if self.bbox_extra_artists:
self.fig.savefig(filename, dpi=dpi, bbox_inches=bbox_inches,
pad_inches=pad_inches,\
bbox_extra_artists= self.bbox_extra_artists);
else:
self.fig.savefig(filename, dpi=dpi, bbox_inches=bbox_inches,
pad_inches=pad_inches);
############################################################
##
def getPlot(self):
"""Returns a handle to the current figure
Args:
| None
Returns:
| A handle to the current figure.
Raises:
| No exception is raised.
"""
return self.fig
############################################################
##
def labelSubplot(self, spax, ptitle=None, xlabel=None, ylabel=None, zlabel=None,
titlefsize=10, labelfsize=10, ):
"""Set the sub-figure title and axes labels (cartesian plots only).
Args:
| spax (handle): subplot axis handle where labels must be drawn
| ptitle (string): plot title (optional)
| xlabel (string): x-axis label (optional)
| ylabel (string): y-axis label (optional)
| zlabel (string): z axis label (optional)
| titlefsize (float): title fontsize (optional)
| labelfsize (float): x,y,z label fontsize (optional)
Returns:
| None.
Raises:
| No exception is raised.
"""
if xlabel is not None:
spax.set_xlabel(xlabel,fontsize=labelfsize)
if ylabel is not None:
spax.set_ylabel(ylabel,fontsize=labelfsize)
if zlabel is not None:
spax.set_ylabel(zlabel,fontsize=labelfsize)
if ptitle is not None:
spax.set_title(ptitle,fontsize=titlefsize)
############################################################
##
def getSubPlot(self, subplotNum = 1):
"""Returns a handle to the subplot, as requested per subplot number.
Subplot numbers range from 1 upwards.
Args:
| subplotNumer (int) : number of the subplot
Returns:
| A handle to the requested subplot or None if not found.
Raises:
| No exception is raised.
"""
if (self.nrow,self.ncol, subplotNum) in list(self.subplots.keys()):
return self.subplots[(self.nrow,self.ncol, subplotNum)]
else:
return None
############################################################
##
def getXLim(self, subplotNum = 1):
"""Returns the x limits of the current subplot.
Subplot numbers range from 1 upwards.
Args:
| subplotNumer (int) : number of the subplot
Returns:
| An array with the two limits
Raises:
| No exception is raised.
"""
if (self.nrow,self.ncol, subplotNum) in list(self.subplots.keys()):
return np.asarray(self.subplots[(self.nrow,self.ncol, subplotNum)].get_xlim())
else:
return None
############################################################
##
def getYLim(self, subplotNum = 1):
"""Returns the y limits of the current subplot.
Subplot numbers range from 1 upwards.
Args:
| subplotNumer (int) : number of the subplot
Returns:
| An array with the two limits
Raises:
| No exception is raised.
"""
if (self.nrow,self.ncol, subplotNum) in list(self.subplots.keys()):
return np.asarray(self.subplots[(self.nrow,self.ncol, subplotNum)].get_ylim())
else:
return None
############################################################
##
def verticalLineCoords(self,subplotNum=1,x=0):
"""Returns two arrays for vertical line at x in the specific subplot.
The line is drawn at specified x, with current y limits in subplot.
Subplot numbers range from 1 upwards.
Use as follows to draw a vertical line in plot:
p.plot(1,*p.verticalLineCoords(subplotNum=1,x=freq),plotCol=['k'])
Args:
| subplotNumer (int) : number of the subplot
| x (double): horizontal value used for line
Returns:
| A tuple with two arrays for line (x-coords,y-coords)
Raises:
| No exception is raised.
"""
if (self.nrow,self.ncol, subplotNum) in list(self.subplots.keys()):
handle = self.subplots[(self.nrow,self.ncol, subplotNum)]
x = np.asarray((x,x))
y = self.getYLim(subplotNum)
return x,y
else:
return None
############################################################
##
def horizontalLineCoords(self,subplotNum=1,y=0):
"""Returns two arrays for horizontal line at y in the specific subplot.
The line is drawn at specified y, with current x limits in subplot.
Subplot numbers range from 1 upwards.
Use as follows to draw a horizontal line in plot:
p.plot(1,*p.horizontalLineCoords(subplotNum=1,x=freq),plotCol=['k'])
Args:
| subplotNumer (int) : number of the subplot
| y (double): horizontal value used for line
Returns:
| A tuple with two arrays for line (x-coords,y-coords)
Raises:
| No exception is raised.
"""
if (self.nrow,self.ncol, subplotNum) in list(self.subplots.keys()):
handle = self.subplots[(self.nrow,self.ncol, subplotNum)]
y = np.asarray((y,y))
x = self.getXLim(subplotNum)
return x,y
else:
return None
############################################################
##
def plot(self, plotnum, x, y, ptitle=None, xlabel=None, ylabel=None,
plotCol=[], linewidths=None, label=[], legendAlpha=0.0,
legendLoc='best',
pltaxis=None, maxNX=10, maxNY=10, linestyle=None,
powerLimits = [-4, 2, -4, 2], titlefsize = 12,
xylabelfsize = 12, xytickfsize = 10, labelfsize=10,
xScientific=False, yScientific=False,
yInvert=False, xInvert=False, drawGrid=True,xIsDate=False,
xTicks=None, xtickRotation=0,
markers=[], markevery=None, markerfacecolor=True,markeredgecolor=True,
zorders=None, clip_on=True,axesequal=False,
xAxisFmt=None, yAxisFmt=None,
PLcolor=None,
PLwidth=None, PLdash=None, PLyAxisSide=None, PLyAxisOverlaying=None,
PLmultipleYAxis=False, PLmultiAxisTitle=None, PLxAxisSide=None,
PLxAxisOverlaying=None, PLmultipleXAxis=False ): #Plotly initialization parameters
"""Cartesian plot on linear scales for abscissa and ordinates.
Given an existing figure, this function plots in a specified subplot position.
The function arguments are described below in some detail. Note that the y-values
or ordinates can be more than one column, each column representing a different
line in the plot. This is convenient if large arrays of data must be plotted. If more
than one column is present, the label argument can contain the legend labels for
each of the columns/lines. The pltaxis argument defines the min/max scale values
for the x and y axes.
Args:
| plotnum (int): subplot number, 1-based index
| x (np.array[N,] or [N,1]): abscissa
| y (np.array[N,] or [N,M]): ordinates - could be M columns
| ptitle (string): plot title (optional)
| xlabel (string): x-axis label (optional)
| ylabel (string): y-axis label (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if [] (optional)
| linewidths ([float]): plot line width in points, list with M entries, use default if None (optional)
| label ([strings]): legend label for ordinate, list with M entries (optional)
| legendAlpha (float): transparency for legend box (optional)
| legendLoc (string): location for legend box (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None. (optional)
| maxNX (int): draw maxNX+1 tick labels on x axis (optional)
| maxNY (int): draw maxNY+1 tick labels on y axis (optional)
| linestyle (string): linestyle for this plot (optional)
| powerLimits[float]: scientific tick label power limits [x-low, x-high, y-low, y-high] (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis tick font size, default 10pt (optional)
| labelfsize (int): label/legend font size, default 10pt (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| yInvert (bool): invert the y-axis (optional)
| xInvert (bool): invert the x-axis (optional)
| xIsDate (bool): convert the datetime x-values to dates (optional)
| xTicks ({tick:label}): dict of x-axis tick locations and associated labels (optional)
| xtickRotation (float) x-axis tick label rotation angle (optional)
| markers ([string]) markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)) subsample when using markers (optional)
| markerfacecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| markeredgecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| zorders ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| axesequal (bool) force scaling on x and y axes to be equal (optional)
| xAxisFmt (string) x-axis format string, e.g., '%.2f', default None (optional)
| yAxisFmt (string) y-axis format string, e.g., '%.2e',, default None (optional)
| PLcolor (string): graph color scheme. Format 'rgb(r,g,b)'
| PLwidth
| PLdash (string): Line stlye
| PLyAxisSide (string): Sets the location of the y-axis (left/right)
| PLyAxisOverlaying (string): Sets the overlaying
| PLmultipleYAxis (bool): Indicates presence of multiple axis
| PLmultiAxisTitle (string): Sets the title of the multiple axis
| PLxAxisSide (string): Sets the location of the x-axis (top/bottom)
| PLxAxisOverlaying (string): Sets the overlaying
| PLmultipleXAxis (bool): Indicates presence of multiple axis
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
#Plotly variables initialization
self.PLcolor=PLcolor
self.PLwidth=PLwidth
self.PLdash=PLdash
self.PLmultipleYAxis=PLmultipleYAxis
self.PLmultiAxisTitle=PLmultiAxisTitle
self.PLyAxisSide=PLyAxisSide
self.PLyAxisOverlaying=PLyAxisOverlaying
self.PLmultipleXAxis=PLmultipleXAxis
self.PLxAxisSide=PLxAxisSide
self.PLxAxisOverlaying=PLxAxisOverlaying
## see self.MyPlot for parameter details.
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum)
ax = self.subplots[pkey]
self.myPlot(ax.plot, plotnum, x, y, ptitle, xlabel, ylabel,
plotCol, linewidths, label,legendAlpha, legendLoc,
pltaxis, maxNX, maxNY, linestyle,
powerLimits,titlefsize,
xylabelfsize, xytickfsize,
labelfsize, drawGrid,
xScientific, yScientific,
yInvert, xInvert, xIsDate,
xTicks, xtickRotation,
markers, markevery, markerfacecolor,markeredgecolor,
zorders, clip_on,axesequal,
xAxisFmt,yAxisFmt)
return ax
############################################################
##
def logLog(self, plotnum, x, y, ptitle=None, xlabel=None, ylabel=None,
plotCol=[], linewidths=None, label=[],legendAlpha=0.0,
legendLoc='best',
pltaxis=None, maxNX=10, maxNY=10, linestyle=None,
powerLimits = [-4, 2, -4, 2], titlefsize = 12,
xylabelfsize = 12, xytickfsize = 10,labelfsize=10,
xScientific=False, yScientific=False,
yInvert=False, xInvert=False, drawGrid=True,xIsDate=False,
xTicks=None, xtickRotation=0,
markers=[], markevery=None, markerfacecolor=True,markeredgecolor=True,
zorders=None, clip_on=True,axesequal=False,
xAxisFmt=None, yAxisFmt=None,
PLcolor=None,
PLwidth=None, PLdash=None, PLyAxisSide=None, PLyAxisOverlaying=None,
PLmultipleYAxis=False, PLmultiAxisTitle=None):
"""Plot data on logarithmic scales for abscissa and ordinates.
Given an existing figure, this function plots in a specified subplot position.
The function arguments are described below in some detail. Note that the y-values
or ordinates can be more than one column, each column representing a different
line in the plot. This is convenient if large arrays of data must be plotted. If more
than one column is present, the label argument can contain the legend labels for
each of the columns/lines. The pltaxis argument defines the min/max scale values
for the x and y axes.
Args:
| plotnum (int): subplot number, 1-based index
| x (np.array[N,] or [N,1]): abscissa
| y (np.array[N,] or [N,M]): ordinates - could be M columns
| ptitle (string): plot title (optional)
| xlabel (string): x-axis label (optional)
| ylabel (string): y-axis label (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if [] (optional)
| linewidths ([float]): plot line width in points, list with M entries, use default if None (optional)
| label ([strings]): legend label for ordinate, list with M entries (optional)
| legendAlpha (float): transparency for legend box (optional)
| legendLoc (string): location for legend box (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None. (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None. (optional)
| maxNX (int): draw maxNX+1 tick labels on x axis (optional)
| maxNY (int): draw maxNY+1 tick labels on y axis (optional)
| linestyle (string): linestyle for this plot (optional)
| powerLimits[float]: scientific tick label power limits [x-low, x-high, y-low, y-high] (optional) (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis tick font size, default 10pt (optional)
| labelfsize (int): label/legend font size, default 10pt (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| yInvert (bool): invert the y-axis (optional)
| xInvert (bool): invert the x-axis (optional)
| xIsDate (bool): convert the datetime x-values to dates (optional)
| xTicks ({tick:label}): dict of x-axis tick locations and associated labels (optional)
| xtickRotation (float) x-axis tick label rotation angle (optional)
| markers ([string]) markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)) subsample when using markers (optional)
| markerfacecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| markeredgecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| zorders ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| axesequal (bool) force scaling on x and y axes to be equal (optional)
| xAxisFmt (string) x-axis format string, e.g., '%.2f', default None (optional)
| yAxisFmt (string) y-axis format string, e.g., '%.2e',, default None (optional)
| PLcolor (string): graph color scheme. Format 'rgb(r,g,b)'
| PLwidth
| PLdash (string): Line stlye
| PLyAxisSide (string): Sets the location of the y-axis (left/right)
| PLyAxisOverlaying (string): Sets the overlaying
| PLmultipleYAxis (bool): Indicates presence of multiple axis
| PLmultiAxisTitle (string): Sets the title of the multiple axis
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
# Plotly variables initialization
self.PLcolor=PLcolor
self.PLwidth=PLwidth
self.PLdash=PLdash
self.PLmultipleYAxis=PLmultipleYAxis
self.PLmultiAxisTitle=PLmultiAxisTitle
self.PLyAxisSide=PLyAxisSide
self.PLyAxisOverlaying=PLyAxisOverlaying
## see self.MyPlot for parameter details.
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum)
ax = self.subplots[pkey]
# self.myPlot(ax.loglog, plotnum, x, y, ptitle, xlabel,ylabel,\
# plotCol, label,legendAlpha, pltaxis, \
# maxNX, maxNY, linestyle, powerLimits,titlefsize,xylabelfsize,
# xytickfsize,labelfsize, drawGrid
# xTicks, xtickRotation,
# markers=markers)
self.myPlot(ax.loglog, plotnum, x, y, ptitle, xlabel, ylabel,
plotCol, linewidths, label,legendAlpha, legendLoc,
pltaxis, maxNX, maxNY, linestyle,
powerLimits,titlefsize,
xylabelfsize, xytickfsize,
labelfsize, drawGrid,
xScientific, yScientific,
yInvert, xInvert, xIsDate,
xTicks, xtickRotation,
markers, markevery, markerfacecolor,markeredgecolor,
zorders, clip_on,axesequal,
xAxisFmt,yAxisFmt)
return ax
############################################################
##
def semilogX(self, plotnum, x, y, ptitle=None, xlabel=None, ylabel=None,
plotCol=[], linewidths=None, label=[],legendAlpha=0.0,
legendLoc='best',
pltaxis=None, maxNX=10, maxNY=10, linestyle=None,
powerLimits = [-4, 2, -4, 2], titlefsize = 12,
xylabelfsize = 12, xytickfsize = 10,labelfsize=10,
xScientific=False, yScientific=False,
yInvert=False, xInvert=False, drawGrid=True,xIsDate=False,
xTicks=None, xtickRotation=0,
markers=[], markevery=None, markerfacecolor=True,markeredgecolor=True,
zorders=None, clip_on=True, axesequal=False,
xAxisFmt=None, yAxisFmt=None,
PLcolor=None,
PLwidth=None, PLdash=None, PLyAxisSide=None, PLyAxisOverlaying=None,
PLmultipleYAxis=False, PLmultiAxisTitle=None):
"""Plot data on logarithmic scales for abscissa and linear scale for ordinates.
Given an existing figure, this function plots in a specified subplot position.
The function arguments are described below in some detail. Note that the y-values
or ordinates can be more than one column, each column representing a different
line in the plot. This is convenient if large arrays of data must be plotted. If more
than one column is present, the label argument can contain the legend labels for
each of the columns/lines. The pltaxis argument defines the min/max scale values
for the x and y axes.
Args:
| plotnum (int): subplot number, 1-based index
| x (np.array[N,] or [N,1]): abscissa
| y (np.array[N,] or [N,M]): ordinates - could be M columns
| ptitle (string): plot title (optional)
| xlabel (string): x-axis label (optional)
| ylabel (string): y-axis label (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if [] (optional)
| linewidths ([float]): plot line width in points, list with M entries, use default if None (optional)
| label ([strings]): legend label for ordinate, list with M entries (optional)
| legendAlpha (float): transparency for legend box (optional)
| legendLoc (string): location for legend box (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None. (optional)
| maxNX (int): draw maxNX+1 tick labels on x axis (optional)
| maxNY (int): draw maxNY+1 tick labels on y axis (optional)
| linestyle (string): linestyle for this plot (optional)
| powerLimits[float]: scientific tick label notation power limits [x-low, x-high, y-low, y-high] (optional) (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis tick font size, default 10pt (optional)
| labelfsize (int): label/legend font size, default 10pt (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| yInvert (bool): invert the y-axis (optional)
| xInvert (bool): invert the x-axis (optional)
| xIsDate (bool): convert the datetime x-values to dates (optional)
| xTicks ({tick:label}): dict of x-axis tick locations and associated labels (optional)
| xtickRotation (float) x-axis tick label rotation angle (optional)
| markers ([string]) markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)) subsample when using markers (optional)
| markerfacecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| markeredgecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| zorders ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| axesequal (bool) force scaling on x and y axes to be equal (optional)
| xAxisFmt (string) x-axis format string, e.g., '%.2f', default None (optional)
| yAxisFmt (string) y-axis format string, e.g., '%.2e',, default None (optional)
| PLcolor (string): graph color scheme. Format 'rgb(r,g,b)'
| PLwidth
| PLdash (string): Line stlye
| PLyAxisSide (string): Sets the location of the y-axis (left/right)
| PLyAxisOverlaying (string): Sets the overlaying
| PLmultipleYAxis (bool): Indicates presence of multiple axis
| PLmultiAxisTitle (string): Sets the title of the multiple axis
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
#Plotly variables initialization
self.PLcolor=PLcolor
self.PLwidth=PLwidth
self.PLdash=PLdash
self.PLmultipleYAxis=PLmultipleYAxis
self.PLmultiAxisTitle=PLmultiAxisTitle
self.PLyAxisSide=PLyAxisSide
self.PLyAxisOverlaying=PLyAxisOverlaying
## see self.MyPlot for parameter details.
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum)
ax = self.subplots[pkey]
self.myPlot(ax.semilogx, plotnum, x, y, ptitle, xlabel, ylabel,\
plotCol, linewidths, label,legendAlpha, legendLoc,
pltaxis, maxNX, maxNY, linestyle,
powerLimits, titlefsize,
xylabelfsize, xytickfsize,
labelfsize, drawGrid,
xScientific, yScientific,
yInvert, xInvert, xIsDate,
xTicks, xtickRotation,
markers, markevery, markerfacecolor,markeredgecolor,
zorders, clip_on,axesequal,
xAxisFmt,yAxisFmt)
return ax
############################################################
##
def semilogY(self, plotnum, x, y, ptitle=None, xlabel=None, ylabel=None,
plotCol=[], linewidths=None, label=[],legendAlpha=0.0,
legendLoc='best',
pltaxis=None, maxNX=10, maxNY=10, linestyle=None,
powerLimits = [-4, 2, -4, 2], titlefsize = 12,
xylabelfsize = 12, xytickfsize = 10, labelfsize=10,
xScientific=False, yScientific=False,
yInvert=False, xInvert=False, drawGrid=True,xIsDate=False,
xTicks=None, xtickRotation=0,
markers=[], markevery=None, markerfacecolor=True,markeredgecolor=True,
zorders=None, clip_on=True,axesequal=False,
xAxisFmt=None, yAxisFmt=None,
PLcolor=None,
PLwidth=None, PLdash=None, PLyAxisSide=None, PLyAxisOverlaying=None,
PLmultipleYAxis=False, PLmultiAxisTitle=None):
"""Plot data on linear scales for abscissa and logarithmic scale for ordinates.
Given an existing figure, this function plots in a specified subplot position.
The function arguments are described below in some detail. Note that the y-values
or ordinates can be more than one column, each column representing a different
line in the plot. This is convenient if large arrays of data must be plotted. If more
than one column is present, the label argument can contain the legend labels for
each of the columns/lines. The pltaxis argument defines the min/max scale values
for the x and y axes.
Args:
| plotnum (int): subplot number, 1-based index
| x (np.array[N,] or [N,1]): abscissa
| y (np.array[N,] or [N,M]): ordinates - could be M columns
| ptitle (string): plot title (optional)
| xlabel (string): x-axis label (optional)
| ylabel (string): y-axis label (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if [] (optional)
| linewidths ([float]): plot line width in points, list with M entries, use default if None (optional)
| label ([strings]): legend label for ordinate, list withM entries (optional)
| legendAlpha (float): transparency for legend box (optional)
| legendLoc (string): location for legend box (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None. (optional)
| maxNX (int): draw maxNX+1 tick labels on x axis (optional)
| maxNY (int): draw maxNY+1 tick labels on y axis (optional)
| linestyle (string): linestyle for this plot (optional)
| powerLimits[float]: scientific tick label power limits [x-low, x-high, y-low, y-high] (optional) (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis tick font size, default 10pt (optional)
| labelfsize (int): label/legend font size, default 10pt (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| yInvert (bool): invert the y-axis (optional)
| xInvert (bool): invert the x-axis (optional)
| xIsDate (bool): convert the datetime x-values to dates (optional)
| xTicks ({tick:label}): dict of x-axis tick locations and associated labels (optional)
| xtickRotation (float) x-axis tick label rotation angle (optional)
| markers ([string]) markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)) subsample when using markers (optional)
| markerfacecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| markeredgecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| zorders ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| axesequal (bool) force scaling on x and y axes to be equal (optional)
| xAxisFmt (string) x-axis format string, e.g., '%.2f', default None (optional)
| yAxisFmt (string) y-axis format string, e.g., '%.2e',, default None (optional)
| PLcolor (string): graph color scheme. Format 'rgb(r,g,b)'
| PLwidth
| PLdash (string): Line stlye
| PLyAxisSide (string): Sets the location of the y-axis (left/right)
| PLyAxisOverlaying (string): Sets the overlaying
| PLmultipleYAxis (bool): Indicates presence of multiple axis
| PLmultiAxisTitle (string): Sets the title of the multiple axis
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
#Plotly variables initialization
self.PLcolor=PLcolor
self.PLwidth=PLwidth
self.PLdash=PLdash
self.PLmultipleYAxis=PLmultipleYAxis
self.PLmultiAxisTitle=PLmultiAxisTitle
self.PLyAxisSide=PLyAxisSide
self.PLyAxisOverlaying=PLyAxisOverlaying
## see self.MyPlot for parameter details.
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum)
ax = self.subplots[pkey]
self.myPlot(ax.semilogy, plotnum, x, y, ptitle,xlabel,ylabel,
plotCol, linewidths, label,legendAlpha, legendLoc,
pltaxis, maxNX, maxNY, linestyle,
powerLimits, titlefsize,
xylabelfsize, xytickfsize,
labelfsize, drawGrid,
xScientific, yScientific,
yInvert, xInvert, xIsDate,
xTicks, xtickRotation,
markers, markevery, markerfacecolor,markeredgecolor,
zorders, clip_on,
axesequal,xAxisFmt,yAxisFmt)
return ax
############################################################
##
def stackplot(self, plotnum, x, y, ptitle=None, xlabel=None, ylabel=None,
plotCol=[], linewidths=None, label=[],legendAlpha=0.0,
legendLoc='best',
pltaxis=None, maxNX=10, maxNY=10, linestyle=None,
powerLimits = [-4, 2, -4, 2], titlefsize = 12,
xylabelfsize = 12, xytickfsize = 10, labelfsize=10,
xScientific=False, yScientific=False,
yInvert=False, xInvert=False, drawGrid=True,xIsDate=False,
xTicks=None, xtickRotation=0,
markers=[], markevery=None, markerfacecolor=True,markeredgecolor=True,
zorders=None, clip_on=True, axesequal=False,
xAxisFmt=None, yAxisFmt=None,
PLcolor=None,
PLwidth=None, PLdash=None, PLyAxisSide=None, PLyAxisOverlaying=None,
PLmultipleYAxis=False, PLmultiAxisTitle=None):
"""Plot stacked data on linear scales for abscissa and ordinates.
Given an existing figure, this function plots in a specified subplot position.
The function arguments are described below in some detail. Note that the y-values
or ordinates can be more than one column, each column representing a different
line in the plot. If more
than one column is present, the label argument can contain the legend labels for
each of the columns/lines. The pltaxis argument defines the min/max scale values
for the x and y axes.
Args:
| plotnum (int): subplot number, 1-based index
| x (np.array[N,] or [N,1]): abscissa
| y (np.array[N,] or [N,M]): ordinates - could be M columns
| ptitle (string): plot title (optional)
| xlabel (string): x-axis label (optional)
| ylabel (string): y-axis label (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if [] (optional)
| linewidths ([float]): plot line width in points, list with M entries, use default if None (optional)
| label ([strings]): legend label for ordinate, list withM entries (optional)
| legendAlpha (float): transparency for legend box (optional)
| legendLoc (string): location for legend box (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None. (optional)
| maxNX (int): draw maxNX+1 tick labels on x axis (optional)
| maxNY (int): draw maxNY+1 tick labels on y axis (optional)
| linestyle (string): linestyle for this plot (optional)
| powerLimits[float]: scientific tick label power limits [x-low, x-high, y-low, y-high] (optional) (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis tick font size, default 10pt (optional)
| labelfsize (int): label/legend font size, default 10pt (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| yInvert (bool): invert the y-axis (optional)
| xInvert (bool): invert the x-axis (optional)
| xIsDate (bool): convert the datetime x-values to dates (optional)
| xTicks ({tick:label}): dict of x-axis tick locations and associated labels (optional)
| xtickRotation (float) x-axis tick label rotation angle (optional)
| markers ([string]) markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)) subsample when using markers (optional)
| markerfacecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| markeredgecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| zorders ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| axesequal (bool) force scaling on x and y axes to be equal (optional)
| xAxisFmt (string) x-axis format string, e.g., '%.2f', default None (optional)
| yAxisFmt (string) y-axis format string, e.g., '%.2e',, default None (optional)
| PLcolor (string): graph color scheme. Format 'rgb(r,g,b)'
| PLwidth
| PLdash (string): Line stlye
| PLyAxisSide (string): Sets the location of the y-axis (left/right)
| PLyAxisOverlaying (string): Sets the overlaying
| PLmultipleYAxis (bool): Indicates presence of multiple axis
| PLmultiAxisTitle (string): Sets the title of the multiple axis
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
#Plotly variables initialization
self.PLcolor=PLcolor
self.PLwidth=PLwidth
self.PLdash=PLdash
self.PLmultipleYAxis=PLmultipleYAxis
self.PLmultiAxisTitle=PLmultiAxisTitle
self.PLyAxisSide=PLyAxisSide
self.PLyAxisOverlaying=PLyAxisOverlaying
## see self.MyPlot for parameter details.
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum)
ax = self.subplots[pkey]
self.myPlot(ax.stackplot, plotnum, x, y.T, ptitle,xlabel,ylabel,
plotCol, linewidths, label,legendAlpha, legendLoc,
pltaxis, maxNX, maxNY, linestyle,
powerLimits, titlefsize,
xylabelfsize, xytickfsize,
labelfsize, drawGrid,
xScientific, yScientific,
yInvert, xInvert, xIsDate,
xTicks, xtickRotation,
markers, markevery, markerfacecolor,markeredgecolor,
zorders, clip_on,
axesequal,xAxisFmt,yAxisFmt)
return ax
############################################################
##
def myPlot(self, plotcommand,plotnum, x, y, ptitle=None,xlabel=None, ylabel=None,
plotCol=[], linewidths=None, label=[], legendAlpha=0.0,
legendLoc='best',
pltaxis=None, maxNX=0, maxNY=0, linestyle=None,
powerLimits = [-4, 2, -4, 2], titlefsize = 12,
xylabelfsize = 12, xytickfsize = 10,
labelfsize=10, drawGrid=True,
xScientific=False, yScientific=False,
yInvert=False, xInvert=False, xIsDate=False,
xTicks=None, xtickRotation=0,
markers=[], markevery=None,
markerfacecolor=True,markeredgecolor=True,
zorders=None,clip_on=True,axesequal=False,
xAxisFmt=None,yAxisFmt=None,
PLyStatic=[0]
):
"""Low level helper function to create a subplot and plot the data as required.
This function does the actual plotting, labelling etc. It uses the plotting
function provided by its user functions.
lineStyles = {
'': '_draw_nothing',
' ': '_draw_nothing',
'None': '_draw_nothing',
'--': '_draw_dashed',
'-.': '_draw_dash_dot',
'-': '_draw_solid',
':': '_draw_dotted'}
Args:
| plotcommand: name of a MatplotLib plotting function
| plotnum (int): subplot number, 1-based index
| ptitle (string): plot title
| xlabel (string): x axis label
| ylabel (string): y axis label
| x (np.array[N,] or [N,1]): abscissa
| y (np.array[N,] or [N,M]): ordinates - could be M columns
| plotCol ([strings]): plot colour and line style, list with M entries, use default if []
| linewidths ([float]): plot line width in points, list with M entries, use default if None (optional)
| label ([strings]): legend label for ordinate, list with M entries
| legendAlpha (float): transparency for legend box
| legendLoc (string): location for legend box (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None.
| maxNX (int): draw maxNX+1 tick labels on x axis
| maxNY (int): draw maxNY+1 tick labels on y axis
| linestyle (string): linestyle for this plot (optional)
| powerLimits[float]: scientific tick label power limits [x-low, x-high, y-low, y-high] (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis tick font size, default 10pt (optional)
| labelfsize (int): label/legend font size, default 10pt (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| yInvert (bool): invert the y-axis (optional)
| xInvert (bool): invert the x-axis (optional)
| xIsDate (bool): convert the datetime x-values to dates (optional)
| xTicks ({tick:label}): dict of x-axis tick locations and associated labels (optional)
| xtickRotation (float) x-axis tick label rotation angle (optional)
| markers ([string]) markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)) subsample when using markers (optional)
| markerfacecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| markeredgecolor (True|None|str) if True same as plotCol, if None empty, otherwise str is colour (optional)
| zorders ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| axesequal (bool) force scaling on x and y axes to be equal (optional)
| xAxisFmt (string) x-axis format string, e.g., '%.2f', default None (optional)
| yAxisFmt (string) y-axis format string, e.g., '%.2e',, default None (optional)
| PLyStatic ([int]) the guy that added this did not document it properly
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
#Initialize plotlyPlot call when Plotly is activated
if self.useplotly:
self.PlotlyPlotCalls = self.PlotlyPlotCalls + 1
if x.ndim>1:
xx=x
else:
if type(x)==type(pd.Series()):
x = x.values
xx=x.reshape(-1, 1)
if y.ndim>1:
yy=y
else:
if type(y)==type(pd.Series()):
y = y.values
yy=y.reshape(-1, 1)
# plotCol = self.buildPlotCol(plotCol, yy.shape[1])
pkey = (self.nrow, self.ncol, plotnum)
ax = self.subplots[pkey]
if drawGrid:
ax.grid(True)
else:
ax.grid(False)
# use scientific format on axes
#yfm = sbp.yaxis.get_major_formatter()
#yfm.set_powerlimits([ -3, 3])
if xlabel is not None:
ax.set_xlabel(xlabel, fontsize=xylabelfsize)
if ylabel is not None:
ax.set_ylabel(ylabel, fontsize=xylabelfsize)
if xIsDate:
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%Y-%m-%d'))
ax.xaxis.set_major_locator(mdates.DayLocator())
if maxNX >0:
ax.xaxis.set_major_locator(mpl.ticker.MaxNLocator(maxNX))
if maxNY >0:
ax.yaxis.set_major_locator(mpl.ticker.MaxNLocator(maxNY))
if xScientific:
# formx = plt.FormatStrFormatter('%.3e')
formx = plt.ScalarFormatter()
formx.set_powerlimits([powerLimits[0], powerLimits[1]])
formx.set_scientific(True)
ax.xaxis.set_major_formatter(formx)
# http://matplotlib.1069221.n5.nabble.com/ScalarFormatter-td28042.html
# http://matplotlib.org/api/ticker_api.html
# http://matplotlib.org/examples/pylab_examples/newscalarformatter_demo.html
# ax.xaxis.set_major_formatter( plt.FormatStrFormatter('%d'))
# http://matplotlib.org/1.3.1/api/axes_api.html#matplotlib.axes.Axes.ticklabel_format
# plt.ticklabel_format(style='sci', axis='x',
# scilimits=(powerLimits[0], powerLimits[1]))
if yScientific:
formy = plt.ScalarFormatter()
formy.set_powerlimits([powerLimits[2], powerLimits[3]])
formy.set_scientific(True)
ax.yaxis.set_major_formatter(formy)
# this user-defined format setting is given at the end of the function.
# # override the format with user defined
# if xAxisFmt is not None:
# ax.xaxis.set_major_formatter(FormatStrFormatter(xAxisFmt))
# if yAxisFmt is not None:
# ax.yaxis.set_major_formatter(FormatStrFormatter(yAxisFmt))
###############################stacked plot #######################
if plotcommand==ax.stackplot:
if not self.useplotly:
if not plotCol:
plotCol = [self.nextPlotCol() for col in range(0,yy.shape[0])]
ax.stackplot(xx.reshape(-1), yy, colors=plotCol)
ax.margins(0, 0) # Set margins to avoid "whitespace"
# creating the legend manually
ax.legend([mpl.patches.Patch(color=col) for col in plotCol], label,
loc=legendLoc, framealpha=legendAlpha)
else: #Plotly stacked plot
#Plotly stacked plot variables
PLXAxis = 0
PLYAxis = 0
for i in range(yy.shape[0]):
PLXAxis = dict(type='category',)
PLYAxis = dict(type='linear')
try:
if len(y[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x, y=y[i,:]+PLyStatic[0],mode='lines', name = label,fill='tonexty',line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
PLyStatic[0] += y[i,:]
elif len(x[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,i], y=y,mode='lines', name = label,fill='tonexty',line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
except:
self.Plotlydata.append(Scatter(x=x, y=y, name = label,fill='tonexty',line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
###############################line plot #######################
else: # not a stacked plot
for i in range(yy.shape[1]):
#set up the line style, either given or next in sequence
mmrk = ''
if markers:
if i >= len(markers):
mmrk = markers[-1]
else:
mmrk = markers[i]
if plotCol:
if i >= len(plotCol):
col = plotCol[-1]
else:
col = plotCol[i]
else:
col = self.nextPlotCol()
if markerfacecolor==True:
markerfacecolor = col
elif markerfacecolor is None:
markerfacecolor='none'
else:
pass # keep as is
if markeredgecolor==True:
markeredgecolor = col
elif markeredgecolor is None:
markeredgecolor='none'
else:
pass # keep as is
if linestyle is None:
linestyleL = '-'
else:
if type(linestyle) == type([1]):
linestyleL = linestyle[i]
else:
linestyleL = linestyle
if zorders:
if len(zorders) > 1:
zorder = zorders[i]
else:
zorder = zorders[0]
else:
zorder = 2
if not self.useplotly:
if not label:
if linewidths is not None:
plotcommand(xx, yy[:, i], col, label=None, linestyle=linestyleL,
markerfacecolor=markerfacecolor,markeredgecolor=markeredgecolor,
marker=mmrk, markevery=markevery, linewidth=linewidths[i],
clip_on=clip_on, zorder=zorder)
else:
plotcommand(xx, yy[:, i], col, label=None, linestyle=linestyleL,
markerfacecolor=markerfacecolor,markeredgecolor=markeredgecolor,
marker=mmrk, markevery=markevery,
clip_on=clip_on, zorder=zorder)
else:
if linewidths is not None:
# print('***************',linewidths)
line, = plotcommand(xx,yy[:,i],col,#label=label[i],
linestyle=linestyleL,
markerfacecolor=markerfacecolor,markeredgecolor=markeredgecolor,
marker=mmrk, markevery=markevery, linewidth=linewidths[i],
clip_on=clip_on, zorder=zorder)
else:
line, = plotcommand(xx,yy[:,i],col,#label=label[i],
linestyle=linestyleL,
markerfacecolor=markerfacecolor,markeredgecolor=markeredgecolor,
marker=mmrk, markevery=markevery,
clip_on=clip_on, zorder=zorder)
line.set_label(label[i])
leg = ax.legend( loc=legendLoc, fancybox=True,fontsize=labelfsize)
leg.get_frame().set_alpha(legendAlpha)
# ax.legend()
self.bbox_extra_artists.append(leg)
else:#Plotly plots
if 'loglog' in str(plotcommand):
PLXAxis = dict(type='log',showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=xlabel,mirror='all')
PLYAxis = dict(type='log',showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=ylabel,mirror='all')
# Assuming that either y or x has to 1
try:
if len(x[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,i], y=y, name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
elif len(y[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,0], y=y[:,i], name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
except:
self.Plotlydata.append(Scatter(x=x, y=y, name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
# Append axis and plot titles
if self.ncol > 1:
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif self.nrow > 1 :
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif 'semilogx' in str(plotcommand):
PLXAxis = dict(type='log',showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=xlabel,mirror='all')
PLYAxis = dict(showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=ylabel,mirror='all')
# Assuming that either y or x has to 1
try:
if len(x[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,i], y=y, name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
elif len(y[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,0], y=y[:,i], name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
except:
self.Plotlydata.append(Scatter(x=x, y=y, name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
# Append axis and plot titles
if self.ncol > 1:
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif self.nrow > 1 :
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif 'semilogy' in str(plotcommand):
PLXAxis = dict(showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=xlabel,mirror='all')
PLYAxis = dict(type='log',showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=ylabel,mirror='all')
# Assuming that either y or x has to 1
try:
if len(x[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,i], y=y, name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
elif len(y[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,0], y=y[:,i], name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
except:
self.Plotlydata.append(Scatter(x=x, y=y, name = label,line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
# Append axis and plot titles
if self.ncol > 1:
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif self.nrow > 1 :
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
else:
PLXAxis = dict(showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=xlabel,mirror='all')
PLYAxis = dict(showgrid=drawGrid,zeroline=False,nticks=20,showline=True,title=ylabel,mirror='all')
# Assuming that either y or x has to 1
try:
if len(x[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,i], y=y, name = label,xaxis='x1',
line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
elif len(y[0,:]) > 1:
self.Plotlydata.append(Scatter(x=x[:,0], y=y[:,i], name = label,xaxis='x1',
line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
except:
self.Plotlydata.append(Scatter(x=x, y=y, name = label,xaxis='x1',
line = dict(color = self.PLcolor, width = self.PLwidth, dash = self.PLdash)))
# Append axis and plot titles
if self.ncol > 1:
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif self.nrow > 1 :
self.PlotlySubPlotNumbers.append(plotnum)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
#Plotly plots setup
if self.useplotly:
if self.PLmultipleYAxis:
self.Plotlylayout.append(Layout(showlegend = True,title = ptitle,xaxis = PLXAxis,yaxis=PLYAxis,yaxis2=dict(title=self.PLmultiAxisTitle,side=self.PLyAxisSide,overlaying=self.PLyAxisOverlaying)))
elif self.PLmultipleXAxis:
self.Plotlylayout.append(Layout(showlegend = True,title = ptitle,yaxis=PLYAxis,xaxis = PLXAxis,xaxis2=dict(title=self.PLmultiAxisTitle,side=self.PLxAxisSide,overlaying=self.PLxAxisOverlaying)))
else:
self.Plotlylayout.append(Layout(showlegend = True,title = ptitle,xaxis = PLXAxis,yaxis=PLYAxis))
if self.ncol > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlySubPlotLabels.append(label)
elif self.nrow > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlySubPlotLabels.append(label)
if xIsDate:
plt.gcf().autofmt_xdate()
#scale the axes
if pltaxis is not None:
# ax.axis(pltaxis)
if not xIsDate:
ax.set_xlim(pltaxis[0],pltaxis[1])
ax.set_ylim(pltaxis[2],pltaxis[3])
if xTicks is not None:
ticks = ax.set_xticks(list(xTicks.keys()))
ax.set_xticklabels([xTicks[key] for key in xTicks],
rotation=xtickRotation, fontsize=xytickfsize)
if xTicks is None and xtickRotation is not None:
ticks = ax.get_xticks()
if xIsDate:
from datetime import date
ticks = [date.fromordinal(int(tick)).strftime('%Y-%m-%d') for tick in ticks]
ax.set_xticks(ticks) # this is workaround for bug in matplotlib
ax.set_xticklabels(ticks,
rotation=xtickRotation, fontsize=xytickfsize)
if(ptitle is not None):
ax.set_title(ptitle, fontsize=titlefsize)
# minor ticks are two points smaller than major
ax.tick_params(axis='both', which='major', labelsize=xytickfsize)
ax.tick_params(axis='both', which='minor', labelsize=xytickfsize-2)
if yInvert:
ax.set_ylim(ax.get_ylim()[::-1])
if xInvert:
ax.set_xlim(ax.get_xlim()[::-1])
if axesequal:
ax.axis('equal')
# override the format with user defined
if xAxisFmt is not None:
ax.xaxis.set_major_formatter(FormatStrFormatter(xAxisFmt))
if yAxisFmt is not None:
ax.yaxis.set_major_formatter(FormatStrFormatter(yAxisFmt))
return ax
############################################################
#Before this function is called, plot data is accumulated in runtime variables
#At the call of this function the Plotly plots are plotted using the accumulated data.
def plotlyPlot(self,filename=None,image=None,image_filename=None,auto_open=True):
if ((self.nrow == self.ncol) & self.ncol == 1 & self.nrow == 1 ): #No subplots
fig = Figure(data=self.Plotlydata,layout=self.Plotlylayout[0])
fig['layout'].update(title=str(self.figuretitle))
else:
dataFormatCatch = 0
try:
len(self.Plotlydata[0].y[1,:])
dataFormatCatch = 0
except:
dataFormatCatch = 1
if self.PLIs3D:
specRow = []
specCol = []
for r in range(int(self.nrow)):
specRow.append({'is_3d': True})
for r in range(int(self.ncol)):
specCol.append({'is_3d': True})
fig = tools.make_subplots(rows=int(self.nrow), cols=int(self.nrow), specs=[specRow,specCol])#[[{'is_3d': True}, {'is_3d': True}], [{'is_3d': True}, {'is_3d': True}]])
else:
fig = tools.make_subplots(int(self.nrow), int(self.ncol), subplot_titles=self.PlotlySubPlotTitles)
# make row and column formats
rowFormat = []
colFormat = []
countRows = 1
rowCount = 1
colCount = 1
for tmp in range(int(self.nrow)*int(self.ncol)):
if int(self.nrow) == int(self.ncol):
if countRows == int(self.nrow):
rowFormat.append(rowCount)
rowCount = rowCount + 1
if rowCount > int(self.nrow):
rowCount = 1
countRows = 1
elif countRows < int(self.nrow) :
rowFormat.append(rowCount)
countRows = countRows + 1
if colCount == int(self.ncol):
colFormat.append(colCount)
colCount = 1
elif colCount < int(self.ncol):
colFormat.append(colCount)
colCount = colCount + 1
else:
if rowCount > int(self.nrow):
rowCount = 1
rowFormat.append(rowCount)
rowCount = rowCount + 1
else:
rowFormat.append(rowCount)
rowCount = rowCount + 1
if colCount > int(self.ncol):
colCount = 1
colFormat.append(colCount)
colCount = colCount + 1
else:
colFormat.append(colCount)
colCount = colCount + 1
if dataFormatCatch == 0:
for tmp in range(self.PlotlyPlotCalls):
if self.PLIs3D:
if str(self.PLType) == "plot3d":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,mode=self.Plotlydata[i].mode), rowFormat[tmp], colFormat[tmp])
elif str(self.PLType) == "mesh3D":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,color=self.Plotlydata[i].color), rowFormat[tmp], colFormat[tmp])
else:
if str(self.PLType) == "meshContour":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,PLcolorscale=self.Plotlydata[i].PLcolorscale), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
fig.append_trace(self.Plotlydata, rowFormat[tmp], colFormat[tmp])
else:
rCntrl = 1
rIndex = 1
cIndex = 1
cCntrl = 1
rStpVal = int(len(self.Plotlydata)/len(rowFormat))
cStpVal = int(len(self.Plotlydata)/len(colFormat))
for i in range(len(self.Plotlydata)):
if rCntrl > rStpVal:
rCntrl = 1
rIndex = rIndex+1
if cCntrl > cStpVal:
cCntrl = 1
cIndex = cIndex+1
if self.PLIs3D:
if str(self.PLType) == "plot3d":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,mode=self.Plotlydata[i].mode), rowFormat[rIndex-1], colFormat[cIndex-1])
elif str(self.PLType) == "mesh3D":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,color=self.Plotlydata[i].color), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
if str(self.PLType) == "meshContour":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,PLcolorscale=self.Plotlydata[i].PLcolorscale), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
if(len(self.Plotlydata) == len(rowFormat)):
fig.append_trace(self.Plotlydata[i], rowFormat[i], colFormat[i])
else:
fig.append_trace(self.Plotlydata[i], rowFormat[self.PlotlySubPlotNumbers[i]-1], colFormat[self.PlotlySubPlotNumbers[i]-1])
cCntrl = cCntrl + 1
else:
if self.PLIs3D:
if str(self.PLType) == "plot3d":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,mode=self.Plotlydata[i].mode), rowFormat[rIndex-1], colFormat[cIndex-1])
elif str(self.PLType) == "mesh3D":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,color=self.Plotlydata[i].color), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
if str(self.PLType) == "meshContour":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,PLcolorscale=self.Plotlydata[i].PLcolorscale), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
fig.append_trace(self.Plotlydata[i], rowFormat[rIndex-1], colFormat[cIndex-1])
rCntrl = rCntrl + 1
elif cCntrl > cStpVal:
cCntrl = 1
cIndex = cIndex+1
if rCntrl > rStpVal:
rCntrl = 1
rIndex = rIndex+1
if self.PLIs3D:
if str(self.PLType) == "plot3d":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,mode=self.Plotlydata[i].mode), rowFormat[rIndex-1], colFormat[cIndex-1])
elif str(self.PLType) == "mesh3D":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,color=self.Plotlydata[i].color), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
if str(self.PLType) == "meshContour":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,PLcolorscale=self.Plotlydata[i].PLcolorscale), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
fig.append_trace(self.Plotlydata[i], rowFormat[rIndex-1], colFormat[cIndex-1])
rCntrl = rCntrl + 1
else:
if self.PLIs3D:
if str(self.PLType) == "plot3d":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,mode=self.Plotlydata[i].mode), rowFormat[rIndex-1], colFormat[cIndex-1])
elif str(self.PLType) == "mesh3D":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,color=self.Plotlydata[i].color), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
if str(self.PLType) == "meshContour":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,PLcolorscale=self.Plotlydata[i].PLcolorscale), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
fig.append_trace(self.Plotlydata[i], rowFormat[rIndex-1], colFormat[cIndex-1])
cCntrl = cCntrl + 1
else:
if self.PLIs3D:
if str(self.PLType) == "plot3d":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,mode=self.Plotlydata[i].mode), rowFormat[rIndex-1], colFormat[cIndex-1])
elif str(self.PLType) == "mesh3D":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,color=self.Plotlydata[i].color), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
if str(self.PLType) == "meshContour":
fig.append_trace(dict(type=self.Plotlydata[i].type,x=self.Plotlydata[i].x, y=self.Plotlydata[i].y, z=self.Plotlydata[i].z,name=self.Plotlydata[i].name,PLcolorscale=self.Plotlydata[i].PLcolorscale), rowFormat[rIndex-1], colFormat[cIndex-1])
else:
fig.append_trace(self.Plotlydata[i], rowFormat[rIndex-1], colFormat[cIndex-1])
rCntrl = rCntrl + 1
cCntrl = cCntrl + 1
fig['layout'].update(title=str(self.figuretitle))
for j in range(self.PlotlyPlotCalls):
if j < len(self.PlotlyXaxisTitles):
fig['layout']['xaxis'+str(j+1)].update(title=self.PlotlyXaxisTitles[j],type=self.Plotlylayout[j].xaxis.type)
else:
fig['layout']['xaxis'+str(j+1)].update(type=self.Plotlylayout[j].xaxis.type)
if j < len(self.PlotlyYaxisTitles):
fig['layout']['yaxis'+str(j+1)].update(title=self.PlotlyYaxisTitles[j],type=self.Plotlylayout[j].yaxis.type)
else:
fig['layout']['yaxis'+str(j+1)].update(type=self.Plotlylayout[j].yaxis.type)
if filename:
offline.plot(fig,filename=filename)
elif image:
offline.plot(fig,image_filename=image_filename,image=image,auto_open=auto_open)
else:
offline.plot(fig)
############################################################
##
def emptyPlot(self,plotnum,projection='rectilinear'):
"""Returns a handler to an empty plot.
This function does not do any plotting, the use must add plots using
the standard MatPlotLib means.
Args:
| plotnum (int): subplot number, 1-based index
| rectilinear (str): type of axes projection, from
['aitoff', 'hammer', 'lambert', 'mollweide', 'polar', 'rectilinear.].
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum,projection=projection)
ax = self.subplots[pkey]
return ax
############################################################
##
def meshContour(self, plotnum, xvals, yvals, zvals, levels=10,
ptitle=None, xlabel=None, ylabel=None, shading='flat',
plotCol=[], pltaxis=None, maxNX=0, maxNY=0,
xScientific=False, yScientific=False,
powerLimits=[-4, 2, -4, 2], titlefsize=12,
xylabelfsize=12, xytickfsize=10,
meshCmap=cm.rainbow, cbarshow=False, cbarorientation='vertical',
cbarcustomticks=[], cbarfontsize=12,
drawGrid=False, yInvert=False, xInvert=False,
contourFill=True, contourLine=True, logScale=False,
negativeSolid=False, zeroContourLine=None,
contLabel=False, contFmt='%.2f', contCol='k', contFonSz=8, contLinWid=0.5,
zorders=None, PLcolorscale='' ):
"""XY colour mesh countour plot for (xvals, yvals, zvals) input sets.
The data values must be given on a fixed mesh grid of three-dimensional
$(x,y,z)$ array input sets. The mesh grid is defined in $(x,y)$, while the height
of the mesh is the $z$ value.
Given an existing figure, this function plots in a specified subplot position.
Only one contour plot is drawn at a time. Future contours in the same subplot
will cover any previous contours.
The data set must have three two dimensional arrays, each for x, y, and z.
The data in x, y, and z arrays must have matching data points. The x and y arrays
each define the grid in terms of x and y values, i.e., the x array contains the
x values for the data set, while the y array contains the y values. The z array
contains the z values for the corresponding x and y values in the contour mesh.
Z-values can be plotted on a log scale, in which case the colourbar is adjusted
to show true values, but on the nonlinear scale.
The current version only saves png files, since there appears to be a problem
saving eps files.
The xvals and yvals vectors may have non-constant grid-intervals, i.e., they do not
have to be on regular intervals.
Args:
| plotnum (int): subplot number, 1-based index
| xvals (np.array[N,M]): array of x values
| yvals (np.array[N,M]): array of y values
| zvals (np.array[N,M]): values on a (x,y) grid
| levels (int or [float]): number of contour levels or a list of levels (optional)
| ptitle (string): plot title (optional)
| xlabel (string): x axis label (optional)
| ylabel (string): y axis label (optional)
| shading (string): not used currently (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if [] (optional)
| pltaxis ([xmin, xmax, ymin,ymax]): scale for x,y axes. Let Matplotlib decide if None. (optional)
| maxNX (int): draw maxNX+1 tick labels on x axis (optional)
| maxNY (int): draw maxNY+1 tick labels on y axis (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| powerLimits[float]: scientific tick label power limits [x-low, x-high, y-low, y-high] (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis tick font size, default 10pt (optional)
| meshCmap (cm): colour map for the mesh (optional)
| cbarshow (bool): if true, the show a colour bar (optional)
| cbarorientation (string): 'vertical' (right) or 'horizontal' (below) (optional)
| cbarcustomticks zip([z values/float],[tick labels/string])` define custom colourbar ticks locations for given z values(optional)
| cbarfontsize (int): font size for colour bar (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| yInvert (bool): invert the y-axis. Flip the y-axis up-down (optional)
| xInvert (bool): invert the x-axis. Flip the x-axis left-right (optional)
| contourFill (bool): fill contours with colour (optional)
| contourLine (bool): draw a series of contour lines (optional)
| logScale (bool): do Z values on log scale, recompute colourbar values (optional)
| negativeSolid (bool): draw negative contours in solid lines, dashed otherwise (optional)
| zeroContourLine (double): draw a single contour at given value (optional)
| contLabel (bool): label the contours with values (optional)
| contFmt (string): contour label c-printf format (optional)
| contCol (string): contour label colour, e.g., 'k' (optional)
| contFonSz (float): contour label fontsize (optional)
| contLinWid (float): contour line width in points (optional)
| zorders ([int]) list of zorders for drawing sequence, highest is last (optional)
| PLcolorscale (?) Plotly parameter ? (optional)
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
#to rank 2
xx=xvals.reshape(-1, 1)
yy=yvals.reshape(-1, 1)
#if this is a log scale plot
if logScale is True:
zvals = np.log10(zvals)
contour_negative_linestyle = plt.rcParams['contour.negative_linestyle']
if contourLine:
if negativeSolid:
plt.rcParams['contour.negative_linestyle'] = 'solid'
else:
plt.rcParams['contour.negative_linestyle'] = 'dashed'
#create subplot if not existing
if (self.nrow,self.ncol, plotnum) not in list(self.subplots.keys()):
self.subplots[(self.nrow,self.ncol, plotnum)] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum)
#get axis
ax = self.subplots[(self.nrow,self.ncol, plotnum)]
if drawGrid:
ax.grid(True)
else:
ax.grid(False)
if xlabel is not None:
ax.set_xlabel(xlabel, fontsize=xylabelfsize)
if xScientific:
formx = plt.ScalarFormatter()
formx.set_scientific(True)
formx.set_powerlimits([powerLimits[0], powerLimits[1]])
ax.xaxis.set_major_formatter(formx)
if ylabel is not None:
ax.set_ylabel(ylabel, fontsize=xylabelfsize)
if yScientific:
formy = plt.ScalarFormatter()
formy.set_powerlimits([powerLimits[2], powerLimits[3]])
formy.set_scientific(True)
ax.yaxis.set_major_formatter(formy)
if maxNX >0:
ax.xaxis.set_major_locator(mpl.ticker.MaxNLocator(maxNX))
if maxNY >0:
ax.yaxis.set_major_locator(mpl.ticker.MaxNLocator(maxNY))
if plotCol:
col = plotCol[0]
else:
col = self.nextPlotCol()
if zorders is not None:
if len(zorders) > 1:
zorder = zorders[i]
else:
zorder = zorders[0]
else:
zorder = 2
if self.useplotly:
self.PlotlyPlotCalls = self.PlotlyPlotCalls + 1
self.PLType = "meshContour"
if cbarshow:
self.Plotlydata.append(Contour(x=list(itertools.chain.from_iterable(xvals)),
y=list(itertools.chain.from_iterable(yvals)),
z=list(itertools.chain.from_iterable(zvals)),
PLcolorscale=PLcolorscale))
#,color=color,colorbar = ColorBar(PLtickmode=PLtickmode,nticks=PLnticks,
# PLtick0=PLtick0,PLdtick=PLdtick,PLtickvals=PLtickvals,PLticktext=PLticktext),
# PLcolorscale = PLcolorScale,intensity = PLintensity))
else:
self.Plotlydata.append(Contour(x=list(itertools.chain.from_iterable(xvals)),
y=list(itertools.chain.from_iterable(yvals)),
z=list(itertools.chain.from_iterable(zvals)),PLcolorscale=PLcolorscale))
#,color=color))
# Append axis and plot titles
if self.ncol > 1:
self.PlotlySubPlotNumbers.append(plotnum)
elif self.nrow > 1 :
self.PlotlySubPlotNumbers.append(plotnum)
#do the plot
if contourFill:
pmplotcf = ax.contourf(xvals, yvals, zvals, levels,
cmap=meshCmap, zorder=zorder)
if contourLine:
pmplot = ax.contour(xvals, yvals, zvals, levels, cmap=None, linewidths=contLinWid,
colors=col, zorder=zorder)
if zeroContourLine:
pmplot = ax.contour(xvals, yvals, zvals, (zeroContourLine,), cmap=None, linewidths=contLinWid,
colors=col, zorder=zorder)
if contLabel: # and not contourFill:
plt.clabel(pmplot, fmt = contFmt, colors = contCol, fontsize=contFonSz) #, zorder=zorder)
if cbarshow and (contourFill):
#http://matplotlib.org/mpl_toolkits/axes_grid/users/overview.html#colorbar-whose-height-or-width-in-sync-with-the-master-axes
divider = make_axes_locatable(ax)
if cbarorientation == 'vertical':
cax = divider.append_axes("right", size="5%", pad=0.05)
else:
cax = divider.append_axes("bottom", size="5%", pad=0.1)
if not cbarcustomticks:
# cbar = self.fig.colorbar(pmplotcf,orientation=cbarorientation)
cbar = self.fig.colorbar(pmplotcf,cax=cax)
if logScale:
cbartickvals = cbar.ax.yaxis.get_ticklabels()
tickVals = []
# need this roundabout trick to handle minus sign in unicode
for item in cbartickvals:
valstr = float(item.get_text().replace(u'\N{MINUS SIGN}', '-').replace('$',''))
# valstr = item.get_text().replace('\u2212', '-').replace('$','')
val = 10**float(valstr)
if abs(val) < 1000:
str = '{0:f}'.format(val)
else:
str = '{0:e}'.format(val)
tickVals.append(str)
cbartickvals = cbar.ax.yaxis.set_ticklabels(tickVals)
else:
ticks, ticklabels = list(zip(*cbarcustomticks))
# cbar = self.fig.colorbar(pmplotcf,ticks=ticks, orientation=cbarorientation)
cbar = self.fig.colorbar(pmplotcf,ticks=ticks, cax=cax)
if cbarorientation == 'vertical':
cbar.ax.set_yticklabels(ticklabels)
else:
cbar.ax.set_xticklabels(ticklabels)
if cbarorientation == 'vertical':
for t in cbar.ax.get_yticklabels():
t.set_fontsize(cbarfontsize)
else:
for t in cbar.ax.get_xticklabels():
t.set_fontsize(cbarfontsize)
#scale the axes
if pltaxis is not None:
ax.axis(pltaxis)
if(ptitle is not None):
ax.set_title(ptitle, fontsize=titlefsize)
# minor ticks are two points smaller than major
ax.tick_params(axis='both', which='major', labelsize=xytickfsize)
ax.tick_params(axis='both', which='minor', labelsize=xytickfsize-2)
if yInvert:
ax.set_ylim(ax.get_ylim()[::-1])
if xInvert:
ax.set_xlim(ax.get_xlim()[::-1])
plt.rcParams['contour.negative_linestyle'] = contour_negative_linestyle
if self.useplotly:
if self.PLmultipleYAxis:
if yInvert:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel),yaxis=dict(title=ylabel,autorange='reversed')))
elif xInvert:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel,autorange='reversed'),yaxis=dict(title=ylabel)))
else:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel),yaxis=dict(title=ylabel)))#,font=dict(title=self.PLmultiAxisTitle,side=self.PLyAxisSide,overlaying=self.PLyAxisOverlaying)))
elif self.PLmultipleXAxis:
if yInvert:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel),yaxis=dict(title=ylabel,autorange='reversed')))
elif xInvert:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel,autorange='reversed'),yaxis=dict(title=ylabel)))
else:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel),yaxis=dict(title=ylabel)))#,yaxis=PLYAxis,xaxis = PLXAxis,xaxis2=dict(title=self.PLmultiAxisTitle,side=self.PLxAxisSide,overlaying=self.PLxAxisOverlaying)))
else:
if yInvert:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel),yaxis=dict(title=ylabel,autorange='reversed')))
elif xInvert:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel,autorange='reversed'),yaxis=dict(title=ylabel)))
else:
self.Plotlylayout.append(Layout(title = ptitle,xaxis=dict(title=xlabel),yaxis=dict(title=ylabel)))#,xaxis = PLXAxis,yaxis=PLYAxis))
if self.ncol > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif self.nrow > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
return ax
############################################################
##
def mesh3D(self, plotnum, xvals, yvals, zvals,
ptitle=None, xlabel=None, ylabel=None, zlabel=None,
rstride=1, cstride=1, linewidth=0,
plotCol=None, edgeCol=None, pltaxis=None, maxNX=0, maxNY=0, maxNZ=0,
xScientific=False, yScientific=False, zScientific=False,
powerLimits=[-4, 2, -4, 2, -2, 2], titlefsize=12,
xylabelfsize=12, xytickfsize=10, wireframe=False, surface=True,
cmap=cm.rainbow, cbarshow=False,
cbarorientation = 'vertical', cbarcustomticks=[], cbarfontsize = 12,
drawGrid=True, xInvert=False, yInvert=False, zInvert=False,
logScale=False, alpha=1, alphawire=1,
azim=45, elev=30, distance=10, zorders=None, clip_on=True,
PLcolor=None,
PLcolorScale=None, PLtickmode=None, PLnticks=None, PLtick0=None, PLdtick=None,
PLtickvals=None, PLticktext=None, PLintensity = None
):
"""XY colour mesh plot for (xvals, yvals, zvals) input sets.
Given an existing figure, this function plots in a specified subplot position.
Only one mesh is drawn at a time. Future meshes in the same subplot
will cover any previous meshes.
The mesh grid is defined in (x,y), while the height of the mesh is the z value.
The data set must have three two dimensional arrays, each for x, y, and z.
The data in x, y, and z arrays must have matching data points.
The x and y arrays each define the grid in terms of x and y values, i.e.,
the x array contains the x values for the data set, while the y array
contains the y values. The z array contains the z values for the
corresponding x and y values in the mesh.
Use wireframe=True to obtain a wireframe plot.
Use surface=True to obtain a surface plot with fill colours.
Z-values can be plotted on a log scale, in which case the colourbar is adjusted
to show true values, but on the nonlinear scale.
The xvals and yvals vectors may have non-constant grid-intervals, i.e.,
they do not have to be on regular intervals, but z array must correspond
to the (x,y) grid.
Args:
| plotnum (int): subplot number, 1-based index
| xvals (np.array[N,M]): array of x values, corresponding to (x,y) grid
| yvals (np.array[N,M]): array of y values, corresponding to (x,y) grid
| zvals (np.array[N,M]): array of z values, corresponding to (x,y) grid
| ptitle (string): plot title (optional)
| xlabel (string): x axis label (optional)
| ylabel (string): y axis label (optional)
| zlabel (string): z axis label (optional)
| rstride (int): mesh line row (y axis) stride, every rstride value along y axis (optional)
| cstride (int): mesh line column (x axis) stride, every cstride value along x axis (optional)
| linewidth (float): mesh line width in points (optional)
| plotCol ([strings]): fill colour, list with M=1 entries, use default if None (optional)
| edgeCol ([strings]): mesh line colour , list with M=1 entries, use default if None (optional)
| pltaxis ([xmin, xmax, ymin, ymax]): scale for x,y axes. z scale is not settable. Let Matplotlib decide if None (optional)
| maxNX (int): draw maxNX+1 tick labels on x axis (optional)
| maxNY (int): draw maxNY+1 tick labels on y axis (optional)
| maxNZ (int): draw maxNY+1 tick labels on z axis (optional)
| xScientific (bool): use scientific notation on x axis (optional)
| yScientific (bool): use scientific notation on y axis (optional)
| zScientific (bool): use scientific notation on z-axis (optional)
| powerLimits[float]: scientific tick label power limits [x-neg, x-pos, y-neg, y-pos, z-neg, z-pos] (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis, z-axis label font size, default 12pt (optional)
| xytickfsize (int): x-axis, y-axis, z-axis tick font size, default 10pt (optional)
| wireframe (bool): If True, do a wireframe plot, (optional)
| surface (bool): If True, do a surface plot, (optional)
| cmap (cm): color map for the mesh (optional)
| cbarshow (bool): if true, the show a color bar (optional)
| cbarorientation (string): 'vertical' (right) or 'horizontal' (below) (optional)
| cbarcustomticks zip([z values/float],[tick labels/string]): define custom colourbar ticks locations for given z values(optional)
| cbarfontsize (int): font size for color bar (optional)
| drawGrid (bool): draw the grid on the plot (optional)
| xInvert (bool): invert the x-axis. Flip the x-axis left-right (optional)
| yInvert (bool): invert the y-axis. Flip the y-axis left-right (optional)
| zInvert (bool): invert the z-axis. Flip the z-axis up-down (optional)
| logScale (bool): do Z values on log scale, recompute colourbar vals (optional)
| alpha (float): surface transparency (optional)
| alphawire (float): mesh transparency (optional)
| azim (float): graph view azimuth angle [degrees] (optional)
| elev (float): graph view evelation angle [degrees] (optional)
| distance (float): distance between viewer and plot (optional)
| zorder ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| PLcolor (string): Graph colors e.g 'FFFFFF'
| PLcolorScale ([int,string]): Color scale for mesh graphs e.g [0, 'rgb(0, 0, 0)']
| PLtickmode (string): Plot mode
| PLnticks (int): number of ticks
| PLtick0 (int): First tick value
| PLdtick (int):
| PLtickvals [int]: Plot intervals
| PLticktext [string]: Plot text
| PLintensity
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
from mpl_toolkits.mplot3d.axes3d import Axes3D
#if this is a log scale plot
if logScale is True:
zvals = np.log10(zvals)
#create subplot if not existing
if (self.nrow,self.ncol, plotnum) not in list(self.subplots.keys()):
self.subplots[(self.nrow,self.ncol, plotnum)] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum, projection='3d')
#get axis
ax = self.subplots[(self.nrow,self.ncol, plotnum)]
if drawGrid:
ax.grid(True)
else:
ax.grid(False)
if xlabel is not None:
ax.set_xlabel(xlabel, fontsize=xylabelfsize)
if xScientific:
formx = plt.ScalarFormatter()
formx.set_scientific(True)
formx.set_powerlimits([powerLimits[0], powerLimits[1]])
ax.xaxis.set_major_formatter(formx)
if ylabel is not None:
ax.set_ylabel(ylabel, fontsize=xylabelfsize)
if yScientific:
formy = plt.ScalarFormatter()
formy.set_powerlimits([powerLimits[2], powerLimits[3]])
formy.set_scientific(True)
ax.yaxis.set_major_formatter(formy)
if zlabel is not None:
ax.set_zlabel(zlabel, fontsize=xylabelfsize)
if zScientific:
formz = plt.ScalarFormatter()
formz.set_powerlimits([powerLimits[4], powerLimits[5]])
formz.set_scientific(True)
ax.zaxis.set_major_formatter(formz)
if maxNX >0:
ax.xaxis.set_major_locator(mpl.ticker.MaxNLocator(maxNX))
if maxNY >0:
ax.yaxis.set_major_locator(mpl.ticker.MaxNLocator(maxNY))
if maxNZ >0:
ax.zaxis.set_major_locator(mpl.ticker.MaxNLocator(maxNZ))
if plotCol:
col = plotCol[0]
else:
col = self.nextPlotCol()
if edgeCol:
edcol = edgeCol[0]
else:
edcol = self.nextPlotCol()
if zorders:
if len(zorders) > 1:
zorder = zorders[i]
else:
zorder = zorders[0]
else:
zorder = 1
if self.useplotly:
self.PlotlyPlotCalls = self.PlotlyPlotCalls + 1
self.PLIs3D = True
self.PLType = "mesh3D"
if cbarshow:
self.Plotlydata.append(Mesh3d(x=list(itertools.chain.from_iterable(xvals)),
y=list(itertools.chain.from_iterable(yvals)),
z=list(itertools.chain.from_iterable(zvals)),color=PLcolor,
colorbar = ColorBar(PLtickmode=PLtickmode,nticks=PLnticks,
PLtick0=PLtick0,PLdtick=PLdtick,PLtickvals=PLtickvals,PLticktext=PLticktext),
PLcolorscale=PLcolorScale,intensity=PLintensity))
else:
self.Plotlydata.append(Mesh3d(x=list(itertools.chain.from_iterable(xvals)),
y=list(itertools.chain.from_iterable(yvals)),
z=list(itertools.chain.from_iterable(zvals)),color=PLcolor))
# Append axis and plot titles
if self.ncol > 1:
self.PlotlySubPlotNumbers.append(plotnum)
elif self.nrow > 1 :
self.PlotlySubPlotNumbers.append(plotnum)
#do the plot
if surface:
pmplot = ax.plot_surface(xvals, yvals, zvals, rstride=rstride, cstride=cstride,
edgecolor=edcol, cmap=cmap, linewidth=linewidth, alpha=alpha,
zorder=zorder, clip_on=clip_on)
if wireframe:
pmplot = ax.plot_wireframe(xvals, yvals, zvals, rstride=rstride, cstride=cstride,
color=col, edgecolor=edcol, linewidth=linewidth, alpha=alphawire,
zorder=zorder, clip_on=clip_on)
ax.view_init(azim=azim, elev=elev)
ax.dist = distance
if cbarshow is True and cmap is not None:
#http://matplotlib.org/mpl_toolkits/axes_grid/users/overview.html#colorbar-whose-height-or-width-in-sync-with-the-master-axes
# divider = make_axes_locatable(ax)
# if cbarorientation == 'vertical':
# cax = divider.append_axes("right", size="5%", pad=0.05)
# else:
# cax = divider.append_axes("bottom", size="5%", pad=0.1)
if not cbarcustomticks:
cbar = self.fig.colorbar(pmplot,orientation=cbarorientation)
# cbar = self.fig.colorbar(pmplot,cax=cax)
if logScale:
cbartickvals = cbar.ax.yaxis.get_ticklabels()
tickVals = []
# need this roundabout trick to handle minus sign in unicode
for item in cbartickvals:
valstr = item.get_text().replace('\u2212', '-').replace('$','')
val = 10**float(valstr)
if abs(val) < 1000:
str = '{0:f}'.format(val)
else:
str = '{0:e}'.format(val)
tickVals.append(str)
cbartickvals = cbar.ax.yaxis.set_ticklabels(tickVals)
else:
ticks, ticklabels = list(zip(*cbarcustomticks))
cbar = self.fig.colorbar(pmplot,ticks=ticks, orientation=cbarorientation)
# cbar = self.fig.colorbar(pmplot,ticks=ticks, cax=cax)
if cbarorientation == 'vertical':
cbar.ax.set_yticklabels(ticklabels)
else:
cbar.ax.set_xticklabels(ticklabels)
if cbarorientation == 'vertical':
for t in cbar.ax.get_yticklabels():
t.set_fontsize(cbarfontsize)
else:
for t in cbar.ax.get_xticklabels():
t.set_fontsize(cbarfontsize)
if(ptitle is not None):
plt.title(ptitle, fontsize=titlefsize)
#scale the axes
if pltaxis is not None:
# ax.axis(pltaxis)
ax.set_xlim(pltaxis[0], pltaxis[1])
ax.set_ylim(pltaxis[2], pltaxis[3])
ax.set_zlim(pltaxis[4], pltaxis[5])
if(ptitle is not None):
ax.set_title(ptitle, fontsize=titlefsize)
# minor ticks are two points smaller than major
ax.tick_params(axis='both', which='major', labelsize=xytickfsize)
ax.tick_params(axis='both', which='minor', labelsize=xytickfsize-2)
if xInvert:
ax.set_xlim(ax.get_xlim()[::-1])
if yInvert:
ax.set_ylim(ax.get_ylim()[::-1])
if zInvert:
ax.set_zlim(ax.get_zlim()[::-1])
if self.useplotly:
if self.PLmultipleYAxis:
self.Plotlylayout.append(Layout(title = ptitle))
elif self.PLmultipleXAxis:
self.Plotlylayout.append(Layout(title = ptitle))
else:
self.Plotlylayout.append(Layout(title = ptitle))
if self.ncol > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
elif self.nrow > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlyXaxisTitles.append(xlabel)
self.PlotlyYaxisTitles.append(ylabel)
return ax
############################################################
##
def polar(self, plotnum, theta, r, ptitle=None, \
plotCol=None, label=[],labelLocation=[-0.1, 0.1], \
highlightNegative=True, highlightCol='#ffff00', highlightWidth=4,\
legendAlpha=0.0, linestyle=None,\
rscale=None, rgrid=[0,5], thetagrid=[30], \
direction='counterclockwise', zerooffset=0, titlefsize=12, drawGrid=True,
zorders=None, clip_on=True, markers=[], markevery=None,
):
"""Create a subplot and plot the data in polar coordinates (linear radial orginates only).
Given an existing figure, this function plots in a specified subplot position.
The function arguments are described below in some detail. Note that the radial values
or ordinates can be more than one column, each column representing a different
line in the plot. This is convenient if large arrays of data must be plotted. If more
than one column is present, the label argument can contain the legend labels for
each of the columns/lines. The scale for the radial ordinates can be set with rscale.
The number of radial grid circles can be set with rgrid - this provides a somewhat
better control over the built-in radial grid in matplotlib. thetagrids defines the angular
grid interval. The angular rotation direction can be set to be clockwise or
counterclockwise. Likewise, the rotation offset where the plot zero angle must be,
is set with `zerooffset`.
For some obscure reason Matplitlib version 1.13 does not plot negative values on the
polar plot. We therefore force the plot by making the values positive and then highlight it as negative.
Args:
| plotnum (int): subplot number, 1-based index
| theta (np.array[N,] or [N,1]): angular abscissa in radians
| r (np.array[N,] or [N,M]): radial ordinates - could be M columns
| ptitle (string): plot title (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if None (optional)
| label ([strings]): legend label, list with M entries (optional)
| labelLocation ([x,y]): where the legend should located (optional)
| highlightNegative (bool): indicate if negative data must be highlighted (optional)
| highlightCol (string): negative highlight colour string (optional)
| highlightWidth (int): negative highlight line width(optional)
| legendAlpha (float): transparency for legend box (optional)
| linestyle ([str]): line style to be used in plot
| rscale ([rmin, rmax]): radial plotting limits. use default setting if None.
If rmin is negative the zero is a circle and rmin is at the centre of the graph (optional)
| rgrid ([rinc, numinc]): radial grid, use default is [0,5].
If rgrid is None don't show. If rinc=0 then numinc is number of intervals.
If rinc is not zero then rinc is the increment and numinc is ignored (optional)
| thetagrid (float): theta grid interval [degrees], if None don't show (optional)
| direction (string): direction in increasing angle, 'counterclockwise' or 'clockwise' (optional)
| zerooffset (float): rotation offset where zero should be [rad]. Positive
zero-offset rotation is counterclockwise from 3'o'clock (optional)
| titlefsize (int): title font size, default 12pt (optional)
| drawGrid (bool): draw a grid on the graph (optional)
| zorder ([int]) list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool) clips objects to drawing axes (optional)
| markers ([string]) markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)) subsample when using markers (optional)
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
if theta.ndim>1:
tt=theta
else:
if type(theta)==type(pd.Series()):
theta = theta.values
tt=theta.reshape(-1, 1)
if r.ndim>1:
rr=r
else:
if type(r)==type(pd.Series()):
r = r.values
rr=r.reshape(-1, 1)
MakeAbs = True
if rscale is not None:
if rscale[0] < 0:
MakeAbs = False
else:
highlightNegative=True #override the function value
else:
highlightNegative=True #override the function value
#plotCol = self.buildPlotCol(plotCol, rr.shape[1])
ax = None
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum, polar=True)
ax = self.subplots[pkey]
ax.grid(drawGrid)
rmax=0
for i in range(rr.shape[1]):
# negative val :forcing positive and phase shifting
# if forceAbsolute:
# ttt = tt + np.pi*(rr[:, i] < 0).reshape(-1, 1)
# rrr = np.abs(rr[:, i])
# else:
ttt = tt.reshape(-1,)
rrr = rr[:, i].reshape(-1,)
#print(rrr)
if highlightNegative:
#find zero crossings in data
zero_crossings = np.where(np.diff(np.sign(rr),axis=0))[0] + 1
#split the input into different subarrays according to crossings
negrrr = np.split(rr,zero_crossings)
negttt = np.split(tt,zero_crossings)
# print('zero crossing',zero_crossings)
# print(len(negrrr))
# print(negrrr)
mmrk = ''
if markers:
if i >= len(markers):
mmrk = markers[-1]
else:
mmrk = markers[i]
#set up the line style, either given or next in sequence
if plotCol:
col = plotCol[i]
else:
col = self.nextPlotCol()
if linestyle is None:
linestyleL = '-'
else:
if type(linestyle) == type([1]):
linestyleL = linestyle[i]
else:
linestyleL = linestyle
# print('p',ttt.shape)
# print('p',rrr.shape)
if zorders:
if len(zorders) > 1:
zorder = zorders[i]
else:
zorder = zorders[0]
else:
zorder = 2
if not label:
if highlightNegative:
lines = ax.plot(ttt, rrr, col, clip_on=clip_on, zorder=zorder,marker=mmrk, markevery=markevery,linestyle=linestyleL)
neglinewith = highlightWidth*plt.getp(lines[0],'linewidth')
for ii in range(0,len(negrrr)):
if len(negrrr[ii]) > 0:
if negrrr[ii][0] < 0:
if MakeAbs:
ax.plot(negttt[ii], np.abs(negrrr[ii]), highlightCol,
linewidth=neglinewith, clip_on=clip_on, zorder=zorder,
marker=mmrk, markevery=markevery,linestyle=linestyleL)
else:
ax.plot(negttt[ii], negrrr[ii], highlightCol,
linewidth=neglinewith, clip_on=clip_on, zorder=zorder,
marker=mmrk, markevery=markevery,linestyle=linestyleL)
ax.plot(ttt, rrr, col, clip_on=clip_on, zorder=zorder,marker=mmrk, markevery=markevery,linestyle=linestyleL)
rmax = np.maximum(np.abs(rrr).max(), rmax)
rmin = 0
else:
if highlightNegative:
lines = ax.plot(ttt, rrr, col, clip_on=clip_on, zorder=zorder,marker=mmrk, markevery=markevery,linestyle=linestyleL)
neglinewith = highlightWidth*plt.getp(lines[0],'linewidth')
for ii in range(0,len(negrrr)):
if len(negrrr[ii]) > 0:
# print(len(negrrr[ii]))
# if negrrr[ii][0] < 0:
if negrrr[ii][0][0] < 0:
if MakeAbs:
ax.plot(negttt[ii], np.abs(negrrr[ii]), highlightCol,
linewidth=neglinewith, clip_on=clip_on, zorder=zorder,
marker=mmrk, markevery=markevery,linestyle=linestyleL)
else:
ax.plot(negttt[ii], negrrr[ii], highlightCol,
linewidth=neglinewith, clip_on=clip_on, zorder=zorder,
marker=mmrk, markevery=markevery,linestyle=linestyleL)
ax.plot(ttt, rrr, col,label=label[i], clip_on=clip_on, zorder=zorder,marker=mmrk, markevery=markevery,linestyle=linestyleL)
rmax=np.maximum(np.abs(rrr).max(), rmax)
rmin = 0
if MakeAbs:
ax.plot(ttt, np.abs(rrr), col, clip_on=clip_on, zorder=zorder,marker=mmrk, markevery=markevery,linestyle=linestyleL)
else:
ax.plot(ttt, rrr, col, clip_on=clip_on, zorder=zorder,marker=mmrk, markevery=markevery,linestyle=linestyleL)
#Plotly polar setup
if self.useplotly:
# Assuming that either y or x has to 1
if thetagrid is None:
tt=tt*(180.0/(np.pi))
else:
tt=tt*(180.0/(np.pi*(thetagrid[0]/(-4.62*i+5))))
try:
if len(r[0,:]) > 1:
self.Plotlydata.append(Scatter(r=rr[:,i], t=tt[:,0], name = label,mode='lines'))
elif len(theta[0,:]) > 1:
self.Plotlydata.append(Scatter(r=rr[:,0], t=tt[:,i], name = label,mode='lines'))
else:
self.Plotlydata.append(Scatter(r=rr[:,0], t=tt[:,0], name = label,mode='lines'))
except:
self.Plotlydata.append(Scatter(r=rr[:,0], t=tt[:,0], name = label))
# Append axis and plot titles
if self.ncol > 1:
self.PlotlySubPlotNumbers.append(plotnum)
elif self.nrow > 1 :
self.PlotlySubPlotNumbers.append(plotnum)
if label:
fontP = mpl.font_manager.FontProperties()
fontP.set_size('small')
leg = ax.legend(loc='upper left',
bbox_to_anchor=(labelLocation[0], labelLocation[1]),
prop = fontP, fancybox=True)
leg.get_frame().set_alpha(legendAlpha)
self.bbox_extra_artists.append(leg)
ax.set_theta_direction(direction)
ax.set_theta_offset(zerooffset)
#set up the grids
if thetagrid is None:
ax.set_xticklabels([])
else:
plt.thetagrids(list(range(0, 360, thetagrid[0])))
#Set increment and maximum radial limits
if rscale is None:
rscale = [rmin, rmax]
if rgrid is None:
ax.set_yticklabels([])
else:
if rgrid[0] == 0:
ax.set_yticks(np.linspace(rscale[0],rscale[1],int(rgrid[1])))
if rgrid[0] != 0:
numrgrid = (rscale[1] - rscale[0] ) / rgrid[0]
ax.set_yticks(np.linspace(rscale[0],rscale[1],int(numrgrid+1.000001)))
ax.set_ylim(rscale[0],rscale[1])
if(ptitle is not None):
ax.set_title(ptitle, fontsize=titlefsize, \
verticalalignment ='bottom', horizontalalignment='center')
if self.useplotly:
if self.PLmultipleYAxis:
self.Plotlylayout.append(Layout(showlegend = True,title = ptitle,orientation=+90))
elif self.PLmultipleXAxis:
self.Plotlylayout.append(Layout(showlegend = True,title = ptitle,orientation=+90))
else:
self.Plotlylayout.append(Layout(showlegend = True,title = ptitle,orientation=+90))
if self.ncol > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlySubPlotLabels.append(label)
elif self.nrow > 1:
self.PlotlySubPlotTitles.append(ptitle)
self.PlotlySubPlotLabels.append(label)
return ax
############################################################
##
def showImage(self, plotnum, img, ptitle=None, xlabel=None, ylabel=None,
cmap=plt.cm.gray, titlefsize=12, cbarshow=False,
cbarorientation = 'vertical', cbarcustomticks=[], cbarfontsize = 12,
labelfsize=10, xylabelfsize = 12,interpolation=None):
"""Creates a subplot and show the image using the colormap provided.
Args:
| plotnum (int): subplot number, 1-based index
| img (np.ndarray): numpy 2d array containing the image
| ptitle (string): plot title (optional)
| xlabel (string): x axis label (optional)
| ylabel (string): y axis label (optional)
| cmap: matplotlib colormap, default gray (optional)
| fsize (int): title font size, default 12pt (optional)
| cbarshow (bool): if true, the show a colour bar (optional)
| cbarorientation (string): 'vertical' (right) or 'horizontal' (below) (optional)
| cbarcustomticks zip([tick locations/float],[tick labels/string]): locations in image grey levels (optional)
| cbarfontsize (int): font size for colour bar (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x-axis, y-axis label font size, default 12pt (optional)
| interpolation (str): 'none', 'nearest', 'bilinear', 'bicubic', 'spline16', 'spline36', 'hanning', 'hamming', 'hermite', 'kaiser', 'quadric', 'catrom', 'gaussian', 'bessel', 'mitchell', 'sinc', 'lanczos'(optional, see pyplot.imshow)
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
#http://matplotlib.sourceforge.net/examples/pylab_examples/colorbar_tick_labelling_demo.html
#http://matplotlib.1069221.n5.nabble.com/Colorbar-Ticks-td21289.html
pkey = (self.nrow, self.ncol, plotnum)
if pkey not in list(self.subplots.keys()):
self.subplots[pkey] = \
self.fig.add_subplot(self.nrow,self.ncol, plotnum)
ax = self.subplots[pkey]
cimage = ax.imshow(img, cmap,interpolation=interpolation)
if xlabel is not None:
ax.set_xlabel(xlabel, fontsize=xylabelfsize)
if ylabel is not None:
ax.set_ylabel(ylabel, fontsize=xylabelfsize)
ax.axis('off')
if cbarshow is True:
#http://matplotlib.org/mpl_toolkits/axes_grid/users/overview.html#colorbar-whose-height-or-width-in-sync-with-the-master-axes
divider = make_axes_locatable(ax)
if cbarorientation == 'vertical':
cax = divider.append_axes("right", size="5%", pad=0.05)
# else:
# cay = divider.append_axes("bottom", size="5%", pad=0.1)
if not cbarcustomticks:
if cbarorientation == 'vertical':
cbar = self.fig.colorbar(cimage,cax=cax)
else:
cbar = self.fig.colorbar(cimage,orientation=cbarorientation)
else:
ticks, ticklabels = list(zip(*cbarcustomticks))
if cbarorientation == 'vertical':
cbar = self.fig.colorbar(cimage,ticks=ticks, cax=cax)
else:
cbar = self.fig.colorbar(cimage,ticks=ticks, orientation=cbarorientation)
if cbarorientation == 'vertical':
cbar.ax.set_yticklabels(ticklabels)
else:
cbar.ax.set_xticklabels(ticklabels)
if cbarorientation == 'vertical':
for t in cbar.ax.get_yticklabels():
t.set_fontsize(cbarfontsize)
else:
for t in cbar.ax.get_xticklabels():
t.set_fontsize(cbarfontsize)
if(ptitle is not None):
ax.set_title(ptitle, fontsize=titlefsize)
return ax
############################################################
##
def plot3d(self, plotnum, x, y, z, ptitle=None, xlabel=None, ylabel=None, zlabel=None,
plotCol=[], linewidths=None, pltaxis=None, label=None, legendAlpha=0.0, titlefsize=12,
xylabelfsize = 12, xInvert=False, yInvert=False, zInvert=False,scatter=False,
markers=None, markevery=None, azim=45, elev=30, zorders=None, clip_on=True, edgeCol=None,
linestyle='-'):
"""3D plot on linear scales for x y z input sets.
Given an existing figure, this function plots in a specified subplot position.
The function arguments are described below in some detail.
Note that multiple 3D data sets can be plotted simultaneously by adding additional
columns to the input coordinates of the (x,y,z) arrays, each set of columns representing
a different line in the plot. This is convenient if large arrays of data must
be plotted. If more than one column is present, the label argument can contain the
legend labels for each of the columns/lines.
Args:
| plotnum (int): subplot number, 1-based index
| x (np.array[N,] or [N,M]) x coordinates of each line.
| y (np.array[N,] or [N,M]) y coordinates of each line.
| z (np.array[N,] or [N,M]) z coordinates of each line.
| ptitle (string): plot title (optional)
| xlabel (string): x-axis label (optional)
| ylabel (string): y-axis label (optional)
| zlabel (string): z axis label (optional)
| plotCol ([strings]): plot colour and line style, list with M entries, use default if None (optional)
| linewidths ([float]): plot line width in points, list with M entries, use default if None (optional)
| pltaxis ([xmin, xmax, ymin, ymax, zmin, zmax]) scale for x,y,z axes. Let Matplotlib decide if None. (optional)
| label ([strings]): legend label for ordinate, list with M entries (optional)
| legendAlpha (float): transparency for legend box (optional)
| titlefsize (int): title font size, default 12pt (optional)
| xylabelfsize (int): x, y, z label font size, default 12pt (optional)
| xInvert (bool): invert the x-axis (optional)
| yInvert (bool): invert the y-axis (optional)
| zInvert (bool): invert the z-axis (optional)
| scatter (bool): draw only the points, no lines (optional)
| markers ([string]): markers to be used for plotting data points (optional)
| markevery (int | (startind, stride)): subsample when using markers (optional)
| azim (float): graph view azimuth angle [degrees] (optional)
| elev (float): graph view evelation angle [degrees] (optional)
| zorder ([int]): list of zorder for drawing sequence, highest is last (optional)
| clip_on (bool): clips objects to drawing axes (optional)
| edgeCol ([int]): list of colour specs, value at [0] used for edge colour (optional).
| linestyle (string): linestyle for this plot (optional)
Returns:
| the axis object for the plot
Raises:
| No exception is raised.
"""
# if required convert 1D arrays into 2D arrays
if type(x)==type(
|
pd.Series()
|
pandas.Series
|
#!/usr/bin/env python
# coding: utf-8
# In[ ]:
from google.colab import drive # Import a library named google.colab
drive.mount('/content/drive', force_remount=True) # mount the content to the directory `/content/drive`
# In[ ]:
get_ipython().run_line_magic('cd', '/content/drive/MyDrive/Netflix_Appetency_Prediction')
# In[ ]:
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
import warnings
warnings.filterwarnings('ignore')
# In[ ]:
train_df=pd.read_csv('train.csv')
# In[ ]:
test_df=pd.read_csv('test.csv')
# In[ ]:
id_list=test_df['id']
# In[ ]:
train_df.info()
# In[ ]:
test_df.info()
# #1.Find categorical variables and numerical variables
# In[ ]:
categorical_var=[column for column in train_df.columns if (train_df[column].dtype=='object') | (len(train_df[column].unique())<=20) ]
# In[ ]:
numerical_var=train_df.columns.drop(categorical_var)
# In[ ]:
print("categorical_var length:", len(categorical_var), '\n' "numerical_var length:", len(numerical_var))
# #2.Read target feature
# In[ ]:
train_df['target'].unique()
# In[ ]:
sns.countplot(train_df['target'])
# #3.Observe percentage null values
#
# In[ ]:
null_values_columns=[]
percent_null_values=[]
for column in train_df.columns:
percent_null_value=((train_df[column].isna().sum())/(len(train_df))) * 100
if(percent_null_value > 0):
null_values_columns.append(column)
percent_null_values.append(percent_null_value)
df_null=
|
pd.DataFrame(null_values_columns,columns=['column'])
|
pandas.DataFrame
|
'''
Copyright 2022 Airbus SAS
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
'''
'''
mode: python; py-indent-offset: 4; tab-width: 4; coding: utf-8
'''
import unittest
from numpy.testing import assert_array_equal
from pandas._testing import assert_frame_equal
import pprint
import numpy as np
import pandas as pd
from time import sleep
from shutil import rmtree
from pathlib import Path
from os.path import join
from sos_trades_core.execution_engine.execution_engine import ExecutionEngine
from copy import deepcopy
from tempfile import gettempdir
class TestMultiScenarioOfDoeEval(unittest.TestCase):
"""
MultiScenario and doe_eval processes test class
"""
def setUp(self):
'''
Initialize third data needed for testing
'''
self.namespace = 'MyCase'
self.study_name = f'{self.namespace}'
self.repo = 'sos_trades_core.sos_processes.test'
self.base_path = 'sos_trades_core.sos_wrapping.test_discs'
self.exec_eng = ExecutionEngine(self.namespace)
self.factory = self.exec_eng.factory
def setup_my_usecase(self):
'''
'''
######### Numerical values ####
x_1 = 2
x_2_a = 4
x_2_b = 5
a_1 = 3
b_1 = 4
a_2 = 6
b_2 = 2
constant = 3
power = 2
z_1 = 1.2
z_2 = 1.5
my_doe_algo = "lhs"
n_samples = 4
######### Selection of variables and DS ####
input_selection_z_scenario_1 = {
'selected_input': [False, False, False, True, False, False],
'full_name': ['x', 'a','b','multi_scenarios.scenario_1.Disc3.z','constant','power']}
input_selection_z_scenario_1 = pd.DataFrame(input_selection_z_scenario_1)
input_selection_z_scenario_2 = {
'selected_input': [False, False, False, True, False, False],
'full_name': ['x', 'a','b','multi_scenarios.scenario_2.Disc3.z','constant','power']}
input_selection_z_scenario_2 = pd.DataFrame(input_selection_z_scenario_2)
output_selection_o_scenario_1 = {
'selected_output': [False, False, True],
'full_name': ['indicator', 'y', 'multi_scenarios.scenario_1.o']}
output_selection_o_scenario_1 = pd.DataFrame(output_selection_o_scenario_1)
output_selection_o_scenario_2 = {
'selected_output': [False, False, True],
'full_name': ['indicator', 'y', 'multi_scenarios.scenario_2.o']}
output_selection_o_scenario_2 =
|
pd.DataFrame(output_selection_o_scenario_2)
|
pandas.DataFrame
|
# -*- coding: utf-8 -*-
## Copyright 2015-2021 PyPSA Developers
## You can find the list of PyPSA Developers at
## https://pypsa.readthedocs.io/en/latest/developers.html
## PyPSA is released under the open source MIT License, see
## https://github.com/PyPSA/PyPSA/blob/master/LICENSE.txt
"""
Power flow functionality.
"""
__author__ = (
"PyPSA Developers, see https://pypsa.readthedocs.io/en/latest/developers.html"
)
__copyright__ = (
"Copyright 2015-2021 PyPSA Developers, see https://pypsa.readthedocs.io/en/latest/developers.html, "
"MIT License"
)
import logging
logger = logging.getLogger(__name__)
import time
from operator import itemgetter
import networkx as nx
import numpy as np
import pandas as pd
from numpy import ones, r_
from numpy.linalg import norm
from pandas.api.types import is_list_like
from scipy.sparse import csc_matrix, csr_matrix, dok_matrix
from scipy.sparse import hstack as shstack
from scipy.sparse import issparse
from scipy.sparse import vstack as svstack
from scipy.sparse.linalg import spsolve
from pypsa.descriptors import (
Dict,
allocate_series_dataframes,
degree,
get_switchable_as_dense,
zsum,
)
pd.Series.zsum = zsum
def normed(s):
return s / s.sum()
def real(X):
return np.real(X.to_numpy())
def imag(X):
return np.imag(X.to_numpy())
def _as_snapshots(network, snapshots):
if snapshots is None:
snapshots = network.snapshots
if not is_list_like(snapshots):
snapshots = pd.Index([snapshots])
if not isinstance(snapshots, pd.MultiIndex):
snapshots = pd.Index(snapshots)
assert snapshots.isin(network.snapshots).all()
snapshots.name = "snapshot"
return snapshots
def _allocate_pf_outputs(network, linear=False):
to_allocate = {
"Generator": ["p"],
"Load": ["p"],
"StorageUnit": ["p"],
"Store": ["p"],
"ShuntImpedance": ["p"],
"Bus": ["p", "v_ang", "v_mag_pu"],
"Line": ["p0", "p1"],
"Transformer": ["p0", "p1"],
"Link": ["p" + col[3:] for col in network.links.columns if col[:3] == "bus"],
}
if not linear:
for component, attrs in to_allocate.items():
if "p" in attrs:
attrs.append("q")
if "p0" in attrs and component != "Link":
attrs.extend(["q0", "q1"])
allocate_series_dataframes(network, to_allocate)
def _calculate_controllable_nodal_power_balance(
sub_network, network, snapshots, buses_o
):
for n in ("q", "p"):
# allow all one ports to dispatch as set
for c in sub_network.iterate_components(
network.controllable_one_port_components
):
c_n_set = get_switchable_as_dense(
network, c.name, n + "_set", snapshots, c.ind
)
network.pnl(c.name)[n].loc[snapshots, c.ind] = c_n_set
# set the power injection at each node from controllable components
network.buses_t[n].loc[snapshots, buses_o] = sum(
[
(
(c.pnl[n].loc[snapshots, c.ind] * c.df.loc[c.ind, "sign"])
.groupby(c.df.loc[c.ind, "bus"], axis=1)
.sum()
.reindex(columns=buses_o, fill_value=0.0)
)
for c in sub_network.iterate_components(
network.controllable_one_port_components
)
]
)
if n == "p":
network.buses_t[n].loc[snapshots, buses_o] += sum(
[
(
-c.pnl[n + str(i)]
.loc[snapshots]
.groupby(c.df["bus" + str(i)], axis=1)
.sum()
.reindex(columns=buses_o, fill_value=0)
)
for c in network.iterate_components(
network.controllable_branch_components
)
for i in [int(col[3:]) for col in c.df.columns if col[:3] == "bus"]
]
)
def _network_prepare_and_run_pf(
network,
snapshots,
skip_pre,
linear=False,
distribute_slack=False,
slack_weights="p_set",
**kwargs
):
if linear:
sub_network_pf_fun = sub_network_lpf
sub_network_prepare_fun = calculate_B_H
else:
sub_network_pf_fun = sub_network_pf
sub_network_prepare_fun = calculate_Y
if not skip_pre:
network.determine_network_topology()
calculate_dependent_values(network)
_allocate_pf_outputs(network, linear)
snapshots = _as_snapshots(network, snapshots)
# deal with links
if not network.links.empty:
p_set = get_switchable_as_dense(network, "Link", "p_set", snapshots)
network.links_t.p0.loc[snapshots] = p_set.loc[snapshots]
for i in [
int(col[3:])
for col in network.links.columns
if col[:3] == "bus" and col != "bus0"
]:
eff_name = "efficiency" if i == 1 else "efficiency{}".format(i)
efficiency = get_switchable_as_dense(network, "Link", eff_name, snapshots)
links = network.links.index[network.links["bus{}".format(i)] != ""]
network.links_t["p{}".format(i)].loc[snapshots, links] = (
-network.links_t.p0.loc[snapshots, links]
* efficiency.loc[snapshots, links]
)
itdf = pd.DataFrame(index=snapshots, columns=network.sub_networks.index, dtype=int)
difdf = pd.DataFrame(index=snapshots, columns=network.sub_networks.index)
cnvdf = pd.DataFrame(
index=snapshots, columns=network.sub_networks.index, dtype=bool
)
for sub_network in network.sub_networks.obj:
if not skip_pre:
find_bus_controls(sub_network)
branches_i = sub_network.branches_i()
if len(branches_i) > 0:
sub_network_prepare_fun(sub_network, skip_pre=True)
if isinstance(slack_weights, dict):
sn_slack_weights = slack_weights[sub_network.name]
else:
sn_slack_weights = slack_weights
if isinstance(sn_slack_weights, dict):
sn_slack_weights = pd.Series(sn_slack_weights)
if not linear:
# escape for single-bus sub-network
if len(sub_network.buses()) <= 1:
(
itdf[sub_network.name],
difdf[sub_network.name],
cnvdf[sub_network.name],
) = sub_network_pf_singlebus(
sub_network,
snapshots=snapshots,
skip_pre=True,
distribute_slack=distribute_slack,
slack_weights=sn_slack_weights,
)
else:
(
itdf[sub_network.name],
difdf[sub_network.name],
cnvdf[sub_network.name],
) = sub_network_pf_fun(
sub_network,
snapshots=snapshots,
skip_pre=True,
distribute_slack=distribute_slack,
slack_weights=sn_slack_weights,
**kwargs
)
else:
sub_network_pf_fun(
sub_network, snapshots=snapshots, skip_pre=True, **kwargs
)
if not linear:
return Dict({"n_iter": itdf, "error": difdf, "converged": cnvdf})
def network_pf(
network,
snapshots=None,
skip_pre=False,
x_tol=1e-6,
use_seed=False,
distribute_slack=False,
slack_weights="p_set",
):
"""
Full non-linear power flow for generic network.
Parameters
----------
snapshots : list-like|single snapshot
A subset or an elements of network.snapshots on which to run
the power flow, defaults to network.snapshots
skip_pre : bool, default False
Skip the preliminary steps of computing topology, calculating dependent values and finding bus controls.
x_tol: float
Tolerance for Newton-Raphson power flow.
use_seed : bool, default False
Use a seed for the initial guess for the Newton-Raphson algorithm.
distribute_slack : bool, default False
If ``True``, distribute the slack power across generators proportional to generator dispatch by default
or according to the distribution scheme provided in ``slack_weights``.
If ``False`` only the slack generator takes up the slack.
slack_weights : dict|str, default 'p_set'
Distribution scheme describing how to determine the fraction of the total slack power
(of each sub network individually) a bus of the subnetwork takes up.
Default is to distribute proportional to generator dispatch ('p_set').
Another option is to distribute proportional to (optimised) nominal capacity ('p_nom' or 'p_nom_opt').
Custom weights can be specified via a dictionary that has a key for each
subnetwork index (``network.sub_networks.index``) and a
pandas.Series/dict with buses or generators of the
corresponding subnetwork as index/keys.
When specifying custom weights with buses as index/keys the slack power of a bus is distributed
among its generators in proportion to their nominal capacity (``p_nom``) if given, otherwise evenly.
Returns
-------
dict
Dictionary with keys 'n_iter', 'converged', 'error' and dataframe
values indicating number of iterations, convergence status, and
iteration error for each snapshot (rows) and sub_network (columns)
"""
return _network_prepare_and_run_pf(
network,
snapshots,
skip_pre,
linear=False,
x_tol=x_tol,
use_seed=use_seed,
distribute_slack=distribute_slack,
slack_weights=slack_weights,
)
def newton_raphson_sparse(
f,
guess,
dfdx,
x_tol=1e-10,
lim_iter=100,
distribute_slack=False,
slack_weights=None,
):
"""Solve f(x) = 0 with initial guess for x and dfdx(x). dfdx(x) should
return a sparse Jacobian. Terminate if error on norm of f(x) is <
x_tol or there were more than lim_iter iterations.
"""
slack_args = {"distribute_slack": distribute_slack, "slack_weights": slack_weights}
converged = False
n_iter = 0
F = f(guess, **slack_args)
diff = norm(F, np.Inf)
logger.debug("Error at iteration %d: %f", n_iter, diff)
while diff > x_tol and n_iter < lim_iter:
n_iter += 1
guess = guess - spsolve(dfdx(guess, **slack_args), F)
F = f(guess, **slack_args)
diff = norm(F, np.Inf)
logger.debug("Error at iteration %d: %f", n_iter, diff)
if diff > x_tol:
logger.warning(
'Warning, we didn\'t reach the required tolerance within %d iterations, error is at %f. See the section "Troubleshooting" in the documentation for tips to fix this. ',
n_iter,
diff,
)
elif not np.isnan(diff):
converged = True
return guess, n_iter, diff, converged
def sub_network_pf_singlebus(
sub_network,
snapshots=None,
skip_pre=False,
distribute_slack=False,
slack_weights="p_set",
linear=False,
):
"""
Non-linear power flow for a sub-network consiting of a single bus.
Parameters
----------
snapshots : list-like|single snapshot
A subset or an elements of network.snapshots on which to run
the power flow, defaults to network.snapshots
skip_pre: bool, default False
Skip the preliminary steps of computing topology, calculating dependent values and finding bus controls.
distribute_slack : bool, default False
If ``True``, distribute the slack power across generators proportional to generator dispatch by default
or according to the distribution scheme provided in ``slack_weights``.
If ``False`` only the slack generator takes up the slack.
slack_weights : pandas.Series|str, default 'p_set'
Distribution scheme describing how to determine the fraction of the total slack power
a bus of the subnetwork takes up. Default is to distribute proportional to generator dispatch
('p_set'). Another option is to distribute proportional to (optimised) nominal capacity ('p_nom' or 'p_nom_opt').
Custom weights can be provided via a pandas.Series/dict
that has the generators of the single bus as index/keys.
"""
snapshots = _as_snapshots(sub_network.network, snapshots)
network = sub_network.network
logger.info(
"Balancing power on single-bus sub-network {} for snapshots {}".format(
sub_network, snapshots
)
)
if not skip_pre:
find_bus_controls(sub_network)
_allocate_pf_outputs(network, linear=False)
if isinstance(slack_weights, dict):
slack_weights = pd.Series(slack_weights)
buses_o = sub_network.buses_o
_calculate_controllable_nodal_power_balance(
sub_network, network, snapshots, buses_o
)
v_mag_pu_set = get_switchable_as_dense(network, "Bus", "v_mag_pu_set", snapshots)
network.buses_t.v_mag_pu.loc[snapshots, sub_network.slack_bus] = v_mag_pu_set.loc[
:, sub_network.slack_bus
]
network.buses_t.v_ang.loc[snapshots, sub_network.slack_bus] = 0.0
if distribute_slack:
for bus, group in sub_network.generators().groupby("bus"):
if slack_weights in ["p_nom", "p_nom_opt"]:
assert (
not all(network.generators[slack_weights]) == 0
), "Invalid slack weights! Generator attribute {} is always zero.".format(
slack_weights
)
bus_generator_shares = (
network.generators[slack_weights]
.loc[group.index]
.pipe(normed)
.fillna(0)
)
elif slack_weights == "p_set":
generators_t_p_choice = get_switchable_as_dense(
network, "Generator", slack_weights, snapshots
)
assert (
not generators_t_p_choice.isna().all().all()
), "Invalid slack weights! Generator attribute {} is always NaN.".format(
slack_weights
)
assert (
not (generators_t_p_choice == 0).all().all()
), "Invalid slack weights! Generator attribute {} is always zero.".format(
slack_weights
)
bus_generator_shares = (
generators_t_p_choice.loc[snapshots, group.index]
.apply(normed, axis=1)
.fillna(0)
)
else:
bus_generator_shares = slack_weights.pipe(normed).fillna(0)
network.generators_t.p.loc[
snapshots, group.index
] += bus_generator_shares.multiply(
-network.buses_t.p.loc[snapshots, bus], axis=0
)
else:
network.generators_t.p.loc[
snapshots, sub_network.slack_generator
] -= network.buses_t.p.loc[snapshots, sub_network.slack_bus]
network.generators_t.q.loc[
snapshots, sub_network.slack_generator
] -= network.buses_t.q.loc[snapshots, sub_network.slack_bus]
network.buses_t.p.loc[snapshots, sub_network.slack_bus] = 0.0
network.buses_t.q.loc[snapshots, sub_network.slack_bus] = 0.0
return 0, 0.0, True # dummy substitute for newton raphson output
def sub_network_pf(
sub_network,
snapshots=None,
skip_pre=False,
x_tol=1e-6,
use_seed=False,
distribute_slack=False,
slack_weights="p_set",
):
"""
Non-linear power flow for connected sub-network.
Parameters
----------
snapshots : list-like|single snapshot
A subset or an elements of network.snapshots on which to run
the power flow, defaults to network.snapshots
skip_pre: bool, default False
Skip the preliminary steps of computing topology, calculating dependent values and finding bus controls.
x_tol: float
Tolerance for Newton-Raphson power flow.
use_seed : bool, default False
Use a seed for the initial guess for the Newton-Raphson algorithm.
distribute_slack : bool, default False
If ``True``, distribute the slack power across generators proportional to generator dispatch by default
or according to the distribution scheme provided in ``slack_weights``.
If ``False`` only the slack generator takes up the slack.
slack_weights : pandas.Series|str, default 'p_set'
Distribution scheme describing how to determine the fraction of the total slack power
a bus of the subnetwork takes up. Default is to distribute proportional to generator dispatch
('p_set'). Another option is to distribute proportional to (optimised) nominal capacity ('p_nom' or 'p_nom_opt').
Custom weights can be provided via a pandas.Series/dict
that has the buses or the generators of the subnetwork as index/keys.
When using custom weights with buses as index/keys the slack power of a bus is distributed
among its generators in proportion to their nominal capacity (``p_nom``) if given, otherwise evenly.
Returns
-------
Tuple of three pandas.Series indicating number of iterations,
remaining error, and convergence status for each snapshot
"""
assert isinstance(
slack_weights, (str, pd.Series, dict)
), "Type of 'slack_weights' must be string, pd.Series or dict. Is {}.".format(
type(slack_weights)
)
if isinstance(slack_weights, dict):
slack_weights = pd.Series(slack_weights)
elif isinstance(slack_weights, str):
valid_strings = ["p_nom", "p_nom_opt", "p_set"]
assert (
slack_weights in valid_strings
), "String value for 'slack_weights' must be one of {}. Is {}.".format(
valid_strings, slack_weights
)
snapshots = _as_snapshots(sub_network.network, snapshots)
logger.info(
"Performing non-linear load-flow on {} sub-network {} for snapshots {}".format(
sub_network.network.sub_networks.at[sub_network.name, "carrier"],
sub_network,
snapshots,
)
)
# _sub_network_prepare_pf(sub_network, snapshots, skip_pre, calculate_Y)
network = sub_network.network
if not skip_pre:
calculate_dependent_values(network)
find_bus_controls(sub_network)
_allocate_pf_outputs(network, linear=False)
# get indices for the components on this subnetwork
branches_i = sub_network.branches_i()
buses_o = sub_network.buses_o
sn_buses = sub_network.buses().index
sn_generators = sub_network.generators().index
generator_slack_weights_b = False
bus_slack_weights_b = False
if isinstance(slack_weights, pd.Series):
if all(i in sn_generators for i in slack_weights.index):
generator_slack_weights_b = True
elif all(i in sn_buses for i in slack_weights.index):
bus_slack_weights_b = True
else:
raise AssertionError(
"Custom slack weights pd.Series/dict must only have the",
"generators or buses of the subnetwork as index/keys.",
)
if not skip_pre and len(branches_i) > 0:
calculate_Y(sub_network, skip_pre=True)
_calculate_controllable_nodal_power_balance(
sub_network, network, snapshots, buses_o
)
def f(guess, distribute_slack=False, slack_weights=None):
last_pq = -1 if distribute_slack else None
network.buses_t.v_ang.loc[now, sub_network.pvpqs] = guess[
: len(sub_network.pvpqs)
]
network.buses_t.v_mag_pu.loc[now, sub_network.pqs] = guess[
len(sub_network.pvpqs) : last_pq
]
v_mag_pu = network.buses_t.v_mag_pu.loc[now, buses_o]
v_ang = network.buses_t.v_ang.loc[now, buses_o]
V = v_mag_pu * np.exp(1j * v_ang)
if distribute_slack:
slack_power = slack_weights * guess[-1]
mismatch = V * np.conj(sub_network.Y * V) - s + slack_power
else:
mismatch = V * np.conj(sub_network.Y * V) - s
if distribute_slack:
F = r_[real(mismatch)[:], imag(mismatch)[1 + len(sub_network.pvs) :]]
else:
F = r_[real(mismatch)[1:], imag(mismatch)[1 + len(sub_network.pvs) :]]
return F
def dfdx(guess, distribute_slack=False, slack_weights=None):
last_pq = -1 if distribute_slack else None
network.buses_t.v_ang.loc[now, sub_network.pvpqs] = guess[
: len(sub_network.pvpqs)
]
network.buses_t.v_mag_pu.loc[now, sub_network.pqs] = guess[
len(sub_network.pvpqs) : last_pq
]
v_mag_pu = network.buses_t.v_mag_pu.loc[now, buses_o]
v_ang = network.buses_t.v_ang.loc[now, buses_o]
V = v_mag_pu * np.exp(1j * v_ang)
index = r_[: len(buses_o)]
# make sparse diagonal matrices
V_diag = csr_matrix((V, (index, index)))
V_norm_diag = csr_matrix((V / abs(V), (index, index)))
I_diag = csr_matrix((sub_network.Y * V, (index, index)))
dS_dVa = 1j * V_diag * np.conj(I_diag - sub_network.Y * V_diag)
dS_dVm = V_norm_diag * np.conj(I_diag) + V_diag * np.conj(
sub_network.Y * V_norm_diag
)
J10 = dS_dVa[1 + len(sub_network.pvs) :, 1:].imag
J11 = dS_dVm[1 + len(sub_network.pvs) :, 1 + len(sub_network.pvs) :].imag
if distribute_slack:
J00 = dS_dVa[:, 1:].real
J01 = dS_dVm[:, 1 + len(sub_network.pvs) :].real
J02 = csr_matrix(slack_weights, (1, 1 + len(sub_network.pvpqs))).T
J12 = csr_matrix((1, len(sub_network.pqs))).T
J_P_blocks = [J00, J01, J02]
J_Q_blocks = [J10, J11, J12]
else:
J00 = dS_dVa[1:, 1:].real
J01 = dS_dVm[1:, 1 + len(sub_network.pvs) :].real
J_P_blocks = [J00, J01]
J_Q_blocks = [J10, J11]
J = svstack([shstack(J_P_blocks), shstack(J_Q_blocks)], format="csr")
return J
# Set what we know: slack V and v_mag_pu for PV buses
v_mag_pu_set = get_switchable_as_dense(network, "Bus", "v_mag_pu_set", snapshots)
network.buses_t.v_mag_pu.loc[snapshots, sub_network.pvs] = v_mag_pu_set.loc[
:, sub_network.pvs
]
network.buses_t.v_mag_pu.loc[snapshots, sub_network.slack_bus] = v_mag_pu_set.loc[
:, sub_network.slack_bus
]
network.buses_t.v_ang.loc[snapshots, sub_network.slack_bus] = 0.0
if not use_seed:
network.buses_t.v_mag_pu.loc[snapshots, sub_network.pqs] = 1.0
network.buses_t.v_ang.loc[snapshots, sub_network.pvpqs] = 0.0
slack_args = {"distribute_slack": distribute_slack}
slack_variable_b = 1 if distribute_slack else 0
if distribute_slack:
if isinstance(slack_weights, str) and slack_weights == "p_set":
generators_t_p_choice = get_switchable_as_dense(
network, "Generator", slack_weights, snapshots
)
bus_generation = generators_t_p_choice.rename(
columns=network.generators.bus
)
slack_weights_calc = (
pd.DataFrame(
bus_generation.groupby(bus_generation.columns, axis=1).sum(),
columns=buses_o,
)
.apply(normed, axis=1)
.fillna(0)
)
elif isinstance(slack_weights, str) and slack_weights in ["p_nom", "p_nom_opt"]:
assert (
not all(network.generators[slack_weights]) == 0
), "Invalid slack weights! Generator attribute {} is always zero.".format(
slack_weights
)
slack_weights_calc = (
network.generators.groupby("bus")
.sum()[slack_weights]
.reindex(buses_o)
.pipe(normed)
.fillna(0)
)
elif generator_slack_weights_b:
# convert generator-based slack weights to bus-based slack weights
slack_weights_calc = (
slack_weights.rename(network.generators.bus)
.groupby(slack_weights.index.name)
.sum()
.reindex(buses_o)
.pipe(normed)
.fillna(0)
)
elif bus_slack_weights_b:
# take bus-based slack weights
slack_weights_calc = slack_weights.reindex(buses_o).pipe(normed).fillna(0)
ss = np.empty((len(snapshots), len(buses_o)), dtype=complex)
roots = np.empty(
(
len(snapshots),
len(sub_network.pvpqs) + len(sub_network.pqs) + slack_variable_b,
)
)
iters =
|
pd.Series(0, index=snapshots)
|
pandas.Series
|
import pandas as pd
# FIXME: Not realistic as would like to adjust positions everyday
def trade_summary(df_input, security_name, position_name):
"""
For static positions only, i.e. at any time a fixed unit of positions are live.
i.e. if on day 0, 100 unit is bought, the unit will be kept at 100 throughout live signal
Take a dataframe with timestamp index, security, and security_pos (position) and calculate PnL trade by trade.
"""
df = df_input.copy()
df["long_short"] = (df[position_name] > 0) * 1 - (df[position_name] < 0) * 1
trade_detail = []
def update_trade(_trade_count, _position, _open_date, _open_price, _close_date, _close_price):
trade_detail.append({"trade": _trade_count,
"position": _position,
"open_date": _open_date,
"open_price": _open_price,
"close_date": _close_date,
"close_price": _close_price,
"realized_pnl": _position * (_close_price - open_price)})
trade_count = 0
long_short = 0
for i, data_slice in enumerate(df.iterrows()):
s = data_slice[1] # Slice
if i > 0 and s.long_short != df.iloc[i - 1].long_short:
if long_short != 0:
close_price, close_date = s[security_name], s.name
update_trade(trade_count, position, open_date, open_price, close_date, close_price)
long_short = 0
if s.long_short != 0:
open_price = s[security_name]
position = s[position_name]
open_date = s.name # date/time from index
trade_count += 1
long_short = s.long_short
if s.long_short != long_short:
close_price, close_date = s[security_name], s.name
close_date = s.name
update_trade(trade_count, position, open_date, open_price, close_date, close_price)
trade_summary_df = pd.DataFrame(trade_detail)
# Merge realized PnL onto original time_series. TODO: Can consider returning only one single series
trade_time_series = trade_summary_df[["close_date", "realized_pnl"]]
trade_time_series = trade_time_series.set_index("close_date")
trade_time_series.index.name = df_input.index.name
# TODO: AMEND DATETIME FORMAT WHEN NECESSARY
trade_time_series.index =
|
pd.to_datetime(trade_time_series.index, format="%d/%m/%Y")
|
pandas.to_datetime
|
import pandas as pd
import tqdm, os, glob, json, re, time
import numpy as np
from nltk.corpus import stopwords
from sklearn.model_selection import train_test_split
import enchant
import pickle as pkl
BASEPATH = '/Users/Janjua/Desktop/Projects/Octofying-COVID19-Literature/dataset'
stop_words = set(stopwords.words('english'))
engDict = enchant.Dict("en_US")
root_path = '/Users/Janjua/Desktop/Projects/Octofying-COVID19-Literature/dataset/CORD-19-research-challenge/'
def retrieve_data_from_json(file_path):
"""
Reads the json file and returns the necessary items.
# Arguments:
file_path: the path to the .json file
"""
with open(file_path) as file:
data = json.loads(file.read())
abstract, full_text = [], []
abstract = str([x['text'] for x in data['abstract']])
full_text = str([x['text'] for x in data['body_text']])
paper_id = data['paper_id']
return (paper_id, abstract, full_text)
def prepare_dataset():
"""
Reads the downloaded .csv file and performs some pre-processing on the data.
# Returns: A dataframe file is returned which has cleaned data columns
by removing the un-necessary information from the previous csv file.
# Credits:
Some aspects of code borrowed from:
https://www.kaggle.com/ivanegapratama/covid-eda-initial-exploration-tool
"""
data = pd.read_csv(BASEPATH + "/CORD-19-research-challenge/metadata.csv")
json_files = glob.glob(BASEPATH + "/CORD-19-research-challenge/*/*/*.json", recursive=True)
covid_data_dict = {'paper_id': [],
'abstract': [],
'body_text': [],
'authors': [],
'title': [],
'journal': []}
for idx, entry in enumerate(json_files):
if idx % (len(json_files) // 10) == 0:
print('Processing: {} of {}'.format(idx, len(json_files)))
paper_id, abstract, full_text = retrieve_data_from_json(entry)
meta = data.loc[data['sha'] == paper_id]
if len(meta) == 0:
continue
covid_data_dict['paper_id'].append(paper_id)
covid_data_dict['abstract'].append(abstract)
covid_data_dict['body_text'].append(full_text)
try:
authors = meta['authors'].values[0].split(';')
if len(authors) > 2:
covid_data_dict['authors'].append(authors[:1] + "...")
else:
covid_data_dict['authors'].append(". ".join(authors))
except:
covid_data_dict['authors'].append(". ".join(authors))
covid_data_dict['title'].append(meta['title'].values[0])
covid_data_dict['journal'].append(meta['journal'].values[0])
covid_df = pd.DataFrame(covid_data_dict, columns=['paper_id', 'abstract', 'body_text', \
'authors', 'title', 'journal'])
covid_df['abstract_word_count'] = covid_df['abstract'].apply(lambda x: len(x.strip().split()))
covid_df['body_text_word_count'] = covid_df['body_text'].apply(lambda x: len(x.strip().split()))
# Removing preposition marks
covid_df['body_text'] = covid_df['body_text'].apply(lambda x: re.sub('[^a-zA-z0-9\s]','', x))
covid_df['abstract'] = covid_df['abstract'].apply(lambda x: re.sub('[^a-zA-z0-9\s]','', x))
# Convert to lower case
covid_df['body_text'] = covid_df['body_text'].apply(lambda x: x.lower())
covid_df['abstract'] = covid_df['abstract'].apply(lambda x: x.lower())
covid_df.to_csv(BASEPATH + '/COVID_19_Lit.csv', encoding='utf-8', index=False)
print(covid_df.head())
print("Written dataframe to .csv file.")
def to_one_hot(data_point_index, vocab_size):
"""
Converts numbers to one hot vectors
# Returns: a one hot vector temp
# Credits:
Function taken from:
https://gist.github.com/aneesh-joshi/c8a451502958fa367d84bf038081ee4b
"""
temp = np.zeros(vocab_size)
temp[data_point_index] = 1
return temp
def load_data_for_training_w2v():
"""
Loads the data for training and testing for the word2vec model.
"""
data = pd.read_csv(BASEPATH + '/COVID_19_Lit.csv')
corpus = data.drop(["paper_id", "abstract", "abstract_word_count", "body_text_word_count", "authors", "title", "journal"], axis=1)
print(corpus.head(1))
words, n_gram = [], []
print(len(corpus))
start = time.time()
for ix in range(0, len(corpus)):
words.append(str(corpus.iloc[ix]['body_text'][1:-1]).split(" "))
print('Word Length: ', len(words))
for word in words:
for i in range(len(word)-2+1):
word1, word2 = word[i:i+2]
if word1 != "" and word2 != "":
if engDict.check(word1) == True and engDict.check(word2) == True:
n_gram.append("".join(word[i:i+2]))
end = time.time()
print("Prepared n-grams in: {}s".format(end-start))
print("N-gram length: ", len(n_gram))
n_gram = n_gram[:100000]
print("Reducing size to: ", len(n_gram))
word2int, int2word = {}, {}
print("N-gram length: ", len(n_gram))
start = time.time()
for i, word in enumerate(n_gram):
word2int[word] = i
int2word[i] = word
word_with_neighbor = list(map(list, zip(n_gram, n_gram[1:])))
end = time.time()
print("Computed neighbours in: {}s".format(end-start))
X, y = [], []
vocab_size = max(word2int.values()) + 1
print("Vocab size: ", vocab_size)
start = time.time()
for idx, word_neigh in enumerate(word_with_neighbor):
if idx % (len(word_with_neighbor) // 10) == 0:
print('Processing: {} of {}'.format(idx, len(word_with_neighbor)))
X.append(to_one_hot(word2int[word_neigh[0]], vocab_size))
y.append(to_one_hot(word2int[word_neigh[1]], vocab_size))
X = np.asarray(X)
y = np.asarray(y)
end = time.time()
print("Prepared the data vectors: {}s".format(end-start))
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
X_train = np.asarray(X_train)
y_train = np.asarray(y_train)
X_test = np.asarray(X_test)
y_test = np.asarray(y_test)
print("Shapes: \nX_train: {}\ny_train: {}\nX_test: {}\ny_test: {}".format(X_train.shape, y_train.shape, X_test.shape, y_test.shape))
np.save('arrays/X_train_w2v.npy', X_train)
np.save('arrays/y_train_w2v.npy', y_train)
np.save('arrays/X_test_w2v.npy', X_test)
np.save('arrays/y_test_w2v.npy', y_test)
print("Saved arrays!")
def read_arrays_and_return():
"""
Reads the prepared numpy arrays
# Returns: the read np arrays
"""
X_train = np.load('arrays/X_train_w2v.npy')
y_train = np.load('arrays/y_train_w2v.npy')
X_test = np.load('arrays/X_test_w2v.npy')
y_test = np.load('arrays/y_test_w2v.npy')
return (X_train, X_test, y_train, y_test)
class FileReader:
def __init__(self, file_path):
with open(file_path) as file:
content = json.load(file)
try:
self.paper_id = content['paper_id']
except:
pass
#self.paper_id = str(content['paper_id'])
self.abstract = []
self.body_text = []
# Abstract
try:
for entry in content['abstract']:
self.abstract.append(entry['text'])
except:
pass
# Body text
for entry in content['body_text']:
self.body_text.append(entry['text'])
self.abstract = '\n'.join(self.abstract)
self.body_text = '\n'.join(self.body_text)
def __repr__(self):
return f'{self.paper_id}: {self.abstract[:200]}... {self.body_text[:200]}...'
def get_breaks(content, length):
data = ""
words = content.split(' ')
total_chars = 0
# add break every length characters
for i in range(len(words)):
total_chars += len(words[i])
if total_chars > length:
data = data + "<br>" + words[i]
total_chars = 0
else:
data = data + " " + words[i]
return data
def prepare_dataset_for_BERT():
print("Preparing dataset for BERT!")
all_json = glob.glob(f'{root_path}/**/*.json', recursive=True)
print('Len of all files: ', len(all_json))
metadata_path = f'{root_path}/metadata.csv'
meta_df = pd.read_csv(metadata_path, dtype={
'pubmed_id': str,
'Microsoft Academic Paper ID': str,
'doi': str
})
dict_ = {'paper_id': [], 'abstract': [], 'body_text': [], 'authors': [], 'title': [], 'journal': [], 'abstract_summary': []}
for idx, entry in enumerate(all_json):
if idx % (len(all_json) // 10) == 0:
print(f'Processing index: {idx} of {len(all_json)}')
content = FileReader(entry)
meta_data = meta_df.loc[meta_df['sha'] == content.paper_id]
if len(meta_data) == 0:
continue
dict_['paper_id'].append(content.paper_id)
dict_['abstract'].append(content.abstract)
dict_['body_text'].append(content.body_text)
if len(content.abstract) == 0:
dict_['abstract_summary'].append("Not provided.")
elif len(content.abstract.split(' ')) > 100:
info = content.abstract.split(' ')[:100]
summary = get_breaks(' '.join(info), 40)
dict_['abstract_summary'].append(summary + "...")
else:
summary = get_breaks(content.abstract, 40)
dict_['abstract_summary'].append(summary)
meta_data = meta_df.loc[meta_df['sha'] == content.paper_id]
try:
authors = meta_data['authors'].values[0].split(';')
if len(authors) > 2:
dict_['authors'].append(". ".join(authors[:2]) + "...")
else:
dict_['authors'].append(". ".join(authors))
except Exception as e:
dict_['authors'].append(meta_data['authors'].values[0])
try:
title = get_breaks(meta_data['title'].values[0], 40)
dict_['title'].append(title)
except Exception as e:
dict_['title'].append(meta_data['title'].values[0])
dict_['journal'].append(meta_data['journal'].values[0])
print('Paper ID len: ', len(dict_['paper_id']))
print('Abstract len: ', len(dict_['abstract']))
print('Text len: ', len(dict_['body_text']))
print('Title len: ', len(dict_['title']))
print('Journal len: ', len(dict_['journal']))
print('Abstract Summary len: ', len(dict_['abstract_summary']))
df_covid = pd.DataFrame(dict_, columns=['paper_id', 'abstract', 'body_text', 'authors', 'title', 'journal', 'abstract_summary'])
print("Data Cleaning!")
df_covid.drop_duplicates(['abstract', 'body_text'], inplace=True)
df_covid.dropna(inplace=True)
df_covid['body_text'] = df_covid['body_text'].apply(lambda x: re.sub('[^a-zA-z0-9\s]','',x))
df_covid['abstract'] = df_covid['abstract'].apply(lambda x: re.sub('[^a-zA-z0-9\s]','',x))
df_covid['body_text'] = df_covid['body_text'].apply(lambda x: x.lower())
df_covid['abstract'] = df_covid['abstract'].apply(lambda x: x.lower())
df_covid.to_csv(root_path + "covid.csv")
def preprocess_for_BERT():
if os.path.exists(root_path + "covid.csv"):
df_covid_test = pd.read_csv(root_path + "covid.csv")
text = df_covid_test.drop(["authors", "journal", "Unnamed: 0"], axis=1)
text_dict = text.to_dict()
len_text = len(text_dict["paper_id"])
print('Text Len: ', len_text)
paper_id_list, body_text_list = [], []
title_list, abstract_list, abstract_summary_list = [], [], []
for i in tqdm.tqdm(range(0, len_text)):
paper_id = text_dict["paper_id"][i]
body_text = text_dict["body_text"][i].split("\n")
title = text_dict["title"][i]
abstract = text_dict["abstract"][i]
abstract_summary = text_dict["abstract_summary"][i]
for b in body_text:
paper_id_list.append(paper_id)
body_text_list.append(b)
title_list.append(title)
abstract_list.append(abstract)
abstract_summary_list.append(abstract_summary)
print('Writing initial sentences to CSV file!')
df_sentences = pd.DataFrame({"paper_id": paper_id_list}, index=body_text_list)
df_sentences.to_csv(root_path + "covid_sentences.csv")
print('Writing complete sentences to CSV file!')
df_sentences =
|
pd.DataFrame({"paper_id":paper_id_list,"title":title_list,"abstract":abstract_list,"abstract_summary":abstract_summary_list},index=body_text_list)
|
pandas.DataFrame
|
import pandas as pd
import numpy as np
from functions import fao_regions as regions
data = 'data/'
def data_build(crop_proxie, diet_div_crop, diet_source_crop, diet_ls_only, diet_ls_only_source, min_waste):
"""*** Import of country data to build national diets ***"""
WPR_height = pd.read_csv(r"data/worldpopulationreview_height_data.csv")
WPR_height.loc[WPR_height.Area == "North Korea", "Area"] = "Democratic People's Republic of Korea"
Countrycodes = pd.read_csv(r"data/countrycodes.csv", sep = ";")
#FAO_pop = pd.read_excel(data+"/FAOSTAT_Population_v3.xlsx")
FAO_pop = pd.read_excel(data+"/FAOSTAT_2018_population.xlsx")
FAO_pop.loc[FAO_pop.Area == "Cote d'Ivoire", "Area"] = "Côte d'Ivoire"
FAO_pop.loc[FAO_pop.Area == "French Guyana", "Area"] = "French Guiana"
FAO_pop.loc[FAO_pop.Area == "Réunion", "Area"] = "Réunion"
"""*** Import and sorting of data ***"""
FAO_crops = pd.read_csv(data+"/FAOSTAT_crop Production.csv")
FAO_crops["group"] = FAO_crops.apply(lambda x: regions.group(x["Item Code"]), axis=1)
FAO_crops = FAO_crops.rename(columns={"Value" : "Production"})
FAO_crops["Production"] = FAO_crops["Production"] / 1000
FAO_crops["Unit"] = "1000 tonnes"
FAO_animals = pd.read_csv(data+"/FAO_animal_prod_2016.csv")
FAO_animals["group"] = FAO_animals.apply(lambda x: regions.group(x["Item Code"]), axis=1)
FAO_animals.loc[FAO_animals.Area == "United Kingdom of Great Britain and Northern Ireland", "Area"] = "United Kingdom"
FAO_animals = FAO_animals.rename(columns={"Value" : "Production"})
FAO_animals.drop(FAO_animals[FAO_animals.Unit != 'tonnes'].index, inplace = True)
FAO_animals["Production"] = FAO_animals["Production"] / 1000
FAO_animals["Unit"] = "1000 tonnes"
FAO_animals_5 = pd.read_csv(data+"/FAOSTAT_animal_prod_5.csv")
FAO_animals_5["group"] = FAO_animals_5.apply(lambda x: regions.group(x["Item Code (FAO)"]), axis=1)
FAO_animals_5.loc[FAO_animals_5.Area == "United Kingdom of Great Britain and Northern Ireland", "Area"] = "United Kingdom"
FAO_animals_5 = FAO_animals_5.rename(columns={"Value" : "Production"})
FAO_animals_5.drop(FAO_animals_5[FAO_animals_5.Unit != 'tonnes'].index, inplace = True)
FAO_animals_5["Production"] = FAO_animals_5["Production"] / 1000
FAO_animals_5["Unit"] = "1000 tonnes"
FAO_animals_5 = FAO_animals_5.groupby(['Area', 'Item']).mean().reset_index()
FAO_animals = pd.merge(FAO_animals, FAO_animals_5[['Area', 'Item', 'Production']], on = ["Area", "Item"], how = 'left')
FAO_animals["Production"] = FAO_animals["Production_y"]
FAO_animals = FAO_animals.drop(columns = ["Production_x", "Production_y"])
FAO_fish = pd.read_csv(data+"FAOSTAT_Fish.csv")
FAO_fish = FAO_fish.rename(columns={"Value" : "Production"})
FAO_fish["group"] = FAO_fish.apply(lambda x: regions.group(x["Item Code"]), axis=1)
meat_products = ["eggs", "beef and lamb", "chicken and other poultry",\
"pork", "whole milk or derivative equivalents"]
fish_products = ["Freshwater Fish", "Demersal Fish", "Pelagic Fish",\
"Marine Fish, Other", "Crustaceans", "Cephalopods",\
"Molluscs, Other", "Meat, Aquatic Mammals", "Aquatic Animals, Others",
"Aquatic Plants", "Fish, Body Oil", "Fish, Liver Oil"]
other_items = ["Honey, natural", "Beeswax", "Silk-worm cocoons, reelable"]
other_items = ["Beeswax", "Silk-worm cocoons, reelable"]
"""*** Import of protein data ***"""
FAO_Protein =
|
pd.read_csv(data+"protein.csv")
|
pandas.read_csv
|
"""Standardize predictor values in a given tip attributes data frame using the mean and standard deviation from a fixed interval of training data and output the standardized attributes data frame and a tab-delimited file of the summary statistics used for standardization of each predictor.
"""
import argparse
import pandas as pd
from sklearn.preprocessing import StandardScaler
import sys
if __name__ == '__main__':
parser = argparse.ArgumentParser(
description="Standardize predictors",
formatter_class=argparse.ArgumentDefaultsHelpFormatter
)
parser.add_argument("--tip-attributes", required=True, help="a tab-delimited file describing tip attributes at one or more timepoints")
parser.add_argument("--standardized-attributes", required=True, help="tab-delimited output file with standardized values for each given predictor")
parser.add_argument("--statistics", required=True, help="tab-delimited output file with mean and standard deviation used to standardize each predictor")
parser.add_argument("--start-date", required=True, help="the earliest timepoint to calculate means and standard deviations from (YYYY-MM-DD format)")
parser.add_argument("--end-date", required=True, help="the latest timepoint to calculate means and standard deviations from (YYYY-MM-DD format)")
parser.add_argument("--predictors", nargs="+", help="a list of columns names for predictors whose values should be standardized")
args = parser.parse_args()
# Load tip attributes.
df = pd.read_csv(args.tip_attributes, sep="\t")
# Confirm presence of all requested predictor columns.
missing_columns = set(args.predictors) - set(df.columns)
if len(missing_columns) > 0:
print("Error: Could not find the following columns in the given attributes table:", file=sys.stderr)
for column in missing_columns:
print(f" - {column}", file=sys.stderr)
sys.exit(1)
# Confirm that timepoints are defined.
if not "timepoint" in df.columns:
print("Error: The given attributes table is missing a 'timepoint' column", file=sys.stderr)
sys.exit(1)
# Convert string timepoints to datetime instances.
df["timepoint"] = pd.to_datetime(df["timepoint"])
# Confirm availability of timepoints for calculating summary statistics.
start_date = pd.to_datetime(args.start_date)
end_date =
|
pd.to_datetime(args.end_date)
|
pandas.to_datetime
|
import networkx as nx
import numpy as np
import pandas as pd
import math
import numbers
import os
from collections import namedtuple
from graph_nets import utils_np
Point = namedtuple('Point', ['x', 'y', 'z'])
Pos = namedtuple('Pos', ['x', 'y', 'z', 'eta', 'phi', 'theta', 'r3', 'r'])
def calc_dphi(phi1, phi2):
"""Computes phi2-phi1 given in range [-pi,pi]"""
dphi = phi2 - phi1
if dphi > np.pi:
dphi -= 2*np.pi
if dphi < -np.pi:
dphi += 2*np.pi
return dphi
def pos_transform(r, phi, z):
x = r * math.cos(phi)
y = r * math.sin(phi)
r3 = math.sqrt(r**2 + z**2)
theta = math.acos(z/r3)
eta = -math.log(math.tan(theta*0.5))
return Pos(x, y, z, eta, phi, theta, r3, r)
def dist(x, y):
return math.sqrt(x**2 + y**2)
def wdist(a, d, w):
pp = a.x*a.x + a.y*a.y + a.z*a.z*w
pd = a.x*d.x + a.y*d.y + a.z*d.z*w
dd = d.x*d.x + d.y*d.y + d.z*d.z*w
return math.sqrt(abs(pp - pd*pd/dd))
def wdistr(r1, dr, az, dz, w):
pp = r1*r1+az*az*w
pd = r1*dr+az*dz*w
dd = dr*dr+dz*dz*w
return math.sqrt(abs(pp-pd*pd/dd))
def circle(a, b, c):
ax = a.x-c.x
ay = a.y-c.y
bx = b.x-c.x
by = b.y-c.y
aa = ax*ax + ay*ay
bb = bx*bx + by*by
idet = 0.5/(ax*by-ay*bx)
p0 = Point(x=(aa*by-bb*ay)*idet, y=(ax*bb-bx*aa)*idet, z=0)
r = math.sqrt(p0.x*p0.x + p0.y*p0.y)
p = Point(x=p0.x+c.x, y=p0.y+c.y, z=p0.z)
return p, r
def zdists(a, b):
origin = Point(x=0, y=0, z=0)
p, r = circle(origin, a, b)
ang_ab = 2*math.asin(dist(a.x-b.x, a.y-b.y)*0.5/r)
ang_a = 2*math.asin(dist(a.x, a.y)*0.5/r)
return abs(b.z-a.z-a.z*ang_ab/ang_a)
def get_edge_features2(in_node, out_node, add_angles=False):
# input are the features of incoming and outgoing nodes
# they are ordered as [r, phi, z]
v_in = pos_transform(*in_node)
v_out = pos_transform(*out_node)
deta = v_out.eta - v_in.eta
dphi = calc_dphi(v_out.phi, v_in.phi)
dR = np.sqrt(deta**2 + dphi**2)
#dZ = v_out.z - v_in.z
dZ = v_in.z - v_out.z #
results = {"distance": np.array([deta, dphi, dR, dZ])}
if add_angles:
pa = Point(x=v_out.x, y=v_out.y, z=v_out.z)
pb = Point(x=v_in.x, y=v_in.y, z=v_in.z)
pd = Point(x=pa.x-pb.x, y=pa.y-pb.y, z=pa.z-pb.z)
wd0 = wdist(pa, pd, 0)
wd1 = wdist(pa, pd, 1)
zd0 = zdists(pa, pb)
wdr = wdistr(v_out.r, v_in.r-v_out.r, pa.z, pd.z, 1)
results['angles'] = np.array([wd0, wd1, zd0, wdr])
return results
def get_edge_features(in_node, out_node):
# input are the features of incoming and outgoing nodes
# they are ordered as [r, phi, z]
in_r, in_phi, in_z = in_node
out_r, out_phi, out_z = out_node
in_r3 = np.sqrt(in_r**2 + in_z**2)
out_r3 = np.sqrt(out_r**2 + out_z**2)
in_theta = np.arccos(in_z/in_r3)
in_eta = -np.log(np.tan(in_theta/2.0))
out_theta = np.arccos(out_z/out_r3)
out_eta = -np.log(np.tan(out_theta/2.0))
deta = out_eta - in_eta
dphi = calc_dphi(out_phi, in_phi)
dR = np.sqrt(deta**2 + dphi**2)
dZ = in_z - out_z
return np.array([deta, dphi, dR, dZ])
def data_dict_to_nx(dd_input, dd_target, use_digraph=True, bidirection=True):
input_nx = utils_np.data_dict_to_networkx(dd_input)
target_nx = utils_np.data_dict_to_networkx(dd_target)
G = nx.DiGraph() if use_digraph else nx.Graph()
for node_index, node_features in input_nx.nodes(data=True):
G.add_node(node_index, pos=node_features['features'])
for sender, receiver, features in target_nx.edges(data=True):
G.add_edge(sender, receiver, solution=features['features'])
if use_digraph and bidirection:
G.add_edge(receiver, sender, solution=features['features'])
return G
def correct_networkx(Gi, isec, n_phi_sections=8, n_eta_sections=2):
G = Gi.copy()
phi_range = (-np.pi, np.pi)
phi_edges = np.linspace(*phi_range, num=n_phi_sections+1)
scale = [1000, np.pi/n_phi_sections, 1000]
# update phi
phi_min = phi_edges[isec//n_eta_sections]
phi_max = phi_edges[isec//n_eta_sections+1]
for node_id, features in G.nodes(data=True):
new_feature = features['pos']*scale
new_feature[1] = new_feature[1] + (phi_min + phi_max) / 2
if new_feature[1] > np.pi:
new_feature[1] -= 2*np.pi
if new_feature[1] < -np.pi:
new_feature[1]+= 2*np.pi
G.node[node_id].update(pos=new_feature)
return G
def networkx_graph_to_hitsgraph(G, is_digraph=True):
n_nodes = len(G.nodes())
n_edges = len(G.edges())//2 if is_digraph else len(G.edges())
n_features = len(G.node[0]['pos'])
X = np.zeros((n_nodes, n_features), dtype=np.float32)
Ri = np.zeros((n_nodes, n_edges), dtype=np.uint8)
Ro = np.zeros((n_nodes, n_edges), dtype=np.uint8)
for node,features in G.nodes(data=True):
X[node, :] = features['pos']
## build relations
segments = []
y = []
for n, nbrsdict in G.adjacency():
for nbr, eattr in nbrsdict.items():
## as hitsgraph is a directed graph from inner-most to outer-most
## so assume sender < receiver;
if n > nbr and is_digraph:
continue
segments.append((n, nbr))
y.append(int(eattr['solution'][0]))
if len(y) != n_edges:
print(len(y),"not equals to # of edges", n_edges)
segments = np.array(segments)
Ro[segments[:, 0], np.arange(n_edges)] = 1
Ri[segments[:, 1], np.arange(n_edges)] = 1
y = np.array(y, dtype=np.float32)
return (X, Ri, Ro, y)
def is_diff_networkx(G1, G2):
"""
G1,G2, networkx graphs
Return True if they are different, False otherwise
note that edge features are not checked!
"""
# check node features first
GRAPH_NX_FEATURES_KEY = 'pos'
node_id1 = np.array([
x[1][GRAPH_NX_FEATURES_KEY]
for x in G1.nodes(data=True)
if x[1][GRAPH_NX_FEATURES_KEY] is not None])
node_id2 = np.array([
x[1][GRAPH_NX_FEATURES_KEY]
for x in G2.nodes(data=True)
if x[1][GRAPH_NX_FEATURES_KEY] is not None])
# check edges
diff = np.any(node_id1 != node_id2)
for sender, receiver, _ in G1.edges(data=True):
try:
_ = G2.edges[(sender, receiver)]
except KeyError:
diff = True
break
return diff
## predefined group info
vlids = [(7,2), (7,4), (7,6), (7,8), (7,10), (7,12), (7,14),
(8,2), (8,4), (8,6), (8,8),
(9,2), (9,4), (9,6), (9,8), (9,10), (9,12), (9,14),
(12,2), (12,4), (12,6), (12,8), (12,10), (12,12),
(13,2), (13,4), (13,6), (13,8),
(14,2), (14,4), (14,6), (14,8), (14,10), (14,12),
(16,2), (16,4), (16,6), (16,8), (16,10), (16,12),
(17,2), (17,4),
(18,2), (18,4), (18,6), (18,8), (18,10), (18,12)]
n_det_layers = len(vlids)
def merge_truth_info_to_hits(hits, particles, truth):
if 'pt' not in particles.columns:
px = particles.px
py = particles.py
pt = np.sqrt(px**2 + py**2)
particles = particles.assign(pt=pt)
hits = hits.merge(truth, on='hit_id', how='left')
hits = hits.merge(particles, on='particle_id', how='left')
# selective information
# noise hits does not have particle info
hits = hits.fillna(value=0)
# Assign convenient layer number [0-47]
vlid_groups = hits.groupby(['volume_id', 'layer_id'])
hits = pd.concat([vlid_groups.get_group(vlids[i]).assign(layer=i)
for i in range(n_det_layers)])
# add new features
x = hits.x
y = hits.y
z = hits.z
absz = np.abs(z)
r = np.sqrt(x**2 + y**2) # distance from origin in transverse plane
r3 = np.sqrt(r**2 + z**2) # in 3D
phi = np.arctan2(hits.y, hits.x)
theta = np.arccos(z/r3)
eta = -np.log(np.tan(theta/2.))
tpx = hits.tpx
tpy = hits.tpy
tpt = np.sqrt(tpx**2 + tpy**2)
hits = hits.assign(r=r, phi=phi, eta=eta, r3=r3, absZ=absz, tpt=tpt)
# add hit indexes to column hit_idx
hits = hits.rename_axis('hit_idx').reset_index()
return hits
def pairs_to_df(pairs, hits):
"""pairs is np.array, each row is a pair. columns are incoming and outgoing nodes
return a DataFrame with columns,
['hit_id_in', 'hit_idx_in', 'layer_in', 'hit_id_out', 'hit_idx_out', 'layer_out']
"""
# form a DataFrame
in_nodes = pd.DataFrame(pairs[:, 0], columns=['hit_id'])
out_nodes =
|
pd.DataFrame(pairs[:, 1], columns=['hit_id'])
|
pandas.DataFrame
|
import pandas as pd
import os
# using the merged_medianData.csv create dose-response files for each compound
infolder = '../data/processed_data/01/LISS/01/'
outfolder = '../data/processed_data/01/LISS/02/'
if not os.path.exists(outfolder):
os.makedirs(outfolder)
geneList = pd.read_csv('../data/lists/gene_list.csv')
genes = geneList['Gene'].tolist()
data = pd.read_csv(infolder + 'merged_medianData.csv')
df0 = data
cmpMetas_cols = ['CMP', 'DOSES']
cmpMetas = pd.DataFrame(columns=cmpMetas_cols)
compounds = data['CMP'].unique().tolist()
times = [6, 24]
df0 = data
cmpMetas_cols = ['CMP', 'DOSES']
cmpMetas = pd.DataFrame(columns=cmpMetas_cols)
compounds = data['CMP'].unique().tolist()
times = [6, 24]
# the control compounds do not have a dose responses
# this is because there is only one 'dose' for each compound
# the dose-response files must still be generated for future computations
control_compounds = ['DMSO_0.5pct', 'DMSO_0.1pct', 'Medium']
# generate dose responses for all compounds except controls
for i in times:
df1 = df0[df0['TIME'] == i]
df1 = df1[~df1['CMP'].isin(control_compounds)]
for j in compounds:
df2 = df1[df1['CMP'] == j]
df2 = df2.sort_values('DOSE', axis=0, ascending=True, inplace=False,
kind='quicksort', na_position='last')
tempMetas = pd.DataFrame(columns=cmpMetas_cols)
tempMetas['CMP'] = [j]
doses = df2['DOSE'].tolist()
tempMetas['DOSES'] = [doses]
cmpMetas = cmpMetas.append(tempMetas)
dosecourse_cols = ['GENE', '1', '2', '3', '4', '5', '6']
dosecourse = pd.DataFrame(columns=dosecourse_cols)
for m in genes:
dosecourseTemp = pd.DataFrame(columns=dosecourse_cols)
dosecourseTemp['GENE'] = [m]
geneValues = df2[m].tolist()
geneValueDoseDict = dict(zip(['1', '2', '3', '4', '5', '6'],
geneValues))
for k in geneValueDoseDict:
dosecourseTemp[k] = [geneValueDoseDict[k]]
dosecourse = dosecourse.append(dosecourseTemp)
file = j + '_' + str(i) + 'h'
dosecourse.to_csv((outfolder + file + '.csv'), index=False)
cmpMetas = cmpMetas.drop_duplicates('CMP')
cmpMetas.to_csv(outfolder + 'cmpMetas.csv', index=False)
# generate dose responses for control compounds
for i in times:
df1 = df0[df0['TIME'] == i]
for j in (control_compounds):
df2 = df1[df1['CMP'] == j]
df2 = df2.sort_values('DOSE', axis=0, ascending=True, inplace=False,
kind='quicksort', na_position='last')
dosecourse_cols = ['GENE', '1', '2', '3', '4', '5', '6']
dosecourse =
|
pd.DataFrame(columns=dosecourse_cols)
|
pandas.DataFrame
|
import logging
import pandas as pd
from src.utils import get_adj_matrix
from src.hits import get_hits
from src.pagerank import get_pagerank
from src.simrank import get_simrank
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(name)s - %(levelname)s - %(message)s')
logger = logging.getLogger(__name__)
logger.setLevel(level=logging.INFO)
def main(args):
with open(args.input, 'r') as input:
links = input.readlines()
adj_matrix = get_adj_matrix(links)
hubs, authorities = get_hits(adj_matrix)
pagerank = get_pagerank(adj_matrix)
# simRank calculation
compare_nodes = (1, 2)
simrank = get_simrank(compare_nodes, adj_matrix)
logger.info(f'hubs: {hubs}, authorities: {authorities}')
logger.info(f'pageRank: {pagerank}')
logger.info(f'simRank: {simrank}')
# Output values to files
results =
|
pd.DataFrame({"hubs": hubs, "authorities": authorities, "pagerank": pagerank})
|
pandas.DataFrame
|
from czsc.extend.utils import push_text
from datetime import datetime
from czsc.extend.analyzeExtend import JKCzscTraderExtend as CzscTrader
import traceback
import time
import datetime
import shutil
import os
from czsc.objects import Signal, Factor, Event, Operate
from czsc.data.jq import get_kline
import pandas as pd
# 基础参数配置
ct_path = os.path.join("d:\\data", "czsc_traders")
os.makedirs(ct_path, exist_ok=True)
symbol = '399006.XSHE'
my_dic_container = {}
def start():
moni_path = os.path.join(ct_path, "monitor")
if os.path.exists(moni_path):
shutil.rmtree(moni_path)
os.makedirs(moni_path, exist_ok=True)
events_monitor = [
# 开多
Event(name="一买", operate=Operate.LO, factors=[
Factor(name="5分钟类一买", signals_all=[Signal("5分钟_倒1笔_类买卖点_类一买_任意_任意_0")]),
Factor(name="5分钟形一买", signals_all=[Signal("5分钟_倒1笔_基础形态_类一买_任意_任意_0")]),
Factor(name="15分钟类一买", signals_all=[Signal("15分钟_倒1笔_类买卖点_类一买_任意_任意_0")]),
Factor(name="15分钟形一买", signals_all=[Signal("15分钟_倒1笔_基础形态_类一买_任意_任意_0")]),
Factor(name="30分钟类一买", signals_all=[Signal("30分钟_倒1笔_类买卖点_类一买_任意_任意_0")]),
Factor(name="30分钟形一买", signals_all=[Signal("30分钟_倒1笔_基础形态_类一买_任意_任意_0")]),
]),
Event(name="二买", operate=Operate.LO, factors=[
Factor(name="5分钟类二买", signals_all=[Signal("5分钟_倒1笔_类买卖点_类二买_任意_任意_0")]),
Factor(name="5分钟形二买", signals_all=[Signal("5分钟_倒1笔_基础形态_类二买_任意_任意_0")]),
Factor(name="15分钟类二买", signals_all=[Signal("15分钟_倒1笔_类买卖点_类二买_任意_任意_0")]),
Factor(name="15分钟形二买", signals_all=[Signal("15分钟_倒1笔_基础形态_类二买_任意_任意_0")]),
Factor(name="30分钟类二买", signals_all=[Signal("30分钟_倒1笔_类买卖点_类二买_任意_任意_0")]),
Factor(name="30分钟形二买", signals_all=[Signal("30分钟_倒1笔_基础形态_类二买_任意_任意_0")]),
]),
Event(name="三买", operate=Operate.LO, factors=[
Factor(name="5分钟类三买", signals_all=[Signal("5分钟_倒1笔_类买卖点_类三买_任意_任意_0")]),
Factor(name="5分钟形三买", signals_all=[Signal("5分钟_倒1笔_基础形态_类三买_任意_任意_0")]),
Factor(name="15分钟类三买", signals_all=[Signal("15分钟_倒1笔_类买卖点_类三买_任意_任意_0")]),
Factor(name="15分钟形三买", signals_all=[Signal("15分钟_倒1笔_基础形态_类三买_任意_任意_0")]),
Factor(name="30分钟类三买", signals_all=[Signal("30分钟_倒1笔_类买卖点_类三买_任意_任意_0")]),
Factor(name="30分钟形三买", signals_all=[Signal("30分钟_倒1笔_基础形态_类三买_任意_任意_0")]),
]),
# 平多
Event(name="一卖", operate=Operate.LE, factors=[
Factor(name="5分钟类一卖", signals_all=[Signal("5分钟_倒1笔_类买卖点_类一卖_任意_任意_0")]),
Factor(name="5分钟形一卖", signals_all=[Signal("5分钟_倒1笔_基础形态_类一卖_任意_任意_0")]),
Factor(name="15分钟类一卖", signals_all=[Signal("15分钟_倒1笔_类买卖点_类一卖_任意_任意_0")]),
Factor(name="15分钟形一卖", signals_all=[Signal("15分钟_倒1笔_基础形态_类一卖_任意_任意_0")]),
Factor(name="30分钟类一卖", signals_all=[Signal("30分钟_倒1笔_类买卖点_类一卖_任意_任意_0")]),
Factor(name="30分钟形一卖", signals_all=[Signal("30分钟_倒1笔_基础形态_类一卖_任意_任意_0")]),
]),
Event(name="二卖", operate=Operate.LE, factors=[
Factor(name="5分钟类二卖", signals_all=[Signal("5分钟_倒1笔_类买卖点_类二卖_任意_任意_0")]),
Factor(name="5分钟形二卖", signals_all=[Signal("5分钟_倒1笔_基础形态_类二卖_任意_任意_0")]),
Factor(name="15分钟类二卖", signals_all=[Signal("15分钟_倒1笔_类买卖点_类二卖_任意_任意_0")]),
Factor(name="15分钟形二卖", signals_all=[Signal("15分钟_倒1笔_基础形态_类二卖_任意_任意_0")]),
Factor(name="30分钟类二卖", signals_all=[Signal("30分钟_倒1笔_类买卖点_类二卖_任意_任意_0")]),
Factor(name="30分钟形二卖", signals_all=[Signal("30分钟_倒1笔_基础形态_类二卖_任意_任意_0")]),
]),
Event(name="三卖", operate=Operate.LE, factors=[
Factor(name="5分钟类三卖", signals_all=[Signal("5分钟_倒1笔_类买卖点_类三卖_任意_任意_0")]),
Factor(name="5分钟形三卖", signals_all=[Signal("5分钟_倒1笔_基础形态_类三卖_任意_任意_0")]),
Factor(name="15分钟类三卖", signals_all=[Signal("15分钟_倒1笔_类买卖点_类三卖_任意_任意_0")]),
Factor(name="15分钟形三卖", signals_all=[Signal("15分钟_倒1笔_基础形态_类三卖_任意_任意_0")]),
Factor(name="30分钟类三卖", signals_all=[Signal("30分钟_倒1笔_类买卖点_类三卖_任意_任意_0")]),
Factor(name="30分钟形三卖", signals_all=[Signal("30分钟_倒1笔_基础形态_类三卖_任意_任意_0")]),
]),
]
try:
current_date: datetime = pd.to_datetime('2021-08-10')
end_date =
|
pd.to_datetime("2021-08-20")
|
pandas.to_datetime
|
from typing import Dict, List, Tuple
from multiprocessing import Pool
from scipy.signal import find_peaks
import numpy as np
import pandas as pd
import pyarrow.parquet as pq
import numpy as np
class DataPipeline:
"""
Top level class which takes in the parquet file and converts it to a low dimensional dataframe.
"""
def __init__(
self,
parquet_fname: str,
data_processor_args,
start_row_num: int,
num_rows: int,
concurrency: int = 100,
):
self._fname = parquet_fname
self._concurrency = concurrency
self._nrows = num_rows
self._start_row_num = start_row_num
self._processor_args = data_processor_args
self._process_count = 4
@staticmethod
def run_one_chunk(arg_tuple):
fname, processor_args, start_row, end_row, num_rows = arg_tuple
cols = [str(i) for i in range(start_row, end_row)]
data = pq.read_pandas(fname, columns=cols)
data_df = data.to_pandas()
processor = DataProcessor(**processor_args)
output_df = processor.transform(data_df)
print('Another ', round((end_row - start_row) / num_rows * 100, 2), '% Complete')
del processor
del data_df
del data
return output_df
def run(self):
outputs = []
args_list = []
for s_index in range(self._start_row_num, self._start_row_num + self._nrows, self._concurrency):
e_index = s_index + self._concurrency
args_this_chunk = [self._fname, self._processor_args, s_index, e_index, self._nrows]
args_list.append(args_this_chunk)
pool = Pool(self._process_count)
outputs = pool.map(DataPipeline.run_one_chunk, args_list)
pool.close()
pool.join()
final_output_df = pd.concat(outputs, axis=1)
return final_output_df
class DataProcessor:
def __init__(
self,
intended_time_steps: int,
original_time_steps: int,
peak_threshold: int,
smoothing_window: int = 3,
remove_corona=False,
corona_max_distance=5,
corona_max_height_ratio=0.8,
corona_cleanup_distance=10,
num_processes: int = 7,
):
self._o_steps = original_time_steps
self._steps = intended_time_steps
# 50 is a confident smoothed version
# resulting signal after subtracting the smoothened version has some noise along with signal.
# with 10, smoothened version seems to have some signals as well.
self._smoothing_window = smoothing_window
self._num_processes = num_processes
self._peak_threshold = peak_threshold
self._remove_corona = remove_corona
self._corona_max_distance = corona_max_distance
self._corona_max_height_ratio = corona_max_height_ratio
self._corona_cleanup_distance = corona_cleanup_distance
def get_noise(self, X_df: pd.DataFrame):
"""
TODO: we need to keep the noise. However, we don't want very high freq jitter.
band pass filter is what is needed.
"""
msg = 'Expected len:{}, found:{}'.format(self._o_steps, len(X_df))
assert self._o_steps == X_df.shape[0], msg
smoothe_df = X_df.rolling(self._smoothing_window, min_periods=1).mean()
noise_df = X_df - smoothe_df
del smoothe_df
return noise_df
@staticmethod
def peak_data(
ser: pd.Series,
threshold: float,
quantiles=[0, 0.25, 0.5, 0.75, 1],
) -> Dict[str, np.array]:
maxima_peak_indices, maxima_data_dict = find_peaks(ser, threshold=threshold, width=0)
maxima_width = maxima_data_dict['widths']
maxima_height = maxima_data_dict['prominences']
minima_peak_indices, minima_data_dict = find_peaks(-1 * ser, threshold=threshold, width=0)
minima_width = minima_data_dict['widths']
minima_height = minima_data_dict['prominences']
peak_indices = np.concatenate([maxima_peak_indices, minima_peak_indices])
peak_width = np.concatenate([maxima_width, minima_width])
peak_height = np.concatenate([maxima_height, minima_height])
maxima_minima = np.concatenate([np.array([1] * len(maxima_height)), np.array([-1] * len(minima_height))])
index_ordering = np.argsort(peak_indices)
peak_width = peak_width[index_ordering]
peak_height = peak_height[index_ordering]
peak_indices = peak_indices[index_ordering]
maxima_minima = maxima_minima[index_ordering]
return {
'width': peak_width,
'height': peak_height,
'maxima_minima': maxima_minima,
'indices': peak_indices,
}
@staticmethod
def corona_discharge_index_pairs(
ser: pd.Series,
peak_threshold: float,
corona_max_distance: int,
corona_max_height_ratio: float,
) -> List[Tuple[int, int]]:
"""
Args:
ser: time series data.
peak_threshold: for detecting peaks, if elevation is more than this value, then consider it a peak.
corona_max_distance: maximum distance between consequitive alternative peaks for it to be a corona discharge.
corona_max_height_ratio: the alternate peaks should have similar peak heights.
Returns:
List of (peak1, peak2) indices. Note that these peaks are consequitive and have opposite sign
"""
data = DataProcessor.peak_data(ser, peak_threshold)
corona_indices = []
for index, data_index in enumerate(data['indices']):
if index < len(data['indices']) - 1:
opposite_peaks = data['maxima_minima'][index] * data['maxima_minima'][index + 1] == -1
nearby_peaks = data['indices'][index + 1] - data['indices'][index] < corona_max_distance
# for height ratio, divide smaller by larger height
h1 = data['height'][index]
h2 = data['height'][index + 1]
height_ratio = (h1 / h2 if h1 < h2 else h2 / h1)
similar_height = height_ratio > corona_max_height_ratio
if opposite_peaks and nearby_peaks and similar_height:
corona_indices.append((data_index, data['indices'][index + 1]))
return corona_indices
@staticmethod
def remove_corona_discharge(
ser: pd.Series,
peak_threshold: float,
corona_max_distance: int,
corona_max_height_ratio: float,
corona_cleanup_distance: int,
) -> pd.Series:
"""
Args:
ser: time series data.
peak_threshold: for detecting peaks, if elevation is more than this value, then consider it a peak.
corona_max_distance: maximum distance between consequitive alternative peaks for it to be a corona discharge.
corona_max_height_ratio: the alternate peaks should have similar peak heights.
corona_cleanup_distance: how many indices after the corona discharge should the data be removed.
Returns:
ser: cleaned up time series data.
"""
pairs = DataProcessor.corona_discharge_index_pairs(
ser,
peak_threshold,
corona_max_distance,
corona_max_height_ratio,
)
ser = ser.copy()
for start_index, end_index in pairs:
smoothing_start_index = max(0, start_index - 1)
smoothing_end_index = min(end_index + corona_cleanup_distance, ser.index[-1])
start_val = ser.iloc[smoothing_start_index]
end_val = ser.iloc[smoothing_end_index]
count = smoothing_end_index - smoothing_start_index
ser.iloc[smoothing_start_index:smoothing_end_index] = np.linspace(start_val, end_val, count)
return ser
@staticmethod
def peak_stats(ser: pd.Series, threshold, quantiles=[0, 0.25, 0.5, 0.75, 1]):
"""
Returns quantiles of peak width, height, distances from next peak.
"""
data = DataProcessor.peak_data(ser, threshold, quantiles=quantiles)
peak_indices = data['indices']
peak_width = data['width']
peak_height = data['height']
if len(peak_indices) == 0:
# no peaks
width_stats = [0] * len(quantiles)
height_stats = [0] * len(quantiles)
distance_stats = [0] * len(quantiles)
else:
peak_distances = np.diff(peak_indices)
peak_width[peak_width > 100] = 100
width_stats = np.quantile(peak_width, quantiles)
height_stats = np.quantile(peak_height, quantiles)
# for just one peak, distance will be empty array.
if len(peak_distances) == 0:
assert len(peak_indices) == 1
distance_stats = [ser.shape[0]] * len(quantiles)
else:
distance_stats = np.quantile(peak_distances, quantiles)
width_names = ['peak_width_' + str(i) for i in quantiles]
height_names = ['peak_height_' + str(i) for i in quantiles]
distance_names = ['peak_distances_' + str(i) for i in quantiles]
index = width_names + height_names + distance_names + ['peak_count']
data = np.concatenate([width_stats, height_stats, distance_stats, [len(peak_indices)]])
return pd.Series(data, index=index)
@staticmethod
def get_peak_stats_df(df, peak_threshold):
"""
Args:
df:
columns are different examples.
axis is time series.
"""
return df.apply(lambda x: DataProcessor.peak_stats(x, peak_threshold), axis=0)
@staticmethod
def pandas_describe(df):
output_df = df.quantile([0, 0.25, 0.5, 0.75, 1], axis=0)
output_df.index = list(map(lambda x: 'Quant-' + str(x), output_df.index.tolist()))
abs_mean_df = df.abs().mean().to_frame('abs_mean')
mean_df = df.mean().to_frame('mean')
std_df = df.std().to_frame('std')
return pd.concat([output_df, abs_mean_df.T, mean_df.T, std_df.T])
@staticmethod
def transform_chunk(signal_time_series_df: pd.DataFrame, peak_threshold: float) -> pd.DataFrame:
"""
It sqashes the time series to a single point multi featured vector.
"""
df = signal_time_series_df
# mean, var, percentile.
# NOTE pandas.describe() is the costliest computation with 95% time of the function.
metrics_df = DataProcessor.pandas_describe(df)
peak_metrics_df = DataProcessor.get_peak_stats_df(df, peak_threshold)
metrics_df.index = list(map(lambda x: 'signal_' + x, metrics_df.index))
temp_metrics = [metrics_df, peak_metrics_df]
for smoothener in [1, 2, 4, 8, 16, 32]:
diff_df = df.rolling(smoothener).mean()[::smoothener].diff().abs()
temp_df = DataProcessor.pandas_describe(diff_df)
temp_df.index = list(map(lambda x: 'diff_smoothend_by_' + str(smoothener) + ' ' + x, temp_df.index))
temp_metrics.append(temp_df)
df = pd.concat(temp_metrics)
df.index.name = 'features'
return df
def transform(self, X_df: pd.DataFrame):
"""
Args:
X_df: dataframe with each column being one data point. Rows are timestamps.
"""
def cleanup_corona(x: pd.Series):
return DataProcessor.remove_corona_discharge(
x,
self._peak_threshold,
self._corona_max_distance,
self._corona_max_height_ratio,
self._corona_cleanup_distance,
)
# Corona removed.
if self._remove_corona:
X_df = X_df.apply(cleanup_corona)
# Remove the smoothened version of the data so as to work with noise.
if self._smoothing_window > 0:
X_df = self.get_noise(X_df)
# stepsize many consequitive timestamps are compressed to form one timestamp.
# this will ensure we are left with self._steps many timestamps.
stepsize = self._o_steps // self._steps
transformed_data = []
for s_tm_index in range(0, self._o_steps, stepsize):
e_tm_index = s_tm_index + stepsize
# NOTE: dask was leading to memory leak.
#one_data_point = delayed(DataProcessor.transform_chunk)(X_df.iloc[s_tm_index:e_tm_index, :])
one_data_point = DataProcessor.transform_chunk(X_df.iloc[s_tm_index:e_tm_index, :], self._peak_threshold)
transformed_data.append(one_data_point)
# transformed_data = dd.compute(*transformed_data, scheduler='processes', num_workers=self._num_processes)
# Add timestamp
for ts in range(0, len(transformed_data)):
transformed_data[ts]['ts'] = ts
df =
|
pd.concat(transformed_data, axis=0)
|
pandas.concat
|
import re
import pandas as pd
# import matplotlib
# matplotlib.use('Agg')
import matplotlib.pyplot as plt
from matplotlib.figure import Figure
import matplotlib.ticker as ticker
import matplotlib.dates as mdates
import numpy as np
import seaborn as sns; sns.set()
from scipy.spatial.distance import squareform
from scipy.spatial.distance import pdist, euclidean
from sklearn.preprocessing import MinMaxScaler
from datetime import datetime, timedelta
from io import StringIO, BytesIO
from app.models import Country, CountryStatus
import base64
import plotly.figure_factory as ff
data_dir = 'data/'
def get_all_places(level='countries'):
# df_places = pd.read_csv(data_dir + 'all_{}_compare.csv'.format(level))
df_places = Country.all_countries_names_as_df()
return list(df_places['Name'])
def get_all_countries_response():
df_places = pd.read_csv(data_dir + 'all_countries_response.csv')
return list(df_places['Country'])
def get_df_similar_places(place, level = 'countries'):
# if level == 'cities':
# df_sim = pd.read_csv(data_dir + 'all_{}_similarity.csv'.format(level))
# df_sim = df_sim[df_sim['CityBase'] == place]
# df_sim = df_sim[['Name', 'gap', 'dist', 'Similarity']].set_index('Name')
# return df_sim
# df_orig = pd.read_csv(data_dir + 'total_cases_{}_normalized.csv'.format(level))
df_orig = Country.all_countries_as_df()
df_orig_piv_day = df_orig.pivot(index='Name', columns='Day', values='TotalDeaths')
df_orig_piv_day = df_orig_piv_day.fillna(0)
sr_place = df_orig_piv_day.loc[place,]
place_start = (sr_place > 0).idxmax()
# place_start_cases = (df_orig.set_index('Name').loc[place,].set_index('Day')['Total'] > 0).idxmax()
days_ahead = 14 #if level == 'countries' else 5
df_places_ahead = df_orig_piv_day[df_orig_piv_day.loc[:, max(place_start - days_ahead,0)] > 0.0]
df_places_rate_norm = df_orig_piv_day.loc[df_places_ahead.index, :]
# df_places_rate_norm = df_orig_piv_day.loc[['France', 'Italy'], :]
df_places_rate_norm = df_places_rate_norm.append(df_orig_piv_day.loc[place,])
# reverse order to keep base place on top
df_places_rate_norm = df_places_rate_norm.iloc[::-1]
sr_place = df_orig_piv_day.loc[place,]
# place_start = (sr_place > 0).idxmax()
# sr_place_compare = sr_place.loc[place_start:].dropna()
sr_place = df_orig_piv_day.loc[place,]
place_start = (sr_place > 0).idxmax()
sr_place_compare = sr_place.loc[place_start:].dropna()
df_places_gap = pd.DataFrame({'Name': [], 'gap': [], 'dist': []})
df_places_gap = df_places_gap.append(pd.Series([place, 0.0, -1], index=df_places_gap.columns),
ignore_index=True)
for other_place in df_places_rate_norm.index[1:]:
sr_other_place = df_places_rate_norm.loc[other_place,].fillna(0)
min_dist = np.inf
min_pos = 0
for i in range(0, 1 + len(sr_other_place) - len(sr_place_compare)):
sr_other_place_compare = sr_other_place[i: i + len(sr_place_compare)]
dist = euclidean(sr_place_compare, sr_other_place_compare)
if (dist < min_dist):
min_dist = dist
min_pos = i
day_place2 = sr_other_place.index[min_pos]
gap = day_place2 - place_start
df_places_gap = df_places_gap.append(
pd.Series([other_place, gap, min_dist], index=df_places_gap.columns),
ignore_index=True)
df_places_gap = df_places_gap.set_index('Name')
similar_places = df_places_gap.sort_values('dist')
dist_max = euclidean(sr_place_compare, np.zeros(len(sr_place_compare)))
similar_places['Similarity'] = similar_places['dist'].apply(lambda x: (1.0 - x / dist_max) if x >= 0 else 1)
return similar_places
# get similar places based on alighment of death curve
def get_similar_places(place, level = 'countries'):
similar_places = get_df_similar_places(place, level = level)
# print(similar_places)
tuples = [tuple(x) for x in similar_places[1:8].reset_index().to_numpy()]
return tuples
#get similar places based on socioeconomic features
def get_similar_places_socio(place, level = 'countries'):
df_socio_stats_orig = pd.read_csv(data_dir + 'socio_stats_{}.csv'.format(level)).drop('score', axis=1)
if not len(df_socio_stats_orig.query('Name == "{}"'.format(place))): return []
df_socio_stats_orig_piv = df_socio_stats_orig.pivot(index='Name', columns='variable')
df_socio_stats_orig_piv = df_socio_stats_orig_piv.fillna(df_socio_stats_orig_piv.mean())
scaler = MinMaxScaler() # feature_range=(-1, 1)
df_socio_stats_orig_piv_norm = pd.DataFrame(scaler.fit_transform(df_socio_stats_orig_piv),
columns=df_socio_stats_orig_piv.columns,
index=df_socio_stats_orig_piv.index)
df_dist = pd.DataFrame(squareform(pdist(df_socio_stats_orig_piv_norm)), index=df_socio_stats_orig_piv_norm.index,
columns=df_socio_stats_orig_piv_norm.index)
df_sim = df_dist.loc[:, place].to_frame(name='dist')
df_sim['similarity'] = 1 - (df_sim['dist'] / df_sim['dist'].max())
df_sim = df_sim.sort_values('similarity', ascending=False).drop('dist', axis=1)
tuples = [tuple(x) for x in df_sim[1:11].reset_index().to_numpy()]
return tuples
def get_places_by_variable(type = 'socio', level = 'countries', variable = 'Population', ascending = False):
if type == 'socio':
df_orig = pd.read_csv(data_dir + 'socio_stats_{}.csv'.format(level)).drop('score', axis=1)
else:
df_orig = pd.read_csv(data_dir + 'live_stats_{}.csv'.format(level))
# df_orig = df_orig.groupby(['Name', 'Date']).tail(1)
df_orig = df_orig[df_orig['variable'] == variable].pivot(index='Name', columns='variable', values='value').reset_index()
df_orig = df_orig[['Name', variable]].sort_values(variable, ascending = ascending).head(10)
tuples = [tuple(x) for x in df_orig.reset_index(drop=True).to_numpy()]
return tuples
def get_fig_compare_rates(place, place2, level = 'countries', scale='log', y='total', mode='static', priority = 'now'):
df_places_to_show = get_place_comparison_df(place, place2, level = level, priority = priority)
fig = make_chart_comparison(df_places_to_show, level = level, scale=scale, y=y, mode=mode)
return fig
def get_html_compare_response(place, place2, level = 'countries', scale='log', y='total', mode='static', priority = 'now'):
# df_places_to_show = get_place_comparison_df(place, place2, level = level, priority = priority, type = 'response')
data_dir = 'data/'
df_orig =
|
pd.read_csv(data_dir + 'response/official_response_countries.csv', parse_dates=['Date'])
|
pandas.read_csv
|
#!/usr/bin/env python
# coding: utf-8
# # Introduction
#
# 1. [EDA (Exploratory Data Analysis)](#1)
# 1. [Line Plot](#2)
# 1. [Histogram](#3)
# 1. [Scatter Plot](#4)
# 1. [Bar Plot](#5)
# 1. [Point Plot](#6)
# 1. [Count Plot](#7)
# 1. [Pie Chart](#8)
# 1. [Pair Plot](#9)
#
# 1. [MACHINE LEARNING](#11)
# 1. [Logistic Regression Classification](#12)
# 1. [KNN (K-Nearest Neighbour) Classification](#13)
# 1. [Support Vector Machine( SVM) Classification](#14)
# 1. [Naive Bayes Classification](#15)
# 1. [Decision Tree Classification](#16)
# 1. [Random Forest Classification](#17)
# 1. [Confusion Matrix](#18)
#
# 1. [Conclusion](#19)
#
# In[ ]:
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# plotly library
import plotly.plotly as py
from plotly.offline import init_notebook_mode, iplot
init_notebook_mode(connected=True)
import plotly.graph_objs as go
# Input data files are available in the "../../../input/aljarah_xAPI-Edu-Data/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../../../input/aljarah_xAPI-Edu-Data"))
# Any results you write to the current directory are saved as output.
# In[ ]:
data=pd.read_csv("../../../input/aljarah_xAPI-Edu-Data/xAPI-Edu-Data.csv")
# In[ ]:
data.head()
# In[ ]:
data.info()
# In[ ]:
data.describe()
# In[ ]:
#heatmap
f,ax=plt.subplots(figsize=(8,8))
print()
print()
# <a id="1"></a>
# # EDA (Exploratory Data Analysis)
# In[ ]:
data.head()
# <a id="2"></a>
# # Line Plot
# In[ ]:
# Line Plot
data.raisedhands.plot(kind="line",color="g",label = 'raisedhands',linewidth=1,alpha = 0.5,grid = True,linestyle = ':',figsize=(10,10))
plt.xlabel('x axis') # label = name of label
plt.ylabel('y axis')
plt.legend(loc="upper right")
plt.title('Line Plot') # title = title of plot
print()
# In[ ]:
plt.subplots(figsize=(10,10))
plt.plot(data.raisedhands[:100],linestyle="-.")
plt.plot(data.VisITedResources[:100],linestyle="-")
plt.xlabel("x axis")
plt.ylabel("y axis")
plt.title("Raidehands and VisITedResources Line Plot")
plt.legend(loc="upper right")
print()
# In[ ]:
#subplots
raisedhands=data.raisedhands
VisITedResources=data.VisITedResources
plt.subplots(figsize=(10,10))
plt.subplot(2,1,1)
plt.title("raisedhands-VisITedResources subplot")
plt.plot(raisedhands[:100],color="r",label="raisedhands")
plt.legend()
plt.grid()
plt.subplot(2,1,2)
plt.plot(VisITedResources[:100],color="b",label="VisITedResources")
plt.legend()
plt.grid()
print()
# In[ ]:
# discussion and raisedhands line plot in plotly
# import graph objects as "go"
import plotly.graph_objs as go
# Creating trace1
trace1 = go.Scatter( x = np.arange(0,82), y = data.Discussion, mode = "lines", name = "discussion", marker = dict(color = 'rgba(16, 112, 2, 0.8)'), )
# Creating trace2
trace2 = go.Scatter( x =np.arange(0,82) , y = data.raisedhands, mode = "lines", name = "raisedhands", marker = dict(color = 'rgba(80, 26, 80, 0.8)'), )
df = [trace1, trace2]
layout = dict(title = 'Discussion and Raisedhands of Students', xaxis= dict(title= 'raisedhands',ticklen= 5,zeroline= False) )
fig = dict(data = df, layout = layout)
iplot(fig)
# <a id="3"></a>
# # Histogram Plot
# In[ ]:
#histogram of raisedhands
data.raisedhands.plot(kind="hist",bins=10,figsize=(10,10),color="b",grid="True")
plt.xlabel("raisedhands")
plt.legend(loc="upper right")
plt.title("raisedhands Histogram")
print()
# In[ ]:
# histogram subplot with non cumulative and cumulative
fig, axes = plt.subplots(nrows=2,ncols=1)
#data.plot(kind="hist",y="raisedhands",bins = 50,range= (0,50),normed = True,ax = axes[0])
#data.plot(kind = "hist",y = "raisedhands",bins = 50,range= (0,50),normed = True,ax = axes[1],cumulative = True)
plt.savefig('graph.png')
print()
# <a id="4"></a>
# # Scatter Plot
# In[ ]:
#raidehands vs Discussion scatter plot
plt.subplots(figsize=(10,10))
plt.scatter(data.raisedhands,data.Discussion,color="green")
plt.xlabel("raisedhands")
plt.ylabel("Discussion")
plt.grid()
plt.title("Raidehands vs Discussion Scatter Plot",color="red")
print()
# In[ ]:
#raidehands vs AnnouncementsView scatter plot
color_list1 = ['red' if i=='M' else 'blue' for i in data.gender]
plt.subplots(figsize=(10,10))
plt.scatter(data.raisedhands,data.AnnouncementsView,color=color_list1, alpha=0.8)
plt.xlabel("raisedhands")
plt.ylabel("AnnouncementsView")
plt.grid()
plt.title("Raidehands vs Announcements View Scatter Plot",color="black",fontsize=15)
print()
# # Scatter Plot in Plotly
#
# * Import graph_objs as *go*
# * Creating traces
# * x = x axis
# * y = y axis
# * mode = type of plot like marker, line or line + markers
# * name = name of the plots
# * marker = marker is used with dictionary.
# * color = color of lines. It takes RGB (red, green, blue) and opacity (alpha)
# * text = The hover text (hover is curser)
# * data = is a list that we add traces into it
# * layout = it is dictionary.
# * title = title of layout
# * x axis = it is dictionary
# * title = label of x axis
# * ticklen = length of x axis ticks
# * zeroline = showing zero line or not
# * y axis = it is dictionary and same with x axis
# * fig = it includes data and layout
# * iplot() = plots the figure(fig) that is created by data and layout
# In[ ]:
len(data.raisedhands.unique())
# In[ ]:
# raisedhands in terms of gender
# import graph objects as "go"
import plotly.graph_objs as go
# creating trace1
trace1 =go.Scatter( x = np.arange(0,82), y = data[data.gender=='M'].raisedhands, mode = "markers", name = "male", marker = dict(color = 'rgba(0, 100, 255, 0.8)'), )
# creating trace2
trace2 =go.Scatter( x = np.arange(0,82), y = data[data.gender=="F"].raisedhands, mode = "markers", name = "female", marker = dict(color = 'rgba(255, 128, 255, 0.8)'), )
df = [trace1, trace2]
layout = dict(title = 'raisedhands', xaxis= dict(title= 'index',ticklen= 5,zeroline= False), yaxis= dict(title= 'Values',ticklen= 5,zeroline= False) )
fig = dict(data = df, layout = layout)
iplot(fig)
# In[ ]:
# Discussion in terms of gender
# import graph objects as "go"
import plotly.graph_objs as go
# creating trace1
trace1 =go.Scatter( x = np.arange(0,82), y = data[data.gender=='M'].Discussion, mode = "markers", name = "male", marker = dict(color = 'rgba(0, 100, 255, 0.8)'), text= data[data.gender=="M"].gender)
# creating trace2
trace2 =go.Scatter( x = np.arange(0,82), y = data[data.gender=="F"].Discussion, mode = "markers", name = "female", marker = dict(color = 'rgba(200, 50, 150, 0.8)'), text= data[data.gender=="F"].gender)
df = [trace1, trace2]
layout = dict(title = 'Discussion', xaxis= dict(title= 'index',ticklen= 5,zeroline= False), yaxis= dict(title= 'Values',ticklen= 5,zeroline= False) )
fig = dict(data = df, layout = layout)
iplot(fig)
# In[ ]:
# Plotting Scatter Matrix
color_list = ['red' if i=='M' else 'green' for i in data.gender]
pd.plotting.scatter_matrix(data.loc[:, data.columns != 'gender'], c=color_list, figsize= [15,15], diagonal='hist', alpha=0.8, s = 200, marker = '.', edgecolor= "black")
print()
# <a id="5"></a>
# # Bar Plot
# In[ ]:
# Raisehands Average in terms of Topic
# we will create a data containing averages of the numerical values of our data.
topic_list=list(data.Topic.unique())
rh_av=[]
d_av=[]
aview_av=[]
vr_av=[]
for i in topic_list:
rh_av.append(sum(data[data["Topic"]==i].raisedhands)/len(data[data["Topic"]==i].raisedhands))
d_av.append(sum(data[data["Topic"]==i].Discussion)/len(data[data["Topic"]==i].Discussion))
aview_av.append(sum(data[data["Topic"]==i].AnnouncementsView)/len(data[data["Topic"]==i].AnnouncementsView))
vr_av.append(sum(data[data["Topic"]==i].VisITedResources)/len(data[data["Topic"]==i].VisITedResources))
data2=pd.DataFrame({"topic":topic_list,"raisedhands_avg":rh_av,"discussion_avg":d_av,"AnnouncementsView_avg":aview_av, "VisITedResources_avg":vr_av})
# we will sort data2 interms of index of raisedhands_avg in ascending order
new_index2 = (data2['raisedhands_avg'].sort_values(ascending=True)).index.values
sorted_data2 = data2.reindex(new_index2)
sorted_data2.head()
# visualization
plt.figure(figsize=(15,10))
sns.barplot(x=sorted_data2['topic'], y=sorted_data2['raisedhands_avg'])
plt.xticks(rotation= 90)
plt.xlabel('Topics')
plt.ylabel('Raisehands Average')
plt.title("Raisehands Average in terms of Topic")
# In[ ]:
# horizontal bar plot
# Raised hands, Discussion and Announcements View averages acording to topics
f,ax = plt.subplots(figsize = (9,15)) #create a figure of 9x15 .
sns.barplot(x=rh_av,y=topic_list,color='cyan',alpha = 0.5,label='Raised hands' )
sns.barplot(x=d_av,y=topic_list,color='blue',alpha = 0.7,label='Discussion')
sns.barplot(x=aview_av,y=topic_list,color='red',alpha = 0.6,label='Announcements View')
ax.legend(loc='upper right',frameon = True)
ax.set(xlabel='Average ', ylabel='Topics',title = "Average of Numerical Values of Data According to Topics ")
# # Bar Plot in Plotly
#
# * Import graph_objs as *go*
# * Creating traces
# * x = x axis
# * y = y axis
# * mode = type of plot like marker, line or line + markers
# * name = name of the plots
# * marker = marker is used with dictionary.
# * color = color of lines. It takes RGB (red, green, blue) and opacity (alpha)
# * line = It is dictionary. line between bars
# * color = line color around bars
# * text = The hover text (hover is curser)
# * data = is a list that we add traces into it
# * layout = it is dictionary.
# * barmode = bar mode of bars like grouped
# * fig = it includes data and layout
# * iplot() = plots the figure(fig) that is created by data and layout
# In[ ]:
# raisehands and discussion average acording to topic
# we will sort data2 interms of index of raisedhands_avg in descending order
new_index3 = (data2['raisedhands_avg'].sort_values(ascending=False)).index.values
sorted_data3 = data2.reindex(new_index3)
# create trace1
trace1 = go.Bar( x = sorted_data3.topic, y = sorted_data3.raisedhands_avg, name = "raisedhands average", marker = dict(color = 'rgba(255, 174, 155, 0.5)', line=dict(color='rgb(0,0,0)',width=1.5)), text = sorted_data3.topic)
# create trace2
trace2 = go.Bar( x = sorted_data3.topic, y = sorted_data3.discussion_avg, name = "discussion average", marker = dict(color = 'rgba(255, 255, 128, 0.5)', line=dict(color='rgb(0,0,0)',width=1.5)), text = sorted_data3.topic)
df= [trace1, trace2]
layout = go.Layout(barmode = "group",title= "Discussion and Raisedhands Average of Each Topic")
fig = go.Figure(data = df, layout = layout)
iplot(fig)
# In[ ]:
# raisehands and discussion average acording to PlaceofBirth
#prepare data
place_list=list(data.PlaceofBirth.unique())
rh_av=[]
d_av=[]
aview_av=[]
vr_av=[]
for i in place_list:
rh_av.append(sum(data[data["PlaceofBirth"]==i].raisedhands)/len(data[data["PlaceofBirth"]==i].raisedhands))
d_av.append(sum(data[data["PlaceofBirth"]==i].Discussion)/len(data[data["PlaceofBirth"]==i].Discussion))
aview_av.append(sum(data[data["PlaceofBirth"]==i].AnnouncementsView)/len(data[data["PlaceofBirth"]==i].AnnouncementsView))
vr_av.append(sum(data[data["PlaceofBirth"]==i].VisITedResources)/len(data[data["PlaceofBirth"]==i].VisITedResources))
data4=pd.DataFrame({"PlaceofBirth":place_list,"raisedhands_avg":rh_av,"discussion_avg":d_av,"AnnouncementsView_avg":aview_av, "VisITedResources_avg":vr_av})
new_index4=data4["raisedhands_avg"].sort_values(ascending=False).index.values
sorted_data4=data4.reindex(new_index4)
# create trace1
trace1 = go.Bar( x = sorted_data4.PlaceofBirth, y = sorted_data4.raisedhands_avg, name = "raisedhands average", marker = dict(color = 'rgba(200, 125, 200, 0.5)', line=dict(color='rgb(0,0,0)',width=1.5)), text = sorted_data4.PlaceofBirth)
# create trace2
trace2 = go.Bar( x = sorted_data4.PlaceofBirth, y = sorted_data4.discussion_avg, name = "discussion average", marker = dict(color = 'rgba(128, 255, 128, 0.5)', line=dict(color='rgb(0,0,0)',width=1.5)), text = sorted_data4.PlaceofBirth)
df= [trace1, trace2]
layout = go.Layout(barmode = "group",title= "Discussion and Raisedhands Average acording to PlaceofBirth")
fig = go.Figure(data = df, layout = layout)
iplot(fig)
# In[ ]:
trace1 = { 'x': sorted_data4.PlaceofBirth, 'y': sorted_data4.raisedhands_avg, 'name': 'raisedhands average', 'type': 'bar' };
trace2 = { 'x': sorted_data4.PlaceofBirth, 'y': sorted_data4.discussion_avg, 'name': 'discussion average', 'type': 'bar' };
df = [trace1, trace2];
layout = { 'xaxis': {'title': 'PlaceofBirth'}, 'barmode': 'relative', 'title': 'Raisedhands and Discussion Average Acording to Place of Birth' };
fig = go.Figure(data = df, layout = layout)
iplot(fig)
# <a id="6"></a>
# # Point Plot
# In[ ]:
# Raisedhands vs Discussion Rate point plot
#normalize the values of discussion_avg and raisedhands_avg
data3=sorted_data2.copy()
data3["raisedhands_avg"]=data3['raisedhands_avg']/max( data3['raisedhands_avg'])
data3["discussion_avg"]=data3['discussion_avg']/max( data3['discussion_avg'])
# visualize
f,ax1 = plt.subplots(figsize =(12,10))
sns.pointplot(x='topic',y='raisedhands_avg',data=data3,color='lime',alpha=0.8)
sns.pointplot(x='topic',y='discussion_avg',data=data3,color='red',alpha=0.8)
plt.text(5,0.50,'Raised hands Average',color='red',fontsize = 17,style = 'italic')
plt.text(5,0.46,'Discussion Average',color='lime',fontsize = 18,style = 'italic')
plt.xlabel('Topics',fontsize = 15,color='blue')
plt.ylabel('Values',fontsize = 15,color='blue')
plt.title('Raisedhands vs Discussion Rate',fontsize = 20,color='blue')
plt.grid()
# In[ ]:
# Raisedhands vs Discussion Rate point plot acording to place of birth
#normalize the values of discussion_avg and raisedhands_avg
data5=sorted_data4.copy()
data5["raisedhands_avg"]=data5['raisedhands_avg']/max( data5['raisedhands_avg'])
data5["discussion_avg"]=data5['discussion_avg']/max( data5['discussion_avg'])
# visualize
f,ax1 = plt.subplots(figsize =(12,10))
sns.pointplot(x='PlaceofBirth',y='raisedhands_avg',data=data5,color='red',alpha=0.8)
sns.pointplot(x='PlaceofBirth',y='discussion_avg',data=data5,color='blue',alpha=0.8)
plt.text(3,0.30,'Raised hands Average',color='red',fontsize = 17,style = 'italic')
plt.text(3,0.36,'Discussion Average',color='blue',fontsize = 18,style = 'italic')
plt.xlabel('PlaceofBirth',fontsize = 15,color='purple')
plt.ylabel('Values',fontsize = 15,color='purple')
plt.title('Raisedhands vs Discussion Rate',fontsize = 20,color='purple')
plt.grid()
# <a id="7"></a>
# # Count Plot
# In[ ]:
data.gender.value_counts()
# In[ ]:
plt.subplots(figsize=(8,5))
sns.countplot(data.gender)
plt.xlabel("gender",fontsize="15")
plt.ylabel("numbers",fontsize="15")
plt.title("Number of Genders in Data", color="red",fontsize="18")
print()
# In[ ]:
#StageID unique values
data.StageID.value_counts()
# In[ ]:
sns.countplot(data.StageID)
plt.xlabel("StageID")
plt.ylabel("numbers")
plt.title("Number of StageID in Data", color="red",fontsize="18")
print()
# <a id="8"></a>
# # Pie Chart
#
# * fig: create figures
# * data: plot type
# * values: values of plot
# * labels: labels of plot
# * name: name of plots
# * hoverinfo: information in hover
# * hole: hole width
# * type: plot type like pie
# * layout: layout of plot
# * title: title of layout
# * annotations: font, showarrow, text, x, y
# In[ ]:
labels=data.StageID.value_counts()
colors=["grey","blue","green"]
explode=[0,0,0]
sizes=data.StageID.value_counts().values
plt.figure(figsize = (7,7))
plt.pie(sizes, explode=explode, labels=labels, colors=colors, autopct='%1.1f%%')
plt.title("StageID in Data",color = 'blue',fontsize = 15)
print()
# In[ ]:
# StageID piechart in plotly
pie1_list=data["StageID"].value_counts().values
labels = data["StageID"].value_counts().index
# figure
fig = { "data": [ { "values": pie1_list, "labels": labels, "domain": {"x": [0, .5]}, "name": "StageID", "hoverinfo":"label+percent", "hole": .3, "type": "pie" },], "layout": { "title":"StageID Type", "annotations": [ { "font": { "size": 20}, "showarrow": False, "text": "StageID", "x": 0.20, "y": 1 }, ] } }
iplot(fig)
# <a id="9"></a>
# # Pair Plot
# In[ ]:
data.head()
# In[ ]:
# raisedhands and VisITedResources pair plot
print()
print()
# <a id="11"></a>
# # MACHINE LEARNING
#
# <a href="https://ibb.co/hgB9j10"><img src="https://i.ibb.co/YNcQMTg/ml1.jpg" alt="ml1" border="0">
# In this part, we will use Machine Learning algoithms in our data. Machine Learning Classification algorithms have the following steps:
# * Split data
# * Fit data
# * Predict Data
# * Find Accuracy
# In[ ]:
data.head()
# We need only numerical features. So create new data containing only numerical values.
# In[ ]:
data_new=data.loc[:,["gender","raisedhands","VisITedResources","AnnouncementsView","Discussion"]]
# We will write 1 for male and 0 for female for classification.
# In[ ]:
data_new.gender=[1 if i=="M" else 0 for i in data_new.gender]
# In[ ]:
data_new.head()
# <a id="12"></a>
# # Logistic Regression Classification
#
# * When we talk about binary classification( 0 and 1 outputs) what comes to mind first is logistic regression.
# * Logistic regression is actually a very simple neural network.
# We need to prepare our data for classificaiton.
# * We will determine x and y values.
# * y: binary output (0 and 1)
# * x_data: rest of the data (i.e. features of data except gender)
# In[ ]:
y=data_new.gender.values
x_data=data_new.drop("gender",axis=1)
# In[ ]:
# normalize the values in x_data
x=(x_data-np.min(x_data))/(np.max(x_data)-np.min(x_data))
# * create x_train, y_train, x_test and y_test arrays with train_test_split method.
# In[ ]:
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(x,y,test_size=0.2,random_state=52)
# In[ ]:
#Logistic Regression
from sklearn.linear_model import LogisticRegression
#fit
lr=LogisticRegression()
lr.fit(x_train,y_train)
#accuracy
print("test accuracy is {}".format(lr.score(x_test,y_test)))
# <a id="13"></a>
# # KNN (K-Nearest Neighbour) Classification
# 1. Choose K value.
# 1. Find the K nearest data points.
# 1. Find the number of data points for each class between K nearest neighbour.
# 1. Identify the class of data or point we tested.
# Assume that we have a graph and we want to determine the class of black points(i.e. they are in class green or red. )
# <a href="https://ibb.co/NmzLJF6"><img src="https://i.ibb.co/MGLRtgD/2.png" alt="2" border="0">
#
#
#
#
#
# * First split data
# * Fit data
# * Predict Data
# * Find Accuracy
# * Find the convenient k value for the highest accuracy.
# In[ ]:
#split data
from sklearn.neighbors import KNeighborsClassifier
x_train,x_test,y_train,y_test=train_test_split(x,y,test_size=0.2,random_state=1)
knn=KNeighborsClassifier(n_neighbors=3)
#fit
knn.fit(x_train,y_train)
#prediction
prediction=knn.predict(x_test)
# In[ ]:
#prediction score (accuracy)
print('KNN (K=3) accuracy is: ',knn.score(x_test,y_test))
# In[ ]:
# find the convenient k value for range (1,31)
score_list=[]
train_accuracy=[]
for i in range(1,31):
knn2=KNeighborsClassifier(n_neighbors=i)
knn2.fit(x_train,y_train)
score_list.append(knn2.score(x_test,y_test))
train_accuracy.append(knn2.score(x_train,y_train))
plt.figure(figsize=(15,10))
plt.plot(range(1,31),score_list,label="testing accuracy",color="blue",linewidth=3)
plt.plot(range(1,31),train_accuracy,label="training accuracy",color="orange",linewidth=3)
plt.xlabel("k values in KNN")
plt.ylabel("accuracy")
plt.title("Accuracy results with respect to k values")
plt.legend()
plt.grid()
print()
print("Maximum value of testing accuracy is {} when k= {}.".format(np.max(score_list),1+score_list.index(np.max(score_list))))
# <a id="14"></a>
# # Support Vector Machine (SVM) Classification
# In[ ]:
from sklearn.svm import SVC
svm=SVC(random_state=1)
svm.fit(x_train,y_train)
#accuracy
print("accuracy of svm algorithm: ",svm.score(x_test,y_test))
# <a id="15"></a>
# # Naive Bayes Classification
# In[ ]:
from sklearn.naive_bayes import GaussianNB
nb=GaussianNB()
nb.fit(x_train,y_train)
# test accuracy
print("Accuracy of naive bayees algorithm: ",nb.score(x_test,y_test))
# <a id="16"></a>
# # Decision Tree Classification
#
# We have points as seen in the figure and we want to classify these points.
#
# <a href="https://ibb.co/n8CYzwN"><img src="https://i.ibb.co/G3T8cd4/d11-640x538.jpg" alt="d11-640x538" border="0"></a>
#
#
# We will classify these points by using 3 splits.
#
# <a href="https://ibb.co/y4n4rRk"><img src="https://i.ibb.co/FHbHFWY/d22-690x569.jpg" alt="d22-690x569" border="0"></a>
# In[ ]:
from sklearn.tree import DecisionTreeClassifier
dt=DecisionTreeClassifier()
dt.fit(x_train,y_train)
print("Accuracy score for Decision Tree Classification: " ,dt.score(x_test,y_test))
# <a id="17"></a>
# # Random Forest Classification
#
# Random Forest divides the train data into n samples, and for each n sample applies Decision Tree algorithm. At he end of n Decision Tree algorithm, it takes the answer which is more.
# In[ ]:
from sklearn.ensemble import RandomForestClassifier
rf=RandomForestClassifier(n_estimators=100,random_state=1)
rf.fit(x_train,y_train)
print("random forest algorithm accuracy: ",rf.score(x_test,y_test))
# In[ ]:
score_list1=[]
for i in range(100,501,100):
rf2=RandomForestClassifier(n_estimators=i,random_state=1)
rf2.fit(x_train,y_train)
score_list1.append(rf2.score(x_test,y_test))
plt.figure(figsize=(10,10))
plt.plot(range(100,501,100),score_list1)
plt.xlabel("number of estimators")
plt.ylabel("accuracy")
plt.grid()
print()
print("Maximum value of accuracy is {} \nwhen n_estimators= {}.".format(max(score_list1),(1+score_list1.index(max(score_list1)))*100))
# As it seen in the graph, it is convenient to choose n_estimators=100 for the best accuracy result.
# Let's look at between 100 and 130.
# In[ ]:
score_list2=[]
for i in range(100,131):
rf3=RandomForestClassifier(n_estimators=i,random_state=1)
rf3.fit(x_train,y_train)
score_list2.append(rf3.score(x_test,y_test))
plt.figure(figsize=(10,10))
plt.plot(range(100,131),score_list2)
plt.xlabel("number of estimators")
plt.ylabel("accuracy")
plt.grid()
print()
print("Maximum value of accuracy is {} when number of estimators between 100 and 131 ".format(max(score_list2)))
# Actually, this graph says that, if n_estimators is between 100 and 122, then the value of accuracy is not changing.
# <a id="18"></a>
# # Confusion Matrix
# Confusion matrix gives the number of true and false predicitons in our classificaiton. It is more reliable than accuracy.
# <br> Here,
# * y_pred: results that we predict.
# * y_test: our real values.
# In[ ]:
#Confusion matrix of Random Forest Classf.
y_pred=rf.predict(x_test)
y_true=y_test
#cm
from sklearn.metrics import confusion_matrix
cm=confusion_matrix(y_true,y_pred)
#cm visualization
f,ax=plt.subplots(figsize=(8,8))
print()
plt.xlabel("predicted value")
plt.ylabel("real value")
print()
# We know that our accuracy is 0.7291, which is the best result, when number of estimators is 100 . But here we predicted
# * 21 true for label 0=TN
# * 49 true for label 1=TP
# * 6 wrong for the label 1 =FN ( I predicted 6 label 0 but they are label 1)
# * 20 wrong for label 0= FP ( I predicted 20 label 1 but they are label 0)
#
# In[ ]:
#Confusion matrix of KNN Classf.
y_pred1=knn.predict(x_test)
y_true=y_test
#cm
cm1=confusion_matrix(y_true,y_pred1)
#cm visualization
f,ax=plt.subplots(figsize=(8,8))
print()
plt.xlabel("predicted value")
plt.ylabel("real value")
print()
# We know that our accuracy is 0.645 when k=3 . We predicted;
# * 15 true for label 0=TN
# * 47 true for label 1=TP
# * 8 wrong for the label 1 =FN ( I predicted 8 label 0 but they are label 1)
# * 26 wrong for label 0= FP ( I predicted 26 label 1 but they are label 0)
# In[ ]:
#Confusion matrix of Decision Tree Classf.
y_pred2=dt.predict(x_test)
y_true=y_test
#cm
cm2=confusion_matrix(y_true,y_pred2)
#cm visualization
f,ax=plt.subplots(figsize=(8,8))
print()
plt.xlabel("predicted value")
plt.ylabel("real value")
print()
# <a id="19"></a>
# # Conclusion
#
# As it seen from confusion matrices, the number of wrong predictions are:
# * KNN Classif: 8, 26
# * Decision Tree Classif: 16, 16
# * Random Forest Classif: 6, 20
#
# It seems that Random Forest Classification is more effective which can also be seen from accuracy scores. Now lets check this by visualizing the scores.
# In[ ]:
dictionary={"model":["LR","KNN","SVM","NB","DT","RF"],"score":[lr.score(x_test,y_test),knn.score(x_test,y_test),svm.score(x_test,y_test),nb.score(x_test,y_test),dt.score(x_test,y_test),rf.score(x_test,y_test)]}
df1=
|
pd.DataFrame(dictionary)
|
pandas.DataFrame
|
import numpy as np
import pandas as pd
import os
import sys
from subprocess import call
import matplotlib
matplotlib.use('TkAgg')
import matplotlib.pyplot as plt
from matplotlib.ticker import LinearLocator
import scipy
import json
from sklearn.decomposition import PCA as skPCA
from scipy.spatial import distance
from scipy.cluster import hierarchy
import seaborn as sns
from matplotlib.colors import rgb2hex, colorConverter
from pprint import pprint
import difflib
from operator import itemgetter
import itertools
from functools import reduce
import matplotlib.ticker as ticker
import math
import matplotlib.patches as patches
from collections import defaultdict
import collections
from sklearn.manifold import TSNE
from sklearn.decomposition import TruncatedSVD
from sklearn.cluster import KMeans
import matplotlib.mlab as mlab
def make_new_matrix_gene(org_matrix_by_gene, gene_list_source, exclude_list=""):
if isinstance(gene_list_source,str):
gene_df = pd.read_csv(open(gene_list_source,'rU'), sep=None, engine='python')
try:
gene_list = list(set(gene_df['GeneID'].tolist()))
if exclude_list != "":
gene_list = [g for g in gene_list if g not in exclude_list]
except KeyError:
sys.exit("Error: Please provide Gene list file with 'GeneID' as header.")
try:
group_list = gene_df['GroupID'].tolist()
except KeyError:
sys.exit("Error: Please provide Gene list file with 'GroupID' as header.")
try:
gmatrix_df = org_matrix_by_gene[gene_list]
except KeyError as error_gene:
cause1 = error_gene.args[0].strip(' not in index')
cause = [v.strip('\n\' ') for v in cause1.strip('[]').split(' ')]
absent_gene = cause
print(' '.join(absent_gene)+' not in matrix file.')
new_list = [x for x in gene_list if x not in absent_gene]
gmatrix_df = org_matrix_by_gene[new_list]
cmatrix_df = gmatrix_df.transpose()
cell_list1 = cmatrix_df.columns.values
new_cmatrix_df = cmatrix_df[cell_list1]
new_gmatrix_df = new_cmatrix_df.transpose()
return new_cmatrix_df, new_gmatrix_df
elif isinstance(gene_list_source,list):
if exclude_list == "":
gene_list = gene_list_source
else:
gene_list = [g for g in gene_list_source if g not in exclude_list]
try:
gmatrix_df = org_matrix_by_gene[gene_list]
except KeyError as error_gene:
cause = error_gene.args[0]
absent_gene = cause.split('\'')[1]
print(absent_gene+' not in matrix file.')
new_list = [x for x in gene_list if x not in [absent_gene]]
gmatrix_df = org_matrix_by_gene[new_list]
cmatrix_df = gmatrix_df.transpose()
cell_list1 = cmatrix_df.columns.values
new_cmatrix_df = cmatrix_df[cell_list1]
new_gmatrix_df = new_cmatrix_df.transpose()
return new_cmatrix_df, new_gmatrix_df
else:
sys.exit("Error: gene list must be filepath or a list.")
def make_new_matrix_cell(org_matrix_by_cell, cell_list_file):
cell_df = pd.read_csv(open(cell_list_file,'rU'), sep=None, engine='python')
cell_list_new = list(set([cell.strip('\n') for cell in cell_df['SampleID'].tolist()]))
cell_list_old = org_matrix_by_cell.columns.tolist()
overlap = [c for c in cell_list_new if c in cell_list_old]
not_in_matrix = [c for c in cell_list_new if c not in cell_list_old]
if not_in_matrix != []:
print('These cells were in the cell list provided, but not found in the matrix provided:')
print(not_in_matrix)
new_cmatrix_df = org_matrix_by_cell[overlap]
new_gmatrix_df = new_cmatrix_df.transpose()
return new_cmatrix_df, new_gmatrix_df
def threshold_genes(by_gene, number_expressed=1, gene_express_cutoff=1.0):
by_gene.apply(lambda column: (column >= 1).sum())
return
def find_top_common_genes(log2_df_by_cell, num_common=25):
top_common_list = []
count = 0
done = False
log2_df_by_gene = log2_df_by_cell.transpose()
log2_df2_gene = log2_df_by_gene.apply(pd.to_numeric,errors='coerce')
log_mean = log2_df2_gene.mean(axis=0).sort_values(ascending=False)
try:
log2_sorted_gene = log2_df_by_gene.reindex_axis(log2_df_by_gene.mean(axis=0).sort_values(ascending=False).index, axis=1)
except ValueError:
overlap_list = [item for item, count in collections.Counter(log2_df_by_cell.index).items() if count > 1]
print(overlap_list, len(overlap_list))
sys.exit('Error: Duplicate GeneIDs are present.')
for gene in log2_sorted_gene.columns.tolist():
if sum(genes < 1 for genes in log2_df_by_gene[gene])<6:
if count < num_common:
count+=1
top_common_list.append(gene)
if count == num_common:
done = True
break
if done:
return log2_df_by_gene[top_common_list].transpose()
else:
return [0]
def log2_oulierfilter(df_by_cell, plot=False, already_log2=False):
if not already_log2:
log2_df = np.log2(df_by_cell+1)
else:
log2_df = df_by_cell
top_log2 = find_top_common_genes(log2_df)
if all(top_log2) != 0:
log2_df2= log2_df.apply(pd.to_numeric,errors='coerce')
log_mean = top_log2.mean(axis=0).sort_values(ascending=False)
log2_sorted = top_log2.reindex_axis(top_log2.mean(axis=0).sort_values(ascending=False).index, axis=1)
xticks = []
keep_col= []
log2_cutoff = np.average(np.average(log2_sorted))-2*np.average(np.std(log2_sorted))
for col, m in zip(log2_sorted.columns.tolist(),log2_sorted.mean()):
if m > log2_cutoff:
keep_col.append(col)
xticks.append(col+' '+str("%.2f" % m))
excluded_cells = [x for x in log2_sorted.columns.tolist() if x not in keep_col]
filtered_df_by_cell = df_by_cell[keep_col]
filtered_df_by_gene = filtered_df_by_cell.transpose()
if not already_log2:
filtered_log2 = np.log2(filtered_df_by_cell[filtered_df_by_cell>0])
else:
filtered_log2 = filtered_df_by_cell[filtered_df_by_cell>0]
if plot:
ax = sns.boxplot(data=filtered_log2, whis= .75, notch=True)
ax = sns.stripplot(x=filtered_log2.columns.values, y=filtered_log2.mean(axis=0), size=4, jitter=True, edgecolor="gray")
xtickNames = plt.setp(ax, xticklabels=xticks)
plt.setp(xtickNames, rotation=90, fontsize=9)
plt.show()
plt.clf()
sns.distplot(filtered_log2.mean())
plt.show()
if not already_log2:
log2_expdf_cell = np.log2(filtered_df_by_cell+1)
else:
log2_expdf_cell = filtered_df_by_cell
log2_expdf_gene = log2_expdf_cell.transpose()
return log2_expdf_cell, log2_expdf_gene
else:
print("no common genes found")
return log2_df, log2_df.transpose()
def augmented_dendrogram(*args, **kwargs):
plt.clf()
ddata = dendrogram(*args, **kwargs)
if not kwargs.get('no_plot', False):
for i, d in zip(ddata['icoord'], ddata['dcoord'], ):
x = 0.5 * sum(i[1:3])
y = d[1]
if y >= 200000:
plt.plot(x, y, 'ro')
plt.annotate("%.3g" % y, (x, y), xytext=(0, -8),
textcoords='offset points',
va='top', ha='center')
plt.show()
plt.savefig(os.path.join(new_file,'augmented_dendrogram.png'))
def cluster_indices(cluster_assignments):
n = cluster_assignments.max()
indices = []
for cluster_number in range(1, n + 1):
indices.append(np.where(cluster_assignments == cluster_number)[0])
return indices
def clust_members(r_link, cutoff):
clust = fcluster(r_link,cutoff)
num_clusters = clust.max()
indices = cluster_indices(clust)
return num_clusters, indices
def print_clust_membs(indices, cell_list):
for k, ind in enumerate(indices):
print("cluster", k + 1, "is", [cell_list[x] for x in ind])
def plot_tree(dendr, path_filename, pos=None, save=False):
icoord = scipy.array(dendr['icoord'])
dcoord = scipy.array(dendr['dcoord'])
color_list = scipy.array(dendr['color_list'])
xmin, xmax = icoord.min(), icoord.max()
ymin, ymax = dcoord.min(), dcoord.max()
if pos:
icoord = icoord[pos]
dcoord = dcoord[pos]
for xs, ys, color in zip(icoord, dcoord, color_list):
plt.plot(xs, ys, color)
plt.xlim(xmin-10, xmax + 0.1*abs(xmax))
plt.ylim(ymin, ymax + 0.1*abs(ymax))
if save:
plt.savefig(os.path.join(path_filename,'plot_dendrogram.png'))
plt.show()
# Create a nested dictionary from the ClusterNode's returned by SciPy
def add_node(node, parent):
# First create the new node and append it to its parent's children
newNode = dict( node_id=node.id, children=[] )
parent["children"].append( newNode )
# Recursively add the current node's children
if node.left: add_node( node.left, newNode )
if node.right: add_node( node.right, newNode )
cc = []
# Label each node with the names of each leaf in its subtree
def label_tree(n, id2name):
# If the node is a leaf, then we have its name
if len(n["children"]) == 0:
leafNames = [ id2name[n["node_id"]] ]
# If not, flatten all the leaves in the node's subtree
else:
leafNames = reduce(lambda ls, c: ls + label_tree(c,id2name), n["children"], [])
cc.append((len(leafNames), [x.strip('\n') for x in leafNames]))
cc.sort(key=lambda tup: tup[0], reverse = True)
# Delete the node id since we don't need it anymore and
# it makes for cleaner JSON
del n["node_id"]
# Labeling convention: "-"-separated leaf names
n["name"] = name = "-".join(sorted(map(str, leafNames)))
return leafNames
#Makes labeled json tree for visulaization in d3, makes and returns cc object within label_tree
def make_tree_json(row_clusters, df_by_gene, path_filename):
T= hierarchy.to_tree(row_clusters)
# Create dictionary for labeling nodes by their IDs
labels = list(df_by_gene.index)
id2name = dict(zip(range(len(labels)), labels))
# Initialize nested dictionary for d3, then recursively iterate through tree
d3Dendro = dict(children=[], name="Root1")
add_node( T, d3Dendro )
label_tree( d3Dendro["children"][0], id2name )
# Output to JSON
json.dump(d3Dendro, open(os.path.join(path_filename,"d3-dendrogram.json"), "w"), sort_keys=True, indent=4)
return cc
#finds significant genes between subclusters
def find_twobytwo(cc, df_by_cell, full_by_cell_df, path_filename, cluster_size=20):
gene_list = full_by_cell_df.index.tolist()
by_gene_df = full_by_cell_df.transpose()
pair_dict = {}
parent = cc[0][1]
p_num = cc[0][0]
l_nums = [x[0] for x in cc]
c_lists = [c[1] for c in cc[1:]]
unique_count = 1
pair_list = []
for i, c in enumerate(c_lists):
for i2, c2 in enumerate(c_lists):
overlap = [i for i in c if i in c2]
if not overlap and len(c)>=cluster_size and len(c2)>=cluster_size:
if (c,c2) not in pair_list:
pair_list.append((c,c2))
pair_list.append((c2,c))
pair_dict[str(len(c))+'cells_vs_'+str(len(c2))+'cells'+str(unique_count)]= [c, c2]
unique_count+=1
for v, k in pair_dict.items():
g_pvalue_dict = {}
index_list = []
sig_gene_list = []
cell_list1 = [x.strip('\n') for x in k[0]]
cell_list2 = [xx.strip('\n') for xx in k[1]]
group1 = str(len(cell_list1))
group2 = str(len(cell_list2))
df_by_cell_1 = full_by_cell_df[cell_list1]
df_by_cell_2 = full_by_cell_df[cell_list2]
df_by_gene_1 = df_by_cell_1.transpose()
df_by_gene_2 = df_by_cell_2.transpose()
for g in gene_list:
g_pvalue = scipy.stats.f_oneway(df_by_gene_1[g], df_by_gene_2[g])
if g_pvalue[0] > 0 and g_pvalue[1] <= 1:
g_pvalue_dict[g] = g_pvalue
if g not in [s[0] for s in sig_gene_list]:
sig_gene_list.append([g, g_pvalue[1]])
sig_gene_list.sort(key=lambda tup: tup[1])
pvalues = [p[1] for p in sig_gene_list]
gene_index = [ge[0] for ge in sig_gene_list]
mean_log2_exp_list = []
sig_1_2_list = []
mean1_list = []
mean2_list = []
for sig_gene in gene_index:
sig_gene_df = by_gene_df[sig_gene]
mean_log2_exp_list.append(sig_gene_df.mean())
sig_cell_df = sig_gene_df.transpose()
mean_cell1 = sig_cell_df[cell_list1].mean()
mean1_list.append(mean_cell1)
mean_cell2 = sig_cell_df[cell_list2].mean()
mean2_list.append(mean_cell2)
ratio_1_2 = (mean_cell1+1)/(mean_cell2+1)
sig_1_2_list.append(ratio_1_2)
sig_df = pd.DataFrame({'pvalues':pvalues,'mean_all':mean_log2_exp_list,'mean_group1':mean1_list, 'mean_group2':mean2_list, 'ratio_1_2':sig_1_2_list}, index=gene_index)
cell_names_df = pd.DataFrame({'cells1':pd.Series(cell_list1, index=range(len(cell_list1))), 'cells2':pd.Series(cell_list2, index=range(len(cell_list2)))})
sig_df.to_csv(os.path.join(path_filename,'sig_'+v+'_pvalues.txt'), sep = '\t')
cell_names_df.to_csv(os.path.join(path_filename,'sig_'+v+'_cells.txt'), sep = '\t')
def ellip_enclose(points, color, inc=1, lw=2, nst=2):
"""
Plot the minimum ellipse around a set of points.
Based on:
https://github.com/joferkington/oost_paper_code/blob/master/error_ellipse.py
"""
def eigsorted(cov):
vals, vecs = np.linalg.eigh(cov)
order = vals.argsort()[::-1]
return vals[order], vecs[:,order]
x = points[:,0]
y = points[:,1]
cov = np.cov(x, y)
vals, vecs = eigsorted(cov)
theta = np.degrees(np.arctan2(*vecs[:,0][::-1]))
w, h = 2 * nst * np.sqrt(vals)
center = np.mean(points, 0)
ell = patches.Ellipse(center, width=inc*w, height=inc*h, angle=theta,
facecolor=color, alpha=0.2, lw=0)
edge = patches.Ellipse(center, width=inc*w, height=inc*h, angle=theta,
facecolor='none', edgecolor=color, lw=lw)
return ell, edge
def return_top_pca_gene(df_by_gene, num_genes=100):
gene_pca = skPCA(n_components=3)
np_by_gene = np.asarray(df_by_gene)
by_gene_trans = gene_pca.fit_transform(np_by_gene)
Pc_df = pd.DataFrame(gene_pca.components_.T, columns=['PC-1', 'PC-2', 'PC-3'], index=df_by_gene.columns.tolist())
pca_rank_df = Pc_df.abs().sum(axis=1)
Pc_sort_df = pca_rank_df.nlargest(len(df_by_gene.columns.tolist()))
top_pca_list = Pc_sort_df.index.tolist()
new_gene_matrix = df_by_gene[top_pca_list[0:num_genes]]
return new_gene_matrix, top_pca_list[0:num_genes]
def plot_PCA(args, df_by_gene, path_filename, num_genes=100, gene_list_filter=False, title='', plot=False, label_map=False, gene_map = False):
gene_list = df_by_gene.columns.tolist()
sns.set_palette("RdBu_r", 10, 1)
if gene_list_filter:
sig_by_gene = df_by_gene[gene_list_filter]
sig_by_cell = sig_by_gene.transpose()
else:
sig_by_gene = df_by_gene
sig_by_cell = sig_by_gene.transpose()
gene_pca = skPCA(n_components=3)
np_by_gene = np.asarray(sig_by_gene)
by_gene_trans = gene_pca.fit_transform(np_by_gene)
Pc_df = pd.DataFrame(gene_pca.components_.T, columns=['PC-1', 'PC-2', 'PC-3'], index=sig_by_gene.columns.tolist())
pca_rank_df = Pc_df.abs().sum(axis=1)
Pc_sort_df = pca_rank_df.nlargest(len(sig_by_gene.columns.tolist()))
top_pca_list = Pc_sort_df.index.tolist()
top_by_gene = df_by_gene[top_pca_list[0:num_genes]]
gene_top = skPCA(n_components=2)
cell_pca = skPCA(n_components=2)
top_by_cell = top_by_gene.transpose()
np_top_gene = np.asarray(top_by_cell)
np_top_cell = np.asarray(top_by_gene)
top_cell_trans = cell_pca.fit_transform(np_top_cell)
top_gene_trans = gene_top.fit_transform(np_top_gene)
if not np.isnan(top_cell_trans).any():
fig, (ax_cell, ax_gene) = plt.subplots(2, 1, figsize=(15, 30), sharex=False)
rect_cell = ax_cell.patch
rect_gene = ax_gene.patch
rect_cell.set_facecolor('white')
rect_gene.set_facecolor('white')
ax_cell.grid(b=True, which='major', color='grey', linestyle='--', linewidth=0.3)
ax_gene.grid(b=True, which='major', color='grey', linestyle='--', linewidth=0.3)
if label_map:
annotate = args.annotate_cell_pca
X = [x for x in top_cell_trans[:, 0]]
Y = [y for y in top_cell_trans[:, 1]]
labels = [label_map[cell][2] for cell in top_by_cell.columns.tolist()]
markers = [label_map[cell][1] for cell in top_by_cell.columns.tolist()]
colors = [label_map[cell][0] for cell in top_by_cell.columns.tolist()]
label_done = []
xy_by_color_dict = {}
for c in set(colors):
xy_by_color_dict[c] = []
for X_pos, Y_pos, m, color, l in zip(X, Y, markers, colors, labels):
if l in label_done:
lab = ''
else:
lab= l
label_done.append(l)
xy_by_color_dict[color].append([X_pos,Y_pos])
ax_cell.scatter(X_pos, Y_pos, marker=m, c=color, label=lab, s=30)
if args.add_ellipse:
for c in set(colors):
ell, edge = ellip_enclose(np.asarray(xy_by_color_dict[c]), c)
ax_cell.add_artist(ell)
ax_cell.add_artist(edge)
else:
ax_cell.scatter(top_cell_trans[:, 0], top_cell_trans[:, 1], alpha=0.75)
annotate = args.annotate_cell_pca
ax_cell.set_xlim([min(top_cell_trans[:, 0])-1, max(top_cell_trans[:, 0]+1)])
ax_cell.set_ylim([min(top_cell_trans[:, 1])-1, max(top_cell_trans[:, 1]+2)])
ax_cell.set_title(title+'_cell')
if label_map:
handles, labs = ax_cell.get_legend_handles_labels()
# sort both labels and handles by labels
labs, handles = zip(*sorted(zip(labs, handles), key=lambda t: t[0]))
ax_cell.legend(handles, labs, loc='best', ncol=1, prop={'size':12}, markerscale=1.5, frameon=True)
ax_cell.set_xlabel('PC1')
ax_cell.set_ylabel('PC2')
if annotate:
for label, x, y in zip(top_by_cell.columns, top_cell_trans[:, 0], top_cell_trans[:, 1]):
ax_cell.annotate(label, (x+0.1, y+0.1))
if gene_map:
X = [x for x in top_gene_trans[:, 0]]
Y = [y for y in top_gene_trans[:, 1]]
labels = top_by_gene.columns.tolist()
markers = [gene_map[gene][1] for gene in top_by_gene.columns.tolist()]
colors = [gene_map[gene][0] for gene in top_by_gene.columns.tolist()]
xy_by_color_dict = {}
for c in set(colors):
xy_by_color_dict[c] = []
for X_pos, Y_pos, m, color, l in zip(X, Y, markers, colors, labels):
xy_by_color_dict[color].append([X_pos,Y_pos])
ax_gene.scatter(X_pos, Y_pos, marker=m, c=color, label = l, s=30)
if args.add_ellipse:
for c in set(colors):
ell, edge = ellip_enclose(np.asarray(xy_by_color_dict[c]), c)
ax_gene.add_artist(ell)
ax_gene.add_artist(edge)
else:
ax_gene.scatter(top_gene_trans[:, 0], top_gene_trans[:, 1], alpha=0.75)
ax_gene.set_xlim([min(top_gene_trans[:, 0])-1, max(top_gene_trans[:, 0])+1])
ax_gene.set_ylim([min(top_gene_trans[:, 1])-1, max(top_gene_trans[:, 1])+2])
ax_gene.set_title(title+'_gene')
ax_gene.set_xlabel('PC1')
ax_gene.set_ylabel('PC2')
if args.annotate_gene_subset:
plot_subset_path = os.path.join(os.path.dirname(args.filepath),args.annotate_gene_subset)
genes_plot = pd.read_csv(plot_subset_path, sep='\t', index_col=False)
for label, x, y in zip(top_by_gene.columns, top_gene_trans[:, 0], top_gene_trans[:, 1]):
if label in genes_plot['GeneID'].tolist():
if '_' in label:
label = label.split('_')[0]
ax_gene.annotate(label, (x+0.1, y+0.1))
else:
for label, x, y in zip(top_by_gene.columns, top_gene_trans[:, 0], top_gene_trans[:, 1]):
if '_' in label:
label = label.split('_')[0]
ax_gene.annotate(label, (x+0.1, y+0.1))
if plot:
plt.show()
if title != '':
save_name = '_'.join(title.split(' ')[0:2])
plt.savefig(os.path.join(path_filename,save_name+'_skpca.pdf'), bbox_inches='tight')
else:
plt.savefig(os.path.join(path_filename,'Group0_skpca.pdf'), bbox_inches='tight')
plt.close('all')
return top_pca_list
else:
return []
def plot_SVD(args,df_by_gene, path_filename, num_genes=100, gene_list_filter=False, title='', plot=False, label_map=False, gene_map = False):
gene_list = df_by_gene.columns.tolist()
sns.set_palette("RdBu_r", 10, 1)
if gene_list_filter:
sig_by_gene = df_by_gene[gene_list_filter]
sig_by_cell = sig_by_gene.transpose()
else:
sig_by_gene = df_by_gene
sig_by_cell = sig_by_gene.transpose()
gene_pca = TruncatedSVD(n_components=3)
np_by_gene = np.asarray(sig_by_gene)
by_gene_trans = gene_pca.fit_transform(np_by_gene)
Pc_df = pd.DataFrame(gene_pca.components_.T, columns=['PC-1', 'PC-2', 'PC-3'], index=sig_by_gene.columns.tolist())
pca_rank_df = Pc_df.abs().sum(axis=1)
Pc_sort_df = pca_rank_df.nlargest(len(sig_by_gene.columns.tolist()))
top_pca_list = Pc_sort_df.index.tolist()
top_by_gene = df_by_gene[top_pca_list[0:num_genes]]
gene_top = TruncatedSVD(n_components=2)
cell_pca = TruncatedSVD(n_components=2)
top_by_cell = top_by_gene.transpose()
np_top_gene = np.asarray(top_by_cell)
np_top_cell = np.asarray(top_by_gene)
top_cell_trans = cell_pca.fit_transform(np_top_cell)
top_gene_trans = gene_top.fit_transform(np_top_gene)
if not np.isnan(top_cell_trans).any():
fig, (ax_cell, ax_gene) = plt.subplots(2, 1, figsize=(15, 30), sharex=False)
rect_cell = ax_cell.patch
rect_gene = ax_gene.patch
rect_cell.set_facecolor('white')
rect_gene.set_facecolor('white')
ax_cell.grid(b=True, which='major', color='grey', linestyle='--', linewidth=0.3)
ax_gene.grid(b=True, which='major', color='grey', linestyle='--', linewidth=0.3)
if label_map:
annotate = args.annotate_cell_pca
X = [x for x in top_cell_trans[:, 0]]
Y = [y for y in top_cell_trans[:, 1]]
labels = [label_map[cell][2] for cell in top_by_cell.columns.tolist()]
markers = [label_map[cell][1] for cell in top_by_cell.columns.tolist()]
colors = [label_map[cell][0] for cell in top_by_cell.columns.tolist()]
label_done = []
xy_by_color_dict = {}
for c in set(colors):
xy_by_color_dict[c] = []
for X_pos, Y_pos, m, color, l in zip(X, Y, markers, colors, labels):
if l in label_done:
lab = ''
else:
lab= l
label_done.append(l)
xy_by_color_dict[color].append([X_pos,Y_pos])
ax_cell.scatter(X_pos, Y_pos, marker=m, c=color, label=lab, s=30)
if args.add_ellipse:
for c in set(colors):
ell, edge = ellip_enclose(np.asarray(xy_by_color_dict[c]), c)
ax_cell.add_artist(ell)
ax_cell.add_artist(edge)
else:
ax_cell.scatter(top_cell_trans[:, 0], top_cell_trans[:, 1], alpha=0.75)
annotate = args.annotate_cell_pca
ax_cell.set_xlim([min(top_cell_trans[:, 0])-1, max(top_cell_trans[:, 0]+1)])
ax_cell.set_ylim([min(top_cell_trans[:, 1])-1, max(top_cell_trans[:, 1]+2)])
ax_cell.set_title(title+'_cell')
if label_map:
handles, labs = ax_cell.get_legend_handles_labels()
# sort both labels and handles by labels
labs, handles = zip(*sorted(zip(labs, handles), key=lambda t: t[0]))
ax_cell.legend(handles, labs, loc='best', ncol=1, prop={'size':12}, markerscale=1.5, frameon=True)
ax_cell.set_xlabel('PC1')
ax_cell.set_ylabel('PC2')
if annotate:
for label, x, y in zip(top_by_cell.columns, top_cell_trans[:, 0], top_cell_trans[:, 1]):
ax_cell.annotate(label, (x+0.1, y+0.1))
if gene_map:
X = [x for x in top_gene_trans[:, 0]]
Y = [y for y in top_gene_trans[:, 1]]
labels = top_by_gene.columns.tolist()
markers = [gene_map[gene][1] for gene in top_by_gene.columns.tolist()]
colors = [gene_map[gene][0] for gene in top_by_gene.columns.tolist()]
xy_by_color_dict = {}
for c in set(colors):
xy_by_color_dict[c] = []
for X_pos, Y_pos, m, color, l in zip(X, Y, markers, colors, labels):
xy_by_color_dict[color].append([X_pos,Y_pos])
ax_gene.scatter(X_pos, Y_pos, marker=m, c=color, label = l, s=30)
if args.add_ellipse:
for c in set(colors):
ell, edge = ellip_enclose(np.asarray(xy_by_color_dict[c]), c)
ax_gene.add_artist(ell)
ax_gene.add_artist(edge)
else:
ax_gene.scatter(top_gene_trans[:, 0], top_gene_trans[:, 1], alpha=0.75)
ax_gene.set_xlim([min(top_gene_trans[:, 0])-1, max(top_gene_trans[:, 0])+1])
ax_gene.set_ylim([min(top_gene_trans[:, 1])-1, max(top_gene_trans[:, 1])+2])
ax_gene.set_title(title+'_gene')
ax_gene.set_xlabel('PC1')
ax_gene.set_ylabel('PC2')
if args.annotate_gene_subset:
plot_subset_path = os.path.join(os.path.dirname(args.filepath),args.annotate_gene_subset)
genes_plot = pd.read_csv(plot_subset_path, sep='\t', index_col=False)
for label, x, y in zip(top_by_gene.columns, top_gene_trans[:, 0], top_gene_trans[:, 1]):
if label in genes_plot['GeneID'].tolist():
if '_' in label:
label = label.split('_')[0]
ax_gene.annotate(label, (x+0.1, y+0.1))
else:
for label, x, y in zip(top_by_gene.columns, top_gene_trans[:, 0], top_gene_trans[:, 1]):
if '_' in label:
label = label.split('_')[0]
ax_gene.annotate(label, (x+0.1, y+0.1))
if plot:
plt.show()
if title != '':
save_name = '_'.join(title.split(' ')[0:2])
plt.savefig(os.path.join(path_filename,save_name+'_TruncatedSVD.pdf'), bbox_inches='tight')
#plot_url = py.plot_mpl(fig)
else:
#plot_url = py.plot_mpl(fig)
plt.savefig(os.path.join(path_filename,'Group0_TruncatedSVD.pdf'), bbox_inches='tight')
plt.close('all')
return top_pca_list
else:
return []
#create cell and gene TSNE scatter plots (one pdf)
def plot_TSNE(args,df_by_gene, path_filename, num_genes=100, gene_list_filter=False, title='', plot=False, label_map=False, gene_map = False):
gene_list = df_by_gene.columns.tolist()
sns.set_palette("RdBu_r", 10, 1)
if gene_list_filter:
sig_by_gene = df_by_gene[gene_list_filter]
sig_by_cell = sig_by_gene.transpose()
else:
sig_by_gene = df_by_gene
sig_by_cell = sig_by_gene.transpose()
gene_pca = TruncatedSVD(n_components=3)
np_by_gene = np.asarray(sig_by_gene)
by_gene_trans = gene_pca.fit_transform(np_by_gene)
Pc_df = pd.DataFrame(gene_pca.components_.T, columns=['PC-1', 'PC-2', 'PC-3'], index=sig_by_gene.columns.tolist())
pca_rank_df = Pc_df.abs().sum(axis=1)
Pc_sort_df = pca_rank_df.nlargest(len(sig_by_gene.columns.tolist()))
top_pca_list = Pc_sort_df.index.tolist()
top_by_gene = df_by_gene[top_pca_list[0:num_genes]]
gene_top = TSNE(n_components=2, init='pca', random_state=0)
cell_pca = TSNE(n_components=2, init='pca', random_state=0)
top_by_cell = top_by_gene.transpose()
np_top_gene = np.asarray(top_by_cell)
np_top_cell = np.asarray(top_by_gene)
top_cell_trans = cell_pca.fit_transform(np_top_cell)
top_gene_trans = gene_top.fit_transform(np_top_gene)
if not np.isnan(top_cell_trans).any():
fig, (ax_cell, ax_gene) = plt.subplots(2, 1, figsize=(15, 30), sharex=False)
rect_cell = ax_cell.patch
rect_gene = ax_gene.patch
rect_cell.set_facecolor('white')
rect_gene.set_facecolor('white')
ax_cell.grid(b=True, which='major', color='grey', linestyle='--', linewidth=0.3)
ax_gene.grid(b=True, which='major', color='grey', linestyle='--', linewidth=0.3)
if label_map:
annotate = args.annotate_cell_pca
X = [x for x in top_cell_trans[:, 0]]
Y = [y for y in top_cell_trans[:, 1]]
labels = [label_map[cell][2] for cell in top_by_cell.columns.tolist()]
markers = [label_map[cell][1] for cell in top_by_cell.columns.tolist()]
colors = [label_map[cell][0] for cell in top_by_cell.columns.tolist()]
label_done = []
xy_by_color_dict = {}
for c in set(colors):
xy_by_color_dict[c] = []
for X_pos, Y_pos, m, color, l in zip(X, Y, markers, colors, labels):
if l in label_done:
lab = ''
else:
lab= l
label_done.append(l)
xy_by_color_dict[color].append([X_pos,Y_pos])
ax_cell.scatter(X_pos, Y_pos, marker=m, c=color, label=lab, s=30)
if args.add_ellipse:
for c in set(colors):
ell, edge = ellip_enclose(np.asarray(xy_by_color_dict[c]), c)
ax_cell.add_artist(ell)
ax_cell.add_artist(edge)
else:
ax_cell.scatter(top_cell_trans[:, 0], top_cell_trans[:, 1], alpha=0.75)
annotate = args.annotate_cell_pca
ax_cell.set_xlim([min(top_cell_trans[:, 0])-1, max(top_cell_trans[:, 0]+1)])
ax_cell.set_ylim([min(top_cell_trans[:, 1])-1, max(top_cell_trans[:, 1]+2)])
ax_cell.set_title(title+'_cell')
if label_map:
handles, labs = ax_cell.get_legend_handles_labels()
# sort both labels and handles by labels
labs, handles = zip(*sorted(zip(labs, handles), key=lambda t: t[0]))
ax_cell.legend(handles, labs, loc='best', ncol=1, prop={'size':12}, markerscale=1.5, frameon=True)
ax_cell.set_xlabel('PC1')
ax_cell.set_ylabel('PC2')
if annotate:
for label, x, y in zip(top_by_cell.columns, top_cell_trans[:, 0], top_cell_trans[:, 1]):
ax_cell.annotate(label, (x+0.1, y+0.1))
if gene_map:
X = [x for x in top_gene_trans[:, 0]]
Y = [y for y in top_gene_trans[:, 1]]
labels = top_by_gene.columns.tolist()
markers = [gene_map[gene][1] for gene in top_by_gene.columns.tolist()]
colors = [gene_map[gene][0] for gene in top_by_gene.columns.tolist()]
xy_by_color_dict = {}
for c in set(colors):
xy_by_color_dict[c] = []
for X_pos, Y_pos, m, color, l in zip(X, Y, markers, colors, labels):
xy_by_color_dict[color].append([X_pos,Y_pos])
ax_gene.scatter(X_pos, Y_pos, marker=m, c=color, label = l, s=30)
if args.add_ellipse:
for c in set(colors):
ell, edge = ellip_enclose(np.asarray(xy_by_color_dict[c]), c)
ax_gene.add_artist(ell)
ax_gene.add_artist(edge)
else:
ax_gene.scatter(top_gene_trans[:, 0], top_gene_trans[:, 1], alpha=0.75)
ax_gene.set_xlim([min(top_gene_trans[:, 0])-1, max(top_gene_trans[:, 0])+1])
ax_gene.set_ylim([min(top_gene_trans[:, 1])-1, max(top_gene_trans[:, 1])+2])
ax_gene.set_title(title+'_gene')
ax_gene.set_xlabel('PC1')
ax_gene.set_ylabel('PC2')
if args.annotate_gene_subset:
plot_subset_path = os.path.join(os.path.dirname(args.filepath),args.annotate_gene_subset)
genes_plot = pd.read_csv(plot_subset_path, sep='\t', index_col=False)
for label, x, y in zip(top_by_gene.columns, top_gene_trans[:, 0], top_gene_trans[:, 1]):
if label in genes_plot['GeneID'].tolist():
if '_' in label:
label = label.split('_')[0]
ax_gene.annotate(label, (x+0.1, y+0.1))
else:
for label, x, y in zip(top_by_gene.columns, top_gene_trans[:, 0], top_gene_trans[:, 1]):
if '_' in label:
label = label.split('_')[0]
ax_gene.annotate(label, (x+0.1, y+0.1))
if plot:
plt.show()
if title != '':
save_name = '_'.join(title.split(' ')[0:2])
plt.savefig(os.path.join(path_filename,save_name+'_TSNE.pdf'), bbox_inches='tight')
else:
plt.savefig(os.path.join(path_filename,'Group0_TSNE.pdf'), bbox_inches='tight')
plt.close('all')
return top_pca_list
else:
return []
#create cell and gene TSNE scatter plots (one pdf)
def plot_kmeans(args, df_by_gene, path_filename, kmeans_range, num_genes=100, gene_list_filter=False, title='', plot=False, label_map=False, gene_map = False, run_sig_test=False):
from sklearn.metrics import silhouette_samples, silhouette_score
import matplotlib.cm as cm
gene_list = df_by_gene.columns.tolist()
sns.set_palette("RdBu_r", 10, 1)
if gene_list_filter:
sig_by_gene = df_by_gene[gene_list_filter]
sig_by_cell = sig_by_gene.transpose()
else:
sig_by_gene = df_by_gene
sig_by_cell = sig_by_gene.transpose()
gene_pca = TruncatedSVD(n_components=3)
np_by_gene = np.asarray(sig_by_gene)
by_gene_trans = gene_pca.fit_transform(np_by_gene)
Pc_df = pd.DataFrame(gene_pca.components_.T, columns=['PC-1', 'PC-2', 'PC-3'], index=sig_by_gene.columns.tolist())
pca_rank_df = Pc_df.abs().sum(axis=1)
Pc_sort_df = pca_rank_df.nlargest(len(sig_by_gene.columns.tolist()))
top_pca_list = Pc_sort_df.index.tolist()
top_by_gene = df_by_gene[top_pca_list[0:num_genes]]
gene_top = TruncatedSVD(n_components=2)
cell_pca = TruncatedSVD(n_components=2)
top_by_cell = top_by_gene.transpose()
np_top_gene = np.asarray(top_by_cell)
np_top_cell = np.asarray(top_by_gene)
top_cell_trans = cell_pca.fit_transform(np_top_cell)
top_gene_trans = gene_top.fit_transform(np_top_gene)
range_n_clusters = range(kmeans_range[0],kmeans_range[1])
for n_clusters in range_n_clusters:
# Create a subplot with 1 row and 2 columns
fig, (ax1, ax2) = plt.subplots(1, 2)
fig.set_size_inches(18, 7)
ax1.set_xlim([-0.1, 1])
# The (n_clusters+1)*10 is for inserting blank space between silhouette
# plots of individual clusters, to demarcate them clearly.
ax1.set_ylim([0, len(np_top_cell) + (n_clusters + 1) * 10])
#cluster cell PCA
cell_clusterer = KMeans(n_clusters=n_clusters)
top_cell_pred = cell_clusterer.fit_predict(top_cell_trans)
#cluster gene PCA
gene_clusterer = KMeans(n_clusters=n_clusters)
top_gene_pred = gene_clusterer.fit_predict(top_gene_trans)
pred_dict = {'SampleID':top_by_cell.columns, 'GroupID':['kmeans_'+str(p) for p in top_cell_pred]}
df_pred = pd.DataFrame(pred_dict)
cell_group_path = os.path.join(path_filename,'kmeans_cell_groups_'+str(n_clusters)+'.txt')
df_pred.to_csv(cell_group_path, sep = '\t')
#compute silouette averages and values
silhouette_avg_cell = silhouette_score(top_cell_trans, top_cell_pred)
print("For n_clusters =", n_clusters,
"The average silhouette_score is :", silhouette_avg_cell)
silhouette_avg_gene = silhouette_score(top_gene_trans, top_gene_pred)
sample_silhouette_values_cell = silhouette_samples(top_cell_trans, top_cell_pred)
sample_silhouette_values_gene = silhouette_samples(top_gene_trans, top_gene_pred)
y_lower = 10
for i in range(n_clusters):
# Aggregate the silhouette scores for samples belonging to
# cluster i, and sort them
ith_cluster_silhouette_values = \
sample_silhouette_values_cell[top_cell_pred == i]
ith_cluster_silhouette_values.sort()
size_cluster_i = ith_cluster_silhouette_values.shape[0]
y_upper = y_lower + size_cluster_i
color = cm.spectral(float(i) / n_clusters)
ax1.fill_betweenx(np.arange(y_lower, y_upper),
0, ith_cluster_silhouette_values,
facecolor=color, edgecolor=color, alpha=0.7)
# Label the silhouette plots with their cluster numbers at the middle
ax1.text(-0.05, y_lower + 0.5 * size_cluster_i, str(i))
# Compute the new y_lower for next plot
y_lower = y_upper + 10 # 10 for the 0 samples
ax1.set_title("The silhouette plot for the various clusters.")
ax1.set_xlabel("The silhouette coefficient values")
ax1.set_ylabel("Cluster label")
# The vertical line for average silhoutte score of all the values
ax1.axvline(x=silhouette_avg_cell, color="red", linestyle="--")
ax1.set_yticks([]) # Clear the yaxis labels / ticks
ax1.set_xticks([-0.1, 0, 0.2, 0.4, 0.6, 0.8, 1])
# 2nd Plot showing the actual clusters formed
colors = cm.spectral(top_cell_pred.astype(float) / n_clusters)
ax2.scatter(top_cell_trans[:, 0], top_cell_trans[:, 1], marker='.', s=30, lw=0, alpha=0.7,
c=colors)
# Labeling the clusters
centers = cell_clusterer.cluster_centers_
# Draw white circles at cluster centers
ax2.scatter(centers[:, 0], centers[:, 1],
marker='o', c="white", alpha=1, s=200)
for i, c in enumerate(centers):
ax2.scatter(c[0], c[1], marker='$%d$' % i, alpha=1, s=50)
ax2.set_title("The visualization of the clustered data.")
ax2.set_xlabel("Feature space for the 1st feature")
ax2.set_ylabel("Feature space for the 2nd feature")
plt.suptitle(("Silhouette analysis for KMeans clustering on sample data "
"with n_clusters = %d" % n_clusters),
fontsize=14, fontweight='bold')
plt.savefig(os.path.join(path_filename,'Group0_kmeans_'+str(n_clusters)+'_clusters.pdf'), bbox_inches='tight')
plt.close('all')
#use colors to make label map compatable with heatmap
color_dict ={}
markers = ['o', 'v','D','*','x','h', 's','p','8','^','>','<', 'd','o', 'v','D','*','x','h', 's','p','8','^','>','<', 'd']
color_dict =dict(zip(top_by_cell.columns, zip(colors,[markers[pred] for pred in top_cell_pred],['kmeans_'+str(p) for p in top_cell_pred])))
group_color_dict = dict(zip(['kmeans_'+str(p) for p in top_cell_pred],zip(colors,[markers[pred] for pred in top_cell_pred])))
#run heatmap with kmeans clustering and colors
top_pca_by_gene, top_pca = return_top_pca_gene(df_by_gene, num_genes=args.gene_number)
top_pca_by_cell = top_pca_by_gene.transpose()
cell_linkage, plotted_df_by_gene, col_order = clust_heatmap(args, top_pca, top_pca_by_gene, path_filename, num_to_plot=args.gene_number, title= 'kmeans_label_with_'+str(n_clusters)+'_clusters',label_map=color_dict)
if run_sig_test:
multi_group_sig(args, df_by_gene.transpose(), cell_group_path, path_filename, group_color_dict, from_kmeans=str(n_clusters))
def clust_heatmap(args, gene_list, df_by_gene, path_filename, num_to_plot, title='', plot=False, label_map=False, gene_map=False, fontsize=18):
cell_list = df_by_gene.index.tolist()
cell_num =len(cell_list)
longest_side = max(num_to_plot,cell_num*2)
if longest_side == num_to_plot:
sns.set(context= 'poster', font_scale = .4*(num_to_plot/100))
width_heatmap = min(28+round(cell_num/50),42+round(cell_num/40))
len_heatmap = min(43+round(num_to_plot/10),58+round(num_to_plot/30))
title_set = 1.15
else:
sns.set(context= 'poster', font_scale = .6*(cell_num/120))
width_heatmap = min(42+round(cell_num/9),68+round(cell_num/40))
len_heatmap = min(47+round(num_to_plot/8),50+round(num_to_plot/30))
title_set = 1.12
font = {'size' : fontsize}
plt.rc('font', **font)
if len(str(args.z_direction)) > 1:
z_choice = str(args.z_direction)
if z_choice != 'None':
sys.exit('Please enter a valid option (0, 1, or None) for z_direction')
else:
z_choice = int(args.z_direction)
if z_choice != 0 and z_choice != 1:
sys.exit('Please enter a valid option (0, 1, or None) for z_direction')
cmap = sns.diverging_palette(255, 10, s=99, sep=1, as_cmap=True)
cluster_df = df_by_gene[gene_list[0:num_to_plot]].transpose()
cluster_df[abs(cluster_df)<3e-12] = 0.0
try:
cg = sns.clustermap(cluster_df, method=args.method, metric=args.metric, z_score=z_choice, figsize=(width_heatmap, len_heatmap), cmap =cmap)
col_order = cg.dendrogram_col.reordered_ind
row_order = cg.dendrogram_row.reordered_ind
if label_map and gene_map:
Xlabs = [cell_list[i] for i in col_order]
Xcolors = [label_map[cell][0] for cell in Xlabs]
col_colors = pd.DataFrame({'Cell Groups': Xcolors},index=Xlabs)
Xgroup_labels = [label_map[cell][2] for cell in Xlabs]
Ylabs = [gene_list[i] for i in row_order]
Ycolors = [gene_map[gene][0] for gene in Ylabs]
Ygroup_labels= [gene_map[gene][2] for gene in Ylabs]
row_colors = pd.DataFrame({'Gene Groups': Ycolors},index=Ylabs)
cg = sns.clustermap(cluster_df, method=args.method, metric=args.metric, z_score=z_choice,row_colors=row_colors, col_colors=col_colors, figsize=(width_heatmap, len_heatmap), cmap =cmap)
elif label_map:
Xlabs = [cell_list[i] for i in col_order]
Xcolors = [label_map[cell][0] for cell in Xlabs]
Xgroup_labels = [label_map[cell][2] for cell in Xlabs]
col_colors = pd.DataFrame({'Cell Groups': Xcolors},index=Xlabs)
cg = sns.clustermap(cluster_df, method=args.method, metric=args.metric, z_score=z_choice, col_colors=col_colors, figsize=(width_heatmap, len_heatmap), cmap =cmap)
elif gene_map:
Ylabs = [gene_list[i] for i in row_order]
Ycolors = [gene_map[gene][0] for gene in Ylabs]
Ygroup_labels= [gene_map[gene][2] for gene in Ylabs]
row_colors = pd.DataFrame({'Gene Groups': Ycolors},index=Ylabs)
cg = sns.clustermap(cluster_df, method=args.method, metric=args.metric, z_score=z_choice,row_colors=row_colors, figsize=(width_heatmap, len_heatmap), cmap =cmap)
cg.ax_heatmap.set_title(title, y=title_set)
cg.cax.set_title('Z-score')
if label_map:
leg_handles_cell =[]
group_seen_cell = []
for xtick, xcolor, xgroup_name in zip(cg.ax_heatmap.get_xticklabels(), Xcolors, Xgroup_labels):
xtick.set_color(xcolor)
xtick.set_rotation(270)
xtick.set_fontsize(fontsize)
if xgroup_name not in group_seen_cell:
leg_handles_cell.append(patches.Patch(color=xcolor, label=xgroup_name))
group_seen_cell.append(xgroup_name)
else:
for xtick in cg.ax_heatmap.get_xticklabels():
xtick.set_rotation(270)
xtick.set_fontsize(fontsize)
if gene_map:
leg_handles_gene =[]
group_seen_gene = []
for ytick, ycolor, ygroup_name in zip(cg.ax_heatmap.get_yticklabels(), list(reversed(Ycolors)), list(reversed(Ygroup_labels))):
ytick.set_color(ycolor)
ytick.set_rotation(0)
ytick.set_fontsize(fontsize)
if ygroup_name not in group_seen_gene:
leg_handles_gene.append(patches.Patch(color=ycolor, label=ygroup_name))
group_seen_gene.append(ygroup_name)
else:
for ytick in cg.ax_heatmap.get_yticklabels():
ytick.set_rotation(0)
ytick.set_fontsize(fontsize)
if gene_map and label_map:
gene_legend = cg.ax_heatmap.legend(handles=leg_handles_gene, loc=2, bbox_to_anchor=(1.04, 0.8), title='Gene groups', prop={'size':fontsize})
plt.setp(gene_legend.get_title(),fontsize=fontsize)
cg.ax_heatmap.add_artist(gene_legend)
cell_legend = cg.ax_heatmap.legend(handles=leg_handles_cell, loc=2, bbox_to_anchor=(1.04, 1), title='Cell groups', prop={'size':fontsize})
plt.setp(cell_legend.get_title(),fontsize=fontsize)
#cg.ax_heatmap.add_artist(cell_legend)
elif label_map:
cell_legend = cg.ax_heatmap.legend(handles=leg_handles_cell, loc=2, bbox_to_anchor=(1.04, 1), title='Cell groups', prop={'size':fontsize})
plt.setp(cell_legend.get_title(),fontsize=fontsize)
elif gene_map:
gene_legend = cg.ax_heatmap.legend(handles=leg_handles_gene, loc=2, bbox_to_anchor=(1.04, 0.8), title='Gene groups', prop={'size':fontsize})
plt.setp(gene_legend.get_title(),fontsize=fontsize)
if plot:
plt.show()
cell_linkage = cg.dendrogram_col.linkage
link_mat = pd.DataFrame(cell_linkage,
columns=['row label 1', 'row label 2', 'distance', 'no. of items in clust.'],
index=['cluster %d' %(i+1) for i in range(cell_linkage.shape[0])])
if title != '':
save_name = '_'.join(title.split(' ')[0:2])
#plot_url = py.plot_mpl(cg)
cg.savefig(os.path.join(path_filename, save_name+'_heatmap.pdf'), bbox_inches='tight')
else:
#plot_url = py.plot_mpl(cg)
cg.savefig(os.path.join(path_filename,'Group0_Heatmap_all_cells.pdf'), bbox_inches='tight')
plt.close('all')
return cell_linkage, df_by_gene[gene_list[0:num_to_plot]], col_order
except FloatingPointError:
print('Linkage distance has too many zeros. Filter to remove non-expressed genes in order to produce heatmap. Heatmap with '+ str(len(cell_list))+' will not be created.')
return False, False, False
def make_subclusters(args, cc, log2_expdf_cell, log2_expdf_cell_full, path_filename, base_name, gene_corr_list, label_map=False, gene_map=False, cluster_size=20, group_colors=False):
'''
Walks a histogram branch map 'cc' and does PCA (SVD), heatmap and correlation search for each non-overlapping
tree branch. Stops at defined cluster_size (default is 20).
'''
#initial cell group is parent
parent = cc[0][1]
#p_num is the number of cells in the parent group
p_num = cc[0][0]
#l_nums is number of members of each leaf of tree
l_nums = [x[0] for x in cc]
#cell list is the list of list of cells in each leaf of tree
c_lists = [c[1] for c in cc]
#Group ID will increment with each group so that each subcluter has a unique ID
group_ID = 0
for num_members, cell_list in zip(l_nums, c_lists):
#run all cell groups that are subgroups of the parent and greater than or equal to the cutoff cluster_size
if num_members < p_num and num_members >= cluster_size:
group_ID+=1
#save name for all files generated within this cluster i.e. 'Group_2_with_105_cells_heatmap.pdf'
current_title = 'Group_'+str(group_ID)+'_with_'+str(num_members)+'_cells'
cell_subset = log2_expdf_cell[list(set(cell_list))]
gene_subset = cell_subset.transpose()
gene_subset = gene_subset.loc[:,(gene_subset!=0).any()]
full_cell_subset = log2_expdf_cell_full[list(set(cell_list))]
full_gene_subset = full_cell_subset.transpose()
full_gene_subset = full_gene_subset.loc[:,(full_gene_subset!=0).any()]
norm_df_cell1 = np.exp2(full_cell_subset)
norm_df_cell = norm_df_cell1 -1
norm_df_cell.to_csv(os.path.join(path_filename, base_name+'_'+current_title+'_matrix.txt'), sep = '\t', index_col=0)
if gene_map:
top_pca_by_gene, top_pca = return_top_pca_gene(gene_subset, num_genes=args.gene_number)
plot_SVD(args,gene_subset, path_filename, num_genes=len(gene_subset.columns.tolist()), title=current_title, plot=False, label_map=label_map, gene_map=gene_map)
else:
top_pca_by_gene, top_pca = return_top_pca_gene(full_gene_subset, num_genes=args.gene_number)
plot_SVD(args,full_gene_subset, path_filename, num_genes=int(args.gene_number), title=current_title, plot=False, label_map=label_map)
if len(top_pca)<args.gene_number:
plot_num = len(top_pca)
else:
plot_num = args.gene_number
if top_pca != []:
top_pca_by_cell = top_pca_by_gene.transpose()
#if no_corr flag is provided (False) no correlation plots will be made
if args.no_corr:
if gene_corr_list != []:
top_genes_search = top_pca[0:50]
corr_plot(top_genes_search, full_gene_subset, path_filename, num_to_plot=3, gene_corr_list= gene_corr_list, title = current_title, label_map=label_map)
else:
top_genes_search = top_pca[0:50]
corr_plot(top_genes_search, full_gene_subset, path_filename, num_to_plot=3, title = current_title, label_map=label_map)
if gene_map:
cell_linkage, plotted_df_by_gene, col_order = clust_heatmap(args, top_pca, top_pca_by_gene, path_filename, num_to_plot=plot_num, title=current_title, plot=False, label_map=label_map, gene_map = gene_map)
else:
cell_linkage, plotted_df_by_gene, col_order = clust_heatmap(args, top_pca, top_pca_by_gene, path_filename,num_to_plot=plot_num, title=current_title, plot=False, label_map=label_map)
plt.close('all')
else:
print('Search for top genes by PCA failed in '+current_title+'. No plots will be generated for this subcluster. ')
pass
def clust_stability(args, log2_expdf_gene, path_filename, iterations, label_map=False):
sns.set(context='poster', font_scale = 1)
sns.set_palette("RdBu_r")
stability_ratio = []
total_genes = len(log2_expdf_gene.columns.tolist())
end_num = 1000
iter_list = range(100,int(round(end_num)),int(round(end_num/iterations)))
for gene_number in iter_list:
title= str(gene_number)+' genes plot.'
top_pca_by_gene, top_pca = return_top_pca_gene(df_by_gene, num_genes=gene_number)
top_pca_by_cell = top_pca_by_gene.transpose()
cell_linkage, plotted_df_by_gene, col_order = clust_heatmap(args, top_pca, top_pca_by_gene, num_to_plot=gene_number, title=title, label_map=label_map)
if gene_number == 100:
s1 = col_order
s0 = col_order
else:
s2= col_order
sm_running = difflib.SequenceMatcher(None,s1,s2)
sm_first = difflib.SequenceMatcher(None,s0,s2)
stability_ratio.append((sm_running.ratio(), sm_first.ratio()))
s1=col_order
plt.close()
x= iter_list[1:]
f, (ax1, ax2) = plt.subplots(2, 1, figsize=(10, 8), sharex=True)
y1= [m[0] for m in stability_ratio]
y2= [m[1] for m in stability_ratio]
sns.barplot(x, y1, palette="RdBu_r", ax=ax1)
ax1.set_ylabel('Running ratio (new/last)')
sns.barplot(x, y2, palette="RdBu_r", ax=ax2)
ax2.set_ylabel('Ratio to 100')
plt.savefig(os.path.join(path_filename,'clustering_stability.pdf'), bbox_inches='tight')
plt.show()
plt.close('all')
return stability_ratio
#run correlation matrix and save only those above threshold
def run_corr(df_by_gene, title, path_filename, method_name='pearson', sig_threshold= 0.5, run_new=True, min_period=3, save_corrs=False):
if run_new:
if len(df_by_gene.columns.tolist())>5000:
df_by_gene, top_pca_list = return_top_pca_gene(df_by_gene, num_genes=5000)
if method_name != 'kendall':
corr_by_gene = df_by_gene.corr(method=method_name, min_periods=min_period)
else:
corr_by_gene = df_by_gene.corr(method=method_name)
df_by_cell = df_by_gene.transpose()
corr_by_cell = df_by_cell.corr()
cor = corr_by_gene
cor.loc[:,:] = np.tril(cor.values, k=-1)
cor = cor.stack()
corr_by_gene_pos = cor[cor >=sig_threshold]
corr_by_gene_neg = cor[cor <=(sig_threshold*-1)]
else:
corr_by_g_pos = open(os.path.join(path_filename,'gene_correlations_sig_pos_'+method_name+'.p'), 'rb')
corr_by_g_neg = open(os.path.join(path_filename,'gene_correlations_sig_neg_'+method_name+'.p'), 'rb')
corr_by_gene_pos = pickle.load(corr_by_g_pos)
corr_by_gene_neg = pickle.load(corr_by_g_neg)
if save_corrs:
with open(os.path.join(path_filename,'gene_correlations_sig_neg_'+method_name+'.p'), 'wb') as fp:
pickle.dump(corr_by_gene_neg, fp)
with open(os.path.join(path_filename,'gene_correlations_sig_pos_'+method_name+'.p'), 'wb') as fp0:
pickle.dump(corr_by_gene_pos, fp0)
with open(os.path.join(path_filename,'by_gene_corr.p'), 'wb') as fp1:
pickle.dump(corr_by_gene, fp1)
with open(os.path.join(path_filename,'by_cell_corr.p'), 'wb') as fp2:
pickle.dump(corr_by_cell, fp2)
cor_pos_df = pd.DataFrame(corr_by_gene_pos)
cor_neg_df = pd.DataFrame(corr_by_gene_neg)
sig_corr = cor_pos_df.append(cor_neg_df)
sig_corrs = pd.DataFrame(sig_corr[0], columns=["corr"])
if run_new:
sig_corrs.to_csv(os.path.join(path_filename, title+'_counts_corr_sig_'+method_name+'.txt'), sep = '\t')
return sig_corrs
#finds most correlated gene groups that are not overlapping
def find_top_corrs(terms_to_search, sig_corrs, num_to_return, gene_corr_list = []):
all_corrs_list = []
best_corrs_list = []
for term_to_search in terms_to_search:
corr_tup = [(term_to_search, 1)]
for index, row in sig_corrs.iterrows():
if term_to_search in index:
if index[0]==term_to_search:
corr_tup.append((index[1],row['corr']))
else:
corr_tup.append((index[0],row['corr']))
all_corrs_list.append(corr_tup)
all_corrs_list.sort(key=len, reverse=True)
good_count = 0
corr_genes_seen = []
while good_count <= num_to_return:
for i, corrs in enumerate(all_corrs_list):
if corrs[0][0] not in corr_genes_seen:
best_corrs_list.append(corrs)
good_count+=1
for g, c in corrs:
if g not in corr_genes_seen and '-' not in str(c):
corr_genes_seen.append(g)
if gene_corr_list != []:
search_corrs = []
for term in gene_corr_list:
corr_tup = [(term, 1)]
for index, row in sig_corrs.iterrows():
if term in index:
if index[0]==term:
corr_tup.append((index[1],row['corr']))
else:
corr_tup.append((index[0],row['corr']))
search_corrs.append(corr_tup)
best_corrs_list = search_corrs+best_corrs_list
return best_corrs_list[0:num_to_return+len(gene_corr_list)+1]
else:
return best_corrs_list[0:num_to_return]
#corr_plot finds and plots all correlated genes, log turns on log scale, sort plots the genes in the rank order of the gene searched
def corr_plot(terms_to_search, df_by_gene_corr, path_filename, title, num_to_plot, gene_corr_list = [], label_map=False, log=False, sort=True, sig_threshold=0.5):
size_cells = len(df_by_gene_corr.index.tolist())
figlen=int(size_cells/12)
if figlen < 15:
figlen = 15
ncol = int(figlen/3.2)
if size_cells <100:
sig_threshold = -0.137*math.log(size_cells)+1.1322
sig_corrs = run_corr(df_by_gene_corr, title, path_filename, sig_threshold=sig_threshold)
corr_list = find_top_corrs(terms_to_search, sig_corrs, num_to_plot, gene_corr_list=gene_corr_list)
for corr_tup in corr_list:
term_to_search = corr_tup[0][0]
corr_tup.sort(key=itemgetter(1), reverse=True)
corr_df =
|
pd.DataFrame(corr_tup, columns=['GeneID', 'Correlation'])
|
pandas.DataFrame
|
# coding: utf-8
# # Logistic Regression
# Logistic regression is a statistical method for predicting binary classes. The outcome or target variable is dichotomous in nature. Dichotomous means there are only two possible classes. For example, it can be used for cancer detection problems. It computes the probability of an event occurrence.
#
# It is a special case of linear regression where the target variable is categorical in nature. It uses a log of odds as the dependent variable. Logistic Regression predicts the probability of occurrence of a binary event utilizing a logit function.
#
# Linear Regression Equation:
# 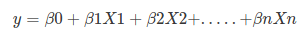
# Sigmoid Function
# 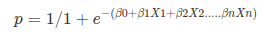
# The last equation is called the Logistic Equation which is responsible for the calculation of Logistic Regression
# ### Linear Regression Vs. Logistic Regression
# Linear regression gives you a continuous output, but logistic regression provides a constant output. An example of the continuous output is house price and stock price. Example's of the discrete output is predicting whether a patient has cancer or not, predicting whether the customer will churn. Linear regression is estimated using Ordinary Least Squares (OLS) while logistic regression is estimated using Maximum Likelihood Estimation (MLE) approach.
# 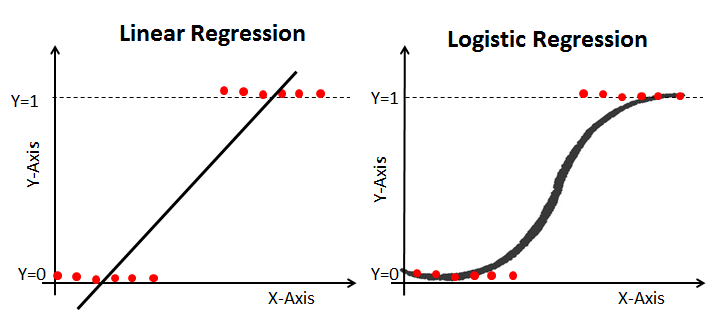
# ### Sigmoid Function
#
# The sigmoid function, also called logistic function gives an ‘S’ shaped curve that can take any real-valued number and map it into a value between 0 and 1. If the curve goes to positive infinity, y predicted will become 1, and if the curve goes to negative infinity, y predicted will become 0. If the output of the sigmoid function is more than 0.5, we can classify the outcome as 1 or YES, and if it is less than 0.5, we can classify it as 0 or NO. The outputcannotFor example: If the output is 0.75, we can say in terms of probability as: There is a 75 percent chance that patient will suffer from cancer.
# 
# 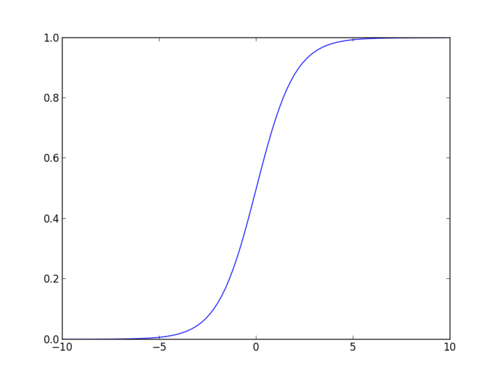
# ### class sklearn.linear_model.LogisticRegression(penalty=’l2’, dual=False, tol=0.0001, C=1.0, fit_intercept=True, intercept_scaling=1, class_weight=None, random_state=None, solver=’warn’, max_iter=100, multi_class=’warn’, verbose=0, warm_start=False, n_jobs=None, l1_ratio=None)
#
# penalty : str, ‘l1’, ‘l2’, ‘elasticnet’ or ‘none’, optional (default=’l2’)
#
# Used to specify the norm used in the penalization. The ‘newton-cg’, ‘sag’ and ‘lbfgs’ solvers support only l2 penalties. ‘elasticnet’ is only supported by the ‘saga’ solver. If ‘none’ (not supported by the liblinear solver), no regularization is applied.
#
# New in version 0.19: l1 penalty with SAGA solver (allowing ‘multinomial’ + L1)
# dual : bool, optional (default=False)
#
# Dual or primal formulation. Dual formulation is only implemented for l2 penalty with liblinear solver. Prefer dual=False when n_samples > n_features.
# tol : float, optional (default=1e-4)
#
# Tolerance for stopping criteria.
# C : float, optional (default=1.0)
#
# Inverse of regularization strength; must be a positive float. Like in support vector machines, smaller values specify stronger regularization.
# fit_intercept : bool, optional (default=True)
#
# Specifies if a constant (a.k.a. bias or intercept) should be added to the decision function.
# intercept_scaling : float, optional (default=1)
#
# Useful only when the solver ‘liblinear’ is used and self.fit_intercept is set to True. In this case, x becomes [x, self.intercept_scaling], i.e. a “synthetic” feature with constant value equal to intercept_scaling is appended to the instance vector. The intercept becomes intercept_scaling * synthetic_feature_weight.
#
# Note! the synthetic feature weight is subject to l1/l2 regularization as all other features. To lessen the effect of regularization on synthetic feature weight (and therefore on the intercept) intercept_scaling has to be increased.
# class_weight : dict or ‘balanced’, optional (default=None)
#
# Weights associated with classes in the form {class_label: weight}. If not given, all classes are supposed to have weight one.
#
# The “balanced” mode uses the values of y to automatically adjust weights inversely proportional to class frequencies in the input data as n_samples / (n_classes * np.bincount(y)).
#
# Note that these weights will be multiplied with sample_weight (passed through the fit method) if sample_weight is specified.
#
# New in version 0.17: class_weight=’balanced’
# random_state : int, RandomState instance or None, optional (default=None)
#
# The seed of the pseudo random number generator to use when shuffling the data. If int, random_state is the seed used by the random number generator; If RandomState instance, random_state is the random number generator; If None, the random number generator is the RandomState instance used by np.random. Used when solver == ‘sag’ or ‘liblinear’.
# solver : str, {‘newton-cg’, ‘lbfgs’, ‘liblinear’, ‘sag’, ‘saga’}, optional (default=’liblinear’).
#
# Algorithm to use in the optimization problem.
#
# For small datasets, ‘liblinear’ is a good choice, whereas ‘sag’ and ‘saga’ are faster for large ones.
# For multiclass problems, only ‘newton-cg’, ‘sag’, ‘saga’ and ‘lbfgs’ handle multinomial loss; ‘liblinear’ is limited to one-versus-rest schemes.
# ‘newton-cg’, ‘lbfgs’, ‘sag’ and ‘saga’ handle L2 or no penalty
# ‘liblinear’ and ‘saga’ also handle L1 penalty
# ‘saga’ also supports ‘elasticnet’ penalty
# ‘liblinear’ does not handle no penalty
#
# Note that ‘sag’ and ‘saga’ fast convergence is only guaranteed on features with approximately the same scale. You can preprocess the data with a scaler from sklearn.preprocessing.
#
# New in version 0.17: Stochastic Average Gradient descent solver.
#
# New in version 0.19: SAGA solver.
#
# Changed in version 0.20: Default will change from ‘liblinear’ to ‘lbfgs’ in 0.22.
# max_iter : int, optional (default=100)
#
# Maximum number of iterations taken for the solvers to converge.
# multi_class : str, {‘ovr’, ‘multinomial’, ‘auto’}, optional (default=’ovr’)
#
# If the option chosen is ‘ovr’, then a binary problem is fit for each label. For ‘multinomial’ the loss minimised is the multinomial loss fit across the entire probability distribution, even when the data is binary. ‘multinomial’ is unavailable when solver=’liblinear’. ‘auto’ selects ‘ovr’ if the data is binary, or if solver=’liblinear’, and otherwise selects ‘multinomial’.
#
# New in version 0.18: Stochastic Average Gradient descent solver for ‘multinomial’ case.
#
# Changed in version 0.20: Default will change from ‘ovr’ to ‘auto’ in 0.22.
# verbose : int, optional (default=0)
#
# For the liblinear and lbfgs solvers set verbose to any positive number for verbosity.
# warm_start : bool, optional (default=False)
#
# When set to True, reuse the solution of the previous call to fit as initialization, otherwise, just erase the previous solution. Useless for liblinear solver. See the Glossary.
#
# New in version 0.17: warm_start to support lbfgs, newton-cg, sag, saga solvers.
# n_jobs : int or None, optional (default=None)
#
# Number of CPU cores used when parallelizing over classes if multi_class=’ovr’”. This parameter is ignored when the solver is set to ‘liblinear’ regardless of whether ‘multi_class’ is specified or not. None means 1 unless in a joblib.parallel_backend context. -1 means using all processors. See Glossary for more details.
# l1_ratio : float or None, optional (default=None)
#
# The Elastic-Net mixing parameter, with 0 <= l1_ratio <= 1. Only used if penalty='elasticnet'`. Setting ``l1_ratio=0 is equivalent to using penalty='l2', while setting l1_ratio=1 is equivalent to using penalty='l1'. For 0 < l1_ratio <1, the penalty is a combination of L1 and L2.
#
# ## Simplified Implmentation
# In[147]:
from sklearn.datasets import load_iris
from sklearn.linear_model import LogisticRegression
X, y = load_iris(return_X_y=True)
clf = LogisticRegression(random_state=0, solver='lbfgs',
multi_class='multinomial').fit(X, y)
clf.predict(X[:2, :])
clf.predict_proba(X[:2, :])
clf.score(X, y)
# ## Implementation in Python
# ### Importing Libraries
# In[2]:
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
get_ipython().run_line_magic('matplotlib', 'inline')
# ### Importing Dataset
# In[3]:
link="D:/As a Trainer/Course Material/Machine Learning with Python/All Special/Logistic Regression/"
df = pd.read_csv(link+'diabetes.csv')
# In[4]:
df.head()
# In[5]:
df.info()
# In[6]:
df.describe()
# ### Exploratory Data Analysis
# In[7]:
sns.pairplot(df)
# In[8]:
df.isna()
# In[9]:
df[df.isna().any(axis=1)]
# In[13]:
df.isna().sum()
# ### Creating profile Report
# In[10]:
import pandas_profiling
pandas_profiling.ProfileReport(df)
# In[15]:
sns.distplot(df['BMI'],hist_kws=dict(edgecolor="black", linewidth=1),color='red')
# ### Correlation Plot
# In[16]:
df.corr()
# In[17]:
plt.figure(figsize=(8,8))
sns.heatmap(df.corr(), annot = True)
# In[21]:
sns.set_style('whitegrid')
sns.countplot(x='Outcome',hue='Outcome',data=df,palette='RdBu_r')
# ### Checking Distribution of Age for Diabates
# In[23]:
sns.distplot(df['Age'],kde=False,color='darkblue',bins=20)
# ### Splitting Dataset into Train and Test
# In[25]:
from sklearn.model_selection import train_test_split
# #### Taking Featured Columns
# In[34]:
X = ['Pregnancies','Glucose','BloodPressure','SkinThickness','Insulin','BMI','DiabetesPedigreeFunction','Age']
y = ['Output']
# In[26]:
df2 =
|
pd.DataFrame(data=df)
|
pandas.DataFrame
|
# qt5模块
from PyQt5 import QtCore, QtGui, QtWidgets
from PyQt5.QtWidgets import *
from PyQt5.QtGui import *
from PyQt5.QtWebEngineWidgets import *
from PyQt5.QtCore import *
# 自定义模块
import mainpage
import addpage
import gridlayout
import editpage
import pinyintool
# 内置模块
import sys
import requests, base64, json
import collections
import os
import re
# 第三方模块
import pandas as pd
from pandas import DataFrame
from PIL import Image
import phone
from pyecharts.charts import Pie
# 获取文件的路径
cdir = os.getcwd()
# 文件路径
path=cdir+'/res/datafile/'
# 读取路径 判断是否创建了文件
if not os.path.exists(path):
# 根据路径建立文件夹
os.makedirs(path)
# 姓名 公司 电话 手机 邮件 地址 城市 分类 name comp tel mobile email addr city type
cardfile = pd.DataFrame(columns=['name', 'comp', 'tel', 'mobile', 'email', 'addr', 'city', 'type'])
# 生成xlsx文件
cardfile.to_excel(path+'名片信息表.xlsx', sheet_name='data', index=None)
# 编辑页面
class editWindow(QWidget,editpage.Ui_Form):
# 初始化方法
def __init__(self):
# 找到父类 首页面
super(editWindow, self).__init__()
# 初始化页面方法
self.setupUi(self)
# 为保存按钮添加事件
self.pushButton_2.clicked.connect(self.editkeep)
# 显示添加名片页面
def OPEN(self):
# 显示页面
self.show()
# 保存编辑内容
def editkeep(self):
# 获取按钮名称
indexName = self.pushButton_2.objectName()
# 获取表
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
# 获取控件信息 # 'name', 'comp', 'tel', 'mobile', 'email', 'addr', 'city', 'type'
name = self.lineEdit.text()
comp = self.lineEdit_2.text()
tel = self.lineEdit_3.text()
mobile = self.lineEdit_4.text()
email = self.lineEdit_5.text()
addr = self.lineEdit_6.text()
# 判断手机号是否为空
if mobile.strip():
# 根据手机号判断区域
try:
info = phone.Phone().find(int(mobile))
except Exception as e:
print("根据手机号判断区域时出错", e)
QMessageBox.critical(self, "错误:", "手机号码不正确!", QMessageBox.Ok) # 弹出提示对话框
self.lineEdit_4.setFocus() # 让手机文本框获得焦点
return
# 判断手机号是否正确返回信息
if info == None:
city = '其他'
else:
# 正确返回信息获取省
city = info['province']
else:
city = '其他'
# 判断姓名是否为空
if name.strip():
# 获取首字母拼音
type = pinyintool.getPinyin(name[0])
# 根据行号删除数据
datas = pi_table.drop(index=[int(indexName)], axis=0)
data = datas.append({'name': name,
'comp': comp,
'tel': tel,
'mobile': mobile,
'email': email,
'addr': addr,
'city': city,
'type': type
}, ignore_index=True)
# 更新xlsx文件
DataFrame(data).to_excel(path + '名片信息表.xlsx',
sheet_name='data', index=False)
self.close()
window.dataall()
else:
QMessageBox.information(self, '提示信息', '姓名不能为空')
pass
#首页列表样式
class griditem(QWidget,gridlayout.Ui_Form):
# 初始化方法
def __init__(self):
# 找到父类 首页面
super(griditem, self).__init__()
# 初始化页面方法
self.setupUi(self)
# 显示饼图类
class HtmlWindows(QMainWindow):
def __init__(self):
super(QMainWindow,self).__init__()
self.setGeometry(200, 200, 850, 500)
self.browser = QWebEngineView()
def set(self,title,hurl):
self.setWindowTitle(title)
d = os.path.dirname(os.path.realpath(sys.argv[0])) # 获取当前文件所在路径
d=re.sub(r'\\','/',d) # 将路径中的分隔符\替换为/
url=d+'/res/datafile/'+hurl
self.browser.load(QUrl(url))
self.setCentralWidget(self.browser)
# 首页页面
class parentWindow(QWidget,mainpage.Ui_Form):
# 初始化方法
def __init__(self):
# 找到父类 首页面
super(parentWindow, self).__init__()
# 初始化页面方法
self.setupUi(self)
# 标签按钮绑定事件
self.pushButton_3.clicked.connect(self.dataall)
self.pushButton_4.clicked.connect(lambda:self.dataother('A'))
self.pushButton_6.clicked.connect(lambda:self.dataother('B'))
self.pushButton_5.clicked.connect(lambda:self.dataother('C'))
self.pushButton_7.clicked.connect(lambda:self.dataother('D'))
self.pushButton_8.clicked.connect(lambda:self.dataother('E'))
self.pushButton_9.clicked.connect(lambda:self.dataother('F'))
self.pushButton_10.clicked.connect(lambda:self.dataother('G'))
self.pushButton_11.clicked.connect(lambda:self.dataother('H'))
self.pushButton_12.clicked.connect(lambda:self.dataother('I'))
self.pushButton_13.clicked.connect(lambda:self.dataother('J'))
self.pushButton_14.clicked.connect(lambda:self.dataother('K'))
self.pushButton_15.clicked.connect(lambda:self.dataother('L'))
self.pushButton_16.clicked.connect(lambda:self.dataother('M'))
self.pushButton_17.clicked.connect(lambda:self.dataother('N'))
self.pushButton_18.clicked.connect(lambda:self.dataother('O'))
self.pushButton_19.clicked.connect(lambda:self.dataother('P'))
self.pushButton_20.clicked.connect(lambda:self.dataother('Q'))
self.pushButton_21.clicked.connect(lambda:self.dataother('R'))
self.pushButton_22.clicked.connect(lambda:self.dataother('S'))
self.pushButton_23.clicked.connect(lambda:self.dataother('T'))
self.pushButton_24.clicked.connect(lambda:self.dataother('U'))
self.pushButton_25.clicked.connect(lambda:self.dataother('V'))
self.pushButton_26.clicked.connect(lambda:self.dataother('W'))
self.pushButton_27.clicked.connect(lambda:self.dataother('X'))
self.pushButton_28.clicked.connect(lambda:self.dataother('Y'))
self.pushButton_29.clicked.connect(lambda:self.dataother('Z'))
# 搜索按钮绑定事件
self.pushButton.clicked.connect(self.seachbtn)
self.pushButton_30.clicked.connect(self.lookpie)
# 显示全部数据
self.dataall()
# 显示饼图
def lookpie(self):
# 获取表数据
# 读取文件内容
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
# 获取城市信息
citattr = pi_table['city'].values
# 判断是否有城市信息
if len(citattr)==0:
# 没有信息提示
QMessageBox.information(self, '提示信息', '没有联系人')
return
attr = []
v1 = []
# 循环表里city项
for i in citattr:
# 判断city项是否包含在attr里
if not i in attr:
# 不包含在attr表里时候添加到attr里
attr.append(i)
# Counter(计数器)是对字典的补充,用于追踪值的出现次数。
d = collections.Counter(citattr)
# 循环城市列表
for k in attr:
# d[k] 是k在列表d中出现的次数
v1.append(d[k])
# 生成饼图
pie = Pie("联系人分布")
pie.add("", attr, v1, is_label_show=True)
pie.show_config()
pie.render(path+'联系人分布饼图.html')
# 显示饼图
htmlwidows.set('联系人分布','联系人分布饼图.html')
htmlwidows.show()
pass
#搜索功能
def seachbtn(self):
# 读取文件内容
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
seachk=self.lineEdit.text()
# 查询数据 用户名和公司如果有一个包含搜索内容筛选出来
cardArray = pi_table[(pi_table['name'].str.contains(seachk)) | (pi_table['comp'].str.contains(seachk))].values
tb = pi_table[(pi_table['name'].str.contains(seachk)) | (pi_table['comp'].str.contains(seachk))]
if len(cardArray) == 0:
QMessageBox.information(self, '提示信息', '没有搜索内容')
else:
# 每次点循环删除管理器的组件
while self.gridLayout.count():
# 获取第一个组件
item = self.gridLayout.takeAt(0)
widget = item.widget()
# 删除组件
widget.deleteLater()
i = -1
for n in range(len(cardArray)):
# x 确定每行显示的个数 0,1,2 每行3个
x = n % 3
# 当x为0的时候设置换行 行数+1
if x == 0:
i += 1
item = griditem()
item.label_8.setText('姓名:' + str(cardArray[n][0]))
item.label_9.setText('公司:' + str(cardArray[n][1]))
item.label_10.setText('电话:' + str(cardArray[n][2]))
item.label_11.setText('手机:' + str(cardArray[n][3]))
item.label_12.setText('邮箱:' + str(cardArray[n][4]))
item.label_13.setText('地址:' + str(cardArray[n][5]))
# 设置名称 为获取项目行数
item.pushButton.setObjectName(str(tb.index.tolist()[n]))
item.pushButton_3.setObjectName(str(tb.index.tolist()[n]))
# 为按钮绑定点击事件
item.pushButton.clicked.connect(self.edit)
item.pushButton_3.clicked.connect(self.deletedata)
# 动态给gridlayout添加布局
self.gridLayout.addWidget(item, i, x)
# 设置上下滑动控件可以滑动 把scrollAreaWidgetContents_2添加到scrollArea中
self.scrollAreaWidgetContents.setMinimumHeight(i * 200)
# girdlayout 添加到滑动控件中
self.scrollAreaWidgetContents.setLayout(self.gridLayout)
#显示全部数据
def dataall(self):
# 每次点循环删除管理器的组件
while self.gridLayout.count():
# 获取第一个组件
item = self.gridLayout.takeAt(0)
widget = item.widget()
# 删除组件
widget.deleteLater()
i=-1
# 读取文件内容
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
# 获取所有数据
cardArray = pi_table.values
# 循环数据
for n in range(len(cardArray)):
# x 确定每行显示的个数 0,1,2 每行3个
x = n % 3
# 当x为0的时候设置换行,即行数+1
if x == 0:
i += 1
item = griditem()
item.label_8.setText('姓名:'+str(cardArray[n][0]))
item.label_9.setText('公司:'+str(cardArray[n][1]))
item.label_10.setText('电话:'+str(cardArray[n][2]))
item.label_11.setText('手机:'+str(cardArray[n][3]))
item.label_12.setText('邮箱:'+str(cardArray[n][4]))
item.label_13.setText('地址:'+str(cardArray[n][5]))
# 设置名称 为获取项目行数
item.pushButton.setObjectName(str(pi_table.index.tolist()[n]))
item.pushButton_3.setObjectName(str(pi_table.index.tolist()[n]))
# 为按钮绑定点击事件
item.pushButton.clicked.connect(self.edit)
item.pushButton_3.clicked.connect(self.deletedata)
# 动态添加控件到gridlayout中
self.gridLayout.addWidget(item,i, x)
# 设置上下滑动控件可以滑动 把scrollAreaWidgetContents_2添加到scrollArea中
self.scrollAreaWidgetContents.setMinimumHeight(i*200)
# 设置gridlayout到滑动控件中
self.scrollAreaWidgetContents.setLayout(self.gridLayout)
pass
# 删除数据方法
def deletedata(self):
# 获取信号源 点击的按钮
sender = self.gridLayout.sender()
# 获取按钮名称
indexName = sender.objectName()
# 获取表信息
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
# 根据行号删除数据
data=pi_table.drop(index=[int(indexName)], axis=0)
# 更新xlsx文件
DataFrame(data).to_excel(path + '名片信息表.xlsx',
sheet_name='data', index=False)
# 显示全部数据
self.dataall()
# 编辑数据
def edit(self):
# 获取信号源 点击的按钮
sender = self.gridLayout.sender()
# 获取按钮名称
indexName=sender.objectName()
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
# 根据行号 获取数据
cardArray =pi_table.iloc[[indexName]].values
# 打开编辑页面
editWindow.OPEN()
# 设置数据
editWindow.lineEdit.setText(str(cardArray[0][0]))
editWindow.lineEdit_2.setText(str(cardArray[0][1]))
editWindow.lineEdit_3.setText(str(cardArray[0][2]))
editWindow.lineEdit_4.setText(str(cardArray[0][3]))
editWindow.lineEdit_5.setText(str(cardArray[0][4]))
editWindow.lineEdit_6.setText(str(cardArray[0][5]))
# 设置按钮名称
editWindow.pushButton_2.setObjectName(str(indexName))
pass
# 显示部分数据
def dataother(self,typeAZ):
# 每次点循环删除管理器的组件
while self.gridLayout.count():
# 获取第一个组件
item = self.gridLayout.takeAt(0)
# 获取布局
widget = item.widget()
# 删除组件
widget.deleteLater()
pass
# 读取文件内容
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
i = -1
# 筛选内容
cardArray = pi_table[pi_table['type'] == typeAZ].values
tb= pi_table[pi_table['type'] == typeAZ]
for n in range(len(cardArray)):
# x 确定每行显示的个数 0,1,2 每行3个
x = n % 3
# 当x为0的时候设置换行 行数+1
if x == 0:
i += 1
item = griditem()
item.label_8.setText('姓名:' + str(cardArray[n][0]))
item.label_9.setText('公司:' + str(cardArray[n][1]))
item.label_10.setText('电话:' + str(cardArray[n][2]))
item.label_11.setText('手机:' + str(cardArray[n][3]))
item.label_12.setText('邮箱:' + str(cardArray[n][4]))
item.label_13.setText('地址:' + str(cardArray[n][5]))
# 设置名称 为获取项目行数
item.pushButton.setObjectName(str(tb.index.tolist()[n]))
item.pushButton_3.setObjectName(str(tb.index.tolist()[n]))
# 为按钮绑定点击事件
item.pushButton.clicked.connect(self.edit)
item.pushButton_3.clicked.connect(self.deletedata)
# 动态设置控件
self.gridLayout.addWidget(item, i, x)
# 动态设置滑动控件滑动高度
self.scrollAreaWidgetContents.setMinimumHeight(i * 200)
# giridLayout 添加到滑动控件中
self.scrollAreaWidgetContents.setLayout(self.gridLayout)
#添加名片页面
class childWindow(QWidget,addpage.Ui_Form):
# 初始化方法
def __init__(self):
# 找到父类 添加名片页面
super(childWindow, self).__init__()
# 初始化页面方法
self.setupUi(self)
# 给选择名片按钮添加事件
self.pushButton.clicked.connect(self.openfile)
# 给保存按钮添加事件
self.pushButton_2.clicked.connect(self.keep)
#保存名片信息到文档
def keep(self):
pi_table = pd.read_excel(path + '名片信息表.xlsx', sheet_name='data')
# 获取输出框内容
name =self.lineEdit.text()
comp = self.lineEdit_2.text()
tel= self.lineEdit_3.text()
mobile= self.lineEdit_4.text()
email= self.lineEdit_5.text()
addr= self.lineEdit_6.text()
# 判断手机号是否为空
if mobile.strip():
# 根据手机号判断区域
try:
info = phone.Phone().find(int(mobile))
except Exception as e:
print("根据手机号判断区域时出错",e)
QMessageBox.critical(self,"错误:","手机号码不正确!",QMessageBox.Ok) # 弹出提示对话框
self.lineEdit_4.setFocus() # 让手机文本框获得焦点
return
# 判断手机号是否正确返回信息
if info==None:
city = '其他'
else:
# 正确返回信息获取省
city = info['province']
else:
city = '其他'
# 判断姓名是否为空
if name.strip():
# 获取首字母拼音
type=pinyintool.getPinyin(name[0])
# 添加数据
data = pi_table.append({'name': name,
'comp': comp,
'tel': tel,
'mobile': mobile,
'email': email,
'addr': addr,
'city': city,
'type': type,
}, ignore_index=True)
# 更新xlsx文件
|
DataFrame(data)
|
pandas.DataFrame
|
from kiwoom import *
from pandas import DataFrame
kiwoom = Kiwoom()
kiwoom.CommConnect()
kospi = kiwoom.GetCodeListByMarket('0')
kosdaq = kiwoom.GetCodeListByMarket('10')
total = kospi + kosdaq
rows = []
for code in total:
name = kiwoom.GetMasterCodeName(code)
rows.append((code, name))
columns = ['code', 'name']
df =
|
DataFrame(data=rows, columns=columns)
|
pandas.DataFrame
|
################################################################################
# This module counts overlapping lemmas among an item stem and the options from
# the input lemma count columns. If the lemma count columns of passage sections
# are also specified as the other input, the overlapping lemmas with
# the corresponded item stem or the options are also counted.
# Parameters df_ac_q: input pandas.DataFrame of questions, it should have,
# at least, lemma count columns with the 'AC_Doc_ID's
# as the index of the DataFrame, the question ID column, and
# stem/option identifier column, if the DataFrame of
# passages are also specified as the other input,
# the DataFrame of questions should also have corresponded
# passage name and the section columns, the module assumes
# that the stem and options which have the same question
# ID share the same passage name and the section(s)
# question_id_clm: column name of question IDs which are shared by
# the item stem and the options of each question
# stem_option_name_clm: column name of stem/option identifier
# lemma_start_q: integer column number (starting from zero)
# specifying the starting point of lemma count
# columns in the question DataFrame, from the point
# to the end, all the columns should be the lemma
# count columns
# stop_words = None: list of lemmas to specify stop words, they
# should all include in the question and passage
# DataFrames
# passage_name_clm_q = None: column name of the passage names
# in the question DataFrame
# passage_sec_clm_q = None: column name of the passage sections
# in the question DataFrame
# df_ac_p = None: input pandas.DataFrame of passages, it should have,
# at least, lemma count columns, passage name
# and the section columns
# passage_name_clm_p = None: column name of the passage names
# in the passage DataFrame
# passage_sec_clm_p = None: column name of the passage sections
# in the passage DataFrame
# lemma_start_p = None: integer column number (starting from zero)
# specifying the starting point of lemma count
# columns in the passage DataFrame, from the point
# to the end, all the columns should be the lemma
# count columns
# Returns Result: pandas.DataFrame as a result of overlapping lemma counts
################################################################################
def ac_overlapping_lemma(df_ac_q, question_id_clm, stem_option_name_clm,
lemma_start_q, stop_words = None,
passage_name_clm_q = None, passage_sec_clm_q = None,
df_ac_p = None, passage_name_clm_p = None,
passage_sec_clm_p = None, lemma_start_p = None):
import pandas as pd
import numpy as np
df_ac_buf = df_ac_q.copy()
df_ac_id_buf = df_ac_buf[question_id_clm]
df_ac_id = df_ac_id_buf.drop_duplicates()
ac_buf_index_name = df_ac_buf.index.name
ac_buf_index = df_ac_buf.index
df_ac_buf = df_ac_buf.set_index([question_id_clm, stem_option_name_clm])
df_ac_buf_lemma = df_ac_buf.iloc[:, (lemma_start_q -2):]
if stop_words != None:
df_ac_buf_lemma = df_ac_buf_lemma.drop(stop_words, axis=1)
if df_ac_p is not None:
df_ac_buf_p = df_ac_p.copy()
df_ac_buf_p = df_ac_buf_p.set_index([passage_name_clm_p, passage_sec_clm_p])
# modified by <EMAIL> 09/22/2020
if stop_words != None:
df_ac_buf_p = df_ac_buf_p.drop(stop_words, axis=1)
df_ac_buf_p_lemma = df_ac_buf_p.iloc[:, (lemma_start_p -2):]
# modified by <EMAIL> 09/22/2020
# In order to avoid overhead of the appending operation for each row,
# the passage name and passage section name are compounded as a temporal index name
df_ac_buf_p_lemma[question_id_clm] = [x[0] + ';' + x[1] for x in df_ac_buf_p_lemma.index]
df_ac_buf_p_lemma[stem_option_name_clm] = 'Passage'
df_ac_buf_p_lemma = df_ac_buf_p_lemma.set_index([question_id_clm, stem_option_name_clm])
row_lgth = df_ac_buf_lemma.shape[0]
df_ac_buf_q_p_lemma = df_ac_buf_lemma.append(df_ac_buf_p_lemma)
df_ac_buf_lemma = df_ac_buf_q_p_lemma.iloc[:row_lgth, :]
df_ac_buf_p_lemma = df_ac_buf_q_p_lemma.iloc[row_lgth:, :]
df_res = pd.DataFrame()
for x in df_ac_id:
print('Question:' + str(x))
if df_ac_p is not None:
df_q_x = df_ac_buf.xs(x)
passage_name = df_q_x[passage_name_clm_q][0]
passage_sections = (df_q_x[passage_sec_clm_q][0]).split(';')
print('Passage:' + passage_name)
# modified by <EMAIL> 09/22/2020
# df_p_x = df_ac_buf_p.xs(passage_name)
# df_p_x = df_p_x.loc[passage_sections]
# df_ac_buf_p_lemma_x = df_p_x.iloc[:, (lemma_start_p -2):]
# modified by <EMAIL> 09/22/2020
# In order to avoid overhead of the appending operation for each row,
# the passage name and passage section name are compounded as a temporal index name
# df_ac_buf_p_lemma_x = df_ac_buf_p_lemma.xs(passage_name)
# df_ac_buf_p_lemma_x = df_ac_buf_p_lemma_x.loc[passage_sections]
df_ac_buf_p_lemma_x = df_ac_buf_p_lemma[[x[0].startswith(passage_name) for x in df_ac_buf_p_lemma.index]]
df_ac_buf_p_lemma_x = df_ac_buf_p_lemma_x[[x[0].endswith(tuple(passage_sections)) for x in df_ac_buf_p_lemma_x.index]]
# modified by <EMAIL> 09/22/2020
# if stop_words != None:
# df_ac_buf_p_lemma_x = df_ac_buf_p_lemma_x.drop(stop_words, axis=1)
df_ac_buf_p_lemma_x_sum = pd.DataFrame({ 'Passage' : df_ac_buf_p_lemma_x.sum() })
df_ac_buf_lemma_x = (df_ac_buf_p_lemma_x_sum.transpose()).append(df_ac_buf_lemma.xs(x))
index_arr = df_ac_buf_lemma_x.index.values
index_arr = np.append(index_arr[1:], index_arr[0])
df_ac_buf_lemma_x = df_ac_buf_lemma_x.reindex(index_arr)
df_ac_buf_lemma_x.index.name = stem_option_name_clm
df_ac_overlap_doc = ac_overlapping_terms(df_ac_buf_lemma_x)
df_ac_overlap_doc = df_ac_overlap_doc.drop('Passage', axis=0)
else:
df_ac_overlap_doc = ac_overlapping_terms(df_ac_buf_lemma.xs(x))
df_ac_overlap_doc = df_ac_overlap_doc.reset_index()
df_doc = pd.DataFrame({ question_id_clm : np.array([x] *
len(df_ac_overlap_doc)) })
df_ac_overlap_doc[question_id_clm] = df_doc[question_id_clm]
df_res = df_res.append(df_ac_overlap_doc, ignore_index=True)
df_doc_id = pd.DataFrame({ ac_buf_index_name : ac_buf_index })
df_res[ac_buf_index_name] = df_doc_id[ac_buf_index_name]
df_res = df_res.set_index(ac_buf_index_name)
return df_res
################################################################################
# This module counts overlapping lemmas among the all input records
# Parameters df_ac_q: input pandas.DataFrame it should only have lemma count
# columns with an index of the DataFrame
# Returns Result: pandas.DataFrame as a result of overlapping lemma counts
################################################################################
def ac_overlapping_terms(df_doc_term_matrix):
import pandas as pd
import numpy as np
t = df_doc_term_matrix.shape
row_lgth = t[0]
col_lgth = t[1]
doc_term_matrix_clm_count = []
doc_term_matrix_clm_terms = []
doc_term_matrix_index = df_doc_term_matrix.index
doc_term_matrix_clms = df_doc_term_matrix.columns
for x in doc_term_matrix_index:
s = 'Count_' + x
doc_term_matrix_clm_count.append(s)
for x in doc_term_matrix_index:
s = 'Terms_' + x
doc_term_matrix_clm_terms.append(s)
# df_overlapping_matrix = pd.DataFrame(np.empty((row_lgth, row_lgth * 2),
# dtype=object), doc_term_matrix_index,
# doc_term_matrix_clm_count + doc_term_matrix_clm_terms)
df_overlapping_count_matrix = pd.DataFrame(np.empty((row_lgth, row_lgth),
dtype=np.int64), doc_term_matrix_index,
doc_term_matrix_clm_count)
df_overlapping_term_matrix = pd.DataFrame(np.empty((row_lgth, row_lgth),
dtype=object), doc_term_matrix_index,
doc_term_matrix_clm_terms)
# modified by <EMAIL> 09/20/2020
'''
df_overlapping_matrix = pd.concat([df_overlapping_count_matrix, df_overlapping_term_matrix], axis=1)
for k, z in enumerate(doc_term_matrix_index):
for i, x in enumerate(doc_term_matrix_clm_count):
if k == i:
#df_overlapping_matrix.iloc[k, i] = ''
df_overlapping_matrix.iloc[k, i] = np.nan
else:
df_overlapping_matrix.iloc[k, i] = 0
df_overlapping_matrix.iloc[k, i + len(doc_term_matrix_clm_count)] = ''
for j, y in enumerate(df_doc_term_matrix.iloc[i, :]):
if y > 0:
if df_doc_term_matrix.iloc[k, j] > 0:
df_overlapping_matrix.iloc[k, i] += 1
s = df_overlapping_matrix.iloc[k, i + len(doc_term_matrix_clm_count)]
if s == '':
s = doc_term_matrix_clms[j]
else:
s = ';'.join([s, doc_term_matrix_clms[j]])
df_overlapping_matrix.iloc[k, i + len(doc_term_matrix_clm_count)] = s
'''
for k, z in enumerate(doc_term_matrix_index):
for i, x in enumerate(doc_term_matrix_clm_count):
if k == i:
df_overlapping_count_matrix.iloc[k, i] = np.nan
else:
se_multiply = df_doc_term_matrix.iloc[k] * df_doc_term_matrix.iloc[i]
se_match = se_multiply / se_multiply
df_overlapping_count_matrix.iloc[k, i] = int(se_match.sum())
se_match.index = df_doc_term_matrix.columns
s = ';'.join(se_match[se_match > 0].index)
df_overlapping_term_matrix.iloc[k, i] = s
df_overlapping_matrix =
|
pd.concat([df_overlapping_count_matrix, df_overlapping_term_matrix], axis=1)
|
pandas.concat
|
from __future__ import division
import copy
import bt
from bt.core import Node, StrategyBase, SecurityBase, AlgoStack, Strategy
import pandas as pd
import numpy as np
from nose.tools import assert_almost_equal as aae
import sys
if sys.version_info < (3, 3):
import mock
else:
from unittest import mock
def test_node_tree():
c1 = Node('c1')
c2 = Node('c2')
p = Node('p', children=[c1, c2])
c1 = p['c1']
c2 = p['c2']
assert len(p.children) == 2
assert 'c1' in p.children
assert 'c2' in p.children
assert p == c1.parent
assert p == c2.parent
m = Node('m', children=[p])
p = m['p']
c1 = p['c1']
c2 = p['c2']
assert len(m.children) == 1
assert 'p' in m.children
assert p.parent == m
assert len(p.children) == 2
assert 'c1' in p.children
assert 'c2' in p.children
assert p == c1.parent
assert p == c2.parent
def test_strategybase_tree():
s1 = SecurityBase('s1')
s2 = SecurityBase('s2')
s = StrategyBase('p', [s1, s2])
s1 = s['s1']
s2 = s['s2']
assert len(s.children) == 2
assert 's1' in s.children
assert 's2' in s.children
assert s == s1.parent
assert s == s2.parent
def test_node_members():
s1 = SecurityBase('s1')
s2 = SecurityBase('s2')
s = StrategyBase('p', [s1, s2])
s1 = s['s1']
s2 = s['s2']
actual = s.members
assert len(actual) == 3
assert s1 in actual
assert s2 in actual
assert s in actual
actual = s1.members
assert len(actual) == 1
assert s1 in actual
actual = s2.members
assert len(actual) == 1
assert s2 in actual
def test_node_full_name():
s1 = SecurityBase('s1')
s2 = SecurityBase('s2')
s = StrategyBase('p', [s1, s2])
# we cannot access s1 and s2 directly since they are copied
# we must therefore access through s
assert s.full_name == 'p'
assert s['s1'].full_name == 'p>s1'
assert s['s2'].full_name == 'p>s2'
def test_security_setup_prices():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[0]] = 105
data['c2'][dts[0]] = 95
s.setup(data)
i = 0
s.update(dts[i], data.ix[dts[i]])
assert c1.price == 105
assert len(c1.prices) == 1
assert c1.prices[0] == 105
assert c2.price == 95
assert len(c2.prices) == 1
assert c2.prices[0] == 95
# now with setup
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[0]] = 105
data['c2'][dts[0]] = 95
s.setup(data)
i = 0
s.update(dts[i], data.ix[dts[i]])
assert c1.price == 105
assert len(c1.prices) == 1
assert c1.prices[0] == 105
assert c2.price == 95
assert len(c2.prices) == 1
assert c2.prices[0] == 95
def test_strategybase_tree_setup():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
s.setup(data)
assert len(s.data) == 3
assert len(c1.data) == 3
assert len(c2.data) == 3
assert len(s._prices) == 3
assert len(c1._prices) == 3
assert len(c2._prices) == 3
assert len(s._values) == 3
assert len(c1._values) == 3
assert len(c2._values) == 3
def test_strategybase_tree_adjust():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
s.setup(data)
s.adjust(1000)
assert s.capital == 1000
assert s.value == 1000
assert c1.value == 0
assert c2.value == 0
assert c1.weight == 0
assert c2.weight == 0
def test_strategybase_tree_update():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
s.setup(data)
i = 0
s.update(dts[i], data.ix[dts[i]])
c1.price == 100
c2.price == 100
i = 1
s.update(dts[i], data.ix[dts[i]])
c1.price == 105
c2.price == 95
i = 2
s.update(dts[i], data.ix[dts[i]])
c1.price == 100
c2.price == 100
def test_update_fails_if_price_is_nan_and_position_open():
c1 = SecurityBase('c1')
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1'], data=100)
data['c1'][dts[1]] = np.nan
c1.setup(data)
i = 0
# mock in position
c1._position = 100
c1.update(dts[i], data.ix[dts[i]])
# test normal case - position & non-nan price
assert c1._value == 100 * 100
i = 1
# this should fail, because we have non-zero position, and price is nan, so
# bt has no way of updating the _value
try:
c1.update(dts[i], data.ix[dts[i]])
assert False
except Exception as e:
assert str(e).startswith('Position is open')
# on the other hand, if position was 0, this should be fine, and update
# value to 0
c1._position = 0
c1.update(dts[i], data.ix[dts[i]])
assert c1._value == 0
def test_strategybase_tree_allocate():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
s.setup(data)
i = 0
s.update(dts[i], data.ix[dts[i]])
s.adjust(1000)
# since children have w == 0 this should stay in s
s.allocate(1000)
assert s.value == 1000
assert s.capital == 1000
assert c1.value == 0
assert c2.value == 0
# now allocate directly to child
c1.allocate(500)
assert c1.position == 5
assert c1.value == 500
assert s.capital == 1000 - 500
assert s.value == 1000
assert c1.weight == 500.0 / 1000
assert c2.weight == 0
def test_strategybase_tree_allocate_child_from_strategy():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
s.setup(data)
i = 0
s.update(dts[i], data.ix[dts[i]])
s.adjust(1000)
# since children have w == 0 this should stay in s
s.allocate(1000)
assert s.value == 1000
assert s.capital == 1000
assert c1.value == 0
assert c2.value == 0
# now allocate to c1
s.allocate(500, 'c1')
assert c1.position == 5
assert c1.value == 500
assert s.capital == 1000 - 500
assert s.value == 1000
assert c1.weight == 500.0 / 1000
assert c2.weight == 0
def test_strategybase_tree_allocate_level2():
c1 = SecurityBase('c1')
c12 = copy.deepcopy(c1)
c2 = SecurityBase('c2')
c22 = copy.deepcopy(c2)
s1 = StrategyBase('s1', [c1, c2])
s2 = StrategyBase('s2', [c12, c22])
m = StrategyBase('m', [s1, s2])
s1 = m['s1']
s2 = m['s2']
c1 = s1['c1']
c2 = s1['c2']
c12 = s2['c1']
c22 = s2['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
m.setup(data)
i = 0
m.update(dts[i], data.ix[dts[i]])
m.adjust(1000)
# since children have w == 0 this should stay in s
m.allocate(1000)
assert m.value == 1000
assert m.capital == 1000
assert s1.value == 0
assert s2.value == 0
assert c1.value == 0
assert c2.value == 0
# now allocate directly to child
s1.allocate(500)
assert s1.value == 500
assert m.capital == 1000 - 500
assert m.value == 1000
assert s1.weight == 500.0 / 1000
assert s2.weight == 0
# now allocate directly to child of child
c1.allocate(200)
assert s1.value == 500
assert s1.capital == 500 - 200
assert c1.value == 200
assert c1.weight == 200.0 / 500
assert c1.position == 2
assert m.capital == 1000 - 500
assert m.value == 1000
assert s1.weight == 500.0 / 1000
assert s2.weight == 0
assert c12.value == 0
def test_strategybase_tree_allocate_long_short():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
s.setup(data)
i = 0
s.update(dts[i], data.ix[dts[i]])
s.adjust(1000)
c1.allocate(500)
assert c1.position == 5
assert c1.value == 500
assert c1.weight == 500.0 / 1000
assert s.capital == 1000 - 500
assert s.value == 1000
c1.allocate(-200)
assert c1.position == 3
assert c1.value == 300
assert c1.weight == 300.0 / 1000
assert s.capital == 1000 - 500 + 200
assert s.value == 1000
c1.allocate(-400)
assert c1.position == -1
assert c1.value == -100
assert c1.weight == -100.0 / 1000
assert s.capital == 1000 - 500 + 200 + 400
assert s.value == 1000
# close up
c1.allocate(-c1.value)
assert c1.position == 0
assert c1.value == 0
assert c1.weight == 0
assert s.capital == 1000 - 500 + 200 + 400 - 100
assert s.value == 1000
def test_strategybase_tree_allocate_update():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('p', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[1]] = 105
data['c2'][dts[1]] = 95
s.setup(data)
i = 0
s.update(dts[i], data.ix[dts[i]])
assert s.price == 100
s.adjust(1000)
assert s.price == 100
assert s.value == 1000
assert s._value == 1000
c1.allocate(500)
assert c1.position == 5
assert c1.value == 500
assert c1.weight == 500.0 / 1000
assert s.capital == 1000 - 500
assert s.value == 1000
assert s.price == 100
i = 1
s.update(dts[i], data.ix[dts[i]])
assert c1.position == 5
assert c1.value == 525
assert c1.weight == 525.0 / 1025
assert s.capital == 1000 - 500
assert s.value == 1025
assert np.allclose(s.price, 102.5)
def test_strategybase_universe():
s = StrategyBase('s')
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[0]] = 105
data['c2'][dts[0]] = 95
s.setup(data)
i = 0
s.update(dts[i])
assert len(s.universe) == 1
assert 'c1' in s.universe
assert 'c2' in s.universe
assert s.universe['c1'][dts[i]] == 105
assert s.universe['c2'][dts[i]] == 95
# should not have children unless allocated
assert len(s.children) == 0
def test_strategybase_allocate():
s = StrategyBase('s')
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data['c1'][dts[0]] = 100
data['c2'][dts[0]] = 95
s.setup(data)
i = 0
s.update(dts[i])
s.adjust(1000)
s.allocate(100, 'c1')
c1 = s['c1']
assert c1.position == 1
assert c1.value == 100
assert s.value == 1000
def test_strategybase_close():
s = StrategyBase('s')
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
s.setup(data)
i = 0
s.update(dts[i])
s.adjust(1000)
s.allocate(100, 'c1')
c1 = s['c1']
assert c1.position == 1
assert c1.value == 100
assert s.value == 1000
s.close('c1')
assert c1.position == 0
assert c1.value == 0
assert s.value == 1000
def test_strategybase_flatten():
s = StrategyBase('s')
dts = pd.date_range('2010-01-01', periods=3)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
s.setup(data)
i = 0
s.update(dts[i])
s.adjust(1000)
s.allocate(100, 'c1')
c1 = s['c1']
s.allocate(100, 'c2')
c2 = s['c2']
assert c1.position == 1
assert c1.value == 100
assert c2.position == 1
assert c2.value == 100
assert s.value == 1000
s.flatten()
assert c1.position == 0
assert c1.value == 0
assert s.value == 1000
def test_strategybase_multiple_calls():
s = StrategyBase('s')
dts = pd.date_range('2010-01-01', periods=5)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
data.c2[dts[0]] = 95
data.c1[dts[1]] = 95
data.c2[dts[2]] = 95
data.c2[dts[3]] = 95
data.c2[dts[4]] = 95
data.c1[dts[4]] = 105
s.setup(data)
# define strategy logic
def algo(target):
# close out any open positions
target.flatten()
# get stock w/ lowest price
c = target.universe.ix[target.now].idxmin()
# allocate all capital to that stock
target.allocate(target.value, c)
# replace run logic
s.run = algo
# start w/ 1000
s.adjust(1000)
# loop through dates manually
i = 0
# update t0
s.update(dts[i])
assert len(s.children) == 0
assert s.value == 1000
# run t0
s.run(s)
assert len(s.children) == 1
assert s.value == 1000
assert s.capital == 50
c2 = s['c2']
assert c2.value == 950
assert c2.weight == 950.0 / 1000
assert c2.price == 95
# update out t0
s.update(dts[i])
c2 == s['c2']
assert len(s.children) == 1
assert s.value == 1000
assert s.capital == 50
assert c2.value == 950
assert c2.weight == 950.0 / 1000
assert c2.price == 95
# update t1
i = 1
s.update(dts[i])
assert s.value == 1050
assert s.capital == 50
assert len(s.children) == 1
assert 'c2' in s.children
c2 == s['c2']
assert c2.value == 1000
assert c2.weight == 1000.0 / 1050.0
assert c2.price == 100
# run t1 - close out c2, open c1
s.run(s)
assert len(s.children) == 2
assert s.value == 1050
assert s.capital == 5
c1 = s['c1']
assert c1.value == 1045
assert c1.weight == 1045.0 / 1050
assert c1.price == 95
assert c2.value == 0
assert c2.weight == 0
assert c2.price == 100
# update out t1
s.update(dts[i])
assert len(s.children) == 2
assert s.value == 1050
assert s.capital == 5
assert c1 == s['c1']
assert c1.value == 1045
assert c1.weight == 1045.0 / 1050
assert c1.price == 95
assert c2.value == 0
assert c2.weight == 0
assert c2.price == 100
# update t2
i = 2
s.update(dts[i])
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 5
assert c1.value == 1100
assert c1.weight == 1100.0 / 1105
assert c1.price == 100
assert c2.value == 0
assert c2.weight == 0
assert c2.price == 95
# run t2
s.run(s)
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
assert c1.price == 100
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
# update out t2
s.update(dts[i])
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
assert c1.price == 100
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
# update t3
i = 3
s.update(dts[i])
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
assert c1.price == 100
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
# run t3
s.run(s)
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
assert c1.price == 100
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
# update out t3
s.update(dts[i])
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
assert c1.price == 100
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
# update t4
i = 4
s.update(dts[i])
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
# accessing price should refresh - this child has been idle for a while -
# must make sure we can still have a fresh prices
assert c1.price == 105
assert len(c1.prices) == 5
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
# run t4
s.run(s)
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
assert c1.price == 105
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
# update out t4
s.update(dts[i])
assert len(s.children) == 2
assert s.value == 1105
assert s.capital == 60
assert c1.value == 0
assert c1.weight == 0
assert c1.price == 105
assert c2.value == 1045
assert c2.weight == 1045.0 / 1105
assert c2.price == 95
def test_strategybase_multiple_calls_preset_secs():
c1 = SecurityBase('c1')
c2 = SecurityBase('c2')
s = StrategyBase('s', [c1, c2])
c1 = s['c1']
c2 = s['c2']
dts =
|
pd.date_range('2010-01-01', periods=5)
|
pandas.date_range
|
#!/usr/bin/env python
# coding: utf-8
# In[10]:
import pandas as pd
from alphacast import Alphacast
from dotenv import dotenv_values
API_KEY = dotenv_values(".env").get("API_KEY")
alphacast = Alphacast(API_KEY)
def replaceMonthByNumber(x):
x = x.str.replace('*','')
meses = { 'Enero': 1,
'Febrero':2,
'Marzo': 3,
'Abril': 4,
'Mayo':5,
'Junio':6,
'Julio':7,
'Agosto':8,
'Septiembre':9,
'Octubre':10,
'Noviembre':11,
'Diciembre':12}
for mes in meses:
x = x.replace(mes,meses[mes])
return x
def fix_data(df, year_col, month_col, join_col=True):
if join_col:
df.columns = df.columns.map(' - '.join)
df["year"] = pd.to_numeric(df[year_col].ffill(), errors='coerce')
df["month"] = replaceMonthByNumber(df[month_col])
df["day"] = 1
df["Date"] = pd.to_datetime(df[["year", "month", "day"]], errors="coerce")
df = df[df["Date"].notnull()]
df = df.set_index("Date")
del df["year"]
del df["day"]
del df["month"]
del df[year_col]
del df[month_col]
return df
df_merge = pd.DataFrame()
df = pd.read_excel('https://www.indec.gob.ar/ftp/cuadros/economia/sh_super_mayoristas.xls', sheet_name="Cuadro 1", skiprows=2, header=[0,1])
df = fix_data(df, "Período - Unnamed: 0_level_1", "Período - Unnamed: 1_level_1")
df_merge = df_merge.merge(df, how="outer", left_index=True, right_index=True)
df =
|
pd.read_excel('https://www.indec.gob.ar/ftp/cuadros/economia/sh_super_mayoristas.xls', sheet_name="Cuadro 2", skiprows=2, header=[0,1])
|
pandas.read_excel
|
#!/usr/bin/env python
# -*- coding: utf-8 -*-
import pandas as pd
from datetime import datetime, timedelta
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.ticker as tck
import matplotlib.font_manager as fm
import math as m
import matplotlib.dates as mdates
import netCDF4 as nc
from netCDF4 import Dataset
id
import itertools
import datetime
from scipy.stats import ks_2samp
import matplotlib.colors as colors
"Codigo que permite la porderación de la nubosidad por la ponderación de sus horas"
## ----------------------LECTURA DE DATOS DE GOES CH02----------------------- ##
ds = Dataset('/home/nacorreasa/Maestria/Datos_Tesis/GOES/GOES_nc_CREADOS/GOES_VA_C2_2019_0320_0822.nc')
## -----------------INCORPORANDO LOS DATOS DE RADIACIÓN Y DE LOS EXPERIMENTOS----------------- ##
df_P975 = pd.read_csv('/home/nacorreasa/Maestria/Datos_Tesis/Experimentos_Panel/Panel975.txt', sep=',', index_col =0)
df_P350 = pd.read_csv('/home/nacorreasa/Maestria/Datos_Tesis/Experimentos_Panel/Panel350.txt', sep=',', index_col =0)
df_P348 = pd.read_csv('/home/nacorreasa/Maestria/Datos_Tesis/Experimentos_Panel/Panel348.txt', sep=',', index_col =0)
df_P975['Fecha_hora'] = df_P975.index
df_P350['Fecha_hora'] = df_P350.index
df_P348['Fecha_hora'] = df_P348.index
df_P975.index = pd.to_datetime(df_P975.index, format="%Y-%m-%d %H:%M:%S", errors='coerce')
df_P350.index = pd.to_datetime(df_P350.index, format="%Y-%m-%d %H:%M:%S", errors='coerce')
df_P348.index = pd.to_datetime(df_P348.index, format="%Y-%m-%d %H:%M:%S", errors='coerce')
## ----------------ACOTANDO LOS DATOS A VALORES VÁLIDOS---------------- ##
'Como en este caso lo que interesa es la radiacion, para la filtración de los datos, se'
'considerarán los datos de potencia mayores o iguales a 0, los que parecen generarse una'
'hora despues de cuando empieza a incidir la radiación.'
df_P975 = df_P975[(df_P975['radiacion'] > 0) & (df_P975['strength'] >=0) & (df_P975['NI'] >=0)]
df_P350 = df_P350[(df_P350['radiacion'] > 0) & (df_P975['strength'] >=0) & (df_P975['NI'] >=0)]
df_P348 = df_P348[(df_P348['radiacion'] > 0) & (df_P975['strength'] >=0) & (df_P975['NI'] >=0)]
df_P975_h = df_P975.groupby(pd.Grouper(level='fecha_hora', freq='1H')).mean()
df_P350_h = df_P350.groupby(pd.Grouper(level='fecha_hora', freq='1H')).mean()
df_P348_h = df_P348.groupby(pd.Grouper(level='fecha_hora', freq='1H')).mean()
df_P975_h = df_P975_h.between_time('06:00', '17:00')
df_P350_h = df_P350_h.between_time('06:00', '17:00')
df_P348_h = df_P348_h.between_time('06:00', '17:00')
#-----------------------------------------------------------------------------
# Rutas para las fuentes -----------------------------------------------------
prop = fm.FontProperties(fname='/home/nacorreasa/SIATA/Cod_Califi/AvenirLTStd-Heavy.otf' )
prop_1 = fm.FontProperties(fname='/home/nacorreasa/SIATA/Cod_Califi/AvenirLTStd-Book.otf')
prop_2 = fm.FontProperties(fname='/home/nacorreasa/SIATA/Cod_Califi/AvenirLTStd-Black.otf')
## -------------------------------------------------------------------------- ##
Umbral_up_348 = 46.26875
Umbral_down_348 = 22.19776
Umbrales_348 = [Umbral_down_348, Umbral_up_348]
Umbral_up_350 = 49.4412
Umbral_down_350 = 26.4400
Umbrales_350 = [Umbral_down_350, Umbral_up_350]
Umbral_up_975 = 49.4867
Umbral_down_975 = 17.3913
Umbrales_975 = [Umbral_down_975, Umbral_up_975]
lat = ds.variables['lat'][:, :]
lon = ds.variables['lon'][:, :]
Rad = ds.variables['Radiancias'][:, :, :]
## -- Obtener el tiempo para cada valor
tiempo = ds.variables['time']
fechas_horas = nc.num2date(tiempo[:], units=tiempo.units)
for i in range(len(fechas_horas)):
fechas_horas[i] = fechas_horas[i].strftime('%Y-%m-%d %H:%M')
## -- Selección del pixel de la TS y creación de DF
lat_index_975 = np.where((lat[:, 0] > 6.25) & (lat[:, 0] < 6.26))[0][0]
lon_index_975 = np.where((lon[0, :] < -75.58) & (lon[0, :] > -75.59))[0][0]
Rad_pixel_975 = Rad[:, lat_index_975, lon_index_975]
Rad_df_975 = pd.DataFrame()
Rad_df_975['Fecha_Hora'] = fechas_horas
Rad_df_975['Radiacias'] = Rad_pixel_975
Rad_df_975['Fecha_Hora'] = pd.to_datetime(Rad_df_975['Fecha_Hora'], format="%Y-%m-%d %H:%M", errors='coerce')
Rad_df_975.index = Rad_df_975['Fecha_Hora']
Rad_df_975 = Rad_df_975.drop(['Fecha_Hora'], axis=1)
## -- Selección del pixel de la CI
lat_index_350 = np.where((lat[:, 0] > 6.16) & (lat[:, 0] < 6.17))[0][0]
lon_index_350 = np.where((lon[0, :] < -75.64) & (lon[0, :] > -75.65))[0][0]
Rad_pixel_350 = Rad[:, lat_index_350, lon_index_350]
Rad_df_350 = pd.DataFrame()
Rad_df_350['Fecha_Hora'] = fechas_horas
Rad_df_350['Radiacias'] = Rad_pixel_350
Rad_df_350['Fecha_Hora'] = pd.to_datetime(Rad_df_350['Fecha_Hora'], format="%Y-%m-%d %H:%M", errors='coerce')
Rad_df_350.index = Rad_df_350['Fecha_Hora']
Rad_df_350 = Rad_df_350.drop(['Fecha_Hora'], axis=1)
## -- Selección del pixel de la JV
lat_index_348 = np.where((lat[:, 0] > 6.25) & (lat[:, 0] < 6.26))[0][0]
lon_index_348 = np.where((lon[0, :] < -75.54) & (lon[0, :] > -75.55))[0][0]
Rad_pixel_348 = Rad[:, lat_index_348, lon_index_348]
Rad_df_348 = pd.DataFrame()
Rad_df_348['Fecha_Hora'] = fechas_horas
Rad_df_348['Radiacias'] = Rad_pixel_348
Rad_df_348['Fecha_Hora'] = pd.to_datetime(Rad_df_348['Fecha_Hora'], format="%Y-%m-%d %H:%M", errors='coerce')
Rad_df_348.index = Rad_df_348['Fecha_Hora']
Rad_df_348 = Rad_df_348.drop(['Fecha_Hora'], axis=1)
'OJOOOO DESDE ACÁ-----------------------------------------------------------------------------------------'
'Se comenta porque se estaba perdiendo la utilidad de la información cada 10 minutos al suavizar la serie.'
## ------------------------CAMBIANDO LOS DATOS HORARIOS POR LOS ORIGINALES---------------------- ##
Rad_df_348_h = Rad_df_348
Rad_df_350_h = Rad_df_350
Rad_df_975_h = Rad_df_975
## ------------------------------------DATOS HORARIOS DE REFLECTANCIAS------------------------- ##
# Rad_df_348_h = Rad_df_348.groupby(pd.Grouper(freq="H")).mean()
# Rad_df_350_h = Rad_df_350.groupby(pd.Grouper(freq="H")).mean()
# Rad_df_975_h = Rad_df_975.groupby(pd.Grouper(freq="H")).mean()
'OJOOOO HASTA ACÁ-----------------------------------------------------------------------------------------'
Rad_df_348_h = Rad_df_348_h.between_time('06:00', '17:00')
Rad_df_350_h = Rad_df_350_h.between_time('06:00', '17:00')
Rad_df_975_h = Rad_df_975_h.between_time('06:00', '17:00')
## --------------------------------------FDP COMO NP.ARRAY----- ------------------------------ ##
Hist_348 = np.histogram(Rad_df_348_h['Radiacias'].values[~np.isnan(Rad_df_348_h['Radiacias'].values)])
Hist_350 = np.histogram(Rad_df_350_h['Radiacias'].values[~np.isnan(Rad_df_350_h['Radiacias'].values)])
Hist_975 = np.histogram(Rad_df_975_h['Radiacias'].values[~np.isnan(Rad_df_975_h['Radiacias'].values)])
## ---------------------------------FDP COMO GRÁFICA----------------------------------------- ##
fig = plt.figure(figsize=[10, 6])
plt.rc('axes', edgecolor='gray')
ax1 = fig.add_subplot(1, 3, 1)
ax1.spines['top'].set_visible(False)
ax1.spines['right'].set_visible(False)
ax1.hist(Rad_df_348_h['Radiacias'].values[~np.isnan(Rad_df_348_h['Radiacias'].values)], bins='auto', alpha = 0.5)
Umbrales_line1 = [ax1.axvline(x=xc, color='k', linestyle='--') for xc in Umbrales_348]
ax1.set_title(u'Distribución del FR en JV', fontproperties=prop, fontsize = 13)
ax1.set_ylabel(u'Frecuencia', fontproperties=prop_1)
ax1.set_xlabel(u'Reflectancia', fontproperties=prop_1)
ax2 = fig.add_subplot(1, 3, 2)
ax2.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
ax2.hist(Rad_df_350_h['Radiacias'].values[~np.isnan(Rad_df_350_h['Radiacias'].values)], bins='auto', alpha = 0.5)
Umbrales_line2 = [ax2.axvline(x=xc, color='k', linestyle='--') for xc in Umbrales_350]
ax2.set_title(u'Distribución del FR en CI', fontproperties=prop, fontsize = 13)
ax2.set_ylabel(u'Frecuencia', fontproperties=prop_1)
ax2.set_xlabel(u'Reflectancia', fontproperties=prop_1)
ax3 = fig.add_subplot(1, 3, 3)
ax3.spines['top'].set_visible(False)
ax3.spines['right'].set_visible(False)
ax3.hist(Rad_df_975_h['Radiacias'].values[~np.isnan(Rad_df_975_h['Radiacias'].values)], bins='auto', alpha = 0.5)
Umbrales_line3 = [ax3.axvline(x=xc, color='k', linestyle='--') for xc in Umbrales_975]
ax3.set_title(u'Distribución del FR en TS', fontproperties=prop, fontsize = 13)
ax3.set_ylabel(u'Frecuencia', fontproperties=prop_1)
ax3.set_xlabel(u'Reflectancia', fontproperties=prop_1)
plt.savefig('/home/nacorreasa/Escritorio/Figuras/HistoFRUmbral.png')
plt.show()
## -------------------------OBTENER EL DF DEL ESCENARIO DESPEJADO---------------------------- ##
df_348_desp = Rad_df_348_h[Rad_df_348_h['Radiacias'] < Umbral_down_348]
df_350_desp = Rad_df_350_h[Rad_df_350_h['Radiacias'] < Umbral_down_350]
df_975_desp = Rad_df_975_h[Rad_df_975_h['Radiacias'] < Umbral_down_975]
## --------------------------OBTENER EL DF DEL ESCENARIO NUBADO------------------------------ ##
df_348_nuba = Rad_df_348_h[Rad_df_348_h['Radiacias'] > Umbral_up_348]
df_350_nuba = Rad_df_350_h[Rad_df_350_h['Radiacias'] > Umbral_up_350]
df_975_nuba = Rad_df_975_h[Rad_df_975_h['Radiacias'] > Umbral_up_975]
## -------------------------OBTENER LAS HORAS Y FECHAS DESPEJADAS---------------------------- ##
Hora_desp_348 = df_348_desp.index.hour
Fecha_desp_348 = df_348_desp.index.date
Hora_desp_350 = df_350_desp.index.hour
Fecha_desp_350 = df_350_desp.index.date
Hora_desp_975 = df_975_desp.index.hour
Fecha_desp_975 = df_975_desp.index.date
## ----------------------------OBTENER LAS HORAS Y FECHAS NUBADAS---------------------------- ##
Hora_nuba_348 = df_348_nuba.index.hour
Fecha_nuba_348 = df_348_nuba.index.date
Hora_nuba_350 = df_350_nuba.index.hour
Fecha_nuba_350 = df_350_nuba.index.date
Hora_nuba_975 = df_975_nuba.index.hour
Fecha_nuba_975 = df_975_nuba.index.date
## -----------------------------DIBUJAR LOS HISTOGRAMAS DE LAS HORAS ------ ----------------------- #
fig = plt.figure(figsize=[10, 6])
plt.rc('axes', edgecolor='gray')
ax1 = fig.add_subplot(1, 3, 1)
ax1.spines['top'].set_visible(False)
ax1.spines['right'].set_visible(False)
ax1.hist(Hora_desp_348, bins='auto', alpha = 0.5, color = 'orange', label = 'Desp')
ax1.hist(Hora_nuba_348, bins='auto', alpha = 0.5, label = 'Nub')
ax1.set_title(u'Distribución de nubes por horas en JV', fontproperties=prop, fontsize = 8)
ax1.set_ylabel(u'Frecuencia', fontproperties=prop_1)
ax1.set_xlabel(u'Horas', fontproperties=prop_1)
ax1.legend()
ax2 = fig.add_subplot(1, 3, 2)
ax2.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
ax2.hist(Hora_desp_350, bins='auto', alpha = 0.5, color = 'orange', label = 'Desp')
ax2.hist(Hora_nuba_350, bins='auto', alpha = 0.5, label = 'Nub')
ax2.set_title(u'Distribución de nubes por horas en CI', fontproperties=prop, fontsize = 8)
ax2.set_ylabel(u'Frecuencia', fontproperties=prop_1)
ax2.set_xlabel(u'Horas', fontproperties=prop_1)
ax2.legend()
ax3 = fig.add_subplot(1, 3, 3)
ax3.spines['top'].set_visible(False)
ax3.spines['right'].set_visible(False)
ax3.hist(Hora_desp_975, bins='auto', alpha = 0.5, color = 'orange', label = 'Desp')
ax3.hist(Hora_nuba_975, bins='auto', alpha = 0.5, label = 'Nub')
ax3.set_title(u'Distribución de nubes por horas en TS', fontproperties=prop, fontsize = 8)
ax3.set_ylabel(u'Frecuencia', fontproperties=prop_1)
ax3.set_xlabel(u'Horas', fontproperties=prop_1)
ax3.legend()
plt.savefig('/home/nacorreasa/Escritorio/Figuras/HistoNubaDesp.png')
plt.show()
##----------ENCONTRANDO LAS RADIACIONES CORRESPONDIENTES A LAS HORAS NUBOSAS----------##
df_FH_nuba_348 = pd.DataFrame()
df_FH_nuba_348 ['Fechas'] = Fecha_nuba_348
df_FH_nuba_348 ['Horas'] = Hora_nuba_348
df_FH_nuba_350 = pd.DataFrame()
df_FH_nuba_350 ['Fechas'] = Fecha_nuba_350
df_FH_nuba_350 ['Horas'] = Hora_nuba_350
df_FH_nuba_975 = pd.DataFrame()
df_FH_nuba_975 ['Fechas'] = Fecha_nuba_975
df_FH_nuba_975 ['Horas'] = Hora_nuba_975
df_FH_nuba_348_groupH = df_FH_nuba_348.groupby('Horas')['Fechas'].unique()
df_nuba_348_groupH = pd.DataFrame(df_FH_nuba_348_groupH[df_FH_nuba_348_groupH.apply(lambda x: len(x)>1)]) ##NO entiendo bien acá que se está haciendo
df_FH_nuba_350_groupH = df_FH_nuba_350.groupby('Horas')['Fechas'].unique()
df_nuba_350_groupH = pd.DataFrame(df_FH_nuba_350_groupH[df_FH_nuba_350_groupH.apply(lambda x: len(x)>1)])
df_FH_nuba_975_groupH = df_FH_nuba_975.groupby('Horas')['Fechas'].unique()
df_nuba_975_groupH = pd.DataFrame(df_FH_nuba_975_groupH[df_FH_nuba_975_groupH.apply(lambda x: len(x)>1)])
c = np.arange(6, 18, 1)
Sk_Nuba_stat_975 = {}
Sk_Nuba_pvalue_975 = {}
Composites_Nuba_975 = {}
for i in df_FH_nuba_975_groupH.index:
H = str(i)
if len(df_FH_nuba_975_groupH.loc[i]) == 1 :
list = df_P975_h[df_P975_h.index.date == df_FH_nuba_975_groupH.loc[i][0]]['radiacion'].values
list_sk_stat = np.ones(12)*np.nan
list_sk_pvalue = np.ones(12)*np.nan
elif len(df_FH_nuba_975_groupH.loc[i]) > 1 :
temporal = pd.DataFrame()
for j in range(len(df_FH_nuba_975_groupH.loc[i])):
temporal = temporal.append(pd.DataFrame(df_P975_h[df_P975_h.index.date == df_FH_nuba_975_groupH.loc[i][j]]['radiacion']))
stat_975 = []
pvalue_975 = []
for k in c:
temporal_sk = temporal[temporal.index.hour == k].radiacion.values
Rad_sk = df_P975_h['radiacion'][df_P975_h.index.hour == k].values
try:
SK = ks_2samp(temporal_sk,Rad_sk)
stat_975.append(SK[0])
pvalue_975.append(SK[1])
except ValueError:
stat_975.append(np.nan)
pvalue_975.append(np.nan)
temporal_CD = temporal.groupby(by=[temporal.index.hour]).mean()
list = temporal_CD['radiacion'].values
list_sk_stat = stat_975
list_sk_pvalue = pvalue_975
Composites_Nuba_975[H] = list
Sk_Nuba_stat_975 [H] = list_sk_stat
Sk_Nuba_pvalue_975 [H] = list_sk_pvalue
del H
Comp_Nuba_975_df = pd.DataFrame(Composites_Nuba_975, index = c)
Sk_Nuba_stat_975_df = pd.DataFrame(Sk_Nuba_stat_975, index = c)
Sk_Nuba_pvalue_975_df = pd.DataFrame(Sk_Nuba_pvalue_975, index = c)
Sk_Nuba_stat_350 = {}
Sk_Nuba_pvalue_350 = {}
Composites_Nuba_350 = {}
for i in df_FH_nuba_350_groupH.index:
H = str(i)
if len(df_FH_nuba_350_groupH.loc[i]) == 1 :
list = df_P350_h[df_P350_h.index.date == df_FH_nuba_350_groupH.loc[i][0]]['radiacion'].values
list_sk_stat = np.ones(12)*np.nan
list_sk_pvalue = np.ones(12)*np.nan
elif len(df_FH_nuba_350_groupH.loc[i]) > 1 :
temporal = pd.DataFrame()
for j in range(len(df_FH_nuba_350_groupH.loc[i])):
temporal = temporal.append(pd.DataFrame(df_P350_h[df_P350_h.index.date == df_FH_nuba_350_groupH.loc[i][j]]['radiacion']))
stat_350 = []
pvalue_350 = []
for k in c:
temporal_sk = temporal[temporal.index.hour == k].radiacion.values
Rad_sk = df_P350_h['radiacion'][df_P350_h.index.hour == k].values
try:
SK = ks_2samp(temporal_sk,Rad_sk)
stat_350.append(SK[0])
pvalue_350.append(SK[1])
except ValueError:
stat_350.append(np.nan)
pvalue_350.append(np.nan)
temporal_CD = temporal.groupby(by=[temporal.index.hour]).mean()
list = temporal_CD['radiacion'].values
list_sk_stat = stat_350
list_sk_pvalue = pvalue_350
Composites_Nuba_350[H] = list
Sk_Nuba_stat_350 [H] = list_sk_stat
Sk_Nuba_pvalue_350 [H] = list_sk_pvalue
del H
Comp_Nuba_350_df = pd.DataFrame(Composites_Nuba_350, index = c)
Sk_Nuba_stat_350_df = pd.DataFrame(Sk_Nuba_stat_350, index = c)
Sk_Nuba_pvalue_350_df = pd.DataFrame(Sk_Nuba_pvalue_350, index = c)
Sk_Nuba_stat_348 = {}
Sk_Nuba_pvalue_348 = {}
Composites_Nuba_348 = {}
for i in df_FH_nuba_348_groupH.index:
H = str(i)
if len(df_FH_nuba_348_groupH.loc[i]) == 1 :
list = df_P348_h[df_P348_h.index.date == df_FH_nuba_348_groupH.loc[i][0]]['radiacion'].values
list_sk_stat = np.ones(12)*np.nan
list_sk_pvalue = np.ones(12)*np.nan
elif len(df_FH_nuba_348_groupH.loc[i]) > 1 :
temporal = pd.DataFrame()
for j in range(len(df_FH_nuba_348_groupH.loc[i])):
temporal = temporal.append(pd.DataFrame(df_P348_h[df_P348_h.index.date == df_FH_nuba_348_groupH.loc[i][j]]['radiacion']))
stat_348 = []
pvalue_348 = []
for k in c:
temporal_sk = temporal[temporal.index.hour == k].radiacion.values
Rad_sk = df_P348_h['radiacion'][df_P348_h.index.hour == k].values
try:
SK = ks_2samp(temporal_sk,Rad_sk)
stat_348.append(SK[0])
pvalue_348.append(SK[1])
except ValueError:
stat_348.append(np.nan)
pvalue_348.append(np.nan)
temporal_CD = temporal.groupby(by=[temporal.index.hour]).mean()
list = temporal_CD['radiacion'].values
list_sk_stat = stat_348
list_sk_pvalue = pvalue_348
Composites_Nuba_348[H] = list
Sk_Nuba_stat_348 [H] = list_sk_stat
Sk_Nuba_pvalue_348 [H] = list_sk_pvalue
del H
Comp_Nuba_348_df = pd.DataFrame(Composites_Nuba_348, index = c)
Sk_Nuba_stat_348_df = pd.DataFrame(Sk_Nuba_stat_348, index = c)
Sk_Nuba_pvalue_348_df = pd.DataFrame(Sk_Nuba_pvalue_348, index = c)
##----------ENCONTRANDO LAS RADIACIONES CORRESPONDIENTES A LAS HORAS DESPEJADAS----------##
df_FH_desp_348 = pd.DataFrame()
df_FH_desp_348 ['Fechas'] = Fecha_desp_348
df_FH_desp_348 ['Horas'] = Hora_desp_348
df_FH_desp_350 = pd.DataFrame()
df_FH_desp_350 ['Fechas'] = Fecha_desp_350
df_FH_desp_350 ['Horas'] = Hora_desp_350
df_FH_desp_975 = pd.DataFrame()
df_FH_desp_975 ['Fechas'] = Fecha_desp_975
df_FH_desp_975 ['Horas'] = Hora_desp_975
df_FH_desp_348_groupH = df_FH_desp_348.groupby('Horas')['Fechas'].unique()
df_desp_348_groupH = pd.DataFrame(df_FH_desp_348_groupH[df_FH_desp_348_groupH.apply(lambda x: len(x)>1)]) ##NO entiendo bien acá que se está haciendo
df_FH_desp_350_groupH = df_FH_desp_350.groupby('Horas')['Fechas'].unique()
df_desp_350_groupH = pd.DataFrame(df_FH_desp_350_groupH[df_FH_desp_350_groupH.apply(lambda x: len(x)>1)])
df_FH_desp_975_groupH = df_FH_desp_975.groupby('Horas')['Fechas'].unique()
df_desp_975_groupH = pd.DataFrame(df_FH_desp_975_groupH[df_FH_desp_975_groupH.apply(lambda x: len(x)>1)])
Sk_Desp_stat_975 = {}
Sk_Desp_pvalue_975 = {}
Composites_Desp_975 = {}
for i in df_FH_desp_975_groupH.index:
H = str(i)
if len(df_FH_desp_975_groupH.loc[i]) == 1 :
list = df_P975_h[df_P975_h.index.date == df_FH_desp_975_groupH.loc[i][0]]['radiacion'].values
list_sk_stat = np.ones(12)*np.nan
list_sk_pvalue = np.ones(12)*np.nan
elif len(df_FH_desp_975_groupH.loc[i]) > 1 :
temporal = pd.DataFrame()
for j in range(len(df_FH_desp_975_groupH.loc[i])):
temporal = temporal.append(pd.DataFrame(df_P975_h[df_P975_h.index.date == df_FH_desp_975_groupH.loc[i][j]]['radiacion']))
stat_975 = []
pvalue_975 = []
for k in c:
temporal_sk = temporal[temporal.index.hour == k].radiacion.values
Rad_sk = df_P975_h['radiacion'][df_P975_h.index.hour == k].values
try:
SK = ks_2samp(temporal_sk,Rad_sk)
stat_975.append(SK[0])
pvalue_975.append(SK[1])
except ValueError:
stat_975.append(np.nan)
pvalue_975.append(np.nan)
temporal_CD = temporal.groupby(by=[temporal.index.hour]).mean()
list = temporal_CD['radiacion'].values
list_sk_stat = stat_975
list_sk_pvalue = pvalue_975
Composites_Desp_975[H] = list
Sk_Desp_stat_975 [H] = list_sk_stat
Sk_Desp_pvalue_975 [H] = list_sk_pvalue
del H
Comp_Desp_975_df = pd.DataFrame(Composites_Desp_975, index = c)
Sk_Desp_stat_975_df = pd.DataFrame(Sk_Desp_stat_975, index = c)
Sk_Desp_pvalue_975_df = pd.DataFrame(Sk_Desp_pvalue_975, index = c)
Sk_Desp_stat_350 = {}
Sk_Desp_pvalue_350 = {}
Composites_Desp_350 = {}
for i in df_FH_desp_350_groupH.index:
H = str(i)
if len(df_FH_desp_350_groupH.loc[i]) == 1 :
list = df_P350_h[df_P350_h.index.date == df_FH_desp_350_groupH.loc[i][0]]['radiacion'].values
list_sk_stat = np.ones(12)*np.nan
list_sk_pvalue = np.ones(12)*np.nan
elif len(df_FH_desp_350_groupH.loc[i]) > 1 :
temporal = pd.DataFrame()
for j in range(len(df_FH_desp_350_groupH.loc[i])):
temporal = temporal.append(pd.DataFrame(df_P350_h[df_P350_h.index.date == df_FH_desp_350_groupH.loc[i][j]]['radiacion']))
stat_350 = []
pvalue_350 = []
for k in c:
temporal_sk = temporal[temporal.index.hour == k].radiacion.values
Rad_sk = df_P350_h['radiacion'][df_P350_h.index.hour == k].values
try:
SK = ks_2samp(temporal_sk,Rad_sk)
stat_350.append(SK[0])
pvalue_350.append(SK[1])
except ValueError:
stat_350.append(np.nan)
pvalue_350.append(np.nan)
temporal_CD = temporal.groupby(by=[temporal.index.hour]).mean()
list = temporal_CD['radiacion'].values
list_sk_stat = stat_350
list_sk_pvalue = pvalue_350
Composites_Desp_350[H] = list
Sk_Desp_stat_350 [H] = list_sk_stat
Sk_Desp_pvalue_350 [H] = list_sk_pvalue
del H
Comp_Desp_350_df = pd.DataFrame(Composites_Desp_350, index = c)
Sk_Desp_stat_350_df = pd.DataFrame(Sk_Desp_stat_350, index = c)
Sk_Desp_pvalue_350_df = pd.DataFrame(Sk_Desp_pvalue_350, index = c)
Sk_Desp_stat_348 = {}
Sk_Desp_pvalue_348 = {}
Composites_Desp_348 = {}
for i in df_FH_desp_348_groupH.index:
H = str(i)
if len(df_FH_desp_348_groupH.loc[i]) == 1 :
list = df_P348_h[df_P348_h.index.date == df_FH_desp_348_groupH.loc[i][0]]['radiacion'].values
list_sk_stat = np.ones(12)*np.nan
list_sk_pvalue = np.ones(12)*np.nan
elif len(df_FH_desp_348_groupH.loc[i]) > 1 :
temporal = pd.DataFrame()
for j in range(len(df_FH_desp_348_groupH.loc[i])):
temporal = temporal.append(pd.DataFrame(df_P348_h[df_P348_h.index.date == df_FH_desp_348_groupH.loc[i][j]]['radiacion']))
stat_348 = []
pvalue_348 = []
for k in c:
temporal_sk = temporal[temporal.index.hour == k].radiacion.values
Rad_sk = df_P348_h['radiacion'][df_P348_h.index.hour == k].values
try:
SK = ks_2samp(temporal_sk,Rad_sk)
stat_348.append(SK[0])
pvalue_348.append(SK[1])
except ValueError:
stat_348.append(np.nan)
pvalue_348.append(np.nan)
temporal_CD = temporal.groupby(by=[temporal.index.hour]).mean()
list = temporal_CD['radiacion'].values
list_sk_stat = stat_348
list_sk_pvalue = pvalue_348
Composites_Desp_348[H] = list
Sk_Desp_stat_348 [H] = list_sk_stat
Sk_Desp_pvalue_348 [H] = list_sk_pvalue
del H
Comp_Desp_348_df = pd.DataFrame(Composites_Desp_348, index = c)
Sk_Desp_stat_348_df = pd.DataFrame(Sk_Desp_stat_348, index = c)
Sk_Desp_pvalue_348_df = pd.DataFrame(Sk_Desp_pvalue_348, index = c)
##-------------------ESTANDARIZANDO LAS FORMAS DE LOS DATAFRAMES A LAS HORAS CASO DESPEJADO----------------##
Comp_Desp_348_df = Comp_Desp_348_df[(Comp_Desp_348_df.index >= 6)&(Comp_Desp_348_df.index <18)]
Comp_Desp_350_df = Comp_Desp_350_df[(Comp_Desp_350_df.index >= 6)&(Comp_Desp_350_df.index <18)]
Comp_Desp_975_df = Comp_Desp_975_df[(Comp_Desp_975_df.index >= 6)&(Comp_Desp_975_df.index <18)]
s = [str(i) for i in Comp_Nuba_348_df.index.values]
ListNan = np.empty((1,len(Comp_Desp_348_df)))
ListNan [:] = np.nan
def convert(set):
return [*set, ]
a_Desp_348 = convert(set(s).difference(Comp_Desp_348_df.columns.values))
a_Desp_348.sort(key=int)
if len(a_Desp_348) > 0:
idx = [i for i,x in enumerate(s) if x in a_Desp_348]
for i in range(len(a_Desp_348)):
Comp_Desp_348_df.insert(loc = idx[i], column = a_Desp_348[i], value=ListNan[0])
del idx
a_Desp_350 = convert(set(s).difference(Comp_Desp_350_df.columns.values))
a_Desp_350.sort(key=int)
if len(a_Desp_350) > 0:
idx = [i for i,x in enumerate(s) if x in a_Desp_350]
for i in range(len(a_Desp_350)):
Comp_Desp_350_df.insert(loc = idx[i], column = a_Desp_350[i], value=ListNan[0])
del idx
a_Desp_975 = convert(set(s).difference(Comp_Desp_975_df.columns.values))
a_Desp_975.sort(key=int)
if len(a_Desp_975) > 0:
idx = [i for i,x in enumerate(s) if x in a_Desp_975]
for i in range(len(a_Desp_975)):
Comp_Desp_975_df.insert(loc = idx[i], column = a_Desp_975[i], value=ListNan[0])
del idx
s = [str(i) for i in Comp_Desp_348_df.index.values]
Comp_Desp_348_df = Comp_Desp_348_df[s]
Comp_Desp_350_df = Comp_Desp_350_df[s]
Comp_Desp_975_df = Comp_Desp_975_df[s]
##-------------------ESTANDARIZANDO LAS FORMAS DE LOS DATAFRAMES A LAS HORAS CASO NUBADO----------------##
Comp_Nuba_348_df = Comp_Nuba_348_df[(Comp_Nuba_348_df.index >= 6)&(Comp_Nuba_348_df.index <18)]
Comp_Nuba_350_df = Comp_Nuba_350_df[(Comp_Nuba_350_df.index >= 6)&(Comp_Nuba_350_df.index <18)]
Comp_Nuba_975_df = Comp_Nuba_975_df[(Comp_Nuba_975_df.index >= 6)&(Comp_Nuba_975_df.index <18)]
s = [str(i) for i in Comp_Nuba_348_df.index.values]
ListNan = np.empty((1,len(Comp_Nuba_348_df)))
ListNan [:] = np.nan
def convert(set):
return [*set, ]
a_Nuba_348 = convert(set(s).difference(Comp_Nuba_348_df.columns.values))
a_Nuba_348.sort(key=int)
if len(a_Nuba_348) > 0:
idx = [i for i,x in enumerate(s) if x in a_Nuba_348]
for i in range(len(a_Nuba_348)):
Comp_Nuba_348_df.insert(loc = idx[i], column = a_Nuba_348[i], value=ListNan[0])
del idx
a_Nuba_350 = convert(set(s).difference(Comp_Nuba_350_df.columns.values))
a_Nuba_350.sort(key=int)
if len(a_Nuba_350) > 0:
idx = [i for i,x in enumerate(s) if x in a_Nuba_350]
for i in range(len(a_Nuba_350)):
Comp_Nuba_350_df.insert(loc = idx[i], column = a_Nuba_350[i], value=ListNan[0])
del idx
a_Nuba_975 = convert(set(s).difference(Comp_Nuba_975_df.columns.values))
a_Nuba_975.sort(key=int)
if len(a_Nuba_975) > 0:
idx = [i for i,x in enumerate(s) if x in a_Nuba_975]
for i in range(len(a_Nuba_975)):
Comp_Nuba_975_df.insert(loc = idx[i], column = a_Nuba_975[i], value=ListNan[0])
del idx
Comp_Nuba_348_df = Comp_Nuba_348_df[s]
Comp_Nuba_350_df = Comp_Nuba_350_df[s]
Comp_Nuba_975_df = Comp_Nuba_975_df[s]
##-------------------CONTEO DE LA CANTIDAD DE DÍAS CONSIDERADOS NUBADOS Y DESPEJADOS----------------##
Cant_Days_Nuba_348 = []
for i in range(len(s)):
try:
Cant_Days_Nuba_348.append(len(df_FH_nuba_348_groupH[df_FH_nuba_348_groupH .index == int(s[i])].values[0]))
except IndexError:
Cant_Days_Nuba_348.append(0)
Cant_Days_Nuba_350 = []
for i in range(len(s)):
try:
Cant_Days_Nuba_350.append(len(df_FH_nuba_350_groupH[df_FH_nuba_350_groupH .index == int(s[i])].values[0]))
except IndexError:
Cant_Days_Nuba_350.append(0)
Cant_Days_Nuba_975 = []
for i in range(len(s)):
try:
Cant_Days_Nuba_975.append(len(df_FH_nuba_975_groupH[df_FH_nuba_975_groupH .index == int(s[i])].values[0]))
except IndexError:
Cant_Days_Nuba_975.append(0)
Cant_Days_Desp_348 = []
for i in range(len(s)):
try:
Cant_Days_Desp_348.append(len(df_FH_desp_348_groupH[df_FH_desp_348_groupH .index == int(s[i])].values[0]))
except IndexError:
Cant_Days_Desp_348.append(0)
Cant_Days_Desp_350 = []
for i in range(len(s)):
try:
Cant_Days_Desp_350.append(len(df_FH_desp_350_groupH[df_FH_desp_350_groupH .index == int(s[i])].values[0]))
except IndexError:
Cant_Days_Desp_350.append(0)
Cant_Days_Desp_975 = []
for i in range(len(s)):
try:
Cant_Days_Desp_975.append(len(df_FH_desp_975_groupH[df_FH_desp_975_groupH .index == int(s[i])].values[0]))
except IndexError:
Cant_Days_Desp_975.append(0)
##-------------------AJUSTADO LOS DATAFRAMES DE LOS ESTADÍSTICOS Y DEL VALOR P----------------##
for i in range(len(c)):
if str(c[i]) not in Sk_Desp_pvalue_975_df.columns:
Sk_Desp_pvalue_975_df.insert(int(c[i]-6), str(c[i]), np.ones(12)*np.nan)
if str(c[i]) not in Sk_Desp_pvalue_350_df.columns:
Sk_Desp_pvalue_350_df.insert(int(c[i]-6), str(c[i]), np.ones(12)*np.nan)
if str(c[i]) not in Sk_Desp_pvalue_348_df.columns:
Sk_Desp_pvalue_348_df.insert(int(c[i]-6), str(c[i]), np.ones(12)*np.nan)
if str(c[i]) not in Sk_Nuba_pvalue_350_df.columns:
Sk_Nuba_pvalue_350_df.insert(int(c[i]-6), str(c[i]), np.ones(12)*np.nan)
if str(c[i]) not in Sk_Nuba_pvalue_348_df.columns:
Sk_Nuba_pvalue_348_df.insert(int(c[i]-6), str(c[i]), np.ones(12)*np.nan)
if str(c[i]) not in Sk_Nuba_pvalue_975_df.columns:
Sk_Nuba_pvalue_975_df.insert(int(c[i]-6), str(c[i]), np.ones(12)*np.nan)
Significancia = 0.05
for i in c:
Sk_Desp_pvalue_348_df.loc[Sk_Desp_pvalue_348_df[str(i)]< Significancia, str(i)] = 100
Sk_Desp_pvalue_350_df.loc[Sk_Desp_pvalue_350_df[str(i)]< Significancia, str(i)] = 100
Sk_Desp_pvalue_975_df.loc[Sk_Desp_pvalue_975_df[str(i)]< Significancia, str(i)] = 100
Sk_Nuba_pvalue_348_df.loc[Sk_Nuba_pvalue_348_df[str(i)]< Significancia, str(i)] = 100
Sk_Nuba_pvalue_350_df.loc[Sk_Nuba_pvalue_350_df[str(i)]< Significancia, str(i)] = 100
Sk_Nuba_pvalue_975_df.loc[Sk_Nuba_pvalue_975_df[str(i)]< Significancia, str(i)] = 100
row_Desp_348 = []
col_Desp_348 = []
for row in range(Sk_Desp_pvalue_348_df.shape[0]):
for col in range(Sk_Desp_pvalue_348_df.shape[1]):
if Sk_Desp_pvalue_348_df.get_value((row+6),str(col+6)) == 100:
row_Desp_348.append(row)
col_Desp_348.append(col)
#print(row+6, col+6)
row_Desp_350 = []
col_Desp_350 = []
for row in range(Sk_Desp_pvalue_350_df.shape[0]):
for col in range(Sk_Desp_pvalue_350_df.shape[1]):
if Sk_Desp_pvalue_350_df.get_value((row+6),str(col+6)) == 100:
row_Desp_350.append(row)
col_Desp_350.append(col)
row_Desp_975 = []
col_Desp_975 = []
for row in range(Sk_Desp_pvalue_975_df.shape[0]):
for col in range(Sk_Desp_pvalue_975_df.shape[1]):
if Sk_Desp_pvalue_975_df.get_value((row+6),str(col+6)) == 100:
row_Desp_975.append(row)
col_Desp_975.append(col)
row_Nuba_348 = []
col_Nuba_348 = []
for row in range(Sk_Nuba_pvalue_348_df.shape[0]):
for col in range(Sk_Nuba_pvalue_348_df.shape[1]):
if Sk_Nuba_pvalue_348_df.get_value((row+6),str(col+6)) == 100:
row_Nuba_348.append(row)
col_Nuba_348.append(col)
#print(row+6, col+6)
row_Nuba_350 = []
col_Nuba_350 = []
for row in range(Sk_Nuba_pvalue_350_df.shape[0]):
for col in range(Sk_Nuba_pvalue_350_df.shape[1]):
if Sk_Nuba_pvalue_350_df.get_value((row+6),str(col+6)) == 100:
row_Nuba_350.append(row)
col_Nuba_350.append(col)
row_Nuba_975 = []
col_Nuba_975 = []
for row in range(Sk_Nuba_pvalue_975_df.shape[0]):
for col in range(Sk_Nuba_pvalue_975_df.shape[1]):
if Sk_Nuba_pvalue_975_df.get_value((row+6),str(col+6)) == 100:
row_Nuba_975.append(row)
col_Nuba_975.append(col)
##-------------------GRÁFICO DEL COMPOSITE NUBADO DE LA RADIACIÓN PARA CADA PUNTO Y LA CANT DE DÍAS----------------##
plt.close("all")
fig = plt.figure(figsize=(10., 8.),facecolor='w',edgecolor='w')
ax1=fig.add_subplot(2,3,1)
mapa = ax1.imshow(Comp_Nuba_348_df, interpolation = 'none', cmap = 'Spectral_r')
ax1.set_yticks(range(0,12), minor=False)
ax1.set_yticklabels(s, minor=False)
ax1.set_xticks(range(0,12), minor=False)
ax1.set_xticklabels(s, minor=False, rotation = 20)
ax1.set_xlabel('Hora', fontsize=10, fontproperties = prop_1)
ax1.set_ylabel('Hora', fontsize=10, fontproperties = prop_1)
ax1.scatter(range(0,12),range(0,12), marker='x', facecolor = 'k', edgecolor = 'k', linewidth='1.', s=30)
ax1.set_title(' x = Horas nubadas en JV', loc = 'center', fontsize=9)
ax2=fig.add_subplot(2,3,2)
mapa = ax2.imshow(Comp_Nuba_350_df, interpolation = 'none', cmap = 'Spectral_r')
ax2.set_yticks(range(0,12), minor=False)
ax2.set_yticklabels(s, minor=False)
ax2.set_xticks(range(0,12), minor=False)
ax2.set_xticklabels(s, minor=False, rotation = 20)
ax2.set_xlabel('Hora', fontsize=10, fontproperties = prop_1)
ax2.set_ylabel('Hora', fontsize=10, fontproperties = prop_1)
ax2.scatter(range(0,12),range(0,12), marker='x', facecolor = 'k', edgecolor = 'k', linewidth='1.', s=30)
ax2.set_title(' x = Horas nubadas en CI', loc = 'center', fontsize=9)
ax3 = fig.add_subplot(2,3,3)
mapa = ax3.imshow(Comp_Nuba_975_df, interpolation = 'none', cmap = 'Spectral_r')
ax3.set_yticks(range(0,12), minor=False)
ax3.set_yticklabels(s, minor=False)
ax3.set_xticks(range(0,12), minor=False)
ax3.set_xticklabels(s, minor=False, rotation = 20)
ax3.set_xlabel('Hora', fontsize=10, fontproperties = prop_1)
ax3.set_ylabel('Hora', fontsize=10, fontproperties = prop_1)
ax3.scatter(range(0,12),range(0,12), marker='x', facecolor = 'k', edgecolor = 'k', linewidth='1.', s=30)
ax3.set_title(' x = Horas nubadas en TS', loc = 'center', fontsize=9)
cbar_ax = fig.add_axes([0.11, 0.93, 0.78, 0.008])
cbar = fig.colorbar(mapa, cax=cbar_ax, orientation='horizontal', format="%.2f")
cbar.set_label(u"Insensidad de la radiación", fontsize=8, fontproperties=prop)
ax4 = fig.add_subplot(2,3,4)
ax4.spines['top'].set_visible(False)
ax4.spines['right'].set_visible(False)
ax4.bar(np.array(s), Cant_Days_Nuba_348, color='orange', align='center', alpha=0.5)
ax4.set_xlabel(u'Hora', fontproperties = prop_1)
ax4.set_ylabel(r"Cantidad de días", fontproperties = prop_1)
ax4.set_xticks(range(0,12), minor=False)
ax4.set_xticklabels(s, minor=False, rotation = 20)
ax4.set_title(u' Cantidad de días en JV', loc = 'center', fontsize=9)
ax5 = fig.add_subplot(2,3,5)
ax5.spines['top'].set_visible(False)
ax5.spines['right'].set_visible(False)
ax5.bar(np.array(s), Cant_Days_Nuba_350, color='orange', align='center', alpha=0.5)
ax5.set_xlabel(u'Hora', fontproperties = prop_1)
ax5.set_ylabel(r"Cantidad de días", fontproperties = prop_1)
ax5.set_xticks(range(0,12), minor=False)
ax5.set_xticklabels(s, minor=False, rotation = 20)
ax5.set_title(u' Cantidad de días en CI', loc = 'center', fontsize=9)
ax6 = fig.add_subplot(2,3,6)
ax6.spines['top'].set_visible(False)
ax6.spines['right'].set_visible(False)
ax6.bar(np.array(s), Cant_Days_Nuba_975, color='orange', align='center', alpha=0.5)
ax6.set_xlabel(u'Hora', fontproperties = prop_1)
ax6.set_ylabel(r"Cantidad de días", fontproperties = prop_1)
ax6.set_xticks(range(0,12), minor=False)
ax6.set_xticklabels(s, minor=False, rotation = 20)
ax6.set_title(u' Cantidad de días en TS', loc = 'center', fontsize=9)
plt.subplots_adjust(wspace=0.3, hspace=0.3)
plt.savefig('/home/nacorreasa/Escritorio/Figuras/Composites_Nuba_Cant_Dias_R.png')
plt.show()
##-------------------GRÁFICO DEL COMPOSITE DESPEJADO DE LA RADIACIÓN PARA CADA PUNTO Y LA CANT DE DÍAS----------------##
plt.close("all")
fig = plt.figure(figsize=(10., 8.),facecolor='w',edgecolor='w')
ax1=fig.add_subplot(2,3,1)
mapa = ax1.imshow(Comp_Desp_348_df, interpolation = 'none', cmap = 'Spectral_r')
ax1.set_yticks(range(0,12), minor=False)
ax1.set_yticklabels(s, minor=False)
ax1.set_xticks(range(0,12), minor=False)
ax1.set_xticklabels(s, minor=False, rotation = 20)
ax1.set_xlabel('Hora', fontsize=10, fontproperties = prop_1)
ax1.set_ylabel('Hora', fontsize=10, fontproperties = prop_1)
ax1.scatter(range(0,12),range(0,12), marker='x', facecolor = 'k', edgecolor = 'k', linewidth='1.', s=30)
ax1.set_title(' x = Horas despejadas en JV', loc = 'center', fontsize=9)
ax2=fig.add_subplot(2,3,2)
mapa = ax2.imshow(Comp_Desp_350_df, interpolation = 'none', cmap = 'Spectral_r')
ax2.set_yticks(range(0,12), minor=False)
ax2.set_yticklabels(s, minor=False)
ax2.set_xticks(range(0,12), minor=False)
ax2.set_xticklabels(s, minor=False, rotation = 20)
ax2.set_xlabel('Hora', fontsize=10, fontproperties = prop_1)
ax2.set_ylabel('Hora', fontsize=10, fontproperties = prop_1)
ax2.scatter(range(0,12),range(0,12), marker='x', facecolor = 'k', edgecolor = 'k', linewidth='1.', s=30)
ax2.set_title(' x = Horas despejadas en CI', loc = 'center', fontsize=9)
ax3 = fig.add_subplot(2,3,3)
mapa = ax3.imshow(Comp_Desp_975_df, interpolation = 'none', cmap = 'Spectral_r')
ax3.set_yticks(range(0,12), minor=False)
ax3.set_yticklabels(s, minor=False)
ax3.set_xticks(range(0,12), minor=False)
ax3.set_xticklabels(s, minor=False, rotation = 20)
ax3.set_xlabel('Hora', fontsize=10, fontproperties = prop_1)
ax3.set_ylabel('Hora', fontsize=10, fontproperties = prop_1)
ax3.scatter(range(0,12),range(0,12), marker='x', facecolor = 'k', edgecolor = 'k', linewidth='1.', s=30)
ax3.set_title(' x = Horas despejadas en TS', loc = 'center', fontsize=9)
cbar_ax = fig.add_axes([0.11, 0.93, 0.78, 0.008])
cbar = fig.colorbar(mapa, cax=cbar_ax, orientation='horizontal', format="%.2f")
cbar.set_label(u"Insensidad de la radiación", fontsize=8, fontproperties=prop)
ax4 = fig.add_subplot(2,3,4)
ax4.spines['top'].set_visible(False)
ax4.spines['right'].set_visible(False)
ax4.bar(np.array(s), Cant_Days_Desp_348, color='orange', align='center', alpha=0.5)
ax4.set_xlabel(u'Hora', fontproperties = prop_1)
ax4.set_ylabel(r"Cantidad de días", fontproperties = prop_1)
ax4.set_xticks(range(0,12), minor=False)
ax4.set_xticklabels(s, minor=False, rotation = 20)
ax4.set_title(u' Cantidad de días en JV', loc = 'center', fontsize=9)
ax5 = fig.add_subplot(2,3,5)
ax5.spines['top'].set_visible(False)
ax5.spines['right'].set_visible(False)
ax5.bar(np.array(s), Cant_Days_Desp_350, color='orange', align='center', alpha=0.5)
ax5.set_xlabel(u'Hora', fontproperties = prop_1)
ax5.set_ylabel(r"Cantidad de días", fontproperties = prop_1)
ax5.set_xticks(range(0,12), minor=False)
ax5.set_xticklabels(s, minor=False, rotation = 20)
ax5.set_title(u' Cantidad de días en CI', loc = 'center', fontsize=9)
ax6 = fig.add_subplot(2,3,6)
ax6.spines['top'].set_visible(False)
ax6.spines['right'].set_visible(False)
ax6.bar(np.array(s), Cant_Days_Desp_975, color='orange', align='center', alpha=0.5)
ax6.set_xlabel(u'Hora', fontproperties = prop_1)
ax6.set_ylabel(r"Cantidad de días", fontproperties = prop_1)
ax6.set_xticks(range(0,12), minor=False)
ax6.set_xticklabels(s, minor=False, rotation = 20)
ax6.set_title(u' Cantidad de días en TS', loc = 'center', fontsize=9)
plt.subplots_adjust(wspace=0.3, hspace=0.3)
plt.savefig('/home/nacorreasa/Escritorio/Figuras/Composites_Desp_Cant_Dias_R.png')
plt.title(u'Composites caso nubado', fontproperties=prop, fontsize = 8)
plt.show()
##--------------------------TOTAL DE DÍAS DE REGISTRO---------------------##
Total_dias_348 = len(Rad_df_348.groupby(pd.Grouper(freq="D")).mean())
Total_dias_350 = len(Rad_df_350.groupby(pd.Grouper(freq="D")).mean())
Total_dias_975 = len(Rad_df_975.groupby(
|
pd.Grouper(freq="D")
|
pandas.Grouper
|
# -*- coding: utf-8 -*-
import re,pandas as pd
import os
pt=('\\txt')
pathDir=os.listdir(pt)
# csv_pt=os.getcwd()+"\\csv"
# if not os.path.isdir(csv_pt): # 如果 _path 目录不存在,则创建
# os.makedirs(csv_pt)
cols=['工单编号','上级工单编号','项目编号','工单描述','上级工单描述','施工单位','合同号','计划服务费','开工日期','完工日期','作业类型','通知单创建','通知单批准','计划','待审','下达','验收确认','完工确认','完工时间','打印者','打印日期','工序号','工作中心','控制码','工序内容','计划量','签证','物料编码','物料描述','单位计划量','出库量','签证']
l=[]
x=0
l1=[]
dfb = pd.DataFrame(columns=['工单编号', '上级工单编号', '项目编号', '工单描述', '上级工单描述', '施工单位', '合同号', '计划服务费','开工日期', '完工日期', '作业类型', '通知单创建', '通知单批准', '计划', '待审', '下达', '验收确认','完工确认', '完工时间', '打印者', '打印日期', '工序号', '工作中心', '控制码', '工序内容', '计划量',
'签证', '物料编码', '物料描述', '单位计划量', '出库量', '签证', '单位', '数量确认'])
for filename in pathDir:
x=x+1
df = pd.DataFrame(index=range(30), columns=cols)
def gg(rg,n):
e=[]
f = open(pt + '\\' + filename, encoding='gbk')
for line in f:
d=re.search(rg,line)
if d:
d=str(d.group())
e.append(d)
print(e)
df[n]=pd.Series(e)
f.close()
desc=gg('工单描述\s\S+','工单描述')#desc = re.findall('工单描述\s\S+', line)
n=gg('工单编号\s\d+','工单编号')
up_n=gg('上级工单编号\s\d+','上级工单编号') #sup_desc = re.findall('上级工单描述\s\d+', line)
pro_n=gg('项目编号\s\d+','项目编号') #pro_co=re.findall('项目编号\s\d+',line)
unit=gg('施工单位\s\S+','施工单位')#unit= re.findall('施工单位\s\S+', line)
contr_co=gg('合同号\s\d+','合同号') #contr_co = re.findall('合同号\s\d+', line)
cost=gg('计划服务费\s+\d+\,*\d*\.\d+','计划服务费')#cost = re.findall('计划服务费\s+\d+\,*\d*\.\d+', line)
#if len(cost)>0:
# money=cost[0].split()[1]
start_d=gg('开工日期\s\S+','开工日期')#start_d = re.findall('开工日期\s\S+', line)
over_d=gg('完工日期\s\S+','完工日期')#over_d = re.findall('完工日期\s\S+', line)
worktp = gg('作业类型\s\S+', '作业类型')#worktp = re.findall('作业类型\s\S+', line)
#ntc_crt = re.findall('通知单创建\s\S+', line)
#ntc_pmt = re.findall('通知单批准\s\S+', line)
#plan = re.findall('计划\s\S+', line)
#ass= re.findall('待审\s\S+', line)
#order= re.findall('下达\s\S+', line)
#acpt_ck = re.findall('验收确认\s\S+', line)
#fns_ck = re.findall('完工确认\s\S+', line)
#fns_tm = re.findall('完工时间\s\S+', line)
#printer = re.findall('打印者:\S+', line)
#prt_d = re.findall('打印日期:\d+-\d+-\d+', line)
ntc_crt = gg('通知单创建\s\S+', '通知单创建')
ntc_pmt = gg('通知单批准\s\S+', '通知单批准')
plan = gg('计划\s\S+', '计划')
ass= gg('待审\s\S+', '待审')
order= gg('下达\s\S+', '下达')
acpt_ck = gg('验收确认\s\S+', '验收确认')
fns_ck = gg('完工确认\s\S+', '完工确认')
fns_tm = gg('完工时间\s\S+', '完工时间')
printer = gg('打印者:\S+', '打印者')
prt_d = gg('打印日期:\d+-\d+-\d+', '打印日期')
wp_num = []
wk_ctr = []
ctr_code = []
wp_contts = []
cert = []
f = open(pt + '\\' + filename, encoding='gbk')
for line in f:
proc_set = re.findall('(^\d+)\s(\D+\d*)(\D+\d*)\s((\S*\d*\s*\.*)+)(\d+\.*\d*\D+)+\n', line)#426
if proc_set:# 工序号/工作中心/控制码/工序内容/签证
sets=list(proc_set[0])
wp_num.append(sets[0])
wk_ctr.append (sets[1])
ctr_code.append (sets[2])
wp_contts.append (sets[3])
cert.append (sets[5])
df['工序号']=pd.Series(wp_num)
df['工作中心']=pd.Series(wk_ctr)
df['控制码']=pd.Series(ctr_code)
df['工序内容']=pd.Series(wp_contts)
df['签证']=pd.Series(cert)
wp_num = []
mat_code = []
mat_descr = []
msr_unit = []
all_num = []
cert=[]
f.close()
f = open(pt + '\\' + filename, encoding='gbk')
for line in f:
mat_set = re.findall('(^\d+)\s(\d+)\s((\S*\s*)+)\s(\D)\s((\d\.*\d*\s*)+)\n', line) # 140
if mat_set: # 工序号/物料编码/物料描述/单位/数量确认/计划量/出库量/签证
sets = list(mat_set[0])
wp_num.append(sets[0])
mat_code.append(sets[1])
mat_descr.append(sets[2])
msr_unit.append(sets[4])
all_num.append(sets[5])
cert.append(sets[6])
df['工序号']=pd.Series(wp_num)
df['物料编码']=pd.Series(mat_code)
df['物料描述']=pd.Series(mat_descr)
df['单位']=pd.Series(msr_unit)
df['数量确认']=pd.Series(all_num)
df['签证']=pd.S
|
eries(cert)
|
pandas.Series
|
import streamlit as st
import pandas as pd
import numpy as np
from sklearn import datasets
from sklearn.ensemble import RandomForestClassifier
import os
st.write("""
# iris dataset flower prediction
the app prdoict iris flower dataset
## by **<NAME>** www.github.com/akashAD98
""")
st.sidebar.header("User input parameter")
def user_input_data():
sepal_length=st.sidebar.slider('sepal lengrh1',4.3,7.9,5.4)
sepal_width=st.sidebar.slider(' sepal width',2.0,4.4,3.4)
petal_length=st.sidebar.slider('petal length',1.0,6.0,1.3)
petal_width=st.sidebar.slider('petal width ',0.1,2.5,0.2)
data={'sepal_length':sepal_length,
'sepal_width':sepal_width,
'petal_length':petal_length,
'petal_width':petal_width}
features=
|
pd.DataFrame(data,index=[0])
|
pandas.DataFrame
|
# pylint: disable=redefined-outer-name,protected-access
# pylint: disable=missing-function-docstring,missing-module-docstring,missing-class-docstring
"""This module contains tests of the tabulator Data Grid"""
# http://tabulator.info/docs/4.7/quickstart
# https://github.com/paulhodel/jexcel
import pandas as pd
import panel as pn
import param
import pytest
from _pytest._code.code import TerminalRepr
from bokeh.models import ColumnDataSource
from awesome_panel_extensions.developer_tools.designer import Designer
from awesome_panel_extensions.developer_tools.designer.services.component_reloader import (
ComponentReloader,
)
from awesome_panel_extensions.widgets.tabulator import CSS_HREFS, Tabulator, TabulatorStylesheet
def _data_records():
return [
{"id": 1, "name": "<NAME>", "age": 12, "col": "red", "dob": pd.Timestamp("14/05/1982")},
{"id": 2, "name": "<NAME>", "age": 1, "col": "blue", "dob": pd.Timestamp("14/05/1982")},
{
"id": 3,
"name": "<NAME>",
"age": 42,
"col": "green",
"dob":
|
pd.Timestamp("22/05/1982")
|
pandas.Timestamp
|
#!/usr/bin/env python3
from __future__ import print_function
import pandas as pd
import tensorflow as tf
import numpy as np
import gzip
import time
from collections import OrderedDict
from datetime import datetime
import sys
import os
import subprocess
import glob
import argparse
import scipy.optimize
import scipy.stats as stats
from scipy.special import loggamma
sys.path.insert(1, os.path.dirname(__file__))
import genotypeio
has_rpy2 = False
e = subprocess.call('which R', shell=True, stdout=subprocess.DEVNULL, stderr=subprocess.DEVNULL)
try:
import rpy2
import rfunc
if e==0:
has_rpy2 = True
except:
pass
if not has_rpy2:
print("Warning: 'rfunc' cannot be imported. R and the 'rpy2' Python package are needed.")
class SimpleLogger(object):
def __init__(self, logfile=None, verbose=True):
self.console = sys.stdout
self.verbose = verbose
if logfile is not None:
self.log = open(logfile, 'w')
else:
self.log = None
def write(self, message):
if self.verbose:
self.console.write(message+'\n')
if self.log is not None:
self.log.write(message+'\n')
self.log.flush()
output_dtype_dict = {
'num_var':np.int32,
'beta_shape1':np.float32,
'beta_shape2':np.float32,
'true_df':np.float32,
'pval_true_df':np.float64,
'variant_id':str,
'tss_distance':np.int32,
'ma_samples':np.int32,
'ma_count':np.int32,
'maf':np.float32,
'ref_factor':np.int32,
'pval_nominal':np.float64,
'slope':np.float32,
'slope_se':np.float32,
'pval_perm':np.float64,
'pval_beta':np.float64,
}
#------------------------------------------------------------------------------
# Core functions for mapping associations on GPU
#------------------------------------------------------------------------------
class Residualizer(object):
def __init__(self, C_t):
# center and orthogonalize
self.Q_t, _ = tf.qr(C_t - tf.reduce_mean(C_t, 0), full_matrices=False, name='qr')
def transform(self, M_t, center=True):
"""Residualize rows of M wrt columns of C"""
if center:
M0_t = M_t - tf.reduce_mean(M_t, axis=1, keepdims=True)
else:
M0_t = M_t
return M_t - tf.matmul(tf.matmul(M0_t, self.Q_t), self.Q_t, transpose_b=True) # keep original mean
def residualize(M_t, C_t):
"""Residualize M wrt columns of C"""
# center and orthogonalize
Q_t, _ = tf.qr(C_t - tf.reduce_mean(C_t, 0), full_matrices=False, name='qr')
# residualize M relative to C
M0_t = M_t - tf.reduce_mean(M_t, axis=1, keepdims=True)
return M_t - tf.matmul(tf.matmul(M0_t, Q_t), Q_t, transpose_b=True) # keep original mean
def center_normalize(M_t, axis=0):
"""Center and normalize M"""
if axis == 0:
N_t = M_t - tf.reduce_mean(M_t, 0)
return tf.divide(N_t, tf.sqrt(tf.reduce_sum(tf.pow(N_t, 2), 0)))
elif axis == 1:
N_t = M_t - tf.reduce_mean(M_t, axis=1, keepdims=True)
return tf.divide(N_t, tf.sqrt(tf.reduce_sum(tf.pow(N_t, 2), axis=1, keepdims=True)))
def calculate_maf(genotype_t):
"""Calculate minor allele frequency"""
af_t = tf.reduce_sum(genotype_t,1) / (2*tf.cast(tf.shape(genotype_t)[1], tf.float32))
return tf.where(af_t>0.5, 1-af_t, af_t)
def _calculate_corr(genotype_t, phenotype_t, covariates_t, return_sd=False):
"""Calculate correlation between normalized residual genotypes and phenotypes"""
# residualize
genotype_res_t = residualize(genotype_t, covariates_t) # variants x samples
phenotype_res_t = residualize(phenotype_t, covariates_t) # phenotypes x samples
if return_sd:
_, gstd = tf.nn.moments(genotype_res_t, axes=1)
_, pstd = tf.nn.moments(phenotype_res_t, axes=1)
# center and normalize
genotype_res_t = center_normalize(genotype_res_t, axis=1)
phenotype_res_t = center_normalize(phenotype_res_t, axis=1)
# correlation
if return_sd:
return tf.squeeze(tf.matmul(genotype_res_t, phenotype_res_t, transpose_b=True)), tf.sqrt(pstd / gstd)
else:
return tf.squeeze(tf.matmul(genotype_res_t, phenotype_res_t, transpose_b=True))
def _calculate_max_r2(genotypes_t, phenotypes_t, permutations_t, covariates_t, maf_threshold=0.05):
maf_t = calculate_maf(genotypes_t)
ix = tf.where(maf_t>=maf_threshold)
g2 = tf.squeeze(tf.gather(genotypes_t, ix))
r2_nom_t = tf.pow(_calculate_corr(g2, phenotypes_t, covariates_t), 2)
r2_emp_t = tf.pow(_calculate_corr(g2, permutations_t, covariates_t), 2)
return tf.squeeze(tf.reduce_max(r2_nom_t, axis=0)), tf.squeeze(tf.reduce_max(r2_emp_t, axis=0)), tf.gather(tf.squeeze(ix), tf.argmax(r2_nom_t, axis=0))
def calculate_pval(r2_t, dof, maf_t=None, return_sparse=True, r2_threshold=0, return_r2=False):
"""Calculate p-values from squared correlations"""
dims = r2_t.get_shape()
if return_sparse:
ix = tf.where(r2_t>=r2_threshold, name='threshold_r2')
r2_t = tf.gather_nd(r2_t, ix)
r2_t = tf.cast(r2_t, tf.float64)
tstat = tf.sqrt(tf.divide(tf.scalar_mul(dof, r2_t), 1 - r2_t), name='tstat')
tdist = tf.contrib.distributions.StudentT(np.float64(dof), loc=np.float64(0.0), scale=np.float64(1.0))
if return_sparse:
pval_t = tf.SparseTensor(ix, tf.scalar_mul(2, tdist.cdf(-tf.abs(tstat))), dims)
if maf_t is not None:
maf_t = tf.gather(maf_t, ix[:,0])
else:
pval_t = tf.scalar_mul(2, tdist.cdf(-tf.abs(tstat)))
if maf_t is not None:
if return_r2:
return pval_t, maf_t, r2_t
else:
return pval_t, maf_t
else:
return pval_t
def _interaction_assoc_row(genotype_t, phenotype_t, icovariates_t, return_sd=False):
"""
genotype_t must be a 1D tensor
icovariates_t: [covariates_t, interaction_t]
"""
gi_covariates_t = tf.concat([icovariates_t, tf.reshape(genotype_t, [-1,1])], axis=1)
ix_t = tf.reshape(tf.multiply(genotype_t, icovariates_t[:,-1]), [1,-1]) # must be 1 x N
return _calculate_corr(ix_t, phenotype_t, gi_covariates_t, return_sd=return_sd)
def calculate_association(genotype_t, phenotype_t, covariates_t, interaction_t=None, return_sparse=True, r2_threshold=None, return_r2=False):
"""Calculate genotype-phenotype associations"""
maf_t = calculate_maf(genotype_t)
if interaction_t is None:
r2_t = tf.pow(_calculate_corr(genotype_t, phenotype_t, covariates_t), 2)
dof = genotype_t.shape[1].value - 2 - covariates_t.shape[1].value
else:
icovariates_t = tf.concat([covariates_t, interaction_t], axis=1)
r2_t = tf.pow(tf.map_fn(lambda x: _interaction_assoc_row(x, phenotype_t, icovariates_t), genotype_t, infer_shape=False), 2)
dof = genotype_t.shape[1].value - 4 - covariates_t.shape[1].value
return calculate_pval(r2_t, dof, maf_t, return_sparse=return_sparse, r2_threshold=r2_threshold, return_r2=return_r2)
def get_sample_indexes(vcf_sample_ids, phenotype_df):
"""Get index of sample IDs in VCF"""
return tf.constant([vcf_sample_ids.index(i) for i in phenotype_df.columns])
def initialize_data(phenotype_df, covariates_df, batch_size, interaction_s=None, dtype=tf.float32):
"""Generate placeholders"""
num_samples = phenotype_df.shape[1]
genotype_t = tf.placeholder(dtype, shape=[batch_size, num_samples])
phenotype_t = tf.constant(phenotype_df.values, dtype=dtype)
phenotype_t = tf.reshape(phenotype_t, shape=[-1, num_samples])
covariates_t = tf.constant(covariates_df.values, dtype=dtype)
covariates_t = tf.reshape(covariates_t, shape=[-1, covariates_df.shape[1]])
if interaction_s is None:
return genotype_t, phenotype_t, covariates_t
else:
interaction_t = tf.constant(interaction_s.values, dtype=dtype)
interaction_t = tf.reshape(interaction_t, [-1,1])
return genotype_t, phenotype_t, covariates_t, interaction_t
#------------------------------------------------------------------------------
# Functions for beta-approximating empirical p-values
#------------------------------------------------------------------------------
def pval_from_corr(r2, dof):
tstat2 = dof * r2 / (1 - r2)
return 2*stats.t.cdf(-np.abs(np.sqrt(tstat2)), dof)
def df_cost(r2, dof):
"""minimize abs(1-alpha) as a function of M_eff"""
pval = pval_from_corr(r2, dof)
mean = np.mean(pval)
var = np.var(pval)
return mean * (mean * (1.0-mean) / var - 1.0) - 1.0
def beta_log_likelihood(x, shape1, shape2):
"""negative log-likelihood of beta distribution"""
logbeta = loggamma(shape1) + loggamma(shape2) - loggamma(shape1+shape2)
return (1.0-shape1)*np.sum(np.log(x)) + (1.0-shape2)*np.sum(np.log(1.0-x)) + len(x)*logbeta
def fit_beta_parameters(r2_perm, dof, tol=1e-4, return_minp=False):
"""
r2_perm: array of max. r2 values from permutations
dof: degrees of freedom
"""
try:
true_dof = scipy.optimize.newton(lambda x: df_cost(r2_perm, x), dof, tol=tol, maxiter=50)
except:
print('WARNING: scipy.optimize.newton failed to converge (running scipy.optimize.minimize)')
res = scipy.optimize.minimize(lambda x: np.abs(df_cost(r2_perm, x)), dof, method='Nelder-Mead', tol=tol)
true_dof = res.x[0]
pval = pval_from_corr(r2_perm, true_dof)
mean, var = np.mean(pval), np.var(pval)
beta_shape1 = mean * (mean * (1 - mean) / var - 1)
beta_shape2 = beta_shape1 * (1/mean - 1)
res = scipy.optimize.minimize(lambda s: beta_log_likelihood(pval, s[0], s[1]), [beta_shape1, beta_shape2], method='Nelder-Mead', tol=tol)
beta_shape1, beta_shape2 = res.x
if return_minp:
return beta_shape1, beta_shape2, true_dof, pval
else:
return beta_shape1, beta_shape2, true_dof
def calculate_beta_approx_pval(r2_perm, r2_nominal, dof, tol=1e-4):
"""
r2_nominal: nominal max. r2 (scalar or array)
r2_perm: array of max. r2 values from permutations
dof: degrees of freedom
"""
beta_shape1, beta_shape2, true_dof = fit_beta_parameters(r2_perm, dof, tol)
pval_true_dof = pval_from_corr(r2_nominal, true_dof)
pval_beta = stats.beta.cdf(pval_true_dof, beta_shape1, beta_shape2)
return pval_beta, beta_shape1, beta_shape2, true_dof, pval_true_dof
#------------------------------------------------------------------------------
# Top-level functions for running cis-/trans-QTL mapping
#------------------------------------------------------------------------------
def calculate_cis_permutations(genotypes_t, range_t, phenotype_t, covariates_t, permutation_ix_t):
"""Calculate nominal and empirical correlations"""
permutations_t = tf.gather(phenotype_t, permutation_ix_t)
r_nominal_t, std_ratio_t = _calculate_corr(genotypes_t, tf.reshape(phenotype_t, [1,-1]), covariates_t, return_sd=True)
corr_t = tf.pow(_calculate_corr(genotypes_t, permutations_t, covariates_t), 2)
corr_t.set_shape([None,None])
r2_perm_t = tf.cast(tf.reduce_max(tf.boolean_mask(corr_t, ~tf.reduce_any(tf.is_nan(corr_t), 1)), axis=0), tf.float64)
ix = tf.argmax(tf.pow(r_nominal_t, 2))
return r_nominal_t[ix], std_ratio_t[ix], range_t[ix], r2_perm_t, genotypes_t[ix], tf.shape(r_nominal_t)[0]
def process_cis_permutations(r2_perm, r_nominal, std_ratio, g, num_var, dof, n_samples, nperm=10000):
"""Calculate beta-approximated empirical p-value and annotate phenotype"""
r2_nominal = r_nominal*r_nominal
pval_perm = (np.sum(r2_perm>=r2_nominal)+1) / (nperm+1)
pval_beta, beta_shape1, beta_shape2, true_dof, pval_true_dof = calculate_beta_approx_pval(r2_perm, r2_nominal, dof)
maf = np.sum(g) / (2*n_samples)
if maf <= 0.5:
ref_factor = 1
ma_samples = np.sum(g>0.5)
ma_count = np.sum(g[g>0.5])
else:
maf = 1-maf
ref_factor = -1
ma_samples = np.sum(g<1.5)
ma_count = np.sum(g[g<1.5])
slope = r_nominal * std_ratio
tstat2 = dof * r2_nominal / (1 - r2_nominal)
slope_se = np.abs(slope) / np.sqrt(tstat2)
return pd.Series(OrderedDict([
('num_var', num_var),
('beta_shape1', beta_shape1),
('beta_shape2', beta_shape2),
('true_df', true_dof),
('pval_true_df', pval_true_dof),
('variant_id', np.NaN),
('tss_distance', np.NaN),
('ma_samples', ma_samples),
('ma_count', ma_count),
('maf', maf),
('ref_factor', ref_factor),
('pval_nominal', pval_from_corr(r2_nominal, dof)),
('slope', slope),
('slope_se', slope_se),
('pval_perm', pval_perm),
('pval_beta', pval_beta),
]))
def _process_group_permutations(buf):
"""
Merge results for grouped phenotypes
buf: [r_nom, s_r, var_ix, r2_perm, g, ng, nid]
"""
# select phenotype with strongest nominal association
max_ix = np.argmax(np.abs([b[0] for b in buf]))
r_nom, s_r, var_ix = buf[max_ix][:3]
g, ng, nid = buf[max_ix][4:]
# select best phenotype correlation for each permutation
r2_perm = np.max([b[3] for b in buf], 0)
return r_nom, s_r, var_ix, r2_perm, g, ng, nid
def map_cis(plink_reader, phenotype_df, phenotype_pos_df, covariates_df, group_s=None, nperm=10000, logger=None):
"""Run cis-QTL mapping"""
assert np.all(phenotype_df.columns==covariates_df.index)
if logger is None:
logger = SimpleLogger()
logger.write('cis-QTL mapping: empirical p-values for phenotypes')
logger.write(' * {} samples'.format(phenotype_df.shape[1]))
logger.write(' * {} phenotypes'.format(phenotype_df.shape[0]))
if group_s is not None:
logger.write(' * {} phenotype groups'.format(len(group_s.unique())))
group_dict = group_s.to_dict()
logger.write(' * {} covariates'.format(covariates_df.shape[1]))
logger.write(' * {} variants'.format(plink_reader.bed.shape[0]))
dof = phenotype_df.shape[1] - 2 - covariates_df.shape[1]
# permutation indices
n_samples = phenotype_df.shape[1]
ix = np.arange(n_samples)
permutation_ix_t = tf.convert_to_tensor(np.array([np.random.permutation(ix) for i in range(nperm)]))
# placeholders
covariates_t = tf.constant(covariates_df.values, dtype=tf.float32)
genotype_t = tf.placeholder(dtype=tf.float32, shape=(None))
phenotype_t = tf.placeholder(dtype=tf.float32, shape=(None))
# iterate over chromosomes
res_df = []
start_time = time.time()
with tf.Session() as sess:
for chrom in phenotype_pos_df.loc[phenotype_df.index, 'chr'].unique():
logger.write(' Mapping chromosome {}'.format(chrom))
igc = genotypeio.InputGeneratorCis(plink_reader, phenotype_df.loc[phenotype_pos_df['chr']==chrom], phenotype_pos_df)
dataset = tf.data.Dataset.from_generator(igc.generate_data, output_types=(tf.float32, tf.float32, tf.int32, tf.string))
dataset = dataset.prefetch(1)
iterator = dataset.make_one_shot_iterator()
next_phenotype, next_genotypes, next_range, next_id = iterator.get_next()
r_nominal_t, std_ratio_t, varpos_t, r2_perm_t, g_t, ng_t = calculate_cis_permutations(
next_genotypes, next_range, next_phenotype, covariates_t, permutation_ix_t)
if group_s is None:
for i in range(1, igc.n_phenotypes+1):
r_nom, s_r, var_ix, r2_perm, g, ng, nid = sess.run([r_nominal_t, std_ratio_t, varpos_t, r2_perm_t, g_t, ng_t, next_id])
# post-processing (on CPU)
res_s = process_cis_permutations(r2_perm, r_nom, s_r, g, ng, dof, phenotype_df.shape[1], nperm=nperm)
res_s.name = nid.decode()
res_s['variant_id'] = igc.chr_variant_pos.index[var_ix]
res_s['tss_distance'] = igc.chr_variant_pos[res_s['variant_id']] - igc.phenotype_tss[res_s.name]
res_df.append(res_s)
print('\r * computing permutations for phenotype {}/{}'.format(i, igc.n_phenotypes), end='')
print()
else:
n_groups = len(igc.phenotype_df.index.map(group_dict).unique())
buf = []
processed_groups = 0
previous_group = None
for i in range(0, igc.n_phenotypes):
group_id = group_dict[igc.phenotype_df.index[i]]
ires = sess.run([r_nominal_t, std_ratio_t, varpos_t, r2_perm_t, g_t, ng_t, next_id])
if (group_id != previous_group and len(buf)>0): # new group, process previous
# post-processing (on CPU)
r_nom, s_r, var_ix, r2_perm, g, ng, nid = _process_group_permutations(buf)
res_s = process_cis_permutations(r2_perm, r_nom, s_r, g, ng, dof, phenotype_df.shape[1], nperm=nperm)
res_s.name = nid.decode()
res_s['variant_id'] = igc.chr_variant_pos.index[var_ix]
res_s['tss_distance'] = igc.chr_variant_pos[res_s['variant_id']] - igc.phenotype_tss[res_s.name]
res_s['group_id'] = group_id
res_s['group_size'] = len(buf)
res_df.append(res_s)
processed_groups += 1
print('\r * computing permutations for phenotype group {}/{}'.format(processed_groups, n_groups), end='')
# reset
buf = [ires]
else:
buf.append(ires)
previous_group = group_id
# process last group
r_nom, s_r, var_ix, r2_perm, g, ng, nid = _process_group_permutations(buf)
res_s = process_cis_permutations(r2_perm, r_nom, s_r, g, ng, dof, phenotype_df.shape[1], nperm=nperm)
res_s.name = nid.decode()
res_s['variant_id'] = igc.chr_variant_pos.index[var_ix]
res_s['tss_distance'] = igc.chr_variant_pos[res_s['variant_id']] - igc.phenotype_tss[res_s.name]
res_s['group_id'] = group_id
res_s['group_size'] = len(buf)
res_df.append(res_s)
processed_groups += 1
print('\r * computing permutations for phenotype group {}/{}'.format(processed_groups, n_groups), end='\n')
res_df = pd.concat(res_df, axis=1).T
res_df.index.name = 'phenotype_id'
logger.write(' Time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
logger.write('done.')
return res_df.astype(output_dtype_dict)
def map_cis_independent(plink_reader, summary_df, phenotype_df, phenotype_pos_df, covariates_df, fdr=0.05, fdr_col='qval', nperm=10000, logger=None):
"""
Run independent cis-QTL mapping (forward-backward regression)
summary_df: output from map_cis, annotated with q-values (calculate_qvalues)
"""
assert np.all(phenotype_df.index==phenotype_pos_df.index)
if logger is None:
logger = SimpleLogger()
signif_df = summary_df[summary_df[fdr_col]<=fdr].copy()
signif_df = signif_df[[
'num_var', 'beta_shape1', 'beta_shape2', 'true_df', 'pval_true_df',
'variant_id', 'tss_distance', 'ma_samples', 'ma_count', 'maf', 'ref_factor',
'pval_nominal', 'slope', 'slope_se', 'pval_perm', 'pval_beta',
]]
signif_threshold = signif_df['pval_beta'].max()
# subset significant phenotypes
ix = signif_df.index[signif_df.index.isin(phenotype_df.index)]
phenotype_df = phenotype_df.loc[ix]
phenotype_pos_df = phenotype_pos_df.loc[ix]
logger.write('cis-QTL mapping: conditionally independent variants')
logger.write(' * {} samples'.format(phenotype_df.shape[1]))
logger.write(' * {} significant phenotypes'.format(signif_df.shape[0]))
logger.write(' * {} covariates'.format(covariates_df.shape[1]))
logger.write(' * {} variants'.format(plink_reader.bed.shape[0]))
# print('Significance threshold: {}'.format(signif_threshold))
# permutation indices
n_samples = phenotype_df.shape[1]
ix = np.arange(n_samples)
permutation_ix_t = tf.convert_to_tensor(np.array([np.random.permutation(ix) for i in range(nperm)]))
# placeholders
genotypes_t = tf.placeholder(dtype=tf.float32, shape=[None,None])
range_t = tf.placeholder(dtype=tf.int32, shape=[None])
phenotype_t = tf.placeholder(dtype=tf.float32, shape=[None])
covariates_t = tf.placeholder(dtype=tf.float32, shape=[None, None])
# graph
r_nominal_t, std_ratio_t, varpos_t, r2_perm_t, g_t, ng_t = calculate_cis_permutations(
genotypes_t, range_t, phenotype_t, covariates_t, permutation_ix_t)
# iterate over chromosomes
res_df = []
start_time = time.time()
with tf.Session() as sess:
for chrom in phenotype_pos_df['chr'].unique():
logger.write(' Mapping chromosome {}'.format(chrom))
igc = genotypeio.InputGeneratorCis(plink_reader, phenotype_df.loc[phenotype_pos_df['chr']==chrom], phenotype_pos_df)
ix_dict = {i:k for k,i in enumerate(plink_reader.bim.loc[plink_reader.bim['chrom']==chrom, 'snp'])}
igc.chr_genotypes, igc.chr_variant_pos = igc.plink_reader.get_region(chrom, verbose=False)
igc.loaded_chrom = chrom
# iterate through significant phenotypes, fetch associated genotypes
for j, (phenotype, genotypes, genotype_range, phenotype_id) in enumerate(igc.generate_data(), 1):
# 1) forward pass
forward_df = [signif_df.loc[phenotype_id]] # initialize results with top variant
covariates = covariates_df.values.copy() # initialize covariates
dosage_dict = {}
while True:
# add variant to covariates
variant_id = forward_df[-1]['variant_id']
ig = igc.chr_genotypes[ix_dict[variant_id]]
dosage_dict[variant_id] = ig
covariates = np.hstack([covariates, ig.reshape(-1,1)]).astype(np.float32)
dof = phenotype_df.shape[1] - 2 - covariates.shape[1]
# find next variant
r_nom, s_r, var_ix, r2_perm, g, ng = sess.run([r_nominal_t, std_ratio_t, varpos_t, r2_perm_t, g_t, ng_t],
feed_dict={genotypes_t:genotypes, range_t:genotype_range, phenotype_t:phenotype, covariates_t:covariates})
res_s = process_cis_permutations(r2_perm, r_nom, s_r, g, ng, dof, phenotype_df.shape[1], nperm=nperm)
# add to list if significant
if res_s['pval_beta'] <= signif_threshold:
res_s.name = phenotype_id
res_s['variant_id'] = igc.chr_variant_pos.index[var_ix]
res_s['tss_distance'] = igc.chr_variant_pos[res_s['variant_id']] - igc.phenotype_tss[res_s.name]
forward_df.append(res_s)
else:
break
forward_df = pd.concat(forward_df, axis=1).T
dosage_df = pd.DataFrame(dosage_dict)
# print(forward_df)
# 2) backward pass
if forward_df.shape[0]>1:
back_df = []
variant_set = set()
for k,i in enumerate(forward_df['variant_id'], 1):
covariates = np.hstack([
covariates_df.values,
dosage_df[np.setdiff1d(forward_df['variant_id'], i)].values,
])
r_nom, s_r, var_ix, r2_perm, g, ng = sess.run([r_nominal_t, std_ratio_t, varpos_t, r2_perm_t, g_t, ng_t],
feed_dict={genotypes_t:genotypes, range_t:genotype_range, phenotype_t:phenotype, covariates_t:covariates})
dof = phenotype_df.shape[1] - 2 - covariates.shape[1]
res_s = process_cis_permutations(r2_perm, r_nom, s_r, g, ng, dof, phenotype_df.shape[1], nperm=nperm)
res_s['variant_id'] = igc.chr_variant_pos.index[var_ix]
if res_s['pval_beta'] <= signif_threshold and res_s['variant_id'] not in variant_set:
res_s.name = phenotype_id
res_s['tss_distance'] = igc.chr_variant_pos[res_s['variant_id']] - igc.phenotype_tss[res_s.name]
res_s['rank'] = k
back_df.append(res_s)
variant_set.add(res_s['variant_id'])
if len(back_df)>0:
res_df.append(pd.concat(back_df, axis=1).T)
# print('back')
# print(pd.concat(back_df, axis=1).T)
else: # single variant
forward_df['rank'] = 1
res_df.append(forward_df)
print('\r * computing independent QTL for phenotype {}/{}'.format(j, igc.n_phenotypes), end='')
print()
res_df = pd.concat(res_df, axis=0)
res_df.index.name = 'phenotype_id'
logger.write(' Time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
logger.write('done.')
return res_df.reset_index().astype(output_dtype_dict)
def calculate_qvalues(res_df, fdr=0.05, qvalue_lambda=None):
"""Annotate permutation results with q-values, p-value threshold"""
print('Computing q-values')
print(' * Number of phenotypes tested: {}'.format(res_df.shape[0]))
print(' * Correlation between Beta-approximated and empirical p-values: : {:.4f}'.format(
stats.pearsonr(res_df['pval_perm'], res_df['pval_beta'])[0]))
# calculate q-values
if qvalue_lambda is None:
qval, pi0 = rfunc.qvalue(res_df['pval_beta'])
else:
print(' * Calculating q-values with lambda = {:.3f}'.format(qvalue_lambda))
qval, pi0 = rfunc.qvalue(res_df['pval_beta'], qvalue_lambda)
res_df['qval'] = qval
print(' * Proportion of significant phenotypes (1-pi0): {:.2f}'.format(1 - pi0))
print(' * QTL phenotypes @ FDR {:.2f}: {}'.format(fdr, np.sum(res_df['qval']<=fdr)))
# determine global min(p) significance threshold and calculate nominal p-value threshold for each gene
ub = res_df.loc[res_df['qval']>fdr, 'pval_beta'].sort_values()[0]
lb = res_df.loc[res_df['qval']<=fdr, 'pval_beta'].sort_values()[-1]
pthreshold = (lb+ub)/2
print(' * min p-value threshold @ FDR {}: {:.6g}'.format(fdr, pthreshold))
res_df['pval_nominal_threshold'] = stats.beta.ppf(pthreshold, res_df['beta_shape1'], res_df['beta_shape2'])
def annotate_genes(gene_df, annotation_gtf, lookup_df=None):
"""
Add gene and variant annotations (e.g., gene_name, rs_id, etc.) to gene-level output
gene_df: output from map_cis()
annotation_gtf: gene annotation in GTF format
lookup_df: DataFrame with variant annotations, indexed by 'variant_id'
"""
gene_dict = {}
print('['+datetime.now().strftime("%b %d %H:%M:%S")+'] Adding gene and variant annotations', flush=True)
print(' * parsing GTF', flush=True)
with open(annotation_gtf) as gtf:
for row in gtf:
row = row.strip().split('\t')
if row[0][0]=='#' or row[2]!='gene': continue
# get gene_id and gene_name from attributes
attr = dict([i.split() for i in row[8].replace('"','').split(';') if i!=''])
# gene_name, gene_chr, gene_start, gene_end, strand
gene_dict[attr['gene_id']] = [attr['gene_name'], row[0], row[3], row[4], row[6]]
print(' * annotating genes', flush=True)
if 'group_id' in gene_df:
gene_info = pd.DataFrame(data=[gene_dict[i] for i in gene_df['group_id']],
columns=['gene_name', 'gene_chr', 'gene_start', 'gene_end', 'strand'],
index=gene_df.index)
else:
gene_info = pd.DataFrame(data=[gene_dict[i] for i in gene_df.index],
columns=['gene_name', 'gene_chr', 'gene_start', 'gene_end', 'strand'],
index=gene_df.index)
gene_df = pd.concat([gene_info, gene_df], axis=1)
assert np.all(gene_df.index==gene_info.index)
col_order = ['gene_name', 'gene_chr', 'gene_start', 'gene_end', 'strand',
'num_var', 'beta_shape1', 'beta_shape2', 'true_df', 'pval_true_df', 'variant_id', 'tss_distance']
if lookup_df is not None:
print(' * adding variant annotations from lookup table', flush=True)
gene_df = gene_df.join(lookup_df, on='variant_id') # add variant information
col_order += list(lookup_df.columns)
col_order += ['ma_samples', 'ma_count', 'maf', 'ref_factor',
'pval_nominal', 'slope', 'slope_se', 'pval_perm', 'pval_beta']
if 'group_id' in gene_df:
col_order += ['group_id', 'group_size']
col_order += ['qval', 'pval_nominal_threshold']
gene_df = gene_df[col_order]
print('done.', flush=True)
return gene_df
def get_significant_pairs(res_df, nominal_prefix, fdr=0.05):
"""Significant variant-phenotype pairs based on nominal p-value threshold for each phenotype"""
print('['+datetime.now().strftime("%b %d %H:%M:%S")+'] tensorQTL: filtering significant variant-phenotype pairs', flush=True)
assert 'qval' in res_df
# significant phenotypes (apply FDR threshold)
df = res_df.loc[res_df['qval']<=fdr, ['pval_nominal_threshold', 'pval_nominal', 'pval_beta']].copy()
df.rename(columns={'pval_nominal': 'min_pval_nominal'}, inplace=True)
signif_phenotype_ids = set(df.index)
threshold_dict = df['pval_nominal_threshold'].to_dict()
nominal_files = {os.path.basename(i).split('.')[-2]:i for i in glob.glob(nominal_prefix+'*.parquet')}
chroms = sorted(nominal_files.keys())
signif_df = []
for k,c in enumerate(chroms, 1):
print(' * parsing significant variant-phenotype pairs for chr. {}/{}'.format(k, len(chroms)), end='\r', flush=True)
nominal_df = pd.read_parquet(nominal_files[c])
nominal_df = nominal_df[nominal_df['phenotype_id'].isin(signif_phenotype_ids)]
m = nominal_df['pval_nominal']<nominal_df['phenotype_id'].apply(lambda x: threshold_dict[x])
signif_df.append(nominal_df[m])
print()
signif_df = pd.concat(signif_df, axis=0)
signif_df = signif_df.merge(df, left_on='phenotype_id', right_index=True)
print('['+datetime.now().strftime("%b %d %H:%M:%S")+'] done', flush=True)
return signif_df
# signif_df.to_parquet(nominal_prefix.rsplit('.',1)[0]+'.cis_qtl_significant_pairs.parquet')
def calculate_cis_nominal(genotypes_t, phenotype_t, covariates_t, dof):
"""
Calculate nominal associations
genotypes_t: genotypes x samples
phenotype_t: single phenotype
covariates_t: covariates matrix, samples x covariates
"""
p = tf.reshape(phenotype_t, [1,-1])
r_nominal_t, std_ratio_t = _calculate_corr(genotypes_t, p, covariates_t, return_sd=True)
r2_nominal_t = tf.pow(r_nominal_t, 2)
pval_t = calculate_pval(r2_nominal_t, dof, maf_t=None, return_sparse=False)
slope_t = tf.multiply(r_nominal_t, std_ratio_t)
slope_se_t = tf.divide(tf.abs(slope_t), tf.sqrt(tf.divide(tf.scalar_mul(dof, r2_nominal_t), 1 - r2_nominal_t)))
# calculate MAF
n = covariates_t.shape[0].value
n2 = 2*n
af_t = tf.reduce_sum(genotypes_t,1) / n2
ix_t = af_t<=0.5
maf_t = tf.where(ix_t, af_t, 1-af_t)
# calculate MA samples and counts
m = tf.cast(genotypes_t>0.5, tf.float32)
a = tf.reduce_sum(m, 1)
b = tf.reduce_sum(tf.cast(genotypes_t<1.5, tf.float32), 1)
ma_samples_t = tf.where(ix_t, a, b)
m = tf.multiply(m, genotypes_t)
a = tf.reduce_sum(m, 1)
ma_count_t = tf.where(ix_t, a, n2-a)
return pval_t, slope_t, slope_se_t, maf_t, ma_samples_t, ma_count_t
def map_cis_nominal(plink_reader, phenotype_df, phenotype_pos_df, covariates_df, prefix,
output_dir='.', logger=None):
"""
cis-QTL mapping: nominal associations for all variant-phenotype pairs
Association results for each chromosome are written to parquet files
in the format <output_dir>/<prefix>.cis_qtl_pairs.<chr>.parquet
"""
if logger is None:
logger = SimpleLogger()
logger.write('cis-QTL mapping: nominal associations for all variant-phenotype pairs')
logger.write(' * {} samples'.format(phenotype_df.shape[1]))
logger.write(' * {} phenotypes'.format(phenotype_df.shape[0]))
logger.write(' * {} covariates'.format(covariates_df.shape[1]))
logger.write(' * {} variants'.format(plink_reader.bed.shape[0]))
# placeholders
covariates_t = tf.constant(covariates_df.values, dtype=tf.float32)
genotype_t = tf.placeholder(dtype=tf.float32, shape=(None))
phenotype_t = tf.placeholder(dtype=tf.float32, shape=(None))
dof = phenotype_df.shape[1] - 2 - covariates_df.shape[1]
with tf.Session() as sess:
# iterate over chromosomes
start_time = time.time()
for chrom in phenotype_pos_df.loc[phenotype_df.index, 'chr'].unique():
logger.write(' Mapping chromosome {}'.format(chrom))
igc = genotypeio.InputGeneratorCis(plink_reader, phenotype_df.loc[phenotype_pos_df['chr']==chrom], phenotype_pos_df)
dataset = tf.data.Dataset.from_generator(igc.generate_data, output_types=(tf.float32, tf.float32, tf.int32, tf.string))
dataset = dataset.prefetch(1)
iterator = dataset.make_one_shot_iterator()
next_phenotype, next_genotypes, _, next_id = iterator.get_next()
x = calculate_cis_nominal(next_genotypes, next_phenotype, covariates_t, dof)
chr_res_df = []
for i in range(1, igc.n_phenotypes+1):
(pval_nominal, slope, slope_se, maf, ma_samples, ma_count), phenotype_id = sess.run([x, next_id])
phenotype_id = phenotype_id.decode()
r = igc.cis_ranges[phenotype_id]
variant_ids = plink_reader.variant_pos[chrom].index[r[0]:r[1]+1]
nv = len(variant_ids)
tss_distance = np.int32(plink_reader.variant_pos[chrom].values[r[0]:r[1]+1] - igc.phenotype_tss[phenotype_id])
chr_res_df.append(pd.DataFrame(OrderedDict([
('phenotype_id', [phenotype_id]*nv),
('variant_id', variant_ids),
('tss_distance', tss_distance),
('maf', maf),
('ma_samples', np.int32(ma_samples)),
('ma_count', np.int32(ma_count)),
('pval_nominal', pval_nominal),
('slope', slope),
('slope_se', slope_se),
])))
print('\r computing associations for phenotype {}/{}'.format(i, igc.n_phenotypes), end='')
print()
logger.write(' time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
print(' * writing output')
pd.concat(chr_res_df, copy=False).to_parquet(os.path.join(output_dir, '{}.cis_qtl_pairs.{}.parquet'.format(prefix, chrom)))
logger.write('done.')
def calculate_nominal_interaction(genotypes_t, phenotype_t, interaction_t, dof, residualizer,
interaction_mask_t=None, maf_threshold_interaction=0.05,
return_sparse=False, tstat_threshold=None):
""""""
phenotype_t = tf.reshape(phenotype_t, [1,-1])
# filter monomorphic sites (to avoid colinearity in X)
mask_t = ~(tf.reduce_all(tf.equal(genotypes_t, 0), axis=1) |
tf.reduce_all(tf.equal(genotypes_t, 1), axis=1) |
tf.reduce_all(tf.equal(genotypes_t, 2), axis=1)
)
if interaction_mask_t is not None:
upper_t = tf.boolean_mask(genotypes_t, interaction_mask_t, axis=1)
lower_t = tf.boolean_mask(genotypes_t, ~interaction_mask_t, axis=1)
mask_t = (mask_t &
(calculate_maf(upper_t) >= maf_threshold_interaction) &
(calculate_maf(lower_t) >= maf_threshold_interaction)
)
mask_t.set_shape([None]) # required for tf.boolean_mask
genotypes_t = tf.boolean_mask(genotypes_t, mask_t)
s = tf.shape(genotypes_t)
ng = s[0]
ns = tf.cast(s[1], tf.float32)
g0_t = genotypes_t - tf.reduce_mean(genotypes_t, axis=1, keepdims=True)
gi_t = tf.multiply(genotypes_t, interaction_t)
gi0_t = gi_t - tf.reduce_mean(gi_t, axis=1, keepdims=True)
i0_t = interaction_t - tf.reduce_mean(interaction_t)
p_t = tf.reshape(phenotype_t, [1,-1])
p0_t = p_t - tf.reduce_mean(p_t, axis=1, keepdims=True)
# residualize rows
g0_t = residualizer.transform(g0_t, center=False)
gi0_t = residualizer.transform(gi0_t, center=False)
p0_t = residualizer.transform(p0_t, center=False)
i0_t = residualizer.transform(i0_t, center=False)
i0_t = tf.tile(i0_t, [ng, 1])
# regression
X_t = tf.stack([g0_t, i0_t, gi0_t], axis=2)
Xinv = tf.linalg.inv(tf.matmul(X_t, X_t, transpose_a=True))
b_t = tf.matmul(tf.matmul(Xinv, X_t, transpose_b=True), tf.tile(tf.reshape(p0_t, [1,1,-1]), [ng,1,1]), transpose_b=True)
# calculate b, b_se
r_t = tf.squeeze(tf.matmul(X_t, b_t)) - p0_t
rss_t = tf.reduce_sum(tf.multiply(r_t, r_t), axis=1)
Cx = Xinv * tf.reshape(rss_t, [-1,1,1]) / dof
b_se_t = tf.sqrt(tf.matrix_diag_part(Cx))
b_t = tf.squeeze(b_t)
tstat_t = tf.divide(tf.cast(b_t, tf.float64), tf.cast(b_se_t, tf.float64))
# weird tf bug? without cast/copy, divide appears to modify b_se_t??
# calculate pval
tdist = tf.contrib.distributions.StudentT(np.float64(dof), loc=np.float64(0.0), scale=np.float64(1.0))
pval_t = tf.scalar_mul(2, tdist.cdf(-tf.abs(tstat_t)))
# calculate MAF
n2 = 2*ns
af_t = tf.reduce_sum(genotypes_t,1) / n2
ix_t = af_t<=0.5
maf_t = tf.where(ix_t, af_t, 1-af_t)
# calculate MA samples and counts
m = tf.cast(genotypes_t>0.5, tf.float32)
a = tf.reduce_sum(m, 1)
b = tf.reduce_sum(tf.cast(genotypes_t<1.5, tf.float32), 1)
ma_samples_t = tf.where(ix_t, a, b)
m = tf.multiply(m, genotypes_t)
a = tf.reduce_sum(m, 1)
ma_count_t = tf.where(ix_t, a, n2-a)
return pval_t, b_t, b_se_t, maf_t, ma_samples_t, ma_count_t, mask_t
def map_cis_interaction_nominal(plink_reader, phenotype_df, phenotype_pos_df, covariates_df, interaction_s,
prefix, maf_threshold_interaction=0.05, best_only=False, output_dir='.', logger=None):
"""
cis-QTL mapping: nominal associations for all variant-phenotype pairs
Association results for each chromosome are written to parquet files
in the format <output_dir>/<prefix>.cis_qtl_pairs.<chr>.parquet
"""
if logger is None:
logger = SimpleLogger()
logger.write('cis-QTL mapping: nominal associations for all variant-phenotype pairs')
logger.write(' * {} samples'.format(phenotype_df.shape[1]))
logger.write(' * {} phenotypes'.format(phenotype_df.shape[0]))
logger.write(' * {} covariates'.format(covariates_df.shape[1]))
logger.write(' * {} variants'.format(plink_reader.bed.shape[0]))
logger.write(' * including interaction term')
covariates_t = tf.constant(covariates_df.values, dtype=tf.float32)
dof = phenotype_df.shape[1] - 4 - covariates_df.shape[1]
interaction_t = tf.constant(interaction_s.values.reshape(1,-1), dtype=tf.float32) # 1 x n
interaction_mask_t = tf.constant(interaction_s >= interaction_s.median())
residualizer = Residualizer(covariates_t)
with tf.Session() as sess:
# iterate over chromosomes
start_time = time.time()
best_assoc = []
for chrom in phenotype_pos_df.loc[phenotype_df.index, 'chr'].unique():
logger.write(' Mapping chromosome {}'.format(chrom))
igc = genotypeio.InputGeneratorCis(plink_reader, phenotype_df.loc[phenotype_pos_df['chr']==chrom], phenotype_pos_df)
dataset = tf.data.Dataset.from_generator(igc.generate_data, output_types=(tf.float32, tf.float32, tf.int32, tf.string))
dataset = dataset.prefetch(1)
iterator = dataset.make_one_shot_iterator()
next_phenotype, next_genotypes, _, next_id = iterator.get_next()
x = calculate_nominal_interaction(next_genotypes, next_phenotype, interaction_t, dof, residualizer,
interaction_mask_t=interaction_mask_t, maf_threshold_interaction=0.05)
chr_res_df = []
for i in range(1, igc.n_phenotypes+1):
(pval, b, b_se, maf, ma_samples, ma_count, maf_mask), phenotype_id = sess.run([x, next_id])
phenotype_id = phenotype_id.decode()
r = igc.cis_ranges[phenotype_id]
variant_ids = plink_reader.variant_pos[chrom].index[r[0]:r[1]+1]
tss_distance = np.int32(plink_reader.variant_pos[chrom].values[r[0]:r[1]+1] - igc.phenotype_tss[phenotype_id])
if maf_mask is not None:
variant_ids = variant_ids[maf_mask]
tss_distance = tss_distance[maf_mask]
nv = len(variant_ids)
df = pd.DataFrame(OrderedDict([
('phenotype_id', [phenotype_id]*nv),
('variant_id', variant_ids),
('tss_distance', tss_distance),
('maf', maf),
('ma_samples', np.int32(ma_samples)),
('ma_count', np.int32(ma_count)),
('pval_g', pval[:,0]),
('b_g', b[:,0]),
('b_g_se', b_se[:,0]),
('pval_i', pval[:,1]),
('b_i', b[:,1]),
('b_i_se', b_se[:,1]),
('pval_gi', pval[:,2]),
('b_gi', b[:,2]),
('b_gi_se', b_se[:,2]),
]))
if best_only:
best_assoc.append(df.loc[df['pval_gi'].idxmin()])
else:
chr_res_df.append(df)
print('\r computing associations for phenotype {}/{}'.format(i, igc.n_phenotypes), end='')
print()
logger.write(' time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
if not best_only:
print(' * writing output')
pd.concat(chr_res_df, copy=False).to_parquet(os.path.join(output_dir, '{}.cis_qtl_pairs.{}.parquet'.format(prefix, chrom)))
if best_only:
pd.concat(best_assoc, axis=1).T.set_index('phenotype_id').to_csv(
os.path.join(output_dir, '{}.cis_qtl_top_assoc.txt.gz'.format(prefix)),
sep='\t', float_format='%.6g', compression='gzip'
)
logger.write('done.')
def map_trans(genotype_df, phenotype_df, covariates_df, interaction_s=None,
return_sparse=True, pval_threshold=1e-5, maf_threshold=0.05,
return_r2=False, batch_size=20000, logger=None, verbose=True):
"""Run trans-QTL mapping from genotypes in memory"""
if logger is None:
logger = SimpleLogger(verbose=verbose)
assert np.all(phenotype_df.columns==covariates_df.index)
variant_ids = genotype_df.index.tolist()
variant_dict = {i:j for i,j in enumerate(variant_ids)}
n_variants = len(variant_ids)
n_samples = phenotype_df.shape[1]
logger.write('trans-QTL mapping')
logger.write(' * {} samples'.format(n_samples))
logger.write(' * {} phenotypes'.format(phenotype_df.shape[0]))
logger.write(' * {} covariates'.format(covariates_df.shape[1]))
logger.write(' * {} variants'.format(n_variants))
if interaction_s is not None:
logger.write(' * including interaction term')
# with tf.device('/cpu:0'):
ggt = genotypeio.GenotypeGeneratorTrans(genotype_df.values, batch_size=batch_size, dtype=np.float32)
dataset_genotypes = tf.data.Dataset.from_generator(ggt.generate_data, output_types=tf.float32)
dataset_genotypes = dataset_genotypes.prefetch(10)
iterator = dataset_genotypes.make_one_shot_iterator()
next_element = iterator.get_next()
# index of VCF samples corresponding to phenotypes
ix_t = get_sample_indexes(genotype_df.columns.tolist(), phenotype_df)
next_element = tf.gather(next_element, ix_t, axis=1)
# calculate correlation threshold for sparse output
if return_sparse:
dof = n_samples - 2 - covariates_df.shape[1]
tstat_threshold = stats.t.ppf(pval_threshold/2, dof)
r2_threshold = tstat_threshold**2 / (dof + tstat_threshold**2)
else:
r2_threshold = None
tstat_threshold = None
if interaction_s is None:
genotypes, phenotypes, covariates = initialize_data(phenotype_df, covariates_df,
batch_size=batch_size, dtype=tf.float32)
# with tf.device('/gpu:0'):
x = calculate_association(genotypes, phenotypes, covariates, return_sparse=return_sparse,
r2_threshold=r2_threshold, return_r2=return_r2)
else:
genotypes, phenotypes, covariates, interaction = initialize_data(phenotype_df, covariates_df,
batch_size=batch_size, interaction_s=interaction_s)
x = calculate_association(genotypes, phenotypes, covariates, interaction_t=interaction,
return_sparse=return_sparse, r2_threshold=r2_threshold)
init_op = tf.group(tf.global_variables_initializer(), tf.local_variables_initializer())
start_time = time.time()
if verbose:
print(' Mapping batches')
with tf.Session() as sess:
# run_options = tf.RunOptions(trace_level=tf.RunOptions.FULL_TRACE)
# run_metadata = tf.RunMetadata()
# writer = tf.summary.FileWriter('logs', sess.graph, session=sess)
sess.run(init_op)
pval_list = []
maf_list = []
r2_list = []
for i in range(1, ggt.num_batches+1):
if verbose:
sys.stdout.write('\r * processing batch {}/{}'.format(i, ggt.num_batches))
sys.stdout.flush()
g_iter = sess.run(next_element)
# x: p_values, maf, {r2}
p_ = sess.run(x, feed_dict={genotypes:g_iter})#, options=run_options, run_metadata=run_metadata)
# writer.add_run_metadata(run_metadata, 'batch{}'.format(i))
pval_list.append(p_[0])
maf_list.append(p_[1])
if return_r2:
r2_list.append(p_[2])
if return_sparse:
pval = tf.sparse_concat(0, pval_list).eval()
else:
pval = tf.concat(pval_list, 0).eval()
maf = tf.concat(maf_list, 0).eval()
if return_r2:
r2 = tf.concat(r2_list, 0).eval()
if verbose:
print()
# writer.close()
logger.write(' time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
if return_sparse:
ix = pval.indices[:,0]<n_variants # truncate last batch
v = [variant_dict[i] for i in pval.indices[ix,0]]
if phenotype_df.shape[0]>1:
phenotype_ids = phenotype_df.index[pval.indices[ix,1]]
else:
phenotype_ids = phenotype_df.index.tolist()*len(pval.values)
pval_df = pd.DataFrame(
np.array([v, phenotype_ids, pval.values[ix], maf[ix]]).T,
columns=['variant_id', 'phenotype_id', 'pval', 'maf']
)
pval_df['pval'] = pval_df['pval'].astype(np.float64)
pval_df['maf'] = pval_df['maf'].astype(np.float32)
if return_r2:
pval_df['r2'] = r2[ix].astype(np.float32)
else:
# truncate last batch
pval = pval[:n_variants]
maf = maf[:n_variants]
# add indices
pval_df = pd.DataFrame(pval, index=variant_ids, columns=[i for i in phenotype_df.index])
pval_df['maf'] = maf
pval_df.index.name = 'variant_id'
if maf_threshold is not None and maf_threshold>0:
logger.write(' * filtering output by MAF >= {}'.format(maf_threshold))
pval_df = pval_df[pval_df['maf']>=maf_threshold]
logger.write('done.')
return pval_df
def map_trans_permutations(genotype_input, phenotype_df, covariates_df, permutations=None,
split_chr=True, pval_df=None, nperms=10000, maf_threshold=0.05,
batch_size=20000, logger=None):
"""
Warning: this function requires that all phenotypes are normally distributed,
e.g., inverse normal transformed
"""
if logger is None:
logger = SimpleLogger()
assert np.all(phenotype_df.columns==covariates_df.index)
if split_chr:
plink_reader = genotype_input
# assert isinstance(plink_reader, genotypeio.PlinkReader)
if pval_df is not None:
assert 'phenotype_chr' in pval_df and 'pval' in pval_df and np.all(pval_df.index==phenotype_df.index)
variant_ids = plink_reader.bim['snp'].tolist()
# index of VCF samples corresponding to phenotypes
ix_t = get_sample_indexes(plink_reader.fam['iid'].tolist(), phenotype_df)
else:
genotype_df = genotype_input
assert isinstance(genotype_df, pd.DataFrame)
variant_ids = genotype_df.index.tolist()
# index of VCF samples corresponding to phenotypes
ix_t = get_sample_indexes(genotype_df.columns.tolist(), phenotype_df)
n_variants = len(variant_ids)
n_samples = phenotype_df.shape[1]
dof = phenotype_df.shape[1] - 2 - covariates_df.shape[1]
logger.write('trans-QTL mapping (FDR)')
logger.write(' * {} samples'.format(n_samples))
logger.write(' * {} phenotypes'.format(phenotype_df.shape[0]))
logger.write(' * {} covariates'.format(covariates_df.shape[1]))
logger.write(' * {} variants'.format(n_variants))
if permutations is None: # generate permutations
q = stats.norm.ppf(np.arange(1,n_samples+1)/(n_samples+1))
qv = np.tile(q,[nperms,1])
for i in np.arange(nperms):
np.random.shuffle(qv[i,:])
else:
assert permutations.shape[1]==n_samples
nperms = permutations.shape[0]
qv = permutations
logger.write(' * {} permutations'.format(nperms))
permutations_t = tf.constant(qv, dtype=tf.float32)
permutations_t = tf.reshape(permutations_t, shape=[-1, n_samples])
genotypes_t, phenotypes_t, covariates_t = initialize_data(phenotype_df, covariates_df, batch_size=batch_size, dtype=tf.float32)
max_r2_nominal_t, max_r2_permuted_t, idxmax_t = _calculate_max_r2(genotypes_t, phenotypes_t, permutations_t, covariates_t, maf_threshold=maf_threshold)
init_op = tf.group(tf.global_variables_initializer(), tf.local_variables_initializer())
if split_chr:
start_time = time.time()
chr_max_r2_nominal = OrderedDict()
chr_max_r2_empirical = OrderedDict()
print(' Mapping batches')
with tf.Session() as sess:
sess.run(init_op)
for chrom in plink_reader.bim['chrom'].unique():
genotypes, variant_pos = plink_reader.get_region(chrom, verbose=True)
ggt = genotypeio.GenotypeGeneratorTrans(genotypes, batch_size=batch_size, dtype=np.float32)
dataset_genotypes = tf.data.Dataset.from_generator(ggt.generate_data, output_types=tf.float32)
dataset_genotypes = dataset_genotypes.prefetch(10)
iterator = dataset_genotypes.make_one_shot_iterator()
next_element = iterator.get_next()
next_element = tf.gather(next_element, ix_t, axis=1)
nominal_list = []
perms_list = []
nominal_idx_list = []
for i in range(1, ggt.num_batches+1):
sys.stdout.write('\r * {}: processing batch {}/{} '.format(chrom, i, ggt.num_batches))
sys.stdout.flush()
g_iter = sess.run(next_element)
max_r2_nominal, max_r2_permuted = sess.run([max_r2_nominal_t, max_r2_permuted_t], feed_dict={genotypes_t:g_iter})
nominal_list.append(max_r2_nominal)
perms_list.append(max_r2_permuted)
chr_max_r2_nominal[chrom] = np.max(np.array(nominal_list), 0)
chr_max_r2_empirical[chrom] = np.max(np.array(perms_list), 0)
logger.write(' time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
chr_max_r2_nominal = pd.DataFrame(chr_max_r2_nominal, index=phenotype_df.index)
chr_max_r2_empirical = pd.DataFrame(chr_max_r2_empirical)
# compute leave-one-out max
max_r2_nominal = OrderedDict()
max_r2_empirical = OrderedDict()
for c in chr_max_r2_nominal:
max_r2_nominal[c] = chr_max_r2_nominal[np.setdiff1d(chr_max_r2_nominal.columns, c)].max(1)
max_r2_empirical[c] = chr_max_r2_empirical[np.setdiff1d(chr_max_r2_empirical.columns, c)].max(1)
max_r2_nominal = pd.DataFrame(max_r2_nominal, index=phenotype_df.index)
max_r2_empirical = pd.DataFrame(max_r2_empirical) # nperms x chrs
if pval_df is not None:
# nominal p-value (sanity check, matches pval_df['pval'])
r2_nominal = max_r2_nominal.lookup(pval_df['phenotype_id'], pval_df['phenotype_chr'])
tstat = np.sqrt( dof*r2_nominal / (1-r2_nominal) )
minp_nominal = pd.Series(2*stats.t.cdf(-np.abs(tstat), dof), index=pval_df['phenotype_id'])
# empirical p-values
tstat = np.sqrt( dof*max_r2_empirical / (1-max_r2_empirical) )
minp_empirical = pd.DataFrame(2*stats.t.cdf(-np.abs(tstat), dof), columns=tstat.columns)
if pval_df is not None:
pval_perm = np.array([(np.sum(minp_empirical[chrom]<=p)+1)/(nperms+1) for p, chrom in zip(pval_df['pval'], pval_df['phenotype_chr'])])
beta_shape1 = {}
beta_shape2 = {}
true_dof = {}
for c in max_r2_empirical:
beta_shape1[c], beta_shape2[c], true_dof[c] = fit_beta_parameters(max_r2_empirical[c], dof)
if pval_df is not None:
beta_shape1 = [beta_shape1[c] for c in pval_df['phenotype_chr']]
beta_shape2 = [beta_shape2[c] for c in pval_df['phenotype_chr']]
true_dof = [true_dof[c] for c in pval_df['phenotype_chr']]
else:
chroms = plink_reader.bim['chrom'].unique()
beta_shape1 = [beta_shape1[c] for c in chroms]
beta_shape2 = [beta_shape2[c] for c in chroms]
true_dof = [true_dof[c] for c in chroms]
if pval_df is not None:
pval_true_dof = pval_from_corr(r2_nominal, true_dof)
pval_beta = stats.beta.cdf(pval_true_dof, beta_shape1, beta_shape2)
variant_id = [np.NaN]*len(pval_beta)
else:
ggt = genotypeio.GenotypeGeneratorTrans(genotype_df.values, batch_size=batch_size, dtype=np.float32)
dataset_genotypes = tf.data.Dataset.from_generator(ggt.generate_data, output_types=tf.float32)
dataset_genotypes = dataset_genotypes.prefetch(10)
iterator = dataset_genotypes.make_one_shot_iterator()
next_element = iterator.get_next()
next_element = tf.gather(next_element, ix_t, axis=1)
start_time = time.time()
max_r2_nominal = []
max_r2_nominal_idx = []
max_r2_empirical = []
with tf.Session() as sess:
sess.run(init_op)
for i in range(ggt.num_batches):
sys.stdout.write('\rProcessing batch {}/{}'.format(i+1, ggt.num_batches))
sys.stdout.flush()
g_iter = sess.run(next_element)
res = sess.run([max_r2_nominal_t, max_r2_permuted_t, idxmax_t], feed_dict={genotypes_t:g_iter})
max_r2_nominal.append(res[0])
max_r2_nominal_idx.append(res[2] + i*batch_size)
max_r2_empirical.append(res[1])
print()
max_r2_nominal = np.array(max_r2_nominal)
max_r2_nominal_idx = np.array(max_r2_nominal_idx)
max_r2_empirical = np.array(max_r2_empirical)
logger.write(' time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
if len(max_r2_nominal_idx.shape)==1:
max_r2_nominal = max_r2_nominal.reshape(-1,1)
max_r2_nominal_idx = max_r2_nominal_idx.reshape(-1,1)
idxmax = np.argmax(max_r2_nominal, 0)
variant_ix = [max_r2_nominal_idx[i,k] for k,i in enumerate(idxmax)]
variant_id = genotype_df.index[variant_ix]
max_r2_nominal = np.max(max_r2_nominal, 0)
tstat = np.sqrt( dof*max_r2_nominal / (1-max_r2_nominal) )
minp_nominal = 2*stats.t.cdf(-np.abs(tstat), dof)
max_r2_empirical = np.max(max_r2_empirical, 0)
tstat = np.sqrt( dof*max_r2_empirical / (1-max_r2_empirical) )
minp_empirical = 2*stats.t.cdf(-np.abs(tstat), dof)
pval_perm = np.array([(np.sum(minp_empirical<=p)+1)/(nperms+1) for p in minp_nominal])
# calculate beta p-values for each phenotype:
beta_shape1, beta_shape2, true_dof, minp_vec = fit_beta_parameters(max_r2_empirical, dof, tol=1e-4, return_minp=True)
pval_true_dof = pval_from_corr(max_r2_nominal, true_dof)
pval_beta = stats.beta.cdf(pval_true_dof, beta_shape1, beta_shape2)
if not split_chr or pval_df is not None:
fit_df = pd.DataFrame(OrderedDict([
('beta_shape1', beta_shape1),
('beta_shape2', beta_shape2),
('true_df', true_dof),
('min_pval_true_df', pval_true_dof),
('variant_id', variant_id),
('min_pval_nominal', minp_nominal),
('pval_perm', pval_perm),
('pval_beta', pval_beta),
]), index=phenotype_df.index)
else:
fit_df = pd.DataFrame(OrderedDict([
('beta_shape1', beta_shape1),
('beta_shape2', beta_shape2),
('true_df', true_dof),
]), index=chroms)
if split_chr:
return fit_df, minp_empirical
else:
return fit_df, minp_vec
def map_trans_tfrecord(vcf_tfrecord, phenotype_df, covariates_df, interaction_s=None, return_sparse=True, pval_threshold=1e-5, maf_threshold=0.05, batch_size=50000, logger=None):
"""Run trans-QTL mapping from genotypes in tfrecord"""
if logger is None:
logger = SimpleLogger()
assert np.all(phenotype_df.columns==covariates_df.index)
with open(vcf_tfrecord+'.samples') as f:
vcf_sample_ids = f.read().strip().split('\n')
n_samples_vcf = len(vcf_sample_ids)
with gzip.open(vcf_tfrecord+'.variants.gz', 'rt') as f:
variant_ids = f.read().strip().split('\n')
variant_dict = {i:j for i,j in enumerate(variant_ids)}
n_variants = len(variant_ids)
# index of VCF samples corresponding to phenotypes
ix_t = get_sample_indexes(vcf_sample_ids, phenotype_df)
n_samples = phenotype_df.shape[1]
# batched_dataset = dataset.apply(tf.contrib.data.padded_batch(batch_size, padded_shapes=[[batch_size], [None]]))
# batched_dataset = dataset.padded_batch(batch_size, padded_shapes=(batch_size,n_samples), padding_values=0)
with tf.device('/cpu:0'):
batched_dataset = genotypeio.make_batched_dataset(vcf_tfrecord, batch_size, ix_t=ix_t)
iterator = batched_dataset.make_one_shot_iterator()
next_element = iterator.get_next()
next_element = genotypeio.pad_up_to(next_element, [batch_size, n_samples]) # not clear if right/best way to do this
logger.write(' * {} samples'.format(n_samples))
logger.write(' * {} phenotypes'.format(phenotype_df.shape[0]))
logger.write(' * {} covariates'.format(covariates_df.shape[1]))
logger.write(' * {} variants'.format(n_variants))
if interaction_s is not None:
logger.write(' * including interaction term')
num_batches = int(np.ceil(np.true_divide(n_variants, batch_size)))
# calculate correlation threshold for sparse output
if return_sparse:
dof = n_samples - 2 - covariates_df.shape[1]
t = stats.t.ppf(pval_threshold/2, dof)**2 / dof
r2_threshold = t / (1+t)
else:
r2_threshold = None
if interaction_s is None:
genotypes, phenotypes, covariates = initialize_data(phenotype_df, covariates_df, batch_size=batch_size)
with tf.device('/gpu:0'):
p_values, maf = calculate_association(genotypes, phenotypes, covariates, return_sparse=return_sparse, r2_threshold=r2_threshold)
else:
genotypes, phenotypes, covariates, interaction = initialize_data(phenotype_df, covariates_df, batch_size=batch_size, interaction_s=interaction_s)
p_values, maf = calculate_association(genotypes, phenotypes, covariates, interaction_t=interaction, return_sparse=return_sparse, r2_threshold=r2_threshold)
# g = _parse_function(next_element, batch_size, n_samples, ix_t)
init_op = tf.group(tf.global_variables_initializer(), tf.local_variables_initializer())
start_time = time.time()
with tf.Session() as sess:
# run_options = tf.RunOptions(trace_level=tf.RunOptions.FULL_TRACE)
# run_metadata = tf.RunMetadata()
# writer = tf.summary.FileWriter('logs', sess.graph, session=sess)
sess.run(init_op)
pval_list = []
maf_list = []
for i in range(num_batches):
sys.stdout.write('\rProcessing batch {}/{}'.format(i+1, num_batches))
sys.stdout.flush()
g_iter = sess.run(next_element)
# g_iter = sess.run(g)
p_ = sess.run([p_values, maf], feed_dict={genotypes:g_iter})#, options=run_options, run_metadata=run_metadata)
# writer.add_run_metadata(run_metadata, 'batch%d' % i)
pval_list.append(p_[0])
maf_list.append(p_[1])
if return_sparse:
pval = tf.sparse_concat(0, pval_list).eval()
else:
pval = tf.concat(pval_list, 0).eval()
maf = tf.concat(maf_list, 0).eval()
print()
# writer.close()
logger.write('Time elapsed: {:.2f} min'.format((time.time()-start_time)/60))
if return_sparse:
ix = pval.indices[:,0]<n_variants # truncate last batch
v = [variant_dict[i] for i in pval.indices[ix,0]]
pval_df = pd.DataFrame(
np.array([v, phenotype_df.index[pval.indices[ix,1]], pval.values[ix], maf[ix]]).T,
columns=['variant_id', 'phenotype_id', 'pval', 'maf']
)
pval_df['pval'] = pval_df['pval'].astype(np.float64)
pval_df['maf'] = pval_df['maf'].astype(np.float32)
else:
# truncate last batch
pval = pval[:n_variants]
maf = maf[:n_variants]
# add indices
pval_df = pd.DataFrame(pval, index=variant_ids, columns=[i for i in phenotype_df.index])
pval_df['maf'] = maf
pval_df.index.name = 'variant_id'
if maf_threshold is not None and maf_threshold>0:
logger.write(' * filtering output by MAF >= {}'.format(maf_threshold))
pval_df = pval_df[pval_df['maf']>=maf_threshold]
return pval_df
def _in_cis(chrom, pos, gene_id, tss_dict, window=1000000):
"""Test if a variant-gene pair is in cis"""
if chrom==tss_dict[gene_id]['chr']:
tss = tss_dict[gene_id]['tss']
if pos>=tss-window and pos<=tss+window:
return True
else:
return False
else:
return False
def filter_cis(pval_df, tss_dict, window=1000000):
"""Filter out cis-QTLs"""
drop_ix = []
for k,gene_id,variant_id in zip(pval_df['phenotype_id'].index, pval_df['phenotype_id'], pval_df['variant_id']):
chrom, pos = variant_id.split('_',2)[:2]
pos = int(pos)
if _in_cis(chrom, pos, gene_id, tss_dict, window=window):
drop_ix.append(k)
return pval_df.drop(drop_ix)
#------------------------------------------------------------------------------
# Input parsers
#------------------------------------------------------------------------------
def read_phenotype_bed(phenotype_bed):
"""Load phenotype BED file as phenotype and TSS DataFrames"""
if phenotype_bed.endswith('.bed.gz'):
phenotype_df = pd.read_csv(phenotype_bed, sep='\t', index_col=3, dtype={'#chr':str, '#Chr':str})
elif phenotype_bed.endswith('.parquet'):
phenotype_df =
|
pd.read_parquet(phenotype_bed)
|
pandas.read_parquet
|
from datetime import datetime
import numpy as np
import pytest
import pandas as pd
from pandas import (
DataFrame,
Index,
MultiIndex,
Series,
_testing as tm,
)
def test_split(any_string_dtype):
values = Series(["a_b_c", "c_d_e", np.nan, "f_g_h"], dtype=any_string_dtype)
result = values.str.split("_")
exp = Series([["a", "b", "c"], ["c", "d", "e"], np.nan, ["f", "g", "h"]])
tm.assert_series_equal(result, exp)
# more than one char
values = Series(["a__b__c", "c__d__e", np.nan, "f__g__h"], dtype=any_string_dtype)
result = values.str.split("__")
tm.assert_series_equal(result, exp)
result = values.str.split("__", expand=False)
tm.assert_series_equal(result, exp)
# regex split
values = Series(["a,b_c", "c_d,e", np.nan, "f,g,h"], dtype=any_string_dtype)
result = values.str.split("[,_]")
exp = Series([["a", "b", "c"], ["c", "d", "e"], np.nan, ["f", "g", "h"]])
tm.assert_series_equal(result, exp)
def test_split_object_mixed():
mixed = Series(["a_b_c", np.nan, "d_e_f", True, datetime.today(), None, 1, 2.0])
result = mixed.str.split("_")
exp = Series(
[
["a", "b", "c"],
np.nan,
["d", "e", "f"],
np.nan,
np.nan,
np.nan,
np.nan,
np.nan,
]
)
assert isinstance(result, Series)
tm.assert_almost_equal(result, exp)
result = mixed.str.split("_", expand=False)
assert isinstance(result, Series)
tm.assert_almost_equal(result, exp)
@pytest.mark.parametrize("method", ["split", "rsplit"])
def test_split_n(any_string_dtype, method):
s = Series(["a b", pd.NA, "b c"], dtype=any_string_dtype)
expected = Series([["a", "b"], pd.NA, ["b", "c"]])
result = getattr(s.str, method)(" ", n=None)
tm.assert_series_equal(result, expected)
result = getattr(s.str, method)(" ", n=0)
tm.assert_series_equal(result, expected)
def test_rsplit(any_string_dtype):
values = Series(["a_b_c", "c_d_e", np.nan, "f_g_h"], dtype=any_string_dtype)
result = values.str.rsplit("_")
exp = Series([["a", "b", "c"], ["c", "d", "e"], np.nan, ["f", "g", "h"]])
tm.assert_series_equal(result, exp)
# more than one char
values = Series(["a__b__c", "c__d__e", np.nan, "f__g__h"], dtype=any_string_dtype)
result = values.str.rsplit("__")
tm.assert_series_equal(result, exp)
result = values.str.rsplit("__", expand=False)
tm.assert_series_equal(result, exp)
# regex split is not supported by rsplit
values = Series(["a,b_c", "c_d,e", np.nan, "f,g,h"], dtype=any_string_dtype)
result = values.str.rsplit("[,_]")
exp = Series([["a,b_c"], ["c_d,e"], np.nan, ["f,g,h"]])
tm.assert_series_equal(result, exp)
# setting max number of splits, make sure it's from reverse
values = Series(["a_b_c", "c_d_e", np.nan, "f_g_h"], dtype=any_string_dtype)
result = values.str.rsplit("_", n=1)
exp = Series([["a_b", "c"], ["c_d", "e"], np.nan, ["f_g", "h"]])
tm.assert_series_equal(result, exp)
def test_rsplit_object_mixed():
# mixed
mixed = Series(["a_b_c", np.nan, "d_e_f", True, datetime.today(), None, 1, 2.0])
result = mixed.str.rsplit("_")
exp = Series(
[
["a", "b", "c"],
np.nan,
["d", "e", "f"],
np.nan,
np.nan,
np.nan,
np.nan,
np.nan,
]
)
assert isinstance(result, Series)
tm.assert_almost_equal(result, exp)
result = mixed.str.rsplit("_", expand=False)
assert isinstance(result, Series)
tm.assert_almost_equal(result, exp)
def test_split_blank_string(any_string_dtype):
# expand blank split GH 20067
values = Series([""], name="test", dtype=any_string_dtype)
result = values.str.split(expand=True)
exp = DataFrame([[]], dtype=any_string_dtype) # NOTE: this is NOT an empty df
tm.assert_frame_equal(result, exp)
values = Series(["a b c", "a b", "", " "], name="test", dtype=any_string_dtype)
result = values.str.split(expand=True)
exp = DataFrame(
[
["a", "b", "c"],
["a", "b", np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
],
dtype=any_string_dtype,
)
|
tm.assert_frame_equal(result, exp)
|
pandas._testing.assert_frame_equal
|
'''
/*******************************************************************************
* Copyright 2016-2019 Exactpro (Exactpro Systems Limited)
*
* Licensed under the Apache License, Version 2.0 (the "License");
* you may not use this file except in compliance with the License.
* You may obtain a copy of the License at
*
* http://www.apache.org/licenses/LICENSE-2.0
*
* Unless required by applicable law or agreed to in writing, software
* distributed under the License is distributed on an "AS IS" BASIS,
* WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
* See the License for the specific language governing permissions and
* limitations under the License.
******************************************************************************/
'''
import numpy
import matplotlib.pyplot as plt
import scipy.stats as stats
import pandas
import datetime
import calendar
class RelativeFrequencyChart:
# returns coordinates for each chart column
def get_coordinates(self, data, bins): # bins - chart columns count
self.btt = numpy.array(list(data))
self.y, self.x, self.bars = plt.hist(self.btt, weights=numpy.zeros_like(self.btt) + 1. / self.btt.size, bins=bins)
return self.x, self.y
class FrequencyDensityChart:
def get_coordinates_histogram(self, data, bins):
self.btt = numpy.array(list(data))
self.y, self.x, self.bars = plt.hist(self.btt, bins=bins, density=True)
return self.x, self.y
def get_coordinates_line(self, data):
try:
self.btt = numpy.array(list(data))
self.density = stats.kde.gaussian_kde(list(data))
self.x_den = numpy.linspace(0, data.max(), data.count())
self.density = self.density(self.x_den)
return self.x_den, self.density
except numpy.linalg.linalg.LinAlgError:
return [-1], [-1]
class DynamicChart:
def get_coordinates(self, frame, step_size):
self.plot = {} # chart coordinates
self.dynamic_bugs = []
self.x = []
self.y = []
self.plot['period'] = step_size
if step_size == 'W-SUN':
self.periods = DynamicChart.get_periods(self, frame, step_size) # separates DataFrame to the specified periods
if len(self.periods) == 0:
return 'error'
self.cumulative = 0 # cumulative total of defect submission for specific period
for self.period in self.periods:
# checks whether the first day of period is Monday (if not then we change first day to Monday)
if pandas.to_datetime(self.period[0]) < pandas.to_datetime(frame['Created_tr']).min():
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >=
pandas.to_datetime(frame['Created_tr']).min()) &
(pandas.to_datetime(frame['Created_tr']) <= pandas.to_datetime(self.period[1]))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(datetime.datetime.date(pandas.to_datetime(frame['Created_tr'], format='%Y-%m-%d').min())))
self.y.append(self.cumulative)
else:
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(self.period[0]))
& (pandas.to_datetime(frame['Created_tr']) <= pandas.to_datetime(self.period[1]))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str((self.period[0])))
self.y.append(self.cumulative)
# check whether the date from new DataFrame is greater than date which is specified in settings
if pandas.to_datetime(frame['Created_tr']).max() > pandas.to_datetime(self.periods[-1][1]):
# processing of days which are out of full period set
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) > pandas.to_datetime(self.periods[-1][1]))
& (pandas.to_datetime(frame['Created_tr']) <=
pandas.to_datetime(frame['Created_tr']).max())]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(datetime.datetime.date(pandas.to_datetime(self.periods[-1][1], format='%Y-%m-%d')) + datetime.timedelta(days=1)))
self.y.append(self.cumulative)
self.dynamic_bugs.append(self.x)
self.dynamic_bugs.append(self.y)
self.plot['dynamic bugs'] = self.dynamic_bugs
self.cumulative = 0
return self.plot
if step_size in ['7D', '10D', '3M', '6M', 'A-DEC']:
self.count0 = 0
self.count1 = 1
self.periods = DynamicChart.get_periods(self, frame, step_size) # DataFrame separation by the specified periods
if len(self.periods) == 0:
return 'error'
self.cumulative = 0
self.countPeriodsList = len(self.periods) # count of calculated periods
self.count = 1
if self.countPeriodsList == 1:
if step_size == '7D':
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >=
pandas.to_datetime(frame['Created_tr']).min())
& (pandas.to_datetime(frame['Created_tr'])
< pandas.to_datetime(datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())
+datetime.timedelta(days=7)))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr'], format='%Y-%m-%d').min()), step_size)))
self.y.append(self.cumulative)
if pandas.to_datetime(frame['Created_tr']).max() > pandas.to_datetime(datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())+datetime.timedelta(days=7)):
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >=
pandas.to_datetime(datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())+
datetime.timedelta(days=7))) & (pandas.to_datetime(frame['Created_tr'])
<= pandas.to_datetime(frame['Created_tr']).max())]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())+datetime.timedelta(days=7), step_size)))
self.y.append(self.cumulative)
self.cumulative = 0
if step_size == '10D':
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(frame['Created_tr']).min()) & (pandas.to_datetime(frame['Created_tr']) < pandas.to_datetime(datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())+datetime.timedelta(days=10)))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr'], format='%Y-%m-%d').min()), step_size)))
self.y.append(self.cumulative)
if pandas.to_datetime(frame['Created_tr']).max() > pandas.to_datetime(datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())+datetime.timedelta(days=10)):
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())+datetime.timedelta(days=10))) & (pandas.to_datetime(frame['Created_tr']) <= pandas.to_datetime(frame['Created_tr']).max())]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min())+datetime.timedelta(days=10), step_size)))
self.y.append(self.cumulative)
self.cumulative = 0
if step_size == '3M':
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(frame['Created_tr']).min())
& (pandas.to_datetime(frame['Created_tr']) <
pandas.to_datetime(DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 3)))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr'], format='%Y-%m-%d').min()), step_size)))
self.y.append(self.cumulative)
if pandas.to_datetime(frame['Created_tr']).max() > pandas.to_datetime(DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 3)):
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 3))) & (pandas.to_datetime(frame['Created_tr']) <= pandas.to_datetime(frame['Created_tr']).max())]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 3), step_size)))
self.y.append(self.cumulative)
self.cumulative = 0
if step_size == '6M':
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(frame['Created_tr']).min())
& (pandas.to_datetime(frame['Created_tr']) <
pandas.to_datetime(DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 6)))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr'], format='%Y-%m-%d').min()), step_size)))
self.y.append(self.cumulative)
if pandas.to_datetime(frame['Created_tr']).max() > pandas.to_datetime(DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 6)):
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 6))) & (pandas.to_datetime(frame['Created_tr']) <= pandas.to_datetime(frame['Created_tr']).max())]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, DynamicChart.add_months(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr']).min()), 6), step_size)))
self.y.append(self.cumulative)
self.cumulative = 0
if step_size == 'A-DEC':
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(frame['Created_tr']).min())
& (pandas.to_datetime(frame['Created_tr']) < pandas.to_datetime(str(int(self.periods[0])+1)))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(frame['Created_tr'], format='%Y-%m-%d').min()), step_size)))
self.y.append(self.cumulative)
if(pandas.to_datetime(frame['Created_tr']).max() > pandas.to_datetime(str(int(self.periods[0])+1))):
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(str(int(self.periods[0])+1)))
& (pandas.to_datetime(frame['Created_tr']) <= pandas.to_datetime(frame['Created_tr']).max())]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(str(int(self.periods[0])+1))), step_size)))
self.y.append(self.cumulative)
self.cumulative = 0
else:
while self.count < self.countPeriodsList:
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(self.periods[self.count0])) &
(pandas.to_datetime(frame['Created_tr']) < pandas.to_datetime(self.periods[self.count1]))]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(self.periods[self.count0], format='%Y-%m-%d')), step_size)))
self.y.append(self.cumulative)
self.count0 = self.count0 + 1
self.count1 = self.count1 + 1
self.count = self.count + 1
if pandas.to_datetime(frame['Created_tr']).max() >= pandas.to_datetime(self.periods[-1]):
self.newFrame = frame[(pandas.to_datetime(frame['Created_tr']) >= pandas.to_datetime(self.periods[-1])) &
(pandas.to_datetime(frame['Created_tr']) <= pandas.to_datetime(frame['Created_tr']).max())]
self.cumulative = self.cumulative + int(self.newFrame['Issue_key_tr'].count())
self.x.append(str(DynamicChart.get_date_for_dynamic_my(self, datetime.datetime.date(pandas.to_datetime(self.periods[-1], format='%Y-%m-%d')), step_size)))
self.y.append(self.cumulative)
self.cumulative = 0
self.dynamic_bugs.append(self.x)
self.dynamic_bugs.append(self.y)
self.plot['dynamic bugs'] = self.dynamic_bugs
return self.plot
# DataFrame separation (by periods)
def get_periods(self, frame, period):
self.periods = []
self.periodsFrame = pandas.period_range(start=
|
pandas.to_datetime(frame['Created_tr'])
|
pandas.to_datetime
|
from __future__ import division #brings in Python 3.0 mixed type calculation rules
import datetime
import inspect
import numpy as np
import numpy.testing as npt
import os.path
import pandas as pd
import sys
from tabulate import tabulate
import unittest
##find parent directory and import model
#parentddir = os.path.abspath(os.path.join(os.path.dirname(__file__), os.path.pardir))
#sys.path.append(parentddir)
from ..agdrift_exe import Agdrift
test = {}
class TestAgdrift(unittest.TestCase):
"""
IEC unit tests.
"""
def setUp(self):
"""
setup the test as needed
e.g. pandas to open agdrift qaqc csv
Read qaqc csv and create pandas DataFrames for inputs and expected outputs
:return:
"""
pass
def tearDown(self):
"""
teardown called after each test
e.g. maybe write test results to some text file
:return:
"""
pass
def create_agdrift_object(self):
# create empty pandas dataframes to create empty object for testing
df_empty = pd.DataFrame()
# create an empty agdrift object
agdrift_empty = Agdrift(df_empty, df_empty)
return agdrift_empty
def test_validate_sim_scenarios(self):
"""
:description determines if user defined scenarios are valid for processing
:param application_method: type of Tier I application method employed
:param aquatic_body_def: type of endpoint of concern (e.g., pond, wetland); implies whether
: endpoint of concern parameters (e.g.,, pond width) are set (i.e., by user or EPA standard)
:param drop_size_*: qualitative description of spray droplet size for aerial & ground applications
:param boom_height: qualitative height above ground of spray boom
:param airblast_type: type of orchard being sprayed
:NOTE we perform an additional validation check related to distances later in the code just before integration
:return
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
agdrift_empty.out_sim_scenario_chk = pd.Series([], dtype='object')
expected_result = pd.Series([
'Valid Tier I Aquatic Aerial Scenario',
'Valid Tier I Terrestrial Aerial Scenario',
'Valid Tier I Aquatic Aerial Scenario',
'Valid Tier I Terrestrial Aerial Scenario',
'Valid Tier I Aquatic Aerial Scenario',
'Valid Tier I Terrestrial Ground Scenario',
'Valid Tier I Aquatic Ground Scenario',
'Valid Tier I Terrestrial Ground Scenario',
'Valid Tier I Aquatic Ground Scenario',
'Valid Tier I Terrestrial Airblast Scenario',
'Valid Tier I Aquatic Airblast Scenario',
'Valid Tier I Terrestrial Airblast Scenario',
'Valid Tier I Aquatic Airblast Scenario',
'Valid Tier I Terrestrial Airblast Scenario',
'Invalid Tier I Aquatic Aerial Scenario',
'Invalid Tier I Aquatic Ground Scenario',
'Invalid Tier I Aquatic Airblast Scenario',
'Invalid Tier I Terrestrial Aerial Scenario',
'Valid Tier I Terrestrial Ground Scenario',
'Valid Tier I Terrestrial Airblast Scenario',
'Invalid scenario ecosystem_type',
'Invalid Tier I Aquatic Assessment application method',
'Invalid Tier I Terrestrial Assessment application method'],dtype='object')
try:
#set test data
agdrift_empty.num_simulations = len(expected_result)
agdrift_empty.application_method = pd.Series(
['tier_1_aerial',
'tier_1_aerial',
'tier_1_aerial',
'tier_1_aerial',
'tier_1_aerial',
'tier_1_ground',
'tier_1_ground',
'tier_1_ground',
'tier_1_ground',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_aerial',
'tier_1_ground',
'tier_1_airblast',
'tier_1_aerial',
'tier_1_ground',
'tier_1_airblast',
'tier_1_aerial',
'Tier II Aerial',
'Tier III Aerial'], dtype='object')
agdrift_empty.ecosystem_type = pd.Series(
['aquatic_assessment',
'terrestrial_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'aquatic_assessment',
'aquatic_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'terrestrial_assessment',
'terrestrial_assessment',
'Field Assessment',
'aquatic_assessment',
'terrestrial_assessment'], dtype='object')
agdrift_empty.aquatic_body_type = pd.Series(
['epa_defined_pond',
'NaN',
'epa_defined_wetland',
'NaN',
'user_defined_pond',
'NaN',
'user_defined_wetland',
'NaN',
'epa_defined_wetland',
'NaN',
'user_defined_pond',
'NaN',
'user_defined_wetland',
'NaN',
'Defined Pond',
'user_defined_pond',
'epa_defined_pond',
'NaN',
'NaN',
'NaN',
'epa_defined_pond',
'user_defined_wetland',
'user_defined_pond'], dtype='object')
agdrift_empty.terrestrial_field_type = pd.Series(
['NaN',
'user_defined_terrestrial',
'NaN',
'epa_defined_terrestrial',
'NaN',
'user_defined_terrestrial',
'NaN',
'user_defined_terrestrial',
'NaN',
'epa_defined_terrestrial',
'NaN',
'user_defined_terrestrial',
'NaN',
'user_defined_terrestrial',
'NaN',
'NaN',
'NaN',
'user_defined_terrestrial',
'user_defined_terrestrial',
'user_defined_terrestrial',
'NaN',
'NaN',
'user_defined_terrestrial'], dtype='object')
agdrift_empty.drop_size_aerial = pd.Series(
['very_fine_to_fine',
'fine_to_medium',
'medium_to_coarse',
'coarse_to_very_coarse',
'fine_to_medium',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'medium_to_coarse',
'NaN',
'very_fine_to_medium',
'NaN',
'very_fine Indeed',
'NaN',
'very_fine_to_medium',
'medium_to_coarse',
'NaN'], dtype='object')
agdrift_empty.drop_size_ground = pd.Series(
['NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'very_fine',
'fine_to_medium-coarse',
'very_fine',
'fine_to_medium-coarse',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'very_fine',
'NaN',
'fine_to_medium-coarse',
'very_fine',
'NaN',
'very_fine_to_medium',
'NaN',
'very_fine'], dtype='object')
agdrift_empty.boom_height = pd.Series(
['NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'high',
'low',
'high',
'low',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'high',
'NaN',
'NaN',
'NaN',
'NaN'],dtype='object')
agdrift_empty.airblast_type = pd.Series(
['NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'normal',
'dense',
'sparse',
'orchard',
'vineyard',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'vineyard',
'NaN',
'NaN',
'NaN'], dtype='object')
agdrift_empty.validate_sim_scenarios()
result = agdrift_empty.out_sim_scenario_chk
npt.assert_array_equal(result, expected_result, err_msg="", verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_set_sim_scenario_id(self):
"""
:description provides scenario ids per simulation that match scenario names (i.e., column_names) from SQL database
:param out_sim_scenario_id: scenario name as assigned to individual simulations
:param num_simulations: number of simulations to assign scenario names
:param out_sim_scenario_chk: from previous method where scenarios were checked for validity
:param application_method: application method of scenario
:param drop_size_*: qualitative description of spray droplet size for aerial and ground applications
:param boom_height: qualitative height above ground of spray boom
:param airblast_type: type of airblast application (e.g., vineyard, orchard)
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series(['aerial_vf2f',
'aerial_f2m',
'aerial_m2c',
'aerial_c2vc',
'ground_low_vf',
'ground_low_fmc',
'ground_high_vf',
'ground_high_fmc',
'airblast_normal',
'airblast_dense',
'airblast_sparse',
'airblast_vineyard',
'airblast_orchard',
'Invalid'], dtype='object')
try:
agdrift_empty.num_simulations = len(expected_result)
agdrift_empty.out_sim_scenario_chk = pd.Series(['Valid Tier I Aerial',
'Valid Tier I Aerial',
'Valid Tier I Aerial',
'Valid Tier I Aerial',
'Valid Tier I Ground',
'Valid Tier I Ground',
'Valid Tier I Ground',
'Valid Tier I Ground',
'Valid Tier I Airblast',
'Valid Tier I Airblast',
'Valid Tier I Airblast',
'Valid Tier I Airblast',
'Valid Tier I Airblast',
'Invalid Scenario'], dtype='object')
agdrift_empty.application_method = pd.Series(['tier_1_aerial',
'tier_1_aerial',
'tier_1_aerial',
'tier_1_aerial',
'tier_1_ground',
'tier_1_ground',
'tier_1_ground',
'tier_1_ground',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_airblast',
'tier_1_aerial'], dtype='object')
agdrift_empty.drop_size_aerial = pd.Series(['very_fine_to_fine',
'fine_to_medium',
'medium_to_coarse',
'coarse_to_very_coarse',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN'], dtype='object')
agdrift_empty.drop_size_ground = pd.Series(['NaN',
'NaN',
'NaN',
'NaN',
'very_fine',
'fine_to_medium-coarse',
'very_fine',
'fine_to_medium-coarse',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN'], dtype='object')
agdrift_empty.boom_height = pd.Series(['NaN',
'NaN',
'NaN',
'NaN',
'low',
'low',
'high',
'high',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN'], dtype='object')
agdrift_empty.airblast_type = pd.Series(['NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'NaN',
'normal',
'dense',
'sparse',
'vineyard',
'orchard',
'NaN'], dtype='object')
agdrift_empty.set_sim_scenario_id()
result = agdrift_empty.out_sim_scenario_id
npt.assert_array_equal(result, expected_result, err_msg="", verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_assign_column_names(self):
"""
:description assigns column names (except distaqnce column) from sql database to internal scenario names
:param column_name: short name for pesiticide application scenario for which distance vs deposition data is provided
:param scenario_name: internal variable for holding scenario names
:param scenario_number: index for scenario_name (this method assumes the distance values could occur in any column
:param distance_name: internal name for the column holding distance data
:NOTE to test both outputs of this method I simply appended them together
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
agdrift_empty.scenario_name = pd.Series([], dtype='object')
expected_result = pd.Series(['aerial_vf2f', 'aerial_f2m', 'aerial_m2c', 'aerial_c2vc',
'ground_low_vf', 'ground_low_fmc',
'ground_high_vf', 'ground_high_fmc',
'airblast_normal', 'airblast_dense', 'airblast_sparse',
'airblast_vineyard', 'airblast_orchard'], dtype='object')
try:
agdrift_empty.column_names = pd.Series(['aerial_vf2f', 'aerial_f2m', 'aerial_m2c', 'aerial_c2vc',
'ground_low_vf', 'ground_low_fmc',
'ground_high_vf', 'ground_high_fmc',
'airblast_normal', 'airblast_dense', 'airblast_sparse',
'airblast_vineyard', 'airblast_orchard', 'distance_ft'])
#call method to assign scenario names
agdrift_empty.assign_column_names()
result = agdrift_empty.scenario_name
npt.assert_array_equal(result, expected_result, err_msg="", verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_get_distances(self):
"""
:description retrieves distance values for deposition scenario datasets
: all scenarios use same distances
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE any blank fields are filled with 'nan'
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
location = os.path.realpath(os.path.join(os.getcwd(), os.path.dirname(__file__)))
agdrift_empty.db_name = os.path.join(location, 'sqlite_agdrift_distance.db')
agdrift_empty.db_table = 'output'
expected_result = pd.Series([], dtype='float')
try:
expected_result = [0.,0.102525,0.20505,0.4101,0.8202,1.6404,3.2808,4.9212,6.5616,9.8424,13.1232,19.6848,26.2464,
32.808,39.3696,45.9312,52.4928,59.0544,65.616,72.1776,78.7392,85.3008,91.8624,98.424,104.9856,
111.5472,118.1088,124.6704,131.232,137.7936,144.3552,150.9168,157.4784,164.04,170.6016,177.1632,
183.7248,190.2864,196.848,203.4096,209.9712,216.5328,223.0944,229.656,236.2176,242.7792,249.3408,
255.9024,262.464,269.0256,275.5872,282.1488,288.7104,295.272,301.8336,308.3952,314.9568,321.5184,
328.08,334.6416,341.2032,347.7648,354.3264,360.888,367.4496,374.0112,380.5728,387.1344,393.696,
400.2576,406.8192,413.3808,419.9424,426.504,433.0656,439.6272,446.1888,452.7504,459.312,465.8736,
472.4352,478.9968,485.5584,492.12,498.6816,505.2432,511.8048,518.3664,524.928,531.4896,538.0512,
544.6128,551.1744,557.736,564.2976,570.8592,577.4208,583.9824,590.544,597.1056,603.6672,610.2288,
616.7904,623.352,629.9136,636.4752,643.0368,649.5984,656.16,662.7216,669.2832,675.8448,682.4064,
688.968,695.5296,702.0912,708.6528,715.2144,721.776,728.3376,734.8992,741.4608,748.0224,754.584,
761.1456,767.7072,774.2688,780.8304,787.392,793.9536,800.5152,807.0768,813.6384,820.2,826.7616,
833.3232,839.8848,846.4464,853.008,859.5696,866.1312,872.6928,879.2544,885.816,892.3776,898.9392,
905.5008,912.0624,918.624,925.1856,931.7472,938.3088,944.8704,951.432,957.9936,964.5552,971.1168,
977.6784,984.24,990.8016,997.3632]
agdrift_empty.distance_name = 'distance_ft'
agdrift_empty.num_db_values = len(expected_result)
result = agdrift_empty.get_distances(agdrift_empty.num_db_values)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_get_scenario_deposition_data(self):
"""
:description retrieves deposition data for all scenarios from sql database
: and checks that for each the first, last, and total number of values
: are correct
:param scenario: name of scenario for which data is to be retrieved
:param num_values: number of values included in scenario datasets
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
#scenario_data = pd.Series([[]], dtype='float')
result = pd.Series([], dtype='float')
#changing expected values to the 161st
expected_result = [0.50013,0.041273,161.0, #aerial_vf2f
0.49997,0.011741,161.0, #aerial_f2m
0.4999,0.0053241,161.0, #aerial_m2c
0.49988,0.0031189,161.0, #aerial_c2vc
1.019339,9.66E-04,161.0, #ground_low_vf
1.007885,6.13E-04,161.0, #ground_low_fmc
1.055205,1.41E-03,161.0, #ground_high_vf
1.012828,7.72E-04,161.0, #ground_high_fmc
8.91E-03,3.87E-05,161.0, #airblast_normal
0.1155276,4.66E-04,161.0, #airblast_dense
0.4762651,5.14E-05,161.0, #airblast_sparse
3.76E-02,3.10E-05,161.0, #airblast_vineyard
0.2223051,3.58E-04,161.0] #airblast_orchard
try:
agdrift_empty.num_db_values = 161 #set number of data values in sql db
location = os.path.realpath(os.path.join(os.getcwd(), os.path.dirname(__file__)))
agdrift_empty.db_name = os.path.join(location, 'sqlite_agdrift_distance.db')
agdrift_empty.db_table = 'output'
#agdrift_empty.db_name = 'sqlite_agdrift_distance.db'
#this is the list of scenario names (column names) in sql db (the order here is important because
#the expected values are ordered in this manner
agdrift_empty.scenario_name = ['aerial_vf2f', 'aerial_f2m', 'aerial_m2c', 'aerial_c2vc',
'ground_low_vf', 'ground_low_fmc', 'ground_high_vf', 'ground_high_fmc',
'airblast_normal', 'airblast_dense', 'airblast_sparse', 'airblast_vineyard',
'airblast_orchard']
#cycle through reading scenarios and building result list
for i in range(len(agdrift_empty.scenario_name)):
#get scenario data
scenario_data = agdrift_empty.get_scenario_deposition_data(agdrift_empty.scenario_name[i],
agdrift_empty.num_db_values)
print(scenario_data)
#extract 1st and last values of scenario data and build result list (including how many values are
#retrieved for each scenario
if i == 0:
#fix this
result = [scenario_data[0], scenario_data[agdrift_empty.num_db_values - 1],
float(len(scenario_data))]
else:
result.extend([scenario_data[0], scenario_data[agdrift_empty.num_db_values - 1],
float(len(scenario_data))])
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_get_column_names(self):
"""
:description retrieves column names from sql database (sqlite_agdrift_distance.db)
: (each column name refers to a specific deposition scenario;
: the scenario name is used later to retrieve the deposition data)
:parameter output name of sql database table from which to retrieve requested data
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
location = os.path.realpath(os.path.join(os.getcwd(), os.path.dirname(__file__)))
agdrift_empty.db_name = os.path.join(location, 'sqlite_agdrift_distance.db')
agdrift_empty.db_table = 'output'
result = pd.Series([], dtype='object')
expected_result = ['distance_ft','aerial_vf2f', 'aerial_f2m', 'aerial_m2c', 'aerial_c2vc',
'ground_low_vf', 'ground_low_fmc', 'ground_high_vf', 'ground_high_fmc',
'airblast_normal', 'airblast_dense', 'airblast_sparse', 'airblast_vineyard',
'airblast_orchard']
try:
result = agdrift_empty.get_column_names()
npt.assert_array_equal(result, expected_result, err_msg="", verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_filter_arrays(self):
"""
:description eliminate blank data cells (i.e., distances for which no deposition value is provided)
(and thus reduce the number of x,y values to be used)
:parameter x_in: array of distance values associated with values for a deposition scenario (e.g., Aerial/EPA Defined Pond)
:parameter y_in: array of deposition values associated with a deposition scenario (e.g., Aerial/EPA Defined Pond)
:parameter x_out: processed array of x_in values eliminating indices of blank distance/deposition values
:parameter y_out: processed array of y_in values eliminating indices of blank distance/deposition values
:NOTE y_in array is assumed to be populated by values >= 0. except for the blanks as 'nan' entries
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([0.,1.,4.,5.,6.,7.], dtype='float')
expected_result_y = pd.Series([10.,11.,14.,15.,16.,17.], dtype='float')
try:
x_in = pd.Series([0.,1.,2.,3.,4.,5.,6.,7.], dtype='float')
y_in = pd.Series([10.,11.,'nan','nan',14.,15.,16.,17.], dtype='float')
x_out, y_out = agdrift_empty.filter_arrays(x_in, y_in)
result_x = x_out
result_y = y_out
npt.assert_allclose(result_x, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(result_y, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result_x, expected_result_x]
tab = [result_y, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_list_sims_per_scenario(self):
"""
:description scan simulations and count number and indices of simulations that apply to each scenario
:parameter num_scenarios number of deposition scenarios included in SQL database
:parameter num_simulations number of simulations included in this model execution
:parameter scenario_name name of deposition scenario as recorded in SQL database
:parameter out_sim_scenario_id identification of deposition scenario specified per model run simulation
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_num_sims = pd.Series([2,2,2,2,2,2,2,2,2,2,2,2,2], dtype='int')
expected_sim_indices = pd.Series([[0,13,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[1,14,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[2,15,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[3,16,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[4,17,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[5,18,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[6,19,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[7,20,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[8,21,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[9,22,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[10,23,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[11,24,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[12,25,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0]], dtype='int')
try:
agdrift_empty.scenario_name = pd.Series(['aerial_vf2f', 'aerial_f2m', 'aerial_m2c', 'aerial_c2vc',
'ground_low_vf', 'ground_low_fmc', 'ground_high_vf', 'ground_high_fmc',
'airblast_normal', 'airblast_dense', 'airblast_sparse', 'airblast_vineyard',
'airblast_orchard'], dtype='object')
agdrift_empty.out_sim_scenario_id = pd.Series(['aerial_vf2f', 'aerial_f2m', 'aerial_m2c', 'aerial_c2vc',
'ground_low_vf', 'ground_low_fmc', 'ground_high_vf', 'ground_high_fmc',
'airblast_normal', 'airblast_dense', 'airblast_sparse', 'airblast_vineyard',
'airblast_orchard','aerial_vf2f', 'aerial_f2m', 'aerial_m2c', 'aerial_c2vc',
'ground_low_vf', 'ground_low_fmc', 'ground_high_vf', 'ground_high_fmc',
'airblast_normal', 'airblast_dense', 'airblast_sparse', 'airblast_vineyard',
'airblast_orchard'], dtype='object')
agdrift_empty.num_simulations = len(agdrift_empty.out_sim_scenario_id)
agdrift_empty.num_scenarios = len(agdrift_empty.scenario_name)
result_num_sims, result_sim_indices = agdrift_empty.list_sims_per_scenario()
npt.assert_array_equal(result_num_sims, expected_num_sims, err_msg='', verbose=True)
npt.assert_array_equal(result_sim_indices, expected_sim_indices, err_msg='', verbose=True)
finally:
tab = [result_num_sims, expected_num_sims, result_sim_indices, expected_sim_indices]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_determine_area_dimensions(self):
"""
:description determine relevant area/length/depth of waterbody or terrestrial area
:param i: simulation number
:param ecosystem_type: type of assessment to be conducted
:param aquatic_body_type: source of dimensional data for area (EPA or User defined)
:param terrestrial_field_type: source of dimensional data for area (EPA or User defined)
:param *_width: default or user specified width of waterbody or terrestrial field
:param *_length: default or user specified length of waterbody or terrestrial field
:param *_depth: default or user specified depth of waterbody or terrestrial field
:NOTE all areas, i.e., ponds, wetlands, and terrestrial fields are of 1 hectare size; the user can elect
to specify a width other than the default width but it won't change the area size; thus for
user specified areas the length is calculated and not specified by the user)
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_width = pd.Series([208.7, 208.7, 100., 400., 150., 0.], dtype='float')
expected_length = pd.Series([515.8, 515.8, 1076.39, 269.098, 717.593, 0.], dtype='float')
expected_depth = pd.Series([6.56, 0.4921, 7., 23., 0., 0.], dtype='float')
try:
agdrift_empty.ecosystem_type = pd.Series(['aquatic_assessment',
'aquatic_assessment',
'aquatic_assessment',
'aquatic_assessment',
'terrestrial_assessment',
'terrestrial_assessment'], dtype='object')
agdrift_empty.aquatic_body_type = pd.Series(['epa_defined_pond',
'epa_defined_wetland',
'user_defined_pond',
'user_defined_wetland',
'NaN',
'NaN'], dtype='object')
agdrift_empty.terrestrial_field_type = pd.Series(['NaN',
'NaN',
'NaN',
'NaN',
'user_defined_terrestrial',
'epa_defined_terrestrial'], dtype='object')
num_simulations = len(agdrift_empty.ecosystem_type)
agdrift_empty.default_width = 208.7
agdrift_empty.default_length = 515.8
agdrift_empty.default_pond_depth = 6.56
agdrift_empty.default_wetland_depth = 0.4921
agdrift_empty.user_pond_width = pd.Series(['NaN', 'NaN', 100., 'NaN', 'NaN', 'NaN'], dtype='float')
agdrift_empty.user_pond_depth = pd.Series(['NaN', 'NaN', 7., 'NaN', 'NaN', 'NaN'], dtype='float')
agdrift_empty.user_wetland_width = pd.Series(['NaN', 'NaN', 'NaN', 400., 'NaN', 'NaN'], dtype='float')
agdrift_empty.user_wetland_depth = pd.Series(['NaN','NaN', 'NaN', 23., 'NaN', 'NaN'], dtype='float')
agdrift_empty.user_terrestrial_width = pd.Series(['NaN', 'NaN', 'NaN', 'NaN', 150., 'NaN'], dtype='float')
width_result = pd.Series(num_simulations * ['NaN'], dtype='float')
length_result = pd.Series(num_simulations * ['NaN'], dtype='float')
depth_result = pd.Series(num_simulations * ['NaN'], dtype='float')
agdrift_empty.out_area_width = pd.Series(num_simulations * ['nan'], dtype='float')
agdrift_empty.out_area_length = pd.Series(num_simulations * ['nan'], dtype='float')
agdrift_empty.out_area_depth = pd.Series(num_simulations * ['nan'], dtype='float')
agdrift_empty.sqft_per_hectare = 107639
for i in range(num_simulations):
width_result[i], length_result[i], depth_result[i] = agdrift_empty.determine_area_dimensions(i)
npt.assert_allclose(width_result, expected_width, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(length_result, expected_length, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(depth_result, expected_depth, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [width_result, expected_width, length_result, expected_length, depth_result, expected_depth]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_dep_foa(self):
"""
:description calculation of average deposition over width of water body
:param integration_result result of integration of deposition curve across the distance
: beginning at the near distance and extending to the far distance of the water body
:param integration_distance effectively the width of the water body
:param avg_dep_foa average deposition rate across the width of the water body
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([0.1538462, 0.5, 240.])
try:
integration_result = pd.Series([1.,125.,3e5], dtype='float')
integration_distance = pd.Series([6.5,250.,1250.], dtype='float')
result = agdrift_empty.calc_avg_dep_foa(integration_result, integration_distance)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_dep_lbac(self):
"""
Deposition calculation.
:param avg_dep_foa: average deposition over width of water body as fraction of applied
:param application_rate: actual application rate
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([6.5, 3.125e4, 3.75e8])
try:
avg_dep_foa = pd.Series([1.,125.,3e5], dtype='float')
application_rate = pd.Series([6.5,250.,1250.], dtype='float')
result = agdrift_empty.calc_avg_dep_lbac(avg_dep_foa, application_rate)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_dep_foa_from_lbac(self):
"""
Deposition calculation.
:param avg_dep_foa: average deposition over width of water body as fraction of applied
:param application_rate: actual application rate
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([1.553846e-01, 8.8e-06, 4.e-08])
try:
avg_dep_lbac = pd.Series([1.01, 0.0022, 0.00005], dtype='float')
application_rate = pd.Series([6.5,250.,1250.], dtype='float')
result = agdrift_empty.calc_avg_dep_foa_from_lbac(avg_dep_lbac, application_rate)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_dep_lbac_from_gha(self):
"""
Deposition calculation.
:param avg_dep_gha: average deposition over width of water body in units of grams/hectare
:param gms_per_lb: conversion factor to convert lbs to grams
:param acres_per_hectare: conversion factor to convert hectares to acres
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([0.01516739, 0.111524, 0.267659])
try:
avg_dep_gha = pd.Series([17., 125., 3e2], dtype='float')
agdrift_empty.gms_per_lb = 453.592
agdrift_empty.acres_per_hectare = 2.471
result = agdrift_empty.calc_avg_dep_lbac_from_gha(avg_dep_gha)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_dep_lbac_from_waterconc_ngl(self):
"""
:description calculate the average deposition onto the pond/wetland/field
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:param area_width: average width of water body
:parem area_length: average length of water body
:param area_depth: average depth of water body
:param gms_per_lb: conversion factor to convert lbs to grams
:param ng_per_gram conversion factor
:param sqft_per_acre conversion factor
:param liters_per_ft3 conversion factor
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([2.311455e-05, 2.209479e-03, 2.447423e-03])
try:
avg_waterconc_ngl = pd.Series([17., 125., 3e2], dtype='float')
area_width = pd.Series([50., 200., 500.], dtype='float')
area_length = pd.Series([6331., 538., 215.], dtype='float')
area_depth = pd.Series([0.5, 6.5, 3.], dtype='float')
agdrift_empty.liters_per_ft3 = 28.3168
agdrift_empty.sqft_per_acre = 43560.
agdrift_empty.ng_per_gram = 1.e9
agdrift_empty.gms_per_lb = 453.592
agdrift_empty.acres_per_hectare = 2.471
result = agdrift_empty.calc_avg_dep_lbac_from_waterconc_ngl(avg_waterconc_ngl, area_width,
area_length, area_depth)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_dep_lbac_from_mgcm2(self):
"""
:description calculate the average deposition of pesticide over the terrestrial field in lbs/acre
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:param area_depth: average depth of water body
:param gms_per_lb: conversion factor to convert lbs to grams
:param mg_per_gram conversion factor
:param sqft_per_acre conversion factor
:param cm2_per_ft2 conversion factor
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([2.676538e-02, 2.2304486, 44.608973])
try:
avg_fielddep_mgcm2 = pd.Series([3.e-4, 2.5e-2, 5.e-01])
agdrift_empty.sqft_per_acre = 43560.
agdrift_empty.gms_per_lb = 453.592
agdrift_empty.cm2_per_ft2 = 929.03
agdrift_empty.mg_per_gram = 1.e3
result = agdrift_empty.calc_avg_dep_lbac_from_mgcm2(avg_fielddep_mgcm2)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_dep_gha(self):
"""
:description average deposition over width of water body in grams per acre
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:param gms_per_lb: conversion factor to convert lbs to grams
:param acres_per_hectare: conversion factor to convert acres to hectares
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([1.401061, 0.3648362, 0.03362546])
try:
avg_dep_lbac = pd.Series([1.25e-3,3.255e-4,3e-5], dtype='float')
agdrift_empty.gms_per_lb = 453.592
agdrift_empty.acres_per_hectare = 2.47105
result = agdrift_empty.calc_avg_dep_gha(avg_dep_lbac)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_waterconc_ngl(self):
"""
:description calculate the average concentration of pesticide in the pond/wetland
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:param area_width: average width of water body
:parem area_length: average length of water body
:param area_depth: average depth of water body
:param gms_per_lb: conversion factor to convert lbs to grams
:param ng_per_gram conversion factor
:param sqft_per_acre conversion factor
:param liters_per_ft3 conversion factor
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([70.07119, 18.24654, 22.41823])
try:
avg_dep_lbac = pd.Series([1.25e-3,3.255e-4,3e-5], dtype='float')
area_width = pd.Series([6.56, 208.7, 997.], dtype='float')
area_length = pd.Series([1.640838e4, 515.7595, 107.9629], dtype='float')
area_depth = pd.Series([6.56, 6.56, 0.4921], dtype='float')
agdrift_empty.ng_per_gram = 1.e9
agdrift_empty.liters_per_ft3 = 28.3168
agdrift_empty.gms_per_lb = 453.592
agdrift_empty.sqft_per_acre = 43560.
result = agdrift_empty.calc_avg_waterconc_ngl(avg_dep_lbac ,area_width, area_length, area_depth)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_calc_avg_fielddep_mgcm2(self):
"""
:description calculate the average deposition of pesticide over the terrestrial field
:param avg_dep_lbac: average deposition over width of water body in lbs per acre
:param area_depth: average depth of water body
:param gms_per_lb: conversion factor to convert lbs to grams
:param mg_per_gram conversion factor
:param sqft_per_acre conversion factor
:param cm2_per_ft2 conversion factor
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result = pd.Series([1.401063e-5, 3.648369e-6, 3.362552e-7])
try:
avg_dep_lbac = pd.Series([1.25e-3,3.255e-4,3e-5], dtype='float')
agdrift_empty.gms_per_lb = 453.592
agdrift_empty.sqft_per_acre = 43560.
agdrift_empty.mg_per_gram = 1.e3
agdrift_empty.cm2_per_ft2 = 929.03
result = agdrift_empty.calc_avg_fielddep_mgcm2(avg_dep_lbac)
npt.assert_allclose(result, expected_result, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_generate_running_avg(self):
"""
:description retrieves values for distance and the first deposition scenario from the sql database
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE any blank fields are filled with 'nan'
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
location = os.path.realpath(os.path.join(os.getcwd(), os.path.dirname(__file__)))
agdrift_empty.db_name = os.path.join(location, 'sqlite_agdrift_distance.db')
agdrift_empty.db_table = 'output'
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
expected_result_npts = pd.Series([], dtype='object')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,0.102525,0.20505,0.4101,0.8202,1.6404,3.2808,4.9212,6.5616,9.8424,13.1232,19.6848,26.2464,
32.808,39.3696,45.9312,52.4928,59.0544,65.616,72.1776,78.7392,85.3008,91.8624,98.424,104.9856,
111.5472,118.1088,124.6704,131.232,137.7936,144.3552,150.9168,157.4784,164.04,170.6016,177.1632,
183.7248,190.2864,196.848,203.4096,209.9712,216.5328,223.0944,229.656,236.2176,242.7792,249.3408,
255.9024,262.464,269.0256,275.5872,282.1488,288.7104,295.272,301.8336,308.3952,314.9568,321.5184,
328.08,334.6416,341.2032,347.7648,354.3264,360.888,367.4496,374.0112,380.5728,387.1344,393.696,
400.2576,406.8192,413.3808,419.9424,426.504,433.0656,439.6272,446.1888,452.7504,459.312,465.8736,
472.4352,478.9968,485.5584,492.12,498.6816,505.2432,511.8048,518.3664,524.928,531.4896,538.0512,
544.6128,551.1744,557.736,564.2976,570.8592,577.4208,583.9824,590.544,597.1056,603.6672,610.2288,
616.7904,623.352,629.9136,636.4752,643.0368,649.5984,656.16,662.7216,669.2832,675.8448,682.4064,
688.968,695.5296,702.0912,708.6528,715.2144,721.776,728.3376,734.8992,741.4608,748.0224,754.584,
761.1456,767.7072,774.2688,780.8304,787.392,793.9536,800.5152,807.0768,813.6384,820.2,826.7616,
833.3232,839.8848,846.4464,853.008,859.5696,866.1312,872.6928,879.2544,885.816,892.3776,898.9392,
905.5008,912.0624,918.624,925.1856,931.7472,938.3088,944.8704,951.432,957.9936,964.5552,971.1168,
977.6784,984.24,990.8016]
expected_result_y = [0.364712246,0.351507467,0.339214283,0.316974687,0.279954504,0.225948786,0.159949625,
0.123048839,0.099781801,0.071666234,0.056352938,0.03860139,0.029600805,0.024150524,
0.020550354,0.01795028,0.015967703,0.014467663,0.013200146,0.01215011,0.011300098,
0.010550085,0.009905072,0.009345065,0.008845057,0.008400051,0.008000046,0.007635043,
0.007300039,0.007000034,0.006725033,0.00646503,0.006230027,0.006010027,0.005805023,
0.005615023,0.005435021,0.00527002,0.00511002,0.004960017,0.004820017,0.004685016,
0.004560015,0.004440015,0.004325013,0.004220012,0.004120012,0.004020012,0.003925011,
0.003835011,0.00375001,0.00367001,0.00359001,0.00351001,0.003435009,0.003365009,
0.003300007,0.003235009,0.003170007,0.003110007,0.003055006,0.003000007,0.002945006,
0.002895006,0.002845006,0.002795006,0.002745006,0.002695006,0.002650005,0.002610005,
0.002570005,0.002525006,0.002485004,0.002450005,0.002410005,0.002370005,0.002335004,
0.002300005,0.002265004,0.002235004,0.002205004,0.002175004,0.002145004,0.002115004,
0.002085004,0.002055004,0.002025004,0.002000002,0.001975004,0.001945004,0.001920002,
0.001900002,0.001875004,0.001850002,0.001830002,0.001805004,0.001780002,0.001760002,
0.001740002,0.001720002,0.001700002,0.001680002,0.001660002,0.001640002,0.001620002,
0.001605001,0.001590002,0.001570002,0.001550002,0.001535001,0.001520002,0.001500002,
0.001485001,0.001470002,0.001455001,0.001440002,0.001425001,0.001410002,0.001395001,
0.001385001,0.001370002,0.001355001,0.001340002,0.001325001,0.001315001,0.001305001,
0.001290002,0.001275001,0.001265001,0.001255001,0.001245001,0.001230002,0.001215001,
0.001205001,0.001195001,0.001185001,0.001175001,0.001165001,0.001155001,0.001145001,
0.001135001,0.001125001,0.001115001,0.001105001,0.001095001,0.001085001,0.001075001,
0.001065001,0.00106,0.001055001,0.001045001,0.001035001,0.001025001,0.001015001,
0.001005001,0.0009985,0.000993001,0.000985001,0.000977001,0.000969501]
expected_result_npts = 160
x_dist = 6.56
agdrift_empty.distance_name = 'distance_ft'
agdrift_empty.scenario_name = 'ground_low_vf'
agdrift_empty.num_db_values = 161
x_array_in = agdrift_empty.get_distances(agdrift_empty.num_db_values)
y_array_in = agdrift_empty.get_scenario_deposition_data(agdrift_empty.scenario_name, agdrift_empty.num_db_values)
x_array_out, y_array_out, npts_out = agdrift_empty.generate_running_avg(agdrift_empty.num_db_values,
x_array_in, y_array_in, x_dist)
# write output arrays to excel file -- just for debugging
agdrift_empty.write_arrays_to_csv(x_array_out, y_array_out, "output_array_generate.csv")
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print("x_array result/x_array_expected")
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print("y_array result/y_array_expected")
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_generate_running_avg1(self):
"""
:description creates a running average for a specified x axis width (e.g., 7-day average values of an array)
:param x_array_in: array of x-axis values
:param y_array_in: array of y-axis values
:param num_db_values: number of points in the input arrays
:param x_array_out: array of x-zxis values in output array
:param y_array_out: array of y-axis values in output array
:param npts_out: number of points in the output array
:param x_dist: width in x_axis units of running weighted average
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE This test uses a uniformly spaced x_array and monotonically increasing y_array
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
expected_result_npts = pd.Series([], dtype='object')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.]
expected_result_y = [2.5,3.5,4.5,5.5,6.5,7.5,8.5,9.5,10.5,11.5,
12.5,13.5,14.5,15.5,16.5,17.5,18.5,19.5,20.5,21.5,
22.5,23.5,24.5,25.5,26.5,27.5,28.5,29.5,30.5,31.5,
32.5,33.5,34.5,35.5,36.5,37.5,38.5,39.5,40.5,41.5,
42.5,43.5,44.5,45.5, 46.5]
expected_result_npts = 45
x_dist = 5.
num_db_values = 51
x_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.]
y_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.]
x_array_out, y_array_out, npts_out = agdrift_empty.generate_running_avg(num_db_values, x_array_in,
y_array_in, x_dist)
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_generate_running_avg2(self):
"""
:description creates a running average for a specified x axis width (e.g., 7-day average values of an array)
:param x_array_in: array of x-axis values
:param y_array_in: array of y-axis values
:param num_db_values: number of points in the input arrays
:param x_array_out: array of x-zxis values in output array
:param y_array_out: array of y-axis values in output array
:param npts_out: number of points in the output array
:param x_dist: width in x_axis units of running weighted average
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE This test uses a non-uniformly spaced x_array and monotonically increasing y_array
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
expected_result_npts = pd.Series([], dtype='object')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.5,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.5,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.5,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.5,42.,43.,44.]
expected_result_y = [2.5,3.5,4.5,5.5,6.5,7.5,8.4666667,9.4,10.4,11.4,
12.4,13.975,14.5,15.5,16.5,17.5,18.466666667,19.4,20.4,21.4,
22.4,23.975,24.5,25.5,26.5,27.5,28.46666667,29.4,30.4,31.4,
32.4,33.975,34.5,35.5,36.5,37.5,38.466666667,39.4,40.4,41.4,
42.4,43.975,44.5,45.5, 46.5]
expected_result_npts = 45
x_dist = 5.
agdrift_empty.num_db_values = 51
x_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.5,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.5,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.5,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.5,42.,43.,44.,45.,46.,47.,48.,49.,50.]
y_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.]
x_array_out, y_array_out, npts_out = agdrift_empty.generate_running_avg(agdrift_empty.num_db_values,
x_array_in, y_array_in, x_dist)
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_generate_running_avg3(self):
"""
:description creates a running average for a specified x axis width (e.g., 7-day average values of an array);
averages reflect weighted average assuming linearity between x points;
average is calculated as the area under the y-curve beginning at each x point and extending out x_dist
divided by x_dist (which yields the weighted average y between the relevant x points)
:param x_array_in: array of x-axis values
:param y_array_in: array of y-axis values
:param num_db_values: number of points in the input arrays
:param x_array_out: array of x-zxis values in output array
:param y_array_out: array of y-axis values in output array
:param npts_out: number of points in the output array
:param x_dist: width in x_axis units of running weighted average
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE This test uses a monotonically increasing y_array and inserts a gap in the x values
that is greater than x_dist
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
expected_result_npts = pd.Series([], dtype='object')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,1.,2.,3.,4.,5.,6.,7.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.,51.,52.]
expected_result_y = [2.5,3.5,4.5,5.4111111,6.14444444,6.7,7.07777777,7.277777777,10.5,11.5,
12.5,13.5,14.5,15.5,16.5,17.5,18.5,19.5,20.5,21.5,
22.5,23.5,24.5,25.5,26.5,27.5,28.5,29.5,30.5,31.5,
32.5,33.5,34.5,35.5,36.5,37.5,38.5,39.5,40.5,41.5,
42.5,43.5,44.5,45.5, 46.5]
expected_result_npts = 45
x_dist = 5.
num_db_values = 51
x_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.,
51.,52.,53.,54.,55.,56.,57.,58.]
y_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.]
x_array_out, y_array_out, npts_out = agdrift_empty.generate_running_avg(num_db_values, x_array_in,
y_array_in, x_dist)
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_locate_integrated_avg(self):
"""
:description retrieves values for distance and the first deposition scenario from the sql database
and generates running weighted averages from the first x,y value until it locates the user
specified integrated average of interest
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE any blank fields are filled with 'nan'
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
location = os.path.realpath(os.path.join(os.getcwd(), os.path.dirname(__file__)))
agdrift_empty.db_name = os.path.join(location, 'sqlite_agdrift_distance.db')
agdrift_empty.db_table = 'output'
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
expected_result_npts = pd.Series([], dtype='object')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,0.102525,0.20505,0.4101,0.8202,1.6404,3.2808,4.9212,6.5616,9.8424,13.1232,19.6848,26.2464,
32.808,39.3696,45.9312,52.4928,59.0544,65.616,72.1776,78.7392,85.3008,91.8624,98.424,104.9856,
111.5472,118.1088,124.6704,131.232,137.7936,144.3552,150.9168,157.4784,164.04,170.6016,177.1632,
183.7248,190.2864,196.848,203.4096,209.9712,216.5328,223.0944,229.656,236.2176,242.7792,249.3408,
255.9024,262.464,269.0256,275.5872,282.1488,288.7104,295.272,301.8336,308.3952,314.9568,321.5184,
328.08,334.6416,341.2032,347.7648,354.3264,360.888,367.4496,374.0112,380.5728,387.1344,393.696,
400.2576,406.8192,413.3808,419.9424,426.504,433.0656,439.6272,446.1888,452.7504,459.312,465.8736,
472.4352,478.9968,485.5584,492.12,498.6816,505.2432,511.8048,518.3664,524.928,531.4896,538.0512,
544.6128,551.1744,557.736,564.2976,570.8592,577.4208,583.9824,590.544,597.1056,603.6672,610.2288,
616.7904,623.352,629.9136,636.4752,643.0368,649.5984,656.16,662.7216,669.2832,675.8448,682.4064,
688.968,695.5296,702.0912,708.6528,715.2144,721.776,728.3376,734.8992,741.4608,748.0224,754.584,
761.1456,767.7072,774.2688,780.8304,787.392,793.9536,800.5152,807.0768,813.6384,820.2,826.7616,
833.3232,839.8848,846.4464,853.008,859.5696,866.1312,872.6928,879.2544,885.816,892.3776,898.9392,
905.5008,912.0624,918.624,925.1856,931.7472,938.3088,944.8704,951.432,957.9936,964.5552,971.1168,
977.6784,984.24,990.8016]
expected_result_y = [0.364712246,0.351507467,0.339214283,0.316974687,0.279954504,0.225948786,0.159949625,
0.123048839,0.099781801,0.071666234,0.056352938,0.03860139,0.029600805,0.024150524,
0.020550354,0.01795028,0.015967703,0.014467663,0.013200146,0.01215011,0.011300098,
0.010550085,0.009905072,0.009345065,0.008845057,0.008400051,0.008000046,0.007635043,
0.007300039,0.007000034,0.006725033,0.00646503,0.006230027,0.006010027,0.005805023,
0.005615023,0.005435021,0.00527002,0.00511002,0.004960017,0.004820017,0.004685016,
0.004560015,0.004440015,0.004325013,0.004220012,0.004120012,0.004020012,0.003925011,
0.003835011,0.00375001,0.00367001,0.00359001,0.00351001,0.003435009,0.003365009,
0.003300007,0.003235009,0.003170007,0.003110007,0.003055006,0.003000007,0.002945006,
0.002895006,0.002845006,0.002795006,0.002745006,0.002695006,0.002650005,0.002610005,
0.002570005,0.002525006,0.002485004,0.002450005,0.002410005,0.002370005,0.002335004,
0.002300005,0.002265004,0.002235004,0.002205004,0.002175004,0.002145004,0.002115004,
0.002085004,0.002055004,0.002025004,0.002000002,0.001975004,0.001945004,0.001920002,
0.001900002,0.001875004,0.001850002,0.001830002,0.001805004,0.001780002,0.001760002,
0.001740002,0.001720002,0.001700002,0.001680002,0.001660002,0.001640002,0.001620002,
0.001605001,0.001590002,0.001570002,0.001550002,0.001535001,0.001520002,0.001500002,
0.001485001,0.001470002,0.001455001,0.001440002,0.001425001,0.001410002,0.001395001,
0.001385001,0.001370002,0.001355001,0.001340002,0.001325001,0.001315001,0.001305001,
0.001290002,0.001275001,0.001265001,0.001255001,0.001245001,0.001230002,0.001215001,
0.001205001,0.001195001,0.001185001,0.001175001,0.001165001,0.001155001,0.001145001,
0.001135001,0.001125001,0.001115001,0.001105001,0.001095001,0.001085001,0.001075001,
0.001065001,0.00106,0.001055001,0.001045001,0.001035001,0.001025001,0.001015001,
0.001005001,0.0009985,0.000993001,0.000985001,0.000977001,0.000969501]
expected_result_npts = 160
expected_x_dist_of_interest = 990.8016
x_dist = 6.56
weighted_avg = 0.0009697 #this is the running average value we're looking for
agdrift_empty.distance_name = 'distance_ft'
agdrift_empty.scenario_name = 'ground_low_vf'
agdrift_empty.num_db_values = 161
agdrift_empty.find_nearest_x = True
x_array_in = agdrift_empty.get_distances(agdrift_empty.num_db_values)
y_array_in = agdrift_empty.get_scenario_deposition_data(agdrift_empty.scenario_name, agdrift_empty.num_db_values)
x_array_out, y_array_out, npts_out, x_dist_of_interest, range_chk = \
agdrift_empty.locate_integrated_avg(agdrift_empty.num_db_values, x_array_in, y_array_in, x_dist, weighted_avg)
npt.assert_array_equal(expected_x_dist_of_interest, x_dist_of_interest, verbose=True)
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} x-units to area and got {1} '.format(expected_x_dist_of_interest, x_dist_of_interest))
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print("x_array result/x_array_expected")
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print("y_array result/y_array_expected")
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_locate_integrated_avg1(self):
"""
:description retrieves values for distance and the first deposition scenario from the sql database
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE this test is for a monotonically increasing function with some irregularity in x-axis points
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,7.0,16.0,17.0,18.0,19.0,20.0,28.0,29.0,30.0,31.]
expected_result_y = [0.357143,1.27778,4.4125,5.15,5.7125,6.1,6.3125,9.5,10.5,11.5,12.5]
expected_result_npts = 11
expected_x_dist_of_interest = 30.5
x_dist = 5.
weighted_avg = 12.
num_db_values = 51
x_array_in = [0.,7.,16.,17.,18.,19.,20.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.,
51.,52.,53.,54.,55.,56.,57.,58.,59.,60.,
61.,62.,63.,64.,65.,66.,67.,68.,69.,70.,
71.]
y_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,10.,
11.,12.,13.,14.,15.,16.,17.,18.,19.,20.,
21.,22.,23.,24.,25.,26.,27.,28.,29.,30.,
31.,32.,33.,34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.]
agdrift_empty.find_nearest_x = True
x_array_out, y_array_out, npts_out, x_dist_of_interest, range_chk = \
agdrift_empty.locate_integrated_avg(num_db_values, x_array_in, y_array_in, x_dist, weighted_avg)
npt.assert_array_equal(expected_x_dist_of_interest, x_dist_of_interest, verbose=True)
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} x-units to area and got {1} '.format(expected_x_dist_of_interest, x_dist_of_interest))
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print("x_array result/x_array_expected")
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print("y_array result/y_array_expected")
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_locate_integrated_avg2(self):
"""
:description retrieves values for distance and the first deposition scenario from the sql database
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE This test is for a monotonically decreasing function with irregular x-axis spacing
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,7.,16.,17.,18.,19.,20.,28.,29.,30.,
34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.,
51.,52.,53.,54.,55.,56.,57.,58.,59.,60.]
expected_result_y = [49.6429,48.7222,45.5875,44.85,44.2875,43.9,43.6875,41.175,40.7,40.3,
37.5,36.5,35.5,34.5,33.5,32.5,31.5,30.5,29.5,28.5,
27.5,26.5,25.5,24.5,23.5,22.5,21.5,20.5,19.5,18.5,
17.5,16.5,15.5,14.5,13.5,12.5,11.5]
expected_result_npts = 37
expected_x_dist_of_interest = 60.
x_dist = 5.
weighted_avg = 12.
num_db_values = 51
agdrift_empty.find_nearest_x = True
x_array_in = [0.,7.,16.,17.,18.,19.,20.,28.,29.,30.,
34.,35.,36.,37.,38.,39.,40.,
41.,42.,43.,44.,45.,46.,47.,48.,49.,50.,
51.,52.,53.,54.,55.,56.,57.,58.,59.,60.,
61.,62.,63.,64.,65.,66.,67.,68.,69.,70.,
71.,72.,73.,74. ]
y_array_in = [50.,49.,48.,47.,46.,45.,44.,43.,42.,41.,
40.,39.,38.,37.,36.,35.,34.,33.,32.,31.,
30.,29.,28.,27.,26.,25.,24.,23.,22.,21.,
20.,19.,18.,17.,16.,15.,14.,13.,12.,11.,
10.,9.,8.,7.,6.,5.,4.,3.,2.,1.,0.]
x_array_out, y_array_out, npts_out, x_dist_of_interest, range_chk = \
agdrift_empty.locate_integrated_avg(num_db_values, x_array_in, y_array_in, x_dist, weighted_avg)
npt.assert_array_equal(expected_x_dist_of_interest, x_dist_of_interest, verbose=True)
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} x-units to area and got {1} '.format(expected_x_dist_of_interest, x_dist_of_interest))
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print("x_array result/x_array_expected")
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print("y_array result/y_array_expected")
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_locate_integrated_avg3(self):
"""
:description retrieves values for distance and the first deposition scenario from the sql database
:param num_db_values: number of distance values to be retrieved
:param distance_name: name of column in sql database that contains the distance values
:NOTE this test is for a monotonically decreasing function with regular x-axis spacing
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
x_array_in = pd.Series([], dtype='float')
y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
expected_result_x_dist = pd.Series([], dtype='float')
try:
expected_result_x = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,
10.,11.,12.,13.,14.,15.,16.,17.,18.,19.,
20.,21.,22.,23.,24.,25.,26.,27.,28.,29.,
30.,31.,32.,33.,34.,35.,36.]
expected_result_y = [47.5,46.5,45.5,44.5,43.5,42.5,41.5,40.5,39.5,38.5,
37.5,36.5,35.5,34.5,33.5,32.5,31.5,30.5,29.5,28.5,
27.5,26.5,25.5,24.5,23.5,22.5,21.5,20.5,19.5,18.5,
17.5,16.5,15.5,14.5,13.5,12.5,11.5]
expected_result_npts = 37
expected_x_dist_of_interest = 36.
x_dist = 5.
weighted_avg = 12.
num_db_values = 51
agdrift_empty.find_nearest_x = True
x_array_in = [0.,1.,2.,3.,4.,5.,6.,7.,8.,9.,
10.,11.,12.,13.,14.,15.,16.,17.,18.,19.,
20.,21.,22.,23.,24.,25.,26.,27.,28.,29.,
30.,31.,32.,33.,34.,35.,36.,37.,38.,39.,
40.,41.,42.,43.,44.,45.,46.,47.,48.,49.,
50.]
y_array_in = [50.,49.,48.,47.,46.,45.,44.,43.,42.,41.,
40.,39.,38.,37.,36.,35.,34.,33.,32.,31.,
30.,29.,28.,27.,26.,25.,24.,23.,22.,21.,
20.,19.,18.,17.,16.,15.,14.,13.,12.,11.,
10.,9.,8.,7.,6.,5.,4.,3.,2.,1.,0.]
x_array_out, y_array_out, npts_out, x_dist_of_interest, range_chk = \
agdrift_empty.locate_integrated_avg(num_db_values, x_array_in, y_array_in, x_dist, weighted_avg)
npt.assert_array_equal(expected_x_dist_of_interest, x_dist_of_interest, verbose=True )
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True )
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} x-units to area and got {1} '.format(expected_x_dist_of_interest, x_dist_of_interest))
print('expected {0} number of points and got {1} points'.format(expected_result_npts, npts_out))
print("x_array result/x_array_expected")
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print("y_array result/y_array_expected")
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_round_model_outputs(self):
"""
:description round output variable values (and place in output variable series) so that they can be directly
compared to expected results (which were limited in terms of their output format from the OPP AGDRIFT
model (V2.1.1) interface (we don't have the AGDRIFT code so we cannot change the output format to
agree with this model
:param avg_dep_foa:
:param avg_dep_lbac:
:param avg_dep_gha:
:param avg_waterconc_ngl:
:param avg_field_dep_mgcm2:
:param num_sims: number of simulations
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
num_sims = 3
num_args = 5
agdrift_empty.out_avg_dep_foa = pd.Series(num_sims * [np.nan], dtype='float')
agdrift_empty.out_avg_dep_lbac = pd.Series(num_sims * [np.nan], dtype='float')
agdrift_empty.out_avg_dep_gha = pd.Series(num_sims * [np.nan], dtype='float')
agdrift_empty.out_avg_waterconc_ngl = pd.Series(num_sims * [np.nan], dtype='float')
agdrift_empty.out_avg_field_dep_mgcm2 = pd.Series(num_sims * [np.nan], dtype='float')
result = pd.Series(num_sims * [num_args*[np.nan]], dtype='float')
expected_result = pd.Series(num_sims * [num_args*[np.nan]], dtype='float')
expected_result[0] = [1.26,1.26,1.26,1.26,1.26]
expected_result[1] = [0.0004,0.0004,0.0004,0.0004,0.0004]
expected_result[2] = [3.45e-05,3.45e-05,3.45e-05,3.45e-05,3.45e-05]
try:
#setting each variable to same values, each value tests a separate pathway through rounding method
avg_dep_lbac = pd.Series([1.2567,3.55e-4,3.454e-5], dtype='float')
avg_dep_foa = pd.Series([1.2567,3.55e-4,3.454e-5], dtype='float')
avg_dep_gha = pd.Series([1.2567,3.55e-4,3.454e-5], dtype='float')
avg_waterconc_ngl = pd.Series([1.2567,3.55e-4,3.454e-5], dtype='float')
avg_field_dep_mgcm2 = pd.Series([1.2567,3.55e-4,3.454e-5], dtype='float')
for i in range(num_sims):
lbac = avg_dep_lbac[i]
foa = avg_dep_foa[i]
gha = avg_dep_gha[i]
ngl = avg_waterconc_ngl[i]
mgcm2 = avg_field_dep_mgcm2[i]
agdrift_empty.round_model_outputs(foa, lbac, gha, ngl, mgcm2, i)
result[i] = [agdrift_empty.out_avg_dep_foa[i], agdrift_empty.out_avg_dep_lbac[i],
agdrift_empty.out_avg_dep_gha[i], agdrift_empty.out_avg_waterconc_ngl[i],
agdrift_empty.out_avg_field_dep_mgcm2[i]]
npt.assert_allclose(result[0], expected_result[0], rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(result[1], expected_result[1], rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(result[2], expected_result[2], rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_find_dep_pt_location(self):
"""
:description this method locates the downwind distance associated with a specific deposition rate
:param x_array: array of distance values
:param y_array: array of deposition values
:param npts: number of values in x/y arrays
:param foa: value of deposition (y value) of interest
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
result = [[],[],[],[]]
expected_result = [(0.0, 'in range'), (259.1832, 'in range'), (997.3632, 'in range'), (np.nan, 'out of range')]
try:
x_array = [0.,0.102525,0.20505,0.4101,0.8202,1.6404,3.2808,4.9212,6.5616,9.8424,13.1232,19.6848,26.2464,
32.808,39.3696,45.9312,52.4928,59.0544,65.616,72.1776,78.7392,85.3008,91.8624,98.424,104.9856,
111.5472,118.1088,124.6704,131.232,137.7936,144.3552,150.9168,157.4784,164.04,170.6016,177.1632,
183.7248,190.2864,196.848,203.4096,209.9712,216.5328,223.0944,229.656,236.2176,242.7792,249.3408,
255.9024,262.464,269.0256,275.5872,282.1488,288.7104,295.272,301.8336,308.3952,314.9568,321.5184,
328.08,334.6416,341.2032,347.7648,354.3264,360.888,367.4496,374.0112,380.5728,387.1344,393.696,
400.2576,406.8192,413.3808,419.9424,426.504,433.0656,439.6272,446.1888,452.7504,459.312,465.8736,
472.4352,478.9968,485.5584,492.12,498.6816,505.2432,511.8048,518.3664,524.928,531.4896,538.0512,
544.6128,551.1744,557.736,564.2976,570.8592,577.4208,583.9824,590.544,597.1056,603.6672,610.2288,
616.7904,623.352,629.9136,636.4752,643.0368,649.5984,656.16,662.7216,669.2832,675.8448,682.4064,
688.968,695.5296,702.0912,708.6528,715.2144,721.776,728.3376,734.8992,741.4608,748.0224,754.584,
761.1456,767.7072,774.2688,780.8304,787.392,793.9536,800.5152,807.0768,813.6384,820.2,826.7616,
833.3232,839.8848,846.4464,853.008,859.5696,866.1312,872.6928,879.2544,885.816,892.3776,898.9392,
905.5008,912.0624,918.624,925.1856,931.7472,938.3088,944.8704,951.432,957.9936,964.5552,971.1168,
977.6784,984.24,990.8016, 997.3632]
y_array = [0.364706389,0.351133211,0.338484161,0.315606383,0.277604029,0.222810736,0.159943507,
0.121479708,0.099778741,0.068653,0.05635,0.0386,0.0296,0.02415,0.02055,0.01795,
0.0159675,0.0144675,0.0132,0.01215,0.0113,0.01055,0.009905,0.009345,0.008845,0.0084,
0.008,0.007635,0.0073,0.007,0.006725,0.006465,0.00623,0.00601,0.005805,0.005615,
0.005435,0.00527,0.00511,0.00496,0.00482,0.004685,0.00456,0.00444,0.004325,0.00422,
0.00412,0.00402,0.003925,0.003835,0.00375,0.00367,0.00359,0.00351,0.003435,0.003365,
0.0033,0.003235,0.00317,0.00311,0.003055,0.003,0.002945,0.002895,0.002845,0.002795,
0.002745,0.002695,0.00265,0.00261,0.00257,0.002525,0.002485,0.00245,0.00241,0.00237,
0.002335,0.0023,0.002265,0.002235,0.002205,0.002175,0.002145,0.002115,0.002085,
0.002055,0.002025,0.002,0.001975,0.001945,0.00192,0.0019,0.001875,0.00185,0.00183,
0.001805,0.00178,0.00176,0.00174,0.00172,0.0017,0.00168,0.00166,0.00164,0.00162,
0.001605,0.00159,0.00157,0.00155,0.001535,0.00152,0.0015,0.001485,0.00147,0.001455,
0.00144,0.001425,0.00141,0.001395,0.001385,0.00137,0.001355,0.00134,0.001325,0.001315,
0.001305,0.00129,0.001275,0.001265,0.001255,0.001245,0.00123,0.001215,0.001205,
0.001195,0.001185,0.001175,0.001165,0.001155,0.001145,0.001135,0.001125,0.001115,
0.001105,0.001095,0.001085,0.001075,0.001065,0.00106,0.001055,0.001045,0.001035,
0.001025,0.001015,0.001005,0.0009985,0.000993,0.000985,0.000977,0.0009695,0.0009612]
npts = len(x_array)
num_sims = 4
foa = [0.37, 0.004, 0.0009613, 0.0008]
for i in range(num_sims):
result[i] = agdrift_empty.find_dep_pt_location(x_array, y_array, npts, foa[i])
npt.assert_equal(expected_result, result, verbose=True)
finally:
tab = [result, expected_result]
print("\n")
print(inspect.currentframe().f_code.co_name)
print(tabulate(tab, headers='keys', tablefmt='rst'))
return
def test_extend_curve_opp(self):
"""
:description extends/extrapolates an x,y array of data points that reflect a ln ln relationship by selecting
a number of points near the end of the x,y arrays and fitting a line to the points
ln ln transforms (two ln ln transforms can by applied; on using the straight natural log of
each selected x,y point and one using a 'relative' value of each of the selected points --
the relative values are calculated by establishing a zero point closest to the selected
points
For AGDRIFT: extends distance vs deposition (fraction of applied) curve to enable model calculations
when area of interest (pond, wetland, terrestrial field) lie partially outside the original
curve (whose extent is 997 feet). The extension is achieved by fitting a line of best fit
to the last 16 points of the original curve. The x,y values representing the last 16 points
are natural log transforms of the distance and deposition values at the 16 points. Two long
transforms are coded here, reflecting the fact that the AGDRIFT model (v2.1.1) uses each of them
under different circumstandes (which I believe is not the intention but is the way the model
functions -- my guess is that one of the transforms was used and then a second one was coded
to increase the degree of conservativeness -- but the code was changed in only one of the two
places where the transformation occurs.
Finally, the AGDRIFT model extends the curve only when necessary (i.e., when it determines that
the area of intereest lies partially beyond the last point of the origanal curve (997 ft). In
this code all the curves are extended out to 1994 ft, which represents the furthest distance that
the downwind edge of an area of concern can be specified. All scenario curves are extended here
because we are running multiple simulations (e.g., monte carlo) and instead of extending the
curves each time a simulation requires it (which may be multiple time for the same scenario
curve) we just do it for all curves up front. There is a case to be made that the
curves should be extended external to this code and simply provide the full curve in the SQLite
database containing the original curve.
:param x_array: array of x values to be extended (must be at least 17 data points in original array)
:param y_array: array of y values to be extended
:param max_dist: maximum distance (ft) associated with unextended x values
:param dist_inc: increment (ft) for each extended data point
:param num_pts_ext: number of points at end of original x,y arrays to be used for extending the curve
:param ln_ln_trans: form of transformation to perform (True: straight ln ln, False: relative ln ln)
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y = pd.Series([], dtype='float')
# x_array_in = pd.Series([], dtype='float')
# y_array_in = pd.Series([], dtype='float')
x_array_out = pd.Series([], dtype='float')
y_array_out = pd.Series([], dtype='float')
try:
expected_result_x = [0.,6.5616,13.1232,19.6848,26.2464,
32.808,39.3696,45.9312,52.4928,59.0544,65.616,72.1776,78.7392,85.3008,91.8624,98.424,104.9856,
111.5472,118.1088,124.6704,131.232,137.7936,144.3552,150.9168,157.4784,164.04,170.6016,177.1632,
183.7248,190.2864,196.848,203.4096,209.9712,216.5328,223.0944,229.656,236.2176,242.7792,249.3408,
255.9024,262.464,269.0256,275.5872,282.1488,288.7104,295.272,301.8336,308.3952,314.9568,321.5184,
328.08,334.6416,341.2032,347.7648,354.3264,360.888,367.4496,374.0112,380.5728,387.1344,393.696,
400.2576,406.8192,413.3808,419.9424,426.504,433.0656,439.6272,446.1888,452.7504,459.312,465.8736,
472.4352,478.9968,485.5584,492.12,498.6816,505.2432,511.8048,518.3664,524.928,531.4896,538.0512,
544.6128,551.1744,557.736,564.2976,570.8592,577.4208,583.9824,590.544,597.1056,603.6672,610.2288,
616.7904,623.352,629.9136,636.4752,643.0368,649.5984,656.16,662.7216,669.2832,675.8448,682.4064,
688.968,695.5296,702.0912,708.6528,715.2144,721.776,728.3376,734.8992,741.4608,748.0224,754.584,
761.1456,767.7072,774.2688,780.8304,787.392,793.9536,800.5152,807.0768,813.6384,820.2,826.7616,
833.3232,839.8848,846.4464,853.008,859.5696,866.1312,872.6928,879.2544,885.816,892.3776,898.9392,
905.5008,912.0624,918.624,925.1856,931.7472,938.3088,944.8704,951.432,957.9936,964.5552,971.1168,
977.6784,984.24,990.8016,997.3632,
1003.9232,1010.4832,1017.0432,1023.6032,1030.1632,1036.7232,1043.2832,1049.8432,1056.4032,
1062.9632,1069.5232,1076.0832,1082.6432,1089.2032,1095.7632,1102.3232,1108.8832,1115.4432,
1122.0032,1128.5632,1135.1232,1141.6832,1148.2432,1154.8032,1161.3632,1167.9232,1174.4832,
1181.0432,1187.6032,1194.1632,1200.7232,1207.2832,1213.8432,1220.4032,1226.9632,1233.5232,
1240.0832,1246.6432,1253.2032,1259.7632,1266.3232,1272.8832,1279.4432,1286.0032,1292.5632,
1299.1232,1305.6832,1312.2432,1318.8032,1325.3632,1331.9232,1338.4832,1345.0432,1351.6032,
1358.1632,1364.7232,1371.2832,1377.8432,1384.4032,1390.9632,1397.5232,1404.0832,1410.6432,
1417.2032,1423.7632,1430.3232,1436.8832,1443.4432,1450.0032,1456.5632,1463.1232,1469.6832,
1476.2432,1482.8032,1489.3632,1495.9232,1502.4832,1509.0432,1515.6032,1522.1632,1528.7232,
1535.2832,1541.8432,1548.4032,1554.9632,1561.5232,1568.0832,1574.6432,1581.2032,1587.7632,
1594.3232,1600.8832,1607.4432,1614.0032,1620.5632,1627.1232,1633.6832,1640.2432,1646.8032,
1653.3632,1659.9232,1666.4832,1673.0432,1679.6032,1686.1632,1692.7232,1699.2832,1705.8432,
1712.4032,1718.9632,1725.5232,1732.0832,1738.6432,1745.2032,1751.7632,1758.3232,1764.8832,
1771.4432,1778.0032,1784.5632,1791.1232,1797.6832,1804.2432,1810.8032,1817.3632,1823.9232,
1830.4832,1837.0432,1843.6032,1850.1632,1856.7232,1863.2832,1869.8432,1876.4032,1882.9632,
1889.5232,1896.0832,1902.6432,1909.2032,1915.7632,1922.3232,1928.8832,1935.4432,1942.0032,
1948.5632,1955.1232,1961.6832,1968.2432,1974.8032,1981.3632,1987.9232,1994.4832]
expected_result_y = [0.49997,0.37451,0.29849,0.25004,0.2138,0.19455,0.18448,0.17591,0.1678,0.15421,0.1401,
0.12693,0.11785,0.11144,0.10675,0.099496,0.092323,0.085695,0.079234,0.074253,0.070316,
0.067191,0.064594,0.062337,0.060348,0.058192,0.055224,0.051972,0.049283,0.04757,
0.046226,0.044969,0.043922,0.043027,0.041934,0.040528,0.039018,0.037744,0.036762,
0.035923,0.035071,0.034267,0.033456,0.032629,0.03184,0.031078,0.030363,0.02968,0.029028,
0.028399,0.027788,0.027199,0.026642,0.026124,0.025635,0.02517,0.024719,0.024287,0.023867,
0.023457,0.023061,0.022685,0.022334,0.021998,0.021675,0.02136,0.021055,0.020758,0.020467,
0.020186,0.019919,0.019665,0.019421,0.019184,0.018951,0.018727,0.018514,0.018311,
0.018118,0.017929,0.017745,0.017564,0.017387,0.017214,0.017046,0.016886,0.016732,
0.016587,0.016446,0.016309,0.016174,0.016039,0.015906,0.015777,0.015653,0.015532,
0.015418,0.015308,0.015202,0.015097,0.014991,0.014885,0.014782,0.014683,0.014588,0.0145,
0.014415,0.014334,0.014254,0.014172,0.01409,0.014007,0.013926,0.013846,0.01377,0.013697,
0.013628,0.013559,0.013491,0.013423,0.013354,0.013288,0.013223,0.01316,0.013099,0.01304,
0.012983,0.012926,0.01287,0.012814,0.012758,0.012703,0.012649,0.012597,0.012547,0.012499,
0.01245,0.012402,0.012352,0.012302,0.012254,0.012205,0.012158,0.012113,0.012068,0.012025,
0.011982,0.01194,0.011899,0.011859,0.011819,0.01178,0.011741,1.1826345E-02,1.1812256E-02,
1.1798945E-02,1.1786331E-02,1.1774344E-02,1.1762927E-02,1.1752028E-02,1.1741602E-02,
1.1731610E-02,1.1722019E-02,1.1712796E-02,1.1703917E-02,1.1695355E-02,1.1687089E-02,
1.1679100E-02,1.1671370E-02,1.1663883E-02,1.1656623E-02,1.1649579E-02,1.1642737E-02,
1.1636087E-02,1.1629617E-02,1.1623319E-02,1.1617184E-02,1.1611203E-02,1.1605369E-02,
1.1599676E-02,1.1594116E-02,1.1588684E-02,1.1583373E-02,1.1578179E-02,1.1573097E-02,
1.1568122E-02,1.1563249E-02,1.1558475E-02,1.1553795E-02,1.1549206E-02,1.1544705E-02,
1.1540288E-02,1.1535953E-02,1.1531695E-02,1.1527514E-02,1.1523405E-02,1.1519367E-02,
1.1515397E-02,1.1511493E-02,1.1507652E-02,1.1503873E-02,1.1500154E-02,1.1496493E-02,
1.1492889E-02,1.1489338E-02,1.1485841E-02,1.1482395E-02,1.1478999E-02,1.1475651E-02,
1.1472351E-02,1.1469096E-02,1.1465886E-02,1.1462720E-02,1.1459595E-02,1.1456512E-02,
1.1453469E-02,1.1450465E-02,1.1447499E-02,1.1444570E-02,1.1441677E-02,1.1438820E-02,
1.1435997E-02,1.1433208E-02,1.1430452E-02,1.1427728E-02,1.1425036E-02,1.1422374E-02,
1.1419742E-02,1.1417139E-02,1.1414566E-02,1.1412020E-02,1.1409502E-02,1.1407011E-02,
1.1404546E-02,1.1402107E-02,1.1399693E-02,1.1397304E-02,1.1394939E-02,1.1392598E-02,
1.1390281E-02,1.1387986E-02,1.1385713E-02,1.1383463E-02,1.1381234E-02,1.1379026E-02,
1.1376840E-02,1.1374673E-02,1.1372527E-02,1.1370400E-02,1.1368292E-02,1.1366204E-02,
1.1364134E-02,1.1362082E-02,1.1360048E-02,1.1358032E-02,1.1356033E-02,1.1354052E-02,
1.1352087E-02,1.1350139E-02,1.1348207E-02,1.1346291E-02,1.1344390E-02,1.1342505E-02,
1.1340635E-02,1.1338781E-02,1.1336941E-02,1.1335115E-02,1.1333304E-02,1.1331507E-02,
1.1329723E-02,1.1327954E-02,1.1326197E-02,1.1324454E-02,1.1322724E-02,1.1321007E-02,
1.1319303E-02,1.1317611E-02,1.1315931E-02,1.1314263E-02,1.1312608E-02,1.1310964E-02,
1.1309332E-02,1.1307711E-02,1.1306101E-02,1.1304503E-02,1.1302915E-02,1.1301339E-02,
1.1299773E-02,1.1298218E-02,1.1296673E-02,1.1295138E-02,1.1293614E-02,1.1292099E-02,
1.1290594E-02,1.1289100E-02,1.1287614E-02,1.1286139E-02,1.1284672E-02,1.1283215E-02,
1.1281767E-02,1.1280328E-02,1.1278898E-02,1.1277477E-02,1.1276065E-02,1.1274661E-02]
expected_result_npts = [305]
max_dist = 997.3632
dist_inc = 6.56
num_pts_ext = 16
ln_ln_trans = False #using the relative ln ln transformation in this test
agdrift_empty.meters_per_ft = 0.3048
x_array_in = pd.Series([0.,6.5616,13.1232,19.6848,26.2464,
32.808,39.3696,45.9312,52.4928,59.0544,65.616,72.1776,78.7392,85.3008,91.8624,98.424,104.9856,
111.5472,118.1088,124.6704,131.232,137.7936,144.3552,150.9168,157.4784,164.04,170.6016,177.1632,
183.7248,190.2864,196.848,203.4096,209.9712,216.5328,223.0944,229.656,236.2176,242.7792,249.3408,
255.9024,262.464,269.0256,275.5872,282.1488,288.7104,295.272,301.8336,308.3952,314.9568,321.5184,
328.08,334.6416,341.2032,347.7648,354.3264,360.888,367.4496,374.0112,380.5728,387.1344,393.696,
400.2576,406.8192,413.3808,419.9424,426.504,433.0656,439.6272,446.1888,452.7504,459.312,465.8736,
472.4352,478.9968,485.5584,492.12,498.6816,505.2432,511.8048,518.3664,524.928,531.4896,538.0512,
544.6128,551.1744,557.736,564.2976,570.8592,577.4208,583.9824,590.544,597.1056,603.6672,610.2288,
616.7904,623.352,629.9136,636.4752,643.0368,649.5984,656.16,662.7216,669.2832,675.8448,682.4064,
688.968,695.5296,702.0912,708.6528,715.2144,721.776,728.3376,734.8992,741.4608,748.0224,754.584,
761.1456,767.7072,774.2688,780.8304,787.392,793.9536,800.5152,807.0768,813.6384,820.2,826.7616,
833.3232,839.8848,846.4464,853.008,859.5696,866.1312,872.6928,879.2544,885.816,892.3776,898.9392,
905.5008,912.0624,918.624,925.1856,931.7472,938.3088,944.8704,951.432,957.9936,964.5552,971.1168,
977.6784,984.24,990.8016,997.3632])
y_array_in = pd.Series([0.49997,0.37451,0.29849,0.25004,0.2138,0.19455,0.18448,0.17591,0.1678,0.15421,0.1401,
0.12693,0.11785,0.11144,0.10675,0.099496,0.092323,0.085695,0.079234,0.074253,0.070316,
0.067191,0.064594,0.062337,0.060348,0.058192,0.055224,0.051972,0.049283,0.04757,
0.046226,0.044969,0.043922,0.043027,0.041934,0.040528,0.039018,0.037744,0.036762,
0.035923,0.035071,0.034267,0.033456,0.032629,0.03184,0.031078,0.030363,0.02968,0.029028,
0.028399,0.027788,0.027199,0.026642,0.026124,0.025635,0.02517,0.024719,0.024287,0.023867,
0.023457 ,0.023061,0.022685,0.022334,0.021998,0.021675,0.02136,0.021055,0.020758,0.020467,
0.020186,0.019919,0.019665,0.019421,0.019184,0.018951,0.018727,0.018514,0.018311,
0.018118,0.017929,0.017745,0.017564,0.017387,0.017214,0.017046,0.016886,0.016732,
0.016587,0.016446,0.016309,0.016174,0.016039,0.015906,0.015777,0.015653,0.015532,
0.015418,0.015308,0.015202,0.015097,0.014991,0.014885,0.014782,0.014683,0.014588,0.0145,
0.014415,0.014334,0.014254,0.014172,0.01409,0.014007,0.013926,0.013846,0.01377,0.013697,
0.013628,0.013559,0.013491,0.013423,0.013354,0.013288,0.013223,0.01316,0.013099,0.01304,
0.012983,0.012926,0.01287,0.012814,0.012758,0.012703,0.012649,0.012597,0.012547,0.012499,
0.01245,0.012402,0.012352,0.012302,0.012254,0.012205,0.012158,0.012113,0.012068,0.012025,
0.011982,0.01194,0.011899,0.011859,0.011819,0.01178,0.011741])
x_array_out, y_array_out = agdrift_empty.extend_curve_opp(x_array_in, y_array_in, max_dist, dist_inc, num_pts_ext,
ln_ln_trans)
npts_out = [len(y_array_out)]
#
#agdrift_empty.write_arrays_to_csv(x_array_out, y_array_out, "extend_data.csv")
npt.assert_array_equal(expected_result_npts, npts_out, verbose=True)
npt.assert_allclose(x_array_out, expected_result_x, rtol=1e-5, atol=0, err_msg='', verbose=True)
npt.assert_allclose(y_array_out, expected_result_y, rtol=1e-5, atol=0, err_msg='', verbose=True)
finally:
pass
tab1 = [x_array_out, expected_result_x]
tab2 = [y_array_out, expected_result_y]
print("\n")
print(inspect.currentframe().f_code.co_name)
print('expected {0} number of points and got {1} points'.format(expected_result_npts[0], npts_out[0]))
print("x_array result/x_array_expected")
print(tabulate(tab1, headers='keys', tablefmt='rst'))
print("y_array result/y_array_expected")
print(tabulate(tab2, headers='keys', tablefmt='rst'))
return
def test_extend_curve_opp1(self):
"""
:description extends/extrapolates an x,y array of data points that reflect a ln ln relationship by selecting
a number of points near the end of the x,y arrays and fitting a line to the points
ln ln transforms (two ln ln transforms can by applied; on using the straight natural log of
each selected x,y point and one using a 'relative' value of each of the selected points --
the relative values are calculated by establishing a zero point closest to the selected
points
For AGDRIFT: extends distance vs deposition (fraction of applied) curve to enable model calculations
when area of interest (pond, wetland, terrestrial field) lie partially outside the original
curve (whose extent is 997 feet). The extension is achieved by fitting a line of best fit
to the last 16 points of the original curve. The x,y values representing the last 16 points
are natural log transforms of the distance and deposition values at the 16 points. Two long
transforms are coded here, reflecting the fact that the AGDRIFT model (v2.1.1) uses each of them
under different circumstandes (which I believe is not the intention but is the way the model
functions -- my guess is that one of the transforms was used and then a second one was coded
to increase the degree of conservativeness -- but the code was changed in only one of the two
places where the transformation occurs.
Finally, the AGDRIFT model extends the curve only when necessary (i.e., when it determines that
the area of intereest lies partially beyond the last point of the origanal curve (997 ft). In
this code all the curves are extended out to 1994 ft, which represents the furthest distance that
the downwind edge of an area of concern can be specified. All scenario curves are extended here
because we are running multiple simulations (e.g., monte carlo) and instead of extending the
curves each time a simulation requires it (which may be multiple time for the same scenario
curve) we just do it for all curves up front. There is a case to be made that the
curves should be extended external to this code and simply provide the full curve in the SQLite
database containing the original curve.
:param x_array: array of x values to be extended (must be at least 17 data points in original array)
:param y_array: array of y values to be extended
:param max_dist: maximum distance (ft) associated with unextended x values
:param dist_inc: increment (ft) for each extended data point
:param num_pts_ext: number of points at end of original x,y arrays to be used for extending the curve
:param ln_ln_trans: form of transformation to perform (True: straight ln ln, False: relative ln ln)
:return:
"""
# create empty pandas dataframes to create empty object for this unittest
agdrift_empty = self.create_agdrift_object()
expected_result_x = pd.Series([], dtype='float')
expected_result_y =
|
pd.Series([], dtype='float')
|
pandas.Series
|
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# Copyright Toolkit Authors
"""V3DBHandler."""
from copy import deepcopy
from datetime import datetime
import hashlib
import importlib
import logging
import os
from migrate import create_column
import numpy as np
import pandas as pd
from sqlalchemy import sql, Column, Table, MetaData, inspect
from sqlalchemy.exc import OperationalError, ProgrammingError
from sqlalchemy.types import VARCHAR, DECIMAL, BOOLEAN, TEXT
from sqlalchemy.dialects.mysql import DOUBLE
from sqlalchemy.dialects.postgresql import DOUBLE_PRECISION
from tqdm import tqdm
from pydtk.db import V2BaseDBHandler as _V2BaseDBHandler
from pydtk.utils.utils import load_config
from pydtk.utils.utils import dicts_to_listed_dict_2d
from pydtk.utils.utils import dtype_string_to_dtype_object
from pydtk.utils.utils import serialize_dict_1d
from pydtk.utils.utils import deserialize_dict_1d
DB_HANDLERS = {} # key: db_class, value: dict( key: db_engine, value: handler )
def map_dtype(dtype, db_engine='general'):
"""Mapper for dtype.
Args:
dtype (str): dtype in dataframe
db_engine (str): DB engine
Returns:
(dict): { 'df': dtype for Pandas.DataFrame, 'sql': dtype for SQL table }
"""
if dtype in ['int', 'int32', 'int64', 'float', 'float32', 'float64', 'double']:
if db_engine in ['mysql', 'mariadb']:
return {'df': 'double', 'sql': DOUBLE}
elif db_engine in ['postgresql', 'timescaledb']:
return {'df': 'double', 'sql': DOUBLE_PRECISION}
else:
return {'df': 'double', 'sql': DECIMAL}
if dtype in ['bool']:
return {'df': 'boolean', 'sql': BOOLEAN}
if dtype in ['object', 'string', 'text']:
return {'df': 'text', 'sql': TEXT}
raise ValueError('Unsupported dtype: {}'.format(dtype))
def register_handlers():
"""Register handlers."""
for filename in os.listdir(os.path.join(os.path.dirname(__file__))):
if not os.path.isfile(os.path.join(os.path.dirname(__file__), filename)):
continue
if filename == '__init__.py':
continue
try:
importlib.import_module(
os.path.join('pydtk.db.v3.handlers',
os.path.splitext(filename)[0]).replace(os.sep, '.')
)
except ModuleNotFoundError:
logging.debug('Failed to load handlers in {}'.format(filename))
def register_handler(db_classes, db_engines):
"""Register a DB-handler.
Args:
db_classes (list): list of db_class names (e.g. ['meta'])
db_engines (list): list of supported db_engines (e.g. ['sqlite', 'mysql'])
"""
def decorator(cls):
for db_class in db_classes:
if db_class not in DB_HANDLERS.keys():
DB_HANDLERS.update({db_class: {}})
for db_engine in db_engines:
if db_engine not in DB_HANDLERS[db_class].keys():
DB_HANDLERS[db_class].update({db_engine: cls})
return cls
return decorator
class BaseDBHandler(_V2BaseDBHandler):
"""Base handler for db."""
__version__ = 'v3'
_df_class = 'base_df'
def __new__(cls, db_class: str = None, db_engine: str = None, **kwargs) -> object:
"""Create object.
Args:
db_class (str): database class (e.g. 'meta')
db_engine (str): database engine (e.g. 'sqlite')
**kwargs: DB-handler specific arguments
Returns:
(object): the corresponding handler object
"""
if cls is BaseDBHandler:
handler = cls._get_handler(db_class, db_engine)
return super(BaseDBHandler, cls).__new__(handler)
else:
return super(BaseDBHandler, cls).__new__(cls)
@classmethod
def _get_handler(cls, db_class, db_engine=None):
"""Returns an appropriate handler.
Args:
db_class (str): database class (e.g. 'meta')
db_engine (str): database engine (e.g. 'sqlite')
Returns:
(handler): database handler object
"""
# Load default config
config = load_config(cls.__version__)
# Check if the db_class is available
if db_class not in DB_HANDLERS.keys():
raise ValueError('Unsupported db_class: {}'.format(db_class))
# Get db_engine from environment variable if not specified
if db_engine is None:
db_engine = os.environ.get('PYDTK_{}_DB_ENGINE'.format(db_class.upper()), None)
# Get the default engine if not specified
if db_engine is None:
try:
db_defaults = getattr(config.sql, db_class)
db_engine = db_defaults.engine
except (ValueError, AttributeError):
raise ValueError('Could not find the default value')
# Check if the corresponding handler is registered
if db_engine not in DB_HANDLERS[db_class].keys():
raise ValueError('Unsupported db_engine: {}'.format(db_engine))
# Get a DB-handler supporting the engine
return DB_HANDLERS[db_class][db_engine]
def __init__(self, **kwargs):
"""Initialize BaseDBHandler.
Args:
**kwargs: kwargs
"""
self._count_total = 0
if 'db_class' in kwargs.keys():
del kwargs['db_class']
super(BaseDBHandler, self).__init__(**kwargs)
def __next__(self):
"""Return the next item."""
if self._cursor >= len(self.df):
self._cursor = 0
raise StopIteration()
# Grab data
data = self.df.take([self._cursor]).to_dict(orient='records')[0]
# Delete internal column
if 'uuid_in_df' in data.keys():
del data['uuid_in_df']
if 'creation_time_in_df' in data.keys():
del data['creation_time_in_df']
# Post-processes
data = deserialize_dict_1d(data)
# Increment
self._cursor += 1
return data
def _initialize_df(self):
"""Initialize DF."""
df = pd.concat(
[pd.Series(name=c['name'],
dtype=dtype_string_to_dtype_object(c['dtype'])) for c in self.columns if c['name'] != 'uuid_in_df' and c['name'] != 'creation_time_in_df'] # noqa: E501
+ [
|
pd.Series(name='uuid_in_df', dtype=str)
|
pandas.Series
|
# pylint: disable=no-member,redefined-outer-name,unused-argument
# pylint: disable=unused-variable
"""Unit tests for pandera API extensions."""
from typing import Any, Optional
import pandas as pd
import pytest
import pandera as pa
import pandera.strategies as st
from pandera import PandasDtype, extensions
from pandera.checks import Check
@pytest.fixture(scope="function")
def custom_check_teardown():
"""Remove all custom checks after execution of each pytest function."""
yield
for check_name in list(pa.Check.REGISTERED_CUSTOM_CHECKS):
delattr(pa.Check, check_name)
del pa.Check.REGISTERED_CUSTOM_CHECKS[check_name]
@pytest.mark.parametrize(
"data",
[
pd.Series([10, 10, 10]),
pd.DataFrame([[10, 10, 10], [10, 10, 10]]),
],
)
def test_register_vectorized_custom_check(custom_check_teardown, data):
"""Test registering a vectorized custom check."""
@extensions.register_check_method(
statistics=["val"],
supported_types=(pd.Series, pd.DataFrame),
check_type="vectorized",
)
def custom_check(pandas_obj, *, val):
return pandas_obj == val
check = Check.custom_check(val=10)
check_result = check(data)
assert check_result.check_passed
for kwargs in [
{"element_wise": True},
{"element_wise": False},
{"groupby": "column"},
{"groups": ["group1", "group2"]},
]:
with pytest.warns(UserWarning):
Check.custom_check(val=10, **kwargs)
with pytest.raises(
ValueError,
match="method with name 'custom_check' already defined",
):
# pylint: disable=function-redefined
@extensions.register_check_method(statistics=["val"])
def custom_check(pandas_obj, val): # noqa
return pandas_obj != val
@pytest.mark.parametrize(
"data",
[
pd.Series([10, 10, 10]),
pd.DataFrame([[10, 10, 10], [10, 10, 10]]),
],
)
def test_register_element_wise_custom_check(custom_check_teardown, data):
"""Test registering an element-wise custom check."""
@extensions.register_check_method(
statistics=["val"],
supported_types=(pd.Series, pd.DataFrame),
check_type="element_wise",
)
def custom_check(element, *, val):
return element == val
check = Check.custom_check(val=10)
check_result = check(data)
assert check_result.check_passed
for kwargs in [
{"element_wise": True},
{"element_wise": False},
{"groupby": "column"},
{"groups": ["group1", "group2"]},
]:
with pytest.warns(UserWarning):
Check.custom_check(val=10, **kwargs)
with pytest.raises(
ValueError,
match="Element-wise checks should support DataFrame and Series "
"validation",
):
@extensions.register_check_method(
supported_types=pd.Series,
check_type="element_wise",
)
def invalid_custom_check(*args):
pass
def test_register_custom_groupby_check(custom_check_teardown):
"""Test registering a custom groupby check."""
@extensions.register_check_method(
statistics=["group_a", "group_b"],
supported_types=(pd.Series, pd.DataFrame),
check_type="groupby",
)
def custom_check(dict_groups, *, group_a, group_b):
"""
Test that the mean values in group A is larger than that of group B.
Note that this function can handle groups of both dataframes and
series.
"""
return (
dict_groups[group_a].values.mean()
> dict_groups[group_b].values.mean()
)
# column groupby check
data_column_check = pd.DataFrame(
{
"col1": [20, 20, 10, 10],
"col2": list("aabb"),
}
)
schema_column_check = pa.DataFrameSchema(
{
"col1": pa.Column(
int,
Check.custom_check(group_a="a", group_b="b", groupby="col2"),
),
"col2": pa.Column(str),
}
)
assert isinstance(schema_column_check(data_column_check), pd.DataFrame)
# dataframe groupby check
data_df_check = pd.DataFrame(
{
"col1": [20, 20, 10, 10],
"col2": [30, 30, 5, 5],
"col3": [10, 10, 1, 1],
},
index=pd.Index(list("aabb"), name="my_index"),
)
schema_df_check = pa.DataFrameSchema(
columns={
"col1": pa.Column(int),
"col2": pa.Column(int),
"col3": pa.Column(int),
},
index=pa.Index(str, name="my_index"),
checks=Check.custom_check(
group_a="a", group_b="b", groupby="my_index"
),
)
assert isinstance(schema_df_check(data_df_check), pd.DataFrame)
for kwargs in [{"element_wise": True}, {"element_wise": False}]:
with pytest.warns(UserWarning):
Check.custom_check(val=10, **kwargs)
@pytest.mark.parametrize(
"supported_types",
[
1,
10.0,
"foo",
{"foo": "bar"},
{1: 10},
["foo", "bar"],
[1, 10],
("foo", "bar"),
(1, 10),
],
)
def test_register_check_invalid_supported_types(supported_types):
"""Test that TypeError is raised on invalid supported_types arg."""
with pytest.raises(TypeError):
@extensions.register_check_method(supported_types=supported_types)
def custom_check(*args, **kwargs):
pass
@pytest.mark.skipif(
not st.HAS_HYPOTHESIS, reason='needs "strategies" module dependencies'
)
def test_register_check_with_strategy(custom_check_teardown):
"""Test registering a custom check with a data generation strategy."""
import hypothesis # pylint: disable=import-outside-toplevel,import-error
def custom_ge_strategy(
pandas_dtype: PandasDtype,
strategy: Optional[st.SearchStrategy] = None,
*,
min_value: Any,
) -> st.SearchStrategy:
if strategy is None:
return st.pandas_dtype_strategy(
pandas_dtype,
min_value=min_value,
exclude_min=False,
)
return strategy.filter(lambda x: x > min_value)
@extensions.register_check_method(
statistics=["min_value"], strategy=custom_ge_strategy
)
def custom_ge_check(pandas_obj, *, min_value):
return pandas_obj >= min_value
check = Check.custom_ge_check(min_value=0)
strat = check.strategy(PandasDtype.Int)
with pytest.warns(hypothesis.errors.NonInteractiveExampleWarning):
assert strat.example() >= 0
def test_schema_model_field_kwarg(custom_check_teardown):
"""Test that registered checks can be specified in a Field."""
# pylint: disable=missing-class-docstring,too-few-public-methods
@extensions.register_check_method(
statistics=["val"],
supported_types=(pd.Series, pd.DataFrame),
check_type="vectorized",
)
def custom_gt(pandas_obj, val):
return pandas_obj > val
@extensions.register_check_method(
statistics=["min_value", "max_value"],
supported_types=(pd.Series, pd.DataFrame),
check_type="vectorized",
)
def custom_in_range(pandas_obj, min_value, max_value):
return (min_value <= pandas_obj) & (pandas_obj <= max_value)
class Schema(pa.SchemaModel):
"""Schema that uses registered checks in Field."""
col1: pa.typing.Series[int] = pa.Field(custom_gt=100)
col2: pa.typing.Series[float] = pa.Field(
custom_in_range={"min_value": -10, "max_value": 10}
)
class Config:
coerce = True
data = pd.DataFrame(
{
"col1": [101, 1000, 2000],
"col2": [-5.0, 0.0, 6.0],
}
)
Schema.validate(data)
for invalid_data in [
pd.DataFrame({"col1": [0], "col2": [-10.0]}),
|
pd.DataFrame({"col1": [1000], "col2": [-100.0]})
|
pandas.DataFrame
|
import pandas as pd
from business_rules.operators import (DataframeType, StringType,
NumericType, BooleanType, SelectType,
SelectMultipleType, GenericType)
from . import TestCase
from decimal import Decimal
import sys
import pandas
class StringOperatorTests(TestCase):
def test_operator_decorator(self):
self.assertTrue(StringType("foo").equal_to.is_operator)
def test_string_equal_to(self):
self.assertTrue(StringType("foo").equal_to("foo"))
self.assertFalse(StringType("foo").equal_to("Foo"))
def test_string_not_equal_to(self):
self.assertTrue(StringType("foo").not_equal_to("Foo"))
self.assertTrue(StringType("foo").not_equal_to("boo"))
self.assertFalse(StringType("foo").not_equal_to("foo"))
def test_string_equal_to_case_insensitive(self):
self.assertTrue(StringType("foo").equal_to_case_insensitive("FOo"))
self.assertTrue(StringType("foo").equal_to_case_insensitive("foo"))
self.assertFalse(StringType("foo").equal_to_case_insensitive("blah"))
def test_string_starts_with(self):
self.assertTrue(StringType("hello").starts_with("he"))
self.assertFalse(StringType("hello").starts_with("hey"))
self.assertFalse(StringType("hello").starts_with("He"))
def test_string_ends_with(self):
self.assertTrue(StringType("hello").ends_with("lo"))
self.assertFalse(StringType("hello").ends_with("boom"))
self.assertFalse(StringType("hello").ends_with("Lo"))
def test_string_contains(self):
self.assertTrue(StringType("hello").contains("ell"))
self.assertTrue(StringType("hello").contains("he"))
self.assertTrue(StringType("hello").contains("lo"))
self.assertFalse(StringType("hello").contains("asdf"))
self.assertFalse(StringType("hello").contains("ElL"))
def test_string_matches_regex(self):
self.assertTrue(StringType("hello").matches_regex(r"^h"))
self.assertFalse(StringType("hello").matches_regex(r"^sh"))
def test_non_empty(self):
self.assertTrue(StringType("hello").non_empty())
self.assertFalse(StringType("").non_empty())
self.assertFalse(StringType(None).non_empty())
class NumericOperatorTests(TestCase):
def test_instantiate(self):
err_string = "foo is not a valid numeric type"
with self.assertRaisesRegexp(AssertionError, err_string):
NumericType("foo")
def test_numeric_type_validates_and_casts_decimal(self):
ten_dec = Decimal(10)
ten_int = 10
ten_float = 10.0
if sys.version_info[0] == 2:
ten_long = long(10)
else:
ten_long = int(10) # long and int are same in python3
ten_var_dec = NumericType(ten_dec) # this should not throw an exception
ten_var_int = NumericType(ten_int)
ten_var_float = NumericType(ten_float)
ten_var_long = NumericType(ten_long)
self.assertTrue(isinstance(ten_var_dec.value, Decimal))
self.assertTrue(isinstance(ten_var_int.value, Decimal))
self.assertTrue(isinstance(ten_var_float.value, Decimal))
self.assertTrue(isinstance(ten_var_long.value, Decimal))
def test_numeric_equal_to(self):
self.assertTrue(NumericType(10).equal_to(10))
self.assertTrue(NumericType(10).equal_to(10.0))
self.assertTrue(NumericType(10).equal_to(10.000001))
self.assertTrue(NumericType(10.000001).equal_to(10))
self.assertTrue(NumericType(Decimal('10.0')).equal_to(10))
self.assertTrue(NumericType(10).equal_to(Decimal('10.0')))
self.assertFalse(NumericType(10).equal_to(10.00001))
self.assertFalse(NumericType(10).equal_to(11))
def test_numeric_not_equal_to(self):
self.assertTrue(NumericType(10).not_equal_to(10.00001))
self.assertTrue(NumericType(10).not_equal_to(11))
self.assertTrue(NumericType(Decimal('10.0')).not_equal_to(Decimal('10.1')))
self.assertFalse(NumericType(10).not_equal_to(10))
self.assertFalse(NumericType(10).not_equal_to(10.0))
self.assertFalse(NumericType(Decimal('10.0')).not_equal_to(Decimal('10.0')))
def test_other_value_not_numeric(self):
error_string = "10 is not a valid numeric type"
with self.assertRaisesRegexp(AssertionError, error_string):
NumericType(10).equal_to("10")
def test_numeric_greater_than(self):
self.assertTrue(NumericType(10).greater_than(1))
self.assertFalse(NumericType(10).greater_than(11))
self.assertTrue(NumericType(10.1).greater_than(10))
self.assertFalse(NumericType(10.000001).greater_than(10))
self.assertTrue(NumericType(10.000002).greater_than(10))
def test_numeric_greater_than_or_equal_to(self):
self.assertTrue(NumericType(10).greater_than_or_equal_to(1))
self.assertFalse(NumericType(10).greater_than_or_equal_to(11))
self.assertTrue(NumericType(10.1).greater_than_or_equal_to(10))
self.assertTrue(NumericType(10.000001).greater_than_or_equal_to(10))
self.assertTrue(NumericType(10.000002).greater_than_or_equal_to(10))
self.assertTrue(NumericType(10).greater_than_or_equal_to(10))
def test_numeric_less_than(self):
self.assertTrue(NumericType(1).less_than(10))
self.assertFalse(NumericType(11).less_than(10))
self.assertTrue(NumericType(10).less_than(10.1))
self.assertFalse(NumericType(10).less_than(10.000001))
self.assertTrue(NumericType(10).less_than(10.000002))
def test_numeric_less_than_or_equal_to(self):
self.assertTrue(NumericType(1).less_than_or_equal_to(10))
self.assertFalse(NumericType(11).less_than_or_equal_to(10))
self.assertTrue(NumericType(10).less_than_or_equal_to(10.1))
self.assertTrue(NumericType(10).less_than_or_equal_to(10.000001))
self.assertTrue(NumericType(10).less_than_or_equal_to(10.000002))
self.assertTrue(NumericType(10).less_than_or_equal_to(10))
class BooleanOperatorTests(TestCase):
def test_instantiate(self):
err_string = "foo is not a valid boolean type"
with self.assertRaisesRegexp(AssertionError, err_string):
BooleanType("foo")
err_string = "None is not a valid boolean type"
with self.assertRaisesRegexp(AssertionError, err_string):
BooleanType(None)
def test_boolean_is_true_and_is_false(self):
self.assertTrue(BooleanType(True).is_true())
self.assertFalse(BooleanType(True).is_false())
self.assertFalse(BooleanType(False).is_true())
self.assertTrue(BooleanType(False).is_false())
class SelectOperatorTests(TestCase):
def test_contains(self):
self.assertTrue(SelectType([1, 2]).contains(2))
self.assertFalse(SelectType([1, 2]).contains(3))
self.assertTrue(SelectType([1, 2, "a"]).contains("A"))
def test_does_not_contain(self):
self.assertTrue(SelectType([1, 2]).does_not_contain(3))
self.assertFalse(SelectType([1, 2]).does_not_contain(2))
self.assertFalse(SelectType([1, 2, "a"]).does_not_contain("A"))
class SelectMultipleOperatorTests(TestCase):
def test_contains_all(self):
self.assertTrue(SelectMultipleType([1, 2]).
contains_all([2, 1]))
self.assertFalse(SelectMultipleType([1, 2]).
contains_all([2, 3]))
self.assertTrue(SelectMultipleType([1, 2, "a"]).
contains_all([2, 1, "A"]))
def test_is_contained_by(self):
self.assertTrue(SelectMultipleType([1, 2]).
is_contained_by([2, 1, 3]))
self.assertFalse(SelectMultipleType([1, 2]).
is_contained_by([2, 3, 4]))
self.assertTrue(SelectMultipleType([1, 2, "a"]).
is_contained_by([2, 1, "A"]))
def test_shares_at_least_one_element_with(self):
self.assertTrue(SelectMultipleType([1, 2]).
shares_at_least_one_element_with([2, 3]))
self.assertFalse(SelectMultipleType([1, 2]).
shares_at_least_one_element_with([4, 3]))
self.assertTrue(SelectMultipleType([1, 2, "a"]).
shares_at_least_one_element_with([4, "A"]))
def test_shares_exactly_one_element_with(self):
self.assertTrue(SelectMultipleType([1, 2]).
shares_exactly_one_element_with([2, 3]))
self.assertFalse(SelectMultipleType([1, 2]).
shares_exactly_one_element_with([4, 3]))
self.assertTrue(SelectMultipleType([1, 2, "a"]).
shares_exactly_one_element_with([4, "A"]))
self.assertFalse(SelectMultipleType([1, 2, 3]).
shares_exactly_one_element_with([2, 3, "a"]))
def test_shares_no_elements_with(self):
self.assertTrue(SelectMultipleType([1, 2]).
shares_no_elements_with([4, 3]))
self.assertFalse(SelectMultipleType([1, 2]).
shares_no_elements_with([2, 3]))
self.assertFalse(SelectMultipleType([1, 2, "a"]).
shares_no_elements_with([4, "A"]))
class DataframeOperatorTests(TestCase):
def test_exists(self):
df = pandas.DataFrame.from_dict({
"var1": [1, 2, 4, ],
"var2": [3, 5, 6, ],
})
result: pd.Series = DataframeType({"value": df}).exists({"target": "var1"})
self.assertTrue(result.equals(pd.Series([True, True, True, ])))
result: pd.Series = DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).exists({"target": "--r1"})
self.assertTrue(result.equals(pd.Series([True, True, True, ])))
result: pd.Series = DataframeType({"value": df}).exists({"target": "invalid"})
self.assertTrue(result.equals(pd.Series([False, False, False, ])))
def test_not_exists(self):
df = pandas.DataFrame.from_dict({
"var1": [1, 2, 4, ],
"var2": [3, 5, 6, ]
})
result: pd.Series = DataframeType({"value": df}).not_exists({"target": "invalid"})
self.assertTrue(result.equals(pd.Series([True, True, True, ])))
result: pd.Series = DataframeType({"value": df}).not_exists({"target": "var1"})
self.assertTrue(result.equals(pd.Series([False, False, False, ])))
result: pd.Series = DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).not_exists({"target": "--r1"})
self.assertTrue(result.equals(pd.Series([False, False, False, ])))
def test_equal_to(self):
df = pandas.DataFrame.from_dict({
"var1": [1, 2, 4, "", 7, ],
"var2": [3, 5, 6, "", 2, ],
"var3": [1, 3, 8, "", 7, ],
"var4": ["test", "issue", "one", "", "two", ]
})
self.assertTrue(DataframeType({"value": df}).equal_to({
"target": "var1",
"comparator": ""
}).equals(pandas.Series([False, False, False, False, False, ])))
self.assertTrue(DataframeType({"value": df}).equal_to({
"target": "var1",
"comparator": 2
}).equals(pandas.Series([False, True, False, False, False, ])))
self.assertTrue(DataframeType({"value": df}).equal_to({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([True, False, False, False, True, ])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).equal_to({
"target": "--r1",
"comparator": "--r3"
}).equals(pandas.Series([True, False, False, False, True, ])))
self.assertTrue(DataframeType({"value": df}).equal_to({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([False, False, False, False, False, ])))
self.assertTrue(DataframeType({"value": df}).equal_to({
"target": "var1",
"comparator": 20
}).equals(pandas.Series([False, False, False, False, False, ])))
self.assertTrue(DataframeType({"value": df}).equal_to({
"target": "var4",
"comparator": "var1",
"value_is_literal": True
}).equals(pandas.Series([False, False, False, False, False, ])))
def test_not_equal_to(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4]
})
self.assertTrue(DataframeType({"value": df}).not_equal_to({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).not_equal_to({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).not_equal_to({
"target": "--r1",
"comparator": "--r2"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).not_equal_to({
"target": "--r1",
"comparator": 20
}).equals(pandas.Series([True, True, True])))
def test_equal_to_case_insensitive(self):
df = pandas.DataFrame.from_dict({
"var1": ["word", "", "new", "val"],
"var2": ["WORD", "", "test", "VAL"],
"var3": ["LET", "", "GO", "read"]
})
self.assertTrue(DataframeType({"value": df}).equal_to_case_insensitive({
"target": "var1",
"comparator": "NEW"
}).equals(pandas.Series([False, False, True, False])))
self.assertTrue(DataframeType({"value": df}).equal_to_case_insensitive({
"target": "var1",
"comparator": ""
}).equals(pandas.Series([False, False, False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).equal_to_case_insensitive({
"target": "--r1",
"comparator": "--r2"
}).equals(pandas.Series([True, False, False, True])))
self.assertTrue(DataframeType({"value": df}).equal_to_case_insensitive({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([True, False, False, True])))
self.assertTrue(DataframeType({"value": df}).equal_to_case_insensitive({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).equal_to_case_insensitive({
"target": "var1",
"comparator": "var1",
"value_is_literal": True
}).equals(pandas.Series([False, False, False, False])))
def test_not_equal_to_case_insensitive(self):
df = pandas.DataFrame.from_dict({
"var1": ["word", "new", "val"],
"var2": ["WORD", "test", "VAL"],
"var3": ["LET", "GO", "read"],
"var4": ["WORD", "NEW", "VAL"]
})
self.assertTrue(DataframeType({"value": df}).not_equal_to_case_insensitive({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).not_equal_to_case_insensitive({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([False, True, False])))
self.assertTrue(DataframeType({"value": df}).not_equal_to_case_insensitive({
"target": "var1",
"comparator": "var1",
"value_is_literal": True
}).equals(pandas.Series([True, True, True])))
def test_less_than(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4]
})
self.assertTrue(DataframeType({"value": df}).less_than({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).less_than({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([False, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).less_than({
"target": "--r1",
"comparator": "var3"
}).equals(pandas.Series([False, True, True])))
self.assertTrue(DataframeType({"value": df}).less_than({
"target": "var2",
"comparator": 2
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).less_than({
"target": "var1",
"comparator": 3
}).equals(pandas.Series([True, True, False])))
another_df = pandas.DataFrame.from_dict(
{
"LBDY": [4, None, None, None, None]
}
)
self.assertTrue(DataframeType({"value": another_df}).less_than({
"target": "LBDY",
"comparator": 5
}).equals(pandas.Series([True, False, False, False, False, ])))
def test_less_than_or_equal_to(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4]
})
self.assertTrue(DataframeType({"value": df}).less_than_or_equal_to({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).less_than_or_equal_to({
"target": "--r1",
"comparator": "var4"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).less_than_or_equal_to({
"target": "var2",
"comparator": "var1"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).less_than_or_equal_to({
"target": "var2",
"comparator": 2
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).less_than_or_equal_to({
"target": "var2",
"comparator": "var3"
}).equals(pandas.Series([False, False, True])))
another_df = pandas.DataFrame.from_dict(
{
"LBDY": [4, 5, None, None, None]
}
)
self.assertTrue(DataframeType({"value": another_df}).less_than_or_equal_to({
"target": "LBDY",
"comparator": 5
}).equals(pandas.Series([True, True, False, False, False, ])))
def test_greater_than(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4]
})
self.assertTrue(DataframeType({"value": df}).greater_than({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).greater_than({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).greater_than({
"target": "var1",
"comparator": "--r3"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).greater_than({
"target": "var2",
"comparator": 2
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).greater_than({
"target": "var1",
"comparator": 5000
}).equals(pandas.Series([False, False, False])))
another_df = pandas.DataFrame.from_dict(
{
"LBDY": [4, None, None, None, None]
}
)
self.assertTrue(DataframeType({"value": another_df}).greater_than({
"target": "LBDY",
"comparator": 3
}).equals(pandas.Series([True, False, False, False, False, ])))
def test_greater_than_or_equal_to(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4]
})
self.assertTrue(DataframeType({"value": df}).greater_than_or_equal_to({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).greater_than_or_equal_to({
"target": "var1",
"comparator": "--r4"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).greater_than_or_equal_to({
"target": "var2",
"comparator": "var3"
}).equals(pandas.Series([True, True, False])))
self.assertTrue(DataframeType({"value": df}).greater_than_or_equal_to({
"target": "var2",
"comparator": 2
}).equals(pandas.Series([True, True, True])))
another_df = pandas.DataFrame.from_dict(
{
"LBDY": [4, 3, None, None, None]
}
)
self.assertTrue(DataframeType({"value": another_df}).greater_than_or_equal_to({
"target": "LBDY",
"comparator": 3
}).equals(pandas.Series([True, True, False, False, False, ])))
def test_contains(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4],
"string_var": ["hj", "word", "c"],
"var5": [[1,3,5],[1,3,5], [1,3,5]]
})
self.assertTrue(DataframeType({"value": df}).contains({
"target": "var1",
"comparator": 2
}).equals(pandas.Series([False, True, False])))
self.assertTrue(DataframeType({"value": df}).contains({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([True, False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).contains({
"target": "var1",
"comparator": "--r3"
}).equals(pandas.Series([True, False, False])))
self.assertTrue(DataframeType({"value": df}).contains({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).contains({
"target": "string_var",
"comparator": "string_var"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).contains({
"target": "string_var",
"comparator": "string_var",
"value_is_literal": True
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).contains({
"target": "var5",
"comparator": "var1"
}).equals(pandas.Series([True, False, False])))
def test_does_not_contain(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4],
"string_var": ["hj", "word", "c"],
"var5": [[1,3,5],[1,3,5], [1,3,5]]
})
self.assertTrue(DataframeType({"value": df}).does_not_contain({
"target": "var1",
"comparator": 5
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).does_not_contain({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([False, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).does_not_contain({
"target": "var1",
"comparator": "--r3"
}).equals(pandas.Series([False, True, True])))
self.assertTrue(DataframeType({"value": df}).does_not_contain({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).does_not_contain({
"target": "string_var",
"comparator": "string_var",
"value_is_literal": True
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).does_not_contain({
"target": "string_var",
"comparator": "string_var"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).does_not_contain({
"target": "var5",
"comparator": "var1"
}).equals(pandas.Series([False, True, True])))
def test_contains_case_insensitive(self):
df = pandas.DataFrame.from_dict({
"var1": ["pikachu", "charmander", "squirtle"],
"var2": ["PIKACHU", "CHARIZARD", "BULBASAUR"],
"var3": ["POKEMON", "CHARIZARD", "BULBASAUR"],
"var4": [
["pikachu", "charizard", "bulbasaur"],
["chikorita", "cyndaquil", "totodile"],
["chikorita", "cyndaquil", "totodile"]
]
})
self.assertTrue(DataframeType({"value": df}).contains_case_insensitive({
"target": "var1",
"comparator": "PIKACHU"
}).equals(pandas.Series([True, False, False])))
self.assertTrue(DataframeType({"value": df}).contains_case_insensitive({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([True, False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).contains_case_insensitive({
"target": "--r1",
"comparator": "--r2"
}).equals(pandas.Series([True, False, False])))
self.assertTrue(DataframeType({"value": df}).contains_case_insensitive({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).contains_case_insensitive({
"target": "var3",
"comparator": "var3"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).contains_case_insensitive({
"target": "var3",
"comparator": "var3",
"value_is_literal": True
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).contains_case_insensitive({
"target": "var4",
"comparator": "var2"
}).equals(pandas.Series([True, False, False])))
def test_does_not_contain_case_insensitive(self):
df = pandas.DataFrame.from_dict({
"var1": ["pikachu", "charmander", "squirtle"],
"var2": ["PIKACHU", "CHARIZARD", "BULBASAUR"],
"var3": ["pikachu", "charizard", "bulbasaur"],
"var4": [
["pikachu", "charizard", "bulbasaur"],
["chikorita", "cyndaquil", "totodile"],
["chikorita", "cyndaquil", "totodile"]
]
})
self.assertTrue(DataframeType({"value": df}).does_not_contain_case_insensitive({
"target": "var1",
"comparator": "IVYSAUR"
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).does_not_contain_case_insensitive({
"target": "var3",
"comparator": "var2"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).does_not_contain_case_insensitive({
"target": "var3",
"comparator": "var3",
"value_is_literal": True
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).does_not_contain_case_insensitive({
"target": "var3",
"comparator": "var3"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).does_not_contain_case_insensitive({
"target": "var4",
"comparator": "var2"
}).equals(pandas.Series([False, True, True])))
def test_is_contained_by(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4],
"var5": [[1,2,3], [1,2], [17]]
})
self.assertTrue(DataframeType({"value": df}).is_contained_by({
"target": "var1",
"comparator": [4,5,6]
}).equals(pandas.Series([False, False, True])))
self.assertTrue(DataframeType({"value": df}).is_contained_by({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([True, False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).is_contained_by({
"target": "var1",
"comparator": "--r3"
}).equals(pandas.Series([True, False, False])))
self.assertTrue(DataframeType({"value": df}).is_contained_by({
"target": "var1",
"comparator": [9, 10, 11]
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).is_contained_by({
"target": "var1",
"comparator": "var2"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).is_contained_by({
"target": "var1",
"comparator": "var5"
}).equals(pandas.Series([True, True, False])))
def test_is_not_contained_by(self):
df = pandas.DataFrame.from_dict({
"var1": [1,2,4],
"var2": [3,5,6],
"var3": [1,3,8],
"var4": [1,2,4],
"var5": [[1,2,3], [1,2], [17]]
})
self.assertTrue(DataframeType({"value": df}).is_not_contained_by({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([False, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).is_not_contained_by({
"target": "var1",
"comparator": "--r3"
}).equals(pandas.Series([False, True, True])))
self.assertTrue(DataframeType({"value": df}).is_not_contained_by({
"target": "var1",
"comparator": [9, 10, 11]
}).equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).is_not_contained_by({
"target": "var1",
"comparator": "var1"
}).equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).is_not_contained_by({
"target": "var1",
"comparator": "var5"
}).equals(pandas.Series([False, False, True])))
def test_is_contained_by_case_insensitive(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"],
"var4": [set(["word"]), set(["test"])]
})
self.assertTrue(DataframeType({"value": df}).is_contained_by_case_insensitive({
"target": "var1",
"comparator": ["word", "TEST"]
}).equals(pandas.Series([True, True])))
self.assertTrue(DataframeType({"value": df}).is_contained_by_case_insensitive({
"target": "var1",
"comparator": "var1"
}).equals(pandas.Series([True, True])))
self.assertTrue(DataframeType({"value": df}).is_contained_by_case_insensitive({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).is_contained_by_case_insensitive({
"target": "var1",
"comparator": "--r3"
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df}).is_contained_by_case_insensitive({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([True, True])))
self.assertTrue(DataframeType({"value": df}).is_contained_by_case_insensitive({
"target": "var3",
"comparator": "var4"
}).equals(pandas.Series([False, False])))
def test_is_not_contained_by_case_insensitive(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"],
"var4": [set(["word"]), set(["test"])]
})
self.assertTrue(DataframeType({"value": df}).is_not_contained_by_case_insensitive({
"target": "var1",
"comparator": ["word", "TEST"]
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df}).is_not_contained_by_case_insensitive({
"target": "var1",
"comparator": "var3"
}).equals(pandas.Series([True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).is_not_contained_by_case_insensitive({
"target": "var1",
"comparator": "--r3"
}).equals(pandas.Series([True, True])))
self.assertTrue(DataframeType({"value": df}).is_not_contained_by_case_insensitive({
"target": "var1",
"comparator": "var4"
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df}).is_not_contained_by_case_insensitive({
"target": "var3",
"comparator": "var4"
}).equals(pandas.Series([True, True])))
def test_prefix_matches_regex(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).prefix_matches_regex({
"target": "--r2",
"comparator": "w.*",
"prefix": 2
}).equals(pandas.Series([True, False])))
self.assertTrue(DataframeType({"value": df}).prefix_matches_regex({
"target": "var2",
"comparator": "[0-9].*",
"prefix": 2
}).equals(pandas.Series([False, False])))
def test_suffix_matches_regex(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).suffix_matches_regex({
"target": "--r1",
"comparator": "es.*",
"suffix": 3
}).equals(pandas.Series([False, True])))
self.assertTrue(DataframeType({"value": df}).suffix_matches_regex({
"target": "var1",
"comparator": "[0-9].*",
"suffix": 3
}).equals(pandas.Series([False, False])))
def test_not_prefix_matches_suffix(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).not_prefix_matches_regex({
"target": "--r1",
"comparator": ".*",
"prefix": 2
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df}).not_prefix_matches_regex({
"target": "var2",
"comparator": "[0-9].*",
"prefix": 2
}).equals(pandas.Series([True, True])))
def test_not_suffix_matches_regex(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df}).not_suffix_matches_regex({
"target": "var1",
"comparator": ".*",
"suffix": 3
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).not_suffix_matches_regex({
"target": "--r1",
"comparator": "[0-9].*",
"suffix": 3
}).equals(pandas.Series([True, True])))
def test_matches_suffix(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).matches_regex({
"target": "--r1",
"comparator": ".*",
}).equals(pandas.Series([True, True])))
self.assertTrue(DataframeType({"value": df}).matches_regex({
"target": "var2",
"comparator": "[0-9].*",
}).equals(pandas.Series([False, False])))
def test_not_matches_regex(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df}).not_matches_regex({
"target": "var1",
"comparator": ".*",
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).not_matches_regex({
"target": "--r1",
"comparator": "[0-9].*",
}).equals(pandas.Series([True, True])))
def test_starts_with(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).starts_with({
"target": "--r1",
"comparator": "WO",
}).equals(pandas.Series([True, False])))
self.assertTrue(DataframeType({"value": df}).starts_with({
"target": "var2",
"comparator": "ABC",
}).equals(pandas.Series([False, False])))
def test_ends_with(self):
df = pandas.DataFrame.from_dict({
"var1": ["WORD", "test"],
"var2": ["word", "TEST"],
"var3": ["another", "item"]
})
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).ends_with({
"target": "--r1",
"comparator": "abc",
}).equals(pandas.Series([False, False])))
self.assertTrue(DataframeType({"value": df}).ends_with({
"target": "var1",
"comparator": "est",
}).equals(pandas.Series([False, True])))
def test_has_equal_length(self):
df = pandas.DataFrame.from_dict(
{
"var_1": ['test', 'value']
}
)
df_operator = DataframeType({"value": df, "column_prefix_map": {"--": "va"}})
result = df_operator.has_equal_length({"target": "--r_1", "comparator": 4})
self.assertTrue(result.equals(pandas.Series([True, False])))
def test_has_not_equal_length(self):
df = pandas.DataFrame.from_dict(
{
"var_1": ['test', 'value']
}
)
df_operator = DataframeType({"value": df, "column_prefix_map": {"--": "va"}})
result = df_operator.has_not_equal_length({"target": "--r_1", "comparator": 4})
self.assertTrue(result.equals(pandas.Series([False, True])))
def test_longer_than(self):
df = pandas.DataFrame.from_dict(
{
"var_1": ['test', 'value']
}
)
df_operator = DataframeType({"value": df, "column_prefix_map": {"--": "va"}})
self.assertTrue(df_operator.longer_than({"target": "--r_1", "comparator": 3}).equals(pandas.Series([True, True])))
def test_longer_than_or_equal_to(self):
df = pandas.DataFrame.from_dict(
{
"var_1": ['test', 'alex']
}
)
df_operator = DataframeType({"value": df, "column_prefix_map": {"--": "va"}})
self.assertTrue(df_operator.longer_than_or_equal_to({"target": "--r_1", "comparator": 3}).equals(pandas.Series([True, True])))
self.assertTrue(df_operator.longer_than_or_equal_to({"target": "var_1", "comparator": 4}).equals(pandas.Series([True, True])))
def test_shorter_than(self):
df = pandas.DataFrame.from_dict(
{
"var_1": ['test', 'val']
}
)
df_operator = DataframeType({"value": df, "column_prefix_map": {"--": "va"}})
self.assertTrue(df_operator.shorter_than({"target": "--r_1", "comparator": 5}).equals(pandas.Series([True, True])))
def test_shorter_than_or_equal_to(self):
df = pandas.DataFrame.from_dict(
{
"var_1": ['test', 'alex']
}
)
df_operator = DataframeType({"value": df, "column_prefix_map": {"--": "va"}})
self.assertTrue(df_operator.shorter_than_or_equal_to({"target": "--r_1", "comparator": 5}).equals(pandas.Series([True, True])))
self.assertTrue(df_operator.shorter_than_or_equal_to({"target": "var_1", "comparator": 4}).equals(pandas.Series([True, True])))
def test_contains_all(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['test', 'value', 'word'],
"var2": ["test", "value", "test"]
}
)
self.assertTrue(DataframeType({"value": df}).contains_all({
"target": "var1",
"comparator": "var2",
}))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).contains_all({
"target": "--r1",
"comparator": "--r2",
}))
self.assertFalse(DataframeType({"value": df}).contains_all({
"target": "var2",
"comparator": "var1",
}))
self.assertTrue(DataframeType({"value": df}).contains_all({
"target": "var2",
"comparator": ["test", "value"],
}))
def test_not_contains_all(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['test', 'value', 'word'],
"var2": ["test", "value", "test"]
}
)
self.assertTrue(DataframeType({"value": df}).contains_all({
"target": "var1",
"comparator": "var2",
}))
self.assertFalse(DataframeType({"value": df}).contains_all({
"target": "var2",
"comparator": "var1",
}))
self.assertTrue(DataframeType({"value": df}).contains_all({
"target": "var2",
"comparator": ["test", "value"],
}))
def test_invalid_date(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['2021', '2021', '2021', '2021', '2099'],
"var2": ["2099", "2022", "2034", "90999", "20999"],
"var3": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
}
)
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).invalid_date({"target": "--r1"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).invalid_date({"target": "var3"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).invalid_date({"target": "var2"})
.equals(pandas.Series([False, False, False, True, True])))
def test_date_equal_to(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['2021', '2021', '2021', '2021', '2021'],
"var2": ["2099", "2022", "2034", "90999", "20999"],
"var3": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var4": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var5": ["1997-08", "1997-08-16", "1997-08-16T19:20:30.45+01:00", "1997-08-16T19:20:30+01:00", "1997-08-16T19:20+01:00"],
"var6": ["1998-08", "1998-08-11", "1998-08-17T20:21:31.46+01:00", "1998-08-17T20:21:31+01:00", "1998-08-17T20:21+01:00"],
"var7": ["", None, "", "", ""]
}
)
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var1", "comparator": '2021'})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "1997-07-16T19:20:30.45+01:00"})
.equals(pandas.Series([False, False, True, False, False])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var4"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df, "column_prefix_map": {"--": "va"}}).date_equal_to({"target": "--r3", "comparator": "--r4", "date_component": "year"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var5", "date_component": "hour"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var5", "date_component": "minute"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var5", "date_component": "second"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var5", "date_component": "microsecond"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var5", "date_component": "year"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var5", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var6", "date_component": "year"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var6", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var3", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_equal_to({"target": "var7", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
def test_date_not_equal_to(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['2021', '2021', '2021', '2021', '2021'],
"var2": ["2099", "2022", "2034", "90999", "20999"],
"var3": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var4": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var5": ["1997-08", "1997-08-16", "1997-08-16T19:20:30.45+01:00", "1997-08-16T19:20:30+01:00", "1997-08-16T19:20+01:00"],
"var6": ["1998-08", "1998-08-11", "1998-08-17T20:21:31.46+01:00", "1998-08-17T20:21:31+01:00", "1998-08-17T20:21+01:00"],
"var7": ["", None, "", "", ""]
}
)
self.assertTrue(DataframeType({"value": df}).date_not_equal_to({"target": "var1", "comparator": '2022'})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_not_equal_to({"target": "var3", "comparator": "1998-07-16T19:20:30.45+01:00"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_not_equal_to({"target": "var3", "comparator": "var4"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_not_equal_to({"target": "var3", "comparator": "var4", "date_component": "year"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_not_equal_to({"target": "var3", "comparator": "var6", "date_component": "hour"})
.equals(pandas.Series([False, False, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_not_equal_to({"target": "var3", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_not_equal_to({"target": "var7", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
def test_date_less_than(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['2021', '2021', '2021', '2021', '2021'],
"var2": ["2099", "2022", "2034", "90999", "20999"],
"var3": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var4": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var5": ["1997-08", "1997-08-16", "1997-08-16T19:20:30.45+01:00", "1997-08-16T19:20:30+01:00", "1997-08-16T19:20+01:00"],
"var6": ["1998-08", "1998-08-11", "1998-08-17T20:21:31.46+01:00", "1998-08-17T20:21:31+01:00", "1998-08-17T20:21+01:00"],
"var7": ["", None, "", "", ""]
}
)
self.assertTrue(DataframeType({"value": df}).date_less_than({"target": "var1", "comparator": '2022'})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_less_than({"target": "var3", "comparator": "1998-07-16T19:20:30.45+01:00"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_less_than({"target": "var3", "comparator": "var4"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_less_than({"target": "var3", "comparator": "var4", "date_component": "year"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_less_than({"target": "var3", "comparator": "var6", "date_component": "hour"})
.equals(pandas.Series([False, False, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_less_than({"target": "var3", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_less_than({"target": "var7", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
def test_date_greater_than(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['2021', '2021', '2021', '2021', '2021'],
"var2": ["2099", "2022", "2034", "90999", "20999"],
"var3": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var4": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var5": ["1997-08", "1997-08-16", "1997-08-16T19:20:30.45+01:00", "1997-08-16T19:20:30+01:00", "1997-08-16T19:20+01:00"],
"var6": ["1998-08", "1998-08-11", "1998-08-17T20:21:31.46+01:00", "1998-08-17T20:21:31+01:00", "1998-08-17T20:21+01:00"],
"var7": ["", None, "", "", ""]
}
)
self.assertTrue(DataframeType({"value": df}).date_greater_than({"target": "var1", "comparator": '2020'})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than({"target": "var3", "comparator": "1996-07-16T19:20:30.45+01:00"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than({"target": "var3", "comparator": "var4"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_greater_than({"target": "var3", "comparator": "var4", "date_component": "year"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_greater_than({"target": "var6", "comparator": "var3", "date_component": "hour"})
.equals(pandas.Series([False, False, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than({"target": "var3", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_greater_than({"target": "var7", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
def test_date_greater_than_or_equal_to(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['2021', '2021', '2021', '2021', '2021'],
"var2": ["2099", "2022", "2034", "90999", "20999"],
"var3": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var4": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var5": ["1997-08", "1997-08-16", "1997-08-16T19:20:30.45+01:00", "1997-08-16T19:20:30+01:00", "1997-08-16T19:20+01:00"],
"var6": ["1998-08", "1998-08-11", "1998-08-17T20:21:31.46+01:00", "1998-08-17T20:21:31+01:00", "1998-08-17T20:21+01:00"],
"var7": ["", None, "", "", ""]
}
)
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var1", "comparator": '2020'})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var1", "comparator": '2023'})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var3", "comparator": "1996-07-16T19:20:30.45+01:00"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var3", "comparator": "var4"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var3", "comparator": "var4", "date_component": "year"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var6", "comparator": "var3", "date_component": "hour"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var3", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_greater_than_or_equal_to({"target": "var7", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
def test_date_less_than_or_equal_to(self):
df = pandas.DataFrame.from_dict(
{
"var1": ['2021', '2021', '2021', '2021', '2021'],
"var2": ["2099", "2022", "2034", "90999", "20999"],
"var3": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var4": ["1997-07", "1997-07-16", "1997-07-16T19:20:30.45+01:00", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
"var5": ["1997-08", "1997-08-16", "1997-08-16T19:20:30.45+01:00", "1997-08-16T19:20:30+01:00", "1997-08-16T19:20+01:00"],
"var6": ["1998-08", "1998-08-11", "1998-08-17T20:21:31.46+01:00", "1998-08-17T20:21:31+01:00", "1998-08-17T20:21+01:00"],
"var7": ["", None, "", "", ""]
}
)
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var1", "comparator": '2022'})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var1", "comparator": '2020'})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var3", "comparator": "1998-07-16T19:20:30.45+01:00"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var3", "comparator": "var4"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var3", "comparator": "var4", "date_component": "year"})
.equals(pandas.Series([True, True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var6", "comparator": "var3", "date_component": "hour"})
.equals(pandas.Series([True, True, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var3", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).date_less_than_or_equal_to({"target": "var7", "comparator": "var7", "date_component": "month"})
.equals(pandas.Series([False, False, False, False, False])))
def test_is_incomplete_date(self):
df = pandas.DataFrame.from_dict(
{
"var1": [ '2021', '2021', '2099'],
"var2": [ "1997-07-16", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
}
)
self.assertTrue(DataframeType({"value": df}).is_incomplete_date({"target" : "var1"})
.equals(pandas.Series([True, True, True])))
self.assertTrue(DataframeType({"value": df}).is_incomplete_date({"target" : "var2"})
.equals(pandas.Series([False, False, False])))
def test_is_complete_date(self):
df = pandas.DataFrame.from_dict(
{
"var1": ["2021", "2021", "2099"],
"var2": ["1997-07-16", "1997-07-16T19:20:30+01:00", "1997-07-16T19:20+01:00"],
}
)
self.assertTrue(DataframeType({"value": df}).is_complete_date({"target": "var1"})
.equals(pandas.Series([False, False, False])))
self.assertTrue(DataframeType({"value": df}).is_complete_date({"target": "var2"})
.equals(pandas.Series([True, True, True])))
def test_is_unique_set(self):
df = pandas.DataFrame.from_dict( {"ARM": ["PLACEBO", "PLACEBO", "A", "A"], "TAE": [1,1,1,2], "LAE": [1,2,1,2], "ARF": [1,2,3,4]})
self.assertTrue(DataframeType({"value": df}).is_unique_set({"target" : "ARM", "comparator": "LAE"})
.equals(pandas.Series([True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).is_unique_set({"target" : "ARM", "comparator": ["LAE"]})
.equals(pandas.Series([True, True, True, True])))
self.assertTrue(DataframeType({"value": df}).is_unique_set({"target" : "ARM", "comparator": ["TAE"]})
.equals(pandas.Series([False, False, True, True])))
self.assertTrue(DataframeType({"value": df}).is_unique_set({"target" : "ARM", "comparator": "TAE"})
.equals(pandas.Series([False, False, True, True])))
self.assertTrue(DataframeType({"value":df, "column_prefix_map": {"--": "AR"}}).is_unique_set({"target" : "--M", "comparator": "--F"})
.equals(pandas.Series([True, True, True, True])))
self.assertTrue(DataframeType({"value":df, "column_prefix_map": {"--": "AR"}}).is_unique_set({"target" : "--M", "comparator": ["--F"]})
.equals(pandas.Series([True, True, True, True])))
def test_is_not_unique_set(self):
df = pandas.DataFrame.from_dict( {"ARM": ["PLACEBO", "PLACEBO", "A", "A"], "TAE": [1,1,1,2], "LAE": [1,2,1,2], "ARF": [1,2,3,4]})
self.assertTrue(DataframeType({"value": df}).is_not_unique_set({"target" : "ARM", "comparator": "LAE"})
.equals(pandas.Series([False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).is_not_unique_set({"target" : "ARM", "comparator": ["LAE"]})
.equals(pandas.Series([False, False, False, False])))
self.assertTrue(DataframeType({"value": df}).is_not_unique_set({"target" : "ARM", "comparator": ["TAE"]})
.equals(pandas.Series([True, True, False, False])))
self.assertTrue(DataframeType({"value": df}).is_not_unique_set({"target" : "ARM", "comparator": "TAE"})
.equals(pandas.Series([True, True, False, False])))
self.assertTrue(DataframeType({"value":df, "column_prefix_map": {"--": "AR"}}).is_not_unique_set({"target" : "--M", "comparator": "--F"})
.equals(pandas.Series([False, False, False, False])))
self.assertTrue(DataframeType({"value":df, "column_prefix_map": {"--": "AR"}}).is_not_unique_set({"target" : "--M", "comparator": ["--F"]})
.equals(pandas.Series([False, False, False, False])))
def test_is_ordered_set(self):
df = pandas.DataFrame.from_dict( {"USUBJID": [1,2,1,2], "SESEQ": [1,1,2,2] })
self.assertTrue(DataframeType({"value": df}).is_ordered_set({"target" : "SESEQ", "comparator": "USUBJID"}))
self.assertTrue(DataframeType({"value":df, "column_prefix_map": {"--": "SE"}}).is_ordered_set({"target" : "--SEQ", "comparator": "USUBJID"}))
df2 = pandas.DataFrame.from_dict( {"USUBJID": [1,2,1,2], "SESEQ": [3,1,2,2] })
self.assertFalse(DataframeType({"value": df2}).is_ordered_set({"target" : "SESEQ", "comparator": "USUBJID"}))
self.assertFalse(DataframeType({"value":df2, "column_prefix_map": {"--": "SE"}}).is_ordered_set({"target" : "--SEQ", "comparator": "USUBJID"}))
def test_is_not_ordered_set(self):
df = pandas.DataFrame.from_dict( {"USUBJID": [1,2,1,2], "SESEQ": [3,1,2,2] })
self.assertTrue(DataframeType({"value": df}).is_not_ordered_set({"target" : "SESEQ", "comparator": "USUBJID"}))
self.assertTrue(DataframeType({"value":df, "column_prefix_map": {"--": "SE"}}).is_not_ordered_set({"target" : "--SEQ", "comparator": "USUBJID"}))
df2 = pandas.DataFrame.from_dict( {"USUBJID": [1,2,1,2], "SESEQ": [1,1,2,2] })
self.assertFalse(DataframeType({"value": df2}).is_not_ordered_set({"target" : "SESEQ", "comparator": "USUBJID"}))
self.assertFalse(DataframeType({"value":df2, "column_prefix_map": {"--": "SE"}}).is_not_ordered_set({"target" : "--SEQ", "comparator": "USUBJID"}))
def test_is_unique_relationship(self):
"""
Test validates one-to-one relationship against a dataset.
One-to-one means that a pair of columns can be duplicated
but its integrity should not be violated.
"""
one_to_one_related_df = pandas.DataFrame.from_dict(
{
"STUDYID": [1, 2, 3, 1, 2],
"USUBJID": ["TEST", "TEST-1", "TEST-2", "TEST-3", "TEST-4", ],
"STUDYDESC": ["Russia", "USA", "China", "Russia", "USA", ],
}
)
self.assertTrue(
DataframeType({"value": one_to_one_related_df}).is_unique_relationship(
{"target": "STUDYID", "comparator": "STUDYDESC"}
).equals(pandas.Series([True, True, True, True, True]))
)
self.assertTrue(
DataframeType({"value": one_to_one_related_df}).is_unique_relationship(
{"target": "STUDYDESC", "comparator": "STUDYID"}
).equals(pandas.Series([True, True, True, True, True]))
)
self.assertTrue(
DataframeType({"value": one_to_one_related_df, "column_prefix_map":{"--": "STUDY"}}).is_unique_relationship(
{"target": "--ID", "comparator": "--DESC"}
).equals(pandas.Series([True, True, True, True, True]))
)
self.assertTrue(
DataframeType({"value": one_to_one_related_df, "column_prefix_map":{"--": "STUDY"}}).is_unique_relationship(
{"target": "--DESC", "comparator": "--ID"}
).equals(pandas.Series([True, True, True, True, True]))
)
df_violates_one_to_one = pandas.DataFrame.from_dict(
{
"STUDYID": ["TEST", "TEST-1", "TEST-2", "TEST-3", ],
"TESTID": [1, 2, 1, 3],
"TESTNAME": ["Functional", "Stress", "Functional", "Stress", ],
}
)
self.assertTrue(DataframeType({"value": df_violates_one_to_one}).is_unique_relationship(
{"target": "TESTID", "comparator": "TESTNAME"}).equals(pandas.Series([True, False, True, False]))
)
self.assertTrue(DataframeType({"value": df_violates_one_to_one}).is_unique_relationship(
{"target": "TESTNAME", "comparator": "TESTID"}).equals(pandas.Series([True, False, True, False]))
)
def test_is_not_unique_relationship(self):
"""
Test validates one-to-one relationship against a dataset.
One-to-one means that a pair of columns can be duplicated
but its integrity should not be violated.
"""
valid_df = pandas.DataFrame.from_dict(
{
"STUDYID": ["TEST", "TEST-1", "TEST-2", "TEST-3", ],
"VISITNUM": [1, 2, 1, 3],
"VISIT": ["Consulting", "Surgery", "Consulting", "Treatment", ],
}
)
self.assertTrue(DataframeType({"value": valid_df}).is_not_unique_relationship(
{"target": "VISITNUM", "comparator": "VISIT"}).equals(pandas.Series([False, False, False, False]))
)
self.assertTrue(DataframeType({"value": valid_df}).is_not_unique_relationship(
{"target": "VISIT", "comparator": "VISITNUM"}).equals(pandas.Series([False, False, False, False]))
)
valid_df_1 = pandas.DataFrame.from_dict(
{
"STUDYID": ["TEST", "TEST-1", "TEST-2", "TEST-3", ],
"VISIT": ["Consulting", "Surgery", "Consulting", "Treatment", ],
"VISITDESC": [
"Doctor Consultation", "Heart Surgery", "Doctor Consultation", "Long Lasting Treatment",
],
}
)
self.assertTrue(DataframeType({"value": valid_df_1}).is_not_unique_relationship(
{"target": "VISIT", "comparator": "VISITDESC"}).equals(pandas.Series([False, False, False, False]))
)
self.assertTrue(DataframeType({"value": valid_df_1}).is_not_unique_relationship(
{"target": "VISITDESC", "comparator": "VISIT"}).equals(pandas.Series([False, False, False, False]))
)
df_violates_one_to_one = pandas.DataFrame.from_dict(
{
"STUDYID": ["TEST", "TEST-1", "TEST-2", "TEST-3", ],
"VISITNUM": [1, 2, 1, 3],
"VISIT": ["Consulting", "Surgery", "Consulting", "Consulting", ],
}
)
self.assertTrue(DataframeType({"value": df_violates_one_to_one}).is_not_unique_relationship(
{"target": "VISITNUM", "comparator": "VISIT"}).equals(pandas.Series([True, False, True, True]))
)
self.assertTrue(DataframeType({"value": df_violates_one_to_one}).is_not_unique_relationship(
{"target": "VISIT", "comparator": "VISITNUM"}).equals(pandas.Series([True, False, True, True]))
)
df_violates_one_to_one_1 = pandas.DataFrame.from_dict(
{
"STUDYID": ["TEST", "TEST-1", "TEST-2", "TEST-3", "TEST-4", ],
"VISIT": ["Consulting", "Consulting", "Surgery", "Consulting", "Treatment", ],
"VISITDESC": ["Doctor Consultation", "Doctor Consultation", "Heart Surgery", "Heart Surgery", "Long Lasting Treatment", ],
}
)
self.assertTrue(DataframeType({"value": df_violates_one_to_one_1}).is_not_unique_relationship(
{"target": "VISIT", "comparator": "VISITDESC"}).equals(
|
pandas.Series([True, True, True, True, False])
|
pandas.Series
|
# -*- coding: utf-8 -*-
"""
Created on Mon May 29 13:56:29 2017
@author: ning
"""
import pandas as pd
import os
import numpy as np
#from sklearn.model_selection import StratifiedKFold,KFold
#from sklearn.pipeline import Pipeline
#from sklearn.preprocessing import StandardScaler
#from sklearn.linear_model import LogisticRegressionCV
#from sklearn.metrics import roc_curve,precision_recall_curve,auc,average_precision_score,confusion_matrix
import matplotlib.pyplot as plt
import matplotlib
matplotlib.rcParams.update({'font.size':22})
matplotlib.rcParams['legend.numpoints'] = 1
import seaborn as sns
sns.set_style('white')
try:
function_dir = 'D:\\NING - spindle\\Spindle_by_Graphical_Features'
os.chdir(function_dir)
except:
function_dir = 'C:\\Users\\ning\\OneDrive\\python works\\Spindle_by_Graphical_Features'
os.chdir(function_dir)
#import eegPipelineFunctions
try:
file_dir = 'D:\\NING - spindle\\training set\\road_trip\\'
# file_dir = 'D:\\NING - spindle\\training set\\road_trip_more_channels\\'
os.chdir(file_dir)
except:
file_dir = 'C:\\Users\\ning\\Downloads\\road_trip\\'
# file_dir = 'C:\\Users\\ning\\Downloads\\road_trip_more_channels\\'
os.chdir(file_dir)
def average(x):
x = x[1:-1].split(', ')
x = np.array(x,dtype=float)
return np.nanmean(x)
figsize = 6
signal_features_indivisual_results_RF=
|
pd.read_csv(file_dir+'individual_signal_feature_RF.csv')
|
pandas.read_csv
|
import numpy as np
import pandas as pd
import datetime as dt
import pickle
import bz2
from .analyzer import summarize_returns
DATA_PATH = '../backtest/'
class Portfolio():
"""
Portfolio is the core class for event-driven backtesting. It conducts the
backtesting in the following order:
1. Initialization:
Set the capital base we invest and the securities we
want to trade.
2. Receive the price information with .receive_price():
Insert the new price information of each securities so that the
Portfolio class will calculated and updated the relevant status such
as the portfolio value and position weights.
3. Rebalance with .rebalance():
Depending on the signal, we can choose to change the position
on each securities.
4. Keep position with .keep_position():
If we don't rebalance the portfolio, we need to tell it to keep
current position at the end of the market.
Example
-------
see Vol_MA.ipynb, Vol_MA_test_robustness.ipynb
Parameters
----------
capital: numeric
capital base we put into the porfolio
inception: datetime.datetime
the time when we start backtesting
components: list of str
tikers of securities to trade, such as ['AAPL', 'MSFT', 'AMZN]
name: str
name of the portfolio
is_share_integer: boolean
If true, the shares of securities will be rounded to integers.
"""
def __init__(self, capital, inception, components,
name='portfolio', is_share_integer=False):
# -----------------------------------------------
# initialize parameters
# -----------------------------------------------
self.capital = capital # initial money invested
if isinstance(components, str):
components = [components] # should be list
self.components = components # equities in the portfolio
# self.commission_rate = commission_rate
self.inception = inception
self.component_prices = pd.DataFrame(columns=self.components)
self.name = name
self.is_share_integer = is_share_integer
# self.benchmark = benchmark
# -----------------------------------------------
# record portfolio status to series and dataFrames
# -----------------------------------------------
# temoprary values
self._nav = pd.Series(capital,index=[inception])
self._cash = pd.Series(capital,index=[inception])
self._security = pd.Series(0,index=[inception])
self._component_prices = pd.DataFrame(columns=self.components) # empty
self._shares = pd.DataFrame(0, index=[inception], columns=self.components)
self._positions = pd.DataFrame(0, index=[inception], columns=self.components)
self._weights = pd.DataFrame(0, index=[inception], columns=self.components)
self._share_changes = pd.DataFrame(columns=self.components) # empty
self._now = self.inception
self._max_nav = pd.Series(capital,index=[inception])
self._drawdown = pd.Series(0, index=[inception])
self._relative_drawdown = pd.Series(0, index=[inception])
# series
self.nav_open = pd.Series()
self.nav_close = pd.Series()
self.cash_open = pd.Series()
self.cash_close = pd.Series()
self.security_open = pd.Series()
self.security_close = pd.Series()
self.max_nav = pd.Series()
self.drawdown_open = pd.Series()
self.drawdown_close = pd.Series()
self.relative_drawdown_open = pd.Series()
self.relative_drawdown_close = pd.Series()
# dataframes
self.shares_open =
|
pd.DataFrame(columns=self.components)
|
pandas.DataFrame
|
"""App run class."""
from functools import partial
from tkinter import *
from tkinter import filedialog
import os
import tkinter as tk
import tkinter.messagebox
import requests
import json
from requests.auth import HTTPBasicAuth
import numpy as np
import pandas as pd
def auth_api(username, password):
"""Auth function to validate API."""
res = requests.get('https://api.quickbutik.com/v1/products', auth=HTTPBasicAuth(username.get(), password.get()))
print(res.status_code)
if res.status_code == 200:
tkinter.messagebox.showinfo("Welcome to quickbutik.", "Login successfully.")
new_frame.pack_forget()
path_frame.pack()
else:
tkinter.messagebox.showinfo("Login failed.", "Wrong user name or password.")
def check_path(json_path):
"""Check path."""
if os.path.exists(json_path):
tkinter.messagebox.showinfo("File name error.", "File already existed.")
return True
else:
return False
def myconverter(obj):
"""Define customized json.dump function."""
if isinstance(obj, np.integer):
return int(obj)
elif isinstance(obj, np.floating):
return float(obj)
elif isinstance(obj, np.ndarray):
return obj.tolist()
def csv_to_json(csvFile, json_path):
"""Get csv file and export to json."""
jsonArray = []
with open(csvFile, 'r', encoding='latin-1') as csvf:
data = pd.read_csv(csvf)
sku = data.sku
title = data.title
description = data.description
price = data.price
before_price = data.before_price
tax_rate = data.tax_rate
weight = data.weight
stock = data.stock
gtin = data.gtin
images = data.images
headcategory_name = data.headcategory_name
visible = data.visible
x = 0
while x < len(sku):
dict_list = {}
# print(images[x])
image_list = []
image_dic = {}
image_dic["url"] = images[x]
image_list.append(image_dic)
dict_list["sku"] = sku[x]
dict_list["title"] = title[x]
dict_list["description"] = description[x]
dict_list["price"] = int(price[x])
# check if before price has any empty values
before_price_data = pd.DataFrame(before_price)
if before_price_data["before_price"].isnull().values.any():
dict_list["before_price"] = " "
else:
dict_list["before_price"] = before_price[x]
# check if tax rate has nan
tax_rate_data =
|
pd.DataFrame(tax_rate)
|
pandas.DataFrame
|
#!/usr/bin/env python
import numpy as np
import pandas as pd
def _canonicalize(dtype):
try:
# put Series first so that, eg, 'Int8' properly gets mapped to the
# nullable type rather than the numpy non-nullable 'int8' type
s = pd.Series(data=[], dtype=dtype)
return s.dtype
except TypeError:
pass
try:
# don't think this should handle anything a Series can't, but leave
# this here to make sure
a = np.empty(1, dtype=dtype)
return a.dtype
except TypeError:
pass
raise ValueError(f"Dtype '{dtype}' ({type(dtype)}) invalid for both "
"Numpy arrays and Pandas series.")
# _FLOAT_TYPES = [np.float16, np.float32, np.float64]
_c = _canonicalize # shorten below definitions
_FLOAT_TYPES = [_c(np.float16), _c(np.float32), _c(np.float64)]
# _UNSIGNED_INT_TYPES = [np.uint8, np.uint16, np.uint32, np.uint64]
# _SIGNED_INT_TYPES = [np.int8, np.int16, np.int32, np.int64]
# _UNSIGNED_INT_TYPES = [np.uint8(), np.uint16(), np.uint32(), np.uint64()]
# _SIGNED_INT_TYPES = [np.int8(), np.int16(), np.int32(), np.int64()]
_UNSIGNED_INT_TYPES = [_c(np.uint8), _c(np.uint16),
_c(np.uint32), _c(np.uint64)]
_SIGNED_INT_TYPES = [_c(np.int8), _c(np.int16), _c(np.int32), _c(np.int64)]
_NONNULLABLE_INT_TYPES = _UNSIGNED_INT_TYPES + _SIGNED_INT_TYPES
# _NULLABLE_SIGNED_INT_TYPES = [
# pd.Int8Dtype(), pd.Int16Dtype(), pd.Int32Dtype(), pd.Int64Dtype()]
# _NULLABLE_UNSIGNED_INT_TYPES = [
# pd.UInt8Dtype(), pd.UInt16Dtype(), pd.UInt32Dtype(), pd.UInt64Dtype()]
# _NULLABLE_SIGNED_INT_TYPES = [_c(pd.Int8Dtype), _c(pd.Int16Dtype),
# _c(pd.Int32Dtype), _c(pd.Int64Dtype)]
# _NULLABLE_UNSIGNED_INT_TYPES = [_c(pd.UInt8Dtype), _c(pd.UInt16Dtype),
# _c(pd.UInt32Dtype), _c(pd.UInt64Dtype)]
# have to use string dtype codes, rather than, eg, pd.Int8Dtype or
# pd.Int8Dtype() or resulting dtype is np.object; because pandas dtypes are
# insane
_NULLABLE_SIGNED_INT_TYPES = [
_c('Int8'), _c('Int16'), _c('Int32'), _c('Int64')]
_NULLABLE_UNSIGNED_INT_TYPES = [
_c('UInt8'), _c('UInt16'), _c('UInt32'), _c('UInt64')]
_NULLABLE_INT_TYPES = _NULLABLE_UNSIGNED_INT_TYPES + _NULLABLE_SIGNED_INT_TYPES
_NULLABLE_TO_NONNULLABLE_INT_DTYPE = dict(zip(
_NULLABLE_INT_TYPES, _NONNULLABLE_INT_TYPES))
_NONNULLABLE_TO_NULLABLE_INT_DTYPE = dict(zip(
_NONNULLABLE_INT_TYPES, _NULLABLE_INT_TYPES))
# _NUMERIC_DTYPES = _NONNULLABLE_INT_TYPES + _NULLABLE_INT_TYPES + _FLOAT_TYPES
# NOTE: np.bool is alias of python bool, while np.bool_ is custom a numpy type
# _BOOLEAN_DTYPES = [np.dtype(np.bool), np.dtype(np.bool_), pd.BooleanDtype()]
_BOOLEAN_DTYPES = [_c(np.bool), _c(np.bool_), _c('boolean')]
_SIGNED_EQUIVALENT = {
_c(np.uint8): _c(np.int8), _c(np.int8): _c(np.int8), # noqa
_c(np.uint16): _c(np.int16), _c(np.int16): _c(np.int16),
_c(np.uint32): _c(np.int32), _c(np.int32): _c(np.int32),
_c(np.uint64): _c(np.int64), _c(np.int64): _c(np.int64),
_c(pd.UInt8Dtype): _c(pd.Int8Dtype), # noqa
_c(pd.Int8Dtype): _c(pd.Int8Dtype), # noqa
_c(pd.UInt16Dtype): _c(pd.Int16Dtype), # noqa
_c(pd.Int16Dtype): _c(pd.Int16Dtype), # noqa
_c(pd.UInt32Dtype): _c(pd.Int32Dtype), # noqa
_c(pd.Int32Dtype): _c(pd.Int32Dtype), # noqa
_c(pd.UInt64Dtype): _c(pd.Int64Dtype), # noqa
_c(pd.Int64Dtype): _c(pd.Int64Dtype)} # noqa
_UNSIGNED_EQUIVALENT = {
_c(np.uint8): _c(np.uint8), _c(np.int8): _c(np.uint8), # noqa
_c(np.uint16): _c(np.uint16), _c(np.int16): _c(np.uint16),
_c(np.uint32): _c(np.uint32), _c(np.int32): _c(np.uint32),
_c(np.uint64): _c(np.uint64), _c(np.int64): _c(np.uint64),
_c(pd.UInt8Dtype): _c(pd.UInt8Dtype), # noqa
_c(pd.Int8Dtype): _c(pd.UInt8Dtype), # noqa
_c(pd.UInt16Dtype): _c(pd.UInt16Dtype), # noqa
_c(pd.Int16Dtype): _c(pd.UInt16Dtype), # noqa
_c(pd.UInt32Dtype): _c(pd.UInt32Dtype), # noqa
_c(pd.Int32Dtype): _c(pd.UInt32Dtype), # noqa
_c(pd.UInt64Dtype): _c(pd.UInt64Dtype), # noqa
_c(pd.Int64Dtype): _c(pd.UInt64Dtype)} # noqa
# _SIGNED_EQUIVALENT = {
# np.uint8(): np.int8(), np.int8(): np.int8(), # noqa
# np.uint16(): np.int16(), np.int16(): np.int16(),
# np.uint32(): np.int32(), np.int32(): np.int32(),
# np.uint64(): np.int64(), np.int64(): np.int64(),
# pd.UInt8Dtype(): pd.Int8Dtype(), pd.Int8Dtype(): pd.Int8Dtype(), # noqa
# pd.UInt16Dtype(): pd.Int16Dtype(), pd.Int16Dtype(): pd.Int16Dtype(),
# pd.UInt32Dtype(): pd.Int32Dtype(), pd.Int32Dtype(): pd.Int32Dtype(),
# pd.UInt64Dtype(): pd.Int64Dtype(), pd.Int64Dtype(): pd.Int64Dtype()}
# _UNSIGNED_EQUIVALENT = {
# np.uint8(): np.uint8(), np.int8(): np.uint8(), # noqa
# np.uint16(): np.uint16(), np.int16(): np.uint16(),
# np.uint32(): np.uint32(), np.int32(): np.uint32(),
# np.uint64(): np.uint64(), np.int64(): np.uint64(),
# pd.UInt8Dtype(): pd.UInt8Dtype(), pd.Int8Dtype(): pd.UInt8Dtype(),# noqa
# pd.UInt16Dtype(): pd.UInt16Dtype(), pd.Int16Dtype(): pd.UInt16Dtype(),
# pd.UInt32Dtype(): pd.UInt32Dtype(), pd.Int32Dtype(): pd.UInt32Dtype(),
# pd.UInt64Dtype(): pd.UInt64Dtype(), pd.Int64Dtype(): pd.UInt64Dtype()}
# _PANDAS_NULLABLE_DTYPES_INSTANTIATED = [t() for t in _NULLABLE_INT_TYPES] +
# _SUPPORTED_PANDAS_DTYPES = _NULLABLE_INT_TYPES + [pd.BooleanDtype()]
def nullable_equivalent(dtype):
if is_nullable(dtype):
return dtype
# TODO support nullable strings and other pandas dtypes
# if dtype in _FLOAT_TYPES:
if is_boolean(dtype):
return _c('boolean')
if is_float(dtype):
return _c(dtype)
dtype = _canonicalize(dtype)
# if dtype in _NULLABLE_INT_TYPES:
# return dtype
return _NULLABLE_TO_NONNULLABLE_INT_DTYPE[dtype]
def nonnullable_equivalent(dtype):
if not is_nullable(dtype):
return dtype
if is_boolean(dtype):
return _c(np.bool_)
if is_float(dtype):
return _c(dtype)
dtype = _canonicalize(dtype)
# if dtype in _NONNULLABLE_INT_TYPES:
# return dtype
# print("dtype: ", dtype, type(dtype))
# print("nullable int dtypes: ")
# for t in _NULLABLE_INT_TYPES:
# print(t, type(t))
# print("dtype in nullable ints: ", dtype in _NULLABLE_INT_TYPES)
return _NULLABLE_TO_NONNULLABLE_INT_DTYPE[dtype]
def signed_equivalent(dtype):
dtype = _canonicalize(dtype)
return _SIGNED_EQUIVALENT[dtype]
def unsigned_equivalent(dtype):
dtype = _canonicalize(dtype)
return _UNSIGNED_EQUIVALENT[dtype]
def is_complex(dtype):
return pd.api.types.is_complex_dtype(dtype)
def is_float(dtype):
return pd.api.types.is_float_dtype(dtype)
# return _canonicalize(dtype) in _FLOAT_TYPES
def is_numeric(dtype):
# print("is_numeric: checking dtype: ", dtype)
if is_boolean(dtype):
return False
# dtype = _canonicalize(dtype)
# if _canonicalize(dtype) in _BOOLEAN_DTYPES:
# _ = _BOOLEAN_DTYPES
# for btype in _BOOLEAN_DTYPES:
# print("dtype, btype: ", dtype, btype)
# if btype == dtype:
# return False
# if dtype in _BOOLEAN_DTYPES:
# # exclude bools since they mess up quantization and just generally
# # don't act like numbers
# return False
return
|
pd.api.types.is_numeric_dtype(dtype)
|
pandas.api.types.is_numeric_dtype
|
# Import Libraries to be used in this code module
import pandas_datareader.data as dr # pandas library for data manipulation and analysis
import pandas as pd
from datetime import datetime, timedelta, date # library for date and time calculations
import sqlalchemy as sal # SQL toolkit, Object-Relational Mapper for Python
from pandas.tseries.holiday import get_calendar, HolidayCalendarFactory, GoodFriday # calendar module to use a pre-configured calendar
import quandl
from fredapi import Fred
fred = Fred(api_key='<KEY>')
# Declaration and Definition of DataFetch class
class DataFetch:
def __init__(self, engine, table_name):
"""
Get raw data for each ticker symbol
:param engine: provides connection to MySQL Server
:param table_name: table name where ticker symbols are stored
"""
self.engine = engine
self.table_name = table_name
self.datasource = 'yahoo'
self.datalength = 2192 # last 6 years, based on actual calendar days of 365
def get_datasources(self):
"""
Method to query MySQL database for ticker symbols
Use pandas read_sql_query function to pass query
:return symbols: pandas data frame object containing the ticker symbols
"""
query = 'SELECT * FROM %s' % self.table_name
symbols = pd.read_sql_query(query, self.engine)
return symbols
def get_data(self, sources):
"""
Get raw data from Yahoo! Finance for each ticker symbol
Store data in MySQL database
:param sources: provides ticker symbols of instruments being tracked
"""
now = datetime.now() # Date Variables
start = datetime.now()-timedelta(days=self.datalength) # get date value from 3 years ago
end = now.strftime("%Y-%m-%d")
# Cycle through each ticker symbol
for n in range(len(sources)):
# data will be a 2D Pandas Dataframe
data = dr.DataReader(sources.iat[n, sources.columns.get_loc('instrumentname')], self.datasource, start, end)
symbol = [sources['instrumentid'][n]] * len(data) # add column to identify instrument id number
data['instrumentid'] = symbol
data = data.reset_index() # no designated index - easier to work with mysql database
# Yahoo! Finance columns to match column names in MySQL database.
# Column names are kept same to avoid any ambiguity.
# Column names are not case-sensitive.
data.rename(columns={'Date': 'date', 'High': 'high', 'Low': 'low', 'Open': 'open', 'Close': 'close',
'Adj Close': 'adj close', 'Volume': 'volume'}, inplace=True)
data.sort_values(by=['date']) # make sure data is ordered by trade date
# send data to database
# replace data each time program is run
data.to_sql('dbo_instrumentstatistics', self.engine, if_exists=('replace' if n == 0 else 'append'),
index=False, dtype={'date': sal.Date, 'open': sal.FLOAT, 'high': sal.FLOAT, 'low': sal.FLOAT,
'close': sal.FLOAT, 'adj close': sal.FLOAT, 'volume': sal.FLOAT})
def get_calendar(self):
"""
Get date data to track weekends, holidays, quarter, etc
Store in database table dbo_DateDim
"""
# drop data from table each time program is run
truncate_query = 'TRUNCATE TABLE dbo_datedim'
self.engine.execute(truncate_query)
# 3 years of past data and up to 1 year of future forecasts
begin = date.today() - timedelta(days=self.datalength)
end = date.today() + timedelta(days=365)
# list of US holidays
cal = get_calendar('USFederalHolidayCalendar') # Create calendar instance
cal.rules.pop(7) # Remove Veteran's Day
cal.rules.pop(6) # Remove Columbus Day
tradingCal =
|
HolidayCalendarFactory('TradingCalendar', cal, GoodFriday)
|
pandas.tseries.holiday.HolidayCalendarFactory
|
from sklearn.neighbors import KNeighborsClassifier
import pandas as pd
import numpy as np
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split, GridSearchCV
# 网格搜索 + 最近邻分类器
def main():
# 导入数据集
data = pd.read_csv("D://A//data//FBlocation//train.csv")
# 缩小给定数据集,缩小x/y范围,多条件查询
data = data.query("x > 1.0 & x < 1.25 & y > 2.5 & y < 2.75")
# 格式化日期字段
date_value =
|
pd.to_datetime(data['time'], unit='s')
|
pandas.to_datetime
|
# -*- coding: utf-8 -*-
"""
Created on Tue Oct 3 13:44:55 2017
@author: rgryan
#=====================================================================================================================
# This code takes a folder, with subfolders containing .std spectra (outputted from DOASIS), and converts them all
# to line-format files. Each line in the file is one spectrum.
# The line formatting is appropriate for reading in by QDOAS.
# This code has been updated so it handle calibration and direct sun spectra
# It has now been updated SO THAT IT CORRECTLY SUBTRACTS THE OFFSET AND DARK CURRENT SPECTRA.
#=====================================================================================================================
# Updated 03-10-2017,
# For the Broady MAX-DOAS intercomparison campaign
# RGRyan
#=====================================================================================================================
# data save in the following format:
# st_ddmmyyyy_Uxxxxxxx
# where st = spectrum type (sc = scattered light, ds = direct sun, dc = dark current cal, oc = offset cal)
# ddmmyyyy = date
# U (or V) indicates UV (or Visible) spectrum
# xxxxxxx is the 7 digit folder number from DOASIS
# (this is needed for the iteration thru folders, rather than strictly needed for naming purposes)
#=====================================================================================================================
# What needs to be varied?
# 1. the folderpath and folderdate, specific to the main folder you're looking in
# 2. The foldernumber (this needs to be the number of the first subfolder you want the program to go to)
# 3. The lastfolder number (this tells the program when to stop looking and converting)
# 4. The folder letter. Once all the "U"s are converted, then you have to change this in order to convert all
# the "V"s
# 5. Whether you want to do the offset and dark current correction
"""
# Section of things to check or change
#=====================================================================================================================
folderpath = 'C:/Users/rgryan/Google Drive/Documents/PhD/Data/Broady_data_backup/UWollongong/SP2017a/SP1703/'
foldernumber = 0 # The initial subfolder number, where the program will start
lastfolder = 100 # The final subfolder number, after which the program will stop converting
folders0indexed = True # folder numbers starting with zero?
folderletter = 'U' # 'U' for UV spectra, 'V' for visible spectra
correct_dc = True # 'False' turns off the dark current correction
correct_os = True # 'False' turns off the offset correction
only_save_hs_int = True # True if wanting to save an abridged version of the horizon scan data, with only
# intensity values, not all the spectral data
calcCI = True # Calculate colour index?
CIn = 330 # Numerator for color index
CId = 390 # denominator for color index
saveSC = True # save scattered light spectra?
saveDS = False # save direct sun spectra?
saveHS = True # save horizon scan spectra?
saveDC = True # save dark current calibrations?
saveOS = True # save offset calibrations?
saveCI = True # save colour index results?
inst = 'UW' # The data from which institution is being plotted? <just for saving purposes!>
# UM = Uni. of Melb, UW = Wollongong Uni, NZ = NIWA New Zealand,
# BM = Bureau of Meteorology Broadmeadows
# Date format
date_format_1 = True # For date format in UniMelb MS-DOAS STD spectra, MM/DD/YYYY
date_format_2 = False # For date format in UW'gong MS-DOAS STD spectra, DD-Mon-YYYY
# settings for saving
end = ".txt"
path2save = folderpath[3:]+folderletter+'\\'
# Import section
#=====================================================================================================================
import numpy as np
import glob
import pandas as pd
# Section to deal with dark and offset calibration spectra
#=====================================================================================================================
if inst == 'UM':
UVoc__ = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UM_calfiles\\UM_UV_offset.std'
visoc__ = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UM_calfiles\\UM_vis_offset.std'
UVdc__= 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UM_calfiles\\UM_UV_darkcurrent.std'
visdc__ = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UM_calfiles\\UM_vis_darkcurrent.std'
Uwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UM_calfiles\\UM_UVcal.txt'
Vwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UM_calfiles\\UM_viscal.txt'
elif inst == 'BM':
UVoc__ = 'E:/PhD/Broady_data_backup/TIMTAM_ref_files/BM_calfiles/ofsuv_U0000003.std'
visoc__ = 'E:/PhD/Broady_data_backup/TIMTAM_ref_files/BM_calfiles/ofsvis_V0000003.std'
visdc__ = 'E:/PhD/Broady_data_backup/TIMTAM_ref_files/BM_calfiles/dcvis_V0000005.std'
UVdc__ = 'E:/PhD/Broady_data_backup/TIMTAM_ref_files/BM_calfiles/dcuv_U0000005.std'
Uwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\BM_calfiles\\BM_UVcal.txt'
Vwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\BM_calfiles\\BM_viscal.txt'
elif inst == 'UW':
UVoc__ = 'E:/PhD/Broady_data_backup/UWollongong/Cals/offset_U_UW.std'
visoc__ = 'E:/PhD/Broady_data_backup/UWollongong/Cals/offset_V_UW.std'
visdc__ = 'E:/PhD/Broady_data_backup/UWollongong/Cals/dc_V_UW.std'
UVdc__ = 'E:/PhD/Broady_data_backup/UWollongong/Cals/dc_U_UW.std'
Uwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UW_calfiles\\UW_UVcal.txt'
Vwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\UW_calfiles\\UW_viscal.txt'
elif inst == 'NZ':
UVoc__ = 'E:/PhD/Broady_data_backup/NIWA/NIWA cal files/OFS_U0060764.std'
UVdc__ = 'E:/PhD/Broady_data_backup/NIWA/NIWA cal files/DC_U0060763.std'
visoc__ = 'C:\\Users\\rgryan\\Google Drive\\Documents\\PhD\\Data\\Broady_data_backup\\NIWA\\spectra\\NZ_STD_Spectra_V\\OFS_V0060764.std'
visdc__ = 'C:\\Users\\rgryan\\Google Drive\\Documents\\PhD\\Data\\Broady_data_backup\\NIWA\\spectra\\NZ_STD_Spectra_V\\DC_V0060763.std'
Uwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\NZ_calfiles\\NZ_UVcal.txt'
Vwlcal = 'C:\\Users\\rgryan\\Google Drive\\TIMTAM\\NZ_calfiles\\NZ_viscal.txt'
else:
print('Error - Offset or DC cal files not defined')
# Read in Offset calibration for UV and Vis
# ==========================================
UVoc_path = open(UVoc__, 'r')
UVoc_data = UVoc_path.readlines()
UVoc_data_strpd = [(UVoc_data[i].strip('\n')) for i in range(len(UVoc_data))]
visoc_path = open(visoc__, 'r')
visoc_data = visoc_path.readlines()
visoc_data_strpd = [(visoc_data[i].strip('\n')) for i in range(len(visoc_data))]
# Find the data in the offset calibration spectrum
# ==========================================
if folderletter == 'U':
ocCal_ = UVoc_data_strpd[3:2051]
elif folderletter == 'V':
ocCal_ = visoc_data_strpd[3:2051]
ocCal = [float(i) for i in ocCal_]
# Dark current calibration readin
# ==========================================
UVdc_path = open(UVdc__, 'r')
UVdc_data = UVdc_path.readlines()
UVdc_data_strpd = [(UVdc_data[i].strip('\n')) for i in range(len(UVdc_data))]
visdc_path = open(visdc__, 'r')
visdc_data = visdc_path.readlines()
visdc_data_strpd = [(visdc_data[i].strip('\n')) for i in range(len(visdc_data))]
if folderletter == 'U':
dcCal_ = UVdc_data_strpd[3:2051]
elif folderletter == 'V':
dcCal_ = visdc_data_strpd[3:2051]
dcCal = [float(i) for i in dcCal_]
# Find the number of scans and the exposure time for the calibration spectra
#===================================================================
oc_numscans_ = UVoc_data_strpd[2059]
oc_numscansX = oc_numscans_.split()
oc_numscans = float(oc_numscansX[1])
oc_texp_ = UVoc_data_strpd[2072] # time in ms
oc_texpX = oc_texp_.split()
oc_texp = float(oc_texpX[2])
oc_inttime = oc_texp*oc_numscans
dc_numscans_ = UVdc_data_strpd[2059]
dc_numscansX = dc_numscans_.split()
dc_numscans = float(dc_numscansX[1])
dc_texp_ = UVdc_data_strpd[2072] # time in ms
dc_texpX = dc_texp_.split()
dc_texp = float(dc_texpX[2])
dc_inttime = dc_numscans*dc_texp
#===================================================================
# Calibration spectra process
# 1. Offset spectrum is proportional to number of scans. Therefore need to divide by number of scans
if correct_os == True:
ocCal_c1 = [(ocCal[i]/oc_numscans) for i in range(len(ocCal))] # This has units of counts/scan
else:
ocCal_c1 = [(0) for i in range(len(ocCal))]
# 2. Correct dark-current spectrum for the offset
if correct_dc == True:
dcCal_c1 = [(dcCal[i] - ((ocCal_c1[i])*dc_numscans)) for i in range(len(dcCal))] # this has units of counts
dcCal_c = [((dcCal_c1[i]/dc_inttime)) for i in range(len(dcCal))] # this has units of counts/ms
else:
dcCal_c = [(0) for i in range(len(dcCal))]
# 3. Correct offset spectrum using corrected dark current spectrum
if correct_os == True:
ocCal_c2 = [(ocCal[i] - (dcCal_c[i]*oc_inttime)) for i in range(len(ocCal))] # this has units of counts
ocCal_c = [(ocCal_c2[i]/oc_numscans) for i in range(len(ocCal_c2))] # this has units of counts/scan
else:
ocCal_c = [(0) for i in range(len(ocCal))]
# corrected dark current passed to the next stage in units of counts/ms
# corrected offeset spectrum passed to the next stage in units of counts/scan
# Create wavelength cal dataframe so we only need to do this once, only to be
# used if Colour Index calculation is performed
if folderletter == 'U':
w = open(Uwlcal, 'r')
else:
w = open(Vwlcal, 'r')
wl_data = w.readlines()
wl_data_strpd = []
for i in range(len(wl_data)):
wl_data_strpd.append(wl_data[i].strip('\n'))
#%%
lastfolderplus1 = lastfolder+1
while foldernumber < lastfolderplus1:
# Empty lists and data frames to write to;
sc_list = [] # for scattered light measurements
ds_list = [] # for direct sun measurements
dc_list = [] # for dark current cakibration measurements
oc_list = [] # for offset calibration measurements
hs_list = [] # for horizon scan measurements
ci_list = []
sc_frame_to_fill = pd.DataFrame()
ds_frame_to_fill = pd.DataFrame()
oc_frame_to_fill = pd.DataFrame()
dc_frame_to_fill = pd.DataFrame()
hs_frame_to_fill = pd.DataFrame()
if folders0indexed:
if len(str(foldernumber)) < 2:
foldername = 'STD_'+folderletter+'000000'+str(foldernumber)
elif len(str(foldernumber)) < 3:
foldername = 'STD_'+folderletter+'00000'+str(foldernumber)
elif len(str(foldernumber)) < 4:
foldername = 'STD_'+folderletter+'0000'+str(foldernumber)
elif len(str(foldernumber)) < 5:
foldername = 'STD_'+folderletter+'000'+str(foldernumber)
elif len(str(foldernumber)) < 6:
foldername = 'STD_'+folderletter+'00'+str(foldernumber)
elif len(str(foldernumber)) < 7:
foldername = 'STD_'+folderletter+'0'+str(foldernumber)
else:
foldername = 'STD_'+folderletter+str(foldernumber)
#total_path = folderpath+folderdate+foldername
total_path = folderpath+foldername
allFiles = glob.glob(total_path + "/*.std")
print("Now converting: ", folderletter,foldernumber)
for file_ in allFiles:
f = open(file_, 'r')
file_data = f.readlines()
file_data_strpd = []
for i in range(len(file_data)):
file_data_strpd.append(file_data[i].strip('\n'))
# This section deals with the time and date
#===================================================================
hhmmss = file_data_strpd[2056] # This is the measurement start time
[hours, mins, secs] = [int(x) for x in hhmmss.split(':')]
dec_time_ = float(hours+(mins/60)+(secs/3600)) # This is now decimal time, appropriate for QDOAS
dec_time = round(dec_time_, 5)
# Find and convert the date
#===================================================================
if date_format_1 == True:
date_MDY = file_data_strpd[2054]
[month, day, year] = [int(info) for info in date_MDY.split("/")]
day_str = str(day)
month_str = str(month)
year_str = str(int(year))
if day<10:
day_str = "0"+day_str
if month<10:
month_str = "0"+month_str
else:
date_MDY = file_data_strpd[2054]
[day, month, year] = [info for info in date_MDY.split("-")]
day_str = str(int(day))
year_str = str(int(year))
if month == 'Jan':
month_str = '01'
elif month == 'Feb':
month_str = '02'
elif month == 'Mar':
month_str = '03'
elif month == 'Apr':
month_str = '04'
elif month == 'May':
month_str = '05'
elif month == 'Jun':
month_str = '06'
elif month == 'Jul':
month_str = '07'
elif month == 'Aug':
month_str = '08'
elif month == 'Sep':
month_str = '09'
elif month == 'Oct':
month_str = '10'
elif month == 'Nov':
month_str = '11'
else:
month_str = '12'
date_DMY = day_str +"/"+ month_str+"/" + year_str
# This section finds the data
#===================================================================
md_data = file_data_strpd[3:2051]
md_data_int_ = [int(i) for i in md_data]
# Find the number of scans and the exp time
#===================================================================
numscans_ = file_data_strpd[2059]
numscansX = numscans_.split()
numscans = float(numscansX[1])
texp_ = file_data_strpd[2072] # time in ms
texpX = texp_.split()
texp = float(texpX[2])
inttime = numscans*texp # int time in ms
inttime_sec = inttime/1000
# Calculate the Dark current spectrum to subtract;
dc_ts = [(dcCal_c[i]*(inttime)) for i in range(len(md_data))] # dcCal_c comes in as counts/ms, so *by measurement
# exposure time (ms)
# Calculate the offset spectrum to subract;
oc_ts = [(ocCal_c[i]*(numscans)) for i in range(len(md_data_int_))] # ocCal_c comes in as counts/scan, so *by
# measurement number of scans
# Total calibration spectrum to subtract
cal_ts = [(oc_ts[i]+dc_ts[i]) for i in range(len(md_data_int_))] # cal_ts now has units of counts
# The calibration-corrected data list to pass to the next stage:
md_data_int = [(md_data_int_[i] - cal_ts[i]) for i in range(len(md_data_int_))]
# Now we need to differentiate between DS, calibration and scattered light spectra
# First, define the names for the different options;
#=================================================================================
ds = False # Direct sun
oc = False # offset calibration spectrum
dc = False # dark current calibration spectrum
sc = False # scattered light (MAX) measurement
hs = False # horizon scan measurements
other_calib = False # To handle other (Hg lamp) calibrations which are run but don't actually work!
angle_data = file_data_strpd[2051]
#print(angle_data)
if angle_data[0:2] == 'DS':
ds = True
elif angle_data[:] == 'ofs':
oc = True
elif angle_data[:] == 'dc':
dc = True
elif angle_data[0] == 'h':
other_calib = True
elif inttime_sec < 5:
hs = True
else:
sc = True
#else:
# hs=True
#=================================================================================
# Direct sun spectrum case;
#=================================================================================
if ds:
#find the elevation angle;
[other1, EA_real_, other2] = [angle for angle in angle_data[3:].split(" ")]
#[EAactual, azim] = [float(angle) for angle in angle_data[3:].split(" ")]
EA_real = round(float(EA_real_), 2)
# find the SZA;
scan_geom = file_data_strpd[2099]
scan_geom_split = scan_geom.split(" ")
SZA_ = float(scan_geom_split[5])
SZA = round(SZA_, 2)
# For direct sun measurements, the azimuth angle is the solar azimuth angle, which is also given
# in the scan geometry section used to find the SZA;
AzA_ = float(scan_geom_split[3])
AzA = round(AzA_, 2)
# This section finds the calibration-corrected data
#===================================================================
ds_data_flt = md_data_int
# Put everything in the right order for QDOAS...
ds_data_flt.insert(0, dec_time) # .append adds things to the end of a list
ds_data_flt.insert(0, date_DMY) # .insert(0, x) adds x to the top of a list
ds_data_flt.insert(0, EA_real) # Values added on top in reverse order to
ds_data_flt.insert(0, AzA) # ensure they are in correct order!
ds_data_flt.insert(0, SZA)
# Prepare the new data frame for saving
ds_data_flt_a = np.array(ds_data_flt)
ds_data_flt_a.transpose()
ds_data_df= pd.DataFrame(ds_data_flt_a)
ds_data_dft = ds_data_df.transpose()
ds_list.append(ds_data_dft)
#=================================================================================
# Scattered light spectrum case;
#=================================================================================
elif sc:
#[EAset, EAactual, azipos] = [float(angle) for angle in angle_data.split(" ") if angle]
[EAset, EAactual, azim] = [float(angle) for angle in angle_data.split(" ") if angle]
#EA_real_ = float(EAset)
if EAactual > 80:
EA_real = 90
else:
EA_real = round(EAactual, 2)
# find the SZA;
scan_geom = file_data_strpd[2099]
scan_geom_split = scan_geom.split(" ")
SZA = float(scan_geom_split[5])
SZA = round(SZA, 2)
# We know the azimuth angle because we just use a compass to find it
AzA = round(float(210), 1)
# This section finds the calibration-corrected data
#===================================================================
sc_data_flt = md_data_int
if calcCI == True:
sc_data_forCI = pd.DataFrame(md_data_int_)
sc_data_forCI.columns = ['int']
sc_data_forCI['wl'] = pd.DataFrame(wl_data_strpd)
sc_data_forCI = sc_data_forCI.astype('float')
sc_data_forCI['wl_n_diff'] = abs(sc_data_forCI['wl'] - CIn)
sc_data_forCI['wl_d_diff'] = abs(sc_data_forCI['wl'] - CId)
numidx = sc_data_forCI['wl_n_diff'].idxmin()
denidx = sc_data_forCI['wl_d_diff'].idxmin()
CI = (sc_data_forCI['int'][numidx])/(sc_data_forCI['int'][denidx])
ave_int__ = file_data_strpd[2065]
ave_int_split = ave_int__.split(" ")
ave_int_ = float(ave_int_split[2])
ave_int = round(ave_int_, 2)
if texp > 0:
norm_ave_int = (ave_int/texp)
else:
norm_ave_int = 0
ci_data_flt = []
ci_data_flt.insert(0, CI)
ci_data_flt.insert(0, norm_ave_int)
ci_data_flt.insert(0, hhmmss)
ci_data_flt.insert(0, date_DMY)
ci_data_flt.insert(0, EAset)
ci_data_flt.insert(0, AzA)
ci_data_flt.insert(0, SZA)
# Prepare the new data frame for saving
ci_data_flt_a = np.array(ci_data_flt)
ci_data_flt_a.transpose()
ci_data_df= pd.DataFrame(ci_data_flt_a)
ci_data_dft = ci_data_df.transpose()
ci_list.append(ci_data_dft)
# Put everything in the right order for QDOAS...
sc_data_flt.insert(0, dec_time) # .append adds things to the end of a list
sc_data_flt.insert(0, date_DMY) # .insert(0, x) adds x to the top of a list
sc_data_flt.insert(0, EAset) # Values added on top in reverse order to
sc_data_flt.insert(0, AzA) # ensure they are in correct order!
sc_data_flt.insert(0, SZA)
# Prepare the new data frame for saving
sc_data_flt_a = np.array(sc_data_flt)
sc_data_flt_a.transpose()
sc_data_df= pd.DataFrame(sc_data_flt_a)
sc_data_dft = sc_data_df.transpose()
sc_list.append(sc_data_dft)
#=================================================================================
# Horizon scan case;
#=================================================================================
elif hs:
[EAsupposed, EAactual, azim] = [float(angle) for angle in angle_data.split(" ")[:3] if angle]
#[EAset, azipos] = [float(angle) for angle in angle_data.split(" ") if angle]
#EA_real_ = float(EAstart)
if EAactual > 80:
EA_real = 90
else:
EA_real = round(EAactual, 2)
# find the SZA;
scan_geom = file_data_strpd[2099]
scan_geom_split = scan_geom.split(" ")
SZA = float(scan_geom_split[5])
SZA = round(SZA, 2)
# find intensity values
int_1138 = float(file_data_strpd[1138])
int_1094 = float(file_data_strpd[1094])
ave_int__ = file_data_strpd[2065]
ave_int_split = ave_int__.split(" ")
ave_int_ = float(ave_int_split[2])
ave_int = round(ave_int_, 2)
if texp > 0:
norm_ave_int = (ave_int/texp)
norm_int_1094 = int_1094/texp
else:
norm_ave_int = 0
norm_int_1094 = 0
# We know the azimuth angle because we just use a compass to find it
AzA = float(200)
AzA = round(AzA, 1)
# This section finds the calibration-corrected data
#===================================================================
if only_save_hs_int == True:
hs_data_flt = []
hs_data_flt.insert(0, ave_int)
hs_data_flt.insert(0, norm_int_1094) # second intensity point
hs_data_flt.insert(0, norm_ave_int) # relative intensity between the two
hs_data_flt.insert(0, dec_time) # .append adds things to the end of a list
hs_data_flt.insert(0, date_DMY) # .insert(0, x) adds x to the top of a list
hs_data_flt.insert(0, EA_real) # Values added on top in reverse order to
hs_data_flt.insert(0, AzA) # ensure they are in correct order!
hs_data_flt.insert(0, SZA)
else:
hs_data_flt = md_data_int # Appropriate for QDOAS!
hs_data_flt.insert(0, dec_time) # .append adds things to the end of a list
hs_data_flt.insert(0, date_DMY) # .insert(0, x) adds x to the top of a list
hs_data_flt.insert(0, EA_real) # Values added on top in reverse order to
hs_data_flt.insert(0, AzA) # ensure they are in correct order!
hs_data_flt.insert(0, SZA)
# Prepare the new data frame for saving
hs_data_flt_a = np.array(hs_data_flt)
hs_data_flt_a.transpose()
hs_data_df= pd.DataFrame(hs_data_flt_a)
hs_data_dft = hs_data_df.transpose()
hs_list.append(hs_data_dft)
#=================================================================================
# Offset calibration spectrum case;
#=================================================================================
elif oc:
SZA = 0
EA_real = 0
AzA = 0
# Put everything in the right order for QDOAS...
# Don't want calibration corrected data here since this is the calibration!
oc_data_flt = md_data_int_
oc_data_flt.insert(0, dec_time)
oc_data_flt.insert(0, date_DMY)
oc_data_flt.insert(0, 'oc')
oc_data_flt.insert(0, numscans)
oc_data_flt.insert(0, texp)
# Prepare the new data frame for saving
oc_data_flt_a = np.array(oc_data_flt)
oc_data_flt_a.transpose()
oc_data_df= pd.DataFrame(oc_data_flt_a)
oc_data_dft = oc_data_df.transpose()
oc_list.append(oc_data_dft)
#=================================================================================
# Dark current calibration spectrum case;
#=================================================================================
elif dc:
SZA = 0
EA_real = 0
AzA = 0
# again don't want the calibration-corrected data so take md_data_int_
dc_data_flt = md_data_int_
# Put everything in the right order for QDOAS...
dc_data_flt.insert(0, dec_time)
dc_data_flt.insert(0, date_DMY)
dc_data_flt.insert(0, 'dc')
dc_data_flt.insert(0, numscans)
dc_data_flt.insert(0, texp)
# Prepare the new data frame for saving
dc_data_flt_a = np.array(dc_data_flt)
dc_data_flt_a.transpose()
dc_data_df= pd.DataFrame(dc_data_flt_a)
dc_data_dft = dc_data_df.transpose()
dc_list.append(dc_data_dft)
elif other_calib:
print('Found calibration spectra in file ', foldernumber)
else:
print("oh dear, something has gone wrong :(")
#print(date_DMY)
# Saving section
#=================================================================================
date2save = day_str + month_str+ year_str
# Save ds to file;
#=================================================================================
if len(ds_list)>0:
if saveDS == True:
ds_frame_to_fill = pd.concat(ds_list)
ds_frame_to_fill.to_csv(r'C:/'+path2save+inst+'_DS_'+date2save+'-'+folderletter+str(foldernumber)+end,
sep = ' ', header =None, index=None)
#else:
#print("There's no direct sun data in this folder")
# Save sc to file;
#=================================================================================
if len(sc_list)>0:
if saveSC == True:
sc_frame_to_fill = pd.concat(sc_list)
sc_frame_to_fill.to_csv(r'C:/'+path2save+inst+'_SC_'+date2save+'-'+folderletter+str(foldernumber)+end,
sep = ' ', header =None, index=None)
# Save hs to file;
#=================================================================================
if len(hs_list)>0:
if saveHS == True:
hs_frame_to_fill =
|
pd.concat(hs_list)
|
pandas.concat
|
import pandas as pd
from .utils import *
from ..smartapi_kg import MetaKG
from ..call_apis import APIQueryDispatcher
from ..config_new import BTE_FILTERS
from .filter.nodeDegree import NodeDegreeFilter
from .filter import Filter
from ..expand import Expander
from .printer import Print
class Predict:
def __init__(self, input_objs, intermediate_nodes, output_types, config=None):
self.input_objs = input_objs
self.intermediate_nodes = intermediate_nodes
if isinstance(intermediate_nodes, str):
intermediate_nodes = [intermediate_nodes]
self.output_types = output_types
self.intermediate_nodes.append(self.output_types)
validate_max_intermediate_nodes(self.intermediate_nodes)
self.steps_results = {}
self.steps_nodes = {}
self.kg = MetaKG()
self.kg.constructMetaKG(source="local")
self.query_completes = False
self.config = config
self.ep = Expander()
self.pt = Print()
def _expand_inputs(self, inputs, verbose=True):
"""
Expand inputs to its descendants.
:param inputs: list of input bioentities with resolved ids.
:param verbose: verbose
"""
if (
self.config
and self.config.get("expand")
and self.config.get("expand") is True
):
if verbose:
self.pt.print_expand_begin_message()
expandedInputs = self.ep.expand(inputs.values())
if verbose:
self.pt.print_expand_summary_message(expandedInputs)
if expandedInputs:
inputs.update(expandedInputs)
return inputs
def _annotate_edges_with_filters(self, edges, step):
"""
Add filter information to each edge.
:param edges: list of edges.
"""
if (
self.config
and self.config.get("filters")
and isinstance(self.config["filters"], list)
and step < len(self.config["filters"])
and isinstance(self.config["filters"][step], dict)
and self.config["filters"][step]
):
if len(set(self.config["filters"][step].keys()) - set(BTE_FILTERS)) == 0:
return
for edge in edges:
output_nodes = self.intermediate_nodes[step]
if isinstance(output_nodes, str):
edge["filter"] = self.config["filters"][step]
if isinstance(output_nodes, list):
for i, node in enumerate(output_nodes):
if node == edge["association"]["output_type"]:
if isinstance(self.config["filters"][step], list):
edge["filter"] = self.config["filters"][step][i]
else:
edge["filter"] = self.config["filters"][step]
def _get_predicates_from_config(self, path):
"""
Get information on predicates from config.
"""
if (
self.config
and self.config.get("predicates")
and isinstance(self.config["predicates"], list)
and path < len(self.config["predicates"])
):
predicates = self.config["predicates"][path]
else:
predicates = None
return predicates
def _annotate_results(self, step, source_types):
# print("annotating results with NodeDegree!")
# ft = NodeDegreeFilter(self.steps_results[step], {})
# ft.annotateNodeDegree()
# self.steps_results[step] = ft.stepResult
if "annotate" in self.config and isinstance(self.config["annotate"], list):
ft = Filter(self.steps_results[step], self.config["annotate"], source_types)
ft.annotate()
self.steps_results[step] = ft.stepResult
return ft
return None
def _filter_results(self, step, ft=None):
if (
self.config
and self.config.get("filters")
and step < len(self.config["filters"])
):
if not ft:
ft = Filter(self.steps_results[step], self.config["filters"][step])
else:
ft.criteria = self.config["filters"][step]
self.steps_results[step] = ft.filter_response()
self.steps_nodes[step] = extractAllResolvedOutputIDs(
self.steps_results[step]
)
print(
"\nAfter applying post-query filter, BTE retrieved {} unique output nodes.".format(
len(self.steps_nodes[step])
)
)
return self.steps_nodes[step]
def connect(self, verbose=True):
if not validate_max_intermediate_nodes(self.intermediate_nodes):
return
self.query_completes = False
inputs = restructureHintOutput(self.input_objs)
inputs = self._expand_inputs(inputs, verbose=verbose)
if verbose:
self.pt.print_query_parameter_summary(inputs, self.intermediate_nodes)
for i, node in enumerate(self.intermediate_nodes):
if verbose:
self.pt.print_query_plan_begin_message(i + 1)
inputs = groupsIDsbySemanticType(inputs)
source_types = list(inputs.keys())
predicates = self._get_predicates_from_config(i)
if verbose:
print(
"Input Types: {}\nOutput Types: {}\nPredicates: {}\n".format(
",".join(list(inputs.keys())), node, predicates
)
)
edges = getEdges(inputs, node, predicates, self.kg)
if len(edges) == 0:
self.pt.print_query_failure_message("APIs", i + 1)
return
annotatedEdges = []
for e in edges:
annotatedEdges += annotateEdgesWithInput(
e.get("edges"), e.get("inputs")
)
if not annotatedEdges:
self.pt.print_query_failure_message("APIs", i + 1)
return
self._annotate_edges_with_filters(annotatedEdges, i)
dp = APIQueryDispatcher(annotatedEdges, verbose=verbose)
self.steps_results[i] = dp.syncQuery()
if not self.steps_results[i] or len(self.steps_results[i]) == 0:
self.pt.print_query_failure_message("results", i + 1)
return
inputs = self.steps_nodes[i] = extractAllResolvedOutputIDs(
self.steps_results[i]
)
if verbose:
self.pt.print_individual_query_step_summary(inputs)
ft = self._annotate_results(i, source_types)
inputs = self._filter_results(i, ft)
self.query_completes = True
if verbose:
self.pt.print_final_result_summary(self.steps_nodes)
def annotate(self, step=0, criteria=[]):
ft = Filter(self.steps_results[step], criteria=criteria)
self.steps_results[step] = ft.annotate()
def display_table_view(self, extra_fields=[]):
if not self.query_completes:
print("Your query fails. Unable to display results!")
return
for step, step_result in self.steps_results.items():
df = stepResult2PandasTable(
step_result, step, len(self.steps_results), extra_fields
)
if step == 0:
result = df
continue
join_columns = [
"node" + str(step) + item for item in ["_id", "_type", "_label"]
]
result =
|
pd.merge(result, df, on=join_columns, how="inner")
|
pandas.merge
|
import pytest
from doltpy.core.dolt import Dolt, _execute, DoltException
from doltpy.core.write import UPDATE, import_df
from doltpy.core.read import pandas_read_sql, read_table
import shutil
import pandas as pd
import uuid
import os
from typing import Tuple, List
from doltpy.core.tests.helpers import get_repo_path_tmp_path
import sqlalchemy
from retry import retry
from sqlalchemy.engine import Engine
BASE_TEST_ROWS = [
{'name': 'Rafael', 'id': 1},
{'name': 'Novak', 'id': 2}
]
@pytest.fixture
def create_test_data(tmp_path) -> str:
path = os.path.join(tmp_path, str(uuid.uuid4()))
pd.DataFrame(BASE_TEST_ROWS).to_csv(path, index_label=False)
yield path
os.remove(path)
@pytest.fixture
def create_test_table(init_empty_test_repo, create_test_data) -> Tuple[Dolt, str]:
repo, test_data_path = init_empty_test_repo, create_test_data
repo.sql(query='''
CREATE TABLE `test_players` (
`name` LONGTEXT NOT NULL COMMENT 'tag:0',
`id` BIGINT NOT NULL COMMENT 'tag:1',
PRIMARY KEY (`id`)
);
''')
import_df(repo, 'test_players',
|
pd.read_csv(test_data_path)
|
pandas.read_csv
|
# @Author: <NAME><Nareshvrao>
# @Date: 2020-12-22, 12:44:08
# @Last modified by: Naresh
# @Last modified time: 2019-12-22, 1:13:26
import warnings
warnings.filterwarnings("ignore")
from pathlib import Path
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import tqdm
from util.utils import *
def compute_detail_score(df, dice):
res = []
res.append(df[dice].mean())
#c1 -> c4 dice
for label in ['Fish', 'Flower', 'Gravel', 'Sugar']:
df_tmp = df[df['cls'] == label]
res.append(df_tmp[dice].mean())
# neg & pos dice
res.append(df[df['truth'] == ''][dice].mean())
res.append(df[df['truth'] != ''][dice].mean())
# c1 -> c4 pos
for label in ['Fish', 'Flower', 'Gravel', 'Sugar']:
df_tmp = df[df['cls'] == label]
res.append(df_tmp[df_tmp['truth'] != ''][dice].mean())
return res
def ensemble_rles(rles1, rles2, mode='intersect'):
res = []
for rle1, rle2 in tqdm.tqdm(zip(rles1, rles2)):
m1 = rle2mask(rle1, height=350, width=525, fill_value=1)
m2 = rle2mask(rle2, height=350, width=525, fill_value=1)
if mode == 'intersect':
mask = ((m1+m2) == 2).astype(int)
elif mode == 'union':
mask = ((m1+m2) > 0).astype(int)
else:
RuntimeError('%s not implemented.'%mode)
rle = mask2rle(mask)
res.append(rle)
return res
def load_stacking(seg_name, tta, ts=0.5):
df_seg_val = pd.read_csv('../output/'+seg_name+'/valid_5fold_tta%d.csv'%tta)
df_seg_test = pd.read_csv('../output/'+seg_name+'/test_5fold_tta%d.csv'%tta)
df_seg_val['s1'], df_seg_test['s1'] = np.nan, np.nan
df_seg_val['s1'].loc[df_seg_val.pred >= ts] = '1 1'
df_seg_test['s1'].loc[df_seg_test.pred >= ts] = '1 1'
return df_seg_val[['Image_Label', 's1']], df_seg_test[['Image_Label', 's1']]
def load_seg_pred(seg_name, name, tta):
#load val
df_val = []
try:
for fold in range(5):
if tta <= 1:
df_val.append(pd.read_csv('../output/'+ seg_name + '/' + 'valid_fold%d.csv'%fold))
else:
df_val.append(pd.read_csv('../output/'+ seg_name + '/' + 'valid_fold%d_tta%d.csv'%(fold, tta)))
df_val = pd.concat(df_val)
except:
df_val = pd.read_csv('../output/'+ seg_name + '/' + 'valid_5fold_tta%d.csv'%(tta))
df_val = df_val[['Image_Label', 'EncodedPixels']]
#df_val.rename(columns={'s3': 'EncodedPixels'}, inplace=True)
df_test = pd.read_csv('../output/'+ seg_name + '/' + 'test_5fold_tta%d.csv'%tta)
df_val.rename(columns={'EncodedPixels': name}, inplace=True)
df_test.rename(columns={'EncodedPixels': name}, inplace=True)
return df_val, df_test
def load_seg_cls_pred(seg_name, name, tta, ts):
#load val
df_val = []
try:
for fold in range(5):
if tta <= 1:
df_val.append(pd.read_csv('../output/'+ seg_name + '/' + 'valid_cls_fold%d.csv'%fold))
else:
df_val.append(pd.read_csv('../output/'+ seg_name + '/' + 'valid_cls_fold%d_tta%d.csv'%(fold, tta)))
df_val = pd.concat(df_val)
except:
df_val = pd.read_csv('../output/'+ seg_name + '/' + 'valid_5fold_tta%d.csv'%(tta))
df_val = df_val[['Image_Label', 'EncodedPixels']]
#df_val.rename(columns={'s3': 'EncodedPixels'}, inplace=True)
df_test = pd.read_csv('../output/'+ seg_name + '/' + 'test_cls_5fold_tta%d.csv'%tta)
df_val['EncodedPixels'] = '1 1'
df_val['EncodedPixels'].loc[df_val['0'] < ts] = np.nan
df_test['EncodedPixels'] = '1 1'
df_test['EncodedPixels'].loc[df_test['0'] < ts] = np.nan
df_val.rename(columns={'EncodedPixels': name}, inplace=True)
df_test.rename(columns={'EncodedPixels': name}, inplace=True)
return df_val, df_test
def load_classifier(classifier, tta):
try:
df_cls_val = []
df_cls_test = []
for fold in range(5):
if tta <= 1:
df_cls_val.append(pd.read_csv('../output/'+ classifier + '/' + 'valid_cls_fold%d.csv'%fold))
df_cls_test.append(pd.read_csv('../output/'+ classifier + '/' + 'test_cls_fold%d.csv'%fold))
else:
df_cls_val.append(pd.read_csv('../output/'+ classifier + '/' + 'valid_cls_fold%d_tta%d.csv'%(fold, tta)))
df_cls_test.append(pd.read_csv('../output/'+ classifier + '/' + 'test_cls_fold%d_tta%d.csv'%(fold, tta)))
df_cls_val = pd.concat(df_cls_val)
df_tmp = df_cls_test[0]
for i in range(1, 5):
assert(np.sum(df_tmp['Image_Label'] != df_cls_test[i]['Image_Label']) == 0)
df_tmp['0'] += df_cls_test[i]['0']
df_tmp['0'] /= 5
df_cls_test = df_tmp
except:
df_cls_val = pd.read_csv('../output/'+ classifier + '/' + 'valid_cls_5fold_tta%d.csv'%tta)
df_cls_test = pd.read_csv('../output/'+ classifier + '/' + 'test_cls_5fold_tta%d.csv'%tta)
df_cls_val.rename(columns={'0': 'prob'}, inplace=True)
df_cls_test.rename(columns={'0': 'prob'}, inplace=True)
return df_cls_val, df_cls_test
df_train = pd.read_csv('../input/train_350.csv')
df_train.rename(columns={'EncodedPixels': 'truth'}, inplace=True)
_save=1
tta=3
seg1 = 'densenet121-FPN-BCE-warmRestart-10x3-bs16'
seg2 = 'b5-Unet-inception-FPN-b7-Unet-b7-FPN-b7-FPNPL'
classifier = 'efficientnetb1-cls-BCE-reduceLR-bs16-PL'
# load classifier results
if classifier:
if 'stacking' in classifier:
df_cls_val = pd.read_csv('../output/'+classifier+'/valid_5fold_tta%d.csv'%tta).rename(columns={'pred': 'prob'})
df_cls_test = pd.read_csv('../output/'+classifier+'/test_5fold_tta%d.csv'%tta).rename(columns={'pred': 'prob'})
else:
df_cls_val, df_cls_test = load_classifier(classifier, tta)
# load seg results
if isinstance(seg1, list):
df_seg1_val, df_seg1_test = load_seg_pred(seg1[0], 's1', tta)
for i in range(1, len(seg1)):
d1, d2 = load_seg_pred(seg1[i], 's1', tta)
df_seg1_val['s1'].loc[d1.s1.isnull()] = np.nan
df_seg1_test['s1'].loc[d2.s1.isnull()] = np.nan
elif 'stacking' in seg1:
df_seg1_val, df_seg1_test = load_stacking(seg1, 3, ts=0.54)
else:
df_seg1_val, df_seg1_test = load_seg_pred(seg1, 's1', 1)
df_seg2_val, df_seg2_test = load_seg_pred(seg2, 's2', tta)
# merge seg valid
df_seg_val = pd.merge(df_seg1_val, df_seg2_val, how='left')
df_seg_val = pd.merge(df_seg_val, df_train, how='left')
if classifier:
df_seg_val = pd.merge(df_seg_val, df_cls_val[['Image_Label', 'prob']], how='left')
df_seg_val['s3'] = df_seg_val['s1'].copy()
df_seg_val['s3'].loc[df_seg_val['s1'].notnull()] = df_seg_val['s2'].loc[df_seg_val['s1'].notnull()]
#df_seg_val['area'] = df_seg_val['s3'].apply(lambda x: rle2mask(x, height=350, width=525).sum())
df_seg_test = pd.merge(df_seg1_test, df_seg2_test, how='left')
df_seg_test =
|
pd.merge(df_seg_test, df_cls_test[['Image_Label', 'prob']], how='left')
|
pandas.merge
|
#%% Import require packages
import h5py
import pandas as pd
import tarfile
import docker
import time
import re
from knmy import knmy
from timeloop import Timeloop
from datetime import timedelta
from dateutil.parser import parse
from io import BytesIO
#%% Helper functions.
def to_writeable_timestamp(timestamp):
timestamp_string = str(timestamp)
cleaned_timestamp_string = re.sub(r'[ :]', '-', re.sub(r'\+.*', '',timestamp_string))
return(cleaned_timestamp_string)
def write_beamformed(data_part_df):
data_part_string = data_part_df.to_csv(sep=",", date_format="%Y-%m-%d %H:%M:%S", index=False)
tarstream = BytesIO()
tar = tarfile.TarFile(fileobj=tarstream, mode='w')
file_data = data_part_string.encode('utf8')
tarinfo = tarfile.TarInfo(name="measurement"+str(measurement_index)+"-"+str(measurement_index+index_delta)+".csv")
tarinfo.size = len(file_data)
tarinfo.mtime = time.time()
tar.addfile(tarinfo, BytesIO(file_data))
tar.close()
tarstream.seek(0)
spark_master.put_archive("/opt/spark-data/beamformed", tarstream)
def fetch_and_write_weather(last_timestamp, last_hourly_measurement):
#Correct for datetime handling in knmy function by adding hour offset
_, _, _, knmi_df = knmy.get_hourly_data(stations=[279], start=last_hourly_measurement-timedelta(hours=1), end=last_timestamp-timedelta(hours=1), parse=True)
knmi_df = knmi_df.drop(knmi_df.index[0]) #drop first row, which contains a duplicate header
knmi_df["timestamp"] = [(parse(date) + timedelta(hours=int(hour))) for date, hour in zip(knmi_df["YYYYMMDD"], knmi_df["HH"])]
knmi_df = knmi_df.drop(["STN", "YYYYMMDD", "HH"], axis=1)
weather_string = knmi_df.to_csv(sep=",", date_format="%Y-%m-%d %H:%M:%S", index=False)
filename = "weather"+to_writeable_timestamp(last_hourly_measurement)+"-to-"+to_writeable_timestamp(last_timestamp)+".csv"
tarstream = BytesIO()
tar = tarfile.TarFile(fileobj=tarstream, mode='w')
file_data = weather_string.encode('utf8')
tarinfo = tarfile.TarInfo(name=filename)
tarinfo.size = len(file_data)
tarinfo.mtime = time.time()
tar.addfile(tarinfo, BytesIO(file_data))
tar.close()
tarstream.seek(0)
spark_master.put_archive("/opt/spark-data/weather", tarstream)
#%% initialize variables and initialize hdf5 access and docker containers
filename = 'L701913_SAP000_B000_S0_P000_bf.h5'
h5 = h5py.File(filename, "r")
stokes = h5["/SUB_ARRAY_POINTING_000/BEAM_000/STOKES_0"]
client = docker.from_env()
spark_master = client.containers.get("spark-master")
tl = Timeloop()
time_start = parse(h5.attrs['OBSERVATION_START_UTC']) #Start of measurements as datetime object
measurement_index = 102984 #Which measurement should streaming start with?
# For when to test the hourly change:
# 18*60*(10**6)/time_delta.microseconds
# Out[172]: 102994.46881556361
# with 102984 first batch should have weather from hour 11, second batch from hour 12
index_delta = 100 #How many measurements should be sent each second
time_delta = timedelta(seconds=h5["/SUB_ARRAY_POINTING_000/BEAM_000/COORDINATES/COORDINATE_0"].attrs["INCREMENT"]) #The time between two consecutive measurements
# calculate first timestamp and set it to 1 hour before measurements start
last_hourly_measurement = (time_start - timedelta(hours=1)).replace(minute=0, second=0, microsecond=0)
#%% define jobs
@tl.job(interval=timedelta(seconds=5))
def read_and_write():
global measurement_index
global last_hourly_measurement
#measurements -and- timestamp
data_part_df =
|
pd.DataFrame(stokes[measurement_index:(measurement_index+index_delta),:])
|
pandas.DataFrame
|
# Based on https://www.kaggle.com/currie32/predicting-fraud-with-tensorflow
import os
import pandas as pd
import numpy as np
import tensorflow as tf
from tensorflow.python.tools import freeze_graph
from tensorflow.python.framework.graph_util import convert_variables_to_constants
# from sklearn.cross_validation import train_test_split
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.metrics import confusion_matrix
import seaborn as sns
import matplotlib.gridspec as gridspec
from sklearn.preprocessing import StandardScaler
from sklearn.manifold import TSNE
df = pd.read_csv("./input/creditcard.csv")
print("Head: \n{}".format(df.head()))
print("Describe: \n{}".format(df.describe()))
print("Nulls: \n{}".format(df.isnull().sum()))
# Let's see how time compares across fraudulent and normal transactions.
print("Fraud")
print(df.Time[df.Class == 1].describe())
print()
print("Normal")
print(df.Time[df.Class == 0].describe())
# f, (ax1, ax2) = plt.subplots(2, 1, sharex=True, figsize=(12,4))
# bins = 50
# ax1.hist(df.Time[df.Class == 1], bins = bins)
# ax1.set_title('Fraud')
# ax2.hist(df.Time[df.Class == 0], bins = bins)
# ax2.set_title('Normal')
# plt.xlabel('Time (in Seconds)')
# plt.ylabel('Number of Transactions')
# plt.show()
# see if the transaction amount differs between the two types.
print("Fraud")
print(df.Amount[df.Class == 1].describe())
print()
print("Normal")
print(df.Amount[df.Class == 0].describe())
# f, (ax1, ax2) = plt.subplots(2, 1, sharex=True, figsize=(12,4))
# bins = 30
# ax1.hist(df.Amount[df.Class == 1], bins = bins)
# ax1.set_title('Fraud')
# ax2.hist(df.Amount[df.Class == 0], bins = bins)
# ax2.set_title('Normal')
# plt.xlabel('Amount ($)')
# plt.ylabel('Number of Transactions')
# plt.yscale('log')
# plt.show()
df['Amount_max_fraud'] = 1
df.loc[df.Amount <= 2125.87, 'Amount_max_fraud'] = 0
# Let's compare Time with Amount and see if we can learn anything new.
# f, (ax1, ax2) = plt.subplots(2, 1, sharex=True, figsize=(12,6))
#
# ax1.scatter(df.Time[df.Class == 1], df.Amount[df.Class == 1])
# ax1.set_title('Fraud')
#
# ax2.scatter(df.Time[df.Class == 0], df.Amount[df.Class == 0])
# ax2.set_title('Normal')
#
# plt.xlabel('Time (in Seconds)')
# plt.ylabel('Amount')
# plt.show()
# take a look at the anonymized features.
# Select only the anonymized features.
# v_features = df.ix[:,1:29].columns
# plt.figure(figsize=(20, 5*2))
# gs = gridspec.GridSpec(5, 6)
# for i, cn in enumerate(df[v_features]):
# ax = plt.subplot(gs[i])
# sns.distplot(df[cn][df.Class == 1], bins=50)
# sns.distplot(df[cn][df.Class == 0], bins=50)
# ax.set_xlabel('')
# ax.set_title('histogram of feature: ' + str(cn))
# plt.show()
# Drop all of the features that have very similar distributions between the two types of transactions.
df = df.drop(['V28', 'V27', 'V26', 'V25', 'V24', 'V23', 'V22', 'V20', 'V15', 'V13', 'V8'], axis=1)
# Based on the plots above, these features are created to identify values where fraudulent transaction are more common.
df['V1_'] = df.V1.map(lambda x: 1 if x < -3 else 0)
df['V2_'] = df.V2.map(lambda x: 1 if x > 2.5 else 0)
df['V3_'] = df.V3.map(lambda x: 1 if x < -4 else 0)
df['V4_'] = df.V4.map(lambda x: 1 if x > 2.5 else 0)
df['V5_'] = df.V5.map(lambda x: 1 if x < -4.5 else 0)
df['V6_'] = df.V6.map(lambda x: 1 if x < -2.5 else 0)
df['V7_'] = df.V7.map(lambda x: 1 if x < -3 else 0)
df['V9_'] = df.V9.map(lambda x: 1 if x < -2 else 0)
df['V10_'] = df.V10.map(lambda x: 1 if x < -2.5 else 0)
df['V11_'] = df.V11.map(lambda x: 1 if x > 2 else 0)
df['V12_'] = df.V12.map(lambda x: 1 if x < -2 else 0)
df['V14_'] = df.V14.map(lambda x: 1 if x < -2.5 else 0)
df['V16_'] = df.V16.map(lambda x: 1 if x < -2 else 0)
df['V17_'] = df.V17.map(lambda x: 1 if x < -2 else 0)
df['V18_'] = df.V18.map(lambda x: 1 if x < -2 else 0)
df['V19_'] = df.V19.map(lambda x: 1 if x > 1.5 else 0)
df['V21_'] = df.V21.map(lambda x: 1 if x > 0.6 else 0)
print("Boza")
print(df.loc[1,:])
print("Boza2")
bla = df.loc[df.Class == 1]
print(bla.loc[541,:])
print("Head: \n{}".format(df.head()))
print("Describe: \n{}".format(df.describe()))
# Create a new feature for normal (non-fraudulent) transactions.
df.loc[df.Class == 0, 'Normal'] = 1
df.loc[df.Class == 1, 'Normal'] = 0
# Rename 'Class' to 'Fraud'.
df = df.rename(columns={'Class': 'Fraud'})
# 492 fraudulent transactions, 284,315 normal transactions.
# 0.172% of transactions were fraud.
print(df.Normal.value_counts())
print()
print(df.Fraud.value_counts())
|
pd.set_option("display.max_columns", 101)
|
pandas.set_option
|
# -*- coding: utf-8 -*-
"""
@file:utils.py
@time:2019/6/1 21:57
@author:Tangj
@software:Pycharm
@Desc
"""
import os
import numpy as np
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.metrics import roc_auc_score
from time import time
import random
import pandas as pd
def frame_to_dict(train):
train_dict = {}
for col in train.columns:
train_dict[col] = train[col].columns
return trian_dict
def del_adSize(ad_Size):
ad_size_mean = []
ad_size_max = []
ad_size_min = []
for adSize in ad_Size:
if not isinstance(adSize, str):
# print(adSize)
ad_size_mean.append(adSize)
ad_size_max.append(adSize)
ad_size_min.append(adSize)
continue
size = adSize.split(',')
s = []
for i in size:
s.append(int(i))
ad_size_mean.append(np.mean(s))
ad_size_max.append(np.max(s))
ad_size_min.append(np.min(s))
return ad_size_mean, ad_size_max, ad_size_max
def write_data_into_parts(data, root_path, nums=5100000):
l = data.shape[0] // nums
for i in range(l + 1):
begin = i * nums
end = min(nums * (i + 1), data.shape[0])
t_data = data[begin:end]
t_data.tofile(root_path + '.bin')
def write_dict(path, data):
fw = open(path, 'w')
for key in data:
fw.write(str(key) + ',' + str(data[key]) + '\n')
fw.close()
def read_allfea(path):
f = open(path, 'r')
fea = '0'
for i in f:
fea = i
fea_val = fea.split(',')
index_dict = {}
for i, fea in enumerate(fea_val):
index_dict[fea] = i + 1
if '-1' not in index_dict:
index_dict['-1'] = len(fea_val)
return fea, index_dict
def one_hot_feature_concat(train, test, fea1, fea2, filter_num=100):
train1 = train[fea1].values
train2 = train[fea2].values
test1 = test[fea1].values
test2 = test[fea2].values
train_data = []
test_data = []
train_res = []
test_res = []
for i, values in enumerate(train1):
new = str(values) + '|' + str(train2[i])
train_data.append(new)
for i, values in enumerate(test1):
new = str(values) + '|' + str(test2[i])
# print(new)
test_data.append(new)
count_dict = {}
for d in train_data:
if d not in count_dict:
count_dict[d] = 0
count_dict[d] += 1
filter_set = []
for i in count_dict:
if count_dict[i] < 1:
filter_set.append(i)
index_dict = {}
begin_index = 1
for d in train_data:
# 给出现的value赋予一个index指引
if d in filter_set:
d = '-1'
if d not in index_dict:
index_dict[d] = begin_index
begin_index += 1
train_res.append(index_dict[d])
if '-1' not in index_dict:
index_dict['-1'] = begin_index
for d in test_data:
if d not in index_dict or d in filter_set:
d = '-1'
test_res.append(index_dict[d])
print(test_res)
return np.array(train_res), np.array(test_res)
def one_hot_feature_process(train_data, val_data, test2_data, begin_num, filter_num=0):
index_dict = {}
begin_index = begin_num
train_res = []
for d in train_data:
# print(d)
# 给出现的value赋予一个index指引
if d not in index_dict:
index_dict[d] = begin_index
begin_index += 1
# print(index_dict[d])
train_res.append(index_dict[d])
if '-1' not in index_dict:
index_dict['-1'] = begin_index
val_res = []
for d in val_data:
if d not in index_dict:
index_dict[d] = begin_index
begin_index += 1
val_res.append(index_dict[d])
test2_res = []
for d in test2_data:
if d not in index_dict:
d = '-1'
test2_res.append(index_dict[d])
# print(np.array(train_res))
return np.array(train_res), np.array(val_res), np.array(test2_res), index_dict
def vector_feature_process(train_data, val_data, test2_data, begin_num, max_len, index_dict):
train_res = []
train_res2 = []
val_res2 = []
test2_res2 = []
train_rate = []
val_rate = []
test2_rate = []
for d in train_data:
lx = d.split(',')
row = [0] * max_len
row2 = [0] * max_len
if len(lx) > max_len or d == 'all':
j = 0
for i in index_dict:
if j >= max_len:
break
row[j] = index_dict[i]
j += 1
train_res.append(row)
row2 = [1] * max_len
train_res2.append(row2)
train_rate.append(1)
continue
for i, x in enumerate(lx):
if x not in index_dict:
x = '-1'
row[i] = index_dict[x]
row2[row[i]] = 1
train_res.append(row)
train_res2.append(row2)
train_rate.append(len(lx) / max_len)
val_res = []
for d in val_data:
lx = d.split(',')
row = [0] * max_len
row2 = [0] * max_len
if len(lx) > max_len or d == 'all':
j = 0
for i in index_dict:
if j >= max_len:
break
row[j] = index_dict[i]
j += 1
val_res.append(row)
row2 = [1] * max_len
val_res2.append(row2)
val_rate.append(1)
continue
for i, x in enumerate(lx):
if x not in index_dict:
x = '-1'
row[i] = index_dict[x]
row2[row[i]] = 1
val_res.append(row)
val_res2.append(row2)
val_rate.append(len(lx) / max_len)
test2_res = []
for d in test2_data:
lx = d.split(',')
row = [0] * max_len
row2 = [0] * max_len
if len(lx) > max_len or d == 'all':
j = 0
for i in index_dict:
if j >= max_len:
break
row[j] = index_dict[i]
j += 1
test2_res.append(row)
row2 = [1] * max_len
test2_res2.append(row2)
test2_rate.append(1)
continue
for i, x in enumerate(lx):
if x not in index_dict:
x = '-1'
row[i] = index_dict[x]
row2[row[i]] = 1
test2_res.append(row)
test2_res2.append(row2)
test2_rate.append(len(lx) / max_len)
return np.array(train_res), np.array(val_res), np.array(test2_res), index_dict, np.array(train_res2), np.array(
val_res2), np.array(test2_res2), np.array(train_rate), np.array(val_rate), np.array(test2_rate),
def count_one_feature_times(train, test, fea):
count_dict = {}
test_res = []
train_res = []
for val in train[fea].values:
if val not in count_dict:
count_dict[val] = 0
count_dict[val] += 1
if '-1' not in count_dict:
count_dict['-1'] = 1
for i in train[fea].values:
train_res.append(count_dict[i])
for i in test:
if i not in count_dict:
i = '-1'
test_res.append(count_dict[i])
return np.array(train_res), np.array(test_res)
def count_vector_feature_times(train, val_data, test, fea):
count_dict = {}
val_res = []
test_res = []
train_res = []
Train = pd.concat([train, val_data])
for val in Train[fea].values:
vals = val.split(',')
for i in vals:
if i not in count_dict:
count_dict[i] = 0
count_dict[i] += 1
if '-1' not in count_dict:
count_dict['-1'] = 1
for val in train[fea].values:
vals = val.split(',')
l = []
for i in vals:
l.append(count_dict[i])
# ['max', 'mean', 'min', 'median']
max_l = np.max(l)
mean_l = np.mean(l)
min_l = np.min(l)
median_l = np.median(l)
train_res.append([max_l, mean_l, min_l, median_l])
for val in val_data[fea].values:
vals = val.split(',')
l = []
for i in vals:
l.append(count_dict[i])
# ['max', 'mean', 'min', 'median']
max_l = np.max(l)
mean_l = np.mean(l)
min_l = np.min(l)
median_l = np.median(l)
val_res.append([max_l, mean_l, min_l, median_l])
for val in test:
vals = val.split(',')
l = []
for i in vals:
if i not in count_dict:
i = '-1'
l.append(count_dict[i])
# ['max', 'mean', 'min', 'median']
max_l = np.max(l)
mean_l = np.mean(l)
min_l = np.min(l)
median_l = np.median(l)
test_res.append([max_l, mean_l, min_l, median_l])
return np.array(train_res), np.array(val_res), np.array(test_res)
# 对曝光、pctr和ecpm和bid的特征
def one_feature_exposure2(Train, test, fea, date):
# 返回曝光的最大值,最小值,均值,中位数四个值,
# 返回bid的最大值,最小值,均值,中位数四个值,
test_res = []
train_res = []
id_res = []
reqday_res = []
train = Train
num1 = train[train['day'] == 20190410].shape[0]
id_res.extend(train[train['day'] == 20190410]['ad_id'].values)
reqday_res.extend(train[train['day'] == 20190410]['day'].values)
for i in range(num1):
train_res.append([0, 0, 0, 0])
for i in range(len(date) - 1):
day = int(date[i + 1])
train_compute = Train[Train['day'] == day]
train_count = Train[Train['day'] < day]
id_res.extend(train_compute['ad_id'].values)
reqday_res.extend(train_compute['day'].values)
exposure_dict = {}
for value in train_count[fea].values:
if value not in exposure_dict:
exposure_dict[value] = []
train1 = train_count[train_count[fea] == value]['sucess_rate'].values
exposure_dict[value].append(np.max(train1))
exposure_dict[value].append(np.min(train1))
exposure_dict[value].append(np.mean(train1))
exposure_dict[value].append(np.median(train1))
if '-1' not in exposure_dict:
exposure_dict['-1'] = [0, 0, 0, 0]
for value in train_compute[fea].values:
if value not in exposure_dict:
value = '-1'
train_res.append(exposure_dict[value])
train_count = Train[Train['day'] > 20190414]
exposure_dict = {}
for value in train_count[fea].values:
if value not in exposure_dict:
train1 = train_count[train_count[fea] == value]['sucess_rate'].values
exposure_dict[value] = []
exposure_dict[value].append(np.max(train1))
exposure_dict[value].append(np.min(train1))
exposure_dict[value].append(np.mean(train1))
exposure_dict[value].append(np.median(train1))
if '-1' not in exposure_dict:
exposure_dict['-1'] = [0, 0, 0, 0]
for value in test:
if value not in exposure_dict:
value = '-1'
test_res.append(exposure_dict[value])
return np.array(train_res), np.array(test_res), np.array(id_res), np.array(reqday_res)
def one_feature_exposure4(Train, test, fea, date):
test_res = []
train_res = []
id_res = []
reqday_res = []
train = Train
train_count = train[train['day'] == 20190410]
train_compute = train[train['day'] == 20190410]
id_res.extend(train_compute['ad_id'].values)
reqday_res.extend(train_compute['day'].values)
exposure_dict = {}
for value in train_count[fea].values:
if value not in exposure_dict:
train1 = train_count[train_count[fea] == value]['ex'].values
exposure_dict[value] = []
exposure_dict[value].append(np.mean(train1))
exposure_dict[value].append(np.median(train1))
if '-1' not in exposure_dict:
exposure_dict['-1'] = [0.9, 0.9]
for value in train_compute[fea].values:
if value not in exposure_dict:
value = '-1'
train_res.append(exposure_dict[value])
train_count = train[train['day'] == 20190410]
train_compute = train[train['day'] == 20190411]
id_res.extend(train_compute['ad_id'].values)
reqday_res.extend(train_compute['day'].values)
exposure_dict = {}
for value in train_count[fea].values:
if value not in exposure_dict:
train1 = train_count[train_count[fea] == value]['ex'].values
exposure_dict[value] = []
exposure_dict[value].append(np.mean(train1))
exposure_dict[value].append(np.median(train1))
if '-1' not in exposure_dict:
exposure_dict['-1'] = [0.9, 0.9]
for value in train_compute[fea].values:
if value not in exposure_dict:
value = '-1'
train_res.append(exposure_dict[value])
for i in range(len(date) - 2):
day1 = int(date[i + 2])
day2 = int(date[i + 1])
day3 = int(date[i])
train1 = Train[Train['day'] == day3]
train2 = Train[Train['day'] == day2]
train_compute = Train[Train['day'] == day1]
id_res.extend(train_compute['ad_id'].values)
reqday_res.extend(train_compute['day'].values)
train_count =
|
pd.concat([train1, train2])
|
pandas.concat
|
"""
Grab stocks from cad tickers
"""
import pandas as pd
class TickerControllerV2:
"""
Grabs cad_tickers dataframes and normalized them
"""
def __init__(self, cfg: dict):
"""
Extract yahoo finance tickers from website
Consider using hardcoded csvs sheets for the tickers to
increase speed, no need to grab all data dynamically.
"""
self.yf_tickers = []
# import csv from github
ticker_df = pd.read_csv(
"https://raw.githubusercontent.com/FriendlyUser/cad_tickers_list/main/static/latest/stocks.csv"
)
tickers_config = cfg.get("tickers_config")
us_df = pd.DataFrame()
if tickers_config != None:
industries = tickers_config.get("industries")
if industries != None:
ticker_df = ticker_df[ticker_df["industry"].isin(industries)]
us_cfg = tickers_config.get("us_tickers")
if us_cfg != None:
# apply filters
# same format as above
us_tickers_url = us_cfg.get("url")
us_df =
|
pd.read_csv(us_tickers_url)
|
pandas.read_csv
|
import numpy as np
import pandas as pd
from utils import data_generator
## data dimension
N_train = 110 # sum of training and valiadation set
dim = 24
## initialize parameters
c1_value = round(np.random.uniform(0, 20),2)
c2_value = round(np.random.uniform(0, 20),2)
duration = round(np.random.uniform(1, 4))
eta = round(np.random.uniform(0.8, 1),2)
paras = pd.DataFrame([[c1_value, c2_value, duration, eta]],columns=("c1", "c2", "P", "eta"))
print(
"Generating data!",
"P1=",
0.5,
"E1=",
0.5 * duration,
"c1 =",
c1_value,
"c2 =",
c2_value,
"eta =",
eta,
)
## load price data
price_hist =
|
pd.read_csv("./ESID_data/price.csv")
|
pandas.read_csv
|
# THIS SCRIPT SERVES TO TRANSLATE INDIVIDUAL DATASET ANNOTATIONS TO CELL TYPE ANNOTATIONS AS IMPLEMENTED BY SASHA AND MARTIJN! :)
import pandas as pd
import numpy as np
import scanpy as sc
import utils # this is a custom script from me
def nan_checker(entry):
"""replaces entry that is not a cell type label, but rather some other
entry that exists in the table (e.g. |, >, nan) with None. Retains
labels that look like proper labels."""
if entry == '|':
new_entry = None
elif entry == '>':
new_entry = None
elif pd.isna(entry):
new_entry = None
else:
new_entry = entry
return new_entry
def load_harmonizing_table(path_to_csv):
"""Loads the csv download version of the google doc in which
cell types for each dataset are assigned a consensus equivalent.
Returns the cleaned dataframe"""
cell_type_harm_df = pd.read_csv(
path_to_csv,
header=1
)
cell_type_harm_df = cell_type_harm_df.applymap(nan_checker)
# strip white spaces
cell_type_harm_df = cell_type_harm_df.apply(lambda x: x.str.strip() if x.dtype == "object" else x)
return cell_type_harm_df
def consensus_index_renamer(consensus_df, idx):
highest_res = int(float(consensus_df.loc[idx, "highest_res"]))
new_index = (
str(highest_res) + "_" + consensus_df.loc[idx, "level_" + str(highest_res)]
)
return new_index
def create_consensus_table(harmonizing_df, max_level=5):
"""creates a clean consensus table based on the harmonizing_df that also
includes dataset level info. The output dataframe contains, for each consensus
cell type, it's 'parent cell types', an indication of whether the cell
type is considered new, and the level of the annotation.
max_level - maximum level of annotation available"""
# set up empty consensus df, that will be filled later on
consensus_df = pd.DataFrame(
columns=[
'level_' + str(levnum) for levnum in range(1,max_level + 1)
] + [
'highest_res','new','harmonizing_df_index'
]
)
# store the index of the harmonizing df, so that we can match rows later on
consensus_df["harmonizing_df_index"] = harmonizing_df.index
# create dictionary storing the most recent instance of every level,
# so that we always track the 'mother levels' of each instance
most_recent_level_instance = dict()
# loop through rows
for idx in harmonizing_df.index:
# set 'new' to false. This variable indicates if the cell type annotation
# is new according to Sasha and Martijn. It will be checked/changed later.
new = False
# create a dictionary where we story the entries of this row,
# for each level
original_entries_per_level = dict()
# create a dictionary where we store the final entries of this row,
# for each level. That is, including the mother levels.
final_entries_per_level = dict()
# if all entries in this row are None, continue to next row:
if (
sum(
[
pd.isna(x)
for x in harmonizing_df.loc[
idx, ["Level_" + str(levnum) for levnum in range(1, max_level + 1)]
]
]
)
== max_level
):
continue
# for each level, check if we need to store the current entry,
# the mother level entry, or no entry (we do not want to store daughters)
for level in range(1, max_level + 1):
original_entry = harmonizing_df.loc[idx, "Level_" + str(level)]
# if the current level has an entry in the original dataframe,
# update the 'most_recent_level_instance'
# and store the entry as the final output entry for this level
if original_entry != None:
# store the lowest level annotated for this row:
lowest_level = level
# check if entry says 'New!', then we need to obtain actual label
# from the dataset where the label appears
if original_entry == "New!":
# set new to true now
new = True
# get all the unique entries from this row
# this should be only one entry aside from 'None' and 'New!'
actual_label = list(set(harmonizing_df.loc[idx, :]))
# filter out 'New!' and 'None'
actual_label = [
x for x in actual_label if not pd.isna(x) and x != "New!"
][0]
original_entry = actual_label
# update most recent instance of level:
most_recent_level_instance[level] = original_entry
# store entry as final entry for this level
final_entries_per_level[level] = original_entry
# Moreover, store the most recent instances of the lower (parent)
# levels as final output:
for parent_level in range(1, level):
final_entries_per_level[parent_level] = most_recent_level_instance[
parent_level
]
# and set the daughter levels to None:
for daughter_level in range(level + 1, max_level + 1):
final_entries_per_level[daughter_level] = None
break
# if none of the columns have an entry,
# set all of them to None:
if bool(final_entries_per_level) == False:
for level in range(1, 5):
final_entries_per_level[level] = None
# store the final outputs in the dataframe:
consensus_df.loc[idx, "highest_res"] = int(lowest_level)
consensus_df.loc[idx, "new"] = new
consensus_df.loc[idx, consensus_df.columns[:max_level]] = [
final_entries_per_level[level] for level in range(1, max_level + 1)
]
rows_to_keep = (
idx
for idx in consensus_df.index
if not consensus_df.loc[
idx, ["level_" + str(levnum) for levnum in range(1, max_level + 1)]
]
.isna()
.all()
)
consensus_df = consensus_df.loc[rows_to_keep, :]
# rename indices so that they also specify the level of origin
# this allows for indexing by cell type while retaining uniqueness
# (i.e. we're able to distinguis 'Epithelial' level 1, and level 2)
consensus_df.index = [
consensus_index_renamer(consensus_df, idx) for idx in consensus_df.index
]
# Now check if there are double index names. Sasha added multiple rows
# for the same consensus type to accomodate his cluster names. We
# should therefore merge these rows:
# First create an empty dictionary in which to store the harmonizing_df row
# indices of the multiple identical rows:
harmonizing_df_index_lists = dict()
for unique_celltype in set(consensus_df.index):
# if there are multiple rows, the only way in which they should differ
# is their harmonizing_df_index. Check this:
celltype_sub_df = consensus_df.loc[unique_celltype]
if type(celltype_sub_df) == pd.DataFrame and celltype_sub_df.shape[0] > 1:
# check if all levels align:
for level in range(1, max_level + 1):
if len(set(celltype_sub_df["level_" + str(level)])) > 1:
print(
"WARNING: {} has different annotations in different rows at level {}. Look into this!!".format(
unique_celltype, str(level)
)
)
harmonizing_df_index_list = celltype_sub_df["harmonizing_df_index"].values
# if there was only one index equal to the unique celltype, the
# celltype_sub_df is actually a pandas series and we need to index
# it diffently
else:
harmonizing_df_index_list = [celltype_sub_df["harmonizing_df_index"]]
# store the harmonizing_df_index_list in the consensus df
# we use the 'at' function to be able to store a list in a single
# cell/entry of the dataframe.
harmonizing_df_index_lists[unique_celltype] = harmonizing_df_index_list
# now that we have a list per cell type, we can drop the duplicate columns
consensus_df.drop_duplicates(
subset=["level_" + str(levnum) for levnum in range(1, max_level + 1)], inplace=True
)
# and replace the harmonizing_df_index column with the generated complete lists
consensus_df["harmonizing_df_index"] = None
for celltype in consensus_df.index:
consensus_df.at[celltype, "harmonizing_df_index"] = harmonizing_df_index_lists[
celltype
]
# forward propagate coarser annotations
# add a prefix (matching with the original level of coarseness) to the
# forward-propagated annotations
for celltype in consensus_df.index:
highest_res = consensus_df.loc[celltype, "highest_res"]
# go to next level of loop if the highest_res is nan
if not type(highest_res) == pd.Series:
if pd.isna(highest_res):
continue
for i in range(highest_res + 1, max_level + 1):
consensus_df.loc[celltype, "level_" + str(i)] = celltype
return consensus_df
def create_orig_ann_to_consensus_translation_df(
adata,
consensus_df,
harmonizing_df,
verbose=True,
number_of_levels=5,
ontology_type="cell_type",
):
"""returns a dataframe. The indices of the dataframe correspond to original
labels from the dataset, with the study name as a prefix (as it appears in
the input adata.obs.study), the columns contain information on the
consensus translation. I.e. a translation at each level, whether or not the
cell type is 'new', and what the level of the annotation is.
ontology_type <"cell_type","anatomical_region_coarse","anatomical_region_fine"> - type of ontology
"""
# list the studies of interest. This name should correspond to the
# study name in the anndata object, 'study' column.
# store the column name in the "cell type harmonization table" (google doc)
# that corresponds to the study, in a dictionary:
study_cat = "study"
studies = set(adata.obs[study_cat])
if ontology_type == "cell_type":
harm_colnames = {study:study for study in studies}
elif ontology_type == "anatomical_region_coarse":
harm_colnames = {study: study + "_coarse" for study in studies}
elif ontology_type == "anatomical_region_fine":
harm_colnames = {study: study + "_fine" for study in studies}
else:
raise ValueError(
"ontology_type must be set to either cell_type, anatomical_region_coarse or anatomical_region_fine. Exiting"
)
# store set of studies
original_annotation_names = {
"cell_type": "original_celltype_ann",
"anatomical_region_coarse": "anatomical_region_coarse",
"anatomical_region_fine": "anatomical_region_detailed",
}
original_annotation_name = original_annotation_names[ontology_type]
original_annotations_prefixed = sorted(
set(
[
cell_study + "_" + ann
for cell_study, ann in zip(
adata.obs[study_cat], adata.obs[original_annotation_name]
)
if str(ann) != "nan"
]
)
)
level_names = [
"level" + "_" + str(level_number)
for level_number in range(1, number_of_levels + 1)
]
translation_df = pd.DataFrame(
index=original_annotations_prefixed,
columns=level_names + ["new", "highest_res"],
)
for study in studies:
harm_colname = harm_colnames[study]
if verbose:
print("working on study " + study + "...")
# get cell type names, remove nan and None from list:
cell_types_original = sorted(
set(
[
cell
for cell, cell_study in zip(
adata.obs[original_annotation_name], adata.obs[study_cat]
)
if cell_study == study
and str(cell) != "nan"
and str(cell) != "None"
]
)
)
for label in cell_types_original:
if verbose:
print(label)
# add study prefix for output, in that way we can distinguish
# between identical labels in different studies
label_prefixed = study + "_" + label
# if the label is 'nan', skip this row
if label == "nan":
continue
# get the rows in which this label appears in the study column
# of the harmonizing_df:
ref_rows = harmonizing_df[harm_colname][
harmonizing_df[harm_colname] == label
].index.tolist()
# if the label does not occur, there is a discrepancy between the
# labels in adata and the labels in the harmonizing table made
# by Martijn and Sasha.
if len(ref_rows) == 0:
print(
"HEY THERE ARE NO ROWS IN THE harmonizing_df WITH THIS LABEL ({}) AS ENTRY!".format(
label
)
)
# if the label occurs twice, there is some ambiguity as to how to
# translate this label to a consensus label. In that case, translate
# to the finest level that is identical in consensus translation
# between the multiple rows.
elif len(ref_rows) > 1:
print(
"HEY THE NUMBER OF ROWS WITH ENTRY {} IS NOT 1 but {}!".format(
label, str(len(ref_rows))
)
)
# get matching indices from consensus_df:
consensus_idc = list()
for i in range(len(ref_rows)):
consensus_idc.append(
consensus_df[
[
ref_rows[i] in index_list
for index_list in consensus_df["harmonizing_df_index"]
]
].index[0]
)
# now get the translations for both cases:
df_label_subset = consensus_df.loc[consensus_idc, :]
# store only labels that are common among all rows with this label:
for level in range(1, number_of_levels + 1):
col = "level_" + str(level)
n_translations = len(set(df_label_subset.loc[:, col]))
if n_translations == 1:
# update finest annotation
finest_annotation = df_label_subset.loc[consensus_idc[0], col]
# update finest annotation level
finest_level = level
# store current annotation at current level
translation_df.loc[label_prefixed, col] = finest_annotation
# if level labels differ between instances, store None for this level
else:
translation_df.loc[label_prefixed, col] = (
str(finest_level) + "_" + finest_annotation
)
translation_df.loc[label_prefixed, "highest_res"] = finest_level
# set "new" to false, since we fell back to a common annotation:
translation_df.loc[label_prefixed, "new"] = False
# add ref rows to harmonizing_df_index?
# ...
# if there is only one row with this label, copy the consensus labels
# from the same row to the translation df.
else:
consensus_idx = consensus_df[
[
ref_rows[0] in index_list
for index_list in consensus_df["harmonizing_df_index"]
]
]
if len(consensus_idx) == 0:
raise ValueError(f"label {label} does not have any reference label in your harmonizing df! Exiting.")
consensus_idx = consensus_idx.index[
0
] # loc[label,'harmonizing_df_index'][0]
translation_df.loc[label_prefixed, :] = consensus_df.loc[
consensus_idx, :
]
if verbose:
print("Done!")
return translation_df
def consensus_annotate_anndata(
adata, translation_df, verbose=True, max_ann_level=5, ontology_type="cell_type"
):
"""annotates cells in adata with consensus annotation. Returns adata."""
# list the studies of interest. This name should correspond to the
# study name in the anndata object, 'study' column.
# get name of adata.obs column with original annotations:
original_annotation_names = {
"cell_type": "original_celltype_ann",
"anatomical_region_coarse": "anatomical_region_coarse",
"anatomical_region_fine": "anatomical_region_detailed",
}
original_annotation_name = original_annotation_names[ontology_type]
# add prefix to original annotations, so that we can distinguish between
# datasets:
adata.obs[original_annotation_name + "_prefixed"] = [
cell_study + "_" + ann if str(ann) != "nan" else np.nan
for cell_study, ann in zip(adata.obs.study, adata.obs[original_annotation_name])
]
# specify the columns to copy:
col_to_copy = [
"level_" + str(level_number) for level_number in range(1, max_ann_level + 1)
]
col_to_copy = col_to_copy + ["highest_res", "new"]
# add prefix for in adata.obs:
prefixes = {
"cell_type": "ann_",
"anatomical_region_coarse": "region_coarse_",
"anatomical_region_fine": "region_fine_",
}
prefix = prefixes[ontology_type]
col_to_copy_new_names = [prefix + name for name in col_to_copy]
# add these columns to adata:
for col_name_old, col_name_new in zip(col_to_copy, col_to_copy_new_names):
translation_dict = dict(zip(translation_df.index, translation_df[col_name_old]))
adata.obs[col_name_new] = adata.obs[original_annotation_name + "_prefixed"].map(
translation_dict
)
adata.obs.drop([original_annotation_name + "_prefixed"], axis=1, inplace=True)
return adata
# ANATOMICAL REGION HARMONIZATION:
def merge_anatomical_annotations(ann_coarse, ann_fine):
"""Takes in two same-level annotation, for coarse and
fine annotation, returns the finest annotation.
To use on vectors, do:
np.vectorize(merge_anatomical_annotations)(
vector_coarse, vector_fine
)
"""
if utils.check_if_nan(ann_coarse):
ann_coarse_annotated = False
elif ann_coarse[0].isdigit() and ann_coarse[1] == "_":
ann_coarse_annotated = ann_coarse[0]
else:
ann_coarse_annotated = True
if utils.check_if_nan(ann_fine):
ann_fine_annotated = False
elif ann_fine[0].isdigit() and ann_fine[1] == "_":
ann_fine_annotated = ann_fine[0]
else:
ann_fine_annotated = True
# if finely annotated, return fine annotation
if ann_fine_annotated == True:
return ann_fine
# if only coarse is annotated, return coarse annotation
elif ann_coarse_annotated == True:
return ann_coarse
# if both are not annotated, return np.nan
elif ann_coarse_annotated == False and ann_fine_annotated == False:
return np.nan
# if only one is not annotated, return the other:
elif ann_coarse_annotated == False:
return ann_fine
elif ann_fine_annotated == False:
return ann_coarse
# if one or both are under-annotated (i.e. have
# forward propagated annotations from higher levels),
# choose the one with the highest prefix
elif ann_coarse_annotated > ann_fine_annotated:
return ann_coarse
elif ann_fine_annotated > ann_coarse_annotated:
return ann_fine
elif ann_fine_annotated == ann_coarse_annotated:
if ann_coarse == ann_fine:
return ann_fine
else:
raise ValueError(
"Contradicting annotations. ann_coarse: {}, ann_fine: {}".format(
ann_coarse, ann_fine
)
)
else:
raise ValueError(
"Something most have gone wrong. ann_coarse: {}, ann_fine: {}".format(
ann_coarse, ann_fine
)
)
def merge_coarse_and_fine_anatomical_ontology_anns(
adata, remove_harm_coarse_and_fine_original=False, n_levels=3
):
"""takes in an adata with in its obs: anatomical_region_coarse_level_[n]
and anatomical_region_fine_level_[n] and merges those for n_levels.
Returns adata with merged annotation under anatomical_region_level_[n].
Removes coarse and fine original harmonizations if
remove_harm_coarse_and_fine_original is set to True."""
for lev in range(1, n_levels + 1):
adata.obs["anatomical_region_level_" + str(lev)] = np.vectorize(
merge_anatomical_annotations
)(
adata.obs["region_coarse_level_" + str(lev)],
adata.obs["region_fine_level_" + str(lev)],
)
adata.obs["anatomical_region_highest_res"] = np.vectorize(max)(
adata.obs["region_coarse_highest_res"], adata.obs["region_fine_highest_res"]
)
if remove_harm_coarse_and_fine_original:
for lev in range(1, n_levels + 1):
del adata.obs["region_coarse_level_" + str(lev)]
del adata.obs["region_fine_level_" + str(lev)]
del adata.obs["region_coarse_highest_res"]
del adata.obs["region_fine_highest_res"]
del adata.obs["region_coarse_new"]
del adata.obs["region_fine_new"]
return adata
def add_clean_annotation(adata, max_level=5):
"""converts ann_level_[annotation level] to annotation without label
propagation from lower levels. I.e. in level 2, we will not have 1_Epithelial
anymore; instead cells without level 2 annotations will have annotation None.
Returns adata with extra annotation levels.
"""
for level in range(1, max_level + 1):
level_name = "ann_level_" + str(level)
anns = sorted(set(adata.obs[level_name]))
ann2pureann = dict(zip(anns, anns))
for ann_name in ann2pureann.keys():
if ann_name[:2] in ["1_", "2_", "3_", "4_"]:
ann2pureann[ann_name] = None
adata.obs[level_name + "_clean"] = adata.obs[level_name].map(ann2pureann)
return adata
def add_anatomical_region_ccf_score(adata, harmonizing_df):
"""
Adds ccf score according to mapping in harmoinzing_df. Uses
adata.obs.anatomical_region_level_1 and adata.obs.anatomical_region_level_2
for mapping.
Returns annotated adata
"""
an_region_l1_to_ccf_score = dict()
an_region_l2_to_ccf_score = dict()
for row, score in enumerate(harmonizing_df.continuous_score_upper_and_lower):
if not pd.isnull(score):
level_1_label = harmonizing_df["Level_1"][row]
level_2_label = harmonizing_df["Level_2"][row]
if not pd.isnull(level_2_label):
an_region_l2_to_ccf_score[level_2_label] = score
elif not pd.isnull(level_1_label):
an_region_l1_to_ccf_score[level_1_label] = score
l1_ccf_scores = adata.obs.anatomical_region_level_1.map(an_region_l1_to_ccf_score)
l2_ccf_scores = adata.obs.anatomical_region_level_2.map(an_region_l2_to_ccf_score)
# sanity checks:
n_unannotated = sum(
[
pd.isnull(l1_ccf) and pd.isnull(l2_ccf)
for l1_ccf, l2_ccf in zip(l1_ccf_scores, l2_ccf_scores)
]
)
n_annotated_double = sum(
[
(pd.isnull(l1_ccf) == False and pd.isnull(l2_ccf) == False)
for l1_ccf, l2_ccf in zip(l1_ccf_scores, l2_ccf_scores)
]
)
if n_unannotated > 0:
raise ValueError(
"There are cells whose anatomical region don't correspond to any of your ccf keys. Exiting."
)
if n_annotated_double > 0:
raise ValueError(
"There are cells that map to two different ccf values. Exiting."
)
adata.obs["anatomical_region_ccf_score"] = l1_ccf_scores
adata.obs.loc[
pd.isnull(l1_ccf_scores), "anatomical_region_ccf_score"
] = l2_ccf_scores[
|
pd.isnull(l1_ccf_scores)
|
pandas.isnull
|
from Functions.ParseFunctions import *
import Functions.AdminFunctions as AF
import pandas as pd
#-------------- NESTED PATH CORRECTION --------------------------------#
import os, re, sys
# For all script files, we add the parent directory to the system path
cwd = re.sub(r"[\\]", "/", os.getcwd())
cwd_list = cwd.split("/")
path = sys.argv[0]
path_list = path.split("/")
# either the entire filepath is entered as command i python
if cwd_list[0:3] == path_list[0:3]:
full_path = path
# or a relative path is entered, in which case we append the path to the cwd_path
else:
full_path = cwd + "/" + path
# remove the overlap
root_dir = re.search(r"(^.+HTML-projektet)", full_path).group(1)
sys.path.append(root_dir)
#----------------------------------------------------------------------#
class Journal():
def __init__(self, journal_abb):
self.name = AF.get_journal_name(journal_abb)
self.abbreviation = journal_abb
self._fp = AF.get_journal_dir(journal_abb)
self.ref_df = pd.read_excel(f"{self._fp}/article_references.xlsx")
def _get_toc_filenames(self):
return os.listdir(f"{self._fp}/Tables_of_Contents")
def list_years(self, type="df"):
l = sorted(extract_from_filenames(self._get_toc_filenames(), "year"))
if type in ['df']:
return pd.DataFrame({'year': l})
elif type in ['list']:
return l
else:
list_method_format_error()
def list_volumes(self, type="df"):
l = sorted(extract_from_filenames(self._get_toc_filenames(), "volume"))
if type in ['df']:
return
|
pd.DataFrame({'volume': l})
|
pandas.DataFrame
|
from flask import Flask, render_template, request, jsonify, redirect
import pandas as pd
import numpy as np
from scipy import sparse
import pickle5 as pickle
## For CB model
def weighted_star_rating(x):
v = x['review_count']
R = x['stars']
return (v/(v+m) * R) + (m/(m+v) * C)
df = pd.read_csv('data/CA_trails.csv')
m = df['review_count'].quantile(q=0.95)
C = df['stars'].mean()
df_Q = df[df['review_count'] > m].copy()
df_Q['WSR'] = df_Q.apply(weighted_star_rating, axis=1)
top_10 = df_Q.sort_values('WSR', ascending = False)[:10] ## Return this for top 10 in CA
def pop_chart_per_region(region):
df_region = df[df['location']==region]
m = df['review_count'].quantile(q=0.70)
C = df['stars'].mean()
df_Q = df_region[ df_region['review_count'] > m]
df_Q['WSR'] = df_Q.apply(weighted_star_rating, axis=1)
top_5_in_region = df_Q.sort_values('WSR', ascending = False)[:5]
return top_5_in_region
PT_feature_cosine_sim = np.loadtxt('data/feature_sim.txt', delimiter =',')
cosine_sim = np.loadtxt('data/text_sim.txt', delimiter =',')
def get_recommendations_by_text_sim(idx, top_n=5):
sim_scores = list(enumerate(cosine_sim[idx]))
sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)
sim_scores = sim_scores[1:top_n+1]
trail_indices = [i[0] for i in sim_scores]
result = df.iloc[trail_indices]
return result
def get_recommendations_by_feature_sim(idx, top_n=5):
sim_scores = list(enumerate(PT_feature_cosine_sim[idx]))
sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)
sim_scores = sim_scores[1:top_n+1]
trail_indices = [i[0] for i in sim_scores]
result = df.iloc[trail_indices]
return result
def get_recommendations_by_hybrid_sim(idx, top_n=5):
hybrid_cosine_sim = 0.5*PT_feature_cosine_sim + 0.5*cosine_sim
sim_scores = list(enumerate(hybrid_cosine_sim[idx]))
sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)
sim_scores = sim_scores[1:top_n+1]
trail_indices = [i[0] for i in sim_scores]
result = df.iloc[trail_indices]
return result
order = ['name', 'location', 'stars','distance','elevation','duration','difficulty']
## CF model
trail_indices =
|
pd.read_csv('data/knn_trail_indices.csv')
|
pandas.read_csv
|
import sys
import csv
import pandas as pd
import numpy as np
def score (keyFileName, responseFileName):
with open(keyFileName, 'r') as keyFile:
reader = csv.reader(keyFile)
key = []
for line in reader:
key.append(line)
with open(responseFileName, 'r') as responseFile:
response = responseFile.readlines()
if len(key) != len(response):
print("length mismatch between key and submitted file")
exit()
categories = ["World", "Sports", "Business", "Sci/Tech"]
correct = 0
incorrect = 0
matrix = np.zeros((len(categories), len(categories)), dtype=np.int8)
confusion_matrix =
|
pd.DataFrame(matrix, columns=categories, index=categories)
|
pandas.DataFrame
|
""" Test cases for DataFrame.plot """
import string
import warnings
import numpy as np
import pytest
import pandas.util._test_decorators as td
import pandas as pd
from pandas import (
DataFrame,
Series,
date_range,
)
import pandas._testing as tm
from pandas.tests.plotting.common import TestPlotBase
from pandas.io.formats.printing import pprint_thing
pytestmark = pytest.mark.slow
@td.skip_if_no_mpl
class TestDataFramePlotsSubplots(TestPlotBase):
def test_subplots(self):
df = DataFrame(np.random.rand(10, 3), index=list(string.ascii_letters[:10]))
for kind in ["bar", "barh", "line", "area"]:
axes = df.plot(kind=kind, subplots=True, sharex=True, legend=True)
self._check_axes_shape(axes, axes_num=3, layout=(3, 1))
assert axes.shape == (3,)
for ax, column in zip(axes, df.columns):
self._check_legend_labels(ax, labels=[pprint_thing(column)])
for ax in axes[:-2]:
self._check_visible(ax.xaxis) # xaxis must be visible for grid
self._check_visible(ax.get_xticklabels(), visible=False)
if kind != "bar":
# change https://github.com/pandas-dev/pandas/issues/26714
self._check_visible(ax.get_xticklabels(minor=True), visible=False)
self._check_visible(ax.xaxis.get_label(), visible=False)
self._check_visible(ax.get_yticklabels())
self._check_visible(axes[-1].xaxis)
self._check_visible(axes[-1].get_xticklabels())
self._check_visible(axes[-1].get_xticklabels(minor=True))
self._check_visible(axes[-1].xaxis.get_label())
self._check_visible(axes[-1].get_yticklabels())
axes = df.plot(kind=kind, subplots=True, sharex=False)
for ax in axes:
self._check_visible(ax.xaxis)
self._check_visible(ax.get_xticklabels())
self._check_visible(ax.get_xticklabels(minor=True))
self._check_visible(ax.xaxis.get_label())
self._check_visible(ax.get_yticklabels())
axes = df.plot(kind=kind, subplots=True, legend=False)
for ax in axes:
assert ax.get_legend() is None
def test_subplots_timeseries(self):
idx = date_range(start="2014-07-01", freq="M", periods=10)
df = DataFrame(np.random.rand(10, 3), index=idx)
for kind in ["line", "area"]:
axes = df.plot(kind=kind, subplots=True, sharex=True)
self._check_axes_shape(axes, axes_num=3, layout=(3, 1))
for ax in axes[:-2]:
# GH 7801
self._check_visible(ax.xaxis) # xaxis must be visible for grid
self._check_visible(ax.get_xticklabels(), visible=False)
self._check_visible(ax.get_xticklabels(minor=True), visible=False)
self._check_visible(ax.xaxis.get_label(), visible=False)
self._check_visible(ax.get_yticklabels())
self._check_visible(axes[-1].xaxis)
self._check_visible(axes[-1].get_xticklabels())
self._check_visible(axes[-1].get_xticklabels(minor=True))
self._check_visible(axes[-1].xaxis.get_label())
self._check_visible(axes[-1].get_yticklabels())
self._check_ticks_props(axes, xrot=0)
axes = df.plot(kind=kind, subplots=True, sharex=False, rot=45, fontsize=7)
for ax in axes:
self._check_visible(ax.xaxis)
self._check_visible(ax.get_xticklabels())
self._check_visible(ax.get_xticklabels(minor=True))
self._check_visible(ax.xaxis.get_label())
self._check_visible(ax.get_yticklabels())
self._check_ticks_props(ax, xlabelsize=7, xrot=45, ylabelsize=7)
def test_subplots_timeseries_y_axis(self):
# GH16953
data = {
"numeric": np.array([1, 2, 5]),
"timedelta": [
|
pd.Timedelta(-10, unit="s")
|
pandas.Timedelta
|
from sklearn.metrics import matthews_corrcoef
import pandas as pd
infile_preds = "preds_mod_v1.csv"
preds = pd.read_csv(infile_preds,index_col=0)
actual = pd.read_csv("data/test.csv",index_col=0)
preds["predictions"][preds["predictions"] > 0.5] = 1.0
preds["predictions"][preds["predictions"] < 0.51] = 0.0
all_df = pd.concat([preds,actual],axis=1)
print("MCC for %s: %.3f" % (infile_preds,matthews_corrcoef(all_df["predictions"],all_df["target"])))
infile_preds = "preds_mod_v2.csv"
preds = pd.read_csv(infile_preds,index_col=0)
actual = pd.read_csv("data/test.csv",index_col=0)
preds["predictions"][preds["predictions"] > 0.5] = 1.0
preds["predictions"][preds["predictions"] < 0.51] = 0.0
all_df = pd.concat([preds,actual],axis=1)
print("MCC for %s: %.3f" % (infile_preds,matthews_corrcoef(all_df["predictions"],all_df["target"])))
infile_preds = "preds_mod_v3.csv"
preds =
|
pd.read_csv(infile_preds,index_col=0)
|
pandas.read_csv
|
"""Provides access to *one* setting of the the IBM benchmarking data.
The challenge data and further explanations of the corresponding
data can be found in [2]. The authors also provide a rough evaluation guideline
in their paper [1], which unfortunately has not been maintained since publication.
The DGP is based on the same covariates as the ACIC2018 [3] challenge, which we hope to
implement as a data set in a future version.
References:
[1] <NAME>, <NAME>, <NAME>, and <NAME>,
“Benchmarking Framework for Performance-Evaluation of Causal Inference Analysis,”
2018.
[2] Data Set Download: https://www.synapse.org/#!Synapse:syn11738767/wiki/512854
[3] ACIC2018 challenge: https://www.synapse.org/#!Synapse:syn11294478/wiki/494269
"""
from typing import List, Optional
import pandas as pd
from ..frames import CausalFrame, Col
from ..transport import get_covariates_df, get_outcomes_df
from ..utils import (
Indices,
add_pot_outcomes_if_missing,
select_replication,
to_rep_list,
)
DATASET_NAME: str = "ibm"
def load_ibm(select_rep: Optional[Indices] = None) -> List[CausalFrame]:
"""Provides the IBM benchmarking data in the common JustCause format.
BEWARE: the replications have different sizes and should be used with caution.
Args:
select_rep: the desired replications
Returns:
data: list of CausalFrames, one for each replication
"""
covariates = get_covariates_df(DATASET_NAME)
outcomes = get_outcomes_df(DATASET_NAME)
if select_rep is not None:
outcomes = select_replication(outcomes, select_rep)
full =
|
pd.merge(covariates, outcomes, on=Col.sample_id)
|
pandas.merge
|
"""
Copyright 2022 HSBC Global Asset Management (Deutschland) GmbH
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
"""
import numpy as np
import pandas as pd
import pytest
import pyratings as rtg
from tests import conftest
@pytest.fixture(scope="session")
def rtg_inputs_longterm():
return pd.DataFrame(
data={
"rtg_sp": ["AAA", "AA-", "AA+", "BB-", "C", np.nan, "BBB+", "AA"],
"rtg_moody": ["Aa1", "Aa3", "Aa2", "Ba3", "Ca", np.nan, np.nan, "Aa2"],
"rtg_fitch": ["AA-", np.nan, "AA-", "B+", "C", np.nan, np.nan, "AA"],
}
)
@pytest.fixture(scope="session")
def rtg_inputs_shortterm():
return pd.DataFrame(
data={
"rtg_sp": ["A-1", "A-3", "A-1+", "D", "B", np.nan, "A-2", "A-3"],
"rtg_moody": ["P-2", "NP", "P-1", "NP", "P-3", np.nan, np.nan, "P-3"],
"rtg_fitch": ["F1", np.nan, "F1", "F3", "F3", np.nan, np.nan, "F3"],
}
)
def test_get_best_rating_longterm_with_explicit_rating_provider(rtg_inputs_longterm):
"""Test computation of best ratings on a security (line-by-line) basis."""
actual = rtg.get_best_ratings(
rtg_inputs_longterm,
rating_provider_input=["SP", "Moody", "Fitch"],
tenor="long-term",
)
expectations = pd.Series(
data=["AAA", "AA-", "AA+", "BB-", "CC", np.nan, "BBB+", "AA"], name="best_rtg"
)
pd.testing.assert_series_equal(actual, expectations)
def test_get_best_rating_longterm_with_inferring_rating_provider(rtg_inputs_longterm):
"""Test computation of best ratings on a security (line-by-line) basis."""
actual = rtg.get_best_ratings(rtg_inputs_longterm, tenor="long-term")
expectations = pd.Series(
data=["AAA", "AA-", "AA+", "BB-", "CC", np.nan, "BBB+", "AA"], name="best_rtg"
)
pd.testing.assert_series_equal(actual, expectations)
def test_get_best_rating_shortterm_with_explicit_rating_provider(rtg_inputs_shortterm):
"""Test computation of best ratings on a security (line-by-line) basis."""
actual = rtg.get_best_ratings(
rtg_inputs_shortterm,
rating_provider_input=["SP", "Moody", "Fitch"],
tenor="short-term",
)
expectations = pd.Series(
data=["A-1", "A-3", "A-1+", "A-3", "A-3", np.nan, "A-2", "A-3"], name="best_rtg"
)
pd.testing.assert_series_equal(actual, expectations)
def test_get_best_rating_shortterm_with_inferring_rating_provider(rtg_inputs_shortterm):
"""Test computation of best ratings on a security (line-by-line) basis."""
actual = rtg.get_best_ratings(rtg_inputs_shortterm, tenor="short-term")
expectations = pd.Series(
data=["A-1", "A-3", "A-1+", "A-3", "A-3", np.nan, "A-2", "A-3"], name="best_rtg"
)
pd.testing.assert_series_equal(actual, expectations)
def test_get_second_best_rating_longterm_with_explicit_rating_provider(
rtg_inputs_longterm,
):
"""Test computation of second-best ratings on a security (line-by-line) basis."""
actual = rtg.get_second_best_ratings(
rtg_inputs_longterm,
rating_provider_input=["SP", "Moody", "Fitch"],
tenor="long-term",
)
expectations = pd.Series(
data=["AA+", "AA-", "AA", "BB-", "C", np.nan, "BBB+", "AA"],
name="second_best_rtg",
)
pd.testing.assert_series_equal(actual, expectations)
def test_get_second_best_rating_longterm_with_inferring_rating_provider(
rtg_inputs_longterm,
):
"""Test computation of best ratings on a security (line-by-line) basis."""
actual = rtg.get_second_best_ratings(rtg_inputs_longterm, tenor="long-term")
expectations = pd.Series(
data=["AA+", "AA-", "AA", "BB-", "C", np.nan, "BBB+", "AA"],
name="second_best_rtg",
)
pd.testing.assert_series_equal(actual, expectations)
def test_get_second_best_rating_shortterm_with_explicit_rating_provider(
rtg_inputs_shortterm,
):
"""Test computation of best ratings on a security (line-by-line) basis."""
actual = rtg.get_second_best_ratings(
rtg_inputs_shortterm,
rating_provider_input=["SP", "Moody", "Fitch"],
tenor="short-term",
)
expectations = pd.Series(
data=["A-1", "B", "A-1+", "B", "A-3", np.nan, "A-2", "A-3"],
name="second_best_rtg",
)
pd.testing.assert_series_equal(actual, expectations)
def test_get_second_best_rating_shortterm_with_inferring_rating_provider(
rtg_inputs_shortterm,
):
"""Test computation of best ratings on a security (line-by-line) basis."""
actual = rtg.get_second_best_ratings(rtg_inputs_shortterm, tenor="short-term")
expectations = pd.Series(
data=["A-1", "B", "A-1+", "B", "A-3", np.nan, "A-2", "A-3"],
name="second_best_rtg",
)
|
pd.testing.assert_series_equal(actual, expectations)
|
pandas.testing.assert_series_equal
|
import tempfile
import copy
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import pandas as pd
try:
from scipy.spatial import distance
from scipy.cluster import hierarchy
_no_scipy = False
except ImportError:
_no_scipy = True
try:
import fastcluster
assert fastcluster
_no_fastcluster = False
except ImportError:
_no_fastcluster = True
import numpy.testing as npt
try:
import pandas.testing as pdt
except ImportError:
import pandas.util.testing as pdt
import pytest
from .. import matrix as mat
from .. import color_palette
from .._testing import assert_colors_equal
class TestHeatmap:
rs = np.random.RandomState(sum(map(ord, "heatmap")))
x_norm = rs.randn(4, 8)
letters = pd.Series(["A", "B", "C", "D"], name="letters")
df_norm = pd.DataFrame(x_norm, index=letters)
x_unif = rs.rand(20, 13)
df_unif = pd.DataFrame(x_unif)
default_kws = dict(vmin=None, vmax=None, cmap=None, center=None,
robust=False, annot=False, fmt=".2f", annot_kws=None,
cbar=True, cbar_kws=None, mask=None)
def test_ndarray_input(self):
p = mat._HeatMapper(self.x_norm, **self.default_kws)
npt.assert_array_equal(p.plot_data, self.x_norm)
pdt.assert_frame_equal(p.data, pd.DataFrame(self.x_norm))
npt.assert_array_equal(p.xticklabels, np.arange(8))
npt.assert_array_equal(p.yticklabels, np.arange(4))
assert p.xlabel == ""
assert p.ylabel == ""
def test_df_input(self):
p = mat._HeatMapper(self.df_norm, **self.default_kws)
npt.assert_array_equal(p.plot_data, self.x_norm)
pdt.assert_frame_equal(p.data, self.df_norm)
npt.assert_array_equal(p.xticklabels, np.arange(8))
npt.assert_array_equal(p.yticklabels, self.letters.values)
assert p.xlabel == ""
assert p.ylabel == "letters"
def test_df_multindex_input(self):
df = self.df_norm.copy()
index = pd.MultiIndex.from_tuples([("A", 1), ("B", 2),
("C", 3), ("D", 4)],
names=["letter", "number"])
index.name = "letter-number"
df.index = index
p = mat._HeatMapper(df, **self.default_kws)
combined_tick_labels = ["A-1", "B-2", "C-3", "D-4"]
npt.assert_array_equal(p.yticklabels, combined_tick_labels)
assert p.ylabel == "letter-number"
p = mat._HeatMapper(df.T, **self.default_kws)
npt.assert_array_equal(p.xticklabels, combined_tick_labels)
assert p.xlabel == "letter-number"
@pytest.mark.parametrize("dtype", [float, np.int64, object])
def test_mask_input(self, dtype):
kws = self.default_kws.copy()
mask = self.x_norm > 0
kws['mask'] = mask
data = self.x_norm.astype(dtype)
p = mat._HeatMapper(data, **kws)
plot_data = np.ma.masked_where(mask, data)
npt.assert_array_equal(p.plot_data, plot_data)
def test_mask_limits(self):
"""Make sure masked cells are not used to calculate extremes"""
kws = self.default_kws.copy()
mask = self.x_norm > 0
kws['mask'] = mask
p = mat._HeatMapper(self.x_norm, **kws)
assert p.vmax == np.ma.array(self.x_norm, mask=mask).max()
assert p.vmin == np.ma.array(self.x_norm, mask=mask).min()
mask = self.x_norm < 0
kws['mask'] = mask
p = mat._HeatMapper(self.x_norm, **kws)
assert p.vmin == np.ma.array(self.x_norm, mask=mask).min()
assert p.vmax == np.ma.array(self.x_norm, mask=mask).max()
def test_default_vlims(self):
p = mat._HeatMapper(self.df_unif, **self.default_kws)
assert p.vmin == self.x_unif.min()
assert p.vmax == self.x_unif.max()
def test_robust_vlims(self):
kws = self.default_kws.copy()
kws["robust"] = True
p = mat._HeatMapper(self.df_unif, **kws)
assert p.vmin == np.percentile(self.x_unif, 2)
assert p.vmax == np.percentile(self.x_unif, 98)
def test_custom_sequential_vlims(self):
kws = self.default_kws.copy()
kws["vmin"] = 0
kws["vmax"] = 1
p = mat._HeatMapper(self.df_unif, **kws)
assert p.vmin == 0
assert p.vmax == 1
def test_custom_diverging_vlims(self):
kws = self.default_kws.copy()
kws["vmin"] = -4
kws["vmax"] = 5
kws["center"] = 0
p = mat._HeatMapper(self.df_norm, **kws)
assert p.vmin == -4
assert p.vmax == 5
def test_array_with_nans(self):
x1 = self.rs.rand(10, 10)
nulls = np.zeros(10) * np.nan
x2 = np.c_[x1, nulls]
m1 = mat._HeatMapper(x1, **self.default_kws)
m2 = mat._HeatMapper(x2, **self.default_kws)
assert m1.vmin == m2.vmin
assert m1.vmax == m2.vmax
def test_mask(self):
df = pd.DataFrame(data={'a': [1, 1, 1],
'b': [2, np.nan, 2],
'c': [3, 3, np.nan]})
kws = self.default_kws.copy()
kws["mask"] = np.isnan(df.values)
m = mat._HeatMapper(df, **kws)
npt.assert_array_equal(np.isnan(m.plot_data.data),
m.plot_data.mask)
def test_custom_cmap(self):
kws = self.default_kws.copy()
kws["cmap"] = "BuGn"
p = mat._HeatMapper(self.df_unif, **kws)
assert p.cmap == mpl.cm.BuGn
def test_centered_vlims(self):
kws = self.default_kws.copy()
kws["center"] = .5
p = mat._HeatMapper(self.df_unif, **kws)
assert p.vmin == self.df_unif.values.min()
assert p.vmax == self.df_unif.values.max()
def test_default_colors(self):
vals = np.linspace(.2, 1, 9)
cmap = mpl.cm.binary
ax = mat.heatmap([vals], cmap=cmap)
fc = ax.collections[0].get_facecolors()
cvals = np.linspace(0, 1, 9)
npt.assert_array_almost_equal(fc, cmap(cvals), 2)
def test_custom_vlim_colors(self):
vals = np.linspace(.2, 1, 9)
cmap = mpl.cm.binary
ax = mat.heatmap([vals], vmin=0, cmap=cmap)
fc = ax.collections[0].get_facecolors()
npt.assert_array_almost_equal(fc, cmap(vals), 2)
def test_custom_center_colors(self):
vals = np.linspace(.2, 1, 9)
cmap = mpl.cm.binary
ax = mat.heatmap([vals], center=.5, cmap=cmap)
fc = ax.collections[0].get_facecolors()
npt.assert_array_almost_equal(fc, cmap(vals), 2)
def test_cmap_with_properties(self):
kws = self.default_kws.copy()
cmap = copy.copy(mpl.cm.get_cmap("BrBG"))
cmap.set_bad("red")
kws["cmap"] = cmap
hm = mat._HeatMapper(self.df_unif, **kws)
npt.assert_array_equal(
cmap(np.ma.masked_invalid([np.nan])),
hm.cmap(np.ma.masked_invalid([np.nan])))
kws["center"] = 0.5
hm = mat._HeatMapper(self.df_unif, **kws)
npt.assert_array_equal(
cmap(np.ma.masked_invalid([np.nan])),
hm.cmap(np.ma.masked_invalid([np.nan])))
kws = self.default_kws.copy()
cmap = copy.copy(mpl.cm.get_cmap("BrBG"))
cmap.set_under("red")
kws["cmap"] = cmap
hm = mat._HeatMapper(self.df_unif, **kws)
npt.assert_array_equal(cmap(-np.inf), hm.cmap(-np.inf))
kws["center"] = .5
hm = mat._HeatMapper(self.df_unif, **kws)
npt.assert_array_equal(cmap(-np.inf), hm.cmap(-np.inf))
kws = self.default_kws.copy()
cmap = copy.copy(mpl.cm.get_cmap("BrBG"))
cmap.set_over("red")
kws["cmap"] = cmap
hm = mat._HeatMapper(self.df_unif, **kws)
npt.assert_array_equal(cmap(-np.inf), hm.cmap(-np.inf))
kws["center"] = .5
hm = mat._HeatMapper(self.df_unif, **kws)
npt.assert_array_equal(cmap(np.inf), hm.cmap(np.inf))
def test_tickabels_off(self):
kws = self.default_kws.copy()
kws['xticklabels'] = False
kws['yticklabels'] = False
p = mat._HeatMapper(self.df_norm, **kws)
assert p.xticklabels == []
assert p.yticklabels == []
def test_custom_ticklabels(self):
kws = self.default_kws.copy()
xticklabels = list('iheartheatmaps'[:self.df_norm.shape[1]])
yticklabels = list('heatmapsarecool'[:self.df_norm.shape[0]])
kws['xticklabels'] = xticklabels
kws['yticklabels'] = yticklabels
p = mat._HeatMapper(self.df_norm, **kws)
assert p.xticklabels == xticklabels
assert p.yticklabels == yticklabels
def test_custom_ticklabel_interval(self):
kws = self.default_kws.copy()
xstep, ystep = 2, 3
kws['xticklabels'] = xstep
kws['yticklabels'] = ystep
p = mat._HeatMapper(self.df_norm, **kws)
nx, ny = self.df_norm.T.shape
npt.assert_array_equal(p.xticks, np.arange(0, nx, xstep) + .5)
npt.assert_array_equal(p.yticks, np.arange(0, ny, ystep) + .5)
npt.assert_array_equal(p.xticklabels,
self.df_norm.columns[0:nx:xstep])
npt.assert_array_equal(p.yticklabels,
self.df_norm.index[0:ny:ystep])
def test_heatmap_annotation(self):
ax = mat.heatmap(self.df_norm, annot=True, fmt=".1f",
annot_kws={"fontsize": 14})
for val, text in zip(self.x_norm.flat, ax.texts):
assert text.get_text() == "{:.1f}".format(val)
assert text.get_fontsize() == 14
def test_heatmap_annotation_overwrite_kws(self):
annot_kws = dict(color="0.3", va="bottom", ha="left")
ax = mat.heatmap(self.df_norm, annot=True, fmt=".1f",
annot_kws=annot_kws)
for text in ax.texts:
assert text.get_color() == "0.3"
assert text.get_ha() == "left"
assert text.get_va() == "bottom"
def test_heatmap_annotation_with_mask(self):
df = pd.DataFrame(data={'a': [1, 1, 1],
'b': [2, np.nan, 2],
'c': [3, 3, np.nan]})
mask = np.isnan(df.values)
df_masked = np.ma.masked_where(mask, df)
ax = mat.heatmap(df, annot=True, fmt='.1f', mask=mask)
assert len(df_masked.compressed()) == len(ax.texts)
for val, text in zip(df_masked.compressed(), ax.texts):
assert "{:.1f}".format(val) == text.get_text()
def test_heatmap_annotation_mesh_colors(self):
ax = mat.heatmap(self.df_norm, annot=True)
mesh = ax.collections[0]
assert len(mesh.get_facecolors()) == self.df_norm.values.size
plt.close("all")
def test_heatmap_annotation_other_data(self):
annot_data = self.df_norm + 10
ax = mat.heatmap(self.df_norm, annot=annot_data, fmt=".1f",
annot_kws={"fontsize": 14})
for val, text in zip(annot_data.values.flat, ax.texts):
assert text.get_text() == "{:.1f}".format(val)
assert text.get_fontsize() == 14
def test_heatmap_annotation_with_limited_ticklabels(self):
ax = mat.heatmap(self.df_norm, fmt=".2f", annot=True,
xticklabels=False, yticklabels=False)
for val, text in zip(self.x_norm.flat, ax.texts):
assert text.get_text() == "{:.2f}".format(val)
def test_heatmap_cbar(self):
f = plt.figure()
mat.heatmap(self.df_norm)
assert len(f.axes) == 2
plt.close(f)
f = plt.figure()
mat.heatmap(self.df_norm, cbar=False)
assert len(f.axes) == 1
plt.close(f)
f, (ax1, ax2) = plt.subplots(2)
mat.heatmap(self.df_norm, ax=ax1, cbar_ax=ax2)
assert len(f.axes) == 2
plt.close(f)
@pytest.mark.xfail(mpl.__version__ == "3.1.1",
reason="matplotlib 3.1.1 bug")
def test_heatmap_axes(self):
ax = mat.heatmap(self.df_norm)
xtl = [int(l.get_text()) for l in ax.get_xticklabels()]
assert xtl == list(self.df_norm.columns)
ytl = [l.get_text() for l in ax.get_yticklabels()]
assert ytl == list(self.df_norm.index)
assert ax.get_xlabel() == ""
assert ax.get_ylabel() == "letters"
assert ax.get_xlim() == (0, 8)
assert ax.get_ylim() == (4, 0)
def test_heatmap_ticklabel_rotation(self):
f, ax = plt.subplots(figsize=(2, 2))
mat.heatmap(self.df_norm, xticklabels=1, yticklabels=1, ax=ax)
for t in ax.get_xticklabels():
assert t.get_rotation() == 0
for t in ax.get_yticklabels():
assert t.get_rotation() == 90
plt.close(f)
df = self.df_norm.copy()
df.columns = [str(c) * 10 for c in df.columns]
df.index = [i * 10 for i in df.index]
f, ax = plt.subplots(figsize=(2, 2))
mat.heatmap(df, xticklabels=1, yticklabels=1, ax=ax)
for t in ax.get_xticklabels():
assert t.get_rotation() == 90
for t in ax.get_yticklabels():
assert t.get_rotation() == 0
plt.close(f)
def test_heatmap_inner_lines(self):
c = (0, 0, 1, 1)
ax = mat.heatmap(self.df_norm, linewidths=2, linecolor=c)
mesh = ax.collections[0]
assert mesh.get_linewidths()[0] == 2
assert tuple(mesh.get_edgecolor()[0]) == c
def test_square_aspect(self):
ax = mat.heatmap(self.df_norm, square=True)
obs_aspect = ax.get_aspect()
# mpl>3.3 returns 1 for setting "equal" aspect
# so test for the two possible equal outcomes
assert obs_aspect == "equal" or obs_aspect == 1
def test_mask_validation(self):
mask = mat._matrix_mask(self.df_norm, None)
assert mask.shape == self.df_norm.shape
assert mask.values.sum() == 0
with pytest.raises(ValueError):
bad_array_mask = self.rs.randn(3, 6) > 0
mat._matrix_mask(self.df_norm, bad_array_mask)
with pytest.raises(ValueError):
bad_df_mask = pd.DataFrame(self.rs.randn(4, 8) > 0)
mat._matrix_mask(self.df_norm, bad_df_mask)
def test_missing_data_mask(self):
data = pd.DataFrame(np.arange(4, dtype=float).reshape(2, 2))
data.loc[0, 0] = np.nan
mask = mat._matrix_mask(data, None)
npt.assert_array_equal(mask, [[True, False], [False, False]])
mask_in = np.array([[False, True], [False, False]])
mask_out = mat._matrix_mask(data, mask_in)
npt.assert_array_equal(mask_out, [[True, True], [False, False]])
def test_cbar_ticks(self):
f, (ax1, ax2) = plt.subplots(2)
mat.heatmap(self.df_norm, ax=ax1, cbar_ax=ax2,
cbar_kws=dict(drawedges=True))
assert len(ax2.collections) == 2
@pytest.mark.skipif(_no_scipy, reason="Test requires scipy")
class TestDendrogram:
rs = np.random.RandomState(sum(map(ord, "dendrogram")))
default_kws = dict(linkage=None, metric='euclidean', method='single',
axis=1, label=True, rotate=False)
x_norm = rs.randn(4, 8) + np.arange(8)
x_norm = (x_norm.T + np.arange(4)).T
letters = pd.Series(["A", "B", "C", "D", "E", "F", "G", "H"],
name="letters")
df_norm = pd.DataFrame(x_norm, columns=letters)
if not _no_scipy:
if _no_fastcluster:
x_norm_distances = distance.pdist(x_norm.T, metric='euclidean')
x_norm_linkage = hierarchy.linkage(x_norm_distances, method='single')
else:
x_norm_linkage = fastcluster.linkage_vector(x_norm.T,
metric='euclidean',
method='single')
x_norm_dendrogram = hierarchy.dendrogram(x_norm_linkage, no_plot=True,
color_threshold=-np.inf)
x_norm_leaves = x_norm_dendrogram['leaves']
df_norm_leaves = np.asarray(df_norm.columns[x_norm_leaves])
def test_ndarray_input(self):
p = mat._DendrogramPlotter(self.x_norm, **self.default_kws)
npt.assert_array_equal(p.array.T, self.x_norm)
pdt.assert_frame_equal(p.data.T, pd.DataFrame(self.x_norm))
npt.assert_array_equal(p.linkage, self.x_norm_linkage)
assert p.dendrogram == self.x_norm_dendrogram
npt.assert_array_equal(p.reordered_ind, self.x_norm_leaves)
npt.assert_array_equal(p.xticklabels, self.x_norm_leaves)
npt.assert_array_equal(p.yticklabels, [])
assert p.xlabel is None
assert p.ylabel == ''
def test_df_input(self):
p = mat._DendrogramPlotter(self.df_norm, **self.default_kws)
npt.assert_array_equal(p.array.T, np.asarray(self.df_norm))
pdt.assert_frame_equal(p.data.T, self.df_norm)
npt.assert_array_equal(p.linkage, self.x_norm_linkage)
assert p.dendrogram == self.x_norm_dendrogram
npt.assert_array_equal(p.xticklabels,
np.asarray(self.df_norm.columns)[
self.x_norm_leaves])
npt.assert_array_equal(p.yticklabels, [])
assert p.xlabel == 'letters'
assert p.ylabel == ''
def test_df_multindex_input(self):
df = self.df_norm.copy()
index = pd.MultiIndex.from_tuples([("A", 1), ("B", 2),
("C", 3), ("D", 4)],
names=["letter", "number"])
index.name = "letter-number"
df.index = index
kws = self.default_kws.copy()
kws['label'] = True
p = mat._DendrogramPlotter(df.T, **kws)
xticklabels = ["A-1", "B-2", "C-3", "D-4"]
xticklabels = [xticklabels[i] for i in p.reordered_ind]
npt.assert_array_equal(p.xticklabels, xticklabels)
npt.assert_array_equal(p.yticklabels, [])
assert p.xlabel == "letter-number"
def test_axis0_input(self):
kws = self.default_kws.copy()
kws['axis'] = 0
p = mat._DendrogramPlotter(self.df_norm.T, **kws)
npt.assert_array_equal(p.array, np.asarray(self.df_norm.T))
pdt.assert_frame_equal(p.data, self.df_norm.T)
npt.assert_array_equal(p.linkage, self.x_norm_linkage)
assert p.dendrogram == self.x_norm_dendrogram
npt.assert_array_equal(p.xticklabels, self.df_norm_leaves)
npt.assert_array_equal(p.yticklabels, [])
assert p.xlabel == 'letters'
assert p.ylabel == ''
def test_rotate_input(self):
kws = self.default_kws.copy()
kws['rotate'] = True
p = mat._DendrogramPlotter(self.df_norm, **kws)
npt.assert_array_equal(p.array.T, np.asarray(self.df_norm))
pdt.assert_frame_equal(p.data.T, self.df_norm)
npt.assert_array_equal(p.xticklabels, [])
npt.assert_array_equal(p.yticklabels, self.df_norm_leaves)
assert p.xlabel == ''
assert p.ylabel == 'letters'
def test_rotate_axis0_input(self):
kws = self.default_kws.copy()
kws['rotate'] = True
kws['axis'] = 0
p = mat._DendrogramPlotter(self.df_norm.T, **kws)
npt.assert_array_equal(p.reordered_ind, self.x_norm_leaves)
def test_custom_linkage(self):
kws = self.default_kws.copy()
try:
import fastcluster
linkage = fastcluster.linkage_vector(self.x_norm, method='single',
metric='euclidean')
except ImportError:
d = distance.pdist(self.x_norm, metric='euclidean')
linkage = hierarchy.linkage(d, method='single')
dendrogram = hierarchy.dendrogram(linkage, no_plot=True,
color_threshold=-np.inf)
kws['linkage'] = linkage
p = mat._DendrogramPlotter(self.df_norm, **kws)
npt.assert_array_equal(p.linkage, linkage)
assert p.dendrogram == dendrogram
def test_label_false(self):
kws = self.default_kws.copy()
kws['label'] = False
p = mat._DendrogramPlotter(self.df_norm, **kws)
assert p.xticks == []
assert p.yticks == []
assert p.xticklabels == []
assert p.yticklabels == []
assert p.xlabel == ""
assert p.ylabel == ""
def test_linkage_scipy(self):
p = mat._DendrogramPlotter(self.x_norm, **self.default_kws)
scipy_linkage = p._calculate_linkage_scipy()
from scipy.spatial import distance
from scipy.cluster import hierarchy
dists = distance.pdist(self.x_norm.T,
metric=self.default_kws['metric'])
linkage = hierarchy.linkage(dists, method=self.default_kws['method'])
npt.assert_array_equal(scipy_linkage, linkage)
@pytest.mark.skipif(_no_fastcluster, reason="fastcluster not installed")
def test_fastcluster_other_method(self):
import fastcluster
kws = self.default_kws.copy()
kws['method'] = 'average'
linkage = fastcluster.linkage(self.x_norm.T, method='average',
metric='euclidean')
p = mat._DendrogramPlotter(self.x_norm, **kws)
npt.assert_array_equal(p.linkage, linkage)
@pytest.mark.skipif(_no_fastcluster, reason="fastcluster not installed")
def test_fastcluster_non_euclidean(self):
import fastcluster
kws = self.default_kws.copy()
kws['metric'] = 'cosine'
kws['method'] = 'average'
linkage = fastcluster.linkage(self.x_norm.T, method=kws['method'],
metric=kws['metric'])
p = mat._DendrogramPlotter(self.x_norm, **kws)
npt.assert_array_equal(p.linkage, linkage)
def test_dendrogram_plot(self):
d = mat.dendrogram(self.x_norm, **self.default_kws)
ax = plt.gca()
xlim = ax.get_xlim()
# 10 comes from _plot_dendrogram in scipy.cluster.hierarchy
xmax = len(d.reordered_ind) * 10
assert xlim[0] == 0
assert xlim[1] == xmax
assert len(ax.collections[0].get_paths()) == len(d.dependent_coord)
@pytest.mark.xfail(mpl.__version__ == "3.1.1",
reason="matplotlib 3.1.1 bug")
def test_dendrogram_rotate(self):
kws = self.default_kws.copy()
kws['rotate'] = True
d = mat.dendrogram(self.x_norm, **kws)
ax = plt.gca()
ylim = ax.get_ylim()
# 10 comes from _plot_dendrogram in scipy.cluster.hierarchy
ymax = len(d.reordered_ind) * 10
# Since y axis is inverted, ylim is (80, 0)
# and therefore not (0, 80) as usual:
assert ylim[1] == 0
assert ylim[0] == ymax
def test_dendrogram_ticklabel_rotation(self):
f, ax = plt.subplots(figsize=(2, 2))
mat.dendrogram(self.df_norm, ax=ax)
for t in ax.get_xticklabels():
assert t.get_rotation() == 0
plt.close(f)
df = self.df_norm.copy()
df.columns = [str(c) * 10 for c in df.columns]
df.index = [i * 10 for i in df.index]
f, ax = plt.subplots(figsize=(2, 2))
mat.dendrogram(df, ax=ax)
for t in ax.get_xticklabels():
assert t.get_rotation() == 90
plt.close(f)
f, ax = plt.subplots(figsize=(2, 2))
mat.dendrogram(df.T, axis=0, rotate=True)
for t in ax.get_yticklabels():
assert t.get_rotation() == 0
plt.close(f)
@pytest.mark.skipif(_no_scipy, reason="Test requires scipy")
class TestClustermap:
rs = np.random.RandomState(sum(map(ord, "clustermap")))
x_norm = rs.randn(4, 8) + np.arange(8)
x_norm = (x_norm.T + np.arange(4)).T
letters = pd.Series(["A", "B", "C", "D", "E", "F", "G", "H"],
name="letters")
df_norm = pd.DataFrame(x_norm, columns=letters)
default_kws = dict(pivot_kws=None, z_score=None, standard_scale=None,
figsize=(10, 10), row_colors=None, col_colors=None,
dendrogram_ratio=.2, colors_ratio=.03,
cbar_pos=(0, .8, .05, .2))
default_plot_kws = dict(metric='euclidean', method='average',
colorbar_kws=None,
row_cluster=True, col_cluster=True,
row_linkage=None, col_linkage=None,
tree_kws=None)
row_colors = color_palette('Set2', df_norm.shape[0])
col_colors = color_palette('Dark2', df_norm.shape[1])
if not _no_scipy:
if _no_fastcluster:
x_norm_distances = distance.pdist(x_norm.T, metric='euclidean')
x_norm_linkage = hierarchy.linkage(x_norm_distances, method='single')
else:
x_norm_linkage = fastcluster.linkage_vector(x_norm.T,
metric='euclidean',
method='single')
x_norm_dendrogram = hierarchy.dendrogram(x_norm_linkage, no_plot=True,
color_threshold=-np.inf)
x_norm_leaves = x_norm_dendrogram['leaves']
df_norm_leaves = np.asarray(df_norm.columns[x_norm_leaves])
def test_ndarray_input(self):
cg = mat.ClusterGrid(self.x_norm, **self.default_kws)
pdt.assert_frame_equal(cg.data, pd.DataFrame(self.x_norm))
assert len(cg.fig.axes) == 4
assert cg.ax_row_colors is None
assert cg.ax_col_colors is None
def test_df_input(self):
cg = mat.ClusterGrid(self.df_norm, **self.default_kws)
pdt.assert_frame_equal(cg.data, self.df_norm)
def test_corr_df_input(self):
df = self.df_norm.corr()
cg = mat.ClusterGrid(df, **self.default_kws)
cg.plot(**self.default_plot_kws)
diag = cg.data2d.values[np.diag_indices_from(cg.data2d)]
npt.assert_array_almost_equal(diag, np.ones(cg.data2d.shape[0]))
def test_pivot_input(self):
df_norm = self.df_norm.copy()
df_norm.index.name = 'numbers'
df_long = pd.melt(df_norm.reset_index(), var_name='letters',
id_vars='numbers')
kws = self.default_kws.copy()
kws['pivot_kws'] = dict(index='numbers', columns='letters',
values='value')
cg = mat.ClusterGrid(df_long, **kws)
pdt.assert_frame_equal(cg.data2d, df_norm)
def test_colors_input(self):
kws = self.default_kws.copy()
kws['row_colors'] = self.row_colors
kws['col_colors'] = self.col_colors
cg = mat.ClusterGrid(self.df_norm, **kws)
npt.assert_array_equal(cg.row_colors, self.row_colors)
npt.assert_array_equal(cg.col_colors, self.col_colors)
assert len(cg.fig.axes) == 6
def test_categorical_colors_input(self):
kws = self.default_kws.copy()
row_colors = pd.Series(self.row_colors, dtype="category")
col_colors = pd.Series(
self.col_colors, dtype="category", index=self.df_norm.columns
)
kws['row_colors'] = row_colors
kws['col_colors'] = col_colors
exp_row_colors = list(map(mpl.colors.to_rgb, row_colors))
exp_col_colors = list(map(mpl.colors.to_rgb, col_colors))
cg = mat.ClusterGrid(self.df_norm, **kws)
npt.assert_array_equal(cg.row_colors, exp_row_colors)
npt.assert_array_equal(cg.col_colors, exp_col_colors)
assert len(cg.fig.axes) == 6
def test_nested_colors_input(self):
kws = self.default_kws.copy()
row_colors = [self.row_colors, self.row_colors]
col_colors = [self.col_colors, self.col_colors]
kws['row_colors'] = row_colors
kws['col_colors'] = col_colors
cm = mat.ClusterGrid(self.df_norm, **kws)
npt.assert_array_equal(cm.row_colors, row_colors)
npt.assert_array_equal(cm.col_colors, col_colors)
assert len(cm.fig.axes) == 6
def test_colors_input_custom_cmap(self):
kws = self.default_kws.copy()
kws['cmap'] = mpl.cm.PRGn
kws['row_colors'] = self.row_colors
kws['col_colors'] = self.col_colors
cg = mat.clustermap(self.df_norm, **kws)
npt.assert_array_equal(cg.row_colors, self.row_colors)
npt.assert_array_equal(cg.col_colors, self.col_colors)
assert len(cg.fig.axes) == 6
def test_z_score(self):
df = self.df_norm.copy()
df = (df - df.mean()) / df.std()
kws = self.default_kws.copy()
kws['z_score'] = 1
cg = mat.ClusterGrid(self.df_norm, **kws)
pdt.assert_frame_equal(cg.data2d, df)
def test_z_score_axis0(self):
df = self.df_norm.copy()
df = df.T
df = (df - df.mean()) / df.std()
df = df.T
kws = self.default_kws.copy()
kws['z_score'] = 0
cg = mat.ClusterGrid(self.df_norm, **kws)
pdt.assert_frame_equal(cg.data2d, df)
def test_standard_scale(self):
df = self.df_norm.copy()
df = (df - df.min()) / (df.max() - df.min())
kws = self.default_kws.copy()
kws['standard_scale'] = 1
cg = mat.ClusterGrid(self.df_norm, **kws)
|
pdt.assert_frame_equal(cg.data2d, df)
|
pandas.util.testing.assert_frame_equal
|
import pandas as pd
import numpy as np
from keras.models import load_model
from sklearn.metrics import roc_curve, roc_auc_score, auc, precision_recall_curve, average_precision_score
import os
import pickle
from scipy.special import softmax
from prg import prg
class MetricsGenerator(object):
def __init__(self, dataset_dir, model_dir, metrics_dir):
self._model_dir = model_dir
self._metrics_dir = metrics_dir
self._train_x = pd.read_csv(dataset_dir + "train_x.csv")
self._test_x = pd.read_csv(dataset_dir + "test_x.csv")
self._train_x = self._train_x.drop(self._train_x.columns[0], axis=1)
self._test_x = self._test_x.drop(self._test_x.columns[0], axis=1)
self._train_y = pd.read_csv(dataset_dir + "train_y.csv")
self._test_y = pd.read_csv(dataset_dir + "test_y.csv")
def generate_metrics_for_model(self, model):
error_df = self.get_error_df(model)
roc_df, roc_auc_df = self.get_roc_and_auc_df(error_df)
precision_recall_df, precision_recall_auc_df, average_precision_score_df = self.get_precision_recall_and_auc_df(error_df)
prg_df, prg_auc_df = self.get_prg_and_auc_df(error_df)
history_df = self.get_history_df(model)
self.create(self._metrics_dir + "model" + str(model))
self.store_df("error_df", model,error_df)
self.store_df("roc_df", model, roc_df)
self.store_df("roc_auc_df", model, roc_auc_df)
self.store_df("precision_recall_df", model, precision_recall_df)
self.store_df("precision_recall_auc_df", model, precision_recall_auc_df)
self.store_df("average_precision_score_df", model, average_precision_score_df)
self.store_df("prg_df", model, prg_df)
self.store_df("prg_auc_df", model, prg_auc_df)
self.store_df("history_df", model, history_df)
def get_error_df(self, model):
model = load_model(self._model_dir + "model" + str(model) + ".h5")
test_x_predicted = model.predict(self._test_x)
mse = np.mean(np.power(self._test_x - test_x_predicted, 2), axis = 1)
error_df = pd.DataFrame({'Reconstruction_error':mse, 'True_values': self._test_y['target']})
return error_df
def get_roc_and_auc_df(self, error_df):
false_pos_rate, true_pos_rate, thresholds = roc_curve(error_df.True_values, error_df.Reconstruction_error)
i = np.arange(len(true_pos_rate))
roc_df = pd.DataFrame({'FPR': pd.Series(false_pos_rate, index=i), 'TPR': pd.Series(true_pos_rate, index=i), 'Threshold':
|
pd.Series(thresholds, index=i)
|
pandas.Series
|
import pyarrow.parquet as pq
import pandas as pd
import json
from typing import List, Callable, Iterator, Union, Optional
from sportsdataverse.config import WBB_BASE_URL, WBB_TEAM_BOX_URL, WBB_PLAYER_BOX_URL, WBB_TEAM_SCHEDULE_URL
from sportsdataverse.errors import SeasonNotFoundError
from sportsdataverse.dl_utils import download
def load_wbb_pbp(seasons: List[int]) -> pd.DataFrame:
"""Load women's college basketball play by play data going back to 2002
Example:
`wbb_df = sportsdataverse.wbb.load_wbb_pbp(seasons=range(2002,2022))`
Args:
seasons (list): Used to define different seasons. 2002 is the earliest available season.
Returns:
pd.DataFrame: Pandas dataframe containing the
play-by-plays available for the requested seasons.
Raises:
ValueError: If `season` is less than 2002.
"""
data =
|
pd.DataFrame()
|
pandas.DataFrame
|
import pandas as pd
import os
import sys
from docx import Document
from pathlib import Path
from nbdev.showdoc import doc
import ipywidgets as widgets
from ipywidgets import interact, fixed, FileUpload
import numpy as np
import xlsxwriter
from openpyxl import Workbook
from openpyxl import load_workbook
from IPython.display import display
from ipywidgets import HTML
from functools import partial
import openpyxl
from openpyxl.utils.dataframe import dataframe_to_rows
from copy import copy
import shutil
from database import *
# TODO fix formulas: overall satisfaction should go from B_scorerow:lastcolletter_scorerows
# TODO fixe dived by 0 error in formulas
# TODO put document links
# TODO create script that retrieves document links
class XLSXDoc:
def __init__(self, db, xlsx_path=xlsx_path, month_year="JUN2021", ws_name='HR3-4 ', template_path=template_path, t_ws_name='HR3-4 '):
self.xlsx_path = xlsx_path
self.month_year = month_year
self.file_name = f'NET UK - QMS - {month_year} - Indicators HR3-HR4.xlsx'
self.file_path = self.xlsx_path/self.file_name
self.xlsx_path.mkdir(parents=True, exist_ok=True)
self.ws_name = ws_name
self.template_path = template_path
self.t_ws_name = t_ws_name
self.db = db
db.listen(self)
self.regenerate()
def regenerate(self):
'''
Regenerate the excel according to the database
'''
wb = Workbook()
ws = wb.active
ws.title = self.ws_name
db_dict = self.db.get_db()
scores = db_dict['scores']
n_employees = db_dict['n_employees']
xlsx_structure = {
'A1': 'Provided courses 2020-2021',
'A2': 'for each course is reported the average of the opinions expressed with rating from 0 to 5'
}
for k, v in xlsx_structure.items():
ws[k] = v
# add the scores
for i,r in enumerate(dataframe_to_rows(scores, index=True, header=True)):
if(i != 1):
ws.append(r)
ws['A3'] = 'HR4'
# popualate the score calcuation (with eqatuions)
score_table_end = 2 + len(list(dataframe_to_rows(scores, index=True, header=True)))
ws.cell(score_table_end,1, value = 'Course average rating')
ws = self.__regenerate_formulas(ws)
# add the docx links, a list of links for each course
courses_docx = self.db.get_courses_docx()
ws.cell(score_table_end + 1, 1, value='General Notes on the effectiveness of the course')
for i, c in enumerate(scores.columns):
if c in courses_docx.keys():
urls = courses_docx[c]
cell_value = ''
for url in urls:
fname = url.stem
path = 'docx'
fpath = path + '/' + fname + '.docx'
cell_value = cell_value + str(fpath) + ' ; '
cell_value = cell_value[:-3]
ws.cell(score_table_end + 1, 2+i, value=cell_value)
hr3_row = score_table_end + 3
ws.cell(hr3_row, 1, value='HR3')
# add
course_nr_row = hr3_row+1
ws.cell(course_nr_row, 1, value='provided courses (nr.)')
ws.cell(course_nr_row, 2, value='=20-COUNTIFS(3:3, "Column*")')
n_people_row = hr3_row+2
ws.cell(n_people_row, 1, value='Number of people affected')
ws.cell(n_people_row, 2, value='=COUNTA(Tabella2[HR4])')
n_employees_row = hr3_row+3
ws.cell(n_employees_row, 1, value='Number of employees')
ws.cell(n_employees_row, 2, value=n_employees)
ws.cell(n_employees_row, 3, value='* taken as the current total')
summary_row = hr3_row + 5
year = 2020
ws.cell(summary_row, 1, value=f'Summary {year}')
ws.cell(summary_row, 2, value='Indexes')
ws.cell(summary_row, 3, value='Threshold')
ws.cell(summary_row+1, 1, value='HR3 - coverage of staff')
ws.cell(summary_row+1, 2, value='=B15/B16')
ws.cell(summary_row+2, 1, value='HR4 - overall satisfaction')
ws.cell(summary_row+2, 2, value='=AVERAGE(B10:R10)')
self.save_changes_to_file(wb)
self.__aplly_template_style()
self.regenerate_download_direcotry()
return wb, ws
def regenerate_download_direcotry(self):
# TODO put data in download_temp
docx_paths = self.db.get_courses_docx()
for courses in docx_paths.values():
for src in courses:
dest = download_temp_docx/(str(src.stem) + '.docx')
dest.touch()
shutil.copy(src, dest)
xlsx_dest = download_temp/(str(self.file_path.stem) + '.xlsx')
shutil.copy(self.file_path, xlsx_dest)
shutil.make_archive(download_dir, 'zip', download_temp)
def get_wb_ws(self):
'''
returns a the workbook corresponding to the actual file
and the worksheet view of it
'''
wb = load_workbook(self.file_path)
ws = wb[self.ws_name]
return wb, ws
def get_courses(self):
wb, ws = self.get_wb_ws()
df = pd.DataFrame(ws.values).set_index(0)
courses = df.iloc[2,:].dropna()
return list(courses)
def user_exists(self, name):
wb, ws = self.get_wb_ws()
df = pd.DataFrame(ws.values).set_index(0)
row_n = df.index.get_loc('Course average rating') + 1
if(name in list(df.index)[3:row_n]):
return True
return False
def course_exists(self, name):
if(name in self.get_courses()):
return True
return False
def add_user(self, name):
wb, ws = self.get_wb_ws()
if self.user_exists(name):
raise ValueError('user already exists')
ws.insert_rows(4)
ws['A4'] = name
self.__regenerate_formulas(ws)
self.save_changes_to_file(wb)
return ws
def add_score(self, user, course, score):
if course not in self.get_courses():
raise ValueError('Course does not exist')
if not self.user_exists(user):
self.add_user(user)
wb, ws = self.get_wb_ws()
df = pd.DataFrame(ws.values).set_index(0)
row_n = df.index.get_loc(user) + 1
col_n =
|
pd.Index(df.iloc[2])
|
pandas.Index
|
import pandas as pd
import random
import pickle
from tqdm import tqdm
import seaborn as sns
from sklearn.metrics import *
from matplotlib import pyplot as plt
from preferences import notas_pref
from ahp import ahp
from data_preparation import create_subsample
from fine_tunning import fine_tunning
from data_preparation import merge_matrices
from tau_distance import normalised_kendall_tau_distance
len_Q = 5 # n_samples to be evaluated
CV = 5 # number of cross-validation
test_size = 0.2 # 80% train and 20% test
accepted_error = .05 # max tau distance accepted between current ranking and the predicted one
df_var = pd.read_csv("dec_5obj_p2.csv", header=None) # decision variables
# df_var = df_var.iloc[0:55, :].round(5)
df_obj = pd.read_csv('obj_5obj_p2.csv', header=None) # values in Pareto front
# df_obj = df_obj.iloc[0:55, :].round(5)
npop, nvar = df_var.shape
nobj = df_obj.shape[1]
# Generate the preferences
df_obj = df_obj.to_numpy()
df_pref = notas_pref(df_obj)
# AHP from the original alternatives
rank_ahp = ahp(df_pref).index
# Generate the index to be evaluated
index = list(df_var.index)
# Aleatory ranking
aleatory = index.copy()
random.shuffle(aleatory)
# Start an aleatory ranking
rank_aleatory = aleatory.copy()
# Distances
current_previous = []
current_ahp = []
# Metrics
mse = []
rmse = []
r2 = []
mape = []
# Iterations
iteration = []
cont = 0
temp = 1
for aux in tqdm(range(len_Q, npop, len_Q)):
cont += 1
# Define Q and N-Q indexes
Q_index = aleatory[0:aux]
N_Q_index = [x for x in index if x not in Q_index]
# Train
df_Q = create_subsample(df_var=df_var, df_pref=df_pref, nobj=nobj, index=Q_index)
X_train = df_Q.iloc[:, :-nobj] # to predict
y_train = df_Q.iloc[:, -nobj:] # real targets
# Test
df_N_Q = create_subsample(df_var=df_var, df_pref=df_pref, nobj=nobj, index=N_Q_index)
X_test = df_N_Q.iloc[:, :-nobj] # to predict
y_test = df_N_Q.iloc[:, -nobj:] # real targets
# Model training
if temp > accepted_error:
tuned_model = fine_tunning(CV, X_train, y_train)
with open("tuned_model_cbic_5obj.pkl", 'wb') as arq: # Save best model
pickle.dump(tuned_model, arq)
tuned_model.fit(X_train, y_train)
else:
with open("tuned_model_cbic_5obj.pkl", "rb") as fp: # Load trained model
tuned_model = pickle.load(fp)
# Model evaluation
y_pred = tuned_model.predict(X_test)
y_pred =
|
pd.DataFrame(y_pred)
|
pandas.DataFrame
|
# Copyright (c) 2020, Xilinx
# All rights reserved.
#
# Redistribution and use in source and binary forms, with or without
# modification, are permitted provided that the following conditions are met:
#
# * Redistributions of source code must retain the above copyright notice, this
# list of conditions and the following disclaimer.
#
# * Redistributions in binary form must reproduce the above copyright notice,
# this list of conditions and the following disclaimer in the documentation
# and/or other materials provided with the distribution.
#
# * Neither the name of FINN nor the names of its
# contributors may be used to endorse or promote products derived from
# this software without specific prior written permission.
#
# THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
# AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
# IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
# DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
# FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
# DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
# SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
# CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
# OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
# OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
import math
import numpy as np
import pandas as pd
import torch
from sklearn import preprocessing
from sklearn.preprocessing import OneHotEncoder
# quantize the UNSW_NB15 dataset and convert it to binary vectors
# reimplementation
# paper: https://ev.fe.uni-lj.si/1-2-2019/Murovic.pdf
# original matlab code: https://git.io/JLLdN
class UNSW_NB15_quantized(torch.utils.data.Dataset):
def __init__(
self,
file_path_train,
file_path_test,
quantization=True,
onehot=False,
train=True,
):
self.dataframe = (
pd.concat([pd.read_csv(file_path_train),
|
pd.read_csv(file_path_test)
|
pandas.read_csv
|
import re
import numpy as np
import pytest
from pandas import Categorical, CategoricalIndex, DataFrame, Index, Series
import pandas._testing as tm
from pandas.core.arrays.categorical import recode_for_categories
from pandas.tests.arrays.categorical.common import TestCategorical
class TestCategoricalAPI:
def test_ordered_api(self):
# GH 9347
cat1 = Categorical(list("acb"), ordered=False)
tm.assert_index_equal(cat1.categories, Index(["a", "b", "c"]))
assert not cat1.ordered
cat2 = Categorical(list("acb"), categories=list("bca"), ordered=False)
tm.assert_index_equal(cat2.categories, Index(["b", "c", "a"]))
assert not cat2.ordered
cat3 = Categorical(list("acb"), ordered=True)
tm.assert_index_equal(cat3.categories, Index(["a", "b", "c"]))
assert cat3.ordered
cat4 = Categorical(list("acb"), categories=list("bca"), ordered=True)
tm.assert_index_equal(cat4.categories, Index(["b", "c", "a"]))
assert cat4.ordered
def test_set_ordered(self):
cat = Categorical(["a", "b", "c", "a"], ordered=True)
cat2 = cat.as_unordered()
assert not cat2.ordered
cat2 = cat.as_ordered()
assert cat2.ordered
cat2.as_unordered(inplace=True)
assert not cat2.ordered
cat2.as_ordered(inplace=True)
assert cat2.ordered
assert cat2.set_ordered(True).ordered
assert not cat2.set_ordered(False).ordered
cat2.set_ordered(True, inplace=True)
assert cat2.ordered
cat2.set_ordered(False, inplace=True)
assert not cat2.ordered
# removed in 0.19.0
msg = "can't set attribute"
with pytest.raises(AttributeError, match=msg):
cat.ordered = True
with pytest.raises(AttributeError, match=msg):
cat.ordered = False
def test_rename_categories(self):
cat = Categorical(["a", "b", "c", "a"])
# inplace=False: the old one must not be changed
res = cat.rename_categories([1, 2, 3])
tm.assert_numpy_array_equal(
res.__array__(), np.array([1, 2, 3, 1], dtype=np.int64)
)
tm.assert_index_equal(res.categories, Index([1, 2, 3]))
exp_cat = np.array(["a", "b", "c", "a"], dtype=np.object_)
tm.assert_numpy_array_equal(cat.__array__(), exp_cat)
exp_cat = Index(["a", "b", "c"])
tm.assert_index_equal(cat.categories, exp_cat)
# GH18862 (let rename_categories take callables)
result = cat.rename_categories(lambda x: x.upper())
expected = Categorical(["A", "B", "C", "A"])
tm.assert_categorical_equal(result, expected)
# and now inplace
res = cat.rename_categories([1, 2, 3], inplace=True)
assert res is None
tm.assert_numpy_array_equal(
cat.__array__(), np.array([1, 2, 3, 1], dtype=np.int64)
)
tm.assert_index_equal(cat.categories, Index([1, 2, 3]))
@pytest.mark.parametrize("new_categories", [[1, 2, 3, 4], [1, 2]])
def test_rename_categories_wrong_length_raises(self, new_categories):
cat = Categorical(["a", "b", "c", "a"])
msg = (
"new categories need to have the same number of items as the "
"old categories!"
)
with pytest.raises(ValueError, match=msg):
cat.rename_categories(new_categories)
def test_rename_categories_series(self):
# https://github.com/pandas-dev/pandas/issues/17981
c = Categorical(["a", "b"])
result = c.rename_categories(Series([0, 1], index=["a", "b"]))
expected = Categorical([0, 1])
tm.assert_categorical_equal(result, expected)
def test_rename_categories_dict(self):
# GH 17336
cat = Categorical(["a", "b", "c", "d"])
res = cat.rename_categories({"a": 4, "b": 3, "c": 2, "d": 1})
expected = Index([4, 3, 2, 1])
tm.assert_index_equal(res.categories, expected)
# Test for inplace
res = cat.rename_categories({"a": 4, "b": 3, "c": 2, "d": 1}, inplace=True)
assert res is None
tm.assert_index_equal(cat.categories, expected)
# Test for dicts of smaller length
cat = Categorical(["a", "b", "c", "d"])
res = cat.rename_categories({"a": 1, "c": 3})
expected = Index([1, "b", 3, "d"])
tm.assert_index_equal(res.categories, expected)
# Test for dicts with bigger length
cat = Categorical(["a", "b", "c", "d"])
res = cat.rename_categories({"a": 1, "b": 2, "c": 3, "d": 4, "e": 5, "f": 6})
expected = Index([1, 2, 3, 4])
tm.assert_index_equal(res.categories, expected)
# Test for dicts with no items from old categories
cat = Categorical(["a", "b", "c", "d"])
res = cat.rename_categories({"f": 1, "g": 3})
expected = Index(["a", "b", "c", "d"])
tm.assert_index_equal(res.categories, expected)
def test_reorder_categories(self):
cat = Categorical(["a", "b", "c", "a"], ordered=True)
old = cat.copy()
new = Categorical(
["a", "b", "c", "a"], categories=["c", "b", "a"], ordered=True
)
# first inplace == False
res = cat.reorder_categories(["c", "b", "a"])
# cat must be the same as before
tm.assert_categorical_equal(cat, old)
# only res is changed
tm.assert_categorical_equal(res, new)
# inplace == True
res = cat.reorder_categories(["c", "b", "a"], inplace=True)
assert res is None
tm.assert_categorical_equal(cat, new)
@pytest.mark.parametrize(
"new_categories",
[
["a"], # not all "old" included in "new"
["a", "b", "d"], # still not all "old" in "new"
["a", "b", "c", "d"], # all "old" included in "new", but too long
],
)
def test_reorder_categories_raises(self, new_categories):
cat = Categorical(["a", "b", "c", "a"], ordered=True)
msg = "items in new_categories are not the same as in old categories"
with pytest.raises(ValueError, match=msg):
cat.reorder_categories(new_categories)
def test_add_categories(self):
cat = Categorical(["a", "b", "c", "a"], ordered=True)
old = cat.copy()
new = Categorical(
["a", "b", "c", "a"], categories=["a", "b", "c", "d"], ordered=True
)
# first inplace == False
res = cat.add_categories("d")
tm.assert_categorical_equal(cat, old)
tm.assert_categorical_equal(res, new)
res = cat.add_categories(["d"])
tm.assert_categorical_equal(cat, old)
tm.assert_categorical_equal(res, new)
# inplace == True
res = cat.add_categories("d", inplace=True)
tm.assert_categorical_equal(cat, new)
assert res is None
# GH 9927
cat = Categorical(list("abc"), ordered=True)
expected = Categorical(list("abc"), categories=list("abcde"), ordered=True)
# test with Series, np.array, index, list
res = cat.add_categories(Series(["d", "e"]))
tm.assert_categorical_equal(res, expected)
res = cat.add_categories(np.array(["d", "e"]))
tm.assert_categorical_equal(res, expected)
res = cat.add_categories(Index(["d", "e"]))
tm.assert_categorical_equal(res, expected)
res = cat.add_categories(["d", "e"])
tm.assert_categorical_equal(res, expected)
def test_add_categories_existing_raises(self):
# new is in old categories
cat = Categorical(["a", "b", "c", "d"], ordered=True)
msg = re.escape("new categories must not include old categories: {'d'}")
with pytest.raises(ValueError, match=msg):
cat.add_categories(["d"])
def test_set_categories(self):
cat = Categorical(["a", "b", "c", "a"], ordered=True)
exp_categories = Index(["c", "b", "a"])
exp_values = np.array(["a", "b", "c", "a"], dtype=np.object_)
res = cat.set_categories(["c", "b", "a"], inplace=True)
tm.assert_index_equal(cat.categories, exp_categories)
tm.assert_numpy_array_equal(cat.__array__(), exp_values)
assert res is None
res = cat.set_categories(["a", "b", "c"])
# cat must be the same as before
tm.assert_index_equal(cat.categories, exp_categories)
tm.assert_numpy_array_equal(cat.__array__(), exp_values)
# only res is changed
exp_categories_back = Index(["a", "b", "c"])
tm.assert_index_equal(res.categories, exp_categories_back)
tm.assert_numpy_array_equal(res.__array__(), exp_values)
# not all "old" included in "new" -> all not included ones are now
# np.nan
cat = Categorical(["a", "b", "c", "a"], ordered=True)
res = cat.set_categories(["a"])
tm.assert_numpy_array_equal(res.codes, np.array([0, -1, -1, 0], dtype=np.int8))
# still not all "old" in "new"
res = cat.set_categories(["a", "b", "d"])
tm.assert_numpy_array_equal(res.codes, np.array([0, 1, -1, 0], dtype=np.int8))
tm.assert_index_equal(res.categories, Index(["a", "b", "d"]))
# all "old" included in "new"
cat = cat.set_categories(["a", "b", "c", "d"])
exp_categories = Index(["a", "b", "c", "d"])
tm.assert_index_equal(cat.categories, exp_categories)
# internals...
c = Categorical([1, 2, 3, 4, 1], categories=[1, 2, 3, 4], ordered=True)
tm.assert_numpy_array_equal(c._codes, np.array([0, 1, 2, 3, 0], dtype=np.int8))
tm.assert_index_equal(c.categories, Index([1, 2, 3, 4]))
exp = np.array([1, 2, 3, 4, 1], dtype=np.int64)
tm.assert_numpy_array_equal(np.asarray(c), exp)
# all "pointers" to '4' must be changed from 3 to 0,...
c = c.set_categories([4, 3, 2, 1])
# positions are changed
tm.assert_numpy_array_equal(c._codes, np.array([3, 2, 1, 0, 3], dtype=np.int8))
# categories are now in new order
tm.assert_index_equal(c.categories, Index([4, 3, 2, 1]))
# output is the same
exp = np.array([1, 2, 3, 4, 1], dtype=np.int64)
tm.assert_numpy_array_equal(np.asarray(c), exp)
assert c.min() == 4
assert c.max() == 1
# set_categories should set the ordering if specified
c2 = c.set_categories([4, 3, 2, 1], ordered=False)
assert not c2.ordered
tm.assert_numpy_array_equal(np.asarray(c), np.asarray(c2))
# set_categories should pass thru the ordering
c2 = c.set_ordered(False).set_categories([4, 3, 2, 1])
assert not c2.ordered
tm.assert_numpy_array_equal(np.asarray(c), np.asarray(c2))
def test_to_dense_deprecated(self):
cat = Categorical(["a", "b", "c", "a"], ordered=True)
with
|
tm.assert_produces_warning(FutureWarning)
|
pandas._testing.assert_produces_warning
|
import pandas as pd
import world_bank_data as wb
import plotly.graph_objs as go
def sundial_plot(metric='SP.POP.TOTL', title='World Population', year=2000):
"""Plot the given metric as a sundial plot"""
countries = wb.get_countries()
values = wb.get_series(metric, date=year, id_or_value='id', simplify_index=True)
df = countries[['region', 'name']].rename(columns={'name': 'country'}).loc[
countries.region != 'Aggregates']
df['values'] = values
# The sunburst plot requires weights (values), labels, and parent (region, or World)
# We build the corresponding table here
columns = ['parents', 'labels', 'values']
level1 = df.copy()
level1.columns = columns
level1['text'] = level1['values'].apply(lambda pop: '{:,.0f}'.format(pop))
level2 = df.groupby('region')['values'].sum().reset_index()[['region', 'region', 'values']]
level2.columns = columns
level2['parents'] = 'World'
# move value to text for this level
level2['text'] = level2['values'].apply(lambda pop: '{:,.0f}'.format(pop))
level2['values'] = 0
level3 = pd.DataFrame({'parents': [''], 'labels': ['World'],
'values': [0.0], 'text': ['{:,.0f}'.format(values.loc['WLD'])]})
all_levels =
|
pd.concat([level1, level2, level3], axis=0)
|
pandas.concat
|
"""
The :mod:`hillmaker.bydatetime` module includes functions for computing occupancy,
arrival, and departure statistics by time bin of day and date.
"""
# Copyright 2022 <NAME>
#
import logging
import numpy as np
import pandas as pd
from pandas import DataFrame
from pandas import Series
from pandas import Timestamp
from datetime import datetime
from pandas.tseries.offsets import Minute
import hillmaker.hmlib as hmlib
CONST_FAKE_OCCWEIGHT_FIELDNAME = 'FakeOccWeightField'
CONST_FAKE_CATFIELD_NAME = 'FakeCatForTotals'
OCC_TOLERANCE = 0.02
# This should inherit level from root logger
logger = logging.getLogger(__name__)
def make_bydatetime(stops_df, infield, outfield,
start_analysis_np, end_analysis_np, catfield=None,
bin_size_minutes=60,
cat_to_exclude=None,
totals=1,
occ_weight_field=None,
edge_bins=1,
verbose=0):
"""
Create bydatetime table based on user inputs.
This is the table from which summary statistics can be computed.
Parameters
----------
stops_df: DataFrame
Stop data
infield: string
Name of column in stops_df to use as arrival datetime
outfield: string
Name of column in stops_df to use as departure datetime
start_analysis_np: numpy datetime64[ns]
Start date for the analysis
end_analysis_np: numpy datetime64[ns]
End date for the analysis
catfield : string, optional
Column name corresponding to the categories. If none is specified, then only overall occupancy is analyzed.
bin_size_minutes: int, default 60
Bin size in minutes. Should divide evenly into 1440.
cat_to_exclude: list of strings, default None
Categories to ignore
edge_bins: int, default 1
Occupancy contribution method for arrival and departure bins. 1=fractional, 2=whole bin
totals: int, default 1
0=no totals, 1=totals by datetime, 2=totals bydatetime as well as totals for each field in the
catfields (only relevant for > 1 category field)
occ_weight_field : string, optional (default=1.0)
Column name corresponding to the weights to use for occupancy incrementing.
verbose : int, default 0
The verbosity level. The default, zero, means silent mode.
Returns
-------
Dict of DataFrames
Occupancy, arrivals, departures by category by datetime bin
Examples
--------
bydt_dfs = make_bydatetime(stops_df, 'InTime', 'OutTime',
... datetime(2014, 3, 1), datetime(2014, 6, 30), 'PatientType', 60)
TODO
----
* Sanity checks on date ranges
* Formal test using short stay data
* Flow conservation checks
Notes
-----
References
----------
See Also
--------
"""
# Number of bins in analysis span
num_bins = hmlib.bin_of_span(end_analysis_np, start_analysis_np, bin_size_minutes) + 1
# Compute min and max of in and out times
min_intime = stops_df[infield].min()
max_intime = stops_df[infield].max()
min_outtime = stops_df[outfield].min()
max_outtime = stops_df[outfield].max()
logger.info(f"min of intime: {min_intime}")
logger.info(f"max of intime: {max_intime}")
logger.info(f"min of outtime: {min_outtime}")
logger.info(f"max of outtime: {max_outtime}")
# TODO - Add warnings here related to min and maxes out of whack with analysis range
# Occupancy weights
# If no occ weight field specified, create fake one containing 1.0 as values.
# Avoids having to check during dataframe iteration whether or not to use
# default occupancy weight.
if occ_weight_field is None:
occ_weight_vec = np.ones(len(stops_df.index), dtype=np.float64)
occ_weight_df = DataFrame({CONST_FAKE_OCCWEIGHT_FIELDNAME: occ_weight_vec})
stops_df = pd.concat([stops_df, occ_weight_df], axis=1)
occ_weight_field = CONST_FAKE_OCCWEIGHT_FIELDNAME
# Handle cases of no catfield, or a single fieldname, (no longer supporting a list of fieldnames)
# If no category, add a temporary dummy column populated with a totals str
total_str = 'total'
do_totals = True
if catfield is not None:
# If it's a string, it's a single cat field --> convert to list
# Keeping catfield as a list in case I change mind about multiple category fields
if isinstance(catfield, str):
catfield = [catfield]
else:
totlist = [total_str] * len(stops_df)
totseries = Series(totlist, dtype=str, name=CONST_FAKE_CATFIELD_NAME)
totfield_df =
|
DataFrame({CONST_FAKE_CATFIELD_NAME: totseries})
|
pandas.DataFrame
|
'''
inference.py: part of pybraincompare package
Functions to calculate reverse inference
'''
from __future__ import print_function
from __future__ import division
from builtins import range
from past.utils import old_div
from pybraincompare.ontology.graph import get_node_fields, get_node_by_name
from pybraincompare.compare.maths import calculate_pairwise_correlation
from pybraincompare.compare.mrutils import get_images_df
from glob import glob
import nibabel
import pickle
import pandas
import numpy
import math
import re
import os
def likelihood_groups_from_tree(tree,standard_mask,input_folder,image_pattern="[0]+%s[.]",
output_folder=None,node_pattern="[0-9]+",):
'''likelihood_groups_from_tree
Function to generate likelihood groups from a pybraincompare.ontology.tree
object. These groups can then be used to calculate likelihoods (eg,
p(activation|cognitive process). The groups are output as pickle objects.
This is done because it is ideal to calculate likelihoods on a cluster.
:param tree: dict
a dictionary of nodes, with base nodes matching a particular pattern
assumed to be image (.nii.gz) files.
:param standard_mask: nifti image (nibabel)
standard image mask that images are registered to
:param output_folder: path
a folder path to save likelihood groups
:param input_folder: path
the folder of images to be matched to the nodes of the tree.
:param pattern: str
the pattern to match to find the base image nodes. Default is a number
of any length [neurovault image primary keys].
:param image_pattern: str
a regular expression to find image files in images_folder. Default will
match any number of leading zeros, any number, and any extension.
:param node_pattern: str
a regular expression to find image nodes in the tree, matched to name
:return groups: pickle
a pickle with the following
..note::
pbc_likelihood_groups_trm_12345.pkl
group["nid"] = "trm_12345"
group["in"] = ["path1","path2",..."pathN"]
group["out"] = ["path3","path4",..."pathM"]
group["meta"]: meta data for the node
group["range_table"]: a data frame of ranges with "start" and "stop"
to calculate the range is based on the mins and max of
the entire set of images
'''
# Find all nodes in the tree, match to images in folder
nodes = get_node_fields(tree,field="name",nodes=[])
contender_files = glob("%s/*" %input_folder)
# Images will match the specified pattern
find_nodes = re.compile(node_pattern)
image_nodes = [node for node in nodes if find_nodes.match(node)]
image_nodes = numpy.unique(image_nodes).tolist()
# Node names must now be matched to files
file_lookup = dict()
file_names = [os.path.split(path)[-1] for path in contender_files]
for node in image_nodes:
find_file = re.compile(image_pattern %node)
idx = [file_names.index(x) for x in file_names if find_file.match(x)]
if len(idx) > 1:
raise ValueError("ERROR: found %s images that match pattern %s." %len(idx),find_file.pattern)
elif len(idx) == 0:
print("Did not find file for %s, will not be included in analysis." %(node))
else:
file_lookup[node] = contender_files[idx[0]]
# Use pandas dataframe to not risk weird dictionary iteration orders
files = pandas.DataFrame(list(file_lookup.values()),columns=["path"])
files.index = list(file_lookup.keys())
# The remaining nodes in the tree (that are not images) will have a RI score
concept_nodes = [x for x in nodes if x not in image_nodes]
# create table of voxels for all images (the top node)
mr = get_images_df(file_paths=files.path,mask=standard_mask)
mr.index = files.index
range_table = make_range_table(mr)
# GROUPS ----------------------------------------------------
# Find groups for image sets at each node (**node names must be unique)
# This is images at (and in lower levels) of node vs. everything else
# will be used to calculate p([activation in range | region (voxel)]
groups = []
for concept_node in concept_nodes:
node = get_node_by_name(tree,concept_node)
node_id = node["nid"] # for save image
node_meta = node["meta"]
if node:
all_children = get_node_fields(node,"name",[])
children_in = [c for c in all_children if c in files.index]
children_out = [c for c in files.index if c not in children_in]
if len(children_in) > 0 and len(children_out) > 0:
print("Generating group for concept node %s" %(concept_node))
group = {"in": files.path.loc[children_in].unique().tolist(),
"out": files.path.loc[children_out].unique().tolist(),
"range_table": range_table,
"meta": node_meta,
"nid": node_id,
"name": concept_node}
groups.append(group)
if output_folder != None:
outpkl = "%s/pbc_group_%s.pkl" %(output_folder,node_id)
pickle.dump(group, open(outpkl,"wb"))
return groups
def make_range_table(mr,ranges=None):
'''make_range_table
Generate a table of ranges, in format
..note::
start stop
[-28.0,-27.5] -28.0 -27.5
[-27.5,-27.0] -27.5 -27.0
[-27.0,-26.5] -27.0 -26.5
[-26.5,-26.0] -26.5 -26.0
from a table of values, where images/objects are expected in rows,
and regions/voxels in columns
ranges, if defined, should be a list of lists of ranges to
extract. eg, [[start,stop],[start,stop]] where start is min, stop is max
The absolute min and max are used to generate a table of ranges
in the format above from min to max
'''
range_table = pandas.DataFrame(columns=["start","stop"])
if ranges:
error = "ERROR: ranges should be a list of ranges [[start,stop], ...]"
if not isinstance(ranges,list):
raise ValueError(error)
for range_group in ranges:
if not isinstance(range_group,list) or len(range_group) != 2:
raise ValueError(error)
name = "[%s,%s]" %(range_group[0],range_group[1])
range_table.loc[name] = [range_group[0],range_group[1]]
# If no start or stop defined, get from min and max of data
else:
mins = mr.min(axis=0)
maxs = mr.max(axis=0)
absmin = math.floor(numpy.min(mins))
absmax = math.ceil(numpy.max(maxs))
steps = ((abs(absmin)+absmax)*2)+1
breaks = numpy.linspace(absmin,absmax,num=steps,retstep=True)[0]
for s in range(0,len(breaks)-1):
start = breaks[s]
stop = breaks[s+1]
name = "[%s,%s]" %(start,stop)
range_table.loc[name] = [start,stop]
return range_table
def get_likelihood_df(nid,in_images,out_images,standard_mask,range_table,
threshold=2.96,output_folder=None,method=["binary"]):
'''get_likelihood_df
will calculate likelihoods and save to a pandas df pickle.
The user must specify the method [default is binary].
Method details:
ranges:
- likelihood in all thresholds defined (calculate_priors in ranges)
binary
- likelihood above / below a certain level [threshold, default=2.96]
Note: you do not need to calculate likelihoods in advance for the mean metric
(using a derivation of the distance from a mean image as a probability score)
In this case, use calculate_reverse_inference_distance
:param nid: str
a unique identifier, a node ID from pybraincompare.ontology.tree
:param in_images: list
a list of files for the "in" group relevant to some concept
:param out_images: list
the rest
:param standard_mask: nibabel.Nifti1Image object
the standard mask images are in space of
:param range_table: pandas data frame
a data frame of ranges with "start" and "stop" to calculate
the range is based on the mins and max of the entire set of images
can be generated with pybraincompare.inference.make_range_table
:param output_folder: path
folder to save likelihood pickles [default is None]
If output_folder is not specified, the df objects are returned.
If specified, will return paths to saved pickle objects:
pbc_likelihood_trm12345_df_in.pkl
EACH VOXEL IS p(activation in voxel is in threshold)
'''
# Read all images into one data frame
if len(numpy.intersect1d(in_images,out_images)) > 0:
raise ValueError("ERROR: in_images and out_images should not share images!")
all_images = in_images + out_images
mr = get_images_df(file_paths=all_images,mask=standard_mask)
mr.index = all_images
in_subset = mr.loc[in_images]
out_subset = mr.loc[out_images]
# Calculate likelihood for user defined methods
df = dict()
if "ranges" in method:
df["out_ranges"] = calculate_likelihood_in_ranges(in_subset,range_table)
df["in_ranges"] = calculate_likelihood_in_ranges(out_subset,range_table)
if output_folder:
df["in_ranges"] = save_likelihood_pickle(df["in_ranges"],
output_folder,nid,"in_ranges")
df["out_ranges"] = save_likelihood_pickle(df["out_ranges"],
output_folder,nid,"out_ranges")
if "binary" in method:
df["in_bin"] = calculate_likelihood_binary(in_subset,threshold)
df["out_bin"] = calculate_likelihood_binary(out_subset,threshold)
if output_folder:
df["in_bin"] = save_likelihood_pickle(df["in_bin"],
output_folder,
nid, "in_bin_%s" %threshold)
df["in_out"] = save_likelihood_pickle(df["out_bin"],
output_folder,
nid, "out_bin_%s" %threshold)
return df
def save_likelihood_pickle(likelihood_df,output_folder,nid,suffix):
outfile = "%s/pbc_likelihood_%s_df_%s.pkl" %(output_folder,nid,suffix)
likelihood_df.to_pickle(outfile)
return outfile
def save_likelihood_nii(input_pkl,output_folder,standard_mask):
'''save_likelihood_nii
save a nii image for each threshold (column) across all voxels (rows)
:param input_pkl: pickle object
the input pickle with likelihood saved by
pybraincompare.ontology.inference.get_likelihood_df
:param output_folder: path
folder for output nifti, one per threshold range
pbc_likelihood_trm12345_df_in_[start]_[stop].nii
EACH VOXEL IS p(activation in voxel is in threshold) for group [in or out]
'''
likelihood =
|
pandas.read_pickle(input_pkl)
|
pandas.read_pickle
|
#!/usr/bin/env python
"""
Download Interface for HADS data
"""
import sys
import cgi
import os
from io import StringIO
import pandas as pd
from pandas.io.sql import read_sql
from pyiem.util import get_dbconn, utc, ssw
PGCONN = get_dbconn('hads')
DELIMITERS = {'comma': ',', 'space': ' ', 'tab': '\t'}
def get_time(form):
""" Get timestamps """
y1 = int(form.getfirst('year'))
m1 = int(form.getfirst('month1'))
m2 = int(form.getfirst('month2'))
d1 = int(form.getfirst('day1'))
d2 = int(form.getfirst('day2'))
h1 = int(form.getfirst('hour1'))
h2 = int(form.getfirst('hour2'))
mi1 = int(form.getfirst('minute1'))
mi2 = int(form.getfirst('minute2'))
sts = utc(y1, m1, d1, h1, mi1)
ets = utc(y1, m2, d2, h2, mi2)
return sts, ets
def threshold_search(table, threshold, thresholdvar, delimiter):
""" Do the threshold searching magic """
cols = list(table.columns.values)
searchfor = "HGI%s" % (thresholdvar.upper(),)
cols5 = [s[:5] for s in cols]
if searchfor not in cols5:
error("Could not find %s variable for this site!" % (searchfor,))
return
mycol = cols[cols5.index(searchfor)]
above = False
maxrunning = -99
maxvalid = None
found = False
res = []
for (station, valid), row in table.iterrows():
val = row[mycol]
if val > threshold and not above:
found = True
res.append(dict(station=station, utc_valid=valid, event='START',
value=val, varname=mycol))
above = True
if val > threshold and above:
if val > maxrunning:
maxrunning = val
maxvalid = valid
if val < threshold and above:
res.append(dict(station=station, utc_valid=maxvalid, event='MAX',
value=maxrunning, varname=mycol))
res.append(dict(station=station, utc_valid=valid, event='END',
value=val, varname=mycol))
above = False
maxrunning = -99
maxvalid = None
if found is False:
error("# OOPS, did not find any exceedance!")
return pd.DataFrame(res)
def error(msg):
""" send back an error """
ssw("Content-type: text/plain\n\n")
ssw(msg)
sys.exit(0)
def main():
""" Go do something """
form = cgi.FieldStorage()
# network = form.getfirst('network')
delimiter = DELIMITERS.get(form.getfirst('delim', 'comma'))
what = form.getfirst('what', 'dl')
threshold = float(form.getfirst('threshold', -99))
thresholdvar = form.getfirst('threshold-var', 'RG')
sts, ets = get_time(form)
stations = form.getlist('stations')
if not stations:
ssw("Content-type: text/plain\n\n")
ssw("Error, no stations specified for the query!")
return
if len(stations) == 1:
stations.append('XXXXXXX')
table = "raw%s" % (sts.year,)
sql = """SELECT station, valid at time zone 'UTC' as utc_valid,
key, value from """+table+"""
WHERE station in %s and valid BETWEEN '%s' and '%s'
and value > -999""" % (tuple(stations), sts, ets)
df =
|
read_sql(sql, PGCONN)
|
pandas.io.sql.read_sql
|
""" Format data """
from __future__ import division, print_function
import pandas as pd
import numpy as np
import re
from os.path import dirname, join
from copy import deepcopy
import lawstructural.lawstructural.constants as lc
import lawstructural.lawstructural.utils as lu
#TODO: Take out entrant stuff from lawData
class Format(object):
""" Basic class for formatting dataset """
def __init__(self):
self.dpath = join(dirname(dirname(__file__)), 'data')
self.data_sets = self.data_imports()
self.data = self.data_sets['usn']
self.ent_data = pd.DataFrame([])
@staticmethod
def _col_fix(col):
""" Fix column strings to be R-readable as well and to be consistent
with older datasets. Think about changing name through rest of program
instead.
"""
col = re.sub('%', 'Percent', col)
col = re.sub('[() -/]', '', col)
if col[0].isdigit():
col = re.sub('thpercentile', '', col)
col = 'p' + col
if col == 'Name':
col = 'school'
if col == 'Issueyear':
col = 'year'
if col == 'Overallrank':
col = 'OverallRank'
return col
@staticmethod
def _fix_bad_values(data):
""" Fix known USN data typos """
data.loc[(data['school'] == 'University of Miami') &
(data['year'] == 2000), 'Tuitionandfeesfulltime'] = 21000
data.loc[(data['school'] == 'Emory University') &
(data['year'] == 2006), 'Employmentrateatgraduation'] = 72.4
data.loc[(data['school'] == 'Northwestern University') &
(data['year'] == 2006),
'EmploymentRate9MonthsafterGraduation'] = 99.5
data.loc[(data['school'] == 'Michigan State University') &
(data['year'] == 2001), 'BarpassageRateinJurisdiction'] = 75
data.loc[(data['school'] == 'Mississippi College') &
(data['year'] == 2001), 'BarpassageRateinJurisdiction'] = 80
return data
def usn_format(self):
""" Basic USN import and format """
#data = pd.read_csv(join(self.dpath, 'usn2015.csv'))
data = pd.read_csv(join(self.dpath, 'Law1988-2015.csv'))
data = data[['Name', 'Value', 'Metric description', 'Issue year']]
data = pd.pivot_table(data, values='Value',
index=['Name', 'Issue year'],
columns='Metric description')
data = data.reset_index()
names = data.columns.tolist()
data.columns = [self._col_fix(el) for el in names]
data = self._fix_bad_values(data)
data = data.sort(['school', 'year'])
data['year'] = data['year'].astype(int)
return data
def cpi_format(self):
""" Basic CPI import and format """
data = pd.read_csv(join(self.dpath, 'lawCPI.csv'))
# Make up for reporting vs data year in USNews and BLS
data['year'] = data['year'] + 2
data = data[data['year'] <= 2015]
data = data.reset_index(drop=True)
return data
@staticmethod
def _id_name_fix(col):
""" Fix outdated names of schools from id dataset """
#FIXME: Find out why this doesn't work for Drexel, Cath U
old = ['Phoenix School of Law',
'Chapman University',
'Drexel University (Mack)',
'Indiana University--Indianapolis',
'Texas Wesleyan University',
'Catholic University of America (Columbus)',
'John Marshall Law School']
new = ['Arizona Summit Law School',
'Chapman University (Fowler)',
'Drexel University',
'Indiana University--Indianapolis (McKinney)',
'Texas A&M University',
'The Catholic University of America',
'The John Marshall Law School']
for i in xrange(len(old)):
col = re.sub(old[i], new[i], col)
return col
def id_format(self):
""" Import LSAC id's. Note that Irvine doesn't have an id. """
data = pd.read_csv(join(self.dpath, 'USNewsNameStateID.csv'))
data['name'] = [self._id_name_fix(col) for col in data['name']]
return data
def elec_format(self):
""" Import yearly electricity prices """
data = pd.read_csv(join(self.dpath, 'lawElectricity.csv'))
states = pd.read_csv(join(self.dpath, 'lawStateAbbr.csv'))
# Change state names to abbreviations
data = pd.merge(data, states)
data = data.drop('state', 1)
columns = data.columns.tolist()
index = columns.index('abbr')
columns[index] = 'state'
data.columns = columns
data['year'] = data['year'] + 2
return data
def data_imports(self):
""" Import dictionary of initially formatted datasets
Datasets are as follows with corresponding sources/scrapers
usn
---
- Data: US News and World Report
- Source: ai.usnews.com
cpi
---
- Data: CPI data from BLS
- Source: http://data.bls.gov/cgi-bin/dsrv?cu
Series Id: CUUR0000SA0,CUUS0000SA0
Not Seasonally Adjusted
Area: U.S. city average
Item: All items
Base Period: 1982-84=100
Years: 1986 to 2015
- Used to be data.bls.gov/timeseries/LNS14000000
wage
----
- Data: Market wages for lawyers from BLS
- Source: bls.gov/oes
states
------
- Data: US News name/state combinations
- Source: US News Top Law Schools
- Scraper: StateScraper.py
id
--
- Data: US News names and LSAC ID combinations
- Source: http://www.lsac.org/lsacresources/publications/
official-guide-archives
- Scraper: NameScraperLSAC.py
entrants
--------
- Data: School entrants, with id's and dates
- Source: http://www.americanbar.org/groups/legal_education/
resources/aba_approved_law_schools/in_alphabetical_order.html
via
http://en.wikipedia.org/
wiki/List_of_law_schools_in_the_United_States
- Scraper: entryscraper.py
electricity
-----------
- Data: State/Country level electricity prices
- Source: eia.gov/electricity/monthly/backissues.html
- Scraper: ElecScraper.py
Returns
-------
data_sets: dict; data sets from specified sources
"""
data_sets = {
'usn': self.usn_format(),
'cpi': self.cpi_format(),
'states': pd.read_csv(join(self.dpath, 'lawNameState.csv')),
'id': self.id_format(),
'entrants': pd.read_csv(join(self.dpath, 'lawEntrants.csv')),
'electricity': self.elec_format(),
'stateregions': pd.read_csv(join(self.dpath, 'StateRegions.csv')),
'aaup_comp_region': pd.read_csv(join(self.dpath,
'aaup_comp_region.csv')),
'aaup_comp': pd.read_csv(join(self.dpath, 'aaup_comp.csv')),
'aaup_salary_region': pd.read_csv(join(self.dpath,
'aaup_salary_region.csv')),
'aaup_salary': pd.read_csv(join(self.dpath, 'aaup_salary.csv'))
}
return data_sets
def fill_ranks(self):
""" Generate top/bottom/inside/squared rank variables,
fill in unranked schools
"""
# Indicate top/bottom ranked schools
self.data['TopRanked'] = 1 * (self.data['OverallRank'] == 1)
self.data['BottomRanked'] = 1 * (self.data['OverallRank'] ==
np.nanmax(self.data['OverallRank']))
self.data['InsideRanked'] = 1 * ((self.data['OverallRank'] > 1) &
(self.data['OverallRank'] <
np.nanmax(self.data['OverallRank'])))
# Squared rank
self.data['OverallRankSquared'] = self.data['OverallRank']**2
# Fill in un-ranked schools as max(rank) + 1 or lc.UNRANKED
mask =
|
pd.isnull(self.data['OverallRank'])
|
pandas.isnull
|
#!/usr/bin/env python
# coding: utf-8
#!pip install sidrapy --upgrade
import sidrapy
import pandas as pd
def get_estimated_population(cod_ibge):
# Get Table
df = sidrapy.get_table(
table_code='6579',
territorial_level='6',
ibge_territorial_code=cod_ibge,
period='all',
header='n',
)
# Dict
dict_col = {
'D1C': 'id_municipio',
'D1N': 'municipio_nome',
'V': 'n_habitantes',
'D2N': 'ano'
}
# Remane Columns
df.rename(
dict_col,
axis=1,
inplace=True
)
# Select Columns
df = df[[v for k,v in dict_col.items()]]
# Adjust Columns
df.sort_values(by=['ano'], inplace=True)
df['id_municipio'] = pd.to_numeric(df['id_municipio'], errors='coerce')
df['n_habitantes'] =
|
pd.to_numeric(df['n_habitantes'], errors='coerce')
|
pandas.to_numeric
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.